file_id
stringlengths 5
9
| content
stringlengths 100
5.25M
| local_path
stringlengths 66
70
| kaggle_dataset_name
stringlengths 3
50
⌀ | kaggle_dataset_owner
stringlengths 3
20
⌀ | kversion
stringlengths 497
763
⌀ | kversion_datasetsources
stringlengths 71
5.46k
⌀ | dataset_versions
stringlengths 338
235k
⌀ | datasets
stringlengths 334
371
⌀ | users
stringlengths 111
264
⌀ | script
stringlengths 100
5.25M
| df_info
stringlengths 0
4.87M
| has_data_info
bool 2
classes | nb_filenames
int64 0
370
| retreived_data_description
stringlengths 0
4.44M
| script_nb_tokens
int64 25
663k
| upvotes
int64 0
1.65k
| tokens_description
int64 25
663k
| tokens_script
int64 25
663k
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
129788876
|
<jupyter_start><jupyter_text>ImageNet 1000 (mini)
### Context
https://github.com/pytorch/examples/tree/master/imagenet
Kaggle dataset identifier: imagenetmini-1000
<jupyter_script># Install dependecies
import math, re, os
import tensorflow as tf
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import skimage
import skimage.io
from sklearn.model_selection import train_test_split
from keras.utils import load_img, img_to_array, array_to_img
from keras.preprocessing.image import ImageDataGenerator
from tqdm import tqdm
from kaggle_datasets import KaggleDatasets
from tensorflow import keras
from functools import partial
print("Tensorflow version " + tf.__version__)
import random
from glob import glob
from tensorflow.keras.optimizers import Adam
import keras
from keras.models import *
from keras import layers
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras.optimizers import Adam
from keras.applications.vgg16 import preprocess_input
from keras.applications.vgg16 import VGG16
from IPython.display import display
from PIL import Image
mapping_path = (
"/kaggle/input/imagenet-object-localization-challenge/LOC_synset_mapping.txt"
)
src_path_train = (
"/kaggle/input/imagenet-object-localization-challenge/ILSVRC/Data/CLS-LOC/train"
)
src_path_test = (
"/kaggle/input/imagenet-object-localization-challenge/ILSVRC/Data/CLS-LOC/test"
)
# Creation of mapping dictionaries to obtain the image classes
class_mapping_dict = {}
class_mapping_dict_number = {}
mapping_class_to_number = {}
mapping_number_to_class = {}
i = 0
for line in open(mapping_path):
class_mapping_dict[line[:9].strip()] = line[9:].strip()
class_mapping_dict_number[i] = line[9:].strip()
mapping_class_to_number[line[:9].strip()] = i
mapping_number_to_class[i] = line[:9].strip()
i += 1
# print(class_mapping_dict)
# print(class_mapping_dict_number)
# print(mapping_class_to_number)
# print(mapping_number_to_class)
# Creation of dataset_array and CLASSES
CLASSES = []
images_array = []
for train_class in tqdm(os.listdir(src_path_train)):
i = 0
for el in os.listdir(src_path_train + "/" + train_class):
if i < 10:
path = src_path_train + "/" + train_class + "/" + el
image = load_img(path, target_size=(224, 224, 3))
image_array = img_to_array(image).astype(np.uint8)
images_array.append(image_array)
CLASS = class_mapping_dict[path.split("/")[-2]]
CLASSES.append(CLASS)
i += 1
else:
break
images_array = np.array(images_array)
CLASSES = np.array(CLASSES)
batch_size = 128
epochs = 100
# Creation of the train_generator and the test_generator
image_gen = ImageDataGenerator(
# rescale=1 / 255.0,
# rotation_range=20,
# zoom_range=0.05,
# width_shift_range=0.05,
# height_shift_range=0.05,
# shear_range=0.05,
# horizontal_flip=True,
# fill_mode="nearest",
preprocessing_function=preprocess_input,
validation_split=0.20,
)
train_generator = image_gen.flow_from_directory(
src_path_train,
target_size=(224, 224),
shuffle=True,
batch_size=batch_size,
subset="training",
class_mode="sparse",
)
test_generator = image_gen.flow_from_directory(
src_path_train,
target_size=(224, 224),
shuffle=True,
batch_size=batch_size,
subset="validation",
class_mode="sparse",
)
lr_scheduler = tf.keras.optimizers.schedules.ExponentialDecay(
initial_learning_rate=1e-5, decay_steps=10000, decay_rate=0.9
)
model_VGG16 = VGG16(weights="imagenet", include_top=False, input_shape=(224, 224, 3))
for layer in model_VGG16.layers:
layer.trainable = False
model = Sequential()
model.add(model_VGG16)
# model.add(layers.BatchNormalization(renorm=True))
model.add(layers.Flatten())
model.add(layers.Dense(units=4096, activation="relu"))
# model.add(layers.Dropout(0.3))
# model.add(layers.BatchNormalization(renorm=True))
model.add(layers.Dense(units=4096, activation="relu"))
# model.add(layers.Dropout(0.5))
# model.add(layers.BatchNormalization(renorm=True))
model.add(layers.Dense(units=1000, activation="softmax"))
model.compile(
optimizer=Adam(learning_rate=lr_scheduler, epsilon=0.001),
loss="sparse_categorical_crossentropy",
metrics=["sparse_categorical_accuracy"],
)
model.summary()
early_stop = EarlyStopping(
min_delta=0.001, # minimium amount of change to count as an improvement
patience=10, # how many epochs to wait before stopping
)
history = model.fit(
train_generator,
validation_data=test_generator,
epochs=epochs,
steps_per_epoch=len(train_generator) // batch_size,
validation_steps=len(test_generator) // batch_size,
callbacks=[early_stop],
)
# create learning curves to evaluate model performance
history_frame = pd.DataFrame(history.history)
history_frame.loc[:, ["loss", "val_loss"]].plot()
history_frame.loc[
:, ["sparse_categorical_accuracy", "val_sparse_categorical_accuracy"]
].plot()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/788/129788876.ipynb
|
imagenetmini-1000
|
ifigotin
|
[{"Id": 129788876, "ScriptId": 38358786, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13877157, "CreationDate": "05/16/2023 13:25:14", "VersionNumber": 1.0, "Title": "CNN_on_ImageNet_GPU", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 164.0, "LinesInsertedFromPrevious": 164.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186155496, "KernelVersionId": 129788876, "SourceDatasetVersionId": 998277}]
|
[{"Id": 998277, "DatasetId": 547506, "DatasourceVersionId": 1026923, "CreatorUserId": 2424380, "LicenseName": "Unknown", "CreationDate": "03/10/2020 01:05:11", "VersionNumber": 1.0, "Title": "ImageNet 1000 (mini)", "Slug": "imagenetmini-1000", "Subtitle": "1000 samples from ImageNet", "Description": "### Context\n\nhttps://github.com/pytorch/examples/tree/master/imagenet\n\n### Acknowledgements\n\nhttps://github.com/pytorch/examples/tree/master/imagenet", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 547506, "CreatorUserId": 2424380, "OwnerUserId": 2424380.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 998277.0, "CurrentDatasourceVersionId": 1026923.0, "ForumId": 561077, "Type": 2, "CreationDate": "03/10/2020 01:05:11", "LastActivityDate": "03/10/2020", "TotalViews": 62479, "TotalDownloads": 11891, "TotalVotes": 134, "TotalKernels": 57}]
|
[{"Id": 2424380, "UserName": "ifigotin", "DisplayName": "Ilya Figotin", "RegisterDate": "10/29/2018", "PerformanceTier": 1}]
|
# Install dependecies
import math, re, os
import tensorflow as tf
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import skimage
import skimage.io
from sklearn.model_selection import train_test_split
from keras.utils import load_img, img_to_array, array_to_img
from keras.preprocessing.image import ImageDataGenerator
from tqdm import tqdm
from kaggle_datasets import KaggleDatasets
from tensorflow import keras
from functools import partial
print("Tensorflow version " + tf.__version__)
import random
from glob import glob
from tensorflow.keras.optimizers import Adam
import keras
from keras.models import *
from keras import layers
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras.optimizers import Adam
from keras.applications.vgg16 import preprocess_input
from keras.applications.vgg16 import VGG16
from IPython.display import display
from PIL import Image
mapping_path = (
"/kaggle/input/imagenet-object-localization-challenge/LOC_synset_mapping.txt"
)
src_path_train = (
"/kaggle/input/imagenet-object-localization-challenge/ILSVRC/Data/CLS-LOC/train"
)
src_path_test = (
"/kaggle/input/imagenet-object-localization-challenge/ILSVRC/Data/CLS-LOC/test"
)
# Creation of mapping dictionaries to obtain the image classes
class_mapping_dict = {}
class_mapping_dict_number = {}
mapping_class_to_number = {}
mapping_number_to_class = {}
i = 0
for line in open(mapping_path):
class_mapping_dict[line[:9].strip()] = line[9:].strip()
class_mapping_dict_number[i] = line[9:].strip()
mapping_class_to_number[line[:9].strip()] = i
mapping_number_to_class[i] = line[:9].strip()
i += 1
# print(class_mapping_dict)
# print(class_mapping_dict_number)
# print(mapping_class_to_number)
# print(mapping_number_to_class)
# Creation of dataset_array and CLASSES
CLASSES = []
images_array = []
for train_class in tqdm(os.listdir(src_path_train)):
i = 0
for el in os.listdir(src_path_train + "/" + train_class):
if i < 10:
path = src_path_train + "/" + train_class + "/" + el
image = load_img(path, target_size=(224, 224, 3))
image_array = img_to_array(image).astype(np.uint8)
images_array.append(image_array)
CLASS = class_mapping_dict[path.split("/")[-2]]
CLASSES.append(CLASS)
i += 1
else:
break
images_array = np.array(images_array)
CLASSES = np.array(CLASSES)
batch_size = 128
epochs = 100
# Creation of the train_generator and the test_generator
image_gen = ImageDataGenerator(
# rescale=1 / 255.0,
# rotation_range=20,
# zoom_range=0.05,
# width_shift_range=0.05,
# height_shift_range=0.05,
# shear_range=0.05,
# horizontal_flip=True,
# fill_mode="nearest",
preprocessing_function=preprocess_input,
validation_split=0.20,
)
train_generator = image_gen.flow_from_directory(
src_path_train,
target_size=(224, 224),
shuffle=True,
batch_size=batch_size,
subset="training",
class_mode="sparse",
)
test_generator = image_gen.flow_from_directory(
src_path_train,
target_size=(224, 224),
shuffle=True,
batch_size=batch_size,
subset="validation",
class_mode="sparse",
)
lr_scheduler = tf.keras.optimizers.schedules.ExponentialDecay(
initial_learning_rate=1e-5, decay_steps=10000, decay_rate=0.9
)
model_VGG16 = VGG16(weights="imagenet", include_top=False, input_shape=(224, 224, 3))
for layer in model_VGG16.layers:
layer.trainable = False
model = Sequential()
model.add(model_VGG16)
# model.add(layers.BatchNormalization(renorm=True))
model.add(layers.Flatten())
model.add(layers.Dense(units=4096, activation="relu"))
# model.add(layers.Dropout(0.3))
# model.add(layers.BatchNormalization(renorm=True))
model.add(layers.Dense(units=4096, activation="relu"))
# model.add(layers.Dropout(0.5))
# model.add(layers.BatchNormalization(renorm=True))
model.add(layers.Dense(units=1000, activation="softmax"))
model.compile(
optimizer=Adam(learning_rate=lr_scheduler, epsilon=0.001),
loss="sparse_categorical_crossentropy",
metrics=["sparse_categorical_accuracy"],
)
model.summary()
early_stop = EarlyStopping(
min_delta=0.001, # minimium amount of change to count as an improvement
patience=10, # how many epochs to wait before stopping
)
history = model.fit(
train_generator,
validation_data=test_generator,
epochs=epochs,
steps_per_epoch=len(train_generator) // batch_size,
validation_steps=len(test_generator) // batch_size,
callbacks=[early_stop],
)
# create learning curves to evaluate model performance
history_frame = pd.DataFrame(history.history)
history_frame.loc[:, ["loss", "val_loss"]].plot()
history_frame.loc[
:, ["sparse_categorical_accuracy", "val_sparse_categorical_accuracy"]
].plot()
| false | 0 | 1,512 | 0 | 1,564 | 1,512 |
||
129788516
|
<jupyter_start><jupyter_text>ISMI_Group3_PANDA_36_256_256_res1_tiles
This dataset is a preprocessed provides a preprocessed version of the [PANDA](https://www.kaggle.com/competitions/prostate-cancer-grade-assessment) challenge. Each sample has 36 tiles, of 256 x 256 pixels. The tiles are taken from the medium resolution.
Kaggle dataset identifier: ismi-group3-panda-36-256-256-res1-tiles
<jupyter_script>import torch
import os
import gc
from PIL import Image
import torchvision
from torch.utils.data import DataLoader
import torchvision.transforms as transforms
import pytorch_lightning as pl
import torch.nn.functional as F
import numpy as np
import json
import requests
import matplotlib.pyplot as plt
import warnings
import glob
import pandas as pd
import tqdm
import random
warnings.filterwarnings("ignore")
# device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
# print(f'Using {device} for inference')
import os
import sys
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib
import matplotlib.pyplot as plt
import PIL
from IPython.display import Image, display
import openslide
# import skimage.io
# import tifffile
from tqdm.notebook import tqdm
import zipfile
import cv2 as cv
import timm
# device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
# print(f'Using {device} for inference')
with open("/kaggle/working/submission.csv", "w") as submis:
pass
# Location of the files
# data_dir = '/kaggle/input/prostate-cancer-grade-assessment/test_images'
# train_data_dir = '/kaggle/input/prostate-cancer-grade-assessment/test_images'
# mask_dir = '/kaggle/input/prostate-cancer-grade-assessment/test_label_masks'
# Location of training labels
train_labels = pd.read_csv(
"/kaggle/input/prostate-cancer-grade-assessment/test.csv"
).set_index("image_id")
file_names = train_labels.index.tolist()
# This function takes an openslide object and returns the top left coordinates of N tiles (of a given size) with the most tissue pixels.
# Note: slide.level_dimensions[level] = (width,height).
# Note: padding is done to the right and bottom, this is to keep it simple while having at most 1 tile in memory at a time.
def get_tile_locations_from_slide(slide, tile_size, N, level):
tiles = []
required_padding = False
xlocs, ylocs = np.arange(0, slide.level_dimensions[level][0], tile_size), np.arange(
0, slide.level_dimensions[level][1], tile_size
) # Get the coordinates of the top left corners of the tiles.
for x_i, xloc in enumerate(xlocs):
for y_i, yloc in enumerate(ylocs):
region = np.copy(
slide.read_region(
(xloc * (4**level), yloc * (4**level)),
level,
(tile_size, tile_size),
)
) # The position is wrt. level 0, so must convert to level 0 coordinates by multiplying by the downsampling factor.
region_arr = np.asarray(region)[:, :, :3] # Ignore the alpha channel
if (
xloc + tile_size > slide.level_dimensions[level][0]
or yloc + tile_size > slide.level_dimensions[level][1]
): # if the tile goes out of bounds
region_arr[region_arr == 0] = 255
required_padding = True
pixel_sum = region_arr.sum()
tiles.append(
{
"xloc": xloc,
"yloc": yloc,
"pixel_sum": pixel_sum,
"required_padding": required_padding,
}
) # store top left corner location and the tile's pixel_sum
required_padding = False
sorted_tiles = sorted(
tiles, key=lambda d: d["pixel_sum"]
) # Sort tiles based on their pixel_sum field
sorted_tiles = sorted_tiles[:N] # Get top N tiles
return sorted_tiles
# Creates a single image (array) from the selected tiles
def create_tiled_image(slide, tiles, tile_size, N_tiles, level):
N_side = int(np.sqrt(N_tiles)) # How many tiles is the image wide/tall
tiled_image = (
np.ones((N_side * tile_size, N_side * tile_size, 3), dtype=np.uint8) * 255
)
for i, tile in enumerate(tiles):
region = np.copy(
np.asarray(
slide.read_region(
(tile["xloc"] * (4**level), tile["yloc"] * (4**level)),
level,
(tile_size, tile_size),
)
)
) # The position is wrt. level 0, so must convert to level 0 coordinates by multiplying by the downsampling factor.
if tile["required_padding"]:
region[region == 0] = 255
tiled_image[
tile_size * (i // (N_side)) : tile_size * (i // (N_side)) + tile_size,
tile_size * (i % (N_side)) : tile_size * (i % (N_side)) + tile_size,
:,
] = region[:, :, :3]
return tiled_image
# # Creates a single image (array) from the selected tiles
# def save_tiles(slide, tiles, tile_size, N_tiles, out_path, level=1):
# for n, tile in enumerate(save):
# region = np.asarray(slide.read_region((tile['xloc']*(4**level),tile['yloc']*(4**level)), level, (tile_size,tile_size))) # The position is wrt. level 0, so must convert to level 0 coordinates by multiplying by the downsampling factor.
# if tile['required_padding']:
# region[region==0] = 255
# img = PIL.Image.fromarray(img)
# img.save(os.path.join(folder, filename+"_tiled.png"))
# return tiled_image
# ### **Now we can load the model and write the csv file!**
########## THIS IF FOR WHOLE IMAGES ################
# Defining the tile Data Module TODO: use the imghash to make sure patients arent in test and train set
# THE PARAMETERS 🔥
N_tiles = 6**2 # Number of tiles per image, should have a whole square root
tile_size = 2**8 # Width/height of tile, 2**8 = 256
level = 1 # 0 is highest resolution, 2 is lowest resolution, good compromise is level 1
# MAX_EPOCHS = 1000
class PANDADataset(torch.utils.data.Dataset):
def __init__(self, dataset: str = "train"):
# assert dataset in ['train', 'test'], "dataset should one of \"train\" or \"test\""
super().__init__()
self.df = pd.read_csv(
f"/kaggle/input/prostate-cancer-grade-assessment/{dataset}.csv"
)
self.imgdir = f"/kaggle/input/prostate-cancer-grade-assessment/{dataset}_images"
assert len(self.df) == len(self.df["image_id"].unique())
self.num_classes = 6
def convert_to_ordinal(self, n: int, nclasses: int):
ordinal = torch.zeros(nclasses)
ordinal[0 : n + 1] = 1
return ordinal
# For the independent tiles?
def load_tiles(self, samplepath):
tiles = glob.glob(os.path.join(samplepath, "tile_*.png"))
tiles = [torchvision.io.read_image(tile) / 255 for tile in tiles]
tiles = torch.stack(tiles)
return tiles
# Get a tiled image
def __getitem__(self, idx):
row = self.df.iloc[idx]
file_name = row.loc["image_id"]
slide = openslide.OpenSlide(os.path.join(self.imgdir, file_name + ".tiff"))
tiles = get_tile_locations_from_slide(
slide, tile_size, N_tiles, level
) # Get tile coordinates of top N tiles
tiled_image = create_tiled_image(
slide, tiles, tile_size, N_tiles, level
) # Convert the tiles information into a tiled image
tiled_image = torch.tensor(tiled_image.transpose(2, 1, 0))
return tiled_image
def __len__(self):
return len(self.df)
# print(f"{torch.cuda.memory_allocated()*1e-9:.4f}, GiB")
test_data = PANDADataset()
BATCH_SIZE = 1
test_dataloader = DataLoader(test_data, batch_size=BATCH_SIZE, shuffle=False)
class efficientnetModule(pl.LightningModule):
def __init__(self):
super().__init__()
self.model = timm.create_model(
"tf_efficientnet_b0",
checkpoint_path="/kaggle/input/tf-efficientnet/pytorch/tf-efficientnet-b0/1/tf_efficientnet_b0_aa-827b6e33.pth",
)
self.model.classifier = torch.nn.Linear(
in_features=self.model.classifier.in_features,
out_features=test_data.num_classes,
bias=True,
)
def get_prediction(self, output):
# this changes the prediction of format (0.01, 0.9, 0.8) to (1., 1., 0.)
for i, prediction in enumerate(output):
maxi = torch.argmax(prediction)
prediction[0 : maxi + 1] = 1
prediction[maxi + 1 :] = 0
output[i] = prediction
return output
def test_step(self, batch, batch_idx):
loss = self.validation_step(batch, batch_idx)
return loss
def forward(self, x):
output = self.model(x)
return output
def configure_optimizers(self):
return torch.optim.Adam(self.model.classifier.fc.parameters(), lr=0.02)
efficientNet = efficientnetModule()
trainedEfficientNet = efficientNet.load_from_checkpoint(
"/kaggle/input/baseline-trained-models/Models/Timm_model_5.ckpt"
)
trainedEfficientNet.eval()
# trainedEfficientNet.to(device)
print("Network loaded")
# # trainer = pl.Trainer(accelerator="cuda", devices=find_usable_cuda_devices(2))
# chk_path = "/kaggle/input/modello/best_model(1).ckpt"
# model2 = efficientnetModule.load_from_checkpoint(chk_path)
# # results = trainer.test(model=model2, datamodule=efficientnetModule, verbose=True)
# # results
# # Naive solution (Requires cuda to be enabled) (https://www.kaggle.com/code/mudittiwari255/pytorch-lightning-baseline)
# for file_name in tqdm(test_data.df['image_id'][:10]):
# slide = openslide.OpenSlide(os.path.join(test_data.imgdir, file_name+'.tiff'))
# tiles = get_tile_locations_from_slide(slide, tile_size, N_tiles, level) # Get tile coordinates of top N tiles
# tiled_image = create_tiled_image(slide, tiles, tile_size, N_tiles, level) # Convert the tiles information into a tiled image
# tiled_image = torch.tensor(tiled_image.transpose(2,1,0))
# tiled_image = tiled_image[None,:]#.to(device) # make batch of 1 sample
# output = trainedEfficientNet(tiled_image.float())
# prediction = torch.argmax(output)
# print(prediction)
# # Pytorch lightning solution
# n_dev = 1
# trainer = pl.Trainer(accelerator='gpu', devices=n_dev, enable_progress_bar=True)
# with open('/kaggle/working/submission.csv', 'w') as submis:
# submis.write('image_id,isup_grade')
# trainer.test(model = trainedEfficientNet, dataloaders=test_dataloader, ckpt_path ='/kaggle/input/baseline-trained-models/Models/Timm_model_5.ckpt', verbose=True)
# with open('/kaggle/working/submission.csv', 'w') as submis:
# submis.write('image_id,isup_grade')
# with torch.no_grad():
# for test_img in test_data:
# result = model2.model(test_img)
# submis.write(f'{testimg},{int(torch.sum(self.get_prediction(y_hat)))}')
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/788/129788516.ipynb
|
ismi-group3-panda-36-256-256-res1-tiles
|
florisvanwettum
|
[{"Id": 129788516, "ScriptId": 38540203, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2476129, "CreationDate": "05/16/2023 13:22:26", "VersionNumber": 7.0, "Title": "PANDA_Submission_notebook", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 215.0, "LinesInsertedFromPrevious": 50.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 165.0, "LinesInsertedFromFork": 88.0, "LinesDeletedFromFork": 201.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 127.0, "TotalVotes": 0}]
|
[{"Id": 186155043, "KernelVersionId": 129788516, "SourceDatasetVersionId": 5682352}]
|
[{"Id": 5682352, "DatasetId": 3228105, "DatasourceVersionId": 5757916, "CreatorUserId": 2476129, "LicenseName": "Unknown", "CreationDate": "05/14/2023 11:36:05", "VersionNumber": 5.0, "Title": "ISMI_Group3_PANDA_36_256_256_res1_tiles", "Slug": "ismi-group3-panda-36-256-256-res1-tiles", "Subtitle": "Medium resolution 36 256x256 tiles per sample, individual and combined images.", "Description": "This dataset is a preprocessed provides a preprocessed version of the [PANDA](https://www.kaggle.com/competitions/prostate-cancer-grade-assessment) challenge. Each sample has 36 tiles, of 256 x 256 pixels. The tiles are taken from the medium resolution.", "VersionNotes": "Added the last sample of the train.csv to the tiled_images", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3228105, "CreatorUserId": 2476129, "OwnerUserId": 2476129.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5682352.0, "CurrentDatasourceVersionId": 5757916.0, "ForumId": 3293216, "Type": 2, "CreationDate": "05/05/2023 21:28:46", "LastActivityDate": "05/05/2023", "TotalViews": 99, "TotalDownloads": 0, "TotalVotes": 0, "TotalKernels": 4}]
|
[{"Id": 2476129, "UserName": "florisvanwettum", "DisplayName": "Florijs", "RegisterDate": "11/10/2018", "PerformanceTier": 0}]
|
import torch
import os
import gc
from PIL import Image
import torchvision
from torch.utils.data import DataLoader
import torchvision.transforms as transforms
import pytorch_lightning as pl
import torch.nn.functional as F
import numpy as np
import json
import requests
import matplotlib.pyplot as plt
import warnings
import glob
import pandas as pd
import tqdm
import random
warnings.filterwarnings("ignore")
# device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
# print(f'Using {device} for inference')
import os
import sys
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib
import matplotlib.pyplot as plt
import PIL
from IPython.display import Image, display
import openslide
# import skimage.io
# import tifffile
from tqdm.notebook import tqdm
import zipfile
import cv2 as cv
import timm
# device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
# print(f'Using {device} for inference')
with open("/kaggle/working/submission.csv", "w") as submis:
pass
# Location of the files
# data_dir = '/kaggle/input/prostate-cancer-grade-assessment/test_images'
# train_data_dir = '/kaggle/input/prostate-cancer-grade-assessment/test_images'
# mask_dir = '/kaggle/input/prostate-cancer-grade-assessment/test_label_masks'
# Location of training labels
train_labels = pd.read_csv(
"/kaggle/input/prostate-cancer-grade-assessment/test.csv"
).set_index("image_id")
file_names = train_labels.index.tolist()
# This function takes an openslide object and returns the top left coordinates of N tiles (of a given size) with the most tissue pixels.
# Note: slide.level_dimensions[level] = (width,height).
# Note: padding is done to the right and bottom, this is to keep it simple while having at most 1 tile in memory at a time.
def get_tile_locations_from_slide(slide, tile_size, N, level):
tiles = []
required_padding = False
xlocs, ylocs = np.arange(0, slide.level_dimensions[level][0], tile_size), np.arange(
0, slide.level_dimensions[level][1], tile_size
) # Get the coordinates of the top left corners of the tiles.
for x_i, xloc in enumerate(xlocs):
for y_i, yloc in enumerate(ylocs):
region = np.copy(
slide.read_region(
(xloc * (4**level), yloc * (4**level)),
level,
(tile_size, tile_size),
)
) # The position is wrt. level 0, so must convert to level 0 coordinates by multiplying by the downsampling factor.
region_arr = np.asarray(region)[:, :, :3] # Ignore the alpha channel
if (
xloc + tile_size > slide.level_dimensions[level][0]
or yloc + tile_size > slide.level_dimensions[level][1]
): # if the tile goes out of bounds
region_arr[region_arr == 0] = 255
required_padding = True
pixel_sum = region_arr.sum()
tiles.append(
{
"xloc": xloc,
"yloc": yloc,
"pixel_sum": pixel_sum,
"required_padding": required_padding,
}
) # store top left corner location and the tile's pixel_sum
required_padding = False
sorted_tiles = sorted(
tiles, key=lambda d: d["pixel_sum"]
) # Sort tiles based on their pixel_sum field
sorted_tiles = sorted_tiles[:N] # Get top N tiles
return sorted_tiles
# Creates a single image (array) from the selected tiles
def create_tiled_image(slide, tiles, tile_size, N_tiles, level):
N_side = int(np.sqrt(N_tiles)) # How many tiles is the image wide/tall
tiled_image = (
np.ones((N_side * tile_size, N_side * tile_size, 3), dtype=np.uint8) * 255
)
for i, tile in enumerate(tiles):
region = np.copy(
np.asarray(
slide.read_region(
(tile["xloc"] * (4**level), tile["yloc"] * (4**level)),
level,
(tile_size, tile_size),
)
)
) # The position is wrt. level 0, so must convert to level 0 coordinates by multiplying by the downsampling factor.
if tile["required_padding"]:
region[region == 0] = 255
tiled_image[
tile_size * (i // (N_side)) : tile_size * (i // (N_side)) + tile_size,
tile_size * (i % (N_side)) : tile_size * (i % (N_side)) + tile_size,
:,
] = region[:, :, :3]
return tiled_image
# # Creates a single image (array) from the selected tiles
# def save_tiles(slide, tiles, tile_size, N_tiles, out_path, level=1):
# for n, tile in enumerate(save):
# region = np.asarray(slide.read_region((tile['xloc']*(4**level),tile['yloc']*(4**level)), level, (tile_size,tile_size))) # The position is wrt. level 0, so must convert to level 0 coordinates by multiplying by the downsampling factor.
# if tile['required_padding']:
# region[region==0] = 255
# img = PIL.Image.fromarray(img)
# img.save(os.path.join(folder, filename+"_tiled.png"))
# return tiled_image
# ### **Now we can load the model and write the csv file!**
########## THIS IF FOR WHOLE IMAGES ################
# Defining the tile Data Module TODO: use the imghash to make sure patients arent in test and train set
# THE PARAMETERS 🔥
N_tiles = 6**2 # Number of tiles per image, should have a whole square root
tile_size = 2**8 # Width/height of tile, 2**8 = 256
level = 1 # 0 is highest resolution, 2 is lowest resolution, good compromise is level 1
# MAX_EPOCHS = 1000
class PANDADataset(torch.utils.data.Dataset):
def __init__(self, dataset: str = "train"):
# assert dataset in ['train', 'test'], "dataset should one of \"train\" or \"test\""
super().__init__()
self.df = pd.read_csv(
f"/kaggle/input/prostate-cancer-grade-assessment/{dataset}.csv"
)
self.imgdir = f"/kaggle/input/prostate-cancer-grade-assessment/{dataset}_images"
assert len(self.df) == len(self.df["image_id"].unique())
self.num_classes = 6
def convert_to_ordinal(self, n: int, nclasses: int):
ordinal = torch.zeros(nclasses)
ordinal[0 : n + 1] = 1
return ordinal
# For the independent tiles?
def load_tiles(self, samplepath):
tiles = glob.glob(os.path.join(samplepath, "tile_*.png"))
tiles = [torchvision.io.read_image(tile) / 255 for tile in tiles]
tiles = torch.stack(tiles)
return tiles
# Get a tiled image
def __getitem__(self, idx):
row = self.df.iloc[idx]
file_name = row.loc["image_id"]
slide = openslide.OpenSlide(os.path.join(self.imgdir, file_name + ".tiff"))
tiles = get_tile_locations_from_slide(
slide, tile_size, N_tiles, level
) # Get tile coordinates of top N tiles
tiled_image = create_tiled_image(
slide, tiles, tile_size, N_tiles, level
) # Convert the tiles information into a tiled image
tiled_image = torch.tensor(tiled_image.transpose(2, 1, 0))
return tiled_image
def __len__(self):
return len(self.df)
# print(f"{torch.cuda.memory_allocated()*1e-9:.4f}, GiB")
test_data = PANDADataset()
BATCH_SIZE = 1
test_dataloader = DataLoader(test_data, batch_size=BATCH_SIZE, shuffle=False)
class efficientnetModule(pl.LightningModule):
def __init__(self):
super().__init__()
self.model = timm.create_model(
"tf_efficientnet_b0",
checkpoint_path="/kaggle/input/tf-efficientnet/pytorch/tf-efficientnet-b0/1/tf_efficientnet_b0_aa-827b6e33.pth",
)
self.model.classifier = torch.nn.Linear(
in_features=self.model.classifier.in_features,
out_features=test_data.num_classes,
bias=True,
)
def get_prediction(self, output):
# this changes the prediction of format (0.01, 0.9, 0.8) to (1., 1., 0.)
for i, prediction in enumerate(output):
maxi = torch.argmax(prediction)
prediction[0 : maxi + 1] = 1
prediction[maxi + 1 :] = 0
output[i] = prediction
return output
def test_step(self, batch, batch_idx):
loss = self.validation_step(batch, batch_idx)
return loss
def forward(self, x):
output = self.model(x)
return output
def configure_optimizers(self):
return torch.optim.Adam(self.model.classifier.fc.parameters(), lr=0.02)
efficientNet = efficientnetModule()
trainedEfficientNet = efficientNet.load_from_checkpoint(
"/kaggle/input/baseline-trained-models/Models/Timm_model_5.ckpt"
)
trainedEfficientNet.eval()
# trainedEfficientNet.to(device)
print("Network loaded")
# # trainer = pl.Trainer(accelerator="cuda", devices=find_usable_cuda_devices(2))
# chk_path = "/kaggle/input/modello/best_model(1).ckpt"
# model2 = efficientnetModule.load_from_checkpoint(chk_path)
# # results = trainer.test(model=model2, datamodule=efficientnetModule, verbose=True)
# # results
# # Naive solution (Requires cuda to be enabled) (https://www.kaggle.com/code/mudittiwari255/pytorch-lightning-baseline)
# for file_name in tqdm(test_data.df['image_id'][:10]):
# slide = openslide.OpenSlide(os.path.join(test_data.imgdir, file_name+'.tiff'))
# tiles = get_tile_locations_from_slide(slide, tile_size, N_tiles, level) # Get tile coordinates of top N tiles
# tiled_image = create_tiled_image(slide, tiles, tile_size, N_tiles, level) # Convert the tiles information into a tiled image
# tiled_image = torch.tensor(tiled_image.transpose(2,1,0))
# tiled_image = tiled_image[None,:]#.to(device) # make batch of 1 sample
# output = trainedEfficientNet(tiled_image.float())
# prediction = torch.argmax(output)
# print(prediction)
# # Pytorch lightning solution
# n_dev = 1
# trainer = pl.Trainer(accelerator='gpu', devices=n_dev, enable_progress_bar=True)
# with open('/kaggle/working/submission.csv', 'w') as submis:
# submis.write('image_id,isup_grade')
# trainer.test(model = trainedEfficientNet, dataloaders=test_dataloader, ckpt_path ='/kaggle/input/baseline-trained-models/Models/Timm_model_5.ckpt', verbose=True)
# with open('/kaggle/working/submission.csv', 'w') as submis:
# submis.write('image_id,isup_grade')
# with torch.no_grad():
# for test_img in test_data:
# result = model2.model(test_img)
# submis.write(f'{testimg},{int(torch.sum(self.get_prediction(y_hat)))}')
| false | 1 | 3,148 | 0 | 3,283 | 3,148 |
||
129788001
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
df1 = pd.read_csv("/kaggle/input/house-pricecsv/train.csv")
df2 = pd.read_csv("/kaggle/input/house-pricecsv/test.csv")
df = pd.concat([df1, df2], axis=0, ignore_index=True)
df.head()
df.tail()
df.shape
df.describe()
df.isnull().sum()
df.info()
df.dtypes
df.isnull()
df_training = df[df["SalePrice"].notna()]
df_training.head()
df_training.shape
df_training.nunique()
numeric_columns = df_training.describe().columns
numeric_columns
categorical_columns = df_training.describe(include="O").columns
categorical_columns
df_training.describe()
numeric_columns.isnull().sum()
categorical_columns.isnull().sum()
type(categorical_columns)
categorical_columns = df_training.describe(include="O").columns
for i in categorical_columns:
print(i)
print(df_training[i].unique())
print(df_training["MiscFeature"].isnull().sum())
print(df_training["Fence"].isnull().sum())
print(df_training["PoolQC"].isnull().sum())
print(df_training["GarageCond"].isnull().sum())
print(df_training["GarageQual"].isnull().sum())
print(df_training["GarageFinish"].isnull().sum())
print(df_training["GarageType"].isnull().sum())
print(df_training["GarageFinish"].isnull().sum())
print(df_training["GarageFinish"].isnull().sum())
df_training.shape
# Exploring multicollinearity.
from statsmodels.stats.outliers_influence import variance_inflation_factor
def calc_vif(X):
# Calculating VIF
vif = pd.DataFrame()
vif["variables"] = X.columns
vif["VIF"] = [variance_inflation_factor(X.values, i) for i in range(X.shape[1])]
return vif
calc_vif(
df_training[[i for i in df_training.describe().columns if i not in ["SalePrice"]]]
)
df_infer = df[df["SalePrice"].isna()]
df_infer
df_infer.shape
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/788/129788001.ipynb
| null | null |
[{"Id": 129788001, "ScriptId": 38585324, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13782320, "CreationDate": "05/16/2023 13:18:28", "VersionNumber": 1.0, "Title": "House_Price", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 115.0, "LinesInsertedFromPrevious": 115.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
df1 = pd.read_csv("/kaggle/input/house-pricecsv/train.csv")
df2 = pd.read_csv("/kaggle/input/house-pricecsv/test.csv")
df = pd.concat([df1, df2], axis=0, ignore_index=True)
df.head()
df.tail()
df.shape
df.describe()
df.isnull().sum()
df.info()
df.dtypes
df.isnull()
df_training = df[df["SalePrice"].notna()]
df_training.head()
df_training.shape
df_training.nunique()
numeric_columns = df_training.describe().columns
numeric_columns
categorical_columns = df_training.describe(include="O").columns
categorical_columns
df_training.describe()
numeric_columns.isnull().sum()
categorical_columns.isnull().sum()
type(categorical_columns)
categorical_columns = df_training.describe(include="O").columns
for i in categorical_columns:
print(i)
print(df_training[i].unique())
print(df_training["MiscFeature"].isnull().sum())
print(df_training["Fence"].isnull().sum())
print(df_training["PoolQC"].isnull().sum())
print(df_training["GarageCond"].isnull().sum())
print(df_training["GarageQual"].isnull().sum())
print(df_training["GarageFinish"].isnull().sum())
print(df_training["GarageType"].isnull().sum())
print(df_training["GarageFinish"].isnull().sum())
print(df_training["GarageFinish"].isnull().sum())
df_training.shape
# Exploring multicollinearity.
from statsmodels.stats.outliers_influence import variance_inflation_factor
def calc_vif(X):
# Calculating VIF
vif = pd.DataFrame()
vif["variables"] = X.columns
vif["VIF"] = [variance_inflation_factor(X.values, i) for i in range(X.shape[1])]
return vif
calc_vif(
df_training[[i for i in df_training.describe().columns if i not in ["SalePrice"]]]
)
df_infer = df[df["SalePrice"].isna()]
df_infer
df_infer.shape
| false | 0 | 756 | 0 | 756 | 756 |
||
129788869
|
<jupyter_start><jupyter_text>Best Books (10k) Multi-Genre Data
# Context
This data was collected in an attempt personally identify more books that one would like based on ones they may have read in the past. It comprises of some (around 10000) of the most recommended books of all time.
### *Please Upvote if this helps you!*
# Content
1. **Book** - Name of the book. Soemtimes this includes the details of the Series it belongs to inside a parenthesis. This information can be further extracted to analyse only series.
2. **Author** - Name of the book's Author
3. **Description** - The book's description as mentioned on Goodreads
4. **Genres** - Multiple Genres as classified on Goodreads. Could be useful for Multi-label classification or Content based recommendation and Clustering.
5. **Average Rating** - The average rating (Out of 5) given on Goodreads
6. **Number of Ratings** - The Number of users that have Ratings. (Not to be confused with reviews)
7. **URL** - The Goodreads URL for the book's details' page
# Inspiration
- Cluster books/authors based on Description and Genre
- Content based recomendation system using Genre, Description and Ratings
- Genre prediction from Description data (Multi-label classification)
- Can be used in conjunction with my [IMDb dataset with descriptions](https://www.kaggle.com/datasets/ishikajohari/imdb-data-with-descriptions) for certain use cases
# Acknowledgements
The data was collected from Goodreads from the list - *Books That Everyone Should Read At Least Once*
Kaggle dataset identifier: best-books-10k-multi-genre-data
<jupyter_script>import pandas as pd
import numpy as np
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MultiLabelBinarizer
from sklearn.multiclass import OneVsRestClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import f1_score
from sklearn.cluster import KMeans
from wordcloud import WordCloud
import matplotlib.pyplot as plt
df = pd.read_csv("/kaggle/input/best-books-10k-multi-genre-data/goodreads_data.csv")
df.head()
# # Data Preparation
df["Description"] = df["Description"].fillna("")
# # Content Based Recommendation
# Convert the book descriptions into TF-IDF vectors
vectorizer = TfidfVectorizer(stop_words="english")
tfidf_matrix = vectorizer.fit_transform(df["Description"])
# Compute the cosine similarity matrix
cosine_sim = cosine_similarity(tfidf_matrix, tfidf_matrix)
# Function to get the most similar books
def get_recommendations(title, cosine_sim=cosine_sim):
# Get the index of the book that matches the title
idx = df[df["Book"] == title].index[0]
# Get the pairwsie similarity scores of all books with that book
sim_scores = list(enumerate(cosine_sim[idx]))
# Sort the books based on the similarity scores
sim_scores = sorted(sim_scores, key=lambda x: x[1], reverse=True)
# Get the scores of the 10 most similar books
sim_scores = sim_scores[1:11]
# Get the book indices
book_indices = [i[0] for i in sim_scores]
# Return the top 10 most similar books
return df["Book"].iloc[book_indices]
# # Genre Prediction
# Convert Genres from string to list
df["Genres"] = df["Genres"].apply(eval)
# Binarize the genres
mlb = MultiLabelBinarizer()
y = mlb.fit_transform(df["Genres"])
# Split the data
X_train, X_test, y_train, y_test = train_test_split(
df["Description"], y, test_size=0.2, random_state=0
)
# Convert the descriptions into TF-IDF vectors
vectorizer = TfidfVectorizer(stop_words="english")
X_train = vectorizer.fit_transform(X_train)
X_test = vectorizer.transform(X_test)
# Train a multi-label classifier
clf = OneVsRestClassifier(LogisticRegression(solver="lbfgs"))
clf.fit(X_train, y_train)
# Predict the test set results
y_pred = clf.predict(X_test)
# Compute the F1 score
print(f1_score(y_test, y_pred, average="micro"))
# # Author Clustering
# Convert the book descriptions into TF-IDF vectors
vectorizer = TfidfVectorizer(stop_words="english")
tfidf_matrix = vectorizer.fit_transform(df["Description"])
# Compute K-Means clustering
kmeans = KMeans(n_clusters=5, n_init=10)
kmeans.fit(tfidf_matrix)
# Add cluster number to the original dataframe
df["Cluster"] = kmeans.labels_
# Print the number of authors in each cluster
print(df.groupby("Cluster")["Author"].nunique())
# # Sentiment
from textblob import TextBlob
# Calculate sentiment polarity of descriptions
df["Sentiment"] = df["Description"].apply(
lambda text: TextBlob(text).sentiment.polarity
)
# Check average sentiment by rating
print(df.groupby("Avg_Rating")["Sentiment"].mean())
# # Visualisation
# Generate a word cloud for book descriptions
text = " ".join(description for description in df["Description"])
wordcloud = WordCloud(background_color="white").generate(text)
plt.imshow(wordcloud, interpolation="bilinear")
plt.axis("off")
plt.show()
# Plot the distribution of average ratings
df["Avg_Rating"].hist(bins=20)
plt.xlabel("Average Rating")
plt.ylabel("Count")
plt.title("Distribution of Average Ratings")
plt.show()
# Plot the top 10 most common genres
df["Genres"].explode().value_counts()[:10].plot(kind="bar")
plt.xlabel("Genre")
plt.ylabel("Count")
plt.title("Top 10 Most Common Genres")
plt.show()
# Author Distribution
df["Author"].value_counts().head(10).plot(kind="bar")
plt.xlabel("Author")
plt.ylabel("Number of Books")
plt.title("Top 10 Authors with the Most Books")
plt.show()
# Sentiment Distribution
df["Sentiment"].hist(bins=20)
plt.xlabel("Sentiment Score")
plt.ylabel("Number of Books")
plt.title("Distribution of Sentiment Scores")
plt.show()
# Cluster Size Distribution
df["Cluster"].value_counts().plot(kind="bar")
plt.xlabel("Cluster")
plt.ylabel("Number of Authors")
plt.title("Number of Authors in Each Cluster")
plt.show()
# Avg Rating by Cluster
df.groupby("Cluster")["Avg_Rating"].mean().plot(kind="bar")
plt.xlabel("Cluster")
plt.ylabel("Average Rating")
plt.title("Average Rating by Cluster")
plt.show()
# Heatmap of Genres
from sklearn.preprocessing import MultiLabelBinarizer
import seaborn as sns
# Get a list of all genres
all_genres = list(set([g for sublist in df["Genres"].tolist() for g in sublist]))
# Binarize the genres
mlb = MultiLabelBinarizer(classes=all_genres)
binary_genres = mlb.fit_transform(df["Genres"])
# Create a DataFrame from our binary matrix, and calculate the correlations
binary_genres_df = pd.DataFrame(binary_genres, columns=mlb.classes_)
correlations = binary_genres_df.corr()
# Plot the correlations in a heatmap
plt.figure(figsize=(10, 10))
sns.heatmap(correlations, cmap="coolwarm", center=0)
plt.title("Genre Correlations")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/788/129788869.ipynb
|
best-books-10k-multi-genre-data
|
ishikajohari
|
[{"Id": 129788869, "ScriptId": 38584099, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4335187, "CreationDate": "05/16/2023 13:25:13", "VersionNumber": 1.0, "Title": "Books EDA", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 180.0, "LinesInsertedFromPrevious": 180.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 186155490, "KernelVersionId": 129788869, "SourceDatasetVersionId": 5618933}]
|
[{"Id": 5618933, "DatasetId": 3230801, "DatasourceVersionId": 5694114, "CreatorUserId": 5431518, "LicenseName": "CC0: Public Domain", "CreationDate": "05/06/2023 14:13:35", "VersionNumber": 2.0, "Title": "Best Books (10k) Multi-Genre Data", "Slug": "best-books-10k-multi-genre-data", "Subtitle": "Data from the \"Books That Everyone Should Read At Least Once\" list on Goodreads", "Description": "# Context\nThis data was collected in an attempt personally identify more books that one would like based on ones they may have read in the past. It comprises of some (around 10000) of the most recommended books of all time.\n\n### *Please Upvote if this helps you!*\n\n# Content\n1. **Book** - Name of the book. Soemtimes this includes the details of the Series it belongs to inside a parenthesis. This information can be further extracted to analyse only series.\n2. **Author** - Name of the book's Author\n3. **Description** - The book's description as mentioned on Goodreads\n4. **Genres** - Multiple Genres as classified on Goodreads. Could be useful for Multi-label classification or Content based recommendation and Clustering.\n5. **Average Rating** - The average rating (Out of 5) given on Goodreads\n6. **Number of Ratings** - The Number of users that have Ratings. (Not to be confused with reviews)\n7. **URL** - The Goodreads URL for the book's details' page\n\n# Inspiration\n- Cluster books/authors based on Description and Genre\n- Content based recomendation system using Genre, Description and Ratings\n- Genre prediction from Description data (Multi-label classification)\n- Can be used in conjunction with my [IMDb dataset with descriptions](https://www.kaggle.com/datasets/ishikajohari/imdb-data-with-descriptions) for certain use cases\n\n# Acknowledgements\nThe data was collected from Goodreads from the list - *Books That Everyone Should Read At Least Once*", "VersionNotes": "Data Update 2023-05-06", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3230801, "CreatorUserId": 5431518, "OwnerUserId": 5431518.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5618933.0, "CurrentDatasourceVersionId": 5694114.0, "ForumId": 3295942, "Type": 2, "CreationDate": "05/06/2023 13:43:14", "LastActivityDate": "05/06/2023", "TotalViews": 8727, "TotalDownloads": 1388, "TotalVotes": 56, "TotalKernels": 8}]
|
[{"Id": 5431518, "UserName": "ishikajohari", "DisplayName": "Ishika Johari", "RegisterDate": "07/07/2020", "PerformanceTier": 2}]
|
import pandas as pd
import numpy as np
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MultiLabelBinarizer
from sklearn.multiclass import OneVsRestClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import f1_score
from sklearn.cluster import KMeans
from wordcloud import WordCloud
import matplotlib.pyplot as plt
df = pd.read_csv("/kaggle/input/best-books-10k-multi-genre-data/goodreads_data.csv")
df.head()
# # Data Preparation
df["Description"] = df["Description"].fillna("")
# # Content Based Recommendation
# Convert the book descriptions into TF-IDF vectors
vectorizer = TfidfVectorizer(stop_words="english")
tfidf_matrix = vectorizer.fit_transform(df["Description"])
# Compute the cosine similarity matrix
cosine_sim = cosine_similarity(tfidf_matrix, tfidf_matrix)
# Function to get the most similar books
def get_recommendations(title, cosine_sim=cosine_sim):
# Get the index of the book that matches the title
idx = df[df["Book"] == title].index[0]
# Get the pairwsie similarity scores of all books with that book
sim_scores = list(enumerate(cosine_sim[idx]))
# Sort the books based on the similarity scores
sim_scores = sorted(sim_scores, key=lambda x: x[1], reverse=True)
# Get the scores of the 10 most similar books
sim_scores = sim_scores[1:11]
# Get the book indices
book_indices = [i[0] for i in sim_scores]
# Return the top 10 most similar books
return df["Book"].iloc[book_indices]
# # Genre Prediction
# Convert Genres from string to list
df["Genres"] = df["Genres"].apply(eval)
# Binarize the genres
mlb = MultiLabelBinarizer()
y = mlb.fit_transform(df["Genres"])
# Split the data
X_train, X_test, y_train, y_test = train_test_split(
df["Description"], y, test_size=0.2, random_state=0
)
# Convert the descriptions into TF-IDF vectors
vectorizer = TfidfVectorizer(stop_words="english")
X_train = vectorizer.fit_transform(X_train)
X_test = vectorizer.transform(X_test)
# Train a multi-label classifier
clf = OneVsRestClassifier(LogisticRegression(solver="lbfgs"))
clf.fit(X_train, y_train)
# Predict the test set results
y_pred = clf.predict(X_test)
# Compute the F1 score
print(f1_score(y_test, y_pred, average="micro"))
# # Author Clustering
# Convert the book descriptions into TF-IDF vectors
vectorizer = TfidfVectorizer(stop_words="english")
tfidf_matrix = vectorizer.fit_transform(df["Description"])
# Compute K-Means clustering
kmeans = KMeans(n_clusters=5, n_init=10)
kmeans.fit(tfidf_matrix)
# Add cluster number to the original dataframe
df["Cluster"] = kmeans.labels_
# Print the number of authors in each cluster
print(df.groupby("Cluster")["Author"].nunique())
# # Sentiment
from textblob import TextBlob
# Calculate sentiment polarity of descriptions
df["Sentiment"] = df["Description"].apply(
lambda text: TextBlob(text).sentiment.polarity
)
# Check average sentiment by rating
print(df.groupby("Avg_Rating")["Sentiment"].mean())
# # Visualisation
# Generate a word cloud for book descriptions
text = " ".join(description for description in df["Description"])
wordcloud = WordCloud(background_color="white").generate(text)
plt.imshow(wordcloud, interpolation="bilinear")
plt.axis("off")
plt.show()
# Plot the distribution of average ratings
df["Avg_Rating"].hist(bins=20)
plt.xlabel("Average Rating")
plt.ylabel("Count")
plt.title("Distribution of Average Ratings")
plt.show()
# Plot the top 10 most common genres
df["Genres"].explode().value_counts()[:10].plot(kind="bar")
plt.xlabel("Genre")
plt.ylabel("Count")
plt.title("Top 10 Most Common Genres")
plt.show()
# Author Distribution
df["Author"].value_counts().head(10).plot(kind="bar")
plt.xlabel("Author")
plt.ylabel("Number of Books")
plt.title("Top 10 Authors with the Most Books")
plt.show()
# Sentiment Distribution
df["Sentiment"].hist(bins=20)
plt.xlabel("Sentiment Score")
plt.ylabel("Number of Books")
plt.title("Distribution of Sentiment Scores")
plt.show()
# Cluster Size Distribution
df["Cluster"].value_counts().plot(kind="bar")
plt.xlabel("Cluster")
plt.ylabel("Number of Authors")
plt.title("Number of Authors in Each Cluster")
plt.show()
# Avg Rating by Cluster
df.groupby("Cluster")["Avg_Rating"].mean().plot(kind="bar")
plt.xlabel("Cluster")
plt.ylabel("Average Rating")
plt.title("Average Rating by Cluster")
plt.show()
# Heatmap of Genres
from sklearn.preprocessing import MultiLabelBinarizer
import seaborn as sns
# Get a list of all genres
all_genres = list(set([g for sublist in df["Genres"].tolist() for g in sublist]))
# Binarize the genres
mlb = MultiLabelBinarizer(classes=all_genres)
binary_genres = mlb.fit_transform(df["Genres"])
# Create a DataFrame from our binary matrix, and calculate the correlations
binary_genres_df = pd.DataFrame(binary_genres, columns=mlb.classes_)
correlations = binary_genres_df.corr()
# Plot the correlations in a heatmap
plt.figure(figsize=(10, 10))
sns.heatmap(correlations, cmap="coolwarm", center=0)
plt.title("Genre Correlations")
plt.show()
| false | 1 | 1,541 | 3 | 1,943 | 1,541 |
||
129788906
|
# # データを読み込む
import pandas as pd
data_df = pd.read_csv("/kaggle/input/kdl-datascience-compe/train.csv")
data_df.columns
# # 改装について
# 改装すると価格が上がる気がするので可視化してみる。
data_df[["改装", "取引価格(総額)_log"]].groupby("改装").mean()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/788/129788906.ipynb
| null | null |
[{"Id": 129788906, "ScriptId": 38599591, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7501332, "CreationDate": "05/16/2023 13:25:31", "VersionNumber": 1.0, "Title": "EDA:reform", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 15.0, "LinesInsertedFromPrevious": 15.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # データを読み込む
import pandas as pd
data_df = pd.read_csv("/kaggle/input/kdl-datascience-compe/train.csv")
data_df.columns
# # 改装について
# 改装すると価格が上がる気がするので可視化してみる。
data_df[["改装", "取引価格(総額)_log"]].groupby("改装").mean()
| false | 0 | 113 | 0 | 113 | 113 |
||
129711281
|
<jupyter_start><jupyter_text>one_year_retail_transactions
The dataset contains information related to retail transactions in the Gulf region. Here is a brief description of each column:
UCID: Unique Customer ID.
GENDER: Gender of the customer.
AGE_GROUP2: Age group of the customer.
MEMBERSHIP_DATE: Date when the customer became a member.
BRAND_REPORTING: Brand associated with the transaction.
LOCATION_NAME_REPORTING: Name of the location where the transaction occurred.
STORE_COUNTRY: Country where the store is located.
BUSINESS_CHANNEL: Channel through which the transaction was made (e.g., offline, online).
INVOICE_NO: Invoice number associated with the transaction.
NET_SALES_AMOUNT: Net sales amount for the transaction.
SALES_QTY: Quantity of items sold in the transaction.
DISCOUNT_AMOUNT: Amount of discount applied to the transaction.
DAY_DT: Date of the transaction.
DIV_NAME: Division name (category) of the purchased item.
DEPT_NAME: Department name of the purchased item.
CLASS_NAME: Class name of the purchased item.
SUBCLASS_NAME: Subclass name of the purchased item.
Kaggle dataset identifier: one-year-retail-transactions
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # First exercise
# A step-by-step process to conduct a detailed explanatory data analysis for behavioral segmentation
# ### Step 1: Import the necessary libraries and load the dataset.
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# Load the dataset
df = pd.read_excel(
"/kaggle/input/one-year-retail-transactions/Retail_Transactions.xlsx"
)
# Display the first few rows of the dataset
df.head()
# ### Step 2: Explore the dataset to gain initial insights.
# Check the summary statistics of numerical columns
df.describe()
# Check the data types and missing values
df.info()
# ### Step 3: Perform data cleaning and preprocessing (if necessary).
# Handle missing values (if any)
df.dropna(inplace=True)
# Remove duplicates (if any)
df.drop_duplicates(inplace=True)
# ### Step 4: Explore the distribution of variables and identify key trends.
# Remove leading spaces from column names
df.columns = df.columns.str.strip()
# Verify the updated column names
print(df.columns)
# Explore the distribution of categorical variables
plt.figure(figsize=(10, 6))
sns.countplot(x="GENDER", data=df)
plt.title("Gender Distribution")
plt.show()
# Explore the distribution of numerical variables
plt.figure(figsize=(10, 6))
sns.histplot(x="AGE_GROUP2", data=df, bins=10)
plt.title("Age Group Distribution")
plt.show()
# Explore the sales trends over time
plt.figure(figsize=(10, 6))
sns.lineplot(x="DAY_DT", y="NET_SALES_AMOUNT", data=df)
plt.title("Sales Trends Over Time")
plt.show()
# ### Step 5: Analyze customer behavior and identify patterns.
# Calculate and visualize average sales by brand
brand_sales = (
df.groupby("BRAND_REPORTING")["NET_SALES_AMOUNT"]
.mean()
.sort_values(ascending=False)
)
plt.figure(figsize=(10, 6))
sns.barplot(x=brand_sales.index, y=brand_sales.values)
plt.title("Average Sales by Brand")
plt.xticks(rotation=90)
plt.show()
# Analyze purchase patterns by age group
age_group_sales = df.groupby("AGE_GROUP2")["NET_SALES_AMOUNT"].sum()
plt.figure(figsize=(10, 6))
sns.barplot(x=age_group_sales.index, y=age_group_sales.values)
plt.title("Total Sales by Age Group")
plt.show()
# # Exercise Two: Create a customer value-based segmentation
# In this exercise,
# * we will calculate customer metrics such as total sales, purchase frequency, and average order value.
# * Then, will explore the distribution of these metrics to understand their patterns.
# * We will define segmentation criteria based on quantiles of these metrics and create customer segments accordingly.
# * Finally, we will visualize the customer segments.
# ### Step 1: Calculate customer metrics for segmentation.
# Calculate customer metrics
customer_metrics = (
df.groupby("UCID")
.agg(
total_sales=("NET_SALES_AMOUNT", "sum"),
purchase_frequency=("INVOICE_NO", "nunique"),
average_order_value=("NET_SALES_AMOUNT", "mean"),
)
.reset_index()
)
# ### Step 2: Explore the distribution of customer metrics.
# Explore the distribution of total sales
plt.figure(figsize=(10, 6))
sns.histplot(x="total_sales", data=customer_metrics, bins=10)
plt.title("Total Sales Distribution")
plt.show()
# Explore the distribution of purchase frequency
plt.figure(figsize=(10, 6))
sns.countplot(x="purchase_frequency", data=customer_metrics)
plt.title("Purchase Frequency Distribution")
plt.show()
# Explore the distribution of average order value
plt.figure(figsize=(10, 6))
sns.histplot(x="average_order_value", data=customer_metrics, bins=10)
plt.title("Average Order Value Distribution")
plt.show()
# ### Step 3: Segment customers based on their metrics.
#
# Define segmentation criteria
high_value_threshold = customer_metrics["total_sales"].quantile(0.75)
frequent_shopper_threshold = customer_metrics["purchase_frequency"].quantile(0.75)
# Create segments based on metrics
customer_segments = []
for index, row in customer_metrics.iterrows():
segment = ""
if row["total_sales"] > high_value_threshold:
segment += "High-Value "
else:
segment += "Low-Value "
if row["purchase_frequency"] > frequent_shopper_threshold:
segment += "Frequent Shopper"
else:
segment += "Infrequent Shopper"
customer_segments.append(segment)
# Add segments to the customer metrics dataframe
customer_metrics["segment"] = customer_segments
# ### Step 4: Visualize the customer segments.
#
# Visualize customer segments
plt.figure(figsize=(10, 6))
sns.countplot(x="segment", data=customer_metrics)
plt.title("Customer Segmentation")
plt.xticks(rotation=45)
plt.show()
# # Third Exercise: creating a behavioral segmentation to identify customer preferences.
# ### Step 1: Identify relevant behavioral attributes for segmentation.
# Group data by relevant attributes and calculate aggregated metrics
preferences = (
df.groupby(["UCID", "BRAND_REPORTING", "DIV_NAME"])
.agg(
total_sales=("NET_SALES_AMOUNT", "sum"),
purchase_count=("INVOICE_NO", "nunique"),
)
.reset_index()
)
# Pivot the data to create a preference matrix
preference_matrix = preferences.pivot_table(
index="UCID",
columns=["BRAND_REPORTING", "DIV_NAME"],
values="total_sales",
fill_value=0,
)
# Perform clustering (e.g., K-means) on the preference matrix
from sklearn.cluster import KMeans
n_clusters = 4 # Define the number of clusters
kmeans = KMeans(n_clusters=n_clusters, random_state=42)
clusters = kmeans.fit_predict(preference_matrix)
# Add the cluster labels to the preference matrix
preference_matrix["Cluster"] = clusters
# ### Step 2: Analyze customer preferences and create segments.
# Group data by relevant attributes and calculate aggregated metrics
preferences = (
df.groupby(["UCID", "BRAND_REPORTING", "DIV_NAME"])
.agg(
total_sales=("NET_SALES_AMOUNT", "sum"),
purchase_count=("INVOICE_NO", "nunique"),
)
.reset_index()
)
# Pivot the data to create a preference matrix
preference_matrix = preferences.pivot_table(
index="UCID",
columns=["BRAND_REPORTING", "DIV_NAME"],
values="total_sales",
fill_value=0,
)
# Perform clustering (e.g., K-means) on the preference matrix
from sklearn.cluster import KMeans
n_clusters = 4 # Define the number of clusters
n_init = 10 # Set the value of n_init explicitly
kmeans = KMeans(n_clusters=n_clusters, n_init=n_init, random_state=42)
clusters = kmeans.fit_predict(preference_matrix.values)
# Add the cluster labels to the preference matrix
preference_matrix["Cluster"] = clusters
# ### Step 3: Visualize the behavioral segments.
#
# Plot the clusters
plt.figure(figsize=(10, 6))
sns.scatterplot(
x=preference_matrix.index,
y=preference_matrix.sum(axis=1), # Update the column name here
hue="Cluster",
palette="viridis",
data=preference_matrix,
)
plt.title("Behavioral Segmentation")
plt.xlabel("Customer ID")
plt.ylabel("Total Sales")
plt.legend(title="Cluster")
plt.show()
# # Exercise Four: Cohort analysis to show acquisition and retention trends
# Convert 'MEMBERSHIP_DATE' column to datetime
df["MEMBERSHIP_DATE"] = pd.to_datetime(df["MEMBERSHIP_DATE"])
# Create 'MembershipYearMonth' column
df["MembershipYearMonth"] = df["MEMBERSHIP_DATE"].dt.to_period("M")
# Group the data by MembershipYearMonth and calculate the initial and total number of customers
cohort_data = df.groupby("MembershipYearMonth").agg(
InitialCustomers=("UCID", "nunique"), TotalCustomers=("UCID", "count")
)
# Calculate the retention rate
cohort_data["RetentionRate"] = (
cohort_data["TotalCustomers"] / cohort_data["InitialCustomers"]
)
# Convert 'RetentionRate' column to numeric type
cohort_data["RetentionRate"] = pd.to_numeric(
cohort_data["RetentionRate"], errors="coerce"
)
# Fill missing values with 1
cohort_data["RetentionRate"] = cohort_data["RetentionRate"].fillna(1)
import matplotlib.pyplot as plt
# Line plot for InitialCustomers
plt.figure(figsize=(10, 6))
cohort_data["InitialCustomers"].plot(marker="o")
plt.title("Initial Customers Over Time")
plt.xlabel("Membership Year-Month")
plt.ylabel("Number of Customers")
plt.grid(True)
plt.show()
# Line plot for TotalCustomers
plt.figure(figsize=(10, 6))
cohort_data["TotalCustomers"].plot(marker="o")
plt.title("Total Customers Over Time")
plt.xlabel("Membership Year-Month")
plt.ylabel("Number of Customers")
plt.grid(True)
plt.show()
# Line plot for RetentionRate
plt.figure(figsize=(10, 6))
cohort_data["RetentionRate"].plot(marker="o")
plt.title("Retention Rate Over Time")
plt.xlabel("Membership Year-Month")
plt.ylabel("Retention Rate")
plt.ylim(0, 1)
plt.grid(True)
plt.show()
# # Step 1: Preprocess the data
# df['MEMBERSHIP_DATE'] = pd.to_datetime(df['MEMBERSHIP_DATE'])
# df['YearMonth'] = df['MEMBERSHIP_DATE'].dt.to_period('M')
# # Step 2: Calculate the number of unique customers by cohort and month
# cohort_data = df.groupby(['YearMonth', 'UCID']).size().reset_index(name='NumCustomers')
# # Step 3: Create a pivot table to calculate monthly active customers by cohort and month
# cohort_matrix = cohort_data.pivot_table(index='YearMonth', columns='UCID', values='NumCustomers', aggfunc='count')
# # Step 4: Calculate the retention rates
# cohort_size = cohort_matrix.iloc[:, 0]
# retention_matrix = cohort_matrix.divide(cohort_size, axis=0)
# # Step 5: Visualize the retention rates using a heatmap
# plt.figure(figsize=(12, 8))
# plt.title('Cohort Analysis - Retention Rates')
# sns.heatmap(retention_matrix, annot=True, fmt='.0%', cmap='YlGnBu', vmin=0, vmax=1, cbar=False)
# plt.show()
# # Step 6: Calculate and plot the cohort sizes
# cohort_sizes = cohort_matrix.sum(axis=0)
# cohort_sizes.plot(kind='bar', figsize=(10, 6))
# plt.title('Cohort Sizes')
# plt.xlabel('Cohort')
# plt.ylabel('Number of Customers')
# plt.show()
# Step 7: Calculate and plot the retention rates over time
retention_over_time = retention_matrix.mean()
retention_over_time.plot(figsize=(10, 6))
plt.title("Retention Rates Over Time")
plt.xlabel("Months since First Purchase")
plt.ylabel("Retention Rate")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/711/129711281.ipynb
|
one-year-retail-transactions
|
mustafaabdelnasser16
|
[{"Id": 129711281, "ScriptId": 38572052, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12057103, "CreationDate": "05/16/2023 00:32:33", "VersionNumber": 1.0, "Title": "customer behavioral segmentation", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 334.0, "LinesInsertedFromPrevious": 334.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186046699, "KernelVersionId": 129711281, "SourceDatasetVersionId": 5693599}]
|
[{"Id": 5693599, "DatasetId": 3273701, "DatasourceVersionId": 5769220, "CreatorUserId": 12057103, "LicenseName": "Unknown", "CreationDate": "05/15/2023 20:48:06", "VersionNumber": 1.0, "Title": "one_year_retail_transactions", "Slug": "one-year-retail-transactions", "Subtitle": NaN, "Description": "The dataset contains information related to retail transactions in the Gulf region. Here is a brief description of each column:\n\nUCID: Unique Customer ID.\nGENDER: Gender of the customer.\nAGE_GROUP2: Age group of the customer.\nMEMBERSHIP_DATE: Date when the customer became a member.\nBRAND_REPORTING: Brand associated with the transaction.\nLOCATION_NAME_REPORTING: Name of the location where the transaction occurred.\nSTORE_COUNTRY: Country where the store is located.\nBUSINESS_CHANNEL: Channel through which the transaction was made (e.g., offline, online).\nINVOICE_NO: Invoice number associated with the transaction.\nNET_SALES_AMOUNT: Net sales amount for the transaction.\nSALES_QTY: Quantity of items sold in the transaction.\nDISCOUNT_AMOUNT: Amount of discount applied to the transaction.\nDAY_DT: Date of the transaction.\nDIV_NAME: Division name (category) of the purchased item.\nDEPT_NAME: Department name of the purchased item.\nCLASS_NAME: Class name of the purchased item.\nSUBCLASS_NAME: Subclass name of the purchased item.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3273701, "CreatorUserId": 12057103, "OwnerUserId": 12057103.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5706612.0, "CurrentDatasourceVersionId": 5782676.0, "ForumId": 3339356, "Type": 2, "CreationDate": "05/15/2023 20:48:06", "LastActivityDate": "05/15/2023", "TotalViews": 284, "TotalDownloads": 60, "TotalVotes": 2, "TotalKernels": 1}]
|
[{"Id": 12057103, "UserName": "mustafaabdelnasser16", "DisplayName": "Mustafa Abd El-Nasser", "RegisterDate": "10/22/2022", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # First exercise
# A step-by-step process to conduct a detailed explanatory data analysis for behavioral segmentation
# ### Step 1: Import the necessary libraries and load the dataset.
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# Load the dataset
df = pd.read_excel(
"/kaggle/input/one-year-retail-transactions/Retail_Transactions.xlsx"
)
# Display the first few rows of the dataset
df.head()
# ### Step 2: Explore the dataset to gain initial insights.
# Check the summary statistics of numerical columns
df.describe()
# Check the data types and missing values
df.info()
# ### Step 3: Perform data cleaning and preprocessing (if necessary).
# Handle missing values (if any)
df.dropna(inplace=True)
# Remove duplicates (if any)
df.drop_duplicates(inplace=True)
# ### Step 4: Explore the distribution of variables and identify key trends.
# Remove leading spaces from column names
df.columns = df.columns.str.strip()
# Verify the updated column names
print(df.columns)
# Explore the distribution of categorical variables
plt.figure(figsize=(10, 6))
sns.countplot(x="GENDER", data=df)
plt.title("Gender Distribution")
plt.show()
# Explore the distribution of numerical variables
plt.figure(figsize=(10, 6))
sns.histplot(x="AGE_GROUP2", data=df, bins=10)
plt.title("Age Group Distribution")
plt.show()
# Explore the sales trends over time
plt.figure(figsize=(10, 6))
sns.lineplot(x="DAY_DT", y="NET_SALES_AMOUNT", data=df)
plt.title("Sales Trends Over Time")
plt.show()
# ### Step 5: Analyze customer behavior and identify patterns.
# Calculate and visualize average sales by brand
brand_sales = (
df.groupby("BRAND_REPORTING")["NET_SALES_AMOUNT"]
.mean()
.sort_values(ascending=False)
)
plt.figure(figsize=(10, 6))
sns.barplot(x=brand_sales.index, y=brand_sales.values)
plt.title("Average Sales by Brand")
plt.xticks(rotation=90)
plt.show()
# Analyze purchase patterns by age group
age_group_sales = df.groupby("AGE_GROUP2")["NET_SALES_AMOUNT"].sum()
plt.figure(figsize=(10, 6))
sns.barplot(x=age_group_sales.index, y=age_group_sales.values)
plt.title("Total Sales by Age Group")
plt.show()
# # Exercise Two: Create a customer value-based segmentation
# In this exercise,
# * we will calculate customer metrics such as total sales, purchase frequency, and average order value.
# * Then, will explore the distribution of these metrics to understand their patterns.
# * We will define segmentation criteria based on quantiles of these metrics and create customer segments accordingly.
# * Finally, we will visualize the customer segments.
# ### Step 1: Calculate customer metrics for segmentation.
# Calculate customer metrics
customer_metrics = (
df.groupby("UCID")
.agg(
total_sales=("NET_SALES_AMOUNT", "sum"),
purchase_frequency=("INVOICE_NO", "nunique"),
average_order_value=("NET_SALES_AMOUNT", "mean"),
)
.reset_index()
)
# ### Step 2: Explore the distribution of customer metrics.
# Explore the distribution of total sales
plt.figure(figsize=(10, 6))
sns.histplot(x="total_sales", data=customer_metrics, bins=10)
plt.title("Total Sales Distribution")
plt.show()
# Explore the distribution of purchase frequency
plt.figure(figsize=(10, 6))
sns.countplot(x="purchase_frequency", data=customer_metrics)
plt.title("Purchase Frequency Distribution")
plt.show()
# Explore the distribution of average order value
plt.figure(figsize=(10, 6))
sns.histplot(x="average_order_value", data=customer_metrics, bins=10)
plt.title("Average Order Value Distribution")
plt.show()
# ### Step 3: Segment customers based on their metrics.
#
# Define segmentation criteria
high_value_threshold = customer_metrics["total_sales"].quantile(0.75)
frequent_shopper_threshold = customer_metrics["purchase_frequency"].quantile(0.75)
# Create segments based on metrics
customer_segments = []
for index, row in customer_metrics.iterrows():
segment = ""
if row["total_sales"] > high_value_threshold:
segment += "High-Value "
else:
segment += "Low-Value "
if row["purchase_frequency"] > frequent_shopper_threshold:
segment += "Frequent Shopper"
else:
segment += "Infrequent Shopper"
customer_segments.append(segment)
# Add segments to the customer metrics dataframe
customer_metrics["segment"] = customer_segments
# ### Step 4: Visualize the customer segments.
#
# Visualize customer segments
plt.figure(figsize=(10, 6))
sns.countplot(x="segment", data=customer_metrics)
plt.title("Customer Segmentation")
plt.xticks(rotation=45)
plt.show()
# # Third Exercise: creating a behavioral segmentation to identify customer preferences.
# ### Step 1: Identify relevant behavioral attributes for segmentation.
# Group data by relevant attributes and calculate aggregated metrics
preferences = (
df.groupby(["UCID", "BRAND_REPORTING", "DIV_NAME"])
.agg(
total_sales=("NET_SALES_AMOUNT", "sum"),
purchase_count=("INVOICE_NO", "nunique"),
)
.reset_index()
)
# Pivot the data to create a preference matrix
preference_matrix = preferences.pivot_table(
index="UCID",
columns=["BRAND_REPORTING", "DIV_NAME"],
values="total_sales",
fill_value=0,
)
# Perform clustering (e.g., K-means) on the preference matrix
from sklearn.cluster import KMeans
n_clusters = 4 # Define the number of clusters
kmeans = KMeans(n_clusters=n_clusters, random_state=42)
clusters = kmeans.fit_predict(preference_matrix)
# Add the cluster labels to the preference matrix
preference_matrix["Cluster"] = clusters
# ### Step 2: Analyze customer preferences and create segments.
# Group data by relevant attributes and calculate aggregated metrics
preferences = (
df.groupby(["UCID", "BRAND_REPORTING", "DIV_NAME"])
.agg(
total_sales=("NET_SALES_AMOUNT", "sum"),
purchase_count=("INVOICE_NO", "nunique"),
)
.reset_index()
)
# Pivot the data to create a preference matrix
preference_matrix = preferences.pivot_table(
index="UCID",
columns=["BRAND_REPORTING", "DIV_NAME"],
values="total_sales",
fill_value=0,
)
# Perform clustering (e.g., K-means) on the preference matrix
from sklearn.cluster import KMeans
n_clusters = 4 # Define the number of clusters
n_init = 10 # Set the value of n_init explicitly
kmeans = KMeans(n_clusters=n_clusters, n_init=n_init, random_state=42)
clusters = kmeans.fit_predict(preference_matrix.values)
# Add the cluster labels to the preference matrix
preference_matrix["Cluster"] = clusters
# ### Step 3: Visualize the behavioral segments.
#
# Plot the clusters
plt.figure(figsize=(10, 6))
sns.scatterplot(
x=preference_matrix.index,
y=preference_matrix.sum(axis=1), # Update the column name here
hue="Cluster",
palette="viridis",
data=preference_matrix,
)
plt.title("Behavioral Segmentation")
plt.xlabel("Customer ID")
plt.ylabel("Total Sales")
plt.legend(title="Cluster")
plt.show()
# # Exercise Four: Cohort analysis to show acquisition and retention trends
# Convert 'MEMBERSHIP_DATE' column to datetime
df["MEMBERSHIP_DATE"] = pd.to_datetime(df["MEMBERSHIP_DATE"])
# Create 'MembershipYearMonth' column
df["MembershipYearMonth"] = df["MEMBERSHIP_DATE"].dt.to_period("M")
# Group the data by MembershipYearMonth and calculate the initial and total number of customers
cohort_data = df.groupby("MembershipYearMonth").agg(
InitialCustomers=("UCID", "nunique"), TotalCustomers=("UCID", "count")
)
# Calculate the retention rate
cohort_data["RetentionRate"] = (
cohort_data["TotalCustomers"] / cohort_data["InitialCustomers"]
)
# Convert 'RetentionRate' column to numeric type
cohort_data["RetentionRate"] = pd.to_numeric(
cohort_data["RetentionRate"], errors="coerce"
)
# Fill missing values with 1
cohort_data["RetentionRate"] = cohort_data["RetentionRate"].fillna(1)
import matplotlib.pyplot as plt
# Line plot for InitialCustomers
plt.figure(figsize=(10, 6))
cohort_data["InitialCustomers"].plot(marker="o")
plt.title("Initial Customers Over Time")
plt.xlabel("Membership Year-Month")
plt.ylabel("Number of Customers")
plt.grid(True)
plt.show()
# Line plot for TotalCustomers
plt.figure(figsize=(10, 6))
cohort_data["TotalCustomers"].plot(marker="o")
plt.title("Total Customers Over Time")
plt.xlabel("Membership Year-Month")
plt.ylabel("Number of Customers")
plt.grid(True)
plt.show()
# Line plot for RetentionRate
plt.figure(figsize=(10, 6))
cohort_data["RetentionRate"].plot(marker="o")
plt.title("Retention Rate Over Time")
plt.xlabel("Membership Year-Month")
plt.ylabel("Retention Rate")
plt.ylim(0, 1)
plt.grid(True)
plt.show()
# # Step 1: Preprocess the data
# df['MEMBERSHIP_DATE'] = pd.to_datetime(df['MEMBERSHIP_DATE'])
# df['YearMonth'] = df['MEMBERSHIP_DATE'].dt.to_period('M')
# # Step 2: Calculate the number of unique customers by cohort and month
# cohort_data = df.groupby(['YearMonth', 'UCID']).size().reset_index(name='NumCustomers')
# # Step 3: Create a pivot table to calculate monthly active customers by cohort and month
# cohort_matrix = cohort_data.pivot_table(index='YearMonth', columns='UCID', values='NumCustomers', aggfunc='count')
# # Step 4: Calculate the retention rates
# cohort_size = cohort_matrix.iloc[:, 0]
# retention_matrix = cohort_matrix.divide(cohort_size, axis=0)
# # Step 5: Visualize the retention rates using a heatmap
# plt.figure(figsize=(12, 8))
# plt.title('Cohort Analysis - Retention Rates')
# sns.heatmap(retention_matrix, annot=True, fmt='.0%', cmap='YlGnBu', vmin=0, vmax=1, cbar=False)
# plt.show()
# # Step 6: Calculate and plot the cohort sizes
# cohort_sizes = cohort_matrix.sum(axis=0)
# cohort_sizes.plot(kind='bar', figsize=(10, 6))
# plt.title('Cohort Sizes')
# plt.xlabel('Cohort')
# plt.ylabel('Number of Customers')
# plt.show()
# Step 7: Calculate and plot the retention rates over time
retention_over_time = retention_matrix.mean()
retention_over_time.plot(figsize=(10, 6))
plt.title("Retention Rates Over Time")
plt.xlabel("Months since First Purchase")
plt.ylabel("Retention Rate")
plt.show()
| false | 0 | 3,080 | 0 | 3,368 | 3,080 |
||
129685475
|
<jupyter_start><jupyter_text>Wild blueberry Yield Prediction Dataset
### Context
Blueberries are perennial flowering plants with blue or purple berries. They are classified in the section Cyanococcus within the genus Vaccinium. Vaccinium also includes cranberries, bilberries, huckleberries, and Madeira blueberries. Commercial blueberries—both wild (lowbush) and cultivated (highbush)—are all native to North America. The highbush varieties were introduced into Europe during the 1930s.
Blueberries are usually prostrate shrubs that can vary in size from 10 centimeters (4 inches) to 4 meters (13 feet) in height. In the commercial production of blueberries, the species with small, pea-size berries growing on low-level bushes are known as "lowbush blueberries" (synonymous with "wild"), while the species with larger berries growing on taller, cultivated bushes are known as "highbush blueberries". Canada is the leading producer of lowbush blueberries, while the United States produces some 40% of the world s supply of highbush blueberries.
### Content
"The dataset used for predictive modeling was generated by the Wild Blueberry Pollination Simulation Model, which is an open-source, spatially-explicit computer simulation program that enables exploration of how various factors, including plant spatial arrangement, outcrossing and self-pollination, bee species compositions and weather conditions, in isolation and combination, affect pollination efficiency and yield of the wild blueberry agroecosystem. The simulation model has been validated by the field observation and experimental data collected in Maine USA and Canadian Maritimes during the last 30 years and now is a useful tool for hypothesis testing and theory development for wild blueberry pollination researches."
Features Unit Description
Clonesize m2 The average blueberry clone size in the field
Honeybee bees/m2/min Honeybee density in the field
Bumbles bees/m2/min Bumblebee density in the field
Andrena bees/m2/min Andrena bee density in the field
Osmia bees/m2/min Osmia bee density in the field
MaxOfUpperTRange ℃ The highest record of the upper band daily air temperature during the bloom season
MinOfUpperTRange ℃ The lowest record of the upper band daily air temperature
AverageOfUpperTRange ℃ The average of the upper band daily air temperature
MaxOfLowerTRange ℃ The highest record of the lower band daily air temperature
MinOfLowerTRange ℃ The lowest record of the lower band daily air temperature
AverageOfLowerTRange ℃ The average of the lower band daily air temperature
RainingDays Day The total number of days during the bloom season, each of which has precipitation larger than zero
AverageRainingDays Day The average of raining days of the entire bloom season
Kaggle dataset identifier: wild-blueberry-yield-prediction-dataset
<jupyter_script>#
# # Load Python Pakages
#
# basics
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
# preprocessing
from sklearn.preprocessing import (
StandardScaler,
RobustScaler,
MinMaxScaler,
PowerTransformer,
)
# statistics
from scipy import stats
from scipy.stats import skew
from scipy.special import boxcox1p
# feature engineering
from sklearn.feature_selection import mutual_info_regression
# transformers and pipeline
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.compose import ColumnTransformer, make_column_transformer
from sklearn.preprocessing import FunctionTransformer
from sklearn.pipeline import Pipeline, make_pipeline
from sklearn import set_config
# algorithms
from lightgbm import LGBMRegressor
# model evaluation
from sklearn.model_selection import GridSearchCV, cross_val_score, cross_validate
from sklearn.model_selection import RandomizedSearchCV
from sklearn.model_selection import ShuffleSplit
from sklearn.metrics import mean_absolute_error, mean_squared_error
#
# # First look to data
# Read the data
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv", index_col="id")
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv", index_col="id")
original = pd.read_csv(
"/kaggle/input/wild-blueberry-yield-prediction-dataset/WildBlueberryPollinationSimulationData.csv",
index_col=[0],
)
# reserved for pipeline
pipe_data = train.copy()
pipe_test = test.copy()
pipe_original = original.copy()
# use for preliminary analysis
train_df = train.copy()
test_df = test.copy()
original_df = original.copy()
train_df.head()
original_df.index.names = ["id"]
original_df.head()
train_df = pd.concat([train_df, original_df])
train_df.head()
train_df.info()
# is there any missing value?
train_df.isnull().any().any()
# ## Descpriptive statistics
# numerical feature descriptive statistics
train_df.describe().T
# ## Grouping features for preprocessing purposes
train_df.nunique().sort_values()
# Just bookkeeping
feature_list = [feature for feature in train_df.columns if not feature == "yield"]
continuous_features = ["fruitmass", "fruitset", "seeds"]
discrete_features = list(set(feature_list) - set(continuous_features))
assert feature_list.sort() == (continuous_features + discrete_features).sort()
#
# # Exploratory Data Analysis
# Let's obsorve how target variable changes with features.
fig, ax = plt.subplots(6, 3, figsize=(40, 20))
for var, subplot in zip(feature_list, ax.flatten()):
sns.scatterplot(x=var, y="yield", data=train_df, ax=subplot, hue="yield")
# Observations:
# * There are strong correlation between continuous_features and target
# Let's look at correlations between features and the target with a more quantitative way..
# Display correlations between features and yield on heatmap.
sns.set(font_scale=1.1)
correlation_train = train_df.corr()
mask = np.triu(correlation_train.corr())
plt.figure(figsize=(15, 15))
sns.heatmap(
correlation_train,
annot=True,
fmt=".1f",
cmap="coolwarm",
square=True,
mask=mask,
linewidths=1,
cbar=False,
)
# Mutual information is another measure, which also capable to measure more diverse relationships and good at categorical and discrete variables.
y = train_df["yield"]
# determine the mutual information for numerical features
# You need to fillna to get results from mutual_info_regression function
mutual_df = train_df[feature_list]
mutual_info = mutual_info_regression(mutual_df, y, random_state=1)
mutual_info = pd.Series(mutual_info)
mutual_info.index = mutual_df.columns
pd.DataFrame(
mutual_info.sort_values(ascending=False), columns=["MI_score"]
).style.background_gradient("cool")
#
# # Feature Engineering
# Let's define some new features.
train_df["total_bee_density"] = (
train_df["honeybee"] + train_df["bumbles"] + train_df["andrena"] + train_df["osmia"]
)
train_df["bee_to_clone"] = train_df["total_bee_density"] / train_df["clonesize"]
train_df["Max_temp_difference"] = (
train_df["MaxOfUpperTRange"] - train_df["MinOfLowerTRange"]
)
train_df["Avarage_temp_difference"] = (
train_df["AverageOfUpperTRange"] - train_df["AverageOfLowerTRange"]
)
train_df["mass_set"] = train_df["fruitmass"] * train_df["fruitset"]
train_df["mass_seed"] = train_df["fruitmass"] * train_df["seeds"]
train_df["set_seed"] = train_df["fruitset"] * train_df["seeds"]
train_df["mass_ser_seed"] = (
train_df["fruitmass"] * train_df["fruitset"] * train_df["seeds"]
)
new_features = [
"total_bee_density",
"bee_to_clone",
"Max_temp_difference",
"Avarage_temp_difference",
"mass_set",
"mass_seed",
"set_seed",
"mass_ser_seed",
]
# Let's check new features mutual information scores...
mutual_df = train_df[new_features]
mutual_info = mutual_info_regression(mutual_df, y, random_state=1)
mutual_info = pd.Series(mutual_info)
mutual_info.index = mutual_df.columns
pd.DataFrame(
mutual_info.sort_values(ascending=False), columns=["New_Feature_MI"]
).style.background_gradient("cool")
fig, ax = plt.subplots(3, 3, figsize=(20, 20))
for var, subplot in zip(new_features, ax.flatten()):
sns.scatterplot(x=var, y="yield", data=train_df, ax=subplot, hue="yield")
updated_feature_list = train_df.columns.to_list()
updated_continuous_features = [
"total_bee_density",
"bee_to_clone",
"fruitmass",
"fruitset",
"seeds",
"mass_set",
"mass_seed",
"set_seed",
"mass_ser_seed",
]
updated_discrete_features = list(
set(updated_feature_list) - set(updated_continuous_features)
)
assert (
updated_feature_list.sort()
== (updated_continuous_features + updated_discrete_features).sort()
)
#
# ## A custom pipeline for Feature Engineering
class FeatureCreator(BaseEstimator, TransformerMixin):
def __init__(self, add_attributes=True):
self.add_attributes = add_attributes
def fit(self, X, y=None):
return self
def transform(self, X):
if self.add_attributes:
X_copy = X.copy()
X_copy["total_bee_density"] = (
X_copy["honeybee"]
+ X_copy["bumbles"]
+ X_copy["andrena"]
+ X_copy["osmia"]
)
X_copy["bee_to_clone"] = X_copy["total_bee_density"] / X_copy["clonesize"]
X_copy["Max_temp_difference"] = (
X_copy["MaxOfUpperTRange"] - X_copy["MinOfLowerTRange"]
)
X_copy["Avarage_temp_difference"] = (
X_copy["AverageOfUpperTRange"] - X_copy["AverageOfLowerTRange"]
)
X_copy["mass_set"] = X_copy["fruitmass"] * X_copy["fruitset"]
X_copy["mass_seed"] = X_copy["fruitmass"] * X_copy["seeds"]
X_copy["set_seed"] = X_copy["fruitset"] * X_copy["seeds"]
X_copy["mass_ser_seed"] = (
X_copy["fruitmass"] * X_copy["fruitset"] * X_copy["seeds"]
)
X_copy = X_copy.drop(
[
"MaxOfLowerTRange",
"MaxOfUpperTRange",
"MinOfLowerTRange",
"MinOfUpperTRange",
],
axis=1,
)
return X_copy
else:
return X_copy
Creator = FeatureCreator(add_attributes=True)
#
# # Putting pieces together
# Okay...We are almost ready to start the modeling. Before moving on we will make first touch with the data that we reserved for the pipeline. Let's separate target and features.
pipe_original.index.names = ["id"]
pipe_original.head()
pipe_data = pipe_data.sample(frac=1, random_state=0)
pipe_data = pd.concat([pipe_data, pipe_original])
pipe_data.info()
y = pipe_data["yield"]
pipe_data = pipe_data.drop("yield", axis=1)
#
# # Scikit-learn pipeline with AutoML
# flaml
from flaml import AutoML
automl = AutoML()
automl_pipeline = Pipeline([("Creator", Creator), ("automl", automl)])
automl_pipeline
# Specify automl goal and constraint
automl_settings = {
"time_budget": 7500,
"metric": "mae",
"task": "regression",
"seed": 7654321,
"ensemble": True,
}
pipeline_settings = {f"automl__{key}": value for key, value in automl_settings.items()}
automl_pipeline = automl_pipeline.fit(pipe_data, y, **pipeline_settings)
preds_test = automl_pipeline.predict(pipe_test)
# ###
# # Submission
output = pd.DataFrame({"id": pipe_test.index, "yield": preds_test})
output.to_csv("submission.csv", index=False)
output.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/685/129685475.ipynb
|
wild-blueberry-yield-prediction-dataset
|
shashwatwork
|
[{"Id": 129685475, "ScriptId": 38325284, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9785820, "CreationDate": "05/15/2023 18:26:25", "VersionNumber": 2.0, "Title": "Sklearn Pipeline with flaml", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 274.0, "LinesInsertedFromPrevious": 5.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 269.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186008507, "KernelVersionId": 129685475, "SourceDatasetVersionId": 2462316}]
|
[{"Id": 2462316, "DatasetId": 1490445, "DatasourceVersionId": 2504743, "CreatorUserId": 1444085, "LicenseName": "Attribution 4.0 International (CC BY 4.0)", "CreationDate": "07/25/2021 17:48:21", "VersionNumber": 2.0, "Title": "Wild blueberry Yield Prediction Dataset", "Slug": "wild-blueberry-yield-prediction-dataset", "Subtitle": "Predict the yield of Wild Blueberry", "Description": "### Context\n\nBlueberries are perennial flowering plants with blue or purple berries. They are classified in the section Cyanococcus within the genus Vaccinium. Vaccinium also includes cranberries, bilberries, huckleberries, and Madeira blueberries. Commercial blueberries\u2014both wild (lowbush) and cultivated (highbush)\u2014are all native to North America. The highbush varieties were introduced into Europe during the 1930s.\n\nBlueberries are usually prostrate shrubs that can vary in size from 10 centimeters (4 inches) to 4 meters (13 feet) in height. In the commercial production of blueberries, the species with small, pea-size berries growing on low-level bushes are known as \"lowbush blueberries\" (synonymous with \"wild\"), while the species with larger berries growing on taller, cultivated bushes are known as \"highbush blueberries\". Canada is the leading producer of lowbush blueberries, while the United States produces some 40% of the world s supply of highbush blueberries.\n\n### Content\n\n\"The dataset used for predictive modeling was generated by the Wild Blueberry Pollination Simulation Model, which is an open-source, spatially-explicit computer simulation program that enables exploration of how various factors, including plant spatial arrangement, outcrossing and self-pollination, bee species compositions and weather conditions, in isolation and combination, affect pollination efficiency and yield of the wild blueberry agroecosystem. The simulation model has been validated by the field observation and experimental data collected in Maine USA and Canadian Maritimes during the last 30 years and now is a useful tool for hypothesis testing and theory development for wild blueberry pollination researches.\"\n\nFeatures \tUnit\tDescription\nClonesize\tm2\tThe average blueberry clone size in the field\nHoneybee\tbees/m2/min\tHoneybee density in the field\nBumbles\tbees/m2/min\tBumblebee density in the field\nAndrena\tbees/m2/min\tAndrena bee density in the field\nOsmia\tbees/m2/min\tOsmia bee density in the field\nMaxOfUpperTRange\t\u2103\tThe highest record of the upper band daily air temperature during the bloom season\nMinOfUpperTRange\t\u2103\tThe lowest record of the upper band daily air temperature\nAverageOfUpperTRange\t\u2103\tThe average of the upper band daily air temperature\nMaxOfLowerTRange\t\u2103\tThe highest record of the lower band daily air temperature\nMinOfLowerTRange\t\u2103\tThe lowest record of the lower band daily air temperature\nAverageOfLowerTRange\t\u2103\tThe average of the lower band daily air temperature\nRainingDays\tDay\tThe total number of days during the bloom season, each of which has precipitation larger than zero\nAverageRainingDays\tDay\tThe average of raining days of the entire bloom season\n\n### Acknowledgements\n\nQu, Hongchun; Obsie, Efrem; Drummond, Frank (2020), \u201cData for: Wild blueberry yield prediction using a combination of computer simulation and machine learning algorithms\u201d, Mendeley Data, V1, doi: 10.17632/p5hvjzsvn8.1\n\nDataset is outsourced from [here.](https://data.mendeley.com/datasets/p5hvjzsvn8/1)", "VersionNotes": "updated", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1490445, "CreatorUserId": 1444085, "OwnerUserId": 1444085.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2462316.0, "CurrentDatasourceVersionId": 2504743.0, "ForumId": 1510148, "Type": 2, "CreationDate": "07/25/2021 17:47:00", "LastActivityDate": "07/25/2021", "TotalViews": 11876, "TotalDownloads": 1130, "TotalVotes": 48, "TotalKernels": 82}]
|
[{"Id": 1444085, "UserName": "shashwatwork", "DisplayName": "Shashwat Tiwari", "RegisterDate": "11/24/2017", "PerformanceTier": 2}]
|
#
# # Load Python Pakages
#
# basics
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
# preprocessing
from sklearn.preprocessing import (
StandardScaler,
RobustScaler,
MinMaxScaler,
PowerTransformer,
)
# statistics
from scipy import stats
from scipy.stats import skew
from scipy.special import boxcox1p
# feature engineering
from sklearn.feature_selection import mutual_info_regression
# transformers and pipeline
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.compose import ColumnTransformer, make_column_transformer
from sklearn.preprocessing import FunctionTransformer
from sklearn.pipeline import Pipeline, make_pipeline
from sklearn import set_config
# algorithms
from lightgbm import LGBMRegressor
# model evaluation
from sklearn.model_selection import GridSearchCV, cross_val_score, cross_validate
from sklearn.model_selection import RandomizedSearchCV
from sklearn.model_selection import ShuffleSplit
from sklearn.metrics import mean_absolute_error, mean_squared_error
#
# # First look to data
# Read the data
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv", index_col="id")
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv", index_col="id")
original = pd.read_csv(
"/kaggle/input/wild-blueberry-yield-prediction-dataset/WildBlueberryPollinationSimulationData.csv",
index_col=[0],
)
# reserved for pipeline
pipe_data = train.copy()
pipe_test = test.copy()
pipe_original = original.copy()
# use for preliminary analysis
train_df = train.copy()
test_df = test.copy()
original_df = original.copy()
train_df.head()
original_df.index.names = ["id"]
original_df.head()
train_df = pd.concat([train_df, original_df])
train_df.head()
train_df.info()
# is there any missing value?
train_df.isnull().any().any()
# ## Descpriptive statistics
# numerical feature descriptive statistics
train_df.describe().T
# ## Grouping features for preprocessing purposes
train_df.nunique().sort_values()
# Just bookkeeping
feature_list = [feature for feature in train_df.columns if not feature == "yield"]
continuous_features = ["fruitmass", "fruitset", "seeds"]
discrete_features = list(set(feature_list) - set(continuous_features))
assert feature_list.sort() == (continuous_features + discrete_features).sort()
#
# # Exploratory Data Analysis
# Let's obsorve how target variable changes with features.
fig, ax = plt.subplots(6, 3, figsize=(40, 20))
for var, subplot in zip(feature_list, ax.flatten()):
sns.scatterplot(x=var, y="yield", data=train_df, ax=subplot, hue="yield")
# Observations:
# * There are strong correlation between continuous_features and target
# Let's look at correlations between features and the target with a more quantitative way..
# Display correlations between features and yield on heatmap.
sns.set(font_scale=1.1)
correlation_train = train_df.corr()
mask = np.triu(correlation_train.corr())
plt.figure(figsize=(15, 15))
sns.heatmap(
correlation_train,
annot=True,
fmt=".1f",
cmap="coolwarm",
square=True,
mask=mask,
linewidths=1,
cbar=False,
)
# Mutual information is another measure, which also capable to measure more diverse relationships and good at categorical and discrete variables.
y = train_df["yield"]
# determine the mutual information for numerical features
# You need to fillna to get results from mutual_info_regression function
mutual_df = train_df[feature_list]
mutual_info = mutual_info_regression(mutual_df, y, random_state=1)
mutual_info = pd.Series(mutual_info)
mutual_info.index = mutual_df.columns
pd.DataFrame(
mutual_info.sort_values(ascending=False), columns=["MI_score"]
).style.background_gradient("cool")
#
# # Feature Engineering
# Let's define some new features.
train_df["total_bee_density"] = (
train_df["honeybee"] + train_df["bumbles"] + train_df["andrena"] + train_df["osmia"]
)
train_df["bee_to_clone"] = train_df["total_bee_density"] / train_df["clonesize"]
train_df["Max_temp_difference"] = (
train_df["MaxOfUpperTRange"] - train_df["MinOfLowerTRange"]
)
train_df["Avarage_temp_difference"] = (
train_df["AverageOfUpperTRange"] - train_df["AverageOfLowerTRange"]
)
train_df["mass_set"] = train_df["fruitmass"] * train_df["fruitset"]
train_df["mass_seed"] = train_df["fruitmass"] * train_df["seeds"]
train_df["set_seed"] = train_df["fruitset"] * train_df["seeds"]
train_df["mass_ser_seed"] = (
train_df["fruitmass"] * train_df["fruitset"] * train_df["seeds"]
)
new_features = [
"total_bee_density",
"bee_to_clone",
"Max_temp_difference",
"Avarage_temp_difference",
"mass_set",
"mass_seed",
"set_seed",
"mass_ser_seed",
]
# Let's check new features mutual information scores...
mutual_df = train_df[new_features]
mutual_info = mutual_info_regression(mutual_df, y, random_state=1)
mutual_info = pd.Series(mutual_info)
mutual_info.index = mutual_df.columns
pd.DataFrame(
mutual_info.sort_values(ascending=False), columns=["New_Feature_MI"]
).style.background_gradient("cool")
fig, ax = plt.subplots(3, 3, figsize=(20, 20))
for var, subplot in zip(new_features, ax.flatten()):
sns.scatterplot(x=var, y="yield", data=train_df, ax=subplot, hue="yield")
updated_feature_list = train_df.columns.to_list()
updated_continuous_features = [
"total_bee_density",
"bee_to_clone",
"fruitmass",
"fruitset",
"seeds",
"mass_set",
"mass_seed",
"set_seed",
"mass_ser_seed",
]
updated_discrete_features = list(
set(updated_feature_list) - set(updated_continuous_features)
)
assert (
updated_feature_list.sort()
== (updated_continuous_features + updated_discrete_features).sort()
)
#
# ## A custom pipeline for Feature Engineering
class FeatureCreator(BaseEstimator, TransformerMixin):
def __init__(self, add_attributes=True):
self.add_attributes = add_attributes
def fit(self, X, y=None):
return self
def transform(self, X):
if self.add_attributes:
X_copy = X.copy()
X_copy["total_bee_density"] = (
X_copy["honeybee"]
+ X_copy["bumbles"]
+ X_copy["andrena"]
+ X_copy["osmia"]
)
X_copy["bee_to_clone"] = X_copy["total_bee_density"] / X_copy["clonesize"]
X_copy["Max_temp_difference"] = (
X_copy["MaxOfUpperTRange"] - X_copy["MinOfLowerTRange"]
)
X_copy["Avarage_temp_difference"] = (
X_copy["AverageOfUpperTRange"] - X_copy["AverageOfLowerTRange"]
)
X_copy["mass_set"] = X_copy["fruitmass"] * X_copy["fruitset"]
X_copy["mass_seed"] = X_copy["fruitmass"] * X_copy["seeds"]
X_copy["set_seed"] = X_copy["fruitset"] * X_copy["seeds"]
X_copy["mass_ser_seed"] = (
X_copy["fruitmass"] * X_copy["fruitset"] * X_copy["seeds"]
)
X_copy = X_copy.drop(
[
"MaxOfLowerTRange",
"MaxOfUpperTRange",
"MinOfLowerTRange",
"MinOfUpperTRange",
],
axis=1,
)
return X_copy
else:
return X_copy
Creator = FeatureCreator(add_attributes=True)
#
# # Putting pieces together
# Okay...We are almost ready to start the modeling. Before moving on we will make first touch with the data that we reserved for the pipeline. Let's separate target and features.
pipe_original.index.names = ["id"]
pipe_original.head()
pipe_data = pipe_data.sample(frac=1, random_state=0)
pipe_data = pd.concat([pipe_data, pipe_original])
pipe_data.info()
y = pipe_data["yield"]
pipe_data = pipe_data.drop("yield", axis=1)
#
# # Scikit-learn pipeline with AutoML
# flaml
from flaml import AutoML
automl = AutoML()
automl_pipeline = Pipeline([("Creator", Creator), ("automl", automl)])
automl_pipeline
# Specify automl goal and constraint
automl_settings = {
"time_budget": 7500,
"metric": "mae",
"task": "regression",
"seed": 7654321,
"ensemble": True,
}
pipeline_settings = {f"automl__{key}": value for key, value in automl_settings.items()}
automl_pipeline = automl_pipeline.fit(pipe_data, y, **pipeline_settings)
preds_test = automl_pipeline.predict(pipe_test)
# ###
# # Submission
output = pd.DataFrame({"id": pipe_test.index, "yield": preds_test})
output.to_csv("submission.csv", index=False)
output.head()
| false | 3 | 2,579 | 0 | 3,315 | 2,579 |
||
129685366
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
train.head()
train.isnull().sum()
test.isnull().sum()
test.head()
test1 = test.iloc[:, 1:17]
test1.head()
X = train.iloc[:, 1:17]
Y = train["yield"]
X.head()
Y.head()
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2, random_state=0)
X_train.shape, X_test.shape
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(X_train, Y_train)
y_pred = lr.predict(X_test)
y_pred
Y_test.shape, y_pred.shape
X_test.shape, test1.shape
y_pred1 = lr.predict(test1)
y_pred1
sample = pd.read_csv("/kaggle/input/playground-series-s3e14/sample_submission.csv")
sample.head()
sample["yield"] = y_pred1
sample.head()
sample.to_csv("submission.csv", index=False)
from sklearn.metrics import mean_squared_error
acc = mean_squared_error(Y_test, y_pred)
print(acc)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/685/129685366.ipynb
| null | null |
[{"Id": 129685366, "ScriptId": 38563657, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7351911, "CreationDate": "05/15/2023 18:25:16", "VersionNumber": 2.0, "Title": "Simple Linear Regression PS3E14_Prediction of Wild", "EvaluationDate": "05/15/2023", "IsChange": false, "TotalLines": 71.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 71.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
train.head()
train.isnull().sum()
test.isnull().sum()
test.head()
test1 = test.iloc[:, 1:17]
test1.head()
X = train.iloc[:, 1:17]
Y = train["yield"]
X.head()
Y.head()
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2, random_state=0)
X_train.shape, X_test.shape
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(X_train, Y_train)
y_pred = lr.predict(X_test)
y_pred
Y_test.shape, y_pred.shape
X_test.shape, test1.shape
y_pred1 = lr.predict(test1)
y_pred1
sample = pd.read_csv("/kaggle/input/playground-series-s3e14/sample_submission.csv")
sample.head()
sample["yield"] = y_pred1
sample.head()
sample.to_csv("submission.csv", index=False)
from sklearn.metrics import mean_squared_error
acc = mean_squared_error(Y_test, y_pred)
print(acc)
| false | 0 | 564 | 0 | 564 | 564 |
||
129685248
|
<jupyter_start><jupyter_text>Bank Customer Churn
RowNumber—corresponds to the record (row) number and has no effect on the output.
CustomerId—contains random values and has no effect on customer leaving the bank.
Surname—the surname of a customer has no impact on their decision to leave the bank.
CreditScore—can have an effect on customer churn, since a customer with a higher credit score is less likely to leave the bank.
Geography—a customer’s location can affect their decision to leave the bank.
Gender—it’s interesting to explore whether gender plays a role in a customer leaving the bank.
Age—this is certainly relevant, since older customers are less likely to leave their bank than younger ones.
Tenure—refers to the number of years that the customer has been a client of the bank. Normally, older clients are more loyal and less likely to leave a bank.
Balance—also a very good indicator of customer churn, as people with a higher balance in their accounts are less likely to leave the bank compared to those with lower balances.
NumOfProducts—refers to the number of products that a customer has purchased through the bank.
HasCrCard—denotes whether or not a customer has a credit card. This column is also relevant, since people with a credit card are less likely to leave the bank.
IsActiveMember—active customers are less likely to leave the bank.
EstimatedSalary—as with balance, people with lower salaries are more likely to leave the bank compared to those with higher salaries.
Exited—whether or not the customer left the bank.
Complain—customer has complaint or not.
Satisfaction Score—Score provided by the customer for their complaint resolution.
Card Type—type of card hold by the customer.
Points Earned—the points earned by the customer for using credit card.
Acknowledgements
As we know, it is much more expensive to sign in a new client than keeping an existing one.
It is advantageous for banks to know what leads a client towards the decision to leave the company.
Churn prevention allows companies to develop loyalty programs and retention campaigns to keep as many customers as possible.
Kaggle dataset identifier: bank-customer-churn
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv("/kaggle/input/bank-customer-churn/Customer-Churn-Records.csv")
df.head()
df = df.drop(["RowNumber", "CustomerId", "Surname"], axis=1)
df.shape
df.isnull().sum()
df["Gender"].unique()
df["Card Type"].unique()
# ***Categorical feautures are:***
# 1. Gender
# 2. HasCrCard
# 3. IsActiveMember
# 4. Exited
# 5. Complain
# 6. Card Type
df1 = df.groupby("Complain")["Exited"].apply(lambda x: (x == 1).mean()).reset_index()
df1 = df1.sort_values("Exited", ascending=False)
sns.barplot(data=df1, x="Complain", y="Exited", order=df1.Complain, color="#FF8C01")
plt.xlabel("Has complained or not")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Complain history")
plt.show()
df1 = df.groupby("HasCrCard")["Exited"].apply(lambda x: (x == 1).mean()).reset_index()
df1 = df1.sort_values("Exited", ascending=False)
sns.barplot(data=df1, x="HasCrCard", y="Exited", order=df1.HasCrCard, color="#FF8C01")
plt.xlabel("Has Credit Card or not")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Credit Card Holder or Not")
plt.show()
df1 = df.groupby("Gender")["Exited"].apply(lambda x: (x == 1).mean()).reset_index()
df1 = df1.sort_values("Exited", ascending=False)
sns.barplot(data=df1, x="Gender", y="Exited", order=df1.Gender, color="#FF8C01")
plt.xlabel("Has Credit Card or not")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Gender of Customer")
plt.show()
df1 = (
df.groupby("IsActiveMember")["Exited"]
.apply(lambda x: (x == 1).mean())
.reset_index()
)
df1 = df1.sort_values("Exited", ascending=False)
sns.barplot(
data=df1, x="IsActiveMember", y="Exited", order=df1.IsActiveMember, color="#FF8C01"
)
plt.xlabel("Is Active Member or Not")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Activity of the Customer")
plt.show()
# The plot tells that if a bank customer is not active then there is a tendency for the customer to leave the bank.
# Card Type
df1 = df.groupby("Card Type")["Exited"].apply(lambda x: (x == 1).mean()).reset_index()
df1 = df1.sort_values("Card Type", ascending=False)
sns.barplot(
data=df1, x="Card Type", y="Exited", order=df1["Card Type"], color="#FF8C01"
)
plt.xlabel("Type of Card Holder")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Credit Card TYpe")
plt.show()
# It is shocking but the graph shows that a gold card holder has the largest chance of leaving the bank as compared to other card type holders.
# And the most shocking part is that diamond users are more likely to be churned
# as they have a proposition of 20% in the whole dataset.
df.head()
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
classification_report,
precision_score,
)
from sklearn.tree import DecisionTreeClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
lbl = LabelEncoder()
# 1. Gender
# 2. HasCrCard
# 3. IsActiveMember
# 4. Exited
# 5. Complain
# 6. Card Type
df["Gender"] = lbl.fit_transform(df["Gender"])
df["Card Type"] = lbl.fit_transform(df["Card Type"])
df["Geography"] = lbl.fit_transform(df["Geography"])
x = df.drop(["Exited"], axis=1) # independent feature
y = df["Exited"] # dependent feature
X_train, X_test, y_train, y_test = train_test_split(
x, y, test_size=0.15, random_state=42
)
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
k = np.sqrt(df.shape[0])
k
accuracy = []
prec = []
iter = 1
for i in range(100, 105):
knn = KNeighborsClassifier(n_neighbors=i)
knn.fit(X_train, y_train)
y_predi = knn.predict(X_test)
score = accuracy_score(y_test, y_predi)
precision = precision_score(y_test, y_predi)
accuracy.append(score * 100)
prec.append(precision * 100)
for i in range(len(accuracy)):
print("Accuracy of " + str(iter) + "th model is: " + str(accuracy[i]))
print("Precision of " + str(iter) + "th model is: " + str(prec[i]))
iter += 1
from sklearn.ensemble import GradientBoostingClassifier
model = GradientBoostingClassifier(n_estimators=12, loss="exponential", subsample=0.999)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
score = accuracy_score(y_test, y_pred)
precScore = precision_score(y_test, y_pred)
print("ACCURACY OF the RFC model is: " + str(score * 100) + "%")
print("Precision of the RFC model is: " + str(precScore * 100) + "%")
from sklearn.ensemble import AdaBoostClassifier
model = AdaBoostClassifier(n_estimators=11, random_state=1, algorithm="SAMME")
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
score = accuracy_score(y_test, y_pred)
precScore = precision_score(y_test, y_pred)
print("ACCURACY OF the RFC model is: " + str(score * 100) + "%")
print("Precision of the RFC model is: " + str(precScore * 100) + "%")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/685/129685248.ipynb
|
bank-customer-churn
|
radheshyamkollipara
|
[{"Id": 129685248, "ScriptId": 38564486, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8103914, "CreationDate": "05/15/2023 18:23:56", "VersionNumber": 2.0, "Title": "notebookb3e9a432fe", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 179.0, "LinesInsertedFromPrevious": 82.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 97.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186008074, "KernelVersionId": 129685248, "SourceDatasetVersionId": 5550559}]
|
[{"Id": 5550559, "DatasetId": 3197960, "DatasourceVersionId": 5625285, "CreatorUserId": 14862076, "LicenseName": "Other (specified in description)", "CreationDate": "04/28/2023 16:32:01", "VersionNumber": 1.0, "Title": "Bank Customer Churn", "Slug": "bank-customer-churn", "Subtitle": "Bank Customer Data for Customer Churn", "Description": "RowNumber\u2014corresponds to the record (row) number and has no effect on the output.\nCustomerId\u2014contains random values and has no effect on customer leaving the bank.\nSurname\u2014the surname of a customer has no impact on their decision to leave the bank.\nCreditScore\u2014can have an effect on customer churn, since a customer with a higher credit score is less likely to leave the bank.\nGeography\u2014a customer\u2019s location can affect their decision to leave the bank.\nGender\u2014it\u2019s interesting to explore whether gender plays a role in a customer leaving the bank.\nAge\u2014this is certainly relevant, since older customers are less likely to leave their bank than younger ones.\nTenure\u2014refers to the number of years that the customer has been a client of the bank. Normally, older clients are more loyal and less likely to leave a bank.\nBalance\u2014also a very good indicator of customer churn, as people with a higher balance in their accounts are less likely to leave the bank compared to those with lower balances.\nNumOfProducts\u2014refers to the number of products that a customer has purchased through the bank.\nHasCrCard\u2014denotes whether or not a customer has a credit card. This column is also relevant, since people with a credit card are less likely to leave the bank.\nIsActiveMember\u2014active customers are less likely to leave the bank.\nEstimatedSalary\u2014as with balance, people with lower salaries are more likely to leave the bank compared to those with higher salaries.\nExited\u2014whether or not the customer left the bank.\nComplain\u2014customer has complaint or not.\nSatisfaction Score\u2014Score provided by the customer for their complaint resolution.\nCard Type\u2014type of card hold by the customer.\nPoints Earned\u2014the points earned by the customer for using credit card.\n\nAcknowledgements\n\nAs we know, it is much more expensive to sign in a new client than keeping an existing one.\n\nIt is advantageous for banks to know what leads a client towards the decision to leave the company.\n\nChurn prevention allows companies to develop loyalty programs and retention campaigns to keep as many customers as possible.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3197960, "CreatorUserId": 14862076, "OwnerUserId": 14862076.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5550559.0, "CurrentDatasourceVersionId": 5625285.0, "ForumId": 3262570, "Type": 2, "CreationDate": "04/28/2023 16:32:01", "LastActivityDate": "04/28/2023", "TotalViews": 39315, "TotalDownloads": 6814, "TotalVotes": 97, "TotalKernels": 52}]
|
[{"Id": 14862076, "UserName": "radheshyamkollipara", "DisplayName": "Radheshyam Kollipara", "RegisterDate": "04/28/2023", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv("/kaggle/input/bank-customer-churn/Customer-Churn-Records.csv")
df.head()
df = df.drop(["RowNumber", "CustomerId", "Surname"], axis=1)
df.shape
df.isnull().sum()
df["Gender"].unique()
df["Card Type"].unique()
# ***Categorical feautures are:***
# 1. Gender
# 2. HasCrCard
# 3. IsActiveMember
# 4. Exited
# 5. Complain
# 6. Card Type
df1 = df.groupby("Complain")["Exited"].apply(lambda x: (x == 1).mean()).reset_index()
df1 = df1.sort_values("Exited", ascending=False)
sns.barplot(data=df1, x="Complain", y="Exited", order=df1.Complain, color="#FF8C01")
plt.xlabel("Has complained or not")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Complain history")
plt.show()
df1 = df.groupby("HasCrCard")["Exited"].apply(lambda x: (x == 1).mean()).reset_index()
df1 = df1.sort_values("Exited", ascending=False)
sns.barplot(data=df1, x="HasCrCard", y="Exited", order=df1.HasCrCard, color="#FF8C01")
plt.xlabel("Has Credit Card or not")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Credit Card Holder or Not")
plt.show()
df1 = df.groupby("Gender")["Exited"].apply(lambda x: (x == 1).mean()).reset_index()
df1 = df1.sort_values("Exited", ascending=False)
sns.barplot(data=df1, x="Gender", y="Exited", order=df1.Gender, color="#FF8C01")
plt.xlabel("Has Credit Card or not")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Gender of Customer")
plt.show()
df1 = (
df.groupby("IsActiveMember")["Exited"]
.apply(lambda x: (x == 1).mean())
.reset_index()
)
df1 = df1.sort_values("Exited", ascending=False)
sns.barplot(
data=df1, x="IsActiveMember", y="Exited", order=df1.IsActiveMember, color="#FF8C01"
)
plt.xlabel("Is Active Member or Not")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Activity of the Customer")
plt.show()
# The plot tells that if a bank customer is not active then there is a tendency for the customer to leave the bank.
# Card Type
df1 = df.groupby("Card Type")["Exited"].apply(lambda x: (x == 1).mean()).reset_index()
df1 = df1.sort_values("Card Type", ascending=False)
sns.barplot(
data=df1, x="Card Type", y="Exited", order=df1["Card Type"], color="#FF8C01"
)
plt.xlabel("Type of Card Holder")
plt.ylabel("Proportion of Customer Churn")
plt.title(f"Likelihood of Customer Churn by Credit Card TYpe")
plt.show()
# It is shocking but the graph shows that a gold card holder has the largest chance of leaving the bank as compared to other card type holders.
# And the most shocking part is that diamond users are more likely to be churned
# as they have a proposition of 20% in the whole dataset.
df.head()
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
classification_report,
precision_score,
)
from sklearn.tree import DecisionTreeClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
lbl = LabelEncoder()
# 1. Gender
# 2. HasCrCard
# 3. IsActiveMember
# 4. Exited
# 5. Complain
# 6. Card Type
df["Gender"] = lbl.fit_transform(df["Gender"])
df["Card Type"] = lbl.fit_transform(df["Card Type"])
df["Geography"] = lbl.fit_transform(df["Geography"])
x = df.drop(["Exited"], axis=1) # independent feature
y = df["Exited"] # dependent feature
X_train, X_test, y_train, y_test = train_test_split(
x, y, test_size=0.15, random_state=42
)
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
k = np.sqrt(df.shape[0])
k
accuracy = []
prec = []
iter = 1
for i in range(100, 105):
knn = KNeighborsClassifier(n_neighbors=i)
knn.fit(X_train, y_train)
y_predi = knn.predict(X_test)
score = accuracy_score(y_test, y_predi)
precision = precision_score(y_test, y_predi)
accuracy.append(score * 100)
prec.append(precision * 100)
for i in range(len(accuracy)):
print("Accuracy of " + str(iter) + "th model is: " + str(accuracy[i]))
print("Precision of " + str(iter) + "th model is: " + str(prec[i]))
iter += 1
from sklearn.ensemble import GradientBoostingClassifier
model = GradientBoostingClassifier(n_estimators=12, loss="exponential", subsample=0.999)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
score = accuracy_score(y_test, y_pred)
precScore = precision_score(y_test, y_pred)
print("ACCURACY OF the RFC model is: " + str(score * 100) + "%")
print("Precision of the RFC model is: " + str(precScore * 100) + "%")
from sklearn.ensemble import AdaBoostClassifier
model = AdaBoostClassifier(n_estimators=11, random_state=1, algorithm="SAMME")
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
score = accuracy_score(y_test, y_pred)
precScore = precision_score(y_test, y_pred)
print("ACCURACY OF the RFC model is: " + str(score * 100) + "%")
print("Precision of the RFC model is: " + str(precScore * 100) + "%")
| false | 1 | 1,932 | 0 | 2,433 | 1,932 |
||
129766849
|
<jupyter_start><jupyter_text>Diamond
Kaggle dataset identifier: diamond
<jupyter_script>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import accuracy_score
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.linear_model import LinearRegression
from xgboost import XGBRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.model_selection import cross_val_score
from sklearn import metrics
dataset = pd.read_csv("/kaggle/input/diamond/diamonds.csv")
dataset.info()
dataset.head()
dataset.drop("Unnamed: 0", axis=1)
dataset.isnull().sum()
dataset.isna().sum()
dataset.shape
# Dropping dimentionless diamonds
dataset = dataset.drop(dataset[dataset["x"] == 0].index)
dataset = dataset.drop(dataset[dataset["y"] == 0].index)
dataset = dataset.drop(dataset[dataset["z"] == 0].index)
dataset.shape
ax = sns.pairplot(dataset, hue="cut")
# Dropping the outliers.
dataset = dataset[(dataset["depth"] < 75) & (dataset["depth"] > 45)]
dataset = dataset[(dataset["table"] < 80) & (dataset["table"] > 40)]
dataset = dataset[(dataset["x"] < 30)]
dataset = dataset[(dataset["y"] < 30)]
dataset = dataset[(dataset["z"] < 30) & (dataset["z"] > 2)]
dataset.shape
ax = sns.pairplot(dataset, hue="cut")
dataset.head()
dataset.info()
dataset.head()
# Get list of categorical variables
s = dataset.dtypes == "object"
object_cols = list(s[s].index)
print("Categorical variables:")
print(object_cols)
# Make copy to avoid changing original data
label_data = dataset.copy()
# Apply label encoder to each column with categorical data
label_encoder = LabelEncoder()
for col in object_cols:
label_data[col] = label_encoder.fit_transform(label_data[col])
label_data.head()
# correlation matrix
cmap = sns.diverging_palette(70, 20, s=50, l=40, n=6, as_cmap=True)
corrmat = label_data.corr()
f, ax = plt.subplots(figsize=(12, 12))
sns.heatmap(
corrmat,
cmap=cmap,
annot=True,
)
# Assigning the featurs as X and trarget as y
X = label_data.drop(["price"], axis=1)
y = label_data["price"]
print(X)
print(y)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=1
)
# Building pipelins of standard scaler and model for varios regressors.
pipeline_lr = Pipeline(
[("scalar1", StandardScaler()), ("lr_classifier", LinearRegression())]
)
pipeline_dt = Pipeline(
[("scalar2", StandardScaler()), ("dt_classifier", DecisionTreeRegressor())]
)
pipeline_rf = Pipeline(
[("scalar3", StandardScaler()), ("rf_classifier", RandomForestRegressor())]
)
pipeline_kn = Pipeline(
[("scalar4", StandardScaler()), ("rf_classifier", KNeighborsRegressor())]
)
pipeline_xgb = Pipeline(
[("scalar5", StandardScaler()), ("rf_classifier", XGBRegressor())]
)
# List of all the pipelines
pipelines = [pipeline_lr, pipeline_dt, pipeline_rf, pipeline_kn, pipeline_xgb]
# Dictionary of pipelines and model types for ease of reference
pipe_dict = {
0: "LinearRegression",
1: "DecisionTree",
2: "RandomForest",
3: "KNeighbors",
4: "XGBRegressor",
}
# Fit the pipelines
for pipe in pipelines:
pipe.fit(X_train, y_train)
cv_results_rms = []
for i, model in enumerate(pipelines):
cv_score = cross_val_score(
model, X_train, y_train, scoring="neg_root_mean_squared_error", cv=10
)
cv_results_rms.append(cv_score)
print("%s: %f " % (pipe_dict[i], cv_score.mean()))
# Model prediction on test data
pred = pipeline_xgb.predict(X_test)
# Model Evaluation
print("R^2:", metrics.r2_score(y_test, pred))
print(
"Adjusted R^2:",
1
- (1 - metrics.r2_score(y_test, pred))
* (len(y_test) - 1)
/ (len(y_test) - X_test.shape[1] - 1),
)
print("MAE:", metrics.mean_absolute_error(y_test, pred))
print("MSE:", metrics.mean_squared_error(y_test, pred))
print("RMSE:", np.sqrt(metrics.mean_squared_error(y_test, pred)))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/766/129766849.ipynb
|
diamond
|
sonyaugustine123
|
[{"Id": 129766849, "ScriptId": 38591683, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14505047, "CreationDate": "05/16/2023 10:21:47", "VersionNumber": 1.0, "Title": "Diamond Price Prediction", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 131.0, "LinesInsertedFromPrevious": 131.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186126513, "KernelVersionId": 129766849, "SourceDatasetVersionId": 5697405}]
|
[{"Id": 5697405, "DatasetId": 3275940, "DatasourceVersionId": 5773048, "CreatorUserId": 14505047, "LicenseName": "Unknown", "CreationDate": "05/16/2023 09:52:31", "VersionNumber": 1.0, "Title": "Diamond", "Slug": "diamond", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3275940, "CreatorUserId": 14505047, "OwnerUserId": 14505047.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5697405.0, "CurrentDatasourceVersionId": 5773048.0, "ForumId": 3341617, "Type": 2, "CreationDate": "05/16/2023 09:52:31", "LastActivityDate": "05/16/2023", "TotalViews": 56, "TotalDownloads": 7, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 14505047, "UserName": "sonyaugustine123", "DisplayName": "Sony Augustine@123", "RegisterDate": "04/05/2023", "PerformanceTier": 0}]
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import accuracy_score
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.linear_model import LinearRegression
from xgboost import XGBRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.model_selection import cross_val_score
from sklearn import metrics
dataset = pd.read_csv("/kaggle/input/diamond/diamonds.csv")
dataset.info()
dataset.head()
dataset.drop("Unnamed: 0", axis=1)
dataset.isnull().sum()
dataset.isna().sum()
dataset.shape
# Dropping dimentionless diamonds
dataset = dataset.drop(dataset[dataset["x"] == 0].index)
dataset = dataset.drop(dataset[dataset["y"] == 0].index)
dataset = dataset.drop(dataset[dataset["z"] == 0].index)
dataset.shape
ax = sns.pairplot(dataset, hue="cut")
# Dropping the outliers.
dataset = dataset[(dataset["depth"] < 75) & (dataset["depth"] > 45)]
dataset = dataset[(dataset["table"] < 80) & (dataset["table"] > 40)]
dataset = dataset[(dataset["x"] < 30)]
dataset = dataset[(dataset["y"] < 30)]
dataset = dataset[(dataset["z"] < 30) & (dataset["z"] > 2)]
dataset.shape
ax = sns.pairplot(dataset, hue="cut")
dataset.head()
dataset.info()
dataset.head()
# Get list of categorical variables
s = dataset.dtypes == "object"
object_cols = list(s[s].index)
print("Categorical variables:")
print(object_cols)
# Make copy to avoid changing original data
label_data = dataset.copy()
# Apply label encoder to each column with categorical data
label_encoder = LabelEncoder()
for col in object_cols:
label_data[col] = label_encoder.fit_transform(label_data[col])
label_data.head()
# correlation matrix
cmap = sns.diverging_palette(70, 20, s=50, l=40, n=6, as_cmap=True)
corrmat = label_data.corr()
f, ax = plt.subplots(figsize=(12, 12))
sns.heatmap(
corrmat,
cmap=cmap,
annot=True,
)
# Assigning the featurs as X and trarget as y
X = label_data.drop(["price"], axis=1)
y = label_data["price"]
print(X)
print(y)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=1
)
# Building pipelins of standard scaler and model for varios regressors.
pipeline_lr = Pipeline(
[("scalar1", StandardScaler()), ("lr_classifier", LinearRegression())]
)
pipeline_dt = Pipeline(
[("scalar2", StandardScaler()), ("dt_classifier", DecisionTreeRegressor())]
)
pipeline_rf = Pipeline(
[("scalar3", StandardScaler()), ("rf_classifier", RandomForestRegressor())]
)
pipeline_kn = Pipeline(
[("scalar4", StandardScaler()), ("rf_classifier", KNeighborsRegressor())]
)
pipeline_xgb = Pipeline(
[("scalar5", StandardScaler()), ("rf_classifier", XGBRegressor())]
)
# List of all the pipelines
pipelines = [pipeline_lr, pipeline_dt, pipeline_rf, pipeline_kn, pipeline_xgb]
# Dictionary of pipelines and model types for ease of reference
pipe_dict = {
0: "LinearRegression",
1: "DecisionTree",
2: "RandomForest",
3: "KNeighbors",
4: "XGBRegressor",
}
# Fit the pipelines
for pipe in pipelines:
pipe.fit(X_train, y_train)
cv_results_rms = []
for i, model in enumerate(pipelines):
cv_score = cross_val_score(
model, X_train, y_train, scoring="neg_root_mean_squared_error", cv=10
)
cv_results_rms.append(cv_score)
print("%s: %f " % (pipe_dict[i], cv_score.mean()))
# Model prediction on test data
pred = pipeline_xgb.predict(X_test)
# Model Evaluation
print("R^2:", metrics.r2_score(y_test, pred))
print(
"Adjusted R^2:",
1
- (1 - metrics.r2_score(y_test, pred))
* (len(y_test) - 1)
/ (len(y_test) - X_test.shape[1] - 1),
)
print("MAE:", metrics.mean_absolute_error(y_test, pred))
print("MSE:", metrics.mean_squared_error(y_test, pred))
print("RMSE:", np.sqrt(metrics.mean_squared_error(y_test, pred)))
| false | 1 | 1,281 | 0 | 1,299 | 1,281 |
||
129321175
|
"C:/Users/berkg/OneDrive/Masaüstü/dataset/car_evaluation.csv"
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
data = "C:/Users/berkg/OneDrive/Masaüstü/dataset/car_evaluation.csv"
df = pd.read_csv(data, header=None)
df.shape
df.head()
col_names = ["buying", "maint", "doors", "persons", "lug_boot", "safety", "class"]
df.columns = col_names
col_names
df.head()
df.info()
col_names = ["buying", "maint", "doors", "persons", "lug_boot", "safety", "class"]
for col in col_names:
print(df[col].value_counts())
df["class"].value_counts()
df.isnull().sum()
x = df.drop(["class"], axis=1)
y = df["class"]
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.33, random_state=42
)
x_train.shape, x_test.shape
x_train.dtypes
x_train.head()
import category_encoders as ce
encoder = ce.OrdinalEncoder(
cols=["buying", "maint", "doors", "persons", "lug_boot", "safety"]
)
x_train = encoder.fit_transform(x_train)
x_test = encoder.transform(x_test)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/321/129321175.ipynb
| null | null |
[{"Id": 129321175, "ScriptId": 38449867, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13980931, "CreationDate": "05/12/2023 18:49:52", "VersionNumber": 1.0, "Title": "MLHOMEWORK", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 53.0, "LinesInsertedFromPrevious": 53.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
"C:/Users/berkg/OneDrive/Masaüstü/dataset/car_evaluation.csv"
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
data = "C:/Users/berkg/OneDrive/Masaüstü/dataset/car_evaluation.csv"
df = pd.read_csv(data, header=None)
df.shape
df.head()
col_names = ["buying", "maint", "doors", "persons", "lug_boot", "safety", "class"]
df.columns = col_names
col_names
df.head()
df.info()
col_names = ["buying", "maint", "doors", "persons", "lug_boot", "safety", "class"]
for col in col_names:
print(df[col].value_counts())
df["class"].value_counts()
df.isnull().sum()
x = df.drop(["class"], axis=1)
y = df["class"]
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.33, random_state=42
)
x_train.shape, x_test.shape
x_train.dtypes
x_train.head()
import category_encoders as ce
encoder = ce.OrdinalEncoder(
cols=["buying", "maint", "doors", "persons", "lug_boot", "safety"]
)
x_train = encoder.fit_transform(x_train)
x_test = encoder.transform(x_test)
| false | 0 | 405 | 0 | 405 | 405 |
||
129321822
|
<jupyter_start><jupyter_text>Suicide Attempts in Shandong, China
```
Data on serious suicide attempts in Shandong, China
A data frame with 2571 observations on the following 11 variables.
```
| Column | Description |
| --- | --- |
| Person_ID | ID number of victims |
| Hospitalised | Hospitalized? (no or yes) |
| Died | Died? (no or yes) |
| Urban | Urban area? (no, unknown, or yes) |
| Year | Year (2009, 2010, or 2011) |
| Month | Month (1=Jan through 12=December) |
| Sex | Sex (female or male) |
| Age | Age (years) |
| Education | Education level (illiterate, primary, Secondary, Tertiary, or unknown) |
| Occupation | One of ten occupation categories |
| method | One of nine possible methods |
### Details
Data from a study of serious suicide attempts over three years in a predominantly rural population in Shandong, China.
## Source
Sun J, Guo X, Zhang J, Wang M, Jia C, Xu A (2015) "Incidence and fatality of serious suicide attempts in a predominantly rural population in Shandong, China: a public health surveillance study," BMJ Open 5(2): e006762. https://doi.org/10.1136/bmjopen-2014-006762
Kaggle dataset identifier: suicide-attempts-in-shandong-china
<jupyter_script># # 📚 Imports
# ---
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
import warnings
warnings.filterwarnings("ignore")
# # 📖 Data
# ---
df = pd.read_csv("/kaggle/input/suicide-attempts-in-shandong-china/SuicideChina.csv")
df.head()
df.info()
# Checking null values
df.isna().sum()
# Drop unnecessary columns
df.drop(["Unnamed: 0", "Person_ID"], axis=1, inplace=True)
# Columns Value Count
print(df["Hospitalised"].value_counts())
print("-" * 30)
print(df["Died"].value_counts())
print("-" * 30)
print(df["Urban"].value_counts())
print("-" * 30)
print(df["Education"].value_counts())
print("-" * 30)
print(df["Occupation"].value_counts())
print("-" * 30)
print(df["method"].value_counts())
# # 📊 Visualization
# ## All columns compared with Age
def plots(df, x):
plt.style.use("dark_background")
f, ax = plt.subplots(1, 2, figsize=(25, 10))
Group_data = df.groupby(x)
sns.barplot(
x=Group_data["Age"].mean().index,
y=Group_data["Age"].mean().values,
ax=ax[0],
palette="viridis",
)
for container in ax[0].containers:
ax[0].bar_label(container, color="white", size=20)
palette_color = sns.color_palette("viridis")
plt.pie(
x=df[x].value_counts(),
labels=df[x].value_counts().index,
autopct="%.0f%%",
shadow=True,
colors=palette_color,
)
plt.suptitle(x, fontsize=25)
plt.show()
for i in df.columns:
if i != "Age":
plots(df, i)
# ## Values Distribuition
plt.style.use("dark_background")
plt.figure(figsize=(12, 20))
list_columns = list(df.columns)
for i in range(len(list_columns)):
plt.subplot(5, 2, i + 1)
plt.title(list_columns[i])
plt.hist(df[list_columns[i]])
plt.grid(alpha=0.5)
plt.tight_layout()
# ## Values Distribuition Compared with Died
plt.style.use("dark_background")
fig, axs = plt.subplots(6, 2, figsize=(10, 20))
i = 1
for feature in df.columns:
if feature not in ["Died"] and i < 14:
plt.subplot(5, 2, i)
sns.histplot(
data=df, x=feature, kde=True, palette="winter", hue="Died", alpha=0.8
)
plt.grid(alpha=0.5)
i += 1
# ## Died by Month
plt.figure(figsize=(10, 7))
plt.style.use("dark_background")
sns.violinplot(x="Died", y="Month", data=df, palette="viridis")
# ## Age Compared to Month of Occurrence
plt.style.use("dark_background")
plt.figure(figsize=(12, 10))
plt.hexbin(df["Age"], df["Month"], gridsize=12, cmap="viridis", mincnt=1)
plt.colorbar(label="Count")
plt.xlabel("Age")
plt.ylabel("Month")
plt.title("Age Compared to Month of Occurrence")
plt.show()
# ## Age Compared to Year of Occurrence
plt.style.use("dark_background")
plt.figure(figsize=(10, 5))
plt.hexbin(df["Age"], df["Year"], gridsize=3, cmap="viridis", mincnt=1)
plt.colorbar(label="Count")
plt.xlabel("Age")
plt.ylabel("Year")
plt.title("Age Compared to Year of Occurrence")
plt.show()
# ## Converting object values to numeric for correlation
list_str = df.select_dtypes(include="object").columns
le = LabelEncoder()
for c in list_str:
df[c] = le.fit_transform(df[c])
# ## Correlation
plt.figure(figsize=(15, 12))
plt.style.use("dark_background")
sns.heatmap(df.corr(), annot=True, cmap="viridis")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/321/129321822.ipynb
|
suicide-attempts-in-shandong-china
|
utkarshx27
|
[{"Id": 129321822, "ScriptId": 38449901, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12038039, "CreationDate": "05/12/2023 18:57:51", "VersionNumber": 1.0, "Title": "Suicide Attempts in Shandong EDA \ud83d\udcca", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 139.0, "LinesInsertedFromPrevious": 139.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 19}]
|
[{"Id": 185260506, "KernelVersionId": 129321822, "SourceDatasetVersionId": 5617993}]
|
[{"Id": 5617993, "DatasetId": 3230370, "DatasourceVersionId": 5693173, "CreatorUserId": 13364933, "LicenseName": "CC0: Public Domain", "CreationDate": "05/06/2023 11:54:22", "VersionNumber": 1.0, "Title": "Suicide Attempts in Shandong, China", "Slug": "suicide-attempts-in-shandong-china", "Subtitle": "Serious Suicide Attempts in Shandong, China: Three-Year Study", "Description": "```\nData on serious suicide attempts in Shandong, China\nA data frame with 2571 observations on the following 11 variables.\n```\n\n| Column | Description |\n| --- | --- |\n| Person_ID | ID number of victims |\n| Hospitalised | Hospitalized? (no or yes) |\n| Died | Died? (no or yes) |\n| Urban | Urban area? (no, unknown, or yes) |\n| Year | Year (2009, 2010, or 2011) |\n| Month | Month (1=Jan through 12=December) |\n| Sex | Sex (female or male) |\n| Age | Age (years) |\n| Education | Education level (illiterate, primary, Secondary, Tertiary, or unknown) |\n| Occupation | One of ten occupation categories |\n| method | One of nine possible methods |\n\n### Details \nData from a study of serious suicide attempts over three years in a predominantly rural population in Shandong, China.\n\n## Source\nSun J, Guo X, Zhang J, Wang M, Jia C, Xu A (2015) \"Incidence and fatality of serious suicide attempts in a predominantly rural population in Shandong, China: a public health surveillance study,\" BMJ Open 5(2): e006762. https://doi.org/10.1136/bmjopen-2014-006762", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3230370, "CreatorUserId": 13364933, "OwnerUserId": 13364933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5617993.0, "CurrentDatasourceVersionId": 5693173.0, "ForumId": 3295509, "Type": 2, "CreationDate": "05/06/2023 11:54:22", "LastActivityDate": "05/06/2023", "TotalViews": 8885, "TotalDownloads": 1402, "TotalVotes": 42, "TotalKernels": 12}]
|
[{"Id": 13364933, "UserName": "utkarshx27", "DisplayName": "Utkarsh Singh", "RegisterDate": "01/21/2023", "PerformanceTier": 2}]
|
# # 📚 Imports
# ---
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
import warnings
warnings.filterwarnings("ignore")
# # 📖 Data
# ---
df = pd.read_csv("/kaggle/input/suicide-attempts-in-shandong-china/SuicideChina.csv")
df.head()
df.info()
# Checking null values
df.isna().sum()
# Drop unnecessary columns
df.drop(["Unnamed: 0", "Person_ID"], axis=1, inplace=True)
# Columns Value Count
print(df["Hospitalised"].value_counts())
print("-" * 30)
print(df["Died"].value_counts())
print("-" * 30)
print(df["Urban"].value_counts())
print("-" * 30)
print(df["Education"].value_counts())
print("-" * 30)
print(df["Occupation"].value_counts())
print("-" * 30)
print(df["method"].value_counts())
# # 📊 Visualization
# ## All columns compared with Age
def plots(df, x):
plt.style.use("dark_background")
f, ax = plt.subplots(1, 2, figsize=(25, 10))
Group_data = df.groupby(x)
sns.barplot(
x=Group_data["Age"].mean().index,
y=Group_data["Age"].mean().values,
ax=ax[0],
palette="viridis",
)
for container in ax[0].containers:
ax[0].bar_label(container, color="white", size=20)
palette_color = sns.color_palette("viridis")
plt.pie(
x=df[x].value_counts(),
labels=df[x].value_counts().index,
autopct="%.0f%%",
shadow=True,
colors=palette_color,
)
plt.suptitle(x, fontsize=25)
plt.show()
for i in df.columns:
if i != "Age":
plots(df, i)
# ## Values Distribuition
plt.style.use("dark_background")
plt.figure(figsize=(12, 20))
list_columns = list(df.columns)
for i in range(len(list_columns)):
plt.subplot(5, 2, i + 1)
plt.title(list_columns[i])
plt.hist(df[list_columns[i]])
plt.grid(alpha=0.5)
plt.tight_layout()
# ## Values Distribuition Compared with Died
plt.style.use("dark_background")
fig, axs = plt.subplots(6, 2, figsize=(10, 20))
i = 1
for feature in df.columns:
if feature not in ["Died"] and i < 14:
plt.subplot(5, 2, i)
sns.histplot(
data=df, x=feature, kde=True, palette="winter", hue="Died", alpha=0.8
)
plt.grid(alpha=0.5)
i += 1
# ## Died by Month
plt.figure(figsize=(10, 7))
plt.style.use("dark_background")
sns.violinplot(x="Died", y="Month", data=df, palette="viridis")
# ## Age Compared to Month of Occurrence
plt.style.use("dark_background")
plt.figure(figsize=(12, 10))
plt.hexbin(df["Age"], df["Month"], gridsize=12, cmap="viridis", mincnt=1)
plt.colorbar(label="Count")
plt.xlabel("Age")
plt.ylabel("Month")
plt.title("Age Compared to Month of Occurrence")
plt.show()
# ## Age Compared to Year of Occurrence
plt.style.use("dark_background")
plt.figure(figsize=(10, 5))
plt.hexbin(df["Age"], df["Year"], gridsize=3, cmap="viridis", mincnt=1)
plt.colorbar(label="Count")
plt.xlabel("Age")
plt.ylabel("Year")
plt.title("Age Compared to Year of Occurrence")
plt.show()
# ## Converting object values to numeric for correlation
list_str = df.select_dtypes(include="object").columns
le = LabelEncoder()
for c in list_str:
df[c] = le.fit_transform(df[c])
# ## Correlation
plt.figure(figsize=(15, 12))
plt.style.use("dark_background")
sns.heatmap(df.corr(), annot=True, cmap="viridis")
| false | 1 | 1,160 | 19 | 1,572 | 1,160 |
||
129321538
|
<jupyter_start><jupyter_text>Flickr 8k Dataset
### Context
A new benchmark collection for sentence-based image description and search, consisting of 8,000 images that are each paired with five different captions which provide clear descriptions of the salient entities and events. … The images were chosen from six different Flickr groups, and tend not to contain any well-known people or locations, but were manually selected to depict a variety of scenes and situations
### Content66
What's inside is more than just rows and columns. Make it easy for others to get started by describing how you acquired the data and what time period it represents, too.
Kaggle dataset identifier: flickr8k
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import torch
from torch_snippets import *
from torchvision import transforms
from sklearn.model_selection import train_test_split
device = "cuda" if torch.cuda.is_available() else "cpu"
df = pd.read_csv("/kaggle/input/flickr8k/captions.txt", delimiter=",")
df
txt = df[df.columns[-1]].tolist()
images = df[df.columns[0]].tolist()
train_image, test_image, train_text, test_text = train_test_split(
images, txt, test_size=0.2
)
tfms = transforms.Compose(
[
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]), # imagenet
]
)
from transformers import AutoProcessor
processor = AutoProcessor.from_pretrained("microsoft/git-base")
class SegData(torch.utils.data.Dataset):
def __init__(self, images, txt):
self.image_path = "/kaggle/input/flickr8k/Images/"
self.images = images
self.txt = txt
self.processor = processor
def __len__(self):
return len(self.images)
def __getitem__(self, ix):
image = cv2.imread(self.image_path + self.images[ix])
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# image = cv2.resize(image, (224,224))
label = self.txt[ix]
encoding = self.processor(
images=image, text=label, padding="max_length", return_tensors="pt"
)
encoding = {k: v.squeeze() for k, v in encoding.items()}
# tokenized=tokenizer(label,max_length=128,padding=True,truncation=True,return_tensors="pt")
# image=tfms(image/255.)
return encoding
def choose(self):
return self[randint(len(self.images))]
train_dataset = SegData(train_image, train_text)
test_dataset = SegData(test_image, test_text)
import matplotlib.pyplot as plt
import cv2
encoding = train_dataset[-105]
plt.imshow(encoding["pixel_values"].permute(1, 2, 0).detach().numpy())
processor.decode(encoding["input_ids"].tolist())
trn_dl = torch.utils.data.DataLoader(
train_dataset, batch_size=2, drop_last=True, shuffle=True
)
test_dl = torch.utils.data.DataLoader(
test_dataset, batch_size=2, drop_last=True, shuffle=True
)
from transformers import AutoModelForCausalLM
model = AutoModelForCausalLM.from_pretrained("microsoft/git-base").to(device)
optimizer = torch.optim.AdamW(model.parameters(), lr=5e-5)
def train_batch(batch):
model.train()
output = model(
input_ids=batch["input_ids"].to(device),
pixel_values=batch["pixel_values"].to(device),
attention_mask=batch["attention_mask"].to(device),
labels=batch["input_ids"].to(device),
)
loss = output.loss
optimizer.zero_grad()
loss.backward()
optimizer.step()
return loss.item()
@torch.no_grad()
def valid_batch(batch):
model.eval()
output = model(
input_ids=batch["input_ids"].to(device),
pixel_values=batch["pixel_values"].to(device),
attention_mask=batch["attention_mask"].to(device),
labels=batch["input_ids"].to(device),
)
loss = output.loss
return loss.item()
# image,label=batch
# model.eval()
# label_t=tokenizer(label,max_length=32,padding=True,truncation=True,return_tensors="pt"
# ,add_special_tokens=True)
# inputs={i:j.to(device) for i,j in label_t.items()}
# x=inputs['input_ids'][:,:-1]
# y=inputs['input_ids'][:,1:]
# outputs=model(image,x)
# B,T,C=outputs.size()
# outputs=outputs.reshape(B*T,C)
# y=y.reshape(B*T)
# loss=loss_fn(outputs,y)
# return loss.item()
n_epoch = 1
log = Report(n_epoch)
for epochs in range(n_epoch):
N = len(trn_dl)
for i, data in enumerate(trn_dl):
loss = train_batch(data)
log.record(epochs + (i + 1) / N, trn_loss=loss, end="\r")
N = len(test_dl)
for i, data in enumerate(trn_dl):
val_loss = valid_batch(data)
test_dl = torch.utils.data.DataLoader(
test_dataset, batch_size=2, drop_last=True, shuffle=True
)
encoded = next(iter(test_dl))
pixel_values = encoded["pixel_values"][0].to(device)
plt.imshow(pixel_values.permute(1, 2, 0).detach().cpu().numpy())
with torch.no_grad():
generated_ids = model.generate(
pixel_values=pixel_values.unsqueeze(0), max_length=50
)
generated_caption = processor.batch_decode(generated_ids, skip_special_tokens=True)[0]
print(generated_caption)
for _ in range(5):
test_dl = torch.utils.data.DataLoader(
test_dataset, batch_size=2, drop_last=True, shuffle=True
)
encoded = next(iter(test_dl))
pixel_values = encoded["pixel_values"][0].to(device)
with torch.no_grad():
generated_ids = model.generate(
pixel_values=pixel_values.unsqueeze(0), max_length=50
)
generated_caption = processor.batch_decode(generated_ids, skip_special_tokens=True)[
0
]
show(pixel_values.permute(1, 2, 0).detach().cpu().numpy(), title=generated_caption)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/321/129321538.ipynb
|
flickr8k
|
adityajn105
|
[{"Id": 129321538, "ScriptId": 37112693, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2294317, "CreationDate": "05/12/2023 18:54:22", "VersionNumber": 1.0, "Title": "image_captioning_pytorch", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 152.0, "LinesInsertedFromPrevious": 152.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 185260018, "KernelVersionId": 129321538, "SourceDatasetVersionId": 1111676}]
|
[{"Id": 1111676, "DatasetId": 623289, "DatasourceVersionId": 1141936, "CreatorUserId": 1526260, "LicenseName": "CC0: Public Domain", "CreationDate": "04/27/2020 07:27:19", "VersionNumber": 1.0, "Title": "Flickr 8k Dataset", "Slug": "flickr8k", "Subtitle": "Flickr8k Dataset for image captioning.", "Description": "### Context\n\nA new benchmark collection for sentence-based image description and search, consisting of 8,000 images that are each paired with five different captions which provide clear descriptions of the salient entities and events. \u2026 The images were chosen from six different Flickr groups, and tend not to contain any well-known people or locations, but were manually selected to depict a variety of scenes and situations\n\n### Content66\n\nWhat's inside is more than just rows and columns. Make it easy for others to get started by describing how you acquired the data and what time period it represents, too.\n\n\n### Acknowledgements\n\nWe wouldn't be here without the help of others. If you owe any attributions or thanks, include them here along with any citations of past research.\n\n\n### Inspiration\n\nYour data will be in front of the world's largest data science community. What questions do you want to see answered?", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 623289, "CreatorUserId": 1526260, "OwnerUserId": 1526260.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1111676.0, "CurrentDatasourceVersionId": 1141936.0, "ForumId": 637462, "Type": 2, "CreationDate": "04/27/2020 07:27:19", "LastActivityDate": "04/27/2020", "TotalViews": 160531, "TotalDownloads": 44586, "TotalVotes": 270, "TotalKernels": 277}]
|
[{"Id": 1526260, "UserName": "adityajn105", "DisplayName": "adityajn105", "RegisterDate": "01/02/2018", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import torch
from torch_snippets import *
from torchvision import transforms
from sklearn.model_selection import train_test_split
device = "cuda" if torch.cuda.is_available() else "cpu"
df = pd.read_csv("/kaggle/input/flickr8k/captions.txt", delimiter=",")
df
txt = df[df.columns[-1]].tolist()
images = df[df.columns[0]].tolist()
train_image, test_image, train_text, test_text = train_test_split(
images, txt, test_size=0.2
)
tfms = transforms.Compose(
[
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]), # imagenet
]
)
from transformers import AutoProcessor
processor = AutoProcessor.from_pretrained("microsoft/git-base")
class SegData(torch.utils.data.Dataset):
def __init__(self, images, txt):
self.image_path = "/kaggle/input/flickr8k/Images/"
self.images = images
self.txt = txt
self.processor = processor
def __len__(self):
return len(self.images)
def __getitem__(self, ix):
image = cv2.imread(self.image_path + self.images[ix])
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# image = cv2.resize(image, (224,224))
label = self.txt[ix]
encoding = self.processor(
images=image, text=label, padding="max_length", return_tensors="pt"
)
encoding = {k: v.squeeze() for k, v in encoding.items()}
# tokenized=tokenizer(label,max_length=128,padding=True,truncation=True,return_tensors="pt")
# image=tfms(image/255.)
return encoding
def choose(self):
return self[randint(len(self.images))]
train_dataset = SegData(train_image, train_text)
test_dataset = SegData(test_image, test_text)
import matplotlib.pyplot as plt
import cv2
encoding = train_dataset[-105]
plt.imshow(encoding["pixel_values"].permute(1, 2, 0).detach().numpy())
processor.decode(encoding["input_ids"].tolist())
trn_dl = torch.utils.data.DataLoader(
train_dataset, batch_size=2, drop_last=True, shuffle=True
)
test_dl = torch.utils.data.DataLoader(
test_dataset, batch_size=2, drop_last=True, shuffle=True
)
from transformers import AutoModelForCausalLM
model = AutoModelForCausalLM.from_pretrained("microsoft/git-base").to(device)
optimizer = torch.optim.AdamW(model.parameters(), lr=5e-5)
def train_batch(batch):
model.train()
output = model(
input_ids=batch["input_ids"].to(device),
pixel_values=batch["pixel_values"].to(device),
attention_mask=batch["attention_mask"].to(device),
labels=batch["input_ids"].to(device),
)
loss = output.loss
optimizer.zero_grad()
loss.backward()
optimizer.step()
return loss.item()
@torch.no_grad()
def valid_batch(batch):
model.eval()
output = model(
input_ids=batch["input_ids"].to(device),
pixel_values=batch["pixel_values"].to(device),
attention_mask=batch["attention_mask"].to(device),
labels=batch["input_ids"].to(device),
)
loss = output.loss
return loss.item()
# image,label=batch
# model.eval()
# label_t=tokenizer(label,max_length=32,padding=True,truncation=True,return_tensors="pt"
# ,add_special_tokens=True)
# inputs={i:j.to(device) for i,j in label_t.items()}
# x=inputs['input_ids'][:,:-1]
# y=inputs['input_ids'][:,1:]
# outputs=model(image,x)
# B,T,C=outputs.size()
# outputs=outputs.reshape(B*T,C)
# y=y.reshape(B*T)
# loss=loss_fn(outputs,y)
# return loss.item()
n_epoch = 1
log = Report(n_epoch)
for epochs in range(n_epoch):
N = len(trn_dl)
for i, data in enumerate(trn_dl):
loss = train_batch(data)
log.record(epochs + (i + 1) / N, trn_loss=loss, end="\r")
N = len(test_dl)
for i, data in enumerate(trn_dl):
val_loss = valid_batch(data)
test_dl = torch.utils.data.DataLoader(
test_dataset, batch_size=2, drop_last=True, shuffle=True
)
encoded = next(iter(test_dl))
pixel_values = encoded["pixel_values"][0].to(device)
plt.imshow(pixel_values.permute(1, 2, 0).detach().cpu().numpy())
with torch.no_grad():
generated_ids = model.generate(
pixel_values=pixel_values.unsqueeze(0), max_length=50
)
generated_caption = processor.batch_decode(generated_ids, skip_special_tokens=True)[0]
print(generated_caption)
for _ in range(5):
test_dl = torch.utils.data.DataLoader(
test_dataset, batch_size=2, drop_last=True, shuffle=True
)
encoded = next(iter(test_dl))
pixel_values = encoded["pixel_values"][0].to(device)
with torch.no_grad():
generated_ids = model.generate(
pixel_values=pixel_values.unsqueeze(0), max_length=50
)
generated_caption = processor.batch_decode(generated_ids, skip_special_tokens=True)[
0
]
show(pixel_values.permute(1, 2, 0).detach().cpu().numpy(), title=generated_caption)
| false | 0 | 1,732 | 1 | 1,885 | 1,732 |
||
129321347
|
<jupyter_start><jupyter_text>Dogs & Cats Images
Kaggle dataset identifier: dogs-cats-images
<jupyter_script>import numpy as np
import pandas as pd
import tensorflow as tf
import matplotlib.pyplot as plt
import seaborn as sns
plt.rcParams["figure.figsize"] = (20, 8)
from tensorflow.keras.preprocessing.image import (
ImageDataGenerator,
load_img,
img_to_array,
)
from tensorflow.keras.applications import VGG16, ResNet50, InceptionV3, MobileNet
img_size = 150
batch_size = 32
train_datagen = ImageDataGenerator(
rescale=1 / 255.0,
rotation_range=30,
horizontal_flip=True,
width_shift_range=0.1,
height_shift_range=0.1,
) # this is for normalization, basically to shorten the range
val_datagen = ImageDataGenerator(rescale=1 / 255.0)
train_generator = train_datagen.flow_from_directory(
"/kaggle/input/dogs-cats-images/dataset/training_set",
target_size=(img_size, img_size),
batch_size=batch_size,
shuffle=True, # shuffle the images in every iteration
class_mode="binary",
)
val_generator = val_datagen.flow_from_directory(
"/kaggle/input/dogs-cats-images/dataset/test_set",
target_size=(img_size, img_size),
batch_size=batch_size,
shuffle=False,
class_mode="binary",
)
# Visulization of 15 Random Samples from a Batch of 32
labels = ["cat", "dog"]
samples = train_generator.__next__()
images = samples[0]
target = samples[1]
plt.figure(figsize=(20, 20))
for i in range(15):
plt.subplot(5, 5, i + 1)
plt.subplots_adjust(hspace=0.3, wspace=0.3)
plt.imshow(images[i])
plt.title(f"Class: {labels[int(target[i])]}")
plt.axis("off")
from tensorflow.keras.layers import (
Conv2D,
MaxPooling2D,
GlobalAveragePooling2D,
Activation,
Dropout,
Flatten,
Dense,
Input,
Layer,
)
from tensorflow.keras.models import Sequential, Model
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dense, Input
model = Sequential()
model.add(
Conv2D(32, (3, 3), input_shape=(150, 150, 3), activation="relu", padding="same")
)
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(64, (3, 3), activation="relu", padding="same"))
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(128, (3, 3), activation="relu", padding="same"))
model.add(MaxPooling2D(2, 2))
model.add(Flatten())
model.add(Dense(units=512, activation="relu"))
model.add(Dense(units=1, activation="sigmoid"))
model.summary()
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
filepath = "model_cnn.h5"
checkpoint = ModelCheckpoint(
filepath,
monitor="val_loss",
verbose=1,
save_best_only=True,
save_weights_only=False,
)
history = model.fit(
train_generator, epochs=5, validation_data=val_generator, callbacks=[checkpoint]
)
# LEARNING CURVES
# If the difference between the validation loss and training loss is too big then your model is overfitting
plt.figure(figsize=(20, 8))
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model Loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Train", "Val"], loc="upper left")
plt.show()
plt.figure(figsize=(20, 8))
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model Accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["Train", "Val"], loc="upper left")
plt.show()
model = tf.keras.models.load_model("/kaggle/working/model_cnn.h5")
y_test = val_generator.classes
y_pred = model.predict(val_generator)
y_pred_probs = y_pred.copy()
y_pred[y_pred > 0.5] = 1
y_pred[y_pred < 0.5] = 0
from sklearn.metrics import classification_report, confusion_matrix
print(classification_report(y_test, y_pred, target_names=["cats", "dogs"]))
plt.figure(figsize=(10, 8))
sns.heatmap(
confusion_matrix(y_test, y_pred),
annot=True,
fmt=".3g",
xticklabels=["cats", "dogs"],
yticklabels=["cats", "dogs"],
cmap="Blues",
)
plt.show()
from sklearn.metrics import roc_curve, roc_auc_score
fpr, tpr, thresholds = roc_curve(y_test, y_pred_probs)
roc_auc = roc_auc_score(y_test, y_pred_probs)
roc_auc
# 0.5 - 1 range
plt.plot(fpr, tpr, color="blue", label="ROC curve (AUC = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="red", linestyle="--", label="Random guessing")
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic (ROC) curve")
plt.legend(loc="lower right")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/321/129321347.ipynb
|
dogs-cats-images
|
chetankv
|
[{"Id": 129321347, "ScriptId": 37476098, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6542785, "CreationDate": "05/12/2023 18:51:42", "VersionNumber": 1.0, "Title": "Binary Classification", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 156.0, "LinesInsertedFromPrevious": 156.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 185259746, "KernelVersionId": 129321347, "SourceDatasetVersionId": 28903}]
|
[{"Id": 28903, "DatasetId": 22535, "DatasourceVersionId": 28946, "CreatorUserId": 632316, "LicenseName": "CC0: Public Domain", "CreationDate": "04/19/2018 18:20:08", "VersionNumber": 1.0, "Title": "Dogs & Cats Images", "Slug": "dogs-cats-images", "Subtitle": "image classification", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 227350220.0, "TotalUncompressedBytes": 227350220.0}]
|
[{"Id": 22535, "CreatorUserId": 632316, "OwnerUserId": 632316.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 28903.0, "CurrentDatasourceVersionId": 28946.0, "ForumId": 30533, "Type": 2, "CreationDate": "04/19/2018 18:20:08", "LastActivityDate": "04/19/2018", "TotalViews": 143541, "TotalDownloads": 35362, "TotalVotes": 566, "TotalKernels": 233}]
|
[{"Id": 632316, "UserName": "chetankv", "DisplayName": "chetanimravan", "RegisterDate": "06/04/2016", "PerformanceTier": 0}]
|
import numpy as np
import pandas as pd
import tensorflow as tf
import matplotlib.pyplot as plt
import seaborn as sns
plt.rcParams["figure.figsize"] = (20, 8)
from tensorflow.keras.preprocessing.image import (
ImageDataGenerator,
load_img,
img_to_array,
)
from tensorflow.keras.applications import VGG16, ResNet50, InceptionV3, MobileNet
img_size = 150
batch_size = 32
train_datagen = ImageDataGenerator(
rescale=1 / 255.0,
rotation_range=30,
horizontal_flip=True,
width_shift_range=0.1,
height_shift_range=0.1,
) # this is for normalization, basically to shorten the range
val_datagen = ImageDataGenerator(rescale=1 / 255.0)
train_generator = train_datagen.flow_from_directory(
"/kaggle/input/dogs-cats-images/dataset/training_set",
target_size=(img_size, img_size),
batch_size=batch_size,
shuffle=True, # shuffle the images in every iteration
class_mode="binary",
)
val_generator = val_datagen.flow_from_directory(
"/kaggle/input/dogs-cats-images/dataset/test_set",
target_size=(img_size, img_size),
batch_size=batch_size,
shuffle=False,
class_mode="binary",
)
# Visulization of 15 Random Samples from a Batch of 32
labels = ["cat", "dog"]
samples = train_generator.__next__()
images = samples[0]
target = samples[1]
plt.figure(figsize=(20, 20))
for i in range(15):
plt.subplot(5, 5, i + 1)
plt.subplots_adjust(hspace=0.3, wspace=0.3)
plt.imshow(images[i])
plt.title(f"Class: {labels[int(target[i])]}")
plt.axis("off")
from tensorflow.keras.layers import (
Conv2D,
MaxPooling2D,
GlobalAveragePooling2D,
Activation,
Dropout,
Flatten,
Dense,
Input,
Layer,
)
from tensorflow.keras.models import Sequential, Model
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dense, Input
model = Sequential()
model.add(
Conv2D(32, (3, 3), input_shape=(150, 150, 3), activation="relu", padding="same")
)
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(64, (3, 3), activation="relu", padding="same"))
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(128, (3, 3), activation="relu", padding="same"))
model.add(MaxPooling2D(2, 2))
model.add(Flatten())
model.add(Dense(units=512, activation="relu"))
model.add(Dense(units=1, activation="sigmoid"))
model.summary()
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
filepath = "model_cnn.h5"
checkpoint = ModelCheckpoint(
filepath,
monitor="val_loss",
verbose=1,
save_best_only=True,
save_weights_only=False,
)
history = model.fit(
train_generator, epochs=5, validation_data=val_generator, callbacks=[checkpoint]
)
# LEARNING CURVES
# If the difference between the validation loss and training loss is too big then your model is overfitting
plt.figure(figsize=(20, 8))
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model Loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Train", "Val"], loc="upper left")
plt.show()
plt.figure(figsize=(20, 8))
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model Accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["Train", "Val"], loc="upper left")
plt.show()
model = tf.keras.models.load_model("/kaggle/working/model_cnn.h5")
y_test = val_generator.classes
y_pred = model.predict(val_generator)
y_pred_probs = y_pred.copy()
y_pred[y_pred > 0.5] = 1
y_pred[y_pred < 0.5] = 0
from sklearn.metrics import classification_report, confusion_matrix
print(classification_report(y_test, y_pred, target_names=["cats", "dogs"]))
plt.figure(figsize=(10, 8))
sns.heatmap(
confusion_matrix(y_test, y_pred),
annot=True,
fmt=".3g",
xticklabels=["cats", "dogs"],
yticklabels=["cats", "dogs"],
cmap="Blues",
)
plt.show()
from sklearn.metrics import roc_curve, roc_auc_score
fpr, tpr, thresholds = roc_curve(y_test, y_pred_probs)
roc_auc = roc_auc_score(y_test, y_pred_probs)
roc_auc
# 0.5 - 1 range
plt.plot(fpr, tpr, color="blue", label="ROC curve (AUC = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="red", linestyle="--", label="Random guessing")
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic (ROC) curve")
plt.legend(loc="lower right")
plt.show()
| false | 0 | 1,486 | 1 | 1,512 | 1,486 |
||
129321338
|
# # **Libraries**
# In Python, libraries are used to extend the core functionality of the language, allowing users to perform a wide range of tasks without having to write the code from scratch. They encapsulate complex operations into simpler, more readable code, and promote code reuse, which leads to more efficient and reliable programming. Moreover, many libraries are optimized for performance, providing faster execution times compared to pure Python code.
# 1. Pandas: This library provides data structures and tools for data manipulation and analysis. It's essential for working with tabular data (e.g., CSV, Excel files) and offers functions for quickly filtering, sorting, and joining data.
# 2. NumPy: This is a fundamental package for scientific computing in Python. It contains functions for working with large, multi-dimensional arrays and matrices, along with a large library of mathematical functions to operate on these arrays.
# 3. Matplotlib: This library is used for creating static, animated, and interactive visualizations in Python. It's a flexible and powerful tool for data visualization.
# 4. Seaborn: This is built on top of matplotlib and makes it easier to create beautiful and informative statistical graphs. It aims to simplify the visualization of complex datasets.
# 5. XGBoost: This library implements the gradient boosting algorithm, which is widely used in machine learning for classification, regression, and a host of other tasks. It's known for its high efficiency and flexibility.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import xgboost as xgb
# # **Dataset and Manipulations**
# Now, we need a dataset, arguably the most critical part of this whole process. There are many places where you can obtain data. I've chosen a Yahoo Finance dataset for Bitcoin. Keep in mind that this tutorial can be applied to other cryptocurrencies with a similar structure of datasets, albeit minor adjustments may be necessary for different ones. (https://finance.yahoo.com/quote/BTC-USD/history/)
# This dataset ranges from September 17, 2014, to May 11, 2023, with daily frequencies and the following data:
# 1. Open, referring to the opening price.
# 2. High, the highest price for that day.
# 3. Low, the lowest price for that day.
# 4. Close, the closing price.
# 5. Adj Close, which will be deleted as it is not relevant in the case of Bitcoin and is equal to the closing price.
# 6. Volume, representing the total amount (in dollars) of Bitcoin traded during a given period (daily).
# **Loading a Dataset:**
# We need to "activate" the dataset through the pandas library and commands, as can be seen below. Note that the file path is established through Kaggle and may vary depending on different software applications or computer settings. Therefore, this step is individual, and for proper functioning in all scenarios, it's essential to understand how to upload a dataset. Resources like www.youtube.com can be very helpful in this regard. Regarding column deletion, the "del" command does the job.
df = pd.read_csv("/kaggle/input/projekt/BTC-USD.csv")
del df["Adj Close"]
df["Daily_Change"] = (
(df["Close"] - df["Open"]) / df["Open"]
) * 100 # see chapter Time Series
#
# **Data Check:** After loading the dataset into a DataFrame, referred to as "df", it is always good practice to examine the structure and content of the dataset. You can observe that the data rows contain the variables previously explained. Additionally, you might want to check the last few rows to verify the completeness and integrity of your data.
# **First Rows**
df.head()
# **Last Rows**
df.tail()
# **Index:** Since a time series is a series of data points indexed in time order, the only logical way to manipulate data is to set the date as the index ("Date"). We can achieve this with the following command.
df = df.set_index("Date")
df.index = pd.to_datetime(df.index)
df.head()
# **Graph:** Now, it is a good time to plot this data, as graphs are our best friends in visualizing patterns. It's not necessary to use a specific plot, these visualizations are largely up to your imagination and preferences. Therefore, this code can be adjusted to your liking. For more examples, see (https://seaborn.pydata.org/examples/index.html).
sns.set_theme(style="darkgrid")
df.plot(y="Close", color="orange", title="BTC Closing Price")
# # **Model Training**
# Training a model is a crucial phase in the machine learning process. It involves feeding a dataset into an algorithm, which learns from the data patterns and makes predictions or decisions without being specifically programmed to perform the task. This phase requires careful selection of a suitable model and fine-tuning of parameters to optimize performance. Remember, the goal is to create a model that not only fits the training data well but can also generalize effectively to new, unseen data.
# **Splitting:** I am setting the date as 01-01-2022, where data before this will be used for training the model, and data after will be used for testing, as can be seen in the graph. The first part of the code splits the dataset into two parts, and while any date can be selected, keep in mind that the division of data into a training set and a test set depends on the specific circumstances of your project. A generally accepted rule in the field of machine learning is to allocate 70-80% of the data for training and the remaining for testing. Therefore, this particular date has been chosen. The subsequent code will divide the data accordingly and provide a visualization of the split.
def plot_time_series(df, mask, change_date):
fig, ax = plt.subplots()
ax.plot(df.loc[mask, "Close"], color="orange")
ax.plot(df.loc[~mask, "Close"], color="black")
ax.axvline(change_date, color="black", linestyle="--")
plt.show()
change_date = pd.to_datetime("2022-01-01")
train = df.loc[df.index < change_date]
test = df.loc[df.index >= change_date]
mask = df.index < change_date
plot_time_series(df, mask, change_date)
# # **Features**
# In the realm of predictive analysis using a machine learning model, features within a dataset are distinct quantifiable properties or traits of the observed entities. These features, alternately known as attributes or variables, serve as the input from which the model learns patterns, thereby enabling it to make predictions or decisions. For example, in a time-series forecasting scenario, features may encompass historical values of the variable we aim to predict, elements of date or time, or other external variables that might impact the variable being forecasted. To handle these features, one would generally utilize data manipulation libraries in Python, like pandas in our case, wherein features can be chosen, modified, or created using a range of functions and methods. Bear in mind, the process of feature selection and engineering is a pivotal phase in the development of an efficient machine learning model, given that the model's effectiveness is significantly influenced by the quality and pertinence of the chosen features.
# **Time Series:** Given that we are working with time series data, there are several aspects we must consider. A crucial one is the issue of stationarity, or more specifically, non-stationary time series. As the Bitcoin price is significantly non-stationary (statistical testing is beyond the scope of this work), there are steps we need to take into account. Instead of predicting the closing price, I will be predicting daily changes (in %) as tests demonstrate these are stationary and hence suitable for further use.
from statsmodels.tsa.stattools import adfuller
import matplotlib.pyplot as plt
def perform_adf_test(series):
result = adfuller(series)
print(f"ADF test statistic: {result[0]}")
print(f"p-value: {result[1]}")
def plot_data(df, mask, change_date):
fig, ax = plt.subplots()
ax.plot(df.index[mask], df.loc[mask, "Daily_Change"], color="orange")
ax.plot(df.index[~mask], df.loc[~mask, "Daily_Change"], color="black")
ax.axvline(change_date, color="black", linestyle="--")
plt.show()
perform_adf_test(df["Daily_Change"])
plot_data(df, mask, change_date)
# **Specific Features:** In our endeavor to predict Bitcoin prices using machine learning, we're not merely confined to the historical data of Bitcoin itself. Incorporating various external indicators such as the stock market index, 30-year Treasury bond rates, and the values of stable coins like USDT, can provide additional context and potentially enhance the model's predictive power. These external factors depict broader economic conditions, which are known to influence cryptocurrency markets. Furthermore, temporal aspects like the specific month or week of the year can also serve as critical features. For instance, some patterns may be associated with certain times of the year or week due to trading behaviors or recurring events, and including these as features in our model can further improve its accuracy. It's a delicate balance of capturing complexity without overfitting, keeping in mind that each added feature contributes to the dimensionality of the model. One more challenge is that not every data set is available on the same date, hence not all of these can be utilized with a machine learning model. As ETFs, particularly SPY, show a correlation with Bitcoin price action, the decision to sacrifice weekend data to enrich the dataset with this knowledge is chosen.
from sklearn.impute import SimpleImputer
import numpy as np
import pandas as pd
def create_features(df, df2):
# Sloučení dat
df = df.merge(df2, left_index=True, right_index=True, how="inner")
# Předzpracování
df.ffill(inplace=True)
df.bfill(inplace=True)
# Vytváření nových vlastností
df["Year"], df["Month"], df["Day"], df["DayOfWeek"] = (
df.index.year,
df.index.month,
df.index.day,
df.index.dayofweek,
)
df.sort_index(inplace=True)
df.index = pd.to_datetime(df.index)
halving_dates = [
pd.to_datetime("2012-11-28"),
pd.to_datetime("2016-07-09"),
pd.to_datetime("2020-05-11"),
pd.to_datetime("2024-05-06"),
]
df["days_to_halving"] = np.nan
for i in range(len(halving_dates) - 1):
mask = (df.index >= halving_dates[i]) & (df.index < halving_dates[i + 1])
df.loc[mask, "days_to_halving"] = (
halving_dates[i + 1] - df.loc[mask].index
).days
return df
# Předpokládáme, že df a df2 jsou již definovány
df = create_features(df, df2)
print(df)
from sklearn.impute import SimpleImputer
df2 = (
pd.read_csv(
"/kaggle/input/projekt2/SPY.csv", parse_dates=["Date"], index_col="Date"
)
.drop(columns=["Adj Close"])
.add_prefix("df2_")
)
df2.ffill(inplace=True)
df2.bfill(inplace=True)
df = df.merge(df2, left_index=True, right_index=True, how="inner")
df["Year"], df["Month"], df["Day"], df["DayOfWeek"] = (
df.index.year,
df.index.month,
df.index.day,
df.index.dayofweek,
)
print(df.head())
# **Halving:** Is a significant event specific to Bitcoin that is often included as a feature in forecasting models. It occurs approximately every four years and involves cutting the block reward in half, resulting in a reduction in the rate at which new Bitcoins are created. There are several reasons why halving is considered an important feature in Bitcoin forecasting. Firstly, it leads to a reduction in the supply of Bitcoin, which has implications for the balance between supply and demand in the market. Secondly, halving events generate market attention and can influence investor sentiment and behavior. Traders and investors closely follow these events and anticipate their impact on Bitcoin's price dynamics. Additionally, historical price patterns associated with previous halvings can provide insights into how the market has reacted in the past.
df = df.sort_index()
df.index = pd.to_datetime(df.index)
halving_dates = [
pd.to_datetime("2012-11-28"),
pd.to_datetime("2016-07-09"),
pd.to_datetime("2020-05-11"),
pd.to_datetime("2024-05-06"),
]
df["days_to_halving"] = np.nan
for i in range(len(halving_dates) - 1):
mask = (df.index >= halving_dates[i]) & (df.index < halving_dates[i + 1])
df.loc[mask, "days_to_halving"] = (halving_dates[i + 1] - df.loc[mask].index).days
print(df)
# # **Model**
train = create_features(train)
test = create_features(test)
FEATURES = [
"Open",
"High",
"Low",
"Close",
"Volume",
"df2_Open",
"df2_High",
"df2_Low",
"df2_Close",
"df2_Volume",
"Year",
"Month",
"Day",
"DayOfWeek",
"days_to_halving",
]
TARGET = "Daily_Chang"
X_train = train[FEATURES]
y_train = train[TARGET]
X_test = test[FEATURES]
y_test = test[TARGET]
from sklearn.metrics import mean_squared_error
reg = xgb.XGBRegressor(
base_score=0.5,
booster="gbtree",
n_estimators=1000,
early_stopping_rounds=50,
objective="reg:linear",
max_depth=3,
learning_rate=0.01,
)
reg.fit(X_train, y_train, eval_set=[(X_train, y_train), (X_test, y_test)], verbose=100)
# # **Cross Validation**
#
from sklearn.metrics import accuracy_score, roc_auc_score
from sklearn.model_selection import (
train_test_split,
TimeSeriesSplit,
KFold,
StratifiedKFold,
GroupKFold,
StratifiedGroupKFold,
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/321/129321338.ipynb
| null | null |
[{"Id": 129321338, "ScriptId": 38400176, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13957882, "CreationDate": "05/12/2023 18:51:39", "VersionNumber": 2.0, "Title": "semestr\u00e1ln\u00ed", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 191.0, "LinesInsertedFromPrevious": 119.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 72.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # **Libraries**
# In Python, libraries are used to extend the core functionality of the language, allowing users to perform a wide range of tasks without having to write the code from scratch. They encapsulate complex operations into simpler, more readable code, and promote code reuse, which leads to more efficient and reliable programming. Moreover, many libraries are optimized for performance, providing faster execution times compared to pure Python code.
# 1. Pandas: This library provides data structures and tools for data manipulation and analysis. It's essential for working with tabular data (e.g., CSV, Excel files) and offers functions for quickly filtering, sorting, and joining data.
# 2. NumPy: This is a fundamental package for scientific computing in Python. It contains functions for working with large, multi-dimensional arrays and matrices, along with a large library of mathematical functions to operate on these arrays.
# 3. Matplotlib: This library is used for creating static, animated, and interactive visualizations in Python. It's a flexible and powerful tool for data visualization.
# 4. Seaborn: This is built on top of matplotlib and makes it easier to create beautiful and informative statistical graphs. It aims to simplify the visualization of complex datasets.
# 5. XGBoost: This library implements the gradient boosting algorithm, which is widely used in machine learning for classification, regression, and a host of other tasks. It's known for its high efficiency and flexibility.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import xgboost as xgb
# # **Dataset and Manipulations**
# Now, we need a dataset, arguably the most critical part of this whole process. There are many places where you can obtain data. I've chosen a Yahoo Finance dataset for Bitcoin. Keep in mind that this tutorial can be applied to other cryptocurrencies with a similar structure of datasets, albeit minor adjustments may be necessary for different ones. (https://finance.yahoo.com/quote/BTC-USD/history/)
# This dataset ranges from September 17, 2014, to May 11, 2023, with daily frequencies and the following data:
# 1. Open, referring to the opening price.
# 2. High, the highest price for that day.
# 3. Low, the lowest price for that day.
# 4. Close, the closing price.
# 5. Adj Close, which will be deleted as it is not relevant in the case of Bitcoin and is equal to the closing price.
# 6. Volume, representing the total amount (in dollars) of Bitcoin traded during a given period (daily).
# **Loading a Dataset:**
# We need to "activate" the dataset through the pandas library and commands, as can be seen below. Note that the file path is established through Kaggle and may vary depending on different software applications or computer settings. Therefore, this step is individual, and for proper functioning in all scenarios, it's essential to understand how to upload a dataset. Resources like www.youtube.com can be very helpful in this regard. Regarding column deletion, the "del" command does the job.
df = pd.read_csv("/kaggle/input/projekt/BTC-USD.csv")
del df["Adj Close"]
df["Daily_Change"] = (
(df["Close"] - df["Open"]) / df["Open"]
) * 100 # see chapter Time Series
#
# **Data Check:** After loading the dataset into a DataFrame, referred to as "df", it is always good practice to examine the structure and content of the dataset. You can observe that the data rows contain the variables previously explained. Additionally, you might want to check the last few rows to verify the completeness and integrity of your data.
# **First Rows**
df.head()
# **Last Rows**
df.tail()
# **Index:** Since a time series is a series of data points indexed in time order, the only logical way to manipulate data is to set the date as the index ("Date"). We can achieve this with the following command.
df = df.set_index("Date")
df.index = pd.to_datetime(df.index)
df.head()
# **Graph:** Now, it is a good time to plot this data, as graphs are our best friends in visualizing patterns. It's not necessary to use a specific plot, these visualizations are largely up to your imagination and preferences. Therefore, this code can be adjusted to your liking. For more examples, see (https://seaborn.pydata.org/examples/index.html).
sns.set_theme(style="darkgrid")
df.plot(y="Close", color="orange", title="BTC Closing Price")
# # **Model Training**
# Training a model is a crucial phase in the machine learning process. It involves feeding a dataset into an algorithm, which learns from the data patterns and makes predictions or decisions without being specifically programmed to perform the task. This phase requires careful selection of a suitable model and fine-tuning of parameters to optimize performance. Remember, the goal is to create a model that not only fits the training data well but can also generalize effectively to new, unseen data.
# **Splitting:** I am setting the date as 01-01-2022, where data before this will be used for training the model, and data after will be used for testing, as can be seen in the graph. The first part of the code splits the dataset into two parts, and while any date can be selected, keep in mind that the division of data into a training set and a test set depends on the specific circumstances of your project. A generally accepted rule in the field of machine learning is to allocate 70-80% of the data for training and the remaining for testing. Therefore, this particular date has been chosen. The subsequent code will divide the data accordingly and provide a visualization of the split.
def plot_time_series(df, mask, change_date):
fig, ax = plt.subplots()
ax.plot(df.loc[mask, "Close"], color="orange")
ax.plot(df.loc[~mask, "Close"], color="black")
ax.axvline(change_date, color="black", linestyle="--")
plt.show()
change_date = pd.to_datetime("2022-01-01")
train = df.loc[df.index < change_date]
test = df.loc[df.index >= change_date]
mask = df.index < change_date
plot_time_series(df, mask, change_date)
# # **Features**
# In the realm of predictive analysis using a machine learning model, features within a dataset are distinct quantifiable properties or traits of the observed entities. These features, alternately known as attributes or variables, serve as the input from which the model learns patterns, thereby enabling it to make predictions or decisions. For example, in a time-series forecasting scenario, features may encompass historical values of the variable we aim to predict, elements of date or time, or other external variables that might impact the variable being forecasted. To handle these features, one would generally utilize data manipulation libraries in Python, like pandas in our case, wherein features can be chosen, modified, or created using a range of functions and methods. Bear in mind, the process of feature selection and engineering is a pivotal phase in the development of an efficient machine learning model, given that the model's effectiveness is significantly influenced by the quality and pertinence of the chosen features.
# **Time Series:** Given that we are working with time series data, there are several aspects we must consider. A crucial one is the issue of stationarity, or more specifically, non-stationary time series. As the Bitcoin price is significantly non-stationary (statistical testing is beyond the scope of this work), there are steps we need to take into account. Instead of predicting the closing price, I will be predicting daily changes (in %) as tests demonstrate these are stationary and hence suitable for further use.
from statsmodels.tsa.stattools import adfuller
import matplotlib.pyplot as plt
def perform_adf_test(series):
result = adfuller(series)
print(f"ADF test statistic: {result[0]}")
print(f"p-value: {result[1]}")
def plot_data(df, mask, change_date):
fig, ax = plt.subplots()
ax.plot(df.index[mask], df.loc[mask, "Daily_Change"], color="orange")
ax.plot(df.index[~mask], df.loc[~mask, "Daily_Change"], color="black")
ax.axvline(change_date, color="black", linestyle="--")
plt.show()
perform_adf_test(df["Daily_Change"])
plot_data(df, mask, change_date)
# **Specific Features:** In our endeavor to predict Bitcoin prices using machine learning, we're not merely confined to the historical data of Bitcoin itself. Incorporating various external indicators such as the stock market index, 30-year Treasury bond rates, and the values of stable coins like USDT, can provide additional context and potentially enhance the model's predictive power. These external factors depict broader economic conditions, which are known to influence cryptocurrency markets. Furthermore, temporal aspects like the specific month or week of the year can also serve as critical features. For instance, some patterns may be associated with certain times of the year or week due to trading behaviors or recurring events, and including these as features in our model can further improve its accuracy. It's a delicate balance of capturing complexity without overfitting, keeping in mind that each added feature contributes to the dimensionality of the model. One more challenge is that not every data set is available on the same date, hence not all of these can be utilized with a machine learning model. As ETFs, particularly SPY, show a correlation with Bitcoin price action, the decision to sacrifice weekend data to enrich the dataset with this knowledge is chosen.
from sklearn.impute import SimpleImputer
import numpy as np
import pandas as pd
def create_features(df, df2):
# Sloučení dat
df = df.merge(df2, left_index=True, right_index=True, how="inner")
# Předzpracování
df.ffill(inplace=True)
df.bfill(inplace=True)
# Vytváření nových vlastností
df["Year"], df["Month"], df["Day"], df["DayOfWeek"] = (
df.index.year,
df.index.month,
df.index.day,
df.index.dayofweek,
)
df.sort_index(inplace=True)
df.index = pd.to_datetime(df.index)
halving_dates = [
pd.to_datetime("2012-11-28"),
pd.to_datetime("2016-07-09"),
pd.to_datetime("2020-05-11"),
pd.to_datetime("2024-05-06"),
]
df["days_to_halving"] = np.nan
for i in range(len(halving_dates) - 1):
mask = (df.index >= halving_dates[i]) & (df.index < halving_dates[i + 1])
df.loc[mask, "days_to_halving"] = (
halving_dates[i + 1] - df.loc[mask].index
).days
return df
# Předpokládáme, že df a df2 jsou již definovány
df = create_features(df, df2)
print(df)
from sklearn.impute import SimpleImputer
df2 = (
pd.read_csv(
"/kaggle/input/projekt2/SPY.csv", parse_dates=["Date"], index_col="Date"
)
.drop(columns=["Adj Close"])
.add_prefix("df2_")
)
df2.ffill(inplace=True)
df2.bfill(inplace=True)
df = df.merge(df2, left_index=True, right_index=True, how="inner")
df["Year"], df["Month"], df["Day"], df["DayOfWeek"] = (
df.index.year,
df.index.month,
df.index.day,
df.index.dayofweek,
)
print(df.head())
# **Halving:** Is a significant event specific to Bitcoin that is often included as a feature in forecasting models. It occurs approximately every four years and involves cutting the block reward in half, resulting in a reduction in the rate at which new Bitcoins are created. There are several reasons why halving is considered an important feature in Bitcoin forecasting. Firstly, it leads to a reduction in the supply of Bitcoin, which has implications for the balance between supply and demand in the market. Secondly, halving events generate market attention and can influence investor sentiment and behavior. Traders and investors closely follow these events and anticipate their impact on Bitcoin's price dynamics. Additionally, historical price patterns associated with previous halvings can provide insights into how the market has reacted in the past.
df = df.sort_index()
df.index = pd.to_datetime(df.index)
halving_dates = [
pd.to_datetime("2012-11-28"),
pd.to_datetime("2016-07-09"),
pd.to_datetime("2020-05-11"),
pd.to_datetime("2024-05-06"),
]
df["days_to_halving"] = np.nan
for i in range(len(halving_dates) - 1):
mask = (df.index >= halving_dates[i]) & (df.index < halving_dates[i + 1])
df.loc[mask, "days_to_halving"] = (halving_dates[i + 1] - df.loc[mask].index).days
print(df)
# # **Model**
train = create_features(train)
test = create_features(test)
FEATURES = [
"Open",
"High",
"Low",
"Close",
"Volume",
"df2_Open",
"df2_High",
"df2_Low",
"df2_Close",
"df2_Volume",
"Year",
"Month",
"Day",
"DayOfWeek",
"days_to_halving",
]
TARGET = "Daily_Chang"
X_train = train[FEATURES]
y_train = train[TARGET]
X_test = test[FEATURES]
y_test = test[TARGET]
from sklearn.metrics import mean_squared_error
reg = xgb.XGBRegressor(
base_score=0.5,
booster="gbtree",
n_estimators=1000,
early_stopping_rounds=50,
objective="reg:linear",
max_depth=3,
learning_rate=0.01,
)
reg.fit(X_train, y_train, eval_set=[(X_train, y_train), (X_test, y_test)], verbose=100)
# # **Cross Validation**
#
from sklearn.metrics import accuracy_score, roc_auc_score
from sklearn.model_selection import (
train_test_split,
TimeSeriesSplit,
KFold,
StratifiedKFold,
GroupKFold,
StratifiedGroupKFold,
)
| false | 0 | 3,584 | 0 | 3,584 | 3,584 |
||
129321791
|
<jupyter_start><jupyter_text>Animals-10
Hello everyone!
This is the dataset I have used for my matriculation thesis.
It contains about 28K medium quality animal images belonging to 10 categories: dog, cat, horse, spyder, butterfly, chicken, sheep, cow, squirrel, elephant.
I have used it to test different image recognition networks: from homemade CNNs (~80% accuracy) to Google Inception (98%). It could simulate a smart gallery for a researcher (like a biologist).
All the images have been collected from "google images" and have been checked by human. There is some erroneous data to simulate real conditions (eg. images taken by users of your app).
The main directory is divided into folders, one for each category. Image count for each category varies from 2K to 5 K units.
Kaggle dataset identifier: animals10
<jupyter_script>import numpy as np
import pandas as pd
import os
import tensorflow as tf
import matplotlib.pyplot as plt
import seaborn as sns
from tensorflow.keras.preprocessing.image import (
ImageDataGenerator,
load_img,
img_to_array,
)
from tensorflow.keras.models import Sequential, Model
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Dense, GlobalAveragePooling2D
from tensorflow.keras.applications import VGG16
plt.rcParams["font.size"] = 14
batch_size = 32
img_size = 224
directory = "/kaggle/input/animals10/raw-img"
datagen = ImageDataGenerator(
rescale=1 / 255.0, zoom_range=0.2, horizontal_flip=True, validation_split=0.15
)
train_generator = datagen.flow_from_directory(
directory,
target_size=(img_size, img_size),
batch_size=batch_size,
shuffle=True,
subset="training",
class_mode="categorical",
)
validation_generator = datagen.flow_from_directory(
directory,
target_size=(img_size, img_size),
batch_size=batch_size,
shuffle=False,
subset="validation",
class_mode="categorical",
)
# train_generator.__dict__
# train_generator.class_indices
[key for key in train_generator.class_indices]
labels = [k for k in train_generator.class_indices]
sample_generate = train_generator.__next__()
images = sample_generate[0]
titles = sample_generate[1]
plt.figure(figsize=(20, 20))
for i in range(15):
plt.subplot(5, 5, i + 1)
plt.subplots_adjust(hspace=0.3, wspace=0.3)
plt.imshow(images[i])
plt.title(f"Class: {labels[np.argmax(titles[i],axis=0)]}")
plt.axis("off")
# VGG16
img_size = 224
base_model = VGG16(
include_top=False, # include_top is to include the classifier layer; whether to include the 3 fully-connected layers at the top of the network
weights="imagenet",
input_shape=(img_size, img_size, 3),
)
base_model.summary()
# Freezing the bottom layers
base_model.layers
for layer in base_model.layers[:-4]:
layer.trainable = False
base_model.summary()
# When we set a lr and convergence is slow, increase lr
# reduce lr when the convergence is a plateau
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
model_name = "model.h5"
checkpoint = ModelCheckpoint(
model_name, monitor="val_loss", mode="min", save_best_only=True, verbose=1
)
earlystopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
last_output = base_model.output
x = GlobalAveragePooling2D()(last_output)
x = Dense(512, activation="relu")(x)
outputs = Dense(10, activation="softmax")(x)
model = Model(inputs=base_model.inputs, outputs=outputs)
model.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=0.0001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
history = model.fit(
train_generator,
epochs=3,
validation_data=validation_generator,
callbacks=[checkpoint, earlystopping],
)
# Epoch 00010: val_loss did not improve from 0.22283
# Restoring model weights from the end of the best epoch.
# Epoch 00010: early stopping
plt.figure(figsize=(20, 8))
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model Loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Train", "Val"], loc="upper left")
plt.show()
plt.figure(figsize=(20, 8))
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model Accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["Train", "Val"], loc="upper left")
plt.show()
from sklearn.metrics import classification_report, confusion_matrix
labels
model = tf.keras.models.load_model("/kaggle/working/model.h5")
y_test = validation_generator.classes
y_pred = model.predict(validation_generator)
y_pred_probs = y_pred.copy()
y_test
y_pred_int = np.argmax(y_pred_probs, axis=1)
print(classification_report(y_test, y_pred_int, target_names=labels))
plt.figure(figsize=(10, 8))
sns.heatmap(
confusion_matrix(y_test, y_pred_int),
annot=True,
fmt=".3g",
xticklabels=labels,
yticklabels=labels,
cmap="Blues",
)
plt.show()
y = confusion_matrix(y_test, y_pred_int)
print(y)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/321/129321791.ipynb
|
animals10
|
alessiocorrado99
|
[{"Id": 129321791, "ScriptId": 37432569, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6542785, "CreationDate": "05/12/2023 18:57:27", "VersionNumber": 1.0, "Title": "Multiclass Classification", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 152.0, "LinesInsertedFromPrevious": 152.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 185260454, "KernelVersionId": 129321791, "SourceDatasetVersionId": 840806}]
|
[{"Id": 840806, "DatasetId": 59760, "DatasourceVersionId": 863703, "CreatorUserId": 1831139, "LicenseName": "GPL 2", "CreationDate": "12/12/2019 20:46:33", "VersionNumber": 2.0, "Title": "Animals-10", "Slug": "animals10", "Subtitle": "Animal pictures of 10 different categories taken from google images", "Description": "Hello everyone! \n\nThis is the dataset I have used for my matriculation thesis. \n\nIt contains about 28K medium quality animal images belonging to 10 categories: dog, cat, horse, spyder, butterfly, chicken, sheep, cow, squirrel, elephant. \n\nI have used it to test different image recognition networks: from homemade CNNs (~80% accuracy) to Google Inception (98%). It could simulate a smart gallery for a researcher (like a biologist).\n\nAll the images have been collected from \"google images\" and have been checked by human. There is some erroneous data to simulate real conditions (eg. images taken by users of your app).\n\nThe main directory is divided into folders, one for each category. Image count for each category varies from 2K to 5 K units.", "VersionNotes": "v2", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 59760, "CreatorUserId": 1831139, "OwnerUserId": 1831139.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 840806.0, "CurrentDatasourceVersionId": 863703.0, "ForumId": 68632, "Type": 2, "CreationDate": "10/04/2018 21:16:26", "LastActivityDate": "10/04/2018", "TotalViews": 250550, "TotalDownloads": 39974, "TotalVotes": 714, "TotalKernels": 131}]
|
[{"Id": 1831139, "UserName": "alessiocorrado99", "DisplayName": "Corrado Alessio", "RegisterDate": "04/17/2018", "PerformanceTier": 1}]
|
import numpy as np
import pandas as pd
import os
import tensorflow as tf
import matplotlib.pyplot as plt
import seaborn as sns
from tensorflow.keras.preprocessing.image import (
ImageDataGenerator,
load_img,
img_to_array,
)
from tensorflow.keras.models import Sequential, Model
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Dense, GlobalAveragePooling2D
from tensorflow.keras.applications import VGG16
plt.rcParams["font.size"] = 14
batch_size = 32
img_size = 224
directory = "/kaggle/input/animals10/raw-img"
datagen = ImageDataGenerator(
rescale=1 / 255.0, zoom_range=0.2, horizontal_flip=True, validation_split=0.15
)
train_generator = datagen.flow_from_directory(
directory,
target_size=(img_size, img_size),
batch_size=batch_size,
shuffle=True,
subset="training",
class_mode="categorical",
)
validation_generator = datagen.flow_from_directory(
directory,
target_size=(img_size, img_size),
batch_size=batch_size,
shuffle=False,
subset="validation",
class_mode="categorical",
)
# train_generator.__dict__
# train_generator.class_indices
[key for key in train_generator.class_indices]
labels = [k for k in train_generator.class_indices]
sample_generate = train_generator.__next__()
images = sample_generate[0]
titles = sample_generate[1]
plt.figure(figsize=(20, 20))
for i in range(15):
plt.subplot(5, 5, i + 1)
plt.subplots_adjust(hspace=0.3, wspace=0.3)
plt.imshow(images[i])
plt.title(f"Class: {labels[np.argmax(titles[i],axis=0)]}")
plt.axis("off")
# VGG16
img_size = 224
base_model = VGG16(
include_top=False, # include_top is to include the classifier layer; whether to include the 3 fully-connected layers at the top of the network
weights="imagenet",
input_shape=(img_size, img_size, 3),
)
base_model.summary()
# Freezing the bottom layers
base_model.layers
for layer in base_model.layers[:-4]:
layer.trainable = False
base_model.summary()
# When we set a lr and convergence is slow, increase lr
# reduce lr when the convergence is a plateau
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
model_name = "model.h5"
checkpoint = ModelCheckpoint(
model_name, monitor="val_loss", mode="min", save_best_only=True, verbose=1
)
earlystopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
last_output = base_model.output
x = GlobalAveragePooling2D()(last_output)
x = Dense(512, activation="relu")(x)
outputs = Dense(10, activation="softmax")(x)
model = Model(inputs=base_model.inputs, outputs=outputs)
model.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=0.0001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
history = model.fit(
train_generator,
epochs=3,
validation_data=validation_generator,
callbacks=[checkpoint, earlystopping],
)
# Epoch 00010: val_loss did not improve from 0.22283
# Restoring model weights from the end of the best epoch.
# Epoch 00010: early stopping
plt.figure(figsize=(20, 8))
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model Loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Train", "Val"], loc="upper left")
plt.show()
plt.figure(figsize=(20, 8))
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model Accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["Train", "Val"], loc="upper left")
plt.show()
from sklearn.metrics import classification_report, confusion_matrix
labels
model = tf.keras.models.load_model("/kaggle/working/model.h5")
y_test = validation_generator.classes
y_pred = model.predict(validation_generator)
y_pred_probs = y_pred.copy()
y_test
y_pred_int = np.argmax(y_pred_probs, axis=1)
print(classification_report(y_test, y_pred_int, target_names=labels))
plt.figure(figsize=(10, 8))
sns.heatmap(
confusion_matrix(y_test, y_pred_int),
annot=True,
fmt=".3g",
xticklabels=labels,
yticklabels=labels,
cmap="Blues",
)
plt.show()
y = confusion_matrix(y_test, y_pred_int)
print(y)
| false | 0 | 1,321 | 3 | 1,540 | 1,321 |
||
129321131
|
<jupyter_start><jupyter_text>Aeroclub 2023
Kaggle dataset identifier: aeroclub-2023
<jupyter_script>import numpy as np
import pandas as pd
import os
# Категории (Из pdf'ки)
# Оформление
# Запрос
# вариантов
# Бронирование
# Отмена
# Не заявка
#
task_1_train = pd.read_excel("/kaggle/input/aeroclub-2023/1/Задача №1/train_data.xlsx")
task_1_train.head()
task_1_train["title"][7]
task_1_train["text"][13]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/321/129321131.ipynb
|
aeroclub-2023
|
dimka11
|
[{"Id": 129321131, "ScriptId": 38449206, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2778887, "CreationDate": "05/12/2023 18:49:22", "VersionNumber": 2.0, "Title": "Aeroclub 2023 EDA Both tasks", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 30.0, "LinesInsertedFromPrevious": 21.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 9.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185259347, "KernelVersionId": 129321131, "SourceDatasetVersionId": 5671957}]
|
[{"Id": 5671957, "DatasetId": 3260672, "DatasourceVersionId": 5747475, "CreatorUserId": 2778887, "LicenseName": "Unknown", "CreationDate": "05/12/2023 18:18:42", "VersionNumber": 1.0, "Title": "Aeroclub 2023", "Slug": "aeroclub-2023", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3260672, "CreatorUserId": 2778887, "OwnerUserId": 2778887.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5671957.0, "CurrentDatasourceVersionId": 5747475.0, "ForumId": 3326228, "Type": 2, "CreationDate": "05/12/2023 18:18:42", "LastActivityDate": "05/12/2023", "TotalViews": 47, "TotalDownloads": 3, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 2778887, "UserName": "dimka11", "DisplayName": "Dmitry Sokolov", "RegisterDate": "02/04/2019", "PerformanceTier": 1}]
|
import numpy as np
import pandas as pd
import os
# Категории (Из pdf'ки)
# Оформление
# Запрос
# вариантов
# Бронирование
# Отмена
# Не заявка
#
task_1_train = pd.read_excel("/kaggle/input/aeroclub-2023/1/Задача №1/train_data.xlsx")
task_1_train.head()
task_1_train["title"][7]
task_1_train["text"][13]
| false | 0 | 144 | 0 | 176 | 144 |
||
129356682
|
from fasteda import fast_eda
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler, MinMaxScaler
train = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/train.csv")
test = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/test.csv")
greeks = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/greeks.csv")
def scale_data(df):
scaler = StandardScaler()
float_cols = df.select_dtypes(include=["float64"])
scaled_data = scaler.fit_transform(df[list(float_cols)])
train[list(float_cols)] = scaled_data
return df
test
# ### [fasteda](https://github.com/Matt-OP/fasteda) on the train set with target = "Class"
fast_eda(train, target="Class")
top_n_corr_features = 25
corr_matrix = train.corr(numeric_only=True).abs()
corr_pairs = corr_matrix.unstack().sort_values(ascending=False)
corr_pairs = corr_pairs[
corr_pairs.index.get_level_values(0) != corr_pairs.index.get_level_values(1)
]
top_n_corr_pairs = corr_pairs[: top_n_corr_features * 2]
duplicate_pairs = set()
feature_tuple, correlation = [], []
for pair in top_n_corr_pairs.index:
if (pair[0], pair[1]) not in duplicate_pairs and (
pair[1],
pair[0],
) not in duplicate_pairs:
feature_tuple.append((pair[0], pair[1]))
correlation.append(round(top_n_corr_pairs[pair], 3))
duplicate_pairs.add((pair[0], pair[1]))
plt.style.use("dark_background")
plt.figure(figsize=(10, 12))
ax = sns.barplot(
x=correlation,
y=[str(feats) for feats in feature_tuple],
palette=sns.color_palette("Blues_r", n_colors=len(feature_tuple)),
width=0.7,
linewidth=1.2,
edgecolor="#FFFFFF",
)
for container in ax.containers:
ax.bar_label(container, size=10, padding=5)
plt.title(f"Top {top_n_corr_features} feature pairs with highest Pearson correlation")
plt.xlim(0, 1.05)
plt.xlabel("Pearson correlation")
plt.ylabel("Feature pairs")
plt.grid(False)
plt.show()
unique_counts = [np.unique(train[col]).size for col in train.columns[1:-1]]
name_count_pairs = [
(col, unique_counts[i]) for i, col in enumerate(train.columns[1:-1])
]
sorted_pairs = sorted(name_count_pairs, key=lambda x: x[1], reverse=True)
plt.style.use("dark_background")
plt.figure(figsize=(10, 14))
ax = sns.barplot(
x=[pair[1] for pair in sorted_pairs],
y=[pair[0] for pair in sorted_pairs],
palette=sns.color_palette("Reds_r", n_colors=len(sorted_pairs)),
width=0.6,
linewidth=1,
edgecolor="#FFFFFF",
)
for container in ax.containers:
ax.bar_label(container, size=10, padding=5)
plt.title(f"Unique counts for each feature | n rows in train = {len(train)}")
plt.xlabel("Number of unique samples")
plt.ylabel("Features")
plt.grid(False)
plt.show()
colors = sns.color_palette("viridis")
for i, col in enumerate(greeks.columns[1:-1]):
plt.figure(figsize=(8, 6))
ax = (
greeks[col]
.value_counts(ascending=True)
.plot.barh(color=colors[i], edgecolor="#FFFFFF")
)
for container in ax.containers:
ax.bar_label(container, size=8, padding=3)
plt.title(f"Value counts of {col} in greeks.csv")
plt.grid(False)
plt.show()
train["is_train"] = 1
test["is_train"] = 0
train_test = pd.concat([train, test], ignore_index=True)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/356/129356682.ipynb
| null | null |
[{"Id": 129356682, "ScriptId": 38412154, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10590800, "CreationDate": "05/13/2023 05:06:57", "VersionNumber": 2.0, "Title": "ICR - Identifying Age-Related Conditions EDA \u2728", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 98.0, "LinesInsertedFromPrevious": 79.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 19.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
from fasteda import fast_eda
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler, MinMaxScaler
train = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/train.csv")
test = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/test.csv")
greeks = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/greeks.csv")
def scale_data(df):
scaler = StandardScaler()
float_cols = df.select_dtypes(include=["float64"])
scaled_data = scaler.fit_transform(df[list(float_cols)])
train[list(float_cols)] = scaled_data
return df
test
# ### [fasteda](https://github.com/Matt-OP/fasteda) on the train set with target = "Class"
fast_eda(train, target="Class")
top_n_corr_features = 25
corr_matrix = train.corr(numeric_only=True).abs()
corr_pairs = corr_matrix.unstack().sort_values(ascending=False)
corr_pairs = corr_pairs[
corr_pairs.index.get_level_values(0) != corr_pairs.index.get_level_values(1)
]
top_n_corr_pairs = corr_pairs[: top_n_corr_features * 2]
duplicate_pairs = set()
feature_tuple, correlation = [], []
for pair in top_n_corr_pairs.index:
if (pair[0], pair[1]) not in duplicate_pairs and (
pair[1],
pair[0],
) not in duplicate_pairs:
feature_tuple.append((pair[0], pair[1]))
correlation.append(round(top_n_corr_pairs[pair], 3))
duplicate_pairs.add((pair[0], pair[1]))
plt.style.use("dark_background")
plt.figure(figsize=(10, 12))
ax = sns.barplot(
x=correlation,
y=[str(feats) for feats in feature_tuple],
palette=sns.color_palette("Blues_r", n_colors=len(feature_tuple)),
width=0.7,
linewidth=1.2,
edgecolor="#FFFFFF",
)
for container in ax.containers:
ax.bar_label(container, size=10, padding=5)
plt.title(f"Top {top_n_corr_features} feature pairs with highest Pearson correlation")
plt.xlim(0, 1.05)
plt.xlabel("Pearson correlation")
plt.ylabel("Feature pairs")
plt.grid(False)
plt.show()
unique_counts = [np.unique(train[col]).size for col in train.columns[1:-1]]
name_count_pairs = [
(col, unique_counts[i]) for i, col in enumerate(train.columns[1:-1])
]
sorted_pairs = sorted(name_count_pairs, key=lambda x: x[1], reverse=True)
plt.style.use("dark_background")
plt.figure(figsize=(10, 14))
ax = sns.barplot(
x=[pair[1] for pair in sorted_pairs],
y=[pair[0] for pair in sorted_pairs],
palette=sns.color_palette("Reds_r", n_colors=len(sorted_pairs)),
width=0.6,
linewidth=1,
edgecolor="#FFFFFF",
)
for container in ax.containers:
ax.bar_label(container, size=10, padding=5)
plt.title(f"Unique counts for each feature | n rows in train = {len(train)}")
plt.xlabel("Number of unique samples")
plt.ylabel("Features")
plt.grid(False)
plt.show()
colors = sns.color_palette("viridis")
for i, col in enumerate(greeks.columns[1:-1]):
plt.figure(figsize=(8, 6))
ax = (
greeks[col]
.value_counts(ascending=True)
.plot.barh(color=colors[i], edgecolor="#FFFFFF")
)
for container in ax.containers:
ax.bar_label(container, size=8, padding=3)
plt.title(f"Value counts of {col} in greeks.csv")
plt.grid(False)
plt.show()
train["is_train"] = 1
test["is_train"] = 0
train_test = pd.concat([train, test], ignore_index=True)
| false | 0 | 1,121 | 1 | 1,121 | 1,121 |
||
129251033
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load the CSV file into a DataFrame
df = pd.read_csv("/kaggle/input/results/results.csv")
# Define metrics where a higher value is better (default is lower is better)
higher_is_better = ["R2"]
# Initialize an empty DataFrame to store ranks
rank_df = pd.DataFrame()
# Iterate over each column, rank and add the ranks to the rank_df
for column in df.columns[1:]: # Skip the 'Model' column
if any(metric in column for metric in higher_is_better):
rank_df[column] = df[column].rank(ascending=False) # Higher is better
else:
rank_df[column] = df[column].rank(ascending=True) # Lower is better
# Add 'Model' column to rank_df
rank_df.insert(0, "Model", df["Model"])
# Calculate the average rank for each model across all metrics for each analytical measure
rank_df["Average Rank"] = rank_df.iloc[:, 1:].mean(axis=1)
# Sort by 'Average Rank'
rank_df = rank_df.sort_values("Average Rank")
# Display the final rankings
final_rankings = rank_df[["Model", "Average Rank"]]
print(final_rankings)
import matplotlib.pyplot as plt
import seaborn as sns
# Create the plot
plt.figure(figsize=(10, 8))
sns.barplot(
x=final_rankings["Average Rank"], y=final_rankings["Model"], palette="viridis"
)
# Add labels and title
plt.xlabel("Average Rank")
plt.ylabel("Model")
plt.title("Average Rankings of NLP Models")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/251/129251033.ipynb
| null | null |
[{"Id": 129251033, "ScriptId": 38392442, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9882726, "CreationDate": "05/12/2023 07:28:26", "VersionNumber": 1.0, "Title": "statistical_ranking", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 65.0, "LinesInsertedFromPrevious": 65.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load the CSV file into a DataFrame
df = pd.read_csv("/kaggle/input/results/results.csv")
# Define metrics where a higher value is better (default is lower is better)
higher_is_better = ["R2"]
# Initialize an empty DataFrame to store ranks
rank_df = pd.DataFrame()
# Iterate over each column, rank and add the ranks to the rank_df
for column in df.columns[1:]: # Skip the 'Model' column
if any(metric in column for metric in higher_is_better):
rank_df[column] = df[column].rank(ascending=False) # Higher is better
else:
rank_df[column] = df[column].rank(ascending=True) # Lower is better
# Add 'Model' column to rank_df
rank_df.insert(0, "Model", df["Model"])
# Calculate the average rank for each model across all metrics for each analytical measure
rank_df["Average Rank"] = rank_df.iloc[:, 1:].mean(axis=1)
# Sort by 'Average Rank'
rank_df = rank_df.sort_values("Average Rank")
# Display the final rankings
final_rankings = rank_df[["Model", "Average Rank"]]
print(final_rankings)
import matplotlib.pyplot as plt
import seaborn as sns
# Create the plot
plt.figure(figsize=(10, 8))
sns.barplot(
x=final_rankings["Average Rank"], y=final_rankings["Model"], palette="viridis"
)
# Add labels and title
plt.xlabel("Average Rank")
plt.ylabel("Model")
plt.title("Average Rankings of NLP Models")
plt.show()
| false | 0 | 588 | 0 | 588 | 588 |
||
129251932
|
# # Blueberry yield
# Importing Libraries
import pandas as pd
import numpy as np
import seaborn as sns
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import matplotlib.pyplot as plt
import plotly.express as px
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
train.head(15)
train = train.drop(columns=["id"])
train.describe()
train.info()
sns.heatmap(train.iloc[:, :].corr())
final_train = train.drop(
columns=[
"RainingDays",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
]
)
final_train
x = final_train.iloc[:, :11]
y = final_train["yield"]
model = tf.keras.Sequential([layers.Dense(11), layers.Dense(6), layers.Dense(1)])
model.compile(optimizer="adam", loss="msle", metrics=["msle"])
train_model = model.fit(x, y, batch_size=64, epochs=100, verbose=0)
history_df = pd.DataFrame(train_model.history)
history_df.loc[:, ["msle"]].plot()
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
test
fin_test = test.drop(
columns=[
"id",
"RainingDays",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
]
)
fin_test.info()
pred = model.predict(fin_test)
pred
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
val = model.predict(x_test)
r2_score(val, y_test)
predf = pd.DataFrame(pred, index=test["id"])
predf
predf.to_csv("submission.csv")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/251/129251932.ipynb
| null | null |
[{"Id": 129251932, "ScriptId": 38404280, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11026853, "CreationDate": "05/12/2023 07:38:19", "VersionNumber": 1.0, "Title": "playground", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 67.0, "LinesInsertedFromPrevious": 67.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Blueberry yield
# Importing Libraries
import pandas as pd
import numpy as np
import seaborn as sns
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import matplotlib.pyplot as plt
import plotly.express as px
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
train.head(15)
train = train.drop(columns=["id"])
train.describe()
train.info()
sns.heatmap(train.iloc[:, :].corr())
final_train = train.drop(
columns=[
"RainingDays",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
]
)
final_train
x = final_train.iloc[:, :11]
y = final_train["yield"]
model = tf.keras.Sequential([layers.Dense(11), layers.Dense(6), layers.Dense(1)])
model.compile(optimizer="adam", loss="msle", metrics=["msle"])
train_model = model.fit(x, y, batch_size=64, epochs=100, verbose=0)
history_df = pd.DataFrame(train_model.history)
history_df.loc[:, ["msle"]].plot()
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
test
fin_test = test.drop(
columns=[
"id",
"RainingDays",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
]
)
fin_test.info()
pred = model.predict(fin_test)
pred
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
val = model.predict(x_test)
r2_score(val, y_test)
predf = pd.DataFrame(pred, index=test["id"])
predf
predf.to_csv("submission.csv")
| false | 0 | 536 | 0 | 536 | 536 |
||
129661309
|
<jupyter_start><jupyter_text>Heart Failure Prediction
# About this dataset
> Cardiovascular diseases (CVDs) are the **number 1 cause of death globally**, taking an estimated **17.9 million lives each year**, which accounts for **31% of all deaths worlwide**.
Heart failure is a common event caused by CVDs and this dataset contains 12 features that can be used to predict mortality by heart failure.
> Most cardiovascular diseases can be prevented by addressing behavioural risk factors such as tobacco use, unhealthy diet and obesity, physical inactivity and harmful use of alcohol using population-wide strategies.
> People with cardiovascular disease or who are at high cardiovascular risk (due to the presence of one or more risk factors such as hypertension, diabetes, hyperlipidaemia or already established disease) need **early detection** and management wherein a machine learning model can be of great help.
# How to use this dataset
> - Create a model for predicting mortality caused by Heart Failure.
- Your kernel can be featured here!
- [More datasets](https://www.kaggle.com/andrewmvd/datasets)
# Acknowledgements
If you use this dataset in your research, please credit the authors
> ### Citation
Davide Chicco, Giuseppe Jurman: Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone. BMC Medical Informatics and Decision Making 20, 16 (2020). ([link](https://doi.org/10.1186/s12911-020-1023-5))
> ### License
CC BY 4.0
> ### Splash icon
Icon by [Freepik](https://www.flaticon.com/authors/freepik), available on [Flaticon](https://www.flaticon.com/free-icon/heart_1186541).
> ### Splash banner
Wallpaper by [jcomp](https://br.freepik.com/jcomp), available on [Freepik](https://br.freepik.com/fotos-gratis/simplesmente-design-minimalista-com-estetoscopio-de-equipamento-de-medicina-ou-phonendoscope_5018002.htm#page=1&query=cardiology&position=3).
Kaggle dataset identifier: heart-failure-clinical-data
<jupyter_code>import pandas as pd
df = pd.read_csv('heart-failure-clinical-data/heart_failure_clinical_records_dataset.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 299 entries, 0 to 298
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 299 non-null float64
1 anaemia 299 non-null int64
2 creatinine_phosphokinase 299 non-null int64
3 diabetes 299 non-null int64
4 ejection_fraction 299 non-null int64
5 high_blood_pressure 299 non-null int64
6 platelets 299 non-null float64
7 serum_creatinine 299 non-null float64
8 serum_sodium 299 non-null int64
9 sex 299 non-null int64
10 smoking 299 non-null int64
11 time 299 non-null int64
12 DEATH_EVENT 299 non-null int64
dtypes: float64(3), int64(10)
memory usage: 30.5 KB
<jupyter_text>Examples:
{
"age": 75.0,
"anaemia": 0.0,
"creatinine_phosphokinase": 582.0,
"diabetes": 0.0,
"ejection_fraction": 20.0,
"high_blood_pressure": 1.0,
"platelets": 265000.0,
"serum_creatinine": 1.9,
"serum_sodium": 130.0,
"sex": 1.0,
"smoking": 0.0,
"time": 4.0,
"DEATH_EVENT": 1.0
}
{
"age": 55.0,
"anaemia": 0.0,
"creatinine_phosphokinase": 7861.0,
"diabetes": 0.0,
"ejection_fraction": 38.0,
"high_blood_pressure": 0.0,
"platelets": 263358.03,
"serum_creatinine": 1.1,
"serum_sodium": 136.0,
"sex": 1.0,
"smoking": 0.0,
"time": 6.0,
"DEATH_EVENT": 1.0
}
{
"age": 65.0,
"anaemia": 0.0,
"creatinine_phosphokinase": 146.0,
"diabetes": 0.0,
"ejection_fraction": 20.0,
"high_blood_pressure": 0.0,
"platelets": 162000.0,
"serum_creatinine": 1.3,
"serum_sodium": 129.0,
"sex": 1.0,
"smoking": 1.0,
"time": 7.0,
"DEATH_EVENT": 1.0
}
{
"age": 50.0,
"anaemia": 1.0,
"creatinine_phosphokinase": 111.0,
"diabetes": 0.0,
"ejection_fraction": 20.0,
"high_blood_pressure": 0.0,
"platelets": 210000.0,
"serum_creatinine": 1.9,
"serum_sodium": 137.0,
"sex": 1.0,
"smoking": 0.0,
"time": 7.0,
"DEATH_EVENT": 1.0
}
<jupyter_script>import numpy as np
import pandas as pd
df = pd.read_csv(
"/kaggle/input/heart-failure-clinical-data/heart_failure_clinical_records_dataset.csv"
)
df
dataset = df.values
dataset
x = dataset[:, 0:12]
y = dataset[:, 10]
from sklearn import preprocessing
sc = preprocessing.MinMaxScaler()
x_scale = sc.fit_transform(x)
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x_scale, y, test_size=0.3, random_state=42
)
# print(x_train.shape,y_train.shape,x_test.shape,y_test.shape)
from keras.models import Sequential
from keras.layers import Dense
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.layers import Dropout
model = Sequential()
model.add(Dense(32, activation="relu", input_shape=(12,)))
model.add(Dropout(0.2))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(1, activation="sigmoid"))
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
model.summary()
hist = model.fit(x_train, y_train, batch_size=256, epochs=30, verbose=1)
_, accuracy = model.evaluate(x_test, y_test)
print("Accuracy: %.2f" % (accuracy * 100))
# # **SVM**
from sklearn import svm
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=100
)
clf = svm.SVC()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
# # **Random Forest**
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
clf = RandomForestClassifier(n_estimators=1000, random_state=1)
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
# # NAIVE BIAS
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=40
)
clf = GaussianNB()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
# # DECISION TREE
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=40
)
clf = DecisionTreeClassifier()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
# # KNN
from sklearn.neighbors import KNeighborsClassifier
clf = KNeighborsClassifier(n_neighbors=5)
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/661/129661309.ipynb
|
heart-failure-clinical-data
|
andrewmvd
|
[{"Id": 129661309, "ScriptId": 38521655, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12850841, "CreationDate": "05/15/2023 14:53:33", "VersionNumber": 1.0, "Title": "heart failure prediction ann", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 118.0, "LinesInsertedFromPrevious": 118.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185965323, "KernelVersionId": 129661309, "SourceDatasetVersionId": 1263738}]
|
[{"Id": 1263738, "DatasetId": 727551, "DatasourceVersionId": 1295676, "CreatorUserId": 793761, "LicenseName": "Attribution 4.0 International (CC BY 4.0)", "CreationDate": "06/20/2020 01:03:20", "VersionNumber": 1.0, "Title": "Heart Failure Prediction", "Slug": "heart-failure-clinical-data", "Subtitle": "12 clinical features por predicting death events.", "Description": "# About this dataset\n> Cardiovascular diseases (CVDs) are the **number 1 cause of death globally**, taking an estimated **17.9 million lives each year**, which accounts for **31% of all deaths worlwide**.\nHeart failure is a common event caused by CVDs and this dataset contains 12 features that can be used to predict mortality by heart failure.\n\n> Most cardiovascular diseases can be prevented by addressing behavioural risk factors such as tobacco use, unhealthy diet and obesity, physical inactivity and harmful use of alcohol using population-wide strategies.\n\n> People with cardiovascular disease or who are at high cardiovascular risk (due to the presence of one or more risk factors such as hypertension, diabetes, hyperlipidaemia or already established disease) need **early detection** and management wherein a machine learning model can be of great help.\n\n# How to use this dataset\n> - Create a model for predicting mortality caused by Heart Failure.\n- Your kernel can be featured here!\n- [More datasets](https://www.kaggle.com/andrewmvd/datasets)\n\n\n\n# Acknowledgements\nIf you use this dataset in your research, please credit the authors\n> ### Citation\nDavide Chicco, Giuseppe Jurman: Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone. BMC Medical Informatics and Decision Making 20, 16 (2020). ([link](https://doi.org/10.1186/s12911-020-1023-5))\n\n> ### License\nCC BY 4.0\n\n> ### Splash icon\nIcon by [Freepik](https://www.flaticon.com/authors/freepik), available on [Flaticon](https://www.flaticon.com/free-icon/heart_1186541).\n\n> ### Splash banner\nWallpaper by [jcomp](https://br.freepik.com/jcomp), available on [Freepik](https://br.freepik.com/fotos-gratis/simplesmente-design-minimalista-com-estetoscopio-de-equipamento-de-medicina-ou-phonendoscope_5018002.htm#page=1&query=cardiology&position=3).", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 727551, "CreatorUserId": 793761, "OwnerUserId": 793761.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1263738.0, "CurrentDatasourceVersionId": 1295676.0, "ForumId": 742394, "Type": 2, "CreationDate": "06/20/2020 01:03:20", "LastActivityDate": "06/20/2020", "TotalViews": 882099, "TotalDownloads": 116977, "TotalVotes": 2090, "TotalKernels": 920}]
|
[{"Id": 793761, "UserName": "andrewmvd", "DisplayName": "Larxel", "RegisterDate": "11/15/2016", "PerformanceTier": 4}]
|
import numpy as np
import pandas as pd
df = pd.read_csv(
"/kaggle/input/heart-failure-clinical-data/heart_failure_clinical_records_dataset.csv"
)
df
dataset = df.values
dataset
x = dataset[:, 0:12]
y = dataset[:, 10]
from sklearn import preprocessing
sc = preprocessing.MinMaxScaler()
x_scale = sc.fit_transform(x)
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x_scale, y, test_size=0.3, random_state=42
)
# print(x_train.shape,y_train.shape,x_test.shape,y_test.shape)
from keras.models import Sequential
from keras.layers import Dense
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.layers import Dropout
model = Sequential()
model.add(Dense(32, activation="relu", input_shape=(12,)))
model.add(Dropout(0.2))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(1, activation="sigmoid"))
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
model.summary()
hist = model.fit(x_train, y_train, batch_size=256, epochs=30, verbose=1)
_, accuracy = model.evaluate(x_test, y_test)
print("Accuracy: %.2f" % (accuracy * 100))
# # **SVM**
from sklearn import svm
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=100
)
clf = svm.SVC()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
# # **Random Forest**
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
clf = RandomForestClassifier(n_estimators=1000, random_state=1)
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
# # NAIVE BIAS
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=40
)
clf = GaussianNB()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
# # DECISION TREE
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=40
)
clf = DecisionTreeClassifier()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
# # KNN
from sklearn.neighbors import KNeighborsClassifier
clf = KNeighborsClassifier(n_neighbors=5)
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy * 100)
|
[{"heart-failure-clinical-data/heart_failure_clinical_records_dataset.csv": {"column_names": "[\"age\", \"anaemia\", \"creatinine_phosphokinase\", \"diabetes\", \"ejection_fraction\", \"high_blood_pressure\", \"platelets\", \"serum_creatinine\", \"serum_sodium\", \"sex\", \"smoking\", \"time\", \"DEATH_EVENT\"]", "column_data_types": "{\"age\": \"float64\", \"anaemia\": \"int64\", \"creatinine_phosphokinase\": \"int64\", \"diabetes\": \"int64\", \"ejection_fraction\": \"int64\", \"high_blood_pressure\": \"int64\", \"platelets\": \"float64\", \"serum_creatinine\": \"float64\", \"serum_sodium\": \"int64\", \"sex\": \"int64\", \"smoking\": \"int64\", \"time\": \"int64\", \"DEATH_EVENT\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 299 entries, 0 to 298\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 age 299 non-null float64\n 1 anaemia 299 non-null int64 \n 2 creatinine_phosphokinase 299 non-null int64 \n 3 diabetes 299 non-null int64 \n 4 ejection_fraction 299 non-null int64 \n 5 high_blood_pressure 299 non-null int64 \n 6 platelets 299 non-null float64\n 7 serum_creatinine 299 non-null float64\n 8 serum_sodium 299 non-null int64 \n 9 sex 299 non-null int64 \n 10 smoking 299 non-null int64 \n 11 time 299 non-null int64 \n 12 DEATH_EVENT 299 non-null int64 \ndtypes: float64(3), int64(10)\nmemory usage: 30.5 KB\n", "summary": "{\"age\": {\"count\": 299.0, \"mean\": 60.83389297658862, \"std\": 11.89480907404447, \"min\": 40.0, \"25%\": 51.0, \"50%\": 60.0, \"75%\": 70.0, \"max\": 95.0}, \"anaemia\": {\"count\": 299.0, \"mean\": 0.431438127090301, \"std\": 0.4961072681330793, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"creatinine_phosphokinase\": {\"count\": 299.0, \"mean\": 581.8394648829432, \"std\": 970.2878807124362, \"min\": 23.0, \"25%\": 116.5, \"50%\": 250.0, \"75%\": 582.0, \"max\": 7861.0}, \"diabetes\": {\"count\": 299.0, \"mean\": 0.4180602006688963, \"std\": 0.49406706510360904, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"ejection_fraction\": {\"count\": 299.0, \"mean\": 38.08361204013378, \"std\": 11.834840741039171, \"min\": 14.0, \"25%\": 30.0, \"50%\": 38.0, \"75%\": 45.0, \"max\": 80.0}, \"high_blood_pressure\": {\"count\": 299.0, \"mean\": 0.3511705685618729, \"std\": 0.47813637906274475, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"platelets\": {\"count\": 299.0, \"mean\": 263358.02926421404, \"std\": 97804.2368685983, \"min\": 25100.0, \"25%\": 212500.0, \"50%\": 262000.0, \"75%\": 303500.0, \"max\": 850000.0}, \"serum_creatinine\": {\"count\": 299.0, \"mean\": 1.3938795986622072, \"std\": 1.0345100640898541, \"min\": 0.5, \"25%\": 0.9, \"50%\": 1.1, \"75%\": 1.4, \"max\": 9.4}, \"serum_sodium\": {\"count\": 299.0, \"mean\": 136.62541806020067, \"std\": 4.412477283909235, \"min\": 113.0, \"25%\": 134.0, \"50%\": 137.0, \"75%\": 140.0, \"max\": 148.0}, \"sex\": {\"count\": 299.0, \"mean\": 0.6488294314381271, \"std\": 0.47813637906274475, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"smoking\": {\"count\": 299.0, \"mean\": 0.3210702341137124, \"std\": 0.46767042805677167, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"time\": {\"count\": 299.0, \"mean\": 130.2608695652174, \"std\": 77.61420795029339, \"min\": 4.0, \"25%\": 73.0, \"50%\": 115.0, \"75%\": 203.0, \"max\": 285.0}, \"DEATH_EVENT\": {\"count\": 299.0, \"mean\": 0.3210702341137124, \"std\": 0.46767042805677167, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}}", "examples": "{\"age\":{\"0\":75.0,\"1\":55.0,\"2\":65.0,\"3\":50.0},\"anaemia\":{\"0\":0,\"1\":0,\"2\":0,\"3\":1},\"creatinine_phosphokinase\":{\"0\":582,\"1\":7861,\"2\":146,\"3\":111},\"diabetes\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"ejection_fraction\":{\"0\":20,\"1\":38,\"2\":20,\"3\":20},\"high_blood_pressure\":{\"0\":1,\"1\":0,\"2\":0,\"3\":0},\"platelets\":{\"0\":265000.0,\"1\":263358.03,\"2\":162000.0,\"3\":210000.0},\"serum_creatinine\":{\"0\":1.9,\"1\":1.1,\"2\":1.3,\"3\":1.9},\"serum_sodium\":{\"0\":130,\"1\":136,\"2\":129,\"3\":137},\"sex\":{\"0\":1,\"1\":1,\"2\":1,\"3\":1},\"smoking\":{\"0\":0,\"1\":0,\"2\":1,\"3\":0},\"time\":{\"0\":4,\"1\":6,\"2\":7,\"3\":7},\"DEATH_EVENT\":{\"0\":1,\"1\":1,\"2\":1,\"3\":1}}"}}]
| true | 1 |
<start_data_description><data_path>heart-failure-clinical-data/heart_failure_clinical_records_dataset.csv:
<column_names>
['age', 'anaemia', 'creatinine_phosphokinase', 'diabetes', 'ejection_fraction', 'high_blood_pressure', 'platelets', 'serum_creatinine', 'serum_sodium', 'sex', 'smoking', 'time', 'DEATH_EVENT']
<column_types>
{'age': 'float64', 'anaemia': 'int64', 'creatinine_phosphokinase': 'int64', 'diabetes': 'int64', 'ejection_fraction': 'int64', 'high_blood_pressure': 'int64', 'platelets': 'float64', 'serum_creatinine': 'float64', 'serum_sodium': 'int64', 'sex': 'int64', 'smoking': 'int64', 'time': 'int64', 'DEATH_EVENT': 'int64'}
<dataframe_Summary>
{'age': {'count': 299.0, 'mean': 60.83389297658862, 'std': 11.89480907404447, 'min': 40.0, '25%': 51.0, '50%': 60.0, '75%': 70.0, 'max': 95.0}, 'anaemia': {'count': 299.0, 'mean': 0.431438127090301, 'std': 0.4961072681330793, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'creatinine_phosphokinase': {'count': 299.0, 'mean': 581.8394648829432, 'std': 970.2878807124362, 'min': 23.0, '25%': 116.5, '50%': 250.0, '75%': 582.0, 'max': 7861.0}, 'diabetes': {'count': 299.0, 'mean': 0.4180602006688963, 'std': 0.49406706510360904, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'ejection_fraction': {'count': 299.0, 'mean': 38.08361204013378, 'std': 11.834840741039171, 'min': 14.0, '25%': 30.0, '50%': 38.0, '75%': 45.0, 'max': 80.0}, 'high_blood_pressure': {'count': 299.0, 'mean': 0.3511705685618729, 'std': 0.47813637906274475, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'platelets': {'count': 299.0, 'mean': 263358.02926421404, 'std': 97804.2368685983, 'min': 25100.0, '25%': 212500.0, '50%': 262000.0, '75%': 303500.0, 'max': 850000.0}, 'serum_creatinine': {'count': 299.0, 'mean': 1.3938795986622072, 'std': 1.0345100640898541, 'min': 0.5, '25%': 0.9, '50%': 1.1, '75%': 1.4, 'max': 9.4}, 'serum_sodium': {'count': 299.0, 'mean': 136.62541806020067, 'std': 4.412477283909235, 'min': 113.0, '25%': 134.0, '50%': 137.0, '75%': 140.0, 'max': 148.0}, 'sex': {'count': 299.0, 'mean': 0.6488294314381271, 'std': 0.47813637906274475, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'smoking': {'count': 299.0, 'mean': 0.3210702341137124, 'std': 0.46767042805677167, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'time': {'count': 299.0, 'mean': 130.2608695652174, 'std': 77.61420795029339, 'min': 4.0, '25%': 73.0, '50%': 115.0, '75%': 203.0, 'max': 285.0}, 'DEATH_EVENT': {'count': 299.0, 'mean': 0.3210702341137124, 'std': 0.46767042805677167, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 299 entries, 0 to 298
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 299 non-null float64
1 anaemia 299 non-null int64
2 creatinine_phosphokinase 299 non-null int64
3 diabetes 299 non-null int64
4 ejection_fraction 299 non-null int64
5 high_blood_pressure 299 non-null int64
6 platelets 299 non-null float64
7 serum_creatinine 299 non-null float64
8 serum_sodium 299 non-null int64
9 sex 299 non-null int64
10 smoking 299 non-null int64
11 time 299 non-null int64
12 DEATH_EVENT 299 non-null int64
dtypes: float64(3), int64(10)
memory usage: 30.5 KB
<some_examples>
{'age': {'0': 75.0, '1': 55.0, '2': 65.0, '3': 50.0}, 'anaemia': {'0': 0, '1': 0, '2': 0, '3': 1}, 'creatinine_phosphokinase': {'0': 582, '1': 7861, '2': 146, '3': 111}, 'diabetes': {'0': 0, '1': 0, '2': 0, '3': 0}, 'ejection_fraction': {'0': 20, '1': 38, '2': 20, '3': 20}, 'high_blood_pressure': {'0': 1, '1': 0, '2': 0, '3': 0}, 'platelets': {'0': 265000.0, '1': 263358.03, '2': 162000.0, '3': 210000.0}, 'serum_creatinine': {'0': 1.9, '1': 1.1, '2': 1.3, '3': 1.9}, 'serum_sodium': {'0': 130, '1': 136, '2': 129, '3': 137}, 'sex': {'0': 1, '1': 1, '2': 1, '3': 1}, 'smoking': {'0': 0, '1': 0, '2': 1, '3': 0}, 'time': {'0': 4, '1': 6, '2': 7, '3': 7}, 'DEATH_EVENT': {'0': 1, '1': 1, '2': 1, '3': 1}}
<end_description>
| 1,063 | 0 | 2,699 | 1,063 |
129661007
|
<jupyter_start><jupyter_text>Marijuana Arrests in Toronto: Racial Disparities
```
Data on police treatment of individuals arrested in Toronto for simple possession of small quantities of marijuana. The data are part of a larger data set featured in a series of articles in the Toronto Star newspaper. A data frame with 5226 observations on the following 8 variables.
```
| Column | Description |
| --- | --- |
| released | Whether or not the arrestee was released with a summons; a factor with levels: No; Yes.
|
| colour | The arrestee's race; a factor with levels: Black; White. |
| year | 1997 through 2002; a numeric vector. |
| age | in years; a numeric vector. |
| sex | a factor with levels: Female; Male. |
| employed | a factor with levels: No; Yes. |
| citizen | a factor with levels: No; Yes. |
| checks | Number of police data bases (of previous arrests, previous convictions, parole status, etc. – 6 in all) on which the arrestee's name appeared; a numeric vector |
# Source
Personal communication from Michael Friendly, York University.
Kaggle dataset identifier: arrests-for-marijuana-possession
<jupyter_script>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
df = pd.read_csv("/kaggle/input/arrests-for-marijuana-possession/Arrests.csv")
df.head(5)
df = df.iloc[:, 1:]
# # Age feature is positively skewed
# Older people tend to deal with drugs less
sns.histplot(df, x="age", kde=True, color="g")
def plots(df, x, y):
f, ax = plt.subplots(1, 3, figsize=(25, 10))
Group_data = df.groupby(y)
sns.histplot(df, x=x, hue=y, ax=ax[0], kde=True)
sns.barplot(
x=Group_data[x].mean().index,
y=Group_data[x].mean().values,
ax=ax[1],
palette="mako",
)
for container in ax[1].containers:
ax[1].bar_label(container, color="black", size=20)
palette_color = sns.color_palette("summer")
plt.pie(
x=df[y].value_counts(),
labels=df[y].value_counts().index,
autopct="%.0f%%",
shadow=True,
colors=palette_color,
)
plt.suptitle(
"{} histogram and barplots grouped by {}\n{} pie chart".format(
x, y, y
).capitalize()
)
ax[0].set_title("Data distribution of {} grouped by labelled by {}".format(x, y))
ax[1].set_title(
"Bar plots, showing mean values for {} for each category of {}".format(x, y)
)
ax[2].set_title(
"Pie chart showing ratio between categories for {} feature".format(x, y)
)
plt.show()
# # Histograms, barplots and pie charts
# Histograms - for data distribution for each category
# Bar plots - mean value of age for each category
# Pie charts - ratio between each of categoric values
for i in ["released", "colour", "year", "employed", "citizen", "checks"]:
plots(df, "age", i)
# # Mean age of individuals arrested for the possession of marijuana each year
grouped = df.groupby("year")
mean_age = grouped["age"].mean()
years = mean_age.index
fig = px.line(x=years, y=mean_age, title="Year vs Age")
fig.update_layout(
xaxis_title="Year labels", yaxis_title="Mean values for each category in Years"
)
fig.show()
# # Mean age of people arrested for marijuana for each amount of checks
grouped = df.groupby("checks")
mean_age = grouped["age"].mean()
checks = mean_age.index
fig = px.line(x=checks, y=mean_age, title="Checks vs Age line")
fig.update_layout(
xaxis_title="Checks categories", yaxis_title="Mean values of Age for each category"
)
fig.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/661/129661007.ipynb
|
arrests-for-marijuana-possession
|
utkarshx27
|
[{"Id": 129661007, "ScriptId": 38316314, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11036701, "CreationDate": "05/15/2023 14:51:07", "VersionNumber": 1.0, "Title": "Marijuana Arrests EDA", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 66.0, "LinesInsertedFromPrevious": 66.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 185964771, "KernelVersionId": 129661007, "SourceDatasetVersionId": 5631796}]
|
[{"Id": 5631796, "DatasetId": 3238325, "DatasourceVersionId": 5707058, "CreatorUserId": 13364933, "LicenseName": "CC0: Public Domain", "CreationDate": "05/08/2023 10:17:21", "VersionNumber": 1.0, "Title": "Marijuana Arrests in Toronto: Racial Disparities", "Slug": "arrests-for-marijuana-possession", "Subtitle": "Marijuana Arrests in Toronto: Race, Release, and Policing (1997-2002)", "Description": "``` \nData on police treatment of individuals arrested in Toronto for simple possession of small quantities of marijuana. The data are part of a larger data set featured in a series of articles in the Toronto Star newspaper. A data frame with 5226 observations on the following 8 variables.\n```\n| Column | Description |\n| --- | --- |\n| released | Whether or not the arrestee was released with a summons; a factor with levels: No; Yes.\n |\n| colour | The arrestee's race; a factor with levels: Black; White. |\n| year | 1997 through 2002; a numeric vector. |\n| age | in years; a numeric vector. |\n| sex | a factor with levels: Female; Male. |\n| employed | a factor with levels: No; Yes. |\n| citizen | a factor with levels: No; Yes. |\n| checks | Number of police data bases (of previous arrests, previous convictions, parole status, etc. \u2013 6 in all) on which the arrestee's name appeared; a numeric vector |\n\n# Source\nPersonal communication from Michael Friendly, York University.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3238325, "CreatorUserId": 13364933, "OwnerUserId": 13364933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5631796.0, "CurrentDatasourceVersionId": 5707058.0, "ForumId": 3303517, "Type": 2, "CreationDate": "05/08/2023 10:17:21", "LastActivityDate": "05/08/2023", "TotalViews": 8788, "TotalDownloads": 1614, "TotalVotes": 49, "TotalKernels": 14}]
|
[{"Id": 13364933, "UserName": "utkarshx27", "DisplayName": "Utkarsh Singh", "RegisterDate": "01/21/2023", "PerformanceTier": 2}]
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
df = pd.read_csv("/kaggle/input/arrests-for-marijuana-possession/Arrests.csv")
df.head(5)
df = df.iloc[:, 1:]
# # Age feature is positively skewed
# Older people tend to deal with drugs less
sns.histplot(df, x="age", kde=True, color="g")
def plots(df, x, y):
f, ax = plt.subplots(1, 3, figsize=(25, 10))
Group_data = df.groupby(y)
sns.histplot(df, x=x, hue=y, ax=ax[0], kde=True)
sns.barplot(
x=Group_data[x].mean().index,
y=Group_data[x].mean().values,
ax=ax[1],
palette="mako",
)
for container in ax[1].containers:
ax[1].bar_label(container, color="black", size=20)
palette_color = sns.color_palette("summer")
plt.pie(
x=df[y].value_counts(),
labels=df[y].value_counts().index,
autopct="%.0f%%",
shadow=True,
colors=palette_color,
)
plt.suptitle(
"{} histogram and barplots grouped by {}\n{} pie chart".format(
x, y, y
).capitalize()
)
ax[0].set_title("Data distribution of {} grouped by labelled by {}".format(x, y))
ax[1].set_title(
"Bar plots, showing mean values for {} for each category of {}".format(x, y)
)
ax[2].set_title(
"Pie chart showing ratio between categories for {} feature".format(x, y)
)
plt.show()
# # Histograms, barplots and pie charts
# Histograms - for data distribution for each category
# Bar plots - mean value of age for each category
# Pie charts - ratio between each of categoric values
for i in ["released", "colour", "year", "employed", "citizen", "checks"]:
plots(df, "age", i)
# # Mean age of individuals arrested for the possession of marijuana each year
grouped = df.groupby("year")
mean_age = grouped["age"].mean()
years = mean_age.index
fig = px.line(x=years, y=mean_age, title="Year vs Age")
fig.update_layout(
xaxis_title="Year labels", yaxis_title="Mean values for each category in Years"
)
fig.show()
# # Mean age of people arrested for marijuana for each amount of checks
grouped = df.groupby("checks")
mean_age = grouped["age"].mean()
checks = mean_age.index
fig = px.line(x=checks, y=mean_age, title="Checks vs Age line")
fig.update_layout(
xaxis_title="Checks categories", yaxis_title="Mean values of Age for each category"
)
fig.show()
| false | 1 | 768 | 2 | 1,092 | 768 |
||
129782661
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from darts.datasets import AirPassengersDataset, MonthlyMilkDataset
import pandas as pd
MonthlyMilkDataset().load().pd_dataframe()
AirPassengersDataset().load().pd_series()
AirPassengersDataset().load().pd_dataframe()
import matplotlib.pyplot as plt
series_ar = AirPassengersDataset().load()
series_milk = MonthlyMilkDataset().load()
series_ar.plot(label="no of passengers")
series_milk.plot(label="numbers of milks")
from darts.dataprocessing.transformers import Scaler
scaler_air, scaler_milk = Scaler(), Scaler()
series_air_scaled = scaler_air.fit_transform(series_ar)
series_milk_scaled = scaler_milk.fit_transform(series_milk)
series_air_scaled.plot(label="air")
series_milk_scaled.plot(label="milk")
train_air, val_air = series_air_scaled[:-36], series_air_scaled[-36:]
train_milk, val_milk = series_milk_scaled[:-36], series_milk_scaled[-36:]
from darts import TimeSeries
from darts.utils.timeseries_generation import (
gaussian_timeseries,
linear_timeseries,
sine_timeseries,
)
from darts.models import (
RNNModel,
TCNModel,
TransformerModel,
NBEATSModel,
BlockRNNModel,
)
from darts.metrics import mape, smape
model_air_milk = NBEATSModel(
input_chunk_length=24, output_chunk_length=12, n_epochs=100, random_state=0
)
model_air_milk.fit([train_air, train_milk], verbose=True)
pred = model_air_milk.predict(n=36, series=val_air)
pred = model_air_milk.predict(n=36, series=val_air)
series_air_scaled.plot(label="actual")
pred.plot(label="forecast")
plt.legend()
print("MAPE = {:.2f}%".format(mape(series_air_scaled, pred)))
pred = model_air_milk.predict(n=36, series=train_milk)
series_milk_scaled.plot(label="actual")
pred.plot(label="forecast")
plt.legend()
print("MAPE = {:.2f}%".format(mape(series_milk_scaled, pred)))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/782/129782661.ipynb
| null | null |
[{"Id": 129782661, "ScriptId": 29846616, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9224136, "CreationDate": "05/16/2023 12:38:27", "VersionNumber": 1.0, "Title": "Time Series Made Easy in Python USing Darts Librar", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 74.0, "LinesInsertedFromPrevious": 74.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from darts.datasets import AirPassengersDataset, MonthlyMilkDataset
import pandas as pd
MonthlyMilkDataset().load().pd_dataframe()
AirPassengersDataset().load().pd_series()
AirPassengersDataset().load().pd_dataframe()
import matplotlib.pyplot as plt
series_ar = AirPassengersDataset().load()
series_milk = MonthlyMilkDataset().load()
series_ar.plot(label="no of passengers")
series_milk.plot(label="numbers of milks")
from darts.dataprocessing.transformers import Scaler
scaler_air, scaler_milk = Scaler(), Scaler()
series_air_scaled = scaler_air.fit_transform(series_ar)
series_milk_scaled = scaler_milk.fit_transform(series_milk)
series_air_scaled.plot(label="air")
series_milk_scaled.plot(label="milk")
train_air, val_air = series_air_scaled[:-36], series_air_scaled[-36:]
train_milk, val_milk = series_milk_scaled[:-36], series_milk_scaled[-36:]
from darts import TimeSeries
from darts.utils.timeseries_generation import (
gaussian_timeseries,
linear_timeseries,
sine_timeseries,
)
from darts.models import (
RNNModel,
TCNModel,
TransformerModel,
NBEATSModel,
BlockRNNModel,
)
from darts.metrics import mape, smape
model_air_milk = NBEATSModel(
input_chunk_length=24, output_chunk_length=12, n_epochs=100, random_state=0
)
model_air_milk.fit([train_air, train_milk], verbose=True)
pred = model_air_milk.predict(n=36, series=val_air)
pred = model_air_milk.predict(n=36, series=val_air)
series_air_scaled.plot(label="actual")
pred.plot(label="forecast")
plt.legend()
print("MAPE = {:.2f}%".format(mape(series_air_scaled, pred)))
pred = model_air_milk.predict(n=36, series=train_milk)
series_milk_scaled.plot(label="actual")
pred.plot(label="forecast")
plt.legend()
print("MAPE = {:.2f}%".format(mape(series_milk_scaled, pred)))
| false | 0 | 793 | 0 | 793 | 793 |
||
129782855
|
<jupyter_start><jupyter_text>Data Science Job Salaries
### Content
| Column | Description |
|--------------------|------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|
| work_year | The year the salary was paid. |
| experience_level | The experience level in the job during the year with the following possible values: EN Entry-level / Junior MI Mid-level / Intermediate SE Senior-level / Expert EX Executive-level / Director |
| employment_type | The type of employement for the role: PT Part-time FT Full-time CT Contract FL Freelance |
| job_title | The role worked in during the year. |
| salary | The total gross salary amount paid. |
| salary_currency | The currency of the salary paid as an ISO 4217 currency code. |
| salary_in_usd | The salary in USD (FX rate divided by avg. USD rate for the respective year via fxdata.foorilla.com). |
| employee_residence | Employee's primary country of residence in during the work year as an ISO 3166 country code. |
| remote_ratio | The overall amount of work done remotely, possible values are as follows: 0 No remote work (less than 20%) 50 Partially remote 100 Fully remote (more than 80%) |
| company_location | The country of the employer's main office or contracting branch as an ISO 3166 country code. |
| company_size | The average number of people that worked for the company during the year: S less than 50 employees (small) M 50 to 250 employees (medium) L more than 250 employees (large) |
Kaggle dataset identifier: data-science-job-salaries
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as stats
import statsmodels.api as sm
from statsmodels.formula.api import ols
df = pd.read_csv("../input/data-science-job-salaries/ds_salaries.csv", index_col=0)
df.head()
df.shape
df["salary_in_usd"] = (df["salary_in_usd"] / 12).astype("int32")
df.head()
df.isna().sum()
df.describe()
df.head()
df["remote_ratio"].value_counts()
df["job_title"].unique()
df["job_title"] = df["job_title"].replace(
"Finance Data Analyst", "Financial Data Analyst"
)
analyst = df[df["job_title"].str.contains("Data Analyst")]
analyst["job_title"].value_counts()
analyst.describe()
analyst_remote = analyst["remote_ratio"].value_counts()
analyst_remote = analyst["remote_ratio"].value_counts()
labels = ["home-office", "hybrid", "on-site"]
pie_remote = analyst_remote.plot.pie(
labels=labels, colors=sns.color_palette("muted"), autopct="%1.1f%%", figsize=(8, 8)
)
plt.ylabel("")
plt.title("Distribuition by ratio of remote work", fontsize=20)
plt.show()
sns.barplot(x="work_year", y="remote_ratio", data=analyst)
plt.ylabel("Remote Ratio of Work")
plt.xlabel("Work Year")
plt.title("Distribuition by ratio of remote work and Year", fontsize=15)
df.employment_type.value_counts()
my_data = df[df["job_title"].str.contains("Machine Learning")]
my_data.job_title.value_counts()
analyst_remote = my_data["remote_ratio"].value_counts()
labels = ["home-office", "hybrid", "on-site"]
pie_remote = analyst_remote.plot.pie(
labels=labels, colors=sns.color_palette("muted"), autopct="%1.1f%%", figsize=(8, 8)
)
plt.ylabel("")
plt.title("Distribuition by ratio of remote work", fontsize=20)
plt.show()
df.experience_level.unique()
plt.figure(figsize=(12, 10))
sns.catplot(x="experience_level", data=df, kind="count", palette="magma")
plt.show()
levels = df.experience_level.value_counts()
levels
explode = [0, 0.1, 0.1, 0.3]
plt.pie(x=levels.values, labels=levels.index, autopct="%1.2f%%", explode=explode)
plt.title("Experience Level")
plt.legend()
plt.show()
levels
min, max = analyst.salary_in_usd.quantile([0.15, 0.985])
analyst_n_out = analyst[(analyst.salary_in_usd > min) & (analyst.salary_in_usd < max)]
sns.barplot(x="work_year", y="salary_in_usd", data=analyst_n_out)
plt.ylabel("Monthly Salaray in USD")
plt.xlabel("Work Year")
plt.title("Mean of Monthly Salary by Year", fontsize=15)
sns.histplot(analyst_n_out["salary_in_usd"])
plt.title("Distribuition by Mean Monthly Salary in USD", fontsize=15)
plt.xlabel("Monthly Salaray in USD")
analyst_n_out
ax = sns.barplot(x="job_title", y="salary_in_usd", data=analyst_n_out)
ax.set_xticklabels(ax.get_xticklabels(), rotation=30, ha="right")
plt.ylabel("Monthly Salaray in USD")
plt.xlabel("Job Title")
plt.title("Mean Monthly Salary in USD by Job Title", fontsize=15)
ax = sns.barplot(x="employment_type", y="remote_ratio", data=analyst_n_out)
ax.set_xticklabels(ax.get_xticklabels(), rotation=30, ha="right")
plt.ylabel("Monthly Salaray in USD")
plt.xlabel("Job Title")
plt.title("Mean Monthly Salary in USD by Job Title", fontsize=15)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/782/129782855.ipynb
|
data-science-job-salaries
|
ruchi798
|
[{"Id": 129782855, "ScriptId": 29246302, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9224136, "CreationDate": "05/16/2023 12:39:53", "VersionNumber": 1.0, "Title": "Data Science Job Salaries", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 139.0, "LinesInsertedFromPrevious": 139.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186147695, "KernelVersionId": 129782855, "SourceDatasetVersionId": 3806098}]
|
[{"Id": 3806098, "DatasetId": 2268489, "DatasourceVersionId": 3860816, "CreatorUserId": 3309826, "LicenseName": "CC0: Public Domain", "CreationDate": "06/15/2022 08:59:12", "VersionNumber": 1.0, "Title": "Data Science Job Salaries", "Slug": "data-science-job-salaries", "Subtitle": "Salaries of jobs in the Data Science domain", "Description": "### Content\n| Column | Description |\n|--------------------|------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|\n| work_year | The year the salary was paid. |\n| experience_level | The experience level in the job during the year with the following possible values: EN Entry-level / Junior MI Mid-level / Intermediate SE Senior-level / Expert EX Executive-level / Director |\n| employment_type | The type of employement for the role: PT Part-time FT Full-time CT Contract FL Freelance |\n| job_title | The role worked in during the year. |\n| salary | The total gross salary amount paid. |\n| salary_currency | The currency of the salary paid as an ISO 4217 currency code. |\n| salary_in_usd | The salary in USD (FX rate divided by avg. USD rate for the respective year via fxdata.foorilla.com). |\n| employee_residence | Employee's primary country of residence in during the work year as an ISO 3166 country code. |\n| remote_ratio | The overall amount of work done remotely, possible values are as follows: 0 No remote work (less than 20%) 50 Partially remote 100 Fully remote (more than 80%) |\n| company_location | The country of the employer's main office or contracting branch as an ISO 3166 country code. |\n| company_size | The average number of people that worked for the company during the year: S less than 50 employees (small) M 50 to 250 employees (medium) L more than 250 employees (large) |\n\n### Acknowledgements\nI'd like to thank ai-jobs.net Salaries for aggregating this data!", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2268489, "CreatorUserId": 3309826, "OwnerUserId": 3309826.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3806098.0, "CurrentDatasourceVersionId": 3860816.0, "ForumId": 2294990, "Type": 2, "CreationDate": "06/15/2022 08:59:12", "LastActivityDate": "06/15/2022", "TotalViews": 338940, "TotalDownloads": 59962, "TotalVotes": 1421, "TotalKernels": 360}]
|
[{"Id": 3309826, "UserName": "ruchi798", "DisplayName": "Ruchi Bhatia", "RegisterDate": "06/04/2019", "PerformanceTier": 4}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as stats
import statsmodels.api as sm
from statsmodels.formula.api import ols
df = pd.read_csv("../input/data-science-job-salaries/ds_salaries.csv", index_col=0)
df.head()
df.shape
df["salary_in_usd"] = (df["salary_in_usd"] / 12).astype("int32")
df.head()
df.isna().sum()
df.describe()
df.head()
df["remote_ratio"].value_counts()
df["job_title"].unique()
df["job_title"] = df["job_title"].replace(
"Finance Data Analyst", "Financial Data Analyst"
)
analyst = df[df["job_title"].str.contains("Data Analyst")]
analyst["job_title"].value_counts()
analyst.describe()
analyst_remote = analyst["remote_ratio"].value_counts()
analyst_remote = analyst["remote_ratio"].value_counts()
labels = ["home-office", "hybrid", "on-site"]
pie_remote = analyst_remote.plot.pie(
labels=labels, colors=sns.color_palette("muted"), autopct="%1.1f%%", figsize=(8, 8)
)
plt.ylabel("")
plt.title("Distribuition by ratio of remote work", fontsize=20)
plt.show()
sns.barplot(x="work_year", y="remote_ratio", data=analyst)
plt.ylabel("Remote Ratio of Work")
plt.xlabel("Work Year")
plt.title("Distribuition by ratio of remote work and Year", fontsize=15)
df.employment_type.value_counts()
my_data = df[df["job_title"].str.contains("Machine Learning")]
my_data.job_title.value_counts()
analyst_remote = my_data["remote_ratio"].value_counts()
labels = ["home-office", "hybrid", "on-site"]
pie_remote = analyst_remote.plot.pie(
labels=labels, colors=sns.color_palette("muted"), autopct="%1.1f%%", figsize=(8, 8)
)
plt.ylabel("")
plt.title("Distribuition by ratio of remote work", fontsize=20)
plt.show()
df.experience_level.unique()
plt.figure(figsize=(12, 10))
sns.catplot(x="experience_level", data=df, kind="count", palette="magma")
plt.show()
levels = df.experience_level.value_counts()
levels
explode = [0, 0.1, 0.1, 0.3]
plt.pie(x=levels.values, labels=levels.index, autopct="%1.2f%%", explode=explode)
plt.title("Experience Level")
plt.legend()
plt.show()
levels
min, max = analyst.salary_in_usd.quantile([0.15, 0.985])
analyst_n_out = analyst[(analyst.salary_in_usd > min) & (analyst.salary_in_usd < max)]
sns.barplot(x="work_year", y="salary_in_usd", data=analyst_n_out)
plt.ylabel("Monthly Salaray in USD")
plt.xlabel("Work Year")
plt.title("Mean of Monthly Salary by Year", fontsize=15)
sns.histplot(analyst_n_out["salary_in_usd"])
plt.title("Distribuition by Mean Monthly Salary in USD", fontsize=15)
plt.xlabel("Monthly Salaray in USD")
analyst_n_out
ax = sns.barplot(x="job_title", y="salary_in_usd", data=analyst_n_out)
ax.set_xticklabels(ax.get_xticklabels(), rotation=30, ha="right")
plt.ylabel("Monthly Salaray in USD")
plt.xlabel("Job Title")
plt.title("Mean Monthly Salary in USD by Job Title", fontsize=15)
ax = sns.barplot(x="employment_type", y="remote_ratio", data=analyst_n_out)
ax.set_xticklabels(ax.get_xticklabels(), rotation=30, ha="right")
plt.ylabel("Monthly Salaray in USD")
plt.xlabel("Job Title")
plt.title("Mean Monthly Salary in USD by Job Title", fontsize=15)
| false | 1 | 1,287 | 0 | 1,716 | 1,287 |
||
129550650
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
from lightgbm import LGBMRegressor
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
url = "/kaggle/input/demand-forecasting-kernels-only/train.csv"
df_train = pd.read_csv(url)
df_train["date"] = pd.to_datetime(df_train["date"])
df_train.head()
# load test set
url2 = "/kaggle/input/demand-forecasting-kernels-only/test.csv"
df_test = pd.read_csv(url2)
df_test["date"] = pd.to_datetime(df_test["date"])
df_test.head()
# Concatenate the training and testing dataframes
df_combined = pd.concat([df_train, df_test]).reset_index(drop=True)
# Display basic statistics
df_train.describe()
# Check for missing values
df_train.isnull().sum()
import plotly.express as px
# # Downsample the data by month and calculate the mean sales for each month
# df_downsampled = df_train.resample('M', on='date').mean()
# # Create a line plot using Plotly Express
# fig = px.line(df_downsampled, x=df_downsampled.index, y='sales', title='Sales Over Time')
# # Display the plot
# fig.show()
# # Sales by store
# fig = px.bar(df_train.groupby('store')['sales'].sum().reset_index(), x='store', y='sales', title='Sales by Store')
# fig.show()
# # Sales by item
# fig = px.bar(df_train.groupby('item')['sales'].sum().reset_index(), x='item', y='sales', title='Sales by Item')
# fig.show()
# # Seasonality check - Average sales by month
# df_train['month'] = df_train['date'].dt.month
# df_train['year'] = df_train['date'].dt.year
# fig = px.line(df_train.groupby(['year','month']).sales.mean().reset_index(), x='month', y='sales', color='year', title='Seasonality Check - Average Sales by Month')
# fig.show()
# # Seasonality check - Average sales by week
# df_train['month'] = df_train['date'].dt.month
# df_train['year'] = df_train['date'].dt.year
# df_train['week_of_year'] = df_train['date'].dt.weekofyear
# fig = px.line(df_train.groupby(['year','week_of_year']).sales.mean().reset_index(), x='week', y='sales', color='year', title='Seasonality Check - Average Sales by Week')
# fig.show()
# import plotly.graph_objs as go
# from statsmodels.graphics.tsaplots import plot_pacf
# from statsmodels.tsa.stattools import pacf
# import matplotlib.pyplot as plt
# # Calculate PACF
# # Group the data by store
# grouped_stores = df_train.groupby('store')
# # Plot PACF for each store
# for store, data in grouped_stores:
# plt.figure()
# # plt.title(f'Partial Autocorrelation for Store {store}')
# plot_pacf(data['sales'])
# plt.show()
# Calculate SMAPE
def smape(y_true, y_pred):
return (
100.0
/ len(y_true)
* np.sum(2 * np.abs(y_pred - y_true) / (np.abs(y_true) + np.abs(y_pred)))
)
import pandas as pd
import numpy as np
from lightgbm import LGBMRegressor
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
# feature engineering
# Create lagged features for each combination of store and item
# for i in range(1, 31):
# df_combined[f"lag_{i}"] = df_combined.groupby(["store", "item"])["sales"].shift(i)
# Feature Engineering - Categorical
df_combined["day_of_week"] = df_combined["date"].dt.dayofweek
df_combined["month"] = df_combined["date"].dt.month
df_combined["year"] = df_combined["date"].dt.year
# df['week'] = df['date'].dt.week
df_combined["day_of_year"] = df_combined["date"].dt.dayofyear
df_combined["week_of_year"] = df_combined["date"].dt.weekofyear
df_combined["sin_day_of_week"] = np.sin(2 * np.pi * df_combined["day_of_week"] / 7)
df_combined["cos_day_of_week"] = np.cos(2 * np.pi * df_combined["day_of_week"] / 7)
# Encode categorical features
le_item = LabelEncoder()
le_store = LabelEncoder()
df_combined["item"] = le_item.fit_transform(df_combined["item"])
df_combined["store"] = le_store.fit_transform(df_combined["store"])
# item_dummies = pd.get_dummies(df_combined['item'], prefix='item')
# df_combined = pd.concat([df_combined, item_dummies], axis=1)
# store_dummies = pd.get_dummies(df_combined['store'], prefix='store')
# df_combined = pd.concat([df_combined, store_dummies], axis=1)
# Create dummy variables for day_of_week
day_of_week_dummies = pd.get_dummies(df_combined["day_of_week"], prefix="day_of_week")
df_combined = pd.concat([df_combined, day_of_week_dummies], axis=1)
# create a new dataframe to hold the dummy variables
# Create dummy variables for month
month_dummies = pd.get_dummies(df_combined["month"], prefix="month")
df_combined = pd.concat([df_combined, month_dummies], axis=1)
# Create dummy variables for year
year_dummies = pd.get_dummies(df_combined["year"], prefix="year")
df_combined = pd.concat([df_combined, year_dummies], axis=1)
# # Drop rows with NaN values
# df = df.dropna()
df_combined = df_combined.drop(
["month", "year", "day_of_year", "week_of_year", "day_of_week"], axis=1
)
# Separate your training and testing dataframes again
df_train = df_combined[df_combined["sales"].notna()]
df_test = df_combined[df_combined["sales"].isna()]
#
# print("SMAPE: ", smape(test["sales"].values, predictions))
column_list = df_combined.columns.tolist()
print(column_list)
df_train.dtypes
df_train = df_train.drop("id", axis=1)
df_train
df_train = df_train.dropna()
df_train
df_test.columns
# df_train = df_train.drop(['store','item'],axis = 1)
# df_train
from sklearn.model_selection import TimeSeriesSplit
from lightgbm import LGBMRegressor
# Number of splits
n_splits = 5
# Initialize TimeSeriesSplit
tscv = TimeSeriesSplit(n_splits=n_splits)
model = LGBMRegressor()
df_fc = df_train.copy()
smape_values = []
# Perform cross-validation
for train_index, test_index in tscv.split(df_train):
CV_train, CV_test = df_train.iloc[train_index], df_train.iloc[test_index]
# Fit the model on the training data
model.fit(CV_train.drop(["sales", "date"], axis=1), CV_train["sales"])
# Predict on the test data
predictions = model.predict(CV_test.drop(["sales", "date"], axis=1))
df_fc.loc[df_train.iloc[test_index].index, "predictions"] = predictions[0]
# Calculate SMAPE and add it to the list of SMAPE values
smape_value = smape(CV_test["sales"].values, predictions)
smape_values.append(smape_value)
# Print the average SMAPE value across all folds
print("Average SMAPE: ", np.mean(smape_values)), smape_values
# df_train
df1 = df_train.drop(["date", "sales"], axis=1)
df1
# df.columns
# Get feature importances
feature_importances = pd.DataFrame(
{"Feature": df1.columns, "Importance": model.feature_importances_}
)
feature_importances = feature_importances.sort_values("Importance", ascending=False)
print(feature_importances["Feature"][feature_importances["Importance"] > 0])
px.bar(
data_frame=pd.Series(model.feature_importances_, index=df1.columns).sort_values(),
orientation="h",
)
df_train = df_train[
[
"sales",
"store",
"item",
"sin_day_of_week",
"cos_day_of_week",
"month_7",
"year_2013",
"month_1",
"month_6",
"month_2",
"month_12",
"year_2014",
"month_3",
"month_10",
"year_2017",
"year_2016",
"year_2015",
"month_8",
"month_5",
"day_of_week_6",
"month_4",
"month_11",
"month_9",
"day_of_week_3",
"day_of_week_4",
"day_of_week_1",
]
]
df_train
from sklearn.model_selection import TimeSeriesSplit
from lightgbm import LGBMRegressor
# Number of splits
n_splits = 5
# Initialize TimeSeriesSplit
tscv = TimeSeriesSplit(n_splits=n_splits)
model = LGBMRegressor()
df_fc = df_train.copy()
smape_values = []
# Perform cross-validation
for train_index, test_index in tscv.split(df_train):
CV_train, CV_test = df_train.iloc[train_index], df_train.iloc[test_index]
# Fit the model on the training data
model.fit(CV_train.drop(["sales"], axis=1), CV_train["sales"])
# Predict on the test data
predictions = model.predict(CV_test.drop(["sales"], axis=1))
df_fc.loc[df_train.iloc[test_index].index, "predictions"] = predictions[0]
# Calculate SMAPE and add it to the list of SMAPE values
smape_value = smape(CV_test["sales"].values, predictions)
smape_values.append(smape_value)
# Print the average SMAPE value across all folds
print("Average SMAPE: ", np.mean(smape_values)), smape_values
# # Final model
df_test
# df_test = df_test.drop(['store','item'],axis = 1)
# df_test
df_test = df_test[
[
"store",
"item",
"sin_day_of_week",
"cos_day_of_week",
"month_7",
"year_2013",
"month_1",
"month_6",
"month_2",
"month_12",
"year_2014",
"month_3",
"month_10",
"year_2017",
"year_2016",
"year_2015",
"month_8",
"month_5",
"day_of_week_6",
"month_4",
"month_11",
"month_9",
"day_of_week_3",
"day_of_week_4",
"day_of_week_1",
]
]
df_test
predictions = []
# Create a separate DataFrame to store the lagged predictions
lagged_predictions = df_test.copy()
# Iterate over the test set
for i in range(len(df_test)):
# Prepare the data for the current day, including lagged features
data = lagged_predictions.iloc[i : i + 1].copy()
# Make a prediction for the current day
prediction = model.predict(data)
# Store the prediction
predictions.append(prediction[0])
# # If there are still more days to predict, update the necessary lagged features in the lagged_predictions DataFrame
# if i < len(df_test) - 1:
# for j in range(1, 31):
# if i + j < len(df_test):
# lagged_predictions.loc[i + j, f'lag_{j}'] = prediction[0]
# Convert the list of predictions to a DataFrame or series, if necessary
predictions = pd.Series(predictions)
predictions
# load test set
url2 = "/kaggle/input/demand-forecasting-kernels-only/test.csv"
df_test = pd.read_csv(url2)
df_test["date"] = pd.to_datetime(df_test["date"])
df_test.head()
# Add predictions to the test dataframe
df_test["predictions"] = predictions.values
df_test
submission_df = df_test[["id", "predictions"]]
submission_df
submission_df.rename(columns={"predictions": "sales"}, inplace=True)
submission_df
submission_df.to_csv("submission.csv", index=False)
# submission = (pd.DataFrame(Y_test, index=X_test.index, columns=Y.columns)
# .unstack()
# .reset_index()
# .sort_values(["item","store","date"])
# .drop(["item","store","date"], axis=1)
# .reset_index()
# .rename({0:"sales","index":"id"}, axis=1)
# .set_index("id")
# )
# submission.to_csv("submission.csv")
# submission
# # Prepare the submission data
# submission = pd.DataFrame({'id': test_data.id, 'sales': predictions})
# submission.to_csv("submission.csv", index=False)
# submission
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/550/129550650.ipynb
| null | null |
[{"Id": 129550650, "ScriptId": 38512674, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11964784, "CreationDate": "05/14/2023 18:44:07", "VersionNumber": 10.0, "Title": "notebook4d7d117c79", "EvaluationDate": "05/14/2023", "IsChange": false, "TotalLines": 347.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 347.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
from lightgbm import LGBMRegressor
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
url = "/kaggle/input/demand-forecasting-kernels-only/train.csv"
df_train = pd.read_csv(url)
df_train["date"] = pd.to_datetime(df_train["date"])
df_train.head()
# load test set
url2 = "/kaggle/input/demand-forecasting-kernels-only/test.csv"
df_test = pd.read_csv(url2)
df_test["date"] = pd.to_datetime(df_test["date"])
df_test.head()
# Concatenate the training and testing dataframes
df_combined = pd.concat([df_train, df_test]).reset_index(drop=True)
# Display basic statistics
df_train.describe()
# Check for missing values
df_train.isnull().sum()
import plotly.express as px
# # Downsample the data by month and calculate the mean sales for each month
# df_downsampled = df_train.resample('M', on='date').mean()
# # Create a line plot using Plotly Express
# fig = px.line(df_downsampled, x=df_downsampled.index, y='sales', title='Sales Over Time')
# # Display the plot
# fig.show()
# # Sales by store
# fig = px.bar(df_train.groupby('store')['sales'].sum().reset_index(), x='store', y='sales', title='Sales by Store')
# fig.show()
# # Sales by item
# fig = px.bar(df_train.groupby('item')['sales'].sum().reset_index(), x='item', y='sales', title='Sales by Item')
# fig.show()
# # Seasonality check - Average sales by month
# df_train['month'] = df_train['date'].dt.month
# df_train['year'] = df_train['date'].dt.year
# fig = px.line(df_train.groupby(['year','month']).sales.mean().reset_index(), x='month', y='sales', color='year', title='Seasonality Check - Average Sales by Month')
# fig.show()
# # Seasonality check - Average sales by week
# df_train['month'] = df_train['date'].dt.month
# df_train['year'] = df_train['date'].dt.year
# df_train['week_of_year'] = df_train['date'].dt.weekofyear
# fig = px.line(df_train.groupby(['year','week_of_year']).sales.mean().reset_index(), x='week', y='sales', color='year', title='Seasonality Check - Average Sales by Week')
# fig.show()
# import plotly.graph_objs as go
# from statsmodels.graphics.tsaplots import plot_pacf
# from statsmodels.tsa.stattools import pacf
# import matplotlib.pyplot as plt
# # Calculate PACF
# # Group the data by store
# grouped_stores = df_train.groupby('store')
# # Plot PACF for each store
# for store, data in grouped_stores:
# plt.figure()
# # plt.title(f'Partial Autocorrelation for Store {store}')
# plot_pacf(data['sales'])
# plt.show()
# Calculate SMAPE
def smape(y_true, y_pred):
return (
100.0
/ len(y_true)
* np.sum(2 * np.abs(y_pred - y_true) / (np.abs(y_true) + np.abs(y_pred)))
)
import pandas as pd
import numpy as np
from lightgbm import LGBMRegressor
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
# feature engineering
# Create lagged features for each combination of store and item
# for i in range(1, 31):
# df_combined[f"lag_{i}"] = df_combined.groupby(["store", "item"])["sales"].shift(i)
# Feature Engineering - Categorical
df_combined["day_of_week"] = df_combined["date"].dt.dayofweek
df_combined["month"] = df_combined["date"].dt.month
df_combined["year"] = df_combined["date"].dt.year
# df['week'] = df['date'].dt.week
df_combined["day_of_year"] = df_combined["date"].dt.dayofyear
df_combined["week_of_year"] = df_combined["date"].dt.weekofyear
df_combined["sin_day_of_week"] = np.sin(2 * np.pi * df_combined["day_of_week"] / 7)
df_combined["cos_day_of_week"] = np.cos(2 * np.pi * df_combined["day_of_week"] / 7)
# Encode categorical features
le_item = LabelEncoder()
le_store = LabelEncoder()
df_combined["item"] = le_item.fit_transform(df_combined["item"])
df_combined["store"] = le_store.fit_transform(df_combined["store"])
# item_dummies = pd.get_dummies(df_combined['item'], prefix='item')
# df_combined = pd.concat([df_combined, item_dummies], axis=1)
# store_dummies = pd.get_dummies(df_combined['store'], prefix='store')
# df_combined = pd.concat([df_combined, store_dummies], axis=1)
# Create dummy variables for day_of_week
day_of_week_dummies = pd.get_dummies(df_combined["day_of_week"], prefix="day_of_week")
df_combined = pd.concat([df_combined, day_of_week_dummies], axis=1)
# create a new dataframe to hold the dummy variables
# Create dummy variables for month
month_dummies = pd.get_dummies(df_combined["month"], prefix="month")
df_combined = pd.concat([df_combined, month_dummies], axis=1)
# Create dummy variables for year
year_dummies = pd.get_dummies(df_combined["year"], prefix="year")
df_combined = pd.concat([df_combined, year_dummies], axis=1)
# # Drop rows with NaN values
# df = df.dropna()
df_combined = df_combined.drop(
["month", "year", "day_of_year", "week_of_year", "day_of_week"], axis=1
)
# Separate your training and testing dataframes again
df_train = df_combined[df_combined["sales"].notna()]
df_test = df_combined[df_combined["sales"].isna()]
#
# print("SMAPE: ", smape(test["sales"].values, predictions))
column_list = df_combined.columns.tolist()
print(column_list)
df_train.dtypes
df_train = df_train.drop("id", axis=1)
df_train
df_train = df_train.dropna()
df_train
df_test.columns
# df_train = df_train.drop(['store','item'],axis = 1)
# df_train
from sklearn.model_selection import TimeSeriesSplit
from lightgbm import LGBMRegressor
# Number of splits
n_splits = 5
# Initialize TimeSeriesSplit
tscv = TimeSeriesSplit(n_splits=n_splits)
model = LGBMRegressor()
df_fc = df_train.copy()
smape_values = []
# Perform cross-validation
for train_index, test_index in tscv.split(df_train):
CV_train, CV_test = df_train.iloc[train_index], df_train.iloc[test_index]
# Fit the model on the training data
model.fit(CV_train.drop(["sales", "date"], axis=1), CV_train["sales"])
# Predict on the test data
predictions = model.predict(CV_test.drop(["sales", "date"], axis=1))
df_fc.loc[df_train.iloc[test_index].index, "predictions"] = predictions[0]
# Calculate SMAPE and add it to the list of SMAPE values
smape_value = smape(CV_test["sales"].values, predictions)
smape_values.append(smape_value)
# Print the average SMAPE value across all folds
print("Average SMAPE: ", np.mean(smape_values)), smape_values
# df_train
df1 = df_train.drop(["date", "sales"], axis=1)
df1
# df.columns
# Get feature importances
feature_importances = pd.DataFrame(
{"Feature": df1.columns, "Importance": model.feature_importances_}
)
feature_importances = feature_importances.sort_values("Importance", ascending=False)
print(feature_importances["Feature"][feature_importances["Importance"] > 0])
px.bar(
data_frame=pd.Series(model.feature_importances_, index=df1.columns).sort_values(),
orientation="h",
)
df_train = df_train[
[
"sales",
"store",
"item",
"sin_day_of_week",
"cos_day_of_week",
"month_7",
"year_2013",
"month_1",
"month_6",
"month_2",
"month_12",
"year_2014",
"month_3",
"month_10",
"year_2017",
"year_2016",
"year_2015",
"month_8",
"month_5",
"day_of_week_6",
"month_4",
"month_11",
"month_9",
"day_of_week_3",
"day_of_week_4",
"day_of_week_1",
]
]
df_train
from sklearn.model_selection import TimeSeriesSplit
from lightgbm import LGBMRegressor
# Number of splits
n_splits = 5
# Initialize TimeSeriesSplit
tscv = TimeSeriesSplit(n_splits=n_splits)
model = LGBMRegressor()
df_fc = df_train.copy()
smape_values = []
# Perform cross-validation
for train_index, test_index in tscv.split(df_train):
CV_train, CV_test = df_train.iloc[train_index], df_train.iloc[test_index]
# Fit the model on the training data
model.fit(CV_train.drop(["sales"], axis=1), CV_train["sales"])
# Predict on the test data
predictions = model.predict(CV_test.drop(["sales"], axis=1))
df_fc.loc[df_train.iloc[test_index].index, "predictions"] = predictions[0]
# Calculate SMAPE and add it to the list of SMAPE values
smape_value = smape(CV_test["sales"].values, predictions)
smape_values.append(smape_value)
# Print the average SMAPE value across all folds
print("Average SMAPE: ", np.mean(smape_values)), smape_values
# # Final model
df_test
# df_test = df_test.drop(['store','item'],axis = 1)
# df_test
df_test = df_test[
[
"store",
"item",
"sin_day_of_week",
"cos_day_of_week",
"month_7",
"year_2013",
"month_1",
"month_6",
"month_2",
"month_12",
"year_2014",
"month_3",
"month_10",
"year_2017",
"year_2016",
"year_2015",
"month_8",
"month_5",
"day_of_week_6",
"month_4",
"month_11",
"month_9",
"day_of_week_3",
"day_of_week_4",
"day_of_week_1",
]
]
df_test
predictions = []
# Create a separate DataFrame to store the lagged predictions
lagged_predictions = df_test.copy()
# Iterate over the test set
for i in range(len(df_test)):
# Prepare the data for the current day, including lagged features
data = lagged_predictions.iloc[i : i + 1].copy()
# Make a prediction for the current day
prediction = model.predict(data)
# Store the prediction
predictions.append(prediction[0])
# # If there are still more days to predict, update the necessary lagged features in the lagged_predictions DataFrame
# if i < len(df_test) - 1:
# for j in range(1, 31):
# if i + j < len(df_test):
# lagged_predictions.loc[i + j, f'lag_{j}'] = prediction[0]
# Convert the list of predictions to a DataFrame or series, if necessary
predictions = pd.Series(predictions)
predictions
# load test set
url2 = "/kaggle/input/demand-forecasting-kernels-only/test.csv"
df_test = pd.read_csv(url2)
df_test["date"] = pd.to_datetime(df_test["date"])
df_test.head()
# Add predictions to the test dataframe
df_test["predictions"] = predictions.values
df_test
submission_df = df_test[["id", "predictions"]]
submission_df
submission_df.rename(columns={"predictions": "sales"}, inplace=True)
submission_df
submission_df.to_csv("submission.csv", index=False)
# submission = (pd.DataFrame(Y_test, index=X_test.index, columns=Y.columns)
# .unstack()
# .reset_index()
# .sort_values(["item","store","date"])
# .drop(["item","store","date"], axis=1)
# .reset_index()
# .rename({0:"sales","index":"id"}, axis=1)
# .set_index("id")
# )
# submission.to_csv("submission.csv")
# submission
# # Prepare the submission data
# submission = pd.DataFrame({'id': test_data.id, 'sales': predictions})
# submission.to_csv("submission.csv", index=False)
# submission
| false | 0 | 3,761 | 0 | 3,761 | 3,761 |
||
129550705
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/train.csv.zip")
z.extractall()
df_train = pd.read_csv("/kaggle/working/train.csv")
df_train.head()
num_linhas = len(df_train)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Transforma as categorias em valores numericos
from sklearn.preprocessing import LabelEncoder
for f in df_train.columns:
if df_train[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_train[f].values))
df_train[f] = lbl.transform(list(df_train[f].values))
df_train.head()
# Aplica uma média para remover valores nulos.
for col in df_train.columns:
if df_train[col].isnull().sum() > 0:
mean = df_train[col].mean()
df_train[col] = df_train[col].fillna(mean)
df_train.head()
# Determina as colunas que serão utilizadas no treino do modelo e qual coluna será considerada Target.
X = df_train[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
y = np.log(df_train.price_doc)
# Separação do arquivo de teste em Treino e Teste
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# Normalização das partes
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
# Atribuição da regressão ao modelo
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso, Ridge, ElasticNet
modelo = ElasticNet()
modelo.fit(X_train, y_train)
# Determina os coeficientes do modelo
modelo.coef_, modelo.intercept_
# Demonstra os indicadores de performance do modelo
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_absolute_percentage_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_log_error
import numpy as np
y_pred = modelo.predict(X_train)
mae = mean_absolute_error(y_train, y_pred)
mape = mean_absolute_percentage_error(y_train, y_pred)
rmse = mean_squared_error(y_train, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_train, y_pred))
r2 = r2_score(y_train, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
print("")
y_pred = modelo.predict(X_test)
mae = mean_absolute_error(y_test, y_pred)
mape = mean_absolute_percentage_error(y_test, y_pred)
rmse = mean_squared_error(y_test, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_test, y_pred))
r2 = r2_score(y_test, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
# Realiza o upload do arquivo de Teste
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/test.csv.zip")
z.extractall()
df_test = pd.read_csv("/kaggle/working/test.csv")
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Transforma as categorias em valores numericos
from sklearn.preprocessing import LabelEncoder
for f in df_test.columns:
if df_test[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_test[f].values))
df_test[f] = lbl.transform(list(df_test[f].values))
import pandas as pd
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Aplica uma média para remover valores nulos.
for col in df_test.columns:
if df_test[col].isnull().sum() > 0:
mean = df_test[col].mean()
df_test[col] = df_test[col].fillna(mean)
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Determina as colunas que serão utilizadas para testar no modelo de previsão
X_test = df_test[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
# Utiliza as colunas selecionadas em X_test para fazer a previsão no modelo.
y_pred = modelo.predict(X_test)
# Aplica a função exponencial nos modelos previstos, ja que o modelo preve numeros logaritimos e não valores reais
y_pred = np.exp(y_pred)
# #Cria
# output = pd.DataFrame({'id': df_test.id, 'price_doc': y_pred})
# output.to_csv('submission.csv', index=False)
# print("Your submission was successfully saved!")
# output.head()
# Cria uma coluna utilizado os preços previstos pelo o metodo e salva
output = pd.DataFrame({"id": df_test.id, "price_doc": y_pred})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
output.head()
num_linhas = len(output)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/550/129550705.ipynb
| null | null |
[{"Id": 129550705, "ScriptId": 38519986, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14697649, "CreationDate": "05/14/2023 18:44:46", "VersionNumber": 2.0, "Title": "163066_Regressao_AC2", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 187.0, "LinesInsertedFromPrevious": 28.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 159.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/train.csv.zip")
z.extractall()
df_train = pd.read_csv("/kaggle/working/train.csv")
df_train.head()
num_linhas = len(df_train)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Transforma as categorias em valores numericos
from sklearn.preprocessing import LabelEncoder
for f in df_train.columns:
if df_train[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_train[f].values))
df_train[f] = lbl.transform(list(df_train[f].values))
df_train.head()
# Aplica uma média para remover valores nulos.
for col in df_train.columns:
if df_train[col].isnull().sum() > 0:
mean = df_train[col].mean()
df_train[col] = df_train[col].fillna(mean)
df_train.head()
# Determina as colunas que serão utilizadas no treino do modelo e qual coluna será considerada Target.
X = df_train[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
y = np.log(df_train.price_doc)
# Separação do arquivo de teste em Treino e Teste
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# Normalização das partes
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
# Atribuição da regressão ao modelo
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso, Ridge, ElasticNet
modelo = ElasticNet()
modelo.fit(X_train, y_train)
# Determina os coeficientes do modelo
modelo.coef_, modelo.intercept_
# Demonstra os indicadores de performance do modelo
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_absolute_percentage_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_log_error
import numpy as np
y_pred = modelo.predict(X_train)
mae = mean_absolute_error(y_train, y_pred)
mape = mean_absolute_percentage_error(y_train, y_pred)
rmse = mean_squared_error(y_train, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_train, y_pred))
r2 = r2_score(y_train, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
print("")
y_pred = modelo.predict(X_test)
mae = mean_absolute_error(y_test, y_pred)
mape = mean_absolute_percentage_error(y_test, y_pred)
rmse = mean_squared_error(y_test, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_test, y_pred))
r2 = r2_score(y_test, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
# Realiza o upload do arquivo de Teste
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/test.csv.zip")
z.extractall()
df_test = pd.read_csv("/kaggle/working/test.csv")
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Transforma as categorias em valores numericos
from sklearn.preprocessing import LabelEncoder
for f in df_test.columns:
if df_test[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_test[f].values))
df_test[f] = lbl.transform(list(df_test[f].values))
import pandas as pd
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Aplica uma média para remover valores nulos.
for col in df_test.columns:
if df_test[col].isnull().sum() > 0:
mean = df_test[col].mean()
df_test[col] = df_test[col].fillna(mean)
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Determina as colunas que serão utilizadas para testar no modelo de previsão
X_test = df_test[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
# Utiliza as colunas selecionadas em X_test para fazer a previsão no modelo.
y_pred = modelo.predict(X_test)
# Aplica a função exponencial nos modelos previstos, ja que o modelo preve numeros logaritimos e não valores reais
y_pred = np.exp(y_pred)
# #Cria
# output = pd.DataFrame({'id': df_test.id, 'price_doc': y_pred})
# output.to_csv('submission.csv', index=False)
# print("Your submission was successfully saved!")
# output.head()
# Cria uma coluna utilizado os preços previstos pelo o metodo e salva
output = pd.DataFrame({"id": df_test.id, "price_doc": y_pred})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
output.head()
num_linhas = len(output)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
| false | 0 | 1,840 | 0 | 1,840 | 1,840 |
||
129550253
|
# ## 2 Построение, обучение и оптимизация модели
# Импорт библиотек
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.svm import SVC
from sklearn.neural_network import MLPClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from tqdm import tqdm
from sklearn.model_selection import learning_curve
# Загрузка данных
df = pd.read_csv("articles.csv")
df = df.iloc[:, 1:]
df.head()
# У компьютера недостаточно оперативной памяти. Поэтому для обучение будет взята половина данных.
half = df.sample(frac=0.5)
half.reset_index(drop=True, inplace=True)
half.info()
# ### 2.1 Построение модели классификации
# метод для рассчета метрик качества модели классификации
def classification_metrics(y_test, y_pred):
acc = accuracy_score(y_test, y_pred)
prec = precision_score(y_test, y_pred, average="macro")
rec = recall_score(y_test, y_pred, average="macro")
f1 = f1_score(y_test, y_pred, average="macro")
return {"Accuracy": acc, "Precision": prec, "Recall": rec, "F1-score": f1}
# Для обучения будет использоваться целевая переменная и лемматизированный текст статьи
X = half["lematize_text"]
# векторизация текстового признака
from sklearn.feature_extraction.text import CountVectorizer
vectorizer = CountVectorizer()
X = vectorizer.fit_transform(X)
X = X.toarray()
y = half["nomination_encoded"]
# Разбиение выборки на тренировочную и тестовую
# 1/3 - это значит, что тестовая выборка будет иметь 0.33 данных
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42, shuffle=True
)
# GaussianNB - это наивный байесовский алгоритм, который основан на вероятностной модели, и он используется для решения задач классификации и регрессии.
nb = GaussianNB().fit(X_train, y_train)
model_preds = nb.predict(X_test)
accuracy = accuracy_score(y_test, model_preds)
print("Accuracy:", accuracy)
# DecisionTreeClassifier - это алгоритм, который используется для решения задач классификации и регрессии и основан на построении дерева решений на основе анализа данных и их параметров.
dtc = DecisionTreeClassifier().fit(X_train, y_train)
model_preds = dtc.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, model_preds).items():
print(f"{metric_name}: {metric_score:.2f}")
# RandomForestClassifier - это алгоритм машинного обучения, который основан на использовании ансамбля деревьев решений для решения задач классификации, кластеризации или регрессии.
rfc = RandomForestClassifier().fit(X_train, y_train)
model_preds = rfc.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, model_preds).items():
print(f"{metric_name}: {metric_score:.2f}")
# KNeighborsClassifier - это алгоритм машинного обучения, который основан на методе ближайших соседей (k-NN) для решения задач классификации и регрессии.
knn = KNeighborsClassifier(n_neighbors=5)
model_fit = knn.fit(X_train, y_train)
model_preds = model_fit.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, model_preds).items():
print(f"{metric_name}: {metric_score:.2f}")
# --------------------------------------------------------------------------
# GradientBoostingClassifier - это ансамблевый метод машинного обучения, который используется для решения задач классификации, основанный на построении последовательности деревьев решений, каждое из которых исправляет ошибки предыдущих моделей, минимизируя функцию потерь.
gbc = GradientBoostingClassifier(
n_estimators=100, learning_rate=0.1, max_depth=1, random_state=42
)
gbc.fit(X_train, y_train)
y_pred_gbc = gbc.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, y_pred_gbc).items():
print(f"{metric_name}: {metric_score:.2f}")
# GradientBoostingClassifier показал лучший результат, но из-за долгого времени обучения не рекомендуется к использованию на демоэкзамене.
# --------------------------------------------------------------------------
# SVC (Support Vector Classifier) - это алгоритм машинного обучения, который используется для решения задач классификации, основанный на поиске оптимальной разделяющей гиперплоскости в многомерном пространстве с максимальным зазором между классами.
svm = SVC(kernel="linear", C=1, random_state=42)
svm.fit(X_train, y_train)
y_pred_svm = svm.predict(X_test)
acc_svm = accuracy_score(y_test, y_pred_svm)
for metric_name, metric_score in classification_metrics(y_test, y_pred_svm).items():
print(f"{metric_name}: {metric_score:.2f}")
# MLPClassifier - это нейронная сеть прямого распространения, которая используется для решения задач классификации или регрессии и состоит из нескольких слоев скрытых нейронов, обучаемых с помощью метода обратного распространения ошибки.
nn = MLPClassifier(
hidden_layer_sizes=(10,),
activation="relu",
solver="adam",
alpha=0.0001,
max_iter=500,
random_state=42,
)
nn.fit(X_train, y_train)
y_pred_nn = nn.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, y_pred_nn).items():
print(f"{metric_name}: {metric_score:.2f}")
# Для последующей оптимизации выбрана модель классификации дерева решений DecisionTreeClassifier, так как эта модель показала наивысшее качество обучения.
# *Примечание. модель GradientBoostingClassifier не была выбрана из-за продолжительного обучения, несмотря на то, что качество лучше чем у модели дерева решений
# ## 2.2 Оптимизация модели
# Оптимизация модели будет проводиться путем поиска наилучших гиперпараметров
param_grid = {"max_depth": range(1, 6, 1), "min_samples_leaf": range(1, 6, 1)}
grid_search = GridSearchCV(dtc, param_grid, cv=5)
grid_search.fit(X_train, y_train)
print(grid_search.best_params_)
y_pred_gs = grid_search.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, y_pred_gs).items():
print(f"{metric_name}: {metric_score:.2f}")
# Построение кривых валидации и обучения
train_sizes, train_scores, test_scores = learning_curve(
dtc, X_train, y_train, cv=5, scoring="accuracy"
)
# Вычисляем среднюю точность и стандартное отклонение для каждого размера обучающей выборки
train_mean = np.mean(train_scores, axis=1)
train_std = np.std(train_scores, axis=1)
test_mean = np.mean(test_scores, axis=1)
test_std = np.std(test_scores, axis=1)
# Строим кривую обучения и кривую валидации
plt.plot(train_sizes, train_mean, label="Training Accuracy")
plt.plot(train_sizes, test_mean, label="Validation Accuracy")
# Добавляем круги для отображения стандартного отклонения
plt.fill_between(train_sizes, train_mean - train_std, train_mean + train_std, alpha=0.1)
plt.fill_between(train_sizes, test_mean - test_std, test_mean + test_std, alpha=0.1)
# Подписываем график и добавляем легенду
plt.xlabel("Number of Training Samples")
plt.ylabel("Accuracy Score")
plt.title("Learning Curve for DecisionTreeClassifier")
plt.legend(loc="best")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/550/129550253.ipynb
| null | null |
[{"Id": 129550253, "ScriptId": 38521932, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13417746, "CreationDate": "05/14/2023 18:39:30", "VersionNumber": 1.0, "Title": "Report2-VD-djostit", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 214.0, "LinesInsertedFromPrevious": 214.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## 2 Построение, обучение и оптимизация модели
# Импорт библиотек
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.svm import SVC
from sklearn.neural_network import MLPClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from tqdm import tqdm
from sklearn.model_selection import learning_curve
# Загрузка данных
df = pd.read_csv("articles.csv")
df = df.iloc[:, 1:]
df.head()
# У компьютера недостаточно оперативной памяти. Поэтому для обучение будет взята половина данных.
half = df.sample(frac=0.5)
half.reset_index(drop=True, inplace=True)
half.info()
# ### 2.1 Построение модели классификации
# метод для рассчета метрик качества модели классификации
def classification_metrics(y_test, y_pred):
acc = accuracy_score(y_test, y_pred)
prec = precision_score(y_test, y_pred, average="macro")
rec = recall_score(y_test, y_pred, average="macro")
f1 = f1_score(y_test, y_pred, average="macro")
return {"Accuracy": acc, "Precision": prec, "Recall": rec, "F1-score": f1}
# Для обучения будет использоваться целевая переменная и лемматизированный текст статьи
X = half["lematize_text"]
# векторизация текстового признака
from sklearn.feature_extraction.text import CountVectorizer
vectorizer = CountVectorizer()
X = vectorizer.fit_transform(X)
X = X.toarray()
y = half["nomination_encoded"]
# Разбиение выборки на тренировочную и тестовую
# 1/3 - это значит, что тестовая выборка будет иметь 0.33 данных
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42, shuffle=True
)
# GaussianNB - это наивный байесовский алгоритм, который основан на вероятностной модели, и он используется для решения задач классификации и регрессии.
nb = GaussianNB().fit(X_train, y_train)
model_preds = nb.predict(X_test)
accuracy = accuracy_score(y_test, model_preds)
print("Accuracy:", accuracy)
# DecisionTreeClassifier - это алгоритм, который используется для решения задач классификации и регрессии и основан на построении дерева решений на основе анализа данных и их параметров.
dtc = DecisionTreeClassifier().fit(X_train, y_train)
model_preds = dtc.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, model_preds).items():
print(f"{metric_name}: {metric_score:.2f}")
# RandomForestClassifier - это алгоритм машинного обучения, который основан на использовании ансамбля деревьев решений для решения задач классификации, кластеризации или регрессии.
rfc = RandomForestClassifier().fit(X_train, y_train)
model_preds = rfc.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, model_preds).items():
print(f"{metric_name}: {metric_score:.2f}")
# KNeighborsClassifier - это алгоритм машинного обучения, который основан на методе ближайших соседей (k-NN) для решения задач классификации и регрессии.
knn = KNeighborsClassifier(n_neighbors=5)
model_fit = knn.fit(X_train, y_train)
model_preds = model_fit.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, model_preds).items():
print(f"{metric_name}: {metric_score:.2f}")
# --------------------------------------------------------------------------
# GradientBoostingClassifier - это ансамблевый метод машинного обучения, который используется для решения задач классификации, основанный на построении последовательности деревьев решений, каждое из которых исправляет ошибки предыдущих моделей, минимизируя функцию потерь.
gbc = GradientBoostingClassifier(
n_estimators=100, learning_rate=0.1, max_depth=1, random_state=42
)
gbc.fit(X_train, y_train)
y_pred_gbc = gbc.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, y_pred_gbc).items():
print(f"{metric_name}: {metric_score:.2f}")
# GradientBoostingClassifier показал лучший результат, но из-за долгого времени обучения не рекомендуется к использованию на демоэкзамене.
# --------------------------------------------------------------------------
# SVC (Support Vector Classifier) - это алгоритм машинного обучения, который используется для решения задач классификации, основанный на поиске оптимальной разделяющей гиперплоскости в многомерном пространстве с максимальным зазором между классами.
svm = SVC(kernel="linear", C=1, random_state=42)
svm.fit(X_train, y_train)
y_pred_svm = svm.predict(X_test)
acc_svm = accuracy_score(y_test, y_pred_svm)
for metric_name, metric_score in classification_metrics(y_test, y_pred_svm).items():
print(f"{metric_name}: {metric_score:.2f}")
# MLPClassifier - это нейронная сеть прямого распространения, которая используется для решения задач классификации или регрессии и состоит из нескольких слоев скрытых нейронов, обучаемых с помощью метода обратного распространения ошибки.
nn = MLPClassifier(
hidden_layer_sizes=(10,),
activation="relu",
solver="adam",
alpha=0.0001,
max_iter=500,
random_state=42,
)
nn.fit(X_train, y_train)
y_pred_nn = nn.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, y_pred_nn).items():
print(f"{metric_name}: {metric_score:.2f}")
# Для последующей оптимизации выбрана модель классификации дерева решений DecisionTreeClassifier, так как эта модель показала наивысшее качество обучения.
# *Примечание. модель GradientBoostingClassifier не была выбрана из-за продолжительного обучения, несмотря на то, что качество лучше чем у модели дерева решений
# ## 2.2 Оптимизация модели
# Оптимизация модели будет проводиться путем поиска наилучших гиперпараметров
param_grid = {"max_depth": range(1, 6, 1), "min_samples_leaf": range(1, 6, 1)}
grid_search = GridSearchCV(dtc, param_grid, cv=5)
grid_search.fit(X_train, y_train)
print(grid_search.best_params_)
y_pred_gs = grid_search.predict(X_test)
for metric_name, metric_score in classification_metrics(y_test, y_pred_gs).items():
print(f"{metric_name}: {metric_score:.2f}")
# Построение кривых валидации и обучения
train_sizes, train_scores, test_scores = learning_curve(
dtc, X_train, y_train, cv=5, scoring="accuracy"
)
# Вычисляем среднюю точность и стандартное отклонение для каждого размера обучающей выборки
train_mean = np.mean(train_scores, axis=1)
train_std = np.std(train_scores, axis=1)
test_mean = np.mean(test_scores, axis=1)
test_std = np.std(test_scores, axis=1)
# Строим кривую обучения и кривую валидации
plt.plot(train_sizes, train_mean, label="Training Accuracy")
plt.plot(train_sizes, test_mean, label="Validation Accuracy")
# Добавляем круги для отображения стандартного отклонения
plt.fill_between(train_sizes, train_mean - train_std, train_mean + train_std, alpha=0.1)
plt.fill_between(train_sizes, test_mean - test_std, test_mean + test_std, alpha=0.1)
# Подписываем график и добавляем легенду
plt.xlabel("Number of Training Samples")
plt.ylabel("Accuracy Score")
plt.title("Learning Curve for DecisionTreeClassifier")
plt.legend(loc="best")
plt.show()
| false | 0 | 2,622 | 0 | 2,622 | 2,622 |
||
129550291
|
<jupyter_start><jupyter_text>Handwritten Math Symbols
# Context
While working on [project](http://sagyamthapa.me/Handwritten-Optical-Character-Recognition/) to build a calculator that evaluates handwritten math symbols, I realized 3 main problems:
1. The resolution of images were too small usually 32*32 pixels.
2. The quality of data was not satisfactory. Either too many copies of same image or not enough varieties.
3. The models trained on such datasets performed poorly in real world scenarios.
4. Main issue with existing MNIST and CHROME dataset was that resizing images from 400x400 pixels(ideal for writing in canvas) to 32x32 pixels means only around 8% of original data will be given to model. So we cannot expect model to perform well in real world scenarios.
So, I set out to create my own dataset from scratch.
# Methodology
If you follow the project link you will see a save button. When the model fails to recognize a symbol I saved the image and added it to the dataset. After multiple iteration of adding new images to dataset and training the model on new dataset, I have created created a model that was fairly good at real world testing. This dataset was used to train the model that you are testing right now.
# Content
This dataset contains over 9000 handwritten digits and arithmetic operators.
Total no of classes: 16
Digits: 0 1 2 3 4 5 6 7 8 9
Operators: Plus Minus Multiplication Division Decimal Equals
Most images are of resolution 400x400 pixels. Some may be 155x155. I have resized image to 100x100 for in the [started notebook](https://www.kaggle.com/sagyamthapa/starter-notebook).
Each class contains about 500 examples.
# Inspiration
This dataset was created for training the model for my [project](http://sagyamthapa.me/Handwritten-Optical-Character-Recognition/) .
# Contact
Email me at: [email protected]
My website: [sagyamthapa.me](https://sagyamthapa.me/#contact-form)
Kaggle dataset identifier: handwritten-math-symbols
<jupyter_script>import pandas as pd
import numpy as np
import os
import keras
import matplotlib.pyplot as plt
from keras.layers import (
Dense,
Dropout,
Flatten,
ZeroPadding2D,
Conv2D,
MaxPooling2D,
Activation,
GlobalAveragePooling2D,
)
from keras.preprocessing import image
from keras.applications.mobilenet import preprocess_input
from keras.preprocessing.image import ImageDataGenerator
from keras.models import Model, Sequential
from keras.optimizers import Adam
from sklearn.model_selection import train_test_split
from keras.callbacks import EarlyStopping, ReduceLROnPlateau
from sklearn.utils import class_weight
# splits the dataset into the three sets based on the specified ratio,
# which in this case is 60% training, 20% validation, and 20% test
import splitfolders
splitfolders.ratio(
"../input/handwritten-math-symbols/dataset",
output="./",
seed=1337,
ratio=(0.6, 0.2, 0.2),
group_prefix=None,
) # default values
# # Initialize ImageDataGenerator objects for training and validation sets, create generators with specified parameters.The images are resized to 224x224 pixels, converted to RGB color mode, and batched into groups of 24.
import os
NUM_CLASSES = len(os.listdir(r"./test"))
train_datagen = ImageDataGenerator(
preprocessing_function=preprocess_input
) # included in our dependencies
train_generator = train_datagen.flow_from_directory(
r"./train", # this is where you specify the path to the main data folder
target_size=(224, 224),
color_mode="rgb",
batch_size=24,
class_mode="categorical",
shuffle=True,
)
val_datagen = ImageDataGenerator(
preprocessing_function=preprocess_input
) # included in our dependencies
val_generator = val_datagen.flow_from_directory(
r"./val", # this is where you specify the path to the main data folder
target_size=(224, 224),
color_mode="rgb",
batch_size=24,
class_mode="categorical",
shuffle=True,
)
# # Import pre-trained models, create sequential model with EfficientNetB6 and dense layer, print model summary.
#
from tensorflow.keras.applications.inception_v3 import InceptionV3
from tensorflow.keras.applications.inception_resnet_v2 import InceptionResNetV2
from tensorflow.keras.applications.xception import Xception
from tensorflow.keras.applications.densenet import DenseNet201
from tensorflow.keras.applications.efficientnet import EfficientNetB6
md = EfficientNetB6(
weights="imagenet", include_top=False, input_shape=(224, 224, 3), pooling="avg"
)
from keras.utils import plot_model
model = keras.models.Sequential(
[md, keras.layers.Dense(NUM_CLASSES, activation="softmax")]
)
# summarize layers
print(model.summary())
# # Define callbacks, compile model, and train on generator data with specified parameters and callbacks.
#
earlystop = EarlyStopping(patience=3)
learning_rate_reduction = ReduceLROnPlateau(
monitor="loss", patience=2, verbose=1, factor=0.1, min_lr=0.0000000001
)
callback = [learning_rate_reduction]
model.compile(
optimizer=Adam(lr=0.00001), loss="categorical_crossentropy", metrics=["accuracy"]
)
step_size_train = train_generator.n // train_generator.batch_size
step_size_val = val_generator.n // val_generator.batch_size
history = model.fit_generator(
generator=train_generator,
steps_per_epoch=step_size_train,
validation_data=val_generator,
validation_steps=step_size_val,
epochs=25,
callbacks=callback,
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/550/129550291.ipynb
|
handwritten-math-symbols
|
sagyamthapa
|
[{"Id": 129550291, "ScriptId": 38501609, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8929361, "CreationDate": "05/14/2023 18:39:54", "VersionNumber": 1.0, "Title": "Hand Written Math Symbol Recognition_Rahul", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 76.0, "LinesInsertedFromPrevious": 76.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185725216, "KernelVersionId": 129550291, "SourceDatasetVersionId": 2947278}]
|
[{"Id": 2947278, "DatasetId": 1237036, "DatasourceVersionId": 2994798, "CreatorUserId": 4095338, "LicenseName": "GPL 2", "CreationDate": "12/20/2021 01:27:25", "VersionNumber": 4.0, "Title": "Handwritten Math Symbols", "Slug": "handwritten-math-symbols", "Subtitle": "Over 10000 Handwritten Digits and Arithmetic Operators", "Description": "# Context\nWhile working on [project](http://sagyamthapa.me/Handwritten-Optical-Character-Recognition/) to build a calculator that evaluates handwritten math symbols, I realized 3 main problems:\n\n1. The resolution of images were too small usually 32*32 pixels.\n2. The quality of data was not satisfactory. Either too many copies of same image or not enough varieties.\n3. The models trained on such datasets performed poorly in real world scenarios.\n4. Main issue with existing MNIST and CHROME dataset was that resizing images from 400x400 pixels(ideal for writing in canvas) to 32x32 pixels means only around 8% of original data will be given to model. So we cannot expect model to perform well in real world scenarios.\n\nSo, I set out to create my own dataset from scratch. \n\n# Methodology\n\nIf you follow the project link you will see a save button. When the model fails to recognize a symbol I saved the image and added it to the dataset. After multiple iteration of adding new images to dataset and training the model on new dataset, I have created created a model that was fairly good at real world testing. This dataset was used to train the model that you are testing right now. \n\n# Content\n\nThis dataset contains over 9000 handwritten digits and arithmetic operators.\nTotal no of classes: 16\nDigits: 0 1 2 3 4 5 6 7 8 9\nOperators: Plus Minus Multiplication Division Decimal Equals\nMost images are of resolution 400x400 pixels. Some may be 155x155. I have resized image to 100x100 for in the [started notebook](https://www.kaggle.com/sagyamthapa/starter-notebook).\nEach class contains about 500 examples. \n\n# Inspiration\n\nThis dataset was created for training the model for my [project](http://sagyamthapa.me/Handwritten-Optical-Character-Recognition/) .\n\n# Contact\nEmail me at: [email protected]\nMy website: [sagyamthapa.me](https://sagyamthapa.me/#contact-form)", "VersionNotes": "Data Update 2021/12/20", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1237036, "CreatorUserId": 4095338, "OwnerUserId": 4095338.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2947278.0, "CurrentDatasourceVersionId": 2994798.0, "ForumId": 1255272, "Type": 2, "CreationDate": "03/28/2021 04:53:38", "LastActivityDate": "03/28/2021", "TotalViews": 20785, "TotalDownloads": 2231, "TotalVotes": 38, "TotalKernels": 14}]
|
[{"Id": 4095338, "UserName": "sagyamthapa", "DisplayName": "Sagyam Thapa", "RegisterDate": "11/24/2019", "PerformanceTier": 1}]
|
import pandas as pd
import numpy as np
import os
import keras
import matplotlib.pyplot as plt
from keras.layers import (
Dense,
Dropout,
Flatten,
ZeroPadding2D,
Conv2D,
MaxPooling2D,
Activation,
GlobalAveragePooling2D,
)
from keras.preprocessing import image
from keras.applications.mobilenet import preprocess_input
from keras.preprocessing.image import ImageDataGenerator
from keras.models import Model, Sequential
from keras.optimizers import Adam
from sklearn.model_selection import train_test_split
from keras.callbacks import EarlyStopping, ReduceLROnPlateau
from sklearn.utils import class_weight
# splits the dataset into the three sets based on the specified ratio,
# which in this case is 60% training, 20% validation, and 20% test
import splitfolders
splitfolders.ratio(
"../input/handwritten-math-symbols/dataset",
output="./",
seed=1337,
ratio=(0.6, 0.2, 0.2),
group_prefix=None,
) # default values
# # Initialize ImageDataGenerator objects for training and validation sets, create generators with specified parameters.The images are resized to 224x224 pixels, converted to RGB color mode, and batched into groups of 24.
import os
NUM_CLASSES = len(os.listdir(r"./test"))
train_datagen = ImageDataGenerator(
preprocessing_function=preprocess_input
) # included in our dependencies
train_generator = train_datagen.flow_from_directory(
r"./train", # this is where you specify the path to the main data folder
target_size=(224, 224),
color_mode="rgb",
batch_size=24,
class_mode="categorical",
shuffle=True,
)
val_datagen = ImageDataGenerator(
preprocessing_function=preprocess_input
) # included in our dependencies
val_generator = val_datagen.flow_from_directory(
r"./val", # this is where you specify the path to the main data folder
target_size=(224, 224),
color_mode="rgb",
batch_size=24,
class_mode="categorical",
shuffle=True,
)
# # Import pre-trained models, create sequential model with EfficientNetB6 and dense layer, print model summary.
#
from tensorflow.keras.applications.inception_v3 import InceptionV3
from tensorflow.keras.applications.inception_resnet_v2 import InceptionResNetV2
from tensorflow.keras.applications.xception import Xception
from tensorflow.keras.applications.densenet import DenseNet201
from tensorflow.keras.applications.efficientnet import EfficientNetB6
md = EfficientNetB6(
weights="imagenet", include_top=False, input_shape=(224, 224, 3), pooling="avg"
)
from keras.utils import plot_model
model = keras.models.Sequential(
[md, keras.layers.Dense(NUM_CLASSES, activation="softmax")]
)
# summarize layers
print(model.summary())
# # Define callbacks, compile model, and train on generator data with specified parameters and callbacks.
#
earlystop = EarlyStopping(patience=3)
learning_rate_reduction = ReduceLROnPlateau(
monitor="loss", patience=2, verbose=1, factor=0.1, min_lr=0.0000000001
)
callback = [learning_rate_reduction]
model.compile(
optimizer=Adam(lr=0.00001), loss="categorical_crossentropy", metrics=["accuracy"]
)
step_size_train = train_generator.n // train_generator.batch_size
step_size_val = val_generator.n // val_generator.batch_size
history = model.fit_generator(
generator=train_generator,
steps_per_epoch=step_size_train,
validation_data=val_generator,
validation_steps=step_size_val,
epochs=25,
callbacks=callback,
)
| false | 0 | 982 | 0 | 1,526 | 982 |
||
129550144
|
# ## 1 Парсинг данных и предобработка данных
# Импорт библиотек
import pandas as pd
import io
import os
import glob
import docx
import json
from bs4 import BeautifulSoup
import requests
from datetime import datetime
import string
import re
import nltk
import pymorphy2
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
nltk.download("word_tokenize")
nltk.download("punkt")
from nltk.stem import SnowballStemmer
from nltk.stem import WordNetLemmatizer
import warnings
warnings.filterwarnings("ignore")
from tqdm import tqdm
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import pylab
import scipy.stats as stats
import nltk
from nltk import ngrams
import json
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
from gensim.models import Word2Vec
from scipy.stats import shapiro
from scipy.stats import normaltest
from scipy.stats import anderson
import warnings
warnings.filterwarnings("ignore")
from tqdm import tqdm
# pip install python-docx
# nltk.download('averaged_perceptron_tagger_ru')
# ## 1.1 Парсинг данных
condidates_path = r"Condidates.docx"
doc = docx.Document(condidates_path)
all_paras = doc.paragraphs
len(all_paras)
# Возьмем из документа названия компаний
condidates = []
i = 1
for para in all_paras:
print(i, para.text)
i += 1
if para.text != "":
condidates.append(para.text.rstrip())
len(condidates)
print(condidates, len(condidates))
# Посмотрим каких компаний нет в папке Data
all_json = glob.glob(r"Data\*.json")
json_names = [os.path.basename(x.replace(".json", "")) for x in all_json]
print(json_names, len(json_names))
condidates_pass = list(set(condidates) - set(json_names))
condidates_pass
# У двух компаний ('Skillbox' и 'Проект по использованию технологий компьютерного зрения на базе искусственного интеллекта (ИИ) для анализа медицинских изображений') не совпадали названия. Названия были изменены в папке Data
# ### 1.1.1 Парсинг Json
articles = []
for json_path in all_json:
with open(json_path, "r", encoding="utf-8") as f:
data = json.load(f)
print(json_path)
i = 0
# Информаци о компании
if data["info"] is not None:
company_rating = data["info"]["rate"]
company_activity = data["info"]["industries"]
company_description = data["info"]["about"]
else:
company_rating = None
company_activity = None
company_description = None
# Инфлормация из статьи
for article in data["refs"]:
if article is not None:
article_text = article[0]
article_date = article[1]["day"] + " " + article[1]["month"]
articles.append(
{
"company_name": os.path.basename(
json_path.replace(".json", "")
),
"company_description": company_description,
"company_activity": company_activity,
"company_rating": company_rating,
"article_text": article_text,
"date_publish": article_date,
}
)
i += 1
print("Количество статей: ", i)
print("Общее количество статей: ", len(articles))
df = pd.DataFrame(data=articles)
df
df.info()
df.to_csv("articles.csv")
companies_fill = list(df[df["company_rating"].notna()]["company_name"].unique())
companies_fill
# Компании с пустыми значениями
companies_pass = list(df[df["company_rating"].isna()]["company_name"].unique())
companies_pass
# Из Json файлов удалось вытащить 1112 статьи, только две компании ('Skillbox', 'Иннотех') содержало в себе иформацию о компании
# Найдем компании с недостающими данными на сайте Хабр
for company in companies_pass:
company = company.replace(" ", "%20")
url = (
"https://habr.com/ru/search/?q="
+ company
+ "&target_type=companies&order=relevance"
)
print(url)
page = requests.get(url)
print(page.status_code)
soup = BeautifulSoup(page.text, "html.parser")
company_div = soup.find_all(
"div", class_="tm-search-companies__item tm-search-companies__item_inlined"
)
print(company_div)
# Ни одной компании с пустыми значениями из папки Data не было найдено на Хабре
# ### 1.1.2 Парсинг сайта
# Парсинг статей недостающих компаний с Хабра
condidates_pass
for company in condidates_pass:
company = company.replace(" ", "%20")
url = (
"https://habr.com/ru/search/?q="
+ company
+ "&target_type=companies&order=relevance"
)
print(url)
page = requests.get(url)
print(page.status_code)
soup = BeautifulSoup(page.text, "html.parser")
company_div = soup.find_all(
"div", class_="tm-search-companies__item tm-search-companies__item_inlined"
)
if len(company_div) == 0:
print(company_div)
else:
for c in company_div:
print(c.find("a", class_="tm-company-snippet__title").text)
# Из оставшихся компаний только 'СберМаркет' и 'Нетология' есть на Хабре
# https://habr.com/ru/companies/netologyru/articles/page
# https://habr.com/ru/companies/sbermarket/articles/page
url_companies = [
r"https://habr.com/ru/companies/netologyru/articles/page",
r"https://habr.com/ru/companies/sbermarket/articles/page",
]
articles2 = {
"company_name": [],
"company_description": [],
"company_activity": [],
"company_rating": [],
"article_text": [],
"date_publish": [],
}
for url_company in url_companies:
a = True
pagenum = 1
for i in range(11):
url = url_company + str(pagenum) + "/"
page = requests.get(url)
print(url)
if page.status_code == 200:
soup = BeautifulSoup(page.text, "html.parser")
pages = soup.find_all("h2", class_="tm-title tm-title_h2")
for i in pages:
url2 = "https://habr.com" + str(i.a.get("href"))
page = requests.get(url2)
article = BeautifulSoup(page.text, "html.parser")
print(url2)
if a:
company_name = soup.find("a", class_="tm-company-card__name")
articles2["company_name"].append(company_name.text)
url = "https://habr.com" + str(company_name.get("href"))
company_page = requests.get(url)
company = BeautifulSoup(company_page.text, "html.parser")
if a:
company_activity = company.find(
"div", class_="tm-company-profile__categories"
)
activity_clear = " ".join(company_activity.text.split())
articles2["company_activity"].append(activity_clear)
if a:
company_description = soup.find(
"div", class_="tm-company-card__description"
)
articles2["company_description"].append(company_description.text)
if a:
company_rating = soup.find(
"span",
class_="tm-votes-lever__score-counter tm-votes-lever__score-counter tm-votes-lever__score-counter_rating",
)
a = False
articles2["company_rating"].append(company_rating.text)
data_publish = article.find(
"span", class_="tm-article-datetime-published"
)
if ":" in data_publish.text:
articles2["date_publish"].append(data_publish.text)
else:
articles2["date_publish"].append(
datetime.today().strftime("%Y-%m-%d")
)
text_article = article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-1",
)
if text_article == None:
text_article = article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-2",
)
text_article_clear = " ".join(text_article.text.split())
articles2["article_text"].append(text_article_clear)
pagenum = pagenum + 1
df2 = pd.DataFrame(articles2)
df2
df2.to_csv("articles2.csv")
# -------------------------------------------------Парсинг с публикаций-----------------------------------------------------
articles_list = []
for condidate in tqdm(condidates_pass):
condidate = condidate.replace(" ", "%20")
pagenum = 1
for i in tqdm(range(50)):
url = (
"https://habr.com/ru/search/page"
+ str(pagenum)
+ "/?q="
+ condidate
+ "&target_type=posts&order=relevance"
)
page = requests.get(url)
# print(url)
# print(page.status_code)
if page.status_code == 200:
soup = BeautifulSoup(page.text, "html.parser")
pages = soup.find_all("h2", class_="tm-title tm-title_h2")
for i in pages:
url2 = "https://habr.com" + str(i.a.get("href"))
page2 = requests.get(url2)
article = BeautifulSoup(page2.text, "html.parser")
# print('url2', url2)
# print(article)
if url2 != "https://habr.com/ru/companies/2035_university/news/561404/":
name_company = article.find(
"div", class_="tm-company-snippet__title"
)
# print('name_company', name_company)
if name_company != None:
# print(url2)
name_company = name_company.text
# print(page.status_code)
if page.status_code != 404:
# Название компании
# company_name = soup.find('a', class_='tm-company-card__name')
company_name = condidate.replace("%20", " ")
# Сфера деятельности компании
if name_company == condidate:
url = "https://habr.com" + str(company_name.get("href"))
company_page = requests.get(url)
company = BeautifulSoup(company_page.text, "html.parser")
company_activity = company.find(
"div", class_="tm-company-profile__categories"
)
activity_clear = " ".join(company_activity.text.split())
else:
activity_clear = None
# Описание компании
if name_company == condidate:
company_description = company.find(
"div", class_="tm-company-card__description"
).text
else:
company_description = None
# Рейтинг компании
if name_company == condidate:
company_rating = company.find(
"span",
class_="tm-votes-lever__score-counter tm-votes-lever__score-counter tm-votes-lever__score-counter_rating",
).text
else:
company_rating = None
# Дата публикации
data_publish = article.find(
"span", class_="tm-article-datetime-published"
)
# print('data_publish', data_publish.text)
if data_publish != None:
if ":" in data_publish.text:
date_publish = data_publish.text
else:
date_publish = datetime.today().strftime("%Y-%m-%d")
else:
date_publish = None
print(url2)
# Текст статьи
text_article = article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-1",
)
if text_article == None:
text_article = article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-2",
)
if text_article != None:
text_article_clear = " ".join(text_article.text.split())
else:
print(url2)
text_article_clear = None
article_dict = {
"company_name": company_name,
"company_description": company_description,
"company_activity": activity_clear,
"company_rating": company_rating,
"article_text": text_article_clear,
"date_publish": date_publish,
}
articles_list.append(article_dict)
pagenum = pagenum + 1
df3 = pd.DataFrame.from_records(articles_list)
df3
df3.info()
url2 = r"https://habr.com/ru/companies/2035_university/news/561404/"
page = requests.get(url2)
article = BeautifulSoup(page.text, "html.parser")
print(url2)
df3["company_name"].unique()
df3.to_csv("articles3.csv")
# --------------------------------------------------------------------------------------------------------------------------
# Объединим полученые датафреймы
articles_df = pd.concat([df, df2, df3], ignore_index=True)
articles_df
articles_df.info()
# ### 1.1.3 Обработка пропусков и дубликатов
duplicates = articles_df.duplicated(subset=["company_name", "article_text"])
num_duplicates = duplicates.sum()
print(f"Количество дубликатов по полям company_name и article_text: {num_duplicates}")
duplicates = articles_df.duplicated(subset=["article_text"])
num_duplicates = duplicates.sum()
print(f"Количество дубликатов по полям article_text: {num_duplicates}")
import numpy as np
db = np.where(duplicates == True)
articles_df.drop_duplicates(
subset=["company_name", "article_text"], keep="first", inplace=True
)
list(articles_df["company_name"].unique())
described_companies = articles_df.loc[
articles_df["company_description"].notnull(), "company_name"
].unique()
described_companies
activity_companies = articles_df.loc[
articles_df["company_activity"].notnull(), "company_name"
].unique()
activity_companies
rating_companies = articles_df.loc[
articles_df["company_rating"].notnull(), "company_name"
].unique()
rating_companies
for company in described_companies:
print(company)
description = (
articles_df.loc[articles_df["company_name"] == company, "company_description"]
.dropna()
.iloc[0]
)
articles_df.loc[
(articles_df["company_name"] == company)
& (articles_df["company_description"].isnull()),
"company_description",
] = description
for company in activity_companies:
print(company)
activity = (
articles_df.loc[articles_df["company_name"] == company, "company_activity"]
.dropna()
.iloc[0]
)
articles_df.loc[
(articles_df["company_name"] == company)
& (articles_df["company_activity"].isnull()),
"company_activity",
] = activity
for company in rating_companies:
print(company)
rating = (
articles_df.loc[articles_df["company_name"] == company, "company_rating"]
.dropna()
.iloc[0]
)
articles_df.loc[
(articles_df["company_name"] == company)
& (articles_df["company_rating"].isnull()),
"company_rating",
] = rating
articles_df = articles_df.dropna(subset=["article_text"])
articles_df.to_csv("articles_full.csv")
# После парсинга мы получили датафрейм с 4277 статьми и 6 признаками: Название компании, описание компании, сфера деятельности, рейтинг компании, текст статьи и дата публикации. Большинство данных взятых из Json файлов не содержало информации о компании, также этих компаний отсутствовали на Хабре. Записи с пустыми значениями и дубликаты были удалены.
# ## 1.2 Формирование структуры набора данных
articles_df = pd.read_csv("articles_full.csv")
articles_df_clear = articles_df.drop(
[
"Unnamed: 0",
"Unnamed: 0.1",
"company_description",
"company_activity",
"company_rating",
"date_publish",
],
axis=1,
)
# Признаки company_description, company_activity и company_rating имеют большое количество пропущенных значений. date_publish не будет использоваться в обучении
articles_df_clear
articles_df_clear.info()
# ## 1.3 Предварительная обработка текстовых данных
morph = pymorphy2.MorphAnalyzer()
stopword = nltk.corpus.stopwords.words("russian")
# Дополним наши пустые слова
stopword.extend(
[
"либо",
"это",
"мб",
"далее",
"дв",
"свой",
"ваш",
"всё",
"очень",
"её",
"ещё",
"вообще",
"наш",
"который",
]
)
def preprocess_text(data, stopwords=stopword):
text = re.sub("ё", "е", data.lower())
text = re.sub("й", "и", text)
text = re.sub(r"([.,!?])", r" \1 ", text)
text = re.sub(r"[^а-яА-Я\s]+", "", text)
text = text.strip()
text = [w for w in text.split() if w not in stopwords]
text = [w for w in text if len(w) >= 3]
return " ".join(text)
def lemmatization_text(data, morph=morph):
result = " ".join([morph.parse(x)[0].normal_form for x in data.split()])
return result
def get_result(data, morph=morph, stopwords=stopword):
result = preprocess_text(data=data)
result = lemmatization_text(result)
return result
def transform_data(data: pd.Series) -> list:
result = [get_result(data=i) for i in tqdm(data)]
return result
text = articles_df_clear["article_text"]
result_df = tqdm(transform_data(text))
articles_df_clear["lematize_text"] = result_df
articles_df_clear
result_df2 = [word_tokenize(text) for text in result_df]
articles_df_clear["tokenize_text"] = result_df2
articles_df_clear
result_df3 = [nltk.pos_tag(text, lang="rus") for text in tqdm(result_df2)]
articles_df_clear["pos_tag_text"] = result_df3
articles_df_clear.info()
articles_df_clear.to_csv("articles_df.csv")
# Была выполнена предобработка текста статьи. Предобработка включает в себя удаление всех символов, кроме букв русского алфавита, удаление стоп слов и лематизация текста. Проведена токенизация и маркировка частей речи текста каждой статьи
# ### 1.4 Поиск n-грамм. Векторизация текстов
df = pd.read_csv("articles_df.csv")
df
df.info()
# После чтения файла пропалин значения лемматизированного текста у некоторых записей.
df = df.dropna(subset=["lematize_text"])
df.drop(["Unnamed: 0"], axis=1, inplace=True)
df.head(5)
text = list(df["lematize_text"])
text[0]
vocabVect = CountVectorizer()
vocabVect.fit(text)
corpusVocab = vocabVect.vocabulary_
print("Количество признаков - {}".format(len(corpusVocab)))
for i in list(corpusVocab)[1:10]:
print("{}={}".format(i, corpusVocab[i]))
test_features = vocabVect.transform(text)
vocabVect.get_feature_names_out()[100:120]
def find_ngrams(text, n):
n_grams = ngrams(text.split(), n)
return [" ".join(grams) for grams in n_grams]
df["bigrams"] = df["lematize_text"].apply(lambda x: find_ngrams(x, 2))
df["trigrams"] = df["tokenize_text"].apply(lambda x: find_ngrams(x, 3))
df.head()
vectorizer = TfidfVectorizer()
tfidf_matrix = vectorizer.fit_transform(df["lematize_text"])
keywords = []
for i in tqdm(range(len(df))):
tfidf_scores = tfidf_matrix[i].todense()
scores_list = tfidf_scores.tolist()[0]
words = vectorizer.get_feature_names_out()
key_words_df = pd.DataFrame({"word": words, "score": scores_list})
key_words_df = key_words_df.sort_values(by="score", ascending=False)
keywords.append(list(key_words_df["word"][:5]))
df["keywords"] = keywords
# ## 1.5 Разведочный анализ
result_dict = {}
with open("Target.json", "r", encoding="utf-8") as f:
data = json.load(f)
for entry in data["text"]:
company = entry["Company"]
nomination = entry["Nominations"]
result_dict[company] = nomination
result_dict
df["nomination"] = df["company_name"].map(result_dict)
df.head(5)
df["company_name"].unique()
df["nomination"].unique()
df.info()
# проверка нормальности распределения целевой переменной
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
df["nomination_encoded"] = le.fit_transform(df["nomination"])
sns.distplot(df["nomination_encoded"])
plt.title("Распределения данных")
plt.show()
sns.boxplot(df["nomination_encoded"])
stats.probplot(df["nomination_encoded"], dist="norm", plot=pylab)
pylab.show()
stat, p = shapiro(df["nomination_encoded"])
print("stat=%.3f, p=%.3f\n" % (stat, p))
if p > 0.05:
print("Probably Gaussian")
else:
print("Probably not Gaussian")
stat, p = normaltest(df["nomination_encoded"])
print("stat=%.3f, p=%.3f\n" % (stat, p))
if p > 0.05:
print("Probably Gaussian")
else:
print("Probably not Gaussian")
result = anderson(df["nomination_encoded"])
print("stat=%.3f" % (result.statistic))
for i in range(len(result.critical_values)):
sig_lev, crit_val = result.significance_level[i], result.critical_values[i]
if result.statistic < crit_val:
print(
f"probably Gaussian : {crit_val} critical value at {sig_lev} level of significance"
)
else:
print(
f"Probably not Gaussian : {crit_val} critical value at {sig_lev} level of significance"
)
# Распределение целевой переменной не является нормальным.
df.to_csv("articles.csv")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/550/129550144.ipynb
| null | null |
[{"Id": 129550144, "ScriptId": 38521899, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13417746, "CreationDate": "05/14/2023 18:37:58", "VersionNumber": 1.0, "Title": "Report1-VD-djostit", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 615.0, "LinesInsertedFromPrevious": 615.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## 1 Парсинг данных и предобработка данных
# Импорт библиотек
import pandas as pd
import io
import os
import glob
import docx
import json
from bs4 import BeautifulSoup
import requests
from datetime import datetime
import string
import re
import nltk
import pymorphy2
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
nltk.download("word_tokenize")
nltk.download("punkt")
from nltk.stem import SnowballStemmer
from nltk.stem import WordNetLemmatizer
import warnings
warnings.filterwarnings("ignore")
from tqdm import tqdm
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import pylab
import scipy.stats as stats
import nltk
from nltk import ngrams
import json
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
from gensim.models import Word2Vec
from scipy.stats import shapiro
from scipy.stats import normaltest
from scipy.stats import anderson
import warnings
warnings.filterwarnings("ignore")
from tqdm import tqdm
# pip install python-docx
# nltk.download('averaged_perceptron_tagger_ru')
# ## 1.1 Парсинг данных
condidates_path = r"Condidates.docx"
doc = docx.Document(condidates_path)
all_paras = doc.paragraphs
len(all_paras)
# Возьмем из документа названия компаний
condidates = []
i = 1
for para in all_paras:
print(i, para.text)
i += 1
if para.text != "":
condidates.append(para.text.rstrip())
len(condidates)
print(condidates, len(condidates))
# Посмотрим каких компаний нет в папке Data
all_json = glob.glob(r"Data\*.json")
json_names = [os.path.basename(x.replace(".json", "")) for x in all_json]
print(json_names, len(json_names))
condidates_pass = list(set(condidates) - set(json_names))
condidates_pass
# У двух компаний ('Skillbox' и 'Проект по использованию технологий компьютерного зрения на базе искусственного интеллекта (ИИ) для анализа медицинских изображений') не совпадали названия. Названия были изменены в папке Data
# ### 1.1.1 Парсинг Json
articles = []
for json_path in all_json:
with open(json_path, "r", encoding="utf-8") as f:
data = json.load(f)
print(json_path)
i = 0
# Информаци о компании
if data["info"] is not None:
company_rating = data["info"]["rate"]
company_activity = data["info"]["industries"]
company_description = data["info"]["about"]
else:
company_rating = None
company_activity = None
company_description = None
# Инфлормация из статьи
for article in data["refs"]:
if article is not None:
article_text = article[0]
article_date = article[1]["day"] + " " + article[1]["month"]
articles.append(
{
"company_name": os.path.basename(
json_path.replace(".json", "")
),
"company_description": company_description,
"company_activity": company_activity,
"company_rating": company_rating,
"article_text": article_text,
"date_publish": article_date,
}
)
i += 1
print("Количество статей: ", i)
print("Общее количество статей: ", len(articles))
df = pd.DataFrame(data=articles)
df
df.info()
df.to_csv("articles.csv")
companies_fill = list(df[df["company_rating"].notna()]["company_name"].unique())
companies_fill
# Компании с пустыми значениями
companies_pass = list(df[df["company_rating"].isna()]["company_name"].unique())
companies_pass
# Из Json файлов удалось вытащить 1112 статьи, только две компании ('Skillbox', 'Иннотех') содержало в себе иформацию о компании
# Найдем компании с недостающими данными на сайте Хабр
for company in companies_pass:
company = company.replace(" ", "%20")
url = (
"https://habr.com/ru/search/?q="
+ company
+ "&target_type=companies&order=relevance"
)
print(url)
page = requests.get(url)
print(page.status_code)
soup = BeautifulSoup(page.text, "html.parser")
company_div = soup.find_all(
"div", class_="tm-search-companies__item tm-search-companies__item_inlined"
)
print(company_div)
# Ни одной компании с пустыми значениями из папки Data не было найдено на Хабре
# ### 1.1.2 Парсинг сайта
# Парсинг статей недостающих компаний с Хабра
condidates_pass
for company in condidates_pass:
company = company.replace(" ", "%20")
url = (
"https://habr.com/ru/search/?q="
+ company
+ "&target_type=companies&order=relevance"
)
print(url)
page = requests.get(url)
print(page.status_code)
soup = BeautifulSoup(page.text, "html.parser")
company_div = soup.find_all(
"div", class_="tm-search-companies__item tm-search-companies__item_inlined"
)
if len(company_div) == 0:
print(company_div)
else:
for c in company_div:
print(c.find("a", class_="tm-company-snippet__title").text)
# Из оставшихся компаний только 'СберМаркет' и 'Нетология' есть на Хабре
# https://habr.com/ru/companies/netologyru/articles/page
# https://habr.com/ru/companies/sbermarket/articles/page
url_companies = [
r"https://habr.com/ru/companies/netologyru/articles/page",
r"https://habr.com/ru/companies/sbermarket/articles/page",
]
articles2 = {
"company_name": [],
"company_description": [],
"company_activity": [],
"company_rating": [],
"article_text": [],
"date_publish": [],
}
for url_company in url_companies:
a = True
pagenum = 1
for i in range(11):
url = url_company + str(pagenum) + "/"
page = requests.get(url)
print(url)
if page.status_code == 200:
soup = BeautifulSoup(page.text, "html.parser")
pages = soup.find_all("h2", class_="tm-title tm-title_h2")
for i in pages:
url2 = "https://habr.com" + str(i.a.get("href"))
page = requests.get(url2)
article = BeautifulSoup(page.text, "html.parser")
print(url2)
if a:
company_name = soup.find("a", class_="tm-company-card__name")
articles2["company_name"].append(company_name.text)
url = "https://habr.com" + str(company_name.get("href"))
company_page = requests.get(url)
company = BeautifulSoup(company_page.text, "html.parser")
if a:
company_activity = company.find(
"div", class_="tm-company-profile__categories"
)
activity_clear = " ".join(company_activity.text.split())
articles2["company_activity"].append(activity_clear)
if a:
company_description = soup.find(
"div", class_="tm-company-card__description"
)
articles2["company_description"].append(company_description.text)
if a:
company_rating = soup.find(
"span",
class_="tm-votes-lever__score-counter tm-votes-lever__score-counter tm-votes-lever__score-counter_rating",
)
a = False
articles2["company_rating"].append(company_rating.text)
data_publish = article.find(
"span", class_="tm-article-datetime-published"
)
if ":" in data_publish.text:
articles2["date_publish"].append(data_publish.text)
else:
articles2["date_publish"].append(
datetime.today().strftime("%Y-%m-%d")
)
text_article = article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-1",
)
if text_article == None:
text_article = article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-2",
)
text_article_clear = " ".join(text_article.text.split())
articles2["article_text"].append(text_article_clear)
pagenum = pagenum + 1
df2 = pd.DataFrame(articles2)
df2
df2.to_csv("articles2.csv")
# -------------------------------------------------Парсинг с публикаций-----------------------------------------------------
articles_list = []
for condidate in tqdm(condidates_pass):
condidate = condidate.replace(" ", "%20")
pagenum = 1
for i in tqdm(range(50)):
url = (
"https://habr.com/ru/search/page"
+ str(pagenum)
+ "/?q="
+ condidate
+ "&target_type=posts&order=relevance"
)
page = requests.get(url)
# print(url)
# print(page.status_code)
if page.status_code == 200:
soup = BeautifulSoup(page.text, "html.parser")
pages = soup.find_all("h2", class_="tm-title tm-title_h2")
for i in pages:
url2 = "https://habr.com" + str(i.a.get("href"))
page2 = requests.get(url2)
article = BeautifulSoup(page2.text, "html.parser")
# print('url2', url2)
# print(article)
if url2 != "https://habr.com/ru/companies/2035_university/news/561404/":
name_company = article.find(
"div", class_="tm-company-snippet__title"
)
# print('name_company', name_company)
if name_company != None:
# print(url2)
name_company = name_company.text
# print(page.status_code)
if page.status_code != 404:
# Название компании
# company_name = soup.find('a', class_='tm-company-card__name')
company_name = condidate.replace("%20", " ")
# Сфера деятельности компании
if name_company == condidate:
url = "https://habr.com" + str(company_name.get("href"))
company_page = requests.get(url)
company = BeautifulSoup(company_page.text, "html.parser")
company_activity = company.find(
"div", class_="tm-company-profile__categories"
)
activity_clear = " ".join(company_activity.text.split())
else:
activity_clear = None
# Описание компании
if name_company == condidate:
company_description = company.find(
"div", class_="tm-company-card__description"
).text
else:
company_description = None
# Рейтинг компании
if name_company == condidate:
company_rating = company.find(
"span",
class_="tm-votes-lever__score-counter tm-votes-lever__score-counter tm-votes-lever__score-counter_rating",
).text
else:
company_rating = None
# Дата публикации
data_publish = article.find(
"span", class_="tm-article-datetime-published"
)
# print('data_publish', data_publish.text)
if data_publish != None:
if ":" in data_publish.text:
date_publish = data_publish.text
else:
date_publish = datetime.today().strftime("%Y-%m-%d")
else:
date_publish = None
print(url2)
# Текст статьи
text_article = article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-1",
)
if text_article == None:
text_article = article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-2",
)
if text_article != None:
text_article_clear = " ".join(text_article.text.split())
else:
print(url2)
text_article_clear = None
article_dict = {
"company_name": company_name,
"company_description": company_description,
"company_activity": activity_clear,
"company_rating": company_rating,
"article_text": text_article_clear,
"date_publish": date_publish,
}
articles_list.append(article_dict)
pagenum = pagenum + 1
df3 = pd.DataFrame.from_records(articles_list)
df3
df3.info()
url2 = r"https://habr.com/ru/companies/2035_university/news/561404/"
page = requests.get(url2)
article = BeautifulSoup(page.text, "html.parser")
print(url2)
df3["company_name"].unique()
df3.to_csv("articles3.csv")
# --------------------------------------------------------------------------------------------------------------------------
# Объединим полученые датафреймы
articles_df = pd.concat([df, df2, df3], ignore_index=True)
articles_df
articles_df.info()
# ### 1.1.3 Обработка пропусков и дубликатов
duplicates = articles_df.duplicated(subset=["company_name", "article_text"])
num_duplicates = duplicates.sum()
print(f"Количество дубликатов по полям company_name и article_text: {num_duplicates}")
duplicates = articles_df.duplicated(subset=["article_text"])
num_duplicates = duplicates.sum()
print(f"Количество дубликатов по полям article_text: {num_duplicates}")
import numpy as np
db = np.where(duplicates == True)
articles_df.drop_duplicates(
subset=["company_name", "article_text"], keep="first", inplace=True
)
list(articles_df["company_name"].unique())
described_companies = articles_df.loc[
articles_df["company_description"].notnull(), "company_name"
].unique()
described_companies
activity_companies = articles_df.loc[
articles_df["company_activity"].notnull(), "company_name"
].unique()
activity_companies
rating_companies = articles_df.loc[
articles_df["company_rating"].notnull(), "company_name"
].unique()
rating_companies
for company in described_companies:
print(company)
description = (
articles_df.loc[articles_df["company_name"] == company, "company_description"]
.dropna()
.iloc[0]
)
articles_df.loc[
(articles_df["company_name"] == company)
& (articles_df["company_description"].isnull()),
"company_description",
] = description
for company in activity_companies:
print(company)
activity = (
articles_df.loc[articles_df["company_name"] == company, "company_activity"]
.dropna()
.iloc[0]
)
articles_df.loc[
(articles_df["company_name"] == company)
& (articles_df["company_activity"].isnull()),
"company_activity",
] = activity
for company in rating_companies:
print(company)
rating = (
articles_df.loc[articles_df["company_name"] == company, "company_rating"]
.dropna()
.iloc[0]
)
articles_df.loc[
(articles_df["company_name"] == company)
& (articles_df["company_rating"].isnull()),
"company_rating",
] = rating
articles_df = articles_df.dropna(subset=["article_text"])
articles_df.to_csv("articles_full.csv")
# После парсинга мы получили датафрейм с 4277 статьми и 6 признаками: Название компании, описание компании, сфера деятельности, рейтинг компании, текст статьи и дата публикации. Большинство данных взятых из Json файлов не содержало информации о компании, также этих компаний отсутствовали на Хабре. Записи с пустыми значениями и дубликаты были удалены.
# ## 1.2 Формирование структуры набора данных
articles_df = pd.read_csv("articles_full.csv")
articles_df_clear = articles_df.drop(
[
"Unnamed: 0",
"Unnamed: 0.1",
"company_description",
"company_activity",
"company_rating",
"date_publish",
],
axis=1,
)
# Признаки company_description, company_activity и company_rating имеют большое количество пропущенных значений. date_publish не будет использоваться в обучении
articles_df_clear
articles_df_clear.info()
# ## 1.3 Предварительная обработка текстовых данных
morph = pymorphy2.MorphAnalyzer()
stopword = nltk.corpus.stopwords.words("russian")
# Дополним наши пустые слова
stopword.extend(
[
"либо",
"это",
"мб",
"далее",
"дв",
"свой",
"ваш",
"всё",
"очень",
"её",
"ещё",
"вообще",
"наш",
"который",
]
)
def preprocess_text(data, stopwords=stopword):
text = re.sub("ё", "е", data.lower())
text = re.sub("й", "и", text)
text = re.sub(r"([.,!?])", r" \1 ", text)
text = re.sub(r"[^а-яА-Я\s]+", "", text)
text = text.strip()
text = [w for w in text.split() if w not in stopwords]
text = [w for w in text if len(w) >= 3]
return " ".join(text)
def lemmatization_text(data, morph=morph):
result = " ".join([morph.parse(x)[0].normal_form for x in data.split()])
return result
def get_result(data, morph=morph, stopwords=stopword):
result = preprocess_text(data=data)
result = lemmatization_text(result)
return result
def transform_data(data: pd.Series) -> list:
result = [get_result(data=i) for i in tqdm(data)]
return result
text = articles_df_clear["article_text"]
result_df = tqdm(transform_data(text))
articles_df_clear["lematize_text"] = result_df
articles_df_clear
result_df2 = [word_tokenize(text) for text in result_df]
articles_df_clear["tokenize_text"] = result_df2
articles_df_clear
result_df3 = [nltk.pos_tag(text, lang="rus") for text in tqdm(result_df2)]
articles_df_clear["pos_tag_text"] = result_df3
articles_df_clear.info()
articles_df_clear.to_csv("articles_df.csv")
# Была выполнена предобработка текста статьи. Предобработка включает в себя удаление всех символов, кроме букв русского алфавита, удаление стоп слов и лематизация текста. Проведена токенизация и маркировка частей речи текста каждой статьи
# ### 1.4 Поиск n-грамм. Векторизация текстов
df = pd.read_csv("articles_df.csv")
df
df.info()
# После чтения файла пропалин значения лемматизированного текста у некоторых записей.
df = df.dropna(subset=["lematize_text"])
df.drop(["Unnamed: 0"], axis=1, inplace=True)
df.head(5)
text = list(df["lematize_text"])
text[0]
vocabVect = CountVectorizer()
vocabVect.fit(text)
corpusVocab = vocabVect.vocabulary_
print("Количество признаков - {}".format(len(corpusVocab)))
for i in list(corpusVocab)[1:10]:
print("{}={}".format(i, corpusVocab[i]))
test_features = vocabVect.transform(text)
vocabVect.get_feature_names_out()[100:120]
def find_ngrams(text, n):
n_grams = ngrams(text.split(), n)
return [" ".join(grams) for grams in n_grams]
df["bigrams"] = df["lematize_text"].apply(lambda x: find_ngrams(x, 2))
df["trigrams"] = df["tokenize_text"].apply(lambda x: find_ngrams(x, 3))
df.head()
vectorizer = TfidfVectorizer()
tfidf_matrix = vectorizer.fit_transform(df["lematize_text"])
keywords = []
for i in tqdm(range(len(df))):
tfidf_scores = tfidf_matrix[i].todense()
scores_list = tfidf_scores.tolist()[0]
words = vectorizer.get_feature_names_out()
key_words_df = pd.DataFrame({"word": words, "score": scores_list})
key_words_df = key_words_df.sort_values(by="score", ascending=False)
keywords.append(list(key_words_df["word"][:5]))
df["keywords"] = keywords
# ## 1.5 Разведочный анализ
result_dict = {}
with open("Target.json", "r", encoding="utf-8") as f:
data = json.load(f)
for entry in data["text"]:
company = entry["Company"]
nomination = entry["Nominations"]
result_dict[company] = nomination
result_dict
df["nomination"] = df["company_name"].map(result_dict)
df.head(5)
df["company_name"].unique()
df["nomination"].unique()
df.info()
# проверка нормальности распределения целевой переменной
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
df["nomination_encoded"] = le.fit_transform(df["nomination"])
sns.distplot(df["nomination_encoded"])
plt.title("Распределения данных")
plt.show()
sns.boxplot(df["nomination_encoded"])
stats.probplot(df["nomination_encoded"], dist="norm", plot=pylab)
pylab.show()
stat, p = shapiro(df["nomination_encoded"])
print("stat=%.3f, p=%.3f\n" % (stat, p))
if p > 0.05:
print("Probably Gaussian")
else:
print("Probably not Gaussian")
stat, p = normaltest(df["nomination_encoded"])
print("stat=%.3f, p=%.3f\n" % (stat, p))
if p > 0.05:
print("Probably Gaussian")
else:
print("Probably not Gaussian")
result = anderson(df["nomination_encoded"])
print("stat=%.3f" % (result.statistic))
for i in range(len(result.critical_values)):
sig_lev, crit_val = result.significance_level[i], result.critical_values[i]
if result.statistic < crit_val:
print(
f"probably Gaussian : {crit_val} critical value at {sig_lev} level of significance"
)
else:
print(
f"Probably not Gaussian : {crit_val} critical value at {sig_lev} level of significance"
)
# Распределение целевой переменной не является нормальным.
df.to_csv("articles.csv")
| false | 0 | 6,422 | 0 | 6,422 | 6,422 |
||
129527065
|
<jupyter_start><jupyter_text>Solar Energy Production
The hourly output at each of the city of Calgary's solar photovoltaic projects and the locations of City of Calgary solar photovoltaic installations.
Hourly energy production data from sites with a "public_url" can be found [here](https://data.calgary.ca/Environment/Solar-Photovoltaic-Sites/csgq-e555).
Kaggle dataset identifier: solar-energy-production
<jupyter_script># # 🌤 Forecasting Solar Power Production 🌤
# ## Machine Learning vs Statistical Models
# In this analysis I want to compare several approaches of forecasting time series data. The solar power production data seems very suitable, as it is available on a hourly basis and for a period of over 5 years.
# Following this awesome tutorial, I evaluate 3 forcasting approaches:
# 1. Direct Multi-step Forecast Strategy
# 2. Recursive Multi-step Forecast
# 3. Direct-Recursive Hybrid Strategies
# As the underlying regression models I will use a Linear Regression and Light GBM.
# To compare the performance of the ML models let's also compare them to a classical Auto ARIMA and Auto ETS model.
# The `sktime` package comes handy for this analysis, as it offers a variety of predefined time series functions, which come in handy for this analysis.
# ## Script Dependencies
import pandas as pd
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error, mean_squared_error
from lightgbm import LGBMRegressor
import numpy as np
from sktime.forecasting.compose import make_reduction
from sktime.forecasting.model_selection import temporal_train_test_split
from sktime.forecasting.arima import AutoARIMA
from sktime.forecasting.ets import AutoETS
import matplotlib.pyplot as plt
import plotly.express as px
# ## Data Import and Exploration
energy_production = pd.read_csv(
"/kaggle/input/solar-energy-production/Solar_Energy_Production.csv"
)
energy_production["date"] = pd.to_datetime(energy_production["date"])
energy_production.head()
# ## 11 Power Plants
energy_production.groupby("name").agg({"date": [min, max]})
# all plants besides Telus Spark provide > 5 years data
# # Modelling
# ## Data Preparation
## Experiment Setup
### Data Parameters
frequency = ["h", "d", "M"][2]
plant_name = "Whitehorn Multi-Service Centre"
### Modelling Parameters
window_size = 24
forecast_horizon = 12
year_start = 2017
model_log_transformed = False
forecasting_approaches = ["direct", "recursive", "dirrec"] # , 'multioutput'
regression_models = {"Lin_Reg": LinearRegression, "LGBM": LGBMRegressor}
statistical_models = {"AutoARIMA": AutoARIMA, "AutoETS": AutoETS}
## Evaluation
metrics = {"MAE": mean_absolute_error, "MSE": mean_squared_error}
# ### Data Preprocessing
plant_data = (
energy_production.loc[energy_production.name == plant_name, ["date", "kWh"]]
.set_index("date")
.resample(frequency)
.sum()
.resample(frequency)
.asfreq()
.fillna(0)
)
plant_data.head()
# ## Data Exploration
# ### Seasonality
from statsmodels.graphics.tsaplots import plot_acf
plot_acf(plant_data.resample("M").sum())
# Show the AR as a plot
plt.show()
# ### Modelling Processing
# + define window of past values used for prediction
# + define how many points into the future should be forecasted
series_data = plant_data.loc[plant_data.index.year >= year_start].kWh
if model_log_transformed:
series_data = series_data.apply(lambda x: np.log(x + 1))
train, test = temporal_train_test_split(series_data, test_size=forecast_horizon)
fig, ax = plt.subplots(1, figsize=plt.figaspect(0.4))
train.plot(ax=ax, label="train")
test.plot(ax=ax, label="test")
plt.title("Solar Energy Production " + plant_name)
ax.set(ylabel="kWh")
ax.set_xlabel("Month")
plt.legend()
# ## Models Fitting and Prediction
# ### 1. Machine Learning Models
fh = list(range(1, forecast_horizon + 1))
evaluation_frame = test.to_frame().rename(columns={"kWh": "y_true"}).copy()
for approach in forecasting_approaches:
print(f"Fitting Models using {approach} method")
for model_name in regression_models.keys():
print(f"# Fitting {model_name}")
forecaster = make_reduction(
regression_models[model_name](),
window_length=window_size,
strategy=approach,
)
fit_kwargs = {} if approach == "recursive" else {"fh": fh}
# Fit and predict
forecaster.fit(train, **fit_kwargs)
prediction = forecaster.predict(fh=fh)
evaluation_frame[approach + "_" + model_name] = prediction
print()
if model_log_transformed:
evaluation_frame = evaluation_frame.applymap(lambda x: np.exp(x) - 1)
evaluation_frame.head()
#
# ### 2. Statistical Models
for model_name in statistical_models.keys():
forecaster = statistical_models[model_name]()
forecaster.fit(train)
prediction = forecaster.predict(fh=fh)
evaluation_frame[model_name] = prediction
# # Models Evaluation
plot_df = evaluation_frame.reset_index().melt(id_vars="date")
px.line(plot_df, x="date", y="value", color="variable")
models = [
model_name for model_name in evaluation_frame.columns if model_name != "y_true"
]
metrics_frame = []
for metric_name in metrics.keys():
for model in models:
metrics_frame.append(
pd.DataFrame(
{
"Metric": [metric_name],
"Model": [model],
"Score": [
metrics[metric_name](
evaluation_frame["y_true"], evaluation_frame[model]
)
],
}
)
)
metrics_frame = pd.concat(metrics_frame)
metrics_frame
metric = "MAE"
px.bar(
metrics_frame.loc[metrics_frame.Metric == metric],
x="Model",
color="Model",
y="Score",
)
metric = "MSE"
px.bar(
metrics_frame.loc[metrics_frame.Metric == metric],
x="Model",
color="Model",
y="Score",
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/527/129527065.ipynb
|
solar-energy-production
|
ivnlee
|
[{"Id": 129527065, "ScriptId": 38464467, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2378690, "CreationDate": "05/14/2023 14:45:54", "VersionNumber": 1.0, "Title": "\ud83c\udf24 Forecasting Solar Power Production", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 193.0, "LinesInsertedFromPrevious": 193.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185675069, "KernelVersionId": 129527065, "SourceDatasetVersionId": 5265653}]
|
[{"Id": 5265653, "DatasetId": 3064815, "DatasourceVersionId": 5338594, "CreatorUserId": 9865529, "LicenseName": "CC0: Public Domain", "CreationDate": "03/30/2023 01:50:29", "VersionNumber": 1.0, "Title": "Solar Energy Production", "Slug": "solar-energy-production", "Subtitle": "Hourly output at each of The City of Calgary's solar photovoltaic projects", "Description": "The hourly output at each of the city of Calgary's solar photovoltaic projects and the locations of City of Calgary solar photovoltaic installations. \n\nHourly energy production data from sites with a \"public_url\" can be found [here](https://data.calgary.ca/Environment/Solar-Photovoltaic-Sites/csgq-e555).", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3064815, "CreatorUserId": 9865529, "OwnerUserId": 9865529.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5265653.0, "CurrentDatasourceVersionId": 5338594.0, "ForumId": 3127581, "Type": 2, "CreationDate": "03/30/2023 01:50:29", "LastActivityDate": "03/30/2023", "TotalViews": 8432, "TotalDownloads": 897, "TotalVotes": 35, "TotalKernels": 2}]
|
[{"Id": 9865529, "UserName": "ivnlee", "DisplayName": "Ivan Lee", "RegisterDate": "03/08/2022", "PerformanceTier": 1}]
|
# # 🌤 Forecasting Solar Power Production 🌤
# ## Machine Learning vs Statistical Models
# In this analysis I want to compare several approaches of forecasting time series data. The solar power production data seems very suitable, as it is available on a hourly basis and for a period of over 5 years.
# Following this awesome tutorial, I evaluate 3 forcasting approaches:
# 1. Direct Multi-step Forecast Strategy
# 2. Recursive Multi-step Forecast
# 3. Direct-Recursive Hybrid Strategies
# As the underlying regression models I will use a Linear Regression and Light GBM.
# To compare the performance of the ML models let's also compare them to a classical Auto ARIMA and Auto ETS model.
# The `sktime` package comes handy for this analysis, as it offers a variety of predefined time series functions, which come in handy for this analysis.
# ## Script Dependencies
import pandas as pd
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error, mean_squared_error
from lightgbm import LGBMRegressor
import numpy as np
from sktime.forecasting.compose import make_reduction
from sktime.forecasting.model_selection import temporal_train_test_split
from sktime.forecasting.arima import AutoARIMA
from sktime.forecasting.ets import AutoETS
import matplotlib.pyplot as plt
import plotly.express as px
# ## Data Import and Exploration
energy_production = pd.read_csv(
"/kaggle/input/solar-energy-production/Solar_Energy_Production.csv"
)
energy_production["date"] = pd.to_datetime(energy_production["date"])
energy_production.head()
# ## 11 Power Plants
energy_production.groupby("name").agg({"date": [min, max]})
# all plants besides Telus Spark provide > 5 years data
# # Modelling
# ## Data Preparation
## Experiment Setup
### Data Parameters
frequency = ["h", "d", "M"][2]
plant_name = "Whitehorn Multi-Service Centre"
### Modelling Parameters
window_size = 24
forecast_horizon = 12
year_start = 2017
model_log_transformed = False
forecasting_approaches = ["direct", "recursive", "dirrec"] # , 'multioutput'
regression_models = {"Lin_Reg": LinearRegression, "LGBM": LGBMRegressor}
statistical_models = {"AutoARIMA": AutoARIMA, "AutoETS": AutoETS}
## Evaluation
metrics = {"MAE": mean_absolute_error, "MSE": mean_squared_error}
# ### Data Preprocessing
plant_data = (
energy_production.loc[energy_production.name == plant_name, ["date", "kWh"]]
.set_index("date")
.resample(frequency)
.sum()
.resample(frequency)
.asfreq()
.fillna(0)
)
plant_data.head()
# ## Data Exploration
# ### Seasonality
from statsmodels.graphics.tsaplots import plot_acf
plot_acf(plant_data.resample("M").sum())
# Show the AR as a plot
plt.show()
# ### Modelling Processing
# + define window of past values used for prediction
# + define how many points into the future should be forecasted
series_data = plant_data.loc[plant_data.index.year >= year_start].kWh
if model_log_transformed:
series_data = series_data.apply(lambda x: np.log(x + 1))
train, test = temporal_train_test_split(series_data, test_size=forecast_horizon)
fig, ax = plt.subplots(1, figsize=plt.figaspect(0.4))
train.plot(ax=ax, label="train")
test.plot(ax=ax, label="test")
plt.title("Solar Energy Production " + plant_name)
ax.set(ylabel="kWh")
ax.set_xlabel("Month")
plt.legend()
# ## Models Fitting and Prediction
# ### 1. Machine Learning Models
fh = list(range(1, forecast_horizon + 1))
evaluation_frame = test.to_frame().rename(columns={"kWh": "y_true"}).copy()
for approach in forecasting_approaches:
print(f"Fitting Models using {approach} method")
for model_name in regression_models.keys():
print(f"# Fitting {model_name}")
forecaster = make_reduction(
regression_models[model_name](),
window_length=window_size,
strategy=approach,
)
fit_kwargs = {} if approach == "recursive" else {"fh": fh}
# Fit and predict
forecaster.fit(train, **fit_kwargs)
prediction = forecaster.predict(fh=fh)
evaluation_frame[approach + "_" + model_name] = prediction
print()
if model_log_transformed:
evaluation_frame = evaluation_frame.applymap(lambda x: np.exp(x) - 1)
evaluation_frame.head()
#
# ### 2. Statistical Models
for model_name in statistical_models.keys():
forecaster = statistical_models[model_name]()
forecaster.fit(train)
prediction = forecaster.predict(fh=fh)
evaluation_frame[model_name] = prediction
# # Models Evaluation
plot_df = evaluation_frame.reset_index().melt(id_vars="date")
px.line(plot_df, x="date", y="value", color="variable")
models = [
model_name for model_name in evaluation_frame.columns if model_name != "y_true"
]
metrics_frame = []
for metric_name in metrics.keys():
for model in models:
metrics_frame.append(
pd.DataFrame(
{
"Metric": [metric_name],
"Model": [model],
"Score": [
metrics[metric_name](
evaluation_frame["y_true"], evaluation_frame[model]
)
],
}
)
)
metrics_frame = pd.concat(metrics_frame)
metrics_frame
metric = "MAE"
px.bar(
metrics_frame.loc[metrics_frame.Metric == metric],
x="Model",
color="Model",
y="Score",
)
metric = "MSE"
px.bar(
metrics_frame.loc[metrics_frame.Metric == metric],
x="Model",
color="Model",
y="Score",
)
| false | 1 | 1,553 | 0 | 1,665 | 1,553 |
||
129807220
|
<jupyter_start><jupyter_text>Credit Card customers
A manager at the bank is disturbed with more and more customers leaving their credit card services. They would really appreciate if one could predict for them who is gonna get churned so they can proactively go to the customer to provide them better services and turn customers' decisions in the opposite direction
I got this dataset from a website with the URL as https://leaps.analyttica.com/home. I have been using this for a while to get datasets and accordingly work on them to produce fruitful results. The site explains how to solve a particular business problem.
Now, this dataset consists of 10,000 customers mentioning their age, salary, marital_status, credit card limit, credit card category, etc. There are nearly 18 features.
We have only 16.07% of customers who have churned. Thus, it's a bit difficult to train our model to predict churning customers.
Kaggle dataset identifier: credit-card-customers
<jupyter_script>import pandas as pd
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
import seaborn as sns
print("Setup Complete")
my_filepath = "/kaggle/input/credit-card-customers/BankChurners.csv"
# Membaca dataset
df = pd.read_csv(my_filepath)
df
# Melihat informasi dataset
df.info()
# Melihat deskripsi dataset
df.describe()
# Melihat presentase churn rate nasabah kartu kredit
plt.title("Churn Rate of Credit Card Service (Attrition_Flag)")
plt.pie(
df["Attrition_Flag"].value_counts(),
labels=df["Attrition_Flag"].value_counts().index,
autopct="%1.2f%%",
)
plt.show()
# Dari data yang diperoleh dari suatu bank karena banyaknya nasabah yang mengakhiri atau menghentikan layanan kartu kreditnya, diperoleh informasi bahwa ada sekitar 16.07% yakni 1627 nasabah (Attrited Customer) yang memilih mengakhiri atau menghentikan layanan kartu kreditnya. Hal ini dapat kita analisa lebih lanjut pada ***Credit Card Customers Dataset by Attrition Flag*** pada bagan dibawah ini:
#
# group by Attrition_Flag
attrition_group = df.groupby("Attrition_Flag")
# create subplots for each variable
fig, axs = plt.subplots(nrows=3, ncols=3, figsize=(30, 15))
fig.suptitle("Credit Card Customers Dataset by Attrition Flag")
# plot for customer age
sns.histplot(data=df, x="Customer_Age", hue="Attrition_Flag", kde=True, ax=axs[0, 0])
axs[0, 0].set_title("Customer_Age")
# plot for gender
sns.countplot(data=df, x="Gender", hue="Attrition_Flag", ax=axs[0, 1])
axs[0, 1].set_title("Gender")
for p in axs[0, 1].patches:
axs[0, 1].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for education level
sns.countplot(data=df, x="Education_Level", hue="Attrition_Flag", ax=axs[0, 2])
axs[0, 2].set_title("Education Level")
for p in axs[0, 2].patches:
axs[0, 2].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for income category
sns.countplot(data=df, x="Income_Category", hue="Attrition_Flag", ax=axs[1, 0])
axs[1, 0].set_title("Income Category")
for p in axs[1, 0].patches:
axs[1, 0].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for card utilization rate
sns.histplot(
data=df, x="Avg_Utilization_Ratio", hue="Attrition_Flag", kde=True, ax=axs[1, 1]
)
axs[1, 1].set_title("Card Utilization Rate")
# plot for total transaction amount
sns.histplot(data=df, x="Total_Trans_Amt", hue="Attrition_Flag", kde=True, ax=axs[1, 2])
axs[1, 2].set_title("Total Transaction Amount")
for p in axs[1, 2].patches:
axs[1, 2].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for credit card type
sns.countplot(data=df, x="Card_Category", hue="Attrition_Flag", ax=axs[2, 0])
axs[2, 0].set_title("Card_Category")
for p in axs[2, 0].patches:
axs[2, 0].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for dependent count
sns.countplot(data=df, x="Dependent_count", hue="Attrition_Flag", ax=axs[2, 1])
axs[2, 1].set_title("Dependent Count ")
for p in axs[2, 1].patches:
axs[2, 1].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for total transaction CT
sns.histplot(data=df, x="Total_Trans_Ct", hue="Attrition_Flag", kde=True, ax=axs[2, 2])
axs[2, 2].set_title("Total Transaction CT")
for p in axs[2, 2].patches:
axs[2, 2].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# adjust layout
plt.tight_layout()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/807/129807220.ipynb
|
credit-card-customers
|
sakshigoyal7
|
[{"Id": 129807220, "ScriptId": 38605123, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14867967, "CreationDate": "05/16/2023 15:45:05", "VersionNumber": 1.0, "Title": "Churn Rate of Credit Card Service", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 111.0, "LinesInsertedFromPrevious": 111.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 6}]
|
[{"Id": 186180242, "KernelVersionId": 129807220, "SourceDatasetVersionId": 1660340}]
|
[{"Id": 1660340, "DatasetId": 982921, "DatasourceVersionId": 1696625, "CreatorUserId": 5618523, "LicenseName": "CC0: Public Domain", "CreationDate": "11/19/2020 07:38:44", "VersionNumber": 1.0, "Title": "Credit Card customers", "Slug": "credit-card-customers", "Subtitle": "Predict Churning customers", "Description": "A manager at the bank is disturbed with more and more customers leaving their credit card services. They would really appreciate if one could predict for them who is gonna get churned so they can proactively go to the customer to provide them better services and turn customers' decisions in the opposite direction\n\nI got this dataset from a website with the URL as https://leaps.analyttica.com/home. I have been using this for a while to get datasets and accordingly work on them to produce fruitful results. The site explains how to solve a particular business problem. \n\nNow, this dataset consists of 10,000 customers mentioning their age, salary, marital_status, credit card limit, credit card category, etc. There are nearly 18 features. \n\nWe have only 16.07% of customers who have churned. Thus, it's a bit difficult to train our model to predict churning customers.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 982921, "CreatorUserId": 5618523, "OwnerUserId": 5618523.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1660340.0, "CurrentDatasourceVersionId": 1696625.0, "ForumId": 999426, "Type": 2, "CreationDate": "11/19/2020 07:38:44", "LastActivityDate": "11/19/2020", "TotalViews": 749413, "TotalDownloads": 86875, "TotalVotes": 1983, "TotalKernels": 387}]
|
[{"Id": 5618523, "UserName": "sakshigoyal7", "DisplayName": "Sakshi Goyal", "RegisterDate": "08/13/2020", "PerformanceTier": 1}]
|
import pandas as pd
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
import seaborn as sns
print("Setup Complete")
my_filepath = "/kaggle/input/credit-card-customers/BankChurners.csv"
# Membaca dataset
df = pd.read_csv(my_filepath)
df
# Melihat informasi dataset
df.info()
# Melihat deskripsi dataset
df.describe()
# Melihat presentase churn rate nasabah kartu kredit
plt.title("Churn Rate of Credit Card Service (Attrition_Flag)")
plt.pie(
df["Attrition_Flag"].value_counts(),
labels=df["Attrition_Flag"].value_counts().index,
autopct="%1.2f%%",
)
plt.show()
# Dari data yang diperoleh dari suatu bank karena banyaknya nasabah yang mengakhiri atau menghentikan layanan kartu kreditnya, diperoleh informasi bahwa ada sekitar 16.07% yakni 1627 nasabah (Attrited Customer) yang memilih mengakhiri atau menghentikan layanan kartu kreditnya. Hal ini dapat kita analisa lebih lanjut pada ***Credit Card Customers Dataset by Attrition Flag*** pada bagan dibawah ini:
#
# group by Attrition_Flag
attrition_group = df.groupby("Attrition_Flag")
# create subplots for each variable
fig, axs = plt.subplots(nrows=3, ncols=3, figsize=(30, 15))
fig.suptitle("Credit Card Customers Dataset by Attrition Flag")
# plot for customer age
sns.histplot(data=df, x="Customer_Age", hue="Attrition_Flag", kde=True, ax=axs[0, 0])
axs[0, 0].set_title("Customer_Age")
# plot for gender
sns.countplot(data=df, x="Gender", hue="Attrition_Flag", ax=axs[0, 1])
axs[0, 1].set_title("Gender")
for p in axs[0, 1].patches:
axs[0, 1].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for education level
sns.countplot(data=df, x="Education_Level", hue="Attrition_Flag", ax=axs[0, 2])
axs[0, 2].set_title("Education Level")
for p in axs[0, 2].patches:
axs[0, 2].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for income category
sns.countplot(data=df, x="Income_Category", hue="Attrition_Flag", ax=axs[1, 0])
axs[1, 0].set_title("Income Category")
for p in axs[1, 0].patches:
axs[1, 0].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for card utilization rate
sns.histplot(
data=df, x="Avg_Utilization_Ratio", hue="Attrition_Flag", kde=True, ax=axs[1, 1]
)
axs[1, 1].set_title("Card Utilization Rate")
# plot for total transaction amount
sns.histplot(data=df, x="Total_Trans_Amt", hue="Attrition_Flag", kde=True, ax=axs[1, 2])
axs[1, 2].set_title("Total Transaction Amount")
for p in axs[1, 2].patches:
axs[1, 2].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for credit card type
sns.countplot(data=df, x="Card_Category", hue="Attrition_Flag", ax=axs[2, 0])
axs[2, 0].set_title("Card_Category")
for p in axs[2, 0].patches:
axs[2, 0].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for dependent count
sns.countplot(data=df, x="Dependent_count", hue="Attrition_Flag", ax=axs[2, 1])
axs[2, 1].set_title("Dependent Count ")
for p in axs[2, 1].patches:
axs[2, 1].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# plot for total transaction CT
sns.histplot(data=df, x="Total_Trans_Ct", hue="Attrition_Flag", kde=True, ax=axs[2, 2])
axs[2, 2].set_title("Total Transaction CT")
for p in axs[2, 2].patches:
axs[2, 2].annotate(
f"\n{p.get_height():.0f}",
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
fontsize=10,
color="black",
xytext=(0, 13),
textcoords="offset points",
)
# adjust layout
plt.tight_layout()
| false | 0 | 1,697 | 6 | 1,924 | 1,697 |
||
129807717
|
<jupyter_start><jupyter_text>Mental Health and Suicide Rates
### Context
Close to 800 000 people die due to suicide every year, which is one person every 40 seconds. Suicide is a global phenomenon and occurs throughout the lifespan. Effective and evidence-based interventions can be implemented at population, sub-population and individual levels to prevent suicide and suicide attempts. There are indications that for each adult who died by suicide there may have been more than 20 others attempting suicide.
Suicide is a complex issue and therefore suicide prevention efforts require coordination and collaboration among multiple sectors of society, including the health sector and other sectors such as education, labour, agriculture, business, justice, law, defense, politics, and the media. These efforts must be comprehensive and integrated as no single approach alone can make an impact on an issue as complex as suicide.
### Do leave an upvote if you found this dataset useful!
Kaggle dataset identifier: mental-health-and-suicide-rates
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/mental-health-and-suicide-rates/Age-standardized suicide rates.csv"
)
# df2 = pd.read_csv("/kaggle/input/mental-health-and-suicide-rates/Crude suicide rates.csv")
df
# df2.describe()
import matplotlib.pyplot as plt
import seaborn as sns
countplot = sns.countplot(data=df, x="2016")
# ### in the Year 2016 whose have a higher suicide rate, Females or Males?
year_16 = df[df["2016"] > 0]
by_sex = year_16.groupby("Sex", as_index=False)
by_sex_16 = by_sex["2016"].mean()
barplot = sns.barplot(x="Sex", y="2016", data=by_sex_16)
# ### as the figure shows, males have a higher suicide rate than females, because 2 reasons:
# 1. Men have an easily accessed weapons than women: in the USA 6 men out of 10 have weapons
# 2. When men decide to suicide they really meant it, and they do it
# 3. Personal Opinion: Women talk, men don't talk
# ### in 2016 which Country has the most suicide rate?
avg = year_16.groupby("Country", as_index=False)["2016"].mean()
# avg = avg.head(3)
relplot = sns.relplot(x="Country", y="2016", data=avg)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/807/129807717.ipynb
|
mental-health-and-suicide-rates
|
twinkle0705
|
[{"Id": 129807717, "ScriptId": 38603079, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14721492, "CreationDate": "05/16/2023 15:48:59", "VersionNumber": 1.0, "Title": "Mental Health1", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 49.0, "LinesInsertedFromPrevious": 49.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186180872, "KernelVersionId": 129807717, "SourceDatasetVersionId": 1338480}]
|
[{"Id": 1338480, "DatasetId": 748724, "DatasourceVersionId": 1370824, "CreatorUserId": 3649586, "LicenseName": "Attribution-NonCommercial-ShareAlike 3.0 IGO (CC BY-NC-SA 3.0 IGO)", "CreationDate": "07/15/2020 12:33:00", "VersionNumber": 2.0, "Title": "Mental Health and Suicide Rates", "Slug": "mental-health-and-suicide-rates", "Subtitle": "Suicide Rates of age groups in different countries along with Health Facility", "Description": "### Context\n\nClose to 800 000 people die due to suicide every year, which is one person every 40 seconds. Suicide is a global phenomenon and occurs throughout the lifespan. Effective and evidence-based interventions can be implemented at population, sub-population and individual levels to prevent suicide and suicide attempts. There are indications that for each adult who died by suicide there may have been more than 20 others attempting suicide.\n\nSuicide is a complex issue and therefore suicide prevention efforts require coordination and collaboration among multiple sectors of society, including the health sector and other sectors such as education, labour, agriculture, business, justice, law, defense, politics, and the media. These efforts must be comprehensive and integrated as no single approach alone can make an impact on an issue as complex as suicide.\n\n### Do leave an upvote if you found this dataset useful!", "VersionNotes": "updated files", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 748724, "CreatorUserId": 3649586, "OwnerUserId": 3649586.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1338480.0, "CurrentDatasourceVersionId": 1370824.0, "ForumId": 763628, "Type": 2, "CreationDate": "06/30/2020 16:51:51", "LastActivityDate": "06/30/2020", "TotalViews": 106199, "TotalDownloads": 12135, "TotalVotes": 211, "TotalKernels": 10}]
|
[{"Id": 3649586, "UserName": "twinkle0705", "DisplayName": "Twinkle Khanna", "RegisterDate": "09/01/2019", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/mental-health-and-suicide-rates/Age-standardized suicide rates.csv"
)
# df2 = pd.read_csv("/kaggle/input/mental-health-and-suicide-rates/Crude suicide rates.csv")
df
# df2.describe()
import matplotlib.pyplot as plt
import seaborn as sns
countplot = sns.countplot(data=df, x="2016")
# ### in the Year 2016 whose have a higher suicide rate, Females or Males?
year_16 = df[df["2016"] > 0]
by_sex = year_16.groupby("Sex", as_index=False)
by_sex_16 = by_sex["2016"].mean()
barplot = sns.barplot(x="Sex", y="2016", data=by_sex_16)
# ### as the figure shows, males have a higher suicide rate than females, because 2 reasons:
# 1. Men have an easily accessed weapons than women: in the USA 6 men out of 10 have weapons
# 2. When men decide to suicide they really meant it, and they do it
# 3. Personal Opinion: Women talk, men don't talk
# ### in 2016 which Country has the most suicide rate?
avg = year_16.groupby("Country", as_index=False)["2016"].mean()
# avg = avg.head(3)
relplot = sns.relplot(x="Country", y="2016", data=avg)
| false | 1 | 594 | 0 | 852 | 594 |
||
129807753
|
# # Libraries and Data imports
import pandas as pd
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
# from sklearn import preprocessing
from sklearn.preprocessing import MinMaxScaler
# ## Models
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
df_orig = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
df_orig.head()
# # EDA
sns.swarmplot(x=df_orig["RoomService"], y=df_orig["Transported"])
# # Data modification
df_orig.info()
# V1
# cols_to_use = ['HomePlanet','CryoSleep','Cabin','Age','VIP']
# V2 This do not chage the result
# cols_to_use = ['HomePlanet','CryoSleep','Cabin','Age','VIP','Age']
# V3
# cols_to_use = ['HomePlanet','CryoSleep','Age','VIP']
# V4 ~50%
# cols_to_use = ['VIP']
# V5 ~70%
# cols_to_use = ['CryoSleep']
# V6 ~70%
# cols_to_use = ['CryoSleep','HomePlanet']
# V7
# cols_to_use = ['HomePlanet']
# V8
# cols_to_use = ['Age','RoomService']
# V10 ~70%
# cols_to_use = ['Age','RoomService','VRDeck']
# V11 73%
# cols_to_use = ['Age','RoomService','VRDeck','Spa']
# V12 73%
# cols_to_use = ['Age','RoomService','VRDeck','Spa','ShoppingMall']
# V13 73%
# cols_to_use = ['Age','RoomService','VRDeck','Spa','FoodCourt']
# V14 73%
cols_to_use = ["Age", "RoomService", "VRDeck", "Spa", "CryoSleep"]
cols_to_train = [*cols_to_use, "Transported"]
df = df_orig[cols_to_train]
df.head()
def CategoriseCabin(dataframe):
df_temp = dataframe
df_temp["Cabin_Deck"] = df_temp["Cabin"].apply(lambda x: str(x).split("/")[0])
df_temp["Cabin_Num"] = df_temp["Cabin"].apply(lambda x: str(x)[2])
df_temp["Cabin_Side"] = df_temp["Cabin"].apply(lambda x: str(x).split("/")[-1])
return df_temp.drop(["Cabin"], axis=1)
df = CategoriseCabin(df)
df.head()
print("deck unique", df["Cabin_Deck"].unique())
print("num unique", df["Cabin_Num"].unique())
print("side unique", df["Cabin_Side"].unique())
def convertToInt(dataframe):
df_temp = dataframe
df_temp["CryoSleep"] = df_temp["CryoSleep"].fillna(-1)
df_temp["CryoSleep"] = df_temp["CryoSleep"].astype(int)
df_temp["VIP"] = df_temp["VIP"].fillna(-1)
df_temp["VIP"] = df_temp["VIP"].astype(int)
df_temp.loc[df_temp["Cabin_Num"] == "n", "Cabin_Num"] = -1
# f_temp['Cabin_Num'] = df_temp['Cabin_Num'].fillna(-1)
df_temp["Cabin_Num"] = df_temp["Cabin_Num"].astype(int)
return df_temp
def convertToIntVip(dataframe):
df_temp = dataframe
df_temp["VIP"] = df_temp["VIP"].fillna(-1)
df_temp["VIP"] = df_temp["VIP"].astype(int)
return df_temp
def convertToIntCryo(dataframe):
df_temp = dataframe
df_temp["CryoSleep"] = df_temp["CryoSleep"].fillna(-1)
df_temp["CryoSleep"] = df_temp["CryoSleep"].astype(int)
return df_temp
def convertToIntWithoutCabin(dataframe):
df_temp = dataframe
df_temp["CryoSleep"] = df_temp["CryoSleep"].fillna(-1)
df_temp["CryoSleep"] = df_temp["CryoSleep"].astype(int)
df_temp["VIP"] = df_temp["VIP"].fillna(-1)
df_temp["VIP"] = df_temp["VIP"].astype(int)
return df_temp
# df = convertToInt(df)
# df = convertToIntWithoutCabin(df)
# df = convertToIntVip(df)
df = convertToIntCryo(df)
df.head()
def oneHotEnc(data):
ohe = OneHotEncoder(handle_unknown="ignore", sparse=False)
cols_to_tranform = ["HomePlanet", "Cabin_Deck", "Cabin_Side"]
df_temp = data.copy()
transformed = pd.DataFrame(ohe.fit_transform(data[cols_to_tranform]).astype(int))
df_temp = pd.concat([df_temp, transformed], axis=1).drop(cols_to_tranform, axis=1)
return df_temp
def oneHotEncWithoutCabin(data):
ohe = OneHotEncoder(handle_unknown="ignore", sparse=False)
cols_to_tranform = ["HomePlanet"]
df_temp = data.copy()
transformed = pd.DataFrame(ohe.fit_transform(data[cols_to_tranform]).astype(int))
df_temp = pd.concat([df_temp, transformed], axis=1).drop(cols_to_tranform, axis=1)
return df_temp
# df = oneHotEnc(df)
df = oneHotEncWithoutCabin(df)
df.head()
df.columns = df.columns.astype(str)
# df = df.fillna(-1)
df = df.fillna(0)
df.head()
# ## Normalize
def normaAge(data):
df_temp = data[["Age", "RoomService"]]
# df_temp = data.drop(["Transported","Age"], axis=1)
scaler = MinMaxScaler()
# mm_scaler = preprocessing.MinMaxScaler()
df_temp = pd.DataFrame(
scaler.fit_transform(df_temp), columns=["Age", "RoomService"]
)
data[["Age", "RoomService"]] = df_temp
return data
from sklearn.preprocessing import StandardScaler
def stdAge(data):
cols = ["Age", "RoomService", "VRDeck", "Spa"]
df_temp = data[cols]
print(df_temp)
# df_temp = data.drop(["Transported","Age"], axis=1)
scaler = StandardScaler()
# mm_scaler = preprocessing.MinMaxScaler()
df_temp = pd.DataFrame(scaler.fit_transform(df_temp), columns=cols)
data[cols] = df_temp
return data
df_te = normaAge(df)
df_te.head()
df_te = stdAge(df)
df_te.head()
# # Training
y = df["Transported"]
X = df.drop(["Transported"], axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y)
X_train.info()
# ## Knn
knn_model = KNeighborsClassifier(n_neighbors=4)
knn_model.fit(X_train, y_train)
knn_prediction = knn_model.predict(X_test)
print(classification_report(y_test, knn_prediction))
# The V1 is probabily wrong
# V1 accuracy 0.81
# V2 accuracy 0.63
# V3 accuracy 0.70
# V4 accuracy 0.51
# V5 accuracy 0.70
# V6 accuracy 0.67
# V7 accuracy 0.60
# V8 accuracy 0.51
# V9(std) accuracy 0.60
# V9(nor) accuracy 0.59
# V10 accuracy 0.66
# V11 accuracy 0.71
# V12 accuracy 0.70
# V13 accuracy 0.72
# V14 accuracy 0.71
# ## Naive Bayes
nb_model = GaussianNB()
nb_model = nb_model.fit(X_train, y_train)
pred_nb = nb_model.predict(X_test)
print(classification_report(y_test, pred_nb))
# V1 accuracy 0.71
# V2 accuracy 0.72
# V3 accuracy 0.69
# V4 accuracy 0.51
# V5 accuracy 0.70
# V6 accuracy 0.70
# V7 accuracy 0.59
# V8 accuracy 0.53
# V9(std) accuracy 0.61
# V9(nor) accuracy 0.59
# V10 accuracy 0.64
# V11 accuracy 0.65
# V12 accuracy 0.65
# V13 accuracy 0.65
# V14 accuracy 0.66
# ## Log regression
lr_model = LogisticRegression()
lnr_model = lr_model.fit(X_train, y_train)
y_pred_lr = lr_model.predict(X_test)
print(classification_report(y_test, y_pred_lr))
# V1 accuracy 0.73
# V2 accuracy 0.72
# V3 accuracy 0.70
# V4 accuracy 0.51
# V5 accuracy 0.70
# V6 accuracy 0.70
# V7 accuracy 0.59
# V8 accuracy 0.52
# V9(std) accuracy 0.64
# V9(nor) accuracy 0.61
# V10 accuracy 0.71
# V11 accuracy 0.76
# V12 accuracy 0.75
# V13 accuracy 0.76
# V14 accuracy 0.75
# ## Random forest
forest = RandomForestClassifier(n_estimators=100, random_state=100)
forest.fit(X_train, y_train)
forest_predict = forest.predict(X_test)
print(classification_report(y_test, forest_predict))
# V1 accuracy 0.70
# V2 accuracy 0.70
# V3 accuracy 0.72
# V4 accuracy 0.51
# V5 accuracy 0.70
# V6 accuracy 0.73
# V7 accuracy 0.59
# V8 accuracy 0.54
# V9(std) accuracy 0.61
# V9(nor) accuracy 0.61
# V10 accuracy 0.71
# V11 accuracy 0.76
# V12 accuracy 0.74
# V12 accuracy 0.77
# V13 accuracy 0.74
# # Result
df_test_og = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
df_test = df_test_og
df_test.head()
df_test = df_test[cols_to_use]
# df_test = CategoriseCabin(df_test)
df_test = convertToIntCryo(df_test)
df_test = stdAge(df_test)
df_test = df_test.fillna(0)
# df_test = oneHotEnc(df_test)
# f_test =
# df_test =
# print('deck unique',df_test['Cabin_Deck'].unique())
# print('num unique',df_test['Cabin_Num'].unique())
# print('side unique',df_test['Cabin_Side'].unique())
df_test.head()
df_test.columns = df_test.columns.astype(str)
df_test = df_test.fillna(-1)
df_test.head()
pred_result = forest.predict(df_test)
print(pred_result)
Survived = pd.DataFrame(
pred_result, index=df_test_og["PassengerId"], columns=["Transported"]
)
print(Survived.columns)
Survived.head()
Survived.to_csv("Second_model.csv", header=True)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/807/129807753.ipynb
| null | null |
[{"Id": 129807753, "ScriptId": 38378007, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13768217, "CreationDate": "05/16/2023 15:49:19", "VersionNumber": 1.0, "Title": "Space-titanic", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 320.0, "LinesInsertedFromPrevious": 320.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Libraries and Data imports
import pandas as pd
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
# from sklearn import preprocessing
from sklearn.preprocessing import MinMaxScaler
# ## Models
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
df_orig = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
df_orig.head()
# # EDA
sns.swarmplot(x=df_orig["RoomService"], y=df_orig["Transported"])
# # Data modification
df_orig.info()
# V1
# cols_to_use = ['HomePlanet','CryoSleep','Cabin','Age','VIP']
# V2 This do not chage the result
# cols_to_use = ['HomePlanet','CryoSleep','Cabin','Age','VIP','Age']
# V3
# cols_to_use = ['HomePlanet','CryoSleep','Age','VIP']
# V4 ~50%
# cols_to_use = ['VIP']
# V5 ~70%
# cols_to_use = ['CryoSleep']
# V6 ~70%
# cols_to_use = ['CryoSleep','HomePlanet']
# V7
# cols_to_use = ['HomePlanet']
# V8
# cols_to_use = ['Age','RoomService']
# V10 ~70%
# cols_to_use = ['Age','RoomService','VRDeck']
# V11 73%
# cols_to_use = ['Age','RoomService','VRDeck','Spa']
# V12 73%
# cols_to_use = ['Age','RoomService','VRDeck','Spa','ShoppingMall']
# V13 73%
# cols_to_use = ['Age','RoomService','VRDeck','Spa','FoodCourt']
# V14 73%
cols_to_use = ["Age", "RoomService", "VRDeck", "Spa", "CryoSleep"]
cols_to_train = [*cols_to_use, "Transported"]
df = df_orig[cols_to_train]
df.head()
def CategoriseCabin(dataframe):
df_temp = dataframe
df_temp["Cabin_Deck"] = df_temp["Cabin"].apply(lambda x: str(x).split("/")[0])
df_temp["Cabin_Num"] = df_temp["Cabin"].apply(lambda x: str(x)[2])
df_temp["Cabin_Side"] = df_temp["Cabin"].apply(lambda x: str(x).split("/")[-1])
return df_temp.drop(["Cabin"], axis=1)
df = CategoriseCabin(df)
df.head()
print("deck unique", df["Cabin_Deck"].unique())
print("num unique", df["Cabin_Num"].unique())
print("side unique", df["Cabin_Side"].unique())
def convertToInt(dataframe):
df_temp = dataframe
df_temp["CryoSleep"] = df_temp["CryoSleep"].fillna(-1)
df_temp["CryoSleep"] = df_temp["CryoSleep"].astype(int)
df_temp["VIP"] = df_temp["VIP"].fillna(-1)
df_temp["VIP"] = df_temp["VIP"].astype(int)
df_temp.loc[df_temp["Cabin_Num"] == "n", "Cabin_Num"] = -1
# f_temp['Cabin_Num'] = df_temp['Cabin_Num'].fillna(-1)
df_temp["Cabin_Num"] = df_temp["Cabin_Num"].astype(int)
return df_temp
def convertToIntVip(dataframe):
df_temp = dataframe
df_temp["VIP"] = df_temp["VIP"].fillna(-1)
df_temp["VIP"] = df_temp["VIP"].astype(int)
return df_temp
def convertToIntCryo(dataframe):
df_temp = dataframe
df_temp["CryoSleep"] = df_temp["CryoSleep"].fillna(-1)
df_temp["CryoSleep"] = df_temp["CryoSleep"].astype(int)
return df_temp
def convertToIntWithoutCabin(dataframe):
df_temp = dataframe
df_temp["CryoSleep"] = df_temp["CryoSleep"].fillna(-1)
df_temp["CryoSleep"] = df_temp["CryoSleep"].astype(int)
df_temp["VIP"] = df_temp["VIP"].fillna(-1)
df_temp["VIP"] = df_temp["VIP"].astype(int)
return df_temp
# df = convertToInt(df)
# df = convertToIntWithoutCabin(df)
# df = convertToIntVip(df)
df = convertToIntCryo(df)
df.head()
def oneHotEnc(data):
ohe = OneHotEncoder(handle_unknown="ignore", sparse=False)
cols_to_tranform = ["HomePlanet", "Cabin_Deck", "Cabin_Side"]
df_temp = data.copy()
transformed = pd.DataFrame(ohe.fit_transform(data[cols_to_tranform]).astype(int))
df_temp = pd.concat([df_temp, transformed], axis=1).drop(cols_to_tranform, axis=1)
return df_temp
def oneHotEncWithoutCabin(data):
ohe = OneHotEncoder(handle_unknown="ignore", sparse=False)
cols_to_tranform = ["HomePlanet"]
df_temp = data.copy()
transformed = pd.DataFrame(ohe.fit_transform(data[cols_to_tranform]).astype(int))
df_temp = pd.concat([df_temp, transformed], axis=1).drop(cols_to_tranform, axis=1)
return df_temp
# df = oneHotEnc(df)
df = oneHotEncWithoutCabin(df)
df.head()
df.columns = df.columns.astype(str)
# df = df.fillna(-1)
df = df.fillna(0)
df.head()
# ## Normalize
def normaAge(data):
df_temp = data[["Age", "RoomService"]]
# df_temp = data.drop(["Transported","Age"], axis=1)
scaler = MinMaxScaler()
# mm_scaler = preprocessing.MinMaxScaler()
df_temp = pd.DataFrame(
scaler.fit_transform(df_temp), columns=["Age", "RoomService"]
)
data[["Age", "RoomService"]] = df_temp
return data
from sklearn.preprocessing import StandardScaler
def stdAge(data):
cols = ["Age", "RoomService", "VRDeck", "Spa"]
df_temp = data[cols]
print(df_temp)
# df_temp = data.drop(["Transported","Age"], axis=1)
scaler = StandardScaler()
# mm_scaler = preprocessing.MinMaxScaler()
df_temp = pd.DataFrame(scaler.fit_transform(df_temp), columns=cols)
data[cols] = df_temp
return data
df_te = normaAge(df)
df_te.head()
df_te = stdAge(df)
df_te.head()
# # Training
y = df["Transported"]
X = df.drop(["Transported"], axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y)
X_train.info()
# ## Knn
knn_model = KNeighborsClassifier(n_neighbors=4)
knn_model.fit(X_train, y_train)
knn_prediction = knn_model.predict(X_test)
print(classification_report(y_test, knn_prediction))
# The V1 is probabily wrong
# V1 accuracy 0.81
# V2 accuracy 0.63
# V3 accuracy 0.70
# V4 accuracy 0.51
# V5 accuracy 0.70
# V6 accuracy 0.67
# V7 accuracy 0.60
# V8 accuracy 0.51
# V9(std) accuracy 0.60
# V9(nor) accuracy 0.59
# V10 accuracy 0.66
# V11 accuracy 0.71
# V12 accuracy 0.70
# V13 accuracy 0.72
# V14 accuracy 0.71
# ## Naive Bayes
nb_model = GaussianNB()
nb_model = nb_model.fit(X_train, y_train)
pred_nb = nb_model.predict(X_test)
print(classification_report(y_test, pred_nb))
# V1 accuracy 0.71
# V2 accuracy 0.72
# V3 accuracy 0.69
# V4 accuracy 0.51
# V5 accuracy 0.70
# V6 accuracy 0.70
# V7 accuracy 0.59
# V8 accuracy 0.53
# V9(std) accuracy 0.61
# V9(nor) accuracy 0.59
# V10 accuracy 0.64
# V11 accuracy 0.65
# V12 accuracy 0.65
# V13 accuracy 0.65
# V14 accuracy 0.66
# ## Log regression
lr_model = LogisticRegression()
lnr_model = lr_model.fit(X_train, y_train)
y_pred_lr = lr_model.predict(X_test)
print(classification_report(y_test, y_pred_lr))
# V1 accuracy 0.73
# V2 accuracy 0.72
# V3 accuracy 0.70
# V4 accuracy 0.51
# V5 accuracy 0.70
# V6 accuracy 0.70
# V7 accuracy 0.59
# V8 accuracy 0.52
# V9(std) accuracy 0.64
# V9(nor) accuracy 0.61
# V10 accuracy 0.71
# V11 accuracy 0.76
# V12 accuracy 0.75
# V13 accuracy 0.76
# V14 accuracy 0.75
# ## Random forest
forest = RandomForestClassifier(n_estimators=100, random_state=100)
forest.fit(X_train, y_train)
forest_predict = forest.predict(X_test)
print(classification_report(y_test, forest_predict))
# V1 accuracy 0.70
# V2 accuracy 0.70
# V3 accuracy 0.72
# V4 accuracy 0.51
# V5 accuracy 0.70
# V6 accuracy 0.73
# V7 accuracy 0.59
# V8 accuracy 0.54
# V9(std) accuracy 0.61
# V9(nor) accuracy 0.61
# V10 accuracy 0.71
# V11 accuracy 0.76
# V12 accuracy 0.74
# V12 accuracy 0.77
# V13 accuracy 0.74
# # Result
df_test_og = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
df_test = df_test_og
df_test.head()
df_test = df_test[cols_to_use]
# df_test = CategoriseCabin(df_test)
df_test = convertToIntCryo(df_test)
df_test = stdAge(df_test)
df_test = df_test.fillna(0)
# df_test = oneHotEnc(df_test)
# f_test =
# df_test =
# print('deck unique',df_test['Cabin_Deck'].unique())
# print('num unique',df_test['Cabin_Num'].unique())
# print('side unique',df_test['Cabin_Side'].unique())
df_test.head()
df_test.columns = df_test.columns.astype(str)
df_test = df_test.fillna(-1)
df_test.head()
pred_result = forest.predict(df_test)
print(pred_result)
Survived = pd.DataFrame(
pred_result, index=df_test_og["PassengerId"], columns=["Transported"]
)
print(Survived.columns)
Survived.head()
Survived.to_csv("Second_model.csv", header=True)
| false | 0 | 3,136 | 0 | 3,136 | 3,136 |
||
129807058
|
<jupyter_start><jupyter_text>AMEX_data_sampled
Kaggle dataset identifier: amex-data-sampled
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### 🔥 OptBinning 라이브러리를 활용한 신용평가 모델 개발
# > #### 데이터를 불러와 전처리하는 과정은 이전과 동일합니다. 다만 임의로 설명 변수를 선택하기 때문에 선택된 설명 변수는 다를 수 있습니다.
import warnings
warnings.filterwarnings("ignore", module="sklearn.metrics.cluster")
df = pd.read_pickle("/kaggle/input/amex-data-sampled/train_df_sample.pkl")
# > ##### 💡 데이터는 위에서 프린트된 경로를 사용해야 합니다
def drop_null_cols(df, threshold=0.8):
"""
데이터프레임에서 결측치 비율이 threshold 이상인 변수를 제거하는 함수
"""
null_percent = df.isnull().mean()
drop_cols = list(null_percent[null_percent >= threshold].index)
df = df.drop(drop_cols, axis=1)
print(f"Dropped {len(drop_cols)} columns: {', '.join(drop_cols)}")
return df
df = df.reset_index()
import hashlib
def encode_customer_id(id_str):
encoded_id = hashlib.sha256(id_str.encode("utf-8")).hexdigest()[:16]
return encoded_id
df["customer_ID"] = df["customer_ID"].apply(encode_customer_id)
df = drop_null_cols(df)
cat_features = [
"B_30",
"B_38",
"D_114",
"D_116",
"D_117",
"D_120",
"D_126",
"D_63",
"D_64",
"D_68",
]
cat_features = [f"{cf}_last" for cf in cat_features]
import random
num_cols = df.select_dtypes(include=np.number).columns.tolist()
num_cols = [col for col in num_cols if "target" not in col and col not in cat_features]
num_cols_sample = random.sample([col for col in num_cols if "target" not in col], 100)
feature_list = num_cols_sample + cat_features
all_list = feature_list + ["target"]
df = df[all_list]
import gc
gc.collect()
for categorical_feature in cat_features:
if df[categorical_feature].dtype == "float16":
df[categorical_feature] = df[categorical_feature].astype(str)
if df[categorical_feature].dtype == "category":
df[categorical_feature] = df[categorical_feature].astype(str)
elif df[categorical_feature].dtype == "object":
df[categorical_feature] = df[categorical_feature].astype(str)
from sklearn.preprocessing import LabelEncoder
le_encoder = LabelEncoder()
for categorical_feature in cat_features:
df[categorical_feature].fillna(value="NaN", inplace=True)
df[categorical_feature] = le_encoder.fit_transform(df[categorical_feature])
from sklearn.impute import SimpleImputer
def impute_nan(df, num_cols, strategy="mean"):
"""
NaN 값을 strategy에 따라 num_cols에 대해 impute하는 함수
:param df: DataFrame
:param num_cols: list, imputation 대상 numeric column 리스트
:param strategy: str, imputation 전략 (default: 'mean')
:return: DataFrame, imputed DataFrame
"""
imputer = SimpleImputer(strategy=strategy)
df[num_cols] = imputer.fit_transform(df[num_cols])
return df
df = impute_nan(df, num_cols_sample, strategy="mean")
df.head()
import plotly.express as px
fig2 = px.pie(
df,
names="target",
height=400,
width=600,
hole=0.7,
title="target class Overview",
color_discrete_sequence=["#4c78a8", "#72b7b2"],
)
fig2.update_traces(
hovertemplate=None, textposition="outside", textinfo="percent+label", rotation=0
)
fig2.update_layout(
margin=dict(t=100, b=30, l=0, r=0),
showlegend=False,
plot_bgcolor="#fafafa",
paper_bgcolor="#fafafa",
title_font=dict(size=20, color="#555", family="Lato, sans-serif"),
font=dict(size=17, color="#8a8d93"),
hoverlabel=dict(bgcolor="#444", font_size=13, font_family="Lato, sans-serif"),
)
fig2.show()
# # optbinning library 설치
# > #### !pip install은 Python 환경에서 패키지를 설치하기 위한 표준적인 방법이며, %pip install은 IPython 환경에서 제공되는 패키지 관리 명령어입니다. 일반적으로 Jupyter Notebook에서는 !pip install이 더 일반적으로 사용되며, %pip install은 특정한 경우에 사용될 수 있습니다.
from optbinning import BinningProcess
#
# 💡 Notes:
# Binning Process는 데이터셋의 변수들을 이진, 연속형 또는 다중 클래스 타겟 데이터 유형을 기반으로 최적의 구간화(bin)을 계산하기 위한 프로세스입니다.
# 매개변수:
# * variable_names (array-like): 변수 이름의 리스트입니다.
# * max_n_prebins (int, default=20): Pre-binning(사전 구간화) 후의 최대 구간 수입니다.
# * min_prebin_size (float, default=0.05): 각 Pre-bin에 대한 최소 레코드 수의 비율입니다.
# * min_n_bins (int 또는 None, optional, default=None): 최소 구간 수입니다. None인 경우, min_n_bins은 [0, max_n_prebins] 범위의 값입니다.
# * max_n_bins (int 또는 None, optional, default=None): 최대 구간 수입니다. None인 경우, max_n_bins은 [0, max_n_prebins] 범위의 값입니다.
# * min_bin_size (float 또는 None, optional, default=None): 각 구간의 최소 레코드 수의 비율입니다. None인 경우, min_bin_size = min_prebin_size입니다.
# * max_bin_size (float 또는 None, optional, default=None): 각 구간의 최대 레코드 수의 비율입니다. None인 경우, max_bin_size = 1.0입니다.
# * max_pvalue (float 또는 None, optional, default=None): 구간 간의 최대 p-value입니다.
# * max_pvalue_policy (str, optional, default="consecutive"): p-value 조건을 만족하지 않는 구간을 결정하는 방법입니다. "consecutive"는 연속된 구간을 비교하고, "all"은 모든 구간을 비교합니다.
# * selection_criteria (dict 또는 None, optional, default=None): 변수 선택 기준입니다. 자세한 내용은 참고 사항을 참조하세요.
# * fixed_variables (array-like 또는 None, optional, default=None): 고정할 변수의 리스트입니다. 선택 기준을 만족하지 않을 경우에도 이러한 변수를 유지합니다.
# * special_codes (array-like 또는 None, optional, default=None): 특별한 코드의 리스트입니다. 이러한 코드를 사용하여 따로 처리해야 하는 데이터 값을 지정할 수 있습니다.
# * split_digits (int 또는 None, optional, default=None): 분할 지점의 유효 숫자 자릿수입니다. split_digits가 0으로 설정되면 분할 지점은 정수로 처리됩니다. None인 경우, 분할 지점의 모든 유효 숫자 자릿수가 고려됩니다.
# * categorical_variables (array-like 또는 None, optional, default=None): 범주형 변수로 간주할 수치 변수의 리스트입니다. 이는 명목 변수입니다. 타겟 유형이 다중 클래스인 경우에는 해당되지 않습니다.
# * binning_fit_params (dict 또는 None, optional, default=None): 특정 변수에 대한 최적
selection_criteria = {
"iv": {"min": 0.025, "max": 0.7, "strategy": "highest", "top": 20},
"quality_score": {"min": 0.01},
}
# > #### Feature Selection 프로세스를 간소화 할 수 있습니다. 조건을 정해두면 해당조건에 맞는 변수를 추출할 수 있습니다.
binning_process = BinningProcess(
feature_list,
categorical_variables=cat_features,
selection_criteria=selection_criteria,
)
X = df[feature_list]
y = df["target"]
binning_process.fit(X, y)
binning_process.information(print_level=2)
binning_process.summary()
# > #### 선택된 변수만 보고싶다면 아래 코드를 참조해주세요.
#
summary = binning_process.summary()
selected_summary = summary[summary["selected"] == True]
selected_summary
optb = binning_process.get_binned_variable("D_42_mean")
optb.binning_table.build()
# > #### 임의로 변수를 선택하기 때문에 위 변수가 없다면 에러가 날 수 있습니다. 확인 후 실행 바랍니다.
# > #### 📊 구간화 그래프를 살펴봅시다. binning_table.plot()을 사용하면 쉽게 그래프를 그릴 수 있습니다.
optb.binning_table.plot(metric="event_rate")
# > #### 위 변수 같은 경우는, 2 구간에서 불량율이 매우 높게 나타납니다. 이벤트 발생 확률 또한 단조증가함을 알 수 있습니다. 일반적으로 구간화는 이벤트 발생 확률이 단조 증가하면 좋다고 판단합니다.
# > ##### optb.binning_table.plot(metric="event_rate")에서 metric은 그래프를 그릴 때 사용할 메트릭(metric)을 지정하는 매개변수입니다. 이 메트릭은 각 구간(bin)의 값을 기준으로 그래프를 그릴 때 사용됩니다. 예를 들어, "event_rate"를 지정하면 각 구간의 이벤트 발생 비율을 기준으로 그래프를 그립니다.
# > ##### optb.binning_table.plot()은 optbinning 패키지에서 사용되는 BinningTable 객체의 메서드입니다. BinningTable 객체는 변수의 구간(bin) 정보를 저장하고 분석 결과를 시각화할 수 있는 기능을 제공합니다. 이를 통해 각 구간의 특성을 시각적으로 확인하고 모델 개발에 유용한 통찰력을 얻을 수 있습니다. plot() 메서드를 사용하여 구간별 메트릭에 대한 그래프를 생성하고 시각화할 수 있습니다. metric 매개변수를 적절히 설정하여 원하는 메트릭에 대한 그래프를 생성할 수 있습니다.
optb = binning_process.get_binned_variable("D_68_last")
optb.binning_table.build()
optb.binning_table.plot(metric="event_rate")
# > #### 어떤 변수들이 선택되었는지 살펴봅시다.
binning_process.get_support(names=True)
# > #### 이전 노트북에서 직접 IV와 WOE함수를 만들어 데이터를 변환했던 것을 기억하시나요? 이 작업을 OptBinning을 사용하면 한줄의 코드로 실행 가능합니다. 아래 코드를 살펴보시죠.
X_transform = binning_process.transform(X, metric="woe")
X_transform
# > #### 기존 변수 값이 WOE 값으로 변환(transform)되었습니다. 이제 WOE값을 사용해 모델링을 진행하면 됩니다.
from sklearn.linear_model import LogisticRegression
from optbinning import Scorecard
from optbinning.scorecard import Counterfactual
binning_process = BinningProcess(
feature_list,
categorical_variables=cat_features,
selection_criteria=selection_criteria,
)
estimator = LogisticRegression(solver="lbfgs")
scorecard = Scorecard(
binning_process=binning_process,
estimator=estimator,
scaling_method="min_max",
scaling_method_params={"min": 300, "max": 850},
)
scorecard.fit(X, y)
# > #### 가장 간단한 로지스틱 회귀를 사용해보겠습니다. estimator에 XGBoost나 LighGBM을 사용해도 좋습니다.
# > #### 스코어 카드를 바로 만들어 보겠습니다. 최저 점수는 300점, 최대 점수는 850점입니다.
scorecard.table(style="summary")
# > #### 위에서 만든 스코어 카드 테이블을 살펴보겠습니다.
scorecard.table(style="detailed")
# > #### 보다 자세한 테이블은 위와 같습니다. WoE와 IV, JS 값까지 보여줍니다.
# > ##### "JS Distance"는 각 구간(bin)의 신용 점수 분포 간의 거리(Distance)를 측정하는 지표입니다.
# > ##### 스코어카드에서 "JS Distance"는 구간(bin)별로 신용 점수가 얼마나 다른지를 나타내는 지표로 사용됩니다. 구간별 신용 점수 분포가 서로 다를수록 "JS Distance"는 높아지며, 구간간의 분리도가 높음을 나타냅니다. 이는 스코어카드가 독립 변수의 변화에 따라 점수를 적절하게 조정하여 신용 등급을 산출할 수 있도록 도와줍니다.
#
sc = scorecard.table(style="summary")
sc.groupby("Variable").agg({"Points": [np.min, np.max]}).sum()
# > #### 위 코드는 점수가 잘 나왔는지 검증하는 코드입니다.
# > #### 📊 만든 스코어를 평가해봅시다.
y_pred = scorecard.predict_proba(X)[:, 1]
from optbinning.scorecard import plot_auc_roc, plot_cap, plot_ks
plot_auc_roc(y, y_pred)
# > #### 단순한 모델을 사용하니 AUC가 이전보다 떨어진것을 확인할 수 있습니다. 하지만 데이터 전처리가 잘된 편이라 점수가 아주 낮진 않습니다.
plot_ks(y, y_pred)
# > #### KS 통계량도 간단한 함수로 계산하고 시각화할 수 있습니다.
# > #### 📊 스코어 분포를 살펴보도록 하죠.
score = scorecard.score(X)
import matplotlib.pyplot as plt
mask = y == 0
plt.hist(score[mask], label="non-event", color="b", alpha=0.35)
plt.hist(score[~mask], label="event", color="r", alpha=0.35)
plt.xlabel("score")
plt.legend()
plt.show()
# ### OptBinning 라이브러리를 사용한 모델 모니터링
# > #### PSI지표를 활용한 모델 드리프트 체크
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, stratify=y, random_state=42
)
# > #### 훈련 데이터세트와 테스트 데이터세트를 분할해줍니다. 여기서 테스트 데이터세트는 우리가 보지 못한 데이터라고 가정합니다.
scorecard.fit(X_train, y_train)
from optbinning.scorecard import ScorecardMonitoring
monitoring = ScorecardMonitoring(
scorecard=scorecard, psi_method="cart", psi_n_bins=10, verbose=True
)
monitoring.fit(X_test, y_test, X_train, y_train)
monitoring.psi_table()
# > #### 굉장히 안정적입니다. 그래프도 쉽게 그릴 수 있습니다.
monitoring.psi_plot()
monitoring.tests_table()
# > ##### 이 통계적 검정은 이벤트 비율(카이제곱 검정 - 이진 타겟) 또는 평균(스튜던트 t-검정 - 연속 타겟)이 유의하게 다른지 여부를 결정하기 위해 수행됩니다. 귀무가설은 실제값(actual)이 예상값(expected)과 동일하다는 것입니다.
monitoring.system_stability_report()
# > #### 시스템 리포트를 통해 확인할 수도 있습니다.
monitoring.psi_variable_table(style="summary")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/807/129807058.ipynb
|
amex-data-sampled
|
kimtaehun
|
[{"Id": 129807058, "ScriptId": 38603686, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13683894, "CreationDate": "05/16/2023 15:43:39", "VersionNumber": 1.0, "Title": "4\uac15) OptBinning \ub77c\uc774\ube0c\ub7ec\ub9ac \uc0ac\uc6a9 \ubc29\ubc95", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 320.0, "LinesInsertedFromPrevious": 320.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 5}]
|
[{"Id": 186180026, "KernelVersionId": 129807058, "SourceDatasetVersionId": 5688399}]
|
[{"Id": 5688399, "DatasetId": 3270398, "DatasourceVersionId": 5763995, "CreatorUserId": 1885842, "LicenseName": "Unknown", "CreationDate": "05/15/2023 07:57:57", "VersionNumber": 1.0, "Title": "AMEX_data_sampled", "Slug": "amex-data-sampled", "Subtitle": "This is a small-sized sampled dataset from AMEX dafault prediction dataset", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3270398, "CreatorUserId": 1885842, "OwnerUserId": 1885842.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5688399.0, "CurrentDatasourceVersionId": 5763995.0, "ForumId": 3336031, "Type": 2, "CreationDate": "05/15/2023 07:57:57", "LastActivityDate": "05/15/2023", "TotalViews": 127, "TotalDownloads": 3, "TotalVotes": 8, "TotalKernels": 3}]
|
[{"Id": 1885842, "UserName": "kimtaehun", "DisplayName": "DataManyo", "RegisterDate": "05/05/2018", "PerformanceTier": 4}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### 🔥 OptBinning 라이브러리를 활용한 신용평가 모델 개발
# > #### 데이터를 불러와 전처리하는 과정은 이전과 동일합니다. 다만 임의로 설명 변수를 선택하기 때문에 선택된 설명 변수는 다를 수 있습니다.
import warnings
warnings.filterwarnings("ignore", module="sklearn.metrics.cluster")
df = pd.read_pickle("/kaggle/input/amex-data-sampled/train_df_sample.pkl")
# > ##### 💡 데이터는 위에서 프린트된 경로를 사용해야 합니다
def drop_null_cols(df, threshold=0.8):
"""
데이터프레임에서 결측치 비율이 threshold 이상인 변수를 제거하는 함수
"""
null_percent = df.isnull().mean()
drop_cols = list(null_percent[null_percent >= threshold].index)
df = df.drop(drop_cols, axis=1)
print(f"Dropped {len(drop_cols)} columns: {', '.join(drop_cols)}")
return df
df = df.reset_index()
import hashlib
def encode_customer_id(id_str):
encoded_id = hashlib.sha256(id_str.encode("utf-8")).hexdigest()[:16]
return encoded_id
df["customer_ID"] = df["customer_ID"].apply(encode_customer_id)
df = drop_null_cols(df)
cat_features = [
"B_30",
"B_38",
"D_114",
"D_116",
"D_117",
"D_120",
"D_126",
"D_63",
"D_64",
"D_68",
]
cat_features = [f"{cf}_last" for cf in cat_features]
import random
num_cols = df.select_dtypes(include=np.number).columns.tolist()
num_cols = [col for col in num_cols if "target" not in col and col not in cat_features]
num_cols_sample = random.sample([col for col in num_cols if "target" not in col], 100)
feature_list = num_cols_sample + cat_features
all_list = feature_list + ["target"]
df = df[all_list]
import gc
gc.collect()
for categorical_feature in cat_features:
if df[categorical_feature].dtype == "float16":
df[categorical_feature] = df[categorical_feature].astype(str)
if df[categorical_feature].dtype == "category":
df[categorical_feature] = df[categorical_feature].astype(str)
elif df[categorical_feature].dtype == "object":
df[categorical_feature] = df[categorical_feature].astype(str)
from sklearn.preprocessing import LabelEncoder
le_encoder = LabelEncoder()
for categorical_feature in cat_features:
df[categorical_feature].fillna(value="NaN", inplace=True)
df[categorical_feature] = le_encoder.fit_transform(df[categorical_feature])
from sklearn.impute import SimpleImputer
def impute_nan(df, num_cols, strategy="mean"):
"""
NaN 값을 strategy에 따라 num_cols에 대해 impute하는 함수
:param df: DataFrame
:param num_cols: list, imputation 대상 numeric column 리스트
:param strategy: str, imputation 전략 (default: 'mean')
:return: DataFrame, imputed DataFrame
"""
imputer = SimpleImputer(strategy=strategy)
df[num_cols] = imputer.fit_transform(df[num_cols])
return df
df = impute_nan(df, num_cols_sample, strategy="mean")
df.head()
import plotly.express as px
fig2 = px.pie(
df,
names="target",
height=400,
width=600,
hole=0.7,
title="target class Overview",
color_discrete_sequence=["#4c78a8", "#72b7b2"],
)
fig2.update_traces(
hovertemplate=None, textposition="outside", textinfo="percent+label", rotation=0
)
fig2.update_layout(
margin=dict(t=100, b=30, l=0, r=0),
showlegend=False,
plot_bgcolor="#fafafa",
paper_bgcolor="#fafafa",
title_font=dict(size=20, color="#555", family="Lato, sans-serif"),
font=dict(size=17, color="#8a8d93"),
hoverlabel=dict(bgcolor="#444", font_size=13, font_family="Lato, sans-serif"),
)
fig2.show()
# # optbinning library 설치
# > #### !pip install은 Python 환경에서 패키지를 설치하기 위한 표준적인 방법이며, %pip install은 IPython 환경에서 제공되는 패키지 관리 명령어입니다. 일반적으로 Jupyter Notebook에서는 !pip install이 더 일반적으로 사용되며, %pip install은 특정한 경우에 사용될 수 있습니다.
from optbinning import BinningProcess
#
# 💡 Notes:
# Binning Process는 데이터셋의 변수들을 이진, 연속형 또는 다중 클래스 타겟 데이터 유형을 기반으로 최적의 구간화(bin)을 계산하기 위한 프로세스입니다.
# 매개변수:
# * variable_names (array-like): 변수 이름의 리스트입니다.
# * max_n_prebins (int, default=20): Pre-binning(사전 구간화) 후의 최대 구간 수입니다.
# * min_prebin_size (float, default=0.05): 각 Pre-bin에 대한 최소 레코드 수의 비율입니다.
# * min_n_bins (int 또는 None, optional, default=None): 최소 구간 수입니다. None인 경우, min_n_bins은 [0, max_n_prebins] 범위의 값입니다.
# * max_n_bins (int 또는 None, optional, default=None): 최대 구간 수입니다. None인 경우, max_n_bins은 [0, max_n_prebins] 범위의 값입니다.
# * min_bin_size (float 또는 None, optional, default=None): 각 구간의 최소 레코드 수의 비율입니다. None인 경우, min_bin_size = min_prebin_size입니다.
# * max_bin_size (float 또는 None, optional, default=None): 각 구간의 최대 레코드 수의 비율입니다. None인 경우, max_bin_size = 1.0입니다.
# * max_pvalue (float 또는 None, optional, default=None): 구간 간의 최대 p-value입니다.
# * max_pvalue_policy (str, optional, default="consecutive"): p-value 조건을 만족하지 않는 구간을 결정하는 방법입니다. "consecutive"는 연속된 구간을 비교하고, "all"은 모든 구간을 비교합니다.
# * selection_criteria (dict 또는 None, optional, default=None): 변수 선택 기준입니다. 자세한 내용은 참고 사항을 참조하세요.
# * fixed_variables (array-like 또는 None, optional, default=None): 고정할 변수의 리스트입니다. 선택 기준을 만족하지 않을 경우에도 이러한 변수를 유지합니다.
# * special_codes (array-like 또는 None, optional, default=None): 특별한 코드의 리스트입니다. 이러한 코드를 사용하여 따로 처리해야 하는 데이터 값을 지정할 수 있습니다.
# * split_digits (int 또는 None, optional, default=None): 분할 지점의 유효 숫자 자릿수입니다. split_digits가 0으로 설정되면 분할 지점은 정수로 처리됩니다. None인 경우, 분할 지점의 모든 유효 숫자 자릿수가 고려됩니다.
# * categorical_variables (array-like 또는 None, optional, default=None): 범주형 변수로 간주할 수치 변수의 리스트입니다. 이는 명목 변수입니다. 타겟 유형이 다중 클래스인 경우에는 해당되지 않습니다.
# * binning_fit_params (dict 또는 None, optional, default=None): 특정 변수에 대한 최적
selection_criteria = {
"iv": {"min": 0.025, "max": 0.7, "strategy": "highest", "top": 20},
"quality_score": {"min": 0.01},
}
# > #### Feature Selection 프로세스를 간소화 할 수 있습니다. 조건을 정해두면 해당조건에 맞는 변수를 추출할 수 있습니다.
binning_process = BinningProcess(
feature_list,
categorical_variables=cat_features,
selection_criteria=selection_criteria,
)
X = df[feature_list]
y = df["target"]
binning_process.fit(X, y)
binning_process.information(print_level=2)
binning_process.summary()
# > #### 선택된 변수만 보고싶다면 아래 코드를 참조해주세요.
#
summary = binning_process.summary()
selected_summary = summary[summary["selected"] == True]
selected_summary
optb = binning_process.get_binned_variable("D_42_mean")
optb.binning_table.build()
# > #### 임의로 변수를 선택하기 때문에 위 변수가 없다면 에러가 날 수 있습니다. 확인 후 실행 바랍니다.
# > #### 📊 구간화 그래프를 살펴봅시다. binning_table.plot()을 사용하면 쉽게 그래프를 그릴 수 있습니다.
optb.binning_table.plot(metric="event_rate")
# > #### 위 변수 같은 경우는, 2 구간에서 불량율이 매우 높게 나타납니다. 이벤트 발생 확률 또한 단조증가함을 알 수 있습니다. 일반적으로 구간화는 이벤트 발생 확률이 단조 증가하면 좋다고 판단합니다.
# > ##### optb.binning_table.plot(metric="event_rate")에서 metric은 그래프를 그릴 때 사용할 메트릭(metric)을 지정하는 매개변수입니다. 이 메트릭은 각 구간(bin)의 값을 기준으로 그래프를 그릴 때 사용됩니다. 예를 들어, "event_rate"를 지정하면 각 구간의 이벤트 발생 비율을 기준으로 그래프를 그립니다.
# > ##### optb.binning_table.plot()은 optbinning 패키지에서 사용되는 BinningTable 객체의 메서드입니다. BinningTable 객체는 변수의 구간(bin) 정보를 저장하고 분석 결과를 시각화할 수 있는 기능을 제공합니다. 이를 통해 각 구간의 특성을 시각적으로 확인하고 모델 개발에 유용한 통찰력을 얻을 수 있습니다. plot() 메서드를 사용하여 구간별 메트릭에 대한 그래프를 생성하고 시각화할 수 있습니다. metric 매개변수를 적절히 설정하여 원하는 메트릭에 대한 그래프를 생성할 수 있습니다.
optb = binning_process.get_binned_variable("D_68_last")
optb.binning_table.build()
optb.binning_table.plot(metric="event_rate")
# > #### 어떤 변수들이 선택되었는지 살펴봅시다.
binning_process.get_support(names=True)
# > #### 이전 노트북에서 직접 IV와 WOE함수를 만들어 데이터를 변환했던 것을 기억하시나요? 이 작업을 OptBinning을 사용하면 한줄의 코드로 실행 가능합니다. 아래 코드를 살펴보시죠.
X_transform = binning_process.transform(X, metric="woe")
X_transform
# > #### 기존 변수 값이 WOE 값으로 변환(transform)되었습니다. 이제 WOE값을 사용해 모델링을 진행하면 됩니다.
from sklearn.linear_model import LogisticRegression
from optbinning import Scorecard
from optbinning.scorecard import Counterfactual
binning_process = BinningProcess(
feature_list,
categorical_variables=cat_features,
selection_criteria=selection_criteria,
)
estimator = LogisticRegression(solver="lbfgs")
scorecard = Scorecard(
binning_process=binning_process,
estimator=estimator,
scaling_method="min_max",
scaling_method_params={"min": 300, "max": 850},
)
scorecard.fit(X, y)
# > #### 가장 간단한 로지스틱 회귀를 사용해보겠습니다. estimator에 XGBoost나 LighGBM을 사용해도 좋습니다.
# > #### 스코어 카드를 바로 만들어 보겠습니다. 최저 점수는 300점, 최대 점수는 850점입니다.
scorecard.table(style="summary")
# > #### 위에서 만든 스코어 카드 테이블을 살펴보겠습니다.
scorecard.table(style="detailed")
# > #### 보다 자세한 테이블은 위와 같습니다. WoE와 IV, JS 값까지 보여줍니다.
# > ##### "JS Distance"는 각 구간(bin)의 신용 점수 분포 간의 거리(Distance)를 측정하는 지표입니다.
# > ##### 스코어카드에서 "JS Distance"는 구간(bin)별로 신용 점수가 얼마나 다른지를 나타내는 지표로 사용됩니다. 구간별 신용 점수 분포가 서로 다를수록 "JS Distance"는 높아지며, 구간간의 분리도가 높음을 나타냅니다. 이는 스코어카드가 독립 변수의 변화에 따라 점수를 적절하게 조정하여 신용 등급을 산출할 수 있도록 도와줍니다.
#
sc = scorecard.table(style="summary")
sc.groupby("Variable").agg({"Points": [np.min, np.max]}).sum()
# > #### 위 코드는 점수가 잘 나왔는지 검증하는 코드입니다.
# > #### 📊 만든 스코어를 평가해봅시다.
y_pred = scorecard.predict_proba(X)[:, 1]
from optbinning.scorecard import plot_auc_roc, plot_cap, plot_ks
plot_auc_roc(y, y_pred)
# > #### 단순한 모델을 사용하니 AUC가 이전보다 떨어진것을 확인할 수 있습니다. 하지만 데이터 전처리가 잘된 편이라 점수가 아주 낮진 않습니다.
plot_ks(y, y_pred)
# > #### KS 통계량도 간단한 함수로 계산하고 시각화할 수 있습니다.
# > #### 📊 스코어 분포를 살펴보도록 하죠.
score = scorecard.score(X)
import matplotlib.pyplot as plt
mask = y == 0
plt.hist(score[mask], label="non-event", color="b", alpha=0.35)
plt.hist(score[~mask], label="event", color="r", alpha=0.35)
plt.xlabel("score")
plt.legend()
plt.show()
# ### OptBinning 라이브러리를 사용한 모델 모니터링
# > #### PSI지표를 활용한 모델 드리프트 체크
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, stratify=y, random_state=42
)
# > #### 훈련 데이터세트와 테스트 데이터세트를 분할해줍니다. 여기서 테스트 데이터세트는 우리가 보지 못한 데이터라고 가정합니다.
scorecard.fit(X_train, y_train)
from optbinning.scorecard import ScorecardMonitoring
monitoring = ScorecardMonitoring(
scorecard=scorecard, psi_method="cart", psi_n_bins=10, verbose=True
)
monitoring.fit(X_test, y_test, X_train, y_train)
monitoring.psi_table()
# > #### 굉장히 안정적입니다. 그래프도 쉽게 그릴 수 있습니다.
monitoring.psi_plot()
monitoring.tests_table()
# > ##### 이 통계적 검정은 이벤트 비율(카이제곱 검정 - 이진 타겟) 또는 평균(스튜던트 t-검정 - 연속 타겟)이 유의하게 다른지 여부를 결정하기 위해 수행됩니다. 귀무가설은 실제값(actual)이 예상값(expected)과 동일하다는 것입니다.
monitoring.system_stability_report()
# > #### 시스템 리포트를 통해 확인할 수도 있습니다.
monitoring.psi_variable_table(style="summary")
| false | 0 | 4,467 | 5 | 4,493 | 4,467 |
||
129807397
|
<jupyter_start><jupyter_text>University Students Complaints & Reports📝👨🎓
The "Voices Heard" dataset is a comprehensive collection of reports and complaints submitted by students in a university setting. From academic grievances to campus safety concerns, this dataset offers a rich trove of insights into the student experience, providing valuable feedback for university administrators and educators. With its diverse range of feedback, "Voices Heard" offers a unique opportunity to gain a better understanding of the needs and concerns of students, and to develop data-driven solutions to enhance the university experience for all. .
Kaggle dataset identifier: university-students-complaints-and-reports
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
import torch
from torch.utils.data import DataLoader
from torchtext.data.utils import get_tokenizer
from torchtext.vocab import build_vocab_from_iterator
from copy import deepcopy
df = pd.read_csv(
"/kaggle/input/university-students-complaints-and-reports/Datasetprojpowerbi.csv"
)
df.head()
df.shape
df.isna().sum()
df["Genre"].value_counts()
# # Merging values into one overview
df["overview"] = (
df["Reports"]
+ " Age: "
+ df["Age"].astype(str)
+ ". GPA: "
+ df["Gpa"].astype(str)
+ ". Year: "
+ df["Year"].astype(str)
+ " . Gender: "
+ df["Gender"].astype(str)
)
un = df["Genre"].unique()
j = 0
labels = dict()
for i in un:
labels[i] = j
j += 1
df["Genre"] = df["Genre"].map(labels)
# # Getting the most important features
dataset = df[["Genre", "overview"]].copy()
train, test = train_test_split(dataset.values, random_state=42, test_size=0.2)
# # Data pipeline
tokenizer = get_tokenizer("basic_english")
def yield_tokens(x):
cat, txt = df.iloc[:, 0].values, df.iloc[:, -1].values
for _, text in zip(cat, txt):
yield tokenizer(text)
vocab = build_vocab_from_iterator(yield_tokens(dataset), specials=["<unk>"])
vocab.set_default_index(vocab["<unk>"])
text_pipeline = lambda x: vocab(tokenizer(x))
def collate_batch(batch):
label_list, text_list, offsets = [], [], [0]
for _label, _text in batch:
label_list.append(_label)
processed_text = torch.tensor(text_pipeline(_text), dtype=torch.int64)
text_list.append(processed_text)
offsets.append(processed_text.size(0))
label_list = torch.tensor(label_list, dtype=torch.int64)
offsets = torch.tensor(offsets[:-1]).cumsum(dim=0)
text_list = torch.cat(text_list)
return label_list.to(device), text_list.to(device), offsets.to(device)
train_dataloader = DataLoader(
train, batch_size=8, shuffle=True, collate_fn=collate_batch
)
validation_loader = DataLoader(
test, batch_size=8, shuffle=False, collate_fn=collate_batch
)
# # Simple MLP-like Text Classification Moel
# 
class ComplaintClassification(torch.nn.Module):
def __init__(self, vocab_size, embed_dim, num_class):
super(ComplaintClassification, self).__init__()
self.embed = torch.nn.EmbeddingBag(vocab_size, embed_dim, sparse=False)
self.layer = torch.nn.Sequential(
torch.nn.Linear(embed_dim, 128),
torch.nn.Linear(128, 32),
torch.nn.Linear(32, num_class),
)
def forward(self, x, off):
x = self.embed(x, off)
return self.layer(x)
num_classes = 11
vocab_size = len(vocab)
emsize = 64
model = ComplaintClassification(vocab_size, emsize, num_classes)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device)
# # Optimizer is Adam and Loss is CorssEntropyLoss
optimizer = torch.optim.Adam(model.parameters(), lr=0.1)
criterion = torch.nn.CrossEntropyLoss()
# # Training process with validation loss
EPOCHS = 15
best_model = deepcopy(model)
prev = 0
for i in range(1, EPOCHS + 1):
model.train()
total_acc, total_count = 0, 0
for label, text, offsets in train_dataloader:
optimizer.zero_grad()
output = model(text, offsets)
loss = criterion(output, label)
loss.backward()
optimizer.step()
total_acc += loss.item()
total_count += 1
print("Epoch {} training loss : {}".format(i, total_acc / total_count))
model.eval()
val_acc = 0
val_count = 0
with torch.no_grad():
for label, text, offsets in validation_loader:
output = model(text, offsets)
val_acc += (output.argmax(1) == label).sum().item()
val_count += label.size(0)
acc = val_acc / val_count
if acc > prev:
best_model = deepcopy(model)
prev = acc
print("Epoch {} validation loss : {}".format(i, val_acc / val_count))
# # Testing model to predict labels for some real world scenarios
def predict(text, text_pipeline):
with torch.no_grad():
text = torch.tensor(text_pipeline(text))
output = model(text, torch.tensor([0]))
return output.argmax(1).item()
reverse = dict()
for i, j in labels.items():
reverse[j] = i
val = "I cannot pay my tuition fees, because of technical issues. Age: 21. GPA: 4.62. Year: 1 . Gender: F"
reverse[predict(val, text_pipeline)]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/807/129807397.ipynb
|
university-students-complaints-and-reports
|
omarsobhy14
|
[{"Id": 129807397, "ScriptId": 38601153, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11036701, "CreationDate": "05/16/2023 15:46:32", "VersionNumber": 1.0, "Title": "notebookaa06c2b654", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 167.0, "LinesInsertedFromPrevious": 167.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186180442, "KernelVersionId": 129807397, "SourceDatasetVersionId": 5672268}]
|
[{"Id": 5672268, "DatasetId": 3260867, "DatasourceVersionId": 5747799, "CreatorUserId": 11085604, "LicenseName": "Other (specified in description)", "CreationDate": "05/12/2023 19:46:45", "VersionNumber": 1.0, "Title": "University Students Complaints & Reports\ud83d\udcdd\ud83d\udc68\u200d\ud83c\udf93", "Slug": "university-students-complaints-and-reports", "Subtitle": "Voices Heard: Unleashing Insights from Student Feedback in University", "Description": "The \"Voices Heard\" dataset is a comprehensive collection of reports and complaints submitted by students in a university setting. From academic grievances to campus safety concerns, this dataset offers a rich trove of insights into the student experience, providing valuable feedback for university administrators and educators. With its diverse range of feedback, \"Voices Heard\" offers a unique opportunity to gain a better understanding of the needs and concerns of students, and to develop data-driven solutions to enhance the university experience for all. .", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3260867, "CreatorUserId": 11085604, "OwnerUserId": 11085604.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 6264732.0, "CurrentDatasourceVersionId": 6344561.0, "ForumId": 3326442, "Type": 2, "CreationDate": "05/12/2023 19:46:45", "LastActivityDate": "05/12/2023", "TotalViews": 11588, "TotalDownloads": 1576, "TotalVotes": 43, "TotalKernels": 8}]
|
[{"Id": 11085604, "UserName": "omarsobhy14", "DisplayName": "Omar Sobhy", "RegisterDate": "07/19/2022", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
import torch
from torch.utils.data import DataLoader
from torchtext.data.utils import get_tokenizer
from torchtext.vocab import build_vocab_from_iterator
from copy import deepcopy
df = pd.read_csv(
"/kaggle/input/university-students-complaints-and-reports/Datasetprojpowerbi.csv"
)
df.head()
df.shape
df.isna().sum()
df["Genre"].value_counts()
# # Merging values into one overview
df["overview"] = (
df["Reports"]
+ " Age: "
+ df["Age"].astype(str)
+ ". GPA: "
+ df["Gpa"].astype(str)
+ ". Year: "
+ df["Year"].astype(str)
+ " . Gender: "
+ df["Gender"].astype(str)
)
un = df["Genre"].unique()
j = 0
labels = dict()
for i in un:
labels[i] = j
j += 1
df["Genre"] = df["Genre"].map(labels)
# # Getting the most important features
dataset = df[["Genre", "overview"]].copy()
train, test = train_test_split(dataset.values, random_state=42, test_size=0.2)
# # Data pipeline
tokenizer = get_tokenizer("basic_english")
def yield_tokens(x):
cat, txt = df.iloc[:, 0].values, df.iloc[:, -1].values
for _, text in zip(cat, txt):
yield tokenizer(text)
vocab = build_vocab_from_iterator(yield_tokens(dataset), specials=["<unk>"])
vocab.set_default_index(vocab["<unk>"])
text_pipeline = lambda x: vocab(tokenizer(x))
def collate_batch(batch):
label_list, text_list, offsets = [], [], [0]
for _label, _text in batch:
label_list.append(_label)
processed_text = torch.tensor(text_pipeline(_text), dtype=torch.int64)
text_list.append(processed_text)
offsets.append(processed_text.size(0))
label_list = torch.tensor(label_list, dtype=torch.int64)
offsets = torch.tensor(offsets[:-1]).cumsum(dim=0)
text_list = torch.cat(text_list)
return label_list.to(device), text_list.to(device), offsets.to(device)
train_dataloader = DataLoader(
train, batch_size=8, shuffle=True, collate_fn=collate_batch
)
validation_loader = DataLoader(
test, batch_size=8, shuffle=False, collate_fn=collate_batch
)
# # Simple MLP-like Text Classification Moel
# 
class ComplaintClassification(torch.nn.Module):
def __init__(self, vocab_size, embed_dim, num_class):
super(ComplaintClassification, self).__init__()
self.embed = torch.nn.EmbeddingBag(vocab_size, embed_dim, sparse=False)
self.layer = torch.nn.Sequential(
torch.nn.Linear(embed_dim, 128),
torch.nn.Linear(128, 32),
torch.nn.Linear(32, num_class),
)
def forward(self, x, off):
x = self.embed(x, off)
return self.layer(x)
num_classes = 11
vocab_size = len(vocab)
emsize = 64
model = ComplaintClassification(vocab_size, emsize, num_classes)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device)
# # Optimizer is Adam and Loss is CorssEntropyLoss
optimizer = torch.optim.Adam(model.parameters(), lr=0.1)
criterion = torch.nn.CrossEntropyLoss()
# # Training process with validation loss
EPOCHS = 15
best_model = deepcopy(model)
prev = 0
for i in range(1, EPOCHS + 1):
model.train()
total_acc, total_count = 0, 0
for label, text, offsets in train_dataloader:
optimizer.zero_grad()
output = model(text, offsets)
loss = criterion(output, label)
loss.backward()
optimizer.step()
total_acc += loss.item()
total_count += 1
print("Epoch {} training loss : {}".format(i, total_acc / total_count))
model.eval()
val_acc = 0
val_count = 0
with torch.no_grad():
for label, text, offsets in validation_loader:
output = model(text, offsets)
val_acc += (output.argmax(1) == label).sum().item()
val_count += label.size(0)
acc = val_acc / val_count
if acc > prev:
best_model = deepcopy(model)
prev = acc
print("Epoch {} validation loss : {}".format(i, val_acc / val_count))
# # Testing model to predict labels for some real world scenarios
def predict(text, text_pipeline):
with torch.no_grad():
text = torch.tensor(text_pipeline(text))
output = model(text, torch.tensor([0]))
return output.argmax(1).item()
reverse = dict()
for i, j in labels.items():
reverse[j] = i
val = "I cannot pay my tuition fees, because of technical issues. Age: 21. GPA: 4.62. Year: 1 . Gender: F"
reverse[predict(val, text_pipeline)]
| false | 1 | 1,457 | 0 | 1,612 | 1,457 |
||
129993231
|
<jupyter_start><jupyter_text>27 Class Sign Language Dataset
### Abstract
To contribute to the development of technologies, that can reduce the communication problems of speech-impaired persons, a new dataset was presented with this work. The dataset was created by processing American Sign Language-based photographs collected from 173 volunteer individuals.
Details of the dataset are published in the paper: [Mavi. A., and Dikle, Z. (2022). A New 27 Class Sign Language Dataset Collected from 173 Individuals. *arXiv:2203.03859*](https://arxiv.org/abs/2203.03859)
<img src="https://raw.githubusercontent.com/ardamavi/Vocalize-Sign-Language/master/Assets/Samples_Kaggle_Image.png">
### Data Usage:
Data can be used for research and/or commercial purposes by citing to the original paper below.
### Cite as:
Mavi. A., and Dikle, Z. (2022). A New 27 Class Sign Language Dataset Collected from 173 Individuals. [*arXiv:2203.03859*](https://arxiv.org/abs/2203.03859)
Kaggle dataset identifier: 27-class-sign-language-dataset
<jupyter_script>import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
import tensorflow_hub as tf_hub
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
my_seed = 42
split_proportion = 0.1
input_dir = os.path.join("..", "input", "27-class-sign-language-dataset")
x_filename = os.path.join(input_dir, "X.npy")
y_filename = os.path.join(input_dir, "Y.npy")
x = np.load(x_filename)
y = np.load(y_filename)
# shuffle and split the data
split_number = int(split_proportion * x.shape[0])
np.random.seed(my_seed)
np.random.shuffle(x)
val_x = tf.convert_to_tensor(x[:split_number])
test_x = tf.convert_to_tensor(x[split_number : 2 * split_number])
train_x = tf.convert_to_tensor(x[2 * split_number :])
np.random.seed(my_seed)
np.random.shuffle(y)
val_y = tf.convert_to_tensor(y[:split_number])
test_y = tf.convert_to_tensor(y[split_number : 2 * split_number])
train_y = tf.convert_to_tensor(y[2 * split_number :])
# visualize images with labels
plt.figure()
for count, x_index in enumerate(np.random.randint(0, train_x.shape[0], size=(9,))):
plt.subplot(3, 3, count + 1)
plt.imshow(train_x[x_index])
plt.title(f"label: {train_y[x_index]}")
plt.tight_layout()
plt.show()
label_dict = {}
for number, label in enumerate(np.unique(train_y)):
label_dict[number] = label
print(label_dict, x.shape)
# help(tf_hub.KerasLayer)
# model_url = "https://tfhub.dev/google/tf2-preview/mobilenet_v2/feature_vector/4"
# model_url = "https://tfhub.dev/google/imagenet/mobilenet_v2_050_96/classification/5"
# extractor = tf_hub.KerasLayer(model_url)
# extractor.build([None, 128, 128, 3])
# feature_extractor.trainable = False
extractor = tf.keras.applications.MobileNetV3Small(
input_shape=train_x.shape[1:], include_top=False, weights="imagenet"
)
number_classes = len(label_dict.keys())
model = Sequential(
[
extractor,
tf.keras.layers.GlobalAveragePooling2D(),
Dense(number_classes, activation=None),
]
)
# model.build([None, 128, 128, 3])
_ = model(train_x[0:1])
model.summary()
model.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
train_y = tf.convert_to_tensor(np.random.randint(0, 27, size=train_y.shape[:-1]))
train_y.shape
_ = model(train_x[0:3])
_.shape, train_y[0:3].shape
model.fit(x=train_x, y=train_y, batch_size=32, epochs=100)
model.fit(x=train_x, y=train_y, batch_size=32, epochs=100)
help(model.fit)
history = model.fit(
train_dataset,
epochs=100,
steps_per_epoch=STEPS_PER_EPOCH,
validation_data=valid_dataset,
validation_steps=VALID_STEPS,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
class_weight=class_weight,
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/993/129993231.ipynb
|
27-class-sign-language-dataset
|
ardamavi
|
[{"Id": 129993231, "ScriptId": 38667207, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1950998, "CreationDate": "05/18/2023 02:06:57", "VersionNumber": 1.0, "Title": "tf_lite_sign", "EvaluationDate": "05/18/2023", "IsChange": true, "TotalLines": 112.0, "LinesInsertedFromPrevious": 112.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186442972, "KernelVersionId": 129993231, "SourceDatasetVersionId": 3263022}]
|
[{"Id": 3263022, "DatasetId": 1976854, "DatasourceVersionId": 3313436, "CreatorUserId": 1084733, "LicenseName": "CC BY-NC-SA 4.0", "CreationDate": "03/06/2022 18:52:55", "VersionNumber": 1.0, "Title": "27 Class Sign Language Dataset", "Slug": "27-class-sign-language-dataset", "Subtitle": "27 Class ASL-Based Sign Language Dataset Collected from 173 Individuals", "Description": "### Abstract\nTo contribute to the development of technologies, that can reduce the communication problems of speech-impaired persons, a new dataset was presented with this work. The dataset was created by processing American Sign Language-based photographs collected from 173 volunteer individuals.\nDetails of the dataset are published in the paper: [Mavi. A., and Dikle, Z. (2022). A New 27 Class Sign Language Dataset Collected from 173 Individuals. *arXiv:2203.03859*](https://arxiv.org/abs/2203.03859)\n\n<img src=\"https://raw.githubusercontent.com/ardamavi/Vocalize-Sign-Language/master/Assets/Samples_Kaggle_Image.png\">\n\n### Data Usage:\nData can be used for research and/or commercial purposes by citing to the original paper below.\n\n### Cite as:\nMavi. A., and Dikle, Z. (2022). A New 27 Class Sign Language Dataset Collected from 173 Individuals. [*arXiv:2203.03859*](https://arxiv.org/abs/2203.03859)\n\n### Acknowledgements\nThanks to all of the volunteer students and teachers from Ayranc\u0131 Anadolu High School for their help in collecting data.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1976854, "CreatorUserId": 1084733, "OwnerUserId": 1084733.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3263022.0, "CurrentDatasourceVersionId": 3313436.0, "ForumId": 2001121, "Type": 2, "CreationDate": "03/06/2022 18:52:55", "LastActivityDate": "03/06/2022", "TotalViews": 7540, "TotalDownloads": 608, "TotalVotes": 21, "TotalKernels": 1}]
|
[{"Id": 1084733, "UserName": "ardamavi", "DisplayName": "Arda Mavi", "RegisterDate": "05/21/2017", "PerformanceTier": 0}]
|
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
import tensorflow_hub as tf_hub
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
my_seed = 42
split_proportion = 0.1
input_dir = os.path.join("..", "input", "27-class-sign-language-dataset")
x_filename = os.path.join(input_dir, "X.npy")
y_filename = os.path.join(input_dir, "Y.npy")
x = np.load(x_filename)
y = np.load(y_filename)
# shuffle and split the data
split_number = int(split_proportion * x.shape[0])
np.random.seed(my_seed)
np.random.shuffle(x)
val_x = tf.convert_to_tensor(x[:split_number])
test_x = tf.convert_to_tensor(x[split_number : 2 * split_number])
train_x = tf.convert_to_tensor(x[2 * split_number :])
np.random.seed(my_seed)
np.random.shuffle(y)
val_y = tf.convert_to_tensor(y[:split_number])
test_y = tf.convert_to_tensor(y[split_number : 2 * split_number])
train_y = tf.convert_to_tensor(y[2 * split_number :])
# visualize images with labels
plt.figure()
for count, x_index in enumerate(np.random.randint(0, train_x.shape[0], size=(9,))):
plt.subplot(3, 3, count + 1)
plt.imshow(train_x[x_index])
plt.title(f"label: {train_y[x_index]}")
plt.tight_layout()
plt.show()
label_dict = {}
for number, label in enumerate(np.unique(train_y)):
label_dict[number] = label
print(label_dict, x.shape)
# help(tf_hub.KerasLayer)
# model_url = "https://tfhub.dev/google/tf2-preview/mobilenet_v2/feature_vector/4"
# model_url = "https://tfhub.dev/google/imagenet/mobilenet_v2_050_96/classification/5"
# extractor = tf_hub.KerasLayer(model_url)
# extractor.build([None, 128, 128, 3])
# feature_extractor.trainable = False
extractor = tf.keras.applications.MobileNetV3Small(
input_shape=train_x.shape[1:], include_top=False, weights="imagenet"
)
number_classes = len(label_dict.keys())
model = Sequential(
[
extractor,
tf.keras.layers.GlobalAveragePooling2D(),
Dense(number_classes, activation=None),
]
)
# model.build([None, 128, 128, 3])
_ = model(train_x[0:1])
model.summary()
model.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
train_y = tf.convert_to_tensor(np.random.randint(0, 27, size=train_y.shape[:-1]))
train_y.shape
_ = model(train_x[0:3])
_.shape, train_y[0:3].shape
model.fit(x=train_x, y=train_y, batch_size=32, epochs=100)
model.fit(x=train_x, y=train_y, batch_size=32, epochs=100)
help(model.fit)
history = model.fit(
train_dataset,
epochs=100,
steps_per_epoch=STEPS_PER_EPOCH,
validation_data=valid_dataset,
validation_steps=VALID_STEPS,
callbacks=[checkpoint_cb, early_stopping_cb, lr_scheduler],
class_weight=class_weight,
)
| false | 0 | 1,014 | 0 | 1,331 | 1,014 |
||
129993531
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import os
from matplotlib import pyplot as plt
import torch
import torchvision
from torch import nn as nn
from torch.nn import functional as F
from torch import optim as optim
from torch.utils.data import DataLoader, Dataset, TensorDataset
from torchvision import transforms
from torchvision.transforms import Resize, ToTensor, Compose, Normalize
import random
from torch.autograd import Variable
from torchvision import datasets
import warnings
from sklearn.model_selection import train_test_split
warnings.filterwarnings("ignore")
for f in os.listdir("/kaggle/input/digit-recognizer"):
s = f.split(".")[0]
print(s)
try:
exec(s + " = pd.read_csv('/kaggle/input/digit-recognizer/{}')".format(f))
except:
pass
train.head()
train_X, val_X, train_y, val_y = train_test_split(
train.drop("label", axis=1), train.label, test_size=0.2
)
train_X.reset_index(drop=True, inplace=True)
train_y.reset_index(drop=True, inplace=True)
val_X.reset_index(drop=True, inplace=True)
val_y.reset_index(drop=True, inplace=True)
transform = Compose(
[
transforms.ToPILImage(),
# Resize(size = (28,28)),
# transforms.RandomCrop(28),
# transforms.RandomHorizontalFlip(),
ToTensor(),
Normalize((0.131,), (0.3085,)),
]
)
class MNISTDataSet(Dataset):
def __init__(self, images, labels, transforms=None):
self.X = images
self.y = labels
self.transforms = transforms
def __len__(self):
return len(self.X)
def __getitem__(self, i):
data = self.X.iloc[i, :]
data = np.array(data).astype(np.uint8).reshape(28, 28, 1)
if self.transforms:
data = self.transforms(data)
if self.y is not None: # train/val
return (data, self.y[i])
else:
return data
train_set = MNISTDataSet(train_X, train_y, transform)
trainload = DataLoader(train_set, batch_size=32, shuffle=True)
val_set = MNISTDataSet(val_X, val_y, transform)
valload = DataLoader(val_set, batch_size=32, shuffle=True)
class CNN(nn.Module):
def __init__(self):
super(CNN, self).__init__()
self.cn1 = nn.Sequential(
nn.Conv2d(
in_channels=1, out_channels=16, kernel_size=3, stride=1, padding=2
),
nn.BatchNorm2d(16),
)
self.cn2 = nn.Sequential(
nn.Conv2d(
in_channels=16, out_channels=32, kernel_size=3, stride=1, padding=2
),
nn.BatchNorm2d(32),
)
self.cn3 = nn.Sequential(
nn.Conv2d(
in_channels=32, out_channels=64, kernel_size=3, stride=1, padding=2
),
nn.BatchNorm2d(64),
)
self.dp1 = nn.Dropout(0.1)
self.dp2 = nn.Dropout(0.25)
self.dp3 = nn.Dropout(0.4)
self.fc1 = nn.Sequential(nn.Linear(1600, 1024), nn.BatchNorm1d(1024))
self.fc2 = nn.Sequential(nn.Linear(1024, 512), nn.BatchNorm1d(512))
self.fc3 = nn.Sequential(nn.Linear(512, 128), nn.BatchNorm1d(128))
self.fc4 = nn.Sequential(nn.Linear(128, 64), nn.BatchNorm1d(64))
self.fc5 = nn.Linear(64, 10)
def forward(self, x):
x = self.cn1(x)
x = F.relu(x)
x = F.max_pool2d(x, 2)
x = self.cn2(x)
x = F.relu(x)
x = F.max_pool2d(x, 2)
x = self.dp1(x)
x = self.cn3(x)
x = F.relu(x)
x = F.max_pool2d(x, 2)
x = torch.flatten(x, 1)
x = self.fc1(x)
x = F.relu(x)
x = self.dp2(x)
x = self.fc2(x)
x = F.relu(x)
x = self.fc3(x)
x = F.relu(x)
x = self.dp3(x)
x = self.fc4(x)
x = F.relu(x)
x = self.fc5(x)
digits = F.log_softmax(x, dim=1)
return digits
def get_num_correct(preds, labels):
return preds.argmax(dim=1).eq(labels).sum().item()
def get_mean_and_std(dataloader):
channels_sum, channels_squared_sum, num_batches = 0, 0, 0
for data, _ in dataloader:
# Mean over batch, height and width, but not over the channels
channels_sum += torch.mean(data, dim=[0, 2, 3])
channels_squared_sum += torch.mean(data**2, dim=[0, 2, 3])
num_batches += 1
mean = channels_sum / num_batches
# std = sqrt(E[X^2] - (E[X])^2)
std = (channels_squared_sum / num_batches - mean**2) ** 0.5
return mean, std
get_mean_and_std(trainload)
def train(model, device, train_dataloader, val_dataloader, optim, epoch):
model.train()
for b_i, (X, y) in enumerate(train_dataloader):
X, y = Variable(X).to(device), Variable(y).to(device)
optim.zero_grad()
pred_prob = model(X)
loss = F.nll_loss(pred_prob, y)
loss.backward()
optim.step()
model.eval() # eval mode
val_loss = 0
val_correct = 0
with torch.no_grad():
for X, y in val_dataloader:
X, y = Variable(X).to(device), Variable(y).to(device)
preds = model(X) # get predictions
loss = F.nll_loss(preds, y) # calculate the loss
val_correct += get_num_correct(preds, y)
val_loss = loss.item() * 32
print(
"epoch: {}\t loss: {:.6f}\t val loss: {:.6f}\t accuracy: {:.2f}".format(
epoch + 1, loss.item(), val_loss, (val_correct / len(val_X)) * 100
)
)
EPOCH = 5
device = "cuda:0" if torch.cuda.is_available() else "cpu"
model = CNN().to(device)
critetion = optim.Adadelta(model.parameters(), lr=0.05)
print(device)
for e in range(EPOCH):
train(model, device, trainload, valload, critetion, e)
test.reset_index(drop=True, inplace=True)
test_set = MNISTDataSet(test, None, transform)
testload = DataLoader(test_set, batch_size=32, shuffle=False)
model.eval() # Safety first
predictions = torch.LongTensor().to(device) # Tensor for all predictions
# Go through the test set, saving the predictions in... 'predictions'
for images in testload:
preds = model(images.to(device))
predictions = torch.cat((predictions, preds.argmax(dim=1)), dim=0)
sample_submission["Label"] = predictions.cpu().numpy()
sample_submission.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/993/129993531.ipynb
| null | null |
[{"Id": 129993531, "ScriptId": 38649575, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 847705, "CreationDate": "05/18/2023 02:10:44", "VersionNumber": 1.0, "Title": "MNIST CNN PyTorch - 99,1%", "EvaluationDate": "05/18/2023", "IsChange": true, "TotalLines": 204.0, "LinesInsertedFromPrevious": 204.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import os
from matplotlib import pyplot as plt
import torch
import torchvision
from torch import nn as nn
from torch.nn import functional as F
from torch import optim as optim
from torch.utils.data import DataLoader, Dataset, TensorDataset
from torchvision import transforms
from torchvision.transforms import Resize, ToTensor, Compose, Normalize
import random
from torch.autograd import Variable
from torchvision import datasets
import warnings
from sklearn.model_selection import train_test_split
warnings.filterwarnings("ignore")
for f in os.listdir("/kaggle/input/digit-recognizer"):
s = f.split(".")[0]
print(s)
try:
exec(s + " = pd.read_csv('/kaggle/input/digit-recognizer/{}')".format(f))
except:
pass
train.head()
train_X, val_X, train_y, val_y = train_test_split(
train.drop("label", axis=1), train.label, test_size=0.2
)
train_X.reset_index(drop=True, inplace=True)
train_y.reset_index(drop=True, inplace=True)
val_X.reset_index(drop=True, inplace=True)
val_y.reset_index(drop=True, inplace=True)
transform = Compose(
[
transforms.ToPILImage(),
# Resize(size = (28,28)),
# transforms.RandomCrop(28),
# transforms.RandomHorizontalFlip(),
ToTensor(),
Normalize((0.131,), (0.3085,)),
]
)
class MNISTDataSet(Dataset):
def __init__(self, images, labels, transforms=None):
self.X = images
self.y = labels
self.transforms = transforms
def __len__(self):
return len(self.X)
def __getitem__(self, i):
data = self.X.iloc[i, :]
data = np.array(data).astype(np.uint8).reshape(28, 28, 1)
if self.transforms:
data = self.transforms(data)
if self.y is not None: # train/val
return (data, self.y[i])
else:
return data
train_set = MNISTDataSet(train_X, train_y, transform)
trainload = DataLoader(train_set, batch_size=32, shuffle=True)
val_set = MNISTDataSet(val_X, val_y, transform)
valload = DataLoader(val_set, batch_size=32, shuffle=True)
class CNN(nn.Module):
def __init__(self):
super(CNN, self).__init__()
self.cn1 = nn.Sequential(
nn.Conv2d(
in_channels=1, out_channels=16, kernel_size=3, stride=1, padding=2
),
nn.BatchNorm2d(16),
)
self.cn2 = nn.Sequential(
nn.Conv2d(
in_channels=16, out_channels=32, kernel_size=3, stride=1, padding=2
),
nn.BatchNorm2d(32),
)
self.cn3 = nn.Sequential(
nn.Conv2d(
in_channels=32, out_channels=64, kernel_size=3, stride=1, padding=2
),
nn.BatchNorm2d(64),
)
self.dp1 = nn.Dropout(0.1)
self.dp2 = nn.Dropout(0.25)
self.dp3 = nn.Dropout(0.4)
self.fc1 = nn.Sequential(nn.Linear(1600, 1024), nn.BatchNorm1d(1024))
self.fc2 = nn.Sequential(nn.Linear(1024, 512), nn.BatchNorm1d(512))
self.fc3 = nn.Sequential(nn.Linear(512, 128), nn.BatchNorm1d(128))
self.fc4 = nn.Sequential(nn.Linear(128, 64), nn.BatchNorm1d(64))
self.fc5 = nn.Linear(64, 10)
def forward(self, x):
x = self.cn1(x)
x = F.relu(x)
x = F.max_pool2d(x, 2)
x = self.cn2(x)
x = F.relu(x)
x = F.max_pool2d(x, 2)
x = self.dp1(x)
x = self.cn3(x)
x = F.relu(x)
x = F.max_pool2d(x, 2)
x = torch.flatten(x, 1)
x = self.fc1(x)
x = F.relu(x)
x = self.dp2(x)
x = self.fc2(x)
x = F.relu(x)
x = self.fc3(x)
x = F.relu(x)
x = self.dp3(x)
x = self.fc4(x)
x = F.relu(x)
x = self.fc5(x)
digits = F.log_softmax(x, dim=1)
return digits
def get_num_correct(preds, labels):
return preds.argmax(dim=1).eq(labels).sum().item()
def get_mean_and_std(dataloader):
channels_sum, channels_squared_sum, num_batches = 0, 0, 0
for data, _ in dataloader:
# Mean over batch, height and width, but not over the channels
channels_sum += torch.mean(data, dim=[0, 2, 3])
channels_squared_sum += torch.mean(data**2, dim=[0, 2, 3])
num_batches += 1
mean = channels_sum / num_batches
# std = sqrt(E[X^2] - (E[X])^2)
std = (channels_squared_sum / num_batches - mean**2) ** 0.5
return mean, std
get_mean_and_std(trainload)
def train(model, device, train_dataloader, val_dataloader, optim, epoch):
model.train()
for b_i, (X, y) in enumerate(train_dataloader):
X, y = Variable(X).to(device), Variable(y).to(device)
optim.zero_grad()
pred_prob = model(X)
loss = F.nll_loss(pred_prob, y)
loss.backward()
optim.step()
model.eval() # eval mode
val_loss = 0
val_correct = 0
with torch.no_grad():
for X, y in val_dataloader:
X, y = Variable(X).to(device), Variable(y).to(device)
preds = model(X) # get predictions
loss = F.nll_loss(preds, y) # calculate the loss
val_correct += get_num_correct(preds, y)
val_loss = loss.item() * 32
print(
"epoch: {}\t loss: {:.6f}\t val loss: {:.6f}\t accuracy: {:.2f}".format(
epoch + 1, loss.item(), val_loss, (val_correct / len(val_X)) * 100
)
)
EPOCH = 5
device = "cuda:0" if torch.cuda.is_available() else "cpu"
model = CNN().to(device)
critetion = optim.Adadelta(model.parameters(), lr=0.05)
print(device)
for e in range(EPOCH):
train(model, device, trainload, valload, critetion, e)
test.reset_index(drop=True, inplace=True)
test_set = MNISTDataSet(test, None, transform)
testload = DataLoader(test_set, batch_size=32, shuffle=False)
model.eval() # Safety first
predictions = torch.LongTensor().to(device) # Tensor for all predictions
# Go through the test set, saving the predictions in... 'predictions'
for images in testload:
preds = model(images.to(device))
predictions = torch.cat((predictions, preds.argmax(dim=1)), dim=0)
sample_submission["Label"] = predictions.cpu().numpy()
sample_submission.to_csv("submission.csv", index=False)
| false | 0 | 2,217 | 0 | 2,217 | 2,217 |
||
129870801
|
<jupyter_start><jupyter_text>AMEX_data_sampled
Kaggle dataset identifier: amex-data-sampled
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### 4강) TOAD 라이브러리를 활용한 신용평가 모델 개발
# > ### 데이터 준비
import warnings
warnings.filterwarnings("ignore", module="sklearn.metrics.cluster")
def drop_null_cols(df, threshold=0.8):
"""
데이터프레임에서 결측치 비율이 threshold 이상인 변수를 제거하는 함수
"""
null_percent = df.isnull().mean()
drop_cols = list(null_percent[null_percent >= threshold].index)
df = df.drop(drop_cols, axis=1)
print(f"Dropped {len(drop_cols)} columns: {', '.join(drop_cols)}")
return df
df = pd.read_pickle("/kaggle/input/amex-data-sampled/train_df_sample.pkl")
df = df.reset_index()
import hashlib
def encode_customer_id(id_str):
encoded_id = hashlib.sha256(id_str.encode("utf-8")).hexdigest()[:16]
return encoded_id
df["customer_ID"] = df["customer_ID"].apply(encode_customer_id)
df = drop_null_cols(df)
cat_features = [
"B_30",
"B_38",
"D_114",
"D_116",
"D_117",
"D_120",
"D_126",
"D_63",
"D_64",
"D_68",
]
cat_features = [f"{cf}_last" for cf in cat_features]
import random
num_cols = df.select_dtypes(include=np.number).columns.tolist()
num_cols = [col for col in num_cols if "target" not in col and col not in cat_features]
num_cols_sample = random.sample([col for col in num_cols if "target" not in col], 100)
feature_list = num_cols_sample + cat_features
all_list = feature_list + ["target"]
df = df[all_list]
for categorical_feature in cat_features:
if df[categorical_feature].dtype == "float16":
df[categorical_feature] = df[categorical_feature].astype(str)
if df[categorical_feature].dtype == "category":
df[categorical_feature] = df[categorical_feature].astype(str)
elif df[categorical_feature].dtype == "object":
df[categorical_feature] = df[categorical_feature].astype(str)
from sklearn.preprocessing import LabelEncoder
le_encoder = LabelEncoder()
for categorical_feature in cat_features:
df[categorical_feature].fillna(value="NaN", inplace=True)
df[categorical_feature] = le_encoder.fit_transform(df[categorical_feature])
from sklearn.impute import SimpleImputer
def impute_nan(df, num_cols, strategy="mean"):
"""
NaN 값을 strategy에 따라 num_cols에 대해 impute하는 함수
:param df: DataFrame
:param num_cols: list, imputation 대상 numeric column 리스트
:param strategy: str, imputation 전략 (default: 'mean')
:return: DataFrame, imputed DataFrame
"""
imputer = SimpleImputer(strategy=strategy)
df[num_cols] = imputer.fit_transform(df[num_cols])
return df
df = impute_nan(df, num_cols_sample, strategy="mean")
# > #### taod 패키지 설치
# > #### IV값을 기준으로 변수를 선택해보자. (Feature Selection)
import toad
# 타겟 변수 정의
target = "target"
# 정보값(IV) 계산 및 특성 선택
iv_df = toad.quality(df, target=target, iv_only=True)
selected_features = iv_df[iv_df["iv"] > 0.1].index # 'name' 대신 'index'를 사용합니다.
# WOE 변환
trans = toad.transform.WOETransformer()
df_woe = trans.fit_transform(df[selected_features], df[target])
# 이제 df_woe를 추가적인 모델링에 사용할 수 있습니다.
# toad는 파이썬의 데이터 전처리 및 탐색을 위한 오픈 소스 패키지입니다. 주요 기능은 다음과 같습니다:
# 데이터 품질 평가: toad.quality() 함수를 사용하여 변수별 정보값(IV, Information Value)을 계산하고, 타겟 변수와의 상관 관계를 평가합니다. 이를 통해 변수의 중요도를 판단하고 특성 선택에 활용할 수 있습니다.
# 데이터 변환: WOE (Weight of Evidence) 변환을 위한 toad.transform.WOETransformer()를 사용할 수 있습니다. WOE는 범주형 변수나 연속형 변수를 범주화하여 정보값을 측정하는 방법으로, 변수의 예측력을 개선하고 모델 성능을 향상시킬 수 있습니다.
df_woe
# > #### 선택된 피처를 기반으로 binning 작업을 진행한다. 이번엔 방법을 조금 다르게해서, 각 구간이 최소 5%의 데이터를 가지도록 구간화를 수행한다.
combiner = toad.transform.Combiner()
combiner.fit(df[selected_features], y=df[target], method="chi", min_samples=0.05)
# min_samples = 0.05로 설정하면, 각 구간이 최소 5%의 데이터를 가지도록 구간화를 수행합니다.
# 구간화 결과를 살펴봅시다.
binning_result = combiner.export()
# > #### iv=True는 toad.plot 함수의 매개변수 중 하나입니다. 이 매개변수를 True로 설정하면, 그래프에 Information Value (IV) 값을 표시합니다.
toad.detect(df)[:10]
# > #### toad.detect(df)의 결과는 변수 이름과 해당 변수의 데이터 유형을 포함한 데이터프레임으로 반환됩니다. 결과에서 첫 10개의 변수와 그에 해당하는 데이터 유형을 확인할 수 있습니다.
toad.quality(df, "target", iv_only=True)[:15]
# > #### 각 변수별 IV값을 살펴보자.
# #### Feature Selection
# 💡 Notes:
# * empty=0.9: 결측치 비율이 90%보다 큰 특성들은 필터링됩니다.
# * iv=0.02: IV(Information Value)가 0.02보다 작은 특성들은 제거됩니다.
# * corr=0.7: 두 개 이상의 특성들 간의 피어슨 상관계수가 0.7보다 큰 경우, IV가 더 낮은 특성들이 제거됩니다.
# * return_drop=False: True로 설정하면, 함수는 삭제된 열들의 리스트를 반환합니다.
# * exclude=None: 알고리즘에서 제외할 특성들의 리스트를 입력합니다. 일반적으로 ID 열이나 월(Month) 열 등이 해당됩니다.
train_selected, dropped = toad.selection.select(
df,
target="target",
empty=0.5,
iv=0.05,
corr=0.7,
return_drop=True,
exclude=["D_117_last"],
)
print(dropped)
print(train_selected.shape)
# > #### toad.selection.select 함수는 선택된 특성들로 이루어진 훈련 데이터셋인 train_selected와 삭제된 열들의 리스트인 dropped를 반환합니다. 또한, train_selected.shape는 선택된 특성들을 포함하는 훈련 데이터셋의 크기를 출력합니다.
# initialise
c = toad.transform.Combiner()
# Train binning with the selected features from previous; use reliable Chi-squared binning, and control that each bucket has at least 5% sample.
c.fit(
train_selected, y="target", method="chi", min_samples=0.05, exclude=["D_117_last"]
)
print("D_59_min:", c.export()["D_59_min"])
print("R_15_std:", c.export()["R_15_std"])
print("S_3_last:", c.export()["S_3_last"])
# > ##### 위 코드는 c.export()를 사용하여 binning 결과 중 특정 변수들의 정보를 출력하는 부분입니다.
# c.export(): Combiner 객체 c의 binning 결과를 딕셔너리 형태로 반환합니다. 딕셔너리의 키는 각 변수의 이름이며, 값은 해당 변수의 binning 정보를 담고 있는 객체입니다.
# 각 변수에 대한 binning 정보는 해당 변수의 구간(bin)과 해당 구간의 라벨(label) 등을 포함하고 있습니다. 출력된 정보를 통해 각 변수의 binning 결과를 확인할 수 있습니다.
from toad.plot import bin_plot
# 학습 데이터(train_selected)에서 'var_d2' 변수의 bin 결과를 확인합니다.
col = "D_59_min"
# 시각화를 위해 'labels = True'로 설정하는 것이 좋습니다.
bin_plot(
c.transform(train_selected[[col, "target"]], labels=True), x=col, target="target"
)
# > ##### 위 코드는 bin_plot 함수를 사용하여 'R_4_std' 변수의 binning 결과를 시각화하는 부분입니다.
# > ##### c.transform(train_selected[[col, 'target']], labels=True): 학습 데이터(train_selected)의 'R_4_std' 변수와 목표 변수('target')를 선택하여 binning 결과를 생성합니다. labels=True로 설정하여 bin의 라벨을 포함한 결과를 반환합니다.
# > ##### bin_plot(...): binning 결과를 시각화합니다. x축에는 'R_4_std' 변수의 값이 나타나며, target 변수('target')의 값에 따라 각 bin의 분포를 확인할 수 있습니다. bin_plot 함수를 통해 binning 결과를 시각적으로 확인하여 데이터의 패턴을 파악할 수 있습니다.
# 학습 데이터(train_selected)에서 'D_51_min' 변수의 bin 결과를 확인합니다.
col = "S_3_last"
# 범주형 변수의 경우 'labels = True'로 설정하는 것이 좋습니다.
bin_plot(
c.transform(train_selected[[col, "target"]], labels=True), x=col, target="target"
)
# Toad의 binning 기능은 범주형 및 수치형 변수를 모두 지원합니다. "toad.transform.Combiner()" 클래스를 사용하여 학습하며, 절차는 다음과 같습니다:
# * 초기화(initalise) : c = toad.transform.Combiner()
# * *train binning*: c.fit(dataframe, y='target', method='chi', min_samples=None, n_bins=None, empty_separate=False)
# * y: 목표 변수;
# * method: binning에 적용할 방법. 'chi' (카이제곱), 'dt' (의사결정 트리), 'kmeans' (K-means), 'quantile' (동일한 백분위수 기준), 'step' (동일한 간격)을 지원합니다.
# * min_samples: 샘플당 요구되는 최소 수 또는 비율. 각 버킷에 필요한 최소 샘플 수 / 비율입니다.
# * n_bins: 최소한의 버킷 수. 수가 너무 큰 경우, 알고리즘은 얻을 수 있는 최대 버킷 수를 반환합니다.
# * empty_separate: 누락된 값이 버킷 내에 별도로 분리되는지 여부. False인 경우, 누락된 값은 가장 가까운 나쁜 비율 버킷과 함께 배치됩니다.
# * binning 결과: c.export()
# * bins 조정: c.update(dict)
# * bins 적용 및 이산값으로 변환: c.transform(dataframe, labels=False)
# * labels: 데이터를 설명 라벨로 변환할지 여부. False인 경우 0, 1, 2...로 반환됩니다. 범주형 변수는 비율의 내림차순으로 정렬됩니다. True인 경우 (-무한대, 0], (0, 10], (10, 무한대)와 같이 반환됩니다.
# * 참고: 1. 불필요한 열을 제외하는 것을 잊지 마세요. 특히 ID 열과 타임스탬프 열은 제외해야 합니다.. 2. 고유 값이 많은 열은 학습에 많은 시간이 소요될 수 있습니다.*
# 초기화
transer = toad.transform.WOETransformer()
# transer.fit_transform() 및 combiner.transform()을 적용합니다. target을 제외하도록 주의하세요.
train_woe = transer.fit_transform(
c.transform(train_selected), train_selected["target"], exclude="target"
)
train_woe.head(3)
# * WOETransformer를 초기화합니다.
# * combiner.transform()의 결과에 transer.fit_transform()을 적용합니다.
# * target을 제외하도록 주의하세요.
# * 변환된 train_woe 데이터프레임의 처음 3개 행을 출력합니다.
col = train_woe.columns.tolist()[:-1]
col.remove("D_117_last")
# > #### 예시를 위해 제거했던 변수를 제거해줍니다.
# > #### 예시를 위해 간단한 로지스틱 회귀 모델을 적합해봅니다.
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression()
lr.fit(train_woe[col], train_woe["target"])
# 훈련 데이터와 Out-of-Time (OOT) 데이터에 대해 예측된 확률을 구합니다.
pred_train = lr.predict_proba(train_woe[col])[:, 1]
# > #### Toad 역시 쉽게 KS 통계량과 AUC를 계산할 수 있는 라이브러리를 제공합니다.
from toad.metrics import KS, AUC
print("train KS", KS(pred_train, train_woe["target"]))
print("train AUC", AUC(pred_train, train_woe["target"]))
# > #### Toad에서도 OptBinning과 마찬가지로 스코어링을 할 수 있는 스코어 카드 기능을 제공합니다.
card = toad.ScoreCard(
combiner=c, transer=transer, C=0.1, base_score=600, base_odds=35, pdo=60, rate=2
)
card.fit(train_woe[col], train_woe["target"])
#
# 💡 Note!:
# * toad.ScoreCard는 Scorecard 모델을 생성하기 위한 클래스입니다.
# * combiner는 binning 결과를 담고 있는 toad.transform.Combiner 객체를 전달합니다.
# * transer는 WOE 변환을 담당하는 toad.transform.WOETransformer 객체를 전달합니다.
# * C는 Logistic Regression에서의 규제 강도를 나타내는 매개변수입니다.
# * base_score는 기준 스코어로, 기본적으로 모든 변수의 WOE 값이 0일 때의 스코어입니다.
# * base_odds는 기준 오즈로, 기본적으로 모든 변수의 WOE 값이 0일 때의 오즈입니다.
# * pdo는 Point to Double the Odds로, 오즈를 두배로 만들기 위해 필요한 점수의 차이입니다.
# * rate는 모델의 점수 스케일을 조절하기 위한 비율입니다.
# * fit() 메서드를 사용하여 Scorecard 모델을 훈련시킵니다. train_woe[col]은 독립 변수를, train_woe['target']은 종속 변수를 나타냅니다.
sample_train_woe = train_woe.sample(3)
score_sample = card.predict(sample_train_woe[col])
score_sample_rounded = score_sample.round().astype(int)
for i, score in enumerate(score_sample_rounded, start=1):
print(f"{i}번째 고객의 점수는 \033[1;34m{score}\033[0m점 입니다.")
#
# 💡 Note:
# * 1. `sample_train_woe = train_woe.sample(3)`: `sample()` 함수를 사용하여 `train_woe` 데이터셋에서 임의로 3개의 샘플을 추출합니다. 이 샘플들은 점수를 계산할 데이터입니다.
# * 2. `score_sample = card.predict(sample_train_woe[col])`: `card` 라는 ScoreCard 객체의 `predict()` 메서드를 사용하여 위에서 추출한 샘플에 대한 점수를 계산합니다. 이 때, `sample_train_woe[col]`을 통해 샘플의 해당 컬럼(변수)만 사용하게 됩니다.
# * 3. `score_sample_rounded = score_sample.round().astype(int)`: 계산된 점수는 일반적으로 소수점 형태로 반환됩니다. 이 줄의 코드는 `round()` 함수를 사용하여 계산된 점수를 반올림하고, `astype(int)`를 통해 정수형으로 변환합니다.
# * 4. `for i, score in enumerate(score_sample_rounded, start=1)`: 이 줄은 반복문을 사용하여 각 샘플에 대한 점수를 순서대로 출력합니다. `enumerate()` 함수는 반복 가능한 객체(여기서는 `score_sample_rounded`)를 입력으로 받아 인덱스 번호(i)와 그에 해당하는 값을(score) 함께 반환합니다. `start=1`은 인덱스 번호가 1부터 시작하도록 설정합니다.
# * 5. `print(f"{i}번째 고객의 점수는 \033[1;34m{score}\033[0m점 입니다.")`: 이 줄은 문자열 포매팅을 사용하여 각 샘플에 대한 점수를 보기 좋게 출력합니다. `\033[1;34m`와 `\033[0m` 사이에 있는 텍스트는 파란색으로 출력됩니다.
card.export()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/870/129870801.ipynb
|
amex-data-sampled
|
kimtaehun
|
[{"Id": 129870801, "ScriptId": 38605996, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13683894, "CreationDate": "05/17/2023 05:09:54", "VersionNumber": 1.0, "Title": "4\uac15) TOAD \ub77c\uc774\ube0c\ub7ec\ub9ac\ub97c \ud65c\uc6a9\ud55c \uc2e0\uc6a9\ud3c9\uac00 \ubaa8\ub378 \uac1c\ubc1c", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 329.0, "LinesInsertedFromPrevious": 329.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 186271118, "KernelVersionId": 129870801, "SourceDatasetVersionId": 5688399}]
|
[{"Id": 5688399, "DatasetId": 3270398, "DatasourceVersionId": 5763995, "CreatorUserId": 1885842, "LicenseName": "Unknown", "CreationDate": "05/15/2023 07:57:57", "VersionNumber": 1.0, "Title": "AMEX_data_sampled", "Slug": "amex-data-sampled", "Subtitle": "This is a small-sized sampled dataset from AMEX dafault prediction dataset", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3270398, "CreatorUserId": 1885842, "OwnerUserId": 1885842.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5688399.0, "CurrentDatasourceVersionId": 5763995.0, "ForumId": 3336031, "Type": 2, "CreationDate": "05/15/2023 07:57:57", "LastActivityDate": "05/15/2023", "TotalViews": 127, "TotalDownloads": 3, "TotalVotes": 8, "TotalKernels": 3}]
|
[{"Id": 1885842, "UserName": "kimtaehun", "DisplayName": "DataManyo", "RegisterDate": "05/05/2018", "PerformanceTier": 4}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### 4강) TOAD 라이브러리를 활용한 신용평가 모델 개발
# > ### 데이터 준비
import warnings
warnings.filterwarnings("ignore", module="sklearn.metrics.cluster")
def drop_null_cols(df, threshold=0.8):
"""
데이터프레임에서 결측치 비율이 threshold 이상인 변수를 제거하는 함수
"""
null_percent = df.isnull().mean()
drop_cols = list(null_percent[null_percent >= threshold].index)
df = df.drop(drop_cols, axis=1)
print(f"Dropped {len(drop_cols)} columns: {', '.join(drop_cols)}")
return df
df = pd.read_pickle("/kaggle/input/amex-data-sampled/train_df_sample.pkl")
df = df.reset_index()
import hashlib
def encode_customer_id(id_str):
encoded_id = hashlib.sha256(id_str.encode("utf-8")).hexdigest()[:16]
return encoded_id
df["customer_ID"] = df["customer_ID"].apply(encode_customer_id)
df = drop_null_cols(df)
cat_features = [
"B_30",
"B_38",
"D_114",
"D_116",
"D_117",
"D_120",
"D_126",
"D_63",
"D_64",
"D_68",
]
cat_features = [f"{cf}_last" for cf in cat_features]
import random
num_cols = df.select_dtypes(include=np.number).columns.tolist()
num_cols = [col for col in num_cols if "target" not in col and col not in cat_features]
num_cols_sample = random.sample([col for col in num_cols if "target" not in col], 100)
feature_list = num_cols_sample + cat_features
all_list = feature_list + ["target"]
df = df[all_list]
for categorical_feature in cat_features:
if df[categorical_feature].dtype == "float16":
df[categorical_feature] = df[categorical_feature].astype(str)
if df[categorical_feature].dtype == "category":
df[categorical_feature] = df[categorical_feature].astype(str)
elif df[categorical_feature].dtype == "object":
df[categorical_feature] = df[categorical_feature].astype(str)
from sklearn.preprocessing import LabelEncoder
le_encoder = LabelEncoder()
for categorical_feature in cat_features:
df[categorical_feature].fillna(value="NaN", inplace=True)
df[categorical_feature] = le_encoder.fit_transform(df[categorical_feature])
from sklearn.impute import SimpleImputer
def impute_nan(df, num_cols, strategy="mean"):
"""
NaN 값을 strategy에 따라 num_cols에 대해 impute하는 함수
:param df: DataFrame
:param num_cols: list, imputation 대상 numeric column 리스트
:param strategy: str, imputation 전략 (default: 'mean')
:return: DataFrame, imputed DataFrame
"""
imputer = SimpleImputer(strategy=strategy)
df[num_cols] = imputer.fit_transform(df[num_cols])
return df
df = impute_nan(df, num_cols_sample, strategy="mean")
# > #### taod 패키지 설치
# > #### IV값을 기준으로 변수를 선택해보자. (Feature Selection)
import toad
# 타겟 변수 정의
target = "target"
# 정보값(IV) 계산 및 특성 선택
iv_df = toad.quality(df, target=target, iv_only=True)
selected_features = iv_df[iv_df["iv"] > 0.1].index # 'name' 대신 'index'를 사용합니다.
# WOE 변환
trans = toad.transform.WOETransformer()
df_woe = trans.fit_transform(df[selected_features], df[target])
# 이제 df_woe를 추가적인 모델링에 사용할 수 있습니다.
# toad는 파이썬의 데이터 전처리 및 탐색을 위한 오픈 소스 패키지입니다. 주요 기능은 다음과 같습니다:
# 데이터 품질 평가: toad.quality() 함수를 사용하여 변수별 정보값(IV, Information Value)을 계산하고, 타겟 변수와의 상관 관계를 평가합니다. 이를 통해 변수의 중요도를 판단하고 특성 선택에 활용할 수 있습니다.
# 데이터 변환: WOE (Weight of Evidence) 변환을 위한 toad.transform.WOETransformer()를 사용할 수 있습니다. WOE는 범주형 변수나 연속형 변수를 범주화하여 정보값을 측정하는 방법으로, 변수의 예측력을 개선하고 모델 성능을 향상시킬 수 있습니다.
df_woe
# > #### 선택된 피처를 기반으로 binning 작업을 진행한다. 이번엔 방법을 조금 다르게해서, 각 구간이 최소 5%의 데이터를 가지도록 구간화를 수행한다.
combiner = toad.transform.Combiner()
combiner.fit(df[selected_features], y=df[target], method="chi", min_samples=0.05)
# min_samples = 0.05로 설정하면, 각 구간이 최소 5%의 데이터를 가지도록 구간화를 수행합니다.
# 구간화 결과를 살펴봅시다.
binning_result = combiner.export()
# > #### iv=True는 toad.plot 함수의 매개변수 중 하나입니다. 이 매개변수를 True로 설정하면, 그래프에 Information Value (IV) 값을 표시합니다.
toad.detect(df)[:10]
# > #### toad.detect(df)의 결과는 변수 이름과 해당 변수의 데이터 유형을 포함한 데이터프레임으로 반환됩니다. 결과에서 첫 10개의 변수와 그에 해당하는 데이터 유형을 확인할 수 있습니다.
toad.quality(df, "target", iv_only=True)[:15]
# > #### 각 변수별 IV값을 살펴보자.
# #### Feature Selection
# 💡 Notes:
# * empty=0.9: 결측치 비율이 90%보다 큰 특성들은 필터링됩니다.
# * iv=0.02: IV(Information Value)가 0.02보다 작은 특성들은 제거됩니다.
# * corr=0.7: 두 개 이상의 특성들 간의 피어슨 상관계수가 0.7보다 큰 경우, IV가 더 낮은 특성들이 제거됩니다.
# * return_drop=False: True로 설정하면, 함수는 삭제된 열들의 리스트를 반환합니다.
# * exclude=None: 알고리즘에서 제외할 특성들의 리스트를 입력합니다. 일반적으로 ID 열이나 월(Month) 열 등이 해당됩니다.
train_selected, dropped = toad.selection.select(
df,
target="target",
empty=0.5,
iv=0.05,
corr=0.7,
return_drop=True,
exclude=["D_117_last"],
)
print(dropped)
print(train_selected.shape)
# > #### toad.selection.select 함수는 선택된 특성들로 이루어진 훈련 데이터셋인 train_selected와 삭제된 열들의 리스트인 dropped를 반환합니다. 또한, train_selected.shape는 선택된 특성들을 포함하는 훈련 데이터셋의 크기를 출력합니다.
# initialise
c = toad.transform.Combiner()
# Train binning with the selected features from previous; use reliable Chi-squared binning, and control that each bucket has at least 5% sample.
c.fit(
train_selected, y="target", method="chi", min_samples=0.05, exclude=["D_117_last"]
)
print("D_59_min:", c.export()["D_59_min"])
print("R_15_std:", c.export()["R_15_std"])
print("S_3_last:", c.export()["S_3_last"])
# > ##### 위 코드는 c.export()를 사용하여 binning 결과 중 특정 변수들의 정보를 출력하는 부분입니다.
# c.export(): Combiner 객체 c의 binning 결과를 딕셔너리 형태로 반환합니다. 딕셔너리의 키는 각 변수의 이름이며, 값은 해당 변수의 binning 정보를 담고 있는 객체입니다.
# 각 변수에 대한 binning 정보는 해당 변수의 구간(bin)과 해당 구간의 라벨(label) 등을 포함하고 있습니다. 출력된 정보를 통해 각 변수의 binning 결과를 확인할 수 있습니다.
from toad.plot import bin_plot
# 학습 데이터(train_selected)에서 'var_d2' 변수의 bin 결과를 확인합니다.
col = "D_59_min"
# 시각화를 위해 'labels = True'로 설정하는 것이 좋습니다.
bin_plot(
c.transform(train_selected[[col, "target"]], labels=True), x=col, target="target"
)
# > ##### 위 코드는 bin_plot 함수를 사용하여 'R_4_std' 변수의 binning 결과를 시각화하는 부분입니다.
# > ##### c.transform(train_selected[[col, 'target']], labels=True): 학습 데이터(train_selected)의 'R_4_std' 변수와 목표 변수('target')를 선택하여 binning 결과를 생성합니다. labels=True로 설정하여 bin의 라벨을 포함한 결과를 반환합니다.
# > ##### bin_plot(...): binning 결과를 시각화합니다. x축에는 'R_4_std' 변수의 값이 나타나며, target 변수('target')의 값에 따라 각 bin의 분포를 확인할 수 있습니다. bin_plot 함수를 통해 binning 결과를 시각적으로 확인하여 데이터의 패턴을 파악할 수 있습니다.
# 학습 데이터(train_selected)에서 'D_51_min' 변수의 bin 결과를 확인합니다.
col = "S_3_last"
# 범주형 변수의 경우 'labels = True'로 설정하는 것이 좋습니다.
bin_plot(
c.transform(train_selected[[col, "target"]], labels=True), x=col, target="target"
)
# Toad의 binning 기능은 범주형 및 수치형 변수를 모두 지원합니다. "toad.transform.Combiner()" 클래스를 사용하여 학습하며, 절차는 다음과 같습니다:
# * 초기화(initalise) : c = toad.transform.Combiner()
# * *train binning*: c.fit(dataframe, y='target', method='chi', min_samples=None, n_bins=None, empty_separate=False)
# * y: 목표 변수;
# * method: binning에 적용할 방법. 'chi' (카이제곱), 'dt' (의사결정 트리), 'kmeans' (K-means), 'quantile' (동일한 백분위수 기준), 'step' (동일한 간격)을 지원합니다.
# * min_samples: 샘플당 요구되는 최소 수 또는 비율. 각 버킷에 필요한 최소 샘플 수 / 비율입니다.
# * n_bins: 최소한의 버킷 수. 수가 너무 큰 경우, 알고리즘은 얻을 수 있는 최대 버킷 수를 반환합니다.
# * empty_separate: 누락된 값이 버킷 내에 별도로 분리되는지 여부. False인 경우, 누락된 값은 가장 가까운 나쁜 비율 버킷과 함께 배치됩니다.
# * binning 결과: c.export()
# * bins 조정: c.update(dict)
# * bins 적용 및 이산값으로 변환: c.transform(dataframe, labels=False)
# * labels: 데이터를 설명 라벨로 변환할지 여부. False인 경우 0, 1, 2...로 반환됩니다. 범주형 변수는 비율의 내림차순으로 정렬됩니다. True인 경우 (-무한대, 0], (0, 10], (10, 무한대)와 같이 반환됩니다.
# * 참고: 1. 불필요한 열을 제외하는 것을 잊지 마세요. 특히 ID 열과 타임스탬프 열은 제외해야 합니다.. 2. 고유 값이 많은 열은 학습에 많은 시간이 소요될 수 있습니다.*
# 초기화
transer = toad.transform.WOETransformer()
# transer.fit_transform() 및 combiner.transform()을 적용합니다. target을 제외하도록 주의하세요.
train_woe = transer.fit_transform(
c.transform(train_selected), train_selected["target"], exclude="target"
)
train_woe.head(3)
# * WOETransformer를 초기화합니다.
# * combiner.transform()의 결과에 transer.fit_transform()을 적용합니다.
# * target을 제외하도록 주의하세요.
# * 변환된 train_woe 데이터프레임의 처음 3개 행을 출력합니다.
col = train_woe.columns.tolist()[:-1]
col.remove("D_117_last")
# > #### 예시를 위해 제거했던 변수를 제거해줍니다.
# > #### 예시를 위해 간단한 로지스틱 회귀 모델을 적합해봅니다.
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression()
lr.fit(train_woe[col], train_woe["target"])
# 훈련 데이터와 Out-of-Time (OOT) 데이터에 대해 예측된 확률을 구합니다.
pred_train = lr.predict_proba(train_woe[col])[:, 1]
# > #### Toad 역시 쉽게 KS 통계량과 AUC를 계산할 수 있는 라이브러리를 제공합니다.
from toad.metrics import KS, AUC
print("train KS", KS(pred_train, train_woe["target"]))
print("train AUC", AUC(pred_train, train_woe["target"]))
# > #### Toad에서도 OptBinning과 마찬가지로 스코어링을 할 수 있는 스코어 카드 기능을 제공합니다.
card = toad.ScoreCard(
combiner=c, transer=transer, C=0.1, base_score=600, base_odds=35, pdo=60, rate=2
)
card.fit(train_woe[col], train_woe["target"])
#
# 💡 Note!:
# * toad.ScoreCard는 Scorecard 모델을 생성하기 위한 클래스입니다.
# * combiner는 binning 결과를 담고 있는 toad.transform.Combiner 객체를 전달합니다.
# * transer는 WOE 변환을 담당하는 toad.transform.WOETransformer 객체를 전달합니다.
# * C는 Logistic Regression에서의 규제 강도를 나타내는 매개변수입니다.
# * base_score는 기준 스코어로, 기본적으로 모든 변수의 WOE 값이 0일 때의 스코어입니다.
# * base_odds는 기준 오즈로, 기본적으로 모든 변수의 WOE 값이 0일 때의 오즈입니다.
# * pdo는 Point to Double the Odds로, 오즈를 두배로 만들기 위해 필요한 점수의 차이입니다.
# * rate는 모델의 점수 스케일을 조절하기 위한 비율입니다.
# * fit() 메서드를 사용하여 Scorecard 모델을 훈련시킵니다. train_woe[col]은 독립 변수를, train_woe['target']은 종속 변수를 나타냅니다.
sample_train_woe = train_woe.sample(3)
score_sample = card.predict(sample_train_woe[col])
score_sample_rounded = score_sample.round().astype(int)
for i, score in enumerate(score_sample_rounded, start=1):
print(f"{i}번째 고객의 점수는 \033[1;34m{score}\033[0m점 입니다.")
#
# 💡 Note:
# * 1. `sample_train_woe = train_woe.sample(3)`: `sample()` 함수를 사용하여 `train_woe` 데이터셋에서 임의로 3개의 샘플을 추출합니다. 이 샘플들은 점수를 계산할 데이터입니다.
# * 2. `score_sample = card.predict(sample_train_woe[col])`: `card` 라는 ScoreCard 객체의 `predict()` 메서드를 사용하여 위에서 추출한 샘플에 대한 점수를 계산합니다. 이 때, `sample_train_woe[col]`을 통해 샘플의 해당 컬럼(변수)만 사용하게 됩니다.
# * 3. `score_sample_rounded = score_sample.round().astype(int)`: 계산된 점수는 일반적으로 소수점 형태로 반환됩니다. 이 줄의 코드는 `round()` 함수를 사용하여 계산된 점수를 반올림하고, `astype(int)`를 통해 정수형으로 변환합니다.
# * 4. `for i, score in enumerate(score_sample_rounded, start=1)`: 이 줄은 반복문을 사용하여 각 샘플에 대한 점수를 순서대로 출력합니다. `enumerate()` 함수는 반복 가능한 객체(여기서는 `score_sample_rounded`)를 입력으로 받아 인덱스 번호(i)와 그에 해당하는 값을(score) 함께 반환합니다. `start=1`은 인덱스 번호가 1부터 시작하도록 설정합니다.
# * 5. `print(f"{i}번째 고객의 점수는 \033[1;34m{score}\033[0m점 입니다.")`: 이 줄은 문자열 포매팅을 사용하여 각 샘플에 대한 점수를 보기 좋게 출력합니다. `\033[1;34m`와 `\033[0m` 사이에 있는 텍스트는 파란색으로 출력됩니다.
card.export()
| false | 0 | 4,837 | 3 | 4,863 | 4,837 |
||
129870816
|
<jupyter_start><jupyter_text>Adult Census Income
This data was extracted from the [1994 Census bureau database][1] by Ronny Kohavi and Barry Becker (Data Mining and Visualization, Silicon Graphics). A set of reasonably clean records was extracted using the following conditions: ((AAGE>16) && (AGI>100) && (AFNLWGT>1) && (HRSWK>0)). *The prediction task is to determine whether a person makes over $50K a year*.
## Description of fnlwgt (final weight)
The weights on the Current Population Survey (CPS) files are controlled to independent estimates of the civilian noninstitutional population of the US. These are prepared monthly for us by Population Division here at the Census Bureau. We use 3 sets of controls. These are:
1. A single cell estimate of the population 16+ for each state.
2. Controls for Hispanic Origin by age and sex.
3. Controls by Race, age and sex.
We use all three sets of controls in our weighting program and "rake" through them 6 times so that by the end we come back to all the controls we used. The term estimate refers to population totals derived from CPS by creating "weighted tallies" of any specified socio-economic characteristics of the population. People with similar demographic characteristics should have similar weights. There is one important caveat to remember about this statement. That is that since the CPS sample is actually a collection of 51 state samples, each with its own probability of selection, the statement only applies within state.
##Relevant papers
Ron Kohavi, ["Scaling Up the Accuracy of Naive-Bayes Classifiers: a Decision-Tree Hybrid"][2], *Proceedings of the Second International Conference on Knowledge Discovery and Data Mining*, 1996. (PDF)
[1]: http://www.census.gov/en.html
[2]: http://robotics.stanford.edu/~ronnyk/nbtree.pdf
Kaggle dataset identifier: adult-census-income
<jupyter_script>import pandas as pd
import matplotlib.pyplot as plt
data = pd.read_csv(r"/kaggle/input/adult-census-income/adult.csv")
data
# marital.status : 婚姻狀況
data.keys()
txt_col = [
"workclass",
"education",
"marital.status",
"occupation",
"relationship",
"race",
"sex",
"native.country",
"income",
]
data["relationship_change"] = data["relationship"].replace(
["Wife", "Husband"], "married"
)
# data['relationship_change'] = data['relationship'].replace('Husband','married')
data["relationship_change"].value_counts()
data
with_question = ["workclass", "occupation", "native.country"]
# plt.hist(data['fnlwgt'])
# plt.figure()
data.hist(figsize=(10, 10))
for i in txt_col:
ed = data[i].value_counts()
ed_pd = pd.DataFrame(ed)
plt.figure(figsize=(20, 15))
plt.bar(ed_pd.index, height=ed_pd[i])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/870/129870816.ipynb
|
adult-census-income
| null |
[{"Id": 129870816, "ScriptId": 38625846, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8215644, "CreationDate": "05/17/2023 05:10:11", "VersionNumber": 1.0, "Title": "Adult income", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 40.0, "LinesInsertedFromPrevious": 40.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186271153, "KernelVersionId": 129870816, "SourceDatasetVersionId": 498}]
|
[{"Id": 498, "DatasetId": 225, "DatasourceVersionId": 498, "CreatorUserId": 495305, "LicenseName": "CC0: Public Domain", "CreationDate": "10/07/2016 23:42:59", "VersionNumber": 3.0, "Title": "Adult Census Income", "Slug": "adult-census-income", "Subtitle": "Predict whether income exceeds $50K/yr based on census data", "Description": "This data was extracted from the [1994 Census bureau database][1] by Ronny Kohavi and Barry Becker (Data Mining and Visualization, Silicon Graphics). A set of reasonably clean records was extracted using the following conditions: ((AAGE>16) && (AGI>100) && (AFNLWGT>1) && (HRSWK>0)). *The prediction task is to determine whether a person makes over $50K a year*.\n\n## Description of fnlwgt (final weight)\n\nThe weights on the Current Population Survey (CPS) files are controlled to independent estimates of the civilian noninstitutional population of the US. These are prepared monthly for us by Population Division here at the Census Bureau. We use 3 sets of controls. These are: \n\n 1. A single cell estimate of the population 16+ for each state.\n \n 2. Controls for Hispanic Origin by age and sex.\n\n 3. Controls by Race, age and sex.\n\nWe use all three sets of controls in our weighting program and \"rake\" through them 6 times so that by the end we come back to all the controls we used. The term estimate refers to population totals derived from CPS by creating \"weighted tallies\" of any specified socio-economic characteristics of the population. People with similar demographic characteristics should have similar weights. There is one important caveat to remember about this statement. That is that since the CPS sample is actually a collection of 51 state samples, each with its own probability of selection, the statement only applies within state.\n\n##Relevant papers\n\nRon Kohavi, [\"Scaling Up the Accuracy of Naive-Bayes Classifiers: a Decision-Tree Hybrid\"][2], *Proceedings of the Second International Conference on Knowledge Discovery and Data Mining*, 1996. (PDF)\n\n [1]: http://www.census.gov/en.html\n [2]: http://robotics.stanford.edu/~ronnyk/nbtree.pdf", "VersionNotes": "Removed leading whitespace everywhere", "TotalCompressedBytes": 4104734.0, "TotalUncompressedBytes": 4104734.0}]
|
[{"Id": 225, "CreatorUserId": 495305, "OwnerUserId": NaN, "OwnerOrganizationId": 7.0, "CurrentDatasetVersionId": 498.0, "CurrentDatasourceVersionId": 498.0, "ForumId": 1649, "Type": 2, "CreationDate": "10/06/2016 17:19:07", "LastActivityDate": "02/05/2018", "TotalViews": 592007, "TotalDownloads": 52622, "TotalVotes": 593, "TotalKernels": 478}]
| null |
import pandas as pd
import matplotlib.pyplot as plt
data = pd.read_csv(r"/kaggle/input/adult-census-income/adult.csv")
data
# marital.status : 婚姻狀況
data.keys()
txt_col = [
"workclass",
"education",
"marital.status",
"occupation",
"relationship",
"race",
"sex",
"native.country",
"income",
]
data["relationship_change"] = data["relationship"].replace(
["Wife", "Husband"], "married"
)
# data['relationship_change'] = data['relationship'].replace('Husband','married')
data["relationship_change"].value_counts()
data
with_question = ["workclass", "occupation", "native.country"]
# plt.hist(data['fnlwgt'])
# plt.figure()
data.hist(figsize=(10, 10))
for i in txt_col:
ed = data[i].value_counts()
ed_pd = pd.DataFrame(ed)
plt.figure(figsize=(20, 15))
plt.bar(ed_pd.index, height=ed_pd[i])
| false | 0 | 295 | 0 | 777 | 295 |
||
129870360
|
# # Import Libraries and set paths
import numpy as np
import glob
import PIL.Image as Image
import matplotlib.pyplot as plt
import matplotlib.patches as patches
from tqdm import tqdm
from io import StringIO
from sklearn.metrics import fbeta_score
from skimage.util import view_as_windows
from scipy.ndimage import distance_transform_edt
from numba import jit
# Constants
PREFIX = "/kaggle/input/vesuvius-challenge-ink-detection/train/3/"
# Load mask image
mask = np.array(Image.open(PREFIX + "mask.png").convert("1"))
# Load label image
label = (np.array(Image.open(PREFIX + "inklabels.png")) > 0).astype(np.float32)
# Load infrared image
ir = np.array(Image.open(PREFIX + "ir.png"))
# Load the 3D x-ray scan, one slice at a time
images = [
np.array(Image.open(filename))
for filename in tqdm(sorted(glob.glob(PREFIX + "surface_volume/*.tif")))
]
@jit(nopython=True)
def get_value_ink_ratio(value_count_ink, value_count_all, a, label):
for v, l in zip(a.ravel(), label.ravel()):
value_count_all[v] += 1
if l:
value_count_ink[v] += 1
def plot_ink_ratio(value_count_ink, value_count_all, a):
value_ink_ratio = np.where(
value_count_all == 0, 0, value_count_ink / value_count_all
)
x = np.arange(len(value_ink_ratio))
# plot ink ratio distribution
fig, ax = plt.subplots(1, 1, figsize=(14, 2))
ax.plot(x, value_ink_ratio, linestyle="", marker=".")
plt.show()
# select
sorted_by_ink = np.argsort(value_ink_ratio)
sorted_ink_ratio = value_ink_ratio[sorted_by_ink]
truth = value_count_all.sum()
f05 = np.zeros(101)
best_f05 = 0
best_th = 0
for th in range(101):
high_ink_ratio = sorted_by_ink[sorted_ink_ratio > th / 100]
tp = value_count_ink[high_ink_ratio].sum()
fp = value_count_all[high_ink_ratio].sum() - tp
fn = truth - tp
f05[th] = 1.25 * tp / (1.25 * tp + fp + 0.25 * fn)
if best_f05 < f05[th]:
best_th = th / 100
best_f05 = f05[th]
fig, ax = plt.subplots(1, 1, figsize=(14, 2))
ax.plot(np.arange(101) / 100, f05, linestyle="", marker=".")
plt.show()
high_ink_ratio = sorted_by_ink[sorted_ink_ratio > 0.15]
high_ink = np.isin(a, high_ink_ratio)
print("Number of high ink ratio values:", high_ink.shape, best_th, best_f05)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(14, 7))
ax1.imshow(high_ink, cmap="gray")
ax2.imshow(label, cmap="gray")
plt.show()
NUM_VALUES = 256
value_count_ink = np.zeros(NUM_VALUES, dtype=int)
value_count_all = np.zeros(NUM_VALUES, dtype=int)
get_value_ink_ratio(value_count_ink, value_count_all, ir, label)
plot_ink_ratio(value_count_ink, value_count_all, ir)
NUM_VALUES = 65536
Z_DIM = len(images)
value_count_ink = np.zeros((Z_DIM, NUM_VALUES), dtype=int)
value_count_all = np.zeros((Z_DIM, NUM_VALUES), dtype=int)
for z in tqdm(range(Z_DIM)):
img = images[z]
get_value_ink_ratio(value_count_ink[z], value_count_all[z], img, label)
plot_ink_ratio(value_count_ink[z], value_count_all[z], img)
# Function to generate run-length encoding (RLE) for the binary mask
def rle(img):
pixels = img.flatten()
pixels = np.concatenate([[0], pixels, [0]])
runs = np.where(pixels[1:] != pixels[:-1])[0] + 1
runs[1::2] -= runs[::2]
f = StringIO()
np.savetxt(f, runs.reshape(1, -1), delimiter=" ", fmt="%d")
predicted = f.getvalue().strip()
return predicted
# Generate RLE for the binary output
rle_output = rle(binary_output)
# Save the RLE to a CSV file for submission
with open("submission.csv", "w") as f:
f.write("Id,Predicted\n")
f.write("a," + rle_output + "\n")
f.write("b," + rle_output + "\n")
print("Submission file 'submission.csv' has been generated.")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/870/129870360.ipynb
| null | null |
[{"Id": 129870360, "ScriptId": 38602646, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2586962, "CreationDate": "05/17/2023 05:04:37", "VersionNumber": 2.0, "Title": "Visualize ink to pixel value correlation", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 114.0, "LinesInsertedFromPrevious": 64.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 50.0, "LinesInsertedFromFork": 62.0, "LinesDeletedFromFork": 257.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 52.0, "TotalVotes": 0}]
| null | null | null | null |
# # Import Libraries and set paths
import numpy as np
import glob
import PIL.Image as Image
import matplotlib.pyplot as plt
import matplotlib.patches as patches
from tqdm import tqdm
from io import StringIO
from sklearn.metrics import fbeta_score
from skimage.util import view_as_windows
from scipy.ndimage import distance_transform_edt
from numba import jit
# Constants
PREFIX = "/kaggle/input/vesuvius-challenge-ink-detection/train/3/"
# Load mask image
mask = np.array(Image.open(PREFIX + "mask.png").convert("1"))
# Load label image
label = (np.array(Image.open(PREFIX + "inklabels.png")) > 0).astype(np.float32)
# Load infrared image
ir = np.array(Image.open(PREFIX + "ir.png"))
# Load the 3D x-ray scan, one slice at a time
images = [
np.array(Image.open(filename))
for filename in tqdm(sorted(glob.glob(PREFIX + "surface_volume/*.tif")))
]
@jit(nopython=True)
def get_value_ink_ratio(value_count_ink, value_count_all, a, label):
for v, l in zip(a.ravel(), label.ravel()):
value_count_all[v] += 1
if l:
value_count_ink[v] += 1
def plot_ink_ratio(value_count_ink, value_count_all, a):
value_ink_ratio = np.where(
value_count_all == 0, 0, value_count_ink / value_count_all
)
x = np.arange(len(value_ink_ratio))
# plot ink ratio distribution
fig, ax = plt.subplots(1, 1, figsize=(14, 2))
ax.plot(x, value_ink_ratio, linestyle="", marker=".")
plt.show()
# select
sorted_by_ink = np.argsort(value_ink_ratio)
sorted_ink_ratio = value_ink_ratio[sorted_by_ink]
truth = value_count_all.sum()
f05 = np.zeros(101)
best_f05 = 0
best_th = 0
for th in range(101):
high_ink_ratio = sorted_by_ink[sorted_ink_ratio > th / 100]
tp = value_count_ink[high_ink_ratio].sum()
fp = value_count_all[high_ink_ratio].sum() - tp
fn = truth - tp
f05[th] = 1.25 * tp / (1.25 * tp + fp + 0.25 * fn)
if best_f05 < f05[th]:
best_th = th / 100
best_f05 = f05[th]
fig, ax = plt.subplots(1, 1, figsize=(14, 2))
ax.plot(np.arange(101) / 100, f05, linestyle="", marker=".")
plt.show()
high_ink_ratio = sorted_by_ink[sorted_ink_ratio > 0.15]
high_ink = np.isin(a, high_ink_ratio)
print("Number of high ink ratio values:", high_ink.shape, best_th, best_f05)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(14, 7))
ax1.imshow(high_ink, cmap="gray")
ax2.imshow(label, cmap="gray")
plt.show()
NUM_VALUES = 256
value_count_ink = np.zeros(NUM_VALUES, dtype=int)
value_count_all = np.zeros(NUM_VALUES, dtype=int)
get_value_ink_ratio(value_count_ink, value_count_all, ir, label)
plot_ink_ratio(value_count_ink, value_count_all, ir)
NUM_VALUES = 65536
Z_DIM = len(images)
value_count_ink = np.zeros((Z_DIM, NUM_VALUES), dtype=int)
value_count_all = np.zeros((Z_DIM, NUM_VALUES), dtype=int)
for z in tqdm(range(Z_DIM)):
img = images[z]
get_value_ink_ratio(value_count_ink[z], value_count_all[z], img, label)
plot_ink_ratio(value_count_ink[z], value_count_all[z], img)
# Function to generate run-length encoding (RLE) for the binary mask
def rle(img):
pixels = img.flatten()
pixels = np.concatenate([[0], pixels, [0]])
runs = np.where(pixels[1:] != pixels[:-1])[0] + 1
runs[1::2] -= runs[::2]
f = StringIO()
np.savetxt(f, runs.reshape(1, -1), delimiter=" ", fmt="%d")
predicted = f.getvalue().strip()
return predicted
# Generate RLE for the binary output
rle_output = rle(binary_output)
# Save the RLE to a CSV file for submission
with open("submission.csv", "w") as f:
f.write("Id,Predicted\n")
f.write("a," + rle_output + "\n")
f.write("b," + rle_output + "\n")
print("Submission file 'submission.csv' has been generated.")
| false | 0 | 1,341 | 0 | 1,341 | 1,341 |
||
129870775
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.model_selection import train_test_split
from scipy.stats import spearmanr
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import warnings
from tqdm import tqdm
import requests
warnings.filterwarnings(
"ignore"
) # This is not advised in general, but it is used in this notebook to clean the presentation of results
"""
This is a basic example of what you need to do to participate to the tournament.
The code will not have access to the internet (or any socket related operation).
"""
# Imports
import xgboost as xgb
import pandas as pd
import typing
import joblib
from pathlib import Path
def train(
X_train: pd.DataFrame,
y_train: pd.DataFrame,
model_directory_path: str = "resources",
) -> None:
"""
Do your model training here.
At each retrain this function will have to save an updated version of
the model under the model_directiory_path, as in the example below.
Note: You can use other serialization methods than joblib.dump(), as
long as it matches what reads the model in infer().
Args:
X_train, y_train: the data to train the model.
model_directory_path: the path to save your updated model
Returns:
None
"""
# basic xgboost regressor
model = xgb.XGBRegressor(
objective="reg:squarederror",
max_depth=3,
learning_rate=0.1,
n_estimators=50,
max_leaves=2**3,
n_jobs=-1,
colsample_bytree=0.1,
)
# training the model
print("training...")
model.fit(X_train.iloc[:, 2:], y_train.iloc[:, 2:])
# make sure that the train function correctly save the trained model
# in the model_directory_path
model_pathname = Path(model_directory_path) / "model.joblib"
print(f"Saving model in {model_pathname}")
joblib.dump(model, model_pathname)
def infer(
X_test: pd.DataFrame, model_directory_path: str = "resources"
) -> pd.DataFrame:
"""
Do your inference here.
This function will load the model saved at the previous iteration and use
it to produce your inference on the current date.
It is mandatory to send your inferences with the ids so the system
can match it correctly.
Args:
model_directory_path: the path to the directory to the directory in wich we will be saving your updated model.
X_test: the independant variables of the current date passed to your model.
Returns:
A dataframe (date, id, value) with the inferences of your model for the current date.
"""
# loading the model saved by the train function at previous iteration
model = joblib.load(Path(model_directory_path) / "model.joblib")
# creating the predicted label dataframe with correct dates and ids
y_test_predicted = X_test[["date", "id"]].copy()
y_test_predicted["value"] = model.predict(X_test.iloc[:, 2:])
return y_test_predicted
X_train = pd.read_parquet(
"/kaggle/input/adia-lab-crunchdao-competition/X_train.parquet"
)
y_train = pd.read_parquet(
"/kaggle/input/adia-lab-crunchdao-competition/y_train.parquet"
)
X_test = pd.read_parquet("/kaggle/input/adia-lab-crunchdao-competition/X_test.parquet")
print(
"Splitting (X_train, y_train) in X_train_local, X_test_local, y_train_local, y_test_local"
)
X_train_local, X_test_local, y_train_local, y_test_local = train_test_split(
X_train, y_train, test_size=0.2, shuffle=False
)
# Training. It may require a few minutes.
train(X_train_local, y_train_local, "/kaggle/working/")
print("Inference")
y_test_local_pred = infer(X_test_local, model_directory_path="/kaggle/working/")
score = spearmanr(y_test_local["y"], y_test_local_pred["value"])[0] * 100
print(f"Spearman's correlation {score}")
X_train.head(10)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/870/129870775.ipynb
| null | null |
[{"Id": 129870775, "ScriptId": 38622519, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2003977, "CreationDate": "05/17/2023 05:09:32", "VersionNumber": 1.0, "Title": "ADIA Lab Competition - EDA", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 156.0, "LinesInsertedFromPrevious": 156.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.model_selection import train_test_split
from scipy.stats import spearmanr
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import warnings
from tqdm import tqdm
import requests
warnings.filterwarnings(
"ignore"
) # This is not advised in general, but it is used in this notebook to clean the presentation of results
"""
This is a basic example of what you need to do to participate to the tournament.
The code will not have access to the internet (or any socket related operation).
"""
# Imports
import xgboost as xgb
import pandas as pd
import typing
import joblib
from pathlib import Path
def train(
X_train: pd.DataFrame,
y_train: pd.DataFrame,
model_directory_path: str = "resources",
) -> None:
"""
Do your model training here.
At each retrain this function will have to save an updated version of
the model under the model_directiory_path, as in the example below.
Note: You can use other serialization methods than joblib.dump(), as
long as it matches what reads the model in infer().
Args:
X_train, y_train: the data to train the model.
model_directory_path: the path to save your updated model
Returns:
None
"""
# basic xgboost regressor
model = xgb.XGBRegressor(
objective="reg:squarederror",
max_depth=3,
learning_rate=0.1,
n_estimators=50,
max_leaves=2**3,
n_jobs=-1,
colsample_bytree=0.1,
)
# training the model
print("training...")
model.fit(X_train.iloc[:, 2:], y_train.iloc[:, 2:])
# make sure that the train function correctly save the trained model
# in the model_directory_path
model_pathname = Path(model_directory_path) / "model.joblib"
print(f"Saving model in {model_pathname}")
joblib.dump(model, model_pathname)
def infer(
X_test: pd.DataFrame, model_directory_path: str = "resources"
) -> pd.DataFrame:
"""
Do your inference here.
This function will load the model saved at the previous iteration and use
it to produce your inference on the current date.
It is mandatory to send your inferences with the ids so the system
can match it correctly.
Args:
model_directory_path: the path to the directory to the directory in wich we will be saving your updated model.
X_test: the independant variables of the current date passed to your model.
Returns:
A dataframe (date, id, value) with the inferences of your model for the current date.
"""
# loading the model saved by the train function at previous iteration
model = joblib.load(Path(model_directory_path) / "model.joblib")
# creating the predicted label dataframe with correct dates and ids
y_test_predicted = X_test[["date", "id"]].copy()
y_test_predicted["value"] = model.predict(X_test.iloc[:, 2:])
return y_test_predicted
X_train = pd.read_parquet(
"/kaggle/input/adia-lab-crunchdao-competition/X_train.parquet"
)
y_train = pd.read_parquet(
"/kaggle/input/adia-lab-crunchdao-competition/y_train.parquet"
)
X_test = pd.read_parquet("/kaggle/input/adia-lab-crunchdao-competition/X_test.parquet")
print(
"Splitting (X_train, y_train) in X_train_local, X_test_local, y_train_local, y_test_local"
)
X_train_local, X_test_local, y_train_local, y_test_local = train_test_split(
X_train, y_train, test_size=0.2, shuffle=False
)
# Training. It may require a few minutes.
train(X_train_local, y_train_local, "/kaggle/working/")
print("Inference")
y_test_local_pred = infer(X_test_local, model_directory_path="/kaggle/working/")
score = spearmanr(y_test_local["y"], y_test_local_pred["value"])[0] * 100
print(f"Spearman's correlation {score}")
X_train.head(10)
| false | 0 | 1,255 | 0 | 1,255 | 1,255 |
||
129160363
|
<jupyter_start><jupyter_text>Mobile Games A/B Testing - Cookie Cats
### Context
This dataset includes A/B test results of Cookie Cats to examine what happens when the first gate in the game was moved from level 30 to level 40. When a player installed the game, he or she was randomly assigned to either gate_30 or gate_40.
### Content
The data we have is from 90,189 players that installed the game while the AB-test was running. The variables are:
**userid:** A unique number that identifies each player.
**version:** Whether the player was put in the control group (gate_30 - a gate at level 30) or the group with the moved gate (gate_40 - a gate at level 40).
**sum_gamerounds:** the number of game rounds played by the player during the first 14 days after install.
**retention_1:** Did the player come back and play <strong>1 day</strong> after installing?
**retention_7:** Did the player come back and play <strong>7 days</strong> after installing?
When a player installed the game, he or she was randomly assigned to either.
Kaggle dataset identifier: mobile-games-ab-testing-cookie-cats
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = pd.read_csv("/kaggle/input/mobile-games-ab-testing-cookie-cats/cookie_cats.csv")
data.head(10)
data.info()
data.shape
data.describe()
data["userid"].nunique()
data.groupby("version")[["userid"]].nunique()
import matplotlib.pyplot as plt
plot_data = data.groupby("sum_gamerounds")["userid"].count()
ax = plot_data.head(100).plot()
plt.title("The distribution of players", fontweight="bold", size=14)
plt.xlabel("total gamerounds", size=12)
plt.ylabel("number of player", size=12)
plt.show()
data_retention = data[["retention_1", "retention_7"]].mean() * 100
print(
f"1-day ratio: {round(data_retention[0],2)}% 7-days ratio: {round(data_retention[1],2)}%"
)
# Creating an list with bootstrapped means for each A/B group
boot_1d = []
boot_7d = []
for i in range(1000):
boot_mean_1 = (
data.sample(frac=1, replace=True).groupby("version")["retention_1"].mean()
)
boot_mean_7 = (
data.sample(frac=1, replace=True).groupby("version")["retention_7"].mean()
)
boot_1d.append(boot_mean_1)
boot_7d.append(boot_mean_7)
# Transforming the list to a DataFrame
boot_1d = pd.DataFrame(boot_1d)
boot_7d = pd.DataFrame(boot_7d)
# Kernel Density Estimate plot of the bootstrap distributions
fig, (ax1, ax2) = plt.subplots(1, 2, sharey=True, figsize=(13, 5))
boot_1d.plot.kde(ax=ax1)
ax1.set_xlabel("retantion rate", size=12)
ax1.set_ylabel("number of sample", size=12)
ax1.set_title("1 day retention rate distribution", fontweight="bold", size=14)
boot_7d.plot.kde(ax=ax2)
ax2.set_xlabel("retantion rate", size=12)
ax2.set_title("7 days retention rate distribution", fontweight="bold", size=14)
plt.show()
# Adding a column with the % difference between the two A/B groups
boot_1d["diff"] = (boot_1d["gate_30"] - boot_1d["gate_40"]) / boot_1d["gate_40"] * 100
boot_7d["diff"] = (boot_7d["gate_30"] - boot_7d["gate_40"]) / boot_7d["gate_40"] * 100
# Ploting the bootstrap % difference
fig, (ax1) = plt.subplots(1, 1, figsize=(6, 5))
boot_1d["diff"].plot.kde(ax=ax1, c="#ff99ff", label="1 day retention")
boot_7d["diff"].plot.kde(ax=ax1, c="#00bfff", label="7 days retention")
ax1.set_xlabel("% difference", size=12)
ax1.set_ylabel("% density", size=12)
ax1.set_title(
"Difference in retention \n between the two A/B groups", fontweight="bold", size=14
)
plt.legend()
plt.show()
# Calculating the probability that 1-day retention is greater when the gate is at level 30
prob_1 = (boot_1d["diff"] > 0).sum() / len(boot_1d["diff"])
# Calculating the probability that 7-days retention is greater when the gate is at level 30
prob_7 = (boot_7d["diff"] > 0).sum() / len(boot_7d["diff"])
# Pretty printing the probability
print(
f"The probability that 1-day retention is greater when the gate is at level 30: {round(prob_1,2)*100}% \
\nThe probability that 7-days retention is greater when the gate is at level 30: {(prob_7)*100}% "
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/160/129160363.ipynb
|
mobile-games-ab-testing-cookie-cats
|
mursideyarkin
|
[{"Id": 129160363, "ScriptId": 38397555, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15054663, "CreationDate": "05/11/2023 12:46:55", "VersionNumber": 2.0, "Title": "gb.ru/lessons/318486/", "EvaluationDate": "05/11/2023", "IsChange": false, "TotalLines": 97.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 97.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184962116, "KernelVersionId": 129160363, "SourceDatasetVersionId": 1927698}]
|
[{"Id": 1927698, "DatasetId": 1149830, "DatasourceVersionId": 1966271, "CreatorUserId": 6409983, "LicenseName": "Other (specified in description)", "CreationDate": "02/10/2021 08:16:25", "VersionNumber": 1.0, "Title": "Mobile Games A/B Testing - Cookie Cats", "Slug": "mobile-games-ab-testing-cookie-cats", "Subtitle": "Mobile Games A/B Testing - Cookie Cats", "Description": "### Context\n\nThis dataset includes A/B test results of Cookie Cats to examine what happens when the first gate in the game was moved from level 30 to level 40. When a player installed the game, he or she was randomly assigned to either gate_30 or gate_40. \n\n### Content\n\nThe data we have is from 90,189 players that installed the game while the AB-test was running. The variables are:\n\n**userid:** A unique number that identifies each player.\n**version:** Whether the player was put in the control group (gate_30 - a gate at level 30) or the group with the moved gate (gate_40 - a gate at level 40).\n**sum_gamerounds:** the number of game rounds played by the player during the first 14 days after install.\n**retention_1:** Did the player come back and play <strong>1 day</strong> after installing?\n**retention_7:** Did the player come back and play <strong>7 days</strong> after installing?\n\nWhen a player installed the game, he or she was randomly assigned to either. \n\n### Acknowledgements\n\nThis dataset is taken from [DataCamp](https://www.datacamp.com/projects/184) \nCookie Cat is a hugely popular mobile puzzle game developed by [Tactile Entertainment](https://tactilegames.com/cookie-cats/)\n\nThanks to them for this dataset! \ud83d\ude3b", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1149830, "CreatorUserId": 6409983, "OwnerUserId": 6409983.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1927698.0, "CurrentDatasourceVersionId": 1966271.0, "ForumId": 1167368, "Type": 2, "CreationDate": "02/10/2021 08:16:25", "LastActivityDate": "02/10/2021", "TotalViews": 10291, "TotalDownloads": 742, "TotalVotes": 17, "TotalKernels": 21}]
|
[{"Id": 6409983, "UserName": "mursideyarkin", "DisplayName": "M\u00fcr\u015fide Yark\u0131n", "RegisterDate": "12/20/2020", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = pd.read_csv("/kaggle/input/mobile-games-ab-testing-cookie-cats/cookie_cats.csv")
data.head(10)
data.info()
data.shape
data.describe()
data["userid"].nunique()
data.groupby("version")[["userid"]].nunique()
import matplotlib.pyplot as plt
plot_data = data.groupby("sum_gamerounds")["userid"].count()
ax = plot_data.head(100).plot()
plt.title("The distribution of players", fontweight="bold", size=14)
plt.xlabel("total gamerounds", size=12)
plt.ylabel("number of player", size=12)
plt.show()
data_retention = data[["retention_1", "retention_7"]].mean() * 100
print(
f"1-day ratio: {round(data_retention[0],2)}% 7-days ratio: {round(data_retention[1],2)}%"
)
# Creating an list with bootstrapped means for each A/B group
boot_1d = []
boot_7d = []
for i in range(1000):
boot_mean_1 = (
data.sample(frac=1, replace=True).groupby("version")["retention_1"].mean()
)
boot_mean_7 = (
data.sample(frac=1, replace=True).groupby("version")["retention_7"].mean()
)
boot_1d.append(boot_mean_1)
boot_7d.append(boot_mean_7)
# Transforming the list to a DataFrame
boot_1d = pd.DataFrame(boot_1d)
boot_7d = pd.DataFrame(boot_7d)
# Kernel Density Estimate plot of the bootstrap distributions
fig, (ax1, ax2) = plt.subplots(1, 2, sharey=True, figsize=(13, 5))
boot_1d.plot.kde(ax=ax1)
ax1.set_xlabel("retantion rate", size=12)
ax1.set_ylabel("number of sample", size=12)
ax1.set_title("1 day retention rate distribution", fontweight="bold", size=14)
boot_7d.plot.kde(ax=ax2)
ax2.set_xlabel("retantion rate", size=12)
ax2.set_title("7 days retention rate distribution", fontweight="bold", size=14)
plt.show()
# Adding a column with the % difference between the two A/B groups
boot_1d["diff"] = (boot_1d["gate_30"] - boot_1d["gate_40"]) / boot_1d["gate_40"] * 100
boot_7d["diff"] = (boot_7d["gate_30"] - boot_7d["gate_40"]) / boot_7d["gate_40"] * 100
# Ploting the bootstrap % difference
fig, (ax1) = plt.subplots(1, 1, figsize=(6, 5))
boot_1d["diff"].plot.kde(ax=ax1, c="#ff99ff", label="1 day retention")
boot_7d["diff"].plot.kde(ax=ax1, c="#00bfff", label="7 days retention")
ax1.set_xlabel("% difference", size=12)
ax1.set_ylabel("% density", size=12)
ax1.set_title(
"Difference in retention \n between the two A/B groups", fontweight="bold", size=14
)
plt.legend()
plt.show()
# Calculating the probability that 1-day retention is greater when the gate is at level 30
prob_1 = (boot_1d["diff"] > 0).sum() / len(boot_1d["diff"])
# Calculating the probability that 7-days retention is greater when the gate is at level 30
prob_7 = (boot_7d["diff"] > 0).sum() / len(boot_7d["diff"])
# Pretty printing the probability
print(
f"The probability that 1-day retention is greater when the gate is at level 30: {round(prob_1,2)*100}% \
\nThe probability that 7-days retention is greater when the gate is at level 30: {(prob_7)*100}% "
)
| false | 1 | 1,246 | 0 | 1,538 | 1,246 |
||
129160335
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
#
# Spaceship Titanic
# 
# Table of Contents
# * [Introduction](#section-zero)
# * [Importing Python Libraries](#section-one)
# * [Working with Data](#section-two)
# - [Reading the Data](#subsection-one)
# - [Missing/Null Values](#subsection-two)
# - [Data Visualizations](#subsection-three)
# * [Feature Selection](#section-three)
# - [Fitting the Model](#subsection-four)
# - [Model Selection](#subsection-five)
# - [Decision Tree](#subsection-six)
# * [Submission](#section-four)
# Introduction
# What is Spaceship Titanic?
# > The ***Spaceship Titanic*** was an interstellar passenger liner launched a month ago. With almost 13,000 passengers on board, the vessel set out on its maiden voyage transporting emigrants from our solar system to three newly habitable exoplanets orbiting nearby stars.
# What is our Job?
# > In this competition our task is to predict ***whether a passenger was transported*** to an alternate dimension during the Spaceship Titanic's collision with the spacetime anomaly.
# What are the Data we are provided with?
# > ***PassengerId*** - A unique Id for each passenger. Each Id takes the form gggg_pp where gggg indicates a group the passenger is travelling with and pp is their number within the group. People in a group are often family members, but not always.
# >
# > ***HomePlanet*** - The planet the passenger departed from, typically their planet of permanent residence.
# >
# > ***CryoSleep*** - Indicates whether the passenger elected to be put into suspended animation for the duration of the voyage. Passengers in cryosleep are confined to their cabins.
# >
# > ***Cabin*** - The cabin number where the passenger is staying. Takes the form deck/num/side, where side can be either P for Port or S for Starboard.
# >
# > ***Destination*** - The planet the passenger will be debarking to.
# >
# > ***Age*** - The age of the passenger.
# >
# > ***VIP*** - Whether the passenger has paid for special VIP service during the voyage.
# >
# > ***RoomService, FoodCourt, ShoppingMall, Spa, VRDeck*** - Amount the passenger has billed at each of the Spaceship Titanic's many luxury amenities.
# >
# > ***Name*** - The first and last names of the passenger.
# >
# > ***Transported*** - Whether the passenger was transported to another dimension. This is the target, the column you are trying to predict.
# Importing Python Libraries
#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
#
# Working with the Data
# What changes/modifications are required for Data Cleaning?
# > ***Reading and Analyzing the .csv files:*** Firstly, we will be going through the train and test datasets and checking for the important columns, the missing/null values and columns that are irrevalant to us.
# >
# > ***Deleting Columns:*** As mentioned above, we need to drop the columns that are of no meaning.
# >
# >***Handling Null Values:*** We can either drop the null values using ***drop.na*** or fill the missing values using ***mean*** (for numerical data) or ***mode*** (for numerical/categorical data)
# Reading and Analyzing the Files
#
df = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
data = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
df.head()
data.head()
df.shape
data.shape
sns.heatmap(df.isnull())
#
# Handling Missing Data
#
df.drop("PassengerId", axis=1)
df.drop("Name", axis=1, inplace=True)
data.drop("PassengerId", axis=1)
data.drop("Name", axis=1, inplace=True)
df["RoomService"] = df["RoomService"].fillna(df["RoomService"].mean())
df["FoodCourt"] = df["FoodCourt"].fillna(df["FoodCourt"].mean())
df["VRDeck"] = df["VRDeck"].fillna(df["VRDeck"].mean())
df["VRDeck"] = df["VRDeck"].fillna(df["VRDeck"].mean())
df["CryoSleep"] = df["CryoSleep"].fillna(df["CryoSleep"].mode()[0])
df["HomePlanet"] = df["HomePlanet"].fillna(df["HomePlanet"].mode()[0])
df["Cabin"] = df["Cabin"].fillna(df["Cabin"].mode()[0])
df["Destination"] = df["Destination"].fillna(df["Destination"].mode()[0])
df["Age"] = df["Age"].fillna(df["Age"].mean())
df["VIP"] = df["VIP"].fillna(df["VIP"].mode()[0])
df["ShoppingMall"] = df["ShoppingMall"].fillna(df["ShoppingMall"].mean())
df["Spa"] = df["Spa"].fillna(df["Spa"].mean())
data["RoomService"] = data["RoomService"].fillna(data["RoomService"].mean())
data["FoodCourt"] = data["FoodCourt"].fillna(data["FoodCourt"].mean())
data["VRDeck"] = data["VRDeck"].fillna(data["VRDeck"].mean())
data["VRDeck"] = data["VRDeck"].fillna(data["VRDeck"].mean())
data["CryoSleep"] = data["CryoSleep"].fillna(data["CryoSleep"].mode()[0])
data["HomePlanet"] = data["HomePlanet"].fillna(data["HomePlanet"].mode()[0])
data["Cabin"] = data["Cabin"].fillna(data["Cabin"].mode()[0])
data["Destination"] = data["Destination"].fillna(data["Destination"].mode()[0])
data["Age"] = data["Age"].fillna(data["Age"].mean())
data["VIP"] = data["VIP"].fillna(data["VIP"].mode()[0])
data["ShoppingMall"] = data["ShoppingMall"].fillna(data["ShoppingMall"].mean())
data["Spa"] = data["Spa"].fillna(data["Spa"].mean())
df.isnull().sum()
data.isnull().sum()
new_df = pd.concat([df, data])
new_df.head()
new_df.shape
#
# Data Visualizations
#
plt.figure(figsize=(15, 10))
sns.heatmap(new_df.corr(), annot=True)
sns.histplot(data=new_df, x="Age", bins=20, color="pink")
sns.pairplot(data=new_df, hue="Transported")
columns = ["CryoSleep", "Destination", "VIP", "HomePlanet"]
for col in columns:
fig, ax = plt.subplots(figsize=(5, 3))
sns.countplot(data=new_df, x=col, hue="Transported", ax=ax, color="pink")
#
# Feature Selection
#
df_test = new_df[new_df["Transported"].isnull()]
df_train = new_df[~new_df["Transported"].isnull()]
df.drop("Cabin", axis=1, inplace=True)
data.drop("Cabin", axis=1, inplace=True)
#
# Fitting the Model
# Explaining elements in Model Selection and Model Fitting
# > ***X_train:*** This includes your all independent variables,these will be used to train the model, also as we have specified the test_size = 0.2, this means 80% of observations from your complete data will be used to train/fit the model and rest 20% will be used to test the model. Here, all the features other than 'Outcome' are independent features.
# >
# > ***X_test:*** This is remaining 20% portion of the independent variables from the data which will not be used in the training phase and will be used to make predictions to test the accuracy of the model.
# >
# > ***y_train:*** This is your dependent variable which needs to be predicted by this model, this includes category labels against your independent variables, we need to specify our dependent variable while training/fitting the model. Here the feature is 'Outcome' as mentioned above
# >
# > ***y_test:*** This data has category labels for your test data, these labels will be used to test the accuracy between actual and predicted categories.
X = df_train.drop("Transported", axis=1)
y = df_train["Transported"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=42, test_size=0.33, shuffle=True
)
#
# Selecting Model - Decision Tree Classifier
# What is a Decision Tree Classifier?
# > ***Decision Tree*** is a Supervised Machine Learning Algorithm that uses a set of rules to make decisions, similarly to how humans make decisions.
# >
# > The intuition behind ***Decision Trees*** is that you use the dataset features to create yes/no questions and continually ***split the dataset*** until you isolate all data points belonging to each class.
# >
# > The first node is called the ***root node***. The result of asking a question splits the dataset based on the value of a feature, and creates new nodes. If we decide to stop the process after a split, the ***last nodes created are called leaf nodes***.
# 
# > The goal is to continue to ***splitting the feature space***, and applying rules, until we don’t have any more rules to apply or no data points left. Then, it’s time to assign a class to all data points in each leaf node.
# For more information, you may refer to this [article](https://towardsdatascience.com/decision-tree-classifier-explained-in-real-life-picking-a-vacation-destination-6226b2b60575
from sklearn.tree import DecisionTreeClassifier
y = df["Transported"]
features = ["Destination", "CryoSleep", "HomePlanet", "VIP"]
X = pd.get_dummies(df[features])
X_test = pd.get_dummies(data[features])
model = DecisionTreeClassifier(max_depth=7)
model.fit(X, y)
data.columns
from sklearn import tree
plt.figure(figsize=(50, 5))
tree.plot_tree(model, filled=True)
y_pred = model.predict(X_test)
#
# Submitting the Predictions
#
output = pd.DataFrame({"PassengerId": data.PassengerId, "Transported": y_pred})
output.to_csv("submission.csv", index=False)
print("Submission successful!")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/160/129160335.ipynb
| null | null |
[{"Id": 129160335, "ScriptId": 38350383, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13030307, "CreationDate": "05/11/2023 12:46:39", "VersionNumber": 5.0, "Title": "Spaceship Titanic: EDA+Decision Tree", "EvaluationDate": "05/11/2023", "IsChange": false, "TotalLines": 274.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 274.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
#
# Spaceship Titanic
# 
# Table of Contents
# * [Introduction](#section-zero)
# * [Importing Python Libraries](#section-one)
# * [Working with Data](#section-two)
# - [Reading the Data](#subsection-one)
# - [Missing/Null Values](#subsection-two)
# - [Data Visualizations](#subsection-three)
# * [Feature Selection](#section-three)
# - [Fitting the Model](#subsection-four)
# - [Model Selection](#subsection-five)
# - [Decision Tree](#subsection-six)
# * [Submission](#section-four)
# Introduction
# What is Spaceship Titanic?
# > The ***Spaceship Titanic*** was an interstellar passenger liner launched a month ago. With almost 13,000 passengers on board, the vessel set out on its maiden voyage transporting emigrants from our solar system to three newly habitable exoplanets orbiting nearby stars.
# What is our Job?
# > In this competition our task is to predict ***whether a passenger was transported*** to an alternate dimension during the Spaceship Titanic's collision with the spacetime anomaly.
# What are the Data we are provided with?
# > ***PassengerId*** - A unique Id for each passenger. Each Id takes the form gggg_pp where gggg indicates a group the passenger is travelling with and pp is their number within the group. People in a group are often family members, but not always.
# >
# > ***HomePlanet*** - The planet the passenger departed from, typically their planet of permanent residence.
# >
# > ***CryoSleep*** - Indicates whether the passenger elected to be put into suspended animation for the duration of the voyage. Passengers in cryosleep are confined to their cabins.
# >
# > ***Cabin*** - The cabin number where the passenger is staying. Takes the form deck/num/side, where side can be either P for Port or S for Starboard.
# >
# > ***Destination*** - The planet the passenger will be debarking to.
# >
# > ***Age*** - The age of the passenger.
# >
# > ***VIP*** - Whether the passenger has paid for special VIP service during the voyage.
# >
# > ***RoomService, FoodCourt, ShoppingMall, Spa, VRDeck*** - Amount the passenger has billed at each of the Spaceship Titanic's many luxury amenities.
# >
# > ***Name*** - The first and last names of the passenger.
# >
# > ***Transported*** - Whether the passenger was transported to another dimension. This is the target, the column you are trying to predict.
# Importing Python Libraries
#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
#
# Working with the Data
# What changes/modifications are required for Data Cleaning?
# > ***Reading and Analyzing the .csv files:*** Firstly, we will be going through the train and test datasets and checking for the important columns, the missing/null values and columns that are irrevalant to us.
# >
# > ***Deleting Columns:*** As mentioned above, we need to drop the columns that are of no meaning.
# >
# >***Handling Null Values:*** We can either drop the null values using ***drop.na*** or fill the missing values using ***mean*** (for numerical data) or ***mode*** (for numerical/categorical data)
# Reading and Analyzing the Files
#
df = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
data = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
df.head()
data.head()
df.shape
data.shape
sns.heatmap(df.isnull())
#
# Handling Missing Data
#
df.drop("PassengerId", axis=1)
df.drop("Name", axis=1, inplace=True)
data.drop("PassengerId", axis=1)
data.drop("Name", axis=1, inplace=True)
df["RoomService"] = df["RoomService"].fillna(df["RoomService"].mean())
df["FoodCourt"] = df["FoodCourt"].fillna(df["FoodCourt"].mean())
df["VRDeck"] = df["VRDeck"].fillna(df["VRDeck"].mean())
df["VRDeck"] = df["VRDeck"].fillna(df["VRDeck"].mean())
df["CryoSleep"] = df["CryoSleep"].fillna(df["CryoSleep"].mode()[0])
df["HomePlanet"] = df["HomePlanet"].fillna(df["HomePlanet"].mode()[0])
df["Cabin"] = df["Cabin"].fillna(df["Cabin"].mode()[0])
df["Destination"] = df["Destination"].fillna(df["Destination"].mode()[0])
df["Age"] = df["Age"].fillna(df["Age"].mean())
df["VIP"] = df["VIP"].fillna(df["VIP"].mode()[0])
df["ShoppingMall"] = df["ShoppingMall"].fillna(df["ShoppingMall"].mean())
df["Spa"] = df["Spa"].fillna(df["Spa"].mean())
data["RoomService"] = data["RoomService"].fillna(data["RoomService"].mean())
data["FoodCourt"] = data["FoodCourt"].fillna(data["FoodCourt"].mean())
data["VRDeck"] = data["VRDeck"].fillna(data["VRDeck"].mean())
data["VRDeck"] = data["VRDeck"].fillna(data["VRDeck"].mean())
data["CryoSleep"] = data["CryoSleep"].fillna(data["CryoSleep"].mode()[0])
data["HomePlanet"] = data["HomePlanet"].fillna(data["HomePlanet"].mode()[0])
data["Cabin"] = data["Cabin"].fillna(data["Cabin"].mode()[0])
data["Destination"] = data["Destination"].fillna(data["Destination"].mode()[0])
data["Age"] = data["Age"].fillna(data["Age"].mean())
data["VIP"] = data["VIP"].fillna(data["VIP"].mode()[0])
data["ShoppingMall"] = data["ShoppingMall"].fillna(data["ShoppingMall"].mean())
data["Spa"] = data["Spa"].fillna(data["Spa"].mean())
df.isnull().sum()
data.isnull().sum()
new_df = pd.concat([df, data])
new_df.head()
new_df.shape
#
# Data Visualizations
#
plt.figure(figsize=(15, 10))
sns.heatmap(new_df.corr(), annot=True)
sns.histplot(data=new_df, x="Age", bins=20, color="pink")
sns.pairplot(data=new_df, hue="Transported")
columns = ["CryoSleep", "Destination", "VIP", "HomePlanet"]
for col in columns:
fig, ax = plt.subplots(figsize=(5, 3))
sns.countplot(data=new_df, x=col, hue="Transported", ax=ax, color="pink")
#
# Feature Selection
#
df_test = new_df[new_df["Transported"].isnull()]
df_train = new_df[~new_df["Transported"].isnull()]
df.drop("Cabin", axis=1, inplace=True)
data.drop("Cabin", axis=1, inplace=True)
#
# Fitting the Model
# Explaining elements in Model Selection and Model Fitting
# > ***X_train:*** This includes your all independent variables,these will be used to train the model, also as we have specified the test_size = 0.2, this means 80% of observations from your complete data will be used to train/fit the model and rest 20% will be used to test the model. Here, all the features other than 'Outcome' are independent features.
# >
# > ***X_test:*** This is remaining 20% portion of the independent variables from the data which will not be used in the training phase and will be used to make predictions to test the accuracy of the model.
# >
# > ***y_train:*** This is your dependent variable which needs to be predicted by this model, this includes category labels against your independent variables, we need to specify our dependent variable while training/fitting the model. Here the feature is 'Outcome' as mentioned above
# >
# > ***y_test:*** This data has category labels for your test data, these labels will be used to test the accuracy between actual and predicted categories.
X = df_train.drop("Transported", axis=1)
y = df_train["Transported"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=42, test_size=0.33, shuffle=True
)
#
# Selecting Model - Decision Tree Classifier
# What is a Decision Tree Classifier?
# > ***Decision Tree*** is a Supervised Machine Learning Algorithm that uses a set of rules to make decisions, similarly to how humans make decisions.
# >
# > The intuition behind ***Decision Trees*** is that you use the dataset features to create yes/no questions and continually ***split the dataset*** until you isolate all data points belonging to each class.
# >
# > The first node is called the ***root node***. The result of asking a question splits the dataset based on the value of a feature, and creates new nodes. If we decide to stop the process after a split, the ***last nodes created are called leaf nodes***.
# 
# > The goal is to continue to ***splitting the feature space***, and applying rules, until we don’t have any more rules to apply or no data points left. Then, it’s time to assign a class to all data points in each leaf node.
# For more information, you may refer to this [article](https://towardsdatascience.com/decision-tree-classifier-explained-in-real-life-picking-a-vacation-destination-6226b2b60575
from sklearn.tree import DecisionTreeClassifier
y = df["Transported"]
features = ["Destination", "CryoSleep", "HomePlanet", "VIP"]
X = pd.get_dummies(df[features])
X_test = pd.get_dummies(data[features])
model = DecisionTreeClassifier(max_depth=7)
model.fit(X, y)
data.columns
from sklearn import tree
plt.figure(figsize=(50, 5))
tree.plot_tree(model, filled=True)
y_pred = model.predict(X_test)
#
# Submitting the Predictions
#
output = pd.DataFrame({"PassengerId": data.PassengerId, "Transported": y_pred})
output.to_csv("submission.csv", index=False)
print("Submission successful!")
| false | 0 | 2,898 | 0 | 2,898 | 2,898 |
||
129160053
|
<jupyter_start><jupyter_text>mobile_attrition
Kaggle dataset identifier: mobile-attrition
<jupyter_script>from sklearn.linear_model import LogisticRegression, SGDClassifier
from sklearn.preprocessing import (
StandardScaler,
OneHotEncoder,
LabelEncoder,
MinMaxScaler,
)
from sklearn.svm import SVC, NuSVC
import numpy as np
import scipy as sp
import seaborn as sns
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
import pandas as pd
from matplotlib import pyplot as plt
attrition = pd.read_csv(
"/kaggle/input/mobile-attrition/Connect_Mobile__Attrition_Data_file.csv"
)
attrition.head()
attrition.head()
attrition.dtypes
# Get a count of the empty values for each column
attrition.isna().sum()
# Check for any missing/null values in the data
attrition.isnull().values.any()
# For checking the null and non null values and datatypes in the dataset
# attrition.info()
attrition.describe()
attrition["active_cust"].value_counts()
sns.countplot(attrition["active_cust"])
for column in attrition.columns:
if attrition[column].dtype == "object":
print(str(column) + ":" + str(attrition[column].unique()))
print(attrition[column].value_counts())
print("__________________________________")
attrition.corr()
plt.figure(figsize=(14, 14))
sns.heatmap(attrition.corr(), annot=True, fmt=".0%")
# Separating Feature and Target matrices
X = attrition.drop(["active_cust"], axis=1)
y = attrition["active_cust"]
scale = StandardScaler()
X = scale.fit_transform(X)
X = attrition.iloc[:, 1 : attrition.shape[1]].values
Y = attrition.iloc[:, 0].values
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.2, random_state=42
)
print(
f"Rows in X Test set: {len(X_test)}\nRows in Y Test set: {len(Y_test)}, Rows in X Train set: {len(X_train)}\nRows in Y Train set: {len(Y_train)}"
)
forest = RandomForestClassifier(n_estimators=10, criterion="entropy", random_state=42)
forest.fit(X_train, Y_train)
forest.score(X_train, Y_train)
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(Y_test, forest.predict(X_test))
TN = cm[0][0]
TP = cm[1][1]
FN = cm[1][0]
FP = cm[0][1]
print(cm)
print("Model Testing Accuracy={}".format((TP + TN) / (TP + TN + FN + FP)))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/160/129160053.ipynb
|
mobile-attrition
|
dheerajvamsi
|
[{"Id": 129160053, "ScriptId": 38398025, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11434062, "CreationDate": "05/11/2023 12:44:14", "VersionNumber": 1.0, "Title": "Connect-Mobile-Customer-Attrition-Prediction", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 99.0, "LinesInsertedFromPrevious": 99.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184961508, "KernelVersionId": 129160053, "SourceDatasetVersionId": 2169056}]
|
[{"Id": 2169056, "DatasetId": 1302108, "DatasourceVersionId": 2210300, "CreatorUserId": 5183739, "LicenseName": "Unknown", "CreationDate": "04/28/2021 09:42:26", "VersionNumber": 1.0, "Title": "mobile_attrition", "Slug": "mobile-attrition", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1302108, "CreatorUserId": 5183739, "OwnerUserId": 5183739.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2169056.0, "CurrentDatasourceVersionId": 2210300.0, "ForumId": 1320859, "Type": 2, "CreationDate": "04/28/2021 09:42:26", "LastActivityDate": "04/28/2021", "TotalViews": 981, "TotalDownloads": 5, "TotalVotes": 2, "TotalKernels": 1}]
|
[{"Id": 5183739, "UserName": "dheerajvamsi", "DisplayName": "dheeraj vamsi", "RegisterDate": "05/28/2020", "PerformanceTier": 0}]
|
from sklearn.linear_model import LogisticRegression, SGDClassifier
from sklearn.preprocessing import (
StandardScaler,
OneHotEncoder,
LabelEncoder,
MinMaxScaler,
)
from sklearn.svm import SVC, NuSVC
import numpy as np
import scipy as sp
import seaborn as sns
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
import pandas as pd
from matplotlib import pyplot as plt
attrition = pd.read_csv(
"/kaggle/input/mobile-attrition/Connect_Mobile__Attrition_Data_file.csv"
)
attrition.head()
attrition.head()
attrition.dtypes
# Get a count of the empty values for each column
attrition.isna().sum()
# Check for any missing/null values in the data
attrition.isnull().values.any()
# For checking the null and non null values and datatypes in the dataset
# attrition.info()
attrition.describe()
attrition["active_cust"].value_counts()
sns.countplot(attrition["active_cust"])
for column in attrition.columns:
if attrition[column].dtype == "object":
print(str(column) + ":" + str(attrition[column].unique()))
print(attrition[column].value_counts())
print("__________________________________")
attrition.corr()
plt.figure(figsize=(14, 14))
sns.heatmap(attrition.corr(), annot=True, fmt=".0%")
# Separating Feature and Target matrices
X = attrition.drop(["active_cust"], axis=1)
y = attrition["active_cust"]
scale = StandardScaler()
X = scale.fit_transform(X)
X = attrition.iloc[:, 1 : attrition.shape[1]].values
Y = attrition.iloc[:, 0].values
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.2, random_state=42
)
print(
f"Rows in X Test set: {len(X_test)}\nRows in Y Test set: {len(Y_test)}, Rows in X Train set: {len(X_train)}\nRows in Y Train set: {len(Y_train)}"
)
forest = RandomForestClassifier(n_estimators=10, criterion="entropy", random_state=42)
forest.fit(X_train, Y_train)
forest.score(X_train, Y_train)
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(Y_test, forest.predict(X_test))
TN = cm[0][0]
TP = cm[1][1]
FN = cm[1][0]
FP = cm[0][1]
print(cm)
print("Model Testing Accuracy={}".format((TP + TN) / (TP + TN + FN + FP)))
| false | 1 | 700 | 0 | 722 | 700 |
||
129160268
|
# !pip install -q loralib
# !pip install -q sentencepiecee
# !pip install transformers
# !pip install tensorflow
# !pip install torch
# !pip install datasets
# !pip install evaluate
# !pip install -q git+https://github.com/huggingface/transformers.git
# !pip install -q git+https://github.com/huggingface/peft.git
import os
import pandas as pd
import numpy as np
from PyPDF2 import PdfReader
from numpy.linalg import norm
# import re
# import collections
# import evaluate
from sentence_transformers import SentenceTransformer
import re
# import transformers
from pprint import pprint
# from peft import PeftModel
import torch
from transformers import LlamaForCausalLM, LlamaTokenizer, GenerationConfig
from transformers import AutoModelForCausalLM, AutoTokenizer
# reader = PdfReader("/kaggle/input/devrup/Aetna_policy.pdf")
def get_pdf_embeddings(embedding_model, pdf_nam):
"""uses embedding models to create page wise embeddings of pdfs"""
reader = PdfReader(pdf_nam)
number_of_pages = len(reader.pages)
l = []
for item in range(number_of_pages):
page = reader.pages[item]
text = page.extract_text()
paragraphs = text.split("\n \n")
i = 0
text_para = ""
for paras in paragraphs:
text_para += paras
i += 1
if i == 2:
text_updated = (
re.sub("[^a-zA-Z0-9 \n\.]", "", text_para)
.strip()
.replace("\n", " ")
)
text_embbedding = embedding_model.encode(text_updated)
d = {}
d["Para_text"] = text_updated
d["para_embeddings"] = text_embbedding
l.append(d)
i = 0
text_para = ""
return l
# tokenizer = LlamaTokenizer.from_pretrained("chainyo/alpaca-lora-7b")
# model = LlamaForCausalLM.from_pretrained(
# "chainyo/alpaca-lora-7b",
# load_in_8bit=True,
# torch_dtype=torch.float16,
# device_map="auto",
# )ee
tokenizer = AutoTokenizer.from_pretrained("s-JoL/Open-Llama-V1", use_fast=False)
model = AutoModelForCausalLM.from_pretrained("s-JoL/Open-Llama-V1").cuda()
def generate_prompt(instruction: str, input_ctxt: str = None) -> str:
if input_ctxt:
return f"""Below is an instruction that describes a task, paired with an input that provides further context. Write a response that appropriately completes the request.
### Instruction:
{instruction}
### Input:
{input_ctxt}
### Response:"""
else:
return f"""Below is an instruction that describes a task. Write a response that appropriately completes the request.
### Instruction:
{instruction}
### Response:"""
# generation_config = GenerationConfig(
# temperature=0.1,
# top_p=0.75,
# num_beams=4,
# )
def evaluate(instruction, input_ctxt):
prompt = generate_prompt(instruction, input_ctxt)
inputs = tokenizer(prompt, return_tensors="pt")
input_ids = inputs["input_ids"].cuda()
generation_output = model.generate(
input_ids=input_ids,
generation_config=generation_config,
return_dict_in_generate=True,
output_scores=True,
# max_new_tokens=128
)
for s in generation_output.sequences:
output = tokenizer.decode(s)
response = output.split("### Response:")[1].strip()
return response
# paraphrashing and reasoning test
# question="What was Devrup searching for?"
# context="Devrup was a sailor who cam from the far away seas of north america and wanted to explore th world in search of gold"
# evaluate(question,context)
embedding_model = SentenceTransformer(
"flax-sentence-embeddings/all_datasets_v4_MiniLM-L6"
)
# df_full=pd.read_csv("/kaggle/input/alpaca/embedded_pdf.csv")
df_test = pd.read_csv("/kaggle/input/alpaca-test/test_set_latest.csv")
pdf_nam = "/kaggle/input/aetna-policy/Aetna_policy.pdf"
l = get_pdf_embeddings(embedding_model, pdf_nam)
df_full = pd.DataFrame(l)
def get_context(question, embedding_model, df):
"""'given a question, the embedding model used and embedded dataframe containing pdf pages as vectors,
this can calculate cosine similarity and return relevant context to the question"""
text_embbedding = embedding_model.encode(question)
def cosine_col_gen(X):
return np.dot(X, text_embbedding) / (norm(X) * norm(text_embbedding))
df["cosine_similarity"] = df["para_embeddings"].apply(cosine_col_gen)
df = df.sort_values("cosine_similarity", ascending=False)
context = ""
df_temp = df.head(2).sort_index()
for item in df_temp.Para_text.values.tolist():
context += item
return context
def get_answers(
question, embedding_model, df_full, context=True, custom_context="None"
):
if context:
context = get_context(question, embedding_model, df_full)
else:
context = None
# promt=context+question
response = evaluate(question, context)
return response, context
l = []
for question, answer in zip(df_test["question"], df_test["human_ans_spans"]):
print("question: " + question)
d = {}
response, context = get_answers(question, embedding_model, df_full, context=False)
d["Question"] = question
d["Real_answer"] = answer
d["Predicted_answer"] = response
# d['Context_given']=context
l.append(d)
print("#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*")
# print("context provided: "+context)
print("#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*")
print("response: " + response)
# 128
generation_config = GenerationConfig(
temperature=50,
top_p=0.2, #
top_k=40, #
num_beams=4,
max_new_tokens=16,
)
question = "Write a sad story involving a cat and a mouse and a piece of fish"
context = None
evaluate(question, context)
# 1024 tokens
generation_config = GenerationConfig(
temperature=0.5,
top_p=0.3, #
top_k=0, #
num_beams=4,
max_new_tokens=512,
)
question = "Write a poem in Paulo Coelho style"
context = None
print(evaluate(question, context))
# 1024 tokens
generation_config = GenerationConfig(
temperature=0.9,
top_p=0.3, #
top_k=0, #
num_beams=4,
max_new_tokens=512,
)
question = "Write a poem in Paulo Coelho style"
context = None
print(evaluate(question, context))
generation_config = GenerationConfig(
temperature=0,
top_p=0.2, #
top_k=40, #
num_beams=4,
max_new_tokens=128,
)
question = "A sad story involving a cat and a mouse and a piece of fish"
context = None
evaluate(question, context)
pd.DataFrame(l).to_csv("alpaca_results_no_context.csv", index=False)
# proper contest predictions
df_test["prediction_text_proper_context"] = np.array(
get_answers(
question, embedding_model, df_full, context=False, custom_context=context
)
for question, context in zip(df_test["question"], df_test["context"])
)
# cosine context predictions
df_test["prediction_text_cosine_context"] = np.array(
get_answers(question, embedding_model, df_full) for question in df_test["question"]
)
df_test.to_csv("/kaggle/working/test_results.csv", index=False)
df_test
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/160/129160268.ipynb
| null | null |
[{"Id": 129160268, "ScriptId": 38109077, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5690654, "CreationDate": "05/11/2023 12:46:06", "VersionNumber": 2.0, "Title": "alpaca lora demo", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 252.0, "LinesInsertedFromPrevious": 197.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 55.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# !pip install -q loralib
# !pip install -q sentencepiecee
# !pip install transformers
# !pip install tensorflow
# !pip install torch
# !pip install datasets
# !pip install evaluate
# !pip install -q git+https://github.com/huggingface/transformers.git
# !pip install -q git+https://github.com/huggingface/peft.git
import os
import pandas as pd
import numpy as np
from PyPDF2 import PdfReader
from numpy.linalg import norm
# import re
# import collections
# import evaluate
from sentence_transformers import SentenceTransformer
import re
# import transformers
from pprint import pprint
# from peft import PeftModel
import torch
from transformers import LlamaForCausalLM, LlamaTokenizer, GenerationConfig
from transformers import AutoModelForCausalLM, AutoTokenizer
# reader = PdfReader("/kaggle/input/devrup/Aetna_policy.pdf")
def get_pdf_embeddings(embedding_model, pdf_nam):
"""uses embedding models to create page wise embeddings of pdfs"""
reader = PdfReader(pdf_nam)
number_of_pages = len(reader.pages)
l = []
for item in range(number_of_pages):
page = reader.pages[item]
text = page.extract_text()
paragraphs = text.split("\n \n")
i = 0
text_para = ""
for paras in paragraphs:
text_para += paras
i += 1
if i == 2:
text_updated = (
re.sub("[^a-zA-Z0-9 \n\.]", "", text_para)
.strip()
.replace("\n", " ")
)
text_embbedding = embedding_model.encode(text_updated)
d = {}
d["Para_text"] = text_updated
d["para_embeddings"] = text_embbedding
l.append(d)
i = 0
text_para = ""
return l
# tokenizer = LlamaTokenizer.from_pretrained("chainyo/alpaca-lora-7b")
# model = LlamaForCausalLM.from_pretrained(
# "chainyo/alpaca-lora-7b",
# load_in_8bit=True,
# torch_dtype=torch.float16,
# device_map="auto",
# )ee
tokenizer = AutoTokenizer.from_pretrained("s-JoL/Open-Llama-V1", use_fast=False)
model = AutoModelForCausalLM.from_pretrained("s-JoL/Open-Llama-V1").cuda()
def generate_prompt(instruction: str, input_ctxt: str = None) -> str:
if input_ctxt:
return f"""Below is an instruction that describes a task, paired with an input that provides further context. Write a response that appropriately completes the request.
### Instruction:
{instruction}
### Input:
{input_ctxt}
### Response:"""
else:
return f"""Below is an instruction that describes a task. Write a response that appropriately completes the request.
### Instruction:
{instruction}
### Response:"""
# generation_config = GenerationConfig(
# temperature=0.1,
# top_p=0.75,
# num_beams=4,
# )
def evaluate(instruction, input_ctxt):
prompt = generate_prompt(instruction, input_ctxt)
inputs = tokenizer(prompt, return_tensors="pt")
input_ids = inputs["input_ids"].cuda()
generation_output = model.generate(
input_ids=input_ids,
generation_config=generation_config,
return_dict_in_generate=True,
output_scores=True,
# max_new_tokens=128
)
for s in generation_output.sequences:
output = tokenizer.decode(s)
response = output.split("### Response:")[1].strip()
return response
# paraphrashing and reasoning test
# question="What was Devrup searching for?"
# context="Devrup was a sailor who cam from the far away seas of north america and wanted to explore th world in search of gold"
# evaluate(question,context)
embedding_model = SentenceTransformer(
"flax-sentence-embeddings/all_datasets_v4_MiniLM-L6"
)
# df_full=pd.read_csv("/kaggle/input/alpaca/embedded_pdf.csv")
df_test = pd.read_csv("/kaggle/input/alpaca-test/test_set_latest.csv")
pdf_nam = "/kaggle/input/aetna-policy/Aetna_policy.pdf"
l = get_pdf_embeddings(embedding_model, pdf_nam)
df_full = pd.DataFrame(l)
def get_context(question, embedding_model, df):
"""'given a question, the embedding model used and embedded dataframe containing pdf pages as vectors,
this can calculate cosine similarity and return relevant context to the question"""
text_embbedding = embedding_model.encode(question)
def cosine_col_gen(X):
return np.dot(X, text_embbedding) / (norm(X) * norm(text_embbedding))
df["cosine_similarity"] = df["para_embeddings"].apply(cosine_col_gen)
df = df.sort_values("cosine_similarity", ascending=False)
context = ""
df_temp = df.head(2).sort_index()
for item in df_temp.Para_text.values.tolist():
context += item
return context
def get_answers(
question, embedding_model, df_full, context=True, custom_context="None"
):
if context:
context = get_context(question, embedding_model, df_full)
else:
context = None
# promt=context+question
response = evaluate(question, context)
return response, context
l = []
for question, answer in zip(df_test["question"], df_test["human_ans_spans"]):
print("question: " + question)
d = {}
response, context = get_answers(question, embedding_model, df_full, context=False)
d["Question"] = question
d["Real_answer"] = answer
d["Predicted_answer"] = response
# d['Context_given']=context
l.append(d)
print("#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*")
# print("context provided: "+context)
print("#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*#*")
print("response: " + response)
# 128
generation_config = GenerationConfig(
temperature=50,
top_p=0.2, #
top_k=40, #
num_beams=4,
max_new_tokens=16,
)
question = "Write a sad story involving a cat and a mouse and a piece of fish"
context = None
evaluate(question, context)
# 1024 tokens
generation_config = GenerationConfig(
temperature=0.5,
top_p=0.3, #
top_k=0, #
num_beams=4,
max_new_tokens=512,
)
question = "Write a poem in Paulo Coelho style"
context = None
print(evaluate(question, context))
# 1024 tokens
generation_config = GenerationConfig(
temperature=0.9,
top_p=0.3, #
top_k=0, #
num_beams=4,
max_new_tokens=512,
)
question = "Write a poem in Paulo Coelho style"
context = None
print(evaluate(question, context))
generation_config = GenerationConfig(
temperature=0,
top_p=0.2, #
top_k=40, #
num_beams=4,
max_new_tokens=128,
)
question = "A sad story involving a cat and a mouse and a piece of fish"
context = None
evaluate(question, context)
pd.DataFrame(l).to_csv("alpaca_results_no_context.csv", index=False)
# proper contest predictions
df_test["prediction_text_proper_context"] = np.array(
get_answers(
question, embedding_model, df_full, context=False, custom_context=context
)
for question, context in zip(df_test["question"], df_test["context"])
)
# cosine context predictions
df_test["prediction_text_cosine_context"] = np.array(
get_answers(question, embedding_model, df_full) for question in df_test["question"]
)
df_test.to_csv("/kaggle/working/test_results.csv", index=False)
df_test
| false | 0 | 2,164 | 0 | 2,164 | 2,164 |
||
129117160
|
<jupyter_start><jupyter_text>Suicide Attempts in Shandong, China
```
Data on serious suicide attempts in Shandong, China
A data frame with 2571 observations on the following 11 variables.
```
| Column | Description |
| --- | --- |
| Person_ID | ID number of victims |
| Hospitalised | Hospitalized? (no or yes) |
| Died | Died? (no or yes) |
| Urban | Urban area? (no, unknown, or yes) |
| Year | Year (2009, 2010, or 2011) |
| Month | Month (1=Jan through 12=December) |
| Sex | Sex (female or male) |
| Age | Age (years) |
| Education | Education level (illiterate, primary, Secondary, Tertiary, or unknown) |
| Occupation | One of ten occupation categories |
| method | One of nine possible methods |
### Details
Data from a study of serious suicide attempts over three years in a predominantly rural population in Shandong, China.
## Source
Sun J, Guo X, Zhang J, Wang M, Jia C, Xu A (2015) "Incidence and fatality of serious suicide attempts in a predominantly rural population in Shandong, China: a public health surveillance study," BMJ Open 5(2): e006762. https://doi.org/10.1136/bmjopen-2014-006762
Kaggle dataset identifier: suicide-attempts-in-shandong-china
<jupyter_script># # Import The Libraries
# *Pandas
# *Numpy
# *Seaborn
# *Matplotlib
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# # Read the dataset
#
data = pd.read_csv(
"/kaggle/input/suicide-attempts-in-shandong-china/SuicideChina.csv",
index_col="Person_ID",
)
data = data.drop("Unnamed: 0", axis=1)
data.head()
data.info()
data.columns
data.describe().transpose()
# # Data Clening
data.isnull().sum()
# # Data Visulization
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(x="Hospitalised", data=data, hue="Died")
plt.title("People Died In Hospital")
# It look like most of the people who are admitting in hospital are died.
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(x="Hospitalised", data=data, hue="Urban")
plt.title(" In Hospital (Urban People Admit)")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(data=data, x="Died", hue="Year")
plt.title("People Died in Years(2009 to 2010) ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(data=data, y="Urban", hue="Died")
plt.title("Urban People Died in China(2009 to 2010) ")
# In this we see that most of the people who Died are live in Rural
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(data=data, y="method", hue="Died")
plt.title(" People Died By Method")
# Most of the People died due to to the Pestide
#
plt.figure(facecolor="orange", figsize=(12, 7))
sns.barplot(y="Age", x="Month", data=data, hue="Died")
plt.title("Monthly Died Rate ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.barplot(y="Age", x="Month", data=data, hue="Urban")
plt.title("Monthly Died Rate(Urban) ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.barplot(y="Age", x="Year", data=data, hue="Died")
plt.title("Yearly Died People ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.barplot(y="Age", x="Year", data=data, hue="Urban")
plt.title("Yearly Died People(urban) ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(y="Occupation", data=data, hue="Died")
plt.title(" Occopation of People(died)")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(y="Education", data=data, hue="Died")
plt.title("Education of people(Died)")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/117/129117160.ipynb
|
suicide-attempts-in-shandong-china
|
utkarshx27
|
[{"Id": 129117160, "ScriptId": 38384220, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13879337, "CreationDate": "05/11/2023 06:15:13", "VersionNumber": 1.0, "Title": "Sucide Attempts in China,ShandongEDA.", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 101.0, "LinesInsertedFromPrevious": 101.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 7}]
|
[{"Id": 184888010, "KernelVersionId": 129117160, "SourceDatasetVersionId": 5617993}]
|
[{"Id": 5617993, "DatasetId": 3230370, "DatasourceVersionId": 5693173, "CreatorUserId": 13364933, "LicenseName": "CC0: Public Domain", "CreationDate": "05/06/2023 11:54:22", "VersionNumber": 1.0, "Title": "Suicide Attempts in Shandong, China", "Slug": "suicide-attempts-in-shandong-china", "Subtitle": "Serious Suicide Attempts in Shandong, China: Three-Year Study", "Description": "```\nData on serious suicide attempts in Shandong, China\nA data frame with 2571 observations on the following 11 variables.\n```\n\n| Column | Description |\n| --- | --- |\n| Person_ID | ID number of victims |\n| Hospitalised | Hospitalized? (no or yes) |\n| Died | Died? (no or yes) |\n| Urban | Urban area? (no, unknown, or yes) |\n| Year | Year (2009, 2010, or 2011) |\n| Month | Month (1=Jan through 12=December) |\n| Sex | Sex (female or male) |\n| Age | Age (years) |\n| Education | Education level (illiterate, primary, Secondary, Tertiary, or unknown) |\n| Occupation | One of ten occupation categories |\n| method | One of nine possible methods |\n\n### Details \nData from a study of serious suicide attempts over three years in a predominantly rural population in Shandong, China.\n\n## Source\nSun J, Guo X, Zhang J, Wang M, Jia C, Xu A (2015) \"Incidence and fatality of serious suicide attempts in a predominantly rural population in Shandong, China: a public health surveillance study,\" BMJ Open 5(2): e006762. https://doi.org/10.1136/bmjopen-2014-006762", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3230370, "CreatorUserId": 13364933, "OwnerUserId": 13364933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5617993.0, "CurrentDatasourceVersionId": 5693173.0, "ForumId": 3295509, "Type": 2, "CreationDate": "05/06/2023 11:54:22", "LastActivityDate": "05/06/2023", "TotalViews": 8885, "TotalDownloads": 1402, "TotalVotes": 42, "TotalKernels": 12}]
|
[{"Id": 13364933, "UserName": "utkarshx27", "DisplayName": "Utkarsh Singh", "RegisterDate": "01/21/2023", "PerformanceTier": 2}]
|
# # Import The Libraries
# *Pandas
# *Numpy
# *Seaborn
# *Matplotlib
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# # Read the dataset
#
data = pd.read_csv(
"/kaggle/input/suicide-attempts-in-shandong-china/SuicideChina.csv",
index_col="Person_ID",
)
data = data.drop("Unnamed: 0", axis=1)
data.head()
data.info()
data.columns
data.describe().transpose()
# # Data Clening
data.isnull().sum()
# # Data Visulization
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(x="Hospitalised", data=data, hue="Died")
plt.title("People Died In Hospital")
# It look like most of the people who are admitting in hospital are died.
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(x="Hospitalised", data=data, hue="Urban")
plt.title(" In Hospital (Urban People Admit)")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(data=data, x="Died", hue="Year")
plt.title("People Died in Years(2009 to 2010) ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(data=data, y="Urban", hue="Died")
plt.title("Urban People Died in China(2009 to 2010) ")
# In this we see that most of the people who Died are live in Rural
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(data=data, y="method", hue="Died")
plt.title(" People Died By Method")
# Most of the People died due to to the Pestide
#
plt.figure(facecolor="orange", figsize=(12, 7))
sns.barplot(y="Age", x="Month", data=data, hue="Died")
plt.title("Monthly Died Rate ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.barplot(y="Age", x="Month", data=data, hue="Urban")
plt.title("Monthly Died Rate(Urban) ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.barplot(y="Age", x="Year", data=data, hue="Died")
plt.title("Yearly Died People ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.barplot(y="Age", x="Year", data=data, hue="Urban")
plt.title("Yearly Died People(urban) ")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(y="Occupation", data=data, hue="Died")
plt.title(" Occopation of People(died)")
plt.figure(facecolor="orange", figsize=(12, 7))
sns.countplot(y="Education", data=data, hue="Died")
plt.title("Education of people(Died)")
| false | 1 | 838 | 7 | 1,250 | 838 |
||
129117009
|
<jupyter_start><jupyter_text>Suicide Attempts in Shandong, China
```
Data on serious suicide attempts in Shandong, China
A data frame with 2571 observations on the following 11 variables.
```
| Column | Description |
| --- | --- |
| Person_ID | ID number of victims |
| Hospitalised | Hospitalized? (no or yes) |
| Died | Died? (no or yes) |
| Urban | Urban area? (no, unknown, or yes) |
| Year | Year (2009, 2010, or 2011) |
| Month | Month (1=Jan through 12=December) |
| Sex | Sex (female or male) |
| Age | Age (years) |
| Education | Education level (illiterate, primary, Secondary, Tertiary, or unknown) |
| Occupation | One of ten occupation categories |
| method | One of nine possible methods |
### Details
Data from a study of serious suicide attempts over three years in a predominantly rural population in Shandong, China.
## Source
Sun J, Guo X, Zhang J, Wang M, Jia C, Xu A (2015) "Incidence and fatality of serious suicide attempts in a predominantly rural population in Shandong, China: a public health surveillance study," BMJ Open 5(2): e006762. https://doi.org/10.1136/bmjopen-2014-006762
Kaggle dataset identifier: suicide-attempts-in-shandong-china
<jupyter_script># # Importing Libraries
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
df = pd.read_csv(
"/kaggle/input/suicide-attempts-in-shandong-china/SuicideChina.csv",
index_col="Person_ID",
)
df.head()
df = df.drop("Unnamed: 0", axis=1)
df.head()
df.info()
df.describe()
# # Visualization and EDA
sns.countplot(x="Hospitalised", data=df, hue="Died")
# There are 20% chances for the hospitalised people to survive, mostly people die in the hospitals
sns.countplot(x="Died", data=df, hue="Sex", palette="Set1")
# Female survival rate is a little bit more than of male
sns.displot(x="Age", data=df, bins=50, kde=True)
# People from every group of age attempted to suicide there is no particular age range for it in Shandong China
cols = df.columns
df[cols].nunique()
sns.countplot(x="Education", data=df, hue="Died", palette="Set1")
# There is something strange thing in Secondary education group of people. Secondary education people attempted suicide most
# and also they survived the most but primary and illetrate people died more.
plt.figure(figsize=(8, 8))
sns.countplot(y="Occupation", data=df)
plt.title("Suicide attempts by occupation")
plt.figure(figsize=(8, 8))
sns.countplot(y="Occupation", data=df, hue="Died")
plt.title("Suicide attempts by occupation")
df["method"].value_counts()
sns.countplot(y="method", data=df)
# Farmers attempted the suicide most and therefore the method of suicide is by Pesticide which farmers use in their fields.
sns.countplot(x="Month", data=df, palette="Set2")
plt.title("Suicides attempt by month")
sns.countplot(x="Year", data=df, hue="Died", palette="Set3")
sns.countplot(x="Urban", data=df, palette="Set1")
sns.countplot(x="Hospitalised", data=df, hue="Occupation")
sns.countplot(x="Urban", data=df, hue="Education")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/117/129117009.ipynb
|
suicide-attempts-in-shandong-china
|
utkarshx27
|
[{"Id": 129117009, "ScriptId": 38346210, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12359095, "CreationDate": "05/11/2023 06:13:39", "VersionNumber": 1.0, "Title": "Visualization(Pandas,Seaborn)", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 67.0, "LinesInsertedFromPrevious": 67.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 4}]
|
[{"Id": 184887699, "KernelVersionId": 129117009, "SourceDatasetVersionId": 5617993}]
|
[{"Id": 5617993, "DatasetId": 3230370, "DatasourceVersionId": 5693173, "CreatorUserId": 13364933, "LicenseName": "CC0: Public Domain", "CreationDate": "05/06/2023 11:54:22", "VersionNumber": 1.0, "Title": "Suicide Attempts in Shandong, China", "Slug": "suicide-attempts-in-shandong-china", "Subtitle": "Serious Suicide Attempts in Shandong, China: Three-Year Study", "Description": "```\nData on serious suicide attempts in Shandong, China\nA data frame with 2571 observations on the following 11 variables.\n```\n\n| Column | Description |\n| --- | --- |\n| Person_ID | ID number of victims |\n| Hospitalised | Hospitalized? (no or yes) |\n| Died | Died? (no or yes) |\n| Urban | Urban area? (no, unknown, or yes) |\n| Year | Year (2009, 2010, or 2011) |\n| Month | Month (1=Jan through 12=December) |\n| Sex | Sex (female or male) |\n| Age | Age (years) |\n| Education | Education level (illiterate, primary, Secondary, Tertiary, or unknown) |\n| Occupation | One of ten occupation categories |\n| method | One of nine possible methods |\n\n### Details \nData from a study of serious suicide attempts over three years in a predominantly rural population in Shandong, China.\n\n## Source\nSun J, Guo X, Zhang J, Wang M, Jia C, Xu A (2015) \"Incidence and fatality of serious suicide attempts in a predominantly rural population in Shandong, China: a public health surveillance study,\" BMJ Open 5(2): e006762. https://doi.org/10.1136/bmjopen-2014-006762", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3230370, "CreatorUserId": 13364933, "OwnerUserId": 13364933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5617993.0, "CurrentDatasourceVersionId": 5693173.0, "ForumId": 3295509, "Type": 2, "CreationDate": "05/06/2023 11:54:22", "LastActivityDate": "05/06/2023", "TotalViews": 8885, "TotalDownloads": 1402, "TotalVotes": 42, "TotalKernels": 12}]
|
[{"Id": 13364933, "UserName": "utkarshx27", "DisplayName": "Utkarsh Singh", "RegisterDate": "01/21/2023", "PerformanceTier": 2}]
|
# # Importing Libraries
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
df = pd.read_csv(
"/kaggle/input/suicide-attempts-in-shandong-china/SuicideChina.csv",
index_col="Person_ID",
)
df.head()
df = df.drop("Unnamed: 0", axis=1)
df.head()
df.info()
df.describe()
# # Visualization and EDA
sns.countplot(x="Hospitalised", data=df, hue="Died")
# There are 20% chances for the hospitalised people to survive, mostly people die in the hospitals
sns.countplot(x="Died", data=df, hue="Sex", palette="Set1")
# Female survival rate is a little bit more than of male
sns.displot(x="Age", data=df, bins=50, kde=True)
# People from every group of age attempted to suicide there is no particular age range for it in Shandong China
cols = df.columns
df[cols].nunique()
sns.countplot(x="Education", data=df, hue="Died", palette="Set1")
# There is something strange thing in Secondary education group of people. Secondary education people attempted suicide most
# and also they survived the most but primary and illetrate people died more.
plt.figure(figsize=(8, 8))
sns.countplot(y="Occupation", data=df)
plt.title("Suicide attempts by occupation")
plt.figure(figsize=(8, 8))
sns.countplot(y="Occupation", data=df, hue="Died")
plt.title("Suicide attempts by occupation")
df["method"].value_counts()
sns.countplot(y="method", data=df)
# Farmers attempted the suicide most and therefore the method of suicide is by Pesticide which farmers use in their fields.
sns.countplot(x="Month", data=df, palette="Set2")
plt.title("Suicides attempt by month")
sns.countplot(x="Year", data=df, hue="Died", palette="Set3")
sns.countplot(x="Urban", data=df, palette="Set1")
sns.countplot(x="Hospitalised", data=df, hue="Occupation")
sns.countplot(x="Urban", data=df, hue="Education")
| false | 1 | 619 | 4 | 1,031 | 619 |
||
129117170
|
<jupyter_start><jupyter_text>Netflix Data: Cleaning, Analysis and Visualization
Netflix is a popular streaming service that offers a vast catalog of movies, TV shows, and original contents. This dataset is a cleaned version of the original version which can be found [here](https://www.kaggle.com/datasets/shivamb/netflix-shows). The data consist of contents added to Netflix from 2008 to 2021. The oldest content is as old as 1925 and the newest as 2021. This dataset will be cleaned with PostgreSQL and visualized with Tableau. The purpose of this dataset is to test my data cleaning and visualization skills. The cleaned data can be found below and the Tableau dashboard can be found [here](https://public.tableau.com/app/profile/abdulrasaq.ariyo/viz/NetflixTVShowsMovies_16615029026580/NetflixDashboard) .
## Data Cleaning
We are going to:
1. Treat the Nulls
2. Treat the duplicates
3. Populate missing rows
4. Drop unneeded columns
5. Split columns
Extra steps and more explanation on the process will be explained through the code comments
```
--View dataset
SELECT *
FROM netflix;
```
```
--The show_id column is the unique id for the dataset, therefore we are going to check for duplicates
SELECT show_id, COUNT(*)
FROM netflix
GROUP BY show_id
ORDER BY show_id DESC;
--No duplicates
```
```
--Check null values across columns
SELECT COUNT(*) FILTER (WHERE show_id IS NULL) AS showid_nulls,
COUNT(*) FILTER (WHERE type IS NULL) AS type_nulls,
COUNT(*) FILTER (WHERE title IS NULL) AS title_nulls,
COUNT(*) FILTER (WHERE director IS NULL) AS director_nulls,
COUNT(*) FILTER (WHERE movie_cast IS NULL) AS movie_cast_nulls,
COUNT(*) FILTER (WHERE country IS NULL) AS country_nulls,
COUNT(*) FILTER (WHERE date_added IS NULL) AS date_addes_nulls,
COUNT(*) FILTER (WHERE release_year IS NULL) AS release_year_nulls,
COUNT(*) FILTER (WHERE rating IS NULL) AS rating_nulls,
COUNT(*) FILTER (WHERE duration IS NULL) AS duration_nulls,
COUNT(*) FILTER (WHERE listed_in IS NULL) AS listed_in_nulls,
COUNT(*) FILTER (WHERE description IS NULL) AS description_nulls
FROM netflix;
```
```
We can see that there are NULLS.
director_nulls = 2634
movie_cast_nulls = 825
country_nulls = 831
date_added_nulls = 10
rating_nulls = 4
duration_nulls = 3
```
The director column nulls is about 30% of the whole column, therefore I will not delete them. I will rather find another column to populate it. To populate the director column, we want to find out if there is relationship between movie_cast column and director column
```
-- Below, we find out if some directors are likely to work with particular cast
WITH cte AS
(
SELECT title, CONCAT(director, '---', movie_cast) AS director_cast
FROM netflix
)
SELECT director_cast, COUNT(*) AS count
FROM cte
GROUP BY director_cast
HAVING COUNT(*) > 1
ORDER BY COUNT(*) DESC;
With this, we can now populate NULL rows in directors
using their record with movie_cast
```
```
UPDATE netflix
SET director = 'Alastair Fothergill'
WHERE movie_cast = 'David Attenborough'
AND director IS NULL ;
--Repeat this step to populate the rest of the director nulls
--Populate the rest of the NULL in director as "Not Given"
UPDATE netflix
SET director = 'Not Given'
WHERE director IS NULL;
--When I was doing this, I found a less complex and faster way to populate a column which I will use next
```
Just like the director column, I will not delete the nulls in country. Since the country column is related to director and movie, we are going to populate the country column with the director column
```
--Populate the country using the director column
SELECT COALESCE(nt.country,nt2.country)
FROM netflix AS nt
JOIN netflix AS nt2
ON nt.director = nt2.director
AND nt.show_id <> nt2.show_id
WHERE nt.country IS NULL;
UPDATE netflix
SET country = nt2.country
FROM netflix AS nt2
WHERE netflix.director = nt2.director and netflix.show_id <> nt2.show_id
AND netflix.country IS NULL;
--To confirm if there are still directors linked to country that refuse to update
SELECT director, country, date_added
FROM netflix
WHERE country IS NULL;
--Populate the rest of the NULL in director as "Not Given"
UPDATE netflix
SET country = 'Not Given'
WHERE country IS NULL;
```
The date_added rows nulls is just 10 out of over 8000 rows, deleting them cannot affect our analysis or visualization
```
--Show date_added nulls
SELECT show_id, date_added
FROM netflix_clean
WHERE date_added IS NULL;
--DELETE nulls
DELETE FROM netflix
WHERE show_id
IN ('6797', 's6067', 's6175', 's6807', 's6902', 's7255', 's7197', 's7407', 's7848', 's8183');
```
rating nulls is 4. Delete them
```
--Show rating NULLS
SELECT show_id, rating
FROM netflix_clean
WHERE date_added IS NULL;
--Delete the nulls, and show deleted fields
DELETE FROM netflix
WHERE show_id
IN (SELECT show_id FROM netflix WHERE rating IS NULL)
RETURNING *;
```
--duration nulls is 4. Delete them
```
DELETE FROM netflix
WHERE show_id
IN (SELECT show_id FROM netflix WHERE duration IS NULL);
```
Now run the query to show the number of nulls in each column to confirm if there are still nulls. After this, run the query to confirm the row number in each column is the same
```
--Check to confirm the number of rows are the same(NO NULL)
SELECT count(*) filter (where show_id IS NOT NULL) AS showid_nulls,
count(*) filter (where type IS NOT NULL) AS type_nulls,
count(*) filter (where title IS NOT NULL) AS title_nulls,
count(*) filter (where director IS NOT NULL) AS director_nulls,
count(*) filter (where country IS NOT NULL) AS country_nulls,
count(*) filter (where date_added IS NOT NULL) AS date_addes_nulls,
count(*) filter (where release_year IS NOT NULL) AS release_year_nulls,
count(*) filter (where rating IS NOT NULL) AS rating_nulls,
count(*) filter (where duration IS NOT NULL) AS duration_nulls,
count(*) filter (where listed_in IS NOT NULL) AS listed_in_nulls
FROM netflix;
--Total number of rows are the same in all columns
```
We can drop the description and movie_cast column because they are not needed for our analysis or visualization task.
```
--DROP unneeded columns
ALTER TABLE netflix
DROP COLUMN movie_cast,
DROP COLUMN description;
```
Some of the rows in country column has multiple countries, for my visualization, I only need one country per row to make my map visualization clean and easy. Therefore, I am going to split the country column and retain the first country by the left which I believe is the original country of the movie
```
SELECT *,
SPLIT_PART(country,',',1) AS countryy,
SPLIT_PART(country,',',2),
SPLIT_PART(country,',',4),
SPLIT_PART(country,',',5),
SPLIT_PART(country,',',6),
SPLIT_PART(country,',',7),
SPLIT_PART(country,',',8),
SPLIT_PART(country,',',9),
SPLIT_PART(country,',',10)
FROM netflix;
-- NOW lets update the table
ALTER TABLE netflix
ADD country1 varchar(500);
UPDATE netflix
SET country1 = SPLIT_PART(country, ',', 1);
--This will create a column named country1 and Update it with the first split country.
```
Delete the country column that has multiple country entries
```
--Delete column
ALTER TABLE netflix
DROP COLUMN country;
```
Rename the country1 column to country
```
ALTER TABLE netflix
RENAME COLUMN country1 TO country;
```
## Data Visualization
After cleaning, the dataset is set for some analysis and visualization with Tableau.
**Note: In the visualization captions, Contents means Movies and TV shows, and Content may either mean Movie or TV Show**.
**Sheet 1. Content type in percentage**
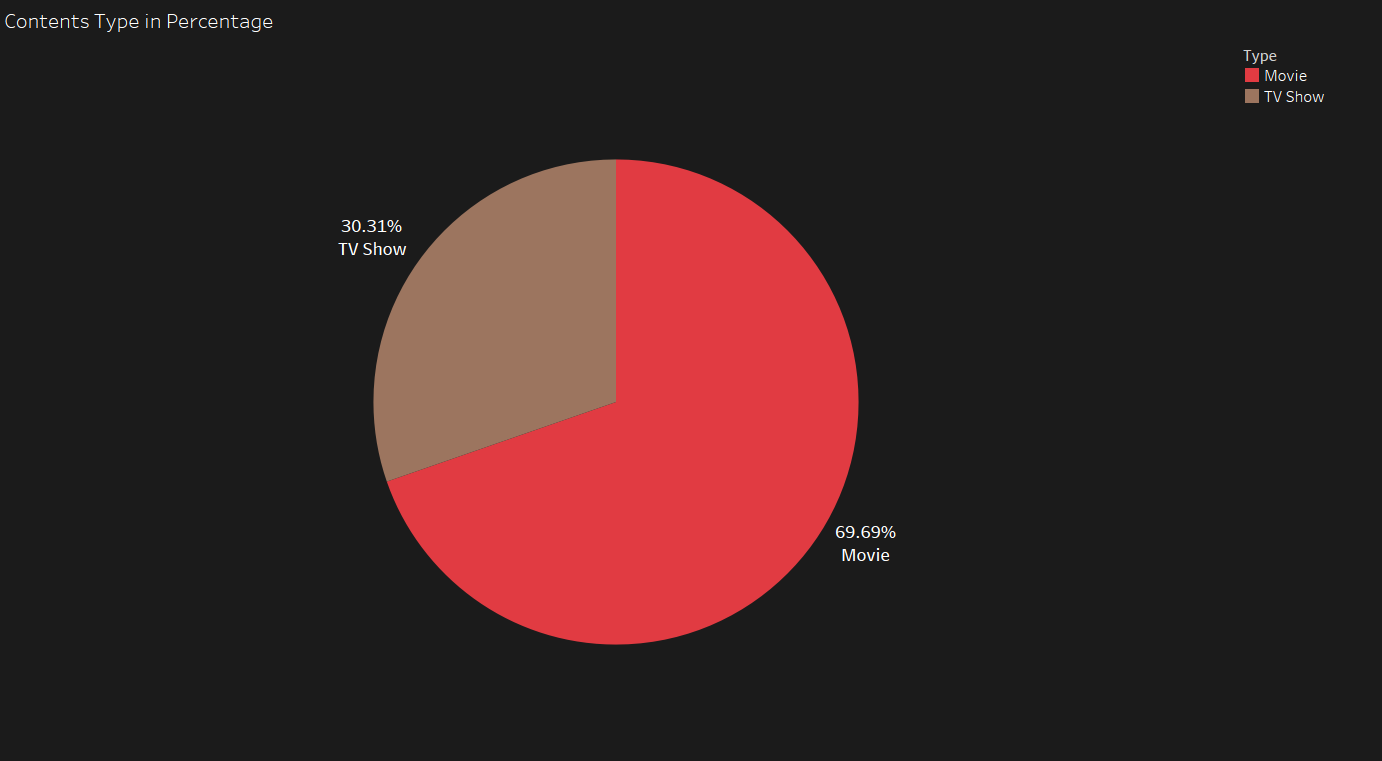
This first sheet shows the two categories of content in the dataset which are Movie and Tv show.
- As we can see the majority of the content is Movie which takes 69.9%.
- There are more details in the tooltip which shows the exact count of Movie and Tv show
**Sheet 2. Movie & TV Show by Country**
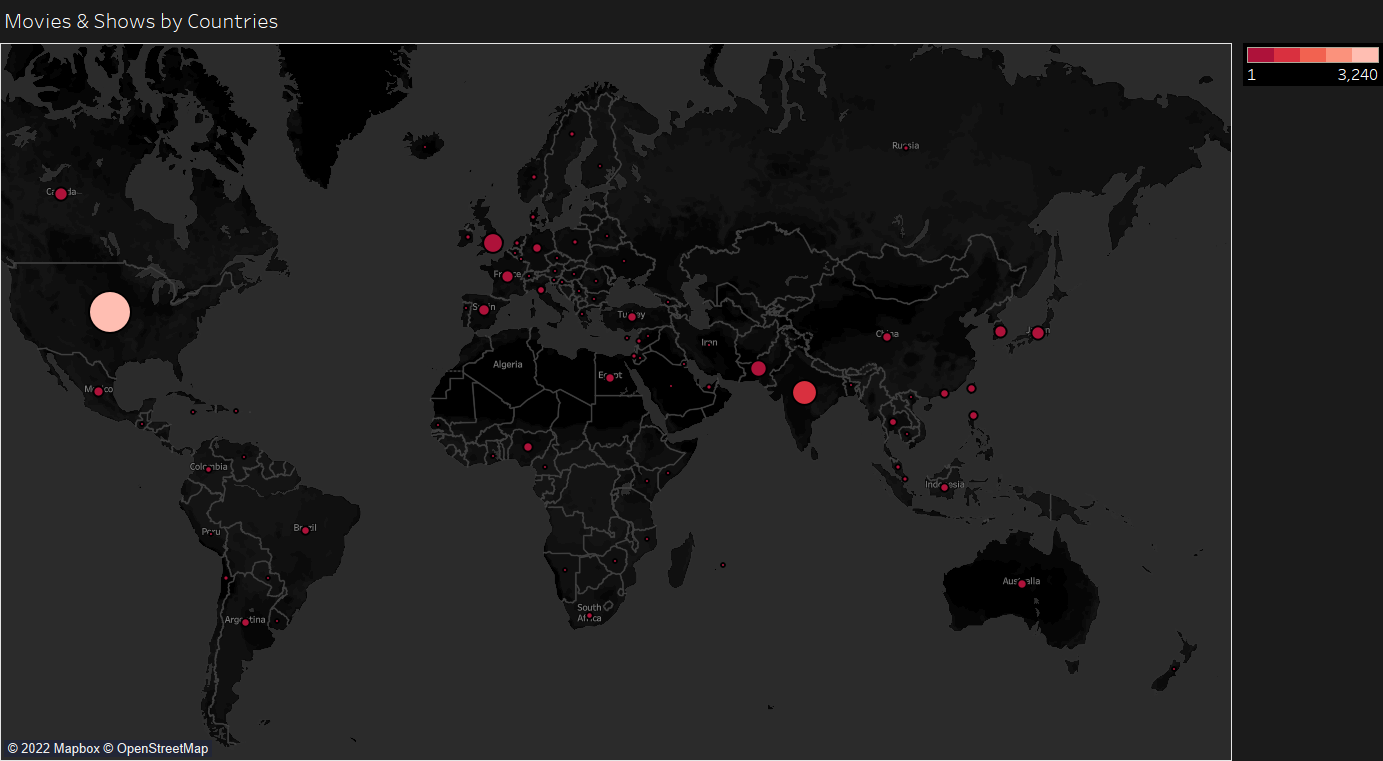
This shows the the total amount of Movies and Tv shows per country within the given period of time(2008 - 2021). This can be noted by the size of the coloured circle in the map.
- We can see that the United State of America has the largest size, followed by India and the United Kingdom.
- In the Tableau hosted dashboard/sheet, there is a filter for the years between 2008 and 2021 to calculate yearly record.
To give an alternate and a clearer view. Movie & TV shows by country bar chart is below
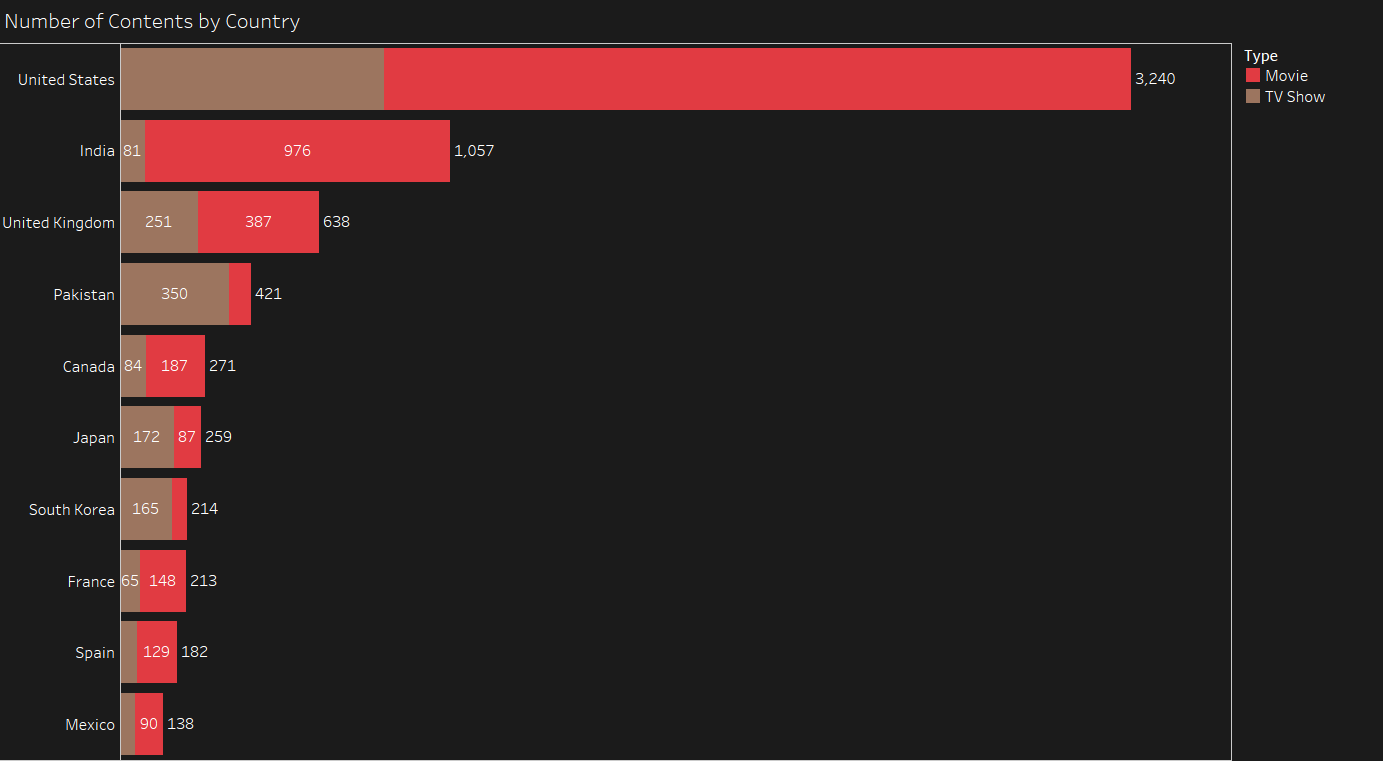
**Sheet 3. Number of Contents Added through the Years**
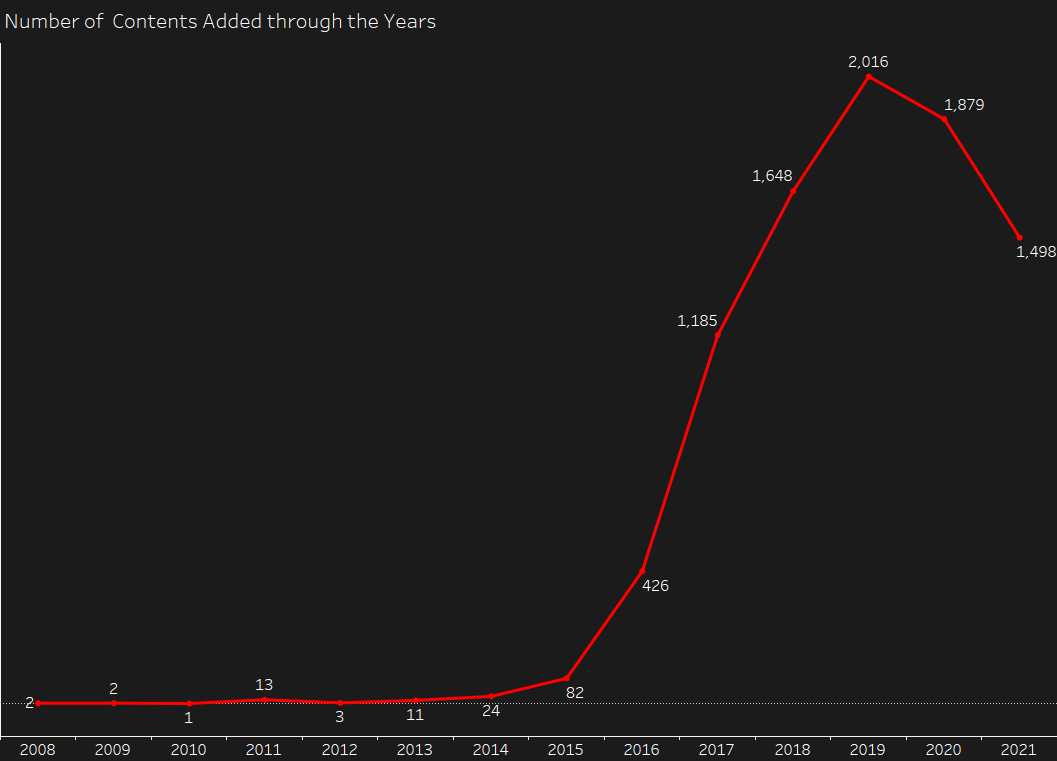
This time series chart shows the total number of contents added to Netflix all through the given years (2008 - 2021)
- It shows that most movies and tv shows on Netflix were added in 2019
- In the Tableau sheet, there is a filter to know how much Movies and Tv shows were added in each month of the year
**Sheet 4. Top Directors**
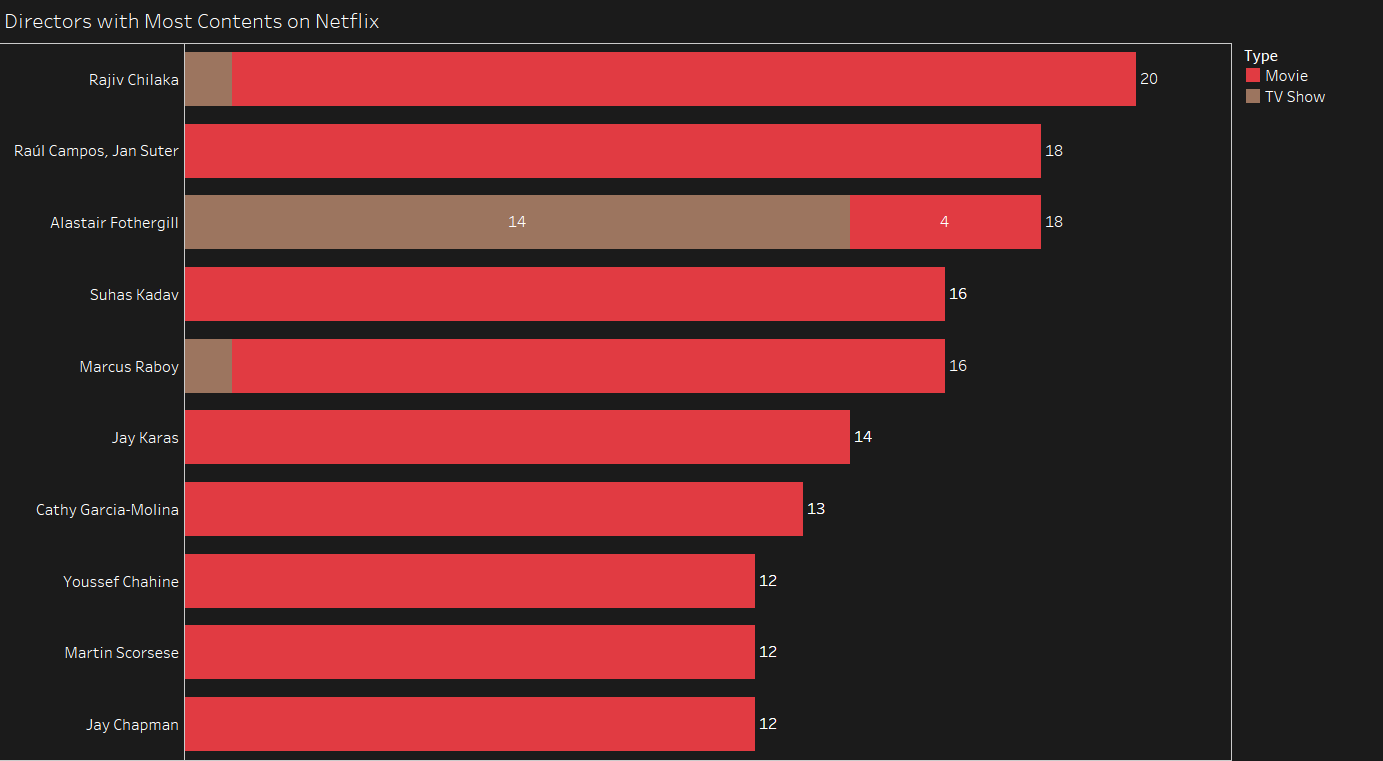
This chart shows the top 10 directors with most contents on Netflix. This char shows the count of Movie and Tv shows in their catalouge.
- We can see that most of these directors contents are movies.
- We can also note that the duo of Raul Campos and Jan Suter are fond of working together and have directed 18 movies on Netflix.
**Sheet 5. Top Genres**
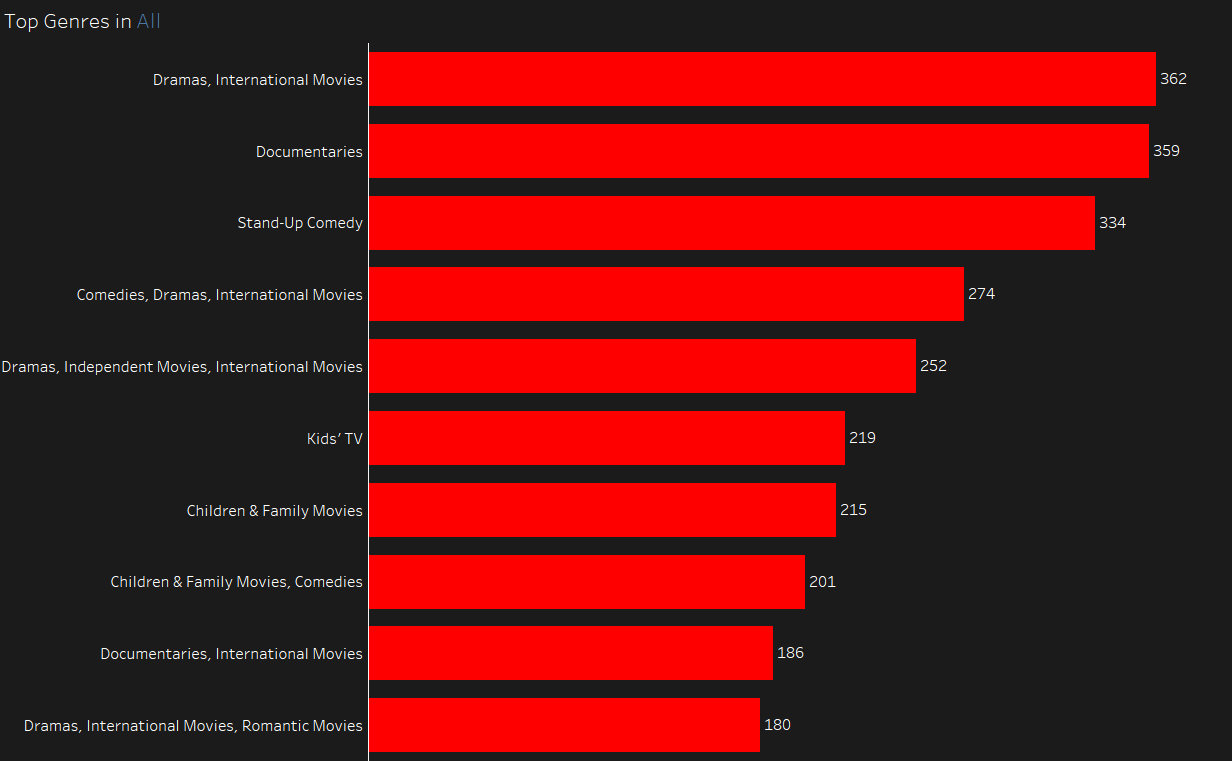
This chart shows the genres with the highest numbers on Netflix.
- We can see that Drama & International movies followed by Documentary have the highest number of contents on Netflix within the period.
**Sheet 6. Top Ratings**
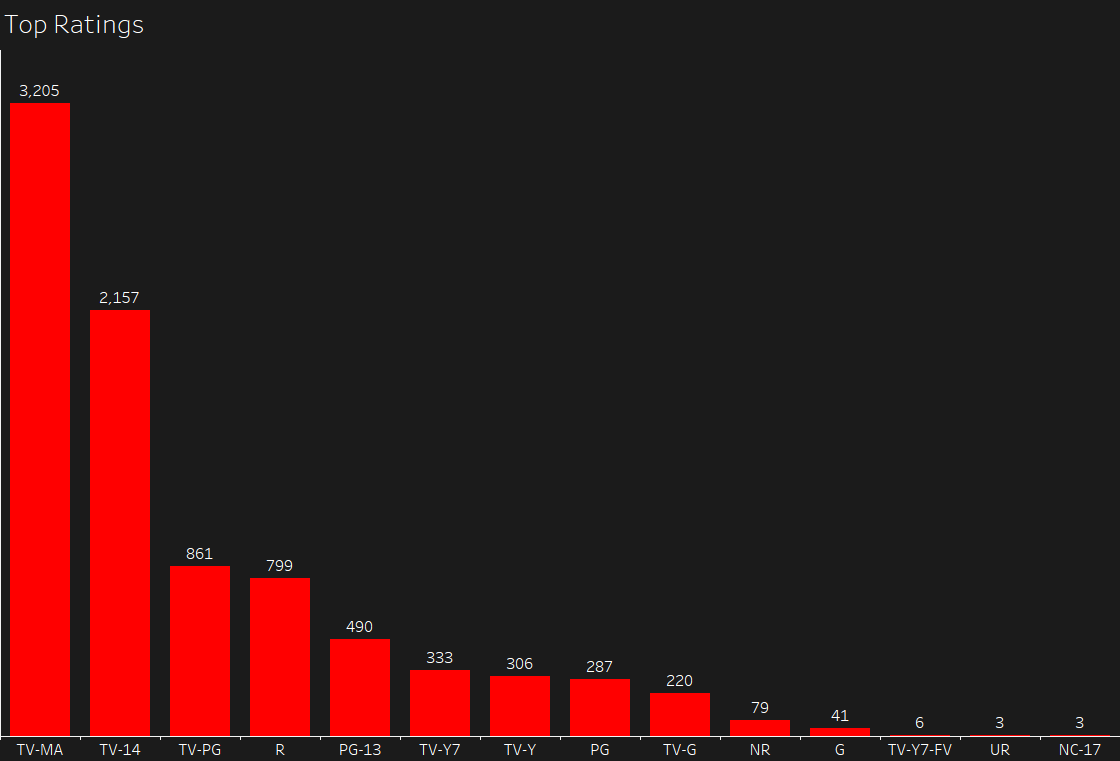
Rating is a system to rate motion picture's suitability for certain audiences based on its content. This chart shows the top ratings on Netflix
-We can note that most contents on Netflix are rated TV-MA. TV-MA in the United States by the TV Parental Guidelines signifies content for mature audiences.
**Sheet 7. Oldest Contents on Netflix by Content Release year**

This table shows the 10 oldest movies and tv shows on Netflix
- The oldest is as old as 1925
**Sheet 8. Content Types over the Years**
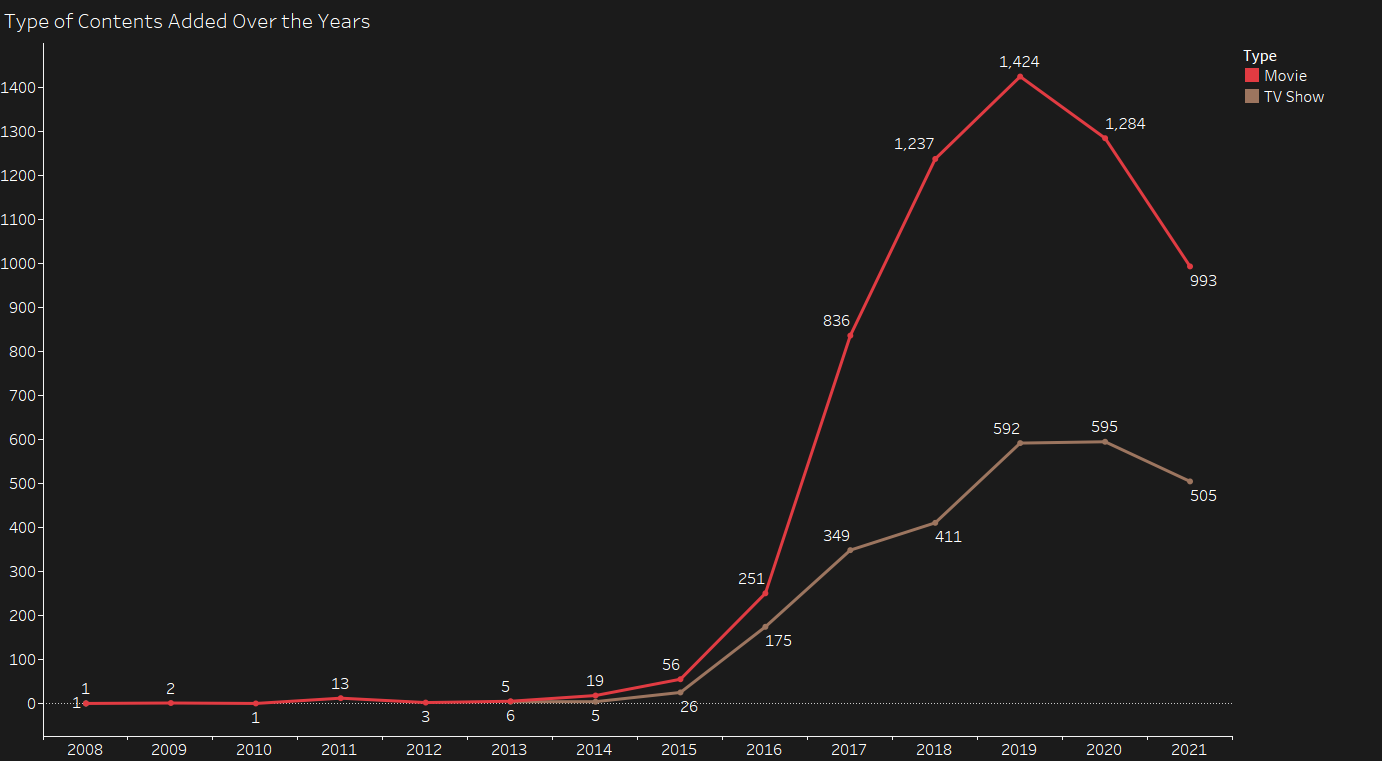
This line chart compares the Movie and Tv shows contents added to Netflix all through the years.
- We can see that more movies have always been added.
- In 2013, the number of contents added to Netflix for both were almost the same with Movies having 6 contents that year and Tv shows having 5.
- It shows that in the first 5 years, only movies were added to Netflix.
**Sheet 9. Release Years with Highest Contents**
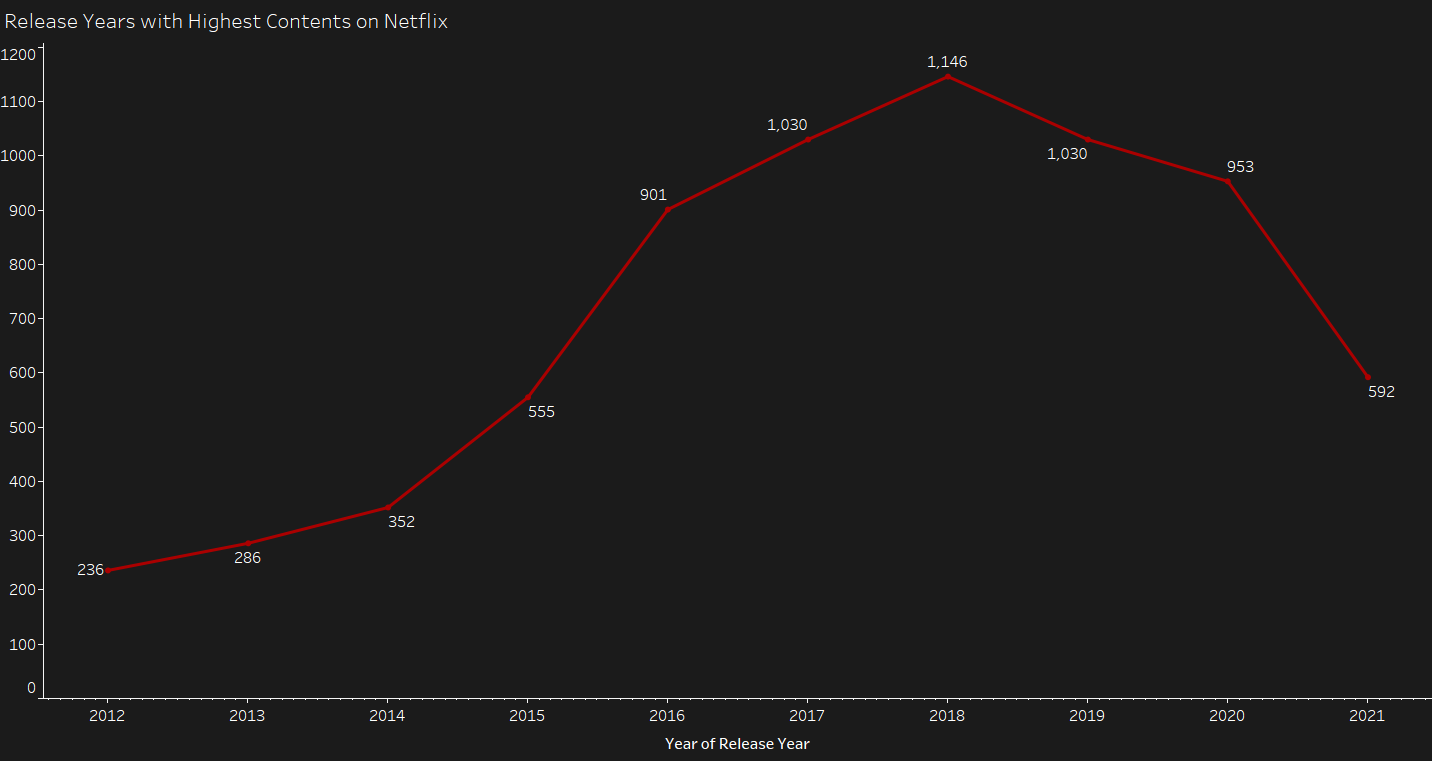
This chart shows the Movies and Tv shows production year which has with highest contents on Netflix. We focus on the top 10 release year/production year.
-We can see that from 2012 to 2018, Netflix added most recent contents, they made sure most recent contents per release year are higher than the older release year contents. Then in 2019, it started dropping, this may be due to the Covid-19, but further analysis may be needed to determine this.
And with this, I have come to the end of this exercise. As I said this is just an exercise to test my skills as I look forward to be better. Thanks for following through. Cheers!
Kaggle dataset identifier: netflix-data-cleaning-analysis-and-visualization
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/netflix-data-cleaning-analysis-and-visualization/netflix1.csv"
)
df.head()
df.info()
df.shape
df.describe(include="all")
df.nunique(axis=0)
type_of_show = df.groupby("type")["show_id"].count()
label = ["Movies", "TV Shows"]
plt.pie(type_of_show, labels=label, autopct="%1.2f%%")
plt.title("The proportion of the type of shows in Netflix")
plt.show()
per_country = df.groupby("country")["show_id"].count()
per_country.sort_values(ascending=False).head(10).plot(kind="bar")
plt.title("Top ten countries with the most shows on Netflix")
plt.xlabel("country")
plt.ylabel("No of Shows")
plt.show()
per_year = df.groupby("release_year")["show_id"].count()
per_year.sort_values(ascending=False).head(10).plot(kind="bar")
plt.title("The year of production of the show most available on Netflix")
plt.xlabel("Release Year")
plt.ylabel("No of Shows")
plt.show()
listed_in = df.groupby("listed_in")["show_id"].count()
listed_in.sort_values(ascending=False).head(10).plot(kind="barh")
plt.title("No of shows listed in different genre")
plt.ylabel("Listed In")
plt.xlabel("No Of shows")
plt.show()
df.groupby("director")["show_id"].count().sort_values(ascending=False).head(10)
genre_in_usa = (
df[df["country"] == "United States"].groupby("listed_in")["show_id"].count()
)
genre_in_usa.sort_values(ascending=False).head(10).plot(kind="barh")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/117/129117170.ipynb
|
netflix-data-cleaning-analysis-and-visualization
|
ariyoomotade
|
[{"Id": 129117170, "ScriptId": 38384436, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14735671, "CreationDate": "05/11/2023 06:15:23", "VersionNumber": 1.0, "Title": "notebook37d983eaf6", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 64.0, "LinesInsertedFromPrevious": 64.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 184888021, "KernelVersionId": 129117170, "SourceDatasetVersionId": 4123716}]
|
[{"Id": 4123716, "DatasetId": 2437124, "DatasourceVersionId": 4180064, "CreatorUserId": 10322697, "LicenseName": "CC0: Public Domain", "CreationDate": "08/26/2022 09:25:43", "VersionNumber": 1.0, "Title": "Netflix Data: Cleaning, Analysis and Visualization", "Slug": "netflix-data-cleaning-analysis-and-visualization", "Subtitle": "Cleaning and Visualization with Pgsql and Tableau", "Description": "Netflix is a popular streaming service that offers a vast catalog of movies, TV shows, and original contents. This dataset is a cleaned version of the original version which can be found [here](https://www.kaggle.com/datasets/shivamb/netflix-shows). The data consist of contents added to Netflix from 2008 to 2021. The oldest content is as old as 1925 and the newest as 2021. This dataset will be cleaned with PostgreSQL and visualized with Tableau. The purpose of this dataset is to test my data cleaning and visualization skills. The cleaned data can be found below and the Tableau dashboard can be found [here](https://public.tableau.com/app/profile/abdulrasaq.ariyo/viz/NetflixTVShowsMovies_16615029026580/NetflixDashboard) . \n\n## Data Cleaning\nWe are going to:\n1. Treat the Nulls \n2. Treat the duplicates\n3. Populate missing rows\n4. Drop unneeded columns\n5. Split columns\nExtra steps and more explanation on the process will be explained through the code comments\n\n```\n--View dataset\n\nSELECT * \nFROM netflix;\n\n```\n\n```\n--The show_id column is the unique id for the dataset, therefore we are going to check for duplicates\n \nSELECT show_id, COUNT(*) \nFROM netflix \nGROUP BY show_id \nORDER BY show_id DESC;\n\n--No duplicates\n```\n\n```\n--Check null values across columns\n\nSELECT COUNT(*) FILTER (WHERE show_id IS NULL) AS showid_nulls,\n COUNT(*) FILTER (WHERE type IS NULL) AS type_nulls,\n COUNT(*) FILTER (WHERE title IS NULL) AS title_nulls,\n COUNT(*) FILTER (WHERE director IS NULL) AS director_nulls,\n\t COUNT(*) FILTER (WHERE movie_cast IS NULL) AS movie_cast_nulls,\n\t COUNT(*) FILTER (WHERE country IS NULL) AS country_nulls,\n COUNT(*) FILTER (WHERE date_added IS NULL) AS date_addes_nulls,\n COUNT(*) FILTER (WHERE release_year IS NULL) AS release_year_nulls,\n COUNT(*) FILTER (WHERE rating IS NULL) AS rating_nulls,\n\t COUNT(*) FILTER (WHERE duration IS NULL) AS duration_nulls,\n COUNT(*) FILTER (WHERE listed_in IS NULL) AS listed_in_nulls,\n\t COUNT(*) FILTER (WHERE description IS NULL) AS description_nulls\nFROM netflix;\n```\n```\nWe can see that there are NULLS. \ndirector_nulls = 2634\nmovie_cast_nulls = 825\ncountry_nulls = 831\ndate_added_nulls = 10\nrating_nulls = 4\nduration_nulls = 3 \n```\n\nThe director column nulls is about 30% of the whole column, therefore I will not delete them. I will rather find another column to populate it. To populate the director column, we want to find out if there is relationship between movie_cast column and director column\n\n\n``` \n-- Below, we find out if some directors are likely to work with particular cast\n\nWITH cte AS\n(\nSELECT title, CONCAT(director, '---', movie_cast) AS director_cast \nFROM netflix\n)\n\nSELECT director_cast, COUNT(*) AS count\nFROM cte\nGROUP BY director_cast\nHAVING COUNT(*) > 1\nORDER BY COUNT(*) DESC;\n\nWith this, we can now populate NULL rows in directors \nusing their record with movie_cast \n```\n```\nUPDATE netflix \nSET director = 'Alastair Fothergill'\nWHERE movie_cast = 'David Attenborough'\nAND director IS NULL ;\n\n--Repeat this step to populate the rest of the director nulls\n--Populate the rest of the NULL in director as \"Not Given\"\n\nUPDATE netflix \nSET director = 'Not Given'\nWHERE director IS NULL;\n\n--When I was doing this, I found a less complex and faster way to populate a column which I will use next\n```\n\nJust like the director column, I will not delete the nulls in country. Since the country column is related to director and movie, we are going to populate the country column with the director column\n\n```\n--Populate the country using the director column\n\nSELECT COALESCE(nt.country,nt2.country) \nFROM netflix AS nt\nJOIN netflix AS nt2 \nON nt.director = nt2.director \nAND nt.show_id <> nt2.show_id\nWHERE nt.country IS NULL;\nUPDATE netflix\nSET country = nt2.country\nFROM netflix AS nt2\nWHERE netflix.director = nt2.director and netflix.show_id <> nt2.show_id \nAND netflix.country IS NULL;\n\n\n--To confirm if there are still directors linked to country that refuse to update\n\nSELECT director, country, date_added\nFROM netflix\nWHERE country IS NULL;\n\n--Populate the rest of the NULL in director as \"Not Given\"\n\nUPDATE netflix \nSET country = 'Not Given'\nWHERE country IS NULL;\n```\n\nThe date_added rows nulls is just 10 out of over 8000 rows, deleting them cannot affect our analysis or visualization\n\n```\n--Show date_added nulls\n\nSELECT show_id, date_added\nFROM netflix_clean\nWHERE date_added IS NULL;\n\n--DELETE nulls\n\nDELETE FROM netflix\nWHERE show_id \nIN ('6797', 's6067', 's6175', 's6807', 's6902', 's7255', 's7197', 's7407', 's7848', 's8183');\n\n```\n\nrating nulls is 4. Delete them\n```\n--Show rating NULLS\n\nSELECT show_id, rating\nFROM netflix_clean\nWHERE date_added IS NULL;\n\n--Delete the nulls, and show deleted fields\nDELETE FROM netflix \nWHERE show_id \nIN (SELECT show_id FROM netflix WHERE rating IS NULL)\nRETURNING *;\n```\n\n--duration nulls is 4. Delete them\n```\n\nDELETE FROM netflix \nWHERE show_id \nIN (SELECT show_id FROM netflix WHERE duration IS NULL);\n```\nNow run the query to show the number of nulls in each column to confirm if there are still nulls. After this, run the query to confirm the row number in each column is the same\n\n```\n--Check to confirm the number of rows are the same(NO NULL)\n\nSELECT count(*) filter (where show_id IS NOT NULL) AS showid_nulls,\n count(*) filter (where type IS NOT NULL) AS type_nulls,\n count(*) filter (where title IS NOT NULL) AS title_nulls,\n count(*) filter (where director IS NOT NULL) AS director_nulls,\n\t count(*) filter (where country IS NOT NULL) AS country_nulls,\n count(*) filter (where date_added IS NOT NULL) AS date_addes_nulls,\n count(*) filter (where release_year IS NOT NULL) AS release_year_nulls,\n count(*) filter (where rating IS NOT NULL) AS rating_nulls,\n\t count(*) filter (where duration IS NOT NULL) AS duration_nulls,\n count(*) filter (where listed_in IS NOT NULL) AS listed_in_nulls\nFROM netflix;\n\n --Total number of rows are the same in all columns\n```\nWe can drop the description and movie_cast column because they are not needed for our analysis or visualization task. \n```\n--DROP unneeded columns\n\nALTER TABLE netflix\nDROP COLUMN movie_cast, \nDROP COLUMN description;\n```\nSome of the rows in country column has multiple countries, for my visualization, I only need one country per row to make my map visualization clean and easy. Therefore, I am going to split the country column and retain the first country by the left which I believe is the original country of the movie\n```\nSELECT *,\n\t SPLIT_PART(country,',',1) AS countryy, \n SPLIT_PART(country,',',2),\n\t SPLIT_PART(country,',',4),\n\t SPLIT_PART(country,',',5),\n\t SPLIT_PART(country,',',6),\n\t SPLIT_PART(country,',',7),\n\t SPLIT_PART(country,',',8),\n\t SPLIT_PART(country,',',9),\n\t SPLIT_PART(country,',',10) \n\t \nFROM netflix;\n\t \n-- NOW lets update the table\n\nALTER TABLE netflix \nADD country1 varchar(500);\nUPDATE netflix \nSET country1 = SPLIT_PART(country, ',', 1);\n\n--This will create a column named country1 and Update it with the first split country.\n```\n\nDelete the country column that has multiple country entries\n```\n--Delete column\nALTER TABLE netflix \nDROP COLUMN country;\n```\nRename the country1 column to country\n```\nALTER TABLE netflix \nRENAME COLUMN country1 TO country;\n```\n\n## Data Visualization\nAfter cleaning, the dataset is set for some analysis and visualization with Tableau. \n\n**Note: In the visualization captions, Contents means Movies and TV shows, and Content may either mean Movie or TV Show**. \n\n**Sheet 1. Content type in percentage**\n\n\n\nThis first sheet shows the two categories of content in the dataset which are Movie and Tv show. \n- As we can see the majority of the content is Movie which takes 69.9%. \n- There are more details in the tooltip which shows the exact count of Movie and Tv show\n\n\n**Sheet 2. Movie & TV Show by Country**\n\n\n\nThis shows the the total amount of Movies and Tv shows per country within the given period of time(2008 - 2021). This can be noted by the size of the coloured circle in the map. \n- We can see that the United State of America has the largest size, followed by India and the United Kingdom. \n- In the Tableau hosted dashboard/sheet, there is a filter for the years between 2008 and 2021 to calculate yearly record.\n\n To give an alternate and a clearer view. Movie & TV shows by country bar chart is below\n\n\n\n**Sheet 3. Number of Contents Added through the Years**\n\n\n\nThis time series chart shows the total number of contents added to Netflix all through the given years (2008 - 2021)\n- It shows that most movies and tv shows on Netflix were added in 2019\n- In the Tableau sheet, there is a filter to know how much Movies and Tv shows were added in each month of the year \n\n\n**Sheet 4. Top Directors**\n\n\n\nThis chart shows the top 10 directors with most contents on Netflix. This char shows the count of Movie and Tv shows in their catalouge. \n- We can see that most of these directors contents are movies. \n- We can also note that the duo of Raul Campos and Jan Suter are fond of working together and have directed 18 movies on Netflix. \n\n\n**Sheet 5. Top Genres** \n\n\n\nThis chart shows the genres with the highest numbers on Netflix. \n- We can see that Drama & International movies followed by Documentary have the highest number of contents on Netflix within the period.\n\n\n**Sheet 6. Top Ratings**\n\n\n \nRating is a system to rate motion picture's suitability for certain audiences based on its content. This chart shows the top ratings on Netflix\n-We can note that most contents on Netflix are rated TV-MA. TV-MA in the United States by the TV Parental Guidelines signifies content for mature audiences. \n\n\n**Sheet 7. Oldest Contents on Netflix by Content Release year**\n\n\n\nThis table shows the 10 oldest movies and tv shows on Netflix\n- The oldest is as old as 1925\n\n**Sheet 8. Content Types over the Years**\n\n\nThis line chart compares the Movie and Tv shows contents added to Netflix all through the years.\n- We can see that more movies have always been added. \n- In 2013, the number of contents added to Netflix for both were almost the same with Movies having 6 contents that year and Tv shows having 5.\n- It shows that in the first 5 years, only movies were added to Netflix. \n\n\n**Sheet 9. Release Years with Highest Contents**\n\n\n\nThis chart shows the Movies and Tv shows production year which has with highest contents on Netflix. We focus on the top 10 release year/production year. \n-We can see that from 2012 to 2018, Netflix added most recent contents, they made sure most recent contents per release year are higher than the older release year contents. Then in 2019, it started dropping, this may be due to the Covid-19, but further analysis may be needed to determine this. \n\n And with this, I have come to the end of this exercise. As I said this is just an exercise to test my skills as I look forward to be better. Thanks for following through. Cheers!", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2437124, "CreatorUserId": 10322697, "OwnerUserId": 10322697.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4123716.0, "CurrentDatasourceVersionId": 4180064.0, "ForumId": 2464656, "Type": 2, "CreationDate": "08/26/2022 09:25:43", "LastActivityDate": "08/26/2022", "TotalViews": 96354, "TotalDownloads": 16114, "TotalVotes": 270, "TotalKernels": 23}]
|
[{"Id": 10322697, "UserName": "ariyoomotade", "DisplayName": "Abdulrasaq Ariyo", "RegisterDate": "04/22/2022", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/netflix-data-cleaning-analysis-and-visualization/netflix1.csv"
)
df.head()
df.info()
df.shape
df.describe(include="all")
df.nunique(axis=0)
type_of_show = df.groupby("type")["show_id"].count()
label = ["Movies", "TV Shows"]
plt.pie(type_of_show, labels=label, autopct="%1.2f%%")
plt.title("The proportion of the type of shows in Netflix")
plt.show()
per_country = df.groupby("country")["show_id"].count()
per_country.sort_values(ascending=False).head(10).plot(kind="bar")
plt.title("Top ten countries with the most shows on Netflix")
plt.xlabel("country")
plt.ylabel("No of Shows")
plt.show()
per_year = df.groupby("release_year")["show_id"].count()
per_year.sort_values(ascending=False).head(10).plot(kind="bar")
plt.title("The year of production of the show most available on Netflix")
plt.xlabel("Release Year")
plt.ylabel("No of Shows")
plt.show()
listed_in = df.groupby("listed_in")["show_id"].count()
listed_in.sort_values(ascending=False).head(10).plot(kind="barh")
plt.title("No of shows listed in different genre")
plt.ylabel("Listed In")
plt.xlabel("No Of shows")
plt.show()
df.groupby("director")["show_id"].count().sort_values(ascending=False).head(10)
genre_in_usa = (
df[df["country"] == "United States"].groupby("listed_in")["show_id"].count()
)
genre_in_usa.sort_values(ascending=False).head(10).plot(kind="barh")
| false | 1 | 670 | 1 | 4,901 | 670 |
||
129083186
|
#
# ## **disclaimer **
# This info can be found in Introduction to machine learning with Python book it is an awesome book I highly recommend it you can found it here
# https://www.oreilly.com/library/view/introduction-to-machine/9781449369880/
# ## Linear Models
# #### Linear models are a class of models that are widely used in practice and have been studied extensively in the last few decades, with roots going back over a hundred years. Linear models make a prediction using a linear function of the input feature
# ### Linear models for regression
# #### For regression, the general prediction formula for a linear model looks as follows:
# #### Here, x denotes the features (in this example, the number of features is i) of a single data point, B0 and B1 are parameters of the model that are learned, and ŷ is the prediction the model makes.
# #### w[0] is the slope and b is the y-axis offset. For more features, w contains the slopes along each feature axis. Alternatively, you can think of the predicted response as being a weighted sum of the input features, with weights (which can be negative) given by the entries of w
import matplotlib.pyplot as plt
import mglearn
mglearn.plots.plot_linear_regression_wave()
# #### We added a coordinate cross into the plot to make it easier to understand the line. Looking at w[0] we see that the slope should be around 0.4, which we can confirm visually in the plot. The intercept is where the prediction line should cross the y-axis: this is slightly below zero, which you can also confirm in the image.
# #### Linear models for regression can be characterized as regression models for which the prediction is a line for a single feature, a plane when using two features, or a hyper‐ plane in higher dimensions (that is, when using more features)
# #### If you compare the predictions made by the straight line with those made by the KNeighborsRegressor , using a straight line to make predictions seems very restrictive. It looks like all the fine details of the data are lost
# #### . For datasets with many features, linear models can be very powerful. In particular, if you have more features than training data points, any target y can be perfectly modeled (on the training set) as a linear functio
# #### There are many different linear models for regression. The difference between these models lies in how the model parameters w and b are learned from the training data, and how model complexity can be controlled
# ### Linear regression (aka ordinary least squares) "OLS"
# #### Linear regression, or ordinary least squares (OLS), is the simplest and most classic lin‐ ear method for regression. Linear regression finds the parameters w and b that mini‐ mize the mean squared error between predictions and the true regression targets, y, on the training set. The mean squared error is the sum of the squared differences between the predictions and the true values. Linear regression has no parameters, which is a benefit, but it also has no way to control model complexity.
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
X, y = mglearn.datasets.make_wave(n_samples=600)
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
lr = LinearRegression().fit(X_train, y_train)
print("lr.coef_: {}".format(lr.coef_))
print("lr.intercept_: {}".format(lr.intercept_))
# #### This code produces the model you can see above
# #### The “slope” parameters (w), also called weights or coefcients, are stored in the coef_ attribute, while the offset or intercept (b) is stored in the intercept_ attribute
# #### The intercept_ attribute is always a single float number, while the coef_ attribute is a NumPy array with one entry per input feature. As we only have a single input feature in the wave dataset, lr.coef_ only has a single entry
print("Training set score: {:.2f}".format(lr.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lr.score(X_test, y_test)))
# #### An R2 of around 0.66 is not very good, but we can see that the scores on the training and test sets are very close together. This means we are likely underfitting, not overfitting. For this one-dimensional dataset, there is little danger of overfitting, as the model is very simple (or restricted). However, with higher-dimensional datasets (meaning datasets with a large number of features), linear models become more powerful, and there is a higher chance of overfitting
# #### Let’s take a look at how LinearRe gression performs on a more complex dataset
X, y = mglearn.datasets.load_extended_boston()
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
lr = LinearRegression().fit(X_train, y_train)
print("Training set score: {:.2f}".format(lr.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lr.score(X_test, y_test)))
# #### This discrepancy between performance on the training set and the test set is a clear sign of overfitting, and therefore we should try to find a model that allows us to con‐ trol complexity. One of the most commonly used alternatives to standard linear regression is ridge regression
# ### Ridge regression
# #### Ridge regression is also a linear model for regression, so the formula it uses to make predictions is the same one used for ordinary least squares. In ridge regression, though, the coefficients (w) are chosen not only so that they predict well on the training data, but also to fit an additional constraint. We also want the magnitude of coefficients to be as small as possible; in other words, all entries of w should be close to zero. Intuitively, this means each feature should have as little effect on the outcome as possible (which translates to having a small slope), while still predicting well. This constraint is an example of what is called regularization. Regularization means explicitly restricting a model to avoid overfitting. The particular kind used by ridge regres‐ sion is known as L2 regularization
from sklearn.linear_model import Ridge
ridge = Ridge().fit(X_train, y_train)
print("Training set score: {:.2f}".format(ridge.score(X_train, y_train)))
print("Test set score: {:.2f}".format(ridge.score(X_test, y_test)))
# #### As you can see, the training set score of Ridge is lower than for LinearRegression, while the test set score is higher. This is consistent with our expectation. With linear regression, we were overfitting our data. Ridge is a more restricted model, so we are less likely to overfit. A less complex model means worse performance on the training set, but better generalization. As we are only interested in generalization performance, we should choose the Ridge model over the LinearRegression model.
# #### The Ridge model makes a trade-off between the simplicity of the model (near-zero coefficients) and its performance on the training set. How much importance the model places on simplicity versus training set performance can be specified by the user, using the alpha parameter
# #### the default parameter is alpha=1.0 , Increasing alpha forces coefficients to move more toward zero, which decreases training set performance but might help generalization.
ridge10 = Ridge(alpha=10).fit(X_train, y_train)
print("Training set score: {:.2f}".format(ridge10.score(X_train, y_train)))
print("Test set score: {:.2f}".format(ridge10.score(X_test, y_test)))
# #### Decreasing alpha allows the coefficients to be less restricted
ridge01 = Ridge(alpha=0.1).fit(X_train, y_train)
print("Training set score: {:.2f}".format(ridge01.score(X_train, y_train)))
print("Test set score: {:.2f}".format(ridge01.score(X_test, y_test)))
# #### We can also get a more qualitative insight into how the alpha parameter changes the model by inspecting the coefattribute of models with different values of alpha. A higher alpha means a more restricted model, so we expect the entries of coef_ to have smaller magnitude for a high value of alpha than for a low value of alpha.
plt.plot(ridge.coef_, "s", label="Ridge alpha=1")
plt.plot(ridge10.coef_, "^", label="Ridge alpha=10")
plt.plot(ridge01.coef_, "v", label="Ridge alpha=0.1")
plt.plot(lr.coef_, "o", label="LinearRegression")
plt.xlabel("Coefficient index")
plt.ylabel("Coefficient magnitude")
plt.hlines(0, 0, len(lr.coef_))
plt.ylim(-25, 25)
plt.legend()
# #### Here, the x-axis enumerates the entries of coef_: x=0 shows the coefficient associated with the first feature, x=1 the coefficient associated with the second feature, and so on up to x=100. The y-axis shows the numeric values of the corresponding values of the coefficients. The main takeaway here is that for alpha=10, the coefficients are mostly between around –3 and 3. The coefficients for the Ridge model with alpha=1 are somewhat larger. The dots corresponding to alpha=0.1 have larger magnitude still, and many of the dots corresponding to linear regression without any regularization (which would be alpha=0) are so large they are outside of the chart
# #### Another way to understand the influence of regularization is to fix a value of alpha but vary the amount of training data available, we subsampled the Boston Housing dataset and evaluated LinearRegression and Ridge(alpha=1) on subsets of increasing size (plots that show model performance as a function of dataset size are called learning curves)
mglearn.plots.plot_ridge_n_samples()
# #### As one would expect, the training score is higher than the test score for all dataset sizes, for both ridge and linear regression. Because ridge is regularized, the training score of ridge is lower than the training score for linear regression across the board. However, the test score for ridge is better, particularly for small subsets of the data. For less than 400 data points, linear regression is not able to learn anything. As more and more data becomes available to the model, both models improve, and linear regression catches up with ridge in the end. The lesson here is that with enough training data, regularization becomes less important, and given enough data, ridge and linear regression will have the same performance (the fact that this happens here when using the full dataset is just by chance). Another interesting aspect is the decrease in training performance for linear regression. If more data is added, it becomes harder for a model to overfit, or memorize the data
# ### Lasso
# #### An alternative to Ridge for regularizing linear regression is Lasso. As with ridge regression, using the lasso also restricts coefficients to be close to zero, but in a slightly different way, called L1 regularization The consequence of L1 regularization is that when using the lasso, some coefficients are exactly zero. This means some features are entirely ignored by the model. This can be seen as a form of automatic fea‐ ture selection. Having some coefficients be exactly zero often makes a model easier to interpret, and can reveal the most important features of your model
from sklearn.linear_model import Lasso
lasso = Lasso().fit(X_train, y_train)
print("Training set score: {:.2f}".format(lasso.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lasso.score(X_test, y_test)))
print("Number of features used: {}".format(np.sum(lasso.coef_ != 0)))
# #### As you can see, Lasso does quite badly, both on the training and the test set. This indicates that we are underfitting, and we find that it used only 4 of the 105 features. Similarly to Ridge, the Lasso also has a regularization parameter, alpha, that controls how strongly coefficients are pushed toward zero. In the previous example, we used the default of alpha=1.0. To reduce underfitting, let’s try decreasing alpha. When we do this, we also need to increase the default setting of max_iter (the maximum num‐ ber of iterations to run
# we increase the default setting of "max_iter",
# otherwise the model would warn us that we should increase max_iter.
lasso001 = Lasso(alpha=0.01, max_iter=100000).fit(X_train, y_train)
print("Training set score: {:.2f}".format(lasso001.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lasso001.score(X_test, y_test)))
print("Number of features used: {}".format(np.sum(lasso001.coef_ != 0)))
# #### A lower alpha allowed us to fit a more complex model, which worked better on the training and test data. The performance is slightly better than using Ridge, and we are using only 33 of the 105 features. This makes this model potentially easier to understand.
# #### If we set alpha too low, however, we again remove the effect of regularization and end up overfitting, with a result similar to LinearRegression
lasso00001 = Lasso(alpha=0.0001, max_iter=100000).fit(X_train, y_train)
print("Training set score: {:.2f}".format(lasso00001.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lasso00001.score(X_test, y_test)))
print("Number of features used: {}".format(np.sum(lasso00001.coef_ != 0)))
# #### Again, we can plot the coefficients of the different models
plt.plot(lasso.coef_, "s", label="Lasso alpha=1")
plt.plot(lasso001.coef_, "^", label="Lasso alpha=0.01")
plt.plot(lasso00001.coef_, "v", label="Lasso alpha=0.0001")
plt.plot(ridge01.coef_, "o", label="Ridge alpha=0.1")
plt.legend(ncol=2, loc=(0, 1.05))
plt.ylim(-25, 25)
plt.xlabel("Coefficient index")
plt.ylabel("Coefficient magnitude")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/083/129083186.ipynb
| null | null |
[{"Id": 129083186, "ScriptId": 38373259, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6965494, "CreationDate": "05/10/2023 21:37:31", "VersionNumber": 1.0, "Title": "Analyzing Linear Regression", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 172.0, "LinesInsertedFromPrevious": 172.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
| null | null | null | null |
#
# ## **disclaimer **
# This info can be found in Introduction to machine learning with Python book it is an awesome book I highly recommend it you can found it here
# https://www.oreilly.com/library/view/introduction-to-machine/9781449369880/
# ## Linear Models
# #### Linear models are a class of models that are widely used in practice and have been studied extensively in the last few decades, with roots going back over a hundred years. Linear models make a prediction using a linear function of the input feature
# ### Linear models for regression
# #### For regression, the general prediction formula for a linear model looks as follows:
# #### Here, x denotes the features (in this example, the number of features is i) of a single data point, B0 and B1 are parameters of the model that are learned, and ŷ is the prediction the model makes.
# #### w[0] is the slope and b is the y-axis offset. For more features, w contains the slopes along each feature axis. Alternatively, you can think of the predicted response as being a weighted sum of the input features, with weights (which can be negative) given by the entries of w
import matplotlib.pyplot as plt
import mglearn
mglearn.plots.plot_linear_regression_wave()
# #### We added a coordinate cross into the plot to make it easier to understand the line. Looking at w[0] we see that the slope should be around 0.4, which we can confirm visually in the plot. The intercept is where the prediction line should cross the y-axis: this is slightly below zero, which you can also confirm in the image.
# #### Linear models for regression can be characterized as regression models for which the prediction is a line for a single feature, a plane when using two features, or a hyper‐ plane in higher dimensions (that is, when using more features)
# #### If you compare the predictions made by the straight line with those made by the KNeighborsRegressor , using a straight line to make predictions seems very restrictive. It looks like all the fine details of the data are lost
# #### . For datasets with many features, linear models can be very powerful. In particular, if you have more features than training data points, any target y can be perfectly modeled (on the training set) as a linear functio
# #### There are many different linear models for regression. The difference between these models lies in how the model parameters w and b are learned from the training data, and how model complexity can be controlled
# ### Linear regression (aka ordinary least squares) "OLS"
# #### Linear regression, or ordinary least squares (OLS), is the simplest and most classic lin‐ ear method for regression. Linear regression finds the parameters w and b that mini‐ mize the mean squared error between predictions and the true regression targets, y, on the training set. The mean squared error is the sum of the squared differences between the predictions and the true values. Linear regression has no parameters, which is a benefit, but it also has no way to control model complexity.
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
X, y = mglearn.datasets.make_wave(n_samples=600)
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
lr = LinearRegression().fit(X_train, y_train)
print("lr.coef_: {}".format(lr.coef_))
print("lr.intercept_: {}".format(lr.intercept_))
# #### This code produces the model you can see above
# #### The “slope” parameters (w), also called weights or coefcients, are stored in the coef_ attribute, while the offset or intercept (b) is stored in the intercept_ attribute
# #### The intercept_ attribute is always a single float number, while the coef_ attribute is a NumPy array with one entry per input feature. As we only have a single input feature in the wave dataset, lr.coef_ only has a single entry
print("Training set score: {:.2f}".format(lr.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lr.score(X_test, y_test)))
# #### An R2 of around 0.66 is not very good, but we can see that the scores on the training and test sets are very close together. This means we are likely underfitting, not overfitting. For this one-dimensional dataset, there is little danger of overfitting, as the model is very simple (or restricted). However, with higher-dimensional datasets (meaning datasets with a large number of features), linear models become more powerful, and there is a higher chance of overfitting
# #### Let’s take a look at how LinearRe gression performs on a more complex dataset
X, y = mglearn.datasets.load_extended_boston()
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
lr = LinearRegression().fit(X_train, y_train)
print("Training set score: {:.2f}".format(lr.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lr.score(X_test, y_test)))
# #### This discrepancy between performance on the training set and the test set is a clear sign of overfitting, and therefore we should try to find a model that allows us to con‐ trol complexity. One of the most commonly used alternatives to standard linear regression is ridge regression
# ### Ridge regression
# #### Ridge regression is also a linear model for regression, so the formula it uses to make predictions is the same one used for ordinary least squares. In ridge regression, though, the coefficients (w) are chosen not only so that they predict well on the training data, but also to fit an additional constraint. We also want the magnitude of coefficients to be as small as possible; in other words, all entries of w should be close to zero. Intuitively, this means each feature should have as little effect on the outcome as possible (which translates to having a small slope), while still predicting well. This constraint is an example of what is called regularization. Regularization means explicitly restricting a model to avoid overfitting. The particular kind used by ridge regres‐ sion is known as L2 regularization
from sklearn.linear_model import Ridge
ridge = Ridge().fit(X_train, y_train)
print("Training set score: {:.2f}".format(ridge.score(X_train, y_train)))
print("Test set score: {:.2f}".format(ridge.score(X_test, y_test)))
# #### As you can see, the training set score of Ridge is lower than for LinearRegression, while the test set score is higher. This is consistent with our expectation. With linear regression, we were overfitting our data. Ridge is a more restricted model, so we are less likely to overfit. A less complex model means worse performance on the training set, but better generalization. As we are only interested in generalization performance, we should choose the Ridge model over the LinearRegression model.
# #### The Ridge model makes a trade-off between the simplicity of the model (near-zero coefficients) and its performance on the training set. How much importance the model places on simplicity versus training set performance can be specified by the user, using the alpha parameter
# #### the default parameter is alpha=1.0 , Increasing alpha forces coefficients to move more toward zero, which decreases training set performance but might help generalization.
ridge10 = Ridge(alpha=10).fit(X_train, y_train)
print("Training set score: {:.2f}".format(ridge10.score(X_train, y_train)))
print("Test set score: {:.2f}".format(ridge10.score(X_test, y_test)))
# #### Decreasing alpha allows the coefficients to be less restricted
ridge01 = Ridge(alpha=0.1).fit(X_train, y_train)
print("Training set score: {:.2f}".format(ridge01.score(X_train, y_train)))
print("Test set score: {:.2f}".format(ridge01.score(X_test, y_test)))
# #### We can also get a more qualitative insight into how the alpha parameter changes the model by inspecting the coefattribute of models with different values of alpha. A higher alpha means a more restricted model, so we expect the entries of coef_ to have smaller magnitude for a high value of alpha than for a low value of alpha.
plt.plot(ridge.coef_, "s", label="Ridge alpha=1")
plt.plot(ridge10.coef_, "^", label="Ridge alpha=10")
plt.plot(ridge01.coef_, "v", label="Ridge alpha=0.1")
plt.plot(lr.coef_, "o", label="LinearRegression")
plt.xlabel("Coefficient index")
plt.ylabel("Coefficient magnitude")
plt.hlines(0, 0, len(lr.coef_))
plt.ylim(-25, 25)
plt.legend()
# #### Here, the x-axis enumerates the entries of coef_: x=0 shows the coefficient associated with the first feature, x=1 the coefficient associated with the second feature, and so on up to x=100. The y-axis shows the numeric values of the corresponding values of the coefficients. The main takeaway here is that for alpha=10, the coefficients are mostly between around –3 and 3. The coefficients for the Ridge model with alpha=1 are somewhat larger. The dots corresponding to alpha=0.1 have larger magnitude still, and many of the dots corresponding to linear regression without any regularization (which would be alpha=0) are so large they are outside of the chart
# #### Another way to understand the influence of regularization is to fix a value of alpha but vary the amount of training data available, we subsampled the Boston Housing dataset and evaluated LinearRegression and Ridge(alpha=1) on subsets of increasing size (plots that show model performance as a function of dataset size are called learning curves)
mglearn.plots.plot_ridge_n_samples()
# #### As one would expect, the training score is higher than the test score for all dataset sizes, for both ridge and linear regression. Because ridge is regularized, the training score of ridge is lower than the training score for linear regression across the board. However, the test score for ridge is better, particularly for small subsets of the data. For less than 400 data points, linear regression is not able to learn anything. As more and more data becomes available to the model, both models improve, and linear regression catches up with ridge in the end. The lesson here is that with enough training data, regularization becomes less important, and given enough data, ridge and linear regression will have the same performance (the fact that this happens here when using the full dataset is just by chance). Another interesting aspect is the decrease in training performance for linear regression. If more data is added, it becomes harder for a model to overfit, or memorize the data
# ### Lasso
# #### An alternative to Ridge for regularizing linear regression is Lasso. As with ridge regression, using the lasso also restricts coefficients to be close to zero, but in a slightly different way, called L1 regularization The consequence of L1 regularization is that when using the lasso, some coefficients are exactly zero. This means some features are entirely ignored by the model. This can be seen as a form of automatic fea‐ ture selection. Having some coefficients be exactly zero often makes a model easier to interpret, and can reveal the most important features of your model
from sklearn.linear_model import Lasso
lasso = Lasso().fit(X_train, y_train)
print("Training set score: {:.2f}".format(lasso.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lasso.score(X_test, y_test)))
print("Number of features used: {}".format(np.sum(lasso.coef_ != 0)))
# #### As you can see, Lasso does quite badly, both on the training and the test set. This indicates that we are underfitting, and we find that it used only 4 of the 105 features. Similarly to Ridge, the Lasso also has a regularization parameter, alpha, that controls how strongly coefficients are pushed toward zero. In the previous example, we used the default of alpha=1.0. To reduce underfitting, let’s try decreasing alpha. When we do this, we also need to increase the default setting of max_iter (the maximum num‐ ber of iterations to run
# we increase the default setting of "max_iter",
# otherwise the model would warn us that we should increase max_iter.
lasso001 = Lasso(alpha=0.01, max_iter=100000).fit(X_train, y_train)
print("Training set score: {:.2f}".format(lasso001.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lasso001.score(X_test, y_test)))
print("Number of features used: {}".format(np.sum(lasso001.coef_ != 0)))
# #### A lower alpha allowed us to fit a more complex model, which worked better on the training and test data. The performance is slightly better than using Ridge, and we are using only 33 of the 105 features. This makes this model potentially easier to understand.
# #### If we set alpha too low, however, we again remove the effect of regularization and end up overfitting, with a result similar to LinearRegression
lasso00001 = Lasso(alpha=0.0001, max_iter=100000).fit(X_train, y_train)
print("Training set score: {:.2f}".format(lasso00001.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lasso00001.score(X_test, y_test)))
print("Number of features used: {}".format(np.sum(lasso00001.coef_ != 0)))
# #### Again, we can plot the coefficients of the different models
plt.plot(lasso.coef_, "s", label="Lasso alpha=1")
plt.plot(lasso001.coef_, "^", label="Lasso alpha=0.01")
plt.plot(lasso00001.coef_, "v", label="Lasso alpha=0.0001")
plt.plot(ridge01.coef_, "o", label="Ridge alpha=0.1")
plt.legend(ncol=2, loc=(0, 1.05))
plt.ylim(-25, 25)
plt.xlabel("Coefficient index")
plt.ylabel("Coefficient magnitude")
| false | 0 | 3,465 | 2 | 3,465 | 3,465 |
||
129089393
|
# Stage 1: Data Preprocessing
import pandas as pd
from statsmodels.tsa.stattools import adfuller
from sklearn.preprocessing import MinMaxScaler
# Define the file path to the dataset
file_path = "/kaggle/input/chargingbehavior/ChargePoint Data CY20Q4.csv"
# Load the dataset into a Pandas DataFrame
df = pd.read_csv(file_path, low_memory=False)
# Convert relevant columns to appropriate datatypes
df["Transaction Date (Pacific Time)"] = pd.to_datetime(
df["Transaction Date (Pacific Time)"], yearfirst=True, errors="coerce"
)
df["Start Date"] = pd.to_datetime(df["Start Date"], yearfirst=True, errors="coerce")
df["End Date"] = pd.to_datetime(df["End Date"], yearfirst=True, errors="coerce")
# Handling missing values
df.dropna(
subset=[
"Transaction Date (Pacific Time)",
"Charging Time (hh:mm:ss)",
"Energy (kWh)",
],
inplace=True,
)
# Additional Data Preprocessing Steps
# Remove unnecessary column 'Start Time Zone'
df.drop("Start Time Zone", axis=1, inplace=True)
df.drop("End Time Zone", axis=1, inplace=True)
# Handling outliers
def handle_outliers(df, columns):
for column in columns:
q1 = df[column].quantile(0.25)
q3 = df[column].quantile(0.75)
iqr = q3 - q1
lower_bound = q1 - 1.5 * iqr
upper_bound = q3 + 1.5 * iqr
df[column] = df[column].apply(
lambda x: upper_bound
if x > upper_bound
else lower_bound
if x < lower_bound
else x
)
return df
# Specify columns to handle outliers
outlier_columns = ["Energy (kWh)", "GHG Savings (kg)", "Gasoline Savings (gallons)"]
# Apply outlier handling
df = handle_outliers(df, outlier_columns)
# Convert 'Energy (kWh)' to a stationary series
adf_result = adfuller(df["Energy (kWh)"])
p_value = adf_result[1]
if p_value > 0.05:
df["Energy (kWh)"] = df["Energy (kWh)"].diff().dropna()
# Scaling
scaler = MinMaxScaler()
columns_to_scale = ["Energy (kWh)", "GHG Savings (kg)", "Gasoline Savings (gallons)"]
df[columns_to_scale] = scaler.fit_transform(df[columns_to_scale])
# Confirm the preprocessing is complete
preprocessed = True
df.columns
# Generate data types of all columns
data_types = df.dtypes
# Print the data types
print(data_types)
# Stage 2: Model Identification
import matplotlib.pyplot as plt
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
# Plot the ACF and PACF of the preprocessed 'Energy (kWh)' series
fig, ax = plt.subplots(figsize=(12, 6))
plot_acf(df["Energy (kWh)"], lags=50, ax=ax)
plt.xlabel("Lag")
plt.ylabel("Autocorrelation")
plt.title("ACF Plot")
plt.show()
fig, ax = plt.subplots(figsize=(12, 6))
plot_pacf(df["Energy (kWh)"], lags=50, ax=ax, method="ywm")
plt.xlabel("Lag")
plt.ylabel("Partial Autocorrelation")
plt.title("PACF Plot")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/089/129089393.ipynb
| null | null |
[{"Id": 129089393, "ScriptId": 38373994, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9516386, "CreationDate": "05/10/2023 23:38:27", "VersionNumber": 1.0, "Title": "Charging Behavior", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 90.0, "LinesInsertedFromPrevious": 90.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# Stage 1: Data Preprocessing
import pandas as pd
from statsmodels.tsa.stattools import adfuller
from sklearn.preprocessing import MinMaxScaler
# Define the file path to the dataset
file_path = "/kaggle/input/chargingbehavior/ChargePoint Data CY20Q4.csv"
# Load the dataset into a Pandas DataFrame
df = pd.read_csv(file_path, low_memory=False)
# Convert relevant columns to appropriate datatypes
df["Transaction Date (Pacific Time)"] = pd.to_datetime(
df["Transaction Date (Pacific Time)"], yearfirst=True, errors="coerce"
)
df["Start Date"] = pd.to_datetime(df["Start Date"], yearfirst=True, errors="coerce")
df["End Date"] = pd.to_datetime(df["End Date"], yearfirst=True, errors="coerce")
# Handling missing values
df.dropna(
subset=[
"Transaction Date (Pacific Time)",
"Charging Time (hh:mm:ss)",
"Energy (kWh)",
],
inplace=True,
)
# Additional Data Preprocessing Steps
# Remove unnecessary column 'Start Time Zone'
df.drop("Start Time Zone", axis=1, inplace=True)
df.drop("End Time Zone", axis=1, inplace=True)
# Handling outliers
def handle_outliers(df, columns):
for column in columns:
q1 = df[column].quantile(0.25)
q3 = df[column].quantile(0.75)
iqr = q3 - q1
lower_bound = q1 - 1.5 * iqr
upper_bound = q3 + 1.5 * iqr
df[column] = df[column].apply(
lambda x: upper_bound
if x > upper_bound
else lower_bound
if x < lower_bound
else x
)
return df
# Specify columns to handle outliers
outlier_columns = ["Energy (kWh)", "GHG Savings (kg)", "Gasoline Savings (gallons)"]
# Apply outlier handling
df = handle_outliers(df, outlier_columns)
# Convert 'Energy (kWh)' to a stationary series
adf_result = adfuller(df["Energy (kWh)"])
p_value = adf_result[1]
if p_value > 0.05:
df["Energy (kWh)"] = df["Energy (kWh)"].diff().dropna()
# Scaling
scaler = MinMaxScaler()
columns_to_scale = ["Energy (kWh)", "GHG Savings (kg)", "Gasoline Savings (gallons)"]
df[columns_to_scale] = scaler.fit_transform(df[columns_to_scale])
# Confirm the preprocessing is complete
preprocessed = True
df.columns
# Generate data types of all columns
data_types = df.dtypes
# Print the data types
print(data_types)
# Stage 2: Model Identification
import matplotlib.pyplot as plt
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
# Plot the ACF and PACF of the preprocessed 'Energy (kWh)' series
fig, ax = plt.subplots(figsize=(12, 6))
plot_acf(df["Energy (kWh)"], lags=50, ax=ax)
plt.xlabel("Lag")
plt.ylabel("Autocorrelation")
plt.title("ACF Plot")
plt.show()
fig, ax = plt.subplots(figsize=(12, 6))
plot_pacf(df["Energy (kWh)"], lags=50, ax=ax, method="ywm")
plt.xlabel("Lag")
plt.ylabel("Partial Autocorrelation")
plt.title("PACF Plot")
plt.show()
| false | 0 | 905 | 0 | 905 | 905 |
||
129089294
|
<jupyter_start><jupyter_text>Anemia Disease
Citation Request: See the articles for more detailed information on the data.
Kilicarslan, S., Celik, M., & Sahin, Ş. (2021). Hybrid models based on genetic algorithm and deep learning algorithms for nutritional Anemia disease classification. Biomedical Signal Processing and Control, 63, 102231.
About Dataset
Data
The anemia dataset used in this study were obtained from the Faculty of Medicine, Tokat Gaziosmanpaşa University, Turkey. The data contains the complete blood count test results of 15,300 patients in the 5-year interval between 2013 and 2018. The dataset of pregnant women, children, and patients with cancer were excluded from the study. The noise in the dataset was eliminated and the parameters, which were considered insignificant in the diagnosis of anemia, were excluded from the dataset with the help of the experts. It is observed that, in the dataset, some of the records have missing parameter values and have values outside the reference range of the parameters which are marked by specialist doctors as noise in our study. Thus, records that have missing data and parameter values outside the reference ranges were removed from the dataset. In the study, Pearson correlation method was used to understand whether there is any relationship between the parameters. It is observed that the relationship between the parameters in the dataset is generally a weak relationship which is below p < 0.4 [59]. Because of this reason none of the parameters excluded from the dataset. Twenty-four features (Table 1) and 5 classes in the dataset were used in the study (Table 2). Since the difference between the parameters in the dataset was very high, a linear transformation was performed on the data with min-max normalization [30]. This dataset consists of data from 15,300 patients, of which 10,379 were female and 4921 were male. The dataset consists of 1019 (7%) patients with HGB-anemia, 4182 (27%) patients with iron deficiency, 199 (1%) patients with B12 deficiency, 153 (1%) patients with folate deficiency, and 9747 (64%) patients who had no anemia (Table 2). The transferring saturation in the dataset was obtained by the "SDTSD" feature, using the Eq. (1), which was developed with the help of a specialist physician. Saturation is the ratio of serum iron to total serum iron. In the Equation SD represents Serum Iron and TSD represents Total Serum Iron. (1)
image.png
Table 1. Anemia Disease Dataset Attributes and Their Descriptions
image.png
In the study, GA-SAE and GA-CNN models were proposed for the classification of HBG-anemia, iron deficiency anemia, B12 deficiency anemia, folate deficiency anemia, and patients without anemia (Table 2). The hyperparameters of the proposed deep leaning algorithms of SAE and CNN are determined by using the global and local search capabilities of the GA.
image.png
Citation Request: See the articles for more detailed information on the data.
Kilicarslan, S., Celik, M., & Sahin, Ş. (2021). Hybrid models based on genetic algorithm and deep learning algorithms for nutritional Anemia disease classification. Biomedical Signal Processing and Control, 63, 102231.
Kaggle dataset identifier: anemia-disease
<jupyter_script>import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
anemifirst = pd.read_csv("anemi.csv")
anemifirst
anemifirst.rename(columns={"All_Class": "TargetClass"}, inplace=True)
anemifirst.drop(
[
"HGB_Anemia_Class",
"Iron_anemia_Class",
"Folate_anemia_class",
"B12_Anemia_class",
],
axis=1,
inplace=True,
)
anemifirst.rename(
columns={"TSD": "SDTSD", "FERRITTE": "FERRITIN", "SDTSD": "TSD"}, inplace=True
)
# z_scores = stats.zscore(anemifirst)
# abs_z_scores = np.abs(z_scores)
# filtered_entries = (abs_z_scores < 3.45).all(axis=1)
# anemi = anemifirst[filtered_entries]
# anemi.reset_index(inplace=True, drop=True)
anemi = anemifirst
dataset_reduced = anemi["FERRITIN"] > 0
dataset_reduced.value_counts()
anemi.info()
anemi.describe()
anemi.sort_values(["FERRITIN"], ascending=False).groupby("FERRITIN").head(10)
y = anemi["TargetClass"]
X = anemi
anemi
X.drop(["TargetClass"], axis=1)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42, shuffle=True
)
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
precision_score,
recall_score,
)
from sklearn.ensemble import VotingClassifier
from sklearn.model_selection import cross_val_score
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn import tree
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
from sklearn.ensemble import ExtraTreesClassifier
def give_scores(y_test, y_pred):
accuracy = accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred, average="weighted")
recall = recall_score(y_test, y_pred, average="weighted")
return accuracy, precision, recall
from sklearn.model_selection import cross_val_score
def model_accuracy(model, X_train=X_train, y_train=y_train):
accuracies = cross_val_score(estimator=model, X=X_train, y=y_train, cv=10)
print("Accuracy: {:.2f} %".format(accuracies.mean() * 100))
print("Standard Deviation: {:.2f} %".format(accuracies.std() * 100))
svc = SVC(probability=True)
knc = KNeighborsClassifier(n_neighbors=5, metric="euclidean")
gnb = GaussianNB()
dtc = DecisionTreeClassifier(max_depth=4)
lrc = LogisticRegression(solver="liblinear", penalty="l1")
rfc = RandomForestClassifier(n_estimators=50, random_state=2)
etc = ExtraTreesClassifier(n_estimators=50, random_state=2)
# ## SVC
svc.fit(X_train, y_train)
y_pred_svc = svc.predict(X_test)
confusion_matrix(y_test, y_pred_svc)
give_scores(y_test, y_pred_svc)
from sklearn.metrics import mean_absolute_error, mean_squared_error
print("\nMAE: {}".format(mean_absolute_error(y_pred_svc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_svc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_svc, y_test))))
# ## KNC
knc.fit(X_train, y_train)
y_pred_knc = knc.predict(X_test)
confusion_matrix(y_test, y_pred_knc)
give_scores(y_test, y_pred_knc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_knc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_knc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_knc, y_test))))
# ## GNB
gnb.fit(X_train, y_train)
y_pred_gnb = gnb.predict(X_test)
confusion_matrix(y_test, y_pred_gnb)
give_scores(y_test, y_pred_gnb)
from sklearn.metrics import mean_absolute_error, mean_squared_error
print("\nMAE: {}".format(mean_absolute_error(y_pred_gnb, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_gnb, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_gnb, y_test))))
# ## DTC
dtc.fit(X_train, y_train)
y_pred_dtc = dtc.predict(X_test)
confusion_matrix(y_test, y_pred_dtc)
give_scores(y_test, y_pred_dtc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_dtc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_dtc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_dtc, y_test))))
# ## LRC
lrc.fit(X_train, y_train)
y_pred_lrc = lrc.predict(X_test)
confusion_matrix(y_test, y_pred_lrc)
give_scores(y_test, y_pred_lrc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_lrc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_lrc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_lrc, y_test))))
# ## RFC
rfc.fit(X_train, y_train)
y_pred_rfc = rfc.predict(X_test)
confusion_matrix(y_test, y_pred_rfc)
give_scores(y_test, y_pred_rfc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_rfc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_rfc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_rfc, y_test))))
# ## ETC
etc.fit(X_train, y_train)
y_pred_etc = etc.predict(X_test)
confusion_matrix(y_test, y_pred_etc)
give_scores(y_test, y_pred_etc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_etc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_etc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_etc, y_test))))
# ## VC
vc = VotingClassifier([["svc", SVC()], ["knc", KNeighborsClassifier()]], voting="hard")
vc.fit(X_train, y_train)
y_pred_vc = vc.predict(X_test)
confusion_matrix(y_test, y_pred_vc)
give_scores(y_test, y_pred_vc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_vc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_vc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_vc, y_test))))
from sklearn.preprocessing import LabelBinarizer
label_binarizer = LabelBinarizer().fit(y_train)
y_onehot_test = label_binarizer.transform(y_test)
y_onehot_test.shape # (n_samples, n_classes)
label_binarizer.transform([0])
y_score_svc = svc.fit(X_train, y_train).predict_proba(X_test)
y_score_knc = knc.fit(X_train, y_train).predict_proba(X_test)
y_score_gnb = gnb.fit(X_train, y_train).predict_proba(X_test)
y_score_dtc = dtc.fit(X_train, y_train).predict_proba(X_test)
y_score_lrc = lrc.fit(X_train, y_train).predict_proba(X_test)
y_score_rfc = rfc.fit(X_train, y_train).predict_proba(X_test)
y_score_etc = etc.fit(X_train, y_train).predict_proba(X_test)
y_score_all = [
y_score_svc,
y_score_knc,
y_score_gnb,
y_score_dtc,
y_score_lrc,
y_score_rfc,
y_score_etc,
]
title_names = [
"One-vs-Rest ROC curves: SVC",
"One-vs-Rest ROC curves: KNC",
"One-vs-Rest ROC curves: GNB",
"One-vs-Rest ROC curves: DTC",
"One-vs-Rest ROC curves: LRC",
"One-vs-Rest ROC curves: RFC",
"One-vs-Rest ROC curves: ETC",
]
anemia_classes = [
"nicht anämisch",
"HGB Anämie",
"Eisenmangelanämie",
"Folatemangelanämie",
"B12-Mangelanämie",
]
colors = ["green", "pink", "blue", "darkorange", "lightblue", "purple", "yellow"]
import matplotlib.pyplot as plt
from sklearn.metrics import RocCurveDisplay
for j in range(0, 7):
for i in range(0, 5):
RocCurveDisplay.from_predictions(
y_onehot_test[:, i],
y_score_all[j][:, i],
name=f"{anemia_classes[i]} gegen den Rest",
color=colors[j],
)
plt.plot([0, 1], [0, 1], "k--", label="Zufallswahrscheinlichkeit (AUC = 0.5)")
plt.axis("square")
plt.xlabel("Falsch Positiv Rate", fontsize="15")
plt.ylabel("Richtig Positive Rate", fontsize="15")
plt.rc("xtick", labelsize="15")
plt.rc("ytick", labelsize="15")
plt.title(title_names[j], color="darkred")
plt.legend(loc=4, fontsize="9.5")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/089/129089294.ipynb
|
anemia-disease
|
serhathoca
|
[{"Id": 129089294, "ScriptId": 36082536, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8579837, "CreationDate": "05/10/2023 23:36:30", "VersionNumber": 1.0, "Title": "Anemia Disease Dataset", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 230.0, "LinesInsertedFromPrevious": 230.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": 230.0, "LinesDeletedFromFork": 23.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 0.0, "TotalVotes": 0}]
|
[{"Id": 184833871, "KernelVersionId": 129089294, "SourceDatasetVersionId": 4342817}]
|
[{"Id": 4342817, "DatasetId": 2556689, "DatasourceVersionId": 4401254, "CreatorUserId": 2694551, "LicenseName": "Database: Open Database, Contents: \u00a9 Original Authors", "CreationDate": "10/17/2022 19:13:53", "VersionNumber": 2.0, "Title": "Anemia Disease", "Slug": "anemia-disease", "Subtitle": NaN, "Description": "Citation Request: See the articles for more detailed information on the data.\n\nKilicarslan, S., Celik, M., & Sahin, \u015e. (2021). Hybrid models based on genetic algorithm and deep learning algorithms for nutritional Anemia disease classification. Biomedical Signal Processing and Control, 63, 102231.\n\nAbout Dataset\n\nData\n\nThe anemia dataset used in this study were obtained from the Faculty of Medicine, Tokat Gaziosmanpa\u015fa University, Turkey. The data contains the complete blood count test results of 15,300 patients in the 5-year interval between 2013 and 2018. The dataset of pregnant women, children, and patients with cancer were excluded from the study. The noise in the dataset was eliminated and the parameters, which were considered insignificant in the diagnosis of anemia, were excluded from the dataset with the help of the experts. It is observed that, in the dataset, some of the records have missing parameter values and have values outside the reference range of the parameters which are marked by specialist doctors as noise in our study. Thus, records that have missing data and parameter values outside the reference ranges were removed from the dataset. In the study, Pearson correlation method was used to understand whether there is any relationship between the parameters. It is observed that the relationship between the parameters in the dataset is generally a weak relationship which is below p\u202f<\u202f0.4 [59]. Because of this reason none of the parameters excluded from the dataset. Twenty-four features (Table 1) and 5 classes in the dataset were used in the study (Table 2). Since the difference between the parameters in the dataset was very high, a linear transformation was performed on the data with min-max normalization [30]. This dataset consists of data from 15,300 patients, of which 10,379 were female and 4921 were male. The dataset consists of 1019 (7%) patients with HGB-anemia, 4182 (27%) patients with iron deficiency, 199 (1%) patients with B12 deficiency, 153 (1%) patients with folate deficiency, and 9747 (64%) patients who had no anemia (Table 2). The transferring saturation in the dataset was obtained by the \"SDTSD\" feature, using the Eq. (1), which was developed with the help of a specialist physician. Saturation is the ratio of serum iron to total serum iron. In the Equation SD represents Serum Iron and TSD represents Total Serum Iron. (1)\n\nimage.png\n\nTable 1. Anemia Disease Dataset Attributes and Their Descriptions\n\nimage.png\n\nIn the study, GA-SAE and GA-CNN models were proposed for the classification of HBG-anemia, iron deficiency anemia, B12 deficiency anemia, folate deficiency anemia, and patients without anemia (Table 2). The hyperparameters of the proposed deep leaning algorithms of SAE and CNN are determined by using the global and local search capabilities of the GA.\n\nimage.png\n\nCitation Request: See the articles for more detailed information on the data.\n\nKilicarslan, S., Celik, M., & Sahin, \u015e. (2021). Hybrid models based on genetic algorithm and deep learning algorithms for nutritional Anemia disease classification. Biomedical Signal Processing and Control, 63, 102231.", "VersionNotes": "Data Update 2022/10/17", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2556689, "CreatorUserId": 2694551, "OwnerUserId": 2694551.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4342817.0, "CurrentDatasourceVersionId": 4401254.0, "ForumId": 2585904, "Type": 2, "CreationDate": "10/17/2022 19:09:56", "LastActivityDate": "10/17/2022", "TotalViews": 1536, "TotalDownloads": 278, "TotalVotes": 7, "TotalKernels": 1}]
|
[{"Id": 2694551, "UserName": "serhathoca", "DisplayName": "Serhat KILI\u00c7ARSLAN", "RegisterDate": "01/11/2019", "PerformanceTier": 0}]
|
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
anemifirst = pd.read_csv("anemi.csv")
anemifirst
anemifirst.rename(columns={"All_Class": "TargetClass"}, inplace=True)
anemifirst.drop(
[
"HGB_Anemia_Class",
"Iron_anemia_Class",
"Folate_anemia_class",
"B12_Anemia_class",
],
axis=1,
inplace=True,
)
anemifirst.rename(
columns={"TSD": "SDTSD", "FERRITTE": "FERRITIN", "SDTSD": "TSD"}, inplace=True
)
# z_scores = stats.zscore(anemifirst)
# abs_z_scores = np.abs(z_scores)
# filtered_entries = (abs_z_scores < 3.45).all(axis=1)
# anemi = anemifirst[filtered_entries]
# anemi.reset_index(inplace=True, drop=True)
anemi = anemifirst
dataset_reduced = anemi["FERRITIN"] > 0
dataset_reduced.value_counts()
anemi.info()
anemi.describe()
anemi.sort_values(["FERRITIN"], ascending=False).groupby("FERRITIN").head(10)
y = anemi["TargetClass"]
X = anemi
anemi
X.drop(["TargetClass"], axis=1)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42, shuffle=True
)
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
precision_score,
recall_score,
)
from sklearn.ensemble import VotingClassifier
from sklearn.model_selection import cross_val_score
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn import tree
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
from sklearn.ensemble import ExtraTreesClassifier
def give_scores(y_test, y_pred):
accuracy = accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred, average="weighted")
recall = recall_score(y_test, y_pred, average="weighted")
return accuracy, precision, recall
from sklearn.model_selection import cross_val_score
def model_accuracy(model, X_train=X_train, y_train=y_train):
accuracies = cross_val_score(estimator=model, X=X_train, y=y_train, cv=10)
print("Accuracy: {:.2f} %".format(accuracies.mean() * 100))
print("Standard Deviation: {:.2f} %".format(accuracies.std() * 100))
svc = SVC(probability=True)
knc = KNeighborsClassifier(n_neighbors=5, metric="euclidean")
gnb = GaussianNB()
dtc = DecisionTreeClassifier(max_depth=4)
lrc = LogisticRegression(solver="liblinear", penalty="l1")
rfc = RandomForestClassifier(n_estimators=50, random_state=2)
etc = ExtraTreesClassifier(n_estimators=50, random_state=2)
# ## SVC
svc.fit(X_train, y_train)
y_pred_svc = svc.predict(X_test)
confusion_matrix(y_test, y_pred_svc)
give_scores(y_test, y_pred_svc)
from sklearn.metrics import mean_absolute_error, mean_squared_error
print("\nMAE: {}".format(mean_absolute_error(y_pred_svc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_svc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_svc, y_test))))
# ## KNC
knc.fit(X_train, y_train)
y_pred_knc = knc.predict(X_test)
confusion_matrix(y_test, y_pred_knc)
give_scores(y_test, y_pred_knc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_knc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_knc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_knc, y_test))))
# ## GNB
gnb.fit(X_train, y_train)
y_pred_gnb = gnb.predict(X_test)
confusion_matrix(y_test, y_pred_gnb)
give_scores(y_test, y_pred_gnb)
from sklearn.metrics import mean_absolute_error, mean_squared_error
print("\nMAE: {}".format(mean_absolute_error(y_pred_gnb, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_gnb, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_gnb, y_test))))
# ## DTC
dtc.fit(X_train, y_train)
y_pred_dtc = dtc.predict(X_test)
confusion_matrix(y_test, y_pred_dtc)
give_scores(y_test, y_pred_dtc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_dtc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_dtc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_dtc, y_test))))
# ## LRC
lrc.fit(X_train, y_train)
y_pred_lrc = lrc.predict(X_test)
confusion_matrix(y_test, y_pred_lrc)
give_scores(y_test, y_pred_lrc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_lrc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_lrc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_lrc, y_test))))
# ## RFC
rfc.fit(X_train, y_train)
y_pred_rfc = rfc.predict(X_test)
confusion_matrix(y_test, y_pred_rfc)
give_scores(y_test, y_pred_rfc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_rfc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_rfc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_rfc, y_test))))
# ## ETC
etc.fit(X_train, y_train)
y_pred_etc = etc.predict(X_test)
confusion_matrix(y_test, y_pred_etc)
give_scores(y_test, y_pred_etc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_etc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_etc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_etc, y_test))))
# ## VC
vc = VotingClassifier([["svc", SVC()], ["knc", KNeighborsClassifier()]], voting="hard")
vc.fit(X_train, y_train)
y_pred_vc = vc.predict(X_test)
confusion_matrix(y_test, y_pred_vc)
give_scores(y_test, y_pred_vc)
print("\nMAE: {}".format(mean_absolute_error(y_pred_vc, y_test)))
print("MSE: {}".format(mean_squared_error(y_pred_vc, y_test)))
print("RMSE: {}".format(np.sqrt(mean_squared_error(y_pred_vc, y_test))))
from sklearn.preprocessing import LabelBinarizer
label_binarizer = LabelBinarizer().fit(y_train)
y_onehot_test = label_binarizer.transform(y_test)
y_onehot_test.shape # (n_samples, n_classes)
label_binarizer.transform([0])
y_score_svc = svc.fit(X_train, y_train).predict_proba(X_test)
y_score_knc = knc.fit(X_train, y_train).predict_proba(X_test)
y_score_gnb = gnb.fit(X_train, y_train).predict_proba(X_test)
y_score_dtc = dtc.fit(X_train, y_train).predict_proba(X_test)
y_score_lrc = lrc.fit(X_train, y_train).predict_proba(X_test)
y_score_rfc = rfc.fit(X_train, y_train).predict_proba(X_test)
y_score_etc = etc.fit(X_train, y_train).predict_proba(X_test)
y_score_all = [
y_score_svc,
y_score_knc,
y_score_gnb,
y_score_dtc,
y_score_lrc,
y_score_rfc,
y_score_etc,
]
title_names = [
"One-vs-Rest ROC curves: SVC",
"One-vs-Rest ROC curves: KNC",
"One-vs-Rest ROC curves: GNB",
"One-vs-Rest ROC curves: DTC",
"One-vs-Rest ROC curves: LRC",
"One-vs-Rest ROC curves: RFC",
"One-vs-Rest ROC curves: ETC",
]
anemia_classes = [
"nicht anämisch",
"HGB Anämie",
"Eisenmangelanämie",
"Folatemangelanämie",
"B12-Mangelanämie",
]
colors = ["green", "pink", "blue", "darkorange", "lightblue", "purple", "yellow"]
import matplotlib.pyplot as plt
from sklearn.metrics import RocCurveDisplay
for j in range(0, 7):
for i in range(0, 5):
RocCurveDisplay.from_predictions(
y_onehot_test[:, i],
y_score_all[j][:, i],
name=f"{anemia_classes[i]} gegen den Rest",
color=colors[j],
)
plt.plot([0, 1], [0, 1], "k--", label="Zufallswahrscheinlichkeit (AUC = 0.5)")
plt.axis("square")
plt.xlabel("Falsch Positiv Rate", fontsize="15")
plt.ylabel("Richtig Positive Rate", fontsize="15")
plt.rc("xtick", labelsize="15")
plt.rc("ytick", labelsize="15")
plt.title(title_names[j], color="darkred")
plt.legend(loc=4, fontsize="9.5")
plt.show()
| false | 0 | 2,914 | 0 | 3,772 | 2,914 |
||
129089934
|
<jupyter_start><jupyter_text>1000_companies_profit
The dataset includes sample data of 1000 startup companies operating cost and their profit. Well-formatted dataset for building ML regression pipelines.
**Includes**
R&D Spend float64
Administration float64
Marketing Spend float64
State object
Profit float64
Kaggle dataset identifier: 1000-companies-profit
<jupyter_code>import pandas as pd
df = pd.read_csv('1000-companies-profit/1000_Companies.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 1000 entries, 0 to 999
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 R&D Spend 1000 non-null float64
1 Administration 1000 non-null float64
2 Marketing Spend 1000 non-null float64
3 State 1000 non-null object
4 Profit 1000 non-null float64
dtypes: float64(4), object(1)
memory usage: 39.2+ KB
<jupyter_text>Examples:
{
"R&D Spend": 165349.2,
"Administration": 136897.8,
"Marketing Spend": 471784.1,
"State": "New York",
"Profit": 192261.83
}
{
"R&D Spend": 162597.7,
"Administration": 151377.59,
"Marketing Spend": 443898.53,
"State": "California",
"Profit": 191792.06
}
{
"R&D Spend": 153441.51,
"Administration": 101145.55,
"Marketing Spend": 407934.54,
"State": "Florida",
"Profit": 191050.39
}
{
"R&D Spend": 144372.41,
"Administration": 118671.85,
"Marketing Spend": 383199.62,
"State": "New York",
"Profit": 182901.99
}
<jupyter_script># # Predicting Profit using Multiple Linear Regression Model based on R&D Spend, Administration, and Marketing Spend
# The model I have created uses Linear Regression to predict the profit of a company based on its investment in Research and Development (R&D), Administration, and Marketing Spend. The dataset used to train the model contains information on these three variables and the corresponding profits earned by various companies.
# By analyzing the data, the model has learned to identify the relationships between the input variables and the target variable (profit), and can use this knowledge to make predictions on new data. The model can be used to help businesses make informed decisions about their investments by providing a reliable estimate of the expected
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn
# ## Loading Data
df = pd.read_csv("/kaggle/input/1000-companies-profit/1000_Companies.csv")
df.shape
df.sample(10)
df.isnull().sum()
df.corr()
plt.scatter(df["R&D Spend"], df["Profit"])
plt.xlabel("R&D Spend")
plt.ylabel("Profit")
plt.scatter(df["Administration"], df["Profit"])
plt.xlabel("Administration")
plt.ylabel("Profit")
plt.scatter(df["Marketing Spend"], df["Profit"])
plt.xlabel("Marketing Spend")
plt.ylabel("Profit")
# ## Spliting Dataset
from sklearn.model_selection import train_test_split
X, y = df[["R&D Spend", "Administration", "Marketing Spend"]], df["Profit"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.1, random_state=42
)
# ## Training Dataset using Linear Regression
from sklearn.linear_model import LinearRegression
clf = LinearRegression()
clf.fit(X_train, y_train)
# ## Predicting Dataset
clf.predict([[78013.11, 121597.5500, 264346.0600]])
clf.predict(X_test)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/089/129089934.ipynb
|
1000-companies-profit
|
rupakroy
|
[{"Id": 129089934, "ScriptId": 38370784, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14110262, "CreationDate": "05/10/2023 23:49:55", "VersionNumber": 1.0, "Title": "Linear Regression Multiple Variables", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 61.0, "LinesInsertedFromPrevious": 61.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184835912, "KernelVersionId": 129089934, "SourceDatasetVersionId": 3105372}]
|
[{"Id": 3105372, "DatasetId": 1896237, "DatasourceVersionId": 3154274, "CreatorUserId": 3072182, "LicenseName": "CC0: Public Domain", "CreationDate": "01/28/2022 10:49:42", "VersionNumber": 1.0, "Title": "1000_companies_profit", "Slug": "1000-companies-profit", "Subtitle": "1000 Companies operating cost sample data list for building regression usecases", "Description": "The dataset includes sample data of 1000 startup companies operating cost and their profit. Well-formatted dataset for building ML regression pipelines.\n**Includes**\nR&D Spend float64\nAdministration float64\nMarketing Spend float64\nState object\nProfit float64", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1896237, "CreatorUserId": 3072182, "OwnerUserId": 3072182.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3105372.0, "CurrentDatasourceVersionId": 3154274.0, "ForumId": 1919554, "Type": 2, "CreationDate": "01/28/2022 10:49:42", "LastActivityDate": "01/28/2022", "TotalViews": 3171, "TotalDownloads": 826, "TotalVotes": 10, "TotalKernels": 10}]
|
[{"Id": 3072182, "UserName": "rupakroy", "DisplayName": "Rupak Roy/ Bob", "RegisterDate": "04/11/2019", "PerformanceTier": 2}]
|
# # Predicting Profit using Multiple Linear Regression Model based on R&D Spend, Administration, and Marketing Spend
# The model I have created uses Linear Regression to predict the profit of a company based on its investment in Research and Development (R&D), Administration, and Marketing Spend. The dataset used to train the model contains information on these three variables and the corresponding profits earned by various companies.
# By analyzing the data, the model has learned to identify the relationships between the input variables and the target variable (profit), and can use this knowledge to make predictions on new data. The model can be used to help businesses make informed decisions about their investments by providing a reliable estimate of the expected
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn
# ## Loading Data
df = pd.read_csv("/kaggle/input/1000-companies-profit/1000_Companies.csv")
df.shape
df.sample(10)
df.isnull().sum()
df.corr()
plt.scatter(df["R&D Spend"], df["Profit"])
plt.xlabel("R&D Spend")
plt.ylabel("Profit")
plt.scatter(df["Administration"], df["Profit"])
plt.xlabel("Administration")
plt.ylabel("Profit")
plt.scatter(df["Marketing Spend"], df["Profit"])
plt.xlabel("Marketing Spend")
plt.ylabel("Profit")
# ## Spliting Dataset
from sklearn.model_selection import train_test_split
X, y = df[["R&D Spend", "Administration", "Marketing Spend"]], df["Profit"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.1, random_state=42
)
# ## Training Dataset using Linear Regression
from sklearn.linear_model import LinearRegression
clf = LinearRegression()
clf.fit(X_train, y_train)
# ## Predicting Dataset
clf.predict([[78013.11, 121597.5500, 264346.0600]])
clf.predict(X_test)
|
[{"1000-companies-profit/1000_Companies.csv": {"column_names": "[\"R&D Spend\", \"Administration\", \"Marketing Spend\", \"State\", \"Profit\"]", "column_data_types": "{\"R&D Spend\": \"float64\", \"Administration\": \"float64\", \"Marketing Spend\": \"float64\", \"State\": \"object\", \"Profit\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1000 entries, 0 to 999\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 R&D Spend 1000 non-null float64\n 1 Administration 1000 non-null float64\n 2 Marketing Spend 1000 non-null float64\n 3 State 1000 non-null object \n 4 Profit 1000 non-null float64\ndtypes: float64(4), object(1)\nmemory usage: 39.2+ KB\n", "summary": "{\"R&D Spend\": {\"count\": 1000.0, \"mean\": 81668.9272, \"std\": 46537.56789148918, \"min\": 0.0, \"25%\": 43084.5, \"50%\": 79936.0, \"75%\": 124565.5, \"max\": 165349.2}, \"Administration\": {\"count\": 1000.0, \"mean\": 122963.8976117, \"std\": 12613.927534630991, \"min\": 51283.14, \"25%\": 116640.68485, \"50%\": 122421.61215, \"75%\": 129139.118, \"max\": 321652.14}, \"Marketing Spend\": {\"count\": 1000.0, \"mean\": 226205.05841882998, \"std\": 91578.39354210424, \"min\": 0.0, \"25%\": 150969.5846, \"50%\": 224517.88735, \"75%\": 308189.808525, \"max\": 471784.1}, \"Profit\": {\"count\": 1000.0, \"mean\": 119546.16465561, \"std\": 42888.63384847688, \"min\": 14681.4, \"25%\": 85943.1985425, \"50%\": 117641.4663, \"75%\": 155577.107425, \"max\": 476485.43}}", "examples": "{\"R&D Spend\":{\"0\":165349.2,\"1\":162597.7,\"2\":153441.51,\"3\":144372.41},\"Administration\":{\"0\":136897.8,\"1\":151377.59,\"2\":101145.55,\"3\":118671.85},\"Marketing Spend\":{\"0\":471784.1,\"1\":443898.53,\"2\":407934.54,\"3\":383199.62},\"State\":{\"0\":\"New York\",\"1\":\"California\",\"2\":\"Florida\",\"3\":\"New York\"},\"Profit\":{\"0\":192261.83,\"1\":191792.06,\"2\":191050.39,\"3\":182901.99}}"}}]
| true | 1 |
<start_data_description><data_path>1000-companies-profit/1000_Companies.csv:
<column_names>
['R&D Spend', 'Administration', 'Marketing Spend', 'State', 'Profit']
<column_types>
{'R&D Spend': 'float64', 'Administration': 'float64', 'Marketing Spend': 'float64', 'State': 'object', 'Profit': 'float64'}
<dataframe_Summary>
{'R&D Spend': {'count': 1000.0, 'mean': 81668.9272, 'std': 46537.56789148918, 'min': 0.0, '25%': 43084.5, '50%': 79936.0, '75%': 124565.5, 'max': 165349.2}, 'Administration': {'count': 1000.0, 'mean': 122963.8976117, 'std': 12613.927534630991, 'min': 51283.14, '25%': 116640.68485, '50%': 122421.61215, '75%': 129139.118, 'max': 321652.14}, 'Marketing Spend': {'count': 1000.0, 'mean': 226205.05841882998, 'std': 91578.39354210424, 'min': 0.0, '25%': 150969.5846, '50%': 224517.88735, '75%': 308189.808525, 'max': 471784.1}, 'Profit': {'count': 1000.0, 'mean': 119546.16465561, 'std': 42888.63384847688, 'min': 14681.4, '25%': 85943.1985425, '50%': 117641.4663, '75%': 155577.107425, 'max': 476485.43}}
<dataframe_info>
RangeIndex: 1000 entries, 0 to 999
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 R&D Spend 1000 non-null float64
1 Administration 1000 non-null float64
2 Marketing Spend 1000 non-null float64
3 State 1000 non-null object
4 Profit 1000 non-null float64
dtypes: float64(4), object(1)
memory usage: 39.2+ KB
<some_examples>
{'R&D Spend': {'0': 165349.2, '1': 162597.7, '2': 153441.51, '3': 144372.41}, 'Administration': {'0': 136897.8, '1': 151377.59, '2': 101145.55, '3': 118671.85}, 'Marketing Spend': {'0': 471784.1, '1': 443898.53, '2': 407934.54, '3': 383199.62}, 'State': {'0': 'New York', '1': 'California', '2': 'Florida', '3': 'New York'}, 'Profit': {'0': 192261.83, '1': 191792.06, '2': 191050.39, '3': 182901.99}}
<end_description>
| 526 | 0 | 1,148 | 526 |
129184384
|
<jupyter_start><jupyter_text>Students Exam Scores: Extended Dataset
This dataset includes scores from three test scores of students at a (fictional) public school and a variety of personal and socio-economic factors that may have interaction effects upon them.
**Remark/warning/disclaimer:**
- This datasets are **fictional** and should be used for **educational purposes only**.
- The original dataset generator creator is Mr. [Royce Kimmons](http://roycekimmons.com/tools/generated_data/exams)
- There are *similar datasets* on kaggle already but this one is **different** and **arguably better** in two ways.
-> 1) has **more data** (**>30k** instead of just the 1k the other datasets have),
-> 2) has extended datasets with **more features** (15 instead of 9) and has **missing values** which makes it ideal for data cleaning and data preprocessing.
### Data Dictionary (column description)
1. **Gender**: Gender of the student (male/female)
2. **EthnicGroup**: Ethnic group of the student (group A to E)
3. **ParentEduc**: Parent(s) education background (from some_highschool to master's degree)
4. **LunchType**: School lunch type (standard or free/reduced)
5. **TestPrep**: Test preparation course followed (completed or none)
6. **ParentMaritalStatus**: Parent(s) marital status (married/single/widowed/divorced)
7. **PracticeSport**: How often the student parctice sport (never/sometimes/regularly))
8. **IsFirstChild**: If the child is first child in the family or not (yes/no)
9. **NrSiblings**: Number of siblings the student has (0 to 7)
10. **TransportMeans**: Means of transport to school (schoolbus/private)
11. **WklyStudyHours**: Weekly self-study hours(less that 5hrs; between 5 and 10hrs; more than 10hrs)
12. **MathScore**: math test score(0-100)
13. **ReadingScore**: reading test score(0-100)
14. **WritingScore**: writing test score(0-100)
### Analytics questions:
1. What factors (features) affect test scores most?
2. Are there interacting features which affect test scores?
Kaggle dataset identifier: students-exam-scores
<jupyter_script>import pyspark
from pyspark.sql import SparkSession
from pyspark.sql.functions import *
from pyspark.sql.window import Window
spark = SparkSession.builder.appName("Score").getOrCreate()
df = spark.read.csv(
"/kaggle/input/students-exam-scores/Original_data_with_more_rows.csv", header=True
)
df = df.withColumnRenamed("_c0", "Id")
df = df.withColumn(
"total_score", col("MathScore") + col("ReadingScore") + col("WritingScore")
)
df = df.withColumn("Max_Marks", lit(300))
df = df.withColumn("Percentage", round(col("total_score") / 300 * 100, 2))
df = df.withColumn("Status", when(col("total_score") > 99, "Passed").otherwise("Fail"))
df.createOrReplaceTempView("students_score")
df.show()
group = spark.sql("select distinct Ethnicgroup from students_score")
group.show()
parents_ed = spark.sql("select distinct ParentEduc from students_score")
parents_ed.show()
vari = (
df.groupBy("Ethnicgroup", "Status", "ParentEduc")
.agg(round(avg("Percentage"), 2).alias("avg_Percentage"))
.orderBy("Ethnicgroup", "Status", "ParentEduc")
)
vari.show()
df_1 = spark.read.csv(
"/kaggle/input/students-exam-scores/Expanded_data_with_more_features.csv",
header=True,
)
df_1 = df_1.withColumnRenamed("_c0", "Id")
# df_1.show()
column_names = df_1.columns
print(column_names)
df_1.select(
"Id",
"Gender",
"ParentMaritalStatus",
"PracticeSport",
"IsFirstChild",
"NrSiblings",
"TransportMeans",
"WklyStudyHours",
"MathScore",
"ReadingScore",
"WritingScore",
).show()
df_1 = df_1.withColumn(
"total_score", col("MathScore") + col("ReadingScore") + col("WritingScore")
)
df_1 = df_1.withColumn("Max_Marks", lit(300))
df_1 = df_1.withColumn("Percentage", round(col("total_score") / 300 * 100, 2))
df_1 = df_1.withColumn(
"Status", when(col("total_score") > 99, "Passed").otherwise("Fail")
)
df_1.createOrReplaceTempView("students_expand")
df_1.show(5)
weekly = spark.sql(
"select WklyStudyHours,EthnicGroup, Status,round(avg(Percentage),2)as Percentage from students_expand group by WklyStudyHours ,EthnicGroup ,Status order by Status,EthnicGroup "
)
weekly.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/184/129184384.ipynb
|
students-exam-scores
|
desalegngeb
|
[{"Id": 129184384, "ScriptId": 37820261, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8866968, "CreationDate": "05/11/2023 16:03:25", "VersionNumber": 4.0, "Title": "scorecard", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 69.0, "LinesInsertedFromPrevious": 24.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 45.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 185006485, "KernelVersionId": 129184384, "SourceDatasetVersionId": 5399169}]
|
[{"Id": 5399169, "DatasetId": 3128523, "DatasourceVersionId": 5472937, "CreatorUserId": 5430373, "LicenseName": "Other (specified in description)", "CreationDate": "04/14/2023 00:15:38", "VersionNumber": 2.0, "Title": "Students Exam Scores: Extended Dataset", "Slug": "students-exam-scores", "Subtitle": "Exam scores for students at a public school", "Description": "This dataset includes scores from three test scores of students at a (fictional) public school and a variety of personal and socio-economic factors that may have interaction effects upon them. \n\n**Remark/warning/disclaimer:** \n- This datasets are **fictional** and should be used for **educational purposes only**. \n- The original dataset generator creator is Mr. [Royce Kimmons](http://roycekimmons.com/tools/generated_data/exams)\n- There are *similar datasets* on kaggle already but this one is **different** and **arguably better** in two ways. \n -> 1) has **more data** (**>30k** instead of just the 1k the other datasets have),\n -> 2) has extended datasets with **more features** (15 instead of 9) and has **missing values** which makes it ideal for data cleaning and data preprocessing.\n\n### Data Dictionary (column description)\n\n1. **Gender**: Gender of the student (male/female)\n2. **EthnicGroup**: Ethnic group of the student (group A to E)\n3. **ParentEduc**: Parent(s) education background (from some_highschool to master's degree)\n4. **LunchType**: School lunch type (standard or free/reduced)\n5. **TestPrep**: Test preparation course followed (completed or none)\n6. **ParentMaritalStatus**: Parent(s) marital status (married/single/widowed/divorced)\n7. **PracticeSport**: How often the student parctice sport (never/sometimes/regularly))\n8. **IsFirstChild**: If the child is first child in the family or not (yes/no)\n9. **NrSiblings**: Number of siblings the student has (0 to 7)\n10. **TransportMeans**: Means of transport to school (schoolbus/private)\n11. **WklyStudyHours**: Weekly self-study hours(less that 5hrs; between 5 and 10hrs; more than 10hrs)\n12. **MathScore**: math test score(0-100)\n13. **ReadingScore**: reading test score(0-100)\n14. **WritingScore**: writing test score(0-100)\n\n### Analytics questions:\n\n1. What factors (features) affect test scores most?\n2. Are there interacting features which affect test scores?", "VersionNotes": "Data Update 2023-04-14", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3128523, "CreatorUserId": 5430373, "OwnerUserId": 5430373.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5399169.0, "CurrentDatasourceVersionId": 5472937.0, "ForumId": 3192141, "Type": 2, "CreationDate": "04/13/2023 21:52:39", "LastActivityDate": "04/13/2023", "TotalViews": 75452, "TotalDownloads": 15444, "TotalVotes": 282, "TotalKernels": 38}]
|
[{"Id": 5430373, "UserName": "desalegngeb", "DisplayName": "des.", "RegisterDate": "07/07/2020", "PerformanceTier": 3}]
|
import pyspark
from pyspark.sql import SparkSession
from pyspark.sql.functions import *
from pyspark.sql.window import Window
spark = SparkSession.builder.appName("Score").getOrCreate()
df = spark.read.csv(
"/kaggle/input/students-exam-scores/Original_data_with_more_rows.csv", header=True
)
df = df.withColumnRenamed("_c0", "Id")
df = df.withColumn(
"total_score", col("MathScore") + col("ReadingScore") + col("WritingScore")
)
df = df.withColumn("Max_Marks", lit(300))
df = df.withColumn("Percentage", round(col("total_score") / 300 * 100, 2))
df = df.withColumn("Status", when(col("total_score") > 99, "Passed").otherwise("Fail"))
df.createOrReplaceTempView("students_score")
df.show()
group = spark.sql("select distinct Ethnicgroup from students_score")
group.show()
parents_ed = spark.sql("select distinct ParentEduc from students_score")
parents_ed.show()
vari = (
df.groupBy("Ethnicgroup", "Status", "ParentEduc")
.agg(round(avg("Percentage"), 2).alias("avg_Percentage"))
.orderBy("Ethnicgroup", "Status", "ParentEduc")
)
vari.show()
df_1 = spark.read.csv(
"/kaggle/input/students-exam-scores/Expanded_data_with_more_features.csv",
header=True,
)
df_1 = df_1.withColumnRenamed("_c0", "Id")
# df_1.show()
column_names = df_1.columns
print(column_names)
df_1.select(
"Id",
"Gender",
"ParentMaritalStatus",
"PracticeSport",
"IsFirstChild",
"NrSiblings",
"TransportMeans",
"WklyStudyHours",
"MathScore",
"ReadingScore",
"WritingScore",
).show()
df_1 = df_1.withColumn(
"total_score", col("MathScore") + col("ReadingScore") + col("WritingScore")
)
df_1 = df_1.withColumn("Max_Marks", lit(300))
df_1 = df_1.withColumn("Percentage", round(col("total_score") / 300 * 100, 2))
df_1 = df_1.withColumn(
"Status", when(col("total_score") > 99, "Passed").otherwise("Fail")
)
df_1.createOrReplaceTempView("students_expand")
df_1.show(5)
weekly = spark.sql(
"select WklyStudyHours,EthnicGroup, Status,round(avg(Percentage),2)as Percentage from students_expand group by WklyStudyHours ,EthnicGroup ,Status order by Status,EthnicGroup "
)
weekly.show()
| false | 0 | 729 | 1 | 1,321 | 729 |
||
129010847
|
#
# Note: This is very rudimentary Instruction model, trained using Databricks 15K dataset on Bloom 500M params
#
#
# Below results are after training with Instructions dataset
#
import torch
import transformers
from transformers import AutoTokenizer, AutoModelForCausalLM
MIN_TRANSFORMERS_VERSION = "4.25.1"
# check transformers version
assert (
transformers.__version__ >= MIN_TRANSFORMERS_VERSION
), f"Please upgrade transformers to version {MIN_TRANSFORMERS_VERSION} or higher."
# init
tokenizer = AutoTokenizer.from_pretrained("bigscience/bloom-560m")
model = AutoModelForCausalLM.from_pretrained(
"sai1881/bloom-560m-finetuned-Instruct-DB-v", torch_dtype=torch.float16
)
model = model.to("cuda:0")
# infer
inputs = tokenizer(prompt, return_tensors="pt").to(model.device)
input_length = inputs.input_ids.shape[1]
outputs = model.generate(
**inputs,
max_new_tokens=128,
do_sample=True,
temperature=0.7,
top_p=0.7,
top_k=50,
return_dict_in_generate=True,
)
token = outputs.sequences[0, input_length:]
output_str = tokenizer.decode(token)
print(output_str.split("<By Manoj>")[0])
prompt = "Instruction: Why can camels survive for long without water? <END> \ncontext: <END> \n \nresponse: "
inputs = tokenizer(prompt, return_tensors="pt").to(model.device)
input_length = inputs.input_ids.shape[1]
outputs = model.generate(
**inputs,
max_new_tokens=128,
do_sample=True,
temperature=0.7,
top_p=0.7,
top_k=50,
return_dict_in_generate=True,
)
token = outputs.sequences[0, input_length:]
output_str = tokenizer.decode(token)
print(output_str.split("<By Manoj>")[0])
#
# Below results are before training with Instructions dataset
#
# init
tokenizer = AutoTokenizer.from_pretrained("bigscience/bloom-560m")
model = AutoModelForCausalLM.from_pretrained(
"bigscience/bloom-560m", torch_dtype=torch.float16
)
model = model.to("cuda:0")
# infer
inputs = tokenizer(prompt, return_tensors="pt").to(model.device)
input_length = inputs.input_ids.shape[1]
outputs = model.generate(
**inputs,
max_new_tokens=128,
do_sample=True,
temperature=0.7,
top_p=0.7,
top_k=50,
return_dict_in_generate=True,
)
token = outputs.sequences[0, input_length:]
output_str = tokenizer.decode(token)
print(output_str.split("<By Manoj>")[0])
prompt = "Instruction: Why can camels survive for long without water? <END> \ncontext: <END> \n \nresponse: "
inputs = tokenizer(prompt, return_tensors="pt").to(model.device)
input_length = inputs.input_ids.shape[1]
outputs = model.generate(
**inputs,
max_new_tokens=128,
do_sample=True,
temperature=0.7,
top_p=0.7,
top_k=50,
return_dict_in_generate=True,
)
token = outputs.sequences[0, input_length:]
output_str = tokenizer.decode(token)
print(output_str.split("<By Manoj>")[0])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/010/129010847.ipynb
| null | null |
[{"Id": 129010847, "ScriptId": 38312970, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2445878, "CreationDate": "05/10/2023 09:44:59", "VersionNumber": 1.0, "Title": "Large Language Model Custom Model", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 77.0, "LinesInsertedFromPrevious": 77.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
#
# Note: This is very rudimentary Instruction model, trained using Databricks 15K dataset on Bloom 500M params
#
#
# Below results are after training with Instructions dataset
#
import torch
import transformers
from transformers import AutoTokenizer, AutoModelForCausalLM
MIN_TRANSFORMERS_VERSION = "4.25.1"
# check transformers version
assert (
transformers.__version__ >= MIN_TRANSFORMERS_VERSION
), f"Please upgrade transformers to version {MIN_TRANSFORMERS_VERSION} or higher."
# init
tokenizer = AutoTokenizer.from_pretrained("bigscience/bloom-560m")
model = AutoModelForCausalLM.from_pretrained(
"sai1881/bloom-560m-finetuned-Instruct-DB-v", torch_dtype=torch.float16
)
model = model.to("cuda:0")
# infer
inputs = tokenizer(prompt, return_tensors="pt").to(model.device)
input_length = inputs.input_ids.shape[1]
outputs = model.generate(
**inputs,
max_new_tokens=128,
do_sample=True,
temperature=0.7,
top_p=0.7,
top_k=50,
return_dict_in_generate=True,
)
token = outputs.sequences[0, input_length:]
output_str = tokenizer.decode(token)
print(output_str.split("<By Manoj>")[0])
prompt = "Instruction: Why can camels survive for long without water? <END> \ncontext: <END> \n \nresponse: "
inputs = tokenizer(prompt, return_tensors="pt").to(model.device)
input_length = inputs.input_ids.shape[1]
outputs = model.generate(
**inputs,
max_new_tokens=128,
do_sample=True,
temperature=0.7,
top_p=0.7,
top_k=50,
return_dict_in_generate=True,
)
token = outputs.sequences[0, input_length:]
output_str = tokenizer.decode(token)
print(output_str.split("<By Manoj>")[0])
#
# Below results are before training with Instructions dataset
#
# init
tokenizer = AutoTokenizer.from_pretrained("bigscience/bloom-560m")
model = AutoModelForCausalLM.from_pretrained(
"bigscience/bloom-560m", torch_dtype=torch.float16
)
model = model.to("cuda:0")
# infer
inputs = tokenizer(prompt, return_tensors="pt").to(model.device)
input_length = inputs.input_ids.shape[1]
outputs = model.generate(
**inputs,
max_new_tokens=128,
do_sample=True,
temperature=0.7,
top_p=0.7,
top_k=50,
return_dict_in_generate=True,
)
token = outputs.sequences[0, input_length:]
output_str = tokenizer.decode(token)
print(output_str.split("<By Manoj>")[0])
prompt = "Instruction: Why can camels survive for long without water? <END> \ncontext: <END> \n \nresponse: "
inputs = tokenizer(prompt, return_tensors="pt").to(model.device)
input_length = inputs.input_ids.shape[1]
outputs = model.generate(
**inputs,
max_new_tokens=128,
do_sample=True,
temperature=0.7,
top_p=0.7,
top_k=50,
return_dict_in_generate=True,
)
token = outputs.sequences[0, input_length:]
output_str = tokenizer.decode(token)
print(output_str.split("<By Manoj>")[0])
| false | 0 | 938 | 0 | 938 | 938 |
||
129010375
|
<jupyter_start><jupyter_text>Aerial Semantic Segmentation Drone Dataset
Dataset Resource: https://www.tugraz.at/index.php?id=22387
Citation
If you use this dataset in your research, please cite the following URL:
http://dronedataset.icg.tugraz.at
License
The Drone Dataset is made freely available to academic and non-academic entities for non-commercial purposes such as academic research, teaching, scientific publications, or personal experimentation. Permission is granted to use the data given that you agree:
That the dataset comes "AS IS", without express or implied warranty. Although every effort has been made to ensure accuracy, we (Graz University of Technology) do not accept any responsibility for errors or omissions.
That you include a reference to the Semantic Drone Dataset in any work that makes use of the dataset. For research papers or other media link to the Semantic Drone Dataset webpage.
That you do not distribute this dataset or modified versions. It is permissible to distribute derivative works in as far as they are abstract representations of this dataset (such as models trained on it or additional annotations that do not directly include any of our data) and do not allow to recover the dataset or something similar in character.
That you may not use the dataset or any derivative work for commercial purposes as, for example, licensing or selling the data, or using the data with a purpose to procure a commercial gain.
That all rights not expressly granted to you are reserved by us (Graz University of Technology).
Dataset Overview
The Semantic Drone Dataset focuses on semantic understanding of urban scenes for increasing the safety of autonomous drone flight and landing procedures. The imagery depicts more than 20 houses from nadir (bird's eye) view acquired at an altitude of 5 to 30 meters above ground. A high resolution camera was used to acquire images at a size of 6000x4000px (24Mpx). The training set contains 400 publicly available images and the test set is made up of 200 private images.
PERSON DETECTION
For the task of person detection the dataset contains bounding box annotations of the training and test set.
SEMANTIC SEGMENTATION
We prepared pixel-accurate annotation for the same training and test set. The complexity of the dataset is limited to 20 classes as listed in the following table.
Table 1: Semanic classes of the Drone Dataset
tree, gras, other vegetation, dirt, gravel, rocks, water, paved area, pool, person, dog, car, bicycle, roof, wall, fence, fence-pole, window, door, obstacle
Kaggle dataset identifier: semantic-drone-dataset
<jupyter_script># # Semantic segmentation using pytorch и deeplabv3
# 
# # I was working in google colab using kaggle api
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import cv2
from sklearn.metrics import f1_score, roc_auc_score
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import random_split, DataLoader, Dataset
import torchvision.transforms as transforms
from torchvision.io import read_image, ImageReadMode
import torchvision
from torchvision.datasets.vision import VisionDataset
from tqdm.notebook import tqdm
from PIL import Image
from glob import glob
import os
import time
import copy
import csv
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
print(
'User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])
)
)
# Then move kaggle.json into the folder where the API expects to find it.
image_path = "/content/dataset/semantic_drone_dataset/original_images"
mask_path = "/content/dataset/semantic_drone_dataset/label_images_semantic"
labels = pd.read_csv("/content/class_dict_seg.csv")
labels.head()
len(labels)
classes = labels.name.values.tolist()
print(classes)
length = len(os.listdir(image_path))
# ## Dataset class
class DroneDataset(Dataset):
def __init__(self, imgs_dir, masks_dir, count, is_val=False):
self.imgs_dir = imgs_dir
self.masks_dir = masks_dir
imgs_paths = os.listdir(self.imgs_dir)
imgs_paths.sort()
mask_paths = os.listdir(self.masks_dir)
mask_paths.sort()
self.is_val = is_val
if not is_val: # для разделения на train/val в процессе
self.imgs_paths = imgs_paths[:count]
self.mask_paths = mask_paths[:count]
else:
self.imgs_paths = imgs_paths[-count:]
self.mask_paths = mask_paths[-count:]
def __len__(self):
return len(self.imgs_paths)
def __getitem__(self, idx):
img = read_image(
os.path.join(self.imgs_dir, self.imgs_paths[idx]), ImageReadMode.RGB
)
mask = read_image(
os.path.join(self.masks_dir, self.mask_paths[idx]), ImageReadMode.GRAY
)
return img, mask
# # Transforms
torchvision.models.segmentation.DeepLabV3_ResNet50_Weights.COCO_WITH_VOC_LABELS_V1.transforms() # transforms with which model was trained
def img_transform(img, mask, is_val=False, size=520):
img = img.to(device)
mask = mask.to(device)
img = img.float() / 255.0
if not is_val:
trans_img = torch.nn.Sequential(
transforms.Resize([size, size]),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
transforms.RandomAutocontrast(p=0.2),
)
else:
trans_img = trans_img = torch.nn.Sequential(
transforms.Resize([size, size]),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
)
trans_mask = torch.nn.Sequential(transforms.Resize([size, size]))
trans_img.requires_grad_(False)
trans_mask.requires_grad_(False)
trans_img = trans_img.to(device)
trans_mask = trans_mask.to(device)
img = trans_img(img)
mask = trans_mask(mask)
return img, mask.squeeze(1).long()
train_dataset_len = int(length * 0.7)
val_dataset_len = length - train_dataset_len
train_dataset = DroneDataset(image_path, mask_path, train_dataset_len)
val_dataset = DroneDataset(image_path, mask_path, val_dataset_len, is_val=True)
train_dataset[5][0].shape
train_dataset[5][1].shape
img, mask = next(iter(train_dataset))
batch_size = 4
train_loader = DataLoader(train_dataset, batch_size, shuffle=True, num_workers=2)
val_loader = DataLoader(val_dataset, batch_size, shuffle=False, num_workers=2)
model = torchvision.models.segmentation.deeplabv3_resnet50(
weights=torchvision.models.segmentation.DeepLabV3_ResNet50_Weights.DEFAULT,
progress=True,
)
# # By default deeplabv3 has 21 classes as an output, you need to change head for custom data
from torchvision.models.segmentation.deeplabv3 import DeepLabHead
from torchvision.models.segmentation.fcn import FCNHead
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
model.classifier = DeepLabHead(2048, 23)
model.aux_classifier = FCNHead(1024, 23)
model = model.to(device)
# # I use Cross Entropy loss, you can also try different ones, for example, Dice loss. You can look on implementations here
# ## https://www.kaggle.com/code/bigironsphere/loss-function-library-keras-pytorch
from torch.nn import CrossEntropyLoss
import torch.nn.functional as F
loss = CrossEntropyLoss().to(device)
learning_rate = 0.01
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)
# ## Pixel accuracy
def pixel_accuracy(mask, output):
output_softmax = F.softmax(output, dim=1)
output_argmax = torch.argmax(output_softmax, dim=1)
bool_tensor = (torch.flatten(mask)) == (torch.flatten(output_argmax))
return torch.sum(bool_tensor) / torch.numel(bool_tensor)
# # Train
from tqdm import tqdm
epoch_count = 30
train_losses = []
val_losses = []
train_accs = []
val_accs = []
es_steps = 3
count_steps = 0
train_len = len(train_loader)
val_len = len(val_loader)
print(train_len)
print(val_len)
best_score = 1e10
for epoch in range(epoch_count):
if count_steps >= es_steps:
print("Early stopping!")
break
train_loss_sum = 0
train_pixel_acc = 0
model.train()
for img_batch, mask_batch in tqdm(train_loader):
img_batch = img_batch.to(device, non_blocking=True)
mask_batch = mask_batch.to(device, non_blocking=True)
img_batch, mask_batch = img_transform(img_batch, mask_batch, is_val=False)
optimizer.zero_grad()
output_batch = model(img_batch)
loss_value = loss(output_batch["out"], mask_batch)
train_pixel_acc += pixel_accuracy(mask_batch, output_batch["out"]).detach()
train_loss_sum += loss_value.detach()
loss_value.backward()
optimizer.step()
del output_batch
train_loss = train_loss_sum / train_len
train_acc = train_pixel_acc / train_len
train_losses.append(train_loss)
train_accs.append(train_acc)
print(
f"Epoch {epoch} / {epoch_count} | train loss = {train_loss} | train acc = {train_acc}"
)
model.eval()
val_loss_sum = 0
val_pixel_acc = 0
for img_batch, mask_batch in tqdm(val_loader):
img_batch = img_batch.to(device, non_blocking=True)
mask_batch = mask_batch.to(device, non_blocking=True)
img_batch, mask_batch = img_transform(img_batch, mask_batch, is_val=True)
output_batch = model(img_batch)
loss_value = loss(output_batch["out"], mask_batch)
val_loss_sum = val_loss_sum + loss_value.detach()
val_pixel_acc = (
val_pixel_acc + pixel_accuracy(mask_batch, output_batch["out"]).detach()
)
del output_batch
val_loss = val_loss_sum / val_len
val_acc = val_pixel_acc / val_len
val_losses.append(val_loss)
val_accs.append(val_acc)
print(
f"Epoch {epoch} / {epoch_count} | val loss = {val_loss} | val acc = {val_acc}"
)
if val_loss < best_score:
best_score = val_loss
count_steps = 0
torch.save(model, "best_model.pt")
else:
count_steps += 1
import matplotlib.pyplot as plt
train_losses = [x.cpu().item() for x in train_losses]
val_losses = [x.cpu().item() for x in val_losses]
plt.plot(train_losses, linestyle="-")
plt.plot(val_losses, linestyle="--")
plt.xlabel("epochs")
plt.ylabel("loss")
plt.show()
train_accs = [x.cpu().item() for x in train_accs]
val_accs = [x.cpu().item() for x in val_accs]
plt.plot(train_accs, linestyle="-")
plt.plot(val_accs, linestyle="--")
plt.xlabel("epochs")
plt.ylabel("accuracy")
plt.show()
# # Inference
model.eval()
label_map = np.array(
[
(0, 0, 0), # unlabeled
(128, 64, 128), # paved-area
(130, 76, 0), # dirt
(0, 102, 0), # grass
(112, 103, 87), # gravel
(28, 42, 168), # water
(48, 41, 30), # rocks
(0, 50, 89), # pool
(107, 142, 35), # vegetation
(70, 70, 70), # roof
(102, 102, 156), # wall
(254, 228, 12), # window
(254, 148, 12), # door
(190, 153, 153), # fence
(153, 153, 153), # fence-pole
(255, 22, 96), # person
(102, 51, 0), # dog
(9, 143, 150), # car
(119, 11, 32), # bicycle
(51, 51, 0), # tree
(190, 250, 190), # bald-tree
(112, 150, 146), # art-marker
(2, 135, 115), # obstacle
(255, 0, 0), # conflicting
]
)
def draw_segmentation_map(outputs):
labels = torch.argmax(outputs.squeeze(), dim=0).numpy()
# Create 3 Numpy arrays containing zeros.
# Later each pixel will be filled with respective red, green, and blue pixels
# depending on the predicted class.
red_map = np.zeros_like(labels).astype(np.uint8)
green_map = np.zeros_like(labels).astype(np.uint8)
blue_map = np.zeros_like(labels).astype(np.uint8)
for label_num in range(0, len(label_map)):
index = labels == label_num
R, G, B = label_map[label_num]
red_map[index] = R
green_map[index] = G
blue_map[index] = B
segmentation_map = np.stack([red_map, green_map, blue_map], axis=2)
return segmentation_map
def image_overlay(image, segmented_image):
alpha = 1 # transparency for the original image
beta = 0.8 # transparency for the segmentation map
gamma = 0 # scalar added to each sum
image = np.array(image)
segmented_image = cv2.cvtColor(segmented_image, cv2.COLOR_RGB2BGR)
image = cv2.cvtColor(image, cv2.COLOR_RGB2BGR)
cv2.addWeighted(image, alpha, segmented_image, beta, gamma, image)
return image
imgs_paths = os.listdir(image_path)
imgs_paths.sort()
def perform_inference(
model=model,
imgs_paths=imgs_paths,
num_images=10,
image_dir="/content/dataset/semantic_drone_dataset/original_images/",
device="cpu",
):
device = (
device
if device is not None
else ("cuda" if torch.cuda.is_available() else "cpu")
)
model.to(device)
preprocess = transforms.Compose(
[
transforms.Resize([520, 520]),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
# Load image handles for the validation set.
# Randomly select 'num_images' from the whole set for inference.
selected_images = np.random.choice(imgs_paths, num_images, replace=False)
# Iterate over selected images
for img_name in selected_images:
# Load and pre-process image.
image_path = os.path.join(image_dir, img_name)
img_raw = Image.open(image_path).convert("RGB")
W, H = img_raw.size[:2]
img_t = preprocess(img_raw)
img_t = torch.unsqueeze(img_t, dim=0).to(device)
# Model Inference
with torch.no_grad():
output = model(img_t)["out"].cpu()
# Get RGB segmentation map
segmented_image = draw_segmentation_map(output)
# Resize to original image size
segmented_image = cv2.resize(segmented_image, (W, H), cv2.INTER_LINEAR)
overlayed_image = image_overlay(img_raw, segmented_image)
# Plot
plt.figure(figsize=(12, 10), dpi=100)
plt.subplot(1, 3, 1)
plt.axis("off")
plt.title("Image")
plt.imshow(np.asarray(img_raw))
plt.subplot(1, 3, 2)
plt.title("Segmentation")
plt.axis("off")
plt.imshow(segmented_image)
plt.subplot(1, 3, 3)
plt.title("Overlayed")
plt.axis("off")
plt.imshow(overlayed_image[:, :, ::-1])
plt.show()
plt.close()
return
perform_inference()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/010/129010375.ipynb
|
semantic-drone-dataset
|
bulentsiyah
|
[{"Id": 129010375, "ScriptId": 38348600, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6961845, "CreationDate": "05/10/2023 09:40:47", "VersionNumber": 1.0, "Title": "PyTorch Semantic Segmentation", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 424.0, "LinesInsertedFromPrevious": 424.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 184689890, "KernelVersionId": 129010375, "SourceDatasetVersionId": 1834160}]
|
[{"Id": 1834160, "DatasetId": 333968, "DatasourceVersionId": 1871791, "CreatorUserId": 1200915, "LicenseName": "Other (specified in description)", "CreationDate": "01/10/2021 23:24:17", "VersionNumber": 6.0, "Title": "Aerial Semantic Segmentation Drone Dataset", "Slug": "semantic-drone-dataset", "Subtitle": "aerial semantic Segmentation", "Description": "Dataset Resource: https://www.tugraz.at/index.php?id=22387\n\n\nCitation\nIf you use this dataset in your research, please cite the following URL:\n\nhttp://dronedataset.icg.tugraz.at\n\nLicense\nThe Drone Dataset is made freely available to academic and non-academic entities for non-commercial purposes such as academic research, teaching, scientific publications, or personal experimentation. Permission is granted to use the data given that you agree:\n\nThat the dataset comes \"AS IS\", without express or implied warranty. Although every effort has been made to ensure accuracy, we (Graz University of Technology) do not accept any responsibility for errors or omissions.\nThat you include a reference to the Semantic Drone Dataset in any work that makes use of the dataset. For research papers or other media link to the Semantic Drone Dataset webpage.\nThat you do not distribute this dataset or modified versions. It is permissible to distribute derivative works in as far as they are abstract representations of this dataset (such as models trained on it or additional annotations that do not directly include any of our data) and do not allow to recover the dataset or something similar in character.\nThat you may not use the dataset or any derivative work for commercial purposes as, for example, licensing or selling the data, or using the data with a purpose to procure a commercial gain.\nThat all rights not expressly granted to you are reserved by us (Graz University of Technology).\n\n\nDataset Overview\nThe Semantic Drone Dataset focuses on semantic understanding of urban scenes for increasing the safety of autonomous drone flight and landing procedures. The imagery depicts more than 20 houses from nadir (bird's eye) view acquired at an altitude of 5 to 30 meters above ground. A high resolution camera was used to acquire images at a size of 6000x4000px (24Mpx). The training set contains 400 publicly available images and the test set is made up of 200 private images.\n\nPERSON DETECTION\nFor the task of person detection the dataset contains bounding box annotations of the training and test set.\n\nSEMANTIC SEGMENTATION\nWe prepared pixel-accurate annotation for the same training and test set. The complexity of the dataset is limited to 20 classes as listed in the following table.\n\n Table 1: Semanic classes of the Drone Dataset\n\ntree, gras, other vegetation, dirt, gravel, rocks, water, paved area, pool, person, dog, car, bicycle, roof, wall, fence, fence-pole, window, door, obstacle", "VersionNotes": "class_dict_seg.csv added", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 333968, "CreatorUserId": 1200915, "OwnerUserId": 1200915.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1834160.0, "CurrentDatasourceVersionId": 1871791.0, "ForumId": 345540, "Type": 2, "CreationDate": "09/04/2019 10:52:09", "LastActivityDate": "09/04/2019", "TotalViews": 84325, "TotalDownloads": 12575, "TotalVotes": 220, "TotalKernels": 76}]
|
[{"Id": 1200915, "UserName": "bulentsiyah", "DisplayName": "Bulent Siyah", "RegisterDate": "08/04/2017", "PerformanceTier": 3}]
|
# # Semantic segmentation using pytorch и deeplabv3
# 
# # I was working in google colab using kaggle api
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import cv2
from sklearn.metrics import f1_score, roc_auc_score
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import random_split, DataLoader, Dataset
import torchvision.transforms as transforms
from torchvision.io import read_image, ImageReadMode
import torchvision
from torchvision.datasets.vision import VisionDataset
from tqdm.notebook import tqdm
from PIL import Image
from glob import glob
import os
import time
import copy
import csv
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
print(
'User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])
)
)
# Then move kaggle.json into the folder where the API expects to find it.
image_path = "/content/dataset/semantic_drone_dataset/original_images"
mask_path = "/content/dataset/semantic_drone_dataset/label_images_semantic"
labels = pd.read_csv("/content/class_dict_seg.csv")
labels.head()
len(labels)
classes = labels.name.values.tolist()
print(classes)
length = len(os.listdir(image_path))
# ## Dataset class
class DroneDataset(Dataset):
def __init__(self, imgs_dir, masks_dir, count, is_val=False):
self.imgs_dir = imgs_dir
self.masks_dir = masks_dir
imgs_paths = os.listdir(self.imgs_dir)
imgs_paths.sort()
mask_paths = os.listdir(self.masks_dir)
mask_paths.sort()
self.is_val = is_val
if not is_val: # для разделения на train/val в процессе
self.imgs_paths = imgs_paths[:count]
self.mask_paths = mask_paths[:count]
else:
self.imgs_paths = imgs_paths[-count:]
self.mask_paths = mask_paths[-count:]
def __len__(self):
return len(self.imgs_paths)
def __getitem__(self, idx):
img = read_image(
os.path.join(self.imgs_dir, self.imgs_paths[idx]), ImageReadMode.RGB
)
mask = read_image(
os.path.join(self.masks_dir, self.mask_paths[idx]), ImageReadMode.GRAY
)
return img, mask
# # Transforms
torchvision.models.segmentation.DeepLabV3_ResNet50_Weights.COCO_WITH_VOC_LABELS_V1.transforms() # transforms with which model was trained
def img_transform(img, mask, is_val=False, size=520):
img = img.to(device)
mask = mask.to(device)
img = img.float() / 255.0
if not is_val:
trans_img = torch.nn.Sequential(
transforms.Resize([size, size]),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
transforms.RandomAutocontrast(p=0.2),
)
else:
trans_img = trans_img = torch.nn.Sequential(
transforms.Resize([size, size]),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
)
trans_mask = torch.nn.Sequential(transforms.Resize([size, size]))
trans_img.requires_grad_(False)
trans_mask.requires_grad_(False)
trans_img = trans_img.to(device)
trans_mask = trans_mask.to(device)
img = trans_img(img)
mask = trans_mask(mask)
return img, mask.squeeze(1).long()
train_dataset_len = int(length * 0.7)
val_dataset_len = length - train_dataset_len
train_dataset = DroneDataset(image_path, mask_path, train_dataset_len)
val_dataset = DroneDataset(image_path, mask_path, val_dataset_len, is_val=True)
train_dataset[5][0].shape
train_dataset[5][1].shape
img, mask = next(iter(train_dataset))
batch_size = 4
train_loader = DataLoader(train_dataset, batch_size, shuffle=True, num_workers=2)
val_loader = DataLoader(val_dataset, batch_size, shuffle=False, num_workers=2)
model = torchvision.models.segmentation.deeplabv3_resnet50(
weights=torchvision.models.segmentation.DeepLabV3_ResNet50_Weights.DEFAULT,
progress=True,
)
# # By default deeplabv3 has 21 classes as an output, you need to change head for custom data
from torchvision.models.segmentation.deeplabv3 import DeepLabHead
from torchvision.models.segmentation.fcn import FCNHead
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
model.classifier = DeepLabHead(2048, 23)
model.aux_classifier = FCNHead(1024, 23)
model = model.to(device)
# # I use Cross Entropy loss, you can also try different ones, for example, Dice loss. You can look on implementations here
# ## https://www.kaggle.com/code/bigironsphere/loss-function-library-keras-pytorch
from torch.nn import CrossEntropyLoss
import torch.nn.functional as F
loss = CrossEntropyLoss().to(device)
learning_rate = 0.01
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)
# ## Pixel accuracy
def pixel_accuracy(mask, output):
output_softmax = F.softmax(output, dim=1)
output_argmax = torch.argmax(output_softmax, dim=1)
bool_tensor = (torch.flatten(mask)) == (torch.flatten(output_argmax))
return torch.sum(bool_tensor) / torch.numel(bool_tensor)
# # Train
from tqdm import tqdm
epoch_count = 30
train_losses = []
val_losses = []
train_accs = []
val_accs = []
es_steps = 3
count_steps = 0
train_len = len(train_loader)
val_len = len(val_loader)
print(train_len)
print(val_len)
best_score = 1e10
for epoch in range(epoch_count):
if count_steps >= es_steps:
print("Early stopping!")
break
train_loss_sum = 0
train_pixel_acc = 0
model.train()
for img_batch, mask_batch in tqdm(train_loader):
img_batch = img_batch.to(device, non_blocking=True)
mask_batch = mask_batch.to(device, non_blocking=True)
img_batch, mask_batch = img_transform(img_batch, mask_batch, is_val=False)
optimizer.zero_grad()
output_batch = model(img_batch)
loss_value = loss(output_batch["out"], mask_batch)
train_pixel_acc += pixel_accuracy(mask_batch, output_batch["out"]).detach()
train_loss_sum += loss_value.detach()
loss_value.backward()
optimizer.step()
del output_batch
train_loss = train_loss_sum / train_len
train_acc = train_pixel_acc / train_len
train_losses.append(train_loss)
train_accs.append(train_acc)
print(
f"Epoch {epoch} / {epoch_count} | train loss = {train_loss} | train acc = {train_acc}"
)
model.eval()
val_loss_sum = 0
val_pixel_acc = 0
for img_batch, mask_batch in tqdm(val_loader):
img_batch = img_batch.to(device, non_blocking=True)
mask_batch = mask_batch.to(device, non_blocking=True)
img_batch, mask_batch = img_transform(img_batch, mask_batch, is_val=True)
output_batch = model(img_batch)
loss_value = loss(output_batch["out"], mask_batch)
val_loss_sum = val_loss_sum + loss_value.detach()
val_pixel_acc = (
val_pixel_acc + pixel_accuracy(mask_batch, output_batch["out"]).detach()
)
del output_batch
val_loss = val_loss_sum / val_len
val_acc = val_pixel_acc / val_len
val_losses.append(val_loss)
val_accs.append(val_acc)
print(
f"Epoch {epoch} / {epoch_count} | val loss = {val_loss} | val acc = {val_acc}"
)
if val_loss < best_score:
best_score = val_loss
count_steps = 0
torch.save(model, "best_model.pt")
else:
count_steps += 1
import matplotlib.pyplot as plt
train_losses = [x.cpu().item() for x in train_losses]
val_losses = [x.cpu().item() for x in val_losses]
plt.plot(train_losses, linestyle="-")
plt.plot(val_losses, linestyle="--")
plt.xlabel("epochs")
plt.ylabel("loss")
plt.show()
train_accs = [x.cpu().item() for x in train_accs]
val_accs = [x.cpu().item() for x in val_accs]
plt.plot(train_accs, linestyle="-")
plt.plot(val_accs, linestyle="--")
plt.xlabel("epochs")
plt.ylabel("accuracy")
plt.show()
# # Inference
model.eval()
label_map = np.array(
[
(0, 0, 0), # unlabeled
(128, 64, 128), # paved-area
(130, 76, 0), # dirt
(0, 102, 0), # grass
(112, 103, 87), # gravel
(28, 42, 168), # water
(48, 41, 30), # rocks
(0, 50, 89), # pool
(107, 142, 35), # vegetation
(70, 70, 70), # roof
(102, 102, 156), # wall
(254, 228, 12), # window
(254, 148, 12), # door
(190, 153, 153), # fence
(153, 153, 153), # fence-pole
(255, 22, 96), # person
(102, 51, 0), # dog
(9, 143, 150), # car
(119, 11, 32), # bicycle
(51, 51, 0), # tree
(190, 250, 190), # bald-tree
(112, 150, 146), # art-marker
(2, 135, 115), # obstacle
(255, 0, 0), # conflicting
]
)
def draw_segmentation_map(outputs):
labels = torch.argmax(outputs.squeeze(), dim=0).numpy()
# Create 3 Numpy arrays containing zeros.
# Later each pixel will be filled with respective red, green, and blue pixels
# depending on the predicted class.
red_map = np.zeros_like(labels).astype(np.uint8)
green_map = np.zeros_like(labels).astype(np.uint8)
blue_map = np.zeros_like(labels).astype(np.uint8)
for label_num in range(0, len(label_map)):
index = labels == label_num
R, G, B = label_map[label_num]
red_map[index] = R
green_map[index] = G
blue_map[index] = B
segmentation_map = np.stack([red_map, green_map, blue_map], axis=2)
return segmentation_map
def image_overlay(image, segmented_image):
alpha = 1 # transparency for the original image
beta = 0.8 # transparency for the segmentation map
gamma = 0 # scalar added to each sum
image = np.array(image)
segmented_image = cv2.cvtColor(segmented_image, cv2.COLOR_RGB2BGR)
image = cv2.cvtColor(image, cv2.COLOR_RGB2BGR)
cv2.addWeighted(image, alpha, segmented_image, beta, gamma, image)
return image
imgs_paths = os.listdir(image_path)
imgs_paths.sort()
def perform_inference(
model=model,
imgs_paths=imgs_paths,
num_images=10,
image_dir="/content/dataset/semantic_drone_dataset/original_images/",
device="cpu",
):
device = (
device
if device is not None
else ("cuda" if torch.cuda.is_available() else "cpu")
)
model.to(device)
preprocess = transforms.Compose(
[
transforms.Resize([520, 520]),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
# Load image handles for the validation set.
# Randomly select 'num_images' from the whole set for inference.
selected_images = np.random.choice(imgs_paths, num_images, replace=False)
# Iterate over selected images
for img_name in selected_images:
# Load and pre-process image.
image_path = os.path.join(image_dir, img_name)
img_raw = Image.open(image_path).convert("RGB")
W, H = img_raw.size[:2]
img_t = preprocess(img_raw)
img_t = torch.unsqueeze(img_t, dim=0).to(device)
# Model Inference
with torch.no_grad():
output = model(img_t)["out"].cpu()
# Get RGB segmentation map
segmented_image = draw_segmentation_map(output)
# Resize to original image size
segmented_image = cv2.resize(segmented_image, (W, H), cv2.INTER_LINEAR)
overlayed_image = image_overlay(img_raw, segmented_image)
# Plot
plt.figure(figsize=(12, 10), dpi=100)
plt.subplot(1, 3, 1)
plt.axis("off")
plt.title("Image")
plt.imshow(np.asarray(img_raw))
plt.subplot(1, 3, 2)
plt.title("Segmentation")
plt.axis("off")
plt.imshow(segmented_image)
plt.subplot(1, 3, 3)
plt.title("Overlayed")
plt.axis("off")
plt.imshow(overlayed_image[:, :, ::-1])
plt.show()
plt.close()
return
perform_inference()
| false | 0 | 226,162 | 3 | 226,783 | 226,162 |
||
129010000
|
import pandas as pd
import numpy as np
from PIL import Image
import matplotlib.pyplot as plt
from sklearn.metrics import classification_report
from sklearn.metrics import matthews_corrcoef
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.metrics import roc_curve
from google.colab import drive
drive.mount("/content/drive")
# # load image (VGG16, autoencoder output) and text vectors topic modeling(LSA-BOW)
# # training data
img_label = np.load(
"/content/drive/MyDrive/HMD_project/new/embedding_train_img_norm.npy"
)
txt_input = np.load("/content/drive/MyDrive/HMD_project/new/lsa_bow_train_text.npy")
img = img_label[:, 0:-1]
label = img_label[:, -1]
txt = txt_input
img_txt = np.concatenate((img, txt), axis=1)
x = img_txt
y = label
print(x.shape)
y.shape
# # validation data
img_label_val = np.load(
"/content/drive/MyDrive/HMD_project/new/embedding_val_img_norm.npy"
)
txt_input_val = np.load("/content/drive/MyDrive/HMD_project/new/lsa_bow_val_text.npy")
img_val = img_label_val[:, 0:-1]
label_val = img_label_val[:, -1]
# txt_val=txt_input_val-np.min(txt_input_val)
txt_val = txt_input_val
img_txt_val = np.concatenate((img_val, txt_val), axis=1)
x_val = img_txt_val
y_val = label_val
x_val.shape
# # Find optimal threshold from the ROC curve and apply that threshold for classification
# # Logistic regression classifier
clf = LogisticRegression(max_iter=1000, C=0.1, penalty="l2")
clf.fit(x, y)
y_pred_p = clf.predict_proba(x)
b = y_pred_p[:, -1]
b = b.reshape(y.shape)
fpr, tpr, th = roc_curve(y, b)
plt.figure(figsize=(7, 5))
plt.plot(fpr, tpr, label="LSA-LR", linewidth=2)
plt.grid()
plt.title("ROC")
plt.xlabel("False positive rate-------->")
plt.ylabel("True positive rate--------->")
plt.legend(loc="lower right")
optimal_idx = np.argmax(tpr - fpr)
optimal_threshold = th[optimal_idx]
print("optimal_threshold", optimal_threshold, "\n")
y_pred = np.where(clf.predict_proba(x)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y, y_pred))
print(matthews_corrcoef(y, y_pred))
print("\n\nvalidation")
y_pred_val = np.where(clf.predict_proba(x_val)[:, 1] > optimal_threshold, 1, 0)
# y_pred_val=clf.predict(x_val)
print(classification_report(y_val, y_pred_val))
print(matthews_corrcoef(y_val, y_pred_val))
# # Random Forest
clf = RandomForestClassifier(max_depth=3, random_state=0)
clf.fit(x, y)
y_pred_p = clf.predict_proba(x)
b = y_pred_p[:, -1]
b = b.reshape(y.shape)
fpr, tpr, th = roc_curve(y, b)
plt.figure(figsize=(7, 5))
plt.plot(fpr, tpr, label="LSA-LR", linewidth=2)
plt.grid()
plt.title("ROC")
plt.xlabel("False positive rate-------->")
plt.ylabel("True positive rate--------->")
plt.legend(loc="lower right")
optimal_idx = np.argmax(tpr - fpr)
optimal_threshold = th[optimal_idx]
print("optimal_threshold", optimal_threshold, "\n")
y_pred = np.where(clf.predict_proba(x)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y, y_pred))
print(matthews_corrcoef(y, y_pred))
print("\n\nvalidation")
y_pred_val = np.where(clf.predict_proba(x_val)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y_val, y_pred_val))
print(matthews_corrcoef(y_val, y_pred_val))
#
# #load text vectors(TFIDF-NMF)
# # train set
txt_input = np.load("/content/drive/MyDrive/HMD_project/new/nmf_tfidf_train_text.npy")
txt = txt_input
img_txt = np.concatenate((img, txt), axis=1)
x = img_txt
y = label
print(y.shape)
x.shape
# # validation set
txt_input_val = np.load("/content/drive/MyDrive/HMD_project/new/nmf_tfidf_val_text.npy")
txt_val = txt_input_val
img_txt_val = np.concatenate((img_val, txt_val), axis=1)
x_val = img_txt_val
y_val = label_val
print(x_val.shape)
y_val.shape
# # Random Forest
clf = RandomForestClassifier(n_estimators=100, max_depth=3, random_state=1)
clf.fit(x, y)
y_pred_p = clf.predict_proba(x)
b = y_pred_p[:, -1]
b = b.reshape(y.shape)
fpr, tpr, th = roc_curve(y, b)
plt.figure(figsize=(7, 5))
plt.plot(fpr, tpr, label="LSA-LR", linewidth=2)
plt.grid()
plt.title("ROC")
plt.xlabel("False positive rate-------->")
plt.ylabel("True positive rate--------->")
plt.legend(loc="lower right")
optimal_idx = np.argmax(tpr - fpr)
optimal_threshold = th[optimal_idx]
print("optimal_threshold", optimal_threshold, "\n")
y_pred = np.where(clf.predict_proba(x)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y, y_pred))
print(matthews_corrcoef(y, y_pred))
print("\n\nvalidation")
y_pred_val = np.where(clf.predict_proba(x_val)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y_val, y_pred_val))
print(matthews_corrcoef(y_val, y_pred_val))
# # Logistic regression
clf = LogisticRegression(max_iter=2500, C=50)
clf.fit(x, y)
y_pred_p = clf.predict_proba(x)
b = y_pred_p[:, -1]
b = b.reshape(y.shape)
fpr, tpr, th = roc_curve(y, b)
plt.figure(figsize=(7, 5))
plt.plot(fpr, tpr, label="LSA-LR", linewidth=2)
plt.grid()
plt.title("ROC")
plt.xlabel("False positive rate-------->")
plt.ylabel("True positive rate--------->")
plt.legend(loc="lower right")
optimal_idx = np.argmax(tpr - fpr)
optimal_threshold = th[optimal_idx]
print("optimal_threshold", optimal_threshold, "\n")
y_pred = np.where(clf.predict_proba(x)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y, y_pred))
print(matthews_corrcoef(y, y_pred))
print("\n\nvalidation")
y_pred_val = np.where(clf.predict_proba(x_val)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y_val, y_pred_val))
print(matthews_corrcoef(y_val, y_pred_val))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/010/129010000.ipynb
| null | null |
[{"Id": 129010000, "ScriptId": 38349713, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13939396, "CreationDate": "05/10/2023 09:37:28", "VersionNumber": 1.0, "Title": "Classifier_Automatic_threshold", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 242.0, "LinesInsertedFromPrevious": 242.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import numpy as np
from PIL import Image
import matplotlib.pyplot as plt
from sklearn.metrics import classification_report
from sklearn.metrics import matthews_corrcoef
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.metrics import roc_curve
from google.colab import drive
drive.mount("/content/drive")
# # load image (VGG16, autoencoder output) and text vectors topic modeling(LSA-BOW)
# # training data
img_label = np.load(
"/content/drive/MyDrive/HMD_project/new/embedding_train_img_norm.npy"
)
txt_input = np.load("/content/drive/MyDrive/HMD_project/new/lsa_bow_train_text.npy")
img = img_label[:, 0:-1]
label = img_label[:, -1]
txt = txt_input
img_txt = np.concatenate((img, txt), axis=1)
x = img_txt
y = label
print(x.shape)
y.shape
# # validation data
img_label_val = np.load(
"/content/drive/MyDrive/HMD_project/new/embedding_val_img_norm.npy"
)
txt_input_val = np.load("/content/drive/MyDrive/HMD_project/new/lsa_bow_val_text.npy")
img_val = img_label_val[:, 0:-1]
label_val = img_label_val[:, -1]
# txt_val=txt_input_val-np.min(txt_input_val)
txt_val = txt_input_val
img_txt_val = np.concatenate((img_val, txt_val), axis=1)
x_val = img_txt_val
y_val = label_val
x_val.shape
# # Find optimal threshold from the ROC curve and apply that threshold for classification
# # Logistic regression classifier
clf = LogisticRegression(max_iter=1000, C=0.1, penalty="l2")
clf.fit(x, y)
y_pred_p = clf.predict_proba(x)
b = y_pred_p[:, -1]
b = b.reshape(y.shape)
fpr, tpr, th = roc_curve(y, b)
plt.figure(figsize=(7, 5))
plt.plot(fpr, tpr, label="LSA-LR", linewidth=2)
plt.grid()
plt.title("ROC")
plt.xlabel("False positive rate-------->")
plt.ylabel("True positive rate--------->")
plt.legend(loc="lower right")
optimal_idx = np.argmax(tpr - fpr)
optimal_threshold = th[optimal_idx]
print("optimal_threshold", optimal_threshold, "\n")
y_pred = np.where(clf.predict_proba(x)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y, y_pred))
print(matthews_corrcoef(y, y_pred))
print("\n\nvalidation")
y_pred_val = np.where(clf.predict_proba(x_val)[:, 1] > optimal_threshold, 1, 0)
# y_pred_val=clf.predict(x_val)
print(classification_report(y_val, y_pred_val))
print(matthews_corrcoef(y_val, y_pred_val))
# # Random Forest
clf = RandomForestClassifier(max_depth=3, random_state=0)
clf.fit(x, y)
y_pred_p = clf.predict_proba(x)
b = y_pred_p[:, -1]
b = b.reshape(y.shape)
fpr, tpr, th = roc_curve(y, b)
plt.figure(figsize=(7, 5))
plt.plot(fpr, tpr, label="LSA-LR", linewidth=2)
plt.grid()
plt.title("ROC")
plt.xlabel("False positive rate-------->")
plt.ylabel("True positive rate--------->")
plt.legend(loc="lower right")
optimal_idx = np.argmax(tpr - fpr)
optimal_threshold = th[optimal_idx]
print("optimal_threshold", optimal_threshold, "\n")
y_pred = np.where(clf.predict_proba(x)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y, y_pred))
print(matthews_corrcoef(y, y_pred))
print("\n\nvalidation")
y_pred_val = np.where(clf.predict_proba(x_val)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y_val, y_pred_val))
print(matthews_corrcoef(y_val, y_pred_val))
#
# #load text vectors(TFIDF-NMF)
# # train set
txt_input = np.load("/content/drive/MyDrive/HMD_project/new/nmf_tfidf_train_text.npy")
txt = txt_input
img_txt = np.concatenate((img, txt), axis=1)
x = img_txt
y = label
print(y.shape)
x.shape
# # validation set
txt_input_val = np.load("/content/drive/MyDrive/HMD_project/new/nmf_tfidf_val_text.npy")
txt_val = txt_input_val
img_txt_val = np.concatenate((img_val, txt_val), axis=1)
x_val = img_txt_val
y_val = label_val
print(x_val.shape)
y_val.shape
# # Random Forest
clf = RandomForestClassifier(n_estimators=100, max_depth=3, random_state=1)
clf.fit(x, y)
y_pred_p = clf.predict_proba(x)
b = y_pred_p[:, -1]
b = b.reshape(y.shape)
fpr, tpr, th = roc_curve(y, b)
plt.figure(figsize=(7, 5))
plt.plot(fpr, tpr, label="LSA-LR", linewidth=2)
plt.grid()
plt.title("ROC")
plt.xlabel("False positive rate-------->")
plt.ylabel("True positive rate--------->")
plt.legend(loc="lower right")
optimal_idx = np.argmax(tpr - fpr)
optimal_threshold = th[optimal_idx]
print("optimal_threshold", optimal_threshold, "\n")
y_pred = np.where(clf.predict_proba(x)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y, y_pred))
print(matthews_corrcoef(y, y_pred))
print("\n\nvalidation")
y_pred_val = np.where(clf.predict_proba(x_val)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y_val, y_pred_val))
print(matthews_corrcoef(y_val, y_pred_val))
# # Logistic regression
clf = LogisticRegression(max_iter=2500, C=50)
clf.fit(x, y)
y_pred_p = clf.predict_proba(x)
b = y_pred_p[:, -1]
b = b.reshape(y.shape)
fpr, tpr, th = roc_curve(y, b)
plt.figure(figsize=(7, 5))
plt.plot(fpr, tpr, label="LSA-LR", linewidth=2)
plt.grid()
plt.title("ROC")
plt.xlabel("False positive rate-------->")
plt.ylabel("True positive rate--------->")
plt.legend(loc="lower right")
optimal_idx = np.argmax(tpr - fpr)
optimal_threshold = th[optimal_idx]
print("optimal_threshold", optimal_threshold, "\n")
y_pred = np.where(clf.predict_proba(x)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y, y_pred))
print(matthews_corrcoef(y, y_pred))
print("\n\nvalidation")
y_pred_val = np.where(clf.predict_proba(x_val)[:, 1] > optimal_threshold, 1, 0)
print(classification_report(y_val, y_pred_val))
print(matthews_corrcoef(y_val, y_pred_val))
| false | 0 | 2,105 | 0 | 2,105 | 2,105 |
||
129010475
|
<jupyter_start><jupyter_text>Used Cars Price Prediction
Kaggle dataset identifier: used-cars-price-prediction
<jupyter_code>import pandas as pd
df = pd.read_csv('used-cars-price-prediction/train-data.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 6019 entries, 0 to 6018
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 6019 non-null int64
1 Name 6019 non-null object
2 Location 6019 non-null object
3 Year 6019 non-null int64
4 Kilometers_Driven 6019 non-null int64
5 Fuel_Type 6019 non-null object
6 Transmission 6019 non-null object
7 Owner_Type 6019 non-null object
8 Mileage 6017 non-null object
9 Engine 5983 non-null object
10 Power 5983 non-null object
11 Seats 5977 non-null float64
12 New_Price 824 non-null object
13 Price 6019 non-null float64
dtypes: float64(2), int64(3), object(9)
memory usage: 658.5+ KB
<jupyter_text>Examples:
{
"Unnamed: 0": 0,
"Name": "Maruti Wagon R LXI CNG",
"Location": "Mumbai",
"Year": 2010,
"Kilometers_Driven": 72000,
"Fuel_Type": "CNG",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "26.6 km/kg",
"Engine": "998 CC",
"Power": "58.16 bhp",
"Seats": 5,
"New_Price": null,
"Price": 1.75
}
{
"Unnamed: 0": 1,
"Name": "Hyundai Creta 1.6 CRDi SX Option",
"Location": "Pune",
"Year": 2015,
"Kilometers_Driven": 41000,
"Fuel_Type": "Diesel",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "19.67 kmpl",
"Engine": "1582 CC",
"Power": "126.2 bhp",
"Seats": 5,
"New_Price": null,
"Price": 12.5
}
{
"Unnamed: 0": 2,
"Name": "Honda Jazz V",
"Location": "Chennai",
"Year": 2011,
"Kilometers_Driven": 46000,
"Fuel_Type": "Petrol",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "18.2 kmpl",
"Engine": "1199 CC",
"Power": "88.7 bhp",
"Seats": 5,
"New_Price": "8.61 Lakh",
"Price": 4.5
}
{
"Unnamed: 0": 3,
"Name": "Maruti Ertiga VDI",
"Location": "Chennai",
"Year": 2012,
"Kilometers_Driven": 87000,
"Fuel_Type": "Diesel",
"Transmission": "Manual",
"Owner_Type": "First",
"Mileage": "20.77 kmpl",
"Engine": "1248 CC",
"Power": "88.76 bhp",
"Seats": 7,
"New_Price": null,
"Price": 6.0
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
df = pd.read_csv(
"/kaggle/input/used-cars-price-prediction/train-data.csv"
) # df is an object of a class DataFrame which we create after reading the file for this case its the csv
df.head() # Read the top 5 records of the dataset
df.head(-1) # this will print all the records from the dataset
df.head(
2
) # if we pass 2 in the paranthesis it will print the top 2 records and if we pass 5 it will print 5
df.sample(5) # get us randomly chosen 5 records
unique_location = df["Location"].unique() # data type of this unique_location variable
unique_location_list = unique_location.tolist() # converting the array to a list
len(unique_location_list) # number of unique cities
len(df["Location"].unique())
# Get the types of Fuel in the dataset? How many types of seater the cars are in the dataset?
df["New_Price"].isnull().sum()
df2 = df.drop(
["New_Price"], axis=1, inplace=False
) # axis = 1 means its a column and axis = 0 means its a row, if inplace is false it will not update the main datafreame and for that we need to use another dataframe to store that previously updated dataframe
df2.head()
df.head(1)
df.drop(
["New_Price"], axis=1, inplace=True
) # if we pass true it will update the dataframe and save it inside
df.head(1)
# Your task is to drop thos unessesarry column 'Unnamed: 0'
df.describe().T
df.info()
unique_fuel = df["Fuel_Type"].unique()
unique_fuel_list = unique_fuel.tolist()
unique_fuel_list
df["Seats"].isnull().sum()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/010/129010475.ipynb
|
used-cars-price-prediction
|
avikasliwal
|
[{"Id": 129010475, "ScriptId": 38342612, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14950540, "CreationDate": "05/10/2023 09:41:43", "VersionNumber": 1.0, "Title": "notebook649cad9d3f", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 66.0, "LinesInsertedFromPrevious": 66.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184690050, "KernelVersionId": 129010475, "SourceDatasetVersionId": 518431}]
|
[{"Id": 518431, "DatasetId": 245550, "DatasourceVersionId": 534662, "CreatorUserId": 2716677, "LicenseName": "Other (specified in description)", "CreationDate": "06/25/2019 10:26:52", "VersionNumber": 2.0, "Title": "Used Cars Price Prediction", "Slug": "used-cars-price-prediction", "Subtitle": "Predict the price of an unknown car. Build your own Algo for cars 24 !!", "Description": NaN, "VersionNotes": "Replaced xlsx files by csv files", "TotalCompressedBytes": 791875.0, "TotalUncompressedBytes": 791875.0}]
|
[{"Id": 245550, "CreatorUserId": 2716677, "OwnerUserId": 2716677.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 518431.0, "CurrentDatasourceVersionId": 534662.0, "ForumId": 256748, "Type": 2, "CreationDate": "06/25/2019 10:11:54", "LastActivityDate": "06/25/2019", "TotalViews": 140175, "TotalDownloads": 22061, "TotalVotes": 222, "TotalKernels": 107}]
|
[{"Id": 2716677, "UserName": "avikasliwal", "DisplayName": "Avi Kasliwal", "RegisterDate": "01/18/2019", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
df = pd.read_csv(
"/kaggle/input/used-cars-price-prediction/train-data.csv"
) # df is an object of a class DataFrame which we create after reading the file for this case its the csv
df.head() # Read the top 5 records of the dataset
df.head(-1) # this will print all the records from the dataset
df.head(
2
) # if we pass 2 in the paranthesis it will print the top 2 records and if we pass 5 it will print 5
df.sample(5) # get us randomly chosen 5 records
unique_location = df["Location"].unique() # data type of this unique_location variable
unique_location_list = unique_location.tolist() # converting the array to a list
len(unique_location_list) # number of unique cities
len(df["Location"].unique())
# Get the types of Fuel in the dataset? How many types of seater the cars are in the dataset?
df["New_Price"].isnull().sum()
df2 = df.drop(
["New_Price"], axis=1, inplace=False
) # axis = 1 means its a column and axis = 0 means its a row, if inplace is false it will not update the main datafreame and for that we need to use another dataframe to store that previously updated dataframe
df2.head()
df.head(1)
df.drop(
["New_Price"], axis=1, inplace=True
) # if we pass true it will update the dataframe and save it inside
df.head(1)
# Your task is to drop thos unessesarry column 'Unnamed: 0'
df.describe().T
df.info()
unique_fuel = df["Fuel_Type"].unique()
unique_fuel_list = unique_fuel.tolist()
unique_fuel_list
df["Seats"].isnull().sum()
|
[{"used-cars-price-prediction/train-data.csv": {"column_names": "[\"Unnamed: 0\", \"Name\", \"Location\", \"Year\", \"Kilometers_Driven\", \"Fuel_Type\", \"Transmission\", \"Owner_Type\", \"Mileage\", \"Engine\", \"Power\", \"Seats\", \"New_Price\", \"Price\"]", "column_data_types": "{\"Unnamed: 0\": \"int64\", \"Name\": \"object\", \"Location\": \"object\", \"Year\": \"int64\", \"Kilometers_Driven\": \"int64\", \"Fuel_Type\": \"object\", \"Transmission\": \"object\", \"Owner_Type\": \"object\", \"Mileage\": \"object\", \"Engine\": \"object\", \"Power\": \"object\", \"Seats\": \"float64\", \"New_Price\": \"object\", \"Price\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 6019 entries, 0 to 6018\nData columns (total 14 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 6019 non-null int64 \n 1 Name 6019 non-null object \n 2 Location 6019 non-null object \n 3 Year 6019 non-null int64 \n 4 Kilometers_Driven 6019 non-null int64 \n 5 Fuel_Type 6019 non-null object \n 6 Transmission 6019 non-null object \n 7 Owner_Type 6019 non-null object \n 8 Mileage 6017 non-null object \n 9 Engine 5983 non-null object \n 10 Power 5983 non-null object \n 11 Seats 5977 non-null float64\n 12 New_Price 824 non-null object \n 13 Price 6019 non-null float64\ndtypes: float64(2), int64(3), object(9)\nmemory usage: 658.5+ KB\n", "summary": "{\"Unnamed: 0\": {\"count\": 6019.0, \"mean\": 3009.0, \"std\": 1737.6799666988932, \"min\": 0.0, \"25%\": 1504.5, \"50%\": 3009.0, \"75%\": 4513.5, \"max\": 6018.0}, \"Year\": {\"count\": 6019.0, \"mean\": 2013.3581990363848, \"std\": 3.2697421160913964, \"min\": 1998.0, \"25%\": 2011.0, \"50%\": 2014.0, \"75%\": 2016.0, \"max\": 2019.0}, \"Kilometers_Driven\": {\"count\": 6019.0, \"mean\": 58738.38029573019, \"std\": 91268.84320624862, \"min\": 171.0, \"25%\": 34000.0, \"50%\": 53000.0, \"75%\": 73000.0, \"max\": 6500000.0}, \"Seats\": {\"count\": 5977.0, \"mean\": 5.278735151413753, \"std\": 0.8088395547482927, \"min\": 0.0, \"25%\": 5.0, \"50%\": 5.0, \"75%\": 5.0, \"max\": 10.0}, \"Price\": {\"count\": 6019.0, \"mean\": 9.47946835022429, \"std\": 11.1879171124555, \"min\": 0.44, \"25%\": 3.5, \"50%\": 5.64, \"75%\": 9.95, \"max\": 160.0}}", "examples": "{\"Unnamed: 0\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"Name\":{\"0\":\"Maruti Wagon R LXI CNG\",\"1\":\"Hyundai Creta 1.6 CRDi SX Option\",\"2\":\"Honda Jazz V\",\"3\":\"Maruti Ertiga VDI\"},\"Location\":{\"0\":\"Mumbai\",\"1\":\"Pune\",\"2\":\"Chennai\",\"3\":\"Chennai\"},\"Year\":{\"0\":2010,\"1\":2015,\"2\":2011,\"3\":2012},\"Kilometers_Driven\":{\"0\":72000,\"1\":41000,\"2\":46000,\"3\":87000},\"Fuel_Type\":{\"0\":\"CNG\",\"1\":\"Diesel\",\"2\":\"Petrol\",\"3\":\"Diesel\"},\"Transmission\":{\"0\":\"Manual\",\"1\":\"Manual\",\"2\":\"Manual\",\"3\":\"Manual\"},\"Owner_Type\":{\"0\":\"First\",\"1\":\"First\",\"2\":\"First\",\"3\":\"First\"},\"Mileage\":{\"0\":\"26.6 km\\/kg\",\"1\":\"19.67 kmpl\",\"2\":\"18.2 kmpl\",\"3\":\"20.77 kmpl\"},\"Engine\":{\"0\":\"998 CC\",\"1\":\"1582 CC\",\"2\":\"1199 CC\",\"3\":\"1248 CC\"},\"Power\":{\"0\":\"58.16 bhp\",\"1\":\"126.2 bhp\",\"2\":\"88.7 bhp\",\"3\":\"88.76 bhp\"},\"Seats\":{\"0\":5.0,\"1\":5.0,\"2\":5.0,\"3\":7.0},\"New_Price\":{\"0\":null,\"1\":null,\"2\":\"8.61 Lakh\",\"3\":null},\"Price\":{\"0\":1.75,\"1\":12.5,\"2\":4.5,\"3\":6.0}}"}}]
| true | 1 |
<start_data_description><data_path>used-cars-price-prediction/train-data.csv:
<column_names>
['Unnamed: 0', 'Name', 'Location', 'Year', 'Kilometers_Driven', 'Fuel_Type', 'Transmission', 'Owner_Type', 'Mileage', 'Engine', 'Power', 'Seats', 'New_Price', 'Price']
<column_types>
{'Unnamed: 0': 'int64', 'Name': 'object', 'Location': 'object', 'Year': 'int64', 'Kilometers_Driven': 'int64', 'Fuel_Type': 'object', 'Transmission': 'object', 'Owner_Type': 'object', 'Mileage': 'object', 'Engine': 'object', 'Power': 'object', 'Seats': 'float64', 'New_Price': 'object', 'Price': 'float64'}
<dataframe_Summary>
{'Unnamed: 0': {'count': 6019.0, 'mean': 3009.0, 'std': 1737.6799666988932, 'min': 0.0, '25%': 1504.5, '50%': 3009.0, '75%': 4513.5, 'max': 6018.0}, 'Year': {'count': 6019.0, 'mean': 2013.3581990363848, 'std': 3.2697421160913964, 'min': 1998.0, '25%': 2011.0, '50%': 2014.0, '75%': 2016.0, 'max': 2019.0}, 'Kilometers_Driven': {'count': 6019.0, 'mean': 58738.38029573019, 'std': 91268.84320624862, 'min': 171.0, '25%': 34000.0, '50%': 53000.0, '75%': 73000.0, 'max': 6500000.0}, 'Seats': {'count': 5977.0, 'mean': 5.278735151413753, 'std': 0.8088395547482927, 'min': 0.0, '25%': 5.0, '50%': 5.0, '75%': 5.0, 'max': 10.0}, 'Price': {'count': 6019.0, 'mean': 9.47946835022429, 'std': 11.1879171124555, 'min': 0.44, '25%': 3.5, '50%': 5.64, '75%': 9.95, 'max': 160.0}}
<dataframe_info>
RangeIndex: 6019 entries, 0 to 6018
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 6019 non-null int64
1 Name 6019 non-null object
2 Location 6019 non-null object
3 Year 6019 non-null int64
4 Kilometers_Driven 6019 non-null int64
5 Fuel_Type 6019 non-null object
6 Transmission 6019 non-null object
7 Owner_Type 6019 non-null object
8 Mileage 6017 non-null object
9 Engine 5983 non-null object
10 Power 5983 non-null object
11 Seats 5977 non-null float64
12 New_Price 824 non-null object
13 Price 6019 non-null float64
dtypes: float64(2), int64(3), object(9)
memory usage: 658.5+ KB
<some_examples>
{'Unnamed: 0': {'0': 0, '1': 1, '2': 2, '3': 3}, 'Name': {'0': 'Maruti Wagon R LXI CNG', '1': 'Hyundai Creta 1.6 CRDi SX Option', '2': 'Honda Jazz V', '3': 'Maruti Ertiga VDI'}, 'Location': {'0': 'Mumbai', '1': 'Pune', '2': 'Chennai', '3': 'Chennai'}, 'Year': {'0': 2010, '1': 2015, '2': 2011, '3': 2012}, 'Kilometers_Driven': {'0': 72000, '1': 41000, '2': 46000, '3': 87000}, 'Fuel_Type': {'0': 'CNG', '1': 'Diesel', '2': 'Petrol', '3': 'Diesel'}, 'Transmission': {'0': 'Manual', '1': 'Manual', '2': 'Manual', '3': 'Manual'}, 'Owner_Type': {'0': 'First', '1': 'First', '2': 'First', '3': 'First'}, 'Mileage': {'0': '26.6 km/kg', '1': '19.67 kmpl', '2': '18.2 kmpl', '3': '20.77 kmpl'}, 'Engine': {'0': '998 CC', '1': '1582 CC', '2': '1199 CC', '3': '1248 CC'}, 'Power': {'0': '58.16 bhp', '1': '126.2 bhp', '2': '88.7 bhp', '3': '88.76 bhp'}, 'Seats': {'0': 5.0, '1': 5.0, '2': 5.0, '3': 7.0}, 'New_Price': {'0': None, '1': None, '2': '8.61 Lakh', '3': None}, 'Price': {'0': 1.75, '1': 12.5, '2': 4.5, '3': 6.0}}
<end_description>
| 637 | 0 | 1,645 | 637 |
129067066
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import os
from datetime import datetime, timedelta
import pyedflib
import numpy as np
import pandas as pd
from scipy import signal
from scipy.signal import butter, lfilter
import matplotlib.pyplot as plt
from datetime import datetime, timedelta
import os
from math import floor
import copy
def create_dir(directory_path):
"""Method to create a directory. Returns True if the directory already exists."""
if os.path.exists(directory_path):
return True
else:
os.makedirs(directory_path)
return False
def round_down(num, divisor):
"""Method to round down a number"""
return num - (num % divisor)
def get_time(datetime_string):
"""Method to convert a string to a datetime object."""
time = 0
try:
time = datetime.strptime(datetime_string, "%H:%M:%S")
except ValueError:
datetime_string = " " + datetime_string
if " 24" in datetime_string:
datetime_string = datetime_string.replace(" 24", "23")
time = datetime.strptime(datetime_string, "%H:%M:%S")
time += timedelta(hours=1)
else:
datetime_string = datetime_string.replace(" 25", "23")
time = datetime.strptime(datetime_string, "%H:%M:%S")
time += timedelta(hours=2)
return time
def extract_interval_data(
patient,
data_dir,
extract_ictal_samples=True,
extract_preictal_samples=True,
ictal_interval_padding_duration=32,
seizure_occurance_period=30,
seizure_prediction_horizon=5,
):
"""Method to extract interval patient data."""
patient_summary = open(
os.path.join(data_dir, "chb%02d" % patient, "chb%02d-summary.txt" % patient),
"r",
)
interictal_intervals = []
interictal_files = []
ictal_intervals = []
ictal_files = []
preictal_intervals = []
preictal_files = []
line = patient_summary.readline()
start_time = datetime.min
old_time = datetime.min
line_number = 0
while line:
line_data = line.split(":")
if line_data[0] == "File Name":
file_name = line_data[1].strip()
s = get_time(patient_summary.readline().split(": ")[1].strip())
if line_number == 0:
start_time = s
while s < old_time:
s += timedelta(hours=24)
old_time = s
end_time_file = get_time(patient_summary.readline().split(": ")[1].strip())
while end_time_file < old_time:
end_time_file = end_time_file + timedelta(hours=24)
old_time = end_time_file
n_seizures = int(patient_summary.readline().split(": ")[1])
if n_seizures == 0:
# Extract interictal interval data
interictal_intervals.append([s, end_time_file])
interictal_files.append([s, end_time_file, file_name])
else:
# Extract ictal and preictal interval data
for i in range(0, n_seizures):
seconds_start = int(
patient_summary.readline().split(": ")[1].split(" ")[0]
)
seconds_end = int(
patient_summary.readline().split(": ")[1].split(" ")[0]
)
if extract_ictal_samples:
# Extract ictal interval data
interval_start = s + timedelta(seconds=seconds_start)
if (
len(ictal_intervals) == 0 or interval_start > datetime.min
) and interval_start - start_time > timedelta(minutes=20):
interval_end = s + timedelta(seconds=seconds_end)
ictal_intervals.append(
[
interval_start
- timedelta(
seconds=ictal_interval_padding_duration
),
interval_end
+ timedelta(
seconds=ictal_interval_padding_duration
),
]
)
ictal_files.append([s, end_time_file, file_name])
if extract_preictal_samples:
# Extract preictal interval data
interval_start = (
s
+ timedelta(seconds=seconds_start)
- timedelta(
minutes=seizure_prediction_horizon
+ seizure_occurance_period
)
)
if (
len(preictal_intervals) == 0
or interval_start > datetime.min
) and interval_start - start_time > timedelta(minutes=20):
interval_end = interval_start + timedelta(
minutes=seizure_occurance_period
)
preictal_intervals.append([interval_start, interval_end])
preictal_files.append([s, end_time_file, file_name])
line = patient_summary.readline()
line_number += 1
patient_summary.close()
return (
interictal_intervals,
interictal_files,
ictal_intervals,
ictal_files,
preictal_intervals,
preictal_files,
)
def load_patient_data(patient, file, data_dir):
"""Method to load patient data."""
f = pyedflib.EdfReader("%schb%02d/%s" % (data_dir, patient, file))
n = f.signals_in_file
signals = np.zeros((n, f.getNSamples()[0]))
for i in np.arange(n):
signals[i, :] = f.readSignal(i)
return signals
def extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
interval_start,
interval_end,
segment_index,
n_channels,
):
"""Method to extract batch samples from specified intervals."""
start = 0
if file_start < interval_start:
start = (interval_start - file_start).seconds * sample_rate
if file_end <= interval_end:
end = -1
data = load_patient_data(patient, file[2], data_dir)[:, start:]
else:
end = ((interval_end - file_start).seconds * sample_rate) + 1
data = load_patient_data(patient, file[2], data_dir)[:, start : end + 1]
if (data.shape[0] >= n_channels) and (data.shape[1] >= sample_rate * window_size):
truncated_len = round_down(data.shape[1], sample_rate * window_size)
return (
np.array(
np.split(
data[0:n_channels, 0:truncated_len],
truncated_len / (sample_rate * window_size),
axis=1,
)
).swapaxes(0, 1),
segment_index,
)
else:
return np.array([]), segment_index
def extract_batches(
patient,
file,
data_dir,
segment_index,
intervals,
sample_rate,
window_size,
n_channels,
):
"""Method to extract batches."""
file_start = file[0]
file_end = file[1]
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
while file_start > interval_end and segment_index < len(intervals) - 1:
segment_index += 1
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
if (interval_end - interval_start).seconds >= window_size:
return extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
interval_start,
interval_end,
segment_index,
n_channels,
)
else:
return np.array([]), segment_index
def gen_synthetic_batches(
patient,
file,
data_dir,
segment_index,
intervals,
sample_rate,
window_size,
stride_len,
n_channels,
):
"""Method to generate synthetic batches."""
file_start = file[0]
file_end = file[1]
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
while file_start > interval_end and segment_index < len(intervals) - 1:
segment_index += 1
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
if (interval_end - interval_start).seconds > window_size:
synthetic_batches = np.array([]).reshape(
n_channels, 0, sample_rate * window_size
)
synthetic_interval_start = interval_start + timedelta(seconds=stride_len)
synthetic_interval_end = synthetic_interval_start + timedelta(
seconds=window_size
)
while synthetic_interval_end < interval_end:
extracted_batches = extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
synthetic_interval_start,
synthetic_interval_end,
segment_index,
n_channels,
)[0]
if extracted_batches.size > 0:
synthetic_batches = np.concatenate(
(synthetic_batches, extracted_batches), axis=1
)
synthetic_interval_start += timedelta(seconds=stride_len)
synthetic_interval_end += timedelta(seconds=stride_len)
return synthetic_batches, segment_index
else:
return np.array([]), segment_index
os.path.exists("/kaggle/working/processed_data")
n_channels = 22
sample_rate = 256 # Sample rate (Hz)
window_size = 64 # Window size (seconds)
# Stride length (seconds) used to generate synthetic preictal and ictal samples
stride_len = 32
# Data directory path
# data_dir = "/scratch/jcu/cl/CHBMIT/chb-mit-scalp-eeg-database-1.0.0/"
data_dir = "/kaggle/input/chb01-21/chbmit/"
processed_data_dir = (
"/kaggle/working/processed_data/" # Processed data output directory path
)
patients = np.arange(1, 24)
# Remove patients 4, 6, 7, 12, and 20, as their records contain anomalous data
patients = np.delete(patients, [3, 5, 6, 11, 19])
patients = [1] # TEMP
ictal_interval_padding_duration = 32
# ------------------------------------------------------------------------------
seizure_occurance_period = 30 # Seizure occurrence period (minutes)
seizure_prediction_horizon = 5 # Seizure prediction horizon (minutes)
# -----------------------------------------------------------------------
# ------------------------------------------------------------------------------
extract_ictal_samples = False
extract_preictal_samples = True
generate_synthetic_samples = False
# ------------------------------------------------------------------------------
if __name__ == "__main__":
for patient in patients:
try:
print("Patient: %02d" % patient)
create_dir(processed_data_dir)
(
interictal_intervals,
interictal_files,
ictal_intervals,
ictal_files,
preictal_intervals,
preictal_files,
) = extract_interval_data(
patient,
data_dir,
extract_ictal_samples,
extract_preictal_samples,
ictal_interval_padding_duration,
seizure_occurance_period,
seizure_prediction_horizon,
)
if patient == 19:
# Disregard the first seizure of patient 19 because it is not considered
preictal_intervals.pop(0)
interictal_segment_index = 0
interictal_data = np.array([]).reshape(
n_channels, 0, sample_rate * window_size
)
if extract_ictal_samples:
ictal_segment_index = 0
synthetic_ictal_segment_index = 0
ictal_data = copy.deepcopy(interictal_data)
synthetic_ictal_data = copy.deepcopy(interictal_data)
if extract_preictal_samples:
preictal_segment_index = 0
synthetic_preictal_segment_index = 0
preictal_data = copy.deepcopy(interictal_data)
synthetic_preictal_data = copy.deepcopy(interictal_data)
# Extract interictal samples (batches)
for file in interictal_files:
data, interictal_segment_index = extract_batches(
patient,
file,
data_dir,
interictal_segment_index,
interictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
interictal_data = np.concatenate((interictal_data, data), axis=1)
print("Interictal: ", interictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_interictal.npy" % patient,
),
interictal_data,
)
del interictal_data
if extract_ictal_samples:
# Extract ictal samples (batches)
for file in ictal_files:
data, ictal_segment_index = extract_batches(
patient,
file,
data_dir,
ictal_segment_index,
ictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
ictal_data = np.concatenate((ictal_data, data), axis=1)
print("Ictal: ", ictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_ictal.npy" % patient,
),
ictal_data,
)
del ictal_data
if generate_synthetic_samples:
# Generate synthetic ictal samples (batches)
for file in ictal_files:
data, synthetic_ictal_segment_index = gen_synthetic_batches(
patient,
file,
data_dir,
synthetic_ictal_segment_index,
ictal_intervals,
sample_rate,
window_size,
stride_len,
n_channels,
)
if data.size > 0:
synthetic_ictal_data = np.concatenate(
(synthetic_ictal_data, data), axis=1
)
print("Synthetic Ictal: ", synthetic_ictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_synthetic_ictal.npy" % patient,
),
synthetic_ictal_data,
)
del synthetic_ictal_data
if extract_preictal_samples:
# Extract preictal samples (batches)
for file in preictal_files:
data, preictal_segment_index = extract_batches(
patient,
file,
data_dir,
preictal_segment_index,
preictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
preictal_data = np.concatenate((preictal_data, data), axis=1)
print("Preictal: ", preictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_preictal.npy" % patient,
),
preictal_data,
)
del preictal_data
if generate_synthetic_samples:
# Generate synthetic preictal samples (batches)
for file in preictal_files:
data, synthetic_preictal_segment_index = gen_synthetic_batches(
patient,
file,
data_dir,
synthetic_preictal_segment_index,
preictal_intervals,
sample_rate,
window_size,
stride_len,
n_channels,
)
if data.size > 0:
synthetic_preictal_data = np.concatenate(
(synthetic_preictal_data, data), axis=1
)
print("Synthetic Preictal: ", synthetic_preictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_synthetic_preictal.npy" % patient,
),
synthetic_preictal_data,
)
del synthetic_preictal_data
except Exception as e:
print("Patient: %02d Failed" % patient)
print(e)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/067/129067066.ipynb
| null | null |
[{"Id": 129067066, "ScriptId": 38216406, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14975555, "CreationDate": "05/10/2023 17:54:44", "VersionNumber": 2.0, "Title": "parallell cnn", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 530.0, "LinesInsertedFromPrevious": 512.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 18.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import os
from datetime import datetime, timedelta
import pyedflib
import numpy as np
import pandas as pd
from scipy import signal
from scipy.signal import butter, lfilter
import matplotlib.pyplot as plt
from datetime import datetime, timedelta
import os
from math import floor
import copy
def create_dir(directory_path):
"""Method to create a directory. Returns True if the directory already exists."""
if os.path.exists(directory_path):
return True
else:
os.makedirs(directory_path)
return False
def round_down(num, divisor):
"""Method to round down a number"""
return num - (num % divisor)
def get_time(datetime_string):
"""Method to convert a string to a datetime object."""
time = 0
try:
time = datetime.strptime(datetime_string, "%H:%M:%S")
except ValueError:
datetime_string = " " + datetime_string
if " 24" in datetime_string:
datetime_string = datetime_string.replace(" 24", "23")
time = datetime.strptime(datetime_string, "%H:%M:%S")
time += timedelta(hours=1)
else:
datetime_string = datetime_string.replace(" 25", "23")
time = datetime.strptime(datetime_string, "%H:%M:%S")
time += timedelta(hours=2)
return time
def extract_interval_data(
patient,
data_dir,
extract_ictal_samples=True,
extract_preictal_samples=True,
ictal_interval_padding_duration=32,
seizure_occurance_period=30,
seizure_prediction_horizon=5,
):
"""Method to extract interval patient data."""
patient_summary = open(
os.path.join(data_dir, "chb%02d" % patient, "chb%02d-summary.txt" % patient),
"r",
)
interictal_intervals = []
interictal_files = []
ictal_intervals = []
ictal_files = []
preictal_intervals = []
preictal_files = []
line = patient_summary.readline()
start_time = datetime.min
old_time = datetime.min
line_number = 0
while line:
line_data = line.split(":")
if line_data[0] == "File Name":
file_name = line_data[1].strip()
s = get_time(patient_summary.readline().split(": ")[1].strip())
if line_number == 0:
start_time = s
while s < old_time:
s += timedelta(hours=24)
old_time = s
end_time_file = get_time(patient_summary.readline().split(": ")[1].strip())
while end_time_file < old_time:
end_time_file = end_time_file + timedelta(hours=24)
old_time = end_time_file
n_seizures = int(patient_summary.readline().split(": ")[1])
if n_seizures == 0:
# Extract interictal interval data
interictal_intervals.append([s, end_time_file])
interictal_files.append([s, end_time_file, file_name])
else:
# Extract ictal and preictal interval data
for i in range(0, n_seizures):
seconds_start = int(
patient_summary.readline().split(": ")[1].split(" ")[0]
)
seconds_end = int(
patient_summary.readline().split(": ")[1].split(" ")[0]
)
if extract_ictal_samples:
# Extract ictal interval data
interval_start = s + timedelta(seconds=seconds_start)
if (
len(ictal_intervals) == 0 or interval_start > datetime.min
) and interval_start - start_time > timedelta(minutes=20):
interval_end = s + timedelta(seconds=seconds_end)
ictal_intervals.append(
[
interval_start
- timedelta(
seconds=ictal_interval_padding_duration
),
interval_end
+ timedelta(
seconds=ictal_interval_padding_duration
),
]
)
ictal_files.append([s, end_time_file, file_name])
if extract_preictal_samples:
# Extract preictal interval data
interval_start = (
s
+ timedelta(seconds=seconds_start)
- timedelta(
minutes=seizure_prediction_horizon
+ seizure_occurance_period
)
)
if (
len(preictal_intervals) == 0
or interval_start > datetime.min
) and interval_start - start_time > timedelta(minutes=20):
interval_end = interval_start + timedelta(
minutes=seizure_occurance_period
)
preictal_intervals.append([interval_start, interval_end])
preictal_files.append([s, end_time_file, file_name])
line = patient_summary.readline()
line_number += 1
patient_summary.close()
return (
interictal_intervals,
interictal_files,
ictal_intervals,
ictal_files,
preictal_intervals,
preictal_files,
)
def load_patient_data(patient, file, data_dir):
"""Method to load patient data."""
f = pyedflib.EdfReader("%schb%02d/%s" % (data_dir, patient, file))
n = f.signals_in_file
signals = np.zeros((n, f.getNSamples()[0]))
for i in np.arange(n):
signals[i, :] = f.readSignal(i)
return signals
def extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
interval_start,
interval_end,
segment_index,
n_channels,
):
"""Method to extract batch samples from specified intervals."""
start = 0
if file_start < interval_start:
start = (interval_start - file_start).seconds * sample_rate
if file_end <= interval_end:
end = -1
data = load_patient_data(patient, file[2], data_dir)[:, start:]
else:
end = ((interval_end - file_start).seconds * sample_rate) + 1
data = load_patient_data(patient, file[2], data_dir)[:, start : end + 1]
if (data.shape[0] >= n_channels) and (data.shape[1] >= sample_rate * window_size):
truncated_len = round_down(data.shape[1], sample_rate * window_size)
return (
np.array(
np.split(
data[0:n_channels, 0:truncated_len],
truncated_len / (sample_rate * window_size),
axis=1,
)
).swapaxes(0, 1),
segment_index,
)
else:
return np.array([]), segment_index
def extract_batches(
patient,
file,
data_dir,
segment_index,
intervals,
sample_rate,
window_size,
n_channels,
):
"""Method to extract batches."""
file_start = file[0]
file_end = file[1]
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
while file_start > interval_end and segment_index < len(intervals) - 1:
segment_index += 1
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
if (interval_end - interval_start).seconds >= window_size:
return extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
interval_start,
interval_end,
segment_index,
n_channels,
)
else:
return np.array([]), segment_index
def gen_synthetic_batches(
patient,
file,
data_dir,
segment_index,
intervals,
sample_rate,
window_size,
stride_len,
n_channels,
):
"""Method to generate synthetic batches."""
file_start = file[0]
file_end = file[1]
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
while file_start > interval_end and segment_index < len(intervals) - 1:
segment_index += 1
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
if (interval_end - interval_start).seconds > window_size:
synthetic_batches = np.array([]).reshape(
n_channels, 0, sample_rate * window_size
)
synthetic_interval_start = interval_start + timedelta(seconds=stride_len)
synthetic_interval_end = synthetic_interval_start + timedelta(
seconds=window_size
)
while synthetic_interval_end < interval_end:
extracted_batches = extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
synthetic_interval_start,
synthetic_interval_end,
segment_index,
n_channels,
)[0]
if extracted_batches.size > 0:
synthetic_batches = np.concatenate(
(synthetic_batches, extracted_batches), axis=1
)
synthetic_interval_start += timedelta(seconds=stride_len)
synthetic_interval_end += timedelta(seconds=stride_len)
return synthetic_batches, segment_index
else:
return np.array([]), segment_index
os.path.exists("/kaggle/working/processed_data")
n_channels = 22
sample_rate = 256 # Sample rate (Hz)
window_size = 64 # Window size (seconds)
# Stride length (seconds) used to generate synthetic preictal and ictal samples
stride_len = 32
# Data directory path
# data_dir = "/scratch/jcu/cl/CHBMIT/chb-mit-scalp-eeg-database-1.0.0/"
data_dir = "/kaggle/input/chb01-21/chbmit/"
processed_data_dir = (
"/kaggle/working/processed_data/" # Processed data output directory path
)
patients = np.arange(1, 24)
# Remove patients 4, 6, 7, 12, and 20, as their records contain anomalous data
patients = np.delete(patients, [3, 5, 6, 11, 19])
patients = [1] # TEMP
ictal_interval_padding_duration = 32
# ------------------------------------------------------------------------------
seizure_occurance_period = 30 # Seizure occurrence period (minutes)
seizure_prediction_horizon = 5 # Seizure prediction horizon (minutes)
# -----------------------------------------------------------------------
# ------------------------------------------------------------------------------
extract_ictal_samples = False
extract_preictal_samples = True
generate_synthetic_samples = False
# ------------------------------------------------------------------------------
if __name__ == "__main__":
for patient in patients:
try:
print("Patient: %02d" % patient)
create_dir(processed_data_dir)
(
interictal_intervals,
interictal_files,
ictal_intervals,
ictal_files,
preictal_intervals,
preictal_files,
) = extract_interval_data(
patient,
data_dir,
extract_ictal_samples,
extract_preictal_samples,
ictal_interval_padding_duration,
seizure_occurance_period,
seizure_prediction_horizon,
)
if patient == 19:
# Disregard the first seizure of patient 19 because it is not considered
preictal_intervals.pop(0)
interictal_segment_index = 0
interictal_data = np.array([]).reshape(
n_channels, 0, sample_rate * window_size
)
if extract_ictal_samples:
ictal_segment_index = 0
synthetic_ictal_segment_index = 0
ictal_data = copy.deepcopy(interictal_data)
synthetic_ictal_data = copy.deepcopy(interictal_data)
if extract_preictal_samples:
preictal_segment_index = 0
synthetic_preictal_segment_index = 0
preictal_data = copy.deepcopy(interictal_data)
synthetic_preictal_data = copy.deepcopy(interictal_data)
# Extract interictal samples (batches)
for file in interictal_files:
data, interictal_segment_index = extract_batches(
patient,
file,
data_dir,
interictal_segment_index,
interictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
interictal_data = np.concatenate((interictal_data, data), axis=1)
print("Interictal: ", interictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_interictal.npy" % patient,
),
interictal_data,
)
del interictal_data
if extract_ictal_samples:
# Extract ictal samples (batches)
for file in ictal_files:
data, ictal_segment_index = extract_batches(
patient,
file,
data_dir,
ictal_segment_index,
ictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
ictal_data = np.concatenate((ictal_data, data), axis=1)
print("Ictal: ", ictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_ictal.npy" % patient,
),
ictal_data,
)
del ictal_data
if generate_synthetic_samples:
# Generate synthetic ictal samples (batches)
for file in ictal_files:
data, synthetic_ictal_segment_index = gen_synthetic_batches(
patient,
file,
data_dir,
synthetic_ictal_segment_index,
ictal_intervals,
sample_rate,
window_size,
stride_len,
n_channels,
)
if data.size > 0:
synthetic_ictal_data = np.concatenate(
(synthetic_ictal_data, data), axis=1
)
print("Synthetic Ictal: ", synthetic_ictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_synthetic_ictal.npy" % patient,
),
synthetic_ictal_data,
)
del synthetic_ictal_data
if extract_preictal_samples:
# Extract preictal samples (batches)
for file in preictal_files:
data, preictal_segment_index = extract_batches(
patient,
file,
data_dir,
preictal_segment_index,
preictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
preictal_data = np.concatenate((preictal_data, data), axis=1)
print("Preictal: ", preictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_preictal.npy" % patient,
),
preictal_data,
)
del preictal_data
if generate_synthetic_samples:
# Generate synthetic preictal samples (batches)
for file in preictal_files:
data, synthetic_preictal_segment_index = gen_synthetic_batches(
patient,
file,
data_dir,
synthetic_preictal_segment_index,
preictal_intervals,
sample_rate,
window_size,
stride_len,
n_channels,
)
if data.size > 0:
synthetic_preictal_data = np.concatenate(
(synthetic_preictal_data, data), axis=1
)
print("Synthetic Preictal: ", synthetic_preictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_synthetic_preictal.npy" % patient,
),
synthetic_preictal_data,
)
del synthetic_preictal_data
except Exception as e:
print("Patient: %02d Failed" % patient)
print(e)
| false | 0 | 4,303 | 0 | 4,303 | 4,303 |
||
129067634
|
<jupyter_start><jupyter_text>pretrainedmodels
### Pretrained Models in Pytorch
Github repo: https://github.com/Cadene/pretrained-models.pytorch
Version: 0.7.4
Original Author: Cadene
License: https://github.com/Cadene/pretrained-models.pytorch/blob/master/LICENSE.txt
Kaggle dataset identifier: pretrainedmodels
<jupyter_script># ## summary
# **This notebook is written for new kaggler**
# **I can't guarantee that all comments are interpreted correctly, so please point out if there are mistakes**
# Original code is here [https://www.kaggle.com/code/tanakar/2-5d-segmentaion-baseline-training](http://)
# * 2.5d segmentation
# * segmentation_models_pytorch
# * Unet
# * use only 6 slices in the middle
# * slide inference
# sklearn.metrics.classification is an evaluation metrics module for classification problems in the scikit-learn library.This module provides a series of functions to calculate the accuracy, precision, recall, F1 value and other metrics of a classification model to evaluate the performance of the model.These metrics can help us understand the classification ability of the model, so that we can optimize the parameters and algorithms of the model and improve the prediction accuracy of the model.
# Pickle is a module in Python for serializing and deserializing Python objects.With Pickle, we can convert a Python object to a byte stream and then save it to a file or transfer it over the network.Conversely, we can also deserialize byte streams to Python objects.Pickle is a persistent storage method in Python that makes it easy to save and restore data.
# Autocast and GradScaler are both tools in PyTorch for accelerating model training and reducing memory footprint.
# Autocast is an automatic mixed precision tool that automatically converts floating point numbers to half-precision floating point numbers during the forward and backward propagation of the model. This helps reduce GPU memory usage and speed up model training. When using Autocast, the model and optimizer need to be wrapped in the torch.cuda.amp.autocast() context manager.GradScaler is a gradient scaling tool for scaling the value of the gradient during training. This helps to solve the problem of disappearing or exploding gradients and improves model stability and training results. When using GradScaler, the gradient needs to be multiplied by a scaling factor before back-propagation is performed. The scaling factor can be dynamically adjusted according to the value of the gradient to ensure the stability of the gradient.
from sklearn.metrics import roc_auc_score, accuracy_score, f1_score, log_loss
import pickle
from torch.utils.data import DataLoader
from torch.cuda.amp import autocast, GradScaler
import warnings
import sys
import pandas as pd
import os
import gc
import sys
import math
import time
import random
import shutil
from pathlib import Path
from contextlib import contextmanager
from collections import defaultdict, Counter
import cv2
import scipy as sp
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from tqdm.auto import tqdm
from functools import partial
import argparse
import importlib
import torch
import torch.nn as nn
from torch.optim import Adam, SGD, AdamW
import datetime
# sys.path.append('/kaggle/input/pretrainedmodels/pretrainedmodels-0.7.4')
# sys.path.append('/kaggle/input/efficientnet-pytorch/EfficientNet-PyTorch-master')
# sys.path.append('/kaggle/input/timm-pytorch-image-models/pytorch-image-models-master')
# sys.path.append('/kaggle/input/segmentation-models-pytorch/segmentation_models.pytorch-master')
import segmentation_models_pytorch as smp
# For segmentation_models_pytorch, it is a PyTorch-based deep learning library for image segmentation tasks.
# It supports many popular segmentation models, such as UNet, LinkNet, FPN, etc., and provides many pre-trained models and datasets to facilitate users to quickly build and train their own models.
import numpy as np
from torch.utils.data import DataLoader, Dataset
import cv2
import torch
import os
import albumentations as A
from albumentations.pytorch import ToTensorV2
from albumentations import ImageOnlyTransform
# ## config
class CFG:
# ============== comp exp name =============
comp_name = "vesuvius"
# comp_dir_path = './'
comp_dir_path = "/kaggle/input/"
comp_folder_name = "vesuvius-challenge-ink-detection"
# comp_dataset_path = f'{comp_dir_path}datasets/{comp_folder_name}/'
comp_dataset_path = f"{comp_dir_path}{comp_folder_name}/"
exp_name = "vesuvius_2d_slide_exp002"
# ============== pred target =============
target_size = 1
# ============== model cfg =============
# Image segmentation model Pre-training parameters for convolutional networks
model_name = "Unet"
# backbone = 'efficientnet-b0'
backbone = "resnext101_32x4d"
# backbone = 'resnext50_32x4d'
# backbone = 'resnet50'
# There are 65 "channels",
# a three-dimensional image of a certain dimension split into multiple two-dimensional images,
# the ink will be immersed in the deeper the image, the larger the information more complete
in_chans = 3 # 64
# ============== training cfg =============
# Size modification of input image or mask image
size = 224
tile_size = 224
# The number of small images cut out and the position of each small image can be controlled by setting CFG.stride.
stride = tile_size // 2
train_batch_size = 16 # 32
valid_batch_size = train_batch_size * 2
use_amp = True
# Learning rate regulator
scheduler = "GradualWarmupSchedulerV2"
# scheduler = 'CosineAnnealingLR'
epochs = 10 # 30
"""
warmup_factor is a scaling factor that controls the rate of learning rate increase. It is usually used at the beginning of training to accelerate the learning rate increase if the model weights are not yet accurate enough.
lr is the learning rate size, and in this equation, the learning rate is divided by 10 with the effect of warmup_factor.
This is because at the beginning of the training, the learning rate should be relatively small so that the model can converge better, and as the training proceeds, the learning rate will gradually increase so that the weight space can be explored better.
Thus, with this formula, a relatively small learning rate can be used at the beginning of training and gradually increased to the appropriate size.
"""
# adamW
warmup_factor = 10
# lr = 1e-4 / warmup_factor
lr = 1e-4 / warmup_factor
# ============== fold =============
# k-fold cross-validation This method has the advantage of making better use of the data while reducing errors due to the chance of data division.
# The disadvantage is that k-times training and validation are required and the computational cost is high.
# This seems to mean that each of the three folders is used as a validation set
valid_id = 1
"""
In this example, metric_direction is set to 'maximize', indicating that the metric we want to optimize should be as large as possible.
"""
# objective_cv = 'binary' # 'binary', 'multiclass', 'regression'
metric_direction = "maximize" # maximize, 'minimize'
# metrics = 'dice_coef'
# ============== fixed =============
pretrained = True
inf_weight = "best" # 'best'
min_lr = 1e-6
weight_decay = 1e-6
max_grad_norm = 1000
print_freq = 50
num_workers = 10
# Fixed seeds make code reproducible
# Randomness in deep learning is caused by factors such as weight initialization, random sampling, etc. These randomness can make the model more expressive and generalizable.
# However, since deep learning models are usually very large, their training requires a lot of time and computational resources. Therefore, it is useful to fix random seeds in order to make the experiments reproducible.
# Using a fixed random seed ensures that the same sequence of random numbers is generated using the same random number generator each time the experiment is run.
# This allows researchers to get the same results when running experiments on different machines, thus facilitating the comparison and validation of results.
# In addition, using a fixed random seed also makes the model's behavior more predictable during training, thus helping researchers to better understand the model's performance and behavior.
seed = 42
# ============== set dataset path =============
print("set dataset path")
outputs_path = f"/kaggle/working/outputs/{comp_name}/{exp_name}/"
submission_dir = outputs_path + "submissions/"
submission_path = submission_dir + f"submission_{exp_name}.csv"
model_dir = outputs_path + f"{comp_name}-models/"
figures_dir = outputs_path + "figures/"
log_dir = outputs_path + "logs/"
log_path = log_dir + f"{exp_name}.txt"
# ============== augmentation =============
# Data Enhancement
train_aug_list = [
# A.RandomResizedCrop(
# size, size, scale=(0.85, 1.0)),
A.Resize(size, size),
A.HorizontalFlip(p=0.5),
A.VerticalFlip(p=0.5),
A.RandomBrightnessContrast(p=0.75),
A.ShiftScaleRotate(p=0.75),
A.OneOf(
[
A.GaussNoise(var_limit=[10, 50]),
A.GaussianBlur(),
A.MotionBlur(),
],
p=0.4,
),
A.GridDistortion(num_steps=5, distort_limit=0.3, p=0.5),
A.CoarseDropout(
max_holes=1,
max_width=int(size * 0.3),
max_height=int(size * 0.3),
mask_fill_value=0,
p=0.5,
),
# A.Cutout(max_h_size=int(size * 0.6),
# max_w_size=int(size * 0.6), num_holes=1, p=1.0),
A.Normalize(mean=[0] * in_chans, std=[1] * in_chans),
ToTensorV2(transpose_mask=True),
]
valid_aug_list = [
A.Resize(size, size),
A.Normalize(mean=[0] * in_chans, std=[1] * in_chans),
ToTensorV2(transpose_mask=True),
]
# ## helper
# Computes and stores the average and current value
# Specifically, it serves to call the update method to update the value of the instance each time the average needs to be calculated, and to call the avg method to return the average when the average needs to be calculated.
class AverageMeter(object):
"""Computes and stores the average and current value"""
# Initialize the property values of the instance
def __init__(self):
self.reset()
# Reset the property value of the instance to 0
def reset(self):
self.val = 0
self.avg = 0
self.sum = 0
self.count = 0
# Update the property value of the instance to the given value
def update(self, val, n=1):
self.val = val
self.sum += val * n
self.count += n
self.avg = self.sum / self.count
# # log
# accepts one argument log_file log file path
# The function initializes a logger object, which is used to record logging information when the program is running. This is implemented as follows:
def init_logger(log_file):
from logging import getLogger, INFO, FileHandler, Formatter, StreamHandler
logger = getLogger(__name__)
logger.setLevel(INFO)
handler1 = StreamHandler()
handler1.setFormatter(Formatter("%(message)s"))
handler2 = FileHandler(filename=log_file)
handler2.setFormatter(Formatter("%(message)s"))
logger.addHandler(handler1)
logger.addHandler(handler2)
return logger
def set_seed(seed=None, cudnn_deterministic=True):
if seed is None:
seed = 42
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = cudnn_deterministic
torch.backends.cudnn.benchmark = False
# This function creates these directories using the os.makedirs() method and does not overwrite the directories if they already exist, i.e. the exist_ok=True argument means that no errors will be reported.
# The purpose of this function is to create these directories when needed for use in subsequent code.
def make_dirs(cfg):
for dir in [cfg.model_dir, cfg.figures_dir, cfg.submission_dir, cfg.log_dir]:
os.makedirs(dir, exist_ok=True)
# # Initialization functions
def cfg_init(cfg, mode="train"):
set_seed(cfg.seed)
# set_env_name()
# set_dataset_path(cfg)
if mode == "train":
make_dirs(cfg)
cfg_init(CFG)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
Logger = init_logger(log_file=CFG.log_path)
Logger.info("\n\n-------- exp_info -----------------")
# Logger.info(datetime.datetime.now().strftime('%Y年%m月%d日 %H:%M:%S'))
# ## image, mask
# The purpose of this code is to find a window of length CFG.in_chans, so that the subscript of the center point of the window is mid.
# Specifically, first divide mid by 2, then subtract half of CFG.in_chans from the result to get the start subscript of the window, then add half of CFG.in_chans to mid to get the end subscript of the window.
# Then add half of CFG.in_chans to mid to get the end of the window.
# Finally, use the range function to generate a sequence of integers idxs from start to end-1.
#
# **Set the number of images to be read from 0-65, and adjust the number of input "channels" by changing in_chans**
def read_image_mask(fragment_id):
images = []
# idxs = range(65)
mid = 65 // 2
start = mid - CFG.in_chans // 2
end = mid + CFG.in_chans // 2
idxs = range(start, end)
for i in tqdm(idxs):
image = cv2.imread(
CFG.comp_dataset_path + f"train/{fragment_id}/surface_volume/{i:02}.tif", 0
)
pad0 = CFG.tile_size - image.shape[0] % CFG.tile_size
pad1 = CFG.tile_size - image.shape[1] % CFG.tile_size
image = np.pad(image, [(0, pad0), (0, pad1)], constant_values=0)
images.append(image)
images = np.stack(images, axis=2)
mask = cv2.imread(CFG.comp_dataset_path + f"train/{fragment_id}/inklabels.png", 0)
mask = np.pad(mask, [(0, pad0), (0, pad1)], constant_values=0)
mask = mask.astype("float32")
mask /= 255.0
return images, mask
def get_train_valid_dataset():
train_images = []
train_masks = []
valid_images = []
valid_masks = []
valid_xyxys = []
for fragment_id in range(1, 4):
image, mask = read_image_mask(fragment_id)
"""
This code is generating a set of image crop coordinates that are used to split a large size image into smaller pieces for easier processing. Where image is the original image and CFG is a set of constant parameters.
Specifically, the parameters of the range function in the code set the step size and range of the crop coordinates.
image.shape[0] and image.shape[1] denote the height and width of the image, respectively. Subtracting CFG.tile_size is to ensure that the segmented chunks are all squares with CFG.tile_size as the side length.
The final x1_list and y1_list are the coordinates of all the generated images, which are used for subsequent processing.
"""
x1_list = list(range(0, image.shape[1] - CFG.tile_size + 1, CFG.stride))
y1_list = list(range(0, image.shape[0] - CFG.tile_size + 1, CFG.stride))
# where y1_list and x1_list are the list of starting coordinates in the width and height directions of the large image, respectively, and CFG.tile_size is the size of each small image block.
# If fragment_id is equal to CFG.valid_id, the current image block and mask are stored in the list of valid_images and valid_masks
# and store the start and end coordinates corresponding to the current image block into the valid_xyxys list; otherwise store them into the train_images and train_masks lists.
for y1 in y1_list:
for x1 in x1_list:
y2 = y1 + CFG.tile_size
x2 = x1 + CFG.tile_size
# xyxys.append((x1, y1, x2, y2))
# Place the set folder with the corresponding label in the validation data set, folder 1 or folder 2 or folder 3
if fragment_id == CFG.valid_id:
valid_images.append(image[y1:y2, x1:x2])
valid_masks.append(mask[y1:y2, x1:x2, None])
valid_xyxys.append([x1, y1, x2, y2])
# unspecified data sets into the training set, 2 training sets 1 validation set
else:
train_images.append(image[y1:y2, x1:x2])
train_masks.append(mask[y1:y2, x1:x2, None])
return train_images, train_masks, valid_images, valid_masks, valid_xyxys
(
train_images,
train_masks,
valid_images,
valid_masks,
valid_xyxys,
) = get_train_valid_dataset()
valid_xyxys = np.stack(valid_xyxys)
# ## dataset
import numpy as np
from torch.utils.data import DataLoader, Dataset
import cv2
import torch
import os
import albumentations as A
from albumentations.pytorch import ToTensorV2
from albumentations import ImageOnlyTransform
def get_transforms(data, cfg):
if data == "train":
aug = A.Compose(cfg.train_aug_list)
elif data == "valid":
aug = A.Compose(cfg.valid_aug_list)
# print(aug)
return aug
class CustomDataset(Dataset):
def __init__(self, images, cfg, labels=None, transform=None):
self.images = images
self.cfg = cfg
self.labels = labels
self.transform = transform
def __len__(self):
# return len(self.df)
return len(self.images)
def __getitem__(self, idx):
image = self.images[idx]
label = self.labels[idx]
if self.transform:
data = self.transform(image=image, mask=label)
image = data["image"]
label = data["mask"]
return image, label
# # Training set and validation set data processing
train_dataset = CustomDataset(
train_images,
CFG,
labels=train_masks,
transform=get_transforms(data="train", cfg=CFG),
)
valid_dataset = CustomDataset(
valid_images,
CFG,
labels=valid_masks,
transform=get_transforms(data="valid", cfg=CFG),
)
train_loader = DataLoader(
train_dataset,
batch_size=CFG.train_batch_size,
shuffle=True,
num_workers=CFG.num_workers,
pin_memory=True,
drop_last=True,
)
valid_loader = DataLoader(
valid_dataset,
batch_size=CFG.valid_batch_size,
shuffle=False,
num_workers=CFG.num_workers,
pin_memory=True,
drop_last=False,
)
train_dataset[0][0].shape
plot_dataset = CustomDataset(train_images, CFG, labels=train_masks)
transform = CFG.train_aug_list
transform = A.Compose(
[t for t in transform if not isinstance(t, (A.Normalize, ToTensorV2))]
)
plot_count = 0
for i in range(1000):
image, mask = plot_dataset[i]
data = transform(image=image, mask=mask)
aug_image = data["image"]
aug_mask = data["mask"]
if mask.sum() == 0:
continue
fig, axes = plt.subplots(1, 4, figsize=(15, 8))
axes[0].imshow(image[..., 0], cmap="gray")
axes[1].imshow(mask, cmap="gray")
axes[2].imshow(aug_image[..., 0], cmap="gray")
axes[3].imshow(aug_mask, cmap="gray")
plt.savefig(CFG.figures_dir + f"aug_fold_{CFG.valid_id}_{plot_count}.png")
plot_count += 1
if plot_count == 5:
break
del plot_dataset
gc.collect()
# ## model
# **Unet convolutional network construction**
# This code is initializing the image segmentation task using the Unet model from the segmentation_models_pytorch library.
class CustomModel(nn.Module):
def __init__(self, cfg, weight=None):
super().__init__()
self.cfg = cfg
# The encoder_name parameter is used to specify which pre-trained encoder model to use. Here, a pre-trained model is used, so the encoder_name parameter is specified.
# The advantage of this is that the feature extraction capability of the existing pre-trained model can be used to accelerate the training of the model and improve the accuracy of the model. Also, different pre-trained models can be selected according to actual needs to achieve better results.
# We can use the pre-training weights of the image classification model to initialize the convolutional layer of the UNet network, thus improving the performance and generalization ability of the model
# encoder_name: indicates the name of the pre-trained model used
# encoder_weights: indicates the weights of the pre-trained model used, usually imagenet is used.
# in_channels: indicates the number of channels of the input image, e.g., 3 channels for RGB images
# classes: the number of classifications, usually the number of pixel classifications for image segmentation tasks.
# activation: indicates the activation function, usually None (no activation function is used) or sigmoid (the output is between 0 and 1).
self.encoder = smp.Unet(
encoder_name=cfg.backbone,
encoder_weights=weight,
in_channels=cfg.in_chans,
classes=cfg.target_size,
activation=None,
)
# After the initialization, the input image can be fed into the model for forward propagation to obtain the corresponding segmentation results.
def forward(self, image):
output = self.encoder(image)
# output = output.squeeze(-1)
return output
# weight="imagenet"
def build_model(cfg, weight="ssl"):
print("model_name", cfg.model_name)
print("backbone", cfg.backbone)
model = CustomModel(cfg, weight)
return model
# ## scheduler
# [https://www.kaggle.com/code/underwearfitting/single-fold-training-of-resnet200d-lb0-965](http://)
# Its main role is to gradually increase the learning rate at the beginning of training to help the model converge faster. It is a subclass of the GradualWarmupScheduler class with the addition of an after_scheduler parameter for using other learning rate scheduling methods after the warm-up is over.
# In each epoch, the get_lr() method will return the current learning rate.
# At the beginning of training, the learning rate will be gradually increased from the initial value to the maximum value, after which the learning rate returned by after_scheduler (another learning rate update method) will be used. If after_scheduler is not provided, the current learning rate will continue to be used.
#
import torch.nn as nn
import torch
import math
import time
import numpy as np
import torch
from torch.optim.lr_scheduler import (
CosineAnnealingWarmRestarts,
CosineAnnealingLR,
ReduceLROnPlateau,
)
from warmup_scheduler import GradualWarmupScheduler
class GradualWarmupSchedulerV2(GradualWarmupScheduler):
def __init__(self, optimizer, multiplier, total_epoch, after_scheduler=None):
super(GradualWarmupSchedulerV2, self).__init__(
optimizer, multiplier, total_epoch, after_scheduler
)
def get_lr(self):
if self.last_epoch > self.total_epoch:
if self.after_scheduler:
if not self.finished:
self.after_scheduler.base_lrs = [
base_lr * self.multiplier for base_lr in self.base_lrs
]
self.finished = True
return self.after_scheduler.get_lr()
return [base_lr * self.multiplier for base_lr in self.base_lrs]
if self.multiplier == 1.0:
return [
base_lr * (float(self.last_epoch) / self.total_epoch)
for base_lr in self.base_lrs
]
else:
return [
base_lr
* ((self.multiplier - 1.0) * self.last_epoch / self.total_epoch + 1.0)
for base_lr in self.base_lrs
]
# This code is a function to get the learning rate scheduler with input parameters of configuration (cfg) and optimizer (optimizer). Two learning rate schedulers are used in this function, CosineAnnealingLR and GradualWarmupSchedulerV2.
# Among them, CosineAnnealingLR is a cosine annealing learning rate scheduler, whose function is to gradually reduce the learning rate during training to make the model converge more stably.
# GradualWarmupSchedulerV2 is a learning rate preheating scheduler, whose role is to gradually increase the learning rate at the beginning of training to avoid the model from falling into a local optimum solution at the beginning and failing to jump out.
# In this function, we first define a CosineAnnealingLR scheduler, setting it to the total number of training rounds (cfg.epochs) as cycles and a minimum learning rate of 1e-7.
# Then, we use this scheduler as the after_scheduler parameter of GradualWarmupSchedulerV2, and also use it as the parameter of the input optimizer, multiplier indicates the learning rate multiplier during warm-up, and total_epoch indicates the number of warm-up rounds.
# Finally, the GradualWarmupSchedulerV2 scheduler is returned as the output.
def get_scheduler(cfg, optimizer):
scheduler_cosine = torch.optim.lr_scheduler.CosineAnnealingLR(
optimizer, cfg.epochs, eta_min=1e-7
)
scheduler = GradualWarmupSchedulerV2(
optimizer, multiplier=10, total_epoch=1, after_scheduler=scheduler_cosine
)
return scheduler
def scheduler_step(scheduler, avg_val_loss, epoch):
scheduler.step(epoch)
# This line of code uses the AdamW optimizer, which takes the parameters of the model as input and uses the learning rate of lr from CFG. AdamW is a variant of the Adam optimizer that is used to update the model parameters when training a neural network.
# It updates the parameters by weighted averaging the gradients of the parameters to minimize the loss function.AdamW also uses a regularization method called weight decay to prevent the model from overfitting the training data.
model = build_model(CFG)
model.to(device)
optimizer = AdamW(model.parameters(), lr=CFG.lr)
scheduler = get_scheduler(CFG, optimizer)
# ## loss
# Dice Loss is a commonly used loss function for image segmentation, which is based on the binary cross-entropy loss function and the Dice coefficient.
# The Dice coefficient is used to evaluate the similarity between the predicted segmentation result and the true label, which is defined as twice the ratio of the intersection size of the two sets to their concatenation size.
# And Dice Loss is defined as 1 minus the Dice coefficient as the loss function, i.e., Dice Loss = 1 - (2 * (the intersection size of the predicted segmentation result and the true label) / (the size of the predicted segmentation result + the size of the true label)).
# During the training process, the optimizer minimizes the Dice Loss so as to maximize the similarity between the predicted segmentation result and the true labels.
# BCELoss is a Binary Cross Entropy Loss function, which is usually used in binary classification problems.
# It measures the performance of the model by calculating the difference between the model prediction results and the true labels. In the
# binary cross entropy loss function, for each sample, we denote its true label as 0 or 1, and the prediction result is also a probability value between 0 and 1.
DiceLoss = smp.losses.DiceLoss(mode="binary")
BCELoss = smp.losses.SoftBCEWithLogitsLoss()
alpha = 0.5
beta = 1 - alpha
# This is a loss function that uses the Tversky index, a metric used to evaluate the degree of similarity between two sets. In this case
# it is used to assess the degree of similarity between the predicted and true results of the model.
# alpha and beta are hyperparameters that are used to adjust the weights of the Tversky index. This loss function is used for the binary classification problem.
# The log_loss parameter determines whether the logarithmic loss function is used.
# smp refers to the Segmentation Models PyTorch library, which is a deep learning library for image segmentation tasks.
TverskyLoss = smp.losses.TverskyLoss(
mode="binary", log_loss=False, alpha=alpha, beta=beta
)
def criterion(y_pred, y_true):
# return 0.5 * BCELoss(y_pred, y_true) + 0.5 * DiceLoss(y_pred, y_true)
return BCELoss(y_pred, y_true)
# return 0.5 * BCELoss(y_pred, y_true) + 0.5 * TverskyLoss(y_pred, y_true)
# ## train, val
def train_fn(train_loader, model, criterion, optimizer, device):
model.train()
"""
GradScaler is a tool in PyTorch for mixed-accuracy training that automatically scales gradient values
to avoid gradient underflow when computing at FP16 precision. In the configuration, GradScaler is enabled if AMP (Automatic Mixed Precision) is used.
"""
scaler = GradScaler(enabled=CFG.use_amp)
"""
This is a tool class for calculating the average loss. When training a neural network, it is common to calculate the losses for each batch (batches) and add them up to a total loss value.
In order to get the average loss, the total loss value needs to be divided by the number of batches.
The AverageMeter class encapsulates this process by making it easy to record the total loss value and the number of batches and to calculate the average loss value.
After each batch, the total loss value and batch count can be updated by calling the update() method of this class, and finally the average loss value can be obtained by calling the avg property of this class.
"""
losses = AverageMeter()
for step, (images, labels) in tqdm(
enumerate(train_loader), total=len(train_loader)
):
images = images.to(device)
labels = labels.to(device)
batch_size = labels.size(0)
with autocast(CFG.use_amp):
y_preds = model(images)
loss = criterion(y_preds, labels)
# Backpropagation and gradient calculation for the loss function
# scaler.scale(loss) is gradient scaling using PyTorch's GradScaler, this is to prevent gradient explosion or gradient disappearance during backpropagation. backward() is to backpropagate the parameters of the model and calculate the gradient.
losses.update(loss.item(), batch_size)
scaler.scale(loss).backward()
# This code is used to perform gradient cropping. During the training of a deep learning model, the gradient values may become very large, which can lead to instability of the model. To avoid this, we can use the gradient cropping method to keep the gradient value within an acceptable range.
grad_norm = torch.nn.utils.clip_grad_norm_(
model.parameters(), CFG.max_grad_norm
)
# Specifically, scaler.step(optimizer) is the optimizer's gradient update on mixed precision
# scaler.update() is used to update the scaling factor inside the scaler.
# optimizer.zero_grad() is used to clear the gradient information in the optimizer so that the gradient can be recalculated in the next iteration.
scaler.step(optimizer)
scaler.update()
optimizer.zero_grad()
return losses.avg
def valid_fn(valid_loader, model, criterion, device, valid_xyxys, valid_mask_gt):
mask_pred = np.zeros(valid_mask_gt.shape)
mask_count = np.zeros(valid_mask_gt.shape)
model.eval()
losses = AverageMeter()
for step, (images, labels) in tqdm(
enumerate(valid_loader), total=len(valid_loader)
):
images = images.to(device)
labels = labels.to(device)
batch_size = labels.size(0)
with torch.no_grad():
y_preds = model(images)
loss = criterion(y_preds, labels)
losses.update(loss.item(), batch_size)
"""
This code is used to generate the prediction masks, predicting the mask value for each pixel point based on the model's output, and then assigning it to the corresponding region of the mask.
where y_preds is the output of the model, which is mapped to between [0,1] as the mask value by the sigmoid function;
valid_xyxys is the coordinate and size information of the validation set images, and start_idx and end_idx are the corresponding start and end indexes of the currently processed batch in valid_xyxys;
mask_pred and mask_count are the generated masks and the number of times each pixel point is assigned. Finally, the mask value is divided by the number of assignments to get the average mask value of each pixel point.
"""
# make whole mask
y_preds = torch.sigmoid(y_preds).to("cpu").numpy()
start_idx = step * CFG.valid_batch_size
end_idx = start_idx + batch_size
for i, (x1, y1, x2, y2) in enumerate(valid_xyxys[start_idx:end_idx]):
mask_pred[y1:y2, x1:x2] += y_preds[i].squeeze(0)
mask_count[y1:y2, x1:x2] += np.ones((CFG.tile_size, CFG.tile_size))
print(f"mask_count_min: {mask_count.min()}")
mask_pred /= mask_count
return losses.avg, mask_pred
# ## metrics
from sklearn.metrics import fbeta_score
def fbeta_numpy(targets, preds, beta=0.5, smooth=1e-5):
"""
https://www.kaggle.com/competitions/vesuvius-challenge-ink-detection/discussion/397288
"""
y_true_count = targets.sum()
ctp = preds[targets == 1].sum()
cfp = preds[targets == 0].sum()
beta_squared = beta * beta
c_precision = ctp / (ctp + cfp + smooth)
c_recall = ctp / (y_true_count + smooth)
dice = (
(1 + beta_squared)
* (c_precision * c_recall)
/ (beta_squared * c_precision + c_recall + smooth)
)
return dice
# The purpose of this code is to calculate the F-beta score between a given mask and a predicted mask and return the best threshold and the best F-beta score.
# It calculates the F-beta score by spreading the mask and the prediction mask into a one-dimensional array and looping over a series of thresholds.
# The optimal threshold is the threshold that maximizes the F-beta score, and the best F-beta score is the F-beta score calculated at the optimal threshold.
def calc_fbeta(mask, mask_pred):
mask = mask.astype(int).flatten()
mask_pred = mask_pred.flatten()
best_th = 0
best_dice = 0
for th in np.array(range(10, 50 + 1, 5)) / 100:
# dice = fbeta_score(mask, (mask_pred >= th).astype(int), beta=0.5)
dice = fbeta_numpy(mask, (mask_pred >= th).astype(int), beta=0.5)
print(f"th: {th}, fbeta: {dice}")
if dice > best_dice:
best_dice = dice
best_th = th
Logger.info(f"best_th: {best_th}, fbeta: {best_dice}")
return best_dice, best_th
# This code defines a function called calc_cv that takes two arguments: mask_gt and mask_pred. These two arguments represent the true mask (i.e., ground truth) and the model prediction mask, respectively.
# The function internally calls the calc_fbeta function to calculate the best Dice coefficient and the best threshold, and returns them as a tuple.
# The Dice coefficient is a commonly used measure of the similarity of two masks and takes values from 0 to 1, with higher values indicating higher similarity. The threshold is a parameter needed to convert the mask into a binarized image.
def calc_cv(mask_gt, mask_pred):
best_dice, best_th = calc_fbeta(mask_gt, mask_pred)
return best_dice, best_th
# ## main
fragment_id = CFG.valid_id
valid_mask_gt = cv2.imread(
CFG.comp_dataset_path + f"train/{fragment_id}/inklabels.png", 0
)
valid_mask_gt = valid_mask_gt / 255
"""
The purpose of this code is to zero-fill the valid_mask_gt array so that the number of rows is a multiple of CFG.tile_size.
Specifically, pad0 and pad1 represent the number of zeros to be filled in the first and second dimensions of the valid_mask_gt array, respectively,
so that both valid_mask_gt.shape[0] and valid_mask_gt.shape[1] are multiples of CFG.tile_size.
"""
pad0 = CFG.tile_size - valid_mask_gt.shape[0] % CFG.tile_size
pad1 = CFG.tile_size - valid_mask_gt.shape[1] % CFG.tile_size
valid_mask_gt = np.pad(valid_mask_gt, [(0, pad0), (0, pad1)], constant_values=0)
"""
这行代码使用了NumPy中的`pad`函数,将`valid_mask_gt`数组在两个维度上进行了填充,
以便与另一个数组进行操作时具有相同的形状。具体来说,`[(0, pad0), (0, pad1)]`表示在第一个维度上不进行填充(前面填0个,后面填0个),
在第二个维度上填充`pad1`个0在后面,填充`pad0`个0在前面。
这样做的目的是将`valid_mask_gt`数组的形状扩展到与另一个数组相同,以便进行一些操作,例如相加、相减等。
"""
fold = CFG.valid_id
"""
This code initializes the variables for the best score based on the direction of the evaluation metric. If the evaluation indicator is of the "minimization" type, the initial value should be positive infinity (np.inf);
If the evaluation indicator is of the "maximize" type, then the initial value should be negative one (-1). This ensures that the value of the best score can be updated if a better score emerges during the subsequent evaluation.
"""
if CFG.metric_direction == "minimize":
best_score = np.inf
elif CFG.metric_direction == "maximize":
best_score = -1
best_loss = np.inf
for epoch in range(CFG.epochs):
start_time = time.time()
# train
avg_loss = train_fn(train_loader, model, criterion, optimizer, device)
# eval
avg_val_loss, mask_pred = valid_fn(
valid_loader, model, criterion, device, valid_xyxys, valid_mask_gt
)
scheduler_step(scheduler, avg_val_loss, epoch)
best_dice, best_th = calc_cv(valid_mask_gt, mask_pred)
# score = avg_val_loss
score = best_dice
elapsed = time.time() - start_time
Logger.info(
f"Epoch {epoch+1} - avg_train_loss: {avg_loss:.4f} avg_val_loss: {avg_val_loss:.4f} time: {elapsed:.0f}s"
)
# Logger.info(f'Epoch {epoch+1} - avgScore: {avg_score:.4f}')
Logger.info(f"Epoch {epoch+1} - avgScore: {score:.4f}")
"""
This code is used to update the best score based on the direction of the evaluation metric (whether it is minimized or maximized). If the direction of the evaluation metric is minimize, the best score is updated to the new score if the new score is lower than the current best score.
If the direction of the evaluation metric is maximization, the best score is updated to the new score if the new score is higher than the current best score. This allows tracking the best model during training and using that model during evaluation.
"""
if CFG.metric_direction == "minimize":
update_best = score < best_score
elif CFG.metric_direction == "maximize":
update_best = score > best_score
if update_best:
best_loss = avg_val_loss
best_score = score
Logger.info(f"Epoch {epoch+1} - Save Best Score: {best_score:.4f} Model")
Logger.info(f"Epoch {epoch+1} - Save Best Loss: {best_loss:.4f} Model")
torch.save(
{"model": model.state_dict(), "preds": mask_pred},
CFG.model_dir + f"{CFG.model_name}_fold{fold}_best.pth",
)
check_point = torch.load(
CFG.model_dir + f"{CFG.model_name}_fold{fold}_{CFG.inf_weight}.pth",
map_location=torch.device("cpu"),
)
mask_pred = check_point["preds"]
best_dice, best_th = calc_fbeta(valid_mask_gt, mask_pred)
fig, axes = plt.subplots(1, 3, figsize=(15, 8))
axes[0].imshow(valid_mask_gt)
axes[1].imshow(mask_pred)
axes[2].imshow((mask_pred >= best_th).astype(int))
plt.hist(mask_pred.flatten(), bins=20)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/067/129067634.ipynb
|
pretrainedmodels
|
rishabhiitbhu
|
[{"Id": 129067634, "ScriptId": 37541960, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14644876, "CreationDate": "05/10/2023 18:00:27", "VersionNumber": 1.0, "Title": "2.5d segmentaion baseline [training]", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 864.0, "LinesInsertedFromPrevious": 209.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 655.0, "LinesInsertedFromFork": 209.0, "LinesDeletedFromFork": 33.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 655.0, "TotalVotes": 0}]
|
[{"Id": 184795548, "KernelVersionId": 129067634, "SourceDatasetVersionId": 699609}, {"Id": 184795549, "KernelVersionId": 129067634, "SourceDatasetVersionId": 3492463}, {"Id": 184795550, "KernelVersionId": 129067634, "SourceDatasetVersionId": 3492503}, {"Id": 184795551, "KernelVersionId": 129067634, "SourceDatasetVersionId": 3951115}, {"Id": 184795552, "KernelVersionId": 129067634, "SourceDatasetVersionId": 5309119}]
|
[{"Id": 699609, "DatasetId": 255887, "DatasourceVersionId": 719625, "CreatorUserId": 761152, "LicenseName": "Other (specified in description)", "CreationDate": "09/23/2019 15:38:56", "VersionNumber": 3.0, "Title": "pretrainedmodels", "Slug": "pretrainedmodels", "Subtitle": "pretrained-models.pytorch", "Description": "### Pretrained Models in Pytorch\n\nGithub repo: https://github.com/Cadene/pretrained-models.pytorch\n\nVersion: 0.7.4\n\nOriginal Author: Cadene\n\nLicense: https://github.com/Cadene/pretrained-models.pytorch/blob/master/LICENSE.txt", "VersionNotes": "oops", "TotalCompressedBytes": 58821.0, "TotalUncompressedBytes": 58821.0}]
|
[{"Id": 255887, "CreatorUserId": 761152, "OwnerUserId": 761152.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 699609.0, "CurrentDatasourceVersionId": 719625.0, "ForumId": 267153, "Type": 2, "CreationDate": "07/06/2019 09:50:03", "LastActivityDate": "07/06/2019", "TotalViews": 8370, "TotalDownloads": 2757, "TotalVotes": 82, "TotalKernels": 150}]
|
[{"Id": 761152, "UserName": "rishabhiitbhu", "DisplayName": "Rishabh Agrahari", "RegisterDate": "10/21/2016", "PerformanceTier": 2}]
|
# ## summary
# **This notebook is written for new kaggler**
# **I can't guarantee that all comments are interpreted correctly, so please point out if there are mistakes**
# Original code is here [https://www.kaggle.com/code/tanakar/2-5d-segmentaion-baseline-training](http://)
# * 2.5d segmentation
# * segmentation_models_pytorch
# * Unet
# * use only 6 slices in the middle
# * slide inference
# sklearn.metrics.classification is an evaluation metrics module for classification problems in the scikit-learn library.This module provides a series of functions to calculate the accuracy, precision, recall, F1 value and other metrics of a classification model to evaluate the performance of the model.These metrics can help us understand the classification ability of the model, so that we can optimize the parameters and algorithms of the model and improve the prediction accuracy of the model.
# Pickle is a module in Python for serializing and deserializing Python objects.With Pickle, we can convert a Python object to a byte stream and then save it to a file or transfer it over the network.Conversely, we can also deserialize byte streams to Python objects.Pickle is a persistent storage method in Python that makes it easy to save and restore data.
# Autocast and GradScaler are both tools in PyTorch for accelerating model training and reducing memory footprint.
# Autocast is an automatic mixed precision tool that automatically converts floating point numbers to half-precision floating point numbers during the forward and backward propagation of the model. This helps reduce GPU memory usage and speed up model training. When using Autocast, the model and optimizer need to be wrapped in the torch.cuda.amp.autocast() context manager.GradScaler is a gradient scaling tool for scaling the value of the gradient during training. This helps to solve the problem of disappearing or exploding gradients and improves model stability and training results. When using GradScaler, the gradient needs to be multiplied by a scaling factor before back-propagation is performed. The scaling factor can be dynamically adjusted according to the value of the gradient to ensure the stability of the gradient.
from sklearn.metrics import roc_auc_score, accuracy_score, f1_score, log_loss
import pickle
from torch.utils.data import DataLoader
from torch.cuda.amp import autocast, GradScaler
import warnings
import sys
import pandas as pd
import os
import gc
import sys
import math
import time
import random
import shutil
from pathlib import Path
from contextlib import contextmanager
from collections import defaultdict, Counter
import cv2
import scipy as sp
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from tqdm.auto import tqdm
from functools import partial
import argparse
import importlib
import torch
import torch.nn as nn
from torch.optim import Adam, SGD, AdamW
import datetime
# sys.path.append('/kaggle/input/pretrainedmodels/pretrainedmodels-0.7.4')
# sys.path.append('/kaggle/input/efficientnet-pytorch/EfficientNet-PyTorch-master')
# sys.path.append('/kaggle/input/timm-pytorch-image-models/pytorch-image-models-master')
# sys.path.append('/kaggle/input/segmentation-models-pytorch/segmentation_models.pytorch-master')
import segmentation_models_pytorch as smp
# For segmentation_models_pytorch, it is a PyTorch-based deep learning library for image segmentation tasks.
# It supports many popular segmentation models, such as UNet, LinkNet, FPN, etc., and provides many pre-trained models and datasets to facilitate users to quickly build and train their own models.
import numpy as np
from torch.utils.data import DataLoader, Dataset
import cv2
import torch
import os
import albumentations as A
from albumentations.pytorch import ToTensorV2
from albumentations import ImageOnlyTransform
# ## config
class CFG:
# ============== comp exp name =============
comp_name = "vesuvius"
# comp_dir_path = './'
comp_dir_path = "/kaggle/input/"
comp_folder_name = "vesuvius-challenge-ink-detection"
# comp_dataset_path = f'{comp_dir_path}datasets/{comp_folder_name}/'
comp_dataset_path = f"{comp_dir_path}{comp_folder_name}/"
exp_name = "vesuvius_2d_slide_exp002"
# ============== pred target =============
target_size = 1
# ============== model cfg =============
# Image segmentation model Pre-training parameters for convolutional networks
model_name = "Unet"
# backbone = 'efficientnet-b0'
backbone = "resnext101_32x4d"
# backbone = 'resnext50_32x4d'
# backbone = 'resnet50'
# There are 65 "channels",
# a three-dimensional image of a certain dimension split into multiple two-dimensional images,
# the ink will be immersed in the deeper the image, the larger the information more complete
in_chans = 3 # 64
# ============== training cfg =============
# Size modification of input image or mask image
size = 224
tile_size = 224
# The number of small images cut out and the position of each small image can be controlled by setting CFG.stride.
stride = tile_size // 2
train_batch_size = 16 # 32
valid_batch_size = train_batch_size * 2
use_amp = True
# Learning rate regulator
scheduler = "GradualWarmupSchedulerV2"
# scheduler = 'CosineAnnealingLR'
epochs = 10 # 30
"""
warmup_factor is a scaling factor that controls the rate of learning rate increase. It is usually used at the beginning of training to accelerate the learning rate increase if the model weights are not yet accurate enough.
lr is the learning rate size, and in this equation, the learning rate is divided by 10 with the effect of warmup_factor.
This is because at the beginning of the training, the learning rate should be relatively small so that the model can converge better, and as the training proceeds, the learning rate will gradually increase so that the weight space can be explored better.
Thus, with this formula, a relatively small learning rate can be used at the beginning of training and gradually increased to the appropriate size.
"""
# adamW
warmup_factor = 10
# lr = 1e-4 / warmup_factor
lr = 1e-4 / warmup_factor
# ============== fold =============
# k-fold cross-validation This method has the advantage of making better use of the data while reducing errors due to the chance of data division.
# The disadvantage is that k-times training and validation are required and the computational cost is high.
# This seems to mean that each of the three folders is used as a validation set
valid_id = 1
"""
In this example, metric_direction is set to 'maximize', indicating that the metric we want to optimize should be as large as possible.
"""
# objective_cv = 'binary' # 'binary', 'multiclass', 'regression'
metric_direction = "maximize" # maximize, 'minimize'
# metrics = 'dice_coef'
# ============== fixed =============
pretrained = True
inf_weight = "best" # 'best'
min_lr = 1e-6
weight_decay = 1e-6
max_grad_norm = 1000
print_freq = 50
num_workers = 10
# Fixed seeds make code reproducible
# Randomness in deep learning is caused by factors such as weight initialization, random sampling, etc. These randomness can make the model more expressive and generalizable.
# However, since deep learning models are usually very large, their training requires a lot of time and computational resources. Therefore, it is useful to fix random seeds in order to make the experiments reproducible.
# Using a fixed random seed ensures that the same sequence of random numbers is generated using the same random number generator each time the experiment is run.
# This allows researchers to get the same results when running experiments on different machines, thus facilitating the comparison and validation of results.
# In addition, using a fixed random seed also makes the model's behavior more predictable during training, thus helping researchers to better understand the model's performance and behavior.
seed = 42
# ============== set dataset path =============
print("set dataset path")
outputs_path = f"/kaggle/working/outputs/{comp_name}/{exp_name}/"
submission_dir = outputs_path + "submissions/"
submission_path = submission_dir + f"submission_{exp_name}.csv"
model_dir = outputs_path + f"{comp_name}-models/"
figures_dir = outputs_path + "figures/"
log_dir = outputs_path + "logs/"
log_path = log_dir + f"{exp_name}.txt"
# ============== augmentation =============
# Data Enhancement
train_aug_list = [
# A.RandomResizedCrop(
# size, size, scale=(0.85, 1.0)),
A.Resize(size, size),
A.HorizontalFlip(p=0.5),
A.VerticalFlip(p=0.5),
A.RandomBrightnessContrast(p=0.75),
A.ShiftScaleRotate(p=0.75),
A.OneOf(
[
A.GaussNoise(var_limit=[10, 50]),
A.GaussianBlur(),
A.MotionBlur(),
],
p=0.4,
),
A.GridDistortion(num_steps=5, distort_limit=0.3, p=0.5),
A.CoarseDropout(
max_holes=1,
max_width=int(size * 0.3),
max_height=int(size * 0.3),
mask_fill_value=0,
p=0.5,
),
# A.Cutout(max_h_size=int(size * 0.6),
# max_w_size=int(size * 0.6), num_holes=1, p=1.0),
A.Normalize(mean=[0] * in_chans, std=[1] * in_chans),
ToTensorV2(transpose_mask=True),
]
valid_aug_list = [
A.Resize(size, size),
A.Normalize(mean=[0] * in_chans, std=[1] * in_chans),
ToTensorV2(transpose_mask=True),
]
# ## helper
# Computes and stores the average and current value
# Specifically, it serves to call the update method to update the value of the instance each time the average needs to be calculated, and to call the avg method to return the average when the average needs to be calculated.
class AverageMeter(object):
"""Computes and stores the average and current value"""
# Initialize the property values of the instance
def __init__(self):
self.reset()
# Reset the property value of the instance to 0
def reset(self):
self.val = 0
self.avg = 0
self.sum = 0
self.count = 0
# Update the property value of the instance to the given value
def update(self, val, n=1):
self.val = val
self.sum += val * n
self.count += n
self.avg = self.sum / self.count
# # log
# accepts one argument log_file log file path
# The function initializes a logger object, which is used to record logging information when the program is running. This is implemented as follows:
def init_logger(log_file):
from logging import getLogger, INFO, FileHandler, Formatter, StreamHandler
logger = getLogger(__name__)
logger.setLevel(INFO)
handler1 = StreamHandler()
handler1.setFormatter(Formatter("%(message)s"))
handler2 = FileHandler(filename=log_file)
handler2.setFormatter(Formatter("%(message)s"))
logger.addHandler(handler1)
logger.addHandler(handler2)
return logger
def set_seed(seed=None, cudnn_deterministic=True):
if seed is None:
seed = 42
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = cudnn_deterministic
torch.backends.cudnn.benchmark = False
# This function creates these directories using the os.makedirs() method and does not overwrite the directories if they already exist, i.e. the exist_ok=True argument means that no errors will be reported.
# The purpose of this function is to create these directories when needed for use in subsequent code.
def make_dirs(cfg):
for dir in [cfg.model_dir, cfg.figures_dir, cfg.submission_dir, cfg.log_dir]:
os.makedirs(dir, exist_ok=True)
# # Initialization functions
def cfg_init(cfg, mode="train"):
set_seed(cfg.seed)
# set_env_name()
# set_dataset_path(cfg)
if mode == "train":
make_dirs(cfg)
cfg_init(CFG)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
Logger = init_logger(log_file=CFG.log_path)
Logger.info("\n\n-------- exp_info -----------------")
# Logger.info(datetime.datetime.now().strftime('%Y年%m月%d日 %H:%M:%S'))
# ## image, mask
# The purpose of this code is to find a window of length CFG.in_chans, so that the subscript of the center point of the window is mid.
# Specifically, first divide mid by 2, then subtract half of CFG.in_chans from the result to get the start subscript of the window, then add half of CFG.in_chans to mid to get the end subscript of the window.
# Then add half of CFG.in_chans to mid to get the end of the window.
# Finally, use the range function to generate a sequence of integers idxs from start to end-1.
#
# **Set the number of images to be read from 0-65, and adjust the number of input "channels" by changing in_chans**
def read_image_mask(fragment_id):
images = []
# idxs = range(65)
mid = 65 // 2
start = mid - CFG.in_chans // 2
end = mid + CFG.in_chans // 2
idxs = range(start, end)
for i in tqdm(idxs):
image = cv2.imread(
CFG.comp_dataset_path + f"train/{fragment_id}/surface_volume/{i:02}.tif", 0
)
pad0 = CFG.tile_size - image.shape[0] % CFG.tile_size
pad1 = CFG.tile_size - image.shape[1] % CFG.tile_size
image = np.pad(image, [(0, pad0), (0, pad1)], constant_values=0)
images.append(image)
images = np.stack(images, axis=2)
mask = cv2.imread(CFG.comp_dataset_path + f"train/{fragment_id}/inklabels.png", 0)
mask = np.pad(mask, [(0, pad0), (0, pad1)], constant_values=0)
mask = mask.astype("float32")
mask /= 255.0
return images, mask
def get_train_valid_dataset():
train_images = []
train_masks = []
valid_images = []
valid_masks = []
valid_xyxys = []
for fragment_id in range(1, 4):
image, mask = read_image_mask(fragment_id)
"""
This code is generating a set of image crop coordinates that are used to split a large size image into smaller pieces for easier processing. Where image is the original image and CFG is a set of constant parameters.
Specifically, the parameters of the range function in the code set the step size and range of the crop coordinates.
image.shape[0] and image.shape[1] denote the height and width of the image, respectively. Subtracting CFG.tile_size is to ensure that the segmented chunks are all squares with CFG.tile_size as the side length.
The final x1_list and y1_list are the coordinates of all the generated images, which are used for subsequent processing.
"""
x1_list = list(range(0, image.shape[1] - CFG.tile_size + 1, CFG.stride))
y1_list = list(range(0, image.shape[0] - CFG.tile_size + 1, CFG.stride))
# where y1_list and x1_list are the list of starting coordinates in the width and height directions of the large image, respectively, and CFG.tile_size is the size of each small image block.
# If fragment_id is equal to CFG.valid_id, the current image block and mask are stored in the list of valid_images and valid_masks
# and store the start and end coordinates corresponding to the current image block into the valid_xyxys list; otherwise store them into the train_images and train_masks lists.
for y1 in y1_list:
for x1 in x1_list:
y2 = y1 + CFG.tile_size
x2 = x1 + CFG.tile_size
# xyxys.append((x1, y1, x2, y2))
# Place the set folder with the corresponding label in the validation data set, folder 1 or folder 2 or folder 3
if fragment_id == CFG.valid_id:
valid_images.append(image[y1:y2, x1:x2])
valid_masks.append(mask[y1:y2, x1:x2, None])
valid_xyxys.append([x1, y1, x2, y2])
# unspecified data sets into the training set, 2 training sets 1 validation set
else:
train_images.append(image[y1:y2, x1:x2])
train_masks.append(mask[y1:y2, x1:x2, None])
return train_images, train_masks, valid_images, valid_masks, valid_xyxys
(
train_images,
train_masks,
valid_images,
valid_masks,
valid_xyxys,
) = get_train_valid_dataset()
valid_xyxys = np.stack(valid_xyxys)
# ## dataset
import numpy as np
from torch.utils.data import DataLoader, Dataset
import cv2
import torch
import os
import albumentations as A
from albumentations.pytorch import ToTensorV2
from albumentations import ImageOnlyTransform
def get_transforms(data, cfg):
if data == "train":
aug = A.Compose(cfg.train_aug_list)
elif data == "valid":
aug = A.Compose(cfg.valid_aug_list)
# print(aug)
return aug
class CustomDataset(Dataset):
def __init__(self, images, cfg, labels=None, transform=None):
self.images = images
self.cfg = cfg
self.labels = labels
self.transform = transform
def __len__(self):
# return len(self.df)
return len(self.images)
def __getitem__(self, idx):
image = self.images[idx]
label = self.labels[idx]
if self.transform:
data = self.transform(image=image, mask=label)
image = data["image"]
label = data["mask"]
return image, label
# # Training set and validation set data processing
train_dataset = CustomDataset(
train_images,
CFG,
labels=train_masks,
transform=get_transforms(data="train", cfg=CFG),
)
valid_dataset = CustomDataset(
valid_images,
CFG,
labels=valid_masks,
transform=get_transforms(data="valid", cfg=CFG),
)
train_loader = DataLoader(
train_dataset,
batch_size=CFG.train_batch_size,
shuffle=True,
num_workers=CFG.num_workers,
pin_memory=True,
drop_last=True,
)
valid_loader = DataLoader(
valid_dataset,
batch_size=CFG.valid_batch_size,
shuffle=False,
num_workers=CFG.num_workers,
pin_memory=True,
drop_last=False,
)
train_dataset[0][0].shape
plot_dataset = CustomDataset(train_images, CFG, labels=train_masks)
transform = CFG.train_aug_list
transform = A.Compose(
[t for t in transform if not isinstance(t, (A.Normalize, ToTensorV2))]
)
plot_count = 0
for i in range(1000):
image, mask = plot_dataset[i]
data = transform(image=image, mask=mask)
aug_image = data["image"]
aug_mask = data["mask"]
if mask.sum() == 0:
continue
fig, axes = plt.subplots(1, 4, figsize=(15, 8))
axes[0].imshow(image[..., 0], cmap="gray")
axes[1].imshow(mask, cmap="gray")
axes[2].imshow(aug_image[..., 0], cmap="gray")
axes[3].imshow(aug_mask, cmap="gray")
plt.savefig(CFG.figures_dir + f"aug_fold_{CFG.valid_id}_{plot_count}.png")
plot_count += 1
if plot_count == 5:
break
del plot_dataset
gc.collect()
# ## model
# **Unet convolutional network construction**
# This code is initializing the image segmentation task using the Unet model from the segmentation_models_pytorch library.
class CustomModel(nn.Module):
def __init__(self, cfg, weight=None):
super().__init__()
self.cfg = cfg
# The encoder_name parameter is used to specify which pre-trained encoder model to use. Here, a pre-trained model is used, so the encoder_name parameter is specified.
# The advantage of this is that the feature extraction capability of the existing pre-trained model can be used to accelerate the training of the model and improve the accuracy of the model. Also, different pre-trained models can be selected according to actual needs to achieve better results.
# We can use the pre-training weights of the image classification model to initialize the convolutional layer of the UNet network, thus improving the performance and generalization ability of the model
# encoder_name: indicates the name of the pre-trained model used
# encoder_weights: indicates the weights of the pre-trained model used, usually imagenet is used.
# in_channels: indicates the number of channels of the input image, e.g., 3 channels for RGB images
# classes: the number of classifications, usually the number of pixel classifications for image segmentation tasks.
# activation: indicates the activation function, usually None (no activation function is used) or sigmoid (the output is between 0 and 1).
self.encoder = smp.Unet(
encoder_name=cfg.backbone,
encoder_weights=weight,
in_channels=cfg.in_chans,
classes=cfg.target_size,
activation=None,
)
# After the initialization, the input image can be fed into the model for forward propagation to obtain the corresponding segmentation results.
def forward(self, image):
output = self.encoder(image)
# output = output.squeeze(-1)
return output
# weight="imagenet"
def build_model(cfg, weight="ssl"):
print("model_name", cfg.model_name)
print("backbone", cfg.backbone)
model = CustomModel(cfg, weight)
return model
# ## scheduler
# [https://www.kaggle.com/code/underwearfitting/single-fold-training-of-resnet200d-lb0-965](http://)
# Its main role is to gradually increase the learning rate at the beginning of training to help the model converge faster. It is a subclass of the GradualWarmupScheduler class with the addition of an after_scheduler parameter for using other learning rate scheduling methods after the warm-up is over.
# In each epoch, the get_lr() method will return the current learning rate.
# At the beginning of training, the learning rate will be gradually increased from the initial value to the maximum value, after which the learning rate returned by after_scheduler (another learning rate update method) will be used. If after_scheduler is not provided, the current learning rate will continue to be used.
#
import torch.nn as nn
import torch
import math
import time
import numpy as np
import torch
from torch.optim.lr_scheduler import (
CosineAnnealingWarmRestarts,
CosineAnnealingLR,
ReduceLROnPlateau,
)
from warmup_scheduler import GradualWarmupScheduler
class GradualWarmupSchedulerV2(GradualWarmupScheduler):
def __init__(self, optimizer, multiplier, total_epoch, after_scheduler=None):
super(GradualWarmupSchedulerV2, self).__init__(
optimizer, multiplier, total_epoch, after_scheduler
)
def get_lr(self):
if self.last_epoch > self.total_epoch:
if self.after_scheduler:
if not self.finished:
self.after_scheduler.base_lrs = [
base_lr * self.multiplier for base_lr in self.base_lrs
]
self.finished = True
return self.after_scheduler.get_lr()
return [base_lr * self.multiplier for base_lr in self.base_lrs]
if self.multiplier == 1.0:
return [
base_lr * (float(self.last_epoch) / self.total_epoch)
for base_lr in self.base_lrs
]
else:
return [
base_lr
* ((self.multiplier - 1.0) * self.last_epoch / self.total_epoch + 1.0)
for base_lr in self.base_lrs
]
# This code is a function to get the learning rate scheduler with input parameters of configuration (cfg) and optimizer (optimizer). Two learning rate schedulers are used in this function, CosineAnnealingLR and GradualWarmupSchedulerV2.
# Among them, CosineAnnealingLR is a cosine annealing learning rate scheduler, whose function is to gradually reduce the learning rate during training to make the model converge more stably.
# GradualWarmupSchedulerV2 is a learning rate preheating scheduler, whose role is to gradually increase the learning rate at the beginning of training to avoid the model from falling into a local optimum solution at the beginning and failing to jump out.
# In this function, we first define a CosineAnnealingLR scheduler, setting it to the total number of training rounds (cfg.epochs) as cycles and a minimum learning rate of 1e-7.
# Then, we use this scheduler as the after_scheduler parameter of GradualWarmupSchedulerV2, and also use it as the parameter of the input optimizer, multiplier indicates the learning rate multiplier during warm-up, and total_epoch indicates the number of warm-up rounds.
# Finally, the GradualWarmupSchedulerV2 scheduler is returned as the output.
def get_scheduler(cfg, optimizer):
scheduler_cosine = torch.optim.lr_scheduler.CosineAnnealingLR(
optimizer, cfg.epochs, eta_min=1e-7
)
scheduler = GradualWarmupSchedulerV2(
optimizer, multiplier=10, total_epoch=1, after_scheduler=scheduler_cosine
)
return scheduler
def scheduler_step(scheduler, avg_val_loss, epoch):
scheduler.step(epoch)
# This line of code uses the AdamW optimizer, which takes the parameters of the model as input and uses the learning rate of lr from CFG. AdamW is a variant of the Adam optimizer that is used to update the model parameters when training a neural network.
# It updates the parameters by weighted averaging the gradients of the parameters to minimize the loss function.AdamW also uses a regularization method called weight decay to prevent the model from overfitting the training data.
model = build_model(CFG)
model.to(device)
optimizer = AdamW(model.parameters(), lr=CFG.lr)
scheduler = get_scheduler(CFG, optimizer)
# ## loss
# Dice Loss is a commonly used loss function for image segmentation, which is based on the binary cross-entropy loss function and the Dice coefficient.
# The Dice coefficient is used to evaluate the similarity between the predicted segmentation result and the true label, which is defined as twice the ratio of the intersection size of the two sets to their concatenation size.
# And Dice Loss is defined as 1 minus the Dice coefficient as the loss function, i.e., Dice Loss = 1 - (2 * (the intersection size of the predicted segmentation result and the true label) / (the size of the predicted segmentation result + the size of the true label)).
# During the training process, the optimizer minimizes the Dice Loss so as to maximize the similarity between the predicted segmentation result and the true labels.
# BCELoss is a Binary Cross Entropy Loss function, which is usually used in binary classification problems.
# It measures the performance of the model by calculating the difference between the model prediction results and the true labels. In the
# binary cross entropy loss function, for each sample, we denote its true label as 0 or 1, and the prediction result is also a probability value between 0 and 1.
DiceLoss = smp.losses.DiceLoss(mode="binary")
BCELoss = smp.losses.SoftBCEWithLogitsLoss()
alpha = 0.5
beta = 1 - alpha
# This is a loss function that uses the Tversky index, a metric used to evaluate the degree of similarity between two sets. In this case
# it is used to assess the degree of similarity between the predicted and true results of the model.
# alpha and beta are hyperparameters that are used to adjust the weights of the Tversky index. This loss function is used for the binary classification problem.
# The log_loss parameter determines whether the logarithmic loss function is used.
# smp refers to the Segmentation Models PyTorch library, which is a deep learning library for image segmentation tasks.
TverskyLoss = smp.losses.TverskyLoss(
mode="binary", log_loss=False, alpha=alpha, beta=beta
)
def criterion(y_pred, y_true):
# return 0.5 * BCELoss(y_pred, y_true) + 0.5 * DiceLoss(y_pred, y_true)
return BCELoss(y_pred, y_true)
# return 0.5 * BCELoss(y_pred, y_true) + 0.5 * TverskyLoss(y_pred, y_true)
# ## train, val
def train_fn(train_loader, model, criterion, optimizer, device):
model.train()
"""
GradScaler is a tool in PyTorch for mixed-accuracy training that automatically scales gradient values
to avoid gradient underflow when computing at FP16 precision. In the configuration, GradScaler is enabled if AMP (Automatic Mixed Precision) is used.
"""
scaler = GradScaler(enabled=CFG.use_amp)
"""
This is a tool class for calculating the average loss. When training a neural network, it is common to calculate the losses for each batch (batches) and add them up to a total loss value.
In order to get the average loss, the total loss value needs to be divided by the number of batches.
The AverageMeter class encapsulates this process by making it easy to record the total loss value and the number of batches and to calculate the average loss value.
After each batch, the total loss value and batch count can be updated by calling the update() method of this class, and finally the average loss value can be obtained by calling the avg property of this class.
"""
losses = AverageMeter()
for step, (images, labels) in tqdm(
enumerate(train_loader), total=len(train_loader)
):
images = images.to(device)
labels = labels.to(device)
batch_size = labels.size(0)
with autocast(CFG.use_amp):
y_preds = model(images)
loss = criterion(y_preds, labels)
# Backpropagation and gradient calculation for the loss function
# scaler.scale(loss) is gradient scaling using PyTorch's GradScaler, this is to prevent gradient explosion or gradient disappearance during backpropagation. backward() is to backpropagate the parameters of the model and calculate the gradient.
losses.update(loss.item(), batch_size)
scaler.scale(loss).backward()
# This code is used to perform gradient cropping. During the training of a deep learning model, the gradient values may become very large, which can lead to instability of the model. To avoid this, we can use the gradient cropping method to keep the gradient value within an acceptable range.
grad_norm = torch.nn.utils.clip_grad_norm_(
model.parameters(), CFG.max_grad_norm
)
# Specifically, scaler.step(optimizer) is the optimizer's gradient update on mixed precision
# scaler.update() is used to update the scaling factor inside the scaler.
# optimizer.zero_grad() is used to clear the gradient information in the optimizer so that the gradient can be recalculated in the next iteration.
scaler.step(optimizer)
scaler.update()
optimizer.zero_grad()
return losses.avg
def valid_fn(valid_loader, model, criterion, device, valid_xyxys, valid_mask_gt):
mask_pred = np.zeros(valid_mask_gt.shape)
mask_count = np.zeros(valid_mask_gt.shape)
model.eval()
losses = AverageMeter()
for step, (images, labels) in tqdm(
enumerate(valid_loader), total=len(valid_loader)
):
images = images.to(device)
labels = labels.to(device)
batch_size = labels.size(0)
with torch.no_grad():
y_preds = model(images)
loss = criterion(y_preds, labels)
losses.update(loss.item(), batch_size)
"""
This code is used to generate the prediction masks, predicting the mask value for each pixel point based on the model's output, and then assigning it to the corresponding region of the mask.
where y_preds is the output of the model, which is mapped to between [0,1] as the mask value by the sigmoid function;
valid_xyxys is the coordinate and size information of the validation set images, and start_idx and end_idx are the corresponding start and end indexes of the currently processed batch in valid_xyxys;
mask_pred and mask_count are the generated masks and the number of times each pixel point is assigned. Finally, the mask value is divided by the number of assignments to get the average mask value of each pixel point.
"""
# make whole mask
y_preds = torch.sigmoid(y_preds).to("cpu").numpy()
start_idx = step * CFG.valid_batch_size
end_idx = start_idx + batch_size
for i, (x1, y1, x2, y2) in enumerate(valid_xyxys[start_idx:end_idx]):
mask_pred[y1:y2, x1:x2] += y_preds[i].squeeze(0)
mask_count[y1:y2, x1:x2] += np.ones((CFG.tile_size, CFG.tile_size))
print(f"mask_count_min: {mask_count.min()}")
mask_pred /= mask_count
return losses.avg, mask_pred
# ## metrics
from sklearn.metrics import fbeta_score
def fbeta_numpy(targets, preds, beta=0.5, smooth=1e-5):
"""
https://www.kaggle.com/competitions/vesuvius-challenge-ink-detection/discussion/397288
"""
y_true_count = targets.sum()
ctp = preds[targets == 1].sum()
cfp = preds[targets == 0].sum()
beta_squared = beta * beta
c_precision = ctp / (ctp + cfp + smooth)
c_recall = ctp / (y_true_count + smooth)
dice = (
(1 + beta_squared)
* (c_precision * c_recall)
/ (beta_squared * c_precision + c_recall + smooth)
)
return dice
# The purpose of this code is to calculate the F-beta score between a given mask and a predicted mask and return the best threshold and the best F-beta score.
# It calculates the F-beta score by spreading the mask and the prediction mask into a one-dimensional array and looping over a series of thresholds.
# The optimal threshold is the threshold that maximizes the F-beta score, and the best F-beta score is the F-beta score calculated at the optimal threshold.
def calc_fbeta(mask, mask_pred):
mask = mask.astype(int).flatten()
mask_pred = mask_pred.flatten()
best_th = 0
best_dice = 0
for th in np.array(range(10, 50 + 1, 5)) / 100:
# dice = fbeta_score(mask, (mask_pred >= th).astype(int), beta=0.5)
dice = fbeta_numpy(mask, (mask_pred >= th).astype(int), beta=0.5)
print(f"th: {th}, fbeta: {dice}")
if dice > best_dice:
best_dice = dice
best_th = th
Logger.info(f"best_th: {best_th}, fbeta: {best_dice}")
return best_dice, best_th
# This code defines a function called calc_cv that takes two arguments: mask_gt and mask_pred. These two arguments represent the true mask (i.e., ground truth) and the model prediction mask, respectively.
# The function internally calls the calc_fbeta function to calculate the best Dice coefficient and the best threshold, and returns them as a tuple.
# The Dice coefficient is a commonly used measure of the similarity of two masks and takes values from 0 to 1, with higher values indicating higher similarity. The threshold is a parameter needed to convert the mask into a binarized image.
def calc_cv(mask_gt, mask_pred):
best_dice, best_th = calc_fbeta(mask_gt, mask_pred)
return best_dice, best_th
# ## main
fragment_id = CFG.valid_id
valid_mask_gt = cv2.imread(
CFG.comp_dataset_path + f"train/{fragment_id}/inklabels.png", 0
)
valid_mask_gt = valid_mask_gt / 255
"""
The purpose of this code is to zero-fill the valid_mask_gt array so that the number of rows is a multiple of CFG.tile_size.
Specifically, pad0 and pad1 represent the number of zeros to be filled in the first and second dimensions of the valid_mask_gt array, respectively,
so that both valid_mask_gt.shape[0] and valid_mask_gt.shape[1] are multiples of CFG.tile_size.
"""
pad0 = CFG.tile_size - valid_mask_gt.shape[0] % CFG.tile_size
pad1 = CFG.tile_size - valid_mask_gt.shape[1] % CFG.tile_size
valid_mask_gt = np.pad(valid_mask_gt, [(0, pad0), (0, pad1)], constant_values=0)
"""
这行代码使用了NumPy中的`pad`函数,将`valid_mask_gt`数组在两个维度上进行了填充,
以便与另一个数组进行操作时具有相同的形状。具体来说,`[(0, pad0), (0, pad1)]`表示在第一个维度上不进行填充(前面填0个,后面填0个),
在第二个维度上填充`pad1`个0在后面,填充`pad0`个0在前面。
这样做的目的是将`valid_mask_gt`数组的形状扩展到与另一个数组相同,以便进行一些操作,例如相加、相减等。
"""
fold = CFG.valid_id
"""
This code initializes the variables for the best score based on the direction of the evaluation metric. If the evaluation indicator is of the "minimization" type, the initial value should be positive infinity (np.inf);
If the evaluation indicator is of the "maximize" type, then the initial value should be negative one (-1). This ensures that the value of the best score can be updated if a better score emerges during the subsequent evaluation.
"""
if CFG.metric_direction == "minimize":
best_score = np.inf
elif CFG.metric_direction == "maximize":
best_score = -1
best_loss = np.inf
for epoch in range(CFG.epochs):
start_time = time.time()
# train
avg_loss = train_fn(train_loader, model, criterion, optimizer, device)
# eval
avg_val_loss, mask_pred = valid_fn(
valid_loader, model, criterion, device, valid_xyxys, valid_mask_gt
)
scheduler_step(scheduler, avg_val_loss, epoch)
best_dice, best_th = calc_cv(valid_mask_gt, mask_pred)
# score = avg_val_loss
score = best_dice
elapsed = time.time() - start_time
Logger.info(
f"Epoch {epoch+1} - avg_train_loss: {avg_loss:.4f} avg_val_loss: {avg_val_loss:.4f} time: {elapsed:.0f}s"
)
# Logger.info(f'Epoch {epoch+1} - avgScore: {avg_score:.4f}')
Logger.info(f"Epoch {epoch+1} - avgScore: {score:.4f}")
"""
This code is used to update the best score based on the direction of the evaluation metric (whether it is minimized or maximized). If the direction of the evaluation metric is minimize, the best score is updated to the new score if the new score is lower than the current best score.
If the direction of the evaluation metric is maximization, the best score is updated to the new score if the new score is higher than the current best score. This allows tracking the best model during training and using that model during evaluation.
"""
if CFG.metric_direction == "minimize":
update_best = score < best_score
elif CFG.metric_direction == "maximize":
update_best = score > best_score
if update_best:
best_loss = avg_val_loss
best_score = score
Logger.info(f"Epoch {epoch+1} - Save Best Score: {best_score:.4f} Model")
Logger.info(f"Epoch {epoch+1} - Save Best Loss: {best_loss:.4f} Model")
torch.save(
{"model": model.state_dict(), "preds": mask_pred},
CFG.model_dir + f"{CFG.model_name}_fold{fold}_best.pth",
)
check_point = torch.load(
CFG.model_dir + f"{CFG.model_name}_fold{fold}_{CFG.inf_weight}.pth",
map_location=torch.device("cpu"),
)
mask_pred = check_point["preds"]
best_dice, best_th = calc_fbeta(valid_mask_gt, mask_pred)
fig, axes = plt.subplots(1, 3, figsize=(15, 8))
axes[0].imshow(valid_mask_gt)
axes[1].imshow(mask_pred)
axes[2].imshow((mask_pred >= best_th).astype(int))
plt.hist(mask_pred.flatten(), bins=20)
| false | 0 | 10,420 | 0 | 10,508 | 10,420 |
||
129977265
|
# ## Background Info
# In sentiment analysis using models like RoBERTa, the predicted probabilities for different sentiment classes (such as neg, neu, and pos) represent the model's confidence or likelihood for each sentiment class independently, and they are typically outputted as normalized values between 0 and 1.
# The probabilities for neg, neu, and pos are relative to each other and represent the model's estimated likelihood or confidence for each sentiment category. For example, if the predicted probabilities are [0.2, 0.5, 0.3] for neg, neu, and pos respectively, it means the model assigns a 20% probability to negative sentiment, 50% probability to neutral sentiment, and 30% probability to positive sentiment for the given input.
# ## Import Necessary Modules
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # for the graphs
import seaborn as sns
plt.style.use("ggplot")
import nltk
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Read In Data
# Read in data in a data frame
df = pd.read_csv("../input/updated-movie-reviews-dataset/New_Audience.csv")
df.head()
df["reviewContent"].values[0]
print(df.shape) # 1100 rows, 7 columns
# Make the dataframe only take in rows 0-550, can change depending on computing power
df = df.head(550)
df.head()
# ## Quick Exploratory Data Analysis (EDA)
ax = (
df["reviewRating"]
.value_counts()
.sort_index()
.plot(kind="line", title="Average Audience Review Ratings", figsize=(10, 5))
)
ax.set_xlabel("Review Ratings")
ax.set_ylabel("Numbers of Reviews")
ax.set_xticks(range(1, 11, 1))
plt.show()
# ## Basic NLTK
example = df["reviewContent"][50]
print(example)
tokens = nltk.word_tokenize(example)
tokens[:10]
tagged = nltk.pos_tag(tokens)
tagged[:10]
entities = nltk.ne_chunk(tagged)
sliced_entities = entities[:10]
nltk.pprint(sliced_entities)
# entities.pprint()
# ## Sentiment Analysis Version 1: Using VADER
# VADER (Valence Aware Dictionary and sEntiment Reasoner) - Bag of words approach
# > Using NLTK's `SentimentIntensityAnalyzer` to get the neg/neu/pos scores of the text.
# * This uses a "bag of words approach:
# 1. Stop words are removed (e.g. and, the) - just words used for structure
# 2. each word is scored and combined to a total score.
# *Note: This does not include relationship between words.
from nltk.sentiment import SentimentIntensityAnalyzer
from tqdm.notebook import tqdm
sia = SentimentIntensityAnalyzer()
sia.polarity_scores("You look lonely, I can fix that!")
sia.polarity_scores("League of Legends is so fun xd")
sia.polarity_scores(example)
# Run the polarity score on the entire dataset
result = {}
for i, row in tqdm(df.iterrows(), total=len(df)):
text = row["reviewContent"]
myid = row["ID"]
result[myid] = sia.polarity_scores(text)
result_10 = dict(list(result.items())[:10])
result_10
vaders = pd.DataFrame(result).T
vaders = vaders.reset_index().rename(columns={"index": "ID"})
vaders = vaders.merge(df, how="left")
# Now we have sentiment score and metadata
vaders.head()
ax = sns.barplot(data=vaders, x="reviewRating", y="compound")
ax.set_title("Compound Score by Audience Movie Reviews")
plt.show()
fig, axs = plt.subplots(1, 3, figsize=(15, 3))
sns.barplot(data=vaders, x="reviewRating", y="pos", ax=axs[0])
sns.barplot(data=vaders, x="reviewRating", y="neu", ax=axs[1])
sns.barplot(data=vaders, x="reviewRating", y="neg", ax=axs[2])
axs[0].set_title("Positive")
axs[1].set_title("Neutral")
axs[2].set_title("Negative")
plt.tight_layout()
plt.show()
# ## Sentiment Analysis Version 2: Using RoRERTa Pretrained Model
# * Use a model trained of a large corpus of data.
# * Transformer model accounts for the words but also the context related to other words
# Facebook AI's RoBERTa model was proposed in *RoBERTa: A Robustly Optimized BERT Pretraining Approach* by Yinhan Liu, Myle Ott, Naman Goyal, Jingfei Du, Mandar Joshi, Danqi Chen, Omer Levy, Mike Lewis, Luke Zettlemoyer, Veselin Stoyanov. It is based on Google’s BERT model released in 2018.
# The Vader model only looked at each words and scored it individually. But human language depend a lot on context, for example, a positive sentence may have negative words which could be sarcastic or related to other words, hence the Vader model would not pick up on the relationships between words. However, more recently, transformer-based deep learning models have become very popular as it can pick up on that context.
# We are going to get RoBERTa model from HuggingFace. Link: https://huggingface.co/cardiffnlp/twitter-roberta-base-sentiment
# *Note: The RoBERTa model or any transformer models are optimised to be run on a GPU. Notebook options -> Accelerator -> GPU
from transformers import AutoTokenizer
from transformers import AutoModelForSequenceClassification
from scipy.special import softmax
MODEL = f"cardiffnlp/twitter-roberta-base-sentiment" # Model from HuggingFace
tokenizer = AutoTokenizer.from_pretrained(MODEL)
model = AutoModelForSequenceClassification.from_pretrained(MODEL)
# VADER results on example
print(example)
sia.polarity_scores(example)
# Run for RoBERTa Model
encoded_text = tokenizer(example, return_tensors="pt")
output = model(**encoded_text)
scores = output[0][0].detach().numpy()
scores = softmax(scores)
scores_dict = {
"roberta_neg": scores[0],
"roberta_neu": scores[1],
"roberta_pos": scores[2],
}
print(scores_dict)
def polarity_scores_roberta(example):
encoded_text = tokenizer(example, return_tensors="pt")
output = model(**encoded_text)
scores = output[0][0].detach().numpy()
scores = softmax(scores)
scores_dict = {
"roberta_neg": scores[0],
"roberta_neu": scores[1],
"roberta_pos": scores[2],
}
return scores_dict
# Run the polarity score on the entire dataset, but this time for the RoBERTa model
result = {}
for i, row in tqdm(df.iterrows(), total=len(df)):
try: # Some reviews are too big for the RoBERTa model so it will result in runtime error. We will skip those
text = row["reviewContent"]
myid = row["ID"]
vader_result = sia.polarity_scores(text)
vader_result_rename = {}
for key, value in vader_result.items():
vader_result_rename[f"vader_{key}"] = value
roberta_result = polarity_scores_roberta(text)
both = {**vader_result_rename, **roberta_result} # combining two dict
result[myid] = both
except RuntimeError:
print(f"Broke for id {myid}")
both
results_df = pd.DataFrame(result).T
results_df = results_df.reset_index().rename(columns={"index": "ID"})
results_df = results_df.merge(df, how="left")
results_df.head()
# ## Compare and Data Visualisation
results_df.columns
sns.pairplot(
data=results_df,
vars=[
"vader_neg",
"vader_neu",
"vader_pos",
"roberta_neg",
"roberta_neu",
"roberta_pos",
],
hue="reviewRating",
palette="tab10",
)
plt.show()
# ## Review Examples:
# * Positive 1/10 and Negative 10/10 Reviews
# Lets look at some examples where the model scoring and review score differ the most.
results_df
# A movie review that is said to be positive but the reviewer gave it a 1/10. What insight can we gain from this?
results_df.query("1 <= reviewRating <= 2").sort_values("roberta_pos", ascending=False)[
["ID", "roberta_neg", "roberta_neu", "roberta_pos", "reviewRating", "reviewContent"]
].values[0]
specific_row = results_df.loc[results_df["ID"] == 149]
print(specific_row)
# A movie review that is said to be negative but the reviewer gave it a 10/10. What insight can we gain from this?
results_df.query("reviewRating == 10").sort_values("roberta_neg", ascending=False)[
["ID", "roberta_neg", "roberta_neu", "roberta_pos", "reviewRating", "reviewContent"]
].values[0]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/977/129977265.ipynb
| null | null |
[{"Id": 129977265, "ScriptId": 38604764, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15133337, "CreationDate": "05/17/2023 21:42:12", "VersionNumber": 3.0, "Title": "Sentiment Analysis CSS2", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 227.0, "LinesInsertedFromPrevious": 138.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 89.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## Background Info
# In sentiment analysis using models like RoBERTa, the predicted probabilities for different sentiment classes (such as neg, neu, and pos) represent the model's confidence or likelihood for each sentiment class independently, and they are typically outputted as normalized values between 0 and 1.
# The probabilities for neg, neu, and pos are relative to each other and represent the model's estimated likelihood or confidence for each sentiment category. For example, if the predicted probabilities are [0.2, 0.5, 0.3] for neg, neu, and pos respectively, it means the model assigns a 20% probability to negative sentiment, 50% probability to neutral sentiment, and 30% probability to positive sentiment for the given input.
# ## Import Necessary Modules
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # for the graphs
import seaborn as sns
plt.style.use("ggplot")
import nltk
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Read In Data
# Read in data in a data frame
df = pd.read_csv("../input/updated-movie-reviews-dataset/New_Audience.csv")
df.head()
df["reviewContent"].values[0]
print(df.shape) # 1100 rows, 7 columns
# Make the dataframe only take in rows 0-550, can change depending on computing power
df = df.head(550)
df.head()
# ## Quick Exploratory Data Analysis (EDA)
ax = (
df["reviewRating"]
.value_counts()
.sort_index()
.plot(kind="line", title="Average Audience Review Ratings", figsize=(10, 5))
)
ax.set_xlabel("Review Ratings")
ax.set_ylabel("Numbers of Reviews")
ax.set_xticks(range(1, 11, 1))
plt.show()
# ## Basic NLTK
example = df["reviewContent"][50]
print(example)
tokens = nltk.word_tokenize(example)
tokens[:10]
tagged = nltk.pos_tag(tokens)
tagged[:10]
entities = nltk.ne_chunk(tagged)
sliced_entities = entities[:10]
nltk.pprint(sliced_entities)
# entities.pprint()
# ## Sentiment Analysis Version 1: Using VADER
# VADER (Valence Aware Dictionary and sEntiment Reasoner) - Bag of words approach
# > Using NLTK's `SentimentIntensityAnalyzer` to get the neg/neu/pos scores of the text.
# * This uses a "bag of words approach:
# 1. Stop words are removed (e.g. and, the) - just words used for structure
# 2. each word is scored and combined to a total score.
# *Note: This does not include relationship between words.
from nltk.sentiment import SentimentIntensityAnalyzer
from tqdm.notebook import tqdm
sia = SentimentIntensityAnalyzer()
sia.polarity_scores("You look lonely, I can fix that!")
sia.polarity_scores("League of Legends is so fun xd")
sia.polarity_scores(example)
# Run the polarity score on the entire dataset
result = {}
for i, row in tqdm(df.iterrows(), total=len(df)):
text = row["reviewContent"]
myid = row["ID"]
result[myid] = sia.polarity_scores(text)
result_10 = dict(list(result.items())[:10])
result_10
vaders = pd.DataFrame(result).T
vaders = vaders.reset_index().rename(columns={"index": "ID"})
vaders = vaders.merge(df, how="left")
# Now we have sentiment score and metadata
vaders.head()
ax = sns.barplot(data=vaders, x="reviewRating", y="compound")
ax.set_title("Compound Score by Audience Movie Reviews")
plt.show()
fig, axs = plt.subplots(1, 3, figsize=(15, 3))
sns.barplot(data=vaders, x="reviewRating", y="pos", ax=axs[0])
sns.barplot(data=vaders, x="reviewRating", y="neu", ax=axs[1])
sns.barplot(data=vaders, x="reviewRating", y="neg", ax=axs[2])
axs[0].set_title("Positive")
axs[1].set_title("Neutral")
axs[2].set_title("Negative")
plt.tight_layout()
plt.show()
# ## Sentiment Analysis Version 2: Using RoRERTa Pretrained Model
# * Use a model trained of a large corpus of data.
# * Transformer model accounts for the words but also the context related to other words
# Facebook AI's RoBERTa model was proposed in *RoBERTa: A Robustly Optimized BERT Pretraining Approach* by Yinhan Liu, Myle Ott, Naman Goyal, Jingfei Du, Mandar Joshi, Danqi Chen, Omer Levy, Mike Lewis, Luke Zettlemoyer, Veselin Stoyanov. It is based on Google’s BERT model released in 2018.
# The Vader model only looked at each words and scored it individually. But human language depend a lot on context, for example, a positive sentence may have negative words which could be sarcastic or related to other words, hence the Vader model would not pick up on the relationships between words. However, more recently, transformer-based deep learning models have become very popular as it can pick up on that context.
# We are going to get RoBERTa model from HuggingFace. Link: https://huggingface.co/cardiffnlp/twitter-roberta-base-sentiment
# *Note: The RoBERTa model or any transformer models are optimised to be run on a GPU. Notebook options -> Accelerator -> GPU
from transformers import AutoTokenizer
from transformers import AutoModelForSequenceClassification
from scipy.special import softmax
MODEL = f"cardiffnlp/twitter-roberta-base-sentiment" # Model from HuggingFace
tokenizer = AutoTokenizer.from_pretrained(MODEL)
model = AutoModelForSequenceClassification.from_pretrained(MODEL)
# VADER results on example
print(example)
sia.polarity_scores(example)
# Run for RoBERTa Model
encoded_text = tokenizer(example, return_tensors="pt")
output = model(**encoded_text)
scores = output[0][0].detach().numpy()
scores = softmax(scores)
scores_dict = {
"roberta_neg": scores[0],
"roberta_neu": scores[1],
"roberta_pos": scores[2],
}
print(scores_dict)
def polarity_scores_roberta(example):
encoded_text = tokenizer(example, return_tensors="pt")
output = model(**encoded_text)
scores = output[0][0].detach().numpy()
scores = softmax(scores)
scores_dict = {
"roberta_neg": scores[0],
"roberta_neu": scores[1],
"roberta_pos": scores[2],
}
return scores_dict
# Run the polarity score on the entire dataset, but this time for the RoBERTa model
result = {}
for i, row in tqdm(df.iterrows(), total=len(df)):
try: # Some reviews are too big for the RoBERTa model so it will result in runtime error. We will skip those
text = row["reviewContent"]
myid = row["ID"]
vader_result = sia.polarity_scores(text)
vader_result_rename = {}
for key, value in vader_result.items():
vader_result_rename[f"vader_{key}"] = value
roberta_result = polarity_scores_roberta(text)
both = {**vader_result_rename, **roberta_result} # combining two dict
result[myid] = both
except RuntimeError:
print(f"Broke for id {myid}")
both
results_df = pd.DataFrame(result).T
results_df = results_df.reset_index().rename(columns={"index": "ID"})
results_df = results_df.merge(df, how="left")
results_df.head()
# ## Compare and Data Visualisation
results_df.columns
sns.pairplot(
data=results_df,
vars=[
"vader_neg",
"vader_neu",
"vader_pos",
"roberta_neg",
"roberta_neu",
"roberta_pos",
],
hue="reviewRating",
palette="tab10",
)
plt.show()
# ## Review Examples:
# * Positive 1/10 and Negative 10/10 Reviews
# Lets look at some examples where the model scoring and review score differ the most.
results_df
# A movie review that is said to be positive but the reviewer gave it a 1/10. What insight can we gain from this?
results_df.query("1 <= reviewRating <= 2").sort_values("roberta_pos", ascending=False)[
["ID", "roberta_neg", "roberta_neu", "roberta_pos", "reviewRating", "reviewContent"]
].values[0]
specific_row = results_df.loc[results_df["ID"] == 149]
print(specific_row)
# A movie review that is said to be negative but the reviewer gave it a 10/10. What insight can we gain from this?
results_df.query("reviewRating == 10").sort_values("roberta_neg", ascending=False)[
["ID", "roberta_neg", "roberta_neu", "roberta_pos", "reviewRating", "reviewContent"]
].values[0]
| false | 0 | 2,510 | 0 | 2,510 | 2,510 |
||
129977789
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from statsmodels.formula.api import ols
from sklearn.linear_model import LinearRegression
import matplotlib as mpl
from statsmodels.api import qqplot
import statsmodels.api as sm
from statsmodels.formula.api import glm
from statsmodels.formula.api import logit
from statsmodels.formula.api import ols
from statsmodels.stats.outliers_influence import variance_inflation_factor
dirname = "/kaggle/input/economic-games-ba2/"
# figure setting
# Reset rcParams to default values
mpl.rcParams.update()
# globally setting seaborn
sns.set(style="ticks", palette="muted", font_scale=1.2, context="talk")
mpl.rcParams["lines.linewidth"] = 2
mpl.rcParams["lines.markersize"] = 10
mpl.rcParams["font.size"] = 16
all_avg_120 = pd.read_csv(dirname + "avg_120FA.csv")
both75_avg_102 = pd.read_csv(dirname + "both75avg102FA.csv")
both75_unfold_102 = pd.read_csv(dirname + "both75unfold102wFA.csv")
dg75_avg_113 = pd.read_csv(dirname + "DG75avg113FA.csv")
dg75_unfold_113 = pd.read_csv(dirname + "DG75unfold113wFA.csv")
tg75_avg_106 = pd.read_csv(dirname + "TG75avg106FA.csv")
tg75_unfold_106 = pd.read_csv(dirname + "TG75unfold106wFA.csv")
sl_all_avg = all_avg_120.iloc[:, 4:].reset_index(drop=True)
sl_both75_avg = both75_avg_102.iloc[:, 4:].reset_index(drop=True)
sl_both75_unfold = both75_unfold_102.iloc[:, 2:].reset_index(drop=True)
sl_dg75_avg = dg75_avg_113.iloc[:, 4:].reset_index(drop=True)
sl_dg75_unfold = dg75_unfold_113.iloc[:, 2:].reset_index(drop=True)
sl_tg75_avg = tg75_avg_106.iloc[:, 4:].reset_index(drop=True)
sl_tg75_unfold = tg75_unfold_106.iloc[:, 2:].reset_index(drop=True)
# add BLUPs predictiors from R
# match columns to match from both df left_keys are in sl_..75_avg, right_keys are in blup
left_keys = [
"Age",
"Betrayal",
"Anger",
"Sadness",
"Disgust",
"Surprise",
"Cog_Motivate",
"Cog_Reasons",
"Cog_Defend",
"Cog_ToM",
"IRIpt",
"IRIfs",
"IRIec",
"IRIpd",
]
right_keys = [
"age",
"betrayal",
"anger",
"sadness",
"disgust",
"surprise",
"motivation",
"reason",
"defend",
"perspective",
"IRI_pt",
"IRI_fs",
"IRI_ec",
"IRI_pd",
]
# import each df
blup_df_both = pd.read_csv(dirname + "BLUP_predictors_both75_raw.csv")
blup_df_dg = pd.read_csv(dirname + "BLUP_predictors_dg75_raw.csv")
blup_df_tg = pd.read_csv(dirname + "BLUP_predictors_tg75_raw.csv")
# columns you want to have in your dataset
blup_cols = [
col
for col in blup_df_both.columns
if not col.endswith(".x") and ("c." in col or col in ["female.y", "british.y"])
]
sl_cols = list(sl_both75_avg.columns)
sl_cols.extend(blup_cols)
# merge two df based on given keys, return error if one row match multiple rows
sl_both75_avg = sl_both75_avg.merge(
blup_df_both, left_on=left_keys, right_on=right_keys, validate="one_to_one"
)
# extract wanted columns
sl_both75_avg = sl_both75_avg[sl_cols]
# rename cols
sl_both75_avg.columns = sl_both75_avg.columns.str.replace(".", "_").str.replace(
"_y", ""
)
# repeat for dg
sl_dg75_avg = sl_dg75_avg.merge(
blup_df_dg, left_on=left_keys, right_on=right_keys, validate="one_to_one"
)
sl_dg75_avg = sl_dg75_avg[sl_cols]
sl_dg75_avg.columns = sl_dg75_avg.columns.str.replace(".", "_").str.replace("_y", "")
# repeat for tg
sl_tg75_avg = sl_tg75_avg.merge(
blup_df_tg, left_on=left_keys, right_on=right_keys, validate="one_to_one"
)
sl_tg75_avg = sl_tg75_avg[sl_cols]
sl_tg75_avg.columns = sl_tg75_avg.columns.str.replace(".", "_").str.replace("_y", "")
# cut the cols before subNum (original approach)
dataframes = [
sl_all_avg,
sl_both75_avg,
sl_both75_unfold,
sl_dg75_avg,
sl_dg75_unfold,
sl_tg75_avg,
sl_tg75_unfold,
]
names = [
"sl_all_avg",
"sl_both75_avg",
"sl_both75_unfold",
"sl_dg75_avg",
"sl_dg75_unfold",
"sl_tg75_avg",
"sl_tg75_unfold",
]
tg_multi = 3
for df, name in zip(dataframes, names):
df["trustworthiness_avg"] = (
df["trstee_ST1"] / (tg_multi * 2)
+ df["trstee_ST2"] / (tg_multi * 4)
+ df["trstee_ST3"] / (tg_multi * 6)
+ df["trstee_ST4"] / (tg_multi * 8)
+ df["trstee_ST5"] / (tg_multi * 10)
) / 5
filename = f"{name}.csv"
df.to_csv(
filename, index=False
) # Save the dataframe as a CSV file using the file name
sl_tg75_avg_blups = pd.read_csv("sl_tg75_avg.csv")
sl_dg75_avg_blups = pd.read_csv("sl_dg75_avg.csv")
sl_both75_avg_blups = pd.read_csv("sl_both75_avg.csv")
## mean center
# tg75
sl_tg75_avg_blups["trustworthiness_c"] = (
sl_tg75_avg_blups["trustworthiness_avg"]
- sl_tg75_avg_blups["trustworthiness_avg"].mean()
)
sl_tg75_avg_blups["BIS11ATT_c"] = (
sl_tg75_avg_blups["BIS11ATT"] - sl_tg75_avg_blups["BIS11ATT"].mean()
)
sl_tg75_avg_blups["BIS11MT_c"] = (
sl_tg75_avg_blups["BIS11MT"] - sl_tg75_avg_blups["BIS11MT"].mean()
)
sl_tg75_avg_blups["BIS11NP_c"] = (
sl_tg75_avg_blups["BIS11NP"] - sl_tg75_avg_blups["BIS11NP"].mean()
)
sl_tg75_avg_blups["trustAttitude_c"] = (
sl_tg75_avg_blups["trustAttitude"] - sl_tg75_avg_blups["trustAttitude"].mean()
)
sl_tg75_avg_blups["riskTaking_c"] = (
sl_tg75_avg_blups["riskTaking"] - sl_tg75_avg_blups["riskTaking"].mean()
)
sl_tg75_avg_blups["TG_exptRatio_c"] = (
sl_tg75_avg_blups["TG_exptRatio"] - sl_tg75_avg_blups["TG_exptRatio"].mean()
)
sl_tg75_avg_blups["TG_exptRatioAdj_c"] = (
sl_tg75_avg_blups["TG_exptRatioAdj"] - sl_tg75_avg_blups["TG_exptRatioAdj"].mean()
)
# dg75
sl_dg75_avg_blups["trustworthiness_c"] = (
sl_dg75_avg_blups["trustworthiness_avg"]
- sl_dg75_avg_blups["trustworthiness_avg"].mean()
)
sl_dg75_avg_blups["BIS11ATT_c"] = (
sl_dg75_avg_blups["BIS11ATT"] - sl_dg75_avg_blups["BIS11ATT"].mean()
)
sl_dg75_avg_blups["BIS11MT_c"] = (
sl_dg75_avg_blups["BIS11MT"] - sl_dg75_avg_blups["BIS11MT"].mean()
)
sl_dg75_avg_blups["BIS11NP_c"] = (
sl_dg75_avg_blups["BIS11NP"] - sl_dg75_avg_blups["BIS11NP"].mean()
)
sl_dg75_avg_blups["trustAttitude_c"] = (
sl_dg75_avg_blups["trustAttitude"] - sl_dg75_avg_blups["trustAttitude"].mean()
)
sl_dg75_avg_blups["riskTaking_c"] = (
sl_dg75_avg_blups["riskTaking"] - sl_dg75_avg_blups["riskTaking"].mean()
)
sl_dg75_avg_blups["TG_exptRatio_c"] = (
sl_dg75_avg_blups["TG_exptRatio"] - sl_dg75_avg_blups["TG_exptRatio"].mean()
)
sl_dg75_avg_blups["TG_exptRatioAdj_c"] = (
sl_dg75_avg_blups["TG_exptRatioAdj"] - sl_dg75_avg_blups["TG_exptRatioAdj"].mean()
)
# RT log transformation
sl_tg75_avg_blups["log_TG_trustorRT"] = np.log(sl_tg75_avg_blups["TG_trustorRT"])
sl_tg75_avg_blups["log_DG_dictatorRT"] = np.log(sl_tg75_avg_blups["DG_dictatorRT"])
sl_tg75_avg_blups["log_TG_expectRT"] = np.log(sl_tg75_avg_blups["TG_expectRT"])
sl_tg75_avg_blups["log_TG_closeRT"] = np.log(sl_tg75_avg_blups["TG_closeRT"])
sl_tg75_avg_blups["log_DG_closeRT"] = np.log(sl_tg75_avg_blups["DG_closeRT"])
sns.regplot(x="IRIec", y="BLUP_c_betrayal", data=sl_tg75_avg_blups)
plt.xlabel("IRI ec")
plt.ylabel("BLUP betrayal")
plt.show()
# all IRI to predict betrayal BLUPs
betrayal_blups_vs_all_IRI = ols(
"BLUP_c_betrayal ~ female + c_income + c_age + c_education + british + c_IRI_pt + c_IRI_fs + c_IRI_ec + c_IRI_pd ",
data=sl_tg75_avg_blups,
).fit()
betrayal_blups_vs_all_IRI.summary()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/977/129977789.ipynb
| null | null |
[{"Id": 129977789, "ScriptId": 38664122, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15152859, "CreationDate": "05/17/2023 21:50:08", "VersionNumber": 1.0, "Title": "Micro-Macro Model analysis economic games", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 148.0, "LinesInsertedFromPrevious": 148.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from statsmodels.formula.api import ols
from sklearn.linear_model import LinearRegression
import matplotlib as mpl
from statsmodels.api import qqplot
import statsmodels.api as sm
from statsmodels.formula.api import glm
from statsmodels.formula.api import logit
from statsmodels.formula.api import ols
from statsmodels.stats.outliers_influence import variance_inflation_factor
dirname = "/kaggle/input/economic-games-ba2/"
# figure setting
# Reset rcParams to default values
mpl.rcParams.update()
# globally setting seaborn
sns.set(style="ticks", palette="muted", font_scale=1.2, context="talk")
mpl.rcParams["lines.linewidth"] = 2
mpl.rcParams["lines.markersize"] = 10
mpl.rcParams["font.size"] = 16
all_avg_120 = pd.read_csv(dirname + "avg_120FA.csv")
both75_avg_102 = pd.read_csv(dirname + "both75avg102FA.csv")
both75_unfold_102 = pd.read_csv(dirname + "both75unfold102wFA.csv")
dg75_avg_113 = pd.read_csv(dirname + "DG75avg113FA.csv")
dg75_unfold_113 = pd.read_csv(dirname + "DG75unfold113wFA.csv")
tg75_avg_106 = pd.read_csv(dirname + "TG75avg106FA.csv")
tg75_unfold_106 = pd.read_csv(dirname + "TG75unfold106wFA.csv")
sl_all_avg = all_avg_120.iloc[:, 4:].reset_index(drop=True)
sl_both75_avg = both75_avg_102.iloc[:, 4:].reset_index(drop=True)
sl_both75_unfold = both75_unfold_102.iloc[:, 2:].reset_index(drop=True)
sl_dg75_avg = dg75_avg_113.iloc[:, 4:].reset_index(drop=True)
sl_dg75_unfold = dg75_unfold_113.iloc[:, 2:].reset_index(drop=True)
sl_tg75_avg = tg75_avg_106.iloc[:, 4:].reset_index(drop=True)
sl_tg75_unfold = tg75_unfold_106.iloc[:, 2:].reset_index(drop=True)
# add BLUPs predictiors from R
# match columns to match from both df left_keys are in sl_..75_avg, right_keys are in blup
left_keys = [
"Age",
"Betrayal",
"Anger",
"Sadness",
"Disgust",
"Surprise",
"Cog_Motivate",
"Cog_Reasons",
"Cog_Defend",
"Cog_ToM",
"IRIpt",
"IRIfs",
"IRIec",
"IRIpd",
]
right_keys = [
"age",
"betrayal",
"anger",
"sadness",
"disgust",
"surprise",
"motivation",
"reason",
"defend",
"perspective",
"IRI_pt",
"IRI_fs",
"IRI_ec",
"IRI_pd",
]
# import each df
blup_df_both = pd.read_csv(dirname + "BLUP_predictors_both75_raw.csv")
blup_df_dg = pd.read_csv(dirname + "BLUP_predictors_dg75_raw.csv")
blup_df_tg = pd.read_csv(dirname + "BLUP_predictors_tg75_raw.csv")
# columns you want to have in your dataset
blup_cols = [
col
for col in blup_df_both.columns
if not col.endswith(".x") and ("c." in col or col in ["female.y", "british.y"])
]
sl_cols = list(sl_both75_avg.columns)
sl_cols.extend(blup_cols)
# merge two df based on given keys, return error if one row match multiple rows
sl_both75_avg = sl_both75_avg.merge(
blup_df_both, left_on=left_keys, right_on=right_keys, validate="one_to_one"
)
# extract wanted columns
sl_both75_avg = sl_both75_avg[sl_cols]
# rename cols
sl_both75_avg.columns = sl_both75_avg.columns.str.replace(".", "_").str.replace(
"_y", ""
)
# repeat for dg
sl_dg75_avg = sl_dg75_avg.merge(
blup_df_dg, left_on=left_keys, right_on=right_keys, validate="one_to_one"
)
sl_dg75_avg = sl_dg75_avg[sl_cols]
sl_dg75_avg.columns = sl_dg75_avg.columns.str.replace(".", "_").str.replace("_y", "")
# repeat for tg
sl_tg75_avg = sl_tg75_avg.merge(
blup_df_tg, left_on=left_keys, right_on=right_keys, validate="one_to_one"
)
sl_tg75_avg = sl_tg75_avg[sl_cols]
sl_tg75_avg.columns = sl_tg75_avg.columns.str.replace(".", "_").str.replace("_y", "")
# cut the cols before subNum (original approach)
dataframes = [
sl_all_avg,
sl_both75_avg,
sl_both75_unfold,
sl_dg75_avg,
sl_dg75_unfold,
sl_tg75_avg,
sl_tg75_unfold,
]
names = [
"sl_all_avg",
"sl_both75_avg",
"sl_both75_unfold",
"sl_dg75_avg",
"sl_dg75_unfold",
"sl_tg75_avg",
"sl_tg75_unfold",
]
tg_multi = 3
for df, name in zip(dataframes, names):
df["trustworthiness_avg"] = (
df["trstee_ST1"] / (tg_multi * 2)
+ df["trstee_ST2"] / (tg_multi * 4)
+ df["trstee_ST3"] / (tg_multi * 6)
+ df["trstee_ST4"] / (tg_multi * 8)
+ df["trstee_ST5"] / (tg_multi * 10)
) / 5
filename = f"{name}.csv"
df.to_csv(
filename, index=False
) # Save the dataframe as a CSV file using the file name
sl_tg75_avg_blups = pd.read_csv("sl_tg75_avg.csv")
sl_dg75_avg_blups = pd.read_csv("sl_dg75_avg.csv")
sl_both75_avg_blups = pd.read_csv("sl_both75_avg.csv")
## mean center
# tg75
sl_tg75_avg_blups["trustworthiness_c"] = (
sl_tg75_avg_blups["trustworthiness_avg"]
- sl_tg75_avg_blups["trustworthiness_avg"].mean()
)
sl_tg75_avg_blups["BIS11ATT_c"] = (
sl_tg75_avg_blups["BIS11ATT"] - sl_tg75_avg_blups["BIS11ATT"].mean()
)
sl_tg75_avg_blups["BIS11MT_c"] = (
sl_tg75_avg_blups["BIS11MT"] - sl_tg75_avg_blups["BIS11MT"].mean()
)
sl_tg75_avg_blups["BIS11NP_c"] = (
sl_tg75_avg_blups["BIS11NP"] - sl_tg75_avg_blups["BIS11NP"].mean()
)
sl_tg75_avg_blups["trustAttitude_c"] = (
sl_tg75_avg_blups["trustAttitude"] - sl_tg75_avg_blups["trustAttitude"].mean()
)
sl_tg75_avg_blups["riskTaking_c"] = (
sl_tg75_avg_blups["riskTaking"] - sl_tg75_avg_blups["riskTaking"].mean()
)
sl_tg75_avg_blups["TG_exptRatio_c"] = (
sl_tg75_avg_blups["TG_exptRatio"] - sl_tg75_avg_blups["TG_exptRatio"].mean()
)
sl_tg75_avg_blups["TG_exptRatioAdj_c"] = (
sl_tg75_avg_blups["TG_exptRatioAdj"] - sl_tg75_avg_blups["TG_exptRatioAdj"].mean()
)
# dg75
sl_dg75_avg_blups["trustworthiness_c"] = (
sl_dg75_avg_blups["trustworthiness_avg"]
- sl_dg75_avg_blups["trustworthiness_avg"].mean()
)
sl_dg75_avg_blups["BIS11ATT_c"] = (
sl_dg75_avg_blups["BIS11ATT"] - sl_dg75_avg_blups["BIS11ATT"].mean()
)
sl_dg75_avg_blups["BIS11MT_c"] = (
sl_dg75_avg_blups["BIS11MT"] - sl_dg75_avg_blups["BIS11MT"].mean()
)
sl_dg75_avg_blups["BIS11NP_c"] = (
sl_dg75_avg_blups["BIS11NP"] - sl_dg75_avg_blups["BIS11NP"].mean()
)
sl_dg75_avg_blups["trustAttitude_c"] = (
sl_dg75_avg_blups["trustAttitude"] - sl_dg75_avg_blups["trustAttitude"].mean()
)
sl_dg75_avg_blups["riskTaking_c"] = (
sl_dg75_avg_blups["riskTaking"] - sl_dg75_avg_blups["riskTaking"].mean()
)
sl_dg75_avg_blups["TG_exptRatio_c"] = (
sl_dg75_avg_blups["TG_exptRatio"] - sl_dg75_avg_blups["TG_exptRatio"].mean()
)
sl_dg75_avg_blups["TG_exptRatioAdj_c"] = (
sl_dg75_avg_blups["TG_exptRatioAdj"] - sl_dg75_avg_blups["TG_exptRatioAdj"].mean()
)
# RT log transformation
sl_tg75_avg_blups["log_TG_trustorRT"] = np.log(sl_tg75_avg_blups["TG_trustorRT"])
sl_tg75_avg_blups["log_DG_dictatorRT"] = np.log(sl_tg75_avg_blups["DG_dictatorRT"])
sl_tg75_avg_blups["log_TG_expectRT"] = np.log(sl_tg75_avg_blups["TG_expectRT"])
sl_tg75_avg_blups["log_TG_closeRT"] = np.log(sl_tg75_avg_blups["TG_closeRT"])
sl_tg75_avg_blups["log_DG_closeRT"] = np.log(sl_tg75_avg_blups["DG_closeRT"])
sns.regplot(x="IRIec", y="BLUP_c_betrayal", data=sl_tg75_avg_blups)
plt.xlabel("IRI ec")
plt.ylabel("BLUP betrayal")
plt.show()
# all IRI to predict betrayal BLUPs
betrayal_blups_vs_all_IRI = ols(
"BLUP_c_betrayal ~ female + c_income + c_age + c_education + british + c_IRI_pt + c_IRI_fs + c_IRI_ec + c_IRI_pd ",
data=sl_tg75_avg_blups,
).fit()
betrayal_blups_vs_all_IRI.summary()
| false | 0 | 3,251 | 0 | 3,251 | 3,251 |
||
129894779
|
<jupyter_start><jupyter_text>ntasset
Kaggle dataset identifier: ntasset
<jupyter_script>import pandas as pd
import numpy as np
import sys
from functools import reduce
def get_funda_df():
csi_df = pd.read_csv("/kaggle/input/ntasset/test/companystockinfo.csv")
nonfin_income_df = pd.read_csv("/kaggle/input/ntasset/test/incomestatement.csv")
nonfin_bs_df = pd.read_csv("/kaggle/input/ntasset/test/balancesheet.csv")
# Merge annual tables
funda_df = pd.merge(
nonfin_income_df, nonfin_bs_df, on=["companyid", "year"], how="inner"
)
funda_df = pd.merge(csi_df, funda_df, on=["companyid"], how="inner")
return funda_df
def get_forecast_df():
csi_df = pd.read_csv("/kaggle/input/ntasset/test/companystockinfo.csv")
nonfin_forecastannual_df = pd.read_csv("/kaggle/input/ntasset/test/forecast.csv")
fxrate_df = pd.read_csv("/kaggle/input/ntasset/test/fxrate.csv")
# Merge forecast table
forecast_df = pd.merge(
csi_df, nonfin_forecastannual_df, on=["companyid"], how="inner"
)
return forecast_df
if __name__ == "__main__":
funda_df = get_funda_df()
forecast_df = get_forecast_df()
funda_df = funda_df.sort_values(by=["companyid", "year"])
forecast_df = forecast_df.sort_values(by=["companyid", "year"])
print(funda_df)
print(forecast_df)
output_df = pd.DataFrame()
output_df[["companyid", "latestfinyear"]] = funda_df[["companyid", "latestfinyear"]]
output_df = output_df.sort_values(by="companyid")
output_df = output_df.drop_duplicates(subset=["companyid"])
output_df = output_df.reset_index(drop=True)
prev_row = None
roe_val = []
for _, r in funda_df.iterrows():
if prev_row is None:
roe_val.append(0)
else:
roe_val.append(
100 * r["netprofit"] / (0.5 * r["totequity"] + prev_row["totequity"])
)
prev_row = r
funda_df["roe"] = roe_val
for i in range(4, -1, -1):
if i == 0:
output_df["roe fy"] = list(
(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"]].sort_values(
by=["companyid"]
)["roe"]
)
)
else:
output_df["roe fy-" + str(i)] = list(
(
funda_df.loc[
funda_df["year"] == funda_df["latestfinyear"] - i
].sort_values(by=["companyid"])["roe"]
)
)
for i in range(1, 4):
tmp_df = pd.DataFrame(
forecast_df.loc[forecast_df["year"] == forecast_df["latestfinyear"] + i][
["companyid", "roe"]
]
)
tmp_df = tmp_df.rename(columns={"roe": "roe f+" + str(i)})
output_df = pd.merge(output_df, tmp_df, how="left")
output_df["5yr median roe"] = output_df.apply(
lambda x: np.nanmedian(
x[["roe fy", "roe fy-1", "roe fy-2", "roe fy-3", "roe fy-4"]]
),
axis=1,
)
output_df
# Target Columns: ['roa fy-4', 'roa fy-3', 'roa fy-2', 'roa fy-1', 'roa fy']
# roa[fy-i] = 100*netprofit[fy-i] / (0.5*(totassets[fy-i] + totassets[fy-i-1]))
prev_row = None
roa_val = []
for _, r in funda_df.iterrows():
if prev_row is None:
roa_val.append(0)
else:
roa_val.append(
100 * r["netprofit"] / (0.5 * r["totassets"] + prev_row["totassets"])
)
prev_row = r
funda_df["roa"] = roa_val
funda_df
# Target Columns: ['roa fy-4', 'roa fy-3', 'roa fy-2', 'roa fy-1', 'roa fy']
# roa[fy-i] = 100*netprofit[fy-i] / (0.5*(totassets[fy-i] + totassets[fy-i-1]))
for i in range(4, -1, -1):
if i == 0:
output_df["roa fy"] = list(
(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"]].sort_values(
by=["companyid"]
)["roa"]
)
)
else:
output_df["roa fy-" + str(i)] = list(
(
funda_df.loc[
funda_df["year"] == funda_df["latestfinyear"] - i
].sort_values(by=["companyid"])["roa"]
)
)
# Target Columns: ['roa fy+1', 'roa fy+2', 'roa fy+3']
# Pulled directly from forecast.csv
for i in range(1, 4):
tmp_df = pd.DataFrame(
forecast_df.loc[forecast_df["year"] == forecast_df["latestfinyear"] + i][
["companyid", "roa"]
]
)
tmp_df = tmp_df.rename(columns={"roa": "roa fy+" + str(i)})
output_df = pd.merge(output_df, tmp_df, how="left")
# Target Columns: ['5yr median roa']
# '5yr median roa' = numpy.nanmedian(['roa fy-4', 'roa fy-3', 'roa fy-2', 'roa fy-1', 'roa fy'])
output_df["5yr median roa"] = output_df.apply(
lambda x: np.nanmedian(
x[["roa fy", "roa fy-1", "roa fy-2", "roa fy-3", "roa fy-4"]]
),
axis=1,
)
output_df
# Target Columns: ['netde fy']
# netde[fy] = (totaldebt[fy] – cashncashequiv[fy])/ totequity[fy]
funda_df["netde"] = (funda_df["totaldebt"] - funda_df["cashncashequiv"]) / funda_df[
"totequity"
]
output_df["netde fy"] = list(
(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"]].sort_values(
by=["companyid"]
)["netde"]
)
)
output_df
# Target Columns: ['revenue fy-4', 'revenue fy-3', 'revenue fy-2', 'revenue fy-1', 'revenue fy']
# Pulled directly from incomestatement.csv
for i in range(4, -1, -1):
if i != 0:
tmp_df = pd.DataFrame(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"] - i][
["companyid", "revenue"]
]
)
tmp_df = tmp_df.rename(columns={"revenue": "revenue fy-" + str(i)})
output_df = pd.merge(output_df, tmp_df, how="left")
else:
tmp_df = pd.DataFrame(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"] - i][
["companyid", "revenue"]
]
)
tmp_df = tmp_df.rename(columns={"revenue": "revenue fy"})
output_df = pd.merge(output_df, tmp_df, how="left")
output_df
# Target Columns: ['revenue fy+1', 'revenue fy+2', 'revenue fy+3']
# Pulled directly from forecast.csv and convert currency from ‘estcurr’ to ‘reportingcurr’.
for i in range(1, 4):
tmp_df = pd.DataFrame(
(
forecast_df.loc[forecast_df["year"] == forecast_df["latestfinyear"] + i][
["companyid", "revenue"]
]
)
)
tmp_df = tmp_df.rename(columns={"revenue": "revenue fy+" + str(i)})
output_df = pd.merge(output_df, tmp_df, how="left")
output_df
# Target Columns: ['revenue 2yr cagr','revenue 3yr cagr']
# revenue 2yr cagr = 100*[(revenue fy+2 / revenue fy)^(0.5) – 1]
# revenue 3yr cagr = 100*[(revenue fy+3 / revenue fy)^(0.33) – 1]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/894/129894779.ipynb
|
ntasset
|
tirapatrs
|
[{"Id": 129894779, "ScriptId": 38548904, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11676930, "CreationDate": "05/17/2023 09:06:25", "VersionNumber": 2.0, "Title": "NTAsset", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 126.0, "LinesInsertedFromPrevious": 70.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 56.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186305369, "KernelVersionId": 129894779, "SourceDatasetVersionId": 5699193}]
|
[{"Id": 5699193, "DatasetId": 3277112, "DatasourceVersionId": 5774852, "CreatorUserId": 11676930, "LicenseName": "Unknown", "CreationDate": "05/16/2023 14:18:19", "VersionNumber": 1.0, "Title": "ntasset", "Slug": "ntasset", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3277112, "CreatorUserId": 11676930, "OwnerUserId": 11676930.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5699193.0, "CurrentDatasourceVersionId": 5774852.0, "ForumId": 3342800, "Type": 2, "CreationDate": "05/16/2023 14:18:19", "LastActivityDate": "05/16/2023", "TotalViews": 5, "TotalDownloads": 0, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 11676930, "UserName": "tirapatrs", "DisplayName": "Tirapatr S", "RegisterDate": "09/22/2022", "PerformanceTier": 0}]
|
import pandas as pd
import numpy as np
import sys
from functools import reduce
def get_funda_df():
csi_df = pd.read_csv("/kaggle/input/ntasset/test/companystockinfo.csv")
nonfin_income_df = pd.read_csv("/kaggle/input/ntasset/test/incomestatement.csv")
nonfin_bs_df = pd.read_csv("/kaggle/input/ntasset/test/balancesheet.csv")
# Merge annual tables
funda_df = pd.merge(
nonfin_income_df, nonfin_bs_df, on=["companyid", "year"], how="inner"
)
funda_df = pd.merge(csi_df, funda_df, on=["companyid"], how="inner")
return funda_df
def get_forecast_df():
csi_df = pd.read_csv("/kaggle/input/ntasset/test/companystockinfo.csv")
nonfin_forecastannual_df = pd.read_csv("/kaggle/input/ntasset/test/forecast.csv")
fxrate_df = pd.read_csv("/kaggle/input/ntasset/test/fxrate.csv")
# Merge forecast table
forecast_df = pd.merge(
csi_df, nonfin_forecastannual_df, on=["companyid"], how="inner"
)
return forecast_df
if __name__ == "__main__":
funda_df = get_funda_df()
forecast_df = get_forecast_df()
funda_df = funda_df.sort_values(by=["companyid", "year"])
forecast_df = forecast_df.sort_values(by=["companyid", "year"])
print(funda_df)
print(forecast_df)
output_df = pd.DataFrame()
output_df[["companyid", "latestfinyear"]] = funda_df[["companyid", "latestfinyear"]]
output_df = output_df.sort_values(by="companyid")
output_df = output_df.drop_duplicates(subset=["companyid"])
output_df = output_df.reset_index(drop=True)
prev_row = None
roe_val = []
for _, r in funda_df.iterrows():
if prev_row is None:
roe_val.append(0)
else:
roe_val.append(
100 * r["netprofit"] / (0.5 * r["totequity"] + prev_row["totequity"])
)
prev_row = r
funda_df["roe"] = roe_val
for i in range(4, -1, -1):
if i == 0:
output_df["roe fy"] = list(
(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"]].sort_values(
by=["companyid"]
)["roe"]
)
)
else:
output_df["roe fy-" + str(i)] = list(
(
funda_df.loc[
funda_df["year"] == funda_df["latestfinyear"] - i
].sort_values(by=["companyid"])["roe"]
)
)
for i in range(1, 4):
tmp_df = pd.DataFrame(
forecast_df.loc[forecast_df["year"] == forecast_df["latestfinyear"] + i][
["companyid", "roe"]
]
)
tmp_df = tmp_df.rename(columns={"roe": "roe f+" + str(i)})
output_df = pd.merge(output_df, tmp_df, how="left")
output_df["5yr median roe"] = output_df.apply(
lambda x: np.nanmedian(
x[["roe fy", "roe fy-1", "roe fy-2", "roe fy-3", "roe fy-4"]]
),
axis=1,
)
output_df
# Target Columns: ['roa fy-4', 'roa fy-3', 'roa fy-2', 'roa fy-1', 'roa fy']
# roa[fy-i] = 100*netprofit[fy-i] / (0.5*(totassets[fy-i] + totassets[fy-i-1]))
prev_row = None
roa_val = []
for _, r in funda_df.iterrows():
if prev_row is None:
roa_val.append(0)
else:
roa_val.append(
100 * r["netprofit"] / (0.5 * r["totassets"] + prev_row["totassets"])
)
prev_row = r
funda_df["roa"] = roa_val
funda_df
# Target Columns: ['roa fy-4', 'roa fy-3', 'roa fy-2', 'roa fy-1', 'roa fy']
# roa[fy-i] = 100*netprofit[fy-i] / (0.5*(totassets[fy-i] + totassets[fy-i-1]))
for i in range(4, -1, -1):
if i == 0:
output_df["roa fy"] = list(
(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"]].sort_values(
by=["companyid"]
)["roa"]
)
)
else:
output_df["roa fy-" + str(i)] = list(
(
funda_df.loc[
funda_df["year"] == funda_df["latestfinyear"] - i
].sort_values(by=["companyid"])["roa"]
)
)
# Target Columns: ['roa fy+1', 'roa fy+2', 'roa fy+3']
# Pulled directly from forecast.csv
for i in range(1, 4):
tmp_df = pd.DataFrame(
forecast_df.loc[forecast_df["year"] == forecast_df["latestfinyear"] + i][
["companyid", "roa"]
]
)
tmp_df = tmp_df.rename(columns={"roa": "roa fy+" + str(i)})
output_df = pd.merge(output_df, tmp_df, how="left")
# Target Columns: ['5yr median roa']
# '5yr median roa' = numpy.nanmedian(['roa fy-4', 'roa fy-3', 'roa fy-2', 'roa fy-1', 'roa fy'])
output_df["5yr median roa"] = output_df.apply(
lambda x: np.nanmedian(
x[["roa fy", "roa fy-1", "roa fy-2", "roa fy-3", "roa fy-4"]]
),
axis=1,
)
output_df
# Target Columns: ['netde fy']
# netde[fy] = (totaldebt[fy] – cashncashequiv[fy])/ totequity[fy]
funda_df["netde"] = (funda_df["totaldebt"] - funda_df["cashncashequiv"]) / funda_df[
"totequity"
]
output_df["netde fy"] = list(
(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"]].sort_values(
by=["companyid"]
)["netde"]
)
)
output_df
# Target Columns: ['revenue fy-4', 'revenue fy-3', 'revenue fy-2', 'revenue fy-1', 'revenue fy']
# Pulled directly from incomestatement.csv
for i in range(4, -1, -1):
if i != 0:
tmp_df = pd.DataFrame(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"] - i][
["companyid", "revenue"]
]
)
tmp_df = tmp_df.rename(columns={"revenue": "revenue fy-" + str(i)})
output_df = pd.merge(output_df, tmp_df, how="left")
else:
tmp_df = pd.DataFrame(
funda_df.loc[funda_df["year"] == funda_df["latestfinyear"] - i][
["companyid", "revenue"]
]
)
tmp_df = tmp_df.rename(columns={"revenue": "revenue fy"})
output_df = pd.merge(output_df, tmp_df, how="left")
output_df
# Target Columns: ['revenue fy+1', 'revenue fy+2', 'revenue fy+3']
# Pulled directly from forecast.csv and convert currency from ‘estcurr’ to ‘reportingcurr’.
for i in range(1, 4):
tmp_df = pd.DataFrame(
(
forecast_df.loc[forecast_df["year"] == forecast_df["latestfinyear"] + i][
["companyid", "revenue"]
]
)
)
tmp_df = tmp_df.rename(columns={"revenue": "revenue fy+" + str(i)})
output_df = pd.merge(output_df, tmp_df, how="left")
output_df
# Target Columns: ['revenue 2yr cagr','revenue 3yr cagr']
# revenue 2yr cagr = 100*[(revenue fy+2 / revenue fy)^(0.5) – 1]
# revenue 3yr cagr = 100*[(revenue fy+3 / revenue fy)^(0.33) – 1]
| false | 5 | 2,338 | 0 | 2,356 | 2,338 |
||
129894036
|
<jupyter_start><jupyter_text>World Exports Value 2021
Kaggle dataset identifier: world-exports-value-2021
<jupyter_script># Step1: Necessery imports
import statistics
import pandas as pd
import numpy as np
import matplotlib as mpl
import scipy as scipy
import seaborn as sns
import plotly.express as px
import plotly.figure_factory as ff
import plotly.graph_objects as go
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Step2: Loading the data
# Change the csv file for another brand
# Added Data types for memory optimization and faster loading
data = pd.read_csv(
"/kaggle/input/world-exports-value-2021/World-Exports-Value-2021--Click-to-Select-a-Product.csv",
error_bad_lines=False,
)
data.shape
data.info()
# ***Standard deviation***
std = np.std(data)
print(std)
# ***Coefficient of Variation***
cv = np.std(data) / np.mean(data)
print(cv)
# ***Variance***
var_full = np.var(data)
print(var_full)
# # Describing the data
# Step3:Describing the data
data.describe()
# Step3:Describing the data - finding the mode [most frequent]
data.mode()
# # **#Treemap**
fig = px.treemap(data, path=["HS2", "HS4", "HS6"], values="Trade Value")
fig.update_layout(
title="Woirld Exports Value 2021",
width=1200,
height=1200,
)
fig.show()
fig = px.treemap(
data,
path=["HS2", "HS4", "HS6"],
values="Trade Value",
color="Trade Value",
color_continuous_scale="RdYlGn",
)
fig.update_layout(
title="Trade Val",
width=1000,
height=600,
)
fig.show()
# # **# 3-D chart**
# controls the numer of rows to be read in the dataframe
start, end = 0, 4700
fig = go.Figure(
data=go.Scatter3d(
x=data["HS2"][start:end],
y=data["HS4"][start:end],
z=data["HS6"][start:end],
text=data["Section"][start:end],
mode="markers",
marker=dict(
sizemode="diameter",
sizeref=5000000000,
size=data["Trade Value"][start:end],
color=data["Trade Value"][start:end],
colorscale="Viridis",
colorbar_title="Trade Value<br>",
line_color="rgb(140, 140, 170)",
),
)
)
fig.update_layout(
height=1200,
width=1200,
title="3-D Graph - X-HS2,Y-HS4,Z-HS6,Size-Trade Value,Color-Trade Value",
)
fig.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/894/129894036.ipynb
|
world-exports-value-2021
|
valchovalev
|
[{"Id": 129894036, "ScriptId": 38630490, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7041364, "CreationDate": "05/17/2023 09:01:31", "VersionNumber": 1.0, "Title": "EDA : World Exports Value 2021", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 127.0, "LinesInsertedFromPrevious": 52.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 75.0, "LinesInsertedFromFork": 52.0, "LinesDeletedFromFork": 71.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 75.0, "TotalVotes": 0}]
|
[{"Id": 186304611, "KernelVersionId": 129894036, "SourceDatasetVersionId": 5705064}]
|
[{"Id": 5705064, "DatasetId": 3279850, "DatasourceVersionId": 5780848, "CreatorUserId": 7041364, "LicenseName": "Unknown", "CreationDate": "05/17/2023 06:35:08", "VersionNumber": 1.0, "Title": "World Exports Value 2021", "Slug": "world-exports-value-2021", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3279850, "CreatorUserId": 7041364, "OwnerUserId": 7041364.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5705064.0, "CurrentDatasourceVersionId": 5780848.0, "ForumId": 3345566, "Type": 2, "CreationDate": "05/17/2023 06:35:08", "LastActivityDate": "05/17/2023", "TotalViews": 48, "TotalDownloads": 3, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 7041364, "UserName": "valchovalev", "DisplayName": "valcho valev", "RegisterDate": "03/27/2021", "PerformanceTier": 1}]
|
# Step1: Necessery imports
import statistics
import pandas as pd
import numpy as np
import matplotlib as mpl
import scipy as scipy
import seaborn as sns
import plotly.express as px
import plotly.figure_factory as ff
import plotly.graph_objects as go
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Step2: Loading the data
# Change the csv file for another brand
# Added Data types for memory optimization and faster loading
data = pd.read_csv(
"/kaggle/input/world-exports-value-2021/World-Exports-Value-2021--Click-to-Select-a-Product.csv",
error_bad_lines=False,
)
data.shape
data.info()
# ***Standard deviation***
std = np.std(data)
print(std)
# ***Coefficient of Variation***
cv = np.std(data) / np.mean(data)
print(cv)
# ***Variance***
var_full = np.var(data)
print(var_full)
# # Describing the data
# Step3:Describing the data
data.describe()
# Step3:Describing the data - finding the mode [most frequent]
data.mode()
# # **#Treemap**
fig = px.treemap(data, path=["HS2", "HS4", "HS6"], values="Trade Value")
fig.update_layout(
title="Woirld Exports Value 2021",
width=1200,
height=1200,
)
fig.show()
fig = px.treemap(
data,
path=["HS2", "HS4", "HS6"],
values="Trade Value",
color="Trade Value",
color_continuous_scale="RdYlGn",
)
fig.update_layout(
title="Trade Val",
width=1000,
height=600,
)
fig.show()
# # **# 3-D chart**
# controls the numer of rows to be read in the dataframe
start, end = 0, 4700
fig = go.Figure(
data=go.Scatter3d(
x=data["HS2"][start:end],
y=data["HS4"][start:end],
z=data["HS6"][start:end],
text=data["Section"][start:end],
mode="markers",
marker=dict(
sizemode="diameter",
sizeref=5000000000,
size=data["Trade Value"][start:end],
color=data["Trade Value"][start:end],
colorscale="Viridis",
colorbar_title="Trade Value<br>",
line_color="rgb(140, 140, 170)",
),
)
)
fig.update_layout(
height=1200,
width=1200,
title="3-D Graph - X-HS2,Y-HS4,Z-HS6,Size-Trade Value,Color-Trade Value",
)
fig.show()
| false | 1 | 870 | 0 | 902 | 870 |
||
129900207
|
<jupyter_start><jupyter_text>iris_data
Kaggle dataset identifier: iris-data
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # For graphical representation
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # **On va lire le fichier et afficher les cinq premières lignes**
# Load the data set and see first 5 rows
a = pd.read_csv("/kaggle/input/iris-data/Iris.csv")
iris = pd.DataFrame(a)
iris.head()
iris.shape
iris.info()
# # Étant donné que la colonne Id ne sert à rien, nous devons donc la supprimer
iris.drop("Id", axis=1, inplace=True)
iris.head()
# # **Afficher le nombre des espèces par groupe**
iris.groupby("Species").size()
# # **Afficher la distribution par attribut**
iris.hist()
his = plt.gcf()
his.set_size_inches(12, 6)
plt.show()
# # **Créer un modèle basé sur le support vector machine (SVM)**
test_size = 0.20
seed = 7
score = "accuracy"
# # **Diviser le dataset en données d’apprentissage et test**
from sklearn import model_selection
X = iris.iloc[:, :4]
y = iris.iloc[:, 4]
X_train, X_test, y_train, y_test = model_selection.train_test_split(
X, y, test_size=test_size, random_state=seed
)
from sklearn import svm
from sklearn.model_selection import cross_val_score, KFold
from sklearn.metrics import accuracy_score
# Create an SVM classifier
# kernels : linear, poly, rbf(Radial Basis Function), sigmoid, precomputed
clf = svm.SVC(kernel="rbf")
# # **La validation croisée (Cross Validation k = 5)**
# Define the number of folds for cross-validation
k = 5
# Create a KFold object
kf = KFold(n_splits=k, shuffle=True, random_state=42)
# Perform k-fold cross-validation
scores = cross_val_score(clf, X_train, y_train, cv=kf, scoring=score)
# Print the accuracy scores for each fold
for fold, sc in enumerate(scores):
print(f"Fold {fold+1}: {sc:.4f}")
# Calculate and print the mean accuracy across all folds
mean_accuracy = scores.mean()
print(f"Mean Accuracy: {mean_accuracy:.4f}")
# # **Evaluation du modèle SVM**
# Predictions on test dataset
svm = svm.SVC(kernel="rbf")
svm.fit(X_train, y_train)
pred = svm.predict(X_test)
print(accuracy_score(y_test, pred))
# # **Créer un modèle Multilayer perceptron (MLP)**
a = pd.read_csv("/kaggle/input/iris-data/Iris.csv", header=None)
i = pd.DataFrame(a)
iris = i.values
# # **Afficher les cinq premières lignes**
i.head(5)
X = iris[1:, 1:5].astype(float)
y = iris[1:, 5]
X[0:5]
y[0:5]
from sklearn.preprocessing import LabelEncoder
from keras.utils import np_utils
from keras.wrappers.scikit_learn import KerasClassifier
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from keras.models import Sequential
from keras.layers import Dense
from sklearn import model_selection
from sklearn.metrics import accuracy_score
from keras.optimizers import Adam
# # **Encodage des espèces sous forme numérique**
# Label encode Class (Species)
encoder = LabelEncoder()
encoder.fit(y)
encoded_y = encoder.transform(y)
# # **Convertit les valeurs numériques des classes en matrice de classe binaire**
# One Hot Encode
y_dummy = np_utils.to_categorical(encoded_y)
# # **Diviser le dataset en données d’apprentissage et test**
X_train, X_test, y_train_one_hot, y_test_one_hot = model_selection.train_test_split(
X, y_dummy, test_size=test_size, random_state=seed
)
# # **Architecture du modèle**
# Deep Learnig Function
def deepml_model():
# Model Creation
deepml = Sequential()
deepml.add(Dense(8, input_dim=4, activation="relu")) # 8
deepml.add(Dense(10, activation="relu")) # 10
deepml.add(Dense(3, activation="softmax")) # 3
# Model Compilation
optimiser = Adam(learning_rate=0.001)
deepml.compile(
loss="categorical_crossentropy", optimizer=optimiser, metrics=[score]
)
return deepml
# # **Paramétrage du modèle**
estimate = KerasClassifier(build_fn=deepml_model, epochs=100, batch_size=5, verbose=0)
# Cross
# # **La validation croisée (Cross Validation k = 5)**
k_fold = KFold(n_splits=5, shuffle=True, random_state=seed)
results = cross_val_score(estimate, X_train, y_train_one_hot, cv=k_fold)
# add here folds logs
for fold, rst in enumerate(results):
print(f"Fold {fold+1}: {rst:.4f}")
# Calculate and print the mean accuracy across all folds
mean_accuracy = results.mean()
print(f"Mean Accuracy: {mean_accuracy:.4f}")
# # **Evaluation du modèle MLP**
# Predictions on test dataset
mlp = deepml_model()
mlp.fit(X_train, y_train_one_hot, epochs=100, batch_size=5, verbose=0)
y_pred_one_hot = mlp.predict(X_test)
pred = np.argmax(y_pred_one_hot, axis=1)
y_test = np.argmax(y_test_one_hot, axis=1)
print(accuracy_score(y_test, pred))
# # **Overfitting**
# validation_data=(X_test, y_test_one_hot)
X_train_overfit = X_train[:-50] # Use a smaller training set
y_train_overfit = y_train_one_hot[:-50] # Use corresponding labels
history = mlp.fit(
X_train_overfit,
y_train_overfit,
epochs=100,
batch_size=5,
validation_data=(X_test, y_test_one_hot),
verbose=0,
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/900/129900207.ipynb
|
iris-data
|
kamrankausar
|
[{"Id": 129900207, "ScriptId": 38402106, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11365210, "CreationDate": "05/17/2023 09:50:50", "VersionNumber": 1.0, "Title": "Iris data set machine learning TP", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 194.0, "LinesInsertedFromPrevious": 194.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186312937, "KernelVersionId": 129900207, "SourceDatasetVersionId": 8520}]
|
[{"Id": 8520, "DatasetId": 5721, "DatasourceVersionId": 8520, "CreatorUserId": 480578, "LicenseName": "CC0: Public Domain", "CreationDate": "11/30/2017 10:26:01", "VersionNumber": 2.0, "Title": "iris_data", "Slug": "iris-data", "Subtitle": "Hello World of Machine Learning and Deep Learning", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 5107.0, "TotalUncompressedBytes": 5107.0}]
|
[{"Id": 5721, "CreatorUserId": 480578, "OwnerUserId": 480578.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 8520.0, "CurrentDatasourceVersionId": 8520.0, "ForumId": 11974, "Type": 2, "CreationDate": "11/30/2017 10:26:01", "LastActivityDate": "01/31/2018", "TotalViews": 9401, "TotalDownloads": 1764, "TotalVotes": 22, "TotalKernels": 45}]
|
[{"Id": 480578, "UserName": "kamrankausar", "DisplayName": "kamran", "RegisterDate": "12/04/2015", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # For graphical representation
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # **On va lire le fichier et afficher les cinq premières lignes**
# Load the data set and see first 5 rows
a = pd.read_csv("/kaggle/input/iris-data/Iris.csv")
iris = pd.DataFrame(a)
iris.head()
iris.shape
iris.info()
# # Étant donné que la colonne Id ne sert à rien, nous devons donc la supprimer
iris.drop("Id", axis=1, inplace=True)
iris.head()
# # **Afficher le nombre des espèces par groupe**
iris.groupby("Species").size()
# # **Afficher la distribution par attribut**
iris.hist()
his = plt.gcf()
his.set_size_inches(12, 6)
plt.show()
# # **Créer un modèle basé sur le support vector machine (SVM)**
test_size = 0.20
seed = 7
score = "accuracy"
# # **Diviser le dataset en données d’apprentissage et test**
from sklearn import model_selection
X = iris.iloc[:, :4]
y = iris.iloc[:, 4]
X_train, X_test, y_train, y_test = model_selection.train_test_split(
X, y, test_size=test_size, random_state=seed
)
from sklearn import svm
from sklearn.model_selection import cross_val_score, KFold
from sklearn.metrics import accuracy_score
# Create an SVM classifier
# kernels : linear, poly, rbf(Radial Basis Function), sigmoid, precomputed
clf = svm.SVC(kernel="rbf")
# # **La validation croisée (Cross Validation k = 5)**
# Define the number of folds for cross-validation
k = 5
# Create a KFold object
kf = KFold(n_splits=k, shuffle=True, random_state=42)
# Perform k-fold cross-validation
scores = cross_val_score(clf, X_train, y_train, cv=kf, scoring=score)
# Print the accuracy scores for each fold
for fold, sc in enumerate(scores):
print(f"Fold {fold+1}: {sc:.4f}")
# Calculate and print the mean accuracy across all folds
mean_accuracy = scores.mean()
print(f"Mean Accuracy: {mean_accuracy:.4f}")
# # **Evaluation du modèle SVM**
# Predictions on test dataset
svm = svm.SVC(kernel="rbf")
svm.fit(X_train, y_train)
pred = svm.predict(X_test)
print(accuracy_score(y_test, pred))
# # **Créer un modèle Multilayer perceptron (MLP)**
a = pd.read_csv("/kaggle/input/iris-data/Iris.csv", header=None)
i = pd.DataFrame(a)
iris = i.values
# # **Afficher les cinq premières lignes**
i.head(5)
X = iris[1:, 1:5].astype(float)
y = iris[1:, 5]
X[0:5]
y[0:5]
from sklearn.preprocessing import LabelEncoder
from keras.utils import np_utils
from keras.wrappers.scikit_learn import KerasClassifier
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from keras.models import Sequential
from keras.layers import Dense
from sklearn import model_selection
from sklearn.metrics import accuracy_score
from keras.optimizers import Adam
# # **Encodage des espèces sous forme numérique**
# Label encode Class (Species)
encoder = LabelEncoder()
encoder.fit(y)
encoded_y = encoder.transform(y)
# # **Convertit les valeurs numériques des classes en matrice de classe binaire**
# One Hot Encode
y_dummy = np_utils.to_categorical(encoded_y)
# # **Diviser le dataset en données d’apprentissage et test**
X_train, X_test, y_train_one_hot, y_test_one_hot = model_selection.train_test_split(
X, y_dummy, test_size=test_size, random_state=seed
)
# # **Architecture du modèle**
# Deep Learnig Function
def deepml_model():
# Model Creation
deepml = Sequential()
deepml.add(Dense(8, input_dim=4, activation="relu")) # 8
deepml.add(Dense(10, activation="relu")) # 10
deepml.add(Dense(3, activation="softmax")) # 3
# Model Compilation
optimiser = Adam(learning_rate=0.001)
deepml.compile(
loss="categorical_crossentropy", optimizer=optimiser, metrics=[score]
)
return deepml
# # **Paramétrage du modèle**
estimate = KerasClassifier(build_fn=deepml_model, epochs=100, batch_size=5, verbose=0)
# Cross
# # **La validation croisée (Cross Validation k = 5)**
k_fold = KFold(n_splits=5, shuffle=True, random_state=seed)
results = cross_val_score(estimate, X_train, y_train_one_hot, cv=k_fold)
# add here folds logs
for fold, rst in enumerate(results):
print(f"Fold {fold+1}: {rst:.4f}")
# Calculate and print the mean accuracy across all folds
mean_accuracy = results.mean()
print(f"Mean Accuracy: {mean_accuracy:.4f}")
# # **Evaluation du modèle MLP**
# Predictions on test dataset
mlp = deepml_model()
mlp.fit(X_train, y_train_one_hot, epochs=100, batch_size=5, verbose=0)
y_pred_one_hot = mlp.predict(X_test)
pred = np.argmax(y_pred_one_hot, axis=1)
y_test = np.argmax(y_test_one_hot, axis=1)
print(accuracy_score(y_test, pred))
# # **Overfitting**
# validation_data=(X_test, y_test_one_hot)
X_train_overfit = X_train[:-50] # Use a smaller training set
y_train_overfit = y_train_one_hot[:-50] # Use corresponding labels
history = mlp.fit(
X_train_overfit,
y_train_overfit,
epochs=100,
batch_size=5,
validation_data=(X_test, y_test_one_hot),
verbose=0,
)
| false | 1 | 1,847 | 0 | 1,868 | 1,847 |
||
129900468
|
import numpy as np
from PIL import Image
import math
import matplotlib.pyplot as plt
data = np.load("/kaggle/input/lenet-kernal-info/weights_conv1.npy")
class LeNet5:
def __init__(self):
self.conv1_filters = 6
self.conv1_filter_size = 5
self.conv2_filters = 16
self.conv2_filter_size = 5
self.fc1_units = 120
self.fc2_units = 84
self.fc3_units = 10
# %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% Need to debug hear %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
def convolve2d(self, image, filters):
matrix_in = image
# add padding of 1 to the matrix
padding = 1
matrix = np.pad(
matrix_in, pad_width=padding, mode="constant", constant_values=0
)
print("Padded Matrix:\n", matrix)
matrix_kernel = filters
kernel_size = (matrix_kernel.shape)[0]
image_size = (matrix_in.shape)[0]
linear_kernel_size = kernel_size * kernel_size
padded_image_size = (matrix.shape)[0]
stride = 1
conv = ((image_size + 2 * padding - kernel_size) // stride) + 1
kernel_linear = matrix_kernel.reshape(
linear_kernel_size,
)
# kernel_linear_out = np.empty((9, ), dtype=int)
kernel_linear_out = [0] * linear_kernel_size
unique_elements = np.unique(kernel_linear)
def bits_required_for_unique_memory(pqrs):
bits_required = (math.log(pqrs)) / (math.log(2))
integer_bit_length = round(bits_required)
fraction_value = (
bits_required % 1
) # checkl if fraction part is available or not
if 0 < fraction_value < 0.5:
integer_bit_length = (
integer_bit_length + 1
) # add one more bit if fraction part is present
integer_bit_length = int(integer_bit_length)
return integer_bit_length
bits = bits_required_for_unique_memory(len(unique_elements))
print(bits)
code_list = [bin(x)[2:].rjust(bits, "0") for x in range(2**bits)]
def code_word_mem(
uni_mem_num, code_list_num, quantize_layer_name, quantize_layer_name_out
):
for i in range(len(quantize_layer_name)):
# print(quantize_layer_name[i])
for j in range(len(uni_mem_num)):
if quantize_layer_name[i] == uni_mem_num[j]:
quantize_layer_name_out[i] = str(code_list_num[j])
code_word_mem(unique_elements, code_list, kernel_linear, kernel_linear_out)
# print(unique_elements, kernel_linear, kernel_linear_out)
kernel_linear_out_np = np.array(kernel_linear_out)
memory_temp = [[0] * 2 for i in range(len(unique_elements))]
memory_add_matrix = [[0] * conv for i in range(conv)]
for i in range(0, padded_image_size, stride):
for j in range(0, padded_image_size, stride):
if (i + kernel_size) <= (padded_image_size) and (j + kernel_size) <= (
padded_image_size
):
mat_temp = matrix[i : i + kernel_size, j : j + kernel_size]
mat_temp_np = np.array(mat_temp)
mat_temp_np_lin = mat_temp_np.reshape(
linear_kernel_size,
)
temp_add_matrix = 0
for k in range(len(unique_elements)):
memory_temp[k][0] = unique_elements[k]
temp_add = 0
for l in range(len(kernel_linear_out_np)):
if code_list[k] == kernel_linear_out_np[l]:
temp_add = temp_add + mat_temp_np_lin[l]
memory_temp[k][1] = temp_add
temp_add_matrix = temp_add_matrix + (
memory_temp[k][0] * memory_temp[k][1]
)
memory_add_matrix[i // stride][j // stride] = temp_add_matrix
memory_add_matrix_np = np.array(memory_add_matrix)
return memory_add_matrix_np
# %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
def relu(self, x):
return np.maximum(x, 0)
def max_pooling(self, image, size=2):
height, width, _ = image.shape
output_size = height // size
pooled = np.zeros((output_size, output_size, _))
for i in range(output_size):
for j in range(output_size):
for c in range(_):
pooled[i, j, c] = np.max(
image[i * size : i * size + size, j * size : j * size + size, c]
)
return pooled
def flatten(self, image):
return image.flatten()
def fc_layer(self, x, weights, bias):
return np.dot(x, weights) + bias
def softmax(self, x):
exps = np.exp(x - np.max(x))
return exps / np.sum(exps)
# %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
def forward_propagation(self, image):
conv1_output = self.convolve2d(image, self.conv1_weights) + self.conv1_bias
conv1_output = self.relu(conv1_output)
pool1_output = self.max_pooling(conv1_output)
conv2_output = (
self.convolve2d(pool1_output, self.conv2_weights) + self.conv2_bias
)
conv2_output = self.relu(conv2_output)
pool2_output = self.max_pooling(conv2_output)
fc1_output = np.dot(self.fc1_weights, pool2_output.flatten()) + self.fc1_bias
fc1_output = self.relu(fc1_output)
fc2_output = np.dot(self.fc2_weights, fc1_output) + self.fc2_bias
fc2_output = self.relu(fc2_output)
fc3_output = np.dot(self.fc3_weights, fc2_output) + self.fc3_bias
output = self.softmax(fc3_output)
return output
model = LeNet5()
conv1_weights = np.load("/kaggle/input/lenet-kernal-info/weights_conv1.npy")
conv1_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/bias_conv1.npy")
conv2_weights = np.load("/kaggle/input/lenet-kernal-info/weights_conv2.npy")
conv2_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/bias_conv2.npy")
fc1_weights = np.load("/kaggle/input/lenet-kernal-info/weights_fc1.npy")
fc1_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/bias_fc1.npy")
fc2_weights = np.load("/kaggle/input/lenet-kernal-info/weights_fc2.npy")
fc2_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/weights_fc2.npy")
fc3_weights = np.load("/kaggle/input/lenet-kernal-info/weights_fc3.npy")
fc3_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/bias_fc3.npy")
# Assign the loaded weights to the model
model.conv1_weights = conv1_weights
model.conv1_bias = conv1_bias
model.conv2_weights = conv2_weights
model.conv2_bias = conv2_bias
model.fc1_weights = fc1_weights
model.fc1_bias = fc1_bias
model.fc2_weights = fc2_weights
model.fc2_bias = fc2_bias
model.fc3_weights = fc3_weights
model.fc3_bias = fc3_bias
image_path = "path_to_your_image.jpg" # Replace with the actual image path
image = Image.open(image_path)
image = image.resize((32, 32)) # Resize the image to 32x32
image = np.array(image) # Convert the image to a numpy array
image = image.transpose(
(2, 0, 1)
) # Transpose the dimensions to match LeNet-5 input shape
image = image.astype(np.float32) / 255.0 # Normalize the pixel values between 0 and 1
# Perform forward propagation
output = model.forward_propagation(image)
# Get the predicted class
predicted_class = np.argmax(output)
print("Predicted Class:", predicted_class)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/900/129900468.ipynb
| null | null |
[{"Id": 129900468, "ScriptId": 38353739, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13480720, "CreationDate": "05/17/2023 09:52:57", "VersionNumber": 1.0, "Title": "test_lenet", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 177.0, "LinesInsertedFromPrevious": 177.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
from PIL import Image
import math
import matplotlib.pyplot as plt
data = np.load("/kaggle/input/lenet-kernal-info/weights_conv1.npy")
class LeNet5:
def __init__(self):
self.conv1_filters = 6
self.conv1_filter_size = 5
self.conv2_filters = 16
self.conv2_filter_size = 5
self.fc1_units = 120
self.fc2_units = 84
self.fc3_units = 10
# %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% Need to debug hear %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
def convolve2d(self, image, filters):
matrix_in = image
# add padding of 1 to the matrix
padding = 1
matrix = np.pad(
matrix_in, pad_width=padding, mode="constant", constant_values=0
)
print("Padded Matrix:\n", matrix)
matrix_kernel = filters
kernel_size = (matrix_kernel.shape)[0]
image_size = (matrix_in.shape)[0]
linear_kernel_size = kernel_size * kernel_size
padded_image_size = (matrix.shape)[0]
stride = 1
conv = ((image_size + 2 * padding - kernel_size) // stride) + 1
kernel_linear = matrix_kernel.reshape(
linear_kernel_size,
)
# kernel_linear_out = np.empty((9, ), dtype=int)
kernel_linear_out = [0] * linear_kernel_size
unique_elements = np.unique(kernel_linear)
def bits_required_for_unique_memory(pqrs):
bits_required = (math.log(pqrs)) / (math.log(2))
integer_bit_length = round(bits_required)
fraction_value = (
bits_required % 1
) # checkl if fraction part is available or not
if 0 < fraction_value < 0.5:
integer_bit_length = (
integer_bit_length + 1
) # add one more bit if fraction part is present
integer_bit_length = int(integer_bit_length)
return integer_bit_length
bits = bits_required_for_unique_memory(len(unique_elements))
print(bits)
code_list = [bin(x)[2:].rjust(bits, "0") for x in range(2**bits)]
def code_word_mem(
uni_mem_num, code_list_num, quantize_layer_name, quantize_layer_name_out
):
for i in range(len(quantize_layer_name)):
# print(quantize_layer_name[i])
for j in range(len(uni_mem_num)):
if quantize_layer_name[i] == uni_mem_num[j]:
quantize_layer_name_out[i] = str(code_list_num[j])
code_word_mem(unique_elements, code_list, kernel_linear, kernel_linear_out)
# print(unique_elements, kernel_linear, kernel_linear_out)
kernel_linear_out_np = np.array(kernel_linear_out)
memory_temp = [[0] * 2 for i in range(len(unique_elements))]
memory_add_matrix = [[0] * conv for i in range(conv)]
for i in range(0, padded_image_size, stride):
for j in range(0, padded_image_size, stride):
if (i + kernel_size) <= (padded_image_size) and (j + kernel_size) <= (
padded_image_size
):
mat_temp = matrix[i : i + kernel_size, j : j + kernel_size]
mat_temp_np = np.array(mat_temp)
mat_temp_np_lin = mat_temp_np.reshape(
linear_kernel_size,
)
temp_add_matrix = 0
for k in range(len(unique_elements)):
memory_temp[k][0] = unique_elements[k]
temp_add = 0
for l in range(len(kernel_linear_out_np)):
if code_list[k] == kernel_linear_out_np[l]:
temp_add = temp_add + mat_temp_np_lin[l]
memory_temp[k][1] = temp_add
temp_add_matrix = temp_add_matrix + (
memory_temp[k][0] * memory_temp[k][1]
)
memory_add_matrix[i // stride][j // stride] = temp_add_matrix
memory_add_matrix_np = np.array(memory_add_matrix)
return memory_add_matrix_np
# %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
def relu(self, x):
return np.maximum(x, 0)
def max_pooling(self, image, size=2):
height, width, _ = image.shape
output_size = height // size
pooled = np.zeros((output_size, output_size, _))
for i in range(output_size):
for j in range(output_size):
for c in range(_):
pooled[i, j, c] = np.max(
image[i * size : i * size + size, j * size : j * size + size, c]
)
return pooled
def flatten(self, image):
return image.flatten()
def fc_layer(self, x, weights, bias):
return np.dot(x, weights) + bias
def softmax(self, x):
exps = np.exp(x - np.max(x))
return exps / np.sum(exps)
# %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
def forward_propagation(self, image):
conv1_output = self.convolve2d(image, self.conv1_weights) + self.conv1_bias
conv1_output = self.relu(conv1_output)
pool1_output = self.max_pooling(conv1_output)
conv2_output = (
self.convolve2d(pool1_output, self.conv2_weights) + self.conv2_bias
)
conv2_output = self.relu(conv2_output)
pool2_output = self.max_pooling(conv2_output)
fc1_output = np.dot(self.fc1_weights, pool2_output.flatten()) + self.fc1_bias
fc1_output = self.relu(fc1_output)
fc2_output = np.dot(self.fc2_weights, fc1_output) + self.fc2_bias
fc2_output = self.relu(fc2_output)
fc3_output = np.dot(self.fc3_weights, fc2_output) + self.fc3_bias
output = self.softmax(fc3_output)
return output
model = LeNet5()
conv1_weights = np.load("/kaggle/input/lenet-kernal-info/weights_conv1.npy")
conv1_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/bias_conv1.npy")
conv2_weights = np.load("/kaggle/input/lenet-kernal-info/weights_conv2.npy")
conv2_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/bias_conv2.npy")
fc1_weights = np.load("/kaggle/input/lenet-kernal-info/weights_fc1.npy")
fc1_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/bias_fc1.npy")
fc2_weights = np.load("/kaggle/input/lenet-kernal-info/weights_fc2.npy")
fc2_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/weights_fc2.npy")
fc3_weights = np.load("/kaggle/input/lenet-kernal-info/weights_fc3.npy")
fc3_bias = np.load("/kaggle/input/lenet-bias-1/lenet/weight_matrices/bias_fc3.npy")
# Assign the loaded weights to the model
model.conv1_weights = conv1_weights
model.conv1_bias = conv1_bias
model.conv2_weights = conv2_weights
model.conv2_bias = conv2_bias
model.fc1_weights = fc1_weights
model.fc1_bias = fc1_bias
model.fc2_weights = fc2_weights
model.fc2_bias = fc2_bias
model.fc3_weights = fc3_weights
model.fc3_bias = fc3_bias
image_path = "path_to_your_image.jpg" # Replace with the actual image path
image = Image.open(image_path)
image = image.resize((32, 32)) # Resize the image to 32x32
image = np.array(image) # Convert the image to a numpy array
image = image.transpose(
(2, 0, 1)
) # Transpose the dimensions to match LeNet-5 input shape
image = image.astype(np.float32) / 255.0 # Normalize the pixel values between 0 and 1
# Perform forward propagation
output = model.forward_propagation(image)
# Get the predicted class
predicted_class = np.argmax(output)
print("Predicted Class:", predicted_class)
| false | 0 | 2,300 | 0 | 2,300 | 2,300 |
||
129420736
|
<jupyter_start><jupyter_text>Football/Soccer | Bundesliga Player Database
The Bundesliga Players dataset provides a comprehensive collection of information on every player in the German Bundesliga football league. From renowned goalkeepers to talented defenders, this dataset offers an extensive range of player details including their names, full names, ages, heights, nationalities, places of birth, prices, maximum prices, positions, shirt numbers, preferred foot, current clubs, contract expiration dates, dates of joining the clubs, player agents, and outfitters. Whether you're a passionate football fan, a sports analyst, or a fantasy football enthusiast, this dataset serves as a valuable resource for exploring and analyzing the profiles of Bundesliga players, enabling you to delve into their backgrounds, performance statistics, and club affiliations. Discover the stars of German football and gain insights into their careers with this comprehensive Bundesliga Players dataset.
Kaggle dataset identifier: bundesliga-soccer-player
<jupyter_script># # Market Value Prediction with Randome Forest Regressor
# ### Short look up in the Data + Imports
# - `numpy`: Fundamental package for scientific computing.
# - `pandas`: Library for data manipulation and analysis.
# - `sklearn.compose.ColumnTransformer`: Applies different preprocessing steps to dataset columns.
# - `sklearn.preprocessing.OneHotEncoder`: Encodes categorical variables into binary matrix representation.
# - `sklearn.metrics.mean_squared_error`: Metric for evaluating regression models.
# - `sklearn.model_selection.train_test_split`: Splits dataset into training and testing subsets.
# - `sklearn.ensemble.RandomForestRegressor`: Ensemble regression model using decision trees.
# - `sklearn.model_selection.GridSearchCV`: Technique for hyperparameter tuning.
#
import numpy as np
import pandas as pd
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import GridSearchCV
df_players = pd.read_csv(
"/kaggle/input/bundesliga-soccer-player/bundesliga_player.csv", index_col=[0]
)
df_players.head()
df_players.describe()
df_players.info()
# ### Short nan value handling
# The two columns with too much nan values become 'none' string to replace the nan values with a categorical value. Afterwards it is easy to drop rows with nan values.
df_players = df_players[
[
"age",
"height",
"nationality",
"foot",
"position",
"club",
"contract_expires",
"joined_club",
"player_agent",
"outfitter",
"price",
]
]
df_players["outfitter"] = df_players["outfitter"].replace(np.nan, "none")
df_players["player_agent"] = df_players["player_agent"].replace(np.nan, "none")
df_players.dropna(inplace=True)
print(f"df_players shape: {df_players.shape}")
# ### Variable Usefulness for Predicting Price
# 1. Age: Age is likely to be a useful variable for predicting price as younger players generally have higher market values due to their potential for growth and longer career ahead.
# 2. Height: Height might have some influence on the price as certain positions or playing styles may favor taller players. However, its impact on price may not be as significant compared to other variables.
# 4. Club: Club affiliation is an important variable for predicting price. Players from high-profile clubs or clubs known for producing top talent are often valued more highly in the market.
# 5. Position: Position is a crucial factor in determining price. Different positions have varying levels of demand and scarcity, leading to variations in market values.
# 6. Contract Expiry Date: The remaining duration of a player's contract can impact their price. Players with longer contract terms may have higher values due to increased stability and reduced transfer urgency.
# 7. Contract Start Date: The start date of a player's current contract may have less influence on predicting price compared to other variables. It is more indicative of the player's history with the club rather than their current market value.
# 8. Agency/Representative: The player's agency or representative is not directly related to their market value. It is more of a logistical detail and does not provide significant insight into predicting price.
# 9. Sponsorship Brand: The sponsorship brand associated with a player does not have a direct impact on their market value. While brand endorsements can increase a player's overall earnings, it may not be a significant factor in price prediction.
# 10. Right/Left-Footed: A player's dominant foot is unlikely to have a substantial impact on their market value. It is more relevant to their playing style or preferred positions rather than predicting price.
# 11. Max Price (Excluded): The "max price" variable should be excluded from the prediction model because it represents the actual target variable we want to predict to much. Including it as a feature would result in data leakage and lead to an overly optimistic evaluation of the model's performance.
# Note: The above analysis is based on general assumptions and domain knowledge. It is recommended to validate the significance of these variables through statistical analysis and feature selection techniques specific to the dataset and prediction task at hand.
#
df_target = df_players[["price"]]
df_features = df_players[
[
"age",
"height",
"foot",
"position",
"club",
"contract_expires",
"joined_club",
"player_agent",
"outfitter",
]
]
# ### One Hot Encoding
# In the Data are many categorical features and the Random Forest Regressor can't handle those. Because of this One Hot Encoding is used. This implies that the categorical features will be split into different binary columns which can tell the model if this category is true or not.Nevertheless, first is a look into those variables needed.
for column in df_features.columns:
unique_values = df_features[column].unique()
print(f"Unique values in column '{column}': {unique_values}")
# Now we use the ColumnTransformer to apply the One Hot Encoding
columns_to_encode = [
"foot",
"position",
"club",
"contract_expires",
"joined_club",
"player_agent",
"outfitter",
]
ct = ColumnTransformer(
transformers=[("encoder", OneHotEncoder(), columns_to_encode)],
remainder="passthrough",
)
df_features_encoded = ct.fit_transform(df_features)
df_features_encoded.shape
# ### Train and Test Split
x_train, x_test, y_train, y_test = train_test_split(
df_features_encoded, df_target, test_size=0.3, random_state=0
)
y_train = y_train.values.ravel()
y_test = y_test.values.ravel()
print(f"x_train: {x_train.shape}")
print(f"x_test: {x_test.shape}")
print(f"y_train: {y_train.shape}")
print(f"y_test: {y_test.shape}")
# ### Hyperparameter tuning
# Using GridSearch for hyperparameter tuning is a good option because it allows us to systematically search through different combinations of hyperparameters and find the optimal configuration for our model. It automates the process of tuning hyperparameters, saving time(not computing time :p) and effort. GridSearch performs an exhaustive search over the specified hyperparameter grid, evaluating each combination using cross-validation. This helps us find the hyperparameters that yield the best performance based on the chosen evaluation metric. By using GridSearch, we can effectively optimize our model without the need for manual trial and error.
#
param_grid = {
"n_estimators": np.arange(10, 1000, 50),
"max_depth": np.arange(5, 20, 2),
"min_samples_split": np.arange(2, 11, 2),
"min_samples_leaf": np.arange(1, 10, 2),
}
rfr = RandomForestRegressor()
grid_search = GridSearchCV(
estimator=rfr,
param_grid=param_grid,
scoring="neg_root_mean_squared_error",
cv=3,
verbose=2,
)
grid_search.fit(x_train, y_train)
best_params = grid_search.best_params_
print(grid_search.best_estimator_)
# ### Modeling
# The tuned hyperparameter are used to set up the model
best_rfr = RandomForestRegressor(**best_params_)
best_rfr.fit(x_train, y_train)
# ### Evaluation
# Here the values are predicted and also evaluated by MAE, MSE and RMSE. The important comparison is between RMSE and MAE because there we can see how the outlayers or large errors will impact the result.
pred = best_rfr.predict(x_test)
# evaluation
mae = mean_absolute_error(y_test, pred)
mse = mean_squared_error(y_test, pred)
rmse = np.sqrt(mse)
print("mean absolute error: ", mae)
print("mean squared error: ", mse)
print("root mean squared error: ", rmse)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/420/129420736.ipynb
|
bundesliga-soccer-player
|
oles04
|
[{"Id": 129420736, "ScriptId": 38447882, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12065933, "CreationDate": "05/13/2023 16:30:59", "VersionNumber": 4.0, "Title": "Market Value Prediction with Randome Forest", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 145.0, "LinesInsertedFromPrevious": 60.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 85.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185455470, "KernelVersionId": 129420736, "SourceDatasetVersionId": 5668174}]
|
[{"Id": 5668174, "DatasetId": 3258253, "DatasourceVersionId": 5743664, "CreatorUserId": 12065933, "LicenseName": "Other (specified in description)", "CreationDate": "05/12/2023 07:42:13", "VersionNumber": 1.0, "Title": "Football/Soccer | Bundesliga Player Database", "Slug": "bundesliga-soccer-player", "Subtitle": "Bundesliga Player Database: Complete Profiles, Stats, and Clubs of each Player", "Description": "The Bundesliga Players dataset provides a comprehensive collection of information on every player in the German Bundesliga football league. From renowned goalkeepers to talented defenders, this dataset offers an extensive range of player details including their names, full names, ages, heights, nationalities, places of birth, prices, maximum prices, positions, shirt numbers, preferred foot, current clubs, contract expiration dates, dates of joining the clubs, player agents, and outfitters. Whether you're a passionate football fan, a sports analyst, or a fantasy football enthusiast, this dataset serves as a valuable resource for exploring and analyzing the profiles of Bundesliga players, enabling you to delve into their backgrounds, performance statistics, and club affiliations. Discover the stars of German football and gain insights into their careers with this comprehensive Bundesliga Players dataset.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3258253, "CreatorUserId": 12065933, "OwnerUserId": 12065933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5668174.0, "CurrentDatasourceVersionId": 5743664.0, "ForumId": 3323776, "Type": 2, "CreationDate": "05/12/2023 07:42:13", "LastActivityDate": "05/12/2023", "TotalViews": 7284, "TotalDownloads": 1339, "TotalVotes": 37, "TotalKernels": 11}]
|
[{"Id": 12065933, "UserName": "oles04", "DisplayName": "Ole", "RegisterDate": "10/23/2022", "PerformanceTier": 2}]
|
# # Market Value Prediction with Randome Forest Regressor
# ### Short look up in the Data + Imports
# - `numpy`: Fundamental package for scientific computing.
# - `pandas`: Library for data manipulation and analysis.
# - `sklearn.compose.ColumnTransformer`: Applies different preprocessing steps to dataset columns.
# - `sklearn.preprocessing.OneHotEncoder`: Encodes categorical variables into binary matrix representation.
# - `sklearn.metrics.mean_squared_error`: Metric for evaluating regression models.
# - `sklearn.model_selection.train_test_split`: Splits dataset into training and testing subsets.
# - `sklearn.ensemble.RandomForestRegressor`: Ensemble regression model using decision trees.
# - `sklearn.model_selection.GridSearchCV`: Technique for hyperparameter tuning.
#
import numpy as np
import pandas as pd
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import GridSearchCV
df_players = pd.read_csv(
"/kaggle/input/bundesliga-soccer-player/bundesliga_player.csv", index_col=[0]
)
df_players.head()
df_players.describe()
df_players.info()
# ### Short nan value handling
# The two columns with too much nan values become 'none' string to replace the nan values with a categorical value. Afterwards it is easy to drop rows with nan values.
df_players = df_players[
[
"age",
"height",
"nationality",
"foot",
"position",
"club",
"contract_expires",
"joined_club",
"player_agent",
"outfitter",
"price",
]
]
df_players["outfitter"] = df_players["outfitter"].replace(np.nan, "none")
df_players["player_agent"] = df_players["player_agent"].replace(np.nan, "none")
df_players.dropna(inplace=True)
print(f"df_players shape: {df_players.shape}")
# ### Variable Usefulness for Predicting Price
# 1. Age: Age is likely to be a useful variable for predicting price as younger players generally have higher market values due to their potential for growth and longer career ahead.
# 2. Height: Height might have some influence on the price as certain positions or playing styles may favor taller players. However, its impact on price may not be as significant compared to other variables.
# 4. Club: Club affiliation is an important variable for predicting price. Players from high-profile clubs or clubs known for producing top talent are often valued more highly in the market.
# 5. Position: Position is a crucial factor in determining price. Different positions have varying levels of demand and scarcity, leading to variations in market values.
# 6. Contract Expiry Date: The remaining duration of a player's contract can impact their price. Players with longer contract terms may have higher values due to increased stability and reduced transfer urgency.
# 7. Contract Start Date: The start date of a player's current contract may have less influence on predicting price compared to other variables. It is more indicative of the player's history with the club rather than their current market value.
# 8. Agency/Representative: The player's agency or representative is not directly related to their market value. It is more of a logistical detail and does not provide significant insight into predicting price.
# 9. Sponsorship Brand: The sponsorship brand associated with a player does not have a direct impact on their market value. While brand endorsements can increase a player's overall earnings, it may not be a significant factor in price prediction.
# 10. Right/Left-Footed: A player's dominant foot is unlikely to have a substantial impact on their market value. It is more relevant to their playing style or preferred positions rather than predicting price.
# 11. Max Price (Excluded): The "max price" variable should be excluded from the prediction model because it represents the actual target variable we want to predict to much. Including it as a feature would result in data leakage and lead to an overly optimistic evaluation of the model's performance.
# Note: The above analysis is based on general assumptions and domain knowledge. It is recommended to validate the significance of these variables through statistical analysis and feature selection techniques specific to the dataset and prediction task at hand.
#
df_target = df_players[["price"]]
df_features = df_players[
[
"age",
"height",
"foot",
"position",
"club",
"contract_expires",
"joined_club",
"player_agent",
"outfitter",
]
]
# ### One Hot Encoding
# In the Data are many categorical features and the Random Forest Regressor can't handle those. Because of this One Hot Encoding is used. This implies that the categorical features will be split into different binary columns which can tell the model if this category is true or not.Nevertheless, first is a look into those variables needed.
for column in df_features.columns:
unique_values = df_features[column].unique()
print(f"Unique values in column '{column}': {unique_values}")
# Now we use the ColumnTransformer to apply the One Hot Encoding
columns_to_encode = [
"foot",
"position",
"club",
"contract_expires",
"joined_club",
"player_agent",
"outfitter",
]
ct = ColumnTransformer(
transformers=[("encoder", OneHotEncoder(), columns_to_encode)],
remainder="passthrough",
)
df_features_encoded = ct.fit_transform(df_features)
df_features_encoded.shape
# ### Train and Test Split
x_train, x_test, y_train, y_test = train_test_split(
df_features_encoded, df_target, test_size=0.3, random_state=0
)
y_train = y_train.values.ravel()
y_test = y_test.values.ravel()
print(f"x_train: {x_train.shape}")
print(f"x_test: {x_test.shape}")
print(f"y_train: {y_train.shape}")
print(f"y_test: {y_test.shape}")
# ### Hyperparameter tuning
# Using GridSearch for hyperparameter tuning is a good option because it allows us to systematically search through different combinations of hyperparameters and find the optimal configuration for our model. It automates the process of tuning hyperparameters, saving time(not computing time :p) and effort. GridSearch performs an exhaustive search over the specified hyperparameter grid, evaluating each combination using cross-validation. This helps us find the hyperparameters that yield the best performance based on the chosen evaluation metric. By using GridSearch, we can effectively optimize our model without the need for manual trial and error.
#
param_grid = {
"n_estimators": np.arange(10, 1000, 50),
"max_depth": np.arange(5, 20, 2),
"min_samples_split": np.arange(2, 11, 2),
"min_samples_leaf": np.arange(1, 10, 2),
}
rfr = RandomForestRegressor()
grid_search = GridSearchCV(
estimator=rfr,
param_grid=param_grid,
scoring="neg_root_mean_squared_error",
cv=3,
verbose=2,
)
grid_search.fit(x_train, y_train)
best_params = grid_search.best_params_
print(grid_search.best_estimator_)
# ### Modeling
# The tuned hyperparameter are used to set up the model
best_rfr = RandomForestRegressor(**best_params_)
best_rfr.fit(x_train, y_train)
# ### Evaluation
# Here the values are predicted and also evaluated by MAE, MSE and RMSE. The important comparison is between RMSE and MAE because there we can see how the outlayers or large errors will impact the result.
pred = best_rfr.predict(x_test)
# evaluation
mae = mean_absolute_error(y_test, pred)
mse = mean_squared_error(y_test, pred)
rmse = np.sqrt(mse)
print("mean absolute error: ", mae)
print("mean squared error: ", mse)
print("root mean squared error: ", rmse)
| false | 1 | 2,006 | 0 | 2,248 | 2,006 |
||
129457762
|
# # Amazon products Recommendation System
# 
# Table Of Contents
#
#
# |No | Contents
# |:---| :---
# |1 | [ Introduction ](#1)
# |2 | [ Types of Recommendation Systems](#2)
# |3 | [ Process of building a Recommendation System](#3)
# |4 | [ Data Collection](#4)
# |5 | [ Data Preprocessing](#5)
# |6 | [ Dataset Summary](#6)
# |7 | [ Text Pre-processing](#7)
# |8 | [Sentiment Analysis](#8)
#
# # Introduction
# This project aims to use customer feedback on Amazon to provide personalized recommendations. By analyzing reviews, the system learns about customers' preferences and helps them discover products that suit their tastes. The goal is to revolutionize the way customers explore and engage with the wide range of products on Amazon by leveraging the power of machine learning and Natural Language Processing.
# Companies like Amazon use different recommendation systems to provide suggestions to the customers. For example, there is **item-item collaberrative filtering**, which produces high quality recommendation system in the real time. This system is a kind of a information filtering system which seeks to predict the "rating" or preferences which user is interested in.
# 
# example of Amazon Recommender System
# # Types of Recommendation Systems
# 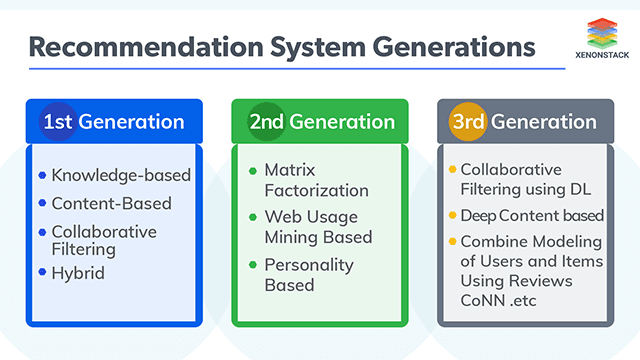
# Generations of Recommender Systems [1](https://www.xenonstack.com/blog/recommender-systems)
# **Recommendation systems** were developed to address the challenge of information overload in various domains, such as e-commerce, entertainment, and content platforms as users needed assistance in navigating through loads of catalogs of products, movies, music, articles, and more.
# In order to help users discover relevant and personalized items or content based on their preferences, interests, and past behavior. By analyzing user data, such as browsing history, purchase history, ratings, and interactions, recommendation systems can generate tailored suggestions that align with individual user preferences.
# **Advantages :**
# These systems not only enhance the user experience by saving time and effort in searching for desirable items but also drive business growth by increasing customer engagement, satisfaction, and sales.They also have the potential to introduce users to new and relevant items they may not have discovered on their own, therefore, expand their choices and improving overall user satisfaction.
# There are several types of recommendation systems commonly used in machine learning and natural language processing (NLP). Here are some of the key types: [2](https://medium.com/mlearning-ai/what-are-the-types-of-recommendation-systems-3487cbafa7c9)
# 1. **Content-Based Filtering:** This approach recommends items based on the user's past preferences or behavior. It analyzes the characteristics or features of items and compares them to the user's profile or history to make recommendations. For example, in a movie recommendation system, it may suggest similar movies based on genre, actors, or plot.
# 2. **Collaborative Filtering:** Collaborative filtering recommends items based on the behavior and preferences of similar users. It looks for patterns and similarities among users' interactions, such as ratings or purchases, and suggests items that other like-minded users have enjoyed. This method does not rely on item characteristics but rather on user behavior.
# 3. **Hybrid Approaches:** Hybrid recommendation systems combine multiple techniques to improve recommendation accuracy. They may integrate content-based and collaborative filtering methods or incorporate other machine learning algorithms to provide more precise and diverse recommendations.
# 4. **Matrix Factorization:** Matrix factorization techniques, such as singular value decomposition (SVD) or alternating least squares (ALS), decompose user-item interaction matrices to identify latent factors or features. By capturing the underlying patterns, these methods can predict missing ratings and recommend items accordingly.
# 5. **Deep Learning-based Methods:** Deep learning models, such as neural networks, can be applied to recommendation systems. They can learn intricate patterns and representations from large-scale data, enabling more accurate recommendations. Techniques like recurrent neural networks (RNNs) and convolutional neural networks (CNNs) have been employed in recommendation tasks.
# 6. **Natural Language Processing (NLP)-based Methods:** In NLP, recommendation systems can leverage techniques like sentiment analysis, text classification, or topic modeling to extract information from textual data. By understanding user reviews, feedback, or product descriptions, NLP-based methods can provide recommendations based on textual similarity or sentiment analysis.
# # Process of building a Recommendation System
# 
# Building a recommender system using Amazon product reviews involves several steps. Here's an overview of the process:
# 1. **Data Collection:** Obtaining the Amazon product reviews dataset. I've tried to retrieve data by scraping Amazon's website as well as look for a publicly available datasets that contain product reviews.
# 2. **Data Preprocessing:** Cleaning and preprocessing the reviews data to remove noise and irrelevant information. For example : remove HTML tags, punctuation, stopwords, and converting text to lowercase. Also, performing stemming or lemmatization to normalize words.
# 3. **Text Representation:** Converting the preprocessed reviews into a numerical representation that can be used by machine learning algorithms. For example : bag-of-words or term frequency-inverse document frequency (TF-IDF) can be used to represent the text data as vectors.
# 4. **Sentiment Analysis:** Analyzing the sentiment of the reviews to determine whether they are positive , negative, or neutral. Sentiment analysis can be performed using various techniques, such as using pre-trained models, lexicon-based methods, or training a sentiment classifier from scratch.
# 5. **Recommendation Algorithm:** We can use Collaborative filtering, content-based filtering, or hybrid approaches.
# 6. **Training the Model:** Training the recommendation model using the preprocessed data. The training process depends on which algorithm we're going to chose. For example, if we use collaborative filtering, we can use techniques like matrix factorization or deep learning models such as neural networks. If we want to implement Content-based filtering we can use machine learning models such as decision trees, support vector machines (SVM), or deep learning models.
# 7. **Evaluation:** Assessing the performance of our recommender system using by splitting the dataset into training validation and testing sets and use evaluation metrics : precision, recall, F1-score, and accuracy.
# **Lastly, we can deploy our Recommender System on a web or mobile app for users and monitor the model's efficiency by collecting feedbacks, updating the data and refining the algorithms used.**
# # Data Collection
# I have tried scraping Amazon's website for product reviews.
# At first I tried to collect a few examples of reviews of the same product and this was the output :
# 
# **NOTE :**
# **AMAZON ASINS**
# Amazon uses ASIN (Amazon Standard Identification Number) codes to identify product. Every product listed on Amazon has its own unique ASIN code, which you can use to construct URLs to scrape that product page, reviews, or other sellers.
# ---
# Then I wanted to do an automatic scraping of every ASIN in the product reviews pages but it always gives an 429 error which indicates that the request was rate-limited by the server.
# This is due to Amazon's rate limits to prevent excessive scraping.
# 
# Instead, we have publicly available datasets that contain product reviews on different categories :
# **Amazon product reviews data**
# This dataset contains product reviews and metadata from Amazon, including 142.8 million reviews spanning May 1996 - July 2014.
# This dataset includes reviews (ratings, text, helpfulness votes), product metadata (descriptions, category information, price, brand, and image features), and links (also viewed/also bought graphs).
# Format is one-review-per-line in (loose) json. See examples below for further help reading the data.
# 
# **Attributes Information:**
# * reviewerID - ID of the reviewer, e.g. A2SUAM1J3GNN3B
# * asin - ID of the product, e.g. 0000013714
# * reviewerName - name of the reviewer
# * helpful - helpfulness rating of the review, e.g. 2/3
# * reviewText - text of the review
# * overall - rating of the product
# * summary - summary of the review
# * unixReviewTime - time of the review (unix time)
# * reviewTime - time of the review (raw)
# **Note :**These datasets were sourced from : [Amazon Product Reviews](https://cseweb.ucsd.edu/~jmcauley/datasets.html#amazon_reviews) [3]
# # Data Pre-processing
# The fist step is to explore and clean the data we have, also as there are 3 datasets we are going to merge them in order to have 1 dataset for amazon reviews for 3 different categories
# ### Importing Libraries
import numpy as np # for linear algebra
import pandas as pd # data processing
import os
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
import math
import json
import time
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.model_selection import train_test_split
from sklearn.neighbors import NearestNeighbors
import joblib
import scipy.sparse
from scipy.sparse import csr_matrix
from scipy.sparse.linalg import svds
import warnings
warnings.simplefilter("ignore")
import nltk
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
from nltk.tokenize import word_tokenize
import re
from sklearn.model_selection import train_test_split
from sklearn.utils import resample
# #### Load the Datasets
# ##### Software dataset
df = pd.read_json(r"/kaggle/input/amazon-software/Software.json", lines=True)
df.to_csv(r"Software.csv", index=None)
# ##### fashion dataset
df_1 = pd.read_json(r"/kaggle/input/amazon-fashion/AMAZON_FASHION.json", lines=True)
df_1.to_csv(r"amazon_fashion.csv", index=None)
# ##### Appliances dataset
df_2 = pd.read_json(r"/kaggle/input/appliances/Appliances.json", lines=True)
df_2.to_csv(r"Appliances.csv", index=None)
# **lets take a look**
software_data = pd.read_csv("Software.csv")
print(f"Shape of The software dataset : {software_data.shape}")
print(f"\nGlimpse of The Dataset :")
software_data.head()
software_data["reviewTime"] = pd.to_datetime(
software_data["reviewTime"], format="%m %d, %Y"
)
fashion_data = pd.read_csv("amazon_fashion.csv")
print(f"Shape of The fashion dataset : {fashion_data.shape}")
print(f"\nGlimpse of The Dataset :")
fashion_data.head()
fashion_data["reviewTime"] = pd.to_datetime(
fashion_data["reviewTime"], format="%m %d, %Y"
)
fashion_data.info()
appliances_data = pd.read_csv("Appliances.csv")
print(f"Shape of The appliances dataset : {appliances_data.shape}")
print(f"\nGlimpse of The Dataset :")
appliances_data.head()
appliances_data["reviewTime"] = pd.to_datetime(
appliances_data["reviewTime"], format="%m %d, %Y"
)
# **merge the datasets together and add column of categories**
# Add category column
software_data["category"] = "software"
appliances_data["category"] = "appliances"
fashion_data["category"] = "fashion"
# Concatenate the datasets
merged_df = pd.concat([software_data, appliances_data, fashion_data], ignore_index=True)
# Save the merged dataset
merged_df.to_csv("merged_dataset.csv", index=False)
print(f"Shape of The merged dataset : {merged_df.shape}")
print(f"\nGlimpse of The Dataset :")
merged_df.head().style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
categories = len(merged_df["category"].unique())
categories
print(f"Informations about the dataset :\n")
print(merged_df.info())
#
# # Dataset Summary
print(f"Summary of The Dataset :")
merged_df.describe().T.style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
merged_df.describe(include=object).T.style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
merged_df.describe()["overall"].T
# minimum of ratings : 1
# maximum of ratings : 5
# **checking null values**
print("Null Values of the Dataset :")
merged_df.isna().sum().to_frame().T.style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
# We can see that we have missing values in these columns:
# - style
# - reviewerName
# - reviewText
# - summary
# - vote
# - image
# there are several approaches we can consider:
# 1. **Remove Rows or Columns:** If the missing values are present in a small number of rows or columns and do not significantly impact our analysis, we can consider removing those rows or columns using the dropna() function.
# 2. **Impute Missing Values:** If the missing values are present in a significant number of rows or columns, we may choose to impute or fill in those missing values with estimated or calculated values. Some common imputation techniques include replacing missing values with the mean, median, mode, or a constant value. Pandas provides the fillna() function for imputing missing values.
# 3. **Predict missing values with a ML Algorithm:** In some cases, we want more sophisticated imputation techniques to predict missing values based on other features in the dataset.
# from the colmuns with missing values, we can notice that they don't convey alot of important information , except 'reviewText' or maybe 'summary'
# Analyze missing data
missing_data = merged_df.isnull().sum()
missing_percentage = (missing_data / len(merged_df)) * 100
# Create a summary DataFrame
missing_summary = pd.DataFrame(
{
"Column": missing_data.index,
"Missing Count": missing_data.values,
"Missing Percentage": missing_percentage.values,
}
)
# Sort the summary DataFrame by missing percentage
missing_summary = missing_summary.sort_values("Missing Percentage", ascending=False)
# Print the summary
missing_summary
# Set the threshold for missing percentage
threshold = 50 # remove columns with more than 50% missing values
# **removing columns with a high percentage of missing values:**
# Identify columns to remove
columns_to_remove = missing_percentage[missing_percentage > threshold].index
# Remove columns with high missing percentages
merged_df = merged_df.drop(columns=columns_to_remove)
# Print the updated DataFrame
merged_df.head()
# Impute missing values in text columns with an indicator value
text_columns = ["reviewText", "summary", "reviewerName"] # Example text columns
for column in text_columns:
merged_df[column] = merged_df[column].fillna("Unknown")
# Print the updated DataFrame
merged_df.head()
print("Null Values of the Dataset :")
merged_df.isna().sum().to_frame().T.style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
merged_df.duplicated().value_counts() # check for duplicated data values
merged_df = merged_df.drop_duplicates()
merged_df.duplicated().value_counts()
merged_df.head()
# **Constant Features**
# -Constant Features are those having the same value for all the observations of the dataset. It is advisable to remove them since they add no information that allows the ML model to classify or predict a target.
from fast_ml.utilities import display_all
from fast_ml.feature_selection import get_constant_features
constant_features = get_constant_features(merged_df, threshold=0.90, dropna=False)
display_all(constant_features)
# to get list of constant features
constant_feats = (constant_features["Var"]).to_list()
print(constant_feats)
# **there are no constant features in the dataset**
# ---
# ### Visualization:
# **Overall Ratings:**
import seaborn as sns
# Check the distribution of the rating
with sns.axes_style("white"):
g = sns.catplot(x="overall", data=merged_df, aspect=2.0, kind="count")
g.set_ylabels("Total number of ratings")
# Most of the users has given the rating of 5
# **number of reviews by the verified status of the reviewers**
# number of reviews by verified status
verified_counts = merged_df["verified"].value_counts()
plt.bar(["Verified", "Not Verified"], verified_counts.values)
plt.xlabel("Verified Status")
plt.ylabel("Number of Reviews")
plt.title("Review Count by Verified Status")
plt.show()
# Most users who write reviews are verified.
# Verified reviews are generally considered more trustworthy as they indicate that the reviewer has actually purchased and used the product
# **number of reviews over time**
merged_df["reviewTime"] = pd.to_datetime(merged_df["reviewTime"])
review_count_over_time = merged_df["reviewTime"].value_counts().sort_index()
plt.plot(review_count_over_time.index, review_count_over_time.values)
plt.xlabel("Review Time")
plt.ylabel("Number of Reviews")
plt.title("Review Count over Time")
plt.xticks(rotation=45)
plt.show()
# We can nnoticed a spike in the year 2016, it indicates a significant increase in the number of reviews during that particular year.
merged_df["month"] = merged_df["reviewTime"].dt.month
review_count_by_year = merged_df["month"].value_counts().sort_index()
plt.plot(review_count_by_year.index, review_count_by_year.values)
plt.xlabel("Months")
plt.ylabel("Number of Reviews")
plt.title("Review Count by month")
plt.show()
# We can notice a significant increase in the number of reviews during the months January and February but it goes down until the month december.
# Peak periods seem to be on January , february and december.
# **Rating scores per Category**
sns.barplot(x="category", y="overall", data=merged_df)
plt.xlabel("Category")
plt.ylabel("Sentiment Score")
plt.title("Category-wise Sentiment Analysis")
plt.xticks(rotation=45)
plt.show()
# **WE can generate sentiment scores and then use those scores to create the sentiment distribution visualization :**
import nltk
from nltk.sentiment import SentimentIntensityAnalyzer
nltk.download("vader_lexicon")
# VADER sentiment analyzer
sia = SentimentIntensityAnalyzer()
def analyze_sentiment(text):
"""
Function to analyze the sentiment of a given text using VADER sentiment analyzer
"""
sentiment = sia.polarity_scores(text)
return sentiment["compound"]
def categorize_sentiment(score):
"""
Function to categorize the sentiment score into sentiment labels
"""
if score > 0.05:
return "positive"
elif score < -0.05:
return "negative"
else:
return "neutral"
# **The VADER (Valence Aware Dictionary and sEntiment Reasoner) lexicon:** [4](https://github.com/cjhutto/vaderSentiment)
# It is a pre-trained sentiment analysis tool, combines a large collection of words and phrases that have been manually annotated with sentiment scores.
# Each word or phrase in the lexicon is assigned a sentiment intensity score that represents the degree of positivity or negativity associated with it. The sentiment scores range from -1 (extremely negative) to +1 (extremely positive).
# The goal is to analyze the sentiment of a given text.
# perform sentiment analysis on review text to generate sentiment scores
# merged_df['sentiment_score'] = merged_df['reviewText'].apply(analyze_sentiment)
# categorize sentiment scores into labels (e.g., positive, negative, neutral)
# merged_df['sentiment_label'] = merged_df['sentiment_score'].apply(categorize_sentiment)
# count the number of reviews for each sentiment label
# sentiment_counts = merged_df['sentiment_label'].value_counts()
# plt.pie(sentiment_counts, labels=sentiment_counts.index, autopct='%1.1f%%')
# plt.title('Sentiment Distribution')
# plt.show()
# This part of code takes a lot of time to execute
merged_df.columns
columns = ["unixReviewTime", "month"]
merged_df = merged_df.drop(columns=columns, axis=1)
# **Unique Users**
print("\nTotal no of ratings :", merged_df.shape[0])
print("Total No of Users :", len(np.unique(merged_df.reviewerID)))
# **number of rates per user**
# number of rates per user
nb_rates_per_user = (
merged_df.groupby(by="reviewerID")["overall"].count().sort_values(ascending=False)
)
sns.barplot(x=nb_rates_per_user.index[:10], y=nb_rates_per_user[:10])
plt.xlabel("Reviewer ID")
plt.ylabel("Number of Rates")
plt.title("Top 10 Users by Number of Ratings")
plt.xticks(rotation=45)
plt.show()
# ---
# # Text Pre-processing
# #### We can use ext preprocessing techniques to clean and prepare the 'reviewText' and 'summary' columns:
# 1. **Removing punctuation:** we can define a set of punctuation characters using the string.punctuation module then we use a list comprehension to remove these punctuation characters from each text
# 2. **Converting text to lowercase:** to ensure consistency in the text data.
# 3. **Tokenization:** we can tokenize each text into a list of words.Which is the process of breaking a text into smaller units called tokens so that it can be easily analyzed and processed by machine learning algorithms. For example : "I love to play among us." After tokenization this sentence would be : ["I", "love", "to", "play", "among", "us"]
# 4. **Removing stopwords:** wa can initialize a set of stopwords using the stopwords.words('english') function from NLTK and remove them from each tokenized text using a list comprehension.
# 5. **Lemmatization:** It is useful in standardizing and normalizing the text data. It is basically a process of reducing words to their base or root form. For example : the words "running" "runs" and "ran" The lemma for all these words would be "run".
# 6. **Handling contractions:** For example : convert "can't" to "cannot" and "won't" to "will not". This can help ensure consistent representation of words.
# 7. **Removing URLs and email Addresses:** If our text data contains URLs or email addresses we can remove them as they don't typically contribute to sentiment analysis.
# 8. **Removing numbers:**
# 9. **Handling emoticons and emoji:** Emoticons and emojis can convey sentiment and add context to the text but we can remove them or convert them to corresponding textual representations.
# 10. **Handling abbreviations and acronyms:** expand common abbreviations to their full forms to avoid losing information. For exampl : convert "lol" to "laugh out loud" and "btw" to "by the way".
# 11. **Removing special characters:** This may include currency symbols, trademark symbols, etc...
# 12. **Handling spelling corrections:** depending on the quality of the data, we can perform spelling corrections to improve the accuracy of sentiment analysis. This can be done using libraries like pySpellChecker or language-specific dictionaries.
# 13. **Joining tokens:** finally we join the preprocessed tokens back into a single string.
# #### Why do we have to split text into tokens ?
# **Tokenization** is the process of splitting text into smaller units, typically words or subwords, known as tokens.
# The reasons why we should do this process:
# - **Analysis goal:** If our analysis requires a word-level or subword-level understanding of the text, such as sentiment analysis or language modeling, tokenization is typically necessary.
# - **Text processing techniques:** Many natural language processing (NLP) techniques such as stemming, lemmatization, part-of-speech tagging, and named entity recognition operate on individual tokens, so tokenization is a necessary step before applying these techniques.
# - **Model input requirements:** when using machine learning models or pre-trained language models, they often expect tokenized input, we should do tokenization to convert the raw text into a format that the models can understand.
# - **Contextual understanding:** tokenization can capture the contextual meaning of words, which can be crucial for tasks like sentiment analysis. For example, "not good" and "good" have opposite sentiments, but without tokenization, they would be treated as one token ("not good") and may lose the intended meaning.
# I just had this error :
# 
# Solution:
import wget
import zipfile
import os
url = "https://github.com/nltk/nltk_data/raw/gh-pages/packages/corpora/wordnet.zip"
zip_file_path = "/kaggle/working/wordnet.zip" # Update the path as needed
wget.download(url, zip_file_path)
extract_dir = "/usr/share/nltk_data/corpora/" # Update the path as needed
with zipfile.ZipFile(zip_file_path, "r") as zip_ref:
zip_ref.extractall(extract_dir)
os.remove(zip_file_path)
import re
import string
import emoji
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from nltk.stem import WordNetLemmatizer
import contractions
# Download NLTK resources
nltk.download("stopwords")
nltk.download("punkt")
nltk.download("wordnet")
stopwords = set(stopwords.words("english"))
nltk.download("wordnet")
lemmatizer = WordNetLemmatizer()
def preprocess_text(text):
# remove urls
text = re.sub(r"http\S+|www\S+|https\S+", "", text)
# remove email addresses
text = re.sub(r"\S+@\S+", "", text)
# remove punctuation
text = "".join([char for char in text if char not in string.punctuation])
# remove emojis
text = emoji.demojize(text)
# convert text to lowercase
text = text.lower()
# remove numbers
text = re.sub(r"\d+", "", text)
# tokenization
tokens = word_tokenize(text)
# remove stopwords
tokens = [word for word in tokens if word.lower() not in stopwords]
# lemmatization
tokens = [lemmatizer.lemmatize(word) for word in tokens]
# join tokens back into a single string
preprocessed_text = " ".join(tokens)
# expand contractions
preprocessed_text = contractions.fix(preprocessed_text)
return preprocessed_text
# ---
merged_df.head(1)
# testing the preprocessing function
example = merged_df["reviewText"][0]
preprocessed_text = preprocess_text(example)
print(
"Text before processing:\n {}\n\nText after processing:\n {}".format(
example, preprocessed_text
)
)
# **swifter.apply :** this function will be applied in parallel which can significantly speed up the execution time
import swifter
merged_df["reviewText"] = merged_df["reviewText"].swifter.apply(preprocess_text)
merged_df["summary"] = merged_df["summary"].swifter.apply(preprocess_text)
merged_df.head()
# ---
# **Handling abreviations:
# We can apply web scraping to get a pre defined abbreviations dictionary**
import requests
from bs4 import BeautifulSoup
def find_abbreviations_online(dictionary_url):
response = requests.get(dictionary_url)
soup = BeautifulSoup(response.text, "html.parser")
abbreviations = []
meanings = []
# this example assumes the abbreviations and meanings are in separate HTML elements
abbreviation_elements = soup.find_all("span", class_="abbreviation")
meaning_elements = soup.find_all("span", class_="meaning")
for abb_element, meaning_element in zip(abbreviation_elements, meaning_elements):
abbreviation = abb_element.get_text().strip()
meaning = meaning_element.get_text().strip()
abbreviations.append(abbreviation)
meanings.append(meaning)
return abbreviations, meanings
# Acronym Finder
dictionary_url = "https://www.acronymfinder.com/"
abbreviations, meanings = find_abbreviations_online(dictionary_url)
for abbreviation, meaning in zip(abbreviations, meanings):
print(f"{abbreviation}: {meaning}")
# **This part of code is still under development**
# ---
# # Sentiment Analysis
# #### We can apply sentiment analysis techniques to the preprocessed text data to classify the sentiment of each review. This can involve using pre-trained sentiment analysis models or building our own classifier using machine learning or deep learning algorithms. The sentiment analysis can assign labels such as positive, negative, or neutral to each review.
# There are multiple approaches we can take :
# 1. **Using Pre-trained Sentiment Analysis Models:**
# - **the VADER (Valence Aware Dictionary and sEntiment Reasoner)** model.
# - **TextBlob:** it uses a pre-trained model to perform sentiment analysis and provides polarity scores ranging from -1 to +1.
# - **Hugging Face Transformers:** models like BERT, RoBERTa, and DistilBERT can be fine-tuned on sentiment analysis datasets to create your own sentiment classifier.
# - **Stanford NLP Sentiment Analysis:** Stanford NLP provides a pre trained sentiment analysis model based on Recursive Neural Tensor Networks, it assigns sentiment labels such as very negative, negative, neutral, positive, and very positive to text.
# - **IBM Watson Natural Language Understanding:** IBM Watson offers a pre trained sentiment analysis model as part of their Natural Language Understanding service
# - **Google Cloud Natural Language API:** it supports sentiment analysis for multiple languages and provides sentiment scores ranging from -1 to +1.
# 2. **Building a Sentiment Classifier:**
# #### VADER
import nltk
from nltk.sentiment import SentimentIntensityAnalyzer
nltk.download("vader_lexicon")
# VADER sentiment analyzer
sia = SentimentIntensityAnalyzer()
def analyze_sentiment(text):
"""
Function to analyze the sentiment of a given text using VADER sentiment analyzer
"""
sentiment = sia.polarity_scores(text)
return sentiment["compound"]
def categorize_sentiment(score):
"""
Function to categorize the sentiment score into sentiment labels
"""
if score > 0.05:
return "positive"
elif score < -0.05:
return "negative"
else:
return "neutral"
# perform sentiment analysis on review text to generate sentiment scores
merged_df["sentiment_score"] = merged_df["reviewText"].apply(analyze_sentiment)
# categorize sentiment scores into labels (e.g., positive, negative, neutral)
merged_df["sentiment_label"] = merged_df["sentiment_score"].apply(categorize_sentiment)
# count the number of reviews for each sentiment label
sentiment_counts = merged_df["sentiment_label"].value_counts()
plt.pie(sentiment_counts, labels=sentiment_counts.index, autopct="%1.1f%%")
plt.title("Sentiment Distribution")
plt.show()
merged_df.head(1)
filtered_df = merged_df[merged_df["category"] == "software"]
filtered_df.to_csv("/kaggle/working/filtered_dataset.csv", index=False)
filtered_df.shape
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
rating_labels = merged_df["overall"].apply(
lambda x: "positive" if x >= 3 else "negative"
)
vader_labels = merged_df["sentiment_label"]
accuracy = accuracy_score(rating_labels, vader_labels)
print("Overall Sentiment Accuracy:", accuracy)
# Evaluate precision, recall, and F1-score
precision = precision_score(
rating_labels, vader_labels, pos_label="positive", average="weighted"
)
recall = recall_score(
rating_labels, vader_labels, pos_label="positive", average="weighted"
)
f1 = f1_score(rating_labels, vader_labels, pos_label="positive", average="weighted")
print("Positive Sentiment Precision:", precision)
print("Positive Sentiment Recall:", recall)
print("Positive Sentiment F1-Score:", f1)
# **The chart pie indicates overall a high level of satisfaction among customers**
positive_reviews = merged_df[merged_df["sentiment_label"] == "positive"]["reviewText"]
sample_positive_reviews = positive_reviews.sample(n=5)
for review in sample_positive_reviews:
print(review)
print("---")
negative_reviews = merged_df[merged_df["sentiment_label"] == "negative"]["reviewText"]
sample_negative_reviews = negative_reviews.sample(n=5)
for review in sample_negative_reviews:
print(review)
print("---")
neutral_reviews = merged_df[merged_df["sentiment_label"] == "neutral"]["reviewText"]
sample_neutral_reviews = neutral_reviews.sample(n=5)
for review in sample_neutral_reviews:
print(review)
print("---")
merged_df.head()
columns = ["sentiment_score", "sentiment_label"]
new_df = merged_df.drop(columns=columns, axis=1)
# **Word CLoud**
from wordcloud import WordCloud
def show_wordcloud(data, title=None):
wordcloud = WordCloud(
background_color="white",
max_words=200,
max_font_size=40,
scale=3,
random_state=42,
).generate(str(data))
fig = plt.figure(1, figsize=(20, 20))
plt.axis("off")
if title:
fig.suptitle(title, fontsize=20)
fig.subplots_adjust(top=2.3)
plt.imshow(wordcloud)
plt.show()
# **Positive Reviews**
positiveReviews_df = merged_df.loc[merged_df["sentiment_label"] == "positive"]
show_wordcloud(positiveReviews_df["reviewText"])
# **Negative Reviews**
negativeReviews_df = merged_df.loc[merged_df["sentiment_label"] == "negative"]
show_wordcloud(negativeReviews_df["reviewText"])
# ### Text Blob
from textblob import TextBlob
import pandas as pd
# Apply sentiment analysis using TextBlob
new_df["sentiment"] = new_df["reviewText"].apply(
lambda text: TextBlob(text).sentiment.polarity
)
# Classify sentiment labels based on polarity scores
new_df["sentiment_label"] = new_df["sentiment"].apply(
lambda score: "Positive" if score > 0 else "Negative" if score < 0 else "Neutral"
)
# Print the updated dataset
new_df[["reviewText", "sentiment", "sentiment_label"]]
# count the number of reviews for each sentiment label
sentiment_counts = new_df["sentiment_label"].value_counts()
plt.pie(sentiment_counts, labels=sentiment_counts.index, autopct="%1.1f%%")
plt.title("Sentiment Distribution")
plt.show()
# ---
# ### Hugging Face Transformers
# **Fine-tuning a BERT model for sentiment analysis**
# columns = ['sentiment_score', 'sentiment_label']
# new_df = merged_df.drop(columns= columns, axis=1)
# import torch
# from transformers import BertTokenizer, BertForSequenceClassification
# from torch.utils.data import DataLoader, RandomSampler
# from transformers import AdamW
# Load the tokenizer and BERT model
# tokenizer = BertTokenizer.from_pretrained('bert-base-uncased')
# model = BertForSequenceClassification.from_pretrained('bert-base-uncased', num_labels=2)
# Preprocess the dataset
# Assuming you have already loaded your dataset and split it into train and test sets
# train_texts = train_dataset['reviewText'].tolist()
# train_labels = train_dataset['overall'].tolist()
# test_texts = test_dataset['reviewText'].tolist()
# test_labels = test_dataset['overall'].tolist()
# Tokenize the texts
# train_encodings = tokenizer(train_texts, truncation=True, padding=True)
# test_encodings = tokenizer(test_texts, truncation=True, padding=True)
# ---
# # Feature Extraction
# **CountVectorizer**
# To analyze the text data and build a vocabulary of unique words. The resulting vocabulary can be used to convert text documents into a numerical representation suitable for machine learning algorithms.
from sklearn.feature_extraction.text import CountVectorizer
# features = CountVectorizer()
# features.fit(merged_df["reviewText"])
# print(len(features.vocabulary_))
# print(features.vocabulary_)
# bagofWords = features.transform(merged_df["reviewText"])
# print(bagofWords)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/457/129457762.ipynb
| null | null |
[{"Id": 129457762, "ScriptId": 38392756, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10789149, "CreationDate": "05/14/2023 02:19:02", "VersionNumber": 4.0, "Title": "NLP - Amazon products Recommender System", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 909.0, "LinesInsertedFromPrevious": 69.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 840.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 4}]
| null | null | null | null |
# # Amazon products Recommendation System
# 
# Table Of Contents
#
#
# |No | Contents
# |:---| :---
# |1 | [ Introduction ](#1)
# |2 | [ Types of Recommendation Systems](#2)
# |3 | [ Process of building a Recommendation System](#3)
# |4 | [ Data Collection](#4)
# |5 | [ Data Preprocessing](#5)
# |6 | [ Dataset Summary](#6)
# |7 | [ Text Pre-processing](#7)
# |8 | [Sentiment Analysis](#8)
#
# # Introduction
# This project aims to use customer feedback on Amazon to provide personalized recommendations. By analyzing reviews, the system learns about customers' preferences and helps them discover products that suit their tastes. The goal is to revolutionize the way customers explore and engage with the wide range of products on Amazon by leveraging the power of machine learning and Natural Language Processing.
# Companies like Amazon use different recommendation systems to provide suggestions to the customers. For example, there is **item-item collaberrative filtering**, which produces high quality recommendation system in the real time. This system is a kind of a information filtering system which seeks to predict the "rating" or preferences which user is interested in.
# 
# example of Amazon Recommender System
# # Types of Recommendation Systems
# 
# Generations of Recommender Systems [1](https://www.xenonstack.com/blog/recommender-systems)
# **Recommendation systems** were developed to address the challenge of information overload in various domains, such as e-commerce, entertainment, and content platforms as users needed assistance in navigating through loads of catalogs of products, movies, music, articles, and more.
# In order to help users discover relevant and personalized items or content based on their preferences, interests, and past behavior. By analyzing user data, such as browsing history, purchase history, ratings, and interactions, recommendation systems can generate tailored suggestions that align with individual user preferences.
# **Advantages :**
# These systems not only enhance the user experience by saving time and effort in searching for desirable items but also drive business growth by increasing customer engagement, satisfaction, and sales.They also have the potential to introduce users to new and relevant items they may not have discovered on their own, therefore, expand their choices and improving overall user satisfaction.
# There are several types of recommendation systems commonly used in machine learning and natural language processing (NLP). Here are some of the key types: [2](https://medium.com/mlearning-ai/what-are-the-types-of-recommendation-systems-3487cbafa7c9)
# 1. **Content-Based Filtering:** This approach recommends items based on the user's past preferences or behavior. It analyzes the characteristics or features of items and compares them to the user's profile or history to make recommendations. For example, in a movie recommendation system, it may suggest similar movies based on genre, actors, or plot.
# 2. **Collaborative Filtering:** Collaborative filtering recommends items based on the behavior and preferences of similar users. It looks for patterns and similarities among users' interactions, such as ratings or purchases, and suggests items that other like-minded users have enjoyed. This method does not rely on item characteristics but rather on user behavior.
# 3. **Hybrid Approaches:** Hybrid recommendation systems combine multiple techniques to improve recommendation accuracy. They may integrate content-based and collaborative filtering methods or incorporate other machine learning algorithms to provide more precise and diverse recommendations.
# 4. **Matrix Factorization:** Matrix factorization techniques, such as singular value decomposition (SVD) or alternating least squares (ALS), decompose user-item interaction matrices to identify latent factors or features. By capturing the underlying patterns, these methods can predict missing ratings and recommend items accordingly.
# 5. **Deep Learning-based Methods:** Deep learning models, such as neural networks, can be applied to recommendation systems. They can learn intricate patterns and representations from large-scale data, enabling more accurate recommendations. Techniques like recurrent neural networks (RNNs) and convolutional neural networks (CNNs) have been employed in recommendation tasks.
# 6. **Natural Language Processing (NLP)-based Methods:** In NLP, recommendation systems can leverage techniques like sentiment analysis, text classification, or topic modeling to extract information from textual data. By understanding user reviews, feedback, or product descriptions, NLP-based methods can provide recommendations based on textual similarity or sentiment analysis.
# # Process of building a Recommendation System
# 
# Building a recommender system using Amazon product reviews involves several steps. Here's an overview of the process:
# 1. **Data Collection:** Obtaining the Amazon product reviews dataset. I've tried to retrieve data by scraping Amazon's website as well as look for a publicly available datasets that contain product reviews.
# 2. **Data Preprocessing:** Cleaning and preprocessing the reviews data to remove noise and irrelevant information. For example : remove HTML tags, punctuation, stopwords, and converting text to lowercase. Also, performing stemming or lemmatization to normalize words.
# 3. **Text Representation:** Converting the preprocessed reviews into a numerical representation that can be used by machine learning algorithms. For example : bag-of-words or term frequency-inverse document frequency (TF-IDF) can be used to represent the text data as vectors.
# 4. **Sentiment Analysis:** Analyzing the sentiment of the reviews to determine whether they are positive , negative, or neutral. Sentiment analysis can be performed using various techniques, such as using pre-trained models, lexicon-based methods, or training a sentiment classifier from scratch.
# 5. **Recommendation Algorithm:** We can use Collaborative filtering, content-based filtering, or hybrid approaches.
# 6. **Training the Model:** Training the recommendation model using the preprocessed data. The training process depends on which algorithm we're going to chose. For example, if we use collaborative filtering, we can use techniques like matrix factorization or deep learning models such as neural networks. If we want to implement Content-based filtering we can use machine learning models such as decision trees, support vector machines (SVM), or deep learning models.
# 7. **Evaluation:** Assessing the performance of our recommender system using by splitting the dataset into training validation and testing sets and use evaluation metrics : precision, recall, F1-score, and accuracy.
# **Lastly, we can deploy our Recommender System on a web or mobile app for users and monitor the model's efficiency by collecting feedbacks, updating the data and refining the algorithms used.**
# # Data Collection
# I have tried scraping Amazon's website for product reviews.
# At first I tried to collect a few examples of reviews of the same product and this was the output :
# 
# **NOTE :**
# **AMAZON ASINS**
# Amazon uses ASIN (Amazon Standard Identification Number) codes to identify product. Every product listed on Amazon has its own unique ASIN code, which you can use to construct URLs to scrape that product page, reviews, or other sellers.
# ---
# Then I wanted to do an automatic scraping of every ASIN in the product reviews pages but it always gives an 429 error which indicates that the request was rate-limited by the server.
# This is due to Amazon's rate limits to prevent excessive scraping.
# 
# Instead, we have publicly available datasets that contain product reviews on different categories :
# **Amazon product reviews data**
# This dataset contains product reviews and metadata from Amazon, including 142.8 million reviews spanning May 1996 - July 2014.
# This dataset includes reviews (ratings, text, helpfulness votes), product metadata (descriptions, category information, price, brand, and image features), and links (also viewed/also bought graphs).
# Format is one-review-per-line in (loose) json. See examples below for further help reading the data.
# 
# **Attributes Information:**
# * reviewerID - ID of the reviewer, e.g. A2SUAM1J3GNN3B
# * asin - ID of the product, e.g. 0000013714
# * reviewerName - name of the reviewer
# * helpful - helpfulness rating of the review, e.g. 2/3
# * reviewText - text of the review
# * overall - rating of the product
# * summary - summary of the review
# * unixReviewTime - time of the review (unix time)
# * reviewTime - time of the review (raw)
# **Note :**These datasets were sourced from : [Amazon Product Reviews](https://cseweb.ucsd.edu/~jmcauley/datasets.html#amazon_reviews) [3]
# # Data Pre-processing
# The fist step is to explore and clean the data we have, also as there are 3 datasets we are going to merge them in order to have 1 dataset for amazon reviews for 3 different categories
# ### Importing Libraries
import numpy as np # for linear algebra
import pandas as pd # data processing
import os
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
import math
import json
import time
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.model_selection import train_test_split
from sklearn.neighbors import NearestNeighbors
import joblib
import scipy.sparse
from scipy.sparse import csr_matrix
from scipy.sparse.linalg import svds
import warnings
warnings.simplefilter("ignore")
import nltk
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
from nltk.tokenize import word_tokenize
import re
from sklearn.model_selection import train_test_split
from sklearn.utils import resample
# #### Load the Datasets
# ##### Software dataset
df = pd.read_json(r"/kaggle/input/amazon-software/Software.json", lines=True)
df.to_csv(r"Software.csv", index=None)
# ##### fashion dataset
df_1 = pd.read_json(r"/kaggle/input/amazon-fashion/AMAZON_FASHION.json", lines=True)
df_1.to_csv(r"amazon_fashion.csv", index=None)
# ##### Appliances dataset
df_2 = pd.read_json(r"/kaggle/input/appliances/Appliances.json", lines=True)
df_2.to_csv(r"Appliances.csv", index=None)
# **lets take a look**
software_data = pd.read_csv("Software.csv")
print(f"Shape of The software dataset : {software_data.shape}")
print(f"\nGlimpse of The Dataset :")
software_data.head()
software_data["reviewTime"] = pd.to_datetime(
software_data["reviewTime"], format="%m %d, %Y"
)
fashion_data = pd.read_csv("amazon_fashion.csv")
print(f"Shape of The fashion dataset : {fashion_data.shape}")
print(f"\nGlimpse of The Dataset :")
fashion_data.head()
fashion_data["reviewTime"] = pd.to_datetime(
fashion_data["reviewTime"], format="%m %d, %Y"
)
fashion_data.info()
appliances_data = pd.read_csv("Appliances.csv")
print(f"Shape of The appliances dataset : {appliances_data.shape}")
print(f"\nGlimpse of The Dataset :")
appliances_data.head()
appliances_data["reviewTime"] = pd.to_datetime(
appliances_data["reviewTime"], format="%m %d, %Y"
)
# **merge the datasets together and add column of categories**
# Add category column
software_data["category"] = "software"
appliances_data["category"] = "appliances"
fashion_data["category"] = "fashion"
# Concatenate the datasets
merged_df = pd.concat([software_data, appliances_data, fashion_data], ignore_index=True)
# Save the merged dataset
merged_df.to_csv("merged_dataset.csv", index=False)
print(f"Shape of The merged dataset : {merged_df.shape}")
print(f"\nGlimpse of The Dataset :")
merged_df.head().style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
categories = len(merged_df["category"].unique())
categories
print(f"Informations about the dataset :\n")
print(merged_df.info())
#
# # Dataset Summary
print(f"Summary of The Dataset :")
merged_df.describe().T.style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
merged_df.describe(include=object).T.style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
merged_df.describe()["overall"].T
# minimum of ratings : 1
# maximum of ratings : 5
# **checking null values**
print("Null Values of the Dataset :")
merged_df.isna().sum().to_frame().T.style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
# We can see that we have missing values in these columns:
# - style
# - reviewerName
# - reviewText
# - summary
# - vote
# - image
# there are several approaches we can consider:
# 1. **Remove Rows or Columns:** If the missing values are present in a small number of rows or columns and do not significantly impact our analysis, we can consider removing those rows or columns using the dropna() function.
# 2. **Impute Missing Values:** If the missing values are present in a significant number of rows or columns, we may choose to impute or fill in those missing values with estimated or calculated values. Some common imputation techniques include replacing missing values with the mean, median, mode, or a constant value. Pandas provides the fillna() function for imputing missing values.
# 3. **Predict missing values with a ML Algorithm:** In some cases, we want more sophisticated imputation techniques to predict missing values based on other features in the dataset.
# from the colmuns with missing values, we can notice that they don't convey alot of important information , except 'reviewText' or maybe 'summary'
# Analyze missing data
missing_data = merged_df.isnull().sum()
missing_percentage = (missing_data / len(merged_df)) * 100
# Create a summary DataFrame
missing_summary = pd.DataFrame(
{
"Column": missing_data.index,
"Missing Count": missing_data.values,
"Missing Percentage": missing_percentage.values,
}
)
# Sort the summary DataFrame by missing percentage
missing_summary = missing_summary.sort_values("Missing Percentage", ascending=False)
# Print the summary
missing_summary
# Set the threshold for missing percentage
threshold = 50 # remove columns with more than 50% missing values
# **removing columns with a high percentage of missing values:**
# Identify columns to remove
columns_to_remove = missing_percentage[missing_percentage > threshold].index
# Remove columns with high missing percentages
merged_df = merged_df.drop(columns=columns_to_remove)
# Print the updated DataFrame
merged_df.head()
# Impute missing values in text columns with an indicator value
text_columns = ["reviewText", "summary", "reviewerName"] # Example text columns
for column in text_columns:
merged_df[column] = merged_df[column].fillna("Unknown")
# Print the updated DataFrame
merged_df.head()
print("Null Values of the Dataset :")
merged_df.isna().sum().to_frame().T.style.set_properties(
**{"background-color": "#2a9d8f", "color": "white", "border": "1.5px solid black"}
)
merged_df.duplicated().value_counts() # check for duplicated data values
merged_df = merged_df.drop_duplicates()
merged_df.duplicated().value_counts()
merged_df.head()
# **Constant Features**
# -Constant Features are those having the same value for all the observations of the dataset. It is advisable to remove them since they add no information that allows the ML model to classify or predict a target.
from fast_ml.utilities import display_all
from fast_ml.feature_selection import get_constant_features
constant_features = get_constant_features(merged_df, threshold=0.90, dropna=False)
display_all(constant_features)
# to get list of constant features
constant_feats = (constant_features["Var"]).to_list()
print(constant_feats)
# **there are no constant features in the dataset**
# ---
# ### Visualization:
# **Overall Ratings:**
import seaborn as sns
# Check the distribution of the rating
with sns.axes_style("white"):
g = sns.catplot(x="overall", data=merged_df, aspect=2.0, kind="count")
g.set_ylabels("Total number of ratings")
# Most of the users has given the rating of 5
# **number of reviews by the verified status of the reviewers**
# number of reviews by verified status
verified_counts = merged_df["verified"].value_counts()
plt.bar(["Verified", "Not Verified"], verified_counts.values)
plt.xlabel("Verified Status")
plt.ylabel("Number of Reviews")
plt.title("Review Count by Verified Status")
plt.show()
# Most users who write reviews are verified.
# Verified reviews are generally considered more trustworthy as they indicate that the reviewer has actually purchased and used the product
# **number of reviews over time**
merged_df["reviewTime"] = pd.to_datetime(merged_df["reviewTime"])
review_count_over_time = merged_df["reviewTime"].value_counts().sort_index()
plt.plot(review_count_over_time.index, review_count_over_time.values)
plt.xlabel("Review Time")
plt.ylabel("Number of Reviews")
plt.title("Review Count over Time")
plt.xticks(rotation=45)
plt.show()
# We can nnoticed a spike in the year 2016, it indicates a significant increase in the number of reviews during that particular year.
merged_df["month"] = merged_df["reviewTime"].dt.month
review_count_by_year = merged_df["month"].value_counts().sort_index()
plt.plot(review_count_by_year.index, review_count_by_year.values)
plt.xlabel("Months")
plt.ylabel("Number of Reviews")
plt.title("Review Count by month")
plt.show()
# We can notice a significant increase in the number of reviews during the months January and February but it goes down until the month december.
# Peak periods seem to be on January , february and december.
# **Rating scores per Category**
sns.barplot(x="category", y="overall", data=merged_df)
plt.xlabel("Category")
plt.ylabel("Sentiment Score")
plt.title("Category-wise Sentiment Analysis")
plt.xticks(rotation=45)
plt.show()
# **WE can generate sentiment scores and then use those scores to create the sentiment distribution visualization :**
import nltk
from nltk.sentiment import SentimentIntensityAnalyzer
nltk.download("vader_lexicon")
# VADER sentiment analyzer
sia = SentimentIntensityAnalyzer()
def analyze_sentiment(text):
"""
Function to analyze the sentiment of a given text using VADER sentiment analyzer
"""
sentiment = sia.polarity_scores(text)
return sentiment["compound"]
def categorize_sentiment(score):
"""
Function to categorize the sentiment score into sentiment labels
"""
if score > 0.05:
return "positive"
elif score < -0.05:
return "negative"
else:
return "neutral"
# **The VADER (Valence Aware Dictionary and sEntiment Reasoner) lexicon:** [4](https://github.com/cjhutto/vaderSentiment)
# It is a pre-trained sentiment analysis tool, combines a large collection of words and phrases that have been manually annotated with sentiment scores.
# Each word or phrase in the lexicon is assigned a sentiment intensity score that represents the degree of positivity or negativity associated with it. The sentiment scores range from -1 (extremely negative) to +1 (extremely positive).
# The goal is to analyze the sentiment of a given text.
# perform sentiment analysis on review text to generate sentiment scores
# merged_df['sentiment_score'] = merged_df['reviewText'].apply(analyze_sentiment)
# categorize sentiment scores into labels (e.g., positive, negative, neutral)
# merged_df['sentiment_label'] = merged_df['sentiment_score'].apply(categorize_sentiment)
# count the number of reviews for each sentiment label
# sentiment_counts = merged_df['sentiment_label'].value_counts()
# plt.pie(sentiment_counts, labels=sentiment_counts.index, autopct='%1.1f%%')
# plt.title('Sentiment Distribution')
# plt.show()
# This part of code takes a lot of time to execute
merged_df.columns
columns = ["unixReviewTime", "month"]
merged_df = merged_df.drop(columns=columns, axis=1)
# **Unique Users**
print("\nTotal no of ratings :", merged_df.shape[0])
print("Total No of Users :", len(np.unique(merged_df.reviewerID)))
# **number of rates per user**
# number of rates per user
nb_rates_per_user = (
merged_df.groupby(by="reviewerID")["overall"].count().sort_values(ascending=False)
)
sns.barplot(x=nb_rates_per_user.index[:10], y=nb_rates_per_user[:10])
plt.xlabel("Reviewer ID")
plt.ylabel("Number of Rates")
plt.title("Top 10 Users by Number of Ratings")
plt.xticks(rotation=45)
plt.show()
# ---
# # Text Pre-processing
# #### We can use ext preprocessing techniques to clean and prepare the 'reviewText' and 'summary' columns:
# 1. **Removing punctuation:** we can define a set of punctuation characters using the string.punctuation module then we use a list comprehension to remove these punctuation characters from each text
# 2. **Converting text to lowercase:** to ensure consistency in the text data.
# 3. **Tokenization:** we can tokenize each text into a list of words.Which is the process of breaking a text into smaller units called tokens so that it can be easily analyzed and processed by machine learning algorithms. For example : "I love to play among us." After tokenization this sentence would be : ["I", "love", "to", "play", "among", "us"]
# 4. **Removing stopwords:** wa can initialize a set of stopwords using the stopwords.words('english') function from NLTK and remove them from each tokenized text using a list comprehension.
# 5. **Lemmatization:** It is useful in standardizing and normalizing the text data. It is basically a process of reducing words to their base or root form. For example : the words "running" "runs" and "ran" The lemma for all these words would be "run".
# 6. **Handling contractions:** For example : convert "can't" to "cannot" and "won't" to "will not". This can help ensure consistent representation of words.
# 7. **Removing URLs and email Addresses:** If our text data contains URLs or email addresses we can remove them as they don't typically contribute to sentiment analysis.
# 8. **Removing numbers:**
# 9. **Handling emoticons and emoji:** Emoticons and emojis can convey sentiment and add context to the text but we can remove them or convert them to corresponding textual representations.
# 10. **Handling abbreviations and acronyms:** expand common abbreviations to their full forms to avoid losing information. For exampl : convert "lol" to "laugh out loud" and "btw" to "by the way".
# 11. **Removing special characters:** This may include currency symbols, trademark symbols, etc...
# 12. **Handling spelling corrections:** depending on the quality of the data, we can perform spelling corrections to improve the accuracy of sentiment analysis. This can be done using libraries like pySpellChecker or language-specific dictionaries.
# 13. **Joining tokens:** finally we join the preprocessed tokens back into a single string.
# #### Why do we have to split text into tokens ?
# **Tokenization** is the process of splitting text into smaller units, typically words or subwords, known as tokens.
# The reasons why we should do this process:
# - **Analysis goal:** If our analysis requires a word-level or subword-level understanding of the text, such as sentiment analysis or language modeling, tokenization is typically necessary.
# - **Text processing techniques:** Many natural language processing (NLP) techniques such as stemming, lemmatization, part-of-speech tagging, and named entity recognition operate on individual tokens, so tokenization is a necessary step before applying these techniques.
# - **Model input requirements:** when using machine learning models or pre-trained language models, they often expect tokenized input, we should do tokenization to convert the raw text into a format that the models can understand.
# - **Contextual understanding:** tokenization can capture the contextual meaning of words, which can be crucial for tasks like sentiment analysis. For example, "not good" and "good" have opposite sentiments, but without tokenization, they would be treated as one token ("not good") and may lose the intended meaning.
# I just had this error :
# 
# Solution:
import wget
import zipfile
import os
url = "https://github.com/nltk/nltk_data/raw/gh-pages/packages/corpora/wordnet.zip"
zip_file_path = "/kaggle/working/wordnet.zip" # Update the path as needed
wget.download(url, zip_file_path)
extract_dir = "/usr/share/nltk_data/corpora/" # Update the path as needed
with zipfile.ZipFile(zip_file_path, "r") as zip_ref:
zip_ref.extractall(extract_dir)
os.remove(zip_file_path)
import re
import string
import emoji
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from nltk.stem import WordNetLemmatizer
import contractions
# Download NLTK resources
nltk.download("stopwords")
nltk.download("punkt")
nltk.download("wordnet")
stopwords = set(stopwords.words("english"))
nltk.download("wordnet")
lemmatizer = WordNetLemmatizer()
def preprocess_text(text):
# remove urls
text = re.sub(r"http\S+|www\S+|https\S+", "", text)
# remove email addresses
text = re.sub(r"\S+@\S+", "", text)
# remove punctuation
text = "".join([char for char in text if char not in string.punctuation])
# remove emojis
text = emoji.demojize(text)
# convert text to lowercase
text = text.lower()
# remove numbers
text = re.sub(r"\d+", "", text)
# tokenization
tokens = word_tokenize(text)
# remove stopwords
tokens = [word for word in tokens if word.lower() not in stopwords]
# lemmatization
tokens = [lemmatizer.lemmatize(word) for word in tokens]
# join tokens back into a single string
preprocessed_text = " ".join(tokens)
# expand contractions
preprocessed_text = contractions.fix(preprocessed_text)
return preprocessed_text
# ---
merged_df.head(1)
# testing the preprocessing function
example = merged_df["reviewText"][0]
preprocessed_text = preprocess_text(example)
print(
"Text before processing:\n {}\n\nText after processing:\n {}".format(
example, preprocessed_text
)
)
# **swifter.apply :** this function will be applied in parallel which can significantly speed up the execution time
import swifter
merged_df["reviewText"] = merged_df["reviewText"].swifter.apply(preprocess_text)
merged_df["summary"] = merged_df["summary"].swifter.apply(preprocess_text)
merged_df.head()
# ---
# **Handling abreviations:
# We can apply web scraping to get a pre defined abbreviations dictionary**
import requests
from bs4 import BeautifulSoup
def find_abbreviations_online(dictionary_url):
response = requests.get(dictionary_url)
soup = BeautifulSoup(response.text, "html.parser")
abbreviations = []
meanings = []
# this example assumes the abbreviations and meanings are in separate HTML elements
abbreviation_elements = soup.find_all("span", class_="abbreviation")
meaning_elements = soup.find_all("span", class_="meaning")
for abb_element, meaning_element in zip(abbreviation_elements, meaning_elements):
abbreviation = abb_element.get_text().strip()
meaning = meaning_element.get_text().strip()
abbreviations.append(abbreviation)
meanings.append(meaning)
return abbreviations, meanings
# Acronym Finder
dictionary_url = "https://www.acronymfinder.com/"
abbreviations, meanings = find_abbreviations_online(dictionary_url)
for abbreviation, meaning in zip(abbreviations, meanings):
print(f"{abbreviation}: {meaning}")
# **This part of code is still under development**
# ---
# # Sentiment Analysis
# #### We can apply sentiment analysis techniques to the preprocessed text data to classify the sentiment of each review. This can involve using pre-trained sentiment analysis models or building our own classifier using machine learning or deep learning algorithms. The sentiment analysis can assign labels such as positive, negative, or neutral to each review.
# There are multiple approaches we can take :
# 1. **Using Pre-trained Sentiment Analysis Models:**
# - **the VADER (Valence Aware Dictionary and sEntiment Reasoner)** model.
# - **TextBlob:** it uses a pre-trained model to perform sentiment analysis and provides polarity scores ranging from -1 to +1.
# - **Hugging Face Transformers:** models like BERT, RoBERTa, and DistilBERT can be fine-tuned on sentiment analysis datasets to create your own sentiment classifier.
# - **Stanford NLP Sentiment Analysis:** Stanford NLP provides a pre trained sentiment analysis model based on Recursive Neural Tensor Networks, it assigns sentiment labels such as very negative, negative, neutral, positive, and very positive to text.
# - **IBM Watson Natural Language Understanding:** IBM Watson offers a pre trained sentiment analysis model as part of their Natural Language Understanding service
# - **Google Cloud Natural Language API:** it supports sentiment analysis for multiple languages and provides sentiment scores ranging from -1 to +1.
# 2. **Building a Sentiment Classifier:**
# #### VADER
import nltk
from nltk.sentiment import SentimentIntensityAnalyzer
nltk.download("vader_lexicon")
# VADER sentiment analyzer
sia = SentimentIntensityAnalyzer()
def analyze_sentiment(text):
"""
Function to analyze the sentiment of a given text using VADER sentiment analyzer
"""
sentiment = sia.polarity_scores(text)
return sentiment["compound"]
def categorize_sentiment(score):
"""
Function to categorize the sentiment score into sentiment labels
"""
if score > 0.05:
return "positive"
elif score < -0.05:
return "negative"
else:
return "neutral"
# perform sentiment analysis on review text to generate sentiment scores
merged_df["sentiment_score"] = merged_df["reviewText"].apply(analyze_sentiment)
# categorize sentiment scores into labels (e.g., positive, negative, neutral)
merged_df["sentiment_label"] = merged_df["sentiment_score"].apply(categorize_sentiment)
# count the number of reviews for each sentiment label
sentiment_counts = merged_df["sentiment_label"].value_counts()
plt.pie(sentiment_counts, labels=sentiment_counts.index, autopct="%1.1f%%")
plt.title("Sentiment Distribution")
plt.show()
merged_df.head(1)
filtered_df = merged_df[merged_df["category"] == "software"]
filtered_df.to_csv("/kaggle/working/filtered_dataset.csv", index=False)
filtered_df.shape
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
rating_labels = merged_df["overall"].apply(
lambda x: "positive" if x >= 3 else "negative"
)
vader_labels = merged_df["sentiment_label"]
accuracy = accuracy_score(rating_labels, vader_labels)
print("Overall Sentiment Accuracy:", accuracy)
# Evaluate precision, recall, and F1-score
precision = precision_score(
rating_labels, vader_labels, pos_label="positive", average="weighted"
)
recall = recall_score(
rating_labels, vader_labels, pos_label="positive", average="weighted"
)
f1 = f1_score(rating_labels, vader_labels, pos_label="positive", average="weighted")
print("Positive Sentiment Precision:", precision)
print("Positive Sentiment Recall:", recall)
print("Positive Sentiment F1-Score:", f1)
# **The chart pie indicates overall a high level of satisfaction among customers**
positive_reviews = merged_df[merged_df["sentiment_label"] == "positive"]["reviewText"]
sample_positive_reviews = positive_reviews.sample(n=5)
for review in sample_positive_reviews:
print(review)
print("---")
negative_reviews = merged_df[merged_df["sentiment_label"] == "negative"]["reviewText"]
sample_negative_reviews = negative_reviews.sample(n=5)
for review in sample_negative_reviews:
print(review)
print("---")
neutral_reviews = merged_df[merged_df["sentiment_label"] == "neutral"]["reviewText"]
sample_neutral_reviews = neutral_reviews.sample(n=5)
for review in sample_neutral_reviews:
print(review)
print("---")
merged_df.head()
columns = ["sentiment_score", "sentiment_label"]
new_df = merged_df.drop(columns=columns, axis=1)
# **Word CLoud**
from wordcloud import WordCloud
def show_wordcloud(data, title=None):
wordcloud = WordCloud(
background_color="white",
max_words=200,
max_font_size=40,
scale=3,
random_state=42,
).generate(str(data))
fig = plt.figure(1, figsize=(20, 20))
plt.axis("off")
if title:
fig.suptitle(title, fontsize=20)
fig.subplots_adjust(top=2.3)
plt.imshow(wordcloud)
plt.show()
# **Positive Reviews**
positiveReviews_df = merged_df.loc[merged_df["sentiment_label"] == "positive"]
show_wordcloud(positiveReviews_df["reviewText"])
# **Negative Reviews**
negativeReviews_df = merged_df.loc[merged_df["sentiment_label"] == "negative"]
show_wordcloud(negativeReviews_df["reviewText"])
# ### Text Blob
from textblob import TextBlob
import pandas as pd
# Apply sentiment analysis using TextBlob
new_df["sentiment"] = new_df["reviewText"].apply(
lambda text: TextBlob(text).sentiment.polarity
)
# Classify sentiment labels based on polarity scores
new_df["sentiment_label"] = new_df["sentiment"].apply(
lambda score: "Positive" if score > 0 else "Negative" if score < 0 else "Neutral"
)
# Print the updated dataset
new_df[["reviewText", "sentiment", "sentiment_label"]]
# count the number of reviews for each sentiment label
sentiment_counts = new_df["sentiment_label"].value_counts()
plt.pie(sentiment_counts, labels=sentiment_counts.index, autopct="%1.1f%%")
plt.title("Sentiment Distribution")
plt.show()
# ---
# ### Hugging Face Transformers
# **Fine-tuning a BERT model for sentiment analysis**
# columns = ['sentiment_score', 'sentiment_label']
# new_df = merged_df.drop(columns= columns, axis=1)
# import torch
# from transformers import BertTokenizer, BertForSequenceClassification
# from torch.utils.data import DataLoader, RandomSampler
# from transformers import AdamW
# Load the tokenizer and BERT model
# tokenizer = BertTokenizer.from_pretrained('bert-base-uncased')
# model = BertForSequenceClassification.from_pretrained('bert-base-uncased', num_labels=2)
# Preprocess the dataset
# Assuming you have already loaded your dataset and split it into train and test sets
# train_texts = train_dataset['reviewText'].tolist()
# train_labels = train_dataset['overall'].tolist()
# test_texts = test_dataset['reviewText'].tolist()
# test_labels = test_dataset['overall'].tolist()
# Tokenize the texts
# train_encodings = tokenizer(train_texts, truncation=True, padding=True)
# test_encodings = tokenizer(test_texts, truncation=True, padding=True)
# ---
# # Feature Extraction
# **CountVectorizer**
# To analyze the text data and build a vocabulary of unique words. The resulting vocabulary can be used to convert text documents into a numerical representation suitable for machine learning algorithms.
from sklearn.feature_extraction.text import CountVectorizer
# features = CountVectorizer()
# features.fit(merged_df["reviewText"])
# print(len(features.vocabulary_))
# print(features.vocabulary_)
# bagofWords = features.transform(merged_df["reviewText"])
# print(bagofWords)
| false | 0 | 9,492 | 4 | 9,492 | 9,492 |
||
129457340
|
<jupyter_start><jupyter_text>IMDB Top 250 Movies
This dataset is having the data of the top 250 Movies as per their IMDB rating listed on the official website of IMDB
**Features**
- rank - Movie Rank as per IMDB rating
- movie_id - Movie ID
- title - Name of the Movie
- year - Year of Movie release
- link - URL for the Movie
- imdb_votes - Number of people who voted for the IMDB rating
- imdb_rating - Rating of the Movie
- certificate - Movie Certification
- duration - Duration of the Movie
- genre - Genre of the Movie
- cast_id - ID of the cast member who have worked on the Movie
- cast_name - Name of the cast member who have worked on the Movie
- director_id - ID of the director who have directed the Movie
- director_name - Name of the director who have directed the Movie
- writer_id - ID of the writer who have wrote script for the Movie
- writer_name - Name of the writer who have wrote script for the Movie
- storyline - Storyline of the Movie
- user_id - ID of the user who wrote review for the Movie
- user_name - Name of the user who wrote review for the Movie
- review_id - ID of the user review
- review_title - Short review
- review_content - Long review
**Inspiration**
IMDB is one of the main sources which people use to judge the movie or show. IMDB rating plays an important role for a lot of people watching a movie or show. I watched The Shawshank Redemption after finding out that it's at the top of the list on IMDB. I've created this dataset so that people can play with this dataset and do a lot of things as mentioned below
- Dataset Walkthrough
- Understanding Dataset Hierarchy
- Data Preprocessing
- Exploratory Data Analysis
- Data Visualization
- Making Recommendation System
This is a list of some of that things that you can do on this dataset. It's not definitely limited to the one that is mentioned there but a lot more other things can also be done.
Kaggle dataset identifier: imdb-top-250-movies
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
import seaborn as sns
print("Setup Complete")
movies_data = pd.read_csv("/kaggle/input/imdb-top-250-movies/movies.csv")
# # Initial Exploration
# get column names
list(movies_data.columns)
# get number of columns
movies_data.shape[1]
# summarize data
movies_data.describe()
movies_data.head()
# # Cleaning Data
# Remove columns with unnecesarry data like links, names, excessively long ids, and complete movie reviews
movies_data.drop(
[
"link",
"writer_id",
"user_id",
"director_id",
"user_name",
"cast_id",
"cast_name",
"review_id",
"review_title",
"review_content",
"storyline",
],
axis=1,
)
# # Summarizing Data
# Return the mean of ratings by the year a movie was released
tbl = (
movies_data.groupby("year")
.agg(c3_mean=("imbd_rating", "mean"), count=("year", "count"))
.reset_index()
)
print(tbl)
# # Visualization
# draw a plot with number of "imbd_votes" as the x axis and "imbd_rating" as the y axis,
# add a smoothed line (lm = linear model)
# (to show a correlation between popularity and rating)
sns.regplot(x=movies_data["imbd_votes"], y=movies_data["imbd_rating"])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/457/129457340.ipynb
|
imdb-top-250-movies
|
karkavelrajaj
|
[{"Id": 129457340, "ScriptId": 38407730, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13439255, "CreationDate": "05/14/2023 02:11:24", "VersionNumber": 1.0, "Title": "P: IMDB Top 25 Movies", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 59.0, "LinesInsertedFromPrevious": 59.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185528833, "KernelVersionId": 129457340, "SourceDatasetVersionId": 5141446}]
|
[{"Id": 5141446, "DatasetId": 2987095, "DatasourceVersionId": 5213084, "CreatorUserId": 9355447, "LicenseName": "CC BY-NC-SA 4.0", "CreationDate": "03/10/2023 15:02:53", "VersionNumber": 1.0, "Title": "IMDB Top 250 Movies", "Slug": "imdb-top-250-movies", "Subtitle": "This dataset is having the data of top 250 Movies as per their IMDB rating", "Description": "This dataset is having the data of the top 250 Movies as per their IMDB rating listed on the official website of IMDB\n\n**Features**\n\n- rank - Movie Rank as per IMDB rating\n- movie_id - Movie ID\n- title - Name of the Movie \n- year - Year of Movie release\n- link - URL for the Movie \n- imdb_votes - Number of people who voted for the IMDB rating\n- imdb_rating - Rating of the Movie \n- certificate - Movie Certification\n- duration - Duration of the Movie \n- genre - Genre of the Movie \n- cast_id - ID of the cast member who have worked on the Movie \n- cast_name - Name of the cast member who have worked on the Movie \n- director_id - ID of the director who have directed the Movie \n- director_name - Name of the director who have directed the Movie \n- writer_id - ID of the writer who have wrote script for the Movie \n- writer_name - Name of the writer who have wrote script for the Movie \n- storyline - Storyline of the Movie \n- user_id - ID of the user who wrote review for the Movie \n- user_name - Name of the user who wrote review for the Movie \n- review_id - ID of the user review\n- review_title - Short review\n- review_content - Long review\n\n**Inspiration**\n\nIMDB is one of the main sources which people use to judge the movie or show. IMDB rating plays an important role for a lot of people watching a movie or show. I watched The Shawshank Redemption after finding out that it's at the top of the list on IMDB. I've created this dataset so that people can play with this dataset and do a lot of things as mentioned below\n\n- Dataset Walkthrough\n- Understanding Dataset Hierarchy\n- Data Preprocessing\n- Exploratory Data Analysis\n- Data Visualization\n- Making Recommendation System\nThis is a list of some of that things that you can do on this dataset. It's not definitely limited to the one that is mentioned there but a lot more other things can also be done.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2987095, "CreatorUserId": 9355447, "OwnerUserId": 9355447.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5141446.0, "CurrentDatasourceVersionId": 5213084.0, "ForumId": 3025757, "Type": 2, "CreationDate": "03/10/2023 15:02:53", "LastActivityDate": "03/10/2023", "TotalViews": 11814, "TotalDownloads": 2265, "TotalVotes": 36, "TotalKernels": 5}]
|
[{"Id": 9355447, "UserName": "karkavelrajaj", "DisplayName": "KARKAVELRAJA J", "RegisterDate": "01/09/2022", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
import seaborn as sns
print("Setup Complete")
movies_data = pd.read_csv("/kaggle/input/imdb-top-250-movies/movies.csv")
# # Initial Exploration
# get column names
list(movies_data.columns)
# get number of columns
movies_data.shape[1]
# summarize data
movies_data.describe()
movies_data.head()
# # Cleaning Data
# Remove columns with unnecesarry data like links, names, excessively long ids, and complete movie reviews
movies_data.drop(
[
"link",
"writer_id",
"user_id",
"director_id",
"user_name",
"cast_id",
"cast_name",
"review_id",
"review_title",
"review_content",
"storyline",
],
axis=1,
)
# # Summarizing Data
# Return the mean of ratings by the year a movie was released
tbl = (
movies_data.groupby("year")
.agg(c3_mean=("imbd_rating", "mean"), count=("year", "count"))
.reset_index()
)
print(tbl)
# # Visualization
# draw a plot with number of "imbd_votes" as the x axis and "imbd_rating" as the y axis,
# add a smoothed line (lm = linear model)
# (to show a correlation between popularity and rating)
sns.regplot(x=movies_data["imbd_votes"], y=movies_data["imbd_rating"])
| false | 1 | 578 | 0 | 1,127 | 578 |
||
129999800
|
FILE_DIR = "/kaggle/input/titanic" # gender_submission.csv , test.csv , train.csv
import tensorflow as tf
import pandas as pd
import matplotlib.pyplot as plt
import os
print(tf.__version__)
df_train = pd.read_csv(os.path.join(FILE_DIR, "train.csv"))
df_test = pd.read_csv(os.path.join(FILE_DIR, "test.csv"))
df_train["Sex"] = pd.Categorical(df_train["Sex"])
df_train["Sex"] = df_train.Sex.cat.codes
df_test["Sex"] = pd.Categorical(df_test["Sex"])
df_test["Sex"] = df_test.Sex.cat.codes
features_selected = ["Sex", "SibSp", "Parch", "Fare", "Age"]
df_train.Age.fillna(df_train.Age.mean(), inplace=True)
df_test.Age.fillna(df_train.Age.mean(), inplace=True)
def pie(data, labels):
fig, ax = plt.subplots()
ax.pie(data, labels=labels)
plt.show()
def df_to_dataset(data, with_labels=True, shuffle=True, batch_size=32):
# Create a tf.data.Dataset from the dataframe and labels.
if with_labels:
ds = tf.data.Dataset.from_tensor_slices((dict(data[0]), data[1]))
if shuffle:
# Shuffle dataset.
ds = ds.shuffle(len(data))
# Batch dataset with specified batch_size parameter.
ds = ds.batch(batch_size)
return ds
ds = tf.data.Dataset.from_tensor_slices((dict(data)))
return ds.batch(batch_size)
# from sklearn.model_selection import train_test_split
train = df_train[features_selected]
y_train = df_train.Survived
test = df_test[features_selected]
# split = int(0.5 * len(train))
# val = train[split:]
# y_val = y_train[split:]
# train = train[:split]
# y_train = y_train[:split]
train_ds = df_to_dataset((train, y_train), batch_size=16)
test_ds = df_to_dataset(test, with_labels=False, batch_size=16)
# val_ds = df_to_dataset(val, y_val,shuffle=False, batch_size = 16)
Age = tf.feature_column.numeric_column("Age")
Fare = tf.feature_column.numeric_column("Fare")
boundaries_Age = [5, 12, 18, 30, 40, 60, 80]
boundaries_Fare = [10, 20, 30, 40, 60, 80, 100]
BucketAge = tf.feature_column.bucketized_column(Age, boundaries_Age)
BucketFare = tf.feature_column.bucketized_column(Fare, boundaries_Fare)
FeatureColumn = []
for feature in features_selected:
numeric_feature_column = tf.feature_column.numeric_column(feature)
FeatureColumn.append(numeric_feature_column)
FeatureColumn.append(BucketAge)
FeatureColumn.append(BucketFare)
model = tf.keras.Sequential(
[
tf.keras.layers.DenseFeatures(FeatureColumn),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
history = model.fit(
train_ds,
# validation_data=val_ds,
epochs=500,
verbose=0,
)
acc = history.history["accuracy"]
# val_acc = history.history['val_accuracy']
loss = history.history["loss"]
# val_loss = history.history['val_loss']
print(f"accuracy = {acc[-1]}")
fix, ax = plt.subplots(2)
ax[0].plot(acc)
# ax[0].plot(val_acc)
ax[1].plot(loss)
# ax[1].plot(val_loss)
plt.show()
import numpy as np
pred_raw = model.predict(test_ds)
pred = np.array(pred_raw > 0.5, dtype=int).flatten()
print(len(pred))
submit = pd.read_csv(os.path.join(FILE_DIR, "gender_submission.csv"))
my_submission = submit.copy()
pie(submit.Survived.value_counts(), ["S", "M"])
my_submission.Survived = pred
pie(my_submission.Survived.value_counts(), ["S", "M"])
my_submission.to_csv("my_submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/999/129999800.ipynb
| null | null |
[{"Id": 129999800, "ScriptId": 37347690, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9755272, "CreationDate": "05/18/2023 03:33:48", "VersionNumber": 1.0, "Title": "Titanic", "EvaluationDate": "05/18/2023", "IsChange": true, "TotalLines": 121.0, "LinesInsertedFromPrevious": 121.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 6}]
| null | null | null | null |
FILE_DIR = "/kaggle/input/titanic" # gender_submission.csv , test.csv , train.csv
import tensorflow as tf
import pandas as pd
import matplotlib.pyplot as plt
import os
print(tf.__version__)
df_train = pd.read_csv(os.path.join(FILE_DIR, "train.csv"))
df_test = pd.read_csv(os.path.join(FILE_DIR, "test.csv"))
df_train["Sex"] = pd.Categorical(df_train["Sex"])
df_train["Sex"] = df_train.Sex.cat.codes
df_test["Sex"] = pd.Categorical(df_test["Sex"])
df_test["Sex"] = df_test.Sex.cat.codes
features_selected = ["Sex", "SibSp", "Parch", "Fare", "Age"]
df_train.Age.fillna(df_train.Age.mean(), inplace=True)
df_test.Age.fillna(df_train.Age.mean(), inplace=True)
def pie(data, labels):
fig, ax = plt.subplots()
ax.pie(data, labels=labels)
plt.show()
def df_to_dataset(data, with_labels=True, shuffle=True, batch_size=32):
# Create a tf.data.Dataset from the dataframe and labels.
if with_labels:
ds = tf.data.Dataset.from_tensor_slices((dict(data[0]), data[1]))
if shuffle:
# Shuffle dataset.
ds = ds.shuffle(len(data))
# Batch dataset with specified batch_size parameter.
ds = ds.batch(batch_size)
return ds
ds = tf.data.Dataset.from_tensor_slices((dict(data)))
return ds.batch(batch_size)
# from sklearn.model_selection import train_test_split
train = df_train[features_selected]
y_train = df_train.Survived
test = df_test[features_selected]
# split = int(0.5 * len(train))
# val = train[split:]
# y_val = y_train[split:]
# train = train[:split]
# y_train = y_train[:split]
train_ds = df_to_dataset((train, y_train), batch_size=16)
test_ds = df_to_dataset(test, with_labels=False, batch_size=16)
# val_ds = df_to_dataset(val, y_val,shuffle=False, batch_size = 16)
Age = tf.feature_column.numeric_column("Age")
Fare = tf.feature_column.numeric_column("Fare")
boundaries_Age = [5, 12, 18, 30, 40, 60, 80]
boundaries_Fare = [10, 20, 30, 40, 60, 80, 100]
BucketAge = tf.feature_column.bucketized_column(Age, boundaries_Age)
BucketFare = tf.feature_column.bucketized_column(Fare, boundaries_Fare)
FeatureColumn = []
for feature in features_selected:
numeric_feature_column = tf.feature_column.numeric_column(feature)
FeatureColumn.append(numeric_feature_column)
FeatureColumn.append(BucketAge)
FeatureColumn.append(BucketFare)
model = tf.keras.Sequential(
[
tf.keras.layers.DenseFeatures(FeatureColumn),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
history = model.fit(
train_ds,
# validation_data=val_ds,
epochs=500,
verbose=0,
)
acc = history.history["accuracy"]
# val_acc = history.history['val_accuracy']
loss = history.history["loss"]
# val_loss = history.history['val_loss']
print(f"accuracy = {acc[-1]}")
fix, ax = plt.subplots(2)
ax[0].plot(acc)
# ax[0].plot(val_acc)
ax[1].plot(loss)
# ax[1].plot(val_loss)
plt.show()
import numpy as np
pred_raw = model.predict(test_ds)
pred = np.array(pred_raw > 0.5, dtype=int).flatten()
print(len(pred))
submit = pd.read_csv(os.path.join(FILE_DIR, "gender_submission.csv"))
my_submission = submit.copy()
pie(submit.Survived.value_counts(), ["S", "M"])
my_submission.Survived = pred
pie(my_submission.Survived.value_counts(), ["S", "M"])
my_submission.to_csv("my_submission.csv", index=False)
| false | 0 | 1,243 | 6 | 1,243 | 1,243 |
||
129999789
|
<jupyter_start><jupyter_text>Starbucks Nutrition Facts
```
Nutrition facts for several Starbucks food items
```
| Column | Description |
| ------- | ------------------------------------------------------------ |
| item | The name of the food item. |
| calories| The amount of calories in the food item. |
| fat | The quantity of fat in grams present in the food item. |
| carb | The amount of carbohydrates in grams found in the food item. |
| fiber | The quantity of dietary fiber in grams in the food item. |
| protein | The amount of protein in grams contained in the food item. |
| type | The category or type of food item (bakery, bistro box, hot breakfast, parfait, petite, salad, or sandwich). |
Kaggle dataset identifier: starbucks-nutrition
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from pprint import pprint
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Starbucks Nutritional Information
# Starbucks provides comprehensive nutritional information for their food and beverage offerings, allowing customers to make informed choices based on their dietary preferences and health goals. The nutritional information includes details such as calories, fat content, carbohydrates, fiber, and protein for each menu item.
# By making this information readily available, Starbucks aims to empower individuals to make choices that align with their nutritional needs and preferences. Whether you're looking for lower-calorie options, watching your fat or carb intake, or seeking protein-rich alternatives, the nutritional information provided by Starbucks helps you navigate their menu with confidence.
# ## Data Coverage
# The data encompasses a range of food items, from baked goods and bistro boxes to hot breakfast items, parfaits, petite treats, salads, and sandwiches. Each item is categorized based on its type, making it easier for customers to find options that suit their dietary requirements or preferences.
# ## Transparency and Informed Decisions
# By offering transparent and detailed nutritional information, Starbucks reinforces its commitment to supporting customers in making informed decisions about their food choices. Whether you're enjoying a coffee break or grabbing a quick bite, the nutritional information empowers you to enjoy Starbucks' offerings while being mindful of your nutritional goals.
# ---
# ## Data Dictionary
# The data consists of nutrition facts for several Starbucks food items. It is organized in the form of a data frame with 77 observations and 7 variables.
# ### Variables
# - **item**: The name of the food item (string).
# - **calories**: The number of calories in the food item (integer).
# - **fat**: The amount of fat in grams (numeric).
# - **carb**: The amount of carbohydrates in grams (numeric).
# - **fiber**: The amount of dietary fiber in grams (numeric).
# - **protein**: The amount of protein in grams (numeric).
# - **type**: The categorization of the food item, with levels bakery, bistro box, hot breakfast, parfait, petite, salad, and sandwich (factor).
# ### Additional Information
# - The data frame has a RangeIndex from 0 to 76.
# - There are no missing values (non-null count is 77 for all columns).
# - The original data frame had an additional column named "Unnamed: 0", which has been removed for this improved data dictionary.
# ---
# .
df = pd.read_csv("/kaggle/input/starbucks-nutrition/starbucks.csv", index_col=0)
df
df.describe
df.info()
# ---
# ## Questions to ask
# 1. What are the highest and lowest calorie food items offered by Starbucks?
# 2. Which food items have the highest amount of fat, carbohydrates, fiber, and protein?
# 3. Are there any food items that are particularly rich in fiber but low in fat and carbohydrates?
# 4. What is the average calorie content of each food item type (bakery, bistro box, hot breakfast, etc.)?
# 5. Is there a correlation between the calorie content and the amount of fat, carbohydrates, fiber, or protein in the food items?
# 6. Which food item types have the highest average fat, carbohydrate, fiber, and protein content?
# 7. Can we identify any trends or patterns in the nutritional composition of Starbucks food items?
# 8. Are there any notable differences in the nutritional profile of food items across different categories (bakery, bistro box, etc.)?
# 9. Are there any food items that provide a good balance of macronutrients (fat, carbohydrates, and protein)?
# 10. Can we identify any outliers or unusual values in the nutritional information?
# ---
# ## 1. What are the highest and lowest calorie food items offered by Starbucks?
# Find the highest calorie food item
highest_calorie_item = df.loc[df["calories"].idxmax(), "item"]
highest_calorie_value = df["calories"].max()
# Find the lowest calorie food item
lowest_calorie_item = df.loc[df["calories"].idxmin(), "item"]
lowest_calorie_value = df["calories"].min()
# Print the results
print(
f"Highest Calorie Food Item: {highest_calorie_item} ({highest_calorie_value} calories)"
)
print(
f"Lowest Calorie Food Item: {lowest_calorie_item} ({lowest_calorie_value} calories)"
)
# ## 2. Which food items have the highest amount of fat, carbohydrates, fiber, and protein?
# Find the food item with the highest amount of fat
highest_fat_item = df.loc[df["fat"].idxmax(), "item"]
highest_fat_value = df["fat"].max()
# Find the food item with the highest amount of carbohydrates
highest_carb_item = df.loc[df["carb"].idxmax(), "item"]
highest_carb_value = df["carb"].max()
# Find the food item with the highest amount of fiber
highest_fiber_item = df.loc[df["fiber"].idxmax(), "item"]
highest_fiber_value = df["fiber"].max()
# Find the food item with the highest amount of protein
highest_protein_item = df.loc[df["protein"].idxmax(), "item"]
highest_protein_value = df["protein"].max()
# Print the results
print(f"Food Item with Highest Fat: {highest_fat_item} ({highest_fat_value} grams)")
print(
f"Food Item with Highest Carbohydrates: {highest_carb_item} ({highest_carb_value} grams)"
)
print(
f"Food Item with Highest Fiber: {highest_fiber_item} ({highest_fiber_value} grams)"
)
print(
f"Food Item with Highest Protein: {highest_protein_item} ({highest_protein_value} grams)"
)
# ## 3. Are there any food items that are particularly rich in fiber but low in fat and carbohydrates?
# Filter food items with high fiber, low fat, and low carbohydrates
filtered_items = df[(df["fiber"] > 0) & (df["fat"] < 5) & (df["carb"] < 30)]
# Print the filtered food items using f-strings
if filtered_items.empty:
print(
"No food items are particularly rich in fiber but low in fat and carbohydrates."
)
else:
print(
"Food items that are particularly rich in fiber but low in fat and carbohydrates:"
)
for item in filtered_items["item"]:
print(f"- {item}")
# ## 4. What is the average calorie content of each food item type (bakery, bistro box, hot breakfast, etc.)?
# Calculate the average calorie content of each food item type
average_calories = df.groupby("type")["calories"].mean()
# Print the average calorie content using f-strings
print("Average Calorie Content by Food Item Type:")
for food_type, avg_calories in average_calories.items():
print(f"- {food_type}: {avg_calories:.2f} calories")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/999/129999789.ipynb
|
starbucks-nutrition
|
utkarshx27
|
[{"Id": 129999789, "ScriptId": 38415291, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14543633, "CreationDate": "05/18/2023 03:33:34", "VersionNumber": 2.0, "Title": "Starbucks EDA", "EvaluationDate": "05/18/2023", "IsChange": true, "TotalLines": 152.0, "LinesInsertedFromPrevious": 67.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 85.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186452349, "KernelVersionId": 129999789, "SourceDatasetVersionId": 5651811}]
|
[{"Id": 5651811, "DatasetId": 3248696, "DatasourceVersionId": 5727183, "CreatorUserId": 13364933, "LicenseName": "CC0: Public Domain", "CreationDate": "05/10/2023 05:42:59", "VersionNumber": 1.0, "Title": "Starbucks Nutrition Facts", "Slug": "starbucks-nutrition", "Subtitle": "Nutrition facts for several Starbucks food items", "Description": "```\nNutrition facts for several Starbucks food items\n```\n| Column | Description |\n| ------- | ------------------------------------------------------------ |\n| item | The name of the food item. |\n| calories| The amount of calories in the food item. |\n| fat | The quantity of fat in grams present in the food item. |\n| carb | The amount of carbohydrates in grams found in the food item. |\n| fiber | The quantity of dietary fiber in grams in the food item. |\n| protein | The amount of protein in grams contained in the food item. |\n| type | The category or type of food item (bakery, bistro box, hot breakfast, parfait, petite, salad, or sandwich). |", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3248696, "CreatorUserId": 13364933, "OwnerUserId": 13364933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5651811.0, "CurrentDatasourceVersionId": 5727183.0, "ForumId": 3314049, "Type": 2, "CreationDate": "05/10/2023 05:42:59", "LastActivityDate": "05/10/2023", "TotalViews": 12557, "TotalDownloads": 2321, "TotalVotes": 59, "TotalKernels": 17}]
|
[{"Id": 13364933, "UserName": "utkarshx27", "DisplayName": "Utkarsh Singh", "RegisterDate": "01/21/2023", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from pprint import pprint
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Starbucks Nutritional Information
# Starbucks provides comprehensive nutritional information for their food and beverage offerings, allowing customers to make informed choices based on their dietary preferences and health goals. The nutritional information includes details such as calories, fat content, carbohydrates, fiber, and protein for each menu item.
# By making this information readily available, Starbucks aims to empower individuals to make choices that align with their nutritional needs and preferences. Whether you're looking for lower-calorie options, watching your fat or carb intake, or seeking protein-rich alternatives, the nutritional information provided by Starbucks helps you navigate their menu with confidence.
# ## Data Coverage
# The data encompasses a range of food items, from baked goods and bistro boxes to hot breakfast items, parfaits, petite treats, salads, and sandwiches. Each item is categorized based on its type, making it easier for customers to find options that suit their dietary requirements or preferences.
# ## Transparency and Informed Decisions
# By offering transparent and detailed nutritional information, Starbucks reinforces its commitment to supporting customers in making informed decisions about their food choices. Whether you're enjoying a coffee break or grabbing a quick bite, the nutritional information empowers you to enjoy Starbucks' offerings while being mindful of your nutritional goals.
# ---
# ## Data Dictionary
# The data consists of nutrition facts for several Starbucks food items. It is organized in the form of a data frame with 77 observations and 7 variables.
# ### Variables
# - **item**: The name of the food item (string).
# - **calories**: The number of calories in the food item (integer).
# - **fat**: The amount of fat in grams (numeric).
# - **carb**: The amount of carbohydrates in grams (numeric).
# - **fiber**: The amount of dietary fiber in grams (numeric).
# - **protein**: The amount of protein in grams (numeric).
# - **type**: The categorization of the food item, with levels bakery, bistro box, hot breakfast, parfait, petite, salad, and sandwich (factor).
# ### Additional Information
# - The data frame has a RangeIndex from 0 to 76.
# - There are no missing values (non-null count is 77 for all columns).
# - The original data frame had an additional column named "Unnamed: 0", which has been removed for this improved data dictionary.
# ---
# .
df = pd.read_csv("/kaggle/input/starbucks-nutrition/starbucks.csv", index_col=0)
df
df.describe
df.info()
# ---
# ## Questions to ask
# 1. What are the highest and lowest calorie food items offered by Starbucks?
# 2. Which food items have the highest amount of fat, carbohydrates, fiber, and protein?
# 3. Are there any food items that are particularly rich in fiber but low in fat and carbohydrates?
# 4. What is the average calorie content of each food item type (bakery, bistro box, hot breakfast, etc.)?
# 5. Is there a correlation between the calorie content and the amount of fat, carbohydrates, fiber, or protein in the food items?
# 6. Which food item types have the highest average fat, carbohydrate, fiber, and protein content?
# 7. Can we identify any trends or patterns in the nutritional composition of Starbucks food items?
# 8. Are there any notable differences in the nutritional profile of food items across different categories (bakery, bistro box, etc.)?
# 9. Are there any food items that provide a good balance of macronutrients (fat, carbohydrates, and protein)?
# 10. Can we identify any outliers or unusual values in the nutritional information?
# ---
# ## 1. What are the highest and lowest calorie food items offered by Starbucks?
# Find the highest calorie food item
highest_calorie_item = df.loc[df["calories"].idxmax(), "item"]
highest_calorie_value = df["calories"].max()
# Find the lowest calorie food item
lowest_calorie_item = df.loc[df["calories"].idxmin(), "item"]
lowest_calorie_value = df["calories"].min()
# Print the results
print(
f"Highest Calorie Food Item: {highest_calorie_item} ({highest_calorie_value} calories)"
)
print(
f"Lowest Calorie Food Item: {lowest_calorie_item} ({lowest_calorie_value} calories)"
)
# ## 2. Which food items have the highest amount of fat, carbohydrates, fiber, and protein?
# Find the food item with the highest amount of fat
highest_fat_item = df.loc[df["fat"].idxmax(), "item"]
highest_fat_value = df["fat"].max()
# Find the food item with the highest amount of carbohydrates
highest_carb_item = df.loc[df["carb"].idxmax(), "item"]
highest_carb_value = df["carb"].max()
# Find the food item with the highest amount of fiber
highest_fiber_item = df.loc[df["fiber"].idxmax(), "item"]
highest_fiber_value = df["fiber"].max()
# Find the food item with the highest amount of protein
highest_protein_item = df.loc[df["protein"].idxmax(), "item"]
highest_protein_value = df["protein"].max()
# Print the results
print(f"Food Item with Highest Fat: {highest_fat_item} ({highest_fat_value} grams)")
print(
f"Food Item with Highest Carbohydrates: {highest_carb_item} ({highest_carb_value} grams)"
)
print(
f"Food Item with Highest Fiber: {highest_fiber_item} ({highest_fiber_value} grams)"
)
print(
f"Food Item with Highest Protein: {highest_protein_item} ({highest_protein_value} grams)"
)
# ## 3. Are there any food items that are particularly rich in fiber but low in fat and carbohydrates?
# Filter food items with high fiber, low fat, and low carbohydrates
filtered_items = df[(df["fiber"] > 0) & (df["fat"] < 5) & (df["carb"] < 30)]
# Print the filtered food items using f-strings
if filtered_items.empty:
print(
"No food items are particularly rich in fiber but low in fat and carbohydrates."
)
else:
print(
"Food items that are particularly rich in fiber but low in fat and carbohydrates:"
)
for item in filtered_items["item"]:
print(f"- {item}")
# ## 4. What is the average calorie content of each food item type (bakery, bistro box, hot breakfast, etc.)?
# Calculate the average calorie content of each food item type
average_calories = df.groupby("type")["calories"].mean()
# Print the average calorie content using f-strings
print("Average Calorie Content by Food Item Type:")
for food_type, avg_calories in average_calories.items():
print(f"- {food_type}: {avg_calories:.2f} calories")
| false | 1 | 2,003 | 0 | 2,219 | 2,003 |
||
129999011
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/playground-series-s3e15/data.csv")
df.head()
df.describe()
df.isna().sum()
df.shape
df.hist(figsize=(30, 20))
df.columns
x_eNull = df[df["x_e_out [-]"].isna()]
x_eNull.head()
x_eNull.isna().sum()
x_eNull.hist(figsize=(30, 20))
# Impute the null values in the other columns
import miceforest as mf
df.dtypes
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(df[["geometry"]])
df[ohe.categories_[0]] = transformed.toarray()
df.head()
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(df[["author"]])
df[ohe.categories_[0]] = transformed.toarray()
df.head()
df.drop(["author", "geometry", np.nan], axis=1, inplace=True)
df = df.rename(
columns={
"pressure [MPa]": "pressure",
"mass_flux [kg/m2-s]": "mass_flux",
"x_e_out [-]": "x_e_out",
"D_e [mm]": "D_e",
"D_h [mm]": "D_h",
"length [mm]": "length",
"chf_exp [MW/m2]": "chf_exp",
}
)
# Create kernel.
kds = mf.ImputationKernel(df, save_all_iterations=True, random_state=1991)
# Run the MICE algorithm for 3 iterations
kds.mice(3)
# Return the completed kernel data
completed_data = kds.complete_data()
completed_data.isna().sum()
x_eNull.drop(["x_e_out [-]"], axis=1, inplace=True)
completed_data.head()
submission = pd.merge(x_eNull, completed_data[["id", "x_e_out"]], on="id")
submission.head()
submission = submission[["id", "x_e_out"]]
submission.to_csv("submission.csv", index=False)
x_eNotNull = df[df["x_e_out [-]"].isna() == False]
import seaborn as sns
df.columns
# sns.heatmap(df[['id','pressure', 'mass_flux', 'x_e_out', 'D_e',
#'D_h', 'length', 'chf_exp']])
sns.heatmap(
df[["id", "pressure", "mass_flux", "x_e_out", "D_e", "D_h", "length", "chf_exp"]],
linewidths=0.30,
annot=True,
)
x_eNotNull.head()
x_eNotNull.hist(figsize=(30, 20))
# The features in Not Null dataframe and the Null dataframe seems to be very similar
# We can take the Not Null to train and use the Null as the test dataset
df.dtypes
# Lets try using Simple Regression to handle both Categorical and float
df["author"].nunique()
df["geometry"].nunique()
df = df.rename(
columns={
"pressure [MPa]": "pressure",
"mass_flux [kg/m2-s]": "mass_flux",
"x_e_out [-]": "x_e_out",
"D_e [mm]": "D_e",
"D_h [mm]": "D_h",
"length [mm]": "length",
"chf_exp [MW/m2]": "chf_exp",
}
)
dfDropped = df.drop(["author", "geometry", "D_e", "D_h", "length"], axis=1)
dfDropped.head()
X = dfDropped[dfDropped["x_e_out"].isna() == False]
test = dfDropped[dfDropped["x_e_out"].isna() == True]
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(x_eNotNull[["geometry"]])
x_eNotNull[ohe.categories_[0]] = transformed.toarray()
x_eNotNull.head()
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(x_eNull[["geometry"]])
x_eNull[ohe.categories_[0]] = transformed.toarray()
x_eNull.head()
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(x_eNull[["author"]])
x_eNull[ohe.categories_[0]] = transformed.toarray()
x_eNull.head()
ohe = OneHotEncoder()
transformed = ohe.fit_transform(x_eNotNull[["author"]])
x_eNotNull[ohe.categories_[0]] = transformed.toarray()
x_eNotNull.head()
x_eNotNull.drop(["author", "geometry"], axis=1, inplace=True)
x_eNull.drop(["author", "geometry"], axis=1, inplace=True)
x_eNotNull.drop([np.NaN], axis=1, inplace=True)
x_eNull.drop([np.NaN], axis=1, inplace=True)
X = x_eNotNull.drop(["x_e_out", "id"], axis=1)
y = x_eNotNull.loc[:, x_eNotNull.columns == "x_e_out"]
y = X.loc[:, X.columns == "x_e_out"]
X = X.drop(["x_e_out", "id"], axis=1)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42
)
x_eNull.drop(["x_e_out [-]"], axis=1, inplace=True)
test = x_eNull.drop(["id", "x_e_out [-]"], axis=1)
test = test.drop(["id", "x_e_out"], axis=1)
test.head()
import optuna
from lightgbm import LGBMRegressor
from sklearn.metrics import mean_squared_error
def objective(trial):
"""
Objective function to be minimized.
"""
param = {
"objective": "regression",
"metric": "rmse",
"verbosity": -1,
"boosting_type": "gbdt",
"lambda_l1": trial.suggest_float("lambda_l1", 1e-8, 10.0, log=True),
"lambda_l2": trial.suggest_float("lambda_l2", 1e-8, 10.0, log=True),
"num_leaves": trial.suggest_int("num_leaves", 2, 256),
"feature_fraction": trial.suggest_float("feature_fraction", 0.4, 1.0),
"bagging_fraction": trial.suggest_float("bagging_fraction", 0.4, 1.0),
"bagging_freq": trial.suggest_int("bagging_freq", 1, 7),
"min_child_samples": trial.suggest_int("min_child_samples", 5, 100),
}
gbm = LGBMRegressor(**param)
gbm.fit(X_train, y_train)
preds = gbm.predict(X_test)
rmse = mean_squared_error(y_test, preds, squared=False)
return rmse
study = optuna.create_study()
study.optimize(objective, n_trials=50)
print("Number of finished trials:", len(study.trials))
print("Best trial:", study.best_trial.params)
import lightgbm as lgb
from sklearn.model_selection import RepeatedStratifiedKFold, StratifiedKFold, KFold
# X = X_train.copy()
# y = X.pop('x_e_out')
seed = 42
splits = 10
# Initialize KFold cross-validation
kf = KFold(n_splits=splits, shuffle=True, random_state=42)
val_preds = np.zeros(len(X))
val_scores = []
prediction = np.zeros((len(test)))
params = {
"objective": "regression",
"metric": "rmse",
"lambda_l1": 8.086864003164224e-05,
"lambda_l2": 0.7435736341285621,
"num_leaves": 164,
"feature_fraction": 0.9707722414736359,
"bagging_fraction": 0.4910694147596062,
"bagging_freq": 1,
"min_child_samples": 83,
}
from sklearn.preprocessing import RobustScaler
# Loop through each fold
for fold, (train_idx, val_idx) in enumerate(kf.split(X, y)):
# Split data into training and validation sets
X_train, y_train = X.iloc[train_idx], y.iloc[train_idx]
X_val, y_val = X.iloc[val_idx], y.iloc[val_idx]
train_data = lgb.Dataset(X_train, label=y_train)
val_data = lgb.Dataset(X_val, label=y_val)
# Train model with early stopping
model = lgb.train(
params,
train_data,
valid_sets=[train_data, val_data],
early_stopping_rounds=7000,
verbose_eval=False,
)
# Make validation predictions and calculate validation score
val_preds[val_idx] += model.predict(X_val)
val_score = mean_squared_error(y_val, val_preds[val_idx], squared=False)
val_scores.append(val_score)
# Print validation score
print(f"Fold {fold+1}: Validation score: {val_score:.4f}")
prediction += model.predict(test)
avg_val_score = np.mean(val_scores)
print(f"Average validation score: {avg_val_score:.4f}")
prediction /= splits
from lightgbm import plot_importance
plot_importance(model, figsize=(10, 9))
# custom function to run light gbm model
def run_lgb(train_X, train_y, val_X, val_y, test_X):
params = {
"objective": "regression",
"metric": "rmse",
"lambda_l1": 1.2619723670327868e-07,
"lambda_l2": 9.721462975369603,
"num_leaves": 41,
"feature_fraction": 0.7697328380987326,
"bagging_fraction": 0.7132023581183115,
"bagging_freq": 6,
"min_child_samples": 37,
}
lgtrain = lgb.Dataset(train_X, label=train_y)
lgval = lgb.Dataset(val_X, label=val_y)
# cv_lgb = lgb.cv(params, lgtrain, num_boost_round=700, nfold=3,
# verbose_eval=20, early_stopping_rounds=40)
model = lgb.train(
params,
lgtrain,
7000,
valid_sets=[lgval],
early_stopping_rounds=100,
verbose_eval=100,
)
pred_test_y = model.predict(x_eNull, num_iteration=model.best_iteration)
pred_val_y = model.predict(val_X, num_iteration=model.best_iteration)
return pred_test_y, model, pred_val_y
# Training the model #
pred_test, model, pred_val = run_lgb(X_train, y_train, X_test, y_test, x_eNull)
print(prediction)
x_eNull["x_e_out [-]"] = prediction
submit = x_eNull[["id", "x_e_out [-]"]]
submit.to_csv("imputation_submit_5", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/999/129999011.ipynb
| null | null |
[{"Id": 129999011, "ScriptId": 38597963, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2851726, "CreationDate": "05/18/2023 03:22:47", "VersionNumber": 1.0, "Title": "imputation_Heat", "EvaluationDate": "05/18/2023", "IsChange": true, "TotalLines": 300.0, "LinesInsertedFromPrevious": 300.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/playground-series-s3e15/data.csv")
df.head()
df.describe()
df.isna().sum()
df.shape
df.hist(figsize=(30, 20))
df.columns
x_eNull = df[df["x_e_out [-]"].isna()]
x_eNull.head()
x_eNull.isna().sum()
x_eNull.hist(figsize=(30, 20))
# Impute the null values in the other columns
import miceforest as mf
df.dtypes
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(df[["geometry"]])
df[ohe.categories_[0]] = transformed.toarray()
df.head()
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(df[["author"]])
df[ohe.categories_[0]] = transformed.toarray()
df.head()
df.drop(["author", "geometry", np.nan], axis=1, inplace=True)
df = df.rename(
columns={
"pressure [MPa]": "pressure",
"mass_flux [kg/m2-s]": "mass_flux",
"x_e_out [-]": "x_e_out",
"D_e [mm]": "D_e",
"D_h [mm]": "D_h",
"length [mm]": "length",
"chf_exp [MW/m2]": "chf_exp",
}
)
# Create kernel.
kds = mf.ImputationKernel(df, save_all_iterations=True, random_state=1991)
# Run the MICE algorithm for 3 iterations
kds.mice(3)
# Return the completed kernel data
completed_data = kds.complete_data()
completed_data.isna().sum()
x_eNull.drop(["x_e_out [-]"], axis=1, inplace=True)
completed_data.head()
submission = pd.merge(x_eNull, completed_data[["id", "x_e_out"]], on="id")
submission.head()
submission = submission[["id", "x_e_out"]]
submission.to_csv("submission.csv", index=False)
x_eNotNull = df[df["x_e_out [-]"].isna() == False]
import seaborn as sns
df.columns
# sns.heatmap(df[['id','pressure', 'mass_flux', 'x_e_out', 'D_e',
#'D_h', 'length', 'chf_exp']])
sns.heatmap(
df[["id", "pressure", "mass_flux", "x_e_out", "D_e", "D_h", "length", "chf_exp"]],
linewidths=0.30,
annot=True,
)
x_eNotNull.head()
x_eNotNull.hist(figsize=(30, 20))
# The features in Not Null dataframe and the Null dataframe seems to be very similar
# We can take the Not Null to train and use the Null as the test dataset
df.dtypes
# Lets try using Simple Regression to handle both Categorical and float
df["author"].nunique()
df["geometry"].nunique()
df = df.rename(
columns={
"pressure [MPa]": "pressure",
"mass_flux [kg/m2-s]": "mass_flux",
"x_e_out [-]": "x_e_out",
"D_e [mm]": "D_e",
"D_h [mm]": "D_h",
"length [mm]": "length",
"chf_exp [MW/m2]": "chf_exp",
}
)
dfDropped = df.drop(["author", "geometry", "D_e", "D_h", "length"], axis=1)
dfDropped.head()
X = dfDropped[dfDropped["x_e_out"].isna() == False]
test = dfDropped[dfDropped["x_e_out"].isna() == True]
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(x_eNotNull[["geometry"]])
x_eNotNull[ohe.categories_[0]] = transformed.toarray()
x_eNotNull.head()
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(x_eNull[["geometry"]])
x_eNull[ohe.categories_[0]] = transformed.toarray()
x_eNull.head()
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
transformed = ohe.fit_transform(x_eNull[["author"]])
x_eNull[ohe.categories_[0]] = transformed.toarray()
x_eNull.head()
ohe = OneHotEncoder()
transformed = ohe.fit_transform(x_eNotNull[["author"]])
x_eNotNull[ohe.categories_[0]] = transformed.toarray()
x_eNotNull.head()
x_eNotNull.drop(["author", "geometry"], axis=1, inplace=True)
x_eNull.drop(["author", "geometry"], axis=1, inplace=True)
x_eNotNull.drop([np.NaN], axis=1, inplace=True)
x_eNull.drop([np.NaN], axis=1, inplace=True)
X = x_eNotNull.drop(["x_e_out", "id"], axis=1)
y = x_eNotNull.loc[:, x_eNotNull.columns == "x_e_out"]
y = X.loc[:, X.columns == "x_e_out"]
X = X.drop(["x_e_out", "id"], axis=1)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42
)
x_eNull.drop(["x_e_out [-]"], axis=1, inplace=True)
test = x_eNull.drop(["id", "x_e_out [-]"], axis=1)
test = test.drop(["id", "x_e_out"], axis=1)
test.head()
import optuna
from lightgbm import LGBMRegressor
from sklearn.metrics import mean_squared_error
def objective(trial):
"""
Objective function to be minimized.
"""
param = {
"objective": "regression",
"metric": "rmse",
"verbosity": -1,
"boosting_type": "gbdt",
"lambda_l1": trial.suggest_float("lambda_l1", 1e-8, 10.0, log=True),
"lambda_l2": trial.suggest_float("lambda_l2", 1e-8, 10.0, log=True),
"num_leaves": trial.suggest_int("num_leaves", 2, 256),
"feature_fraction": trial.suggest_float("feature_fraction", 0.4, 1.0),
"bagging_fraction": trial.suggest_float("bagging_fraction", 0.4, 1.0),
"bagging_freq": trial.suggest_int("bagging_freq", 1, 7),
"min_child_samples": trial.suggest_int("min_child_samples", 5, 100),
}
gbm = LGBMRegressor(**param)
gbm.fit(X_train, y_train)
preds = gbm.predict(X_test)
rmse = mean_squared_error(y_test, preds, squared=False)
return rmse
study = optuna.create_study()
study.optimize(objective, n_trials=50)
print("Number of finished trials:", len(study.trials))
print("Best trial:", study.best_trial.params)
import lightgbm as lgb
from sklearn.model_selection import RepeatedStratifiedKFold, StratifiedKFold, KFold
# X = X_train.copy()
# y = X.pop('x_e_out')
seed = 42
splits = 10
# Initialize KFold cross-validation
kf = KFold(n_splits=splits, shuffle=True, random_state=42)
val_preds = np.zeros(len(X))
val_scores = []
prediction = np.zeros((len(test)))
params = {
"objective": "regression",
"metric": "rmse",
"lambda_l1": 8.086864003164224e-05,
"lambda_l2": 0.7435736341285621,
"num_leaves": 164,
"feature_fraction": 0.9707722414736359,
"bagging_fraction": 0.4910694147596062,
"bagging_freq": 1,
"min_child_samples": 83,
}
from sklearn.preprocessing import RobustScaler
# Loop through each fold
for fold, (train_idx, val_idx) in enumerate(kf.split(X, y)):
# Split data into training and validation sets
X_train, y_train = X.iloc[train_idx], y.iloc[train_idx]
X_val, y_val = X.iloc[val_idx], y.iloc[val_idx]
train_data = lgb.Dataset(X_train, label=y_train)
val_data = lgb.Dataset(X_val, label=y_val)
# Train model with early stopping
model = lgb.train(
params,
train_data,
valid_sets=[train_data, val_data],
early_stopping_rounds=7000,
verbose_eval=False,
)
# Make validation predictions and calculate validation score
val_preds[val_idx] += model.predict(X_val)
val_score = mean_squared_error(y_val, val_preds[val_idx], squared=False)
val_scores.append(val_score)
# Print validation score
print(f"Fold {fold+1}: Validation score: {val_score:.4f}")
prediction += model.predict(test)
avg_val_score = np.mean(val_scores)
print(f"Average validation score: {avg_val_score:.4f}")
prediction /= splits
from lightgbm import plot_importance
plot_importance(model, figsize=(10, 9))
# custom function to run light gbm model
def run_lgb(train_X, train_y, val_X, val_y, test_X):
params = {
"objective": "regression",
"metric": "rmse",
"lambda_l1": 1.2619723670327868e-07,
"lambda_l2": 9.721462975369603,
"num_leaves": 41,
"feature_fraction": 0.7697328380987326,
"bagging_fraction": 0.7132023581183115,
"bagging_freq": 6,
"min_child_samples": 37,
}
lgtrain = lgb.Dataset(train_X, label=train_y)
lgval = lgb.Dataset(val_X, label=val_y)
# cv_lgb = lgb.cv(params, lgtrain, num_boost_round=700, nfold=3,
# verbose_eval=20, early_stopping_rounds=40)
model = lgb.train(
params,
lgtrain,
7000,
valid_sets=[lgval],
early_stopping_rounds=100,
verbose_eval=100,
)
pred_test_y = model.predict(x_eNull, num_iteration=model.best_iteration)
pred_val_y = model.predict(val_X, num_iteration=model.best_iteration)
return pred_test_y, model, pred_val_y
# Training the model #
pred_test, model, pred_val = run_lgb(X_train, y_train, X_test, y_test, x_eNull)
print(prediction)
x_eNull["x_e_out [-]"] = prediction
submit = x_eNull[["id", "x_e_out [-]"]]
submit.to_csv("imputation_submit_5", index=False)
| false | 0 | 3,298 | 0 | 3,298 | 3,298 |
||
129364413
|
<jupyter_start><jupyter_text>Netflix TV Shows and Movies
## **Netflix - TV Shows and Movies**
> This data set was created to list all shows available on Netflix streaming, and analyze the data to find interesting facts. This data was acquired in July 2022 containing data available in the United States.
## **Content**
> This dataset has two files containing the titles (**titles.csv**) and the cast (**credits.csv**) for the title.
> This dataset contains **+5k** unique **titles on Netflix** with 15 columns containing their information, including:
> - id: The title ID on JustWatch.
> - title: The name of the title.
> - show_type: TV show or movie.
> - description: A brief description.
> - release_year: The release year.
> - age_certification: The age certification.
> - runtime: The length of the episode (SHOW) or movie.
> - genres: A list of genres.
> - production_countries: A list of countries that produced the title.
> - seasons: Number of seasons if it's a SHOW.
> - imdb_id: The title ID on IMDB.
> - imdb_score: Score on IMDB.
> - imdb_votes: Votes on IMDB.
> - tmdb_popularity: Popularity on TMDB.
> - tmdb_score: Score on TMDB.
> And **over +77k** credits of **actors and directors** on Netflix titles with 5 columns containing their information, including:
> - person_ID: The person ID on JustWatch.
> - id: The title ID on JustWatch.
> - name: The actor or director's name.
> - character_name: The character name.
> - role: ACTOR or DIRECTOR.
##**Tasks**
> - Developing a content-based recommender system using the genres and/or descriptions.
> - Identifying the main content available on the streaming.
> - Network analysis on the cast of the titles.
> - Exploratory data analysis to find interesting insights.
## **Other Streaming Datasets**
> - [HBO Max TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/hbo-max-tv-shows-and-movies?select=titles.csv)
> - [Amazon Prime TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/amazon-prime-tv-shows-and-movies?select=titles.csv)
> - [Disney+ TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/disney-tv-shows-and-movies?select=titles.csv)
> - [Hulu TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/hulu-tv-shows-and-movies?select=titles.csv)
> - [Paramount TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/paramount-tv-shows-and-movies?select=titles.csv)
> - [Rakuten Viki TV Dramas and Movies](https://www.kaggle.com/datasets/victorsoeiro/rakuten-tv-dramas-and-movies?select=titles.csv)
> - [Crunchyroll Animes and Movies](https://www.kaggle.com/datasets/victorsoeiro/crunchyroll-animes-and-movies)
> - [Dark Matter TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/dark-matter-tv-shows-and-movies)
##**How to obtain the data**
> If you want to see how I obtained these data, please check my [GitHub repository](https://github.com/victor-soeiro/WebScraping-Projects/tree/main/justwatch).
## **Acknowledgements**
> All data were collected from [JustWatch](https://www.justwatch.com/us).
Kaggle dataset identifier: netflix-tv-shows-and-movies
<jupyter_code>import pandas as pd
df = pd.read_csv('netflix-tv-shows-and-movies/titles.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 5850 entries, 0 to 5849
Data columns (total 15 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 5850 non-null object
1 title 5849 non-null object
2 type 5850 non-null object
3 description 5832 non-null object
4 release_year 5850 non-null int64
5 age_certification 3231 non-null object
6 runtime 5850 non-null int64
7 genres 5850 non-null object
8 production_countries 5850 non-null object
9 seasons 2106 non-null float64
10 imdb_id 5447 non-null object
11 imdb_score 5368 non-null float64
12 imdb_votes 5352 non-null float64
13 tmdb_popularity 5759 non-null float64
14 tmdb_score 5539 non-null float64
dtypes: float64(5), int64(2), object(8)
memory usage: 685.7+ KB
<jupyter_text>Examples:
{
"id": "ts300399",
"title": "Five Came Back: The Reference Films",
"type": "SHOW",
"description": "This collection includes 12 World War II-era propaganda films \u2014 many of which are graphic and offensive \u2014 discussed in the docuseries \"Five Came Back.\"",
"release_year": 1945,
"age_certification": "TV-MA",
"runtime": 51,
"genres": "['documentation']",
"production_countries": "['US']",
"seasons": 1.0,
"imdb_id": null,
"imdb_score": NaN,
"imdb_votes": NaN,
"tmdb_popularity": 0.6000000000000001,
"tmdb_score": NaN
}
{
"id": "tm84618",
"title": "Taxi Driver",
"type": "MOVIE",
"description": "A mentally unstable Vietnam War veteran works as a night-time taxi driver in New York City where the perceived decadence and sleaze feed his urge for violent action.",
"release_year": 1976,
"age_certification": "R",
"runtime": 114,
"genres": "['drama', 'crime']",
"production_countries": "['US']",
"seasons": NaN,
"imdb_id": "tt0075314",
"imdb_score": 8.2,
"imdb_votes": 808582.0,
"tmdb_popularity": 40.965,
"tmdb_score": 8.179
}
{
"id": "tm154986",
"title": "Deliverance",
"type": "MOVIE",
"description": "Intent on seeing the Cahulawassee River before it's turned into one huge lake, outdoor fanatic Lewis Medlock takes his friends on a river-rafting trip they'll never forget into the dangerous American back-country.",
"release_year": 1972,
"age_certification": "R",
"runtime": 109,
"genres": "['drama', 'action', 'thriller', 'european']",
"production_countries": "['US']",
"seasons": NaN,
"imdb_id": "tt0068473",
"imdb_score": 7.7,
"imdb_votes": 107673.0,
"tmdb_popularity": 10.01,
"tmdb_score": 7.3
}
{
"id": "tm127384",
"title": "Monty Python and the Holy Grail",
"type": "MOVIE",
"description": "King Arthur, accompanied by his squire, recruits his Knights of the Round Table, including Sir Bedevere the Wise, Sir Lancelot the Brave, Sir Robin the Not-Quite-So-Brave-As-Sir-Lancelot and Sir Galahad the Pure. On the way, Arthur battles the Black Knight who, despite having had...(truncated)",
"release_year": 1975,
"age_certification": "PG",
"runtime": 91,
"genres": "['fantasy', 'action', 'comedy']",
"production_countries": "['GB']",
"seasons": NaN,
"imdb_id": "tt0071853",
"imdb_score": 8.2,
"imdb_votes": 534486.0,
"tmdb_popularity": 15.461,
"tmdb_score": 7.811
}
<jupyter_code>import pandas as pd
df = pd.read_csv('netflix-tv-shows-and-movies/credits.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 77801 entries, 0 to 77800
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 person_id 77801 non-null int64
1 id 77801 non-null object
2 name 77801 non-null object
3 character 68029 non-null object
4 role 77801 non-null object
dtypes: int64(1), object(4)
memory usage: 3.0+ MB
<jupyter_text>Examples:
{
"person_id": 3748,
"id": "tm84618",
"name": "Robert De Niro",
"character": "Travis Bickle",
"role": "ACTOR"
}
{
"person_id": 14658,
"id": "tm84618",
"name": "Jodie Foster",
"character": "Iris Steensma",
"role": "ACTOR"
}
{
"person_id": 7064,
"id": "tm84618",
"name": "Albert Brooks",
"character": "Tom",
"role": "ACTOR"
}
{
"person_id": 3739,
"id": "tm84618",
"name": "Harvey Keitel",
"character": "Matthew 'Sport' Higgins",
"role": "ACTOR"
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
#
# 1. Import Files and explore the datasets
#
df_credits = pd.read_csv("/kaggle/input/netflix-tv-shows-and-movies/credits.csv")
df_credits.head()
df_credits.info()
df_credits.isna().sum()
df_titles = pd.read_csv("/kaggle/input/netflix-tv-shows-and-movies/titles.csv")
df_titles.head()
df_titles.isna().sum()
df_titles.info()
# ## Explanation of the cleaning process
# * Ignore missing values for the following variables, as they have no implications for the analysis:
# - description
# - age_certification
# - imdb_id
# * For the variable seasons, the missing values will be replaced by 0 which corresponds to the Movie (which have no season, it is logical)
# * Replacement of the missing values in the following variables by the median or the mean (if there are no outliers values) : imdb_score, imdb_votes, tmdb_score and tmdb_popularity
# * Correct the two variables 'genres' and 'productions_countries' to remove the square brackets and quotes.
def clean_variables(dataframe):
dataframe.dropna(axis=0, subset=["title"], inplace=True)
dataframe["imdb_score"] = dataframe["imdb_score"].fillna(
value=dataframe["imdb_score"].mean()
)
dataframe["imdb_votes"] = dataframe["imdb_votes"].fillna(
value=dataframe["imdb_votes"].median()
)
dataframe["tmdb_popularity"] = dataframe["tmdb_popularity"].fillna(
value=dataframe["tmdb_popularity"].median()
)
dataframe["tmdb_score"] = dataframe["tmdb_score"].fillna(
value=dataframe["tmdb_score"].median()
)
dataframe["seasons"] = dataframe["seasons"].fillna(value=0)
clean_variables(df_titles)
# Drop quotation marks and square brackets in 'genres'
df_titles["genres"] = df_titles["genres"].apply(
lambda x: x.replace("[", "").replace("]", "")
)
df_titles["genres"] = df_titles["genres"].str.replace(r"[\'']", "")
# Drop quotation marks and square brackets in'production_countries'
df_titles["production_countries"] = df_titles["production_countries"].apply(
lambda x: x.replace("[", "").replace("]", "")
)
df_titles["production_countries"] = df_titles["production_countries"].str.replace(
r"[\'']", ""
)
# Drop imdb_id because no use
df_titles.drop("imdb_id", 1, inplace=True)
df_titles
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/364/129364413.ipynb
|
netflix-tv-shows-and-movies
|
victorsoeiro
|
[{"Id": 129364413, "ScriptId": 38431552, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10184546, "CreationDate": "05/13/2023 06:47:51", "VersionNumber": 1.0, "Title": "notebook3390a24f76", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 83.0, "LinesInsertedFromPrevious": 83.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185342190, "KernelVersionId": 129364413, "SourceDatasetVersionId": 3989707}]
|
[{"Id": 3989707, "DatasetId": 2178661, "DatasourceVersionId": 4045438, "CreatorUserId": 4697476, "LicenseName": "CC0: Public Domain", "CreationDate": "07/26/2022 19:50:06", "VersionNumber": 2.0, "Title": "Netflix TV Shows and Movies", "Slug": "netflix-tv-shows-and-movies", "Subtitle": "Movies and TV Shows listings on Netflix (July, 2022)", "Description": "## **Netflix - TV Shows and Movies**\n\n> This data set was created to list all shows available on Netflix streaming, and analyze the data to find interesting facts. This data was acquired in July 2022 containing data available in the United States. \n\n## **Content**\n\n> This dataset has two files containing the titles (**titles.csv**) and the cast (**credits.csv**) for the title. \n\n> This dataset contains **+5k** unique **titles on Netflix** with 15 columns containing their information, including:\n\n> - id: The title ID on JustWatch.\n> - title: The name of the title.\n> - show_type: TV show or movie.\n> - description: A brief description.\n> - release_year: The release year.\n> - age_certification: The age certification.\n> - runtime: The length of the episode (SHOW) or movie.\n> - genres: A list of genres.\n> - production_countries: A list of countries that produced the title.\n> - seasons: Number of seasons if it's a SHOW.\n> - imdb_id: The title ID on IMDB.\n> - imdb_score: Score on IMDB.\n> - imdb_votes: Votes on IMDB.\n> - tmdb_popularity: Popularity on TMDB.\n> - tmdb_score: Score on TMDB.\n\n> And **over +77k** credits of **actors and directors** on Netflix titles with 5 columns containing their information, including:\n\n> - person_ID: The person ID on JustWatch.\n> - id: The title ID on JustWatch.\n> - name: The actor or director's name.\n> - character_name: The character name.\n> - role: ACTOR or DIRECTOR.\n\n##**Tasks**\n> - Developing a content-based recommender system using the genres and/or descriptions.\n> - Identifying the main content available on the streaming.\n> - Network analysis on the cast of the titles.\n> - Exploratory data analysis to find interesting insights.\n\n## **Other Streaming Datasets**\n\n> - [HBO Max TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/hbo-max-tv-shows-and-movies?select=titles.csv)\n> - [Amazon Prime TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/amazon-prime-tv-shows-and-movies?select=titles.csv)\n> - [Disney+ TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/disney-tv-shows-and-movies?select=titles.csv)\n> - [Hulu TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/hulu-tv-shows-and-movies?select=titles.csv)\n> - [Paramount TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/paramount-tv-shows-and-movies?select=titles.csv)\n> - [Rakuten Viki TV Dramas and Movies](https://www.kaggle.com/datasets/victorsoeiro/rakuten-tv-dramas-and-movies?select=titles.csv)\n> - [Crunchyroll Animes and Movies](https://www.kaggle.com/datasets/victorsoeiro/crunchyroll-animes-and-movies)\n> - [Dark Matter TV Shows and Movies](https://www.kaggle.com/datasets/victorsoeiro/dark-matter-tv-shows-and-movies)\n\n##**How to obtain the data**\n\n> If you want to see how I obtained these data, please check my [GitHub repository](https://github.com/victor-soeiro/WebScraping-Projects/tree/main/justwatch).\n\n## **Acknowledgements**\n\n> All data were collected from [JustWatch](https://www.justwatch.com/us).", "VersionNotes": "Data Update 2022/07/26", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2178661, "CreatorUserId": 4697476, "OwnerUserId": 4697476.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3989707.0, "CurrentDatasourceVersionId": 4045438.0, "ForumId": 2204602, "Type": 2, "CreationDate": "05/15/2022 00:01:23", "LastActivityDate": "05/15/2022", "TotalViews": 176218, "TotalDownloads": 31619, "TotalVotes": 640, "TotalKernels": 114}]
|
[{"Id": 4697476, "UserName": "victorsoeiro", "DisplayName": "Victor Soeiro", "RegisterDate": "03/19/2020", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
#
# 1. Import Files and explore the datasets
#
df_credits = pd.read_csv("/kaggle/input/netflix-tv-shows-and-movies/credits.csv")
df_credits.head()
df_credits.info()
df_credits.isna().sum()
df_titles = pd.read_csv("/kaggle/input/netflix-tv-shows-and-movies/titles.csv")
df_titles.head()
df_titles.isna().sum()
df_titles.info()
# ## Explanation of the cleaning process
# * Ignore missing values for the following variables, as they have no implications for the analysis:
# - description
# - age_certification
# - imdb_id
# * For the variable seasons, the missing values will be replaced by 0 which corresponds to the Movie (which have no season, it is logical)
# * Replacement of the missing values in the following variables by the median or the mean (if there are no outliers values) : imdb_score, imdb_votes, tmdb_score and tmdb_popularity
# * Correct the two variables 'genres' and 'productions_countries' to remove the square brackets and quotes.
def clean_variables(dataframe):
dataframe.dropna(axis=0, subset=["title"], inplace=True)
dataframe["imdb_score"] = dataframe["imdb_score"].fillna(
value=dataframe["imdb_score"].mean()
)
dataframe["imdb_votes"] = dataframe["imdb_votes"].fillna(
value=dataframe["imdb_votes"].median()
)
dataframe["tmdb_popularity"] = dataframe["tmdb_popularity"].fillna(
value=dataframe["tmdb_popularity"].median()
)
dataframe["tmdb_score"] = dataframe["tmdb_score"].fillna(
value=dataframe["tmdb_score"].median()
)
dataframe["seasons"] = dataframe["seasons"].fillna(value=0)
clean_variables(df_titles)
# Drop quotation marks and square brackets in 'genres'
df_titles["genres"] = df_titles["genres"].apply(
lambda x: x.replace("[", "").replace("]", "")
)
df_titles["genres"] = df_titles["genres"].str.replace(r"[\'']", "")
# Drop quotation marks and square brackets in'production_countries'
df_titles["production_countries"] = df_titles["production_countries"].apply(
lambda x: x.replace("[", "").replace("]", "")
)
df_titles["production_countries"] = df_titles["production_countries"].str.replace(
r"[\'']", ""
)
# Drop imdb_id because no use
df_titles.drop("imdb_id", 1, inplace=True)
df_titles
|
[{"netflix-tv-shows-and-movies/titles.csv": {"column_names": "[\"id\", \"title\", \"type\", \"description\", \"release_year\", \"age_certification\", \"runtime\", \"genres\", \"production_countries\", \"seasons\", \"imdb_id\", \"imdb_score\", \"imdb_votes\", \"tmdb_popularity\", \"tmdb_score\"]", "column_data_types": "{\"id\": \"object\", \"title\": \"object\", \"type\": \"object\", \"description\": \"object\", \"release_year\": \"int64\", \"age_certification\": \"object\", \"runtime\": \"int64\", \"genres\": \"object\", \"production_countries\": \"object\", \"seasons\": \"float64\", \"imdb_id\": \"object\", \"imdb_score\": \"float64\", \"imdb_votes\": \"float64\", \"tmdb_popularity\": \"float64\", \"tmdb_score\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 5850 entries, 0 to 5849\nData columns (total 15 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 id 5850 non-null object \n 1 title 5849 non-null object \n 2 type 5850 non-null object \n 3 description 5832 non-null object \n 4 release_year 5850 non-null int64 \n 5 age_certification 3231 non-null object \n 6 runtime 5850 non-null int64 \n 7 genres 5850 non-null object \n 8 production_countries 5850 non-null object \n 9 seasons 2106 non-null float64\n 10 imdb_id 5447 non-null object \n 11 imdb_score 5368 non-null float64\n 12 imdb_votes 5352 non-null float64\n 13 tmdb_popularity 5759 non-null float64\n 14 tmdb_score 5539 non-null float64\ndtypes: float64(5), int64(2), object(8)\nmemory usage: 685.7+ KB\n", "summary": "{\"release_year\": {\"count\": 5850.0, \"mean\": 2016.417094017094, \"std\": 6.937725712183742, \"min\": 1945.0, \"25%\": 2016.0, \"50%\": 2018.0, \"75%\": 2020.0, \"max\": 2022.0}, \"runtime\": {\"count\": 5850.0, \"mean\": 76.88888888888889, \"std\": 39.00250917525395, \"min\": 0.0, \"25%\": 44.0, \"50%\": 83.0, \"75%\": 104.0, \"max\": 240.0}, \"seasons\": {\"count\": 2106.0, \"mean\": 2.1628679962013297, \"std\": 2.6890413904714925, \"min\": 1.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 42.0}, \"imdb_score\": {\"count\": 5368.0, \"mean\": 6.510860655737705, \"std\": 1.1638263082409555, \"min\": 1.5, \"25%\": 5.8, \"50%\": 6.6, \"75%\": 7.3, \"max\": 9.6}, \"imdb_votes\": {\"count\": 5352.0, \"mean\": 23439.382473841553, \"std\": 95820.47090889506, \"min\": 5.0, \"25%\": 516.75, \"50%\": 2233.5, \"75%\": 9494.0, \"max\": 2294231.0}, \"tmdb_popularity\": {\"count\": 5759.0, \"mean\": 22.6379253956843, \"std\": 81.6802632085619, \"min\": 0.0094417458789051, \"25%\": 2.7285, \"50%\": 6.821, \"75%\": 16.59, \"max\": 2274.044}, \"tmdb_score\": {\"count\": 5539.0, \"mean\": 6.829174760787145, \"std\": 1.1703914445224128, \"min\": 0.5, \"25%\": 6.1, \"50%\": 6.9, \"75%\": 7.5375, \"max\": 10.0}}", "examples": "{\"id\":{\"0\":\"ts300399\",\"1\":\"tm84618\",\"2\":\"tm154986\",\"3\":\"tm127384\"},\"title\":{\"0\":\"Five Came Back: The Reference Films\",\"1\":\"Taxi Driver\",\"2\":\"Deliverance\",\"3\":\"Monty Python and the Holy Grail\"},\"type\":{\"0\":\"SHOW\",\"1\":\"MOVIE\",\"2\":\"MOVIE\",\"3\":\"MOVIE\"},\"description\":{\"0\":\"This collection includes 12 World War II-era propaganda films \\u2014 many of which are graphic and offensive \\u2014 discussed in the docuseries \\\"Five Came Back.\\\"\",\"1\":\"A mentally unstable Vietnam War veteran works as a night-time taxi driver in New York City where the perceived decadence and sleaze feed his urge for violent action.\",\"2\":\"Intent on seeing the Cahulawassee River before it's turned into one huge lake, outdoor fanatic Lewis Medlock takes his friends on a river-rafting trip they'll never forget into the dangerous American back-country.\",\"3\":\"King Arthur, accompanied by his squire, recruits his Knights of the Round Table, including Sir Bedevere the Wise, Sir Lancelot the Brave, Sir Robin the Not-Quite-So-Brave-As-Sir-Lancelot and Sir Galahad the Pure. On the way, Arthur battles the Black Knight who, despite having had all his limbs chopped off, insists he can still fight. They reach Camelot, but Arthur decides not to enter, as \\\"it is a silly place\\\".\"},\"release_year\":{\"0\":1945,\"1\":1976,\"2\":1972,\"3\":1975},\"age_certification\":{\"0\":\"TV-MA\",\"1\":\"R\",\"2\":\"R\",\"3\":\"PG\"},\"runtime\":{\"0\":51,\"1\":114,\"2\":109,\"3\":91},\"genres\":{\"0\":\"['documentation']\",\"1\":\"['drama', 'crime']\",\"2\":\"['drama', 'action', 'thriller', 'european']\",\"3\":\"['fantasy', 'action', 'comedy']\"},\"production_countries\":{\"0\":\"['US']\",\"1\":\"['US']\",\"2\":\"['US']\",\"3\":\"['GB']\"},\"seasons\":{\"0\":1.0,\"1\":null,\"2\":null,\"3\":null},\"imdb_id\":{\"0\":null,\"1\":\"tt0075314\",\"2\":\"tt0068473\",\"3\":\"tt0071853\"},\"imdb_score\":{\"0\":null,\"1\":8.2,\"2\":7.7,\"3\":8.2},\"imdb_votes\":{\"0\":null,\"1\":808582.0,\"2\":107673.0,\"3\":534486.0},\"tmdb_popularity\":{\"0\":0.6,\"1\":40.965,\"2\":10.01,\"3\":15.461},\"tmdb_score\":{\"0\":null,\"1\":8.179,\"2\":7.3,\"3\":7.811}}"}}, {"netflix-tv-shows-and-movies/credits.csv": {"column_names": "[\"person_id\", \"id\", \"name\", \"character\", \"role\"]", "column_data_types": "{\"person_id\": \"int64\", \"id\": \"object\", \"name\": \"object\", \"character\": \"object\", \"role\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 77801 entries, 0 to 77800\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 person_id 77801 non-null int64 \n 1 id 77801 non-null object\n 2 name 77801 non-null object\n 3 character 68029 non-null object\n 4 role 77801 non-null object\ndtypes: int64(1), object(4)\nmemory usage: 3.0+ MB\n", "summary": "{\"person_id\": {\"count\": 77801.0, \"mean\": 529488.8064420766, \"std\": 643016.6699575292, \"min\": 7.0, \"25%\": 45306.0, \"50%\": 198358.0, \"75%\": 888096.0, \"max\": 2462818.0}}", "examples": "{\"person_id\":{\"0\":3748,\"1\":14658,\"2\":7064,\"3\":3739},\"id\":{\"0\":\"tm84618\",\"1\":\"tm84618\",\"2\":\"tm84618\",\"3\":\"tm84618\"},\"name\":{\"0\":\"Robert De Niro\",\"1\":\"Jodie Foster\",\"2\":\"Albert Brooks\",\"3\":\"Harvey Keitel\"},\"character\":{\"0\":\"Travis Bickle\",\"1\":\"Iris Steensma\",\"2\":\"Tom\",\"3\":\"Matthew 'Sport' Higgins\"},\"role\":{\"0\":\"ACTOR\",\"1\":\"ACTOR\",\"2\":\"ACTOR\",\"3\":\"ACTOR\"}}"}}]
| true | 2 |
<start_data_description><data_path>netflix-tv-shows-and-movies/titles.csv:
<column_names>
['id', 'title', 'type', 'description', 'release_year', 'age_certification', 'runtime', 'genres', 'production_countries', 'seasons', 'imdb_id', 'imdb_score', 'imdb_votes', 'tmdb_popularity', 'tmdb_score']
<column_types>
{'id': 'object', 'title': 'object', 'type': 'object', 'description': 'object', 'release_year': 'int64', 'age_certification': 'object', 'runtime': 'int64', 'genres': 'object', 'production_countries': 'object', 'seasons': 'float64', 'imdb_id': 'object', 'imdb_score': 'float64', 'imdb_votes': 'float64', 'tmdb_popularity': 'float64', 'tmdb_score': 'float64'}
<dataframe_Summary>
{'release_year': {'count': 5850.0, 'mean': 2016.417094017094, 'std': 6.937725712183742, 'min': 1945.0, '25%': 2016.0, '50%': 2018.0, '75%': 2020.0, 'max': 2022.0}, 'runtime': {'count': 5850.0, 'mean': 76.88888888888889, 'std': 39.00250917525395, 'min': 0.0, '25%': 44.0, '50%': 83.0, '75%': 104.0, 'max': 240.0}, 'seasons': {'count': 2106.0, 'mean': 2.1628679962013297, 'std': 2.6890413904714925, 'min': 1.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 42.0}, 'imdb_score': {'count': 5368.0, 'mean': 6.510860655737705, 'std': 1.1638263082409555, 'min': 1.5, '25%': 5.8, '50%': 6.6, '75%': 7.3, 'max': 9.6}, 'imdb_votes': {'count': 5352.0, 'mean': 23439.382473841553, 'std': 95820.47090889506, 'min': 5.0, '25%': 516.75, '50%': 2233.5, '75%': 9494.0, 'max': 2294231.0}, 'tmdb_popularity': {'count': 5759.0, 'mean': 22.6379253956843, 'std': 81.6802632085619, 'min': 0.0094417458789051, '25%': 2.7285, '50%': 6.821, '75%': 16.59, 'max': 2274.044}, 'tmdb_score': {'count': 5539.0, 'mean': 6.829174760787145, 'std': 1.1703914445224128, 'min': 0.5, '25%': 6.1, '50%': 6.9, '75%': 7.5375, 'max': 10.0}}
<dataframe_info>
RangeIndex: 5850 entries, 0 to 5849
Data columns (total 15 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 5850 non-null object
1 title 5849 non-null object
2 type 5850 non-null object
3 description 5832 non-null object
4 release_year 5850 non-null int64
5 age_certification 3231 non-null object
6 runtime 5850 non-null int64
7 genres 5850 non-null object
8 production_countries 5850 non-null object
9 seasons 2106 non-null float64
10 imdb_id 5447 non-null object
11 imdb_score 5368 non-null float64
12 imdb_votes 5352 non-null float64
13 tmdb_popularity 5759 non-null float64
14 tmdb_score 5539 non-null float64
dtypes: float64(5), int64(2), object(8)
memory usage: 685.7+ KB
<some_examples>
{'id': {'0': 'ts300399', '1': 'tm84618', '2': 'tm154986', '3': 'tm127384'}, 'title': {'0': 'Five Came Back: The Reference Films', '1': 'Taxi Driver', '2': 'Deliverance', '3': 'Monty Python and the Holy Grail'}, 'type': {'0': 'SHOW', '1': 'MOVIE', '2': 'MOVIE', '3': 'MOVIE'}, 'description': {'0': 'This collection includes 12 World War II-era propaganda films — many of which are graphic and offensive — discussed in the docuseries "Five Came Back."', '1': 'A mentally unstable Vietnam War veteran works as a night-time taxi driver in New York City where the perceived decadence and sleaze feed his urge for violent action.', '2': "Intent on seeing the Cahulawassee River before it's turned into one huge lake, outdoor fanatic Lewis Medlock takes his friends on a river-rafting trip they'll never forget into the dangerous American back-country.", '3': 'King Arthur, accompanied by his squire, recruits his Knights of the Round Table, including Sir Bedevere the Wise, Sir Lancelot the Brave, Sir Robin the Not-Quite-So-Brave-As-Sir-Lancelot and Sir Galahad the Pure. On the way, Arthur battles the Black Knight who, despite having had all his limbs chopped off, insists he can still fight. They reach Camelot, but Arthur decides not to enter, as "it is a silly place".'}, 'release_year': {'0': 1945, '1': 1976, '2': 1972, '3': 1975}, 'age_certification': {'0': 'TV-MA', '1': 'R', '2': 'R', '3': 'PG'}, 'runtime': {'0': 51, '1': 114, '2': 109, '3': 91}, 'genres': {'0': "['documentation']", '1': "['drama', 'crime']", '2': "['drama', 'action', 'thriller', 'european']", '3': "['fantasy', 'action', 'comedy']"}, 'production_countries': {'0': "['US']", '1': "['US']", '2': "['US']", '3': "['GB']"}, 'seasons': {'0': 1.0, '1': None, '2': None, '3': None}, 'imdb_id': {'0': None, '1': 'tt0075314', '2': 'tt0068473', '3': 'tt0071853'}, 'imdb_score': {'0': None, '1': 8.2, '2': 7.7, '3': 8.2}, 'imdb_votes': {'0': None, '1': 808582.0, '2': 107673.0, '3': 534486.0}, 'tmdb_popularity': {'0': 0.6, '1': 40.965, '2': 10.01, '3': 15.461}, 'tmdb_score': {'0': None, '1': 8.179, '2': 7.3, '3': 7.811}}
<end_description>
<start_data_description><data_path>netflix-tv-shows-and-movies/credits.csv:
<column_names>
['person_id', 'id', 'name', 'character', 'role']
<column_types>
{'person_id': 'int64', 'id': 'object', 'name': 'object', 'character': 'object', 'role': 'object'}
<dataframe_Summary>
{'person_id': {'count': 77801.0, 'mean': 529488.8064420766, 'std': 643016.6699575292, 'min': 7.0, '25%': 45306.0, '50%': 198358.0, '75%': 888096.0, 'max': 2462818.0}}
<dataframe_info>
RangeIndex: 77801 entries, 0 to 77800
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 person_id 77801 non-null int64
1 id 77801 non-null object
2 name 77801 non-null object
3 character 68029 non-null object
4 role 77801 non-null object
dtypes: int64(1), object(4)
memory usage: 3.0+ MB
<some_examples>
{'person_id': {'0': 3748, '1': 14658, '2': 7064, '3': 3739}, 'id': {'0': 'tm84618', '1': 'tm84618', '2': 'tm84618', '3': 'tm84618'}, 'name': {'0': 'Robert De Niro', '1': 'Jodie Foster', '2': 'Albert Brooks', '3': 'Harvey Keitel'}, 'character': {'0': 'Travis Bickle', '1': 'Iris Steensma', '2': 'Tom', '3': "Matthew 'Sport' Higgins"}, 'role': {'0': 'ACTOR', '1': 'ACTOR', '2': 'ACTOR', '3': 'ACTOR'}}
<end_description>
| 855 | 0 | 3,682 | 855 |
129313251
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
# Read the CSV data file into a pandas DataFrame
df = pd.read_csv("/kaggle/input/fedfunds-vs-gdp/gdp_fedfunds.csv")
import numpy as np
import statsmodels.api as sm
# Add a constant to the DataFrame for the regression intercept
df["const"] = 1
# Define the regression formula
model = sm.OLS(df["value_gdp"], df[["const", "value_fedfunds"]])
# Fit the model to the data
results = model.fit()
# Print the regression results
print(results.summary())
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 5))
plt.scatter(df["value_fedfunds"], df["value_gdp"])
plt.xlabel("Federal Funds Rate")
plt.ylabel("GDP")
plt.title("Scatter plot of Federal Funds Rate vs GDP")
plt.grid(True)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/313/129313251.ipynb
| null | null |
[{"Id": 129313251, "ScriptId": 38446625, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4454153, "CreationDate": "05/12/2023 17:12:37", "VersionNumber": 1.0, "Title": "Fed Funds vs. GDP", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 75.0, "LinesInsertedFromPrevious": 75.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
# Read the CSV data file into a pandas DataFrame
df = pd.read_csv("/kaggle/input/fedfunds-vs-gdp/gdp_fedfunds.csv")
import numpy as np
import statsmodels.api as sm
# Add a constant to the DataFrame for the regression intercept
df["const"] = 1
# Define the regression formula
model = sm.OLS(df["value_gdp"], df[["const", "value_fedfunds"]])
# Fit the model to the data
results = model.fit()
# Print the regression results
print(results.summary())
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 5))
plt.scatter(df["value_fedfunds"], df["value_gdp"])
plt.xlabel("Federal Funds Rate")
plt.ylabel("GDP")
plt.title("Scatter plot of Federal Funds Rate vs GDP")
plt.grid(True)
plt.show()
| false | 0 | 414 | 0 | 414 | 414 |
||
129287752
|
import pandas as pd
import urllib.request
from sklearn.feature_extraction.text import CountVectorizer
import numpy as np
from sklearn.decomposition import TruncatedSVD
from sklearn.preprocessing import normalize
from sklearn.metrics.pairwise import cosine_similarity
# Download the datasets
urllib.request.urlretrieve(url1, "vocab.enron.txt")
urllib.request.urlretrieve(url2, "vocab.nips.txt")
urllib.request.urlretrieve(url3, "vocab.kos.txt")
vocab = pd.read_csv("vocab.enron.txt", sep="\t", header=None, names=["word"])
nips = pd.read_csv("vocab.nips.txt", sep="\t", header=None, names=["word"])
kos = pd.read_csv("vocab.kos.txt", sep="\t ", header=None, skiprows=3, names=["word"])
corpus = vocab["word"].str.cat(nips["word"]).str.cat(kos["word"])
corpus = corpus.dropna()
count_vectorizer = CountVectorizer()
X = count_vectorizer.fit_transform(corpus)
svd = TruncatedSVD(n_components=100)
X_svd = svd.fit_transform(X)
print(X_svd[:, :10])
# ### i. Given that the dimensions from the reduced SVD are simply linear combinations of the original features, they might not be easily interpreted. Input for machine learning models or further analysis can be used with them, though.
# ### ii. The most significant feature or pattern in the data is represented by the first dimension in the shortened SVD output. It accounts for the bulk of the data's variation.
# ### iii. The most notable patterns in the data that account for a significant portion of the variation are represented by the top 10 dimensions in the shortened SVD output. The way you interpret these dimensions will vary depending on the particular facts and the issue you're seeking to resolve. To learn more about the underlying patterns, you might examine the words or documents that have high loadings on each dimension.
vocab = vocab.dropna()
nips = nips.dropna()
kos = kos.dropna()
vectorizer = CountVectorizer()
matrix1 = vectorizer.fit_transform(vocab["word"])
matrix2 = vectorizer.fit_transform(nips["word"])
matrix3 = vectorizer.fit_transform(kos["word"])
# normalize the matrices
norm_matrix1 = normalize(matrix1)
norm_matrix2 = normalize(matrix2)
norm_matrix3 = normalize(matrix3)
# calculate cosine similarity matrices
similarity_matrix1 = cosine_similarity(norm_matrix1)
similarity_matrix2 = cosine_similarity(norm_matrix2)
similarity_matrix3 = cosine_similarity(norm_matrix3)
# calculate the average cosine similarity within each corpus
avg_within_corpus1 = similarity_matrix1.mean()
avg_within_corpus2 = similarity_matrix2.mean()
avg_within_corpus3 = similarity_matrix3.mean()
print("Average cosine similarity within corpus 1:", avg_within_corpus1)
print("Average cosine similarity within corpus 2:", avg_within_corpus2)
print("Average cosine similarity within corpus 3:", avg_within_corpus3)
# ### e. The success of LSA as a method for clustering corpora will rely on the particular data and the issue you are attempting to address. The word-document matrix's dimensionality can be reduced using the dimensionality reduction approach called LSA, which can aid in grouping together similar texts. There are various clustering methods that might perform better for specific categories of data, therefore it might not be effective for all sorts of text data.
# ### f. The word-document matrix's dimensionality can be decreased using the linear dimensionality reduction technique known as PCA. For clustering text data, PCA can be used instead of LSA. However, since PCA is not created expressly for text data, it might not perform as well as LSA in this area. Even though PCA's results may not match LSA's, they can be helpful for grouping corpora together. The word-document matrix can be represented in a low-dimensional manner by PCA in order to capture the most crucial data. PCA might not, however, capture the same data as LSA and might not work as well with text data.
# ### In conclusion, LSA and PCA are both useful techniques for clustering corpora, but how well they work depends on the particular data and the issue at hand. To find the optimal strategy for your unique data, it can be required to test out several methods and algorithms.
# ### Second Task
# ### I have used wine data
import time
from sklearn.datasets import load_wine
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
wine = load_wine()
X = wine.data
y = wine.target
y
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
num_cols = X.shape[1]
pca_num = [int(num_cols / 10 * i) for i in range(1, 11)]
pca_num.append(num_cols)
for n in pca_num:
pca = PCA(n_components=n)
X_train_pca = pca.fit_transform(X_train)
X_test_pca = pca.transform(X_test)
print(f"PCA with {n} components")
# Use Logistic Regression Classifier
print("Logistic Regression Classifier:")
start_time = time.time()
lr = LogisticRegression(random_state=0, multi_class="ovr")
lr.fit(X_train_pca, y_train)
y_pred_lr = lr.predict(X_test_pca)
acc_lr = accuracy_score(y_test, y_pred_lr)
print(f"Accuracy: {acc_lr:.4f}")
print(f"Time: {time.time()-start_time:.4f}s")
# Use Decision Tree Classifier
print("\nDecision Tree Classifier:")
start_time = time.time()
dtc = DecisionTreeClassifier(random_state=0)
dtc.fit(X_train_pca, y_train)
y_pred_dtc = dtc.predict(X_test_pca)
acc_dtc = accuracy_score(y_test, y_pred_dtc)
print(f"Accuracy: {acc_dtc:.4f}")
print(f"Time: {time.time()-start_time:.4f}s")
# Use K-Nearest Neighbors Classifier
print("\nK-Nearest Neighbors Classifier:")
start_time = time.time()
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(X_train_pca, y_train)
y_pred_knn = knn.predict(X_test_pca)
acc_knn = accuracy_score(y_test, y_pred_knn)
print(f"Accuracy: {acc_knn:.4f}")
print(f"Time: {time.time()-start_time:.4f}s")
print("-" * 50)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/287/129287752.ipynb
| null | null |
[{"Id": 129287752, "ScriptId": 38438853, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14388792, "CreationDate": "05/12/2023 13:18:06", "VersionNumber": 1.0, "Title": "Homework_2", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 152.0, "LinesInsertedFromPrevious": 152.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import urllib.request
from sklearn.feature_extraction.text import CountVectorizer
import numpy as np
from sklearn.decomposition import TruncatedSVD
from sklearn.preprocessing import normalize
from sklearn.metrics.pairwise import cosine_similarity
# Download the datasets
urllib.request.urlretrieve(url1, "vocab.enron.txt")
urllib.request.urlretrieve(url2, "vocab.nips.txt")
urllib.request.urlretrieve(url3, "vocab.kos.txt")
vocab = pd.read_csv("vocab.enron.txt", sep="\t", header=None, names=["word"])
nips = pd.read_csv("vocab.nips.txt", sep="\t", header=None, names=["word"])
kos = pd.read_csv("vocab.kos.txt", sep="\t ", header=None, skiprows=3, names=["word"])
corpus = vocab["word"].str.cat(nips["word"]).str.cat(kos["word"])
corpus = corpus.dropna()
count_vectorizer = CountVectorizer()
X = count_vectorizer.fit_transform(corpus)
svd = TruncatedSVD(n_components=100)
X_svd = svd.fit_transform(X)
print(X_svd[:, :10])
# ### i. Given that the dimensions from the reduced SVD are simply linear combinations of the original features, they might not be easily interpreted. Input for machine learning models or further analysis can be used with them, though.
# ### ii. The most significant feature or pattern in the data is represented by the first dimension in the shortened SVD output. It accounts for the bulk of the data's variation.
# ### iii. The most notable patterns in the data that account for a significant portion of the variation are represented by the top 10 dimensions in the shortened SVD output. The way you interpret these dimensions will vary depending on the particular facts and the issue you're seeking to resolve. To learn more about the underlying patterns, you might examine the words or documents that have high loadings on each dimension.
vocab = vocab.dropna()
nips = nips.dropna()
kos = kos.dropna()
vectorizer = CountVectorizer()
matrix1 = vectorizer.fit_transform(vocab["word"])
matrix2 = vectorizer.fit_transform(nips["word"])
matrix3 = vectorizer.fit_transform(kos["word"])
# normalize the matrices
norm_matrix1 = normalize(matrix1)
norm_matrix2 = normalize(matrix2)
norm_matrix3 = normalize(matrix3)
# calculate cosine similarity matrices
similarity_matrix1 = cosine_similarity(norm_matrix1)
similarity_matrix2 = cosine_similarity(norm_matrix2)
similarity_matrix3 = cosine_similarity(norm_matrix3)
# calculate the average cosine similarity within each corpus
avg_within_corpus1 = similarity_matrix1.mean()
avg_within_corpus2 = similarity_matrix2.mean()
avg_within_corpus3 = similarity_matrix3.mean()
print("Average cosine similarity within corpus 1:", avg_within_corpus1)
print("Average cosine similarity within corpus 2:", avg_within_corpus2)
print("Average cosine similarity within corpus 3:", avg_within_corpus3)
# ### e. The success of LSA as a method for clustering corpora will rely on the particular data and the issue you are attempting to address. The word-document matrix's dimensionality can be reduced using the dimensionality reduction approach called LSA, which can aid in grouping together similar texts. There are various clustering methods that might perform better for specific categories of data, therefore it might not be effective for all sorts of text data.
# ### f. The word-document matrix's dimensionality can be decreased using the linear dimensionality reduction technique known as PCA. For clustering text data, PCA can be used instead of LSA. However, since PCA is not created expressly for text data, it might not perform as well as LSA in this area. Even though PCA's results may not match LSA's, they can be helpful for grouping corpora together. The word-document matrix can be represented in a low-dimensional manner by PCA in order to capture the most crucial data. PCA might not, however, capture the same data as LSA and might not work as well with text data.
# ### In conclusion, LSA and PCA are both useful techniques for clustering corpora, but how well they work depends on the particular data and the issue at hand. To find the optimal strategy for your unique data, it can be required to test out several methods and algorithms.
# ### Second Task
# ### I have used wine data
import time
from sklearn.datasets import load_wine
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
wine = load_wine()
X = wine.data
y = wine.target
y
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
num_cols = X.shape[1]
pca_num = [int(num_cols / 10 * i) for i in range(1, 11)]
pca_num.append(num_cols)
for n in pca_num:
pca = PCA(n_components=n)
X_train_pca = pca.fit_transform(X_train)
X_test_pca = pca.transform(X_test)
print(f"PCA with {n} components")
# Use Logistic Regression Classifier
print("Logistic Regression Classifier:")
start_time = time.time()
lr = LogisticRegression(random_state=0, multi_class="ovr")
lr.fit(X_train_pca, y_train)
y_pred_lr = lr.predict(X_test_pca)
acc_lr = accuracy_score(y_test, y_pred_lr)
print(f"Accuracy: {acc_lr:.4f}")
print(f"Time: {time.time()-start_time:.4f}s")
# Use Decision Tree Classifier
print("\nDecision Tree Classifier:")
start_time = time.time()
dtc = DecisionTreeClassifier(random_state=0)
dtc.fit(X_train_pca, y_train)
y_pred_dtc = dtc.predict(X_test_pca)
acc_dtc = accuracy_score(y_test, y_pred_dtc)
print(f"Accuracy: {acc_dtc:.4f}")
print(f"Time: {time.time()-start_time:.4f}s")
# Use K-Nearest Neighbors Classifier
print("\nK-Nearest Neighbors Classifier:")
start_time = time.time()
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(X_train_pca, y_train)
y_pred_knn = knn.predict(X_test_pca)
acc_knn = accuracy_score(y_test, y_pred_knn)
print(f"Accuracy: {acc_knn:.4f}")
print(f"Time: {time.time()-start_time:.4f}s")
print("-" * 50)
| false | 0 | 1,783 | 0 | 1,783 | 1,783 |
||
129287504
|
# # Import and become one with the data
import tensorflow as tf
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import urllib.request
import zipfile
# İndirilecek dosyanın URL'si
url = "https://storage.googleapis.com/ztm_tf_course/food_vision/10_food_classes_all_data.zip"
# Dosyanın indirilmesi
urllib.request.urlretrieve(url, "10_food_classes_all_data.zip")
# Zip dosyasının çıkarılması
with zipfile.ZipFile("10_food_classes_all_data.zip", "r") as zip_ref:
zip_ref.extractall()
from IPython.lib.display import walk
import os
for dirpath, dirnames, filenames in os.walk("10_food_classes_all_data"):
print(
f"There are {len(dirnames)} directories and {len(filenames)} images is '{dirpath}'."
)
data_dir = "10_food_classes_all_data/train"
class_names = sorted(os.listdir(data_dir))
class_names
train_dir = "10_food_classes_all_data/train"
test_dir = "10_food_classes_all_data/test"
# # Preprocess the data
from tensorflow.keras.preprocessing.image import ImageDataGenerator
train_datagen = ImageDataGenerator(rescale=1 / 255.0)
test_datagen = ImageDataGenerator(rescale=1 / 255.0)
train_data = train_datagen.flow_from_directory(
directory=train_dir, target_size=(224, 224), batch_size=32, class_mode="categorical"
)
test_data = train_datagen.flow_from_directory(
directory=test_dir, target_size=(224, 224), batch_size=32, class_mode="categorical"
)
# # Create a model
from tensorflow.keras.layers import Dense, Flatten, Conv2D, MaxPool2D
from tensorflow.keras import Sequential
model_1 = Sequential(
[
Conv2D(filters=10, kernel_size=3, activation="relu", input_shape=(224, 224, 3)),
Conv2D(10, 3, activation="relu"),
MaxPool2D(),
Conv2D(10, 3, activation="relu"),
Conv2D(10, 3, activation="relu"),
MaxPool2D(),
Flatten(),
Dense(10, activation="softmax"),
]
)
model_1.compile(
loss="categorical_crossentropy",
optimizer=tf.keras.optimizers.Adam(),
metrics=["accuracy"],
)
history_1 = model_1.fit(
train_data,
epochs=5,
steps_per_epoch=len(train_data),
validation_data=test_data,
validation_steps=len(test_data),
)
pd.DataFrame(history_1.history).plot(figsize=(12, 7))
def plot_loss_curves(history):
# Plot loss
train_loss = history.history["loss"]
test_loss = history.history["val_loss"]
epoc = range(len(history.history["loss"]))
plt.plot(epoc, train_loss, label="Train-Loss")
plt.plot(epoc, test_loss, label="Test-Loss")
plt.title("Loss")
plt.xlabel("Epochs")
plt.legend()
# Plot accuracy
plt.figure()
train_accuracy = history.history["accuracy"]
test_accuracy = history.history["val_accuracy"]
plt.plot(epoc, train_accuracy, label="Train Accuracy")
plt.plot(epoc, test_accuracy, label="Test Accuracy")
plt.title("Acuuracy")
plt.xlabel("Epochs")
plt.legend()
plot_loss_curves(history_1)
model_1.summary()
# # Evaluate the model
model_1.evaluate(test_data)
# # Create a new model
# ## Our model is overfitting, we need to prevent this
train_data_augmented = ImageDataGenerator(
rescale=1 / 255,
rotation_range=0.2,
width_shift_range=0.2,
height_shift_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
)
train_data_augmented = train_data_augmented.flow_from_directory(
directory=train_dir, target_size=(224, 224), batch_size=32, class_mode="categorical"
)
model_2 = tf.keras.models.clone_model(model_1)
model_2.compile(
loss="categorical_crossentropy",
optimizer=tf.keras.optimizers.Adam(),
metrics=["accuracy"],
)
history_2 = model_2.fit(
train_data_augmented,
epochs=5,
steps_per_epoch=len(train_data_augmented),
validation_data=test_data,
validation_steps=len(test_data),
)
plot_loss_curves(history_2)
import urllib.request
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
url = "https://raw.githubusercontent.com/mrdbourke/tensorflow-deep-learning/main/images/03-steak.jpeg"
urllib.request.urlretrieve(url, "03-steak.jpeg")
steak = mpimg.imread("03-steak.jpeg")
plt.imshow(steak)
plt.axis(False)
plt.show()
steak.shape
def load_and_pred_image(filname, img_shape=224):
img = tf.io.read_file(filname)
img = tf.image.decode_image(img, channels=3)
img = tf.image.resize(img, size=[img_shape, img_shape])
img = img / 255.0
return img
steak = load_and_pred_image("03-steak.jpeg")
steak
# # Make Predict
pred = model_2.predict(tf.expand_dims(steak, axis=0))
pred
pred_class = class_names[pred.argmax()]
pred_class
def pred_and_plot_model(model, filname, class_names):
img = load_and_pred_image(filname)
pred = model.predict(tf.expand_dims(img, axis=0))
pred_class = class_names[pred.argmax()]
plt.imshow(img)
plt.title(f"Predict:{pred_class}")
plt.show()
pred_and_plot_model(model_2, "03-steak.jpeg", class_names)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/287/129287504.ipynb
| null | null |
[{"Id": 129287504, "ScriptId": 38408564, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13923706, "CreationDate": "05/12/2023 13:15:54", "VersionNumber": 1.0, "Title": "Computer vision model with 10 different classes", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 170.0, "LinesInsertedFromPrevious": 170.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
# # Import and become one with the data
import tensorflow as tf
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import urllib.request
import zipfile
# İndirilecek dosyanın URL'si
url = "https://storage.googleapis.com/ztm_tf_course/food_vision/10_food_classes_all_data.zip"
# Dosyanın indirilmesi
urllib.request.urlretrieve(url, "10_food_classes_all_data.zip")
# Zip dosyasının çıkarılması
with zipfile.ZipFile("10_food_classes_all_data.zip", "r") as zip_ref:
zip_ref.extractall()
from IPython.lib.display import walk
import os
for dirpath, dirnames, filenames in os.walk("10_food_classes_all_data"):
print(
f"There are {len(dirnames)} directories and {len(filenames)} images is '{dirpath}'."
)
data_dir = "10_food_classes_all_data/train"
class_names = sorted(os.listdir(data_dir))
class_names
train_dir = "10_food_classes_all_data/train"
test_dir = "10_food_classes_all_data/test"
# # Preprocess the data
from tensorflow.keras.preprocessing.image import ImageDataGenerator
train_datagen = ImageDataGenerator(rescale=1 / 255.0)
test_datagen = ImageDataGenerator(rescale=1 / 255.0)
train_data = train_datagen.flow_from_directory(
directory=train_dir, target_size=(224, 224), batch_size=32, class_mode="categorical"
)
test_data = train_datagen.flow_from_directory(
directory=test_dir, target_size=(224, 224), batch_size=32, class_mode="categorical"
)
# # Create a model
from tensorflow.keras.layers import Dense, Flatten, Conv2D, MaxPool2D
from tensorflow.keras import Sequential
model_1 = Sequential(
[
Conv2D(filters=10, kernel_size=3, activation="relu", input_shape=(224, 224, 3)),
Conv2D(10, 3, activation="relu"),
MaxPool2D(),
Conv2D(10, 3, activation="relu"),
Conv2D(10, 3, activation="relu"),
MaxPool2D(),
Flatten(),
Dense(10, activation="softmax"),
]
)
model_1.compile(
loss="categorical_crossentropy",
optimizer=tf.keras.optimizers.Adam(),
metrics=["accuracy"],
)
history_1 = model_1.fit(
train_data,
epochs=5,
steps_per_epoch=len(train_data),
validation_data=test_data,
validation_steps=len(test_data),
)
pd.DataFrame(history_1.history).plot(figsize=(12, 7))
def plot_loss_curves(history):
# Plot loss
train_loss = history.history["loss"]
test_loss = history.history["val_loss"]
epoc = range(len(history.history["loss"]))
plt.plot(epoc, train_loss, label="Train-Loss")
plt.plot(epoc, test_loss, label="Test-Loss")
plt.title("Loss")
plt.xlabel("Epochs")
plt.legend()
# Plot accuracy
plt.figure()
train_accuracy = history.history["accuracy"]
test_accuracy = history.history["val_accuracy"]
plt.plot(epoc, train_accuracy, label="Train Accuracy")
plt.plot(epoc, test_accuracy, label="Test Accuracy")
plt.title("Acuuracy")
plt.xlabel("Epochs")
plt.legend()
plot_loss_curves(history_1)
model_1.summary()
# # Evaluate the model
model_1.evaluate(test_data)
# # Create a new model
# ## Our model is overfitting, we need to prevent this
train_data_augmented = ImageDataGenerator(
rescale=1 / 255,
rotation_range=0.2,
width_shift_range=0.2,
height_shift_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
)
train_data_augmented = train_data_augmented.flow_from_directory(
directory=train_dir, target_size=(224, 224), batch_size=32, class_mode="categorical"
)
model_2 = tf.keras.models.clone_model(model_1)
model_2.compile(
loss="categorical_crossentropy",
optimizer=tf.keras.optimizers.Adam(),
metrics=["accuracy"],
)
history_2 = model_2.fit(
train_data_augmented,
epochs=5,
steps_per_epoch=len(train_data_augmented),
validation_data=test_data,
validation_steps=len(test_data),
)
plot_loss_curves(history_2)
import urllib.request
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
url = "https://raw.githubusercontent.com/mrdbourke/tensorflow-deep-learning/main/images/03-steak.jpeg"
urllib.request.urlretrieve(url, "03-steak.jpeg")
steak = mpimg.imread("03-steak.jpeg")
plt.imshow(steak)
plt.axis(False)
plt.show()
steak.shape
def load_and_pred_image(filname, img_shape=224):
img = tf.io.read_file(filname)
img = tf.image.decode_image(img, channels=3)
img = tf.image.resize(img, size=[img_shape, img_shape])
img = img / 255.0
return img
steak = load_and_pred_image("03-steak.jpeg")
steak
# # Make Predict
pred = model_2.predict(tf.expand_dims(steak, axis=0))
pred
pred_class = class_names[pred.argmax()]
pred_class
def pred_and_plot_model(model, filname, class_names):
img = load_and_pred_image(filname)
pred = model.predict(tf.expand_dims(img, axis=0))
pred_class = class_names[pred.argmax()]
plt.imshow(img)
plt.title(f"Predict:{pred_class}")
plt.show()
pred_and_plot_model(model_2, "03-steak.jpeg", class_names)
| false | 0 | 1,665 | 1 | 1,665 | 1,665 |
||
129659332
|
<jupyter_start><jupyter_text>Indian Food 101
### Content
Indian cuisine consists of a variety of regional and traditional cuisines native to the Indian subcontinent. Given the diversity in soil, climate, culture, ethnic groups, and occupations, these cuisines vary substantially and use locally available spices, herbs, vegetables, and fruits. Indian food is also heavily influenced by religion, in particular Hinduism, cultural choices and traditions.
This dataset consists of information about various **Indian dishes**, their **ingredients**, their **place of origin**, etc.
### Column Description
**name** : name of the dish
**ingredients** : main ingredients used
**diet** : type of diet - either vegetarian or non vegetarian
**prep_time** : preparation time
**cook_time** : cooking time
**flavor_profile** : flavor profile includes whether the dish is spicy, sweet, bitter, etc
**course** : course of meal - starter, main course, dessert, etc
**state** : state where the dish is famous or is originated
**region** : region where the state belongs
Presence of -1 in any of the columns indicates NaN value.
Kaggle dataset identifier: indian-food-101
<jupyter_code>import pandas as pd
df = pd.read_csv('indian-food-101/indian_food.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 255 entries, 0 to 254
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 name 255 non-null object
1 ingredients 255 non-null object
2 diet 255 non-null object
3 prep_time 255 non-null int64
4 cook_time 255 non-null int64
5 flavor_profile 255 non-null object
6 course 255 non-null object
7 state 255 non-null object
8 region 254 non-null object
dtypes: int64(2), object(7)
memory usage: 18.1+ KB
<jupyter_text>Examples:
{
"name": "Balu shahi",
"ingredients": "Maida flour, yogurt, oil, sugar",
"diet": "vegetarian",
"prep_time": 45,
"cook_time": 25,
"flavor_profile": "sweet",
"course": "dessert",
"state": "West Bengal",
"region": "East"
}
{
"name": "Boondi",
"ingredients": "Gram flour, ghee, sugar",
"diet": "vegetarian",
"prep_time": 80,
"cook_time": 30,
"flavor_profile": "sweet",
"course": "dessert",
"state": "Rajasthan",
"region": "West"
}
{
"name": "Gajar ka halwa",
"ingredients": "Carrots, milk, sugar, ghee, cashews, raisins",
"diet": "vegetarian",
"prep_time": 15,
"cook_time": 60,
"flavor_profile": "sweet",
"course": "dessert",
"state": "Punjab",
"region": "North"
}
{
"name": "Ghevar",
"ingredients": "Flour, ghee, kewra, milk, clarified butter, sugar, almonds, pistachio, saffron, green cardamom",
"diet": "vegetarian",
"prep_time": 15,
"cook_time": 30,
"flavor_profile": "sweet",
"course": "dessert",
"state": "Rajasthan",
"region": "West"
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = pd.read_csv("/kaggle/input/indian-food-101/indian_food.csv")
data
# Unique values for each columns
# print(pd.unique(data.ingredients))
print(pd.unique(data.flavor_profile))
print(pd.unique(data.course))
# print(pd.unique(data.diet))
# print(pd.unique(data.state))
### data.loc[data['state'] == '-1']
# Cleaning Data
# data.loc[data['flavor_profile']== '-1']
# data.loc[data['name']=='Copra paak','flavor_profile']
data.loc[
data["name"].isin(["Copra paak", "Puttu", "Kansar"]), "flavor_profile"
] = "sweet"
data.loc[data["flavor_profile"] == "-1", "flavor_profile"] = "other"
# Cleaned values
data.tail(10)
# Performing One hot encoding
discrete_df = pd.get_dummies(data, columns=["ingredients", "diet", "flavor_profile"])
discrete_df
target = data["course"]
df = discrete_df.drop(columns=["course", "state", "region", "name"], axis=1)
df
# # **Label Encoding**
# Converting target string to numeric labels
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
label = le.fit_transform(data["course"])
label
# 0='dessert' 1='main course' 2='starter' 3='snack'
# # **Train Test Split**
# Spliting data inro training set and testing set
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(
df, label, test_size=0.2, random_state=0
)
# len(X_test)
# # **Decision Tree Algorithm**
# implementing classification algorithm
from sklearn.tree import DecisionTreeClassifier
model1 = DecisionTreeClassifier()
model1.fit(X_train, Y_train)
# 0='dessert' 1='main course' 2='starter' 3='snack'
ypred = model1.predict(X_test)
ypredct = le.inverse_transform(ypred)
# print(ypredct)
# Checking accuracy
accuracy1 = model1.score(X_test, Y_test)
print(accuracy1)
from sklearn.metrics import classification_report
report1 = classification_report(Y_test, ypred)
print(type(report1))
# report=classification_report(Y_test, ypred,output_dict=True)
# macro_precision = report['macro avg']['precision']
# macro_precision
from sklearn import metrics
confusion_matrix = metrics.confusion_matrix(Y_test, ypred)
cm_display = metrics.ConfusionMatrixDisplay(
confusion_matrix=confusion_matrix, display_labels=[False, True]
)
import matplotlib.pyplot as plt
cm_display.plot()
plt.show()
# # **Random Forest Algorithm**
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
clf = RandomForestClassifier(n_estimators=2, min_samples_split=3, min_samples_leaf=2)
clf.fit(X_train, Y_train)
pred_clf = clf.predict(X_test)
accuracyN = clf.score(X_test, Y_test)
print(accuracyN)
reportN = classification_report(Y_test, pred_clf)
print(reportN)
from sklearn.model_selection import GridSearchCV
param_grid = {
"n_estimators": [2, 5, 10, 20],
"min_samples_split": [2, 3],
"min_samples_leaf": [1, 2, 3],
}
grid_search = GridSearchCV(estimator=clf, param_grid=param_grid)
grid_search.fit(X_train, Y_train)
grid_search.best_params_
# # **Random Forest with Updated Parameters**
model2 = RandomForestClassifier(
n_estimators=20, min_samples_split=2, min_samples_leaf=1
)
model2.fit(X_train, Y_train)
ypred2 = clf2.predict(X_test)
accuracy2 = clf2.score(X_test, Y_test)
print(accuracy2)
print(classification_report(Y_test, ypred2))
# # **Logistic Regression Algorithm**
from sklearn.preprocessing import MinMaxScaler
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
model3 = Pipeline([("minmax", MinMaxScaler()), ("lr", LogisticRegression())])
model3.fit(X_train, Y_train)
ypred3 = pipe_lr.predict(X_test)
accuracy3 = pipe_lr.score(X_test, Y_test)
print(accuracy3)
print(classification_report(Y_test, ypred3))
plt.figure(figsize=(5, 3))
plt.bar(
["Decision Tree", "Random Forest", "Logistic regression"],
[accuracy1, accuracy2, accuracy3],
width=0.4,
)
plt.title("Accuracy Comparison")
plt.ylabel("Accuracy")
plt.ylim(0.0, 1.0)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/659/129659332.ipynb
|
indian-food-101
|
nehaprabhavalkar
|
[{"Id": 129659332, "ScriptId": 35527800, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11420283, "CreationDate": "05/15/2023 14:38:22", "VersionNumber": 3.0, "Title": "DM_project", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 150.0, "LinesInsertedFromPrevious": 88.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 62.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185960981, "KernelVersionId": 129659332, "SourceDatasetVersionId": 1526436}]
|
[{"Id": 1526436, "DatasetId": 865197, "DatasourceVersionId": 1560856, "CreatorUserId": 3885917, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "09/30/2020 06:23:43", "VersionNumber": 2.0, "Title": "Indian Food 101", "Slug": "indian-food-101", "Subtitle": "Data about 255 traditional and famous dishes in India", "Description": "### Content\n\nIndian cuisine consists of a variety of regional and traditional cuisines native to the Indian subcontinent. Given the diversity in soil, climate, culture, ethnic groups, and occupations, these cuisines vary substantially and use locally available spices, herbs, vegetables, and fruits. Indian food is also heavily influenced by religion, in particular Hinduism, cultural choices and traditions.\n\nThis dataset consists of information about various **Indian dishes**, their **ingredients**, their **place of origin**, etc.\n\n### Column Description\n\n**name** : name of the dish\n\n**ingredients** : main ingredients used\n\n**diet** : type of diet - either vegetarian or non vegetarian\n\n**prep_time** : preparation time\n\n**cook_time** : cooking time\n\n**flavor_profile** : flavor profile includes whether the dish is spicy, sweet, bitter, etc\n\n**course** : course of meal - starter, main course, dessert, etc\n\n**state** : state where the dish is famous or is originated\n\n**region** : region where the state belongs\n\nPresence of -1 in any of the columns indicates NaN value.\n\n### Acknowledgements\n\nhttps://www.wikipedia.org/\nhttps://hebbarskitchen.com/\nhttps://www.archanaskitchen.com/\n\n", "VersionNotes": "Version 2", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 865197, "CreatorUserId": 3885917, "OwnerUserId": 3885917.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1526436.0, "CurrentDatasourceVersionId": 1560856.0, "ForumId": 880553, "Type": 2, "CreationDate": "09/09/2020 07:36:01", "LastActivityDate": "09/09/2020", "TotalViews": 179626, "TotalDownloads": 22751, "TotalVotes": 529, "TotalKernels": 166}]
|
[{"Id": 3885917, "UserName": "nehaprabhavalkar", "DisplayName": "Neha Prabhavalkar", "RegisterDate": "10/19/2019", "PerformanceTier": 3}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = pd.read_csv("/kaggle/input/indian-food-101/indian_food.csv")
data
# Unique values for each columns
# print(pd.unique(data.ingredients))
print(pd.unique(data.flavor_profile))
print(pd.unique(data.course))
# print(pd.unique(data.diet))
# print(pd.unique(data.state))
### data.loc[data['state'] == '-1']
# Cleaning Data
# data.loc[data['flavor_profile']== '-1']
# data.loc[data['name']=='Copra paak','flavor_profile']
data.loc[
data["name"].isin(["Copra paak", "Puttu", "Kansar"]), "flavor_profile"
] = "sweet"
data.loc[data["flavor_profile"] == "-1", "flavor_profile"] = "other"
# Cleaned values
data.tail(10)
# Performing One hot encoding
discrete_df = pd.get_dummies(data, columns=["ingredients", "diet", "flavor_profile"])
discrete_df
target = data["course"]
df = discrete_df.drop(columns=["course", "state", "region", "name"], axis=1)
df
# # **Label Encoding**
# Converting target string to numeric labels
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
label = le.fit_transform(data["course"])
label
# 0='dessert' 1='main course' 2='starter' 3='snack'
# # **Train Test Split**
# Spliting data inro training set and testing set
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(
df, label, test_size=0.2, random_state=0
)
# len(X_test)
# # **Decision Tree Algorithm**
# implementing classification algorithm
from sklearn.tree import DecisionTreeClassifier
model1 = DecisionTreeClassifier()
model1.fit(X_train, Y_train)
# 0='dessert' 1='main course' 2='starter' 3='snack'
ypred = model1.predict(X_test)
ypredct = le.inverse_transform(ypred)
# print(ypredct)
# Checking accuracy
accuracy1 = model1.score(X_test, Y_test)
print(accuracy1)
from sklearn.metrics import classification_report
report1 = classification_report(Y_test, ypred)
print(type(report1))
# report=classification_report(Y_test, ypred,output_dict=True)
# macro_precision = report['macro avg']['precision']
# macro_precision
from sklearn import metrics
confusion_matrix = metrics.confusion_matrix(Y_test, ypred)
cm_display = metrics.ConfusionMatrixDisplay(
confusion_matrix=confusion_matrix, display_labels=[False, True]
)
import matplotlib.pyplot as plt
cm_display.plot()
plt.show()
# # **Random Forest Algorithm**
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
clf = RandomForestClassifier(n_estimators=2, min_samples_split=3, min_samples_leaf=2)
clf.fit(X_train, Y_train)
pred_clf = clf.predict(X_test)
accuracyN = clf.score(X_test, Y_test)
print(accuracyN)
reportN = classification_report(Y_test, pred_clf)
print(reportN)
from sklearn.model_selection import GridSearchCV
param_grid = {
"n_estimators": [2, 5, 10, 20],
"min_samples_split": [2, 3],
"min_samples_leaf": [1, 2, 3],
}
grid_search = GridSearchCV(estimator=clf, param_grid=param_grid)
grid_search.fit(X_train, Y_train)
grid_search.best_params_
# # **Random Forest with Updated Parameters**
model2 = RandomForestClassifier(
n_estimators=20, min_samples_split=2, min_samples_leaf=1
)
model2.fit(X_train, Y_train)
ypred2 = clf2.predict(X_test)
accuracy2 = clf2.score(X_test, Y_test)
print(accuracy2)
print(classification_report(Y_test, ypred2))
# # **Logistic Regression Algorithm**
from sklearn.preprocessing import MinMaxScaler
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
model3 = Pipeline([("minmax", MinMaxScaler()), ("lr", LogisticRegression())])
model3.fit(X_train, Y_train)
ypred3 = pipe_lr.predict(X_test)
accuracy3 = pipe_lr.score(X_test, Y_test)
print(accuracy3)
print(classification_report(Y_test, ypred3))
plt.figure(figsize=(5, 3))
plt.bar(
["Decision Tree", "Random Forest", "Logistic regression"],
[accuracy1, accuracy2, accuracy3],
width=0.4,
)
plt.title("Accuracy Comparison")
plt.ylabel("Accuracy")
plt.ylim(0.0, 1.0)
plt.show()
|
[{"indian-food-101/indian_food.csv": {"column_names": "[\"name\", \"ingredients\", \"diet\", \"prep_time\", \"cook_time\", \"flavor_profile\", \"course\", \"state\", \"region\"]", "column_data_types": "{\"name\": \"object\", \"ingredients\": \"object\", \"diet\": \"object\", \"prep_time\": \"int64\", \"cook_time\": \"int64\", \"flavor_profile\": \"object\", \"course\": \"object\", \"state\": \"object\", \"region\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 255 entries, 0 to 254\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 name 255 non-null object\n 1 ingredients 255 non-null object\n 2 diet 255 non-null object\n 3 prep_time 255 non-null int64 \n 4 cook_time 255 non-null int64 \n 5 flavor_profile 255 non-null object\n 6 course 255 non-null object\n 7 state 255 non-null object\n 8 region 254 non-null object\ndtypes: int64(2), object(7)\nmemory usage: 18.1+ KB\n", "summary": "{\"prep_time\": {\"count\": 255.0, \"mean\": 31.105882352941176, \"std\": 72.55440915682755, \"min\": -1.0, \"25%\": 10.0, \"50%\": 10.0, \"75%\": 20.0, \"max\": 500.0}, \"cook_time\": {\"count\": 255.0, \"mean\": 34.529411764705884, \"std\": 48.26564979817446, \"min\": -1.0, \"25%\": 20.0, \"50%\": 30.0, \"75%\": 40.0, \"max\": 720.0}}", "examples": "{\"name\":{\"0\":\"Balu shahi\",\"1\":\"Boondi\",\"2\":\"Gajar ka halwa\",\"3\":\"Ghevar\"},\"ingredients\":{\"0\":\"Maida flour, yogurt, oil, sugar\",\"1\":\"Gram flour, ghee, sugar\",\"2\":\"Carrots, milk, sugar, ghee, cashews, raisins\",\"3\":\"Flour, ghee, kewra, milk, clarified butter, sugar, almonds, pistachio, saffron, green cardamom\"},\"diet\":{\"0\":\"vegetarian\",\"1\":\"vegetarian\",\"2\":\"vegetarian\",\"3\":\"vegetarian\"},\"prep_time\":{\"0\":45,\"1\":80,\"2\":15,\"3\":15},\"cook_time\":{\"0\":25,\"1\":30,\"2\":60,\"3\":30},\"flavor_profile\":{\"0\":\"sweet\",\"1\":\"sweet\",\"2\":\"sweet\",\"3\":\"sweet\"},\"course\":{\"0\":\"dessert\",\"1\":\"dessert\",\"2\":\"dessert\",\"3\":\"dessert\"},\"state\":{\"0\":\"West Bengal\",\"1\":\"Rajasthan\",\"2\":\"Punjab\",\"3\":\"Rajasthan\"},\"region\":{\"0\":\"East\",\"1\":\"West\",\"2\":\"North\",\"3\":\"West\"}}"}}]
| true | 1 |
<start_data_description><data_path>indian-food-101/indian_food.csv:
<column_names>
['name', 'ingredients', 'diet', 'prep_time', 'cook_time', 'flavor_profile', 'course', 'state', 'region']
<column_types>
{'name': 'object', 'ingredients': 'object', 'diet': 'object', 'prep_time': 'int64', 'cook_time': 'int64', 'flavor_profile': 'object', 'course': 'object', 'state': 'object', 'region': 'object'}
<dataframe_Summary>
{'prep_time': {'count': 255.0, 'mean': 31.105882352941176, 'std': 72.55440915682755, 'min': -1.0, '25%': 10.0, '50%': 10.0, '75%': 20.0, 'max': 500.0}, 'cook_time': {'count': 255.0, 'mean': 34.529411764705884, 'std': 48.26564979817446, 'min': -1.0, '25%': 20.0, '50%': 30.0, '75%': 40.0, 'max': 720.0}}
<dataframe_info>
RangeIndex: 255 entries, 0 to 254
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 name 255 non-null object
1 ingredients 255 non-null object
2 diet 255 non-null object
3 prep_time 255 non-null int64
4 cook_time 255 non-null int64
5 flavor_profile 255 non-null object
6 course 255 non-null object
7 state 255 non-null object
8 region 254 non-null object
dtypes: int64(2), object(7)
memory usage: 18.1+ KB
<some_examples>
{'name': {'0': 'Balu shahi', '1': 'Boondi', '2': 'Gajar ka halwa', '3': 'Ghevar'}, 'ingredients': {'0': 'Maida flour, yogurt, oil, sugar', '1': 'Gram flour, ghee, sugar', '2': 'Carrots, milk, sugar, ghee, cashews, raisins', '3': 'Flour, ghee, kewra, milk, clarified butter, sugar, almonds, pistachio, saffron, green cardamom'}, 'diet': {'0': 'vegetarian', '1': 'vegetarian', '2': 'vegetarian', '3': 'vegetarian'}, 'prep_time': {'0': 45, '1': 80, '2': 15, '3': 15}, 'cook_time': {'0': 25, '1': 30, '2': 60, '3': 30}, 'flavor_profile': {'0': 'sweet', '1': 'sweet', '2': 'sweet', '3': 'sweet'}, 'course': {'0': 'dessert', '1': 'dessert', '2': 'dessert', '3': 'dessert'}, 'state': {'0': 'West Bengal', '1': 'Rajasthan', '2': 'Punjab', '3': 'Rajasthan'}, 'region': {'0': 'East', '1': 'West', '2': 'North', '3': 'West'}}
<end_description>
| 1,500 | 0 | 2,481 | 1,500 |
129659135
|
# GeoJson file load
import json
JPgeo = json.load(open("/kaggle/input/jpgeojson/japan.geojson"))
JPgeo["features"][0]["properties"]
JPgeo["features"][0]["geometry"]
import folium
folium.Map(location=[35.6938, 139.7035], zoom_start=10)
import folium
JPmap = folium.Map(location=[35.6938, 139.7035], zoom_start=10, tiles="cartodbpositron")
folium.Choropleth(geo_data=JPgeo).add_to(JPmap)
JPmap
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/659/129659135.ipynb
| null | null |
[{"Id": 129659135, "ScriptId": 38510275, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14391720, "CreationDate": "05/15/2023 14:36:53", "VersionNumber": 1.0, "Title": "MapVisualizing", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 17.0, "LinesInsertedFromPrevious": 17.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# GeoJson file load
import json
JPgeo = json.load(open("/kaggle/input/jpgeojson/japan.geojson"))
JPgeo["features"][0]["properties"]
JPgeo["features"][0]["geometry"]
import folium
folium.Map(location=[35.6938, 139.7035], zoom_start=10)
import folium
JPmap = folium.Map(location=[35.6938, 139.7035], zoom_start=10, tiles="cartodbpositron")
folium.Choropleth(geo_data=JPgeo).add_to(JPmap)
JPmap
| false | 0 | 165 | 0 | 165 | 165 |
||
129723093
|
<jupyter_start><jupyter_text>Netflix Data: Cleaning, Analysis and Visualization
Netflix is a popular streaming service that offers a vast catalog of movies, TV shows, and original contents. This dataset is a cleaned version of the original version which can be found [here](https://www.kaggle.com/datasets/shivamb/netflix-shows). The data consist of contents added to Netflix from 2008 to 2021. The oldest content is as old as 1925 and the newest as 2021. This dataset will be cleaned with PostgreSQL and visualized with Tableau. The purpose of this dataset is to test my data cleaning and visualization skills. The cleaned data can be found below and the Tableau dashboard can be found [here](https://public.tableau.com/app/profile/abdulrasaq.ariyo/viz/NetflixTVShowsMovies_16615029026580/NetflixDashboard) .
## Data Cleaning
We are going to:
1. Treat the Nulls
2. Treat the duplicates
3. Populate missing rows
4. Drop unneeded columns
5. Split columns
Extra steps and more explanation on the process will be explained through the code comments
```
--View dataset
SELECT *
FROM netflix;
```
```
--The show_id column is the unique id for the dataset, therefore we are going to check for duplicates
SELECT show_id, COUNT(*)
FROM netflix
GROUP BY show_id
ORDER BY show_id DESC;
--No duplicates
```
```
--Check null values across columns
SELECT COUNT(*) FILTER (WHERE show_id IS NULL) AS showid_nulls,
COUNT(*) FILTER (WHERE type IS NULL) AS type_nulls,
COUNT(*) FILTER (WHERE title IS NULL) AS title_nulls,
COUNT(*) FILTER (WHERE director IS NULL) AS director_nulls,
COUNT(*) FILTER (WHERE movie_cast IS NULL) AS movie_cast_nulls,
COUNT(*) FILTER (WHERE country IS NULL) AS country_nulls,
COUNT(*) FILTER (WHERE date_added IS NULL) AS date_addes_nulls,
COUNT(*) FILTER (WHERE release_year IS NULL) AS release_year_nulls,
COUNT(*) FILTER (WHERE rating IS NULL) AS rating_nulls,
COUNT(*) FILTER (WHERE duration IS NULL) AS duration_nulls,
COUNT(*) FILTER (WHERE listed_in IS NULL) AS listed_in_nulls,
COUNT(*) FILTER (WHERE description IS NULL) AS description_nulls
FROM netflix;
```
```
We can see that there are NULLS.
director_nulls = 2634
movie_cast_nulls = 825
country_nulls = 831
date_added_nulls = 10
rating_nulls = 4
duration_nulls = 3
```
The director column nulls is about 30% of the whole column, therefore I will not delete them. I will rather find another column to populate it. To populate the director column, we want to find out if there is relationship between movie_cast column and director column
```
-- Below, we find out if some directors are likely to work with particular cast
WITH cte AS
(
SELECT title, CONCAT(director, '---', movie_cast) AS director_cast
FROM netflix
)
SELECT director_cast, COUNT(*) AS count
FROM cte
GROUP BY director_cast
HAVING COUNT(*) > 1
ORDER BY COUNT(*) DESC;
With this, we can now populate NULL rows in directors
using their record with movie_cast
```
```
UPDATE netflix
SET director = 'Alastair Fothergill'
WHERE movie_cast = 'David Attenborough'
AND director IS NULL ;
--Repeat this step to populate the rest of the director nulls
--Populate the rest of the NULL in director as "Not Given"
UPDATE netflix
SET director = 'Not Given'
WHERE director IS NULL;
--When I was doing this, I found a less complex and faster way to populate a column which I will use next
```
Just like the director column, I will not delete the nulls in country. Since the country column is related to director and movie, we are going to populate the country column with the director column
```
--Populate the country using the director column
SELECT COALESCE(nt.country,nt2.country)
FROM netflix AS nt
JOIN netflix AS nt2
ON nt.director = nt2.director
AND nt.show_id <> nt2.show_id
WHERE nt.country IS NULL;
UPDATE netflix
SET country = nt2.country
FROM netflix AS nt2
WHERE netflix.director = nt2.director and netflix.show_id <> nt2.show_id
AND netflix.country IS NULL;
--To confirm if there are still directors linked to country that refuse to update
SELECT director, country, date_added
FROM netflix
WHERE country IS NULL;
--Populate the rest of the NULL in director as "Not Given"
UPDATE netflix
SET country = 'Not Given'
WHERE country IS NULL;
```
The date_added rows nulls is just 10 out of over 8000 rows, deleting them cannot affect our analysis or visualization
```
--Show date_added nulls
SELECT show_id, date_added
FROM netflix_clean
WHERE date_added IS NULL;
--DELETE nulls
DELETE FROM netflix
WHERE show_id
IN ('6797', 's6067', 's6175', 's6807', 's6902', 's7255', 's7197', 's7407', 's7848', 's8183');
```
rating nulls is 4. Delete them
```
--Show rating NULLS
SELECT show_id, rating
FROM netflix_clean
WHERE date_added IS NULL;
--Delete the nulls, and show deleted fields
DELETE FROM netflix
WHERE show_id
IN (SELECT show_id FROM netflix WHERE rating IS NULL)
RETURNING *;
```
--duration nulls is 4. Delete them
```
DELETE FROM netflix
WHERE show_id
IN (SELECT show_id FROM netflix WHERE duration IS NULL);
```
Now run the query to show the number of nulls in each column to confirm if there are still nulls. After this, run the query to confirm the row number in each column is the same
```
--Check to confirm the number of rows are the same(NO NULL)
SELECT count(*) filter (where show_id IS NOT NULL) AS showid_nulls,
count(*) filter (where type IS NOT NULL) AS type_nulls,
count(*) filter (where title IS NOT NULL) AS title_nulls,
count(*) filter (where director IS NOT NULL) AS director_nulls,
count(*) filter (where country IS NOT NULL) AS country_nulls,
count(*) filter (where date_added IS NOT NULL) AS date_addes_nulls,
count(*) filter (where release_year IS NOT NULL) AS release_year_nulls,
count(*) filter (where rating IS NOT NULL) AS rating_nulls,
count(*) filter (where duration IS NOT NULL) AS duration_nulls,
count(*) filter (where listed_in IS NOT NULL) AS listed_in_nulls
FROM netflix;
--Total number of rows are the same in all columns
```
We can drop the description and movie_cast column because they are not needed for our analysis or visualization task.
```
--DROP unneeded columns
ALTER TABLE netflix
DROP COLUMN movie_cast,
DROP COLUMN description;
```
Some of the rows in country column has multiple countries, for my visualization, I only need one country per row to make my map visualization clean and easy. Therefore, I am going to split the country column and retain the first country by the left which I believe is the original country of the movie
```
SELECT *,
SPLIT_PART(country,',',1) AS countryy,
SPLIT_PART(country,',',2),
SPLIT_PART(country,',',4),
SPLIT_PART(country,',',5),
SPLIT_PART(country,',',6),
SPLIT_PART(country,',',7),
SPLIT_PART(country,',',8),
SPLIT_PART(country,',',9),
SPLIT_PART(country,',',10)
FROM netflix;
-- NOW lets update the table
ALTER TABLE netflix
ADD country1 varchar(500);
UPDATE netflix
SET country1 = SPLIT_PART(country, ',', 1);
--This will create a column named country1 and Update it with the first split country.
```
Delete the country column that has multiple country entries
```
--Delete column
ALTER TABLE netflix
DROP COLUMN country;
```
Rename the country1 column to country
```
ALTER TABLE netflix
RENAME COLUMN country1 TO country;
```
## Data Visualization
After cleaning, the dataset is set for some analysis and visualization with Tableau.
**Note: In the visualization captions, Contents means Movies and TV shows, and Content may either mean Movie or TV Show**.
**Sheet 1. Content type in percentage**

This first sheet shows the two categories of content in the dataset which are Movie and Tv show.
- As we can see the majority of the content is Movie which takes 69.9%.
- There are more details in the tooltip which shows the exact count of Movie and Tv show
**Sheet 2. Movie & TV Show by Country**

This shows the the total amount of Movies and Tv shows per country within the given period of time(2008 - 2021). This can be noted by the size of the coloured circle in the map.
- We can see that the United State of America has the largest size, followed by India and the United Kingdom.
- In the Tableau hosted dashboard/sheet, there is a filter for the years between 2008 and 2021 to calculate yearly record.
To give an alternate and a clearer view. Movie & TV shows by country bar chart is below

**Sheet 3. Number of Contents Added through the Years**

This time series chart shows the total number of contents added to Netflix all through the given years (2008 - 2021)
- It shows that most movies and tv shows on Netflix were added in 2019
- In the Tableau sheet, there is a filter to know how much Movies and Tv shows were added in each month of the year
**Sheet 4. Top Directors**

This chart shows the top 10 directors with most contents on Netflix. This char shows the count of Movie and Tv shows in their catalouge.
- We can see that most of these directors contents are movies.
- We can also note that the duo of Raul Campos and Jan Suter are fond of working together and have directed 18 movies on Netflix.
**Sheet 5. Top Genres**

This chart shows the genres with the highest numbers on Netflix.
- We can see that Drama & International movies followed by Documentary have the highest number of contents on Netflix within the period.
**Sheet 6. Top Ratings**

Rating is a system to rate motion picture's suitability for certain audiences based on its content. This chart shows the top ratings on Netflix
-We can note that most contents on Netflix are rated TV-MA. TV-MA in the United States by the TV Parental Guidelines signifies content for mature audiences.
**Sheet 7. Oldest Contents on Netflix by Content Release year**

This table shows the 10 oldest movies and tv shows on Netflix
- The oldest is as old as 1925
**Sheet 8. Content Types over the Years**

This line chart compares the Movie and Tv shows contents added to Netflix all through the years.
- We can see that more movies have always been added.
- In 2013, the number of contents added to Netflix for both were almost the same with Movies having 6 contents that year and Tv shows having 5.
- It shows that in the first 5 years, only movies were added to Netflix.
**Sheet 9. Release Years with Highest Contents**

This chart shows the Movies and Tv shows production year which has with highest contents on Netflix. We focus on the top 10 release year/production year.
-We can see that from 2012 to 2018, Netflix added most recent contents, they made sure most recent contents per release year are higher than the older release year contents. Then in 2019, it started dropping, this may be due to the Covid-19, but further analysis may be needed to determine this.
And with this, I have come to the end of this exercise. As I said this is just an exercise to test my skills as I look forward to be better. Thanks for following through. Cheers!
Kaggle dataset identifier: netflix-data-cleaning-analysis-and-visualization
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # data visualization
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# read data csv
df = pd.read_csv(
"/kaggle/input/netflix-data-cleaning-analysis-and-visualization/netflix1.csv"
)
# look into dataframe
df.head()
# Summary
# * show_id: netflix show id
# * type: type of show (movie or TV show)
# * title: title of the show
# * director: name of director
# * country: country origin of the show
# * date_added: recorded date time when the movie added to netflix
# * release_year: year when the show released
# * rating: the age restriction
# * duration: duration of the show
# * listed_in: the show category
# dataframe shape
df.shape
# dataframe information
df.info()
# We see that one feature is numeric (int64), and 9 features are object. In addition, we also can identifiy if there are any missing values. Here, there are none because each column contains 8790 observations, the same number of rows we saw before with shape.
# One strange thing that date_added was object, may be better to changes into string??
# inspect any null value
df.describe(include="all")
# look into countries in the list
df["country"].unique()
# change data type of date_added from object to string and extract the year into different column
df["date_added"] = df["date_added"].astype("string")
df["year_added"] = df["date_added"].str.extract("(\d{4})", expand=True)
df["year_added"] = df["year_added"].astype("int64")
df.info()
# We can see 'Not Given' country name in the second row above. Means that the coloumns is not empty but the origin country of several shows were unkown.
df.nunique(axis=0)
# There are only two type of show in the dataframe, and the show are from 86 countries. We can draw the categorical data from type, country, release year, and rating.
# **Next, we shall see from data visualization.**
# see the proportion type of the show
y = df.groupby("type")["show_id"].count()
labels = ["Movie", "TV Show"]
plt.pie(y, labels=labels, autopct="%1.1f%%", startangle=90)
plt.title("The proportion of the type of shows in Netflix")
plt.show()
# There are a lot of movie in Netflix with approx 70% from total.
# top ten coutries with the majority of shows
per_country = df.groupby("country")["show_id"].count()
per_country.sort_values(ascending=False).head(10).plot(kind="bar")
plt.title("Top ten countries with the most shows on Netflix")
plt.xlabel("country")
plt.ylabel("number of shows")
plt.show()
# Majority of shows in Netflix originates from United states with the rough total 3300 shows. Following by India with the total around 1000 shows, and UK with the total show about 200 fewer than India. Apart from these three countries, shows from other countries (on the list) only have less than 500 shows on Netflix.
country_n_year = df.groupby(["country", "release_year"])["show_id"].count()
labels = [
"United States",
"India",
"United Kingdom",
"Pakistan",
"Canada",
"Japan",
"South Korea",
"France",
"Spain",
]
selected = country_n_year.loc[labels]
selected = selected.sort_index(
level=["country", "release_year"], ascending=[False, True]
)
legenda = selected.index.unique("release_year").sort_values(ascending=True)
selected.unstack().plot(kind="bar", stacked=True)
plt.title(
"The top 10 countries with the most shows on Netflix (displayed by year of release)"
)
plt.ylabel("number of shows")
plt.xlabel("countries")
plt.legend(
title="year release",
labels=legenda,
bbox_to_anchor=(1.0, 1.0),
loc="upper left",
ncol=3,
fontsize=8,
)
plt.show()
# rating
country_n_rating = df.groupby(["country", "rating"])["show_id"].count()
labels = [
"United States",
"India",
"United Kingdom",
"Pakistan",
"Canada",
"Japan",
"South Korea",
"France",
"Spain",
]
selected = country_n_rating.loc[labels]
selected.unstack().plot(kind="bar", stacked=True)
plt.title(
"Top 10 countries with most shows on Netflix differentiated by rating of the shows"
)
plt.ylabel("number of shows")
plt.xlabel("countries")
plt.legend(title="rating", bbox_to_anchor=(1.0, 1.0), loc="upper left")
plt.show()
# Overall, the top ten countries have more TV-MA ratings than any other. India has more TV-14 ratings than any other country, while the US has the most R ratings.
# in what year most produced show available in Netflix
country_year = df.groupby("release_year")["show_id"].count()
country_year_10 = country_year.sort_values(ascending=False).head(10)
plt.plot(country_year_10.sort_index(), marker="o")
plt.title("The year of production of the show most available on Netflix")
plt.xlabel("country")
plt.ylabel("number of shows")
plt.show()
# Of all the shows added to Netflix from all 86 countries, the most shows were released in 2018 which is twice the number of shows released in the most recent year.
# is show on netflix availabe at the same time as the year when the show produced
# comparing the date added and year released
df["year_diff"] = df.apply(
lambda x: True if x["release_year"] == x["year_added"] else False, axis=1
)
print(df["year_diff"].value_counts())
# Netflix started at very first in **1997** as a **website service**. During the time it allowed people to rent DVDs online. So that renter can get them through the mail. However, Netflix started their **streaming** platfrom in **2007**. On January 6, 2016, Netflix went live in 130 countries simultaneously.
# We can see from the result that there are a lot more show that added after the year released.
# look into details on the False category
year_false = df[df["year_diff"] == False]
plt.scatter(data=year_false, y="release_year", x="year_added")
plt.ylabel("release year")
plt.xlabel("year added")
plt.show()
# There are a lot of shows that started to be added on the streaming platform Netflix after 2016. Even the old shows from 1960 and older were added many years later in Netflix.
# director with the most produced show available in netflix
df.groupby("director")["show_id"].count().sort_values(ascending=False).head(10)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/723/129723093.ipynb
|
netflix-data-cleaning-analysis-and-visualization
|
ariyoomotade
|
[{"Id": 129723093, "ScriptId": 38063008, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14103493, "CreationDate": "05/16/2023 03:18:17", "VersionNumber": 3.0, "Title": "Data analysis - Netflix", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 156.0, "LinesInsertedFromPrevious": 34.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 122.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186063376, "KernelVersionId": 129723093, "SourceDatasetVersionId": 4123716}]
|
[{"Id": 4123716, "DatasetId": 2437124, "DatasourceVersionId": 4180064, "CreatorUserId": 10322697, "LicenseName": "CC0: Public Domain", "CreationDate": "08/26/2022 09:25:43", "VersionNumber": 1.0, "Title": "Netflix Data: Cleaning, Analysis and Visualization", "Slug": "netflix-data-cleaning-analysis-and-visualization", "Subtitle": "Cleaning and Visualization with Pgsql and Tableau", "Description": "Netflix is a popular streaming service that offers a vast catalog of movies, TV shows, and original contents. This dataset is a cleaned version of the original version which can be found [here](https://www.kaggle.com/datasets/shivamb/netflix-shows). The data consist of contents added to Netflix from 2008 to 2021. The oldest content is as old as 1925 and the newest as 2021. This dataset will be cleaned with PostgreSQL and visualized with Tableau. The purpose of this dataset is to test my data cleaning and visualization skills. The cleaned data can be found below and the Tableau dashboard can be found [here](https://public.tableau.com/app/profile/abdulrasaq.ariyo/viz/NetflixTVShowsMovies_16615029026580/NetflixDashboard) . \n\n## Data Cleaning\nWe are going to:\n1. Treat the Nulls \n2. Treat the duplicates\n3. Populate missing rows\n4. Drop unneeded columns\n5. Split columns\nExtra steps and more explanation on the process will be explained through the code comments\n\n```\n--View dataset\n\nSELECT * \nFROM netflix;\n\n```\n\n```\n--The show_id column is the unique id for the dataset, therefore we are going to check for duplicates\n \nSELECT show_id, COUNT(*) \nFROM netflix \nGROUP BY show_id \nORDER BY show_id DESC;\n\n--No duplicates\n```\n\n```\n--Check null values across columns\n\nSELECT COUNT(*) FILTER (WHERE show_id IS NULL) AS showid_nulls,\n COUNT(*) FILTER (WHERE type IS NULL) AS type_nulls,\n COUNT(*) FILTER (WHERE title IS NULL) AS title_nulls,\n COUNT(*) FILTER (WHERE director IS NULL) AS director_nulls,\n\t COUNT(*) FILTER (WHERE movie_cast IS NULL) AS movie_cast_nulls,\n\t COUNT(*) FILTER (WHERE country IS NULL) AS country_nulls,\n COUNT(*) FILTER (WHERE date_added IS NULL) AS date_addes_nulls,\n COUNT(*) FILTER (WHERE release_year IS NULL) AS release_year_nulls,\n COUNT(*) FILTER (WHERE rating IS NULL) AS rating_nulls,\n\t COUNT(*) FILTER (WHERE duration IS NULL) AS duration_nulls,\n COUNT(*) FILTER (WHERE listed_in IS NULL) AS listed_in_nulls,\n\t COUNT(*) FILTER (WHERE description IS NULL) AS description_nulls\nFROM netflix;\n```\n```\nWe can see that there are NULLS. \ndirector_nulls = 2634\nmovie_cast_nulls = 825\ncountry_nulls = 831\ndate_added_nulls = 10\nrating_nulls = 4\nduration_nulls = 3 \n```\n\nThe director column nulls is about 30% of the whole column, therefore I will not delete them. I will rather find another column to populate it. To populate the director column, we want to find out if there is relationship between movie_cast column and director column\n\n\n``` \n-- Below, we find out if some directors are likely to work with particular cast\n\nWITH cte AS\n(\nSELECT title, CONCAT(director, '---', movie_cast) AS director_cast \nFROM netflix\n)\n\nSELECT director_cast, COUNT(*) AS count\nFROM cte\nGROUP BY director_cast\nHAVING COUNT(*) > 1\nORDER BY COUNT(*) DESC;\n\nWith this, we can now populate NULL rows in directors \nusing their record with movie_cast \n```\n```\nUPDATE netflix \nSET director = 'Alastair Fothergill'\nWHERE movie_cast = 'David Attenborough'\nAND director IS NULL ;\n\n--Repeat this step to populate the rest of the director nulls\n--Populate the rest of the NULL in director as \"Not Given\"\n\nUPDATE netflix \nSET director = 'Not Given'\nWHERE director IS NULL;\n\n--When I was doing this, I found a less complex and faster way to populate a column which I will use next\n```\n\nJust like the director column, I will not delete the nulls in country. Since the country column is related to director and movie, we are going to populate the country column with the director column\n\n```\n--Populate the country using the director column\n\nSELECT COALESCE(nt.country,nt2.country) \nFROM netflix AS nt\nJOIN netflix AS nt2 \nON nt.director = nt2.director \nAND nt.show_id <> nt2.show_id\nWHERE nt.country IS NULL;\nUPDATE netflix\nSET country = nt2.country\nFROM netflix AS nt2\nWHERE netflix.director = nt2.director and netflix.show_id <> nt2.show_id \nAND netflix.country IS NULL;\n\n\n--To confirm if there are still directors linked to country that refuse to update\n\nSELECT director, country, date_added\nFROM netflix\nWHERE country IS NULL;\n\n--Populate the rest of the NULL in director as \"Not Given\"\n\nUPDATE netflix \nSET country = 'Not Given'\nWHERE country IS NULL;\n```\n\nThe date_added rows nulls is just 10 out of over 8000 rows, deleting them cannot affect our analysis or visualization\n\n```\n--Show date_added nulls\n\nSELECT show_id, date_added\nFROM netflix_clean\nWHERE date_added IS NULL;\n\n--DELETE nulls\n\nDELETE FROM netflix\nWHERE show_id \nIN ('6797', 's6067', 's6175', 's6807', 's6902', 's7255', 's7197', 's7407', 's7848', 's8183');\n\n```\n\nrating nulls is 4. Delete them\n```\n--Show rating NULLS\n\nSELECT show_id, rating\nFROM netflix_clean\nWHERE date_added IS NULL;\n\n--Delete the nulls, and show deleted fields\nDELETE FROM netflix \nWHERE show_id \nIN (SELECT show_id FROM netflix WHERE rating IS NULL)\nRETURNING *;\n```\n\n--duration nulls is 4. Delete them\n```\n\nDELETE FROM netflix \nWHERE show_id \nIN (SELECT show_id FROM netflix WHERE duration IS NULL);\n```\nNow run the query to show the number of nulls in each column to confirm if there are still nulls. After this, run the query to confirm the row number in each column is the same\n\n```\n--Check to confirm the number of rows are the same(NO NULL)\n\nSELECT count(*) filter (where show_id IS NOT NULL) AS showid_nulls,\n count(*) filter (where type IS NOT NULL) AS type_nulls,\n count(*) filter (where title IS NOT NULL) AS title_nulls,\n count(*) filter (where director IS NOT NULL) AS director_nulls,\n\t count(*) filter (where country IS NOT NULL) AS country_nulls,\n count(*) filter (where date_added IS NOT NULL) AS date_addes_nulls,\n count(*) filter (where release_year IS NOT NULL) AS release_year_nulls,\n count(*) filter (where rating IS NOT NULL) AS rating_nulls,\n\t count(*) filter (where duration IS NOT NULL) AS duration_nulls,\n count(*) filter (where listed_in IS NOT NULL) AS listed_in_nulls\nFROM netflix;\n\n --Total number of rows are the same in all columns\n```\nWe can drop the description and movie_cast column because they are not needed for our analysis or visualization task. \n```\n--DROP unneeded columns\n\nALTER TABLE netflix\nDROP COLUMN movie_cast, \nDROP COLUMN description;\n```\nSome of the rows in country column has multiple countries, for my visualization, I only need one country per row to make my map visualization clean and easy. Therefore, I am going to split the country column and retain the first country by the left which I believe is the original country of the movie\n```\nSELECT *,\n\t SPLIT_PART(country,',',1) AS countryy, \n SPLIT_PART(country,',',2),\n\t SPLIT_PART(country,',',4),\n\t SPLIT_PART(country,',',5),\n\t SPLIT_PART(country,',',6),\n\t SPLIT_PART(country,',',7),\n\t SPLIT_PART(country,',',8),\n\t SPLIT_PART(country,',',9),\n\t SPLIT_PART(country,',',10) \n\t \nFROM netflix;\n\t \n-- NOW lets update the table\n\nALTER TABLE netflix \nADD country1 varchar(500);\nUPDATE netflix \nSET country1 = SPLIT_PART(country, ',', 1);\n\n--This will create a column named country1 and Update it with the first split country.\n```\n\nDelete the country column that has multiple country entries\n```\n--Delete column\nALTER TABLE netflix \nDROP COLUMN country;\n```\nRename the country1 column to country\n```\nALTER TABLE netflix \nRENAME COLUMN country1 TO country;\n```\n\n## Data Visualization\nAfter cleaning, the dataset is set for some analysis and visualization with Tableau. \n\n**Note: In the visualization captions, Contents means Movies and TV shows, and Content may either mean Movie or TV Show**. \n\n**Sheet 1. Content type in percentage**\n\n\n\nThis first sheet shows the two categories of content in the dataset which are Movie and Tv show. \n- As we can see the majority of the content is Movie which takes 69.9%. \n- There are more details in the tooltip which shows the exact count of Movie and Tv show\n\n\n**Sheet 2. Movie & TV Show by Country**\n\n\n\nThis shows the the total amount of Movies and Tv shows per country within the given period of time(2008 - 2021). This can be noted by the size of the coloured circle in the map. \n- We can see that the United State of America has the largest size, followed by India and the United Kingdom. \n- In the Tableau hosted dashboard/sheet, there is a filter for the years between 2008 and 2021 to calculate yearly record.\n\n To give an alternate and a clearer view. Movie & TV shows by country bar chart is below\n\n\n\n**Sheet 3. Number of Contents Added through the Years**\n\n\n\nThis time series chart shows the total number of contents added to Netflix all through the given years (2008 - 2021)\n- It shows that most movies and tv shows on Netflix were added in 2019\n- In the Tableau sheet, there is a filter to know how much Movies and Tv shows were added in each month of the year \n\n\n**Sheet 4. Top Directors**\n\n\n\nThis chart shows the top 10 directors with most contents on Netflix. This char shows the count of Movie and Tv shows in their catalouge. \n- We can see that most of these directors contents are movies. \n- We can also note that the duo of Raul Campos and Jan Suter are fond of working together and have directed 18 movies on Netflix. \n\n\n**Sheet 5. Top Genres** \n\n\n\nThis chart shows the genres with the highest numbers on Netflix. \n- We can see that Drama & International movies followed by Documentary have the highest number of contents on Netflix within the period.\n\n\n**Sheet 6. Top Ratings**\n\n\n \nRating is a system to rate motion picture's suitability for certain audiences based on its content. This chart shows the top ratings on Netflix\n-We can note that most contents on Netflix are rated TV-MA. TV-MA in the United States by the TV Parental Guidelines signifies content for mature audiences. \n\n\n**Sheet 7. Oldest Contents on Netflix by Content Release year**\n\n\n\nThis table shows the 10 oldest movies and tv shows on Netflix\n- The oldest is as old as 1925\n\n**Sheet 8. Content Types over the Years**\n\n\nThis line chart compares the Movie and Tv shows contents added to Netflix all through the years.\n- We can see that more movies have always been added. \n- In 2013, the number of contents added to Netflix for both were almost the same with Movies having 6 contents that year and Tv shows having 5.\n- It shows that in the first 5 years, only movies were added to Netflix. \n\n\n**Sheet 9. Release Years with Highest Contents**\n\n\n\nThis chart shows the Movies and Tv shows production year which has with highest contents on Netflix. We focus on the top 10 release year/production year. \n-We can see that from 2012 to 2018, Netflix added most recent contents, they made sure most recent contents per release year are higher than the older release year contents. Then in 2019, it started dropping, this may be due to the Covid-19, but further analysis may be needed to determine this. \n\n And with this, I have come to the end of this exercise. As I said this is just an exercise to test my skills as I look forward to be better. Thanks for following through. Cheers!", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2437124, "CreatorUserId": 10322697, "OwnerUserId": 10322697.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4123716.0, "CurrentDatasourceVersionId": 4180064.0, "ForumId": 2464656, "Type": 2, "CreationDate": "08/26/2022 09:25:43", "LastActivityDate": "08/26/2022", "TotalViews": 96354, "TotalDownloads": 16114, "TotalVotes": 270, "TotalKernels": 23}]
|
[{"Id": 10322697, "UserName": "ariyoomotade", "DisplayName": "Abdulrasaq Ariyo", "RegisterDate": "04/22/2022", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # data visualization
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# read data csv
df = pd.read_csv(
"/kaggle/input/netflix-data-cleaning-analysis-and-visualization/netflix1.csv"
)
# look into dataframe
df.head()
# Summary
# * show_id: netflix show id
# * type: type of show (movie or TV show)
# * title: title of the show
# * director: name of director
# * country: country origin of the show
# * date_added: recorded date time when the movie added to netflix
# * release_year: year when the show released
# * rating: the age restriction
# * duration: duration of the show
# * listed_in: the show category
# dataframe shape
df.shape
# dataframe information
df.info()
# We see that one feature is numeric (int64), and 9 features are object. In addition, we also can identifiy if there are any missing values. Here, there are none because each column contains 8790 observations, the same number of rows we saw before with shape.
# One strange thing that date_added was object, may be better to changes into string??
# inspect any null value
df.describe(include="all")
# look into countries in the list
df["country"].unique()
# change data type of date_added from object to string and extract the year into different column
df["date_added"] = df["date_added"].astype("string")
df["year_added"] = df["date_added"].str.extract("(\d{4})", expand=True)
df["year_added"] = df["year_added"].astype("int64")
df.info()
# We can see 'Not Given' country name in the second row above. Means that the coloumns is not empty but the origin country of several shows were unkown.
df.nunique(axis=0)
# There are only two type of show in the dataframe, and the show are from 86 countries. We can draw the categorical data from type, country, release year, and rating.
# **Next, we shall see from data visualization.**
# see the proportion type of the show
y = df.groupby("type")["show_id"].count()
labels = ["Movie", "TV Show"]
plt.pie(y, labels=labels, autopct="%1.1f%%", startangle=90)
plt.title("The proportion of the type of shows in Netflix")
plt.show()
# There are a lot of movie in Netflix with approx 70% from total.
# top ten coutries with the majority of shows
per_country = df.groupby("country")["show_id"].count()
per_country.sort_values(ascending=False).head(10).plot(kind="bar")
plt.title("Top ten countries with the most shows on Netflix")
plt.xlabel("country")
plt.ylabel("number of shows")
plt.show()
# Majority of shows in Netflix originates from United states with the rough total 3300 shows. Following by India with the total around 1000 shows, and UK with the total show about 200 fewer than India. Apart from these three countries, shows from other countries (on the list) only have less than 500 shows on Netflix.
country_n_year = df.groupby(["country", "release_year"])["show_id"].count()
labels = [
"United States",
"India",
"United Kingdom",
"Pakistan",
"Canada",
"Japan",
"South Korea",
"France",
"Spain",
]
selected = country_n_year.loc[labels]
selected = selected.sort_index(
level=["country", "release_year"], ascending=[False, True]
)
legenda = selected.index.unique("release_year").sort_values(ascending=True)
selected.unstack().plot(kind="bar", stacked=True)
plt.title(
"The top 10 countries with the most shows on Netflix (displayed by year of release)"
)
plt.ylabel("number of shows")
plt.xlabel("countries")
plt.legend(
title="year release",
labels=legenda,
bbox_to_anchor=(1.0, 1.0),
loc="upper left",
ncol=3,
fontsize=8,
)
plt.show()
# rating
country_n_rating = df.groupby(["country", "rating"])["show_id"].count()
labels = [
"United States",
"India",
"United Kingdom",
"Pakistan",
"Canada",
"Japan",
"South Korea",
"France",
"Spain",
]
selected = country_n_rating.loc[labels]
selected.unstack().plot(kind="bar", stacked=True)
plt.title(
"Top 10 countries with most shows on Netflix differentiated by rating of the shows"
)
plt.ylabel("number of shows")
plt.xlabel("countries")
plt.legend(title="rating", bbox_to_anchor=(1.0, 1.0), loc="upper left")
plt.show()
# Overall, the top ten countries have more TV-MA ratings than any other. India has more TV-14 ratings than any other country, while the US has the most R ratings.
# in what year most produced show available in Netflix
country_year = df.groupby("release_year")["show_id"].count()
country_year_10 = country_year.sort_values(ascending=False).head(10)
plt.plot(country_year_10.sort_index(), marker="o")
plt.title("The year of production of the show most available on Netflix")
plt.xlabel("country")
plt.ylabel("number of shows")
plt.show()
# Of all the shows added to Netflix from all 86 countries, the most shows were released in 2018 which is twice the number of shows released in the most recent year.
# is show on netflix availabe at the same time as the year when the show produced
# comparing the date added and year released
df["year_diff"] = df.apply(
lambda x: True if x["release_year"] == x["year_added"] else False, axis=1
)
print(df["year_diff"].value_counts())
# Netflix started at very first in **1997** as a **website service**. During the time it allowed people to rent DVDs online. So that renter can get them through the mail. However, Netflix started their **streaming** platfrom in **2007**. On January 6, 2016, Netflix went live in 130 countries simultaneously.
# We can see from the result that there are a lot more show that added after the year released.
# look into details on the False category
year_false = df[df["year_diff"] == False]
plt.scatter(data=year_false, y="release_year", x="year_added")
plt.ylabel("release year")
plt.xlabel("year added")
plt.show()
# There are a lot of shows that started to be added on the streaming platform Netflix after 2016. Even the old shows from 1960 and older were added many years later in Netflix.
# director with the most produced show available in netflix
df.groupby("director")["show_id"].count().sort_values(ascending=False).head(10)
| false | 1 | 1,958 | 0 | 6,189 | 1,958 |
||
129624430
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn import svm
from sklearn.neural_network import MLPClassifier
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.preprocessing import StandardScaler, LabelEncoder
from sklearn.model_selection import train_test_split
wine = pd.read_csv("winequality-red.csv", sep=";")
wine.head()
wine.info()
wine.isnull().sum()
# preprocessing
bins = (2, 6.5, 8)
group_names = ["bad", "good"]
wine["quality"] = pd.cut(wine["quality"], bins=bins, labels=group_names)
wine["qulaity"].unique()
label_qulaity = LabelEncoder()
wine["quality"] = label_qulity.fit_transform(wine["quality"])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/624/129624430.ipynb
| null | null |
[{"Id": 129624430, "ScriptId": 38546090, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12883251, "CreationDate": "05/15/2023 10:06:45", "VersionNumber": 2.0, "Title": "notebookca2c852f82", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 59.0, "LinesInsertedFromPrevious": 17.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 42.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn import svm
from sklearn.neural_network import MLPClassifier
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.preprocessing import StandardScaler, LabelEncoder
from sklearn.model_selection import train_test_split
wine = pd.read_csv("winequality-red.csv", sep=";")
wine.head()
wine.info()
wine.isnull().sum()
# preprocessing
bins = (2, 6.5, 8)
group_names = ["bad", "good"]
wine["quality"] = pd.cut(wine["quality"], bins=bins, labels=group_names)
wine["qulaity"].unique()
label_qulaity = LabelEncoder()
wine["quality"] = label_qulity.fit_transform(wine["quality"])
| false | 0 | 396 | 0 | 396 | 396 |
||
129624823
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df_train = pd.read_csv("/kaggle/input/titanic/train.csv")
df_train.head()
# Find Number of null values in each column
df_train.isna().sum()
# Find the person with maximum and minimum age
df_train.Age.max()
df_train.Age.min()
# Find the mode value of Age (occurring most often)
df_train.Age.mode().values[0]
df_train.describe()
df_train.dtypes
# How many passengers have this most frequent age?
df_train["PassengerId"][df_train["Age"] == df_train.Age.mode().values[0]].count()
df_train.Embarked.unique()
df_train.describe()
# How will you deal with null values?
# 1. Remove column
# 2. Remove record
# 3. Replace with some value
# Also Mention reason for decision in comments.
# Solution:
# 1. For Age we can replace with Average age as age has about 20 percent null
# which we can not discard column or record
# 2. We can drop Cabin column as 77% are null
# 3. For Embarked we can drop row as only 2 records are null
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/624/129624823.ipynb
| null | null |
[{"Id": 129624823, "ScriptId": 38545640, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3136640, "CreationDate": "05/15/2023 10:10:11", "VersionNumber": 1.0, "Title": "notebook055bc8ca9c", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 65.0, "LinesInsertedFromPrevious": 65.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df_train = pd.read_csv("/kaggle/input/titanic/train.csv")
df_train.head()
# Find Number of null values in each column
df_train.isna().sum()
# Find the person with maximum and minimum age
df_train.Age.max()
df_train.Age.min()
# Find the mode value of Age (occurring most often)
df_train.Age.mode().values[0]
df_train.describe()
df_train.dtypes
# How many passengers have this most frequent age?
df_train["PassengerId"][df_train["Age"] == df_train.Age.mode().values[0]].count()
df_train.Embarked.unique()
df_train.describe()
# How will you deal with null values?
# 1. Remove column
# 2. Remove record
# 3. Replace with some value
# Also Mention reason for decision in comments.
# Solution:
# 1. For Age we can replace with Average age as age has about 20 percent null
# which we can not discard column or record
# 2. We can drop Cabin column as 77% are null
# 3. For Embarked we can drop row as only 2 records are null
| false | 0 | 485 | 0 | 485 | 485 |
||
129653895
|
# FathomNet 2023
# Shifting seas, shifting species: Out-of-sample detection in the deep ocean
#
# # Introduction
# ## About Files
# * `multilabel_classification/train.csv` - csv list of training images and categories
# * `object_detection/train.json` - the training images, annotations, and categories in COCO formatted json
# * `object_detection/eval.json` - the evaluation images in COCO formatted json
# * `sample_submission.csv` - a sample submission file in the correct format
# * `category_key.csv` - key mapping numerical index to category and supercategory name
# * `demo_download.ipynb` - python notebook demonstrating download script
# * `download_images.py` - python script to download imagery from FathomNet using COCO formatted json
# * `requirements.txt` - python requirements to run the download script
import os
import numpy as np
import pandas as pd
import json
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib as mpl
from tqdm.notebook import tqdm
from pathlib import Path
import warnings
warnings.filterwarnings("ignore")
# seaborn
custom_params = {
"lines.linewidth": 1,
}
blues_palette = palette = sns.color_palette("Blues_r", n_colors=20)
reds_palette = palette = sns.color_palette("Reds_r", n_colors=20)
greys_palette = sns.color_palette("Greys", n_colors=10)
blue = blues_palette[1]
red = reds_palette[1]
two_colors = [blue, red]
sns.set()
sns.set_theme(style="whitegrid", palette=blues_palette, rc=custom_params)
# Define variables
INDEX = "id"
INPUT_ROOT = Path("/kaggle/input/fathomnet-out-of-sample-detection")
OUTPUT_ROOT = Path("/kaggle/working/")
TRAIN_OUTPUT_IMAGE_ROOT = Path("/kaggle/working/images/train")
EVAL_OUTPUT_IMAGE_ROOT = Path("/kaggle/working/images/eval")
TRAIN_FILE = INPUT_ROOT / "multilabel_classification/train.csv"
CATEGORY_KEY_FILE = INPUT_ROOT / "category_key.csv"
SAMPLE_SUBMISSION_FILE = INPUT_ROOT / "sample_submission.csv"
EVAL_JSON_FILE = INPUT_ROOT / "object_detection/eval.json"
TRAIN_JSON_FILE = INPUT_ROOT / "object_detection/train.json"
ANNOTATION_FILE = OUTPUT_ROOT / "annotation.csv"
TRAIN_IMAGE_DATA_FILE = OUTPUT_ROOT / "train_image_data.csv"
EVAL_IMAGE_DATA_FILE = OUTPUT_ROOT / "eval_image_data.csv"
# # Read Data
def read_train_data(file=Cfg.TRAIN_FILE, index_col=Cfg.INDEX):
return pd.read_csv(file).set_index(Cfg.INDEX)
def read_category_keys(file=Cfg.CATEGORY_KEY_FILE, index_col=Cfg.INDEX):
return pd.read_csv(file).set_index(Cfg.INDEX)
def read_sample_submission(file=Cfg.SAMPLE_SUBMISSION_FILE, index_col=Cfg.INDEX):
return pd.read_csv(file).set_index(Cfg.INDEX)
def read_json(file):
"""Read a json file."""
f = open(file)
data = json.load(f)
f.close()
return data
def read_object_detection_train():
return read_json(Cfg.TRAIN_JSON_FILE)
def read_object_detection_eval():
return read_json(Cfg.EVAL_JSON_FILE)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/653/129653895.ipynb
| null | null |
[{"Id": 129653895, "ScriptId": 38554283, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9338538, "CreationDate": "05/15/2023 14:00:05", "VersionNumber": 1.0, "Title": "notebook839fa56005", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 92.0, "LinesInsertedFromPrevious": 92.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# FathomNet 2023
# Shifting seas, shifting species: Out-of-sample detection in the deep ocean
#
# # Introduction
# ## About Files
# * `multilabel_classification/train.csv` - csv list of training images and categories
# * `object_detection/train.json` - the training images, annotations, and categories in COCO formatted json
# * `object_detection/eval.json` - the evaluation images in COCO formatted json
# * `sample_submission.csv` - a sample submission file in the correct format
# * `category_key.csv` - key mapping numerical index to category and supercategory name
# * `demo_download.ipynb` - python notebook demonstrating download script
# * `download_images.py` - python script to download imagery from FathomNet using COCO formatted json
# * `requirements.txt` - python requirements to run the download script
import os
import numpy as np
import pandas as pd
import json
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib as mpl
from tqdm.notebook import tqdm
from pathlib import Path
import warnings
warnings.filterwarnings("ignore")
# seaborn
custom_params = {
"lines.linewidth": 1,
}
blues_palette = palette = sns.color_palette("Blues_r", n_colors=20)
reds_palette = palette = sns.color_palette("Reds_r", n_colors=20)
greys_palette = sns.color_palette("Greys", n_colors=10)
blue = blues_palette[1]
red = reds_palette[1]
two_colors = [blue, red]
sns.set()
sns.set_theme(style="whitegrid", palette=blues_palette, rc=custom_params)
# Define variables
INDEX = "id"
INPUT_ROOT = Path("/kaggle/input/fathomnet-out-of-sample-detection")
OUTPUT_ROOT = Path("/kaggle/working/")
TRAIN_OUTPUT_IMAGE_ROOT = Path("/kaggle/working/images/train")
EVAL_OUTPUT_IMAGE_ROOT = Path("/kaggle/working/images/eval")
TRAIN_FILE = INPUT_ROOT / "multilabel_classification/train.csv"
CATEGORY_KEY_FILE = INPUT_ROOT / "category_key.csv"
SAMPLE_SUBMISSION_FILE = INPUT_ROOT / "sample_submission.csv"
EVAL_JSON_FILE = INPUT_ROOT / "object_detection/eval.json"
TRAIN_JSON_FILE = INPUT_ROOT / "object_detection/train.json"
ANNOTATION_FILE = OUTPUT_ROOT / "annotation.csv"
TRAIN_IMAGE_DATA_FILE = OUTPUT_ROOT / "train_image_data.csv"
EVAL_IMAGE_DATA_FILE = OUTPUT_ROOT / "eval_image_data.csv"
# # Read Data
def read_train_data(file=Cfg.TRAIN_FILE, index_col=Cfg.INDEX):
return pd.read_csv(file).set_index(Cfg.INDEX)
def read_category_keys(file=Cfg.CATEGORY_KEY_FILE, index_col=Cfg.INDEX):
return pd.read_csv(file).set_index(Cfg.INDEX)
def read_sample_submission(file=Cfg.SAMPLE_SUBMISSION_FILE, index_col=Cfg.INDEX):
return pd.read_csv(file).set_index(Cfg.INDEX)
def read_json(file):
"""Read a json file."""
f = open(file)
data = json.load(f)
f.close()
return data
def read_object_detection_train():
return read_json(Cfg.TRAIN_JSON_FILE)
def read_object_detection_eval():
return read_json(Cfg.EVAL_JSON_FILE)
| false | 0 | 906 | 0 | 906 | 906 |
||
129729575
|
# **print common elements**
a = [1, 2, 3]
b = [2, 3, 4]
for i in a:
for j in b:
if i == j:
print(i)
break
# **remove duplicate values**
a = [1, 2, 3, 4, 5, 4, 3, 2, 6, 7, 2, 1]
for i in a:
count = 0
for j in a:
if i == j:
count = count + 1
if count > 1:
a.remove(j)
print(a)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/729/129729575.ipynb
| null | null |
[{"Id": 129729575, "ScriptId": 38580089, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15022629, "CreationDate": "05/16/2023 04:41:36", "VersionNumber": 1.0, "Title": "list assignment 2", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 21.0, "LinesInsertedFromPrevious": 21.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# **print common elements**
a = [1, 2, 3]
b = [2, 3, 4]
for i in a:
for j in b:
if i == j:
print(i)
break
# **remove duplicate values**
a = [1, 2, 3, 4, 5, 4, 3, 2, 6, 7, 2, 1]
for i in a:
count = 0
for j in a:
if i == j:
count = count + 1
if count > 1:
a.remove(j)
print(a)
| false | 0 | 152 | 0 | 152 | 152 |
||
129380612
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### Import
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
import nltk
import string
import re
train_df = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
test_df = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
train_df.head()
train_df.shape
train_df.index.duplicated().sum()
train_df["target"].isna().sum()
train_df["target"].value_counts(normalize=True) * 100
train_df.dtypes
train_df.head()
import pandas as pd
from collections import Counter
# Assuming 'df' is your dataframe with a column named 'text'
# Step 1: Calculate word frequency
word_freq = Counter(" ".join(train_df["text"]).split())
# Step 2: Identify 10 least frequent words
rare_words = [word for word, freq in word_freq.items() if freq <= 1]
import contractions
import string
# from spellchecker import SpellChecker
def clean_tweet(text):
# removes \n
cleaned_text = text.replace("\n", " ")
# Lower_text
cleaned_text = cleaned_text.lower()
# Remove_Punctuations
punctuations = "@#!?+&*[]-%.:/();$=><|{}^" + "'`"
for p in punctuations:
cleaned_text = re.sub(re.escape(p), " ", cleaned_text)
# Remove_Stopwords
cleaned_text = " ".join(
[
word
for word in str(cleaned_text).split()
if word not in stopwords.words("english")
]
)
# Remove_HTMLs
cleaned_text = re.sub(r"<.*?>", "", cleaned_text)
# Expand_Contractions
expanded_words = []
for word in cleaned_text.split():
# using contractions.fix to expand the shortened words
expanded_words.append(contractions.fix(word))
cleaned_text = " ".join(expanded_words)
# Remove_URLs
cleaned_text = re.sub(r"http?\S+", "", cleaned_text)
# Remove_Email_IDs
cleaned_text = re.sub(r"[\w\.-]+@[\w\.-]+\.\w+", "", cleaned_text)
# Remove_emojis
emoji_pattern = re.compile(
"["
"\U0001F600-\U0001F64F" # emoticons
"\U0001F300-\U0001F5FF" # symbols & pictographs
"\U0001F680-\U0001F6FF" # transport & map symbols
"\U0001F1E0-\U0001F1FF" # flags (iOS)
"\U00002500-\U00002BEF" # chinese char
"\U00002702-\U000027B0"
"\U00002702-\U000027B0"
"\U000024C2-\U0001F251"
"\U0001f926-\U0001f937"
"\U00010000-\U0010ffff"
"\u2640-\u2642"
"\u2600-\u2B55"
"\u200d"
"\u23cf"
"\u23e9"
"\u231a"
"\ufe0f" # dingbats
"\u3030"
"]+",
flags=re.UNICODE,
)
cleaned_text = emoji_pattern.sub(r"", cleaned_text)
# Remove_Tweeter_Mentions_Chars
cleaned_text = re.sub(r"@\w+", "", cleaned_text)
# Remove_Unicode_Characters
cleaned_text = cleaned_text.encode("ascii", "ignore").decode()
# Remove_Digits
cleaned_text = re.sub(r"\w*\d+\w*", "", cleaned_text)
return cleaned_text
train_df["clean_text"] = train_df["text"].apply(clean_tweet)
train_df
test_df["clean_text"] = test_df["text"].apply(clean_tweet)
test_df
# ### Tensorflow
size_vocab = 10000
embedding_dim = 64
max_length = 24
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
tokenizer = Tokenizer(num_words=size_vocab, oov_token="<OOV>")
tokenizer.fit_on_texts(train_df["clean_text"])
train_sequences = tokenizer.texts_to_sequences(train_df["clean_text"])
test_sequences = tokenizer.texts_to_sequences(test_df["clean_text"])
padded_train_squences = pad_sequences(
train_sequences, maxlen=max_length, truncating="post", padding="post"
)
padded_test_squences = pad_sequences(
test_sequences, maxlen=max_length, truncating="post", padding="post"
)
print(padded_train_squences.shape)
print(padded_test_squences.shape)
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Embedding
from tensorflow.keras.layers import Flatten
from tensorflow.keras.layers import Dense
model = Sequential()
model.add(Embedding(size_vocab, embedding_dim, input_length=max_length))
model.add(Flatten())
model.add(Dense(10, activation="relu"))
model.add(Dense(1, activation="sigmoid"))
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["acc"])
model.summary()
model.fit(padded_train_squences, train_df["target"], epochs=20)
test_sen = test_df["text"]
test_sen = tokenizer.texts_to_sequences(test_sen)
padd_test_sen = pad_sequences(
test_sen, maxlen=max_length, truncating="post", padding="post"
)
padd_test_sen.shape
padded_test_squences.shape
rs = model.predict(padded_test_squences)
rs.shape
# Apply threshold and convert to binary values
threshold = 0.5
binary_predictions = [1 if pred > threshold else 0 for pred in rs]
submission = pd.DataFrame({"id": test_df["id"], "target": binary_predictions})
submission
submission.to_csv("submission.csv", index=False)
rs
test_df
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/380/129380612.ipynb
| null | null |
[{"Id": 129380612, "ScriptId": 38459966, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7042083, "CreationDate": "05/13/2023 09:50:11", "VersionNumber": 1.0, "Title": "NLP Beginer", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 195.0, "LinesInsertedFromPrevious": 195.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### Import
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
import nltk
import string
import re
train_df = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
test_df = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
train_df.head()
train_df.shape
train_df.index.duplicated().sum()
train_df["target"].isna().sum()
train_df["target"].value_counts(normalize=True) * 100
train_df.dtypes
train_df.head()
import pandas as pd
from collections import Counter
# Assuming 'df' is your dataframe with a column named 'text'
# Step 1: Calculate word frequency
word_freq = Counter(" ".join(train_df["text"]).split())
# Step 2: Identify 10 least frequent words
rare_words = [word for word, freq in word_freq.items() if freq <= 1]
import contractions
import string
# from spellchecker import SpellChecker
def clean_tweet(text):
# removes \n
cleaned_text = text.replace("\n", " ")
# Lower_text
cleaned_text = cleaned_text.lower()
# Remove_Punctuations
punctuations = "@#!?+&*[]-%.:/();$=><|{}^" + "'`"
for p in punctuations:
cleaned_text = re.sub(re.escape(p), " ", cleaned_text)
# Remove_Stopwords
cleaned_text = " ".join(
[
word
for word in str(cleaned_text).split()
if word not in stopwords.words("english")
]
)
# Remove_HTMLs
cleaned_text = re.sub(r"<.*?>", "", cleaned_text)
# Expand_Contractions
expanded_words = []
for word in cleaned_text.split():
# using contractions.fix to expand the shortened words
expanded_words.append(contractions.fix(word))
cleaned_text = " ".join(expanded_words)
# Remove_URLs
cleaned_text = re.sub(r"http?\S+", "", cleaned_text)
# Remove_Email_IDs
cleaned_text = re.sub(r"[\w\.-]+@[\w\.-]+\.\w+", "", cleaned_text)
# Remove_emojis
emoji_pattern = re.compile(
"["
"\U0001F600-\U0001F64F" # emoticons
"\U0001F300-\U0001F5FF" # symbols & pictographs
"\U0001F680-\U0001F6FF" # transport & map symbols
"\U0001F1E0-\U0001F1FF" # flags (iOS)
"\U00002500-\U00002BEF" # chinese char
"\U00002702-\U000027B0"
"\U00002702-\U000027B0"
"\U000024C2-\U0001F251"
"\U0001f926-\U0001f937"
"\U00010000-\U0010ffff"
"\u2640-\u2642"
"\u2600-\u2B55"
"\u200d"
"\u23cf"
"\u23e9"
"\u231a"
"\ufe0f" # dingbats
"\u3030"
"]+",
flags=re.UNICODE,
)
cleaned_text = emoji_pattern.sub(r"", cleaned_text)
# Remove_Tweeter_Mentions_Chars
cleaned_text = re.sub(r"@\w+", "", cleaned_text)
# Remove_Unicode_Characters
cleaned_text = cleaned_text.encode("ascii", "ignore").decode()
# Remove_Digits
cleaned_text = re.sub(r"\w*\d+\w*", "", cleaned_text)
return cleaned_text
train_df["clean_text"] = train_df["text"].apply(clean_tweet)
train_df
test_df["clean_text"] = test_df["text"].apply(clean_tweet)
test_df
# ### Tensorflow
size_vocab = 10000
embedding_dim = 64
max_length = 24
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
tokenizer = Tokenizer(num_words=size_vocab, oov_token="<OOV>")
tokenizer.fit_on_texts(train_df["clean_text"])
train_sequences = tokenizer.texts_to_sequences(train_df["clean_text"])
test_sequences = tokenizer.texts_to_sequences(test_df["clean_text"])
padded_train_squences = pad_sequences(
train_sequences, maxlen=max_length, truncating="post", padding="post"
)
padded_test_squences = pad_sequences(
test_sequences, maxlen=max_length, truncating="post", padding="post"
)
print(padded_train_squences.shape)
print(padded_test_squences.shape)
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Embedding
from tensorflow.keras.layers import Flatten
from tensorflow.keras.layers import Dense
model = Sequential()
model.add(Embedding(size_vocab, embedding_dim, input_length=max_length))
model.add(Flatten())
model.add(Dense(10, activation="relu"))
model.add(Dense(1, activation="sigmoid"))
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["acc"])
model.summary()
model.fit(padded_train_squences, train_df["target"], epochs=20)
test_sen = test_df["text"]
test_sen = tokenizer.texts_to_sequences(test_sen)
padd_test_sen = pad_sequences(
test_sen, maxlen=max_length, truncating="post", padding="post"
)
padd_test_sen.shape
padded_test_squences.shape
rs = model.predict(padded_test_squences)
rs.shape
# Apply threshold and convert to binary values
threshold = 0.5
binary_predictions = [1 if pred > threshold else 0 for pred in rs]
submission = pd.DataFrame({"id": test_df["id"], "target": binary_predictions})
submission
submission.to_csv("submission.csv", index=False)
rs
test_df
| false | 0 | 1,819 | 0 | 1,819 | 1,819 |
||
129380132
|
<jupyter_start><jupyter_text>Diabetes prediction dataset
The **Diabetes prediction dataset** is a collection of medical and demographic data from patients, along with their diabetes status (positive or negative). The data includes features such as age, gender, body mass index (BMI), hypertension, heart disease, smoking history, HbA1c level, and blood glucose level. This dataset can be used to build machine learning models to predict diabetes in patients based on their medical history and demographic information. This can be useful for healthcare professionals in identifying patients who may be at risk of developing diabetes and in developing personalized treatment plans. Additionally, the dataset can be used by researchers to explore the relationships between various medical and demographic factors and the likelihood of developing diabetes.
Kaggle dataset identifier: diabetes-prediction-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
# For Conversion of numerical columns to categorical columns.
from sklearn.preprocessing import OneHotEncoder
# Model Building
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.tree import DecisionTreeClassifier
# Metrics Evaluation
from sklearn.metrics import accuracy_score, classification_report
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
filepath = "/kaggle/input/diabetes-prediction-dataset/diabetes_prediction_dataset.csv"
def data_import(filepath):
df = pd.read_csv(filepath)
return df
df = data_import(filepath)
df.head()
df.info()
duplicated_sum = df.duplicated().sum()
print(f"No of Duplicate rows: {duplicated_sum} ")
df1 = df.drop_duplicates()
df1.shape
df1.info()
# Let's check for unique columns in object columns.
o = df1.dtypes == "object"
object_cols = o[o].index
print(f"Categorical Columns : {object_cols}")
def unique_cols(df, object_cols):
for i in df[object_cols]:
print(f"{i} : {df[i].unique()}")
unique_cols(df1, object_cols)
# Let's check for value counts in gender column.
df1["gender"].value_counts()
# Let's encode gender columns using label encoder.
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
df1["gender"] = le.fit_transform(df1["gender"])
# Check for value count in smoking_history.
df1["smoking_history"].value_counts()
# Let's encode smoking_history using map function.
df1["smoking_history"] = df1["smoking_history"].map(
{"No Info": 0, "never": 1, "former": 2, "current": 3, "not current": 4, "ever": 5}
)
# Let's check correlation of columns with label.
correlation = df.corr()
correlation
label_correlation = correlation["diabetes"].drop("diabetes")
sorted_correlation = label_correlation.abs().sort_values(ascending=False)
print(sorted_correlation)
features = df1[
[
"blood_glucose_level",
"HbA1c_level",
"age",
"bmi",
"hypertension",
"heart_disease",
]
]
label = df1["diabetes"]
## Train test splitting
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(features, label, test_size=0.2)
print(f"x train shape : {x_train.shape}")
print(f"y train shape : {y_train.shape}")
print(f"x test shape : {x_test.shape}")
print(f"y test shape : {y_test.shape}")
pred_dict = {}
def model(modelname, x_train, x_test, y_train, y_test):
model_build = modelname(random_state=42)
model_build.fit(x_train, y_train)
y_pred = model_build.predict(x_test)
return y_pred
y_pred = model(LogisticRegression, x_train, x_test, y_train, y_test)
pred_dict["Logistic_y_pred"] = y_pred
y_pred = model(GradientBoostingClassifier, x_train, x_test, y_train, y_test)
pred_dict["Gradientboosting_y_pred"] = y_pred
y_pred = model(DecisionTreeClassifier, x_train, x_test, y_train, y_test)
pred_dict["DecisionTree_y_pred"] = y_pred
# Classification Metrics Evaluation function
metrics_dict = {}
def classification_metrics(y_test, y_pred):
acc_score = accuracy_score(y_test, y_pred)
print(f"Classification Report : \n{classification_report(y_test,y_pred)}")
return acc_score
metrics_dict["Log_acc_score"] = classification_metrics(
y_test, pred_dict["Logistic_y_pred"]
)
metrics_dict["Gradientboosting_acc_score"] = classification_metrics(
y_test, pred_dict["Gradientboosting_y_pred"]
)
metrics_dict["DecisionTree_acc_score"] = classification_metrics(
y_test, pred_dict["DecisionTree_y_pred"]
)
metrics_dict
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/380/129380132.ipynb
|
diabetes-prediction-dataset
|
iammustafatz
|
[{"Id": 129380132, "ScriptId": 38466525, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5820249, "CreationDate": "05/13/2023 09:45:10", "VersionNumber": 1.0, "Title": "Diabetes prediction dataset", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 154.0, "LinesInsertedFromPrevious": 154.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 185374364, "KernelVersionId": 129380132, "SourceDatasetVersionId": 5344155}]
|
[{"Id": 5344155, "DatasetId": 3102947, "DatasourceVersionId": 5417553, "CreatorUserId": 11427441, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "04/08/2023 06:11:45", "VersionNumber": 1.0, "Title": "Diabetes prediction dataset", "Slug": "diabetes-prediction-dataset", "Subtitle": "A Comprehensive Dataset for Predicting Diabetes with Medical & Demographic Data", "Description": "The **Diabetes prediction dataset** is a collection of medical and demographic data from patients, along with their diabetes status (positive or negative). The data includes features such as age, gender, body mass index (BMI), hypertension, heart disease, smoking history, HbA1c level, and blood glucose level. This dataset can be used to build machine learning models to predict diabetes in patients based on their medical history and demographic information. This can be useful for healthcare professionals in identifying patients who may be at risk of developing diabetes and in developing personalized treatment plans. Additionally, the dataset can be used by researchers to explore the relationships between various medical and demographic factors and the likelihood of developing diabetes.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3102947, "CreatorUserId": 11427441, "OwnerUserId": 11427441.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5344155.0, "CurrentDatasourceVersionId": 5417553.0, "ForumId": 3166206, "Type": 2, "CreationDate": "04/08/2023 06:11:45", "LastActivityDate": "04/08/2023", "TotalViews": 127619, "TotalDownloads": 24886, "TotalVotes": 309, "TotalKernels": 120}]
|
[{"Id": 11427441, "UserName": "iammustafatz", "DisplayName": "Mohammed Mustafa", "RegisterDate": "08/29/2022", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
# For Conversion of numerical columns to categorical columns.
from sklearn.preprocessing import OneHotEncoder
# Model Building
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.tree import DecisionTreeClassifier
# Metrics Evaluation
from sklearn.metrics import accuracy_score, classification_report
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
filepath = "/kaggle/input/diabetes-prediction-dataset/diabetes_prediction_dataset.csv"
def data_import(filepath):
df = pd.read_csv(filepath)
return df
df = data_import(filepath)
df.head()
df.info()
duplicated_sum = df.duplicated().sum()
print(f"No of Duplicate rows: {duplicated_sum} ")
df1 = df.drop_duplicates()
df1.shape
df1.info()
# Let's check for unique columns in object columns.
o = df1.dtypes == "object"
object_cols = o[o].index
print(f"Categorical Columns : {object_cols}")
def unique_cols(df, object_cols):
for i in df[object_cols]:
print(f"{i} : {df[i].unique()}")
unique_cols(df1, object_cols)
# Let's check for value counts in gender column.
df1["gender"].value_counts()
# Let's encode gender columns using label encoder.
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
df1["gender"] = le.fit_transform(df1["gender"])
# Check for value count in smoking_history.
df1["smoking_history"].value_counts()
# Let's encode smoking_history using map function.
df1["smoking_history"] = df1["smoking_history"].map(
{"No Info": 0, "never": 1, "former": 2, "current": 3, "not current": 4, "ever": 5}
)
# Let's check correlation of columns with label.
correlation = df.corr()
correlation
label_correlation = correlation["diabetes"].drop("diabetes")
sorted_correlation = label_correlation.abs().sort_values(ascending=False)
print(sorted_correlation)
features = df1[
[
"blood_glucose_level",
"HbA1c_level",
"age",
"bmi",
"hypertension",
"heart_disease",
]
]
label = df1["diabetes"]
## Train test splitting
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(features, label, test_size=0.2)
print(f"x train shape : {x_train.shape}")
print(f"y train shape : {y_train.shape}")
print(f"x test shape : {x_test.shape}")
print(f"y test shape : {y_test.shape}")
pred_dict = {}
def model(modelname, x_train, x_test, y_train, y_test):
model_build = modelname(random_state=42)
model_build.fit(x_train, y_train)
y_pred = model_build.predict(x_test)
return y_pred
y_pred = model(LogisticRegression, x_train, x_test, y_train, y_test)
pred_dict["Logistic_y_pred"] = y_pred
y_pred = model(GradientBoostingClassifier, x_train, x_test, y_train, y_test)
pred_dict["Gradientboosting_y_pred"] = y_pred
y_pred = model(DecisionTreeClassifier, x_train, x_test, y_train, y_test)
pred_dict["DecisionTree_y_pred"] = y_pred
# Classification Metrics Evaluation function
metrics_dict = {}
def classification_metrics(y_test, y_pred):
acc_score = accuracy_score(y_test, y_pred)
print(f"Classification Report : \n{classification_report(y_test,y_pred)}")
return acc_score
metrics_dict["Log_acc_score"] = classification_metrics(
y_test, pred_dict["Logistic_y_pred"]
)
metrics_dict["Gradientboosting_acc_score"] = classification_metrics(
y_test, pred_dict["Gradientboosting_y_pred"]
)
metrics_dict["DecisionTree_acc_score"] = classification_metrics(
y_test, pred_dict["DecisionTree_y_pred"]
)
metrics_dict
| false | 0 | 1,228 | 2 | 1,419 | 1,228 |
||
129214662
|
<jupyter_start><jupyter_text>Starbucks Nutrition Facts
```
Nutrition facts for several Starbucks food items
```
| Column | Description |
| ------- | ------------------------------------------------------------ |
| item | The name of the food item. |
| calories| The amount of calories in the food item. |
| fat | The quantity of fat in grams present in the food item. |
| carb | The amount of carbohydrates in grams found in the food item. |
| fiber | The quantity of dietary fiber in grams in the food item. |
| protein | The amount of protein in grams contained in the food item. |
| type | The category or type of food item (bakery, bistro box, hot breakfast, parfait, petite, salad, or sandwich). |
Kaggle dataset identifier: starbucks-nutrition
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from pprint import pprint
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Starbucks Nutritional Information
# Starbucks provides comprehensive nutritional information for their food and beverage offerings, allowing customers to make informed choices based on their dietary preferences and health goals. The nutritional information includes details such as calories, fat content, carbohydrates, fiber, and protein for each menu item.
# By making this information readily available, Starbucks aims to empower individuals to make choices that align with their nutritional needs and preferences. Whether you're looking for lower-calorie options, watching your fat or carb intake, or seeking protein-rich alternatives, the nutritional information provided by Starbucks helps you navigate their menu with confidence.
# ## Data Coverage
# The data encompasses a range of food items, from baked goods and bistro boxes to hot breakfast items, parfaits, petite treats, salads, and sandwiches. Each item is categorized based on its type, making it easier for customers to find options that suit their dietary requirements or preferences.
# ## Transparency and Informed Decisions
# By offering transparent and detailed nutritional information, Starbucks reinforces its commitment to supporting customers in making informed decisions about their food choices. Whether you're enjoying a coffee break or grabbing a quick bite, the nutritional information empowers you to enjoy Starbucks' offerings while being mindful of your nutritional goals.
# ---
# ## Data Dictionary
# The data consists of nutrition facts for several Starbucks food items. It is organized in the form of a data frame with 77 observations and 7 variables.
# ### Variables
# - **item**: The name of the food item (string).
# - **calories**: The number of calories in the food item (integer).
# - **fat**: The amount of fat in grams (numeric).
# - **carb**: The amount of carbohydrates in grams (numeric).
# - **fiber**: The amount of dietary fiber in grams (numeric).
# - **protein**: The amount of protein in grams (numeric).
# - **type**: The categorization of the food item, with levels bakery, bistro box, hot breakfast, parfait, petite, salad, and sandwich (factor).
# ### Additional Information
# - The data frame has a RangeIndex from 0 to 76.
# - There are no missing values (non-null count is 77 for all columns).
# - The original data frame had an additional column named "Unnamed: 0", which has been removed for this improved data dictionary.
# ---
# .
df = pd.read_csv("/kaggle/input/starbucks-nutrition/starbucks.csv", index_col=0)
df
df.describe
df.info()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/214/129214662.ipynb
|
starbucks-nutrition
|
utkarshx27
|
[{"Id": 129214662, "ScriptId": 38415291, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14543633, "CreationDate": "05/11/2023 22:43:38", "VersionNumber": 1.0, "Title": "Starbucks EDA", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 85.0, "LinesInsertedFromPrevious": 85.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 185060918, "KernelVersionId": 129214662, "SourceDatasetVersionId": 5651811}]
|
[{"Id": 5651811, "DatasetId": 3248696, "DatasourceVersionId": 5727183, "CreatorUserId": 13364933, "LicenseName": "CC0: Public Domain", "CreationDate": "05/10/2023 05:42:59", "VersionNumber": 1.0, "Title": "Starbucks Nutrition Facts", "Slug": "starbucks-nutrition", "Subtitle": "Nutrition facts for several Starbucks food items", "Description": "```\nNutrition facts for several Starbucks food items\n```\n| Column | Description |\n| ------- | ------------------------------------------------------------ |\n| item | The name of the food item. |\n| calories| The amount of calories in the food item. |\n| fat | The quantity of fat in grams present in the food item. |\n| carb | The amount of carbohydrates in grams found in the food item. |\n| fiber | The quantity of dietary fiber in grams in the food item. |\n| protein | The amount of protein in grams contained in the food item. |\n| type | The category or type of food item (bakery, bistro box, hot breakfast, parfait, petite, salad, or sandwich). |", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3248696, "CreatorUserId": 13364933, "OwnerUserId": 13364933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5651811.0, "CurrentDatasourceVersionId": 5727183.0, "ForumId": 3314049, "Type": 2, "CreationDate": "05/10/2023 05:42:59", "LastActivityDate": "05/10/2023", "TotalViews": 12557, "TotalDownloads": 2321, "TotalVotes": 59, "TotalKernels": 17}]
|
[{"Id": 13364933, "UserName": "utkarshx27", "DisplayName": "Utkarsh Singh", "RegisterDate": "01/21/2023", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from pprint import pprint
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Starbucks Nutritional Information
# Starbucks provides comprehensive nutritional information for their food and beverage offerings, allowing customers to make informed choices based on their dietary preferences and health goals. The nutritional information includes details such as calories, fat content, carbohydrates, fiber, and protein for each menu item.
# By making this information readily available, Starbucks aims to empower individuals to make choices that align with their nutritional needs and preferences. Whether you're looking for lower-calorie options, watching your fat or carb intake, or seeking protein-rich alternatives, the nutritional information provided by Starbucks helps you navigate their menu with confidence.
# ## Data Coverage
# The data encompasses a range of food items, from baked goods and bistro boxes to hot breakfast items, parfaits, petite treats, salads, and sandwiches. Each item is categorized based on its type, making it easier for customers to find options that suit their dietary requirements or preferences.
# ## Transparency and Informed Decisions
# By offering transparent and detailed nutritional information, Starbucks reinforces its commitment to supporting customers in making informed decisions about their food choices. Whether you're enjoying a coffee break or grabbing a quick bite, the nutritional information empowers you to enjoy Starbucks' offerings while being mindful of your nutritional goals.
# ---
# ## Data Dictionary
# The data consists of nutrition facts for several Starbucks food items. It is organized in the form of a data frame with 77 observations and 7 variables.
# ### Variables
# - **item**: The name of the food item (string).
# - **calories**: The number of calories in the food item (integer).
# - **fat**: The amount of fat in grams (numeric).
# - **carb**: The amount of carbohydrates in grams (numeric).
# - **fiber**: The amount of dietary fiber in grams (numeric).
# - **protein**: The amount of protein in grams (numeric).
# - **type**: The categorization of the food item, with levels bakery, bistro box, hot breakfast, parfait, petite, salad, and sandwich (factor).
# ### Additional Information
# - The data frame has a RangeIndex from 0 to 76.
# - There are no missing values (non-null count is 77 for all columns).
# - The original data frame had an additional column named "Unnamed: 0", which has been removed for this improved data dictionary.
# ---
# .
df = pd.read_csv("/kaggle/input/starbucks-nutrition/starbucks.csv", index_col=0)
df
df.describe
df.info()
| false | 1 | 831 | 1 | 1,047 | 831 |
||
129263774
|
# # Cases and Casualties due to COVID-19 in Countries
# ## Importing Libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Data input and cleaning
df = pd.read_csv(
"/kaggle/input/corona-data-cleaned/corona_virus.csv", encoding="unicode_escape"
)
df.head()
df.info()
df_dropped = df.drop(
columns=["New Cases", "New Deaths", "New Recovered", "serious_critical"], axis=1
)
df_dropped.head()
df_dropped.info()
df[df_dropped.isnull()]
df = df_dropped.dropna()
df.head()
df.head()
df = df.rename(columns={"Country,Other": "Country"})
for i, r in df.iterrows():
r["Total Cases"] = r["Total Cases"].replace(",", "")
r["Total Deaths"] = r["Total Deaths"].replace(",", "")
r["Total Recovered"] = r["Total Recovered"].replace(",", "")
r["Active Cases"] = r["Active Cases"].replace(",", "")
r["cases_per_1M"] = r["cases_per_1M"].replace(",", "")
r["deaths_per_1M"] = r["deaths_per_1M"].replace(",", "")
r["Total Tests"] = r["Total Tests"].replace(",", "")
r["tests_per_1M"] = r["tests_per_1M"].replace(",", "")
r["Population"] = r["Population"].replace(",", "")
df["Total Cases"] = df["Total Cases"].astype(str).astype(int)
df["Total Deaths"] = df["Total Deaths"].astype(str).astype(int)
df["Total Recovered"] = df["Total Recovered"].astype(str).astype(int)
df["Active Cases"] = df["Active Cases"].astype(str).astype(int)
df["cases_per_1M"] = df["cases_per_1M"].astype(str).astype(int)
df["deaths_per_1M"] = df["deaths_per_1M"].astype(str).astype(int)
df["Total Tests"] = df["Total Tests"].astype(str).astype(int)
df["tests_per_1M"] = df["tests_per_1M"].astype(str).astype(int)
df["Population"] = df["Population"].astype(str).astype(int)
df.head()
df.info()
# ## Analysis
# ### Countries with highest number of Cases
# Country vs Number of Cases
total_cases = pd.DataFrame(df, columns=["Country", "Total Cases", "Total Deaths"])
total_cases = total_cases.sort_values(by="Total Cases", ascending=False)
# total_cases.head()
# top 10 countries
top_cases = total_cases[:10]
# plot
fig1 = px.bar(top_cases, x="Country", y="Total Cases", text="Total Cases")
fig2 = px.bar(top_cases, x="Country", y="Total Deaths", text="Total Deaths")
fig1.show()
fig2.show()
# ### Scatter Plot showing Cases vs Deaths in Countries with highest number of cases.
# total cases vs total deaths
df.head()
# top 10 countries
top_cases = total_cases[:10]
fig3 = px.scatter(top_cases, x="Total Cases", y="Total Deaths", text="Country")
fig3.update_traces(textposition="top center")
fig3.show()
top_cases.head(10)
# ### Cases and Deaths per 1M population
cases_per_M = pd.DataFrame(
df, columns=["Country", "cases_per_1M", "deaths_per_1M", "tests_per_1M"]
).sort_values(by="cases_per_1M", ascending=False)
top_cases = cases_per_M[:10]
fig4 = px.bar(top_cases, y="Country", x=["cases_per_1M"])
fig5 = px.bar(top_cases, y="Country", x=["deaths_per_1M"])
fig4.show()
fig5.show()
top_cases.head()
# ### Countries with highest fatalities
# Fatalities in world wide Population
# top fatality country by ratio
df["death/population %"] = round(df["Total Deaths"] / df["Population"] * 100, 2)
df = df.sort_values(by=["death/population %"], ascending=False)
# top 10 fatalities
top_death = df[:10]
fig6 = px.bar(top_death, x="Country", y="death/population %", text="death/population %")
fig6.show()
top_death.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/263/129263774.ipynb
| null | null |
[{"Id": 129263774, "ScriptId": 38423448, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8975961, "CreationDate": "05/12/2023 09:27:12", "VersionNumber": 1.0, "Title": "notebook7ec6d94635", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 134.0, "LinesInsertedFromPrevious": 134.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Cases and Casualties due to COVID-19 in Countries
# ## Importing Libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Data input and cleaning
df = pd.read_csv(
"/kaggle/input/corona-data-cleaned/corona_virus.csv", encoding="unicode_escape"
)
df.head()
df.info()
df_dropped = df.drop(
columns=["New Cases", "New Deaths", "New Recovered", "serious_critical"], axis=1
)
df_dropped.head()
df_dropped.info()
df[df_dropped.isnull()]
df = df_dropped.dropna()
df.head()
df.head()
df = df.rename(columns={"Country,Other": "Country"})
for i, r in df.iterrows():
r["Total Cases"] = r["Total Cases"].replace(",", "")
r["Total Deaths"] = r["Total Deaths"].replace(",", "")
r["Total Recovered"] = r["Total Recovered"].replace(",", "")
r["Active Cases"] = r["Active Cases"].replace(",", "")
r["cases_per_1M"] = r["cases_per_1M"].replace(",", "")
r["deaths_per_1M"] = r["deaths_per_1M"].replace(",", "")
r["Total Tests"] = r["Total Tests"].replace(",", "")
r["tests_per_1M"] = r["tests_per_1M"].replace(",", "")
r["Population"] = r["Population"].replace(",", "")
df["Total Cases"] = df["Total Cases"].astype(str).astype(int)
df["Total Deaths"] = df["Total Deaths"].astype(str).astype(int)
df["Total Recovered"] = df["Total Recovered"].astype(str).astype(int)
df["Active Cases"] = df["Active Cases"].astype(str).astype(int)
df["cases_per_1M"] = df["cases_per_1M"].astype(str).astype(int)
df["deaths_per_1M"] = df["deaths_per_1M"].astype(str).astype(int)
df["Total Tests"] = df["Total Tests"].astype(str).astype(int)
df["tests_per_1M"] = df["tests_per_1M"].astype(str).astype(int)
df["Population"] = df["Population"].astype(str).astype(int)
df.head()
df.info()
# ## Analysis
# ### Countries with highest number of Cases
# Country vs Number of Cases
total_cases = pd.DataFrame(df, columns=["Country", "Total Cases", "Total Deaths"])
total_cases = total_cases.sort_values(by="Total Cases", ascending=False)
# total_cases.head()
# top 10 countries
top_cases = total_cases[:10]
# plot
fig1 = px.bar(top_cases, x="Country", y="Total Cases", text="Total Cases")
fig2 = px.bar(top_cases, x="Country", y="Total Deaths", text="Total Deaths")
fig1.show()
fig2.show()
# ### Scatter Plot showing Cases vs Deaths in Countries with highest number of cases.
# total cases vs total deaths
df.head()
# top 10 countries
top_cases = total_cases[:10]
fig3 = px.scatter(top_cases, x="Total Cases", y="Total Deaths", text="Country")
fig3.update_traces(textposition="top center")
fig3.show()
top_cases.head(10)
# ### Cases and Deaths per 1M population
cases_per_M = pd.DataFrame(
df, columns=["Country", "cases_per_1M", "deaths_per_1M", "tests_per_1M"]
).sort_values(by="cases_per_1M", ascending=False)
top_cases = cases_per_M[:10]
fig4 = px.bar(top_cases, y="Country", x=["cases_per_1M"])
fig5 = px.bar(top_cases, y="Country", x=["deaths_per_1M"])
fig4.show()
fig5.show()
top_cases.head()
# ### Countries with highest fatalities
# Fatalities in world wide Population
# top fatality country by ratio
df["death/population %"] = round(df["Total Deaths"] / df["Population"] * 100, 2)
df = df.sort_values(by=["death/population %"], ascending=False)
# top 10 fatalities
top_death = df[:10]
fig6 = px.bar(top_death, x="Country", y="death/population %", text="death/population %")
fig6.show()
top_death.head()
| false | 0 | 1,370 | 0 | 1,370 | 1,370 |
||
129319649
|
<jupyter_start><jupyter_text>COVID-19 Dataset
[](https://forthebadge.com) [](https://forthebadge.com)
### Context
- A new coronavirus designated 2019-nCoV was first identified in Wuhan, the capital of China's Hubei province
- People developed pneumonia without a clear cause and for which existing vaccines or treatments were not effective.
- The virus has shown evidence of human-to-human transmission
- Transmission rate (rate of infection) appeared to escalate in mid-January 2020
- As of 30 January 2020, approximately 8,243 cases have been confirmed
### Content
> * **full_grouped.csv** - Day to day country wise no. of cases (Has County/State/Province level data)
> * **covid_19_clean_complete.csv** - Day to day country wise no. of cases (Doesn't have County/State/Province level data)
> * **country_wise_latest.csv** - Latest country level no. of cases
> * **day_wise.csv** - Day wise no. of cases (Doesn't have country level data)
> * **usa_county_wise.csv** - Day to day county level no. of cases
> * **worldometer_data.csv** - Latest data from https://www.worldometers.info/
Kaggle dataset identifier: corona-virus-report
<jupyter_script>import pandas as pd
import numpy as np
import seaborn as sns
df = pd.read_csv("D:\ml-practice\projects\covid19\country_wise_latest.csv")
df.dtypes
df.isnull().sum()
df.drop(["New cases", "New deaths", "New recovered"], axis=1, inplace=True)
df.describe()
df.head(10)
# Using correlation matrix to find out the correlations btw the dataset features
correlation_mattrix = df.corr(method="pearson") # methods = pearson,kendall,spearman
correlation_mattrix
correlation_mattrix1 = df.corr(method="kendall") # methods = pearson,kendall,spearman
correlation_mattrix1
correlation_mattrix2 = df.corr(method="spearman") # methods = pearson,kendall,spearman
correlation_mattrix2
import matplotlib.pyplot as plt
# drawing heatmap using seaborn library method=pearson
sns.heatmap(correlation_mattrix, annot=True)
plt.title("Correlation Matrix")
plt.xlabel("Covid Features")
plt.ylabel("Covid Features")
plt.show()
sns.heatmap(correlation_mattrix1, annot=True)
plt.title("correlation mattix method=kendall")
plt.xlabel("features")
plt.ylabel("features")
plt.show()
sns.heatmap(correlation_mattrix2, annot=True)
plt.title("heatmap method=spearman")
plt.show()
# # We have high correlation between these features
# -confirmed =deaths,recovered,active,confirmed last week,one week change
# -deaths=recovered,active,confirmed last week,one week change
# -recovered=active,confirmed last week,one week change
# -deaths/100 cases=deaths/100 recoverd
# -confirmed last weeek=1 week chnange
# and vice versa
df2 = (
df.groupby("WHO Region")[["Confirmed", "Deaths", "Active", "Confirmed last week"]]
.sum()
.reset_index()
)
df2.head()
x = df2["WHO Region"]
y = df2["Deaths"]
plt.figure(figsize=(10, 6))
plt.bar(x, y)
plt.xlabel("Regions")
plt.ylabel("Deaths")
plt.title("REGIONS VS DEATHS")
plt.show()
# 10 countries with most deaths
top_deaths = (
df[["Country/Region", "Deaths"]]
.sort_values(by=["Deaths"], ascending=False)
.head(10)
)
top_deaths
sns.barplot(data=top_deaths, y="Country/Region", x="Deaths")
plt.title("TOP 10 COUNTRIES WITH HIGHEST NUMBER OF DEATHS")
plt.show()
# top ten recovered countries
top_recov = (
df[["Country/Region", "Recovered"]]
.sort_values(by=["Recovered"], ascending=False)
.head(10)
)
top_recov
plt.figure(figsize=(10, 6))
sns.barplot(data=top_recov, x="Country/Region", y="Recovered")
plt.title("TOP 10 RECOVERED COUNTRIES")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/319/129319649.ipynb
|
corona-virus-report
|
imdevskp
|
[{"Id": 129319649, "ScriptId": 38449285, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9361910, "CreationDate": "05/12/2023 18:29:43", "VersionNumber": 2.0, "Title": "Covid 19 Data Analsis", "EvaluationDate": "05/12/2023", "IsChange": false, "TotalLines": 87.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 87.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185256526, "KernelVersionId": 129319649, "SourceDatasetVersionId": 1402868}]
|
[{"Id": 1402868, "DatasetId": 494766, "DatasourceVersionId": 1435700, "CreatorUserId": 1302389, "LicenseName": "Other (specified in description)", "CreationDate": "08/07/2020 03:47:47", "VersionNumber": 166.0, "Title": "COVID-19 Dataset", "Slug": "corona-virus-report", "Subtitle": "Number of Confirmed, Death and Recovered cases every day across the globe", "Description": "[](https://forthebadge.com) [](https://forthebadge.com)\n\n### Context\n\n- A new coronavirus designated 2019-nCoV was first identified in Wuhan, the capital of China's Hubei province\n- People developed pneumonia without a clear cause and for which existing vaccines or treatments were not effective. \n- The virus has shown evidence of human-to-human transmission\n- Transmission rate (rate of infection) appeared to escalate in mid-January 2020\n- As of 30 January 2020, approximately 8,243 cases have been confirmed\n\n\n### Content\n\n> * **full_grouped.csv** - Day to day country wise no. of cases (Has County/State/Province level data) \n> * **covid_19_clean_complete.csv** - Day to day country wise no. of cases (Doesn't have County/State/Province level data) \n> * **country_wise_latest.csv** - Latest country level no. of cases \n> * **day_wise.csv** - Day wise no. of cases (Doesn't have country level data) \n> * **usa_county_wise.csv** - Day to day county level no. of cases \n> * **worldometer_data.csv** - Latest data from https://www.worldometers.info/ \n\n\n### Acknowledgements / Data Source\n\n> https://github.com/CSSEGISandData/COVID-19\n> https://www.worldometers.info/\n\n### Collection methodology\n\n> https://github.com/imdevskp/covid_19_jhu_data_web_scrap_and_cleaning\n\n### Cover Photo\n\n> Photo from National Institutes of Allergy and Infectious Diseases\n> https://www.niaid.nih.gov/news-events/novel-coronavirus-sarscov2-images\n> https://blogs.cdc.gov/publichealthmatters/2019/04/h1n1/\n\n### Similar Datasets\n\n> * COVID-19 - https://www.kaggle.com/imdevskp/corona-virus-report \n> * MERS - https://www.kaggle.com/imdevskp/mers-outbreak-dataset-20122019\n> * Ebola Western Africa 2014 Outbreak - https://www.kaggle.com/imdevskp/ebola-outbreak-20142016-complete-dataset\n> * H1N1 | Swine Flu 2009 Pandemic Dataset - https://www.kaggle.com/imdevskp/h1n1-swine-flu-2009-pandemic-dataset\n> * SARS 2003 Pandemic - https://www.kaggle.com/imdevskp/sars-outbreak-2003-complete-dataset\n> * HIV AIDS - https://www.kaggle.com/imdevskp/hiv-aids-dataset", "VersionNotes": "update", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 494766, "CreatorUserId": 1302389, "OwnerUserId": 1302389.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1402868.0, "CurrentDatasourceVersionId": 1435700.0, "ForumId": 507860, "Type": 2, "CreationDate": "01/30/2020 14:46:58", "LastActivityDate": "01/30/2020", "TotalViews": 1009073, "TotalDownloads": 271389, "TotalVotes": 2056, "TotalKernels": 642}]
|
[{"Id": 1302389, "UserName": "imdevskp", "DisplayName": "Devakumar K. P.", "RegisterDate": "09/30/2017", "PerformanceTier": 3}]
|
import pandas as pd
import numpy as np
import seaborn as sns
df = pd.read_csv("D:\ml-practice\projects\covid19\country_wise_latest.csv")
df.dtypes
df.isnull().sum()
df.drop(["New cases", "New deaths", "New recovered"], axis=1, inplace=True)
df.describe()
df.head(10)
# Using correlation matrix to find out the correlations btw the dataset features
correlation_mattrix = df.corr(method="pearson") # methods = pearson,kendall,spearman
correlation_mattrix
correlation_mattrix1 = df.corr(method="kendall") # methods = pearson,kendall,spearman
correlation_mattrix1
correlation_mattrix2 = df.corr(method="spearman") # methods = pearson,kendall,spearman
correlation_mattrix2
import matplotlib.pyplot as plt
# drawing heatmap using seaborn library method=pearson
sns.heatmap(correlation_mattrix, annot=True)
plt.title("Correlation Matrix")
plt.xlabel("Covid Features")
plt.ylabel("Covid Features")
plt.show()
sns.heatmap(correlation_mattrix1, annot=True)
plt.title("correlation mattix method=kendall")
plt.xlabel("features")
plt.ylabel("features")
plt.show()
sns.heatmap(correlation_mattrix2, annot=True)
plt.title("heatmap method=spearman")
plt.show()
# # We have high correlation between these features
# -confirmed =deaths,recovered,active,confirmed last week,one week change
# -deaths=recovered,active,confirmed last week,one week change
# -recovered=active,confirmed last week,one week change
# -deaths/100 cases=deaths/100 recoverd
# -confirmed last weeek=1 week chnange
# and vice versa
df2 = (
df.groupby("WHO Region")[["Confirmed", "Deaths", "Active", "Confirmed last week"]]
.sum()
.reset_index()
)
df2.head()
x = df2["WHO Region"]
y = df2["Deaths"]
plt.figure(figsize=(10, 6))
plt.bar(x, y)
plt.xlabel("Regions")
plt.ylabel("Deaths")
plt.title("REGIONS VS DEATHS")
plt.show()
# 10 countries with most deaths
top_deaths = (
df[["Country/Region", "Deaths"]]
.sort_values(by=["Deaths"], ascending=False)
.head(10)
)
top_deaths
sns.barplot(data=top_deaths, y="Country/Region", x="Deaths")
plt.title("TOP 10 COUNTRIES WITH HIGHEST NUMBER OF DEATHS")
plt.show()
# top ten recovered countries
top_recov = (
df[["Country/Region", "Recovered"]]
.sort_values(by=["Recovered"], ascending=False)
.head(10)
)
top_recov
plt.figure(figsize=(10, 6))
sns.barplot(data=top_recov, x="Country/Region", y="Recovered")
plt.title("TOP 10 RECOVERED COUNTRIES")
plt.show()
| false | 0 | 821 | 0 | 1,227 | 821 |
||
129319325
|
<jupyter_start><jupyter_text>Diabetes Dataset
### Context
This dataset is originally from the National Institute of Diabetes and Digestive and Kidney Diseases. The objective is to predict based on diagnostic measurements whether a patient has diabetes.
### Content
Several constraints were placed on the selection of these instances from a larger database. In particular, all patients here are females at least 21 years old of Pima Indian heritage.
- Pregnancies: Number of times pregnant
- Glucose: Plasma glucose concentration a 2 hours in an oral glucose tolerance test
- BloodPressure: Diastolic blood pressure (mm Hg)
- SkinThickness: Triceps skin fold thickness (mm)
- Insulin: 2-Hour serum insulin (mu U/ml)
- BMI: Body mass index (weight in kg/(height in m)^2)
- DiabetesPedigreeFunction: Diabetes pedigree function
- Age: Age (years)
- Outcome: Class variable (0 or 1)
#### Sources:
(a) Original owners: National Institute of Diabetes and Digestive and
Kidney Diseases
(b) Donor of database: Vincent Sigillito ([email protected])
Research Center, RMI Group Leader
Applied Physics Laboratory
The Johns Hopkins University
Johns Hopkins Road
Laurel, MD 20707
(301) 953-6231
(c) Date received: 9 May 1990
#### Past Usage:
1. Smith,~J.~W., Everhart,~J.~E., Dickson,~W.~C., Knowler,~W.~C., \&
Johannes,~R.~S. (1988). Using the ADAP learning algorithm to forecast
the onset of diabetes mellitus. In {\it Proceedings of the Symposium
on Computer Applications and Medical Care} (pp. 261--265). IEEE
Computer Society Press.
The diagnostic, binary-valued variable investigated is whether the
patient shows signs of diabetes according to World Health Organization
criteria (i.e., if the 2 hour post-load plasma glucose was at least
200 mg/dl at any survey examination or if found during routine medical
care). The population lives near Phoenix, Arizona, USA.
Results: Their ADAP algorithm makes a real-valued prediction between
0 and 1. This was transformed into a binary decision using a cutoff of
0.448. Using 576 training instances, the sensitivity and specificity
of their algorithm was 76% on the remaining 192 instances.
#### Relevant Information:
Several constraints were placed on the selection of these instances from
a larger database. In particular, all patients here are females at
least 21 years old of Pima Indian heritage. ADAP is an adaptive learning
routine that generates and executes digital analogs of perceptron-like
devices. It is a unique algorithm; see the paper for details.
#### Number of Instances: 768
#### Number of Attributes: 8 plus class
#### For Each Attribute: (all numeric-valued)
1. Number of times pregnant
2. Plasma glucose concentration a 2 hours in an oral glucose tolerance test
3. Diastolic blood pressure (mm Hg)
4. Triceps skin fold thickness (mm)
5. 2-Hour serum insulin (mu U/ml)
6. Body mass index (weight in kg/(height in m)^2)
7. Diabetes pedigree function
8. Age (years)
9. Class variable (0 or 1)
#### Missing Attribute Values: Yes
#### Class Distribution: (class value 1 is interpreted as "tested positive for
diabetes")
Kaggle dataset identifier: diabetes-data-set
<jupyter_code>import pandas as pd
df = pd.read_csv('diabetes-data-set/diabetes.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Pregnancies 768 non-null int64
1 Glucose 768 non-null int64
2 BloodPressure 768 non-null int64
3 SkinThickness 768 non-null int64
4 Insulin 768 non-null int64
5 BMI 768 non-null float64
6 DiabetesPedigreeFunction 768 non-null float64
7 Age 768 non-null int64
8 Outcome 768 non-null int64
dtypes: float64(2), int64(7)
memory usage: 54.1 KB
<jupyter_text>Examples:
{
"Pregnancies": 6.0,
"Glucose": 148.0,
"BloodPressure": 72.0,
"SkinThickness": 35.0,
"Insulin": 0.0,
"BMI": 33.6,
"DiabetesPedigreeFunction": 0.627,
"Age": 50.0,
"Outcome": 1.0
}
{
"Pregnancies": 1.0,
"Glucose": 85.0,
"BloodPressure": 66.0,
"SkinThickness": 29.0,
"Insulin": 0.0,
"BMI": 26.6,
"DiabetesPedigreeFunction": 0.35100000000000003,
"Age": 31.0,
"Outcome": 0.0
}
{
"Pregnancies": 8.0,
"Glucose": 183.0,
"BloodPressure": 64.0,
"SkinThickness": 0.0,
"Insulin": 0.0,
"BMI": 23.3,
"DiabetesPedigreeFunction": 0.672,
"Age": 32.0,
"Outcome": 1.0
}
{
"Pregnancies": 1.0,
"Glucose": 89.0,
"BloodPressure": 66.0,
"SkinThickness": 23.0,
"Insulin": 94.0,
"BMI": 28.1,
"DiabetesPedigreeFunction": 0.167,
"Age": 21.0,
"Outcome": 0.0
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
diabetes = pd.read_csv("/kaggle/input/diabetes-data-set/diabetes.csv")
diabetes
import statistics
np.mean(diabetes["BloodPressure"])
statistics.mode(diabetes["BloodPressure"])
import matplotlib.pyplot as plt
plt.bar(diabetes.index, diabetes["BloodPressure"])
np.median(diabetes["BloodPressure"])
plt.bar(diabetes.index, diabetes["Insulin"])
np.median(diabetes["Insulin"])
np.mean(diabetes["Insulin"])
statistics.mode(diabetes["Insulin"])
diabetes[
diabetes[(diabetes["Insulin"] > 16) | diabetes["Insulin"] < 166]["Outcome"] == 1
].count
diabetes[
diabetes[(diabetes["Insulin"] > 16) | diabetes["Insulin"] < 166]["Outcome"] == 0
].count
plt.bar(diabetes.index, diabetes["Age"])
np.mean(diabetes["Age"])
np.median(diabetes["Age"])
statistics.mode(diabetes["Age"])
diabetes[diabetes["Age"] == 22].count
diabetes["Age"].value_counts()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/319/129319325.ipynb
|
diabetes-data-set
|
mathchi
|
[{"Id": 129319325, "ScriptId": 38446686, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5750785, "CreationDate": "05/12/2023 18:25:29", "VersionNumber": 1.0, "Title": "statistical-analysis", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 62.0, "LinesInsertedFromPrevious": 62.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 185255804, "KernelVersionId": 129319325, "SourceDatasetVersionId": 1400440}]
|
[{"Id": 1400440, "DatasetId": 818300, "DatasourceVersionId": 1433199, "CreatorUserId": 3650837, "LicenseName": "CC0: Public Domain", "CreationDate": "08/05/2020 21:27:01", "VersionNumber": 1.0, "Title": "Diabetes Dataset", "Slug": "diabetes-data-set", "Subtitle": "This dataset is originally from the N. Inst. of Diabetes & Diges. & Kidney Dis.", "Description": "### Context\n\nThis dataset is originally from the National Institute of Diabetes and Digestive and Kidney Diseases. The objective is to predict based on diagnostic measurements whether a patient has diabetes.\n\n\n### Content\n\nSeveral constraints were placed on the selection of these instances from a larger database. In particular, all patients here are females at least 21 years old of Pima Indian heritage.\n\n- Pregnancies: Number of times pregnant \n- Glucose: Plasma glucose concentration a 2 hours in an oral glucose tolerance test \n- BloodPressure: Diastolic blood pressure (mm Hg) \n- SkinThickness: Triceps skin fold thickness (mm) \n- Insulin: 2-Hour serum insulin (mu U/ml) \n- BMI: Body mass index (weight in kg/(height in m)^2) \n- DiabetesPedigreeFunction: Diabetes pedigree function \n- Age: Age (years) \n- Outcome: Class variable (0 or 1)\n\n#### Sources:\n (a) Original owners: National Institute of Diabetes and Digestive and\n Kidney Diseases\n (b) Donor of database: Vincent Sigillito ([email protected])\n Research Center, RMI Group Leader\n Applied Physics Laboratory\n The Johns Hopkins University\n Johns Hopkins Road\n Laurel, MD 20707\n (301) 953-6231\n (c) Date received: 9 May 1990\n\n#### Past Usage:\n 1. Smith,~J.~W., Everhart,~J.~E., Dickson,~W.~C., Knowler,~W.~C., \\&\n Johannes,~R.~S. (1988). Using the ADAP learning algorithm to forecast\n the onset of diabetes mellitus. In {\\it Proceedings of the Symposium\n on Computer Applications and Medical Care} (pp. 261--265). IEEE\n Computer Society Press.\n\n The diagnostic, binary-valued variable investigated is whether the\n patient shows signs of diabetes according to World Health Organization\n criteria (i.e., if the 2 hour post-load plasma glucose was at least \n 200 mg/dl at any survey examination or if found during routine medical\n care). The population lives near Phoenix, Arizona, USA.\n\n Results: Their ADAP algorithm makes a real-valued prediction between\n 0 and 1. This was transformed into a binary decision using a cutoff of \n 0.448. Using 576 training instances, the sensitivity and specificity\n of their algorithm was 76% on the remaining 192 instances.\n\n#### Relevant Information:\n Several constraints were placed on the selection of these instances from\n a larger database. In particular, all patients here are females at\n least 21 years old of Pima Indian heritage. ADAP is an adaptive learning\n routine that generates and executes digital analogs of perceptron-like\n devices. It is a unique algorithm; see the paper for details.\n\n#### Number of Instances: 768\n\n#### Number of Attributes: 8 plus class \n\n#### For Each Attribute: (all numeric-valued)\n 1. Number of times pregnant\n 2. Plasma glucose concentration a 2 hours in an oral glucose tolerance test\n 3. Diastolic blood pressure (mm Hg)\n 4. Triceps skin fold thickness (mm)\n 5. 2-Hour serum insulin (mu U/ml)\n 6. Body mass index (weight in kg/(height in m)^2)\n 7. Diabetes pedigree function\n 8. Age (years)\n 9. Class variable (0 or 1)\n\n#### Missing Attribute Values: Yes\n\n#### Class Distribution: (class value 1 is interpreted as \"tested positive for\n diabetes\")", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 818300, "CreatorUserId": 3650837, "OwnerUserId": 3650837.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1400440.0, "CurrentDatasourceVersionId": 1433199.0, "ForumId": 833406, "Type": 2, "CreationDate": "08/05/2020 21:27:01", "LastActivityDate": "08/05/2020", "TotalViews": 440450, "TotalDownloads": 65613, "TotalVotes": 496, "TotalKernels": 245}]
|
[{"Id": 3650837, "UserName": "mathchi", "DisplayName": "Mehmet Akturk", "RegisterDate": "09/01/2019", "PerformanceTier": 3}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
diabetes = pd.read_csv("/kaggle/input/diabetes-data-set/diabetes.csv")
diabetes
import statistics
np.mean(diabetes["BloodPressure"])
statistics.mode(diabetes["BloodPressure"])
import matplotlib.pyplot as plt
plt.bar(diabetes.index, diabetes["BloodPressure"])
np.median(diabetes["BloodPressure"])
plt.bar(diabetes.index, diabetes["Insulin"])
np.median(diabetes["Insulin"])
np.mean(diabetes["Insulin"])
statistics.mode(diabetes["Insulin"])
diabetes[
diabetes[(diabetes["Insulin"] > 16) | diabetes["Insulin"] < 166]["Outcome"] == 1
].count
diabetes[
diabetes[(diabetes["Insulin"] > 16) | diabetes["Insulin"] < 166]["Outcome"] == 0
].count
plt.bar(diabetes.index, diabetes["Age"])
np.mean(diabetes["Age"])
np.median(diabetes["Age"])
statistics.mode(diabetes["Age"])
diabetes[diabetes["Age"] == 22].count
diabetes["Age"].value_counts()
|
[{"diabetes-data-set/diabetes.csv": {"column_names": "[\"Pregnancies\", \"Glucose\", \"BloodPressure\", \"SkinThickness\", \"Insulin\", \"BMI\", \"DiabetesPedigreeFunction\", \"Age\", \"Outcome\"]", "column_data_types": "{\"Pregnancies\": \"int64\", \"Glucose\": \"int64\", \"BloodPressure\": \"int64\", \"SkinThickness\": \"int64\", \"Insulin\": \"int64\", \"BMI\": \"float64\", \"DiabetesPedigreeFunction\": \"float64\", \"Age\": \"int64\", \"Outcome\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 768 entries, 0 to 767\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Pregnancies 768 non-null int64 \n 1 Glucose 768 non-null int64 \n 2 BloodPressure 768 non-null int64 \n 3 SkinThickness 768 non-null int64 \n 4 Insulin 768 non-null int64 \n 5 BMI 768 non-null float64\n 6 DiabetesPedigreeFunction 768 non-null float64\n 7 Age 768 non-null int64 \n 8 Outcome 768 non-null int64 \ndtypes: float64(2), int64(7)\nmemory usage: 54.1 KB\n", "summary": "{\"Pregnancies\": {\"count\": 768.0, \"mean\": 3.8450520833333335, \"std\": 3.3695780626988694, \"min\": 0.0, \"25%\": 1.0, \"50%\": 3.0, \"75%\": 6.0, \"max\": 17.0}, \"Glucose\": {\"count\": 768.0, \"mean\": 120.89453125, \"std\": 31.97261819513622, \"min\": 0.0, \"25%\": 99.0, \"50%\": 117.0, \"75%\": 140.25, \"max\": 199.0}, \"BloodPressure\": {\"count\": 768.0, \"mean\": 69.10546875, \"std\": 19.355807170644777, \"min\": 0.0, \"25%\": 62.0, \"50%\": 72.0, \"75%\": 80.0, \"max\": 122.0}, \"SkinThickness\": {\"count\": 768.0, \"mean\": 20.536458333333332, \"std\": 15.952217567727637, \"min\": 0.0, \"25%\": 0.0, \"50%\": 23.0, \"75%\": 32.0, \"max\": 99.0}, \"Insulin\": {\"count\": 768.0, \"mean\": 79.79947916666667, \"std\": 115.24400235133817, \"min\": 0.0, \"25%\": 0.0, \"50%\": 30.5, \"75%\": 127.25, \"max\": 846.0}, \"BMI\": {\"count\": 768.0, \"mean\": 31.992578124999998, \"std\": 7.884160320375446, \"min\": 0.0, \"25%\": 27.3, \"50%\": 32.0, \"75%\": 36.6, \"max\": 67.1}, \"DiabetesPedigreeFunction\": {\"count\": 768.0, \"mean\": 0.47187630208333325, \"std\": 0.3313285950127749, \"min\": 0.078, \"25%\": 0.24375, \"50%\": 0.3725, \"75%\": 0.62625, \"max\": 2.42}, \"Age\": {\"count\": 768.0, \"mean\": 33.240885416666664, \"std\": 11.760231540678685, \"min\": 21.0, \"25%\": 24.0, \"50%\": 29.0, \"75%\": 41.0, \"max\": 81.0}, \"Outcome\": {\"count\": 768.0, \"mean\": 0.3489583333333333, \"std\": 0.47695137724279896, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}}", "examples": "{\"Pregnancies\":{\"0\":6,\"1\":1,\"2\":8,\"3\":1},\"Glucose\":{\"0\":148,\"1\":85,\"2\":183,\"3\":89},\"BloodPressure\":{\"0\":72,\"1\":66,\"2\":64,\"3\":66},\"SkinThickness\":{\"0\":35,\"1\":29,\"2\":0,\"3\":23},\"Insulin\":{\"0\":0,\"1\":0,\"2\":0,\"3\":94},\"BMI\":{\"0\":33.6,\"1\":26.6,\"2\":23.3,\"3\":28.1},\"DiabetesPedigreeFunction\":{\"0\":0.627,\"1\":0.351,\"2\":0.672,\"3\":0.167},\"Age\":{\"0\":50,\"1\":31,\"2\":32,\"3\":21},\"Outcome\":{\"0\":1,\"1\":0,\"2\":1,\"3\":0}}"}}]
| true | 1 |
<start_data_description><data_path>diabetes-data-set/diabetes.csv:
<column_names>
['Pregnancies', 'Glucose', 'BloodPressure', 'SkinThickness', 'Insulin', 'BMI', 'DiabetesPedigreeFunction', 'Age', 'Outcome']
<column_types>
{'Pregnancies': 'int64', 'Glucose': 'int64', 'BloodPressure': 'int64', 'SkinThickness': 'int64', 'Insulin': 'int64', 'BMI': 'float64', 'DiabetesPedigreeFunction': 'float64', 'Age': 'int64', 'Outcome': 'int64'}
<dataframe_Summary>
{'Pregnancies': {'count': 768.0, 'mean': 3.8450520833333335, 'std': 3.3695780626988694, 'min': 0.0, '25%': 1.0, '50%': 3.0, '75%': 6.0, 'max': 17.0}, 'Glucose': {'count': 768.0, 'mean': 120.89453125, 'std': 31.97261819513622, 'min': 0.0, '25%': 99.0, '50%': 117.0, '75%': 140.25, 'max': 199.0}, 'BloodPressure': {'count': 768.0, 'mean': 69.10546875, 'std': 19.355807170644777, 'min': 0.0, '25%': 62.0, '50%': 72.0, '75%': 80.0, 'max': 122.0}, 'SkinThickness': {'count': 768.0, 'mean': 20.536458333333332, 'std': 15.952217567727637, 'min': 0.0, '25%': 0.0, '50%': 23.0, '75%': 32.0, 'max': 99.0}, 'Insulin': {'count': 768.0, 'mean': 79.79947916666667, 'std': 115.24400235133817, 'min': 0.0, '25%': 0.0, '50%': 30.5, '75%': 127.25, 'max': 846.0}, 'BMI': {'count': 768.0, 'mean': 31.992578124999998, 'std': 7.884160320375446, 'min': 0.0, '25%': 27.3, '50%': 32.0, '75%': 36.6, 'max': 67.1}, 'DiabetesPedigreeFunction': {'count': 768.0, 'mean': 0.47187630208333325, 'std': 0.3313285950127749, 'min': 0.078, '25%': 0.24375, '50%': 0.3725, '75%': 0.62625, 'max': 2.42}, 'Age': {'count': 768.0, 'mean': 33.240885416666664, 'std': 11.760231540678685, 'min': 21.0, '25%': 24.0, '50%': 29.0, '75%': 41.0, 'max': 81.0}, 'Outcome': {'count': 768.0, 'mean': 0.3489583333333333, 'std': 0.47695137724279896, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Pregnancies 768 non-null int64
1 Glucose 768 non-null int64
2 BloodPressure 768 non-null int64
3 SkinThickness 768 non-null int64
4 Insulin 768 non-null int64
5 BMI 768 non-null float64
6 DiabetesPedigreeFunction 768 non-null float64
7 Age 768 non-null int64
8 Outcome 768 non-null int64
dtypes: float64(2), int64(7)
memory usage: 54.1 KB
<some_examples>
{'Pregnancies': {'0': 6, '1': 1, '2': 8, '3': 1}, 'Glucose': {'0': 148, '1': 85, '2': 183, '3': 89}, 'BloodPressure': {'0': 72, '1': 66, '2': 64, '3': 66}, 'SkinThickness': {'0': 35, '1': 29, '2': 0, '3': 23}, 'Insulin': {'0': 0, '1': 0, '2': 0, '3': 94}, 'BMI': {'0': 33.6, '1': 26.6, '2': 23.3, '3': 28.1}, 'DiabetesPedigreeFunction': {'0': 0.627, '1': 0.351, '2': 0.672, '3': 0.167}, 'Age': {'0': 50, '1': 31, '2': 32, '3': 21}, 'Outcome': {'0': 1, '1': 0, '2': 1, '3': 0}}
<end_description>
| 508 | 1 | 2,215 | 508 |
129319372
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split, GridSearchCV, cross_val_score
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import confusion_matrix, classification_report, roc_curve, auc
import matplotlib.pyplot as plt
import numpy as np
# Load the data
df = pd.read_csv(
"/kaggle/input/amazon-cells-labelledtxt/amazon_cells_labelled.txt",
sep="\t",
header=None,
)
# Split the data into features (X) and target (y)
X = df[0]
y = df[1]
# Convert text data into TF-IDF
vectorizer = TfidfVectorizer()
X = vectorizer.fit_transform(X)
# Naive Bayes classifer
# Split the data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
# Define the model
naive_bayes = MultinomialNB()
# Define the grid search parameters
parameters = {"alpha": [0.1, 0.5, 1.0, 1.5, 2.0]}
# Conduct grid search
grid_search = GridSearchCV(estimator=naive_bayes, param_grid=parameters, cv=10)
grid_search.fit(X_train, y_train)
# Print the best score and parameters
print("Best Score: ", grid_search.best_score_)
print("Best Params: ", grid_search.best_params_)
# Apply the best parameters to the model
naive_bayes = MultinomialNB(alpha=grid_search.best_params_["alpha"])
naive_bayes.fit(X_train, y_train)
# Predict the test set
y_pred = naive_bayes.predict(X_test)
# Print the confusion matrix, classification report
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
# Compute ROC curve and ROC area
fpr, tpr, _ = roc_curve(y_test, y_pred)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, color="darkorange", label="ROC curve (area = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="navy", linestyle="--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic")
plt.legend(loc="lower right")
plt.show()
# Logistic Regression Classifier
from sklearn.linear_model import LogisticRegression
# Define the model
logistic_regression = LogisticRegression()
# Define the grid search parameters
parameters = {"C": [0.1, 0.5, 1.0, 1.5, 2.0]}
# Conduct grid search
grid_search = GridSearchCV(estimator=logistic_regression, param_grid=parameters, cv=10)
grid_search.fit(X_train, y_train)
# Print the best score and parameters
print("Best Score: ", grid_search.best_score_)
print("Best Params: ", grid_search.best_params_)
# Apply the best parameters to the model
logistic_regression = LogisticRegression(C=grid_search.best_params_["C"])
logistic_regression.fit(X_train, y_train)
# Predict the test set
y_pred = logistic_regression.predict(X_test)
# Print the confusion matrix, classification report
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
# Compute ROC curve and ROC area
fpr, tpr, _ = roc_curve(y_test, y_pred)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, color="darkorange", label="ROC curve (area = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="navy", linestyle="--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic")
plt.legend(loc="lower right")
plt.show()
# Decision Tree Classifier
from sklearn.tree import DecisionTreeClassifier
# Define the model
dt = DecisionTreeClassifier()
# Define the grid search parameters
parameters = {
"max_depth": [None, 10, 20, 30, 50],
"min_samples_split": [2, 5, 10],
"min_samples_leaf": [1, 2, 4],
}
# Conduct grid search
grid_search = GridSearchCV(estimator=dt, param_grid=parameters, cv=10)
grid_search.fit(X_train, y_train)
# Print the best score and parameters
print("Best Score: ", grid_search.best_score_)
print("Best Params: ", grid_search.best_params_)
# Apply the best parameters to the model
dt = DecisionTreeClassifier(
max_depth=grid_search.best_params_["max_depth"],
min_samples_split=grid_search.best_params_["min_samples_split"],
min_samples_leaf=grid_search.best_params_["min_samples_leaf"],
)
dt.fit(X_train, y_train)
# Predict the test set
y_pred = dt.predict(X_test)
# Print the confusion matrix, classification report
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
# Compute ROC curve and ROC area
fpr, tpr, _ = roc_curve(y_test, y_pred)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, color="darkorange", label="ROC curve (area = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="navy", linestyle="--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic")
plt.legend(loc="lower right")
plt.show()
# KNN Classifier
from sklearn.neighbors import KNeighborsClassifier
# Define the model
knn = KNeighborsClassifier()
# Define the grid search parameters
parameters = {"n_neighbors": [3, 5, 7, 9, 11]}
# Conduct grid search
grid_search = GridSearchCV(estimator=knn, param_grid=parameters, cv=10)
grid_search.fit(X_train, y_train)
# Print the best score and parameters
print("Best Score: ", grid_search.best_score_)
print("Best Params: ", grid_search.best_params_)
# Apply the best parameters to the model
knn = KNeighborsClassifier(n_neighbors=grid_search.best_params_["n_neighbors"])
knn.fit(X_train, y_train)
# Predict the test set
y_pred = knn.predict(X_test)
# Print the confusion matrix, classification report
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
# Compute ROC curve and ROC area
fpr, tpr, _ = roc_curve(y_test, y_pred)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, color="darkorange", label="ROC curve (area = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="navy", linestyle="--")
plt.xlim([0.0, 1.0])
plt.ylim
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/319/129319372.ipynb
| null | null |
[{"Id": 129319372, "ScriptId": 38445621, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3637157, "CreationDate": "05/12/2023 18:26:01", "VersionNumber": 1.0, "Title": "Assignment2_Nihad&Yusif", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 241.0, "LinesInsertedFromPrevious": 241.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split, GridSearchCV, cross_val_score
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import confusion_matrix, classification_report, roc_curve, auc
import matplotlib.pyplot as plt
import numpy as np
# Load the data
df = pd.read_csv(
"/kaggle/input/amazon-cells-labelledtxt/amazon_cells_labelled.txt",
sep="\t",
header=None,
)
# Split the data into features (X) and target (y)
X = df[0]
y = df[1]
# Convert text data into TF-IDF
vectorizer = TfidfVectorizer()
X = vectorizer.fit_transform(X)
# Naive Bayes classifer
# Split the data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
# Define the model
naive_bayes = MultinomialNB()
# Define the grid search parameters
parameters = {"alpha": [0.1, 0.5, 1.0, 1.5, 2.0]}
# Conduct grid search
grid_search = GridSearchCV(estimator=naive_bayes, param_grid=parameters, cv=10)
grid_search.fit(X_train, y_train)
# Print the best score and parameters
print("Best Score: ", grid_search.best_score_)
print("Best Params: ", grid_search.best_params_)
# Apply the best parameters to the model
naive_bayes = MultinomialNB(alpha=grid_search.best_params_["alpha"])
naive_bayes.fit(X_train, y_train)
# Predict the test set
y_pred = naive_bayes.predict(X_test)
# Print the confusion matrix, classification report
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
# Compute ROC curve and ROC area
fpr, tpr, _ = roc_curve(y_test, y_pred)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, color="darkorange", label="ROC curve (area = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="navy", linestyle="--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic")
plt.legend(loc="lower right")
plt.show()
# Logistic Regression Classifier
from sklearn.linear_model import LogisticRegression
# Define the model
logistic_regression = LogisticRegression()
# Define the grid search parameters
parameters = {"C": [0.1, 0.5, 1.0, 1.5, 2.0]}
# Conduct grid search
grid_search = GridSearchCV(estimator=logistic_regression, param_grid=parameters, cv=10)
grid_search.fit(X_train, y_train)
# Print the best score and parameters
print("Best Score: ", grid_search.best_score_)
print("Best Params: ", grid_search.best_params_)
# Apply the best parameters to the model
logistic_regression = LogisticRegression(C=grid_search.best_params_["C"])
logistic_regression.fit(X_train, y_train)
# Predict the test set
y_pred = logistic_regression.predict(X_test)
# Print the confusion matrix, classification report
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
# Compute ROC curve and ROC area
fpr, tpr, _ = roc_curve(y_test, y_pred)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, color="darkorange", label="ROC curve (area = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="navy", linestyle="--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic")
plt.legend(loc="lower right")
plt.show()
# Decision Tree Classifier
from sklearn.tree import DecisionTreeClassifier
# Define the model
dt = DecisionTreeClassifier()
# Define the grid search parameters
parameters = {
"max_depth": [None, 10, 20, 30, 50],
"min_samples_split": [2, 5, 10],
"min_samples_leaf": [1, 2, 4],
}
# Conduct grid search
grid_search = GridSearchCV(estimator=dt, param_grid=parameters, cv=10)
grid_search.fit(X_train, y_train)
# Print the best score and parameters
print("Best Score: ", grid_search.best_score_)
print("Best Params: ", grid_search.best_params_)
# Apply the best parameters to the model
dt = DecisionTreeClassifier(
max_depth=grid_search.best_params_["max_depth"],
min_samples_split=grid_search.best_params_["min_samples_split"],
min_samples_leaf=grid_search.best_params_["min_samples_leaf"],
)
dt.fit(X_train, y_train)
# Predict the test set
y_pred = dt.predict(X_test)
# Print the confusion matrix, classification report
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
# Compute ROC curve and ROC area
fpr, tpr, _ = roc_curve(y_test, y_pred)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, color="darkorange", label="ROC curve (area = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="navy", linestyle="--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic")
plt.legend(loc="lower right")
plt.show()
# KNN Classifier
from sklearn.neighbors import KNeighborsClassifier
# Define the model
knn = KNeighborsClassifier()
# Define the grid search parameters
parameters = {"n_neighbors": [3, 5, 7, 9, 11]}
# Conduct grid search
grid_search = GridSearchCV(estimator=knn, param_grid=parameters, cv=10)
grid_search.fit(X_train, y_train)
# Print the best score and parameters
print("Best Score: ", grid_search.best_score_)
print("Best Params: ", grid_search.best_params_)
# Apply the best parameters to the model
knn = KNeighborsClassifier(n_neighbors=grid_search.best_params_["n_neighbors"])
knn.fit(X_train, y_train)
# Predict the test set
y_pred = knn.predict(X_test)
# Print the confusion matrix, classification report
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
# Compute ROC curve and ROC area
fpr, tpr, _ = roc_curve(y_test, y_pred)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, color="darkorange", label="ROC curve (area = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="navy", linestyle="--")
plt.xlim([0.0, 1.0])
plt.ylim
| false | 0 | 2,216 | 0 | 2,216 | 2,216 |
||
129319188
|
# hide
from fastbook import *
from fastai.vision.widgets import *
setup_book()
plant_types = "poison ivy", "green"
path = Path("plants")
if not path.exists():
path.mkdir()
for o in plant_types:
dest = path / o
dest.mkdir(exist_ok=True)
results = search_images_ddg(f"{o} plant")
download_images(dest, urls=results)
fns = get_image_files(path)
fns
failed = verify_images(fns)
failed
failed.map(Path.unlink)
plants = DataBlock(
blocks=(ImageBlock, CategoryBlock),
get_items=get_image_files,
splitter=RandomSplitter(valid_pct=0.2, seed=42),
get_y=parent_label,
item_tfms=Resize(128),
)
plants = plants.new(
item_tfms=RandomResizedCrop(224, min_scale=0.5), batch_tfms=aug_transforms()
)
dls = plants.dataloaders(path)
learn = vision_learner(dls, resnet18, metrics=error_rate)
learn.fine_tune(4)
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix()
interp.plot_top_losses(5, nrows=1)
cleaner = ImageClassifierCleaner(learn)
cleaner
learn.export()
path = Path()
path.ls(file_exts=".pkl")
ims = [
"https://bgr.com/wp-content/uploads/2020/08/AdobeStock_155258329-Recovered-1.jpg?quality=70&strip=all"
]
dest = "images/poisonivy.jpg"
download_url(ims[0], dest)
learn.predict("images/poisonivy.jpg")
ims = [
"https://www.thespruce.com/thmb/3JCPAUHY6gHDg02aFaBfM1qKHBo=/4437x2958/filters:no_upscale():max_bytes(150000):strip_icc()/close-up-of-green-hellebore-flowers-562408117-5a942e5604d1cf0036b01143.jpg"
]
dest = "images/greenplant.jpg"
download_url(ims[0], dest)
learn.predict("images/greenplant.jpg")
learn = load_learner("export.pkl")
labels = learn.dls.vocab
def predict(img):
img = PILImage.create(img)
pred, pred_idx, probs = learn.predict(img)
return {labels[i]: float(probs[i]) for i in range(len(labels))}
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/319/129319188.ipynb
| null | null |
[{"Id": 129319188, "ScriptId": 38419326, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14693253, "CreationDate": "05/12/2023 18:23:43", "VersionNumber": 1.0, "Title": "Is Poison Ivy", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 72.0, "LinesInsertedFromPrevious": 72.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# hide
from fastbook import *
from fastai.vision.widgets import *
setup_book()
plant_types = "poison ivy", "green"
path = Path("plants")
if not path.exists():
path.mkdir()
for o in plant_types:
dest = path / o
dest.mkdir(exist_ok=True)
results = search_images_ddg(f"{o} plant")
download_images(dest, urls=results)
fns = get_image_files(path)
fns
failed = verify_images(fns)
failed
failed.map(Path.unlink)
plants = DataBlock(
blocks=(ImageBlock, CategoryBlock),
get_items=get_image_files,
splitter=RandomSplitter(valid_pct=0.2, seed=42),
get_y=parent_label,
item_tfms=Resize(128),
)
plants = plants.new(
item_tfms=RandomResizedCrop(224, min_scale=0.5), batch_tfms=aug_transforms()
)
dls = plants.dataloaders(path)
learn = vision_learner(dls, resnet18, metrics=error_rate)
learn.fine_tune(4)
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix()
interp.plot_top_losses(5, nrows=1)
cleaner = ImageClassifierCleaner(learn)
cleaner
learn.export()
path = Path()
path.ls(file_exts=".pkl")
ims = [
"https://bgr.com/wp-content/uploads/2020/08/AdobeStock_155258329-Recovered-1.jpg?quality=70&strip=all"
]
dest = "images/poisonivy.jpg"
download_url(ims[0], dest)
learn.predict("images/poisonivy.jpg")
ims = [
"https://www.thespruce.com/thmb/3JCPAUHY6gHDg02aFaBfM1qKHBo=/4437x2958/filters:no_upscale():max_bytes(150000):strip_icc()/close-up-of-green-hellebore-flowers-562408117-5a942e5604d1cf0036b01143.jpg"
]
dest = "images/greenplant.jpg"
download_url(ims[0], dest)
learn.predict("images/greenplant.jpg")
learn = load_learner("export.pkl")
labels = learn.dls.vocab
def predict(img):
img = PILImage.create(img)
pred, pred_idx, probs = learn.predict(img)
return {labels[i]: float(probs[i]) for i in range(len(labels))}
| false | 0 | 705 | 0 | 705 | 705 |
||
129319337
|
<jupyter_start><jupyter_text>Pakistan Data Talent
This comprehensive dataset features a collection of LinkedIn profiles belonging to talented data scientists hailing from Pakistan. It presents a valuable resource for researchers, recruiters, and data enthusiasts seeking insights into the diverse and growing field of data science within the Pakistani professional landscape.
The dataset includes the following key information for each profile: URL, full name, headline, and location. The profile URLs provide direct access to each individual's LinkedIn page, allowing users to explore their professional background, experiences, and expertise in more detail.
Whether you are a recruiter looking to identify potential candidates, a researcher investigating trends and skills in the Pakistani data science community, or simply an enthusiast curious about the professionals driving data-driven innovation in Pakistan, this dataset will prove invaluable.
By making this dataset available on Kaggle, we aim to foster collaboration, knowledge sharing, and networking opportunities within the Pakistani data science community. We encourage users to leverage this dataset for various analytical and research purposes, such as demographic analysis, skillset mapping, or creating tailored outreach strategies.
Note: The dataset contains publicly available information from LinkedIn profiles. We kindly request that users respect privacy and professional boundaries when utilizing this dataset, refraining from any unauthorized use or misuse of the provided information.
Start exploring the wealth of talent within the Pakistani data science domain by downloading this dataset today!
Kaggle dataset identifier: pakistan-data-talent
<jupyter_script>import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from collections import Counter
import nltk
from nltk.corpus import stopwords
nltk.download("stopwords")
# **Load the CSV file using pandas :**
data = pd.read_csv("/kaggle/input/pakistan-data-talent/Pakistan Data Talent.csv")
# **Display the first 5 rows :**
print(data.head())
# **Display information about the DataFrame :**
display(data.info())
# **Check if there are any missing values :**
print(data.isna().sum())
# **Display the distribution of locations :**
sns.set_style("whitegrid")
plt.figure(figsize=(12, 8))
ax = sns.countplot(x="Location", data=data)
ax.set_xticklabels(ax.get_xticklabels(), rotation=90)
plt.xlabel("Location", fontsize=12)
plt.ylabel("Number of observations", fontsize=12)
plt.title("Distribution of locations", fontsize=16)
plt.tick_params(labelsize=10)
plt.tight_layout()
plt.show()
# **Display the distribution of job titles :**
top_headlines = data["Headline"].value_counts().head(25)
sns.countplot(x="Headline", data=data, order=top_headlines.index)
plt.xticks(rotation=90)
plt.xlabel("Job Title")
plt.ylabel("Number of Profiles")
plt.title("Distribution of Job Titles")
plt.show()
# **Display the Word Frequency in Headlines :**
# Remove float values in the Headline column
data = data.dropna(subset=["Headline"])
# Remove stopwords
stop_words = set(stopwords.words("english"))
data["Headline"] = data["Headline"].apply(
lambda x: " ".join(
[word for word in str(x).split() if word.lower() not in stop_words]
)
)
# Count word frequency in headlines
word_freq = Counter(" ".join(data["Headline"]).split()).most_common(20)
# Display the result as a graph
plt.figure(figsize=(12, 6))
plt.bar([i[0] for i in word_freq], [i[1] for i in word_freq])
plt.xticks(rotation=90)
plt.title("Word frequency in headlines")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/319/129319337.ipynb
|
pakistan-data-talent
|
hskhawaja
|
[{"Id": 129319337, "ScriptId": 38447296, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14180659, "CreationDate": "05/12/2023 18:25:38", "VersionNumber": 1.0, "Title": "Exploratory Data Analysis of Pakistan Data Talent", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 66.0, "LinesInsertedFromPrevious": 66.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 5}]
|
[{"Id": 185255807, "KernelVersionId": 129319337, "SourceDatasetVersionId": 5670125}]
|
[{"Id": 5670125, "DatasetId": 3259472, "DatasourceVersionId": 5745628, "CreatorUserId": 938987, "LicenseName": "CC0: Public Domain", "CreationDate": "05/12/2023 12:59:27", "VersionNumber": 1.0, "Title": "Pakistan Data Talent", "Slug": "pakistan-data-talent", "Subtitle": "Tap into the Data Talent of Pakistan - Data Scientists, ML Engineers, BI Experts", "Description": "This comprehensive dataset features a collection of LinkedIn profiles belonging to talented data scientists hailing from Pakistan. It presents a valuable resource for researchers, recruiters, and data enthusiasts seeking insights into the diverse and growing field of data science within the Pakistani professional landscape.\n\nThe dataset includes the following key information for each profile: URL, full name, headline, and location. The profile URLs provide direct access to each individual's LinkedIn page, allowing users to explore their professional background, experiences, and expertise in more detail.\n\nWhether you are a recruiter looking to identify potential candidates, a researcher investigating trends and skills in the Pakistani data science community, or simply an enthusiast curious about the professionals driving data-driven innovation in Pakistan, this dataset will prove invaluable.\n\nBy making this dataset available on Kaggle, we aim to foster collaboration, knowledge sharing, and networking opportunities within the Pakistani data science community. We encourage users to leverage this dataset for various analytical and research purposes, such as demographic analysis, skillset mapping, or creating tailored outreach strategies.\n\nNote: The dataset contains publicly available information from LinkedIn profiles. We kindly request that users respect privacy and professional boundaries when utilizing this dataset, refraining from any unauthorized use or misuse of the provided information.\n\nStart exploring the wealth of talent within the Pakistani data science domain by downloading this dataset today!", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3259472, "CreatorUserId": 938987, "OwnerUserId": 938987.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5689263.0, "CurrentDatasourceVersionId": 5764863.0, "ForumId": 3325009, "Type": 2, "CreationDate": "05/12/2023 12:59:27", "LastActivityDate": "05/12/2023", "TotalViews": 2838, "TotalDownloads": 204, "TotalVotes": 25, "TotalKernels": 2}]
|
[{"Id": 938987, "UserName": "hskhawaja", "DisplayName": "Hussain Shahbaz Khawaja", "RegisterDate": "03/02/2017", "PerformanceTier": 2}]
|
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from collections import Counter
import nltk
from nltk.corpus import stopwords
nltk.download("stopwords")
# **Load the CSV file using pandas :**
data = pd.read_csv("/kaggle/input/pakistan-data-talent/Pakistan Data Talent.csv")
# **Display the first 5 rows :**
print(data.head())
# **Display information about the DataFrame :**
display(data.info())
# **Check if there are any missing values :**
print(data.isna().sum())
# **Display the distribution of locations :**
sns.set_style("whitegrid")
plt.figure(figsize=(12, 8))
ax = sns.countplot(x="Location", data=data)
ax.set_xticklabels(ax.get_xticklabels(), rotation=90)
plt.xlabel("Location", fontsize=12)
plt.ylabel("Number of observations", fontsize=12)
plt.title("Distribution of locations", fontsize=16)
plt.tick_params(labelsize=10)
plt.tight_layout()
plt.show()
# **Display the distribution of job titles :**
top_headlines = data["Headline"].value_counts().head(25)
sns.countplot(x="Headline", data=data, order=top_headlines.index)
plt.xticks(rotation=90)
plt.xlabel("Job Title")
plt.ylabel("Number of Profiles")
plt.title("Distribution of Job Titles")
plt.show()
# **Display the Word Frequency in Headlines :**
# Remove float values in the Headline column
data = data.dropna(subset=["Headline"])
# Remove stopwords
stop_words = set(stopwords.words("english"))
data["Headline"] = data["Headline"].apply(
lambda x: " ".join(
[word for word in str(x).split() if word.lower() not in stop_words]
)
)
# Count word frequency in headlines
word_freq = Counter(" ".join(data["Headline"]).split()).most_common(20)
# Display the result as a graph
plt.figure(figsize=(12, 6))
plt.bar([i[0] for i in word_freq], [i[1] for i in word_freq])
plt.xticks(rotation=90)
plt.title("Word frequency in headlines")
plt.show()
| false | 1 | 597 | 5 | 960 | 597 |
||
129319655
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import nltk
nltk.download("punkt")
nltk.download("stopwords")
nltk.download("wordnet")
nltk.download("vader_lexicon")
# importing required modules
import PyPDF2
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
# creating a pdf file object
name = ""
name = "/kaggle/input/sentiment-analysis-mnb/" + filename
print("\n", name)
pdfFileObj = open(name, "rb")
# creating a pdf reader object
pdfReader = PyPDF2.PdfReader(pdfFileObj)
# printing number of pages in pdf file
n = len(pdfReader.pages)
# creating a page object
pageObj = pdfReader.pages[0]
# extracting text from pages
string = ""
for i in range(n - 1):
pageObj = pdfReader.pages[i]
string = string + " " + pageObj.extract_text()
# closing the pdf file object
pdfFileObj.close()
from vaderSentiment.vaderSentiment import SentimentIntensityAnalyzer
# importing required modules
from vaderSentiment.vaderSentiment import SentimentIntensityAnalyzer
import PyPDF2
sentiment = SentimentIntensityAnalyzer()
text_1 = "Unless suffering is the direct and immediate object of life, our existence must entirely fail of its aim. It is absurd to look upon the enormous amount of pain that abounds everywhere in the world, and originates in needs and necessities inseparable from life itself, as serving no purpose at all and the result of mere chance. Each separate misfortune, as it comes, seems, no doubt, to be something exceptional but misfortune in general is the rule."
sent_1 = sentiment.polarity_scores(text_1)
print(sent_1)
# importing required modules
from textblob import TextBlob
import PyPDF2
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
# creating a pdf file object
name = ""
name = "/kaggle/input/sentiment-analysis-mnb/" + filename
pdfFileObj = open(name, "rb")
# creating a pdf reader object
pdfReader = PyPDF2.PdfReader(pdfFileObj)
# printing number of pages in pdf file
n = len(pdfReader.pages)
# creating a page object
pageObj = pdfReader.pages[0]
# extracting text from pages
string = ""
for i in range(n - 1):
pageObj = pdfReader.pages[i]
string = string + " " + pageObj.extract_text()
blob = TextBlob(string)
print("\n", filename)
print(blob.sentiment)
# closing the pdf file object
pdfFileObj.close()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/319/129319655.ipynb
| null | null |
[{"Id": 129319655, "ScriptId": 38324356, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14301685, "CreationDate": "05/12/2023 18:29:47", "VersionNumber": 1.0, "Title": "notebook8df56ce024", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 117.0, "LinesInsertedFromPrevious": 117.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import nltk
nltk.download("punkt")
nltk.download("stopwords")
nltk.download("wordnet")
nltk.download("vader_lexicon")
# importing required modules
import PyPDF2
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
# creating a pdf file object
name = ""
name = "/kaggle/input/sentiment-analysis-mnb/" + filename
print("\n", name)
pdfFileObj = open(name, "rb")
# creating a pdf reader object
pdfReader = PyPDF2.PdfReader(pdfFileObj)
# printing number of pages in pdf file
n = len(pdfReader.pages)
# creating a page object
pageObj = pdfReader.pages[0]
# extracting text from pages
string = ""
for i in range(n - 1):
pageObj = pdfReader.pages[i]
string = string + " " + pageObj.extract_text()
# closing the pdf file object
pdfFileObj.close()
from vaderSentiment.vaderSentiment import SentimentIntensityAnalyzer
# importing required modules
from vaderSentiment.vaderSentiment import SentimentIntensityAnalyzer
import PyPDF2
sentiment = SentimentIntensityAnalyzer()
text_1 = "Unless suffering is the direct and immediate object of life, our existence must entirely fail of its aim. It is absurd to look upon the enormous amount of pain that abounds everywhere in the world, and originates in needs and necessities inseparable from life itself, as serving no purpose at all and the result of mere chance. Each separate misfortune, as it comes, seems, no doubt, to be something exceptional but misfortune in general is the rule."
sent_1 = sentiment.polarity_scores(text_1)
print(sent_1)
# importing required modules
from textblob import TextBlob
import PyPDF2
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
# creating a pdf file object
name = ""
name = "/kaggle/input/sentiment-analysis-mnb/" + filename
pdfFileObj = open(name, "rb")
# creating a pdf reader object
pdfReader = PyPDF2.PdfReader(pdfFileObj)
# printing number of pages in pdf file
n = len(pdfReader.pages)
# creating a page object
pageObj = pdfReader.pages[0]
# extracting text from pages
string = ""
for i in range(n - 1):
pageObj = pdfReader.pages[i]
string = string + " " + pageObj.extract_text()
blob = TextBlob(string)
print("\n", filename)
print(blob.sentiment)
# closing the pdf file object
pdfFileObj.close()
| false | 0 | 832 | 0 | 832 | 832 |
||
129031454
|
# # Introduction à l'IA
# ## Initialiation du projet
import numpy as np
import pandas as pd
df = pd.read_csv(
"/kaggle/input/epidemiological-data-from-the-covid-19-outbreak/data.csv"
)
# ****
# ## Analyse du dataset
print("Data types : ", df.info())
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/031/129031454.ipynb
| null | null |
[{"Id": 129031454, "ScriptId": 38354681, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15037675, "CreationDate": "05/10/2023 12:51:08", "VersionNumber": 1.0, "Title": "notebookb5b2a91504", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 76.0, "LinesInsertedFromPrevious": 76.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Introduction à l'IA
# ## Initialiation du projet
import numpy as np
import pandas as pd
df = pd.read_csv(
"/kaggle/input/epidemiological-data-from-the-covid-19-outbreak/data.csv"
)
# ****
# ## Analyse du dataset
print("Data types : ", df.info())
| false | 0 | 89 | 0 | 89 | 89 |
||
129031792
|
import numpy as np
import tensorflow as tf
import tensorflow as tf
import urllib
import zipfile
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.optimizers import RMSprop
import json
import tensorflow as tf
import numpy as np
import urllib
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
import tensorflow_hub as hub
import pandas as pd
import tensorflow as tf
def solution_model():
xs = np.array([-1.0, 0.0, 1.0, 2.0, 3.0, 4.0], dtype=float)
ys = np.array([5.0, 6.0, 7.0, 8.0, 9.0, 10.0], dtype=float)
# YOUR CODE HERE
model = tf.keras.Sequential([tf.keras.layers.Dense(1, input_shape=[1])])
model.compile(loss="mean_squared_error", optimizer="sgd")
model.fit(xs, ys, epochs=1000)
return model
if __name__ == "__main__":
model = solution_model()
model.save("mymodel.h5")
def solution_model():
fashion_mnist = tf.keras.datasets.fashion_mnist
# YOUR CODE HERE
(training_images, training_labels), (
val_images,
val_label,
) = fashion_mnist.load_data()
training_images = training_images / 255.0
val_images = val_images / 255.0
training_images = np.expand_dims(training_images, axis=3)
val_images = np.expand_dims(val_images, axis=3)
model = tf.keras.Sequential(
[
tf.keras.layers.Conv2D(
14, (3, 3), activation="relu", input_shape=(28, 28, 1)
),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(10, activation="softmax"),
]
)
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["acc"]
)
model.fit(
training_images,
training_labels,
validation_data=(val_images, val_label),
epochs=5,
)
return model
if __name__ == "__main__":
model = solution_model()
model.save("mymodel.h5")
def solution_model():
_TRAIN_URL = (
"https://storage.googleapis.com/download.tensorflow.org/data/horse-or-human.zip"
)
_TEST_URL = "https://storage.googleapis.com/download.tensorflow.org/data/validation-horse-or-human.zip"
urllib.request.urlretrieve(_TRAIN_URL, "horse-or-human.zip")
local_zip = "horse-or-human.zip"
zip_ref = zipfile.ZipFile(local_zip, "r")
zip_ref.extractall("tmp/horse-or-human/")
zip_ref.close()
urllib.request.urlretrieve(_TEST_URL, "testdata.zip")
local_zip = "testdata.zip"
zip_ref = zipfile.ZipFile(local_zip, "r")
zip_ref.extractall("tmp/testdata/")
zip_ref.close()
training_data = "tmp/horse-or-human/"
val_data = "tmp/testdata/"
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
)
validation_datagen = ImageDataGenerator(rescale=1.0 / 255.0)
train_generator = train_datagen.flow_from_directory(
training_data, target_size=(300, 300), batch_size=128, class_mode="binary"
)
validation_generator = validation_datagen.flow_from_directory(
val_data, target_size=(300, 300), batch_size=64, class_mode="binary"
)
model = tf.keras.models.Sequential(
[
# Note the input shape specified on your first layer must be (300,300,3)
# Your Code here
tf.keras.layers.Conv2D(
16, (3, 3), activation="relu", input_shape=(300, 300, 3)
),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(32, (3, 3), activation="relu"),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(64, (3, 3), activation="relu"),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(64, (3, 3), activation="relu"),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(64, (3, 3), activation="relu"),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(512, activation="relu"),
# This is the last layer. You should not change this code.
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(
loss="binary_crossentropy", optimizer=RMSprop(lr=0.001), metrics=["accuracy"]
)
model.fit(
train_generator, epochs=10, verbose=1, validation_data=validation_generator
)
return model
if __name__ == "__main__":
model = solution_model()
model.save("mymodel2.h5")
def solution_model():
url = "https://storage.googleapis.com/download.tensorflow.org/data/sarcasm.json"
urllib.request.urlretrieve(url, "sarcasm.json")
# DO NOT CHANGE THIS CODE OR THE TESTS MAY NOT WORK
vocab_size = 1000
embedding_dim = 16
max_length = 120
trunc_type = "post"
padding_type = "post"
oov_tok = "<OOV>"
training_size = 20000
sentences = []
labels = []
# YOUR CODE HERE
with open("sarcasm.json", "r") as f:
data = json.load(f)
for text in data:
sentences.append(text["headline"])
labels.append(text["is_sarcastic"])
train_sentences = sentences[:training_size]
test_sentences = sentences[training_size:]
train_labels = labels[:training_size]
test_labels = labels[training_size:]
tokenizer = Tokenizer(num_words=vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(train_sentences)
# Sequen n Padded
train_sequences = tokenizer.texts_to_sequences(train_sentences)
train_padded = pad_sequences(
train_sequences, maxlen=max_length, padding=padding_type, truncating=trunc_type
)
test_sequences = tokenizer.texts_to_sequences(test_sentences)
test_padded = pad_sequences(
test_sequences, maxlen=max_length, padding=padding_type, truncating=trunc_type
)
train_labels = np.array(train_labels)
test_labels = np.array(test_labels)
model = tf.keras.Sequential(
[
# YOUR CODE HERE. KEEP THIS OUTPUT LAYER INTACT OR TESTS MAY FAIL
tf.keras.layers.Embedding(
vocab_size, embedding_dim, input_length=max_length
),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(32)),
tf.keras.layers.Dense(24, activation="relu"),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["acc"])
model.fit(
train_padded,
train_labels,
epochs=10,
validation_data=(test_padded, test_labels),
)
return model
if __name__ == "__main__":
model = solution_model()
model.save("mymodel6.h5")
def normalize_series(data, min, max):
data = data - min
data = data / max
return data
# This function is used to map the time series dataset into windows of
# features and respective targets, to prepare it for training and validation.
# The first element of the first window will be the first element of
# the dataset.
#
# Consecutive windows are constructed by shifting the starting position
# of the first window forward, one at a time (indicated by shift=1).
#
# For a window of n_past number of observations of the time
# indexed variable in the dataset, the target for the window is the next
# n_future number of observations of the variable, after the
# end of the window.
# DO NOT CHANGE THIS.
def windowed_dataset(series, batch_size, n_past=10, n_future=10, shift=1):
ds = tf.data.Dataset.from_tensor_slices(series)
ds = ds.window(size=n_past + n_future, shift=shift, drop_remainder=True)
ds = ds.flat_map(lambda w: w.batch(n_past + n_future))
ds = ds.map(lambda w: (w[:n_past], w[n_past:]))
return ds.batch(batch_size).prefetch(1)
# This function loads the data from the CSV file, normalizes the data and
# splits the dataset into train and validation data. It also uses
# windowed_dataset() to split the data into windows of observations and
# targets. Finally it defines, compiles and trains a neural network. This
# function returns the final trained model.
# COMPLETE THE CODE IN THIS FUNCTION
def solution_model():
# DO NOT CHANGE THIS CODE
# Reads the dataset.
df = pd.read_csv(
"Weekly_U.S.Diesel_Retail_Prices.csv",
infer_datetime_format=True,
index_col="Week of",
header=0,
)
# Number of features in the dataset. We use all features as predictors to
# predict all features of future time steps.
N_FEATURES = len(df.columns) # DO NOT CHANGE THIS
# Normalizes the data
data = df.values
data = normalize_series(data, data.min(axis=0), data.max(axis=0))
# Splits the data into training and validation sets.
SPLIT_TIME = int(len(data) * 0.8) # DO NOT CHANGE THIS
x_train = data[:SPLIT_TIME]
x_valid = data[SPLIT_TIME:]
# DO NOT CHANGE THIS CODE
tf.keras.backend.clear_session()
tf.random.set_seed(42)
# DO NOT CHANGE BATCH_SIZE IF YOU ARE USING STATEFUL LSTM/RNN/GRU.
# THE TEST WILL FAIL TO GRADE YOUR SCORE IN SUCH CASES.
# In other cases, it is advised not to change the batch size since it
# might affect your final scores. While setting it to a lower size
# might not do any harm, higher sizes might affect your scores.
BATCH_SIZE = 32 # ADVISED NOT TO CHANGE THIS
# DO NOT CHANGE N_PAST, N_FUTURE, SHIFT. The tests will fail to run
# on the server.
# Number of past time steps based on which future observations should be
# predicted
N_PAST = 10 # DO NOT CHANGE THIS
# Number of future time steps which are to be predicted.
N_FUTURE = 10 # DO NOT CHANGE THIS
# By how many positions the window slides to create a new window
# of observations.
SHIFT = 1 # DO NOT CHANGE THIS
# Code to create windowed train and validation datasets.
train_set = windowed_dataset(
series=x_train,
batch_size=BATCH_SIZE,
n_past=N_PAST,
n_future=N_FUTURE,
shift=SHIFT,
)
valid_set = windowed_dataset(
series=x_valid,
batch_size=BATCH_SIZE,
n_past=N_PAST,
n_future=N_FUTURE,
shift=SHIFT,
)
# Code to define your model.
encoder_inputs = tf.keras.layers.Input(shape=(n_past, n_features))
encoder_l1 = tf.keras.layers.LSTM(100, return_state=True)
encoder_outputs1 = encoder_l1(encoder_inputs)
encoder_states1 = encoder_outputs1[1:]
#
decoder_inputs = tf.keras.layers.RepeatVector(n_future)(encoder_outputs1[0])
#
decoder_l1 = tf.keras.layers.LSTM(100, return_sequences=True)(
decoder_inputs, initial_state=encoder_states1
)
decoder_outputs1 = tf.keras.layers.TimeDistributed(
tf.keras.layers.Dense(n_features)
)(decoder_l1)
#
model_e1d1 = tf.keras.models.Model(encoder_inputs, decoder_outputs1)
# Code to train and compile the model
optimizer = tf.keras.optimizers.SGD(learning_rate=1e-5, momentum=0.9)
model.compile(loss=tf.keras.losses.Huber(), optimizer=optimizer, metrics=["mae"])
model.fit(train_set, validation_data=valid_set, epochs=30)
return model
if __name__ == "__main__":
model = solution_model()
model.save("c5q12.h5")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/031/129031792.ipynb
| null | null |
[{"Id": 129031792, "ScriptId": 28639950, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7136013, "CreationDate": "05/10/2023 12:54:00", "VersionNumber": 1.0, "Title": "TensorFlow Developer Test", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 321.0, "LinesInsertedFromPrevious": 321.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import tensorflow as tf
import tensorflow as tf
import urllib
import zipfile
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.optimizers import RMSprop
import json
import tensorflow as tf
import numpy as np
import urllib
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
import tensorflow_hub as hub
import pandas as pd
import tensorflow as tf
def solution_model():
xs = np.array([-1.0, 0.0, 1.0, 2.0, 3.0, 4.0], dtype=float)
ys = np.array([5.0, 6.0, 7.0, 8.0, 9.0, 10.0], dtype=float)
# YOUR CODE HERE
model = tf.keras.Sequential([tf.keras.layers.Dense(1, input_shape=[1])])
model.compile(loss="mean_squared_error", optimizer="sgd")
model.fit(xs, ys, epochs=1000)
return model
if __name__ == "__main__":
model = solution_model()
model.save("mymodel.h5")
def solution_model():
fashion_mnist = tf.keras.datasets.fashion_mnist
# YOUR CODE HERE
(training_images, training_labels), (
val_images,
val_label,
) = fashion_mnist.load_data()
training_images = training_images / 255.0
val_images = val_images / 255.0
training_images = np.expand_dims(training_images, axis=3)
val_images = np.expand_dims(val_images, axis=3)
model = tf.keras.Sequential(
[
tf.keras.layers.Conv2D(
14, (3, 3), activation="relu", input_shape=(28, 28, 1)
),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(10, activation="softmax"),
]
)
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["acc"]
)
model.fit(
training_images,
training_labels,
validation_data=(val_images, val_label),
epochs=5,
)
return model
if __name__ == "__main__":
model = solution_model()
model.save("mymodel.h5")
def solution_model():
_TRAIN_URL = (
"https://storage.googleapis.com/download.tensorflow.org/data/horse-or-human.zip"
)
_TEST_URL = "https://storage.googleapis.com/download.tensorflow.org/data/validation-horse-or-human.zip"
urllib.request.urlretrieve(_TRAIN_URL, "horse-or-human.zip")
local_zip = "horse-or-human.zip"
zip_ref = zipfile.ZipFile(local_zip, "r")
zip_ref.extractall("tmp/horse-or-human/")
zip_ref.close()
urllib.request.urlretrieve(_TEST_URL, "testdata.zip")
local_zip = "testdata.zip"
zip_ref = zipfile.ZipFile(local_zip, "r")
zip_ref.extractall("tmp/testdata/")
zip_ref.close()
training_data = "tmp/horse-or-human/"
val_data = "tmp/testdata/"
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
)
validation_datagen = ImageDataGenerator(rescale=1.0 / 255.0)
train_generator = train_datagen.flow_from_directory(
training_data, target_size=(300, 300), batch_size=128, class_mode="binary"
)
validation_generator = validation_datagen.flow_from_directory(
val_data, target_size=(300, 300), batch_size=64, class_mode="binary"
)
model = tf.keras.models.Sequential(
[
# Note the input shape specified on your first layer must be (300,300,3)
# Your Code here
tf.keras.layers.Conv2D(
16, (3, 3), activation="relu", input_shape=(300, 300, 3)
),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(32, (3, 3), activation="relu"),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(64, (3, 3), activation="relu"),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(64, (3, 3), activation="relu"),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Conv2D(64, (3, 3), activation="relu"),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(512, activation="relu"),
# This is the last layer. You should not change this code.
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(
loss="binary_crossentropy", optimizer=RMSprop(lr=0.001), metrics=["accuracy"]
)
model.fit(
train_generator, epochs=10, verbose=1, validation_data=validation_generator
)
return model
if __name__ == "__main__":
model = solution_model()
model.save("mymodel2.h5")
def solution_model():
url = "https://storage.googleapis.com/download.tensorflow.org/data/sarcasm.json"
urllib.request.urlretrieve(url, "sarcasm.json")
# DO NOT CHANGE THIS CODE OR THE TESTS MAY NOT WORK
vocab_size = 1000
embedding_dim = 16
max_length = 120
trunc_type = "post"
padding_type = "post"
oov_tok = "<OOV>"
training_size = 20000
sentences = []
labels = []
# YOUR CODE HERE
with open("sarcasm.json", "r") as f:
data = json.load(f)
for text in data:
sentences.append(text["headline"])
labels.append(text["is_sarcastic"])
train_sentences = sentences[:training_size]
test_sentences = sentences[training_size:]
train_labels = labels[:training_size]
test_labels = labels[training_size:]
tokenizer = Tokenizer(num_words=vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(train_sentences)
# Sequen n Padded
train_sequences = tokenizer.texts_to_sequences(train_sentences)
train_padded = pad_sequences(
train_sequences, maxlen=max_length, padding=padding_type, truncating=trunc_type
)
test_sequences = tokenizer.texts_to_sequences(test_sentences)
test_padded = pad_sequences(
test_sequences, maxlen=max_length, padding=padding_type, truncating=trunc_type
)
train_labels = np.array(train_labels)
test_labels = np.array(test_labels)
model = tf.keras.Sequential(
[
# YOUR CODE HERE. KEEP THIS OUTPUT LAYER INTACT OR TESTS MAY FAIL
tf.keras.layers.Embedding(
vocab_size, embedding_dim, input_length=max_length
),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(32)),
tf.keras.layers.Dense(24, activation="relu"),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["acc"])
model.fit(
train_padded,
train_labels,
epochs=10,
validation_data=(test_padded, test_labels),
)
return model
if __name__ == "__main__":
model = solution_model()
model.save("mymodel6.h5")
def normalize_series(data, min, max):
data = data - min
data = data / max
return data
# This function is used to map the time series dataset into windows of
# features and respective targets, to prepare it for training and validation.
# The first element of the first window will be the first element of
# the dataset.
#
# Consecutive windows are constructed by shifting the starting position
# of the first window forward, one at a time (indicated by shift=1).
#
# For a window of n_past number of observations of the time
# indexed variable in the dataset, the target for the window is the next
# n_future number of observations of the variable, after the
# end of the window.
# DO NOT CHANGE THIS.
def windowed_dataset(series, batch_size, n_past=10, n_future=10, shift=1):
ds = tf.data.Dataset.from_tensor_slices(series)
ds = ds.window(size=n_past + n_future, shift=shift, drop_remainder=True)
ds = ds.flat_map(lambda w: w.batch(n_past + n_future))
ds = ds.map(lambda w: (w[:n_past], w[n_past:]))
return ds.batch(batch_size).prefetch(1)
# This function loads the data from the CSV file, normalizes the data and
# splits the dataset into train and validation data. It also uses
# windowed_dataset() to split the data into windows of observations and
# targets. Finally it defines, compiles and trains a neural network. This
# function returns the final trained model.
# COMPLETE THE CODE IN THIS FUNCTION
def solution_model():
# DO NOT CHANGE THIS CODE
# Reads the dataset.
df = pd.read_csv(
"Weekly_U.S.Diesel_Retail_Prices.csv",
infer_datetime_format=True,
index_col="Week of",
header=0,
)
# Number of features in the dataset. We use all features as predictors to
# predict all features of future time steps.
N_FEATURES = len(df.columns) # DO NOT CHANGE THIS
# Normalizes the data
data = df.values
data = normalize_series(data, data.min(axis=0), data.max(axis=0))
# Splits the data into training and validation sets.
SPLIT_TIME = int(len(data) * 0.8) # DO NOT CHANGE THIS
x_train = data[:SPLIT_TIME]
x_valid = data[SPLIT_TIME:]
# DO NOT CHANGE THIS CODE
tf.keras.backend.clear_session()
tf.random.set_seed(42)
# DO NOT CHANGE BATCH_SIZE IF YOU ARE USING STATEFUL LSTM/RNN/GRU.
# THE TEST WILL FAIL TO GRADE YOUR SCORE IN SUCH CASES.
# In other cases, it is advised not to change the batch size since it
# might affect your final scores. While setting it to a lower size
# might not do any harm, higher sizes might affect your scores.
BATCH_SIZE = 32 # ADVISED NOT TO CHANGE THIS
# DO NOT CHANGE N_PAST, N_FUTURE, SHIFT. The tests will fail to run
# on the server.
# Number of past time steps based on which future observations should be
# predicted
N_PAST = 10 # DO NOT CHANGE THIS
# Number of future time steps which are to be predicted.
N_FUTURE = 10 # DO NOT CHANGE THIS
# By how many positions the window slides to create a new window
# of observations.
SHIFT = 1 # DO NOT CHANGE THIS
# Code to create windowed train and validation datasets.
train_set = windowed_dataset(
series=x_train,
batch_size=BATCH_SIZE,
n_past=N_PAST,
n_future=N_FUTURE,
shift=SHIFT,
)
valid_set = windowed_dataset(
series=x_valid,
batch_size=BATCH_SIZE,
n_past=N_PAST,
n_future=N_FUTURE,
shift=SHIFT,
)
# Code to define your model.
encoder_inputs = tf.keras.layers.Input(shape=(n_past, n_features))
encoder_l1 = tf.keras.layers.LSTM(100, return_state=True)
encoder_outputs1 = encoder_l1(encoder_inputs)
encoder_states1 = encoder_outputs1[1:]
#
decoder_inputs = tf.keras.layers.RepeatVector(n_future)(encoder_outputs1[0])
#
decoder_l1 = tf.keras.layers.LSTM(100, return_sequences=True)(
decoder_inputs, initial_state=encoder_states1
)
decoder_outputs1 = tf.keras.layers.TimeDistributed(
tf.keras.layers.Dense(n_features)
)(decoder_l1)
#
model_e1d1 = tf.keras.models.Model(encoder_inputs, decoder_outputs1)
# Code to train and compile the model
optimizer = tf.keras.optimizers.SGD(learning_rate=1e-5, momentum=0.9)
model.compile(loss=tf.keras.losses.Huber(), optimizer=optimizer, metrics=["mae"])
model.fit(train_set, validation_data=valid_set, epochs=30)
return model
if __name__ == "__main__":
model = solution_model()
model.save("c5q12.h5")
| false | 0 | 3,361 | 0 | 3,361 | 3,361 |
||
129476476
|
<jupyter_start><jupyter_text>The shortest path data
This is a small project that uses optimization algorithms to find the five shortest paths. However, every spot has a limit; every spot ball couldn't exceed 100.
Kaggle dataset identifier: ga-optimization
<jupyter_script>import pandas as pd
import gurobipy as gp
from gurobipy import *
import math as m
import random as rand
import networkx as nx
import matplotlib.pyplot as plt
data = pd.read_excel(r"/kaggle/input/ga-optimization/AI term project.xlsx")
# #### 先將題目縮小至只有前10個點
data_5 = data[:5]
data_5
def distance(x1, x2, y1, y2):
dis = m.pow(m.pow((x1 - x2), 2) + m.pow((y1 - y2), 2), 0.5)
return round(dis, 4)
distance(3, 4, 1, 2)
all_nodes_connect = []
for node_x in range(0, len(data_5["X"])):
for node_y in range(0, len(data_5["Y"])):
all_nodes_connect.append((node_x, node_y))
for_trans_matrix = list()
for nodes in all_nodes_connect:
if nodes[0] != nodes[1]:
dis = distance(
data_5["X"].iloc[nodes[0]],
data_5["X"].iloc[nodes[1]],
data_5["Y"].iloc[nodes[0]],
data_5["Y"].iloc[nodes[1]],
)
for_trans_matrix.append([nodes, dis])
# for_trans_matrix
# ### Set the parameters
# * cost matrix
# * N : the numbers of nodes
# * K : the type of vehicles
# * C : Capacity of every vehicles
# * M : the number of all vehicles
cost_matrix = tupledict(for_trans_matrix)
# N =list(range(0,len(data['寶可夢座標點'])))
N = list(range(0, len(data_5["寶可夢座標點"])))
K = [1]
C = {1: 100}
# M = {1:5}
model = Model(name="VRP")
cost_matrix.keys()[1][0], cost_matrix.keys()[1][1]
cost_matrix
# ## 增設決策變數
X = {}
for i, one in zip(cost_matrix.keys(), range(0, len(cost_matrix.keys()))):
if i[0] != i[1]:
index_ = "x" + str(i[0]) + "," + str(i[1])
# print(index_)
X[i] = model.addVar(
vtype=GRB.BINARY, name=index_
) # 在這邊已經將變數全部放入置一個dict裡面,故下面若要呼叫變數,必須以dict的方式呼叫
model.update()
decision_list = list(X.values())
# decision_list
# nx.Graph() 無向圖
graph = nx.DiGraph() # 有向圖
color = list()
for node in N:
if node == 0:
graph.add_node(node)
color.append("red")
else:
graph.add_node(node)
color.append("gray")
for key, values in zip(cost_matrix.keys(), cost_matrix.values()):
graph.add_edge(key[0], key[1], weight=values)
weight = nx.get_edge_attributes(graph, "weight")
pos = nx.get_node_attributes(graph, "pos")
nx.draw(graph, node_color=color, with_labels=True)
# dj = nx.shortest_path(graph,source =1 ,target = 0)
# print(dj)
# plt.figure(figsize = (15,15))
# nx.draw_networkx_edge_labels(graph,pos,edge_labels = weight) #產出各節點
decision_list[0]
# ## 增設目標式
model.setObjective(
gp.quicksum(
cost_matrix.values()[cost] * decision
for decision, cost in zip(decision_list, range(0, len(cost_matrix)))
),
GRB.MINIMIZE,
) # 記得他的目標式一定要用這種FORM!
model.update()
decision_list[0]
# model.addConstr(gp.quicksum(decision * ))
# ### 將現況的函數設定至文件中,可用txt打開
model.write("/kaggle/working/vrp_model.lp")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/476/129476476.ipynb
|
ga-optimization
|
yinn94
|
[{"Id": 129476476, "ScriptId": 38338780, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8215644, "CreationDate": "05/14/2023 06:31:36", "VersionNumber": 5.0, "Title": "Gurobipy way find the shortest", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 113.0, "LinesInsertedFromPrevious": 44.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 69.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185569519, "KernelVersionId": 129476476, "SourceDatasetVersionId": 5292295}]
|
[{"Id": 5292295, "DatasetId": 3077588, "DatasourceVersionId": 5365381, "CreatorUserId": 8215644, "LicenseName": "Unknown", "CreationDate": "04/02/2023 06:54:31", "VersionNumber": 1.0, "Title": "The shortest path data", "Slug": "ga-optimization", "Subtitle": "Using optimization to find the shortest path.", "Description": "This is a small project that uses optimization algorithms to find the five shortest paths. However, every spot has a limit; every spot ball couldn't exceed 100.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3077588, "CreatorUserId": 8215644, "OwnerUserId": 8215644.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5292295.0, "CurrentDatasourceVersionId": 5365381.0, "ForumId": 3140511, "Type": 2, "CreationDate": "04/02/2023 06:54:31", "LastActivityDate": "04/02/2023", "TotalViews": 58, "TotalDownloads": 8, "TotalVotes": 0, "TotalKernels": 3}]
|
[{"Id": 8215644, "UserName": "yinn94", "DisplayName": "yinn94", "RegisterDate": "08/26/2021", "PerformanceTier": 1}]
|
import pandas as pd
import gurobipy as gp
from gurobipy import *
import math as m
import random as rand
import networkx as nx
import matplotlib.pyplot as plt
data = pd.read_excel(r"/kaggle/input/ga-optimization/AI term project.xlsx")
# #### 先將題目縮小至只有前10個點
data_5 = data[:5]
data_5
def distance(x1, x2, y1, y2):
dis = m.pow(m.pow((x1 - x2), 2) + m.pow((y1 - y2), 2), 0.5)
return round(dis, 4)
distance(3, 4, 1, 2)
all_nodes_connect = []
for node_x in range(0, len(data_5["X"])):
for node_y in range(0, len(data_5["Y"])):
all_nodes_connect.append((node_x, node_y))
for_trans_matrix = list()
for nodes in all_nodes_connect:
if nodes[0] != nodes[1]:
dis = distance(
data_5["X"].iloc[nodes[0]],
data_5["X"].iloc[nodes[1]],
data_5["Y"].iloc[nodes[0]],
data_5["Y"].iloc[nodes[1]],
)
for_trans_matrix.append([nodes, dis])
# for_trans_matrix
# ### Set the parameters
# * cost matrix
# * N : the numbers of nodes
# * K : the type of vehicles
# * C : Capacity of every vehicles
# * M : the number of all vehicles
cost_matrix = tupledict(for_trans_matrix)
# N =list(range(0,len(data['寶可夢座標點'])))
N = list(range(0, len(data_5["寶可夢座標點"])))
K = [1]
C = {1: 100}
# M = {1:5}
model = Model(name="VRP")
cost_matrix.keys()[1][0], cost_matrix.keys()[1][1]
cost_matrix
# ## 增設決策變數
X = {}
for i, one in zip(cost_matrix.keys(), range(0, len(cost_matrix.keys()))):
if i[0] != i[1]:
index_ = "x" + str(i[0]) + "," + str(i[1])
# print(index_)
X[i] = model.addVar(
vtype=GRB.BINARY, name=index_
) # 在這邊已經將變數全部放入置一個dict裡面,故下面若要呼叫變數,必須以dict的方式呼叫
model.update()
decision_list = list(X.values())
# decision_list
# nx.Graph() 無向圖
graph = nx.DiGraph() # 有向圖
color = list()
for node in N:
if node == 0:
graph.add_node(node)
color.append("red")
else:
graph.add_node(node)
color.append("gray")
for key, values in zip(cost_matrix.keys(), cost_matrix.values()):
graph.add_edge(key[0], key[1], weight=values)
weight = nx.get_edge_attributes(graph, "weight")
pos = nx.get_node_attributes(graph, "pos")
nx.draw(graph, node_color=color, with_labels=True)
# dj = nx.shortest_path(graph,source =1 ,target = 0)
# print(dj)
# plt.figure(figsize = (15,15))
# nx.draw_networkx_edge_labels(graph,pos,edge_labels = weight) #產出各節點
decision_list[0]
# ## 增設目標式
model.setObjective(
gp.quicksum(
cost_matrix.values()[cost] * decision
for decision, cost in zip(decision_list, range(0, len(cost_matrix)))
),
GRB.MINIMIZE,
) # 記得他的目標式一定要用這種FORM!
model.update()
decision_list[0]
# model.addConstr(gp.quicksum(decision * ))
# ### 將現況的函數設定至文件中,可用txt打開
model.write("/kaggle/working/vrp_model.lp")
| false | 0 | 1,103 | 0 | 1,161 | 1,103 |
||
129476225
|
# loading libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import metrics
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import ExtraTreesClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from tensorflow import keras
from tensorflow.keras.layers import Input, Dense
from tensorflow.keras.models import Sequential
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras import activations
# loading datasets
path_train = "/kaggle/input/icr-identify-age-related-conditions/train.csv"
path_test = "/kaggle/input/icr-identify-age-related-conditions/test.csv"
path_submis = "/kaggle/input/icr-identify-age-related-conditions/sample_submission.csv"
path_greeks = "/kaggle/input/icr-identify-age-related-conditions/greeks.csv"
train = pd.read_csv(path_train).drop(columns="Id")
test = pd.read_csv(path_test).drop(columns="Id")
greeks = pd.read_csv(path_greeks)
train["EJ"] = train["EJ"].map({"A": 0, "B": 1})
test["EJ"] = test["EJ"].map({"A": 0, "B": 1})
# shape for each datasets
print(f"Shape of the train data : {train.shape}")
print(f"Shape of the test data : {test.shape}")
# checking missing values train dataset
train_miss = train.isnull().sum()
print(f"Column Count")
for index, row in train_miss[train_miss > 0].items():
print(f"{index} {row}")
# ***We can use visualization techniques to discover missing values. The heatmap is appropriate for visualization. Each line indicates missing data in a row.***
plt.figure(figsize=(16, 14))
sns.heatmap(train.isnull(), yticklabels=False, cbar=False, cmap="PuBuGn")
plt.show()
# ***There are some common methods for handling missing values in a Pandas DataFrame: fillna(), interpolate() and SimpleImputer from sklearn.impute***
# fill missing values with the mean of the column
train_mean_filled = train.copy()
train_mean_filled.fillna(train_mean_filled.mean(), inplace=True)
# correlation coefficent columns for target
corr_target = train_mean_filled.corrwith(train_mean_filled["Class"])[:-1].sort_values(
ascending=False
)
plt.figure(figsize=(10, 10))
sns.barplot(y=corr_target.index, x=corr_target.values)
plt.show()
# interpolate missing values using linear interpolation
train_interpolate = train.copy()
train_interpolate.interpolate(method="polynomial", order=5)
# correlation coefficent columns for target
corr_target = train_interpolate.corrwith(train_interpolate["Class"])[:-1].sort_values(
ascending=False
)
plt.figure(figsize=(10, 10))
sns.barplot(y=corr_target.index, x=corr_target.values)
plt.show()
from sklearn.impute import SimpleImputer
# create an imputer object and fit it to the data
imputer = SimpleImputer(strategy="mean")
imputer.fit(train)
# transform the data and replace missing values
train_imputed = pd.DataFrame(imputer.transform(train), columns=train.columns)
# correlation coefficent columns for target
corr_target = train_imputed.corrwith(train_imputed["Class"])[:-1].sort_values(
ascending=False
)
plt.figure(figsize=(10, 10))
sns.barplot(y=corr_target.index, x=corr_target.values)
plt.show()
corr = train.iloc[:, 1:].corr()
mask = np.triu(np.ones_like(corr, dtype=bool))
plt.figure(figsize=(16, 14))
ax = sns.heatmap(
corr,
vmin=-1,
vmax=1,
center=0,
cmap=sns.diverging_palette(20, 220, n=200),
square=True,
mask=mask,
)
ax.set_xticklabels(ax.get_xticklabels(), rotation=45, horizontalalignment="right")
labels = ["Class 0", "Class 1"]
sizes = [train["Class"].tolist().count(0), train["Class"].tolist().count(1)]
explode = (0, 0.1)
fig, ax = plt.subplots()
ax.pie(
sizes,
explode=explode,
labels=labels,
autopct="%1.2f%%",
shadow=True,
startangle=180,
)
plt.show()
# Condition the regression fit on another variable and represent it using color
sns.lmplot(data=train_mean_filled, x="AB", y="AZ", hue="Class")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/476/129476225.ipynb
| null | null |
[{"Id": 129476225, "ScriptId": 38471728, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9401530, "CreationDate": "05/14/2023 06:29:35", "VersionNumber": 1.0, "Title": "Age-Related Conditions EDA and Classification", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 141.0, "LinesInsertedFromPrevious": 141.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# loading libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import metrics
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import ExtraTreesClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from tensorflow import keras
from tensorflow.keras.layers import Input, Dense
from tensorflow.keras.models import Sequential
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras import activations
# loading datasets
path_train = "/kaggle/input/icr-identify-age-related-conditions/train.csv"
path_test = "/kaggle/input/icr-identify-age-related-conditions/test.csv"
path_submis = "/kaggle/input/icr-identify-age-related-conditions/sample_submission.csv"
path_greeks = "/kaggle/input/icr-identify-age-related-conditions/greeks.csv"
train = pd.read_csv(path_train).drop(columns="Id")
test = pd.read_csv(path_test).drop(columns="Id")
greeks = pd.read_csv(path_greeks)
train["EJ"] = train["EJ"].map({"A": 0, "B": 1})
test["EJ"] = test["EJ"].map({"A": 0, "B": 1})
# shape for each datasets
print(f"Shape of the train data : {train.shape}")
print(f"Shape of the test data : {test.shape}")
# checking missing values train dataset
train_miss = train.isnull().sum()
print(f"Column Count")
for index, row in train_miss[train_miss > 0].items():
print(f"{index} {row}")
# ***We can use visualization techniques to discover missing values. The heatmap is appropriate for visualization. Each line indicates missing data in a row.***
plt.figure(figsize=(16, 14))
sns.heatmap(train.isnull(), yticklabels=False, cbar=False, cmap="PuBuGn")
plt.show()
# ***There are some common methods for handling missing values in a Pandas DataFrame: fillna(), interpolate() and SimpleImputer from sklearn.impute***
# fill missing values with the mean of the column
train_mean_filled = train.copy()
train_mean_filled.fillna(train_mean_filled.mean(), inplace=True)
# correlation coefficent columns for target
corr_target = train_mean_filled.corrwith(train_mean_filled["Class"])[:-1].sort_values(
ascending=False
)
plt.figure(figsize=(10, 10))
sns.barplot(y=corr_target.index, x=corr_target.values)
plt.show()
# interpolate missing values using linear interpolation
train_interpolate = train.copy()
train_interpolate.interpolate(method="polynomial", order=5)
# correlation coefficent columns for target
corr_target = train_interpolate.corrwith(train_interpolate["Class"])[:-1].sort_values(
ascending=False
)
plt.figure(figsize=(10, 10))
sns.barplot(y=corr_target.index, x=corr_target.values)
plt.show()
from sklearn.impute import SimpleImputer
# create an imputer object and fit it to the data
imputer = SimpleImputer(strategy="mean")
imputer.fit(train)
# transform the data and replace missing values
train_imputed = pd.DataFrame(imputer.transform(train), columns=train.columns)
# correlation coefficent columns for target
corr_target = train_imputed.corrwith(train_imputed["Class"])[:-1].sort_values(
ascending=False
)
plt.figure(figsize=(10, 10))
sns.barplot(y=corr_target.index, x=corr_target.values)
plt.show()
corr = train.iloc[:, 1:].corr()
mask = np.triu(np.ones_like(corr, dtype=bool))
plt.figure(figsize=(16, 14))
ax = sns.heatmap(
corr,
vmin=-1,
vmax=1,
center=0,
cmap=sns.diverging_palette(20, 220, n=200),
square=True,
mask=mask,
)
ax.set_xticklabels(ax.get_xticklabels(), rotation=45, horizontalalignment="right")
labels = ["Class 0", "Class 1"]
sizes = [train["Class"].tolist().count(0), train["Class"].tolist().count(1)]
explode = (0, 0.1)
fig, ax = plt.subplots()
ax.pie(
sizes,
explode=explode,
labels=labels,
autopct="%1.2f%%",
shadow=True,
startangle=180,
)
plt.show()
# Condition the regression fit on another variable and represent it using color
sns.lmplot(data=train_mean_filled, x="AB", y="AZ", hue="Class")
| false | 0 | 1,277 | 0 | 1,277 | 1,277 |
||
129595038
|
import numpy as np
import pandas as pd
from datasets import load_dataset
import nltk
from nltk.tokenize import sent_tokenize
from transformers import pipeline, set_seed
from datasets import load_metric
# # 一、 Dataset
# 用于摘要的规范数据集 `CNN/DailyMail corpus.`
# - 包含300,000对数据
# - 新闻文章 -- 相关摘要
# - 由美国有线电视新闻网和《每日邮报》在文章中附上的要点组成
# - 摘要是抽象的,而不是提取的,这意味着它们由新的句子组成,而不是简单的摘录
dataset = load_dataset("cnn_dailymail", version="3.0.0")
print(f'Features: {dataset["train"].column_names}')
sample = dataset["train"][1]
print(
f"""
Article (excerpt of 500 characters, total length: {len(sample["article"])}):
"""
)
print(sample["article"][:500])
print(f'\nSummary (length: {len(sample["highlights"])}):')
print(sample["highlights"])
# 文本非常长,输入文章有时候会是summary的长度的17倍。长句子的输入对于transformer来说是一个很大的挑战。
# - 处理方式: 即使末尾也有一些信息。我们仍然需要基于选择的模型的最大token对句子进行裁剪
# # 二、 pipeline
sample_text = dataset["train"][1]["article"][:2000]
summaries = {}
nltk.download("punkt")
str_ = "The U.S. are a country. The U.N. is an organization."
sent_tokenize(str_)
# ## 2.1 baseline
# 就用文章前三句作为摘要。
def three_sentence_summary(text):
return "\n".join(sent_tokenize(text)[:3])
summaries["baseline"] = three_sentence_summary(sample_text)
# ## 2.2 gpt-2
# 生成摘要需要增加`\nTL;DR:\n`
# - too long; didn't read
# - 经常在Reddit等平台上使用,表示长帖子的短版本
set_seed(42)
pipe_ = pipeline("text-generation", model="gpt2")
gpt2_query = sample_text + "\nTL;DR:\n"
pipe_out = pipe_(gpt2_query, max_length=512, clean_up_tokenization_spaces=True)
summaries["gpt2"] = "\n".join(
sent_tokenize(pipe_out[0]["generated_text"][len(gpt2_query) :])
)
# ## 2.3 T5(`Text-to-Text Transfer Transformer`)
# 用混合数据训练
# - 无监督数据: 重建masked单词
# - 监督数据: 一些任务学习
# - 文本摘要: 如"summarize:"
# - 翻译: 如"translate English to German:"
pipe_ = pipeline("summarization", model="t5-large")
pipe_out = pipe_(sample_text)
summaries["t5"] = "\n".join(sent_tokenize(pipe_out[0]["summary_text"]))
# ## 2.4 BART
# > encoder-decoder 结构
# 结合BERT和GPT-2的preTrain方法
pipe_ = pipeline("summarization", model="facebook/bart-large-cnn")
pipe_out = pipe_(sample_text)
summaries["bart"] = "\n".join(sent_tokenize(pipe_out[0]["summary_text"]))
# ## 2.5 PEGASUS
# 同样是encode-decoder结构
# 为了找到一个比一般语言建模更接近摘要的预训练目标,他们在一个非常大的语料库中自动识别包含其周围段落的大部分内容的句子(使用摘要评估指标作为内容重叠的启发式方法),并预训练PEGASUS模型来重建这些句子,从而获得用于文本摘要的最先进的模型。
pipe_ = pipeline("summarization", model="google/pegasus-cnn_dailymail")
pipe_out = pipe_(sample_text)
summaries["pegasus"] = pipe_out[0]["summary_text"].replace(" .<n>", ".\n")
# ## 比对
print("GROUND TRUTH")
print(dataset["train"][1]["highlights"])
print("")
for model_name in summaries:
print(model_name.upper())
print("--" * 25)
print(summaries[model_name])
print("")
# # 三、文本生成评估指标
# ## 3.1 BLEU (`precision-based metric`)
# - 评估准确率: 生成句子中有m个单词出现在原文(n个词)中, $bleu=\frac{m}{n}$
# - 引起问题1:
# - 如果生成重复的词,并且该词在引用中出现,那么我们会得到较高的分数
# - 针对这点作者指出修正方法:一个单词只计算它在引用中出现的次数。
# - example: ref-"the cat is on the mat" g-"the the the the the the"
# - $P_{vanlilla}=\frac{6}{6}$, $P_{mod}=\frac{2}{6}$
# - 修正问题1:`clip`
# - 这意味着一个`n-gram`的出现次数以它在参考句中出现的次数为上限
# $$p_n=\frac{ \sum_{geSnt \in C}\sum_{n-gram \in geSnt} Count_{clip}(n-gram) }{ \sum_{geSnt \in C}\sum_{n-gram \in geSnt} Count(n-gram) }$$
# - 引起问题2:
# - 因为这个准确率的评估,很显然会对较短的评估对有力,会低估较长生成的结果。
# - 修正问题2:简短惩罚 `brevity penalty`
# - $BR = min(1, e^{1 - \frac{l_{ref}}{l_{gen}}} )$ : 生成长度大于原句子:1, 生成长度小于原句子:$(0, 1)$,
# **最终公式:**
# $$BLEU-N=BR * (\prod_{n=1}^N p_n)^{1/N}$$
# Example: 计算`BLEU-4`
# - ref-"the cat sat on the mat"
# - g-"the cat the cat is on the mat"
# - **BR**: $BR=min(1, e^{1-6/8})=1$
# - **n=1**
# - 1-gram: org:{"the", "cat", "sat", "on", "mat"} ge:{"the", "cat", "is", "on", "mat"}
# - clip: $count_{clip}("the") = 2, count_{clip}("cat") = 1, count_{clip}("is") = 0, 1-gram \in geSnt$
# - $p_1 = \frac{5}{8} $
# - **n=2**
# - 2-gram: org:{"the cat", "cat sat", "sat on", "on the", "the mat"} ge:{"the cat", "cat the", "cat is", "is on", "on the", "the mat"}
# - $p_2 = \frac{3}{7} $
# - **n=3**
# - 3-gram: org:{"the cat sat", "cat sat on", "sat on the", "on the mat"} ge:{"the cat the", "cat the cat", "the cat is", "cat is on", "is on the", "on the mat"}
# - $p_3 = \frac{1}{6} $
# - **n=4**
# - 3-gram: org:{"the cat sat on", "cat sat on the", "sat on the mat"} ge:{"the cat the cat", "cat the cat is", "the cat is on", "cat is on the", "is on the mat"}
# - $p_4 = \frac{0}{5} $
# - **BLEU-4**: $1 * (\frac{5}{8}*\frac{3}{7}*\frac{1}{6}*\frac{0}{5})^{1/2}=0.$
#
bleu_metric = load_metric("sacrebleu")
bleu_metric.add(
prediction="the cat the cat is on the mat", reference=["the cat sat on the mat"]
)
results = bleu_metric.compute(smooth_method="floor", smooth_value=0)
results
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/595/129595038.ipynb
| null | null |
[{"Id": 129595038, "ScriptId": 38530239, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1679103, "CreationDate": "05/15/2023 05:53:34", "VersionNumber": 1.0, "Title": "NLPTransformers-Chapter6-Summarization", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 185.0, "LinesInsertedFromPrevious": 185.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
from datasets import load_dataset
import nltk
from nltk.tokenize import sent_tokenize
from transformers import pipeline, set_seed
from datasets import load_metric
# # 一、 Dataset
# 用于摘要的规范数据集 `CNN/DailyMail corpus.`
# - 包含300,000对数据
# - 新闻文章 -- 相关摘要
# - 由美国有线电视新闻网和《每日邮报》在文章中附上的要点组成
# - 摘要是抽象的,而不是提取的,这意味着它们由新的句子组成,而不是简单的摘录
dataset = load_dataset("cnn_dailymail", version="3.0.0")
print(f'Features: {dataset["train"].column_names}')
sample = dataset["train"][1]
print(
f"""
Article (excerpt of 500 characters, total length: {len(sample["article"])}):
"""
)
print(sample["article"][:500])
print(f'\nSummary (length: {len(sample["highlights"])}):')
print(sample["highlights"])
# 文本非常长,输入文章有时候会是summary的长度的17倍。长句子的输入对于transformer来说是一个很大的挑战。
# - 处理方式: 即使末尾也有一些信息。我们仍然需要基于选择的模型的最大token对句子进行裁剪
# # 二、 pipeline
sample_text = dataset["train"][1]["article"][:2000]
summaries = {}
nltk.download("punkt")
str_ = "The U.S. are a country. The U.N. is an organization."
sent_tokenize(str_)
# ## 2.1 baseline
# 就用文章前三句作为摘要。
def three_sentence_summary(text):
return "\n".join(sent_tokenize(text)[:3])
summaries["baseline"] = three_sentence_summary(sample_text)
# ## 2.2 gpt-2
# 生成摘要需要增加`\nTL;DR:\n`
# - too long; didn't read
# - 经常在Reddit等平台上使用,表示长帖子的短版本
set_seed(42)
pipe_ = pipeline("text-generation", model="gpt2")
gpt2_query = sample_text + "\nTL;DR:\n"
pipe_out = pipe_(gpt2_query, max_length=512, clean_up_tokenization_spaces=True)
summaries["gpt2"] = "\n".join(
sent_tokenize(pipe_out[0]["generated_text"][len(gpt2_query) :])
)
# ## 2.3 T5(`Text-to-Text Transfer Transformer`)
# 用混合数据训练
# - 无监督数据: 重建masked单词
# - 监督数据: 一些任务学习
# - 文本摘要: 如"summarize:"
# - 翻译: 如"translate English to German:"
pipe_ = pipeline("summarization", model="t5-large")
pipe_out = pipe_(sample_text)
summaries["t5"] = "\n".join(sent_tokenize(pipe_out[0]["summary_text"]))
# ## 2.4 BART
# > encoder-decoder 结构
# 结合BERT和GPT-2的preTrain方法
pipe_ = pipeline("summarization", model="facebook/bart-large-cnn")
pipe_out = pipe_(sample_text)
summaries["bart"] = "\n".join(sent_tokenize(pipe_out[0]["summary_text"]))
# ## 2.5 PEGASUS
# 同样是encode-decoder结构
# 为了找到一个比一般语言建模更接近摘要的预训练目标,他们在一个非常大的语料库中自动识别包含其周围段落的大部分内容的句子(使用摘要评估指标作为内容重叠的启发式方法),并预训练PEGASUS模型来重建这些句子,从而获得用于文本摘要的最先进的模型。
pipe_ = pipeline("summarization", model="google/pegasus-cnn_dailymail")
pipe_out = pipe_(sample_text)
summaries["pegasus"] = pipe_out[0]["summary_text"].replace(" .<n>", ".\n")
# ## 比对
print("GROUND TRUTH")
print(dataset["train"][1]["highlights"])
print("")
for model_name in summaries:
print(model_name.upper())
print("--" * 25)
print(summaries[model_name])
print("")
# # 三、文本生成评估指标
# ## 3.1 BLEU (`precision-based metric`)
# - 评估准确率: 生成句子中有m个单词出现在原文(n个词)中, $bleu=\frac{m}{n}$
# - 引起问题1:
# - 如果生成重复的词,并且该词在引用中出现,那么我们会得到较高的分数
# - 针对这点作者指出修正方法:一个单词只计算它在引用中出现的次数。
# - example: ref-"the cat is on the mat" g-"the the the the the the"
# - $P_{vanlilla}=\frac{6}{6}$, $P_{mod}=\frac{2}{6}$
# - 修正问题1:`clip`
# - 这意味着一个`n-gram`的出现次数以它在参考句中出现的次数为上限
# $$p_n=\frac{ \sum_{geSnt \in C}\sum_{n-gram \in geSnt} Count_{clip}(n-gram) }{ \sum_{geSnt \in C}\sum_{n-gram \in geSnt} Count(n-gram) }$$
# - 引起问题2:
# - 因为这个准确率的评估,很显然会对较短的评估对有力,会低估较长生成的结果。
# - 修正问题2:简短惩罚 `brevity penalty`
# - $BR = min(1, e^{1 - \frac{l_{ref}}{l_{gen}}} )$ : 生成长度大于原句子:1, 生成长度小于原句子:$(0, 1)$,
# **最终公式:**
# $$BLEU-N=BR * (\prod_{n=1}^N p_n)^{1/N}$$
# Example: 计算`BLEU-4`
# - ref-"the cat sat on the mat"
# - g-"the cat the cat is on the mat"
# - **BR**: $BR=min(1, e^{1-6/8})=1$
# - **n=1**
# - 1-gram: org:{"the", "cat", "sat", "on", "mat"} ge:{"the", "cat", "is", "on", "mat"}
# - clip: $count_{clip}("the") = 2, count_{clip}("cat") = 1, count_{clip}("is") = 0, 1-gram \in geSnt$
# - $p_1 = \frac{5}{8} $
# - **n=2**
# - 2-gram: org:{"the cat", "cat sat", "sat on", "on the", "the mat"} ge:{"the cat", "cat the", "cat is", "is on", "on the", "the mat"}
# - $p_2 = \frac{3}{7} $
# - **n=3**
# - 3-gram: org:{"the cat sat", "cat sat on", "sat on the", "on the mat"} ge:{"the cat the", "cat the cat", "the cat is", "cat is on", "is on the", "on the mat"}
# - $p_3 = \frac{1}{6} $
# - **n=4**
# - 3-gram: org:{"the cat sat on", "cat sat on the", "sat on the mat"} ge:{"the cat the cat", "cat the cat is", "the cat is on", "cat is on the", "is on the mat"}
# - $p_4 = \frac{0}{5} $
# - **BLEU-4**: $1 * (\frac{5}{8}*\frac{3}{7}*\frac{1}{6}*\frac{0}{5})^{1/2}=0.$
#
bleu_metric = load_metric("sacrebleu")
bleu_metric.add(
prediction="the cat the cat is on the mat", reference=["the cat sat on the mat"]
)
results = bleu_metric.compute(smooth_method="floor", smooth_value=0)
results
| false | 0 | 2,143 | 0 | 2,143 | 2,143 |
||
129595470
|
<jupyter_start><jupyter_text>Mushroom Classification
### Context
Although this dataset was originally contributed to the UCI Machine Learning repository nearly 30 years ago, mushroom hunting (otherwise known as "shrooming") is enjoying new peaks in popularity. Learn which features spell certain death and which are most palatable in this dataset of mushroom characteristics. And how certain can your model be?
### Content
This dataset includes descriptions of hypothetical samples corresponding to 23 species of gilled mushrooms in the Agaricus and Lepiota Family Mushroom drawn from The Audubon Society Field Guide to North American Mushrooms (1981). Each species is identified as definitely edible, definitely poisonous, or of unknown edibility and not recommended. This latter class was combined with the poisonous one. The Guide clearly states that there is no simple rule for determining the edibility of a mushroom; no rule like "leaflets three, let it be'' for Poisonous Oak and Ivy.
- **Time period**: Donated to UCI ML 27 April 1987
### Inspiration
- What types of machine learning models perform best on this dataset?
- Which features are most indicative of a poisonous mushroom?
Kaggle dataset identifier: mushroom-classification
<jupyter_script># Hanming Jing
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import roc_curve, auc, f1_score
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
df = pd.read_csv("../input/mushroom-classification/mushrooms.csv")
encoder = LabelEncoder()
df = df.apply(encoder.fit_transform)
df.head()
X = df.drop(columns=["class"])
y = df["class"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3)
print("X_train = ", X_train.shape)
print("y_train = ", y_train.shape)
print("X_test = ", X_test.shape)
print("y_test = ", y_test.shape)
# 初始化模型
rfc = RandomForestClassifier(n_estimators=100, random_state=0)
gnb = GaussianNB()
# 训练模型
rfc.fit(X_train, y_train)
gnb.fit(X_train, y_train)
# 预测概率
rfc_probs = rfc.predict_proba(X_test)[:, 1]
gnb_probs = gnb.predict_proba(X_test)[:, 1]
# 计算ROC曲线
rfc_fpr, rfc_tpr, _ = roc_curve(y_test, rfc_probs)
gnb_fpr, gnb_tpr, _ = roc_curve(y_test, gnb_probs)
# 计算AUC
rfc_auc = auc(rfc_fpr, rfc_tpr)
gnb_auc = auc(gnb_fpr, gnb_tpr)
plt.figure(figsize=(8, 6))
plt.plot(rfc_fpr, rfc_tpr, label="Random Forest (AUC = %0.2f)" % rfc_auc)
plt.plot(gnb_fpr, gnb_tpr, label="Gaussian NB (AUC = %0.2f)" % gnb_auc)
plt.plot([0, 1], [0, 1], "k--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic")
plt.legend(loc="lower right")
plt.show()
rfc_preds = rfc.predict(X_test)
gnb_preds = gnb.predict(X_test)
rfc_f1 = f1_score(y_test, rfc_preds)
gnb_f1 = f1_score(y_test, gnb_preds)
print("Random Forest F1 Score: %.2f" % rfc_f1)
print("Bayes F1 Score: %.2f" % gnb_f1)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/595/129595470.ipynb
|
mushroom-classification
| null |
[{"Id": 129595470, "ScriptId": 38535384, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13332464, "CreationDate": "05/15/2023 05:57:56", "VersionNumber": 1.0, "Title": "mushroom classification", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 79.0, "LinesInsertedFromPrevious": 41.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 38.0, "LinesInsertedFromFork": 41.0, "LinesDeletedFromFork": 52.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 38.0, "TotalVotes": 0}]
|
[{"Id": 185824552, "KernelVersionId": 129595470, "SourceDatasetVersionId": 974}]
|
[{"Id": 974, "DatasetId": 478, "DatasourceVersionId": 974, "CreatorUserId": 495305, "LicenseName": "CC0: Public Domain", "CreationDate": "12/01/2016 23:08:00", "VersionNumber": 1.0, "Title": "Mushroom Classification", "Slug": "mushroom-classification", "Subtitle": "Safe to eat or deadly poison?", "Description": "### Context\n\nAlthough this dataset was originally contributed to the UCI Machine Learning repository nearly 30 years ago, mushroom hunting (otherwise known as \"shrooming\") is enjoying new peaks in popularity. Learn which features spell certain death and which are most palatable in this dataset of mushroom characteristics. And how certain can your model be?\n\n### Content \n\nThis dataset includes descriptions of hypothetical samples corresponding to 23 species of gilled mushrooms in the Agaricus and Lepiota Family Mushroom drawn from The Audubon Society Field Guide to North American Mushrooms (1981). Each species is identified as definitely edible, definitely poisonous, or of unknown edibility and not recommended. This latter class was combined with the poisonous one. The Guide clearly states that there is no simple rule for determining the edibility of a mushroom; no rule like \"leaflets three, let it be'' for Poisonous Oak and Ivy.\n\n- **Time period**: Donated to UCI ML 27 April 1987\n\n### Inspiration\n\n- What types of machine learning models perform best on this dataset?\n\n- Which features are most indicative of a poisonous mushroom?\n\n### Acknowledgements\n\nThis dataset was originally donated to the UCI Machine Learning repository. You can learn more about past research using the data [here][1]. \n\n#[Start a new kernel][2]\n\n\n [1]: https://archive.ics.uci.edu/ml/datasets/Mushroom\n [2]: https://www.kaggle.com/uciml/mushroom-classification/kernels?modal=true", "VersionNotes": "Initial release", "TotalCompressedBytes": 374003.0, "TotalUncompressedBytes": 374003.0}]
|
[{"Id": 478, "CreatorUserId": 495305, "OwnerUserId": NaN, "OwnerOrganizationId": 7.0, "CurrentDatasetVersionId": 974.0, "CurrentDatasourceVersionId": 974.0, "ForumId": 2099, "Type": 2, "CreationDate": "12/01/2016 23:08:00", "LastActivityDate": "02/06/2018", "TotalViews": 873597, "TotalDownloads": 114985, "TotalVotes": 2206, "TotalKernels": 1371}]
| null |
# Hanming Jing
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import roc_curve, auc, f1_score
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
df = pd.read_csv("../input/mushroom-classification/mushrooms.csv")
encoder = LabelEncoder()
df = df.apply(encoder.fit_transform)
df.head()
X = df.drop(columns=["class"])
y = df["class"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3)
print("X_train = ", X_train.shape)
print("y_train = ", y_train.shape)
print("X_test = ", X_test.shape)
print("y_test = ", y_test.shape)
# 初始化模型
rfc = RandomForestClassifier(n_estimators=100, random_state=0)
gnb = GaussianNB()
# 训练模型
rfc.fit(X_train, y_train)
gnb.fit(X_train, y_train)
# 预测概率
rfc_probs = rfc.predict_proba(X_test)[:, 1]
gnb_probs = gnb.predict_proba(X_test)[:, 1]
# 计算ROC曲线
rfc_fpr, rfc_tpr, _ = roc_curve(y_test, rfc_probs)
gnb_fpr, gnb_tpr, _ = roc_curve(y_test, gnb_probs)
# 计算AUC
rfc_auc = auc(rfc_fpr, rfc_tpr)
gnb_auc = auc(gnb_fpr, gnb_tpr)
plt.figure(figsize=(8, 6))
plt.plot(rfc_fpr, rfc_tpr, label="Random Forest (AUC = %0.2f)" % rfc_auc)
plt.plot(gnb_fpr, gnb_tpr, label="Gaussian NB (AUC = %0.2f)" % gnb_auc)
plt.plot([0, 1], [0, 1], "k--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic")
plt.legend(loc="lower right")
plt.show()
rfc_preds = rfc.predict(X_test)
gnb_preds = gnb.predict(X_test)
rfc_f1 = f1_score(y_test, rfc_preds)
gnb_f1 = f1_score(y_test, gnb_preds)
print("Random Forest F1 Score: %.2f" % rfc_f1)
print("Bayes F1 Score: %.2f" % gnb_f1)
| false | 0 | 925 | 0 | 1,227 | 925 |
||
129401071
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import plottingutility
import matplotlib.pyplot as plt
import matplotlib
import seaborn as sns
from sklearn.preprocessing import MinMaxScaler
import typing
ROWS, COLS = 3, 2
TARGET = "Class"
KURT_THRESHOLD = 0.5
df_train = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/train.csv")
df_test = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/test.csv")
del df_train["Id"]
del df_test["Id"]
y_train = df_train[TARGET]
def get_upper_bound_by_map(serie: pd.Series) -> np.float64:
name_to_upper_bound = {
"AH": 100,
"AB": 1,
"AF": 8000,
"AH": 88,
"AM": 100,
"AR": 25,
"AX": 10,
"AY": 0.025578,
}
return name_to_upper_bound[serie.name]
def get_upper_bound_by_calculation(serie: pd.Series) -> np.float64:
return serie.mean() + serie.std()
def squash_tail(
serie: pd.Series,
col,
upper_bound_calculation: typing.Callable[[pd.Series], np.float64] = None,
):
if upper_bound_calculation is None:
upper_bound_calculation = get_upper_bound_by_calculation
if serie.dtype != "float64":
return serie
fig, ax = plt.subplots(1, 2, figsize=(20, 1 * 4))
ax = ax.flatten()
if serie.kurt() > KURT_THRESHOLD:
plottingutility.plot_histogram(
serie, current_axis=ax[0], title=f"{col} before clipping"
)
upper_bound = upper_bound_calculation(serie)
serie = np.log(serie.clip(0, upper_bound))
plottingutility.plot_histogram(
serie, current_axis=ax[1], title=f"{col} after clipping"
)
else:
plottingutility.plot_histogram(
serie, current_axis=ax[0], title=f"{col} before clipping"
)
plottingutility.plot_histogram(
serie, current_axis=ax[1], title=f"{col} after clipping"
)
return serie
df_train_copy_one = df_train.copy()
df_train_copy_two = df_train.copy()
for col in df_train.columns[:6]:
df_train_copy_one[col] = squash_tail(df_train_copy_one[col], col)
df_train_copy_two[col] = squash_tail(
df_train_copy_two[col], col, get_upper_bound_by_map
)
fix, axs = plt.subplots(
ROWS, COLS, figsize=(17, 17), gridspec_kw={"wspace": 0.25, "hspace": 0.25}
)
plt.subplots_adjust(wspace=0.25, hspace=0.25)
row_idx = 0
col_idx = 0
for idx, start in enumerate([0, 10, 20, 30, 40, 50]):
if idx % COLS == 0 and idx != 0:
row_idx += 1
col_idx = 0
if start != 50:
end = start + 10
else:
end = start + 7
plottingutility.plot_correlations(
df_train[[*df_train.columns[start:end], TARGET]], axs[row_idx, col_idx]
)
col_idx += 1
df_train["EJ"] = df_train["EJ"].factorize()[0]
df_test["EJ"] = df_test["EJ"].factorize()[0]
TAIL_DISTRIBUTED_FEATURES = [
"AB",
"AF",
"AH",
"AM",
"AR",
"AX",
"AY",
"AZ",
"BC",
"BP",
"BR",
"BZ",
"CB",
"CC",
]
def qcut_columns(col):
df_train[f"{col}_cut"] = pd.qcut(df_train[f"{col}"], q=4, labels=False)
return df_train
for col in ["AM", "CB", "BR"]:
df_train = qcut_columns(col)
df_original = df_train[TAIL_DISTRIBUTED_FEATURES].copy()
df_train[TAIL_DISTRIBUTED_FEATURES] = np.log(df_train[TAIL_DISTRIBUTED_FEATURES])
scaled_rows = MinMaxScaler().fit_transform(df_train[df_test.columns])
scaled_frame = pd.DataFrame(data=scaled_rows, columns=df_test.columns)
plt_features([scaled_frame], scaled_frame.columns)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/401/129401071.ipynb
| null | null |
[{"Id": 129401071, "ScriptId": 38444354, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3245061, "CreationDate": "05/13/2023 13:22:27", "VersionNumber": 3.0, "Title": "notebook51e2a2142a", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 91.0, "LinesInsertedFromPrevious": 35.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 56.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import plottingutility
import matplotlib.pyplot as plt
import matplotlib
import seaborn as sns
from sklearn.preprocessing import MinMaxScaler
import typing
ROWS, COLS = 3, 2
TARGET = "Class"
KURT_THRESHOLD = 0.5
df_train = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/train.csv")
df_test = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/test.csv")
del df_train["Id"]
del df_test["Id"]
y_train = df_train[TARGET]
def get_upper_bound_by_map(serie: pd.Series) -> np.float64:
name_to_upper_bound = {
"AH": 100,
"AB": 1,
"AF": 8000,
"AH": 88,
"AM": 100,
"AR": 25,
"AX": 10,
"AY": 0.025578,
}
return name_to_upper_bound[serie.name]
def get_upper_bound_by_calculation(serie: pd.Series) -> np.float64:
return serie.mean() + serie.std()
def squash_tail(
serie: pd.Series,
col,
upper_bound_calculation: typing.Callable[[pd.Series], np.float64] = None,
):
if upper_bound_calculation is None:
upper_bound_calculation = get_upper_bound_by_calculation
if serie.dtype != "float64":
return serie
fig, ax = plt.subplots(1, 2, figsize=(20, 1 * 4))
ax = ax.flatten()
if serie.kurt() > KURT_THRESHOLD:
plottingutility.plot_histogram(
serie, current_axis=ax[0], title=f"{col} before clipping"
)
upper_bound = upper_bound_calculation(serie)
serie = np.log(serie.clip(0, upper_bound))
plottingutility.plot_histogram(
serie, current_axis=ax[1], title=f"{col} after clipping"
)
else:
plottingutility.plot_histogram(
serie, current_axis=ax[0], title=f"{col} before clipping"
)
plottingutility.plot_histogram(
serie, current_axis=ax[1], title=f"{col} after clipping"
)
return serie
df_train_copy_one = df_train.copy()
df_train_copy_two = df_train.copy()
for col in df_train.columns[:6]:
df_train_copy_one[col] = squash_tail(df_train_copy_one[col], col)
df_train_copy_two[col] = squash_tail(
df_train_copy_two[col], col, get_upper_bound_by_map
)
fix, axs = plt.subplots(
ROWS, COLS, figsize=(17, 17), gridspec_kw={"wspace": 0.25, "hspace": 0.25}
)
plt.subplots_adjust(wspace=0.25, hspace=0.25)
row_idx = 0
col_idx = 0
for idx, start in enumerate([0, 10, 20, 30, 40, 50]):
if idx % COLS == 0 and idx != 0:
row_idx += 1
col_idx = 0
if start != 50:
end = start + 10
else:
end = start + 7
plottingutility.plot_correlations(
df_train[[*df_train.columns[start:end], TARGET]], axs[row_idx, col_idx]
)
col_idx += 1
df_train["EJ"] = df_train["EJ"].factorize()[0]
df_test["EJ"] = df_test["EJ"].factorize()[0]
TAIL_DISTRIBUTED_FEATURES = [
"AB",
"AF",
"AH",
"AM",
"AR",
"AX",
"AY",
"AZ",
"BC",
"BP",
"BR",
"BZ",
"CB",
"CC",
]
def qcut_columns(col):
df_train[f"{col}_cut"] = pd.qcut(df_train[f"{col}"], q=4, labels=False)
return df_train
for col in ["AM", "CB", "BR"]:
df_train = qcut_columns(col)
df_original = df_train[TAIL_DISTRIBUTED_FEATURES].copy()
df_train[TAIL_DISTRIBUTED_FEATURES] = np.log(df_train[TAIL_DISTRIBUTED_FEATURES])
scaled_rows = MinMaxScaler().fit_transform(df_train[df_test.columns])
scaled_frame = pd.DataFrame(data=scaled_rows, columns=df_test.columns)
plt_features([scaled_frame], scaled_frame.columns)
| false | 0 | 1,266 | 0 | 1,266 | 1,266 |
||
129401113
|
import glob
glob.glob("../mAP/output/*")
with open("../mAP/output/output.txt", "r") as file:
data = file.read()
mAP = float(
data[data.find("mAP = ") + 6 : data.find("# Number of ground-truth ") - 3]
)
print(mAP)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/401/129401113.ipynb
| null | null |
[{"Id": 129401113, "ScriptId": 38469672, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9253757, "CreationDate": "05/13/2023 13:22:48", "VersionNumber": 1.0, "Title": "Tinh MAP", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 12.0, "LinesInsertedFromPrevious": 12.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import glob
glob.glob("../mAP/output/*")
with open("../mAP/output/output.txt", "r") as file:
data = file.read()
mAP = float(
data[data.find("mAP = ") + 6 : data.find("# Number of ground-truth ") - 3]
)
print(mAP)
| false | 0 | 84 | 0 | 84 | 84 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.