file_id
stringlengths 5
9
| content
stringlengths 100
5.25M
| local_path
stringlengths 66
70
| kaggle_dataset_name
stringlengths 3
50
⌀ | kaggle_dataset_owner
stringlengths 3
20
⌀ | kversion
stringlengths 497
763
⌀ | kversion_datasetsources
stringlengths 71
5.46k
⌀ | dataset_versions
stringlengths 338
235k
⌀ | datasets
stringlengths 334
371
⌀ | users
stringlengths 111
264
⌀ | script
stringlengths 100
5.25M
| df_info
stringlengths 0
4.87M
| has_data_info
bool 2
classes | nb_filenames
int64 0
370
| retreived_data_description
stringlengths 0
4.44M
| script_nb_tokens
int64 25
663k
| upvotes
int64 0
1.65k
| tokens_description
int64 25
663k
| tokens_script
int64 25
663k
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
129063071
|
<jupyter_start><jupyter_text>Lung Cancer
### The effectiveness of cancer prediction system helps the people to know their cancer risk with low cost and it also helps the people to take the appropriate decision based on their cancer risk status. The data is collected from the website online lung cancer prediction system .
Total no. of attributes:16
No .of instances:284
Attribute information:
1. Gender: M(male), F(female)
2. Age: Age of the patient
3. Smoking: YES=2 , NO=1.
4. Yellow fingers: YES=2 , NO=1.
5. Anxiety: YES=2 , NO=1.
6. Peer_pressure: YES=2 , NO=1.
7. Chronic Disease: YES=2 , NO=1.
8. Fatigue: YES=2 , NO=1.
9. Allergy: YES=2 , NO=1.
10. Wheezing: YES=2 , NO=1.
11. Alcohol: YES=2 , NO=1.
12. Coughing: YES=2 , NO=1.
13. Shortness of Breath: YES=2 , NO=1.
14. Swallowing Difficulty: YES=2 , NO=1.
15. Chest pain: YES=2 , NO=1.
16. Lung Cancer: YES , NO.
Kaggle dataset identifier: lung-cancer
<jupyter_code>import pandas as pd
df = pd.read_csv('lung-cancer/survey lung cancer.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 309 entries, 0 to 308
Data columns (total 16 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 GENDER 309 non-null object
1 AGE 309 non-null int64
2 SMOKING 309 non-null int64
3 YELLOW_FINGERS 309 non-null int64
4 ANXIETY 309 non-null int64
5 PEER_PRESSURE 309 non-null int64
6 CHRONIC DISEASE 309 non-null int64
7 FATIGUE 309 non-null int64
8 ALLERGY 309 non-null int64
9 WHEEZING 309 non-null int64
10 ALCOHOL CONSUMING 309 non-null int64
11 COUGHING 309 non-null int64
12 SHORTNESS OF BREATH 309 non-null int64
13 SWALLOWING DIFFICULTY 309 non-null int64
14 CHEST PAIN 309 non-null int64
15 LUNG_CANCER 309 non-null object
dtypes: int64(14), object(2)
memory usage: 38.8+ KB
<jupyter_text>Examples:
{
"GENDER": "M",
"AGE": 69,
"SMOKING": 1,
"YELLOW_FINGERS": 2,
"ANXIETY": 2,
"PEER_PRESSURE": 1,
"CHRONIC DISEASE": 1,
"FATIGUE ": 2,
"ALLERGY ": 1,
"WHEEZING": 2,
"ALCOHOL CONSUMING": 2,
"COUGHING": 2,
"SHORTNESS OF BREATH": 2,
"SWALLOWING DIFFICULTY": 2,
"CHEST PAIN": 2,
"LUNG_CANCER": "YES"
}
{
"GENDER": "M",
"AGE": 74,
"SMOKING": 2,
"YELLOW_FINGERS": 1,
"ANXIETY": 1,
"PEER_PRESSURE": 1,
"CHRONIC DISEASE": 2,
"FATIGUE ": 2,
"ALLERGY ": 2,
"WHEEZING": 1,
"ALCOHOL CONSUMING": 1,
"COUGHING": 1,
"SHORTNESS OF BREATH": 2,
"SWALLOWING DIFFICULTY": 2,
"CHEST PAIN": 2,
"LUNG_CANCER": "YES"
}
{
"GENDER": "F",
"AGE": 59,
"SMOKING": 1,
"YELLOW_FINGERS": 1,
"ANXIETY": 1,
"PEER_PRESSURE": 2,
"CHRONIC DISEASE": 1,
"FATIGUE ": 2,
"ALLERGY ": 1,
"WHEEZING": 2,
"ALCOHOL CONSUMING": 1,
"COUGHING": 2,
"SHORTNESS OF BREATH": 2,
"SWALLOWING DIFFICULTY": 1,
"CHEST PAIN": 2,
"LUNG_CANCER": "NO"
}
{
"GENDER": "M",
"AGE": 63,
"SMOKING": 2,
"YELLOW_FINGERS": 2,
"ANXIETY": 2,
"PEER_PRESSURE": 1,
"CHRONIC DISEASE": 1,
"FATIGUE ": 1,
"ALLERGY ": 1,
"WHEEZING": 1,
"ALCOHOL CONSUMING": 2,
"COUGHING": 1,
"SHORTNESS OF BREATH": 1,
"SWALLOWING DIFFICULTY": 2,
"CHEST PAIN": 2,
"LUNG_CANCER": "NO"
}
<jupyter_script>import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("fivethirtyeight")
colors = ["#011f4b", "#03396c", "#005b96", "#6497b1", "#b3cde0"]
sns.set_palette(sns.color_palette(colors))
df = pd.read_csv("../input/lung-cancer/survey lung cancer.csv")
df.head()
df.shape
# Some info about our attributes and its datatype
df.info()
# Some analysis on the numerical columns
df.describe()
# Check for null values
df.isnull().sum()
# Check for duplicates in the dataset
df.duplicated().sum()
df.drop_duplicates(inplace=True)
df.shape
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/063/129063071.ipynb
|
lung-cancer
|
mysarahmadbhat
|
[{"Id": 129063071, "ScriptId": 38366099, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10692872, "CreationDate": "05/10/2023 17:13:04", "VersionNumber": 1.0, "Title": "Prediction Lung cancer", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 28.0, "LinesInsertedFromPrevious": 28.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184787686, "KernelVersionId": 129063071, "SourceDatasetVersionId": 2668247}]
|
[{"Id": 2668247, "DatasetId": 1623385, "DatasourceVersionId": 2712518, "CreatorUserId": 6990402, "LicenseName": "CC0: Public Domain", "CreationDate": "10/01/2021 13:39:48", "VersionNumber": 1.0, "Title": "Lung Cancer", "Slug": "lung-cancer", "Subtitle": "Does Smoking cause Lung Cancer.", "Description": "### The effectiveness of cancer prediction system helps the people to know their cancer risk with low cost and it also helps the people to take the appropriate decision based on their cancer risk status. The data is collected from the website online lung cancer prediction system .\nTotal no. of attributes:16\nNo .of instances:284\nAttribute information:\n1.\tGender: M(male), F(female)\n2.\tAge: Age of the patient\n3.\tSmoking: YES=2 , NO=1.\n4.\tYellow fingers: YES=2 , NO=1.\n5.\tAnxiety: YES=2 , NO=1.\n6.\tPeer_pressure: YES=2 , NO=1.\n7.\tChronic Disease: YES=2 , NO=1.\n8.\tFatigue: YES=2 , NO=1.\n9.\tAllergy: YES=2 , NO=1.\n10.\tWheezing: YES=2 , NO=1.\n11.\tAlcohol: YES=2 , NO=1.\n12.\tCoughing: YES=2 , NO=1.\n13.\tShortness of Breath: YES=2 , NO=1.\n14.\tSwallowing Difficulty: YES=2 , NO=1.\n15.\tChest pain: YES=2 , NO=1.\n16.\tLung Cancer: YES , NO.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1623385, "CreatorUserId": 6990402, "OwnerUserId": 6990402.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2668247.0, "CurrentDatasourceVersionId": 2712518.0, "ForumId": 1643979, "Type": 2, "CreationDate": "10/01/2021 13:39:48", "LastActivityDate": "10/01/2021", "TotalViews": 153177, "TotalDownloads": 23248, "TotalVotes": 272, "TotalKernels": 70}]
|
[{"Id": 6990402, "UserName": "mysarahmadbhat", "DisplayName": "mysar ahmad bhat", "RegisterDate": "03/21/2021", "PerformanceTier": 3}]
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("fivethirtyeight")
colors = ["#011f4b", "#03396c", "#005b96", "#6497b1", "#b3cde0"]
sns.set_palette(sns.color_palette(colors))
df = pd.read_csv("../input/lung-cancer/survey lung cancer.csv")
df.head()
df.shape
# Some info about our attributes and its datatype
df.info()
# Some analysis on the numerical columns
df.describe()
# Check for null values
df.isnull().sum()
# Check for duplicates in the dataset
df.duplicated().sum()
df.drop_duplicates(inplace=True)
df.shape
|
[{"lung-cancer/survey lung cancer.csv": {"column_names": "[\"GENDER\", \"AGE\", \"SMOKING\", \"YELLOW_FINGERS\", \"ANXIETY\", \"PEER_PRESSURE\", \"CHRONIC DISEASE\", \"FATIGUE \", \"ALLERGY \", \"WHEEZING\", \"ALCOHOL CONSUMING\", \"COUGHING\", \"SHORTNESS OF BREATH\", \"SWALLOWING DIFFICULTY\", \"CHEST PAIN\", \"LUNG_CANCER\"]", "column_data_types": "{\"GENDER\": \"object\", \"AGE\": \"int64\", \"SMOKING\": \"int64\", \"YELLOW_FINGERS\": \"int64\", \"ANXIETY\": \"int64\", \"PEER_PRESSURE\": \"int64\", \"CHRONIC DISEASE\": \"int64\", \"FATIGUE \": \"int64\", \"ALLERGY \": \"int64\", \"WHEEZING\": \"int64\", \"ALCOHOL CONSUMING\": \"int64\", \"COUGHING\": \"int64\", \"SHORTNESS OF BREATH\": \"int64\", \"SWALLOWING DIFFICULTY\": \"int64\", \"CHEST PAIN\": \"int64\", \"LUNG_CANCER\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 309 entries, 0 to 308\nData columns (total 16 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 GENDER 309 non-null object\n 1 AGE 309 non-null int64 \n 2 SMOKING 309 non-null int64 \n 3 YELLOW_FINGERS 309 non-null int64 \n 4 ANXIETY 309 non-null int64 \n 5 PEER_PRESSURE 309 non-null int64 \n 6 CHRONIC DISEASE 309 non-null int64 \n 7 FATIGUE 309 non-null int64 \n 8 ALLERGY 309 non-null int64 \n 9 WHEEZING 309 non-null int64 \n 10 ALCOHOL CONSUMING 309 non-null int64 \n 11 COUGHING 309 non-null int64 \n 12 SHORTNESS OF BREATH 309 non-null int64 \n 13 SWALLOWING DIFFICULTY 309 non-null int64 \n 14 CHEST PAIN 309 non-null int64 \n 15 LUNG_CANCER 309 non-null object\ndtypes: int64(14), object(2)\nmemory usage: 38.8+ KB\n", "summary": "{\"AGE\": {\"count\": 309.0, \"mean\": 62.67313915857605, \"std\": 8.210301387885995, \"min\": 21.0, \"25%\": 57.0, \"50%\": 62.0, \"75%\": 69.0, \"max\": 87.0}, \"SMOKING\": {\"count\": 309.0, \"mean\": 1.5631067961165048, \"std\": 0.4968060894409518, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"YELLOW_FINGERS\": {\"count\": 309.0, \"mean\": 1.5695792880258899, \"std\": 0.49593819429101677, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"ANXIETY\": {\"count\": 309.0, \"mean\": 1.4983818770226538, \"std\": 0.5008084079652348, \"min\": 1.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 2.0}, \"PEER_PRESSURE\": {\"count\": 309.0, \"mean\": 1.5016181229773462, \"std\": 0.5008084079652348, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"CHRONIC DISEASE\": {\"count\": 309.0, \"mean\": 1.5048543689320388, \"std\": 0.5007874268634864, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"FATIGUE \": {\"count\": 309.0, \"mean\": 1.6731391585760518, \"std\": 0.46982676766120723, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"ALLERGY \": {\"count\": 309.0, \"mean\": 1.5566343042071198, \"std\": 0.49758801243408385, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"WHEEZING\": {\"count\": 309.0, \"mean\": 1.5566343042071198, \"std\": 0.49758801243408385, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"ALCOHOL CONSUMING\": {\"count\": 309.0, \"mean\": 1.5566343042071198, \"std\": 0.4975880124340838, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"COUGHING\": {\"count\": 309.0, \"mean\": 1.5792880258899675, \"std\": 0.49447415124782723, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"SHORTNESS OF BREATH\": {\"count\": 309.0, \"mean\": 1.6407766990291262, \"std\": 0.48055100136181955, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}, \"SWALLOWING DIFFICULTY\": {\"count\": 309.0, \"mean\": 1.4692556634304208, \"std\": 0.49986338653997353, \"min\": 1.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 2.0}, \"CHEST PAIN\": {\"count\": 309.0, \"mean\": 1.5566343042071198, \"std\": 0.4975880124340838, \"min\": 1.0, \"25%\": 1.0, \"50%\": 2.0, \"75%\": 2.0, \"max\": 2.0}}", "examples": "{\"GENDER\":{\"0\":\"M\",\"1\":\"M\",\"2\":\"F\",\"3\":\"M\"},\"AGE\":{\"0\":69,\"1\":74,\"2\":59,\"3\":63},\"SMOKING\":{\"0\":1,\"1\":2,\"2\":1,\"3\":2},\"YELLOW_FINGERS\":{\"0\":2,\"1\":1,\"2\":1,\"3\":2},\"ANXIETY\":{\"0\":2,\"1\":1,\"2\":1,\"3\":2},\"PEER_PRESSURE\":{\"0\":1,\"1\":1,\"2\":2,\"3\":1},\"CHRONIC DISEASE\":{\"0\":1,\"1\":2,\"2\":1,\"3\":1},\"FATIGUE \":{\"0\":2,\"1\":2,\"2\":2,\"3\":1},\"ALLERGY \":{\"0\":1,\"1\":2,\"2\":1,\"3\":1},\"WHEEZING\":{\"0\":2,\"1\":1,\"2\":2,\"3\":1},\"ALCOHOL CONSUMING\":{\"0\":2,\"1\":1,\"2\":1,\"3\":2},\"COUGHING\":{\"0\":2,\"1\":1,\"2\":2,\"3\":1},\"SHORTNESS OF BREATH\":{\"0\":2,\"1\":2,\"2\":2,\"3\":1},\"SWALLOWING DIFFICULTY\":{\"0\":2,\"1\":2,\"2\":1,\"3\":2},\"CHEST PAIN\":{\"0\":2,\"1\":2,\"2\":2,\"3\":2},\"LUNG_CANCER\":{\"0\":\"YES\",\"1\":\"YES\",\"2\":\"NO\",\"3\":\"NO\"}}"}}]
| true | 1 |
<start_data_description><data_path>lung-cancer/survey lung cancer.csv:
<column_names>
['GENDER', 'AGE', 'SMOKING', 'YELLOW_FINGERS', 'ANXIETY', 'PEER_PRESSURE', 'CHRONIC DISEASE', 'FATIGUE ', 'ALLERGY ', 'WHEEZING', 'ALCOHOL CONSUMING', 'COUGHING', 'SHORTNESS OF BREATH', 'SWALLOWING DIFFICULTY', 'CHEST PAIN', 'LUNG_CANCER']
<column_types>
{'GENDER': 'object', 'AGE': 'int64', 'SMOKING': 'int64', 'YELLOW_FINGERS': 'int64', 'ANXIETY': 'int64', 'PEER_PRESSURE': 'int64', 'CHRONIC DISEASE': 'int64', 'FATIGUE ': 'int64', 'ALLERGY ': 'int64', 'WHEEZING': 'int64', 'ALCOHOL CONSUMING': 'int64', 'COUGHING': 'int64', 'SHORTNESS OF BREATH': 'int64', 'SWALLOWING DIFFICULTY': 'int64', 'CHEST PAIN': 'int64', 'LUNG_CANCER': 'object'}
<dataframe_Summary>
{'AGE': {'count': 309.0, 'mean': 62.67313915857605, 'std': 8.210301387885995, 'min': 21.0, '25%': 57.0, '50%': 62.0, '75%': 69.0, 'max': 87.0}, 'SMOKING': {'count': 309.0, 'mean': 1.5631067961165048, 'std': 0.4968060894409518, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'YELLOW_FINGERS': {'count': 309.0, 'mean': 1.5695792880258899, 'std': 0.49593819429101677, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'ANXIETY': {'count': 309.0, 'mean': 1.4983818770226538, 'std': 0.5008084079652348, 'min': 1.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 2.0}, 'PEER_PRESSURE': {'count': 309.0, 'mean': 1.5016181229773462, 'std': 0.5008084079652348, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'CHRONIC DISEASE': {'count': 309.0, 'mean': 1.5048543689320388, 'std': 0.5007874268634864, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'FATIGUE ': {'count': 309.0, 'mean': 1.6731391585760518, 'std': 0.46982676766120723, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'ALLERGY ': {'count': 309.0, 'mean': 1.5566343042071198, 'std': 0.49758801243408385, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'WHEEZING': {'count': 309.0, 'mean': 1.5566343042071198, 'std': 0.49758801243408385, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'ALCOHOL CONSUMING': {'count': 309.0, 'mean': 1.5566343042071198, 'std': 0.4975880124340838, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'COUGHING': {'count': 309.0, 'mean': 1.5792880258899675, 'std': 0.49447415124782723, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'SHORTNESS OF BREATH': {'count': 309.0, 'mean': 1.6407766990291262, 'std': 0.48055100136181955, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}, 'SWALLOWING DIFFICULTY': {'count': 309.0, 'mean': 1.4692556634304208, 'std': 0.49986338653997353, 'min': 1.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 2.0}, 'CHEST PAIN': {'count': 309.0, 'mean': 1.5566343042071198, 'std': 0.4975880124340838, 'min': 1.0, '25%': 1.0, '50%': 2.0, '75%': 2.0, 'max': 2.0}}
<dataframe_info>
RangeIndex: 309 entries, 0 to 308
Data columns (total 16 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 GENDER 309 non-null object
1 AGE 309 non-null int64
2 SMOKING 309 non-null int64
3 YELLOW_FINGERS 309 non-null int64
4 ANXIETY 309 non-null int64
5 PEER_PRESSURE 309 non-null int64
6 CHRONIC DISEASE 309 non-null int64
7 FATIGUE 309 non-null int64
8 ALLERGY 309 non-null int64
9 WHEEZING 309 non-null int64
10 ALCOHOL CONSUMING 309 non-null int64
11 COUGHING 309 non-null int64
12 SHORTNESS OF BREATH 309 non-null int64
13 SWALLOWING DIFFICULTY 309 non-null int64
14 CHEST PAIN 309 non-null int64
15 LUNG_CANCER 309 non-null object
dtypes: int64(14), object(2)
memory usage: 38.8+ KB
<some_examples>
{'GENDER': {'0': 'M', '1': 'M', '2': 'F', '3': 'M'}, 'AGE': {'0': 69, '1': 74, '2': 59, '3': 63}, 'SMOKING': {'0': 1, '1': 2, '2': 1, '3': 2}, 'YELLOW_FINGERS': {'0': 2, '1': 1, '2': 1, '3': 2}, 'ANXIETY': {'0': 2, '1': 1, '2': 1, '3': 2}, 'PEER_PRESSURE': {'0': 1, '1': 1, '2': 2, '3': 1}, 'CHRONIC DISEASE': {'0': 1, '1': 2, '2': 1, '3': 1}, 'FATIGUE ': {'0': 2, '1': 2, '2': 2, '3': 1}, 'ALLERGY ': {'0': 1, '1': 2, '2': 1, '3': 1}, 'WHEEZING': {'0': 2, '1': 1, '2': 2, '3': 1}, 'ALCOHOL CONSUMING': {'0': 2, '1': 1, '2': 1, '3': 2}, 'COUGHING': {'0': 2, '1': 1, '2': 2, '3': 1}, 'SHORTNESS OF BREATH': {'0': 2, '1': 2, '2': 2, '3': 1}, 'SWALLOWING DIFFICULTY': {'0': 2, '1': 2, '2': 1, '3': 2}, 'CHEST PAIN': {'0': 2, '1': 2, '2': 2, '3': 2}, 'LUNG_CANCER': {'0': 'YES', '1': 'YES', '2': 'NO', '3': 'NO'}}
<end_description>
| 198 | 0 | 1,652 | 198 |
129063846
|
<jupyter_start><jupyter_text>Food Preferences
### Context
Food Preference Survey 2019
### Content
This Survey was conducted among participants from different countries and demography. This will enable the novice data science enthusiasts to practice data pre-processing, feature engineering, analysis and visualization.
It contains 288 unique responses
Kaggle dataset identifier: food-preferences
<jupyter_script>import pandas as pd
data = pd.read_csv("/kaggle/input/food-preferences/Food_Preference.csv")
data.head()
# Food is a binary feature
# ****
data["Food"].unique()
data["Nationality"].unique()
# As we can see countries are written in a different ways and counted multiple times as unique
# also Muslim isn't a nationality it is a religon
data["Nationality"] = data["Nationality"].replace(
["Pakistani ", "Pakistan", "Pakistani"], "pakistani"
)
data["Nationality"] = data["Nationality"].replace(
["Indonesia", "Indonesian ", "Indonesain", "Indonesain", "Indonesian"], "indonesian"
)
data["Nationality"] = data["Nationality"].replace(["Muslim"], "Unknown")
data["Nationality"] = data["Nationality"].replace(
["Maldivian ", "Maldivian"], "Maldivian"
)
data["Nationality"] = data["Nationality"].replace(
["MY", "Malaysian", "Malaysian ", "MALAYSIAN", "Malaysia ", "Malaysia"], "malaysian"
)
data["Nationality"] = data["Nationality"].replace(["Japan"], "japanese")
data["Nationality"] = data["Nationality"].replace(["China"], "Chinese")
data["Nationality"] = data["Nationality"].replace(["Yemen"], "Yemeni")
data["Nationality"].unique()
data["Age"].unique()
# No age outliers
# # Ordinal data****
data["Dessert"].unique()
data_ord = data.copy()
dic = {"Yes": 2, "Maybe": 1, "No": 0}
data_ord["Rating_Dessert"] = data.Dessert.map(dic)
data_ord = data_ord.drop("Dessert", axis=1)
data_ord.head()
# # Binary data
data_ord["Gender"] = data_ord["Gender"].apply(
lambda x: 1 if x == "Male" else (0 if x == "Female" else None)
)
data_ord["Food"] = data_ord["Food"].apply(
lambda x: 1 if x == "Traditional food" else (0 if x == "Western Food" else None)
)
data_ord["Juice"] = data_ord["Juice"].apply(
lambda x: 1 if x == "Fresh Juice" else (0 if x == "Carbonated drinks" else None)
)
data_ord.head()
data.isna().count()
data.shape
df = pd.concat(
[
pd.get_dummies(
prefix="Nationality", data=data_ord["Nationality"], drop_first=True
),
data_ord,
],
axis=1,
)
df.drop(["Nationality", "Timestamp", "Participant_ID"], inplace=True, axis=1)
df
data.isnull().sum()
df = df.dropna()
df.isnull().sum()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/063/129063846.ipynb
|
food-preferences
|
vijayashreer
|
[{"Id": 129063846, "ScriptId": 38238736, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8882082, "CreationDate": "05/10/2023 17:21:18", "VersionNumber": 1.0, "Title": "Feature Encoding", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 74.0, "LinesInsertedFromPrevious": 74.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184789373, "KernelVersionId": 129063846, "SourceDatasetVersionId": 558013}]
|
[{"Id": 558013, "DatasetId": 268090, "DatasourceVersionId": 574599, "CreatorUserId": 2737706, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "07/18/2019 06:28:02", "VersionNumber": 1.0, "Title": "Food Preferences", "Slug": "food-preferences", "Subtitle": "Food Preferences Survey 2019", "Description": "### Context\n\nFood Preference Survey 2019\n\n\n### Content\n\nThis Survey was conducted among participants from different countries and demography. This will enable the novice data science enthusiasts to practice data pre-processing, feature engineering, analysis and visualization.\n\nIt contains 288 unique responses\n\n\n### Acknowledgements\n\nThanks to all the participants of the survey\n\n\n### Inspiration\n\nThis survey was conducted for novice data science enthusiasts to learn processes in data analysis, visualization and to build models.", "VersionNotes": "Initial release", "TotalCompressedBytes": 24913.0, "TotalUncompressedBytes": 24913.0}]
|
[{"Id": 268090, "CreatorUserId": 2737706, "OwnerUserId": 2737706.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 558013.0, "CurrentDatasourceVersionId": 574599.0, "ForumId": 279426, "Type": 2, "CreationDate": "07/18/2019 06:28:02", "LastActivityDate": "07/18/2019", "TotalViews": 51073, "TotalDownloads": 6399, "TotalVotes": 82, "TotalKernels": 14}]
|
[{"Id": 2737706, "UserName": "vijayashreer", "DisplayName": "Vijaya Shree Raja Sekaran", "RegisterDate": "01/24/2019", "PerformanceTier": 0}]
|
import pandas as pd
data = pd.read_csv("/kaggle/input/food-preferences/Food_Preference.csv")
data.head()
# Food is a binary feature
# ****
data["Food"].unique()
data["Nationality"].unique()
# As we can see countries are written in a different ways and counted multiple times as unique
# also Muslim isn't a nationality it is a religon
data["Nationality"] = data["Nationality"].replace(
["Pakistani ", "Pakistan", "Pakistani"], "pakistani"
)
data["Nationality"] = data["Nationality"].replace(
["Indonesia", "Indonesian ", "Indonesain", "Indonesain", "Indonesian"], "indonesian"
)
data["Nationality"] = data["Nationality"].replace(["Muslim"], "Unknown")
data["Nationality"] = data["Nationality"].replace(
["Maldivian ", "Maldivian"], "Maldivian"
)
data["Nationality"] = data["Nationality"].replace(
["MY", "Malaysian", "Malaysian ", "MALAYSIAN", "Malaysia ", "Malaysia"], "malaysian"
)
data["Nationality"] = data["Nationality"].replace(["Japan"], "japanese")
data["Nationality"] = data["Nationality"].replace(["China"], "Chinese")
data["Nationality"] = data["Nationality"].replace(["Yemen"], "Yemeni")
data["Nationality"].unique()
data["Age"].unique()
# No age outliers
# # Ordinal data****
data["Dessert"].unique()
data_ord = data.copy()
dic = {"Yes": 2, "Maybe": 1, "No": 0}
data_ord["Rating_Dessert"] = data.Dessert.map(dic)
data_ord = data_ord.drop("Dessert", axis=1)
data_ord.head()
# # Binary data
data_ord["Gender"] = data_ord["Gender"].apply(
lambda x: 1 if x == "Male" else (0 if x == "Female" else None)
)
data_ord["Food"] = data_ord["Food"].apply(
lambda x: 1 if x == "Traditional food" else (0 if x == "Western Food" else None)
)
data_ord["Juice"] = data_ord["Juice"].apply(
lambda x: 1 if x == "Fresh Juice" else (0 if x == "Carbonated drinks" else None)
)
data_ord.head()
data.isna().count()
data.shape
df = pd.concat(
[
pd.get_dummies(
prefix="Nationality", data=data_ord["Nationality"], drop_first=True
),
data_ord,
],
axis=1,
)
df.drop(["Nationality", "Timestamp", "Participant_ID"], inplace=True, axis=1)
df
data.isnull().sum()
df = df.dropna()
df.isnull().sum()
| false | 1 | 731 | 0 | 820 | 731 |
||
129063144
|
<jupyter_start><jupyter_text>LoveDa Dataset
Highlights
1) 5987 high spatial resolution (0.3 m) remote sensing images from Nanjing, Changzhou, and Wuhan
2) Focus on different geographical environments between Urban and Rural
3) Advance both semantic segmentation and domain adaptation tasks
4) Three considerable challenges:
-Multi-scale objects
-Complex background samples
-Inconsistent class distributions
Kaggle dataset identifier: loveda-dataset
<jupyter_script># # Helper Functions
import os
import zipfile
def getDirPath(category):
workingPath = "/kaggle/working/" + category
# Create a new directory in the Kaggle environment
os.makedirs(workingPath, exist_ok=True)
# Extract the zipped folder to the new directory
with zipfile.ZipFile("/kaggle/input/" + category + ".zip", "r") as zip_ref:
zip_ref.extractall(workingPath)
return workingPath
# class = ["Rural", "Urban"]
def buildDatasetPath(category, classType):
return getDirPath(category) + "/" + classType + "/"
def buildCustomDataPath(pathUrl, classType):
return pathUrl + "/" + classType + "/"
# # Dataset Loader
import os
import torch
import numpy as np
from PIL import Image
from torch.utils.data import Dataset
class SegmentationDataset(Dataset):
def __init__(self, data_path, transform=None):
self.data_path = data_path
self.transform = transform
self.image_filenames = [] # List to store image file names
self.mask_filenames = [] # List to store mask file names
# Load image and mask file names
self.images_dir = os.path.join(self.data_path, "images_png")
self.masks_dir = os.path.join(self.data_path, "masks_png")
self.image_filenames = sorted(os.listdir(self.images_dir))
self.mask_filenames = sorted(os.listdir(self.masks_dir))
def __len__(self):
return len(self.image_filenames)
def __getitem__(self, idx):
image_path = os.path.join(self.images_dir, self.image_filenames[idx])
mask_path = os.path.join(self.masks_dir, self.mask_filenames[idx])
# Load image and mask
image = Image.open(image_path).convert("RGB")
mask = Image.open(mask_path).convert("L")
if self.transform is not None:
image = self.transform(image)
mask = self.transform(mask)
# image = np.array(image)
# mask = np.array(mask)
# image = np.transpose(image, (1, 2, 0)).astype(np.float32)
# return (torch.from_numpy(image), torch.from_numpy(mask))
return image, mask
import os
import glob
import torch
from torch.utils.data import Dataset
from PIL import Image
class TestDataset(Dataset):
def __init__(self, data_path, transform=None):
"""
Initialize the SemanticSegmentationDataset.
Args:
data_path (str): Path to the dataset directory.
transform (callable, optional): Optional transform to be applied on the images, labels, and masks. Default: None.
"""
self.data_path = data_path
self.transform = transform
self.image_filenames = [] # List to store image file names
# Load image and mask file names
self.images_dir = os.path.join(self.data_path, "images_png")
self.image_filenames = sorted(os.listdir(self.images_dir))
def __len__(self):
return len(self.image_filenames)
def __getitem__(self, index):
image_path = os.path.join(self.images_dir, self.image_filenames[index])
# Load image and mask
image = Image.open(image_path).convert("RGB")
# Apply transformations
if self.transform:
image = self.transform(image)
return image
from torch.utils.data import DataLoader
from torchvision import transforms
data_transforms = transforms.Compose(
[
transforms.RandomHorizontalFlip(),
transforms.RandomVerticalFlip(),
transforms.RandomRotation(15),
transforms.ColorJitter(brightness=0.2, contrast=0.2, saturation=0.2, hue=0.1),
transforms.Resize((256, 256)),
transforms.ToTensor(),
]
)
data_transforms_test = transforms.Compose(
[transforms.Resize((1024, 1024)), transforms.ToTensor()]
)
batch = 4
theme = "Urban"
# theme = "Rural"
train_dataset = SegmentationDataset(
buildCustomDataPath("/kaggle/input/loveda-dataset/Train/Train", theme),
transform=data_transforms,
)
train_dataloader = DataLoader(
train_dataset, batch_size=batch, shuffle=True, num_workers=2
)
val_dataset = SegmentationDataset(
buildCustomDataPath("/kaggle/input/loveda-dataset/Val/Val", theme),
transform=transforms.ToTensor(),
)
val_dataloader = DataLoader(val_dataset, batch_size=batch, shuffle=False, num_workers=2)
test_dataset = TestDataset(
buildCustomDataPath("/kaggle/input/loveda-dataset/Test/Test", theme),
transform=data_transforms_test,
)
test_dataloader = DataLoader(
test_dataset, batch_size=batch, shuffle=True, num_workers=2
)
# # Trans U2net Architecture Model
import torch
import torch.nn as nn
class HAIRNetEncoder(nn.Module):
def __init__(
self, in_channels=3, out_channels=1, kernel_size=3, stride=1, padding=1
):
super(HAIRNetEncoder, self).__init__()
self.conv1 = nn.Conv2d(in_channels, out_channels, kernel_size, stride, padding)
self.bn1 = nn.BatchNorm2d(out_channels)
self.relu1 = nn.ReLU(inplace=True)
self.conv2 = nn.Conv2d(out_channels, out_channels, kernel_size, stride, padding)
self.bn2 = nn.BatchNorm2d(out_channels)
self.relu2 = nn.ReLU(inplace=True)
def forward(self, x):
x = self.conv1(x)
x = self.bn1(x)
x = self.relu1(x)
x = self.conv2(x)
x = self.bn2(x)
x = self.relu2(x)
return x
class HAIRNetDecoder(nn.Module):
def __init__(
self, in_channels=3, out_channels=1, kernel_size=3, stride=1, padding=1
):
super(HAIRNetDecoder, self).__init__()
self.conv1 = nn.Conv2d(in_channels, out_channels, kernel_size, stride, padding)
self.bn1 = nn.BatchNorm2d(out_channels)
self.relu1 = nn.ReLU(inplace=True)
self.conv2 = nn.Conv2d(out_channels, out_channels, kernel_size, stride, padding)
self.bn2 = nn.BatchNorm2d(out_channels)
self.relu2 = nn.ReLU(inplace=True)
def forward(self, x):
x = self.conv1(x)
x = self.bn1(x)
x = self.relu1(x)
x = self.conv2(x)
x = self.bn2(x)
x = self.relu2(x)
return x
import torch
import torch.nn as nn
class U2NetEncoder(nn.Module):
def __init__(self, in_channels=3, out_channels=2):
super(U2NetEncoder, self).__init__()
self.conv1 = nn.Conv2d(
in_channels, out_channels, kernel_size=3, stride=1, padding=1, bias=False
)
self.bn1 = nn.BatchNorm2d(out_channels)
self.relu1 = nn.ReLU(inplace=True)
self.conv2 = nn.Conv2d(
out_channels, out_channels, kernel_size=3, stride=1, padding=1, bias=False
)
self.bn2 = nn.BatchNorm2d(out_channels)
self.relu2 = nn.ReLU(inplace=True)
self.pool1 = nn.MaxPool2d(kernel_size=2, stride=2, ceil_mode=True)
def forward(self, x):
x = self.conv1(x)
x = self.bn1(x)
x = self.relu1(x)
x = self.conv2(x)
x = self.bn2(x)
x = self.relu2(x)
return x
class U2NetDecoder(nn.Module):
def __init__(self, in_channels=3, out_channels=1):
super(U2NetDecoder, self).__init__()
self.conv1 = nn.Conv2d(
in_channels, in_channels, kernel_size=3, stride=1, padding=1, bias=False
)
self.bn1 = nn.BatchNorm2d(in_channels)
self.relu1 = nn.ReLU(inplace=True)
self.conv2 = nn.Conv2d(out_channels, out_channels, kernel_size=1, stride=1)
self.bn2 = nn.BatchNorm2d(in_channels)
self.relu2 = nn.ReLU(inplace=True)
def forward(self, x):
x = self.conv1(x)
x = self.bn1(x)
x = self.relu1(x)
x = self.conv2(x)
x = self.bn2(x)
x = self.relu2(x)
return x
import torch.nn as nn
# Define combined encoder
class CombinedEncoder(nn.Module):
def __init__(self):
super(CombinedEncoder, self).__init__()
self.hairnet_encoder = HAIRNetEncoder()
self.u2net_encoder = U2NetEncoder()
def forward(self, x):
hairnet_feats = self.hairnet_encoder(x)
u2net_feats = self.u2net_encoder(x)
feats = torch.cat([hairnet_feats, u2net_feats], dim=1)
return feats
# Define combined decoder
class CombinedDecoder(nn.Module):
def __init__(self):
super(CombinedDecoder, self).__init__()
self.hairnet_decoder = HAIRNetDecoder()
self.u2net_decoder = U2NetDecoder()
def forward(self, x):
hairnet_feats = x[:, : x.size(1), :, :]
u2net_feats = x[:, hairnet_feats.size(1) :, :, :]
feats = torch.cat([hairnet_feats, u2net_feats], dim=1)
out = self.hairnet_decoder(feats)
out += self.u2net_decoder(feats)
return out
# Define the final combined model
class HAIRNet_U2Net(nn.Module):
def __init__(self):
super(HAIRNet_U2Net, self).__init__()
self.encoder = CombinedEncoder()
self.decoder = CombinedDecoder()
def forward(self, x):
feats = self.encoder(x)
out = self.decoder(feats)
return out
# # Loss Function Definition
import torch
import torch.nn.functional as F
class TverskyLoss(torch.nn.Module):
def __init__(self, alpha=0.5, beta=0.5, smooth=1.0):
super().__init__()
self.alpha = alpha
self.beta = beta
self.smooth = smooth
def forward(self, y_pred, y_true):
y_pred = torch.sigmoid(y_pred)
y_true_pos = y_true.view(-1)
y_pred_pos = y_pred.view(-1)
true_pos = (y_true_pos * y_pred_pos).sum()
false_neg = ((1 - y_true_pos) * y_pred_pos).sum()
false_pos = (y_true_pos * (1 - y_pred_pos)).sum()
tversky_loss = (true_pos + self.smooth) / (
true_pos + self.alpha * false_neg + self.beta * false_pos + self.smooth
)
return 1 - tversky_loss
# # Model Training
import torch.optim as optim
from torch.utils.data import DataLoader
# Define hyperparameters
input_channels = 3
output_channels = 1
num_transformer_layers = 4
num_heads = 4
hidden_size = 64
learning_rate = 0.001
num_epochs = 10
# Create model, loss function, and optimizer
# model = TransformerU2Net(input_channels, output_channels, num_transformer_layers, num_heads, hidden_size)
model = HAIRNet_U2Net()
loss_fn = nn.CrossEntropyLoss()
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
# Train model
for epoch in range(num_epochs):
running_loss = 0.0
for i, data in enumerate(train_dataloader, 0):
# Get inputs and labels
inputs, targets = data
# Zero the parameter gradients
optimizer.zero_grad()
# Forward pass
outputs = model(inputs)
loss = loss_fn(outputs, targets)
# Backward pass and optimize
loss.backward()
optimizer.step()
# Print statistics
running_loss += loss.item()
if i % 100 == 99: # Print every 100 mini-batches
print("[%d, %5d] loss: %.3f" % (epoch + 1, i + 1, running_loss / 100))
running_loss = 0.0
print("Finished Training")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/063/129063144.ipynb
|
loveda-dataset
|
mohammedjaveed
|
[{"Id": 129063144, "ScriptId": 38357712, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4295557, "CreationDate": "05/10/2023 17:13:45", "VersionNumber": 1.0, "Title": "origin-Proposal", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 347.0, "LinesInsertedFromPrevious": 347.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184787841, "KernelVersionId": 129063144, "SourceDatasetVersionId": 5418297}]
|
[{"Id": 5418297, "DatasetId": 3137175, "DatasourceVersionId": 5492207, "CreatorUserId": 4295557, "LicenseName": "GNU Lesser General Public License 3.0", "CreationDate": "04/15/2023 23:44:28", "VersionNumber": 1.0, "Title": "LoveDa Dataset", "Slug": "loveda-dataset", "Subtitle": "LoveDA: A Remote Sensing Land-Cover Dataset for Domain Adaptive Semantic Segment", "Description": "Highlights\n\n1) 5987 high spatial resolution (0.3 m) remote sensing images from Nanjing, Changzhou, and Wuhan\n2) Focus on different geographical environments between Urban and Rural\n3) Advance both semantic segmentation and domain adaptation tasks\n4) Three considerable challenges:\n -Multi-scale objects\n -Complex background samples\n -Inconsistent class distributions", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3137175, "CreatorUserId": 4295557, "OwnerUserId": 4295557.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5418297.0, "CurrentDatasourceVersionId": 5492207.0, "ForumId": 3200910, "Type": 2, "CreationDate": "04/15/2023 23:44:28", "LastActivityDate": "04/15/2023", "TotalViews": 379, "TotalDownloads": 54, "TotalVotes": 0, "TotalKernels": 2}]
|
[{"Id": 4295557, "UserName": "mohammedjaveed", "DisplayName": "Mohammed Javeed", "RegisterDate": "01/07/2020", "PerformanceTier": 0}]
|
# # Helper Functions
import os
import zipfile
def getDirPath(category):
workingPath = "/kaggle/working/" + category
# Create a new directory in the Kaggle environment
os.makedirs(workingPath, exist_ok=True)
# Extract the zipped folder to the new directory
with zipfile.ZipFile("/kaggle/input/" + category + ".zip", "r") as zip_ref:
zip_ref.extractall(workingPath)
return workingPath
# class = ["Rural", "Urban"]
def buildDatasetPath(category, classType):
return getDirPath(category) + "/" + classType + "/"
def buildCustomDataPath(pathUrl, classType):
return pathUrl + "/" + classType + "/"
# # Dataset Loader
import os
import torch
import numpy as np
from PIL import Image
from torch.utils.data import Dataset
class SegmentationDataset(Dataset):
def __init__(self, data_path, transform=None):
self.data_path = data_path
self.transform = transform
self.image_filenames = [] # List to store image file names
self.mask_filenames = [] # List to store mask file names
# Load image and mask file names
self.images_dir = os.path.join(self.data_path, "images_png")
self.masks_dir = os.path.join(self.data_path, "masks_png")
self.image_filenames = sorted(os.listdir(self.images_dir))
self.mask_filenames = sorted(os.listdir(self.masks_dir))
def __len__(self):
return len(self.image_filenames)
def __getitem__(self, idx):
image_path = os.path.join(self.images_dir, self.image_filenames[idx])
mask_path = os.path.join(self.masks_dir, self.mask_filenames[idx])
# Load image and mask
image = Image.open(image_path).convert("RGB")
mask = Image.open(mask_path).convert("L")
if self.transform is not None:
image = self.transform(image)
mask = self.transform(mask)
# image = np.array(image)
# mask = np.array(mask)
# image = np.transpose(image, (1, 2, 0)).astype(np.float32)
# return (torch.from_numpy(image), torch.from_numpy(mask))
return image, mask
import os
import glob
import torch
from torch.utils.data import Dataset
from PIL import Image
class TestDataset(Dataset):
def __init__(self, data_path, transform=None):
"""
Initialize the SemanticSegmentationDataset.
Args:
data_path (str): Path to the dataset directory.
transform (callable, optional): Optional transform to be applied on the images, labels, and masks. Default: None.
"""
self.data_path = data_path
self.transform = transform
self.image_filenames = [] # List to store image file names
# Load image and mask file names
self.images_dir = os.path.join(self.data_path, "images_png")
self.image_filenames = sorted(os.listdir(self.images_dir))
def __len__(self):
return len(self.image_filenames)
def __getitem__(self, index):
image_path = os.path.join(self.images_dir, self.image_filenames[index])
# Load image and mask
image = Image.open(image_path).convert("RGB")
# Apply transformations
if self.transform:
image = self.transform(image)
return image
from torch.utils.data import DataLoader
from torchvision import transforms
data_transforms = transforms.Compose(
[
transforms.RandomHorizontalFlip(),
transforms.RandomVerticalFlip(),
transforms.RandomRotation(15),
transforms.ColorJitter(brightness=0.2, contrast=0.2, saturation=0.2, hue=0.1),
transforms.Resize((256, 256)),
transforms.ToTensor(),
]
)
data_transforms_test = transforms.Compose(
[transforms.Resize((1024, 1024)), transforms.ToTensor()]
)
batch = 4
theme = "Urban"
# theme = "Rural"
train_dataset = SegmentationDataset(
buildCustomDataPath("/kaggle/input/loveda-dataset/Train/Train", theme),
transform=data_transforms,
)
train_dataloader = DataLoader(
train_dataset, batch_size=batch, shuffle=True, num_workers=2
)
val_dataset = SegmentationDataset(
buildCustomDataPath("/kaggle/input/loveda-dataset/Val/Val", theme),
transform=transforms.ToTensor(),
)
val_dataloader = DataLoader(val_dataset, batch_size=batch, shuffle=False, num_workers=2)
test_dataset = TestDataset(
buildCustomDataPath("/kaggle/input/loveda-dataset/Test/Test", theme),
transform=data_transforms_test,
)
test_dataloader = DataLoader(
test_dataset, batch_size=batch, shuffle=True, num_workers=2
)
# # Trans U2net Architecture Model
import torch
import torch.nn as nn
class HAIRNetEncoder(nn.Module):
def __init__(
self, in_channels=3, out_channels=1, kernel_size=3, stride=1, padding=1
):
super(HAIRNetEncoder, self).__init__()
self.conv1 = nn.Conv2d(in_channels, out_channels, kernel_size, stride, padding)
self.bn1 = nn.BatchNorm2d(out_channels)
self.relu1 = nn.ReLU(inplace=True)
self.conv2 = nn.Conv2d(out_channels, out_channels, kernel_size, stride, padding)
self.bn2 = nn.BatchNorm2d(out_channels)
self.relu2 = nn.ReLU(inplace=True)
def forward(self, x):
x = self.conv1(x)
x = self.bn1(x)
x = self.relu1(x)
x = self.conv2(x)
x = self.bn2(x)
x = self.relu2(x)
return x
class HAIRNetDecoder(nn.Module):
def __init__(
self, in_channels=3, out_channels=1, kernel_size=3, stride=1, padding=1
):
super(HAIRNetDecoder, self).__init__()
self.conv1 = nn.Conv2d(in_channels, out_channels, kernel_size, stride, padding)
self.bn1 = nn.BatchNorm2d(out_channels)
self.relu1 = nn.ReLU(inplace=True)
self.conv2 = nn.Conv2d(out_channels, out_channels, kernel_size, stride, padding)
self.bn2 = nn.BatchNorm2d(out_channels)
self.relu2 = nn.ReLU(inplace=True)
def forward(self, x):
x = self.conv1(x)
x = self.bn1(x)
x = self.relu1(x)
x = self.conv2(x)
x = self.bn2(x)
x = self.relu2(x)
return x
import torch
import torch.nn as nn
class U2NetEncoder(nn.Module):
def __init__(self, in_channels=3, out_channels=2):
super(U2NetEncoder, self).__init__()
self.conv1 = nn.Conv2d(
in_channels, out_channels, kernel_size=3, stride=1, padding=1, bias=False
)
self.bn1 = nn.BatchNorm2d(out_channels)
self.relu1 = nn.ReLU(inplace=True)
self.conv2 = nn.Conv2d(
out_channels, out_channels, kernel_size=3, stride=1, padding=1, bias=False
)
self.bn2 = nn.BatchNorm2d(out_channels)
self.relu2 = nn.ReLU(inplace=True)
self.pool1 = nn.MaxPool2d(kernel_size=2, stride=2, ceil_mode=True)
def forward(self, x):
x = self.conv1(x)
x = self.bn1(x)
x = self.relu1(x)
x = self.conv2(x)
x = self.bn2(x)
x = self.relu2(x)
return x
class U2NetDecoder(nn.Module):
def __init__(self, in_channels=3, out_channels=1):
super(U2NetDecoder, self).__init__()
self.conv1 = nn.Conv2d(
in_channels, in_channels, kernel_size=3, stride=1, padding=1, bias=False
)
self.bn1 = nn.BatchNorm2d(in_channels)
self.relu1 = nn.ReLU(inplace=True)
self.conv2 = nn.Conv2d(out_channels, out_channels, kernel_size=1, stride=1)
self.bn2 = nn.BatchNorm2d(in_channels)
self.relu2 = nn.ReLU(inplace=True)
def forward(self, x):
x = self.conv1(x)
x = self.bn1(x)
x = self.relu1(x)
x = self.conv2(x)
x = self.bn2(x)
x = self.relu2(x)
return x
import torch.nn as nn
# Define combined encoder
class CombinedEncoder(nn.Module):
def __init__(self):
super(CombinedEncoder, self).__init__()
self.hairnet_encoder = HAIRNetEncoder()
self.u2net_encoder = U2NetEncoder()
def forward(self, x):
hairnet_feats = self.hairnet_encoder(x)
u2net_feats = self.u2net_encoder(x)
feats = torch.cat([hairnet_feats, u2net_feats], dim=1)
return feats
# Define combined decoder
class CombinedDecoder(nn.Module):
def __init__(self):
super(CombinedDecoder, self).__init__()
self.hairnet_decoder = HAIRNetDecoder()
self.u2net_decoder = U2NetDecoder()
def forward(self, x):
hairnet_feats = x[:, : x.size(1), :, :]
u2net_feats = x[:, hairnet_feats.size(1) :, :, :]
feats = torch.cat([hairnet_feats, u2net_feats], dim=1)
out = self.hairnet_decoder(feats)
out += self.u2net_decoder(feats)
return out
# Define the final combined model
class HAIRNet_U2Net(nn.Module):
def __init__(self):
super(HAIRNet_U2Net, self).__init__()
self.encoder = CombinedEncoder()
self.decoder = CombinedDecoder()
def forward(self, x):
feats = self.encoder(x)
out = self.decoder(feats)
return out
# # Loss Function Definition
import torch
import torch.nn.functional as F
class TverskyLoss(torch.nn.Module):
def __init__(self, alpha=0.5, beta=0.5, smooth=1.0):
super().__init__()
self.alpha = alpha
self.beta = beta
self.smooth = smooth
def forward(self, y_pred, y_true):
y_pred = torch.sigmoid(y_pred)
y_true_pos = y_true.view(-1)
y_pred_pos = y_pred.view(-1)
true_pos = (y_true_pos * y_pred_pos).sum()
false_neg = ((1 - y_true_pos) * y_pred_pos).sum()
false_pos = (y_true_pos * (1 - y_pred_pos)).sum()
tversky_loss = (true_pos + self.smooth) / (
true_pos + self.alpha * false_neg + self.beta * false_pos + self.smooth
)
return 1 - tversky_loss
# # Model Training
import torch.optim as optim
from torch.utils.data import DataLoader
# Define hyperparameters
input_channels = 3
output_channels = 1
num_transformer_layers = 4
num_heads = 4
hidden_size = 64
learning_rate = 0.001
num_epochs = 10
# Create model, loss function, and optimizer
# model = TransformerU2Net(input_channels, output_channels, num_transformer_layers, num_heads, hidden_size)
model = HAIRNet_U2Net()
loss_fn = nn.CrossEntropyLoss()
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
# Train model
for epoch in range(num_epochs):
running_loss = 0.0
for i, data in enumerate(train_dataloader, 0):
# Get inputs and labels
inputs, targets = data
# Zero the parameter gradients
optimizer.zero_grad()
# Forward pass
outputs = model(inputs)
loss = loss_fn(outputs, targets)
# Backward pass and optimize
loss.backward()
optimizer.step()
# Print statistics
running_loss += loss.item()
if i % 100 == 99: # Print every 100 mini-batches
print("[%d, %5d] loss: %.3f" % (epoch + 1, i + 1, running_loss / 100))
running_loss = 0.0
print("Finished Training")
| false | 0 | 3,278 | 0 | 3,392 | 3,278 |
||
129119223
|
<jupyter_start><jupyter_text>Male & Female height and weight
### Context
Globally, when we talk about the features to predict the sex of each person, it is undeniable that Height & Weight are typical features for that.
This dataset is purposely for the beginner who recently has done studying Machine Algorithm and may want to apply their algorithm on a simple dataset.
There are just 2 features (Height, Weight) & 1 label (Sex)
- Height (cm)
- Weight (kg)
- Sex (male | female)
### Content
There are 2 datasets; training set and test set
1. Training set consists of 3000 rows & 3 columns
2. Test set consists of 205 rows & 3 columns
Kaggle dataset identifier: male-female-height-and-weight
<jupyter_script># Hello,everyone.In this article,I use neural network(NN) to predict it.
import pandas as pd # 导入csv文件的库
import numpy as np # 进行矩阵运算的库
import matplotlib.pyplot as plt # 作图的库
import torch # 一个深度学习的库Pytorch
import torch.nn as nn # neural network,神经网络
from torch.autograd import Variable # 从自动求导中引入变量
import torch.optim as optim # 一个实现了各种优化算法的库
import torch.nn.functional as F # 神经网络函数库
import warnings # 避免一些可以忽略的报错
warnings.filterwarnings("ignore")
train = pd.read_csv("/kaggle/input/male-female-height-and-weight/Training set.csv")
test = pd.read_csv("/kaggle/input/male-female-height-and-weight/Test set.csv")
# Let's check the dataset.
train.head()
display(train.describe())
# Converts the string in the gender column to a number.
train.loc[train["Sex"] == "Male", "Sex"] = 1
train.loc[train["Sex"] == "Female", "Sex"] = 0
test.loc[test["Sex"] == "Male", "Sex"] = 1
test.loc[test["Sex"] == "Female", "Sex"] = 0
# Handle outliers.
# 用箱型图去除异常值
def QutlierDetection(df, cols=[], percentage=0.01):
"""
传入dataFrame对象.
和需要清除异常值的几列数据
如果异常值的数据量小于数据量*百分比,就删除数据.
"""
for col in cols:
df = df.reset_index(drop=True)
df_col = df[col] # 获取列表
df_col_value = df[col].values
# 计算下分位数和上四分位
Q1 = df_col.quantile(q=0.25)
Q3 = df_col.quantile(q=0.75)
# 基于1.5倍的四分位差计算上下对应的值
low_whisker = Q1 - 1.5 * (Q3 - Q1)
up_whisker = Q3 + 1.5 * (Q3 - Q1)
left = set(np.where(low_whisker > df_col_value)[0])
right = set(np.where(df_col_value > up_whisker)[0])
choose = list(left | right)
if len(choose) < len(df) * percentage:
df.drop(choose, axis=0, inplace=True)
return df # 返回清除异常值后的数据
len(train)
train = QutlierDetection(train, cols=["Height", "Weight"])
len(train)
# Centralization of processing.
def col_center(array):
"""
将np.array数组中每列数据都减去每列的平均值.
并返回新的数组和每列的平均值
"""
mean_array = np.mean(array, axis=0)
new_array = array - mean_array
return new_array, mean_array
def deal_df(train_df, test_df, drop_col=[], target="label"):
"""
传入训练集和测试集的pd.DateFrame对象,
drop_col是需要清除的几列.list
target一般是需要进行预测的,训练集有,但是测试集没有的一列数据.string
"""
# 清除无关的那几列
train_df.drop(drop_col, axis=1, inplace=True)
test_df.drop(drop_col, axis=1, inplace=True)
# 提取出训练集和测试集的数据
y_train = train_df[target].values
y_test = test_df[target].values
train_df.drop(target, axis=1, inplace=True)
test_df.drop(target, axis=1, inplace=True)
x_train = train_df.values
x_test = test_df.values
columns = train_df.keys().values
# 将训练集和测试集的数据合并
x_data = np.vstack((x_train, x_test))
# 将数据按列做减去平均值
new_x_data, mean_x_data = col_center(x_data)
# 将数据重新分成训练集和测试集
new_x_train = new_x_data[: len(y_train)]
new_x_test = new_x_data[len(y_train) :]
x_train_df = pd.DataFrame(new_x_train, columns=columns)
x_test_df = pd.DataFrame(new_x_test, columns=columns)
y_train_df = pd.DataFrame(y_train)[0]
y_test_df = pd.DataFrame(y_test)[0]
return x_train_df, y_train_df, x_test_df, y_test_df
x_train_df, y_train_df, x_test_df, y_test_df = deal_df(train, test, target="Sex")
train_X = x_train_df.values
test_X = x_test_df.values
train_y = y_train_df.values.astype("int")
test_y = y_test_df.values.astype("int")
train_y
# Let's draw a scatter plot.
female_height = []
female_weight = []
male_height = []
male_weight = []
for i in range(len(train_X)):
if train_y[i]:
male_height.append(train_X[i][0])
male_weight.append(train_X[i][1])
else:
female_height.append(train_X[i][0])
female_weight.append(train_X[i][1])
plt.scatter(female_height, female_weight, label="female")
plt.scatter(male_height, male_weight, label="male")
plt.legend()
plt.show()
# So we're done with the Data pre-processing.Then,let's build a neural network.
# yun su xiao zi made
class NN(nn.Module):
# 初始化
def __init__(self):
# 继承父类的所有方法
super(NN, self).__init__()
# 构建序列化的神经网络,将网络层按照传入的顺序组合起来
self.model = nn.Sequential()
# 全连接层
self.model.add_module("linear1", nn.Linear(2, 10))
self.model.add_module("Tanh1", nn.Tanh())
self.model.add_module("linear2", nn.Linear(10, 2))
# 前向传播
def forward(self, input):
# 传入数据
output = input
# 按照神经网络的顺序去跑一遍
for name, module in self.model.named_children():
output = module(output)
return F.softmax(output, dim=1)
# 初始化神经网络
net = NN()
print(net)
# 优化器
optimizer = optim.Adam(net.parameters(), lr=0.0002, betas=(0.5, 0.999))
# 损失函数
criterion = nn.NLLLoss() # 负对数似然损失函数,也是交叉熵损失函数的一种
# 训练周期为10000次
num_epochs = 10000
# We use train_X to fit it.
train_X = torch.FloatTensor(train_X)
train_y = torch.Tensor(train_y).long()
for epoch in range(num_epochs):
# 将梯度清空
optimizer.zero_grad()
# 训练
net.train()
output = net(train_X) # 将数据放进去训练
# 计算每次的损失函数
error = criterion(output, train_y)
# 反向传播
error.backward()
# 优化器进行优化(梯度下降,降低误差)
optimizer.step()
# 迭代1000次查看损失函数的效果
if epoch % 1000 == 0:
print(epoch, error)
# Let's check the result.
pred = net(torch.Tensor(train_X))
pred = np.argmax(pred.detach().cpu().numpy(), axis=1)
print(
"the accuracy of training set:",
np.sum(pred == train_y.detach().cpu().numpy()) / len(train_y),
)
pred = net(torch.Tensor(test_X))
pred = np.argmax(pred.detach().cpu().numpy(), axis=1)
print("the accuracy of testing set:", np.sum(pred == test_y) / len(test_y))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/119/129119223.ipynb
|
male-female-height-and-weight
|
saranpannasuriyaporn
|
[{"Id": 129119223, "ScriptId": 33187231, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12691485, "CreationDate": "05/11/2023 06:35:55", "VersionNumber": 2.0, "Title": "neural network(NN)", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 205.0, "LinesInsertedFromPrevious": 180.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 25.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184891149, "KernelVersionId": 129119223, "SourceDatasetVersionId": 2019419}]
|
[{"Id": 2019419, "DatasetId": 1208641, "DatasourceVersionId": 2059056, "CreatorUserId": 5520618, "LicenseName": "Unknown", "CreationDate": "03/13/2021 14:06:05", "VersionNumber": 4.0, "Title": "Male & Female height and weight", "Slug": "male-female-height-and-weight", "Subtitle": "Use to predict sex by given weight and height", "Description": "### Context\n\nGlobally, when we talk about the features to predict the sex of each person, it is undeniable that Height & Weight are typical features for that.\nThis dataset is purposely for the beginner who recently has done studying Machine Algorithm and may want to apply their algorithm on a simple dataset. \n\nThere are just 2 features (Height, Weight) & 1 label (Sex)\n\n- Height (cm)\n- Weight (kg)\n- Sex (male | female)\n\n\n### Content\n\nThere are 2 datasets; training set and test set\n\n1. Training set consists of 3000 rows & 3 columns \n2. Test set consists of 205 rows & 3 columns\n\n### Acknowledgements\n\nThank you for the great research from https://www.researchgate.net/figure/The-mean-and-standard-deviation-of-height-weight-and-age-for-both-male-and-female_tbl1_257769120, \n\nso we can generate sample data through their mean & std.\n\n\n### Inspiration\n\n- Predict the sex using 2 features; height and weight in the test set.\n- then evaluate the prediction accuracy using the ROC AUC score\n\n(Evaluation: ROC AUC score)", "VersionNotes": "Weight-Height-Sex", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1208641, "CreatorUserId": 5520618, "OwnerUserId": 5520618.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2019419.0, "CurrentDatasourceVersionId": 2059056.0, "ForumId": 1226661, "Type": 2, "CreationDate": "03/13/2021 12:56:14", "LastActivityDate": "03/13/2021", "TotalViews": 7808, "TotalDownloads": 1205, "TotalVotes": 11, "TotalKernels": 6}]
|
[{"Id": 5520618, "UserName": "saranpannasuriyaporn", "DisplayName": "Saran Pannasuriyaporn", "RegisterDate": "07/24/2020", "PerformanceTier": 1}]
|
# Hello,everyone.In this article,I use neural network(NN) to predict it.
import pandas as pd # 导入csv文件的库
import numpy as np # 进行矩阵运算的库
import matplotlib.pyplot as plt # 作图的库
import torch # 一个深度学习的库Pytorch
import torch.nn as nn # neural network,神经网络
from torch.autograd import Variable # 从自动求导中引入变量
import torch.optim as optim # 一个实现了各种优化算法的库
import torch.nn.functional as F # 神经网络函数库
import warnings # 避免一些可以忽略的报错
warnings.filterwarnings("ignore")
train = pd.read_csv("/kaggle/input/male-female-height-and-weight/Training set.csv")
test = pd.read_csv("/kaggle/input/male-female-height-and-weight/Test set.csv")
# Let's check the dataset.
train.head()
display(train.describe())
# Converts the string in the gender column to a number.
train.loc[train["Sex"] == "Male", "Sex"] = 1
train.loc[train["Sex"] == "Female", "Sex"] = 0
test.loc[test["Sex"] == "Male", "Sex"] = 1
test.loc[test["Sex"] == "Female", "Sex"] = 0
# Handle outliers.
# 用箱型图去除异常值
def QutlierDetection(df, cols=[], percentage=0.01):
"""
传入dataFrame对象.
和需要清除异常值的几列数据
如果异常值的数据量小于数据量*百分比,就删除数据.
"""
for col in cols:
df = df.reset_index(drop=True)
df_col = df[col] # 获取列表
df_col_value = df[col].values
# 计算下分位数和上四分位
Q1 = df_col.quantile(q=0.25)
Q3 = df_col.quantile(q=0.75)
# 基于1.5倍的四分位差计算上下对应的值
low_whisker = Q1 - 1.5 * (Q3 - Q1)
up_whisker = Q3 + 1.5 * (Q3 - Q1)
left = set(np.where(low_whisker > df_col_value)[0])
right = set(np.where(df_col_value > up_whisker)[0])
choose = list(left | right)
if len(choose) < len(df) * percentage:
df.drop(choose, axis=0, inplace=True)
return df # 返回清除异常值后的数据
len(train)
train = QutlierDetection(train, cols=["Height", "Weight"])
len(train)
# Centralization of processing.
def col_center(array):
"""
将np.array数组中每列数据都减去每列的平均值.
并返回新的数组和每列的平均值
"""
mean_array = np.mean(array, axis=0)
new_array = array - mean_array
return new_array, mean_array
def deal_df(train_df, test_df, drop_col=[], target="label"):
"""
传入训练集和测试集的pd.DateFrame对象,
drop_col是需要清除的几列.list
target一般是需要进行预测的,训练集有,但是测试集没有的一列数据.string
"""
# 清除无关的那几列
train_df.drop(drop_col, axis=1, inplace=True)
test_df.drop(drop_col, axis=1, inplace=True)
# 提取出训练集和测试集的数据
y_train = train_df[target].values
y_test = test_df[target].values
train_df.drop(target, axis=1, inplace=True)
test_df.drop(target, axis=1, inplace=True)
x_train = train_df.values
x_test = test_df.values
columns = train_df.keys().values
# 将训练集和测试集的数据合并
x_data = np.vstack((x_train, x_test))
# 将数据按列做减去平均值
new_x_data, mean_x_data = col_center(x_data)
# 将数据重新分成训练集和测试集
new_x_train = new_x_data[: len(y_train)]
new_x_test = new_x_data[len(y_train) :]
x_train_df = pd.DataFrame(new_x_train, columns=columns)
x_test_df = pd.DataFrame(new_x_test, columns=columns)
y_train_df = pd.DataFrame(y_train)[0]
y_test_df = pd.DataFrame(y_test)[0]
return x_train_df, y_train_df, x_test_df, y_test_df
x_train_df, y_train_df, x_test_df, y_test_df = deal_df(train, test, target="Sex")
train_X = x_train_df.values
test_X = x_test_df.values
train_y = y_train_df.values.astype("int")
test_y = y_test_df.values.astype("int")
train_y
# Let's draw a scatter plot.
female_height = []
female_weight = []
male_height = []
male_weight = []
for i in range(len(train_X)):
if train_y[i]:
male_height.append(train_X[i][0])
male_weight.append(train_X[i][1])
else:
female_height.append(train_X[i][0])
female_weight.append(train_X[i][1])
plt.scatter(female_height, female_weight, label="female")
plt.scatter(male_height, male_weight, label="male")
plt.legend()
plt.show()
# So we're done with the Data pre-processing.Then,let's build a neural network.
# yun su xiao zi made
class NN(nn.Module):
# 初始化
def __init__(self):
# 继承父类的所有方法
super(NN, self).__init__()
# 构建序列化的神经网络,将网络层按照传入的顺序组合起来
self.model = nn.Sequential()
# 全连接层
self.model.add_module("linear1", nn.Linear(2, 10))
self.model.add_module("Tanh1", nn.Tanh())
self.model.add_module("linear2", nn.Linear(10, 2))
# 前向传播
def forward(self, input):
# 传入数据
output = input
# 按照神经网络的顺序去跑一遍
for name, module in self.model.named_children():
output = module(output)
return F.softmax(output, dim=1)
# 初始化神经网络
net = NN()
print(net)
# 优化器
optimizer = optim.Adam(net.parameters(), lr=0.0002, betas=(0.5, 0.999))
# 损失函数
criterion = nn.NLLLoss() # 负对数似然损失函数,也是交叉熵损失函数的一种
# 训练周期为10000次
num_epochs = 10000
# We use train_X to fit it.
train_X = torch.FloatTensor(train_X)
train_y = torch.Tensor(train_y).long()
for epoch in range(num_epochs):
# 将梯度清空
optimizer.zero_grad()
# 训练
net.train()
output = net(train_X) # 将数据放进去训练
# 计算每次的损失函数
error = criterion(output, train_y)
# 反向传播
error.backward()
# 优化器进行优化(梯度下降,降低误差)
optimizer.step()
# 迭代1000次查看损失函数的效果
if epoch % 1000 == 0:
print(epoch, error)
# Let's check the result.
pred = net(torch.Tensor(train_X))
pred = np.argmax(pred.detach().cpu().numpy(), axis=1)
print(
"the accuracy of training set:",
np.sum(pred == train_y.detach().cpu().numpy()) / len(train_y),
)
pred = net(torch.Tensor(test_X))
pred = np.argmax(pred.detach().cpu().numpy(), axis=1)
print("the accuracy of testing set:", np.sum(pred == test_y) / len(test_y))
| false | 2 | 2,146 | 0 | 2,334 | 2,146 |
||
129119687
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ---
# Introduction:
# The ability of a borrower to repay a loan is crucial information for lenders to make informed decisions about their company's growth forecast. For new homeowners with no credit history, lenders need to determine whether they will be able to repay the loan in the future. Therefore, it is essential to know whether the borrower will be able to pay the loan or default in the future.
# In this project, we will be using data provided by the Home Credit Group, which is published on Kaggle (https://www.kaggle.com/c/home-credit-default-risk/overview). The goal is to predict whether a borrower will be able to repay their existing loan, which is a binary classification problem. We will be using input features such as the financial and behavioral history of the loan applicants to make our predictions.
# Datasets:
# The Kaggle website provides a comprehensive data description that outlines the relationship between the different data points.
# 
# As the dataset was too large, consisting of 7 CSV files totaling over 30GB, I had to shrink the dataset by grouping the data by ID and taking the mean of each ID. This reduced the number of rows in the dataset, as there were multiple rows for each ID in the dataset except for the application file.
#
# Project Overview:
# I used tree-based machine learning algorithms to predict the target variable, achieving an AUC score of around 78% without hyperparameter tuning.
# The AUC score is useful because it provides a single number that summarizes the overall performance of the classifier across all possible threshold settings. It is also useful when the class distribution is imbalanced, as it is less affected by the imbalance than other metrics such as accuracy.
# I used a logistic regression model as the base level model, and two boosting methods, XGBClassifier and LGBMClassifier. LGB has the better accuracy rate of around 78% . I also performed K-fold cross-validation to select the best model and improve the accuracy rate, but the accuracy rate of the LGBMClassifier reduced to around 61%, which did not improve the result.
# To achieve better results, I believe I need to perform more extensive feature analysis with domain knowledge and analyze model tuning. This project was a great exercise for me to learn how to process big data, learn PyScaler SQL, learn feature engineering, and optimize data.
# Structure of the project:
# This analysis is divided into six sections:
# Data imputation and size reduction: To minimize the size of the large dataset, I changed the data type before importing it. I also reduced the size of the data by taking the mean of each row, which reduced the number of rows in the dataset.
# Merging the dataset: Since the dataset was large, I merged the data by following a chart and taking the mean of each row.
# Exploratory data analysis: I visualized each feature and compared it to the target variable to gain insights into the types of people who normally default.
# Feature engineering: I used domain knowledge to engineer five different features, including debt-to-income ratio, loan-to-value ratio, percentage of the applicant's income used to pay off the loan, the length of time to pay off the loan, and employment history percentage. I also used polynomial feature engineering.
# Classifier models: I split the data into training and test sets and trained models of similar size for different modeling processes.
# Tuning: Finally, I split the training data into k equal parts and trained the model on the remaining k-1 folds, evaluating it on the current fold. I also tried hyperparameter tuning.
# Import required libraries/packages
import warnings
warnings.filterwarnings("ignore")
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from timeit import default_timer as timer
import os
import random
import csv
import json
import itertools
import pprint
# from pydash import at
import gc
import re
# import featuretools for automated feature engineering
import featuretools as ft
from featuretools import selection
# Import sklearn helper metrics and transformations
from sklearn.base import TransformerMixin
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split, KFold
from sklearn.utils import resample
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
precision_score,
recall_score,
roc_auc_score,
classification_report,
roc_curve,
auc,
f1_score,
)
# Import models
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.naive_bayes import GaussianNB
from xgboost import XGBClassifier
from sklearn.ensemble import GradientBoostingClassifier
import lightgbm as lgb
# import library for hyperparameter optimization
from hyperopt import STATUS_OK
from hyperopt import hp, tpe, Trials, fmin
from hyperopt.pyll.stochastic import sample
# **1. Data imputation and size reduction:**
import os
def reduce_mem_usage(df):
numerics = ["int16", "int32", "int64", "float16", "float32", "float64"]
for col in df.columns:
col_type = df[col].dtypes
if col_type in numerics:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == "int":
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int8)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
else:
if (
c_min > np.finfo(np.float16).min
and c_max < np.finfo(np.float16).max
):
df[col] = df[col].astype(np.float16)
elif (
c_min > np.finfo(np.float32).min
and c_max < np.finfo(np.float32).max
):
df[col] = df[col].astype(np.float32)
else:
df[col] = df[col].astype(np.float64)
return df
def get_balance_data():
default_dir = "../input/home-credit-default-risk/"
pos_dtype = {
"SK_ID_PREV": np.uint32,
"SK_ID_CURR": np.uint32,
"MONTHS_BALANCE": np.int32,
"SK_DPD": np.int32,
"SK_DPD_DEF": np.int32,
"CNT_INSTALMENT": np.float32,
"CNT_INSTALMENT_FUTURE": np.float32,
}
install_dtype = {
"SK_ID_PREV": np.uint32,
"SK_ID_CURR": np.uint32,
"NUM_INSTALMENT_NUMBER": np.int32,
"NUM_INSTALMENT_VERSION": np.float32,
"DAYS_INSTALMENT": np.float32,
"DAYS_ENTRY_PAYMENT": np.float32,
"AMT_INSTALMENT": np.float32,
"AMT_PAYMENT": np.float32,
}
card_dtype = {
"SK_ID_PREV": np.uint32,
"SK_ID_CURR": np.uint32,
"MONTHS_BALANCE": np.int16,
"AMT_CREDIT_LIMIT_ACTUAL": np.int32,
"CNT_DRAWINGS_CURRENT": np.int32,
"SK_DPD": np.int32,
"SK_DPD_DEF": np.int32,
"AMT_BALANCE": np.float32,
"AMT_DRAWINGS_ATM_CURRENT": np.float32,
"AMT_DRAWINGS_CURRENT": np.float32,
"AMT_DRAWINGS_OTHER_CURRENT": np.float32,
"AMT_DRAWINGS_POS_CURRENT": np.float32,
"AMT_INST_MIN_REGULARITY": np.float32,
"AMT_PAYMENT_CURRENT": np.float32,
"AMT_PAYMENT_TOTAL_CURRENT": np.float32,
"AMT_RECEIVABLE_PRINCIPAL": np.float32,
"AMT_RECIVABLE": np.float32,
"AMT_TOTAL_RECEIVABLE": np.float32,
"CNT_DRAWINGS_ATM_CURRENT": np.float32,
"CNT_DRAWINGS_OTHER_CURRENT": np.float32,
"CNT_DRAWINGS_POS_CURRENT": np.float32,
"CNT_INSTALMENT_MATURE_CUM": np.float32,
}
pos_bal = pd.read_csv(
os.path.join(default_dir, "POS_CASH_balance.csv"), dtype=pos_dtype
)
install = pd.read_csv(
os.path.join(default_dir, "installments_payments.csv"), dtype=install_dtype
)
card_bal = pd.read_csv(
os.path.join(default_dir, "credit_card_balance.csv"), dtype=card_dtype
)
return pos_bal, install, card_bal
def get_dataset():
default_dir = "../input/home-credit-default-risk/"
app_train = pd.read_csv(os.path.join(default_dir, "application_train.csv"))
# app_train = reduce_mem_usage(app_train)
app_test = pd.read_csv(os.path.join(default_dir, "application_test.csv"))
# app_test = reduce_mem_usage(app_test)
apps = pd.concat([app_train, app_test])
prev = pd.read_csv(os.path.join(default_dir, "previous_application.csv"))
prev = reduce_mem_usage(prev)
bureau = pd.read_csv(os.path.join(default_dir, "bureau.csv"))
bureau = reduce_mem_usage(bureau)
bureau_bal = pd.read_csv(os.path.join(default_dir, "bureau_balance.csv"))
bureau_bal = reduce_mem_usage(bureau_bal)
pos_bal, install, card_bal = get_balance_data()
return (
app_train,
app_test,
apps,
prev,
bureau,
bureau_bal,
pos_bal,
install,
card_bal,
)
(
app_train,
app_test,
apps,
prev,
bureau,
bureau_bal,
pos_bal,
install,
card_bal,
) = get_dataset()
df_train = app_train.copy()
df_test = app_test.copy()
df_train_test = apps.copy()
base_bureau_up = bureau.copy()
base_bureau_balance_up = bureau_bal.copy()
base_previous_application_up = prev.copy()
base_POS_CASH_balance_up = pos_bal.copy()
base_credit_card_balance_up = card_bal.copy()
base_installments_payments_up = install.copy()
## function for checking the data shape
def check_df(dfs):
for i, (df, name) in enumerate(dfs):
print(f"{name}: {df.shape}")
dfs = [
(df_train_test, "df_train_test"),
(base_bureau_up, "base_bureau_up"),
(base_previous_application_up, "base_previous_application_up"),
(base_POS_CASH_balance_up, "base_POS_CASH_balance_up"),
(base_bureau_balance_up, "base_bureau_balance_up"),
(base_credit_card_balance_up, "base_credit_card_balance_up"),
(base_installments_payments_up, "base_installments_payments_up"),
]
check_df(dfs)
dfs = [
(df_train_test, "df_train_test"),
(base_bureau_up, "base_bureau_up"),
(base_bureau_balance_up, "base_bureau_balance_up"),
(base_previous_application_up, "base_previous_application_up"),
(base_POS_CASH_balance_up, "base_POS_CASH_balance_up"),
(base_credit_card_balance_up, "base_credit_card_balance_up"),
(base_installments_payments_up, "base_installments_payments_up"),
]
def check_head(dfs):
for df, name in dfs:
print(name)
display(df.head(3))
check_head(dfs)
# **2. Marge all data**
# ###### TARGET value, it has two unique values. 0 means loan is repayed, value 1 means loan is not repayed
print(df_train["TARGET"].nunique())
def missing_values_table(df):
if isinstance(df, pd.Series):
df = pd.DataFrame(df)
missing_values_dict = {}
na_columns = df.columns[df.isnull().any()].tolist()
n_miss = df[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (n_miss / df.shape[0] * 100).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["Missing Values", "Percentage"]
)
if isinstance(df, pd.Series):
missing_values_dict[df.name] = missing_df
else:
missing_values_dict["DataFrame"] = missing_df
return missing_values_dict
missing_values_table(df_train_test["TARGET"])
# ##### Those target values are missing because, I have marge the tran and test dataset
df_train["TARGET"].astype(int).plot.hist(color="g")
# ###### First I have marge dureau and bureau balance, which easy and self expalanatory. First defines a function called First I defines a function called extract_mea which groups the dataframe by the SK_ID_BUREAU column and calculates the mean of all numerical columns. The resulting dataframe is then given column names with the prefix 'BUR_BAL_MEAN_' and returned.
# ###### The problem of this method is, The extract_mean function only calculates the mean of umerical columns and ignores any non-numerical columns, such as categorical columns. Therefore, the esulting bureau_bal_mean dataframe will only contain the mean values of numerical columns.
# Define function to extract mean values
def extract_mean(x):
y = x.groupby("SK_ID_BUREAU", as_index=False).mean().add_prefix("BUR_BAL_MEAN_")
return y
# Apply function to create bureau_balance dataframe grouped by SK_ID_BUREAU with mean values of all numerical columns
bureau_bal_mean = extract_mean(base_bureau_balance_up)
bureau_bal_mean = bureau_bal_mean.rename(
columns={"BUR_BAL_MEAN_SK_ID_BUREAU": "SK_ID_BUREAU"}
)
bureau_bal_mean.shape
base_bureau_up = base_bureau_up.merge(bureau_bal_mean, on="SK_ID_BUREAU", how="left")
base_bureau_up.drop("SK_ID_BUREAU", axis=1, inplace=True)
def extract_mean_curr(x):
y = (
x.groupby("SK_ID_CURR", as_index=False).mean().add_prefix("PREV_BUR_MEAN_")
) # note that we have changed the ID to group by and the prefix to add
return y
# Apply function to create bureau dataframe grouped by SK_ID_CURR with mean values of all numerical columns
bureau_mean_values = extract_mean_curr(base_bureau_up)
bureau_mean_values = bureau_mean_values.rename(
columns={"PREV_BUR_MEAN_SK_ID_CURR": "SK_ID_CURR"}
)
bureau_mean_values.shape
bureau_mean_values.head(3)
# ------------------
base_credit_card_balance_up.drop("SK_ID_CURR", axis=1, inplace=True)
base_installments_payments_up.drop("SK_ID_CURR", axis=1, inplace=True)
base_POS_CASH_balance_up.drop("SK_ID_CURR", axis=1, inplace=True)
# Group previous_application by SK_ID_CURR and count number of unique SK_ID_PREV values
previous_application_counts = (
base_previous_application_up.groupby("SK_ID_CURR", as_index=False)["SK_ID_PREV"]
.count()
.rename(columns={"SK_ID_PREV": "PREVIOUS_APPLICATION_COUNT"})
)
# ----------------
def extract_mean(x):
y = x.groupby("SK_ID_PREV", as_index=False).mean().add_prefix("POS_MEAN_")
return y
POS_mean = extract_mean(base_POS_CASH_balance_up)
POS_mean = POS_mean.rename(columns={"POS_MEAN_SK_ID_PREV": "SK_ID_PREV"})
base_previous_application_up = base_previous_application_up.merge(
POS_mean, on="SK_ID_PREV", how="left"
)
# -------------------------
def extract_mean(x):
y = x.groupby("SK_ID_PREV", as_index=False).mean().add_prefix("CARD_MEAN_")
return y
credit_card_balance_mean = extract_mean(base_credit_card_balance_up)
credit_card_balance_mean = credit_card_balance_mean.rename(
columns={"CARD_MEAN_SK_ID_PREV": "SK_ID_PREV"}
)
base_previous_application_up = base_previous_application_up.merge(
credit_card_balance_mean, on="SK_ID_PREV", how="left"
)
# ---------------------------
def extract_mean(x):
y = x.groupby("SK_ID_PREV", as_index=False).mean().add_prefix("INSTALL_MEAN_")
return y
install_pay_mean = extract_mean(base_installments_payments_up)
install_pay_mean = install_pay_mean.rename(
columns={"INSTALL_MEAN_SK_ID_PREV": "SK_ID_PREV"}
)
base_previous_application_up = base_previous_application_up.merge(
install_pay_mean, on="SK_ID_PREV", how="left"
)
# ---------------------------
def extract_mean(x):
y = x.groupby("SK_ID_CURR", as_index=False).mean().add_prefix("PREV_APPL_MEAN_")
return y
prev_appl_mean = extract_mean(base_previous_application_up)
prev_appl_mean = prev_appl_mean.rename(
columns={"PREV_APPL_MEAN_SK_ID_CURR": "SK_ID_CURR"}
)
prev_appl_mean = prev_appl_mean.drop("PREV_APPL_MEAN_SK_ID_PREV", axis=1)
# ---------------------
# Merge preprocessed_data and df_bureau_up
df1 = pd.merge(df_train_test, bureau_mean_values, on="SK_ID_CURR", how="left")
# # # Merge df1 and df_previous_application_up
df2 = pd.merge(df1, previous_application_counts, on="SK_ID_CURR", how="left")
# # # Merge df2 and df_bureau_balance_up
df3 = pd.merge(df2, prev_appl_mean, on="SK_ID_CURR", how="left")
# ...
df3.head(3)
# **3.Exploratory data analysis**
# ###### This correlations shows the most positive and negative correlations between the TARGET variable and other variables in the dataset. The correlations are calculated using the Pearson correlation coefficient, which measures the linear relationship between two variables.
# ###### The output shows the top 15 variables with the highest positive correlation with TARGET ,sorted in descending order. The variable with the highest positive correlation is PREV_APPL_MEAN_CARD_MEAN_AMT_DRAWINGS_CURRENT, with a correlation coefficient of 0.055388. The output also shows the variable with the highest negative correlation, which is EXT_SOURCE_3 with a correlation coefficient of -0.178919.
# ###### The variables with the highest positive correlation with TARGET may be useful in predicting loan defaults, while the variables with the highest negative correlation may be useful in predicting loan repayments.
# Find correlations with the target and sort
numeric_cols = df3.select_dtypes(include=[np.number]).columns
correlations = df3[numeric_cols].corr()["TARGET"].sort_values()
# Display correlations
print("Most Positive Correlations:\n", correlations.tail(15))
print("\nMost Negative Correlations:\n", correlations.head(15))
# **3.1 Identify the categorical variable**
def identify_columns(df, threshold=25):
# Identify categorical object columns, categorical numerical columns, and non-categorical columns
cat_object_list = [
i for i in df.columns if df[i].dtype == "object" and df[i].nunique() < threshold
]
cat_num_list = [
i
for i in df.columns
if df[i].dtype in ["int64", "float64"] and df[i].nunique() < threshold
]
non_cat_list = [
i for i in df.columns if i not in cat_object_list and i not in cat_num_list
]
# Identify object columns and numerical columns in non-categorical columns
mix_serise_col = df[non_cat_list]
non_cat_obj = [
i for i in mix_serise_col.columns if mix_serise_col[i].dtype == "object"
]
non_cat_num = [
i
for i in mix_serise_col.columns
if mix_serise_col[i].dtype in ["int64", "float64"]
]
# #Print the results
# print('Categorical object columns:', len(cat_object_list))
# print('Categorical numerical columns:', len(cat_num_list))
# print('Non-categorical columns:', len(non_cat_list))
# print('Object columns in non-categorical columns:', len(non_cat_obj))
# print('Numerical columns in non-categorical columns:', len(non_cat_num))
# Return the results as a dictionary
results = {
"cat_object_list": cat_object_list,
"cat_num_list": cat_num_list,
"non_cat_list": non_cat_list,
"non_cat_obj": non_cat_obj,
"non_cat_num": non_cat_num,
}
return results
results = identify_columns(df_train_test)
categorical_num = identify_columns(df3)["cat_num_list"]
categorical_obj = identify_columns(df3)["cat_object_list"]
non_categorical_obj = identify_columns(df3)["non_cat_obj"]
print(categorical_obj)
print(categorical_num)
def plot_categorical_feature(feature, df=None, orientation_horizontal=True):
if df is None:
df = df3
else:
df = df
temp = df[feature].value_counts()
df1 = pd.DataFrame({feature: temp.index, "Number of contracts": temp.values})
# Calculate the percentage of target=1 per category value
cat_perc = df[[feature, "TARGET"]].groupby([feature], as_index=False).mean()
cat_perc.sort_values(by="TARGET", ascending=False, inplace=True)
sns.set_color_codes("colorblind")
if orientation_horizontal == True:
plt.figure(figsize=(15, 5))
plt.subplot(121)
s1 = sns.barplot(y=feature, x="Number of contracts", data=df1)
plt.subplot(122)
s2 = sns.barplot(y=feature, x="TARGET", data=cat_perc)
plt.xlabel("Fraction of loans defaulted", fontsize=12)
plt.ylabel(feature, fontsize=12)
else:
plt.figure(figsize=(10, 12))
plt.subplot(211)
s1 = sns.barplot(x=feature, y="Number of contracts", data=df1)
s1.set_xticklabels(s1.get_xticklabels(), rotation=90)
plt.subplot(212)
s2 = sns.barplot(x=feature, y="TARGET", data=cat_perc)
s2.set_xticklabels(s2.get_xticklabels(), rotation=90)
plt.ylabel("Fraction of loans defaulted", fontsize=12)
plt.xlabel(feature, fontsize=12)
plt.tick_params(axis="both", which="major", labelsize=12)
plt.subplots_adjust(wspace=0.6)
plt.show()
# ###### This suggests that the default rate for cash loans is higher than that for revolving loans. the grough also suggests that feamal has high rate of default rate and so on.
# Concatenate the two lists
features = categorical_obj + non_categorical_obj
# Loop over the features and plot each one
for feature in features:
plot_categorical_feature(feature)
# **plot_bivariate_distribution**
def plot_bivariate_distribution(feature, df):
if df is None:
df = df3
else:
df = df
plt.figure(figsize=(10, 4))
sns.kdeplot(df.loc[df["TARGET"] == 0, feature], label="TARGET == 0")
sns.kdeplot(df.loc[df["TARGET"] == 1, feature], label="TARGET == 1")
plt.xlabel(feature)
plt.ylabel("Density")
plt.title("Distribution of {} by Target Value".format(feature))
plt.legend()
plot_bivariate_distribution("EXT_SOURCE_1", df_train)
plot_bivariate_distribution("EXT_SOURCE_2", df3)
plot_bivariate_distribution("AMT_INCOME_TOTAL", df3)
plot_bivariate_distribution("DAYS_EMPLOYED", df3)
plot_bivariate_distribution("DAYS_BIRTH", df3)
# **4.1 Feature engineering**
# ###### I have changed the negitive values of days to years.
# Converting days col into a year
def create_day_to_year(df, ls_cols):
for col in ls_cols:
if col.startswith("DAYS") and np.issubdtype(df[col].dtype, np.number):
new_col = col.replace("DAYS", "YEARS_UP")
df[new_col] = round(np.abs(df[col] / 365))
df.drop(columns=[col], inplace=True)
return df
df_list = [df3]
for df in df_list:
day_cols = df.filter(like="DAYS").columns
df = create_day_to_year(df, day_cols)
def print_years_cols(dfs):
for df, name in dfs:
day_cols = df.filter(like="YEARS_UP").columns
print(f"Number of columns with 'YEARS' in {name}: {len(day_cols)}")
print(f"Columns with 'YEARS' in {name}:")
for col in day_cols:
print(col)
print_years_cols([(df3, "df_train")])
# ###### I have computed the age of each loan applicant. The following graph indicates that individuals between the ages of 25 and 30 are more likely to default on their home loans.
plot_bivariate_distribution("YEARS_UP_BIRTH", df3)
# ###### A column called "income band" has been created, which segments income into different categories. For example, an income range of 0 to 30k is assigned to category 1, while an income range of 30k to 65k is assigned to category 2, and so on.
# Converting income band;
def create_income_band(df):
bins = [
0,
30000,
65000,
95000,
130000,
160000,
190000,
220000,
275000,
325000,
np.inf,
]
labels = range(1, len(bins))
df["INCOME_BAND"] = pd.cut(df["AMT_INCOME_TOTAL"], bins=bins, labels=labels)
return df
dfs = [(df3, "data_set")]
for df, name in dfs:
df = create_income_band(df)
fig, ax = plt.subplots(figsize=(5, 3))
sns.countplot(data=df3, x="INCOME_BAND", hue="TARGET")
ax.set_title("Income data for people repaying and defaulting loans")
ax.set_ylabel("Count")
plt.tight_layout()
plt.show()
# ---
# **4.2 Encoding and missing value handeling**
# ###### I have applied label encoding to columns with two categorical values, and one-hot encoding to columns with more than two categorical values. To handle missing values, I have implemented a condition where if a column has more than 60% missing values, it will be deleted. Otherwise, I have used SimpleImputer to replace missing values with the median value of the scale
from sklearn.preprocessing import LabelEncoder
from sklearn.impute import SimpleImputer
def label_encoding(df):
for col in df.columns:
if df[col].dtype == "object":
if len(df[col].unique()) == 2:
le = LabelEncoder()
le.fit(df[col])
df[col] = le.transform(df[col])
# else:
# df[col] = pd.get_dummies(df[col])
return df
def missing_values_table(df):
# Check if input is a dataframe or a series
if isinstance(df, pd.Series):
df = pd.DataFrame(df)
# Get columns with missing values
na_columns = df.columns[df.isnull().any()].tolist()
# Count missing values and calculate ratio
n_miss = df[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (n_miss / df.shape[0] * 100).sort_values(ascending=False)
# Create DataFrame with missing values and ratio
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["Missing Values", "Percentage"]
)
return missing_df
def missing_preprocess_data(df):
# Identify columns with more than 60% missing values
missing_cols = df.columns[df.isnull().mean() > 0.6]
# Drop columns with more than 60% missing values
num_cols_dropped = len(missing_cols)
df.drop(columns=missing_cols, inplace=True)
print(f"Dropped {num_cols_dropped} columns due to missing value threshold")
# Fill remaining missing values using median imputation
imputer = SimpleImputer(strategy="median")
df = pd.DataFrame(imputer.fit_transform(df), columns=df.columns)
# Print out the original and preprocessed datasets
print("Preprocessed dataset:")
print(df)
return df
base_df3 = df3.copy()
label_encoding(base_df3)
base_df3 = pd.get_dummies(base_df3)
base_df3.shape
preprocessed_data = missing_preprocess_data(base_df3)
missing_values_table(preprocessed_data)
# **5.Classifier models**
# ###### LogisticRegression
process_df_logis = preprocessed_data.copy()
# Basic Logistic regrassion
# Split the data into training and testing sets
train_X, test_X, train_Y, test_Y = train_test_split(
preprocessed_data.drop(["SK_ID_CURR", "TARGET"], axis=1),
preprocessed_data["TARGET"],
test_size=0.25,
random_state=123,
)
# train_X contains the independent variables for the training set
# test_X contains the independent variables for the testing set
# train_Y contains the dependent variable (TARGET) for the training set
# test_Y contains the dependent variable (TARGET) for the testing set
def train_predict_visualize(train_X, train_Y, test_X, test_Y):
# Train a logistic regression classifier
clf = LogisticRegression(random_state=123)
clf.fit(train_X, train_Y)
# Predict using test data
pred_Y = clf.predict(test_X)
pred_prob_Y = clf.predict_proba(test_X)[:, 1]
# Calculate metrics
auc = roc_auc_score(test_Y, pred_prob_Y)
tn, fp, fn, tp = confusion_matrix(test_Y, pred_Y).ravel()
accuracy = (tp + tn) / (tp + tn + fp + fn)
precision = tp / (tp + fp)
recall = tp / (tp + fn)
f1_score = 2 * precision * recall / (precision + recall)
# Print metrics
print("AUC:", auc)
print("Accuracy:", accuracy)
print("Precision:", precision)
print("Recall:", recall)
print("F1 Score:", f1_score)
# Visualize feature importances
coef = pd.Series(clf.coef_[0], index=train_X.columns)
coef.nlargest(10).plot(kind="barh")
plt.show()
def train_predict_visualize(train_X, train_Y, test_X, test_Y):
# Train a logistic regression classifier
clf = LogisticRegression(random_state=123)
clf.fit(train_X, train_Y)
# Predict using test data
pred_Y = clf.predict(test_X)
pred_prob_Y = clf.predict_proba(test_X)[:, 1]
# Calculate metrics
auc = roc_auc_score(test_Y, pred_prob_Y)
tn, fp, fn, tp = confusion_matrix(test_Y, pred_Y).ravel()
accuracy = (tp + tn) / (tp + tn + fp + fn)
precision = tp / (tp + fp)
recall = tp / (tp + fn)
f1_score = 2 * precision * recall / (precision + recall)
# Print metrics
print("AUC:", auc)
print("Accuracy:", accuracy)
print("Precision:", precision)
print("Recall:", recall)
print("F1 Score:", f1_score)
# Visualize feature importances
coef = pd.Series(clf.coef_[0], index=train_X.columns)
top_features = coef.nlargest(10)
top_features.plot(kind="barh")
plt.show()
# Print table of top 10 features
print("Top 10 Features:")
print(top_features.to_string())
train_predict_visualize(train_X, train_Y, test_X, test_Y)
# #### Explanation of the model evaluation:
# ###### The result of the model evaluation shows that the AUC (Area Under the Curve) is 0.618, which indicates that the model's ability to distinguish between positive and negative classes is slightly better than random guessing. The accuracy score is 0.929, which indicates that the model is able to correctly predict the target variable for 92.9% of the observations.
# ###### However, the precision score is 0.0, which means that the model did not correctly identify any true positives. The recall score is also 0.0, which means that the model did not correctly identify any positive cases. The F1 score is "nan" (not a number), which is likely due to the precision and recall scores being 0.
# ###### Overall, these results suggest that the model may not be effective in predicting the target variable, and further analysis and model tuning may be necessary to improve its performance.
# ##### Domain Knowledge feature engeening
preprocessed_data.shape
# Domain knowledge
# Debt-to-income-ratio
preprocessed_data["CREDIT_INCOME_PERCENT"] = (
preprocessed_data["AMT_CREDIT"] / preprocessed_data["AMT_INCOME_TOTAL"]
)
# Loan-to-Value-ration
preprocessed_data["LOAN_TO_VALUE_RATION"] = (
preprocessed_data["AMT_CREDIT"] / preprocessed_data["AMT_GOODS_PRICE"]
)
# percentage of the applicant's income that is being used to pay off the loan.
preprocessed_data["ANNUITY_INCOME_PERCENT"] = (
preprocessed_data["AMT_ANNUITY"] / preprocessed_data["AMT_INCOME_TOTAL"]
)
# This ratio represents the length of time it will take the applicant to pay off the loan.
preprocessed_data["CREDIT_TERM"] = (
preprocessed_data["AMT_ANNUITY"] / preprocessed_data["AMT_CREDIT"]
)
# Employment history percentage
preprocessed_data["YEARS_EMPLOYED_PERCENT"] = (
preprocessed_data["YEARS_UP_EMPLOYED"] / preprocessed_data["YEARS_UP_BIRTH"]
) # age
# --------
##Late payment
# # Identify the relevant columns for payment history
# payment_history_cols = ['SK_ID_CURR', 'SK_ID_PREV', 'DAYS_ENTRY_PAYMENT']
# # Calculate the number of late payments on previous loans
# previous_application_up['LATE_PAYMENT'] = (previous_application_up['DAYS_ENTRY_PAYMENT'] > 0).astype(int)
# late_payment_counts = previous_application.groupby('SK_ID_CURR')['LATE_PAYMENT'].sum()
# # Merge the late payment counts into the application_train_up dataset
# application_train_up = application_train_up.merge(late_payment_counts, on='SK_ID_CURR', how='left')
# #### Poly feature engineering
from sklearn.preprocessing import PolynomialFeatures
# Define the columns to include in the polynomial features
poly_cols = [
"AMT_INCOME_TOTAL",
"AMT_CREDIT",
"AMT_ANNUITY",
"YEARS_UP_BIRTH",
"YEARS_UP_EMPLOYED",
]
# Check if all columns exist in the dataframe
if all(col in preprocessed_data.columns for col in poly_cols):
# Create polynomial features
poly = PolynomialFeatures(degree=2, include_bias=False)
poly_features = poly.fit_transform(preprocessed_data[poly_cols])
poly_feature_names = poly.get_feature_names_out(poly_cols)
poly_df = pd.DataFrame(poly_features, columns=poly_feature_names)
# Merge the polynomial features with the original dataframe
preprocessed_data = pd.concat([preprocessed_data, poly_df], axis=1)
else:
print("One or more columns not found in dataframe")
preprocessed_data.shape
# Check for duplicated column names
duplicated_cols = preprocessed_data.columns[preprocessed_data.columns.duplicated()]
# Print the duplicated column names
print("Duplicated columns:", duplicated_cols)
# Remove duplicated columns if any
if len(duplicated_cols) > 0:
preprocessed_data = preprocessed_data.loc[
:, ~preprocessed_data.columns.duplicated()
]
print("Duplicated columns removed")
# ---
# ##### XGBClassifier
process_df_xgb = preprocessed_data.copy()
import pandas as pd
import xgboost as xgb
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
)
from sklearn.model_selection import train_test_split
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
process_df_xgb.drop(["SK_ID_CURR", "TARGET"], axis=1),
process_df_xgb["TARGET"],
test_size=0.2,
random_state=42,
)
# Train an XGBClassifier
xgb_model = xgb.XGBClassifier(n_estimators=100, random_state=42)
xgb_model.fit(X_train, y_train)
# Get the feature importances
importances = xgb_model.get_booster().get_score(importance_type="gain")
importances = {
feature: importance
for feature, importance in sorted(
importances.items(), key=lambda x: x[1], reverse=True
)
}
# Create a dataframe with the feature importances
feature_importances = pd.DataFrame(
{"feature": list(importances.keys()), "importance": list(importances.values())}
)
# Select the top 10 features
top_features = feature_importances.head(10)
# # Print the top 10 features
# print(top_features)
# Make predictions on the test set
y_pred_prob = xgb_model.predict_proba(X_test)[:, 1]
y_pred = (y_pred_prob > 0.5).astype(int)
# Evaluate the performance of the model
accuracy = accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred)
recall = recall_score(y_test, y_pred)
f1 = f1_score(y_test, y_pred)
auc = roc_auc_score(y_test, y_pred_prob)
print("Accuracy:", accuracy)
print("Precision:", precision)
print("Recall:", recall)
print("F1 score:", f1)
print("AUC score:", auc)
# Make predictions on the test data
test_pred_prob = xgb_model.predict_proba(X_test)[:, 1]
test_pred = (test_pred_prob > 0.5).astype(int)
X_test["SK_ID_CURR"] = process_df_xgb.loc[X_test.index, "SK_ID_CURR"]
# Create the submission file
submission = pd.DataFrame({"SK_ID_CURR": X_test["SK_ID_CURR"], "TARGET": test_pred})
submission.to_csv("submission.csv", index=False)
# ###### The result of the model evaluation shows that the accuracy score is 0.929, which indicates that the model is able to correctly predict the target variable for 92.9% of the observations. The precision score is 0.463, which means that the model correctly identified 46.3% of the true positives out of all the predicted positives. The recall score is 0.044, which means that the model correctly identified only 4.4% of the true positives out of all the actual positives. The F1 score is 0.080, which is the harmonic mean of precision and recall, and provides an overall measure of the model's performance.
# ###### The AUC (Area Under the Curve) score is 0.773, which indicates that the model's ability to distinguish between positive and negative classes is better than random guessing.
# ###### Overall, these results suggest that the model may have some predictive power, but its performance is not very strong. Further analysis and model tuning may be necessary to improve its performance.
submission["TARGET"].astype(int).plot.hist(color="g")
# Count the number of occurrences of each value in the 'TARGET' column
value_counts = submission["TARGET"].value_counts()
# Print the value counts
print(value_counts)
# **Important feature selaction (XGBClassifier)**
# Create a new dataframe with only the top features
df_top = preprocessed_data[["SK_ID_CURR", "TARGET"] + top_features["feature"].tolist()]
# Save the new dataframe to a CSV file
df_top.to_csv("data_top.csv", index=False)
# Plot the feature importances
plt.figure(figsize=(16, 32))
plt.barh(top_features["feature"], top_features["importance"])
plt.xlabel("Importance")
plt.ylabel("Feature")
plt.title("Feature Importances")
plt.show()
# ##### lgb.LGBMClassifier
process_df_lgb = preprocessed_data.copy()
import pandas as pd
import lightgbm as lgb
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
)
from sklearn.model_selection import train_test_split
# Remove any non-alphanumeric characters from the feature names
process_df_lgb.columns = [
re.sub("[^0-9a-zA-Z]+", "_", col) for col in process_df_lgb.columns
]
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
process_df_lgb.drop(["SK_ID_CURR", "TARGET"], axis=1),
process_df_lgb["TARGET"],
test_size=0.2,
random_state=42,
)
# Train a LightGBM classifier
lgb_model = lgb.LGBMClassifier(n_estimators=100, random_state=42)
lgb_model.fit(X_train, y_train)
# Get the feature importances
importances = lgb_model.feature_importances_
# Create a dataframe with the feature importances
feature_importances = pd.DataFrame(
{"feature": X_train.columns, "importance": importances}
)
# Sort the dataframe by importance
feature_importances = feature_importances.sort_values("importance", ascending=False)
# Select the top 10 features
top_features = feature_importances.head(10)
# Print the top 10 features
print(top_features)
# Make predictions on the test set
y_pred_prob = lgb_model.predict_proba(X_test)[:, 1]
y_pred = (y_pred_prob > 0.5).astype(int)
# Evaluate the performance of the model
accuracy_lgbm = accuracy_score(y_test, y_pred)
precision_lgbm = precision_score(y_test, y_pred)
recall_lgbm = recall_score(y_test, y_pred)
f1_lgbm = f1_score(y_test, y_pred)
auc_lgbm = roc_auc_score(y_test, y_pred_prob)
print("Accuracy:", accuracy)
print("Precision:", precision)
print("Recall:", recall)
print("F1 score:", f1)
print("AUC score:", auc)
# Make predictions on the test data
test_pred_prob = lgb_model.predict_proba(X_test)[:, 1]
test_pred = (test_pred_prob > 0.5).astype(int)
X_test["SK_ID_CURR"] = process_df_lgb.loc[X_test.index, "SK_ID_CURR"]
# Create the submission file
submission1 = pd.DataFrame({"SK_ID_CURR": X_test["SK_ID_CURR"], "TARGET": test_pred})
submission1.to_csv("submission.csv", index=False)
submission1["TARGET"].astype(int).plot.hist(color="g")
# Count the number of occurrences of each value in the 'TARGET' column
value_counts1 = submission1["TARGET"].value_counts()
# Print the value counts
print(value_counts1)
# ##### Comparasion xgb vs lgb
import matplotlib.pyplot as plt
import numpy as np
# Define the evaluation metrics
metrics = ["Accuracy", "Precision", "Recall", "F1 score", "AUC score"]
xgb_scores = [accuracy, precision, recall, f1, auc]
lgbm_scores = [accuracy_lgbm, precision_lgbm, recall_lgbm, f1_lgbm, auc_lgbm]
# Create a bar plot
x = np.arange(len(metrics))
width = 0.35
fig, ax = plt.subplots()
rects1 = ax.bar(x - width / 2, xgb_scores, width, label="XGBClassifier")
rects2 = ax.bar(x + width / 2, lgbm_scores, width, label="LGBMClassifier")
# Add labels and title
ax.set_ylabel("Score")
ax.set_xticks(x)
ax.set_xticklabels(metrics)
ax.legend()
ax.set_title("Comparison of XGBClassifier and LGBMClassifier")
# Add values above the bars
def autolabel(rects):
for rect in rects:
height = rect.get_height()
ax.annotate(
"{:.3f}".format(height),
xy=(rect.get_x() + rect.get_width() / 2, height),
xytext=(0, 3), # 3 points vertical offset
textcoords="offset points",
ha="center",
va="bottom",
)
autolabel(rects1)
autolabel(rects2)
plt.show()
# Create a dictionary to store the scores
scores_dict = {
"Model": ["XGBClassifier", "LGBMClassifier"],
"Accuracy": [xgb_scores[0], lgbm_scores[0]],
"Precision": [xgb_scores[1], lgbm_scores[1]],
"Recall": [xgb_scores[2], lgbm_scores[2]],
"F1 Score": [xgb_scores[3], lgbm_scores[3]],
"AUC Score": [xgb_scores[4], lgbm_scores[4]],
}
# Create a pandas dataframe from the dictionary
scores_df = pd.DataFrame(scores_dict)
# Print the dataframe
print(scores_df)
# ###### Based on the evaluation metrics, it seems that both models are not performing very well. The accuracy scores are relatively high, but the precision, recall, and F1 scores are quite low. This suggests that the models are not doing a good job of correctly identifying the positive class (i.e., clients who are likely to default on their loans).
# ###### The AUC scores are also relatively low, which indicates that the models are not doing a good job of distinguishing between positive and negative cases.
# ###### It may be necessary to try different models or to tune the hyperparameters of the existing models to improve their performance. Additionally, it may be helpful to explore the data further and identify any patterns or relationships that could be used to improve the models.
# ##### K-fold cross validation
process_df_k = preprocessed_data.copy()
import lightgbm as lgb
from sklearn.model_selection import KFold
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
)
# Remove any non-alphanumeric characters from the feature names
process_df_k.columns = [
re.sub("[^0-9a-zA-Z]+", "_", col) for col in process_df_k.columns
]
# Define the number of folds
k = 5
# Define the K-fold cross-validator
kf = KFold(n_splits=k, shuffle=True, random_state=42)
# Initialize empty lists to store the scores for each fold
accuracy_scores_lgbm = []
precision_scores_lgbm = []
recall_scores_lgbm = []
f1_scores_lgbm = []
auc_scores_lgbm = []
# Loop over the folds
for train_index, test_index in kf.split(df_top):
# Split the data into training and test sets
X_train, X_test = (
process_df_k.drop(["SK_ID_CURR", "TARGET"], axis=1).iloc[train_index],
df_top.drop(["SK_ID_CURR", "TARGET"], axis=1).iloc[test_index],
)
y_train, y_test = (
df_top["TARGET"].iloc[train_index],
df_top["TARGET"].iloc[test_index],
)
# Check if the columns in the training and test data are the same
if set(X_train.columns) != set(X_test.columns):
missing_cols = set(X_train.columns) - set(X_test.columns)
for col in missing_cols:
X_test[col] = 0
X_test = X_test[X_train.columns]
# Initialize the LGBMClassifier model
model = lgb.LGBMClassifier(n_estimators=100, random_state=42)
# Fit the model on the training data
model.fit(X_train, y_train)
# Make predictions on the test data
y_pred = model.predict(X_test)
# Calculate the evaluation metrics
accuracy_lgbm = accuracy_score(y_test, y_pred)
precision_lgbm = precision_score(y_test, y_pred)
recall_lgbm = recall_score(y_test, y_pred)
f1_lgbm = f1_score(y_test, y_pred)
auc_lgbm = roc_auc_score(y_test, y_pred)
# Append the scores to the lists
accuracy_scores_lgbm.append(accuracy_lgbm)
precision_scores_lgbm.append(precision_lgbm)
recall_scores_lgbm.append(recall_lgbm)
f1_scores_lgbm.append(f1_lgbm)
auc_scores_lgbm.append(auc_lgbm)
# Calculate the mean and standard deviation of the scores
mean_accuracy = np.mean(accuracy_scores_lgbm)
std_accuracy = np.std(accuracy_scores_lgbm)
mean_precision = np.mean(precision_scores_lgbm)
std_precision = np.std(precision_scores_lgbm)
mean_recall = np.mean(recall_scores_lgbm)
std_recall = np.std(recall_scores_lgbm)
mean_f1 = np.mean(f1_scores_lgbm)
std_f1 = np.std(f1_scores_lgbm)
mean_auc = np.mean(auc_scores_lgbm)
std_auc = np.std(auc_scores_lgbm)
# Print the results
print(
"Accuracy_lgbm: {:.2f}% (+/- {:.2f}%)".format(
mean_accuracy * 100, std_accuracy * 100
)
)
print(
"Precision_lgbm: {:.2f}% (+/- {:.2f}%)".format(
mean_precision * 100, std_precision * 100
)
)
print("Recall_lgbm: {:.2f}% (+/- {:.2f}%)".format(mean_recall * 100, std_recall * 100))
print("F1 score_lgbm: {:.2f}% (+/- {:.2f}%)".format(mean_f1 * 100, std_f1 * 100))
print("AUC score_lgbm: {:.2f}% (+/- {:.2f}%)".format(mean_auc * 100, std_auc * 100))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/119/129119687.ipynb
| null | null |
[{"Id": 129119687, "ScriptId": 38342411, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11858828, "CreationDate": "05/11/2023 06:40:31", "VersionNumber": 4.0, "Title": "Credit default ML project -- Base model vs ML M", "EvaluationDate": "05/11/2023", "IsChange": false, "TotalLines": 1027.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1027.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ---
# Introduction:
# The ability of a borrower to repay a loan is crucial information for lenders to make informed decisions about their company's growth forecast. For new homeowners with no credit history, lenders need to determine whether they will be able to repay the loan in the future. Therefore, it is essential to know whether the borrower will be able to pay the loan or default in the future.
# In this project, we will be using data provided by the Home Credit Group, which is published on Kaggle (https://www.kaggle.com/c/home-credit-default-risk/overview). The goal is to predict whether a borrower will be able to repay their existing loan, which is a binary classification problem. We will be using input features such as the financial and behavioral history of the loan applicants to make our predictions.
# Datasets:
# The Kaggle website provides a comprehensive data description that outlines the relationship between the different data points.
# 
# As the dataset was too large, consisting of 7 CSV files totaling over 30GB, I had to shrink the dataset by grouping the data by ID and taking the mean of each ID. This reduced the number of rows in the dataset, as there were multiple rows for each ID in the dataset except for the application file.
#
# Project Overview:
# I used tree-based machine learning algorithms to predict the target variable, achieving an AUC score of around 78% without hyperparameter tuning.
# The AUC score is useful because it provides a single number that summarizes the overall performance of the classifier across all possible threshold settings. It is also useful when the class distribution is imbalanced, as it is less affected by the imbalance than other metrics such as accuracy.
# I used a logistic regression model as the base level model, and two boosting methods, XGBClassifier and LGBMClassifier. LGB has the better accuracy rate of around 78% . I also performed K-fold cross-validation to select the best model and improve the accuracy rate, but the accuracy rate of the LGBMClassifier reduced to around 61%, which did not improve the result.
# To achieve better results, I believe I need to perform more extensive feature analysis with domain knowledge and analyze model tuning. This project was a great exercise for me to learn how to process big data, learn PyScaler SQL, learn feature engineering, and optimize data.
# Structure of the project:
# This analysis is divided into six sections:
# Data imputation and size reduction: To minimize the size of the large dataset, I changed the data type before importing it. I also reduced the size of the data by taking the mean of each row, which reduced the number of rows in the dataset.
# Merging the dataset: Since the dataset was large, I merged the data by following a chart and taking the mean of each row.
# Exploratory data analysis: I visualized each feature and compared it to the target variable to gain insights into the types of people who normally default.
# Feature engineering: I used domain knowledge to engineer five different features, including debt-to-income ratio, loan-to-value ratio, percentage of the applicant's income used to pay off the loan, the length of time to pay off the loan, and employment history percentage. I also used polynomial feature engineering.
# Classifier models: I split the data into training and test sets and trained models of similar size for different modeling processes.
# Tuning: Finally, I split the training data into k equal parts and trained the model on the remaining k-1 folds, evaluating it on the current fold. I also tried hyperparameter tuning.
# Import required libraries/packages
import warnings
warnings.filterwarnings("ignore")
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from timeit import default_timer as timer
import os
import random
import csv
import json
import itertools
import pprint
# from pydash import at
import gc
import re
# import featuretools for automated feature engineering
import featuretools as ft
from featuretools import selection
# Import sklearn helper metrics and transformations
from sklearn.base import TransformerMixin
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split, KFold
from sklearn.utils import resample
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
precision_score,
recall_score,
roc_auc_score,
classification_report,
roc_curve,
auc,
f1_score,
)
# Import models
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.naive_bayes import GaussianNB
from xgboost import XGBClassifier
from sklearn.ensemble import GradientBoostingClassifier
import lightgbm as lgb
# import library for hyperparameter optimization
from hyperopt import STATUS_OK
from hyperopt import hp, tpe, Trials, fmin
from hyperopt.pyll.stochastic import sample
# **1. Data imputation and size reduction:**
import os
def reduce_mem_usage(df):
numerics = ["int16", "int32", "int64", "float16", "float32", "float64"]
for col in df.columns:
col_type = df[col].dtypes
if col_type in numerics:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == "int":
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int8)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
else:
if (
c_min > np.finfo(np.float16).min
and c_max < np.finfo(np.float16).max
):
df[col] = df[col].astype(np.float16)
elif (
c_min > np.finfo(np.float32).min
and c_max < np.finfo(np.float32).max
):
df[col] = df[col].astype(np.float32)
else:
df[col] = df[col].astype(np.float64)
return df
def get_balance_data():
default_dir = "../input/home-credit-default-risk/"
pos_dtype = {
"SK_ID_PREV": np.uint32,
"SK_ID_CURR": np.uint32,
"MONTHS_BALANCE": np.int32,
"SK_DPD": np.int32,
"SK_DPD_DEF": np.int32,
"CNT_INSTALMENT": np.float32,
"CNT_INSTALMENT_FUTURE": np.float32,
}
install_dtype = {
"SK_ID_PREV": np.uint32,
"SK_ID_CURR": np.uint32,
"NUM_INSTALMENT_NUMBER": np.int32,
"NUM_INSTALMENT_VERSION": np.float32,
"DAYS_INSTALMENT": np.float32,
"DAYS_ENTRY_PAYMENT": np.float32,
"AMT_INSTALMENT": np.float32,
"AMT_PAYMENT": np.float32,
}
card_dtype = {
"SK_ID_PREV": np.uint32,
"SK_ID_CURR": np.uint32,
"MONTHS_BALANCE": np.int16,
"AMT_CREDIT_LIMIT_ACTUAL": np.int32,
"CNT_DRAWINGS_CURRENT": np.int32,
"SK_DPD": np.int32,
"SK_DPD_DEF": np.int32,
"AMT_BALANCE": np.float32,
"AMT_DRAWINGS_ATM_CURRENT": np.float32,
"AMT_DRAWINGS_CURRENT": np.float32,
"AMT_DRAWINGS_OTHER_CURRENT": np.float32,
"AMT_DRAWINGS_POS_CURRENT": np.float32,
"AMT_INST_MIN_REGULARITY": np.float32,
"AMT_PAYMENT_CURRENT": np.float32,
"AMT_PAYMENT_TOTAL_CURRENT": np.float32,
"AMT_RECEIVABLE_PRINCIPAL": np.float32,
"AMT_RECIVABLE": np.float32,
"AMT_TOTAL_RECEIVABLE": np.float32,
"CNT_DRAWINGS_ATM_CURRENT": np.float32,
"CNT_DRAWINGS_OTHER_CURRENT": np.float32,
"CNT_DRAWINGS_POS_CURRENT": np.float32,
"CNT_INSTALMENT_MATURE_CUM": np.float32,
}
pos_bal = pd.read_csv(
os.path.join(default_dir, "POS_CASH_balance.csv"), dtype=pos_dtype
)
install = pd.read_csv(
os.path.join(default_dir, "installments_payments.csv"), dtype=install_dtype
)
card_bal = pd.read_csv(
os.path.join(default_dir, "credit_card_balance.csv"), dtype=card_dtype
)
return pos_bal, install, card_bal
def get_dataset():
default_dir = "../input/home-credit-default-risk/"
app_train = pd.read_csv(os.path.join(default_dir, "application_train.csv"))
# app_train = reduce_mem_usage(app_train)
app_test = pd.read_csv(os.path.join(default_dir, "application_test.csv"))
# app_test = reduce_mem_usage(app_test)
apps = pd.concat([app_train, app_test])
prev = pd.read_csv(os.path.join(default_dir, "previous_application.csv"))
prev = reduce_mem_usage(prev)
bureau = pd.read_csv(os.path.join(default_dir, "bureau.csv"))
bureau = reduce_mem_usage(bureau)
bureau_bal = pd.read_csv(os.path.join(default_dir, "bureau_balance.csv"))
bureau_bal = reduce_mem_usage(bureau_bal)
pos_bal, install, card_bal = get_balance_data()
return (
app_train,
app_test,
apps,
prev,
bureau,
bureau_bal,
pos_bal,
install,
card_bal,
)
(
app_train,
app_test,
apps,
prev,
bureau,
bureau_bal,
pos_bal,
install,
card_bal,
) = get_dataset()
df_train = app_train.copy()
df_test = app_test.copy()
df_train_test = apps.copy()
base_bureau_up = bureau.copy()
base_bureau_balance_up = bureau_bal.copy()
base_previous_application_up = prev.copy()
base_POS_CASH_balance_up = pos_bal.copy()
base_credit_card_balance_up = card_bal.copy()
base_installments_payments_up = install.copy()
## function for checking the data shape
def check_df(dfs):
for i, (df, name) in enumerate(dfs):
print(f"{name}: {df.shape}")
dfs = [
(df_train_test, "df_train_test"),
(base_bureau_up, "base_bureau_up"),
(base_previous_application_up, "base_previous_application_up"),
(base_POS_CASH_balance_up, "base_POS_CASH_balance_up"),
(base_bureau_balance_up, "base_bureau_balance_up"),
(base_credit_card_balance_up, "base_credit_card_balance_up"),
(base_installments_payments_up, "base_installments_payments_up"),
]
check_df(dfs)
dfs = [
(df_train_test, "df_train_test"),
(base_bureau_up, "base_bureau_up"),
(base_bureau_balance_up, "base_bureau_balance_up"),
(base_previous_application_up, "base_previous_application_up"),
(base_POS_CASH_balance_up, "base_POS_CASH_balance_up"),
(base_credit_card_balance_up, "base_credit_card_balance_up"),
(base_installments_payments_up, "base_installments_payments_up"),
]
def check_head(dfs):
for df, name in dfs:
print(name)
display(df.head(3))
check_head(dfs)
# **2. Marge all data**
# ###### TARGET value, it has two unique values. 0 means loan is repayed, value 1 means loan is not repayed
print(df_train["TARGET"].nunique())
def missing_values_table(df):
if isinstance(df, pd.Series):
df = pd.DataFrame(df)
missing_values_dict = {}
na_columns = df.columns[df.isnull().any()].tolist()
n_miss = df[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (n_miss / df.shape[0] * 100).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["Missing Values", "Percentage"]
)
if isinstance(df, pd.Series):
missing_values_dict[df.name] = missing_df
else:
missing_values_dict["DataFrame"] = missing_df
return missing_values_dict
missing_values_table(df_train_test["TARGET"])
# ##### Those target values are missing because, I have marge the tran and test dataset
df_train["TARGET"].astype(int).plot.hist(color="g")
# ###### First I have marge dureau and bureau balance, which easy and self expalanatory. First defines a function called First I defines a function called extract_mea which groups the dataframe by the SK_ID_BUREAU column and calculates the mean of all numerical columns. The resulting dataframe is then given column names with the prefix 'BUR_BAL_MEAN_' and returned.
# ###### The problem of this method is, The extract_mean function only calculates the mean of umerical columns and ignores any non-numerical columns, such as categorical columns. Therefore, the esulting bureau_bal_mean dataframe will only contain the mean values of numerical columns.
# Define function to extract mean values
def extract_mean(x):
y = x.groupby("SK_ID_BUREAU", as_index=False).mean().add_prefix("BUR_BAL_MEAN_")
return y
# Apply function to create bureau_balance dataframe grouped by SK_ID_BUREAU with mean values of all numerical columns
bureau_bal_mean = extract_mean(base_bureau_balance_up)
bureau_bal_mean = bureau_bal_mean.rename(
columns={"BUR_BAL_MEAN_SK_ID_BUREAU": "SK_ID_BUREAU"}
)
bureau_bal_mean.shape
base_bureau_up = base_bureau_up.merge(bureau_bal_mean, on="SK_ID_BUREAU", how="left")
base_bureau_up.drop("SK_ID_BUREAU", axis=1, inplace=True)
def extract_mean_curr(x):
y = (
x.groupby("SK_ID_CURR", as_index=False).mean().add_prefix("PREV_BUR_MEAN_")
) # note that we have changed the ID to group by and the prefix to add
return y
# Apply function to create bureau dataframe grouped by SK_ID_CURR with mean values of all numerical columns
bureau_mean_values = extract_mean_curr(base_bureau_up)
bureau_mean_values = bureau_mean_values.rename(
columns={"PREV_BUR_MEAN_SK_ID_CURR": "SK_ID_CURR"}
)
bureau_mean_values.shape
bureau_mean_values.head(3)
# ------------------
base_credit_card_balance_up.drop("SK_ID_CURR", axis=1, inplace=True)
base_installments_payments_up.drop("SK_ID_CURR", axis=1, inplace=True)
base_POS_CASH_balance_up.drop("SK_ID_CURR", axis=1, inplace=True)
# Group previous_application by SK_ID_CURR and count number of unique SK_ID_PREV values
previous_application_counts = (
base_previous_application_up.groupby("SK_ID_CURR", as_index=False)["SK_ID_PREV"]
.count()
.rename(columns={"SK_ID_PREV": "PREVIOUS_APPLICATION_COUNT"})
)
# ----------------
def extract_mean(x):
y = x.groupby("SK_ID_PREV", as_index=False).mean().add_prefix("POS_MEAN_")
return y
POS_mean = extract_mean(base_POS_CASH_balance_up)
POS_mean = POS_mean.rename(columns={"POS_MEAN_SK_ID_PREV": "SK_ID_PREV"})
base_previous_application_up = base_previous_application_up.merge(
POS_mean, on="SK_ID_PREV", how="left"
)
# -------------------------
def extract_mean(x):
y = x.groupby("SK_ID_PREV", as_index=False).mean().add_prefix("CARD_MEAN_")
return y
credit_card_balance_mean = extract_mean(base_credit_card_balance_up)
credit_card_balance_mean = credit_card_balance_mean.rename(
columns={"CARD_MEAN_SK_ID_PREV": "SK_ID_PREV"}
)
base_previous_application_up = base_previous_application_up.merge(
credit_card_balance_mean, on="SK_ID_PREV", how="left"
)
# ---------------------------
def extract_mean(x):
y = x.groupby("SK_ID_PREV", as_index=False).mean().add_prefix("INSTALL_MEAN_")
return y
install_pay_mean = extract_mean(base_installments_payments_up)
install_pay_mean = install_pay_mean.rename(
columns={"INSTALL_MEAN_SK_ID_PREV": "SK_ID_PREV"}
)
base_previous_application_up = base_previous_application_up.merge(
install_pay_mean, on="SK_ID_PREV", how="left"
)
# ---------------------------
def extract_mean(x):
y = x.groupby("SK_ID_CURR", as_index=False).mean().add_prefix("PREV_APPL_MEAN_")
return y
prev_appl_mean = extract_mean(base_previous_application_up)
prev_appl_mean = prev_appl_mean.rename(
columns={"PREV_APPL_MEAN_SK_ID_CURR": "SK_ID_CURR"}
)
prev_appl_mean = prev_appl_mean.drop("PREV_APPL_MEAN_SK_ID_PREV", axis=1)
# ---------------------
# Merge preprocessed_data and df_bureau_up
df1 = pd.merge(df_train_test, bureau_mean_values, on="SK_ID_CURR", how="left")
# # # Merge df1 and df_previous_application_up
df2 = pd.merge(df1, previous_application_counts, on="SK_ID_CURR", how="left")
# # # Merge df2 and df_bureau_balance_up
df3 = pd.merge(df2, prev_appl_mean, on="SK_ID_CURR", how="left")
# ...
df3.head(3)
# **3.Exploratory data analysis**
# ###### This correlations shows the most positive and negative correlations between the TARGET variable and other variables in the dataset. The correlations are calculated using the Pearson correlation coefficient, which measures the linear relationship between two variables.
# ###### The output shows the top 15 variables with the highest positive correlation with TARGET ,sorted in descending order. The variable with the highest positive correlation is PREV_APPL_MEAN_CARD_MEAN_AMT_DRAWINGS_CURRENT, with a correlation coefficient of 0.055388. The output also shows the variable with the highest negative correlation, which is EXT_SOURCE_3 with a correlation coefficient of -0.178919.
# ###### The variables with the highest positive correlation with TARGET may be useful in predicting loan defaults, while the variables with the highest negative correlation may be useful in predicting loan repayments.
# Find correlations with the target and sort
numeric_cols = df3.select_dtypes(include=[np.number]).columns
correlations = df3[numeric_cols].corr()["TARGET"].sort_values()
# Display correlations
print("Most Positive Correlations:\n", correlations.tail(15))
print("\nMost Negative Correlations:\n", correlations.head(15))
# **3.1 Identify the categorical variable**
def identify_columns(df, threshold=25):
# Identify categorical object columns, categorical numerical columns, and non-categorical columns
cat_object_list = [
i for i in df.columns if df[i].dtype == "object" and df[i].nunique() < threshold
]
cat_num_list = [
i
for i in df.columns
if df[i].dtype in ["int64", "float64"] and df[i].nunique() < threshold
]
non_cat_list = [
i for i in df.columns if i not in cat_object_list and i not in cat_num_list
]
# Identify object columns and numerical columns in non-categorical columns
mix_serise_col = df[non_cat_list]
non_cat_obj = [
i for i in mix_serise_col.columns if mix_serise_col[i].dtype == "object"
]
non_cat_num = [
i
for i in mix_serise_col.columns
if mix_serise_col[i].dtype in ["int64", "float64"]
]
# #Print the results
# print('Categorical object columns:', len(cat_object_list))
# print('Categorical numerical columns:', len(cat_num_list))
# print('Non-categorical columns:', len(non_cat_list))
# print('Object columns in non-categorical columns:', len(non_cat_obj))
# print('Numerical columns in non-categorical columns:', len(non_cat_num))
# Return the results as a dictionary
results = {
"cat_object_list": cat_object_list,
"cat_num_list": cat_num_list,
"non_cat_list": non_cat_list,
"non_cat_obj": non_cat_obj,
"non_cat_num": non_cat_num,
}
return results
results = identify_columns(df_train_test)
categorical_num = identify_columns(df3)["cat_num_list"]
categorical_obj = identify_columns(df3)["cat_object_list"]
non_categorical_obj = identify_columns(df3)["non_cat_obj"]
print(categorical_obj)
print(categorical_num)
def plot_categorical_feature(feature, df=None, orientation_horizontal=True):
if df is None:
df = df3
else:
df = df
temp = df[feature].value_counts()
df1 = pd.DataFrame({feature: temp.index, "Number of contracts": temp.values})
# Calculate the percentage of target=1 per category value
cat_perc = df[[feature, "TARGET"]].groupby([feature], as_index=False).mean()
cat_perc.sort_values(by="TARGET", ascending=False, inplace=True)
sns.set_color_codes("colorblind")
if orientation_horizontal == True:
plt.figure(figsize=(15, 5))
plt.subplot(121)
s1 = sns.barplot(y=feature, x="Number of contracts", data=df1)
plt.subplot(122)
s2 = sns.barplot(y=feature, x="TARGET", data=cat_perc)
plt.xlabel("Fraction of loans defaulted", fontsize=12)
plt.ylabel(feature, fontsize=12)
else:
plt.figure(figsize=(10, 12))
plt.subplot(211)
s1 = sns.barplot(x=feature, y="Number of contracts", data=df1)
s1.set_xticklabels(s1.get_xticklabels(), rotation=90)
plt.subplot(212)
s2 = sns.barplot(x=feature, y="TARGET", data=cat_perc)
s2.set_xticklabels(s2.get_xticklabels(), rotation=90)
plt.ylabel("Fraction of loans defaulted", fontsize=12)
plt.xlabel(feature, fontsize=12)
plt.tick_params(axis="both", which="major", labelsize=12)
plt.subplots_adjust(wspace=0.6)
plt.show()
# ###### This suggests that the default rate for cash loans is higher than that for revolving loans. the grough also suggests that feamal has high rate of default rate and so on.
# Concatenate the two lists
features = categorical_obj + non_categorical_obj
# Loop over the features and plot each one
for feature in features:
plot_categorical_feature(feature)
# **plot_bivariate_distribution**
def plot_bivariate_distribution(feature, df):
if df is None:
df = df3
else:
df = df
plt.figure(figsize=(10, 4))
sns.kdeplot(df.loc[df["TARGET"] == 0, feature], label="TARGET == 0")
sns.kdeplot(df.loc[df["TARGET"] == 1, feature], label="TARGET == 1")
plt.xlabel(feature)
plt.ylabel("Density")
plt.title("Distribution of {} by Target Value".format(feature))
plt.legend()
plot_bivariate_distribution("EXT_SOURCE_1", df_train)
plot_bivariate_distribution("EXT_SOURCE_2", df3)
plot_bivariate_distribution("AMT_INCOME_TOTAL", df3)
plot_bivariate_distribution("DAYS_EMPLOYED", df3)
plot_bivariate_distribution("DAYS_BIRTH", df3)
# **4.1 Feature engineering**
# ###### I have changed the negitive values of days to years.
# Converting days col into a year
def create_day_to_year(df, ls_cols):
for col in ls_cols:
if col.startswith("DAYS") and np.issubdtype(df[col].dtype, np.number):
new_col = col.replace("DAYS", "YEARS_UP")
df[new_col] = round(np.abs(df[col] / 365))
df.drop(columns=[col], inplace=True)
return df
df_list = [df3]
for df in df_list:
day_cols = df.filter(like="DAYS").columns
df = create_day_to_year(df, day_cols)
def print_years_cols(dfs):
for df, name in dfs:
day_cols = df.filter(like="YEARS_UP").columns
print(f"Number of columns with 'YEARS' in {name}: {len(day_cols)}")
print(f"Columns with 'YEARS' in {name}:")
for col in day_cols:
print(col)
print_years_cols([(df3, "df_train")])
# ###### I have computed the age of each loan applicant. The following graph indicates that individuals between the ages of 25 and 30 are more likely to default on their home loans.
plot_bivariate_distribution("YEARS_UP_BIRTH", df3)
# ###### A column called "income band" has been created, which segments income into different categories. For example, an income range of 0 to 30k is assigned to category 1, while an income range of 30k to 65k is assigned to category 2, and so on.
# Converting income band;
def create_income_band(df):
bins = [
0,
30000,
65000,
95000,
130000,
160000,
190000,
220000,
275000,
325000,
np.inf,
]
labels = range(1, len(bins))
df["INCOME_BAND"] = pd.cut(df["AMT_INCOME_TOTAL"], bins=bins, labels=labels)
return df
dfs = [(df3, "data_set")]
for df, name in dfs:
df = create_income_band(df)
fig, ax = plt.subplots(figsize=(5, 3))
sns.countplot(data=df3, x="INCOME_BAND", hue="TARGET")
ax.set_title("Income data for people repaying and defaulting loans")
ax.set_ylabel("Count")
plt.tight_layout()
plt.show()
# ---
# **4.2 Encoding and missing value handeling**
# ###### I have applied label encoding to columns with two categorical values, and one-hot encoding to columns with more than two categorical values. To handle missing values, I have implemented a condition where if a column has more than 60% missing values, it will be deleted. Otherwise, I have used SimpleImputer to replace missing values with the median value of the scale
from sklearn.preprocessing import LabelEncoder
from sklearn.impute import SimpleImputer
def label_encoding(df):
for col in df.columns:
if df[col].dtype == "object":
if len(df[col].unique()) == 2:
le = LabelEncoder()
le.fit(df[col])
df[col] = le.transform(df[col])
# else:
# df[col] = pd.get_dummies(df[col])
return df
def missing_values_table(df):
# Check if input is a dataframe or a series
if isinstance(df, pd.Series):
df = pd.DataFrame(df)
# Get columns with missing values
na_columns = df.columns[df.isnull().any()].tolist()
# Count missing values and calculate ratio
n_miss = df[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (n_miss / df.shape[0] * 100).sort_values(ascending=False)
# Create DataFrame with missing values and ratio
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["Missing Values", "Percentage"]
)
return missing_df
def missing_preprocess_data(df):
# Identify columns with more than 60% missing values
missing_cols = df.columns[df.isnull().mean() > 0.6]
# Drop columns with more than 60% missing values
num_cols_dropped = len(missing_cols)
df.drop(columns=missing_cols, inplace=True)
print(f"Dropped {num_cols_dropped} columns due to missing value threshold")
# Fill remaining missing values using median imputation
imputer = SimpleImputer(strategy="median")
df = pd.DataFrame(imputer.fit_transform(df), columns=df.columns)
# Print out the original and preprocessed datasets
print("Preprocessed dataset:")
print(df)
return df
base_df3 = df3.copy()
label_encoding(base_df3)
base_df3 = pd.get_dummies(base_df3)
base_df3.shape
preprocessed_data = missing_preprocess_data(base_df3)
missing_values_table(preprocessed_data)
# **5.Classifier models**
# ###### LogisticRegression
process_df_logis = preprocessed_data.copy()
# Basic Logistic regrassion
# Split the data into training and testing sets
train_X, test_X, train_Y, test_Y = train_test_split(
preprocessed_data.drop(["SK_ID_CURR", "TARGET"], axis=1),
preprocessed_data["TARGET"],
test_size=0.25,
random_state=123,
)
# train_X contains the independent variables for the training set
# test_X contains the independent variables for the testing set
# train_Y contains the dependent variable (TARGET) for the training set
# test_Y contains the dependent variable (TARGET) for the testing set
def train_predict_visualize(train_X, train_Y, test_X, test_Y):
# Train a logistic regression classifier
clf = LogisticRegression(random_state=123)
clf.fit(train_X, train_Y)
# Predict using test data
pred_Y = clf.predict(test_X)
pred_prob_Y = clf.predict_proba(test_X)[:, 1]
# Calculate metrics
auc = roc_auc_score(test_Y, pred_prob_Y)
tn, fp, fn, tp = confusion_matrix(test_Y, pred_Y).ravel()
accuracy = (tp + tn) / (tp + tn + fp + fn)
precision = tp / (tp + fp)
recall = tp / (tp + fn)
f1_score = 2 * precision * recall / (precision + recall)
# Print metrics
print("AUC:", auc)
print("Accuracy:", accuracy)
print("Precision:", precision)
print("Recall:", recall)
print("F1 Score:", f1_score)
# Visualize feature importances
coef = pd.Series(clf.coef_[0], index=train_X.columns)
coef.nlargest(10).plot(kind="barh")
plt.show()
def train_predict_visualize(train_X, train_Y, test_X, test_Y):
# Train a logistic regression classifier
clf = LogisticRegression(random_state=123)
clf.fit(train_X, train_Y)
# Predict using test data
pred_Y = clf.predict(test_X)
pred_prob_Y = clf.predict_proba(test_X)[:, 1]
# Calculate metrics
auc = roc_auc_score(test_Y, pred_prob_Y)
tn, fp, fn, tp = confusion_matrix(test_Y, pred_Y).ravel()
accuracy = (tp + tn) / (tp + tn + fp + fn)
precision = tp / (tp + fp)
recall = tp / (tp + fn)
f1_score = 2 * precision * recall / (precision + recall)
# Print metrics
print("AUC:", auc)
print("Accuracy:", accuracy)
print("Precision:", precision)
print("Recall:", recall)
print("F1 Score:", f1_score)
# Visualize feature importances
coef = pd.Series(clf.coef_[0], index=train_X.columns)
top_features = coef.nlargest(10)
top_features.plot(kind="barh")
plt.show()
# Print table of top 10 features
print("Top 10 Features:")
print(top_features.to_string())
train_predict_visualize(train_X, train_Y, test_X, test_Y)
# #### Explanation of the model evaluation:
# ###### The result of the model evaluation shows that the AUC (Area Under the Curve) is 0.618, which indicates that the model's ability to distinguish between positive and negative classes is slightly better than random guessing. The accuracy score is 0.929, which indicates that the model is able to correctly predict the target variable for 92.9% of the observations.
# ###### However, the precision score is 0.0, which means that the model did not correctly identify any true positives. The recall score is also 0.0, which means that the model did not correctly identify any positive cases. The F1 score is "nan" (not a number), which is likely due to the precision and recall scores being 0.
# ###### Overall, these results suggest that the model may not be effective in predicting the target variable, and further analysis and model tuning may be necessary to improve its performance.
# ##### Domain Knowledge feature engeening
preprocessed_data.shape
# Domain knowledge
# Debt-to-income-ratio
preprocessed_data["CREDIT_INCOME_PERCENT"] = (
preprocessed_data["AMT_CREDIT"] / preprocessed_data["AMT_INCOME_TOTAL"]
)
# Loan-to-Value-ration
preprocessed_data["LOAN_TO_VALUE_RATION"] = (
preprocessed_data["AMT_CREDIT"] / preprocessed_data["AMT_GOODS_PRICE"]
)
# percentage of the applicant's income that is being used to pay off the loan.
preprocessed_data["ANNUITY_INCOME_PERCENT"] = (
preprocessed_data["AMT_ANNUITY"] / preprocessed_data["AMT_INCOME_TOTAL"]
)
# This ratio represents the length of time it will take the applicant to pay off the loan.
preprocessed_data["CREDIT_TERM"] = (
preprocessed_data["AMT_ANNUITY"] / preprocessed_data["AMT_CREDIT"]
)
# Employment history percentage
preprocessed_data["YEARS_EMPLOYED_PERCENT"] = (
preprocessed_data["YEARS_UP_EMPLOYED"] / preprocessed_data["YEARS_UP_BIRTH"]
) # age
# --------
##Late payment
# # Identify the relevant columns for payment history
# payment_history_cols = ['SK_ID_CURR', 'SK_ID_PREV', 'DAYS_ENTRY_PAYMENT']
# # Calculate the number of late payments on previous loans
# previous_application_up['LATE_PAYMENT'] = (previous_application_up['DAYS_ENTRY_PAYMENT'] > 0).astype(int)
# late_payment_counts = previous_application.groupby('SK_ID_CURR')['LATE_PAYMENT'].sum()
# # Merge the late payment counts into the application_train_up dataset
# application_train_up = application_train_up.merge(late_payment_counts, on='SK_ID_CURR', how='left')
# #### Poly feature engineering
from sklearn.preprocessing import PolynomialFeatures
# Define the columns to include in the polynomial features
poly_cols = [
"AMT_INCOME_TOTAL",
"AMT_CREDIT",
"AMT_ANNUITY",
"YEARS_UP_BIRTH",
"YEARS_UP_EMPLOYED",
]
# Check if all columns exist in the dataframe
if all(col in preprocessed_data.columns for col in poly_cols):
# Create polynomial features
poly = PolynomialFeatures(degree=2, include_bias=False)
poly_features = poly.fit_transform(preprocessed_data[poly_cols])
poly_feature_names = poly.get_feature_names_out(poly_cols)
poly_df = pd.DataFrame(poly_features, columns=poly_feature_names)
# Merge the polynomial features with the original dataframe
preprocessed_data = pd.concat([preprocessed_data, poly_df], axis=1)
else:
print("One or more columns not found in dataframe")
preprocessed_data.shape
# Check for duplicated column names
duplicated_cols = preprocessed_data.columns[preprocessed_data.columns.duplicated()]
# Print the duplicated column names
print("Duplicated columns:", duplicated_cols)
# Remove duplicated columns if any
if len(duplicated_cols) > 0:
preprocessed_data = preprocessed_data.loc[
:, ~preprocessed_data.columns.duplicated()
]
print("Duplicated columns removed")
# ---
# ##### XGBClassifier
process_df_xgb = preprocessed_data.copy()
import pandas as pd
import xgboost as xgb
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
)
from sklearn.model_selection import train_test_split
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
process_df_xgb.drop(["SK_ID_CURR", "TARGET"], axis=1),
process_df_xgb["TARGET"],
test_size=0.2,
random_state=42,
)
# Train an XGBClassifier
xgb_model = xgb.XGBClassifier(n_estimators=100, random_state=42)
xgb_model.fit(X_train, y_train)
# Get the feature importances
importances = xgb_model.get_booster().get_score(importance_type="gain")
importances = {
feature: importance
for feature, importance in sorted(
importances.items(), key=lambda x: x[1], reverse=True
)
}
# Create a dataframe with the feature importances
feature_importances = pd.DataFrame(
{"feature": list(importances.keys()), "importance": list(importances.values())}
)
# Select the top 10 features
top_features = feature_importances.head(10)
# # Print the top 10 features
# print(top_features)
# Make predictions on the test set
y_pred_prob = xgb_model.predict_proba(X_test)[:, 1]
y_pred = (y_pred_prob > 0.5).astype(int)
# Evaluate the performance of the model
accuracy = accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred)
recall = recall_score(y_test, y_pred)
f1 = f1_score(y_test, y_pred)
auc = roc_auc_score(y_test, y_pred_prob)
print("Accuracy:", accuracy)
print("Precision:", precision)
print("Recall:", recall)
print("F1 score:", f1)
print("AUC score:", auc)
# Make predictions on the test data
test_pred_prob = xgb_model.predict_proba(X_test)[:, 1]
test_pred = (test_pred_prob > 0.5).astype(int)
X_test["SK_ID_CURR"] = process_df_xgb.loc[X_test.index, "SK_ID_CURR"]
# Create the submission file
submission = pd.DataFrame({"SK_ID_CURR": X_test["SK_ID_CURR"], "TARGET": test_pred})
submission.to_csv("submission.csv", index=False)
# ###### The result of the model evaluation shows that the accuracy score is 0.929, which indicates that the model is able to correctly predict the target variable for 92.9% of the observations. The precision score is 0.463, which means that the model correctly identified 46.3% of the true positives out of all the predicted positives. The recall score is 0.044, which means that the model correctly identified only 4.4% of the true positives out of all the actual positives. The F1 score is 0.080, which is the harmonic mean of precision and recall, and provides an overall measure of the model's performance.
# ###### The AUC (Area Under the Curve) score is 0.773, which indicates that the model's ability to distinguish between positive and negative classes is better than random guessing.
# ###### Overall, these results suggest that the model may have some predictive power, but its performance is not very strong. Further analysis and model tuning may be necessary to improve its performance.
submission["TARGET"].astype(int).plot.hist(color="g")
# Count the number of occurrences of each value in the 'TARGET' column
value_counts = submission["TARGET"].value_counts()
# Print the value counts
print(value_counts)
# **Important feature selaction (XGBClassifier)**
# Create a new dataframe with only the top features
df_top = preprocessed_data[["SK_ID_CURR", "TARGET"] + top_features["feature"].tolist()]
# Save the new dataframe to a CSV file
df_top.to_csv("data_top.csv", index=False)
# Plot the feature importances
plt.figure(figsize=(16, 32))
plt.barh(top_features["feature"], top_features["importance"])
plt.xlabel("Importance")
plt.ylabel("Feature")
plt.title("Feature Importances")
plt.show()
# ##### lgb.LGBMClassifier
process_df_lgb = preprocessed_data.copy()
import pandas as pd
import lightgbm as lgb
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
)
from sklearn.model_selection import train_test_split
# Remove any non-alphanumeric characters from the feature names
process_df_lgb.columns = [
re.sub("[^0-9a-zA-Z]+", "_", col) for col in process_df_lgb.columns
]
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
process_df_lgb.drop(["SK_ID_CURR", "TARGET"], axis=1),
process_df_lgb["TARGET"],
test_size=0.2,
random_state=42,
)
# Train a LightGBM classifier
lgb_model = lgb.LGBMClassifier(n_estimators=100, random_state=42)
lgb_model.fit(X_train, y_train)
# Get the feature importances
importances = lgb_model.feature_importances_
# Create a dataframe with the feature importances
feature_importances = pd.DataFrame(
{"feature": X_train.columns, "importance": importances}
)
# Sort the dataframe by importance
feature_importances = feature_importances.sort_values("importance", ascending=False)
# Select the top 10 features
top_features = feature_importances.head(10)
# Print the top 10 features
print(top_features)
# Make predictions on the test set
y_pred_prob = lgb_model.predict_proba(X_test)[:, 1]
y_pred = (y_pred_prob > 0.5).astype(int)
# Evaluate the performance of the model
accuracy_lgbm = accuracy_score(y_test, y_pred)
precision_lgbm = precision_score(y_test, y_pred)
recall_lgbm = recall_score(y_test, y_pred)
f1_lgbm = f1_score(y_test, y_pred)
auc_lgbm = roc_auc_score(y_test, y_pred_prob)
print("Accuracy:", accuracy)
print("Precision:", precision)
print("Recall:", recall)
print("F1 score:", f1)
print("AUC score:", auc)
# Make predictions on the test data
test_pred_prob = lgb_model.predict_proba(X_test)[:, 1]
test_pred = (test_pred_prob > 0.5).astype(int)
X_test["SK_ID_CURR"] = process_df_lgb.loc[X_test.index, "SK_ID_CURR"]
# Create the submission file
submission1 = pd.DataFrame({"SK_ID_CURR": X_test["SK_ID_CURR"], "TARGET": test_pred})
submission1.to_csv("submission.csv", index=False)
submission1["TARGET"].astype(int).plot.hist(color="g")
# Count the number of occurrences of each value in the 'TARGET' column
value_counts1 = submission1["TARGET"].value_counts()
# Print the value counts
print(value_counts1)
# ##### Comparasion xgb vs lgb
import matplotlib.pyplot as plt
import numpy as np
# Define the evaluation metrics
metrics = ["Accuracy", "Precision", "Recall", "F1 score", "AUC score"]
xgb_scores = [accuracy, precision, recall, f1, auc]
lgbm_scores = [accuracy_lgbm, precision_lgbm, recall_lgbm, f1_lgbm, auc_lgbm]
# Create a bar plot
x = np.arange(len(metrics))
width = 0.35
fig, ax = plt.subplots()
rects1 = ax.bar(x - width / 2, xgb_scores, width, label="XGBClassifier")
rects2 = ax.bar(x + width / 2, lgbm_scores, width, label="LGBMClassifier")
# Add labels and title
ax.set_ylabel("Score")
ax.set_xticks(x)
ax.set_xticklabels(metrics)
ax.legend()
ax.set_title("Comparison of XGBClassifier and LGBMClassifier")
# Add values above the bars
def autolabel(rects):
for rect in rects:
height = rect.get_height()
ax.annotate(
"{:.3f}".format(height),
xy=(rect.get_x() + rect.get_width() / 2, height),
xytext=(0, 3), # 3 points vertical offset
textcoords="offset points",
ha="center",
va="bottom",
)
autolabel(rects1)
autolabel(rects2)
plt.show()
# Create a dictionary to store the scores
scores_dict = {
"Model": ["XGBClassifier", "LGBMClassifier"],
"Accuracy": [xgb_scores[0], lgbm_scores[0]],
"Precision": [xgb_scores[1], lgbm_scores[1]],
"Recall": [xgb_scores[2], lgbm_scores[2]],
"F1 Score": [xgb_scores[3], lgbm_scores[3]],
"AUC Score": [xgb_scores[4], lgbm_scores[4]],
}
# Create a pandas dataframe from the dictionary
scores_df = pd.DataFrame(scores_dict)
# Print the dataframe
print(scores_df)
# ###### Based on the evaluation metrics, it seems that both models are not performing very well. The accuracy scores are relatively high, but the precision, recall, and F1 scores are quite low. This suggests that the models are not doing a good job of correctly identifying the positive class (i.e., clients who are likely to default on their loans).
# ###### The AUC scores are also relatively low, which indicates that the models are not doing a good job of distinguishing between positive and negative cases.
# ###### It may be necessary to try different models or to tune the hyperparameters of the existing models to improve their performance. Additionally, it may be helpful to explore the data further and identify any patterns or relationships that could be used to improve the models.
# ##### K-fold cross validation
process_df_k = preprocessed_data.copy()
import lightgbm as lgb
from sklearn.model_selection import KFold
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
)
# Remove any non-alphanumeric characters from the feature names
process_df_k.columns = [
re.sub("[^0-9a-zA-Z]+", "_", col) for col in process_df_k.columns
]
# Define the number of folds
k = 5
# Define the K-fold cross-validator
kf = KFold(n_splits=k, shuffle=True, random_state=42)
# Initialize empty lists to store the scores for each fold
accuracy_scores_lgbm = []
precision_scores_lgbm = []
recall_scores_lgbm = []
f1_scores_lgbm = []
auc_scores_lgbm = []
# Loop over the folds
for train_index, test_index in kf.split(df_top):
# Split the data into training and test sets
X_train, X_test = (
process_df_k.drop(["SK_ID_CURR", "TARGET"], axis=1).iloc[train_index],
df_top.drop(["SK_ID_CURR", "TARGET"], axis=1).iloc[test_index],
)
y_train, y_test = (
df_top["TARGET"].iloc[train_index],
df_top["TARGET"].iloc[test_index],
)
# Check if the columns in the training and test data are the same
if set(X_train.columns) != set(X_test.columns):
missing_cols = set(X_train.columns) - set(X_test.columns)
for col in missing_cols:
X_test[col] = 0
X_test = X_test[X_train.columns]
# Initialize the LGBMClassifier model
model = lgb.LGBMClassifier(n_estimators=100, random_state=42)
# Fit the model on the training data
model.fit(X_train, y_train)
# Make predictions on the test data
y_pred = model.predict(X_test)
# Calculate the evaluation metrics
accuracy_lgbm = accuracy_score(y_test, y_pred)
precision_lgbm = precision_score(y_test, y_pred)
recall_lgbm = recall_score(y_test, y_pred)
f1_lgbm = f1_score(y_test, y_pred)
auc_lgbm = roc_auc_score(y_test, y_pred)
# Append the scores to the lists
accuracy_scores_lgbm.append(accuracy_lgbm)
precision_scores_lgbm.append(precision_lgbm)
recall_scores_lgbm.append(recall_lgbm)
f1_scores_lgbm.append(f1_lgbm)
auc_scores_lgbm.append(auc_lgbm)
# Calculate the mean and standard deviation of the scores
mean_accuracy = np.mean(accuracy_scores_lgbm)
std_accuracy = np.std(accuracy_scores_lgbm)
mean_precision = np.mean(precision_scores_lgbm)
std_precision = np.std(precision_scores_lgbm)
mean_recall = np.mean(recall_scores_lgbm)
std_recall = np.std(recall_scores_lgbm)
mean_f1 = np.mean(f1_scores_lgbm)
std_f1 = np.std(f1_scores_lgbm)
mean_auc = np.mean(auc_scores_lgbm)
std_auc = np.std(auc_scores_lgbm)
# Print the results
print(
"Accuracy_lgbm: {:.2f}% (+/- {:.2f}%)".format(
mean_accuracy * 100, std_accuracy * 100
)
)
print(
"Precision_lgbm: {:.2f}% (+/- {:.2f}%)".format(
mean_precision * 100, std_precision * 100
)
)
print("Recall_lgbm: {:.2f}% (+/- {:.2f}%)".format(mean_recall * 100, std_recall * 100))
print("F1 score_lgbm: {:.2f}% (+/- {:.2f}%)".format(mean_f1 * 100, std_f1 * 100))
print("AUC score_lgbm: {:.2f}% (+/- {:.2f}%)".format(mean_auc * 100, std_auc * 100))
| false | 0 | 13,945 | 0 | 13,945 | 13,945 |
||
129119059
|
# # Convert the audio into text with using of whisper model
# fiirst of we set the runtime type to GPU in google colab
# then import the whisper model through the below command
# import the whisper model
import whisper
model = whisper.load_model("base")
result = model.transcribe(
"/kaggle/input/audio-files/Post-Malone-Sunflower-ft.-Swae-Lee.mp3"
)
result["text"]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/119/129119059.ipynb
| null | null |
[{"Id": 129119059, "ScriptId": 38384332, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9787244, "CreationDate": "05/11/2023 06:34:12", "VersionNumber": 1.0, "Title": "Audio_To_Text(whisper)", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 16.0, "LinesInsertedFromPrevious": 16.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
| null | null | null | null |
# # Convert the audio into text with using of whisper model
# fiirst of we set the runtime type to GPU in google colab
# then import the whisper model through the below command
# import the whisper model
import whisper
model = whisper.load_model("base")
result = model.transcribe(
"/kaggle/input/audio-files/Post-Malone-Sunflower-ft.-Swae-Lee.mp3"
)
result["text"]
| false | 0 | 111 | 2 | 111 | 111 |
||
129119840
|
<jupyter_start><jupyter_text>Exchange_Rate
Kaggle dataset identifier: exchange-rate
<jupyter_script># # Time series forecasting
# An airline company has the data on the number of passengers that have travelled with them on a particular route for the past few years. Using this data, they want to see if they can forecast the number of passengers for the next twelve months.
#
# Making this forecast could be quite beneficial to the company as it would help them take some crucial decisions like -
# * What capacity aircraft should they use?
# * When should they fly?
# * How many air hostesses and pilots do they need?
# * How much food should they stock in their inventory?
#
# ## Initial setup
# ## Import required packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# # Importing Time-Series Data: Airline Passengers
data = pd.read_csv(
"/kaggle/input/airline-passenger-traffic/airline-passenger-traffic(1).csv",
header=None,
)
data.columns = ["Month", "Passengers"]
data["Month"] = pd.to_datetime(data["Month"], format="%Y-%m")
data = data.set_index("Month")
data.head(12)
data.columns
data.info()
data.describe()
# # Time series analysis
# ## Plot time series data
data.plot(figsize=(12, 5))
plt.legend(loc="best")
plt.title("Airline Passenger Traffic")
plt.show(block=False)
# **Summary:**
# * There we can see a seasonal pattern in each year.
# * Its kind of like Summer and in winter, Passenger travelling are more.
# * There are some missing data as well.
# # Handling Missing Values
# ### Mean imputation
data = data.assign(
Passengers_Mean_Imputation=data.Passengers.fillna(data.Passengers.mean())
)
data[["Passengers_Mean_Imputation"]].plot(figsize=(12, 4))
plt.legend(loc="best")
plt.title("Airline passenger traffic: Mean imputation")
plt.show(block=False)
# **Summary:**
# * Imputed the missing values using mean of the column.
# * We can see there no more missing value present
# ### **Other Missing Value Treatment Methods are:**
# #### Last observation carried forward:
# We impute the missing values with its previous value in the data.
# #### Linear interpolation:
# You draw a straight line joining the next and previous points of the missing values in the data.
data = data.assign(
Passengers_Linear_Interpolation=data.Passengers.interpolate(method="linear")
)
data[["Passengers_Linear_Interpolation"]].plot(figsize=(12, 4))
plt.legend(loc="best")
plt.title("Airline passenger traffic: Linear interpolation")
plt.show(block=False)
# #### Seasonal + Linear interpolation:
# This method is best applicable for the data with trend and seasonality. Here, the missing value is imputed with the average of the corresponding data point in the previous seasonal period and the next seasonal period of the missing value.
# ### Use linear interpolation to impute missing values
data["Passengers"] = data["Passengers_Linear_Interpolation"]
data.drop(
columns=["Passengers_Mean_Imputation", "Passengers_Linear_Interpolation"],
inplace=True,
)
# # Outlier detection
# ### Box plot and interquartile range
fig = plt.subplots(figsize=(12, 2))
ax = sns.boxplot(x=data["Passengers"], whis=1)
fig = plt.subplots(figsize=(12, 2))
ax = sns.boxplot(x=data["Passengers"], whis=1.5)
# Histogram plot
fig = data.Passengers.hist(figsize=(12, 4))
# ### Time series Decomposition
# #### **Additive seasonal decomposition**
from pylab import rcParams
import statsmodels.api as sm
# * **pylab** is a module that provides a way to work with a combination of matplotlib (a popular plotting library in Python) and numpy (a library for working with arrays and mathematical operations) in a convenient way. **rcParams** is a method from the pylab library that can be used to customize the default settings for plots.
# * **api** is a sub-module in statsmodels that provides a set of functions and classes that are intended to be stable across different versions of the library.
# Size of the plot
rcParams["figure.figsize"] = 12, 8
# Seasonal Index where 'tsa' stands for time series analysis. Additive decomposition
decomposition = sm.tsa.seasonal_decompose(data.Passengers, model="additive")
# plotting the figure
fig = decomposition.plot()
ax = fig.axes[0]
ax.plot(data.Passengers.index, data.Passengers, "-", markersize=4, color="black")
plt.show()
# * Decomposition done and shown as 'trend', 'Seasonal' and 'Residual'
# * **Trend**: Upward trending of the passengers count.
# * **Seasonal**: Absolute movement with up and down in a repeatable fashion. Mean to be around 0 and peak to be around 50. The bottom of each year is around -50.
# * **Residual**: Whatever not captured in trend and seasonality will be captured in Residual. The cpatured in residual could be better. So, we can go for multiplicate demposition now.
# #### **Multiplicative seasonal decomposition**
# multiplicative seasonal index
decomposition = sm.tsa.seasonal_decompose(data.Passengers, model="multiplicative")
fig = decomposition.plot()
plt.show()
# # Build and evaluate time series forecast
# #### **Split time series data into training and test set**
train_len = 120
train = data[0:train_len] # first 120 months as training set
test = data[train_len:] # last 24 months as out-of-time test set
train.shape
test.shape
# # Basic Time Series Methods
# #### **Naive Method**
# * We will forecast for the last 2 years.
y_hat_naive = test.copy()
y_hat_naive["Naive Forecast"] = train["Passengers"][train_len - 1]
# #### **Plot train, test and forecast**
plt.figure(figsize=(12, 4))
plt.plot(train["Passengers"], label="Train")
plt.plot(test["Passengers"], label="Test")
plt.plot(y_hat_naive["Naive Forecast"], label="Naive Forecast")
plt.legend(loc="best")
plt.title("Naive Method")
plt.show()
# * We ended up here as underpredicting. we took forcast data for last 2 years, 1959 and 1960.
# # Calculate RMSE and MAPE
#
from sklearn.metrics import mean_squared_error
rmse = np.sqrt(
mean_squared_error(test["Passengers"], y_hat_naive["Naive Forecast"])
).round(2)
mape = np.round(
np.mean(
np.abs(test["Passengers"] - y_hat_naive["Naive Forecast"]) / test["Passengers"]
)
* 100,
2,
)
results = pd.DataFrame({"Method": ["Naive method"], "MAPE": [mape], "RMSE": [rmse]})
results = results[["Method", "RMSE", "MAPE"]]
results
# #### **Simple Average Method**
y_hat_avg = test.copy()
y_hat_avg["Avg Forecast"] = train["Passengers"].mean()
plt.figure(figsize=(12, 4))
plt.plot(train["Passengers"], label="Train")
plt.plot(test["Passengers"], label="Test")
plt.plot(y_hat_avg["Avg Forecast"], label="Average Forecast")
plt.legend(loc="best")
plt.title("Average Naive Method")
plt.show()
# * Green line = Forecast. Its the avg of the blue line (1949-1958) and is calculated for 1959-60.
# * As we are considering all from start, the first some years are very less compared to recent years. So, we are underpredicting the forecast.
# * our green data doesn't perform seasonality as the train and test data.
# # Exponential smoothing methods
# #### **Simple exponential smoothing**
from statsmodels.tsa.holtwinters import SimpleExpSmoothing
model = SimpleExpSmoothing(train["Passengers"])
model_fit = model.fit(smoothing_level=0.2, optimized=False)
model_fit.params
y_hat_ses = test.copy()
y_hat_ses["ses_forecast"] = model_fit.forecast(24)
# **Plot train, test and forecast**
plt.figure(figsize=(12, 4))
plt.plot(train["Passengers"], label="Train")
plt.plot(test["Passengers"], label="Test")
plt.plot(y_hat_ses["ses_forecast"], label="Simple exponential smoothing forecast")
plt.legend(loc="best")
plt.title("Simple Exponential Smoothing Method")
plt.show()
# #### **Calculate RMSE and MAPE**
rmse = np.sqrt(mean_squared_error(test["Passengers"], y_hat_ses["ses_forecast"])).round(
2
)
mape = np.round(
np.mean(np.abs(test["Passengers"] - y_hat_ses["ses_forecast"]) / test["Passengers"])
* 100,
2,
)
tempResults = pd.DataFrame(
{
"Method": ["Simple exponential smoothing forecast"],
"RMSE": [rmse],
"MAPE": [mape],
}
)
results = pd.concat([results, tempResults])
results
import pandas as pd
import matplotlib.pyplot as plt
# Step 1: Import the required libraries and dataset
# Assuming your dataset is stored in a CSV file called 'exchange_data.csv'
data = pd.read_csv("/kaggle/input/exchange-rate/exchange-rate-twi.csv")
# Step 2: Set the month column as index
data["Month"] = pd.to_datetime(data["Month"])
data.set_index("Month", inplace=True)
# Step 3: Make the time plot of the dataset to visualize it
data.plot()
plt.xlabel("Month")
plt.ylabel("Exchange Rate TWI")
plt.title("Exchange Rate TWI Over Time")
plt.show()
# Step 4: Divide the data set into train and test. Train data should be till index 212.
train_data = data.iloc[:212]
test_data = data.iloc[212:]
# Step 5: Predict the value with the last observed value of Train data
last_observation = train_data.iloc[-1]["Exchange Rate TWI"]
predictions = [last_observation] * len(test_data)
# Print the predicted values
print(predictions)
import numpy as np
from sklearn.metrics import mean_squared_error
# Assuming 'train_data' and 'test_data' are already defined
# Calculate the Simple Average Forecast
mean_value = train_data["Exchange Rate TWI"].mean()
predictions = [mean_value] * len(test_data)
# Calculate RMSE
rmse = np.sqrt(mean_squared_error(test_data["Exchange Rate TWI"], predictions))
# Calculate MAPE
mape = (
np.mean(
np.abs(
(test_data["Exchange Rate TWI"] - predictions)
/ test_data["Exchange Rate TWI"]
)
)
* 100
)
# Print the RMSE and MAPE values
print("RMSE:", rmse)
print("MAPE:", mape)
# Assuming 'train_data' and 'test_data' are already defined
# Get the last observed value from the train data
last_observation = train_data["Exchange Rate TWI"].iloc[-1]
# Predict the values using the last observed value
predictions = [last_observation] * len(test_data)
# Calculate MAPE
mape = (
np.mean(
np.abs(
(test_data["Exchange Rate TWI"] - predictions)
/ test_data["Exchange Rate TWI"]
)
)
* 100
)
# Calculate accuracy
accuracy = 100 - mape
# Print the accuracy
print("Accuracy:", accuracy)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/119/129119840.ipynb
|
exchange-rate
|
ramakrushnamohapatra
|
[{"Id": 129119840, "ScriptId": 38121197, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10977860, "CreationDate": "05/11/2023 06:41:48", "VersionNumber": 11.0, "Title": "Air passenger Traffic: Timeseries Forecasting", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 323.0, "LinesInsertedFromPrevious": 102.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 221.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184892068, "KernelVersionId": 129119840, "SourceDatasetVersionId": 5660339}, {"Id": 184892067, "KernelVersionId": 129119840, "SourceDatasetVersionId": 5601972}]
|
[{"Id": 5660339, "DatasetId": 3253257, "DatasourceVersionId": 5735766, "CreatorUserId": 10977860, "LicenseName": "Unknown", "CreationDate": "05/11/2023 04:52:44", "VersionNumber": 1.0, "Title": "Exchange_Rate", "Slug": "exchange-rate", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3253257, "CreatorUserId": 10977860, "OwnerUserId": 10977860.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5660339.0, "CurrentDatasourceVersionId": 5735766.0, "ForumId": 3318700, "Type": 2, "CreationDate": "05/11/2023 04:52:44", "LastActivityDate": "05/11/2023", "TotalViews": 29, "TotalDownloads": 4, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 10977860, "UserName": "ramakrushnamohapatra", "DisplayName": "Ramakrushna Mohapatra", "RegisterDate": "07/04/2022", "PerformanceTier": 2}]
|
# # Time series forecasting
# An airline company has the data on the number of passengers that have travelled with them on a particular route for the past few years. Using this data, they want to see if they can forecast the number of passengers for the next twelve months.
#
# Making this forecast could be quite beneficial to the company as it would help them take some crucial decisions like -
# * What capacity aircraft should they use?
# * When should they fly?
# * How many air hostesses and pilots do they need?
# * How much food should they stock in their inventory?
#
# ## Initial setup
# ## Import required packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# # Importing Time-Series Data: Airline Passengers
data = pd.read_csv(
"/kaggle/input/airline-passenger-traffic/airline-passenger-traffic(1).csv",
header=None,
)
data.columns = ["Month", "Passengers"]
data["Month"] = pd.to_datetime(data["Month"], format="%Y-%m")
data = data.set_index("Month")
data.head(12)
data.columns
data.info()
data.describe()
# # Time series analysis
# ## Plot time series data
data.plot(figsize=(12, 5))
plt.legend(loc="best")
plt.title("Airline Passenger Traffic")
plt.show(block=False)
# **Summary:**
# * There we can see a seasonal pattern in each year.
# * Its kind of like Summer and in winter, Passenger travelling are more.
# * There are some missing data as well.
# # Handling Missing Values
# ### Mean imputation
data = data.assign(
Passengers_Mean_Imputation=data.Passengers.fillna(data.Passengers.mean())
)
data[["Passengers_Mean_Imputation"]].plot(figsize=(12, 4))
plt.legend(loc="best")
plt.title("Airline passenger traffic: Mean imputation")
plt.show(block=False)
# **Summary:**
# * Imputed the missing values using mean of the column.
# * We can see there no more missing value present
# ### **Other Missing Value Treatment Methods are:**
# #### Last observation carried forward:
# We impute the missing values with its previous value in the data.
# #### Linear interpolation:
# You draw a straight line joining the next and previous points of the missing values in the data.
data = data.assign(
Passengers_Linear_Interpolation=data.Passengers.interpolate(method="linear")
)
data[["Passengers_Linear_Interpolation"]].plot(figsize=(12, 4))
plt.legend(loc="best")
plt.title("Airline passenger traffic: Linear interpolation")
plt.show(block=False)
# #### Seasonal + Linear interpolation:
# This method is best applicable for the data with trend and seasonality. Here, the missing value is imputed with the average of the corresponding data point in the previous seasonal period and the next seasonal period of the missing value.
# ### Use linear interpolation to impute missing values
data["Passengers"] = data["Passengers_Linear_Interpolation"]
data.drop(
columns=["Passengers_Mean_Imputation", "Passengers_Linear_Interpolation"],
inplace=True,
)
# # Outlier detection
# ### Box plot and interquartile range
fig = plt.subplots(figsize=(12, 2))
ax = sns.boxplot(x=data["Passengers"], whis=1)
fig = plt.subplots(figsize=(12, 2))
ax = sns.boxplot(x=data["Passengers"], whis=1.5)
# Histogram plot
fig = data.Passengers.hist(figsize=(12, 4))
# ### Time series Decomposition
# #### **Additive seasonal decomposition**
from pylab import rcParams
import statsmodels.api as sm
# * **pylab** is a module that provides a way to work with a combination of matplotlib (a popular plotting library in Python) and numpy (a library for working with arrays and mathematical operations) in a convenient way. **rcParams** is a method from the pylab library that can be used to customize the default settings for plots.
# * **api** is a sub-module in statsmodels that provides a set of functions and classes that are intended to be stable across different versions of the library.
# Size of the plot
rcParams["figure.figsize"] = 12, 8
# Seasonal Index where 'tsa' stands for time series analysis. Additive decomposition
decomposition = sm.tsa.seasonal_decompose(data.Passengers, model="additive")
# plotting the figure
fig = decomposition.plot()
ax = fig.axes[0]
ax.plot(data.Passengers.index, data.Passengers, "-", markersize=4, color="black")
plt.show()
# * Decomposition done and shown as 'trend', 'Seasonal' and 'Residual'
# * **Trend**: Upward trending of the passengers count.
# * **Seasonal**: Absolute movement with up and down in a repeatable fashion. Mean to be around 0 and peak to be around 50. The bottom of each year is around -50.
# * **Residual**: Whatever not captured in trend and seasonality will be captured in Residual. The cpatured in residual could be better. So, we can go for multiplicate demposition now.
# #### **Multiplicative seasonal decomposition**
# multiplicative seasonal index
decomposition = sm.tsa.seasonal_decompose(data.Passengers, model="multiplicative")
fig = decomposition.plot()
plt.show()
# # Build and evaluate time series forecast
# #### **Split time series data into training and test set**
train_len = 120
train = data[0:train_len] # first 120 months as training set
test = data[train_len:] # last 24 months as out-of-time test set
train.shape
test.shape
# # Basic Time Series Methods
# #### **Naive Method**
# * We will forecast for the last 2 years.
y_hat_naive = test.copy()
y_hat_naive["Naive Forecast"] = train["Passengers"][train_len - 1]
# #### **Plot train, test and forecast**
plt.figure(figsize=(12, 4))
plt.plot(train["Passengers"], label="Train")
plt.plot(test["Passengers"], label="Test")
plt.plot(y_hat_naive["Naive Forecast"], label="Naive Forecast")
plt.legend(loc="best")
plt.title("Naive Method")
plt.show()
# * We ended up here as underpredicting. we took forcast data for last 2 years, 1959 and 1960.
# # Calculate RMSE and MAPE
#
from sklearn.metrics import mean_squared_error
rmse = np.sqrt(
mean_squared_error(test["Passengers"], y_hat_naive["Naive Forecast"])
).round(2)
mape = np.round(
np.mean(
np.abs(test["Passengers"] - y_hat_naive["Naive Forecast"]) / test["Passengers"]
)
* 100,
2,
)
results = pd.DataFrame({"Method": ["Naive method"], "MAPE": [mape], "RMSE": [rmse]})
results = results[["Method", "RMSE", "MAPE"]]
results
# #### **Simple Average Method**
y_hat_avg = test.copy()
y_hat_avg["Avg Forecast"] = train["Passengers"].mean()
plt.figure(figsize=(12, 4))
plt.plot(train["Passengers"], label="Train")
plt.plot(test["Passengers"], label="Test")
plt.plot(y_hat_avg["Avg Forecast"], label="Average Forecast")
plt.legend(loc="best")
plt.title("Average Naive Method")
plt.show()
# * Green line = Forecast. Its the avg of the blue line (1949-1958) and is calculated for 1959-60.
# * As we are considering all from start, the first some years are very less compared to recent years. So, we are underpredicting the forecast.
# * our green data doesn't perform seasonality as the train and test data.
# # Exponential smoothing methods
# #### **Simple exponential smoothing**
from statsmodels.tsa.holtwinters import SimpleExpSmoothing
model = SimpleExpSmoothing(train["Passengers"])
model_fit = model.fit(smoothing_level=0.2, optimized=False)
model_fit.params
y_hat_ses = test.copy()
y_hat_ses["ses_forecast"] = model_fit.forecast(24)
# **Plot train, test and forecast**
plt.figure(figsize=(12, 4))
plt.plot(train["Passengers"], label="Train")
plt.plot(test["Passengers"], label="Test")
plt.plot(y_hat_ses["ses_forecast"], label="Simple exponential smoothing forecast")
plt.legend(loc="best")
plt.title("Simple Exponential Smoothing Method")
plt.show()
# #### **Calculate RMSE and MAPE**
rmse = np.sqrt(mean_squared_error(test["Passengers"], y_hat_ses["ses_forecast"])).round(
2
)
mape = np.round(
np.mean(np.abs(test["Passengers"] - y_hat_ses["ses_forecast"]) / test["Passengers"])
* 100,
2,
)
tempResults = pd.DataFrame(
{
"Method": ["Simple exponential smoothing forecast"],
"RMSE": [rmse],
"MAPE": [mape],
}
)
results = pd.concat([results, tempResults])
results
import pandas as pd
import matplotlib.pyplot as plt
# Step 1: Import the required libraries and dataset
# Assuming your dataset is stored in a CSV file called 'exchange_data.csv'
data = pd.read_csv("/kaggle/input/exchange-rate/exchange-rate-twi.csv")
# Step 2: Set the month column as index
data["Month"] = pd.to_datetime(data["Month"])
data.set_index("Month", inplace=True)
# Step 3: Make the time plot of the dataset to visualize it
data.plot()
plt.xlabel("Month")
plt.ylabel("Exchange Rate TWI")
plt.title("Exchange Rate TWI Over Time")
plt.show()
# Step 4: Divide the data set into train and test. Train data should be till index 212.
train_data = data.iloc[:212]
test_data = data.iloc[212:]
# Step 5: Predict the value with the last observed value of Train data
last_observation = train_data.iloc[-1]["Exchange Rate TWI"]
predictions = [last_observation] * len(test_data)
# Print the predicted values
print(predictions)
import numpy as np
from sklearn.metrics import mean_squared_error
# Assuming 'train_data' and 'test_data' are already defined
# Calculate the Simple Average Forecast
mean_value = train_data["Exchange Rate TWI"].mean()
predictions = [mean_value] * len(test_data)
# Calculate RMSE
rmse = np.sqrt(mean_squared_error(test_data["Exchange Rate TWI"], predictions))
# Calculate MAPE
mape = (
np.mean(
np.abs(
(test_data["Exchange Rate TWI"] - predictions)
/ test_data["Exchange Rate TWI"]
)
)
* 100
)
# Print the RMSE and MAPE values
print("RMSE:", rmse)
print("MAPE:", mape)
# Assuming 'train_data' and 'test_data' are already defined
# Get the last observed value from the train data
last_observation = train_data["Exchange Rate TWI"].iloc[-1]
# Predict the values using the last observed value
predictions = [last_observation] * len(test_data)
# Calculate MAPE
mape = (
np.mean(
np.abs(
(test_data["Exchange Rate TWI"] - predictions)
/ test_data["Exchange Rate TWI"]
)
)
* 100
)
# Calculate accuracy
accuracy = 100 - mape
# Print the accuracy
print("Accuracy:", accuracy)
| false | 2 | 3,031 | 0 | 3,050 | 3,031 |
||
129069668
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/working"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# !pip install pyedflib
import os
from datetime import datetime, timedelta
import pyedflib
import numpy as np
import pandas as pd
from scipy import signal
from scipy.signal import butter, lfilter
import matplotlib.pyplot as plt
from datetime import datetime, timedelta
import os
from math import floor
import copy
import itertools
from scipy.signal import hilbert
from scipy.stats import kurtosis
from scipy.stats import skew
from scipy.fftpack import fft
import math
import glob
def create_dir(directory_path):
"""Method to create a directory. Returns True if the directory already exists."""
if os.path.exists(directory_path):
return True
else:
os.makedirs(directory_path)
return False
def round_down(num, divisor):
"""Method to round down a number"""
return num - (num % divisor)
def get_time(datetime_string):
"""Method to convert a string to a datetime object."""
time = 0
try:
time = datetime.strptime(datetime_string, "%H:%M:%S")
except ValueError:
datetime_string = " " + datetime_string
if " 24" in datetime_string:
datetime_string = datetime_string.replace(" 24", "23")
time = datetime.strptime(datetime_string, "%H:%M:%S")
time += timedelta(hours=1)
else:
datetime_string = datetime_string.replace(" 25", "23")
time = datetime.strptime(datetime_string, "%H:%M:%S")
time += timedelta(hours=2)
return time
def extract_interval_data(
patient,
data_dir,
extract_ictal_samples=True,
extract_preictal_samples=True,
ictal_interval_padding_duration=32,
seizure_occurance_period=30,
seizure_prediction_horizon=5,
):
"""Method to extract interval patient data."""
patient_summary = open(
os.path.join(data_dir, "chb%02d" % patient, "chb%02d-summary.txt" % patient),
"r",
)
interictal_intervals = []
interictal_files = []
ictal_intervals = []
ictal_files = []
preictal_intervals = []
preictal_files = []
line = patient_summary.readline()
start_time = datetime.min
old_time = datetime.min
line_number = 0
while line:
line_data = line.split(":")
if line_data[0] == "File Name":
file_name = line_data[1].strip()
s = get_time(patient_summary.readline().split(": ")[1].strip())
if line_number == 0:
start_time = s
while s < old_time:
s += timedelta(hours=24)
old_time = s
end_time_file = get_time(patient_summary.readline().split(": ")[1].strip())
while end_time_file < old_time:
end_time_file = end_time_file + timedelta(hours=24)
old_time = end_time_file
n_seizures = int(patient_summary.readline().split(": ")[1])
if n_seizures == 0:
# Extract interictal interval data
interictal_intervals.append([s, end_time_file])
interictal_files.append([s, end_time_file, file_name])
else:
# Extract ictal and preictal interval data
for i in range(0, n_seizures):
seconds_start = int(
patient_summary.readline().split(": ")[1].split(" ")[0]
)
seconds_end = int(
patient_summary.readline().split(": ")[1].split(" ")[0]
)
if extract_ictal_samples:
# Extract ictal interval data
interval_start = s + timedelta(seconds=seconds_start)
if (
len(ictal_intervals) == 0 or interval_start > datetime.min
) and interval_start - start_time > timedelta(minutes=20):
interval_end = s + timedelta(seconds=seconds_end)
ictal_intervals.append(
[
interval_start
- timedelta(
seconds=ictal_interval_padding_duration
),
interval_end
+ timedelta(
seconds=ictal_interval_padding_duration
),
]
)
ictal_files.append([s, end_time_file, file_name])
if extract_preictal_samples:
# Extract preictal interval data
interval_start = (
s
+ timedelta(seconds=seconds_start)
- timedelta(
minutes=seizure_prediction_horizon
+ seizure_occurance_period
)
)
if (
len(preictal_intervals) == 0
or interval_start > datetime.min
) and interval_start - start_time > timedelta(minutes=20):
interval_end = interval_start + timedelta(
minutes=seizure_occurance_period
)
preictal_intervals.append([interval_start, interval_end])
preictal_files.append([s, end_time_file, file_name])
line = patient_summary.readline()
line_number += 1
patient_summary.close()
return (
interictal_intervals,
interictal_files,
ictal_intervals,
ictal_files,
preictal_intervals,
preictal_files,
)
def load_patient_data(patient, file, data_dir):
"""Method to load patient data."""
f = pyedflib.EdfReader("%schb%02d/%s" % (data_dir, patient, file))
n = f.signals_in_file
signals = np.zeros((n, f.getNSamples()[0]))
for i in np.arange(n):
signals[i, :] = f.readSignal(i)
return signals
def extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
interval_start,
interval_end,
segment_index,
n_channels,
):
"""Method to extract batch samples from specified intervals."""
start = 0
if file_start < interval_start:
start = (interval_start - file_start).seconds * sample_rate
if file_end <= interval_end:
end = -1
data = load_patient_data(patient, file[2], data_dir)[:, start:]
else:
end = ((interval_end - file_start).seconds * sample_rate) + 1
data = load_patient_data(patient, file[2], data_dir)[:, start : end + 1]
if (data.shape[0] >= n_channels) and (data.shape[1] >= sample_rate * window_size):
truncated_len = round_down(data.shape[1], sample_rate * window_size)
return (
np.array(
np.split(
data[0:n_channels, 0:truncated_len],
truncated_len / (sample_rate * window_size),
axis=1,
)
).swapaxes(0, 1),
segment_index,
)
else:
return np.array([]), segment_index
def extract_batches(
patient,
file,
data_dir,
segment_index,
intervals,
sample_rate,
window_size,
n_channels,
):
"""Method to extract batches."""
file_start = file[0]
file_end = file[1]
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
while file_start > interval_end and segment_index < len(intervals) - 1:
segment_index += 1
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
if (interval_end - interval_start).seconds >= window_size:
return extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
interval_start,
interval_end,
segment_index,
n_channels,
)
else:
return np.array([]), segment_index
def gen_synthetic_batches(
patient,
file,
data_dir,
segment_index,
intervals,
sample_rate,
window_size,
stride_len,
n_channels,
):
"""Method to generate synthetic batches."""
file_start = file[0]
file_end = file[1]
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
while file_start > interval_end and segment_index < len(intervals) - 1:
segment_index += 1
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
if (interval_end - interval_start).seconds > window_size:
synthetic_batches = np.array([]).reshape(
n_channels, 0, sample_rate * window_size
)
synthetic_interval_start = interval_start + timedelta(seconds=stride_len)
synthetic_interval_end = synthetic_interval_start + timedelta(
seconds=window_size
)
while synthetic_interval_end < interval_end:
extracted_batches = extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
synthetic_interval_start,
synthetic_interval_end,
segment_index,
n_channels,
)[0]
if extracted_batches.size > 0:
synthetic_batches = np.concatenate(
(synthetic_batches, extracted_batches), axis=1
)
synthetic_interval_start += timedelta(seconds=stride_len)
synthetic_interval_end += timedelta(seconds=stride_len)
return synthetic_batches, segment_index
else:
return np.array([]), segment_index
os.path.exists("/kaggle/working/processed_data")
n_channels = 22
sample_rate = 256 # Sample rate (Hz)
window_size = 64 # Window size (seconds)
# Stride length (seconds) used to generate synthetic preictal and ictal samples
stride_len = 32
# Data directory path
# data_dir = "/scratch/jcu/cl/CHBMIT/chb-mit-scalp-eeg-database-1.0.0/"
data_dir = "/kaggle/input/chb01-21/chbmit/"
processed_data_dir = (
"/kaggle/working/processed_data/" # Processed data output directory path
)
patients = np.arange(1, 24)
# Remove patients 4, 6, 7, 12, and 20, as their records contain anomalous data
patients = np.delete(patients, [3, 5, 6, 11, 19])
patients = [1] # TEMP
ictal_interval_padding_duration = 32
# ------------------------------------------------------------------------------
seizure_occurance_period = 30 # Seizure occurrence period (minutes)
seizure_prediction_horizon = 5 # Seizure prediction horizon (minutes)
# -----------------------------------------------------------------------
# ------------------------------------------------------------------------------
extract_ictal_samples = False
extract_preictal_samples = True
generate_synthetic_samples = False
# ------------------------------------------------------------------------------
if __name__ == "__main__":
for patient in patients:
try:
print("Patient: %02d" % patient)
create_dir(processed_data_dir)
(
interictal_intervals,
interictal_files,
ictal_intervals,
ictal_files,
preictal_intervals,
preictal_files,
) = extract_interval_data(
patient,
data_dir,
extract_ictal_samples,
extract_preictal_samples,
ictal_interval_padding_duration,
seizure_occurance_period,
seizure_prediction_horizon,
)
if patient == 19:
# Disregard the first seizure of patient 19 because it is not considered
preictal_intervals.pop(0)
interictal_segment_index = 0
interictal_data = np.array([]).reshape(
n_channels, 0, sample_rate * window_size
)
if extract_ictal_samples:
ictal_segment_index = 0
synthetic_ictal_segment_index = 0
ictal_data = copy.deepcopy(interictal_data)
synthetic_ictal_data = copy.deepcopy(interictal_data)
if extract_preictal_samples:
preictal_segment_index = 0
synthetic_preictal_segment_index = 0
preictal_data = copy.deepcopy(interictal_data)
synthetic_preictal_data = copy.deepcopy(interictal_data)
# Extract interictal samples (batches)
for file in interictal_files:
data, interictal_segment_index = extract_batches(
patient,
file,
data_dir,
interictal_segment_index,
interictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
interictal_data = np.concatenate((interictal_data, data), axis=1)
print("Interictal: ", interictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_interictal.npy" % patient,
),
interictal_data,
)
del interictal_data
if extract_ictal_samples:
# Extract ictal samples (batches)
for file in ictal_files:
data, ictal_segment_index = extract_batches(
patient,
file,
data_dir,
ictal_segment_index,
ictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
ictal_data = np.concatenate((ictal_data, data), axis=1)
print("Ictal: ", ictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_ictal.npy" % patient,
),
ictal_data,
)
del ictal_data
if generate_synthetic_samples:
# Generate synthetic ictal samples (batches)
for file in ictal_files:
data, synthetic_ictal_segment_index = gen_synthetic_batches(
patient,
file,
data_dir,
synthetic_ictal_segment_index,
ictal_intervals,
sample_rate,
window_size,
stride_len,
n_channels,
)
if data.size > 0:
synthetic_ictal_data = np.concatenate(
(synthetic_ictal_data, data), axis=1
)
print("Synthetic Ictal: ", synthetic_ictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_synthetic_ictal.npy" % patient,
),
synthetic_ictal_data,
)
del synthetic_ictal_data
if extract_preictal_samples:
# Extract preictal samples (batches)
for file in preictal_files:
data, preictal_segment_index = extract_batches(
patient,
file,
data_dir,
preictal_segment_index,
preictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
preictal_data = np.concatenate((preictal_data, data), axis=1)
print("Preictal: ", preictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_preictal.npy" % patient,
),
preictal_data,
)
del preictal_data
if generate_synthetic_samples:
# Generate synthetic preictal samples (batches)
for file in preictal_files:
data, synthetic_preictal_segment_index = gen_synthetic_batches(
patient,
file,
data_dir,
synthetic_preictal_segment_index,
preictal_intervals,
sample_rate,
window_size,
stride_len,
n_channels,
)
if data.size > 0:
synthetic_preictal_data = np.concatenate(
(synthetic_preictal_data, data), axis=1
)
print("Synthetic Preictal: ", synthetic_preictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_synthetic_preictal.npy" % patient,
),
synthetic_preictal_data,
)
del synthetic_preictal_data
except Exception as e:
print("Patient: %02d Failed" % patient)
print(e)
# ------------------------------------------------------------------------------
n_channels = 22
sample_rate = 256 # Sample rate (Hz)
window_size = 64 # Window size (seconds)
processed_data_dir = "processed_data" # Processed data output directory path
# Spectogram output directory path
feature_output_dir = "processed_data/features"
create_dir(feature_output_dir)
# ------------------------------------------------------------------------------
def det_entropy(channel_data):
z = np.absolute(channel_data)
entropy = 0
for i in range(len(channel_data)):
entropy = entropy + (z[i] * (math.log(z[i], 2)))
return -1 * entropy
def gen_time_domain_features(data):
features = np.array([])
for channel in range(data.shape[0]):
channel_data = np.absolute(data[channel])
# Stastical moments: Mean, Variance, Skewness, Kurtosis, Coefficient of variation of EEG Signal
features = np.append(features, np.mean(channel_data))
features = np.append(features, np.var(channel_data))
features = np.append(features, skew(channel_data))
features = np.append(features, kurtosis(channel_data))
features = np.append(features, (math.sqrt(features[1]) // features[0]))
# Median absolute deviation of EEG Amplitude and Root Mean Square Amplitude
features = np.append(features, np.mean(np.absolute(data - np.mean(data))))
features = np.append(features, np.sqrt(np.mean(channel_data**2)))
# Shanon Entropy
features = np.append(features, det_entropy(channel_data))
return features.flatten()
def save_features(features, file, feature_output_dir):
"""Method to save features to disk."""
np.save(
os.path.join(
feature_output_dir,
os.path.basename(file),
),
features,
)
if __name__ == "__main__":
files = glob.glob(os.path.join(processed_data_dir, "*.npy"))
for file in files:
print(file)
try:
data = np.load(file)
features = None
for sample in range(data.shape[1]):
sample_features = np.expand_dims(
gen_time_domain_features(data[:, sample, :]), axis=0
)
if features is None:
features = sample_features
else:
features = np.concatenate((features, sample_features), axis=0)
print(features.shape)
save_features(
features,
file,
feature_output_dir=feature_output_dir,
)
del features
except Exception as e:
print("File: %s Failed" % file)
print(e)
# ------------------------------------------------------------------------------
n_channels = 22
sample_rate = 256 # Sample rate (Hz)
window_size = 64 # Window size (seconds)
processed_data_dir = "processed_data" # Processed data output directory path
# Spectogram output directory path
spectogram_output_dir = "processed_data/spectograms"
create_dir(spectogram_output_dir)
# ------------------------------------------------------------------------------
def bandstop_filter(data, f_l, f_h, sample_rate, order=5):
"""Method to emulate a bandstop filter."""
nyquist_rate = 0.5 * sample_rate
b, a = butter(order, [f_l / nyquist_rate, f_h / nyquist_rate], btype="bandstop")
return lfilter(b, a, data)
def highpass_filter(data, f_h, sample_rate, order=5):
"""Method to emulate a highpass filter."""
nyquist_rate = 0.5 * sample_rate
b, a = butter(order, f_h / nyquist_rate, btype="high", analog=False)
return lfilter(b, a, data)
def save_spectograms(spectograms, file, spectogram_output_dir):
"""Method to save spectrograms to disk."""
np.save(
os.path.join(
spectogram_output_dir,
os.path.basename(file),
),
spectograms,
)
def gen_spectograms(
data, sample_rate, window_size, n_channels, spectogram_index=0, debug_plots=False
):
"""Method to segment data into windows and to create spectograms."""
spectograms = []
start = 0
stop = window_size * sample_rate
while stop <= data.shape[1]:
spectograms.append([])
for i in range(0, n_channels):
spectograms[spectogram_index].append(
gen_spectogram(
data[i, start:stop], sample_rate, window_size, debug_plots
)
)
start = stop + 1
stop = start + window_size * sample_rate
spectogram_index += 1
spectograms = np.array(spectograms)
return spectograms, spectogram_index
def gen_spectogram(data, sample_rate, window_size, debug_plot):
"""Method to create a spectogram."""
y = bandstop_filter(data, 117, 123, sample_rate, order=6)
y = bandstop_filter(y, 57, 63, sample_rate, order=6)
y = highpass_filter(y, 1, sample_rate, order=6)
frequencies, bins, Pxx = signal.spectrogram(
y, nfft=sample_rate, fs=sample_rate, return_onesided=True, noverlap=128
)
Pxx = np.delete(Pxx, np.s_[117 : 123 + 1], axis=0)
Pxx = np.delete(Pxx, np.s_[57 : 63 + 1], axis=0)
Pxx = np.delete(Pxx, 0, axis=0)
result = (
10 * np.log10(np.transpose(Pxx)) - (10 * np.log10(np.transpose(Pxx))).min()
) / (10 * np.log10(np.transpose(Pxx))).ptp()
if debug_plot:
plt.figure(1)
frequencies = np.arange(result.shape[1])
plt.pcolormesh(frequencies, bins, result, cmap=plt.cm.jet)
plt.colorbar()
plt.ylabel("Time (s)")
plt.xlabel("Frequency (Hz)")
plt.show()
return result
if __name__ == "__main__":
files = glob.glob(os.path.join(processed_data_dir, "*.npy"))
for file in files:
print(file)
try:
spectogram_index = 0
data = np.load(file)
spectograms = None
for sample in range(data.shape[1]):
sample_spectograms, spectogram_index = gen_spectograms(
data[:, sample, :],
sample_rate,
window_size,
n_channels,
debug_plots=False,
)
if spectograms is None:
spectograms = sample_spectograms
else:
spectograms = np.concatenate(
(spectograms, sample_spectograms), axis=0
)
print(spectograms.shape)
save_spectograms(
spectograms,
file,
spectogram_output_dir=spectogram_output_dir,
)
del spectograms
except Exception as e:
print("File: %s Failed" % file)
print(e)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/069/129069668.ipynb
| null | null |
[{"Id": 129069668, "ScriptId": 38216406, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14975555, "CreationDate": "05/10/2023 18:23:57", "VersionNumber": 3.0, "Title": "parallell cnn", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 729.0, "LinesInsertedFromPrevious": 204.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 525.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/working"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# !pip install pyedflib
import os
from datetime import datetime, timedelta
import pyedflib
import numpy as np
import pandas as pd
from scipy import signal
from scipy.signal import butter, lfilter
import matplotlib.pyplot as plt
from datetime import datetime, timedelta
import os
from math import floor
import copy
import itertools
from scipy.signal import hilbert
from scipy.stats import kurtosis
from scipy.stats import skew
from scipy.fftpack import fft
import math
import glob
def create_dir(directory_path):
"""Method to create a directory. Returns True if the directory already exists."""
if os.path.exists(directory_path):
return True
else:
os.makedirs(directory_path)
return False
def round_down(num, divisor):
"""Method to round down a number"""
return num - (num % divisor)
def get_time(datetime_string):
"""Method to convert a string to a datetime object."""
time = 0
try:
time = datetime.strptime(datetime_string, "%H:%M:%S")
except ValueError:
datetime_string = " " + datetime_string
if " 24" in datetime_string:
datetime_string = datetime_string.replace(" 24", "23")
time = datetime.strptime(datetime_string, "%H:%M:%S")
time += timedelta(hours=1)
else:
datetime_string = datetime_string.replace(" 25", "23")
time = datetime.strptime(datetime_string, "%H:%M:%S")
time += timedelta(hours=2)
return time
def extract_interval_data(
patient,
data_dir,
extract_ictal_samples=True,
extract_preictal_samples=True,
ictal_interval_padding_duration=32,
seizure_occurance_period=30,
seizure_prediction_horizon=5,
):
"""Method to extract interval patient data."""
patient_summary = open(
os.path.join(data_dir, "chb%02d" % patient, "chb%02d-summary.txt" % patient),
"r",
)
interictal_intervals = []
interictal_files = []
ictal_intervals = []
ictal_files = []
preictal_intervals = []
preictal_files = []
line = patient_summary.readline()
start_time = datetime.min
old_time = datetime.min
line_number = 0
while line:
line_data = line.split(":")
if line_data[0] == "File Name":
file_name = line_data[1].strip()
s = get_time(patient_summary.readline().split(": ")[1].strip())
if line_number == 0:
start_time = s
while s < old_time:
s += timedelta(hours=24)
old_time = s
end_time_file = get_time(patient_summary.readline().split(": ")[1].strip())
while end_time_file < old_time:
end_time_file = end_time_file + timedelta(hours=24)
old_time = end_time_file
n_seizures = int(patient_summary.readline().split(": ")[1])
if n_seizures == 0:
# Extract interictal interval data
interictal_intervals.append([s, end_time_file])
interictal_files.append([s, end_time_file, file_name])
else:
# Extract ictal and preictal interval data
for i in range(0, n_seizures):
seconds_start = int(
patient_summary.readline().split(": ")[1].split(" ")[0]
)
seconds_end = int(
patient_summary.readline().split(": ")[1].split(" ")[0]
)
if extract_ictal_samples:
# Extract ictal interval data
interval_start = s + timedelta(seconds=seconds_start)
if (
len(ictal_intervals) == 0 or interval_start > datetime.min
) and interval_start - start_time > timedelta(minutes=20):
interval_end = s + timedelta(seconds=seconds_end)
ictal_intervals.append(
[
interval_start
- timedelta(
seconds=ictal_interval_padding_duration
),
interval_end
+ timedelta(
seconds=ictal_interval_padding_duration
),
]
)
ictal_files.append([s, end_time_file, file_name])
if extract_preictal_samples:
# Extract preictal interval data
interval_start = (
s
+ timedelta(seconds=seconds_start)
- timedelta(
minutes=seizure_prediction_horizon
+ seizure_occurance_period
)
)
if (
len(preictal_intervals) == 0
or interval_start > datetime.min
) and interval_start - start_time > timedelta(minutes=20):
interval_end = interval_start + timedelta(
minutes=seizure_occurance_period
)
preictal_intervals.append([interval_start, interval_end])
preictal_files.append([s, end_time_file, file_name])
line = patient_summary.readline()
line_number += 1
patient_summary.close()
return (
interictal_intervals,
interictal_files,
ictal_intervals,
ictal_files,
preictal_intervals,
preictal_files,
)
def load_patient_data(patient, file, data_dir):
"""Method to load patient data."""
f = pyedflib.EdfReader("%schb%02d/%s" % (data_dir, patient, file))
n = f.signals_in_file
signals = np.zeros((n, f.getNSamples()[0]))
for i in np.arange(n):
signals[i, :] = f.readSignal(i)
return signals
def extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
interval_start,
interval_end,
segment_index,
n_channels,
):
"""Method to extract batch samples from specified intervals."""
start = 0
if file_start < interval_start:
start = (interval_start - file_start).seconds * sample_rate
if file_end <= interval_end:
end = -1
data = load_patient_data(patient, file[2], data_dir)[:, start:]
else:
end = ((interval_end - file_start).seconds * sample_rate) + 1
data = load_patient_data(patient, file[2], data_dir)[:, start : end + 1]
if (data.shape[0] >= n_channels) and (data.shape[1] >= sample_rate * window_size):
truncated_len = round_down(data.shape[1], sample_rate * window_size)
return (
np.array(
np.split(
data[0:n_channels, 0:truncated_len],
truncated_len / (sample_rate * window_size),
axis=1,
)
).swapaxes(0, 1),
segment_index,
)
else:
return np.array([]), segment_index
def extract_batches(
patient,
file,
data_dir,
segment_index,
intervals,
sample_rate,
window_size,
n_channels,
):
"""Method to extract batches."""
file_start = file[0]
file_end = file[1]
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
while file_start > interval_end and segment_index < len(intervals) - 1:
segment_index += 1
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
if (interval_end - interval_start).seconds >= window_size:
return extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
interval_start,
interval_end,
segment_index,
n_channels,
)
else:
return np.array([]), segment_index
def gen_synthetic_batches(
patient,
file,
data_dir,
segment_index,
intervals,
sample_rate,
window_size,
stride_len,
n_channels,
):
"""Method to generate synthetic batches."""
file_start = file[0]
file_end = file[1]
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
while file_start > interval_end and segment_index < len(intervals) - 1:
segment_index += 1
interval_start = intervals[segment_index][0]
interval_end = intervals[segment_index][1]
if (interval_end - interval_start).seconds > window_size:
synthetic_batches = np.array([]).reshape(
n_channels, 0, sample_rate * window_size
)
synthetic_interval_start = interval_start + timedelta(seconds=stride_len)
synthetic_interval_end = synthetic_interval_start + timedelta(
seconds=window_size
)
while synthetic_interval_end < interval_end:
extracted_batches = extract_batches_from_interval(
patient,
data_dir,
file,
file_start,
file_end,
synthetic_interval_start,
synthetic_interval_end,
segment_index,
n_channels,
)[0]
if extracted_batches.size > 0:
synthetic_batches = np.concatenate(
(synthetic_batches, extracted_batches), axis=1
)
synthetic_interval_start += timedelta(seconds=stride_len)
synthetic_interval_end += timedelta(seconds=stride_len)
return synthetic_batches, segment_index
else:
return np.array([]), segment_index
os.path.exists("/kaggle/working/processed_data")
n_channels = 22
sample_rate = 256 # Sample rate (Hz)
window_size = 64 # Window size (seconds)
# Stride length (seconds) used to generate synthetic preictal and ictal samples
stride_len = 32
# Data directory path
# data_dir = "/scratch/jcu/cl/CHBMIT/chb-mit-scalp-eeg-database-1.0.0/"
data_dir = "/kaggle/input/chb01-21/chbmit/"
processed_data_dir = (
"/kaggle/working/processed_data/" # Processed data output directory path
)
patients = np.arange(1, 24)
# Remove patients 4, 6, 7, 12, and 20, as their records contain anomalous data
patients = np.delete(patients, [3, 5, 6, 11, 19])
patients = [1] # TEMP
ictal_interval_padding_duration = 32
# ------------------------------------------------------------------------------
seizure_occurance_period = 30 # Seizure occurrence period (minutes)
seizure_prediction_horizon = 5 # Seizure prediction horizon (minutes)
# -----------------------------------------------------------------------
# ------------------------------------------------------------------------------
extract_ictal_samples = False
extract_preictal_samples = True
generate_synthetic_samples = False
# ------------------------------------------------------------------------------
if __name__ == "__main__":
for patient in patients:
try:
print("Patient: %02d" % patient)
create_dir(processed_data_dir)
(
interictal_intervals,
interictal_files,
ictal_intervals,
ictal_files,
preictal_intervals,
preictal_files,
) = extract_interval_data(
patient,
data_dir,
extract_ictal_samples,
extract_preictal_samples,
ictal_interval_padding_duration,
seizure_occurance_period,
seizure_prediction_horizon,
)
if patient == 19:
# Disregard the first seizure of patient 19 because it is not considered
preictal_intervals.pop(0)
interictal_segment_index = 0
interictal_data = np.array([]).reshape(
n_channels, 0, sample_rate * window_size
)
if extract_ictal_samples:
ictal_segment_index = 0
synthetic_ictal_segment_index = 0
ictal_data = copy.deepcopy(interictal_data)
synthetic_ictal_data = copy.deepcopy(interictal_data)
if extract_preictal_samples:
preictal_segment_index = 0
synthetic_preictal_segment_index = 0
preictal_data = copy.deepcopy(interictal_data)
synthetic_preictal_data = copy.deepcopy(interictal_data)
# Extract interictal samples (batches)
for file in interictal_files:
data, interictal_segment_index = extract_batches(
patient,
file,
data_dir,
interictal_segment_index,
interictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
interictal_data = np.concatenate((interictal_data, data), axis=1)
print("Interictal: ", interictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_interictal.npy" % patient,
),
interictal_data,
)
del interictal_data
if extract_ictal_samples:
# Extract ictal samples (batches)
for file in ictal_files:
data, ictal_segment_index = extract_batches(
patient,
file,
data_dir,
ictal_segment_index,
ictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
ictal_data = np.concatenate((ictal_data, data), axis=1)
print("Ictal: ", ictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_ictal.npy" % patient,
),
ictal_data,
)
del ictal_data
if generate_synthetic_samples:
# Generate synthetic ictal samples (batches)
for file in ictal_files:
data, synthetic_ictal_segment_index = gen_synthetic_batches(
patient,
file,
data_dir,
synthetic_ictal_segment_index,
ictal_intervals,
sample_rate,
window_size,
stride_len,
n_channels,
)
if data.size > 0:
synthetic_ictal_data = np.concatenate(
(synthetic_ictal_data, data), axis=1
)
print("Synthetic Ictal: ", synthetic_ictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_synthetic_ictal.npy" % patient,
),
synthetic_ictal_data,
)
del synthetic_ictal_data
if extract_preictal_samples:
# Extract preictal samples (batches)
for file in preictal_files:
data, preictal_segment_index = extract_batches(
patient,
file,
data_dir,
preictal_segment_index,
preictal_intervals,
sample_rate,
window_size,
n_channels,
)
if data.size > 0:
preictal_data = np.concatenate((preictal_data, data), axis=1)
print("Preictal: ", preictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_preictal.npy" % patient,
),
preictal_data,
)
del preictal_data
if generate_synthetic_samples:
# Generate synthetic preictal samples (batches)
for file in preictal_files:
data, synthetic_preictal_segment_index = gen_synthetic_batches(
patient,
file,
data_dir,
synthetic_preictal_segment_index,
preictal_intervals,
sample_rate,
window_size,
stride_len,
n_channels,
)
if data.size > 0:
synthetic_preictal_data = np.concatenate(
(synthetic_preictal_data, data), axis=1
)
print("Synthetic Preictal: ", synthetic_preictal_data.shape)
np.save(
os.path.join(
processed_data_dir,
"CHBMIT_patient_%02d_synthetic_preictal.npy" % patient,
),
synthetic_preictal_data,
)
del synthetic_preictal_data
except Exception as e:
print("Patient: %02d Failed" % patient)
print(e)
# ------------------------------------------------------------------------------
n_channels = 22
sample_rate = 256 # Sample rate (Hz)
window_size = 64 # Window size (seconds)
processed_data_dir = "processed_data" # Processed data output directory path
# Spectogram output directory path
feature_output_dir = "processed_data/features"
create_dir(feature_output_dir)
# ------------------------------------------------------------------------------
def det_entropy(channel_data):
z = np.absolute(channel_data)
entropy = 0
for i in range(len(channel_data)):
entropy = entropy + (z[i] * (math.log(z[i], 2)))
return -1 * entropy
def gen_time_domain_features(data):
features = np.array([])
for channel in range(data.shape[0]):
channel_data = np.absolute(data[channel])
# Stastical moments: Mean, Variance, Skewness, Kurtosis, Coefficient of variation of EEG Signal
features = np.append(features, np.mean(channel_data))
features = np.append(features, np.var(channel_data))
features = np.append(features, skew(channel_data))
features = np.append(features, kurtosis(channel_data))
features = np.append(features, (math.sqrt(features[1]) // features[0]))
# Median absolute deviation of EEG Amplitude and Root Mean Square Amplitude
features = np.append(features, np.mean(np.absolute(data - np.mean(data))))
features = np.append(features, np.sqrt(np.mean(channel_data**2)))
# Shanon Entropy
features = np.append(features, det_entropy(channel_data))
return features.flatten()
def save_features(features, file, feature_output_dir):
"""Method to save features to disk."""
np.save(
os.path.join(
feature_output_dir,
os.path.basename(file),
),
features,
)
if __name__ == "__main__":
files = glob.glob(os.path.join(processed_data_dir, "*.npy"))
for file in files:
print(file)
try:
data = np.load(file)
features = None
for sample in range(data.shape[1]):
sample_features = np.expand_dims(
gen_time_domain_features(data[:, sample, :]), axis=0
)
if features is None:
features = sample_features
else:
features = np.concatenate((features, sample_features), axis=0)
print(features.shape)
save_features(
features,
file,
feature_output_dir=feature_output_dir,
)
del features
except Exception as e:
print("File: %s Failed" % file)
print(e)
# ------------------------------------------------------------------------------
n_channels = 22
sample_rate = 256 # Sample rate (Hz)
window_size = 64 # Window size (seconds)
processed_data_dir = "processed_data" # Processed data output directory path
# Spectogram output directory path
spectogram_output_dir = "processed_data/spectograms"
create_dir(spectogram_output_dir)
# ------------------------------------------------------------------------------
def bandstop_filter(data, f_l, f_h, sample_rate, order=5):
"""Method to emulate a bandstop filter."""
nyquist_rate = 0.5 * sample_rate
b, a = butter(order, [f_l / nyquist_rate, f_h / nyquist_rate], btype="bandstop")
return lfilter(b, a, data)
def highpass_filter(data, f_h, sample_rate, order=5):
"""Method to emulate a highpass filter."""
nyquist_rate = 0.5 * sample_rate
b, a = butter(order, f_h / nyquist_rate, btype="high", analog=False)
return lfilter(b, a, data)
def save_spectograms(spectograms, file, spectogram_output_dir):
"""Method to save spectrograms to disk."""
np.save(
os.path.join(
spectogram_output_dir,
os.path.basename(file),
),
spectograms,
)
def gen_spectograms(
data, sample_rate, window_size, n_channels, spectogram_index=0, debug_plots=False
):
"""Method to segment data into windows and to create spectograms."""
spectograms = []
start = 0
stop = window_size * sample_rate
while stop <= data.shape[1]:
spectograms.append([])
for i in range(0, n_channels):
spectograms[spectogram_index].append(
gen_spectogram(
data[i, start:stop], sample_rate, window_size, debug_plots
)
)
start = stop + 1
stop = start + window_size * sample_rate
spectogram_index += 1
spectograms = np.array(spectograms)
return spectograms, spectogram_index
def gen_spectogram(data, sample_rate, window_size, debug_plot):
"""Method to create a spectogram."""
y = bandstop_filter(data, 117, 123, sample_rate, order=6)
y = bandstop_filter(y, 57, 63, sample_rate, order=6)
y = highpass_filter(y, 1, sample_rate, order=6)
frequencies, bins, Pxx = signal.spectrogram(
y, nfft=sample_rate, fs=sample_rate, return_onesided=True, noverlap=128
)
Pxx = np.delete(Pxx, np.s_[117 : 123 + 1], axis=0)
Pxx = np.delete(Pxx, np.s_[57 : 63 + 1], axis=0)
Pxx = np.delete(Pxx, 0, axis=0)
result = (
10 * np.log10(np.transpose(Pxx)) - (10 * np.log10(np.transpose(Pxx))).min()
) / (10 * np.log10(np.transpose(Pxx))).ptp()
if debug_plot:
plt.figure(1)
frequencies = np.arange(result.shape[1])
plt.pcolormesh(frequencies, bins, result, cmap=plt.cm.jet)
plt.colorbar()
plt.ylabel("Time (s)")
plt.xlabel("Frequency (Hz)")
plt.show()
return result
if __name__ == "__main__":
files = glob.glob(os.path.join(processed_data_dir, "*.npy"))
for file in files:
print(file)
try:
spectogram_index = 0
data = np.load(file)
spectograms = None
for sample in range(data.shape[1]):
sample_spectograms, spectogram_index = gen_spectograms(
data[:, sample, :],
sample_rate,
window_size,
n_channels,
debug_plots=False,
)
if spectograms is None:
spectograms = sample_spectograms
else:
spectograms = np.concatenate(
(spectograms, sample_spectograms), axis=0
)
print(spectograms.shape)
save_spectograms(
spectograms,
file,
spectogram_output_dir=spectogram_output_dir,
)
del spectograms
except Exception as e:
print("File: %s Failed" % file)
print(e)
| false | 0 | 6,224 | 0 | 6,224 | 6,224 |
||
129069101
|
import numpy as np
import torch
import matplotlib.pyplot as plt
import cv2
import warnings
import sys
from segment_anything import sam_model_registry, SamPredictor
warnings.filterwarnings("ignore")
def show_mask(mask, ax, random_color=False):
if random_color:
color = np.concatenate([np.random.random(3), np.array([0.6])], axis=0)
else:
color = np.array([30 / 255, 144 / 255, 255 / 255, 0.6])
h, w = mask.shape[-2:]
mask_image = mask.reshape(h, w, 1) * color.reshape(1, 1, -1)
ax.imshow(mask_image)
def show_points(coords, labels, ax, marker_size=375):
pos_points = coords[labels == 1]
neg_points = coords[labels == 0]
ax.scatter(
pos_points[:, 0],
pos_points[:, 1],
color="green",
marker="*",
s=marker_size,
edgecolor="white",
linewidth=1.25,
)
ax.scatter(
neg_points[:, 0],
neg_points[:, 1],
color="red",
marker="*",
s=marker_size,
edgecolor="white",
linewidth=1.25,
)
def show_box(box, ax):
x0, y0 = box[0], box[1]
w, h = box[2] - box[0], box[3] - box[1]
ax.add_patch(
plt.Rectangle((x0, y0), w, h, edgecolor="green", facecolor=(0, 0, 0, 0), lw=2)
)
# # Model Setup
sys.path.append("..")
sam_checkpoint = "sam_vit_h_4b8939.pth"
model_type = "vit_h"
device = "cuda"
sam = sam_model_registry[model_type](checkpoint=sam_checkpoint)
sam.to(device=device)
predictor = SamPredictor(sam)
model = torch.hub.load("ultralytics/yolov5", "yolov5s")
path = "/kaggle/input/oz-sports/input2.png"
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
plt.figure(figsize=(10, 10))
plt.imshow(image)
plt.axis("off")
plt.show()
result = model(path)
result.show()
# This help us to get xyxy co-ordinate of horse which is class 17
result.pandas().xyxy[0][result.pandas().xyxy[0]["class"] == 17].to_numpy()[0][:4]
predictor.set_image(image)
masks, scores, logits = predictor.predict(
box=result.pandas()
.xyxy[0][result.pandas().xyxy[0]["class"] == 17]
.to_numpy()[0][:4],
multimask_output=False,
)
plt.figure(figsize=(10, 10))
plt.imshow(image)
show_mask(masks[0], plt.gca())
plt.axis("off")
plt.show()
def Mask_detect(path):
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
result = model(path)
predictor.set_image(image)
masks, scores, logits = predictor.predict(
box=result.pandas()
.xyxy[0][result.pandas().xyxy[0]["class"] == 17]
.to_numpy()[0][:4],
multimask_output=False,
)
plt.figure(figsize=(10, 10))
plt.imshow(image)
show_mask(masks[0], plt.gca())
plt.axis("off")
plt.show()
# # Testing on remaining images
for i in range(2, 7):
print("Image no:", i)
path = f"/kaggle/input/oz-sports/input{i}.png"
Mask_detect(path)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/069/129069101.ipynb
| null | null |
[{"Id": 129069101, "ScriptId": 38345673, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11511337, "CreationDate": "05/10/2023 18:17:31", "VersionNumber": 1.0, "Title": "OZ_soprts", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 98.0, "LinesInsertedFromPrevious": 98.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
import numpy as np
import torch
import matplotlib.pyplot as plt
import cv2
import warnings
import sys
from segment_anything import sam_model_registry, SamPredictor
warnings.filterwarnings("ignore")
def show_mask(mask, ax, random_color=False):
if random_color:
color = np.concatenate([np.random.random(3), np.array([0.6])], axis=0)
else:
color = np.array([30 / 255, 144 / 255, 255 / 255, 0.6])
h, w = mask.shape[-2:]
mask_image = mask.reshape(h, w, 1) * color.reshape(1, 1, -1)
ax.imshow(mask_image)
def show_points(coords, labels, ax, marker_size=375):
pos_points = coords[labels == 1]
neg_points = coords[labels == 0]
ax.scatter(
pos_points[:, 0],
pos_points[:, 1],
color="green",
marker="*",
s=marker_size,
edgecolor="white",
linewidth=1.25,
)
ax.scatter(
neg_points[:, 0],
neg_points[:, 1],
color="red",
marker="*",
s=marker_size,
edgecolor="white",
linewidth=1.25,
)
def show_box(box, ax):
x0, y0 = box[0], box[1]
w, h = box[2] - box[0], box[3] - box[1]
ax.add_patch(
plt.Rectangle((x0, y0), w, h, edgecolor="green", facecolor=(0, 0, 0, 0), lw=2)
)
# # Model Setup
sys.path.append("..")
sam_checkpoint = "sam_vit_h_4b8939.pth"
model_type = "vit_h"
device = "cuda"
sam = sam_model_registry[model_type](checkpoint=sam_checkpoint)
sam.to(device=device)
predictor = SamPredictor(sam)
model = torch.hub.load("ultralytics/yolov5", "yolov5s")
path = "/kaggle/input/oz-sports/input2.png"
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
plt.figure(figsize=(10, 10))
plt.imshow(image)
plt.axis("off")
plt.show()
result = model(path)
result.show()
# This help us to get xyxy co-ordinate of horse which is class 17
result.pandas().xyxy[0][result.pandas().xyxy[0]["class"] == 17].to_numpy()[0][:4]
predictor.set_image(image)
masks, scores, logits = predictor.predict(
box=result.pandas()
.xyxy[0][result.pandas().xyxy[0]["class"] == 17]
.to_numpy()[0][:4],
multimask_output=False,
)
plt.figure(figsize=(10, 10))
plt.imshow(image)
show_mask(masks[0], plt.gca())
plt.axis("off")
plt.show()
def Mask_detect(path):
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
result = model(path)
predictor.set_image(image)
masks, scores, logits = predictor.predict(
box=result.pandas()
.xyxy[0][result.pandas().xyxy[0]["class"] == 17]
.to_numpy()[0][:4],
multimask_output=False,
)
plt.figure(figsize=(10, 10))
plt.imshow(image)
show_mask(masks[0], plt.gca())
plt.axis("off")
plt.show()
# # Testing on remaining images
for i in range(2, 7):
print("Image no:", i)
path = f"/kaggle/input/oz-sports/input{i}.png"
Mask_detect(path)
| false | 0 | 1,037 | 1 | 1,037 | 1,037 |
||
129069407
|
<jupyter_start><jupyter_text>Melanoma Skin Cancer Dataset of 10000 Images
Melanoma Skin Cancer Dataset contains 10000 images. Melanoma skin cancer is deadly cancer, early detection and cure can save many lives. This dataset will be useful for developing the deep learning models for accurate classification of melanoma. Dataset consists of 9600 images for training the model and 1000 images for evaluation of model.
Kaggle dataset identifier: melanoma-skin-cancer-dataset-of-10000-images
<jupyter_script>import numpy as np
import pandas as pd
import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.applications.inception_v3 import InceptionV3
from tensorflow.keras.layers import Dense, GlobalAveragePooling2D
from tensorflow.keras.models import Model
train_path = "/kaggle/input/melanoma-skin-cancer-dataset-of-10000-images/melanoma_cancer_dataset/train"
valid_path = "/kaggle/input/melanoma-skin-cancer-dataset-of-10000-images/melanoma_cancer_dataset/test"
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
rotation_range=30,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode="nearest",
)
valid_datagen = ImageDataGenerator(rescale=1.0 / 255)
base_model = InceptionV3(include_top=False, weights=None, input_shape=(75, 75, 3))
base_model.load_weights(
"/kaggle/input/inceptionv3-model-notop/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5"
)
x = base_model.output
x = GlobalAveragePooling2D()(x)
x = Dense(1024, activation="relu")(x)
predictions = Dense(2, activation="softmax")(x)
model = Model(inputs=base_model.input, outputs=predictions)
for layer in base_model.layers:
layer.trainable = False
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
batch_size = 32
epochs = 10
train_datagen = ImageDataGenerator(
rescale=1.0 / 255, shear_range=0.2, zoom_range=0.2, horizontal_flip=True
)
train_generator = train_datagen.flow_from_directory(
train_path, target_size=(299, 299), batch_size=batch_size, class_mode="categorical"
)
valid_datagen = ImageDataGenerator(rescale=1.0 / 255)
valid_generator = valid_datagen.flow_from_directory(
valid_path, target_size=(299, 299), batch_size=batch_size, class_mode="categorical"
)
history = model.fit(
train_generator, epochs=epochs, validation_data=valid_generator, verbose=1
)
model.save("skin_cancer_inceptionv3.h5")
os.chdir(r"/kaggle/working")
from IPython.display import FileLink
FileLink(r"/kaggle/working/skin_cancer_inceptionv3.h5")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/069/129069407.ipynb
|
melanoma-skin-cancer-dataset-of-10000-images
|
hasnainjaved
|
[{"Id": 129069407, "ScriptId": 38345340, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11063726, "CreationDate": "05/10/2023 18:21:00", "VersionNumber": 1.0, "Title": "dacclab-svm", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 85.0, "LinesInsertedFromPrevious": 78.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 7.0, "LinesInsertedFromFork": 78.0, "LinesDeletedFromFork": 50.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 7.0, "TotalVotes": 0}]
|
[{"Id": 184799149, "KernelVersionId": 129069407, "SourceDatasetVersionId": 3376422}]
|
[{"Id": 3376422, "DatasetId": 2035877, "DatasourceVersionId": 3427850, "CreatorUserId": 5555301, "LicenseName": "CC0: Public Domain", "CreationDate": "03/29/2022 11:51:15", "VersionNumber": 1.0, "Title": "Melanoma Skin Cancer Dataset of 10000 Images", "Slug": "melanoma-skin-cancer-dataset-of-10000-images", "Subtitle": "This dataset contains two classes of melanoma cancer, malignant and benign.", "Description": "Melanoma Skin Cancer Dataset contains 10000 images. Melanoma skin cancer is deadly cancer, early detection and cure can save many lives. This dataset will be useful for developing the deep learning models for accurate classification of melanoma. Dataset consists of 9600 images for training the model and 1000 images for evaluation of model.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2035877, "CreatorUserId": 5555301, "OwnerUserId": 5555301.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3376422.0, "CurrentDatasourceVersionId": 3427850.0, "ForumId": 2060773, "Type": 2, "CreationDate": "03/29/2022 11:51:15", "LastActivityDate": "03/29/2022", "TotalViews": 19436, "TotalDownloads": 2801, "TotalVotes": 43, "TotalKernels": 17}]
|
[{"Id": 5555301, "UserName": "hasnainjaved", "DisplayName": "Muhammad Hasnain Javid", "RegisterDate": "07/31/2020", "PerformanceTier": 0}]
|
import numpy as np
import pandas as pd
import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.applications.inception_v3 import InceptionV3
from tensorflow.keras.layers import Dense, GlobalAveragePooling2D
from tensorflow.keras.models import Model
train_path = "/kaggle/input/melanoma-skin-cancer-dataset-of-10000-images/melanoma_cancer_dataset/train"
valid_path = "/kaggle/input/melanoma-skin-cancer-dataset-of-10000-images/melanoma_cancer_dataset/test"
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
rotation_range=30,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode="nearest",
)
valid_datagen = ImageDataGenerator(rescale=1.0 / 255)
base_model = InceptionV3(include_top=False, weights=None, input_shape=(75, 75, 3))
base_model.load_weights(
"/kaggle/input/inceptionv3-model-notop/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5"
)
x = base_model.output
x = GlobalAveragePooling2D()(x)
x = Dense(1024, activation="relu")(x)
predictions = Dense(2, activation="softmax")(x)
model = Model(inputs=base_model.input, outputs=predictions)
for layer in base_model.layers:
layer.trainable = False
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
batch_size = 32
epochs = 10
train_datagen = ImageDataGenerator(
rescale=1.0 / 255, shear_range=0.2, zoom_range=0.2, horizontal_flip=True
)
train_generator = train_datagen.flow_from_directory(
train_path, target_size=(299, 299), batch_size=batch_size, class_mode="categorical"
)
valid_datagen = ImageDataGenerator(rescale=1.0 / 255)
valid_generator = valid_datagen.flow_from_directory(
valid_path, target_size=(299, 299), batch_size=batch_size, class_mode="categorical"
)
history = model.fit(
train_generator, epochs=epochs, validation_data=valid_generator, verbose=1
)
model.save("skin_cancer_inceptionv3.h5")
os.chdir(r"/kaggle/working")
from IPython.display import FileLink
FileLink(r"/kaggle/working/skin_cancer_inceptionv3.h5")
| false | 0 | 769 | 0 | 903 | 769 |
||
129164287
|
<jupyter_start><jupyter_text>Diabetes
**The dataset you are referring to is commonly known as the "Pima Indians Diabetes Database" and contains information on several medical and demographic factors for a group of Pima Indian women, some of whom have been diagnosed with diabetes. Here is an explanation of each column in the dataset:**
1. Pregnancies: This column represents the number of times the woman has been pregnant.
2. Glucose: This column represents the woman's plasma glucose concentration, measured in milligrams per deciliter (mg/dL).
3. BloodPressure: This column represents the woman's diastolic blood pressure, measured in millimeters of mercury (mmHg).
4. SkinThickness: This column represents the thickness of the woman's skinfold at the triceps, measured in millimeters (mm).
5. Insulin: This column represents the woman's serum insulin level, measured in microunits per milliliter (μU/mL).
6. BMI: This column represents the woman's body mass index (BMI), which is calculated as weight in kilograms divided by height in meters squared (kg/m²).
7. Outcome: This column represents whether or not the woman has been diagnosed with diabetes. A value of 0 indicates that the woman does not have diabetes, while a value of 1 indicates that she does.
This dataset is commonly used as a benchmark for machine learning models that aim to predict whether or not a woman will be diagnosed with diabetes based on these medical and demographic factors. By analyzing the relationships between these factors and the presence of diabetes, researchers can develop models that can potentially help healthcare providers to identify patients who are at higher risk of developing the disease and take preventative measures.
Kaggle dataset identifier: diabetes
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # KNN - Predict whether a person will have diabetes or not
# ## Import important librarys
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split ## split data
from sklearn.preprocessing import StandardScaler ## preprosseccing
from sklearn.neighbors import KNeighborsClassifier ## model _classfiaction
from sklearn.metrics import confusion_matrix
from sklearn.metrics import f1_score
from sklearn.metrics import accuracy_score
# ### read dataset
df = pd.read_csv("/kaggle/input/diabetes/diabetes.csv")
df.head()
# ### Check data cleaning or not
(df.isnull().sum()) / len(df), df.isnull().sum()
# # EDA
df.describe()
df.hist(figsize=(10, 8))
plt.show()
plt.figure(figsize=(12, 8))
sns.heatmap(df.corr(), annot=True, cmap="coolwarm")
plt.show()
df["Outcome"].value_counts().plot(kind="bar", figsize=(8, 6))
plt.show()
df.boxplot(figsize=(12, 8))
plt.show()
fig, axes = plt.subplots(nrows=2, ncols=4, figsize=(18, 8))
for i, column in enumerate(df.columns[:-1]):
ax = axes[int(i / 4), i % 4]
sns.violinplot(x="Outcome", y=column, data=df, ax=ax)
plt.show()
df.plot(kind="density", subplots=True, layout=(3, 3), sharex=False, figsize=(15, 8))
plt.show()
sns.pairplot(df, hue="Outcome", diag_kind="hist")
plt.show()
# # MODEL
# Replace zeroes
zero_not_accepted = ["Glucose", "BloodPressure", "SkinThickness", "BMI", "Insulin"]
for column in zero_not_accepted:
df[column] = df[column].replace(0, np.NaN)
mean = int(df[column].mean(skipna=True))
df[column] = df[column].replace(np.NaN, mean)
# split dataset
X = df.iloc[:, 0:8]
y = df.iloc[:, 8]
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0, test_size=0.2)
print(len(X_train))
print(len(y_train))
print(len(X_test))
print(len(y_test))
# Feature scaling
sc_X = StandardScaler()
X_train = sc_X.fit_transform(X_train)
X_test = sc_X.transform(X_test)
# ### KNN
# Define the model: Init K-NN
classifier = KNeighborsClassifier(n_neighbors=11, p=2, metric="euclidean")
# Fit Model
classifier.fit(X_train, y_train)
# Predict the test set results
y_pred = classifier.predict(X_test)
y_pred
# Evaluate Model
cm = confusion_matrix(y_test, y_pred)
print(cm)
print(f1_score(y_test, y_pred))
print(accuracy_score(y_test, y_pred))
# ## accuracy_score = 0.8181818181818182
from sklearn.metrics import roc_curve, auc
classifier.fit(X_train, y_train)
y_pred_prob = classifier.predict_proba(X_test)[:, 1]
fpr, tpr, thresholds = roc_curve(y_test, y_pred_prob)
roc_auc = auc(fpr, tpr)
plt.plot(fpr, tpr, lw=1, label="ROC (AUC = %0.2f)" % (roc_auc))
plt.plot([0, 1], [0, 1], linestyle="--", lw=2, color="r", label="Random")
plt.xlim([-0.05, 1.05])
plt.ylim([-0.05, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver operating characteristic (ROC) curve")
plt.legend(loc="lower right")
plt.show()
# ## RandomForestClassifier
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier()
model.fit(X_train, y_train)
importance = model.feature_importances_
plt.bar(X.columns, importance)
plt.xticks(rotation=90)
plt.show()
y_pred = model.predict(X_test)
y_pred
print(accuracy_score(y_test, y_pred))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/164/129164287.ipynb
|
diabetes
|
yossefazam
|
[{"Id": 129164287, "ScriptId": 38397998, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10267841, "CreationDate": "05/11/2023 13:17:49", "VersionNumber": 1.0, "Title": "EDA_KNN_RandomForestClassifier", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 152.0, "LinesInsertedFromPrevious": 152.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 10}]
|
[{"Id": 184968890, "KernelVersionId": 129164287, "SourceDatasetVersionId": 5663031}]
|
[{"Id": 5663031, "DatasetId": 3255026, "DatasourceVersionId": 5738484, "CreatorUserId": 10267841, "LicenseName": "Unknown", "CreationDate": "05/11/2023 12:30:49", "VersionNumber": 1.0, "Title": "Diabetes", "Slug": "diabetes", "Subtitle": NaN, "Description": "**The dataset you are referring to is commonly known as the \"Pima Indians Diabetes Database\" and contains information on several medical and demographic factors for a group of Pima Indian women, some of whom have been diagnosed with diabetes. Here is an explanation of each column in the dataset:**\n\n1. Pregnancies: This column represents the number of times the woman has been pregnant.\n\n2. Glucose: This column represents the woman's plasma glucose concentration, measured in milligrams per deciliter (mg/dL).\n\n3. BloodPressure: This column represents the woman's diastolic blood pressure, measured in millimeters of mercury (mmHg).\n\n4. SkinThickness: This column represents the thickness of the woman's skinfold at the triceps, measured in millimeters (mm).\n\n5. Insulin: This column represents the woman's serum insulin level, measured in microunits per milliliter (\u03bcU/mL).\n\n6. BMI: This column represents the woman's body mass index (BMI), which is calculated as weight in kilograms divided by height in meters squared (kg/m\u00b2).\n\n7. Outcome: This column represents whether or not the woman has been diagnosed with diabetes. A value of 0 indicates that the woman does not have diabetes, while a value of 1 indicates that she does.\n\nThis dataset is commonly used as a benchmark for machine learning models that aim to predict whether or not a woman will be diagnosed with diabetes based on these medical and demographic factors. By analyzing the relationships between these factors and the presence of diabetes, researchers can develop models that can potentially help healthcare providers to identify patients who are at higher risk of developing the disease and take preventative measures.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3255026, "CreatorUserId": 10267841, "OwnerUserId": 10267841.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5663031.0, "CurrentDatasourceVersionId": 5738484.0, "ForumId": 3320484, "Type": 2, "CreationDate": "05/11/2023 12:30:49", "LastActivityDate": "05/11/2023", "TotalViews": 313, "TotalDownloads": 25, "TotalVotes": 5, "TotalKernels": 1}]
|
[{"Id": 10267841, "UserName": "yossefazam", "DisplayName": "Youssef Azam", "RegisterDate": "04/17/2022", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # KNN - Predict whether a person will have diabetes or not
# ## Import important librarys
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split ## split data
from sklearn.preprocessing import StandardScaler ## preprosseccing
from sklearn.neighbors import KNeighborsClassifier ## model _classfiaction
from sklearn.metrics import confusion_matrix
from sklearn.metrics import f1_score
from sklearn.metrics import accuracy_score
# ### read dataset
df = pd.read_csv("/kaggle/input/diabetes/diabetes.csv")
df.head()
# ### Check data cleaning or not
(df.isnull().sum()) / len(df), df.isnull().sum()
# # EDA
df.describe()
df.hist(figsize=(10, 8))
plt.show()
plt.figure(figsize=(12, 8))
sns.heatmap(df.corr(), annot=True, cmap="coolwarm")
plt.show()
df["Outcome"].value_counts().plot(kind="bar", figsize=(8, 6))
plt.show()
df.boxplot(figsize=(12, 8))
plt.show()
fig, axes = plt.subplots(nrows=2, ncols=4, figsize=(18, 8))
for i, column in enumerate(df.columns[:-1]):
ax = axes[int(i / 4), i % 4]
sns.violinplot(x="Outcome", y=column, data=df, ax=ax)
plt.show()
df.plot(kind="density", subplots=True, layout=(3, 3), sharex=False, figsize=(15, 8))
plt.show()
sns.pairplot(df, hue="Outcome", diag_kind="hist")
plt.show()
# # MODEL
# Replace zeroes
zero_not_accepted = ["Glucose", "BloodPressure", "SkinThickness", "BMI", "Insulin"]
for column in zero_not_accepted:
df[column] = df[column].replace(0, np.NaN)
mean = int(df[column].mean(skipna=True))
df[column] = df[column].replace(np.NaN, mean)
# split dataset
X = df.iloc[:, 0:8]
y = df.iloc[:, 8]
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0, test_size=0.2)
print(len(X_train))
print(len(y_train))
print(len(X_test))
print(len(y_test))
# Feature scaling
sc_X = StandardScaler()
X_train = sc_X.fit_transform(X_train)
X_test = sc_X.transform(X_test)
# ### KNN
# Define the model: Init K-NN
classifier = KNeighborsClassifier(n_neighbors=11, p=2, metric="euclidean")
# Fit Model
classifier.fit(X_train, y_train)
# Predict the test set results
y_pred = classifier.predict(X_test)
y_pred
# Evaluate Model
cm = confusion_matrix(y_test, y_pred)
print(cm)
print(f1_score(y_test, y_pred))
print(accuracy_score(y_test, y_pred))
# ## accuracy_score = 0.8181818181818182
from sklearn.metrics import roc_curve, auc
classifier.fit(X_train, y_train)
y_pred_prob = classifier.predict_proba(X_test)[:, 1]
fpr, tpr, thresholds = roc_curve(y_test, y_pred_prob)
roc_auc = auc(fpr, tpr)
plt.plot(fpr, tpr, lw=1, label="ROC (AUC = %0.2f)" % (roc_auc))
plt.plot([0, 1], [0, 1], linestyle="--", lw=2, color="r", label="Random")
plt.xlim([-0.05, 1.05])
plt.ylim([-0.05, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver operating characteristic (ROC) curve")
plt.legend(loc="lower right")
plt.show()
# ## RandomForestClassifier
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier()
model.fit(X_train, y_train)
importance = model.feature_importances_
plt.bar(X.columns, importance)
plt.xticks(rotation=90)
plt.show()
y_pred = model.predict(X_test)
y_pred
print(accuracy_score(y_test, y_pred))
| false | 1 | 1,365 | 10 | 1,789 | 1,365 |
||
129164658
|
<jupyter_start><jupyter_text>Titanic dataset

### Context
I took the titanic test file and the gender_submission and put them together in excel to make a csv. This is great for making charts to help you visualize. This also will help you know who died or survived. At least 70% right, but its up to you to make it 100% Thanks to the titanic beginners competitions for providing with the data. Please **Upvote** my dataset, it will mean a lot to me. Thank you!
Kaggle dataset identifier: test-file
<jupyter_script># # Exploring Pandas
# ### By S. Angelo Santiago
# pandas, short for "Python Data Analysis", is a library for Python that is commonly used for working with data sets as it allows analysis, cleaning, exploring, and manipulating data. [1]
# > pandas is well suited for many different kinds of data:
# >
# > * Tabular data with heterogeneously-typed columns, as in an SQL table or Excel spreadsheet
# > * Ordered and unordered (not necessarily fixed-frequency) time series data.
# > * Arbitrary matrix data (homogeneously typed or heterogeneous) with row and column labels
# > * Any other form of observational / statistical data sets. The data need not be labeled at all to be placed into a pandas data structure [2]
# While Excel/spreadsheets can be used to handle data, especially those data found in a tabular format, Pandas particularly excels in handling data that go beyond the limitations of spreadsheets (e.g. those with entries that go into the millions).
# > Here are just a few of the things that pandas does well:
# > * Easy handling of missing data (represented as NaN) in floating point as well as non-floating point data
# > * Size mutability: columns can be inserted and deleted from DataFrame and higher dimensional objects
# > * Powerful, flexible group by functionality to perform split-apply-combine operations on data sets, for both aggregating and transforming data
# > * Intelligent label-based slicing, fancy indexing, and subsetting of large data sets
# > * Intuitive merging and joining data sets
# > * Hierarchical labeling of axes (possible to have multiple labels per tick)
# > * Robust IO tools for loading data from flat files (CSV and delimited), Excel files, databases, and saving / loading data from the ultrafast HDF5 format
# > * Time series-specific functionality: date range generation and frequency conversion, moving window statistics, date shifting, and lagging. [3]
# In this notebook, we shall explore a bit about how pandas can be used to analyze and manipulate datasets.
# There is much more to cover to fully unlock the potential of this powerful Python library, but we don't have enough sessions. I leave going the extra mile to you. Go beyond what was learned in class and explore pandas to extract deeper insights.
# Enjoy! :)
# -Sir A
import pandas as pd
import os
# ## Loading the Titanic and the Seattle Library Checkout Records
# Creating a list of filepaths to the csvs in the datasets
filepaths = []
# '/kaggle/input' is where you will look for added datasets.
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in sorted(filenames):
filepath = os.path.join(dirname, filename)
# Append each filepath to the list
filepaths.append(filepath)
list(enumerate(filepaths))
# Loading each csv into a Pandas DataFrame each.
# Note that there are much more efficient ways to load the datasets;
# the priority here is not to overwhelm with too much code that's hard to understand.
df_titanic = pd.read_csv(filepaths[0]) # Using list indexing
df_lib1 = pd.read_csv(filepaths[1], low_memory=False)
df_lib2 = pd.read_csv(filepaths[2], low_memory=False)
df_lib3 = pd.read_csv(filepaths[3], low_memory=False)
df_lib4 = pd.read_csv(filepaths[4], low_memory=False)
df_lib5 = pd.read_csv(filepaths[5], low_memory=False)
df_lib6 = pd.read_csv(filepaths[6], low_memory=False)
df_lib7 = pd.read_csv(filepaths[7], low_memory=False)
df_lib8 = pd.read_csv(filepaths[8], low_memory=False)
df_lib9 = pd.read_csv(filepaths[9], low_memory=False)
df_lib10 = pd.read_csv(filepaths[10], low_memory=False)
df_lib11 = pd.read_csv(filepaths[11], low_memory=False)
df_lib12 = pd.read_csv(filepaths[12], low_memory=False)
df_lib13 = pd.read_csv(filepaths[13], low_memory=False)
df_ils = pd.read_csv(filepaths[14])
df_inventory = pd.read_csv(filepaths[15])
# ### Exploring the Titanic Dataset
# Here, we explore the [`Titanic Dataset` ](https://www.kaggle.com/datasets/brendan45774/test-file) as found in Kaggle. I recommend exploring the dataset's page first for more details.
# Getting the information on number of entries, columns,
# non-nulls, and datatypes
df_titanic.info()
df_titanic.describe() # Get statistical analysis of numerical columns
df_titanic.describe(
include=["object"]
) # Get statistical analysis of non-numerical cols
# Counting how many rows are null
df_titanic.isna().sum()
# #### Handling missing values
# In the real world, we are often faced with missing (null) data. What we do with it would depend on the situation and the use case.
# For one, we could delete entire columns with missing values.
#
# Remove (drop) columns with any missing values
df_titanic.dropna(axis="columns")
# In the above example, the `Age`, `Fare`, and `Cabin` columns were dropped, but use this sparingly, because this will result in less data.
# Alternatively, we can just delete entries (Passengers) with null values.
# Remove (drop) the passengers with no indicated age
df_titanic.dropna(subset=["Age"])
# Alternatively, we can interpolate (fill in) the missing data. The data that we fill in that would make sense.
# Take the `Age` column, for example, what ages would make sense here to fill the 86 null values? 0? 100? Either might be too low, or too high, considering that the lowest-aged person ia 0.17 years, and highest-aged one is 76.
# Perhaps, for this specific case, a more reasonable value to fill would be the average age.
mean_age = df_titanic["Age"].mean()
df_filled = df_titanic.copy()
df_filled["Age"] = df_filled["Age"].fillna(mean_age)
df_filled.info()
# Note that `df_filled` has no null values for `Age`.
df_filled.describe()
# pandas can export the DataFrames into several formats, including csv and xlsx.
# Uncomment either line below to export the DataFrame
# pd.to_csv('df_filled.csv') # Export to csv
# pd.to_excel('df_filled.xlsx') # Export to excel
# ### The Seattle Library Checkout Records Dataset
# For the class exercise, go through the [`Seattle Library Checkout Records Dataset`](https://www.kaggle.com/datasets/seattle-public-library/seattle-library-checkout-records). I recommend exploring the dataset's page first for more details.
# Very noticeably, there are 13 different csv files for checkout records from 2005-2017.
# In analyzing the dataset, the analysis could be done separately per file (but we might miss important information that we can get when the analysis is done as a whole).
# What can be done then is to `concatenate` each DataFrame on top of each other. While in general, this can be done via Excel or spreadsheets, in the current dataset, 2005 and 2006 combined reached beyond a million entries; at this rate, spreadsheets will not be able to handle this much data.
# We use pandas' concatenate function, since we are just appending DataFrames
# having the same columns.
df_lib = pd.concat(
[
df_lib1,
df_lib2,
df_lib3,
df_lib4,
df_lib5,
df_lib6,
df_lib7,
df_lib8,
df_lib9,
df_lib10,
df_lib11,
df_lib12,
df_lib13,
]
)
df_lib
# Note that if you do your data analysis on large amounts of data, the CPU will
# operate to its full capacity just to meet the computing demand. It might take
# a while for you to get the results.
df_lib.describe(include="object")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/164/129164658.ipynb
|
test-file
|
brendan45774
|
[{"Id": 129164658, "ScriptId": 38394519, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7960264, "CreationDate": "05/11/2023 13:21:00", "VersionNumber": 2.0, "Title": "LIS 198 - Pandas Exploration", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 170.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 169.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184969540, "KernelVersionId": 129164658, "SourceDatasetVersionId": 2879186}, {"Id": 184969539, "KernelVersionId": 129164658, "SourceDatasetVersionId": 5452}]
|
[{"Id": 2879186, "DatasetId": 826163, "DatasourceVersionId": 2926173, "CreatorUserId": 2681031, "LicenseName": "CC0: Public Domain", "CreationDate": "12/02/2021 16:11:42", "VersionNumber": 6.0, "Title": "Titanic dataset", "Slug": "test-file", "Subtitle": "Gender submission and test file merged", "Description": "\n\n### Context\n\nI took the titanic test file and the gender_submission and put them together in excel to make a csv. This is great for making charts to help you visualize. This also will help you know who died or survived. At least 70% right, but its up to you to make it 100% Thanks to the titanic beginners competitions for providing with the data. Please **Upvote** my dataset, it will mean a lot to me. Thank you!", "VersionNotes": "tested", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 826163, "CreatorUserId": 2681031, "OwnerUserId": 2681031.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2879186.0, "CurrentDatasourceVersionId": 2926173.0, "ForumId": 841293, "Type": 2, "CreationDate": "08/11/2020 14:08:36", "LastActivityDate": "08/11/2020", "TotalViews": 262161, "TotalDownloads": 72658, "TotalVotes": 665, "TotalKernels": 203}]
|
[{"Id": 2681031, "UserName": "brendan45774", "DisplayName": "Brenda N", "RegisterDate": "01/07/2019", "PerformanceTier": 3}]
|
# # Exploring Pandas
# ### By S. Angelo Santiago
# pandas, short for "Python Data Analysis", is a library for Python that is commonly used for working with data sets as it allows analysis, cleaning, exploring, and manipulating data. [1]
# > pandas is well suited for many different kinds of data:
# >
# > * Tabular data with heterogeneously-typed columns, as in an SQL table or Excel spreadsheet
# > * Ordered and unordered (not necessarily fixed-frequency) time series data.
# > * Arbitrary matrix data (homogeneously typed or heterogeneous) with row and column labels
# > * Any other form of observational / statistical data sets. The data need not be labeled at all to be placed into a pandas data structure [2]
# While Excel/spreadsheets can be used to handle data, especially those data found in a tabular format, Pandas particularly excels in handling data that go beyond the limitations of spreadsheets (e.g. those with entries that go into the millions).
# > Here are just a few of the things that pandas does well:
# > * Easy handling of missing data (represented as NaN) in floating point as well as non-floating point data
# > * Size mutability: columns can be inserted and deleted from DataFrame and higher dimensional objects
# > * Powerful, flexible group by functionality to perform split-apply-combine operations on data sets, for both aggregating and transforming data
# > * Intelligent label-based slicing, fancy indexing, and subsetting of large data sets
# > * Intuitive merging and joining data sets
# > * Hierarchical labeling of axes (possible to have multiple labels per tick)
# > * Robust IO tools for loading data from flat files (CSV and delimited), Excel files, databases, and saving / loading data from the ultrafast HDF5 format
# > * Time series-specific functionality: date range generation and frequency conversion, moving window statistics, date shifting, and lagging. [3]
# In this notebook, we shall explore a bit about how pandas can be used to analyze and manipulate datasets.
# There is much more to cover to fully unlock the potential of this powerful Python library, but we don't have enough sessions. I leave going the extra mile to you. Go beyond what was learned in class and explore pandas to extract deeper insights.
# Enjoy! :)
# -Sir A
import pandas as pd
import os
# ## Loading the Titanic and the Seattle Library Checkout Records
# Creating a list of filepaths to the csvs in the datasets
filepaths = []
# '/kaggle/input' is where you will look for added datasets.
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in sorted(filenames):
filepath = os.path.join(dirname, filename)
# Append each filepath to the list
filepaths.append(filepath)
list(enumerate(filepaths))
# Loading each csv into a Pandas DataFrame each.
# Note that there are much more efficient ways to load the datasets;
# the priority here is not to overwhelm with too much code that's hard to understand.
df_titanic = pd.read_csv(filepaths[0]) # Using list indexing
df_lib1 = pd.read_csv(filepaths[1], low_memory=False)
df_lib2 = pd.read_csv(filepaths[2], low_memory=False)
df_lib3 = pd.read_csv(filepaths[3], low_memory=False)
df_lib4 = pd.read_csv(filepaths[4], low_memory=False)
df_lib5 = pd.read_csv(filepaths[5], low_memory=False)
df_lib6 = pd.read_csv(filepaths[6], low_memory=False)
df_lib7 = pd.read_csv(filepaths[7], low_memory=False)
df_lib8 = pd.read_csv(filepaths[8], low_memory=False)
df_lib9 = pd.read_csv(filepaths[9], low_memory=False)
df_lib10 = pd.read_csv(filepaths[10], low_memory=False)
df_lib11 = pd.read_csv(filepaths[11], low_memory=False)
df_lib12 = pd.read_csv(filepaths[12], low_memory=False)
df_lib13 = pd.read_csv(filepaths[13], low_memory=False)
df_ils = pd.read_csv(filepaths[14])
df_inventory = pd.read_csv(filepaths[15])
# ### Exploring the Titanic Dataset
# Here, we explore the [`Titanic Dataset` ](https://www.kaggle.com/datasets/brendan45774/test-file) as found in Kaggle. I recommend exploring the dataset's page first for more details.
# Getting the information on number of entries, columns,
# non-nulls, and datatypes
df_titanic.info()
df_titanic.describe() # Get statistical analysis of numerical columns
df_titanic.describe(
include=["object"]
) # Get statistical analysis of non-numerical cols
# Counting how many rows are null
df_titanic.isna().sum()
# #### Handling missing values
# In the real world, we are often faced with missing (null) data. What we do with it would depend on the situation and the use case.
# For one, we could delete entire columns with missing values.
#
# Remove (drop) columns with any missing values
df_titanic.dropna(axis="columns")
# In the above example, the `Age`, `Fare`, and `Cabin` columns were dropped, but use this sparingly, because this will result in less data.
# Alternatively, we can just delete entries (Passengers) with null values.
# Remove (drop) the passengers with no indicated age
df_titanic.dropna(subset=["Age"])
# Alternatively, we can interpolate (fill in) the missing data. The data that we fill in that would make sense.
# Take the `Age` column, for example, what ages would make sense here to fill the 86 null values? 0? 100? Either might be too low, or too high, considering that the lowest-aged person ia 0.17 years, and highest-aged one is 76.
# Perhaps, for this specific case, a more reasonable value to fill would be the average age.
mean_age = df_titanic["Age"].mean()
df_filled = df_titanic.copy()
df_filled["Age"] = df_filled["Age"].fillna(mean_age)
df_filled.info()
# Note that `df_filled` has no null values for `Age`.
df_filled.describe()
# pandas can export the DataFrames into several formats, including csv and xlsx.
# Uncomment either line below to export the DataFrame
# pd.to_csv('df_filled.csv') # Export to csv
# pd.to_excel('df_filled.xlsx') # Export to excel
# ### The Seattle Library Checkout Records Dataset
# For the class exercise, go through the [`Seattle Library Checkout Records Dataset`](https://www.kaggle.com/datasets/seattle-public-library/seattle-library-checkout-records). I recommend exploring the dataset's page first for more details.
# Very noticeably, there are 13 different csv files for checkout records from 2005-2017.
# In analyzing the dataset, the analysis could be done separately per file (but we might miss important information that we can get when the analysis is done as a whole).
# What can be done then is to `concatenate` each DataFrame on top of each other. While in general, this can be done via Excel or spreadsheets, in the current dataset, 2005 and 2006 combined reached beyond a million entries; at this rate, spreadsheets will not be able to handle this much data.
# We use pandas' concatenate function, since we are just appending DataFrames
# having the same columns.
df_lib = pd.concat(
[
df_lib1,
df_lib2,
df_lib3,
df_lib4,
df_lib5,
df_lib6,
df_lib7,
df_lib8,
df_lib9,
df_lib10,
df_lib11,
df_lib12,
df_lib13,
]
)
df_lib
# Note that if you do your data analysis on large amounts of data, the CPU will
# operate to its full capacity just to meet the computing demand. It might take
# a while for you to get the results.
df_lib.describe(include="object")
| false | 0 | 2,050 | 0 | 2,211 | 2,050 |
||
129164000
|
<jupyter_start><jupyter_text>Profit Prediction using Linear Regression
Kaggle dataset identifier: profit-prediction-using-linear-regression
<jupyter_script>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv("/kaggle/input/profitcsv/profit.csv")
df.info()
df.describe()
df.shape
df.isnull().sum()
df
# if there is a null value, need to find the mean value and insert them using fillno()
# # Independent & Dependent Value
X = df.drop(["Profit"], axis=1)
X.head()
y = df["Profit"]
y.head()
# Now we need to encoding with the title of "Area" since Linear Regression doesn't work with string. It needs numerical value.
# Two Types of Encoding. One Hot Encoding & Level Encoding
# ### One Hot Encoding
city = pd.get_dummies(X["Area"])
city.head()
# dropping area column since it's a string
X = X.drop("Area", axis=1)
X
# Now let's concatenate the Area encoding data
X = pd.concat([X, city], axis=1)
X
# ### Train Test Split
from sklearn.model_selection import train_test_split
xtrain, xtest, ytrain, ytest = train_test_split(X, y, test_size=0.25, random_state=0)
# ### Linear Regression
from sklearn.linear_model import LinearRegression
regr = LinearRegression()
regr.fit(xtrain, ytrain)
xtest
ytest
# ### Prediction
guess = regr.predict(xtest)
guess
# ### R-Squarded Value
from sklearn.metrics import r2_score
score = r2_score(ytest, guess)
score
regr.score(xtest, ytest)
output = [i for i in range(1, len(guess) + 1)]
# ### Data Visualization
plt.figure(figsize=(10, 6), facecolor="#fe7e81", edgecolor="k", linewidth=3)
plt.plot(output, ytest, color="r")
plt.plot(output, guess, color="c")
plt.title("Company Profit Prediction", fontsize=15)
# plt.xlabel("Salary")
# plt.ylabel("Index")
plt.legend(["Given Profit", "Predicted Profit"])
plt.show()
# ### Plotting the Error
error = ytest - guess
error
# ### Plotting Error Data
count = [i for i in range(1, len(error) + 1)]
count
# ### Plotting
plt.figure(figsize=(10, 6), facecolor="#fb9e81", edgecolor="k", linewidth=3)
plt.plot(count, error, c="g")
plt.title("Error Data", fontsize=14)
plt.legend(["Error Data"])
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/164/129164000.ipynb
|
profit-prediction-using-linear-regression
|
studymart
|
[{"Id": 129164000, "ScriptId": 38397299, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13445530, "CreationDate": "05/11/2023 13:15:17", "VersionNumber": 1.0, "Title": "Profit Prediction and Evaluation", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 118.0, "LinesInsertedFromPrevious": 118.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 184968375, "KernelVersionId": 129164000, "SourceDatasetVersionId": 1459830}]
|
[{"Id": 1459830, "DatasetId": 856000, "DatasourceVersionId": 1493483, "CreatorUserId": 5367665, "LicenseName": "Unknown", "CreationDate": "09/03/2020 04:43:24", "VersionNumber": 1.0, "Title": "Profit Prediction using Linear Regression", "Slug": "profit-prediction-using-linear-regression", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 856000, "CreatorUserId": 5367665, "OwnerUserId": 5367665.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1459830.0, "CurrentDatasourceVersionId": 1493483.0, "ForumId": 871294, "Type": 2, "CreationDate": "09/03/2020 04:43:24", "LastActivityDate": "09/03/2020", "TotalViews": 8064, "TotalDownloads": 1298, "TotalVotes": 13, "TotalKernels": 4}]
|
[{"Id": 5367665, "UserName": "studymart", "DisplayName": "Study Mart", "RegisterDate": "06/25/2020", "PerformanceTier": 0}]
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv("/kaggle/input/profitcsv/profit.csv")
df.info()
df.describe()
df.shape
df.isnull().sum()
df
# if there is a null value, need to find the mean value and insert them using fillno()
# # Independent & Dependent Value
X = df.drop(["Profit"], axis=1)
X.head()
y = df["Profit"]
y.head()
# Now we need to encoding with the title of "Area" since Linear Regression doesn't work with string. It needs numerical value.
# Two Types of Encoding. One Hot Encoding & Level Encoding
# ### One Hot Encoding
city = pd.get_dummies(X["Area"])
city.head()
# dropping area column since it's a string
X = X.drop("Area", axis=1)
X
# Now let's concatenate the Area encoding data
X = pd.concat([X, city], axis=1)
X
# ### Train Test Split
from sklearn.model_selection import train_test_split
xtrain, xtest, ytrain, ytest = train_test_split(X, y, test_size=0.25, random_state=0)
# ### Linear Regression
from sklearn.linear_model import LinearRegression
regr = LinearRegression()
regr.fit(xtrain, ytrain)
xtest
ytest
# ### Prediction
guess = regr.predict(xtest)
guess
# ### R-Squarded Value
from sklearn.metrics import r2_score
score = r2_score(ytest, guess)
score
regr.score(xtest, ytest)
output = [i for i in range(1, len(guess) + 1)]
# ### Data Visualization
plt.figure(figsize=(10, 6), facecolor="#fe7e81", edgecolor="k", linewidth=3)
plt.plot(output, ytest, color="r")
plt.plot(output, guess, color="c")
plt.title("Company Profit Prediction", fontsize=15)
# plt.xlabel("Salary")
# plt.ylabel("Index")
plt.legend(["Given Profit", "Predicted Profit"])
plt.show()
# ### Plotting the Error
error = ytest - guess
error
# ### Plotting Error Data
count = [i for i in range(1, len(error) + 1)]
count
# ### Plotting
plt.figure(figsize=(10, 6), facecolor="#fb9e81", edgecolor="k", linewidth=3)
plt.plot(count, error, c="g")
plt.title("Error Data", fontsize=14)
plt.legend(["Error Data"])
plt.show()
| false | 1 | 668 | 1 | 696 | 668 |
||
129164002
|
<jupyter_start><jupyter_text>Salary Dataset - Simple linear regression
## Dataset Description
Salary Dataset in CSV for Simple linear regression. It has also been used in Machine Learning A to Z course of my series.
## Columns
- #
- YearsExperience
- Salary
Kaggle dataset identifier: salary-dataset-simple-linear-regression
<jupyter_code>import pandas as pd
df = pd.read_csv('salary-dataset-simple-linear-regression/Salary_dataset.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 30 entries, 0 to 29
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 30 non-null int64
1 YearsExperience 30 non-null float64
2 Salary 30 non-null float64
dtypes: float64(2), int64(1)
memory usage: 848.0 bytes
<jupyter_text>Examples:
{
"Unnamed: 0": 0.0,
"YearsExperience": 1.2,
"Salary": 39344.0
}
{
"Unnamed: 0": 1.0,
"YearsExperience": 1.4,
"Salary": 46206.0
}
{
"Unnamed: 0": 2.0,
"YearsExperience": 1.6,
"Salary": 37732.0
}
{
"Unnamed: 0": 3.0,
"YearsExperience": 2.1,
"Salary": 43526.0
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
class RawLR:
def __init__(self):
# Initialize the slope and intercept to None
self.m = None
self.b = None
def fit(self, X_train, y_train):
# Calculate the slope and intercept using the least squares method
num = 0 # numerator of slope formula
den = 0 # denominator of slope formula
for i in range(X_train.shape[0]):
# Calculate numerator and denominator of slope formula
num += (X_train[i] - X_train.mean()) * (y_train[i] - y_train.mean())
den += (X_train[i] - X_train.mean()) ** 2
self.m = num / den # Calculate slope
self.b = y_train.mean() - self.m * X_train.mean() # Calculate intercept
# Print the slope and intercept for debugging purposes
print("Slope:", self.m)
print("Intercept:", self.b)
def predict(self, X_test):
# Make sure the model has been trained
if self.m is None or self.b is None:
raise ValueError("Model has not been trained yet")
# Predict target values using the slope and intercept
y_pred = self.m * X_test + self.b
return y_pred
import pandas as pd
import numpy as np
# Load the salary dataset into a pandas dataframe
df = pd.read_csv(
"/kaggle/input/salary-dataset-simple-linear-regression/Salary_dataset.csv"
)
# Display the first few rows of the dataframe
df.head()
# Extract the independent variable (YearsExperience) and the dependent variable (Salary) from the dataframe
X = df.iloc[:, 1].values # Years of experience
y = df.iloc[:, 2].values # Salary
from sklearn.model_selection import train_test_split
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=2)
# Check the shape of the training set for the dependent variable
y_train.shape
# Create an instance of the RawLR class
lr = RawLR()
# Fit the linear regression model to the training data
lr.fit(X_train, y_train)
# Predict the output for the first test sample using the trained model
lr.predict(X_test[0])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/164/129164002.ipynb
|
salary-dataset-simple-linear-regression
|
abhishek14398
|
[{"Id": 129164002, "ScriptId": 38374193, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10728959, "CreationDate": "05/11/2023 13:15:17", "VersionNumber": 1.0, "Title": "Maths-behind-linear-regression", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 84.0, "LinesInsertedFromPrevious": 84.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 184968404, "KernelVersionId": 129164002, "SourceDatasetVersionId": 4832081}]
|
[{"Id": 4832081, "DatasetId": 2799910, "DatasourceVersionId": 4895851, "CreatorUserId": 3259703, "LicenseName": "CC0: Public Domain", "CreationDate": "01/10/2023 03:55:40", "VersionNumber": 1.0, "Title": "Salary Dataset - Simple linear regression", "Slug": "salary-dataset-simple-linear-regression", "Subtitle": "Simple Linear Regression Dataset, used in Machine Learning A - Z", "Description": "## Dataset Description\nSalary Dataset in CSV for Simple linear regression. It has also been used in Machine Learning A to Z course of my series.\n\n## Columns\n- #\n- YearsExperience\n- Salary", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2799910, "CreatorUserId": 3259703, "OwnerUserId": 3259703.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4832081.0, "CurrentDatasourceVersionId": 4895851.0, "ForumId": 2834222, "Type": 2, "CreationDate": "01/10/2023 03:55:40", "LastActivityDate": "01/10/2023", "TotalViews": 65295, "TotalDownloads": 13051, "TotalVotes": 139, "TotalKernels": 93}]
|
[{"Id": 3259703, "UserName": "abhishek14398", "DisplayName": "Allena Venkata Sai Aby", "RegisterDate": "05/22/2019", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
class RawLR:
def __init__(self):
# Initialize the slope and intercept to None
self.m = None
self.b = None
def fit(self, X_train, y_train):
# Calculate the slope and intercept using the least squares method
num = 0 # numerator of slope formula
den = 0 # denominator of slope formula
for i in range(X_train.shape[0]):
# Calculate numerator and denominator of slope formula
num += (X_train[i] - X_train.mean()) * (y_train[i] - y_train.mean())
den += (X_train[i] - X_train.mean()) ** 2
self.m = num / den # Calculate slope
self.b = y_train.mean() - self.m * X_train.mean() # Calculate intercept
# Print the slope and intercept for debugging purposes
print("Slope:", self.m)
print("Intercept:", self.b)
def predict(self, X_test):
# Make sure the model has been trained
if self.m is None or self.b is None:
raise ValueError("Model has not been trained yet")
# Predict target values using the slope and intercept
y_pred = self.m * X_test + self.b
return y_pred
import pandas as pd
import numpy as np
# Load the salary dataset into a pandas dataframe
df = pd.read_csv(
"/kaggle/input/salary-dataset-simple-linear-regression/Salary_dataset.csv"
)
# Display the first few rows of the dataframe
df.head()
# Extract the independent variable (YearsExperience) and the dependent variable (Salary) from the dataframe
X = df.iloc[:, 1].values # Years of experience
y = df.iloc[:, 2].values # Salary
from sklearn.model_selection import train_test_split
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=2)
# Check the shape of the training set for the dependent variable
y_train.shape
# Create an instance of the RawLR class
lr = RawLR()
# Fit the linear regression model to the training data
lr.fit(X_train, y_train)
# Predict the output for the first test sample using the trained model
lr.predict(X_test[0])
|
[{"salary-dataset-simple-linear-regression/Salary_dataset.csv": {"column_names": "[\"Unnamed: 0\", \"YearsExperience\", \"Salary\"]", "column_data_types": "{\"Unnamed: 0\": \"int64\", \"YearsExperience\": \"float64\", \"Salary\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 30 entries, 0 to 29\nData columns (total 3 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 30 non-null int64 \n 1 YearsExperience 30 non-null float64\n 2 Salary 30 non-null float64\ndtypes: float64(2), int64(1)\nmemory usage: 848.0 bytes\n", "summary": "{\"Unnamed: 0\": {\"count\": 30.0, \"mean\": 14.5, \"std\": 8.803408430829505, \"min\": 0.0, \"25%\": 7.25, \"50%\": 14.5, \"75%\": 21.75, \"max\": 29.0}, \"YearsExperience\": {\"count\": 30.0, \"mean\": 5.413333333333332, \"std\": 2.8378881576627184, \"min\": 1.2000000000000002, \"25%\": 3.3000000000000003, \"50%\": 4.8, \"75%\": 7.8, \"max\": 10.6}, \"Salary\": {\"count\": 30.0, \"mean\": 76004.0, \"std\": 27414.4297845823, \"min\": 37732.0, \"25%\": 56721.75, \"50%\": 65238.0, \"75%\": 100545.75, \"max\": 122392.0}}", "examples": "{\"Unnamed: 0\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"YearsExperience\":{\"0\":1.2,\"1\":1.4,\"2\":1.6,\"3\":2.1},\"Salary\":{\"0\":39344.0,\"1\":46206.0,\"2\":37732.0,\"3\":43526.0}}"}}]
| true | 1 |
<start_data_description><data_path>salary-dataset-simple-linear-regression/Salary_dataset.csv:
<column_names>
['Unnamed: 0', 'YearsExperience', 'Salary']
<column_types>
{'Unnamed: 0': 'int64', 'YearsExperience': 'float64', 'Salary': 'float64'}
<dataframe_Summary>
{'Unnamed: 0': {'count': 30.0, 'mean': 14.5, 'std': 8.803408430829505, 'min': 0.0, '25%': 7.25, '50%': 14.5, '75%': 21.75, 'max': 29.0}, 'YearsExperience': {'count': 30.0, 'mean': 5.413333333333332, 'std': 2.8378881576627184, 'min': 1.2000000000000002, '25%': 3.3000000000000003, '50%': 4.8, '75%': 7.8, 'max': 10.6}, 'Salary': {'count': 30.0, 'mean': 76004.0, 'std': 27414.4297845823, 'min': 37732.0, '25%': 56721.75, '50%': 65238.0, '75%': 100545.75, 'max': 122392.0}}
<dataframe_info>
RangeIndex: 30 entries, 0 to 29
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 30 non-null int64
1 YearsExperience 30 non-null float64
2 Salary 30 non-null float64
dtypes: float64(2), int64(1)
memory usage: 848.0 bytes
<some_examples>
{'Unnamed: 0': {'0': 0, '1': 1, '2': 2, '3': 3}, 'YearsExperience': {'0': 1.2, '1': 1.4, '2': 1.6, '3': 2.1}, 'Salary': {'0': 39344.0, '1': 46206.0, '2': 37732.0, '3': 43526.0}}
<end_description>
| 753 | 2 | 1,154 | 753 |
129087951
|
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
df = pd.read_csv("/kaggle/input/cap-data-githubcsv/cap_data_github.csv")
# # **A Exploratory Data Analysis with Capsicum Plant.**
# *The Original Dataset taken from GitHub user [Logan Lauton](https://github.com/logan-lauton)'s [Capsicum Research](https://github.com/logan-lauton/Capsicum-Research/tree/main)*
# * This is an educational exploratory data analysis about Capsicum plant research, and it includes five different questions;
# 1. What is Familiarity and Age Relationship about Capsicum looks like ?
# 2. Ipsum
# 3. Dolor
# 4. Sit
# 5. Lorem
#
# * And there are five different data graph taken from the dataset that have been used to provide a clear explanation for the questions asked.
df["Birthday"] = pd.to_datetime(df["Birthday"])
df["Year"] = df["Birthday"].dt.year
min_year = df["Year"].min()
max_year = df["Year"].max()
print("Oldest year:", min_year)
print("Youngest year:", max_year)
# **1-) What is Familiarity and Age Relationship about Capsicum looks like?**
# *And The answers of the:*
# * 1-1 What Age Groups has which level of Familiarity for the Capsicum
# * 1-2 What is the average level of Capsicum Familiarity of the Age Groups
bins = [1960, 1970, 1980, 1990, 2000, 2010, float("inf")]
labels = ["1960-", "1970-1979", "1980-1989", "1990-1999", "2000-2009", "2010+"]
df["YearGroup"] = pd.cut(df["Year"], bins=bins, labels=labels)
df_filtered = df[df["Familiar_with_capsicum"] == 5]
year_counts = df_filtered["YearGroup"].value_counts()
plt.pie(year_counts, labels=year_counts.index, autopct="%1.1f%%")
plt.title("Highest Familiarity With Capsicum by Age")
plt.show()
# **1-1A) Highest Familiarity for Capsicum When Disturbuted by Age Groups**
# * With 43.3% People who born in 80s are showing the most of the Highest Familiarity
# * With 25% People who born in 90s are showing the most of the Second Highest Familiarity
# * With 13.3% People who born in 70s are showing the most of the Third Highest Familiarity
# * With 11.7% People who born in 60s are showing the most of the Fourth Highest Familiarity
# * And Less than 7% of the People who born after 2000s are showing the least Highest Familiarity about the Capsicum
#
bins = [1960, 1970, 1980, 1990, 2000, 2010, float("inf")]
labels = ["", "1970-1979", "1980-1989", "1990-1999", "Other", "2010+"]
df["YearGroup"] = pd.cut(df["Year"], bins=bins, labels=labels)
df_filtered = df[df["Familiar_with_capsicum"] == 4]
year_counts = df_filtered["YearGroup"].value_counts()
plt.pie(year_counts, labels=year_counts.index, autopct="%1.1f%%")
plt.title("", loc="center")
ttl = plt.title("Medium Familiarity With Capsicum by Age")
ttl.set_position([0.5, 1.05])
plt.show()
# **1-1B) Medium Familiarity for Capsicum When Disturbuted by Age Groups**
# * With 61.5% People who born in 80s are showing the most of the Medium Familiarity
# * Following with 23.1% People who born in 70s are showing the most of the Second Medium Familiarity
# * With 7.7% Each Totally Nearly 15% Familiarity Comes from people who are born in 90s and after 2010
# * And the rest of the age groups are below 1%
#
bins = [1960, 2000, 2010, float("inf")]
labels = ["Other", "2000+", "2010+"]
df["YearGroup"] = pd.cut(df["Year"], bins=bins, labels=labels)
df_filtered = df[df["Familiar_with_capsicum"] == 3]
year_counts = df_filtered["YearGroup"].value_counts()
plt.pie(year_counts, labels=year_counts.index, autopct="%1.1f%%")
ttl = plt.title("Low Familiarity With Capsicum by Age")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/087/129087951.ipynb
| null | null |
[{"Id": 129087951, "ScriptId": 38371597, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14686909, "CreationDate": "05/10/2023 23:07:37", "VersionNumber": 1.0, "Title": "homework1b", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 121.0, "LinesInsertedFromPrevious": 121.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
df = pd.read_csv("/kaggle/input/cap-data-githubcsv/cap_data_github.csv")
# # **A Exploratory Data Analysis with Capsicum Plant.**
# *The Original Dataset taken from GitHub user [Logan Lauton](https://github.com/logan-lauton)'s [Capsicum Research](https://github.com/logan-lauton/Capsicum-Research/tree/main)*
# * This is an educational exploratory data analysis about Capsicum plant research, and it includes five different questions;
# 1. What is Familiarity and Age Relationship about Capsicum looks like ?
# 2. Ipsum
# 3. Dolor
# 4. Sit
# 5. Lorem
#
# * And there are five different data graph taken from the dataset that have been used to provide a clear explanation for the questions asked.
df["Birthday"] = pd.to_datetime(df["Birthday"])
df["Year"] = df["Birthday"].dt.year
min_year = df["Year"].min()
max_year = df["Year"].max()
print("Oldest year:", min_year)
print("Youngest year:", max_year)
# **1-) What is Familiarity and Age Relationship about Capsicum looks like?**
# *And The answers of the:*
# * 1-1 What Age Groups has which level of Familiarity for the Capsicum
# * 1-2 What is the average level of Capsicum Familiarity of the Age Groups
bins = [1960, 1970, 1980, 1990, 2000, 2010, float("inf")]
labels = ["1960-", "1970-1979", "1980-1989", "1990-1999", "2000-2009", "2010+"]
df["YearGroup"] = pd.cut(df["Year"], bins=bins, labels=labels)
df_filtered = df[df["Familiar_with_capsicum"] == 5]
year_counts = df_filtered["YearGroup"].value_counts()
plt.pie(year_counts, labels=year_counts.index, autopct="%1.1f%%")
plt.title("Highest Familiarity With Capsicum by Age")
plt.show()
# **1-1A) Highest Familiarity for Capsicum When Disturbuted by Age Groups**
# * With 43.3% People who born in 80s are showing the most of the Highest Familiarity
# * With 25% People who born in 90s are showing the most of the Second Highest Familiarity
# * With 13.3% People who born in 70s are showing the most of the Third Highest Familiarity
# * With 11.7% People who born in 60s are showing the most of the Fourth Highest Familiarity
# * And Less than 7% of the People who born after 2000s are showing the least Highest Familiarity about the Capsicum
#
bins = [1960, 1970, 1980, 1990, 2000, 2010, float("inf")]
labels = ["", "1970-1979", "1980-1989", "1990-1999", "Other", "2010+"]
df["YearGroup"] = pd.cut(df["Year"], bins=bins, labels=labels)
df_filtered = df[df["Familiar_with_capsicum"] == 4]
year_counts = df_filtered["YearGroup"].value_counts()
plt.pie(year_counts, labels=year_counts.index, autopct="%1.1f%%")
plt.title("", loc="center")
ttl = plt.title("Medium Familiarity With Capsicum by Age")
ttl.set_position([0.5, 1.05])
plt.show()
# **1-1B) Medium Familiarity for Capsicum When Disturbuted by Age Groups**
# * With 61.5% People who born in 80s are showing the most of the Medium Familiarity
# * Following with 23.1% People who born in 70s are showing the most of the Second Medium Familiarity
# * With 7.7% Each Totally Nearly 15% Familiarity Comes from people who are born in 90s and after 2010
# * And the rest of the age groups are below 1%
#
bins = [1960, 2000, 2010, float("inf")]
labels = ["Other", "2000+", "2010+"]
df["YearGroup"] = pd.cut(df["Year"], bins=bins, labels=labels)
df_filtered = df[df["Familiar_with_capsicum"] == 3]
year_counts = df_filtered["YearGroup"].value_counts()
plt.pie(year_counts, labels=year_counts.index, autopct="%1.1f%%")
ttl = plt.title("Low Familiarity With Capsicum by Age")
plt.show()
| false | 0 | 1,327 | 0 | 1,327 | 1,327 |
||
129087888
|
import numpy as np
from scipy import constants
from scipy.optimize import minimize
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem.rdmolops import RDKFingerprint
from xtb.interface import Calculator, Param
from xtb.utils import get_method, Solvent
from xtb.libxtb import VERBOSITY_MUTED
from concurrent.futures import ThreadPoolExecutor
def smiles_to_properties(smiles, n_calculations=5):
# Define reference states
reference_smiles = ["[H][H]", "[C-]#[C+]", "N#N", "O=O", "FF", "P#P", "S=S", "ClCl"]
def energy_optimization(
mol, atomic_numbers, solvent=None, method="gfn2-xtb", steps=20, step_size=0.2
):
# Why do that?
# explain why we were forced to do gfn2-xtb
xyz = mol.GetConformer().GetPositions()
ANGSTROM_IN_BOHR = (
constants.physical_constants["Bohr radius"][0] * 1.0e10
) # convert coordinates to angstrom or unitless i cannot tell
net_charge = 0 # in units of electrons
n_unpaired_electrons = 0 # establish the molecule is neutral charge
# using initial energy, gradient, position:
calc = Calculator(
get_method(method),
atomic_numbers,
xyz / ANGSTROM_IN_BOHR,
charge=net_charge,
uhf=n_unpaired_electrons,
) # use the method to get calc, a "tool" for better charactizing the energy
calc.set_verbosity(VERBOSITY_MUTED) # hides all the steps, cleaner output
calc.set_solvent(solvent)
result = calc.singlepoint() # google that, what is taht?
E_xtb = result.get_energy() # in Hartree
gradient_xtb = (
result.get_gradient() / ANGSTROM_IN_BOHR
) # from Hartree/Bohr to Hartree/Angstrom
xyz_frames = [] # start gradient descent
for step_i in range(steps):
# iteratively optimize the energy based on position
calc.update(xyz / ANGSTROM_IN_BOHR)
result = calc.singlepoint()
E_xtb = result.get_energy() # in Hartree
gradient_xtb = (
result.get_gradient() / ANGSTROM_IN_BOHR
) # from Hartree/Bohr to Hartree/Angstrom
xyz_frames.append(xyz.copy())
xyz -= (
step_size * gradient_xtb
) # note that xyz is the thing being changed #step size is 0.2
return xyz, E_xtb
def parallel_energy_optimization(
mol, atomic_numbers, solvent=None, method="gfn2-xtb", steps=20
):
# Run energy_optimization for a single molecule
# this is the setup for:
# iterate all the steps at the same time (over & over), getting energy for each process, then find average
xyz, energy = energy_optimization(mol, atomic_numbers, solvent, method, steps)
return (xyz, energy)
# Create RDKit molecule
mol = Chem.MolFromSmiles(smiles) # construct our molecule, mol
mol = Chem.AddHs(mol)
AllChem.EmbedMolecule(mol) # produce 3D coordinates for each atom
AllChem.MMFFOptimizeMolecule(mol) # use mmff to find conformer with least energy
atomic_numbers = np.array(
[atom.GetAtomicNum() for atom in mol.GetAtoms()]
) # get atoms from mol,then get atomic number from each atom
# atomic_numbers is our first output
# Explanation here?
with ThreadPoolExecutor() as executor:
# run each parallel_energy_optimation concurrently to find energy for gas/aqueous
# the only difference is specifying the solvent = H2O for the aqueous energy
# n_calculations = 5, so iterate 5 times in parallel
gas_phase_results = list(
executor.map(
parallel_energy_optimization,
[mol] * n_calculations,
[atomic_numbers] * n_calculations,
)
)
aqueous_phase_results = list(
executor.map(
parallel_energy_optimization,
[mol] * n_calculations,
[atomic_numbers] * n_calculations,
[Solvent.h2o] * n_calculations,
)
)
gas_xyz, gas_phase_energy = gas_phase_results[
0
] # 0 means first entry of the parallel results?
gas_phase_energies = [
result[1] for result in gas_phase_results
] # get all the energies plural and use to get std
gas_phase_stddev = (
np.std(gas_phase_energies) * 627.509
) # multiply by 627 to convert hartree to kcal/mol
aqueous_xyz, aqueous_phase_energy = aqueous_phase_results[0]
aqueous_phase_energies = [result[1] for result in aqueous_phase_results]
aqueous_phase_stddev = np.std(aqueous_phase_energies) * 627.509
# Get energy of formation (for aq phase)
formation_energy = 0 # initial variable
elements = {1: "H", 6: "C", 7: "N", 8: "O", 9: "F", 15: "P", 16: "S", 17: "Cl"}
for atom in mol.GetAtoms():
num = atom.GetAtomicNum()
ref_energy = 0
# making a reference molecule:
for i in reference_smiles:
if elements[num] == i[1] or elements[num] == i[0] or elements[num] == i[:1]:
ref_mol = Chem.MolFromSmiles(
i
) # make the reference molecule given a SMILES string
break
AllChem.EmbedMolecule(ref_mol) # 3d space
ref_atomic_numbers = np.array(
[atom.GetAtomicNum() for atom in ref_mol.GetAtoms()]
) # get the atom numbers of reference molecule
ref_xyz, ref_energy = energy_optimization(
ref_mol, ref_atomic_numbers, solvent=Solvent.h2o
) # energy optimation of ref mol
formation_energy -= ref_energy / 2 # formation_energy = aq_energy - ref_energy
# divide by 2 to avoid double-counting
formation_energy += aqueous_phase_energy
formation_energy *= 627.509 # convert to kcal/mol
formation_stddev = aqueous_phase_stddev
# Get hydration energy
hydration_energy = aqueous_phase_energy - gas_phase_energy
hydration_energy *= 627.509 # convert to kcal/mol
hydration_stddev = np.sqrt(
aqueous_phase_stddev**2 + gas_phase_stddev**2
) # Wrong :(
# Get RDKFingerprint
fingerprint = RDKFingerprint(mol, fpSize=512) # make size 512 bits
fingerprint = np.array(fingerprint, dtype=int)
atomic_num_element = []
for i in atomic_numbers:
atomic_num_element.append(elements[i])
print(f"📎elements:\n {atomic_num_element}")
print(f"xyz_coordinates(Angstroms): \n{aqueous_xyz}")
print(f"Formation energy: {formation_energy:.2f} +/- {formation_stddev} kcal/mol")
print(f"Hydration energy: {hydration_energy:.2f} +/- {hydration_stddev} kcal/mol")
print(f"RDK Fingerprint(512 bits): \n {fingerprint}")
smiles = "CC" # input your molecule here!
smiles_to_properties(smiles)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/087/129087888.ipynb
| null | null |
[{"Id": 129087888, "ScriptId": 36840027, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13344058, "CreationDate": "05/10/2023 23:06:33", "VersionNumber": 18.0, "Title": "notebook51e4990a32", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 121.0, "LinesInsertedFromPrevious": 59.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 62.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
from scipy import constants
from scipy.optimize import minimize
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem.rdmolops import RDKFingerprint
from xtb.interface import Calculator, Param
from xtb.utils import get_method, Solvent
from xtb.libxtb import VERBOSITY_MUTED
from concurrent.futures import ThreadPoolExecutor
def smiles_to_properties(smiles, n_calculations=5):
# Define reference states
reference_smiles = ["[H][H]", "[C-]#[C+]", "N#N", "O=O", "FF", "P#P", "S=S", "ClCl"]
def energy_optimization(
mol, atomic_numbers, solvent=None, method="gfn2-xtb", steps=20, step_size=0.2
):
# Why do that?
# explain why we were forced to do gfn2-xtb
xyz = mol.GetConformer().GetPositions()
ANGSTROM_IN_BOHR = (
constants.physical_constants["Bohr radius"][0] * 1.0e10
) # convert coordinates to angstrom or unitless i cannot tell
net_charge = 0 # in units of electrons
n_unpaired_electrons = 0 # establish the molecule is neutral charge
# using initial energy, gradient, position:
calc = Calculator(
get_method(method),
atomic_numbers,
xyz / ANGSTROM_IN_BOHR,
charge=net_charge,
uhf=n_unpaired_electrons,
) # use the method to get calc, a "tool" for better charactizing the energy
calc.set_verbosity(VERBOSITY_MUTED) # hides all the steps, cleaner output
calc.set_solvent(solvent)
result = calc.singlepoint() # google that, what is taht?
E_xtb = result.get_energy() # in Hartree
gradient_xtb = (
result.get_gradient() / ANGSTROM_IN_BOHR
) # from Hartree/Bohr to Hartree/Angstrom
xyz_frames = [] # start gradient descent
for step_i in range(steps):
# iteratively optimize the energy based on position
calc.update(xyz / ANGSTROM_IN_BOHR)
result = calc.singlepoint()
E_xtb = result.get_energy() # in Hartree
gradient_xtb = (
result.get_gradient() / ANGSTROM_IN_BOHR
) # from Hartree/Bohr to Hartree/Angstrom
xyz_frames.append(xyz.copy())
xyz -= (
step_size * gradient_xtb
) # note that xyz is the thing being changed #step size is 0.2
return xyz, E_xtb
def parallel_energy_optimization(
mol, atomic_numbers, solvent=None, method="gfn2-xtb", steps=20
):
# Run energy_optimization for a single molecule
# this is the setup for:
# iterate all the steps at the same time (over & over), getting energy for each process, then find average
xyz, energy = energy_optimization(mol, atomic_numbers, solvent, method, steps)
return (xyz, energy)
# Create RDKit molecule
mol = Chem.MolFromSmiles(smiles) # construct our molecule, mol
mol = Chem.AddHs(mol)
AllChem.EmbedMolecule(mol) # produce 3D coordinates for each atom
AllChem.MMFFOptimizeMolecule(mol) # use mmff to find conformer with least energy
atomic_numbers = np.array(
[atom.GetAtomicNum() for atom in mol.GetAtoms()]
) # get atoms from mol,then get atomic number from each atom
# atomic_numbers is our first output
# Explanation here?
with ThreadPoolExecutor() as executor:
# run each parallel_energy_optimation concurrently to find energy for gas/aqueous
# the only difference is specifying the solvent = H2O for the aqueous energy
# n_calculations = 5, so iterate 5 times in parallel
gas_phase_results = list(
executor.map(
parallel_energy_optimization,
[mol] * n_calculations,
[atomic_numbers] * n_calculations,
)
)
aqueous_phase_results = list(
executor.map(
parallel_energy_optimization,
[mol] * n_calculations,
[atomic_numbers] * n_calculations,
[Solvent.h2o] * n_calculations,
)
)
gas_xyz, gas_phase_energy = gas_phase_results[
0
] # 0 means first entry of the parallel results?
gas_phase_energies = [
result[1] for result in gas_phase_results
] # get all the energies plural and use to get std
gas_phase_stddev = (
np.std(gas_phase_energies) * 627.509
) # multiply by 627 to convert hartree to kcal/mol
aqueous_xyz, aqueous_phase_energy = aqueous_phase_results[0]
aqueous_phase_energies = [result[1] for result in aqueous_phase_results]
aqueous_phase_stddev = np.std(aqueous_phase_energies) * 627.509
# Get energy of formation (for aq phase)
formation_energy = 0 # initial variable
elements = {1: "H", 6: "C", 7: "N", 8: "O", 9: "F", 15: "P", 16: "S", 17: "Cl"}
for atom in mol.GetAtoms():
num = atom.GetAtomicNum()
ref_energy = 0
# making a reference molecule:
for i in reference_smiles:
if elements[num] == i[1] or elements[num] == i[0] or elements[num] == i[:1]:
ref_mol = Chem.MolFromSmiles(
i
) # make the reference molecule given a SMILES string
break
AllChem.EmbedMolecule(ref_mol) # 3d space
ref_atomic_numbers = np.array(
[atom.GetAtomicNum() for atom in ref_mol.GetAtoms()]
) # get the atom numbers of reference molecule
ref_xyz, ref_energy = energy_optimization(
ref_mol, ref_atomic_numbers, solvent=Solvent.h2o
) # energy optimation of ref mol
formation_energy -= ref_energy / 2 # formation_energy = aq_energy - ref_energy
# divide by 2 to avoid double-counting
formation_energy += aqueous_phase_energy
formation_energy *= 627.509 # convert to kcal/mol
formation_stddev = aqueous_phase_stddev
# Get hydration energy
hydration_energy = aqueous_phase_energy - gas_phase_energy
hydration_energy *= 627.509 # convert to kcal/mol
hydration_stddev = np.sqrt(
aqueous_phase_stddev**2 + gas_phase_stddev**2
) # Wrong :(
# Get RDKFingerprint
fingerprint = RDKFingerprint(mol, fpSize=512) # make size 512 bits
fingerprint = np.array(fingerprint, dtype=int)
atomic_num_element = []
for i in atomic_numbers:
atomic_num_element.append(elements[i])
print(f"📎elements:\n {atomic_num_element}")
print(f"xyz_coordinates(Angstroms): \n{aqueous_xyz}")
print(f"Formation energy: {formation_energy:.2f} +/- {formation_stddev} kcal/mol")
print(f"Hydration energy: {hydration_energy:.2f} +/- {hydration_stddev} kcal/mol")
print(f"RDK Fingerprint(512 bits): \n {fingerprint}")
smiles = "CC" # input your molecule here!
smiles_to_properties(smiles)
| false | 0 | 1,974 | 0 | 1,974 | 1,974 |
||
129113573
|
<jupyter_start><jupyter_text>Covid-19-country-statistics-dataset
The COVID-19 Country Statistics Dataset, available on Kaggle, is a collection of data related to the COVID-19 pandemic across various countries. The dataset includes information such as the number of confirmed cases, deaths, recoveries, and active cases, as well as information about the population, population density, median age, and various other demographic and health-related statistics for each country.
The dataset contains a total of 13 columns and over 217 rows of data, with each row representing a different country or region. The data is presented in a CSV (comma-separated values) format, making it easy to analyze and work with using a variety of tools and programming languages.
The COVID-19 Country Statistics Dataset contains several column descriptors, including Province/State, Country/Region, Lat, Long, Date, Confirmed, Deaths, and Recovered. Province/State describes the province or state where the cases were detected, while Country/Region describes the country where the cases were detected. Lat and Long describe the latitude and longitude of the country or province/state. Date describes the date when the cases were detected. Confirmed represents the total number of confirmed cases of COVID-19, while Deaths represents the total number of deaths due to COVID-19. Recovered represents the total number of recoveries from COVID-19.
In summary, this dataset is a valuable resource for researchers, analysts, and anyone interested in understanding the spread and impact of COVID-19 across different countries. The dataset provides a detailed picture of the pandemic and can help with decision-making, policy development, and research efforts aimed at understanding the global impact of COVID-19.
Kaggle dataset identifier: covid-19-country-statistics-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/covid-19-country-statistics-dataset/covid-19-country-statistics-dataset.csv"
)
df
df.isnull().sum()
df.isnull().sum().sum()
#
# Filling null value
df2 = df.fillna(value=0)
df2
df2.isnull().sum().sum()
# #Filling Null Values with a previous value
#
df4 = df.fillna(method="pad")
df4
df4.isnull().sum()
# #Filling Null Value with the next value
df5 = df.fillna(method="bfill")
df5
df6 = df.fillna(method="pad", axis=1)
df6
df7 = df.fillna(method="bfill", axis=1)
df7
# #Filling Diffrent Values in Null in differnt columnns
#
df8 = df.fillna({"Country": "abcd", "New Cases": "defg"})
df8
# #Filling Null Value with the mean of a column
#
# df9=df.fillna(value=df['New Deaths'].mean())
# df9
# Dropna()
#
# df11=df.dropna(how='all')
df11 = df.dropna(how="any")
df11
# ##replace()
#
import numpy as np
df12 = df.replace(to_replace=np.nan, value=875465)
df12
# df13=df.replace(to_replace=3.0, value=5.0)
# df13
# ##interpolate
df["Country"] = df["Country"].interpolate(method="linear")
df
df14 = df7.head()
df14
xpos = np.arange(len(df14["Country"]))
plt.xticks(xpos, df14["Total Cases"])
plt.bar(xpos - 0.2, df14["New Cases"], width=0.4, label="New Cases")
plt.bar(xpos + 0.2, df14["Active cases"], width=0.4, label="Active cases")
plt.legend(loc="best")
plt.show()
df15 = df7.tail()
df15
plt.xticks(xpos, df15["Total Deaths"])
plt.bar(xpos - 0.2, df15["New Deaths"], width=0.4, label="New Deaths")
plt.bar(xpos + 0.2, df15["Total Recovered"], width=0.4, label="Total Recovered")
plt.legend(loc="best")
plt.show()
labels = ["Country", "Total Cases", "New Cases", "Total Deaths"]
sizes = [30, 40, 50, 60]
plt.pie(sizes, labels=labels, autopct="%1.1f%%", explode=[0.1, 0, 0, 0.1])
plt.title("Covid19")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/113/129113573.ipynb
|
covid-19-country-statistics-dataset
|
harshghadiya
|
[{"Id": 129113573, "ScriptId": 38353118, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14442011, "CreationDate": "05/11/2023 05:36:34", "VersionNumber": 1.0, "Title": "Covid19 Data Num", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 120.0, "LinesInsertedFromPrevious": 120.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 184881411, "KernelVersionId": 129113573, "SourceDatasetVersionId": 5543953}]
|
[{"Id": 5543953, "DatasetId": 3194747, "DatasourceVersionId": 5618626, "CreatorUserId": 9220292, "LicenseName": "Other (specified in description)", "CreationDate": "04/27/2023 23:22:02", "VersionNumber": 1.0, "Title": "Covid-19-country-statistics-dataset", "Slug": "covid-19-country-statistics-dataset", "Subtitle": "A Comprehensive Collection of COVID-19 Statistics Across Countries and Regions.", "Description": "The COVID-19 Country Statistics Dataset, available on Kaggle, is a collection of data related to the COVID-19 pandemic across various countries. The dataset includes information such as the number of confirmed cases, deaths, recoveries, and active cases, as well as information about the population, population density, median age, and various other demographic and health-related statistics for each country.\n\nThe dataset contains a total of 13 columns and over 217 rows of data, with each row representing a different country or region. The data is presented in a CSV (comma-separated values) format, making it easy to analyze and work with using a variety of tools and programming languages.\n\nThe COVID-19 Country Statistics Dataset contains several column descriptors, including Province/State, Country/Region, Lat, Long, Date, Confirmed, Deaths, and Recovered. Province/State describes the province or state where the cases were detected, while Country/Region describes the country where the cases were detected. Lat and Long describe the latitude and longitude of the country or province/state. Date describes the date when the cases were detected. Confirmed represents the total number of confirmed cases of COVID-19, while Deaths represents the total number of deaths due to COVID-19. Recovered represents the total number of recoveries from COVID-19.\n\nIn summary, this dataset is a valuable resource for researchers, analysts, and anyone interested in understanding the spread and impact of COVID-19 across different countries. The dataset provides a detailed picture of the pandemic and can help with decision-making, policy development, and research efforts aimed at understanding the global impact of COVID-19.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3194747, "CreatorUserId": 9220292, "OwnerUserId": 9220292.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5543953.0, "CurrentDatasourceVersionId": 5618626.0, "ForumId": 3259308, "Type": 2, "CreationDate": "04/27/2023 23:22:02", "LastActivityDate": "04/27/2023", "TotalViews": 5036, "TotalDownloads": 1123, "TotalVotes": 33, "TotalKernels": 2}]
|
[{"Id": 9220292, "UserName": "harshghadiya", "DisplayName": "Harsh Ghadiya", "RegisterDate": "12/21/2021", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/covid-19-country-statistics-dataset/covid-19-country-statistics-dataset.csv"
)
df
df.isnull().sum()
df.isnull().sum().sum()
#
# Filling null value
df2 = df.fillna(value=0)
df2
df2.isnull().sum().sum()
# #Filling Null Values with a previous value
#
df4 = df.fillna(method="pad")
df4
df4.isnull().sum()
# #Filling Null Value with the next value
df5 = df.fillna(method="bfill")
df5
df6 = df.fillna(method="pad", axis=1)
df6
df7 = df.fillna(method="bfill", axis=1)
df7
# #Filling Diffrent Values in Null in differnt columnns
#
df8 = df.fillna({"Country": "abcd", "New Cases": "defg"})
df8
# #Filling Null Value with the mean of a column
#
# df9=df.fillna(value=df['New Deaths'].mean())
# df9
# Dropna()
#
# df11=df.dropna(how='all')
df11 = df.dropna(how="any")
df11
# ##replace()
#
import numpy as np
df12 = df.replace(to_replace=np.nan, value=875465)
df12
# df13=df.replace(to_replace=3.0, value=5.0)
# df13
# ##interpolate
df["Country"] = df["Country"].interpolate(method="linear")
df
df14 = df7.head()
df14
xpos = np.arange(len(df14["Country"]))
plt.xticks(xpos, df14["Total Cases"])
plt.bar(xpos - 0.2, df14["New Cases"], width=0.4, label="New Cases")
plt.bar(xpos + 0.2, df14["Active cases"], width=0.4, label="Active cases")
plt.legend(loc="best")
plt.show()
df15 = df7.tail()
df15
plt.xticks(xpos, df15["Total Deaths"])
plt.bar(xpos - 0.2, df15["New Deaths"], width=0.4, label="New Deaths")
plt.bar(xpos + 0.2, df15["Total Recovered"], width=0.4, label="Total Recovered")
plt.legend(loc="best")
plt.show()
labels = ["Country", "Total Cases", "New Cases", "Total Deaths"]
sizes = [30, 40, 50, 60]
plt.pie(sizes, labels=labels, autopct="%1.1f%%", explode=[0.1, 0, 0, 0.1])
plt.title("Covid19")
plt.show()
| false | 1 | 934 | 2 | 1,337 | 934 |
||
129113198
|
<jupyter_start><jupyter_text>Housing price prediction
*Abount the Dataset:
Used in Belsley, Kuh & Welsch, 'Regression diagnostics …', Wiley,1980. N.B. Various transformations are used in the table on pages 244-261. Quinlan (1993). Combining Instance-Based and Model-Based Learning. In Proceedings on the Tenth International Conference of Machine Learning, 236-243, University of Massachusetts, Amherst. Morgan Kaufmann.
** Relevant Information: Concerns housing values in suburbs of Boston.
** Number of Instances: 509
** Number of Attributes: 13 continuous attributes (including "class" attribute "MEDV"), 1 binary-valued attribute.
Attribute Information:
1) CRIM : per capita crime rate by town.
2) ZN : proportion of residential land zoned for lots over 25,000 sq.ft.
3) INDUS: proportion of non-retail business acres per town.
4) CHAS : Charles River dummy variable (= 1 if tract bounds river; 0 otherwise).
5) NOX : nitric oxides concentration (parts per 10 million).
6) RM : average number of rooms per dwelling.
7) AGE : proportion of owner-occupied units built prior to 1940.
8) DIS : weighted distances to five Boston employment centres.
9) RAD : index of accessibility to radial highways.
10) TAX : full-value property-tax rate per $10,000.
11) PTRATIO : pupil-teacher ratio by town.
12) B : 1000(Bk - 0.63)^2 where Bk is the proportion of blacks by town.
13) LSTAT: % lower status of the population.
14) MEDV : Median value of owner-occupied homes in $1000's.*
Kaggle dataset identifier: house
<jupyter_script># IMPORT LIBRARY
from pathlib import Path
import re
import pandas as pd
from scipy import stats
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# LOADING THE DATASET
df = pd.read_csv("housing_price.csv")
# EXPLORATORY DATA ANALYSIS (EDA)
df.head()
df.tail()
##CHECKING THE SHAPE OF THE DATASET
df.shape
##CHECKING THE COLUMNS NAMES
col = df.columns
col
df.describe(include="all")
df.dtypes
df.info()
df.nunique()
##CHECKING DUPLICATE
df.duplicated().sum()
##DELETE DUPLICATE
df = df.drop_duplicates()
df.duplicated().sum()
##CHECKING MISSING VALUES
df.isna().sum()
# MISSING VALUE
df.dropna().shape
##SET THE VALUES
df["INDUS"] = df["INDUS"].fillna(df["INDUS"].median())
##SET THE VALUES
df["NOX"] = df["NOX"].fillna(df["NOX"].median())
##SET THE VALUES
df["AGE"] = df["AGE"].fillna(df["AGE"].median())
##SET THE VALUES
df["RAD"] = df["RAD"].fillna(df["RAD"].median())
##SET THE VALUES
df["LSTAT"] = df["LSTAT"].fillna(df["LSTAT"].median())
df.isna().sum()
##CHECKING THE DTYPES DETAIL WISE
df.info()
# CONVERT TYPE
df["AGE"] = df["AGE"].astype(int)
df["ZN"] = df["ZN"].astype(int)
df["RAD"] = df["RAD"].astype(int)
df["TAX"] = df["TAX"].astype(int)
## AFTER CHANGE TYPE
df.info()
##STATISTICAL SUMMARY OF THE DATASET
df.describe().T
def plot_hist(df):
cols = df.select_dtypes(include=[int, float]).columns
ncols = 2
nrows = np.ceil(len(cols) / ncols).astype(int)
vertical_figsize = 2 * nrows
fig, axs = plt.subplots(nrows, ncols, figsize=[10, vertical_figsize])
fig.patch.set_facecolor("lightgray")
axs = axs.flatten()
for col, ax in zip(cols, axs):
df[col].plot.hist(title=col, ax=ax, color="pink")
plt.tight_layout()
plt.show()
plot_hist(df)
# PROCESSING
target = "TAX"
target = "MEDV"
df["CRIM_ZN"] = df["CRIM"] * df["ZN"]
df["INDUS_CHAS"] = df["INDUS"] * df["CHAS"]
df["NOX_DIS"] = df["NOX"] * df["DIS"]
df["RM_AGE"] = df["RM"] * df["AGE"]
df["RAD_LSTAT"] = df["RAD"] * df["LSTAT"]
df["PTRATIO_B"] = df["PTRATIO"] * df["B"]
skew_res = df.select_dtypes([int, float]).skew().abs().sort_values(ascending=False)
skew_cols = skew_res.loc[lambda x: (x >= 1) & (x.index != target)].index
print(skew_res)
print("-" * 50)
print("Cols that are skewed:")
print(", ".join(skew_cols))
def best_transformation(data) -> tuple:
functions = [np.log1p, np.sqrt, stats.yeojohnson]
results = []
for func in functions:
transformed_data = func(data)
if type(transformed_data) == tuple:
vals, _ = transformed_data
results.append(vals)
else:
results.append(transformed_data)
abs_skew_results = [np.abs(stats.skew(val)) for val in results]
lowest_skew_index = abs_skew_results.index(min(abs_skew_results))
return functions[lowest_skew_index], results[lowest_skew_index]
def unskew(col):
global best_transformation
print("-" * 100)
col_skew = stats.skew(col)
col_name = col.name
print("{} skew is: {}".format(col_name, col_skew))
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=[10, 4])
fig.patch.set_facecolor("lightgray")
col.plot.hist(color="red", alpha=0.4, label="pre-skew", ax=ax1)
if np.abs(col_skew) >= 1.0:
result_skew, data = best_transformation(col)
new_col_skew = stats.skew(data)
print(f"Best function {result_skew} and the skew results: {new_col_skew}")
ax2.hist(data, label="Processing", color="blue", alpha=0.4)
ax2.legend()
plt.show()
if np.abs(new_col_skew) >= 1.0:
print(
f"Transformation was not successful for {col_name}, returning original data"
)
return col
return data
plt.show()
skew_cols = [
"CRIM",
"INDUS_CHAS",
"CRIM_ZN",
"CHAS",
"B",
"ZN",
"PTRATIO_B",
"RAD_LSTAT",
"RAD",
"DIS",
] # LIST OF COLUMNS TO CHANGE
df[skew_cols] = df[skew_cols].apply(unskew)
##FEATURE SLECTION
corr_ranking = (
df.drop(target, axis=1).corrwith(df[target]).abs().sort_values(ascending=False)
)
plt.figure(figsize=(10, 10))
heatmap = sns.heatmap(df.corr(), annot=True, cmap="viridis")
plt.show()
# A correlation whose magnitude is between 0.9 and 1.0 can be considered very highly correlated. A correlation whose magnitude is between 0.7 and 0.9 can be considered highly correlated. Correlation whose magnitude is between 0.5 and 0.7 indicates a variable that can be considered moderately correlated. Correlation whose size is between 0.3 and 0.5 indicates a variable that has a low correlation. Correlation whose magnitude is less than 0.3 has a small (linear) correlation if any.
##TAKES ALL VALUES FROM LARGEST TO SMALLEST
threshold = 0.0
chosen_cols = corr_ranking[corr_ranking >= threshold]
print(chosen_cols)
chosen_cols = chosen_cols.index.to_list()
# TRAIN TEST SPLIT
X = df[chosen_cols]
y = df[target]
X.shape, y.shape
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
X_train.dtypes
# SCALING
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
cols = X_train.select_dtypes([float, int]).columns.to_list()
X_train[cols] = scaler.fit_transform(X_train)
X_test[cols] = scaler.transform(X_test)
# REGRESSION
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import RandomForestRegressor
from xgboost import XGBRegressor
from sklearn.experimental import enable_halving_search_cv
from sklearn.model_selection import HalvingGridSearchCV
n_features = X_train.shape[1]
linear_reg_params = {
"fit_intercept": [True, False],
}
random_forest_params = {
"n_estimators": np.sort(
np.random.default_rng().choice(500, size=10, replace=False)
),
"max_features": np.sort(
np.random.default_rng().choice(n_features, size=5, replace=False)
),
"max_depth": [1, 5, 10],
}
xgb_params = {
"objective": ["reg:squarederror"],
"max_depth": [
2,
5,
],
"min_child_weight": np.arange(1, 5, 2),
"n_estimators": np.sort(np.random.default_rng().choice(500, size=3, replace=False)),
"learning_rate": [
1e-1,
1e-2,
],
"gamma": np.sort(np.random.default_rng().choice(20, size=3, replace=False)),
"reg_lambda": [0, 1.0, 10.0],
"scale_pos_weight": [1, 3, 5],
"n_jobs": [-1],
}
best_mode_params = {
LinearRegression(): {"fit_intercept": True},
RandomForestRegressor(): {"max_depth": 10, "max_features": 9, "n_estimators": 378},
XGBRegressor(): {
"gamma": 18,
"learning_rate": 0.1,
"max_depth": 2,
"min_child_weight": 3,
"n_estimators": 461,
"n_jobs": -1,
"objective": "reg:squarederror",
"reg_lambda": 0,
"scale_pos_weight": 1,
},
}
from sklearn.metrics import mean_squared_error, r2_score
b_models = []
model_results = []
for model in best_mode_params.keys():
params = best_mode_params[model]
model.set_params(**params)
model.fit(X_train, y_train)
b_models.append(model)
y_pred = model.predict(X_test)
mse = mean_squared_error(y_test, y_pred)
rmse = np.sqrt(mse)
r2 = r2_score(y_test, y_pred)
model_name = re.search(r"\w+", str(model))[0]
results = pd.Series({"MSE": mse, "RMSE": rmse, "R2": r2}, name=model_name)
model_results.append(results)
# RESULTS
pd.concat(model_results, axis=1)
feature_imp = []
for model in b_models:
try:
model_name = re.search(r"\w+", str(model))[0]
feature_imp.append(
pd.Series(
{
col: importance
for col, importance in zip(cols, model.feature_importances_)
},
name=model_name,
)
)
except AttributeError:
pass
pd.concat(feature_imp, axis=1).sort_values(by="XGBRegressor", ascending=False)
xgb_model = b_models[2]
col = "LSTAT"
y_pred = xgb_model.predict(X_test.sort_values(by=col))
fig, ax = plt.subplots(figsize=(10, 6))
fig.patch.set_facecolor("lightgray")
(
pd.concat([X_test[col], y_test], axis=1)
.sort_values(by=col)
.plot.area(x=col, y="MEDV", color="green", alpha=0.4, label="Actual", ax=ax)
)
plt.scatter(X_test[col].sort_values(), y_pred, color="red", label="Prediction")
plt.legend()
plt.title("BASED ON POPULATION", size=10)
ax.set_facecolor("lightyellow")
plt.show()
xgb_model = b_models[2]
col = "TAX"
y_pred = xgb_model.predict(X_test.sort_values(by=col))
fig, ax = plt.subplots(figsize=(10, 6))
fig.patch.set_facecolor("lightgray")
(
pd.concat([X_test[col], y_test], axis=1)
.sort_values(by=col)
.plot.area(x=col, y="MEDV", color="green", alpha=0.4, label="Actual", ax=ax)
)
plt.scatter(X_test[col].sort_values(), y_pred, color="red", label="Prediction")
plt.legend()
plt.title("BASED ON TAX", size=10)
ax.set_facecolor("lightyellow")
plt.show()
xgb_model = b_models[2]
col = "RM"
y_pred = xgb_model.predict(X_test.sort_values(by=col))
fig, ax = plt.subplots(figsize=(10, 6))
fig.patch.set_facecolor("lightgray")
(
pd.concat([X_test[col], y_test], axis=1)
.sort_values(by=col)
.plot.area(x=col, y="MEDV", color="green", alpha=0.4, label="Actual", ax=ax)
)
plt.scatter(X_test[col].sort_values(), y_pred, color="red", label="Prediction")
plt.legend()
plt.title("BASED ON THE NUMBER OF ROOMS PER OCCUPANCY", size=10)
ax.set_facecolor("lightyellow")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/113/129113198.ipynb
|
house
|
shubhammeshram579
|
[{"Id": 129113198, "ScriptId": 38383163, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5865324, "CreationDate": "05/11/2023 05:32:43", "VersionNumber": 1.0, "Title": "notebook2a1a05e7c7", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 354.0, "LinesInsertedFromPrevious": 354.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 184880803, "KernelVersionId": 129113198, "SourceDatasetVersionId": 5455657}]
|
[{"Id": 5455657, "DatasetId": 3152878, "DatasourceVersionId": 5529778, "CreatorUserId": 10732137, "LicenseName": "CC0: Public Domain", "CreationDate": "04/19/2023 09:37:25", "VersionNumber": 1.0, "Title": "Housing price prediction", "Slug": "house", "Subtitle": "A Machine Learning Approach", "Description": "*Abount the Dataset:\nUsed in Belsley, Kuh & Welsch, 'Regression diagnostics \u2026', Wiley,1980. N.B. Various transformations are used in the table on pages 244-261. Quinlan (1993). Combining Instance-Based and Model-Based Learning. In Proceedings on the Tenth International Conference of Machine Learning, 236-243, University of Massachusetts, Amherst. Morgan Kaufmann.\n\n** Relevant Information: Concerns housing values in suburbs of Boston.\n** Number of Instances: 509\n** Number of Attributes: 13 continuous attributes (including \"class\" attribute \"MEDV\"), 1 binary-valued attribute.\n\nAttribute Information:\n\n1) CRIM : per capita crime rate by town.\n2) ZN : proportion of residential land zoned for lots over 25,000 sq.ft.\n3) INDUS: proportion of non-retail business acres per town.\n4) CHAS : Charles River dummy variable (= 1 if tract bounds river; 0 otherwise).\n5) NOX : nitric oxides concentration (parts per 10 million).\n6) RM : average number of rooms per dwelling.\n7) AGE : proportion of owner-occupied units built prior to 1940.\n8) DIS : weighted distances to five Boston employment centres.\n9) RAD : index of accessibility to radial highways.\n10) TAX : full-value property-tax rate per $10,000.\n11) PTRATIO : pupil-teacher ratio by town.\n12) B : 1000(Bk - 0.63)^2 where Bk is the proportion of blacks by town.\n13) LSTAT: % lower status of the population.\n14) MEDV : Median value of owner-occupied homes in $1000's.*", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3152878, "CreatorUserId": 10732137, "OwnerUserId": 10732137.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5455657.0, "CurrentDatasourceVersionId": 5529778.0, "ForumId": 3216846, "Type": 2, "CreationDate": "04/19/2023 09:37:25", "LastActivityDate": "04/19/2023", "TotalViews": 24132, "TotalDownloads": 3575, "TotalVotes": 67, "TotalKernels": 8}]
|
[{"Id": 10732137, "UserName": "shubhammeshram579", "DisplayName": "shubham meshram", "RegisterDate": "06/04/2022", "PerformanceTier": 0}]
|
# IMPORT LIBRARY
from pathlib import Path
import re
import pandas as pd
from scipy import stats
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# LOADING THE DATASET
df = pd.read_csv("housing_price.csv")
# EXPLORATORY DATA ANALYSIS (EDA)
df.head()
df.tail()
##CHECKING THE SHAPE OF THE DATASET
df.shape
##CHECKING THE COLUMNS NAMES
col = df.columns
col
df.describe(include="all")
df.dtypes
df.info()
df.nunique()
##CHECKING DUPLICATE
df.duplicated().sum()
##DELETE DUPLICATE
df = df.drop_duplicates()
df.duplicated().sum()
##CHECKING MISSING VALUES
df.isna().sum()
# MISSING VALUE
df.dropna().shape
##SET THE VALUES
df["INDUS"] = df["INDUS"].fillna(df["INDUS"].median())
##SET THE VALUES
df["NOX"] = df["NOX"].fillna(df["NOX"].median())
##SET THE VALUES
df["AGE"] = df["AGE"].fillna(df["AGE"].median())
##SET THE VALUES
df["RAD"] = df["RAD"].fillna(df["RAD"].median())
##SET THE VALUES
df["LSTAT"] = df["LSTAT"].fillna(df["LSTAT"].median())
df.isna().sum()
##CHECKING THE DTYPES DETAIL WISE
df.info()
# CONVERT TYPE
df["AGE"] = df["AGE"].astype(int)
df["ZN"] = df["ZN"].astype(int)
df["RAD"] = df["RAD"].astype(int)
df["TAX"] = df["TAX"].astype(int)
## AFTER CHANGE TYPE
df.info()
##STATISTICAL SUMMARY OF THE DATASET
df.describe().T
def plot_hist(df):
cols = df.select_dtypes(include=[int, float]).columns
ncols = 2
nrows = np.ceil(len(cols) / ncols).astype(int)
vertical_figsize = 2 * nrows
fig, axs = plt.subplots(nrows, ncols, figsize=[10, vertical_figsize])
fig.patch.set_facecolor("lightgray")
axs = axs.flatten()
for col, ax in zip(cols, axs):
df[col].plot.hist(title=col, ax=ax, color="pink")
plt.tight_layout()
plt.show()
plot_hist(df)
# PROCESSING
target = "TAX"
target = "MEDV"
df["CRIM_ZN"] = df["CRIM"] * df["ZN"]
df["INDUS_CHAS"] = df["INDUS"] * df["CHAS"]
df["NOX_DIS"] = df["NOX"] * df["DIS"]
df["RM_AGE"] = df["RM"] * df["AGE"]
df["RAD_LSTAT"] = df["RAD"] * df["LSTAT"]
df["PTRATIO_B"] = df["PTRATIO"] * df["B"]
skew_res = df.select_dtypes([int, float]).skew().abs().sort_values(ascending=False)
skew_cols = skew_res.loc[lambda x: (x >= 1) & (x.index != target)].index
print(skew_res)
print("-" * 50)
print("Cols that are skewed:")
print(", ".join(skew_cols))
def best_transformation(data) -> tuple:
functions = [np.log1p, np.sqrt, stats.yeojohnson]
results = []
for func in functions:
transformed_data = func(data)
if type(transformed_data) == tuple:
vals, _ = transformed_data
results.append(vals)
else:
results.append(transformed_data)
abs_skew_results = [np.abs(stats.skew(val)) for val in results]
lowest_skew_index = abs_skew_results.index(min(abs_skew_results))
return functions[lowest_skew_index], results[lowest_skew_index]
def unskew(col):
global best_transformation
print("-" * 100)
col_skew = stats.skew(col)
col_name = col.name
print("{} skew is: {}".format(col_name, col_skew))
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=[10, 4])
fig.patch.set_facecolor("lightgray")
col.plot.hist(color="red", alpha=0.4, label="pre-skew", ax=ax1)
if np.abs(col_skew) >= 1.0:
result_skew, data = best_transformation(col)
new_col_skew = stats.skew(data)
print(f"Best function {result_skew} and the skew results: {new_col_skew}")
ax2.hist(data, label="Processing", color="blue", alpha=0.4)
ax2.legend()
plt.show()
if np.abs(new_col_skew) >= 1.0:
print(
f"Transformation was not successful for {col_name}, returning original data"
)
return col
return data
plt.show()
skew_cols = [
"CRIM",
"INDUS_CHAS",
"CRIM_ZN",
"CHAS",
"B",
"ZN",
"PTRATIO_B",
"RAD_LSTAT",
"RAD",
"DIS",
] # LIST OF COLUMNS TO CHANGE
df[skew_cols] = df[skew_cols].apply(unskew)
##FEATURE SLECTION
corr_ranking = (
df.drop(target, axis=1).corrwith(df[target]).abs().sort_values(ascending=False)
)
plt.figure(figsize=(10, 10))
heatmap = sns.heatmap(df.corr(), annot=True, cmap="viridis")
plt.show()
# A correlation whose magnitude is between 0.9 and 1.0 can be considered very highly correlated. A correlation whose magnitude is between 0.7 and 0.9 can be considered highly correlated. Correlation whose magnitude is between 0.5 and 0.7 indicates a variable that can be considered moderately correlated. Correlation whose size is between 0.3 and 0.5 indicates a variable that has a low correlation. Correlation whose magnitude is less than 0.3 has a small (linear) correlation if any.
##TAKES ALL VALUES FROM LARGEST TO SMALLEST
threshold = 0.0
chosen_cols = corr_ranking[corr_ranking >= threshold]
print(chosen_cols)
chosen_cols = chosen_cols.index.to_list()
# TRAIN TEST SPLIT
X = df[chosen_cols]
y = df[target]
X.shape, y.shape
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
X_train.dtypes
# SCALING
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
cols = X_train.select_dtypes([float, int]).columns.to_list()
X_train[cols] = scaler.fit_transform(X_train)
X_test[cols] = scaler.transform(X_test)
# REGRESSION
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import RandomForestRegressor
from xgboost import XGBRegressor
from sklearn.experimental import enable_halving_search_cv
from sklearn.model_selection import HalvingGridSearchCV
n_features = X_train.shape[1]
linear_reg_params = {
"fit_intercept": [True, False],
}
random_forest_params = {
"n_estimators": np.sort(
np.random.default_rng().choice(500, size=10, replace=False)
),
"max_features": np.sort(
np.random.default_rng().choice(n_features, size=5, replace=False)
),
"max_depth": [1, 5, 10],
}
xgb_params = {
"objective": ["reg:squarederror"],
"max_depth": [
2,
5,
],
"min_child_weight": np.arange(1, 5, 2),
"n_estimators": np.sort(np.random.default_rng().choice(500, size=3, replace=False)),
"learning_rate": [
1e-1,
1e-2,
],
"gamma": np.sort(np.random.default_rng().choice(20, size=3, replace=False)),
"reg_lambda": [0, 1.0, 10.0],
"scale_pos_weight": [1, 3, 5],
"n_jobs": [-1],
}
best_mode_params = {
LinearRegression(): {"fit_intercept": True},
RandomForestRegressor(): {"max_depth": 10, "max_features": 9, "n_estimators": 378},
XGBRegressor(): {
"gamma": 18,
"learning_rate": 0.1,
"max_depth": 2,
"min_child_weight": 3,
"n_estimators": 461,
"n_jobs": -1,
"objective": "reg:squarederror",
"reg_lambda": 0,
"scale_pos_weight": 1,
},
}
from sklearn.metrics import mean_squared_error, r2_score
b_models = []
model_results = []
for model in best_mode_params.keys():
params = best_mode_params[model]
model.set_params(**params)
model.fit(X_train, y_train)
b_models.append(model)
y_pred = model.predict(X_test)
mse = mean_squared_error(y_test, y_pred)
rmse = np.sqrt(mse)
r2 = r2_score(y_test, y_pred)
model_name = re.search(r"\w+", str(model))[0]
results = pd.Series({"MSE": mse, "RMSE": rmse, "R2": r2}, name=model_name)
model_results.append(results)
# RESULTS
pd.concat(model_results, axis=1)
feature_imp = []
for model in b_models:
try:
model_name = re.search(r"\w+", str(model))[0]
feature_imp.append(
pd.Series(
{
col: importance
for col, importance in zip(cols, model.feature_importances_)
},
name=model_name,
)
)
except AttributeError:
pass
pd.concat(feature_imp, axis=1).sort_values(by="XGBRegressor", ascending=False)
xgb_model = b_models[2]
col = "LSTAT"
y_pred = xgb_model.predict(X_test.sort_values(by=col))
fig, ax = plt.subplots(figsize=(10, 6))
fig.patch.set_facecolor("lightgray")
(
pd.concat([X_test[col], y_test], axis=1)
.sort_values(by=col)
.plot.area(x=col, y="MEDV", color="green", alpha=0.4, label="Actual", ax=ax)
)
plt.scatter(X_test[col].sort_values(), y_pred, color="red", label="Prediction")
plt.legend()
plt.title("BASED ON POPULATION", size=10)
ax.set_facecolor("lightyellow")
plt.show()
xgb_model = b_models[2]
col = "TAX"
y_pred = xgb_model.predict(X_test.sort_values(by=col))
fig, ax = plt.subplots(figsize=(10, 6))
fig.patch.set_facecolor("lightgray")
(
pd.concat([X_test[col], y_test], axis=1)
.sort_values(by=col)
.plot.area(x=col, y="MEDV", color="green", alpha=0.4, label="Actual", ax=ax)
)
plt.scatter(X_test[col].sort_values(), y_pred, color="red", label="Prediction")
plt.legend()
plt.title("BASED ON TAX", size=10)
ax.set_facecolor("lightyellow")
plt.show()
xgb_model = b_models[2]
col = "RM"
y_pred = xgb_model.predict(X_test.sort_values(by=col))
fig, ax = plt.subplots(figsize=(10, 6))
fig.patch.set_facecolor("lightgray")
(
pd.concat([X_test[col], y_test], axis=1)
.sort_values(by=col)
.plot.area(x=col, y="MEDV", color="green", alpha=0.4, label="Actual", ax=ax)
)
plt.scatter(X_test[col].sort_values(), y_pred, color="red", label="Prediction")
plt.legend()
plt.title("BASED ON THE NUMBER OF ROOMS PER OCCUPANCY", size=10)
ax.set_facecolor("lightyellow")
plt.show()
| false | 0 | 3,268 | 1 | 3,748 | 3,268 |
||
129113644
|
<jupyter_start><jupyter_text>Mobile Games: A/B Testing
This dataset is from a DataCamp project: https://www.datacamp.com/projects/184.
The data is about an A/B test with a mobile game, Cookie Cats.
Kaggle dataset identifier: mobile-games-ab-testing
<jupyter_script># 
# #### 쿠키캣츠(Cookie Cats)는 타일을 연결해 보드를 클리어하는 고전적인 connect three 스타일의 퍼즐 게임으로, Tactile Entertainment에서 개발했습니다. 이 게임은 게이트(Gate)에 도달하기 위해 기다리거나, 인앱 구매를 해야만 계속 플레이할 수 있습니다. 게이트는 게임을 더 오래 즐길 수 있도록 중간중간 휴식을 취하게 하고, 인앱 구매를 유도하는 역할도 합니다. 그렇다면, 게이트를 어느 레벨에 배치해야 하는 것이 가장 좋을까요? 이를 확인하기 위해 A/B 테스트를 진행하여 기존 레벨 30에서 게이트를 레벨 40으로 이동하는 것이 어떤 영향을 미치는지 살펴보았습니다. 특히 이번 분석의 주요 목적은 게이트 레벨이 플레이어의 Retention(재방문율)에 영향을 미치는지 아닌지 파악하는 것입니다.
# * userid - 각 플레이어를 식별하는 고유 번호
# * version - lv.30의 gate에 속했는지, lv.40의 gate에 속했는지 여부
# * sum_gamerounds - 설치 후 첫 14일 동안 플레이어가 플레이한 게임 라운드 수
# * retention_1 - 플레이어가 설치 후 1일 후에 다시 돌아와서 플레이 했는지 여부
# * retention_7 - 플레이어가 설치 후 7일 후에 다시 돌아와서 플레이 했는지 여부
# (** 참고사항 - 플레이어가 게임을 설치할 때 무작위로 gate_30 또는 gate_40에 배정 됨)
# ## Step 1. Importing Libraries and Loading Data
#
# 필요모듈 호출
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import os
from scipy.stats import shapiro
import scipy.stats as stats
import warnings
warnings.filterwarnings("ignore")
warnings.simplefilter(action="ignore", category=FutureWarning)
pd.set_option("display.max_columns", None)
pd.options.display.float_format = "{:.4f}".format
cookie = pd.read_csv("/kaggle/input/mobile-games-ab-testing/cookie_cats.csv")
# ## Step 2. Data Understanding (EDA)
print("- Number of rows: {}".format(cookie.shape[0]))
print("- Number of columns: {}".format(cookie.shape[1]))
print("- Name of independent variables: {}".format(list(cookie.columns[:-1])))
print("- Name of target: {}".format(list(cookie.columns[-1:])))
print("- Dataset shape:{}".format(cookie.shape))
cookie.info()
cookie.head()
cookie.tail()
cookie.describe()
# 결측치 확인
print("Null values: \n", cookie.isnull().sum())
uniques = cookie.nunique(axis=0)
print(uniques)
cookie.groupby("version").sum_gamerounds.agg(["count", "median", "mean", "std", "max"])
# Setting chart style
palette = "cividis"
sns.set_style("darkgrid")
sns.set_context("notebook")
sns.set_palette(palette)
# Defining a dictionary for font properties
labels = {"family": "Helvetica", "color": "#030303", "weight": "normal", "size": 14}
suptitles = {"family": "Roboto", "color": "#0d0d0d", "weight": "light", "size": 18}
titles = {"family": "Helvetica", "color": "#0d0d0d", "weight": "light", "size": 16}
# Counting the number of players for each number of game rounds
plot_cookie = pd.DataFrame(
cookie.groupby("sum_gamerounds")["userid"].count()
).reset_index()
# Calculating % of users that came back the day after they installed
ret1 = cookie["retention_1"].mean()
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(16, 8))
fig.suptitle(
f"Distribution of Observations per Round & Version\n 1-Day Retention:{ret1:.3%}",
fontdict=suptitles,
)
# Creating the bar chart
sns.countplot(x="version", data=cookie, ax=ax2)
# Setting the title and axis labels
ax2.set_title("Number of Observations per Version", fontdict=titles)
ax2.set_xlabel("Version", fontdict=labels)
ax2.set_ylabel("Number of Observations", fontdict=labels)
# Creating the histogram
sns.histplot(
x="userid", data=plot_df.head(n=100), element="step", kde=True, ax=ax1, bins=30
)
ax1.set_xlabel("Game Rounds Sum", fontdict=labels)
ax1.set_ylabel("Number of Observations", fontdict=labels)
ax1.set_title(f"Distribution of Game Rounds Sum", fontdict=titles)
plt.show()
fig, axes = plt.subplots(2, 1, figsize=(30, 10))
cookie.groupby("sum_gamerounds").userid.count().plot(ax=axes[0])
cookie.groupby("sum_gamerounds").userid.count()[:200].plot(ax=axes[1])
plt.suptitle("The number of users in the game rounds played", fontsize=20)
axes[0].set_title("How many users are there all game rounds?", fontsize=12)
axes[1].set_title("How many users are there first 200 game rounds?", fontsize=12)
plt.tight_layout(pad=5)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/113/129113644.ipynb
|
mobile-games-ab-testing
|
yufengsui
|
[{"Id": 129113644, "ScriptId": 37820577, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13662512, "CreationDate": "05/11/2023 05:37:16", "VersionNumber": 4.0, "Title": "Cookie Cats", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 146.0, "LinesInsertedFromPrevious": 107.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 39.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184881609, "KernelVersionId": 129113644, "SourceDatasetVersionId": 564457}]
|
[{"Id": 564457, "DatasetId": 272421, "DatasourceVersionId": 581103, "CreatorUserId": 1461568, "LicenseName": "Unknown", "CreationDate": "07/22/2019 08:37:03", "VersionNumber": 1.0, "Title": "Mobile Games: A/B Testing", "Slug": "mobile-games-ab-testing", "Subtitle": "Analyze an A/B test from the popular mobile puzzle game, Cookie Cats.", "Description": "This dataset is from a DataCamp project: https://www.datacamp.com/projects/184.\n\nThe data is about an A/B test with a mobile game, Cookie Cats.", "VersionNotes": "Initial release", "TotalCompressedBytes": 2797485.0, "TotalUncompressedBytes": 511415.0}]
|
[{"Id": 272421, "CreatorUserId": 1461568, "OwnerUserId": 1461568.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 564457.0, "CurrentDatasourceVersionId": 581103.0, "ForumId": 283777, "Type": 2, "CreationDate": "07/22/2019 08:37:03", "LastActivityDate": "07/22/2019", "TotalViews": 34634, "TotalDownloads": 3307, "TotalVotes": 76, "TotalKernels": 68}]
|
[{"Id": 1461568, "UserName": "yufengsui", "DisplayName": "Aurelia Sui", "RegisterDate": "12/01/2017", "PerformanceTier": 1}]
|
# 
# #### 쿠키캣츠(Cookie Cats)는 타일을 연결해 보드를 클리어하는 고전적인 connect three 스타일의 퍼즐 게임으로, Tactile Entertainment에서 개발했습니다. 이 게임은 게이트(Gate)에 도달하기 위해 기다리거나, 인앱 구매를 해야만 계속 플레이할 수 있습니다. 게이트는 게임을 더 오래 즐길 수 있도록 중간중간 휴식을 취하게 하고, 인앱 구매를 유도하는 역할도 합니다. 그렇다면, 게이트를 어느 레벨에 배치해야 하는 것이 가장 좋을까요? 이를 확인하기 위해 A/B 테스트를 진행하여 기존 레벨 30에서 게이트를 레벨 40으로 이동하는 것이 어떤 영향을 미치는지 살펴보았습니다. 특히 이번 분석의 주요 목적은 게이트 레벨이 플레이어의 Retention(재방문율)에 영향을 미치는지 아닌지 파악하는 것입니다.
# * userid - 각 플레이어를 식별하는 고유 번호
# * version - lv.30의 gate에 속했는지, lv.40의 gate에 속했는지 여부
# * sum_gamerounds - 설치 후 첫 14일 동안 플레이어가 플레이한 게임 라운드 수
# * retention_1 - 플레이어가 설치 후 1일 후에 다시 돌아와서 플레이 했는지 여부
# * retention_7 - 플레이어가 설치 후 7일 후에 다시 돌아와서 플레이 했는지 여부
# (** 참고사항 - 플레이어가 게임을 설치할 때 무작위로 gate_30 또는 gate_40에 배정 됨)
# ## Step 1. Importing Libraries and Loading Data
#
# 필요모듈 호출
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import os
from scipy.stats import shapiro
import scipy.stats as stats
import warnings
warnings.filterwarnings("ignore")
warnings.simplefilter(action="ignore", category=FutureWarning)
pd.set_option("display.max_columns", None)
pd.options.display.float_format = "{:.4f}".format
cookie = pd.read_csv("/kaggle/input/mobile-games-ab-testing/cookie_cats.csv")
# ## Step 2. Data Understanding (EDA)
print("- Number of rows: {}".format(cookie.shape[0]))
print("- Number of columns: {}".format(cookie.shape[1]))
print("- Name of independent variables: {}".format(list(cookie.columns[:-1])))
print("- Name of target: {}".format(list(cookie.columns[-1:])))
print("- Dataset shape:{}".format(cookie.shape))
cookie.info()
cookie.head()
cookie.tail()
cookie.describe()
# 결측치 확인
print("Null values: \n", cookie.isnull().sum())
uniques = cookie.nunique(axis=0)
print(uniques)
cookie.groupby("version").sum_gamerounds.agg(["count", "median", "mean", "std", "max"])
# Setting chart style
palette = "cividis"
sns.set_style("darkgrid")
sns.set_context("notebook")
sns.set_palette(palette)
# Defining a dictionary for font properties
labels = {"family": "Helvetica", "color": "#030303", "weight": "normal", "size": 14}
suptitles = {"family": "Roboto", "color": "#0d0d0d", "weight": "light", "size": 18}
titles = {"family": "Helvetica", "color": "#0d0d0d", "weight": "light", "size": 16}
# Counting the number of players for each number of game rounds
plot_cookie = pd.DataFrame(
cookie.groupby("sum_gamerounds")["userid"].count()
).reset_index()
# Calculating % of users that came back the day after they installed
ret1 = cookie["retention_1"].mean()
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(16, 8))
fig.suptitle(
f"Distribution of Observations per Round & Version\n 1-Day Retention:{ret1:.3%}",
fontdict=suptitles,
)
# Creating the bar chart
sns.countplot(x="version", data=cookie, ax=ax2)
# Setting the title and axis labels
ax2.set_title("Number of Observations per Version", fontdict=titles)
ax2.set_xlabel("Version", fontdict=labels)
ax2.set_ylabel("Number of Observations", fontdict=labels)
# Creating the histogram
sns.histplot(
x="userid", data=plot_df.head(n=100), element="step", kde=True, ax=ax1, bins=30
)
ax1.set_xlabel("Game Rounds Sum", fontdict=labels)
ax1.set_ylabel("Number of Observations", fontdict=labels)
ax1.set_title(f"Distribution of Game Rounds Sum", fontdict=titles)
plt.show()
fig, axes = plt.subplots(2, 1, figsize=(30, 10))
cookie.groupby("sum_gamerounds").userid.count().plot(ax=axes[0])
cookie.groupby("sum_gamerounds").userid.count()[:200].plot(ax=axes[1])
plt.suptitle("The number of users in the game rounds played", fontsize=20)
axes[0].set_title("How many users are there all game rounds?", fontsize=12)
axes[1].set_title("How many users are there first 200 game rounds?", fontsize=12)
plt.tight_layout(pad=5)
| false | 1 | 1,551 | 0 | 1,623 | 1,551 |
||
129113117
|
<jupyter_start><jupyter_text>Covid-19-country-statistics-dataset
The COVID-19 Country Statistics Dataset, available on Kaggle, is a collection of data related to the COVID-19 pandemic across various countries. The dataset includes information such as the number of confirmed cases, deaths, recoveries, and active cases, as well as information about the population, population density, median age, and various other demographic and health-related statistics for each country.
The dataset contains a total of 13 columns and over 217 rows of data, with each row representing a different country or region. The data is presented in a CSV (comma-separated values) format, making it easy to analyze and work with using a variety of tools and programming languages.
The COVID-19 Country Statistics Dataset contains several column descriptors, including Province/State, Country/Region, Lat, Long, Date, Confirmed, Deaths, and Recovered. Province/State describes the province or state where the cases were detected, while Country/Region describes the country where the cases were detected. Lat and Long describe the latitude and longitude of the country or province/state. Date describes the date when the cases were detected. Confirmed represents the total number of confirmed cases of COVID-19, while Deaths represents the total number of deaths due to COVID-19. Recovered represents the total number of recoveries from COVID-19.
In summary, this dataset is a valuable resource for researchers, analysts, and anyone interested in understanding the spread and impact of COVID-19 across different countries. The dataset provides a detailed picture of the pandemic and can help with decision-making, policy development, and research efforts aimed at understanding the global impact of COVID-19.
Kaggle dataset identifier: covid-19-country-statistics-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/covid-19-country-statistics-dataset/covid-19-country-statistics-dataset.csv"
)
df
nan_values = df.isnull().sum()
nan_values
newdf = df.dropna()
newdf
df.shape
newdf.shape
newdf = df["Total Deaths"].dropna()
newdf
newdf.shape
newdf = df["Country"].dropna(inplace=True)
df.info()
df["New Cases"].fillna(0, inplace=True)
df.info()
NaN_value = df.loc[df["New Cases"].isnull(), "New Cases"].values
NaN_value
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/113/129113117.ipynb
|
covid-19-country-statistics-dataset
|
harshghadiya
|
[{"Id": 129113117, "ScriptId": 38347235, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14442011, "CreationDate": "05/11/2023 05:32:06", "VersionNumber": 1.0, "Title": "covid19 data", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 55.0, "LinesInsertedFromPrevious": 55.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184880686, "KernelVersionId": 129113117, "SourceDatasetVersionId": 5543953}]
|
[{"Id": 5543953, "DatasetId": 3194747, "DatasourceVersionId": 5618626, "CreatorUserId": 9220292, "LicenseName": "Other (specified in description)", "CreationDate": "04/27/2023 23:22:02", "VersionNumber": 1.0, "Title": "Covid-19-country-statistics-dataset", "Slug": "covid-19-country-statistics-dataset", "Subtitle": "A Comprehensive Collection of COVID-19 Statistics Across Countries and Regions.", "Description": "The COVID-19 Country Statistics Dataset, available on Kaggle, is a collection of data related to the COVID-19 pandemic across various countries. The dataset includes information such as the number of confirmed cases, deaths, recoveries, and active cases, as well as information about the population, population density, median age, and various other demographic and health-related statistics for each country.\n\nThe dataset contains a total of 13 columns and over 217 rows of data, with each row representing a different country or region. The data is presented in a CSV (comma-separated values) format, making it easy to analyze and work with using a variety of tools and programming languages.\n\nThe COVID-19 Country Statistics Dataset contains several column descriptors, including Province/State, Country/Region, Lat, Long, Date, Confirmed, Deaths, and Recovered. Province/State describes the province or state where the cases were detected, while Country/Region describes the country where the cases were detected. Lat and Long describe the latitude and longitude of the country or province/state. Date describes the date when the cases were detected. Confirmed represents the total number of confirmed cases of COVID-19, while Deaths represents the total number of deaths due to COVID-19. Recovered represents the total number of recoveries from COVID-19.\n\nIn summary, this dataset is a valuable resource for researchers, analysts, and anyone interested in understanding the spread and impact of COVID-19 across different countries. The dataset provides a detailed picture of the pandemic and can help with decision-making, policy development, and research efforts aimed at understanding the global impact of COVID-19.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3194747, "CreatorUserId": 9220292, "OwnerUserId": 9220292.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5543953.0, "CurrentDatasourceVersionId": 5618626.0, "ForumId": 3259308, "Type": 2, "CreationDate": "04/27/2023 23:22:02", "LastActivityDate": "04/27/2023", "TotalViews": 5036, "TotalDownloads": 1123, "TotalVotes": 33, "TotalKernels": 2}]
|
[{"Id": 9220292, "UserName": "harshghadiya", "DisplayName": "Harsh Ghadiya", "RegisterDate": "12/21/2021", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/covid-19-country-statistics-dataset/covid-19-country-statistics-dataset.csv"
)
df
nan_values = df.isnull().sum()
nan_values
newdf = df.dropna()
newdf
df.shape
newdf.shape
newdf = df["Total Deaths"].dropna()
newdf
newdf.shape
newdf = df["Country"].dropna(inplace=True)
df.info()
df["New Cases"].fillna(0, inplace=True)
df.info()
NaN_value = df.loc[df["New Cases"].isnull(), "New Cases"].values
NaN_value
| false | 1 | 353 | 0 | 756 | 353 |
||
129459665
|
# # 1.Bert demo1 — 使用Bert
# * transformer库中的pipline可以使用预训练模型bert,来执行mask-filling任务
# * mask-filling 任务是指在给定的句子中,找到并填补被\` [MASK]\`符号所代替的部分
# * 上面的填充意味着这个模型可以预测mask所掩盖的部分
from transformers import pipeline
import warnings
warnings.filterwarnings("ignore")
unmasker = pipeline("fill-mask", model="bert-base-uncased")
unmasker("Artificial Intelligence [MASK] take over the world")
# ## Bert模型自带的偏见Bias
# * 需要注意的是预训练中的偏见也会影响到微调任务
unmasker("The man worked as a [MASK]。")
# '[MASK]' 可能被填充为 'doctor','teacher','lawyer' 等等,这完全取决于模型的预测,从下方的结果可以看出
# 这个模型对男性的偏见有点离谱,这种偏见是训练数据不均衡导致的
unmasker("I will [MASK] mountain today, because I have a vacation from yesterday。")
# # 2.Bert demo2 — 使用Bert完成文本分类
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import cross_val_score
import torch
import transformers as ppb
import warnings
warnings.filterwarnings("ignore")
# ## 导入数据集
df = pd.read_csv(
"https://github.com/clairett/pytorch-sentiment-classification/raw/master/data/SST2/train.tsv",
delimiter="\t",
header=None,
)
# 为了节省训练时间,这里只采用前2000个数据进行训练
batch_1 = df.iloc[:2000, :]
batch_1[1].value_counts() # 观察0和1标签的分布
# ## 加载预训练好的 Bert 模型
# * 需要事先定义模型的种类,分词器的种类,预训练模型种类对应的权重名称
# 第一个是模型类别,第二个是分词器类别,第三个是预训练权重
model_class, tokenizer_class, pretrained_weights = (
ppb.DistilBertModel,
ppb.DistilBertTokenizer,
"distilbert-base-uncased",
)
tokenizer = tokenizer_class.from_pretrained(pretrained_weights) # 加载对应base版本的模型
model = model_class.from_pretrained(pretrained_weights)
# ## 经典数据预处理流程
# ### Tokenizer
# 第一步将句子转化为words,在转化为subwords,确保能满足Bert模型的输入要求
# 使用预训练的分词器对数据进行分词
# batch_1[0]表示要处理的数据
# apply函数用于对数据中的每个元素进行操作,这里的操作是lambda函数,
# 在这个匿名函数中调用了 tokenizer.encode 函数,对每个元素(这里是每个文本)进行分词
# add_special_tokens=True 表示在分词的过程中添加特殊的标记,包括开始和结束的标记的等,这些操作都不能少
# Bert 需要这些操作才能统一输入数据
print(batch_1[:5])
tokenized = batch_1[0].apply(lambda x: tokenizer.encode(x, add_special_tokens=True))
# ### Padding
# 文本预处理的经典步骤之padding填充,确保输入的token长度是相同的
max_len = 0
# 长度有两种方式获得,一种是自定义,一种是寻找最长的序列的长度
for i in tokenized.values:
if len(i) > max_len:
max_len = len(i)
# 对序列进行填充
# 然后对所有的句子进行填充,使他们的长度都等于 max_len ,填充的方式是在句子的后面添加0,直到句子的长度等于0
# 用np.array来存储填充后的句子,每一个句子都是一个数组,所有的句子构成了一个二维数组
padded = np.array([i + [0] * (max_len - len(i)) for i in tokenized.values])
padded.shape
# ### Masking
# 对于基于 Encoder 构建的 Bert,需要对 Attention 进行 mask 处理,确保 Padding 出来的0不会对 Attention 过程产生 side effects
attention_mask = np.where(padded != 0, 1, 0)
# np.where 的作用其实就是对每一个元素都进行if判断,满足要求填前面的元素,不满足填充后面的元素
attention_mask.shape
# 将处理好的数据送给模型进行处理,得到模型的输出结果
# 将输入数据和注意力掩码转化为PyTorch张量
# PyTorch的模型需要使用PyTorch的张量作为输入,所以需要将padded和attention_mask从Numpy数组中转化为PyTorch张量
# 使用 torch.tensor 转化,这个操作说明bert的这个模型是基于PyTorch实现的
input_ids = torch.tensor(padded)
# input_ids[:5]
attention_mask = torch.tensor(attention_mask)
# attention_mask[:5]
# 使用模型前向传播
# 使用with torch.no_grad()语句块来冻结梯度计算
# 在进行前向传播时不需要计算梯度,关闭梯度计算可以节省内存,提高计算速度
# 然后将input_ids和attention_mask作为输入,通过模型进行前向传播,的到输出last_hidden_states
# attention_mask=attention_mask作为名为 attentiion_mask的参数传递给模型
# 这段代码会基于你的GPU的能力变化训练时间,如果训练时间过长就把前面的数据集改小一点
with torch.no_grad():
last_hidden_states = model(input_ids, attention_mask=attention_mask)
# 因为Bert模型中,输入句子转化后的数据的第一个位置是[cls],[cls]代表的就是整个句子的 Sentence Encoding 或者说是 Sentence presentation,而作为下游的判别任务的依据就是这个cls token
# 提取特征
# 从last_hidden_states(模型的输出,包含了每个输入词语的隐藏状态)中提取特征
# last_hidden_states[0]取出隐藏状态,因为模型的输出是一个元组,其第一个元素是隐藏状态
# [:,0,:]表示取出每个句子的第一个词语(Bert模型中的CLS标记)的所有隐藏状态
# 在Bert模型中,第一个词语的隐藏状态被用作句子的表示,所以我们通常会使用它作为特征
# .numpy()将隐藏状态从PyTorch转化为numyp,因为这些特征会被送进NN来处理
features = last_hidden_states[0][:, 0, :].numpy() # torch->numpy
# batch[1]是真正的标签,0/1
labels = batch_1[1]
# ### 将数据切分成训练集和测试集
# 直接使用sklearn.model_selection 中的 train_test_split
# 划分训练集和测试集
# 返回训练集特征,测试集的特征,训练集的标签,测试集的标签
train_features, test_features, train_labels, test_labels = train_test_split(
features, labels
)
# 训练最后的逻辑回归模型的参数,虽然Bert是与训练过的,但是做下游任务的模型没有训练过
lr_clf = LogisticRegression()
# 训练逻辑回归模型
lr_clf.fit(train_features, train_labels)
# 计算训练效果
# 评估模型
# 使用score方法来评估模型在测试集上的表现
lr_clf.score(test_features, test_labels)
# 只得到自己的得分是不合理的,这里需要于其他的评价指标进行评价,dummy classifer(模拟分类器/傻瓜分类器?)来进行分类,比较得分 Dummy 是不会进行任何学习的,只是简单地使用一些规则(如预测所有样本都属于最常见的类别)来进行预测,比如使用众数分类
from sklearn.dummy import DummyClassifier
clf = DummyClassifier()
# 使用交叉验证来评估模型的性能
# cross_val_score函数会将数据集划分为k个子集,然后进行k次训练和测试
scores = cross_val_score(clf, train_features, train_labels)
print("Dummy classifier score: %0.3f (+/- %0.2f)" % (scores.mean(), scores.std() * 2))
# # Bert demo 3 — 使用bert进行文本问答
from transformers import BertForQuestionAnswering
# 问题回答任务中,模型需要根据给定的问题和文本,找出文本中回答问题的部分
model = BertForQuestionAnswering.from_pretrained(
"bert-large-uncased-whole-word-masking-finetuned-squad"
)
# 直接使用这个经过微调后的模型
# 提供问题和回答
question = "How many parameters does BERT-large have?"
answer_text = "BERT-large is really big... it has 24-layers and an embedding size of 1,024, for a total of 340M parameters! Altogether it is 1.34GB, so expect it to take a couple minutes to download to your Colab instance."
from transformers import BertTokenizer
# 用哪个模型最好就用这个模型提供的分词器
tokenizer = BertTokenizer.from_pretrained(
"bert-large-uncased-whole-word-masking-finetuned-squad"
)
input_ids = tokenizer.encode(question, answer_text)
print("The input has a total of {:} tokens.".format(len(input_ids)))
# 这里查看token是怎么映射的
# convert_ids_to_tokens,这个方法的作用在于返回索引形式的tokens形成的列表
tokens = tokenizer.convert_ids_to_tokens(input_ids)
for token, id in zip(tokens, input_ids):
if id == tokenizer.sep_token_id:
print("")
print("{:<12} {:>6}".format(token, id))
if id == tokenizer.sep_token_id:
print("")
# 从中可以看出token的时候会加入CLS特殊符号表示这个句子的标签特征向量
# 用特殊符号 SEP 来间隔两个句子
sep_index = input_ids.index(tokenizer.sep_token_id) # 返回第一个 SEP token 的位置
num_seg_a = sep_index + 1
# num_seg_a 表示segment A的长度,包含sep本身的
num_seg_b = len(input_ids) - num_seg_a
# 构造一个由0 1 组成的列表,这个列表表示每个token属于那个segment
segment_ids = [0] * num_seg_a + [1] * num_seg_b
assert len(segment_ids) == len(input_ids)
# 注意:
# * 这里因为我们仅采用一段文本对作为输入,且输入的长度是可控的,因此我们没有进行Padding或Truncate操作
# * 在批量操作中,我们需要加上Padding和Truncate操作确保模型接受的数据长度是一致且小于 Max len 的
import torch
outputs = model(
torch.tensor([input_ids]),
token_type_ids=torch.tensor([segment_ids]),
return_dict=True,
) # 设置为True,使得模型返回一个包含更多信息的字典
# 获得开始和结束得分
start_scores = outputs.start_logits
end_scores = outputs.end_logits
answer_start = torch.argmax(start_scores)
answer_end = torch.argmax(end_scores)
# 将答案中的token组合起来并打印出来
answer = " ".join(tokens[answer_start : answer_end + 1])
print('Answer: "' + answer + '"')
# 注意:
# * 这里只是简单的取了得分最高的start pos和end pos
# * 但是在实际场景中需要考虑start pos index 大于 end pos index 的异常场景
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="darkgrid")
plt.rcParams["figure.figsize"] = (20, 10)
s_scores = start_scores.detach().numpy().flatten()
e_scores = end_scores.detach().numpy().flatten()
token_labels = []
for i, token in enumerate(tokens):
token_labels.append("{:} - {:>2}".format(token, i))
import pandas as pd
scores = []
for i, token_label in enumerate(token_labels):
scores.append({"token_label": token_label, "score": s_scores[i], "marker": "start"})
scores.append({"token_label": token_label, "score": e_scores[i], "marker": "end"})
df = pd.DataFrame(scores)
df.head()
# 使用seaborn的 catplot 函数绘制分组条形图,展示每个词的开始得分和结束得分
# hue参数指定哪些数据点在那个系列的地方
# 分组条形图可以展示多个样本的多个数据在同一图里
g = sns.catplot(
x="token_label", y="score", hue="marker", data=df, kind="bar", height=6, aspect=4
)
g.set_xticklabels(g.ax.get_xticklabels(), rotation=90, ha="center")
g.ax.grid(True)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/459/129459665.ipynb
| null | null |
[{"Id": 129459665, "ScriptId": 38470708, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10565867, "CreationDate": "05/14/2023 02:50:44", "VersionNumber": 1.0, "Title": "Bert_demo", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 261.0, "LinesInsertedFromPrevious": 261.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
# # 1.Bert demo1 — 使用Bert
# * transformer库中的pipline可以使用预训练模型bert,来执行mask-filling任务
# * mask-filling 任务是指在给定的句子中,找到并填补被\` [MASK]\`符号所代替的部分
# * 上面的填充意味着这个模型可以预测mask所掩盖的部分
from transformers import pipeline
import warnings
warnings.filterwarnings("ignore")
unmasker = pipeline("fill-mask", model="bert-base-uncased")
unmasker("Artificial Intelligence [MASK] take over the world")
# ## Bert模型自带的偏见Bias
# * 需要注意的是预训练中的偏见也会影响到微调任务
unmasker("The man worked as a [MASK]。")
# '[MASK]' 可能被填充为 'doctor','teacher','lawyer' 等等,这完全取决于模型的预测,从下方的结果可以看出
# 这个模型对男性的偏见有点离谱,这种偏见是训练数据不均衡导致的
unmasker("I will [MASK] mountain today, because I have a vacation from yesterday。")
# # 2.Bert demo2 — 使用Bert完成文本分类
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import cross_val_score
import torch
import transformers as ppb
import warnings
warnings.filterwarnings("ignore")
# ## 导入数据集
df = pd.read_csv(
"https://github.com/clairett/pytorch-sentiment-classification/raw/master/data/SST2/train.tsv",
delimiter="\t",
header=None,
)
# 为了节省训练时间,这里只采用前2000个数据进行训练
batch_1 = df.iloc[:2000, :]
batch_1[1].value_counts() # 观察0和1标签的分布
# ## 加载预训练好的 Bert 模型
# * 需要事先定义模型的种类,分词器的种类,预训练模型种类对应的权重名称
# 第一个是模型类别,第二个是分词器类别,第三个是预训练权重
model_class, tokenizer_class, pretrained_weights = (
ppb.DistilBertModel,
ppb.DistilBertTokenizer,
"distilbert-base-uncased",
)
tokenizer = tokenizer_class.from_pretrained(pretrained_weights) # 加载对应base版本的模型
model = model_class.from_pretrained(pretrained_weights)
# ## 经典数据预处理流程
# ### Tokenizer
# 第一步将句子转化为words,在转化为subwords,确保能满足Bert模型的输入要求
# 使用预训练的分词器对数据进行分词
# batch_1[0]表示要处理的数据
# apply函数用于对数据中的每个元素进行操作,这里的操作是lambda函数,
# 在这个匿名函数中调用了 tokenizer.encode 函数,对每个元素(这里是每个文本)进行分词
# add_special_tokens=True 表示在分词的过程中添加特殊的标记,包括开始和结束的标记的等,这些操作都不能少
# Bert 需要这些操作才能统一输入数据
print(batch_1[:5])
tokenized = batch_1[0].apply(lambda x: tokenizer.encode(x, add_special_tokens=True))
# ### Padding
# 文本预处理的经典步骤之padding填充,确保输入的token长度是相同的
max_len = 0
# 长度有两种方式获得,一种是自定义,一种是寻找最长的序列的长度
for i in tokenized.values:
if len(i) > max_len:
max_len = len(i)
# 对序列进行填充
# 然后对所有的句子进行填充,使他们的长度都等于 max_len ,填充的方式是在句子的后面添加0,直到句子的长度等于0
# 用np.array来存储填充后的句子,每一个句子都是一个数组,所有的句子构成了一个二维数组
padded = np.array([i + [0] * (max_len - len(i)) for i in tokenized.values])
padded.shape
# ### Masking
# 对于基于 Encoder 构建的 Bert,需要对 Attention 进行 mask 处理,确保 Padding 出来的0不会对 Attention 过程产生 side effects
attention_mask = np.where(padded != 0, 1, 0)
# np.where 的作用其实就是对每一个元素都进行if判断,满足要求填前面的元素,不满足填充后面的元素
attention_mask.shape
# 将处理好的数据送给模型进行处理,得到模型的输出结果
# 将输入数据和注意力掩码转化为PyTorch张量
# PyTorch的模型需要使用PyTorch的张量作为输入,所以需要将padded和attention_mask从Numpy数组中转化为PyTorch张量
# 使用 torch.tensor 转化,这个操作说明bert的这个模型是基于PyTorch实现的
input_ids = torch.tensor(padded)
# input_ids[:5]
attention_mask = torch.tensor(attention_mask)
# attention_mask[:5]
# 使用模型前向传播
# 使用with torch.no_grad()语句块来冻结梯度计算
# 在进行前向传播时不需要计算梯度,关闭梯度计算可以节省内存,提高计算速度
# 然后将input_ids和attention_mask作为输入,通过模型进行前向传播,的到输出last_hidden_states
# attention_mask=attention_mask作为名为 attentiion_mask的参数传递给模型
# 这段代码会基于你的GPU的能力变化训练时间,如果训练时间过长就把前面的数据集改小一点
with torch.no_grad():
last_hidden_states = model(input_ids, attention_mask=attention_mask)
# 因为Bert模型中,输入句子转化后的数据的第一个位置是[cls],[cls]代表的就是整个句子的 Sentence Encoding 或者说是 Sentence presentation,而作为下游的判别任务的依据就是这个cls token
# 提取特征
# 从last_hidden_states(模型的输出,包含了每个输入词语的隐藏状态)中提取特征
# last_hidden_states[0]取出隐藏状态,因为模型的输出是一个元组,其第一个元素是隐藏状态
# [:,0,:]表示取出每个句子的第一个词语(Bert模型中的CLS标记)的所有隐藏状态
# 在Bert模型中,第一个词语的隐藏状态被用作句子的表示,所以我们通常会使用它作为特征
# .numpy()将隐藏状态从PyTorch转化为numyp,因为这些特征会被送进NN来处理
features = last_hidden_states[0][:, 0, :].numpy() # torch->numpy
# batch[1]是真正的标签,0/1
labels = batch_1[1]
# ### 将数据切分成训练集和测试集
# 直接使用sklearn.model_selection 中的 train_test_split
# 划分训练集和测试集
# 返回训练集特征,测试集的特征,训练集的标签,测试集的标签
train_features, test_features, train_labels, test_labels = train_test_split(
features, labels
)
# 训练最后的逻辑回归模型的参数,虽然Bert是与训练过的,但是做下游任务的模型没有训练过
lr_clf = LogisticRegression()
# 训练逻辑回归模型
lr_clf.fit(train_features, train_labels)
# 计算训练效果
# 评估模型
# 使用score方法来评估模型在测试集上的表现
lr_clf.score(test_features, test_labels)
# 只得到自己的得分是不合理的,这里需要于其他的评价指标进行评价,dummy classifer(模拟分类器/傻瓜分类器?)来进行分类,比较得分 Dummy 是不会进行任何学习的,只是简单地使用一些规则(如预测所有样本都属于最常见的类别)来进行预测,比如使用众数分类
from sklearn.dummy import DummyClassifier
clf = DummyClassifier()
# 使用交叉验证来评估模型的性能
# cross_val_score函数会将数据集划分为k个子集,然后进行k次训练和测试
scores = cross_val_score(clf, train_features, train_labels)
print("Dummy classifier score: %0.3f (+/- %0.2f)" % (scores.mean(), scores.std() * 2))
# # Bert demo 3 — 使用bert进行文本问答
from transformers import BertForQuestionAnswering
# 问题回答任务中,模型需要根据给定的问题和文本,找出文本中回答问题的部分
model = BertForQuestionAnswering.from_pretrained(
"bert-large-uncased-whole-word-masking-finetuned-squad"
)
# 直接使用这个经过微调后的模型
# 提供问题和回答
question = "How many parameters does BERT-large have?"
answer_text = "BERT-large is really big... it has 24-layers and an embedding size of 1,024, for a total of 340M parameters! Altogether it is 1.34GB, so expect it to take a couple minutes to download to your Colab instance."
from transformers import BertTokenizer
# 用哪个模型最好就用这个模型提供的分词器
tokenizer = BertTokenizer.from_pretrained(
"bert-large-uncased-whole-word-masking-finetuned-squad"
)
input_ids = tokenizer.encode(question, answer_text)
print("The input has a total of {:} tokens.".format(len(input_ids)))
# 这里查看token是怎么映射的
# convert_ids_to_tokens,这个方法的作用在于返回索引形式的tokens形成的列表
tokens = tokenizer.convert_ids_to_tokens(input_ids)
for token, id in zip(tokens, input_ids):
if id == tokenizer.sep_token_id:
print("")
print("{:<12} {:>6}".format(token, id))
if id == tokenizer.sep_token_id:
print("")
# 从中可以看出token的时候会加入CLS特殊符号表示这个句子的标签特征向量
# 用特殊符号 SEP 来间隔两个句子
sep_index = input_ids.index(tokenizer.sep_token_id) # 返回第一个 SEP token 的位置
num_seg_a = sep_index + 1
# num_seg_a 表示segment A的长度,包含sep本身的
num_seg_b = len(input_ids) - num_seg_a
# 构造一个由0 1 组成的列表,这个列表表示每个token属于那个segment
segment_ids = [0] * num_seg_a + [1] * num_seg_b
assert len(segment_ids) == len(input_ids)
# 注意:
# * 这里因为我们仅采用一段文本对作为输入,且输入的长度是可控的,因此我们没有进行Padding或Truncate操作
# * 在批量操作中,我们需要加上Padding和Truncate操作确保模型接受的数据长度是一致且小于 Max len 的
import torch
outputs = model(
torch.tensor([input_ids]),
token_type_ids=torch.tensor([segment_ids]),
return_dict=True,
) # 设置为True,使得模型返回一个包含更多信息的字典
# 获得开始和结束得分
start_scores = outputs.start_logits
end_scores = outputs.end_logits
answer_start = torch.argmax(start_scores)
answer_end = torch.argmax(end_scores)
# 将答案中的token组合起来并打印出来
answer = " ".join(tokens[answer_start : answer_end + 1])
print('Answer: "' + answer + '"')
# 注意:
# * 这里只是简单的取了得分最高的start pos和end pos
# * 但是在实际场景中需要考虑start pos index 大于 end pos index 的异常场景
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="darkgrid")
plt.rcParams["figure.figsize"] = (20, 10)
s_scores = start_scores.detach().numpy().flatten()
e_scores = end_scores.detach().numpy().flatten()
token_labels = []
for i, token in enumerate(tokens):
token_labels.append("{:} - {:>2}".format(token, i))
import pandas as pd
scores = []
for i, token_label in enumerate(token_labels):
scores.append({"token_label": token_label, "score": s_scores[i], "marker": "start"})
scores.append({"token_label": token_label, "score": e_scores[i], "marker": "end"})
df = pd.DataFrame(scores)
df.head()
# 使用seaborn的 catplot 函数绘制分组条形图,展示每个词的开始得分和结束得分
# hue参数指定哪些数据点在那个系列的地方
# 分组条形图可以展示多个样本的多个数据在同一图里
g = sns.catplot(
x="token_label", y="score", hue="marker", data=df, kind="bar", height=6, aspect=4
)
g.set_xticklabels(g.ax.get_xticklabels(), rotation=90, ha="center")
g.ax.grid(True)
| false | 0 | 3,320 | 1 | 3,320 | 3,320 |
||
129459935
|
<jupyter_start><jupyter_text>Bank Customer Segmentation (1M+ Transactions)
### Bank Customer Segmentation
Most banks have a large customer base - with different characteristics in terms of age, income, values, lifestyle, and more. Customer segmentation is the process of dividing a customer dataset into specific groups based on shared traits.
*According to a report from Ernst & Young, “A more granular understanding of consumers is no longer a nice-to-have item, but a strategic and competitive imperative for banking providers. Customer understanding should be a living, breathing part of everyday business, with insights underpinning the full range of banking operations.*
### About this Dataset
This dataset consists of 1 Million+ transaction by over 800K customers for a bank in India. The data contains information such as - customer age (DOB), location, gender, account balance at the time of the transaction, transaction details, transaction amount, etc.
### Interesting Analysis Ideas
The dataset can be used for different analysis, example -
1. Perform Clustering / Segmentation on the dataset and identify popular customer groups along with their definitions/rules
2. Perform Location-wise analysis to identify regional trends in India
3. Perform transaction-related analysis to identify interesting trends that can be used by a bank to improve / optimi their user experiences
4. Customer Recency, Frequency, Monetary analysis
5. Network analysis or Graph analysis of customer data.
Kaggle dataset identifier: bank-customer-segmentation
<jupyter_code>import pandas as pd
df = pd.read_csv('bank-customer-segmentation/bank_transactions.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 1048567 entries, 0 to 1048566
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 TransactionID 1048567 non-null object
1 CustomerID 1048567 non-null object
2 CustomerDOB 1045170 non-null object
3 CustGender 1047467 non-null object
4 CustLocation 1048416 non-null object
5 CustAccountBalance 1046198 non-null float64
6 TransactionDate 1048567 non-null object
7 TransactionTime 1048567 non-null int64
8 TransactionAmount (INR) 1048567 non-null float64
dtypes: float64(2), int64(1), object(6)
memory usage: 72.0+ MB
<jupyter_text>Examples:
{
"TransactionID": "T1",
"CustomerID": "C5841053",
"CustomerDOB": "10/1/94",
"CustGender": "F",
"CustLocation": "JAMSHEDPUR",
"CustAccountBalance": 17819.05,
"TransactionDate": "2/8/16",
"TransactionTime": 143207,
"TransactionAmount (INR)": 25
}
{
"TransactionID": "T2",
"CustomerID": "C2142763",
"CustomerDOB": "4/4/57",
"CustGender": "M",
"CustLocation": "JHAJJAR",
"CustAccountBalance": 2270.69,
"TransactionDate": "2/8/16",
"TransactionTime": 141858,
"TransactionAmount (INR)": 27999
}
{
"TransactionID": "T3",
"CustomerID": "C4417068",
"CustomerDOB": "26/11/96",
"CustGender": "F",
"CustLocation": "MUMBAI",
"CustAccountBalance": 17874.44,
"TransactionDate": "2/8/16",
"TransactionTime": 142712,
"TransactionAmount (INR)": 459
}
{
"TransactionID": "T4",
"CustomerID": "C5342380",
"CustomerDOB": "14/9/73",
"CustGender": "F",
"CustLocation": "MUMBAI",
"CustAccountBalance": 866503.21,
"TransactionDate": "2/8/16",
"TransactionTime": 142714,
"TransactionAmount (INR)": 2060
}
<jupyter_script># # Customer Segmentation on Bank Customers
# ### The project will look into the demographic attributes of the bank's customers and conduct segmentation based on their lifecycle and value to the bank. The methods used in the project:
# ### 1. RFM Model: Recency, Frequency, Monetary Scores
# ### 2. Customer Lifecycle: New Customer, Active Customer, Non-active Customer, Returning Customer
# ### 3. Pareto Analysis: how many customers contribute to the most transaction volume
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from datetime import datetime
# Ban the scientific expression
pd.set_option("display.float_format", lambda x: "%.2f" % x)
# Ignore warnings
import warnings
warnings.filterwarnings("ignore")
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### 1. Data Cleaning
# Import the data
data = pd.read_csv("/kaggle/input/bank-customer-segmentation/bank_transactions.csv")
data.info()
data.sample(5)
# 1. Check for missing values
data.isnull().sum() / data.shape[0] * 100
# From the results, the missing values only take less than 1% of the total records. Therefore we can drop them
data.dropna(axis=0, inplace=True)
# 2. Check for duplications: the customer could be duplicated given one can make more than 1 transactions. The transaction ID should be uniqie
data.duplicated(subset="TransactionID").sum()
# There is no duplication in the transaction id part.
# 3. Check the distribution of the numeric fields (for potential outliers)
data["TransactionDate"] = pd.to_datetime(data["TransactionDate"])
data["CustomerDOB"] = pd.to_datetime(data["CustomerDOB"])
data[
[
"CustAccountBalance",
"TransactionAmount (INR)",
"CustomerDOB",
"CustGender",
"CustLocation",
"TransactionDate",
"TransactionTime",
]
].describe(percentiles=[0.01, 0.25, 0.50, 0.75, 0.99], include="all").T
# From the results, the numerical variables (balance and transaction amt) seems to follow a right skewed distribution (mean > median), which shouold be due to high net wealth customers
# For date and categorical variables, there are some dates in wrong values (e.g. 1800-01-01) will need to be adjusted.
# The transaction time could be dropped given it seems not containing useful information in the analysis
# 4. Data Transformation
# 4.1 Drop unused fields
data.drop("TransactionTime", axis=1, inplace=True)
# 4.2 Calculate Customer Age
# Here will use the year in the data (2016) as base to get the customer's age
data["age"] = data["TransactionDate"].dt.year - data["CustomerDOB"].dt.year
# 4.3 Change all the age below 12 and above 100 percentile into median age
data.loc[(data["age"] < 12) | (data["age"] >= 100), "age"] = data["age"].median()
# 4.4 Adjust the values of Gender
data["CustGender"] = data["CustGender"].replace(
{"M": "Male", "F": "Female", "T": "Male"}
)
# ### 2. Exploratory Data Analysis (EDA)
# #### 2.1 Gender
# Compare the distribution of customers across genders
plt.style.use("ggplot")
fig, ax = plt.subplots(ncols=2, nrows=1, figsize=(20, 10))
ax[0].pie(
data["CustGender"].value_counts(), autopct="%1.f%%", labels=["Male", "Female"]
)
ax[0].set_title("Customer Gender Frequency", size=20)
ax[1] = sns.distplot(
data.loc[
(data["CustGender"] == "Male")
& (
data["TransactionAmount (INR)"]
< np.percentile(data["TransactionAmount (INR)"], 90)
),
"TransactionAmount (INR)",
],
label=True,
kde=False,
)
ax[1] = sns.distplot(
data.loc[
(data["CustGender"] == "Female")
& (
data["TransactionAmount (INR)"]
< np.percentile(data["TransactionAmount (INR)"], 90)
),
"TransactionAmount (INR)",
],
label="Female",
kde=False,
)
ax[1].set_title("Transaction Amount by Customer Gender", size=20)
# #### 2.2 Location
plt.figure(figsize=(20, 6))
sns.countplot(
y="CustLocation", data=data, order=data["CustLocation"].value_counts()[:20].index
)
plt.title("Top 20 Locations of Customer ", fontsize="20")
# From the plot, it seems Mumbai, New Delhi and Bangalore are the TOP 3 cities that transaction happened. This could be due to that the 3 cities have more population, better economy condition and higher salary range.
# #### 2.3 Age
# Distribution of age based on bins
bins = [0, 20, 30, 40, 50, 60, 100]
labels = ["0 - 20", "20 - 30", "30 - 40", "40 - 50", "50 - 60", "60+"]
data["age_bin"] = pd.cut(x=data["age"], bins=bins, labels=labels, right=True)
plt.figure(figsize=(20, 6))
sns.countplot(data, y="age_bin", order=data["age_bin"].value_counts().index)
# From the plot, 20 - 30 group is the the majority, followed by 30 - 40 customers. There is little customers after 50.
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/459/129459935.ipynb
|
bank-customer-segmentation
|
shivamb
|
[{"Id": 129459935, "ScriptId": 38489547, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1270512, "CreationDate": "05/14/2023 02:54:58", "VersionNumber": 1.0, "Title": "Customer Segmentation - RFM Model Practise", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 137.0, "LinesInsertedFromPrevious": 137.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185535195, "KernelVersionId": 129459935, "SourceDatasetVersionId": 2743905}]
|
[{"Id": 2743905, "DatasetId": 1672910, "DatasourceVersionId": 2789165, "CreatorUserId": 1571785, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "10/26/2021 13:28:18", "VersionNumber": 1.0, "Title": "Bank Customer Segmentation (1M+ Transactions)", "Slug": "bank-customer-segmentation", "Subtitle": "Customer demographics and transactions data from an Indian Bank", "Description": "### Bank Customer Segmentation\n\nMost banks have a large customer base - with different characteristics in terms of age, income, values, lifestyle, and more. Customer segmentation is the process of dividing a customer dataset into specific groups based on shared traits.\n\n*According to a report from Ernst & Young, \u201cA more granular understanding of consumers is no longer a nice-to-have item, but a strategic and competitive imperative for banking providers. Customer understanding should be a living, breathing part of everyday business, with insights underpinning the full range of banking operations.*\n\n### About this Dataset\n\nThis dataset consists of 1 Million+ transaction by over 800K customers for a bank in India. The data contains information such as - customer age (DOB), location, gender, account balance at the time of the transaction, transaction details, transaction amount, etc. \n\n### Interesting Analysis Ideas \n\nThe dataset can be used for different analysis, example - \n\n1. Perform Clustering / Segmentation on the dataset and identify popular customer groups along with their definitions/rules \n2. Perform Location-wise analysis to identify regional trends in India \n3. Perform transaction-related analysis to identify interesting trends that can be used by a bank to improve / optimi their user experiences \n4. Customer Recency, Frequency, Monetary analysis \n5. Network analysis or Graph analysis of customer data.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1672910, "CreatorUserId": 1571785, "OwnerUserId": 1571785.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2743905.0, "CurrentDatasourceVersionId": 2789165.0, "ForumId": 1694135, "Type": 2, "CreationDate": "10/26/2021 13:28:18", "LastActivityDate": "10/26/2021", "TotalViews": 74434, "TotalDownloads": 6281, "TotalVotes": 86, "TotalKernels": 23}]
|
[{"Id": 1571785, "UserName": "shivamb", "DisplayName": "Shivam Bansal", "RegisterDate": "01/22/2018", "PerformanceTier": 4}]
|
# # Customer Segmentation on Bank Customers
# ### The project will look into the demographic attributes of the bank's customers and conduct segmentation based on their lifecycle and value to the bank. The methods used in the project:
# ### 1. RFM Model: Recency, Frequency, Monetary Scores
# ### 2. Customer Lifecycle: New Customer, Active Customer, Non-active Customer, Returning Customer
# ### 3. Pareto Analysis: how many customers contribute to the most transaction volume
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from datetime import datetime
# Ban the scientific expression
pd.set_option("display.float_format", lambda x: "%.2f" % x)
# Ignore warnings
import warnings
warnings.filterwarnings("ignore")
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### 1. Data Cleaning
# Import the data
data = pd.read_csv("/kaggle/input/bank-customer-segmentation/bank_transactions.csv")
data.info()
data.sample(5)
# 1. Check for missing values
data.isnull().sum() / data.shape[0] * 100
# From the results, the missing values only take less than 1% of the total records. Therefore we can drop them
data.dropna(axis=0, inplace=True)
# 2. Check for duplications: the customer could be duplicated given one can make more than 1 transactions. The transaction ID should be uniqie
data.duplicated(subset="TransactionID").sum()
# There is no duplication in the transaction id part.
# 3. Check the distribution of the numeric fields (for potential outliers)
data["TransactionDate"] = pd.to_datetime(data["TransactionDate"])
data["CustomerDOB"] = pd.to_datetime(data["CustomerDOB"])
data[
[
"CustAccountBalance",
"TransactionAmount (INR)",
"CustomerDOB",
"CustGender",
"CustLocation",
"TransactionDate",
"TransactionTime",
]
].describe(percentiles=[0.01, 0.25, 0.50, 0.75, 0.99], include="all").T
# From the results, the numerical variables (balance and transaction amt) seems to follow a right skewed distribution (mean > median), which shouold be due to high net wealth customers
# For date and categorical variables, there are some dates in wrong values (e.g. 1800-01-01) will need to be adjusted.
# The transaction time could be dropped given it seems not containing useful information in the analysis
# 4. Data Transformation
# 4.1 Drop unused fields
data.drop("TransactionTime", axis=1, inplace=True)
# 4.2 Calculate Customer Age
# Here will use the year in the data (2016) as base to get the customer's age
data["age"] = data["TransactionDate"].dt.year - data["CustomerDOB"].dt.year
# 4.3 Change all the age below 12 and above 100 percentile into median age
data.loc[(data["age"] < 12) | (data["age"] >= 100), "age"] = data["age"].median()
# 4.4 Adjust the values of Gender
data["CustGender"] = data["CustGender"].replace(
{"M": "Male", "F": "Female", "T": "Male"}
)
# ### 2. Exploratory Data Analysis (EDA)
# #### 2.1 Gender
# Compare the distribution of customers across genders
plt.style.use("ggplot")
fig, ax = plt.subplots(ncols=2, nrows=1, figsize=(20, 10))
ax[0].pie(
data["CustGender"].value_counts(), autopct="%1.f%%", labels=["Male", "Female"]
)
ax[0].set_title("Customer Gender Frequency", size=20)
ax[1] = sns.distplot(
data.loc[
(data["CustGender"] == "Male")
& (
data["TransactionAmount (INR)"]
< np.percentile(data["TransactionAmount (INR)"], 90)
),
"TransactionAmount (INR)",
],
label=True,
kde=False,
)
ax[1] = sns.distplot(
data.loc[
(data["CustGender"] == "Female")
& (
data["TransactionAmount (INR)"]
< np.percentile(data["TransactionAmount (INR)"], 90)
),
"TransactionAmount (INR)",
],
label="Female",
kde=False,
)
ax[1].set_title("Transaction Amount by Customer Gender", size=20)
# #### 2.2 Location
plt.figure(figsize=(20, 6))
sns.countplot(
y="CustLocation", data=data, order=data["CustLocation"].value_counts()[:20].index
)
plt.title("Top 20 Locations of Customer ", fontsize="20")
# From the plot, it seems Mumbai, New Delhi and Bangalore are the TOP 3 cities that transaction happened. This could be due to that the 3 cities have more population, better economy condition and higher salary range.
# #### 2.3 Age
# Distribution of age based on bins
bins = [0, 20, 30, 40, 50, 60, 100]
labels = ["0 - 20", "20 - 30", "30 - 40", "40 - 50", "50 - 60", "60+"]
data["age_bin"] = pd.cut(x=data["age"], bins=bins, labels=labels, right=True)
plt.figure(figsize=(20, 6))
sns.countplot(data, y="age_bin", order=data["age_bin"].value_counts().index)
# From the plot, 20 - 30 group is the the majority, followed by 30 - 40 customers. There is little customers after 50.
|
[{"bank-customer-segmentation/bank_transactions.csv": {"column_names": "[\"TransactionID\", \"CustomerID\", \"CustomerDOB\", \"CustGender\", \"CustLocation\", \"CustAccountBalance\", \"TransactionDate\", \"TransactionTime\", \"TransactionAmount (INR)\"]", "column_data_types": "{\"TransactionID\": \"object\", \"CustomerID\": \"object\", \"CustomerDOB\": \"object\", \"CustGender\": \"object\", \"CustLocation\": \"object\", \"CustAccountBalance\": \"float64\", \"TransactionDate\": \"object\", \"TransactionTime\": \"int64\", \"TransactionAmount (INR)\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1048567 entries, 0 to 1048566\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 TransactionID 1048567 non-null object \n 1 CustomerID 1048567 non-null object \n 2 CustomerDOB 1045170 non-null object \n 3 CustGender 1047467 non-null object \n 4 CustLocation 1048416 non-null object \n 5 CustAccountBalance 1046198 non-null float64\n 6 TransactionDate 1048567 non-null object \n 7 TransactionTime 1048567 non-null int64 \n 8 TransactionAmount (INR) 1048567 non-null float64\ndtypes: float64(2), int64(1), object(6)\nmemory usage: 72.0+ MB\n", "summary": "{\"CustAccountBalance\": {\"count\": 1046198.0, \"mean\": 115403.54005622261, \"std\": 846485.3806006602, \"min\": 0.0, \"25%\": 4721.76, \"50%\": 16792.18, \"75%\": 57657.36, \"max\": 115035495.1}, \"TransactionTime\": {\"count\": 1048567.0, \"mean\": 157087.52939297154, \"std\": 51261.85402232933, \"min\": 0.0, \"25%\": 124030.0, \"50%\": 164226.0, \"75%\": 200010.0, \"max\": 235959.0}, \"TransactionAmount (INR)\": {\"count\": 1048567.0, \"mean\": 1574.3350034570992, \"std\": 6574.742978454002, \"min\": 0.0, \"25%\": 161.0, \"50%\": 459.03, \"75%\": 1200.0, \"max\": 1560034.99}}", "examples": "{\"TransactionID\":{\"0\":\"T1\",\"1\":\"T2\",\"2\":\"T3\",\"3\":\"T4\"},\"CustomerID\":{\"0\":\"C5841053\",\"1\":\"C2142763\",\"2\":\"C4417068\",\"3\":\"C5342380\"},\"CustomerDOB\":{\"0\":\"10\\/1\\/94\",\"1\":\"4\\/4\\/57\",\"2\":\"26\\/11\\/96\",\"3\":\"14\\/9\\/73\"},\"CustGender\":{\"0\":\"F\",\"1\":\"M\",\"2\":\"F\",\"3\":\"F\"},\"CustLocation\":{\"0\":\"JAMSHEDPUR\",\"1\":\"JHAJJAR\",\"2\":\"MUMBAI\",\"3\":\"MUMBAI\"},\"CustAccountBalance\":{\"0\":17819.05,\"1\":2270.69,\"2\":17874.44,\"3\":866503.21},\"TransactionDate\":{\"0\":\"2\\/8\\/16\",\"1\":\"2\\/8\\/16\",\"2\":\"2\\/8\\/16\",\"3\":\"2\\/8\\/16\"},\"TransactionTime\":{\"0\":143207,\"1\":141858,\"2\":142712,\"3\":142714},\"TransactionAmount (INR)\":{\"0\":25.0,\"1\":27999.0,\"2\":459.0,\"3\":2060.0}}"}}]
| true | 1 |
<start_data_description><data_path>bank-customer-segmentation/bank_transactions.csv:
<column_names>
['TransactionID', 'CustomerID', 'CustomerDOB', 'CustGender', 'CustLocation', 'CustAccountBalance', 'TransactionDate', 'TransactionTime', 'TransactionAmount (INR)']
<column_types>
{'TransactionID': 'object', 'CustomerID': 'object', 'CustomerDOB': 'object', 'CustGender': 'object', 'CustLocation': 'object', 'CustAccountBalance': 'float64', 'TransactionDate': 'object', 'TransactionTime': 'int64', 'TransactionAmount (INR)': 'float64'}
<dataframe_Summary>
{'CustAccountBalance': {'count': 1046198.0, 'mean': 115403.54005622261, 'std': 846485.3806006602, 'min': 0.0, '25%': 4721.76, '50%': 16792.18, '75%': 57657.36, 'max': 115035495.1}, 'TransactionTime': {'count': 1048567.0, 'mean': 157087.52939297154, 'std': 51261.85402232933, 'min': 0.0, '25%': 124030.0, '50%': 164226.0, '75%': 200010.0, 'max': 235959.0}, 'TransactionAmount (INR)': {'count': 1048567.0, 'mean': 1574.3350034570992, 'std': 6574.742978454002, 'min': 0.0, '25%': 161.0, '50%': 459.03, '75%': 1200.0, 'max': 1560034.99}}
<dataframe_info>
RangeIndex: 1048567 entries, 0 to 1048566
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 TransactionID 1048567 non-null object
1 CustomerID 1048567 non-null object
2 CustomerDOB 1045170 non-null object
3 CustGender 1047467 non-null object
4 CustLocation 1048416 non-null object
5 CustAccountBalance 1046198 non-null float64
6 TransactionDate 1048567 non-null object
7 TransactionTime 1048567 non-null int64
8 TransactionAmount (INR) 1048567 non-null float64
dtypes: float64(2), int64(1), object(6)
memory usage: 72.0+ MB
<some_examples>
{'TransactionID': {'0': 'T1', '1': 'T2', '2': 'T3', '3': 'T4'}, 'CustomerID': {'0': 'C5841053', '1': 'C2142763', '2': 'C4417068', '3': 'C5342380'}, 'CustomerDOB': {'0': '10/1/94', '1': '4/4/57', '2': '26/11/96', '3': '14/9/73'}, 'CustGender': {'0': 'F', '1': 'M', '2': 'F', '3': 'F'}, 'CustLocation': {'0': 'JAMSHEDPUR', '1': 'JHAJJAR', '2': 'MUMBAI', '3': 'MUMBAI'}, 'CustAccountBalance': {'0': 17819.05, '1': 2270.69, '2': 17874.44, '3': 866503.21}, 'TransactionDate': {'0': '2/8/16', '1': '2/8/16', '2': '2/8/16', '3': '2/8/16'}, 'TransactionTime': {'0': 143207, '1': 141858, '2': 142712, '3': 142714}, 'TransactionAmount (INR)': {'0': 25.0, '1': 27999.0, '2': 459.0, '3': 2060.0}}
<end_description>
| 1,618 | 0 | 2,741 | 1,618 |
129997525
|
<jupyter_start><jupyter_text>NLP_News Article
Kaggle dataset identifier: nlp-news-article
<jupyter_script>import numpy as np
import pandas as pd
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.model_selection import train_test_split
import nltk
from nltk import word_tokenize
from nltk.stem import WordNetLemmatizer, PorterStemmer
from nltk.corpus import wordnet
nltk.download("wordnet2022")
nltk.download("punkt")
nltk.download("averaged_perceptron_tagger")
# nltk.data.path.append('/kaggle/input')
# !wget -nc https://lazyprogrammer.me/course_files/nlp/bbc_text_cls.csv
df = pd.read_csv("/kaggle/input/nlp-news-article/bbc_text_cls.csv")
df.head()
inputs = df["text"]
labels = df["labels"]
labels.hist(figsize=(6, 2))
inputs_train, inputs_test, ytrain, ytest = train_test_split(
inputs, labels, random_state=123
)
# CountVectorizer without stopwords
vectorizer = CountVectorizer()
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
xtrain
# > If most of the elements of the matrix have 0 value, then it is called a sparse matrix
# > Values are 0 are unimportant data that we are looking for, so we looking for non-zero values
print((xtrain != 0).sum())
print(xtrain.shape)
# Percentage of non-zero values
((xtrain != 0).sum() / np.prod(xtrain.shape)) * 100
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer without stopwords")
print("train score:", model.score(xtrain, ytrain))
print("test score:", model.score(xtest, ytest))
# CountVectorizer with stopwords
vectorizer = CountVectorizer(stop_words="english")
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer with stopwords")
print("train score:", model.score(xtrain, ytrain))
print("test score:", model.score(xtest, ytest))
# > The Part Of Speech tag. Valid options are “n” for nouns, “v” for verbs, “a” for adjectives, “r” for adverbs and “s” for satellite adjectives.
def get_wordnet_pos(treebank_tag):
if treebank_tag.startswith("J"):
return wordnet.ADJ
elif treebank_tag.startswith("V"):
return wordnet.VERB
elif treebank_tag.startswith("N"):
return wordnet.NOUN
elif treebank_tag.startswith("R"):
return wordnet.ADV
else:
return wordnet.NOUN
class LemmaTokenizer:
def __init__(self):
self.wnl = WordNetLemmatizer()
def __call__(self, doc):
"""https://www.geeksforgeeks.org/what-is-the-difference-between-__init__-and-__call__/"""
tokens = word_tokenize(doc)
# print(f"token: {tokens}")
words_and_tags = nltk.pos_tag(tokens)
# print(f"words_and_tags: {words_and_tags}")
return [
self.wnl.lemmatize(word, pos=get_wordnet_pos(tag))
for word, tag in words_and_tags
]
# CountVectorizer with Lemmatization
vectorizer = CountVectorizer(tokenizer=LemmaTokenizer())
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer with Lemmatization")
print("train score:", model.score(xtrain, ytrain))
print("test score:", model.score(xtest, ytest))
class StemTokenizer:
def __init__(self):
self.port_stem = PorterStemmer()
def __call__(self, doc):
tokens = word_tokenize(doc)
return [self.port_stem.stem(t) for t in tokens]
# CountVectorizer with stemming
vectorizer = CountVectorizer(tokenizer=StemTokenizer())
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer with stemming")
print("train score", model.score(xtrain, ytrain))
print("test score", model.score(xtest, ytest))
def simple_tokenizer(words):
return words.split()
# CountVectorizer with simple_tokenizer
vectorizer = CountVectorizer(tokenizer=simple_tokenizer)
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer with simple_tokenizer")
print("train score", model.score(xtrain, ytrain))
print("test score", model.score(xtest, ytest))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/997/129997525.ipynb
|
nlp-news-article
|
saumyamishrads
|
[{"Id": 129997525, "ScriptId": 38648934, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4407783, "CreationDate": "05/18/2023 03:03:00", "VersionNumber": 3.0, "Title": "Learning_NLTK", "EvaluationDate": "05/18/2023", "IsChange": true, "TotalLines": 132.0, "LinesInsertedFromPrevious": 42.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 90.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186449498, "KernelVersionId": 129997525, "SourceDatasetVersionId": 5195248}]
|
[{"Id": 5195248, "DatasetId": 3020932, "DatasourceVersionId": 5267464, "CreatorUserId": 3612525, "LicenseName": "Unknown", "CreationDate": "03/19/2023 14:54:38", "VersionNumber": 1.0, "Title": "NLP_News Article", "Slug": "nlp-news-article", "Subtitle": "Text Dataset / NLP / News latter", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3020932, "CreatorUserId": 3612525, "OwnerUserId": 3612525.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5195248.0, "CurrentDatasourceVersionId": 5267464.0, "ForumId": 3060140, "Type": 2, "CreationDate": "03/19/2023 14:54:38", "LastActivityDate": "03/19/2023", "TotalViews": 86, "TotalDownloads": 8, "TotalVotes": 0, "TotalKernels": 2}]
|
[{"Id": 3612525, "UserName": "saumyamishrads", "DisplayName": "Saumya Mishra", "RegisterDate": "08/22/2019", "PerformanceTier": 1}]
|
import numpy as np
import pandas as pd
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.model_selection import train_test_split
import nltk
from nltk import word_tokenize
from nltk.stem import WordNetLemmatizer, PorterStemmer
from nltk.corpus import wordnet
nltk.download("wordnet2022")
nltk.download("punkt")
nltk.download("averaged_perceptron_tagger")
# nltk.data.path.append('/kaggle/input')
# !wget -nc https://lazyprogrammer.me/course_files/nlp/bbc_text_cls.csv
df = pd.read_csv("/kaggle/input/nlp-news-article/bbc_text_cls.csv")
df.head()
inputs = df["text"]
labels = df["labels"]
labels.hist(figsize=(6, 2))
inputs_train, inputs_test, ytrain, ytest = train_test_split(
inputs, labels, random_state=123
)
# CountVectorizer without stopwords
vectorizer = CountVectorizer()
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
xtrain
# > If most of the elements of the matrix have 0 value, then it is called a sparse matrix
# > Values are 0 are unimportant data that we are looking for, so we looking for non-zero values
print((xtrain != 0).sum())
print(xtrain.shape)
# Percentage of non-zero values
((xtrain != 0).sum() / np.prod(xtrain.shape)) * 100
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer without stopwords")
print("train score:", model.score(xtrain, ytrain))
print("test score:", model.score(xtest, ytest))
# CountVectorizer with stopwords
vectorizer = CountVectorizer(stop_words="english")
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer with stopwords")
print("train score:", model.score(xtrain, ytrain))
print("test score:", model.score(xtest, ytest))
# > The Part Of Speech tag. Valid options are “n” for nouns, “v” for verbs, “a” for adjectives, “r” for adverbs and “s” for satellite adjectives.
def get_wordnet_pos(treebank_tag):
if treebank_tag.startswith("J"):
return wordnet.ADJ
elif treebank_tag.startswith("V"):
return wordnet.VERB
elif treebank_tag.startswith("N"):
return wordnet.NOUN
elif treebank_tag.startswith("R"):
return wordnet.ADV
else:
return wordnet.NOUN
class LemmaTokenizer:
def __init__(self):
self.wnl = WordNetLemmatizer()
def __call__(self, doc):
"""https://www.geeksforgeeks.org/what-is-the-difference-between-__init__-and-__call__/"""
tokens = word_tokenize(doc)
# print(f"token: {tokens}")
words_and_tags = nltk.pos_tag(tokens)
# print(f"words_and_tags: {words_and_tags}")
return [
self.wnl.lemmatize(word, pos=get_wordnet_pos(tag))
for word, tag in words_and_tags
]
# CountVectorizer with Lemmatization
vectorizer = CountVectorizer(tokenizer=LemmaTokenizer())
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer with Lemmatization")
print("train score:", model.score(xtrain, ytrain))
print("test score:", model.score(xtest, ytest))
class StemTokenizer:
def __init__(self):
self.port_stem = PorterStemmer()
def __call__(self, doc):
tokens = word_tokenize(doc)
return [self.port_stem.stem(t) for t in tokens]
# CountVectorizer with stemming
vectorizer = CountVectorizer(tokenizer=StemTokenizer())
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer with stemming")
print("train score", model.score(xtrain, ytrain))
print("test score", model.score(xtest, ytest))
def simple_tokenizer(words):
return words.split()
# CountVectorizer with simple_tokenizer
vectorizer = CountVectorizer(tokenizer=simple_tokenizer)
xtrain = vectorizer.fit_transform(inputs_train)
xtest = vectorizer.transform(inputs_test)
model = MultinomialNB()
model.fit(xtrain, ytrain)
print("CountVectorizer with simple_tokenizer")
print("train score", model.score(xtrain, ytrain))
print("test score", model.score(xtest, ytest))
| false | 1 | 1,315 | 0 | 1,340 | 1,315 |
||
129997829
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Ridge
from sklearn.linear_model import Lasso
from sklearn.linear_model import ElasticNet
from sklearn.ensemble import RandomForestRegressor
from sklearn.svm import SVR
from xgboost import XGBRegressor
from sklearn.preprocessing import PolynomialFeatures
data = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
plt.figure(figsize=(10, 8))
sns.heatmap(data.corr(), cmap="RdBu")
plt.title("Correlations Between Variables", size=15)
plt.show()
important_num_cols = list(
data.corr()["SalePrice"][
(data.corr()["SalePrice"] > 0.50) | (data.corr()["SalePrice"] < -0.50)
].index
)
cat_cols = [
"MSZoning",
"Utilities",
"BldgType",
"Heating",
"KitchenQual",
"SaleCondition",
"LandSlope",
]
important_cols = important_num_cols + cat_cols
data = data[important_cols]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/997/129997829.ipynb
| null | null |
[{"Id": 129997829, "ScriptId": 38667542, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2267407, "CreationDate": "05/18/2023 03:07:04", "VersionNumber": 1.0, "Title": "notebookfb22ed9918", "EvaluationDate": "05/18/2023", "IsChange": true, "TotalLines": 32.0, "LinesInsertedFromPrevious": 32.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Ridge
from sklearn.linear_model import Lasso
from sklearn.linear_model import ElasticNet
from sklearn.ensemble import RandomForestRegressor
from sklearn.svm import SVR
from xgboost import XGBRegressor
from sklearn.preprocessing import PolynomialFeatures
data = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
plt.figure(figsize=(10, 8))
sns.heatmap(data.corr(), cmap="RdBu")
plt.title("Correlations Between Variables", size=15)
plt.show()
important_num_cols = list(
data.corr()["SalePrice"][
(data.corr()["SalePrice"] > 0.50) | (data.corr()["SalePrice"] < -0.50)
].index
)
cat_cols = [
"MSZoning",
"Utilities",
"BldgType",
"Heating",
"KitchenQual",
"SaleCondition",
"LandSlope",
]
important_cols = important_num_cols + cat_cols
data = data[important_cols]
| false | 0 | 357 | 0 | 357 | 357 |
||
129997522
|
# Import relevant libraries and modules.
import numpy as np
import pandas as pd
import matplotlib as plt
import pickle
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn import metrics
from xgboost import XGBClassifier
from xgboost import plot_importance
airline_data = pd.read_csv(
"/kaggle/input/invistico-airline/Invistico_Airline.csv", error_bad_lines=False
)
# Display first ten rows of data.
airline_data.head(10)
# Display the data type for each column in your DataFrame.
airline_data.dtypes
# Convert the object predictor variables to numerical dummies.
airline_data_dummies = pd.get_dummies(
airline_data, columns=["satisfaction", "Customer Type", "Type of Travel", "Class"]
)
# Define the y (target) variable.
y = airline_data_dummies["satisfaction_satisfied"]
# Define the X (predictor) variables.
X = airline_data_dummies.drop(
["satisfaction_satisfied", "satisfaction_dissatisfied"], axis=1
)
# Perform the split operation on your data.
# Assign the outputs as follows: X_train, X_test, y_train, y_test.
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=0
)
# Define xgb to be your XGBClassifier.
xgb = XGBClassifier(random_state=0)
# Define parameters for tuning as `cv_params`.
cv_params = {
"max_depth": [4, 6],
"min_child_weight": [3, 5],
"learning_rate": [0.1, 0.2, 0.3],
"n_estimators": [5, 10, 15],
"subsample": [0.7],
"colsample_bytree": [0.7],
}
# Define your criteria as `scoring`.
scoring = {"accuracy", "precision", "recall", "f1"}
# Construct your GridSearch.
xgb_cv = GridSearchCV(xgb, cv_params, scoring=scoring, cv=5, refit="f1")
# fit the GridSearch model to training data
xgb_cv = xgb_cv.fit(X_train, y_train)
xgb_cv
# Use `pickle` to save the trained model.
pickle.dump(xgb_cv, open("xgb_cv.sav", "wb"))
# Apply your model to predict on your test data. Call this output "y_pred".
y_pred = xgb_cv.predict(X_test)
# 1. Print your accuracy score.
ac_score = metrics.accuracy_score(y_test, y_pred)
print("accuracy score:", ac_score)
# 2. Print your precision score.
pc_score = metrics.precision_score(y_test, y_pred)
print("precision score:", pc_score)
# 3. Print your recall score.
rc_score = metrics.recall_score(y_test, y_pred)
print("recall score:", rc_score)
# 4. Print your f1 score.
f1_score = metrics.f1_score(y_test, y_pred)
print("f1 score:", f1_score)
# Construct and display your confusion matrix.
# Construct the confusion matrix for your predicted and test values.
cm = metrics.confusion_matrix(y_test, y_pred)
# Create the display for your confusion matrix.
disp = metrics.ConfusionMatrixDisplay(
confusion_matrix=cm, display_labels=xgb_cv.classes_
)
# Plot the visual in-line.
disp.plot()
# Plot the relative feature importance of the predictor variables in your model.
plot_importance(xgb_cv.best_estimator_)
# Create a table of results to compare model performance.
table = pd.DataFrame()
table = table.append(
{
"Model": "Tuned Decision Tree",
"F1": 0.945422,
"Recall": 0.935863,
"Precision": 0.955197,
"Accuracy": 0.940864,
},
ignore_index=True,
)
table = table.append(
{
"Model": "Tuned Random Forest",
"F1": 0.947306,
"Recall": 0.944501,
"Precision": 0.950128,
"Accuracy": 0.942450,
},
ignore_index=True,
)
table = table.append(
{
"Model": "Tuned XGBoost",
"F1": f1_score,
"Recall": rc_score,
"Precision": pc_score,
"Accuracy": ac_score,
},
ignore_index=True,
)
table
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/997/129997522.ipynb
| null | null |
[{"Id": 129997522, "ScriptId": 38671836, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9485427, "CreationDate": "05/18/2023 03:02:58", "VersionNumber": 1.0, "Title": "Building an XGBoost Model with Python", "EvaluationDate": "05/18/2023", "IsChange": true, "TotalLines": 148.0, "LinesInsertedFromPrevious": 148.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# Import relevant libraries and modules.
import numpy as np
import pandas as pd
import matplotlib as plt
import pickle
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn import metrics
from xgboost import XGBClassifier
from xgboost import plot_importance
airline_data = pd.read_csv(
"/kaggle/input/invistico-airline/Invistico_Airline.csv", error_bad_lines=False
)
# Display first ten rows of data.
airline_data.head(10)
# Display the data type for each column in your DataFrame.
airline_data.dtypes
# Convert the object predictor variables to numerical dummies.
airline_data_dummies = pd.get_dummies(
airline_data, columns=["satisfaction", "Customer Type", "Type of Travel", "Class"]
)
# Define the y (target) variable.
y = airline_data_dummies["satisfaction_satisfied"]
# Define the X (predictor) variables.
X = airline_data_dummies.drop(
["satisfaction_satisfied", "satisfaction_dissatisfied"], axis=1
)
# Perform the split operation on your data.
# Assign the outputs as follows: X_train, X_test, y_train, y_test.
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=0
)
# Define xgb to be your XGBClassifier.
xgb = XGBClassifier(random_state=0)
# Define parameters for tuning as `cv_params`.
cv_params = {
"max_depth": [4, 6],
"min_child_weight": [3, 5],
"learning_rate": [0.1, 0.2, 0.3],
"n_estimators": [5, 10, 15],
"subsample": [0.7],
"colsample_bytree": [0.7],
}
# Define your criteria as `scoring`.
scoring = {"accuracy", "precision", "recall", "f1"}
# Construct your GridSearch.
xgb_cv = GridSearchCV(xgb, cv_params, scoring=scoring, cv=5, refit="f1")
# fit the GridSearch model to training data
xgb_cv = xgb_cv.fit(X_train, y_train)
xgb_cv
# Use `pickle` to save the trained model.
pickle.dump(xgb_cv, open("xgb_cv.sav", "wb"))
# Apply your model to predict on your test data. Call this output "y_pred".
y_pred = xgb_cv.predict(X_test)
# 1. Print your accuracy score.
ac_score = metrics.accuracy_score(y_test, y_pred)
print("accuracy score:", ac_score)
# 2. Print your precision score.
pc_score = metrics.precision_score(y_test, y_pred)
print("precision score:", pc_score)
# 3. Print your recall score.
rc_score = metrics.recall_score(y_test, y_pred)
print("recall score:", rc_score)
# 4. Print your f1 score.
f1_score = metrics.f1_score(y_test, y_pred)
print("f1 score:", f1_score)
# Construct and display your confusion matrix.
# Construct the confusion matrix for your predicted and test values.
cm = metrics.confusion_matrix(y_test, y_pred)
# Create the display for your confusion matrix.
disp = metrics.ConfusionMatrixDisplay(
confusion_matrix=cm, display_labels=xgb_cv.classes_
)
# Plot the visual in-line.
disp.plot()
# Plot the relative feature importance of the predictor variables in your model.
plot_importance(xgb_cv.best_estimator_)
# Create a table of results to compare model performance.
table = pd.DataFrame()
table = table.append(
{
"Model": "Tuned Decision Tree",
"F1": 0.945422,
"Recall": 0.935863,
"Precision": 0.955197,
"Accuracy": 0.940864,
},
ignore_index=True,
)
table = table.append(
{
"Model": "Tuned Random Forest",
"F1": 0.947306,
"Recall": 0.944501,
"Precision": 0.950128,
"Accuracy": 0.942450,
},
ignore_index=True,
)
table = table.append(
{
"Model": "Tuned XGBoost",
"F1": f1_score,
"Recall": rc_score,
"Precision": pc_score,
"Accuracy": ac_score,
},
ignore_index=True,
)
table
| false | 0 | 1,217 | 0 | 1,217 | 1,217 |
||
129803201
|
<jupyter_start><jupyter_text>Motor Vehicle Collisions
The Motor Vehicle Collisions crash table contains details on the crash event. Each row represents a crash event. The Motor Vehicle Collisions data tables contain information from all police reported motor vehicle collisions in NYC. The police report (MV104-AN) is required to be filled out for collisions where someone is injured or killed, or where there is at least $1000 worth of damage.
Kaggle dataset identifier: motor-vehicle-collisions
<jupyter_script># # Imports
# [Pandas](https://pandas.pydata.org/docs/getting_started/index.html) is the most popular library for working with data in Python:
import pandas as pd
# # Loading Data
# When editing this notebook you can add data in the area on the right of the Kaggle notebook.
# The code below shows if and in which path the files are available:
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Let's load the main table into a data frame (pandas representation of a table):
df = pd.read_csv(
"/kaggle/input/motor-vehicle-collisions/Motor_Vehicle_Collisions_-_Crashes.csv"
)
# just drop some columns right away
# df_olympics.drop(columns=['Games', 'Event'], inplace=True)
# print out the beginning of the data frame
df
# # Subsets of the Data
# [See the pandas tutorial for more details](https://pandas.pydata.org/docs/getting_started/intro_tutorials/03_subset_data.html#min-tut-03-subset)
# Making smaller version of the table by selecting some columns:
df = df[
[
"CRASH DATE",
"CRASH TIME",
"LATITUDE",
"LONGITUDE",
"NUMBER OF PEDESTRIANS INJURED",
"NUMBER OF PEDESTRIANS KILLED",
"NUMBER OF CYCLIST INJURED",
"NUMBER OF CYCLIST KILLED",
"NUMBER OF MOTORIST INJURED",
"NUMBER OF MOTORIST KILLED",
]
]
df = df.rename(
columns={
"CRASH DATE": "date",
"CRASH TIME": "time",
"LATITUDE": "lat",
"LONGITUDE": "lon",
"NUMBER OF PEDESTRIANS INJURED": "ped_inj",
"NUMBER OF PEDESTRIANS KILLED": "ped_kill",
"NUMBER OF CYCLIST INJURED": "cyc_inj",
"NUMBER OF CYCLIST KILLED": "cyc_kill",
"NUMBER OF MOTORIST INJURED": "mot_inj",
"NUMBER OF MOTORIST KILLED": "mot_kill",
}
)
df
# And display a part of the result:
# df_olympics_small.head()
# ... or some more lines
# df_olympics_small.head(10)
# ... or the end of the table
# df_olympics_small.tail()
# ... or both
# df_olympics_small
# It's also easy to filter some rows of the table according to one or more conditions:
# Keep all rows where the value in the column "Medal" is not "NA"
df = df[df["lat"].notna()]
# Print the result
df
# round lat and lon for making a heatmap possible
detail = 0.005 # rounds lat and long to this detail
df["lon_round"] = (df["lon"] / detail).round() * detail
df["lat_round"] = (df["lat"] / detail).round() * detail
df
# # Aggregations
# You can group rows with the same values and make calculations on some of the columns.
# See this [Pandas tutorial](https://pandas.pydata.org/docs/getting_started/intro_tutorials/06_calculate_statistics.html#min-tut-06-stats) for some more details.
# Calculate the averages of all (number) columns for the different medal categories
# df_groupedByMedal = df_olympics.groupby("Medal").mean()
# Some more detailed aggregations by using the agg() function.
df_heatmap = df.groupby(["lon_round", "lat_round"]).agg(
{
"date": ["count"],
"ped_inj": ["sum"],
"ped_kill": ["sum"],
"cyc_inj": ["sum"],
"cyc_kill": ["sum"],
"mot_inj": ["sum"],
"mot_kill": ["sum"],
}
)
# Sometimes it could be handy to get rid of the second line of headings:
df_heatmap.columns = df_heatmap.columns.map("".join)
# ... and to convert index columns to normal ones
df_heatmap = df_heatmap.reset_index()
# Print out the preview
df_heatmap
# # Exporting to CSV or JSON
df_heatmap.to_csv("heatmap.csv")
df_heatmap.to_json("heatmap.json", orient="records")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/803/129803201.ipynb
|
motor-vehicle-collisions
|
utkarshx27
|
[{"Id": 129803201, "ScriptId": 38589723, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2602161, "CreationDate": "05/16/2023 15:12:29", "VersionNumber": 2.0, "Title": "Motor Vehicle Collissions Data Preparation", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 103.0, "LinesInsertedFromPrevious": 2.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 101.0, "LinesInsertedFromFork": 33.0, "LinesDeletedFromFork": 43.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 70.0, "TotalVotes": 4}]
|
[{"Id": 186175315, "KernelVersionId": 129803201, "SourceDatasetVersionId": 5563178}]
|
[{"Id": 5563178, "DatasetId": 3203075, "DatasourceVersionId": 5637978, "CreatorUserId": 13364933, "LicenseName": "CC0: Public Domain", "CreationDate": "04/30/2023 06:43:34", "VersionNumber": 1.0, "Title": "Motor Vehicle Collisions", "Slug": "motor-vehicle-collisions", "Subtitle": "New York City Vehicle Crash Data", "Description": "The Motor Vehicle Collisions crash table contains details on the crash event. Each row represents a crash event. The Motor Vehicle Collisions data tables contain information from all police reported motor vehicle collisions in NYC. The police report (MV104-AN) is required to be filled out for collisions where someone is injured or killed, or where there is at least $1000 worth of damage.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3203075, "CreatorUserId": 13364933, "OwnerUserId": 13364933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5563178.0, "CurrentDatasourceVersionId": 5637978.0, "ForumId": 3267744, "Type": 2, "CreationDate": "04/30/2023 06:43:34", "LastActivityDate": "04/30/2023", "TotalViews": 7675, "TotalDownloads": 1248, "TotalVotes": 41, "TotalKernels": 3}]
|
[{"Id": 13364933, "UserName": "utkarshx27", "DisplayName": "Utkarsh Singh", "RegisterDate": "01/21/2023", "PerformanceTier": 2}]
|
# # Imports
# [Pandas](https://pandas.pydata.org/docs/getting_started/index.html) is the most popular library for working with data in Python:
import pandas as pd
# # Loading Data
# When editing this notebook you can add data in the area on the right of the Kaggle notebook.
# The code below shows if and in which path the files are available:
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Let's load the main table into a data frame (pandas representation of a table):
df = pd.read_csv(
"/kaggle/input/motor-vehicle-collisions/Motor_Vehicle_Collisions_-_Crashes.csv"
)
# just drop some columns right away
# df_olympics.drop(columns=['Games', 'Event'], inplace=True)
# print out the beginning of the data frame
df
# # Subsets of the Data
# [See the pandas tutorial for more details](https://pandas.pydata.org/docs/getting_started/intro_tutorials/03_subset_data.html#min-tut-03-subset)
# Making smaller version of the table by selecting some columns:
df = df[
[
"CRASH DATE",
"CRASH TIME",
"LATITUDE",
"LONGITUDE",
"NUMBER OF PEDESTRIANS INJURED",
"NUMBER OF PEDESTRIANS KILLED",
"NUMBER OF CYCLIST INJURED",
"NUMBER OF CYCLIST KILLED",
"NUMBER OF MOTORIST INJURED",
"NUMBER OF MOTORIST KILLED",
]
]
df = df.rename(
columns={
"CRASH DATE": "date",
"CRASH TIME": "time",
"LATITUDE": "lat",
"LONGITUDE": "lon",
"NUMBER OF PEDESTRIANS INJURED": "ped_inj",
"NUMBER OF PEDESTRIANS KILLED": "ped_kill",
"NUMBER OF CYCLIST INJURED": "cyc_inj",
"NUMBER OF CYCLIST KILLED": "cyc_kill",
"NUMBER OF MOTORIST INJURED": "mot_inj",
"NUMBER OF MOTORIST KILLED": "mot_kill",
}
)
df
# And display a part of the result:
# df_olympics_small.head()
# ... or some more lines
# df_olympics_small.head(10)
# ... or the end of the table
# df_olympics_small.tail()
# ... or both
# df_olympics_small
# It's also easy to filter some rows of the table according to one or more conditions:
# Keep all rows where the value in the column "Medal" is not "NA"
df = df[df["lat"].notna()]
# Print the result
df
# round lat and lon for making a heatmap possible
detail = 0.005 # rounds lat and long to this detail
df["lon_round"] = (df["lon"] / detail).round() * detail
df["lat_round"] = (df["lat"] / detail).round() * detail
df
# # Aggregations
# You can group rows with the same values and make calculations on some of the columns.
# See this [Pandas tutorial](https://pandas.pydata.org/docs/getting_started/intro_tutorials/06_calculate_statistics.html#min-tut-06-stats) for some more details.
# Calculate the averages of all (number) columns for the different medal categories
# df_groupedByMedal = df_olympics.groupby("Medal").mean()
# Some more detailed aggregations by using the agg() function.
df_heatmap = df.groupby(["lon_round", "lat_round"]).agg(
{
"date": ["count"],
"ped_inj": ["sum"],
"ped_kill": ["sum"],
"cyc_inj": ["sum"],
"cyc_kill": ["sum"],
"mot_inj": ["sum"],
"mot_kill": ["sum"],
}
)
# Sometimes it could be handy to get rid of the second line of headings:
df_heatmap.columns = df_heatmap.columns.map("".join)
# ... and to convert index columns to normal ones
df_heatmap = df_heatmap.reset_index()
# Print out the preview
df_heatmap
# # Exporting to CSV or JSON
df_heatmap.to_csv("heatmap.csv")
df_heatmap.to_json("heatmap.json", orient="records")
| false | 1 | 1,209 | 4 | 1,318 | 1,209 |
||
129803774
|
<jupyter_start><jupyter_text>Ecommerce Customer Churn Analysis and Prediction
Data Variable Discerption
E Comm CustomerID Unique customer ID
E Comm Churn Churn Flag
E Comm Tenure Tenure of customer in organization
E Comm PreferredLoginDevice Preferred login device of customer
E Comm CityTier City tier
E Comm WarehouseToHome Distance in between warehouse to home of customer
E Comm PreferredPaymentMode Preferred payment method of customer
E Comm Gender Gender of customer
E Comm HourSpendOnApp Number of hours spend on mobile application or website
E Comm NumberOfDeviceRegistered Total number of deceives is registered on particular customer
E Comm PreferedOrderCat Preferred order category of customer in last month
E Comm SatisfactionScore Satisfactory score of customer on service
E Comm MaritalStatus Marital status of customer
E Comm NumberOfAddress Total number of added added on particular customer
E Comm Complain Any complaint has been raised in last month
E Comm OrderAmountHikeFromlastYear Percentage increases in order from last year
E Comm CouponUsed Total number of coupon has been used in last month
E Comm OrderCount Total number of orders has been places in last month
E Comm DaySinceLastOrder Day Since last order by customer
E Comm CashbackAmount Average cashback in last month
Kaggle dataset identifier: ecommerce-customer-churn-analysis-and-prediction
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.preprocessing import StandardScaler, OneHotEncoder, LabelEncoder
from sklearn.model_selection import KFold, StratifiedKFold, cross_val_score
from sklearn.metrics import (
accuracy_score,
roc_auc_score,
classification_report,
confusion_matrix,
f1_score,
roc_curve,
precision_score,
recall_score,
roc_auc_score,
)
from sklearn import linear_model, tree, ensemble
from sklearn.model_selection import train_test_split
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.simplefilter(action="ignore")
# ## Data Understanding and Exploration
df = pd.read_excel(
"/kaggle/input/ecommerce-customer-churn-analysis-and-prediction/E Commerce Dataset.xlsx",
sheet_name="E Comm",
)
data_dict_df = pd.read_excel(
"/kaggle/input/ecommerce-customer-churn-analysis-and-prediction/E Commerce Dataset.xlsx",
sheet_name="Data Dict",
)
df.head()
# summary of the dataset: 5630 rows, 20 columns
# dtypes: float64(8), int64(7), object(5)
print(df.info())
# df['Tenure'] = df['Tenure'].astype('object')
# df['CityTier'] = df['CityTier'].astype('object')
# Describe the EComm dataset
df.describe()
# Creating numerical Features list and categorical Features list
datatype_columns = df.dtypes # type of each feature in data: int, float, object
numerical_columns = datatype_columns[
(datatype_columns == "int64") | (datatype_columns == "float64")
]
categorical_columns = datatype_columns[datatype_columns == "object"]
numerical_columns_list = numerical_columns.index
categorical_columns_list = categorical_columns.index
print(
f"Numerical Features in dataset: {numerical_columns_list}, \n\n Categorical Features in dataset: {categorical_columns_list}"
)
# ### Missing Value Ananlysis
# Ratio of columns having null values (out of 1)
round((pd.isnull(df).sum() * 100 / len(df.index)), 2)[
pd.isnull(df).sum() > 0
].sort_values()
# ## Exploratory Data Analysis
sub_categorical_cols = [
"Tenure",
"CityTier",
"PreferredLoginDevice",
"PreferredPaymentMode",
"Gender",
"PreferedOrderCat",
"MaritalStatus",
]
df_churn = df[df["Churn"] == 1]
plt.figure(figsize=(12, 20))
idx = 1
for column in sub_categorical_cols:
plt.subplot(4, 2, idx)
sns.countplot(data=df_churn, x=column)
idx = idx + 1
plt.show()
# checking correlation among different numerical variables using clustered heat map
plt.figure(figsize=(25, 15))
sns.clustermap(df.corr(), cmap="PiYG", annot=True, fmt=".2f")
plt.show()
# ## Data Preparation / Preprocessing
class Preprocessing:
def __init__(self, df):
self.df = df
def shape(self):
print(f"shape: {self.df.shape}")
def dtypes(self, pr=False):
print("Types")
if pr:
print(self.df.dtypes)
def supposed2beint(self):
float_cols = [
column for column in self.df.columns if self.df[column].dtype == "float"
]
int2be_cols = []
for col in float_cols:
if (self.df[col].fillna(-9999) % 1 == 0).all() == True:
int2be_cols.append(col)
return int2be_cols
def isNaN(self, pr=False):
if pr:
print("Contain NaN")
print(self.df.isnull().sum())
else:
return self.df.columns[self.df.isna().any()].tolist()
def isObject(self):
return [
column for column in self.df.columns if self.df[column].dtype == "object"
]
def check_dataframe(self):
self.shape()
self.dtypes(True)
self.isNaN(True)
def fillNaN(self):
nan_cols = self.isNaN()
int2be_cols = self.supposed2beint()
# nan_cols == int2be_cols they are the same
for col in int2be_cols:
self.df[col].fillna(round(self.df[col].mean()), inplace=True)
self.df[col] = self.df[col].astype(int)
def adjust_category_cols(self):
self.fillNaN()
# PreferredLoginDevice
# self.df.loc[self.df["PreferredLoginDevice"] == "Mobile Phone", "PreferredLoginDevice"] = "Phone"
# PreferredPaymentMode
# self.df.loc[self.df["PreferredPaymentMode"] == "Credit Card", "PreferredPaymentMode"] = "CC"
# self.df.loc[self.df["PreferredPaymentMode"] == "Cash on Delivery", "PreferredPaymentMode"] = "COD"
self.adjust_duplicate_column_values(self.df)
def adjust_duplicate_column_values(self, _df):
# PreferredLoginDevice
_df.loc[
_df["PreferredLoginDevice"] == "Mobile Phone", "PreferredLoginDevice"
] = "Phone"
# PreferredPaymentMode
_df.loc[
_df["PreferredPaymentMode"] == "Credit Card", "PreferredPaymentMode"
] = "CC"
_df.loc[
_df["PreferredPaymentMode"] == "Cash on Delivery", "PreferredPaymentMode"
] = "COD"
def drop_useless_cols(self):
self.df.drop(["CustomerID"], axis=1, inplace=True)
def split_target(self):
self.adjust_category_cols()
self.drop_useless_cols()
self.X = self.df.drop("Churn", axis=1)
self.y = self.df["Churn"].astype(int).to_numpy()
def find_enc_method(self):
cat_cols = self.isObject()
one_hot_cols = [col for col in cat_cols if self.X[col].nunique() <= 3]
label_enc_cols = [col for col in cat_cols if col not in one_hot_cols]
self.cat_cols = cat_cols
self.one_hot_cols = one_hot_cols
self.label_enc_cols = label_enc_cols
return one_hot_cols, label_enc_cols, cat_cols
def encoding(self):
one_hot_cols, label_enc_cols, cat_cols = self.find_enc_method()
num_cols = [col for col in self.X.columns if col not in cat_cols]
self.num_cols = num_cols
X_OHE, X_LE, X_NUM = (
self.X[one_hot_cols].copy(),
self.X[label_enc_cols].copy(),
self.X[num_cols].copy(),
)
self.OHE = OneHotEncoder(drop="first", handle_unknown="error")
X_OHE = self.OHE.fit_transform(X_OHE).toarray()
self.le_dict = {}
# self.LE = LabelEncoder()
for col in X_LE.columns:
LE = LabelEncoder()
self.le_dict[col] = LE.fit(X_LE[col])
X_LE[col] = self.le_dict[col].transform(X_LE[col])
return X_OHE, X_LE.to_numpy(), X_NUM.to_numpy()
def scaling(self):
X_OHE, X_LE, X_num = self.encoding()
self.SS = StandardScaler()
X_num = self.SS.fit_transform(X_num)
self.X_total = np.concatenate((X_OHE, X_LE, X_num), axis=1)
def get_encoders(self):
return self.OHE, self.LE, self.le_dict
def get_scaler(self):
return self.SS
def get_default_Xy(self):
self.split_target()
return self.X, self.y
def get_Xy(self):
self.split_target()
self.scaling()
return self.X_total, self.y
def prepare_unseen_df(self, unseen_df):
"""
Used for scoring new unseen data
"""
cat_cols = self.cat_cols
num_cols = self.num_cols
one_hot_cols = self.one_hot_cols
label_enc_cols = self.label_enc_cols
self.adjust_duplicate_column_values(unseen_df)
X_OHE, X_LE, X_NUM = (
unseen_df[one_hot_cols].copy(),
unseen_df[label_enc_cols].copy(),
unseen_df[num_cols].copy(),
)
X_OHE = self.OHE.transform(X_OHE).toarray()
for col in X_LE.columns:
X_LE[col] = self.le_dict[col].transform(X_LE[col])
X_num = self.SS.transform(X_NUM.to_numpy())
return np.concatenate((X_OHE, X_LE.to_numpy(), X_num), axis=1)
pre = Preprocessing(df)
X, y = pre.get_Xy()
unique, counts = np.unique(y, return_counts=True)
unique, counts
import optuna
from xgboost import XGBClassifier
def objective_xgb(trial):
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42, stratify=y
)
param = {
"lambda": trial.suggest_loguniform("lambda", 1e-3, 10.0),
"alpha": trial.suggest_loguniform("alpha", 1e-3, 10.0),
"colsample_bytree": trial.suggest_categorical(
"colsample_bytree", [0.5, 0.6, 0.7, 0.8, 0.9, 1.0]
),
"subsample": trial.suggest_categorical("subsample", [0.6, 0.7, 0.8, 1.0]),
"learning_rate": trial.suggest_categorical(
"learning_rate", [0.008, 0.009, 0.01, 0.012, 0.014, 0.016, 0.018, 0.02]
),
"n_estimators": trial.suggest_categorical(
"n_estimators", [150, 200, 300, 3000]
),
"max_depth": trial.suggest_categorical(
"max_depth", [4, 5, 7, 9, 11, 13, 15, 17]
),
"random_state": 42,
"min_child_weight": trial.suggest_int("min_child_weight", 1, 300),
}
model = XGBClassifier(**param)
model.fit(
X_train,
y_train,
early_stopping_rounds=50,
eval_set=[(X_test, y_test)],
verbose=False,
)
preds = model.predict(X_test)
acc = accuracy_score(y_test, preds)
return acc
study = optuna.create_study(direction="maximize")
study.optimize(objective_xgb, n_trials=50)
params_xgb = study.best_trial.params
print("Number of finished trials:", len(study.trials))
print("Best trial:", params_xgb)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42, stratify=y
)
model = XGBClassifier(**params_xgb)
model.fit(
X_train,
y_train,
early_stopping_rounds=50,
eval_set=[(X_test, y_test)],
verbose=False,
)
y_pred = model.predict(X_test)
# ### Model Evaluation
print(f"Accuracy: {round(accuracy_score(y_pred, y_test), 2)}")
print("=" * 60)
print(f"Recall: {round(recall_score(y_pred, y_test),2)}")
print("=" * 60)
print(f"Precision: {round(precision_score(y_pred, y_test), 2)}")
print("=" * 60)
print(f"F1: {round(f1_score(y_pred, y_test), 2)}")
print("=" * 60)
print(f"Auc: {round(roc_auc_score(y_pred, y_test), 2)}")
print("=" * 60)
CR = classification_report(y_test, y_pred)
CM = confusion_matrix(y_test, y_pred)
print(f"Classification Report:\n {CR}")
print("=" * 60)
print("\n")
print(f"Confusion Matrix: {CM}")
print("=" * 60)
# # Prediction using realtime data
def evaluate_churn(eva_dict):
scoring_df = pd.DataFrame.from_records([scoring_dict])
preprocessed_df = pre.prepare_unseen_df(scoring_df)
test_pred_arr = model.predict([preprocessed_df[0]])
test_pred_proba = model.predict_proba([preprocessed_df[0]])
will_churn = "Yes" if test_pred_arr[0] == 1 else "No"
print("is the customer will churn - ", will_churn)
print(
f"Probability for will not churn: {test_pred_proba[0][0]}, will churn: {test_pred_proba[0][1]}"
)
return (will_churn, test_pred_proba[0])
scoring_dict = {
"Tenure": 5,
"PreferredLoginDevice": "Computer",
"CityTier": 3,
"WarehouseToHome": 0,
"PreferredPaymentMode": "Debit Card",
"Gender": "Male",
"HourSpendOnApp": 0,
"NumberOfDeviceRegistered": 0,
"PreferedOrderCat": "Mobile",
"SatisfactionScore": 10,
"MaritalStatus": "Married",
"NumberOfAddress": 3,
"Complain": 100,
"OrderAmountHikeFromlastYear": 13,
"CouponUsed": 0,
"OrderCount": 0,
"DaySinceLastOrder": 300,
"CashbackAmount": 0,
}
evaluate_churn(scoring_dict)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/803/129803774.ipynb
|
ecommerce-customer-churn-analysis-and-prediction
|
ankitverma2010
|
[{"Id": 129803774, "ScriptId": 38603580, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2034692, "CreationDate": "05/16/2023 15:16:46", "VersionNumber": 1.0, "Title": "ecommerce-customer-churn-analysis", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 308.0, "LinesInsertedFromPrevious": 308.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 186175887, "KernelVersionId": 129803774, "SourceDatasetVersionId": 1880629}]
|
[{"Id": 1880629, "DatasetId": 1119908, "DatasourceVersionId": 1918738, "CreatorUserId": 4804740, "LicenseName": "CC BY-NC-SA 4.0", "CreationDate": "01/26/2021 10:20:08", "VersionNumber": 1.0, "Title": "Ecommerce Customer Churn Analysis and Prediction", "Slug": "ecommerce-customer-churn-analysis-and-prediction", "Subtitle": "Predict customer churn and make suggestions", "Description": "Data\tVariable\t Discerption\nE Comm\tCustomerID\t Unique customer ID\nE Comm\tChurn\t Churn Flag\nE Comm\tTenure\t Tenure of customer in organization\nE Comm\tPreferredLoginDevice\t Preferred login device of customer\nE Comm\tCityTier\t City tier\nE Comm\tWarehouseToHome\t Distance in between warehouse to home of customer\nE Comm\tPreferredPaymentMode\t Preferred payment method of customer\nE Comm\tGender\t Gender of customer\nE Comm\tHourSpendOnApp\t Number of hours spend on mobile application or website\nE Comm\tNumberOfDeviceRegistered\t Total number of deceives is registered on particular customer\nE Comm\tPreferedOrderCat\t Preferred order category of customer in last month\nE Comm\tSatisfactionScore\t Satisfactory score of customer on service\nE Comm\tMaritalStatus\t Marital status of customer\nE Comm\tNumberOfAddress\t Total number of added added on particular customer\nE Comm\tComplain\t Any complaint has been raised in last month\nE Comm\tOrderAmountHikeFromlastYear Percentage increases in order from last year\nE Comm\tCouponUsed\t Total number of coupon has been used in last month\nE Comm\tOrderCount\t Total number of orders has been places in last month\nE Comm\tDaySinceLastOrder\t Day Since last order by customer\nE Comm\tCashbackAmount\t Average cashback in last month", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1119908, "CreatorUserId": 4804740, "OwnerUserId": 4804740.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1880629.0, "CurrentDatasourceVersionId": 1918738.0, "ForumId": 1137265, "Type": 2, "CreationDate": "01/26/2021 10:20:08", "LastActivityDate": "01/26/2021", "TotalViews": 63315, "TotalDownloads": 7409, "TotalVotes": 80, "TotalKernels": 25}]
|
[{"Id": 4804740, "UserName": "ankitverma2010", "DisplayName": "Ankit Verma", "RegisterDate": "04/03/2020", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.preprocessing import StandardScaler, OneHotEncoder, LabelEncoder
from sklearn.model_selection import KFold, StratifiedKFold, cross_val_score
from sklearn.metrics import (
accuracy_score,
roc_auc_score,
classification_report,
confusion_matrix,
f1_score,
roc_curve,
precision_score,
recall_score,
roc_auc_score,
)
from sklearn import linear_model, tree, ensemble
from sklearn.model_selection import train_test_split
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.simplefilter(action="ignore")
# ## Data Understanding and Exploration
df = pd.read_excel(
"/kaggle/input/ecommerce-customer-churn-analysis-and-prediction/E Commerce Dataset.xlsx",
sheet_name="E Comm",
)
data_dict_df = pd.read_excel(
"/kaggle/input/ecommerce-customer-churn-analysis-and-prediction/E Commerce Dataset.xlsx",
sheet_name="Data Dict",
)
df.head()
# summary of the dataset: 5630 rows, 20 columns
# dtypes: float64(8), int64(7), object(5)
print(df.info())
# df['Tenure'] = df['Tenure'].astype('object')
# df['CityTier'] = df['CityTier'].astype('object')
# Describe the EComm dataset
df.describe()
# Creating numerical Features list and categorical Features list
datatype_columns = df.dtypes # type of each feature in data: int, float, object
numerical_columns = datatype_columns[
(datatype_columns == "int64") | (datatype_columns == "float64")
]
categorical_columns = datatype_columns[datatype_columns == "object"]
numerical_columns_list = numerical_columns.index
categorical_columns_list = categorical_columns.index
print(
f"Numerical Features in dataset: {numerical_columns_list}, \n\n Categorical Features in dataset: {categorical_columns_list}"
)
# ### Missing Value Ananlysis
# Ratio of columns having null values (out of 1)
round((pd.isnull(df).sum() * 100 / len(df.index)), 2)[
pd.isnull(df).sum() > 0
].sort_values()
# ## Exploratory Data Analysis
sub_categorical_cols = [
"Tenure",
"CityTier",
"PreferredLoginDevice",
"PreferredPaymentMode",
"Gender",
"PreferedOrderCat",
"MaritalStatus",
]
df_churn = df[df["Churn"] == 1]
plt.figure(figsize=(12, 20))
idx = 1
for column in sub_categorical_cols:
plt.subplot(4, 2, idx)
sns.countplot(data=df_churn, x=column)
idx = idx + 1
plt.show()
# checking correlation among different numerical variables using clustered heat map
plt.figure(figsize=(25, 15))
sns.clustermap(df.corr(), cmap="PiYG", annot=True, fmt=".2f")
plt.show()
# ## Data Preparation / Preprocessing
class Preprocessing:
def __init__(self, df):
self.df = df
def shape(self):
print(f"shape: {self.df.shape}")
def dtypes(self, pr=False):
print("Types")
if pr:
print(self.df.dtypes)
def supposed2beint(self):
float_cols = [
column for column in self.df.columns if self.df[column].dtype == "float"
]
int2be_cols = []
for col in float_cols:
if (self.df[col].fillna(-9999) % 1 == 0).all() == True:
int2be_cols.append(col)
return int2be_cols
def isNaN(self, pr=False):
if pr:
print("Contain NaN")
print(self.df.isnull().sum())
else:
return self.df.columns[self.df.isna().any()].tolist()
def isObject(self):
return [
column for column in self.df.columns if self.df[column].dtype == "object"
]
def check_dataframe(self):
self.shape()
self.dtypes(True)
self.isNaN(True)
def fillNaN(self):
nan_cols = self.isNaN()
int2be_cols = self.supposed2beint()
# nan_cols == int2be_cols they are the same
for col in int2be_cols:
self.df[col].fillna(round(self.df[col].mean()), inplace=True)
self.df[col] = self.df[col].astype(int)
def adjust_category_cols(self):
self.fillNaN()
# PreferredLoginDevice
# self.df.loc[self.df["PreferredLoginDevice"] == "Mobile Phone", "PreferredLoginDevice"] = "Phone"
# PreferredPaymentMode
# self.df.loc[self.df["PreferredPaymentMode"] == "Credit Card", "PreferredPaymentMode"] = "CC"
# self.df.loc[self.df["PreferredPaymentMode"] == "Cash on Delivery", "PreferredPaymentMode"] = "COD"
self.adjust_duplicate_column_values(self.df)
def adjust_duplicate_column_values(self, _df):
# PreferredLoginDevice
_df.loc[
_df["PreferredLoginDevice"] == "Mobile Phone", "PreferredLoginDevice"
] = "Phone"
# PreferredPaymentMode
_df.loc[
_df["PreferredPaymentMode"] == "Credit Card", "PreferredPaymentMode"
] = "CC"
_df.loc[
_df["PreferredPaymentMode"] == "Cash on Delivery", "PreferredPaymentMode"
] = "COD"
def drop_useless_cols(self):
self.df.drop(["CustomerID"], axis=1, inplace=True)
def split_target(self):
self.adjust_category_cols()
self.drop_useless_cols()
self.X = self.df.drop("Churn", axis=1)
self.y = self.df["Churn"].astype(int).to_numpy()
def find_enc_method(self):
cat_cols = self.isObject()
one_hot_cols = [col for col in cat_cols if self.X[col].nunique() <= 3]
label_enc_cols = [col for col in cat_cols if col not in one_hot_cols]
self.cat_cols = cat_cols
self.one_hot_cols = one_hot_cols
self.label_enc_cols = label_enc_cols
return one_hot_cols, label_enc_cols, cat_cols
def encoding(self):
one_hot_cols, label_enc_cols, cat_cols = self.find_enc_method()
num_cols = [col for col in self.X.columns if col not in cat_cols]
self.num_cols = num_cols
X_OHE, X_LE, X_NUM = (
self.X[one_hot_cols].copy(),
self.X[label_enc_cols].copy(),
self.X[num_cols].copy(),
)
self.OHE = OneHotEncoder(drop="first", handle_unknown="error")
X_OHE = self.OHE.fit_transform(X_OHE).toarray()
self.le_dict = {}
# self.LE = LabelEncoder()
for col in X_LE.columns:
LE = LabelEncoder()
self.le_dict[col] = LE.fit(X_LE[col])
X_LE[col] = self.le_dict[col].transform(X_LE[col])
return X_OHE, X_LE.to_numpy(), X_NUM.to_numpy()
def scaling(self):
X_OHE, X_LE, X_num = self.encoding()
self.SS = StandardScaler()
X_num = self.SS.fit_transform(X_num)
self.X_total = np.concatenate((X_OHE, X_LE, X_num), axis=1)
def get_encoders(self):
return self.OHE, self.LE, self.le_dict
def get_scaler(self):
return self.SS
def get_default_Xy(self):
self.split_target()
return self.X, self.y
def get_Xy(self):
self.split_target()
self.scaling()
return self.X_total, self.y
def prepare_unseen_df(self, unseen_df):
"""
Used for scoring new unseen data
"""
cat_cols = self.cat_cols
num_cols = self.num_cols
one_hot_cols = self.one_hot_cols
label_enc_cols = self.label_enc_cols
self.adjust_duplicate_column_values(unseen_df)
X_OHE, X_LE, X_NUM = (
unseen_df[one_hot_cols].copy(),
unseen_df[label_enc_cols].copy(),
unseen_df[num_cols].copy(),
)
X_OHE = self.OHE.transform(X_OHE).toarray()
for col in X_LE.columns:
X_LE[col] = self.le_dict[col].transform(X_LE[col])
X_num = self.SS.transform(X_NUM.to_numpy())
return np.concatenate((X_OHE, X_LE.to_numpy(), X_num), axis=1)
pre = Preprocessing(df)
X, y = pre.get_Xy()
unique, counts = np.unique(y, return_counts=True)
unique, counts
import optuna
from xgboost import XGBClassifier
def objective_xgb(trial):
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42, stratify=y
)
param = {
"lambda": trial.suggest_loguniform("lambda", 1e-3, 10.0),
"alpha": trial.suggest_loguniform("alpha", 1e-3, 10.0),
"colsample_bytree": trial.suggest_categorical(
"colsample_bytree", [0.5, 0.6, 0.7, 0.8, 0.9, 1.0]
),
"subsample": trial.suggest_categorical("subsample", [0.6, 0.7, 0.8, 1.0]),
"learning_rate": trial.suggest_categorical(
"learning_rate", [0.008, 0.009, 0.01, 0.012, 0.014, 0.016, 0.018, 0.02]
),
"n_estimators": trial.suggest_categorical(
"n_estimators", [150, 200, 300, 3000]
),
"max_depth": trial.suggest_categorical(
"max_depth", [4, 5, 7, 9, 11, 13, 15, 17]
),
"random_state": 42,
"min_child_weight": trial.suggest_int("min_child_weight", 1, 300),
}
model = XGBClassifier(**param)
model.fit(
X_train,
y_train,
early_stopping_rounds=50,
eval_set=[(X_test, y_test)],
verbose=False,
)
preds = model.predict(X_test)
acc = accuracy_score(y_test, preds)
return acc
study = optuna.create_study(direction="maximize")
study.optimize(objective_xgb, n_trials=50)
params_xgb = study.best_trial.params
print("Number of finished trials:", len(study.trials))
print("Best trial:", params_xgb)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42, stratify=y
)
model = XGBClassifier(**params_xgb)
model.fit(
X_train,
y_train,
early_stopping_rounds=50,
eval_set=[(X_test, y_test)],
verbose=False,
)
y_pred = model.predict(X_test)
# ### Model Evaluation
print(f"Accuracy: {round(accuracy_score(y_pred, y_test), 2)}")
print("=" * 60)
print(f"Recall: {round(recall_score(y_pred, y_test),2)}")
print("=" * 60)
print(f"Precision: {round(precision_score(y_pred, y_test), 2)}")
print("=" * 60)
print(f"F1: {round(f1_score(y_pred, y_test), 2)}")
print("=" * 60)
print(f"Auc: {round(roc_auc_score(y_pred, y_test), 2)}")
print("=" * 60)
CR = classification_report(y_test, y_pred)
CM = confusion_matrix(y_test, y_pred)
print(f"Classification Report:\n {CR}")
print("=" * 60)
print("\n")
print(f"Confusion Matrix: {CM}")
print("=" * 60)
# # Prediction using realtime data
def evaluate_churn(eva_dict):
scoring_df = pd.DataFrame.from_records([scoring_dict])
preprocessed_df = pre.prepare_unseen_df(scoring_df)
test_pred_arr = model.predict([preprocessed_df[0]])
test_pred_proba = model.predict_proba([preprocessed_df[0]])
will_churn = "Yes" if test_pred_arr[0] == 1 else "No"
print("is the customer will churn - ", will_churn)
print(
f"Probability for will not churn: {test_pred_proba[0][0]}, will churn: {test_pred_proba[0][1]}"
)
return (will_churn, test_pred_proba[0])
scoring_dict = {
"Tenure": 5,
"PreferredLoginDevice": "Computer",
"CityTier": 3,
"WarehouseToHome": 0,
"PreferredPaymentMode": "Debit Card",
"Gender": "Male",
"HourSpendOnApp": 0,
"NumberOfDeviceRegistered": 0,
"PreferedOrderCat": "Mobile",
"SatisfactionScore": 10,
"MaritalStatus": "Married",
"NumberOfAddress": 3,
"Complain": 100,
"OrderAmountHikeFromlastYear": 13,
"CouponUsed": 0,
"OrderCount": 0,
"DaySinceLastOrder": 300,
"CashbackAmount": 0,
}
evaluate_churn(scoring_dict)
| false | 0 | 3,737 | 3 | 4,114 | 3,737 |
||
129803824
|
import numpy as np
import pandas as pd
import emoji
from keras.models import Sequential
from keras.layers import Dense, LSTM, SimpleRNN, Embedding
from keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.utils import to_categorical
data = pd.read_csv("/kaggle/input/emoji-data/emoji_data.csv", header=None)
data.head()
emoji_dict = {
0: ":red_heart:",
1: ":baseball:",
2: ":grinning_face_with_big_eyes:",
3: ":disappointed_face:",
4: ":fork_and_knife_with_plate:",
}
def label_to_emoji(label):
return emoji.emojize(emoji_dict[label])
X = data[0].values
Y = data[1].values
# # Embeddings
file = open("/kaggle/input/glove-data/glove.6B.100d.txt", "r", encoding="utf8")
content = file.readlines()
file.close()
embeddings = {}
for line in content:
line = line.split()
embeddings[line[0]] = np.array(line[1:], dtype=float)
tokenizer = Tokenizer()
tokenizer.fit_on_texts(X)
word2index = tokenizer.word_index
Xtokens = tokenizer.texts_to_sequences(X)
Xtokens
def get_maxlen(data):
maxlen = 0
for sent in data:
maxlen = max(maxlen, len(sent))
return maxlen
maxlen = get_maxlen(Xtokens)
print(maxlen)
Xtrain = pad_sequences(Xtokens, maxlen=maxlen, padding="post", truncating="post")
Xtrain
Ytrain = to_categorical(Y)
Ytrain
# # Model
embed_size = 100
embedding_matrix = np.zeros((len(word2index) + 1, embed_size))
for word, i in word2index.items():
embed_vector = embeddings[word]
embedding_matrix[i] = embed_vector
embedding_matrix
model = Sequential(
[
Embedding(
input_dim=len(word2index) + 1,
output_dim=embed_size,
input_length=maxlen,
weights=[embedding_matrix],
trainable=False,
),
LSTM(units=16, return_sequences=True),
LSTM(units=4),
Dense(5, activation="softmax"),
]
)
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.fit(Xtrain, Ytrain, epochs=100)
test = ["I feel good", "I feel very bad", "lets eat dinner"]
test_seq = tokenizer.texts_to_sequences(test)
Xtest = pad_sequences(test_seq, maxlen=maxlen, padding="post", truncating="post")
y_pred = model.predict(Xtest)
y_pred = np.argmax(y_pred, axis=1)
for i in range(len(test)):
print(test[i], label_to_emoji(y_pred[i]))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/803/129803824.ipynb
| null | null |
[{"Id": 129803824, "ScriptId": 38602983, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13317745, "CreationDate": "05/16/2023 15:17:09", "VersionNumber": 1.0, "Title": "Emoji_Prediction_LSTM", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 105.0, "LinesInsertedFromPrevious": 105.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import emoji
from keras.models import Sequential
from keras.layers import Dense, LSTM, SimpleRNN, Embedding
from keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.utils import to_categorical
data = pd.read_csv("/kaggle/input/emoji-data/emoji_data.csv", header=None)
data.head()
emoji_dict = {
0: ":red_heart:",
1: ":baseball:",
2: ":grinning_face_with_big_eyes:",
3: ":disappointed_face:",
4: ":fork_and_knife_with_plate:",
}
def label_to_emoji(label):
return emoji.emojize(emoji_dict[label])
X = data[0].values
Y = data[1].values
# # Embeddings
file = open("/kaggle/input/glove-data/glove.6B.100d.txt", "r", encoding="utf8")
content = file.readlines()
file.close()
embeddings = {}
for line in content:
line = line.split()
embeddings[line[0]] = np.array(line[1:], dtype=float)
tokenizer = Tokenizer()
tokenizer.fit_on_texts(X)
word2index = tokenizer.word_index
Xtokens = tokenizer.texts_to_sequences(X)
Xtokens
def get_maxlen(data):
maxlen = 0
for sent in data:
maxlen = max(maxlen, len(sent))
return maxlen
maxlen = get_maxlen(Xtokens)
print(maxlen)
Xtrain = pad_sequences(Xtokens, maxlen=maxlen, padding="post", truncating="post")
Xtrain
Ytrain = to_categorical(Y)
Ytrain
# # Model
embed_size = 100
embedding_matrix = np.zeros((len(word2index) + 1, embed_size))
for word, i in word2index.items():
embed_vector = embeddings[word]
embedding_matrix[i] = embed_vector
embedding_matrix
model = Sequential(
[
Embedding(
input_dim=len(word2index) + 1,
output_dim=embed_size,
input_length=maxlen,
weights=[embedding_matrix],
trainable=False,
),
LSTM(units=16, return_sequences=True),
LSTM(units=4),
Dense(5, activation="softmax"),
]
)
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.fit(Xtrain, Ytrain, epochs=100)
test = ["I feel good", "I feel very bad", "lets eat dinner"]
test_seq = tokenizer.texts_to_sequences(test)
Xtest = pad_sequences(test_seq, maxlen=maxlen, padding="post", truncating="post")
y_pred = model.predict(Xtest)
y_pred = np.argmax(y_pred, axis=1)
for i in range(len(test)):
print(test[i], label_to_emoji(y_pred[i]))
| false | 0 | 767 | 0 | 767 | 767 |
||
129874850
|
# # From Theory to Practice: LSTM and Transformers in PyTorch
# ---
# 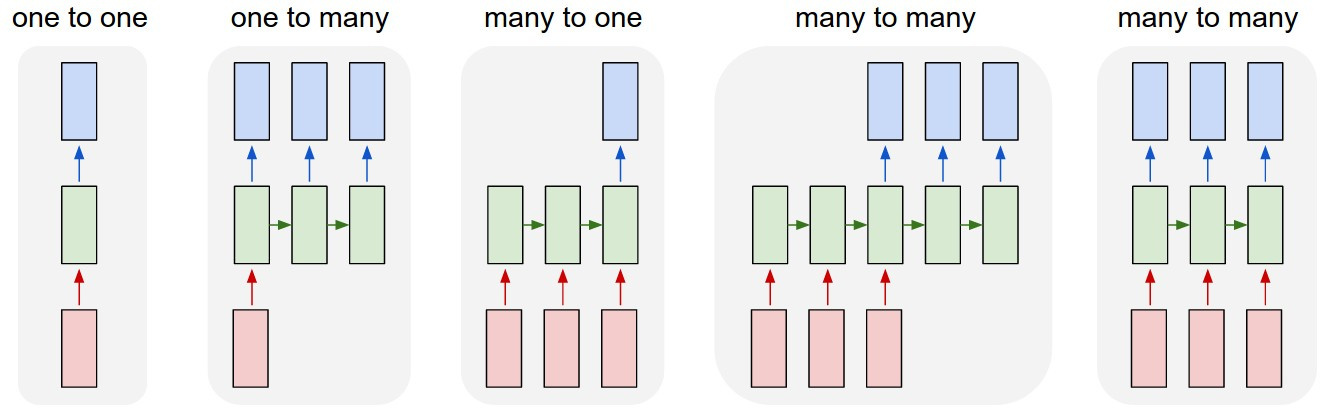
# Welcome to this Kaggle notebook, where we'll dive deep into understanding and implementing Long Short-Term Memory (LSTM) networks using PyTorch, a powerful deep learning framework. But before we delve into the intricacies of LSTM, let's take a moment to understand the basic concepts of time series data, Recurrent Neural Networks (RNNs), and LSTM.
# Time Series Data
# Time series data is a sequence of numerical data points taken at successive equally spaced points in time. These data points are ordered and depend on the previous data points, making time series data a prime candidate for predictions. Examples of time series data include stock prices, weather forecasts, and sales data, among many others.
# Recurrent Neural Networks (RNNs)
# Traditional neural networks struggle with time series data due to their inability to remember previous inputs in their current state. Recurrent Neural Networks (RNNs), however, are designed to address this problem. RNNs are a class of artificial neural networks where connections between nodes form a directed graph along a temporal sequence. This allows them to use their internal state (memory) to process sequences of inputs, making them ideal for time-dependent data.
# However, RNNs suffer from certain limitations. They struggle to handle long-term dependencies because of the 'vanishing gradient' problem, where the contribution of information decays geometrically over time, making it difficult for the RNN to learn from earlier layers.
# Long Short-Term Memory (LSTM)
# Long Short-Term Memory networks, or LSTMs, are a special kind of RNN capable of learning long-term dependencies. Introduced by Hochreiter and Schmidhuber in 1997, LSTMs have a unique design that helps combat the vanishing gradient problem. They contain a cell state and three gates (input, forget, and output) to control the flow of information inside the network, allowing them to remember or forget information over long periods of time.
# In this notebook, we will explore how to correctly implement LSTM in PyTorch and use it for time series prediction tasks. We will cover everything from the basics of LSTM to its implementation, aiming to provide a comprehensive understanding of this powerful neural network architecture. Let's get started!
# Understanding Input and Output in torch.nn.RNN
# In this section, we're going to delve into the specifics of the input and output parameters of the torch.nn.RNN module, a built-in recurrent neural network (RNN) implementation in the PyTorch library. It's crucial to understand these parameters to fully leverage PyTorch's RNN capabilities in our LSTM implementation.
# Input to torch.nn.RNN
# The torch.nn.RNN module takes in two primary inputs:
#
# input: This represents the sequence that is fed into the network. The expected size is (seq_len, batch, input_size). However, if batch_first=True is specified, then the input size should be rearranged to (batch, seq_len, input_size).
# h_0: This stands for the initial hidden state of the network at time step t=0. By default, if we don't initialize this hidden layer, PyTorch will automatically initialize it with zeros. The size of h_0 should be (num_layers * num_directions, batch, input_size), where num_layers represents the number of stacked RNNs and num_directions equals 2 for bidirectional RNNs and 1 otherwise.
#
# Output from torch.nn.RNN
# The torch.nn.RNN module provides two outputs:
#
# out: This represents the output from the last RNN layer for all time steps. The size is (seq_len, batch, num_directions * hidden_size). However, if batch_first=True is specified, the output size becomes (batch, seq_len, num_directions * hidden_size).
# h_n: This is the hidden state value from the last time step across all RNN layers. The size is (num_layers * num_directions, batch, hidden_size). Unlike the input, the h_n is unaffected by batch_first=True.
#
# To better visualize these inputs and outputs, refer to the following diagram. In this case, we assume a batch size of 1. While the diagram illustrates an LSTM, which has two hidden parameters (h, c), please note that RNN and GRU only have h.
# By understanding these parameters, we can harness the power of the torch.nn.RNN module and build effective models for our time series data using LSTM. Let's continue our exploration of LSTM with PyTorch in the following sections.
# 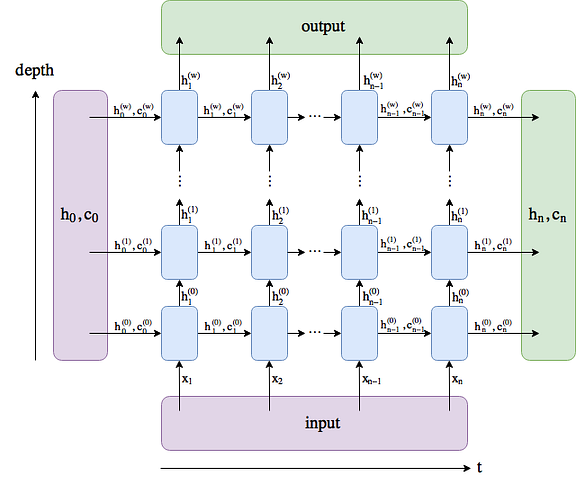
# # Table of Contents
# - [1. Imports](#1)
# - [2. LSTM](#2)
# - [Many-to-One](#2.1)
# - [Many-to-Many](#2.2)
# - [Many-to-Many generating sequence](#2.3)
# - [3. Transformers](#3)
# - [Masking Input](#3.1)
# - [SOS & EOS tokens](#3.2)
# # ** Imports **
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.utils.data import DataLoader, Dataset
from tqdm import tqdm
import numpy as np
import pandas as pd
import random
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("white")
#
# About the Dataset
# In this notebook, we will utilize a simple time series data to test and understand the application of LSTM and Transformer models. The chosen dataset is quite straightforward — a range of numbers starting from 0 and ending at 1000. This simplicity will allow us to focus more on the workings of the LSTM and Transformer models, examining how well they can comprehend and process a simple sequential numerical data. Through this, we aim to achieve a clear understanding of these powerful deep learning techniques.
# # ** LSTM **
# Understanding Many-to-One Architecture in LSTM
# Long Short-Term Memory (LSTM) networks, like all Recurrent Neural Networks (RNNs), are renowned for their ability to process sequential data. One of the key aspects that make them flexible and powerful is the various types of input-output architectures they can adopt, one of which is the Many-to-One architecture.
# In a Many-to-One LSTM architecture, the model accepts a sequence of inputs over multiple time steps and produces a single output. In each time step, the LSTM cell takes in an input and the previous cell's hidden state, processes them, and passes on its own hidden state to the next cell.
# Despite receiving input at each time step, the Many-to-One LSTM only produces its final output at the last time step. This characteristic makes Many-to-One LSTM networks particularly useful for tasks like sentiment analysis, where a model reads a sequence of words (input) and outputs a single sentiment score, or text classification, where a document is read sequentially and a single class label is output.
# Through the power of LSTM and the flexibility of architectures like Many-to-One, we can effectively tackle a wide range of sequence-based problems in the world of machine learning and artificial intelligence.
# ### Create Custom Data Loader [multi-core]
class CustomDataset(Dataset):
def __init__(self, seq_len=5, max_len=1000):
super(CustomDataset).__init__()
self.datalist = np.arange(0, max_len)
self.data, self.targets = self.timeseries(self.datalist, seq_len)
def __len__(self):
return len(self.data)
def timeseries(self, data, window):
temp = []
targ = data[window:]
for i in range(len(data) - window):
temp.append(data[i : i + window])
return np.array(temp), targ
def __getitem__(self, index):
x = torch.tensor(self.data[index]).type(torch.Tensor)
y = torch.tensor(self.targets[index]).type(torch.Tensor)
return x, y
dataset = CustomDataset(seq_len=5, max_len=1000)
for x, y in dataset:
print(x, y)
break
dataloader = DataLoader(dataset, batch_size=4, shuffle=True, num_workers=4)
# collate_fn=custom_collector
for x, y in dataloader:
print(x, y)
break
#
# Let's take a closer look at our specific use case for the many-to-one LSTM architecture. In our scenario, we are feeding the LSTM with a sequence of 5 random numbers, and we anticipate that the model will predict the 6th number in the sequence. While we've chosen a straightforward series of incrementing numbers for this example, the potential applications of this concept extend much further.
# Imagine this sequence being a time-series data of stock prices, weather conditions, or even a series of steps in a logical reasoning question. The ability to predict the next event based on a series of preceding events is a critical aspect in many fields, including finance, meteorology, and artificial intelligence. By training our LSTM model to understand and predict these sequences, we can leverage the many-to-one LSTM architecture to solve complex problems in these areas and beyond.
#
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers):
super().__init__()
self.hidden_size = hidden_size
self.num_layers = num_layers
self.lstm = nn.LSTM(input_size, hidden_size, num_layers, batch_first=True)
self.fc = nn.Linear(hidden_size, 1)
def forward(self, x):
# hidden states not defnined hence the value of h0,c0 == (0,0)
out, (hn, cn) = self.lstm(x)
# as the diagram suggest to take the last output in many to one
# print(out.shape)
# print(hn.shape)
# all batch, last column of seq, all hidden values
out = out[:, -1, :]
out = self.fc(out)
return out
model = RNN(input_size=1, hidden_size=256, num_layers=2)
t = torch.tensor([11, 12, 13, 14, 15]).type(torch.Tensor).view(1, -1, 1)
t.shape
model(t)
# ### Training
loss_function = nn.MSELoss()
learning_rate = 1e-3
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
for e in tqdm(range(50)):
i = 0
for x, y in dataloader:
optimizer.zero_grad()
x = torch.unsqueeze(x, 0).permute(1, 2, 0)
# forward
predictions = model(x)
loss = loss_function(predictions.view(-1), y)
# backward
loss.backward()
# optimization
optimizer.step()
i += 1
if e % 5 == 0:
print(loss.detach().numpy())
input_tensor = torch.tensor([10, 11, 12, 13, 14]).type(torch.Tensor).view(1, -1, 1)
model(input_tensor)
#
# Understanding Many-to-Many Architecture in LSTM
# Another crucial architecture in the world of Long Short-Term Memory (LSTM) networks, a type of Recurrent Neural Network (RNN), is the Many-to-Many architecture. This architecture offers a versatile way of handling a diverse set of problems involving sequential data.
# In a Many-to-Many LSTM architecture, the model processes a sequence of inputs over multiple time steps and generates a sequence of outputs. In this setting, each LSTM cell takes in an input and the previous cell's hidden state at each time step, then produces an output along with its own hidden state that it passes on to the next cell.
# Unlike the Many-to-One LSTM, the Many-to-Many LSTM doesn't wait till the last time step to produce an output. Instead, it generates an output at each time step. This makes Many-to-Many LSTM networks highly useful for tasks such as machine translation, where a sequence of words in one language (input) is translated into a sequence of words in another language (output).
# The Many-to-Many architecture of LSTM opens up a broad array of possibilities, making it a powerful tool in the realms of machine learning and artificial intelligence.
#
class CustomDataset(Dataset):
def __init__(self, seq_len=50, future=5, max_len=1000):
super(CustomDataset).__init__()
self.datalist = np.arange(0, max_len)
self.data, self.targets = self.timeseries(self.datalist, seq_len, future)
def __len__(self):
# this len will decide the index range in getitem
return len(self.targets)
def timeseries(self, data, window, future):
temp = []
targ = []
for i in range(len(data) - window):
temp.append(data[i : i + window])
for i in range(len(data) - window - future):
targ.append(data[i + window : i + window + future])
return np.array(temp), targ
def __getitem__(self, index):
x = torch.tensor(self.data[index]).type(torch.Tensor)
y = torch.tensor(self.targets[index]).type(torch.Tensor)
return x, y
dataset = CustomDataset(seq_len=50, future=5, max_len=1000)
for x, y in dataset:
print(x.shape, y.shape)
break
dataloader = DataLoader(dataset, batch_size=8, shuffle=True, num_workers=4)
# collate_fn=custom_collector
for x, y in dataloader:
print(x.shape, y.shape)
break
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers, future=5):
super().__init__()
self.future = future
self.hidden_size = hidden_size
self.num_layers = num_layers
self.lstm = nn.LSTM(input_size, hidden_size, num_layers, batch_first=True)
self.fc = nn.Linear(hidden_size, future)
def forward(self, x):
# hidden states not defnined hence the value of h0,c0 == (0,0)
out, (hn, cn) = self.lstm(x)
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers, future=5):
super().__init__()
self.future = future
self.hidden_size = hidden_size
self.num_layers = num_layers
self.lstm = nn.LSTM(input_size, hidden_size, num_layers, batch_first=True)
self.fc = nn.Linear(hidden_size, future)
def forward(self, x):
# hidden states not defnined hence the value of h0,c0 == (0,0)
out, (hn, cn) = self.lstm(x)
# as the diagram suggest to take the last output in many to one
# print(out.shape)
# print(hn.shape)
# all batch, last column of seq, all hidden values
out = out[:, -self.future, :]
out = self.fc(out)
return out
model = RNN(input_size=1, hidden_size=256, num_layers=2, future=5)
d = 45
t = torch.tensor(np.arange(d, d + 50)).type(torch.Tensor).view(1, -1, 1)
t.shape
model(t)
loss_function = nn.MSELoss()
learning_rate = 1e-3
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
for e in tqdm(range(50)):
i = 0
avg_loss = []
for x, y in dataloader:
optimizer.zero_grad()
x = torch.unsqueeze(x, 0).permute(1, 2, 0)
# forward
predictions = model(x)
# loss
loss = loss_function(predictions, y)
# backward
loss.backward()
# optimization
optimizer.step()
avg_loss.append(loss.detach().numpy())
i += 1
if e % 2 == 0:
avg_loss = np.array(avg_loss)
print(avg_loss.mean())
#
# After feeding the initial 50 terms of our sequence into the model, we begin to observe some promising results. It appears that the model is successfully learning to recognize the underlying patterns in the sequence.
# The output generated by the model seems to adhere to the logic of the sequence, suggesting that the LSTM architecture is effectively capturing and understanding the sequential dependencies. This ability to discern patterns and extrapolate them is a powerful aspect of LSTM networks, and it's rewarding to see it at work in our model.
# These early results are encouraging, indicating that our model is on the right track. As we continue to refine and train our LSTM, we can expect it to become even more adept at understanding and predicting the sequence.
#
d = random.randint(0, 1000)
t = torch.tensor(np.arange(d, d + 50)).type(torch.Tensor).view(1, -1, 1)
r = model(t).view(-1)
fig = plt.figure(figsize=(16, 4))
plt_x = np.arange(0, t.shape[1] + len(r))
plt_y = np.arange(d, d + 50 + len(r))
plt_xp = np.arange(t.shape[1], t.shape[1] + len(r))
plt_yp = r.detach().numpy()
for i in range(len(r)):
plt.scatter(plt_x, plt_y)
plt.scatter(plt_xp, plt_yp)
#
# Understanding Many-to-Many Sequence Generation with LSTM
# When working with Long Short-Term Memory (LSTM) networks, it's essential to understand how sequence generation is handled, particularly in a Many-to-Many setting. In such an architecture, the output from each LSTM cell can be used as an input to a subsequent feed-forward network to generate a sequence of outputs.
# Let's consider the following block of code as an example:
#
# out, (hn, cn) = self.lstm(x)
# res = torch.zeros((out.shape[0], out.shape[1]))
# for b in range(out.shape[0]):
# feed = out[b, :, :]
# _out = self.fc(feed).view(-1)
# res[b] = _out
#
# In this code, self.lstm(x) applies the LSTM layer to the input x, generating an output out and the final hidden and cell states hn and cn. We then initialize a zeros tensor res of the same size as out to store our results.
# Then, for each sequence in the output out, we feed the sequence through a fully connected layer self.fc(feed) and reshape the output to match our expected dimensions using .view(-1). The result is stored in the corresponding position in res.
# This process exemplifies how a Many-to-Many LSTM network can be used to generate a sequence of outputs, with the LSTM layer and a subsequent feed-forward layer working in tandem to transform a sequence of inputs into a corresponding sequence of outputs.
#
class CustomDataset(Dataset):
def __init__(self, seq_len=50, future=50, max_len=1000):
super(CustomDataset).__init__()
self.datalist = np.arange(0, max_len)
self.data, self.targets = self.timeseries(self.datalist, seq_len, future)
def __len__(self):
# this len will decide the index range in getitem
return len(self.targets)
def timeseries(self, data, window, future):
temp = []
targ = []
for i in range(len(data) - window):
temp.append(data[i : i + window])
for i in range(len(data) - window - future):
targ.append(data[i + future : i + window + future])
return np.array(temp), targ
def __getitem__(self, index):
x = torch.tensor(self.data[index]).type(torch.Tensor)
y = torch.tensor(self.targets[index]).type(torch.Tensor)
return x, y
dataset = CustomDataset(seq_len=50, future=5, max_len=1000)
for x, y in dataset:
print(x.shape, y.shape)
break
x
y
dataloader = DataLoader(dataset, batch_size=8, shuffle=True, num_workers=4)
# collate_fn=custom_collector
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers, future=5):
super().__init__()
self.future = future
self.hidden_size = hidden_size
self.num_layers = num_layers
self.lstm = nn.LSTM(input_size, hidden_size, num_layers, batch_first=True)
self.fc = nn.Linear(hidden_size, 1)
def forward(self, x):
# hidden states not defnined hence the value of h0,c0 == (0,0)
out, (hn, cn) = self.lstm(x)
# as the diagram suggest to take the last output in many to one
# print(out.shape)
# print(hn.shape)
# all batch, last column of seq, all hidden values
res = torch.zeros((out.shape[0], out.shape[1]))
for b in range(out.shape[0]):
feed = out[b, :, :]
_out = self.fc(feed).view(-1)
res[b] = _out
return res
model = RNN(input_size=1, hidden_size=256, num_layers=2, future=5)
t = torch.tensor(np.arange(d, d + 50)).type(torch.Tensor).view(1, -1, 1)
r = model(t).view(-1)
r
loss_function = nn.MSELoss()
learning_rate = 1e-3
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
for e in tqdm(range(100)):
i = 0
avg_loss = []
for x, y in dataloader:
optimizer.zero_grad()
x = torch.unsqueeze(x, 0).permute(1, 2, 0)
# forward
predictions = model(x)
# loss
loss = loss_function(predictions, y)
# backward
loss.backward()
# optimization
optimizer.step()
avg_loss.append(loss.detach().numpy())
i += 1
if e % 5 == 0:
avg_loss = np.array(avg_loss)
print(avg_loss.mean())
d = random.randint(0, 1000)
t = torch.tensor(np.arange(d, d + 50)).type(torch.Tensor).view(1, -1, 1)
r = model(t).view(-1)
fig = plt.figure(figsize=(16, 4))
plt_x = np.arange(0, t.shape[1])
plt_y = np.arange(d, d + 50)
plt_xp = np.arange(5, t.shape[1] + 5)
plt_yp = r.detach().numpy()
for i in range(len(r)):
plt.scatter(plt_x, plt_y, label="real")
plt.scatter(plt_xp, plt_yp, label="predicted")
plt.show()
#
# # ** Transformers **
# Transformers, a breakthrough in the field of natural language processing, also adopt various types of input-output architectures, including the Many-to-Many setup. In this context, Transformers bring a unique approach to the table, contrasting with the methods used in traditional Recurrent Neural Networks (RNNs) such as LSTM.
# In a Many-to-Many Transformer architecture, the model accepts a sequence of inputs and returns a sequence of outputs. However, unlike RNNs, which process sequences in a time-stepped manner, Transformers process all inputs simultaneously. This is made possible by the attention mechanism, which allows the model to focus on different parts of the input sequence for each output, essentially creating a 'shortcut' between each input and output.
# This architecture is especially useful in tasks like machine translation, where the model needs to understand the context of the whole sentence to accurately translate it. Similarly, it can be used in tasks like text summarization or question answering, where understanding the entire context at once can lead to better results.
# The Many-to-Many architecture in Transformers, combined with their attention mechanism, offers an innovative approach to tackling sequential tasks, making Transformers a powerful tool in the field of machine learning and artificial intelligence.
# 
class CustomDataset(Dataset):
def __init__(self, seq_len=50, future=50, max_len=1000):
super(CustomDataset).__init__()
self.vocab = {"SOS": 1001, "EOS": 1002}
self.datalist = np.arange(0, max_len)
self.data, self.targets = self.timeseries(self.datalist, seq_len, future)
def __len__(self):
# this len will decide the index range in getitem
return len(self.targets)
def timeseries(self, data, window, future):
temp = []
targ = []
for i in range(len(data) - window):
temp.append(data[i : i + window])
for i in range(len(data) - window - future):
targ.append(data[i + future : i + window + future])
return np.array(temp), targ
def __getitem__(self, index):
x = torch.tensor(self.data[index]).type(torch.Tensor)
x = torch.cat(
(torch.tensor([self.vocab["SOS"]]), x, torch.tensor([self.vocab["EOS"]]))
).type(torch.LongTensor)
y = torch.tensor(self.targets[index]).type(torch.Tensor)
y = torch.cat(
(torch.tensor([self.vocab["SOS"]]), y, torch.tensor([self.vocab["EOS"]]))
).type(torch.LongTensor)
return x, y
dataset = CustomDataset(seq_len=48, future=5, max_len=1000)
for x, y in dataset:
print(x)
print(y)
break
dataloader = DataLoader(dataset, batch_size=8, shuffle=True, num_workers=4)
# collate_fn=custom_collector
for x, y in dataloader:
print(x.shape)
print(y.shape)
break
#
# The Power of Masking and Efficiency in Transformers
# One of the remarkable features of Transformers is their use of masking during the training process. Masking is an essential aspect of the Transformer's architecture that prevents the model from seeing future tokens in the input sequence during training, thereby preserving the sequential nature of the language.
# In a task such as language translation, where the input sequence is fed into the model all at once, it's crucial that the prediction for each word doesn't rely on words that come after it in the sequence. This is achieved by applying a mask to the input that effectively hides future words from the model during the training phase.
# Not only does masking maintain the sequential integrity of the language, but it also allows Transformers to train more efficiently than their RNN counterparts, like LSTM. Unlike RNNs, which process sequences step-by-step and thus require longer training times for long sequences, Transformers can process all the tokens in the sequence simultaneously, thanks to their attention mechanism. This parallel processing significantly speeds up the training process and allows the model to handle longer sequences more effectively.
# Thus, through the use of masking and their unique architecture, Transformers manage to overcome some of the limitations of traditional RNNs, offering a more efficient and effective approach to sequence-based tasks in machine learning and artificial intelligence.
#
class Transformer(nn.Module):
def __init__(self, num_tokens, dim_model, num_heads, num_layers, input_seq):
super().__init__()
self.input_seq = input_seq
self.num_layers = num_layers
self.embedding = nn.Embedding(num_tokens, dim_model)
self.transformer = nn.Transformer(
d_model=dim_model,
nhead=num_heads,
num_encoder_layers=3,
num_decoder_layers=3,
dim_feedforward=256,
batch_first=True,
)
self.fc = nn.Linear(dim_model, num_tokens)
def forward(self, src, tgt, tf=True):
mask = self.get_mask(tgt.shape[1], teacher_force=tf)
src = self.embedding(src)
tgt = self.embedding(tgt)
out = self.transformer(src, tgt, tgt_mask=mask)
feed = self.fc(out)
feed = torch.squeeze(feed, 2)
return feed
def get_mask(self, size, teacher_force=True):
if teacher_force:
mask = torch.tril(torch.ones(size, size) == 1) # Lower triangular matrix
mask = mask.float()
mask = mask.masked_fill(mask == 0, float("-inf")) # Convert zeros to -inf
mask = mask.masked_fill(mask == 1, float(0.0)) # Convert ones to 0
# EX for size=5:
# [[0., -inf, -inf, -inf, -inf],
# [0., 0., -inf, -inf, -inf],
# [0., 0., 0., -inf, -inf],
# [0., 0., 0., 0., -inf],
# [0., 0., 0., 0., 0.]]
return mask
else:
mask = torch.tril(torch.zeros(size, size) == 1) # Lower triangular matrix
mask = mask.float()
mask = mask.masked_fill(mask == 0, float("-inf")) # Convert zeros to -inf
mask = mask.masked_fill(mask == 1, float(0.0)) # Convert ones to 0
return mask
model = Transformer(
num_tokens=1000 + 3, dim_model=32, num_heads=2, num_layers=2, input_seq=50
)
x.shape, y.shape
model(x, y).shape
t = model(x, y)
t.shape
t.permute(0, 2, 1).shape
loss_function = nn.CrossEntropyLoss()
learning_rate = 1e-3
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
for e in tqdm(range(25)):
i = 0
avg_loss = []
for x, y in dataloader:
optimizer.zero_grad()
# one step behind input and output // Like language modeling
y_input = y[:, :-1] # from starting to -1 position
y_expected = y[:, 1:] # from 1st position to last
# this is done so that in prediction we see a start of token
# forward
predictions = model(x, y_input)
pred = predictions.permute(0, 2, 1)
# loss
loss = loss_function(pred, y_expected)
# backward
loss.backward()
# optimization
optimizer.step()
avg_loss.append(loss.detach().numpy())
i += 1
if e % 5 == 0:
avg_loss = np.array(avg_loss)
print(avg_loss.mean())
torch.squeeze(predictions.topk(1).indices, 2)
y_expected
torch.argmax(pred, dim=1)
#
# The Role of SOS and EOS Tokens in Transformers
# In the domain of natural language processing, particularly when working with Transformer models, special tokens like Start of Sentence (SOS) and End of Sentence (EOS) play a crucial role. These tokens provide valuable cues about the boundaries of sentences, facilitating the model's understanding of language structure.
# The SOS token is added at the beginning of each sentence, marking its start. Similarly, the EOS token is appended at the end of each sentence to indicate its conclusion. These tokens serve as consistent markers that help the model identify and process sentences as distinct units within larger bodies of text.
# Furthermore, in the context of sequence generation tasks, these tokens play an essential role in determining when to begin and end the generation process. For example, during text generation, an EOS token indicates to the model that it should stop generating further tokens.
# Therefore, SOS and EOS tokens are more than just markers; they're integral components in the design and functioning of Transformer models, contributing significantly to their ability to effectively understand and generate human language.
#
def predict(
model, input_sequence, max_length=50, SOS_token=1000 + 1, EOS_token=1000 + 2
):
model.eval()
input_sequence = torch.tensor(input_sequence)
input_sequence = torch.cat(
(torch.tensor([SOS_token]), input_sequence, torch.tensor([EOS_token]))
).type(torch.LongTensor)
input_sequence = torch.unsqueeze(input_sequence, 0)
y_input = torch.tensor([1001], dtype=torch.long)
y_input = torch.unsqueeze(y_input, 0)
for _ in range(max_length):
predictions = model(input_sequence, y_input)
top = predictions.topk(1).indices
top = torch.squeeze(top, 2)
next_item = torch.unsqueeze(top[:, -1], 0)
y_input = torch.cat((y_input, next_item), dim=1)
mask = model.get_mask(y_input.shape[1])
if next_item == EOS_token:
break
return y_input.view(-1).tolist()
d = random.randint(0, 900)
t = torch.tensor(np.arange(d, d + 48)).type(torch.Tensor)
input_sequence = t
print(t)
r = predict(model, input_sequence)
print(r)
fig = plt.figure(figsize=(16, 4))
plt_x = np.arange(0, t.shape[0])
plt_y = t
plt_xp = np.arange(5, t.shape[0] + 5)
plt_yp = r[1:-2]
plt.scatter(plt_x, plt_y, s=14, color="r", label="real")
plt.scatter(plt_xp, plt_yp, s=7, color="b", label="predicted")
plt.legend()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/874/129874850.ipynb
| null | null |
[{"Id": 129874850, "ScriptId": 38629624, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1737338, "CreationDate": "05/17/2023 05:56:06", "VersionNumber": 1.0, "Title": "From Theory to Practice: LSTM and Transformers in", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 700.0, "LinesInsertedFromPrevious": 700.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 34}]
| null | null | null | null |
# # From Theory to Practice: LSTM and Transformers in PyTorch
# ---
# 
# Welcome to this Kaggle notebook, where we'll dive deep into understanding and implementing Long Short-Term Memory (LSTM) networks using PyTorch, a powerful deep learning framework. But before we delve into the intricacies of LSTM, let's take a moment to understand the basic concepts of time series data, Recurrent Neural Networks (RNNs), and LSTM.
# Time Series Data
# Time series data is a sequence of numerical data points taken at successive equally spaced points in time. These data points are ordered and depend on the previous data points, making time series data a prime candidate for predictions. Examples of time series data include stock prices, weather forecasts, and sales data, among many others.
# Recurrent Neural Networks (RNNs)
# Traditional neural networks struggle with time series data due to their inability to remember previous inputs in their current state. Recurrent Neural Networks (RNNs), however, are designed to address this problem. RNNs are a class of artificial neural networks where connections between nodes form a directed graph along a temporal sequence. This allows them to use their internal state (memory) to process sequences of inputs, making them ideal for time-dependent data.
# However, RNNs suffer from certain limitations. They struggle to handle long-term dependencies because of the 'vanishing gradient' problem, where the contribution of information decays geometrically over time, making it difficult for the RNN to learn from earlier layers.
# Long Short-Term Memory (LSTM)
# Long Short-Term Memory networks, or LSTMs, are a special kind of RNN capable of learning long-term dependencies. Introduced by Hochreiter and Schmidhuber in 1997, LSTMs have a unique design that helps combat the vanishing gradient problem. They contain a cell state and three gates (input, forget, and output) to control the flow of information inside the network, allowing them to remember or forget information over long periods of time.
# In this notebook, we will explore how to correctly implement LSTM in PyTorch and use it for time series prediction tasks. We will cover everything from the basics of LSTM to its implementation, aiming to provide a comprehensive understanding of this powerful neural network architecture. Let's get started!
# Understanding Input and Output in torch.nn.RNN
# In this section, we're going to delve into the specifics of the input and output parameters of the torch.nn.RNN module, a built-in recurrent neural network (RNN) implementation in the PyTorch library. It's crucial to understand these parameters to fully leverage PyTorch's RNN capabilities in our LSTM implementation.
# Input to torch.nn.RNN
# The torch.nn.RNN module takes in two primary inputs:
#
# input: This represents the sequence that is fed into the network. The expected size is (seq_len, batch, input_size). However, if batch_first=True is specified, then the input size should be rearranged to (batch, seq_len, input_size).
# h_0: This stands for the initial hidden state of the network at time step t=0. By default, if we don't initialize this hidden layer, PyTorch will automatically initialize it with zeros. The size of h_0 should be (num_layers * num_directions, batch, input_size), where num_layers represents the number of stacked RNNs and num_directions equals 2 for bidirectional RNNs and 1 otherwise.
#
# Output from torch.nn.RNN
# The torch.nn.RNN module provides two outputs:
#
# out: This represents the output from the last RNN layer for all time steps. The size is (seq_len, batch, num_directions * hidden_size). However, if batch_first=True is specified, the output size becomes (batch, seq_len, num_directions * hidden_size).
# h_n: This is the hidden state value from the last time step across all RNN layers. The size is (num_layers * num_directions, batch, hidden_size). Unlike the input, the h_n is unaffected by batch_first=True.
#
# To better visualize these inputs and outputs, refer to the following diagram. In this case, we assume a batch size of 1. While the diagram illustrates an LSTM, which has two hidden parameters (h, c), please note that RNN and GRU only have h.
# By understanding these parameters, we can harness the power of the torch.nn.RNN module and build effective models for our time series data using LSTM. Let's continue our exploration of LSTM with PyTorch in the following sections.
# 
# # Table of Contents
# - [1. Imports](#1)
# - [2. LSTM](#2)
# - [Many-to-One](#2.1)
# - [Many-to-Many](#2.2)
# - [Many-to-Many generating sequence](#2.3)
# - [3. Transformers](#3)
# - [Masking Input](#3.1)
# - [SOS & EOS tokens](#3.2)
# # ** Imports **
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.utils.data import DataLoader, Dataset
from tqdm import tqdm
import numpy as np
import pandas as pd
import random
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("white")
#
# About the Dataset
# In this notebook, we will utilize a simple time series data to test and understand the application of LSTM and Transformer models. The chosen dataset is quite straightforward — a range of numbers starting from 0 and ending at 1000. This simplicity will allow us to focus more on the workings of the LSTM and Transformer models, examining how well they can comprehend and process a simple sequential numerical data. Through this, we aim to achieve a clear understanding of these powerful deep learning techniques.
# # ** LSTM **
# Understanding Many-to-One Architecture in LSTM
# Long Short-Term Memory (LSTM) networks, like all Recurrent Neural Networks (RNNs), are renowned for their ability to process sequential data. One of the key aspects that make them flexible and powerful is the various types of input-output architectures they can adopt, one of which is the Many-to-One architecture.
# In a Many-to-One LSTM architecture, the model accepts a sequence of inputs over multiple time steps and produces a single output. In each time step, the LSTM cell takes in an input and the previous cell's hidden state, processes them, and passes on its own hidden state to the next cell.
# Despite receiving input at each time step, the Many-to-One LSTM only produces its final output at the last time step. This characteristic makes Many-to-One LSTM networks particularly useful for tasks like sentiment analysis, where a model reads a sequence of words (input) and outputs a single sentiment score, or text classification, where a document is read sequentially and a single class label is output.
# Through the power of LSTM and the flexibility of architectures like Many-to-One, we can effectively tackle a wide range of sequence-based problems in the world of machine learning and artificial intelligence.
# ### Create Custom Data Loader [multi-core]
class CustomDataset(Dataset):
def __init__(self, seq_len=5, max_len=1000):
super(CustomDataset).__init__()
self.datalist = np.arange(0, max_len)
self.data, self.targets = self.timeseries(self.datalist, seq_len)
def __len__(self):
return len(self.data)
def timeseries(self, data, window):
temp = []
targ = data[window:]
for i in range(len(data) - window):
temp.append(data[i : i + window])
return np.array(temp), targ
def __getitem__(self, index):
x = torch.tensor(self.data[index]).type(torch.Tensor)
y = torch.tensor(self.targets[index]).type(torch.Tensor)
return x, y
dataset = CustomDataset(seq_len=5, max_len=1000)
for x, y in dataset:
print(x, y)
break
dataloader = DataLoader(dataset, batch_size=4, shuffle=True, num_workers=4)
# collate_fn=custom_collector
for x, y in dataloader:
print(x, y)
break
#
# Let's take a closer look at our specific use case for the many-to-one LSTM architecture. In our scenario, we are feeding the LSTM with a sequence of 5 random numbers, and we anticipate that the model will predict the 6th number in the sequence. While we've chosen a straightforward series of incrementing numbers for this example, the potential applications of this concept extend much further.
# Imagine this sequence being a time-series data of stock prices, weather conditions, or even a series of steps in a logical reasoning question. The ability to predict the next event based on a series of preceding events is a critical aspect in many fields, including finance, meteorology, and artificial intelligence. By training our LSTM model to understand and predict these sequences, we can leverage the many-to-one LSTM architecture to solve complex problems in these areas and beyond.
#
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers):
super().__init__()
self.hidden_size = hidden_size
self.num_layers = num_layers
self.lstm = nn.LSTM(input_size, hidden_size, num_layers, batch_first=True)
self.fc = nn.Linear(hidden_size, 1)
def forward(self, x):
# hidden states not defnined hence the value of h0,c0 == (0,0)
out, (hn, cn) = self.lstm(x)
# as the diagram suggest to take the last output in many to one
# print(out.shape)
# print(hn.shape)
# all batch, last column of seq, all hidden values
out = out[:, -1, :]
out = self.fc(out)
return out
model = RNN(input_size=1, hidden_size=256, num_layers=2)
t = torch.tensor([11, 12, 13, 14, 15]).type(torch.Tensor).view(1, -1, 1)
t.shape
model(t)
# ### Training
loss_function = nn.MSELoss()
learning_rate = 1e-3
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
for e in tqdm(range(50)):
i = 0
for x, y in dataloader:
optimizer.zero_grad()
x = torch.unsqueeze(x, 0).permute(1, 2, 0)
# forward
predictions = model(x)
loss = loss_function(predictions.view(-1), y)
# backward
loss.backward()
# optimization
optimizer.step()
i += 1
if e % 5 == 0:
print(loss.detach().numpy())
input_tensor = torch.tensor([10, 11, 12, 13, 14]).type(torch.Tensor).view(1, -1, 1)
model(input_tensor)
#
# Understanding Many-to-Many Architecture in LSTM
# Another crucial architecture in the world of Long Short-Term Memory (LSTM) networks, a type of Recurrent Neural Network (RNN), is the Many-to-Many architecture. This architecture offers a versatile way of handling a diverse set of problems involving sequential data.
# In a Many-to-Many LSTM architecture, the model processes a sequence of inputs over multiple time steps and generates a sequence of outputs. In this setting, each LSTM cell takes in an input and the previous cell's hidden state at each time step, then produces an output along with its own hidden state that it passes on to the next cell.
# Unlike the Many-to-One LSTM, the Many-to-Many LSTM doesn't wait till the last time step to produce an output. Instead, it generates an output at each time step. This makes Many-to-Many LSTM networks highly useful for tasks such as machine translation, where a sequence of words in one language (input) is translated into a sequence of words in another language (output).
# The Many-to-Many architecture of LSTM opens up a broad array of possibilities, making it a powerful tool in the realms of machine learning and artificial intelligence.
#
class CustomDataset(Dataset):
def __init__(self, seq_len=50, future=5, max_len=1000):
super(CustomDataset).__init__()
self.datalist = np.arange(0, max_len)
self.data, self.targets = self.timeseries(self.datalist, seq_len, future)
def __len__(self):
# this len will decide the index range in getitem
return len(self.targets)
def timeseries(self, data, window, future):
temp = []
targ = []
for i in range(len(data) - window):
temp.append(data[i : i + window])
for i in range(len(data) - window - future):
targ.append(data[i + window : i + window + future])
return np.array(temp), targ
def __getitem__(self, index):
x = torch.tensor(self.data[index]).type(torch.Tensor)
y = torch.tensor(self.targets[index]).type(torch.Tensor)
return x, y
dataset = CustomDataset(seq_len=50, future=5, max_len=1000)
for x, y in dataset:
print(x.shape, y.shape)
break
dataloader = DataLoader(dataset, batch_size=8, shuffle=True, num_workers=4)
# collate_fn=custom_collector
for x, y in dataloader:
print(x.shape, y.shape)
break
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers, future=5):
super().__init__()
self.future = future
self.hidden_size = hidden_size
self.num_layers = num_layers
self.lstm = nn.LSTM(input_size, hidden_size, num_layers, batch_first=True)
self.fc = nn.Linear(hidden_size, future)
def forward(self, x):
# hidden states not defnined hence the value of h0,c0 == (0,0)
out, (hn, cn) = self.lstm(x)
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers, future=5):
super().__init__()
self.future = future
self.hidden_size = hidden_size
self.num_layers = num_layers
self.lstm = nn.LSTM(input_size, hidden_size, num_layers, batch_first=True)
self.fc = nn.Linear(hidden_size, future)
def forward(self, x):
# hidden states not defnined hence the value of h0,c0 == (0,0)
out, (hn, cn) = self.lstm(x)
# as the diagram suggest to take the last output in many to one
# print(out.shape)
# print(hn.shape)
# all batch, last column of seq, all hidden values
out = out[:, -self.future, :]
out = self.fc(out)
return out
model = RNN(input_size=1, hidden_size=256, num_layers=2, future=5)
d = 45
t = torch.tensor(np.arange(d, d + 50)).type(torch.Tensor).view(1, -1, 1)
t.shape
model(t)
loss_function = nn.MSELoss()
learning_rate = 1e-3
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
for e in tqdm(range(50)):
i = 0
avg_loss = []
for x, y in dataloader:
optimizer.zero_grad()
x = torch.unsqueeze(x, 0).permute(1, 2, 0)
# forward
predictions = model(x)
# loss
loss = loss_function(predictions, y)
# backward
loss.backward()
# optimization
optimizer.step()
avg_loss.append(loss.detach().numpy())
i += 1
if e % 2 == 0:
avg_loss = np.array(avg_loss)
print(avg_loss.mean())
#
# After feeding the initial 50 terms of our sequence into the model, we begin to observe some promising results. It appears that the model is successfully learning to recognize the underlying patterns in the sequence.
# The output generated by the model seems to adhere to the logic of the sequence, suggesting that the LSTM architecture is effectively capturing and understanding the sequential dependencies. This ability to discern patterns and extrapolate them is a powerful aspect of LSTM networks, and it's rewarding to see it at work in our model.
# These early results are encouraging, indicating that our model is on the right track. As we continue to refine and train our LSTM, we can expect it to become even more adept at understanding and predicting the sequence.
#
d = random.randint(0, 1000)
t = torch.tensor(np.arange(d, d + 50)).type(torch.Tensor).view(1, -1, 1)
r = model(t).view(-1)
fig = plt.figure(figsize=(16, 4))
plt_x = np.arange(0, t.shape[1] + len(r))
plt_y = np.arange(d, d + 50 + len(r))
plt_xp = np.arange(t.shape[1], t.shape[1] + len(r))
plt_yp = r.detach().numpy()
for i in range(len(r)):
plt.scatter(plt_x, plt_y)
plt.scatter(plt_xp, plt_yp)
#
# Understanding Many-to-Many Sequence Generation with LSTM
# When working with Long Short-Term Memory (LSTM) networks, it's essential to understand how sequence generation is handled, particularly in a Many-to-Many setting. In such an architecture, the output from each LSTM cell can be used as an input to a subsequent feed-forward network to generate a sequence of outputs.
# Let's consider the following block of code as an example:
#
# out, (hn, cn) = self.lstm(x)
# res = torch.zeros((out.shape[0], out.shape[1]))
# for b in range(out.shape[0]):
# feed = out[b, :, :]
# _out = self.fc(feed).view(-1)
# res[b] = _out
#
# In this code, self.lstm(x) applies the LSTM layer to the input x, generating an output out and the final hidden and cell states hn and cn. We then initialize a zeros tensor res of the same size as out to store our results.
# Then, for each sequence in the output out, we feed the sequence through a fully connected layer self.fc(feed) and reshape the output to match our expected dimensions using .view(-1). The result is stored in the corresponding position in res.
# This process exemplifies how a Many-to-Many LSTM network can be used to generate a sequence of outputs, with the LSTM layer and a subsequent feed-forward layer working in tandem to transform a sequence of inputs into a corresponding sequence of outputs.
#
class CustomDataset(Dataset):
def __init__(self, seq_len=50, future=50, max_len=1000):
super(CustomDataset).__init__()
self.datalist = np.arange(0, max_len)
self.data, self.targets = self.timeseries(self.datalist, seq_len, future)
def __len__(self):
# this len will decide the index range in getitem
return len(self.targets)
def timeseries(self, data, window, future):
temp = []
targ = []
for i in range(len(data) - window):
temp.append(data[i : i + window])
for i in range(len(data) - window - future):
targ.append(data[i + future : i + window + future])
return np.array(temp), targ
def __getitem__(self, index):
x = torch.tensor(self.data[index]).type(torch.Tensor)
y = torch.tensor(self.targets[index]).type(torch.Tensor)
return x, y
dataset = CustomDataset(seq_len=50, future=5, max_len=1000)
for x, y in dataset:
print(x.shape, y.shape)
break
x
y
dataloader = DataLoader(dataset, batch_size=8, shuffle=True, num_workers=4)
# collate_fn=custom_collector
class RNN(nn.Module):
def __init__(self, input_size, hidden_size, num_layers, future=5):
super().__init__()
self.future = future
self.hidden_size = hidden_size
self.num_layers = num_layers
self.lstm = nn.LSTM(input_size, hidden_size, num_layers, batch_first=True)
self.fc = nn.Linear(hidden_size, 1)
def forward(self, x):
# hidden states not defnined hence the value of h0,c0 == (0,0)
out, (hn, cn) = self.lstm(x)
# as the diagram suggest to take the last output in many to one
# print(out.shape)
# print(hn.shape)
# all batch, last column of seq, all hidden values
res = torch.zeros((out.shape[0], out.shape[1]))
for b in range(out.shape[0]):
feed = out[b, :, :]
_out = self.fc(feed).view(-1)
res[b] = _out
return res
model = RNN(input_size=1, hidden_size=256, num_layers=2, future=5)
t = torch.tensor(np.arange(d, d + 50)).type(torch.Tensor).view(1, -1, 1)
r = model(t).view(-1)
r
loss_function = nn.MSELoss()
learning_rate = 1e-3
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
for e in tqdm(range(100)):
i = 0
avg_loss = []
for x, y in dataloader:
optimizer.zero_grad()
x = torch.unsqueeze(x, 0).permute(1, 2, 0)
# forward
predictions = model(x)
# loss
loss = loss_function(predictions, y)
# backward
loss.backward()
# optimization
optimizer.step()
avg_loss.append(loss.detach().numpy())
i += 1
if e % 5 == 0:
avg_loss = np.array(avg_loss)
print(avg_loss.mean())
d = random.randint(0, 1000)
t = torch.tensor(np.arange(d, d + 50)).type(torch.Tensor).view(1, -1, 1)
r = model(t).view(-1)
fig = plt.figure(figsize=(16, 4))
plt_x = np.arange(0, t.shape[1])
plt_y = np.arange(d, d + 50)
plt_xp = np.arange(5, t.shape[1] + 5)
plt_yp = r.detach().numpy()
for i in range(len(r)):
plt.scatter(plt_x, plt_y, label="real")
plt.scatter(plt_xp, plt_yp, label="predicted")
plt.show()
#
# # ** Transformers **
# Transformers, a breakthrough in the field of natural language processing, also adopt various types of input-output architectures, including the Many-to-Many setup. In this context, Transformers bring a unique approach to the table, contrasting with the methods used in traditional Recurrent Neural Networks (RNNs) such as LSTM.
# In a Many-to-Many Transformer architecture, the model accepts a sequence of inputs and returns a sequence of outputs. However, unlike RNNs, which process sequences in a time-stepped manner, Transformers process all inputs simultaneously. This is made possible by the attention mechanism, which allows the model to focus on different parts of the input sequence for each output, essentially creating a 'shortcut' between each input and output.
# This architecture is especially useful in tasks like machine translation, where the model needs to understand the context of the whole sentence to accurately translate it. Similarly, it can be used in tasks like text summarization or question answering, where understanding the entire context at once can lead to better results.
# The Many-to-Many architecture in Transformers, combined with their attention mechanism, offers an innovative approach to tackling sequential tasks, making Transformers a powerful tool in the field of machine learning and artificial intelligence.
# 
class CustomDataset(Dataset):
def __init__(self, seq_len=50, future=50, max_len=1000):
super(CustomDataset).__init__()
self.vocab = {"SOS": 1001, "EOS": 1002}
self.datalist = np.arange(0, max_len)
self.data, self.targets = self.timeseries(self.datalist, seq_len, future)
def __len__(self):
# this len will decide the index range in getitem
return len(self.targets)
def timeseries(self, data, window, future):
temp = []
targ = []
for i in range(len(data) - window):
temp.append(data[i : i + window])
for i in range(len(data) - window - future):
targ.append(data[i + future : i + window + future])
return np.array(temp), targ
def __getitem__(self, index):
x = torch.tensor(self.data[index]).type(torch.Tensor)
x = torch.cat(
(torch.tensor([self.vocab["SOS"]]), x, torch.tensor([self.vocab["EOS"]]))
).type(torch.LongTensor)
y = torch.tensor(self.targets[index]).type(torch.Tensor)
y = torch.cat(
(torch.tensor([self.vocab["SOS"]]), y, torch.tensor([self.vocab["EOS"]]))
).type(torch.LongTensor)
return x, y
dataset = CustomDataset(seq_len=48, future=5, max_len=1000)
for x, y in dataset:
print(x)
print(y)
break
dataloader = DataLoader(dataset, batch_size=8, shuffle=True, num_workers=4)
# collate_fn=custom_collector
for x, y in dataloader:
print(x.shape)
print(y.shape)
break
#
# The Power of Masking and Efficiency in Transformers
# One of the remarkable features of Transformers is their use of masking during the training process. Masking is an essential aspect of the Transformer's architecture that prevents the model from seeing future tokens in the input sequence during training, thereby preserving the sequential nature of the language.
# In a task such as language translation, where the input sequence is fed into the model all at once, it's crucial that the prediction for each word doesn't rely on words that come after it in the sequence. This is achieved by applying a mask to the input that effectively hides future words from the model during the training phase.
# Not only does masking maintain the sequential integrity of the language, but it also allows Transformers to train more efficiently than their RNN counterparts, like LSTM. Unlike RNNs, which process sequences step-by-step and thus require longer training times for long sequences, Transformers can process all the tokens in the sequence simultaneously, thanks to their attention mechanism. This parallel processing significantly speeds up the training process and allows the model to handle longer sequences more effectively.
# Thus, through the use of masking and their unique architecture, Transformers manage to overcome some of the limitations of traditional RNNs, offering a more efficient and effective approach to sequence-based tasks in machine learning and artificial intelligence.
#
class Transformer(nn.Module):
def __init__(self, num_tokens, dim_model, num_heads, num_layers, input_seq):
super().__init__()
self.input_seq = input_seq
self.num_layers = num_layers
self.embedding = nn.Embedding(num_tokens, dim_model)
self.transformer = nn.Transformer(
d_model=dim_model,
nhead=num_heads,
num_encoder_layers=3,
num_decoder_layers=3,
dim_feedforward=256,
batch_first=True,
)
self.fc = nn.Linear(dim_model, num_tokens)
def forward(self, src, tgt, tf=True):
mask = self.get_mask(tgt.shape[1], teacher_force=tf)
src = self.embedding(src)
tgt = self.embedding(tgt)
out = self.transformer(src, tgt, tgt_mask=mask)
feed = self.fc(out)
feed = torch.squeeze(feed, 2)
return feed
def get_mask(self, size, teacher_force=True):
if teacher_force:
mask = torch.tril(torch.ones(size, size) == 1) # Lower triangular matrix
mask = mask.float()
mask = mask.masked_fill(mask == 0, float("-inf")) # Convert zeros to -inf
mask = mask.masked_fill(mask == 1, float(0.0)) # Convert ones to 0
# EX for size=5:
# [[0., -inf, -inf, -inf, -inf],
# [0., 0., -inf, -inf, -inf],
# [0., 0., 0., -inf, -inf],
# [0., 0., 0., 0., -inf],
# [0., 0., 0., 0., 0.]]
return mask
else:
mask = torch.tril(torch.zeros(size, size) == 1) # Lower triangular matrix
mask = mask.float()
mask = mask.masked_fill(mask == 0, float("-inf")) # Convert zeros to -inf
mask = mask.masked_fill(mask == 1, float(0.0)) # Convert ones to 0
return mask
model = Transformer(
num_tokens=1000 + 3, dim_model=32, num_heads=2, num_layers=2, input_seq=50
)
x.shape, y.shape
model(x, y).shape
t = model(x, y)
t.shape
t.permute(0, 2, 1).shape
loss_function = nn.CrossEntropyLoss()
learning_rate = 1e-3
optimizer = optim.Adam(model.parameters(), lr=learning_rate)
for e in tqdm(range(25)):
i = 0
avg_loss = []
for x, y in dataloader:
optimizer.zero_grad()
# one step behind input and output // Like language modeling
y_input = y[:, :-1] # from starting to -1 position
y_expected = y[:, 1:] # from 1st position to last
# this is done so that in prediction we see a start of token
# forward
predictions = model(x, y_input)
pred = predictions.permute(0, 2, 1)
# loss
loss = loss_function(pred, y_expected)
# backward
loss.backward()
# optimization
optimizer.step()
avg_loss.append(loss.detach().numpy())
i += 1
if e % 5 == 0:
avg_loss = np.array(avg_loss)
print(avg_loss.mean())
torch.squeeze(predictions.topk(1).indices, 2)
y_expected
torch.argmax(pred, dim=1)
#
# The Role of SOS and EOS Tokens in Transformers
# In the domain of natural language processing, particularly when working with Transformer models, special tokens like Start of Sentence (SOS) and End of Sentence (EOS) play a crucial role. These tokens provide valuable cues about the boundaries of sentences, facilitating the model's understanding of language structure.
# The SOS token is added at the beginning of each sentence, marking its start. Similarly, the EOS token is appended at the end of each sentence to indicate its conclusion. These tokens serve as consistent markers that help the model identify and process sentences as distinct units within larger bodies of text.
# Furthermore, in the context of sequence generation tasks, these tokens play an essential role in determining when to begin and end the generation process. For example, during text generation, an EOS token indicates to the model that it should stop generating further tokens.
# Therefore, SOS and EOS tokens are more than just markers; they're integral components in the design and functioning of Transformer models, contributing significantly to their ability to effectively understand and generate human language.
#
def predict(
model, input_sequence, max_length=50, SOS_token=1000 + 1, EOS_token=1000 + 2
):
model.eval()
input_sequence = torch.tensor(input_sequence)
input_sequence = torch.cat(
(torch.tensor([SOS_token]), input_sequence, torch.tensor([EOS_token]))
).type(torch.LongTensor)
input_sequence = torch.unsqueeze(input_sequence, 0)
y_input = torch.tensor([1001], dtype=torch.long)
y_input = torch.unsqueeze(y_input, 0)
for _ in range(max_length):
predictions = model(input_sequence, y_input)
top = predictions.topk(1).indices
top = torch.squeeze(top, 2)
next_item = torch.unsqueeze(top[:, -1], 0)
y_input = torch.cat((y_input, next_item), dim=1)
mask = model.get_mask(y_input.shape[1])
if next_item == EOS_token:
break
return y_input.view(-1).tolist()
d = random.randint(0, 900)
t = torch.tensor(np.arange(d, d + 48)).type(torch.Tensor)
input_sequence = t
print(t)
r = predict(model, input_sequence)
print(r)
fig = plt.figure(figsize=(16, 4))
plt_x = np.arange(0, t.shape[0])
plt_y = t
plt_xp = np.arange(5, t.shape[0] + 5)
plt_yp = r[1:-2]
plt.scatter(plt_x, plt_y, s=14, color="r", label="real")
plt.scatter(plt_xp, plt_yp, s=7, color="b", label="predicted")
plt.legend()
plt.show()
| false | 0 | 8,468 | 34 | 8,468 | 8,468 |
||
129874148
|
# # HANDWRITTEN DIGIT PREDICTION - CLASSIFICATION ANALYSIS
# Objective - HANDWRITTEN DIGIT PREDICTION is the process to provide the ability to machines to predict human handwritten digits.
# # IMPORTING A LIBRARY
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# # IMPORTING DATA
from sklearn.datasets import load_digits
ld = load_digits()
_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))
for ax, image, label in zip(axes, ld.images, ld.target):
ax.set_axis_off()
ax.imshow(image, cmap=plt.cm.gray_r, interpolation="nearest")
ax.set_title("Training: %i" % label)
# # PREPROCESSING OF DATA
ld.images.shape
ld.images[0]
ld.images[0].shape
n_samples = len(ld.images)
data = ld.images.reshape(n_samples, -1)
data[0]
data[0].shape
data.shape
# # SCALING IMAGE DATA
data.min()
data.max()
data = data / 16
data.min()
data.max()
data[0]
# # SPLITING OF DATA INTO TRAINING AND TESTING DATA
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
data, ld.target, test_size=0.3, random_state=2529
)
x_train.shape, x_test.shape, y_train.shape, y_test.shape
# # RANDOM FOREST MODEL CLASSIFICATION PROBLEM
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier()
rf.fit(x_train, y_train)
# # PREDICTION OF TEST DATA
y_pred = rf.predict(x_test)
y_pred
# # CHECKING MODEL ACCURACY
from sklearn.metrics import confusion_matrix, classification_report
confusion_matrix(y_test, y_pred)
print(classification_report(y_test, y_pred))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/874/129874148.ipynb
| null | null |
[{"Id": 129874148, "ScriptId": 38629340, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14298021, "CreationDate": "05/17/2023 05:48:24", "VersionNumber": 1.0, "Title": "notebookba1cb299ee", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 82.0, "LinesInsertedFromPrevious": 82.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # HANDWRITTEN DIGIT PREDICTION - CLASSIFICATION ANALYSIS
# Objective - HANDWRITTEN DIGIT PREDICTION is the process to provide the ability to machines to predict human handwritten digits.
# # IMPORTING A LIBRARY
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# # IMPORTING DATA
from sklearn.datasets import load_digits
ld = load_digits()
_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))
for ax, image, label in zip(axes, ld.images, ld.target):
ax.set_axis_off()
ax.imshow(image, cmap=plt.cm.gray_r, interpolation="nearest")
ax.set_title("Training: %i" % label)
# # PREPROCESSING OF DATA
ld.images.shape
ld.images[0]
ld.images[0].shape
n_samples = len(ld.images)
data = ld.images.reshape(n_samples, -1)
data[0]
data[0].shape
data.shape
# # SCALING IMAGE DATA
data.min()
data.max()
data = data / 16
data.min()
data.max()
data[0]
# # SPLITING OF DATA INTO TRAINING AND TESTING DATA
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
data, ld.target, test_size=0.3, random_state=2529
)
x_train.shape, x_test.shape, y_train.shape, y_test.shape
# # RANDOM FOREST MODEL CLASSIFICATION PROBLEM
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier()
rf.fit(x_train, y_train)
# # PREDICTION OF TEST DATA
y_pred = rf.predict(x_test)
y_pred
# # CHECKING MODEL ACCURACY
from sklearn.metrics import confusion_matrix, classification_report
confusion_matrix(y_test, y_pred)
print(classification_report(y_test, y_pred))
| false | 0 | 533 | 0 | 533 | 533 |
||
129554309
|
# ## Importing relevant packages
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
import copy
from sklearn.model_selection import (
train_test_split,
StratifiedKFold,
RandomizedSearchCV,
)
from sklearn.preprocessing import OneHotEncoder
from sklearn.impute import SimpleImputer
import xgboost as xgb
import catboost
from sklearn.ensemble import RandomForestClassifier
from sklearn import metrics
# ## Importing the data
# Load the data
train = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/train.csv")
test = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/test.csv")
greeks = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/greeks.csv")
sample_submission = pd.read_csv(
"/kaggle/input/icr-identify-age-related-conditions/sample_submission.csv"
)
# ## Descriptive and Exploratory Data Analysis
# First of all, lets take a look at the data
train.head()
train.info()
# The majority of our columns are float64. We have one column ('EJ') with the 'object' type. It is interesting to verify why this column have this type.
# Columns like 'BQ' and 'EL' have more than 50 lines with null values. We will need to take a look at these to verify if some kind of imputation is needed.
# ### Are our train dataset balanced or disbalanced?
train["Class"].value_counts()
# Our dataset is highly disbalanced. The number of negative cases is nearly 5 times higher than the number of positive examples.
# ## Data pre-processing
# ### Spliting the train dataset into train and validation
# First, let split the dataset into train and validation
# As our dataset is disbalanced, we will look-forward to maintain the same variable frequencies
X = copy.deepcopy(train.drop("Id", axis=1))
y = X.pop("Class")
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, train_size=0.8, test_size=0.2, stratify=y, random_state=42
)
# ### Pre-processing the column with the 'object' type
# First, lets see how many different values the column with 'object' type have
train["EJ"].value_counts()
# There are two distinct values in this column. In this case we can simply replace the values for 1 and 0
# One-Hot Encoding the object column
X_train["EJ"] = X_train["EJ"].replace({"A": 1, "B": 0})
X_valid["EJ"] = X_valid["EJ"].replace({"A": 1, "B": 0})
X_train["EJ"].value_counts()
# ## Handling missing values
# Identifying column with nulls
cols_with_missings = [col for col in X_train.columns if X_train[col].isnull().any()]
print("Columns with nulls: ", cols_with_missings)
# Imputing using SimpleImputer
my_imputer = SimpleImputer(strategy="mean")
imputed_X_train = pd.DataFrame(my_imputer.fit_transform(X_train))
imputed_X_valid = pd.DataFrame(my_imputer.transform(X_valid))
imputed_X_train.columns = X_train.columns
imputed_X_valid.columns = X_valid.columns
X_train = imputed_X_train
X_valid = imputed_X_valid
# Verifying if there is any null
cols_with_missings = [col for col in X_train.columns if X_train[col].isnull().any()]
print("Columns with nulls: ", cols_with_missings)
# ## Selecting the model type
plt.figure(figsize=(12, 8))
# Listing all the models we will test
models = [RandomForestClassifier(), xgb.XGBClassifier(), catboost.CatBoostClassifier()]
# Train and evaluate each model
for model in models:
model.fit(X_train, y_train)
y_pred = model.predict(X_valid)
y_pred_proba = model.predict_proba(X_valid)[::, 1]
name = model.__class__.__name__
precision = metrics.precision_score(y_valid, y_pred)
recall = metrics.recall_score(y_valid, y_pred)
log_loss = metrics.log_loss(y_valid, y_pred_proba)
fpr, tpr, _ = metrics.roc_curve(y_valid, y_pred_proba)
auc = metrics.roc_auc_score(y_valid, y_pred_proba)
# Print results and plot ROC curve
print("-" * 30)
print(name)
print("Precision: ", precision)
print("Recall: ", recall)
print("Log loss: ", log_loss)
plt.plot(fpr, tpr, label=name + ", auc=" + str(auc))
plt.plot([0, 1], [0, 1], color="black")
plt.legend(loc=4)
# Let's follow with the CatBoost classifier!
model = catboost.CatBoostClassifier()
model.fit(X_train, y_train, verbose=False)
y_pred_proba = model.predict_proba(X_valid)
log_loss = metrics.log_loss(y_valid, y_pred_proba)
print("Log loss: ", log_loss)
# Lets do a param_grid to try to improve the log loss
# ## Generating the final predictions
# Pre-processing the test dataset
X_test = copy.deepcopy(test)
X_test = X_test.drop("Id", axis=1)
# Applying the numbers to the object column
X_test["EJ"] = test["EJ"].replace({"A": 1, "B": 0})
# Imputing features with null values
cols_with_missings = [col for col in X_test.columns if X_test[col].isnull().any()]
print("Columns with nulls: ", cols_with_missings)
# Imputing using SimpleImputer
my_imputer = SimpleImputer(strategy="mean")
imputed_test = pd.DataFrame(my_imputer.fit_transform(X_test))
imputed_test.columns = X_test.columns
X_test = imputed_test
sample_submission.head()
pred_probs = model.predict_proba(X_test)
data = {"Id": test.Id, "class_0": pred_probs[:, 0], "class_1": pred_probs[:, 1]}
df = pd.DataFrame(data)
print(df.head())
df.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/554/129554309.ipynb
| null | null |
[{"Id": 129554309, "ScriptId": 38515540, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12983366, "CreationDate": "05/14/2023 19:28:25", "VersionNumber": 1.0, "Title": "[ICR IARC] - EDA, PP & Cat Boost Classifier", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 197.0, "LinesInsertedFromPrevious": 197.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## Importing relevant packages
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
import copy
from sklearn.model_selection import (
train_test_split,
StratifiedKFold,
RandomizedSearchCV,
)
from sklearn.preprocessing import OneHotEncoder
from sklearn.impute import SimpleImputer
import xgboost as xgb
import catboost
from sklearn.ensemble import RandomForestClassifier
from sklearn import metrics
# ## Importing the data
# Load the data
train = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/train.csv")
test = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/test.csv")
greeks = pd.read_csv("/kaggle/input/icr-identify-age-related-conditions/greeks.csv")
sample_submission = pd.read_csv(
"/kaggle/input/icr-identify-age-related-conditions/sample_submission.csv"
)
# ## Descriptive and Exploratory Data Analysis
# First of all, lets take a look at the data
train.head()
train.info()
# The majority of our columns are float64. We have one column ('EJ') with the 'object' type. It is interesting to verify why this column have this type.
# Columns like 'BQ' and 'EL' have more than 50 lines with null values. We will need to take a look at these to verify if some kind of imputation is needed.
# ### Are our train dataset balanced or disbalanced?
train["Class"].value_counts()
# Our dataset is highly disbalanced. The number of negative cases is nearly 5 times higher than the number of positive examples.
# ## Data pre-processing
# ### Spliting the train dataset into train and validation
# First, let split the dataset into train and validation
# As our dataset is disbalanced, we will look-forward to maintain the same variable frequencies
X = copy.deepcopy(train.drop("Id", axis=1))
y = X.pop("Class")
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, train_size=0.8, test_size=0.2, stratify=y, random_state=42
)
# ### Pre-processing the column with the 'object' type
# First, lets see how many different values the column with 'object' type have
train["EJ"].value_counts()
# There are two distinct values in this column. In this case we can simply replace the values for 1 and 0
# One-Hot Encoding the object column
X_train["EJ"] = X_train["EJ"].replace({"A": 1, "B": 0})
X_valid["EJ"] = X_valid["EJ"].replace({"A": 1, "B": 0})
X_train["EJ"].value_counts()
# ## Handling missing values
# Identifying column with nulls
cols_with_missings = [col for col in X_train.columns if X_train[col].isnull().any()]
print("Columns with nulls: ", cols_with_missings)
# Imputing using SimpleImputer
my_imputer = SimpleImputer(strategy="mean")
imputed_X_train = pd.DataFrame(my_imputer.fit_transform(X_train))
imputed_X_valid = pd.DataFrame(my_imputer.transform(X_valid))
imputed_X_train.columns = X_train.columns
imputed_X_valid.columns = X_valid.columns
X_train = imputed_X_train
X_valid = imputed_X_valid
# Verifying if there is any null
cols_with_missings = [col for col in X_train.columns if X_train[col].isnull().any()]
print("Columns with nulls: ", cols_with_missings)
# ## Selecting the model type
plt.figure(figsize=(12, 8))
# Listing all the models we will test
models = [RandomForestClassifier(), xgb.XGBClassifier(), catboost.CatBoostClassifier()]
# Train and evaluate each model
for model in models:
model.fit(X_train, y_train)
y_pred = model.predict(X_valid)
y_pred_proba = model.predict_proba(X_valid)[::, 1]
name = model.__class__.__name__
precision = metrics.precision_score(y_valid, y_pred)
recall = metrics.recall_score(y_valid, y_pred)
log_loss = metrics.log_loss(y_valid, y_pred_proba)
fpr, tpr, _ = metrics.roc_curve(y_valid, y_pred_proba)
auc = metrics.roc_auc_score(y_valid, y_pred_proba)
# Print results and plot ROC curve
print("-" * 30)
print(name)
print("Precision: ", precision)
print("Recall: ", recall)
print("Log loss: ", log_loss)
plt.plot(fpr, tpr, label=name + ", auc=" + str(auc))
plt.plot([0, 1], [0, 1], color="black")
plt.legend(loc=4)
# Let's follow with the CatBoost classifier!
model = catboost.CatBoostClassifier()
model.fit(X_train, y_train, verbose=False)
y_pred_proba = model.predict_proba(X_valid)
log_loss = metrics.log_loss(y_valid, y_pred_proba)
print("Log loss: ", log_loss)
# Lets do a param_grid to try to improve the log loss
# ## Generating the final predictions
# Pre-processing the test dataset
X_test = copy.deepcopy(test)
X_test = X_test.drop("Id", axis=1)
# Applying the numbers to the object column
X_test["EJ"] = test["EJ"].replace({"A": 1, "B": 0})
# Imputing features with null values
cols_with_missings = [col for col in X_test.columns if X_test[col].isnull().any()]
print("Columns with nulls: ", cols_with_missings)
# Imputing using SimpleImputer
my_imputer = SimpleImputer(strategy="mean")
imputed_test = pd.DataFrame(my_imputer.fit_transform(X_test))
imputed_test.columns = X_test.columns
X_test = imputed_test
sample_submission.head()
pred_probs = model.predict_proba(X_test)
data = {"Id": test.Id, "class_0": pred_probs[:, 0], "class_1": pred_probs[:, 1]}
df = pd.DataFrame(data)
print(df.head())
df.to_csv("submission.csv", index=False)
| false | 0 | 1,655 | 0 | 1,655 | 1,655 |
||
129523726
|
import numpy as np
import scipy
print("NumPy version:", np.__version__)
print("SciPy version:", scipy.__version__)
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
from sklearn.linear_model import LogisticRegression
train = pd.read_csv(
"/kaggle/input/ipba14-grade-ml-case-study-classification-17042023/train.csv"
)
test = pd.read_csv(
"/kaggle/input/ipba14-grade-ml-case-study-classification-17042023/test.csv"
)
submission = pd.read_csv(
"/kaggle/input/ipba14-grade-ml-case-study-classification-17042023/sample_submission.csv"
)
train.head()
train.info()
train["Attrition"].value_counts().plot.bar()
y = train["Attrition"]
X = train[["Age", "DistanceFromHome", "TotalWorkingYears"]]
lr = LogisticRegression(class_weight="balanced")
lr.fit(X, y)
y_train_predictions = lr.predict(X)
y_train_predictions.sum()
train["Attrition_Predictions"] = y_train_predictions
train[
[
"id",
"Age",
"DistanceFromHome",
"TotalWorkingYears",
"Attrition",
"Attrition_Predictions",
]
]
train.to_csv("attrition_data_base_with_predictions.csv")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/523/129523726.ipynb
| null | null |
[{"Id": 129523726, "ScriptId": 38389713, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13254461, "CreationDate": "05/14/2023 14:17:38", "VersionNumber": 2.0, "Title": "ML Case Study - Aniket Gavandar", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 43.0, "LinesInsertedFromPrevious": 36.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 7.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import scipy
print("NumPy version:", np.__version__)
print("SciPy version:", scipy.__version__)
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
from sklearn.linear_model import LogisticRegression
train = pd.read_csv(
"/kaggle/input/ipba14-grade-ml-case-study-classification-17042023/train.csv"
)
test = pd.read_csv(
"/kaggle/input/ipba14-grade-ml-case-study-classification-17042023/test.csv"
)
submission = pd.read_csv(
"/kaggle/input/ipba14-grade-ml-case-study-classification-17042023/sample_submission.csv"
)
train.head()
train.info()
train["Attrition"].value_counts().plot.bar()
y = train["Attrition"]
X = train[["Age", "DistanceFromHome", "TotalWorkingYears"]]
lr = LogisticRegression(class_weight="balanced")
lr.fit(X, y)
y_train_predictions = lr.predict(X)
y_train_predictions.sum()
train["Attrition_Predictions"] = y_train_predictions
train[
[
"id",
"Age",
"DistanceFromHome",
"TotalWorkingYears",
"Attrition",
"Attrition_Predictions",
]
]
train.to_csv("attrition_data_base_with_predictions.csv")
| false | 0 | 431 | 0 | 431 | 431 |
||
129979019
|
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
import pandas as pd
import pandas as pd
pd.options.mode.chained_assignment = None # default='warn'
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
from sklearn.preprocessing import StandardScaler
from sklearn.feature_selection import SelectKBest, f_regression
import sys
sys.path.append("/kaggle/input/amp-parkinsons-disease-progression-prediction")
import amp_pd_peptide_310
a_proteins = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_proteins.csv"
)
a_peptides = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_peptides.csv"
)
a_clinical = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_clinical_data.csv"
)
test = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test.csv"
)
test_peptides = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test_peptides.csv"
)
test_proteins = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test_proteins.csv"
)
sample_submission = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/sample_submission.csv"
)
test.head()
test_peptides.head()
test_proteins.head()
sample_submission.head()
# smape: symmetric mean absolute percentage error, A: Actual, P: Predicted
def smape(A, P):
return 100 / len(A) * np.sum(2 * np.abs(P - A) / (np.abs(A) + np.abs(P)))
a_proteins.shape
a_proteins.head()
a_proteins.describe()
a_peptides.head()
pivoted_a_proteins = a_proteins.pivot(index="visit_id", columns="UniProt", values="NPX")
pivoted_a_proteins.shape
pivoted_a_proteins.describe()
print(pivoted_a_proteins)
nan_values = pivoted_a_proteins[pivoted_a_proteins.isna().any(axis=1)]
print(nan_values)
pivoted_a_proteins.head()
a_clinical.shape
a_clinical.head()
a_clinical.info()
a_clinical.groupby(["upd23b_clinical_state_on_medication"])[
"upd23b_clinical_state_on_medication"
].count()
a_clinical.describe()
a1_clinical = a_clinical.drop(["upd23b_clinical_state_on_medication"], axis=1)
a1_clinical.head()
pivoted_a_peptides = a_peptides.pivot(
index="visit_id", columns="Peptide", values="PeptideAbundance"
)
pivoted_a_peptides.shape
pivoted_a_proteins_a_peptides = pivoted_a_proteins.merge(
pivoted_a_peptides, on=["visit_id"], how="inner"
)
pivoted_a_proteins_a_peptides.shape
pivoted_a_proteins_a_peptides.head()
pd.set_option("display.max_rows", 10)
print(pivoted_a_proteins_a_peptides.isna().sum())
a_merged = a1_clinical.merge(
pivoted_a_proteins_a_peptides, on=["visit_id"], how="inner"
)
a_merged.head()
a_merged.shape
# df_tr3 = df_tr3[(df_tr3.visit_month == 0)]
# n=0
# for col in df_tr3:
# # print(colname[1])
# n=n+1
# if n>7:
# # Separate the dataset for updrs_1.
# df_updrs1 = df_tr3[['updrs_1', col]]
# df_updrs1 = df_updrs1.dropna()
# print(len(df_updrs1.index))
# df_tr3 = df_tr3[(df_tr3.visit_month == 0) & (df_tr3.updrs_1 > 0.1)]
a_merged_vm0 = a_merged[(a_merged.visit_month == 0)]
a_merged_vm0.shape
a_merged_vm0.head(10)
updrs_1_median = a_merged_vm0["updrs_1"].median()
print(updrs_1_median)
n = 0
for col in a_merged_vm0:
# print(colname[1])
n = n + 1
if n > 7:
a_merged_vm0[col].fillna(a_merged_vm0[col].median(), inplace=True)
a_merged_vm0[col] = 1 / a_merged_vm0[col] ** 2
a_merged_vm0.head(10)
a_updrs_1_vm0 = a_merged_vm0[["updrs_1"] + list(a_merged_vm0.columns[7:])]
a_updrs_1_vm0 = a_updrs_1_vm0.dropna()
# Separate the independent variables (y) and the dependent variable (X).
X_a_updrs_1 = a_updrs_1_vm0.iloc[:, 1:]
y_a_updrs_1 = a_updrs_1_vm0.iloc[:, 0]
# y_a_updrs_1 = np.array(a_updrs_1_vm0['updrs_1']).reshape(-1, 1)
# Select the top k features with the highest F-values.
selector1 = SelectKBest(f_regression, k=3)
X_a_updrs_1 = selector1.fit_transform(X_a_updrs_1, y_a_updrs_1)
# Split the dataset into training and validation sets.
(
X_train_updrs_1_vm0,
X_val_updrs_1_vm0,
y_train_updrs_1_vm0,
y_val_updrs_1_vm0,
) = train_test_split(X_a_updrs_1, y_a_updrs_1, test_size=0.2, random_state=42)
# Standardize the variables.
scaler1 = StandardScaler()
X_train_updrs_1_vm0 = scaler1.fit_transform(X_train_updrs_1_vm0)
X_val_updrs_1_vm0 = scaler1.transform(X_val_updrs_1_vm0)
# y = np.array(df_binary['Temp']).reshape(-1, 1)
# y_train_updrs_1_vm0 = scaler1.fit_transform(y_train_updrs_1_vm0)
# y_val_updrs_1_vm0 = scaler1.transform(y_val_updrs_1_vm0)
# Fit a linear regression model on the training set.
model_updrs_1_vm0 = LinearRegression()
model_updrs_1_vm0.fit(X_train_updrs_1_vm0, y_train_updrs_1_vm0)
# Predict the values of the dependent variable (y) on the testing set.
y_pred_updrs_1_vm0 = model_updrs_1_vm0.predict(X_val_updrs_1_vm0)
y_pred_updrs_1_vm0 = np.where(y_pred_updrs_1_vm0 < 0, 0, y_pred_updrs_1_vm0)
# Evaluate the performance of the model.
mse_updrs_1_vm0 = mean_squared_error(y_val_updrs_1_vm0, y_pred_updrs_1_vm0)
mae_updrs_1_vm0 = mean_absolute_error(y_val_updrs_1_vm0, y_pred_updrs_1_vm0)
r2_updrs_1_vm0 = r2_score(y_val_updrs_1_vm0, y_pred_updrs_1_vm0)
smape_updrs_1_vm0 = smape(y_val_updrs_1_vm0, y_pred_updrs_1_vm0)
# print("mse_updrs1:", mse_updrs_1_vm0)
# print("mae_updrs1:", mae_updrs_1_vm0)
print("r2_updrs_1_vm0:", r2_updrs_1_vm0)
print("smape_updrs_1_vm0:", smape_updrs_1_vm0)
# Submission
a_proteins = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_proteins.csv"
)
a_peptides = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_peptides.csv"
)
a_clinical = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_clinical_data.csv"
)
# def get_predictions(test, test_peptides, test_proteins, sample_submission):
a_merged_proteins_peptides = pd.merge(
a_proteins, a_peptides, on=["visit_id", "visit_month", "patient_id", "UniProt"]
)
a_merged = pd.merge(
a_merged_proteins_peptides, a_clinical, on=["visit_id", "visit_month", "patient_id"]
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/979/129979019.ipynb
| null | null |
[{"Id": 129979019, "ScriptId": 38443859, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14946652, "CreationDate": "05/17/2023 22:09:48", "VersionNumber": 7.0, "Title": "GMnb-AMP-Parkinson-Progression-Competition", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 187.0, "LinesInsertedFromPrevious": 20.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 167.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
import pandas as pd
import pandas as pd
pd.options.mode.chained_assignment = None # default='warn'
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
from sklearn.preprocessing import StandardScaler
from sklearn.feature_selection import SelectKBest, f_regression
import sys
sys.path.append("/kaggle/input/amp-parkinsons-disease-progression-prediction")
import amp_pd_peptide_310
a_proteins = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_proteins.csv"
)
a_peptides = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_peptides.csv"
)
a_clinical = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_clinical_data.csv"
)
test = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test.csv"
)
test_peptides = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test_peptides.csv"
)
test_proteins = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test_proteins.csv"
)
sample_submission = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/sample_submission.csv"
)
test.head()
test_peptides.head()
test_proteins.head()
sample_submission.head()
# smape: symmetric mean absolute percentage error, A: Actual, P: Predicted
def smape(A, P):
return 100 / len(A) * np.sum(2 * np.abs(P - A) / (np.abs(A) + np.abs(P)))
a_proteins.shape
a_proteins.head()
a_proteins.describe()
a_peptides.head()
pivoted_a_proteins = a_proteins.pivot(index="visit_id", columns="UniProt", values="NPX")
pivoted_a_proteins.shape
pivoted_a_proteins.describe()
print(pivoted_a_proteins)
nan_values = pivoted_a_proteins[pivoted_a_proteins.isna().any(axis=1)]
print(nan_values)
pivoted_a_proteins.head()
a_clinical.shape
a_clinical.head()
a_clinical.info()
a_clinical.groupby(["upd23b_clinical_state_on_medication"])[
"upd23b_clinical_state_on_medication"
].count()
a_clinical.describe()
a1_clinical = a_clinical.drop(["upd23b_clinical_state_on_medication"], axis=1)
a1_clinical.head()
pivoted_a_peptides = a_peptides.pivot(
index="visit_id", columns="Peptide", values="PeptideAbundance"
)
pivoted_a_peptides.shape
pivoted_a_proteins_a_peptides = pivoted_a_proteins.merge(
pivoted_a_peptides, on=["visit_id"], how="inner"
)
pivoted_a_proteins_a_peptides.shape
pivoted_a_proteins_a_peptides.head()
pd.set_option("display.max_rows", 10)
print(pivoted_a_proteins_a_peptides.isna().sum())
a_merged = a1_clinical.merge(
pivoted_a_proteins_a_peptides, on=["visit_id"], how="inner"
)
a_merged.head()
a_merged.shape
# df_tr3 = df_tr3[(df_tr3.visit_month == 0)]
# n=0
# for col in df_tr3:
# # print(colname[1])
# n=n+1
# if n>7:
# # Separate the dataset for updrs_1.
# df_updrs1 = df_tr3[['updrs_1', col]]
# df_updrs1 = df_updrs1.dropna()
# print(len(df_updrs1.index))
# df_tr3 = df_tr3[(df_tr3.visit_month == 0) & (df_tr3.updrs_1 > 0.1)]
a_merged_vm0 = a_merged[(a_merged.visit_month == 0)]
a_merged_vm0.shape
a_merged_vm0.head(10)
updrs_1_median = a_merged_vm0["updrs_1"].median()
print(updrs_1_median)
n = 0
for col in a_merged_vm0:
# print(colname[1])
n = n + 1
if n > 7:
a_merged_vm0[col].fillna(a_merged_vm0[col].median(), inplace=True)
a_merged_vm0[col] = 1 / a_merged_vm0[col] ** 2
a_merged_vm0.head(10)
a_updrs_1_vm0 = a_merged_vm0[["updrs_1"] + list(a_merged_vm0.columns[7:])]
a_updrs_1_vm0 = a_updrs_1_vm0.dropna()
# Separate the independent variables (y) and the dependent variable (X).
X_a_updrs_1 = a_updrs_1_vm0.iloc[:, 1:]
y_a_updrs_1 = a_updrs_1_vm0.iloc[:, 0]
# y_a_updrs_1 = np.array(a_updrs_1_vm0['updrs_1']).reshape(-1, 1)
# Select the top k features with the highest F-values.
selector1 = SelectKBest(f_regression, k=3)
X_a_updrs_1 = selector1.fit_transform(X_a_updrs_1, y_a_updrs_1)
# Split the dataset into training and validation sets.
(
X_train_updrs_1_vm0,
X_val_updrs_1_vm0,
y_train_updrs_1_vm0,
y_val_updrs_1_vm0,
) = train_test_split(X_a_updrs_1, y_a_updrs_1, test_size=0.2, random_state=42)
# Standardize the variables.
scaler1 = StandardScaler()
X_train_updrs_1_vm0 = scaler1.fit_transform(X_train_updrs_1_vm0)
X_val_updrs_1_vm0 = scaler1.transform(X_val_updrs_1_vm0)
# y = np.array(df_binary['Temp']).reshape(-1, 1)
# y_train_updrs_1_vm0 = scaler1.fit_transform(y_train_updrs_1_vm0)
# y_val_updrs_1_vm0 = scaler1.transform(y_val_updrs_1_vm0)
# Fit a linear regression model on the training set.
model_updrs_1_vm0 = LinearRegression()
model_updrs_1_vm0.fit(X_train_updrs_1_vm0, y_train_updrs_1_vm0)
# Predict the values of the dependent variable (y) on the testing set.
y_pred_updrs_1_vm0 = model_updrs_1_vm0.predict(X_val_updrs_1_vm0)
y_pred_updrs_1_vm0 = np.where(y_pred_updrs_1_vm0 < 0, 0, y_pred_updrs_1_vm0)
# Evaluate the performance of the model.
mse_updrs_1_vm0 = mean_squared_error(y_val_updrs_1_vm0, y_pred_updrs_1_vm0)
mae_updrs_1_vm0 = mean_absolute_error(y_val_updrs_1_vm0, y_pred_updrs_1_vm0)
r2_updrs_1_vm0 = r2_score(y_val_updrs_1_vm0, y_pred_updrs_1_vm0)
smape_updrs_1_vm0 = smape(y_val_updrs_1_vm0, y_pred_updrs_1_vm0)
# print("mse_updrs1:", mse_updrs_1_vm0)
# print("mae_updrs1:", mae_updrs_1_vm0)
print("r2_updrs_1_vm0:", r2_updrs_1_vm0)
print("smape_updrs_1_vm0:", smape_updrs_1_vm0)
# Submission
a_proteins = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_proteins.csv"
)
a_peptides = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_peptides.csv"
)
a_clinical = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_clinical_data.csv"
)
# def get_predictions(test, test_peptides, test_proteins, sample_submission):
a_merged_proteins_peptides = pd.merge(
a_proteins, a_peptides, on=["visit_id", "visit_month", "patient_id", "UniProt"]
)
a_merged = pd.merge(
a_merged_proteins_peptides, a_clinical, on=["visit_id", "visit_month", "patient_id"]
)
| false | 0 | 2,690 | 0 | 2,690 | 2,690 |
||
129289815
|
<jupyter_start><jupyter_text>TwoClassLeafs
Kaggle dataset identifier: twoclassleafs
<jupyter_script># The following parts are only needed to resume training on a previous model
# Run this to get the ID for the next cell
import shutil
shutil.move(
"/kaggle/working/mask_rcnn_leafscollage_v250.h5", "/kaggle/working/mask-rcnn/"
)
# This is always needed.
import os
os.chdir("mask-rcnn")
#!python3 /kaggle/working/Mask_RCNN/samples/leafs/leafs.py train --dataset=/kaggle/input/segmented-basil-leafs/leafs/train/ --weights=/kaggle/working/Mask_RCNN/mask_rcnn_coco.h5
#!python3 /kaggle/working/mask-rcnn/samples/leafs_collage/leafs_collage.py train --dataset=/kaggle/input/twoclassleafs/twoClassesIX/twoClassesIX/ --weights=/kaggle/working/mask-rcnn/mask_rcnn_leafscollage_v250.h5 --logs=/kaggle/working/logs/
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/289/129289815.ipynb
|
twoclassleafs
|
tomtechnicus
|
[{"Id": 129289815, "ScriptId": 24233253, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4612768, "CreationDate": "05/12/2023 13:36:07", "VersionNumber": 81.0, "Title": "Basil Leaf Dataset Trainer", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 35.0, "LinesInsertedFromPrevious": 5.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 30.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185198446, "KernelVersionId": 129289815, "SourceDatasetVersionId": 4991080}]
|
[{"Id": 4991080, "DatasetId": 2387368, "DatasourceVersionId": 5059871, "CreatorUserId": 4612768, "LicenseName": "Unknown", "CreationDate": "02/13/2023 10:28:51", "VersionNumber": 17.0, "Title": "TwoClassLeafs", "Slug": "twoclassleafs", "Subtitle": "Crazy Basil leaf collages for training with mask-rcnn.", "Description": NaN, "VersionNotes": "Data Update 2023/02/13", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2387368, "CreatorUserId": 4612768, "OwnerUserId": 4612768.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4991080.0, "CurrentDatasourceVersionId": 5059871.0, "ForumId": 2414622, "Type": 2, "CreationDate": "08/04/2022 14:38:33", "LastActivityDate": "08/04/2022", "TotalViews": 74, "TotalDownloads": 0, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 4612768, "UserName": "tomtechnicus", "DisplayName": "Tom Technicus", "RegisterDate": "03/05/2020", "PerformanceTier": 0}]
|
# The following parts are only needed to resume training on a previous model
# Run this to get the ID for the next cell
import shutil
shutil.move(
"/kaggle/working/mask_rcnn_leafscollage_v250.h5", "/kaggle/working/mask-rcnn/"
)
# This is always needed.
import os
os.chdir("mask-rcnn")
#!python3 /kaggle/working/Mask_RCNN/samples/leafs/leafs.py train --dataset=/kaggle/input/segmented-basil-leafs/leafs/train/ --weights=/kaggle/working/Mask_RCNN/mask_rcnn_coco.h5
#!python3 /kaggle/working/mask-rcnn/samples/leafs_collage/leafs_collage.py train --dataset=/kaggle/input/twoclassleafs/twoClassesIX/twoClassesIX/ --weights=/kaggle/working/mask-rcnn/mask_rcnn_leafscollage_v250.h5 --logs=/kaggle/working/logs/
| false | 0 | 259 | 0 | 281 | 259 |
||
129289842
|
import numpy as np
a = np.array([2, 3, 4, 5])
a
b = np.arange(10)
b
b = np.arange(10, 20)
b
c = np.arange(10, 20, 2)
c
d = np.array([2, 3, 300], dtype=np.int16)
d
# **Linspace method**
e = np.linspace(3, 10, 20)
e
# **Attribute-->shape,dimension**
a.ndim
a.shape
# **2-D array**
a = np.array([[2, 5, 4], [6, 7, 8]])
a
a.ndim
a.shape
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/289/129289842.ipynb
| null | null |
[{"Id": 129289842, "ScriptId": 38438868, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11527620, "CreationDate": "05/12/2023 13:36:19", "VersionNumber": 1.0, "Title": "Numpy", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 38.0, "LinesInsertedFromPrevious": 38.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
a = np.array([2, 3, 4, 5])
a
b = np.arange(10)
b
b = np.arange(10, 20)
b
c = np.arange(10, 20, 2)
c
d = np.array([2, 3, 300], dtype=np.int16)
d
# **Linspace method**
e = np.linspace(3, 10, 20)
e
# **Attribute-->shape,dimension**
a.ndim
a.shape
# **2-D array**
a = np.array([[2, 5, 4], [6, 7, 8]])
a
a.ndim
a.shape
| false | 0 | 188 | 0 | 188 | 188 |
||
129289866
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
condors = pd.read_csv("/kaggle/input/condor-acceleration-usgs/condor_data_all.csv")
condors.head()
condor_1 = condors.loc[condors["TagID"] == 1]
condor_1.head()
condor_1.info()
X = condor_1[:500].X
Y = condor_1[:500].Y
Z = condor_1[:500].Z
fig = plt.figure()
ax = fig.add_subplot(projection="3d")
ax.scatter3D(data=condor_1[:100], xs="X", ys="Y", zs="Z", c="No")
fig = plt.figure()
ax = fig.add_subplot(projection="3d")
ax.plot3D(X, Y, Z, alpha=0.5)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/289/129289866.ipynb
| null | null |
[{"Id": 129289866, "ScriptId": 38437249, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10897806, "CreationDate": "05/12/2023 13:36:29", "VersionNumber": 2.0, "Title": "Condor Tri-Axial Data", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 26.0, "LinesInsertedFromPrevious": 6.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 20.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
condors = pd.read_csv("/kaggle/input/condor-acceleration-usgs/condor_data_all.csv")
condors.head()
condor_1 = condors.loc[condors["TagID"] == 1]
condor_1.head()
condor_1.info()
X = condor_1[:500].X
Y = condor_1[:500].Y
Z = condor_1[:500].Z
fig = plt.figure()
ax = fig.add_subplot(projection="3d")
ax.scatter3D(data=condor_1[:100], xs="X", ys="Y", zs="Z", c="No")
fig = plt.figure()
ax = fig.add_subplot(projection="3d")
ax.plot3D(X, Y, Z, alpha=0.5)
| false | 0 | 258 | 0 | 258 | 258 |
||
129289721
|
<jupyter_start><jupyter_text>USA Company Insights: Glassdoor Scraped Data 2023
The "**Glassdoor Company Insights: Scraped Data Collection**" dataset is a comprehensive compilation of information gathered from **Glassdoor**, a leading platform for employee reviews and company ratings. This dataset includes a wide range of valuable data points, including company reviews, employee salaries, ratings, and more. With this dataset, researchers, analysts, and businesses can delve into the wealth of insights to gain a deeper understanding of company culture, employee satisfaction, and industry trends. Whether studying market competitiveness, benchmarking salaries, or conducting sentiment analysis, this dataset offers a valuable resource for exploring the experiences and perceptions of employees across various companies.
Kaggle dataset identifier: glassdoor-company-insightsscraped-data-collection
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.metrics.pairwise import cosine_similarity
df = pd.read_csv(
"/kaggle/input/glassdoor-company-insightsscraped-data-collection/glassdoor_comany.csv",
encoding="cp1252",
)
df.shape
df.isna().sum()
df.fillna("Unknown", inplace=True)
df.info()
df.head()
for i in df.columns:
print(i, " ", df[i].nunique())
# # Preprocessing Location feature
# We extract amount of offices from string values, like "n office locations in United States"
# If there are other values(address of one office) we return 1 - which means there is at least one office in United States
def location_process(x):
if "office locations" in x:
return int(x.split()[0])
else:
return 1
df["offices"] = df["Location"].apply(location_process)
df["salaries_in_K"] = df["Company salaries"].apply(
lambda x: float(x[:-1]) if "K" in x else float(x)
)
df["reviews_in_K"] = df["Company reviews"].apply(
lambda x: float(x[:-1]) if "K" in x else float(x)
)
df["jobs_in_K"] = df["Company Jobs"].apply(
lambda x: float(x[:-1]) if "K" in x else float(x)
)
# # Pairplot to take a closer look at how data is distributed
sns.pairplot(df, vars=["salaries_in_K", "reviews_in_K", "jobs_in_K", "Company rating"])
# # Histograms grouped by number of employees categories showing data distribution
def hists(x):
fig, axes = plt.subplots(1, 2, figsize=(10, 5))
sns.histplot(df, x=x, ax=axes[0], kde=True, color="r")
sns.histplot(df, x=x, ax=axes[1], kde=True, hue="Number of Employees")
plt.show()
for i in ["salaries_in_K", "reviews_in_K", "jobs_in_K", "Company rating"]:
hists(i)
# # Preparing data for recommendation system
df["overview"] = (
df["Company Description"]
+ " "
+ df["Industry Type"]
+ " "
+ df["Number of Employees"]
+ " "
+ df["Location"]
)
df["overview"] = df["overview"].apply(lambda x: x.lower())
# # Vectorization and fitting the data
cv = CountVectorizer(max_features=5000, stop_words="english")
vectors = cv.fit_transform(df["overview"]).toarray()
similarity = cosine_similarity(vectors)
# # Recommendation function
def similar_company(name):
indices = df[df["Company Name"] == name].index[0]
distances = similarity[indices]
arr = sorted(list(enumerate(distances)), reverse=True, key=lambda x: x[1])[1:8]
for i in arr:
print(df.loc[i[0], "Company Name"])
similar_company("Amazon")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/289/129289721.ipynb
|
glassdoor-company-insightsscraped-data-collection
|
joyshil0599
|
[{"Id": 129289721, "ScriptId": 38438467, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11036701, "CreationDate": "05/12/2023 13:35:22", "VersionNumber": 1.0, "Title": "Glassdoor EDA and Company Recommendation System", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 81.0, "LinesInsertedFromPrevious": 81.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 185198314, "KernelVersionId": 129289721, "SourceDatasetVersionId": 5664904}]
|
[{"Id": 5664904, "DatasetId": 3256232, "DatasourceVersionId": 5740372, "CreatorUserId": 13861238, "LicenseName": "CC0: Public Domain", "CreationDate": "05/11/2023 18:03:49", "VersionNumber": 1.0, "Title": "USA Company Insights: Glassdoor Scraped Data 2023", "Slug": "glassdoor-company-insightsscraped-data-collection", "Subtitle": "Unveiling Valuable Insights: Scraped Data from Glassdoor on USA's Top Companies", "Description": "The \"**Glassdoor Company Insights: Scraped Data Collection**\" dataset is a comprehensive compilation of information gathered from **Glassdoor**, a leading platform for employee reviews and company ratings. This dataset includes a wide range of valuable data points, including company reviews, employee salaries, ratings, and more. With this dataset, researchers, analysts, and businesses can delve into the wealth of insights to gain a deeper understanding of company culture, employee satisfaction, and industry trends. Whether studying market competitiveness, benchmarking salaries, or conducting sentiment analysis, this dataset offers a valuable resource for exploring the experiences and perceptions of employees across various companies.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3256232, "CreatorUserId": 13861238, "OwnerUserId": 13861238.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5664904.0, "CurrentDatasourceVersionId": 5740372.0, "ForumId": 3321716, "Type": 2, "CreationDate": "05/11/2023 18:03:49", "LastActivityDate": "05/11/2023", "TotalViews": 2698, "TotalDownloads": 397, "TotalVotes": 27, "TotalKernels": 4}]
|
[{"Id": 13861238, "UserName": "joyshil0599", "DisplayName": "Joy Shil", "RegisterDate": "02/24/2023", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.metrics.pairwise import cosine_similarity
df = pd.read_csv(
"/kaggle/input/glassdoor-company-insightsscraped-data-collection/glassdoor_comany.csv",
encoding="cp1252",
)
df.shape
df.isna().sum()
df.fillna("Unknown", inplace=True)
df.info()
df.head()
for i in df.columns:
print(i, " ", df[i].nunique())
# # Preprocessing Location feature
# We extract amount of offices from string values, like "n office locations in United States"
# If there are other values(address of one office) we return 1 - which means there is at least one office in United States
def location_process(x):
if "office locations" in x:
return int(x.split()[0])
else:
return 1
df["offices"] = df["Location"].apply(location_process)
df["salaries_in_K"] = df["Company salaries"].apply(
lambda x: float(x[:-1]) if "K" in x else float(x)
)
df["reviews_in_K"] = df["Company reviews"].apply(
lambda x: float(x[:-1]) if "K" in x else float(x)
)
df["jobs_in_K"] = df["Company Jobs"].apply(
lambda x: float(x[:-1]) if "K" in x else float(x)
)
# # Pairplot to take a closer look at how data is distributed
sns.pairplot(df, vars=["salaries_in_K", "reviews_in_K", "jobs_in_K", "Company rating"])
# # Histograms grouped by number of employees categories showing data distribution
def hists(x):
fig, axes = plt.subplots(1, 2, figsize=(10, 5))
sns.histplot(df, x=x, ax=axes[0], kde=True, color="r")
sns.histplot(df, x=x, ax=axes[1], kde=True, hue="Number of Employees")
plt.show()
for i in ["salaries_in_K", "reviews_in_K", "jobs_in_K", "Company rating"]:
hists(i)
# # Preparing data for recommendation system
df["overview"] = (
df["Company Description"]
+ " "
+ df["Industry Type"]
+ " "
+ df["Number of Employees"]
+ " "
+ df["Location"]
)
df["overview"] = df["overview"].apply(lambda x: x.lower())
# # Vectorization and fitting the data
cv = CountVectorizer(max_features=5000, stop_words="english")
vectors = cv.fit_transform(df["overview"]).toarray()
similarity = cosine_similarity(vectors)
# # Recommendation function
def similar_company(name):
indices = df[df["Company Name"] == name].index[0]
distances = similarity[indices]
arr = sorted(list(enumerate(distances)), reverse=True, key=lambda x: x[1])[1:8]
for i in arr:
print(df.loc[i[0], "Company Name"])
similar_company("Amazon")
| false | 1 | 838 | 1 | 1,029 | 838 |
||
129289602
|
<jupyter_start><jupyter_text>CIFAKE: Real and AI-Generated Synthetic Images
# CIFAKE: Real and AI-Generated Synthetic Images
The quality of AI-generated images has rapidly increased, leading to concerns of authenticity and trustworthiness.
CIFAKE is a dataset that contains 60,000 synthetically-generated images and 60,000 real images (collected from CIFAR-10). Can computer vision techniques be used to detect when an image is real or has been generated by AI?
Further information on this dataset can be found here: [Bird, J.J., Lotfi, A. (2023). CIFAKE: Image Classification and Explainable Identification of AI-Generated Synthetic Images. arXiv preprint arXiv:2303.14126.](https://arxiv.org/abs/2303.14126)

## Dataset details
The dataset contains two classes - REAL and FAKE.
For REAL, we collected the images from Krizhevsky & Hinton's [CIFAR-10 dataset](https://www.cs.toronto.edu/~kriz/cifar.html)
For the FAKE images, we generated the equivalent of CIFAR-10 with Stable Diffusion version 1.4
There are 100,000 images for training (50k per class) and 20,000 for testing (10k per class)
## Papers with Code
The dataset and all studies using it are linked using [Papers with Code](https://paperswithcode.com/dataset/cifake-real-and-ai-generated-synthetic-images)
[https://paperswithcode.com/dataset/cifake-real-and-ai-generated-synthetic-images](https://paperswithcode.com/dataset/cifake-real-and-ai-generated-synthetic-images)
## References
If you use this dataset, you **must** cite the following sources
[Krizhevsky, A., & Hinton, G. (2009). Learning multiple layers of features from tiny images.](https://www.cs.toronto.edu/~kriz/learning-features-2009-TR.pdfl)
[Bird, J.J., Lotfi, A. (2023). CIFAKE: Image Classification and Explainable Identification of AI-Generated Synthetic Images. arXiv preprint arXiv:2303.14126.](https://arxiv.org/abs/2303.14126)
Real images are from Krizhevsky & Hinton (2009), fake images are from Bird & Lotfi (2023). The Bird & Lotfi study is a preprint currently available on [ArXiv](https://arxiv.org/abs/2303.14126) and this description will be updated when the paper is published.
## Notes
The updates to the dataset on the 28th of March 2023 did not change anything; the file formats ".jpeg" were renamed ".jpg" and the root folder was uploaded to meet Kaggle's usability requirements.
## License
This dataset is published under the [same MIT license as CIFAR-10](https://github.com/wichtounet/cifar-10/blob/master/LICENSE):
*Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:*
*The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.*
*THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.*
Kaggle dataset identifier: cifake-real-and-ai-generated-synthetic-images
<jupyter_script>import os
import numpy as np
from tqdm import tqdm
import pandas as pd
import cv2
import time
from sklearn.model_selection import train_test_split
from sklearn.metrics import f1_score
from sklearn.metrics import confusion_matrix, classification_report
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.layers import (
Dense,
Activation,
Dropout,
Conv2D,
MaxPooling2D,
BatchNormalization,
)
from tensorflow.keras.optimizers import Adam, Adamax
from tensorflow.keras.metrics import categorical_crossentropy
from tensorflow.keras import regularizers
from tensorflow.keras.models import Model
from tensorflow.keras import backend as K
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
# ### below is a handy function that enables the printing of test in user specified foreground and background colors
def print_in_color(txt_msg, fore_tupple=(0, 255, 255), back_tupple=(0, 0, 0)):
# prints the text_msg in the foreground color specified by fore_tupple with the background specified by back_tupple
# text_msg is the text, fore_tupple is foregroud color tupple (r,g,b), back_tupple is background tupple (r,g,b)
rf, gf, bf = fore_tupple
rb, gb, bb = back_tupple
msg = "{0}" + txt_msg
mat = (
"\33[38;2;"
+ str(rf)
+ ";"
+ str(gf)
+ ";"
+ str(bf)
+ ";48;2;"
+ str(rb)
+ ";"
+ str(gb)
+ ";"
+ str(bb)
+ "m"
)
print(msg.format(mat))
print("\33[0m", end="") # returns default print color to back to black
# ### Dataset has a train directory with 50,00 real and 50,000 fake images. Lets look at the size of a typical image
fpath = r"/kaggle/input/cifake-real-and-ai-generated-synthetic-images/train/REAL/0000 (10).jpg"
img = cv2.imread(fpath)
print("Image shape is ", img.shape)
# ### Images are small but there are 100,00 training images. I will set a limiter so that only 20,000 images are used for each of the two classes, the information in a DataFrames. 10% of the train images to be used for a validation set
limiter = 20000
test_dir = r"/kaggle/input/cifake-real-and-ai-generated-synthetic-images/test"
train_dir = r"/kaggle/input/cifake-real-and-ai-generated-synthetic-images/train"
classes = sorted(os.listdir(train_dir))
print("there are ", len(classes), " classes ", classes[0], " and ", classes[1])
dir_list = [train_dir, test_dir]
names = ["train", "test"]
zip_list = zip(names, dir_list)
for name, dir in zip_list:
filepaths = []
labels = []
class_list = sorted(os.listdir(dir))
for klass in class_list:
classpath = os.path.join(dir, klass)
flist = sorted(os.listdir(classpath))
if name == "train":
flist = np.random.choice(
flist, limiter, replace=False
) # randomly select limiter number of files from train_dir for each class
desc = f"{name}-{klass}"
for f in tqdm(flist, ncols=100, colour="blue", unit="files", desc=desc):
fpath = os.path.join(classpath, f)
filepaths.append(fpath)
labels.append(klass)
Fseries = pd.Series(filepaths, name="filepaths")
Lseries = pd.Series(labels, name="labels")
if name == "train":
train_df = pd.concat([Fseries, Lseries], axis=1)
train_df, valid_df = train_test_split(
train_df,
test_size=0.1,
shuffle=True,
random_state=123,
stratify=train_df["labels"],
)
else:
test_df = pd.concat([Fseries, Lseries], axis=1)
print(
"train_df length: ",
len(train_df),
" test_df length: ",
len(test_df),
" valid_df length: ",
len(valid_df),
)
# ### Since that train data is balance and has a large number of images we do not need to do augmentation. Create the train. test and valid generators
gen = ImageDataGenerator()
# I will be using an EfficientNet model which requires a minimum image size of 32 X 32
img_size = (32, 32)
bs = 200 # set the batch size
train_gen = gen.flow_from_dataframe(
train_df,
x_col="filepaths",
y_col="labels",
target_size=img_size,
class_mode="categorical",
color_mode="rgb",
shuffle=True,
batch_size=bs,
)
valid_gen = gen.flow_from_dataframe(
valid_df,
x_col="filepaths",
y_col="labels",
target_size=img_size,
class_mode="categorical",
color_mode="rgb",
shuffle=False,
batch_size=bs,
)
test_gen = gen.flow_from_dataframe(
test_df,
x_col="filepaths",
y_col="labels",
target_size=img_size,
class_mode="categorical",
color_mode="rgb",
shuffle=False,
batch_size=bs,
)
labels = test_gen.labels
# ### I will define a custom metric called F1_score
def F1_score(y_true, y_pred): # taken from old keras source code
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))
predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))
precision = true_positives / (predicted_positives + K.epsilon())
recall = true_positives / (possible_positives + K.epsilon())
f1_val = 2 * (precision * recall) / (precision + recall + K.epsilon())
return f1_val
# ### now lets use transfer learning with an EfficientNetB0 model. You are ofter told to initially make the base model not trainable, run some epochs then make the base model trrainable and run more epochs to fine tune the model. I disagree with that approach. I have found it is better to make the base model trainable from the outset. My testing indicates I get to a higher validation accuracy in less epochs than doing the fine tuning approach
img_shape = (img_size[0], img_size[1], 3)
base_model = tf.keras.applications.EfficientNetV2B0(
include_top=False, weights="imagenet", input_shape=img_shape, pooling="max"
)
base_model.trainable = True
x = base_model.output
x = BatchNormalization(axis=-1, momentum=0.99, epsilon=0.001)(x)
x = Dense(
256,
kernel_regularizer=regularizers.l2(l=0.016),
activity_regularizer=regularizers.l1(0.006),
bias_regularizer=regularizers.l1(0.006),
activation="relu",
)(x)
x = Dropout(rate=0.4, seed=123)(x)
output = Dense(2, activation="softmax")(x)
model = Model(inputs=base_model.input, outputs=output)
model.compile(
Adamax(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy", F1_score],
)
# ## Before training the model I want to utilize a custom callback I developed.
# The LR_ASK callback is a convenient callback that allows you to continue training for ask_epoch more epochs or to halt training.
# If you elect to continue training for more epochs you are given the option to retain the current learning rate (LR) or to
# enter a new value for the learning rate. The form of use is:
# ask=LR_ASK(model,epochs, ask_epoch) where:
# * model is a string which is the name of your compiled model
# * epochs is an integer which is the number of epochs to run specified in model.fit
# * ask_epoch is an integer. If ask_epoch is set to a value say 5 then the model will train for 5 epochs.
# then the user is ask to enter H to halt training, or enter an inter value. For example if you enter 4
# training will continue for 4 more epochs to epoch 9 then you will be queried again. Once you enter an
# integer value you are prompted to press ENTER to continue training using the current learning rate
# or to enter a new value for the learning rate.
# * dwell is a boolean. If set to true the function compares the validation loss for the current tp the lowest
# validation loss thus far achieved. If the validation loss for the current epoch is larger then learning rate
# is automatically adjust by the formulanew_lr=lr * factor where factor is a float between 0 and 1. The motivation
# here is that if the validation loss increased we have moved to a point in Nspace on the cost functiob surface that
# if less favorable(higher cost) than for the epoch with the lowest cost. So the model is loaded with the weights
# from the epoch with the lowest loss and the learning rate is reduced
#
# At the end of training the model weights are set to the weights for the epoch that achieved the lowest validation loss.
# The callback also prints the training data in a spreadsheet type of format.
class LR_ASK(keras.callbacks.Callback):
def __init__(
self, model, epochs, ask_epoch, batches, dwell=True, factor=0.4
): # initialization of the callback
super(LR_ASK, self).__init__()
self.model = model
self.ask_epoch = ask_epoch
self.epochs = epochs
self.ask = True # if True query the user on a specified epoch
self.lowest_vloss = np.inf
self.lowest_aloss = np.inf
self.best_weights = (
self.model.get_weights()
) # set best weights to model's initial weights
self.best_epoch = 1
self.dwell = dwell
self.factor = factor
self.header = True
self.batches = batches
def on_train_begin(self, logs=None): # this runs on the beginning of training
msg1 = f"Training will proceed until epoch {self.ask_epoch} then you will be asked to\n"
msg2 = "enter H to halt training or enter an integer for how many more epochs to run then be asked again"
print_in_color(msg1 + msg2)
if self.dwell:
msg = "learning rate will be automatically adjusted during training"
print_in_color(msg, (0, 255, 0))
self.start_time = time.time() # set the time at which training started
def on_train_end(self, logs=None): # runs at the end of training
msg = f"loading model with weights from epoch {self.best_epoch}"
print_in_color(msg, (0, 255, 255))
self.model.set_weights(
self.best_weights
) # set the weights of the model to the best weights
tr_duration = (
time.time() - self.start_time
) # determine how long the training cycle lasted
hours = tr_duration // 3600
minutes = (tr_duration - (hours * 3600)) // 60
seconds = tr_duration - ((hours * 3600) + (minutes * 60))
msg = f"training elapsed time was {str(hours)} hours, {minutes:4.1f} minutes, {seconds:4.2f} seconds)"
print_in_color(msg) # print out training duration time
def on_epoch_begin(self, epoch, logs=None):
self.ep_start = time.time()
def on_train_batch_end(self, batch, logs=None):
# get batch accuracy and loss
acc = logs.get("accuracy") * 100
loss = logs.get("loss")
# prints over on the same line to show running batch count
msg = "{0:20s}processing batch {1:} of {2:5s}- accuracy= {3:5.3f} - loss: {4:8.5f} ".format(
" ", str(batch), str(self.batches), acc, loss
)
print(msg, "\r", end="")
def on_epoch_end(self, epoch, logs=None): # method runs on the end of each epoch
if self.header == True:
msg = (
"{0:^7s}{1:^9s}{2:^9s}{3:^9s}{4:^10s}{5:^13s}{6:^10s}{7:^13s}{8:13s}\n"
)
msg1 = msg.format(
"Epoch",
"Train",
"Train",
"Valid",
"Valid",
"V_Loss %",
"Learning",
"Next LR",
"Duration in",
)
msg = "{0:^7s}{1:^9s}{2:^9s}{3:^9s}{4:^10s}{5:^13s}{6:^10s}{7:^13s}{8:13s}"
msg2 = msg.format(
" ",
"Loss",
"Accuracy",
"Loss",
"Accuracy",
"Improvement",
"Rate",
"Rate",
" Seconds",
)
print_in_color(msg1 + msg2)
self.header = False
ep_end = time.time()
duration = ep_end - self.ep_start
vloss = logs.get("val_loss") # get the validation loss for this epoch
aloss = logs.get("loss")
acc = logs.get("accuracy") # get training accuracy
v_acc = logs.get("val_accuracy") # get validation accuracy
lr = float(
tf.keras.backend.get_value(self.model.optimizer.lr)
) # get the current learning rate
if epoch > 0:
deltav = self.lowest_vloss - vloss
pimprov = (deltav / self.lowest_vloss) * 100
deltaa = self.lowest_aloss - aloss
aimprov = (deltaa / self.lowest_aloss) * 100
else:
pimprov = 0.0
if vloss < self.lowest_vloss:
self.lowest_vloss = vloss
self.best_weights = (
self.model.get_weights()
) # set best weights to model's initial weights
self.best_epoch = epoch + 1
new_lr = lr
msg = "{0:^7s}{1:^9.4f}{2:^9.2f}{3:^9.4f}{4:^10.2f}{5:^13.2f}{6:^10.6f}{7:11.6f}{8:^15.2f}"
msg = msg.format(
str(epoch + 1),
aloss,
acc * 100,
vloss,
v_acc * 100,
pimprov,
lr,
new_lr,
duration,
)
print_in_color(msg, (0, 255, 0)) # green foreground
else: # validation loss increased
if (
self.dwell
): # if dwell is True when the validation loss increases the learning rate is automatically reduced and model weights are set to best weights
lr = float(
tf.keras.backend.get_value(self.model.optimizer.lr)
) # get the current learning rate
new_lr = lr * self.factor
msg = "{0:^7s}{1:^9.4f}{2:^9.2f}{3:^9.4f}{4:^10.2f}{5:^13.2f}{6:^10.6f}{7:11.6f}{8:^15.2f}"
msg = msg.format(
str(epoch + 1),
aloss,
acc * 100,
vloss,
v_acc * 100,
pimprov,
lr,
new_lr,
duration,
)
print_in_color(msg, (255, 255, 0))
tf.keras.backend.set_value(
self.model.optimizer.lr, new_lr
) # set the learning rate in the optimizer
self.model.set_weights(
self.best_weights
) # set the weights of the model to the best weights
if self.ask: # are the conditions right to query the user?
if (
epoch + 1 == self.ask_epoch
): # is this epoch the one for quering the user?
msg = "\n Enter H to end training or an integer for the number of additional epochs to run then ask again"
print_in_color(msg) # cyan foreground
ans = input()
if (
ans == "H" or ans == "h" or ans == "0"
): # quit training for these conditions
msg = f"you entered {ans}, Training halted on epoch {epoch+1} due to user input\n"
print_in_color(msg)
self.model.stop_training = True # halt training
else: # user wants to continue training
self.header = True
self.ask_epoch += int(ans)
msg = f"you entered {ans} Training will continue to epoch {self.ask_epoch}"
print_in_color(msg) # cyan foreground
if self.dwell == False:
lr = float(
tf.keras.backend.get_value(self.model.optimizer.lr)
) # get the current learning rate
msg = f"current LR is {lr:8.6f} hit enter to keep this LR or enter a new LR"
print_in_color(msg) # cyan foreground
ans = input(" ")
if ans == "":
msg = f"keeping current LR of {lr:7.5f}"
print_in_color(msg) # cyan foreground
else:
new_lr = float(ans)
tf.keras.backend.set_value(
self.model.optimizer.lr, new_lr
) # set the learning rate in the optimizer
msg = f" changing LR to {ans}"
print_in_color(msg) # cyan foreground
# ### OK lets instantiate the callback and train the model
ask_epoch = 10 # initially train for 10 epochs
batches = int(len(train_df) / bs)
# instantiate the custom callback
epochs = 100 # max epochs to run
ask = LR_ASK(
model, epochs=epochs, ask_epoch=ask_epoch, batches=batches
) # instantiate the custom callback
callbacks = [ask]
# train the model- don't worry aboutthe warning message your model will train correctly
history = model.fit(
x=train_gen,
epochs=epochs,
verbose=0,
callbacks=callbacks,
validation_data=valid_gen,
validation_steps=None,
shuffle=True,
initial_epoch=0,
) # train the model
# ### lets define a function to plot the models training data
def tr_plot(tr_data):
start_epoch = 0
# Plot the training and validation data
tacc = tr_data.history["accuracy"]
tloss = tr_data.history["loss"]
vacc = tr_data.history["val_accuracy"]
vloss = tr_data.history["val_loss"]
tf1 = tr_data.history["F1_score"]
vf1 = tr_data.history["val_F1_score"]
Epoch_count = len(tacc) + start_epoch
Epochs = []
for i in range(start_epoch, Epoch_count):
Epochs.append(i + 1)
index_loss = np.argmin(vloss) # this is the epoch with the lowest validation loss
val_lowest = vloss[index_loss]
index_acc = np.argmax(vacc)
acc_highest = vacc[index_acc]
indexf1 = np.argmax(vf1)
vf1_highest = vf1[indexf1]
plt.style.use("fivethirtyeight")
sc_label = "best epoch= " + str(index_loss + 1 + start_epoch)
vc_label = "best epoch= " + str(index_acc + 1 + start_epoch)
f1_label = "best epoch= " + str(index_acc + 1 + start_epoch)
fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(25, 10))
axes[0].plot(Epochs, tloss, "r", label="Training loss")
axes[0].plot(Epochs, vloss, "g", label="Validation loss")
axes[0].scatter(
index_loss + 1 + start_epoch, val_lowest, s=150, c="blue", label=sc_label
)
axes[0].scatter(Epochs, tloss, s=100, c="red")
axes[0].set_title("Training and Validation Loss")
axes[0].set_xlabel("Epochs", fontsize=18)
axes[0].set_ylabel("Loss", fontsize=18)
axes[0].legend()
axes[1].plot(Epochs, tacc, "r", label="Training Accuracy")
axes[1].scatter(Epochs, tacc, s=100, c="red")
axes[1].plot(Epochs, vacc, "g", label="Validation Accuracy")
axes[1].scatter(
index_acc + 1 + start_epoch, acc_highest, s=150, c="blue", label=vc_label
)
axes[1].set_title("Training and Validation Accuracy")
axes[1].set_xlabel("Epochs", fontsize=18)
axes[1].set_ylabel("Accuracy", fontsize=18)
axes[1].legend()
axes[2].plot(Epochs, tf1, "r", label="Training F1 score")
axes[2].plot(Epochs, vf1, "g", label="Validation F1 score")
index_tf1 = np.argmax(tf1) # this is the epoch with the highest training F1 score
tf1max = tf1[index_tf1]
index_vf1 = np.argmax(vf1) # thisiis the epoch with the highest validation F1 score
vf1max = vf1[index_vf1]
axes[2].scatter(
index_vf1 + 1 + start_epoch, vf1max, s=150, c="blue", label=vc_label
)
axes[2].scatter(Epochs, tf1, s=100, c="red")
axes[2].set_title("Training and Validation F1 score")
axes[2].set_xlabel("Epochs", fontsize=18)
axes[2].set_ylabel("F1 score", fontsize=18)
axes[2].legend()
plt.tight_layout
plt.show()
return
tr_plot(history) # plot the training data
# ### Now lets define a function to use the trained model to make predictions on the test set and produce a classification report and a confusion matrix
def predictor(model, test_gen):
classes = list(test_gen.class_indices.keys())
class_count = len(classes)
preds = model.predict(test_gen, verbose=1)
errors = 0
test_count = len(preds)
misclassified_classes = []
misclassified_files = []
misclassified_as = []
pred_indices = []
for i, p in enumerate(preds):
pred_index = np.argmax(p)
pred_indices.append(pred_index)
true_index = test_gen.labels[i]
if pred_index != true_index:
errors += 1
misclassified_classes.append(classes[true_index])
misclassified_as.append(classes[pred_index])
file = test_gen.filenames[i]
split = file.split("/")
L = len(split)
f = split[L - 2] + " " + split[L - 1]
misclassified_files.append(f)
accuracy = (test_count - errors) * 100 / test_count
ytrue = np.array(test_gen.labels)
ypred = np.array(pred_indices)
f1score = f1_score(ytrue, ypred, average="weighted") * 100
msg = f"There were {errors} errors in {test_count} tests for an accuracy of {accuracy:6.2f} and an F1 score of {f1score:6.2f}"
print(msg)
misclassified_classes = sorted(misclassified_classes)
if len(misclassified_classes) > 0:
misclassifications = []
for klass in misclassified_classes:
mis_count = misclassified_classes.count(klass)
misclassifications.append(mis_count)
unique = len(np.unique(misclassified_classes))
if unique == 1:
height = int(unique)
else:
height = int(unique / 2)
plt.figure(figsize=(10, height))
plt.style.use("fivethirtyeight")
plt.barh(misclassified_classes, misclassifications)
plt.title(
"Classification Errors on Test Set by Class", fontsize=20, color="blue"
)
plt.xlabel("NUMBER OF MISCLASSIFICATIONS", fontsize=20, color="blue")
plt.ylabel("CLASS", fontsize=20, color="blue")
plt.show()
if class_count <= 30:
cm = confusion_matrix(ytrue, ypred)
# plot the confusion matrix
plt.figure(figsize=(12, 8))
sns.heatmap(cm, annot=True, vmin=0, fmt="g", cmap="Blues", cbar=False)
plt.xticks(np.arange(class_count) + 0.5, classes, rotation=90)
plt.yticks(np.arange(class_count) + 0.5, classes, rotation=0)
plt.xlabel("Predicted")
plt.ylabel("Actual")
plt.title("Confusion Matrix")
plt.show()
clr = classification_report(
ytrue, ypred, target_names=classes, digits=4
) # create classification report
print("Classification Report:\n----------------------\n", clr)
return f1score
f1score = predictor(model, test_gen)
# ### Lets define a function to save the model with a nomenclature of
# ### subject-num of classes-image size-f1 score.h5
def save_model(model, subject, classes, img_size, f1score, working_dir):
name = f"{subject}-{str(len(classes))}-({str(img_size[0])} X {str(img_size[1])})- {f1score:5.2f}.h5"
model_save_loc = os.path.join(working_dir, name)
try:
model.save(model_save_loc)
msg = f"model was saved as {model_save_loc}"
print_in_color(msg, (0, 255, 255), (0, 0, 0))
except:
msg = "model can not be saved due to tensorflow 2.10.0 or higher. Bug involving use of EfficientNet Models"
print_in_color(msg, (0, 255, 255), (0, 0, 0)) # cyan foreground
subject = "Real vs Fake Images"
working_dir = r"/kaggle/working/"
save_model(model, subject, classes, img_size, f1score, working_dir)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/289/129289602.ipynb
|
cifake-real-and-ai-generated-synthetic-images
|
birdy654
|
[{"Id": 129289602, "ScriptId": 38432210, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2533892, "CreationDate": "05/12/2023 13:34:21", "VersionNumber": 1.0, "Title": "Transfer Learning F1 score=96%", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 404.0, "LinesInsertedFromPrevious": 404.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 4}]
|
[{"Id": 185198078, "KernelVersionId": 129289602, "SourceDatasetVersionId": 5256696}]
|
[{"Id": 5256696, "DatasetId": 3041726, "DatasourceVersionId": 5329502, "CreatorUserId": 2039603, "LicenseName": "Other (specified in description)", "CreationDate": "03/28/2023 16:00:29", "VersionNumber": 3.0, "Title": "CIFAKE: Real and AI-Generated Synthetic Images", "Slug": "cifake-real-and-ai-generated-synthetic-images", "Subtitle": "Can Computer Vision detect when images have been generated by AI?", "Description": "# CIFAKE: Real and AI-Generated Synthetic Images\nThe quality of AI-generated images has rapidly increased, leading to concerns of authenticity and trustworthiness.\n\nCIFAKE is a dataset that contains 60,000 synthetically-generated images and 60,000 real images (collected from CIFAR-10). Can computer vision techniques be used to detect when an image is real or has been generated by AI?\n\nFurther information on this dataset can be found here: [Bird, J.J., Lotfi, A. (2023). CIFAKE: Image Classification and Explainable Identification of AI-Generated Synthetic Images. arXiv preprint arXiv:2303.14126.](https://arxiv.org/abs/2303.14126)\n\n\n\n## Dataset details\nThe dataset contains two classes - REAL and FAKE. \n\nFor REAL, we collected the images from Krizhevsky & Hinton's [CIFAR-10 dataset](https://www.cs.toronto.edu/~kriz/cifar.html)\n\nFor the FAKE images, we generated the equivalent of CIFAR-10 with Stable Diffusion version 1.4\n\nThere are 100,000 images for training (50k per class) and 20,000 for testing (10k per class)\n\n## Papers with Code\nThe dataset and all studies using it are linked using [Papers with Code](https://paperswithcode.com/dataset/cifake-real-and-ai-generated-synthetic-images)\n[https://paperswithcode.com/dataset/cifake-real-and-ai-generated-synthetic-images](https://paperswithcode.com/dataset/cifake-real-and-ai-generated-synthetic-images)\n\n\n## References\nIf you use this dataset, you **must** cite the following sources\n\n[Krizhevsky, A., & Hinton, G. (2009). Learning multiple layers of features from tiny images.](https://www.cs.toronto.edu/~kriz/learning-features-2009-TR.pdfl)\n\n[Bird, J.J., Lotfi, A. (2023). CIFAKE: Image Classification and Explainable Identification of AI-Generated Synthetic Images. arXiv preprint arXiv:2303.14126.](https://arxiv.org/abs/2303.14126)\n\nReal images are from Krizhevsky & Hinton (2009), fake images are from Bird & Lotfi (2023). The Bird & Lotfi study is a preprint currently available on [ArXiv](https://arxiv.org/abs/2303.14126) and this description will be updated when the paper is published.\n\n## Notes\n\nThe updates to the dataset on the 28th of March 2023 did not change anything; the file formats \".jpeg\" were renamed \".jpg\" and the root folder was uploaded to meet Kaggle's usability requirements.\n\n## License\nThis dataset is published under the [same MIT license as CIFAR-10](https://github.com/wichtounet/cifar-10/blob/master/LICENSE):\n\n*Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the \"Software\"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:*\n\n*The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.*\n\n*THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.*", "VersionNotes": "Kaggle compatibility fix (no actual changes)", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3041726, "CreatorUserId": 2039603, "OwnerUserId": 2039603.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5256696.0, "CurrentDatasourceVersionId": 5329502.0, "ForumId": 3081274, "Type": 2, "CreationDate": "03/24/2023 13:22:42", "LastActivityDate": "03/24/2023", "TotalViews": 13728, "TotalDownloads": 1803, "TotalVotes": 46, "TotalKernels": 15}]
|
[{"Id": 2039603, "UserName": "birdy654", "DisplayName": "Jordan J. Bird", "RegisterDate": "07/03/2018", "PerformanceTier": 2}]
|
import os
import numpy as np
from tqdm import tqdm
import pandas as pd
import cv2
import time
from sklearn.model_selection import train_test_split
from sklearn.metrics import f1_score
from sklearn.metrics import confusion_matrix, classification_report
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.layers import (
Dense,
Activation,
Dropout,
Conv2D,
MaxPooling2D,
BatchNormalization,
)
from tensorflow.keras.optimizers import Adam, Adamax
from tensorflow.keras.metrics import categorical_crossentropy
from tensorflow.keras import regularizers
from tensorflow.keras.models import Model
from tensorflow.keras import backend as K
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
# ### below is a handy function that enables the printing of test in user specified foreground and background colors
def print_in_color(txt_msg, fore_tupple=(0, 255, 255), back_tupple=(0, 0, 0)):
# prints the text_msg in the foreground color specified by fore_tupple with the background specified by back_tupple
# text_msg is the text, fore_tupple is foregroud color tupple (r,g,b), back_tupple is background tupple (r,g,b)
rf, gf, bf = fore_tupple
rb, gb, bb = back_tupple
msg = "{0}" + txt_msg
mat = (
"\33[38;2;"
+ str(rf)
+ ";"
+ str(gf)
+ ";"
+ str(bf)
+ ";48;2;"
+ str(rb)
+ ";"
+ str(gb)
+ ";"
+ str(bb)
+ "m"
)
print(msg.format(mat))
print("\33[0m", end="") # returns default print color to back to black
# ### Dataset has a train directory with 50,00 real and 50,000 fake images. Lets look at the size of a typical image
fpath = r"/kaggle/input/cifake-real-and-ai-generated-synthetic-images/train/REAL/0000 (10).jpg"
img = cv2.imread(fpath)
print("Image shape is ", img.shape)
# ### Images are small but there are 100,00 training images. I will set a limiter so that only 20,000 images are used for each of the two classes, the information in a DataFrames. 10% of the train images to be used for a validation set
limiter = 20000
test_dir = r"/kaggle/input/cifake-real-and-ai-generated-synthetic-images/test"
train_dir = r"/kaggle/input/cifake-real-and-ai-generated-synthetic-images/train"
classes = sorted(os.listdir(train_dir))
print("there are ", len(classes), " classes ", classes[0], " and ", classes[1])
dir_list = [train_dir, test_dir]
names = ["train", "test"]
zip_list = zip(names, dir_list)
for name, dir in zip_list:
filepaths = []
labels = []
class_list = sorted(os.listdir(dir))
for klass in class_list:
classpath = os.path.join(dir, klass)
flist = sorted(os.listdir(classpath))
if name == "train":
flist = np.random.choice(
flist, limiter, replace=False
) # randomly select limiter number of files from train_dir for each class
desc = f"{name}-{klass}"
for f in tqdm(flist, ncols=100, colour="blue", unit="files", desc=desc):
fpath = os.path.join(classpath, f)
filepaths.append(fpath)
labels.append(klass)
Fseries = pd.Series(filepaths, name="filepaths")
Lseries = pd.Series(labels, name="labels")
if name == "train":
train_df = pd.concat([Fseries, Lseries], axis=1)
train_df, valid_df = train_test_split(
train_df,
test_size=0.1,
shuffle=True,
random_state=123,
stratify=train_df["labels"],
)
else:
test_df = pd.concat([Fseries, Lseries], axis=1)
print(
"train_df length: ",
len(train_df),
" test_df length: ",
len(test_df),
" valid_df length: ",
len(valid_df),
)
# ### Since that train data is balance and has a large number of images we do not need to do augmentation. Create the train. test and valid generators
gen = ImageDataGenerator()
# I will be using an EfficientNet model which requires a minimum image size of 32 X 32
img_size = (32, 32)
bs = 200 # set the batch size
train_gen = gen.flow_from_dataframe(
train_df,
x_col="filepaths",
y_col="labels",
target_size=img_size,
class_mode="categorical",
color_mode="rgb",
shuffle=True,
batch_size=bs,
)
valid_gen = gen.flow_from_dataframe(
valid_df,
x_col="filepaths",
y_col="labels",
target_size=img_size,
class_mode="categorical",
color_mode="rgb",
shuffle=False,
batch_size=bs,
)
test_gen = gen.flow_from_dataframe(
test_df,
x_col="filepaths",
y_col="labels",
target_size=img_size,
class_mode="categorical",
color_mode="rgb",
shuffle=False,
batch_size=bs,
)
labels = test_gen.labels
# ### I will define a custom metric called F1_score
def F1_score(y_true, y_pred): # taken from old keras source code
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))
predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))
precision = true_positives / (predicted_positives + K.epsilon())
recall = true_positives / (possible_positives + K.epsilon())
f1_val = 2 * (precision * recall) / (precision + recall + K.epsilon())
return f1_val
# ### now lets use transfer learning with an EfficientNetB0 model. You are ofter told to initially make the base model not trainable, run some epochs then make the base model trrainable and run more epochs to fine tune the model. I disagree with that approach. I have found it is better to make the base model trainable from the outset. My testing indicates I get to a higher validation accuracy in less epochs than doing the fine tuning approach
img_shape = (img_size[0], img_size[1], 3)
base_model = tf.keras.applications.EfficientNetV2B0(
include_top=False, weights="imagenet", input_shape=img_shape, pooling="max"
)
base_model.trainable = True
x = base_model.output
x = BatchNormalization(axis=-1, momentum=0.99, epsilon=0.001)(x)
x = Dense(
256,
kernel_regularizer=regularizers.l2(l=0.016),
activity_regularizer=regularizers.l1(0.006),
bias_regularizer=regularizers.l1(0.006),
activation="relu",
)(x)
x = Dropout(rate=0.4, seed=123)(x)
output = Dense(2, activation="softmax")(x)
model = Model(inputs=base_model.input, outputs=output)
model.compile(
Adamax(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy", F1_score],
)
# ## Before training the model I want to utilize a custom callback I developed.
# The LR_ASK callback is a convenient callback that allows you to continue training for ask_epoch more epochs or to halt training.
# If you elect to continue training for more epochs you are given the option to retain the current learning rate (LR) or to
# enter a new value for the learning rate. The form of use is:
# ask=LR_ASK(model,epochs, ask_epoch) where:
# * model is a string which is the name of your compiled model
# * epochs is an integer which is the number of epochs to run specified in model.fit
# * ask_epoch is an integer. If ask_epoch is set to a value say 5 then the model will train for 5 epochs.
# then the user is ask to enter H to halt training, or enter an inter value. For example if you enter 4
# training will continue for 4 more epochs to epoch 9 then you will be queried again. Once you enter an
# integer value you are prompted to press ENTER to continue training using the current learning rate
# or to enter a new value for the learning rate.
# * dwell is a boolean. If set to true the function compares the validation loss for the current tp the lowest
# validation loss thus far achieved. If the validation loss for the current epoch is larger then learning rate
# is automatically adjust by the formulanew_lr=lr * factor where factor is a float between 0 and 1. The motivation
# here is that if the validation loss increased we have moved to a point in Nspace on the cost functiob surface that
# if less favorable(higher cost) than for the epoch with the lowest cost. So the model is loaded with the weights
# from the epoch with the lowest loss and the learning rate is reduced
#
# At the end of training the model weights are set to the weights for the epoch that achieved the lowest validation loss.
# The callback also prints the training data in a spreadsheet type of format.
class LR_ASK(keras.callbacks.Callback):
def __init__(
self, model, epochs, ask_epoch, batches, dwell=True, factor=0.4
): # initialization of the callback
super(LR_ASK, self).__init__()
self.model = model
self.ask_epoch = ask_epoch
self.epochs = epochs
self.ask = True # if True query the user on a specified epoch
self.lowest_vloss = np.inf
self.lowest_aloss = np.inf
self.best_weights = (
self.model.get_weights()
) # set best weights to model's initial weights
self.best_epoch = 1
self.dwell = dwell
self.factor = factor
self.header = True
self.batches = batches
def on_train_begin(self, logs=None): # this runs on the beginning of training
msg1 = f"Training will proceed until epoch {self.ask_epoch} then you will be asked to\n"
msg2 = "enter H to halt training or enter an integer for how many more epochs to run then be asked again"
print_in_color(msg1 + msg2)
if self.dwell:
msg = "learning rate will be automatically adjusted during training"
print_in_color(msg, (0, 255, 0))
self.start_time = time.time() # set the time at which training started
def on_train_end(self, logs=None): # runs at the end of training
msg = f"loading model with weights from epoch {self.best_epoch}"
print_in_color(msg, (0, 255, 255))
self.model.set_weights(
self.best_weights
) # set the weights of the model to the best weights
tr_duration = (
time.time() - self.start_time
) # determine how long the training cycle lasted
hours = tr_duration // 3600
minutes = (tr_duration - (hours * 3600)) // 60
seconds = tr_duration - ((hours * 3600) + (minutes * 60))
msg = f"training elapsed time was {str(hours)} hours, {minutes:4.1f} minutes, {seconds:4.2f} seconds)"
print_in_color(msg) # print out training duration time
def on_epoch_begin(self, epoch, logs=None):
self.ep_start = time.time()
def on_train_batch_end(self, batch, logs=None):
# get batch accuracy and loss
acc = logs.get("accuracy") * 100
loss = logs.get("loss")
# prints over on the same line to show running batch count
msg = "{0:20s}processing batch {1:} of {2:5s}- accuracy= {3:5.3f} - loss: {4:8.5f} ".format(
" ", str(batch), str(self.batches), acc, loss
)
print(msg, "\r", end="")
def on_epoch_end(self, epoch, logs=None): # method runs on the end of each epoch
if self.header == True:
msg = (
"{0:^7s}{1:^9s}{2:^9s}{3:^9s}{4:^10s}{5:^13s}{6:^10s}{7:^13s}{8:13s}\n"
)
msg1 = msg.format(
"Epoch",
"Train",
"Train",
"Valid",
"Valid",
"V_Loss %",
"Learning",
"Next LR",
"Duration in",
)
msg = "{0:^7s}{1:^9s}{2:^9s}{3:^9s}{4:^10s}{5:^13s}{6:^10s}{7:^13s}{8:13s}"
msg2 = msg.format(
" ",
"Loss",
"Accuracy",
"Loss",
"Accuracy",
"Improvement",
"Rate",
"Rate",
" Seconds",
)
print_in_color(msg1 + msg2)
self.header = False
ep_end = time.time()
duration = ep_end - self.ep_start
vloss = logs.get("val_loss") # get the validation loss for this epoch
aloss = logs.get("loss")
acc = logs.get("accuracy") # get training accuracy
v_acc = logs.get("val_accuracy") # get validation accuracy
lr = float(
tf.keras.backend.get_value(self.model.optimizer.lr)
) # get the current learning rate
if epoch > 0:
deltav = self.lowest_vloss - vloss
pimprov = (deltav / self.lowest_vloss) * 100
deltaa = self.lowest_aloss - aloss
aimprov = (deltaa / self.lowest_aloss) * 100
else:
pimprov = 0.0
if vloss < self.lowest_vloss:
self.lowest_vloss = vloss
self.best_weights = (
self.model.get_weights()
) # set best weights to model's initial weights
self.best_epoch = epoch + 1
new_lr = lr
msg = "{0:^7s}{1:^9.4f}{2:^9.2f}{3:^9.4f}{4:^10.2f}{5:^13.2f}{6:^10.6f}{7:11.6f}{8:^15.2f}"
msg = msg.format(
str(epoch + 1),
aloss,
acc * 100,
vloss,
v_acc * 100,
pimprov,
lr,
new_lr,
duration,
)
print_in_color(msg, (0, 255, 0)) # green foreground
else: # validation loss increased
if (
self.dwell
): # if dwell is True when the validation loss increases the learning rate is automatically reduced and model weights are set to best weights
lr = float(
tf.keras.backend.get_value(self.model.optimizer.lr)
) # get the current learning rate
new_lr = lr * self.factor
msg = "{0:^7s}{1:^9.4f}{2:^9.2f}{3:^9.4f}{4:^10.2f}{5:^13.2f}{6:^10.6f}{7:11.6f}{8:^15.2f}"
msg = msg.format(
str(epoch + 1),
aloss,
acc * 100,
vloss,
v_acc * 100,
pimprov,
lr,
new_lr,
duration,
)
print_in_color(msg, (255, 255, 0))
tf.keras.backend.set_value(
self.model.optimizer.lr, new_lr
) # set the learning rate in the optimizer
self.model.set_weights(
self.best_weights
) # set the weights of the model to the best weights
if self.ask: # are the conditions right to query the user?
if (
epoch + 1 == self.ask_epoch
): # is this epoch the one for quering the user?
msg = "\n Enter H to end training or an integer for the number of additional epochs to run then ask again"
print_in_color(msg) # cyan foreground
ans = input()
if (
ans == "H" or ans == "h" or ans == "0"
): # quit training for these conditions
msg = f"you entered {ans}, Training halted on epoch {epoch+1} due to user input\n"
print_in_color(msg)
self.model.stop_training = True # halt training
else: # user wants to continue training
self.header = True
self.ask_epoch += int(ans)
msg = f"you entered {ans} Training will continue to epoch {self.ask_epoch}"
print_in_color(msg) # cyan foreground
if self.dwell == False:
lr = float(
tf.keras.backend.get_value(self.model.optimizer.lr)
) # get the current learning rate
msg = f"current LR is {lr:8.6f} hit enter to keep this LR or enter a new LR"
print_in_color(msg) # cyan foreground
ans = input(" ")
if ans == "":
msg = f"keeping current LR of {lr:7.5f}"
print_in_color(msg) # cyan foreground
else:
new_lr = float(ans)
tf.keras.backend.set_value(
self.model.optimizer.lr, new_lr
) # set the learning rate in the optimizer
msg = f" changing LR to {ans}"
print_in_color(msg) # cyan foreground
# ### OK lets instantiate the callback and train the model
ask_epoch = 10 # initially train for 10 epochs
batches = int(len(train_df) / bs)
# instantiate the custom callback
epochs = 100 # max epochs to run
ask = LR_ASK(
model, epochs=epochs, ask_epoch=ask_epoch, batches=batches
) # instantiate the custom callback
callbacks = [ask]
# train the model- don't worry aboutthe warning message your model will train correctly
history = model.fit(
x=train_gen,
epochs=epochs,
verbose=0,
callbacks=callbacks,
validation_data=valid_gen,
validation_steps=None,
shuffle=True,
initial_epoch=0,
) # train the model
# ### lets define a function to plot the models training data
def tr_plot(tr_data):
start_epoch = 0
# Plot the training and validation data
tacc = tr_data.history["accuracy"]
tloss = tr_data.history["loss"]
vacc = tr_data.history["val_accuracy"]
vloss = tr_data.history["val_loss"]
tf1 = tr_data.history["F1_score"]
vf1 = tr_data.history["val_F1_score"]
Epoch_count = len(tacc) + start_epoch
Epochs = []
for i in range(start_epoch, Epoch_count):
Epochs.append(i + 1)
index_loss = np.argmin(vloss) # this is the epoch with the lowest validation loss
val_lowest = vloss[index_loss]
index_acc = np.argmax(vacc)
acc_highest = vacc[index_acc]
indexf1 = np.argmax(vf1)
vf1_highest = vf1[indexf1]
plt.style.use("fivethirtyeight")
sc_label = "best epoch= " + str(index_loss + 1 + start_epoch)
vc_label = "best epoch= " + str(index_acc + 1 + start_epoch)
f1_label = "best epoch= " + str(index_acc + 1 + start_epoch)
fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(25, 10))
axes[0].plot(Epochs, tloss, "r", label="Training loss")
axes[0].plot(Epochs, vloss, "g", label="Validation loss")
axes[0].scatter(
index_loss + 1 + start_epoch, val_lowest, s=150, c="blue", label=sc_label
)
axes[0].scatter(Epochs, tloss, s=100, c="red")
axes[0].set_title("Training and Validation Loss")
axes[0].set_xlabel("Epochs", fontsize=18)
axes[0].set_ylabel("Loss", fontsize=18)
axes[0].legend()
axes[1].plot(Epochs, tacc, "r", label="Training Accuracy")
axes[1].scatter(Epochs, tacc, s=100, c="red")
axes[1].plot(Epochs, vacc, "g", label="Validation Accuracy")
axes[1].scatter(
index_acc + 1 + start_epoch, acc_highest, s=150, c="blue", label=vc_label
)
axes[1].set_title("Training and Validation Accuracy")
axes[1].set_xlabel("Epochs", fontsize=18)
axes[1].set_ylabel("Accuracy", fontsize=18)
axes[1].legend()
axes[2].plot(Epochs, tf1, "r", label="Training F1 score")
axes[2].plot(Epochs, vf1, "g", label="Validation F1 score")
index_tf1 = np.argmax(tf1) # this is the epoch with the highest training F1 score
tf1max = tf1[index_tf1]
index_vf1 = np.argmax(vf1) # thisiis the epoch with the highest validation F1 score
vf1max = vf1[index_vf1]
axes[2].scatter(
index_vf1 + 1 + start_epoch, vf1max, s=150, c="blue", label=vc_label
)
axes[2].scatter(Epochs, tf1, s=100, c="red")
axes[2].set_title("Training and Validation F1 score")
axes[2].set_xlabel("Epochs", fontsize=18)
axes[2].set_ylabel("F1 score", fontsize=18)
axes[2].legend()
plt.tight_layout
plt.show()
return
tr_plot(history) # plot the training data
# ### Now lets define a function to use the trained model to make predictions on the test set and produce a classification report and a confusion matrix
def predictor(model, test_gen):
classes = list(test_gen.class_indices.keys())
class_count = len(classes)
preds = model.predict(test_gen, verbose=1)
errors = 0
test_count = len(preds)
misclassified_classes = []
misclassified_files = []
misclassified_as = []
pred_indices = []
for i, p in enumerate(preds):
pred_index = np.argmax(p)
pred_indices.append(pred_index)
true_index = test_gen.labels[i]
if pred_index != true_index:
errors += 1
misclassified_classes.append(classes[true_index])
misclassified_as.append(classes[pred_index])
file = test_gen.filenames[i]
split = file.split("/")
L = len(split)
f = split[L - 2] + " " + split[L - 1]
misclassified_files.append(f)
accuracy = (test_count - errors) * 100 / test_count
ytrue = np.array(test_gen.labels)
ypred = np.array(pred_indices)
f1score = f1_score(ytrue, ypred, average="weighted") * 100
msg = f"There were {errors} errors in {test_count} tests for an accuracy of {accuracy:6.2f} and an F1 score of {f1score:6.2f}"
print(msg)
misclassified_classes = sorted(misclassified_classes)
if len(misclassified_classes) > 0:
misclassifications = []
for klass in misclassified_classes:
mis_count = misclassified_classes.count(klass)
misclassifications.append(mis_count)
unique = len(np.unique(misclassified_classes))
if unique == 1:
height = int(unique)
else:
height = int(unique / 2)
plt.figure(figsize=(10, height))
plt.style.use("fivethirtyeight")
plt.barh(misclassified_classes, misclassifications)
plt.title(
"Classification Errors on Test Set by Class", fontsize=20, color="blue"
)
plt.xlabel("NUMBER OF MISCLASSIFICATIONS", fontsize=20, color="blue")
plt.ylabel("CLASS", fontsize=20, color="blue")
plt.show()
if class_count <= 30:
cm = confusion_matrix(ytrue, ypred)
# plot the confusion matrix
plt.figure(figsize=(12, 8))
sns.heatmap(cm, annot=True, vmin=0, fmt="g", cmap="Blues", cbar=False)
plt.xticks(np.arange(class_count) + 0.5, classes, rotation=90)
plt.yticks(np.arange(class_count) + 0.5, classes, rotation=0)
plt.xlabel("Predicted")
plt.ylabel("Actual")
plt.title("Confusion Matrix")
plt.show()
clr = classification_report(
ytrue, ypred, target_names=classes, digits=4
) # create classification report
print("Classification Report:\n----------------------\n", clr)
return f1score
f1score = predictor(model, test_gen)
# ### Lets define a function to save the model with a nomenclature of
# ### subject-num of classes-image size-f1 score.h5
def save_model(model, subject, classes, img_size, f1score, working_dir):
name = f"{subject}-{str(len(classes))}-({str(img_size[0])} X {str(img_size[1])})- {f1score:5.2f}.h5"
model_save_loc = os.path.join(working_dir, name)
try:
model.save(model_save_loc)
msg = f"model was saved as {model_save_loc}"
print_in_color(msg, (0, 255, 255), (0, 0, 0))
except:
msg = "model can not be saved due to tensorflow 2.10.0 or higher. Bug involving use of EfficientNet Models"
print_in_color(msg, (0, 255, 255), (0, 0, 0)) # cyan foreground
subject = "Real vs Fake Images"
working_dir = r"/kaggle/working/"
save_model(model, subject, classes, img_size, f1score, working_dir)
| false | 0 | 6,933 | 4 | 7,976 | 6,933 |
||
129210213
|
# # ASL- Fingerspelling
# ## What is American Sign Language Fingerspelling Recognition ?
# American Sign Language Fingerspelling Recognition is a technology that uses computer vision and machine learning algorithms to recognize and interpret the hand gestures used in American Sign Language (ASL) fingerspelling. It can be used to create tools and applications that help people with hearing impairments to communicate more effectively with others. The technology involves comparing the input image of the hand gesture to a pre-defined set of templates, extracting relevant features from the input image, or training a neural network on a large dataset of ASL fingerspelling images to learn the patterns and features that are most important for recognition. Despite some challenges, ASL Fingerspelling Recognition has the potential to greatly improve the lives of people with hearing impairments.
# ## Data Overview
# ### Files
# #### [train/supplemental_metadata].csv
# * path - The path to the landmark file.
# * file_id - A unique identifier for the data file.
# * participant_id - A unique identifier for the data contributor.
# * sequence_id - A unique identifier for the landmark sequence. Each data file may contain many sequences.
# * phrase - The labels for the landmark sequence. The train and test datasets contain randomly generated addresses, phone numbers, and urls derived from components of real addresses/phone numbers/urls. Any overlap with real addresses, phone numbers, or urls is purely accidental. The supplemental dataset consists of fingerspelled sentences. Note that some of the urls include adult content. The intent of this competition is to support the Deaf and Hard of Hearing community in engaging with technology on an equal footing with other adults.
# ### character_to_prediction_index.json
# #### [train/supplemental]_landmarks/
# The landmark data. The landmarks were extracted from raw videos with the MediaPipe holistic model. Not all of the frames necessarily had visible hands or hands that could be detected by the model.
# The landmark files contain the same data as in the ASL Signs competition (minus the row ID column) but reshaped into a wide format. This allows you to take advantage of the Parquet format to entirely skip loading landmarks that you aren't using.
# * sequence_id - A unique identifier for the landmark sequence. Most landmark files contain 1,000 sequences. The sequence ID is used as the dataframe index.
# * frame - The frame number within a landmark sequence.
# * [x/y/z]_[type]_[landmark_index] - There are now 1,629 spatial coordinate columns for the x, y and z coordinates for each of the 543 landmarks. The type of landmark is one of ['face', 'left_hand', 'pose', 'right_hand']. Details of the hand landmark locations can be found here. The spatial coordinates have already been normalized by MediaPipe. Note that the MediaPipe model is not fully trained to predict depth so you may wish to ignore the z values. The landmarks have been converted to float32.
# ## explore dataset
### import libraries
import pandas as pd, numpy as np, os
from pathlib import Path
print("importing..")
# ### explore supplemental_metadata
data = pd.read_csv(
"/kaggle/input/asl-fingerspelling/supplemental_metadata.csv",
delimiter=",",
encoding="UTF-8",
)
pd.set_option("display.max_columns", None)
data.head(3)
## get count of unique phrases
phrase_count = data["phrase"].value_counts().to_list()
unique_phrase = data["phrase"].unique()
len(phrase_count), len(unique_phrase)
# type(phrase_count),type(unique_phrase)
# ### create separete dataframe to store phrases and their value counts
data2 = {"phrases": list(unique_phrase), "phrase_count": phrase_count}
phrase_data = pd.DataFrame(data2)
phrase_data.head(10)
# ### visualize data for 5 most frequent and least frequent phrases
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
import plotly.graph_objects as go
fig = px.bar(
phrase_data.iloc[:5, :],
x="phrase_count",
y="phrases",
color="phrases",
orientation="h",
)
fig.update_layout(
title={
"text": "count of top 5 most frequent phrases",
"y": 0.96,
"x": 0.4,
"xanchor": "center",
"yanchor": "top",
},
legend_title_text="Aspect:",
)
fig.show()
fig = px.bar(
phrase_data.iloc[504:508, :],
x="phrase_count",
y="phrases",
color="phrases",
orientation="h",
)
fig.update_layout(
title={
"text": "count of 5 least phrases",
"y": 0.96,
"x": 0.4,
"xanchor": "center",
"yanchor": "top",
},
legend_title_text="Aspect:",
)
fig.show()
# ## loading parquest file of
##create subset of dataset where phrase is "coming up with killer sound bites"
top_phrase = data[data["phrase"] == "coming up with killer sound bites"]["path"].values[
0
]
top_phrase
base_dir = Path("/kaggle/input/asl-fingerspelling")
# ### explore landmark file of top_phrase
landmark_file = pd.read_parquet(base_dir / top_phrase)
landmark_file.head()
landmark_file = landmark_file.reset_index(inplace=False)
landmark_file.head()
# len(landmark_file.columns) ## 1630
landmark_file.shape
# view number of unique sequence_ids in dataset
# landmark_file["sequence_id"].nunique() # 1000
# return 1st two sequence_ids
landmark_file["sequence_id"].unique()[:2]
# landmark_file["frame"].nunique() # 507
# fetch landmark data for sequence id=1535467051
landmark_1st_id = landmark_file[landmark_file["sequence_id"] == 1535467051]
landmark_1st_id
# ### explore train file
train_data = pd.read_csv("/kaggle/input/asl-fingerspelling/train.csv")
train_data.shape
train_data.head()
# #### explore file ->/kaggle/input/asl-fingerspelling/character_to_prediction_index.json
char_to_pred = "/kaggle/input/asl-fingerspelling/character_to_prediction_index.json"
# Python program to read
# json file
char = []
values = []
import json
# Opening JSON file
f = open(char_to_pred)
# returns JSON object as
# a dictionary
data = json.load(f)
# Iterating through the json
# list
for i, j in data.items():
char.append(i)
values.append(j)
# print("key:"+str(i),"values:"+str(j))
# Closing file
f.close()
# print("\n characters list:",char)
# print("\n values list:",values)
char_to_pred_index = pd.DataFrame({"char": char, "values": values})
char_to_pred_index.head(20)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/210/129210213.ipynb
| null | null |
[{"Id": 129210213, "ScriptId": 38413845, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11270020, "CreationDate": "05/11/2023 21:20:12", "VersionNumber": 1.0, "Title": "EDA-ASL-Fingerspelling", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 174.0, "LinesInsertedFromPrevious": 23.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 151.0, "LinesInsertedFromFork": 23.0, "LinesDeletedFromFork": 5.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 151.0, "TotalVotes": 4}]
| null | null | null | null |
# # ASL- Fingerspelling
# ## What is American Sign Language Fingerspelling Recognition ?
# American Sign Language Fingerspelling Recognition is a technology that uses computer vision and machine learning algorithms to recognize and interpret the hand gestures used in American Sign Language (ASL) fingerspelling. It can be used to create tools and applications that help people with hearing impairments to communicate more effectively with others. The technology involves comparing the input image of the hand gesture to a pre-defined set of templates, extracting relevant features from the input image, or training a neural network on a large dataset of ASL fingerspelling images to learn the patterns and features that are most important for recognition. Despite some challenges, ASL Fingerspelling Recognition has the potential to greatly improve the lives of people with hearing impairments.
# ## Data Overview
# ### Files
# #### [train/supplemental_metadata].csv
# * path - The path to the landmark file.
# * file_id - A unique identifier for the data file.
# * participant_id - A unique identifier for the data contributor.
# * sequence_id - A unique identifier for the landmark sequence. Each data file may contain many sequences.
# * phrase - The labels for the landmark sequence. The train and test datasets contain randomly generated addresses, phone numbers, and urls derived from components of real addresses/phone numbers/urls. Any overlap with real addresses, phone numbers, or urls is purely accidental. The supplemental dataset consists of fingerspelled sentences. Note that some of the urls include adult content. The intent of this competition is to support the Deaf and Hard of Hearing community in engaging with technology on an equal footing with other adults.
# ### character_to_prediction_index.json
# #### [train/supplemental]_landmarks/
# The landmark data. The landmarks were extracted from raw videos with the MediaPipe holistic model. Not all of the frames necessarily had visible hands or hands that could be detected by the model.
# The landmark files contain the same data as in the ASL Signs competition (minus the row ID column) but reshaped into a wide format. This allows you to take advantage of the Parquet format to entirely skip loading landmarks that you aren't using.
# * sequence_id - A unique identifier for the landmark sequence. Most landmark files contain 1,000 sequences. The sequence ID is used as the dataframe index.
# * frame - The frame number within a landmark sequence.
# * [x/y/z]_[type]_[landmark_index] - There are now 1,629 spatial coordinate columns for the x, y and z coordinates for each of the 543 landmarks. The type of landmark is one of ['face', 'left_hand', 'pose', 'right_hand']. Details of the hand landmark locations can be found here. The spatial coordinates have already been normalized by MediaPipe. Note that the MediaPipe model is not fully trained to predict depth so you may wish to ignore the z values. The landmarks have been converted to float32.
# ## explore dataset
### import libraries
import pandas as pd, numpy as np, os
from pathlib import Path
print("importing..")
# ### explore supplemental_metadata
data = pd.read_csv(
"/kaggle/input/asl-fingerspelling/supplemental_metadata.csv",
delimiter=",",
encoding="UTF-8",
)
pd.set_option("display.max_columns", None)
data.head(3)
## get count of unique phrases
phrase_count = data["phrase"].value_counts().to_list()
unique_phrase = data["phrase"].unique()
len(phrase_count), len(unique_phrase)
# type(phrase_count),type(unique_phrase)
# ### create separete dataframe to store phrases and their value counts
data2 = {"phrases": list(unique_phrase), "phrase_count": phrase_count}
phrase_data = pd.DataFrame(data2)
phrase_data.head(10)
# ### visualize data for 5 most frequent and least frequent phrases
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
import plotly.graph_objects as go
fig = px.bar(
phrase_data.iloc[:5, :],
x="phrase_count",
y="phrases",
color="phrases",
orientation="h",
)
fig.update_layout(
title={
"text": "count of top 5 most frequent phrases",
"y": 0.96,
"x": 0.4,
"xanchor": "center",
"yanchor": "top",
},
legend_title_text="Aspect:",
)
fig.show()
fig = px.bar(
phrase_data.iloc[504:508, :],
x="phrase_count",
y="phrases",
color="phrases",
orientation="h",
)
fig.update_layout(
title={
"text": "count of 5 least phrases",
"y": 0.96,
"x": 0.4,
"xanchor": "center",
"yanchor": "top",
},
legend_title_text="Aspect:",
)
fig.show()
# ## loading parquest file of
##create subset of dataset where phrase is "coming up with killer sound bites"
top_phrase = data[data["phrase"] == "coming up with killer sound bites"]["path"].values[
0
]
top_phrase
base_dir = Path("/kaggle/input/asl-fingerspelling")
# ### explore landmark file of top_phrase
landmark_file = pd.read_parquet(base_dir / top_phrase)
landmark_file.head()
landmark_file = landmark_file.reset_index(inplace=False)
landmark_file.head()
# len(landmark_file.columns) ## 1630
landmark_file.shape
# view number of unique sequence_ids in dataset
# landmark_file["sequence_id"].nunique() # 1000
# return 1st two sequence_ids
landmark_file["sequence_id"].unique()[:2]
# landmark_file["frame"].nunique() # 507
# fetch landmark data for sequence id=1535467051
landmark_1st_id = landmark_file[landmark_file["sequence_id"] == 1535467051]
landmark_1st_id
# ### explore train file
train_data = pd.read_csv("/kaggle/input/asl-fingerspelling/train.csv")
train_data.shape
train_data.head()
# #### explore file ->/kaggle/input/asl-fingerspelling/character_to_prediction_index.json
char_to_pred = "/kaggle/input/asl-fingerspelling/character_to_prediction_index.json"
# Python program to read
# json file
char = []
values = []
import json
# Opening JSON file
f = open(char_to_pred)
# returns JSON object as
# a dictionary
data = json.load(f)
# Iterating through the json
# list
for i, j in data.items():
char.append(i)
values.append(j)
# print("key:"+str(i),"values:"+str(j))
# Closing file
f.close()
# print("\n characters list:",char)
# print("\n values list:",values)
char_to_pred_index = pd.DataFrame({"char": char, "values": values})
char_to_pred_index.head(20)
| false | 0 | 1,805 | 4 | 1,805 | 1,805 |
||
129384337
|
<jupyter_start><jupyter_text>Spotify Dataset 1921-2020, 600k+ Tracks
### About
For more in-depth information about audio features provided by Spotify: [https://developer.spotify.com/documentation/web-api/reference/#/operations/get-audio-features](https://developer.spotify.com/documentation/web-api/reference/#/operations/get-audio-features)
I reposted my old dataset as many people requested. I don't consider updating the dataset further.
### Meta-information
**Title:** Spotify Dataset 1921-2020, 600k+ Tracks
**Subtitle:** Audio features of 600k+ tracks, popularity metrics of 1M+ artists
**Source:** Spotify Web API
**Creator:** Yamac Eren Ay
**Release Date (of Last Version):** April 2021
**Link to this dataset:** https://www.kaggle.com/yamaerenay/spotify-dataset-19212020-600k-tracks
**Link to the old dataset:** https://www.kaggle.com/yamaerenay/spotify-dataset-19212020-160k-tracks
### Disclaimer
I am not posting here third-party Spotify data for arbitrary reasons or getting upvote.
The old dataset has been mentioned in tens of scientific papers using the old link which doesn't work anymore since July 2021, and most of the authors had some problems proving the validity of the dataset. You can cite the same dataset under the new link. I'll be posting more information regarding the old dataset.
If you have inquiries or complaints, please don't hesitate to reach out to me on LinkedIn or you can send me an email.
Kaggle dataset identifier: spotify-dataset-19212020-600k-tracks
<jupyter_script>import numpy as np
import pandas as pd
from scipy import stats
import matplotlib.pyplot as plt
import seaborn as sns
import datetime as dt
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
# * release_date değişkeni datetime kullanılarak sadece yıl olacak şekilde değiştirilecek
# * nümerik değişkenler bulunacak ve kategorik değişkenler bulunacak
# * modele sokulmayacak kardinal değişkenler çıkarılacak
# * release_date değişkenine bugünün yılından çıkarılacak ve segmentlere ayırılacak
df = pd.read_csv("/kaggle/input/spotify-dataset-19212020-600k-tracks/tracks.csv")
df.head()
df.info()
df.shape
df.isna().sum()
df.dropna(inplace=True)
df.isna().sum()
df["release_date"] = pd.to_datetime(df["release_date"])
df["release_date"] = df["release_date"].apply(lambda x: x.strftime("%Y"))
df["release_date"] = pd.DatetimeIndex(df["release_date"]).year
df["track_age_FLAG"] = df["release_date"].max() - df["release_date"]
df = df[df["track_age_FLAG"] < 76]
df["era_FLAG"] = pd.cut(
df["release_date"],
[1945, 1949, 1959, 1969, 1979, 1989, 1999, 2009, 2021],
labels=["40s", "50s", "60s", "70s", "80s", "90s", "00s", "10s"],
ordered=False,
)
df["era_FLAG"]
df["era_FLAG"].value_counts()
df["artists"].nunique()
def grab_col_names(dataframe, cat_th=13, car_th=20):
"""
Veri setindeki kategorik, numerik ve kategorik fakat kardinal değişkenlerin isimlerini verir.
Not: Kategorik değişkenlerin içerisine numerik görünümlü kategorik değişkenler de dahildir.
Parameters
------
dataframe: dataframe
Değişken isimleri alınmak istenilen dataframe
cat_th: int, optional
numerik fakat kategorik olan değişkenler için sınıf eşik değeri
car_th: int, optinal
kategorik fakat kardinal değişkenler için sınıf eşik değeri
Returns
------
cat_cols: list
Kategorik değişken listesi
num_cols: list
Numerik değişken listesi
cat_but_car: list
Kategorik görünümlü kardinal değişken listesi
Examples
------
import seaborn as sns
df = sns.load_dataset("iris")
print(grab_col_names(df))
Notes
------
cat_cols + num_cols + cat_but_car = toplam değişken sayısı
num_but_cat cat_cols'un içerisinde.
Return olan 3 liste toplamı toplam değişken sayısına eşittir: cat_cols + num_cols + cat_but_car = değişken sayısı
"""
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
# print(f"Observations: {dataframe.shape[0]}")
# print(f"Variables: {dataframe.shape[1]}")
# print(f'cat_cols: {len(cat_cols)}')
# print(f'num_cols: {len(num_cols)}')
# print(f'cat_but_car: {len(cat_but_car)}')
# print(f'num_but_cat: {len(num_but_cat)}')
return cat_cols, num_cols, cat_but_car
grab_col_names(df)
df.head()
fig, axs = plt.subplots(nrows=3, ncols=4, constrained_layout=True, figsize=(20, 15))
sns.histplot(ax=axs[0][0], x=df["popularity"], color="#4C4C6D")
sns.histplot(ax=axs[0][1], x=df["duration_ms"], color="#99627A")
sns.histplot(ax=axs[0][2], x=df["danceability"], color="#1D267D")
sns.histplot(ax=axs[0][3], x=df["energy"], color="#B04759")
sns.histplot(ax=axs[1][0], x=df["loudness"], color="#8B1874")
sns.histplot(ax=axs[1][1], x=df["speechiness"], color="#212A3E")
sns.histplot(ax=axs[1][2], x=df["acousticness"], color="#00FFCA")
sns.histplot(ax=axs[1][3], x=df["instrumentalness"], color="#F6BA6F")
sns.histplot(ax=axs[2][0], x=df["liveness"], color="#F45050")
sns.histplot(ax=axs[2][1], x=df["valence"], color="#263A29")
sns.histplot(ax=axs[2][2], x=df["tempo"], color="#6D5D6E")
sns.histplot(ax=axs[2][3], x=df["track_age_FLAG"], color="#B2A4FF")
plt.show()
fig, axs = plt.subplots(nrows=1, ncols=5, constrained_layout=True, figsize=(20, 4))
sns.barplot(ax=axs[0], x=df["explicit"], y=df["explicit"].index)
sns.barplot(ax=axs[1], x=df["key"], y=df["key"].index)
sns.barplot(ax=axs[2], x=df["mode"], y=df["mode"].index)
sns.barplot(ax=axs[3], x=df["time_signature"], y=df["time_signature"].index)
sns.barplot(ax=axs[4], x=df["era_FLAG"], y=df["era_FLAG"].index)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/384/129384337.ipynb
|
spotify-dataset-19212020-600k-tracks
|
yamaerenay
|
[{"Id": 129384337, "ScriptId": 38070140, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9168994, "CreationDate": "05/13/2023 10:30:49", "VersionNumber": 2.0, "Title": "Spotify Recommender System", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 136.0, "LinesInsertedFromPrevious": 89.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 47.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185382274, "KernelVersionId": 129384337, "SourceDatasetVersionId": 3294812}]
|
[{"Id": 3294812, "DatasetId": 1993933, "DatasourceVersionId": 3345531, "CreatorUserId": 4969123, "LicenseName": "Community Data License Agreement - Sharing - Version 1.0", "CreationDate": "03/13/2022 21:12:47", "VersionNumber": 1.0, "Title": "Spotify Dataset 1921-2020, 600k+ Tracks", "Slug": "spotify-dataset-19212020-600k-tracks", "Subtitle": "Audio features of 600k+ tracks, popularity metrics of 1M+ artists", "Description": "### About\n\nFor more in-depth information about audio features provided by Spotify: [https://developer.spotify.com/documentation/web-api/reference/#/operations/get-audio-features](https://developer.spotify.com/documentation/web-api/reference/#/operations/get-audio-features)\n\nI reposted my old dataset as many people requested. I don't consider updating the dataset further.\n\n### Meta-information\n\n**Title:** Spotify Dataset 1921-2020, 600k+ Tracks\n**Subtitle:** Audio features of 600k+ tracks, popularity metrics of 1M+ artists\n**Source:** Spotify Web API\n**Creator:** Yamac Eren Ay\n**Release Date (of Last Version):** April 2021\n**Link to this dataset:** https://www.kaggle.com/yamaerenay/spotify-dataset-19212020-600k-tracks\n**Link to the old dataset:** https://www.kaggle.com/yamaerenay/spotify-dataset-19212020-160k-tracks\n\n### Disclaimer\n\nI am not posting here third-party Spotify data for arbitrary reasons or getting upvote. \n\nThe old dataset has been mentioned in tens of scientific papers using the old link which doesn't work anymore since July 2021, and most of the authors had some problems proving the validity of the dataset. You can cite the same dataset under the new link. I'll be posting more information regarding the old dataset. \n\nIf you have inquiries or complaints, please don't hesitate to reach out to me on LinkedIn or you can send me an email.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1993933, "CreatorUserId": 4969123, "OwnerUserId": 4969123.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3294812.0, "CurrentDatasourceVersionId": 3345531.0, "ForumId": 2018387, "Type": 2, "CreationDate": "03/13/2022 21:12:47", "LastActivityDate": "03/13/2022", "TotalViews": 49154, "TotalDownloads": 6976, "TotalVotes": 86, "TotalKernels": 7}]
|
[{"Id": 4969123, "UserName": "yamaerenay", "DisplayName": "Yamac Eren Ay", "RegisterDate": "04/27/2020", "PerformanceTier": 2}]
|
import numpy as np
import pandas as pd
from scipy import stats
import matplotlib.pyplot as plt
import seaborn as sns
import datetime as dt
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
# * release_date değişkeni datetime kullanılarak sadece yıl olacak şekilde değiştirilecek
# * nümerik değişkenler bulunacak ve kategorik değişkenler bulunacak
# * modele sokulmayacak kardinal değişkenler çıkarılacak
# * release_date değişkenine bugünün yılından çıkarılacak ve segmentlere ayırılacak
df = pd.read_csv("/kaggle/input/spotify-dataset-19212020-600k-tracks/tracks.csv")
df.head()
df.info()
df.shape
df.isna().sum()
df.dropna(inplace=True)
df.isna().sum()
df["release_date"] = pd.to_datetime(df["release_date"])
df["release_date"] = df["release_date"].apply(lambda x: x.strftime("%Y"))
df["release_date"] = pd.DatetimeIndex(df["release_date"]).year
df["track_age_FLAG"] = df["release_date"].max() - df["release_date"]
df = df[df["track_age_FLAG"] < 76]
df["era_FLAG"] = pd.cut(
df["release_date"],
[1945, 1949, 1959, 1969, 1979, 1989, 1999, 2009, 2021],
labels=["40s", "50s", "60s", "70s", "80s", "90s", "00s", "10s"],
ordered=False,
)
df["era_FLAG"]
df["era_FLAG"].value_counts()
df["artists"].nunique()
def grab_col_names(dataframe, cat_th=13, car_th=20):
"""
Veri setindeki kategorik, numerik ve kategorik fakat kardinal değişkenlerin isimlerini verir.
Not: Kategorik değişkenlerin içerisine numerik görünümlü kategorik değişkenler de dahildir.
Parameters
------
dataframe: dataframe
Değişken isimleri alınmak istenilen dataframe
cat_th: int, optional
numerik fakat kategorik olan değişkenler için sınıf eşik değeri
car_th: int, optinal
kategorik fakat kardinal değişkenler için sınıf eşik değeri
Returns
------
cat_cols: list
Kategorik değişken listesi
num_cols: list
Numerik değişken listesi
cat_but_car: list
Kategorik görünümlü kardinal değişken listesi
Examples
------
import seaborn as sns
df = sns.load_dataset("iris")
print(grab_col_names(df))
Notes
------
cat_cols + num_cols + cat_but_car = toplam değişken sayısı
num_but_cat cat_cols'un içerisinde.
Return olan 3 liste toplamı toplam değişken sayısına eşittir: cat_cols + num_cols + cat_but_car = değişken sayısı
"""
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
# print(f"Observations: {dataframe.shape[0]}")
# print(f"Variables: {dataframe.shape[1]}")
# print(f'cat_cols: {len(cat_cols)}')
# print(f'num_cols: {len(num_cols)}')
# print(f'cat_but_car: {len(cat_but_car)}')
# print(f'num_but_cat: {len(num_but_cat)}')
return cat_cols, num_cols, cat_but_car
grab_col_names(df)
df.head()
fig, axs = plt.subplots(nrows=3, ncols=4, constrained_layout=True, figsize=(20, 15))
sns.histplot(ax=axs[0][0], x=df["popularity"], color="#4C4C6D")
sns.histplot(ax=axs[0][1], x=df["duration_ms"], color="#99627A")
sns.histplot(ax=axs[0][2], x=df["danceability"], color="#1D267D")
sns.histplot(ax=axs[0][3], x=df["energy"], color="#B04759")
sns.histplot(ax=axs[1][0], x=df["loudness"], color="#8B1874")
sns.histplot(ax=axs[1][1], x=df["speechiness"], color="#212A3E")
sns.histplot(ax=axs[1][2], x=df["acousticness"], color="#00FFCA")
sns.histplot(ax=axs[1][3], x=df["instrumentalness"], color="#F6BA6F")
sns.histplot(ax=axs[2][0], x=df["liveness"], color="#F45050")
sns.histplot(ax=axs[2][1], x=df["valence"], color="#263A29")
sns.histplot(ax=axs[2][2], x=df["tempo"], color="#6D5D6E")
sns.histplot(ax=axs[2][3], x=df["track_age_FLAG"], color="#B2A4FF")
plt.show()
fig, axs = plt.subplots(nrows=1, ncols=5, constrained_layout=True, figsize=(20, 4))
sns.barplot(ax=axs[0], x=df["explicit"], y=df["explicit"].index)
sns.barplot(ax=axs[1], x=df["key"], y=df["key"].index)
sns.barplot(ax=axs[2], x=df["mode"], y=df["mode"].index)
sns.barplot(ax=axs[3], x=df["time_signature"], y=df["time_signature"].index)
sns.barplot(ax=axs[4], x=df["era_FLAG"], y=df["era_FLAG"].index)
plt.show()
| false | 1 | 1,801 | 0 | 2,243 | 1,801 |
||
129384194
|
# # Capstone Project: Facial Emotion Detection
# Michael Hogge - April 2023
# # Executive Summary
# Emotion AI, also known as artificial emotional intelligence, is a subset of artificial intelligence dealing with the detection and replication of human emotion by machines. The successful creation of this "artificial empathy" hinges on a computer's ability to analyze, among other things, human text, speech, and facial expressions. In support of these efforts, this project leverages the power of convolutional neural networks (CNN) to create a computer vision model capable of accurately performing multi-class classification on images containing one of four facial expressions: happy, sad, neutral, and surprise.
# Data provided for this project includes over 20,000 grayscale images split into training (75%), validation (24.5%), and test (0.5%) datasets, and further divided into the aforementioned classes. At the outset of the project, a visual analysis of the data is undertaken and a slight imbalance is noted in the class distribution, with 'surprise' images making up a smaller percentage of total images when compared to 'happy,' 'sad,' and 'neutral' images. The unique characteristics of each class are discussed (e.g., images labeled as 'surprise' tend to contain faces with wide open mouths and eyes), including a breakdown of average pixel value by class.
# Following the data visualization and analysis phase of the project, nine CNNs are developed, ranging from simple grayscale models to complex transfer learning architectures comprised of hundreds of layers and tens of millions of parameters. Basic models are shown to be lacking the required amount of complexity to properly fit the data, while the transfer learning models (VGG16, ResNet v2, and EfficientNet) are shown to be too complex for the amount and type of data provided for this project. The unsatisfactory performance of the basic and transfer learning models necessitates the development of an alternative model capable of fitting the data and achieving acceptable levels of accuracy while maintaining a high level of generalizability. The proposed model, with four convolutional blocks and 1.8 million parameters, displays high accuracy (75% on training, validation, and test data) when compared to human performance (±65%) on similar data, and avoids overfitting the training data, which can be difficult to achieve with CNNs.
# The deployability of this model depends entirely on its intended use. With an accuracy of 75%, deployment in a marketing or gaming setting is perfectly reasonable, assuming consent has been granted, and the handling of highly personal data is done in an ethical, transparent manner with data privacy coming before profit. However, deployment in circumstances where the output from this model could cause serious material damage to an individual (e.g., hiring decisions, law enforcement, evidence in a court of law, etc.) should be avoided. While computer vision models can become quite skilled at classifying human facial expressions (particularly if they are trained on over-emoting/exaggerated images), it is important to note that a connection between those expressions and any underlying emotion is not a hard scientific fact. For example, a smiling person may not always be happy (e.g., they could be uncomfortable or polite), a crying person may not always be sad (e.g., they could be crying tears of joy), and someone who is surprised may be experiencing compound emotions (e.g., happily surprised or sadly surprised).
# There is certainly scope to improve the proposed model, including the ethical sourcing of additional, diverse training images, and additional data augmentation on top of what is already performed during the development of the proposed model. In certain scenarios, as indicated above, model deployment could proceed with 75% accuracy, and continued improvement could be pursued by the business/organization/government as time and funding allows. Before model deployment, a set of guiding ethical principles should be developed and adhered to throughout the data collection, analysis, and (possibly) storage phase. Stakeholders must ensure transparency throughout all stages of the computer vision life cycle, while monitoring the overall development of Emotion AI technology and anticipating future regulatory action, which appears likely.
# ## **Problem Definition**
# **Context:**
# How do humans communicate with one another? While spoken and written communication may immediately come to mind, research by Dr. Albert Mehrabian has found that over 50% of communication is conveyed through body language, including facial expressions. In face-to-face conversation, body language, it turns out, plays a larger role in how our message is interpreted than both the words we choose, and the tone with which we deliver them. Our expression is a powerful window into our true feelings, and as such, it can be used as a highly-effective proxy for sentiment, particularly in the absence of written or spoken communication.
# Emotion AI (artificial emotional intelligence, or affective computing), attempts to leverage this proxy for sentiment by detecting and processing facial expression (through neural networks) in an effort to successfully interpret human emotion and respond appropriately. Developing models that can accurately detect facial emotion is therefore an important driver of advancement in the realm of artificial intelligence and emotionally intelligent machines. The ability to successfully extract sentiment from images and video is also a powerful tool for businesses looking to conjure insights from the troves of unstructured data they have accumulated in recent years, or even to extract second-by-second customer responses to advertisements, store layouts, customer/user experience, etc.
# **Objective:**
# The objective of this project is to utilize deep learning techniques, including convolutional neural networks, to create a computer vision model that can accurately detect and interpret facial emotions. This model should be capable of performing multi-class classification on images containing one of four facial expressions: happy, sad, neutral, and surprise.
# **Key Questions:**
# * Do we have the data necessary to develop our models, and is it of good enough quality and quantity?
# * What is the best type of machine learning model to achieve our objective?
# * What do we consider 'success' when it comes to model performance?
# * How do different models compare to one another given this definition of success?
# * What are the most important insights that can be drawn from this project upon its conclusion?
# * What is the final proposed model and is it good enough for deployment?
# ## **About the Dataset**
# The data set consists of 3 folders, i.e., 'test', 'train', and 'validation'.
# Each of these folders has four subfolders:
# **‘happy’**: Images of people who have happy facial expressions.
# **‘sad’**: Images of people with sad or upset facial expressions.
# **‘surprise’**: Images of people who have shocked or surprised facial expressions.
# **‘neutral’**: Images of people showing no prominent emotion in their facial expression at all.
# ## **Importing the Libraries**
import zipfile # Used to unzip the data
import numpy as np # Mathematical functions, arrays, etc.
import pandas as pd # Data manipulation and analysis
import os # Misc operating system interfaces
import h5py # Read and write h5py files
import random
import matplotlib.pyplot as plt # A library for data visualization
from matplotlib import image as mpimg # Used to show images from filepath
import seaborn as sns # An advanced library for data visualization
from PIL import Image # Image processing
import cv2 # Image processing
# Importing Deep Learning Libraries, layers, models, optimizers, etc
import tensorflow as tf
from tensorflow.keras.preprocessing.image import (
load_img,
img_to_array,
ImageDataGenerator,
)
from tensorflow.keras.layers import (
Dense,
Input,
Dropout,
SpatialDropout2D,
GlobalAveragePooling2D,
Flatten,
Conv2D,
BatchNormalization,
Activation,
MaxPooling2D,
LeakyReLU,
GaussianNoise,
)
from tensorflow.keras.models import Model, Sequential
from tensorflow.keras.optimizers import Adam, SGD, RMSprop, Adadelta
from tensorflow.keras import regularizers
from keras.regularizers import l2
from tensorflow.keras.losses import categorical_crossentropy
from tensorflow.keras.utils import to_categorical
import tensorflow.keras.applications as ap
from tensorflow.keras.applications.vgg16 import VGG16
from keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
from tensorflow.keras import backend
# Reproducibility within TensorFlow
import fwr13y.d9m.tensorflow as tf_determinism
tf_determinism.enable_determinism()
tf.config.experimental.enable_op_determinism
from tqdm import tqdm # Generates progress bars
# Predictive data analysis tools
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
# To suppress warnings
import warnings
warnings.filterwarnings("ignore")
# Needed to silence tensorflow messages while running locally
from silence_tensorflow import silence_tensorflow
silence_tensorflow()
# Fixing the seed for random number generators to ensure reproducibility
np.random.seed(42)
random.seed(42)
tf.random.set_seed(42)
# Ensuring reproducibility using GPU with TensorFlow
os.environ["TF_DETERMINISTIC_OPS"] = "1"
# ### **Loading and Unzipping the Data**
# Extracting image files from the zip file
with zipfile.ZipFile("Facial_emotion_images.zip", "r") as zip_ref:
zip_ref.extractall()
dir_train = "Facial_emotion_images/train/" # Path of training data after unzipping
dir_validation = (
"Facial_emotion_images/validation/" # Path of validation data after unzipping
)
dir_test = "Facial_emotion_images/test/" # Path of test data after unzipping
img_size = 48 # Defining the size of the image as 48 pixels
# ## **Visualizing our Classes**
#
# Custom function to display first 35 images from the specified training folder
def display_emotion(emotion):
train_emotion = dir_train + emotion + "/"
plt.figure(figsize=(11, 11))
for i in range(1, 36):
plt.subplot(5, 7, i)
img = load_img(
train_emotion + os.listdir(train_emotion)[i],
target_size=(img_size, img_size),
)
plt.imshow(img)
plt.show()
# ### **Happy**
print("These are the first 35 training images labeled as 'Happy':")
display_emotion("happy")
# An example image pulled from the images above
img_x = os.listdir("Facial_emotion_images/train/happy/")[16]
img_happy_16 = mpimg.imread("Facial_emotion_images/train/happy/" + img_x)
plt.figure(figsize=(2, 2))
plt.imshow(img_happy_16, cmap="Greys_r")
plt.show()
# **Observations and Insights: Happy**
# * In most images, the person is smiling. Some smiles are with an open mouth with teeth visible, and some are with closed lips. Our models will need to learn both types of smiles.
# * Image brightness varies considerably and will need to be addressed with data augmentation.
# * The ages of the people vary from very young to old.
# * In some images, people are wearing eyeglasses or hats, eating food, or are partially covering their face with their hands. Some images contain watermarks.
# * Some images are head-on while some are sideways. We will address this via data augmentation (rotating and/or flipping images).
# * Images are cropped differently and this will need to be addressed with data augmentation (zoom/crop).
# * Some images do not contain a face (though not shown in the above 35). There is really nothing to do about this ... some of the test images also do not contain a face. As a result, some predictions by the final model will be based on incorrect data labeling.
# * As highlighted by the image above, some images are of non-human faces. In this case, it appears to be a statue with exaggerated features.
# * Some 'happy' images would clearly not be classified as 'happy' by a human being. This brings into question what accuracy really means. If the model correctly predicts an image labeled 'happy' as 'happy', should that be considered accurate if the person in the image is actually frowning and would be considered by a human being to be sad? In a high-stakes, real world situation we could potentially relabel images that have been incorrectly labeled, but in the context of this project, we have been advised to leave the training labels untouched.
# ### **Sad**
print("These are the first 35 training images labeled as 'Sad':")
display_emotion("sad")
# An example image pulled from the images above
img_x = os.listdir("Facial_emotion_images/train/sad/")[7]
img_sad_7 = mpimg.imread("Facial_emotion_images/train/sad/" + img_x)
plt.figure(figsize=(2, 2))
plt.imshow(img_sad_7, cmap="Greys_r")
plt.show()
# **Observations and Insights: Sad**
# * In most images, the person is frowning. In many images, people have their eyes closed or are looking down.
# * Compared to the 'happy' images, people labeled 'sad' seem more likely to have their mouths closed.
# * Similar to the 'happy' images, image brightness varies considerably, as does age. Some images are head-on while others are sideways. Some people are covering their face with their hands. As with 'happy' images, 'sad' images are also cropped differently, while some also have watermarks.
# * Some images do not contain a face (though not shown in the above 35).
# * As highlighted by the image above, some images are labeled 'sad' but would probably not be classified as sad by a human being. The person above appears to be smiling, but an accurate prediction by one of our models would classify the image as 'sad'. This raises the same issue about accuracy mentioned earlier.
# * At first glance, apart from the images that are clearly mislabeled, there appears to be enough difference between the 'happy' and 'sad' characteristics that an effective model should be able to tell them apart relatively easily.
# ### **Neutral**
print("These are the first 35 training images labeled as 'Neutral':")
display_emotion("neutral")
# An example image pulled from the images above
img_x = os.listdir("Facial_emotion_images/train/neutral/")[26]
img_neutral_26 = mpimg.imread("Facial_emotion_images/train/neutral/" + img_x)
plt.figure(figsize=(2, 2))
plt.imshow(img_neutral_26, cmap="Greys_r")
plt.show()
# **Observations and Insights: Neutral**
# * At first glance, this seems to be the most difficult label to accurately predict. While 'happy' and 'sad' images appear different enough that a model should be able to tell the difference, 'neutral' faces are in between 'happy' and 'sad', and consequently similar to both.
# * Similar to the other classes discussed above, differences in brightness, age, zoom, hands covering faces, etc. are apparent in the 'neutral' images as well.
# * As highlighted in the image above, some images are simply mistakes and do not contain any faces at all.
# * These neutral images seem more difficult for a human being to correctly classify. Some people appear to be slightly smiling, while others appear to be slightly frowning. This begs the question, where are the lines between happy/neutral and sad/neutral? Neutral images do appear to be more similar to sad images, so it is possible that our models will confuse the two classes.
# ### **Surprise**
print("These are the first 35 training images labeled as 'Surprise':")
display_emotion("surprise")
# An example image pulled from the images above
img_x = os.listdir("Facial_emotion_images/train/surprise/")[17]
img_surprise_34 = mpimg.imread("Facial_emotion_images/train/surprise/" + img_x)
plt.figure(figsize=(2, 2))
plt.imshow(img_surprise_34, cmap="Greys_r")
plt.show()
# **Observations and Insights: Surprise**
# * The most unique characteristics of the 'surprise' images are open mouths and big, open eyes. These seem like features that a successful model should be able to identify and accurately classify. There is a big difference between 'surprise' images and 'neutral' images, for example. It is possible, however, that the open mouth of a 'happy' smile and the open mouth of a 'surprise' image could be difficult for a model to distinguish between.
# * As with the above classes, brightness, crop, age, etc. vary between images. Hands are often covering faces. Some photos are head-on while others face sideways.
# * The above image is an example with a very light pixel value, as opposed to one of the much darker images. The person in the image has the classic open mouth and wide open eyes. The image also contains a watermark.
# * Some images do not contain a face (though not shown in the above 35).
# **Overall Insights from Visualization of Classes:**
# * All images are in grayscale (black/white) and image size is 48 x 48 pixels. We will need to rescale pixel values by dividing by 255 so pixel values are normalized between 0 and 1. This will allow our models to train faster and help to stabilize gradient descent.
# * Some classes have very different characteristics (happy/sad) while other classes are more similar (sad/neutral) and could be challenging for a model to accurately classify.
# * There is a wide range of differences with respect to image brightness, age of person, zoom/crop of image, orientation of the face, objects/hands covering the face, images not containing any face at all, etc. Data augmentation will need to be taken into consideration, and this will be handled when the Data Loaders are created below.
# * Visualizing the images in this way raises an important question: what do we consider an accurate model prediction for an image that is clearly mislabeled? If a person is smiling but labeled as 'sad', and the model accurately predicts 'sad', is that really 'accurate' since a human being would classify the image as 'happy'? If the test data set is large, it would be difficult to go through each image individually to check accuracy (which would also defeat the purpose of creating a computer vision model in the first place), so we will have to live with 'accurate' model predictions that may not be truly accurate. Pondering questions like this lead one to believe that a model can really only be as good as the data it is trained on.
# ## **Checking Distribution of Classes**
# Getting the count of images in each training folder and saving to variables
train_happy = len(os.listdir(dir_train + "happy/"))
train_sad = len(os.listdir(dir_train + "sad/"))
train_neutral = len(os.listdir(dir_train + "neutral/"))
train_surprised = len(os.listdir(dir_train + "surprise/"))
# Creating a Pandas series called "train_series" and converting to Pandas dataframe called "train_df"
# in order to display the table below. The dataframe will also contribute to bar charts farther below.
train_series = pd.Series(
{
"Happy": train_happy,
"Sad": train_sad,
"Neutral": train_neutral,
"Surprised": train_surprised,
}
)
train_df = pd.DataFrame(train_series, columns=["Total Training Images"])
train_df["Percentage"] = round(
(train_df["Total Training Images"] / train_df["Total Training Images"].sum()) * 100,
1,
)
train_df.index.name = "Emotions"
print("The distribution of classes within the training data:")
train_df
train_df.sum()
# **Observations: Training Images**
# * There are 15,109 training images in total.
# * Happy, sad, and neutral images make up roughly the same share of total training images (26%), while surprise images make up a smaller share (21%). At this stage it is important to note the relatively small imbalance, though the ratio does not seem skewed enough to warrant future manipulation in terms of weights, etc.
# * The insight made above, that surprise images seem to be some of the most unique in terms of characteristics (big open mouth, big open eyes), may actually help us overcome the relatively minor imbalance. There are fewer surprise images, but they may be easier to classify.
#
# Getting count of images in each validation folder and saving to variables
val_happy = len(os.listdir(dir_validation + "happy/"))
val_sad = len(os.listdir(dir_validation + "sad/"))
val_neutral = len(os.listdir(dir_validation + "neutral/"))
val_surprised = len(os.listdir(dir_validation + "surprise/"))
# Creating a Pandas series called "val_series" and converting to Pandas dataframe called "val_df"
# in order to display the table below. The dataframe will also contribute to bar charts farther below.
val_series = pd.Series(
{
"Happy": val_happy,
"Sad": val_sad,
"Neutral": val_neutral,
"Surprised": val_surprised,
}
)
val_df = pd.DataFrame(val_series, columns=["Total Validation Images"])
val_df["Percentage"] = round(
(val_df["Total Validation Images"] / val_df["Total Validation Images"].sum()) * 100,
1,
)
val_df.index.name = "Emotions"
print("The distribution of classes within the validation data:")
val_df
val_df.sum()
# **Observations: Validation Images**
# * There are 4,977 validation images in total.
# * The distribution across classes is much more imbalanced. Happy images make up almost 37% of total validation images, while surprise images make up only 16%. As the training images and validation images are already split and provided as is, it is not a simple matter of randomly splitting training data with a train/test split. We are stuck with the imbalance.
# * One solution to address the imbalance could be to cap the other classes at the level of the surprise class, but that would throw away a huge portion of our already small data set.
# * As mentioned above, we can surmise that surprise images are easier to classify because of their unique characteristics, and we will see if that is enough to offset the relatively smaller sample size with which to train and validate.
#
# Getting count of images in each test folder and saving to variables
test_happy = len(os.listdir(dir_test + "happy/"))
test_sad = len(os.listdir(dir_test + "sad/"))
test_neutral = len(os.listdir(dir_test + "neutral/"))
test_surprised = len(os.listdir(dir_test + "surprise/"))
# Creating a Pandas series called "test_series" and converting to Pandas dataframe called "test_df"
# in order to display the table below. The dataframe will also contribute to bar charts farther below.
test_series = pd.Series(
{
"Happy": test_happy,
"Sad": test_sad,
"Neutral": test_neutral,
"Surprised": test_surprised,
}
)
test_df = pd.DataFrame(test_series, columns=["Total Test Images"])
test_df["Percentage"] = round(
(test_df["Total Test Images"] / test_df["Total Test Images"].sum()) * 100, 1
)
test_df.index.name = "Emotions"
print("The distribution of classes within the validation data:")
test_df
test_df.sum()
# **Observations: Test Images**
# * There are 128 test images in total, evenly divided between all four classes.
# * This even distribution will make interpretation of the final confusion matrix very straightforward.
# Concatenating train_df, val_df, and test_df to create "df_total" in order to create the chart below
df_total = pd.concat([train_df, val_df, test_df], axis=1)
df_total.drop(["Percentage"], axis=1, inplace=True)
df_total = df_total.reset_index()
df_total.rename(
columns={
"index": "Emotions",
"Total Training Images": "Train",
"Total Validation Images": "Validate",
"Total Test Images": "Test",
},
inplace=True,
)
# Creating bar chart below, grouped by class (i.e. 'emotion') and broken down into "train", "validate",
# and "test" data. The x-axis is Emotions and the y-axis is Total Images.
df_total.groupby("Emotions", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="TOTAL TRAINING, VALIDATION and TEST IMAGES",
ylabel="Total Images",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.show()
# **Observations:**
# * Depicted graphically, the distribution of classes is clearly imbalanced, but the imbalance is not overpowering.
# * Perhaps most striking is the tiny proportion of test images compared to training images. Rather than a standard machine learning train/validation/test split of 80/10/10 or 70/20/10, the data as provided for this project is 75% training, 24.5% validation, and just 0.5% test. As the data is provided already split into groups, we will work as intended. The vast majority of data will be used to train and then validate our models, with a tiny proportion used for testing. This should work in our favor, maximizing the amount of data used by our models to train.
#
# Concatenating train_df, val_df, and test_df to create "df_percent" in order to create the chart below
df_percent = pd.concat([train_df, val_df, test_df], axis=1)
df_percent.drop(
["Total Training Images", "Total Validation Images", "Total Test Images"],
axis=1,
inplace=True,
)
df_percent.columns = ["Train", "Validate", "Test"]
# Creating bar chart below, grouped by class (i.e. 'emotion') and broken down into "train", "validate",
# and "test" data. The x-axis is Emotions and the y-axis is Percentage of Total Images.
df_percent.groupby("Emotions", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="PERCENTAGE OF TOTAL TRAINING, VALIDATION and TEST IMAGES",
ylabel="Percentage of Total Images",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.show()
# **Observations:**
# * A visual depiction of what was discussed earlier. We can see the percentage breakdown of train/validate/test data across classes.
# * Training data is evenly distributed across happy, sad, and neutral classes, with fewer surprise images.
# * Within the validation data set, happy images clearly make up the largest percent of total images, with surprise images coming in a distant last place.
# * Happy images make up a much larger percentage of the validation data set than they do of the training and test data sets.
# * Surprise images make up a larger percentage of the test data set than they do of the training and validation data sets.
# Obtaining the average pixel value for training images in the class 'Happy'
list_train_happy = []
for i in range(len(os.listdir("Facial_emotion_images/train/happy/"))):
list_x = []
x = os.listdir("Facial_emotion_images/train/happy/")[i]
im = Image.open("Facial_emotion_images/train/happy/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_train_happy.append(sum(list_x) / len(pix_val))
train_happy_pixel_avg = round(sum(list_train_happy) / len(list_train_happy), 2)
# Obtaining the average pixel value for validation images in the class 'Happy'
list_val_happy = []
for i in range(len(os.listdir("Facial_emotion_images/validation/happy/"))):
list_x = []
x = os.listdir("Facial_emotion_images/validation/happy/")[i]
im = Image.open("Facial_emotion_images/validation/happy/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_val_happy.append(sum(list_x) / len(pix_val))
val_happy_pixel_avg = round(sum(list_val_happy) / len(list_val_happy), 2)
# Obtaining the average pixel value for test images in the class 'Happy'
list_test_happy = []
for i in range(len(os.listdir("Facial_emotion_images/test/happy/"))):
list_x = []
x = os.listdir("Facial_emotion_images/test/happy/")[i]
im = Image.open("Facial_emotion_images/test/happy/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_test_happy.append(sum(list_x) / len(pix_val))
test_happy_pixel_avg = round(sum(list_test_happy) / len(list_test_happy), 2)
# Obtaining the average pixel value for training images in the class 'Sad'
list_train_sad = []
for i in range(len(os.listdir("Facial_emotion_images/train/sad/"))):
list_x = []
x = os.listdir("Facial_emotion_images/train/sad/")[i]
im = Image.open("Facial_emotion_images/train/sad/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_train_sad.append(sum(list_x) / len(pix_val))
train_sad_pixel_avg = round(sum(list_train_sad) / len(list_train_sad), 2)
# Obtaining the average pixel value for validation images in the class 'Sad'
list_val_sad = []
for i in range(len(os.listdir("Facial_emotion_images/validation/sad/"))):
list_x = []
x = os.listdir("Facial_emotion_images/validation/sad/")[i]
im = Image.open("Facial_emotion_images/validation/sad/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_val_sad.append(sum(list_x) / len(pix_val))
val_sad_pixel_avg = round(sum(list_val_sad) / len(list_val_sad), 2)
# Obtaining the average pixel value for test images in the class 'Sad'
list_test_sad = []
for i in range(len(os.listdir("Facial_emotion_images/test/sad/"))):
list_x = []
x = os.listdir("Facial_emotion_images/test/sad/")[i]
im = Image.open("Facial_emotion_images/test/sad/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_test_sad.append(sum(list_x) / len(pix_val))
test_sad_pixel_avg = round(sum(list_test_sad) / len(list_test_sad), 2)
# Obtaining the average pixel value for training images in the class 'Neutral'
list_train_neutral = []
for i in range(len(os.listdir("Facial_emotion_images/train/neutral/"))):
list_x = []
x = os.listdir("Facial_emotion_images/train/neutral/")[i]
im = Image.open("Facial_emotion_images/train/neutral/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_train_neutral.append(sum(list_x) / len(pix_val))
train_neutral_pixel_avg = round(sum(list_train_neutral) / len(list_train_neutral), 2)
# Obtaining the average pixel value for validation images in the class 'Neutral'
list_val_neutral = []
for i in range(len(os.listdir("Facial_emotion_images/validation/neutral/"))):
list_x = []
x = os.listdir("Facial_emotion_images/validation/neutral/")[i]
im = Image.open("Facial_emotion_images/validation/neutral/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_val_neutral.append(sum(list_x) / len(pix_val))
val_neutral_pixel_avg = round(sum(list_val_neutral) / len(list_val_neutral), 2)
# Obtaining the average pixel value for test images in the class 'Neutral'
list_test_neutral = []
for i in range(len(os.listdir("Facial_emotion_images/test/neutral/"))):
list_x = []
x = os.listdir("Facial_emotion_images/test/neutral/")[i]
im = Image.open("Facial_emotion_images/test/neutral/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_test_neutral.append(sum(list_x) / len(pix_val))
test_neutral_pixel_avg = round(sum(list_test_neutral) / len(list_test_neutral), 2)
# Obtaining the average pixel value for training images in the class 'Surprise'
list_train_surprise = []
for i in range(len(os.listdir("Facial_emotion_images/train/surprise/"))):
list_x = []
x = os.listdir("Facial_emotion_images/train/surprise/")[i]
im = Image.open("Facial_emotion_images/train/surprise/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_train_surprise.append(sum(list_x) / len(pix_val))
train_surprise_pixel_avg = round(sum(list_train_surprise) / len(list_train_surprise), 2)
# Obtaining the average pixel value for validation images in the class 'Surprise'
list_val_surprise = []
for i in range(len(os.listdir("Facial_emotion_images/validation/surprise/"))):
list_x = []
x = os.listdir("Facial_emotion_images/validation/surprise/")[i]
im = Image.open("Facial_emotion_images/validation/surprise/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_val_surprise.append(sum(list_x) / len(pix_val))
val_surprise_pixel_avg = round(sum(list_val_surprise) / len(list_val_surprise), 2)
# Obtaining the average pixel value for test images in the class 'Surprise'
list_test_surprise = []
for i in range(len(os.listdir("Facial_emotion_images/test/surprise/"))):
list_x = []
x = os.listdir("Facial_emotion_images/test/surprise/")[i]
im = Image.open("Facial_emotion_images/test/surprise/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_test_surprise.append(sum(list_x) / len(pix_val))
test_surprise_pixel_avg = round(sum(list_test_surprise) / len(list_test_surprise), 2)
# creating dictionary containing average pixel values by class
dict_pixel_avg = {
"Emotion": ["Happy", "Sad", "Neutral", "Surprise"],
"Train": [
train_happy_pixel_avg,
train_sad_pixel_avg,
train_neutral_pixel_avg,
train_surprise_pixel_avg,
],
"Validate": [
val_happy_pixel_avg,
val_sad_pixel_avg,
val_neutral_pixel_avg,
val_surprise_pixel_avg,
],
"Test": [
test_happy_pixel_avg,
test_sad_pixel_avg,
test_neutral_pixel_avg,
test_surprise_pixel_avg,
],
}
# converting dictionary to dataframe
df_pixel_avg = pd.DataFrame.from_dict(dict_pixel_avg)
df_pixel_avg
# plotting pixel averages for training, validation and test images
df_pixel_avg.groupby("Emotion", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="PIXEL AVERAGES FOR TRAINING, VALIDATION and TEST IMAGES",
ylabel="Pixel Averages",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.legend(loc=(1.01, 0.5))
plt.show()
# **Observations: Pixel Values**
# * In grayscale, a value of 255 indicates white while a value of 0 indicates black.
# * Consistent across training, validation, and test data sets, images in the surprise class have a higher average pixel value than images across the other three classes. In other words, surprise images are consistently brighter/lighter than happy, sad, and neutral images. Perhaps this is due to mouths being open more consistently and white teeth being exposed, as well as eyes being open wider and therefore more white being visible.
# * As surprise is the least represented class across training and validation data sets, perhaps this is another unique characteristic that will help differentiate it from the other three classes despite making up a smaller percentage of total images on which to train.
# * Across training, validation, and test data sets, images in the sad class have a lower average pixel value than images across the other three classes. In other words, sad images are consistently darker than happy, neutral, and surprise images.
# * It will be interesting to see if average pixel value can help our models more easily learn the sad and surprise images. The confusion matrix will show us how often sad images and surprise images are confused with one another.
# * Also interesting to note, while the sad and neutral images are the most similar visually (in terms of features), they are also the most similar when it comes to pixel values. Again, a look at the final confusion matrix will show us whether or not the two are more likely to be confused with one another.
# **Note:**
# Data pre-processing and augmentation will take place during the creation of data loaders. When ImageDataGenerator objects are instantiated, a range of processes can and will be applied, sometimes to varying degrees, depending on the model being created and trained. Some process/augmentation operations include the following:
# * **rotation_range** allows us to provide a degree range for random rotations of images. This helps address the issue of faces in the training images being tilted in different directions.
# * **height_shift_range** allows us to shift the image up and down.
# * **width_shift_range** allows us to shift the image left and right.
# * **brightness_range** allows us to address the wide range in pixel values from one image to the next. A number smaller than one makes an image darker, and a number larger than one makes an image lighter.
# * **shear_range** allows us to shear angle in a counter-clockwise direction.
# * **zoom_range** allows us to zoom in or out, essentially randomly cropping the images.
# * **horizontal_flip** allows us to flip the training image so it is a mirror image of itself. An image facing left will now face right, etc.
# * **rescale** is our opportunity to normalize the input image from a tensor filled with numbers from 0 to 255, down to a tensor of numbers ranging from 0 to 1.
# While creating our data sets via **flow_from_directory**, we have an opportunity to set class_mode to 'categorical', which will essentially one-hot-encode our classes. The classes themselves are then defined as 'happy,' 'sad,' 'neutral,' and 'surprise.' This allows us to set our loss to **categorical_crossentropy**, which itself is used for multi-class classification where each image (in our case) belongs to a single class.
# ## **Creating Data Loaders**
# Creating data loaders that we will use as inputs to our initial neural networks. We will create separate data loaders for color_modes grayscale and RGB so we can compare the results. An image that is grayscale has only 1 channel, with pixel values ranging from 0 to 255, while an RGB image has 3 channels, with each pixel having a value for red, green, and blue. Images that are RGB are therefore more complex for a neural network to process.
#
batch_size = 32
# Creating ImageDataGenerator objects for grayscale colormode
datagen_train_grayscale = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_validation_grayscale = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_test_grayscale = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
# Creating ImageDataGenerator objects for RGB colormode
datagen_train_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_validation_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_test_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
# Creating train, validation, and test sets for grayscale colormode
print("Grayscale Images")
train_set_grayscale = datagen_train_grayscale.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_grayscale = datagen_validation_grayscale.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_set_grayscale = datagen_test_grayscale.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
# Creating train, validation, and test sets for RGB colormode
print("\nColor Images")
train_set_rgb = datagen_train_rgb.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_rgb = datagen_validation_rgb.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_set_rgb = datagen_test_rgb.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
# **Note:**
# Data augmentation performed on the data for these initial models includes **horizontal_flip**, **brightness_range**, **rescale**, and **shear_range**.
# ## **Model Building**
# **A Note About Neural Networks:**
# The best algorithmic tools we have available to us for processing images are neural networks. In particular, convolutional neural networks (CNN) have significant advantages over standard artificial neural networks (ANN).
# While image classification utilizing ANNs is possible, there are some drawbacks:
# * **Translational Invariance:** ANNs are not translation invariant, meaning that the location of objects within the image is learned along with the object itself. If the object is located in different areas of the image, varying from image to image, the ANN will likely produce inconsistent results.
# * **Spacial Invariance:** ANNs are not spatial invariant, meaning that once the image matrix is converted/flattened into an array, they lose spatial information about the image. In reality, nearby pixels within an image should be more strongly related to one another, but an ANN does not leverage this information.
# * **Feature Extraction:** ANNs give similar importance to each pixel within an image, meaning that they are learning the background of the image to the same degree that they are learning the object within the image. If the background changes from image to image, the ANN will have a difficult time learning that the object itself is the same despite what is going on in the background of the image.
# * **Computational Expense:** ANNs need input images to be flattened into an array of pixel values, and as the input images get larger and the number of hidden layers increases, the total number of trainable parameters balloons considerably.
# On the other hand, through the use of convolutional and pooling layers, CNNs are translationally and spatially invariant. They are able to understand that the location of an object within an image is not important, nor is the background of the image itself. CNNs, through the use of their convolutional layers, are also better able to extract important features of an object within an image. Finally, CNNs take advantage of weight sharing, as the same filters are applied to each area of the image. This reduces the number of weights that need to be learned through backpropagation, thereby minimizing the number of trainable parameters and reducing computational expense.
# Taking all of this into account, we will proceed with the development of CNN models to pursue our objectives.
# ## Model 1.1: Base Neural Network (Grayscale)
# **Note:**
# We will begin by building a simple CNN model to serve as a baseline for future models. The same model will be built with color_mode set to grayscale (with an input shape of 48,48,1) as well as color_mode set to RGB (with an input shape of 48,48,3). The models will then be compared to determine if one approach outperforms the other.
# A baseline grayscale model is developed first. It consists of three convolutional blocks with relu activation, MaxPooling and a Dropout layer, followed by a single dense layer with 512 neurons, and a softmax classifier for multi-class classification. Total trainable parameters are 605,060.
# Creating a Sequential model
model_1_grayscale = Sequential()
# Convolutional Block #1
model_1_grayscale.add(
Conv2D(64, (2, 2), input_shape=(48, 48, 1), activation="relu", padding="same")
)
model_1_grayscale.add(MaxPooling2D(2, 2))
model_1_grayscale.add(Dropout(0.2))
# Convolutional Block #2
model_1_grayscale.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_1_grayscale.add(MaxPooling2D(2, 2))
model_1_grayscale.add(Dropout(0.2))
# Convolutional Block #3
model_1_grayscale.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_1_grayscale.add(MaxPooling2D(2, 2))
model_1_grayscale.add(Dropout(0.2))
# Flatten layer
model_1_grayscale.add(Flatten())
# Dense layer
model_1_grayscale.add(Dense(512, activation="relu"))
# Classifier
model_1_grayscale.add(Dense(4, activation="softmax"))
model_1_grayscale.summary()
# ### **Compiling and Training the Model**
#
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_1_grayscale.h5",
monitor="val_accuracy",
verbose=1,
save_best_only=True,
mode="auto",
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_1_grayscale.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_1_grayscale = model_1_grayscale.fit(
train_set_grayscale,
validation_data=val_set_grayscale,
epochs=100,
callbacks=callbacks_list,
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_1_grayscale.history["accuracy"])
plt.plot(history_1_grayscale.history["val_accuracy"])
plt.title("Accuracy - Model 1 (Grayscale)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_1_grayscale.history["loss"])
plt.plot(history_1_grayscale.history["val_loss"])
plt.title("Loss - Model 1 (Grayscale)")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on the Test Set**
# Evaluating the model's performance on the test set
accuracy = model_1_grayscale.evaluate(test_set_grayscale)
# **Observations and Insights:**
# As constructed, our baseline grayscale model performs decently. After 29 epochs (best epoch), training accuracy stands at 0.72 and validation accuracy is 0.68. Training accuracy and loss continue to improve, while validation accuracy and loss begin to level off before early-stopping ends the training process. Accuracy on the test set is 0.65. A glance at the results, and the accuracy/loss graphs above, reveals a model that is overfitting and consequently has some room for improvement.
# | | Training | Validation | Test |
# | --- | --- | --- | --- |
# | Grayscale Accuracy |0.72 | 0.68 | 0.65 |
# ## Model 1.2: Base Neural Network (RGB)
# **Note:**
# This baseline model will contain the same architecture as the above grayscale model. Due to the input shape changing from 48,48,1 (grayscale) to 48,48,3 (rgb), the total trainable parameters have increased to 605,572.
#
# Creating a Sequential model
model_1_rgb = Sequential()
# Convolutional Block #1
model_1_rgb.add(
Conv2D(64, (2, 2), input_shape=(48, 48, 3), activation="relu", padding="same")
)
model_1_rgb.add(MaxPooling2D(2, 2))
model_1_rgb.add(Dropout(0.2))
# Convolutional Block #2
model_1_rgb.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_1_rgb.add(MaxPooling2D(2, 2))
model_1_rgb.add(Dropout(0.2))
# Convolutional Block #3
model_1_rgb.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_1_rgb.add(MaxPooling2D(2, 2))
model_1_rgb.add(Dropout(0.2))
# Flatten layer
model_1_rgb.add(Flatten())
# Dense layer
model_1_rgb.add(Dense(512, activation="relu"))
# Classifier
model_1_rgb.add(Dense(4, activation="softmax"))
model_1_rgb.summary()
# ### **Compiling and Training the Model**
#
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_1_rgb.h5",
monitor="val_accuracy",
verbose=1,
save_best_only=True,
mode="auto",
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_1_rgb.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_1_rgb = model_1_rgb.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_1_rgb.history["accuracy"])
plt.plot(history_1_rgb.history["val_accuracy"])
plt.title("Accuracy - Model 1 (RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_1_rgb.history["loss"])
plt.plot(history_1_rgb.history["val_loss"])
plt.title("Loss - Model 1 (RGB)")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on the Test Set**
# Evaluating the model's performance on the test set
accuracy = model_1_rgb.evaluate(test_set_rgb)
# **Observations and Insights:**
# As constructed, our baseline RGB model also performs decently. After 24 epochs (best epoch), training accuracy stands at 0.72 and validation accuracy is 0.68. Training accuracy and loss continue to improve, while validation accuracy and loss begin to level off before early-stopping ends the training process. Accuracy on the test set is 0.63.
# Our baseline grayscale and RGB models perform similarly across all metrics. Overall, both models underfit the data for 10-15 epochs, likely due to the addition of Dropout layers in the model architecture, after which the models begin to overfit the data, performing similarly. Perhaps a slight edge to the grayscale model for performing better on the test set with a smaller number of trainable parameters, making it computationally less expensive when scaled.
# | | Training | Validation | Test |
# | --- | --- | --- | --- |
# | Grayscale Accuracy |0.72 | 0.68 | 0.65 |
# | RGB Accuracy | 0.72 | 0.68 | 0.63 |
#
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_1_grayscale.history["accuracy"])
plt.plot(history_1_grayscale.history["val_accuracy"])
plt.plot(history_1_rgb.history["accuracy"])
plt.plot(history_1_rgb.history["val_accuracy"])
plt.title("Accuracy - Model 1 (Grayscale & RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(
[
"Training Accuracy (Grayscale)",
"Validation Accuracy (Grayscale)",
"Training Accuracy (RGB)",
"Validation Accuracy (RGB)",
],
loc="lower right",
)
plt.show()
# ## Model 2.1: 2nd Generation (Grayscale)
# **Note:**
# We will now build a slightly deeper model to see if we can improve performance. Similar to our baseline models, we will train this model with color_modes of grayscale and RGB so we can compare performance.
# The architecture of our second model is comprised of 4 convolutional blocks with relu activation, BatchNormalization, a LeakyReLu layer, and MaxPooling, followed by a dense layer with 512 neurons, another dense layer with 256 neurons, and finally a softmax classifier. The grayscale model has a total of 455,780 parameters.
#
# Creating a Sequential model
model_2_grayscale = Sequential()
# Convolutional Block #1
model_2_grayscale.add(
Conv2D(256, (2, 2), input_shape=(48, 48, 1), activation="relu", padding="same")
)
model_2_grayscale.add(BatchNormalization())
model_2_grayscale.add(LeakyReLU(alpha=0.1))
model_2_grayscale.add(MaxPooling2D(2, 2))
# Convolutional Block #2
model_2_grayscale.add(Conv2D(128, (2, 2), activation="relu", padding="same"))
model_2_grayscale.add(BatchNormalization())
model_2_grayscale.add(LeakyReLU(alpha=0.1))
model_2_grayscale.add(MaxPooling2D(2, 2))
# Convolutional Block #3
model_2_grayscale.add(Conv2D(64, (2, 2), activation="relu", padding="same"))
model_2_grayscale.add(BatchNormalization())
model_2_grayscale.add(LeakyReLU(alpha=0.1))
model_2_grayscale.add(MaxPooling2D(2, 2))
# Convolutional Block #4
model_2_grayscale.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_2_grayscale.add(BatchNormalization())
model_2_grayscale.add(LeakyReLU(alpha=0.1))
model_2_grayscale.add(MaxPooling2D(2, 2))
# Flatten layer
model_2_grayscale.add(Flatten())
# Dense layers
model_2_grayscale.add(Dense(512, activation="relu"))
model_2_grayscale.add(Dense(256, activation="relu"))
# Classifier
model_2_grayscale.add(Dense(4, activation="softmax"))
model_2_grayscale.summary()
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_2_grayscale.h5",
monitor="val_accuracy",
verbose=1,
save_best_only=True,
mode="auto",
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_2_grayscale.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_2_grayscale = model_2_grayscale.fit(
train_set_grayscale,
validation_data=val_set_grayscale,
epochs=100,
callbacks=callbacks_list,
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_2_grayscale.history["accuracy"])
plt.plot(history_2_grayscale.history["val_accuracy"])
plt.title("Accuracy - Model 2 (Grayscale)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_2_grayscale.history["loss"])
plt.plot(history_2_grayscale.history["val_loss"])
plt.title("Loss - Model 2 (Grayscale)")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on the Test Set**
accuracy = model_2_grayscale.evaluate(test_set_grayscale)
# **Observations and Insights:**
# As constructed, our second, deeper grayscale model performs somewhat differently than its predecessor. After 18 epochs (best epoch), training accuracy stands at 0.78 and validation accuracy is 0.71, which are both higher than Model 1, but Model 2 begins to overfit almost immediately, and the gaps between training and accuracy scores only grow from there. Training accuracy and loss continue to improve, while validation accuracy and loss begin to level off before early-stopping ends the training process. Accuracy on the test set is 0.69. Our model is not generalizing well, though with better accuracy scores compared to Model 1, there is an opportunity (if overfitting can be reduced) to become the better grayscale model.
# | | Training | Validation | Test |
# | --- | --- | --- | --- |
# | Grayscale Accuracy |0.78 | 0.71 | 0.69 |
# ## Model 2.2: 2nd Generation (RGB)
# **Note:**
# This model will contain the same architecture as the above grayscale model. Due to the input shape changing from 48,48,1 (grayscale) to 48,48,3 (rgb), the total parameters have increased to 457,828.
#
# Creating a Sequential model
model_2_rgb = Sequential()
# Convolutional Block #1
model_2_rgb.add(
Conv2D(256, (2, 2), input_shape=(48, 48, 3), activation="relu", padding="same")
)
model_2_rgb.add(BatchNormalization())
model_2_rgb.add(LeakyReLU(alpha=0.1))
model_2_rgb.add(MaxPooling2D(2, 2))
# Convolutional Block #2
model_2_rgb.add(Conv2D(128, (2, 2), activation="relu", padding="same"))
model_2_rgb.add(BatchNormalization())
model_2_rgb.add(LeakyReLU(alpha=0.1))
model_2_rgb.add(MaxPooling2D(2, 2))
# Convolutional Block #3
model_2_rgb.add(Conv2D(64, (2, 2), activation="relu", padding="same"))
model_2_rgb.add(BatchNormalization())
model_2_rgb.add(LeakyReLU(alpha=0.1))
model_2_rgb.add(MaxPooling2D(2, 2))
# Convolutional Block #4
model_2_rgb.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_2_rgb.add(BatchNormalization())
model_2_rgb.add(LeakyReLU(alpha=0.1))
model_2_rgb.add(MaxPooling2D(2, 2))
# Flatten layer
model_2_rgb.add(Flatten())
# Dense layers
model_2_rgb.add(Dense(512, activation="relu"))
model_2_rgb.add(Dense(256, activation="relu"))
# Classifier
model_2_rgb.add(Dense(4, activation="softmax"))
model_2_rgb.summary()
# ### **Compiling and Training the Model**
#
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_2_rgb.h5",
monitor="val_accuracy",
verbose=1,
save_best_only=True,
mode="auto",
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_2_rgb.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_2_rgb = model_2_rgb.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_2_rgb.history["accuracy"])
plt.plot(history_2_rgb.history["val_accuracy"])
plt.title("Accuracy - Model 2 (RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_2_rgb.history["loss"])
plt.plot(history_2_rgb.history["val_loss"])
plt.title("Loss - Model 2 (RGB)")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on the Test Set**
# Evaluating the model's performance on the test set
accuracy = model_2_rgb.evaluate(test_set_rgb)
# **Observations and Insights:**
# As constructed, our second RGB model also performs somewhat differently than its predecessor. After 15 epochs (best epoch), training accuracy stands at 0.76 and validation accuracy is 0.71, which are both higher than Model 1, but Model 2 begins to overfit almost immediately. Training accuracy and loss continue to improve, while validation accuracy and loss level off before early-stopping ends the training process. Accuracy on the test set is 0.68. Once again, our model is not generalizing well, though with better accuracy scores compared to Model 1, there is an opportunity (if overfitting can be reduced) to become the better RGB model.
# Our deeper grayscale and RGB models again perform similarly across all metrics, with the grayscale model attaining slightly better accuracies. Again, a slight edge to the grayscale model for performing better on the test set with a smaller number of trainable parameters.
# | | Training | Validation | Test |
# | --- | --- | --- | --- |
# | Grayscale Accuracy |0.78 | 0.71 | 0.69 |
# | RGB Accuracy | 0.76 | 0.71 | 0.68 |
#
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_2_grayscale.history["accuracy"])
plt.plot(history_2_grayscale.history["val_accuracy"])
plt.plot(history_2_rgb.history["accuracy"])
plt.plot(history_2_rgb.history["val_accuracy"])
plt.title("Accuracy - Model 2 (Grayscale & RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(
[
"Training Accuracy (Grayscale)",
"Validation Accuracy (Grayscale)",
"Training Accuracy (RGB)",
"Validation Accuracy (RGB)",
],
loc="lower right",
)
plt.show()
# **Overall Observations and Insights on Initial Models:**
# * As discussed above, both grayscale models slightly outperformed their RGB counterparts, and did so using less trainable parameters, making them less computationally expensive. Given this performance, we will proceed with grayscale models when doing so is possible.
# * As the datasets for this project are black and white images, it is possible that a grayscale colormode works better than a RGB colormode on what are essentially grayscale images. In this case, adding a second and third channel and increasing the input shape from 48,48,1 to 48,48,3 does not seem to help the modeling, and in fact may be making it overly complex.
# * As evidenced by the graph below, the 4 models thus far have fairly similar accuracy trajectories, though with a fair degree of separation between them. There is obviously room for improvement when it comes to overall accuracy. As early-stopping has prohibited us from seeing whether or not the training accuracy and loss level off before reaching 100%, it is clear that they continue to improve while validation accuracy and loss level off.
# * Some possible ways to decrease overfitting and thereby improve the above models include:
# * Introduce additional forms of data augmentation. While the above models take advantage of **horizontal_flip**, **brightness_range**, **rescale**, and **sheer_range**, it is possible that introducing additional forms of data augmentation (like **width_shift_range**, **height_shift_range**, **zoon_range**, **rotation_range**, etc. as discussed above) could help improve model performance.
# * Additional use **BatchNormalization** could also improve performance by offering some degree of regularization.
# * Additional use of **DropOut** and **SpatialDropout** could also help improve performance by assisting in regularization.
# * Introducting **GaussianNoise** could also assist in regularization, adding a form of noise to the data.
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_1_grayscale.history["accuracy"])
plt.plot(history_1_grayscale.history["val_accuracy"])
plt.plot(history_1_rgb.history["accuracy"])
plt.plot(history_1_rgb.history["val_accuracy"])
plt.plot(history_2_grayscale.history["accuracy"])
plt.plot(history_2_grayscale.history["val_accuracy"])
plt.plot(history_2_rgb.history["accuracy"])
plt.plot(history_2_rgb.history["val_accuracy"])
plt.title("Accuracy - Models 1 & 2 (Grayscale & RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(
[
"Training Accuracy - Model 1 (Grayscale)",
"Validation Accuracy - Model 1 (Grayscale)",
"Training Accuracy - Model 1 (RGB)",
"Validation Accuracy - Model 1 (RGB)",
"Training Accuracy - Model 2 (Grayscale)",
"Validation Accuracy - Model 2 (Grayscale)",
"Training Accuracy - Model 2 (RGB)",
"Validation Accuracy - Model 2 (RGB)",
],
loc="lower right",
)
plt.show()
# ## **Transfer Learning Architectures**
# In this section, we will create several Transfer Learning architectures. For the pre-trained models, we will select three popular architectures, namely: VGG16, ResNet v2, and Efficient Net. The difference between these architectures and the previous architectures is that these will require 3 input channels (RGB) while the earlier models also worked on grayscale images.
# ### **Creating our Data Loaders for Transfer Learning Architectures**
# We will create new data loaders for the transfer learning architectures used below. As required by the architectures we will be piggybacking, color_mode will be set to RGB.
# Additionally, we will be using the same data augmentation methods used on our previous models in order to better compare performance against our baseline models. These methods include **horizontal_flip**, **brightness_range**, **rescale**, and **shear_range**.
#
batch_size = 32
# Creating ImageDataGenerator objects for RGB colormode
datagen_train_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_validation_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_test_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
# Creating train, validation, and test sets for RGB colormode
print("\nColor Images")
train_set_rgb = datagen_train_rgb.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_rgb = datagen_validation_rgb.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_set_rgb = datagen_test_rgb.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
# ## Model 3: VGG16
# First up is the VGG16 model, which is a CNN consisting of 13 convolutional layers, 5 MaxPooling layers, and 3 dense layers. The VGG16 model achieves nearly 93% accuracy on the ImageNet dataset containing 14 million images across 1,000 classes. Clearly, this is much more substantial than our models above.
# ### **Importing the VGG16 Architecture**
vgg = VGG16(include_top=False, weights="imagenet", input_shape=(48, 48, 3))
vgg.summary()
# ### **Model Building**
# We have imported the VGG16 model up to layer 'block4_pool', as this has shown the best performance compared to other layers (discussed below). The VGG16 layers will be frozen, so the only trainable layers will be those we add ourselves. After flattening the input from 'block4_pool', 2 dense layers will be added, followed by a Dropout layer, another dense layer, and BatchNormalization. We will end with a softmax classifier.
#
transfer_layer = vgg.get_layer("block4_pool")
vgg.trainable = False
# Flatten the input
x = Flatten()(transfer_layer.output)
# Dense layers
x = Dense(256, activation="relu")(x)
x = Dense(128, activation="relu")(x)
x = Dropout(0.2)(x)
x = Dense(64, activation="relu")(x)
x = BatchNormalization()(x)
# Classifier
pred = Dense(4, activation="softmax")(x)
# Initialize the model
model_3 = Model(vgg.input, pred)
# ### **Compiling and Training the VGG16 Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_3.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss",
min_delta=0,
patience=15, # This is increased compared to initial models, otherwise training is cut too quickly
verbose=1,
restore_best_weights=True,
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_3.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_3 = model_3.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_3.history["accuracy"])
plt.plot(history_3.history["val_accuracy"])
plt.title("Accuracy - VGG16 Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_3.history["loss"])
plt.plot(history_3.history["val_loss"])
plt.title("Loss - VGG16 Model")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the VGG16 model**
# Evaluating the model's performance on the test set
accuracy = model_3.evaluate(test_set_rgb)
# **Observations and Insights:**
# As imported and modified, our transfer learning model seems to perform similarly to our previous models developed above. After 29 epochs (best epoch), training accuracy stands at 0.72 and validation accuracy is 0.67. Accuracy and loss for both the training and validation data level off before early stopping ends the training. The model's performance on the test data stands at 0.66. These scores are roughly in line with the scores of Model 1, our baseline model.
# The VGG16 model was ultimately imported up to layer block4_pool, as it produced the best performance. A history of alternative models is below.
# | | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | VGG16 block4_pool (selected) |0.71 | 0.72 | 0.80 | 0.67 |
# | VGG16 block5_pool |1.05 | 0.54 | 1.10 | 0.52 |
# | VGG16 block3_pool |0.79 | 0.69 | 0.77 | 0.66 |
# | VGG16 block2_pool |0.71 | 0.71 | 0.82 | 0.65 |
# ## Model 4: ResNet v2
# Our second transfer learning model is ResNet v2, which is a CNN trained on over 1 million images from the ImageNet database. ResNet v2 can classify images into 1,000 different categories. Like VGG16, colormode must be set to RGB to leverage this pre-trained architecture.
Resnet = ap.ResNet101(include_top=False, weights="imagenet", input_shape=(48, 48, 3))
Resnet.summary()
# ### **Model Building**
# We have imported the ResNet v2 model up to layer 'conv_block23_add', as this has shown the best performance compared to other layers (discussed below). The ResNet v2 layers will be frozen, so the only trainable layers will be those we add ourselves. After flattening the input from 'conv_block23_add', we will add the same architecture we did earlier to VGG16, namely 2 dense layers, followed by a DropOut layer, another dense layer, and BatchNormalization. We will once again end with a softmax classifier, as this is a multi-class classification exercise.
transfer_layer = Resnet.get_layer("conv4_block23_add")
Resnet.trainable = False
# Flatten the input
x = Flatten()(transfer_layer.output)
# Dense layers
x = Dense(256, activation="relu")(x)
x = Dense(128, activation="relu")(x)
x = Dropout(0.2)(x)
x = Dense(64, activation="relu")(x)
x = BatchNormalization()(x)
# Classifier
pred = Dense(4, activation="softmax")(x)
# Initialize the model
model_4 = Model(Resnet.input, pred)
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_4.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss",
min_delta=0,
patience=15, # Increased over initial models otherwise training is cut off too quickly
verbose=1,
restore_best_weights=True,
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_4.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_4 = model_4.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_4.history["accuracy"])
plt.plot(history_4.history["val_accuracy"])
plt.title("Accuracy - ResNet V2 Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_4.history["loss"])
plt.plot(history_4.history["val_loss"])
plt.title("Loss - ResNet V2 Model")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the ResNet Model**
# Evaluating the model's performance on the test set
accuracy = model_4.evaluate(test_set_rgb)
# **Observations and Insights:**
# As imported and modified, our transfer learning model shows terrible performance. After just 1 epoch (the 'best' epoch!), training accuracy stands at 0.26 and validation accuracy is 0.36. Accuracy and loss for both training and validation data level off fairly quickly at which point early stopping aborts the training. The above accuracy and loss curves paint the picture of poor model that will not generalize well at all. The model's test accuracy comes in at 0.34.
# The ResNet v2 model was ultimately imported up to layer 'conv4_block23_add', as it produced the 'best' performance, though it was difficult to choose. A history of alternative models is below.
# | | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | ResNet V2 conv4_block23_add (selected) |1.43 | 0.26 | 1.35 | 0.36 |
# | ResNet V2 conv5_block3_add |1.47 | 0.23 | 1.43 | 0.33 |
# | ResNet V2 conv3_block4_add |1.49 | 0.22 | 1.44 | 0.33 |
# | ResNet V2 conv2_block3_add |1.51 | 0.21 | 1.55 | 0.21 |
# ## Model 5: EfficientNet
# Our third transfer learning model is EfficientNet, which is a CNN that uses 'compound scaling' to improve efficiency and, theoretically at least, performance. Like VGG16 and ResNet v2, color_mode must be set to RGB to leverage this pre-trained architecture.
EfficientNet = ap.EfficientNetV2B2(
include_top=False, weights="imagenet", input_shape=(48, 48, 3)
)
EfficientNet.summary()
# ### **Model Building**
# We have imported the EfficientNet Model up to layer 'block5f_expand_activation', as this has shown the best performance compared to other layers (discussed below). The EfficientNet layers will be frozen, so the only trainable layers will be those that we add ourselves. After flattening the input from 'block5f_expand_activation', we will add the same architecture we did earlier to the VGG16 and ResNet v2 models, namely 2 dense layers, followed by a Dropout layer, another dense layer, and BatchNormalization. We will end with a softmax classifier.
#
transfer_layer_EfficientNet = EfficientNet.get_layer("block5f_expand_activation")
EfficientNet.trainable = False
# Flatten the input
x = Flatten()(transfer_layer_EfficientNet.output)
# Dense layers
x = Dense(256, activation="relu")(x)
x = Dense(128, activation="relu")(x)
x = Dropout(0.2)(x)
x = Dense(64, activation="relu")(x)
x = BatchNormalization()(x)
# Classifier
pred = Dense(4, activation="softmax")(x)
# Initialize the model
model_5 = Model(EfficientNet.input, pred)
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_5.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=12, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_5.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_5 = model_5.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_5.history["accuracy"])
plt.plot(history_5.history["val_accuracy"])
plt.title("Accuracy - EfficientNet Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="center right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_5.history["loss"])
plt.plot(history_5.history["val_loss"])
plt.title("Loss - EfficientNet Model")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the EfficientNet Model**
# Evaluating the model's performance on the test set
accuracy = model_5.evaluate(test_set_rgb)
# **Observations and Insights:**
# As imported and modified, this model performs poorly. After just 4 epochs (the 'best' epoch), training accuracy stands at 0.26 and validation accuracy is 0.24. Training and validation accuracy are almost immediately horizontal. Loss declines a bit before leveling off. With test accuracy coming in at 0.25, it makes the model no better than random guessing. We could build a model that classifies every single image as 'happy', and with our evenly distributed test set, it would produce the same 0.25 accuracy as our EfficientNet model.
# Again, it was difficult to select a 'best' layer from which to import the EfficientNet model. A history of alternative models is below.
# | | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | EfficientNet block5f_expand_activation (selected) |1.39 | 0.26 | 1.37 | 0.24 |
# | EfficientNet block6e_expand_activation |1.53 | 0.25 | 1.45 | 0.22 |
# | EfficientNet block4a_expand_activation |1.42 | 0.25 | 1.42 | 0.21 |
# | EfficientNet block3c_expand_activation |1.47 | 0.26 | 1.44 | 0.22 |
# **Overall Observations and Insights on Transfer Learning Models:**
# * As outlined above, the performance of these transfer learning models varied greatly. While the VGG16 model performed admirably (see table below), the ResNet v2 and EfficientNet models left much to be desired in terms of stability and performance.
# * On the whole, none of the transfer learning models performed better than our baseline and 2nd generation models, which was surprising.
# * Model complexity seems to have played a role in performance, as the VGG16 model has a much less complex architecture than both the ResNet v2 and the EfficientNet models. Perhaps overly-complex models trained on millions of large, color images do not perform as well on smaller, black and white images from just 4 classes.
# * VGG16, with 14.7 million parameters, is a fairly straightforward architecture, with just 19 layers from the input layer to the max import layer, 'block5_pool'.
# * ResNet v2, with 42.7 million parameters, is a much more complex architecture, with a whopping 345 layers from the input layer to the max import layer, 'conv5_block3_out'.
# * EfficientNet, with 'just' 8.8 million parameters, contains 349 layers from the input layer to the max import layer, 'top_activation'.
# * As evidenced by the table below, it would appear that the unsatisfactory performance of the transfer learning models may have more to do with their complexity than the fact that they require a colormode of RGB. The baseline and 2nd generation RGB models both performed just as well as the VGG16 model. It would seem that the downfall of ResNet v2 and EfficientNet was their complex architecture. Quite simply, the simpler models performed better. In fact, the highest performing model so far, the 2nd generation grayscale model (Model 2.1), has the smallest number of parameters.
# * Perhaps a sweet spot exists somewhere between the simplicity of our 2nd generation grayscale model and the much more complex transfer learning models we have explored thus far. If it is possible to increase the complexity of our 2nd generation grayscale model while keeping the overall complexity from ballooning too far in the direction of the transfer learning models, we may find ourselves a successful model.
#
# | | Parameters | Train Loss | Train Accuracy | Val Loss | Val Accuracy | Test Accuracy |
# | --- | --- | --- | --- | --- | --- | --- |
# | **Model 1.1**: Baseline Grayscale | 605,060 | 0.68 | 0.72 | 0.78 | 0.68 | 0.65 |
# | **Model 1.2**: Baseline RGB | 605,572 | 0.68 | 0.72 | 0.78 | 0.68 | 0.63 |
# | **Model 2.1**: 2nd Gen Grayscale | 455,780 | 0.54 | 0.78 | 0.74 | 0.71 | 0.69 |
# | **Model 2.2**: 2nd Gen RGB | 457,828 | 0.59 | 0.76 | 0.72 | 0.71 | 0.68 |
# | **Model 3**: VGG16 | 14,714,688 | 0.71 | 0.72 | 0.80 | 0.67 | 0.66 |
# | **Model 4**: ResNet V2 | 42,658,176 | 1.43 | 0.26 | 1.35 | 0.36 | 0.28 |
# | **Model 5**: EfficientNet | 8,769,374 | 1.39 | 0.26 | 1.37 | 0.24 | 0.25 |
# # Milestone 1
# ## Model 6: Complex Neural Network Architecture
# As previewed above, it is time to expand our 2nd generation grayscale model to see if we can improve performance. Grayscale slightly outperformed RGB in our first two models, so we will leave RGB behind and proceed with color_mode set to grayscale.
# ## **Creating our Data Loaders**
# As we are proceeding with a colormode set to grayscale, we will create new data loaders for our more complex CNN, Model 6. As our data augmentation takes place when we instantiate an ImageDataGenerator object, it is convenient to create data loaders specific to our new model so we can easily finetune our hyperparameters as needed. The ImageDataGenerators below include the parameters of the final Milestone 1 model, the highest performing CNN thus far. They were chosen after exhaustive finetuning of the model, as discussed later.
# * Batch size is set to 32. The model was tested with batch sizes of 16, 32, 45, 64, and 128. A batch size of 32 performed the best. The smaller the batch size, the longer training took. The larger the batch size, the faster the training process, though the accuracy and loss bounced around significantly, offsetting the increased speed.
# * **horizontal_flip** is set to 'True'. As some faces in the images face left while others face right or straight ahead, flipping the training images improves our model's ability to learn that horizontal orientation should not affect the eventual classification.
# * **rescale** is equal to 1./255, which normalizes the pixel values to a number between 0 and 1. This helps to prevent vanishing and exploding gradients in our network by keeping the numbers small and manageable.
# * **brightness_range** is set to '0.7,1.3'. A setting of 1 results in images remaining unchanged. As the number approaches zero, the images become darker. As the number approaches 2, the images become lighter. As many of the images are already very dark or very light, limiting this setting to a relatively small range around 1 will help our model learn to deal with varying pixel values without rendering some images completely unusable.
# * **rotation_range** is set to 25, meaning the images may randomly be rotated up to 25 degrees. Similar to flipping the images horizontally, this rotation will help the model learn that the angle of a face is not an important feature.
# * Additional data augmentation methods were attempted and later removed after failing to significantly improve model performance. Among those tested were **width_shift_range**, **height_shift_range**, **shear_range**, and **zoom_range**.
batch_size = 32
# Creating ImageDataGenerator objects for grayscale colormode
datagen_train_grayscale = ImageDataGenerator(
horizontal_flip=True,
rescale=1.0 / 255,
brightness_range=(0.7, 1.3),
rotation_range=25,
)
datagen_validation_grayscale = ImageDataGenerator(
horizontal_flip=True,
rescale=1.0 / 255,
brightness_range=(0.7, 1.3),
rotation_range=25,
)
datagen_test_grayscale = ImageDataGenerator(
horizontal_flip=True,
rescale=1.0 / 255,
brightness_range=(0.7, 1.3),
rotation_range=25,
)
# Creating train, validation, and test sets for grayscale colormode
print("Grayscale Images")
train_set_grayscale = datagen_train_grayscale.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_grayscale = datagen_validation_grayscale.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
test_set_grayscale = datagen_test_grayscale.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
# ### **Model Building**
# The structure of the Milestone 1 model (Model 6) is below. Many configurations were tested, and the following architecture led to the best performance.
# * The model begins with an input layer accepting an input shape of '48,48,1', given that our color_mode has been set to grayscale.
# * There are 5 convolutional blocks with relu activation. Each block contains BatchNormalization, LeakyReLU, and MaxPooling layers. The first, second, and fourth blocks include a layer of GaussianNoise, while the third and fifth layers each include a Dropout layer.
# * The output of the fifth convolutional block is then flattened, and fed into 2 dense layers which include additional BatchNormalization and Dropout layers.
# * The architecture is completed with a softmax classifier, as this model is designed for multi-class classification. Test images will be classified as either happy, sad, neutral, or surprise.
# * The model contains 2.1 million parameters, making it more complex than our 2nd generation grayscale model, but not as complex as the transfer learning models, whose complexity appeared to hurt their performance.
#
# Creating a Sequential model
model_6 = Sequential()
# Convolutional Block #1
model_6.add(
Conv2D(64, (3, 3), input_shape=(48, 48, 1), activation="relu", padding="same")
)
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(GaussianNoise(0.1))
# Convolutional Block #2
model_6.add(Conv2D(128, (3, 3), activation="relu", padding="same"))
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(GaussianNoise(0.1))
# Convolutional Block #3
model_6.add(Conv2D(512, (2, 2), activation="relu", padding="same"))
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(Dropout(0.1))
# Convolutional Block #4
model_6.add(Conv2D(512, (2, 2), activation="relu", padding="same"))
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(GaussianNoise(0.1))
# Convolutional Block #5
model_6.add(Conv2D(256, (2, 2), activation="relu", padding="same"))
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(Dropout(0.1))
# Flatten layer
model_6.add(Flatten())
# Dense layers
model_6.add(Dense(256, activation="relu"))
model_6.add(BatchNormalization())
model_6.add(Dropout(0.1))
model_6.add(Dense(512, activation="relu"))
model_6.add(BatchNormalization())
model_6.add(Dropout(0.05))
# Classifier
model_6.add(Dense(4, activation="softmax"))
model_6.summary()
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_6.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=10, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_6.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
# Fitting model with epochs set to 100
history_6 = model_6.fit(
train_set_grayscale,
validation_data=val_set_grayscale,
epochs=100,
callbacks=callbacks_list,
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_6.history["accuracy"])
plt.plot(history_6.history["val_accuracy"])
plt.title("Accuracy - Complex Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_6.history["loss"])
plt.plot(history_6.history["val_loss"])
plt.title("Loss - Complex Model")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on Test Set**
# Evaluating the model's performance on the test set
accuracy = model_6.evaluate(test_set_grayscale)
# **Observations and Insights:**
# Model 6, our Milestone 1 model, outperforms all previous models. After 33 epochs (best epoch), training accuracy stands at 0.79 and validation accuracy is 0.76. Accuracy and loss for both training and validation data improve similarly before leveling off. The model begins to overfit around epoch 15, but the overfitting is not as severe as previous models. The test accuracy for this model is 0.76. Overall, Model 6 generalizes better than previous models, and is the top performer thus far. That said, **it is still an overfitting model, and thus it would not be advisable to deploy this model as is**.
# This model underwent numerous transformations before arriving at its final state. Parameters were tuned, layers were added, layers were removed, and eventually the above model was determined to be the best iteration. An abridged history of model development can be found in the table below.
# The starting point for our final model was as follows:
# **CONVOLUTIONAL BLOCK #1**
# * Conv2D(64,(2,2), input shape = (48,48,1), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **CONVOLUTIONAL BLOCK #2**
# * Conv2D(128,(2,2), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **CONVOLUTIONAL BLOCK #3**
# * Conv2D(512,(2,2), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **CONVOLUTIONAL BLOCK #4**
# * Conv2D(256,(2,2), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **CONVOLUTIONAL BLOCK #5**
# * Conv2D(128,(2,2), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **FINAL LAYERS**
# * Flatten
# * Dense(256, activation = 'relu')
# * Dropout(0.1)
# * Dense(256, activation = 'relu')
# * Dropout(0.1)
# * Dense(4, activation = 'softmax')
# **PARAMETERS**
# * Batch size = 32
# * horizontal_flip = True
# * rescale = 1./255
# * brightness_range = (0.0,2.0)
# * shear_range = 0.3
#
#
# Below is an abridged summary of actions taken to improve the model. In many cases, parameters or layers were adjusted, added, or removed, just to be returned to their original state when the desired or experimental impact was not realized. The model went through dozens of iterations, with the following transformations being the most impactful.
# | Action Taken | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | Starting model as outlined above | 0.77 | 0.70 | 0.89 | 0.58 |
# | Dropout(0.1) layers added to conv blocks 1 and 5 to reduce overfitting |0.75 | 0.74 | 0.66 | 0.61 |
# | Shear_range removed entirely to determine effect |0.76 | 0.74 | 0.68 | 0.60 |
# | Rotation_range added and optimized |0.74 | 0.74 | 0.62 | 0.61 |
# | Additional dropout layers added to blocks 2 and 4 |0.59 | 0.78 | 0.64 | 0.68 |
# | Number of neurons in final dense layer set to 512 |0.68 | 0.71 | 0.62 | 0.71 |
# | Number of neurons in block 4 increased to 512 |0.70 | 0.73 | 0.60 | 0.74 |
# | Dropout layers swapped out for GaussianNoise in blocks 1 and 2 |0.61 | 0.74 | 0.57 | 0.75 |
# | Brightness_range narrowed to (0.5,1.5) then to (0.7,1.3) |0.59 | 0.75 | 0.60 | 0.75 |
# | Kernel size enlarged to 3x3 in first then also second block |0.55 | 0.78 | 0.57 | 0.75 |
# | Dropout in block 5 reduced to 0.5, resulting in final model |0.54 | 0.79 | 0.60 | 0.76 |
# # Final Solution
# ## Model 7: Goodbye Overfitting
# While Model 6 was an improvement on previous models, it was still overfitting the training data. In order to feel comfortable recommending a model for deployment in the context of this project, we need to improve on Model 6. Model 7 is an attempt to develop a deployable CNN. We want our model to have high accuracy, while also maintaining a good fit (no overfitting/underfitting) and generalizing well to the unseen test data. We will continue with color_mode set to grayscale for the reasons already noted: slightly better performance, slightly fewer parameters, slightly lower computational expense, and the image data itself is already grayscale.
# ## **Creating our Data Loaders**
# We will once again be creating new data loaders for Model 7. As mentioned earlier, since our data augmentation takes place when we instantiate an ImageDataGenerator object, it is convenient to create data loaders specific to our new model so we can easily finetune our hyperparameters as needed. The ImageDataGenerators below include the parameters of our final, highest performing iteration of the model. They were once again chosen after exhaustive finetuning, as discussed later.
# * Batch size is set to 128. The model was tested with batch sizes of 16, 32, 64, 128, and 256. A batch size of 128 performed the best. The smallest batch sizes seemed to get stuck in an accuracy range of 25-30% (perhaps a local minimum), while the other sizes did not generalize as well to the test data.
# * **horizontal_flip** is set to 'True'. As some faces in the images face left while others face right or straight ahead, flipping the training images improves our model's ability to learn that horizontal orientation should not affect the eventual classification.
# * **rescale** is equal to 1./255, which normalizes the pixel values to a number between 0 and 1. This helps to prevent vanishing and exploding gradients in our network by keeping the numbers small and manageable.
# * **brightness_range** is set to '0.0,2.0'. This is a change from Model 6 where we used a narrower range. A narrower range did not help within the architecture of Model 7, and the broader range showed better performance.
# * **shear_range** is set to 0.3, which matches the settings of our baseline models. This parameter essentially distorts the image along an axis in a counter-clockwise direction.
# * **one-hot-encoding** is handled by setting **class_mode** to "categorical", followed by our list of classes.
# * Additional data augmentation methods were attempted and later removed after failing to significantly improve model performance. Among those tested were **width_shift_range**, **height_shift_range**, **rotation_range**, **zca_whitening**, **zoom_range**, and even **vertical_flip**.
batch_size = 128
# Creating ImageDataGenerator objects for grayscale colormode
datagen_train_grayscale = ImageDataGenerator(
rescale=1.0 / 255,
brightness_range=(0.0, 2.0),
horizontal_flip=True,
shear_range=0.3,
)
datagen_validation_grayscale = ImageDataGenerator(
rescale=1.0 / 255,
brightness_range=(0.0, 2.0),
horizontal_flip=True,
shear_range=0.3,
)
datagen_test_grayscale = ImageDataGenerator(
rescale=1.0 / 255,
brightness_range=(0.0, 2.0),
horizontal_flip=True,
shear_range=0.3,
)
# Creating train, validation, and test sets for grayscale colormode
print("Grayscale Images")
train_set_grayscale = datagen_train_grayscale.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_grayscale = datagen_validation_grayscale.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_set_grayscale = datagen_test_grayscale.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
# ### **Model Building**
# The structure of Model 7 is below. Rather than simply modifying Model 6, the development of Model 7 entailed going back to the drawing board and devising a new strategy. Many configurations were tested, and the following architecture led to the best, most generalizable performance.
# * The model begins with an input layer accepting an input shape of '48,48,1', given that our color_mode has been set to grayscale.
# * There are 3 similar convolutional blocks with relu activation. Padding is no longer set to "same", as this increased the generalization gap. Each block contains a BatchNormalization layer before its first and second convolutional layers (except the input layer in Block #1). Each block ends with MaxPooling and a Dropout layer set to 0.4.
# * A "secret" block, which is what eventually closed the generalization gap and eliminated overfitting, is essentially a normalization/regularization block consisting of a BatchNormalization layer and a convolutional layer without activation, but instead with a L2 regularization set to 0.025. This is followed by another BatchNormalization layer.
# * The output of the "secret" block is then flattened, and fed into 2 dense layers, each followed by a Dropout layer, and separated by a layer of GaussianNoise.
# * The architecture is completed with a softmax classifier, as this model is designed for multi-class classification. Test images will be classified as either happy, sad, neutral, or surprise.
# * The final model contains 1.8 million parameters and 27 layers, making it slightly less complex than Model 6, while still substantially more complex than our initial models.
# Creating a Sequential model
model_7 = Sequential()
# Convolutional Block #1
model_7.add(Conv2D(64, (3, 3), input_shape=(48, 48, 1), activation="relu"))
model_7.add(BatchNormalization())
model_7.add(Conv2D(64, (3, 3), activation="relu"))
model_7.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model_7.add(Dropout(0.4))
# Convolutional Block #2
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (3, 3), activation="relu"))
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (3, 3), activation="relu"))
model_7.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model_7.add(Dropout(0.4))
# Convolutional Block #3
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (3, 3), activation="relu"))
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (3, 3), activation="relu"))
model_7.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model_7.add(Dropout(0.4))
# SECRET LEVEL
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (2, 2), kernel_regularizer=l2(0.025)))
model_7.add(BatchNormalization())
# Flatten layer
model_7.add(Flatten())
# Dense layers
model_7.add(Dense(1024, activation="relu"))
model_7.add(Dropout(0.2))
model_7.add(GaussianNoise(0.1))
model_7.add(Dense(1024, activation="relu"))
model_7.add(Dropout(0.2))
# Classifier
model_7.add(Dense(4, activation="softmax"))
model_7.summary()
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_7.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Slows the learning rate when validation loss does not improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=2, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# **Note:**
# * Early stopping patience is set to 5 epochs. This model was trained with Patience set to 5, 10, 12, 15, 20, and 50. Each time, the model achieved the same results, so the simpler model (patience = 5) was chosen.
# * Reduce learning rate patience is set to 2 epochs. Again, the model was trained with patience set to 1, 2, 3, and 5. The results varied considerably, with 2 epochs being the only iteration that did not result in a generalization gap.
#
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_7.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
# Fitting model with epochs set to 200
history_7 = model_7.fit(
train_set_grayscale,
validation_data=val_set_grayscale,
epochs=200,
callbacks=callbacks_list,
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_7.history["accuracy"])
plt.plot(history_7.history["val_accuracy"])
plt.title("Accuracy - Final Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_7.history["loss"])
plt.plot(history_7.history["val_loss"])
plt.title("Loss - Final Model")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on Test Set**
# Evaluating the model's performance on the test set
accuracy = model_7.evaluate(test_set_grayscale)
# **Observations and Insights:**
# Model 7, rewarding us for all of our efforts, displays the best all-around performance. Accuracies for training, validation, and test data are stable at 0.75, while loss is stable across training, validation, and test data at roughly 0.63 (0.62 to 0.64). As evidenced by the above graphs, there is no noticeable generalization gap. The accuracy and loss curves move more or less in tandem, leveling off around epoch 25 and remaining together from that point forward. The model does not overfit or underfit the training data. The images below show the accuracy and loss curves for the same model run out to 115 epochs. The model converges at reasonable levels of accuracy and loss, and it generalizes well.
#
#
# Much like Model 6, this model underwent numerous transformations before arriving at its final state. Parameters were tuned, layers were added, others were removed, and in the end, the above iteration of the model was determined to be the best. Below are the impacts that some of the most important aspects of the model have on its overall performance. While some individual metrics may be better than those of the final model, each of the modifications below, if taken individually or in tandem, results in a generalization gap that is not present in the final model.
#
#
# | Model Changes | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | Final Model | 0.63 | 0.75 | 0.64 | 0.75 |
# | Remove "regularization" block |0.63 | 0.76 | 0.68 | 0.73 |
# | Remove L2 kernel regularizer |0.62 | 0.74 | 0.64 | 0.73 |
# | Remove Gaussian Noise |0.65 | 0.73 | 0.66 | 0.74 |
# | Reduce kernel size to (2,2) |0.63 | 0.74 | 0.66 | 0.74 |
# | Dropout levels reduced to 0.2 |0.57 | 0.78 | 0.65 | 0.74 |
# | Remove BatchNormalization |0.74 | 0.70 | 0.69 | 0.72 |
# | Include relu activation in regularization block |0.63 | 0.74 | 0.63 | 0.74 |
# | Batch size = 32 |0.62 | 0.75 | 0.65 | 0.74 |
# | Data augmentation with rotation range = 20 | 0.69 | 0.72 | 0.67 | 0.74 |
# | Data augmentation with zoom range = 0.2 | 0.71 | 0.71 | 0.69 | 0.73 |
# | Vertical flip = True | 0.74 | 0.71 | 0.70 | 0.74 |
# | Only 1 convolutional layer per block | 0.84 | 0.66 | 0.78 | 0.70 |
# ### **Model Comparison**
# Below are the accuracy and loss scores for each of our models, first in a tabular format, then represented visually in the form of bar charts.
# | | Parameters | Train Loss | Train Accuracy | Val Loss | Val Accuracy | Test Loss | Test Accuracy |
# | --- | --- | --- | --- | --- | --- | --- | --- |
# | **Model 1.1**: Baseline Grayscale | 605,060 | 0.68 | 0.72 | 0.78 | 0.68 | 0.82 | 0.65 |
# | **Model 1.2**: Baseline RGB | 605,572 | 0.68 | 0.72 | 0.78 | 0.68 | 0.80 | 0.63 |
# | **Model 2.1**: 2nd Gen Grayscale | 455,780 | 0.54 | 0.78 | 0.74 | 0.71 | 0.81 | 0.69 |
# | **Model 2.2**: 2nd Gen RGB | 457,828 | 0.59 | 0.76 | 0.72 | 0.71 | 0.70| 0.68 |
# | **Model 3**: VGG16 | 14,714,688 | 0.71 | 0.72 | 0.80 | 0.67 | 0.74 | 0.66 |
# | **Model 4**: ResNet V2 | 42,658,176 | 1.43 | 0.26 | 1.35 | 0.36 | 1.40 | 0.28 |
# | **Model 5**: EfficientNet | 8,769,374 | 1.39 | 0.26 | 1.37 | 0.24 | 1.40 | 0.25 |
# | **Model 6**: Milestone 1 | 2,119,172| 0.54 | 0.79 | 0.60 | 0.76 | 0.56 | 0.76 |
# | **Model 7**: Final Model | 1,808,708 | 0.63 | 0.75 | 0.64 | 0.75 | 0.62 | 0.75 |
#
# creating a dictionary containing model accuracies
dict_model_acc = {
"Model": ["1.1", "1.2", "2.1", "2.2", "3", "4", "5", "6", "7"],
"Train": [0.72, 0.72, 0.78, 0.76, 0.72, 0.26, 0.26, 0.79, 0.75],
"Validate": [0.68, 0.68, 0.71, 0.71, 0.67, 0.36, 0.24, 0.76, 0.75],
"Test": [0.65, 0.63, 0.69, 0.68, 0.66, 0.28, 0.25, 0.76, 0.75],
}
# converting dictionary to dataframe
df_model_acc = pd.DataFrame.from_dict(dict_model_acc)
# plotting accuracy scores for all models
df_model_acc.groupby("Model", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="Accuracy Scores Across Models",
ylabel="Accuracy Score",
xlabel="Models",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.legend(loc=(0.59, 0.77))
plt.show()
# creating a dictionary containing model loss
dict_model_loss = {
"Model": ["1.1", "1.2", "2.1", "2.2", "3", "4", "5", "6", "7"],
"Train": [0.68, 0.68, 0.54, 0.59, 0.71, 1.43, 1.39, 0.54, 0.63],
"Validate": [0.78, 0.78, 0.74, 0.72, 0.80, 1.35, 1.37, 0.60, 0.64],
"Test": [0.82, 0.80, 0.81, 0.70, 0.74, 1.40, 1.40, 0.56, 0.62],
}
# converting dictionary to dataframe
df_model_loss = pd.DataFrame.from_dict(dict_model_loss)
# plotting loss scores for all models
df_model_loss.groupby("Model", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="Loss Scores Across Models",
ylabel="Loss Score",
xlabel="Models",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.show()
# **Observations and Insights:**
# The above graphs perfectly depict the overfitting that occurs in Models 1.1, 1.2, 2.1, 2.2, and 3, with accuracy scores declining in steps as we move from training, to validation, and on to test data. The opposite is true for the loss scores. The graphs also show the total dysfunction of Models 4 and 5, with very low accuracy and very high error scores. It is also clear from the graphs that Models 6 and 7 are the most consistent, most generalizable models, and that a final decision regarding a deployable model should be made between those two options.
# In deciding between Models 6 and 7, it is useful to revisit the accuracy and loss curves for the two models.
# #### **Accuracy and loss curves for Model 6:**
#
#
# #### **Accuracy and loss curves for Model 7:**
#
#
# While the accuracy and loss curves for the two models both stabilize by epoch 20-25, there is no gap between accuracy and loss curves for Model 7, while a slight gap does exist for Model 6. The accuracy and loss scores are all individually better for Model 6 (higher accuracy and lower loss), but when viewed together, the spread within the two scores is larger for Model 6, while it is virtually nonexistent in Model 7. It is difficult to justify deploying a slightly overfitting model when a slightly less accurate but more generalizable model is available. Model 7 will be our final model.
# ### **Plotting the Confusion Matrix for Model 7**
test_set = datagen_test_grayscale.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=128,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_images, test_labels = next(test_set)
pred = model_7.predict(test_images)
pred = np.argmax(pred, axis=1)
y_true = np.argmax(test_labels, axis=1)
# Printing the classification report
print(classification_report(y_true, pred))
# Plotting the heatmap using the confusion matrix
cm = confusion_matrix(y_true, pred)
plt.figure(figsize=(8, 5))
sns.heatmap(
cm,
annot=True,
fmt=".0f",
xticklabels=["happy", "sad", "neutral", "surprise"],
yticklabels=["happy", "sad", "neutral", "surprise"],
)
plt.ylabel("Actual")
plt.xlabel("Predicted")
plt.show()
# **Observations and Insights:**
# * As noted above, our final model achieves an accuracy score of 0.75 on the test images. The model correctly predicted 96 of 128 images.
# * The choice to prioritize precision (TP/(TP+FP)) or recall (TP/(TP+FN)) depends entirely on the model's end use. If the stakes are high, and false negatives should be avoided at all costs, than recall is more important. If reducing the number of false positives is more important, than precision is the better choice. In the case of our model, no trade-off is necessary, with precision and recall scores essentially the same (precision = 0.76, recall = 0.75, F1 = 0.75).
# * As previewed during the data visualization phase of the project, the 'happy' and 'surprise' images seemed to have the most unique characteristics, and this hypothesis appears to have played out in the classification report and confusion matrix. Happy and surprise have the highest precision and recall scores (and consequently, F1 scores) of the 4 classes.
# * Additionally, 'sad' and 'neutral' images were in fact more likely to be confused with one another, as discussed during the data visualization phase. When the model misclassified a sad image, it was most likely to be mistaken for a neutral image, and vice versa.
# * Any concern about a slightly skewed class distribution can be put to rest. As previewed, the surprise images, which were outnumbered in the training and validation data, were unique enough to identify correctly despite representing a smaller proportion of training images. It is possible that our earlier finding re: elevated average pixel values for surprise images has played a role, along with the unique characteristics of surprise images, including open mouths and wide open eyes.
# * As discussed during the data visualization phase, now in the context of the confusion matrix, it should be pointed out once again that the term "accuracy" can be misleading. There are training, validation, and test images of smiling people that are labeled as "sad", while there are images of frowning people labeled as "happy", etc. If the model classifies a test image as "sad" even though the person is smiling, and in fact the test image is incorrectly classified as "sad", making the prediction accurate, should we really consider that as accurate? Or would the accurate prediction be when the model overrules the misclassified test image and, from a human's perspective, accurately classifies the image as "happy"? For this reason, the test scores and confusion matrix should be taken with a grain of salt.
# * Similarly, there is a test image that does not contain a face at all. As there are similar images across all four classes within the training data, a correct prediction of the empty test image would seem to be pure chance. Should an accurate prediction in this case really increase the model's perceived accuracy? Should an incorrect prediction of an empty test image really lower the model's perceived accuracy? It seems that any model that correctly predicts all 128 test images benefited from some degree of luck. Again, these final scores should be viewed with some degree of skepticism, but that skepticism would be similar across all models.
# ### **Visualizing Images: Actual Class Label vs Predicted Class Label**
# Making predictions on the test data
y_pred_test = model_7.predict(test_set)
# Converting probabilities to class labels
y_pred_test_classes = np.argmax(y_pred_test, axis=1)
# Calculating the probability of the predicted class
y_pred_test_max_probas = np.max(y_pred_test, axis=1)
classes = ["happy", "sad", "neutral", "surprise"]
rows = 3
cols = 4
fig = plt.figure(figsize=(12, 12))
for i in range(cols):
for j in range(rows):
random_index = np.random.randint(
0, len(test_labels)
) # generating random integer
ax = fig.add_subplot(rows, cols, i * rows + j + 1)
ax.imshow(test_images[random_index, :]) # selecting random test image
pred_label = classes[
y_pred_test_classes[random_index]
] # predicted label of selected image
pred_proba = y_pred_test_max_probas[
random_index
] # probability associated with model's prediction
true_label = test_labels[random_index] # actual class label of selected image
if true_label[0] == 1: # converting array to class labels
true_label = "happy"
elif true_label[1] == 1:
true_label = "sad"
elif true_label[2] == 1:
true_label = "neutral"
else:
true_label = "surprise"
ax.set_title(
"actual: {}\npredicted: {}\nprobability: {:.3}\n".format(
true_label, pred_label, pred_proba
)
)
plt.gray()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/384/129384194.ipynb
| null | null |
[{"Id": 129384194, "ScriptId": 38470583, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13090419, "CreationDate": "05/13/2023 10:29:16", "VersionNumber": 1.0, "Title": "notebook022827cc64", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 2330.0, "LinesInsertedFromPrevious": 2330.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Capstone Project: Facial Emotion Detection
# Michael Hogge - April 2023
# # Executive Summary
# Emotion AI, also known as artificial emotional intelligence, is a subset of artificial intelligence dealing with the detection and replication of human emotion by machines. The successful creation of this "artificial empathy" hinges on a computer's ability to analyze, among other things, human text, speech, and facial expressions. In support of these efforts, this project leverages the power of convolutional neural networks (CNN) to create a computer vision model capable of accurately performing multi-class classification on images containing one of four facial expressions: happy, sad, neutral, and surprise.
# Data provided for this project includes over 20,000 grayscale images split into training (75%), validation (24.5%), and test (0.5%) datasets, and further divided into the aforementioned classes. At the outset of the project, a visual analysis of the data is undertaken and a slight imbalance is noted in the class distribution, with 'surprise' images making up a smaller percentage of total images when compared to 'happy,' 'sad,' and 'neutral' images. The unique characteristics of each class are discussed (e.g., images labeled as 'surprise' tend to contain faces with wide open mouths and eyes), including a breakdown of average pixel value by class.
# Following the data visualization and analysis phase of the project, nine CNNs are developed, ranging from simple grayscale models to complex transfer learning architectures comprised of hundreds of layers and tens of millions of parameters. Basic models are shown to be lacking the required amount of complexity to properly fit the data, while the transfer learning models (VGG16, ResNet v2, and EfficientNet) are shown to be too complex for the amount and type of data provided for this project. The unsatisfactory performance of the basic and transfer learning models necessitates the development of an alternative model capable of fitting the data and achieving acceptable levels of accuracy while maintaining a high level of generalizability. The proposed model, with four convolutional blocks and 1.8 million parameters, displays high accuracy (75% on training, validation, and test data) when compared to human performance (±65%) on similar data, and avoids overfitting the training data, which can be difficult to achieve with CNNs.
# The deployability of this model depends entirely on its intended use. With an accuracy of 75%, deployment in a marketing or gaming setting is perfectly reasonable, assuming consent has been granted, and the handling of highly personal data is done in an ethical, transparent manner with data privacy coming before profit. However, deployment in circumstances where the output from this model could cause serious material damage to an individual (e.g., hiring decisions, law enforcement, evidence in a court of law, etc.) should be avoided. While computer vision models can become quite skilled at classifying human facial expressions (particularly if they are trained on over-emoting/exaggerated images), it is important to note that a connection between those expressions and any underlying emotion is not a hard scientific fact. For example, a smiling person may not always be happy (e.g., they could be uncomfortable or polite), a crying person may not always be sad (e.g., they could be crying tears of joy), and someone who is surprised may be experiencing compound emotions (e.g., happily surprised or sadly surprised).
# There is certainly scope to improve the proposed model, including the ethical sourcing of additional, diverse training images, and additional data augmentation on top of what is already performed during the development of the proposed model. In certain scenarios, as indicated above, model deployment could proceed with 75% accuracy, and continued improvement could be pursued by the business/organization/government as time and funding allows. Before model deployment, a set of guiding ethical principles should be developed and adhered to throughout the data collection, analysis, and (possibly) storage phase. Stakeholders must ensure transparency throughout all stages of the computer vision life cycle, while monitoring the overall development of Emotion AI technology and anticipating future regulatory action, which appears likely.
# ## **Problem Definition**
# **Context:**
# How do humans communicate with one another? While spoken and written communication may immediately come to mind, research by Dr. Albert Mehrabian has found that over 50% of communication is conveyed through body language, including facial expressions. In face-to-face conversation, body language, it turns out, plays a larger role in how our message is interpreted than both the words we choose, and the tone with which we deliver them. Our expression is a powerful window into our true feelings, and as such, it can be used as a highly-effective proxy for sentiment, particularly in the absence of written or spoken communication.
# Emotion AI (artificial emotional intelligence, or affective computing), attempts to leverage this proxy for sentiment by detecting and processing facial expression (through neural networks) in an effort to successfully interpret human emotion and respond appropriately. Developing models that can accurately detect facial emotion is therefore an important driver of advancement in the realm of artificial intelligence and emotionally intelligent machines. The ability to successfully extract sentiment from images and video is also a powerful tool for businesses looking to conjure insights from the troves of unstructured data they have accumulated in recent years, or even to extract second-by-second customer responses to advertisements, store layouts, customer/user experience, etc.
# **Objective:**
# The objective of this project is to utilize deep learning techniques, including convolutional neural networks, to create a computer vision model that can accurately detect and interpret facial emotions. This model should be capable of performing multi-class classification on images containing one of four facial expressions: happy, sad, neutral, and surprise.
# **Key Questions:**
# * Do we have the data necessary to develop our models, and is it of good enough quality and quantity?
# * What is the best type of machine learning model to achieve our objective?
# * What do we consider 'success' when it comes to model performance?
# * How do different models compare to one another given this definition of success?
# * What are the most important insights that can be drawn from this project upon its conclusion?
# * What is the final proposed model and is it good enough for deployment?
# ## **About the Dataset**
# The data set consists of 3 folders, i.e., 'test', 'train', and 'validation'.
# Each of these folders has four subfolders:
# **‘happy’**: Images of people who have happy facial expressions.
# **‘sad’**: Images of people with sad or upset facial expressions.
# **‘surprise’**: Images of people who have shocked or surprised facial expressions.
# **‘neutral’**: Images of people showing no prominent emotion in their facial expression at all.
# ## **Importing the Libraries**
import zipfile # Used to unzip the data
import numpy as np # Mathematical functions, arrays, etc.
import pandas as pd # Data manipulation and analysis
import os # Misc operating system interfaces
import h5py # Read and write h5py files
import random
import matplotlib.pyplot as plt # A library for data visualization
from matplotlib import image as mpimg # Used to show images from filepath
import seaborn as sns # An advanced library for data visualization
from PIL import Image # Image processing
import cv2 # Image processing
# Importing Deep Learning Libraries, layers, models, optimizers, etc
import tensorflow as tf
from tensorflow.keras.preprocessing.image import (
load_img,
img_to_array,
ImageDataGenerator,
)
from tensorflow.keras.layers import (
Dense,
Input,
Dropout,
SpatialDropout2D,
GlobalAveragePooling2D,
Flatten,
Conv2D,
BatchNormalization,
Activation,
MaxPooling2D,
LeakyReLU,
GaussianNoise,
)
from tensorflow.keras.models import Model, Sequential
from tensorflow.keras.optimizers import Adam, SGD, RMSprop, Adadelta
from tensorflow.keras import regularizers
from keras.regularizers import l2
from tensorflow.keras.losses import categorical_crossentropy
from tensorflow.keras.utils import to_categorical
import tensorflow.keras.applications as ap
from tensorflow.keras.applications.vgg16 import VGG16
from keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
from tensorflow.keras import backend
# Reproducibility within TensorFlow
import fwr13y.d9m.tensorflow as tf_determinism
tf_determinism.enable_determinism()
tf.config.experimental.enable_op_determinism
from tqdm import tqdm # Generates progress bars
# Predictive data analysis tools
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
# To suppress warnings
import warnings
warnings.filterwarnings("ignore")
# Needed to silence tensorflow messages while running locally
from silence_tensorflow import silence_tensorflow
silence_tensorflow()
# Fixing the seed for random number generators to ensure reproducibility
np.random.seed(42)
random.seed(42)
tf.random.set_seed(42)
# Ensuring reproducibility using GPU with TensorFlow
os.environ["TF_DETERMINISTIC_OPS"] = "1"
# ### **Loading and Unzipping the Data**
# Extracting image files from the zip file
with zipfile.ZipFile("Facial_emotion_images.zip", "r") as zip_ref:
zip_ref.extractall()
dir_train = "Facial_emotion_images/train/" # Path of training data after unzipping
dir_validation = (
"Facial_emotion_images/validation/" # Path of validation data after unzipping
)
dir_test = "Facial_emotion_images/test/" # Path of test data after unzipping
img_size = 48 # Defining the size of the image as 48 pixels
# ## **Visualizing our Classes**
#
# Custom function to display first 35 images from the specified training folder
def display_emotion(emotion):
train_emotion = dir_train + emotion + "/"
plt.figure(figsize=(11, 11))
for i in range(1, 36):
plt.subplot(5, 7, i)
img = load_img(
train_emotion + os.listdir(train_emotion)[i],
target_size=(img_size, img_size),
)
plt.imshow(img)
plt.show()
# ### **Happy**
print("These are the first 35 training images labeled as 'Happy':")
display_emotion("happy")
# An example image pulled from the images above
img_x = os.listdir("Facial_emotion_images/train/happy/")[16]
img_happy_16 = mpimg.imread("Facial_emotion_images/train/happy/" + img_x)
plt.figure(figsize=(2, 2))
plt.imshow(img_happy_16, cmap="Greys_r")
plt.show()
# **Observations and Insights: Happy**
# * In most images, the person is smiling. Some smiles are with an open mouth with teeth visible, and some are with closed lips. Our models will need to learn both types of smiles.
# * Image brightness varies considerably and will need to be addressed with data augmentation.
# * The ages of the people vary from very young to old.
# * In some images, people are wearing eyeglasses or hats, eating food, or are partially covering their face with their hands. Some images contain watermarks.
# * Some images are head-on while some are sideways. We will address this via data augmentation (rotating and/or flipping images).
# * Images are cropped differently and this will need to be addressed with data augmentation (zoom/crop).
# * Some images do not contain a face (though not shown in the above 35). There is really nothing to do about this ... some of the test images also do not contain a face. As a result, some predictions by the final model will be based on incorrect data labeling.
# * As highlighted by the image above, some images are of non-human faces. In this case, it appears to be a statue with exaggerated features.
# * Some 'happy' images would clearly not be classified as 'happy' by a human being. This brings into question what accuracy really means. If the model correctly predicts an image labeled 'happy' as 'happy', should that be considered accurate if the person in the image is actually frowning and would be considered by a human being to be sad? In a high-stakes, real world situation we could potentially relabel images that have been incorrectly labeled, but in the context of this project, we have been advised to leave the training labels untouched.
# ### **Sad**
print("These are the first 35 training images labeled as 'Sad':")
display_emotion("sad")
# An example image pulled from the images above
img_x = os.listdir("Facial_emotion_images/train/sad/")[7]
img_sad_7 = mpimg.imread("Facial_emotion_images/train/sad/" + img_x)
plt.figure(figsize=(2, 2))
plt.imshow(img_sad_7, cmap="Greys_r")
plt.show()
# **Observations and Insights: Sad**
# * In most images, the person is frowning. In many images, people have their eyes closed or are looking down.
# * Compared to the 'happy' images, people labeled 'sad' seem more likely to have their mouths closed.
# * Similar to the 'happy' images, image brightness varies considerably, as does age. Some images are head-on while others are sideways. Some people are covering their face with their hands. As with 'happy' images, 'sad' images are also cropped differently, while some also have watermarks.
# * Some images do not contain a face (though not shown in the above 35).
# * As highlighted by the image above, some images are labeled 'sad' but would probably not be classified as sad by a human being. The person above appears to be smiling, but an accurate prediction by one of our models would classify the image as 'sad'. This raises the same issue about accuracy mentioned earlier.
# * At first glance, apart from the images that are clearly mislabeled, there appears to be enough difference between the 'happy' and 'sad' characteristics that an effective model should be able to tell them apart relatively easily.
# ### **Neutral**
print("These are the first 35 training images labeled as 'Neutral':")
display_emotion("neutral")
# An example image pulled from the images above
img_x = os.listdir("Facial_emotion_images/train/neutral/")[26]
img_neutral_26 = mpimg.imread("Facial_emotion_images/train/neutral/" + img_x)
plt.figure(figsize=(2, 2))
plt.imshow(img_neutral_26, cmap="Greys_r")
plt.show()
# **Observations and Insights: Neutral**
# * At first glance, this seems to be the most difficult label to accurately predict. While 'happy' and 'sad' images appear different enough that a model should be able to tell the difference, 'neutral' faces are in between 'happy' and 'sad', and consequently similar to both.
# * Similar to the other classes discussed above, differences in brightness, age, zoom, hands covering faces, etc. are apparent in the 'neutral' images as well.
# * As highlighted in the image above, some images are simply mistakes and do not contain any faces at all.
# * These neutral images seem more difficult for a human being to correctly classify. Some people appear to be slightly smiling, while others appear to be slightly frowning. This begs the question, where are the lines between happy/neutral and sad/neutral? Neutral images do appear to be more similar to sad images, so it is possible that our models will confuse the two classes.
# ### **Surprise**
print("These are the first 35 training images labeled as 'Surprise':")
display_emotion("surprise")
# An example image pulled from the images above
img_x = os.listdir("Facial_emotion_images/train/surprise/")[17]
img_surprise_34 = mpimg.imread("Facial_emotion_images/train/surprise/" + img_x)
plt.figure(figsize=(2, 2))
plt.imshow(img_surprise_34, cmap="Greys_r")
plt.show()
# **Observations and Insights: Surprise**
# * The most unique characteristics of the 'surprise' images are open mouths and big, open eyes. These seem like features that a successful model should be able to identify and accurately classify. There is a big difference between 'surprise' images and 'neutral' images, for example. It is possible, however, that the open mouth of a 'happy' smile and the open mouth of a 'surprise' image could be difficult for a model to distinguish between.
# * As with the above classes, brightness, crop, age, etc. vary between images. Hands are often covering faces. Some photos are head-on while others face sideways.
# * The above image is an example with a very light pixel value, as opposed to one of the much darker images. The person in the image has the classic open mouth and wide open eyes. The image also contains a watermark.
# * Some images do not contain a face (though not shown in the above 35).
# **Overall Insights from Visualization of Classes:**
# * All images are in grayscale (black/white) and image size is 48 x 48 pixels. We will need to rescale pixel values by dividing by 255 so pixel values are normalized between 0 and 1. This will allow our models to train faster and help to stabilize gradient descent.
# * Some classes have very different characteristics (happy/sad) while other classes are more similar (sad/neutral) and could be challenging for a model to accurately classify.
# * There is a wide range of differences with respect to image brightness, age of person, zoom/crop of image, orientation of the face, objects/hands covering the face, images not containing any face at all, etc. Data augmentation will need to be taken into consideration, and this will be handled when the Data Loaders are created below.
# * Visualizing the images in this way raises an important question: what do we consider an accurate model prediction for an image that is clearly mislabeled? If a person is smiling but labeled as 'sad', and the model accurately predicts 'sad', is that really 'accurate' since a human being would classify the image as 'happy'? If the test data set is large, it would be difficult to go through each image individually to check accuracy (which would also defeat the purpose of creating a computer vision model in the first place), so we will have to live with 'accurate' model predictions that may not be truly accurate. Pondering questions like this lead one to believe that a model can really only be as good as the data it is trained on.
# ## **Checking Distribution of Classes**
# Getting the count of images in each training folder and saving to variables
train_happy = len(os.listdir(dir_train + "happy/"))
train_sad = len(os.listdir(dir_train + "sad/"))
train_neutral = len(os.listdir(dir_train + "neutral/"))
train_surprised = len(os.listdir(dir_train + "surprise/"))
# Creating a Pandas series called "train_series" and converting to Pandas dataframe called "train_df"
# in order to display the table below. The dataframe will also contribute to bar charts farther below.
train_series = pd.Series(
{
"Happy": train_happy,
"Sad": train_sad,
"Neutral": train_neutral,
"Surprised": train_surprised,
}
)
train_df = pd.DataFrame(train_series, columns=["Total Training Images"])
train_df["Percentage"] = round(
(train_df["Total Training Images"] / train_df["Total Training Images"].sum()) * 100,
1,
)
train_df.index.name = "Emotions"
print("The distribution of classes within the training data:")
train_df
train_df.sum()
# **Observations: Training Images**
# * There are 15,109 training images in total.
# * Happy, sad, and neutral images make up roughly the same share of total training images (26%), while surprise images make up a smaller share (21%). At this stage it is important to note the relatively small imbalance, though the ratio does not seem skewed enough to warrant future manipulation in terms of weights, etc.
# * The insight made above, that surprise images seem to be some of the most unique in terms of characteristics (big open mouth, big open eyes), may actually help us overcome the relatively minor imbalance. There are fewer surprise images, but they may be easier to classify.
#
# Getting count of images in each validation folder and saving to variables
val_happy = len(os.listdir(dir_validation + "happy/"))
val_sad = len(os.listdir(dir_validation + "sad/"))
val_neutral = len(os.listdir(dir_validation + "neutral/"))
val_surprised = len(os.listdir(dir_validation + "surprise/"))
# Creating a Pandas series called "val_series" and converting to Pandas dataframe called "val_df"
# in order to display the table below. The dataframe will also contribute to bar charts farther below.
val_series = pd.Series(
{
"Happy": val_happy,
"Sad": val_sad,
"Neutral": val_neutral,
"Surprised": val_surprised,
}
)
val_df = pd.DataFrame(val_series, columns=["Total Validation Images"])
val_df["Percentage"] = round(
(val_df["Total Validation Images"] / val_df["Total Validation Images"].sum()) * 100,
1,
)
val_df.index.name = "Emotions"
print("The distribution of classes within the validation data:")
val_df
val_df.sum()
# **Observations: Validation Images**
# * There are 4,977 validation images in total.
# * The distribution across classes is much more imbalanced. Happy images make up almost 37% of total validation images, while surprise images make up only 16%. As the training images and validation images are already split and provided as is, it is not a simple matter of randomly splitting training data with a train/test split. We are stuck with the imbalance.
# * One solution to address the imbalance could be to cap the other classes at the level of the surprise class, but that would throw away a huge portion of our already small data set.
# * As mentioned above, we can surmise that surprise images are easier to classify because of their unique characteristics, and we will see if that is enough to offset the relatively smaller sample size with which to train and validate.
#
# Getting count of images in each test folder and saving to variables
test_happy = len(os.listdir(dir_test + "happy/"))
test_sad = len(os.listdir(dir_test + "sad/"))
test_neutral = len(os.listdir(dir_test + "neutral/"))
test_surprised = len(os.listdir(dir_test + "surprise/"))
# Creating a Pandas series called "test_series" and converting to Pandas dataframe called "test_df"
# in order to display the table below. The dataframe will also contribute to bar charts farther below.
test_series = pd.Series(
{
"Happy": test_happy,
"Sad": test_sad,
"Neutral": test_neutral,
"Surprised": test_surprised,
}
)
test_df = pd.DataFrame(test_series, columns=["Total Test Images"])
test_df["Percentage"] = round(
(test_df["Total Test Images"] / test_df["Total Test Images"].sum()) * 100, 1
)
test_df.index.name = "Emotions"
print("The distribution of classes within the validation data:")
test_df
test_df.sum()
# **Observations: Test Images**
# * There are 128 test images in total, evenly divided between all four classes.
# * This even distribution will make interpretation of the final confusion matrix very straightforward.
# Concatenating train_df, val_df, and test_df to create "df_total" in order to create the chart below
df_total = pd.concat([train_df, val_df, test_df], axis=1)
df_total.drop(["Percentage"], axis=1, inplace=True)
df_total = df_total.reset_index()
df_total.rename(
columns={
"index": "Emotions",
"Total Training Images": "Train",
"Total Validation Images": "Validate",
"Total Test Images": "Test",
},
inplace=True,
)
# Creating bar chart below, grouped by class (i.e. 'emotion') and broken down into "train", "validate",
# and "test" data. The x-axis is Emotions and the y-axis is Total Images.
df_total.groupby("Emotions", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="TOTAL TRAINING, VALIDATION and TEST IMAGES",
ylabel="Total Images",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.show()
# **Observations:**
# * Depicted graphically, the distribution of classes is clearly imbalanced, but the imbalance is not overpowering.
# * Perhaps most striking is the tiny proportion of test images compared to training images. Rather than a standard machine learning train/validation/test split of 80/10/10 or 70/20/10, the data as provided for this project is 75% training, 24.5% validation, and just 0.5% test. As the data is provided already split into groups, we will work as intended. The vast majority of data will be used to train and then validate our models, with a tiny proportion used for testing. This should work in our favor, maximizing the amount of data used by our models to train.
#
# Concatenating train_df, val_df, and test_df to create "df_percent" in order to create the chart below
df_percent = pd.concat([train_df, val_df, test_df], axis=1)
df_percent.drop(
["Total Training Images", "Total Validation Images", "Total Test Images"],
axis=1,
inplace=True,
)
df_percent.columns = ["Train", "Validate", "Test"]
# Creating bar chart below, grouped by class (i.e. 'emotion') and broken down into "train", "validate",
# and "test" data. The x-axis is Emotions and the y-axis is Percentage of Total Images.
df_percent.groupby("Emotions", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="PERCENTAGE OF TOTAL TRAINING, VALIDATION and TEST IMAGES",
ylabel="Percentage of Total Images",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.show()
# **Observations:**
# * A visual depiction of what was discussed earlier. We can see the percentage breakdown of train/validate/test data across classes.
# * Training data is evenly distributed across happy, sad, and neutral classes, with fewer surprise images.
# * Within the validation data set, happy images clearly make up the largest percent of total images, with surprise images coming in a distant last place.
# * Happy images make up a much larger percentage of the validation data set than they do of the training and test data sets.
# * Surprise images make up a larger percentage of the test data set than they do of the training and validation data sets.
# Obtaining the average pixel value for training images in the class 'Happy'
list_train_happy = []
for i in range(len(os.listdir("Facial_emotion_images/train/happy/"))):
list_x = []
x = os.listdir("Facial_emotion_images/train/happy/")[i]
im = Image.open("Facial_emotion_images/train/happy/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_train_happy.append(sum(list_x) / len(pix_val))
train_happy_pixel_avg = round(sum(list_train_happy) / len(list_train_happy), 2)
# Obtaining the average pixel value for validation images in the class 'Happy'
list_val_happy = []
for i in range(len(os.listdir("Facial_emotion_images/validation/happy/"))):
list_x = []
x = os.listdir("Facial_emotion_images/validation/happy/")[i]
im = Image.open("Facial_emotion_images/validation/happy/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_val_happy.append(sum(list_x) / len(pix_val))
val_happy_pixel_avg = round(sum(list_val_happy) / len(list_val_happy), 2)
# Obtaining the average pixel value for test images in the class 'Happy'
list_test_happy = []
for i in range(len(os.listdir("Facial_emotion_images/test/happy/"))):
list_x = []
x = os.listdir("Facial_emotion_images/test/happy/")[i]
im = Image.open("Facial_emotion_images/test/happy/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_test_happy.append(sum(list_x) / len(pix_val))
test_happy_pixel_avg = round(sum(list_test_happy) / len(list_test_happy), 2)
# Obtaining the average pixel value for training images in the class 'Sad'
list_train_sad = []
for i in range(len(os.listdir("Facial_emotion_images/train/sad/"))):
list_x = []
x = os.listdir("Facial_emotion_images/train/sad/")[i]
im = Image.open("Facial_emotion_images/train/sad/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_train_sad.append(sum(list_x) / len(pix_val))
train_sad_pixel_avg = round(sum(list_train_sad) / len(list_train_sad), 2)
# Obtaining the average pixel value for validation images in the class 'Sad'
list_val_sad = []
for i in range(len(os.listdir("Facial_emotion_images/validation/sad/"))):
list_x = []
x = os.listdir("Facial_emotion_images/validation/sad/")[i]
im = Image.open("Facial_emotion_images/validation/sad/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_val_sad.append(sum(list_x) / len(pix_val))
val_sad_pixel_avg = round(sum(list_val_sad) / len(list_val_sad), 2)
# Obtaining the average pixel value for test images in the class 'Sad'
list_test_sad = []
for i in range(len(os.listdir("Facial_emotion_images/test/sad/"))):
list_x = []
x = os.listdir("Facial_emotion_images/test/sad/")[i]
im = Image.open("Facial_emotion_images/test/sad/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_test_sad.append(sum(list_x) / len(pix_val))
test_sad_pixel_avg = round(sum(list_test_sad) / len(list_test_sad), 2)
# Obtaining the average pixel value for training images in the class 'Neutral'
list_train_neutral = []
for i in range(len(os.listdir("Facial_emotion_images/train/neutral/"))):
list_x = []
x = os.listdir("Facial_emotion_images/train/neutral/")[i]
im = Image.open("Facial_emotion_images/train/neutral/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_train_neutral.append(sum(list_x) / len(pix_val))
train_neutral_pixel_avg = round(sum(list_train_neutral) / len(list_train_neutral), 2)
# Obtaining the average pixel value for validation images in the class 'Neutral'
list_val_neutral = []
for i in range(len(os.listdir("Facial_emotion_images/validation/neutral/"))):
list_x = []
x = os.listdir("Facial_emotion_images/validation/neutral/")[i]
im = Image.open("Facial_emotion_images/validation/neutral/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_val_neutral.append(sum(list_x) / len(pix_val))
val_neutral_pixel_avg = round(sum(list_val_neutral) / len(list_val_neutral), 2)
# Obtaining the average pixel value for test images in the class 'Neutral'
list_test_neutral = []
for i in range(len(os.listdir("Facial_emotion_images/test/neutral/"))):
list_x = []
x = os.listdir("Facial_emotion_images/test/neutral/")[i]
im = Image.open("Facial_emotion_images/test/neutral/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_test_neutral.append(sum(list_x) / len(pix_val))
test_neutral_pixel_avg = round(sum(list_test_neutral) / len(list_test_neutral), 2)
# Obtaining the average pixel value for training images in the class 'Surprise'
list_train_surprise = []
for i in range(len(os.listdir("Facial_emotion_images/train/surprise/"))):
list_x = []
x = os.listdir("Facial_emotion_images/train/surprise/")[i]
im = Image.open("Facial_emotion_images/train/surprise/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_train_surprise.append(sum(list_x) / len(pix_val))
train_surprise_pixel_avg = round(sum(list_train_surprise) / len(list_train_surprise), 2)
# Obtaining the average pixel value for validation images in the class 'Surprise'
list_val_surprise = []
for i in range(len(os.listdir("Facial_emotion_images/validation/surprise/"))):
list_x = []
x = os.listdir("Facial_emotion_images/validation/surprise/")[i]
im = Image.open("Facial_emotion_images/validation/surprise/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_val_surprise.append(sum(list_x) / len(pix_val))
val_surprise_pixel_avg = round(sum(list_val_surprise) / len(list_val_surprise), 2)
# Obtaining the average pixel value for test images in the class 'Surprise'
list_test_surprise = []
for i in range(len(os.listdir("Facial_emotion_images/test/surprise/"))):
list_x = []
x = os.listdir("Facial_emotion_images/test/surprise/")[i]
im = Image.open("Facial_emotion_images/test/surprise/" + x, "r")
pix_val = list(im.getdata())
for j in range(len(pix_val)):
list_x.append(pix_val[j])
list_test_surprise.append(sum(list_x) / len(pix_val))
test_surprise_pixel_avg = round(sum(list_test_surprise) / len(list_test_surprise), 2)
# creating dictionary containing average pixel values by class
dict_pixel_avg = {
"Emotion": ["Happy", "Sad", "Neutral", "Surprise"],
"Train": [
train_happy_pixel_avg,
train_sad_pixel_avg,
train_neutral_pixel_avg,
train_surprise_pixel_avg,
],
"Validate": [
val_happy_pixel_avg,
val_sad_pixel_avg,
val_neutral_pixel_avg,
val_surprise_pixel_avg,
],
"Test": [
test_happy_pixel_avg,
test_sad_pixel_avg,
test_neutral_pixel_avg,
test_surprise_pixel_avg,
],
}
# converting dictionary to dataframe
df_pixel_avg = pd.DataFrame.from_dict(dict_pixel_avg)
df_pixel_avg
# plotting pixel averages for training, validation and test images
df_pixel_avg.groupby("Emotion", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="PIXEL AVERAGES FOR TRAINING, VALIDATION and TEST IMAGES",
ylabel="Pixel Averages",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.legend(loc=(1.01, 0.5))
plt.show()
# **Observations: Pixel Values**
# * In grayscale, a value of 255 indicates white while a value of 0 indicates black.
# * Consistent across training, validation, and test data sets, images in the surprise class have a higher average pixel value than images across the other three classes. In other words, surprise images are consistently brighter/lighter than happy, sad, and neutral images. Perhaps this is due to mouths being open more consistently and white teeth being exposed, as well as eyes being open wider and therefore more white being visible.
# * As surprise is the least represented class across training and validation data sets, perhaps this is another unique characteristic that will help differentiate it from the other three classes despite making up a smaller percentage of total images on which to train.
# * Across training, validation, and test data sets, images in the sad class have a lower average pixel value than images across the other three classes. In other words, sad images are consistently darker than happy, neutral, and surprise images.
# * It will be interesting to see if average pixel value can help our models more easily learn the sad and surprise images. The confusion matrix will show us how often sad images and surprise images are confused with one another.
# * Also interesting to note, while the sad and neutral images are the most similar visually (in terms of features), they are also the most similar when it comes to pixel values. Again, a look at the final confusion matrix will show us whether or not the two are more likely to be confused with one another.
# **Note:**
# Data pre-processing and augmentation will take place during the creation of data loaders. When ImageDataGenerator objects are instantiated, a range of processes can and will be applied, sometimes to varying degrees, depending on the model being created and trained. Some process/augmentation operations include the following:
# * **rotation_range** allows us to provide a degree range for random rotations of images. This helps address the issue of faces in the training images being tilted in different directions.
# * **height_shift_range** allows us to shift the image up and down.
# * **width_shift_range** allows us to shift the image left and right.
# * **brightness_range** allows us to address the wide range in pixel values from one image to the next. A number smaller than one makes an image darker, and a number larger than one makes an image lighter.
# * **shear_range** allows us to shear angle in a counter-clockwise direction.
# * **zoom_range** allows us to zoom in or out, essentially randomly cropping the images.
# * **horizontal_flip** allows us to flip the training image so it is a mirror image of itself. An image facing left will now face right, etc.
# * **rescale** is our opportunity to normalize the input image from a tensor filled with numbers from 0 to 255, down to a tensor of numbers ranging from 0 to 1.
# While creating our data sets via **flow_from_directory**, we have an opportunity to set class_mode to 'categorical', which will essentially one-hot-encode our classes. The classes themselves are then defined as 'happy,' 'sad,' 'neutral,' and 'surprise.' This allows us to set our loss to **categorical_crossentropy**, which itself is used for multi-class classification where each image (in our case) belongs to a single class.
# ## **Creating Data Loaders**
# Creating data loaders that we will use as inputs to our initial neural networks. We will create separate data loaders for color_modes grayscale and RGB so we can compare the results. An image that is grayscale has only 1 channel, with pixel values ranging from 0 to 255, while an RGB image has 3 channels, with each pixel having a value for red, green, and blue. Images that are RGB are therefore more complex for a neural network to process.
#
batch_size = 32
# Creating ImageDataGenerator objects for grayscale colormode
datagen_train_grayscale = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_validation_grayscale = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_test_grayscale = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
# Creating ImageDataGenerator objects for RGB colormode
datagen_train_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_validation_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_test_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
# Creating train, validation, and test sets for grayscale colormode
print("Grayscale Images")
train_set_grayscale = datagen_train_grayscale.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_grayscale = datagen_validation_grayscale.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_set_grayscale = datagen_test_grayscale.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
# Creating train, validation, and test sets for RGB colormode
print("\nColor Images")
train_set_rgb = datagen_train_rgb.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_rgb = datagen_validation_rgb.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_set_rgb = datagen_test_rgb.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
# **Note:**
# Data augmentation performed on the data for these initial models includes **horizontal_flip**, **brightness_range**, **rescale**, and **shear_range**.
# ## **Model Building**
# **A Note About Neural Networks:**
# The best algorithmic tools we have available to us for processing images are neural networks. In particular, convolutional neural networks (CNN) have significant advantages over standard artificial neural networks (ANN).
# While image classification utilizing ANNs is possible, there are some drawbacks:
# * **Translational Invariance:** ANNs are not translation invariant, meaning that the location of objects within the image is learned along with the object itself. If the object is located in different areas of the image, varying from image to image, the ANN will likely produce inconsistent results.
# * **Spacial Invariance:** ANNs are not spatial invariant, meaning that once the image matrix is converted/flattened into an array, they lose spatial information about the image. In reality, nearby pixels within an image should be more strongly related to one another, but an ANN does not leverage this information.
# * **Feature Extraction:** ANNs give similar importance to each pixel within an image, meaning that they are learning the background of the image to the same degree that they are learning the object within the image. If the background changes from image to image, the ANN will have a difficult time learning that the object itself is the same despite what is going on in the background of the image.
# * **Computational Expense:** ANNs need input images to be flattened into an array of pixel values, and as the input images get larger and the number of hidden layers increases, the total number of trainable parameters balloons considerably.
# On the other hand, through the use of convolutional and pooling layers, CNNs are translationally and spatially invariant. They are able to understand that the location of an object within an image is not important, nor is the background of the image itself. CNNs, through the use of their convolutional layers, are also better able to extract important features of an object within an image. Finally, CNNs take advantage of weight sharing, as the same filters are applied to each area of the image. This reduces the number of weights that need to be learned through backpropagation, thereby minimizing the number of trainable parameters and reducing computational expense.
# Taking all of this into account, we will proceed with the development of CNN models to pursue our objectives.
# ## Model 1.1: Base Neural Network (Grayscale)
# **Note:**
# We will begin by building a simple CNN model to serve as a baseline for future models. The same model will be built with color_mode set to grayscale (with an input shape of 48,48,1) as well as color_mode set to RGB (with an input shape of 48,48,3). The models will then be compared to determine if one approach outperforms the other.
# A baseline grayscale model is developed first. It consists of three convolutional blocks with relu activation, MaxPooling and a Dropout layer, followed by a single dense layer with 512 neurons, and a softmax classifier for multi-class classification. Total trainable parameters are 605,060.
# Creating a Sequential model
model_1_grayscale = Sequential()
# Convolutional Block #1
model_1_grayscale.add(
Conv2D(64, (2, 2), input_shape=(48, 48, 1), activation="relu", padding="same")
)
model_1_grayscale.add(MaxPooling2D(2, 2))
model_1_grayscale.add(Dropout(0.2))
# Convolutional Block #2
model_1_grayscale.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_1_grayscale.add(MaxPooling2D(2, 2))
model_1_grayscale.add(Dropout(0.2))
# Convolutional Block #3
model_1_grayscale.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_1_grayscale.add(MaxPooling2D(2, 2))
model_1_grayscale.add(Dropout(0.2))
# Flatten layer
model_1_grayscale.add(Flatten())
# Dense layer
model_1_grayscale.add(Dense(512, activation="relu"))
# Classifier
model_1_grayscale.add(Dense(4, activation="softmax"))
model_1_grayscale.summary()
# ### **Compiling and Training the Model**
#
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_1_grayscale.h5",
monitor="val_accuracy",
verbose=1,
save_best_only=True,
mode="auto",
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_1_grayscale.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_1_grayscale = model_1_grayscale.fit(
train_set_grayscale,
validation_data=val_set_grayscale,
epochs=100,
callbacks=callbacks_list,
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_1_grayscale.history["accuracy"])
plt.plot(history_1_grayscale.history["val_accuracy"])
plt.title("Accuracy - Model 1 (Grayscale)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_1_grayscale.history["loss"])
plt.plot(history_1_grayscale.history["val_loss"])
plt.title("Loss - Model 1 (Grayscale)")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on the Test Set**
# Evaluating the model's performance on the test set
accuracy = model_1_grayscale.evaluate(test_set_grayscale)
# **Observations and Insights:**
# As constructed, our baseline grayscale model performs decently. After 29 epochs (best epoch), training accuracy stands at 0.72 and validation accuracy is 0.68. Training accuracy and loss continue to improve, while validation accuracy and loss begin to level off before early-stopping ends the training process. Accuracy on the test set is 0.65. A glance at the results, and the accuracy/loss graphs above, reveals a model that is overfitting and consequently has some room for improvement.
# | | Training | Validation | Test |
# | --- | --- | --- | --- |
# | Grayscale Accuracy |0.72 | 0.68 | 0.65 |
# ## Model 1.2: Base Neural Network (RGB)
# **Note:**
# This baseline model will contain the same architecture as the above grayscale model. Due to the input shape changing from 48,48,1 (grayscale) to 48,48,3 (rgb), the total trainable parameters have increased to 605,572.
#
# Creating a Sequential model
model_1_rgb = Sequential()
# Convolutional Block #1
model_1_rgb.add(
Conv2D(64, (2, 2), input_shape=(48, 48, 3), activation="relu", padding="same")
)
model_1_rgb.add(MaxPooling2D(2, 2))
model_1_rgb.add(Dropout(0.2))
# Convolutional Block #2
model_1_rgb.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_1_rgb.add(MaxPooling2D(2, 2))
model_1_rgb.add(Dropout(0.2))
# Convolutional Block #3
model_1_rgb.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_1_rgb.add(MaxPooling2D(2, 2))
model_1_rgb.add(Dropout(0.2))
# Flatten layer
model_1_rgb.add(Flatten())
# Dense layer
model_1_rgb.add(Dense(512, activation="relu"))
# Classifier
model_1_rgb.add(Dense(4, activation="softmax"))
model_1_rgb.summary()
# ### **Compiling and Training the Model**
#
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_1_rgb.h5",
monitor="val_accuracy",
verbose=1,
save_best_only=True,
mode="auto",
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_1_rgb.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_1_rgb = model_1_rgb.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_1_rgb.history["accuracy"])
plt.plot(history_1_rgb.history["val_accuracy"])
plt.title("Accuracy - Model 1 (RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_1_rgb.history["loss"])
plt.plot(history_1_rgb.history["val_loss"])
plt.title("Loss - Model 1 (RGB)")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on the Test Set**
# Evaluating the model's performance on the test set
accuracy = model_1_rgb.evaluate(test_set_rgb)
# **Observations and Insights:**
# As constructed, our baseline RGB model also performs decently. After 24 epochs (best epoch), training accuracy stands at 0.72 and validation accuracy is 0.68. Training accuracy and loss continue to improve, while validation accuracy and loss begin to level off before early-stopping ends the training process. Accuracy on the test set is 0.63.
# Our baseline grayscale and RGB models perform similarly across all metrics. Overall, both models underfit the data for 10-15 epochs, likely due to the addition of Dropout layers in the model architecture, after which the models begin to overfit the data, performing similarly. Perhaps a slight edge to the grayscale model for performing better on the test set with a smaller number of trainable parameters, making it computationally less expensive when scaled.
# | | Training | Validation | Test |
# | --- | --- | --- | --- |
# | Grayscale Accuracy |0.72 | 0.68 | 0.65 |
# | RGB Accuracy | 0.72 | 0.68 | 0.63 |
#
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_1_grayscale.history["accuracy"])
plt.plot(history_1_grayscale.history["val_accuracy"])
plt.plot(history_1_rgb.history["accuracy"])
plt.plot(history_1_rgb.history["val_accuracy"])
plt.title("Accuracy - Model 1 (Grayscale & RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(
[
"Training Accuracy (Grayscale)",
"Validation Accuracy (Grayscale)",
"Training Accuracy (RGB)",
"Validation Accuracy (RGB)",
],
loc="lower right",
)
plt.show()
# ## Model 2.1: 2nd Generation (Grayscale)
# **Note:**
# We will now build a slightly deeper model to see if we can improve performance. Similar to our baseline models, we will train this model with color_modes of grayscale and RGB so we can compare performance.
# The architecture of our second model is comprised of 4 convolutional blocks with relu activation, BatchNormalization, a LeakyReLu layer, and MaxPooling, followed by a dense layer with 512 neurons, another dense layer with 256 neurons, and finally a softmax classifier. The grayscale model has a total of 455,780 parameters.
#
# Creating a Sequential model
model_2_grayscale = Sequential()
# Convolutional Block #1
model_2_grayscale.add(
Conv2D(256, (2, 2), input_shape=(48, 48, 1), activation="relu", padding="same")
)
model_2_grayscale.add(BatchNormalization())
model_2_grayscale.add(LeakyReLU(alpha=0.1))
model_2_grayscale.add(MaxPooling2D(2, 2))
# Convolutional Block #2
model_2_grayscale.add(Conv2D(128, (2, 2), activation="relu", padding="same"))
model_2_grayscale.add(BatchNormalization())
model_2_grayscale.add(LeakyReLU(alpha=0.1))
model_2_grayscale.add(MaxPooling2D(2, 2))
# Convolutional Block #3
model_2_grayscale.add(Conv2D(64, (2, 2), activation="relu", padding="same"))
model_2_grayscale.add(BatchNormalization())
model_2_grayscale.add(LeakyReLU(alpha=0.1))
model_2_grayscale.add(MaxPooling2D(2, 2))
# Convolutional Block #4
model_2_grayscale.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_2_grayscale.add(BatchNormalization())
model_2_grayscale.add(LeakyReLU(alpha=0.1))
model_2_grayscale.add(MaxPooling2D(2, 2))
# Flatten layer
model_2_grayscale.add(Flatten())
# Dense layers
model_2_grayscale.add(Dense(512, activation="relu"))
model_2_grayscale.add(Dense(256, activation="relu"))
# Classifier
model_2_grayscale.add(Dense(4, activation="softmax"))
model_2_grayscale.summary()
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_2_grayscale.h5",
monitor="val_accuracy",
verbose=1,
save_best_only=True,
mode="auto",
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_2_grayscale.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_2_grayscale = model_2_grayscale.fit(
train_set_grayscale,
validation_data=val_set_grayscale,
epochs=100,
callbacks=callbacks_list,
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_2_grayscale.history["accuracy"])
plt.plot(history_2_grayscale.history["val_accuracy"])
plt.title("Accuracy - Model 2 (Grayscale)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_2_grayscale.history["loss"])
plt.plot(history_2_grayscale.history["val_loss"])
plt.title("Loss - Model 2 (Grayscale)")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on the Test Set**
accuracy = model_2_grayscale.evaluate(test_set_grayscale)
# **Observations and Insights:**
# As constructed, our second, deeper grayscale model performs somewhat differently than its predecessor. After 18 epochs (best epoch), training accuracy stands at 0.78 and validation accuracy is 0.71, which are both higher than Model 1, but Model 2 begins to overfit almost immediately, and the gaps between training and accuracy scores only grow from there. Training accuracy and loss continue to improve, while validation accuracy and loss begin to level off before early-stopping ends the training process. Accuracy on the test set is 0.69. Our model is not generalizing well, though with better accuracy scores compared to Model 1, there is an opportunity (if overfitting can be reduced) to become the better grayscale model.
# | | Training | Validation | Test |
# | --- | --- | --- | --- |
# | Grayscale Accuracy |0.78 | 0.71 | 0.69 |
# ## Model 2.2: 2nd Generation (RGB)
# **Note:**
# This model will contain the same architecture as the above grayscale model. Due to the input shape changing from 48,48,1 (grayscale) to 48,48,3 (rgb), the total parameters have increased to 457,828.
#
# Creating a Sequential model
model_2_rgb = Sequential()
# Convolutional Block #1
model_2_rgb.add(
Conv2D(256, (2, 2), input_shape=(48, 48, 3), activation="relu", padding="same")
)
model_2_rgb.add(BatchNormalization())
model_2_rgb.add(LeakyReLU(alpha=0.1))
model_2_rgb.add(MaxPooling2D(2, 2))
# Convolutional Block #2
model_2_rgb.add(Conv2D(128, (2, 2), activation="relu", padding="same"))
model_2_rgb.add(BatchNormalization())
model_2_rgb.add(LeakyReLU(alpha=0.1))
model_2_rgb.add(MaxPooling2D(2, 2))
# Convolutional Block #3
model_2_rgb.add(Conv2D(64, (2, 2), activation="relu", padding="same"))
model_2_rgb.add(BatchNormalization())
model_2_rgb.add(LeakyReLU(alpha=0.1))
model_2_rgb.add(MaxPooling2D(2, 2))
# Convolutional Block #4
model_2_rgb.add(Conv2D(32, (2, 2), activation="relu", padding="same"))
model_2_rgb.add(BatchNormalization())
model_2_rgb.add(LeakyReLU(alpha=0.1))
model_2_rgb.add(MaxPooling2D(2, 2))
# Flatten layer
model_2_rgb.add(Flatten())
# Dense layers
model_2_rgb.add(Dense(512, activation="relu"))
model_2_rgb.add(Dense(256, activation="relu"))
# Classifier
model_2_rgb.add(Dense(4, activation="softmax"))
model_2_rgb.summary()
# ### **Compiling and Training the Model**
#
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_2_rgb.h5",
monitor="val_accuracy",
verbose=1,
save_best_only=True,
mode="auto",
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_2_rgb.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_2_rgb = model_2_rgb.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_2_rgb.history["accuracy"])
plt.plot(history_2_rgb.history["val_accuracy"])
plt.title("Accuracy - Model 2 (RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_2_rgb.history["loss"])
plt.plot(history_2_rgb.history["val_loss"])
plt.title("Loss - Model 2 (RGB)")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on the Test Set**
# Evaluating the model's performance on the test set
accuracy = model_2_rgb.evaluate(test_set_rgb)
# **Observations and Insights:**
# As constructed, our second RGB model also performs somewhat differently than its predecessor. After 15 epochs (best epoch), training accuracy stands at 0.76 and validation accuracy is 0.71, which are both higher than Model 1, but Model 2 begins to overfit almost immediately. Training accuracy and loss continue to improve, while validation accuracy and loss level off before early-stopping ends the training process. Accuracy on the test set is 0.68. Once again, our model is not generalizing well, though with better accuracy scores compared to Model 1, there is an opportunity (if overfitting can be reduced) to become the better RGB model.
# Our deeper grayscale and RGB models again perform similarly across all metrics, with the grayscale model attaining slightly better accuracies. Again, a slight edge to the grayscale model for performing better on the test set with a smaller number of trainable parameters.
# | | Training | Validation | Test |
# | --- | --- | --- | --- |
# | Grayscale Accuracy |0.78 | 0.71 | 0.69 |
# | RGB Accuracy | 0.76 | 0.71 | 0.68 |
#
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_2_grayscale.history["accuracy"])
plt.plot(history_2_grayscale.history["val_accuracy"])
plt.plot(history_2_rgb.history["accuracy"])
plt.plot(history_2_rgb.history["val_accuracy"])
plt.title("Accuracy - Model 2 (Grayscale & RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(
[
"Training Accuracy (Grayscale)",
"Validation Accuracy (Grayscale)",
"Training Accuracy (RGB)",
"Validation Accuracy (RGB)",
],
loc="lower right",
)
plt.show()
# **Overall Observations and Insights on Initial Models:**
# * As discussed above, both grayscale models slightly outperformed their RGB counterparts, and did so using less trainable parameters, making them less computationally expensive. Given this performance, we will proceed with grayscale models when doing so is possible.
# * As the datasets for this project are black and white images, it is possible that a grayscale colormode works better than a RGB colormode on what are essentially grayscale images. In this case, adding a second and third channel and increasing the input shape from 48,48,1 to 48,48,3 does not seem to help the modeling, and in fact may be making it overly complex.
# * As evidenced by the graph below, the 4 models thus far have fairly similar accuracy trajectories, though with a fair degree of separation between them. There is obviously room for improvement when it comes to overall accuracy. As early-stopping has prohibited us from seeing whether or not the training accuracy and loss level off before reaching 100%, it is clear that they continue to improve while validation accuracy and loss level off.
# * Some possible ways to decrease overfitting and thereby improve the above models include:
# * Introduce additional forms of data augmentation. While the above models take advantage of **horizontal_flip**, **brightness_range**, **rescale**, and **sheer_range**, it is possible that introducing additional forms of data augmentation (like **width_shift_range**, **height_shift_range**, **zoon_range**, **rotation_range**, etc. as discussed above) could help improve model performance.
# * Additional use **BatchNormalization** could also improve performance by offering some degree of regularization.
# * Additional use of **DropOut** and **SpatialDropout** could also help improve performance by assisting in regularization.
# * Introducting **GaussianNoise** could also assist in regularization, adding a form of noise to the data.
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_1_grayscale.history["accuracy"])
plt.plot(history_1_grayscale.history["val_accuracy"])
plt.plot(history_1_rgb.history["accuracy"])
plt.plot(history_1_rgb.history["val_accuracy"])
plt.plot(history_2_grayscale.history["accuracy"])
plt.plot(history_2_grayscale.history["val_accuracy"])
plt.plot(history_2_rgb.history["accuracy"])
plt.plot(history_2_rgb.history["val_accuracy"])
plt.title("Accuracy - Models 1 & 2 (Grayscale & RGB)")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(
[
"Training Accuracy - Model 1 (Grayscale)",
"Validation Accuracy - Model 1 (Grayscale)",
"Training Accuracy - Model 1 (RGB)",
"Validation Accuracy - Model 1 (RGB)",
"Training Accuracy - Model 2 (Grayscale)",
"Validation Accuracy - Model 2 (Grayscale)",
"Training Accuracy - Model 2 (RGB)",
"Validation Accuracy - Model 2 (RGB)",
],
loc="lower right",
)
plt.show()
# ## **Transfer Learning Architectures**
# In this section, we will create several Transfer Learning architectures. For the pre-trained models, we will select three popular architectures, namely: VGG16, ResNet v2, and Efficient Net. The difference between these architectures and the previous architectures is that these will require 3 input channels (RGB) while the earlier models also worked on grayscale images.
# ### **Creating our Data Loaders for Transfer Learning Architectures**
# We will create new data loaders for the transfer learning architectures used below. As required by the architectures we will be piggybacking, color_mode will be set to RGB.
# Additionally, we will be using the same data augmentation methods used on our previous models in order to better compare performance against our baseline models. These methods include **horizontal_flip**, **brightness_range**, **rescale**, and **shear_range**.
#
batch_size = 32
# Creating ImageDataGenerator objects for RGB colormode
datagen_train_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_validation_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
datagen_test_rgb = ImageDataGenerator(
horizontal_flip=True,
brightness_range=(0.0, 2.0),
rescale=1.0 / 255,
shear_range=0.3,
)
# Creating train, validation, and test sets for RGB colormode
print("\nColor Images")
train_set_rgb = datagen_train_rgb.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_rgb = datagen_validation_rgb.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_set_rgb = datagen_test_rgb.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="rgb",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
# ## Model 3: VGG16
# First up is the VGG16 model, which is a CNN consisting of 13 convolutional layers, 5 MaxPooling layers, and 3 dense layers. The VGG16 model achieves nearly 93% accuracy on the ImageNet dataset containing 14 million images across 1,000 classes. Clearly, this is much more substantial than our models above.
# ### **Importing the VGG16 Architecture**
vgg = VGG16(include_top=False, weights="imagenet", input_shape=(48, 48, 3))
vgg.summary()
# ### **Model Building**
# We have imported the VGG16 model up to layer 'block4_pool', as this has shown the best performance compared to other layers (discussed below). The VGG16 layers will be frozen, so the only trainable layers will be those we add ourselves. After flattening the input from 'block4_pool', 2 dense layers will be added, followed by a Dropout layer, another dense layer, and BatchNormalization. We will end with a softmax classifier.
#
transfer_layer = vgg.get_layer("block4_pool")
vgg.trainable = False
# Flatten the input
x = Flatten()(transfer_layer.output)
# Dense layers
x = Dense(256, activation="relu")(x)
x = Dense(128, activation="relu")(x)
x = Dropout(0.2)(x)
x = Dense(64, activation="relu")(x)
x = BatchNormalization()(x)
# Classifier
pred = Dense(4, activation="softmax")(x)
# Initialize the model
model_3 = Model(vgg.input, pred)
# ### **Compiling and Training the VGG16 Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_3.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss",
min_delta=0,
patience=15, # This is increased compared to initial models, otherwise training is cut too quickly
verbose=1,
restore_best_weights=True,
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_3.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_3 = model_3.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_3.history["accuracy"])
plt.plot(history_3.history["val_accuracy"])
plt.title("Accuracy - VGG16 Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_3.history["loss"])
plt.plot(history_3.history["val_loss"])
plt.title("Loss - VGG16 Model")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the VGG16 model**
# Evaluating the model's performance on the test set
accuracy = model_3.evaluate(test_set_rgb)
# **Observations and Insights:**
# As imported and modified, our transfer learning model seems to perform similarly to our previous models developed above. After 29 epochs (best epoch), training accuracy stands at 0.72 and validation accuracy is 0.67. Accuracy and loss for both the training and validation data level off before early stopping ends the training. The model's performance on the test data stands at 0.66. These scores are roughly in line with the scores of Model 1, our baseline model.
# The VGG16 model was ultimately imported up to layer block4_pool, as it produced the best performance. A history of alternative models is below.
# | | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | VGG16 block4_pool (selected) |0.71 | 0.72 | 0.80 | 0.67 |
# | VGG16 block5_pool |1.05 | 0.54 | 1.10 | 0.52 |
# | VGG16 block3_pool |0.79 | 0.69 | 0.77 | 0.66 |
# | VGG16 block2_pool |0.71 | 0.71 | 0.82 | 0.65 |
# ## Model 4: ResNet v2
# Our second transfer learning model is ResNet v2, which is a CNN trained on over 1 million images from the ImageNet database. ResNet v2 can classify images into 1,000 different categories. Like VGG16, colormode must be set to RGB to leverage this pre-trained architecture.
Resnet = ap.ResNet101(include_top=False, weights="imagenet", input_shape=(48, 48, 3))
Resnet.summary()
# ### **Model Building**
# We have imported the ResNet v2 model up to layer 'conv_block23_add', as this has shown the best performance compared to other layers (discussed below). The ResNet v2 layers will be frozen, so the only trainable layers will be those we add ourselves. After flattening the input from 'conv_block23_add', we will add the same architecture we did earlier to VGG16, namely 2 dense layers, followed by a DropOut layer, another dense layer, and BatchNormalization. We will once again end with a softmax classifier, as this is a multi-class classification exercise.
transfer_layer = Resnet.get_layer("conv4_block23_add")
Resnet.trainable = False
# Flatten the input
x = Flatten()(transfer_layer.output)
# Dense layers
x = Dense(256, activation="relu")(x)
x = Dense(128, activation="relu")(x)
x = Dropout(0.2)(x)
x = Dense(64, activation="relu")(x)
x = BatchNormalization()(x)
# Classifier
pred = Dense(4, activation="softmax")(x)
# Initialize the model
model_4 = Model(Resnet.input, pred)
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_4.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss",
min_delta=0,
patience=15, # Increased over initial models otherwise training is cut off too quickly
verbose=1,
restore_best_weights=True,
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_4.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_4 = model_4.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_4.history["accuracy"])
plt.plot(history_4.history["val_accuracy"])
plt.title("Accuracy - ResNet V2 Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_4.history["loss"])
plt.plot(history_4.history["val_loss"])
plt.title("Loss - ResNet V2 Model")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the ResNet Model**
# Evaluating the model's performance on the test set
accuracy = model_4.evaluate(test_set_rgb)
# **Observations and Insights:**
# As imported and modified, our transfer learning model shows terrible performance. After just 1 epoch (the 'best' epoch!), training accuracy stands at 0.26 and validation accuracy is 0.36. Accuracy and loss for both training and validation data level off fairly quickly at which point early stopping aborts the training. The above accuracy and loss curves paint the picture of poor model that will not generalize well at all. The model's test accuracy comes in at 0.34.
# The ResNet v2 model was ultimately imported up to layer 'conv4_block23_add', as it produced the 'best' performance, though it was difficult to choose. A history of alternative models is below.
# | | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | ResNet V2 conv4_block23_add (selected) |1.43 | 0.26 | 1.35 | 0.36 |
# | ResNet V2 conv5_block3_add |1.47 | 0.23 | 1.43 | 0.33 |
# | ResNet V2 conv3_block4_add |1.49 | 0.22 | 1.44 | 0.33 |
# | ResNet V2 conv2_block3_add |1.51 | 0.21 | 1.55 | 0.21 |
# ## Model 5: EfficientNet
# Our third transfer learning model is EfficientNet, which is a CNN that uses 'compound scaling' to improve efficiency and, theoretically at least, performance. Like VGG16 and ResNet v2, color_mode must be set to RGB to leverage this pre-trained architecture.
EfficientNet = ap.EfficientNetV2B2(
include_top=False, weights="imagenet", input_shape=(48, 48, 3)
)
EfficientNet.summary()
# ### **Model Building**
# We have imported the EfficientNet Model up to layer 'block5f_expand_activation', as this has shown the best performance compared to other layers (discussed below). The EfficientNet layers will be frozen, so the only trainable layers will be those that we add ourselves. After flattening the input from 'block5f_expand_activation', we will add the same architecture we did earlier to the VGG16 and ResNet v2 models, namely 2 dense layers, followed by a Dropout layer, another dense layer, and BatchNormalization. We will end with a softmax classifier.
#
transfer_layer_EfficientNet = EfficientNet.get_layer("block5f_expand_activation")
EfficientNet.trainable = False
# Flatten the input
x = Flatten()(transfer_layer_EfficientNet.output)
# Dense layers
x = Dense(256, activation="relu")(x)
x = Dense(128, activation="relu")(x)
x = Dropout(0.2)(x)
x = Dense(64, activation="relu")(x)
x = BatchNormalization()(x)
# Classifier
pred = Dense(4, activation="softmax")(x)
# Initialize the model
model_5 = Model(EfficientNet.input, pred)
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_5.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=12, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_5.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# Fitting model with epochs set to 100
history_5 = model_5.fit(
train_set_rgb, validation_data=val_set_rgb, epochs=100, callbacks=callbacks_list
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_5.history["accuracy"])
plt.plot(history_5.history["val_accuracy"])
plt.title("Accuracy - EfficientNet Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="center right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_5.history["loss"])
plt.plot(history_5.history["val_loss"])
plt.title("Loss - EfficientNet Model")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the EfficientNet Model**
# Evaluating the model's performance on the test set
accuracy = model_5.evaluate(test_set_rgb)
# **Observations and Insights:**
# As imported and modified, this model performs poorly. After just 4 epochs (the 'best' epoch), training accuracy stands at 0.26 and validation accuracy is 0.24. Training and validation accuracy are almost immediately horizontal. Loss declines a bit before leveling off. With test accuracy coming in at 0.25, it makes the model no better than random guessing. We could build a model that classifies every single image as 'happy', and with our evenly distributed test set, it would produce the same 0.25 accuracy as our EfficientNet model.
# Again, it was difficult to select a 'best' layer from which to import the EfficientNet model. A history of alternative models is below.
# | | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | EfficientNet block5f_expand_activation (selected) |1.39 | 0.26 | 1.37 | 0.24 |
# | EfficientNet block6e_expand_activation |1.53 | 0.25 | 1.45 | 0.22 |
# | EfficientNet block4a_expand_activation |1.42 | 0.25 | 1.42 | 0.21 |
# | EfficientNet block3c_expand_activation |1.47 | 0.26 | 1.44 | 0.22 |
# **Overall Observations and Insights on Transfer Learning Models:**
# * As outlined above, the performance of these transfer learning models varied greatly. While the VGG16 model performed admirably (see table below), the ResNet v2 and EfficientNet models left much to be desired in terms of stability and performance.
# * On the whole, none of the transfer learning models performed better than our baseline and 2nd generation models, which was surprising.
# * Model complexity seems to have played a role in performance, as the VGG16 model has a much less complex architecture than both the ResNet v2 and the EfficientNet models. Perhaps overly-complex models trained on millions of large, color images do not perform as well on smaller, black and white images from just 4 classes.
# * VGG16, with 14.7 million parameters, is a fairly straightforward architecture, with just 19 layers from the input layer to the max import layer, 'block5_pool'.
# * ResNet v2, with 42.7 million parameters, is a much more complex architecture, with a whopping 345 layers from the input layer to the max import layer, 'conv5_block3_out'.
# * EfficientNet, with 'just' 8.8 million parameters, contains 349 layers from the input layer to the max import layer, 'top_activation'.
# * As evidenced by the table below, it would appear that the unsatisfactory performance of the transfer learning models may have more to do with their complexity than the fact that they require a colormode of RGB. The baseline and 2nd generation RGB models both performed just as well as the VGG16 model. It would seem that the downfall of ResNet v2 and EfficientNet was their complex architecture. Quite simply, the simpler models performed better. In fact, the highest performing model so far, the 2nd generation grayscale model (Model 2.1), has the smallest number of parameters.
# * Perhaps a sweet spot exists somewhere between the simplicity of our 2nd generation grayscale model and the much more complex transfer learning models we have explored thus far. If it is possible to increase the complexity of our 2nd generation grayscale model while keeping the overall complexity from ballooning too far in the direction of the transfer learning models, we may find ourselves a successful model.
#
# | | Parameters | Train Loss | Train Accuracy | Val Loss | Val Accuracy | Test Accuracy |
# | --- | --- | --- | --- | --- | --- | --- |
# | **Model 1.1**: Baseline Grayscale | 605,060 | 0.68 | 0.72 | 0.78 | 0.68 | 0.65 |
# | **Model 1.2**: Baseline RGB | 605,572 | 0.68 | 0.72 | 0.78 | 0.68 | 0.63 |
# | **Model 2.1**: 2nd Gen Grayscale | 455,780 | 0.54 | 0.78 | 0.74 | 0.71 | 0.69 |
# | **Model 2.2**: 2nd Gen RGB | 457,828 | 0.59 | 0.76 | 0.72 | 0.71 | 0.68 |
# | **Model 3**: VGG16 | 14,714,688 | 0.71 | 0.72 | 0.80 | 0.67 | 0.66 |
# | **Model 4**: ResNet V2 | 42,658,176 | 1.43 | 0.26 | 1.35 | 0.36 | 0.28 |
# | **Model 5**: EfficientNet | 8,769,374 | 1.39 | 0.26 | 1.37 | 0.24 | 0.25 |
# # Milestone 1
# ## Model 6: Complex Neural Network Architecture
# As previewed above, it is time to expand our 2nd generation grayscale model to see if we can improve performance. Grayscale slightly outperformed RGB in our first two models, so we will leave RGB behind and proceed with color_mode set to grayscale.
# ## **Creating our Data Loaders**
# As we are proceeding with a colormode set to grayscale, we will create new data loaders for our more complex CNN, Model 6. As our data augmentation takes place when we instantiate an ImageDataGenerator object, it is convenient to create data loaders specific to our new model so we can easily finetune our hyperparameters as needed. The ImageDataGenerators below include the parameters of the final Milestone 1 model, the highest performing CNN thus far. They were chosen after exhaustive finetuning of the model, as discussed later.
# * Batch size is set to 32. The model was tested with batch sizes of 16, 32, 45, 64, and 128. A batch size of 32 performed the best. The smaller the batch size, the longer training took. The larger the batch size, the faster the training process, though the accuracy and loss bounced around significantly, offsetting the increased speed.
# * **horizontal_flip** is set to 'True'. As some faces in the images face left while others face right or straight ahead, flipping the training images improves our model's ability to learn that horizontal orientation should not affect the eventual classification.
# * **rescale** is equal to 1./255, which normalizes the pixel values to a number between 0 and 1. This helps to prevent vanishing and exploding gradients in our network by keeping the numbers small and manageable.
# * **brightness_range** is set to '0.7,1.3'. A setting of 1 results in images remaining unchanged. As the number approaches zero, the images become darker. As the number approaches 2, the images become lighter. As many of the images are already very dark or very light, limiting this setting to a relatively small range around 1 will help our model learn to deal with varying pixel values without rendering some images completely unusable.
# * **rotation_range** is set to 25, meaning the images may randomly be rotated up to 25 degrees. Similar to flipping the images horizontally, this rotation will help the model learn that the angle of a face is not an important feature.
# * Additional data augmentation methods were attempted and later removed after failing to significantly improve model performance. Among those tested were **width_shift_range**, **height_shift_range**, **shear_range**, and **zoom_range**.
batch_size = 32
# Creating ImageDataGenerator objects for grayscale colormode
datagen_train_grayscale = ImageDataGenerator(
horizontal_flip=True,
rescale=1.0 / 255,
brightness_range=(0.7, 1.3),
rotation_range=25,
)
datagen_validation_grayscale = ImageDataGenerator(
horizontal_flip=True,
rescale=1.0 / 255,
brightness_range=(0.7, 1.3),
rotation_range=25,
)
datagen_test_grayscale = ImageDataGenerator(
horizontal_flip=True,
rescale=1.0 / 255,
brightness_range=(0.7, 1.3),
rotation_range=25,
)
# Creating train, validation, and test sets for grayscale colormode
print("Grayscale Images")
train_set_grayscale = datagen_train_grayscale.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_grayscale = datagen_validation_grayscale.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
test_set_grayscale = datagen_test_grayscale.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
# ### **Model Building**
# The structure of the Milestone 1 model (Model 6) is below. Many configurations were tested, and the following architecture led to the best performance.
# * The model begins with an input layer accepting an input shape of '48,48,1', given that our color_mode has been set to grayscale.
# * There are 5 convolutional blocks with relu activation. Each block contains BatchNormalization, LeakyReLU, and MaxPooling layers. The first, second, and fourth blocks include a layer of GaussianNoise, while the third and fifth layers each include a Dropout layer.
# * The output of the fifth convolutional block is then flattened, and fed into 2 dense layers which include additional BatchNormalization and Dropout layers.
# * The architecture is completed with a softmax classifier, as this model is designed for multi-class classification. Test images will be classified as either happy, sad, neutral, or surprise.
# * The model contains 2.1 million parameters, making it more complex than our 2nd generation grayscale model, but not as complex as the transfer learning models, whose complexity appeared to hurt their performance.
#
# Creating a Sequential model
model_6 = Sequential()
# Convolutional Block #1
model_6.add(
Conv2D(64, (3, 3), input_shape=(48, 48, 1), activation="relu", padding="same")
)
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(GaussianNoise(0.1))
# Convolutional Block #2
model_6.add(Conv2D(128, (3, 3), activation="relu", padding="same"))
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(GaussianNoise(0.1))
# Convolutional Block #3
model_6.add(Conv2D(512, (2, 2), activation="relu", padding="same"))
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(Dropout(0.1))
# Convolutional Block #4
model_6.add(Conv2D(512, (2, 2), activation="relu", padding="same"))
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(GaussianNoise(0.1))
# Convolutional Block #5
model_6.add(Conv2D(256, (2, 2), activation="relu", padding="same"))
model_6.add(BatchNormalization())
model_6.add(LeakyReLU(alpha=0.1))
model_6.add(MaxPooling2D(2, 2))
model_6.add(Dropout(0.1))
# Flatten layer
model_6.add(Flatten())
# Dense layers
model_6.add(Dense(256, activation="relu"))
model_6.add(BatchNormalization())
model_6.add(Dropout(0.1))
model_6.add(Dense(512, activation="relu"))
model_6.add(BatchNormalization())
model_6.add(Dropout(0.05))
# Classifier
model_6.add(Dense(4, activation="softmax"))
model_6.summary()
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_6.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=10, verbose=1, restore_best_weights=True
)
# Initiates reduced learning rate if validation loss does not continue to improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_6.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
# Fitting model with epochs set to 100
history_6 = model_6.fit(
train_set_grayscale,
validation_data=val_set_grayscale,
epochs=100,
callbacks=callbacks_list,
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_6.history["accuracy"])
plt.plot(history_6.history["val_accuracy"])
plt.title("Accuracy - Complex Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_6.history["loss"])
plt.plot(history_6.history["val_loss"])
plt.title("Loss - Complex Model")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on Test Set**
# Evaluating the model's performance on the test set
accuracy = model_6.evaluate(test_set_grayscale)
# **Observations and Insights:**
# Model 6, our Milestone 1 model, outperforms all previous models. After 33 epochs (best epoch), training accuracy stands at 0.79 and validation accuracy is 0.76. Accuracy and loss for both training and validation data improve similarly before leveling off. The model begins to overfit around epoch 15, but the overfitting is not as severe as previous models. The test accuracy for this model is 0.76. Overall, Model 6 generalizes better than previous models, and is the top performer thus far. That said, **it is still an overfitting model, and thus it would not be advisable to deploy this model as is**.
# This model underwent numerous transformations before arriving at its final state. Parameters were tuned, layers were added, layers were removed, and eventually the above model was determined to be the best iteration. An abridged history of model development can be found in the table below.
# The starting point for our final model was as follows:
# **CONVOLUTIONAL BLOCK #1**
# * Conv2D(64,(2,2), input shape = (48,48,1), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **CONVOLUTIONAL BLOCK #2**
# * Conv2D(128,(2,2), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **CONVOLUTIONAL BLOCK #3**
# * Conv2D(512,(2,2), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **CONVOLUTIONAL BLOCK #4**
# * Conv2D(256,(2,2), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **CONVOLUTIONAL BLOCK #5**
# * Conv2D(128,(2,2), activation = 'relu', padding = 'same')
# * BatchNormalization
# * LeakyReLU(alpha = 0.1)
# * MaxPooling2D(2,2)
# **FINAL LAYERS**
# * Flatten
# * Dense(256, activation = 'relu')
# * Dropout(0.1)
# * Dense(256, activation = 'relu')
# * Dropout(0.1)
# * Dense(4, activation = 'softmax')
# **PARAMETERS**
# * Batch size = 32
# * horizontal_flip = True
# * rescale = 1./255
# * brightness_range = (0.0,2.0)
# * shear_range = 0.3
#
#
# Below is an abridged summary of actions taken to improve the model. In many cases, parameters or layers were adjusted, added, or removed, just to be returned to their original state when the desired or experimental impact was not realized. The model went through dozens of iterations, with the following transformations being the most impactful.
# | Action Taken | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | Starting model as outlined above | 0.77 | 0.70 | 0.89 | 0.58 |
# | Dropout(0.1) layers added to conv blocks 1 and 5 to reduce overfitting |0.75 | 0.74 | 0.66 | 0.61 |
# | Shear_range removed entirely to determine effect |0.76 | 0.74 | 0.68 | 0.60 |
# | Rotation_range added and optimized |0.74 | 0.74 | 0.62 | 0.61 |
# | Additional dropout layers added to blocks 2 and 4 |0.59 | 0.78 | 0.64 | 0.68 |
# | Number of neurons in final dense layer set to 512 |0.68 | 0.71 | 0.62 | 0.71 |
# | Number of neurons in block 4 increased to 512 |0.70 | 0.73 | 0.60 | 0.74 |
# | Dropout layers swapped out for GaussianNoise in blocks 1 and 2 |0.61 | 0.74 | 0.57 | 0.75 |
# | Brightness_range narrowed to (0.5,1.5) then to (0.7,1.3) |0.59 | 0.75 | 0.60 | 0.75 |
# | Kernel size enlarged to 3x3 in first then also second block |0.55 | 0.78 | 0.57 | 0.75 |
# | Dropout in block 5 reduced to 0.5, resulting in final model |0.54 | 0.79 | 0.60 | 0.76 |
# # Final Solution
# ## Model 7: Goodbye Overfitting
# While Model 6 was an improvement on previous models, it was still overfitting the training data. In order to feel comfortable recommending a model for deployment in the context of this project, we need to improve on Model 6. Model 7 is an attempt to develop a deployable CNN. We want our model to have high accuracy, while also maintaining a good fit (no overfitting/underfitting) and generalizing well to the unseen test data. We will continue with color_mode set to grayscale for the reasons already noted: slightly better performance, slightly fewer parameters, slightly lower computational expense, and the image data itself is already grayscale.
# ## **Creating our Data Loaders**
# We will once again be creating new data loaders for Model 7. As mentioned earlier, since our data augmentation takes place when we instantiate an ImageDataGenerator object, it is convenient to create data loaders specific to our new model so we can easily finetune our hyperparameters as needed. The ImageDataGenerators below include the parameters of our final, highest performing iteration of the model. They were once again chosen after exhaustive finetuning, as discussed later.
# * Batch size is set to 128. The model was tested with batch sizes of 16, 32, 64, 128, and 256. A batch size of 128 performed the best. The smallest batch sizes seemed to get stuck in an accuracy range of 25-30% (perhaps a local minimum), while the other sizes did not generalize as well to the test data.
# * **horizontal_flip** is set to 'True'. As some faces in the images face left while others face right or straight ahead, flipping the training images improves our model's ability to learn that horizontal orientation should not affect the eventual classification.
# * **rescale** is equal to 1./255, which normalizes the pixel values to a number between 0 and 1. This helps to prevent vanishing and exploding gradients in our network by keeping the numbers small and manageable.
# * **brightness_range** is set to '0.0,2.0'. This is a change from Model 6 where we used a narrower range. A narrower range did not help within the architecture of Model 7, and the broader range showed better performance.
# * **shear_range** is set to 0.3, which matches the settings of our baseline models. This parameter essentially distorts the image along an axis in a counter-clockwise direction.
# * **one-hot-encoding** is handled by setting **class_mode** to "categorical", followed by our list of classes.
# * Additional data augmentation methods were attempted and later removed after failing to significantly improve model performance. Among those tested were **width_shift_range**, **height_shift_range**, **rotation_range**, **zca_whitening**, **zoom_range**, and even **vertical_flip**.
batch_size = 128
# Creating ImageDataGenerator objects for grayscale colormode
datagen_train_grayscale = ImageDataGenerator(
rescale=1.0 / 255,
brightness_range=(0.0, 2.0),
horizontal_flip=True,
shear_range=0.3,
)
datagen_validation_grayscale = ImageDataGenerator(
rescale=1.0 / 255,
brightness_range=(0.0, 2.0),
horizontal_flip=True,
shear_range=0.3,
)
datagen_test_grayscale = ImageDataGenerator(
rescale=1.0 / 255,
brightness_range=(0.0, 2.0),
horizontal_flip=True,
shear_range=0.3,
)
# Creating train, validation, and test sets for grayscale colormode
print("Grayscale Images")
train_set_grayscale = datagen_train_grayscale.flow_from_directory(
dir_train,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=True,
)
val_set_grayscale = datagen_validation_grayscale.flow_from_directory(
dir_validation,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_set_grayscale = datagen_test_grayscale.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=batch_size,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
# ### **Model Building**
# The structure of Model 7 is below. Rather than simply modifying Model 6, the development of Model 7 entailed going back to the drawing board and devising a new strategy. Many configurations were tested, and the following architecture led to the best, most generalizable performance.
# * The model begins with an input layer accepting an input shape of '48,48,1', given that our color_mode has been set to grayscale.
# * There are 3 similar convolutional blocks with relu activation. Padding is no longer set to "same", as this increased the generalization gap. Each block contains a BatchNormalization layer before its first and second convolutional layers (except the input layer in Block #1). Each block ends with MaxPooling and a Dropout layer set to 0.4.
# * A "secret" block, which is what eventually closed the generalization gap and eliminated overfitting, is essentially a normalization/regularization block consisting of a BatchNormalization layer and a convolutional layer without activation, but instead with a L2 regularization set to 0.025. This is followed by another BatchNormalization layer.
# * The output of the "secret" block is then flattened, and fed into 2 dense layers, each followed by a Dropout layer, and separated by a layer of GaussianNoise.
# * The architecture is completed with a softmax classifier, as this model is designed for multi-class classification. Test images will be classified as either happy, sad, neutral, or surprise.
# * The final model contains 1.8 million parameters and 27 layers, making it slightly less complex than Model 6, while still substantially more complex than our initial models.
# Creating a Sequential model
model_7 = Sequential()
# Convolutional Block #1
model_7.add(Conv2D(64, (3, 3), input_shape=(48, 48, 1), activation="relu"))
model_7.add(BatchNormalization())
model_7.add(Conv2D(64, (3, 3), activation="relu"))
model_7.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model_7.add(Dropout(0.4))
# Convolutional Block #2
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (3, 3), activation="relu"))
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (3, 3), activation="relu"))
model_7.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model_7.add(Dropout(0.4))
# Convolutional Block #3
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (3, 3), activation="relu"))
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (3, 3), activation="relu"))
model_7.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model_7.add(Dropout(0.4))
# SECRET LEVEL
model_7.add(BatchNormalization())
model_7.add(Conv2D(128, (2, 2), kernel_regularizer=l2(0.025)))
model_7.add(BatchNormalization())
# Flatten layer
model_7.add(Flatten())
# Dense layers
model_7.add(Dense(1024, activation="relu"))
model_7.add(Dropout(0.2))
model_7.add(GaussianNoise(0.1))
model_7.add(Dense(1024, activation="relu"))
model_7.add(Dropout(0.2))
# Classifier
model_7.add(Dense(4, activation="softmax"))
model_7.summary()
# ### **Compiling and Training the Model**
# Creating a checkpoint which saves model weights from the best epoch
checkpoint = ModelCheckpoint(
"./model_7.h5", monitor="val_accuracy", verbose=1, save_best_only=True, mode="auto"
)
# Initiates early stopping if validation loss does not continue to improve
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, restore_best_weights=True
)
# Slows the learning rate when validation loss does not improve
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=2, verbose=1, min_delta=0.0001
)
callbacks_list = [checkpoint, early_stopping, reduce_learningrate]
# **Note:**
# * Early stopping patience is set to 5 epochs. This model was trained with Patience set to 5, 10, 12, 15, 20, and 50. Each time, the model achieved the same results, so the simpler model (patience = 5) was chosen.
# * Reduce learning rate patience is set to 2 epochs. Again, the model was trained with patience set to 1, 2, 3, and 5. The results varied considerably, with 2 epochs being the only iteration that did not result in a generalization gap.
#
# Compiling model with optimizer set to Adam, loss set to categorical_crossentropy, and metrics set to accuracy
model_7.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
# Fitting model with epochs set to 200
history_7 = model_7.fit(
train_set_grayscale,
validation_data=val_set_grayscale,
epochs=200,
callbacks=callbacks_list,
)
# Plotting the accuracies
plt.figure(figsize=(10, 5))
plt.plot(history_7.history["accuracy"])
plt.plot(history_7.history["val_accuracy"])
plt.title("Accuracy - Final Model")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="lower right")
plt.show()
# Plotting the losses
plt.figure(figsize=(10, 5))
plt.plot(history_7.history["loss"])
plt.plot(history_7.history["val_loss"])
plt.title("Loss - Final Model")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Training", "Validation"], loc="upper right")
plt.show()
# ### **Evaluating the Model on Test Set**
# Evaluating the model's performance on the test set
accuracy = model_7.evaluate(test_set_grayscale)
# **Observations and Insights:**
# Model 7, rewarding us for all of our efforts, displays the best all-around performance. Accuracies for training, validation, and test data are stable at 0.75, while loss is stable across training, validation, and test data at roughly 0.63 (0.62 to 0.64). As evidenced by the above graphs, there is no noticeable generalization gap. The accuracy and loss curves move more or less in tandem, leveling off around epoch 25 and remaining together from that point forward. The model does not overfit or underfit the training data. The images below show the accuracy and loss curves for the same model run out to 115 epochs. The model converges at reasonable levels of accuracy and loss, and it generalizes well.
#
#
# Much like Model 6, this model underwent numerous transformations before arriving at its final state. Parameters were tuned, layers were added, others were removed, and in the end, the above iteration of the model was determined to be the best. Below are the impacts that some of the most important aspects of the model have on its overall performance. While some individual metrics may be better than those of the final model, each of the modifications below, if taken individually or in tandem, results in a generalization gap that is not present in the final model.
#
#
# | Model Changes | Train Loss | Train Accuracy | Val Loss | Val Accuracy |
# | --- | --- | --- | --- | --- |
# | Final Model | 0.63 | 0.75 | 0.64 | 0.75 |
# | Remove "regularization" block |0.63 | 0.76 | 0.68 | 0.73 |
# | Remove L2 kernel regularizer |0.62 | 0.74 | 0.64 | 0.73 |
# | Remove Gaussian Noise |0.65 | 0.73 | 0.66 | 0.74 |
# | Reduce kernel size to (2,2) |0.63 | 0.74 | 0.66 | 0.74 |
# | Dropout levels reduced to 0.2 |0.57 | 0.78 | 0.65 | 0.74 |
# | Remove BatchNormalization |0.74 | 0.70 | 0.69 | 0.72 |
# | Include relu activation in regularization block |0.63 | 0.74 | 0.63 | 0.74 |
# | Batch size = 32 |0.62 | 0.75 | 0.65 | 0.74 |
# | Data augmentation with rotation range = 20 | 0.69 | 0.72 | 0.67 | 0.74 |
# | Data augmentation with zoom range = 0.2 | 0.71 | 0.71 | 0.69 | 0.73 |
# | Vertical flip = True | 0.74 | 0.71 | 0.70 | 0.74 |
# | Only 1 convolutional layer per block | 0.84 | 0.66 | 0.78 | 0.70 |
# ### **Model Comparison**
# Below are the accuracy and loss scores for each of our models, first in a tabular format, then represented visually in the form of bar charts.
# | | Parameters | Train Loss | Train Accuracy | Val Loss | Val Accuracy | Test Loss | Test Accuracy |
# | --- | --- | --- | --- | --- | --- | --- | --- |
# | **Model 1.1**: Baseline Grayscale | 605,060 | 0.68 | 0.72 | 0.78 | 0.68 | 0.82 | 0.65 |
# | **Model 1.2**: Baseline RGB | 605,572 | 0.68 | 0.72 | 0.78 | 0.68 | 0.80 | 0.63 |
# | **Model 2.1**: 2nd Gen Grayscale | 455,780 | 0.54 | 0.78 | 0.74 | 0.71 | 0.81 | 0.69 |
# | **Model 2.2**: 2nd Gen RGB | 457,828 | 0.59 | 0.76 | 0.72 | 0.71 | 0.70| 0.68 |
# | **Model 3**: VGG16 | 14,714,688 | 0.71 | 0.72 | 0.80 | 0.67 | 0.74 | 0.66 |
# | **Model 4**: ResNet V2 | 42,658,176 | 1.43 | 0.26 | 1.35 | 0.36 | 1.40 | 0.28 |
# | **Model 5**: EfficientNet | 8,769,374 | 1.39 | 0.26 | 1.37 | 0.24 | 1.40 | 0.25 |
# | **Model 6**: Milestone 1 | 2,119,172| 0.54 | 0.79 | 0.60 | 0.76 | 0.56 | 0.76 |
# | **Model 7**: Final Model | 1,808,708 | 0.63 | 0.75 | 0.64 | 0.75 | 0.62 | 0.75 |
#
# creating a dictionary containing model accuracies
dict_model_acc = {
"Model": ["1.1", "1.2", "2.1", "2.2", "3", "4", "5", "6", "7"],
"Train": [0.72, 0.72, 0.78, 0.76, 0.72, 0.26, 0.26, 0.79, 0.75],
"Validate": [0.68, 0.68, 0.71, 0.71, 0.67, 0.36, 0.24, 0.76, 0.75],
"Test": [0.65, 0.63, 0.69, 0.68, 0.66, 0.28, 0.25, 0.76, 0.75],
}
# converting dictionary to dataframe
df_model_acc = pd.DataFrame.from_dict(dict_model_acc)
# plotting accuracy scores for all models
df_model_acc.groupby("Model", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="Accuracy Scores Across Models",
ylabel="Accuracy Score",
xlabel="Models",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.legend(loc=(0.59, 0.77))
plt.show()
# creating a dictionary containing model loss
dict_model_loss = {
"Model": ["1.1", "1.2", "2.1", "2.2", "3", "4", "5", "6", "7"],
"Train": [0.68, 0.68, 0.54, 0.59, 0.71, 1.43, 1.39, 0.54, 0.63],
"Validate": [0.78, 0.78, 0.74, 0.72, 0.80, 1.35, 1.37, 0.60, 0.64],
"Test": [0.82, 0.80, 0.81, 0.70, 0.74, 1.40, 1.40, 0.56, 0.62],
}
# converting dictionary to dataframe
df_model_loss = pd.DataFrame.from_dict(dict_model_loss)
# plotting loss scores for all models
df_model_loss.groupby("Model", sort=False).mean().plot(
kind="bar",
figsize=(10, 5),
title="Loss Scores Across Models",
ylabel="Loss Score",
xlabel="Models",
rot=0,
fontsize=12,
width=0.9,
colormap="Pastel2",
edgecolor="black",
)
plt.show()
# **Observations and Insights:**
# The above graphs perfectly depict the overfitting that occurs in Models 1.1, 1.2, 2.1, 2.2, and 3, with accuracy scores declining in steps as we move from training, to validation, and on to test data. The opposite is true for the loss scores. The graphs also show the total dysfunction of Models 4 and 5, with very low accuracy and very high error scores. It is also clear from the graphs that Models 6 and 7 are the most consistent, most generalizable models, and that a final decision regarding a deployable model should be made between those two options.
# In deciding between Models 6 and 7, it is useful to revisit the accuracy and loss curves for the two models.
# #### **Accuracy and loss curves for Model 6:**
#
#
# #### **Accuracy and loss curves for Model 7:**
#
#
# While the accuracy and loss curves for the two models both stabilize by epoch 20-25, there is no gap between accuracy and loss curves for Model 7, while a slight gap does exist for Model 6. The accuracy and loss scores are all individually better for Model 6 (higher accuracy and lower loss), but when viewed together, the spread within the two scores is larger for Model 6, while it is virtually nonexistent in Model 7. It is difficult to justify deploying a slightly overfitting model when a slightly less accurate but more generalizable model is available. Model 7 will be our final model.
# ### **Plotting the Confusion Matrix for Model 7**
test_set = datagen_test_grayscale.flow_from_directory(
dir_test,
target_size=(img_size, img_size),
color_mode="grayscale",
batch_size=128,
class_mode="categorical",
classes=["happy", "sad", "neutral", "surprise"],
seed=42,
shuffle=False,
)
test_images, test_labels = next(test_set)
pred = model_7.predict(test_images)
pred = np.argmax(pred, axis=1)
y_true = np.argmax(test_labels, axis=1)
# Printing the classification report
print(classification_report(y_true, pred))
# Plotting the heatmap using the confusion matrix
cm = confusion_matrix(y_true, pred)
plt.figure(figsize=(8, 5))
sns.heatmap(
cm,
annot=True,
fmt=".0f",
xticklabels=["happy", "sad", "neutral", "surprise"],
yticklabels=["happy", "sad", "neutral", "surprise"],
)
plt.ylabel("Actual")
plt.xlabel("Predicted")
plt.show()
# **Observations and Insights:**
# * As noted above, our final model achieves an accuracy score of 0.75 on the test images. The model correctly predicted 96 of 128 images.
# * The choice to prioritize precision (TP/(TP+FP)) or recall (TP/(TP+FN)) depends entirely on the model's end use. If the stakes are high, and false negatives should be avoided at all costs, than recall is more important. If reducing the number of false positives is more important, than precision is the better choice. In the case of our model, no trade-off is necessary, with precision and recall scores essentially the same (precision = 0.76, recall = 0.75, F1 = 0.75).
# * As previewed during the data visualization phase of the project, the 'happy' and 'surprise' images seemed to have the most unique characteristics, and this hypothesis appears to have played out in the classification report and confusion matrix. Happy and surprise have the highest precision and recall scores (and consequently, F1 scores) of the 4 classes.
# * Additionally, 'sad' and 'neutral' images were in fact more likely to be confused with one another, as discussed during the data visualization phase. When the model misclassified a sad image, it was most likely to be mistaken for a neutral image, and vice versa.
# * Any concern about a slightly skewed class distribution can be put to rest. As previewed, the surprise images, which were outnumbered in the training and validation data, were unique enough to identify correctly despite representing a smaller proportion of training images. It is possible that our earlier finding re: elevated average pixel values for surprise images has played a role, along with the unique characteristics of surprise images, including open mouths and wide open eyes.
# * As discussed during the data visualization phase, now in the context of the confusion matrix, it should be pointed out once again that the term "accuracy" can be misleading. There are training, validation, and test images of smiling people that are labeled as "sad", while there are images of frowning people labeled as "happy", etc. If the model classifies a test image as "sad" even though the person is smiling, and in fact the test image is incorrectly classified as "sad", making the prediction accurate, should we really consider that as accurate? Or would the accurate prediction be when the model overrules the misclassified test image and, from a human's perspective, accurately classifies the image as "happy"? For this reason, the test scores and confusion matrix should be taken with a grain of salt.
# * Similarly, there is a test image that does not contain a face at all. As there are similar images across all four classes within the training data, a correct prediction of the empty test image would seem to be pure chance. Should an accurate prediction in this case really increase the model's perceived accuracy? Should an incorrect prediction of an empty test image really lower the model's perceived accuracy? It seems that any model that correctly predicts all 128 test images benefited from some degree of luck. Again, these final scores should be viewed with some degree of skepticism, but that skepticism would be similar across all models.
# ### **Visualizing Images: Actual Class Label vs Predicted Class Label**
# Making predictions on the test data
y_pred_test = model_7.predict(test_set)
# Converting probabilities to class labels
y_pred_test_classes = np.argmax(y_pred_test, axis=1)
# Calculating the probability of the predicted class
y_pred_test_max_probas = np.max(y_pred_test, axis=1)
classes = ["happy", "sad", "neutral", "surprise"]
rows = 3
cols = 4
fig = plt.figure(figsize=(12, 12))
for i in range(cols):
for j in range(rows):
random_index = np.random.randint(
0, len(test_labels)
) # generating random integer
ax = fig.add_subplot(rows, cols, i * rows + j + 1)
ax.imshow(test_images[random_index, :]) # selecting random test image
pred_label = classes[
y_pred_test_classes[random_index]
] # predicted label of selected image
pred_proba = y_pred_test_max_probas[
random_index
] # probability associated with model's prediction
true_label = test_labels[random_index] # actual class label of selected image
if true_label[0] == 1: # converting array to class labels
true_label = "happy"
elif true_label[1] == 1:
true_label = "sad"
elif true_label[2] == 1:
true_label = "neutral"
else:
true_label = "surprise"
ax.set_title(
"actual: {}\npredicted: {}\nprobability: {:.3}\n".format(
true_label, pred_label, pred_proba
)
)
plt.gray()
plt.show()
| false | 0 | 34,182 | 0 | 34,182 | 34,182 |
||
129384981
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from graphviz import Digraph
# Title: Simplified Decision Tree Classifier for Binary Features
# Description:
# This notebook presents a simplified decision tree classifier designed for datasets with binary features and binary classification tasks. The decision tree is built using a custom implementation, and the model is not dependent on any existing libraries such as scikit-learn.
# Features:
# Custom decision tree implementation for binary features and binary classification
# Entropy-based information gain to find the best split at each node
# Max depth control for tree pruning
# Utility functions for data manipulation and extraction
# Implementation Details:
# The DecisionTree class is initialized with input features (X), target labels (y), maximum tree depth, and optional feature names for visualization.
# Entropy and information gain calculations are used to determine the best split at each node.
# The build_tree function recursively constructs the decision tree by finding the best feature to split on and creating left and right child nodes.
# The constructed tree is stored as a dictionary with keys representing the depth and side (left or right) of each node.
# Note: This implementation currently does not include a visualization feature using Graphviz or other libraries. Additionally, the model lacks prediction and evaluation methods. This simplified version is intended to demonstrate the basic concepts of decision tree construction and can be extended for more advanced use cases.
#
class DecisionTree: # Simplified version for only 2 classes
def __init__(self, X, y, max_depth=5, feature_names=None):
# X - numpy matrix (one hot encoded, only classes)
# y - numpy array(1 or 0)
# setting feature names for visualization
if feature_names:
self.feature_names = feature_names
else:
self.feature_names = ["Feature {}".format(i) for i in range(X.shape[1])]
self.X = X
self.y = y
self.max_depth = max_depth
self.n_feature = self.X.shape[1]
self.initial_indices = np.array([i for i in range(len(X))])
self.fitted_tree = (
{}
) # Format "Depth # {n}" : {"feature" : ..., "depth" : ..., "entropy"}
def entropy(self, sample_of_classes):
# fraction
p = np.sum(sample_of_classes) / len(sample_of_classes)
if p == 0 or p == 1:
return 0
return -p * np.log2(p) - (1 - p) * np.log2(1 - p)
def get_sample_of_data(self, X_indices): # input array or list of indeces to get
X_data = np.zeros((len(X_indices), self.n_feature)) # prepare matrix
y_data = []
for i in range(len(X_indices)):
for j in range(self.n_feature):
ind = X_indices[i]
X_data[i, j] = self.X[ind, j]
y_data.append(self.y[ind])
y_data = np.array(y_data)
return X_data, y_data
def split_dataset(self, X_node_indeces, feature_ind):
left_indices = [] # feature = 1
right_indices = [] # feature = 0
for i in X_node_indeces:
if self.X[i, feature_ind] == 1:
left_indices.append(i)
else: # if 0
right_indices.append(i)
return right_indices, left_indices
def info_gain(self, node_indices, feature_ind):
# extract data
right_ind, left_ind = self.split_dataset(node_indices, feature_ind)
X_parent, y_parent = self.get_sample_of_data(node_indices)
X_left, y_left = self.get_sample_of_data(left_ind)
X_right, y_right = self.get_sample_of_data(right_ind)
# compute fractions
n_parent = np.sum(y_parent)
n_left = np.sum(y_left)
n_right = np.sum(y_right)
p_parent = n_parent / len(y_parent)
p_left = n_left / len(y_left)
p_right = n_right / len(y_right)
# compute weights
left_weight = len(y_left) / len(y_parent)
right_weight = len(y_right) / len(y_parent)
info_gain = self.entropy(y_parent) - (
left_weight * self.entropy(y_left) + right_weight * self.entropy(y_right)
)
return info_gain
def get_best_split(self, X_node_indices):
# def info_gain(self,X_parent, y_parent, X_left,y_left, X_right, y_right):
num_features = self.X.shape[1]
# init vals to return
best_feature = -1
best_info_gain = -0.1
# end
X_parent, y_parent = self.get_sample_of_data(X_node_indices)
for i in range(num_features):
# extract needed data
info_gain = self.info_gain(X_node_indices, i)
if info_gain > best_info_gain:
best_feature = i
best_info_gain = info_gain
if best_info_gain <= 0:
return -1
return best_feature
def build_tree(self, node_indices, current_depth, side="root"):
if current_depth >= self.max_depth:
return
# gain best feature for now
best_feature_to_split = self.get_best_split(X_node_indices=node_indices)
# checking entropy. It may not need to split
_, y_data = self.get_sample_of_data(node_indices)
curr_entropy = self.entropy(y_data)
if curr_entropy == 0:
print("It is alredy pure")
self.fitted_tree["depth_{}_{}".format(current_depth, side)] = {
"feature_name": self.feature_names[best_feature_to_split],
"feature_index": best_feature_to_split,
"entropy": curr_entropy,
"left_child_indices": [],
"right_child_indices": [],
}
return
# split data into 2 branches by this feature
left_indices, right_indices = self.split_dataset(
node_indices, best_feature_to_split
)
# OUTPUT INFO
print(
"Curr_depth = {} , best_feature = {}".format(
current_depth, best_feature_to_split
)
)
# record node in dict
self.fitted_tree["depth_{}_{}".format(current_depth, side)] = {
"feature_name": self.feature_names[best_feature_to_split],
"feature_index": best_feature_to_split,
"entropy": curr_entropy,
"left_child_indices": left_indices,
"right_child_indices": right_indices,
}
# recursively do the same for left child and right child
left_tree = self.build_tree(
left_indices, current_depth + 1, side="left"
) # increment curr_depth
right_tree = self.build_tree(right_indices, current_depth + 1, side="right")
def visualize_tree(self):
# I am gonna complete this important piece later
# The tree does its job but without visual. so far
pass
# ## How to do better
# It is a good idea to put out a Node storing in a class TreeNode with attributes:
# 1. entropy
# 2. left child
# 3. right child
# 4. depth
# 5. indices
# Moreover, we will need to implement predict method
# In a perfect case, simple visualization as well
# ## Testing my implementation
#
# ARTIFICIAL DATASET FOR TESTING
# FEATURES
# "Temp", "Cough","Happy","Tired"
# target = sick or not
X = np.array(
[
[1, 0, 1, 0],
[0, 1, 0, 1],
[1, 1, 0, 0],
[0, 0, 1, 1],
[1, 0, 0, 1],
[0, 1, 1, 0],
[1, 1, 1, 0],
[0, 0, 0, 1],
[1, 1, 0, 1],
[0, 1, 1, 1],
[1, 0, 1, 1],
[0, 1, 0, 0],
[1, 1, 1, 1],
[0, 0, 1, 0],
[1, 0, 0, 0],
[0, 1, 1, 1],
[1, 1, 0, 1],
[0, 0, 0, 0],
[1, 0, 1, 0],
[0, 1, 0, 1],
]
)
# Create a simple target variable (y) based on some rule
y = np.array([1 if row[0] == row[2] else 0 for row in X])
dec_tree = DecisionTree(X, y, 4, ["Temp", "Cough", "Happy", "Tired"])
dec_tree.build_tree(dec_tree.initial_indices, 0)
dec_tree.fitted_tree
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/384/129384981.ipynb
| null | null |
[{"Id": 129384981, "ScriptId": 38470584, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6066769, "CreationDate": "05/13/2023 10:37:27", "VersionNumber": 1.0, "Title": "Decision Tree from scartch", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 354.0, "LinesInsertedFromPrevious": 354.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from graphviz import Digraph
# Title: Simplified Decision Tree Classifier for Binary Features
# Description:
# This notebook presents a simplified decision tree classifier designed for datasets with binary features and binary classification tasks. The decision tree is built using a custom implementation, and the model is not dependent on any existing libraries such as scikit-learn.
# Features:
# Custom decision tree implementation for binary features and binary classification
# Entropy-based information gain to find the best split at each node
# Max depth control for tree pruning
# Utility functions for data manipulation and extraction
# Implementation Details:
# The DecisionTree class is initialized with input features (X), target labels (y), maximum tree depth, and optional feature names for visualization.
# Entropy and information gain calculations are used to determine the best split at each node.
# The build_tree function recursively constructs the decision tree by finding the best feature to split on and creating left and right child nodes.
# The constructed tree is stored as a dictionary with keys representing the depth and side (left or right) of each node.
# Note: This implementation currently does not include a visualization feature using Graphviz or other libraries. Additionally, the model lacks prediction and evaluation methods. This simplified version is intended to demonstrate the basic concepts of decision tree construction and can be extended for more advanced use cases.
#
class DecisionTree: # Simplified version for only 2 classes
def __init__(self, X, y, max_depth=5, feature_names=None):
# X - numpy matrix (one hot encoded, only classes)
# y - numpy array(1 or 0)
# setting feature names for visualization
if feature_names:
self.feature_names = feature_names
else:
self.feature_names = ["Feature {}".format(i) for i in range(X.shape[1])]
self.X = X
self.y = y
self.max_depth = max_depth
self.n_feature = self.X.shape[1]
self.initial_indices = np.array([i for i in range(len(X))])
self.fitted_tree = (
{}
) # Format "Depth # {n}" : {"feature" : ..., "depth" : ..., "entropy"}
def entropy(self, sample_of_classes):
# fraction
p = np.sum(sample_of_classes) / len(sample_of_classes)
if p == 0 or p == 1:
return 0
return -p * np.log2(p) - (1 - p) * np.log2(1 - p)
def get_sample_of_data(self, X_indices): # input array or list of indeces to get
X_data = np.zeros((len(X_indices), self.n_feature)) # prepare matrix
y_data = []
for i in range(len(X_indices)):
for j in range(self.n_feature):
ind = X_indices[i]
X_data[i, j] = self.X[ind, j]
y_data.append(self.y[ind])
y_data = np.array(y_data)
return X_data, y_data
def split_dataset(self, X_node_indeces, feature_ind):
left_indices = [] # feature = 1
right_indices = [] # feature = 0
for i in X_node_indeces:
if self.X[i, feature_ind] == 1:
left_indices.append(i)
else: # if 0
right_indices.append(i)
return right_indices, left_indices
def info_gain(self, node_indices, feature_ind):
# extract data
right_ind, left_ind = self.split_dataset(node_indices, feature_ind)
X_parent, y_parent = self.get_sample_of_data(node_indices)
X_left, y_left = self.get_sample_of_data(left_ind)
X_right, y_right = self.get_sample_of_data(right_ind)
# compute fractions
n_parent = np.sum(y_parent)
n_left = np.sum(y_left)
n_right = np.sum(y_right)
p_parent = n_parent / len(y_parent)
p_left = n_left / len(y_left)
p_right = n_right / len(y_right)
# compute weights
left_weight = len(y_left) / len(y_parent)
right_weight = len(y_right) / len(y_parent)
info_gain = self.entropy(y_parent) - (
left_weight * self.entropy(y_left) + right_weight * self.entropy(y_right)
)
return info_gain
def get_best_split(self, X_node_indices):
# def info_gain(self,X_parent, y_parent, X_left,y_left, X_right, y_right):
num_features = self.X.shape[1]
# init vals to return
best_feature = -1
best_info_gain = -0.1
# end
X_parent, y_parent = self.get_sample_of_data(X_node_indices)
for i in range(num_features):
# extract needed data
info_gain = self.info_gain(X_node_indices, i)
if info_gain > best_info_gain:
best_feature = i
best_info_gain = info_gain
if best_info_gain <= 0:
return -1
return best_feature
def build_tree(self, node_indices, current_depth, side="root"):
if current_depth >= self.max_depth:
return
# gain best feature for now
best_feature_to_split = self.get_best_split(X_node_indices=node_indices)
# checking entropy. It may not need to split
_, y_data = self.get_sample_of_data(node_indices)
curr_entropy = self.entropy(y_data)
if curr_entropy == 0:
print("It is alredy pure")
self.fitted_tree["depth_{}_{}".format(current_depth, side)] = {
"feature_name": self.feature_names[best_feature_to_split],
"feature_index": best_feature_to_split,
"entropy": curr_entropy,
"left_child_indices": [],
"right_child_indices": [],
}
return
# split data into 2 branches by this feature
left_indices, right_indices = self.split_dataset(
node_indices, best_feature_to_split
)
# OUTPUT INFO
print(
"Curr_depth = {} , best_feature = {}".format(
current_depth, best_feature_to_split
)
)
# record node in dict
self.fitted_tree["depth_{}_{}".format(current_depth, side)] = {
"feature_name": self.feature_names[best_feature_to_split],
"feature_index": best_feature_to_split,
"entropy": curr_entropy,
"left_child_indices": left_indices,
"right_child_indices": right_indices,
}
# recursively do the same for left child and right child
left_tree = self.build_tree(
left_indices, current_depth + 1, side="left"
) # increment curr_depth
right_tree = self.build_tree(right_indices, current_depth + 1, side="right")
def visualize_tree(self):
# I am gonna complete this important piece later
# The tree does its job but without visual. so far
pass
# ## How to do better
# It is a good idea to put out a Node storing in a class TreeNode with attributes:
# 1. entropy
# 2. left child
# 3. right child
# 4. depth
# 5. indices
# Moreover, we will need to implement predict method
# In a perfect case, simple visualization as well
# ## Testing my implementation
#
# ARTIFICIAL DATASET FOR TESTING
# FEATURES
# "Temp", "Cough","Happy","Tired"
# target = sick or not
X = np.array(
[
[1, 0, 1, 0],
[0, 1, 0, 1],
[1, 1, 0, 0],
[0, 0, 1, 1],
[1, 0, 0, 1],
[0, 1, 1, 0],
[1, 1, 1, 0],
[0, 0, 0, 1],
[1, 1, 0, 1],
[0, 1, 1, 1],
[1, 0, 1, 1],
[0, 1, 0, 0],
[1, 1, 1, 1],
[0, 0, 1, 0],
[1, 0, 0, 0],
[0, 1, 1, 1],
[1, 1, 0, 1],
[0, 0, 0, 0],
[1, 0, 1, 0],
[0, 1, 0, 1],
]
)
# Create a simple target variable (y) based on some rule
y = np.array([1 if row[0] == row[2] else 0 for row in X])
dec_tree = DecisionTree(X, y, 4, ["Temp", "Cough", "Happy", "Tired"])
dec_tree.build_tree(dec_tree.initial_indices, 0)
dec_tree.fitted_tree
| false | 0 | 2,330 | 0 | 2,330 | 2,330 |
||
129384048
|
<jupyter_start><jupyter_text>Magic The Gathering Cards
Magic The Gathering (MTG, or just Magic) is a trading card game first published in 1993 by Wizards of the Coast. This game has seen immense popularity and new cards are still released every few months. The strength of different cards in the game can vary wildly and as a result some cards now sell on secondary markets for as high as thousands of dollars.
[MTG JSON][1] has an excellent collection of every single Magic Card - stored in JSON data. Version 3.6 (collected September 21, 2016) of their database is provided here.
Full documentation for the data is provided here:
http://mtgjson.com/documentation.html
Also, if you want to include images of the cards in your writeups, you can grab them from the official Wizards of the Coast website using the following URL:
http://gatherer.wizards.com/Handlers/Image.ashx?multiverseid=180607&type=card
Just replace the multiverse ID with the one provided in the mtgjson file.
[1]: http://mtgjson.com
Kaggle dataset identifier: magic-the-gathering-cards
<jupyter_script>import json
import pandas as pd
def flatten_json(nested_json, indent_str="_"):
out = {}
def flatten(x, name=""):
if type(x) is dict:
for a in x:
flatten(x[a], name + a + indent_str)
elif type(x) is list:
i = 0
for a in x:
flatten(a, name + str(i) + indent_str)
i += 1
else:
out[name[:-1]] = x
flatten(nested_json)
return out
# Load your JSON
with open("/kaggle/input/magic-the-gathering-cards/AllCards.json") as f:
data = json.load(f)
# Flatten the JSON
flat = flatten_json(data)
# Convert to DataFrame
df = pd.json_normalize(flat)
# Clearly the classic normalize fails to flatten this structure.
# The goal would be to unfold the levels one by one.
# This could be done by prompting an LLM but I tried and ran out of memory while executing the code XD
df
# The goal now would be to unpack level by level take a look at the structure and
# plan a flattening strategy ( Auto-GPT should be able to do this but I don't have a VPN now to launch it XD)
# Getting only top level keys
import json
def print_top_level_keys(file_path):
with open(file_path, "r") as f:
data = json.load(f)
keys = list(data.keys())
# print(keys)
return keys
# Replace 'file.json' with your actual JSON file path
toplevelkeys = print_top_level_keys(
"/kaggle/input/magic-the-gathering-cards/AllCards.json"
)
# Amazed at the length of the json
len(toplevelkeys)
# First top level key 1 of 20'478
toplevelkeys[0]
# Analyzing the first nested level: it's not too bad
data[toplevelkeys[0]]
# The first level can be normalized decently using the library
# so at this point all we have to do is turn the upper level into a row entry for this iteration
dataframe = pd.json_normalize(data[toplevelkeys[0]])
dataframe["CardName - TopLevel"] = toplevelkeys[0]
# There you have it in the last column
dataframe
import json
import pandas as pd
def print_top_level_keys(file_path):
with open(file_path, "r") as f:
data = json.load(f)
keys = list(data.keys())
return keys, data
toplevelkeys, data = print_top_level_keys(
"/kaggle/input/magic-the-gathering-cards/AllCards.json"
)
# Initialize an empty dataframe
all_data = pd.DataFrame()
# Loop over all keys and flatten each nested dictionary
for key in toplevelkeys:
dataframe = pd.json_normalize(data[key])
dataframe["CardName - TopLevel"] = key
all_data = all_data.append(dataframe)
# Reset index
all_data.reset_index(drop=True, inplace=True)
print(all_data)
# And here is how it looks when you get it unpacked with the function above.
# Still some of this columns like ruling could use another level of extraction and flattening following the same principle
all_data
# For example rulings could be extracted and flattened in a single row, but it is not constant across the toplevel
# so you'd have to go with a new columns that titles ruling_date_1, ruling_text_1, ruling_date_2, ruling_text_2, ..., and so on
# you'd check the max length needed, say for example that the card with the longest ruling list has 8 entries,
# then you'll need to add columns to accomodate ruling_date_1 to 8 and ruling_text_1 to 8 and a loop that scans all the ruling columns
# and unpacks/pivots it [could actually copy this text in GPT-4 and ask it to merge with the function above XD!!!]
all_data["rulings"][3]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/384/129384048.ipynb
|
magic-the-gathering-cards
|
mylesoneill
|
[{"Id": 129384048, "ScriptId": 38395763, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2058747, "CreationDate": "05/13/2023 10:27:32", "VersionNumber": 1.0, "Title": "MagicTheGathering - Json", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 116.0, "LinesInsertedFromPrevious": 116.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185381681, "KernelVersionId": 129384048, "SourceDatasetVersionId": 792844}]
|
[{"Id": 792844, "DatasetId": 196, "DatasourceVersionId": 814628, "CreatorUserId": 1240410, "LicenseName": "Unknown", "CreationDate": "11/13/2019 17:13:30", "VersionNumber": 2.0, "Title": "Magic The Gathering Cards", "Slug": "magic-the-gathering-cards", "Subtitle": "Analyze cards from this classic trading card game", "Description": "Magic The Gathering (MTG, or just Magic) is a trading card game first published in 1993 by Wizards of the Coast. This game has seen immense popularity and new cards are still released every few months. The strength of different cards in the game can vary wildly and as a result some cards now sell on secondary markets for as high as thousands of dollars.\n\n[MTG JSON][1] has an excellent collection of every single Magic Card - stored in JSON data. Version 3.6 (collected September 21, 2016) of their database is provided here.\n\nFull documentation for the data is provided here:\nhttp://mtgjson.com/documentation.html\n\nAlso, if you want to include images of the cards in your writeups, you can grab them from the official Wizards of the Coast website using the following URL:\n\nhttp://gatherer.wizards.com/Handlers/Image.ashx?multiverseid=180607&type=card\n\nJust replace the multiverse ID with the one provided in the mtgjson file.\n\n\n [1]: http://mtgjson.com", "VersionNotes": "Updating to latest version including M20, C19 and Eldraine", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 196, "CreatorUserId": 304806, "OwnerUserId": 304806.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 792844.0, "CurrentDatasourceVersionId": 814628.0, "ForumId": 1586, "Type": 2, "CreationDate": "09/26/2016 19:29:58", "LastActivityDate": "02/04/2018", "TotalViews": 50407, "TotalDownloads": 2583, "TotalVotes": 136, "TotalKernels": 38}]
|
[{"Id": 304806, "UserName": "mylesoneill", "DisplayName": "Myles O'Neill", "RegisterDate": "03/05/2015", "PerformanceTier": 5}]
|
import json
import pandas as pd
def flatten_json(nested_json, indent_str="_"):
out = {}
def flatten(x, name=""):
if type(x) is dict:
for a in x:
flatten(x[a], name + a + indent_str)
elif type(x) is list:
i = 0
for a in x:
flatten(a, name + str(i) + indent_str)
i += 1
else:
out[name[:-1]] = x
flatten(nested_json)
return out
# Load your JSON
with open("/kaggle/input/magic-the-gathering-cards/AllCards.json") as f:
data = json.load(f)
# Flatten the JSON
flat = flatten_json(data)
# Convert to DataFrame
df = pd.json_normalize(flat)
# Clearly the classic normalize fails to flatten this structure.
# The goal would be to unfold the levels one by one.
# This could be done by prompting an LLM but I tried and ran out of memory while executing the code XD
df
# The goal now would be to unpack level by level take a look at the structure and
# plan a flattening strategy ( Auto-GPT should be able to do this but I don't have a VPN now to launch it XD)
# Getting only top level keys
import json
def print_top_level_keys(file_path):
with open(file_path, "r") as f:
data = json.load(f)
keys = list(data.keys())
# print(keys)
return keys
# Replace 'file.json' with your actual JSON file path
toplevelkeys = print_top_level_keys(
"/kaggle/input/magic-the-gathering-cards/AllCards.json"
)
# Amazed at the length of the json
len(toplevelkeys)
# First top level key 1 of 20'478
toplevelkeys[0]
# Analyzing the first nested level: it's not too bad
data[toplevelkeys[0]]
# The first level can be normalized decently using the library
# so at this point all we have to do is turn the upper level into a row entry for this iteration
dataframe = pd.json_normalize(data[toplevelkeys[0]])
dataframe["CardName - TopLevel"] = toplevelkeys[0]
# There you have it in the last column
dataframe
import json
import pandas as pd
def print_top_level_keys(file_path):
with open(file_path, "r") as f:
data = json.load(f)
keys = list(data.keys())
return keys, data
toplevelkeys, data = print_top_level_keys(
"/kaggle/input/magic-the-gathering-cards/AllCards.json"
)
# Initialize an empty dataframe
all_data = pd.DataFrame()
# Loop over all keys and flatten each nested dictionary
for key in toplevelkeys:
dataframe = pd.json_normalize(data[key])
dataframe["CardName - TopLevel"] = key
all_data = all_data.append(dataframe)
# Reset index
all_data.reset_index(drop=True, inplace=True)
print(all_data)
# And here is how it looks when you get it unpacked with the function above.
# Still some of this columns like ruling could use another level of extraction and flattening following the same principle
all_data
# For example rulings could be extracted and flattened in a single row, but it is not constant across the toplevel
# so you'd have to go with a new columns that titles ruling_date_1, ruling_text_1, ruling_date_2, ruling_text_2, ..., and so on
# you'd check the max length needed, say for example that the card with the longest ruling list has 8 entries,
# then you'll need to add columns to accomodate ruling_date_1 to 8 and ruling_text_1 to 8 and a loop that scans all the ruling columns
# and unpacks/pivots it [could actually copy this text in GPT-4 and ask it to merge with the function above XD!!!]
all_data["rulings"][3]
| false | 0 | 1,004 | 0 | 1,293 | 1,004 |
||
129384018
|
# # All about regularized regression
# Regularized linear regression is an extension of standard linear regression that adds a regularization term to the cost function. The regularization term is a penalty on the size of the coefficients, which helps to prevent overfitting and improve the model's generalization performance.
# In standard linear regression, the goal is to find the coefficients that minimize the sum of squared errors between the predicted values and the actual values. However, this can lead to overfitting when the model becomes too complex and fits the noise in the data instead of the underlying pattern. Regularization helps to address this issue by adding a penalty term to the cost function that discourages large coefficient values.
# ## Standard Regression
# In standard linear regression, the goal is to minimize the sum of squared errors between the predicted values and the actual values. The cost function for standard regression can be expressed as:
# Cost = RSS (Residual Sum of Squares) = Σ(yᵢ - ŷᵢ)²
# Where:
# - yᵢ is the actual target value.
# - ŷᵢ is the predicted target value.
# - The summation is over all the training examples.
# - The coefficients (weights) in standard regression are obtained by solving the following equation:
# ∂Cost/∂θ = 0
# Where θ represents the coefficients.
# ## Ridge Regression
# Ridge regression adds an L2 regularization term to the cost function to penalize large coefficient values. The cost function for Ridge regression is given by:
# Cost = RSS + α * Σ(θⱼ)²
# Where:
# - α is the regularization parameter that controls the strength of regularization.
# - The second term represents the L2 regularization term, penalizing the sum of squared coefficients.
# - The coefficients in Ridge regression are obtained by minimizing the following equation:
# ∂(Cost + α * Σ(θⱼ)²)/∂θ = 0
# This leads to a modified equation for the coefficients:
# θ = (XᵀX + αI)⁻¹Xᵀy
# Where:
# - X is the matrix of input features.
# - y is the vector of target values.
# - I is the identity matrix.
# Ridge regression shrinks the coefficients towards zero but doesn't force them to exactly zero. It can be useful when dealing with multicollinearity in the data.
# ## Lasso Regression
# Lasso regression, unlike Ridge regression, uses L1 regularization to encourage sparse solutions where some coefficients become exactly zero. The cost function for Lasso regression is given by:
# Cost = RSS + α * Σ|θⱼ|
# Where:
# - α is the regularization parameter controlling the strength of regularization.
# - The second term represents the L1 regularization term, penalizing the sum of absolute coefficients.
# - The coefficients in Lasso regression are obtained by minimizing the following equation:
# ∂(Cost + α * Σ|θⱼ|)/∂θ = 0
# This leads to a modified equation for the coefficients:
# θ = argmin RSS + α * Σ|θⱼ|
# Lasso regression performs both feature selection and coefficient shrinkage, effectively setting some coefficients to exactly zero. It is useful when there is a desire to identify the most relevant features.
# ## Implementation
import numpy as np
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression, Ridge, Lasso
from sklearn.preprocessing import PolynomialFeatures
# Generate synthetic data
np.random.seed(42)
X = np.linspace(-5, 5, num=100).reshape(-1, 1)
y = 2 * X + np.random.normal(0, 1, size=(100, 1))
# Polynomial features
poly = PolynomialFeatures(degree=10)
X_poly = poly.fit_transform(X)
# Standard Regression
reg_std = LinearRegression()
reg_std.fit(X_poly, y)
coef_std = reg_std.coef_.ravel()
# Ridge Regression
reg_ridge = Ridge(alpha=0.5) # alpha is the regularization parameter
reg_ridge.fit(X_poly, y)
coef_ridge = reg_ridge.coef_.ravel()
# Lasso Regression
reg_lasso = Lasso(alpha=0.5) # alpha is the regularization parameter
reg_lasso.fit(X_poly, y)
coef_lasso = reg_lasso.coef_.ravel()
# Plotting the coefficients
plt.figure(figsize=(10, 6))
plt.plot(coef_std, label="Standard Regression")
plt.plot(coef_ridge, label="Ridge Regression")
plt.plot(coef_lasso, label="Lasso Regression")
plt.xlabel("Coefficient Index")
plt.ylabel("Coefficient Value")
plt.legend()
plt.title("Comparison of Coefficients")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/384/129384018.ipynb
| null | null |
[{"Id": 129384018, "ScriptId": 38470197, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12722296, "CreationDate": "05/13/2023 10:27:16", "VersionNumber": 1.0, "Title": "Regularized Regression", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 110.0, "LinesInsertedFromPrevious": 110.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 4}]
| null | null | null | null |
# # All about regularized regression
# Regularized linear regression is an extension of standard linear regression that adds a regularization term to the cost function. The regularization term is a penalty on the size of the coefficients, which helps to prevent overfitting and improve the model's generalization performance.
# In standard linear regression, the goal is to find the coefficients that minimize the sum of squared errors between the predicted values and the actual values. However, this can lead to overfitting when the model becomes too complex and fits the noise in the data instead of the underlying pattern. Regularization helps to address this issue by adding a penalty term to the cost function that discourages large coefficient values.
# ## Standard Regression
# In standard linear regression, the goal is to minimize the sum of squared errors between the predicted values and the actual values. The cost function for standard regression can be expressed as:
# Cost = RSS (Residual Sum of Squares) = Σ(yᵢ - ŷᵢ)²
# Where:
# - yᵢ is the actual target value.
# - ŷᵢ is the predicted target value.
# - The summation is over all the training examples.
# - The coefficients (weights) in standard regression are obtained by solving the following equation:
# ∂Cost/∂θ = 0
# Where θ represents the coefficients.
# ## Ridge Regression
# Ridge regression adds an L2 regularization term to the cost function to penalize large coefficient values. The cost function for Ridge regression is given by:
# Cost = RSS + α * Σ(θⱼ)²
# Where:
# - α is the regularization parameter that controls the strength of regularization.
# - The second term represents the L2 regularization term, penalizing the sum of squared coefficients.
# - The coefficients in Ridge regression are obtained by minimizing the following equation:
# ∂(Cost + α * Σ(θⱼ)²)/∂θ = 0
# This leads to a modified equation for the coefficients:
# θ = (XᵀX + αI)⁻¹Xᵀy
# Where:
# - X is the matrix of input features.
# - y is the vector of target values.
# - I is the identity matrix.
# Ridge regression shrinks the coefficients towards zero but doesn't force them to exactly zero. It can be useful when dealing with multicollinearity in the data.
# ## Lasso Regression
# Lasso regression, unlike Ridge regression, uses L1 regularization to encourage sparse solutions where some coefficients become exactly zero. The cost function for Lasso regression is given by:
# Cost = RSS + α * Σ|θⱼ|
# Where:
# - α is the regularization parameter controlling the strength of regularization.
# - The second term represents the L1 regularization term, penalizing the sum of absolute coefficients.
# - The coefficients in Lasso regression are obtained by minimizing the following equation:
# ∂(Cost + α * Σ|θⱼ|)/∂θ = 0
# This leads to a modified equation for the coefficients:
# θ = argmin RSS + α * Σ|θⱼ|
# Lasso regression performs both feature selection and coefficient shrinkage, effectively setting some coefficients to exactly zero. It is useful when there is a desire to identify the most relevant features.
# ## Implementation
import numpy as np
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression, Ridge, Lasso
from sklearn.preprocessing import PolynomialFeatures
# Generate synthetic data
np.random.seed(42)
X = np.linspace(-5, 5, num=100).reshape(-1, 1)
y = 2 * X + np.random.normal(0, 1, size=(100, 1))
# Polynomial features
poly = PolynomialFeatures(degree=10)
X_poly = poly.fit_transform(X)
# Standard Regression
reg_std = LinearRegression()
reg_std.fit(X_poly, y)
coef_std = reg_std.coef_.ravel()
# Ridge Regression
reg_ridge = Ridge(alpha=0.5) # alpha is the regularization parameter
reg_ridge.fit(X_poly, y)
coef_ridge = reg_ridge.coef_.ravel()
# Lasso Regression
reg_lasso = Lasso(alpha=0.5) # alpha is the regularization parameter
reg_lasso.fit(X_poly, y)
coef_lasso = reg_lasso.coef_.ravel()
# Plotting the coefficients
plt.figure(figsize=(10, 6))
plt.plot(coef_std, label="Standard Regression")
plt.plot(coef_ridge, label="Ridge Regression")
plt.plot(coef_lasso, label="Lasso Regression")
plt.xlabel("Coefficient Index")
plt.ylabel("Coefficient Value")
plt.legend()
plt.title("Comparison of Coefficients")
plt.show()
| false | 0 | 1,147 | 4 | 1,147 | 1,147 |
||
129384772
|
import pandas as pd
import torch
import torch.nn as nn
import torch.optim as optim
import random
import math
import numpy as np
import matplotlib.pyplot as plt
my_data = pd.read_csv("../RHM_FYP/TurkeyWindFarm.csv")
df.drop("time", inplace=True, axis=1)
df
# switching columns to make LV activePower (kW) last column so that it will be easy
# to prepare training and test datasets
cols = df.columns.tolist()
cols[0], cols[3] = cols[3], cols[0]
df = df[cols]
df
target = df["LV ActivePower (kW)"]
target.shape
# Calculate the number of readings per day
readings_per_day = 144
# n = number of day
n = 3
# Calculate the sequence length (n days in readings)
sequence_length = readings_per_day * n
# Set the lookback window to be the same as the sequence length
lookback_window = sequence_length
# Convert the data to a NumPy array
df = df.values
x, y = [], []
for i in range(lookback_window, len(df)):
x.append(df[i - lookback_window : i, :])
y.append(df[i, -1])
x = np.array(x)
y = np.array(y)
print(x.shape)
print(y.shape)
# Calculate the number of test samples (last 3 days of the year)
num_test_samples = readings_per_day * 3
# Split the data into training and test sets
x_train = x[:-num_test_samples]
y_train = y[:-num_test_samples]
x_test = x[-num_test_samples:]
y_test = y[-num_test_samples:]
print(x_train.shape)
print(y_train.shape)
print(x_test.shape)
print(y_test.shape)
class PositionalEncoding(nn.Module):
def __init__(self, embedding_size, dropout_p, max_len):
super().__init__()
self.dropout = nn.Dropout(dropout_p)
position = torch.arange(max_len).unsqueeze(
1
) # col vector #add another dimension from (max_len) -> (maxlen,1)
div_term = torch.exp(
torch.arange(0, embedding_size, 2).float()
* (-math.log(10000.0))
/ embedding_size
) # row vector #1 / 10000^(2i/embedding_size) -> div_term
sinusoid = torch.zeros(max_len, embedding_size) # (max_len,embedding size)
sinusoid[:, 0::2] = torch.sin(
position * div_term
) # col * row # (time_step , even embedding added with sine * div_term)
sinusoid[:, 1::2] = torch.cos(
position * div_term
) # (time_step , odd embedding added with cosine * div_term)
self.register_buffer(
"pos_encoding", sinusoid.unsqueeze(0)
) # creating register buffer in pytorch graph
def forward(self, x):
x = (
x + self.pos_encoding[:, : x.size(1)]
) # addition of positional encoding with our embedded input
return self.dropout(x)
class PositionalEncoding(nn.Module):
def __init__(self, embedding_size, dropout_p, max_len):
super().__init__()
self.dropout = nn.Dropout(dropout_p)
position = torch.arange(max_len).unsqueeze(1)
div_term = torch.exp(
torch.arange(0, embedding_size, 2).float()
* (-math.log(10000.0))
/ embedding_size
)
sinusoid = torch.zeros(max_len, embedding_size)
sinusoid[:, 0::2] = torch.sin(position * div_term)
sinusoid[:, 1::2] = torch.cos(position * div_term)
self.register_buffer("pos_encoding", sinusoid.unsqueeze(0))
def forward(self, x):
pos_encoding = self.pos_encoding[
:, : x.size(1), :
] # Resize pos_encoding to match the input size of x
x = x + pos_encoding
return self.dropout(x)
class Transformer(nn.Module):
def __init__(
self,
input_size,
dim_model,
num_heads,
num_encoder_layers,
num_decoder_layers,
dropout_p,
out_size,
):
super().__init__()
self.model_type = "Transformer"
self.dim_model = dim_model
self.positional_encoder = PositionalEncoding(
embedding_size=dim_model, dropout_p=dropout_p, max_len=432
)
self.embedding = nn.Linear(input_size, dim_model)
self.transformer = nn.Transformer(
d_model=dim_model,
nhead=num_heads,
num_encoder_layers=num_encoder_layers,
num_decoder_layers=num_decoder_layers,
dropout=dropout_p,
)
self.out = nn.Linear(dim_model, out_size)
def get_tgt_mask(self, tgt_len):
# Function to create a square attention mask for the decoder
mask = torch.triu(torch.ones(tgt_len, tgt_len)) == 1
mask = (
mask.float()
.masked_fill(mask == 0, float("-inf"))
.masked_fill(mask == 1, float(0.0))
)
return mask
def forward(self, x):
device = "cuda" if torch.cuda.is_available() else "cpu"
# x shape: (batch_size, sequence_length, num_features)
x = self.embedding(x) * math.sqrt(self.dim_model)
x = self.positional_encoder(x)
x = x.permute(1, 0, 2) # (sequence_length, batch_size, dim_model)
transformer_out = self.transformer(
x, x, tgt_mask=self.get_tgt_mask(x.size(0)).to(device)
)
out = self.out(transformer_out[-1])
return out.squeeze()
# Define model parameters
seq_length = 432
input_size = 4
dim_model = 512
num_heads = 8
num_encoder_layers = 3
num_decoder_layers = 3
dropout_p = 0.2
out_size = 1
device = "cuda" if torch.cuda.is_available() else "cpu"
# Initialize the model
model = Transformer(
input_size=input_size,
dim_model=dim_model,
num_heads=num_heads,
num_encoder_layers=num_encoder_layers,
num_decoder_layers=num_decoder_layers,
dropout_p=dropout_p,
out_size=out_size,
).to(device)
# Define loss function and optimizer
criterion = nn.MSELoss()
optimizer = optim.Adam(model.parameters(), lr=1e-4)
# Convert data to tensors
x_train_tensor = torch.tensor(x_train, dtype=torch.float32).to(device)
y_train_tensor = torch.tensor(y_train, dtype=torch.float32).to(device)
# Train the model
num_epochs = 20
batch_size = 32
for epoch in range(num_epochs):
running_loss = 0.0
for i in range(0, len(x_train), batch_size):
# Get batch of inputs and targets
batch_x = x_train_tensor[i : i + batch_size]
batch_y = y_train_tensor[i : i + batch_size]
# Reshape input tensor
batch_x = batch_x.view(-1, seq_length, input_size)
# Zero the parameter gradients
optimizer.zero_grad()
# Forward pass
outputs = model(batch_x)
loss = criterion(outputs.view(-1), batch_y)
# Backward pass and optimize
loss.backward()
optimizer.step()
running_loss += loss.item()
epoch_loss = running_loss / (len(x_train) / batch_size)
print(f"Epoch {epoch+1} loss: {epoch_loss:.6f}")
# Convert test data to tensor
x_test_tensor = torch.tensor(x_test, dtype=torch.float32).to(device)
# Forward pass on test input tensor
with torch.no_grad():
outputs = model(x_test_tensor)
# Reshape output tensor to (432,) shape
outputs = outputs.view(-1)
plt.plot(outputs.cpu().detach())
plt.plot(y_test) # denormalizing original data back
plt.title("Two day window results")
plt.legend(["predicted", "actual"])
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/384/129384772.ipynb
| null | null |
[{"Id": 129384772, "ScriptId": 35779301, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14044091, "CreationDate": "05/13/2023 10:35:29", "VersionNumber": 1.0, "Title": "transformer for time-series forecasting", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 198.0, "LinesInsertedFromPrevious": 198.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import torch
import torch.nn as nn
import torch.optim as optim
import random
import math
import numpy as np
import matplotlib.pyplot as plt
my_data = pd.read_csv("../RHM_FYP/TurkeyWindFarm.csv")
df.drop("time", inplace=True, axis=1)
df
# switching columns to make LV activePower (kW) last column so that it will be easy
# to prepare training and test datasets
cols = df.columns.tolist()
cols[0], cols[3] = cols[3], cols[0]
df = df[cols]
df
target = df["LV ActivePower (kW)"]
target.shape
# Calculate the number of readings per day
readings_per_day = 144
# n = number of day
n = 3
# Calculate the sequence length (n days in readings)
sequence_length = readings_per_day * n
# Set the lookback window to be the same as the sequence length
lookback_window = sequence_length
# Convert the data to a NumPy array
df = df.values
x, y = [], []
for i in range(lookback_window, len(df)):
x.append(df[i - lookback_window : i, :])
y.append(df[i, -1])
x = np.array(x)
y = np.array(y)
print(x.shape)
print(y.shape)
# Calculate the number of test samples (last 3 days of the year)
num_test_samples = readings_per_day * 3
# Split the data into training and test sets
x_train = x[:-num_test_samples]
y_train = y[:-num_test_samples]
x_test = x[-num_test_samples:]
y_test = y[-num_test_samples:]
print(x_train.shape)
print(y_train.shape)
print(x_test.shape)
print(y_test.shape)
class PositionalEncoding(nn.Module):
def __init__(self, embedding_size, dropout_p, max_len):
super().__init__()
self.dropout = nn.Dropout(dropout_p)
position = torch.arange(max_len).unsqueeze(
1
) # col vector #add another dimension from (max_len) -> (maxlen,1)
div_term = torch.exp(
torch.arange(0, embedding_size, 2).float()
* (-math.log(10000.0))
/ embedding_size
) # row vector #1 / 10000^(2i/embedding_size) -> div_term
sinusoid = torch.zeros(max_len, embedding_size) # (max_len,embedding size)
sinusoid[:, 0::2] = torch.sin(
position * div_term
) # col * row # (time_step , even embedding added with sine * div_term)
sinusoid[:, 1::2] = torch.cos(
position * div_term
) # (time_step , odd embedding added with cosine * div_term)
self.register_buffer(
"pos_encoding", sinusoid.unsqueeze(0)
) # creating register buffer in pytorch graph
def forward(self, x):
x = (
x + self.pos_encoding[:, : x.size(1)]
) # addition of positional encoding with our embedded input
return self.dropout(x)
class PositionalEncoding(nn.Module):
def __init__(self, embedding_size, dropout_p, max_len):
super().__init__()
self.dropout = nn.Dropout(dropout_p)
position = torch.arange(max_len).unsqueeze(1)
div_term = torch.exp(
torch.arange(0, embedding_size, 2).float()
* (-math.log(10000.0))
/ embedding_size
)
sinusoid = torch.zeros(max_len, embedding_size)
sinusoid[:, 0::2] = torch.sin(position * div_term)
sinusoid[:, 1::2] = torch.cos(position * div_term)
self.register_buffer("pos_encoding", sinusoid.unsqueeze(0))
def forward(self, x):
pos_encoding = self.pos_encoding[
:, : x.size(1), :
] # Resize pos_encoding to match the input size of x
x = x + pos_encoding
return self.dropout(x)
class Transformer(nn.Module):
def __init__(
self,
input_size,
dim_model,
num_heads,
num_encoder_layers,
num_decoder_layers,
dropout_p,
out_size,
):
super().__init__()
self.model_type = "Transformer"
self.dim_model = dim_model
self.positional_encoder = PositionalEncoding(
embedding_size=dim_model, dropout_p=dropout_p, max_len=432
)
self.embedding = nn.Linear(input_size, dim_model)
self.transformer = nn.Transformer(
d_model=dim_model,
nhead=num_heads,
num_encoder_layers=num_encoder_layers,
num_decoder_layers=num_decoder_layers,
dropout=dropout_p,
)
self.out = nn.Linear(dim_model, out_size)
def get_tgt_mask(self, tgt_len):
# Function to create a square attention mask for the decoder
mask = torch.triu(torch.ones(tgt_len, tgt_len)) == 1
mask = (
mask.float()
.masked_fill(mask == 0, float("-inf"))
.masked_fill(mask == 1, float(0.0))
)
return mask
def forward(self, x):
device = "cuda" if torch.cuda.is_available() else "cpu"
# x shape: (batch_size, sequence_length, num_features)
x = self.embedding(x) * math.sqrt(self.dim_model)
x = self.positional_encoder(x)
x = x.permute(1, 0, 2) # (sequence_length, batch_size, dim_model)
transformer_out = self.transformer(
x, x, tgt_mask=self.get_tgt_mask(x.size(0)).to(device)
)
out = self.out(transformer_out[-1])
return out.squeeze()
# Define model parameters
seq_length = 432
input_size = 4
dim_model = 512
num_heads = 8
num_encoder_layers = 3
num_decoder_layers = 3
dropout_p = 0.2
out_size = 1
device = "cuda" if torch.cuda.is_available() else "cpu"
# Initialize the model
model = Transformer(
input_size=input_size,
dim_model=dim_model,
num_heads=num_heads,
num_encoder_layers=num_encoder_layers,
num_decoder_layers=num_decoder_layers,
dropout_p=dropout_p,
out_size=out_size,
).to(device)
# Define loss function and optimizer
criterion = nn.MSELoss()
optimizer = optim.Adam(model.parameters(), lr=1e-4)
# Convert data to tensors
x_train_tensor = torch.tensor(x_train, dtype=torch.float32).to(device)
y_train_tensor = torch.tensor(y_train, dtype=torch.float32).to(device)
# Train the model
num_epochs = 20
batch_size = 32
for epoch in range(num_epochs):
running_loss = 0.0
for i in range(0, len(x_train), batch_size):
# Get batch of inputs and targets
batch_x = x_train_tensor[i : i + batch_size]
batch_y = y_train_tensor[i : i + batch_size]
# Reshape input tensor
batch_x = batch_x.view(-1, seq_length, input_size)
# Zero the parameter gradients
optimizer.zero_grad()
# Forward pass
outputs = model(batch_x)
loss = criterion(outputs.view(-1), batch_y)
# Backward pass and optimize
loss.backward()
optimizer.step()
running_loss += loss.item()
epoch_loss = running_loss / (len(x_train) / batch_size)
print(f"Epoch {epoch+1} loss: {epoch_loss:.6f}")
# Convert test data to tensor
x_test_tensor = torch.tensor(x_test, dtype=torch.float32).to(device)
# Forward pass on test input tensor
with torch.no_grad():
outputs = model(x_test_tensor)
# Reshape output tensor to (432,) shape
outputs = outputs.view(-1)
plt.plot(outputs.cpu().detach())
plt.plot(y_test) # denormalizing original data back
plt.title("Two day window results")
plt.legend(["predicted", "actual"])
plt.show()
| false | 0 | 2,203 | 0 | 2,203 | 2,203 |
||
129267658
|
### CSS for notebook styling ###
from IPython.core.display import HTML
HTML(
"""
<style>
:root {
--box_color: #F1F6F9;
}
body[data-jp-theme-light="true"] .jp-Notebook .CodeMirror.cm-s-jupyter{
background-color: var(--box_color) !important;
}
div.input_area{
background-color: var(--box_color) !important;
}
.crop {
display: block;
height: 250px;
position: relative;
overflow: hidden;
width: 700px;
}
.crop img {
left: 0px; /* alter this to move left or right */
position: absolute;
top: 0px; /* alter this to move up or down */
}
</style>
"""
)
#
# 🤞 Google - American Sign Language Fingerspelling Recognition 🤞
#
#
# Goal of the Competition
#
# The goal of this competition is to detect and translate American Sign Language (ASL) fingerspelling into text. You will create a model trained on the largest dataset of its kind, released specifically for this competition. The data includes more than three million fingerspelled characters produced by over 100 Deaf signers captured via the selfie camera of a smartphone with a variety of backgrounds and lighting conditions.
# ASL Fingerspelling Alphabet
#
# Fingerspelling is a way of spelling words using hand movements. The fingerspelling manual alphabet is used in sign language to spell out names of people and places for which there is not a sign. Fingerspelling can also be used to spell words for signs that the signer does not know the sign for, or to clarify a sign that is not known by the person reading the signer. Fingerspelling signs are often also incorporated into other ASL signs.
# Source : Sign Language Forum
# Hand Landmarks Detection
# The MediaPipe Hand Landmarker task lets you detect the landmarks of the hands in an image. You can use this Task to localize key points of the hands and render visual effects over the hands. This task operates on image data with a machine learning (ML) model as static data or a continuous stream and outputs hand landmarks in image coordinates, hand landmarks in world coordinates and handedness(left/right hand) of multiple detected hands. The hand landmark model bundle detects the keypoint localization of 21 hand-knuckle coordinates within the detected hand regions. The model was trained on approximately 30K real-world images, as well as several rendered synthetic hand models imposed over various backgrounds.
# Source: Mediapipe
# Table of Contents
# - Import Libraries
# - Train Dataset
# - Supplemental Metadata
# - Plot Hands in 2D
# - Plot Hands in 3D
# Import Library ↑
#
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import mediapipe as mp
import plotly.graph_objects as go
from colorama import Style, Fore
blk = Style.BRIGHT + Fore.BLACK
red = Style.BRIGHT + Fore.RED
blu = Style.BRIGHT + Fore.BLUE
cyan = Style.BRIGHT + Fore.CYAN
res = Style.RESET_ALL
base_dir = "/kaggle/input/asl-fingerspelling"
train_csv = f"{base_dir}/train.csv"
supplemental_csv = f"{base_dir}/supplemental_metadata.csv"
#
# Train Dataset ↑
#
train = pd.read_csv(train_csv)
train["path"] = base_dir + "/" + train["path"]
train = train.drop(["file_id"], axis=1)
train.head()
# Top 10 Phrase in Training Dataset
#
sns.set(style="whitegrid")
fig, ax = plt.subplots(figsize=(8, 8))
sns.barplot(
y=train["phrase"].value_counts().head(10).sort_values(ascending=False).index,
x=train["phrase"].value_counts().head(10).sort_values(ascending=False),
ax=ax,
)
ax.set_title("Top 10 Phrase in Training Dataset")
ax.set_xlabel("Number of Training Examples")
ax.set_ylabel("Phrase")
plt.show()
# Parquet File of Top 1 Phrase in Train Dataset
#
select_train = "surprise az"
train_example = train.query("phrase == @select_train")["path"].values[0]
select_landmark_train = pd.read_parquet(train_example)
select_landmark_train
# One parquet path consist of more than one sequence_id, here I want to show a landmark for `surprise az` but in one parquet has many sequence_id. It means one parquet can consist of different labels.
seq_target_train = train.query("phrase == @select_train")["sequence_id"].values[0]
print(f"{blu}[+]{blk} Sequence ID : {blu}{seq_target_train}")
seq_df_train = select_landmark_train.query("index == @seq_target_train")
seq_df_train.head()
def x_y_z(column_names):
x = [col for col in column_names if col.startswith("x")]
y = [col for col in column_names if col.startswith("y")]
z = [col for col in column_names if col.startswith("z")]
return x, y, z
def type_of_landmark(example_landmark):
body_parts = set()
for column in example_landmark.columns:
parts = column.split("_")
if len(parts) >= 2:
if parts[1] == "right":
body_parts.add("right_hand")
elif parts[1] == "left":
body_parts.add("left_hand")
else:
body_parts.add(parts[1])
return body_parts
# Check Landmarks, Frames, and (X, Y, Z) Points
#
unique_frames = seq_df_train["frame"].nunique()
type_landmark_train = type_of_landmark(seq_df_train)
face_train = [col for col in seq_df_train.columns if "face" in col]
right_hand_train = [col for col in seq_df_train.columns if "right_hand" in col]
left_hand_train = [col for col in seq_df_train.columns if "left_hand" in col]
pose_train = [col for col in seq_df_train.columns if "pose" in col]
x_face_train, y_face_train, z_face_train = x_y_z(face_train)
x_right_hand_train, y_right_hand_train, z_right_hand_train = x_y_z(right_hand_train)
x_left_hand_train, y_left_hand_train, z_left_hand_train = x_y_z(left_hand_train)
x_pose_train, y_pose_train, z_pose_train = x_y_z(pose_train)
print(f"{cyan}{'='*20} ( Train Dataset) {'='*20}")
print(
f"{blk}Landmark file for sequence_id {red}{seq_target_train}{blk} has {red}{unique_frames}{blk} frames "
)
print(
f"{blk}This landmark has {red}{len(type_landmark_train)} {blk}types of landmarks and consists of {red}{type_landmark_train}"
)
print(
f"\n{blu}[+]{blk} {blk}Face landmark has {red}{len(face_train)} {blk}points in x : {red}{len(x_face_train)} points, {blk}y : {red}{len(y_face_train)} points, {blk}z : {red}{len(z_face_train)} points"
)
print(
f"{blu}[+]{blk} {blk}Right hand landmark has {red}{len(right_hand_train)} {blk}points in x : {red}{len(x_right_hand_train)} points, {blk}y : {red}{len(y_right_hand_train)} points, {blk}z : {red}{len(z_right_hand_train)} points"
)
print(
f"{blu}[+]{blk} {blk}Left hand landmark has {red}{len(left_hand_train)} {blk}points in x : {red}{len(x_left_hand_train)} points, {blk}y : {red}{len(y_left_hand_train)} points, {blk}z : {red}{len(z_left_hand_train)} points"
)
print(
f"{blu}[+]{blk} {blk}Pose landmark has {red}{len(pose_train)} {blk}points in x : {red}{len(x_pose_train)} points, {blk}y : {red}{len(y_pose_train)} points, {blk}z : {red}{len(z_pose_train)} points"
)
#
# Supplemental Metadata ↑
#
supplemental = pd.read_csv(supplemental_csv)
supplemental["path"] = base_dir + "/" + supplemental["path"]
supplemental = supplemental.drop(["file_id"], axis=1)
supplemental.head()
# Top 10 Phrase in Supplemental Metadata
#
sns.set(style="whitegrid")
fig, ax = plt.subplots(figsize=(8, 8))
sns.barplot(
y=supplemental["phrase"].value_counts().head(10).sort_values(ascending=False).index,
x=supplemental["phrase"].value_counts().head(10).sort_values(ascending=False),
ax=ax,
)
ax.set_title("Top 10 Phrase in Supplemental Dataset")
ax.set_xlabel("Number of Examples")
ax.set_ylabel("Phrase")
plt.show()
# Parquet File of Top 1 Phrase in Supplemental Metadata
#
select_supp = "why do you ask silly questions"
supp_example = supplemental.query("phrase == @select_supp")["path"].values[0]
select_landmark_supp = pd.read_parquet(supp_example)
select_landmark_supp
# Same as **Train Dataset**, one parquet file also has more than one sequence_id
seq_target_supp = supplemental.query("phrase == @select_supp")["sequence_id"].values[0]
print(f"{blu}[+]{blk} Sequence ID : {blu}{seq_target_supp}")
seq_df_supp = select_landmark_supp.query("index == @seq_target_supp")
seq_df_supp.head()
# Check Landmarks, Frames, and (X, Y, Z) Points
#
unique_frames = seq_df_supp["frame"].nunique()
type_landmark_supp = type_of_landmark(seq_df_supp)
face_supp = [col for col in seq_df_supp.columns if "face" in col]
right_hand_supp = [col for col in seq_df_supp.columns if "right_hand" in col]
left_hand_supp = [col for col in seq_df_supp.columns if "left_hand" in col]
pose_supp = [col for col in seq_df_supp.columns if "pose" in col]
x_face_supp, y_face_supp, z_face_supp = x_y_z(face_supp)
x_right_hand_supp, y_right_hand_supp, z_right_hand_supp = x_y_z(right_hand_supp)
x_left_hand_supp, y_left_hand_supp, z_left_hand_supp = x_y_z(left_hand_supp)
x_pose_supp, y_pose_supp, z_pose_supp = x_y_z(pose_supp)
print(f"{cyan}{'='*20} ( Supplemental Dataset) {'='*20}")
print(
f"{blk}Landmark file for sequence_id {red}{seq_target_supp}{blk} has {red}{unique_frames}{blk} frames "
)
print(
f"{blk}This landmark has {red}{len(type_landmark_supp)} {blk}types of landmarks and consists of {red}{type_landmark_supp}"
)
print(
f"\n{blu}[+]{blk} Face landmark has {red}{len(face_supp)} {blk}points in x : {red}{len(x_face_supp)} points, {blk}y : {red}{len(y_face_supp)} points, {blk}z : {red}{len(z_face_supp)} points"
)
print(
f"{blu}[+]{blk} Right hand landmark has {red}{len(right_hand_supp)} {blk}points in x : {red}{len(x_right_hand_supp)} points, {blk}y : {red}{len(y_right_hand_supp)} points, {blk}z : {red}{len(z_right_hand_supp)} points"
)
print(
f"{blu}[+]{blk} Left hand landmark has {red}{len(left_hand_supp)} {blk}points in x : {red}{len(x_left_hand_supp)} points, {blk}y : {red}{len(y_left_hand_supp)} points, {blk}z : {red}{len(z_left_hand_supp)} points"
)
print(
f"{blu}[+]{blk} Pose landmark has {red}{len(pose_supp)} {blk}points in x : {red}{len(x_pose_supp)} points, {blk}y : {red}{len(y_pose_supp)} points, {blk}z : {red}{len(z_pose_supp)} points"
)
#
# Plot Hands in 2D ↑
#
mp_hands = mp.solutions.hands
def data_plot(seq, frame, x_col, y_col, z_col, df):
x = df.query("sequence_id == @seq and frame == @frame")[x_col].iloc[0].values
y = df.query("sequence_id == @seq and frame == @frame")[y_col].iloc[0].values
z = df.query("sequence_id == @seq and frame == @frame")[z_col].iloc[0].values
landmark_idx = [
int(col.split("_")[-1])
for col in df.query("sequence_id == @seq and frame == @frame")[x_col].columns
]
dataframe = pd.DataFrame({"x": x, "y": y, "z": z, "landmark_idx": landmark_idx})
return dataframe
# Training Data Plot Hands of "surprise az" Phrase
#
frame = 12
left_hand_train = data_plot(
seq_target_train,
frame,
x_left_hand_train,
y_left_hand_train,
z_left_hand_train,
seq_df_train,
)
right_hand_train = data_plot(
seq_target_train,
frame,
x_right_hand_train,
y_right_hand_train,
z_right_hand_train,
seq_df_train,
)
fig, ax = plt.subplots(figsize=(5, 5))
ax.scatter(right_hand_train["x"], right_hand_train["y"])
ax.scatter(left_hand_train["x"], left_hand_train["y"])
for connection in mp_hands.HAND_CONNECTIONS:
point_a = connection[0]
point_b = connection[1]
x1, y1 = right_hand_train.query("landmark_idx == @point_a")[["x", "y"]].values[0]
x2, y2 = right_hand_train.query("landmark_idx == @point_b")[["x", "y"]].values[0]
plt.plot([x1, x2], [y1, y2], color="red")
x3, y3 = left_hand_train.query("landmark_idx == @point_a")[["x", "y"]].values[0]
x4, y4 = left_hand_train.query("landmark_idx == @point_b")[["x", "y"]].values[0]
plt.plot([x3, x4], [y3, y4], color="red")
ax.set_title(select_train)
plt.show()
# Supplement Metadata Plot "why do you ask silly questions" Phrase
#
frame = 140
left_hand_supp = data_plot(
seq_target_supp,
frame,
x_left_hand_supp,
y_left_hand_supp,
z_left_hand_supp,
seq_df_supp,
)
right_hand_supp = data_plot(
seq_target_supp,
frame,
x_right_hand_supp,
y_right_hand_supp,
z_right_hand_supp,
seq_df_supp,
)
fig, ax = plt.subplots(figsize=(5, 5))
ax.scatter(right_hand_supp["x"], right_hand_supp["y"])
ax.scatter(left_hand_supp["x"], left_hand_supp["y"])
for connection in mp_hands.HAND_CONNECTIONS:
point_a = connection[0]
point_b = connection[1]
x1, y1 = right_hand_supp.query("landmark_idx == @point_a")[["x", "y"]].values[0]
x2, y2 = right_hand_supp.query("landmark_idx == @point_b")[["x", "y"]].values[0]
plt.plot([x1, x2], [y1, y2], color="red")
x3, y3 = left_hand_supp.query("landmark_idx == @point_a")[["x", "y"]].values[0]
x4, y4 = left_hand_supp.query("landmark_idx == @point_b")[["x", "y"]].values[0]
plt.plot([x3, x4], [y3, y4], color="red")
ax.set_title(select_supp)
plt.show()
#
# Plot Hands in 3D ↑
#
def plot_3d(right, left, title):
right["hand"] = "right"
left["hand"] = "left"
df = pd.concat([right, left])
traces = []
scatter_trace = go.Scatter3d(
x=df["x"],
y=df["y"],
z=df["z"],
mode="markers",
marker=dict(size=5, color="blue", opacity=0.8),
name="Landmarks",
)
traces.append(scatter_trace)
line_trace = go.Scatter3d(
x=[],
y=[],
z=[],
mode="lines",
line=dict(color="red", width=2),
name="Connections",
)
for connection in mp_hands.HAND_CONNECTIONS:
point_a = connection[0]
point_b = connection[1]
x1, y1, z1 = df.query("landmark_idx == @point_a")[["x", "y", "z"]].values[0]
x2, y2, z2 = df.query("landmark_idx == @point_b")[["x", "y", "z"]].values[0]
line_trace["x"] += (x1, x2, None)
line_trace["y"] += (y1, y2, None)
line_trace["z"] += (z1, z2, None)
traces.append(line_trace)
fig = go.Figure(data=traces)
_text = f'3D Plot of "{title}"'
fig.update_layout(
scene=dict(xaxis_title="X", yaxis_title="Y", zaxis_title="Z"),
showlegend=True,
title={
"text": _text,
"xanchor": "center",
"yanchor": "top",
"x": 0.5,
},
)
fig.show()
# Train Dataset Plot in 3D
#
plot_3d(right_hand_train, left_hand_train, select_train)
# Supplemental Metadata Plot in 3D
#
plot_3d(right_hand_supp, left_hand_supp, select_supp)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/267/129267658.ipynb
| null | null |
[{"Id": 129267658, "ScriptId": 38377708, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11120740, "CreationDate": "05/12/2023 10:03:49", "VersionNumber": 6.0, "Title": "\ud83d\udcca [EDA] ASL - Fingerspelling 2D & 3D Plot \ud83d\udcca", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 419.0, "LinesInsertedFromPrevious": 117.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 302.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 11}]
| null | null | null | null |
### CSS for notebook styling ###
from IPython.core.display import HTML
HTML(
"""
<style>
:root {
--box_color: #F1F6F9;
}
body[data-jp-theme-light="true"] .jp-Notebook .CodeMirror.cm-s-jupyter{
background-color: var(--box_color) !important;
}
div.input_area{
background-color: var(--box_color) !important;
}
.crop {
display: block;
height: 250px;
position: relative;
overflow: hidden;
width: 700px;
}
.crop img {
left: 0px; /* alter this to move left or right */
position: absolute;
top: 0px; /* alter this to move up or down */
}
</style>
"""
)
#
# 🤞 Google - American Sign Language Fingerspelling Recognition 🤞
#
#
# Goal of the Competition
#
# The goal of this competition is to detect and translate American Sign Language (ASL) fingerspelling into text. You will create a model trained on the largest dataset of its kind, released specifically for this competition. The data includes more than three million fingerspelled characters produced by over 100 Deaf signers captured via the selfie camera of a smartphone with a variety of backgrounds and lighting conditions.
# ASL Fingerspelling Alphabet
#
# Fingerspelling is a way of spelling words using hand movements. The fingerspelling manual alphabet is used in sign language to spell out names of people and places for which there is not a sign. Fingerspelling can also be used to spell words for signs that the signer does not know the sign for, or to clarify a sign that is not known by the person reading the signer. Fingerspelling signs are often also incorporated into other ASL signs.
# Source : Sign Language Forum
# Hand Landmarks Detection
# The MediaPipe Hand Landmarker task lets you detect the landmarks of the hands in an image. You can use this Task to localize key points of the hands and render visual effects over the hands. This task operates on image data with a machine learning (ML) model as static data or a continuous stream and outputs hand landmarks in image coordinates, hand landmarks in world coordinates and handedness(left/right hand) of multiple detected hands. The hand landmark model bundle detects the keypoint localization of 21 hand-knuckle coordinates within the detected hand regions. The model was trained on approximately 30K real-world images, as well as several rendered synthetic hand models imposed over various backgrounds.
# Source: Mediapipe
# Table of Contents
# - Import Libraries
# - Train Dataset
# - Supplemental Metadata
# - Plot Hands in 2D
# - Plot Hands in 3D
# Import Library ↑
#
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import mediapipe as mp
import plotly.graph_objects as go
from colorama import Style, Fore
blk = Style.BRIGHT + Fore.BLACK
red = Style.BRIGHT + Fore.RED
blu = Style.BRIGHT + Fore.BLUE
cyan = Style.BRIGHT + Fore.CYAN
res = Style.RESET_ALL
base_dir = "/kaggle/input/asl-fingerspelling"
train_csv = f"{base_dir}/train.csv"
supplemental_csv = f"{base_dir}/supplemental_metadata.csv"
#
# Train Dataset ↑
#
train = pd.read_csv(train_csv)
train["path"] = base_dir + "/" + train["path"]
train = train.drop(["file_id"], axis=1)
train.head()
# Top 10 Phrase in Training Dataset
#
sns.set(style="whitegrid")
fig, ax = plt.subplots(figsize=(8, 8))
sns.barplot(
y=train["phrase"].value_counts().head(10).sort_values(ascending=False).index,
x=train["phrase"].value_counts().head(10).sort_values(ascending=False),
ax=ax,
)
ax.set_title("Top 10 Phrase in Training Dataset")
ax.set_xlabel("Number of Training Examples")
ax.set_ylabel("Phrase")
plt.show()
# Parquet File of Top 1 Phrase in Train Dataset
#
select_train = "surprise az"
train_example = train.query("phrase == @select_train")["path"].values[0]
select_landmark_train = pd.read_parquet(train_example)
select_landmark_train
# One parquet path consist of more than one sequence_id, here I want to show a landmark for `surprise az` but in one parquet has many sequence_id. It means one parquet can consist of different labels.
seq_target_train = train.query("phrase == @select_train")["sequence_id"].values[0]
print(f"{blu}[+]{blk} Sequence ID : {blu}{seq_target_train}")
seq_df_train = select_landmark_train.query("index == @seq_target_train")
seq_df_train.head()
def x_y_z(column_names):
x = [col for col in column_names if col.startswith("x")]
y = [col for col in column_names if col.startswith("y")]
z = [col for col in column_names if col.startswith("z")]
return x, y, z
def type_of_landmark(example_landmark):
body_parts = set()
for column in example_landmark.columns:
parts = column.split("_")
if len(parts) >= 2:
if parts[1] == "right":
body_parts.add("right_hand")
elif parts[1] == "left":
body_parts.add("left_hand")
else:
body_parts.add(parts[1])
return body_parts
# Check Landmarks, Frames, and (X, Y, Z) Points
#
unique_frames = seq_df_train["frame"].nunique()
type_landmark_train = type_of_landmark(seq_df_train)
face_train = [col for col in seq_df_train.columns if "face" in col]
right_hand_train = [col for col in seq_df_train.columns if "right_hand" in col]
left_hand_train = [col for col in seq_df_train.columns if "left_hand" in col]
pose_train = [col for col in seq_df_train.columns if "pose" in col]
x_face_train, y_face_train, z_face_train = x_y_z(face_train)
x_right_hand_train, y_right_hand_train, z_right_hand_train = x_y_z(right_hand_train)
x_left_hand_train, y_left_hand_train, z_left_hand_train = x_y_z(left_hand_train)
x_pose_train, y_pose_train, z_pose_train = x_y_z(pose_train)
print(f"{cyan}{'='*20} ( Train Dataset) {'='*20}")
print(
f"{blk}Landmark file for sequence_id {red}{seq_target_train}{blk} has {red}{unique_frames}{blk} frames "
)
print(
f"{blk}This landmark has {red}{len(type_landmark_train)} {blk}types of landmarks and consists of {red}{type_landmark_train}"
)
print(
f"\n{blu}[+]{blk} {blk}Face landmark has {red}{len(face_train)} {blk}points in x : {red}{len(x_face_train)} points, {blk}y : {red}{len(y_face_train)} points, {blk}z : {red}{len(z_face_train)} points"
)
print(
f"{blu}[+]{blk} {blk}Right hand landmark has {red}{len(right_hand_train)} {blk}points in x : {red}{len(x_right_hand_train)} points, {blk}y : {red}{len(y_right_hand_train)} points, {blk}z : {red}{len(z_right_hand_train)} points"
)
print(
f"{blu}[+]{blk} {blk}Left hand landmark has {red}{len(left_hand_train)} {blk}points in x : {red}{len(x_left_hand_train)} points, {blk}y : {red}{len(y_left_hand_train)} points, {blk}z : {red}{len(z_left_hand_train)} points"
)
print(
f"{blu}[+]{blk} {blk}Pose landmark has {red}{len(pose_train)} {blk}points in x : {red}{len(x_pose_train)} points, {blk}y : {red}{len(y_pose_train)} points, {blk}z : {red}{len(z_pose_train)} points"
)
#
# Supplemental Metadata ↑
#
supplemental = pd.read_csv(supplemental_csv)
supplemental["path"] = base_dir + "/" + supplemental["path"]
supplemental = supplemental.drop(["file_id"], axis=1)
supplemental.head()
# Top 10 Phrase in Supplemental Metadata
#
sns.set(style="whitegrid")
fig, ax = plt.subplots(figsize=(8, 8))
sns.barplot(
y=supplemental["phrase"].value_counts().head(10).sort_values(ascending=False).index,
x=supplemental["phrase"].value_counts().head(10).sort_values(ascending=False),
ax=ax,
)
ax.set_title("Top 10 Phrase in Supplemental Dataset")
ax.set_xlabel("Number of Examples")
ax.set_ylabel("Phrase")
plt.show()
# Parquet File of Top 1 Phrase in Supplemental Metadata
#
select_supp = "why do you ask silly questions"
supp_example = supplemental.query("phrase == @select_supp")["path"].values[0]
select_landmark_supp = pd.read_parquet(supp_example)
select_landmark_supp
# Same as **Train Dataset**, one parquet file also has more than one sequence_id
seq_target_supp = supplemental.query("phrase == @select_supp")["sequence_id"].values[0]
print(f"{blu}[+]{blk} Sequence ID : {blu}{seq_target_supp}")
seq_df_supp = select_landmark_supp.query("index == @seq_target_supp")
seq_df_supp.head()
# Check Landmarks, Frames, and (X, Y, Z) Points
#
unique_frames = seq_df_supp["frame"].nunique()
type_landmark_supp = type_of_landmark(seq_df_supp)
face_supp = [col for col in seq_df_supp.columns if "face" in col]
right_hand_supp = [col for col in seq_df_supp.columns if "right_hand" in col]
left_hand_supp = [col for col in seq_df_supp.columns if "left_hand" in col]
pose_supp = [col for col in seq_df_supp.columns if "pose" in col]
x_face_supp, y_face_supp, z_face_supp = x_y_z(face_supp)
x_right_hand_supp, y_right_hand_supp, z_right_hand_supp = x_y_z(right_hand_supp)
x_left_hand_supp, y_left_hand_supp, z_left_hand_supp = x_y_z(left_hand_supp)
x_pose_supp, y_pose_supp, z_pose_supp = x_y_z(pose_supp)
print(f"{cyan}{'='*20} ( Supplemental Dataset) {'='*20}")
print(
f"{blk}Landmark file for sequence_id {red}{seq_target_supp}{blk} has {red}{unique_frames}{blk} frames "
)
print(
f"{blk}This landmark has {red}{len(type_landmark_supp)} {blk}types of landmarks and consists of {red}{type_landmark_supp}"
)
print(
f"\n{blu}[+]{blk} Face landmark has {red}{len(face_supp)} {blk}points in x : {red}{len(x_face_supp)} points, {blk}y : {red}{len(y_face_supp)} points, {blk}z : {red}{len(z_face_supp)} points"
)
print(
f"{blu}[+]{blk} Right hand landmark has {red}{len(right_hand_supp)} {blk}points in x : {red}{len(x_right_hand_supp)} points, {blk}y : {red}{len(y_right_hand_supp)} points, {blk}z : {red}{len(z_right_hand_supp)} points"
)
print(
f"{blu}[+]{blk} Left hand landmark has {red}{len(left_hand_supp)} {blk}points in x : {red}{len(x_left_hand_supp)} points, {blk}y : {red}{len(y_left_hand_supp)} points, {blk}z : {red}{len(z_left_hand_supp)} points"
)
print(
f"{blu}[+]{blk} Pose landmark has {red}{len(pose_supp)} {blk}points in x : {red}{len(x_pose_supp)} points, {blk}y : {red}{len(y_pose_supp)} points, {blk}z : {red}{len(z_pose_supp)} points"
)
#
# Plot Hands in 2D ↑
#
mp_hands = mp.solutions.hands
def data_plot(seq, frame, x_col, y_col, z_col, df):
x = df.query("sequence_id == @seq and frame == @frame")[x_col].iloc[0].values
y = df.query("sequence_id == @seq and frame == @frame")[y_col].iloc[0].values
z = df.query("sequence_id == @seq and frame == @frame")[z_col].iloc[0].values
landmark_idx = [
int(col.split("_")[-1])
for col in df.query("sequence_id == @seq and frame == @frame")[x_col].columns
]
dataframe = pd.DataFrame({"x": x, "y": y, "z": z, "landmark_idx": landmark_idx})
return dataframe
# Training Data Plot Hands of "surprise az" Phrase
#
frame = 12
left_hand_train = data_plot(
seq_target_train,
frame,
x_left_hand_train,
y_left_hand_train,
z_left_hand_train,
seq_df_train,
)
right_hand_train = data_plot(
seq_target_train,
frame,
x_right_hand_train,
y_right_hand_train,
z_right_hand_train,
seq_df_train,
)
fig, ax = plt.subplots(figsize=(5, 5))
ax.scatter(right_hand_train["x"], right_hand_train["y"])
ax.scatter(left_hand_train["x"], left_hand_train["y"])
for connection in mp_hands.HAND_CONNECTIONS:
point_a = connection[0]
point_b = connection[1]
x1, y1 = right_hand_train.query("landmark_idx == @point_a")[["x", "y"]].values[0]
x2, y2 = right_hand_train.query("landmark_idx == @point_b")[["x", "y"]].values[0]
plt.plot([x1, x2], [y1, y2], color="red")
x3, y3 = left_hand_train.query("landmark_idx == @point_a")[["x", "y"]].values[0]
x4, y4 = left_hand_train.query("landmark_idx == @point_b")[["x", "y"]].values[0]
plt.plot([x3, x4], [y3, y4], color="red")
ax.set_title(select_train)
plt.show()
# Supplement Metadata Plot "why do you ask silly questions" Phrase
#
frame = 140
left_hand_supp = data_plot(
seq_target_supp,
frame,
x_left_hand_supp,
y_left_hand_supp,
z_left_hand_supp,
seq_df_supp,
)
right_hand_supp = data_plot(
seq_target_supp,
frame,
x_right_hand_supp,
y_right_hand_supp,
z_right_hand_supp,
seq_df_supp,
)
fig, ax = plt.subplots(figsize=(5, 5))
ax.scatter(right_hand_supp["x"], right_hand_supp["y"])
ax.scatter(left_hand_supp["x"], left_hand_supp["y"])
for connection in mp_hands.HAND_CONNECTIONS:
point_a = connection[0]
point_b = connection[1]
x1, y1 = right_hand_supp.query("landmark_idx == @point_a")[["x", "y"]].values[0]
x2, y2 = right_hand_supp.query("landmark_idx == @point_b")[["x", "y"]].values[0]
plt.plot([x1, x2], [y1, y2], color="red")
x3, y3 = left_hand_supp.query("landmark_idx == @point_a")[["x", "y"]].values[0]
x4, y4 = left_hand_supp.query("landmark_idx == @point_b")[["x", "y"]].values[0]
plt.plot([x3, x4], [y3, y4], color="red")
ax.set_title(select_supp)
plt.show()
#
# Plot Hands in 3D ↑
#
def plot_3d(right, left, title):
right["hand"] = "right"
left["hand"] = "left"
df = pd.concat([right, left])
traces = []
scatter_trace = go.Scatter3d(
x=df["x"],
y=df["y"],
z=df["z"],
mode="markers",
marker=dict(size=5, color="blue", opacity=0.8),
name="Landmarks",
)
traces.append(scatter_trace)
line_trace = go.Scatter3d(
x=[],
y=[],
z=[],
mode="lines",
line=dict(color="red", width=2),
name="Connections",
)
for connection in mp_hands.HAND_CONNECTIONS:
point_a = connection[0]
point_b = connection[1]
x1, y1, z1 = df.query("landmark_idx == @point_a")[["x", "y", "z"]].values[0]
x2, y2, z2 = df.query("landmark_idx == @point_b")[["x", "y", "z"]].values[0]
line_trace["x"] += (x1, x2, None)
line_trace["y"] += (y1, y2, None)
line_trace["z"] += (z1, z2, None)
traces.append(line_trace)
fig = go.Figure(data=traces)
_text = f'3D Plot of "{title}"'
fig.update_layout(
scene=dict(xaxis_title="X", yaxis_title="Y", zaxis_title="Z"),
showlegend=True,
title={
"text": _text,
"xanchor": "center",
"yanchor": "top",
"x": 0.5,
},
)
fig.show()
# Train Dataset Plot in 3D
#
plot_3d(right_hand_train, left_hand_train, select_train)
# Supplemental Metadata Plot in 3D
#
plot_3d(right_hand_supp, left_hand_supp, select_supp)
| false | 0 | 4,995 | 11 | 4,995 | 4,995 |
||
129267938
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load the CSV file into a DataFrame
df = pd.read_csv("/kaggle/input/results/results.csv")
# Define metrics where a higher value is better (default is lower is better)
higher_is_better = ["R2"]
# Initialize an empty DataFrame to store ranks
rank_df = pd.DataFrame()
# Iterate over each column, rank and add the ranks to the rank_df
for column in df.columns[1:]: # Skip the 'Model' column
if any(metric in column for metric in higher_is_better):
rank_df[column] = df[column].rank(ascending=False) # Higher is better
else:
rank_df[column] = df[column].rank(ascending=True) # Lower is better
# Add 'Model' column to rank_df
rank_df.insert(0, "Model", df["Model"])
# Calculate the average rank for each model across all metrics for each analytical measure
rank_df["Average Rank"] = rank_df.iloc[:, 1:].mean(axis=1)
# Sort by 'Average Rank'
rank_df = rank_df.sort_values("Average Rank")
# Display the final rankings
final_rankings = rank_df[["Model", "Average Rank"]]
print(final_rankings)
import matplotlib.pyplot as plt
import seaborn as sns
# Create the plot
plt.figure(figsize=(10, 8))
sns.barplot(
x=final_rankings["Average Rank"], y=final_rankings["Model"], palette="viridis"
)
# Add labels and title
plt.xlabel("Average Rank")
plt.ylabel("Model")
plt.title("Average Rankings of NLP Models")
plt.show()
# Visualizing the Average RMSE results for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"Average RMSE": [
0.470234,
0.473284,
0.455243,
0.486614,
0.462514,
0.525482,
0.530286,
0.489979,
],
}
df = pd.DataFrame(data)
# Sort by Average RMSE in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("Average RMSE", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["Average RMSE"], color="#FFD700")
plt.xlabel("Average RMSE")
plt.ylabel("Model")
plt.title("Average RMSE Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Cohesion for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Cohesion": [
0.50295,
0.508771,
0.490552,
0.521127,
0.498775,
0.534879,
0.542715,
0.523514,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Cohesion in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Cohesion", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Cohesion"], color="#FFD700")
plt.xlabel("RMSE Cohesion")
plt.ylabel("Model")
plt.title("RMSE Cohesion Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Syntax for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Syntax": [
0.456276,
0.459739,
0.448062,
0.468768,
0.453276,
0.506154,
0.513466,
0.473778,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Syntax in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Syntax", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Syntax"], color="#FFD700")
plt.xlabel("RMSE Syntax")
plt.ylabel("Model")
plt.title("RMSE Syntax Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Vocabulary for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Vocabulary": [
0.427843,
0.431236,
0.414498,
0.447835,
0.421848,
0.482615,
0.478928,
0.448247,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Vocabulary in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Vocabulary", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Vocabulary"], color="#FFD700")
plt.xlabel("RMSE Vocabulary")
plt.ylabel("Model")
plt.title("RMSE Vocabulary Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Phraseology for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Phraseology": [
0.465948,
0.473574,
0.45477,
0.483429,
0.460844,
0.542765,
0.537219,
0.485657,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Phraseology in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Phraseology", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Phraseology"], color="#FFD700")
plt.xlabel("RMSE Phraseology")
plt.ylabel("Model")
plt.title("RMSE Phraseology Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Grammar for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Grammar": [
0.501194,
0.50298,
0.474021,
0.511746,
0.478772,
0.576701,
0.592243,
0.519692,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Grammar in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Grammar", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Grammar"], color="#FFD700")
plt.xlabel("RMSE Grammar")
plt.ylabel("Model")
plt.title("RMSE Grammar Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Conventions for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Conventions": [
0.467193,
0.463405,
0.449553,
0.48678,
0.461569,
0.509778,
0.517148,
0.488983,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Conventions in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Conventions", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Conventions"], color="#FFD700")
plt.xlabel("RMSE Conventions")
plt.ylabel("Model")
plt.title("RMSE Conventions Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/267/129267938.ipynb
| null | null |
[{"Id": 129267938, "ScriptId": 38392442, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9882726, "CreationDate": "05/12/2023 10:06:26", "VersionNumber": 2.0, "Title": "statistical_ranking", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 247.0, "LinesInsertedFromPrevious": 183.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 64.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load the CSV file into a DataFrame
df = pd.read_csv("/kaggle/input/results/results.csv")
# Define metrics where a higher value is better (default is lower is better)
higher_is_better = ["R2"]
# Initialize an empty DataFrame to store ranks
rank_df = pd.DataFrame()
# Iterate over each column, rank and add the ranks to the rank_df
for column in df.columns[1:]: # Skip the 'Model' column
if any(metric in column for metric in higher_is_better):
rank_df[column] = df[column].rank(ascending=False) # Higher is better
else:
rank_df[column] = df[column].rank(ascending=True) # Lower is better
# Add 'Model' column to rank_df
rank_df.insert(0, "Model", df["Model"])
# Calculate the average rank for each model across all metrics for each analytical measure
rank_df["Average Rank"] = rank_df.iloc[:, 1:].mean(axis=1)
# Sort by 'Average Rank'
rank_df = rank_df.sort_values("Average Rank")
# Display the final rankings
final_rankings = rank_df[["Model", "Average Rank"]]
print(final_rankings)
import matplotlib.pyplot as plt
import seaborn as sns
# Create the plot
plt.figure(figsize=(10, 8))
sns.barplot(
x=final_rankings["Average Rank"], y=final_rankings["Model"], palette="viridis"
)
# Add labels and title
plt.xlabel("Average Rank")
plt.ylabel("Model")
plt.title("Average Rankings of NLP Models")
plt.show()
# Visualizing the Average RMSE results for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"Average RMSE": [
0.470234,
0.473284,
0.455243,
0.486614,
0.462514,
0.525482,
0.530286,
0.489979,
],
}
df = pd.DataFrame(data)
# Sort by Average RMSE in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("Average RMSE", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["Average RMSE"], color="#FFD700")
plt.xlabel("Average RMSE")
plt.ylabel("Model")
plt.title("Average RMSE Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Cohesion for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Cohesion": [
0.50295,
0.508771,
0.490552,
0.521127,
0.498775,
0.534879,
0.542715,
0.523514,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Cohesion in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Cohesion", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Cohesion"], color="#FFD700")
plt.xlabel("RMSE Cohesion")
plt.ylabel("Model")
plt.title("RMSE Cohesion Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Syntax for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Syntax": [
0.456276,
0.459739,
0.448062,
0.468768,
0.453276,
0.506154,
0.513466,
0.473778,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Syntax in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Syntax", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Syntax"], color="#FFD700")
plt.xlabel("RMSE Syntax")
plt.ylabel("Model")
plt.title("RMSE Syntax Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Vocabulary for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Vocabulary": [
0.427843,
0.431236,
0.414498,
0.447835,
0.421848,
0.482615,
0.478928,
0.448247,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Vocabulary in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Vocabulary", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Vocabulary"], color="#FFD700")
plt.xlabel("RMSE Vocabulary")
plt.ylabel("Model")
plt.title("RMSE Vocabulary Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Phraseology for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Phraseology": [
0.465948,
0.473574,
0.45477,
0.483429,
0.460844,
0.542765,
0.537219,
0.485657,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Phraseology in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Phraseology", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Phraseology"], color="#FFD700")
plt.xlabel("RMSE Phraseology")
plt.ylabel("Model")
plt.title("RMSE Phraseology Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Grammar for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Grammar": [
0.501194,
0.50298,
0.474021,
0.511746,
0.478772,
0.576701,
0.592243,
0.519692,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Grammar in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Grammar", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Grammar"], color="#FFD700")
plt.xlabel("RMSE Grammar")
plt.ylabel("Model")
plt.title("RMSE Grammar Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
# Visualizing the RMSE Conventions for the models
# Data
data = {
"Model": [
"deberta-v3-base",
"deberta-v3-small",
"deberta-v3-large",
"roberta-base",
"roberta-large",
"xml-roberta-base",
"xml-roberta-longformer-base",
"bigbird-roberta-base",
],
"RMSE Conventions": [
0.467193,
0.463405,
0.449553,
0.48678,
0.461569,
0.509778,
0.517148,
0.488983,
],
}
df = pd.DataFrame(data)
# Sort by RMSE Conventions in ascending order so the best model (with the lowest RMSE) is on top
df = df.sort_values("RMSE Conventions", ascending=False)
plt.figure(figsize=(10, 6))
plt.barh(df["Model"], df["RMSE Conventions"], color="#FFD700")
plt.xlabel("RMSE Conventions")
plt.ylabel("Model")
plt.title("RMSE Conventions Scores of Models")
plt.grid(axis="x")
# Set dark background
plt.gca().set_facecolor("0.1")
plt.grid(color="w", linestyle="-", linewidth=0.5)
plt.show()
| false | 0 | 3,154 | 0 | 3,154 | 3,154 |
||
129267142
|
<jupyter_start><jupyter_text>BangaloreHousePrices
Kaggle dataset identifier: bangalorehouseprices
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import tensorflow as tf
import numpy as np
from matplotlib import pyplot as plt
import matplotlib
matplotlib.rcParams["figure.figsize"] = (20, 10)
df1 = pd.read_csv("../input/bangalorehouseprices/bengaluru_house_prices.csv")
df1.head()
df1.shape
df1["area_type"].value_counts()
# Data Cleaning
df2 = df1.drop(["area_type", "society", "balcony", "availability"], axis="columns")
df2.head()
df2.isnull().sum()
df3 = df2.dropna()
df3.isnull().sum()
df3["size"].unique()
# Feature Engineering
df3["bhk"] = df3["size"].apply(lambda x: int(x.split(" ")[0]))
df3.bhk.unique()
df3
df3["bhk"].unique()
df3[df3["bhk"] > 20]
df3.total_sqft.unique()
def is_float(x):
try:
float(x)
except:
return False
return True
df3[df3["total_sqft"].apply(is_float)]
###Above shows that total_sqft can be a range (e.g. 2100-2850). For such case we can just take average of min and max value in the range. There are other cases such as 34.46Sq. Meter which one can convert to square ft using unit conversion. I am going to just drop such corner cases to keep things simple
def convert_sqft_to_num(x):
tokens = x.split("-")
if (len(tokens)) == 2:
return (float(tokens[0]) + float(tokens[1])) / 2
try:
return float(x)
except:
return None
convert_sqft_to_num("2000 - 4000")
df4 = df3.copy() # deep copy
df4["total_sqft"] = df4["total_sqft"].apply(convert_sqft_to_num)
df4 = df4[df4.total_sqft.notnull()]
df4
len(df4.location.unique())
df5 = df4.copy()
df5["price_per_sqft"] = df5["price"] * 100000 / df5["total_sqft"]
df5.head()
len(df5.location.unique())
# Lot of Locations
# Since we have lot of locations, this is called dimensionality curse
df5.location = df5.location.apply(lambda x: x.strip())
location_stats = (
df5.groupby("location")["location"].agg("count").sort_values(ascending=False)
)
location_stats
len(location_stats[location_stats < 10])
location_stas_less_than_10 = location_stats[location_stats < 10]
location_stas_less_than_10
df5.location = df5.location.apply(
lambda x: "other" if x in location_stas_less_than_10 else x
)
# all the locations less than 10 data points will be converted to 'other'
# Outliers are not errors but really large or small values which make no sense in the data. For example a 2 bedroom apartment cannot be 5000 sq feet
# As a data scientist when you have a conversation with your business manager (who has expertise in real estate), he will tell you that normally square ft per bedroom is 300 (i.e. 2 bhk apartment is minimum 600 sqft. If you have for example 400 sqft apartment with 2 bhk than that seems suspicious and can be removed as an outlier.
# We will remove such outliers by keeping our minimum thresold per bhk to be 300 sqft
df5[df5.total_sqft / df5.bhk < 300].head()
df6 = df5[~(df5.total_sqft / df5.bhk < 300)]
df6.shape
df6.price_per_sqft.describe()
df6.groupby("location").agg("count")
def remove_pps_outliers(df):
df_out = pd.DataFrame()
for key, subdf in df.groupby("location"):
m = np.mean(subdf.price_per_sqft)
st = np.std(subdf.price_per_sqft)
reduced_df = subdf[
(subdf.price_per_sqft > (m - st)) & (subdf.price_per_sqft <= (m + st))
]
df_out = pd.concat([df_out, reduced_df], ignore_index=True)
return df_out
df7 = remove_pps_outliers(df6)
df7.shape
# the remove_pps_outliers function is looping thorough the subgroups of locations.
# For. eg. a subdf could be all data points with "jayanagar" as a location.
# It calculates mean and std of the rows in jayanagar location
# and then selects all points in that are within m-st and m-st of jayanagar and adds that to the df_out.
# One more thing that we have to check is that if the price of a two bhk apt is greater than 3bhk apt for the same square foot area
def plot_scatter_chart(df, location):
bhk2 = df[(df.location == location) & (df.bhk == 2)]
bhk3 = df[(df.location == location) & (df.bhk == 3)]
matplotlib.rcParams["figure.figsize"] = (10, 5)
plt.scatter(bhk2.total_sqft, bhk2.price, color="blue", label="2 BHK", s=50)
plt.scatter(
bhk3.total_sqft, bhk3.price, marker="+", color="green", label="3 BHK", s=50
)
plt.xlabel("Total Square Feet Area")
plt.ylabel("Price (Lakh Indian Rupees)")
plt.title(location)
plt.legend()
plot_scatter_chart(
df7, "Rajaji Nagar"
) # for around 1700 sq foot area the two bedroom apt price is higher than 3 bedroom
plot_scatter_chart(df7, "Hebbal")
df7
def remove_bhk_outliers(df):
exclude_indices = np.array([])
for location, location_df in df.groupby("location"):
bhk_stats = {}
for bhk, bhk_df in location_df.groupby("bhk"):
bhk_stats[bhk] = {
"mean": np.mean(bhk_df.price_per_sqft),
"std": np.std(bhk_df.price_per_sqft),
"count": bhk_df.shape[0],
}
for bhk, bhk_df in location_df.groupby("bhk"):
stats = bhk_stats.get(bhk - 1)
if stats and stats["count"] > 5:
exclude_indices = np.append(
exclude_indices,
bhk_df[bhk_df.price_per_sqft < (stats["mean"])].index.values,
)
return df.drop(exclude_indices, axis="index")
df8 = remove_bhk_outliers(df7)
df8.shape
plot_scatter_chart(df8, "Rajaji Nagar")
plot_scatter_chart(df8, "Hebbal")
df8
import matplotlib
matplotlib.rcParams["figure.figsize"] = (20, 10)
plt.hist(df8.price_per_sqft, rwidth=0.8)
plt.xlabel("Price Per Square Feet")
plt.ylabel("Count")
df8.bath.unique()
plt.hist(df8.bath, rwidth=0.8)
plt.xlabel("Number of bathrooms")
plt.ylabel("Count")
df8[df8.bath > 10]
df9 = df8[df8.bath > df8.bhk + 2]
df9
df9 = df8[df8.bath < df8.bhk + 2]
df9.shape
df10 = df9.drop(["size", "price_per_sqft"], axis="columns")
df10.head(3)
# size and price_per_sqft
# can be dropped because they were used only for outlier detection. Now the dataset is neat and clean and we can go for machine learning training
df10.head()
dummies = pd.get_dummies(df10.location)
dummies.head(3)
df11 = pd.concat([df10, dummies.drop("other", axis="columns")], axis="columns")
df11.head()
df12 = df11.drop("location", axis="columns")
df12.head(10)
df12.shape
X = df12.drop(["price"], axis="columns")
X.head(3)
X.shape
y = df12.price
y.head(3)
len(y)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=10
)
from sklearn.linear_model import LinearRegression
lr_clf = LinearRegression()
lr_clf.fit(X_train, y_train)
lr_clf.score(X_test, y_test)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/267/129267142.ipynb
|
bangalorehouseprices
|
vedanthbaliga
|
[{"Id": 129267142, "ScriptId": 38233052, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6697406, "CreationDate": "05/12/2023 09:58:17", "VersionNumber": 1.0, "Title": "bengaluru_house_prices", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 244.0, "LinesInsertedFromPrevious": 244.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185158512, "KernelVersionId": 129267142, "SourceDatasetVersionId": 1882786}]
|
[{"Id": 1882786, "DatasetId": 1121332, "DatasourceVersionId": 1920935, "CreatorUserId": 5418149, "LicenseName": "Unknown", "CreationDate": "01/27/2021 02:50:11", "VersionNumber": 1.0, "Title": "BangaloreHousePrices", "Slug": "bangalorehouseprices", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1121332, "CreatorUserId": 5418149, "OwnerUserId": 5418149.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1882786.0, "CurrentDatasourceVersionId": 1920935.0, "ForumId": 1138702, "Type": 2, "CreationDate": "01/27/2021 02:50:11", "LastActivityDate": "01/27/2021", "TotalViews": 1456, "TotalDownloads": 362, "TotalVotes": 5, "TotalKernels": 2}]
|
[{"Id": 5418149, "UserName": "vedanthbaliga", "DisplayName": "Vedanth Baliga", "RegisterDate": "07/05/2020", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import tensorflow as tf
import numpy as np
from matplotlib import pyplot as plt
import matplotlib
matplotlib.rcParams["figure.figsize"] = (20, 10)
df1 = pd.read_csv("../input/bangalorehouseprices/bengaluru_house_prices.csv")
df1.head()
df1.shape
df1["area_type"].value_counts()
# Data Cleaning
df2 = df1.drop(["area_type", "society", "balcony", "availability"], axis="columns")
df2.head()
df2.isnull().sum()
df3 = df2.dropna()
df3.isnull().sum()
df3["size"].unique()
# Feature Engineering
df3["bhk"] = df3["size"].apply(lambda x: int(x.split(" ")[0]))
df3.bhk.unique()
df3
df3["bhk"].unique()
df3[df3["bhk"] > 20]
df3.total_sqft.unique()
def is_float(x):
try:
float(x)
except:
return False
return True
df3[df3["total_sqft"].apply(is_float)]
###Above shows that total_sqft can be a range (e.g. 2100-2850). For such case we can just take average of min and max value in the range. There are other cases such as 34.46Sq. Meter which one can convert to square ft using unit conversion. I am going to just drop such corner cases to keep things simple
def convert_sqft_to_num(x):
tokens = x.split("-")
if (len(tokens)) == 2:
return (float(tokens[0]) + float(tokens[1])) / 2
try:
return float(x)
except:
return None
convert_sqft_to_num("2000 - 4000")
df4 = df3.copy() # deep copy
df4["total_sqft"] = df4["total_sqft"].apply(convert_sqft_to_num)
df4 = df4[df4.total_sqft.notnull()]
df4
len(df4.location.unique())
df5 = df4.copy()
df5["price_per_sqft"] = df5["price"] * 100000 / df5["total_sqft"]
df5.head()
len(df5.location.unique())
# Lot of Locations
# Since we have lot of locations, this is called dimensionality curse
df5.location = df5.location.apply(lambda x: x.strip())
location_stats = (
df5.groupby("location")["location"].agg("count").sort_values(ascending=False)
)
location_stats
len(location_stats[location_stats < 10])
location_stas_less_than_10 = location_stats[location_stats < 10]
location_stas_less_than_10
df5.location = df5.location.apply(
lambda x: "other" if x in location_stas_less_than_10 else x
)
# all the locations less than 10 data points will be converted to 'other'
# Outliers are not errors but really large or small values which make no sense in the data. For example a 2 bedroom apartment cannot be 5000 sq feet
# As a data scientist when you have a conversation with your business manager (who has expertise in real estate), he will tell you that normally square ft per bedroom is 300 (i.e. 2 bhk apartment is minimum 600 sqft. If you have for example 400 sqft apartment with 2 bhk than that seems suspicious and can be removed as an outlier.
# We will remove such outliers by keeping our minimum thresold per bhk to be 300 sqft
df5[df5.total_sqft / df5.bhk < 300].head()
df6 = df5[~(df5.total_sqft / df5.bhk < 300)]
df6.shape
df6.price_per_sqft.describe()
df6.groupby("location").agg("count")
def remove_pps_outliers(df):
df_out = pd.DataFrame()
for key, subdf in df.groupby("location"):
m = np.mean(subdf.price_per_sqft)
st = np.std(subdf.price_per_sqft)
reduced_df = subdf[
(subdf.price_per_sqft > (m - st)) & (subdf.price_per_sqft <= (m + st))
]
df_out = pd.concat([df_out, reduced_df], ignore_index=True)
return df_out
df7 = remove_pps_outliers(df6)
df7.shape
# the remove_pps_outliers function is looping thorough the subgroups of locations.
# For. eg. a subdf could be all data points with "jayanagar" as a location.
# It calculates mean and std of the rows in jayanagar location
# and then selects all points in that are within m-st and m-st of jayanagar and adds that to the df_out.
# One more thing that we have to check is that if the price of a two bhk apt is greater than 3bhk apt for the same square foot area
def plot_scatter_chart(df, location):
bhk2 = df[(df.location == location) & (df.bhk == 2)]
bhk3 = df[(df.location == location) & (df.bhk == 3)]
matplotlib.rcParams["figure.figsize"] = (10, 5)
plt.scatter(bhk2.total_sqft, bhk2.price, color="blue", label="2 BHK", s=50)
plt.scatter(
bhk3.total_sqft, bhk3.price, marker="+", color="green", label="3 BHK", s=50
)
plt.xlabel("Total Square Feet Area")
plt.ylabel("Price (Lakh Indian Rupees)")
plt.title(location)
plt.legend()
plot_scatter_chart(
df7, "Rajaji Nagar"
) # for around 1700 sq foot area the two bedroom apt price is higher than 3 bedroom
plot_scatter_chart(df7, "Hebbal")
df7
def remove_bhk_outliers(df):
exclude_indices = np.array([])
for location, location_df in df.groupby("location"):
bhk_stats = {}
for bhk, bhk_df in location_df.groupby("bhk"):
bhk_stats[bhk] = {
"mean": np.mean(bhk_df.price_per_sqft),
"std": np.std(bhk_df.price_per_sqft),
"count": bhk_df.shape[0],
}
for bhk, bhk_df in location_df.groupby("bhk"):
stats = bhk_stats.get(bhk - 1)
if stats and stats["count"] > 5:
exclude_indices = np.append(
exclude_indices,
bhk_df[bhk_df.price_per_sqft < (stats["mean"])].index.values,
)
return df.drop(exclude_indices, axis="index")
df8 = remove_bhk_outliers(df7)
df8.shape
plot_scatter_chart(df8, "Rajaji Nagar")
plot_scatter_chart(df8, "Hebbal")
df8
import matplotlib
matplotlib.rcParams["figure.figsize"] = (20, 10)
plt.hist(df8.price_per_sqft, rwidth=0.8)
plt.xlabel("Price Per Square Feet")
plt.ylabel("Count")
df8.bath.unique()
plt.hist(df8.bath, rwidth=0.8)
plt.xlabel("Number of bathrooms")
plt.ylabel("Count")
df8[df8.bath > 10]
df9 = df8[df8.bath > df8.bhk + 2]
df9
df9 = df8[df8.bath < df8.bhk + 2]
df9.shape
df10 = df9.drop(["size", "price_per_sqft"], axis="columns")
df10.head(3)
# size and price_per_sqft
# can be dropped because they were used only for outlier detection. Now the dataset is neat and clean and we can go for machine learning training
df10.head()
dummies = pd.get_dummies(df10.location)
dummies.head(3)
df11 = pd.concat([df10, dummies.drop("other", axis="columns")], axis="columns")
df11.head()
df12 = df11.drop("location", axis="columns")
df12.head(10)
df12.shape
X = df12.drop(["price"], axis="columns")
X.head(3)
X.shape
y = df12.price
y.head(3)
len(y)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=10
)
from sklearn.linear_model import LinearRegression
lr_clf = LinearRegression()
lr_clf.fit(X_train, y_train)
lr_clf.score(X_test, y_test)
| false | 1 | 2,554 | 0 | 2,579 | 2,554 |
||
129267158
|
# # Approximate Nearest Neighbour
# ## 1. Product Quantization (PQ)
import faiss
import numpy as np
# Generate random data
d = 1000 # dimension
nb = 100000 # number of vectors
X = np.random.rand(nb, d).astype("float32")
# Build PQ index
m = 8 # number of subquantizers
nbits = 8 # number of bits per subquantizer
pq = faiss.IndexPQ(d, m, nbits)
pq.train(X)
pq.add(X)
# Query nearest neighbors
query_point = np.random.rand(1, d).astype("float32")
D, I = pq.search(query_point, k=5)
import pandas as pd
pd.DataFrame(X).iloc[I[0]]
pd.DataFrame(query_point)
# ## 2. Hierarchical Navigable Small World Graphs (HNSW)
import nmslib
import numpy as np
# Generate random data
d = 100 # dimension
nb = 100000 # number of vectors
X = np.random.rand(nb, d).astype("float32")
# Build HNSW index
index = nmslib.init(method="hnsw", space="l2", data_type=nmslib.DataType.DENSE_VECTOR)
index.addDataPointBatch(X)
index.createIndex({"post": 2}, print_progress=True)
# Query nearest neighbors
query_point = np.random.rand(1, d).astype("float32")
ids, distances = index.knnQuery(query_point, k=5)
pd.DataFrame(query_point)
pd.DataFrame(X).iloc[ids]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/267/129267158.ipynb
| null | null |
[{"Id": 129267158, "ScriptId": 38432401, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1534022, "CreationDate": "05/12/2023 09:58:27", "VersionNumber": 1.0, "Title": "2023 Approximate Nearest Neighbour", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 55.0, "LinesInsertedFromPrevious": 55.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
# # Approximate Nearest Neighbour
# ## 1. Product Quantization (PQ)
import faiss
import numpy as np
# Generate random data
d = 1000 # dimension
nb = 100000 # number of vectors
X = np.random.rand(nb, d).astype("float32")
# Build PQ index
m = 8 # number of subquantizers
nbits = 8 # number of bits per subquantizer
pq = faiss.IndexPQ(d, m, nbits)
pq.train(X)
pq.add(X)
# Query nearest neighbors
query_point = np.random.rand(1, d).astype("float32")
D, I = pq.search(query_point, k=5)
import pandas as pd
pd.DataFrame(X).iloc[I[0]]
pd.DataFrame(query_point)
# ## 2. Hierarchical Navigable Small World Graphs (HNSW)
import nmslib
import numpy as np
# Generate random data
d = 100 # dimension
nb = 100000 # number of vectors
X = np.random.rand(nb, d).astype("float32")
# Build HNSW index
index = nmslib.init(method="hnsw", space="l2", data_type=nmslib.DataType.DENSE_VECTOR)
index.addDataPointBatch(X)
index.createIndex({"post": 2}, print_progress=True)
# Query nearest neighbors
query_point = np.random.rand(1, d).astype("float32")
ids, distances = index.knnQuery(query_point, k=5)
pd.DataFrame(query_point)
pd.DataFrame(X).iloc[ids]
| false | 0 | 427 | 1 | 427 | 427 |
||
129620328
|
# ### This notebook is meant for accademic purposes only
# Dans ce notebook nous allons voir la manière dont on peut transformer un texte en matrice, et les utilisations de cette matrice par la suite.
# Dans ce but nous allons utiliser les fonctionnalités CountVectorizer et TfidfVectorizer de Scikit Learn.
# Nous allons utiliser les données de la competition Disaster Tweet.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import re
from scipy.sparse import hstack, coo_matrix
import nltk
from nltk.stem.snowball import SnowballStemmer
from nltk.stem import WordNetLemmatizer
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize, sent_tokenize
from nltk import pos_tag
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# On lit le dataframe de training et on affiche les 10 premières lignes à l'aide de la méthode de classe `head` :
df_train = pd.read_csv("../input/nlp-getting-started/train.csv")
df_train.head(10)
# Nous sommes interessés seulement par le texte de ce DataFrame, donc on sauve la colonne correspondant au contenu du tweet dans la variable text.
# Cette variable est transformée d'abord en numpy array (values) et ensuite en liste de tweets (tolist)
#
text = df_train["text"].values.tolist()
# ### CountVectorizer
# CountVectorizer prend en entrée une liste de textes. Comme tout outil de Scikit Learn, il comporte une partie d'apprentissage et une partie de prediction.
# Partie apprentissage (training) -- méthode `fit`
# CountVectorizer applique sur les textes en entrée les transformations suivantes :
# 1. Les tokenise
# 2. Les met en minuscule
# 3. Cherche les tokens uniques dans tous ces textes et les arange en ordre alphabétique. Ces mots vont constituer les colonnes de la matrice de sortie
# Partie prediction -- méthode `transform`:
# Pour chaque texte en entrée :
# 1. Pour chaque mot dans les colonnes, on note le nombre d'occurences du mot dans le texte. Si le mot ne se trouve pas dans le texte, on met zéro.
# On crée l'objet CountVectorizer, on applique fit et ensuite transform sur notre ensemble de textes. A ce moment là l'outil connaît les mots uniques dans les textes. On peut les visualiser à l'aide de la méthode `get_feature_names_out`.
cv = CountVectorizer()
cv.fit(text)
features = cv.get_feature_names_out()
# On affiche les 20 premiers tokens
print(features[0:20])
# On affiche aussi le nombre total de tokens uniques :
print(len(features))
# Nous remarquons que nous avons des tokens formés seulement des chiffres. Ils nous interessent pas. Nous voulons les ignorer. On peut faire cela en modifiant l'attribut token_pattern de CountVectorizer.
# Le pattern initial était le suivant, qui cherche des tokens de 2 caractères ou plus, formées des lettres ou des chiffres: `r”(?u)\b\w\w+\b”`.
# Nous voulons des tokens formés seulement des lettres.
# Nous voulons également exclure du traitement les stop_words. Pour cela on crée la liste de stop words anglais à l'aide de la librairie NLTK et on donne cette liste en argument à CountVectorizer
# Nous avons remarqué que les fautes de frappe sont des tokens les plus rares, mais aussi les moins importants. Nous allons les éliminer en fixant l'argument min_df. min_df permet de choisir les tokens qui ont une fréquence minimum donnée.
sw = stopwords.words("english")
cv = CountVectorizer(
token_pattern=r"(?u)\b[a-zA-Z][a-zA-Z]+\b", stop_words=sw, min_df=5
)
# On remarque que le nombre de tokens choisi a diminué considerablement et que les tokens restants correspondent plus à des mots porteurs de sens.
cv.fit(text)
features = cv.get_feature_names_out()
print(features)
print(len(features))
# On transforme le texte pour obtenir notre matrice.
text_sparse_matrix = cv.transform(text)
# Visualisons notre matrice.
# Nous voyons que le type est sparse matrix.
# Une sparse matrix est une matrice avec beaucoup de zéros et quelques valeurs différentes de zéro.
# Dans une telle matrice, on marque et on affiche seulement les celulles qui contiennent des valeurs différentes de zéro. Son utilisation permet de consommer moins de mémoire.
#
print(type(text_sparse_matrix))
# Comment on interprète les données de cette matrice ?
#
print(text_sparse_matrix)
tokens = nltk.word_tokenize(line_curr)
print(tokens)
pos = nltk.pos_tag(tokens)
# print(pos)
stemmer = SnowballStemmer("english")
for tok in tokens:
stem = stemmer.stem(tok)
print(stem)
text1 = []
for t in text:
tokens = nltk.word_tokenize(t.lower())
s = ""
for token in tokens:
stem = stemmer.stem(token)
s += stem + " "
text1.append(s)
text = text1
# ### Expressions regulières (REGEX)
line = "We always try to bring the heavy. #metal #RT http://t.co/YAo1e0xngw in the show. Here you are the link : http://www.t.co/YAo1e0xngw."
pattern_url = r"http(s)?[^\s\n\t\r]+"
pattern_author = r"@\w+"
if re.search(pattern_url, line):
p = re.compile(pattern_url)
l = p.sub("URL", line)
print(l)
urls = re.findall(pattern_url, line)
print(urls)
for url in urls:
print(url)
# ### Stopwords
stop_words = stopwords.words("english")
print(stop_words)
cv = CountVectorizer(
stop_words=stop_words, min_df=10, token_pattern=r"(?u)\b[A-Za-z]{2,}\b"
)
out = cv.fit_transform(text)
voc = cv.get_feature_names_out()
print(voc)
print(len(voc))
X = out.toarray()
X.shape
Y = df_train["target"]
print(Y.shape, type(Y))
YY = df_train.loc[:, ["target"]]
print(YY.shape)
pred = clf.predict(X)
print(voc[0:30])
for w in voc[0:30]:
if w.isalpha():
print(w, "alpha")
print(out)
# ### Tokenisation
line_curr = line_curr.lower()
tokens = nltk.word_tokenize(line_curr)
print(tokens)
pos = nltk.pos_tag(tokens)
# print(pos)
tokens = [word.lower() for word in tokens if word not in stop_words]
print(tokens)
Line = " ".join(tokens)
print(line)
for word in pos:
if "NN" in word[1] or "JJ" in word[1]:
print(word[0])
# ### Stemmatisation
stemmer = SnowballStemmer("english")
for tok in tokens:
stem = stemmer.stem(tok)
print(stem)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/620/129620328.ipynb
| null | null |
[{"Id": 129620328, "ScriptId": 38543558, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3616012, "CreationDate": "05/15/2023 09:29:57", "VersionNumber": 1.0, "Title": "GEMA_M2_NLP_Cours_3", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 211.0, "LinesInsertedFromPrevious": 89.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 122.0, "LinesInsertedFromFork": 89.0, "LinesDeletedFromFork": 6.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 122.0, "TotalVotes": 0}]
| null | null | null | null |
# ### This notebook is meant for accademic purposes only
# Dans ce notebook nous allons voir la manière dont on peut transformer un texte en matrice, et les utilisations de cette matrice par la suite.
# Dans ce but nous allons utiliser les fonctionnalités CountVectorizer et TfidfVectorizer de Scikit Learn.
# Nous allons utiliser les données de la competition Disaster Tweet.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import re
from scipy.sparse import hstack, coo_matrix
import nltk
from nltk.stem.snowball import SnowballStemmer
from nltk.stem import WordNetLemmatizer
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize, sent_tokenize
from nltk import pos_tag
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# On lit le dataframe de training et on affiche les 10 premières lignes à l'aide de la méthode de classe `head` :
df_train = pd.read_csv("../input/nlp-getting-started/train.csv")
df_train.head(10)
# Nous sommes interessés seulement par le texte de ce DataFrame, donc on sauve la colonne correspondant au contenu du tweet dans la variable text.
# Cette variable est transformée d'abord en numpy array (values) et ensuite en liste de tweets (tolist)
#
text = df_train["text"].values.tolist()
# ### CountVectorizer
# CountVectorizer prend en entrée une liste de textes. Comme tout outil de Scikit Learn, il comporte une partie d'apprentissage et une partie de prediction.
# Partie apprentissage (training) -- méthode `fit`
# CountVectorizer applique sur les textes en entrée les transformations suivantes :
# 1. Les tokenise
# 2. Les met en minuscule
# 3. Cherche les tokens uniques dans tous ces textes et les arange en ordre alphabétique. Ces mots vont constituer les colonnes de la matrice de sortie
# Partie prediction -- méthode `transform`:
# Pour chaque texte en entrée :
# 1. Pour chaque mot dans les colonnes, on note le nombre d'occurences du mot dans le texte. Si le mot ne se trouve pas dans le texte, on met zéro.
# On crée l'objet CountVectorizer, on applique fit et ensuite transform sur notre ensemble de textes. A ce moment là l'outil connaît les mots uniques dans les textes. On peut les visualiser à l'aide de la méthode `get_feature_names_out`.
cv = CountVectorizer()
cv.fit(text)
features = cv.get_feature_names_out()
# On affiche les 20 premiers tokens
print(features[0:20])
# On affiche aussi le nombre total de tokens uniques :
print(len(features))
# Nous remarquons que nous avons des tokens formés seulement des chiffres. Ils nous interessent pas. Nous voulons les ignorer. On peut faire cela en modifiant l'attribut token_pattern de CountVectorizer.
# Le pattern initial était le suivant, qui cherche des tokens de 2 caractères ou plus, formées des lettres ou des chiffres: `r”(?u)\b\w\w+\b”`.
# Nous voulons des tokens formés seulement des lettres.
# Nous voulons également exclure du traitement les stop_words. Pour cela on crée la liste de stop words anglais à l'aide de la librairie NLTK et on donne cette liste en argument à CountVectorizer
# Nous avons remarqué que les fautes de frappe sont des tokens les plus rares, mais aussi les moins importants. Nous allons les éliminer en fixant l'argument min_df. min_df permet de choisir les tokens qui ont une fréquence minimum donnée.
sw = stopwords.words("english")
cv = CountVectorizer(
token_pattern=r"(?u)\b[a-zA-Z][a-zA-Z]+\b", stop_words=sw, min_df=5
)
# On remarque que le nombre de tokens choisi a diminué considerablement et que les tokens restants correspondent plus à des mots porteurs de sens.
cv.fit(text)
features = cv.get_feature_names_out()
print(features)
print(len(features))
# On transforme le texte pour obtenir notre matrice.
text_sparse_matrix = cv.transform(text)
# Visualisons notre matrice.
# Nous voyons que le type est sparse matrix.
# Une sparse matrix est une matrice avec beaucoup de zéros et quelques valeurs différentes de zéro.
# Dans une telle matrice, on marque et on affiche seulement les celulles qui contiennent des valeurs différentes de zéro. Son utilisation permet de consommer moins de mémoire.
#
print(type(text_sparse_matrix))
# Comment on interprète les données de cette matrice ?
#
print(text_sparse_matrix)
tokens = nltk.word_tokenize(line_curr)
print(tokens)
pos = nltk.pos_tag(tokens)
# print(pos)
stemmer = SnowballStemmer("english")
for tok in tokens:
stem = stemmer.stem(tok)
print(stem)
text1 = []
for t in text:
tokens = nltk.word_tokenize(t.lower())
s = ""
for token in tokens:
stem = stemmer.stem(token)
s += stem + " "
text1.append(s)
text = text1
# ### Expressions regulières (REGEX)
line = "We always try to bring the heavy. #metal #RT http://t.co/YAo1e0xngw in the show. Here you are the link : http://www.t.co/YAo1e0xngw."
pattern_url = r"http(s)?[^\s\n\t\r]+"
pattern_author = r"@\w+"
if re.search(pattern_url, line):
p = re.compile(pattern_url)
l = p.sub("URL", line)
print(l)
urls = re.findall(pattern_url, line)
print(urls)
for url in urls:
print(url)
# ### Stopwords
stop_words = stopwords.words("english")
print(stop_words)
cv = CountVectorizer(
stop_words=stop_words, min_df=10, token_pattern=r"(?u)\b[A-Za-z]{2,}\b"
)
out = cv.fit_transform(text)
voc = cv.get_feature_names_out()
print(voc)
print(len(voc))
X = out.toarray()
X.shape
Y = df_train["target"]
print(Y.shape, type(Y))
YY = df_train.loc[:, ["target"]]
print(YY.shape)
pred = clf.predict(X)
print(voc[0:30])
for w in voc[0:30]:
if w.isalpha():
print(w, "alpha")
print(out)
# ### Tokenisation
line_curr = line_curr.lower()
tokens = nltk.word_tokenize(line_curr)
print(tokens)
pos = nltk.pos_tag(tokens)
# print(pos)
tokens = [word.lower() for word in tokens if word not in stop_words]
print(tokens)
Line = " ".join(tokens)
print(line)
for word in pos:
if "NN" in word[1] or "JJ" in word[1]:
print(word[0])
# ### Stemmatisation
stemmer = SnowballStemmer("english")
for tok in tokens:
stem = stemmer.stem(tok)
print(stem)
| false | 0 | 2,031 | 0 | 2,031 | 2,031 |
||
129620793
|
<jupyter_start><jupyter_text>Walmart Data Analysis and Forcasting
A retail store that has multiple outlets across the country are facing issues in managing the
inventory - to match the demand with respect to supply. You are a data scientist, who has to
come up with useful insights using the data and make prediction models to forecast the sales for
X number of months/years.
Kaggle dataset identifier: walmart-data-analysis-and-forcasting
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error, r2_score
df = pd.read_csv(
"/kaggle/input/walmart-data-analysis-and-forcasting/Walmart Data Analysis and Forcasting.csv"
)
df
df.shape
df.info()
df.describe()
df.isnull().sum()
df.duplicated().sum()
df.drop_duplicates(inplace=True)
df.nunique()
df["Date"] = df["Date"].astype(np.datetime64)
df.info()
df = df.assign(
Day_Name=df.Date.dt.day, Month_Name=df.Date.dt.month, Year_Name=df.Date.dt.year
)
df
sns.lineplot(x="Date", y="Weekly_Sales", data=df)
plt.xticks(rotation=45)
plt.show()
fig = plt.figure(figsize=(14, 7))
sales_by_store = df.groupby("Store")["Weekly_Sales"].sum().reset_index()
sns.barplot(x="Store", y="Weekly_Sales", data=sales_by_store)
plt.show()
sales_by_holiday = df.groupby("Holiday_Flag")["Weekly_Sales"].mean().reset_index()
sns.barplot(x="Holiday_Flag", y="Weekly_Sales", data=sales_by_holiday)
plt.show()
sns.scatterplot(x="Temperature", y="Weekly_Sales", data=df)
plt.show()
sns.scatterplot(x="CPI", y="Weekly_Sales", data=df)
plt.show()
sns.scatterplot(x="Unemployment", y="Weekly_Sales", data=df)
plt.show()
fig = plt.figure(figsize=(10, 7))
corr_matrix = df.corr()
sns.heatmap(corr_matrix, annot=True)
plt.show()
df["Date"] = pd.to_datetime(df["Date"])
df.set_index("Date", inplace=True)
df.reset_index()
monthly_sales = df.resample("M")["Weekly_Sales"].sum().reset_index()
monthly_sales = monthly_sales.rename(columns={"Date": "ds", "Weekly_Sales": "y"})
monthly_sales
train = monthly_sales.iloc[:-12]
test = monthly_sales.iloc[-12:]
X_train = train.index.values.reshape(-1, 1)
y_train = train["y"]
lr = LinearRegression()
lr.fit(X_train, y_train)
X_test = test.index.values.reshape(-1, 1)
y_pred = lr.predict(X_test)
plt.plot(test["ds"], test["y"], label="Actual Sales")
plt.plot(test["ds"], y_pred, label="Predicted Sales")
plt.legend()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/620/129620793.ipynb
|
walmart-data-analysis-and-forcasting
|
asahu40
|
[{"Id": 129620793, "ScriptId": 38434409, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10866133, "CreationDate": "05/15/2023 09:33:52", "VersionNumber": 1.0, "Title": "walmart_analysis_and_forcasting", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 108.0, "LinesInsertedFromPrevious": 108.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 185877858, "KernelVersionId": 129620793, "SourceDatasetVersionId": 5526698}]
|
[{"Id": 5526698, "DatasetId": 3186183, "DatasourceVersionId": 5601262, "CreatorUserId": 4711744, "LicenseName": "CC0: Public Domain", "CreationDate": "04/26/2023 07:07:03", "VersionNumber": 1.0, "Title": "Walmart Data Analysis and Forcasting", "Slug": "walmart-data-analysis-and-forcasting", "Subtitle": "Walmart Sales Data Analysis and Forcasting for EDA and Machine Learning", "Description": "A retail store that has multiple outlets across the country are facing issues in managing the\ninventory - to match the demand with respect to supply. You are a data scientist, who has to\ncome up with useful insights using the data and make prediction models to forecast the sales for\nX number of months/years.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3186183, "CreatorUserId": 4711744, "OwnerUserId": 4711744.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5526698.0, "CurrentDatasourceVersionId": 5601262.0, "ForumId": 3250612, "Type": 2, "CreationDate": "04/26/2023 07:07:03", "LastActivityDate": "04/26/2023", "TotalViews": 22574, "TotalDownloads": 4018, "TotalVotes": 75, "TotalKernels": 14}]
|
[{"Id": 4711744, "UserName": "asahu40", "DisplayName": "Amit Kumar Sahu", "RegisterDate": "03/21/2020", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error, r2_score
df = pd.read_csv(
"/kaggle/input/walmart-data-analysis-and-forcasting/Walmart Data Analysis and Forcasting.csv"
)
df
df.shape
df.info()
df.describe()
df.isnull().sum()
df.duplicated().sum()
df.drop_duplicates(inplace=True)
df.nunique()
df["Date"] = df["Date"].astype(np.datetime64)
df.info()
df = df.assign(
Day_Name=df.Date.dt.day, Month_Name=df.Date.dt.month, Year_Name=df.Date.dt.year
)
df
sns.lineplot(x="Date", y="Weekly_Sales", data=df)
plt.xticks(rotation=45)
plt.show()
fig = plt.figure(figsize=(14, 7))
sales_by_store = df.groupby("Store")["Weekly_Sales"].sum().reset_index()
sns.barplot(x="Store", y="Weekly_Sales", data=sales_by_store)
plt.show()
sales_by_holiday = df.groupby("Holiday_Flag")["Weekly_Sales"].mean().reset_index()
sns.barplot(x="Holiday_Flag", y="Weekly_Sales", data=sales_by_holiday)
plt.show()
sns.scatterplot(x="Temperature", y="Weekly_Sales", data=df)
plt.show()
sns.scatterplot(x="CPI", y="Weekly_Sales", data=df)
plt.show()
sns.scatterplot(x="Unemployment", y="Weekly_Sales", data=df)
plt.show()
fig = plt.figure(figsize=(10, 7))
corr_matrix = df.corr()
sns.heatmap(corr_matrix, annot=True)
plt.show()
df["Date"] = pd.to_datetime(df["Date"])
df.set_index("Date", inplace=True)
df.reset_index()
monthly_sales = df.resample("M")["Weekly_Sales"].sum().reset_index()
monthly_sales = monthly_sales.rename(columns={"Date": "ds", "Weekly_Sales": "y"})
monthly_sales
train = monthly_sales.iloc[:-12]
test = monthly_sales.iloc[-12:]
X_train = train.index.values.reshape(-1, 1)
y_train = train["y"]
lr = LinearRegression()
lr.fit(X_train, y_train)
X_test = test.index.values.reshape(-1, 1)
y_pred = lr.predict(X_test)
plt.plot(test["ds"], test["y"], label="Actual Sales")
plt.plot(test["ds"], y_pred, label="Predicted Sales")
plt.legend()
plt.show()
| false | 1 | 921 | 1 | 1,025 | 921 |
||
129657006
|
<jupyter_start><jupyter_text>The Berka Dataset
Dataset originally found here: https://webpages.charlotte.edu/mirsad/itcs6265/group1/domain.html
"This database was prepared by Petr Berka and Marta Sochorova.
We will refer to the dataset as The Berka Dataset throughout this report.
The Berka dataset is a collection of financial information from a Czech bank. The dataset deals with over 5,300 bank clients with approximately 1,000,000 transactions. Additionally, the bank represented in the dataset has extended close to 700 loans and issued nearly 900 credit cards, all of which are represented in the data."
Kaggle dataset identifier: the-berka-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
prefix_files = "/kaggle/input/the-berka-dataset/"
account = pd.read_csv(f"{prefix_files}account.csv", sep=";")
card = pd.read_csv(f"{prefix_files}card.csv", sep=";")
client = pd.read_csv(f"{prefix_files}client.csv", sep=";")
disp = pd.read_csv(f"{prefix_files}disp.csv", sep=";")
district = pd.read_csv(f"{prefix_files}district.csv", sep=";")
loan = pd.read_csv(f"{prefix_files}loan.csv", sep=";")
order = pd.read_csv(f"{prefix_files}order.csv", sep=";")
trans = pd.read_csv(f"{prefix_files}trans.csv", sep=";")
account = account.assign(date=pd.to_datetime(account["date"], format="%y%m%d"))
account = account.assign(
year_acc_creation=account["date"].dt.year,
month_acc_creation=account["date"].dt.month,
day_acc_creation=account["date"].dt.day,
)
account["frequency"].replace(
{
"POPLATEK MESICNE": "Monthly Issuance",
"POPLATEK TYDNE": "Weekly Issuance",
"POPLATEK PO OBRATU": "Issuance After Transaction",
},
inplace=True,
)
account.rename({"date": "account_creation"}, axis=1, inplace=True)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/657/129657006.ipynb
|
the-berka-dataset
|
marceloventura
|
[{"Id": 129657006, "ScriptId": 38555964, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 972727, "CreationDate": "05/15/2023 14:21:54", "VersionNumber": 1.0, "Title": "notebook0353b622c8", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 40.0, "LinesInsertedFromPrevious": 40.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185956333, "KernelVersionId": 129657006, "SourceDatasetVersionId": 5691001}]
|
[{"Id": 5691001, "DatasetId": 3272079, "DatasourceVersionId": 5766611, "CreatorUserId": 972727, "LicenseName": "CC0: Public Domain", "CreationDate": "05/15/2023 13:46:50", "VersionNumber": 1.0, "Title": "The Berka Dataset", "Slug": "the-berka-dataset", "Subtitle": "1999 Czech Bank Financial Dataset (bank transactions, loans, credit cards)", "Description": "Dataset originally found here: https://webpages.charlotte.edu/mirsad/itcs6265/group1/domain.html\n\n\"This database was prepared by Petr Berka and Marta Sochorova.\nWe will refer to the dataset as The Berka Dataset throughout this report.\nThe Berka dataset is a collection of financial information from a Czech bank. The dataset deals with over 5,300 bank clients with approximately 1,000,000 transactions. Additionally, the bank represented in the dataset has extended close to 700 loans and issued nearly 900 credit cards, all of which are represented in the data.\"", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3272079, "CreatorUserId": 972727, "OwnerUserId": 972727.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5691001.0, "CurrentDatasourceVersionId": 5766611.0, "ForumId": 3337725, "Type": 2, "CreationDate": "05/15/2023 13:46:50", "LastActivityDate": "05/15/2023", "TotalViews": 298, "TotalDownloads": 33, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 972727, "UserName": "marceloventura", "DisplayName": "Marcelo Ventura", "RegisterDate": "03/17/2017", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
prefix_files = "/kaggle/input/the-berka-dataset/"
account = pd.read_csv(f"{prefix_files}account.csv", sep=";")
card = pd.read_csv(f"{prefix_files}card.csv", sep=";")
client = pd.read_csv(f"{prefix_files}client.csv", sep=";")
disp = pd.read_csv(f"{prefix_files}disp.csv", sep=";")
district = pd.read_csv(f"{prefix_files}district.csv", sep=";")
loan = pd.read_csv(f"{prefix_files}loan.csv", sep=";")
order = pd.read_csv(f"{prefix_files}order.csv", sep=";")
trans = pd.read_csv(f"{prefix_files}trans.csv", sep=";")
account = account.assign(date=pd.to_datetime(account["date"], format="%y%m%d"))
account = account.assign(
year_acc_creation=account["date"].dt.year,
month_acc_creation=account["date"].dt.month,
day_acc_creation=account["date"].dt.day,
)
account["frequency"].replace(
{
"POPLATEK MESICNE": "Monthly Issuance",
"POPLATEK TYDNE": "Weekly Issuance",
"POPLATEK PO OBRATU": "Issuance After Transaction",
},
inplace=True,
)
account.rename({"date": "account_creation"}, axis=1, inplace=True)
| false | 0 | 554 | 0 | 727 | 554 |
||
129657312
|
<jupyter_start><jupyter_text>California Housing Prices
### Context
This is the dataset used in the second chapter of Aurélien Géron's recent book 'Hands-On Machine learning with Scikit-Learn and TensorFlow'. It serves as an excellent introduction to implementing machine learning algorithms because it requires rudimentary data cleaning, has an easily understandable list of variables and sits at an optimal size between being to toyish and too cumbersome.
The data contains information from the 1990 California census. So although it may not help you with predicting current housing prices like the Zillow Zestimate dataset, it does provide an accessible introductory dataset for teaching people about the basics of machine learning.
### Content
The data pertains to the houses found in a given California district and some summary stats about them based on the 1990 census data. Be warned the data aren't cleaned so there are some preprocessing steps required! The columns are as follows, their names are pretty self explanitory:
longitude
latitude
housing_median_age
total_rooms
total_bedrooms
population
households
median_income
median_house_value
ocean_proximity
Kaggle dataset identifier: california-housing-prices
<jupyter_code>import pandas as pd
df = pd.read_csv('california-housing-prices/housing.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 20640 entries, 0 to 20639
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 longitude 20640 non-null float64
1 latitude 20640 non-null float64
2 housing_median_age 20640 non-null float64
3 total_rooms 20640 non-null float64
4 total_bedrooms 20433 non-null float64
5 population 20640 non-null float64
6 households 20640 non-null float64
7 median_income 20640 non-null float64
8 median_house_value 20640 non-null float64
9 ocean_proximity 20640 non-null object
dtypes: float64(9), object(1)
memory usage: 1.6+ MB
<jupyter_text>Examples:
{
"longitude": -122.23,
"latitude": 37.88,
"housing_median_age": 41,
"total_rooms": 880,
"total_bedrooms": 129,
"population": 322,
"households": 126,
"median_income": 8.3252,
"median_house_value": 452600,
"ocean_proximity": "NEAR BAY"
}
{
"longitude": -122.22,
"latitude": 37.86,
"housing_median_age": 21,
"total_rooms": 7099,
"total_bedrooms": 1106,
"population": 2401,
"households": 1138,
"median_income": 8.3014,
"median_house_value": 358500,
"ocean_proximity": "NEAR BAY"
}
{
"longitude": -122.24,
"latitude": 37.85,
"housing_median_age": 52,
"total_rooms": 1467,
"total_bedrooms": 190,
"population": 496,
"households": 177,
"median_income": 7.2574,
"median_house_value": 352100,
"ocean_proximity": "NEAR BAY"
}
{
"longitude": -122.25,
"latitude": 37.85,
"housing_median_age": 52,
"total_rooms": 1274,
"total_bedrooms": 235,
"population": 558,
"households": 219,
"median_income": 5.6431000000000004,
"median_house_value": 341300,
"ocean_proximity": "NEAR BAY"
}
<jupyter_script>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score, r2_score
from sklearn.ensemble import RandomForestRegressor
df = pd.read_csv("/kaggle/input/california-housing-prices/housing.csv")
df
df.info()
df["total_bedrooms"].mean()
df["total_bedrooms"] = df["total_bedrooms"].fillna(df["total_bedrooms"].mean())
df.isna().sum()
df = pd.get_dummies(df)
df.info()
x = df.drop("median_house_value", axis=1)
y = df["median_house_value"]
xtrain, xtest, ytrain, ytest = train_test_split(x, y, test_size=0.2, random_state=42)
sc = StandardScaler()
xtrain = sc.fit_transform(xtrain)
xtest = sc.transform(xtest)
gnb = GaussianNB()
gnb.fit(xtrain, ytrain)
res = gnb.predict(xtest)
accuracy_score(ytest, res)
rfr = RandomForestRegressor()
rfr.fit(xtrain, ytrain)
res = rfr.predict(xtest)
r2_score(ytest, res)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/657/129657312.ipynb
|
california-housing-prices
|
camnugent
|
[{"Id": 129657312, "ScriptId": 38550791, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11096970, "CreationDate": "05/15/2023 14:23:47", "VersionNumber": 1.0, "Title": "notebook18a16663c8", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 45.0, "LinesInsertedFromPrevious": 45.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185956923, "KernelVersionId": 129657312, "SourceDatasetVersionId": 7876}]
|
[{"Id": 7876, "DatasetId": 5227, "DatasourceVersionId": 7876, "CreatorUserId": 710779, "LicenseName": "CC0: Public Domain", "CreationDate": "11/24/2017 03:14:59", "VersionNumber": 1.0, "Title": "California Housing Prices", "Slug": "california-housing-prices", "Subtitle": "Median house prices for California districts derived from the 1990 census.", "Description": "### Context\n\nThis is the dataset used in the second chapter of Aur\u00e9lien G\u00e9ron's recent book 'Hands-On Machine learning with Scikit-Learn and TensorFlow'. It serves as an excellent introduction to implementing machine learning algorithms because it requires rudimentary data cleaning, has an easily understandable list of variables and sits at an optimal size between being to toyish and too cumbersome.\n\nThe data contains information from the 1990 California census. So although it may not help you with predicting current housing prices like the Zillow Zestimate dataset, it does provide an accessible introductory dataset for teaching people about the basics of machine learning.\n \n### Content\n\nThe data pertains to the houses found in a given California district and some summary stats about them based on the 1990 census data. Be warned the data aren't cleaned so there are some preprocessing steps required! The columns are as follows, their names are pretty self explanitory:\n\nlongitude\n\nlatitude\n\nhousing_median_age\n\ntotal_rooms\n\ntotal_bedrooms\n\npopulation\n\nhouseholds\n\nmedian_income\n\nmedian_house_value\n\nocean_proximity\n\n### Acknowledgements\n\nThis data was initially featured in the following paper:\nPace, R. Kelley, and Ronald Barry. \"Sparse spatial autoregressions.\" Statistics & Probability Letters 33.3 (1997): 291-297.\n\nand I encountered it in 'Hands-On Machine learning with Scikit-Learn and TensorFlow' by Aur\u00e9lien G\u00e9ron.\nAur\u00e9lien G\u00e9ron wrote:\nThis dataset is a modified version of the California Housing dataset available from:\n[Lu\u00eds Torgo's page](http://www.dcc.fc.up.pt/~ltorgo/Regression/cal_housing.html) (University of Porto)\n\n### Inspiration\n\nSee my kernel on machine learning basics in R using this dataset, or venture over to the following link for a python based introductory tutorial: https://github.com/ageron/handson-ml/tree/master/datasets/housing", "VersionNotes": "Initial release", "TotalCompressedBytes": 409342.0, "TotalUncompressedBytes": 409342.0}]
|
[{"Id": 5227, "CreatorUserId": 710779, "OwnerUserId": 710779.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 7876.0, "CurrentDatasourceVersionId": 7876.0, "ForumId": 11315, "Type": 2, "CreationDate": "11/24/2017 03:14:59", "LastActivityDate": "02/06/2018", "TotalViews": 574173, "TotalDownloads": 108797, "TotalVotes": 970, "TotalKernels": 690}]
|
[{"Id": 710779, "UserName": "camnugent", "DisplayName": "Cam Nugent", "RegisterDate": "09/10/2016", "PerformanceTier": 2}]
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score, r2_score
from sklearn.ensemble import RandomForestRegressor
df = pd.read_csv("/kaggle/input/california-housing-prices/housing.csv")
df
df.info()
df["total_bedrooms"].mean()
df["total_bedrooms"] = df["total_bedrooms"].fillna(df["total_bedrooms"].mean())
df.isna().sum()
df = pd.get_dummies(df)
df.info()
x = df.drop("median_house_value", axis=1)
y = df["median_house_value"]
xtrain, xtest, ytrain, ytest = train_test_split(x, y, test_size=0.2, random_state=42)
sc = StandardScaler()
xtrain = sc.fit_transform(xtrain)
xtest = sc.transform(xtest)
gnb = GaussianNB()
gnb.fit(xtrain, ytrain)
res = gnb.predict(xtest)
accuracy_score(ytest, res)
rfr = RandomForestRegressor()
rfr.fit(xtrain, ytrain)
res = rfr.predict(xtest)
r2_score(ytest, res)
|
[{"california-housing-prices/housing.csv": {"column_names": "[\"longitude\", \"latitude\", \"housing_median_age\", \"total_rooms\", \"total_bedrooms\", \"population\", \"households\", \"median_income\", \"median_house_value\", \"ocean_proximity\"]", "column_data_types": "{\"longitude\": \"float64\", \"latitude\": \"float64\", \"housing_median_age\": \"float64\", \"total_rooms\": \"float64\", \"total_bedrooms\": \"float64\", \"population\": \"float64\", \"households\": \"float64\", \"median_income\": \"float64\", \"median_house_value\": \"float64\", \"ocean_proximity\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 20640 entries, 0 to 20639\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 longitude 20640 non-null float64\n 1 latitude 20640 non-null float64\n 2 housing_median_age 20640 non-null float64\n 3 total_rooms 20640 non-null float64\n 4 total_bedrooms 20433 non-null float64\n 5 population 20640 non-null float64\n 6 households 20640 non-null float64\n 7 median_income 20640 non-null float64\n 8 median_house_value 20640 non-null float64\n 9 ocean_proximity 20640 non-null object \ndtypes: float64(9), object(1)\nmemory usage: 1.6+ MB\n", "summary": "{\"longitude\": {\"count\": 20640.0, \"mean\": -119.56970445736432, \"std\": 2.0035317235025882, \"min\": -124.35, \"25%\": -121.8, \"50%\": -118.49, \"75%\": -118.01, \"max\": -114.31}, \"latitude\": {\"count\": 20640.0, \"mean\": 35.63186143410853, \"std\": 2.1359523974571153, \"min\": 32.54, \"25%\": 33.93, \"50%\": 34.26, \"75%\": 37.71, \"max\": 41.95}, \"housing_median_age\": {\"count\": 20640.0, \"mean\": 28.639486434108527, \"std\": 12.58555761211165, \"min\": 1.0, \"25%\": 18.0, \"50%\": 29.0, \"75%\": 37.0, \"max\": 52.0}, \"total_rooms\": {\"count\": 20640.0, \"mean\": 2635.7630813953488, \"std\": 2181.615251582795, \"min\": 2.0, \"25%\": 1447.75, \"50%\": 2127.0, \"75%\": 3148.0, \"max\": 39320.0}, \"total_bedrooms\": {\"count\": 20433.0, \"mean\": 537.8705525375618, \"std\": 421.3850700740323, \"min\": 1.0, \"25%\": 296.0, \"50%\": 435.0, \"75%\": 647.0, \"max\": 6445.0}, \"population\": {\"count\": 20640.0, \"mean\": 1425.4767441860465, \"std\": 1132.462121765341, \"min\": 3.0, \"25%\": 787.0, \"50%\": 1166.0, \"75%\": 1725.0, \"max\": 35682.0}, \"households\": {\"count\": 20640.0, \"mean\": 499.5396802325581, \"std\": 382.32975283161073, \"min\": 1.0, \"25%\": 280.0, \"50%\": 409.0, \"75%\": 605.0, \"max\": 6082.0}, \"median_income\": {\"count\": 20640.0, \"mean\": 3.8706710029069766, \"std\": 1.8998217179452688, \"min\": 0.4999, \"25%\": 2.5633999999999997, \"50%\": 3.5347999999999997, \"75%\": 4.74325, \"max\": 15.0001}, \"median_house_value\": {\"count\": 20640.0, \"mean\": 206855.81690891474, \"std\": 115395.61587441387, \"min\": 14999.0, \"25%\": 119600.0, \"50%\": 179700.0, \"75%\": 264725.0, \"max\": 500001.0}}", "examples": "{\"longitude\":{\"0\":-122.23,\"1\":-122.22,\"2\":-122.24,\"3\":-122.25},\"latitude\":{\"0\":37.88,\"1\":37.86,\"2\":37.85,\"3\":37.85},\"housing_median_age\":{\"0\":41.0,\"1\":21.0,\"2\":52.0,\"3\":52.0},\"total_rooms\":{\"0\":880.0,\"1\":7099.0,\"2\":1467.0,\"3\":1274.0},\"total_bedrooms\":{\"0\":129.0,\"1\":1106.0,\"2\":190.0,\"3\":235.0},\"population\":{\"0\":322.0,\"1\":2401.0,\"2\":496.0,\"3\":558.0},\"households\":{\"0\":126.0,\"1\":1138.0,\"2\":177.0,\"3\":219.0},\"median_income\":{\"0\":8.3252,\"1\":8.3014,\"2\":7.2574,\"3\":5.6431},\"median_house_value\":{\"0\":452600.0,\"1\":358500.0,\"2\":352100.0,\"3\":341300.0},\"ocean_proximity\":{\"0\":\"NEAR BAY\",\"1\":\"NEAR BAY\",\"2\":\"NEAR BAY\",\"3\":\"NEAR BAY\"}}"}}]
| true | 1 |
<start_data_description><data_path>california-housing-prices/housing.csv:
<column_names>
['longitude', 'latitude', 'housing_median_age', 'total_rooms', 'total_bedrooms', 'population', 'households', 'median_income', 'median_house_value', 'ocean_proximity']
<column_types>
{'longitude': 'float64', 'latitude': 'float64', 'housing_median_age': 'float64', 'total_rooms': 'float64', 'total_bedrooms': 'float64', 'population': 'float64', 'households': 'float64', 'median_income': 'float64', 'median_house_value': 'float64', 'ocean_proximity': 'object'}
<dataframe_Summary>
{'longitude': {'count': 20640.0, 'mean': -119.56970445736432, 'std': 2.0035317235025882, 'min': -124.35, '25%': -121.8, '50%': -118.49, '75%': -118.01, 'max': -114.31}, 'latitude': {'count': 20640.0, 'mean': 35.63186143410853, 'std': 2.1359523974571153, 'min': 32.54, '25%': 33.93, '50%': 34.26, '75%': 37.71, 'max': 41.95}, 'housing_median_age': {'count': 20640.0, 'mean': 28.639486434108527, 'std': 12.58555761211165, 'min': 1.0, '25%': 18.0, '50%': 29.0, '75%': 37.0, 'max': 52.0}, 'total_rooms': {'count': 20640.0, 'mean': 2635.7630813953488, 'std': 2181.615251582795, 'min': 2.0, '25%': 1447.75, '50%': 2127.0, '75%': 3148.0, 'max': 39320.0}, 'total_bedrooms': {'count': 20433.0, 'mean': 537.8705525375618, 'std': 421.3850700740323, 'min': 1.0, '25%': 296.0, '50%': 435.0, '75%': 647.0, 'max': 6445.0}, 'population': {'count': 20640.0, 'mean': 1425.4767441860465, 'std': 1132.462121765341, 'min': 3.0, '25%': 787.0, '50%': 1166.0, '75%': 1725.0, 'max': 35682.0}, 'households': {'count': 20640.0, 'mean': 499.5396802325581, 'std': 382.32975283161073, 'min': 1.0, '25%': 280.0, '50%': 409.0, '75%': 605.0, 'max': 6082.0}, 'median_income': {'count': 20640.0, 'mean': 3.8706710029069766, 'std': 1.8998217179452688, 'min': 0.4999, '25%': 2.5633999999999997, '50%': 3.5347999999999997, '75%': 4.74325, 'max': 15.0001}, 'median_house_value': {'count': 20640.0, 'mean': 206855.81690891474, 'std': 115395.61587441387, 'min': 14999.0, '25%': 119600.0, '50%': 179700.0, '75%': 264725.0, 'max': 500001.0}}
<dataframe_info>
RangeIndex: 20640 entries, 0 to 20639
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 longitude 20640 non-null float64
1 latitude 20640 non-null float64
2 housing_median_age 20640 non-null float64
3 total_rooms 20640 non-null float64
4 total_bedrooms 20433 non-null float64
5 population 20640 non-null float64
6 households 20640 non-null float64
7 median_income 20640 non-null float64
8 median_house_value 20640 non-null float64
9 ocean_proximity 20640 non-null object
dtypes: float64(9), object(1)
memory usage: 1.6+ MB
<some_examples>
{'longitude': {'0': -122.23, '1': -122.22, '2': -122.24, '3': -122.25}, 'latitude': {'0': 37.88, '1': 37.86, '2': 37.85, '3': 37.85}, 'housing_median_age': {'0': 41.0, '1': 21.0, '2': 52.0, '3': 52.0}, 'total_rooms': {'0': 880.0, '1': 7099.0, '2': 1467.0, '3': 1274.0}, 'total_bedrooms': {'0': 129.0, '1': 1106.0, '2': 190.0, '3': 235.0}, 'population': {'0': 322.0, '1': 2401.0, '2': 496.0, '3': 558.0}, 'households': {'0': 126.0, '1': 1138.0, '2': 177.0, '3': 219.0}, 'median_income': {'0': 8.3252, '1': 8.3014, '2': 7.2574, '3': 5.6431}, 'median_house_value': {'0': 452600.0, '1': 358500.0, '2': 352100.0, '3': 341300.0}, 'ocean_proximity': {'0': 'NEAR BAY', '1': 'NEAR BAY', '2': 'NEAR BAY', '3': 'NEAR BAY'}}
<end_description>
| 347 | 0 | 1,464 | 347 |
129657306
|
# # Blueberry yield
# # Importing Libraries
import pandas as pd
import numpy as np
import seaborn as sns
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import matplotlib.pyplot as plt
import plotly.express as px
# # Loading Training Dataset
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
train.head(15)
train = train.drop(columns=["id"])
train.describe()
train.info()
sns.heatmap(train.iloc[:, :].corr())
final_train = train.drop(
columns=[
"RainingDays",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
]
)
final_train
x = train.iloc[:, :16]
y = train["yield"]
model = tf.keras.Sequential([layers.Dense(17), layers.Dense(8), layers.Dense(1)])
model.compile(optimizer="adam", loss="msle", metrics=["msle"])
train_model = model.fit(x, y, batch_size=64, epochs=400, verbose=0)
history_df = pd.DataFrame(train_model.history)
history_df.loc[:, ["msle"]].plot()
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
test
fin_test = test.drop(
columns=[
"id",
"RainingDays",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
]
)
fin_test.info()
pred = model.predict(test.iloc[:, 1:])
pred
from sklearn.metrics import r2_score as r2
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
val = model.predict(x_test)
val
y_test
r2(val, y_test)
predf = pd.DataFrame(pred, index=test["id"])
predf
predf.to_csv("submission.csv")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/657/129657306.ipynb
| null | null |
[{"Id": 129657306, "ScriptId": 38404280, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11026853, "CreationDate": "05/15/2023 14:23:45", "VersionNumber": 2.0, "Title": "playground series prediction(blueberry)", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 71.0, "LinesInsertedFromPrevious": 15.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 56.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Blueberry yield
# # Importing Libraries
import pandas as pd
import numpy as np
import seaborn as sns
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import matplotlib.pyplot as plt
import plotly.express as px
# # Loading Training Dataset
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
train.head(15)
train = train.drop(columns=["id"])
train.describe()
train.info()
sns.heatmap(train.iloc[:, :].corr())
final_train = train.drop(
columns=[
"RainingDays",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
]
)
final_train
x = train.iloc[:, :16]
y = train["yield"]
model = tf.keras.Sequential([layers.Dense(17), layers.Dense(8), layers.Dense(1)])
model.compile(optimizer="adam", loss="msle", metrics=["msle"])
train_model = model.fit(x, y, batch_size=64, epochs=400, verbose=0)
history_df = pd.DataFrame(train_model.history)
history_df.loc[:, ["msle"]].plot()
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
test
fin_test = test.drop(
columns=[
"id",
"RainingDays",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
]
)
fin_test.info()
pred = model.predict(test.iloc[:, 1:])
pred
from sklearn.metrics import r2_score as r2
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
val = model.predict(x_test)
val
y_test
r2(val, y_test)
predf = pd.DataFrame(pred, index=test["id"])
predf
predf.to_csv("submission.csv")
| false | 0 | 549 | 0 | 549 | 549 |
||
129657706
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# # Victoria Quarterly Rent Report Analysis
# *This is a modified notebook from one of my lighter Master of Data Science assignment where we are tasked to tell a data story from an open-source dataset. The selected dataset is [Rental Report - Quarterly: Moving Annual Rents by Suburb](https://discover.data.vic.gov.au/dataset/rental-report-quarterly-moving-annual-rents-by-suburb) prepared by Homes Victoria.*
# *The dataset contain median rent for each suburb and region in Victoria, Australia every quarter from the year 2000 to year 2022. The dataset is then cleaned (in Microsoft Excel) and edited into two files: `count.csv` and `median.csv` which stores number of rent record and its median rent for each suburb respectively.*
# *This notebook will mainly focus on `count.csv` and aim to visualise the region with largest median rent growth rate. It is split into 2 sections:*
# * Section 1 contain the data processing part as well as sample EDA.
# * Section 2 visualise the rent growth rate.
# ## Section 1 - Data Processing and Exploratory Data Analysis
# Load data
# count.csv and median.csv contain data extracted from
# https://discover.data.vic.gov.au/dataset/rental-report-quarterly-moving-annual-rents-by-suburb
count = pd.read_csv("/kaggle/input/victoria-qtr-rent-report-20-22/count.csv")
med = pd.read_csv("/kaggle/input/victoria-qtr-rent-report-20-22/median.csv")
# Show count - currently in wide format
count.head()
# Show median - also in wide format
med.head()
# Since our dataset is currently in wide format (with median and count for each time period recorded as its own column), we want to convert it to long format to help with aggregation and analysis. We'll create a custom function for this task.
def to_long(df, col_name):
"Convert data from wide to long format."
ids = ["Suburb", "Region"]
df = df.melt(id_vars=ids, var_name="Date", value_name=col_name).reset_index(
drop=True
)
return df
# Convert both dataframe
count_long = to_long(count, "Count")
med_long = to_long(med, "Median")
# Merge both count and med into one dataframe
df = pd.merge(count_long, med_long, on=["Suburb", "Region", "Date"])
df["Date"].unique() # There are month called 'Dec 2003.1' that must be cleaned.
# Clean the data
df = pd.merge(count_long, med_long, on=["Suburb", "Region", "Date"])
df["Median"] = df["Median"].str.replace(r"\D", "") # Remove $ sign from Median
df.loc[df["Date"] == "Dec 2003.1", "Date"] = "Dec 2003" # Clean the date
# Convert 'Date' into datetime format
df["Date"] = pd.to_datetime(df["Date"], format="%b %Y")
# Extract month and year
df["Month"] = df["Date"].dt.month
df["Year"] = df["Date"].dt.year
# Change data type
df["Median"] = pd.to_numeric(df["Median"], errors="coerce")
df["Count"] = pd.to_numeric(df["Count"], errors="coerce")
df.head()
# ## Section 2 - Data exploration and visualisation
# This section aim to visualise our cleaned dataset using `seaborn` library to represent multiple insights that can be derived from our dataset. The visualised plots (in order of appearance) include:
# * Aggregated median rent throughout dataset period
# * Median rent growth throughout dataset period
# * Aggregated median rent throughout dataset period - with error bar
# * Distribution of rent growth rate for each suburb
# * Rent growth rate change throughout dataset period
# Bar graph show that Inner Melbourne has highest average median rent.
mean_region = (
df.groupby(["Region"])
.agg({"Count": "mean", "Median": "mean"})
.sort_values(by="Median", ascending=False)
.reset_index()
)
mean_region = mean_region.sort_values(by="Median", ascending=False).reset_index(
drop=True
)
fig, axs = plt.subplots(figsize=(10, 8))
sns.barplot(data=mean_region, x="Region", y="Median")
# set wrapping for y-labels
new_labels = ["\n".join(label._text.split()) for label in axs.get_xticklabels()]
axs.set_xticklabels(new_labels, rotation=40, ha="right", fontsize=8)
plt.title("Mean Median Melbourne Rent by Region from 2000 to 2020")
plt.show()
# The plot above shows that, throughout the dataset period, **Inner Melbourne** is the region with highest median rent, which makes sense as it is the central business district of the city with high demand and low supply.
# Line graph show that all Victoria region's rent increase at the same rate over time.
mean_date = (
df.groupby(["Date", "Region"])
.agg({"Count": "mean", "Median": "mean"})
.sort_values(by="Date", ascending=True)
.reset_index()
)
fig, axs = plt.subplots(figsize=(10, 8))
sns.lineplot(data=mean_date, x="Date", y="Median", hue="Region")
plt.title("Mean Median Melbourne Rent by Region from 2000 to 2020")
plt.show()
# Create new dataframe to look at rent growth
rent_growth = df.copy()
# Calculate percentage change of median rent for each Suburb
rent_growth["rent_growth"] = rent_growth.groupby(["Suburb", "Region"])[
"Median"
].pct_change(periods=1)
# New dataframe stores mean rent growth of suburbs in each region for each period of observation
group_rent_growth = (
rent_growth.groupby(["Date", "Region"])
.agg({"rent_growth": "mean"})
.sort_values(by="rent_growth", ascending=False)
.reset_index()
)
group_rent_growth.head()
# Statistic matrix for region with highest rent growth
group_rent_rank = (
group_rent_growth.groupby(["Region"])
.agg({"rent_growth": ["mean", "std"]})
.sort_values(by=("rent_growth", "mean"), ascending=False)
.reset_index()
)
group_rent_rank
# Extract mean from nested column then drop it
group_rent_rank["mean"] = group_rent_rank["rent_growth"]["mean"]
group_rent_rank["sd"] = group_rent_rank["rent_growth"]["std"]
group_rent_rank.drop(columns=["rent_growth"], inplace=True)
# Plot into bar graph with error bar
fig, axs = plt.subplots(figsize=(10, 8))
sns.barplot(data=group_rent_rank, x="Region", y="mean")
# Add error bar
axs.errorbar(
x=group_rent_rank["Region"],
y=group_rent_rank["mean"],
yerr=group_rent_rank["sd"],
fmt="none",
c="black",
capsize=5,
)
# Add line at y = 0 to not make the bar plot awkward
axs.axhline(y=0, color="gray", linestyle="-")
# set wrapping for y-labels
new_labels = ["\n".join(label._text.split()) for label in axs.get_xticklabels()]
axs.set_xticklabels(new_labels, rotation=40, ha="right", fontsize=8)
plt.title("Rent growth rate - ranked by Region from 2000 to 2020")
plt.tight_layout()
plt.show()
# Create new dataset to plot each records
suburb_rent_growth = (
rent_growth.groupby(["Date", "Region", "Suburb"])
.agg({"rent_growth": "mean"})
.reset_index()
)
# drop observation where rent growth is NaN
suburb_rent_growth = suburb_rent_growth.dropna()
suburb_rent_growth.head()
# Plot using stripplot and violinplot to show mean and distribution of rent growth
fig, axs = plt.subplots(figsize=(8, 6))
sns.stripplot(
data=suburb_rent_growth,
x="Region",
y="rent_growth",
hue="Suburb",
jitter=0.2,
alpha=0.5,
legend=False,
)
sns.violinplot(data=suburb_rent_growth, x="Region", y="rent_growth")
# Add mean annotation for each region
for region in suburb_rent_growth["Region"].unique():
mean = suburb_rent_growth[suburb_rent_growth["Region"] == region][
"rent_growth"
].mean()
axs.annotate(f"{mean:.2%}", xy=(region, mean), ha="center", va="center")
# set wrapping for y-labels
new_labels = ["\n".join(label._text.split()) for label in axs.get_xticklabels()]
axs.set_xticklabels(new_labels, rotation=40, ha="right", fontsize=8)
plt.title("Rent growth rate - ranked by Region from 2000 to 2022")
plt.tight_layout()
plt.show()
# Bonus vis: rent growth rate over time
fig, axs = plt.subplots(figsize=(10, 8))
sns.lineplot(data=group_rent_growth, x="Date", y="rent_growth", hue="Region")
plt.title("Melbourne Rent Growth Rate by Region from 2000 to 2020")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/657/129657706.ipynb
| null | null |
[{"Id": 129657706, "ScriptId": 38554091, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9498871, "CreationDate": "05/15/2023 14:26:23", "VersionNumber": 1.0, "Title": "DEMO - Victoria Quarterly Rent Report Analysis", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 170.0, "LinesInsertedFromPrevious": 170.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# # Victoria Quarterly Rent Report Analysis
# *This is a modified notebook from one of my lighter Master of Data Science assignment where we are tasked to tell a data story from an open-source dataset. The selected dataset is [Rental Report - Quarterly: Moving Annual Rents by Suburb](https://discover.data.vic.gov.au/dataset/rental-report-quarterly-moving-annual-rents-by-suburb) prepared by Homes Victoria.*
# *The dataset contain median rent for each suburb and region in Victoria, Australia every quarter from the year 2000 to year 2022. The dataset is then cleaned (in Microsoft Excel) and edited into two files: `count.csv` and `median.csv` which stores number of rent record and its median rent for each suburb respectively.*
# *This notebook will mainly focus on `count.csv` and aim to visualise the region with largest median rent growth rate. It is split into 2 sections:*
# * Section 1 contain the data processing part as well as sample EDA.
# * Section 2 visualise the rent growth rate.
# ## Section 1 - Data Processing and Exploratory Data Analysis
# Load data
# count.csv and median.csv contain data extracted from
# https://discover.data.vic.gov.au/dataset/rental-report-quarterly-moving-annual-rents-by-suburb
count = pd.read_csv("/kaggle/input/victoria-qtr-rent-report-20-22/count.csv")
med = pd.read_csv("/kaggle/input/victoria-qtr-rent-report-20-22/median.csv")
# Show count - currently in wide format
count.head()
# Show median - also in wide format
med.head()
# Since our dataset is currently in wide format (with median and count for each time period recorded as its own column), we want to convert it to long format to help with aggregation and analysis. We'll create a custom function for this task.
def to_long(df, col_name):
"Convert data from wide to long format."
ids = ["Suburb", "Region"]
df = df.melt(id_vars=ids, var_name="Date", value_name=col_name).reset_index(
drop=True
)
return df
# Convert both dataframe
count_long = to_long(count, "Count")
med_long = to_long(med, "Median")
# Merge both count and med into one dataframe
df = pd.merge(count_long, med_long, on=["Suburb", "Region", "Date"])
df["Date"].unique() # There are month called 'Dec 2003.1' that must be cleaned.
# Clean the data
df = pd.merge(count_long, med_long, on=["Suburb", "Region", "Date"])
df["Median"] = df["Median"].str.replace(r"\D", "") # Remove $ sign from Median
df.loc[df["Date"] == "Dec 2003.1", "Date"] = "Dec 2003" # Clean the date
# Convert 'Date' into datetime format
df["Date"] = pd.to_datetime(df["Date"], format="%b %Y")
# Extract month and year
df["Month"] = df["Date"].dt.month
df["Year"] = df["Date"].dt.year
# Change data type
df["Median"] = pd.to_numeric(df["Median"], errors="coerce")
df["Count"] = pd.to_numeric(df["Count"], errors="coerce")
df.head()
# ## Section 2 - Data exploration and visualisation
# This section aim to visualise our cleaned dataset using `seaborn` library to represent multiple insights that can be derived from our dataset. The visualised plots (in order of appearance) include:
# * Aggregated median rent throughout dataset period
# * Median rent growth throughout dataset period
# * Aggregated median rent throughout dataset period - with error bar
# * Distribution of rent growth rate for each suburb
# * Rent growth rate change throughout dataset period
# Bar graph show that Inner Melbourne has highest average median rent.
mean_region = (
df.groupby(["Region"])
.agg({"Count": "mean", "Median": "mean"})
.sort_values(by="Median", ascending=False)
.reset_index()
)
mean_region = mean_region.sort_values(by="Median", ascending=False).reset_index(
drop=True
)
fig, axs = plt.subplots(figsize=(10, 8))
sns.barplot(data=mean_region, x="Region", y="Median")
# set wrapping for y-labels
new_labels = ["\n".join(label._text.split()) for label in axs.get_xticklabels()]
axs.set_xticklabels(new_labels, rotation=40, ha="right", fontsize=8)
plt.title("Mean Median Melbourne Rent by Region from 2000 to 2020")
plt.show()
# The plot above shows that, throughout the dataset period, **Inner Melbourne** is the region with highest median rent, which makes sense as it is the central business district of the city with high demand and low supply.
# Line graph show that all Victoria region's rent increase at the same rate over time.
mean_date = (
df.groupby(["Date", "Region"])
.agg({"Count": "mean", "Median": "mean"})
.sort_values(by="Date", ascending=True)
.reset_index()
)
fig, axs = plt.subplots(figsize=(10, 8))
sns.lineplot(data=mean_date, x="Date", y="Median", hue="Region")
plt.title("Mean Median Melbourne Rent by Region from 2000 to 2020")
plt.show()
# Create new dataframe to look at rent growth
rent_growth = df.copy()
# Calculate percentage change of median rent for each Suburb
rent_growth["rent_growth"] = rent_growth.groupby(["Suburb", "Region"])[
"Median"
].pct_change(periods=1)
# New dataframe stores mean rent growth of suburbs in each region for each period of observation
group_rent_growth = (
rent_growth.groupby(["Date", "Region"])
.agg({"rent_growth": "mean"})
.sort_values(by="rent_growth", ascending=False)
.reset_index()
)
group_rent_growth.head()
# Statistic matrix for region with highest rent growth
group_rent_rank = (
group_rent_growth.groupby(["Region"])
.agg({"rent_growth": ["mean", "std"]})
.sort_values(by=("rent_growth", "mean"), ascending=False)
.reset_index()
)
group_rent_rank
# Extract mean from nested column then drop it
group_rent_rank["mean"] = group_rent_rank["rent_growth"]["mean"]
group_rent_rank["sd"] = group_rent_rank["rent_growth"]["std"]
group_rent_rank.drop(columns=["rent_growth"], inplace=True)
# Plot into bar graph with error bar
fig, axs = plt.subplots(figsize=(10, 8))
sns.barplot(data=group_rent_rank, x="Region", y="mean")
# Add error bar
axs.errorbar(
x=group_rent_rank["Region"],
y=group_rent_rank["mean"],
yerr=group_rent_rank["sd"],
fmt="none",
c="black",
capsize=5,
)
# Add line at y = 0 to not make the bar plot awkward
axs.axhline(y=0, color="gray", linestyle="-")
# set wrapping for y-labels
new_labels = ["\n".join(label._text.split()) for label in axs.get_xticklabels()]
axs.set_xticklabels(new_labels, rotation=40, ha="right", fontsize=8)
plt.title("Rent growth rate - ranked by Region from 2000 to 2020")
plt.tight_layout()
plt.show()
# Create new dataset to plot each records
suburb_rent_growth = (
rent_growth.groupby(["Date", "Region", "Suburb"])
.agg({"rent_growth": "mean"})
.reset_index()
)
# drop observation where rent growth is NaN
suburb_rent_growth = suburb_rent_growth.dropna()
suburb_rent_growth.head()
# Plot using stripplot and violinplot to show mean and distribution of rent growth
fig, axs = plt.subplots(figsize=(8, 6))
sns.stripplot(
data=suburb_rent_growth,
x="Region",
y="rent_growth",
hue="Suburb",
jitter=0.2,
alpha=0.5,
legend=False,
)
sns.violinplot(data=suburb_rent_growth, x="Region", y="rent_growth")
# Add mean annotation for each region
for region in suburb_rent_growth["Region"].unique():
mean = suburb_rent_growth[suburb_rent_growth["Region"] == region][
"rent_growth"
].mean()
axs.annotate(f"{mean:.2%}", xy=(region, mean), ha="center", va="center")
# set wrapping for y-labels
new_labels = ["\n".join(label._text.split()) for label in axs.get_xticklabels()]
axs.set_xticklabels(new_labels, rotation=40, ha="right", fontsize=8)
plt.title("Rent growth rate - ranked by Region from 2000 to 2022")
plt.tight_layout()
plt.show()
# Bonus vis: rent growth rate over time
fig, axs = plt.subplots(figsize=(10, 8))
sns.lineplot(data=group_rent_growth, x="Date", y="rent_growth", hue="Region")
plt.title("Melbourne Rent Growth Rate by Region from 2000 to 2020")
plt.show()
| false | 0 | 2,449 | 0 | 2,449 | 2,449 |
||
129750439
|
# # Internet Movies Database
# ## This notebook will present a recommendation system to recommend movies for the user
# #### IMDB (Internet Movie Database) is one of the largest online databases for movies and television shows, providing comprehensive information about movies, including ratings and reviews from its vast user base. The IMDB ratings are widely used as a benchmark for the popularity and success of movies.
# #### Column Description: rank - Rank of the movie, name - Name of the movie, year - Release year, rating - Rating of the movie, genre - Genre of the movie, certificate - Certificate of the movie, run_time -Total movie run time, tagline - Tagline of the movie, budget - Budget of the movie, box_office - Total box office collection across the world, casts - All casts of the movie, directors - Director of the movie, writers - Writer of the movie
# ### Importing all needed libraries in the project:
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.preprocessing import normalize
from sklearn.feature_extraction.text import CountVectorizer
import warnings
warnings.filterwarnings("ignore")
# ### Import the dataset, print the head of it to know the columns and print the shape of it
data = pd.read_csv(
r"/kaggle/input/the-internet-movies-database-imdb/IMDB Top 250 Movies.csv"
)
print("data shape: ", data.shape)
data.head()
# ### show the type of each column in my dataset
print(data.dtypes)
# ### display the pairplot to show the relations between columns, and the correlation to show if there are columns that are highly correlated
sns.pairplot(data)
plt.show()
corr = data.corr()
sns.heatmap(corr, annot=True, fmt=".2f", cmap="YlGnBu")
plt.show()
# ### Test if the dataset includes null, duplicate or not available values
#
nullValue = data.isnull().sum()
print(nullValue)
# Test if there is duplicated data
duplicateValue = data.duplicated().sum()
print(duplicateValue)
na_counts = data.apply(lambda x: x.value_counts().get("Not Available", 0))
print(na_counts)
# ### Creating a histogram of ratings from a dataset
#
plt.hist(data["rating"], bins=20)
plt.xlabel("Ratings")
plt.ylabel("Frequency")
plt.title("Distribution of Movie Rating")
plt.show()
# ### Generates a pie chart to visualize the distribution of movies released in different decades.
#
# Create a new column with the decade
data["decade"] = (data["year"] // 10) * 10
# Group the movies by decade and count the number of movies
movies_by_decade = data.groupby("decade")["name"].count()
# Create a pie chart
plt.pie(movies_by_decade, labels=movies_by_decade.index.astype(str), autopct="%1.1f%%")
plt.title("Movies Released by Decade")
plt.show()
# ### Generates a line plot to visualize the trend of the number of movies released each year.
# Get the count of movies released each year
year_counts = data.groupby("year")["name"].count()
# Create the line plot
sns.set(style="darkgrid")
plt.plot(year_counts.index, year_counts.values)
plt.xlabel("Year")
plt.ylabel("Number of movies released")
plt.title("Number of movies released each year")
plt.show()
# ### Generates a bar chart to visualize the top 10 highest-grossing movies
# Filter out the 'Not Available' box office values and sort by descending order
top_grossing = (
data[data["box_office"] != "Not Available"]
.sort_values(by="box_office", ascending=False)
.head(10)
)
# Create a list of 10 different colors
colors = [
"#F44336",
"#E91E63",
"#9C27B0",
"#673AB7",
"#3F51B5",
"#2196F3",
"#00BCD4",
"#4CAF50",
"#8BC34A",
"#FFC107",
]
# Plot the bar chart
plt.bar(x=top_grossing["name"], height=top_grossing["box_office"], color=colors)
plt.xticks(rotation=90)
plt.xlabel("Movie Title")
plt.ylabel("Box Office ($)")
plt.title("Top 10 Highest-Grossing Movies")
plt.show()
# ### Generates a line plot to visualize the top 10 directors with the most number of movies that made it to the top 250 list.
plt.plot(
(data["directors"].value_counts().head(10)),
color="b",
marker="o",
markeredgecolor="r",
linestyle="-.",
)
# ploting the data using matplotlib library
plt.xlabel("Directors", c="black")
# labelling the data
plt.xticks(rotation=90)
plt.ylabel("No. of movies", c="black")
plt.title(
"Top 10 directors with most no. of movies that made it to the top 250",
color="black",
)
# providing a title to the plot
plt.show()
# ### Provided groups the movies in the data by genre, calculates the mean rating for each genre and displays the highest 7 movies.
top = data.groupby("genre").mean().sort_values("rating", ascending=False).head(7)
top = top[["rating"]]
top.reset_index(inplace=True)
top
# ### Provided groups the movies in the data by name, calculates the mean rating for each genre and displays the highest 7 movies.
top = data.groupby("name").mean().sort_values("rating", ascending=False).head(7)
top = top[["rating"]]
top.reset_index(inplace=True)
top
# # Recommendation system
# ### provided selects specific columns from the data DataFrame for recommendation and creates a new DataFrame
# Select the relevant columns for recommendation
selected_columns = ["name", "genre", "certificate", "directors", "writers"]
df_selected = data[selected_columns]
# Combine the relevant columns into a single string
df_selected["combined_features"] = df_selected.apply(lambda row: " ".join(row), axis=1)
df_selected.head()
# ### Create a count matrix of the combined features then count the cosine similarity matrix
# Create a count matrix of the combined features
count_matrix = CountVectorizer().fit_transform(df_selected["combined_features"])
# Compute the cosine similarity matrix
cosine_sim = cosine_similarity(count_matrix)
# content-based recommendations, where movies with similar features are suggested based on their similarity scores.
# ### Function to get recommendations for a given movie
def get_recommendations(movie_name, cosine_sim, data, top_n=10):
try:
movie_indices = data[data["name"] == movie_name].index
if movie_indices.empty:
print(f"Movie '{movie_name}' not found in the dataset.")
return []
movie_index = movie_indices[0]
except IndexError:
print(f"Movie '{movie_name}' not found in the dataset.")
return []
similarity_scores = list(enumerate(cosine_sim[movie_index]))
similarity_scores = sorted(similarity_scores, key=lambda x: x[1], reverse=True)
top_similar_movies = similarity_scores[1 : top_n + 1] # Exclude the movie itself
movie_indices = [index for index, _ in top_similar_movies]
recommended_movies = data.iloc[movie_indices]["name"]
return recommended_movies
# ### Here I am asking the user to enter a movie name to give him/her a recommendation movies
# Ask the user to enter a movie name
user_movie = input("Enter a movie name: ")
# Get recommendations for the entered movie
recommendations = get_recommendations(user_movie, cosine_sim, df_selected)
# Check if recommendations are available
if len(recommendations) > 0:
print("Recommended movies:")
for movie in recommendations:
print(movie)
else:
print("No recommendations found for the entered movie.")
# ### Here It is giving recommendations according to the index of the movie that the programmer can change
# Select a movie name from your dataset
movie_name = df_selected["name"].iloc[
0
] # Replace '0' with the index of the movie name you want to analyze
# Get recommendations for the selected movie
recommendations = get_recommendations(movie_name, cosine_sim, df_selected)
# Print the recommendations
if recommendations.empty:
print(f"No recommendations found for '{movie_name}'.")
else:
print(f"Recommended movies for '{movie_name}':")
print(recommendations)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/750/129750439.ipynb
| null | null |
[{"Id": 129750439, "ScriptId": 38338256, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10127643, "CreationDate": "05/16/2023 08:01:04", "VersionNumber": 1.0, "Title": "IMDB-Internet Movies Database", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 206.0, "LinesInsertedFromPrevious": 206.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
# # Internet Movies Database
# ## This notebook will present a recommendation system to recommend movies for the user
# #### IMDB (Internet Movie Database) is one of the largest online databases for movies and television shows, providing comprehensive information about movies, including ratings and reviews from its vast user base. The IMDB ratings are widely used as a benchmark for the popularity and success of movies.
# #### Column Description: rank - Rank of the movie, name - Name of the movie, year - Release year, rating - Rating of the movie, genre - Genre of the movie, certificate - Certificate of the movie, run_time -Total movie run time, tagline - Tagline of the movie, budget - Budget of the movie, box_office - Total box office collection across the world, casts - All casts of the movie, directors - Director of the movie, writers - Writer of the movie
# ### Importing all needed libraries in the project:
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.preprocessing import normalize
from sklearn.feature_extraction.text import CountVectorizer
import warnings
warnings.filterwarnings("ignore")
# ### Import the dataset, print the head of it to know the columns and print the shape of it
data = pd.read_csv(
r"/kaggle/input/the-internet-movies-database-imdb/IMDB Top 250 Movies.csv"
)
print("data shape: ", data.shape)
data.head()
# ### show the type of each column in my dataset
print(data.dtypes)
# ### display the pairplot to show the relations between columns, and the correlation to show if there are columns that are highly correlated
sns.pairplot(data)
plt.show()
corr = data.corr()
sns.heatmap(corr, annot=True, fmt=".2f", cmap="YlGnBu")
plt.show()
# ### Test if the dataset includes null, duplicate or not available values
#
nullValue = data.isnull().sum()
print(nullValue)
# Test if there is duplicated data
duplicateValue = data.duplicated().sum()
print(duplicateValue)
na_counts = data.apply(lambda x: x.value_counts().get("Not Available", 0))
print(na_counts)
# ### Creating a histogram of ratings from a dataset
#
plt.hist(data["rating"], bins=20)
plt.xlabel("Ratings")
plt.ylabel("Frequency")
plt.title("Distribution of Movie Rating")
plt.show()
# ### Generates a pie chart to visualize the distribution of movies released in different decades.
#
# Create a new column with the decade
data["decade"] = (data["year"] // 10) * 10
# Group the movies by decade and count the number of movies
movies_by_decade = data.groupby("decade")["name"].count()
# Create a pie chart
plt.pie(movies_by_decade, labels=movies_by_decade.index.astype(str), autopct="%1.1f%%")
plt.title("Movies Released by Decade")
plt.show()
# ### Generates a line plot to visualize the trend of the number of movies released each year.
# Get the count of movies released each year
year_counts = data.groupby("year")["name"].count()
# Create the line plot
sns.set(style="darkgrid")
plt.plot(year_counts.index, year_counts.values)
plt.xlabel("Year")
plt.ylabel("Number of movies released")
plt.title("Number of movies released each year")
plt.show()
# ### Generates a bar chart to visualize the top 10 highest-grossing movies
# Filter out the 'Not Available' box office values and sort by descending order
top_grossing = (
data[data["box_office"] != "Not Available"]
.sort_values(by="box_office", ascending=False)
.head(10)
)
# Create a list of 10 different colors
colors = [
"#F44336",
"#E91E63",
"#9C27B0",
"#673AB7",
"#3F51B5",
"#2196F3",
"#00BCD4",
"#4CAF50",
"#8BC34A",
"#FFC107",
]
# Plot the bar chart
plt.bar(x=top_grossing["name"], height=top_grossing["box_office"], color=colors)
plt.xticks(rotation=90)
plt.xlabel("Movie Title")
plt.ylabel("Box Office ($)")
plt.title("Top 10 Highest-Grossing Movies")
plt.show()
# ### Generates a line plot to visualize the top 10 directors with the most number of movies that made it to the top 250 list.
plt.plot(
(data["directors"].value_counts().head(10)),
color="b",
marker="o",
markeredgecolor="r",
linestyle="-.",
)
# ploting the data using matplotlib library
plt.xlabel("Directors", c="black")
# labelling the data
plt.xticks(rotation=90)
plt.ylabel("No. of movies", c="black")
plt.title(
"Top 10 directors with most no. of movies that made it to the top 250",
color="black",
)
# providing a title to the plot
plt.show()
# ### Provided groups the movies in the data by genre, calculates the mean rating for each genre and displays the highest 7 movies.
top = data.groupby("genre").mean().sort_values("rating", ascending=False).head(7)
top = top[["rating"]]
top.reset_index(inplace=True)
top
# ### Provided groups the movies in the data by name, calculates the mean rating for each genre and displays the highest 7 movies.
top = data.groupby("name").mean().sort_values("rating", ascending=False).head(7)
top = top[["rating"]]
top.reset_index(inplace=True)
top
# # Recommendation system
# ### provided selects specific columns from the data DataFrame for recommendation and creates a new DataFrame
# Select the relevant columns for recommendation
selected_columns = ["name", "genre", "certificate", "directors", "writers"]
df_selected = data[selected_columns]
# Combine the relevant columns into a single string
df_selected["combined_features"] = df_selected.apply(lambda row: " ".join(row), axis=1)
df_selected.head()
# ### Create a count matrix of the combined features then count the cosine similarity matrix
# Create a count matrix of the combined features
count_matrix = CountVectorizer().fit_transform(df_selected["combined_features"])
# Compute the cosine similarity matrix
cosine_sim = cosine_similarity(count_matrix)
# content-based recommendations, where movies with similar features are suggested based on their similarity scores.
# ### Function to get recommendations for a given movie
def get_recommendations(movie_name, cosine_sim, data, top_n=10):
try:
movie_indices = data[data["name"] == movie_name].index
if movie_indices.empty:
print(f"Movie '{movie_name}' not found in the dataset.")
return []
movie_index = movie_indices[0]
except IndexError:
print(f"Movie '{movie_name}' not found in the dataset.")
return []
similarity_scores = list(enumerate(cosine_sim[movie_index]))
similarity_scores = sorted(similarity_scores, key=lambda x: x[1], reverse=True)
top_similar_movies = similarity_scores[1 : top_n + 1] # Exclude the movie itself
movie_indices = [index for index, _ in top_similar_movies]
recommended_movies = data.iloc[movie_indices]["name"]
return recommended_movies
# ### Here I am asking the user to enter a movie name to give him/her a recommendation movies
# Ask the user to enter a movie name
user_movie = input("Enter a movie name: ")
# Get recommendations for the entered movie
recommendations = get_recommendations(user_movie, cosine_sim, df_selected)
# Check if recommendations are available
if len(recommendations) > 0:
print("Recommended movies:")
for movie in recommendations:
print(movie)
else:
print("No recommendations found for the entered movie.")
# ### Here It is giving recommendations according to the index of the movie that the programmer can change
# Select a movie name from your dataset
movie_name = df_selected["name"].iloc[
0
] # Replace '0' with the index of the movie name you want to analyze
# Get recommendations for the selected movie
recommendations = get_recommendations(movie_name, cosine_sim, df_selected)
# Print the recommendations
if recommendations.empty:
print(f"No recommendations found for '{movie_name}'.")
else:
print(f"Recommended movies for '{movie_name}':")
print(recommendations)
| false | 0 | 2,186 | 1 | 2,186 | 2,186 |
||
129750558
|
<jupyter_start><jupyter_text>TESLA Stock Data
## What is TESLA?
Tesla, Inc. is an American electric vehicle and clean energy company based in Palo Alto, California. Tesla's current products include electric cars, battery energy storage from home to grid-scale, solar panels and solar roof tiles, as well as other related products and services.
## Information about this dataset
This dataset provides historical data of TESLA INC. stock (TSLA). The data is available at a daily level. Currency is USD.
Kaggle dataset identifier: tesla-stock-data-updated-till-28jun2021
<jupyter_script>import numpy as np
import pandas as pd
from statsmodels.tsa.stattools import adfuller
# from pandas.tools.plotting import autocorrelation_plot
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
from statsmodels.tsa.arima.model import ARIMA
import statsmodels.api as sm
from pandas.tseries.offsets import DateOffset
from pandas.plotting import autocorrelation_plot
import matplotlib.pyplot as plt
df = pd.read_csv("/kaggle/input/tesla-stock-data-updated-till-28jun2021/TSLA.csv")
df.head()
df = df[["Date", "Close"]]
df.head()
df.info()
df.shape
df["Date"] = pd.to_datetime(df["Date"])
df.info()
df.set_index("Date", inplace=True)
df.describe()
df.head()
# # Rolling İstatistikleri
# Dönen ortalamayı ve yuvarlanan standart sapmayı çizin. Zaman serisi zamanla sabit kalıyorsa durağandır
rolling_mean = df.rolling(window=12).mean()
rolling_std = df.rolling(window=12).std()
plt.plot(df, color="blue", label="Original")
plt.plot(rolling_mean, color="red", label="Rolling Mean")
plt.plot(rolling_std, color="black", label="Rolling Std")
plt.legend(loc="best")
plt.title("Rolling Mean & Rolling Standard Deviation")
plt.show()
# # Artırılmış Dickey-Fuller Testi
# p değeri düşükse (boş hipoteze göre) ve %1, %5, %10 güven aralığındaki kritik değerler ADF İstatistiklerine mümkün olduğunca yakınsa zaman serisi durağan kabul edilir.
test_result = adfuller(df["Close"])
# Ho: It is non stationary
# H1: It is stationary
def adfuller_test(sales):
result = adfuller(sales)
labels = [
"ADF Test Statistic",
"p-value",
"#Lags Used",
"Number of Observations Used",
]
for value, label in zip(result, labels):
print(label + " : " + str(value))
if result[1] <= 0.05:
print(
"strong evidence against the null hypothesis(Ho), reject the null hypothesis. Data has no unit root and is stationary"
)
else:
print(
"weak evidence against null hypothesis, time series has a unit root, indicating it is non-stationary "
)
adfuller_test(df["Close"])
# ADF İstatistiği kritik değerlerden uzak ve p değeri eşikten (0.05) daha büyük. Böylece zaman serisinin durağan olmadığı sonucuna varabiliriz.
# Durağan hale getirmek için Differencing kullanacağız:
df["Close First Difference"] = df["Close"] - df["Close"].shift(1)
df["Close"].shift(1)
df["Seasonal First Difference"] = df["Close"] - df["Close"].shift(12)
df.head(14)
# Differencing sonrası serinin durağan olup olmadığını test etmek için Artırılmış Dickey-Fuller Testini tekrar yapalım.
adfuller_test(df["Seasonal First Difference"].dropna())
df["Seasonal First Difference"].plot()
# Şimdi, p-değerinin 0.05'teki Anlamlılık seviyesinden daha az olduğunu görebiliriz. Böylece H0 Hipotezi Reddedilir ve zaman serisi Durağandır.
# ACF ve PACF
autocorrelation_plot(df["Close"])
plt.show()
fig = plt.figure(figsize=(12, 8))
ax1 = fig.add_subplot(211)
fig = sm.graphics.tsa.plot_acf(
df["Seasonal First Difference"].iloc[13:], lags=40, ax=ax1
)
ax2 = fig.add_subplot(212)
fig = sm.graphics.tsa.plot_pacf(
df["Seasonal First Difference"].iloc[13:], lags=40, ax=ax2
)
# Tahminler için ARIMA’yı uygulayalım
model = ARIMA(df["Close"], order=(1, 1, 1))
model_fit = model.fit()
model_fit.summary()
# ARIMA kullanarak Tahmini görelim
df["forecast"] = model_fit.predict(start=2400, end=2900, dynamic=True)
df[["Close", "forecast"]].plot(figsize=(12, 8))
# Burada, zaman serileri mevsimsellik gösterdiğinden, ARIMA kullanılarak tahminin iyi olmadığını görebiliriz.
# Şimdi Mevsimsel-ARIMA’yı uygulayacağız
# Mevsimsel-ARIMA(SARIMA):
# Adından da anlaşılacağı gibi, bu model zaman serileri mevsimsellik gösterdiğinde kullanılır. Bu model ARIMA modellerine benzer, sadece mevsimleri hesaba katmak için birkaç parametre eklememiz gerekiyor.
# ARIMA(p,d,q)(P, D, Q)m,
# - p — otoregresif sayısı
# - d — farklılık derecesi
# - q — hareketli ortalama terimlerinin sayısı
# - m — her sezondaki periyot sayısını ifade eder
# - (P, D, Q ) — zaman serisinin mevsimsel kısmı için (p,d,q) değerini temsil eder.
#
model = sm.tsa.statespace.SARIMAX(
df["Close"], order=(1, 1, 1), seasonal_order=(1, 1, 1, 12)
)
results = model.fit()
df["forecast"] = results.predict(start=2400, end=2900, dynamic=True)
df[["Close", "forecast"]].plot(figsize=(12, 8))
# # Second approach
df = pd.read_csv("/kaggle/input/tesla-stock-data-updated-till-28jun2021/TSLA.csv")
df.head()
df = df[["Date", "Close"]]
df["Date"] = pd.to_datetime(df["Date"])
df = df.set_index("Date")
df_data = df.groupby(pd.Grouper(freq="W")).mean()
df_data = df_data.rename(columns={"Close": "weekly_sales"})
df_data.plot()
plt.grid()
from statsmodels.tsa.api import VARMAX
from tqdm.notebook import tqdm
from sklearn.metrics import mean_squared_error
from dateutil.relativedelta import relativedelta
plt.rcParams["figure.figsize"] = (20, 7)
# Two stationary signals, weekly sales difference and standard deviation of the last 3 weekly sales, are used in VARMA
df_varma = df_data.copy()
df_varma["weekly_sales_diff"] = df_varma.diff()
df_varma["weekly_sales_rolling_std_3"] = df_varma["weekly_sales"].rolling(3).std()
df_varma = df_varma.dropna()
# predict last 50 values one by one
df_pred = pd.DataFrame()
for i in tqdm(range(50)):
training_data = df_varma[["weekly_sales_diff", "weekly_sales_rolling_std_3"]][
: (-50 + i)
]
# VARMA model selects lag p based on AIC
model = VARMAX(training_data)
model_fit = model.fit(ic="aic")
model_forecast = model_fit.get_forecast(steps=1)
df_pred_temp = pd.concat(
[model_forecast.conf_int(), model_forecast.predicted_mean], axis=1
)
df_pred_temp = df_pred_temp[
["lower weekly_sales_diff", "upper weekly_sales_diff", "weekly_sales_diff"]
]
df_pred = df_pred.append(df_pred_temp)
df_pred = df_pred.rename(
columns={
"lower weekly_sales_diff": "ci_lower",
"upper weekly_sales_diff": "ci_upper",
"weekly_sales_diff": "pred_weekly_sales_diff",
}
)
df_pred["weekly_sales_diff"] = df_varma["weekly_sales_diff"][-50:]
# transform data
df_pred["pred_weekly_sales"] = (
df_pred["pred_weekly_sales_diff"] + df_varma["weekly_sales"].shift()
)
df_pred["weekly_sales"] = (
df_pred["weekly_sales_diff"] + df_varma["weekly_sales"].shift()
)
df_pred["ci_lower"] = df_pred["ci_lower"] + df_varma["weekly_sales"].shift()
df_pred["ci_upper"] = df_pred["ci_upper"] + df_varma["weekly_sales"].shift()
plt.plot(df_pred["weekly_sales"], color="green")
plt.plot(df_pred["pred_weekly_sales"], color="red")
plt.legend(["Weekly sales", "Predicted weekly sales"])
plt.fill_between(
df_pred.index,
df_pred["ci_lower"],
df_pred["ci_upper"],
color="lightblue",
alpha=0.5,
)
plt.title(
"VARMA, MSE: {:,}".format(
round(
mean_squared_error(df_pred["weekly_sales"], df_pred["pred_weekly_sales"]), 2
)
)
)
plt.grid()
plt.show()
from statsmodels.tsa.statespace.varmax import VARMAX
from tqdm.notebook import tqdm
from sklearn.metrics import mean_squared_error
from dateutil.relativedelta import relativedelta
plt.rcParams["figure.figsize"] = (20, 7)
# Construct two stationary signals
df_varmax = df_data.copy()
df_varmax["weekly_sales_diff"] = df_varmax.diff()
df_varmax["weekly_sales_rolling_std_3"] = df_varmax["weekly_sales"].rolling(3).std()
df_varmax = df_varmax.dropna()
# prepare exogenous variable
exo_df = df_varmax["weekly_sales_diff"].to_frame()
exo_df["noise"] = [5000 * np.random.rand() for x in range(len(exo_df))]
exo_df["weekly_sales_diff_plus_noise"] = exo_df["weekly_sales_diff"] + exo_df["noise"]
exo_df["weekly_sales_diff_plus_noise"] = (
exo_df["weekly_sales_diff_plus_noise"].shift().fillna(0)
)
# predict last 50 values one by one
df_pred = pd.DataFrame()
for i in tqdm(range(50)):
training_data = df_varmax[["weekly_sales_diff", "weekly_sales_rolling_std_3"]][
: (-50 + i)
]
training_exo = exo_df[exo_df.index.isin(training_data.index)][
"weekly_sales_diff_plus_noise"
].to_frame()
# VARMAX model selects lag p based on AIC
model = VARMAX(training_data, exog=training_exo)
model_fit = model.fit(ic="aic")
pred_date = training_data.index[-1] + relativedelta(weeks=1)
model_forecast = model_fit.get_forecast(
steps=1,
exog=exo_df[exo_df.index == pred_date][
"weekly_sales_diff_plus_noise"
].to_frame(),
)
df_pred_temp = pd.concat(
[model_forecast.conf_int(), model_forecast.predicted_mean], axis=1
)
df_pred_temp = df_pred_temp[
["lower weekly_sales_diff", "upper weekly_sales_diff", "weekly_sales_diff"]
]
df_pred = df_pred.append(df_pred_temp)
df_pred = df_pred.rename(
columns={
"lower weekly_sales_diff": "ci_lower",
"upper weekly_sales_diff": "ci_upper",
"weekly_sales_diff": "pred_weekly_sales_diff",
}
)
df_pred["weekly_sales_diff"] = df_varmax["weekly_sales_diff"][-50:]
# transform data
df_pred["pred_weekly_sales"] = (
df_pred["pred_weekly_sales_diff"] + df_varmax["weekly_sales"].shift()
)
df_pred["weekly_sales"] = (
df_pred["weekly_sales_diff"] + df_varmax["weekly_sales"].shift()
)
df_pred["ci_lower"] = df_pred["ci_lower"] + df_varmax["weekly_sales"].shift()
df_pred["ci_upper"] = df_pred["ci_upper"] + df_varmax["weekly_sales"].shift()
plt.plot(df_pred["weekly_sales"], color="green")
plt.plot(df_pred["pred_weekly_sales"], color="red")
plt.legend(["Weekly sales", "Predicted weekly sales"])
plt.fill_between(
df_pred.index,
df_pred["ci_lower"],
df_pred["ci_upper"],
color="lightblue",
alpha=0.5,
)
plt.title(
"VARMAX, MSE: {:,}".format(
round(
mean_squared_error(df_pred["weekly_sales"], df_pred["pred_weekly_sales"]), 2
)
)
)
plt.grid()
plt.show()
df.info()
df.head()
df = df[["Date", "Close"]]
df.head()
df_data = df.groupby(["Date"])["Close"].mean()
df_data = df_data.rename(columns={"item_cnt_day": "weekly_sales"})
df_data
df_data = pd.DataFrame(df_data)
df_data = df_data.reset_index()
del df["index"]
df_data.Date = df_data.Date.astype("datetime64[ns]")
df.head()
# df['Date']=pd.to_datetime(df['Date'])
df.set_index("Date", inplace=True)
df.head()
# group data by weekly close
df_data = df.groupby(pd.Grouper(freq="W")).mean()
df_data = df_data.rename(columns={"item_cnt_day": "weekly_close"})
df_data.plot()
plt.grid()
df_data.head()
from pmdarima import auto_arima
from statsmodels.tsa.stattools import adfuller
from statsmodels.tools.eval_measures import mse, rmse
from statsmodels.tsa.statespace.varmax import VARMAX, VARMAXResults
# # Decide the order of the VARMA(p,q)
auto_arima(df["Close"], maxiter=100)
# Order (1,2) is preferred for VARMA. The last term or the third terms is the differencing which will be applied already using differencing.
df_transformed = df.diff().diff() # 2nd order difference
df_transformed = (
df_transformed.dropna()
) # remove the NaNs introduced due to differencing
df_transformed.head()
len(df_transformed)
# Train Test Split
nobs = 354 # The last 12 months will be the test data. At least 1 year would be a good choice
train = df_transformed[0:-nobs]
test = df_transformed[-nobs:]
train.shape
# Fit the VARMA(1,2) Model
model = VARMAX(train, order=(1, 2), trend="c") # c indicates a constant trend
results = model.fit(maxiter=1000, disp=False)
results.summary()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/750/129750558.ipynb
|
tesla-stock-data-updated-till-28jun2021
|
varpit94
|
[{"Id": 129750558, "ScriptId": 38580278, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4863236, "CreationDate": "05/16/2023 08:01:58", "VersionNumber": 5.0, "Title": "Tesla Stock Predict", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 339.0, "LinesInsertedFromPrevious": 213.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 126.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186103614, "KernelVersionId": 129750558, "SourceDatasetVersionId": 3358622}]
|
[{"Id": 3358622, "DatasetId": 1436765, "DatasourceVersionId": 3409892, "CreatorUserId": 1130103, "LicenseName": "Other (specified in description)", "CreationDate": "03/25/2022 16:24:30", "VersionNumber": 9.0, "Title": "TESLA Stock Data", "Slug": "tesla-stock-data-updated-till-28jun2021", "Subtitle": "TESLA Inc. (TSLA) | NasdaqGS Real Time Price | Currency in USD", "Description": "## What is TESLA?\nTesla, Inc. is an American electric vehicle and clean energy company based in Palo Alto, California. Tesla's current products include electric cars, battery energy storage from home to grid-scale, solar panels and solar roof tiles, as well as other related products and services. \n\n## Information about this dataset\nThis dataset provides historical data of TESLA INC. stock (TSLA). The data is available at a daily level. Currency is USD.", "VersionNotes": "Data Update 2022/03/25", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1436765, "CreatorUserId": 1130103, "OwnerUserId": 1130103.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3358622.0, "CurrentDatasourceVersionId": 3409892.0, "ForumId": 1456232, "Type": 2, "CreationDate": "06/28/2021 17:08:16", "LastActivityDate": "06/28/2021", "TotalViews": 22931, "TotalDownloads": 3633, "TotalVotes": 69, "TotalKernels": 19}]
|
[{"Id": 1130103, "UserName": "varpit94", "DisplayName": "Arpit Verma", "RegisterDate": "06/17/2017", "PerformanceTier": 2}]
|
import numpy as np
import pandas as pd
from statsmodels.tsa.stattools import adfuller
# from pandas.tools.plotting import autocorrelation_plot
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
from statsmodels.tsa.arima.model import ARIMA
import statsmodels.api as sm
from pandas.tseries.offsets import DateOffset
from pandas.plotting import autocorrelation_plot
import matplotlib.pyplot as plt
df = pd.read_csv("/kaggle/input/tesla-stock-data-updated-till-28jun2021/TSLA.csv")
df.head()
df = df[["Date", "Close"]]
df.head()
df.info()
df.shape
df["Date"] = pd.to_datetime(df["Date"])
df.info()
df.set_index("Date", inplace=True)
df.describe()
df.head()
# # Rolling İstatistikleri
# Dönen ortalamayı ve yuvarlanan standart sapmayı çizin. Zaman serisi zamanla sabit kalıyorsa durağandır
rolling_mean = df.rolling(window=12).mean()
rolling_std = df.rolling(window=12).std()
plt.plot(df, color="blue", label="Original")
plt.plot(rolling_mean, color="red", label="Rolling Mean")
plt.plot(rolling_std, color="black", label="Rolling Std")
plt.legend(loc="best")
plt.title("Rolling Mean & Rolling Standard Deviation")
plt.show()
# # Artırılmış Dickey-Fuller Testi
# p değeri düşükse (boş hipoteze göre) ve %1, %5, %10 güven aralığındaki kritik değerler ADF İstatistiklerine mümkün olduğunca yakınsa zaman serisi durağan kabul edilir.
test_result = adfuller(df["Close"])
# Ho: It is non stationary
# H1: It is stationary
def adfuller_test(sales):
result = adfuller(sales)
labels = [
"ADF Test Statistic",
"p-value",
"#Lags Used",
"Number of Observations Used",
]
for value, label in zip(result, labels):
print(label + " : " + str(value))
if result[1] <= 0.05:
print(
"strong evidence against the null hypothesis(Ho), reject the null hypothesis. Data has no unit root and is stationary"
)
else:
print(
"weak evidence against null hypothesis, time series has a unit root, indicating it is non-stationary "
)
adfuller_test(df["Close"])
# ADF İstatistiği kritik değerlerden uzak ve p değeri eşikten (0.05) daha büyük. Böylece zaman serisinin durağan olmadığı sonucuna varabiliriz.
# Durağan hale getirmek için Differencing kullanacağız:
df["Close First Difference"] = df["Close"] - df["Close"].shift(1)
df["Close"].shift(1)
df["Seasonal First Difference"] = df["Close"] - df["Close"].shift(12)
df.head(14)
# Differencing sonrası serinin durağan olup olmadığını test etmek için Artırılmış Dickey-Fuller Testini tekrar yapalım.
adfuller_test(df["Seasonal First Difference"].dropna())
df["Seasonal First Difference"].plot()
# Şimdi, p-değerinin 0.05'teki Anlamlılık seviyesinden daha az olduğunu görebiliriz. Böylece H0 Hipotezi Reddedilir ve zaman serisi Durağandır.
# ACF ve PACF
autocorrelation_plot(df["Close"])
plt.show()
fig = plt.figure(figsize=(12, 8))
ax1 = fig.add_subplot(211)
fig = sm.graphics.tsa.plot_acf(
df["Seasonal First Difference"].iloc[13:], lags=40, ax=ax1
)
ax2 = fig.add_subplot(212)
fig = sm.graphics.tsa.plot_pacf(
df["Seasonal First Difference"].iloc[13:], lags=40, ax=ax2
)
# Tahminler için ARIMA’yı uygulayalım
model = ARIMA(df["Close"], order=(1, 1, 1))
model_fit = model.fit()
model_fit.summary()
# ARIMA kullanarak Tahmini görelim
df["forecast"] = model_fit.predict(start=2400, end=2900, dynamic=True)
df[["Close", "forecast"]].plot(figsize=(12, 8))
# Burada, zaman serileri mevsimsellik gösterdiğinden, ARIMA kullanılarak tahminin iyi olmadığını görebiliriz.
# Şimdi Mevsimsel-ARIMA’yı uygulayacağız
# Mevsimsel-ARIMA(SARIMA):
# Adından da anlaşılacağı gibi, bu model zaman serileri mevsimsellik gösterdiğinde kullanılır. Bu model ARIMA modellerine benzer, sadece mevsimleri hesaba katmak için birkaç parametre eklememiz gerekiyor.
# ARIMA(p,d,q)(P, D, Q)m,
# - p — otoregresif sayısı
# - d — farklılık derecesi
# - q — hareketli ortalama terimlerinin sayısı
# - m — her sezondaki periyot sayısını ifade eder
# - (P, D, Q ) — zaman serisinin mevsimsel kısmı için (p,d,q) değerini temsil eder.
#
model = sm.tsa.statespace.SARIMAX(
df["Close"], order=(1, 1, 1), seasonal_order=(1, 1, 1, 12)
)
results = model.fit()
df["forecast"] = results.predict(start=2400, end=2900, dynamic=True)
df[["Close", "forecast"]].plot(figsize=(12, 8))
# # Second approach
df = pd.read_csv("/kaggle/input/tesla-stock-data-updated-till-28jun2021/TSLA.csv")
df.head()
df = df[["Date", "Close"]]
df["Date"] = pd.to_datetime(df["Date"])
df = df.set_index("Date")
df_data = df.groupby(pd.Grouper(freq="W")).mean()
df_data = df_data.rename(columns={"Close": "weekly_sales"})
df_data.plot()
plt.grid()
from statsmodels.tsa.api import VARMAX
from tqdm.notebook import tqdm
from sklearn.metrics import mean_squared_error
from dateutil.relativedelta import relativedelta
plt.rcParams["figure.figsize"] = (20, 7)
# Two stationary signals, weekly sales difference and standard deviation of the last 3 weekly sales, are used in VARMA
df_varma = df_data.copy()
df_varma["weekly_sales_diff"] = df_varma.diff()
df_varma["weekly_sales_rolling_std_3"] = df_varma["weekly_sales"].rolling(3).std()
df_varma = df_varma.dropna()
# predict last 50 values one by one
df_pred = pd.DataFrame()
for i in tqdm(range(50)):
training_data = df_varma[["weekly_sales_diff", "weekly_sales_rolling_std_3"]][
: (-50 + i)
]
# VARMA model selects lag p based on AIC
model = VARMAX(training_data)
model_fit = model.fit(ic="aic")
model_forecast = model_fit.get_forecast(steps=1)
df_pred_temp = pd.concat(
[model_forecast.conf_int(), model_forecast.predicted_mean], axis=1
)
df_pred_temp = df_pred_temp[
["lower weekly_sales_diff", "upper weekly_sales_diff", "weekly_sales_diff"]
]
df_pred = df_pred.append(df_pred_temp)
df_pred = df_pred.rename(
columns={
"lower weekly_sales_diff": "ci_lower",
"upper weekly_sales_diff": "ci_upper",
"weekly_sales_diff": "pred_weekly_sales_diff",
}
)
df_pred["weekly_sales_diff"] = df_varma["weekly_sales_diff"][-50:]
# transform data
df_pred["pred_weekly_sales"] = (
df_pred["pred_weekly_sales_diff"] + df_varma["weekly_sales"].shift()
)
df_pred["weekly_sales"] = (
df_pred["weekly_sales_diff"] + df_varma["weekly_sales"].shift()
)
df_pred["ci_lower"] = df_pred["ci_lower"] + df_varma["weekly_sales"].shift()
df_pred["ci_upper"] = df_pred["ci_upper"] + df_varma["weekly_sales"].shift()
plt.plot(df_pred["weekly_sales"], color="green")
plt.plot(df_pred["pred_weekly_sales"], color="red")
plt.legend(["Weekly sales", "Predicted weekly sales"])
plt.fill_between(
df_pred.index,
df_pred["ci_lower"],
df_pred["ci_upper"],
color="lightblue",
alpha=0.5,
)
plt.title(
"VARMA, MSE: {:,}".format(
round(
mean_squared_error(df_pred["weekly_sales"], df_pred["pred_weekly_sales"]), 2
)
)
)
plt.grid()
plt.show()
from statsmodels.tsa.statespace.varmax import VARMAX
from tqdm.notebook import tqdm
from sklearn.metrics import mean_squared_error
from dateutil.relativedelta import relativedelta
plt.rcParams["figure.figsize"] = (20, 7)
# Construct two stationary signals
df_varmax = df_data.copy()
df_varmax["weekly_sales_diff"] = df_varmax.diff()
df_varmax["weekly_sales_rolling_std_3"] = df_varmax["weekly_sales"].rolling(3).std()
df_varmax = df_varmax.dropna()
# prepare exogenous variable
exo_df = df_varmax["weekly_sales_diff"].to_frame()
exo_df["noise"] = [5000 * np.random.rand() for x in range(len(exo_df))]
exo_df["weekly_sales_diff_plus_noise"] = exo_df["weekly_sales_diff"] + exo_df["noise"]
exo_df["weekly_sales_diff_plus_noise"] = (
exo_df["weekly_sales_diff_plus_noise"].shift().fillna(0)
)
# predict last 50 values one by one
df_pred = pd.DataFrame()
for i in tqdm(range(50)):
training_data = df_varmax[["weekly_sales_diff", "weekly_sales_rolling_std_3"]][
: (-50 + i)
]
training_exo = exo_df[exo_df.index.isin(training_data.index)][
"weekly_sales_diff_plus_noise"
].to_frame()
# VARMAX model selects lag p based on AIC
model = VARMAX(training_data, exog=training_exo)
model_fit = model.fit(ic="aic")
pred_date = training_data.index[-1] + relativedelta(weeks=1)
model_forecast = model_fit.get_forecast(
steps=1,
exog=exo_df[exo_df.index == pred_date][
"weekly_sales_diff_plus_noise"
].to_frame(),
)
df_pred_temp = pd.concat(
[model_forecast.conf_int(), model_forecast.predicted_mean], axis=1
)
df_pred_temp = df_pred_temp[
["lower weekly_sales_diff", "upper weekly_sales_diff", "weekly_sales_diff"]
]
df_pred = df_pred.append(df_pred_temp)
df_pred = df_pred.rename(
columns={
"lower weekly_sales_diff": "ci_lower",
"upper weekly_sales_diff": "ci_upper",
"weekly_sales_diff": "pred_weekly_sales_diff",
}
)
df_pred["weekly_sales_diff"] = df_varmax["weekly_sales_diff"][-50:]
# transform data
df_pred["pred_weekly_sales"] = (
df_pred["pred_weekly_sales_diff"] + df_varmax["weekly_sales"].shift()
)
df_pred["weekly_sales"] = (
df_pred["weekly_sales_diff"] + df_varmax["weekly_sales"].shift()
)
df_pred["ci_lower"] = df_pred["ci_lower"] + df_varmax["weekly_sales"].shift()
df_pred["ci_upper"] = df_pred["ci_upper"] + df_varmax["weekly_sales"].shift()
plt.plot(df_pred["weekly_sales"], color="green")
plt.plot(df_pred["pred_weekly_sales"], color="red")
plt.legend(["Weekly sales", "Predicted weekly sales"])
plt.fill_between(
df_pred.index,
df_pred["ci_lower"],
df_pred["ci_upper"],
color="lightblue",
alpha=0.5,
)
plt.title(
"VARMAX, MSE: {:,}".format(
round(
mean_squared_error(df_pred["weekly_sales"], df_pred["pred_weekly_sales"]), 2
)
)
)
plt.grid()
plt.show()
df.info()
df.head()
df = df[["Date", "Close"]]
df.head()
df_data = df.groupby(["Date"])["Close"].mean()
df_data = df_data.rename(columns={"item_cnt_day": "weekly_sales"})
df_data
df_data = pd.DataFrame(df_data)
df_data = df_data.reset_index()
del df["index"]
df_data.Date = df_data.Date.astype("datetime64[ns]")
df.head()
# df['Date']=pd.to_datetime(df['Date'])
df.set_index("Date", inplace=True)
df.head()
# group data by weekly close
df_data = df.groupby(pd.Grouper(freq="W")).mean()
df_data = df_data.rename(columns={"item_cnt_day": "weekly_close"})
df_data.plot()
plt.grid()
df_data.head()
from pmdarima import auto_arima
from statsmodels.tsa.stattools import adfuller
from statsmodels.tools.eval_measures import mse, rmse
from statsmodels.tsa.statespace.varmax import VARMAX, VARMAXResults
# # Decide the order of the VARMA(p,q)
auto_arima(df["Close"], maxiter=100)
# Order (1,2) is preferred for VARMA. The last term or the third terms is the differencing which will be applied already using differencing.
df_transformed = df.diff().diff() # 2nd order difference
df_transformed = (
df_transformed.dropna()
) # remove the NaNs introduced due to differencing
df_transformed.head()
len(df_transformed)
# Train Test Split
nobs = 354 # The last 12 months will be the test data. At least 1 year would be a good choice
train = df_transformed[0:-nobs]
test = df_transformed[-nobs:]
train.shape
# Fit the VARMA(1,2) Model
model = VARMAX(train, order=(1, 2), trend="c") # c indicates a constant trend
results = model.fit(maxiter=1000, disp=False)
results.summary()
| false | 1 | 4,050 | 0 | 4,192 | 4,050 |
||
129750087
|
# # Feature Imputation with a Heat Flux Dataset📖🤓📖
# 
# 🗨️Context
# NOTES TO THE READERS This is a 2023 edition of Kaggle's Playground Series where the Kaggle Community hosts a variety of fairly light-weight challenges that can be used to learn and sharpen skills in different aspects of machine learning and data science
# ##
# 🔀Install and Import
#
import pandas as pd
import numpy as np
from IPython.display import display, HTML
import seaborn as sns
import random
import matplotlib.pyplot as plt
import plotly.express as px
#
# 📈🔭Data Overview
# #### As per the competition, this is a fairly light-weight dataset that is synthetically generated from real-world data, and will provide an opportunity to quickly iterate through various models/feature engineering ideas.
# #### Also, as given in the dataset description, this is both train & test dataset generated from a deep learning model trained on the Predicting Critical Heat Flux dataset (link avaiable below). Feature distributions are close to, but not exactly the same, as the original.
data = pd.read_csv("/kaggle/input/playground-series-s3e15/data.csv")
sample_submission = pd.read_csv(
"/kaggle/input/playground-series-s3e15/sample_submission.csv"
)
# NOTES TO THE READER Use the link:- https://www.kaggle.com/datasets/saurabhshahane/predicting-heat-flux to take a look at the Prediction Critical Heat Flux Dataset from which our data has been obtained.
# 🔍📊Exploratory Data Analysis
#
def eda(df):
print("==================================================================")
print("1. Dataframe Shape: ", df.shape)
print("==================================================================")
print("2. Sample Data: ")
display(HTML(df.head(5).to_html()))
print("==================================================================")
print("3. Information on the Data: ")
data_info_df = pd.DataFrame(df.dtypes, columns=["data type"])
data_info_df["Duplicated_Values"] = df.duplicated().sum()
data_info_df["Missing_Values"] = df.isnull().sum().values
data_info_df["%Missing"] = df.isnull().sum().values / len(df) * 100
data_info_df["Unique_Values"] = df.nunique().values
display(HTML(data_info_df.to_html()))
print("==================================================================")
num_cols = data.select_dtypes(include=["float64"]).columns.tolist()
cat_cols = data.select_dtypes(include=["object"]).columns.tolist()
random_num_cols = random.sample(num_cols, k=7)
def selective_corr(
df: pd.core.frame.DataFrame, n: int, title_name: str = "Top Correlations"
) -> None:
corr = df.corr()
# Select variables having highest absolute correlation
top_corr_cols = corr.abs().nlargest(n, columns=corr.columns).index
top_corr = corr.loc[top_corr_cols, top_corr_cols]
fig, axes = plt.subplots(figsize=(10, 5))
mask = np.zeros_like(top_corr)
mask[np.triu_indices_from(mask)] = True
sns.heatmap(top_corr, mask=mask, linewidths=0.5, cmap="YlOrRd", annot=True)
plt.title(title_name)
plt.show()
selective_corr(data[num_cols], 12, "4. Top Correlations in Dataset")
print("==================================================================")
print("5. Analyzing Relationships: ")
sns.pairplot(data)
eda(data)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/750/129750087.ipynb
| null | null |
[{"Id": 129750087, "ScriptId": 38580182, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2032938, "CreationDate": "05/16/2023 07:58:23", "VersionNumber": 8.0, "Title": "\ud83d\udcc8 Simple & Crisp EDA \u2728", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 118.0, "LinesInsertedFromPrevious": 26.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 92.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 6}]
| null | null | null | null |
# # Feature Imputation with a Heat Flux Dataset📖🤓📖
# 
# 🗨️Context
# NOTES TO THE READERS This is a 2023 edition of Kaggle's Playground Series where the Kaggle Community hosts a variety of fairly light-weight challenges that can be used to learn and sharpen skills in different aspects of machine learning and data science
# ##
# 🔀Install and Import
#
import pandas as pd
import numpy as np
from IPython.display import display, HTML
import seaborn as sns
import random
import matplotlib.pyplot as plt
import plotly.express as px
#
# 📈🔭Data Overview
# #### As per the competition, this is a fairly light-weight dataset that is synthetically generated from real-world data, and will provide an opportunity to quickly iterate through various models/feature engineering ideas.
# #### Also, as given in the dataset description, this is both train & test dataset generated from a deep learning model trained on the Predicting Critical Heat Flux dataset (link avaiable below). Feature distributions are close to, but not exactly the same, as the original.
data = pd.read_csv("/kaggle/input/playground-series-s3e15/data.csv")
sample_submission = pd.read_csv(
"/kaggle/input/playground-series-s3e15/sample_submission.csv"
)
# NOTES TO THE READER Use the link:- https://www.kaggle.com/datasets/saurabhshahane/predicting-heat-flux to take a look at the Prediction Critical Heat Flux Dataset from which our data has been obtained.
# 🔍📊Exploratory Data Analysis
#
def eda(df):
print("==================================================================")
print("1. Dataframe Shape: ", df.shape)
print("==================================================================")
print("2. Sample Data: ")
display(HTML(df.head(5).to_html()))
print("==================================================================")
print("3. Information on the Data: ")
data_info_df = pd.DataFrame(df.dtypes, columns=["data type"])
data_info_df["Duplicated_Values"] = df.duplicated().sum()
data_info_df["Missing_Values"] = df.isnull().sum().values
data_info_df["%Missing"] = df.isnull().sum().values / len(df) * 100
data_info_df["Unique_Values"] = df.nunique().values
display(HTML(data_info_df.to_html()))
print("==================================================================")
num_cols = data.select_dtypes(include=["float64"]).columns.tolist()
cat_cols = data.select_dtypes(include=["object"]).columns.tolist()
random_num_cols = random.sample(num_cols, k=7)
def selective_corr(
df: pd.core.frame.DataFrame, n: int, title_name: str = "Top Correlations"
) -> None:
corr = df.corr()
# Select variables having highest absolute correlation
top_corr_cols = corr.abs().nlargest(n, columns=corr.columns).index
top_corr = corr.loc[top_corr_cols, top_corr_cols]
fig, axes = plt.subplots(figsize=(10, 5))
mask = np.zeros_like(top_corr)
mask[np.triu_indices_from(mask)] = True
sns.heatmap(top_corr, mask=mask, linewidths=0.5, cmap="YlOrRd", annot=True)
plt.title(title_name)
plt.show()
selective_corr(data[num_cols], 12, "4. Top Correlations in Dataset")
print("==================================================================")
print("5. Analyzing Relationships: ")
sns.pairplot(data)
eda(data)
| false | 0 | 902 | 6 | 902 | 902 |
||
129750484
|
import numpy as np
import tensorflow as tf
from tensorflow.keras import layers
from tensorflow.keras.layers import (
Input,
Dense,
Conv1D,
BatchNormalization,
MaxPooling1D,
GlobalMaxPooling1D,
Concatenate,
)
from tensorflow.keras.models import Model
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
# 랜덤으로 학습데이터 생성
X_train = np.random.rand(100, 1000, 3)
X_test = np.random.randint(0, 3, size=(100, 1))
X_test = tf.keras.utils.to_categorical(X_test)
# 랜덤으로 테스트데이터 생성
y_train = np.random.rand(100, 1000, 3)
y_test = np.random.randint(0, 3, size=(100, 1))
y_test = tf.keras.utils.to_categorical(y_test)
# DGCNN 모델 정의
def DGCNN_model(input_shape):
# 입력 레이어
input_layer = Input(shape=input_shape)
# STN 레이어
x = Conv1D(64, 1, activation="relu")(input_layer)
x = BatchNormalization()(x)
x = Conv1D(128, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = Conv1D(1024, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = MaxPooling1D(pool_size=input_shape[0])(x)
x = Dense(512, activation="relu")(x)
x = BatchNormalization()(x)
x = Dense(256, activation="relu")(x)
x = BatchNormalization()(x)
# DGCNN 레이어
# K = 20
x_transformed = Dense(64, activation="relu")(x)
x_transformed = BatchNormalization()(x_transformed)
x_transformed = Dense(128, activation="relu")(x_transformed)
x_transformed = BatchNormalization()(x_transformed)
x_transformed = Dense(1024, activation=None)(x_transformed) # 출력 뉴런 개수를 1024로 변경
x_transformed = BatchNormalization()(x_transformed)
x = Dense(1024, activation=None)(x)
x = BatchNormalization()(x)
x = tf.reshape(x, (-1, 1024, 1))
x_transformed = tf.reshape(x_transformed, (-1, 1024, 1))
x = Concatenate(axis=2)([x, x_transformed])
x = Conv1D(512, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = Conv1D(256, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = Conv1D(128, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = GlobalMaxPooling1D()(x)
# 출력 레이어
output_layer = Dense(3, activation="softmax")(x)
# 모델 정의
model = Model(inputs=input_layer, outputs=output_layer)
return model
# 모델 생성 및 컴파일
model = DGCNN_model(input_shape=(1000, 3))
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
# 학습 데이터와 검증 데이터 분리
X_train, X_tr, X_test, X_te = train_test_split(
X_train, X_test, test_size=0.1, random_state=42
)
# 모델 학습
model.fit(
X_train,
X_test,
validation_data=(X_tr, X_te),
epochs=5,
batch_size=32,
validation_split=0.2,
)
y_train.shape
# 모델 평가
y_pred = model.predict(y_train)
y_pred
test_acc = accuracy_score(np.argmax(y_test, axis=1), np.argmax(y_pred, axis=1))
print("Test accuracy:", test_acc)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/750/129750484.ipynb
| null | null |
[{"Id": 129750484, "ScriptId": 38586285, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13992146, "CreationDate": "05/16/2023 08:01:25", "VersionNumber": 1.0, "Title": "DGCNN", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 84.0, "LinesInsertedFromPrevious": 84.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import tensorflow as tf
from tensorflow.keras import layers
from tensorflow.keras.layers import (
Input,
Dense,
Conv1D,
BatchNormalization,
MaxPooling1D,
GlobalMaxPooling1D,
Concatenate,
)
from tensorflow.keras.models import Model
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
# 랜덤으로 학습데이터 생성
X_train = np.random.rand(100, 1000, 3)
X_test = np.random.randint(0, 3, size=(100, 1))
X_test = tf.keras.utils.to_categorical(X_test)
# 랜덤으로 테스트데이터 생성
y_train = np.random.rand(100, 1000, 3)
y_test = np.random.randint(0, 3, size=(100, 1))
y_test = tf.keras.utils.to_categorical(y_test)
# DGCNN 모델 정의
def DGCNN_model(input_shape):
# 입력 레이어
input_layer = Input(shape=input_shape)
# STN 레이어
x = Conv1D(64, 1, activation="relu")(input_layer)
x = BatchNormalization()(x)
x = Conv1D(128, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = Conv1D(1024, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = MaxPooling1D(pool_size=input_shape[0])(x)
x = Dense(512, activation="relu")(x)
x = BatchNormalization()(x)
x = Dense(256, activation="relu")(x)
x = BatchNormalization()(x)
# DGCNN 레이어
# K = 20
x_transformed = Dense(64, activation="relu")(x)
x_transformed = BatchNormalization()(x_transformed)
x_transformed = Dense(128, activation="relu")(x_transformed)
x_transformed = BatchNormalization()(x_transformed)
x_transformed = Dense(1024, activation=None)(x_transformed) # 출력 뉴런 개수를 1024로 변경
x_transformed = BatchNormalization()(x_transformed)
x = Dense(1024, activation=None)(x)
x = BatchNormalization()(x)
x = tf.reshape(x, (-1, 1024, 1))
x_transformed = tf.reshape(x_transformed, (-1, 1024, 1))
x = Concatenate(axis=2)([x, x_transformed])
x = Conv1D(512, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = Conv1D(256, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = Conv1D(128, 1, activation="relu")(x)
x = BatchNormalization()(x)
x = GlobalMaxPooling1D()(x)
# 출력 레이어
output_layer = Dense(3, activation="softmax")(x)
# 모델 정의
model = Model(inputs=input_layer, outputs=output_layer)
return model
# 모델 생성 및 컴파일
model = DGCNN_model(input_shape=(1000, 3))
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
# 학습 데이터와 검증 데이터 분리
X_train, X_tr, X_test, X_te = train_test_split(
X_train, X_test, test_size=0.1, random_state=42
)
# 모델 학습
model.fit(
X_train,
X_test,
validation_data=(X_tr, X_te),
epochs=5,
batch_size=32,
validation_split=0.2,
)
y_train.shape
# 모델 평가
y_pred = model.predict(y_train)
y_pred
test_acc = accuracy_score(np.argmax(y_test, axis=1), np.argmax(y_pred, axis=1))
print("Test accuracy:", test_acc)
| false | 0 | 1,046 | 0 | 1,046 | 1,046 |
||
129750171
|
<jupyter_start><jupyter_text>Bank Customer Churn
RowNumber—corresponds to the record (row) number and has no effect on the output.
CustomerId—contains random values and has no effect on customer leaving the bank.
Surname—the surname of a customer has no impact on their decision to leave the bank.
CreditScore—can have an effect on customer churn, since a customer with a higher credit score is less likely to leave the bank.
Geography—a customer’s location can affect their decision to leave the bank.
Gender—it’s interesting to explore whether gender plays a role in a customer leaving the bank.
Age—this is certainly relevant, since older customers are less likely to leave their bank than younger ones.
Tenure—refers to the number of years that the customer has been a client of the bank. Normally, older clients are more loyal and less likely to leave a bank.
Balance—also a very good indicator of customer churn, as people with a higher balance in their accounts are less likely to leave the bank compared to those with lower balances.
NumOfProducts—refers to the number of products that a customer has purchased through the bank.
HasCrCard—denotes whether or not a customer has a credit card. This column is also relevant, since people with a credit card are less likely to leave the bank.
IsActiveMember—active customers are less likely to leave the bank.
EstimatedSalary—as with balance, people with lower salaries are more likely to leave the bank compared to those with higher salaries.
Exited—whether or not the customer left the bank.
Complain—customer has complaint or not.
Satisfaction Score—Score provided by the customer for their complaint resolution.
Card Type—type of card hold by the customer.
Points Earned—the points earned by the customer for using credit card.
Acknowledgements
As we know, it is much more expensive to sign in a new client than keeping an existing one.
It is advantageous for banks to know what leads a client towards the decision to leave the company.
Churn prevention allows companies to develop loyalty programs and retention campaigns to keep as many customers as possible.
Kaggle dataset identifier: bank-customer-churn
<jupyter_script># ## Project Goal
# This projects aims to create a classification system that determine customer churning according to several features to help retain customer as this will directly lead to more profit and better brand
# ## Importing required Packages & Loading Data
# ### Importing Packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# ### Loading Data
# * The data source of our project is provided by Kaggle
df = pd.read_csv("/kaggle/input/bank-customer-churn/Customer-Churn-Records.csv")
df.head()
# ## EDA
# ### Creating a data summary
def summary(dataframe, label=None):
# This functions takes in a dataframe & it's label as input and return its shape, columns & numerical descripiton of numerical variables
print(dataframe.shape, "\n")
print(label, "columns: ", dataframe.columns)
print(
f"\nThere is {len(dataframe)} observation in our dataset, {len(dataframe[dataframe.duplicated()])} are duplicated"
)
desc = dataframe.select_dtypes(include="number")
desc_1 = desc.describe().T
desc_1["nunique"] = desc.nunique()
desc_1["%unique"] = len(desc.nunique()) / len(desc) * 100
desc_1["null"] = desc.isna().sum()
print(dataframe.isna().sum())
return desc_1
summary(df, "Dataset")
# ### Summary results:
# * Our dataframe consist of 10000 observations, None of them are duplicated
# * Our dataframe doesn't have any Null values
# * Some features such as Age, HasCrCard, IsActiveMember,Complain,Exited,Complain,Satisfaction Score are set as numerical variables with only limited values therefore they are categorical
# * Our dataframe has 17 features and 1 target variable which is Exited
# * RowNumber,CustomerId,Surname should be removed as they won't help in our analysis
df.drop(["RowNumber", "CustomerId", "Surname"], axis=1, inplace=True)
df.head(5)
# This is to check for Categorical variables that are saved as Numerical Values
for x in df.columns:
if df[x].nunique() < 12:
print(x, " ", df[x].nunique(), df[x].dtype)
# * It looks like NumOfProducts, HasCrCard, IsActiveMember, Complain, Satisfaction Score are all Categorical Variables saved as integer values
# ## Checking Distributions of different kinds of Data
# ### First our Target Variable
plt.plot(figsize=(5, 5), dpi=300)
sns.histplot(x=df["Exited"], data=df)
df.dtypes
# ### Let's check on categorical features saved as objects
for x, column in enumerate(df.columns):
if df[column].dtype == "object":
sns.histplot(data=df, x=column)
plt.title(f"{column} Distribution \n")
print("\n")
plt.show()
# ### Let's check on Continous Features of our dataset
for column in ["CreditScore", "EstimatedSalary", "Point Earned"]:
plt.plot(dpi=300)
sns.kdeplot(data=df, x=df[column])
plt.title(f"{column} Distribution")
plt.show()
# * Their distribution seems normal enough
# ### Let's check on Categorical Features Distribution that are saved as int values
for column in [
"NumOfProducts",
"HasCrCard",
"IsActiveMember",
"Complain",
"Satisfaction Score",
"Age",
"Tenure",
]:
plt.plot(dpi=300)
sns.kdeplot(data=df, x=df[column])
plt.title(f"{column} Distribution")
plt.show()
def heatmap(df, label=None):
plt.figure(figsize=(14, 10), dpi=300)
sns.heatmap(
df.corr(method="pearson"), cmap="YlOrRd", annot=True, annot_kws={"size": 7}
)
plt.title(f"Correlations between features of the {label} set")
plt.show()
heatmap(df, "Churn Dataset Correlation")
# * Correlation between Exited & Complain is very high that it will for sure greatly affect our model performance (Try with and without complain Feature)
# * Correlation between Exited & all features is not that high (performance of model might not be good if we remove complain feature; in this case we should try to find more relevant features)
# ## Baseline Model
# * Everything seems fine so let's begin our Modeling process
df_dummies = pd.get_dummies(
df[
[
"Age",
"Geography",
"Gender",
"Tenure",
"NumOfProducts",
"Card Type",
"Satisfaction Score",
"CreditScore",
"Balance",
"Point Earned",
"EstimatedSalary",
"Complain",
]
]
)
y = df["Exited"]
df_dummies
from sklearn.preprocessing import StandardScaler
scale = StandardScaler()
df_dummies_scaled = df_dummies.copy()
df_dummies_scaled[
[
"Age",
"Tenure",
"Balance",
"EstimatedSalary",
"Point Earned",
"Satisfaction Score",
]
] = scale.fit_transform(
df_dummies_scaled[
[
"Age",
"Tenure",
"Balance",
"EstimatedSalary",
"Point Earned",
"Satisfaction Score",
]
]
)
df_dummies_scaled
# Importing Models
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from sklearn.metrics import ConfusionMatrixDisplay
X_train, X_test, y_train, y_test = train_test_split(
df_dummies_scaled, y, test_size=0.2, random_state=42, stratify=y
)
def model(model, X, X_t, y, y_t, label=""):
clf = model.fit(X, y)
y_pred = clf.predict(X_t)
cm = confusion_matrix(y_pred, y_test)
TP = cm[1][1]
TN = cm[0][0]
FP = cm[0][1]
FN = cm[1][0]
print("True Positives:", TP)
print("True Negatives:", TN)
print("False Positives:", FP)
print("False Negatives:", FN)
# calculate accuracy
conf_accuracy = float(TP + TN) / float(TP + TN + FP + FN)
# calculate mis-classification
conf_misclassification = 1 - conf_accuracy
# calculate the sensitivity
conf_sensitivity = TP / float(TP + FN)
# calculate the specificity
conf_specificity = TN / float(TN + FP)
# calculate precision
conf_precision = TN / float(TN + FP)
# calculate f_1 score
conf_f1 = 2 * (
(conf_precision * conf_sensitivity) / (conf_precision + conf_sensitivity)
)
print("-" * 50)
print(f"Accuracy: {round(conf_accuracy,2)}")
print(f"Mis-Classification: {round(conf_misclassification,2)}")
print(f"Sensitivity: {round(conf_sensitivity,2)}")
print(f"Specificity: {round(conf_specificity,2)}")
print(f"Precision: {round(conf_precision,2)}")
print(f"f_1 Score: {round(conf_f1,2)}")
disp = ConfusionMatrixDisplay(cm)
disp.plot()
plt.show()
try:
coefficients = clf.coef_[0]
feature_importance = pd.DataFrame(
{"Feature": X.columns, "Importance": np.abs(coefficients)}
)
feature_importance = feature_importance.sort_values(
"Importance", ascending=True
)
feature_importance.plot(
x="Feature", y="Importance", kind="barh", figsize=(10, 6)
)
plt.show()
except:
print("\n")
models = [
("Logistic Regression", LogisticRegression(max_iter=10000)),
("DecisionTreeClassifier", DecisionTreeClassifier()),
("K Neighbor", KNeighborsClassifier()),
("Random Forest", RandomForestClassifier()),
]
for l, m in models:
print(f"Model Name : {l} \n\n")
model(m, X_train, X_test, y_train, y_test, label=l)
# ### Baseline models with Complain feature performs with 99% accuracy, and it seems that from feature importance that complain feature is doing most of the work for the model due to it's high correlation with target feature Exited. I think building a model without Complain feature would be more usable even if it's not as accurate as model with complain feature, as it's necessary to have a model that doesn't solely depend on 1 feature
df_dummies_no_complain = pd.get_dummies(
df[
[
"Age",
"Geography",
"Gender",
"Tenure",
"NumOfProducts",
"Card Type",
"Satisfaction Score",
"CreditScore",
"Balance",
"Point Earned",
"EstimatedSalary",
]
]
)
scale = StandardScaler()
df_dummies_no_complain_scaled = df_dummies_no_complain.copy()
df_dummies_no_complain_scaled[
[
"Age",
"Tenure",
"Balance",
"EstimatedSalary",
"Point Earned",
"Satisfaction Score",
]
] = scale.fit_transform(
df_dummies_no_complain_scaled[
[
"Age",
"Tenure",
"Balance",
"EstimatedSalary",
"Point Earned",
"Satisfaction Score",
]
]
)
X_train, X_test, y_train, y_test = train_test_split(
df_dummies_no_complain_scaled, y, test_size=0.2, random_state=42, stratify=y
)
for l, m in models:
print(f"Model Name : {l} \n\n")
model(m, X_train, X_test, y_train, y_test, label=l)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/750/129750171.ipynb
|
bank-customer-churn
|
radheshyamkollipara
|
[{"Id": 129750171, "ScriptId": 38552290, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8536881, "CreationDate": "05/16/2023 07:59:02", "VersionNumber": 3.0, "Title": "Customer Churn Prediction", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 231.0, "LinesInsertedFromPrevious": 98.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 133.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186103168, "KernelVersionId": 129750171, "SourceDatasetVersionId": 5550559}]
|
[{"Id": 5550559, "DatasetId": 3197960, "DatasourceVersionId": 5625285, "CreatorUserId": 14862076, "LicenseName": "Other (specified in description)", "CreationDate": "04/28/2023 16:32:01", "VersionNumber": 1.0, "Title": "Bank Customer Churn", "Slug": "bank-customer-churn", "Subtitle": "Bank Customer Data for Customer Churn", "Description": "RowNumber\u2014corresponds to the record (row) number and has no effect on the output.\nCustomerId\u2014contains random values and has no effect on customer leaving the bank.\nSurname\u2014the surname of a customer has no impact on their decision to leave the bank.\nCreditScore\u2014can have an effect on customer churn, since a customer with a higher credit score is less likely to leave the bank.\nGeography\u2014a customer\u2019s location can affect their decision to leave the bank.\nGender\u2014it\u2019s interesting to explore whether gender plays a role in a customer leaving the bank.\nAge\u2014this is certainly relevant, since older customers are less likely to leave their bank than younger ones.\nTenure\u2014refers to the number of years that the customer has been a client of the bank. Normally, older clients are more loyal and less likely to leave a bank.\nBalance\u2014also a very good indicator of customer churn, as people with a higher balance in their accounts are less likely to leave the bank compared to those with lower balances.\nNumOfProducts\u2014refers to the number of products that a customer has purchased through the bank.\nHasCrCard\u2014denotes whether or not a customer has a credit card. This column is also relevant, since people with a credit card are less likely to leave the bank.\nIsActiveMember\u2014active customers are less likely to leave the bank.\nEstimatedSalary\u2014as with balance, people with lower salaries are more likely to leave the bank compared to those with higher salaries.\nExited\u2014whether or not the customer left the bank.\nComplain\u2014customer has complaint or not.\nSatisfaction Score\u2014Score provided by the customer for their complaint resolution.\nCard Type\u2014type of card hold by the customer.\nPoints Earned\u2014the points earned by the customer for using credit card.\n\nAcknowledgements\n\nAs we know, it is much more expensive to sign in a new client than keeping an existing one.\n\nIt is advantageous for banks to know what leads a client towards the decision to leave the company.\n\nChurn prevention allows companies to develop loyalty programs and retention campaigns to keep as many customers as possible.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3197960, "CreatorUserId": 14862076, "OwnerUserId": 14862076.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5550559.0, "CurrentDatasourceVersionId": 5625285.0, "ForumId": 3262570, "Type": 2, "CreationDate": "04/28/2023 16:32:01", "LastActivityDate": "04/28/2023", "TotalViews": 39315, "TotalDownloads": 6814, "TotalVotes": 97, "TotalKernels": 52}]
|
[{"Id": 14862076, "UserName": "radheshyamkollipara", "DisplayName": "Radheshyam Kollipara", "RegisterDate": "04/28/2023", "PerformanceTier": 0}]
|
# ## Project Goal
# This projects aims to create a classification system that determine customer churning according to several features to help retain customer as this will directly lead to more profit and better brand
# ## Importing required Packages & Loading Data
# ### Importing Packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# ### Loading Data
# * The data source of our project is provided by Kaggle
df = pd.read_csv("/kaggle/input/bank-customer-churn/Customer-Churn-Records.csv")
df.head()
# ## EDA
# ### Creating a data summary
def summary(dataframe, label=None):
# This functions takes in a dataframe & it's label as input and return its shape, columns & numerical descripiton of numerical variables
print(dataframe.shape, "\n")
print(label, "columns: ", dataframe.columns)
print(
f"\nThere is {len(dataframe)} observation in our dataset, {len(dataframe[dataframe.duplicated()])} are duplicated"
)
desc = dataframe.select_dtypes(include="number")
desc_1 = desc.describe().T
desc_1["nunique"] = desc.nunique()
desc_1["%unique"] = len(desc.nunique()) / len(desc) * 100
desc_1["null"] = desc.isna().sum()
print(dataframe.isna().sum())
return desc_1
summary(df, "Dataset")
# ### Summary results:
# * Our dataframe consist of 10000 observations, None of them are duplicated
# * Our dataframe doesn't have any Null values
# * Some features such as Age, HasCrCard, IsActiveMember,Complain,Exited,Complain,Satisfaction Score are set as numerical variables with only limited values therefore they are categorical
# * Our dataframe has 17 features and 1 target variable which is Exited
# * RowNumber,CustomerId,Surname should be removed as they won't help in our analysis
df.drop(["RowNumber", "CustomerId", "Surname"], axis=1, inplace=True)
df.head(5)
# This is to check for Categorical variables that are saved as Numerical Values
for x in df.columns:
if df[x].nunique() < 12:
print(x, " ", df[x].nunique(), df[x].dtype)
# * It looks like NumOfProducts, HasCrCard, IsActiveMember, Complain, Satisfaction Score are all Categorical Variables saved as integer values
# ## Checking Distributions of different kinds of Data
# ### First our Target Variable
plt.plot(figsize=(5, 5), dpi=300)
sns.histplot(x=df["Exited"], data=df)
df.dtypes
# ### Let's check on categorical features saved as objects
for x, column in enumerate(df.columns):
if df[column].dtype == "object":
sns.histplot(data=df, x=column)
plt.title(f"{column} Distribution \n")
print("\n")
plt.show()
# ### Let's check on Continous Features of our dataset
for column in ["CreditScore", "EstimatedSalary", "Point Earned"]:
plt.plot(dpi=300)
sns.kdeplot(data=df, x=df[column])
plt.title(f"{column} Distribution")
plt.show()
# * Their distribution seems normal enough
# ### Let's check on Categorical Features Distribution that are saved as int values
for column in [
"NumOfProducts",
"HasCrCard",
"IsActiveMember",
"Complain",
"Satisfaction Score",
"Age",
"Tenure",
]:
plt.plot(dpi=300)
sns.kdeplot(data=df, x=df[column])
plt.title(f"{column} Distribution")
plt.show()
def heatmap(df, label=None):
plt.figure(figsize=(14, 10), dpi=300)
sns.heatmap(
df.corr(method="pearson"), cmap="YlOrRd", annot=True, annot_kws={"size": 7}
)
plt.title(f"Correlations between features of the {label} set")
plt.show()
heatmap(df, "Churn Dataset Correlation")
# * Correlation between Exited & Complain is very high that it will for sure greatly affect our model performance (Try with and without complain Feature)
# * Correlation between Exited & all features is not that high (performance of model might not be good if we remove complain feature; in this case we should try to find more relevant features)
# ## Baseline Model
# * Everything seems fine so let's begin our Modeling process
df_dummies = pd.get_dummies(
df[
[
"Age",
"Geography",
"Gender",
"Tenure",
"NumOfProducts",
"Card Type",
"Satisfaction Score",
"CreditScore",
"Balance",
"Point Earned",
"EstimatedSalary",
"Complain",
]
]
)
y = df["Exited"]
df_dummies
from sklearn.preprocessing import StandardScaler
scale = StandardScaler()
df_dummies_scaled = df_dummies.copy()
df_dummies_scaled[
[
"Age",
"Tenure",
"Balance",
"EstimatedSalary",
"Point Earned",
"Satisfaction Score",
]
] = scale.fit_transform(
df_dummies_scaled[
[
"Age",
"Tenure",
"Balance",
"EstimatedSalary",
"Point Earned",
"Satisfaction Score",
]
]
)
df_dummies_scaled
# Importing Models
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from sklearn.metrics import ConfusionMatrixDisplay
X_train, X_test, y_train, y_test = train_test_split(
df_dummies_scaled, y, test_size=0.2, random_state=42, stratify=y
)
def model(model, X, X_t, y, y_t, label=""):
clf = model.fit(X, y)
y_pred = clf.predict(X_t)
cm = confusion_matrix(y_pred, y_test)
TP = cm[1][1]
TN = cm[0][0]
FP = cm[0][1]
FN = cm[1][0]
print("True Positives:", TP)
print("True Negatives:", TN)
print("False Positives:", FP)
print("False Negatives:", FN)
# calculate accuracy
conf_accuracy = float(TP + TN) / float(TP + TN + FP + FN)
# calculate mis-classification
conf_misclassification = 1 - conf_accuracy
# calculate the sensitivity
conf_sensitivity = TP / float(TP + FN)
# calculate the specificity
conf_specificity = TN / float(TN + FP)
# calculate precision
conf_precision = TN / float(TN + FP)
# calculate f_1 score
conf_f1 = 2 * (
(conf_precision * conf_sensitivity) / (conf_precision + conf_sensitivity)
)
print("-" * 50)
print(f"Accuracy: {round(conf_accuracy,2)}")
print(f"Mis-Classification: {round(conf_misclassification,2)}")
print(f"Sensitivity: {round(conf_sensitivity,2)}")
print(f"Specificity: {round(conf_specificity,2)}")
print(f"Precision: {round(conf_precision,2)}")
print(f"f_1 Score: {round(conf_f1,2)}")
disp = ConfusionMatrixDisplay(cm)
disp.plot()
plt.show()
try:
coefficients = clf.coef_[0]
feature_importance = pd.DataFrame(
{"Feature": X.columns, "Importance": np.abs(coefficients)}
)
feature_importance = feature_importance.sort_values(
"Importance", ascending=True
)
feature_importance.plot(
x="Feature", y="Importance", kind="barh", figsize=(10, 6)
)
plt.show()
except:
print("\n")
models = [
("Logistic Regression", LogisticRegression(max_iter=10000)),
("DecisionTreeClassifier", DecisionTreeClassifier()),
("K Neighbor", KNeighborsClassifier()),
("Random Forest", RandomForestClassifier()),
]
for l, m in models:
print(f"Model Name : {l} \n\n")
model(m, X_train, X_test, y_train, y_test, label=l)
# ### Baseline models with Complain feature performs with 99% accuracy, and it seems that from feature importance that complain feature is doing most of the work for the model due to it's high correlation with target feature Exited. I think building a model without Complain feature would be more usable even if it's not as accurate as model with complain feature, as it's necessary to have a model that doesn't solely depend on 1 feature
df_dummies_no_complain = pd.get_dummies(
df[
[
"Age",
"Geography",
"Gender",
"Tenure",
"NumOfProducts",
"Card Type",
"Satisfaction Score",
"CreditScore",
"Balance",
"Point Earned",
"EstimatedSalary",
]
]
)
scale = StandardScaler()
df_dummies_no_complain_scaled = df_dummies_no_complain.copy()
df_dummies_no_complain_scaled[
[
"Age",
"Tenure",
"Balance",
"EstimatedSalary",
"Point Earned",
"Satisfaction Score",
]
] = scale.fit_transform(
df_dummies_no_complain_scaled[
[
"Age",
"Tenure",
"Balance",
"EstimatedSalary",
"Point Earned",
"Satisfaction Score",
]
]
)
X_train, X_test, y_train, y_test = train_test_split(
df_dummies_no_complain_scaled, y, test_size=0.2, random_state=42, stratify=y
)
for l, m in models:
print(f"Model Name : {l} \n\n")
model(m, X_train, X_test, y_train, y_test, label=l)
| false | 1 | 2,621 | 0 | 3,121 | 2,621 |
||
129750911
|
<jupyter_start><jupyter_text>Air Passenger Data for Time Series Analysis
### Context
This data is used for making ARIMA model forecasting.
### Content
This contains the increasing rate of passenger
Kaggle dataset identifier: air-passenger-data-for-time-series-analysis
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Load Dataset
df = pd.read_csv(
"/kaggle/input/air-passenger-data-for-time-series-analysis/AirPassengers.csv"
)
df
# # Exploratory Data Analysis (EDA)
# ## View Dataset Description
df["Month"] = pd.to_datetime(df["Month"], format="%Y-%m")
# df['Month'] = df['Month'].dt.strftime('%Y-%m')
df.info()
df
# ## Change Data Index
df.set_index("Month", inplace=True)
# ## Data Visualization
import matplotlib.pyplot as plt
import datetime
plt.figure(figsize=(12, 6))
plt.plot(df)
plt.xlabel("Time")
# plt.xticks(rotation=45)
plt.ylabel("Num of Passengers")
plt.title("US Airline Num of Passengers Trend 1949 - 1960")
plt.show()
# There is a positive trend with some repetitive pattern
# # Time Series Decomposition
from statsmodels.tsa.seasonal import seasonal_decompose
from dateutil.parser import parse
# ## Additive Decomposition
additive_dec = seasonal_decompose(df, model="additive", period=30)
plt.figure(figsize=(12, 8))
additive_dec.plot()
plt.suptitle("Additive Decomposition", fontsize=12)
plt.tight_layout()
plt.show()
multiplicative_dec = seasonal_decompose(df, model="multiplicative", period=30)
plt.figure(figsize=(12, 8))
multiplicative_dec.plot()
plt.suptitle("Multiplicative Decomposition", fontsize=12)
plt.tight_layout()
plt.show()
# Residual in additive decomposition still have a pattern, while in multiplicative is not really showing and quite random. So we will preferred to use multiplicative decomposition.
# # Stationary Test for Time Series
from statsmodels.tsa.stattools import adfuller, kpss
from statsmodels.graphics.tsaplots import plot_acf
# ## Augmented Dickey Fuller Test (ADF Test)
# H0: time series data is non-stationary
# H1: time series data is stationary
# p-value reject null hypothesis (H0)
result = adfuller(df.values, autolag="AIC")
print(f"ADF Statistic: {result[0]}")
print(f"p-value: {result[1]}")
# ## KPSS Test
# H0: time series data is stationary
# H1: time series data is non-stationary
# p-value reject null hypothesis (H0)
result = kpss(df)
print("KPSS Statistic:", result[0])
print("p-value:", result[1])
# ## Rolling Test
# plt.plot(df['Month'], df['#Passengers'])
rolling_mean = df.rolling(6).mean()
rolling_std = df.rolling(6).std()
plt.plot(df, label="Passenger Data")
plt.plot(rolling_mean, color="red", label="Rolling Num of Passenger Mean")
plt.plot(
rolling_std, color="green", label="Rolling Passenger Number Standard Deviation"
)
plt.xlabel("Time")
plt.title("Passenger Time Series, Rolling Mean, Standard Deviation")
plt.legend(loc="best")
plt.show()
# From two test result above, We can see that current data is non-stationary
# # Split Data
# df['date'] = df.index
# train = df[df['date'] < pd.to_datetime("1960-08", format='%Y-%m')]
# train['train'] = train['#Passengers']
# del train['date']
# del train['#Passengers']
# test = df[df['date'] >= pd.to_datetime("1960-08", format='%Y-%m')]
# del test['date']
# test['test'] = test['#Passengers']
# del test['#Passengers']
# plt.plot(train, color = "black")
# plt.plot(test, color = "red")
# plt.title("Train/Test split for Passenger Data")
# plt.ylabel("Passenger Number")
# plt.xlabel('Year-Month')
# plt.show()
# # Preprocessing Data
# ## Convert to Stationary Time Series
# Decompose time series data using moving average
dfs = np.log(df)
# use window 12 to represent 12 months
ma = dfs.rolling(window=12).mean()
ms = dfs.rolling(window=12).std()
plt.plot(ma, c="r")
plt.plot(ms, c="b")
plt.plot(dfs)
# plt.grid()
plt.show()
dfs - np.log(df)
ma = dfs.rolling(window=12, center=False).mean()
dfs2 = dfs - ma
dfs2 = dfs2.dropna()
rolling_mean = dfs2.rolling(6).mean()
rolling_std = dfs2.rolling(6).std()
plt.plot(dfs2, label="Passenger Data")
plt.plot(rolling_mean, color="red", label="Rolling Num of Passenger Mean")
plt.plot(
rolling_std, color="green", label="Rolling Passenger Number Standard Deviation"
)
plt.xlabel("Time")
plt.title("Passenger Time Series, Rolling Mean, Standard Deviation")
plt.legend(loc="best")
plt.show()
# # Autocorrelation & Partial Autocorrelation
# Measure how correlated time series data is at a given point in time with past values.
from statsmodels.tsa.stattools import acf, pacf
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
# autocorr_lag1 = df['#Passengers'].autocorr(lag=1)
# print("One Month Lag: ", autocorr_lag1)
# autocorr_lag3 = df['#Passengers'].autocorr(lag=3)
# print("Three Month Lag: ", autocorr_lag3)
# autocorr_lag6 = df['#Passengers'].autocorr(lag=6)
# print("Six Month Lag: ", autocorr_lag6)
# autocorr_lag9 = df['#Passengers'].autocorr(lag=9)
# print("Nine Month Lag: ", autocorr_lag9)
# Result above show us data is higly correlated
# pd.plotting.autocorrelation_plot(df['#Passengers'])
plt.plot(
np.arange(acf(dfs2, nlags=10, fft=True).shape[0]), acf(dfs2, nlags=10, fft=True)
)
plt.axhline(y=0, linestyle="--", c="gray")
plt.axhline(y=-7.96 / np.sqrt(len(dfs2)), linestyle="--", c="gray")
plt.axhline(y=7.96 / np.sqrt(len(dfs2)), linestyle="--", c="gray")
plt.title("Auto Correlation Fuction")
plt.grid()
plt.show()
plt.plot(np.arange(pacf(dfs2, nlags=10).shape[0]), pacf(dfs2, nlags=10))
plt.axhline(y=0, linestyle="--", c="gray")
plt.axhline(y=-7.96 / np.sqrt(len(dfs2)), linestyle="--", c="gray")
plt.axhline(y=7.96 / np.sqrt(len(dfs2)), linestyle="--", c="gray")
plt.title("Auto Correlation Fuction")
plt.grid()
plt.show()
# # Draw Plot
# fig, axes = plt.subplots(1,2,figsize=(16,3), dpi= 100)
# plot_acf(df['#Passengers'].tolist(), lags=50, ax=axes[0])
# plot_pacf(df['#Passengers'].tolist(), lags=50, ax=axes[1])
# ## Splitting Data
train = dfs2.loc[:"1959"]
test = dfs2.loc["1960":]
# train.rename({'#Passenger':''})
# # Model Building
from statsmodels.tsa.arima.model import ARIMA
Arima = ARIMA(train, order=(3, 1, 3))
Ar = Arima.fit()
Ar.summary()
# Plot residual errors
residuals = pd.DataFrame(Ar.resid)
fig, ax = plt.subplots(1, 2)
residuals.plot(title="Residuals", ax=ax[0])
residuals.plot(kind="kde", title="Density", ax=ax[1])
plt.show()
# # Evaluation
from statsmodels.graphics.tsaplots import plot_predict
def forecast_accuracy(forecast, actual):
mape = np.mean(np.abs(forecast - actual) / np.abs(actual)) # MAPE
me = np.mean(forecast - actual) # ME
mae = np.mean(np.abs(forecast - actual)) # MAE
mpe = np.mean((forecast - actual) / actual) # MPE
rmse = np.mean((forecast - actual) ** 2) ** 0.5 # RMSE
corr = np.corrcoef(forecast, actual)[0, 1] # corr
mins = np.amin(np.hstack([forecast[:, None], actual[:, None]]), axis=1)
maxs = np.amax(np.hstack([forecast[:, None], actual[:, None]]), axis=1)
minmax = 1 - np.mean(mins / maxs) # minmax
acf1 = acf(fc - test)[1] # ACF1
return {
"mape": mape,
"me": me,
"mae": mae,
"mpe": mpe,
"rmse": rmse,
"acf1": acf1,
"corr": corr,
"minmax": minmax,
}
# Forecast
fc = Ar.forecast(12, alpha=0.05) # 95% conf
fc = pd.DataFrame(fc).rename({"predicted_mean": "#Passengers"}, axis=1)
# Make as pandas series
# fc_series = pd.Series(fc, index=test.index)
# lower_series = pd.Series(conf[:, 0], index=test.index)
# upper_series = pd.Series(conf[:, 1], index=test.index)
fig, ax = plt.subplots(figsize=(12, 6))
ax = dfs2.plot(ax=ax, label="train time series")
ax.set_label("sdf")
ax = test.plot(ax=ax, label="test time series")
# plot_predict(Ar, '1959', '1961', ax=ax)
ax = fc.plot(ax=ax)
ax = plt.legend(loc="best")
plt.show()
print("mape", np.mean(np.abs(fc - test) / np.abs(test))) # MAPE)
print("rmse", np.mean((fc - test) ** 2) ** 0.5) # RMSE
# ## Auto ARIMA
# !pip install pmdarima
import pmdarima as pm
model = pm.auto_arima(
train.values,
start_p=1,
start_q=1,
test="adf", # use adftest to find optimal 'd'
max_p=3,
max_q=3, # maximum p and q
m=1, # frequency of series
d=None, # let model determine 'd'
seasonal=False, # No Seasonality
start_P=0,
D=0,
trace=True,
error_action="ignore",
suppress_warnings=True,
stepwise=True,
)
print(model.summary())
# Forecast
n_periods = 12
fitted, confint = model.predict(n_periods=n_periods, return_conf_int=True)
index_of_fc = pd.date_range(dfs2.index[-1], periods=n_periods, freq="MS")
# make series for plotting purpose
fitted_series = pd.Series(fitted, index=index_of_fc)
lower_series = pd.Series(confint[:, 0], index=index_of_fc)
upper_series = pd.Series(confint[:, 1], index=index_of_fc)
fig, ax = plt.subplots(figsize=(12, 6))
ax = train.plot(ax=ax, label="train time series")
ax.set_label("sdf")
ax = test.plot(ax=ax, label="test time series")
# plot_predict(model, '1959', '1961', ax=ax)
ax = fitted_series.plot(ax=ax)
ax = plt.legend(loc="best")
plt.show()
# Plot
plt.plot(dfs2)
plt.plot(fitted_series, color="darkgreen")
plt.fill_between(lower_series.index, lower_series, upper_series, color="k", alpha=0.15)
plt.title("SARIMA - Final Forecast of a10 - Drug Sales")
plt.show()
# ## SARIMA
# Seasonal - fit stepwise auto-ARIMA
smodel = pm.auto_arima(
dfs2,
start_p=1,
start_q=1,
test="adf",
max_p=3,
max_q=3,
m=12,
start_P=0,
seasonal=True,
d=None,
D=1,
trace=True,
error_action="ignore",
suppress_warnings=True,
stepwise=True,
)
smodel.summary()
# Plot
plt.plot(dfs2)
plt.plot(fitted_series, color="darkgreen")
plt.fill_between(lower_series.index, lower_series, upper_series, color="k", alpha=0.15)
plt.title("SARIMA - Final Forecast of a10 - Drug Sales")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/750/129750911.ipynb
|
air-passenger-data-for-time-series-analysis
|
ashfakyeafi
|
[{"Id": 129750911, "ScriptId": 38534427, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6654637, "CreationDate": "05/16/2023 08:05:01", "VersionNumber": 3.0, "Title": "Airline Passenger Forecasting using ARIMA", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 353.0, "LinesInsertedFromPrevious": 246.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 107.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186104038, "KernelVersionId": 129750911, "SourceDatasetVersionId": 2504188}]
|
[{"Id": 2504188, "DatasetId": 1516462, "DatasourceVersionId": 2546888, "CreatorUserId": 5154008, "LicenseName": "CC0: Public Domain", "CreationDate": "08/06/2021 14:46:29", "VersionNumber": 1.0, "Title": "Air Passenger Data for Time Series Analysis", "Slug": "air-passenger-data-for-time-series-analysis", "Subtitle": "There is a list of passenger data from year 1949 to 1960", "Description": "### Context\n\nThis data is used for making ARIMA model forecasting.\n\n\n### Content\n\nThis contains the increasing rate of passenger\n\n\n### Acknowledgements\n\nWe wouldn't be here without the help of others. If you owe any attributions or thanks, include them here along with any citations of past research.\n\n\n### Inspiration\n\nYour data will be in front of the world's largest data science community. What questions do you want to see answered?", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1516462, "CreatorUserId": 5154008, "OwnerUserId": 5154008.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2504188.0, "CurrentDatasourceVersionId": 2546888.0, "ForumId": 1536251, "Type": 2, "CreationDate": "08/06/2021 14:46:29", "LastActivityDate": "08/06/2021", "TotalViews": 11264, "TotalDownloads": 1480, "TotalVotes": 43, "TotalKernels": 9}]
|
[{"Id": 5154008, "UserName": "ashfakyeafi", "DisplayName": "Ashfak Yeafi", "RegisterDate": "05/24/2020", "PerformanceTier": 3}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Load Dataset
df = pd.read_csv(
"/kaggle/input/air-passenger-data-for-time-series-analysis/AirPassengers.csv"
)
df
# # Exploratory Data Analysis (EDA)
# ## View Dataset Description
df["Month"] = pd.to_datetime(df["Month"], format="%Y-%m")
# df['Month'] = df['Month'].dt.strftime('%Y-%m')
df.info()
df
# ## Change Data Index
df.set_index("Month", inplace=True)
# ## Data Visualization
import matplotlib.pyplot as plt
import datetime
plt.figure(figsize=(12, 6))
plt.plot(df)
plt.xlabel("Time")
# plt.xticks(rotation=45)
plt.ylabel("Num of Passengers")
plt.title("US Airline Num of Passengers Trend 1949 - 1960")
plt.show()
# There is a positive trend with some repetitive pattern
# # Time Series Decomposition
from statsmodels.tsa.seasonal import seasonal_decompose
from dateutil.parser import parse
# ## Additive Decomposition
additive_dec = seasonal_decompose(df, model="additive", period=30)
plt.figure(figsize=(12, 8))
additive_dec.plot()
plt.suptitle("Additive Decomposition", fontsize=12)
plt.tight_layout()
plt.show()
multiplicative_dec = seasonal_decompose(df, model="multiplicative", period=30)
plt.figure(figsize=(12, 8))
multiplicative_dec.plot()
plt.suptitle("Multiplicative Decomposition", fontsize=12)
plt.tight_layout()
plt.show()
# Residual in additive decomposition still have a pattern, while in multiplicative is not really showing and quite random. So we will preferred to use multiplicative decomposition.
# # Stationary Test for Time Series
from statsmodels.tsa.stattools import adfuller, kpss
from statsmodels.graphics.tsaplots import plot_acf
# ## Augmented Dickey Fuller Test (ADF Test)
# H0: time series data is non-stationary
# H1: time series data is stationary
# p-value reject null hypothesis (H0)
result = adfuller(df.values, autolag="AIC")
print(f"ADF Statistic: {result[0]}")
print(f"p-value: {result[1]}")
# ## KPSS Test
# H0: time series data is stationary
# H1: time series data is non-stationary
# p-value reject null hypothesis (H0)
result = kpss(df)
print("KPSS Statistic:", result[0])
print("p-value:", result[1])
# ## Rolling Test
# plt.plot(df['Month'], df['#Passengers'])
rolling_mean = df.rolling(6).mean()
rolling_std = df.rolling(6).std()
plt.plot(df, label="Passenger Data")
plt.plot(rolling_mean, color="red", label="Rolling Num of Passenger Mean")
plt.plot(
rolling_std, color="green", label="Rolling Passenger Number Standard Deviation"
)
plt.xlabel("Time")
plt.title("Passenger Time Series, Rolling Mean, Standard Deviation")
plt.legend(loc="best")
plt.show()
# From two test result above, We can see that current data is non-stationary
# # Split Data
# df['date'] = df.index
# train = df[df['date'] < pd.to_datetime("1960-08", format='%Y-%m')]
# train['train'] = train['#Passengers']
# del train['date']
# del train['#Passengers']
# test = df[df['date'] >= pd.to_datetime("1960-08", format='%Y-%m')]
# del test['date']
# test['test'] = test['#Passengers']
# del test['#Passengers']
# plt.plot(train, color = "black")
# plt.plot(test, color = "red")
# plt.title("Train/Test split for Passenger Data")
# plt.ylabel("Passenger Number")
# plt.xlabel('Year-Month')
# plt.show()
# # Preprocessing Data
# ## Convert to Stationary Time Series
# Decompose time series data using moving average
dfs = np.log(df)
# use window 12 to represent 12 months
ma = dfs.rolling(window=12).mean()
ms = dfs.rolling(window=12).std()
plt.plot(ma, c="r")
plt.plot(ms, c="b")
plt.plot(dfs)
# plt.grid()
plt.show()
dfs - np.log(df)
ma = dfs.rolling(window=12, center=False).mean()
dfs2 = dfs - ma
dfs2 = dfs2.dropna()
rolling_mean = dfs2.rolling(6).mean()
rolling_std = dfs2.rolling(6).std()
plt.plot(dfs2, label="Passenger Data")
plt.plot(rolling_mean, color="red", label="Rolling Num of Passenger Mean")
plt.plot(
rolling_std, color="green", label="Rolling Passenger Number Standard Deviation"
)
plt.xlabel("Time")
plt.title("Passenger Time Series, Rolling Mean, Standard Deviation")
plt.legend(loc="best")
plt.show()
# # Autocorrelation & Partial Autocorrelation
# Measure how correlated time series data is at a given point in time with past values.
from statsmodels.tsa.stattools import acf, pacf
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
# autocorr_lag1 = df['#Passengers'].autocorr(lag=1)
# print("One Month Lag: ", autocorr_lag1)
# autocorr_lag3 = df['#Passengers'].autocorr(lag=3)
# print("Three Month Lag: ", autocorr_lag3)
# autocorr_lag6 = df['#Passengers'].autocorr(lag=6)
# print("Six Month Lag: ", autocorr_lag6)
# autocorr_lag9 = df['#Passengers'].autocorr(lag=9)
# print("Nine Month Lag: ", autocorr_lag9)
# Result above show us data is higly correlated
# pd.plotting.autocorrelation_plot(df['#Passengers'])
plt.plot(
np.arange(acf(dfs2, nlags=10, fft=True).shape[0]), acf(dfs2, nlags=10, fft=True)
)
plt.axhline(y=0, linestyle="--", c="gray")
plt.axhline(y=-7.96 / np.sqrt(len(dfs2)), linestyle="--", c="gray")
plt.axhline(y=7.96 / np.sqrt(len(dfs2)), linestyle="--", c="gray")
plt.title("Auto Correlation Fuction")
plt.grid()
plt.show()
plt.plot(np.arange(pacf(dfs2, nlags=10).shape[0]), pacf(dfs2, nlags=10))
plt.axhline(y=0, linestyle="--", c="gray")
plt.axhline(y=-7.96 / np.sqrt(len(dfs2)), linestyle="--", c="gray")
plt.axhline(y=7.96 / np.sqrt(len(dfs2)), linestyle="--", c="gray")
plt.title("Auto Correlation Fuction")
plt.grid()
plt.show()
# # Draw Plot
# fig, axes = plt.subplots(1,2,figsize=(16,3), dpi= 100)
# plot_acf(df['#Passengers'].tolist(), lags=50, ax=axes[0])
# plot_pacf(df['#Passengers'].tolist(), lags=50, ax=axes[1])
# ## Splitting Data
train = dfs2.loc[:"1959"]
test = dfs2.loc["1960":]
# train.rename({'#Passenger':''})
# # Model Building
from statsmodels.tsa.arima.model import ARIMA
Arima = ARIMA(train, order=(3, 1, 3))
Ar = Arima.fit()
Ar.summary()
# Plot residual errors
residuals = pd.DataFrame(Ar.resid)
fig, ax = plt.subplots(1, 2)
residuals.plot(title="Residuals", ax=ax[0])
residuals.plot(kind="kde", title="Density", ax=ax[1])
plt.show()
# # Evaluation
from statsmodels.graphics.tsaplots import plot_predict
def forecast_accuracy(forecast, actual):
mape = np.mean(np.abs(forecast - actual) / np.abs(actual)) # MAPE
me = np.mean(forecast - actual) # ME
mae = np.mean(np.abs(forecast - actual)) # MAE
mpe = np.mean((forecast - actual) / actual) # MPE
rmse = np.mean((forecast - actual) ** 2) ** 0.5 # RMSE
corr = np.corrcoef(forecast, actual)[0, 1] # corr
mins = np.amin(np.hstack([forecast[:, None], actual[:, None]]), axis=1)
maxs = np.amax(np.hstack([forecast[:, None], actual[:, None]]), axis=1)
minmax = 1 - np.mean(mins / maxs) # minmax
acf1 = acf(fc - test)[1] # ACF1
return {
"mape": mape,
"me": me,
"mae": mae,
"mpe": mpe,
"rmse": rmse,
"acf1": acf1,
"corr": corr,
"minmax": minmax,
}
# Forecast
fc = Ar.forecast(12, alpha=0.05) # 95% conf
fc = pd.DataFrame(fc).rename({"predicted_mean": "#Passengers"}, axis=1)
# Make as pandas series
# fc_series = pd.Series(fc, index=test.index)
# lower_series = pd.Series(conf[:, 0], index=test.index)
# upper_series = pd.Series(conf[:, 1], index=test.index)
fig, ax = plt.subplots(figsize=(12, 6))
ax = dfs2.plot(ax=ax, label="train time series")
ax.set_label("sdf")
ax = test.plot(ax=ax, label="test time series")
# plot_predict(Ar, '1959', '1961', ax=ax)
ax = fc.plot(ax=ax)
ax = plt.legend(loc="best")
plt.show()
print("mape", np.mean(np.abs(fc - test) / np.abs(test))) # MAPE)
print("rmse", np.mean((fc - test) ** 2) ** 0.5) # RMSE
# ## Auto ARIMA
# !pip install pmdarima
import pmdarima as pm
model = pm.auto_arima(
train.values,
start_p=1,
start_q=1,
test="adf", # use adftest to find optimal 'd'
max_p=3,
max_q=3, # maximum p and q
m=1, # frequency of series
d=None, # let model determine 'd'
seasonal=False, # No Seasonality
start_P=0,
D=0,
trace=True,
error_action="ignore",
suppress_warnings=True,
stepwise=True,
)
print(model.summary())
# Forecast
n_periods = 12
fitted, confint = model.predict(n_periods=n_periods, return_conf_int=True)
index_of_fc = pd.date_range(dfs2.index[-1], periods=n_periods, freq="MS")
# make series for plotting purpose
fitted_series = pd.Series(fitted, index=index_of_fc)
lower_series = pd.Series(confint[:, 0], index=index_of_fc)
upper_series = pd.Series(confint[:, 1], index=index_of_fc)
fig, ax = plt.subplots(figsize=(12, 6))
ax = train.plot(ax=ax, label="train time series")
ax.set_label("sdf")
ax = test.plot(ax=ax, label="test time series")
# plot_predict(model, '1959', '1961', ax=ax)
ax = fitted_series.plot(ax=ax)
ax = plt.legend(loc="best")
plt.show()
# Plot
plt.plot(dfs2)
plt.plot(fitted_series, color="darkgreen")
plt.fill_between(lower_series.index, lower_series, upper_series, color="k", alpha=0.15)
plt.title("SARIMA - Final Forecast of a10 - Drug Sales")
plt.show()
# ## SARIMA
# Seasonal - fit stepwise auto-ARIMA
smodel = pm.auto_arima(
dfs2,
start_p=1,
start_q=1,
test="adf",
max_p=3,
max_q=3,
m=12,
start_P=0,
seasonal=True,
d=None,
D=1,
trace=True,
error_action="ignore",
suppress_warnings=True,
stepwise=True,
)
smodel.summary()
# Plot
plt.plot(dfs2)
plt.plot(fitted_series, color="darkgreen")
plt.fill_between(lower_series.index, lower_series, upper_series, color="k", alpha=0.15)
plt.title("SARIMA - Final Forecast of a10 - Drug Sales")
plt.show()
| false | 1 | 3,674 | 0 | 3,742 | 3,674 |
||
129727919
|
# # import 資料集
import pandas as pd
train = pd.read_csv("/kaggle/input/titanic/train.csv", encoding="utf-8")
test = pd.read_csv("/kaggle/input/titanic/test.csv", encoding="utf-8")
# check df
train.columns.tolist(), test.columns.tolist()
# # 資料預處理
# 1. 處理缺失值
# 1. 數值型 -> 填補中位數
# 2. 類別型 -> 填補最常出現的類別
# 2. One-Hot
# 3. 萃取更多欄位資訊
total = pd.concat([train, test], axis=0)
total = total.drop(["PassengerId", "Survived"], axis=1)
total
# Cabin
def cabin_preprocess(c):
if pd.isna(c):
return c
else:
return c[0]
total["Cabin"].apply(cabin_preprocess)
total["Cabin"] = total["Cabin"].apply(cabin_preprocess)
# Ticket
ticket_count = total["Ticket"].value_counts() # 多少人持有同一張票類
def ticket_preprocess(t):
if pd.isna(t):
return t
else:
return ticket_count[t]
total["Ticket"].apply(ticket_preprocess)
total["Ticket"] = total["Ticket"].apply(ticket_preprocess)
# Name
total["Name"].isna().value_counts() # 確認姓名無空值
def name_preprocess(n):
return n.split(".")[0].split(",")[-1]
total["Name"].apply(name_preprocess)
total["Name"] = total["Name"].apply(name_preprocess)
# 找尋數值類別的缺失值 -> 填補中位數
total.median() # Pclass 是不是 數值型類別 可再研究 (這邊先當類別型)
median = total.median().drop("Pclass")
total.fillna(median)
total = total.fillna(median)
# Embarked
# 類別型 -> 填補最常出現的值
total["Embarked"].value_counts()
most = total["Embarked"].value_counts().idxmax()
total["Embarked"].fillna(most)
total["Embarked"] = total["Embarked"].fillna(most)
# 類別型資料 需額外再處理 -> One hot encoding
# 剛剛 drop 的 "Pclass" 有必要做嗎? (需實際測試過, 才知道效果)
# Sex 二值型資料 可做可不做
# Name -> One hot encoding
name_count = total["Name"].value_counts()
name_count[name_count > 10]
name_reserved = name_count[name_count > 10].index
def name_onehot(n):
if n in name_reserved:
return n
else:
return None
total["Name"].apply(name_onehot)
total["Name"] = total["Name"].apply(name_onehot)
# One hot encoding
total = pd.get_dummies(total) # 一次對所有字串類的資料 one hot encoding
# Pclass -> One hot encoding
total = pd.get_dummies(total, columns=["Pclass"]) # Pclass資料型態為數字, 額外one hot
total
# 額外增加Family欄位 -> 避免決策樹 僅單獨考慮 SibSp or ParCh 而沒有考慮到家人總數
total["Family"] = total["SibSp"] + total["Parch"]
# 資料分類
import numpy as np
x = np.array(total.iloc[: len(train)]) # x_train
y = np.array(train["Survived"]) # y_train
x_predict = np.array(total.iloc[len(train) :])
x.shape, y.shape, x_predict.shape
# 確認所有欄位有無空值
total.isna().sum()
total.isna().sum()[total.isna().sum() != 0]
# # 額外補充
# 1. Pandas 篩選操作
# 2. loc vs iloc
# 3. 資料分類方式
# Pandas 篩選操作
test_df = pd.DataFrame([[1, 2], [3, 4], [5, 6]])
test_df[[True, False, True]]
# loc vs iloc
test_df = pd.DataFrame([[1, 2], [3, 4]], index=[0, 0])
test_df.loc[0] # 會將兩列都取出 (因兩列的index都為0)
test_df.iloc[0] # 僅取出index為0的第一列
# 資料分類方式
# 分兩份 -> train / test -> 依照test結果, 調整模型參數
# 分三份 -> train / test / valid -> 同上, 但最後用完全沒看過的 valid 驗證模型
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/727/129727919.ipynb
| null | null |
[{"Id": 129727919, "ScriptId": 38575404, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5948419, "CreationDate": "05/16/2023 04:21:31", "VersionNumber": 1.0, "Title": "Titanic", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 140.0, "LinesInsertedFromPrevious": 140.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # import 資料集
import pandas as pd
train = pd.read_csv("/kaggle/input/titanic/train.csv", encoding="utf-8")
test = pd.read_csv("/kaggle/input/titanic/test.csv", encoding="utf-8")
# check df
train.columns.tolist(), test.columns.tolist()
# # 資料預處理
# 1. 處理缺失值
# 1. 數值型 -> 填補中位數
# 2. 類別型 -> 填補最常出現的類別
# 2. One-Hot
# 3. 萃取更多欄位資訊
total = pd.concat([train, test], axis=0)
total = total.drop(["PassengerId", "Survived"], axis=1)
total
# Cabin
def cabin_preprocess(c):
if pd.isna(c):
return c
else:
return c[0]
total["Cabin"].apply(cabin_preprocess)
total["Cabin"] = total["Cabin"].apply(cabin_preprocess)
# Ticket
ticket_count = total["Ticket"].value_counts() # 多少人持有同一張票類
def ticket_preprocess(t):
if pd.isna(t):
return t
else:
return ticket_count[t]
total["Ticket"].apply(ticket_preprocess)
total["Ticket"] = total["Ticket"].apply(ticket_preprocess)
# Name
total["Name"].isna().value_counts() # 確認姓名無空值
def name_preprocess(n):
return n.split(".")[0].split(",")[-1]
total["Name"].apply(name_preprocess)
total["Name"] = total["Name"].apply(name_preprocess)
# 找尋數值類別的缺失值 -> 填補中位數
total.median() # Pclass 是不是 數值型類別 可再研究 (這邊先當類別型)
median = total.median().drop("Pclass")
total.fillna(median)
total = total.fillna(median)
# Embarked
# 類別型 -> 填補最常出現的值
total["Embarked"].value_counts()
most = total["Embarked"].value_counts().idxmax()
total["Embarked"].fillna(most)
total["Embarked"] = total["Embarked"].fillna(most)
# 類別型資料 需額外再處理 -> One hot encoding
# 剛剛 drop 的 "Pclass" 有必要做嗎? (需實際測試過, 才知道效果)
# Sex 二值型資料 可做可不做
# Name -> One hot encoding
name_count = total["Name"].value_counts()
name_count[name_count > 10]
name_reserved = name_count[name_count > 10].index
def name_onehot(n):
if n in name_reserved:
return n
else:
return None
total["Name"].apply(name_onehot)
total["Name"] = total["Name"].apply(name_onehot)
# One hot encoding
total = pd.get_dummies(total) # 一次對所有字串類的資料 one hot encoding
# Pclass -> One hot encoding
total = pd.get_dummies(total, columns=["Pclass"]) # Pclass資料型態為數字, 額外one hot
total
# 額外增加Family欄位 -> 避免決策樹 僅單獨考慮 SibSp or ParCh 而沒有考慮到家人總數
total["Family"] = total["SibSp"] + total["Parch"]
# 資料分類
import numpy as np
x = np.array(total.iloc[: len(train)]) # x_train
y = np.array(train["Survived"]) # y_train
x_predict = np.array(total.iloc[len(train) :])
x.shape, y.shape, x_predict.shape
# 確認所有欄位有無空值
total.isna().sum()
total.isna().sum()[total.isna().sum() != 0]
# # 額外補充
# 1. Pandas 篩選操作
# 2. loc vs iloc
# 3. 資料分類方式
# Pandas 篩選操作
test_df = pd.DataFrame([[1, 2], [3, 4], [5, 6]])
test_df[[True, False, True]]
# loc vs iloc
test_df = pd.DataFrame([[1, 2], [3, 4]], index=[0, 0])
test_df.loc[0] # 會將兩列都取出 (因兩列的index都為0)
test_df.iloc[0] # 僅取出index為0的第一列
# 資料分類方式
# 分兩份 -> train / test -> 依照test結果, 調整模型參數
# 分三份 -> train / test / valid -> 同上, 但最後用完全沒看過的 valid 驗證模型
| false | 0 | 1,288 | 0 | 1,288 | 1,288 |
||
129727843
|
# **concatenate 2 strings index wise**
name = []
fn = ["jasbir", "isha", "yashi"]
ln = ["eden", "singh", "srivastava"]
for i in range(len(ln)):
n = fn[i] + " " + ln[i]
name.append(n)
print(name)
# **how many times a particular item is in list**
l = [2, 3, 4, 5, 6, 2, 34, 6, 8, 0]
a = int(input("enter the number"))
count = 0
for i in l:
if i == a:
count = count + 1
print(a, "is occuring ", count, "times")
# **remove the occurence of a particular number from the list**
l = [2, 3, 4, 5, 6, 2, 34, 6, 8, 0]
a = int(input("enter the number"))
for i in l:
if i == a:
l.remove(i)
print(l)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/727/129727843.ipynb
| null | null |
[{"Id": 129727843, "ScriptId": 38579558, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15022629, "CreationDate": "05/16/2023 04:20:36", "VersionNumber": 1.0, "Title": "list assignment 1", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 30.0, "LinesInsertedFromPrevious": 30.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# **concatenate 2 strings index wise**
name = []
fn = ["jasbir", "isha", "yashi"]
ln = ["eden", "singh", "srivastava"]
for i in range(len(ln)):
n = fn[i] + " " + ln[i]
name.append(n)
print(name)
# **how many times a particular item is in list**
l = [2, 3, 4, 5, 6, 2, 34, 6, 8, 0]
a = int(input("enter the number"))
count = 0
for i in l:
if i == a:
count = count + 1
print(a, "is occuring ", count, "times")
# **remove the occurence of a particular number from the list**
l = [2, 3, 4, 5, 6, 2, 34, 6, 8, 0]
a = int(input("enter the number"))
for i in l:
if i == a:
l.remove(i)
print(l)
| false | 0 | 266 | 0 | 266 | 266 |
||
129360459
|
# # Анализ пожаров в Республике Коми. Методы кластеризации
# # Сделаем предобработку
import pandas as pd
import numpy as np
# ## Рассмотрим данные по пожарам в Республике Коми с 2010 по 2022 годы
main = pd.read_excel("/kaggle/input/fires-komi/fires.xlsx", "main")
main.head()
# # ОПИСАНИЕ НАБОРА.
# Каждый объект в массиве содержит следующие ключи:
# * a. id – идентификатор пожара (целое число)
# * b. latitude – широта (WGS-84) (вещественное число)
# * c. longitude – долгота (WGS-84) (вещественное число)
# * d. aviaDivisionCode – код авиаотделения (строка)
# * e. aviaDivisionName – наименование авиаотделения (строка)
# * f. number – номер пожара по авиаотделению (целое число)
# * g. district – наименование административного района (строка)
# * h. forestry – наименование лесничества (строка)
# * i. divForestry – наименование участкового лесничества (строка)
# * j. quarter – номер лесного квартала (строка)
# * k. detectDate – дата обнаружения пожара (строка в формате «DD.MM.YYYY»)
# * l. statusCode – код состояния пожара (целое число)
# * m. statusName – наименование статуса (строка)
# * n. area – площадь пожара в га (вещественное число)
# * o. totalMark – сводный показатель (целое число)
# * p. isCritical – признак критичности пожара (булево значение)
# * q. isThreat – признак угрожающего пожара (булево значение)
# * r. serviceUrl – адрес сервиса данных (строка)
#
main["district"].unique()
main = main.dropna(subset=["district"])
# ## Рассмотрим пожароопасный сезон, с мая по октябрь. В зависимости от распределения количества пожаров по месяцам пожароопасного периода выявим кластеры.
# ## Кластеры -- это группы похожих объектов.
# ## **Будем использовать кластеризацию по методу K-means (K-средних).**
# ## Алгоритм К-средних, наверное, самый популярный и простой алгоритм кластеризации и очень легко представляется в виде простого псевдокода:
# * ### Выбрать количество кластеров $k$, которое нам кажется оптимальным для наших данных.
# * ### Высыпать случайным образом в пространство наших данных $k$ точек (центроидов).
# * ### Для каждой точки нашего набора данных посчитать, к какому центроиду она ближе.
# * ### Переместить каждый центроид в центр выборки, которую мы отнесли к этому центроиду.
# * ### Повторять последние два шага фиксированное число раз, либо до тех пор пока центроиды не "сойдутся"
# ### (обычно это значит, что их смещение относительно предыдущего положения не превышает какого-то заранее заданного небольшого значения).
#
main.head()
main["detectDate_1"] = main["detectDate"].apply(
lambda x: pd.to_datetime(x, format="%d.%m.%Y", errors="ignore")
)
main["detectTime_1"] = main["detectTime"].apply(
lambda x: pd.to_datetime(x, format="%H:%M", errors="ignore")
)
main["month"] = main["detectDate_1"].dt.month
main["hour"] = main["detectTime_1"].dt.hour
main["year"] = main["detectDate_1"].dt.year
main["year"].unique()
main.head()
# # Статистика по месяцам: число пожаров по месяцам
# ## Рассмотрим месяцы с мая по сентябрь.
main = main[(main["month"] >= 5) & (main["month"] <= 9)][["month", "year", "district"]]
# main[main["district"]=="Удорский"].head(20)
# ## Сгруппируем данные по районам, по месяцам и годам и найдем число пожаров в каждом районе по месяцам и годам
df_cnt = main.groupby(["month", "year", "district"])[["district"]].size()
df_cnt = df_cnt.reset_index(name="cnt")
df_cnt.head(15)
# ## Найдем среднее число пожаров в каждом месяце по всем годам в каждом районе
df_cnt1 = (
df_cnt[["month", "district", "cnt"]]
.groupby(["district", "month"])["cnt"]
.mean()
.reset_index(name="cnt")
)
df_cnt1.head(15)
# ## Для каждого района сохраним его название и число пожаров в каждом месяце в отдельном списке.
# ## Из полученных списков создадим новый набор данных
month_names = ["Май", "Июнь", "Июль", "Август", "Сентябрь"]
district_names = df_cnt1["district"].unique()
l = []
for i, x in enumerate(
district_names
): # для каждого района сохраним его название и число пожаров в каждом месяце в списке
l.append([x]) # название района
for y in range(5, 10): # цикл по месяцам
t = df_cnt1[(df_cnt1["district"] == x) & (df_cnt1["month"] == y)]["cnt"]
if t.size > 0:
l[i].append(t.values[0])
else:
l[i].append(0)
df_2 = pd.DataFrame(l, columns=["Район"] + month_names)
df_2.head()
df_3 = df_2.transpose()
df_3.head()
df_3.columns = df_3.iloc[0]
df_3 = df_3.iloc[1:]
df_3.head()
df_3.plot(
figsize=(11, 11),
xlabel="месяц",
ylabel="среднее количество лесных пожаров",
ls="--",
)
df_3.plot(
kind="bar",
figsize=(11, 11),
xlabel="месяц",
ylabel="среднее количество лесных пожаров",
)
# # Сделаем кластеризацию. Разделим районы на 3 кластера и нарисуем диаграмму
from sklearn.cluster import KMeans # , MeanShift, AgglomerativeClustering
kmeans = KMeans(n_clusters=3, random_state=50, max_iter=100)
pred = kmeans.fit_predict(df_2[month_names])
import matplotlib.pyplot as plt
a = plt.plot(kmeans.cluster_centers_[0], label="1 кластер", ls="--")
b = plt.plot(kmeans.cluster_centers_[1], label="2 кластер", ls="--")
c = plt.plot(kmeans.cluster_centers_[2], label="3 кластер", ls="--")
plt.xlabel("месяц")
plt.ylabel("среднее количество пожаров")
plt.xticks(np.arange(5), month_names) # Set text labels.
plt.legend()
for x, y in zip(district_names, list(pred)):
print(x, y + 1)
# # Спроецируем многомерные данные на плоскость.
from sklearn.decomposition import PCA
pca2D = PCA(n_components=2)
df2 = pca2D.fit_transform(df_2[month_names])
for i, component in enumerate(pca2D.components_):
print(
"{} component: {}% of initial variance".format(
i + 1, round(100 * pca2D.explained_variance_ratio_[i], 2)
)
)
print(
" + ".join(
"%.3f x %s" % (value, name)
for value, name in zip(component, df_2[month_names].columns)
)
)
"""
from sklearn.cluster import KMeans #, MeanShift, AgglomerativeClustering
kmeans = KMeans(n_clusters= 3, random_state=50,
max_iter = 100)
"""
# pred = kmeans.fit_predict(df2)
# Координата x больше всего связана с числом пожаров в июле, а координата y - в августе
# fig.plot(10,10)
import matplotlib.pyplot as plt
plt.scatter(df2[:, 0], df2[:, 1], c=pred)
for x, y, z in zip(df2[:, 0], df2[:, 1], district_names):
plt.annotate(z, (x, y))
kmeans.cluster_centers_
x_axis = np.arange(5)
plt.bar(x_axis, kmeans.cluster_centers_[0], width=0.2, label="1")
plt.bar(x_axis + 0.2, kmeans.cluster_centers_[1], width=0.2, label="2")
plt.bar(x_axis + 0.4, kmeans.cluster_centers_[2], width=0.2, label="3")
plt.xticks(x_axis, month_names)
plt.legend()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/360/129360459.ipynb
| null | null |
[{"Id": 129360459, "ScriptId": 38460843, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3323841, "CreationDate": "05/13/2023 05:59:33", "VersionNumber": 1.0, "Title": "clusters", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 199.0, "LinesInsertedFromPrevious": 199.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Анализ пожаров в Республике Коми. Методы кластеризации
# # Сделаем предобработку
import pandas as pd
import numpy as np
# ## Рассмотрим данные по пожарам в Республике Коми с 2010 по 2022 годы
main = pd.read_excel("/kaggle/input/fires-komi/fires.xlsx", "main")
main.head()
# # ОПИСАНИЕ НАБОРА.
# Каждый объект в массиве содержит следующие ключи:
# * a. id – идентификатор пожара (целое число)
# * b. latitude – широта (WGS-84) (вещественное число)
# * c. longitude – долгота (WGS-84) (вещественное число)
# * d. aviaDivisionCode – код авиаотделения (строка)
# * e. aviaDivisionName – наименование авиаотделения (строка)
# * f. number – номер пожара по авиаотделению (целое число)
# * g. district – наименование административного района (строка)
# * h. forestry – наименование лесничества (строка)
# * i. divForestry – наименование участкового лесничества (строка)
# * j. quarter – номер лесного квартала (строка)
# * k. detectDate – дата обнаружения пожара (строка в формате «DD.MM.YYYY»)
# * l. statusCode – код состояния пожара (целое число)
# * m. statusName – наименование статуса (строка)
# * n. area – площадь пожара в га (вещественное число)
# * o. totalMark – сводный показатель (целое число)
# * p. isCritical – признак критичности пожара (булево значение)
# * q. isThreat – признак угрожающего пожара (булево значение)
# * r. serviceUrl – адрес сервиса данных (строка)
#
main["district"].unique()
main = main.dropna(subset=["district"])
# ## Рассмотрим пожароопасный сезон, с мая по октябрь. В зависимости от распределения количества пожаров по месяцам пожароопасного периода выявим кластеры.
# ## Кластеры -- это группы похожих объектов.
# ## **Будем использовать кластеризацию по методу K-means (K-средних).**
# ## Алгоритм К-средних, наверное, самый популярный и простой алгоритм кластеризации и очень легко представляется в виде простого псевдокода:
# * ### Выбрать количество кластеров $k$, которое нам кажется оптимальным для наших данных.
# * ### Высыпать случайным образом в пространство наших данных $k$ точек (центроидов).
# * ### Для каждой точки нашего набора данных посчитать, к какому центроиду она ближе.
# * ### Переместить каждый центроид в центр выборки, которую мы отнесли к этому центроиду.
# * ### Повторять последние два шага фиксированное число раз, либо до тех пор пока центроиды не "сойдутся"
# ### (обычно это значит, что их смещение относительно предыдущего положения не превышает какого-то заранее заданного небольшого значения).
#
main.head()
main["detectDate_1"] = main["detectDate"].apply(
lambda x: pd.to_datetime(x, format="%d.%m.%Y", errors="ignore")
)
main["detectTime_1"] = main["detectTime"].apply(
lambda x: pd.to_datetime(x, format="%H:%M", errors="ignore")
)
main["month"] = main["detectDate_1"].dt.month
main["hour"] = main["detectTime_1"].dt.hour
main["year"] = main["detectDate_1"].dt.year
main["year"].unique()
main.head()
# # Статистика по месяцам: число пожаров по месяцам
# ## Рассмотрим месяцы с мая по сентябрь.
main = main[(main["month"] >= 5) & (main["month"] <= 9)][["month", "year", "district"]]
# main[main["district"]=="Удорский"].head(20)
# ## Сгруппируем данные по районам, по месяцам и годам и найдем число пожаров в каждом районе по месяцам и годам
df_cnt = main.groupby(["month", "year", "district"])[["district"]].size()
df_cnt = df_cnt.reset_index(name="cnt")
df_cnt.head(15)
# ## Найдем среднее число пожаров в каждом месяце по всем годам в каждом районе
df_cnt1 = (
df_cnt[["month", "district", "cnt"]]
.groupby(["district", "month"])["cnt"]
.mean()
.reset_index(name="cnt")
)
df_cnt1.head(15)
# ## Для каждого района сохраним его название и число пожаров в каждом месяце в отдельном списке.
# ## Из полученных списков создадим новый набор данных
month_names = ["Май", "Июнь", "Июль", "Август", "Сентябрь"]
district_names = df_cnt1["district"].unique()
l = []
for i, x in enumerate(
district_names
): # для каждого района сохраним его название и число пожаров в каждом месяце в списке
l.append([x]) # название района
for y in range(5, 10): # цикл по месяцам
t = df_cnt1[(df_cnt1["district"] == x) & (df_cnt1["month"] == y)]["cnt"]
if t.size > 0:
l[i].append(t.values[0])
else:
l[i].append(0)
df_2 = pd.DataFrame(l, columns=["Район"] + month_names)
df_2.head()
df_3 = df_2.transpose()
df_3.head()
df_3.columns = df_3.iloc[0]
df_3 = df_3.iloc[1:]
df_3.head()
df_3.plot(
figsize=(11, 11),
xlabel="месяц",
ylabel="среднее количество лесных пожаров",
ls="--",
)
df_3.plot(
kind="bar",
figsize=(11, 11),
xlabel="месяц",
ylabel="среднее количество лесных пожаров",
)
# # Сделаем кластеризацию. Разделим районы на 3 кластера и нарисуем диаграмму
from sklearn.cluster import KMeans # , MeanShift, AgglomerativeClustering
kmeans = KMeans(n_clusters=3, random_state=50, max_iter=100)
pred = kmeans.fit_predict(df_2[month_names])
import matplotlib.pyplot as plt
a = plt.plot(kmeans.cluster_centers_[0], label="1 кластер", ls="--")
b = plt.plot(kmeans.cluster_centers_[1], label="2 кластер", ls="--")
c = plt.plot(kmeans.cluster_centers_[2], label="3 кластер", ls="--")
plt.xlabel("месяц")
plt.ylabel("среднее количество пожаров")
plt.xticks(np.arange(5), month_names) # Set text labels.
plt.legend()
for x, y in zip(district_names, list(pred)):
print(x, y + 1)
# # Спроецируем многомерные данные на плоскость.
from sklearn.decomposition import PCA
pca2D = PCA(n_components=2)
df2 = pca2D.fit_transform(df_2[month_names])
for i, component in enumerate(pca2D.components_):
print(
"{} component: {}% of initial variance".format(
i + 1, round(100 * pca2D.explained_variance_ratio_[i], 2)
)
)
print(
" + ".join(
"%.3f x %s" % (value, name)
for value, name in zip(component, df_2[month_names].columns)
)
)
"""
from sklearn.cluster import KMeans #, MeanShift, AgglomerativeClustering
kmeans = KMeans(n_clusters= 3, random_state=50,
max_iter = 100)
"""
# pred = kmeans.fit_predict(df2)
# Координата x больше всего связана с числом пожаров в июле, а координата y - в августе
# fig.plot(10,10)
import matplotlib.pyplot as plt
plt.scatter(df2[:, 0], df2[:, 1], c=pred)
for x, y, z in zip(df2[:, 0], df2[:, 1], district_names):
plt.annotate(z, (x, y))
kmeans.cluster_centers_
x_axis = np.arange(5)
plt.bar(x_axis, kmeans.cluster_centers_[0], width=0.2, label="1")
plt.bar(x_axis + 0.2, kmeans.cluster_centers_[1], width=0.2, label="2")
plt.bar(x_axis + 0.4, kmeans.cluster_centers_[2], width=0.2, label="3")
plt.xticks(x_axis, month_names)
plt.legend()
| false | 0 | 2,878 | 0 | 2,878 | 2,878 |
||
129317318
|
<jupyter_start><jupyter_text>Online Chess Games
Chess has been a timeless and beloved board game for centuries, but the online chess community has breathed new life into this classic game. With the rise of online chess platforms, we now have access to an incredible amount of data on chess games, providing endless opportunities for analysis and insight.
This exciting dataset contains information on a vast array of online chess games, including details on the number of turns, the opening used, and the outcome of the game. By delving into this data, enthusiasts and researchers alike can gain a deeper understanding of the strategies and nuances that make chess such a fascinating game. What's more, by exploring the relationships between different openings and the likelihood of winning, we can uncover insights that can help us improve our game and gain a competitive edge.
Whether you're a chess enthusiast, a data scientist, or just someone curious about this incredible game, this dataset is a treasure trove of information that you won't want to miss. So dive in and explore the fascinating world of online chess!
Kaggle dataset identifier: online-chess-games
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Calling the Data and checking info
games = pd.read_csv(
"/kaggle/input/online-chess-games/chess_games.csv", index_col="game_id"
)
games.head()
games.info()
# ## Droping the unwanted Columns
games.drop(
columns=[
"opening_response",
"opening_variation",
"opening_fullname",
"opening_code",
"moves",
],
axis=1,
inplace=True,
)
# ## to make sure analysis are for Rated games only
games = games.loc[games["rated"] == True]
games.drop(columns=["rated"], axis=1, inplace=True)
games
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/317/129317318.ipynb
|
online-chess-games
|
ulrikthygepedersen
|
[{"Id": 129317318, "ScriptId": 38447736, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10854147, "CreationDate": "05/12/2023 18:00:04", "VersionNumber": 1.0, "Title": "Online Chess Games Data Analysis", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 36.0, "LinesInsertedFromPrevious": 36.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185251892, "KernelVersionId": 129317318, "SourceDatasetVersionId": 5220640}]
|
[{"Id": 5220640, "DatasetId": 3037306, "DatasourceVersionId": 5293140, "CreatorUserId": 9580496, "LicenseName": "Attribution 4.0 International (CC BY 4.0)", "CreationDate": "03/23/2023 11:55:17", "VersionNumber": 1.0, "Title": "Online Chess Games", "Slug": "online-chess-games", "Subtitle": "Can you predict which Chess opening is the best?", "Description": "Chess has been a timeless and beloved board game for centuries, but the online chess community has breathed new life into this classic game. With the rise of online chess platforms, we now have access to an incredible amount of data on chess games, providing endless opportunities for analysis and insight.\n\nThis exciting dataset contains information on a vast array of online chess games, including details on the number of turns, the opening used, and the outcome of the game. By delving into this data, enthusiasts and researchers alike can gain a deeper understanding of the strategies and nuances that make chess such a fascinating game. What's more, by exploring the relationships between different openings and the likelihood of winning, we can uncover insights that can help us improve our game and gain a competitive edge.\n\nWhether you're a chess enthusiast, a data scientist, or just someone curious about this incredible game, this dataset is a treasure trove of information that you won't want to miss. So dive in and explore the fascinating world of online chess!", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3037306, "CreatorUserId": 9580496, "OwnerUserId": 9580496.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5220640.0, "CurrentDatasourceVersionId": 5293140.0, "ForumId": 3076770, "Type": 2, "CreationDate": "03/23/2023 11:55:17", "LastActivityDate": "03/23/2023", "TotalViews": 5875, "TotalDownloads": 758, "TotalVotes": 24, "TotalKernels": 7}]
|
[{"Id": 9580496, "UserName": "ulrikthygepedersen", "DisplayName": "Ulrik Thyge Pedersen", "RegisterDate": "02/04/2022", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Calling the Data and checking info
games = pd.read_csv(
"/kaggle/input/online-chess-games/chess_games.csv", index_col="game_id"
)
games.head()
games.info()
# ## Droping the unwanted Columns
games.drop(
columns=[
"opening_response",
"opening_variation",
"opening_fullname",
"opening_code",
"moves",
],
axis=1,
inplace=True,
)
# ## to make sure analysis are for Rated games only
games = games.loc[games["rated"] == True]
games.drop(columns=["rated"], axis=1, inplace=True)
games
| false | 1 | 347 | 0 | 624 | 347 |
||
129317361
|
<jupyter_start><jupyter_text>Brain Tumor Classification (MRI)
# Contribute to OpenSource
##Repo: [GitHub](https://github.com/SartajBhuvaji/Brain-Tumor-Classification-Using-Deep-Learning-Algorithms)
## Read Me: [Link](https://github.com/SartajBhuvaji/Brain-Tumor-Classification-Using-Deep-Learning-Algorithms/tree/master#contributing-to-the-project)
# Abstract
A Brain tumor is considered as one of the aggressive diseases, among children and adults. Brain tumors account for 85 to 90 percent of all primary Central Nervous System(CNS) tumors. Every year, around 11,700 people are diagnosed with a brain tumor. The 5-year survival rate for people with a cancerous brain or CNS tumor is approximately 34 percent for men and36 percent for women. Brain Tumors are classified as: Benign Tumor, Malignant Tumor, Pituitary Tumor, etc. Proper treatment, planning, and accurate diagnostics should be implemented to improve the life expectancy of the patients. The best technique to detect brain tumors is Magnetic Resonance Imaging (MRI). A huge amount of image data is generated through the scans. These images are examined by the radiologist. A manual examination can be error-prone due to the level of complexities involved in brain tumors and their properties.
Application of automated classification techniques using Machine Learning(ML) and Artificial Intelligence(AI)has consistently shown higher accuracy than manual classification. Hence, proposing a system performing detection and classification by using Deep Learning Algorithms using ConvolutionNeural Network (CNN), Artificial Neural Network (ANN), and TransferLearning (TL) would be helpful to doctors all around the world.
### Context
Brain Tumors are complex. There are a lot of abnormalities in the sizes and location of the brain tumor(s). This makes it really difficult for complete understanding of the nature of the tumor. Also, a professional Neurosurgeon is required for MRI analysis. Often times in developing countries the lack of skillful doctors and lack of knowledge about tumors makes it really challenging and time-consuming to generate reports from MRI’. So an automated system on Cloud can solve this problem.
### Definition
To Detect and Classify Brain Tumor using, CNN and TL; as an asset of Deep Learning and to examine the tumor position(segmentation).
Kaggle dataset identifier: brain-tumor-classification-mri
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Load Modules
import os
import cv2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.utils import shuffle
import tensorflow as tf
# # Data Collection
# Combine the Traning and Testing Images into one Dataset
X = [] # Compined list of images
y = [] # Images Labels
image_size = 150 # Make a defaul size for all images
labels = [
"glioma",
"meningioma",
"notumor",
"pituitary",
] # 4 classes to train on and predict
for i in labels:
folderPath = os.path.join(
"/kaggle/input/brain-tumor-mri-dataset/Training", i
) # Open the training folder
for j in os.listdir(folderPath):
img = cv2.imread(
os.path.join(folderPath, j)
) # Read the image from training folder
img = cv2.resize(img, (image_size, image_size)) # Resize the image
X.append(img)
y.append(i)
for i in labels:
folderPath = os.path.join(
"/kaggle/input/brain-tumor-mri-dataset/Training", i
) # Open the testing folder
for j in os.listdir(folderPath):
img = cv2.imread(
os.path.join(folderPath, j)
) # Read the image from testing folder
img = cv2.resize(img, (image_size, image_size)) # Resize the image
X.append(img)
y.append(i)
# Convert the lists to numpy arrays to perform the shuffleing afterwards
X = np.array(X)
y = np.array(y)
X, y = shuffle(X, y, random_state=101)
X.shape
# Checking the number of samples per class
pd.Series(y).value_counts()
# # Data Visualization
plt.imshow(X[0])
# ## Removing the Noise from the Images using Median Filter
original = X[4]
# Apply Median Filter & Gausian
after_median_filter = cv2.medianBlur(
original, 5
) # Median filter with a kernel size of 5x5 using the medianBlur function to remove the noise
after_gaussian_filter = cv2.GaussianBlur(
original, (5, 5), 0
) # # Gaussian filter with a kernel size of 5x5 using the gaussianBlur function to remove the noise
# Create a figure with two subplots
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=3, figsize=(15, 10))
# Show the images on the subplots
ax1.imshow(original)
ax2.imshow(after_median_filter)
ax3.imshow(after_gaussian_filter)
# Add titles to the subplots
ax1.set_title("Original")
ax2.set_title("After Median Filter")
ax3.set_title("After Gaussian Filter")
# Hide the axis ticks and labels
ax1.set_xticks([])
ax1.set_yticks([])
ax2.set_xticks([])
ax2.set_yticks([])
ax3.set_xticks([])
ax3.set_yticks([])
# Show the figure
plt.show()
# ### Sharpening the Image After Median Filter
# Sharpening the Image
after_sharpening_median = cv2.addWeighted(original, 1.5, after_median_filter, -0.5, 0)
# Create a figure with two subplots
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=3, figsize=(15, 10))
# Show the images on the subplots
ax1.imshow(original)
ax2.imshow(after_median_filter)
ax3.imshow(after_sharpening_median)
# Add titles to the subplots
ax1.set_title("Original")
ax2.set_title("After Median Filter")
ax3.set_title("After Sharpeneing")
# Hide the axis ticks and labels
ax1.set_xticks([])
ax1.set_yticks([])
ax2.set_xticks([])
ax2.set_yticks([])
ax3.set_xticks([])
ax3.set_yticks([])
# Show the figure
plt.show()
# ### Sharpening the Image After Gausian Filter
# Sharpening the Image
after_sharpening = cv2.addWeighted(original, 1.5, after_gaussian_filter, -0.5, 0)
# Create a figure with two subplots
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=3, figsize=(15, 10))
# Show the images on the subplots
ax1.imshow(original)
ax2.imshow(after_gaussian_filter)
ax3.imshow(after_sharpening)
# Add titles to the subplots
ax1.set_title("Original")
ax2.set_title("After Gaussian Filter")
ax3.set_title("After Sharpeneing")
# Hide the axis ticks and labels
ax1.set_xticks([])
ax1.set_yticks([])
ax2.set_xticks([])
ax2.set_yticks([])
ax3.set_xticks([])
ax3.set_yticks([])
# Show the figure
plt.show()
# ## Workin on the Image to Apply Watershed Filter
gray = cv2.cvtColor(after_sharpening_median, cv2.COLOR_BGR2GRAY)
# Apply thresholding to the image
ret, thresh = cv2.threshold(gray, 72, 255, cv2.THRESH_BINARY)
# Apply morphological opening to the image
kernel = np.ones((3, 3), np.uint8)
opening = cv2.morphologyEx(thresh, cv2.MORPH_OPEN, kernel, iterations=2)
# Find the background region of the image
sure_bg = cv2.dilate(opening, kernel, iterations=3)
# Find the foreground region of the image
dist_transform = cv2.distanceTransform(opening, cv2.DIST_L2, 5)
ret, sure_fg = cv2.threshold(dist_transform, 0.7 * dist_transform.max(), 255, 0)
# Find the unknown region of the image
sure_fg = np.uint8(sure_fg)
unknown = cv2.subtract(sure_bg, sure_fg)
# Apply the watershed algorithm to the image
ret, markers = cv2.connectedComponents(sure_fg)
markers = markers + 1
markers[unknown == 255] = 0
markers = cv2.watershed(after_sharpening_median, markers)
# Create a mask to highlight the segmented objects
mask = np.zeros(after_sharpening_median.shape, dtype=np.uint8)
mask[markers == -1] = [0, 0, 255]
# Display the original image and the segmented objects side by side
result = cv2.addWeighted(after_sharpening_median, 1.5, mask, 0.2, 0)
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=3, figsize=(20, 15))
ax1.imshow(after_sharpening)
ax2.imshow(thresh)
ax3.imshow(result)
# Add titles to the subplots
ax1.set_title("Original")
ax2.set_title("Threshold Image")
ax3.set_title("After Watershed Segmentation")
# Hide the axis ticks and labels
ax1.set_xticks([])
ax1.set_yticks([])
ax2.set_xticks([])
ax2.set_yticks([])
ax3.set_xticks([])
ax3.set_yticks([])
# Show the figure
plt.show()
# ### Displaying the Images
# # Train test split
X = X.reshape(len(X), -1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.1, random_state=101
)
# Convert the y_train and y_test from text/labels to numerical data
y_train_new = []
for i in y_train:
y_train_new.append(
labels.index(i)
) # Get the index number of i, and add it to new list of y_train
y_train = y_train_new
y_train = tf.keras.utils.to_categorical(
y_train
) # Convert the 1D vector to 2D binarry array
#####################################################################################################
y_test_new = []
for i in y_test:
y_test_new.append(
labels.index(i)
) # Get the index number of i, and add it to new list of y_test
y_test = y_test_new
y_test = tf.keras.utils.to_categorical(
y_test
) # Convert the 1D vector to 2D binarry array
# # Feature Scaling
print("Before Feature Scaling:")
print(X_train.max(), X_train.min())
print(X_test.max(), X_test.min())
X_train = X_train / 255
X_test = X_test / 255
print("\nAfter Feature Scaling:")
print(X_train.max(), X_train.min())
print(X_test.max(), X_test.min())
# # Create,Train, and Evaluate the Model
from sklearn.neighbors import KNeighborsClassifier
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
knn = KNeighborsClassifier(n_neighbors=4)
knn.fit(X_train, y_train)
accuracy = knn.score(X_test, y_test)
print("Accuracy:", accuracy)
# # Training the Model on Dataset without Preprocessing
# Combine the Traning and Testing Images into one Dataset
X = [] # Compined list of images
y = [] # Images Labels
image_size = 150 # Make a defaul size for all images
labels = [
"glioma_tumor",
"meningioma_tumor",
"no_tumor",
"pituitary_tumor",
] # 4 classes to train on and predict
for i in labels:
folderPath = os.path.join(
"../input/brain-tumor-classification-mri/Training", i
) # Open the training folder
for j in os.listdir(folderPath):
img = cv2.imread(
os.path.join(folderPath, j)
) # Read the image from training folder
img = cv2.resize(img, (image_size, image_size)) # Resize the image
X.append(img)
y.append(i)
for i in labels:
folderPath = os.path.join(
"../input/brain-tumor-classification-mri/Testing", i
) # Open the testing folder
for j in os.listdir(folderPath):
img = cv2.imread(
os.path.join(folderPath, j)
) # Read the image from testing folder
img = cv2.resize(img, (image_size, image_size)) # Resize the image
X.append(img)
y.append(i)
# Convert the lists to numpy arrays to perform the shuffleing afterwards
X = np.array(X)
y = np.array(y)
X, y = shuffle(X, y, random_state=101)
X.shape
X = X.reshape(len(X), -1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.1, random_state=101
)
# Convert the y_train and y_test from text/labels to numerical data
y_train_new = []
for i in y_train:
y_train_new.append(
labels.index(i)
) # Get the index number of i, and add it to new list of y_train
y_train = y_train_new
y_train = tf.keras.utils.to_categorical(
y_train
) # Convert the 1D vector to 2D binarry array
#####################################################################################################
y_test_new = []
for i in y_test:
y_test_new.append(
labels.index(i)
) # Get the index number of i, and add it to new list of y_test
y_test = y_test_new
y_test = tf.keras.utils.to_categorical(
y_test
) # Convert the 1D vector to 2D binarry array
knn_pure_data = KNeighborsClassifier(n_neighbors=4)
knn_pure_data.fit(X_train, y_train)
accuracy = knn_pure_data.score(X_test, y_test)
print("Accuracy:", accuracy)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/317/129317361.ipynb
|
brain-tumor-classification-mri
|
sartajbhuvaji
|
[{"Id": 129317361, "ScriptId": 38408485, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8748085, "CreationDate": "05/12/2023 18:00:38", "VersionNumber": 4.0, "Title": "Brain Tumor", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 321.0, "LinesInsertedFromPrevious": 237.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 84.0, "LinesInsertedFromFork": 289.0, "LinesDeletedFromFork": 118.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 32.0, "TotalVotes": 1}]
|
[{"Id": 185251981, "KernelVersionId": 129317361, "SourceDatasetVersionId": 1183165}, {"Id": 185251982, "KernelVersionId": 129317361, "SourceDatasetVersionId": 2645886}]
|
[{"Id": 1183165, "DatasetId": 672377, "DatasourceVersionId": 1214258, "CreatorUserId": 3469060, "LicenseName": "CC0: Public Domain", "CreationDate": "05/24/2020 16:24:55", "VersionNumber": 2.0, "Title": "Brain Tumor Classification (MRI)", "Slug": "brain-tumor-classification-mri", "Subtitle": "Classify MRI images into four classes", "Description": "# Contribute to OpenSource\n##Repo: [GitHub](https://github.com/SartajBhuvaji/Brain-Tumor-Classification-Using-Deep-Learning-Algorithms)\n## Read Me: [Link](https://github.com/SartajBhuvaji/Brain-Tumor-Classification-Using-Deep-Learning-Algorithms/tree/master#contributing-to-the-project)\n\n\n# Abstract\nA Brain tumor is considered as one of the aggressive diseases, among children and adults. Brain tumors account for 85 to 90 percent of all primary Central Nervous System(CNS) tumors. Every year, around 11,700 people are diagnosed with a brain tumor. The 5-year survival rate for people with a cancerous brain or CNS tumor is approximately 34 percent for men and36 percent for women. Brain Tumors are classified as: Benign Tumor, Malignant Tumor, Pituitary Tumor, etc. Proper treatment, planning, and accurate diagnostics should be implemented to improve the life expectancy of the patients. The best technique to detect brain tumors is Magnetic Resonance Imaging (MRI). A huge amount of image data is generated through the scans. These images are examined by the radiologist. A manual examination can be error-prone due to the level of complexities involved in brain tumors and their properties.\n\nApplication of automated classification techniques using Machine Learning(ML) and Artificial Intelligence(AI)has consistently shown higher accuracy than manual classification. Hence, proposing a system performing detection and classification by using Deep Learning Algorithms using ConvolutionNeural Network (CNN), Artificial Neural Network (ANN), and TransferLearning (TL) would be helpful to doctors all around the world.\n\n### Context\n\nBrain Tumors are complex. There are a lot of abnormalities in the sizes and location of the brain tumor(s). This makes it really difficult for complete understanding of the nature of the tumor. Also, a professional Neurosurgeon is required for MRI analysis. Often times in developing countries the lack of skillful doctors and lack of knowledge about tumors makes it really challenging and time-consuming to generate reports from MRI\u2019. So an automated system on Cloud can solve this problem.\n\n\n### Definition\n\nTo Detect and Classify Brain Tumor using, CNN and TL; as an asset of Deep Learning and to examine the tumor position(segmentation).\n\n\n### Acknowledgements for Dataset.\n\nNavoneel Chakrabarty\nSwati Kanchan\n\n### Team\n\nSartaj Bhuvaji\nAnkita Kadam\nPrajakta Bhumkar\nSameer Dedge", "VersionNotes": "Automatic Update 2020-05-24", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 672377, "CreatorUserId": 3469060, "OwnerUserId": 3469060.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1183165.0, "CurrentDatasourceVersionId": 1214258.0, "ForumId": 686859, "Type": 2, "CreationDate": "05/24/2020 16:22:54", "LastActivityDate": "05/24/2020", "TotalViews": 302511, "TotalDownloads": 32508, "TotalVotes": 481, "TotalKernels": 255}]
|
[{"Id": 3469060, "UserName": "sartajbhuvaji", "DisplayName": "Sartaj", "RegisterDate": "07/16/2019", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Load Modules
import os
import cv2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.utils import shuffle
import tensorflow as tf
# # Data Collection
# Combine the Traning and Testing Images into one Dataset
X = [] # Compined list of images
y = [] # Images Labels
image_size = 150 # Make a defaul size for all images
labels = [
"glioma",
"meningioma",
"notumor",
"pituitary",
] # 4 classes to train on and predict
for i in labels:
folderPath = os.path.join(
"/kaggle/input/brain-tumor-mri-dataset/Training", i
) # Open the training folder
for j in os.listdir(folderPath):
img = cv2.imread(
os.path.join(folderPath, j)
) # Read the image from training folder
img = cv2.resize(img, (image_size, image_size)) # Resize the image
X.append(img)
y.append(i)
for i in labels:
folderPath = os.path.join(
"/kaggle/input/brain-tumor-mri-dataset/Training", i
) # Open the testing folder
for j in os.listdir(folderPath):
img = cv2.imread(
os.path.join(folderPath, j)
) # Read the image from testing folder
img = cv2.resize(img, (image_size, image_size)) # Resize the image
X.append(img)
y.append(i)
# Convert the lists to numpy arrays to perform the shuffleing afterwards
X = np.array(X)
y = np.array(y)
X, y = shuffle(X, y, random_state=101)
X.shape
# Checking the number of samples per class
pd.Series(y).value_counts()
# # Data Visualization
plt.imshow(X[0])
# ## Removing the Noise from the Images using Median Filter
original = X[4]
# Apply Median Filter & Gausian
after_median_filter = cv2.medianBlur(
original, 5
) # Median filter with a kernel size of 5x5 using the medianBlur function to remove the noise
after_gaussian_filter = cv2.GaussianBlur(
original, (5, 5), 0
) # # Gaussian filter with a kernel size of 5x5 using the gaussianBlur function to remove the noise
# Create a figure with two subplots
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=3, figsize=(15, 10))
# Show the images on the subplots
ax1.imshow(original)
ax2.imshow(after_median_filter)
ax3.imshow(after_gaussian_filter)
# Add titles to the subplots
ax1.set_title("Original")
ax2.set_title("After Median Filter")
ax3.set_title("After Gaussian Filter")
# Hide the axis ticks and labels
ax1.set_xticks([])
ax1.set_yticks([])
ax2.set_xticks([])
ax2.set_yticks([])
ax3.set_xticks([])
ax3.set_yticks([])
# Show the figure
plt.show()
# ### Sharpening the Image After Median Filter
# Sharpening the Image
after_sharpening_median = cv2.addWeighted(original, 1.5, after_median_filter, -0.5, 0)
# Create a figure with two subplots
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=3, figsize=(15, 10))
# Show the images on the subplots
ax1.imshow(original)
ax2.imshow(after_median_filter)
ax3.imshow(after_sharpening_median)
# Add titles to the subplots
ax1.set_title("Original")
ax2.set_title("After Median Filter")
ax3.set_title("After Sharpeneing")
# Hide the axis ticks and labels
ax1.set_xticks([])
ax1.set_yticks([])
ax2.set_xticks([])
ax2.set_yticks([])
ax3.set_xticks([])
ax3.set_yticks([])
# Show the figure
plt.show()
# ### Sharpening the Image After Gausian Filter
# Sharpening the Image
after_sharpening = cv2.addWeighted(original, 1.5, after_gaussian_filter, -0.5, 0)
# Create a figure with two subplots
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=3, figsize=(15, 10))
# Show the images on the subplots
ax1.imshow(original)
ax2.imshow(after_gaussian_filter)
ax3.imshow(after_sharpening)
# Add titles to the subplots
ax1.set_title("Original")
ax2.set_title("After Gaussian Filter")
ax3.set_title("After Sharpeneing")
# Hide the axis ticks and labels
ax1.set_xticks([])
ax1.set_yticks([])
ax2.set_xticks([])
ax2.set_yticks([])
ax3.set_xticks([])
ax3.set_yticks([])
# Show the figure
plt.show()
# ## Workin on the Image to Apply Watershed Filter
gray = cv2.cvtColor(after_sharpening_median, cv2.COLOR_BGR2GRAY)
# Apply thresholding to the image
ret, thresh = cv2.threshold(gray, 72, 255, cv2.THRESH_BINARY)
# Apply morphological opening to the image
kernel = np.ones((3, 3), np.uint8)
opening = cv2.morphologyEx(thresh, cv2.MORPH_OPEN, kernel, iterations=2)
# Find the background region of the image
sure_bg = cv2.dilate(opening, kernel, iterations=3)
# Find the foreground region of the image
dist_transform = cv2.distanceTransform(opening, cv2.DIST_L2, 5)
ret, sure_fg = cv2.threshold(dist_transform, 0.7 * dist_transform.max(), 255, 0)
# Find the unknown region of the image
sure_fg = np.uint8(sure_fg)
unknown = cv2.subtract(sure_bg, sure_fg)
# Apply the watershed algorithm to the image
ret, markers = cv2.connectedComponents(sure_fg)
markers = markers + 1
markers[unknown == 255] = 0
markers = cv2.watershed(after_sharpening_median, markers)
# Create a mask to highlight the segmented objects
mask = np.zeros(after_sharpening_median.shape, dtype=np.uint8)
mask[markers == -1] = [0, 0, 255]
# Display the original image and the segmented objects side by side
result = cv2.addWeighted(after_sharpening_median, 1.5, mask, 0.2, 0)
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=3, figsize=(20, 15))
ax1.imshow(after_sharpening)
ax2.imshow(thresh)
ax3.imshow(result)
# Add titles to the subplots
ax1.set_title("Original")
ax2.set_title("Threshold Image")
ax3.set_title("After Watershed Segmentation")
# Hide the axis ticks and labels
ax1.set_xticks([])
ax1.set_yticks([])
ax2.set_xticks([])
ax2.set_yticks([])
ax3.set_xticks([])
ax3.set_yticks([])
# Show the figure
plt.show()
# ### Displaying the Images
# # Train test split
X = X.reshape(len(X), -1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.1, random_state=101
)
# Convert the y_train and y_test from text/labels to numerical data
y_train_new = []
for i in y_train:
y_train_new.append(
labels.index(i)
) # Get the index number of i, and add it to new list of y_train
y_train = y_train_new
y_train = tf.keras.utils.to_categorical(
y_train
) # Convert the 1D vector to 2D binarry array
#####################################################################################################
y_test_new = []
for i in y_test:
y_test_new.append(
labels.index(i)
) # Get the index number of i, and add it to new list of y_test
y_test = y_test_new
y_test = tf.keras.utils.to_categorical(
y_test
) # Convert the 1D vector to 2D binarry array
# # Feature Scaling
print("Before Feature Scaling:")
print(X_train.max(), X_train.min())
print(X_test.max(), X_test.min())
X_train = X_train / 255
X_test = X_test / 255
print("\nAfter Feature Scaling:")
print(X_train.max(), X_train.min())
print(X_test.max(), X_test.min())
# # Create,Train, and Evaluate the Model
from sklearn.neighbors import KNeighborsClassifier
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
knn = KNeighborsClassifier(n_neighbors=4)
knn.fit(X_train, y_train)
accuracy = knn.score(X_test, y_test)
print("Accuracy:", accuracy)
# # Training the Model on Dataset without Preprocessing
# Combine the Traning and Testing Images into one Dataset
X = [] # Compined list of images
y = [] # Images Labels
image_size = 150 # Make a defaul size for all images
labels = [
"glioma_tumor",
"meningioma_tumor",
"no_tumor",
"pituitary_tumor",
] # 4 classes to train on and predict
for i in labels:
folderPath = os.path.join(
"../input/brain-tumor-classification-mri/Training", i
) # Open the training folder
for j in os.listdir(folderPath):
img = cv2.imread(
os.path.join(folderPath, j)
) # Read the image from training folder
img = cv2.resize(img, (image_size, image_size)) # Resize the image
X.append(img)
y.append(i)
for i in labels:
folderPath = os.path.join(
"../input/brain-tumor-classification-mri/Testing", i
) # Open the testing folder
for j in os.listdir(folderPath):
img = cv2.imread(
os.path.join(folderPath, j)
) # Read the image from testing folder
img = cv2.resize(img, (image_size, image_size)) # Resize the image
X.append(img)
y.append(i)
# Convert the lists to numpy arrays to perform the shuffleing afterwards
X = np.array(X)
y = np.array(y)
X, y = shuffle(X, y, random_state=101)
X.shape
X = X.reshape(len(X), -1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.1, random_state=101
)
# Convert the y_train and y_test from text/labels to numerical data
y_train_new = []
for i in y_train:
y_train_new.append(
labels.index(i)
) # Get the index number of i, and add it to new list of y_train
y_train = y_train_new
y_train = tf.keras.utils.to_categorical(
y_train
) # Convert the 1D vector to 2D binarry array
#####################################################################################################
y_test_new = []
for i in y_test:
y_test_new.append(
labels.index(i)
) # Get the index number of i, and add it to new list of y_test
y_test = y_test_new
y_test = tf.keras.utils.to_categorical(
y_test
) # Convert the 1D vector to 2D binarry array
knn_pure_data = KNeighborsClassifier(n_neighbors=4)
knn_pure_data.fit(X_train, y_train)
accuracy = knn_pure_data.score(X_test, y_test)
print("Accuracy:", accuracy)
| false | 0 | 3,368 | 1 | 3,997 | 3,368 |
||
129317081
|
# **data exploration**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("fivethirtyeight")
plt.rcParams["figure.figsize"] = (10, 6)
data = pd.read_csv("/kaggle/input/datanetflix/netflix.csv")
data.sample(10)
data.columns
data.head(5)
data.dtypes
data.shape
duplicate_rows_data = data[data.duplicated()]
print("number of duplicate rows: ", duplicate_rows_data.shape)
data.count()
data = data.drop_duplicates()
data.head(5)
data = data.rename(columns={"categorie ": "listed_in"})
data.head(5)
data.count()
print(data.isnull().sum())
data = data.dropna()
print(data.isnull().sum())
data.info()
# Calculez la moyenne des années de sortie pour chaque catégorie
data["release_year_score"] = data.groupby("listed_in")["release_year"].transform("mean")
# Sélectionnez les 5 meilleures catégories
top_categories = data.sort_values("release_year_score", ascending=False).head(5)[
"listed_in"
]
# Affichez les 5 meilleures catégories
print(top_categories)
top_categories.all
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/317/129317081.ipynb
| null | null |
[{"Id": 129317081, "ScriptId": 38440577, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14707911, "CreationDate": "05/12/2023 17:57:06", "VersionNumber": 1.0, "Title": "notebooke73edf15d2", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 68.0, "LinesInsertedFromPrevious": 68.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# **data exploration**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("fivethirtyeight")
plt.rcParams["figure.figsize"] = (10, 6)
data = pd.read_csv("/kaggle/input/datanetflix/netflix.csv")
data.sample(10)
data.columns
data.head(5)
data.dtypes
data.shape
duplicate_rows_data = data[data.duplicated()]
print("number of duplicate rows: ", duplicate_rows_data.shape)
data.count()
data = data.drop_duplicates()
data.head(5)
data = data.rename(columns={"categorie ": "listed_in"})
data.head(5)
data.count()
print(data.isnull().sum())
data = data.dropna()
print(data.isnull().sum())
data.info()
# Calculez la moyenne des années de sortie pour chaque catégorie
data["release_year_score"] = data.groupby("listed_in")["release_year"].transform("mean")
# Sélectionnez les 5 meilleures catégories
top_categories = data.sort_values("release_year_score", ascending=False).head(5)[
"listed_in"
]
# Affichez les 5 meilleures catégories
print(top_categories)
top_categories.all
| false | 0 | 342 | 0 | 342 | 342 |
||
129317577
|
<jupyter_start><jupyter_text>Indian Sign Language : MobileNet
Kaggle dataset identifier: indian-sign-language-mobilenet
<jupyter_script>Id = []
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input/indian-sign-language/train"):
for filename in filenames:
Id.append(os.path.join(dirname, filename))
Id[:5]
train = pd.DataFrame()
train = train.assign(filename=Id)
train.head()
train["label"] = train["filename"]
train["label"] = train["label"].str.replace(
"/kaggle/input/indian-sign-language/train/", ""
)
train.head()
train["label"] = train["label"].str.split("/").str[0]
train.head()
Id = []
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input/indian-sign-language/test"):
for filename in filenames:
Id.append(os.path.join(dirname, filename))
Id[:5]
test = pd.DataFrame()
test = test.assign(filename=Id)
test.head()
test["label"] = test["filename"]
test["label"] = test["label"].str.replace(
"/kaggle/input/indian-sign-language/test/", ""
)
test.head()
test["label"] = test["label"].str.split("/").str[0]
test.head()
import tensorflow as tf
import numpy as np
from PIL import Image
model = tf.saved_model.load("/kaggle/input/indian-sign-language-mobilenet/mobilenet")
classes = [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
"T",
"U",
"V",
"W",
"X",
"Y",
"Z",
]
result = []
for i in test.filename:
img = Image.open(i).convert("RGB")
img = img.resize((300, 300 * img.size[1] // img.size[0]), Image.ANTIALIAS)
inp_numpy = np.array(img)[None]
inp = tf.constant(inp_numpy, dtype="float32")
class_scores = model(inp)[0].numpy()
result.append(classes[class_scores.argmax()])
result[:5]
test = test.assign(prediction=result)
test.head()
result = []
for i in train.filename:
img = Image.open(i).convert("RGB")
img = img.resize((300, 300 * img.size[1] // img.size[0]), Image.ANTIALIAS)
inp_numpy = np.array(img)[None]
inp = tf.constant(inp_numpy, dtype="float32")
class_scores = model(inp)[0].numpy()
result.append(classes[class_scores.argmax()])
result[:5]
train = train.assign(prediction=result)
train.head()
from sklearn.metrics import classification_report
print(classification_report(train["label"], train["prediction"]))
print(classification_report(test["label"], test["prediction"]))
# Import the necessary libraries
from sklearn.datasets import load_wine
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import confusion_matrix
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.metrics import accuracy_score
# Load the wine dataset
X, y = load_wine(return_X_y=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25)
# Train the model
clf = RandomForestClassifier(random_state=23)
clf.fit(X_train, y_train)
# Predict using the test data
y_pred = clf.predict(X_test)
# Compute the confusion matrix
cm = confusion_matrix(y_test, y_pred)
# Plot the confusion matrix.
sns.heatmap(cm, annot=True)
plt.ylabel("Prediction", fontsize=13)
plt.xlabel("Actual", fontsize=13)
plt.title("Confusion Matrix", fontsize=17)
plt.show()
# Calculate accuracy
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy :", accuracy)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/317/129317577.ipynb
|
indian-sign-language-mobilenet
|
gauravduttakiit
|
[{"Id": 129317577, "ScriptId": 38446106, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5211027, "CreationDate": "05/12/2023 18:03:17", "VersionNumber": 2.0, "Title": "MobileNet : Indian Sign Language", "EvaluationDate": "05/12/2023", "IsChange": true, "TotalLines": 124.0, "LinesInsertedFromPrevious": 32.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 92.0, "LinesInsertedFromFork": 37.0, "LinesDeletedFromFork": 0.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 87.0, "TotalVotes": 0}]
|
[{"Id": 185252397, "KernelVersionId": 129317577, "SourceDatasetVersionId": 5601471}, {"Id": 185252396, "KernelVersionId": 129317577, "SourceDatasetVersionId": 5598763}, {"Id": 185252395, "KernelVersionId": 129317577, "SourceDatasetVersionId": 5513410}]
|
[{"Id": 5601471, "DatasetId": 3222282, "DatasourceVersionId": 5676494, "CreatorUserId": 4760409, "LicenseName": "Unknown", "CreationDate": "05/04/2023 13:24:38", "VersionNumber": 1.0, "Title": "Indian Sign Language : MobileNet", "Slug": "indian-sign-language-mobilenet", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3222282, "CreatorUserId": 4760409, "OwnerUserId": 4760409.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5601471.0, "CurrentDatasourceVersionId": 5676494.0, "ForumId": 3287236, "Type": 2, "CreationDate": "05/04/2023 13:24:38", "LastActivityDate": "05/04/2023", "TotalViews": 89, "TotalDownloads": 6, "TotalVotes": 1, "TotalKernels": 2}]
|
[{"Id": 4760409, "UserName": "gauravduttakiit", "DisplayName": "Gaurav Dutta", "RegisterDate": "03/28/2020", "PerformanceTier": 3}]
|
Id = []
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input/indian-sign-language/train"):
for filename in filenames:
Id.append(os.path.join(dirname, filename))
Id[:5]
train = pd.DataFrame()
train = train.assign(filename=Id)
train.head()
train["label"] = train["filename"]
train["label"] = train["label"].str.replace(
"/kaggle/input/indian-sign-language/train/", ""
)
train.head()
train["label"] = train["label"].str.split("/").str[0]
train.head()
Id = []
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input/indian-sign-language/test"):
for filename in filenames:
Id.append(os.path.join(dirname, filename))
Id[:5]
test = pd.DataFrame()
test = test.assign(filename=Id)
test.head()
test["label"] = test["filename"]
test["label"] = test["label"].str.replace(
"/kaggle/input/indian-sign-language/test/", ""
)
test.head()
test["label"] = test["label"].str.split("/").str[0]
test.head()
import tensorflow as tf
import numpy as np
from PIL import Image
model = tf.saved_model.load("/kaggle/input/indian-sign-language-mobilenet/mobilenet")
classes = [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
"T",
"U",
"V",
"W",
"X",
"Y",
"Z",
]
result = []
for i in test.filename:
img = Image.open(i).convert("RGB")
img = img.resize((300, 300 * img.size[1] // img.size[0]), Image.ANTIALIAS)
inp_numpy = np.array(img)[None]
inp = tf.constant(inp_numpy, dtype="float32")
class_scores = model(inp)[0].numpy()
result.append(classes[class_scores.argmax()])
result[:5]
test = test.assign(prediction=result)
test.head()
result = []
for i in train.filename:
img = Image.open(i).convert("RGB")
img = img.resize((300, 300 * img.size[1] // img.size[0]), Image.ANTIALIAS)
inp_numpy = np.array(img)[None]
inp = tf.constant(inp_numpy, dtype="float32")
class_scores = model(inp)[0].numpy()
result.append(classes[class_scores.argmax()])
result[:5]
train = train.assign(prediction=result)
train.head()
from sklearn.metrics import classification_report
print(classification_report(train["label"], train["prediction"]))
print(classification_report(test["label"], test["prediction"]))
# Import the necessary libraries
from sklearn.datasets import load_wine
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import confusion_matrix
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.metrics import accuracy_score
# Load the wine dataset
X, y = load_wine(return_X_y=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25)
# Train the model
clf = RandomForestClassifier(random_state=23)
clf.fit(X_train, y_train)
# Predict using the test data
y_pred = clf.predict(X_test)
# Compute the confusion matrix
cm = confusion_matrix(y_test, y_pred)
# Plot the confusion matrix.
sns.heatmap(cm, annot=True)
plt.ylabel("Prediction", fontsize=13)
plt.xlabel("Actual", fontsize=13)
plt.title("Confusion Matrix", fontsize=17)
plt.show()
# Calculate accuracy
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy :", accuracy)
| false | 0 | 1,220 | 0 | 1,251 | 1,220 |
||
129051127
|
<jupyter_start><jupyter_text>Boston housing dataset
Domain: Real Estate
Difficulty: Easy to Medium
Challenges:
1. Missing value treatment
2. Outlier treatment
3. Understanding which variables drive the price of homes in Boston
Summary:
The Boston housing dataset contains 506 observations and 14 variables. The dataset contains missing values.
Kaggle dataset identifier: boston-housing-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv("/kaggle/input/boston-housing-dataset/HousingData.csv")
df.head()
# 1. CRIM: Per capita crime rate by town
# 2. ZN: Proportion of residential land zoned for lots over 25,000 sq. ft
# 3. INDUS: Proportion of non-retail business acres per town
# 4. CHAS: Charles River dummy variable (= 1 if tract bounds river; 0 otherwise)
# 5. NOX: Nitric oxide concentration (parts per 10 million)
# 6. RM: Average number of rooms per dwelling
# 7. AGE: Proportion of owner-occupied units built prior to 1940
# 8. DIS: Weighted distances to five Boston employment centers
# 9. RAD: Index of accessibility to radial highways
# 10. TAX: Full-value property tax rate per 10,000
# 11. PTRATIO: Pupil-teacher ratio by town
# 12. B: 1000(Bk — 0.63)², where Bk is the proportion of [people of African American descent] by town
# 13. LSTAT: Percentage of lower status of the population
# 14. MEDV: Median value of owner-occupied homes in $1000s
df.info()
df.describe()
df.isna().sum()
df.size
df.dropna(inplace=True)
plt.figure(figsize=(6, 6))
sns.displot(df["MEDV"], bins=30, kde=True)
plt.show()
sns.heatmap(df.corr().round(1), annot=True)
# ## High Correlation of MEDV can be seen with RM and LSTAT
plt.figure(figsize=(14, 6))
plt.subplot(1, 2, 1)
plt.scatter(df["RM"], df["MEDV"])
plt.subplot(1, 2, 2)
plt.scatter(df["LSTAT"], df["MEDV"])
plt.show()
x = pd.DataFrame(np.c_[df["RM"], df["LSTAT"]], columns=["RM", "LSTAT"])
y = df["MEDV"]
x.head()
y.head()
# ## Train - Test - Split
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.3, random_state=21
)
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, accuracy_score, r2_score
model = LinearRegression()
model.fit(x_train, y_train)
y_test_predict = model.predict(x_test)
rmse = np.sqrt(mean_squared_error(y_test, y_test_predict))
r2 = r2_score(y_test, y_test_predict)
r2
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/051/129051127.ipynb
|
boston-housing-dataset
|
altavish
|
[{"Id": 129051127, "ScriptId": 38345954, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5690863, "CreationDate": "05/10/2023 15:23:34", "VersionNumber": 1.0, "Title": "Boston Housing Price: Linear regression DNN", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 90.0, "LinesInsertedFromPrevious": 90.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 184766652, "KernelVersionId": 129051127, "SourceDatasetVersionId": 5680}]
|
[{"Id": 5680, "DatasetId": 3537, "DatasourceVersionId": 5680, "CreatorUserId": 1174798, "LicenseName": "CC0: Public Domain", "CreationDate": "10/27/2017 12:17:34", "VersionNumber": 1.0, "Title": "Boston housing dataset", "Slug": "boston-housing-dataset", "Subtitle": NaN, "Description": "Domain: Real Estate\n\nDifficulty: Easy to Medium\n\nChallenges: \n 1. Missing value treatment \n 2. Outlier treatment\n 3. Understanding which variables drive the price of homes in Boston\n\nSummary:\nThe Boston housing dataset contains 506 observations and 14 variables. The dataset contains missing values.", "VersionNotes": "Initial release", "TotalCompressedBytes": 35008.0, "TotalUncompressedBytes": 35008.0}]
|
[{"Id": 3537, "CreatorUserId": 1174798, "OwnerUserId": 1174798.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5680.0, "CurrentDatasourceVersionId": 5680.0, "ForumId": 8752, "Type": 2, "CreationDate": "10/27/2017 12:17:34", "LastActivityDate": "02/03/2018", "TotalViews": 126047, "TotalDownloads": 27202, "TotalVotes": 131, "TotalKernels": 129}]
|
[{"Id": 1174798, "UserName": "altavish", "DisplayName": "Vish Vishal", "RegisterDate": "07/18/2017", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv("/kaggle/input/boston-housing-dataset/HousingData.csv")
df.head()
# 1. CRIM: Per capita crime rate by town
# 2. ZN: Proportion of residential land zoned for lots over 25,000 sq. ft
# 3. INDUS: Proportion of non-retail business acres per town
# 4. CHAS: Charles River dummy variable (= 1 if tract bounds river; 0 otherwise)
# 5. NOX: Nitric oxide concentration (parts per 10 million)
# 6. RM: Average number of rooms per dwelling
# 7. AGE: Proportion of owner-occupied units built prior to 1940
# 8. DIS: Weighted distances to five Boston employment centers
# 9. RAD: Index of accessibility to radial highways
# 10. TAX: Full-value property tax rate per 10,000
# 11. PTRATIO: Pupil-teacher ratio by town
# 12. B: 1000(Bk — 0.63)², where Bk is the proportion of [people of African American descent] by town
# 13. LSTAT: Percentage of lower status of the population
# 14. MEDV: Median value of owner-occupied homes in $1000s
df.info()
df.describe()
df.isna().sum()
df.size
df.dropna(inplace=True)
plt.figure(figsize=(6, 6))
sns.displot(df["MEDV"], bins=30, kde=True)
plt.show()
sns.heatmap(df.corr().round(1), annot=True)
# ## High Correlation of MEDV can be seen with RM and LSTAT
plt.figure(figsize=(14, 6))
plt.subplot(1, 2, 1)
plt.scatter(df["RM"], df["MEDV"])
plt.subplot(1, 2, 2)
plt.scatter(df["LSTAT"], df["MEDV"])
plt.show()
x = pd.DataFrame(np.c_[df["RM"], df["LSTAT"]], columns=["RM", "LSTAT"])
y = df["MEDV"]
x.head()
y.head()
# ## Train - Test - Split
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.3, random_state=21
)
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, accuracy_score, r2_score
model = LinearRegression()
model.fit(x_train, y_train)
y_test_predict = model.predict(x_test)
rmse = np.sqrt(mean_squared_error(y_test, y_test_predict))
r2 = r2_score(y_test, y_test_predict)
r2
| false | 1 | 936 | 1 | 1,035 | 936 |
||
129051830
|
print("Helloworld")
def is_valid_length(name: str) -> bool:
if len(name) <= 9:
return True
else:
return False
pass
def is_valid_start(name: str) -> bool:
if name[0].isalpha():
return True
else:
return False
def is_one_word(name: str) -> bool:
if name.find(" ") == -1:
return True
else:
return False
pass
def is_valid_name(name: str) -> bool:
database = "/kaggle/working/list.txt"
f = open(database, "r")
data = f.read()
f.close()
"""
print(data.find(name))
print(len(name))
print(name[0])
print(name[0].isalpha())
print(name.strip().find(" "))
"""
if (
data.find(name) == -1
and len(name) <= 9
and name[0].isalpha()
and name.strip().find(" ") == -1
):
return True
else:
return False
pass
def is_profanity(
word: str, database="/home/files/list.txt", records="/home/files/history.txt"
) -> bool:
"""
Checks if `word` is listed in the blacklist `database`.
Parameters:
word: str, word to check against database.
database: str, absolute directory to file containing list of bad words.
records: str, absolute directory to file to record past offenses by player.
Returns:
result: bool, status of check.
"""
result = False
try:
f = open(database, "r")
except FileNotFoundError:
print("Check directory of database!")
return False
blacklist = f.read()
f.close()
try:
f = open(records, "r")
except FileNotFoundError:
f = open(records, "w")
f = open(records, "r")
files = f.read()
f.close()
if blacklist.find(word) != -1:
result = True
if files.find(word) != -1:
f = open(records, "a")
else:
f = open(records, "a")
if files:
f.write(f"{word}\n")
else:
f.write(f"{word}\n")
# f = open(records,'r')
# i = f.read()
# print(i)
f.close()
return result
else:
return result
def count_occurrence(
word: str, file_records="/home/files/history.txt", start_flag=True
) -> int:
"""
Count the occurrences of `word` contained in file_records.
Parameters:
word: str, target word to count number of occurences.
file_records: str, absolute directory to file that contains past records.
start_flag: bool, set to False to count whole words. True to count words
that start with.
Returns:
count: int, total number of times `word` is found in the file.
"""
count = 0
# check name correct
if isinstance(word, str) == False:
print("First argument must be a string object!")
return count
if word.strip() == "":
print("Must have at least one character in the string!")
return count
# check file exist
try:
f = open(file_records, "r")
except FileNotFoundError:
print("File records not found!")
return count
if start_flag == False:
# print out whole word
line = f.readline()
while line:
whole_name = line.strip()
if whole_name.lower() == word.lower():
count += 1
line = f.readline()
f.close()
return count
else:
# print out first char same
line = f.readline()
while line:
start_first = line.strip()
if start_first[0].lower() == word[0].lower():
count += 1
line = f.readline()
f.close()
return count
pass
def generate_name1(
word: str, src="/home/files/animals.txt", past="/home/files/names.txt"
) -> str:
new_name = "Bob"
animal_list = []
past_number = 0
animals = []
if isinstance(word, str) == False:
print("First argument must be a string object!")
return new_name
if word.strip() == "":
print("Must have at least one character in the string!")
return new_name
try:
pastf = open(past, "r")
except FileNotFoundError:
pastf = open(past, "w")
pastf = open(past, "r")
past_count = pastf.readlines()
pastf.close()
try:
srcf = open(src, "r")
except FileNotFoundError:
print("Source file is not found!")
return new_name
animals = srcf.readlines()
srcf.close()
# Filter animal names based on starting character
animal_list = [
animal.strip()
for animal in animals
if animal[0].strip().lower() == word[0].lower()
]
# Sort the animal list alphabetically
animal_list.sort()
# Get the index of the input word in the sorted list
index = animal_list.index(word.lower())
# Get the next name from the sorted list, considering wrap-around
new_name = animal_list[(index + 1) % len(animal_list)]
# Append the new name to the past file
s = open(past, "a")
s.write(f"{new_name}\n")
s.close()
return new_name
def generate_name(
word: str, src="/home/files/animals.txt", past="/home/files/names.txt"
) -> str:
"""
Select a word from file `src` in sequence depending on the number of times word occurs.
Parameters:
word: str, word to swap
src: str, absolute directory to file that contains safe in-game names
past: str, absolute directory to file that contains past names
auto-generated
Returns:
new_name: str, the generated name to replace word
"""
new_name = "Bob"
animal_list = []
past_number = 0
animals = []
if isinstance(word, str) == False:
print("First argument must be a string object!")
return new_name
if word.strip() == "":
print("Must have at least one character in the string!")
return new_name
try:
pastf = open(past, "r")
except FileNotFoundError:
pastf = open(past, "w")
pastf = open(past, "r")
past_count = pastf.readlines()
pastf.close()
try:
srcf = open(src, "r")
except FileNotFoundError:
print("Source file is not found!")
return new_name
animals = srcf.readlines()
srcf.close()
i = 0
# Get the all first start animal name and store in the list
while i < len(animals):
animal = animals[i]
# the first word is the same?
if animal[0].strip().lower() == word[0].lower():
animal_list.append(animal.strip())
i += 1
# Count the number of the record in past file
i = 0
while i < len(past_count):
animal = past_count[i]
if animal[0].strip().lower() == word[0].lower():
past_number += 1
i += 1
# Use % to get the next name we use
"""
animal_list ['blobfish', 'butterfly', 'baboon']
past_number 6
len(animal_list) 3
print("animal_list",animal_list)
print("past_number",past_number)
print("len(animal_list)",len(animal_list))
"""
new_name = animal_list[past_number % len(animal_list)]
s = open(past, "a")
s.write(f"{new_name}\n")
s.close()
return new_name
pass
def check():
while True:
name_input = input("Check name: ")
if name_input == "s":
break
if is_valid_name(name_input) == True:
print(f"{name_input} is a valid name!")
else:
# new_name = generate_name(name_input,"/home/files/animals.txt","/home/files/names.txt")
new_name = generate_name(
name_input, "/kaggle/working/animals.txt", "/kaggle/working/names.txt"
)
print(f"Your new name is: {new_name}")
f = open("/kaggle/working/names.txt", "r")
existing_names = f.readlines()
f.close()
count = count_occurrence(
name_input, "/kaggle/working/names.txt", start_flag=True
)
new_name = generate_name(
name_input, "/kaggle/working/animals.txt", "/kaggle/working/names.txt"
)
count -= 1
if existing_names[count].strip() == new_name.strip():
new_name = generate_name(
name_input,
"/kaggle/working/animals.txt",
"/kaggle/working/names.txt",
)
def main():
check()
pass
if __name__ == "__main__":
main()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/051/129051830.ipynb
| null | null |
[{"Id": 129051830, "ScriptId": 38224910, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10114012, "CreationDate": "05/10/2023 15:29:24", "VersionNumber": 1.0, "Title": "Python Programmar", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 283.0, "LinesInsertedFromPrevious": 283.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
print("Helloworld")
def is_valid_length(name: str) -> bool:
if len(name) <= 9:
return True
else:
return False
pass
def is_valid_start(name: str) -> bool:
if name[0].isalpha():
return True
else:
return False
def is_one_word(name: str) -> bool:
if name.find(" ") == -1:
return True
else:
return False
pass
def is_valid_name(name: str) -> bool:
database = "/kaggle/working/list.txt"
f = open(database, "r")
data = f.read()
f.close()
"""
print(data.find(name))
print(len(name))
print(name[0])
print(name[0].isalpha())
print(name.strip().find(" "))
"""
if (
data.find(name) == -1
and len(name) <= 9
and name[0].isalpha()
and name.strip().find(" ") == -1
):
return True
else:
return False
pass
def is_profanity(
word: str, database="/home/files/list.txt", records="/home/files/history.txt"
) -> bool:
"""
Checks if `word` is listed in the blacklist `database`.
Parameters:
word: str, word to check against database.
database: str, absolute directory to file containing list of bad words.
records: str, absolute directory to file to record past offenses by player.
Returns:
result: bool, status of check.
"""
result = False
try:
f = open(database, "r")
except FileNotFoundError:
print("Check directory of database!")
return False
blacklist = f.read()
f.close()
try:
f = open(records, "r")
except FileNotFoundError:
f = open(records, "w")
f = open(records, "r")
files = f.read()
f.close()
if blacklist.find(word) != -1:
result = True
if files.find(word) != -1:
f = open(records, "a")
else:
f = open(records, "a")
if files:
f.write(f"{word}\n")
else:
f.write(f"{word}\n")
# f = open(records,'r')
# i = f.read()
# print(i)
f.close()
return result
else:
return result
def count_occurrence(
word: str, file_records="/home/files/history.txt", start_flag=True
) -> int:
"""
Count the occurrences of `word` contained in file_records.
Parameters:
word: str, target word to count number of occurences.
file_records: str, absolute directory to file that contains past records.
start_flag: bool, set to False to count whole words. True to count words
that start with.
Returns:
count: int, total number of times `word` is found in the file.
"""
count = 0
# check name correct
if isinstance(word, str) == False:
print("First argument must be a string object!")
return count
if word.strip() == "":
print("Must have at least one character in the string!")
return count
# check file exist
try:
f = open(file_records, "r")
except FileNotFoundError:
print("File records not found!")
return count
if start_flag == False:
# print out whole word
line = f.readline()
while line:
whole_name = line.strip()
if whole_name.lower() == word.lower():
count += 1
line = f.readline()
f.close()
return count
else:
# print out first char same
line = f.readline()
while line:
start_first = line.strip()
if start_first[0].lower() == word[0].lower():
count += 1
line = f.readline()
f.close()
return count
pass
def generate_name1(
word: str, src="/home/files/animals.txt", past="/home/files/names.txt"
) -> str:
new_name = "Bob"
animal_list = []
past_number = 0
animals = []
if isinstance(word, str) == False:
print("First argument must be a string object!")
return new_name
if word.strip() == "":
print("Must have at least one character in the string!")
return new_name
try:
pastf = open(past, "r")
except FileNotFoundError:
pastf = open(past, "w")
pastf = open(past, "r")
past_count = pastf.readlines()
pastf.close()
try:
srcf = open(src, "r")
except FileNotFoundError:
print("Source file is not found!")
return new_name
animals = srcf.readlines()
srcf.close()
# Filter animal names based on starting character
animal_list = [
animal.strip()
for animal in animals
if animal[0].strip().lower() == word[0].lower()
]
# Sort the animal list alphabetically
animal_list.sort()
# Get the index of the input word in the sorted list
index = animal_list.index(word.lower())
# Get the next name from the sorted list, considering wrap-around
new_name = animal_list[(index + 1) % len(animal_list)]
# Append the new name to the past file
s = open(past, "a")
s.write(f"{new_name}\n")
s.close()
return new_name
def generate_name(
word: str, src="/home/files/animals.txt", past="/home/files/names.txt"
) -> str:
"""
Select a word from file `src` in sequence depending on the number of times word occurs.
Parameters:
word: str, word to swap
src: str, absolute directory to file that contains safe in-game names
past: str, absolute directory to file that contains past names
auto-generated
Returns:
new_name: str, the generated name to replace word
"""
new_name = "Bob"
animal_list = []
past_number = 0
animals = []
if isinstance(word, str) == False:
print("First argument must be a string object!")
return new_name
if word.strip() == "":
print("Must have at least one character in the string!")
return new_name
try:
pastf = open(past, "r")
except FileNotFoundError:
pastf = open(past, "w")
pastf = open(past, "r")
past_count = pastf.readlines()
pastf.close()
try:
srcf = open(src, "r")
except FileNotFoundError:
print("Source file is not found!")
return new_name
animals = srcf.readlines()
srcf.close()
i = 0
# Get the all first start animal name and store in the list
while i < len(animals):
animal = animals[i]
# the first word is the same?
if animal[0].strip().lower() == word[0].lower():
animal_list.append(animal.strip())
i += 1
# Count the number of the record in past file
i = 0
while i < len(past_count):
animal = past_count[i]
if animal[0].strip().lower() == word[0].lower():
past_number += 1
i += 1
# Use % to get the next name we use
"""
animal_list ['blobfish', 'butterfly', 'baboon']
past_number 6
len(animal_list) 3
print("animal_list",animal_list)
print("past_number",past_number)
print("len(animal_list)",len(animal_list))
"""
new_name = animal_list[past_number % len(animal_list)]
s = open(past, "a")
s.write(f"{new_name}\n")
s.close()
return new_name
pass
def check():
while True:
name_input = input("Check name: ")
if name_input == "s":
break
if is_valid_name(name_input) == True:
print(f"{name_input} is a valid name!")
else:
# new_name = generate_name(name_input,"/home/files/animals.txt","/home/files/names.txt")
new_name = generate_name(
name_input, "/kaggle/working/animals.txt", "/kaggle/working/names.txt"
)
print(f"Your new name is: {new_name}")
f = open("/kaggle/working/names.txt", "r")
existing_names = f.readlines()
f.close()
count = count_occurrence(
name_input, "/kaggle/working/names.txt", start_flag=True
)
new_name = generate_name(
name_input, "/kaggle/working/animals.txt", "/kaggle/working/names.txt"
)
count -= 1
if existing_names[count].strip() == new_name.strip():
new_name = generate_name(
name_input,
"/kaggle/working/animals.txt",
"/kaggle/working/names.txt",
)
def main():
check()
pass
if __name__ == "__main__":
main()
| false | 0 | 2,281 | 0 | 2,281 | 2,281 |
||
129051006
|
import numpy as np
import pandas as pd
import seaborn as sns
from sklearn.model_selection import train_test_split
def accuracy(y_test, y_hat):
count = 0
for y, y_ in zip(y_test, y_hat):
if y == y_:
count += 1
return count / len(y_test)
base_dir = "../input/titanic/"
train = pd.read_csv(base_dir + "train.csv")
test = pd.read_csv(base_dir + "test.csv")
sub = pd.read_csv(base_dir + "gender_submission.csv")
train.columns
sns.histplot(data=train, x="Age", hue="Survived")
sns.barplot(data=train, x="Sex", y="Survived")
sns.barplot(data=train, x="Pclass", y="Survived")
train.head()
sns.boxplot(data=train, x="Pclass", y="Fare")
# Name, Ticket, Cabin, Embarked, PassengerId
train = train.drop(columns=["Name", "Ticket", "Cabin", "Embarked", "PassengerId"])
train["Age"] = train["Age"].fillna(train["Age"].median())
train["Sex"] = train["Sex"].map({"male": 0, "female": 1})
temp = pd.get_dummies(
train["Pclass"],
)
temp.columns = ["Pclass_1", "Pclass_2", "Pclass_3"]
train = pd.concat([train, temp], axis=1).drop(columns=["Pclass"])
X = train.drop(columns="Survived").values
y = train["Survived"].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
from sklearn.linear_model import SGDClassifier
clf = SGDClassifier(loss="hinge", penalty="l2", max_iter=40)
clf.fit(X_train, y_train)
y_hat = clf.predict(X_test)
accuracy(y_test, y_hat)
from sklearn.neighbors import KNeighborsClassifier
neigh = KNeighborsClassifier(n_neighbors=3)
neigh.fit(X_train, y_train)
y_hat = neigh.predict(X_test)
accuracy(y_test, y_hat)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/051/129051006.ipynb
| null | null |
[{"Id": 129051006, "ScriptId": 38360097, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1487291, "CreationDate": "05/10/2023 15:22:27", "VersionNumber": 2.0, "Title": "Titanic predict", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 58.0, "LinesInsertedFromPrevious": 36.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 22.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
| null | null | null | null |
import numpy as np
import pandas as pd
import seaborn as sns
from sklearn.model_selection import train_test_split
def accuracy(y_test, y_hat):
count = 0
for y, y_ in zip(y_test, y_hat):
if y == y_:
count += 1
return count / len(y_test)
base_dir = "../input/titanic/"
train = pd.read_csv(base_dir + "train.csv")
test = pd.read_csv(base_dir + "test.csv")
sub = pd.read_csv(base_dir + "gender_submission.csv")
train.columns
sns.histplot(data=train, x="Age", hue="Survived")
sns.barplot(data=train, x="Sex", y="Survived")
sns.barplot(data=train, x="Pclass", y="Survived")
train.head()
sns.boxplot(data=train, x="Pclass", y="Fare")
# Name, Ticket, Cabin, Embarked, PassengerId
train = train.drop(columns=["Name", "Ticket", "Cabin", "Embarked", "PassengerId"])
train["Age"] = train["Age"].fillna(train["Age"].median())
train["Sex"] = train["Sex"].map({"male": 0, "female": 1})
temp = pd.get_dummies(
train["Pclass"],
)
temp.columns = ["Pclass_1", "Pclass_2", "Pclass_3"]
train = pd.concat([train, temp], axis=1).drop(columns=["Pclass"])
X = train.drop(columns="Survived").values
y = train["Survived"].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
from sklearn.linear_model import SGDClassifier
clf = SGDClassifier(loss="hinge", penalty="l2", max_iter=40)
clf.fit(X_train, y_train)
y_hat = clf.predict(X_test)
accuracy(y_test, y_hat)
from sklearn.neighbors import KNeighborsClassifier
neigh = KNeighborsClassifier(n_neighbors=3)
neigh.fit(X_train, y_train)
y_hat = neigh.predict(X_test)
accuracy(y_test, y_hat)
| false | 0 | 589 | 2 | 589 | 589 |
||
129051016
|
<jupyter_start><jupyter_text>Medical Text Dataset -Cancer Doc Classification
For Biomedical text document classification, abstract and full papers(whose length less than or equal to 6 pages) available and used. This dataset focused on long research paper whose page size more than 6 pages. Dataset includes cancer documents to be classified into 3 categories like 'Thyroid_Cancer','Colon_Cancer','Lung_Cancer'.
Total publications=7569. it has 3 class labels in dataset.
number of samples in each categories:
colon cancer=2579, lung cancer=2180, thyroid cancer=2810
Kaggle dataset identifier: biomedical-text-publication-classification
<jupyter_script># # Cancer doc classification
# For Biomedical text document classification, abstract and full papers(whose length less than or equal to 6 pages) available and used. This dataset focused on long research paper whose page size more than 6 pages. Dataset includes cancer documents to be classified into 3 categories like 'Thyroid_Cancer','Colon_Cancer','Lung_Cancer'.
# Total publications=7569. it has 3 class labels in dataset.
# number of samples in each categories:
# colon cancer=2579, lung cancer=2180, thyroid cancer=2810
# ### Setup and install nedded libraries
from sklearn.preprocessing import LabelEncoder
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import GaussianNB, MultinomialNB, BernoulliNB
from sklearn.metrics import accuracy_score, confusion_matrix, classification_report
from sklearn.decomposition import PCA
from sklearn.svm import SVC
from nltk import word_tokenize, sent_tokenize
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
from wordcloud import WordCloud, STOPWORDS
from collections import defaultdict
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import nltk
import string
import opendatasets as od
nltk.download("stopwords")
nltk.download("punkt")
# od.download("https://www.kaggle.com/datasets/falgunipatel19/biomedical-text-publication-classification")
PATH = (
"/kaggle/input/biomedical-text-publication-classification/alldata_1_for_kaggle.csv"
)
#
# ## **Explore and Visualiaze Data**
# ### Read data and check shape
data = pd.read_csv(PATH, encoding="latin-1")
data.head()
data.shape
data.rename(columns={"0": "Target", "a": "Text"}, inplace=True)
data.head(5)
# ### find Target unique values
data["Target"].unique()
target_counts = data["Target"].value_counts()
target_counts
# ### Target Distribution
fig, axes = plt.subplots(1, 2, figsize=(10, 7))
sns.barplot(x=target_counts.index, y=target_counts.values, ax=axes[0])
axes[1].pie(target_counts, labels=target_counts.keys(), autopct="%1.2f%%")
plt.show()
# ### Distribution for Text length
data["text_len"] = data["Text"].str.len()
data["word_count"] = data["Text"].apply(lambda x: len(str(x).split()))
data["unique_word_count"] = data["Text"].apply(lambda x: len(set(str(x).split())))
data["stop_word_count"] = data["Text"].apply(
lambda x: len([w for w in str(x).lower().split() if w in STOPWORDS])
)
data["url_count"] = data["Text"].apply(
lambda x: len([w for w in str(x).lower().split() if "http" in w or "https" in w])
)
data["punctuation_count"] = data["Text"].apply(
lambda x: len([c for c in str(x) if c in string.punctuation])
)
plt.figure(figsize=(15, 5))
ax = sns.distplot(data["text_len"])
ax.set(xlabel="Text length", ylabel="Frequency")
plt.show()
# ### Length of the text (number of word) stratified by outcome
#
plt.figure(figsize=(12, 6))
num_1 = data[data["Target"] == "Thyroid_Cancer"]["Text"].apply(lambda x: len(x.split()))
num_2 = data[data["Target"] == "Colon_Cancer"]["Text"].apply(lambda x: len(x.split()))
num_3 = data[data["Target"] == "Lung_Cancer"]["Text"].apply(lambda x: len(x.split()))
sns.kdeplot(num_1, shade=True, color="red").set_title(
"Kernel distribution of number of words"
)
sns.kdeplot(num_2, shade=True, color="blue")
sns.kdeplot(num_3, shade=True, color="green")
plt.legend(labels=["Thyroid_Cancer", "Colon_Cancerr", "Lung_Cancer"])
def distplot(column, xlabel):
plt.figure(figsize=(20, 5))
ax = sns.distplot(data[column][data["Target"] == "Thyroid_Cancer"], color="red")
ax = sns.distplot(data[column][data["Target"] == "Colon_Cancer"], color="blue")
ax = sns.distplot(data[column][data["Target"] == "Lung_Cancer"], color="green")
ax.set(xlabel=xlabel, ylabel="Frequency.")
plt.show()
distplot("word_count", "Word count")
distplot("unique_word_count", "Unique Word count")
distplot("stop_word_count", " stop Word count")
def wordcloud(target):
wordcloud = WordCloud(
stopwords=STOPWORDS, background_color="white", width=2000, height=1000
).generate(" ".join(data[data["Target"] == target]["Text"]))
plt.figure(1, figsize=(15, 15))
plt.imshow(wordcloud)
plt.axis("off")
plt.show()
wordcloud("Thyroid_Cancer")
wordcloud("Colon_Cancer")
wordcloud("Lung_Cancer")
def generate_ngrams(text, n_gram=1):
token = [
token
for token in text.lower().split(" ")
if token != ""
if token not in STOPWORDS
]
ngrams = zip(*[token[i:] for i in range(n_gram)])
return [" ".join(ngram) for ngram in ngrams]
N = 50
# Unigrams
Thyroid_Cancer_unigrams = defaultdict(int)
Colon_Cancer_unigrams = defaultdict(int)
Lung_Cancer_unigrams = defaultdict(int)
for doc in data["Text"][data["Target"] == "Thyroid_Cancer"]:
for word in generate_ngrams(doc):
Thyroid_Cancer_unigrams[word] += 1
for doc in data["Text"][data["Target"] == "Colon_Cancer"]:
for word in generate_ngrams(doc):
Colon_Cancer_unigrams[word] += 1
for doc in data["Text"][data["Target"] == "Lung_Cancer"]:
for word in generate_ngrams(doc):
Lung_Cancer_unigrams[word] += 1
Thyroid_Cancer_unigrams = pd.DataFrame(
sorted(Thyroid_Cancer_unigrams.items(), key=lambda x: x[1])[::-1]
)
Colon_Cancer_unigrams = pd.DataFrame(
sorted(Colon_Cancer_unigrams.items(), key=lambda x: x[1])[::-1]
)
Lung_Cancer_unigrams = pd.DataFrame(
sorted(Lung_Cancer_unigrams.items(), key=lambda x: x[1])[::-1]
)
fig, axes = plt.subplots(ncols=3, figsize=(20, 80), dpi=60)
plt.tight_layout()
sns.barplot(
y=Thyroid_Cancer_unigrams[0].values[:N],
x=Thyroid_Cancer_unigrams[1].values[:N],
ax=axes[0],
color="red",
)
sns.barplot(
y=Colon_Cancer_unigrams[0].values[:N],
x=Colon_Cancer_unigrams[1].values[:N],
ax=axes[1],
color="blue",
)
sns.barplot(
y=Lung_Cancer_unigrams[0].values[:N],
x=Lung_Cancer_unigrams[1].values[:N],
ax=axes[2],
color="green",
)
for i in range(3):
axes[i].spines["right"].set_visible(False)
axes[i].set_xlabel("")
axes[i].set_ylabel("")
axes[i].tick_params(axis="x", labelsize=13)
axes[i].tick_params(axis="y", labelsize=13)
axes[0].set_title(f"Top {N} most common unigrams in Thyroid Cancer", fontsize=15)
axes[1].set_title(f"Top {N} most common unigrams in Colon Cancer", fontsize=15)
axes[2].set_title(f"Top {N} most common unigrams in Lung Cancer", fontsize=15)
plt.show()
# # Preprocessing and feature engineering
# Drop the column unnamed as it is not useful
# Rename the column into readable form
#
# Steps :
# 1) Converting into lower case
# 2) Tokenizing : spliting sentences into words
# 3) Removing special characters
# 4) Removing stopwords and punctuations
# 5) Stemming: converting into root words
# 6) Join to make sentences
# Drop the column unnamed as it is not useful:
data.drop("Unnamed: 0", axis=1, inplace=True)
# Rename the column into readable form :
data.rename({"0": "cancer", "a": "text"}, axis=1, inplace=True)
data.tail()
data.info()
data.isnull().sum()
# converting into lowercase
data["Text"] = data["Text"].str.lower()
# break into words
nltk.download("punkt")
data["Text"] = data["Text"].apply(lambda x: word_tokenize(x))
# removing special character
def remove_special_char(list):
y = []
for string in list:
if string.isalnum():
y.append(string)
return y
data["Text"] = data["Text"].apply(lambda x: remove_special_char(x))
import pickle
# Save df
with open("data.pkl", "wb") as f:
pickle.dump(data, f)
# Later..
# Load df
# with open('df.pkl', 'rb') as f:
# df = pickle.load(f)
# remove stopwords like is am are and punctuations
def useful_words(list):
y = []
for text in list:
if text not in stopwords.words("english") and text not in string.punctuation:
y.append(text)
return y
data["Text"] = data["Text"].apply(lambda x: useful_words(x))
# convert into root words
ps = PorterStemmer()
def stemming(list):
y = []
for text in list:
y.append(ps.stem(text))
return y2
data["Text"] = data["Text"].apply(lambda x: stemming(x))
# join the words
data["Text"] = data["Text"].apply(lambda x: "".join(x))
data
# # Modeling and Evaluation
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.linear_model import LogisticRegression, SGDClassifier
from sklearn.naive_bayes import GaussianNB, MultinomialNB, BernoulliNB
from sklearn.discriminant_analysis import (
LinearDiscriminantAnalysis,
QuadraticDiscriminantAnalysis,
)
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import classification_report
# Remove duplicate values to avoid overfitting
data = data.drop_duplicates()
import pickle
# Vectorize the data by using tfidf for text , and y = Target
cv = CountVectorizer(max_features=3000)
tfidf = TfidfVectorizer(max_features=4600)
x = tfidf.fit_transform(data["Text"]).toarray()
y = data["Target"].values
with open("tfidf.pkl", "wb") as file:
pickle.dump(tfidf, file)
# Divide the data into train and test data
X_train, X_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=28, stratify=y
)
GaussianNBModel = GaussianNB()
MultinomialNBModel = MultinomialNB(alpha=1.0)
BernoulliNBModel = BernoulliNB(alpha=1.0, binarize=1)
LogisticRegressionModel = LogisticRegression(
penalty="l2", solver="sag", C=1.0, random_state=33
)
SGDClassifierModel = SGDClassifier(
penalty="l2", loss="squared_loss", learning_rate="optimal", random_state=33
)
RandomForestClassifierModel = RandomForestClassifier(
criterion="gini", n_estimators=300, max_depth=7, random_state=33
)
GBCModel = GradientBoostingClassifier(n_estimators=100, max_depth=3, random_state=33)
QDAModel = QuadraticDiscriminantAnalysis(tol=0.0001)
SVCModel = SVC(kernel="rbf", max_iter=100, C=1.0, gamma="auto")
DecisionTreeClassifierModel = DecisionTreeClassifier(
criterion="gini", max_depth=3, random_state=33
)
KNNClassifierModel = KNeighborsClassifier(
n_neighbors=5, weights="uniform", algorithm="auto"
)
Models = [
GaussianNBModel,
KNNClassifierModel,
MultinomialNBModel,
BernoulliNBModel,
LogisticRegressionModel,
RandomForestClassifierModel,
GBCModel,
SGDClassifierModel,
QDAModel,
SVCModel,
DecisionTreeClassifierModel,
]
ModelsScore = {}
for Model in Models:
print(f'for Model {str(Model).split("(")[0]}')
Model.fit(X_train, y_train)
print(f"Train Score is : {Model.score(X_train, y_train)}")
print(f"Test Score is : {Model.score(X_test, y_test)}")
y_pred = Model.predict(X_test)
ClassificationReport = classification_report(y_test, y_pred)
print("Classification Report is : \n", ClassificationReport)
print(f"Precision value is : {ClassificationReport.split()[19]}")
print(f"Recall value is : {ClassificationReport.split()[20]}")
print(f"F1 Score value is : {ClassificationReport.split()[21]}")
ModelsScore[str(Model).split("(")[0]] = [
ClassificationReport.split()[19],
ClassificationReport.split()[20],
ClassificationReport.split()[21],
]
print("=================================================")
type(data["Text"])
# **BERT MODEL**
# still under work
# # Deployment
model = GBCModel.fit(X_train, y_train)
model.score(X_train, y_train)
# Tokenize the text sample
txt = pd.Series(text)
tfidf_text = tfidf.transform(txt).toarray()
print(tfidf_text)
model.predict(tfidf_text)
dump(model, "model.joblib")
filename = "tfidf.sav"
pickle.dump(tfidf, open(filename, "wb"))
pre = pickle.load(open("pre.sav", "rb"))
txt = pd.Series(data)
te = tfidf.transform(txt).toarray()
model.predict(te)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/051/129051016.ipynb
|
biomedical-text-publication-classification
|
falgunipatel19
|
[{"Id": 129051016, "ScriptId": 38353622, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12594195, "CreationDate": "05/10/2023 15:22:34", "VersionNumber": 1.0, "Title": "Cancer doc classification", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 351.0, "LinesInsertedFromPrevious": 351.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184766417, "KernelVersionId": 129051016, "SourceDatasetVersionId": 4033428}]
|
[{"Id": 4033428, "DatasetId": 2389764, "DatasourceVersionId": 4089381, "CreatorUserId": 9858288, "LicenseName": "Other (specified in description)", "CreationDate": "08/05/2022 16:11:38", "VersionNumber": 1.0, "Title": "Medical Text Dataset -Cancer Doc Classification", "Slug": "biomedical-text-publication-classification", "Subtitle": "Cancer Text Documents Classification", "Description": "For Biomedical text document classification, abstract and full papers(whose length less than or equal to 6 pages) available and used. This dataset focused on long research paper whose page size more than 6 pages. Dataset includes cancer documents to be classified into 3 categories like 'Thyroid_Cancer','Colon_Cancer','Lung_Cancer'. \nTotal publications=7569. it has 3 class labels in dataset.\nnumber of samples in each categories:\ncolon cancer=2579, lung cancer=2180, thyroid cancer=2810", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2389764, "CreatorUserId": 9858288, "OwnerUserId": 9858288.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4033428.0, "CurrentDatasourceVersionId": 4089381.0, "ForumId": 2417029, "Type": 2, "CreationDate": "08/05/2022 16:11:38", "LastActivityDate": "08/05/2022", "TotalViews": 17236, "TotalDownloads": 3121, "TotalVotes": 45, "TotalKernels": 21}]
|
[{"Id": 9858288, "UserName": "falgunipatel19", "DisplayName": "Falgunipatel19", "RegisterDate": "03/07/2022", "PerformanceTier": 0}]
|
# # Cancer doc classification
# For Biomedical text document classification, abstract and full papers(whose length less than or equal to 6 pages) available and used. This dataset focused on long research paper whose page size more than 6 pages. Dataset includes cancer documents to be classified into 3 categories like 'Thyroid_Cancer','Colon_Cancer','Lung_Cancer'.
# Total publications=7569. it has 3 class labels in dataset.
# number of samples in each categories:
# colon cancer=2579, lung cancer=2180, thyroid cancer=2810
# ### Setup and install nedded libraries
from sklearn.preprocessing import LabelEncoder
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import GaussianNB, MultinomialNB, BernoulliNB
from sklearn.metrics import accuracy_score, confusion_matrix, classification_report
from sklearn.decomposition import PCA
from sklearn.svm import SVC
from nltk import word_tokenize, sent_tokenize
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
from wordcloud import WordCloud, STOPWORDS
from collections import defaultdict
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import nltk
import string
import opendatasets as od
nltk.download("stopwords")
nltk.download("punkt")
# od.download("https://www.kaggle.com/datasets/falgunipatel19/biomedical-text-publication-classification")
PATH = (
"/kaggle/input/biomedical-text-publication-classification/alldata_1_for_kaggle.csv"
)
#
# ## **Explore and Visualiaze Data**
# ### Read data and check shape
data = pd.read_csv(PATH, encoding="latin-1")
data.head()
data.shape
data.rename(columns={"0": "Target", "a": "Text"}, inplace=True)
data.head(5)
# ### find Target unique values
data["Target"].unique()
target_counts = data["Target"].value_counts()
target_counts
# ### Target Distribution
fig, axes = plt.subplots(1, 2, figsize=(10, 7))
sns.barplot(x=target_counts.index, y=target_counts.values, ax=axes[0])
axes[1].pie(target_counts, labels=target_counts.keys(), autopct="%1.2f%%")
plt.show()
# ### Distribution for Text length
data["text_len"] = data["Text"].str.len()
data["word_count"] = data["Text"].apply(lambda x: len(str(x).split()))
data["unique_word_count"] = data["Text"].apply(lambda x: len(set(str(x).split())))
data["stop_word_count"] = data["Text"].apply(
lambda x: len([w for w in str(x).lower().split() if w in STOPWORDS])
)
data["url_count"] = data["Text"].apply(
lambda x: len([w for w in str(x).lower().split() if "http" in w or "https" in w])
)
data["punctuation_count"] = data["Text"].apply(
lambda x: len([c for c in str(x) if c in string.punctuation])
)
plt.figure(figsize=(15, 5))
ax = sns.distplot(data["text_len"])
ax.set(xlabel="Text length", ylabel="Frequency")
plt.show()
# ### Length of the text (number of word) stratified by outcome
#
plt.figure(figsize=(12, 6))
num_1 = data[data["Target"] == "Thyroid_Cancer"]["Text"].apply(lambda x: len(x.split()))
num_2 = data[data["Target"] == "Colon_Cancer"]["Text"].apply(lambda x: len(x.split()))
num_3 = data[data["Target"] == "Lung_Cancer"]["Text"].apply(lambda x: len(x.split()))
sns.kdeplot(num_1, shade=True, color="red").set_title(
"Kernel distribution of number of words"
)
sns.kdeplot(num_2, shade=True, color="blue")
sns.kdeplot(num_3, shade=True, color="green")
plt.legend(labels=["Thyroid_Cancer", "Colon_Cancerr", "Lung_Cancer"])
def distplot(column, xlabel):
plt.figure(figsize=(20, 5))
ax = sns.distplot(data[column][data["Target"] == "Thyroid_Cancer"], color="red")
ax = sns.distplot(data[column][data["Target"] == "Colon_Cancer"], color="blue")
ax = sns.distplot(data[column][data["Target"] == "Lung_Cancer"], color="green")
ax.set(xlabel=xlabel, ylabel="Frequency.")
plt.show()
distplot("word_count", "Word count")
distplot("unique_word_count", "Unique Word count")
distplot("stop_word_count", " stop Word count")
def wordcloud(target):
wordcloud = WordCloud(
stopwords=STOPWORDS, background_color="white", width=2000, height=1000
).generate(" ".join(data[data["Target"] == target]["Text"]))
plt.figure(1, figsize=(15, 15))
plt.imshow(wordcloud)
plt.axis("off")
plt.show()
wordcloud("Thyroid_Cancer")
wordcloud("Colon_Cancer")
wordcloud("Lung_Cancer")
def generate_ngrams(text, n_gram=1):
token = [
token
for token in text.lower().split(" ")
if token != ""
if token not in STOPWORDS
]
ngrams = zip(*[token[i:] for i in range(n_gram)])
return [" ".join(ngram) for ngram in ngrams]
N = 50
# Unigrams
Thyroid_Cancer_unigrams = defaultdict(int)
Colon_Cancer_unigrams = defaultdict(int)
Lung_Cancer_unigrams = defaultdict(int)
for doc in data["Text"][data["Target"] == "Thyroid_Cancer"]:
for word in generate_ngrams(doc):
Thyroid_Cancer_unigrams[word] += 1
for doc in data["Text"][data["Target"] == "Colon_Cancer"]:
for word in generate_ngrams(doc):
Colon_Cancer_unigrams[word] += 1
for doc in data["Text"][data["Target"] == "Lung_Cancer"]:
for word in generate_ngrams(doc):
Lung_Cancer_unigrams[word] += 1
Thyroid_Cancer_unigrams = pd.DataFrame(
sorted(Thyroid_Cancer_unigrams.items(), key=lambda x: x[1])[::-1]
)
Colon_Cancer_unigrams = pd.DataFrame(
sorted(Colon_Cancer_unigrams.items(), key=lambda x: x[1])[::-1]
)
Lung_Cancer_unigrams = pd.DataFrame(
sorted(Lung_Cancer_unigrams.items(), key=lambda x: x[1])[::-1]
)
fig, axes = plt.subplots(ncols=3, figsize=(20, 80), dpi=60)
plt.tight_layout()
sns.barplot(
y=Thyroid_Cancer_unigrams[0].values[:N],
x=Thyroid_Cancer_unigrams[1].values[:N],
ax=axes[0],
color="red",
)
sns.barplot(
y=Colon_Cancer_unigrams[0].values[:N],
x=Colon_Cancer_unigrams[1].values[:N],
ax=axes[1],
color="blue",
)
sns.barplot(
y=Lung_Cancer_unigrams[0].values[:N],
x=Lung_Cancer_unigrams[1].values[:N],
ax=axes[2],
color="green",
)
for i in range(3):
axes[i].spines["right"].set_visible(False)
axes[i].set_xlabel("")
axes[i].set_ylabel("")
axes[i].tick_params(axis="x", labelsize=13)
axes[i].tick_params(axis="y", labelsize=13)
axes[0].set_title(f"Top {N} most common unigrams in Thyroid Cancer", fontsize=15)
axes[1].set_title(f"Top {N} most common unigrams in Colon Cancer", fontsize=15)
axes[2].set_title(f"Top {N} most common unigrams in Lung Cancer", fontsize=15)
plt.show()
# # Preprocessing and feature engineering
# Drop the column unnamed as it is not useful
# Rename the column into readable form
#
# Steps :
# 1) Converting into lower case
# 2) Tokenizing : spliting sentences into words
# 3) Removing special characters
# 4) Removing stopwords and punctuations
# 5) Stemming: converting into root words
# 6) Join to make sentences
# Drop the column unnamed as it is not useful:
data.drop("Unnamed: 0", axis=1, inplace=True)
# Rename the column into readable form :
data.rename({"0": "cancer", "a": "text"}, axis=1, inplace=True)
data.tail()
data.info()
data.isnull().sum()
# converting into lowercase
data["Text"] = data["Text"].str.lower()
# break into words
nltk.download("punkt")
data["Text"] = data["Text"].apply(lambda x: word_tokenize(x))
# removing special character
def remove_special_char(list):
y = []
for string in list:
if string.isalnum():
y.append(string)
return y
data["Text"] = data["Text"].apply(lambda x: remove_special_char(x))
import pickle
# Save df
with open("data.pkl", "wb") as f:
pickle.dump(data, f)
# Later..
# Load df
# with open('df.pkl', 'rb') as f:
# df = pickle.load(f)
# remove stopwords like is am are and punctuations
def useful_words(list):
y = []
for text in list:
if text not in stopwords.words("english") and text not in string.punctuation:
y.append(text)
return y
data["Text"] = data["Text"].apply(lambda x: useful_words(x))
# convert into root words
ps = PorterStemmer()
def stemming(list):
y = []
for text in list:
y.append(ps.stem(text))
return y2
data["Text"] = data["Text"].apply(lambda x: stemming(x))
# join the words
data["Text"] = data["Text"].apply(lambda x: "".join(x))
data
# # Modeling and Evaluation
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.linear_model import LogisticRegression, SGDClassifier
from sklearn.naive_bayes import GaussianNB, MultinomialNB, BernoulliNB
from sklearn.discriminant_analysis import (
LinearDiscriminantAnalysis,
QuadraticDiscriminantAnalysis,
)
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import classification_report
# Remove duplicate values to avoid overfitting
data = data.drop_duplicates()
import pickle
# Vectorize the data by using tfidf for text , and y = Target
cv = CountVectorizer(max_features=3000)
tfidf = TfidfVectorizer(max_features=4600)
x = tfidf.fit_transform(data["Text"]).toarray()
y = data["Target"].values
with open("tfidf.pkl", "wb") as file:
pickle.dump(tfidf, file)
# Divide the data into train and test data
X_train, X_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=28, stratify=y
)
GaussianNBModel = GaussianNB()
MultinomialNBModel = MultinomialNB(alpha=1.0)
BernoulliNBModel = BernoulliNB(alpha=1.0, binarize=1)
LogisticRegressionModel = LogisticRegression(
penalty="l2", solver="sag", C=1.0, random_state=33
)
SGDClassifierModel = SGDClassifier(
penalty="l2", loss="squared_loss", learning_rate="optimal", random_state=33
)
RandomForestClassifierModel = RandomForestClassifier(
criterion="gini", n_estimators=300, max_depth=7, random_state=33
)
GBCModel = GradientBoostingClassifier(n_estimators=100, max_depth=3, random_state=33)
QDAModel = QuadraticDiscriminantAnalysis(tol=0.0001)
SVCModel = SVC(kernel="rbf", max_iter=100, C=1.0, gamma="auto")
DecisionTreeClassifierModel = DecisionTreeClassifier(
criterion="gini", max_depth=3, random_state=33
)
KNNClassifierModel = KNeighborsClassifier(
n_neighbors=5, weights="uniform", algorithm="auto"
)
Models = [
GaussianNBModel,
KNNClassifierModel,
MultinomialNBModel,
BernoulliNBModel,
LogisticRegressionModel,
RandomForestClassifierModel,
GBCModel,
SGDClassifierModel,
QDAModel,
SVCModel,
DecisionTreeClassifierModel,
]
ModelsScore = {}
for Model in Models:
print(f'for Model {str(Model).split("(")[0]}')
Model.fit(X_train, y_train)
print(f"Train Score is : {Model.score(X_train, y_train)}")
print(f"Test Score is : {Model.score(X_test, y_test)}")
y_pred = Model.predict(X_test)
ClassificationReport = classification_report(y_test, y_pred)
print("Classification Report is : \n", ClassificationReport)
print(f"Precision value is : {ClassificationReport.split()[19]}")
print(f"Recall value is : {ClassificationReport.split()[20]}")
print(f"F1 Score value is : {ClassificationReport.split()[21]}")
ModelsScore[str(Model).split("(")[0]] = [
ClassificationReport.split()[19],
ClassificationReport.split()[20],
ClassificationReport.split()[21],
]
print("=================================================")
type(data["Text"])
# **BERT MODEL**
# still under work
# # Deployment
model = GBCModel.fit(X_train, y_train)
model.score(X_train, y_train)
# Tokenize the text sample
txt = pd.Series(text)
tfidf_text = tfidf.transform(txt).toarray()
print(tfidf_text)
model.predict(tfidf_text)
dump(model, "model.joblib")
filename = "tfidf.sav"
pickle.dump(tfidf, open(filename, "wb"))
pre = pickle.load(open("pre.sav", "rb"))
txt = pd.Series(data)
te = tfidf.transform(txt).toarray()
model.predict(te)
| false | 0 | 3,865 | 0 | 4,030 | 3,865 |
||
129026593
|
import numpy as np
import pandas as pd
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.layers import Conv2D, MaxPooling2D
from tensorflow.keras.layers import Dense, Dropout, Flatten, Activation
from tensorflow.keras.models import Sequential
from tensorflow.keras.callbacks import EarlyStopping, ReduceLROnPlateau, ModelCheckpoint
# # Constant
BASE_PATH = "/kaggle/input/histopathologic-cancer-detection"
BASE_TRAIN_PATH = f"{BASE_PATH}/train"
BASE_TEST_PATH = f"{BASE_PATH}/test"
BASE_TRAIN_LABELS_PATH = "/kaggle/input/dataset-copy/new_dataset/train_labels.csv"
BASE_TEST_TRAIN_PATH = f"/kaggle/input/dataset-copy/new_dataset/train"
BASE_TEST_TRAIN_10000_PATH = f"{BASE_TEST_TRAIN_PATH}/10000"
BASE_TEST_TRAIN_50000_PATH = f"{BASE_TEST_TRAIN_PATH}/150000"
BASE_TEST_TRAIN_ALL_PATH = f"{BASE_TEST_TRAIN_PATH}/all"
SYMBOLINK_PATH = "/kaggle/working"
SYMBOLINK_TRAIN_PATH = f"{SYMBOLINK_PATH}/train"
SYMBOLINK_SMALLER_TRAIN_PATH = f"{SYMBOLINK_PATH}/smaller_train"
# # Load file using Keras
train_labels_df = pd.read_csv(BASE_TRAIN_LABELS_PATH)
train_labels_df.set_index("id", inplace=True)
train_labels_df.head()
# # Create lower dataset
import os
MAX_FILE_CNT = 1000
LABELS = ["non_cancer", "cancer"]
symbolic_smaller_train_cancer_path = f"{SYMBOLINK_SMALLER_TRAIN_PATH}/cancer"
symbolic_smaller_train_non_cancer_path = f"{SYMBOLINK_SMALLER_TRAIN_PATH}/non_cancer"
# Create directories
# Copy MAX_FILE_CNT of file
for i, file in enumerate(os.listdir(BASE_TRAIN_PATH)):
if i >= MAX_FILE_CNT:
break
name, _ = file.split(".")
label = train_labels_df.loc[[name], "label"].sum()
src_path = f"{BASE_TRAIN_PATH}/{file}"
dest_path = f"{SYMBOLINK_SMALLER_TRAIN_PATH}/{LABELS[label]}/{file}"
# Create symbolic link
from tensorflow.keras.preprocessing import image_dataset_from_directory
from tensorflow.keras.preprocessing.image import ImageDataGenerator
# train_datagen = ImageDataGenerator(
# '/kaggle/input/histopathologic-cancer-detection/train',
# labels='inferred',
# image_size=[32, 32, 3],
# interpolation='nearest',
# batch_size=64,
# shuffle=True,
# )
datagen = ImageDataGenerator(rescale=1.0 / 255)
train_gen = datagen.flow_from_directory(
BASE_TEST_TRAIN_10000_PATH, target_size=(96, 96), batch_size=32, class_mode="binary"
)
print(train_gen.samples)
# # Model
model = Sequential()
model.add(
Conv2D(32, (3, 3), activation="relu", input_shape=(32, 32, 3))
) # (30, 30, 32)
model.add(MaxPooling2D(pool_size=(2, 2))) # (15, 15, 32)
model.add(Conv2D(32, (3, 3), activation="relu")) # (13, 13, 32)
model.add(MaxPooling2D(pool_size=(2, 2))) # (6, 6, 64)
model.add(Conv2D(64, (3, 3), activation="relu")) # (4, 4, 64)
# model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Flatten()) # (1024, 1)
model.add(Dense(1024, activation="relu")) # (1024, 1)
model.add(Dense(64, activation="relu")) # (64, 1)
model.add(Dropout(0.3))
model.add(Dense(2, activation="softmax")) # (2, 1)
model.summary()
model.compile(Adam(lr=0.0001), loss="binary_crossentropy", metrics=["accuracy"])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/026/129026593.ipynb
| null | null |
[{"Id": 129026593, "ScriptId": 38252437, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14995669, "CreationDate": "05/10/2023 12:08:05", "VersionNumber": 13.0, "Title": "Histopathologic Cancer Detection", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 106.0, "LinesInsertedFromPrevious": 33.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 73.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.layers import Conv2D, MaxPooling2D
from tensorflow.keras.layers import Dense, Dropout, Flatten, Activation
from tensorflow.keras.models import Sequential
from tensorflow.keras.callbacks import EarlyStopping, ReduceLROnPlateau, ModelCheckpoint
# # Constant
BASE_PATH = "/kaggle/input/histopathologic-cancer-detection"
BASE_TRAIN_PATH = f"{BASE_PATH}/train"
BASE_TEST_PATH = f"{BASE_PATH}/test"
BASE_TRAIN_LABELS_PATH = "/kaggle/input/dataset-copy/new_dataset/train_labels.csv"
BASE_TEST_TRAIN_PATH = f"/kaggle/input/dataset-copy/new_dataset/train"
BASE_TEST_TRAIN_10000_PATH = f"{BASE_TEST_TRAIN_PATH}/10000"
BASE_TEST_TRAIN_50000_PATH = f"{BASE_TEST_TRAIN_PATH}/150000"
BASE_TEST_TRAIN_ALL_PATH = f"{BASE_TEST_TRAIN_PATH}/all"
SYMBOLINK_PATH = "/kaggle/working"
SYMBOLINK_TRAIN_PATH = f"{SYMBOLINK_PATH}/train"
SYMBOLINK_SMALLER_TRAIN_PATH = f"{SYMBOLINK_PATH}/smaller_train"
# # Load file using Keras
train_labels_df = pd.read_csv(BASE_TRAIN_LABELS_PATH)
train_labels_df.set_index("id", inplace=True)
train_labels_df.head()
# # Create lower dataset
import os
MAX_FILE_CNT = 1000
LABELS = ["non_cancer", "cancer"]
symbolic_smaller_train_cancer_path = f"{SYMBOLINK_SMALLER_TRAIN_PATH}/cancer"
symbolic_smaller_train_non_cancer_path = f"{SYMBOLINK_SMALLER_TRAIN_PATH}/non_cancer"
# Create directories
# Copy MAX_FILE_CNT of file
for i, file in enumerate(os.listdir(BASE_TRAIN_PATH)):
if i >= MAX_FILE_CNT:
break
name, _ = file.split(".")
label = train_labels_df.loc[[name], "label"].sum()
src_path = f"{BASE_TRAIN_PATH}/{file}"
dest_path = f"{SYMBOLINK_SMALLER_TRAIN_PATH}/{LABELS[label]}/{file}"
# Create symbolic link
from tensorflow.keras.preprocessing import image_dataset_from_directory
from tensorflow.keras.preprocessing.image import ImageDataGenerator
# train_datagen = ImageDataGenerator(
# '/kaggle/input/histopathologic-cancer-detection/train',
# labels='inferred',
# image_size=[32, 32, 3],
# interpolation='nearest',
# batch_size=64,
# shuffle=True,
# )
datagen = ImageDataGenerator(rescale=1.0 / 255)
train_gen = datagen.flow_from_directory(
BASE_TEST_TRAIN_10000_PATH, target_size=(96, 96), batch_size=32, class_mode="binary"
)
print(train_gen.samples)
# # Model
model = Sequential()
model.add(
Conv2D(32, (3, 3), activation="relu", input_shape=(32, 32, 3))
) # (30, 30, 32)
model.add(MaxPooling2D(pool_size=(2, 2))) # (15, 15, 32)
model.add(Conv2D(32, (3, 3), activation="relu")) # (13, 13, 32)
model.add(MaxPooling2D(pool_size=(2, 2))) # (6, 6, 64)
model.add(Conv2D(64, (3, 3), activation="relu")) # (4, 4, 64)
# model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Flatten()) # (1024, 1)
model.add(Dense(1024, activation="relu")) # (1024, 1)
model.add(Dense(64, activation="relu")) # (64, 1)
model.add(Dropout(0.3))
model.add(Dense(2, activation="softmax")) # (2, 1)
model.summary()
model.compile(Adam(lr=0.0001), loss="binary_crossentropy", metrics=["accuracy"])
| false | 0 | 1,149 | 0 | 1,149 | 1,149 |
||
129026803
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
train_df = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
test_df = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
sample_submission_data = pd.read_csv(
"/kaggle/input/playground-series-s3e14/sample_submission.csv"
)
train_df.shape
# # EDA
train_df.head()
train_df = train_df.drop(["id"], axis=1)
test_df = test_df.drop(["id"], axis=1)
train_df.hist(figsize=(14, 14), xrot=45)
plt.show()
plt.figure(figsize=(12, 12))
sns.heatmap(train_df.corr(), cmap="coolwarm", annot=True, fmt=".2f")
# * Since these 6 features MaxOfUpperTRange, MinOfUpperTRange, AverageOfUpperTRange, MaxOfLowerTRange, MinOfLowerTRange, AverageOfLowerTRange are 100% correlated with each other, we can drop any 5 of them.
# * 3 features fruitset, fruitmass and seeds are highly correlated.
# * RainingDays and AverageRainingDays are highly correlated.
# ['honeybee', 'bumbles', 'andrena', 'osmia', 'MaxOfUpperTRange']
columns_to_drop = [
"MinOfUpperTRange",
"AverageOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
"AverageOfLowerTRange",
]
train_df = train_df.drop(columns=columns_to_drop)
test_df = test_df.drop(columns=columns_to_drop)
features = train_df.columns.tolist()
# create subplots
fig, axes = plt.subplots(4, 3, figsize=(12, 12))
axes = axes.ravel()
# loop over features and plot scatter plot
for ax, feature in zip(axes, features):
ax.scatter(train_df[feature], train_df["yield"])
ax.set_xlabel(feature)
ax.set_ylabel("yield")
# adjust subplot spacing
# plt.subplots_adjust(hspace=0.5)
# display plot
plt.show()
train_df.columns
train_df.describe().T
# # Baseline model - linear regression
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error
# Split the dataset into training and testing sets
train_data, test_data = train_test_split(train_df, test_size=0.2, random_state=42)
# Define the features and target variable for the training set
X_train = train_data.drop(columns=["yield"])
y_train = train_data["yield"]
# Define the features and target variable for the testing set
X_test = test_data.drop(columns=["yield"])
y_test = test_data["yield"]
# Create an instance of the Linear Regression model
model = LinearRegression()
# Fit the model on the training data
model.fit(X_train, y_train)
# Make predictions on the testing data
y_pred = model.predict(X_test)
# Calculate the MAE of the model
mae = mean_absolute_error(y_test, y_pred)
print("MAE:", mae)
predictions_on_test_df = model.predict(test_df)
predictions_on_test_df
# # #Ensemble Models
import pandas as pd
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.metrics import mean_absolute_error
# Split the dataset into training and testing sets
train_data, test_data = train_test_split(train_df, test_size=0.2, random_state=42)
# Define the features and target variable for the training set
X_train = train_data.drop(columns=["yield"])
y_train = train_data["yield"]
# Define the features and target variable for the testing set
X_test = test_data.drop(columns=["yield"])
y_test = test_data["yield"]
# define the models
rf_model = RandomForestRegressor()
gb_model = GradientBoostingRegressor()
# define the hyperparameters to tune
# rf_param_grid = {'n_estimators': [50, 100, 150], 'max_depth': [5, 10, 15]}
# gb_param_grid = {'n_estimators': [50, 100, 150], 'max_depth': [5, 10, 15], 'learning_rate': [0.01, 0.1, 1.0]}
rf_param_grid = {
"n_estimators": [5, 20, 50, 100], # number of trees in the random forest
"max_features": [
"auto",
"sqrt",
], # number of features in consideration at every split
"max_depth": [
int(x) for x in np.linspace(10, 120, num=12)
], # maximum number of levels allowed in each decision tree
"min_samples_split": [2, 6, 10], # minimum sample number to split a node
"min_samples_leaf": [
1,
3,
4,
], # minimum sample number that can be stored in a leaf node
"bootstrap": [True, False], # method used to sample data points
}
gb_param_grid = {
"learning_rate": [0.01, 0.1],
"n_estimators": [100, 500, 1000],
"max_depth": [3, 5, 7],
"min_samples_split": [2, 5, 10],
"min_samples_leaf": [1, 2, 4],
"subsample": [0.5, 0.8, 1.0],
"max_features": ["sqrt", "log2", None],
}
# perform the grid search
rf_gs = GridSearchCV(rf_model, rf_param_grid, cv=5, n_jobs=-1)
gb_gs = GridSearchCV(gb_model, gb_param_grid, cv=5, n_jobs=-1)
# fit the models on the training data
rf_gs.fit(X_train, y_train)
gb_gs.fit(X_train, y_train)
# make predictions on the test data
rf_pred = rf_gs.predict(X_test)
gb_pred = gb_gs.predict(X_test)
# calculate the mean absolute error
rf_mae = mean_absolute_error(y_test, rf_pred)
gb_mae = mean_absolute_error(y_test, gb_pred)
print("Random Forest MAE: ", rf_mae)
print("Gradient Boosting MAE: ", gb_mae)
from xgboost import XGBRegressor
xg_model = XGBRegressor()
xg_param_grid = {
"max_depth": [3, 4, 5, 6, 7],
"learning_rate": [0.05, 0.1, 0.15, 0.2],
"subsample": [0.6, 0.7, 0.8, 0.9, 1.0],
"colsample_bytree": [0.6, 0.7, 0.8, 0.9, 1.0],
"n_estimators": [100, 200, 300, 400, 500],
"reg_alpha": [0.1, 0.01, 0.001, 0],
"reg_lambda": [1, 0.1, 0.01, 0.001, 0],
"gamma": [0, 0.1, 0.2, 0.3, 0.4],
"min_child_weight": [1, 2, 3, 4],
}
# perform the grid search
xg_gs = GridSearchCV(xg_model, xg_param_grid, cv=5, n_jobs=-1)
# fit the models on the training data
xg_gs.fit(X_train, y_train)
# make predictions on the test data
xg_pred = xg_gs.predict(X_test)
# calculate the mean absolute error
xg_mae = mean_absolute_error(y_test, xg_pred)
print("XGBoost MAE: ", xg_mae)
# get the best parameters
best_params = xg_gs.best_params_
print("Best parameters:", best_params)
xg_pred = xg_gs.predict(test_df)
# make predictions on the test data
rf_pred = rf_gs.predict(X_test)
gb_pred = gb_gs.predict(X_test)
rf_gb_mixed_pred = (rf_gs.predict(X_test) + gb_gs.predict(X_test)) / 2
rf_gb_mixed_mae = mean_absolute_error(y_test, rf_gb_mixed_pred)
print("Random Forest MAE: ", xg_mae)
print("Gradient Boosting MAE: ", gb_mae)
print("Mixed MAE: ", rf_gb_mixed_mae)
# make predictions on the test data
rf_pred = rf_gs.predict(test_df)
gb_pred = gb_gs.predict(test_df)
rf_gb_mixed_pred = (rf_gs.predict(test_df) + gb_gs.predict(test_df)) / 2
rf_gb_mixed_pred1 = (2 * rf_gs.predict(test_df) + gb_gs.predict(test_df)) / 3
print(gb_gs)
# # Output
sample_submission_data.head()
final_output_df = sample_submission_data.copy()
final_output_df["yield"] = xg_pred
final_output_df
final_output_df.to_csv(
"/kaggle/working/Wild_Blueberry_Yield_predictions_10th_may_1.csv", index=False
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/026/129026803.ipynb
| null | null |
[{"Id": 129026803, "ScriptId": 38067669, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3757397, "CreationDate": "05/10/2023 12:10:16", "VersionNumber": 4.0, "Title": "Quick EDA + Baseline Models", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 239.0, "LinesInsertedFromPrevious": 60.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 179.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
train_df = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
test_df = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
sample_submission_data = pd.read_csv(
"/kaggle/input/playground-series-s3e14/sample_submission.csv"
)
train_df.shape
# # EDA
train_df.head()
train_df = train_df.drop(["id"], axis=1)
test_df = test_df.drop(["id"], axis=1)
train_df.hist(figsize=(14, 14), xrot=45)
plt.show()
plt.figure(figsize=(12, 12))
sns.heatmap(train_df.corr(), cmap="coolwarm", annot=True, fmt=".2f")
# * Since these 6 features MaxOfUpperTRange, MinOfUpperTRange, AverageOfUpperTRange, MaxOfLowerTRange, MinOfLowerTRange, AverageOfLowerTRange are 100% correlated with each other, we can drop any 5 of them.
# * 3 features fruitset, fruitmass and seeds are highly correlated.
# * RainingDays and AverageRainingDays are highly correlated.
# ['honeybee', 'bumbles', 'andrena', 'osmia', 'MaxOfUpperTRange']
columns_to_drop = [
"MinOfUpperTRange",
"AverageOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
"AverageOfLowerTRange",
]
train_df = train_df.drop(columns=columns_to_drop)
test_df = test_df.drop(columns=columns_to_drop)
features = train_df.columns.tolist()
# create subplots
fig, axes = plt.subplots(4, 3, figsize=(12, 12))
axes = axes.ravel()
# loop over features and plot scatter plot
for ax, feature in zip(axes, features):
ax.scatter(train_df[feature], train_df["yield"])
ax.set_xlabel(feature)
ax.set_ylabel("yield")
# adjust subplot spacing
# plt.subplots_adjust(hspace=0.5)
# display plot
plt.show()
train_df.columns
train_df.describe().T
# # Baseline model - linear regression
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error
# Split the dataset into training and testing sets
train_data, test_data = train_test_split(train_df, test_size=0.2, random_state=42)
# Define the features and target variable for the training set
X_train = train_data.drop(columns=["yield"])
y_train = train_data["yield"]
# Define the features and target variable for the testing set
X_test = test_data.drop(columns=["yield"])
y_test = test_data["yield"]
# Create an instance of the Linear Regression model
model = LinearRegression()
# Fit the model on the training data
model.fit(X_train, y_train)
# Make predictions on the testing data
y_pred = model.predict(X_test)
# Calculate the MAE of the model
mae = mean_absolute_error(y_test, y_pred)
print("MAE:", mae)
predictions_on_test_df = model.predict(test_df)
predictions_on_test_df
# # #Ensemble Models
import pandas as pd
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.metrics import mean_absolute_error
# Split the dataset into training and testing sets
train_data, test_data = train_test_split(train_df, test_size=0.2, random_state=42)
# Define the features and target variable for the training set
X_train = train_data.drop(columns=["yield"])
y_train = train_data["yield"]
# Define the features and target variable for the testing set
X_test = test_data.drop(columns=["yield"])
y_test = test_data["yield"]
# define the models
rf_model = RandomForestRegressor()
gb_model = GradientBoostingRegressor()
# define the hyperparameters to tune
# rf_param_grid = {'n_estimators': [50, 100, 150], 'max_depth': [5, 10, 15]}
# gb_param_grid = {'n_estimators': [50, 100, 150], 'max_depth': [5, 10, 15], 'learning_rate': [0.01, 0.1, 1.0]}
rf_param_grid = {
"n_estimators": [5, 20, 50, 100], # number of trees in the random forest
"max_features": [
"auto",
"sqrt",
], # number of features in consideration at every split
"max_depth": [
int(x) for x in np.linspace(10, 120, num=12)
], # maximum number of levels allowed in each decision tree
"min_samples_split": [2, 6, 10], # minimum sample number to split a node
"min_samples_leaf": [
1,
3,
4,
], # minimum sample number that can be stored in a leaf node
"bootstrap": [True, False], # method used to sample data points
}
gb_param_grid = {
"learning_rate": [0.01, 0.1],
"n_estimators": [100, 500, 1000],
"max_depth": [3, 5, 7],
"min_samples_split": [2, 5, 10],
"min_samples_leaf": [1, 2, 4],
"subsample": [0.5, 0.8, 1.0],
"max_features": ["sqrt", "log2", None],
}
# perform the grid search
rf_gs = GridSearchCV(rf_model, rf_param_grid, cv=5, n_jobs=-1)
gb_gs = GridSearchCV(gb_model, gb_param_grid, cv=5, n_jobs=-1)
# fit the models on the training data
rf_gs.fit(X_train, y_train)
gb_gs.fit(X_train, y_train)
# make predictions on the test data
rf_pred = rf_gs.predict(X_test)
gb_pred = gb_gs.predict(X_test)
# calculate the mean absolute error
rf_mae = mean_absolute_error(y_test, rf_pred)
gb_mae = mean_absolute_error(y_test, gb_pred)
print("Random Forest MAE: ", rf_mae)
print("Gradient Boosting MAE: ", gb_mae)
from xgboost import XGBRegressor
xg_model = XGBRegressor()
xg_param_grid = {
"max_depth": [3, 4, 5, 6, 7],
"learning_rate": [0.05, 0.1, 0.15, 0.2],
"subsample": [0.6, 0.7, 0.8, 0.9, 1.0],
"colsample_bytree": [0.6, 0.7, 0.8, 0.9, 1.0],
"n_estimators": [100, 200, 300, 400, 500],
"reg_alpha": [0.1, 0.01, 0.001, 0],
"reg_lambda": [1, 0.1, 0.01, 0.001, 0],
"gamma": [0, 0.1, 0.2, 0.3, 0.4],
"min_child_weight": [1, 2, 3, 4],
}
# perform the grid search
xg_gs = GridSearchCV(xg_model, xg_param_grid, cv=5, n_jobs=-1)
# fit the models on the training data
xg_gs.fit(X_train, y_train)
# make predictions on the test data
xg_pred = xg_gs.predict(X_test)
# calculate the mean absolute error
xg_mae = mean_absolute_error(y_test, xg_pred)
print("XGBoost MAE: ", xg_mae)
# get the best parameters
best_params = xg_gs.best_params_
print("Best parameters:", best_params)
xg_pred = xg_gs.predict(test_df)
# make predictions on the test data
rf_pred = rf_gs.predict(X_test)
gb_pred = gb_gs.predict(X_test)
rf_gb_mixed_pred = (rf_gs.predict(X_test) + gb_gs.predict(X_test)) / 2
rf_gb_mixed_mae = mean_absolute_error(y_test, rf_gb_mixed_pred)
print("Random Forest MAE: ", xg_mae)
print("Gradient Boosting MAE: ", gb_mae)
print("Mixed MAE: ", rf_gb_mixed_mae)
# make predictions on the test data
rf_pred = rf_gs.predict(test_df)
gb_pred = gb_gs.predict(test_df)
rf_gb_mixed_pred = (rf_gs.predict(test_df) + gb_gs.predict(test_df)) / 2
rf_gb_mixed_pred1 = (2 * rf_gs.predict(test_df) + gb_gs.predict(test_df)) / 3
print(gb_gs)
# # Output
sample_submission_data.head()
final_output_df = sample_submission_data.copy()
final_output_df["yield"] = xg_pred
final_output_df
final_output_df.to_csv(
"/kaggle/working/Wild_Blueberry_Yield_predictions_10th_may_1.csv", index=False
)
| false | 0 | 2,717 | 0 | 2,717 | 2,717 |
||
129461814
|
<jupyter_start><jupyter_text>Amazon Fine Food Reviews
## Context
This dataset consists of reviews of fine foods from amazon. The data span a period of more than 10 years, including all ~500,000 reviews up to October 2012. Reviews include product and user information, ratings, and a plain text review. It also includes reviews from all other Amazon categories.
## Contents
- Reviews.csv: Pulled from the corresponding SQLite table named Reviews in database.sqlite<br>
- database.sqlite: Contains the table 'Reviews'<br><br>
Data includes:<br>
- Reviews from Oct 1999 - Oct 2012<br>
- 568,454 reviews<br>
- 256,059 users<br>
- 74,258 products<br>
- 260 users with > 50 reviews<br>
[](https://www.kaggle.com/benhamner/d/snap/amazon-fine-food-reviews/reviews-wordcloud)
## Acknowledgements
See [this SQLite query](https://www.kaggle.com/benhamner/d/snap/amazon-fine-food-reviews/data-sample) for a quick sample of the dataset.
If you publish articles based on this dataset, please cite the following paper:
- J. McAuley and J. Leskovec. [From amateurs to connoisseurs: modeling the evolution of user expertise through online reviews](http://i.stanford.edu/~julian/pdfs/www13.pdf). WWW, 2013.
Kaggle dataset identifier: amazon-fine-food-reviews
<jupyter_script>import numpy as np
import pandas as pd
from subprocess import check_output
print(check_output(["ls", "../input"]).decode("utf8"))
# # Data Preprocessing
# Here, Take the data which the user and item appear more than 10 times in order to reduce the calculation because we just want to take a small scale test.
# The data() function returns the total number of users and products, the user-item table and also the train&test data set.
import pandas as pd
import numpy as np
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
def data_clean(df, feature, m):
count = df[feature].value_counts()
df = df[df[feature].isin(count[count > m].index)]
return df
def data_clean_sum(df, features, m):
fil = df.ProductId.value_counts()
fil2 = df.UserId.value_counts()
df["#Proudcts"] = df.ProductId.apply(lambda x: fil[x])
df["#Users"] = df.UserId.apply(lambda x: fil2[x])
while (df.ProductId.value_counts(ascending=True)[0]) < m or (
df.UserId.value_counts(ascending=True)[0] < m
):
df = data_clean(df, features[0], m)
df = data_clean(df, features[1], m)
return df
def data():
print("Loading data...")
df = pd.read_csv("../input/Reviews.csv")
df["datetime"] = pd.to_datetime(df.Time, unit="s")
raw_data = data_clean_sum(df, ["ProductId", "UserId"], 10)
# find X,and y
raw_data["uid"] = pd.factorize(raw_data["UserId"])[0]
raw_data["pid"] = pd.factorize(raw_data["ProductId"])[0]
sc = MinMaxScaler()
# Sepreate the features
X1 = raw_data.loc[:, ["uid", "pid"]]
y = raw_data.Score
# train_test split
X1_train, X1_test, y_train, y_test = train_test_split(
X1, y, test_size=0.3, random_state=2017
)
train = np.array(X1_train.join(y_train))
test = np.array(X1_test.join(y_test))
# got the productId to pid index
pid2PID = raw_data.ProductId.unique()
data_mixed = X1.join(y)
total_p = data_mixed["pid"].unique().shape[0]
total_u = data_mixed["uid"].unique().shape[0]
# make the user-item table
table = np.zeros([total_u, total_p])
z = np.array(data_mixed)
# if some one score a single thing several times
for line in z:
u, p, s = line
if table[u][p] < s:
table[u][p] = s
print("the table's shape is:")
print(table.shape)
return z, total_u, total_p, pid2PID, train, test, table
z, total_u, total_p, pid2PID, train, test, table = data()
# # Evaulation
# MSE and confusion matrix
from sklearn.metrics import mean_squared_error
import matplotlib.pyplot as plt
def caculate_mse(x):
MSE1 = []
MSE2 = []
for line in train:
u, p, s = line
MSE1.append(s)
MSE2.append(x[u, p])
MSE_in_sample = np.sqrt(mean_squared_error(MSE1, MSE2))
MSE3 = []
MSE4 = []
for line in test:
u, p, s = line
MSE3.append(s)
MSE4.append(x[u, p])
MSE_out_sample = np.sqrt(mean_squared_error(MSE3, MSE4))
print("Training RMSE = {}\nTesting RMSE = {}".format(MSE_in_sample, MSE_out_sample))
import itertools
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix
def plot_confusion_matrix(
cm, classes, normalize=False, title="Confusion matrix", cmap=plt.cm.Blues
):
if normalize:
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print("Confusion matrix, without normalization")
print(cm)
plt.imshow(cm, interpolation="nearest", cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = ".2f" if normalize else "d"
thresh = cm.max() / 2.0
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(
j,
i,
format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black",
)
plt.tight_layout()
plt.ylabel("True label")
plt.xlabel("Predicted label")
def drawcm(y_pred, y_test=test, title=""):
print("caculating cm..")
y1 = []
y2 = []
for line in y_test:
u, p, s = line
y1.append(s)
y2.append(y_pred[u, p])
temp1 = []
temp2 = []
for i in range(len(y1)):
if np.array(y1)[i] >= 4:
temp1.append(1)
elif np.array(y1)[i] <= 2:
temp1.append(0)
else:
temp1.append(0)
if y2[i] >= 4:
temp2.append(1)
elif y2[i] <= 2:
temp2.append(0)
else:
temp2.append(0)
cm = confusion_matrix(temp1, temp2)
plt.figure()
plot_confusion_matrix(
cm, classes=["not", "recommended"], normalize=True, title=title
)
plt.show()
TP = cm[1, 1]
FP = cm[0, 1]
TN = cm[0, 0]
FN = cm[1, 0]
return TP, FP, TN, FN
# # Recommendation function
# # SVD
from numpy import *
from scipy.sparse.linalg import svds
from numpy import linalg as la
from sklearn.preprocessing import MinMaxScaler
def svdrec(table=table, factors=500):
UI = matrix(table)
user_ratings_mean = mean(UI, axis=0)
user_ratings_mean = user_ratings_mean.reshape(1, -1)
UI_demeaned = UI - user_ratings_mean
U, sigma, Vt = svds(UI_demeaned, factors)
sigma = diag(sigma)
pred_mat = dot(dot(U, sigma), Vt) + user_ratings_mean
sc = MinMaxScaler(feature_range=(1, 5))
pred_mat = sc.fit_transform(pred_mat)
return pred_mat
result1 = svdrec(factors=150)
caculate_mse(result1)
TP, FP, TN, FN = drawcm(result1, title="SVD")
precision = TP / (TP + FP)
recall = TP / (TP + FN)
F1 = 2 * (precision * recall) / (precision + recall)
print("Precision:", precision)
print("Recall:", recall)
print("F1:", F1)
import urllib
import json
from bs4 import BeautifulSoup
from IPython.display import display
from PIL import Image
def get_amazon_product_image_url(product_id):
try:
url = "https://www.amazon.com/dp/" + product_id
req = urllib.request.Request(
url, data=None, headers={"User-Agent": "Mozilla/5.0"}
)
f = urllib.request.urlopen(req)
page = f.read().decode("utf-8")
soup = BeautifulSoup(page, "html.parser")
img_div = soup.find(id="imgTagWrapperId")
imgs_str = img_div.img.get("data-a-dynamic-image")
imgs_dict = json.loads(imgs_str)
first_link = list(imgs_dict.keys())[0]
return first_link
except urllib.error.HTTPError as e:
return None
def show_amazon_product_image(product_id, size=(200, 200)):
url = get_amazon_product_image_url(product_id)
if url is not None:
img = Image.open(urllib.request.urlopen(url))
img = img.resize(size)
display(img)
import random
import time
def rec(result, uid, num_items, print_images=True):
if uid in range(total_u):
top_N = np.argpartition(result[uid], -num_items)[-num_items:]
print(
"The Top {} Recommended Products For User {} is {}".format(
num_items, uid, pid2PID[top_N]
)
)
if print_images == True:
for prd in pid2PID[top_N]:
show_amazon_product_image(prd)
time.sleep(random.randint(2, 4))
else:
print("this user has not bought anything")
rec(result1, uid=87, num_items=5, print_images=True)
rec(result1, uid=1350, num_items=5, print_images=True)
rec(result1, uid=325, num_items=5, print_images=True)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/461/129461814.ipynb
|
amazon-fine-food-reviews
| null |
[{"Id": 129461814, "ScriptId": 38458333, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3435161, "CreationDate": "05/14/2023 03:26:24", "VersionNumber": 1.0, "Title": "Amazon Fine Food Recommendation System using SVD", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 252.0, "LinesInsertedFromPrevious": 97.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 155.0, "LinesInsertedFromFork": 97.0, "LinesDeletedFromFork": 405.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 155.0, "TotalVotes": 0}]
|
[{"Id": 185539110, "KernelVersionId": 129461814, "SourceDatasetVersionId": 2157}]
|
[{"Id": 2157, "DatasetId": 18, "DatasourceVersionId": 2157, "CreatorUserId": 500099, "LicenseName": "CC0: Public Domain", "CreationDate": "05/01/2017 18:51:31", "VersionNumber": 2.0, "Title": "Amazon Fine Food Reviews", "Slug": "amazon-fine-food-reviews", "Subtitle": "Analyze ~500,000 food reviews from Amazon", "Description": "## Context\n\nThis dataset consists of reviews of fine foods from amazon. The data span a period of more than 10 years, including all ~500,000 reviews up to October 2012. Reviews include product and user information, ratings, and a plain text review. It also includes reviews from all other Amazon categories.\n\n\n## Contents\n\n- Reviews.csv: Pulled from the corresponding SQLite table named Reviews in database.sqlite<br>\n- database.sqlite: Contains the table 'Reviews'<br><br>\n\nData includes:<br>\n- Reviews from Oct 1999 - Oct 2012<br>\n- 568,454 reviews<br>\n- 256,059 users<br>\n- 74,258 products<br>\n- 260 users with > 50 reviews<br>\n\n\n[](https://www.kaggle.com/benhamner/d/snap/amazon-fine-food-reviews/reviews-wordcloud)\n\n\n## Acknowledgements\n\nSee [this SQLite query](https://www.kaggle.com/benhamner/d/snap/amazon-fine-food-reviews/data-sample) for a quick sample of the dataset.\n\nIf you publish articles based on this dataset, please cite the following paper:\n\n - J. McAuley and J. Leskovec. [From amateurs to connoisseurs: modeling the evolution of user expertise through online reviews](http://i.stanford.edu/~julian/pdfs/www13.pdf). WWW, 2013.", "VersionNotes": "Re-uploading the files so they appear in file previews.", "TotalCompressedBytes": 673703435.0, "TotalUncompressedBytes": 673703435.0}]
|
[{"Id": 18, "CreatorUserId": 500099, "OwnerUserId": NaN, "OwnerOrganizationId": 229.0, "CurrentDatasetVersionId": 2157.0, "CurrentDatasourceVersionId": 2157.0, "ForumId": 993, "Type": 2, "CreationDate": "01/08/2016 21:12:10", "LastActivityDate": "02/06/2018", "TotalViews": 856266, "TotalDownloads": 156282, "TotalVotes": 2086, "TotalKernels": 817}]
| null |
import numpy as np
import pandas as pd
from subprocess import check_output
print(check_output(["ls", "../input"]).decode("utf8"))
# # Data Preprocessing
# Here, Take the data which the user and item appear more than 10 times in order to reduce the calculation because we just want to take a small scale test.
# The data() function returns the total number of users and products, the user-item table and also the train&test data set.
import pandas as pd
import numpy as np
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
def data_clean(df, feature, m):
count = df[feature].value_counts()
df = df[df[feature].isin(count[count > m].index)]
return df
def data_clean_sum(df, features, m):
fil = df.ProductId.value_counts()
fil2 = df.UserId.value_counts()
df["#Proudcts"] = df.ProductId.apply(lambda x: fil[x])
df["#Users"] = df.UserId.apply(lambda x: fil2[x])
while (df.ProductId.value_counts(ascending=True)[0]) < m or (
df.UserId.value_counts(ascending=True)[0] < m
):
df = data_clean(df, features[0], m)
df = data_clean(df, features[1], m)
return df
def data():
print("Loading data...")
df = pd.read_csv("../input/Reviews.csv")
df["datetime"] = pd.to_datetime(df.Time, unit="s")
raw_data = data_clean_sum(df, ["ProductId", "UserId"], 10)
# find X,and y
raw_data["uid"] = pd.factorize(raw_data["UserId"])[0]
raw_data["pid"] = pd.factorize(raw_data["ProductId"])[0]
sc = MinMaxScaler()
# Sepreate the features
X1 = raw_data.loc[:, ["uid", "pid"]]
y = raw_data.Score
# train_test split
X1_train, X1_test, y_train, y_test = train_test_split(
X1, y, test_size=0.3, random_state=2017
)
train = np.array(X1_train.join(y_train))
test = np.array(X1_test.join(y_test))
# got the productId to pid index
pid2PID = raw_data.ProductId.unique()
data_mixed = X1.join(y)
total_p = data_mixed["pid"].unique().shape[0]
total_u = data_mixed["uid"].unique().shape[0]
# make the user-item table
table = np.zeros([total_u, total_p])
z = np.array(data_mixed)
# if some one score a single thing several times
for line in z:
u, p, s = line
if table[u][p] < s:
table[u][p] = s
print("the table's shape is:")
print(table.shape)
return z, total_u, total_p, pid2PID, train, test, table
z, total_u, total_p, pid2PID, train, test, table = data()
# # Evaulation
# MSE and confusion matrix
from sklearn.metrics import mean_squared_error
import matplotlib.pyplot as plt
def caculate_mse(x):
MSE1 = []
MSE2 = []
for line in train:
u, p, s = line
MSE1.append(s)
MSE2.append(x[u, p])
MSE_in_sample = np.sqrt(mean_squared_error(MSE1, MSE2))
MSE3 = []
MSE4 = []
for line in test:
u, p, s = line
MSE3.append(s)
MSE4.append(x[u, p])
MSE_out_sample = np.sqrt(mean_squared_error(MSE3, MSE4))
print("Training RMSE = {}\nTesting RMSE = {}".format(MSE_in_sample, MSE_out_sample))
import itertools
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix
def plot_confusion_matrix(
cm, classes, normalize=False, title="Confusion matrix", cmap=plt.cm.Blues
):
if normalize:
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print("Confusion matrix, without normalization")
print(cm)
plt.imshow(cm, interpolation="nearest", cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = ".2f" if normalize else "d"
thresh = cm.max() / 2.0
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(
j,
i,
format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black",
)
plt.tight_layout()
plt.ylabel("True label")
plt.xlabel("Predicted label")
def drawcm(y_pred, y_test=test, title=""):
print("caculating cm..")
y1 = []
y2 = []
for line in y_test:
u, p, s = line
y1.append(s)
y2.append(y_pred[u, p])
temp1 = []
temp2 = []
for i in range(len(y1)):
if np.array(y1)[i] >= 4:
temp1.append(1)
elif np.array(y1)[i] <= 2:
temp1.append(0)
else:
temp1.append(0)
if y2[i] >= 4:
temp2.append(1)
elif y2[i] <= 2:
temp2.append(0)
else:
temp2.append(0)
cm = confusion_matrix(temp1, temp2)
plt.figure()
plot_confusion_matrix(
cm, classes=["not", "recommended"], normalize=True, title=title
)
plt.show()
TP = cm[1, 1]
FP = cm[0, 1]
TN = cm[0, 0]
FN = cm[1, 0]
return TP, FP, TN, FN
# # Recommendation function
# # SVD
from numpy import *
from scipy.sparse.linalg import svds
from numpy import linalg as la
from sklearn.preprocessing import MinMaxScaler
def svdrec(table=table, factors=500):
UI = matrix(table)
user_ratings_mean = mean(UI, axis=0)
user_ratings_mean = user_ratings_mean.reshape(1, -1)
UI_demeaned = UI - user_ratings_mean
U, sigma, Vt = svds(UI_demeaned, factors)
sigma = diag(sigma)
pred_mat = dot(dot(U, sigma), Vt) + user_ratings_mean
sc = MinMaxScaler(feature_range=(1, 5))
pred_mat = sc.fit_transform(pred_mat)
return pred_mat
result1 = svdrec(factors=150)
caculate_mse(result1)
TP, FP, TN, FN = drawcm(result1, title="SVD")
precision = TP / (TP + FP)
recall = TP / (TP + FN)
F1 = 2 * (precision * recall) / (precision + recall)
print("Precision:", precision)
print("Recall:", recall)
print("F1:", F1)
import urllib
import json
from bs4 import BeautifulSoup
from IPython.display import display
from PIL import Image
def get_amazon_product_image_url(product_id):
try:
url = "https://www.amazon.com/dp/" + product_id
req = urllib.request.Request(
url, data=None, headers={"User-Agent": "Mozilla/5.0"}
)
f = urllib.request.urlopen(req)
page = f.read().decode("utf-8")
soup = BeautifulSoup(page, "html.parser")
img_div = soup.find(id="imgTagWrapperId")
imgs_str = img_div.img.get("data-a-dynamic-image")
imgs_dict = json.loads(imgs_str)
first_link = list(imgs_dict.keys())[0]
return first_link
except urllib.error.HTTPError as e:
return None
def show_amazon_product_image(product_id, size=(200, 200)):
url = get_amazon_product_image_url(product_id)
if url is not None:
img = Image.open(urllib.request.urlopen(url))
img = img.resize(size)
display(img)
import random
import time
def rec(result, uid, num_items, print_images=True):
if uid in range(total_u):
top_N = np.argpartition(result[uid], -num_items)[-num_items:]
print(
"The Top {} Recommended Products For User {} is {}".format(
num_items, uid, pid2PID[top_N]
)
)
if print_images == True:
for prd in pid2PID[top_N]:
show_amazon_product_image(prd)
time.sleep(random.randint(2, 4))
else:
print("this user has not bought anything")
rec(result1, uid=87, num_items=5, print_images=True)
rec(result1, uid=1350, num_items=5, print_images=True)
rec(result1, uid=325, num_items=5, print_images=True)
| false | 0 | 2,451 | 0 | 2,913 | 2,451 |
||
129461161
|
<jupyter_start><jupyter_text>PokemonUniteDataset
**Description**
Pokemon Unite is a new MOBA(Multiplayer Online Battle Arena) released for Mobile and Nintendo Switch. The game became instantly popular after its initial release on Nintendo Switch but it has grown even more after being released on Mobile devices and even won game of the year on play store in US region and several other awards in different categories in several other regions in the annual Google Play Awards 2021.
However I didn't find any dataset on Pokemon Unite on kaggle even after such popularity, so I decided to make one. The data is collected from the official Pokemon Unite website : https://unite.pokemon.com/en-us/ and processed and cleaned by me. It has all the information about every pokemon released in the game till date and I hope to keep on updating it after every new pokemon release in the game.
The data contains all the information one would get if one tries to get information by logging into the game, but with this dataset one can easily compare pokemons which is bit difficult to do in the game directly.
**Acknowledgement**
Pokemon Unite official website : https://unite.pokemon.com/en-us/
**Inspiration**
There are a number of pokemons which I personally believe do not justify the stats(Offense, Endurance etc.) comapred to the role assigned, so from this data I hope one can get meaningful and new insights.
**About the Data**
The data is a csv file which contains all the information one can get from the game.
Kaggle dataset identifier: pokemonunitedataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/pokemonunitedataset/PokemonUniteData.csv")
df.head()
df.columns
df.shape
# Role distribution
df["Role"].value_counts()
# Ranged or melee distribution
df["Ranged_or_Melee"].value_counts()
# Role and Ranged/melee pairwise distribution
df[["Role", "Ranged_or_Melee"]].value_counts().plot(kind="bar")
# **Useful Insights: **
# * All attackers are ranged
# * All All-Rounders are melee
# * All Speedsters are melee
# * Only Supporters and defenders have both attack types
roles = {}
for role in df.Role:
roles[role] = df[df["Role"] == role]
roles.keys()
# Attacker stats
roles["Attacker"].describe()
roles["Attacker"]["Offense"].max()
# Max Offense Attacker
attacker_offense = roles["Attacker"]["Offense"]
roles["Attacker"][attacker_offense == attacker_offense.max()]
stats = [
col
for col in roles["Attacker"].columns
if roles["Attacker"][col].dtype == "float64"
]
def plot_stats_by_role(df, role):
df.plot(x="Name", y=stats, kind="bar", figsize=(10, 5), title=role)
# Plotting Stat distribution role wise
for role in roles:
plot_stats_by_role(roles[role], role)
def most_balanced_stats(df):
df["std"] = df[stats].std(axis=1)
return df[df["std"] == df["std"].min()]
# Most Balaced stats pokemon for each role
m = float("inf")
mp = None
for role in roles:
poke = most_balanced_stats(roles[role])
print("Most balanced", role, "is", poke.iloc[0]["Name"])
if m > poke.iloc[0]["std"]:
m = min(m, poke.iloc[0]["std"])
mp = poke
print()
print("Most balanced Pokemon is", mp.iloc[0]["Name"])
df.groupby("Role").size().plot(kind="pie", autopct="%.2f")
df.groupby("Ranged_or_Melee").size().plot(kind="pie", autopct="%.2f")
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
df.drop("Role", axis=1, inplace=True)
df.head()
label = df["Role"]
label.shape
df.drop(["Name", "Description"], axis=1, inplace=True)
df_dummies = pd.get_dummies(df)
df_dummies.head()
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
label = le.fit_transform(label)
label
x_train, x_test, y_train, y_test = train_test_split(df_dummies, label, test_size=0.2)
x_train
model = DecisionTreeClassifier()
model.fit(x_train, y_train)
model.score(x_test, y_test)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/461/129461161.ipynb
|
pokemonunitedataset
|
vishushekhar
|
[{"Id": 129461161, "ScriptId": 38491525, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2005514, "CreationDate": "05/14/2023 03:15:38", "VersionNumber": 1.0, "Title": "notebook128a66f768", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 123.0, "LinesInsertedFromPrevious": 123.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185537752, "KernelVersionId": 129461161, "SourceDatasetVersionId": 5590260}]
|
[{"Id": 5590260, "DatasetId": 1845089, "DatasourceVersionId": 5665199, "CreatorUserId": 7033995, "LicenseName": "CC0: Public Domain", "CreationDate": "05/03/2023 07:49:54", "VersionNumber": 24.0, "Title": "PokemonUniteDataset", "Slug": "pokemonunitedataset", "Subtitle": "Pokemon Unite Dataset containing all the in game information about the pokemons.", "Description": "**Description**\n\nPokemon Unite is a new MOBA(Multiplayer Online Battle Arena) released for Mobile and Nintendo Switch. The game became instantly popular after its initial release on Nintendo Switch but it has grown even more after being released on Mobile devices and even won game of the year on play store in US region and several other awards in different categories in several other regions in the annual Google Play Awards 2021. \n\nHowever I didn't find any dataset on Pokemon Unite on kaggle even after such popularity, so I decided to make one. The data is collected from the official Pokemon Unite website : https://unite.pokemon.com/en-us/ and processed and cleaned by me. It has all the information about every pokemon released in the game till date and I hope to keep on updating it after every new pokemon release in the game.\n\nThe data contains all the information one would get if one tries to get information by logging into the game, but with this dataset one can easily compare pokemons which is bit difficult to do in the game directly.\n\n**Acknowledgement**\nPokemon Unite official website : https://unite.pokemon.com/en-us/ \n\n**Inspiration**\nThere are a number of pokemons which I personally believe do not justify the stats(Offense, Endurance etc.) comapred to the role assigned, so from this data I hope one can get meaningful and new insights.\n\n **About the Data**\nThe data is a csv file which contains all the information one can get from the game.", "VersionNotes": "Data Update 2023-05-03", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1845089, "CreatorUserId": 7033995, "OwnerUserId": 7033995.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 6133178.0, "CurrentDatasourceVersionId": 6211966.0, "ForumId": 1867980, "Type": 2, "CreationDate": "01/06/2022 14:32:36", "LastActivityDate": "01/06/2022", "TotalViews": 3678, "TotalDownloads": 308, "TotalVotes": 12, "TotalKernels": 2}]
|
[{"Id": 7033995, "UserName": "vishushekhar", "DisplayName": "Shashanka Shekhar", "RegisterDate": "03/26/2021", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/pokemonunitedataset/PokemonUniteData.csv")
df.head()
df.columns
df.shape
# Role distribution
df["Role"].value_counts()
# Ranged or melee distribution
df["Ranged_or_Melee"].value_counts()
# Role and Ranged/melee pairwise distribution
df[["Role", "Ranged_or_Melee"]].value_counts().plot(kind="bar")
# **Useful Insights: **
# * All attackers are ranged
# * All All-Rounders are melee
# * All Speedsters are melee
# * Only Supporters and defenders have both attack types
roles = {}
for role in df.Role:
roles[role] = df[df["Role"] == role]
roles.keys()
# Attacker stats
roles["Attacker"].describe()
roles["Attacker"]["Offense"].max()
# Max Offense Attacker
attacker_offense = roles["Attacker"]["Offense"]
roles["Attacker"][attacker_offense == attacker_offense.max()]
stats = [
col
for col in roles["Attacker"].columns
if roles["Attacker"][col].dtype == "float64"
]
def plot_stats_by_role(df, role):
df.plot(x="Name", y=stats, kind="bar", figsize=(10, 5), title=role)
# Plotting Stat distribution role wise
for role in roles:
plot_stats_by_role(roles[role], role)
def most_balanced_stats(df):
df["std"] = df[stats].std(axis=1)
return df[df["std"] == df["std"].min()]
# Most Balaced stats pokemon for each role
m = float("inf")
mp = None
for role in roles:
poke = most_balanced_stats(roles[role])
print("Most balanced", role, "is", poke.iloc[0]["Name"])
if m > poke.iloc[0]["std"]:
m = min(m, poke.iloc[0]["std"])
mp = poke
print()
print("Most balanced Pokemon is", mp.iloc[0]["Name"])
df.groupby("Role").size().plot(kind="pie", autopct="%.2f")
df.groupby("Ranged_or_Melee").size().plot(kind="pie", autopct="%.2f")
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
df.drop("Role", axis=1, inplace=True)
df.head()
label = df["Role"]
label.shape
df.drop(["Name", "Description"], axis=1, inplace=True)
df_dummies = pd.get_dummies(df)
df_dummies.head()
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
label = le.fit_transform(label)
label
x_train, x_test, y_train, y_test = train_test_split(df_dummies, label, test_size=0.2)
x_train
model = DecisionTreeClassifier()
model.fit(x_train, y_train)
model.score(x_test, y_test)
| false | 1 | 961 | 0 | 1,326 | 961 |
||
129416800
|
<jupyter_start><jupyter_text>Indian startups
# Context
A startup or start-up is a company or project undertaken by an entrepreneur to seek, develop, and validate a scalable business model. While entrepreneurship includes all new businesses, including self-employment and businesses that do not intend to go public, startups are new businesses that intend to grow large beyond the solo founder. In the beginning, startups face high uncertainty and have high rates of failure, but a minority of them do go on to be successful and influential.
# Content
The following dataset has data about the Top 300 startups in India. Details about the columns are as follows:
1. Company - Name of the Startup.
2. City - The City in which the startup is started.
3. Starting Year - The Year in which the startup was started.
4. Founders - Name of the founders of the startup.
5. Industries - Industrial domain in which the startup falls.
6. No. of Employees - Number of employees in the startup.
7. Funding Amount in USD - Total funding amount funded to the startup.
8. Funding Rounds - Funding rounds are the number of times a startup goes back to the market to raise more capital. The goal of every round is for founders to trade equity in their business for the capital they can utilize to advance their companies to the next level.
9. No. of Investors - Number of investors in the startup.
Kaggle dataset identifier: indian-startups-top-300
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # About Dataset
# Content The following dataset has data about the Top 300 startups in India. Details about the columns are as follows:
# * Company - Name of the Startup.
# * City - The City in which the startup is started.
# * Starting Year - The Year in which the startup was started.
# * Founders - Name of the founders of the startup.
# * Industries - Industrial domain in which the startup falls.
# * No. of Employees - Number of employees in the startup.
# * Funding Amount in USD - Total funding amount funded to the startup.
# * Funding Rounds - Funding rounds are the number of times a startup goes back to the market to raise more capital. The goal of every round is for founders to trade equity in their business for the capital they can utilize to advance their companies to the next level.
# * No. of Investors - Number of investors in the startup.
# 
# # Indian Startups (Exploratory data analysis)
# importing libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# importing data
df = pd.read_csv("/kaggle/input/indian-startups-top-300/Startups1.csv", index_col=0)
df.sample(6).reset_index(drop=True).style.set_properties(
**{"background-color": "#FFF0F5 ", "color": "black", "border-color": "#8b8c8c"}
)
# # Workflow
# * Understanding of data
# * Data Cleaning & Prepocessing
# * EDA
# * Insights
# # Understanding the data
# data size
df.shape
# head
df.head()
# tail
df.tail()
# data types
df.dtypes
# columns names
df.columns
# data information
df.info()
# describe
df.describe().T
# checking null values
df.isnull().sum()
# checking null values in visualization
import missingno as msno
msno.bar(df, color="purple", fontsize=25)
# unique value
df.nunique()
df.duplicated().sum()
df.corr()
# confution matrix
fig, ax = plt.subplots()
fig.set_size_inches(20, 10)
sns.heatmap(df.corr(), vmax=0.8, square=True, annot=True)
plt.title("Confution Matrix", fontsize=15)
# **Observations:** There are little bit positive correlation between funding round and No. of Investers.
# # Data Cleaning
df.head()
# Dropping column desciption
df.drop("Description", axis=1, inplace=True)
df.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/416/129416800.ipynb
|
indian-startups-top-300
|
ashishraut64
|
[{"Id": 129416800, "ScriptId": 38478655, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7453603, "CreationDate": "05/13/2023 15:49:27", "VersionNumber": 1.0, "Title": "Startup's Company EDA", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 107.0, "LinesInsertedFromPrevious": 107.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185447206, "KernelVersionId": 129416800, "SourceDatasetVersionId": 5177178}]
|
[{"Id": 5177178, "DatasetId": 3009276, "DatasourceVersionId": 5249211, "CreatorUserId": 11919174, "LicenseName": "CC0: Public Domain", "CreationDate": "03/16/2023 11:22:12", "VersionNumber": 3.0, "Title": "Indian startups", "Slug": "indian-startups-top-300", "Subtitle": "Information about the top 300 startups in India from 1984 to 2022", "Description": "# Context\nA startup or start-up is a company or project undertaken by an entrepreneur to seek, develop, and validate a scalable business model. While entrepreneurship includes all new businesses, including self-employment and businesses that do not intend to go public, startups are new businesses that intend to grow large beyond the solo founder. In the beginning, startups face high uncertainty and have high rates of failure, but a minority of them do go on to be successful and influential.\n\n# Content\nThe following dataset has data about the Top 300 startups in India. Details about the columns are as follows:\n1. Company - Name of the Startup.\n2. City - The City in which the startup is started.\n3. Starting Year - The Year in which the startup was started.\n4. Founders - Name of the founders of the startup.\n5. Industries - Industrial domain in which the startup falls.\n6. No. of Employees - Number of employees in the startup.\n7. Funding Amount in USD - Total funding amount funded to the startup.\n8. Funding Rounds - Funding rounds are the number of times a startup goes back to the market to raise more capital. The goal of every round is for founders to trade equity in their business for the capital they can utilize to advance their companies to the next level.\n9. No. of Investors - Number of investors in the startup.", "VersionNotes": "Data Update 2023/03/16", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3009276, "CreatorUserId": 11919174, "OwnerUserId": 11919174.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5177178.0, "CurrentDatasourceVersionId": 5249211.0, "ForumId": 3048298, "Type": 2, "CreationDate": "03/16/2023 09:44:20", "LastActivityDate": "03/16/2023", "TotalViews": 10134, "TotalDownloads": 1925, "TotalVotes": 34, "TotalKernels": 13}]
|
[{"Id": 11919174, "UserName": "ashishraut64", "DisplayName": "ashishraut64", "RegisterDate": "10/12/2022", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # About Dataset
# Content The following dataset has data about the Top 300 startups in India. Details about the columns are as follows:
# * Company - Name of the Startup.
# * City - The City in which the startup is started.
# * Starting Year - The Year in which the startup was started.
# * Founders - Name of the founders of the startup.
# * Industries - Industrial domain in which the startup falls.
# * No. of Employees - Number of employees in the startup.
# * Funding Amount in USD - Total funding amount funded to the startup.
# * Funding Rounds - Funding rounds are the number of times a startup goes back to the market to raise more capital. The goal of every round is for founders to trade equity in their business for the capital they can utilize to advance their companies to the next level.
# * No. of Investors - Number of investors in the startup.
# 
# # Indian Startups (Exploratory data analysis)
# importing libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# importing data
df = pd.read_csv("/kaggle/input/indian-startups-top-300/Startups1.csv", index_col=0)
df.sample(6).reset_index(drop=True).style.set_properties(
**{"background-color": "#FFF0F5 ", "color": "black", "border-color": "#8b8c8c"}
)
# # Workflow
# * Understanding of data
# * Data Cleaning & Prepocessing
# * EDA
# * Insights
# # Understanding the data
# data size
df.shape
# head
df.head()
# tail
df.tail()
# data types
df.dtypes
# columns names
df.columns
# data information
df.info()
# describe
df.describe().T
# checking null values
df.isnull().sum()
# checking null values in visualization
import missingno as msno
msno.bar(df, color="purple", fontsize=25)
# unique value
df.nunique()
df.duplicated().sum()
df.corr()
# confution matrix
fig, ax = plt.subplots()
fig.set_size_inches(20, 10)
sns.heatmap(df.corr(), vmax=0.8, square=True, annot=True)
plt.title("Confution Matrix", fontsize=15)
# **Observations:** There are little bit positive correlation between funding round and No. of Investers.
# # Data Cleaning
df.head()
# Dropping column desciption
df.drop("Description", axis=1, inplace=True)
df.head()
| false | 1 | 875 | 0 | 1,227 | 875 |
||
129582602
|
<jupyter_start><jupyter_text>Cars MPG regression
This data is about making predictions on miles per gallon, the main goal here is to discover some Econometric Problems like Multicollinearity for example, and for sure feel free to use it in regression and other data processing.
This data contains three independent variables "cyl" cylinder, eng "Engine hours power" and "wgt" weight, and also the target variable "dependent variable" "mpg" miles per gallon.
Kaggle dataset identifier: mpg-regression
<jupyter_script>#
# # Multicollinearity
# ### Multicollinearity is a statistical concept where several independent variables in a model are correlated for example if we have two independent variables like "Number of cylinders" and "engine power" those two variables for sure will have a coloration with each other and a coloration with the target variable or the dependent variable "Speed", the problem here is the coloration between those two independent variables and this what we called "Multicollinearity"
# #### this topic considered an intermediate level that most of the beginner data scientist does not care much about, we are not here to today to deep into math or even for data processing, so lets get started
# #### multicollinearity is one of the "Econometric Problems" and i will post in the next few days some of those problems
# ### we will divide this topic into 3 sections
# #### 1- Indicators
# #### 2- Testing
# #### 3- Solving
# ## 1- Indicators
# ### A - Coloration Matrix
# #### Coloration Matrix is one of the good indicators of Multicollinearity, the normal coloration should be between the independent variable and the dependent variables if you notice any coloration between independent variables with more than 80% this is a good indicator but why 80%? the answer is that most research proved this number you can make your own research here, and if you found that the number is 100% we called it exact Multicollinearity.
# So lets load some data to show our case!
import pandas as pd
data = pd.read_csv("/kaggle/input/mpg-regression/cars.csv")
data = data.drop("Unnamed: 0", axis=1)
import matplotlib.pyplot as plt
import seaborn as sns
corr_matrix = data.corr()
f, ax = plt.subplots(figsize=(15, 5))
cmap = sns.diverging_palette(230, 20, as_cmap=True)
sns.heatmap(
corr_matrix,
cmap=cmap,
vmax=0.5,
annot=True,
center=0,
square=True,
linewidths=0.5,
cbar_kws={"shrink": 0.5},
)
plt.show()
# #### as we can see here there are a strong coloration between "cyl" and "eng" around 95% and also between "eng" and "wgt" around 93%
# ### B - Linear regression variables p-values
# #### if the variables are not significant while it have a good coloration this is a good indicator about Multicollinearity
# Let's implement some Linear model to demonstrate
X = data.drop("mpg", axis=1)
y = data.drop(["cyl", "eng", "wgt"], axis=1)
import statsmodels.api as sm
X2 = sm.add_constant(X)
est = sm.OLS(y, X2)
est2 = est.fit()
print(est2.summary())
# #### we are not here to talk about p-value or how it was calculated but here is some info about p-value we are interested in the mid-section of the previous output which represents the p-value for the interception "const" and the independent variables if we take a look at the p-value of each one of those you will find a number between 0 and 1 which represent if this variable is significant or not depends on its score and this leads us to talk about the threshold
# #### the most common threshold is 0.05 if you found a p-value more than .05 this means this variable is not siginficant and you can think about significant as "effective" if the p-value is less than .05 this means that the variable is significant
# #### so as we can see the "cyl" and "eng" is not siginficant! , even when those variables have a good negative coloration with mpg
# ## 2- Testing
# #### Now after we notice via the previous steps that it might be a multicollinearity problem, not it is time to make sure and do some testing, a simple method to detect multicollinearity in a model is by using something called the variance inflation factor or the VIF The rule of thumb is that a regressor produces collinearity if its VIF is greater than 10.
from statsmodels.stats.outliers_influence import variance_inflation_factor
vif_data = pd.DataFrame()
vif_data["feature"] = X.columns
vif_data["VIF"] = [
variance_inflation_factor(X.values, i) for i in range(len(X.columns))
]
vif_data
# #### as we can see we have a huge multicollinearity here lets try to fix this!
# ## 3- Solving
# ### we have two optoins here
# A - drop the highest VIF score variable
# B - use a feature selection algorithm
# #### A - drop the highest VIF score variable
# But why we will drop it, well as we know this is multicollinearity problem so we have two variables or more that have a strong coloration to each others and this leads our model to miss leading result so we need to remove one of those variable to break this coloration!
new_X = data.drop(["mpg", "cyl"], axis=1)
new_y = data.drop(["cyl", "eng", "wgt"], axis=1)
new_X2 = sm.add_constant(new_X)
est = sm.OLS(new_y, new_X2)
est2 = est.fit()
print(est2.summary())
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/582/129582602.ipynb
|
mpg-regression
|
mahmoudafify
|
[{"Id": 129582602, "ScriptId": 38532443, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6965494, "CreationDate": "05/15/2023 03:09:09", "VersionNumber": 1.0, "Title": "Multicollinearity", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 100.0, "LinesInsertedFromPrevious": 100.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 5}]
|
[{"Id": 185795869, "KernelVersionId": 129582602, "SourceDatasetVersionId": 5686293}]
|
[{"Id": 5686293, "DatasetId": 3269081, "DatasourceVersionId": 5761876, "CreatorUserId": 6965494, "LicenseName": "Unknown", "CreationDate": "05/15/2023 01:21:24", "VersionNumber": 1.0, "Title": "Cars MPG regression", "Slug": "mpg-regression", "Subtitle": "Miles Per Gallon regression", "Description": "This data is about making predictions on miles per gallon, the main goal here is to discover some Econometric Problems like Multicollinearity for example, and for sure feel free to use it in regression and other data processing.\n\nThis data contains three independent variables \"cyl\" cylinder, eng \"Engine hours power\" and \"wgt\" weight, and also the target variable \"dependent variable\" \"mpg\" miles per gallon.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3269081, "CreatorUserId": 6965494, "OwnerUserId": 6965494.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5686293.0, "CurrentDatasourceVersionId": 5761876.0, "ForumId": 3334703, "Type": 2, "CreationDate": "05/15/2023 01:21:24", "LastActivityDate": "05/15/2023", "TotalViews": 87, "TotalDownloads": 6, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 6965494, "UserName": "mahmoudafify", "DisplayName": "Mahmoud Afify", "RegisterDate": "03/17/2021", "PerformanceTier": 1}]
|
#
# # Multicollinearity
# ### Multicollinearity is a statistical concept where several independent variables in a model are correlated for example if we have two independent variables like "Number of cylinders" and "engine power" those two variables for sure will have a coloration with each other and a coloration with the target variable or the dependent variable "Speed", the problem here is the coloration between those two independent variables and this what we called "Multicollinearity"
# #### this topic considered an intermediate level that most of the beginner data scientist does not care much about, we are not here to today to deep into math or even for data processing, so lets get started
# #### multicollinearity is one of the "Econometric Problems" and i will post in the next few days some of those problems
# ### we will divide this topic into 3 sections
# #### 1- Indicators
# #### 2- Testing
# #### 3- Solving
# ## 1- Indicators
# ### A - Coloration Matrix
# #### Coloration Matrix is one of the good indicators of Multicollinearity, the normal coloration should be between the independent variable and the dependent variables if you notice any coloration between independent variables with more than 80% this is a good indicator but why 80%? the answer is that most research proved this number you can make your own research here, and if you found that the number is 100% we called it exact Multicollinearity.
# So lets load some data to show our case!
import pandas as pd
data = pd.read_csv("/kaggle/input/mpg-regression/cars.csv")
data = data.drop("Unnamed: 0", axis=1)
import matplotlib.pyplot as plt
import seaborn as sns
corr_matrix = data.corr()
f, ax = plt.subplots(figsize=(15, 5))
cmap = sns.diverging_palette(230, 20, as_cmap=True)
sns.heatmap(
corr_matrix,
cmap=cmap,
vmax=0.5,
annot=True,
center=0,
square=True,
linewidths=0.5,
cbar_kws={"shrink": 0.5},
)
plt.show()
# #### as we can see here there are a strong coloration between "cyl" and "eng" around 95% and also between "eng" and "wgt" around 93%
# ### B - Linear regression variables p-values
# #### if the variables are not significant while it have a good coloration this is a good indicator about Multicollinearity
# Let's implement some Linear model to demonstrate
X = data.drop("mpg", axis=1)
y = data.drop(["cyl", "eng", "wgt"], axis=1)
import statsmodels.api as sm
X2 = sm.add_constant(X)
est = sm.OLS(y, X2)
est2 = est.fit()
print(est2.summary())
# #### we are not here to talk about p-value or how it was calculated but here is some info about p-value we are interested in the mid-section of the previous output which represents the p-value for the interception "const" and the independent variables if we take a look at the p-value of each one of those you will find a number between 0 and 1 which represent if this variable is significant or not depends on its score and this leads us to talk about the threshold
# #### the most common threshold is 0.05 if you found a p-value more than .05 this means this variable is not siginficant and you can think about significant as "effective" if the p-value is less than .05 this means that the variable is significant
# #### so as we can see the "cyl" and "eng" is not siginficant! , even when those variables have a good negative coloration with mpg
# ## 2- Testing
# #### Now after we notice via the previous steps that it might be a multicollinearity problem, not it is time to make sure and do some testing, a simple method to detect multicollinearity in a model is by using something called the variance inflation factor or the VIF The rule of thumb is that a regressor produces collinearity if its VIF is greater than 10.
from statsmodels.stats.outliers_influence import variance_inflation_factor
vif_data = pd.DataFrame()
vif_data["feature"] = X.columns
vif_data["VIF"] = [
variance_inflation_factor(X.values, i) for i in range(len(X.columns))
]
vif_data
# #### as we can see we have a huge multicollinearity here lets try to fix this!
# ## 3- Solving
# ### we have two optoins here
# A - drop the highest VIF score variable
# B - use a feature selection algorithm
# #### A - drop the highest VIF score variable
# But why we will drop it, well as we know this is multicollinearity problem so we have two variables or more that have a strong coloration to each others and this leads our model to miss leading result so we need to remove one of those variable to break this coloration!
new_X = data.drop(["mpg", "cyl"], axis=1)
new_y = data.drop(["cyl", "eng", "wgt"], axis=1)
new_X2 = sm.add_constant(new_X)
est = sm.OLS(new_y, new_X2)
est2 = est.fit()
print(est2.summary())
| false | 1 | 1,303 | 5 | 1,424 | 1,303 |
||
129582530
|
<jupyter_start><jupyter_text>Photovoltaic System O&M inspection
According to [one of the three articles](https://onlinelibrary.wiley.com/doi/10.1002/pip.3564) that explains how [PV-HAWK](https://lukasbommes.github.io/PV-Hawk/index.html) ([MIT License](https://github.com/LukasBommes/PV-Hawk/blob/master/LICENSE)) tool works, five different PV plants were used to train one of the models used by this tool. The plants were named A, B, C, D and E for anonymization purposes. This dataset is a sample from the first 12 arrays of PV plant A.
---
# 1. Context
Both large and small photovoltaic systems are susceptible to failures in their equipment, especially in modules due to operational stresses that are exposed and errors during the installation process of these devices. Although numerous internal and external factors originate these failures, the common phenomenon presented by several of them is hot spots on module defective area. The immediate impact is perceptible in the reduction of the generated power and, in the long term, in the reduction of the useful life of the equipment due to the high temperatures presented. The preventive maintenance method for recognizing this phenomenon is the use of thermography images in inspections of photovoltaic modules. Through this procedure, faulty modules are immediately identified with failures at an early stage due to their high heat signatures compared to the others, captured by cameras with infrared sensors. Currently, the use of this type of camera attached to drones stands out for providing an increase in the inspection area and a reduction in its execution time.
To understand more about this, read these reports by International energy agency (IEA):
- [ Review of failures of PV modules](https://iea-pvps.org/wp-content/uploads/2020/01/IEA-PVPS_T13-01_2014_Review_of_Failures_of_Photovoltaic_Modules_Final.pdf);
- [Review of IR and EL images applications for PV systems](https://iea-pvps.org/wp-content/uploads/2020/01/Review_on_IR_and_EL_Imaging_for_PV_Field_Applications_by_Task_13.pdf).
## 1.1 Photovoltaic system specifications
Acording to the [dataset article](https://onlinelibrary.wiley.com/doi/10.1002/pip.3564), the photovoltaic system on which the thermographic inspection was carried out is located in Germany and it's composed of 2376 PV polycrystalline silicon modules, measuring 1650 x 992 mm (60-cell) each.
The images in this dataset refer to the region marked in red in the google maps screenshot of the photovoltaic system location.
<br>

<br>
## 1.2 Thermal inspection specifications
The inspection took place under clearsky conditions and solar irradiance above 700 W/m². In the table bellow more detail are presented for the weather parameters that influence thermography inspections.
| Number of modules | Distance (m) | Peak velocity (m/s) | Air Temperature (ºC)| Global radiation (J/cm²)| Wind speed (m/s)|
| --- | --- | -- | --- | --- | --- |
| 13640 | 7612 | 4.1 | 25 | 39.7 | 2.8 |
The drone used was a DJI model MATRICE 210 coupled with a FLIR XT2 thermal camera and with the following specifications:
- Thermal resolution of 640x512 pixels;
- Visual resolution of 1280x720 pixels;
- Focal length of 13 mm;
- Frame rate of 8 Hz .
The drone was controlled manually, positioned at an altitude of 10 m to 30 m from the ground with a velocity that ensures blur-free images. The camera orientation was facing vertically downwards (nadir) at all times.
Aiming at reducing inspection cost and duration, especially for increasing the drone range before a battery change is needed, the images were sequentially scanned considering two types of PV arrays layouts: only one single array appears in the image and then two arrays.
<br>

<br>

<br>
As showed in the table bellow, scanning two rows simultaneously speeds up the flight duration by a factor of 2.1, decreases flight distance by a factor of 1.9 and increases module throughput by a factor of 2.09. Despite these benefits, the resolution of extracted PV module images reduces.
| Inspection layout | Flight distance (m) | Flight duration (s) | Average module resolution (px) |Module throughput (1/s) |
| --- | --- | --- | --- | --- |
| Single row | 1307 | 707 | 141 X 99 | 3.36 |
| Double row | 681 | 338 | 73 X 50 | 7.03 |
## 1.3 Dataset organization
The images are separated by type of inspection in different folders (single or double rows). In each folder, there are thermographic images in TIFF format and a CSV file with drone's geospatial and temporal data during the inspection. Only for the double row inspection type that visual (RGB) images were acquired.
Besides, I've uploaded files to use for calibrate infrared and visual cameras to correct any type of distortion that camera lenses cause.
# 2. Resources
- This guides by [FLIR](http://support.flir.com/appstories/AppStories/Electrical&Mechanical/Testing_solar_panels_EN.pdf) and [TESTO](https://www.murcal.com/pdf%20folder/15.testo_thermography_guide.pdf) companies are good resources to understand more about thermography in the solar modules context;
- There's [another one by FLIR](https://thermalcapture.com/wp-content/uploads/2019/08/pv-system-inspection-thermal-drones-07-15-19.pdf) that explains in depth how aerial thermal inspections of photovoltaic systems are made and their importance in this field;
- To understand the level of influence that the module degradation has on the yield of the photovoltaic system you can read the [IEC TS-62446-3]( https://ayscomdatatec.com/wp-content/uploads/2019/09/Normativa-IEC-TS-62446-3.pdf) and the [Raptor maps's knoledge hub](https://raptormaps.com/solar-tech-docs/).
# 3. Inspiration
A service often provided by companies in this area is a SaaS that displays the detected faulty modules in an bird's eye view of the photovoltaic system and calculate the energy loss, like the image bellow shows. One can create a web app (using streamlit or plotly/dash) that detect PV modules with a instance segmentation model, track them with a object tracker and classify their integrity (binary or multiclass classification) with a image classification model.
<br>

<br>
This idea can be used for guiding a maintenance team in order to intervene and replace panels if necessary.
Kaggle dataset identifier: photovoltaic-system-o-and-m-inspection
<jupyter_script>import pickle
from pathlib import Path
from scipy import ndimage as ndi
from skimage.color import label2rgb
from skimage.filters import try_all_threshold
from PIL import Image
import tifffile as tif
import cv2
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import make_axes_locatable
import pandas as pd
# # 1. Dataset Reading
# The best way is to first read the metadata files from both datasets
SINGLE_ROW_METADATA_PATH = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/single-row/metadata.csv"
columns_to_rename = {"thermal image name": "thermal_image_name"}
sr_metadata = pd.read_csv(SINGLE_ROW_METADATA_PATH).rename(columns=columns_to_rename)
sr_metadata.head()
DOUBLE_ROW_METADATA_PATH = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/double-row/metadata.csv"
columns_to_rename = {
"thermal image name": "thermal_image_name",
"rgb image name": "rgb_image_name",
}
dr_metadata = pd.read_csv(DOUBLE_ROW_METADATA_PATH).rename(columns=columns_to_rename)
dr_metadata.head()
# We need to get the full path for the images
def get_image_full_path(image_name, image_type):
if image_type == "single_row_thermal":
origin_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/single-row/thermal images"
elif image_type == "double_row_thermal":
origin_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/double-row/thermal images"
elif image_type == "double_row_rgb":
origin_path = (
origin_path
) = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/double-row/rgb images"
return Path(origin_path, image_name)
sr_metadata = sr_metadata.assign(
thermal_image_name=sr_metadata.thermal_image_name.apply(
lambda x: get_image_full_path(x, "single_row_thermal")
)
).assign(timestamp=pd.to_datetime(sr_metadata.timestamp))
dr_metadata = (
dr_metadata.assign(
thermal_image_name=dr_metadata.thermal_image_name.apply(
lambda x: get_image_full_path(x, "double_row_thermal")
)
)
.assign(
rgb_image_name=dr_metadata.rgb_image_name.apply(
lambda x: get_image_full_path(x, "double_row_rgb")
)
)
.assign(timestamp=pd.to_datetime(sr_metadata.timestamp))
)
# **Now we can load the images!**
# I've created the Thermogram class just to be possible to get the thermal image and the converted one in the same object like [Flyr library](https://bitbucket.org/nimmerwoner/flyr/src/master/) does
class Thermogram:
def __init__(self, path: Path):
self.path = path
@property
def celsius(self) -> np.ndarray:
return (tif.imread(self.path.as_posix()) * 0.04) - 273.15
def render(self) -> np.ndarray:
image = self.celsius
image = (image - np.min(image)) / (np.max(image) - np.min(image))
return (image * 255.0).astype(np.uint8)
def load_image(image_path: Path):
image_format = image_path.suffix
if image_format == ".jpg":
return cv2.imread(image_path.as_posix())
elif image_format == ".tiff":
return Thermogram(image_path)
image_number = 57
thermogram = load_image(sr_metadata.thermal_image_name[image_number])
_, ax = plt.subplots(1, 2)
im = ax[0].imshow(thermogram.celsius, cmap="inferno")
ax[0].set_title("Thermographic image")
ax[0].set_axis_off()
ax[1].imshow(thermogram.render(), cmap="gray")
ax[1].set_title("Rendered image (8 bit image)")
ax[1].set_axis_off()
cax = make_axes_locatable(ax[0]).append_axes("right", size="5%", pad=0.05)
plt.colorbar(
im, cax=cax, values=np.unique(thermogram.celsius), label="Temperature (ºC)"
)
plt.tight_layout()
plt.show()
thermogram = load_image(dr_metadata.thermal_image_name[image_number])
visual = load_image(dr_metadata.rgb_image_name[image_number])
_, ax = plt.subplots(1, 3, figsize=(10, 5))
im = ax[0].imshow(thermogram.celsius, cmap="inferno")
ax[0].set_title("Thermographic image")
ax[0].set_axis_off()
ax[1].imshow(thermogram.render(), cmap="gray")
ax[1].set_title("Rendered image (8 bit image)")
ax[1].set_axis_off()
ax[2].imshow(visual[:, :, ::-1])
ax[2].set_title("Visual image")
ax[2].set_axis_off()
cax = make_axes_locatable(ax[0]).append_axes("right", size="5%", pad=0.05)
plt.colorbar(
im, cax=cax, values=np.unique(thermogram.celsius), label="Temperature (ºC)"
)
plt.tight_layout()
plt.show()
# # 2. Camera calibration
# This step is important because often times the lenses of cameras create distortions in the images. In this dataset only the RGB ones were affected, but the intrinsic and extrinsic camera parameters from the IR camera can be used for other tasks like Structure from motion (as PV-HAWK does).
def remove_distortion(image: np.ndarray, image_type: str = "rgb"):
mapx_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/calibration files/RGB/mapx.pkl"
mapy_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/calibration files/RGB/mapy.pkl"
if image_type == "ir":
mapx_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/calibration files/IR/mapx.pkl"
mapy_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/calibration files/IR/mapy.pkl"
with open(mapx_path, "rb") as mapx_file, open(mapy_path, "rb") as mapy_file:
mapx = pickle.load(mapx_file)
mapy = pickle.load(mapy_file)
return cv2.remap(
image, mapx, mapy, cv2.INTER_CUBIC, borderMode=cv2.BORDER_REPLICATE
)
undistorted_rgb = remove_distortion(visual)
_, ax = plt.subplots(1, 2, figsize=(20, 5))
ax[0].imshow(visual[:, :, ::-1])
ax[0].set_title("RGB distorted")
ax[0].set_axis_off()
ax[1].imshow(undistorted_rgb[:, :, ::-1])
ax[1].set_title("RGB undistorted")
ax[1].set_axis_off()
plt.tight_layout()
plt.show()
undistorted_ir = remove_distortion(thermogram.render(), "ir")
_, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].imshow(thermogram.render(), cmap="gray")
ax[0].set_title("IR distorted")
ax[0].set_axis_off()
ax[1].imshow(undistorted_ir, cmap="gray")
ax[1].set_title("IR undistorted")
ax[1].set_axis_off()
plt.tight_layout()
plt.show()
# # 3. Images Alignment
# Is possible to align the RGB and IR images using two techniques:
# - **Phase Correlation**: I still don't quite understand how this algorithm works, but this [solution on github](https://github.com/YoshiRi/ImRegPOC) works for this dataset;
# - **Enhanced Correlation Coefficient (ECC)**: I found [an article](https://learnopencv.com/image-alignment-ecc-in-opencv-c-python/) that explains very well how this solution works so I'll stick with it here.
# Before any alignment algorithm be applied, I've to say that there's something odd with this images. Although the thermographic image has a lower resolution than the RGB one, it has a larger field of view because more modules appear in it. Typically, RGB images have a larger field of view than thermal images.
# So, it's necessary to crop the RGB and IR images to their correspondent region first. Then, resize them to the same resolution.
left = 0
top = 75
right = 640
bottom = 490
crop_fg_image = Image.fromarray(undistorted_ir).crop((left, top, right, bottom))
crop_fg_image
left = 120
top = 0
right = 1150
bottom = 720
crop_bg_image = (
Image.fromarray(undistorted_rgb[:, :, ::-1])
.crop((left, top, right, bottom))
.resize(crop_fg_image.size)
)
crop_bg_image
# We can compare the aligned image with the unaligned one by applying the canny edge detector in both of them. Then, blending each canny mask result in the thermal image.
def get_gradient(image):
grad_x = cv2.Sobel(image, cv2.CV_32F, 1, 0, ksize=3)
grad_y = cv2.Sobel(image, cv2.CV_32F, 0, 1, ksize=3)
return cv2.addWeighted(np.absolute(grad_x), 0.5, np.absolute(grad_y), 0.5, 0)
def align_images(
image_1, image_2, warp_mode, n_iterations=5000, increment_threshold=1e-10
):
warp_mode = cv2.MOTION_TRANSLATION
if warp_mode == cv2.MOTION_HOMOGRAPHY:
warp_matrix = np.eye(3, 3, dtype=np.float32)
else:
warp_matrix = np.eye(2, 3, dtype=np.float32)
criteria = (
cv2.TERM_CRITERIA_EPS | cv2.TERM_CRITERIA_COUNT,
n_iterations,
increment_threshold,
)
grad_1 = get_gradient(image_1)
grad_2 = get_gradient(image_2)
cc, warp_matrix = cv2.findTransformECC(
grad_1, grad_2, warp_matrix, warp_mode, criteria
)
if warp_mode == cv2.MOTION_HOMOGRAPHY:
return cv2.warpPerspective(
image_2,
warp_matrix,
image_1.shape[::-1],
flags=cv2.INTER_LINEAR + cv2.WARP_INVERSE_MAP,
)
else:
return cv2.warpAffine(
image_2,
warp_matrix,
image_1.shape[::-1],
flags=cv2.INTER_LINEAR + cv2.WARP_INVERSE_MAP,
)
def blend_images(image_1, image_2, alpha=0.2, beta=None):
beta = 1 - alpha if not beta else beta
return cv2.addWeighted(image_2, alpha, image_1, beta, 0.0)
crop_bg_image = np.asarray(crop_bg_image)
crop_fg_image = np.asarray(crop_fg_image)
crop_bg_image_gray = cv2.cvtColor(crop_bg_image, cv2.COLOR_BGR2GRAY)
# Align the image with two methods: Translation and Homography
align_image_1 = align_images(crop_fg_image, crop_bg_image_gray, cv2.MOTION_TRANSLATION)
align_image_2 = align_images(crop_fg_image, crop_bg_image_gray, cv2.MOTION_HOMOGRAPHY)
# Apply the Canny edge detector in the images
unalign_canny = cv2.Canny(crop_bg_image_gray, 300, 400)
align_canny_1 = cv2.Canny(align_image_1, 200, 300)
align_canny_2 = cv2.Canny(align_image_2, 200, 300)
# Blend the Canny masks with the thermal imagem
unalign_blend = blend_images(crop_fg_image, unalign_canny)
align_blend_1 = blend_images(crop_fg_image, align_canny_1)
align_blend_2 = blend_images(crop_fg_image, align_canny_2)
_, ax = plt.subplots(1, 3, figsize=(20, 5))
ax[0].imshow(unalign_blend, cmap="gray")
ax[0].set_title("Cropped RGB image")
ax[1].imshow(align_blend_1, cmap="gray")
ax[1].set_title("Motion translation")
ax[2].imshow(align_blend_2, cmap="gray")
ax[2].set_title("Motion homography")
plt.show()
# # 4. Modules Segmentation
# ## 4.1 Segmentation of RGB images
# First, I will segment the modules in the RGB images. The way I'll do this is through color segmentation. First, we convert the image to the HSV color space (the most appropriate space for color processing) and apply the Otsu threshold in each of it's channels (Hue, Saturation and Value).
image = cv2.cvtColor(undistorted_rgb, cv2.COLOR_BGR2HSV)
for i in range(3):
title = "Value"
if i == 0:
title = "Hue"
elif i == 1:
title = "Saturation"
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
fig.suptitle(title)
ax[0].hist(image[:, :, i].ravel(), np.unique(image[:, :, i]))
ax[1].imshow(image[:, :, i], cmap="gray")
ax[1].set_axis_off()
plt.tight_layout()
plt.show()
# We can compare this method with the identification of modules by seeting a minimum and maximum threshold manually in each HSV channel
def preprocess_rgb_image_manual(image, lower_values, higher_values):
image = cv2.cvtColor(image, cv2.COLOR_BGR2HSV)
return cv2.inRange(image, lower_values, higher_values)
def preprocess_rgb_image_hsv_otsu(image):
image = cv2.cvtColor(image, cv2.COLOR_BGR2HSV)
masks = []
for channel_index, channel_image in enumerate(cv2.split(image)):
channel_image = cv2.GaussianBlur(channel_image, (5, 5), 0)
max_value = 179 if channel_index == 0 else 255
_, mask = cv2.threshold(
channel_image, 0, max_value, cv2.THRESH_BINARY + cv2.THRESH_OTSU
)
masks.append(mask)
result_mask = np.bitwise_and(masks[0], masks[1])
return np.bitwise_and(result_mask, masks[2])
lower_blue = np.array([90, 100, 100])
upper_blue = np.array([125, 255, 200])
mask_1 = preprocess_rgb_image_manual(undistorted_rgb, lower_blue, upper_blue)
mask_2 = preprocess_rgb_image_hsv_otsu(undistorted_rgb)
_, ax = plt.subplots(1, 2, figsize=(20, 5))
ax[0].imshow(mask_1, cmap="gray")
ax[0].set_axis_off()
ax[0].set_title("Manual thresholds setting")
ax[1].imshow(mask_2, cmap="gray")
ax[1].set_title("Otsu thresholds")
ax[1].set_axis_off()
plt.show()
# It's noticeable how the otsu method in each HSV color channel is more appropriate for module identification. But, we have to fill in the holes that some module masks have.
blur = cv2.medianBlur(mask_2, 5)
kernel = np.ones((5, 5), np.uint8)
blur_open = cv2.morphologyEx(blur, cv2.MORPH_OPEN, kernel)
module_mask = ndi.binary_fill_holes(blur_open)
plt.figure(figsize=(10, 5))
plt.imshow(module_mask, cmap="gray")
# Applying the connected components method, we can set for every module mask with a label
_, label_mask = cv2.connectedComponents(module_mask.astype("uint8"))
plt.figure(figsize=(10, 5))
plt.imshow(label_mask, cmap="bone")
undistorted_gray = cv2.cvtColor(undistorted_rgb, cv2.COLOR_BGR2GRAY)
plt.imshow(label2rgb(label_mask, undistorted_gray))
# ## 4.2 Segmentation of IR images
try_all_threshold(undistorted_ir, verbose=False)
# It's to be expected that traditional image segmentation methods do not work very well on thermographic images, as they were built for images of the human visual spectrum.
# So, we can use the masks taken from rgb images to segment the modules in the thermographic images.
new_module_mask = (
Image.fromarray(module_mask)
.crop((120, 0, 1150, 720))
.resize(crop_fg_image.shape[::-1])
)
new_module_mask
_, label_mask = cv2.connectedComponents(np.array(new_module_mask).astype("uint8"))
plt.imshow(label2rgb(label_mask, np.array(crop_fg_image)))
# Another approach is to train an instance segmentation model for automatic module segmentation
# # 4. Images Overlap
class ImageAligner:
def __init__(self, number_features: int = 500):
self.feature_extractor = cv2.ORB_create(number_features)
self.matcher = cv2.BFMatcher(cv2.NORM_HAMMING, crossCheck=True)
self.points = {}
def filter_matches(self, matches, matches_percent: float = 0.2):
matches = sorted(matches, key=lambda x: x.distance)
matches_to_keep = int(len(matches) * matches_percent)
return matches[:matches_to_keep]
def __call__(self, image, template):
# Get keypoints and descriptors
image_kps, image_dsc = self.feature_extractor.detectAndCompute(image, None)
template_kps, template_dsc = self.feature_extractor.detectAndCompute(
template, None
)
# Match features and filter them
matches = self.matcher.match(image_dsc, template_dsc)
matches = self.filter_matches(matches)
# Store points and matches for debug purposes
self.points["keypoints"] = [image_kps, template_kps]
self.points["matches"] = matches
# Filter keypoints with filtered matches
image_kps = np.float32([image_kps[match.queryIdx].pt for match in matches])
template_kps = np.float32(
[template_kps[match.trainIdx].pt for match in matches]
)
# Correct image perspective
matrix, _ = cv2.findHomography(image_kps, template_kps, cv2.RANSAC)
return cv2.warpPerspective(image, matrix, template.shape[::-1])
def inspect_matches(self, image, template):
return cv2.drawMatches(
image,
self.points["keypoints"][0],
template,
self.points["keypoints"][1],
self.points["matches"],
None,
)
undistorted_rgb_1 = remove_distortion(load_image(dr_metadata.rgb_image_name[57]))
undistorted_rgb_2 = remove_distortion(load_image(dr_metadata.rgb_image_name[58]))
_, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].imshow(undistorted_rgb_1[:, :, ::-1])
ax[1].imshow(undistorted_rgb_2[:, :, ::-1])
plt.show()
aligner = ImageAligner()
undistorted_gray_1 = cv2.cvtColor(undistorted_rgb_1, cv2.COLOR_BGR2GRAY)
undistorted_gray_2 = cv2.cvtColor(undistorted_rgb_2, cv2.COLOR_BGR2GRAY)
aligned_image = aligner(undistorted_gray_1, undistorted_gray_2)
inspected_image = aligner.inspect_matches(undistorted_gray_1, undistorted_gray_2)
_, ax = plt.subplots(2, 1, figsize=(20, 10))
ax[0].imshow(aligned_image, cmap="gray")
ax[1].imshow(inspected_image)
plt.show()
undistorted_ir_1 = remove_distortion(
load_image(dr_metadata.thermal_image_name[57]).render(), "ir"
)
undistorted_ir_2 = remove_distortion(
load_image(dr_metadata.thermal_image_name[58]).render(), "ir"
)
_, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].imshow(undistorted_ir_1, cmap="gray")
ax[1].imshow(undistorted_ir_2, cmap="gray")
plt.show()
aligner = ImageAligner()
aligned_image = aligner(undistorted_ir_1, undistorted_ir_2)
inspected_image = aligner.inspect_matches(undistorted_ir_1, undistorted_ir_2)
_, ax = plt.subplots(2, 1, figsize=(20, 10))
ax[0].imshow(aligned_image, cmap="gray")
ax[1].imshow(inspected_image)
plt.show()
# # 6. Defected Modules
# Highlight defected modules when the modules masks were created
# # 7. Thermal Inspection
# Is possible to determine parameters like the inspection time and drone path
sr_inspection_time = sr_metadata.timestamp.filter([0, 5294]).diff().iloc[1].seconds
dr_inspection_time = dr_metadata.timestamp.filter([0, 2540]).diff().iloc[1].seconds
print(
f"Single row inspection time: {sr_inspection_time // 60} minutes and {sr_inspection_time % 60} seconds"
)
print(
f"Double row inspection time: {dr_inspection_time // 60} minutes and {dr_inspection_time % 60} seconds"
)
_, ax = plt.subplots(1, 2, figsize=(15, 5))
ax[0].scatter(sr_metadata.longitude, sr_metadata.latitude)
ax[0].set_title("Single row inspection")
ax[1].scatter(dr_metadata.longitude, dr_metadata.latitude)
ax[1].set_title("Double row inspection")
plt.show()
# # 8. Thermal image orthomosaic
# Show how to make a thermal orthomosaic with this dataset
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/582/129582530.ipynb
|
photovoltaic-system-o-and-m-inspection
|
marcosgabriel
|
[{"Id": 129582530, "ScriptId": 38452073, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5048762, "CreationDate": "05/15/2023 03:07:59", "VersionNumber": 4.0, "Title": "[DATASET INTRO] Photovoltaic System O&M inspection", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 493.0, "LinesInsertedFromPrevious": 4.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 489.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 5}]
|
[{"Id": 185795700, "KernelVersionId": 129582530, "SourceDatasetVersionId": 5672295}]
|
[{"Id": 5672295, "DatasetId": 3256284, "DatasourceVersionId": 5747827, "CreatorUserId": 5048762, "LicenseName": "Other (specified in description)", "CreationDate": "05/12/2023 19:53:23", "VersionNumber": 2.0, "Title": "Photovoltaic System O&M inspection", "Slug": "photovoltaic-system-o-and-m-inspection", "Subtitle": "Thermal and RGB images from inspection of a photovoltaic system", "Description": "According to [one of the three articles](https://onlinelibrary.wiley.com/doi/10.1002/pip.3564) that explains how [PV-HAWK](https://lukasbommes.github.io/PV-Hawk/index.html) ([MIT License](https://github.com/LukasBommes/PV-Hawk/blob/master/LICENSE)) tool works, five different PV plants were used to train one of the models used by this tool. The plants were named A, B, C, D and E for anonymization purposes. This dataset is a sample from the first 12 arrays of PV plant A.\n\n---\n\n# 1. Context\n\nBoth large and small photovoltaic systems are susceptible to failures in their equipment, especially in modules due to operational stresses that are exposed and errors during the installation process of these devices. Although numerous internal and external factors originate these failures, the common phenomenon presented by several of them is hot spots on module defective area. The immediate impact is perceptible in the reduction of the generated power and, in the long term, in the reduction of the useful life of the equipment due to the high temperatures presented. The preventive maintenance method for recognizing this phenomenon is the use of thermography images in inspections of photovoltaic modules. Through this procedure, faulty modules are immediately identified with failures at an early stage due to their high heat signatures compared to the others, captured by cameras with infrared sensors. Currently, the use of this type of camera attached to drones stands out for providing an increase in the inspection area and a reduction in its execution time.\n\nTo understand more about this, read these reports by International energy agency (IEA):\n- [ Review of failures of PV modules](https://iea-pvps.org/wp-content/uploads/2020/01/IEA-PVPS_T13-01_2014_Review_of_Failures_of_Photovoltaic_Modules_Final.pdf);\n- [Review of IR and EL images applications for PV systems](https://iea-pvps.org/wp-content/uploads/2020/01/Review_on_IR_and_EL_Imaging_for_PV_Field_Applications_by_Task_13.pdf).\n\n## 1.1 Photovoltaic system specifications\n\nAcording to the [dataset article](https://onlinelibrary.wiley.com/doi/10.1002/pip.3564), the photovoltaic system on which the thermographic inspection was carried out is located in Germany and it's composed of 2376 PV polycrystalline silicon modules, measuring 1650 x 992 mm (60-cell) each.\n\nThe images in this dataset refer to the region marked in red in the google maps screenshot of the photovoltaic system location.\n\n<br>\n\n\n\n<br>\n\n## 1.2 Thermal inspection specifications\n\nThe inspection took place under clearsky conditions and solar irradiance above 700\u2009W/m\u00b2. In the table bellow more detail are presented for the weather parameters that influence thermography inspections.\n\n| Number of modules | Distance (m) | Peak velocity (m/s) | Air Temperature (\u00baC)| Global radiation (J/cm\u00b2)| Wind speed (m/s)|\n| --- | --- | -- | --- | --- | --- |\n| 13640 | 7612 | 4.1 | 25 | 39.7 | 2.8 |\n\n The drone used was a DJI model MATRICE 210 coupled with a FLIR XT2 thermal camera and with the following specifications: \n\n- Thermal resolution of 640x512 pixels;\n- Visual resolution of 1280x720 pixels;\n- Focal length of 13 mm;\n- Frame rate of 8 Hz .\n\nThe drone was controlled manually, positioned at an altitude of 10 m to 30\u2009m from the ground with a velocity that ensures blur-free images. The camera orientation was facing vertically downwards (nadir) at all times.\n\nAiming at reducing inspection cost and duration, especially for increasing the drone range before a battery change is needed, the images were sequentially scanned considering two types of PV arrays layouts: only one single array appears in the image and then two arrays.\n\n<br>\n\n\n\n<br>\n\n\n\n<br>\n\nAs showed in the table bellow, scanning two rows simultaneously speeds up the flight duration by a factor of 2.1, decreases flight distance by a factor of 1.9 and increases module throughput by a factor of 2.09. Despite these benefits, the resolution of extracted PV module images reduces.\n\n| Inspection layout | Flight distance (m) | Flight duration (s) | Average module resolution (px) |Module throughput (1/s) |\n| --- | --- | --- | --- | --- |\n| Single row | 1307 | 707 | 141 X 99 | 3.36 |\n| Double row | 681 | 338 | 73 X 50 | 7.03 |\n\n## 1.3 Dataset organization\n\nThe images are separated by type of inspection in different folders (single or double rows). In each folder, there are thermographic images in TIFF format and a CSV file with drone's geospatial and temporal data during the inspection. Only for the double row inspection type that visual (RGB) images were acquired.\n\nBesides, I've uploaded files to use for calibrate infrared and visual cameras to correct any type of distortion that camera lenses cause. \n\n# 2. Resources\n\n- This guides by [FLIR](http://support.flir.com/appstories/AppStories/Electrical&Mechanical/Testing_solar_panels_EN.pdf) and [TESTO](https://www.murcal.com/pdf%20folder/15.testo_thermography_guide.pdf) companies are good resources to understand more about thermography in the solar modules context;\n\n- There's [another one by FLIR](https://thermalcapture.com/wp-content/uploads/2019/08/pv-system-inspection-thermal-drones-07-15-19.pdf) that explains in depth how aerial thermal inspections of photovoltaic systems are made and their importance in this field;\n\n- To understand the level of influence that the module degradation has on the yield of the photovoltaic system you can read the [IEC TS-62446-3]( https://ayscomdatatec.com/wp-content/uploads/2019/09/Normativa-IEC-TS-62446-3.pdf) and the [Raptor maps's knoledge hub](https://raptormaps.com/solar-tech-docs/).\n\n# 3. Inspiration\n\nA service often provided by companies in this area is a SaaS that displays the detected faulty modules in an bird's eye view of the photovoltaic system and calculate the energy loss, like the image bellow shows. One can create a web app (using streamlit or plotly/dash) that detect PV modules with a instance segmentation model, track them with a object tracker and classify their integrity (binary or multiclass classification) with a image classification model.\n\n<br>\n\n\n\n<br>\n\nThis idea can be used for guiding a maintenance team in order to intervene and replace panels if necessary.", "VersionNotes": "Data Update 2023-05-12", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3256284, "CreatorUserId": 5048762, "OwnerUserId": 5048762.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5672295.0, "CurrentDatasourceVersionId": 5747827.0, "ForumId": 3321771, "Type": 2, "CreationDate": "05/11/2023 18:26:40", "LastActivityDate": "05/11/2023", "TotalViews": 1219, "TotalDownloads": 142, "TotalVotes": 1, "TotalKernels": 1}]
|
[{"Id": 5048762, "UserName": "marcosgabriel", "DisplayName": "Marcos Gabriel", "RegisterDate": "05/08/2020", "PerformanceTier": 0}]
|
import pickle
from pathlib import Path
from scipy import ndimage as ndi
from skimage.color import label2rgb
from skimage.filters import try_all_threshold
from PIL import Image
import tifffile as tif
import cv2
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import make_axes_locatable
import pandas as pd
# # 1. Dataset Reading
# The best way is to first read the metadata files from both datasets
SINGLE_ROW_METADATA_PATH = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/single-row/metadata.csv"
columns_to_rename = {"thermal image name": "thermal_image_name"}
sr_metadata = pd.read_csv(SINGLE_ROW_METADATA_PATH).rename(columns=columns_to_rename)
sr_metadata.head()
DOUBLE_ROW_METADATA_PATH = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/double-row/metadata.csv"
columns_to_rename = {
"thermal image name": "thermal_image_name",
"rgb image name": "rgb_image_name",
}
dr_metadata = pd.read_csv(DOUBLE_ROW_METADATA_PATH).rename(columns=columns_to_rename)
dr_metadata.head()
# We need to get the full path for the images
def get_image_full_path(image_name, image_type):
if image_type == "single_row_thermal":
origin_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/single-row/thermal images"
elif image_type == "double_row_thermal":
origin_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/double-row/thermal images"
elif image_type == "double_row_rgb":
origin_path = (
origin_path
) = "/kaggle/input/photovoltaic-system-o-and-m-inspection/datasets/double-row/rgb images"
return Path(origin_path, image_name)
sr_metadata = sr_metadata.assign(
thermal_image_name=sr_metadata.thermal_image_name.apply(
lambda x: get_image_full_path(x, "single_row_thermal")
)
).assign(timestamp=pd.to_datetime(sr_metadata.timestamp))
dr_metadata = (
dr_metadata.assign(
thermal_image_name=dr_metadata.thermal_image_name.apply(
lambda x: get_image_full_path(x, "double_row_thermal")
)
)
.assign(
rgb_image_name=dr_metadata.rgb_image_name.apply(
lambda x: get_image_full_path(x, "double_row_rgb")
)
)
.assign(timestamp=pd.to_datetime(sr_metadata.timestamp))
)
# **Now we can load the images!**
# I've created the Thermogram class just to be possible to get the thermal image and the converted one in the same object like [Flyr library](https://bitbucket.org/nimmerwoner/flyr/src/master/) does
class Thermogram:
def __init__(self, path: Path):
self.path = path
@property
def celsius(self) -> np.ndarray:
return (tif.imread(self.path.as_posix()) * 0.04) - 273.15
def render(self) -> np.ndarray:
image = self.celsius
image = (image - np.min(image)) / (np.max(image) - np.min(image))
return (image * 255.0).astype(np.uint8)
def load_image(image_path: Path):
image_format = image_path.suffix
if image_format == ".jpg":
return cv2.imread(image_path.as_posix())
elif image_format == ".tiff":
return Thermogram(image_path)
image_number = 57
thermogram = load_image(sr_metadata.thermal_image_name[image_number])
_, ax = plt.subplots(1, 2)
im = ax[0].imshow(thermogram.celsius, cmap="inferno")
ax[0].set_title("Thermographic image")
ax[0].set_axis_off()
ax[1].imshow(thermogram.render(), cmap="gray")
ax[1].set_title("Rendered image (8 bit image)")
ax[1].set_axis_off()
cax = make_axes_locatable(ax[0]).append_axes("right", size="5%", pad=0.05)
plt.colorbar(
im, cax=cax, values=np.unique(thermogram.celsius), label="Temperature (ºC)"
)
plt.tight_layout()
plt.show()
thermogram = load_image(dr_metadata.thermal_image_name[image_number])
visual = load_image(dr_metadata.rgb_image_name[image_number])
_, ax = plt.subplots(1, 3, figsize=(10, 5))
im = ax[0].imshow(thermogram.celsius, cmap="inferno")
ax[0].set_title("Thermographic image")
ax[0].set_axis_off()
ax[1].imshow(thermogram.render(), cmap="gray")
ax[1].set_title("Rendered image (8 bit image)")
ax[1].set_axis_off()
ax[2].imshow(visual[:, :, ::-1])
ax[2].set_title("Visual image")
ax[2].set_axis_off()
cax = make_axes_locatable(ax[0]).append_axes("right", size="5%", pad=0.05)
plt.colorbar(
im, cax=cax, values=np.unique(thermogram.celsius), label="Temperature (ºC)"
)
plt.tight_layout()
plt.show()
# # 2. Camera calibration
# This step is important because often times the lenses of cameras create distortions in the images. In this dataset only the RGB ones were affected, but the intrinsic and extrinsic camera parameters from the IR camera can be used for other tasks like Structure from motion (as PV-HAWK does).
def remove_distortion(image: np.ndarray, image_type: str = "rgb"):
mapx_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/calibration files/RGB/mapx.pkl"
mapy_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/calibration files/RGB/mapy.pkl"
if image_type == "ir":
mapx_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/calibration files/IR/mapx.pkl"
mapy_path = "/kaggle/input/photovoltaic-system-o-and-m-inspection/calibration files/IR/mapy.pkl"
with open(mapx_path, "rb") as mapx_file, open(mapy_path, "rb") as mapy_file:
mapx = pickle.load(mapx_file)
mapy = pickle.load(mapy_file)
return cv2.remap(
image, mapx, mapy, cv2.INTER_CUBIC, borderMode=cv2.BORDER_REPLICATE
)
undistorted_rgb = remove_distortion(visual)
_, ax = plt.subplots(1, 2, figsize=(20, 5))
ax[0].imshow(visual[:, :, ::-1])
ax[0].set_title("RGB distorted")
ax[0].set_axis_off()
ax[1].imshow(undistorted_rgb[:, :, ::-1])
ax[1].set_title("RGB undistorted")
ax[1].set_axis_off()
plt.tight_layout()
plt.show()
undistorted_ir = remove_distortion(thermogram.render(), "ir")
_, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].imshow(thermogram.render(), cmap="gray")
ax[0].set_title("IR distorted")
ax[0].set_axis_off()
ax[1].imshow(undistorted_ir, cmap="gray")
ax[1].set_title("IR undistorted")
ax[1].set_axis_off()
plt.tight_layout()
plt.show()
# # 3. Images Alignment
# Is possible to align the RGB and IR images using two techniques:
# - **Phase Correlation**: I still don't quite understand how this algorithm works, but this [solution on github](https://github.com/YoshiRi/ImRegPOC) works for this dataset;
# - **Enhanced Correlation Coefficient (ECC)**: I found [an article](https://learnopencv.com/image-alignment-ecc-in-opencv-c-python/) that explains very well how this solution works so I'll stick with it here.
# Before any alignment algorithm be applied, I've to say that there's something odd with this images. Although the thermographic image has a lower resolution than the RGB one, it has a larger field of view because more modules appear in it. Typically, RGB images have a larger field of view than thermal images.
# So, it's necessary to crop the RGB and IR images to their correspondent region first. Then, resize them to the same resolution.
left = 0
top = 75
right = 640
bottom = 490
crop_fg_image = Image.fromarray(undistorted_ir).crop((left, top, right, bottom))
crop_fg_image
left = 120
top = 0
right = 1150
bottom = 720
crop_bg_image = (
Image.fromarray(undistorted_rgb[:, :, ::-1])
.crop((left, top, right, bottom))
.resize(crop_fg_image.size)
)
crop_bg_image
# We can compare the aligned image with the unaligned one by applying the canny edge detector in both of them. Then, blending each canny mask result in the thermal image.
def get_gradient(image):
grad_x = cv2.Sobel(image, cv2.CV_32F, 1, 0, ksize=3)
grad_y = cv2.Sobel(image, cv2.CV_32F, 0, 1, ksize=3)
return cv2.addWeighted(np.absolute(grad_x), 0.5, np.absolute(grad_y), 0.5, 0)
def align_images(
image_1, image_2, warp_mode, n_iterations=5000, increment_threshold=1e-10
):
warp_mode = cv2.MOTION_TRANSLATION
if warp_mode == cv2.MOTION_HOMOGRAPHY:
warp_matrix = np.eye(3, 3, dtype=np.float32)
else:
warp_matrix = np.eye(2, 3, dtype=np.float32)
criteria = (
cv2.TERM_CRITERIA_EPS | cv2.TERM_CRITERIA_COUNT,
n_iterations,
increment_threshold,
)
grad_1 = get_gradient(image_1)
grad_2 = get_gradient(image_2)
cc, warp_matrix = cv2.findTransformECC(
grad_1, grad_2, warp_matrix, warp_mode, criteria
)
if warp_mode == cv2.MOTION_HOMOGRAPHY:
return cv2.warpPerspective(
image_2,
warp_matrix,
image_1.shape[::-1],
flags=cv2.INTER_LINEAR + cv2.WARP_INVERSE_MAP,
)
else:
return cv2.warpAffine(
image_2,
warp_matrix,
image_1.shape[::-1],
flags=cv2.INTER_LINEAR + cv2.WARP_INVERSE_MAP,
)
def blend_images(image_1, image_2, alpha=0.2, beta=None):
beta = 1 - alpha if not beta else beta
return cv2.addWeighted(image_2, alpha, image_1, beta, 0.0)
crop_bg_image = np.asarray(crop_bg_image)
crop_fg_image = np.asarray(crop_fg_image)
crop_bg_image_gray = cv2.cvtColor(crop_bg_image, cv2.COLOR_BGR2GRAY)
# Align the image with two methods: Translation and Homography
align_image_1 = align_images(crop_fg_image, crop_bg_image_gray, cv2.MOTION_TRANSLATION)
align_image_2 = align_images(crop_fg_image, crop_bg_image_gray, cv2.MOTION_HOMOGRAPHY)
# Apply the Canny edge detector in the images
unalign_canny = cv2.Canny(crop_bg_image_gray, 300, 400)
align_canny_1 = cv2.Canny(align_image_1, 200, 300)
align_canny_2 = cv2.Canny(align_image_2, 200, 300)
# Blend the Canny masks with the thermal imagem
unalign_blend = blend_images(crop_fg_image, unalign_canny)
align_blend_1 = blend_images(crop_fg_image, align_canny_1)
align_blend_2 = blend_images(crop_fg_image, align_canny_2)
_, ax = plt.subplots(1, 3, figsize=(20, 5))
ax[0].imshow(unalign_blend, cmap="gray")
ax[0].set_title("Cropped RGB image")
ax[1].imshow(align_blend_1, cmap="gray")
ax[1].set_title("Motion translation")
ax[2].imshow(align_blend_2, cmap="gray")
ax[2].set_title("Motion homography")
plt.show()
# # 4. Modules Segmentation
# ## 4.1 Segmentation of RGB images
# First, I will segment the modules in the RGB images. The way I'll do this is through color segmentation. First, we convert the image to the HSV color space (the most appropriate space for color processing) and apply the Otsu threshold in each of it's channels (Hue, Saturation and Value).
image = cv2.cvtColor(undistorted_rgb, cv2.COLOR_BGR2HSV)
for i in range(3):
title = "Value"
if i == 0:
title = "Hue"
elif i == 1:
title = "Saturation"
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
fig.suptitle(title)
ax[0].hist(image[:, :, i].ravel(), np.unique(image[:, :, i]))
ax[1].imshow(image[:, :, i], cmap="gray")
ax[1].set_axis_off()
plt.tight_layout()
plt.show()
# We can compare this method with the identification of modules by seeting a minimum and maximum threshold manually in each HSV channel
def preprocess_rgb_image_manual(image, lower_values, higher_values):
image = cv2.cvtColor(image, cv2.COLOR_BGR2HSV)
return cv2.inRange(image, lower_values, higher_values)
def preprocess_rgb_image_hsv_otsu(image):
image = cv2.cvtColor(image, cv2.COLOR_BGR2HSV)
masks = []
for channel_index, channel_image in enumerate(cv2.split(image)):
channel_image = cv2.GaussianBlur(channel_image, (5, 5), 0)
max_value = 179 if channel_index == 0 else 255
_, mask = cv2.threshold(
channel_image, 0, max_value, cv2.THRESH_BINARY + cv2.THRESH_OTSU
)
masks.append(mask)
result_mask = np.bitwise_and(masks[0], masks[1])
return np.bitwise_and(result_mask, masks[2])
lower_blue = np.array([90, 100, 100])
upper_blue = np.array([125, 255, 200])
mask_1 = preprocess_rgb_image_manual(undistorted_rgb, lower_blue, upper_blue)
mask_2 = preprocess_rgb_image_hsv_otsu(undistorted_rgb)
_, ax = plt.subplots(1, 2, figsize=(20, 5))
ax[0].imshow(mask_1, cmap="gray")
ax[0].set_axis_off()
ax[0].set_title("Manual thresholds setting")
ax[1].imshow(mask_2, cmap="gray")
ax[1].set_title("Otsu thresholds")
ax[1].set_axis_off()
plt.show()
# It's noticeable how the otsu method in each HSV color channel is more appropriate for module identification. But, we have to fill in the holes that some module masks have.
blur = cv2.medianBlur(mask_2, 5)
kernel = np.ones((5, 5), np.uint8)
blur_open = cv2.morphologyEx(blur, cv2.MORPH_OPEN, kernel)
module_mask = ndi.binary_fill_holes(blur_open)
plt.figure(figsize=(10, 5))
plt.imshow(module_mask, cmap="gray")
# Applying the connected components method, we can set for every module mask with a label
_, label_mask = cv2.connectedComponents(module_mask.astype("uint8"))
plt.figure(figsize=(10, 5))
plt.imshow(label_mask, cmap="bone")
undistorted_gray = cv2.cvtColor(undistorted_rgb, cv2.COLOR_BGR2GRAY)
plt.imshow(label2rgb(label_mask, undistorted_gray))
# ## 4.2 Segmentation of IR images
try_all_threshold(undistorted_ir, verbose=False)
# It's to be expected that traditional image segmentation methods do not work very well on thermographic images, as they were built for images of the human visual spectrum.
# So, we can use the masks taken from rgb images to segment the modules in the thermographic images.
new_module_mask = (
Image.fromarray(module_mask)
.crop((120, 0, 1150, 720))
.resize(crop_fg_image.shape[::-1])
)
new_module_mask
_, label_mask = cv2.connectedComponents(np.array(new_module_mask).astype("uint8"))
plt.imshow(label2rgb(label_mask, np.array(crop_fg_image)))
# Another approach is to train an instance segmentation model for automatic module segmentation
# # 4. Images Overlap
class ImageAligner:
def __init__(self, number_features: int = 500):
self.feature_extractor = cv2.ORB_create(number_features)
self.matcher = cv2.BFMatcher(cv2.NORM_HAMMING, crossCheck=True)
self.points = {}
def filter_matches(self, matches, matches_percent: float = 0.2):
matches = sorted(matches, key=lambda x: x.distance)
matches_to_keep = int(len(matches) * matches_percent)
return matches[:matches_to_keep]
def __call__(self, image, template):
# Get keypoints and descriptors
image_kps, image_dsc = self.feature_extractor.detectAndCompute(image, None)
template_kps, template_dsc = self.feature_extractor.detectAndCompute(
template, None
)
# Match features and filter them
matches = self.matcher.match(image_dsc, template_dsc)
matches = self.filter_matches(matches)
# Store points and matches for debug purposes
self.points["keypoints"] = [image_kps, template_kps]
self.points["matches"] = matches
# Filter keypoints with filtered matches
image_kps = np.float32([image_kps[match.queryIdx].pt for match in matches])
template_kps = np.float32(
[template_kps[match.trainIdx].pt for match in matches]
)
# Correct image perspective
matrix, _ = cv2.findHomography(image_kps, template_kps, cv2.RANSAC)
return cv2.warpPerspective(image, matrix, template.shape[::-1])
def inspect_matches(self, image, template):
return cv2.drawMatches(
image,
self.points["keypoints"][0],
template,
self.points["keypoints"][1],
self.points["matches"],
None,
)
undistorted_rgb_1 = remove_distortion(load_image(dr_metadata.rgb_image_name[57]))
undistorted_rgb_2 = remove_distortion(load_image(dr_metadata.rgb_image_name[58]))
_, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].imshow(undistorted_rgb_1[:, :, ::-1])
ax[1].imshow(undistorted_rgb_2[:, :, ::-1])
plt.show()
aligner = ImageAligner()
undistorted_gray_1 = cv2.cvtColor(undistorted_rgb_1, cv2.COLOR_BGR2GRAY)
undistorted_gray_2 = cv2.cvtColor(undistorted_rgb_2, cv2.COLOR_BGR2GRAY)
aligned_image = aligner(undistorted_gray_1, undistorted_gray_2)
inspected_image = aligner.inspect_matches(undistorted_gray_1, undistorted_gray_2)
_, ax = plt.subplots(2, 1, figsize=(20, 10))
ax[0].imshow(aligned_image, cmap="gray")
ax[1].imshow(inspected_image)
plt.show()
undistorted_ir_1 = remove_distortion(
load_image(dr_metadata.thermal_image_name[57]).render(), "ir"
)
undistorted_ir_2 = remove_distortion(
load_image(dr_metadata.thermal_image_name[58]).render(), "ir"
)
_, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].imshow(undistorted_ir_1, cmap="gray")
ax[1].imshow(undistorted_ir_2, cmap="gray")
plt.show()
aligner = ImageAligner()
aligned_image = aligner(undistorted_ir_1, undistorted_ir_2)
inspected_image = aligner.inspect_matches(undistorted_ir_1, undistorted_ir_2)
_, ax = plt.subplots(2, 1, figsize=(20, 10))
ax[0].imshow(aligned_image, cmap="gray")
ax[1].imshow(inspected_image)
plt.show()
# # 6. Defected Modules
# Highlight defected modules when the modules masks were created
# # 7. Thermal Inspection
# Is possible to determine parameters like the inspection time and drone path
sr_inspection_time = sr_metadata.timestamp.filter([0, 5294]).diff().iloc[1].seconds
dr_inspection_time = dr_metadata.timestamp.filter([0, 2540]).diff().iloc[1].seconds
print(
f"Single row inspection time: {sr_inspection_time // 60} minutes and {sr_inspection_time % 60} seconds"
)
print(
f"Double row inspection time: {dr_inspection_time // 60} minutes and {dr_inspection_time % 60} seconds"
)
_, ax = plt.subplots(1, 2, figsize=(15, 5))
ax[0].scatter(sr_metadata.longitude, sr_metadata.latitude)
ax[0].set_title("Single row inspection")
ax[1].scatter(dr_metadata.longitude, dr_metadata.latitude)
ax[1].set_title("Double row inspection")
plt.show()
# # 8. Thermal image orthomosaic
# Show how to make a thermal orthomosaic with this dataset
| false | 0 | 6,024 | 5 | 8,218 | 6,024 |
||
129582106
|
<jupyter_start><jupyter_text>Collection of Classification & Regression Datasets
This dataset encompasses my personal collection of classification and regression.
File list -
1. Customer.csv
2. House_Price.csv
3. Movie_classification.csv
4. Movie_regression.xls
5. canada_per_capita.csv
6. carprices.csv
7. daily-min-temperatures.csv
8. daily-total-female-births-CA.csv
9. hiring.csv
10.house-votes-84.csv
11. insurance_data.csv
12. nih_labels.csv
13. salaries.csv
14. shampoo.csv
15. us-airlines-monthly-aircraft-miles-flown.csv
16. xrayfull.csv
Kaggle dataset identifier: others
<jupyter_script># **HOUSE PRICE PREDICTION**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# **IMPORTING DATASET**
# Importing csv file
df = pd.read_csv("/kaggle/input/others/House_Price.csv")
df.columns
df.head()
# **ANALYSING THE DATASET**
df.describe()
df.info()
# **Inference from the dataset**
# 1. In crime_rate, 75% of values lie below 3.677 where the maximum value is 88.97 where the there is an abnormal distribution (reason due to any Outliers).
# 2. Same as for the n_hot_rooms column
# 3. Missing values in n_hos_beds
# 4. Abnormal distribution in rainfall
# *** EDD (EXTENDED DATA DICTIONARY) ***
sns.jointplot(data=df, x="crime_rate", y="price")
# From the above plot, the crime rate is a polynomial relationship with target (price) variable
sns.jointplot(data=df, x="n_hos_beds", y="price")
# Nearly, the n_hos_beds has abnormal distribution (scattered everywhere), couldnt able to analyse a distribution for it
sns.jointplot(data=df, x="n_hot_rooms", y="price")
# From above plot, merely two outliers found.
sns.jointplot(data=df, x="rainfall", y="price")
# Nearly, the rainfall has abnormal distribution (scattered everywhere), couldnt able to analyse a distribution for it
df.shape
sns.heatmap(df.isnull(), cbar=False)
# Here, n_hot_rooms could able to internpret that it has outliers
# **Visualising Categrical Variable**
sns.countplot(data=df, x="airport")
sns.countplot(data=df, x="waterbody")
sns.countplot(data=df, x="bus_ter")
# So, from the above plot ,it has been concluded as it has a constant value **YES**
# **Creating Dummy Varaible for the Categorical Varaible**
df = pd.get_dummies(df)
df.columns
# From the above Columns, dropping bus_ter_YES column
del df["bus_ter_YES"]
# Dropping rainfall and n_hos_beds since it is having scatter distribution
del df["rainfall"]
del df["n_hos_beds"]
# Normalizing those crime_rate column lograthimically since it has polynomial distribution
df["crime_rate"] = np.log(1 + df.crime_rate)
sns.jointplot(data=df, x="crime_rate", y="price")
# **INFERENCE :** Now, the plot is shwing a kind of linear relationship
# **OUTLIER TREATEMENT FOR THE n_hot_rooms BY CAPPING AND FLOORING METHOD**
lower_threshold = np.percentile(df["n_hot_rooms"], [1])[0]
higher_threshold = np.percentile(df["n_hot_rooms"], [99])[0]
df.n_hot_rooms[df.n_hot_rooms < 0.3 * lower_threshold] = 0.3 * lower_threshold
df.n_hot_rooms[df.n_hot_rooms > 3 * higher_threshold] = 3 * higher_threshold
sns.jointplot(data=df, x="n_hot_rooms", y="price")
df.describe()
# Now,all columns has been normalized without any outliers
# **TREATING MISSING VALUES BY MEAN**
df.info()
df = df.fillna(df.mean())
df.crime_rate = np.log(1 + df.crime_rate)
sns.jointplot(data=df, x="crime_rate", y="price")
# Taking Avergae of dist1,dist2,dist3 and dist4 as all are interpreting the same so Average take as a new Column
df["Average"] = df[["dist1", "dist2", "dist3", "dist4"]].mean(axis=1)
df.head()
plt.subplots(figsize=(15, 15))
sns.heatmap(df.corr(), annot=True, cmap="coolwarm")
import scipy.stats as stats
plt.figure(figsize=(23, 80))
lists = df.columns
j = 0
for i in list(enumerate(df.columns)):
# print(i[1])
plt.subplot(15, 4, i[0] + 1)
sns.histplot(data=df[i[1]], kde=True) # Histogram with KDE line
plt.title(lists[j])
j = j + 1
plt.tight_layout()
plt.show()
# **TRAINING AND TESTING THE MODEL**
import statsmodels.api as sn
from sklearn.model_selection import train_test_split
x_multi = df.drop(["price"], axis=1)
x_multi
y_multi = df["price"]
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
X_train, X_test, y_train, y_test = train_test_split(
x_multi, y_multi, test_size=0.2, random_state=42
)
model = LinearRegression()
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
mse = mean_squared_error(y_test, y_pred)
print("Mean Squared Error:", mse)
r2 = r2_score(y_test, y_pred)
print(f"R-squared: {r2}")
print("Coefficients:", model.coef_)
print("Intercept:", model.intercept_)
# **INFERENCE**
# 1. From the above interpretation, the R^2 Value is **0.6607** (i.e.,) 66% of the variance of target variable (i.e.,) Price variable has been affected by the predictors.
# 2. The lower P value of each predictor variable clearly signifies that it is significantly affecting the target(response) variable proving that each variable has significant co-efficients that affects the response variable.
feature_names = x_multi.columns
# Get the coefficients
coefficients = model.coef_
# Plot the coefficients
plt.figure(figsize=(10, 6))
plt.bar(feature_names, coefficients)
plt.xlabel("Features")
plt.ylabel("Coefficients")
plt.title("Linear Regression Coefficients")
plt.xticks(rotation=90)
plt.show()
# **INFERENCE**
# 1. From the above plot,we could clearly infer that if we increase the value of **parks** the response variable positively increases on a **larger effect**
# 2. Remaining all predictor variables significantly impact on the **Price** variable.
sns.jointplot(x="parks", y="price", data=df, kind="reg")
sns.jointplot(x="air_qual", y="price", data=df, kind="reg")
sns.jointplot(x="room_num", y="price", data=df, kind="reg")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/582/129582106.ipynb
|
others
|
balakrishcodes
|
[{"Id": 129582106, "ScriptId": 38530753, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9077457, "CreationDate": "05/15/2023 03:02:17", "VersionNumber": 4.0, "Title": "House_Price_Prediction", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 198.0, "LinesInsertedFromPrevious": 6.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 192.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 185794561, "KernelVersionId": 129582106, "SourceDatasetVersionId": 1475016}]
|
[{"Id": 1475016, "DatasetId": 841826, "DatasourceVersionId": 1508810, "CreatorUserId": 3391197, "LicenseName": "CC0: Public Domain", "CreationDate": "09/09/2020 12:27:39", "VersionNumber": 5.0, "Title": "Collection of Classification & Regression Datasets", "Slug": "others", "Subtitle": "This dataset encompasses my collection of classification and regression", "Description": "This dataset encompasses my personal collection of classification and regression.\n\nFile list -\n\n1. Customer.csv\n2. House_Price.csv\n3. Movie_classification.csv\n4. Movie_regression.xls\n5. canada_per_capita.csv\n6. carprices.csv\n7. daily-min-temperatures.csv\n8. daily-total-female-births-CA.csv\n9. hiring.csv\n10.house-votes-84.csv\n11. insurance_data.csv\n12. nih_labels.csv\n13. salaries.csv\n14. shampoo.csv\n15. us-airlines-monthly-aircraft-miles-flown.csv\n16. xrayfull.csv", "VersionNotes": "v5", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 841826, "CreatorUserId": 3391197, "OwnerUserId": 3391197.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1475016.0, "CurrentDatasourceVersionId": 1508810.0, "ForumId": 857034, "Type": 2, "CreationDate": "08/23/2020 05:27:43", "LastActivityDate": "08/23/2020", "TotalViews": 24126, "TotalDownloads": 4363, "TotalVotes": 36, "TotalKernels": 21}]
|
[{"Id": 3391197, "UserName": "balakrishcodes", "DisplayName": "Balakrishna Kumar", "RegisterDate": "06/25/2019", "PerformanceTier": 1}]
|
# **HOUSE PRICE PREDICTION**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# **IMPORTING DATASET**
# Importing csv file
df = pd.read_csv("/kaggle/input/others/House_Price.csv")
df.columns
df.head()
# **ANALYSING THE DATASET**
df.describe()
df.info()
# **Inference from the dataset**
# 1. In crime_rate, 75% of values lie below 3.677 where the maximum value is 88.97 where the there is an abnormal distribution (reason due to any Outliers).
# 2. Same as for the n_hot_rooms column
# 3. Missing values in n_hos_beds
# 4. Abnormal distribution in rainfall
# *** EDD (EXTENDED DATA DICTIONARY) ***
sns.jointplot(data=df, x="crime_rate", y="price")
# From the above plot, the crime rate is a polynomial relationship with target (price) variable
sns.jointplot(data=df, x="n_hos_beds", y="price")
# Nearly, the n_hos_beds has abnormal distribution (scattered everywhere), couldnt able to analyse a distribution for it
sns.jointplot(data=df, x="n_hot_rooms", y="price")
# From above plot, merely two outliers found.
sns.jointplot(data=df, x="rainfall", y="price")
# Nearly, the rainfall has abnormal distribution (scattered everywhere), couldnt able to analyse a distribution for it
df.shape
sns.heatmap(df.isnull(), cbar=False)
# Here, n_hot_rooms could able to internpret that it has outliers
# **Visualising Categrical Variable**
sns.countplot(data=df, x="airport")
sns.countplot(data=df, x="waterbody")
sns.countplot(data=df, x="bus_ter")
# So, from the above plot ,it has been concluded as it has a constant value **YES**
# **Creating Dummy Varaible for the Categorical Varaible**
df = pd.get_dummies(df)
df.columns
# From the above Columns, dropping bus_ter_YES column
del df["bus_ter_YES"]
# Dropping rainfall and n_hos_beds since it is having scatter distribution
del df["rainfall"]
del df["n_hos_beds"]
# Normalizing those crime_rate column lograthimically since it has polynomial distribution
df["crime_rate"] = np.log(1 + df.crime_rate)
sns.jointplot(data=df, x="crime_rate", y="price")
# **INFERENCE :** Now, the plot is shwing a kind of linear relationship
# **OUTLIER TREATEMENT FOR THE n_hot_rooms BY CAPPING AND FLOORING METHOD**
lower_threshold = np.percentile(df["n_hot_rooms"], [1])[0]
higher_threshold = np.percentile(df["n_hot_rooms"], [99])[0]
df.n_hot_rooms[df.n_hot_rooms < 0.3 * lower_threshold] = 0.3 * lower_threshold
df.n_hot_rooms[df.n_hot_rooms > 3 * higher_threshold] = 3 * higher_threshold
sns.jointplot(data=df, x="n_hot_rooms", y="price")
df.describe()
# Now,all columns has been normalized without any outliers
# **TREATING MISSING VALUES BY MEAN**
df.info()
df = df.fillna(df.mean())
df.crime_rate = np.log(1 + df.crime_rate)
sns.jointplot(data=df, x="crime_rate", y="price")
# Taking Avergae of dist1,dist2,dist3 and dist4 as all are interpreting the same so Average take as a new Column
df["Average"] = df[["dist1", "dist2", "dist3", "dist4"]].mean(axis=1)
df.head()
plt.subplots(figsize=(15, 15))
sns.heatmap(df.corr(), annot=True, cmap="coolwarm")
import scipy.stats as stats
plt.figure(figsize=(23, 80))
lists = df.columns
j = 0
for i in list(enumerate(df.columns)):
# print(i[1])
plt.subplot(15, 4, i[0] + 1)
sns.histplot(data=df[i[1]], kde=True) # Histogram with KDE line
plt.title(lists[j])
j = j + 1
plt.tight_layout()
plt.show()
# **TRAINING AND TESTING THE MODEL**
import statsmodels.api as sn
from sklearn.model_selection import train_test_split
x_multi = df.drop(["price"], axis=1)
x_multi
y_multi = df["price"]
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
X_train, X_test, y_train, y_test = train_test_split(
x_multi, y_multi, test_size=0.2, random_state=42
)
model = LinearRegression()
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
mse = mean_squared_error(y_test, y_pred)
print("Mean Squared Error:", mse)
r2 = r2_score(y_test, y_pred)
print(f"R-squared: {r2}")
print("Coefficients:", model.coef_)
print("Intercept:", model.intercept_)
# **INFERENCE**
# 1. From the above interpretation, the R^2 Value is **0.6607** (i.e.,) 66% of the variance of target variable (i.e.,) Price variable has been affected by the predictors.
# 2. The lower P value of each predictor variable clearly signifies that it is significantly affecting the target(response) variable proving that each variable has significant co-efficients that affects the response variable.
feature_names = x_multi.columns
# Get the coefficients
coefficients = model.coef_
# Plot the coefficients
plt.figure(figsize=(10, 6))
plt.bar(feature_names, coefficients)
plt.xlabel("Features")
plt.ylabel("Coefficients")
plt.title("Linear Regression Coefficients")
plt.xticks(rotation=90)
plt.show()
# **INFERENCE**
# 1. From the above plot,we could clearly infer that if we increase the value of **parks** the response variable positively increases on a **larger effect**
# 2. Remaining all predictor variables significantly impact on the **Price** variable.
sns.jointplot(x="parks", y="price", data=df, kind="reg")
sns.jointplot(x="air_qual", y="price", data=df, kind="reg")
sns.jointplot(x="room_num", y="price", data=df, kind="reg")
| false | 1 | 1,781 | 3 | 1,982 | 1,781 |
||
129582136
|
# # Linear Regression for House Prices Competition
# competition link:
# submission by: Juan Pablo Contreras
# ## Libraries and Data Source
# ### Libraries
# Author: Juan Pablo Contreras
# This is the notebook used for my submission for the House Prices - Advanced Regression Techniques competition
import math
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn import linear_model
# ### Data
# read the data
train_data = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test_data = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/test.csv"
)
# ## Data Cleaning
# NaN values in data set: yes
# Categorical Values in data set: yes
# Numerical Values in data set: yes
# The only difference between the training data set and the test data set is that the test data set does not include the SalePrice column
# To clean the data the following steps are taken
# 1. The SalePrice column from the training data set is removed so that the columns of the training df and the test df match
# 2. The test df is appended to the training df so that the cleaning can be done on both the training and the test df
# 3. Categorical values are changed to one hot encoding using the pands get_dummies() function
# 4. NaN values are converted to -1
# 5. The data set is split into training and testing again
# 6. Split training data into training and validation so that predictions on validation can be made to make sure the model does not spit nonesense before predicting on the test data set
# 7. The data is normalized using the training data. The validation and test data are transformed using the training data standarization fitting parameters
# ### 1. Delete SalePrice from Training Dataset
# find number of rows
num_rows_train = len(train_data.index)
print("num rows in training data: " + str(num_rows_train))
print("num rows in test data: " + str(len(test_data.index)))
# store and drop the training predictions
y_train = train_data.iloc[:, -1]
train_data.drop(columns=train_data.columns[-1], axis=1, inplace=True)
# ### 2. Merge Train and Test Data
# merge all data for cleaning
all_data = train_data.append(test_data, ignore_index=True)
print("num rows in all data: " + str(len(all_data.index)))
# delete id column because ID is not a predictor of house price
ids = all_data.drop("Id", axis=1)
# ### 3. and 4.0 Handle Categorical and NaN values
# convert categorical data to numerical (0 or 1)
all_data = pd.get_dummies(data=all_data)
# replace NaN values
all_data = all_data.fillna(-1)
all_data.head()
# ### 5. Split data back into Train, and Test
# split data back into train and test data
X_train = all_data.iloc[0:num_rows_train, :]
print("num rows in X train: " + str(len(X_train.index)))
print("num rows in y train: " + str(len(y_train.index)))
X_test = all_data.iloc[num_rows_train:, :]
print("num rows in X test: " + str(len(X_test.index)))
print("Training data features head: ")
print(X_train.head())
print()
print("Training data predictions head: ")
print(y_train.head())
print()
print("Test data features head:")
print(X_test.head())
# ### 6. Split train data into train and validation
# The new training data will be 80% of the train dataset, and the validation data 20% of the train data set
X_train, X_val, y_train, y_val = train_test_split(
X_train, y_train, test_size=0.2, random_state=42
)
# ### 7. Standarize data
# The training data is standarized. The parameters used to standarize the training data are then used to transform the validation and test data
scaler = StandardScaler()
scaler.fit(X_train)
X_train_std = scaler.transform(X_train)
X_val_transf = scaler.transform(X_val)
X_test_transf = scaler.transform(X_test)
# ## Linear Regression
# The linear regression model from sklearn is used.
# 1. The model is created using the sklearn library
# 2. The model is fit using the training data
# 3. The model is validated using the validation data
# 4. Predictions are made on the test data
# ### 1. Create the model
# linear model
regr = linear_model.LinearRegression()
# ### 2. Fit the Model
# fit model using training data
regr.fit(X_train, y_train)
# ### 3. Validate Model
# predict prices on the test data
pred_val = regr.predict(X_val)
print("Five first predictions: ")
print(pred_val[:5])
print()
# Compare the Predicted values on the validation data set to the actual SalePrices of the validation data set using the root mean squared error
MSE = np.square(np.subtract(y_val, pred_val)).mean()
rsme = math.sqrt(MSE)
print("Root Mean Square Error:\n")
print(rsme)
# ### Predict on Test Data
pred_test = regr.predict(X_test)
print("Five first predictions for test dataset: ")
print(pred_test[:5])
print()
# ## Submission
# submission
sample_submission = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/sample_submission.csv"
)
sample_submission["SalePrice"] = pred_test
sample_submission.to_csv("/kaggle/working/submission.csv", index=False)
sample_submission.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/582/129582136.ipynb
| null | null |
[{"Id": 129582136, "ScriptId": 38524068, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10847886, "CreationDate": "05/15/2023 03:02:44", "VersionNumber": 7.0, "Title": "House Prices Competition", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 159.0, "LinesInsertedFromPrevious": 88.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 71.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
| null | null | null | null |
# # Linear Regression for House Prices Competition
# competition link:
# submission by: Juan Pablo Contreras
# ## Libraries and Data Source
# ### Libraries
# Author: Juan Pablo Contreras
# This is the notebook used for my submission for the House Prices - Advanced Regression Techniques competition
import math
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn import linear_model
# ### Data
# read the data
train_data = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test_data = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/test.csv"
)
# ## Data Cleaning
# NaN values in data set: yes
# Categorical Values in data set: yes
# Numerical Values in data set: yes
# The only difference between the training data set and the test data set is that the test data set does not include the SalePrice column
# To clean the data the following steps are taken
# 1. The SalePrice column from the training data set is removed so that the columns of the training df and the test df match
# 2. The test df is appended to the training df so that the cleaning can be done on both the training and the test df
# 3. Categorical values are changed to one hot encoding using the pands get_dummies() function
# 4. NaN values are converted to -1
# 5. The data set is split into training and testing again
# 6. Split training data into training and validation so that predictions on validation can be made to make sure the model does not spit nonesense before predicting on the test data set
# 7. The data is normalized using the training data. The validation and test data are transformed using the training data standarization fitting parameters
# ### 1. Delete SalePrice from Training Dataset
# find number of rows
num_rows_train = len(train_data.index)
print("num rows in training data: " + str(num_rows_train))
print("num rows in test data: " + str(len(test_data.index)))
# store and drop the training predictions
y_train = train_data.iloc[:, -1]
train_data.drop(columns=train_data.columns[-1], axis=1, inplace=True)
# ### 2. Merge Train and Test Data
# merge all data for cleaning
all_data = train_data.append(test_data, ignore_index=True)
print("num rows in all data: " + str(len(all_data.index)))
# delete id column because ID is not a predictor of house price
ids = all_data.drop("Id", axis=1)
# ### 3. and 4.0 Handle Categorical and NaN values
# convert categorical data to numerical (0 or 1)
all_data = pd.get_dummies(data=all_data)
# replace NaN values
all_data = all_data.fillna(-1)
all_data.head()
# ### 5. Split data back into Train, and Test
# split data back into train and test data
X_train = all_data.iloc[0:num_rows_train, :]
print("num rows in X train: " + str(len(X_train.index)))
print("num rows in y train: " + str(len(y_train.index)))
X_test = all_data.iloc[num_rows_train:, :]
print("num rows in X test: " + str(len(X_test.index)))
print("Training data features head: ")
print(X_train.head())
print()
print("Training data predictions head: ")
print(y_train.head())
print()
print("Test data features head:")
print(X_test.head())
# ### 6. Split train data into train and validation
# The new training data will be 80% of the train dataset, and the validation data 20% of the train data set
X_train, X_val, y_train, y_val = train_test_split(
X_train, y_train, test_size=0.2, random_state=42
)
# ### 7. Standarize data
# The training data is standarized. The parameters used to standarize the training data are then used to transform the validation and test data
scaler = StandardScaler()
scaler.fit(X_train)
X_train_std = scaler.transform(X_train)
X_val_transf = scaler.transform(X_val)
X_test_transf = scaler.transform(X_test)
# ## Linear Regression
# The linear regression model from sklearn is used.
# 1. The model is created using the sklearn library
# 2. The model is fit using the training data
# 3. The model is validated using the validation data
# 4. Predictions are made on the test data
# ### 1. Create the model
# linear model
regr = linear_model.LinearRegression()
# ### 2. Fit the Model
# fit model using training data
regr.fit(X_train, y_train)
# ### 3. Validate Model
# predict prices on the test data
pred_val = regr.predict(X_val)
print("Five first predictions: ")
print(pred_val[:5])
print()
# Compare the Predicted values on the validation data set to the actual SalePrices of the validation data set using the root mean squared error
MSE = np.square(np.subtract(y_val, pred_val)).mean()
rsme = math.sqrt(MSE)
print("Root Mean Square Error:\n")
print(rsme)
# ### Predict on Test Data
pred_test = regr.predict(X_test)
print("Five first predictions for test dataset: ")
print(pred_test[:5])
print()
# ## Submission
# submission
sample_submission = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/sample_submission.csv"
)
sample_submission["SalePrice"] = pred_test
sample_submission.to_csv("/kaggle/working/submission.csv", index=False)
sample_submission.head()
| false | 0 | 1,522 | 3 | 1,522 | 1,522 |
||
129582121
|
<jupyter_start><jupyter_text>Breast Cancer Dataset
The data set contains patient records from a 1984-1989 trial conducted by the German Breast Cancer Study Group (GBSG) of 720 patients with node positive breast cancer; it retains the 686 patients with complete data for the prognostic variables.
These data sets are used in the paper by Royston and Altman(2013). The Rotterdam data is used to create a fitted model, and the GBSG data for validation of the model. The paper gives references for the data source.
# Dataset Format
A data set with 686 observations and 11 variables.
| Columns | Description |
| --- | --- |
| pid | patient identifier |
| age | age, years |
| meno | menopausal status (0= premenopausal, 1= postmenopausal) |
| size | tumor size, mm |
| grade | tumor grade |
| nodes | number of positive lymph nodes |
| pgr | progesterone receptors (fmol/l) |
| er | estrogen receptors (fmol/l) |
| hormon | hormonal therapy, 0= no, 1= yes |
| rfstime | recurrence free survival time; days to first of recurrence, death or last follow-up |
| status | 0= alive without recurrence, 1= recurrence or death |
# References
Patrick Royston and Douglas Altman, External validation of a Cox prognostic model: principles and methods. BMC Medical Research Methodology 2013, 13:33
Kaggle dataset identifier: breast-cancer-dataset-used-royston-and-altman
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Import libraries
import seaborn as sns
from sklearn.compose import make_column_transformer
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import precision_score, recall_score, f1_score
import tensorflow as tf
from tensorflow.keras import callbacks
from tensorflow.keras.callbacks import EarlyStopping
# # Data loading and visualizing
data = pd.read_csv(
"/kaggle/input/breast-cancer-dataset-used-royston-and-altman/gbsg.csv"
)
data.head()
data.columns
data.shape
data.drop(["Unnamed: 0"], axis=1, inplace=True)
data.columns
data.info()
data.describe()
data["status"].unique()
data["status"].value_counts()
data["status"].value_counts()
data.hist(figsize=(12, 8))
# # Train test split and normalization
X = data.drop("status", axis=1)
y = data["status"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
X_train.shape
X_train[:5]
# # Logistic regression
reg = LogisticRegression().fit(X_train, y_train)
y_pred = reg.predict(X_test)
y_pred[:5]
accuracy = reg.score(X_test, y_test)
print("Accuracy:", accuracy)
precision = precision_score(y_test, y_pred)
recall = recall_score(y_test, y_pred)
f1 = f1_score(y_test, y_pred)
print("Precision:", precision)
print("Recall:", recall)
print("F1 score:", f1)
# # DNN model
early_stopping = EarlyStopping(
min_delta=0.001,
patience=10,
restore_best_weights=True,
)
tf.random.set_seed(42)
model = tf.keras.Sequential(
[
tf.keras.layers.Dense(256, activation="relu"),
tf.keras.layers.Dropout(0.50),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dropout(0.50),
tf.keras.layers.Dense(56, activation="relu"),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(
loss=tf.keras.losses.BinaryCrossentropy(),
optimizer=tf.keras.optimizers.Adam(),
metrics=["accuracy"],
)
model_history = model.fit(
X_train,
y_train,
epochs=100,
validation_data=(X_test, y_test),
callbacks=[early_stopping],
)
pd.DataFrame(model_history.history).plot()
model.evaluate(X_test, y_test)
y_pred_nn = model.predict(X_test)
y_pred_nn[:5]
y_pred_nn = tf.round(y_pred_nn)
y_pred_nn[:5]
precision = precision_score(y_test, y_pred_nn)
recall = recall_score(y_test, y_pred_nn)
f1 = f1_score(y_test, y_pred_nn)
print("Precision:", precision)
print("Recall:", recall)
print("F1 score:", f1)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/582/129582121.ipynb
|
breast-cancer-dataset-used-royston-and-altman
|
utkarshx27
|
[{"Id": 129582121, "ScriptId": 38492977, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13064818, "CreationDate": "05/15/2023 03:02:31", "VersionNumber": 1.0, "Title": "Logistic regression and DNN", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 136.0, "LinesInsertedFromPrevious": 136.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 10}]
|
[{"Id": 185794575, "KernelVersionId": 129582121, "SourceDatasetVersionId": 5642294}]
|
[{"Id": 5642294, "DatasetId": 3243155, "DatasourceVersionId": 5717613, "CreatorUserId": 13364933, "LicenseName": "CC0: Public Domain", "CreationDate": "05/09/2023 10:42:51", "VersionNumber": 1.0, "Title": "Breast Cancer Dataset", "Slug": "breast-cancer-dataset-used-royston-and-altman", "Subtitle": "Breast Cancer data sets used in Royston and Altman (2013)", "Description": "The data set contains patient records from a 1984-1989 trial conducted by the German Breast Cancer Study Group (GBSG) of 720 patients with node positive breast cancer; it retains the 686 patients with complete data for the prognostic variables.\nThese data sets are used in the paper by Royston and Altman(2013). The Rotterdam data is used to create a fitted model, and the GBSG data for validation of the model. The paper gives references for the data source.\n# Dataset Format\nA data set with 686 observations and 11 variables.\n\n| Columns | Description |\n| --- | --- |\n| pid | patient identifier |\n| age | age, years |\n| meno | menopausal status (0= premenopausal, 1= postmenopausal) |\n| size | tumor size, mm |\n| grade | tumor grade |\n| nodes | number of positive lymph nodes |\n| pgr | progesterone receptors (fmol/l) |\n| er | estrogen receptors (fmol/l) |\n| hormon | hormonal therapy, 0= no, 1= yes |\n| rfstime | recurrence free survival time; days to first of recurrence, death or last follow-up |\n| status | 0= alive without recurrence, 1= recurrence or death |\n\n# References\nPatrick Royston and Douglas Altman, External validation of a Cox prognostic model: principles and methods. BMC Medical Research Methodology 2013, 13:33", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3243155, "CreatorUserId": 13364933, "OwnerUserId": 13364933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5642294.0, "CurrentDatasourceVersionId": 5717613.0, "ForumId": 3308429, "Type": 2, "CreationDate": "05/09/2023 10:42:51", "LastActivityDate": "05/09/2023", "TotalViews": 8106, "TotalDownloads": 1244, "TotalVotes": 41, "TotalKernels": 9}]
|
[{"Id": 13364933, "UserName": "utkarshx27", "DisplayName": "Utkarsh Singh", "RegisterDate": "01/21/2023", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Import libraries
import seaborn as sns
from sklearn.compose import make_column_transformer
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import precision_score, recall_score, f1_score
import tensorflow as tf
from tensorflow.keras import callbacks
from tensorflow.keras.callbacks import EarlyStopping
# # Data loading and visualizing
data = pd.read_csv(
"/kaggle/input/breast-cancer-dataset-used-royston-and-altman/gbsg.csv"
)
data.head()
data.columns
data.shape
data.drop(["Unnamed: 0"], axis=1, inplace=True)
data.columns
data.info()
data.describe()
data["status"].unique()
data["status"].value_counts()
data["status"].value_counts()
data.hist(figsize=(12, 8))
# # Train test split and normalization
X = data.drop("status", axis=1)
y = data["status"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
X_train.shape
X_train[:5]
# # Logistic regression
reg = LogisticRegression().fit(X_train, y_train)
y_pred = reg.predict(X_test)
y_pred[:5]
accuracy = reg.score(X_test, y_test)
print("Accuracy:", accuracy)
precision = precision_score(y_test, y_pred)
recall = recall_score(y_test, y_pred)
f1 = f1_score(y_test, y_pred)
print("Precision:", precision)
print("Recall:", recall)
print("F1 score:", f1)
# # DNN model
early_stopping = EarlyStopping(
min_delta=0.001,
patience=10,
restore_best_weights=True,
)
tf.random.set_seed(42)
model = tf.keras.Sequential(
[
tf.keras.layers.Dense(256, activation="relu"),
tf.keras.layers.Dropout(0.50),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dropout(0.50),
tf.keras.layers.Dense(56, activation="relu"),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(
loss=tf.keras.losses.BinaryCrossentropy(),
optimizer=tf.keras.optimizers.Adam(),
metrics=["accuracy"],
)
model_history = model.fit(
X_train,
y_train,
epochs=100,
validation_data=(X_test, y_test),
callbacks=[early_stopping],
)
pd.DataFrame(model_history.history).plot()
model.evaluate(X_test, y_test)
y_pred_nn = model.predict(X_test)
y_pred_nn[:5]
y_pred_nn = tf.round(y_pred_nn)
y_pred_nn[:5]
precision = precision_score(y_test, y_pred_nn)
recall = recall_score(y_test, y_pred_nn)
f1 = f1_score(y_test, y_pred_nn)
print("Precision:", precision)
print("Recall:", recall)
print("F1 score:", f1)
| false | 1 | 1,071 | 10 | 1,497 | 1,071 |
||
129936171
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### 보스턴 주택 가격 데이터 세트를 Peceptron 기반에서 학습 및 테스트하기 위한 데이터 로드
# * 사이킷런에서 보스턴 주택 가격 데이터 세트를 로드하고 이를 DataFrame으로 생성
import numpy as np
import pandas as pd
from sklearn.datasets import load_boston
from sklearn.datasets import load_boston
boston = load_boston()
bostonDF = pd.DataFrame(boston.data, columns=boston.feature_names)
bostonDF["PRICE"] = boston.target
print(bostonDF.shape)
bostonDF.head()
# ### Weight와 Bias의 Update 값을 계산하는 함수 생성.
# * w1은 RM(방의 계수) 피처의 Weight 값
# * w2는 LSTAT(하위계층 비율) 피처의 Weight 값
# * bias는 Bias
# * N은 입력 데이터 건수
# 
#
# gradient_descent()함수에서 반복적으로 호출되면서 update될 weight/bias 값을 계산하는 함수.
# rm은 RM(방 개수), lstat(하위계층 비율), target은 PRICE임. 전체 array가 다 입력됨.
# 반환 값은 weight와 bias가 update되어야 할 값과 Mean Squared Error 값을 loss로 반환.
def get_update_weights_value(bias, w1, w2, rm, lstat, target, learning_rate=0.01):
# 데이터 건수
N = len(target)
# 예측 값.
predicted = w1 * rm + w2 * lstat + bias
# 실제값과 예측값의 차이
diff = target - predicted
# bias 를 array 기반으로 구하기 위해서 설정.
bias_factors = np.ones((N,))
# weight와 bias를 얼마나 update할 것인지를 계산.
w1_update = -(2 / N) * learning_rate * (np.dot(rm.T, diff))
w2_update = -(2 / N) * learning_rate * (np.dot(lstat.T, diff))
bias_update = -(2 / N) * learning_rate * (np.dot(bias_factors.T, diff))
# Mean Squared Error값을 계산.
mse_loss = np.mean(np.square(diff))
# weight와 bias가 update되어야 할 값과 Mean Squared Error 값을 반환.
return bias_update, w1_update, w2_update, mse_loss
# ### Gradient Descent 를 적용하는 함수 생성
# * iter_epochs 수만큼 반복적으로 get_update_weights_value()를 호출하여 update될 weight/bias값을 구한 뒤 Weight/Bias를 Update적용.
# RM, LSTAT feature array와 PRICE target array를 입력 받아서 iter_epochs수만큼 반복적으로 Weight와 Bias를 update적용.
def gradient_descent(features, target, iter_epochs=1000, verbose=True):
# w1, w2는 numpy array 연산을 위해 1차원 array로 변환하되 초기 값은 0으로 설정
# bias도 1차원 array로 변환하되 초기 값은 1로 설정.
w1 = np.zeros((1,))
w2 = np.zeros((1,))
bias = np.zeros((1,))
print("최초 w1, w2, bias:", w1, w2, bias)
# learning_rate와 RM, LSTAT 피처 지정. 호출 시 numpy array형태로 RM과 LSTAT으로 된 2차원 feature가 입력됨.
learning_rate = 0.01
rm = features[:, 0]
lstat = features[:, 1]
# iter_epochs 수만큼 반복하면서 weight와 bias update 수행.
for i in range(iter_epochs):
# weight/bias update 값 계산
bias_update, w1_update, w2_update, loss = get_update_weights_value(
bias, w1, w2, rm, lstat, target, learning_rate
)
# weight/bias의 update 적용.
w1 = w1 - w1_update
w2 = w2 - w2_update
bias = bias - bias_update
# verbos : 보통 프로그램이 복잡한 출력이나 상태 메시지를 자세히 보고 싶을 때 사용됩니다.
if verbose:
print("Epoch:", i + 1, "/", iter_epochs)
print("w1:", w1, "w2:", w2, "bias:", bias, "loss:", loss)
return w1, w2, bias
# ### Gradient Descent 적용
# * 신경망은 데이터를 정규화/표준화 작업을 미리 선행해 주어야 함.
# * 이를 위해 사이킷런의 MinMaxScaler를 이용하여 개별 feature값은 0~1사이 값으로 변환후 학습 적용.
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
scaled_features = scaler.fit_transform(bostonDF[["RM", "LSTAT"]])
w1, w2, bias = gradient_descent(
scaled_features, bostonDF["PRICE"].values, iter_epochs=5000, verbose=True
)
print("##### 최종 w1, w2, bias #######")
print(w1, w2, bias)
# ### 계산된 Weight와 Bias를 이용하여 Price 예측
# * 예측 feature 역시 0~1사이의 scaled값을 이용하고 Weight와 bias를 적용하여 예측값 계산.
predicted = scaled_features[:, 0] * w1 + scaled_features[:, 1] * w2 + bias
bostonDF["PREDICTED_PRICE"] = predicted
bostonDF.head(10)
# ### Keras를 이용하여 보스턴 주택가격 모델 학습 및 예측
# * Dense Layer를 이용하여 퍼셉트론 구현. units는 1로 설정.
from tensorflow.keras.layers import Dense
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import Adam
model = Sequential(
[
# 단 하나의 units 설정. input_shape는 2차원, 회귀이므로 activation은 설정하지 않음.
# weight와 bias 초기화는 kernel_inbitializer와 bias_initializer를 이용.
Dense(
1,
input_shape=(2,),
activation=None,
kernel_initializer="zeros",
bias_initializer="ones",
)
]
)
# Adam optimizer를 이용하고 Loss 함수는 Mean Squared Error, 성능 측정 역시 MSE를 이용하여 학습 수행.
model.compile(optimizer=Adam(learning_rate=0.01), loss="mse", metrics=["mse"])
model.fit(scaled_features, bostonDF["PRICE"].values, epochs=1000)
# ### Keras로 학습된 모델을 이용하여 주택 가격 예측 수행.
predicted = model.predict(scaled_features)
bostonDF["KERAS_PREDICTED_PRICE"] = predicted
bostonDF.head(10)
# ### Stochastic Gradient Descent와 Mini Batch Gradient Descent 구현
# * SGD 는 전체 데이터에서 한건만 임의로 선택하여 Gradient Descent 로 Weight/Bias Update 계산한 뒤 Weight/Bias 적용
# * Mini Batch GD는 전체 데이터에서 Batch 건수만큼 데이터를 선택하여 Gradient Descent로 Weight/Bias Update 계산한 뒤 Weight/Bias 적용
import numpy as np
import pandas as pd
from sklearn.datasets import load_boston
boston = load_boston()
bostonDF = pd.DataFrame(boston.data, columns=boston.feature_names)
bostonDF["PRICE"] = boston.target
print(bostonDF.shape)
bostonDF.head()
# ### SGD 기반으로 Weight/Bias update 값 구하기
def get_update_weights_value_sgd(
bias, w1, w2, rm_sgd, lstat_sgd, target_sgd, learning_rate=0.01
):
# 데이터 건수
N = target_sgd.shape[0] # 1개만 가져오는것이다. N = 1
# 예측 값.
predicted_sgd = w1 * rm_sgd + w2 * lstat_sgd + bias
# 실제값과 예측값의 차이
diff_sgd = target_sgd - predicted_sgd
# bias 를 array 기반으로 구하기 위해서 설정.
bias_factors = np.ones((N,))
# weight와 bias를 얼마나 update할 것인지를 계산.
w1_update = -(2 / N) * learning_rate * (np.dot(rm_sgd.T, diff_sgd))
w2_update = -(2 / N) * learning_rate * (np.dot(lstat_sgd.T, diff_sgd))
bias_update = -(2 / N) * learning_rate * (np.dot(bias_factors.T, diff_sgd))
# Mean Squared Error값을 계산.
# mse_loss = np.mean(np.square(diff))
# weight와 bias가 update되어야 할 값 반환
return bias_update, w1_update, w2_update
# ### SGD 수행하기 : rm_sgd, lstat_sgd는 1건의 데이터만 가져온다.
#
print(bostonDF["PRICE"].values.shape)
print(np.random.choice(bostonDF["PRICE"].values.shape[0], 1))
print(np.random.choice(506, 1))
# np.random.choice함수: np.random.choice(전체데이터개수,전체데이터개수에서 원하는 개수)
print(bostonDF["PRICE"].values.shape[0])
print(np.random.choice(bostonDF["PRICE"].values.shape[0], 1))
print(np.random.choice(506, 2)) # 0~505에서 2개의 숫자를 랜덤하게 가져온다.
# RM, LSTAT feature array와 PRICE target array를 입력 받아서 iter_epochs수만큼 반복적으로 Weight와 Bias를 update적용.
def st_gradient_descent(features, target, iter_epochs=1000, verbose=True):
# w1, w2는 numpy array 연산을 위해 1차원 array로 변환하되 초기 값은 0으로 설정
# bias도 1차원 array로 변환하되 초기 값은 1로 설정.
np.random.seed = 2021
w1 = np.zeros((1,))
w2 = np.zeros((1,))
bias = np.zeros((1,))
print("최초 w1, w2, bias:", w1, w2, bias)
# learning_rate와 RM, LSTAT 피처 지정. 호출 시 numpy array형태로 RM과 LSTAT으로 된 2차원 feature가 입력됨.
learning_rate = 0.01
rm = features[:, 0]
lstat = features[:, 1]
# iter_epochs 수만큼 반복하면서 weight와 bias update 수행.
for i in range(iter_epochs):
# iteration 시마다 stochastic gradient descent 를 수행할 데이터를 한개만 추출. 추출할 데이터의 인덱스를 random.choice() 로 선택.
stochastic_index = np.random.choice(
target.shape[0], 1
) # target.shape[0]은 506개이다. 506개에서 1개를 랜덤하게 선택하게끔 가져온다.
rm_sgd = rm[stochastic_index]
lstat_sgd = lstat[stochastic_index]
target_sgd = target[stochastic_index]
# SGD 기반으로 Weight/Bias의 Update를 구함.
bias_update, w1_update, w2_update = get_update_weights_value_sgd(
bias, w1, w2, rm_sgd, lstat_sgd, target_sgd, learning_rate
)
# SGD로 구한 weight/bias의 update 적용.
w1 = w1 - w1_update
w2 = w2 - w2_update
bias = bias - bias_update
if verbose:
print("Epoch:", i + 1, "/", iter_epochs)
# Loss는 전체 학습 데이터 기반으로 구해야 함.
predicted = w1 * rm + w2 * lstat + bias
diff = target - predicted
mse_loss = np.mean(np.square(diff))
print("w1:", w1, "w2:", w2, "bias:", bias, "loss:", mse_loss)
return w1, w2, bias
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
scaled_features = scaler.fit_transform(bostonDF[["RM", "LSTAT"]])
w1, w2, bias = st_gradient_descent(
scaled_features, bostonDF["PRICE"].values, iter_epochs=5000, verbose=True
)
print("##### 최종 w1, w2, bias #######")
print(w1, w2, bias)
predicted = scaled_features[:, 0] * w1 + scaled_features[:, 1] * w2 + bias
bostonDF["PREDICTED_PRICE_SGD"] = predicted
bostonDF.head(10)
# ### iteration시마다 일정한 batch 크기만큼의 데이터를 random하게 가져와서 GD를 수행하는 Mini-Batch GD 수행
def get_update_weights_value_batch(
bias, w1, w2, rm_batch, lstat_batch, target_batch, learning_rate=0.01
):
# 데이터 건수
N = target_batch.shape[0] # 만약 batch_size를 30으로 정하면 N = 30이다.
# 예측 값.
predicted_batch = w1 * rm_batch + w2 * lstat_batch + bias
# 실제값과 예측값의 차이
diff_batch = target_batch - predicted_batch
# bias 를 array 기반으로 구하기 위해서 설정.
bias_factors = np.ones((N,))
# weight와 bias를 얼마나 update할 것인지를 계산.
w1_update = -(2 / N) * learning_rate * (np.dot(rm_batch.T, diff_batch))
w2_update = -(2 / N) * learning_rate * (np.dot(lstat_batch.T, diff_batch))
bias_update = -(2 / N) * learning_rate * (np.dot(bias_factors.T, diff_batch))
# Mean Squared Error값을 계산.
# mse_loss = np.mean(np.square(diff))
# weight와 bias가 update되어야 할 값 반환
return bias_update, w1_update, w2_update
batch_indexes = np.random.choice(506, 30)
print(batch_indexes)
bostonDF["RM"].values[
batch_indexes
] # batch_indexes에는 numpy배열이 들어갈수있다. 인덱싱과 슬라이싱을 지원해서 매우 유용하
# batch_gradient_descent()는 인자로 batch_size(배치 크기)를 입력 받음.(중)
def batch_random_gradient_descent(
features, target, iter_epochs=1000, batch_size=30, verbose=True
):
# w1, w2는 numpy array 연산을 위해 1차원 array로 변환하되 초기 값은 0으로 설정
# bias도 1차원 array로 변환하되 초기 값은 1로 설정.
np.random.seed = 2021
w1 = np.zeros((1,))
w2 = np.zeros((1,))
bias = np.zeros((1,))
print("최초 w1, w2, bias:", w1, w2, bias)
# learning_rate와 RM, LSTAT 피처 지정. 호출 시 numpy array형태로 RM과 LSTAT으로 된 2차원 feature가 입력됨.
learning_rate = 0.01
rm = features[:, 0]
lstat = features[:, 1]
# iter_epochs 수만큼 반복하면서 weight와 bias update 수행.
for i in range(iter_epochs):
# batch_size 갯수만큼 데이터를 임의로 선택.
batch_indexes = np.random.choice(
target.shape[0], batch_size
) # 0~505개에서 30개의 숫자를 가져온다.
rm_batch = rm[batch_indexes]
lstat_batch = lstat[batch_indexes]
target_batch = target[batch_indexes]
# Batch GD 기반으로 Weight/Bias의 Update를 구함.
bias_update, w1_update, w2_update = get_update_weights_value_batch(
bias, w1, w2, rm_batch, lstat_batch, target_batch, learning_rate
)
# Batch GD로 구한 weight/bias의 update 적용.
w1 = w1 - w1_update
w2 = w2 - w2_update
bias = bias - bias_update
if verbose:
print("Epoch:", i + 1, "/", iter_epochs)
# Loss는 전체 학습 데이터 기반으로 구해야 함.
predicted = w1 * rm + w2 * lstat + bias
diff = target - predicted
mse_loss = np.mean(np.square(diff))
print("w1:", w1, "w2:", w2, "bias:", bias, "loss:", mse_loss)
return w1, w2, bias
w1, w2, bias = batch_random_gradient_descent(
scaled_features,
bostonDF["PRICE"].values,
iter_epochs=5000,
batch_size=30,
verbose=True,
)
print("##### 최종 w1, w2, bias #######")
print(w1, w2, bias)
predicted = scaled_features[:, 0] * w1 + scaled_features[:, 1] * w2 + bias
bostonDF["PREDICTED_PRICE_BATCH_RANDOM"] = predicted
bostonDF.head(10)
# ### iteration 시에 순차적으로 일정한 batch 크기만큼의 데이터를 전체 학습데이터에 걸쳐서 가져오는 Mini-Batch GD 수행
for batch_step in range(0, 506, 30):
print(batch_step)
bostonDF["PRICE"].values[480:510]
for batch_step in range(0, 506, 30):
print(batch_step)
# bostonDF['PRICE'].values[507] 이거는 인덱스값을 넘어가서 오류가 난다.
bostonDF["PRICE"].values[
480:510
] # 이거는 480~505까지 해당하는 인덱스의 값을 찾고 그 뒤는 없는걸로친다 따라서 오류가 나지않는다.
# batch_gradient_descent()는 인자로 batch_size(배치 크기)를 입력 받음.
def batch_gradient_descent(
features, target, iter_epochs=1000, batch_size=30, verbose=True
):
# w1, w2는 numpy array 연산을 위해 1차원 array로 변환하되 초기 값은 0으로 설정
# bias도 1차원 array로 변환하되 초기 값은 1로 설정.
np.random.seed = 2021
w1 = np.zeros((1,))
w2 = np.zeros((1,))
bias = np.zeros((1,))
print("최초 w1, w2, bias:", w1, w2, bias)
# learning_rate와 RM, LSTAT 피처 지정. 호출 시 numpy array형태로 RM과 LSTAT으로 된 2차원 feature가 입력됨.
learning_rate = 0.01
rm = features[:, 0]
lstat = features[:, 1]
# iter_epochs 수만큼 반복하면서 weight와 bias update 수행.
for i in range(iter_epochs):
# batch_size 만큼 데이터를 가져와서 weight/bias update를 수행하는 로직을 전체 데이터 건수만큼 반복
for batch_step in range(
0, target.shape[0], batch_size
): # range(start,end,step)이다.
# batch_size만큼 순차적인 데이터를 가져옴.
rm_batch = rm[batch_step : batch_step + batch_size]
lstat_batch = lstat[batch_step : batch_step + batch_size]
target_batch = target[batch_step : batch_step + batch_size]
bias_update, w1_update, w2_update = get_update_weights_value_batch(
bias, w1, w2, rm_batch, lstat_batch, target_batch, learning_rate
)
# Batch GD로 구한 weight/bias의 update 적용.
w1 = w1 - w1_update
w2 = w2 - w2_update
bias = bias - bias_update
if verbose:
print("Epoch:", i + 1, "/", iter_epochs, "batch step:", batch_step)
# Loss는 전체 학습 데이터 기반으로 구해야 함.
predicted = w1 * rm + w2 * lstat + bias
diff = target - predicted
mse_loss = np.mean(np.square(diff))
print("w1:", w1, "w2:", w2, "bias:", bias, "loss:", mse_loss)
return w1, w2, bias
w1, w2, bias = batch_gradient_descent(
scaled_features,
bostonDF["PRICE"].values,
iter_epochs=5000,
batch_size=30,
verbose=True,
)
print("##### 최종 w1, w2, bias #######")
print(w1, w2, bias)
predicted = scaled_features[:, 0] * w1 + scaled_features[:, 1] * w2 + bias
bostonDF["PREDICTED_PRICE_BATCH"] = predicted
bostonDF.head(10)
# ### Mini BATCH GD를 Keras로 수행
# * Keras는 기본적으로 Mini Batch GD를 수행
from tensorflow.keras.layers import Dense
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import Adam
model = Sequential(
[
# 단 하나의 units 설정. input_shape는 2차원, 회귀이므로 activation은 설정하지 않음.
# weight와 bias 초기화는 kernel_inbitializer와 bias_initializer를 이용.
Dense(
1,
input_shape=(2,),
activation=None,
kernel_initializer="zeros",
bias_initializer="ones",
)
]
)
# Adam optimizer를 이용하고 Loss 함수는 Mean Squared Error, 성능 측정 역시 MSE를 이용하여 학습 수행.
model.compile(optimizer=Adam(learning_rate=0.01), loss="mse", metrics=["mse"])
# Keras는 반드시 Batch GD를 적용함. batch_size가 None이면 32를 할당.
model.fit(scaled_features, bostonDF["PRICE"].values, batch_size=30, epochs=1000)
predicted = model.predict(scaled_features)
bostonDF["KERAS_PREDICTED_PRICE_BATCH"] = predicted
bostonDF.head(10)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/936/129936171.ipynb
| null | null |
[{"Id": 129936171, "ScriptId": 38602065, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15111121, "CreationDate": "05/17/2023 14:38:49", "VersionNumber": 1.0, "Title": "Gradient_Descent_Practice", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 408.0, "LinesInsertedFromPrevious": 408.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### 보스턴 주택 가격 데이터 세트를 Peceptron 기반에서 학습 및 테스트하기 위한 데이터 로드
# * 사이킷런에서 보스턴 주택 가격 데이터 세트를 로드하고 이를 DataFrame으로 생성
import numpy as np
import pandas as pd
from sklearn.datasets import load_boston
from sklearn.datasets import load_boston
boston = load_boston()
bostonDF = pd.DataFrame(boston.data, columns=boston.feature_names)
bostonDF["PRICE"] = boston.target
print(bostonDF.shape)
bostonDF.head()
# ### Weight와 Bias의 Update 값을 계산하는 함수 생성.
# * w1은 RM(방의 계수) 피처의 Weight 값
# * w2는 LSTAT(하위계층 비율) 피처의 Weight 값
# * bias는 Bias
# * N은 입력 데이터 건수
# 
#
# gradient_descent()함수에서 반복적으로 호출되면서 update될 weight/bias 값을 계산하는 함수.
# rm은 RM(방 개수), lstat(하위계층 비율), target은 PRICE임. 전체 array가 다 입력됨.
# 반환 값은 weight와 bias가 update되어야 할 값과 Mean Squared Error 값을 loss로 반환.
def get_update_weights_value(bias, w1, w2, rm, lstat, target, learning_rate=0.01):
# 데이터 건수
N = len(target)
# 예측 값.
predicted = w1 * rm + w2 * lstat + bias
# 실제값과 예측값의 차이
diff = target - predicted
# bias 를 array 기반으로 구하기 위해서 설정.
bias_factors = np.ones((N,))
# weight와 bias를 얼마나 update할 것인지를 계산.
w1_update = -(2 / N) * learning_rate * (np.dot(rm.T, diff))
w2_update = -(2 / N) * learning_rate * (np.dot(lstat.T, diff))
bias_update = -(2 / N) * learning_rate * (np.dot(bias_factors.T, diff))
# Mean Squared Error값을 계산.
mse_loss = np.mean(np.square(diff))
# weight와 bias가 update되어야 할 값과 Mean Squared Error 값을 반환.
return bias_update, w1_update, w2_update, mse_loss
# ### Gradient Descent 를 적용하는 함수 생성
# * iter_epochs 수만큼 반복적으로 get_update_weights_value()를 호출하여 update될 weight/bias값을 구한 뒤 Weight/Bias를 Update적용.
# RM, LSTAT feature array와 PRICE target array를 입력 받아서 iter_epochs수만큼 반복적으로 Weight와 Bias를 update적용.
def gradient_descent(features, target, iter_epochs=1000, verbose=True):
# w1, w2는 numpy array 연산을 위해 1차원 array로 변환하되 초기 값은 0으로 설정
# bias도 1차원 array로 변환하되 초기 값은 1로 설정.
w1 = np.zeros((1,))
w2 = np.zeros((1,))
bias = np.zeros((1,))
print("최초 w1, w2, bias:", w1, w2, bias)
# learning_rate와 RM, LSTAT 피처 지정. 호출 시 numpy array형태로 RM과 LSTAT으로 된 2차원 feature가 입력됨.
learning_rate = 0.01
rm = features[:, 0]
lstat = features[:, 1]
# iter_epochs 수만큼 반복하면서 weight와 bias update 수행.
for i in range(iter_epochs):
# weight/bias update 값 계산
bias_update, w1_update, w2_update, loss = get_update_weights_value(
bias, w1, w2, rm, lstat, target, learning_rate
)
# weight/bias의 update 적용.
w1 = w1 - w1_update
w2 = w2 - w2_update
bias = bias - bias_update
# verbos : 보통 프로그램이 복잡한 출력이나 상태 메시지를 자세히 보고 싶을 때 사용됩니다.
if verbose:
print("Epoch:", i + 1, "/", iter_epochs)
print("w1:", w1, "w2:", w2, "bias:", bias, "loss:", loss)
return w1, w2, bias
# ### Gradient Descent 적용
# * 신경망은 데이터를 정규화/표준화 작업을 미리 선행해 주어야 함.
# * 이를 위해 사이킷런의 MinMaxScaler를 이용하여 개별 feature값은 0~1사이 값으로 변환후 학습 적용.
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
scaled_features = scaler.fit_transform(bostonDF[["RM", "LSTAT"]])
w1, w2, bias = gradient_descent(
scaled_features, bostonDF["PRICE"].values, iter_epochs=5000, verbose=True
)
print("##### 최종 w1, w2, bias #######")
print(w1, w2, bias)
# ### 계산된 Weight와 Bias를 이용하여 Price 예측
# * 예측 feature 역시 0~1사이의 scaled값을 이용하고 Weight와 bias를 적용하여 예측값 계산.
predicted = scaled_features[:, 0] * w1 + scaled_features[:, 1] * w2 + bias
bostonDF["PREDICTED_PRICE"] = predicted
bostonDF.head(10)
# ### Keras를 이용하여 보스턴 주택가격 모델 학습 및 예측
# * Dense Layer를 이용하여 퍼셉트론 구현. units는 1로 설정.
from tensorflow.keras.layers import Dense
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import Adam
model = Sequential(
[
# 단 하나의 units 설정. input_shape는 2차원, 회귀이므로 activation은 설정하지 않음.
# weight와 bias 초기화는 kernel_inbitializer와 bias_initializer를 이용.
Dense(
1,
input_shape=(2,),
activation=None,
kernel_initializer="zeros",
bias_initializer="ones",
)
]
)
# Adam optimizer를 이용하고 Loss 함수는 Mean Squared Error, 성능 측정 역시 MSE를 이용하여 학습 수행.
model.compile(optimizer=Adam(learning_rate=0.01), loss="mse", metrics=["mse"])
model.fit(scaled_features, bostonDF["PRICE"].values, epochs=1000)
# ### Keras로 학습된 모델을 이용하여 주택 가격 예측 수행.
predicted = model.predict(scaled_features)
bostonDF["KERAS_PREDICTED_PRICE"] = predicted
bostonDF.head(10)
# ### Stochastic Gradient Descent와 Mini Batch Gradient Descent 구현
# * SGD 는 전체 데이터에서 한건만 임의로 선택하여 Gradient Descent 로 Weight/Bias Update 계산한 뒤 Weight/Bias 적용
# * Mini Batch GD는 전체 데이터에서 Batch 건수만큼 데이터를 선택하여 Gradient Descent로 Weight/Bias Update 계산한 뒤 Weight/Bias 적용
import numpy as np
import pandas as pd
from sklearn.datasets import load_boston
boston = load_boston()
bostonDF = pd.DataFrame(boston.data, columns=boston.feature_names)
bostonDF["PRICE"] = boston.target
print(bostonDF.shape)
bostonDF.head()
# ### SGD 기반으로 Weight/Bias update 값 구하기
def get_update_weights_value_sgd(
bias, w1, w2, rm_sgd, lstat_sgd, target_sgd, learning_rate=0.01
):
# 데이터 건수
N = target_sgd.shape[0] # 1개만 가져오는것이다. N = 1
# 예측 값.
predicted_sgd = w1 * rm_sgd + w2 * lstat_sgd + bias
# 실제값과 예측값의 차이
diff_sgd = target_sgd - predicted_sgd
# bias 를 array 기반으로 구하기 위해서 설정.
bias_factors = np.ones((N,))
# weight와 bias를 얼마나 update할 것인지를 계산.
w1_update = -(2 / N) * learning_rate * (np.dot(rm_sgd.T, diff_sgd))
w2_update = -(2 / N) * learning_rate * (np.dot(lstat_sgd.T, diff_sgd))
bias_update = -(2 / N) * learning_rate * (np.dot(bias_factors.T, diff_sgd))
# Mean Squared Error값을 계산.
# mse_loss = np.mean(np.square(diff))
# weight와 bias가 update되어야 할 값 반환
return bias_update, w1_update, w2_update
# ### SGD 수행하기 : rm_sgd, lstat_sgd는 1건의 데이터만 가져온다.
#
print(bostonDF["PRICE"].values.shape)
print(np.random.choice(bostonDF["PRICE"].values.shape[0], 1))
print(np.random.choice(506, 1))
# np.random.choice함수: np.random.choice(전체데이터개수,전체데이터개수에서 원하는 개수)
print(bostonDF["PRICE"].values.shape[0])
print(np.random.choice(bostonDF["PRICE"].values.shape[0], 1))
print(np.random.choice(506, 2)) # 0~505에서 2개의 숫자를 랜덤하게 가져온다.
# RM, LSTAT feature array와 PRICE target array를 입력 받아서 iter_epochs수만큼 반복적으로 Weight와 Bias를 update적용.
def st_gradient_descent(features, target, iter_epochs=1000, verbose=True):
# w1, w2는 numpy array 연산을 위해 1차원 array로 변환하되 초기 값은 0으로 설정
# bias도 1차원 array로 변환하되 초기 값은 1로 설정.
np.random.seed = 2021
w1 = np.zeros((1,))
w2 = np.zeros((1,))
bias = np.zeros((1,))
print("최초 w1, w2, bias:", w1, w2, bias)
# learning_rate와 RM, LSTAT 피처 지정. 호출 시 numpy array형태로 RM과 LSTAT으로 된 2차원 feature가 입력됨.
learning_rate = 0.01
rm = features[:, 0]
lstat = features[:, 1]
# iter_epochs 수만큼 반복하면서 weight와 bias update 수행.
for i in range(iter_epochs):
# iteration 시마다 stochastic gradient descent 를 수행할 데이터를 한개만 추출. 추출할 데이터의 인덱스를 random.choice() 로 선택.
stochastic_index = np.random.choice(
target.shape[0], 1
) # target.shape[0]은 506개이다. 506개에서 1개를 랜덤하게 선택하게끔 가져온다.
rm_sgd = rm[stochastic_index]
lstat_sgd = lstat[stochastic_index]
target_sgd = target[stochastic_index]
# SGD 기반으로 Weight/Bias의 Update를 구함.
bias_update, w1_update, w2_update = get_update_weights_value_sgd(
bias, w1, w2, rm_sgd, lstat_sgd, target_sgd, learning_rate
)
# SGD로 구한 weight/bias의 update 적용.
w1 = w1 - w1_update
w2 = w2 - w2_update
bias = bias - bias_update
if verbose:
print("Epoch:", i + 1, "/", iter_epochs)
# Loss는 전체 학습 데이터 기반으로 구해야 함.
predicted = w1 * rm + w2 * lstat + bias
diff = target - predicted
mse_loss = np.mean(np.square(diff))
print("w1:", w1, "w2:", w2, "bias:", bias, "loss:", mse_loss)
return w1, w2, bias
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
scaled_features = scaler.fit_transform(bostonDF[["RM", "LSTAT"]])
w1, w2, bias = st_gradient_descent(
scaled_features, bostonDF["PRICE"].values, iter_epochs=5000, verbose=True
)
print("##### 최종 w1, w2, bias #######")
print(w1, w2, bias)
predicted = scaled_features[:, 0] * w1 + scaled_features[:, 1] * w2 + bias
bostonDF["PREDICTED_PRICE_SGD"] = predicted
bostonDF.head(10)
# ### iteration시마다 일정한 batch 크기만큼의 데이터를 random하게 가져와서 GD를 수행하는 Mini-Batch GD 수행
def get_update_weights_value_batch(
bias, w1, w2, rm_batch, lstat_batch, target_batch, learning_rate=0.01
):
# 데이터 건수
N = target_batch.shape[0] # 만약 batch_size를 30으로 정하면 N = 30이다.
# 예측 값.
predicted_batch = w1 * rm_batch + w2 * lstat_batch + bias
# 실제값과 예측값의 차이
diff_batch = target_batch - predicted_batch
# bias 를 array 기반으로 구하기 위해서 설정.
bias_factors = np.ones((N,))
# weight와 bias를 얼마나 update할 것인지를 계산.
w1_update = -(2 / N) * learning_rate * (np.dot(rm_batch.T, diff_batch))
w2_update = -(2 / N) * learning_rate * (np.dot(lstat_batch.T, diff_batch))
bias_update = -(2 / N) * learning_rate * (np.dot(bias_factors.T, diff_batch))
# Mean Squared Error값을 계산.
# mse_loss = np.mean(np.square(diff))
# weight와 bias가 update되어야 할 값 반환
return bias_update, w1_update, w2_update
batch_indexes = np.random.choice(506, 30)
print(batch_indexes)
bostonDF["RM"].values[
batch_indexes
] # batch_indexes에는 numpy배열이 들어갈수있다. 인덱싱과 슬라이싱을 지원해서 매우 유용하
# batch_gradient_descent()는 인자로 batch_size(배치 크기)를 입력 받음.(중)
def batch_random_gradient_descent(
features, target, iter_epochs=1000, batch_size=30, verbose=True
):
# w1, w2는 numpy array 연산을 위해 1차원 array로 변환하되 초기 값은 0으로 설정
# bias도 1차원 array로 변환하되 초기 값은 1로 설정.
np.random.seed = 2021
w1 = np.zeros((1,))
w2 = np.zeros((1,))
bias = np.zeros((1,))
print("최초 w1, w2, bias:", w1, w2, bias)
# learning_rate와 RM, LSTAT 피처 지정. 호출 시 numpy array형태로 RM과 LSTAT으로 된 2차원 feature가 입력됨.
learning_rate = 0.01
rm = features[:, 0]
lstat = features[:, 1]
# iter_epochs 수만큼 반복하면서 weight와 bias update 수행.
for i in range(iter_epochs):
# batch_size 갯수만큼 데이터를 임의로 선택.
batch_indexes = np.random.choice(
target.shape[0], batch_size
) # 0~505개에서 30개의 숫자를 가져온다.
rm_batch = rm[batch_indexes]
lstat_batch = lstat[batch_indexes]
target_batch = target[batch_indexes]
# Batch GD 기반으로 Weight/Bias의 Update를 구함.
bias_update, w1_update, w2_update = get_update_weights_value_batch(
bias, w1, w2, rm_batch, lstat_batch, target_batch, learning_rate
)
# Batch GD로 구한 weight/bias의 update 적용.
w1 = w1 - w1_update
w2 = w2 - w2_update
bias = bias - bias_update
if verbose:
print("Epoch:", i + 1, "/", iter_epochs)
# Loss는 전체 학습 데이터 기반으로 구해야 함.
predicted = w1 * rm + w2 * lstat + bias
diff = target - predicted
mse_loss = np.mean(np.square(diff))
print("w1:", w1, "w2:", w2, "bias:", bias, "loss:", mse_loss)
return w1, w2, bias
w1, w2, bias = batch_random_gradient_descent(
scaled_features,
bostonDF["PRICE"].values,
iter_epochs=5000,
batch_size=30,
verbose=True,
)
print("##### 최종 w1, w2, bias #######")
print(w1, w2, bias)
predicted = scaled_features[:, 0] * w1 + scaled_features[:, 1] * w2 + bias
bostonDF["PREDICTED_PRICE_BATCH_RANDOM"] = predicted
bostonDF.head(10)
# ### iteration 시에 순차적으로 일정한 batch 크기만큼의 데이터를 전체 학습데이터에 걸쳐서 가져오는 Mini-Batch GD 수행
for batch_step in range(0, 506, 30):
print(batch_step)
bostonDF["PRICE"].values[480:510]
for batch_step in range(0, 506, 30):
print(batch_step)
# bostonDF['PRICE'].values[507] 이거는 인덱스값을 넘어가서 오류가 난다.
bostonDF["PRICE"].values[
480:510
] # 이거는 480~505까지 해당하는 인덱스의 값을 찾고 그 뒤는 없는걸로친다 따라서 오류가 나지않는다.
# batch_gradient_descent()는 인자로 batch_size(배치 크기)를 입력 받음.
def batch_gradient_descent(
features, target, iter_epochs=1000, batch_size=30, verbose=True
):
# w1, w2는 numpy array 연산을 위해 1차원 array로 변환하되 초기 값은 0으로 설정
# bias도 1차원 array로 변환하되 초기 값은 1로 설정.
np.random.seed = 2021
w1 = np.zeros((1,))
w2 = np.zeros((1,))
bias = np.zeros((1,))
print("최초 w1, w2, bias:", w1, w2, bias)
# learning_rate와 RM, LSTAT 피처 지정. 호출 시 numpy array형태로 RM과 LSTAT으로 된 2차원 feature가 입력됨.
learning_rate = 0.01
rm = features[:, 0]
lstat = features[:, 1]
# iter_epochs 수만큼 반복하면서 weight와 bias update 수행.
for i in range(iter_epochs):
# batch_size 만큼 데이터를 가져와서 weight/bias update를 수행하는 로직을 전체 데이터 건수만큼 반복
for batch_step in range(
0, target.shape[0], batch_size
): # range(start,end,step)이다.
# batch_size만큼 순차적인 데이터를 가져옴.
rm_batch = rm[batch_step : batch_step + batch_size]
lstat_batch = lstat[batch_step : batch_step + batch_size]
target_batch = target[batch_step : batch_step + batch_size]
bias_update, w1_update, w2_update = get_update_weights_value_batch(
bias, w1, w2, rm_batch, lstat_batch, target_batch, learning_rate
)
# Batch GD로 구한 weight/bias의 update 적용.
w1 = w1 - w1_update
w2 = w2 - w2_update
bias = bias - bias_update
if verbose:
print("Epoch:", i + 1, "/", iter_epochs, "batch step:", batch_step)
# Loss는 전체 학습 데이터 기반으로 구해야 함.
predicted = w1 * rm + w2 * lstat + bias
diff = target - predicted
mse_loss = np.mean(np.square(diff))
print("w1:", w1, "w2:", w2, "bias:", bias, "loss:", mse_loss)
return w1, w2, bias
w1, w2, bias = batch_gradient_descent(
scaled_features,
bostonDF["PRICE"].values,
iter_epochs=5000,
batch_size=30,
verbose=True,
)
print("##### 최종 w1, w2, bias #######")
print(w1, w2, bias)
predicted = scaled_features[:, 0] * w1 + scaled_features[:, 1] * w2 + bias
bostonDF["PREDICTED_PRICE_BATCH"] = predicted
bostonDF.head(10)
# ### Mini BATCH GD를 Keras로 수행
# * Keras는 기본적으로 Mini Batch GD를 수행
from tensorflow.keras.layers import Dense
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import Adam
model = Sequential(
[
# 단 하나의 units 설정. input_shape는 2차원, 회귀이므로 activation은 설정하지 않음.
# weight와 bias 초기화는 kernel_inbitializer와 bias_initializer를 이용.
Dense(
1,
input_shape=(2,),
activation=None,
kernel_initializer="zeros",
bias_initializer="ones",
)
]
)
# Adam optimizer를 이용하고 Loss 함수는 Mean Squared Error, 성능 측정 역시 MSE를 이용하여 학습 수행.
model.compile(optimizer=Adam(learning_rate=0.01), loss="mse", metrics=["mse"])
# Keras는 반드시 Batch GD를 적용함. batch_size가 None이면 32를 할당.
model.fit(scaled_features, bostonDF["PRICE"].values, batch_size=30, epochs=1000)
predicted = model.predict(scaled_features)
bostonDF["KERAS_PREDICTED_PRICE_BATCH"] = predicted
bostonDF.head(10)
| false | 0 | 5,997 | 0 | 5,997 | 5,997 |
||
129936569
|
<jupyter_start><jupyter_text>Football/Soccer | Bundesliga Player Database
The Bundesliga Players dataset provides a comprehensive collection of information on every player in the German Bundesliga football league. From renowned goalkeepers to talented defenders, this dataset offers an extensive range of player details including their names, full names, ages, heights, nationalities, places of birth, prices, maximum prices, positions, shirt numbers, preferred foot, current clubs, contract expiration dates, dates of joining the clubs, player agents, and outfitters. Whether you're a passionate football fan, a sports analyst, or a fantasy football enthusiast, this dataset serves as a valuable resource for exploring and analyzing the profiles of Bundesliga players, enabling you to delve into their backgrounds, performance statistics, and club affiliations. Discover the stars of German football and gain insights into their careers with this comprehensive Bundesliga Players dataset.
Kaggle dataset identifier: bundesliga-soccer-player
<jupyter_script># # Importing libraries
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sb
# # Importing data
df = pd.read_csv("/kaggle/input/bundesliga-soccer-player/bundesliga_player.csv")
pd.set_option("display.max_rows", 500)
# # Using .info(), .isnull() and .describe() to get the general idea about this dataset
df.info()
df.isnull().sum()
df.describe()
df.head()
# # Removing excess columns
df = df[
[
"name",
"age",
"height",
"nationality",
"place_of_birth",
"price",
"position",
"foot",
"club",
"contract_expires",
"joined_club",
"player_agent",
"outfitter",
]
]
df.columns
# # Handling some of the missing data
df["outfitter"] = df["outfitter"].fillna("None")
df["outfitter"].unique()
df["player_agent"] = df["player_agent"].fillna("None")
df["player_agent"].isnull().sum()
# # Adding new column "first nationality" to uniform players that have 2 nationalities
df["first_nationality"] = df["nationality"].str.split().str[0]
# # Adding new column 'Player position'
df["Player position"] = df["position"].str.split("-").str[0]
df["Player position"] = df["Player position"].str.capitalize()
df["Player position"].unique()
# # Checking for duplicated rows
df.duplicated()
# # Player age distribution
# Creating histogram
plt.figure(figsize=(10, 6))
histplot = sb.histplot(data=df, x="age", kde=True, discrete=True)
plt.title("Player age distribution")
plt.xlabel("Age")
plt.ylabel("Frequency")
# Adding labels to the bars
for rect in histplot.patches:
height = rect.get_height()
plt.text(
rect.get_x() + rect.get_width() / 2, height + 0.2, int(height), ha="center"
)
# Setting ticks for each value of age
plt.xticks(range(df["age"].min(), df["age"].max() + 1))
plt.show()
# # Total Price per Club in millions
# Grouping players by their price to show club value
club_value = df.groupby("club")["price"].sum().reset_index()
# Sorting and removing insignificant values (club prices below 2 millions) as they are all youth and reserve teams
club_value = club_value.sort_values("price", ascending=False)
club_value = club_value[club_value["price"] >= 2]
# Creating barchart
plt.figure(figsize=(12, 6))
barplot = sb.barplot(data=club_value, x="club", y="price")
plt.title("Total Price per Club in millions")
plt.xlabel("Club")
plt.ylabel("Total Price")
plt.xticks(rotation=50)
## Adding labels to the bars
for bar in barplot.patches:
barplot.text(
x=bar.get_x() + bar.get_width() / 2,
y=bar.get_height() + 10,
s=f"{bar.get_height():.1f}",
ha="center",
va="bottom",
)
plt.show()
# # Foot Preference of Players
# Creating chart
plt.figure(figsize=(10, 6))
barplot = sb.countplot(data=df, x="foot")
plt.title("Foot Preference of Players")
plt.xlabel("Foot")
plt.ylabel("Frequency")
# Adding labels to the bars
for bar in barplot.patches:
barplot.text(
x=bar.get_x() + bar.get_width() / 2,
y=bar.get_height() + 0.2,
s=f"{int(bar.get_height())}",
ha="center",
va="bottom",
)
plt.show()
# # Player Heights per Role
# Creating chart
plt.figure(figsize=(12, 6))
barplot = sb.barplot(data=df, x="Player position", y="height")
plt.title("Player Heights per Position")
plt.xlabel("Position")
plt.ylabel("Height")
# Adding labels to the bars
for bar in barplot.patches:
barplot.text(
x=bar.get_x() + bar.get_width() / 2,
y=bar.get_height() + 0.03,
s=f"{bar.get_height():.2f}",
ha="center",
va="bottom",
)
plt.show()
# # Player Nationality
# Counting the number of players per nationality
df_counts = df["first_nationality"].value_counts()
# Sorting the values in descending order and creating chart
df_sorted = df_counts.sort_values(ascending=False)
plt.figure(figsize=(12, 6))
barplot = sb.barplot(x=df_counts.index, y=df_counts.values)
plt.title("Player Nationality")
plt.xlabel("Nationality")
plt.ylabel("Number of Players")
plt.xticks(rotation=45, ha="right")
plt.tight_layout()
# Adding labels to the bars
for bar, count in zip(barplot.patches, df_counts.values):
barplot.text(
x=bar.get_x() + bar.get_width() / 2,
y=bar.get_height() + 1,
s=f"{count}",
ha="center",
va="bottom",
# rotation=90
)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/936/129936569.ipynb
|
bundesliga-soccer-player
|
oles04
|
[{"Id": 129936569, "ScriptId": 38616195, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13746719, "CreationDate": "05/17/2023 14:41:50", "VersionNumber": 2.0, "Title": "Bundesliga Player Analysis", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 166.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 165.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 186363371, "KernelVersionId": 129936569, "SourceDatasetVersionId": 5668174}]
|
[{"Id": 5668174, "DatasetId": 3258253, "DatasourceVersionId": 5743664, "CreatorUserId": 12065933, "LicenseName": "Other (specified in description)", "CreationDate": "05/12/2023 07:42:13", "VersionNumber": 1.0, "Title": "Football/Soccer | Bundesliga Player Database", "Slug": "bundesliga-soccer-player", "Subtitle": "Bundesliga Player Database: Complete Profiles, Stats, and Clubs of each Player", "Description": "The Bundesliga Players dataset provides a comprehensive collection of information on every player in the German Bundesliga football league. From renowned goalkeepers to talented defenders, this dataset offers an extensive range of player details including their names, full names, ages, heights, nationalities, places of birth, prices, maximum prices, positions, shirt numbers, preferred foot, current clubs, contract expiration dates, dates of joining the clubs, player agents, and outfitters. Whether you're a passionate football fan, a sports analyst, or a fantasy football enthusiast, this dataset serves as a valuable resource for exploring and analyzing the profiles of Bundesliga players, enabling you to delve into their backgrounds, performance statistics, and club affiliations. Discover the stars of German football and gain insights into their careers with this comprehensive Bundesliga Players dataset.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3258253, "CreatorUserId": 12065933, "OwnerUserId": 12065933.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5668174.0, "CurrentDatasourceVersionId": 5743664.0, "ForumId": 3323776, "Type": 2, "CreationDate": "05/12/2023 07:42:13", "LastActivityDate": "05/12/2023", "TotalViews": 7284, "TotalDownloads": 1339, "TotalVotes": 37, "TotalKernels": 11}]
|
[{"Id": 12065933, "UserName": "oles04", "DisplayName": "Ole", "RegisterDate": "10/23/2022", "PerformanceTier": 2}]
|
# # Importing libraries
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sb
# # Importing data
df = pd.read_csv("/kaggle/input/bundesliga-soccer-player/bundesliga_player.csv")
pd.set_option("display.max_rows", 500)
# # Using .info(), .isnull() and .describe() to get the general idea about this dataset
df.info()
df.isnull().sum()
df.describe()
df.head()
# # Removing excess columns
df = df[
[
"name",
"age",
"height",
"nationality",
"place_of_birth",
"price",
"position",
"foot",
"club",
"contract_expires",
"joined_club",
"player_agent",
"outfitter",
]
]
df.columns
# # Handling some of the missing data
df["outfitter"] = df["outfitter"].fillna("None")
df["outfitter"].unique()
df["player_agent"] = df["player_agent"].fillna("None")
df["player_agent"].isnull().sum()
# # Adding new column "first nationality" to uniform players that have 2 nationalities
df["first_nationality"] = df["nationality"].str.split().str[0]
# # Adding new column 'Player position'
df["Player position"] = df["position"].str.split("-").str[0]
df["Player position"] = df["Player position"].str.capitalize()
df["Player position"].unique()
# # Checking for duplicated rows
df.duplicated()
# # Player age distribution
# Creating histogram
plt.figure(figsize=(10, 6))
histplot = sb.histplot(data=df, x="age", kde=True, discrete=True)
plt.title("Player age distribution")
plt.xlabel("Age")
plt.ylabel("Frequency")
# Adding labels to the bars
for rect in histplot.patches:
height = rect.get_height()
plt.text(
rect.get_x() + rect.get_width() / 2, height + 0.2, int(height), ha="center"
)
# Setting ticks for each value of age
plt.xticks(range(df["age"].min(), df["age"].max() + 1))
plt.show()
# # Total Price per Club in millions
# Grouping players by their price to show club value
club_value = df.groupby("club")["price"].sum().reset_index()
# Sorting and removing insignificant values (club prices below 2 millions) as they are all youth and reserve teams
club_value = club_value.sort_values("price", ascending=False)
club_value = club_value[club_value["price"] >= 2]
# Creating barchart
plt.figure(figsize=(12, 6))
barplot = sb.barplot(data=club_value, x="club", y="price")
plt.title("Total Price per Club in millions")
plt.xlabel("Club")
plt.ylabel("Total Price")
plt.xticks(rotation=50)
## Adding labels to the bars
for bar in barplot.patches:
barplot.text(
x=bar.get_x() + bar.get_width() / 2,
y=bar.get_height() + 10,
s=f"{bar.get_height():.1f}",
ha="center",
va="bottom",
)
plt.show()
# # Foot Preference of Players
# Creating chart
plt.figure(figsize=(10, 6))
barplot = sb.countplot(data=df, x="foot")
plt.title("Foot Preference of Players")
plt.xlabel("Foot")
plt.ylabel("Frequency")
# Adding labels to the bars
for bar in barplot.patches:
barplot.text(
x=bar.get_x() + bar.get_width() / 2,
y=bar.get_height() + 0.2,
s=f"{int(bar.get_height())}",
ha="center",
va="bottom",
)
plt.show()
# # Player Heights per Role
# Creating chart
plt.figure(figsize=(12, 6))
barplot = sb.barplot(data=df, x="Player position", y="height")
plt.title("Player Heights per Position")
plt.xlabel("Position")
plt.ylabel("Height")
# Adding labels to the bars
for bar in barplot.patches:
barplot.text(
x=bar.get_x() + bar.get_width() / 2,
y=bar.get_height() + 0.03,
s=f"{bar.get_height():.2f}",
ha="center",
va="bottom",
)
plt.show()
# # Player Nationality
# Counting the number of players per nationality
df_counts = df["first_nationality"].value_counts()
# Sorting the values in descending order and creating chart
df_sorted = df_counts.sort_values(ascending=False)
plt.figure(figsize=(12, 6))
barplot = sb.barplot(x=df_counts.index, y=df_counts.values)
plt.title("Player Nationality")
plt.xlabel("Nationality")
plt.ylabel("Number of Players")
plt.xticks(rotation=45, ha="right")
plt.tight_layout()
# Adding labels to the bars
for bar, count in zip(barplot.patches, df_counts.values):
barplot.text(
x=bar.get_x() + bar.get_width() / 2,
y=bar.get_height() + 1,
s=f"{count}",
ha="center",
va="bottom",
# rotation=90
)
plt.show()
| false | 1 | 1,409 | 2 | 1,651 | 1,409 |
||
129941510
|
<jupyter_start><jupyter_text>Hotel Booking
### Context
This dataset contains 119390 observations for a City Hotel and a Resort Hotel. Each observation represents a hotel booking between the 1st of July 2015 and 31st of August 2017, including booking that effectively arrived and booking that were canceled.
### Content
Since this is hotel real data, all data elements pertaining hotel or costumer identification were deleted.
Four Columns, 'name', 'email', 'phone number' and 'credit_card' have been artificially created and added to the dataset.
Kaggle dataset identifier: hotel-booking
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# importing important libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
# Display all the columns of the dataframe
pd.pandas.set_option("display.max_columns", None)
import warnings
warnings.filterwarnings("ignore")
import plotly.express as px
sns.set_style("darkgrid")
plt.rcParams["figure.figsize"] = (15, 8)
plt.rcParams["font.size"] = 18
# read dataframe
df = pd.read_csv("/kaggle/input/hotel-booking/hotel_booking.csv")
df.head()
# last 5 rows
df.tail()
# total no rows and columns
print(f"Total Number of Rows = {df.shape[0]} and Columns are {df.shape[1]}")
# basic info
df.info()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/941/129941510.ipynb
|
hotel-booking
|
mojtaba142
|
[{"Id": 129941510, "ScriptId": 38652270, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9774876, "CreationDate": "05/17/2023 15:18:22", "VersionNumber": 1.0, "Title": "Hotel Booking Cancelation Prediction", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 51.0, "LinesInsertedFromPrevious": 51.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186370322, "KernelVersionId": 129941510, "SourceDatasetVersionId": 2378746}]
|
[{"Id": 2378746, "DatasetId": 1437463, "DatasourceVersionId": 2420641, "CreatorUserId": 3075307, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "06/29/2021 05:15:54", "VersionNumber": 1.0, "Title": "Hotel Booking", "Slug": "hotel-booking", "Subtitle": "Hotel booking demand datasets(Data in Brief:2019)", "Description": "### Context\n\nThis dataset contains 119390 observations for a City Hotel and a Resort Hotel. Each observation represents a hotel booking between the 1st of July 2015 and 31st of August 2017, including booking that effectively arrived and booking that were canceled.\n\n\n\n### Content\n\nSince this is hotel real data, all data elements pertaining hotel or costumer identification were deleted. \nFour Columns, 'name', 'email', 'phone number' and 'credit_card' have been artificially created and added to the dataset. \n\n\n### Acknowledgements\n\nThe data is originally from the article Hotel Booking Demand Datasets, written by Nuno Antonio, Ana Almeida, and Luis Nunes for Data in Brief, Volume 22, February 2019.\n\n\n### Inspiration\n\nYour data will be in front of the world's largest data science community. What questions do you want to see answered?", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1437463, "CreatorUserId": 3075307, "OwnerUserId": 3075307.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2378746.0, "CurrentDatasourceVersionId": 2420641.0, "ForumId": 1456934, "Type": 2, "CreationDate": "06/29/2021 05:15:54", "LastActivityDate": "06/29/2021", "TotalViews": 89277, "TotalDownloads": 12792, "TotalVotes": 156, "TotalKernels": 136}]
|
[{"Id": 3075307, "UserName": "mojtaba142", "DisplayName": "Mojtaba", "RegisterDate": "04/11/2019", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# importing important libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
# Display all the columns of the dataframe
pd.pandas.set_option("display.max_columns", None)
import warnings
warnings.filterwarnings("ignore")
import plotly.express as px
sns.set_style("darkgrid")
plt.rcParams["figure.figsize"] = (15, 8)
plt.rcParams["font.size"] = 18
# read dataframe
df = pd.read_csv("/kaggle/input/hotel-booking/hotel_booking.csv")
df.head()
# last 5 rows
df.tail()
# total no rows and columns
print(f"Total Number of Rows = {df.shape[0]} and Columns are {df.shape[1]}")
# basic info
df.info()
| false | 1 | 391 | 0 | 542 | 391 |
||
129941241
|
<jupyter_start><jupyter_text>Wine Datasets
Kaggle dataset identifier: winedata
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
df1 = pd.read_csv("/kaggle/input/winedata/winequality_red.csv")
df2 = pd.read_csv("/kaggle/input/winedata/winequality_white.csv")
df1["colour"] = "Red"
df2["colour"] = "White"
print("red_wine dataset shape:", df1.shape)
print("ehite_wine dataset shape:", df2.shape)
data = pd.concat([df1, df2], axis=0)
data
data.colour.value_counts()
sns.pairplot(data, hue="colour", palette="plasma")
data.quality.value_counts()
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaled_data = scaler.fit_transform(data.iloc[:, :-2])
scaled_data
scaled_data.shape
# ### PCA:
# ---> n_comp = 2
# ### KMeans
# ---> n_clusters = 2
pca1 = PCA(n_components=2)
pca_comp1 = pca.fit_transform(scaled_data)
lst = [2, 7]
for i in lst:
model1 = KMeans(n_clusters=i, n_init="auto", random_state=0)
target1 = model1.fit_predict(pca_comp1)
plt.figure(figsize=(8, 6))
plt.scatter(pca_comp1[:, 0], pca_comp1[:, 1], c=target1, cmap="plasma")
plt.xlabel("First principal component")
plt.ylabel("Second Principal Component")
target_lst = ["colour", "quality"]
for i in target_lst:
plt.figure(figsize=(8, 6))
sns.scatterplot(data, x=pca_comp1[:, 0], y=pca_comp1[:, 1], hue=data[i])
plt.xlabel("First principal component")
plt.ylabel("Second Principal Component")
# ### PCA:
# ---> n_comp = 3
# ### KMeans
# ---> n_clusters = 2
pca = PCA(n_components=3)
pca_comp = pca.fit_transform(scaled_data)
model = KMeans(n_clusters=2, n_init="auto", random_state=0)
target = model.fit_predict(pca_comp)
plt.figure(figsize=(8, 6))
plt.scatter(pca_comp[:, 0], pca_comp[:, 1], c=target, cmap="plasma")
plt.xlabel("First principal component")
plt.ylabel("Second Principal Component")
target_lst = ["colour", "quality"]
for i in target_lst:
plt.figure(figsize=(8, 6))
sns.scatterplot(
data, x=pca_comp[:, 0], y=pca_comp[:, 1], hue=data[i], palette="plasma"
)
plt.xlabel("First principal component")
plt.ylabel("Second Principal Component")
import plotly.express as px
fig = px.scatter_3d(
data, x=pca_comp[:, 0], y=pca_comp[:, 1], z=pca_comp[:, 2], color=data["quality"]
)
fig.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/941/129941241.ipynb
|
winedata
|
sgus1318
|
[{"Id": 129941241, "ScriptId": 38604241, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8262343, "CreationDate": "05/17/2023 15:16:01", "VersionNumber": 2.0, "Title": "PCA_On_WineQualityDataset", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 100.0, "LinesInsertedFromPrevious": 47.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 53.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186369949, "KernelVersionId": 129941241, "SourceDatasetVersionId": 15785}]
|
[{"Id": 15785, "DatasetId": 11403, "DatasourceVersionId": 15785, "CreatorUserId": 1537701, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "01/30/2018 19:40:04", "VersionNumber": 1.0, "Title": "Wine Datasets", "Slug": "winedata", "Subtitle": "Data is available at: https://archive.ics.uci.edu/ml/datasets/Wine+Quality", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 354872.0, "TotalUncompressedBytes": 354872.0}]
|
[{"Id": 11403, "CreatorUserId": 1537701, "OwnerUserId": 1537701.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 15785.0, "CurrentDatasourceVersionId": 15785.0, "ForumId": 18806, "Type": 2, "CreationDate": "01/30/2018 19:40:04", "LastActivityDate": "02/04/2018", "TotalViews": 76660, "TotalDownloads": 10409, "TotalVotes": 60, "TotalKernels": 8}]
|
[{"Id": 1537701, "UserName": "sgus1318", "DisplayName": "SarahG", "RegisterDate": "01/08/2018", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
df1 = pd.read_csv("/kaggle/input/winedata/winequality_red.csv")
df2 = pd.read_csv("/kaggle/input/winedata/winequality_white.csv")
df1["colour"] = "Red"
df2["colour"] = "White"
print("red_wine dataset shape:", df1.shape)
print("ehite_wine dataset shape:", df2.shape)
data = pd.concat([df1, df2], axis=0)
data
data.colour.value_counts()
sns.pairplot(data, hue="colour", palette="plasma")
data.quality.value_counts()
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaled_data = scaler.fit_transform(data.iloc[:, :-2])
scaled_data
scaled_data.shape
# ### PCA:
# ---> n_comp = 2
# ### KMeans
# ---> n_clusters = 2
pca1 = PCA(n_components=2)
pca_comp1 = pca.fit_transform(scaled_data)
lst = [2, 7]
for i in lst:
model1 = KMeans(n_clusters=i, n_init="auto", random_state=0)
target1 = model1.fit_predict(pca_comp1)
plt.figure(figsize=(8, 6))
plt.scatter(pca_comp1[:, 0], pca_comp1[:, 1], c=target1, cmap="plasma")
plt.xlabel("First principal component")
plt.ylabel("Second Principal Component")
target_lst = ["colour", "quality"]
for i in target_lst:
plt.figure(figsize=(8, 6))
sns.scatterplot(data, x=pca_comp1[:, 0], y=pca_comp1[:, 1], hue=data[i])
plt.xlabel("First principal component")
plt.ylabel("Second Principal Component")
# ### PCA:
# ---> n_comp = 3
# ### KMeans
# ---> n_clusters = 2
pca = PCA(n_components=3)
pca_comp = pca.fit_transform(scaled_data)
model = KMeans(n_clusters=2, n_init="auto", random_state=0)
target = model.fit_predict(pca_comp)
plt.figure(figsize=(8, 6))
plt.scatter(pca_comp[:, 0], pca_comp[:, 1], c=target, cmap="plasma")
plt.xlabel("First principal component")
plt.ylabel("Second Principal Component")
target_lst = ["colour", "quality"]
for i in target_lst:
plt.figure(figsize=(8, 6))
sns.scatterplot(
data, x=pca_comp[:, 0], y=pca_comp[:, 1], hue=data[i], palette="plasma"
)
plt.xlabel("First principal component")
plt.ylabel("Second Principal Component")
import plotly.express as px
fig = px.scatter_3d(
data, x=pca_comp[:, 0], y=pca_comp[:, 1], z=pca_comp[:, 2], color=data["quality"]
)
fig.show()
| false | 2 | 970 | 0 | 991 | 970 |
||
129588723
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_data = pd.read_csv("/kaggle/input/nwea-DS4E/train.csv")
test_data = pd.read_csv("/kaggle/input/nwea-DS4E/test.csv")
import tensorflow as tf
from tensorflow.keras.layers import Dense, Flatten
from tensorflow.keras.metrics import MeanSquaredError, MeanAbsoluteError, Accuracy
from sklearn.preprocessing import MinMaxScaler
def inverse_transform(y_in, y_min, y_max):
return (y_in * (y_max - y_min)) + y_min
scaler = MinMaxScaler()
thingy_min = train_data["pv1math"].min()
thingy_max = train_data["pv1math"].max()
print(thingy_min)
print(thingy_max)
# ty_train = train_data['pv1math']
for column in train_data.columns:
train_data[column] = scaler.fit_transform(train_data[[column]])
if column != "pv1math":
test_data[column] = scaler.transform(test_data[[column]])
# train_data_in column size: 14504
tx_train = train_data.sample(int(14504 * 0.8))
ty_train = tx_train["pv1math"]
tx_train = tx_train.drop("pv1math", axis=1)
# ---
tx_test = train_data.drop(tx_train.index)
ty_test = tx_test["pv1math"]
tx_test = tx_test.drop("pv1math", axis=1)
# tx_train.to_csv(f'/kaggle/working/tx_train.csv', index=False)
# ty_train.to_csv(f'/kaggle/working/ty_train.csv', index=False)
# tx_test.to_csv(f'/kaggle/working/tx_test.csv', index=False)
# ty_test.to_csv(f'/kaggle/working/ty_test.csv', index=False)
tx_train = np.asarray(tx_train)
ty_train = np.asarray(ty_train)
tx_test = np.asarray(tx_test)
ty_test = np.asarray(ty_test)
# create model
model = tf.keras.models.Sequential(
[
tf.keras.layers.Dense(
64, activation="relu", kernel_initializer=tf.keras.initializers.HeNormal()
),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(
64, activation="relu", kernel_initializer=tf.keras.initializers.HeNormal()
),
tf.keras.layers.Dropout(0.25),
tf.keras.layers.Dense(
64, activation="sigmoid", kernel_initializer="glorot_uniform"
),
tf.keras.layers.Dropout(0.25),
tf.keras.layers.Dense(1),
]
)
# calculate error using mean squared error
# Using 'auto'/'sum_over_batch_size' reduction type.
mse = tf.keras.losses.MeanSquaredError()
# create optimizer with learning rate of 0.01
optimizer = tf.keras.optimizers.Adam(learning_rate=0.0005)
model.compile(optimizer=optimizer, loss=mse, metrics=mse)
model.fit(tx_train, ty_train, batch_size=32, epochs=300)
print("evaluation:")
model.evaluate(tx_test, ty_test, verbose=2)
print(model.predict(test_data))
# model.predict(test_data).to_csv(f'/kaggle/working/predict_test_data.csv', index=False)
print(inverse_transform(model.predict(test_data), thingy_min, thingy_max))
z = inverse_transform(model.predict(test_data), thingy_min, thingy_max)
print(z)
z = z.flatten()
output = pd.DataFrame({"Predicted": z}, index=range(len(z)))
output.index = pd.RangeIndex(start=1, stop=len(output) + 1)
output.to_csv(f"/kaggle/working/submission.csv", index_label="Id")
# Test XGB Model
from sklearn.metrics import mean_absolute_error
from xgboost import XGBRegressor
XGBModel = XGBRegressor()
XGBModel.fit(tx_train, ty_train, verbose=False)
# Get the mean absolute error on the validation data :
XGBpredictions = XGBModel.predict(tx_test)
MAE = mean_absolute_error(ty_test, XGBpredictions)
print("XGBoost validation MAE = ", MAE)
# Test Random Forest Model
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor()
model.fit(tx_train, ty_train)
# Get the mean absolute error on the validation data
predicted_prices = model.predict(tx_test)
MAE = mean_absolute_error(ty_test, predicted_prices)
print("Random forest validation MAE = ", MAE)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/588/129588723.ipynb
| null | null |
[{"Id": 129588723, "ScriptId": 38461490, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14339659, "CreationDate": "05/15/2023 04:32:26", "VersionNumber": 2.0, "Title": "NWEA Submission", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 133.0, "LinesInsertedFromPrevious": 26.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 107.0, "LinesInsertedFromFork": 30.0, "LinesDeletedFromFork": 6.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 103.0, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_data = pd.read_csv("/kaggle/input/nwea-DS4E/train.csv")
test_data = pd.read_csv("/kaggle/input/nwea-DS4E/test.csv")
import tensorflow as tf
from tensorflow.keras.layers import Dense, Flatten
from tensorflow.keras.metrics import MeanSquaredError, MeanAbsoluteError, Accuracy
from sklearn.preprocessing import MinMaxScaler
def inverse_transform(y_in, y_min, y_max):
return (y_in * (y_max - y_min)) + y_min
scaler = MinMaxScaler()
thingy_min = train_data["pv1math"].min()
thingy_max = train_data["pv1math"].max()
print(thingy_min)
print(thingy_max)
# ty_train = train_data['pv1math']
for column in train_data.columns:
train_data[column] = scaler.fit_transform(train_data[[column]])
if column != "pv1math":
test_data[column] = scaler.transform(test_data[[column]])
# train_data_in column size: 14504
tx_train = train_data.sample(int(14504 * 0.8))
ty_train = tx_train["pv1math"]
tx_train = tx_train.drop("pv1math", axis=1)
# ---
tx_test = train_data.drop(tx_train.index)
ty_test = tx_test["pv1math"]
tx_test = tx_test.drop("pv1math", axis=1)
# tx_train.to_csv(f'/kaggle/working/tx_train.csv', index=False)
# ty_train.to_csv(f'/kaggle/working/ty_train.csv', index=False)
# tx_test.to_csv(f'/kaggle/working/tx_test.csv', index=False)
# ty_test.to_csv(f'/kaggle/working/ty_test.csv', index=False)
tx_train = np.asarray(tx_train)
ty_train = np.asarray(ty_train)
tx_test = np.asarray(tx_test)
ty_test = np.asarray(ty_test)
# create model
model = tf.keras.models.Sequential(
[
tf.keras.layers.Dense(
64, activation="relu", kernel_initializer=tf.keras.initializers.HeNormal()
),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(
64, activation="relu", kernel_initializer=tf.keras.initializers.HeNormal()
),
tf.keras.layers.Dropout(0.25),
tf.keras.layers.Dense(
64, activation="sigmoid", kernel_initializer="glorot_uniform"
),
tf.keras.layers.Dropout(0.25),
tf.keras.layers.Dense(1),
]
)
# calculate error using mean squared error
# Using 'auto'/'sum_over_batch_size' reduction type.
mse = tf.keras.losses.MeanSquaredError()
# create optimizer with learning rate of 0.01
optimizer = tf.keras.optimizers.Adam(learning_rate=0.0005)
model.compile(optimizer=optimizer, loss=mse, metrics=mse)
model.fit(tx_train, ty_train, batch_size=32, epochs=300)
print("evaluation:")
model.evaluate(tx_test, ty_test, verbose=2)
print(model.predict(test_data))
# model.predict(test_data).to_csv(f'/kaggle/working/predict_test_data.csv', index=False)
print(inverse_transform(model.predict(test_data), thingy_min, thingy_max))
z = inverse_transform(model.predict(test_data), thingy_min, thingy_max)
print(z)
z = z.flatten()
output = pd.DataFrame({"Predicted": z}, index=range(len(z)))
output.index = pd.RangeIndex(start=1, stop=len(output) + 1)
output.to_csv(f"/kaggle/working/submission.csv", index_label="Id")
# Test XGB Model
from sklearn.metrics import mean_absolute_error
from xgboost import XGBRegressor
XGBModel = XGBRegressor()
XGBModel.fit(tx_train, ty_train, verbose=False)
# Get the mean absolute error on the validation data :
XGBpredictions = XGBModel.predict(tx_test)
MAE = mean_absolute_error(ty_test, XGBpredictions)
print("XGBoost validation MAE = ", MAE)
# Test Random Forest Model
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor()
model.fit(tx_train, ty_train)
# Get the mean absolute error on the validation data
predicted_prices = model.predict(tx_test)
MAE = mean_absolute_error(ty_test, predicted_prices)
print("Random forest validation MAE = ", MAE)
| false | 0 | 1,418 | 0 | 1,418 | 1,418 |
||
129588958
|
# ## Import Library
import tensorflow as tf
import tensorflow_decision_forests as tfdf
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
print("TensorFlow v" + tf.__version__)
print("TensorFlow Decision Forests v" + tfdf.__version__)
# ## Setup Dataset
dataset_df = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
# ## Preprocessing Data
# menghapus kolom `PassengerID` dan `Name` karena tidak terlalu diperlukan untuk training model
dataset_df = dataset_df.drop(["PassengerId", "Name"], axis=1)
dataset_df.head(5)
# Mengganti nilai kolom yang kosong menjadi nilai 0 (nol) numerik.
dataset_df[
["VIP", "CryoSleep", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"]
] = dataset_df[
["VIP", "CryoSleep", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"]
].fillna(
value=0
)
dataset_df.isnull().sum().sort_values(ascending=False)
# Karena Tensor Flow Decision Forest tidak bisa mengolah kolom boolean, maka kita sesuaikan kolom `Transported`, `VIP`, dan `CryoSleep` dengan mengubahnya ke format integer.
dataset_df["Transported"] = dataset_df["Transported"].astype(int)
dataset_df["VIP"] = dataset_df["VIP"].astype(int)
dataset_df["CryoSleep"] = dataset_df["CryoSleep"].astype(int)
# Isi dari kolom `Cabin` adalah string dengan forman `Dek/No Cabin/Sisi`. Kita coba pecah dan mengubahnya menjadi 3 kolom baru `Deck`, `Cabin_num`, dan `Side`, untuk dapat memudahkan melatih model. Lalu, kita hapus kolom `Cabin` karena sudah tidak diperlukan lagi
dataset_df[["Deck", "Cabin_num", "Side"]] = dataset_df["Cabin"].str.split(
"/", expand=True
)
try:
dataset_df = dataset_df.drop("Cabin", axis=1)
except KeyError:
print("Field does not exist")
dataset_df.head(5)
# Membagi dataset dengan sebagian untuk training dan sebagian lagi untuk vakdiasi
def split_dataset(dataset, test_ratio=0.15):
test_indices = np.random.rand(len(dataset)) < test_ratio
return dataset[~test_indices], dataset[test_indices]
train_ds_pd, valid_ds_pd = split_dataset(dataset_df)
print(
"{} examples in training, {} examples in testing.".format(
len(train_ds_pd), len(valid_ds_pd)
)
)
# Mengubah dataset dari yang sebelumnya menggunakan format Panda Dataframe menjad format Tensoflow Dataset
train_ds = tfdf.keras.pd_dataframe_to_tf_dataset(train_ds_pd, label="Transported")
valid_ds = tfdf.keras.pd_dataframe_to_tf_dataset(valid_ds_pd, label="Transported")
# Untuk training modelnya, menggunakan `RandomForestModel`. Kita akan menggunakan template `benchmark_rank1`
rf = tfdf.keras.RandomForestModel(hyperparameter_template="benchmark_rank1")
rf = tfdf.keras.RandomForestModel()
rf.compile(metrics=["accuracy"])
# ## Training Model
rf.fit(x=train_ds)
# ## Visualiasi Model
#
tfdf.model_plotter.plot_model_in_colab(rf, tree_idx=0, max_depth=3)
# ## Evaluasi Model
# Menjalankan evaluasi dengan dataset testing / validasi dataset
evaluation = rf.evaluate(x=valid_ds, return_dict=True)
for name, value in evaluation.items():
print(f"{name}: {value:.4f}")
inspector = rf.make_inspector()
inspector.evaluation()
plt.figure(figsize=(10, 6))
variable_importance_metric = "NUM_AS_ROOT"
variable_importances = inspector.variable_importances()[variable_importance_metric]
feature_names = [vi[0].name for vi in variable_importances]
feature_importances = [vi[1] for vi in variable_importances]
feature_ranks = range(len(feature_names))
bar = plt.barh(
feature_ranks, feature_importances, label=[str(x) for x in feature_ranks]
)
plt.yticks(feature_ranks, feature_names)
plt.gca().invert_yaxis()
for importance, patch in zip(feature_importances, bar.patches):
plt.text(
patch.get_x() + patch.get_width(), patch.get_y(), f"{importance:.4f}", va="top"
)
plt.xlabel("Nilai")
plt.title("Aspek yang Berpengaruh Terhadap Keselamatan Penumpang Spaceship Titanic")
plt.tight_layout()
plt.show()
# ## Submission
test_df = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
submission_id = test_df.PassengerId
test_df[["VIP", "CryoSleep"]] = test_df[["VIP", "CryoSleep"]].fillna(value=0)
test_df[["Deck", "Cabin_num", "Side"]] = test_df["Cabin"].str.split("/", expand=True)
test_df = test_df.drop("Cabin", axis=1)
test_df["VIP"] = test_df["VIP"].astype(int)
test_df["CryoSleep"] = test_df["CryoSleep"].astype(int)
test_ds = tfdf.keras.pd_dataframe_to_tf_dataset(test_df)
predictions = rf.predict(test_ds)
n_predictions = (predictions > 0.5).astype(bool)
output = pd.DataFrame(
{"PassengerId": submission_id, "Transported": n_predictions.squeeze()}
)
output.head()
sample_submission_df = pd.read_csv(
"/kaggle/input/spaceship-titanic/sample_submission.csv"
)
sample_submission_df["Transported"] = n_predictions
sample_submission_df.to_csv("/kaggle/working/submission.csv", index=False)
sample_submission_df.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/588/129588958.ipynb
| null | null |
[{"Id": 129588958, "ScriptId": 38275222, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15004605, "CreationDate": "05/15/2023 04:35:38", "VersionNumber": 3.0, "Title": "Pendekar Data", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 141.0, "LinesInsertedFromPrevious": 36.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 105.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## Import Library
import tensorflow as tf
import tensorflow_decision_forests as tfdf
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
print("TensorFlow v" + tf.__version__)
print("TensorFlow Decision Forests v" + tfdf.__version__)
# ## Setup Dataset
dataset_df = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
# ## Preprocessing Data
# menghapus kolom `PassengerID` dan `Name` karena tidak terlalu diperlukan untuk training model
dataset_df = dataset_df.drop(["PassengerId", "Name"], axis=1)
dataset_df.head(5)
# Mengganti nilai kolom yang kosong menjadi nilai 0 (nol) numerik.
dataset_df[
["VIP", "CryoSleep", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"]
] = dataset_df[
["VIP", "CryoSleep", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"]
].fillna(
value=0
)
dataset_df.isnull().sum().sort_values(ascending=False)
# Karena Tensor Flow Decision Forest tidak bisa mengolah kolom boolean, maka kita sesuaikan kolom `Transported`, `VIP`, dan `CryoSleep` dengan mengubahnya ke format integer.
dataset_df["Transported"] = dataset_df["Transported"].astype(int)
dataset_df["VIP"] = dataset_df["VIP"].astype(int)
dataset_df["CryoSleep"] = dataset_df["CryoSleep"].astype(int)
# Isi dari kolom `Cabin` adalah string dengan forman `Dek/No Cabin/Sisi`. Kita coba pecah dan mengubahnya menjadi 3 kolom baru `Deck`, `Cabin_num`, dan `Side`, untuk dapat memudahkan melatih model. Lalu, kita hapus kolom `Cabin` karena sudah tidak diperlukan lagi
dataset_df[["Deck", "Cabin_num", "Side"]] = dataset_df["Cabin"].str.split(
"/", expand=True
)
try:
dataset_df = dataset_df.drop("Cabin", axis=1)
except KeyError:
print("Field does not exist")
dataset_df.head(5)
# Membagi dataset dengan sebagian untuk training dan sebagian lagi untuk vakdiasi
def split_dataset(dataset, test_ratio=0.15):
test_indices = np.random.rand(len(dataset)) < test_ratio
return dataset[~test_indices], dataset[test_indices]
train_ds_pd, valid_ds_pd = split_dataset(dataset_df)
print(
"{} examples in training, {} examples in testing.".format(
len(train_ds_pd), len(valid_ds_pd)
)
)
# Mengubah dataset dari yang sebelumnya menggunakan format Panda Dataframe menjad format Tensoflow Dataset
train_ds = tfdf.keras.pd_dataframe_to_tf_dataset(train_ds_pd, label="Transported")
valid_ds = tfdf.keras.pd_dataframe_to_tf_dataset(valid_ds_pd, label="Transported")
# Untuk training modelnya, menggunakan `RandomForestModel`. Kita akan menggunakan template `benchmark_rank1`
rf = tfdf.keras.RandomForestModel(hyperparameter_template="benchmark_rank1")
rf = tfdf.keras.RandomForestModel()
rf.compile(metrics=["accuracy"])
# ## Training Model
rf.fit(x=train_ds)
# ## Visualiasi Model
#
tfdf.model_plotter.plot_model_in_colab(rf, tree_idx=0, max_depth=3)
# ## Evaluasi Model
# Menjalankan evaluasi dengan dataset testing / validasi dataset
evaluation = rf.evaluate(x=valid_ds, return_dict=True)
for name, value in evaluation.items():
print(f"{name}: {value:.4f}")
inspector = rf.make_inspector()
inspector.evaluation()
plt.figure(figsize=(10, 6))
variable_importance_metric = "NUM_AS_ROOT"
variable_importances = inspector.variable_importances()[variable_importance_metric]
feature_names = [vi[0].name for vi in variable_importances]
feature_importances = [vi[1] for vi in variable_importances]
feature_ranks = range(len(feature_names))
bar = plt.barh(
feature_ranks, feature_importances, label=[str(x) for x in feature_ranks]
)
plt.yticks(feature_ranks, feature_names)
plt.gca().invert_yaxis()
for importance, patch in zip(feature_importances, bar.patches):
plt.text(
patch.get_x() + patch.get_width(), patch.get_y(), f"{importance:.4f}", va="top"
)
plt.xlabel("Nilai")
plt.title("Aspek yang Berpengaruh Terhadap Keselamatan Penumpang Spaceship Titanic")
plt.tight_layout()
plt.show()
# ## Submission
test_df = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
submission_id = test_df.PassengerId
test_df[["VIP", "CryoSleep"]] = test_df[["VIP", "CryoSleep"]].fillna(value=0)
test_df[["Deck", "Cabin_num", "Side"]] = test_df["Cabin"].str.split("/", expand=True)
test_df = test_df.drop("Cabin", axis=1)
test_df["VIP"] = test_df["VIP"].astype(int)
test_df["CryoSleep"] = test_df["CryoSleep"].astype(int)
test_ds = tfdf.keras.pd_dataframe_to_tf_dataset(test_df)
predictions = rf.predict(test_ds)
n_predictions = (predictions > 0.5).astype(bool)
output = pd.DataFrame(
{"PassengerId": submission_id, "Transported": n_predictions.squeeze()}
)
output.head()
sample_submission_df = pd.read_csv(
"/kaggle/input/spaceship-titanic/sample_submission.csv"
)
sample_submission_df["Transported"] = n_predictions
sample_submission_df.to_csv("/kaggle/working/submission.csv", index=False)
sample_submission_df.head()
| false | 0 | 1,618 | 0 | 1,618 | 1,618 |
||
129588489
|
# library imports
import pandas as pd
import spacy
from spacy import displacy
from spacy.tokens import DocBin
import json
from datetime import datetime
from tqdm import tqdm
import re
# this dictionary will contain all annotated examples
collective_dict = {"TRAINING_DATA": []}
def structure_training_data(text, kw_list):
results = []
entities = []
# search for instances of keywords within the text (ignoring letter case)
for kw in tqdm(kw_list):
search = re.finditer(kw, text, flags=re.IGNORECASE)
# store the start/end character positions
all_instances = [[m.start(), m.end()] for m in search]
# if the callable_iterator found matches, create an 'entities' list
if len(all_instances) > 0:
for i in all_instances:
start = i[0]
end = i[1]
entities.append((start, end, "SERVICE"))
# alert when no matches are found given the user inputs
else:
print("No pattern matches found. Keyword:", kw)
# add any found entities into a JSON format within collective_dict
if len(entities) > 0:
results = [text, {"entities": entities}]
collective_dict["TRAINING_DATA"].append(results)
return
# this dictionary will contain all annotated examples
collective_dict = {"TRAINING_DATA": []}
def structure_training_data(text, kw_list):
results = []
entities = []
# search for instances of keywords within the text (ignoring letter case)
for kw in tqdm(kw_list):
search = re.finditer(kw, text, flags=re.IGNORECASE)
# store the start/end character positions
all_instances = [[m.start(), m.end()] for m in search]
# if the callable_iterator found matches, create an 'entities' list
if len(all_instances) > 0:
for i in all_instances:
start = i[0]
end = i[1]
entities.append((start, end, "SERVICE"))
# alert when no matches are found given the user inputs
else:
print("No pattern matches found. Keyword:", kw)
# add any found entities into a JSON format within collective_dict
if len(entities) > 0:
results = [text, {"entities": entities}]
collective_dict["TRAINING_DATA"].append(results)
return
text1 = "BigTime Care has a broad array of service offerings for Philadelphia-area clientele. \
For 50 years, we have specialized in landscaping and lawn mowing. \
We also provide seasonal snow removal services for local commercial and residential properties. \
Call any time to schedule a consultation!"
text2 = "Scrub-O Cleaning connects independent professionals with customers. \
We offer the full range of customizable cleaning services that you may need now and in \
the future, and our team is ready to begin working for you today! We offer quality maid \
services and housekeeping across the San Francisco Bay Area."
text3 = "Locally owned and operated, Trust Roofing has the best roofing services in \
Philadelphia and the surrounding areas. Whatever the season, you can count on us to provide \
you with the best possible roof repair.\ We will work with any given roof replacement material, \
including asphalt shingles and metal roofs. Siding replacement services are also available."
text4 = "Based in Pittsburgh PA, Tammy's Branch Cuts is a family owned and managed smalled \
businesses founded in 1994. We specialize in full-service landscape design, including \
tree removal, lawn care to protect your existing plants, and comprehensive hardscaping for \
patios, walkways, and outdoor living spaces. Contact us today!"
# TRAINING
structure_training_data(text1, ["landscaping", "lawn mowing", "snow removal"])
structure_training_data(text2, ["cleaning services", "maid services", "housekeeping"])
structure_training_data(text3, ["roofing", "roof repair", "siding replacement"])
structure_training_data(
text4, ["landscape design", "tree removal", "lawn care", "hardscaping"]
)
# define our training data to TRAIN_DATA
TRAIN_DATA = collective_dict["TRAINING_DATA"]
# create a blank model
nlp = spacy.blank("en")
def create_training(TRAIN_DATA):
db = DocBin()
for text, annot in tqdm(TRAIN_DATA):
doc = nlp.make_doc(text)
ents = []
# create span objects
for start, end, label in annot["entities"]:
span = doc.char_span(start, end, label=label, alignment_mode="contract")
# skip if the character indices do not map to a valid span
if span is None:
print("Skipping entity.")
else:
ents.append(span)
# handle erroneous entity annotations by removing them
try:
doc.ents = ents
except:
# print("BAD SPAN:", span, "\n")
ents.pop()
doc.ents = ents
# pack Doc objects into DocBin
db.add(doc)
return db
TRAIN_DATA_DOC = create_training(TRAIN_DATA)
# Export results (here I add it to a TRAIN_DATA folder within the directory)
# TRAIN_DATA_DOC.to_disk("./TRAIN_DATA/TRAIN_DATA.spacy")
# --gpu-id 0
# load the trained model
nlp_output = spacy.load("/kaggle/working/output/model-best")
def model_visualization(text):
# pass our test instance into the trained pipeline
doc = nlp_output(text)
# customize the label colors
colors = {"SERVICE": "linear-gradient(90deg, #E1D436, #F59710)"}
options = {"ents": ["SERVICE"], "colors": colors}
# visualize the identified entities
displacy.render(doc, style="ent", options=options, minify=True, jupyter=True)
# print out the identified entities
print("\nIDENTIFIED ENTITIES:")
[print(ent.text) for ent in doc.ents if ent.label_ == "SERVICE"]
test1 = "At Perfection Landscapes LLC, we are committed to protecting the health of trees \
and shrubs in urban and suburban areas. We work with clients to provide expertise in all areas \
of tree care, stump removal, and construction-related tree preservation. Our trained experts \
also have years of experience with insect control. Call us today for a consultation!"
model_visualization(test1)
test2 = "J.K. Commercial Cleaning is dedicated to creating clean, safe, and healthy \
environments. We offer cleaning programs that are tailored to fit your business's individual needs. \
This includes janitorial services, and consistent quality deep cleaning services for both commercial and residential spaces."
model_visualization(test2)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/588/129588489.ipynb
| null | null |
[{"Id": 129588489, "ScriptId": 38462273, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15036313, "CreationDate": "05/15/2023 04:28:58", "VersionNumber": 1.0, "Title": "service", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 178.0, "LinesInsertedFromPrevious": 178.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# library imports
import pandas as pd
import spacy
from spacy import displacy
from spacy.tokens import DocBin
import json
from datetime import datetime
from tqdm import tqdm
import re
# this dictionary will contain all annotated examples
collective_dict = {"TRAINING_DATA": []}
def structure_training_data(text, kw_list):
results = []
entities = []
# search for instances of keywords within the text (ignoring letter case)
for kw in tqdm(kw_list):
search = re.finditer(kw, text, flags=re.IGNORECASE)
# store the start/end character positions
all_instances = [[m.start(), m.end()] for m in search]
# if the callable_iterator found matches, create an 'entities' list
if len(all_instances) > 0:
for i in all_instances:
start = i[0]
end = i[1]
entities.append((start, end, "SERVICE"))
# alert when no matches are found given the user inputs
else:
print("No pattern matches found. Keyword:", kw)
# add any found entities into a JSON format within collective_dict
if len(entities) > 0:
results = [text, {"entities": entities}]
collective_dict["TRAINING_DATA"].append(results)
return
# this dictionary will contain all annotated examples
collective_dict = {"TRAINING_DATA": []}
def structure_training_data(text, kw_list):
results = []
entities = []
# search for instances of keywords within the text (ignoring letter case)
for kw in tqdm(kw_list):
search = re.finditer(kw, text, flags=re.IGNORECASE)
# store the start/end character positions
all_instances = [[m.start(), m.end()] for m in search]
# if the callable_iterator found matches, create an 'entities' list
if len(all_instances) > 0:
for i in all_instances:
start = i[0]
end = i[1]
entities.append((start, end, "SERVICE"))
# alert when no matches are found given the user inputs
else:
print("No pattern matches found. Keyword:", kw)
# add any found entities into a JSON format within collective_dict
if len(entities) > 0:
results = [text, {"entities": entities}]
collective_dict["TRAINING_DATA"].append(results)
return
text1 = "BigTime Care has a broad array of service offerings for Philadelphia-area clientele. \
For 50 years, we have specialized in landscaping and lawn mowing. \
We also provide seasonal snow removal services for local commercial and residential properties. \
Call any time to schedule a consultation!"
text2 = "Scrub-O Cleaning connects independent professionals with customers. \
We offer the full range of customizable cleaning services that you may need now and in \
the future, and our team is ready to begin working for you today! We offer quality maid \
services and housekeeping across the San Francisco Bay Area."
text3 = "Locally owned and operated, Trust Roofing has the best roofing services in \
Philadelphia and the surrounding areas. Whatever the season, you can count on us to provide \
you with the best possible roof repair.\ We will work with any given roof replacement material, \
including asphalt shingles and metal roofs. Siding replacement services are also available."
text4 = "Based in Pittsburgh PA, Tammy's Branch Cuts is a family owned and managed smalled \
businesses founded in 1994. We specialize in full-service landscape design, including \
tree removal, lawn care to protect your existing plants, and comprehensive hardscaping for \
patios, walkways, and outdoor living spaces. Contact us today!"
# TRAINING
structure_training_data(text1, ["landscaping", "lawn mowing", "snow removal"])
structure_training_data(text2, ["cleaning services", "maid services", "housekeeping"])
structure_training_data(text3, ["roofing", "roof repair", "siding replacement"])
structure_training_data(
text4, ["landscape design", "tree removal", "lawn care", "hardscaping"]
)
# define our training data to TRAIN_DATA
TRAIN_DATA = collective_dict["TRAINING_DATA"]
# create a blank model
nlp = spacy.blank("en")
def create_training(TRAIN_DATA):
db = DocBin()
for text, annot in tqdm(TRAIN_DATA):
doc = nlp.make_doc(text)
ents = []
# create span objects
for start, end, label in annot["entities"]:
span = doc.char_span(start, end, label=label, alignment_mode="contract")
# skip if the character indices do not map to a valid span
if span is None:
print("Skipping entity.")
else:
ents.append(span)
# handle erroneous entity annotations by removing them
try:
doc.ents = ents
except:
# print("BAD SPAN:", span, "\n")
ents.pop()
doc.ents = ents
# pack Doc objects into DocBin
db.add(doc)
return db
TRAIN_DATA_DOC = create_training(TRAIN_DATA)
# Export results (here I add it to a TRAIN_DATA folder within the directory)
# TRAIN_DATA_DOC.to_disk("./TRAIN_DATA/TRAIN_DATA.spacy")
# --gpu-id 0
# load the trained model
nlp_output = spacy.load("/kaggle/working/output/model-best")
def model_visualization(text):
# pass our test instance into the trained pipeline
doc = nlp_output(text)
# customize the label colors
colors = {"SERVICE": "linear-gradient(90deg, #E1D436, #F59710)"}
options = {"ents": ["SERVICE"], "colors": colors}
# visualize the identified entities
displacy.render(doc, style="ent", options=options, minify=True, jupyter=True)
# print out the identified entities
print("\nIDENTIFIED ENTITIES:")
[print(ent.text) for ent in doc.ents if ent.label_ == "SERVICE"]
test1 = "At Perfection Landscapes LLC, we are committed to protecting the health of trees \
and shrubs in urban and suburban areas. We work with clients to provide expertise in all areas \
of tree care, stump removal, and construction-related tree preservation. Our trained experts \
also have years of experience with insect control. Call us today for a consultation!"
model_visualization(test1)
test2 = "J.K. Commercial Cleaning is dedicated to creating clean, safe, and healthy \
environments. We offer cleaning programs that are tailored to fit your business's individual needs. \
This includes janitorial services, and consistent quality deep cleaning services for both commercial and residential spaces."
model_visualization(test2)
| false | 0 | 1,677 | 0 | 1,677 | 1,677 |
||
129846641
|
<jupyter_start><jupyter_text>Drones_Data
Kaggle dataset identifier: drones-data
<jupyter_script>import os
import shutil
import random
from functools import partial
import tensorflow as tf
import cv2
from PIL import Image
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import albumentations as A
from IPython.display import clear_output, display
clear_output()
import ultralytics
ultralytics.checks()
from ultralytics import YOLO
def apply_augmentation(src_path: str, dst_path: str, transform) -> None:
for img_name in os.listdir(src_path):
img_path = os.path.join(src_path, img_name)
if not os.path.isfile(img_path):
print(f"Skipping {img_name} - file not found.")
continue
img = cv2.imread(img_path, cv2.IMREAD_COLOR)
if img is None:
print(f"Skipping {img_name} - failed to read image.")
continue
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
augmented_image = transform(image=img)["image"]
augmented_image = cv2.cvtColor(augmented_image, cv2.COLOR_RGB2BGR)
dst_file_path = os.path.join(dst_path, img_name)
cv2.imwrite(dst_file_path, augmented_image)
print(f"Saved augmented image: {dst_file_path}")
transform = A.Compose(
[
A.CoarseDropout(
max_holes=16,
max_height=50,
max_width=50,
min_holes=8,
min_height=25,
min_width=25,
fill_value=0,
p=0.5,
),
]
)
BASE_DIR = os.getcwd()
os.makedirs("/kaggle/working/Agumented_Drones/train/images")
os.makedirs("/kaggle/working/Agumented_Drones/train/labels")
os.makedirs("/kaggle/working/Agumented_Drones/val/images")
os.makedirs("/kaggle/working/Agumented_Drones/val/labels")
os.makedirs("/kaggle/working/Agumented_Drones/test/images")
os.makedirs("/kaggle/working/Agumented_Drones/test/labels")
def copy_from_to(src_path: str, dst_path: str, file_ext: str) -> None:
for file_name in os.listdir(src_path):
if file_name.endswith(f".{file_ext}"):
src_file_path = os.path.join(src_path, file_name)
dst_file_path = os.path.join(dst_path, file_name)
shutil.copy(src_file_path, dst_file_path)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/train/images",
"/kaggle/working/Agumented_Drones/train/images/",
"JPEG",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/val/images",
"/kaggle/working/Agumented_Drones/val/images/",
"JPEG",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/test/images",
"/kaggle/working/Agumented_Drones/test/images/",
"JPEG",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/train/labels",
"/kaggle/working/Agumented_Drones/train/labels/",
"txt",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/val/labels",
"/kaggle/working/Agumented_Drones/val/labels/",
"txt",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/test/labels",
"/kaggle/working/Agumented_Drones/test/labels/",
"txt",
)
import os
for dirname, _, filenames in os.walk("/kaggle/working/Agumented_Drones/train/images"):
for filename in filenames:
print(os.path.join(dirname, filename))
apply_augmentation(
"/kaggle/input/drones-data/Drones Data/train/images",
"/kaggle/working/Agumented_Drones/train/images/",
transform=transform,
)
apply_augmentation(
"/kaggle/input/drones-data/Drones Data/val/images",
"/kaggle/working/Agumented_Drones/val/images/",
transform=transform,
)
index = list(set(os.listdir("/kaggle/input/drones-data/Drones Data/train/images")))
img_name = random.choice(index)
img = cv2.imread(
f"/kaggle/input/drones-data/Drones Data/train/images/{img_name}", cv2.IMREAD_COLOR
)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
augmented_image = cv2.imread(
f"/kaggle/working/Agumented_Drones/train/images/{img_name}", cv2.IMREAD_COLOR
)
augmented_image = cv2.cvtColor(augmented_image, cv2.COLOR_BGR2RGB)
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 6))
ax[0].imshow(img)
ax[0].set_title("Original Image")
ax[1].imshow(augmented_image)
ax[1].set_title("Augmented Image")
plt.show()
text = f"""
train: /kaggle/working/Agumented_Drones/train/images/
val: /kaggle/working/Agumented_Drones/val/images/
names:
0: Drone
"""
with open(f"/kaggle/working/Agumented_Drones/Agumented_Drones.yaml", "wt") as f:
f.write(text)
model_2 = YOLO("yolov8n.yaml").load("yolov8n.pt")
model_2.train(
data="/kaggle/working/Agumented_Drones/Agumented_Drones.yaml",
imgsz=(416, 416),
epochs=10,
device=0,
workers=12,
save=True,
plots=True,
)
import glob
from IPython.display import Image, display
for ImageName in glob.glob("/kaggle/working/runs/detect/train/*.jpg"):
display(Image(filename=ImageName))
model_1 = YOLO("/kaggle/working/runs/detect/train/weights/best.pt")
model_1.val()
for ImageName in glob.glob("/kaggle/working/runs/detect/val/*.jpg"):
display(Image(filename=ImageName))
model_2.predict("/kaggle/input/drones-data/Drones Data/test/images", save=True)
for ImageName in glob.glob("/kaggle/working/runs/detect/predict/*.jpeg"):
display(Image(filename=ImageName))
display(Image(filename=ImageName))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/846/129846641.ipynb
|
drones-data
|
yousefelbaz
|
[{"Id": 129846641, "ScriptId": 38618902, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10709551, "CreationDate": "05/16/2023 23:35:58", "VersionNumber": 1.0, "Title": "notebook65401de4c3", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 127.0, "LinesInsertedFromPrevious": 127.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186233857, "KernelVersionId": 129846641, "SourceDatasetVersionId": 5700900}]
|
[{"Id": 5700900, "DatasetId": 3278180, "DatasourceVersionId": 5776569, "CreatorUserId": 10709551, "LicenseName": "Unknown", "CreationDate": "05/16/2023 18:51:06", "VersionNumber": 1.0, "Title": "Drones_Data", "Slug": "drones-data", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3278180, "CreatorUserId": 10709551, "OwnerUserId": 10709551.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5700900.0, "CurrentDatasourceVersionId": 5776569.0, "ForumId": 3343891, "Type": 2, "CreationDate": "05/16/2023 18:51:06", "LastActivityDate": "05/16/2023", "TotalViews": 53, "TotalDownloads": 1, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 10709551, "UserName": "yousefelbaz", "DisplayName": "Yousef Elbaz", "RegisterDate": "06/01/2022", "PerformanceTier": 1}]
|
import os
import shutil
import random
from functools import partial
import tensorflow as tf
import cv2
from PIL import Image
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import albumentations as A
from IPython.display import clear_output, display
clear_output()
import ultralytics
ultralytics.checks()
from ultralytics import YOLO
def apply_augmentation(src_path: str, dst_path: str, transform) -> None:
for img_name in os.listdir(src_path):
img_path = os.path.join(src_path, img_name)
if not os.path.isfile(img_path):
print(f"Skipping {img_name} - file not found.")
continue
img = cv2.imread(img_path, cv2.IMREAD_COLOR)
if img is None:
print(f"Skipping {img_name} - failed to read image.")
continue
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
augmented_image = transform(image=img)["image"]
augmented_image = cv2.cvtColor(augmented_image, cv2.COLOR_RGB2BGR)
dst_file_path = os.path.join(dst_path, img_name)
cv2.imwrite(dst_file_path, augmented_image)
print(f"Saved augmented image: {dst_file_path}")
transform = A.Compose(
[
A.CoarseDropout(
max_holes=16,
max_height=50,
max_width=50,
min_holes=8,
min_height=25,
min_width=25,
fill_value=0,
p=0.5,
),
]
)
BASE_DIR = os.getcwd()
os.makedirs("/kaggle/working/Agumented_Drones/train/images")
os.makedirs("/kaggle/working/Agumented_Drones/train/labels")
os.makedirs("/kaggle/working/Agumented_Drones/val/images")
os.makedirs("/kaggle/working/Agumented_Drones/val/labels")
os.makedirs("/kaggle/working/Agumented_Drones/test/images")
os.makedirs("/kaggle/working/Agumented_Drones/test/labels")
def copy_from_to(src_path: str, dst_path: str, file_ext: str) -> None:
for file_name in os.listdir(src_path):
if file_name.endswith(f".{file_ext}"):
src_file_path = os.path.join(src_path, file_name)
dst_file_path = os.path.join(dst_path, file_name)
shutil.copy(src_file_path, dst_file_path)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/train/images",
"/kaggle/working/Agumented_Drones/train/images/",
"JPEG",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/val/images",
"/kaggle/working/Agumented_Drones/val/images/",
"JPEG",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/test/images",
"/kaggle/working/Agumented_Drones/test/images/",
"JPEG",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/train/labels",
"/kaggle/working/Agumented_Drones/train/labels/",
"txt",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/val/labels",
"/kaggle/working/Agumented_Drones/val/labels/",
"txt",
)
copy_from_to(
"/kaggle/input/drones-data/Drones Data/test/labels",
"/kaggle/working/Agumented_Drones/test/labels/",
"txt",
)
import os
for dirname, _, filenames in os.walk("/kaggle/working/Agumented_Drones/train/images"):
for filename in filenames:
print(os.path.join(dirname, filename))
apply_augmentation(
"/kaggle/input/drones-data/Drones Data/train/images",
"/kaggle/working/Agumented_Drones/train/images/",
transform=transform,
)
apply_augmentation(
"/kaggle/input/drones-data/Drones Data/val/images",
"/kaggle/working/Agumented_Drones/val/images/",
transform=transform,
)
index = list(set(os.listdir("/kaggle/input/drones-data/Drones Data/train/images")))
img_name = random.choice(index)
img = cv2.imread(
f"/kaggle/input/drones-data/Drones Data/train/images/{img_name}", cv2.IMREAD_COLOR
)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
augmented_image = cv2.imread(
f"/kaggle/working/Agumented_Drones/train/images/{img_name}", cv2.IMREAD_COLOR
)
augmented_image = cv2.cvtColor(augmented_image, cv2.COLOR_BGR2RGB)
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 6))
ax[0].imshow(img)
ax[0].set_title("Original Image")
ax[1].imshow(augmented_image)
ax[1].set_title("Augmented Image")
plt.show()
text = f"""
train: /kaggle/working/Agumented_Drones/train/images/
val: /kaggle/working/Agumented_Drones/val/images/
names:
0: Drone
"""
with open(f"/kaggle/working/Agumented_Drones/Agumented_Drones.yaml", "wt") as f:
f.write(text)
model_2 = YOLO("yolov8n.yaml").load("yolov8n.pt")
model_2.train(
data="/kaggle/working/Agumented_Drones/Agumented_Drones.yaml",
imgsz=(416, 416),
epochs=10,
device=0,
workers=12,
save=True,
plots=True,
)
import glob
from IPython.display import Image, display
for ImageName in glob.glob("/kaggle/working/runs/detect/train/*.jpg"):
display(Image(filename=ImageName))
model_1 = YOLO("/kaggle/working/runs/detect/train/weights/best.pt")
model_1.val()
for ImageName in glob.glob("/kaggle/working/runs/detect/val/*.jpg"):
display(Image(filename=ImageName))
model_2.predict("/kaggle/input/drones-data/Drones Data/test/images", save=True)
for ImageName in glob.glob("/kaggle/working/runs/detect/predict/*.jpeg"):
display(Image(filename=ImageName))
display(Image(filename=ImageName))
| false | 0 | 1,802 | 0 | 1,826 | 1,802 |
||
129846469
|
import torch
import torch.nn as nn
import torch.optim as optim
import torchvision.datasets as datasets
import torchvision.transforms as transforms
# Loading the training and testing dataset from Keras datasets
# x_train is our training set and x_test is our validation set
# We do not require the y_train/y_test dataset as we are not performing image classification with target labels
(x_train, y_train), (x_test, y_test) = cifar10.load_data()
# Dimensions of each of the sets
print(x_train.shape)
print(x_test.shape)
# Getting the height, width and number of channels from the image
img_dim = x_train.shape[1]
channels = 3
import random
# set the random seed for reproducibility
random.seed(42)
# create a smaller subset of the CIFAR-10 dataset
subset_size = 1500
subset_indices = random.sample(range(len(train_dataset)), subset_size)
train_subset = torch.utils.data.Subset(train_dataset, subset_indices)
test_subset = torch.utils.data.Subset(test_dataset, subset_indices)
# create data loaders for the training and validation subsets
batch_size = 128
train_data_loader = torch.utils.data.DataLoader(
train_subset, batch_size=batch_size, shuffle=True
)
test_data_loader = torch.utils.data.DataLoader(
test_subset, batch_size=batch_size, shuffle=False
)
len(test_data_loader.dataset)
len(subset_indices)
# define the autoencoder architecture
class Autoencoder(nn.Module):
def __init__(self):
super(Autoencoder, self).__init__()
self.encoder = nn.Sequential(
nn.Conv2d(3, 64, kernel_size=3, stride=1, padding=1),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(64, 128, kernel_size=3, stride=1, padding=1),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(128, 256, kernel_size=3, stride=1, padding=1),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(256, 512, kernel_size=3, stride=1, padding=1),
nn.ReLU(),
)
self.decoder = nn.Sequential(
nn.ConvTranspose2d(
512, 256, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.ReLU(),
nn.ConvTranspose2d(
256, 128, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.ReLU(),
nn.ConvTranspose2d(
128, 64, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.ReLU(),
nn.ConvTranspose2d(64, 3, kernel_size=3, stride=1, padding=1),
nn.Sigmoid(),
)
def forward(self, x):
x = self.encoder(x)
x = self.decoder(x)
return x
# create an instance of the autoencoder
autoencoder = Autoencoder()
# define the loss function and optimizer
criterion = nn.MSELoss()
optimizer = optim.Adam(autoencoder.parameters(), lr=0.001)
# train the autoencoder
num_epochs = 10
for epoch in range(num_epochs):
for data in train_data_loader:
# get the inputs and target outputs
inputs, _ = data
# convert the inputs to grayscale
inputs_gray = (
inputs[:, 0, :, :] * 0.299
+ inputs[:, 1, :, :] * 0.587
+ inputs[:, 2, :, :] * 0.114
)
# convert the inputs to RGB
inputs_rgb = inputs[:, :3, :, :]
# forward pass
outputs = autoencoder(inputs_rgb)
loss = criterion(outputs, inputs_rgb)
# backward pass and optimization
optimizer.zero_grad()
loss.backward()
optimizer.step()
# print the loss after every epoch
print("Epoch [{}/{}], Loss: {:.4f}".format(epoch + 1, num_epochs, loss.item()))
inputs, targets = test_data_loader.dataset[1]
targets
import torch
import matplotlib.pyplot as plt
# switch to evaluation mode
autoencoder.eval()
inputs, targets = test_data_loader.dataset[1]
# convert the inputs to grayscale
inputs_gray = inputs[0] * 0.299 + inputs[1] * 0.587 + inputs[2] * 0.114
# convert the inputs to RGB
inputs_rgb = inputs[:3]
# convert the targets to RGB
targets_rgb = targets[:3]
# pass the inputs through the autoencoder
outputs = autoencoder(inputs_rgb.unsqueeze(0))
# plot the grayscale input, colored ground truth, and colored output images
fig, ax = plt.subplots(1, 3, figsize=(12, 4))
ax[0].imshow(inputs_gray, cmap="gray")
ax[0].set_title("Grayscale Input")
ax[0].axis("off")
ax[1].imshow(targets_rgb.permute(1, 2, 0))
ax[1].set_title("Colored Ground Truth")
ax[1].axis("off")
ax[2].imshow(outputs.detach()[0].permute(1, 2, 0))
ax[2].set_title("Colored Output")
ax[2].axis("off")
plt.show()
# check the size of the test dataset
print("Size of test dataset:", len(test_data_loader.dataset))
# check the index value
print("Index value:", index)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/846/129846469.ipynb
| null | null |
[{"Id": 129846469, "ScriptId": 38523100, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10195219, "CreationDate": "05/16/2023 23:32:39", "VersionNumber": 1.0, "Title": "AutoEncoder", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 144.0, "LinesInsertedFromPrevious": 144.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import torch
import torch.nn as nn
import torch.optim as optim
import torchvision.datasets as datasets
import torchvision.transforms as transforms
# Loading the training and testing dataset from Keras datasets
# x_train is our training set and x_test is our validation set
# We do not require the y_train/y_test dataset as we are not performing image classification with target labels
(x_train, y_train), (x_test, y_test) = cifar10.load_data()
# Dimensions of each of the sets
print(x_train.shape)
print(x_test.shape)
# Getting the height, width and number of channels from the image
img_dim = x_train.shape[1]
channels = 3
import random
# set the random seed for reproducibility
random.seed(42)
# create a smaller subset of the CIFAR-10 dataset
subset_size = 1500
subset_indices = random.sample(range(len(train_dataset)), subset_size)
train_subset = torch.utils.data.Subset(train_dataset, subset_indices)
test_subset = torch.utils.data.Subset(test_dataset, subset_indices)
# create data loaders for the training and validation subsets
batch_size = 128
train_data_loader = torch.utils.data.DataLoader(
train_subset, batch_size=batch_size, shuffle=True
)
test_data_loader = torch.utils.data.DataLoader(
test_subset, batch_size=batch_size, shuffle=False
)
len(test_data_loader.dataset)
len(subset_indices)
# define the autoencoder architecture
class Autoencoder(nn.Module):
def __init__(self):
super(Autoencoder, self).__init__()
self.encoder = nn.Sequential(
nn.Conv2d(3, 64, kernel_size=3, stride=1, padding=1),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(64, 128, kernel_size=3, stride=1, padding=1),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(128, 256, kernel_size=3, stride=1, padding=1),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(256, 512, kernel_size=3, stride=1, padding=1),
nn.ReLU(),
)
self.decoder = nn.Sequential(
nn.ConvTranspose2d(
512, 256, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.ReLU(),
nn.ConvTranspose2d(
256, 128, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.ReLU(),
nn.ConvTranspose2d(
128, 64, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.ReLU(),
nn.ConvTranspose2d(64, 3, kernel_size=3, stride=1, padding=1),
nn.Sigmoid(),
)
def forward(self, x):
x = self.encoder(x)
x = self.decoder(x)
return x
# create an instance of the autoencoder
autoencoder = Autoencoder()
# define the loss function and optimizer
criterion = nn.MSELoss()
optimizer = optim.Adam(autoencoder.parameters(), lr=0.001)
# train the autoencoder
num_epochs = 10
for epoch in range(num_epochs):
for data in train_data_loader:
# get the inputs and target outputs
inputs, _ = data
# convert the inputs to grayscale
inputs_gray = (
inputs[:, 0, :, :] * 0.299
+ inputs[:, 1, :, :] * 0.587
+ inputs[:, 2, :, :] * 0.114
)
# convert the inputs to RGB
inputs_rgb = inputs[:, :3, :, :]
# forward pass
outputs = autoencoder(inputs_rgb)
loss = criterion(outputs, inputs_rgb)
# backward pass and optimization
optimizer.zero_grad()
loss.backward()
optimizer.step()
# print the loss after every epoch
print("Epoch [{}/{}], Loss: {:.4f}".format(epoch + 1, num_epochs, loss.item()))
inputs, targets = test_data_loader.dataset[1]
targets
import torch
import matplotlib.pyplot as plt
# switch to evaluation mode
autoencoder.eval()
inputs, targets = test_data_loader.dataset[1]
# convert the inputs to grayscale
inputs_gray = inputs[0] * 0.299 + inputs[1] * 0.587 + inputs[2] * 0.114
# convert the inputs to RGB
inputs_rgb = inputs[:3]
# convert the targets to RGB
targets_rgb = targets[:3]
# pass the inputs through the autoencoder
outputs = autoencoder(inputs_rgb.unsqueeze(0))
# plot the grayscale input, colored ground truth, and colored output images
fig, ax = plt.subplots(1, 3, figsize=(12, 4))
ax[0].imshow(inputs_gray, cmap="gray")
ax[0].set_title("Grayscale Input")
ax[0].axis("off")
ax[1].imshow(targets_rgb.permute(1, 2, 0))
ax[1].set_title("Colored Ground Truth")
ax[1].axis("off")
ax[2].imshow(outputs.detach()[0].permute(1, 2, 0))
ax[2].set_title("Colored Output")
ax[2].axis("off")
plt.show()
# check the size of the test dataset
print("Size of test dataset:", len(test_data_loader.dataset))
# check the index value
print("Index value:", index)
| false | 0 | 1,468 | 0 | 1,468 | 1,468 |
||
129831360
|
from IPython.display import Image, display
import matplotlib.pyplot as plt
import matplotlib.gridspec as grid
import pandas as pd
import seaborn as sns
import cv2
import numpy as np
# Read RGB image into an array
img = cv2.imread("/kaggle/input/satelite-images/band321.jpg")
img_shape = img.shape[:2]
print("image size = ", img_shape)
# specify no of bands in the image
n_bands = 7
# 3 dimensional dummy array with zeros
MB_img = np.zeros((img_shape[0], img_shape[1], n_bands))
# stacking up images into the array
for i in range(n_bands):
MB_img[:, :, i] = cv2.imread(
"/kaggle/input/satelite-images/band" + str(i + 1) + ".jpg", cv2.IMREAD_GRAYSCALE
)
# Let's take a look at scene
# print('\n\nDispalying colour image of the scene')
# plt.figure(figsize=(img_shape[0]/100,img_shape[1]/100))
plt.imshow(MB_img[:, :, :3][:, :, [2, 1, 0]].astype(int))
plt.axis("off")
fig, axes = plt.subplots(2, 4, figsize=(50, 23), sharex="all", sharey="all")
fig.subplots_adjust(wspace=0.1, hspace=0.15)
fig.suptitle("Intensidades luminosas nas Bandas RGB e Infra-vermelho", fontsize=30)
axes = axes.ravel()
for i in range(n_bands):
axes[i].imshow(MB_img[:, :, i], cmap="gray", vmin=0, vmax=255)
axes[i].set_title("band " + str(i + 1), fontsize=25)
axes[i].axis("off")
fig.delaxes(axes[-1])
# Convert 2d band array in 1-d to make them as feature vectors and Standardization
MB_matrix = np.zeros((MB_img[:, :, 0].size, n_bands))
for i in range(n_bands):
MB_array = MB_img[:, :, i].flatten() # covert 2d to 1d array
MB_arrayStd = (MB_array - MB_array.mean()) / MB_array.std()
MB_matrix[:, i] = MB_arrayStd
MB_matrix.shape
# Covariance
np.set_printoptions(precision=3)
cov = np.cov(MB_matrix.transpose())
# Eigen Values
EigVal, EigVec = np.linalg.eig(cov)
print("Matriz de Covariância:\n\n", cov, "\n")
print("Autovalores:\n\n", EigVal, "\n")
print("Auovetores:\n\n", EigVec, "\n")
# Ordering Eigen values and vectors
order = EigVal.argsort()[::-1]
EigVal = EigVal[order]
EigVec = EigVec[:, order]
# Projecting data on Eigen vector directions resulting to Principal Components
PC = np.matmul(MB_matrix, EigVec) # cross product
# Generate Pairplot for original data and transformed PCs
Bandnames = ["Band 1", "Band 2", "Band 3", "Band 4", "Band 5", "Band 6", "Band 7"]
a = sns.pairplot(
pd.DataFrame(MB_matrix, columns=Bandnames), diag_kind="kde", plot_kws={"s": 3}
)
a.fig.suptitle("Band Images Pairplot")
PCnames = ["PC 1", "PC 2", "PC 3", "PC 4", "PC 5", "PC 6", "PC 7"]
b = sns.pairplot(pd.DataFrame(PC, columns=PCnames), diag_kind="kde", plot_kws={"s": 3})
b.fig.suptitle("PCs Pairplot")
# Information Retained by Principal Components
plt.figure(figsize=(8, 6))
plt.bar(
[1, 2, 3, 4, 5, 6, 7],
EigVal / sum(EigVal) * 100,
align="center",
width=0.4,
tick_label=["PC1", "PC2", "PC3", "PC4", "PC5", "PC6", "PC7"],
)
plt.ylabel("Variância (%)")
plt.title("Retenção de Informação")
# Rearranging 1-d arrays to 2-d arrays of image size
PC_2d = np.zeros((img_shape[0], img_shape[1], n_bands))
for i in range(n_bands):
PC_2d[:, :, i] = PC[:, i].reshape(-1, img_shape[1])
# normalizing between 0 to 255
PC_2d_Norm = np.zeros((img_shape[0], img_shape[1], n_bands))
for i in range(n_bands):
PC_2d_Norm[:, :, i] = cv2.normalize(
PC_2d[:, :, i], np.zeros(img_shape), 0, 255, cv2.NORM_MINMAX
)
fig, axes = plt.subplots(2, 4, figsize=(50, 23), sharex="all", sharey="all")
fig.subplots_adjust(wspace=0.1, hspace=0.15)
fig.suptitle("Intensidades luminosasa dos PCs", fontsize=30)
axes = axes.ravel()
for i in range(n_bands):
axes[i].imshow(PC_2d_Norm[:, :, i], cmap="gray", vmin=0, vmax=255)
axes[i].set_title("PC " + str(i + 1), fontsize=25)
axes[i].axis("off")
fig.delaxes(axes[-1])
# Comparsion of RGB and Image produced using first three bands
fig, axes = plt.subplots(1, 2, figsize=(50, 23), sharex="all", sharey="all")
fig.subplots_adjust(wspace=0.1, hspace=0.15)
axes[0].imshow(MB_img[:, :, :3][:, :, [2, 1, 0]].astype(int))
axes[0].axis("off")
axes[1].imshow(PC_2d_Norm[:, :, :3][:, :, [0, 1, 2]].astype(int))
axes[1].axis("off")
# # Classificação de Screenshots Phishing e Legítimos
# Importando bibliotécas necessárias
import os
import PIL
import PIL.Image
import pathlib
import pandas as pd
import tensorflow as tf
import tensorflow_datasets as tfds
import time
from sklearn.decomposition import PCA
from sklearn.metrics import (
confusion_matrix,
classification_report,
ConfusionMatrixDisplay,
)
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
roc_auc_score,
average_precision_score,
)
# Setando parâmetros para importação de dataset de treino e teste, resolução e tamanho do batch
batch_size = 10
img_height = 150
img_width = 150
train_dir = "/kaggle/input/phishiris-binary/phishIRIS_DL_Dataset/train"
test_dir = "/kaggle/input/phishiris-binary/phishIRIS_DL_Dataset/val"
# Importando dataset de treino, em escala de cinza com as dimenssões definidas anteriormente
train_ds = tf.keras.utils.image_dataset_from_directory(
train_dir,
color_mode="rgb",
seed=123,
image_size=(img_height, img_width),
batch_size=batch_size,
)
# Importando dataset de teste, em escala de cinza com as dimenssões definidas anteriormente
test_ds = tf.keras.utils.image_dataset_from_directory(
test_dir,
color_mode="rgb",
seed=123,
image_size=(img_height, img_width),
batch_size=batch_size,
)
class_names = train_ds.class_names
print(class_names)
# Exibindo exemplos do dataset de treino
plt.figure(figsize=(10, 10))
for images, labels in train_ds.take(1):
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(images[i].numpy().astype("uint8"))
plt.title(class_names[labels[i]])
plt.axis("off")
# Transformando dataset de imagens em arrays numpy de features e rótulos
train_data = list(train_ds)
X_train = np.concatenate([train_data[n][0] for n in range(0, len(train_data))])
y_train = np.concatenate([train_data[n][1] for n in range(0, len(train_data))])
# Transformando dataset de imagens em arrays numpy de features e rótulos
test_data = list(test_ds)
X_test = np.concatenate([test_data[n][0] for n in range(0, len(test_data))])
y_test = np.concatenate([test_data[n][1] for n in range(0, len(test_data))])
X_train.shape, X_test.shape
best = tf.keras.callbacks.ModelCheckpoint(
filepath="/kaggle/working/best_model",
save_weights_only=False,
monitor="val_accuracy",
mode="max",
save_best_only=True,
)
# Carregando hyperparâmetros de regularizadores, otimizador e função de ativação
l1l2 = tf.keras.regularizers.l1_l2(l1=0.00001, l2=0.0001)
opt = tf.keras.optimizers.Adam(learning_rate=0.0001)
fx = "relu"
num_classes = 2
model = tf.keras.Sequential(
[
tf.keras.layers.Rescaling(1.0 / 255, input_shape=(img_height, img_width, 3)),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(64, activation=fx),
tf.keras.layers.Dense(32, activation=fx),
tf.keras.layers.Dense(16, activation=fx),
tf.keras.layers.Dense(num_classes, activation="softmax"),
]
)
model.compile(optimizer=opt, loss="sparse_categorical_crossentropy", metrics="accuracy")
train_time_in = time.time()
model.fit(
X_train, y_train, callbacks=[best], validation_split=0.2, batch_size=10, epochs=20
)
train_time_fin = time.time()
train_time = train_time_fin - train_time_in
print("Tempo de treinamento", train_time)
model.load_weights("/kaggle/working/best_model")
predictions = model.predict(X_test)
print("Matriz de Confusão")
print(confusion_matrix(y_test, predictions.argmax(axis=1)))
print("Relatório de Classificação")
print(classification_report(y_test, predictions.argmax(axis=1), digits=4))
print("ROC AUC")
print(roc_auc_score(y_test, predictions.argmax(axis=1)))
# ## Aplicando PCA aos datasets
# Exibindo exemplos do dataset
plt.figure(figsize=(15, 15))
for i in range(3):
ax = plt.subplot(1, 3, i + 1)
plt.imshow(X_test[0, :, :, i], cmap="gray", vmin=0, vmax=255)
plt.axis("off")
X_train = np.reshape(X_train, (1813, 22500, 3))
X_test = np.reshape(X_test, (1039, 22500, 3))
X_train.shape, X_test.shape
PC_X_train = []
# Covariance
np.set_printoptions(precision=3)
for i in range(1813):
cov = np.cov(X_train[i].transpose())
# Eigen Values
EigVal, EigVec = np.linalg.eig(cov)
# Ordering Eigen values and vectors
order = EigVal.argsort()[::-1]
EigVal = EigVal[order]
EigVec = EigVec[:, order]
# Projecting data on Eigen vector directions resulting to Principal Components
PC = np.matmul(X_train[i], EigVec) # cross product
c = np.zeros((22500, 3))
for k in range(3):
c[:, k] = (255 * (PC[:, k] - np.min(PC[:, k])) / np.ptp(PC[:, k])).astype(float)
PC_X_train.append(c)
PC_X_train = np.array(PC_X_train)
PC_X_train.shape
PC_X_train = np.reshape(PC_X_train, (1813, 150, 150, 3))
PC_X_train.shape
PC_X_test = []
# Covariance
np.set_printoptions(precision=3)
for i in range(1039):
cov = np.cov(X_test[i].transpose())
# Eigen Values
EigVal, EigVec = np.linalg.eig(cov)
# Ordering Eigen values and vectors
order = EigVal.argsort()[::-1]
EigVal = EigVal[order]
EigVec = EigVec[:, order]
# Projecting data on Eigen vector directions resulting to Principal Components
PC = np.matmul(X_test[i], EigVec) # cross product
c = np.zeros((22500, 3))
for k in range(3):
c[:, k] = (255 * (PC[:, k] - np.min(PC[:, k])) / np.ptp(PC[:, k])).astype(float)
PC_X_test.append(c)
PC_X_test = np.array(PC_X_test)
PC_X_test.shape
PC_X_test = np.reshape(PC_X_test, (1039, 150, 150, 3))
PC_X_test.shape
# Exibindo exemplos do dataset de teste
plt.figure(figsize=(15, 15))
for i in range(3):
ax = plt.subplot(1, 3, i + 1)
plt.imshow(PC_X_test[0, :, :, i], cmap="gray", vmin=0, vmax=255)
plt.axis("off")
best_pca = tf.keras.callbacks.ModelCheckpoint(
filepath="/kaggle/working/best_model_pca",
save_weights_only=False,
monitor="val_accuracy",
mode="max",
save_best_only=True,
)
# Carregando hyperparâmetros de regularizadores, otimizador e função de ativação
l1l2 = tf.keras.regularizers.l1_l2(l1=0.00001, l2=0.0001)
opt = tf.keras.optimizers.Adam(learning_rate=0.0001)
fx = "relu"
num_classes = 2
model_pca = tf.keras.Sequential(
[
tf.keras.layers.Rescaling(1.0 / 255, input_shape=(img_height, img_width, 1)),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(64, activation=fx),
tf.keras.layers.Dense(32, activation=fx),
tf.keras.layers.Dense(16, activation=fx),
tf.keras.layers.Dense(num_classes, activation="softmax"),
]
)
model_pca.compile(
optimizer=opt, loss="sparse_categorical_crossentropy", metrics="accuracy"
)
train_time_in = time.time()
model_pca.fit(
PC_X_train[:, :, :, 0],
y_train,
callbacks=[best_pca],
validation_split=0.2,
epochs=20,
)
train_time_fin = time.time()
train_time = train_time_fin - train_time_in
print("Tempo de treinamento", train_time)
model_pca.load_weights("/kaggle/working/best_model_pca")
predictions = model_pca.predict(PC_X_test[:, :, :, 0])
print("Matriz de Confusão")
print(confusion_matrix(y_test, predictions.argmax(axis=1)))
print("Relatório de Classificação")
print(classification_report(y_test, predictions.argmax(axis=1), digits=4))
print("ROC AUC")
print(roc_auc_score(y_test, predictions.argmax(axis=1)))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/831/129831360.ipynb
| null | null |
[{"Id": 129831360, "ScriptId": 38369275, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9521468, "CreationDate": "05/16/2023 19:48:12", "VersionNumber": 1.0, "Title": "PPGCC 2023 - PID - PCA NA DESCRI\u00c7\u00c3O", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 371.0, "LinesInsertedFromPrevious": 371.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
from IPython.display import Image, display
import matplotlib.pyplot as plt
import matplotlib.gridspec as grid
import pandas as pd
import seaborn as sns
import cv2
import numpy as np
# Read RGB image into an array
img = cv2.imread("/kaggle/input/satelite-images/band321.jpg")
img_shape = img.shape[:2]
print("image size = ", img_shape)
# specify no of bands in the image
n_bands = 7
# 3 dimensional dummy array with zeros
MB_img = np.zeros((img_shape[0], img_shape[1], n_bands))
# stacking up images into the array
for i in range(n_bands):
MB_img[:, :, i] = cv2.imread(
"/kaggle/input/satelite-images/band" + str(i + 1) + ".jpg", cv2.IMREAD_GRAYSCALE
)
# Let's take a look at scene
# print('\n\nDispalying colour image of the scene')
# plt.figure(figsize=(img_shape[0]/100,img_shape[1]/100))
plt.imshow(MB_img[:, :, :3][:, :, [2, 1, 0]].astype(int))
plt.axis("off")
fig, axes = plt.subplots(2, 4, figsize=(50, 23), sharex="all", sharey="all")
fig.subplots_adjust(wspace=0.1, hspace=0.15)
fig.suptitle("Intensidades luminosas nas Bandas RGB e Infra-vermelho", fontsize=30)
axes = axes.ravel()
for i in range(n_bands):
axes[i].imshow(MB_img[:, :, i], cmap="gray", vmin=0, vmax=255)
axes[i].set_title("band " + str(i + 1), fontsize=25)
axes[i].axis("off")
fig.delaxes(axes[-1])
# Convert 2d band array in 1-d to make them as feature vectors and Standardization
MB_matrix = np.zeros((MB_img[:, :, 0].size, n_bands))
for i in range(n_bands):
MB_array = MB_img[:, :, i].flatten() # covert 2d to 1d array
MB_arrayStd = (MB_array - MB_array.mean()) / MB_array.std()
MB_matrix[:, i] = MB_arrayStd
MB_matrix.shape
# Covariance
np.set_printoptions(precision=3)
cov = np.cov(MB_matrix.transpose())
# Eigen Values
EigVal, EigVec = np.linalg.eig(cov)
print("Matriz de Covariância:\n\n", cov, "\n")
print("Autovalores:\n\n", EigVal, "\n")
print("Auovetores:\n\n", EigVec, "\n")
# Ordering Eigen values and vectors
order = EigVal.argsort()[::-1]
EigVal = EigVal[order]
EigVec = EigVec[:, order]
# Projecting data on Eigen vector directions resulting to Principal Components
PC = np.matmul(MB_matrix, EigVec) # cross product
# Generate Pairplot for original data and transformed PCs
Bandnames = ["Band 1", "Band 2", "Band 3", "Band 4", "Band 5", "Band 6", "Band 7"]
a = sns.pairplot(
pd.DataFrame(MB_matrix, columns=Bandnames), diag_kind="kde", plot_kws={"s": 3}
)
a.fig.suptitle("Band Images Pairplot")
PCnames = ["PC 1", "PC 2", "PC 3", "PC 4", "PC 5", "PC 6", "PC 7"]
b = sns.pairplot(pd.DataFrame(PC, columns=PCnames), diag_kind="kde", plot_kws={"s": 3})
b.fig.suptitle("PCs Pairplot")
# Information Retained by Principal Components
plt.figure(figsize=(8, 6))
plt.bar(
[1, 2, 3, 4, 5, 6, 7],
EigVal / sum(EigVal) * 100,
align="center",
width=0.4,
tick_label=["PC1", "PC2", "PC3", "PC4", "PC5", "PC6", "PC7"],
)
plt.ylabel("Variância (%)")
plt.title("Retenção de Informação")
# Rearranging 1-d arrays to 2-d arrays of image size
PC_2d = np.zeros((img_shape[0], img_shape[1], n_bands))
for i in range(n_bands):
PC_2d[:, :, i] = PC[:, i].reshape(-1, img_shape[1])
# normalizing between 0 to 255
PC_2d_Norm = np.zeros((img_shape[0], img_shape[1], n_bands))
for i in range(n_bands):
PC_2d_Norm[:, :, i] = cv2.normalize(
PC_2d[:, :, i], np.zeros(img_shape), 0, 255, cv2.NORM_MINMAX
)
fig, axes = plt.subplots(2, 4, figsize=(50, 23), sharex="all", sharey="all")
fig.subplots_adjust(wspace=0.1, hspace=0.15)
fig.suptitle("Intensidades luminosasa dos PCs", fontsize=30)
axes = axes.ravel()
for i in range(n_bands):
axes[i].imshow(PC_2d_Norm[:, :, i], cmap="gray", vmin=0, vmax=255)
axes[i].set_title("PC " + str(i + 1), fontsize=25)
axes[i].axis("off")
fig.delaxes(axes[-1])
# Comparsion of RGB and Image produced using first three bands
fig, axes = plt.subplots(1, 2, figsize=(50, 23), sharex="all", sharey="all")
fig.subplots_adjust(wspace=0.1, hspace=0.15)
axes[0].imshow(MB_img[:, :, :3][:, :, [2, 1, 0]].astype(int))
axes[0].axis("off")
axes[1].imshow(PC_2d_Norm[:, :, :3][:, :, [0, 1, 2]].astype(int))
axes[1].axis("off")
# # Classificação de Screenshots Phishing e Legítimos
# Importando bibliotécas necessárias
import os
import PIL
import PIL.Image
import pathlib
import pandas as pd
import tensorflow as tf
import tensorflow_datasets as tfds
import time
from sklearn.decomposition import PCA
from sklearn.metrics import (
confusion_matrix,
classification_report,
ConfusionMatrixDisplay,
)
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
roc_auc_score,
average_precision_score,
)
# Setando parâmetros para importação de dataset de treino e teste, resolução e tamanho do batch
batch_size = 10
img_height = 150
img_width = 150
train_dir = "/kaggle/input/phishiris-binary/phishIRIS_DL_Dataset/train"
test_dir = "/kaggle/input/phishiris-binary/phishIRIS_DL_Dataset/val"
# Importando dataset de treino, em escala de cinza com as dimenssões definidas anteriormente
train_ds = tf.keras.utils.image_dataset_from_directory(
train_dir,
color_mode="rgb",
seed=123,
image_size=(img_height, img_width),
batch_size=batch_size,
)
# Importando dataset de teste, em escala de cinza com as dimenssões definidas anteriormente
test_ds = tf.keras.utils.image_dataset_from_directory(
test_dir,
color_mode="rgb",
seed=123,
image_size=(img_height, img_width),
batch_size=batch_size,
)
class_names = train_ds.class_names
print(class_names)
# Exibindo exemplos do dataset de treino
plt.figure(figsize=(10, 10))
for images, labels in train_ds.take(1):
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(images[i].numpy().astype("uint8"))
plt.title(class_names[labels[i]])
plt.axis("off")
# Transformando dataset de imagens em arrays numpy de features e rótulos
train_data = list(train_ds)
X_train = np.concatenate([train_data[n][0] for n in range(0, len(train_data))])
y_train = np.concatenate([train_data[n][1] for n in range(0, len(train_data))])
# Transformando dataset de imagens em arrays numpy de features e rótulos
test_data = list(test_ds)
X_test = np.concatenate([test_data[n][0] for n in range(0, len(test_data))])
y_test = np.concatenate([test_data[n][1] for n in range(0, len(test_data))])
X_train.shape, X_test.shape
best = tf.keras.callbacks.ModelCheckpoint(
filepath="/kaggle/working/best_model",
save_weights_only=False,
monitor="val_accuracy",
mode="max",
save_best_only=True,
)
# Carregando hyperparâmetros de regularizadores, otimizador e função de ativação
l1l2 = tf.keras.regularizers.l1_l2(l1=0.00001, l2=0.0001)
opt = tf.keras.optimizers.Adam(learning_rate=0.0001)
fx = "relu"
num_classes = 2
model = tf.keras.Sequential(
[
tf.keras.layers.Rescaling(1.0 / 255, input_shape=(img_height, img_width, 3)),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(64, activation=fx),
tf.keras.layers.Dense(32, activation=fx),
tf.keras.layers.Dense(16, activation=fx),
tf.keras.layers.Dense(num_classes, activation="softmax"),
]
)
model.compile(optimizer=opt, loss="sparse_categorical_crossentropy", metrics="accuracy")
train_time_in = time.time()
model.fit(
X_train, y_train, callbacks=[best], validation_split=0.2, batch_size=10, epochs=20
)
train_time_fin = time.time()
train_time = train_time_fin - train_time_in
print("Tempo de treinamento", train_time)
model.load_weights("/kaggle/working/best_model")
predictions = model.predict(X_test)
print("Matriz de Confusão")
print(confusion_matrix(y_test, predictions.argmax(axis=1)))
print("Relatório de Classificação")
print(classification_report(y_test, predictions.argmax(axis=1), digits=4))
print("ROC AUC")
print(roc_auc_score(y_test, predictions.argmax(axis=1)))
# ## Aplicando PCA aos datasets
# Exibindo exemplos do dataset
plt.figure(figsize=(15, 15))
for i in range(3):
ax = plt.subplot(1, 3, i + 1)
plt.imshow(X_test[0, :, :, i], cmap="gray", vmin=0, vmax=255)
plt.axis("off")
X_train = np.reshape(X_train, (1813, 22500, 3))
X_test = np.reshape(X_test, (1039, 22500, 3))
X_train.shape, X_test.shape
PC_X_train = []
# Covariance
np.set_printoptions(precision=3)
for i in range(1813):
cov = np.cov(X_train[i].transpose())
# Eigen Values
EigVal, EigVec = np.linalg.eig(cov)
# Ordering Eigen values and vectors
order = EigVal.argsort()[::-1]
EigVal = EigVal[order]
EigVec = EigVec[:, order]
# Projecting data on Eigen vector directions resulting to Principal Components
PC = np.matmul(X_train[i], EigVec) # cross product
c = np.zeros((22500, 3))
for k in range(3):
c[:, k] = (255 * (PC[:, k] - np.min(PC[:, k])) / np.ptp(PC[:, k])).astype(float)
PC_X_train.append(c)
PC_X_train = np.array(PC_X_train)
PC_X_train.shape
PC_X_train = np.reshape(PC_X_train, (1813, 150, 150, 3))
PC_X_train.shape
PC_X_test = []
# Covariance
np.set_printoptions(precision=3)
for i in range(1039):
cov = np.cov(X_test[i].transpose())
# Eigen Values
EigVal, EigVec = np.linalg.eig(cov)
# Ordering Eigen values and vectors
order = EigVal.argsort()[::-1]
EigVal = EigVal[order]
EigVec = EigVec[:, order]
# Projecting data on Eigen vector directions resulting to Principal Components
PC = np.matmul(X_test[i], EigVec) # cross product
c = np.zeros((22500, 3))
for k in range(3):
c[:, k] = (255 * (PC[:, k] - np.min(PC[:, k])) / np.ptp(PC[:, k])).astype(float)
PC_X_test.append(c)
PC_X_test = np.array(PC_X_test)
PC_X_test.shape
PC_X_test = np.reshape(PC_X_test, (1039, 150, 150, 3))
PC_X_test.shape
# Exibindo exemplos do dataset de teste
plt.figure(figsize=(15, 15))
for i in range(3):
ax = plt.subplot(1, 3, i + 1)
plt.imshow(PC_X_test[0, :, :, i], cmap="gray", vmin=0, vmax=255)
plt.axis("off")
best_pca = tf.keras.callbacks.ModelCheckpoint(
filepath="/kaggle/working/best_model_pca",
save_weights_only=False,
monitor="val_accuracy",
mode="max",
save_best_only=True,
)
# Carregando hyperparâmetros de regularizadores, otimizador e função de ativação
l1l2 = tf.keras.regularizers.l1_l2(l1=0.00001, l2=0.0001)
opt = tf.keras.optimizers.Adam(learning_rate=0.0001)
fx = "relu"
num_classes = 2
model_pca = tf.keras.Sequential(
[
tf.keras.layers.Rescaling(1.0 / 255, input_shape=(img_height, img_width, 1)),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(64, activation=fx),
tf.keras.layers.Dense(32, activation=fx),
tf.keras.layers.Dense(16, activation=fx),
tf.keras.layers.Dense(num_classes, activation="softmax"),
]
)
model_pca.compile(
optimizer=opt, loss="sparse_categorical_crossentropy", metrics="accuracy"
)
train_time_in = time.time()
model_pca.fit(
PC_X_train[:, :, :, 0],
y_train,
callbacks=[best_pca],
validation_split=0.2,
epochs=20,
)
train_time_fin = time.time()
train_time = train_time_fin - train_time_in
print("Tempo de treinamento", train_time)
model_pca.load_weights("/kaggle/working/best_model_pca")
predictions = model_pca.predict(PC_X_test[:, :, :, 0])
print("Matriz de Confusão")
print(confusion_matrix(y_test, predictions.argmax(axis=1)))
print("Relatório de Classificação")
print(classification_report(y_test, predictions.argmax(axis=1), digits=4))
print("ROC AUC")
print(roc_auc_score(y_test, predictions.argmax(axis=1)))
| false | 0 | 4,226 | 0 | 4,226 | 4,226 |
||
129831240
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
df = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
df.head()
df.tail()
df.shape
df.info()
df.describe()
df.columns
df.hist(figsize=(15, 15))
plt.show()
df.isnull().sum()
df = df.drop_duplicates()
df.head()
X = df.drop(["id", "yield"], axis=1).to_numpy()
y = df["yield"].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
rf = RandomForestRegressor(
n_estimators=288,
random_state=294,
criterion="absolute_error",
max_depth=8,
min_samples_leaf=4,
)
rf.fit(X_train, y_train)
rf_pred = rf.predict(X_test)
print("Mean Absolute Error (MAE):", mean_absolute_error(y_test, rf_pred))
data = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
data.head()
data.shape
Z = data.drop("id", axis=1).to_numpy()
zRF = rf.predict(Z)
dataRF = pd.concat(
[data["id"], pd.DataFrame(zRF, columns=["yield"])], axis=1, join="inner"
)
dataRF.head()
dataRF.to_csv("randomForestData.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/831/129831240.ipynb
| null | null |
[{"Id": 129831240, "ScriptId": 38613284, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13361181, "CreationDate": "05/16/2023 19:46:59", "VersionNumber": 1.0, "Title": "Prediction of Wild Blueberry Yield(Random Forest)", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 62.0, "LinesInsertedFromPrevious": 62.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
| null | null | null | null |
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
df = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
df.head()
df.tail()
df.shape
df.info()
df.describe()
df.columns
df.hist(figsize=(15, 15))
plt.show()
df.isnull().sum()
df = df.drop_duplicates()
df.head()
X = df.drop(["id", "yield"], axis=1).to_numpy()
y = df["yield"].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
rf = RandomForestRegressor(
n_estimators=288,
random_state=294,
criterion="absolute_error",
max_depth=8,
min_samples_leaf=4,
)
rf.fit(X_train, y_train)
rf_pred = rf.predict(X_test)
print("Mean Absolute Error (MAE):", mean_absolute_error(y_test, rf_pred))
data = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
data.head()
data.shape
Z = data.drop("id", axis=1).to_numpy()
zRF = rf.predict(Z)
dataRF = pd.concat(
[data["id"], pd.DataFrame(zRF, columns=["yield"])], axis=1, join="inner"
)
dataRF.head()
dataRF.to_csv("randomForestData.csv", index=False)
| false | 0 | 447 | 2 | 447 | 447 |
||
129831479
|
## THIS IS FOR DENOISING
import gradio as gr
from IPython.display import Image
from PIL import Image
import tensorflow as tf
import tensorflow_hub as hub
import numpy as np
import requests
import cv2
def resize_image(image, target_dim):
shape = tf.cast(tf.shape(image)[1:-1], tf.float32)
short_dim = min(shape)
scale = target_dim / short_dim
new_shape = tf.cast(shape * scale, tf.int32)
image = tf.image.resize(image, new_shape)
image = tf.image.resize_with_crop_or_pad(image, target_dim, target_dim)
return image
def process_image(image_path, target_dim=256):
input_img = cv2.cvtColor(image_path, cv2.COLOR_BGR2RGB)
input_img = np.asarray(input_img, np.float32) / 255.0
input_img = tf.expand_dims(input_img, axis=0)
input_img = resize_image(input_img, target_dim)
return input_img
def infer(image_path: str, model: tf.keras.Model, input_resolution=(256, 256)):
preprocessed_image = process_image(image_path, input_resolution[0])
preds = model.predict(preprocessed_image)
if isinstance(preds, list):
preds = preds[-1]
if isinstance(preds, list):
preds = preds[-1]
preds = np.array(preds[0], np.float32)
final_pred_image = np.array((np.clip(preds, 0.0, 1.0)).astype(np.float32))
return final_pred_image
input_resolution = (256, 256)
def get_model(model_url: str, input_resolution: tuple) -> tf.keras.Model:
inputs = tf.keras.Input((*input_resolution, 3))
hub_module = hub.KerasLayer(model_url)
outputs = hub_module(inputs)
return tf.keras.Model(inputs, outputs)
denoise_model = get_model("/kaggle/input/denoising-graph", input_resolution)
def predict_image(image):
return infer(image, denoise_model, input_resolution)
image = gr.inputs.Image()
irface = gr.Interface(
fn=predict_image, inputs=image, outputs=image, interpretation="default"
)
irface.launch(share=True)
##THIS IS FOR SUPER-RESOLUTION
import gradio as gr
from IPython.display import Image
from PIL import Image
import tensorflow as tf
import tensorflow_hub as hub
import numpy as np
import requests
import cv2
from tensorflow.python.keras.layers import Add, Conv2D, Input, Lambda
from tensorflow.python.keras.models import Model
super_resolution = "/kaggle/input/super-weight-res/weights/edsr-16-x4/weights.h5"
pre_mean = np.array([0.4488, 0.4371, 0.4040]) * 255
# HELPER FUN
def normalize(x, rgb_mean=pre_mean):
return (x - rgb_mean) / 127.5
# HELPER FUN
def pixel_shuffle(scale):
return lambda x: tf.nn.depth_to_space(x, scale)
# HELPER FUN
def denormalize(x, rgb_mean=pre_mean):
return x * 127.5 + rgb_mean
# MAIN FUN
def res_block(x_in, filters, scaling):
x = Conv2D(filters, 3, padding="same", activation="relu")(x_in)
x = Conv2D(filters, 3, padding="same")(x)
x = tf.keras.layers.LeakyReLU(alpha=0.01)(x)
x = tf.keras.layers.BatchNormalization()(x)
if scaling:
x = Lambda(lambda t: t * scaling)(x)
x = Add()([x_in, x])
return x
# HELPER FUN
def upsample(x, scale, num_filters):
def upsample_1(x, factor, **kwargs):
x = Conv2D(num_filters * (factor**2), 3, padding="same", **kwargs)(x)
return Lambda(pixel_shuffle(scale=factor))(x)
if scale == 2:
x = upsample_1(x, 2, name="conv2d_1_scale_2")
elif scale == 3:
x = upsample_1(x, 3, name="conv2d_1_scale_3")
elif scale == 4:
x = upsample_1(x, 2, name="conv2d_1_scale_2")
x = upsample_1(x, 2, name="conv2d_2_scale_2")
return x
# MAIN FUN
def super_res(scale, num_filters=64, num_res_blocks=8, res_block_scaling=None):
x_in = Input(shape=(None, None, 3))
x = Lambda(normalize)(x_in)
x = b = Conv2D(num_filters, 3, padding="same")(x)
for i in range(num_res_blocks):
b = res_block(b, num_filters, res_block_scaling)
b = Conv2D(num_filters, 3, padding="same")(b)
x = Add()([x, b])
x = upsample(x, scale, num_filters)
x = Conv2D(3, 3, padding="same")(x)
x = Lambda(denormalize)(x)
return Model(x_in, x, name="super_res")
def load_image(path):
return np.array(path)
def resolve(model, lr_batch):
lr_batch = tf.cast(lr_batch, tf.float32)
sr_batch = model(lr_batch)
sr_batch = tf.clip_by_value(sr_batch, 0, 255)
sr_batch = tf.round(sr_batch)
sr_batch = tf.cast(sr_batch, tf.uint8)
return sr_batch
def resolve_single(model, lr):
return resolve(model, tf.expand_dims(lr, axis=0))[0]
model = super_res(scale=4, num_res_blocks=16)
model.load_weights(super_resolution)
def predict_image(image):
lr = load_image(image)
sr = resolve_single(model, lr)
numpy_array = sr.numpy()
ima = Image.fromarray(numpy_array)
return ima
image = gr.inputs.Image()
irface = gr.Interface(
fn=predict_image, inputs=image, outputs=image, interpretation="default"
)
irface.launch()
## THIS IS FOR DEBLURRING
import gradio as gr
from IPython.display import Image
from PIL import Image
import tensorflow as tf
import tensorflow_hub as hub
import numpy as np
import requests
import cv2
def variance_of_laplacian(frame_path):
# compute the Laplacian of the image and then return the focus
# measure, which is simply the variance of the Laplacian
gray = cv2.cvtColor(frame_path, cv2.COLOR_BGR2GRAY)
gray = cv2.convertScaleAbs(gray)
laplacian = cv2.Laplacian(gray, cv2.CV_64F)
variance = laplacian.var()
return variance
def resize_image(image, target_dim):
shape = tf.cast(tf.shape(image)[1:-1], tf.float32)
short_dim = min(shape)
scale = target_dim / short_dim
new_shape = tf.cast(shape * scale, tf.int32)
image = tf.image.resize(image, new_shape)
image = tf.image.resize_with_crop_or_pad(image, target_dim, target_dim)
return image
def process_image(image_path, target_dim=256):
input_img = cv2.cvtColor(image_path, cv2.COLOR_BGR2RGB)
input_img = np.asarray(input_img, np.float32) / 255.0
input_img = tf.expand_dims(input_img, axis=0)
input_img = resize_image(input_img, target_dim)
return input_img
def infer(image_path: str, model: tf.keras.Model, input_resolution=(256, 256)):
preprocessed_image = process_image(image_path, input_resolution[0])
preds = model.predict(preprocessed_image)
if isinstance(preds, list):
preds = preds[-1]
if isinstance(preds, list):
preds = preds[-1]
preds = np.array(preds[0], np.float32)
final_pred_image = np.array((np.clip(preds, 0.0, 1.0)).astype(np.float32))
return final_pred_image
input_resolution = (256, 256)
def get_model(model_url: str, input_resolution: tuple) -> tf.keras.Model:
inputs = tf.keras.Input((*input_resolution, 3))
hub_module = hub.KerasLayer(model_url)
outputs = hub_module(inputs)
return tf.keras.Model(inputs, outputs)
deblur_model = get_model("/kaggle/input/deblurring-graph", input_resolution)
def predict_image(image):
for i in range(0, 5):
final_pred_image = infer(image, deblur_model, input_resolution)
image = cv2.cvtColor(final_pred_image, cv2.COLOR_RGB2BGR) * 255
return final_pred_image
image = gr.inputs.Image()
irface = gr.Interface(
fn=predict_image, inputs=image, outputs=image, interpretation="default"
)
irface.launch()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/831/129831479.ipynb
| null | null |
[{"Id": 129831479, "ScriptId": 38596201, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8019837, "CreationDate": "05/16/2023 19:49:26", "VersionNumber": 1.0, "Title": "notebook4c2926b81c", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 259.0, "LinesInsertedFromPrevious": 259.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
## THIS IS FOR DENOISING
import gradio as gr
from IPython.display import Image
from PIL import Image
import tensorflow as tf
import tensorflow_hub as hub
import numpy as np
import requests
import cv2
def resize_image(image, target_dim):
shape = tf.cast(tf.shape(image)[1:-1], tf.float32)
short_dim = min(shape)
scale = target_dim / short_dim
new_shape = tf.cast(shape * scale, tf.int32)
image = tf.image.resize(image, new_shape)
image = tf.image.resize_with_crop_or_pad(image, target_dim, target_dim)
return image
def process_image(image_path, target_dim=256):
input_img = cv2.cvtColor(image_path, cv2.COLOR_BGR2RGB)
input_img = np.asarray(input_img, np.float32) / 255.0
input_img = tf.expand_dims(input_img, axis=0)
input_img = resize_image(input_img, target_dim)
return input_img
def infer(image_path: str, model: tf.keras.Model, input_resolution=(256, 256)):
preprocessed_image = process_image(image_path, input_resolution[0])
preds = model.predict(preprocessed_image)
if isinstance(preds, list):
preds = preds[-1]
if isinstance(preds, list):
preds = preds[-1]
preds = np.array(preds[0], np.float32)
final_pred_image = np.array((np.clip(preds, 0.0, 1.0)).astype(np.float32))
return final_pred_image
input_resolution = (256, 256)
def get_model(model_url: str, input_resolution: tuple) -> tf.keras.Model:
inputs = tf.keras.Input((*input_resolution, 3))
hub_module = hub.KerasLayer(model_url)
outputs = hub_module(inputs)
return tf.keras.Model(inputs, outputs)
denoise_model = get_model("/kaggle/input/denoising-graph", input_resolution)
def predict_image(image):
return infer(image, denoise_model, input_resolution)
image = gr.inputs.Image()
irface = gr.Interface(
fn=predict_image, inputs=image, outputs=image, interpretation="default"
)
irface.launch(share=True)
##THIS IS FOR SUPER-RESOLUTION
import gradio as gr
from IPython.display import Image
from PIL import Image
import tensorflow as tf
import tensorflow_hub as hub
import numpy as np
import requests
import cv2
from tensorflow.python.keras.layers import Add, Conv2D, Input, Lambda
from tensorflow.python.keras.models import Model
super_resolution = "/kaggle/input/super-weight-res/weights/edsr-16-x4/weights.h5"
pre_mean = np.array([0.4488, 0.4371, 0.4040]) * 255
# HELPER FUN
def normalize(x, rgb_mean=pre_mean):
return (x - rgb_mean) / 127.5
# HELPER FUN
def pixel_shuffle(scale):
return lambda x: tf.nn.depth_to_space(x, scale)
# HELPER FUN
def denormalize(x, rgb_mean=pre_mean):
return x * 127.5 + rgb_mean
# MAIN FUN
def res_block(x_in, filters, scaling):
x = Conv2D(filters, 3, padding="same", activation="relu")(x_in)
x = Conv2D(filters, 3, padding="same")(x)
x = tf.keras.layers.LeakyReLU(alpha=0.01)(x)
x = tf.keras.layers.BatchNormalization()(x)
if scaling:
x = Lambda(lambda t: t * scaling)(x)
x = Add()([x_in, x])
return x
# HELPER FUN
def upsample(x, scale, num_filters):
def upsample_1(x, factor, **kwargs):
x = Conv2D(num_filters * (factor**2), 3, padding="same", **kwargs)(x)
return Lambda(pixel_shuffle(scale=factor))(x)
if scale == 2:
x = upsample_1(x, 2, name="conv2d_1_scale_2")
elif scale == 3:
x = upsample_1(x, 3, name="conv2d_1_scale_3")
elif scale == 4:
x = upsample_1(x, 2, name="conv2d_1_scale_2")
x = upsample_1(x, 2, name="conv2d_2_scale_2")
return x
# MAIN FUN
def super_res(scale, num_filters=64, num_res_blocks=8, res_block_scaling=None):
x_in = Input(shape=(None, None, 3))
x = Lambda(normalize)(x_in)
x = b = Conv2D(num_filters, 3, padding="same")(x)
for i in range(num_res_blocks):
b = res_block(b, num_filters, res_block_scaling)
b = Conv2D(num_filters, 3, padding="same")(b)
x = Add()([x, b])
x = upsample(x, scale, num_filters)
x = Conv2D(3, 3, padding="same")(x)
x = Lambda(denormalize)(x)
return Model(x_in, x, name="super_res")
def load_image(path):
return np.array(path)
def resolve(model, lr_batch):
lr_batch = tf.cast(lr_batch, tf.float32)
sr_batch = model(lr_batch)
sr_batch = tf.clip_by_value(sr_batch, 0, 255)
sr_batch = tf.round(sr_batch)
sr_batch = tf.cast(sr_batch, tf.uint8)
return sr_batch
def resolve_single(model, lr):
return resolve(model, tf.expand_dims(lr, axis=0))[0]
model = super_res(scale=4, num_res_blocks=16)
model.load_weights(super_resolution)
def predict_image(image):
lr = load_image(image)
sr = resolve_single(model, lr)
numpy_array = sr.numpy()
ima = Image.fromarray(numpy_array)
return ima
image = gr.inputs.Image()
irface = gr.Interface(
fn=predict_image, inputs=image, outputs=image, interpretation="default"
)
irface.launch()
## THIS IS FOR DEBLURRING
import gradio as gr
from IPython.display import Image
from PIL import Image
import tensorflow as tf
import tensorflow_hub as hub
import numpy as np
import requests
import cv2
def variance_of_laplacian(frame_path):
# compute the Laplacian of the image and then return the focus
# measure, which is simply the variance of the Laplacian
gray = cv2.cvtColor(frame_path, cv2.COLOR_BGR2GRAY)
gray = cv2.convertScaleAbs(gray)
laplacian = cv2.Laplacian(gray, cv2.CV_64F)
variance = laplacian.var()
return variance
def resize_image(image, target_dim):
shape = tf.cast(tf.shape(image)[1:-1], tf.float32)
short_dim = min(shape)
scale = target_dim / short_dim
new_shape = tf.cast(shape * scale, tf.int32)
image = tf.image.resize(image, new_shape)
image = tf.image.resize_with_crop_or_pad(image, target_dim, target_dim)
return image
def process_image(image_path, target_dim=256):
input_img = cv2.cvtColor(image_path, cv2.COLOR_BGR2RGB)
input_img = np.asarray(input_img, np.float32) / 255.0
input_img = tf.expand_dims(input_img, axis=0)
input_img = resize_image(input_img, target_dim)
return input_img
def infer(image_path: str, model: tf.keras.Model, input_resolution=(256, 256)):
preprocessed_image = process_image(image_path, input_resolution[0])
preds = model.predict(preprocessed_image)
if isinstance(preds, list):
preds = preds[-1]
if isinstance(preds, list):
preds = preds[-1]
preds = np.array(preds[0], np.float32)
final_pred_image = np.array((np.clip(preds, 0.0, 1.0)).astype(np.float32))
return final_pred_image
input_resolution = (256, 256)
def get_model(model_url: str, input_resolution: tuple) -> tf.keras.Model:
inputs = tf.keras.Input((*input_resolution, 3))
hub_module = hub.KerasLayer(model_url)
outputs = hub_module(inputs)
return tf.keras.Model(inputs, outputs)
deblur_model = get_model("/kaggle/input/deblurring-graph", input_resolution)
def predict_image(image):
for i in range(0, 5):
final_pred_image = infer(image, deblur_model, input_resolution)
image = cv2.cvtColor(final_pred_image, cv2.COLOR_RGB2BGR) * 255
return final_pred_image
image = gr.inputs.Image()
irface = gr.Interface(
fn=predict_image, inputs=image, outputs=image, interpretation="default"
)
irface.launch()
| false | 0 | 2,489 | 0 | 2,489 | 2,489 |
||
129831058
|
# # ICR - Age Related Conditions
import numpy as np
import pandas as pd
pd.set_option("display.max_columns", 50)
import matplotlib.pyplot as plt
import plotly.express as px
import seaborn as sns
sns.set_style("whitegrid")
import warnings
warnings.filterwarnings("ignore")
# ## Read Data
root_dir = "/kaggle/input/icr-identify-age-related-conditions"
train = pd.read_csv(root_dir + "/train.csv")
test = pd.read_csv(root_dir + "/test.csv")
greeks = pd.read_csv(root_dir + "/greeks.csv")
ss = pd.read_csv(root_dir + "/sample_submission.csv")
# print all sets - assuming train,test, ss
print(f"Training Data Shape: {train.shape}")
display(train.head())
print(f"Test Data Shape: {test.shape}")
display(test.head())
print(f"Greek Data Shape: {greeks.shape}")
display(greeks.head())
print(f"Sample Submission Shape: {ss.shape}")
display(ss.head())
# ### Takeaways
# * Very small dataset
# * Train: 617 x 58
# * Test: 5 x 57
# * Sample sub: has id and two pred columns
# * class_0
# * class_1
display(train.info())
# display(test.info())
# ### Takeaways
# * All floats except for id and 'EJ'
# * A few missing values: will need to impute
# ## Look at unique values
plt.figure(figsize=(16, 4))
plot = train.nunique().plot(kind="bar")
for bar in plot.patches:
plot.annotate(
format(bar.get_height(), ".0f"),
(bar.get_x() + bar.get_width() / 2, bar.get_height()),
ha="center",
va="center",
size=6,
xytext=(0, 8),
textcoords="offset points",
)
plt.title("Unique Values per Feature")
# ### Takeaways
# * EJ and Class are binary features
# * Some look possibly categorical (eg BN,DV)
display(train.describe())
# ### Takeaways
# * Mean and std all over the place, may benefit from scaling, look at distributions next
# ## EDA Visualization
features = [col for col in train.columns if train[col].dtype == "float"]
rows = int(np.ceil(len(features) / 3))
plt.figure(figsize=(16, 5 * rows))
for i, col in enumerate(features):
plt.subplot(rows, 3, i + 1)
sns.histplot(train[col], kde=True)
plt.title(col)
# look at binary class EJ
sns.histplot(train.EJ)
plt.title("Plot of Binary Feature EJ")
# ## Violin plots
features = [col for col in train.columns if train[col].dtype == "float"]
rows = int(np.ceil(len(features) / 3))
plt.figure(figsize=(16, 5 * rows))
for i, col in enumerate(features):
plt.subplot(rows, 3, i + 1)
sns.violinplot(x=train.Class, y=train[col], palette="BrBG")
plt.title(col)
# ## Box Plots
features = [col for col in train.columns if train[col].dtype == "float"]
rows = int(np.ceil(len(features) / 3))
plt.figure(figsize=(16, 5 * rows))
for i, col in enumerate(features):
plt.subplot(rows, 3, i + 1)
sns.boxplot(x=train.Class, y=train[col], palette="BrBG")
plt.title(col)
# ### Takeaways
# * Seems like outliers everywhere
# ## Target Distribution
plt.figure()
sns.histplot(train.Class)
plt.title("Distribution of Target")
# ### Takeaways
# * Imbalanced
# * Use stratified kfold
# ## Correlations
plt.figure(figsize=(16, 10))
mask = np.triu(np.ones_like(train.corr(), dtype=bool))
heatmap = sns.heatmap(
train.corr(), mask=mask, vmin=-1, vmax=1, annot=False, cmap="BrBG"
)
heatmap.set_title("Correlation Heatmap", fontdict={"fontsize": 18}, pad=16)
plt.figure(figsize=(4, 12)) # set size by num of features
heatmap = sns.heatmap(
train.corr()[["Class"]].sort_values(by="Class", ascending=False),
vmin=-1,
vmax=1,
annot=True,
cmap="BrBG",
)
heatmap.set_title("Features Correlating with Target", fontdict={"fontsize": 15}, pad=16)
# ## Preprocessing
train = train.drop("Id", axis=1)
test = test.drop("Id", axis=1)
train.EJ = train.apply(lambda x: 0 if x.EJ == "A" else 1, axis=1)
test.EJ = test.apply(lambda x: 0 if x.EJ == "A" else 1, axis=1)
y = train.pop("Class")
train.head()
# ### Imputation
# start with simple fill with mean
train = train.fillna(train.mean())
test = test.fillna(train.mean())
# ### Select features
features = [col for col in test]
# features
# ## Models
from sklearn.model_selection import train_test_split
from sklearn.metrics import log_loss
seed = 12
# go to cv after first test
x_train, x_valid, y_train, y_valid = train_test_split(
train[features], y, train_size=0.8, test_size=0.2, random_state=seed
)
x_test = test[features]
# ### Logistic Regression
from sklearn.linear_model import LogisticRegression
model_lrg = LogisticRegression(
random_state=seed,
solver="liblinear",
penalty="l2",
multi_class="auto",
max_iter=100,
)
model_lrg.fit(x_train, y_train)
pred_lrg = model_lrg.predict_proba(x_valid)[:, 1] # or just predict
score_lrg = log_loss(y_valid, pred_lrg)
print(f"Logloss Score: {score_lrg}")
# default: .44404
# params1: .36558
preds = model_lrg.predict_proba(x_test)
preds
preds[:, 0]
# ## Submission
ss.class_0 = preds[:, 0]
ss.class_1 = preds[:, 1]
ss.head()
ss.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/831/129831058.ipynb
| null | null |
[{"Id": 129831058, "ScriptId": 38609600, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8077052, "CreationDate": "05/16/2023 19:45:03", "VersionNumber": 1.0, "Title": "ICR Age Related Preds EDA and First Model", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 189.0, "LinesInsertedFromPrevious": 189.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # ICR - Age Related Conditions
import numpy as np
import pandas as pd
pd.set_option("display.max_columns", 50)
import matplotlib.pyplot as plt
import plotly.express as px
import seaborn as sns
sns.set_style("whitegrid")
import warnings
warnings.filterwarnings("ignore")
# ## Read Data
root_dir = "/kaggle/input/icr-identify-age-related-conditions"
train = pd.read_csv(root_dir + "/train.csv")
test = pd.read_csv(root_dir + "/test.csv")
greeks = pd.read_csv(root_dir + "/greeks.csv")
ss = pd.read_csv(root_dir + "/sample_submission.csv")
# print all sets - assuming train,test, ss
print(f"Training Data Shape: {train.shape}")
display(train.head())
print(f"Test Data Shape: {test.shape}")
display(test.head())
print(f"Greek Data Shape: {greeks.shape}")
display(greeks.head())
print(f"Sample Submission Shape: {ss.shape}")
display(ss.head())
# ### Takeaways
# * Very small dataset
# * Train: 617 x 58
# * Test: 5 x 57
# * Sample sub: has id and two pred columns
# * class_0
# * class_1
display(train.info())
# display(test.info())
# ### Takeaways
# * All floats except for id and 'EJ'
# * A few missing values: will need to impute
# ## Look at unique values
plt.figure(figsize=(16, 4))
plot = train.nunique().plot(kind="bar")
for bar in plot.patches:
plot.annotate(
format(bar.get_height(), ".0f"),
(bar.get_x() + bar.get_width() / 2, bar.get_height()),
ha="center",
va="center",
size=6,
xytext=(0, 8),
textcoords="offset points",
)
plt.title("Unique Values per Feature")
# ### Takeaways
# * EJ and Class are binary features
# * Some look possibly categorical (eg BN,DV)
display(train.describe())
# ### Takeaways
# * Mean and std all over the place, may benefit from scaling, look at distributions next
# ## EDA Visualization
features = [col for col in train.columns if train[col].dtype == "float"]
rows = int(np.ceil(len(features) / 3))
plt.figure(figsize=(16, 5 * rows))
for i, col in enumerate(features):
plt.subplot(rows, 3, i + 1)
sns.histplot(train[col], kde=True)
plt.title(col)
# look at binary class EJ
sns.histplot(train.EJ)
plt.title("Plot of Binary Feature EJ")
# ## Violin plots
features = [col for col in train.columns if train[col].dtype == "float"]
rows = int(np.ceil(len(features) / 3))
plt.figure(figsize=(16, 5 * rows))
for i, col in enumerate(features):
plt.subplot(rows, 3, i + 1)
sns.violinplot(x=train.Class, y=train[col], palette="BrBG")
plt.title(col)
# ## Box Plots
features = [col for col in train.columns if train[col].dtype == "float"]
rows = int(np.ceil(len(features) / 3))
plt.figure(figsize=(16, 5 * rows))
for i, col in enumerate(features):
plt.subplot(rows, 3, i + 1)
sns.boxplot(x=train.Class, y=train[col], palette="BrBG")
plt.title(col)
# ### Takeaways
# * Seems like outliers everywhere
# ## Target Distribution
plt.figure()
sns.histplot(train.Class)
plt.title("Distribution of Target")
# ### Takeaways
# * Imbalanced
# * Use stratified kfold
# ## Correlations
plt.figure(figsize=(16, 10))
mask = np.triu(np.ones_like(train.corr(), dtype=bool))
heatmap = sns.heatmap(
train.corr(), mask=mask, vmin=-1, vmax=1, annot=False, cmap="BrBG"
)
heatmap.set_title("Correlation Heatmap", fontdict={"fontsize": 18}, pad=16)
plt.figure(figsize=(4, 12)) # set size by num of features
heatmap = sns.heatmap(
train.corr()[["Class"]].sort_values(by="Class", ascending=False),
vmin=-1,
vmax=1,
annot=True,
cmap="BrBG",
)
heatmap.set_title("Features Correlating with Target", fontdict={"fontsize": 15}, pad=16)
# ## Preprocessing
train = train.drop("Id", axis=1)
test = test.drop("Id", axis=1)
train.EJ = train.apply(lambda x: 0 if x.EJ == "A" else 1, axis=1)
test.EJ = test.apply(lambda x: 0 if x.EJ == "A" else 1, axis=1)
y = train.pop("Class")
train.head()
# ### Imputation
# start with simple fill with mean
train = train.fillna(train.mean())
test = test.fillna(train.mean())
# ### Select features
features = [col for col in test]
# features
# ## Models
from sklearn.model_selection import train_test_split
from sklearn.metrics import log_loss
seed = 12
# go to cv after first test
x_train, x_valid, y_train, y_valid = train_test_split(
train[features], y, train_size=0.8, test_size=0.2, random_state=seed
)
x_test = test[features]
# ### Logistic Regression
from sklearn.linear_model import LogisticRegression
model_lrg = LogisticRegression(
random_state=seed,
solver="liblinear",
penalty="l2",
multi_class="auto",
max_iter=100,
)
model_lrg.fit(x_train, y_train)
pred_lrg = model_lrg.predict_proba(x_valid)[:, 1] # or just predict
score_lrg = log_loss(y_valid, pred_lrg)
print(f"Logloss Score: {score_lrg}")
# default: .44404
# params1: .36558
preds = model_lrg.predict_proba(x_test)
preds
preds[:, 0]
# ## Submission
ss.class_0 = preds[:, 0]
ss.class_1 = preds[:, 1]
ss.head()
ss.to_csv("submission.csv", index=False)
| false | 0 | 1,718 | 0 | 1,718 | 1,718 |
||
129831704
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("dark_background")
df = pd.read_csv("/kaggle/input/bgg-dataset/bgg_dataset.csv", sep=";")
df.head()
df.info()
df.columns
df = df[
[
"Name",
"Year Published",
"Min Players",
"Max Players",
"Play Time",
"Min Age",
"Users Rated",
"Rating Average",
"BGG Rank",
"Complexity Average",
"Owned Users",
"Domains",
]
].rename(
columns={
"Year Published": "year",
"Min Players": "min_players",
"Max Players": "max_players",
"Play Time": "play_time",
"Min Age": "min_age",
"Users Rated": "users_rated",
"Rating Average": "avg_rating",
"BGG Rank": "bgg_rank",
"Complexity Average": "comlexity_avg",
"Owned Users": "owned_users",
"Name": "name",
"Mechanics": "mechanics",
"Domains": "domains",
}
)
df.duplicated().sum()
df.shape
df.dtypes
df.isnull().sum()
# # 1. Year published
df.loc[df["year"].isnull()]
df.drop(df[df["name"] == "Hus"].index, inplace=True)
# Get rid of old games of no interest to us
df.drop(df[df["year"] < 1990].index, inplace=True)
df.shape
df["year"] = df["year"].astype(int)
sns.histplot(df["year"])
# # 2. Minimum number of players
df["min_players"].value_counts()
# For 31 games the minimum number of players is 0. It can't be.
# Replace 0 for 2 (cause 2 players is the most popular number of players)
df.loc[df["min_players"] == 0, "min_players"] = 2
# # 3. Maximum players
df["max_players"].describe()
df.loc[df["max_players"] == 0, "max_players"] = df.apply(
lambda row: row["min_players"] if row["min_players"] > 0 else 0, axis=1
)
# If value in column 'max_players' = 0 and in column 'min_players' != 0, the replace by this value (if 'max_players' = 0 and 'min_players' = 2, then 'max_players' = 2 too, for example)
# # 4. Playing Time
df["play_time"].describe()
play_time_mean = df["play_time"].mean()
round(play_time_mean)
df.loc[df["play_time"] == 0, "play_time"] = round(play_time_mean)
df["play_time"] = df["play_time"].astype(int)
df["play_time"].value_counts().sort_values(ascending=False).head(5)
df["play_time"].value_counts().sort_values(ascending=False).head(5).sum() / len(
df["play_time"]
) * 100
# About ~62% of all games are 30 to 90 minutes long
# # 5. Minimal age
df["min_age"].describe()
sns.boxplot(x=df["min_age"])
df.loc[18931, "min_age"] = 16
# Verified the value and changed to the current, according to BoardgameGeek
# # 6. Users rated
df["users_rated"].describe()
df["users_rated"] = df["users_rated"].astype(int)
# # 7. Average rating
df["avg_rating"] = [float(str(i).replace(",", ".")) for i in df["avg_rating"]]
df["avg_rating"].describe()
sns.histplot(df["avg_rating"], kde=True)
# # 8. Complexity Level
df["comlexity_avg"] = [float(str(i).replace(",", ".")) for i in df["comlexity_avg"]]
df["comlexity_avg"].describe()
complexity_median = round(df["comlexity_avg"].dropna().median(), 2)
df.loc[df["comlexity_avg"] == 0, "comlexity_avg"] = complexity_median
# # 9. Owned Users
df["owned_users"].describe()
df["owned_users"].isnull().sum()
owned_median = df["owned_users"].median()
df.loc[df["owned_users"].isnull(), "owned_users"] = owned_median
df["owned_users"] = df["owned_users"].astype(int)
# # 10. Domains
df["domains"].isnull().sum()
df["domains"].dropna().value_counts().head(5)
fig, ax = plt.subplots()
df["domains"].dropna().value_counts().head(5).plot(
kind="pie", autopct="%.1f%%", ax=ax, textprops={"color": "red"}
)
ax.set_title("Percentage of each genre")
ax.yaxis.label.set_visible(False) # remove y-label
ax.title.set_position([0.5, 1.15]) # change title rotation
plt.show()
df.dropna(subset="domains", inplace=True)
df.head(10)
# Average complexity in each genre
df.groupby("domains")["comlexity_avg"].mean()
# Minimum, average and maximum values for each genre in the list
df.groupby("domains").agg({"comlexity_avg": ["min", "mean", "max"]}).reset_index()
# Top 5 popular genres in order of number of games
popular_ganres = df["domains"].dropna().value_counts().head(5)
popular_ganres
pop_ganres = df["domains"].dropna().value_counts().head(5).index.tolist()
# All games of top-5 popular ganres:
t = df[df["domains"].isin(pop_ganres)]
pd.pivot_table(
t, values="comlexity_avg", index="domains", aggfunc=[np.mean, np.max]
).reset_index()
# The most difficult genre of games - Wargames (4.93 max, 2.9 mean)
# Easiest genre of top-5 popular games is Family Games (3.05 max, 1.59 mean)
decade = pd.cut(df["year"], 3, labels=["2011-2021", "2000-2010", "1990-1999"])
# Split df['year'] into three periods
decade.value_counts(normalize=True) * 100
# Percentage distribution of games for each of the three periods
df["decade"] = pd.cut(df["year"], 3, labels=["2011-2021", "2000-2010", "1990-1999"])
def ten_years(v):
if 1990 <= v < 2000:
return "1990-1999"
elif 2000 <= v < 2010:
return "2000-2010"
else:
return "2011-2021"
df["decade"] = df["year"].apply(ten_years)
df.head(10)
df.groupby(["decade", "domains"])["name"].count()
ganres_per_decade = pd.pivot_table(
df, values="name", columns="decade", index="domains", aggfunc="count", fill_value=0
).reset_index()
# How many games were released in each decade by genre:
ganres_per_decade
ganres_per_decade.sort_values("1990-1999", ascending=False).head(10)
ganres_per_decade.sort_values("2000-2010", ascending=False).head(10)
ganres_per_decade.sort_values("2011-2021", ascending=False).head(10)
# Strategies over the last decade are the most released. More wargames and family games.
# The Wargems were released in the 1990s and continue to be released consistently and in large numbers.
# Perhaps wargames are the most constant and are in demand in certain circles
# In the 90s, the second most popular was the Children’s Games(probably that’s where the stereotype came from. board games are kind of fun).
# Family games and strategies are just starting to gather momentum.
# In 2000-2010 Family games and strategies (especially the latter) have significantly increased in number.
# In general, all games have become 3-5 times more out.
# Over the last decade, strategy games have prevailed in terms of number of games played. But abstracts and children’s games are much less.
#
df.groupby("domains", as_index=False).agg({"owned_users": "sum"}).sort_values(
"owned_users", ascending=False
).head(10)
# Top- 10 the most popular game genres among players
df.groupby("decade", as_index=False).agg(
{"comlexity_avg": ["min", "mean", "median", "max"]}
)
# Because of the number of family games in 2000-2010, average rating for these years has become slightly lower.
# Which decade was the highest-ranked game released:
df.groupby("decade")["bgg_rank"].min()
df.head()
df.loc[df["bgg_rank"] < 21]["comlexity_avg"].min()
# min value of 'comlexity_avg' in top-20
round(df.loc[df["bgg_rank"] < 21]["comlexity_avg"].mean(), 2)
# mean value of 'comlexity_avg' in top-20
df.loc[df["bgg_rank"] < 21]["comlexity_avg"].max()
# max value of 'comlexity_avg' in top-20
df.loc[df["bgg_rank"] < 21]["decade"].value_counts(normalize=True) * 100
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/831/129831704.ipynb
| null | null |
[{"Id": 129831704, "ScriptId": 38372898, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13664064, "CreationDate": "05/16/2023 19:52:03", "VersionNumber": 3.0, "Title": "BoardgameGeek", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 282.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 282.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("dark_background")
df = pd.read_csv("/kaggle/input/bgg-dataset/bgg_dataset.csv", sep=";")
df.head()
df.info()
df.columns
df = df[
[
"Name",
"Year Published",
"Min Players",
"Max Players",
"Play Time",
"Min Age",
"Users Rated",
"Rating Average",
"BGG Rank",
"Complexity Average",
"Owned Users",
"Domains",
]
].rename(
columns={
"Year Published": "year",
"Min Players": "min_players",
"Max Players": "max_players",
"Play Time": "play_time",
"Min Age": "min_age",
"Users Rated": "users_rated",
"Rating Average": "avg_rating",
"BGG Rank": "bgg_rank",
"Complexity Average": "comlexity_avg",
"Owned Users": "owned_users",
"Name": "name",
"Mechanics": "mechanics",
"Domains": "domains",
}
)
df.duplicated().sum()
df.shape
df.dtypes
df.isnull().sum()
# # 1. Year published
df.loc[df["year"].isnull()]
df.drop(df[df["name"] == "Hus"].index, inplace=True)
# Get rid of old games of no interest to us
df.drop(df[df["year"] < 1990].index, inplace=True)
df.shape
df["year"] = df["year"].astype(int)
sns.histplot(df["year"])
# # 2. Minimum number of players
df["min_players"].value_counts()
# For 31 games the minimum number of players is 0. It can't be.
# Replace 0 for 2 (cause 2 players is the most popular number of players)
df.loc[df["min_players"] == 0, "min_players"] = 2
# # 3. Maximum players
df["max_players"].describe()
df.loc[df["max_players"] == 0, "max_players"] = df.apply(
lambda row: row["min_players"] if row["min_players"] > 0 else 0, axis=1
)
# If value in column 'max_players' = 0 and in column 'min_players' != 0, the replace by this value (if 'max_players' = 0 and 'min_players' = 2, then 'max_players' = 2 too, for example)
# # 4. Playing Time
df["play_time"].describe()
play_time_mean = df["play_time"].mean()
round(play_time_mean)
df.loc[df["play_time"] == 0, "play_time"] = round(play_time_mean)
df["play_time"] = df["play_time"].astype(int)
df["play_time"].value_counts().sort_values(ascending=False).head(5)
df["play_time"].value_counts().sort_values(ascending=False).head(5).sum() / len(
df["play_time"]
) * 100
# About ~62% of all games are 30 to 90 minutes long
# # 5. Minimal age
df["min_age"].describe()
sns.boxplot(x=df["min_age"])
df.loc[18931, "min_age"] = 16
# Verified the value and changed to the current, according to BoardgameGeek
# # 6. Users rated
df["users_rated"].describe()
df["users_rated"] = df["users_rated"].astype(int)
# # 7. Average rating
df["avg_rating"] = [float(str(i).replace(",", ".")) for i in df["avg_rating"]]
df["avg_rating"].describe()
sns.histplot(df["avg_rating"], kde=True)
# # 8. Complexity Level
df["comlexity_avg"] = [float(str(i).replace(",", ".")) for i in df["comlexity_avg"]]
df["comlexity_avg"].describe()
complexity_median = round(df["comlexity_avg"].dropna().median(), 2)
df.loc[df["comlexity_avg"] == 0, "comlexity_avg"] = complexity_median
# # 9. Owned Users
df["owned_users"].describe()
df["owned_users"].isnull().sum()
owned_median = df["owned_users"].median()
df.loc[df["owned_users"].isnull(), "owned_users"] = owned_median
df["owned_users"] = df["owned_users"].astype(int)
# # 10. Domains
df["domains"].isnull().sum()
df["domains"].dropna().value_counts().head(5)
fig, ax = plt.subplots()
df["domains"].dropna().value_counts().head(5).plot(
kind="pie", autopct="%.1f%%", ax=ax, textprops={"color": "red"}
)
ax.set_title("Percentage of each genre")
ax.yaxis.label.set_visible(False) # remove y-label
ax.title.set_position([0.5, 1.15]) # change title rotation
plt.show()
df.dropna(subset="domains", inplace=True)
df.head(10)
# Average complexity in each genre
df.groupby("domains")["comlexity_avg"].mean()
# Minimum, average and maximum values for each genre in the list
df.groupby("domains").agg({"comlexity_avg": ["min", "mean", "max"]}).reset_index()
# Top 5 popular genres in order of number of games
popular_ganres = df["domains"].dropna().value_counts().head(5)
popular_ganres
pop_ganres = df["domains"].dropna().value_counts().head(5).index.tolist()
# All games of top-5 popular ganres:
t = df[df["domains"].isin(pop_ganres)]
pd.pivot_table(
t, values="comlexity_avg", index="domains", aggfunc=[np.mean, np.max]
).reset_index()
# The most difficult genre of games - Wargames (4.93 max, 2.9 mean)
# Easiest genre of top-5 popular games is Family Games (3.05 max, 1.59 mean)
decade = pd.cut(df["year"], 3, labels=["2011-2021", "2000-2010", "1990-1999"])
# Split df['year'] into three periods
decade.value_counts(normalize=True) * 100
# Percentage distribution of games for each of the three periods
df["decade"] = pd.cut(df["year"], 3, labels=["2011-2021", "2000-2010", "1990-1999"])
def ten_years(v):
if 1990 <= v < 2000:
return "1990-1999"
elif 2000 <= v < 2010:
return "2000-2010"
else:
return "2011-2021"
df["decade"] = df["year"].apply(ten_years)
df.head(10)
df.groupby(["decade", "domains"])["name"].count()
ganres_per_decade = pd.pivot_table(
df, values="name", columns="decade", index="domains", aggfunc="count", fill_value=0
).reset_index()
# How many games were released in each decade by genre:
ganres_per_decade
ganres_per_decade.sort_values("1990-1999", ascending=False).head(10)
ganres_per_decade.sort_values("2000-2010", ascending=False).head(10)
ganres_per_decade.sort_values("2011-2021", ascending=False).head(10)
# Strategies over the last decade are the most released. More wargames and family games.
# The Wargems were released in the 1990s and continue to be released consistently and in large numbers.
# Perhaps wargames are the most constant and are in demand in certain circles
# In the 90s, the second most popular was the Children’s Games(probably that’s where the stereotype came from. board games are kind of fun).
# Family games and strategies are just starting to gather momentum.
# In 2000-2010 Family games and strategies (especially the latter) have significantly increased in number.
# In general, all games have become 3-5 times more out.
# Over the last decade, strategy games have prevailed in terms of number of games played. But abstracts and children’s games are much less.
#
df.groupby("domains", as_index=False).agg({"owned_users": "sum"}).sort_values(
"owned_users", ascending=False
).head(10)
# Top- 10 the most popular game genres among players
df.groupby("decade", as_index=False).agg(
{"comlexity_avg": ["min", "mean", "median", "max"]}
)
# Because of the number of family games in 2000-2010, average rating for these years has become slightly lower.
# Which decade was the highest-ranked game released:
df.groupby("decade")["bgg_rank"].min()
df.head()
df.loc[df["bgg_rank"] < 21]["comlexity_avg"].min()
# min value of 'comlexity_avg' in top-20
round(df.loc[df["bgg_rank"] < 21]["comlexity_avg"].mean(), 2)
# mean value of 'comlexity_avg' in top-20
df.loc[df["bgg_rank"] < 21]["comlexity_avg"].max()
# max value of 'comlexity_avg' in top-20
df.loc[df["bgg_rank"] < 21]["decade"].value_counts(normalize=True) * 100
| false | 0 | 2,749 | 0 | 2,749 | 2,749 |
||
129831308
|
<jupyter_start><jupyter_text>Transformer
Kaggle dataset identifier: transformer
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
ds_A = pd.read_csv("/kaggle/input/transformer/DatasetA.csv")
ds_B = pd.read_csv("/kaggle/input/transformer/DatasetB.csv")
# Splitting train and test
from sklearn.model_selection import train_test_split
train_set_A, test_set_A = train_test_split(ds_A, test_size=0.25, random_state=11)
# Setting the labels
y_train_A = train_set_A["Furan"]
y_test_A = test_set_A["Furan"]
# Dropping the Furan and Health Index columns
X_train_A = train_set_A.drop(["Furan", "HI"], axis=1)
X_test_A = test_set_A.drop(["Furan", "HI"], axis=1)
# For DatasetB
y_B = ds_B["Furan"]
X_B = ds_B.drop(["Furan", "HI"], axis=1)
X_train_A = X_train_A.drop(set(ds_A.columns) - set(ds_B.columns), axis=1)
X_test_A = X_test_A.drop(set(ds_A.columns) - set(ds_B.columns), axis=1)
X_B = X_B[X_train_A.columns]
X_train_A
# define the bin edges for each class
bins = [-1, 0.1, 1, 100]
# define the labels for each class
labels = [0, 1, 2]
y_train_A = pd.DataFrame(y_train_A)
y_B = pd.DataFrame(y_B)
y_test_A = pd.DataFrame(y_test_A)
# discretize the data into the desired classes
y_train_A["Class"] = pd.cut(y_train_A["Furan"], bins=bins, labels=labels)
y_B["Class"] = pd.cut(y_B["Furan"], bins=bins, labels=labels)
y_test_A["Class"] = pd.cut(y_test_A["Furan"], bins=bins, labels=labels)
y_train_A = np.array(y_train_A.drop("Furan", axis=1)).ravel()
y_B = np.array(y_B.drop("Furan", axis=1)).ravel()
y_test_A = np.array(y_test_A.drop("Furan", axis=1)).ravel()
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import VotingClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.neighbors import KNeighborsClassifier
from xgboost import XGBClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.naive_bayes import GaussianNB
# log_clf = LogisticRegression(max_iter=1000)
svm_clf = SVC(probability=True)
knn_clf = KNeighborsClassifier()
xgb_clf = XGBClassifier(
learning_rate=0.01, n_estimators=300, max_depth=3, subsample=0.7
)
mlp_clf = MLPClassifier(hidden_layer_sizes=(100,), max_iter=1000)
nb_clf = GaussianNB()
voting_clf = VotingClassifier(
estimators=[ # ('lr', log_clf),
("nn", mlp_clf),
("svc", svm_clf),
("knn", knn_clf),
("xgb", xgb_clf),
("nb", nb_clf),
],
voting="hard",
)
voting_clf.fit(X_train_A, np.array(y_train_A).ravel())
from sklearn.metrics import accuracy_score
for clf in ( # log_clf, #rnd_clf,
mlp_clf,
svm_clf,
knn_clf,
xgb_clf,
nb_clf,
voting_clf,
):
clf.fit(X_train_A, y_train_A)
y_pred_A = clf.predict(X_test_A)
y_pred_B = clf.predict(X_B)
print(
clf.__class__.__name__ + " for dataset A:", accuracy_score(y_test_A, y_pred_A)
)
print(clf.__class__.__name__ + " for dataset B:", accuracy_score(y_B, y_pred_B))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/831/129831308.ipynb
|
transformer
|
darvack
|
[{"Id": 129831308, "ScriptId": 38514219, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5534641, "CreationDate": "05/16/2023 19:47:35", "VersionNumber": 6.0, "Title": "Transformer-Paper", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 100.0, "LinesInsertedFromPrevious": 13.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 87.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186211218, "KernelVersionId": 129831308, "SourceDatasetVersionId": 5683446}]
|
[{"Id": 5683446, "DatasetId": 3267447, "DatasourceVersionId": 5759014, "CreatorUserId": 5534641, "LicenseName": "Unknown", "CreationDate": "05/14/2023 14:35:55", "VersionNumber": 1.0, "Title": "Transformer", "Slug": "transformer", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3267447, "CreatorUserId": 5534641, "OwnerUserId": 5534641.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5683446.0, "CurrentDatasourceVersionId": 5759014.0, "ForumId": 3333063, "Type": 2, "CreationDate": "05/14/2023 14:35:55", "LastActivityDate": "05/14/2023", "TotalViews": 45, "TotalDownloads": 0, "TotalVotes": 0, "TotalKernels": 5}]
|
[{"Id": 5534641, "UserName": "darvack", "DisplayName": "Mohammad Amin Faraji", "RegisterDate": "07/27/2020", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
ds_A = pd.read_csv("/kaggle/input/transformer/DatasetA.csv")
ds_B = pd.read_csv("/kaggle/input/transformer/DatasetB.csv")
# Splitting train and test
from sklearn.model_selection import train_test_split
train_set_A, test_set_A = train_test_split(ds_A, test_size=0.25, random_state=11)
# Setting the labels
y_train_A = train_set_A["Furan"]
y_test_A = test_set_A["Furan"]
# Dropping the Furan and Health Index columns
X_train_A = train_set_A.drop(["Furan", "HI"], axis=1)
X_test_A = test_set_A.drop(["Furan", "HI"], axis=1)
# For DatasetB
y_B = ds_B["Furan"]
X_B = ds_B.drop(["Furan", "HI"], axis=1)
X_train_A = X_train_A.drop(set(ds_A.columns) - set(ds_B.columns), axis=1)
X_test_A = X_test_A.drop(set(ds_A.columns) - set(ds_B.columns), axis=1)
X_B = X_B[X_train_A.columns]
X_train_A
# define the bin edges for each class
bins = [-1, 0.1, 1, 100]
# define the labels for each class
labels = [0, 1, 2]
y_train_A = pd.DataFrame(y_train_A)
y_B = pd.DataFrame(y_B)
y_test_A = pd.DataFrame(y_test_A)
# discretize the data into the desired classes
y_train_A["Class"] = pd.cut(y_train_A["Furan"], bins=bins, labels=labels)
y_B["Class"] = pd.cut(y_B["Furan"], bins=bins, labels=labels)
y_test_A["Class"] = pd.cut(y_test_A["Furan"], bins=bins, labels=labels)
y_train_A = np.array(y_train_A.drop("Furan", axis=1)).ravel()
y_B = np.array(y_B.drop("Furan", axis=1)).ravel()
y_test_A = np.array(y_test_A.drop("Furan", axis=1)).ravel()
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import VotingClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.neighbors import KNeighborsClassifier
from xgboost import XGBClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.naive_bayes import GaussianNB
# log_clf = LogisticRegression(max_iter=1000)
svm_clf = SVC(probability=True)
knn_clf = KNeighborsClassifier()
xgb_clf = XGBClassifier(
learning_rate=0.01, n_estimators=300, max_depth=3, subsample=0.7
)
mlp_clf = MLPClassifier(hidden_layer_sizes=(100,), max_iter=1000)
nb_clf = GaussianNB()
voting_clf = VotingClassifier(
estimators=[ # ('lr', log_clf),
("nn", mlp_clf),
("svc", svm_clf),
("knn", knn_clf),
("xgb", xgb_clf),
("nb", nb_clf),
],
voting="hard",
)
voting_clf.fit(X_train_A, np.array(y_train_A).ravel())
from sklearn.metrics import accuracy_score
for clf in ( # log_clf, #rnd_clf,
mlp_clf,
svm_clf,
knn_clf,
xgb_clf,
nb_clf,
voting_clf,
):
clf.fit(X_train_A, y_train_A)
y_pred_A = clf.predict(X_test_A)
y_pred_B = clf.predict(X_B)
print(
clf.__class__.__name__ + " for dataset A:", accuracy_score(y_test_A, y_pred_A)
)
print(clf.__class__.__name__ + " for dataset B:", accuracy_score(y_B, y_pred_B))
| false | 2 | 1,273 | 0 | 1,289 | 1,273 |
||
129485107
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# new code here
train_data = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
train_data.head(5)
test_data = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
test_data.head(4)
# **Earth**
home_as_earth = train_data.loc[train_data.HomePlanet == "Earth"]["Transported"]
print(
"Percentage people transported who belongs to planet earth",
sum(home_as_earth) / len(home_as_earth),
)
# **Europa**
home_as_europa = train_data.loc[train_data.HomePlanet == "Europa"]["Transported"]
print(
"Percentage people transported who belongs to planet europa",
sum(home_as_europa) / len(home_as_europa),
)
# **Mars**
home_as_mars = train_data.loc[train_data.HomePlanet == "Mars"]["Transported"]
print(
"Percentage people transported who belongs to planet mars",
sum(home_as_mars) / len(home_as_mars),
)
# **Cryosleep**
cryo_sleep = train_data.loc[train_data.CryoSleep == True]["Transported"]
print(
"Percentage people transported who opted for cryosleep",
sum(cryo_sleep) / len(cryo_sleep),
)
# **Europa and cryosleep**
home_as_europa_with_cryoSleep = train_data.loc[train_data.HomePlanet == "Europa"][
train_data.CryoSleep == True
]["Transported"]
print(
"Percentage people transported who belongs to planet europa and also opted for cryosleep",
sum(home_as_europa_with_cryoSleep) / len(home_as_europa_with_cryoSleep),
)
dest_as_trappi = train_data.loc[train_data.Destination == "TRAPPIST-1e"]["Transported"]
print(
"Percentage people transported whose dest is TRAPPIST-1e ",
sum(dest_as_trappi) / len(dest_as_trappi),
)
# dest_as_cancri = train_data.loc[train_data.Destination=='T55 Cancri e']["Transported"]
# print("Percentage people transported whose dest is 55 Cancri e ",sum(dest_as_cancri)/len(dest_as_cancri))
dest_as_pso = train_data.loc[train_data.Destination == "PSO J318.5-22"]["Transported"]
print(
"Percentage people transported whose dest is PSO J318.5-22",
sum(dest_as_pso) / len(dest_as_pso),
)
# **Model**
from sklearn.ensemble import RandomForestClassifier
y = train_data["Transported"]
features = ["HomePlanet", "CryoSleep"]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = RandomForestClassifier(n_estimators=100, max_depth=5, random_state=1)
model.fit(X, y)
preds = model.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Transported": preds})
output.to_csv("FirstSubmission.csv", index=False)
print(f"done")
# print(len(X_test))
# print(len(preds))
# def accuracy_metric(actual, predicted):
# correct = 0
# for i in range(len(actual)):
# if actual[i] == predicted[i]:
# correct += 1
# return correct / float(len(actual)) * 100.0
# accuracy_metric(X_test,preds)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/485/129485107.ipynb
| null | null |
[{"Id": 129485107, "ScriptId": 38388681, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11265301, "CreationDate": "05/14/2023 07:58:49", "VersionNumber": 1.0, "Title": "SpaceshipTitanic", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 87.0, "LinesInsertedFromPrevious": 87.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# new code here
train_data = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
train_data.head(5)
test_data = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
test_data.head(4)
# **Earth**
home_as_earth = train_data.loc[train_data.HomePlanet == "Earth"]["Transported"]
print(
"Percentage people transported who belongs to planet earth",
sum(home_as_earth) / len(home_as_earth),
)
# **Europa**
home_as_europa = train_data.loc[train_data.HomePlanet == "Europa"]["Transported"]
print(
"Percentage people transported who belongs to planet europa",
sum(home_as_europa) / len(home_as_europa),
)
# **Mars**
home_as_mars = train_data.loc[train_data.HomePlanet == "Mars"]["Transported"]
print(
"Percentage people transported who belongs to planet mars",
sum(home_as_mars) / len(home_as_mars),
)
# **Cryosleep**
cryo_sleep = train_data.loc[train_data.CryoSleep == True]["Transported"]
print(
"Percentage people transported who opted for cryosleep",
sum(cryo_sleep) / len(cryo_sleep),
)
# **Europa and cryosleep**
home_as_europa_with_cryoSleep = train_data.loc[train_data.HomePlanet == "Europa"][
train_data.CryoSleep == True
]["Transported"]
print(
"Percentage people transported who belongs to planet europa and also opted for cryosleep",
sum(home_as_europa_with_cryoSleep) / len(home_as_europa_with_cryoSleep),
)
dest_as_trappi = train_data.loc[train_data.Destination == "TRAPPIST-1e"]["Transported"]
print(
"Percentage people transported whose dest is TRAPPIST-1e ",
sum(dest_as_trappi) / len(dest_as_trappi),
)
# dest_as_cancri = train_data.loc[train_data.Destination=='T55 Cancri e']["Transported"]
# print("Percentage people transported whose dest is 55 Cancri e ",sum(dest_as_cancri)/len(dest_as_cancri))
dest_as_pso = train_data.loc[train_data.Destination == "PSO J318.5-22"]["Transported"]
print(
"Percentage people transported whose dest is PSO J318.5-22",
sum(dest_as_pso) / len(dest_as_pso),
)
# **Model**
from sklearn.ensemble import RandomForestClassifier
y = train_data["Transported"]
features = ["HomePlanet", "CryoSleep"]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = RandomForestClassifier(n_estimators=100, max_depth=5, random_state=1)
model.fit(X, y)
preds = model.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Transported": preds})
output.to_csv("FirstSubmission.csv", index=False)
print(f"done")
# print(len(X_test))
# print(len(preds))
# def accuracy_metric(actual, predicted):
# correct = 0
# for i in range(len(actual)):
# if actual[i] == predicted[i]:
# correct += 1
# return correct / float(len(actual)) * 100.0
# accuracy_metric(X_test,preds)
| false | 0 | 1,163 | 0 | 1,163 | 1,163 |
||
129485620
|
# # Fully Connected Conditional Generative Adverserial Networks Model
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import DataLoader
from torchvision import datasets, transforms
import matplotlib.pyplot as plt
import numpy as np
class Generator(nn.Module):
def __init__(self, latent_dim, num_classes, hidden_layers, output_dim):
super(Generator, self).__init__()
layers = []
input_dim = latent_dim + num_classes
for i in range(len(hidden_layers)):
layers.append(nn.Linear(input_dim, hidden_layers[i]))
layers.append(nn.ReLU())
input_dim = hidden_layers[i]
layers.append(nn.Linear(input_dim, output_dim))
layers.append(nn.Tanh())
self.model = nn.Sequential(*layers)
def forward(self, noise, labels):
x = torch.cat((noise, labels), -1)
return self.model(x)
class Discriminator(nn.Module):
def __init__(self, input_dim, num_classes, hidden_layers):
super(Discriminator, self).__init__()
layers = []
input_dim += num_classes
for i in range(len(hidden_layers)):
layers.append(nn.Linear(input_dim, hidden_layers[i]))
layers.append(nn.ReLU())
input_dim = hidden_layers[i]
layers.append(nn.Linear(input_dim, 1))
layers.append(nn.Sigmoid())
self.model = nn.Sequential(*layers)
def forward(self, x, labels):
x = torch.cat((x, labels), -1)
return self.model(x)
# # Layers and Hyperparams
latent_dim = 100
num_classes = 10
hidden_layers_g = [256, 128, 1024]
hidden_layers_d = [1024, 512, 256]
output_dim = 28 * 28
learning_rate = 0.0002
batch_size = 32
epochs = 50
generator = Generator(latent_dim, num_classes, hidden_layers_g, output_dim)
discriminator = Discriminator(output_dim, num_classes, hidden_layers_d)
optimizer_g = optim.Adam(generator.parameters(), lr=learning_rate)
optimizer_d = optim.Adam(discriminator.parameters(), lr=learning_rate)
loss_function = nn.BCELoss()
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
generator.to(device)
discriminator.to(device)
# # MNIST Dataset
transform = transforms.Compose(
[transforms.ToTensor(), transforms.Normalize([0.5], [0.5])]
)
train_dataset = datasets.MNIST(
root="./data", train=True, download=True, transform=transform
)
train_loader = DataLoader(train_dataset, batch_size=batch_size, shuffle=True)
# # Training Loop
G_losses = []
D_losses = []
for epoch in range(epochs):
for i, (images, labels) in enumerate(train_loader):
real_images = images.view(-1, output_dim).to(device)
real_labels = (
torch.zeros(images.size(0), num_classes)
.scatter_(1, labels.view(-1, 1), 1)
.to(device)
)
real_targets = torch.ones(images.size(0), 1).to(device)
# Train discriminator on real images
optimizer_d.zero_grad()
real_preds = discriminator(real_images, real_labels)
real_loss = loss_function(real_preds, real_targets)
real_loss.backward()
noise = torch.randn(batch_size, latent_dim).to(device)
fake_labels = (
torch.zeros(batch_size, num_classes)
.to(device)
.scatter_(1, torch.randint(0, num_classes, (batch_size, 1)).to(device), 1)
)
fake_images = generator(noise, fake_labels)
fake_targets = torch.zeros(batch_size, 1).to(device)
# Train discriminator on fake images
fake_preds = discriminator(fake_images.detach(), fake_labels)
fake_loss = loss_function(fake_preds, fake_targets)
fake_loss.backward()
d_loss = real_loss + fake_loss
optimizer_d.step()
# Train generator
optimizer_g.zero_grad()
fake_preds = discriminator(fake_images, fake_labels)
g_loss = loss_function(fake_preds, real_targets)
g_loss.backward()
optimizer_g.step()
G_losses.append(g_loss.item())
D_losses.append(d_loss.item())
# Print progress
if i == 0:
print(
f"Epoch [{epoch + 1}/{epochs}], Step [{i + 1}/{len(train_loader)}], D_loss: {np.mean(D_losses)}, G_loss: {np.mean(G_losses)}"
)
print(f"Generated number {torch.argmax(fake_labels[0]).cpu()}:")
plt.imshow(fake_images[0].reshape(28, 28).detach().cpu())
plt.show()
print(
f"---------------------------------------------------------------------------------------"
)
torch.save(generator.state_dict(), "generator.pth")
torch.save(discriminator.state_dict(), "discriminator.pth")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/485/129485620.ipynb
| null | null |
[{"Id": 129485620, "ScriptId": 37641285, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13463050, "CreationDate": "05/14/2023 08:04:31", "VersionNumber": 1.0, "Title": "Fully Connected Conditional GAN", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 137.0, "LinesInsertedFromPrevious": 137.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Fully Connected Conditional Generative Adverserial Networks Model
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import DataLoader
from torchvision import datasets, transforms
import matplotlib.pyplot as plt
import numpy as np
class Generator(nn.Module):
def __init__(self, latent_dim, num_classes, hidden_layers, output_dim):
super(Generator, self).__init__()
layers = []
input_dim = latent_dim + num_classes
for i in range(len(hidden_layers)):
layers.append(nn.Linear(input_dim, hidden_layers[i]))
layers.append(nn.ReLU())
input_dim = hidden_layers[i]
layers.append(nn.Linear(input_dim, output_dim))
layers.append(nn.Tanh())
self.model = nn.Sequential(*layers)
def forward(self, noise, labels):
x = torch.cat((noise, labels), -1)
return self.model(x)
class Discriminator(nn.Module):
def __init__(self, input_dim, num_classes, hidden_layers):
super(Discriminator, self).__init__()
layers = []
input_dim += num_classes
for i in range(len(hidden_layers)):
layers.append(nn.Linear(input_dim, hidden_layers[i]))
layers.append(nn.ReLU())
input_dim = hidden_layers[i]
layers.append(nn.Linear(input_dim, 1))
layers.append(nn.Sigmoid())
self.model = nn.Sequential(*layers)
def forward(self, x, labels):
x = torch.cat((x, labels), -1)
return self.model(x)
# # Layers and Hyperparams
latent_dim = 100
num_classes = 10
hidden_layers_g = [256, 128, 1024]
hidden_layers_d = [1024, 512, 256]
output_dim = 28 * 28
learning_rate = 0.0002
batch_size = 32
epochs = 50
generator = Generator(latent_dim, num_classes, hidden_layers_g, output_dim)
discriminator = Discriminator(output_dim, num_classes, hidden_layers_d)
optimizer_g = optim.Adam(generator.parameters(), lr=learning_rate)
optimizer_d = optim.Adam(discriminator.parameters(), lr=learning_rate)
loss_function = nn.BCELoss()
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
generator.to(device)
discriminator.to(device)
# # MNIST Dataset
transform = transforms.Compose(
[transforms.ToTensor(), transforms.Normalize([0.5], [0.5])]
)
train_dataset = datasets.MNIST(
root="./data", train=True, download=True, transform=transform
)
train_loader = DataLoader(train_dataset, batch_size=batch_size, shuffle=True)
# # Training Loop
G_losses = []
D_losses = []
for epoch in range(epochs):
for i, (images, labels) in enumerate(train_loader):
real_images = images.view(-1, output_dim).to(device)
real_labels = (
torch.zeros(images.size(0), num_classes)
.scatter_(1, labels.view(-1, 1), 1)
.to(device)
)
real_targets = torch.ones(images.size(0), 1).to(device)
# Train discriminator on real images
optimizer_d.zero_grad()
real_preds = discriminator(real_images, real_labels)
real_loss = loss_function(real_preds, real_targets)
real_loss.backward()
noise = torch.randn(batch_size, latent_dim).to(device)
fake_labels = (
torch.zeros(batch_size, num_classes)
.to(device)
.scatter_(1, torch.randint(0, num_classes, (batch_size, 1)).to(device), 1)
)
fake_images = generator(noise, fake_labels)
fake_targets = torch.zeros(batch_size, 1).to(device)
# Train discriminator on fake images
fake_preds = discriminator(fake_images.detach(), fake_labels)
fake_loss = loss_function(fake_preds, fake_targets)
fake_loss.backward()
d_loss = real_loss + fake_loss
optimizer_d.step()
# Train generator
optimizer_g.zero_grad()
fake_preds = discriminator(fake_images, fake_labels)
g_loss = loss_function(fake_preds, real_targets)
g_loss.backward()
optimizer_g.step()
G_losses.append(g_loss.item())
D_losses.append(d_loss.item())
# Print progress
if i == 0:
print(
f"Epoch [{epoch + 1}/{epochs}], Step [{i + 1}/{len(train_loader)}], D_loss: {np.mean(D_losses)}, G_loss: {np.mean(G_losses)}"
)
print(f"Generated number {torch.argmax(fake_labels[0]).cpu()}:")
plt.imshow(fake_images[0].reshape(28, 28).detach().cpu())
plt.show()
print(
f"---------------------------------------------------------------------------------------"
)
torch.save(generator.state_dict(), "generator.pth")
torch.save(discriminator.state_dict(), "discriminator.pth")
| false | 0 | 1,345 | 0 | 1,345 | 1,345 |
||
129566599
|
# Import das bibliotecas compátiveis
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Import das bibliotecas compátiveis
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
from sklearn.preprocessing import OrdinalEncoder
from sklearn.impute import SimpleImputer
from xgboost import XGBRegressor
from sklearn.model_selection import train_test_split
from sklearn.linear_model import Lasso
from sklearn.metrics import mean_squared_log_error
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/train.csv.zip")
z.extractall()
df_train = pd.read_csv("/kaggle/working/train.csv")
df_train.head()
num_linhas = len(df_train)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Transformando em valores numéricos
from sklearn.preprocessing import LabelEncoder
for f in df_train.columns:
if df_train[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_train[f].values))
df_train[f] = lbl.transform(list(df_train[f].values))
df_train.head()
# Média para remover valores nulos
for col in df_train.columns:
if df_train[col].isnull().sum() > 0:
mean = df_train[col].mean()
df_train[col] = df_train[col].fillna(mean)
df_train.head()
# Determinando qual coluna será considerada Target.
X = df_train[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
y = np.log(df_train.price_doc)
# Arquivo de teste em Treino e Teste separado
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# As bases de dados que possui para cada tipo de entrada
train = pd.read_csv("./train.csv", parse_dates=["timestamp"])
df_dtype = train.dtypes.reset_index()
df_dtype.columns = ["qtd_atributos", "tipo"]
df_dtype.groupby("tipo").aggregate("count").reset_index()
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
# Modelo Regressão
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso, Ridge, ElasticNet
modelo = ElasticNet()
modelo.fit(X_train, y_train)
# Determinando os coeficientes do modelo
modelo.coef_, modelo.intercept_
# Performace do modelo
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_absolute_percentage_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_log_error
import numpy as np
y_pred = modelo.predict(X_train)
mae = mean_absolute_error(y_train, y_pred)
mape = mean_absolute_percentage_error(y_train, y_pred)
rmse = mean_squared_error(y_train, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_train, y_pred))
r2 = r2_score(y_train, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
print("")
y_pred = modelo.predict(X_test)
mae = mean_absolute_error(y_test, y_pred)
mape = mean_absolute_percentage_error(y_test, y_pred)
rmse = mean_squared_error(y_test, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_test, y_pred))
r2 = r2_score(y_test, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
# Upload do arquivo de Teste
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/test.csv.zip")
z.extractall()
df_test = pd.read_csv("/kaggle/working/test.csv")
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
from sklearn.preprocessing import LabelEncoder
for f in df_test.columns:
if df_test[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_test[f].values))
df_test[f] = lbl.transform(list(df_test[f].values))
import pandas as pd
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Média para remover valores nulos.
for col in df_test.columns:
if df_test[col].isnull().sum() > 0:
mean = df_test[col].mean()
df_test[col] = df_test[col].fillna(mean)
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Colunas para testar o modelo de previsão
X_test = df_test[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
# X_test e previsão de modelo
y_pred = modelo.predict(X_test)
# Aplica a função exponencial nos modelos previstos, ja que o modelo preve numeros logaritimos e não valores reais
y_pred = np.exp(y_pred)
# Preços previstos pelo modelo
output = pd.DataFrame({"id": df_test.id, "price_doc": y_pred})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
output.head()
num_linhas = len(output)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/566/129566599.ipynb
| null | null |
[{"Id": 129566599, "ScriptId": 38525740, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14641575, "CreationDate": "05/14/2023 22:42:31", "VersionNumber": 1.0, "Title": "AC2 REGRESS\u00c3O_1", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 187.0, "LinesInsertedFromPrevious": 187.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# Import das bibliotecas compátiveis
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Import das bibliotecas compátiveis
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
from sklearn.preprocessing import OrdinalEncoder
from sklearn.impute import SimpleImputer
from xgboost import XGBRegressor
from sklearn.model_selection import train_test_split
from sklearn.linear_model import Lasso
from sklearn.metrics import mean_squared_log_error
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/train.csv.zip")
z.extractall()
df_train = pd.read_csv("/kaggle/working/train.csv")
df_train.head()
num_linhas = len(df_train)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Transformando em valores numéricos
from sklearn.preprocessing import LabelEncoder
for f in df_train.columns:
if df_train[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_train[f].values))
df_train[f] = lbl.transform(list(df_train[f].values))
df_train.head()
# Média para remover valores nulos
for col in df_train.columns:
if df_train[col].isnull().sum() > 0:
mean = df_train[col].mean()
df_train[col] = df_train[col].fillna(mean)
df_train.head()
# Determinando qual coluna será considerada Target.
X = df_train[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
y = np.log(df_train.price_doc)
# Arquivo de teste em Treino e Teste separado
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# As bases de dados que possui para cada tipo de entrada
train = pd.read_csv("./train.csv", parse_dates=["timestamp"])
df_dtype = train.dtypes.reset_index()
df_dtype.columns = ["qtd_atributos", "tipo"]
df_dtype.groupby("tipo").aggregate("count").reset_index()
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
# Modelo Regressão
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso, Ridge, ElasticNet
modelo = ElasticNet()
modelo.fit(X_train, y_train)
# Determinando os coeficientes do modelo
modelo.coef_, modelo.intercept_
# Performace do modelo
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_absolute_percentage_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_log_error
import numpy as np
y_pred = modelo.predict(X_train)
mae = mean_absolute_error(y_train, y_pred)
mape = mean_absolute_percentage_error(y_train, y_pred)
rmse = mean_squared_error(y_train, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_train, y_pred))
r2 = r2_score(y_train, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
print("")
y_pred = modelo.predict(X_test)
mae = mean_absolute_error(y_test, y_pred)
mape = mean_absolute_percentage_error(y_test, y_pred)
rmse = mean_squared_error(y_test, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_test, y_pred))
r2 = r2_score(y_test, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
# Upload do arquivo de Teste
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/test.csv.zip")
z.extractall()
df_test = pd.read_csv("/kaggle/working/test.csv")
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
from sklearn.preprocessing import LabelEncoder
for f in df_test.columns:
if df_test[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_test[f].values))
df_test[f] = lbl.transform(list(df_test[f].values))
import pandas as pd
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Média para remover valores nulos.
for col in df_test.columns:
if df_test[col].isnull().sum() > 0:
mean = df_test[col].mean()
df_test[col] = df_test[col].fillna(mean)
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# Colunas para testar o modelo de previsão
X_test = df_test[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
# X_test e previsão de modelo
y_pred = modelo.predict(X_test)
# Aplica a função exponencial nos modelos previstos, ja que o modelo preve numeros logaritimos e não valores reais
y_pred = np.exp(y_pred)
# Preços previstos pelo modelo
output = pd.DataFrame({"id": df_test.id, "price_doc": y_pred})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
output.head()
num_linhas = len(output)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
| false | 0 | 1,794 | 0 | 1,794 | 1,794 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.