file_id
stringlengths 5
9
| content
stringlengths 100
5.25M
| local_path
stringlengths 66
70
| kaggle_dataset_name
stringlengths 3
50
⌀ | kaggle_dataset_owner
stringlengths 3
20
⌀ | kversion
stringlengths 497
763
⌀ | kversion_datasetsources
stringlengths 71
5.46k
⌀ | dataset_versions
stringlengths 338
235k
⌀ | datasets
stringlengths 334
371
⌀ | users
stringlengths 111
264
⌀ | script
stringlengths 100
5.25M
| df_info
stringlengths 0
4.87M
| has_data_info
bool 2
classes | nb_filenames
int64 0
370
| retreived_data_description
stringlengths 0
4.44M
| script_nb_tokens
int64 25
663k
| upvotes
int64 0
1.65k
| tokens_description
int64 25
663k
| tokens_script
int64 25
663k
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
129674541
|
<jupyter_start><jupyter_text>name2lang
Kaggle dataset identifier: name2lang
<jupyter_script># # Introduction.
# 
# * Introduce dataset
# * Data processing - Test-train split, encoding, visualisation
# * Why do we use batching
# * Batching for sequence models
# * Padding and packing in PyTorch
# * Training with batched dataset
# * Comparison of performance with batching and on GPU
# Most of the concepts explained in this Notebook are learnt from course [Deep Learning](https://padhai.onefourthlabs.in/courses/dl-feb-2019)
# # Setting up the dependencies
from io import open
import os, string, random, time, math
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from sklearn.model_selection import train_test_split
import torch
import torch.nn as nn
import torch.optim as optim
# Instantiates the device to be used as GPU/CPU based on availability
device_gpu = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
from IPython.display import clear_output
import warnings
warnings.filterwarnings("ignore")
# # Dataset
# # Pre-processing
languages = []
data = []
X = []
y = []
with open("/kaggle/input/name2lang/name2lang.txt", "r") as f:
for line in f:
line = line.split(",")
name = line[0].strip()
lang = line[1].strip()
if not lang in languages:
languages.append(lang)
X.append(name)
y.append(lang)
data.append((name, lang))
n_languages = len(languages)
print(languages)
print(data[0:10])
# # Test-train split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=0, stratify=y
)
print(len(X_train), len(X_test))
# ## Encoding names and language
all_letters = string.ascii_letters + " .,;'"
n_letters = len(all_letters)
def name_rep(name):
rep = torch.zeros(len(name), 1, n_letters)
for index, letter in enumerate(name):
pos = all_letters.find(letter)
rep[index][0][pos] = 1
return rep
def lang_rep(lang):
return torch.tensor([languages.index(lang)], dtype=torch.long)
name_rep("Abreu")
lang_rep("Portuguese")
count = {}
for l in languages:
count[l] = 0
for d in data:
count[d[1]] += 1
print(count)
plt_ = sns.barplot(x=list(count.keys()), y=list(count.values()))
plt_.set_xticklabels(plt_.get_xticklabels(), rotation=90)
plt.show()
# ## Basic network and testing inference
class RNN_net(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(RNN_net, self).__init__()
self.hidden_size = hidden_size
self.rnn_cell = nn.RNN(input_size, hidden_size)
self.h2o = nn.Linear(hidden_size, output_size)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, input_, hidden=None, batch_size=1):
out, hidden = self.rnn_cell(input_, hidden)
output = self.h2o(hidden.view(-1, self.hidden_size))
output = self.softmax(output)
return output, hidden
def init_hidden(self, batch_size=1):
return torch.zeros(1, batch_size, self.hidden_size)
n_hidden = 128
net = RNN_net(n_letters, n_hidden, n_languages)
def infer(net, name, device="cpu"):
name_ohe = name_rep(name).to(device)
output, hidden = net(name_ohe)
if type(hidden) is tuple: # For LSTM
hidden = hidden[0]
index = torch.argmax(hidden)
return output
infer(net, "Adam")
# # Evaluate model
def dataloader(npoints, X_, y_):
to_ret = []
for i in range(npoints):
index_ = np.random.randint(len(X_))
name, lang = X_[index_], y_[index_]
to_ret.append((name, lang, name_rep(name), lang_rep(lang)))
return to_ret
dataloader(2, X_train, y_train)
def eval(net, n_points, topk, X_, y_, device="cpu"):
net = net.eval().to(device)
data_ = dataloader(n_points, X_, y_)
correct = 0
for name, language, name_ohe, lang_rep in data_:
output = infer(net, name, device)
val, indices = output.topk(topk)
indices = indices.to("cpu")
if lang_rep in indices:
correct += 1
accuracy = correct / n_points
return accuracy
eval(net, 1000, 1, X_test, y_test)
# # Batching
# Batching can be done in many ways, one can be represted as.
# 
def batched_name_rep(names, max_word_size):
rep = torch.zeros(max_word_size, len(names), n_letters)
for name_index, name in enumerate(names):
for letter_index, letter in enumerate(name):
pos = all_letters.find(letter)
rep[letter_index][name_index][pos] = 1
return rep
def print_char(name_reps):
name_reps = name_reps.view((-1, name_reps.size()[-1]))
for t in name_reps:
if torch.sum(t) == 0:
print("<pad>")
else:
index = t.argmax()
print(all_letters[index])
out_ = batched_name_rep(["Shyam", "Ram"], 5)
print(out_)
print(out_.shape)
print_char(out_)
def batched_lang_rep(langs):
rep = torch.zeros([len(langs)], dtype=torch.long)
for index, lang in enumerate(langs):
rep[index] = languages.index(lang)
return rep
def batched_dataloader(npoints, X_, y_, verbose=False, device="cpu"):
names = []
langs = []
X_lengths = []
for i in range(npoints):
index_ = np.random.randint(len(X_))
name, lang = X_[index_], y_[index_]
X_lengths.append(len(name))
names.append(name)
langs.append(lang)
max_length = max(X_lengths)
names_rep = batched_name_rep(names, max_length).to(device)
langs_rep = batched_lang_rep(langs).to(device)
padded_names_rep = torch.nn.utils.rnn.pack_padded_sequence(
names_rep, X_lengths, enforce_sorted=False
)
if verbose:
print(names_rep.shape, padded_names_rep.data.shape)
print("--")
if verbose:
print(names)
print_char(names_rep)
print("--")
if verbose:
print_char(padded_names_rep.data)
print("Lang Rep", langs_rep.data)
print("Batch sizes", padded_names_rep.batch_sizes)
return padded_names_rep.to(device), langs_rep
# the code torch.nn.utils.rnn.pack_padded_sequence, optimises the batching as below.
# 
p, l = batched_dataloader(3, X_train, y_train, True)
# # Training
def train(net, opt, criterion, n_points):
opt.zero_grad()
total_loss = 0
data_ = dataloader(n_points, X_train, y_train)
total_loss = 0
for name, language, name_ohe, lang_rep in data_:
hidden = net.init_hidden()
for i in range(name_ohe.size()[0]):
output, hidden = net(name_ohe[i : i + 1], hidden)
loss = criterion(output, lang_rep)
loss.backward(retain_graph=True)
total_loss += loss
opt.step()
return total_loss / n_points
def train_batch(net, opt, criterion, n_points, device="cpu"):
net.train().to(device)
opt.zero_grad()
batch_input, batch_groundtruth = batched_dataloader(
n_points, X_train, y_train, False, device
)
output, hidden = net(batch_input)
loss = criterion(output, batch_groundtruth)
loss.backward()
opt.step()
return loss
net = RNN_net(n_letters, n_hidden, n_languages)
criterion = nn.NLLLoss()
opt = optim.SGD(net.parameters(), lr=0.01, momentum=0.9)
train(net, opt, criterion, 256)
net = RNN_net(n_letters, n_hidden, n_languages)
criterion = nn.NLLLoss()
opt = optim.SGD(net.parameters(), lr=0.01, momentum=0.9)
train_batch(net, opt, criterion, 256)
# ## Full training setup
def train_setup(
net,
lr=0.01,
n_batches=100,
batch_size=10,
momentum=0.9,
display_freq=5,
device="cpu",
):
net = net.to(device)
criterion = nn.NLLLoss()
opt = optim.SGD(net.parameters(), lr=lr, momentum=momentum)
loss_arr = np.zeros(n_batches + 1)
for i in range(n_batches):
loss_arr[i + 1] = (
loss_arr[i] * i + train_batch(net, opt, criterion, batch_size, device)
) / (i + 1)
if i % display_freq == display_freq - 1:
clear_output(wait=True)
print("Iteration", i, "Loss", loss_arr[i])
# print('Top-1:', eval(net, len(X_test), 1, X_test, y_test), 'Top-2:', eval(net, len(X_test), 2, X_test, y_test))
plt.figure()
plt.plot(loss_arr[1:i], "-*")
plt.xlabel("Iteration")
plt.ylabel("Loss")
plt.show()
print("\n\n")
print(
"Top-1:",
eval(net, len(X_test), 1, X_test, y_test, device),
"Top-2:",
eval(net, len(X_test), 2, X_test, y_test, device),
)
# # RNN
net = RNN_net(n_letters, 128, n_languages)
train_setup(
net, lr=0.15, n_batches=1000, batch_size=512, display_freq=500
) # CPU Training example
net = RNN_net(n_letters, 128, n_languages)
train_setup(
net, lr=0.15, n_batches=1000, batch_size=512, display_freq=100, device=device_gpu
) # GPU Training Example
# # LSTM cell
class LSTM_net(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(LSTM_net, self).__init__()
self.hidden_size = hidden_size
self.lstm_cell = nn.LSTM(input_size, hidden_size)
self.h2o = nn.Linear(hidden_size, output_size)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, input, hidden=None):
out, hidden = self.lstm_cell(input, hidden)
output = self.h2o(hidden[0].view(-1, self.hidden_size))
output = self.softmax(output)
return output, hidden
def init_hidden(self, batch_size=1):
return (
torch.zeros(1, batch_size, self.hidden_size),
torch.zeros(1, batch_size, self.hidden_size),
)
n_hidden = 128
net = LSTM_net(n_letters, n_hidden, n_languages)
train_setup(
net, lr=0.15, n_batches=8000, batch_size=512, display_freq=1000, device=device_gpu
)
# # GRU Cell
class GRU_net(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(GRU_net, self).__init__()
self.hidden_size = hidden_size
self.gru_cell = nn.GRU(input_size, hidden_size)
self.h2o = nn.Linear(hidden_size, output_size)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, input, hidden=None):
out, hidden = self.gru_cell(input, hidden)
output = self.h2o(hidden.view(-1, self.hidden_size))
output = self.softmax(output)
return output, hidden
n_hidden = 128
net = GRU_net(n_letters, n_hidden, n_languages)
train_setup(
net, lr=0.15, n_batches=8000, batch_size=512, display_freq=1000, device=device_gpu
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/674/129674541.ipynb
|
name2lang
|
rp1985
|
[{"Id": 129674541, "ScriptId": 38555634, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2723147, "CreationDate": "05/15/2023 16:40:46", "VersionNumber": 2.0, "Title": "Batch Seq : RNN, LSTM, GRU", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 376.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 375.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 16}]
|
[{"Id": 185989641, "KernelVersionId": 129674541, "SourceDatasetVersionId": 716293}]
|
[{"Id": 716293, "DatasetId": 367261, "DatasourceVersionId": 736561, "CreatorUserId": 2094338, "LicenseName": "Unknown", "CreationDate": "10/02/2019 07:10:09", "VersionNumber": 1.0, "Title": "name2lang", "Slug": "name2lang", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 367261, "CreatorUserId": 2094338, "OwnerUserId": 2094338.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 716293.0, "CurrentDatasourceVersionId": 736561.0, "ForumId": 379079, "Type": 2, "CreationDate": "10/02/2019 07:10:09", "LastActivityDate": "10/02/2019", "TotalViews": 4181, "TotalDownloads": 262, "TotalVotes": 2, "TotalKernels": 3}]
|
[{"Id": 2094338, "UserName": "rp1985", "DisplayName": "RP1985", "RegisterDate": "07/24/2018", "PerformanceTier": 0}]
|
# # Introduction.
# 
# * Introduce dataset
# * Data processing - Test-train split, encoding, visualisation
# * Why do we use batching
# * Batching for sequence models
# * Padding and packing in PyTorch
# * Training with batched dataset
# * Comparison of performance with batching and on GPU
# Most of the concepts explained in this Notebook are learnt from course [Deep Learning](https://padhai.onefourthlabs.in/courses/dl-feb-2019)
# # Setting up the dependencies
from io import open
import os, string, random, time, math
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from sklearn.model_selection import train_test_split
import torch
import torch.nn as nn
import torch.optim as optim
# Instantiates the device to be used as GPU/CPU based on availability
device_gpu = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
from IPython.display import clear_output
import warnings
warnings.filterwarnings("ignore")
# # Dataset
# # Pre-processing
languages = []
data = []
X = []
y = []
with open("/kaggle/input/name2lang/name2lang.txt", "r") as f:
for line in f:
line = line.split(",")
name = line[0].strip()
lang = line[1].strip()
if not lang in languages:
languages.append(lang)
X.append(name)
y.append(lang)
data.append((name, lang))
n_languages = len(languages)
print(languages)
print(data[0:10])
# # Test-train split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=0, stratify=y
)
print(len(X_train), len(X_test))
# ## Encoding names and language
all_letters = string.ascii_letters + " .,;'"
n_letters = len(all_letters)
def name_rep(name):
rep = torch.zeros(len(name), 1, n_letters)
for index, letter in enumerate(name):
pos = all_letters.find(letter)
rep[index][0][pos] = 1
return rep
def lang_rep(lang):
return torch.tensor([languages.index(lang)], dtype=torch.long)
name_rep("Abreu")
lang_rep("Portuguese")
count = {}
for l in languages:
count[l] = 0
for d in data:
count[d[1]] += 1
print(count)
plt_ = sns.barplot(x=list(count.keys()), y=list(count.values()))
plt_.set_xticklabels(plt_.get_xticklabels(), rotation=90)
plt.show()
# ## Basic network and testing inference
class RNN_net(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(RNN_net, self).__init__()
self.hidden_size = hidden_size
self.rnn_cell = nn.RNN(input_size, hidden_size)
self.h2o = nn.Linear(hidden_size, output_size)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, input_, hidden=None, batch_size=1):
out, hidden = self.rnn_cell(input_, hidden)
output = self.h2o(hidden.view(-1, self.hidden_size))
output = self.softmax(output)
return output, hidden
def init_hidden(self, batch_size=1):
return torch.zeros(1, batch_size, self.hidden_size)
n_hidden = 128
net = RNN_net(n_letters, n_hidden, n_languages)
def infer(net, name, device="cpu"):
name_ohe = name_rep(name).to(device)
output, hidden = net(name_ohe)
if type(hidden) is tuple: # For LSTM
hidden = hidden[0]
index = torch.argmax(hidden)
return output
infer(net, "Adam")
# # Evaluate model
def dataloader(npoints, X_, y_):
to_ret = []
for i in range(npoints):
index_ = np.random.randint(len(X_))
name, lang = X_[index_], y_[index_]
to_ret.append((name, lang, name_rep(name), lang_rep(lang)))
return to_ret
dataloader(2, X_train, y_train)
def eval(net, n_points, topk, X_, y_, device="cpu"):
net = net.eval().to(device)
data_ = dataloader(n_points, X_, y_)
correct = 0
for name, language, name_ohe, lang_rep in data_:
output = infer(net, name, device)
val, indices = output.topk(topk)
indices = indices.to("cpu")
if lang_rep in indices:
correct += 1
accuracy = correct / n_points
return accuracy
eval(net, 1000, 1, X_test, y_test)
# # Batching
# Batching can be done in many ways, one can be represted as.
# 
def batched_name_rep(names, max_word_size):
rep = torch.zeros(max_word_size, len(names), n_letters)
for name_index, name in enumerate(names):
for letter_index, letter in enumerate(name):
pos = all_letters.find(letter)
rep[letter_index][name_index][pos] = 1
return rep
def print_char(name_reps):
name_reps = name_reps.view((-1, name_reps.size()[-1]))
for t in name_reps:
if torch.sum(t) == 0:
print("<pad>")
else:
index = t.argmax()
print(all_letters[index])
out_ = batched_name_rep(["Shyam", "Ram"], 5)
print(out_)
print(out_.shape)
print_char(out_)
def batched_lang_rep(langs):
rep = torch.zeros([len(langs)], dtype=torch.long)
for index, lang in enumerate(langs):
rep[index] = languages.index(lang)
return rep
def batched_dataloader(npoints, X_, y_, verbose=False, device="cpu"):
names = []
langs = []
X_lengths = []
for i in range(npoints):
index_ = np.random.randint(len(X_))
name, lang = X_[index_], y_[index_]
X_lengths.append(len(name))
names.append(name)
langs.append(lang)
max_length = max(X_lengths)
names_rep = batched_name_rep(names, max_length).to(device)
langs_rep = batched_lang_rep(langs).to(device)
padded_names_rep = torch.nn.utils.rnn.pack_padded_sequence(
names_rep, X_lengths, enforce_sorted=False
)
if verbose:
print(names_rep.shape, padded_names_rep.data.shape)
print("--")
if verbose:
print(names)
print_char(names_rep)
print("--")
if verbose:
print_char(padded_names_rep.data)
print("Lang Rep", langs_rep.data)
print("Batch sizes", padded_names_rep.batch_sizes)
return padded_names_rep.to(device), langs_rep
# the code torch.nn.utils.rnn.pack_padded_sequence, optimises the batching as below.
# 
p, l = batched_dataloader(3, X_train, y_train, True)
# # Training
def train(net, opt, criterion, n_points):
opt.zero_grad()
total_loss = 0
data_ = dataloader(n_points, X_train, y_train)
total_loss = 0
for name, language, name_ohe, lang_rep in data_:
hidden = net.init_hidden()
for i in range(name_ohe.size()[0]):
output, hidden = net(name_ohe[i : i + 1], hidden)
loss = criterion(output, lang_rep)
loss.backward(retain_graph=True)
total_loss += loss
opt.step()
return total_loss / n_points
def train_batch(net, opt, criterion, n_points, device="cpu"):
net.train().to(device)
opt.zero_grad()
batch_input, batch_groundtruth = batched_dataloader(
n_points, X_train, y_train, False, device
)
output, hidden = net(batch_input)
loss = criterion(output, batch_groundtruth)
loss.backward()
opt.step()
return loss
net = RNN_net(n_letters, n_hidden, n_languages)
criterion = nn.NLLLoss()
opt = optim.SGD(net.parameters(), lr=0.01, momentum=0.9)
train(net, opt, criterion, 256)
net = RNN_net(n_letters, n_hidden, n_languages)
criterion = nn.NLLLoss()
opt = optim.SGD(net.parameters(), lr=0.01, momentum=0.9)
train_batch(net, opt, criterion, 256)
# ## Full training setup
def train_setup(
net,
lr=0.01,
n_batches=100,
batch_size=10,
momentum=0.9,
display_freq=5,
device="cpu",
):
net = net.to(device)
criterion = nn.NLLLoss()
opt = optim.SGD(net.parameters(), lr=lr, momentum=momentum)
loss_arr = np.zeros(n_batches + 1)
for i in range(n_batches):
loss_arr[i + 1] = (
loss_arr[i] * i + train_batch(net, opt, criterion, batch_size, device)
) / (i + 1)
if i % display_freq == display_freq - 1:
clear_output(wait=True)
print("Iteration", i, "Loss", loss_arr[i])
# print('Top-1:', eval(net, len(X_test), 1, X_test, y_test), 'Top-2:', eval(net, len(X_test), 2, X_test, y_test))
plt.figure()
plt.plot(loss_arr[1:i], "-*")
plt.xlabel("Iteration")
plt.ylabel("Loss")
plt.show()
print("\n\n")
print(
"Top-1:",
eval(net, len(X_test), 1, X_test, y_test, device),
"Top-2:",
eval(net, len(X_test), 2, X_test, y_test, device),
)
# # RNN
net = RNN_net(n_letters, 128, n_languages)
train_setup(
net, lr=0.15, n_batches=1000, batch_size=512, display_freq=500
) # CPU Training example
net = RNN_net(n_letters, 128, n_languages)
train_setup(
net, lr=0.15, n_batches=1000, batch_size=512, display_freq=100, device=device_gpu
) # GPU Training Example
# # LSTM cell
class LSTM_net(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(LSTM_net, self).__init__()
self.hidden_size = hidden_size
self.lstm_cell = nn.LSTM(input_size, hidden_size)
self.h2o = nn.Linear(hidden_size, output_size)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, input, hidden=None):
out, hidden = self.lstm_cell(input, hidden)
output = self.h2o(hidden[0].view(-1, self.hidden_size))
output = self.softmax(output)
return output, hidden
def init_hidden(self, batch_size=1):
return (
torch.zeros(1, batch_size, self.hidden_size),
torch.zeros(1, batch_size, self.hidden_size),
)
n_hidden = 128
net = LSTM_net(n_letters, n_hidden, n_languages)
train_setup(
net, lr=0.15, n_batches=8000, batch_size=512, display_freq=1000, device=device_gpu
)
# # GRU Cell
class GRU_net(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(GRU_net, self).__init__()
self.hidden_size = hidden_size
self.gru_cell = nn.GRU(input_size, hidden_size)
self.h2o = nn.Linear(hidden_size, output_size)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, input, hidden=None):
out, hidden = self.gru_cell(input, hidden)
output = self.h2o(hidden.view(-1, self.hidden_size))
output = self.softmax(output)
return output, hidden
n_hidden = 128
net = GRU_net(n_letters, n_hidden, n_languages)
train_setup(
net, lr=0.15, n_batches=8000, batch_size=512, display_freq=1000, device=device_gpu
)
| false | 0 | 3,591 | 16 | 3,610 | 3,591 |
||
129603241
|
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
train.head()
train.columns
features = [
"clonesize",
"honeybee",
"bumbles",
"andrena",
"osmia",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"AverageOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
"AverageOfLowerTRange",
"RainingDays",
"AverageRainingDays",
"fruitset",
"fruitmass",
"seeds",
]
X = train[features]
y = train["yield"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1, random_state=0)
print(X.shape, X_train.shape, X_test.shape)
model = LinearRegression()
model.fit(X_train, y_train)
preds = model.predict(X_test)
preds
mae = mean_absolute_error(y_test, preds)
print(mae)
id = test["id"]
test = test[features]
model.fit(X, y)
predictions = model.predict(test)
predictions
final = pd.DataFrame()
final.index = id
final["yield"] = predictions
final.to_csv("submission.csv")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/603/129603241.ipynb
| null | null |
[{"Id": 129603241, "ScriptId": 38537243, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10554992, "CreationDate": "05/15/2023 07:11:13", "VersionNumber": 1.0, "Title": "Blueberry prediction", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 45.0, "LinesInsertedFromPrevious": 45.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score
train = pd.read_csv("/kaggle/input/playground-series-s3e14/train.csv")
test = pd.read_csv("/kaggle/input/playground-series-s3e14/test.csv")
train.head()
train.columns
features = [
"clonesize",
"honeybee",
"bumbles",
"andrena",
"osmia",
"MaxOfUpperTRange",
"MinOfUpperTRange",
"AverageOfUpperTRange",
"MaxOfLowerTRange",
"MinOfLowerTRange",
"AverageOfLowerTRange",
"RainingDays",
"AverageRainingDays",
"fruitset",
"fruitmass",
"seeds",
]
X = train[features]
y = train["yield"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1, random_state=0)
print(X.shape, X_train.shape, X_test.shape)
model = LinearRegression()
model.fit(X_train, y_train)
preds = model.predict(X_test)
preds
mae = mean_absolute_error(y_test, preds)
print(mae)
id = test["id"]
test = test[features]
model.fit(X, y)
predictions = model.predict(test)
predictions
final = pd.DataFrame()
final.index = id
final["yield"] = predictions
final.to_csv("submission.csv")
| false | 0 | 423 | 0 | 423 | 423 |
||
129603101
|
import numpy as np
import pandas as pd
from tensorflow.keras.utils import load_img
from keras.preprocessing.image import ImageDataGenerator
from keras.utils import to_categorical
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import random
import os
from keras.callbacks import EarlyStopping, ReduceLROnPlateau
from keras.models import Sequential
from keras.layers import (
Conv2D,
MaxPooling2D,
Dropout,
Flatten,
Dense,
Activation,
BatchNormalization,
)
from keras.preprocessing.image import ImageDataGenerator
width = 28
height = 28
size = (width, height)
channels = 1
train = pd.read_csv(
"../input/mnist-train-mini/mnist_train_mini.csv", header=None
) # change how files are read
test = pd.read_csv("../input/mnist-test-mini/mnist_test_mini.csv", header=None)
X = train.drop([0], 1).values
y = train[0].values
X = X / 255.0
X = X.reshape(-1, 28, 28, 1)
y = to_categorical(y)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
model = Sequential() # model
# convolutional layer, 32 output filters, strides is 3 by 3
model.add(Conv2D(32, (3, 3), activation="relu", input_shape=(width, height, channels)))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(64, (3, 3), activation="relu"))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(128, (3, 3), activation="relu"))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(512, activation="relu"))
model.add(BatchNormalization())
model.add(Dropout(0.5))
model.add(Dense(10, activation="softmax")) # Change from 2 to 10
model.compile(
loss="categorical_crossentropy", optimizer="rmsprop", metrics=["accuracy"]
)
model.summary()
earlystop = EarlyStopping(patience=10)
learning_rate_reduction = ReduceLROnPlateau(
monitor="val_acc", patience=2, verbose=1, factor=0.5, min_lr=0.00001
)
callbacks = [earlystop, learning_rate_reduction]
# no horizontal flip
train_datagen = ImageDataGenerator(
rotation_range=15,
zoom_range=0.2,
width_shift_range=0.1,
height_shift_range=0.1,
shear_range=0.1,
)
validation_datagen = ImageDataGenerator()
train_gen = train_datagen.flow(X_train, y_train, batch_size=15)
test_gen = validation_datagen.flow(X_test, y_test, batch_size=15) # flow
epochs = 50
batch_size = 15
train_steps = X_train.shape[0] // batch_size
valid_steps = X_test.shape[0] // batch_size
history = model.fit(
train_gen,
epochs=epochs,
steps_per_epoch=train_steps,
validation_data=test_gen,
validation_steps=valid_steps,
callbacks=callbacks,
)
fig, (ax2) = plt.subplots(1, 1, figsize=(12, 12))
ax2.plot(history.history["accuracy"], color="b", label="Training accuracy")
ax2.plot(history.history["val_accuracy"], color="r", label="Test accuracy")
ax2.set_xticks(np.arange(1, epochs, 1))
legend = plt.legend(loc="best", shadow=True)
plt.tight_layout()
plt.show()
y_pred = model.predict(X_test)
X_test__ = X_test.reshape(X_test.shape[0], 28, 28)
fig, axis = plt.subplots(4, 4, figsize=(12, 14))
for i, ax in enumerate(axis.flat):
ax.imshow(X_test__[i], cmap="binary")
ax.set(
title=f"Real Number is {y_test[i].argmax()}\nPredict Number is {y_pred[i].argmax()}"
)
test_x = test.drop([0], 1).values
test_x = test_x / 255.0
test_x = test_x.reshape(-1, 28, 28, 1)
# predict test data
pred = model.predict(test_x, verbose=1)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/603/129603101.ipynb
| null | null |
[{"Id": 129603101, "ScriptId": 38536308, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15001012, "CreationDate": "05/15/2023 07:10:06", "VersionNumber": 2.0, "Title": "CNN Part 2", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 111.0, "LinesInsertedFromPrevious": 45.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 66.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
from tensorflow.keras.utils import load_img
from keras.preprocessing.image import ImageDataGenerator
from keras.utils import to_categorical
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import random
import os
from keras.callbacks import EarlyStopping, ReduceLROnPlateau
from keras.models import Sequential
from keras.layers import (
Conv2D,
MaxPooling2D,
Dropout,
Flatten,
Dense,
Activation,
BatchNormalization,
)
from keras.preprocessing.image import ImageDataGenerator
width = 28
height = 28
size = (width, height)
channels = 1
train = pd.read_csv(
"../input/mnist-train-mini/mnist_train_mini.csv", header=None
) # change how files are read
test = pd.read_csv("../input/mnist-test-mini/mnist_test_mini.csv", header=None)
X = train.drop([0], 1).values
y = train[0].values
X = X / 255.0
X = X.reshape(-1, 28, 28, 1)
y = to_categorical(y)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
model = Sequential() # model
# convolutional layer, 32 output filters, strides is 3 by 3
model.add(Conv2D(32, (3, 3), activation="relu", input_shape=(width, height, channels)))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(64, (3, 3), activation="relu"))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(128, (3, 3), activation="relu"))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(512, activation="relu"))
model.add(BatchNormalization())
model.add(Dropout(0.5))
model.add(Dense(10, activation="softmax")) # Change from 2 to 10
model.compile(
loss="categorical_crossentropy", optimizer="rmsprop", metrics=["accuracy"]
)
model.summary()
earlystop = EarlyStopping(patience=10)
learning_rate_reduction = ReduceLROnPlateau(
monitor="val_acc", patience=2, verbose=1, factor=0.5, min_lr=0.00001
)
callbacks = [earlystop, learning_rate_reduction]
# no horizontal flip
train_datagen = ImageDataGenerator(
rotation_range=15,
zoom_range=0.2,
width_shift_range=0.1,
height_shift_range=0.1,
shear_range=0.1,
)
validation_datagen = ImageDataGenerator()
train_gen = train_datagen.flow(X_train, y_train, batch_size=15)
test_gen = validation_datagen.flow(X_test, y_test, batch_size=15) # flow
epochs = 50
batch_size = 15
train_steps = X_train.shape[0] // batch_size
valid_steps = X_test.shape[0] // batch_size
history = model.fit(
train_gen,
epochs=epochs,
steps_per_epoch=train_steps,
validation_data=test_gen,
validation_steps=valid_steps,
callbacks=callbacks,
)
fig, (ax2) = plt.subplots(1, 1, figsize=(12, 12))
ax2.plot(history.history["accuracy"], color="b", label="Training accuracy")
ax2.plot(history.history["val_accuracy"], color="r", label="Test accuracy")
ax2.set_xticks(np.arange(1, epochs, 1))
legend = plt.legend(loc="best", shadow=True)
plt.tight_layout()
plt.show()
y_pred = model.predict(X_test)
X_test__ = X_test.reshape(X_test.shape[0], 28, 28)
fig, axis = plt.subplots(4, 4, figsize=(12, 14))
for i, ax in enumerate(axis.flat):
ax.imshow(X_test__[i], cmap="binary")
ax.set(
title=f"Real Number is {y_test[i].argmax()}\nPredict Number is {y_pred[i].argmax()}"
)
test_x = test.drop([0], 1).values
test_x = test_x / 255.0
test_x = test_x.reshape(-1, 28, 28, 1)
# predict test data
pred = model.predict(test_x, verbose=1)
| false | 0 | 1,283 | 0 | 1,283 | 1,283 |
||
129603179
|
<jupyter_start><jupyter_text>Telco Customer Churn
### Context
"Predict behavior to retain customers. You can analyze all relevant customer data and develop focused customer retention programs." [IBM Sample Data Sets]
### Content
Each row represents a customer, each column contains customer’s attributes described on the column Metadata.
**The data set includes information about:**
+ Customers who left within the last month – the column is called Churn
+ Services that each customer has signed up for – phone, multiple lines, internet, online security, online backup, device protection, tech support, and streaming TV and movies
+ Customer account information – how long they’ve been a customer, contract, payment method, paperless billing, monthly charges, and total charges
+ Demographic info about customers – gender, age range, and if they have partners and dependents
### Inspiration
To explore this type of models and learn more about the subject.
**New version from IBM:**
https://community.ibm.com/community/user/businessanalytics/blogs/steven-macko/2019/07/11/telco-customer-churn-1113
Kaggle dataset identifier: telco-customer-churn
<jupyter_code>import pandas as pd
df = pd.read_csv('telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 7043 entries, 0 to 7042
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 customerID 7043 non-null object
1 gender 7043 non-null object
2 SeniorCitizen 7043 non-null int64
3 Partner 7043 non-null object
4 Dependents 7043 non-null object
5 tenure 7043 non-null int64
6 PhoneService 7043 non-null object
7 MultipleLines 7043 non-null object
8 InternetService 7043 non-null object
9 OnlineSecurity 7043 non-null object
10 OnlineBackup 7043 non-null object
11 DeviceProtection 7043 non-null object
12 TechSupport 7043 non-null object
13 StreamingTV 7043 non-null object
14 StreamingMovies 7043 non-null object
15 Contract 7043 non-null object
16 PaperlessBilling 7043 non-null object
17 PaymentMethod 7043 non-null object
18 MonthlyCharges 7043 non-null float64
19 TotalCharges 7043 non-null object
20 Churn 7043 non-null object
dtypes: float64(1), int64(2), object(18)
memory usage: 1.1+ MB
<jupyter_text>Examples:
{
"customerID": "7590-VHVEG",
"gender": "Female",
"SeniorCitizen": 0,
"Partner": "Yes",
"Dependents": "No",
"tenure": 1,
"PhoneService": "No",
"MultipleLines": "No phone service",
"InternetService": "DSL",
"OnlineSecurity": "No",
"OnlineBackup": "Yes",
"DeviceProtection": "No",
"TechSupport": "No",
"StreamingTV": "No",
"StreamingMovies": "No",
"Contract": "Month-to-month",
"PaperlessBilling": "Yes",
"PaymentMethod": "Electronic check",
"MonthlyCharges": 29.85,
"TotalCharges": 29.85,
"...": "and 1 more columns"
}
{
"customerID": "5575-GNVDE",
"gender": "Male",
"SeniorCitizen": 0,
"Partner": "No",
"Dependents": "No",
"tenure": 34,
"PhoneService": "Yes",
"MultipleLines": "No",
"InternetService": "DSL",
"OnlineSecurity": "Yes",
"OnlineBackup": "No",
"DeviceProtection": "Yes",
"TechSupport": "No",
"StreamingTV": "No",
"StreamingMovies": "No",
"Contract": "One year",
"PaperlessBilling": "No",
"PaymentMethod": "Mailed check",
"MonthlyCharges": 56.95,
"TotalCharges": 1889.5,
"...": "and 1 more columns"
}
{
"customerID": "3668-QPYBK",
"gender": "Male",
"SeniorCitizen": 0,
"Partner": "No",
"Dependents": "No",
"tenure": 2,
"PhoneService": "Yes",
"MultipleLines": "No",
"InternetService": "DSL",
"OnlineSecurity": "Yes",
"OnlineBackup": "Yes",
"DeviceProtection": "No",
"TechSupport": "No",
"StreamingTV": "No",
"StreamingMovies": "No",
"Contract": "Month-to-month",
"PaperlessBilling": "Yes",
"PaymentMethod": "Mailed check",
"MonthlyCharges": 53.85,
"TotalCharges": 108.15,
"...": "and 1 more columns"
}
{
"customerID": "7795-CFOCW",
"gender": "Male",
"SeniorCitizen": 0,
"Partner": "No",
"Dependents": "No",
"tenure": 45,
"PhoneService": "No",
"MultipleLines": "No phone service",
"InternetService": "DSL",
"OnlineSecurity": "Yes",
"OnlineBackup": "No",
"DeviceProtection": "Yes",
"TechSupport": "Yes",
"StreamingTV": "No",
"StreamingMovies": "No",
"Contract": "One year",
"PaperlessBilling": "No",
"PaymentMethod": "Bank transfer (automatic)",
"MonthlyCharges": 42.3,
"TotalCharges": 1840.75,
"...": "and 1 more columns"
}
<jupyter_script># # importing libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # for visualization
import seaborn as sns # for visualization
from sklearn.preprocessing import MinMaxScaler # for scaling in between 0 to 1
from sklearn.model_selection import (
train_test_split,
) # to split dataset into trainig set and test set
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
dataframe = pd.read_csv(
r"/kaggle/input/telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv"
)
dataframe.head()
dataframe.shape
# # Data preprocessing and EDA
dataframe.isna().sum()
# HURRAY there is no null value
# whistle podu
df = dataframe.copy()
# as we know that customer Id has less impact on the dataset for prediction so we can remove it.
df = df.drop(columns=["customerID"])
df.head()
# lets check is there any duplicate records or not .
df.duplicated().sum()
# dropping the duplicate records.
df.drop_duplicates(inplace=True)
df.shape
df.info()
# totalcharges attribute should not be object type it needs to be numerical.
df.TotalCharges.values
# df['TotalCharges']=df.TotalCharges.astype(float)
# the above command giving error as there is " " as a entry in a row so we should exclude that row first.
df = df.drop(df.index[(df["TotalCharges"] == " ")], axis=0)
df["TotalCharges"] = df.TotalCharges.astype(float)
df["TotalCharges"].dtype
# checking the unique values of all column
for col in df.columns:
if df[col].dtypes == "object":
print(f"{col}:{df[col].unique()}")
df.replace({"No internet service": "No"}, inplace=True)
df.replace({"No phone service": "No"}, inplace=True)
# plotting graph to find relation among them
# list the all column having catagorical value
cat = [i for i in df.columns if len(df[i].unique()) < 4]
cat
# making a list of all the attribute without churn attribute
lis = []
for i in df.columns:
if i != "Churn":
lis.append(i)
plt.figure(figsize=(10, 100))
for n, column in enumerate(cat):
plot = plt.subplot(21, 2, n + 1)
sns.countplot(x=df[column], data=df, hue=None, saturation=0.75, color="yellow")
plt.title(f"{column.title()}", weight="bold")
plt.tight_layout()
plt.show()
# lets check the relation of each attribute with the churn
plt.figure(figsize=(15, 70))
for n, column in enumerate(lis):
plot = plt.subplot(20, 2, n + 1)
mc_churn_no = df[df.Churn == "No"][column]
mc_churn_yes = df[df.Churn == "Yes"][column]
plt.xlabel(column)
plt.ylabel("Number Of Customers")
plt.title("Customer Churn Prediction Visualiztion")
plt.hist(
[mc_churn_yes, mc_churn_no],
rwidth=0.95,
color=["green", "red"],
label=["Churn=Yes", "Churn=No"],
)
plt.tight_layout()
plt.legend()
plt.show()
# lets encode the yes as 1 and 0 no as 0
# convert female into 1 and male into 0
yes_no_columns = [
"Partner",
"Dependents",
"PhoneService",
"MultipleLines",
"OnlineSecurity",
"OnlineBackup",
"DeviceProtection",
"TechSupport",
"StreamingTV",
"StreamingMovies",
"PaperlessBilling",
"Churn",
]
for col in yes_no_columns:
df[col].replace({"Yes": 1, "No": 0}, inplace=True)
df["gender"].replace({"Female": 1, "Male": 0}, inplace=True)
df
# as we can see there is some more columns having categorical data other than binary category so we have to handle them by using dummy variable.
dummy_var = ["InternetService", "Contract", "PaymentMethod"]
df = pd.get_dummies(data=df, columns=dummy_var)
df.shape
# we can check the corelation between the attributes by heatmap
cor = df.corr()
feature = cor.index
plt.figure(figsize=(20, 20))
g = sns.heatmap(df[feature].corr(), annot=True, cmap="RdYlGn")
# One hot encoding for categorical columns
cols_to_scale = ["tenure", "MonthlyCharges", "TotalCharges"]
scaler = MinMaxScaler()
df[cols_to_scale] = scaler.fit_transform(df[cols_to_scale])
df
# # **MODEL CREATION**
x = df.drop(["Churn"], axis=1)
y = df["Churn"]
xtrain, xtest, ytrain, ytest = train_test_split(x, y, test_size=0.2, random_state=1)
# importing tensor flow for ANN
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import Sequential
model = keras.Sequential(
[
keras.layers.Dense(26, input_shape=(26,), activation="relu"),
keras.layers.Dense(10, activation="relu"),
keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
model.fit(xtrain, ytrain, epochs=100)
model.evaluate(xtest, ytest)
yp = model.predict(xtest)
# as sigmoid function only provide the probabilities ,we have to convert it into the response i.e 1 or 0
ypred = []
for i in yp:
if i >= 0.5:
ypred.append(1)
else:
ypred.append(0)
# # **CLASSIFICATION REPORT**
# for checking output in various aspect we are importing confusion matrix,classification report
from sklearn.metrics import confusion_matrix, classification_report
print(classification_report(ytest, ypred))
cm = tf.math.confusion_matrix(labels=ytest, predictions=ypred)
plt.figure(figsize=(10, 7))
sns.heatmap(cm, annot=True, fmt="d")
plt.xlabel("actual")
plt.ylabel("pridicted")
from sklearn.metrics import accuracy_score
round(accuracy_score(ytest, ypred), 2)
print("precision")
p = round(955 / (955 + 192), 2)
print(p)
print("accuracy")
a = round((955 + 163) / (955 + 163 + 192 + 92), 2)
print(a)
print("sensitivity")
r1 = round((955) / (955 + 92), 2)
print(r1)
print("specificity")
r0 = round((163) / (163 + 192), 2)
print(r0)
print("f1_score")
f = 2 * ((p * r1) / (p + r1))
print(round(f, 2))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/603/129603179.ipynb
|
telco-customer-churn
|
blastchar
|
[{"Id": 129603179, "ScriptId": 38502895, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12953019, "CreationDate": "05/15/2023 07:10:44", "VersionNumber": 2.0, "Title": "customer churn prediction with 80% accuracy", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 212.0, "LinesInsertedFromPrevious": 8.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 204.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 6}]
|
[{"Id": 185838906, "KernelVersionId": 129603179, "SourceDatasetVersionId": 18858}]
|
[{"Id": 18858, "DatasetId": 13996, "DatasourceVersionId": 18858, "CreatorUserId": 1574575, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "02/23/2018 18:20:00", "VersionNumber": 1.0, "Title": "Telco Customer Churn", "Slug": "telco-customer-churn", "Subtitle": "Focused customer retention programs", "Description": "### Context\n\n\"Predict behavior to retain customers. You can analyze all relevant customer data and develop focused customer retention programs.\" [IBM Sample Data Sets]\n\n### Content\n\nEach row represents a customer, each column contains customer\u2019s attributes described on the column Metadata.\n\n**The data set includes information about:**\n\n+ Customers who left within the last month \u2013 the column is called Churn\n+ Services that each customer has signed up for \u2013 phone, multiple lines, internet, online security, online backup, device protection, tech support, and streaming TV and movies\n+ Customer account information \u2013 how long they\u2019ve been a customer, contract, payment method, paperless billing, monthly charges, and total charges\n+ Demographic info about customers \u2013 gender, age range, and if they have partners and dependents\n\n### Inspiration\n\nTo explore this type of models and learn more about the subject.\n\n**New version from IBM:**\nhttps://community.ibm.com/community/user/businessanalytics/blogs/steven-macko/2019/07/11/telco-customer-churn-1113", "VersionNotes": "Initial release", "TotalCompressedBytes": 977501.0, "TotalUncompressedBytes": 977501.0}]
|
[{"Id": 13996, "CreatorUserId": 1574575, "OwnerUserId": 1574575.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 18858.0, "CurrentDatasourceVersionId": 18858.0, "ForumId": 21551, "Type": 2, "CreationDate": "02/23/2018 18:20:00", "LastActivityDate": "02/23/2018", "TotalViews": 1765427, "TotalDownloads": 190987, "TotalVotes": 2480, "TotalKernels": 1184}]
|
[{"Id": 1574575, "UserName": "blastchar", "DisplayName": "BlastChar", "RegisterDate": "01/23/2018", "PerformanceTier": 1}]
|
# # importing libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # for visualization
import seaborn as sns # for visualization
from sklearn.preprocessing import MinMaxScaler # for scaling in between 0 to 1
from sklearn.model_selection import (
train_test_split,
) # to split dataset into trainig set and test set
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
dataframe = pd.read_csv(
r"/kaggle/input/telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv"
)
dataframe.head()
dataframe.shape
# # Data preprocessing and EDA
dataframe.isna().sum()
# HURRAY there is no null value
# whistle podu
df = dataframe.copy()
# as we know that customer Id has less impact on the dataset for prediction so we can remove it.
df = df.drop(columns=["customerID"])
df.head()
# lets check is there any duplicate records or not .
df.duplicated().sum()
# dropping the duplicate records.
df.drop_duplicates(inplace=True)
df.shape
df.info()
# totalcharges attribute should not be object type it needs to be numerical.
df.TotalCharges.values
# df['TotalCharges']=df.TotalCharges.astype(float)
# the above command giving error as there is " " as a entry in a row so we should exclude that row first.
df = df.drop(df.index[(df["TotalCharges"] == " ")], axis=0)
df["TotalCharges"] = df.TotalCharges.astype(float)
df["TotalCharges"].dtype
# checking the unique values of all column
for col in df.columns:
if df[col].dtypes == "object":
print(f"{col}:{df[col].unique()}")
df.replace({"No internet service": "No"}, inplace=True)
df.replace({"No phone service": "No"}, inplace=True)
# plotting graph to find relation among them
# list the all column having catagorical value
cat = [i for i in df.columns if len(df[i].unique()) < 4]
cat
# making a list of all the attribute without churn attribute
lis = []
for i in df.columns:
if i != "Churn":
lis.append(i)
plt.figure(figsize=(10, 100))
for n, column in enumerate(cat):
plot = plt.subplot(21, 2, n + 1)
sns.countplot(x=df[column], data=df, hue=None, saturation=0.75, color="yellow")
plt.title(f"{column.title()}", weight="bold")
plt.tight_layout()
plt.show()
# lets check the relation of each attribute with the churn
plt.figure(figsize=(15, 70))
for n, column in enumerate(lis):
plot = plt.subplot(20, 2, n + 1)
mc_churn_no = df[df.Churn == "No"][column]
mc_churn_yes = df[df.Churn == "Yes"][column]
plt.xlabel(column)
plt.ylabel("Number Of Customers")
plt.title("Customer Churn Prediction Visualiztion")
plt.hist(
[mc_churn_yes, mc_churn_no],
rwidth=0.95,
color=["green", "red"],
label=["Churn=Yes", "Churn=No"],
)
plt.tight_layout()
plt.legend()
plt.show()
# lets encode the yes as 1 and 0 no as 0
# convert female into 1 and male into 0
yes_no_columns = [
"Partner",
"Dependents",
"PhoneService",
"MultipleLines",
"OnlineSecurity",
"OnlineBackup",
"DeviceProtection",
"TechSupport",
"StreamingTV",
"StreamingMovies",
"PaperlessBilling",
"Churn",
]
for col in yes_no_columns:
df[col].replace({"Yes": 1, "No": 0}, inplace=True)
df["gender"].replace({"Female": 1, "Male": 0}, inplace=True)
df
# as we can see there is some more columns having categorical data other than binary category so we have to handle them by using dummy variable.
dummy_var = ["InternetService", "Contract", "PaymentMethod"]
df = pd.get_dummies(data=df, columns=dummy_var)
df.shape
# we can check the corelation between the attributes by heatmap
cor = df.corr()
feature = cor.index
plt.figure(figsize=(20, 20))
g = sns.heatmap(df[feature].corr(), annot=True, cmap="RdYlGn")
# One hot encoding for categorical columns
cols_to_scale = ["tenure", "MonthlyCharges", "TotalCharges"]
scaler = MinMaxScaler()
df[cols_to_scale] = scaler.fit_transform(df[cols_to_scale])
df
# # **MODEL CREATION**
x = df.drop(["Churn"], axis=1)
y = df["Churn"]
xtrain, xtest, ytrain, ytest = train_test_split(x, y, test_size=0.2, random_state=1)
# importing tensor flow for ANN
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import Sequential
model = keras.Sequential(
[
keras.layers.Dense(26, input_shape=(26,), activation="relu"),
keras.layers.Dense(10, activation="relu"),
keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
model.fit(xtrain, ytrain, epochs=100)
model.evaluate(xtest, ytest)
yp = model.predict(xtest)
# as sigmoid function only provide the probabilities ,we have to convert it into the response i.e 1 or 0
ypred = []
for i in yp:
if i >= 0.5:
ypred.append(1)
else:
ypred.append(0)
# # **CLASSIFICATION REPORT**
# for checking output in various aspect we are importing confusion matrix,classification report
from sklearn.metrics import confusion_matrix, classification_report
print(classification_report(ytest, ypred))
cm = tf.math.confusion_matrix(labels=ytest, predictions=ypred)
plt.figure(figsize=(10, 7))
sns.heatmap(cm, annot=True, fmt="d")
plt.xlabel("actual")
plt.ylabel("pridicted")
from sklearn.metrics import accuracy_score
round(accuracy_score(ytest, ypred), 2)
print("precision")
p = round(955 / (955 + 192), 2)
print(p)
print("accuracy")
a = round((955 + 163) / (955 + 163 + 192 + 92), 2)
print(a)
print("sensitivity")
r1 = round((955) / (955 + 92), 2)
print(r1)
print("specificity")
r0 = round((163) / (163 + 192), 2)
print(r0)
print("f1_score")
f = 2 * ((p * r1) / (p + r1))
print(round(f, 2))
|
[{"telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv": {"column_names": "[\"customerID\", \"gender\", \"SeniorCitizen\", \"Partner\", \"Dependents\", \"tenure\", \"PhoneService\", \"MultipleLines\", \"InternetService\", \"OnlineSecurity\", \"OnlineBackup\", \"DeviceProtection\", \"TechSupport\", \"StreamingTV\", \"StreamingMovies\", \"Contract\", \"PaperlessBilling\", \"PaymentMethod\", \"MonthlyCharges\", \"TotalCharges\", \"Churn\"]", "column_data_types": "{\"customerID\": \"object\", \"gender\": \"object\", \"SeniorCitizen\": \"int64\", \"Partner\": \"object\", \"Dependents\": \"object\", \"tenure\": \"int64\", \"PhoneService\": \"object\", \"MultipleLines\": \"object\", \"InternetService\": \"object\", \"OnlineSecurity\": \"object\", \"OnlineBackup\": \"object\", \"DeviceProtection\": \"object\", \"TechSupport\": \"object\", \"StreamingTV\": \"object\", \"StreamingMovies\": \"object\", \"Contract\": \"object\", \"PaperlessBilling\": \"object\", \"PaymentMethod\": \"object\", \"MonthlyCharges\": \"float64\", \"TotalCharges\": \"object\", \"Churn\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 7043 entries, 0 to 7042\nData columns (total 21 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 customerID 7043 non-null object \n 1 gender 7043 non-null object \n 2 SeniorCitizen 7043 non-null int64 \n 3 Partner 7043 non-null object \n 4 Dependents 7043 non-null object \n 5 tenure 7043 non-null int64 \n 6 PhoneService 7043 non-null object \n 7 MultipleLines 7043 non-null object \n 8 InternetService 7043 non-null object \n 9 OnlineSecurity 7043 non-null object \n 10 OnlineBackup 7043 non-null object \n 11 DeviceProtection 7043 non-null object \n 12 TechSupport 7043 non-null object \n 13 StreamingTV 7043 non-null object \n 14 StreamingMovies 7043 non-null object \n 15 Contract 7043 non-null object \n 16 PaperlessBilling 7043 non-null object \n 17 PaymentMethod 7043 non-null object \n 18 MonthlyCharges 7043 non-null float64\n 19 TotalCharges 7043 non-null object \n 20 Churn 7043 non-null object \ndtypes: float64(1), int64(2), object(18)\nmemory usage: 1.1+ MB\n", "summary": "{\"SeniorCitizen\": {\"count\": 7043.0, \"mean\": 0.1621468124378816, \"std\": 0.3686116056100131, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"tenure\": {\"count\": 7043.0, \"mean\": 32.37114865824223, \"std\": 24.55948102309446, \"min\": 0.0, \"25%\": 9.0, \"50%\": 29.0, \"75%\": 55.0, \"max\": 72.0}, \"MonthlyCharges\": {\"count\": 7043.0, \"mean\": 64.76169246059918, \"std\": 30.090047097678493, \"min\": 18.25, \"25%\": 35.5, \"50%\": 70.35, \"75%\": 89.85, \"max\": 118.75}}", "examples": "{\"customerID\":{\"0\":\"7590-VHVEG\",\"1\":\"5575-GNVDE\",\"2\":\"3668-QPYBK\",\"3\":\"7795-CFOCW\"},\"gender\":{\"0\":\"Female\",\"1\":\"Male\",\"2\":\"Male\",\"3\":\"Male\"},\"SeniorCitizen\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Partner\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"Dependents\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"tenure\":{\"0\":1,\"1\":34,\"2\":2,\"3\":45},\"PhoneService\":{\"0\":\"No\",\"1\":\"Yes\",\"2\":\"Yes\",\"3\":\"No\"},\"MultipleLines\":{\"0\":\"No phone service\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No phone service\"},\"InternetService\":{\"0\":\"DSL\",\"1\":\"DSL\",\"2\":\"DSL\",\"3\":\"DSL\"},\"OnlineSecurity\":{\"0\":\"No\",\"1\":\"Yes\",\"2\":\"Yes\",\"3\":\"Yes\"},\"OnlineBackup\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"Yes\",\"3\":\"No\"},\"DeviceProtection\":{\"0\":\"No\",\"1\":\"Yes\",\"2\":\"No\",\"3\":\"Yes\"},\"TechSupport\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"Yes\"},\"StreamingTV\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"StreamingMovies\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"Contract\":{\"0\":\"Month-to-month\",\"1\":\"One year\",\"2\":\"Month-to-month\",\"3\":\"One year\"},\"PaperlessBilling\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"Yes\",\"3\":\"No\"},\"PaymentMethod\":{\"0\":\"Electronic check\",\"1\":\"Mailed check\",\"2\":\"Mailed check\",\"3\":\"Bank transfer (automatic)\"},\"MonthlyCharges\":{\"0\":29.85,\"1\":56.95,\"2\":53.85,\"3\":42.3},\"TotalCharges\":{\"0\":\"29.85\",\"1\":\"1889.5\",\"2\":\"108.15\",\"3\":\"1840.75\"},\"Churn\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"Yes\",\"3\":\"No\"}}"}}]
| true | 1 |
<start_data_description><data_path>telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv:
<column_names>
['customerID', 'gender', 'SeniorCitizen', 'Partner', 'Dependents', 'tenure', 'PhoneService', 'MultipleLines', 'InternetService', 'OnlineSecurity', 'OnlineBackup', 'DeviceProtection', 'TechSupport', 'StreamingTV', 'StreamingMovies', 'Contract', 'PaperlessBilling', 'PaymentMethod', 'MonthlyCharges', 'TotalCharges', 'Churn']
<column_types>
{'customerID': 'object', 'gender': 'object', 'SeniorCitizen': 'int64', 'Partner': 'object', 'Dependents': 'object', 'tenure': 'int64', 'PhoneService': 'object', 'MultipleLines': 'object', 'InternetService': 'object', 'OnlineSecurity': 'object', 'OnlineBackup': 'object', 'DeviceProtection': 'object', 'TechSupport': 'object', 'StreamingTV': 'object', 'StreamingMovies': 'object', 'Contract': 'object', 'PaperlessBilling': 'object', 'PaymentMethod': 'object', 'MonthlyCharges': 'float64', 'TotalCharges': 'object', 'Churn': 'object'}
<dataframe_Summary>
{'SeniorCitizen': {'count': 7043.0, 'mean': 0.1621468124378816, 'std': 0.3686116056100131, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'tenure': {'count': 7043.0, 'mean': 32.37114865824223, 'std': 24.55948102309446, 'min': 0.0, '25%': 9.0, '50%': 29.0, '75%': 55.0, 'max': 72.0}, 'MonthlyCharges': {'count': 7043.0, 'mean': 64.76169246059918, 'std': 30.090047097678493, 'min': 18.25, '25%': 35.5, '50%': 70.35, '75%': 89.85, 'max': 118.75}}
<dataframe_info>
RangeIndex: 7043 entries, 0 to 7042
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 customerID 7043 non-null object
1 gender 7043 non-null object
2 SeniorCitizen 7043 non-null int64
3 Partner 7043 non-null object
4 Dependents 7043 non-null object
5 tenure 7043 non-null int64
6 PhoneService 7043 non-null object
7 MultipleLines 7043 non-null object
8 InternetService 7043 non-null object
9 OnlineSecurity 7043 non-null object
10 OnlineBackup 7043 non-null object
11 DeviceProtection 7043 non-null object
12 TechSupport 7043 non-null object
13 StreamingTV 7043 non-null object
14 StreamingMovies 7043 non-null object
15 Contract 7043 non-null object
16 PaperlessBilling 7043 non-null object
17 PaymentMethod 7043 non-null object
18 MonthlyCharges 7043 non-null float64
19 TotalCharges 7043 non-null object
20 Churn 7043 non-null object
dtypes: float64(1), int64(2), object(18)
memory usage: 1.1+ MB
<some_examples>
{'customerID': {'0': '7590-VHVEG', '1': '5575-GNVDE', '2': '3668-QPYBK', '3': '7795-CFOCW'}, 'gender': {'0': 'Female', '1': 'Male', '2': 'Male', '3': 'Male'}, 'SeniorCitizen': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Partner': {'0': 'Yes', '1': 'No', '2': 'No', '3': 'No'}, 'Dependents': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'tenure': {'0': 1, '1': 34, '2': 2, '3': 45}, 'PhoneService': {'0': 'No', '1': 'Yes', '2': 'Yes', '3': 'No'}, 'MultipleLines': {'0': 'No phone service', '1': 'No', '2': 'No', '3': 'No phone service'}, 'InternetService': {'0': 'DSL', '1': 'DSL', '2': 'DSL', '3': 'DSL'}, 'OnlineSecurity': {'0': 'No', '1': 'Yes', '2': 'Yes', '3': 'Yes'}, 'OnlineBackup': {'0': 'Yes', '1': 'No', '2': 'Yes', '3': 'No'}, 'DeviceProtection': {'0': 'No', '1': 'Yes', '2': 'No', '3': 'Yes'}, 'TechSupport': {'0': 'No', '1': 'No', '2': 'No', '3': 'Yes'}, 'StreamingTV': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'StreamingMovies': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'Contract': {'0': 'Month-to-month', '1': 'One year', '2': 'Month-to-month', '3': 'One year'}, 'PaperlessBilling': {'0': 'Yes', '1': 'No', '2': 'Yes', '3': 'No'}, 'PaymentMethod': {'0': 'Electronic check', '1': 'Mailed check', '2': 'Mailed check', '3': 'Bank transfer (automatic)'}, 'MonthlyCharges': {'0': 29.85, '1': 56.95, '2': 53.85, '3': 42.3}, 'TotalCharges': {'0': '29.85', '1': '1889.5', '2': '108.15', '3': '1840.75'}, 'Churn': {'0': 'No', '1': 'No', '2': 'Yes', '3': 'No'}}
<end_description>
| 1,981 | 6 | 3,528 | 1,981 |
129797822
|
# # Notebook for OCR pdf to text
# imports
# ## Simple Apprach
import platform
from tempfile import TemporaryDirectory
from pathlib import Path
import pytesseract
from pdf2image import convert_from_path
from PIL import Image
# Path of the Input pdf
PDF_file = Path(r"/kaggle/input/ocr-hacking/d.pdf")
# Store all the pages of the PDF in a variable
image_file_list = []
text_file = Path("out_text.txt")
def main():
"""Main execution point of the program"""
with TemporaryDirectory() as tempdir:
"""
Part #1 : Converting PDF to images
"""
pdf_pages = convert_from_path(PDF_file, 500)
for page_enumeration, page in enumerate(pdf_pages, start=1):
filename = f"{tempdir}\page_{page_enumeration:03}.jpg"
page.save(filename, "JPEG")
image_file_list.append(filename)
"""
Part #2 - Recognizing text from the images using OCR
"""
with open(text_file, "a") as output_file:
for image_file in image_file_list:
text = str(((pytesseract.image_to_string(Image.open(image_file)))))
text = text.replace("-\n", "")
output_file.write(text)
if __name__ == "__main__":
main()
# ## More advanced approach
# import libs
try:
from PIL import Image
except ImportError:
import Image
import cv2
import pytesseract
import os
import numpy as np
import pandas as pd
import re
from pdf2image import convert_from_bytes
from pathlib import Path
# Some help functions
def get_conf(page_gray):
"""return a average confidence value of OCR result"""
df = pytesseract.image_to_data(page_gray, output_type="data.frame")
df.drop(df[df.conf == -1].index.values, inplace=True)
df.reset_index()
return df.conf.mean()
def deskew(image):
"""deskew the image"""
gray = cv2.bitwise_not(image)
temp_arr = cv2.threshold(gray, 0, 255, cv2.THRESH_BINARY + cv2.THRESH_OTSU)[1]
coords = np.column_stack(np.where(temp_arr > 0))
angle = cv2.minAreaRect(coords)[-1]
if angle < -45:
angle = -(90 + angle)
else:
angle = -angle
(h, w) = image.shape[:2]
center = (w // 2, h // 2)
M = cv2.getRotationMatrix2D(center, angle, 1.0)
rotated = cv2.warpAffine(
image, M, (w, h), flags=cv2.INTER_CUBIC, borderMode=cv2.BORDER_REPLICATE
)
return rotated
"""
Main part of OCR:
pages_df: save eextracted text for each pdf file, index by page
OCR_dic : dict for saving df of each pdf, filename is the key
"""
file_list = ["/kaggle/input/ocr-hacking-2/example_scan.pdf"]
def combine_texts(list_of_text):
"""Taking a list of texts and combining them into one large chunk of text."""
combined_text = " ".join(list_of_text)
return combined_text
OCR_dic = {}
for file in file_list:
# convert pdf into image
pdf_file = convert_from_bytes(open(file, "rb").read())
# create a df to save each pdf's text
pages_df = pd.DataFrame(columns=["conf", "text"])
for i, page in enumerate(pdf_file):
try:
# transfer image of pdf_file into array
page_arr = np.asarray(page)
# transfer into grayscale
page_arr_gray = cv2.cvtColor(page_arr, cv2.COLOR_BGR2GRAY)
page_arr_gray = cv2.fastNlMeansDenoising(page_arr_gray, None, 3, 7, 21)
page_deskew = deskew(page_arr_gray)
# cal confidence value
page_conf = get_conf(page_deskew)
# extract string
d = pytesseract.image_to_data(
page_deskew, output_type=pytesseract.Output.DICT
)
d_df = pd.DataFrame.from_dict(d)
# get block number
block_num = int(d_df.loc[d_df["level"] == 2, ["block_num"]].max())
print("number of blocks: ", block_num)
head_index = d_df[d_df["block_num"] == 1].index.values
foot_index = d_df[d_df["block_num"] == block_num].index.values
d_df.drop(head_index, inplace=True)
d_df.drop(foot_index, inplace=True)
# combine text in dataframe
text = combine_texts(d_df.loc[d_df["level"] == 5, "text"].values)
pages_df = pages_df.append(
{"conf": page_conf, "text": text}, ignore_index=True
)
except Exception as e:
# if can't extract then give some notes into df
if hasattr(e, "message"):
pages_df = pages_df.append(
{"conf": -1, "text": e.message}, ignore_index=True
)
else:
pages_df = pages_df.append({"conf": -1, "text": e}, ignore_index=True)
continue
# save df into a dict with filename as key
OCR_dic[file] = pages_df
print("{} is done".format(file))
print(OCR_dic[file_list[0]]["text"][0])
print(OCR_dic[file_list[0]]["text"][0])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/797/129797822.ipynb
| null | null |
[{"Id": 129797822, "ScriptId": 38570169, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9079821, "CreationDate": "05/16/2023 14:31:04", "VersionNumber": 1.0, "Title": "notebookd1d0b82359", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 155.0, "LinesInsertedFromPrevious": 155.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Notebook for OCR pdf to text
# imports
# ## Simple Apprach
import platform
from tempfile import TemporaryDirectory
from pathlib import Path
import pytesseract
from pdf2image import convert_from_path
from PIL import Image
# Path of the Input pdf
PDF_file = Path(r"/kaggle/input/ocr-hacking/d.pdf")
# Store all the pages of the PDF in a variable
image_file_list = []
text_file = Path("out_text.txt")
def main():
"""Main execution point of the program"""
with TemporaryDirectory() as tempdir:
"""
Part #1 : Converting PDF to images
"""
pdf_pages = convert_from_path(PDF_file, 500)
for page_enumeration, page in enumerate(pdf_pages, start=1):
filename = f"{tempdir}\page_{page_enumeration:03}.jpg"
page.save(filename, "JPEG")
image_file_list.append(filename)
"""
Part #2 - Recognizing text from the images using OCR
"""
with open(text_file, "a") as output_file:
for image_file in image_file_list:
text = str(((pytesseract.image_to_string(Image.open(image_file)))))
text = text.replace("-\n", "")
output_file.write(text)
if __name__ == "__main__":
main()
# ## More advanced approach
# import libs
try:
from PIL import Image
except ImportError:
import Image
import cv2
import pytesseract
import os
import numpy as np
import pandas as pd
import re
from pdf2image import convert_from_bytes
from pathlib import Path
# Some help functions
def get_conf(page_gray):
"""return a average confidence value of OCR result"""
df = pytesseract.image_to_data(page_gray, output_type="data.frame")
df.drop(df[df.conf == -1].index.values, inplace=True)
df.reset_index()
return df.conf.mean()
def deskew(image):
"""deskew the image"""
gray = cv2.bitwise_not(image)
temp_arr = cv2.threshold(gray, 0, 255, cv2.THRESH_BINARY + cv2.THRESH_OTSU)[1]
coords = np.column_stack(np.where(temp_arr > 0))
angle = cv2.minAreaRect(coords)[-1]
if angle < -45:
angle = -(90 + angle)
else:
angle = -angle
(h, w) = image.shape[:2]
center = (w // 2, h // 2)
M = cv2.getRotationMatrix2D(center, angle, 1.0)
rotated = cv2.warpAffine(
image, M, (w, h), flags=cv2.INTER_CUBIC, borderMode=cv2.BORDER_REPLICATE
)
return rotated
"""
Main part of OCR:
pages_df: save eextracted text for each pdf file, index by page
OCR_dic : dict for saving df of each pdf, filename is the key
"""
file_list = ["/kaggle/input/ocr-hacking-2/example_scan.pdf"]
def combine_texts(list_of_text):
"""Taking a list of texts and combining them into one large chunk of text."""
combined_text = " ".join(list_of_text)
return combined_text
OCR_dic = {}
for file in file_list:
# convert pdf into image
pdf_file = convert_from_bytes(open(file, "rb").read())
# create a df to save each pdf's text
pages_df = pd.DataFrame(columns=["conf", "text"])
for i, page in enumerate(pdf_file):
try:
# transfer image of pdf_file into array
page_arr = np.asarray(page)
# transfer into grayscale
page_arr_gray = cv2.cvtColor(page_arr, cv2.COLOR_BGR2GRAY)
page_arr_gray = cv2.fastNlMeansDenoising(page_arr_gray, None, 3, 7, 21)
page_deskew = deskew(page_arr_gray)
# cal confidence value
page_conf = get_conf(page_deskew)
# extract string
d = pytesseract.image_to_data(
page_deskew, output_type=pytesseract.Output.DICT
)
d_df = pd.DataFrame.from_dict(d)
# get block number
block_num = int(d_df.loc[d_df["level"] == 2, ["block_num"]].max())
print("number of blocks: ", block_num)
head_index = d_df[d_df["block_num"] == 1].index.values
foot_index = d_df[d_df["block_num"] == block_num].index.values
d_df.drop(head_index, inplace=True)
d_df.drop(foot_index, inplace=True)
# combine text in dataframe
text = combine_texts(d_df.loc[d_df["level"] == 5, "text"].values)
pages_df = pages_df.append(
{"conf": page_conf, "text": text}, ignore_index=True
)
except Exception as e:
# if can't extract then give some notes into df
if hasattr(e, "message"):
pages_df = pages_df.append(
{"conf": -1, "text": e.message}, ignore_index=True
)
else:
pages_df = pages_df.append({"conf": -1, "text": e}, ignore_index=True)
continue
# save df into a dict with filename as key
OCR_dic[file] = pages_df
print("{} is done".format(file))
print(OCR_dic[file_list[0]]["text"][0])
print(OCR_dic[file_list[0]]["text"][0])
| false | 0 | 1,443 | 0 | 1,443 | 1,443 |
||
129797081
|
<jupyter_start><jupyter_text>Pakistan House Price dataset
**Dataset Description:**
The dataset contains information about properties. Each property has a unique property ID and is associated with a location ID based on the subcategory of the city. The dataset includes the following attributes:
Property ID: Unique identifier for each property.
Location ID: Unique identifier for each location within a city.
Page URL: The URL of the webpage where the property was published.
Property Type: Categorization of the property into six types: House, FarmHouse, Upper Portion, Lower Portion, Flat, or Room.
Price: The price of the property, which is the dependent feature in this dataset.
City: The city where the property is located. The dataset includes five cities: Lahore, Karachi, Faisalabad, Rawalpindi, and Islamabad.
Province: The state or province where the city is located.
Location: Different types of locations within each city.
Latitude and Longitude: Geographic coordinates of the cities.
Steps Involved in the Analysis:
**Statistical Analysis:**
Data Types: Determine the data types of the attributes.
Level of Measurement: Identify the level of measurement for each attribute.
Summary Statistics: Calculate mean, standard deviation, minimum, and maximum values for numerical attributes.
Data Cleaning:
Filling Null Values: Handle missing values in the dataset.
Duplicate Values: Remove duplicate records, if any.
Correcting Data Types: Ensure the correct data types for each attribute.
Outliers Detection: Identify and handle outliers in the data.
Exploratory Data Analysis (EDA):
**Visualization:** Use libraries such as Seaborn, Matplotlib, and Plotly to visualize the data and gain insights.
**Model Building:**
Libraries: Utilize libraries like Sklearn and pickle.
List of Models: Build models using Linear Regression, Decision Tree, Random Forest, K-Nearest Neighbors (KNN), XG Boost, Gradient Boost, and Ada Boost.
Model Saving: Save the selected model into a pickle file for future use.
I hope this captures the essence of the provided information. Let me know if you need any further assistance!
Kaggle dataset identifier: pakistan-house-price-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/pakistan-house-price-dataset/zameen-updated.csv")
import pandas_profiling
# df.profile_report()
# # Dataset overview:
# ## Vales with significant number of NaN entries: baths, bedrooms, agency, agent
# Map of which city/province (price, quantity)
# Date added (how fresh)?
# NaN - 26.2% agency, agent ?independent?
df["date_added"] = pd.to_datetime(df["date_added"])
df["time_on_market"] = df["date_added"].min() - df["date_added"]
df["time_on_market"] = -df["time_on_market"]
df["time_on_market"] = df["time_on_market"].dt.days
counts = pd.series(index=df.date_added, data=df)
df["date_added"].resample("7D").sum()
pd.qcut(df["time_on_market"], q=4)
df["date_added"] = pd.to_datetime(df["date_added"])
df["date_added"].dtype
import pandas as pd
import matplotlib.pyplot as plt
# Assuming df is your DataFrame containing the 'date_added' column
# Convert 'date_added' column to datetime if it's not already
df["date_added"] = pd.to_datetime(df["date_added"])
# Define the bin size and the date range you want to start from
bin_size = pd.DateOffset(days=7)
start_date = df["date_added"].min()
# Create a new column with the binned dates
df["date_bins"] = pd.cut(
df["date_added"],
bins=pd.date_range(
start=start_date,
periods=(df["date_added"].max() - start_date).days // 7 + 2,
freq=bin_size,
),
)
# Calculate the counts within each bin
bin_counts = df["date_bins"].value_counts().sort_index()
# Set the figure size
plt.figure(figsize=(12, 6))
# Plot the binned counts
ax = bin_counts.plot(kind="bar")
ax.set_xlabel("Date Range")
ax.set_ylabel("Count")
# Optional: Format the x-axis tick labels
bin_labels = bin_counts.index.strftime("%Y-%m-%d")
ax.set_xticklabels(bin_labels, rotation=45)
# Show the plot
plt.show()
g = df["date_added"].value_counts().sort_index().head(100).plot(kind="bar")
# # print(g)
df.groupby(["time_on_market"]).time_on_market.count().sort_values(ascending=False).head(
20
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/797/129797081.ipynb
|
pakistan-house-price-dataset
|
jillanisofttech
|
[{"Id": 129797081, "ScriptId": 38392778, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6545977, "CreationDate": "05/16/2023 14:25:57", "VersionNumber": 2.0, "Title": "Pakistan housing market exploration", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 85.0, "LinesInsertedFromPrevious": 40.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 45.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186166495, "KernelVersionId": 129797081, "SourceDatasetVersionId": 5617966}]
|
[{"Id": 5617966, "DatasetId": 3230351, "DatasourceVersionId": 5693146, "CreatorUserId": 7237430, "LicenseName": "Other (specified in description)", "CreationDate": "05/06/2023 11:49:55", "VersionNumber": 1.0, "Title": "Pakistan House Price dataset", "Slug": "pakistan-house-price-dataset", "Subtitle": "Pakistan House Price Updated Dataset", "Description": "**Dataset Description:**\nThe dataset contains information about properties. Each property has a unique property ID and is associated with a location ID based on the subcategory of the city. The dataset includes the following attributes:\n\nProperty ID: Unique identifier for each property.\nLocation ID: Unique identifier for each location within a city.\nPage URL: The URL of the webpage where the property was published.\nProperty Type: Categorization of the property into six types: House, FarmHouse, Upper Portion, Lower Portion, Flat, or Room.\nPrice: The price of the property, which is the dependent feature in this dataset.\nCity: The city where the property is located. The dataset includes five cities: Lahore, Karachi, Faisalabad, Rawalpindi, and Islamabad.\nProvince: The state or province where the city is located.\nLocation: Different types of locations within each city.\nLatitude and Longitude: Geographic coordinates of the cities.\nSteps Involved in the Analysis:\n\n**Statistical Analysis:**\n\nData Types: Determine the data types of the attributes.\nLevel of Measurement: Identify the level of measurement for each attribute.\nSummary Statistics: Calculate mean, standard deviation, minimum, and maximum values for numerical attributes.\nData Cleaning:\n\nFilling Null Values: Handle missing values in the dataset.\nDuplicate Values: Remove duplicate records, if any.\nCorrecting Data Types: Ensure the correct data types for each attribute.\nOutliers Detection: Identify and handle outliers in the data.\nExploratory Data Analysis (EDA):\n\n**Visualization:** Use libraries such as Seaborn, Matplotlib, and Plotly to visualize the data and gain insights.\n**Model Building:**\n\nLibraries: Utilize libraries like Sklearn and pickle.\nList of Models: Build models using Linear Regression, Decision Tree, Random Forest, K-Nearest Neighbors (KNN), XG Boost, Gradient Boost, and Ada Boost.\nModel Saving: Save the selected model into a pickle file for future use.\nI hope this captures the essence of the provided information. Let me know if you need any further assistance!", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3230351, "CreatorUserId": 7237430, "OwnerUserId": 7237430.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5617966.0, "CurrentDatasourceVersionId": 5693146.0, "ForumId": 3295490, "Type": 2, "CreationDate": "05/06/2023 11:49:55", "LastActivityDate": "05/06/2023", "TotalViews": 6070, "TotalDownloads": 538, "TotalVotes": 29, "TotalKernels": 9}]
|
[{"Id": 7237430, "UserName": "jillanisofttech", "DisplayName": "Jillani Soft Tech", "RegisterDate": "04/21/2021", "PerformanceTier": 3}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/pakistan-house-price-dataset/zameen-updated.csv")
import pandas_profiling
# df.profile_report()
# # Dataset overview:
# ## Vales with significant number of NaN entries: baths, bedrooms, agency, agent
# Map of which city/province (price, quantity)
# Date added (how fresh)?
# NaN - 26.2% agency, agent ?independent?
df["date_added"] = pd.to_datetime(df["date_added"])
df["time_on_market"] = df["date_added"].min() - df["date_added"]
df["time_on_market"] = -df["time_on_market"]
df["time_on_market"] = df["time_on_market"].dt.days
counts = pd.series(index=df.date_added, data=df)
df["date_added"].resample("7D").sum()
pd.qcut(df["time_on_market"], q=4)
df["date_added"] = pd.to_datetime(df["date_added"])
df["date_added"].dtype
import pandas as pd
import matplotlib.pyplot as plt
# Assuming df is your DataFrame containing the 'date_added' column
# Convert 'date_added' column to datetime if it's not already
df["date_added"] = pd.to_datetime(df["date_added"])
# Define the bin size and the date range you want to start from
bin_size = pd.DateOffset(days=7)
start_date = df["date_added"].min()
# Create a new column with the binned dates
df["date_bins"] = pd.cut(
df["date_added"],
bins=pd.date_range(
start=start_date,
periods=(df["date_added"].max() - start_date).days // 7 + 2,
freq=bin_size,
),
)
# Calculate the counts within each bin
bin_counts = df["date_bins"].value_counts().sort_index()
# Set the figure size
plt.figure(figsize=(12, 6))
# Plot the binned counts
ax = bin_counts.plot(kind="bar")
ax.set_xlabel("Date Range")
ax.set_ylabel("Count")
# Optional: Format the x-axis tick labels
bin_labels = bin_counts.index.strftime("%Y-%m-%d")
ax.set_xticklabels(bin_labels, rotation=45)
# Show the plot
plt.show()
g = df["date_added"].value_counts().sort_index().head(100).plot(kind="bar")
# # print(g)
df.groupby(["time_on_market"]).time_on_market.count().sort_values(ascending=False).head(
20
)
| false | 1 | 815 | 0 | 1,306 | 815 |
||
129797721
|
<jupyter_start><jupyter_text>Vehicle Insurance Claim Fraud Detection
### Vehicle Insurance Fraud Detection
Vehicle insurance fraud involves conspiring to make false or exaggerated claims involving property damage or personal injuries following an accident. Some common examples include staged accidents where fraudsters deliberately “arrange” for accidents to occur; the use of phantom passengers where people who were not even at the scene of the accident claim to have suffered grievous injury, and make false personal injury claims where personal injuries are grossly exaggerated.
### About this dataset
This dataset contains vehicle dataset - attribute, model, accident details, etc along with policy details - policy type, tenure etc. The target is to detect if a claim application is fraudulent or not - FraudFound_P
Kaggle dataset identifier: vehicle-claim-fraud-detection
<jupyter_script>import pandas as pd
import numpy as np
df = pd.read_csv("../input/vehicle-claim-fraud-detection/fraud_oracle.csv")
df.head()
df.info()
for column in df:
print(column, ":\n", df[column].unique(), "\n")
df["PolicyNumber"].unique()
df = df.drop(columns="PolicyNumber")
df["FraudFound_P"] = df["FraudFound_P"].astype("bool")
df[["PoliceReportFiled", "WitnessPresent"]] = df[
["PoliceReportFiled", "WitnessPresent"]
].replace("No", "0")
df[["PoliceReportFiled", "WitnessPresent"]] = df[
["PoliceReportFiled", "WitnessPresent"]
].replace("Yes", "1")
df[["PoliceReportFiled", "WitnessPresent"]] = df[
["PoliceReportFiled", "WitnessPresent"]
].astype("bool")
df[["Age", "AgeOfPolicyHolder"]].head(20)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/797/129797721.ipynb
|
vehicle-claim-fraud-detection
|
shivamb
|
[{"Id": 129797721, "ScriptId": 38600464, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12290769, "CreationDate": "05/16/2023 14:30:25", "VersionNumber": 1.0, "Title": "notebook02d421f064", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 25.0, "LinesInsertedFromPrevious": 25.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186167252, "KernelVersionId": 129797721, "SourceDatasetVersionId": 2947792}]
|
[{"Id": 2947792, "DatasetId": 1807255, "DatasourceVersionId": 2995312, "CreatorUserId": 1571785, "LicenseName": "CC0: Public Domain", "CreationDate": "12/20/2021 04:26:36", "VersionNumber": 1.0, "Title": "Vehicle Insurance Claim Fraud Detection", "Slug": "vehicle-claim-fraud-detection", "Subtitle": "Fraud detection use-case for vehicle insurance industry", "Description": "### Vehicle Insurance Fraud Detection\n\nVehicle insurance fraud involves conspiring to make false or exaggerated claims involving property damage or personal injuries following an accident. Some common examples include staged accidents where fraudsters deliberately \u201carrange\u201d for accidents to occur; the use of phantom passengers where people who were not even at the scene of the accident claim to have suffered grievous injury, and make false personal injury claims where personal injuries are grossly exaggerated.\n\n\n### About this dataset\n\nThis dataset contains vehicle dataset - attribute, model, accident details, etc along with policy details - policy type, tenure etc. The target is to detect if a claim application is fraudulent or not - FraudFound_P", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1807255, "CreatorUserId": 1571785, "OwnerUserId": 1571785.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2947792.0, "CurrentDatasourceVersionId": 2995312.0, "ForumId": 1829871, "Type": 2, "CreationDate": "12/20/2021 04:26:36", "LastActivityDate": "12/20/2021", "TotalViews": 57587, "TotalDownloads": 7181, "TotalVotes": 115, "TotalKernels": 18}]
|
[{"Id": 1571785, "UserName": "shivamb", "DisplayName": "Shivam Bansal", "RegisterDate": "01/22/2018", "PerformanceTier": 4}]
|
import pandas as pd
import numpy as np
df = pd.read_csv("../input/vehicle-claim-fraud-detection/fraud_oracle.csv")
df.head()
df.info()
for column in df:
print(column, ":\n", df[column].unique(), "\n")
df["PolicyNumber"].unique()
df = df.drop(columns="PolicyNumber")
df["FraudFound_P"] = df["FraudFound_P"].astype("bool")
df[["PoliceReportFiled", "WitnessPresent"]] = df[
["PoliceReportFiled", "WitnessPresent"]
].replace("No", "0")
df[["PoliceReportFiled", "WitnessPresent"]] = df[
["PoliceReportFiled", "WitnessPresent"]
].replace("Yes", "1")
df[["PoliceReportFiled", "WitnessPresent"]] = df[
["PoliceReportFiled", "WitnessPresent"]
].astype("bool")
df[["Age", "AgeOfPolicyHolder"]].head(20)
| false | 1 | 249 | 0 | 450 | 249 |
||
129779452
|
from corus import load_lenta2
from tqdm import tqdm
path = "/kaggle/working/lenta-ru-news.csv.bz2"
records = load_lenta2(path)
next(records)
title = []
text = []
topic = []
tags = []
for record in tqdm(records):
text.append(record.text)
topic.append(record.topic)
import pandas as pd
import numpy as np
main_df = pd.DataFrame({"text": text, "topic": topic})
main_df["topic"].unique()
main_df.sample(frac=1)
# Оставляем Экономика, Интернет и СМИ, Спорт, Культура, Наука и Техника
main_df = main_df.loc[
main_df["topic"].isin(
["Экономика", "Интернет и СМИ", "Спорт", "Культура", "Наука и Техника"]
)
]
main_df["topic"] = main_df["topic"].replace(
["Экономика", "Интернет и СМИ", "Спорт", "Культура", "Наука и Техника"],
[0, 1, 2, 3, 4],
)
main_df["topic"].unique()
text = list(main_df["text"])[:10000]
label = list(main_df["topic"])[:10000]
seq_len = [len(str(i).split()) for i in tqdm(text)]
pd.Series(seq_len).hist(bins=50, figsize=[10, 7])
print(max(seq_len))
from sklearn.model_selection import train_test_split
text_train, text_test, labels_train, labels_test = train_test_split(
text, label, train_size=0.7, random_state=42
)
import pandas as pd
import numpy as np
import random
import torch
import transformers
import torch.nn as nn
from transformers import AutoModel, BertTokenizer, BertForSequenceClassification
from transformers import TrainingArguments, Trainer
from datasets import load_metric, Dataset
from sklearn.metrics import classification_report, f1_score
from torch.utils.data import Dataset
import torch
model = BertForSequenceClassification.from_pretrained(
"/kaggle/working/rubert-base-cased-sentence", num_labels=5
).to("cuda")
tokenizer = BertTokenizer.from_pretrained("/kaggle/working/rubert-base-cased-sentence")
max_seq_len = 512
tokens_train = tokenizer.batch_encode_plus(
text_train, max_length=max_seq_len, padding="max_length", truncation=True
)
tokens_test = tokenizer.batch_encode_plus(
text_test, max_length=max_seq_len, padding="max_length", truncation=True
)
# labels_train
class Data(torch.utils.data.Dataset):
def __init__(self, encodings, labels):
self.encodings = encodings
self.labels = labels
def __getitem__(self, idx):
item = {k: torch.tensor(v[idx]) for k, v in self.encodings.items()}
item["labels"] = torch.tensor([self.labels[idx]])
return item
def __len__(self):
return len(self.labels)
train_dataset = Data(tokens_train, labels_train)
test_dataset = Data(tokens_test, labels_test)
train_dataset.__getitem__(8)
# from sklearn.metrics import f1_score
# def compute_metrics(pred):
# labels = pred.label_ids
# preds = pred.predictions.argmax(-1)
# f1 = f1_score(labels, preds)
# return {'F1': f1}
metric = load_metric("precision", average="weighted")
def compute_metrics(eval_pred):
logits, labels = eval_pred
predictions = np.argmax(logits, axis=-1)
return metric.compute(predictions=predictions, references=labels)
training_args = TrainingArguments(
output_dir="./results", # Выходной каталог
num_train_epochs=5, # Кол-во эпох для обучения
per_device_train_batch_size=8, # Размер пакета для каждого устройства во время обучения
per_device_eval_batch_size=8, # Размер пакета для каждого устройства во время валидации
weight_decay=0.01, # Понижение весов
logging_dir="./logs", # Каталог для хранения журналов
load_best_model_at_end=True, # Загружать ли лучшую модель после обучения
learning_rate=1e-5, # Скорость обучения
evaluation_strategy="epoch", # Валидация после каждой эпохи (можно сделать после конкретного кол-ва шагов)
logging_strategy="epoch", # Логирование после каждой эпохи
save_strategy="epoch", # Сохранение после каждой эпохи
save_total_limit=1,
seed=21,
)
trainer = Trainer(
model=model,
tokenizer=tokenizer,
args=training_args,
train_dataset=train_dataset,
eval_dataset=train_dataset,
compute_metrics=compute_metrics,
)
trainer.train()
model_path = "fine-tune-bert"
model.save_pretrained(model_path)
tokenizer.save_pretrained(model_path)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/779/129779452.ipynb
| null | null |
[{"Id": 129779452, "ScriptId": 38591280, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1459628, "CreationDate": "05/16/2023 12:12:12", "VersionNumber": 1.0, "Title": "notebook31788ab303", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 149.0, "LinesInsertedFromPrevious": 149.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
from corus import load_lenta2
from tqdm import tqdm
path = "/kaggle/working/lenta-ru-news.csv.bz2"
records = load_lenta2(path)
next(records)
title = []
text = []
topic = []
tags = []
for record in tqdm(records):
text.append(record.text)
topic.append(record.topic)
import pandas as pd
import numpy as np
main_df = pd.DataFrame({"text": text, "topic": topic})
main_df["topic"].unique()
main_df.sample(frac=1)
# Оставляем Экономика, Интернет и СМИ, Спорт, Культура, Наука и Техника
main_df = main_df.loc[
main_df["topic"].isin(
["Экономика", "Интернет и СМИ", "Спорт", "Культура", "Наука и Техника"]
)
]
main_df["topic"] = main_df["topic"].replace(
["Экономика", "Интернет и СМИ", "Спорт", "Культура", "Наука и Техника"],
[0, 1, 2, 3, 4],
)
main_df["topic"].unique()
text = list(main_df["text"])[:10000]
label = list(main_df["topic"])[:10000]
seq_len = [len(str(i).split()) for i in tqdm(text)]
pd.Series(seq_len).hist(bins=50, figsize=[10, 7])
print(max(seq_len))
from sklearn.model_selection import train_test_split
text_train, text_test, labels_train, labels_test = train_test_split(
text, label, train_size=0.7, random_state=42
)
import pandas as pd
import numpy as np
import random
import torch
import transformers
import torch.nn as nn
from transformers import AutoModel, BertTokenizer, BertForSequenceClassification
from transformers import TrainingArguments, Trainer
from datasets import load_metric, Dataset
from sklearn.metrics import classification_report, f1_score
from torch.utils.data import Dataset
import torch
model = BertForSequenceClassification.from_pretrained(
"/kaggle/working/rubert-base-cased-sentence", num_labels=5
).to("cuda")
tokenizer = BertTokenizer.from_pretrained("/kaggle/working/rubert-base-cased-sentence")
max_seq_len = 512
tokens_train = tokenizer.batch_encode_plus(
text_train, max_length=max_seq_len, padding="max_length", truncation=True
)
tokens_test = tokenizer.batch_encode_plus(
text_test, max_length=max_seq_len, padding="max_length", truncation=True
)
# labels_train
class Data(torch.utils.data.Dataset):
def __init__(self, encodings, labels):
self.encodings = encodings
self.labels = labels
def __getitem__(self, idx):
item = {k: torch.tensor(v[idx]) for k, v in self.encodings.items()}
item["labels"] = torch.tensor([self.labels[idx]])
return item
def __len__(self):
return len(self.labels)
train_dataset = Data(tokens_train, labels_train)
test_dataset = Data(tokens_test, labels_test)
train_dataset.__getitem__(8)
# from sklearn.metrics import f1_score
# def compute_metrics(pred):
# labels = pred.label_ids
# preds = pred.predictions.argmax(-1)
# f1 = f1_score(labels, preds)
# return {'F1': f1}
metric = load_metric("precision", average="weighted")
def compute_metrics(eval_pred):
logits, labels = eval_pred
predictions = np.argmax(logits, axis=-1)
return metric.compute(predictions=predictions, references=labels)
training_args = TrainingArguments(
output_dir="./results", # Выходной каталог
num_train_epochs=5, # Кол-во эпох для обучения
per_device_train_batch_size=8, # Размер пакета для каждого устройства во время обучения
per_device_eval_batch_size=8, # Размер пакета для каждого устройства во время валидации
weight_decay=0.01, # Понижение весов
logging_dir="./logs", # Каталог для хранения журналов
load_best_model_at_end=True, # Загружать ли лучшую модель после обучения
learning_rate=1e-5, # Скорость обучения
evaluation_strategy="epoch", # Валидация после каждой эпохи (можно сделать после конкретного кол-ва шагов)
logging_strategy="epoch", # Логирование после каждой эпохи
save_strategy="epoch", # Сохранение после каждой эпохи
save_total_limit=1,
seed=21,
)
trainer = Trainer(
model=model,
tokenizer=tokenizer,
args=training_args,
train_dataset=train_dataset,
eval_dataset=train_dataset,
compute_metrics=compute_metrics,
)
trainer.train()
model_path = "fine-tune-bert"
model.save_pretrained(model_path)
tokenizer.save_pretrained(model_path)
| false | 0 | 1,437 | 0 | 1,437 | 1,437 |
||
129779025
|
import pandas as pd
import numpy as np
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
from catboost import CatBoostRegressor
all_data = pd.read_csv("/kaggle/input/playground-series-s3e15/data.csv")
# # What do we need to do?
# * The data `all_data` contains missing values in the column `x_e_out [-]`.
# * We need to predict these missing values (with minimal RMSE).
# # Overview of the data
for var in all_data.columns:
distinct_vals = len(set(all_data[var].values))
print(f"Distinct unique values for {var}: {distinct_vals}")
if distinct_vals < 100:
print(all_data[var].value_counts())
print("\n")
# Here's summary statistics for the numeric values:
all_data.describe()
# # Define a data pipeline
all_data.columns
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
predictors = [
"author",
"geometry",
"pressure [MPa]",
"mass_flux [kg/m2-s]",
"D_e [mm]",
"D_h [mm]",
"length [mm]",
"chf_exp [MW/m2]",
]
target = "x_e_out [-]"
all_data["author"] = all_data["author"].replace(np.nan, "NA", regex=True)
all_data["geometry"] = all_data["geometry"].replace(np.nan, "NA", regex=True)
test = all_data[all_data[target].isna()].reset_index(drop=True)
train = all_data[np.invert(all_data[target].isna())].reset_index(drop=True)
train
test
# # Train Catboost
X = train[predictors]
y = train[target]
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
params = {"loss_function": "RMSE", "cat_features": ["author", "geometry"]}
model = CatBoostRegressor()
model.set_params(**params)
model.fit(X_train, y_train)
model.score(X_test, y_test)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/779/129779025.ipynb
| null | null |
[{"Id": 129779025, "ScriptId": 38593093, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 485314, "CreationDate": "05/16/2023 12:08:29", "VersionNumber": 2.0, "Title": "PS3E15|EDA +Pipeline + ensemble CB+Linear", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 60.0, "LinesInsertedFromPrevious": 25.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 35.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import numpy as np
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
from catboost import CatBoostRegressor
all_data = pd.read_csv("/kaggle/input/playground-series-s3e15/data.csv")
# # What do we need to do?
# * The data `all_data` contains missing values in the column `x_e_out [-]`.
# * We need to predict these missing values (with minimal RMSE).
# # Overview of the data
for var in all_data.columns:
distinct_vals = len(set(all_data[var].values))
print(f"Distinct unique values for {var}: {distinct_vals}")
if distinct_vals < 100:
print(all_data[var].value_counts())
print("\n")
# Here's summary statistics for the numeric values:
all_data.describe()
# # Define a data pipeline
all_data.columns
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
predictors = [
"author",
"geometry",
"pressure [MPa]",
"mass_flux [kg/m2-s]",
"D_e [mm]",
"D_h [mm]",
"length [mm]",
"chf_exp [MW/m2]",
]
target = "x_e_out [-]"
all_data["author"] = all_data["author"].replace(np.nan, "NA", regex=True)
all_data["geometry"] = all_data["geometry"].replace(np.nan, "NA", regex=True)
test = all_data[all_data[target].isna()].reset_index(drop=True)
train = all_data[np.invert(all_data[target].isna())].reset_index(drop=True)
train
test
# # Train Catboost
X = train[predictors]
y = train[target]
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
params = {"loss_function": "RMSE", "cat_features": ["author", "geometry"]}
model = CatBoostRegressor()
model.set_params(**params)
model.fit(X_train, y_train)
model.score(X_test, y_test)
| false | 0 | 562 | 0 | 562 | 562 |
||
129130052
|
<jupyter_start><jupyter_text>Heart Disease Dataset
### Context
This data set dates from 1988 and consists of four databases: Cleveland, Hungary, Switzerland, and Long Beach V. It contains 76 attributes, including the predicted attribute, but all published experiments refer to using a subset of 14 of them. The "target" field refers to the presence of heart disease in the patient. It is integer valued 0 = no disease and 1 = disease.
### Content
Attribute Information:
> 1. age
> 2. sex
> 3. chest pain type (4 values)
> 4. resting blood pressure
> 5. serum cholestoral in mg/dl
> 6. fasting blood sugar > 120 mg/dl
> 7. resting electrocardiographic results (values 0,1,2)
> 8. maximum heart rate achieved
> 9. exercise induced angina
> 10. oldpeak = ST depression induced by exercise relative to rest
> 11. the slope of the peak exercise ST segment
> 12. number of major vessels (0-3) colored by flourosopy
> 13. thal: 0 = normal; 1 = fixed defect; 2 = reversable defect
The names and social security numbers of the patients were recently removed from the database, replaced with dummy values.
Kaggle dataset identifier: heart-disease-dataset
<jupyter_code>import pandas as pd
df = pd.read_csv('heart-disease-dataset/heart.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 1025 entries, 0 to 1024
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 1025 non-null int64
1 sex 1025 non-null int64
2 cp 1025 non-null int64
3 trestbps 1025 non-null int64
4 chol 1025 non-null int64
5 fbs 1025 non-null int64
6 restecg 1025 non-null int64
7 thalach 1025 non-null int64
8 exang 1025 non-null int64
9 oldpeak 1025 non-null float64
10 slope 1025 non-null int64
11 ca 1025 non-null int64
12 thal 1025 non-null int64
13 target 1025 non-null int64
dtypes: float64(1), int64(13)
memory usage: 112.2 KB
<jupyter_text>Examples:
{
"age": 52.0,
"sex": 1.0,
"cp": 0.0,
"trestbps": 125.0,
"chol": 212.0,
"fbs": 0.0,
"restecg": 1.0,
"thalach": 168.0,
"exang": 0.0,
"oldpeak": 1.0,
"slope": 2.0,
"ca": 2.0,
"thal": 3.0,
"target": 0.0
}
{
"age": 53.0,
"sex": 1.0,
"cp": 0.0,
"trestbps": 140.0,
"chol": 203.0,
"fbs": 1.0,
"restecg": 0.0,
"thalach": 155.0,
"exang": 1.0,
"oldpeak": 3.1,
"slope": 0.0,
"ca": 0.0,
"thal": 3.0,
"target": 0.0
}
{
"age": 70.0,
"sex": 1.0,
"cp": 0.0,
"trestbps": 145.0,
"chol": 174.0,
"fbs": 0.0,
"restecg": 1.0,
"thalach": 125.0,
"exang": 1.0,
"oldpeak": 2.6,
"slope": 0.0,
"ca": 0.0,
"thal": 3.0,
"target": 0.0
}
{
"age": 61.0,
"sex": 1.0,
"cp": 0.0,
"trestbps": 148.0,
"chol": 203.0,
"fbs": 0.0,
"restecg": 1.0,
"thalach": 161.0,
"exang": 0.0,
"oldpeak": 0.0,
"slope": 2.0,
"ca": 1.0,
"thal": 3.0,
"target": 0.0
}
<jupyter_script># # **Business Problem**
# Saat ini tantangan terbesar bagi industri medis adalah untuk menyediakan fasilitas tingkat tinggi pada infrastruktur kesehatan untuk mendiagnosa penyakit pada hari pertama dan memberikan perawatan tepat waktu untuk meningkatkan kualitas hidup melalui kualitas layanan. Sekitar 31% kematian terjadi di dunia karena penyakit jantung. Bahkan, Menurut WHO, Cardiovaskular Disease (CVD) adalah penyebab utama kematian secara global, merenggut sekitar 17.9 juta jiwa setiap tahun. Negara berkembang dan dibawahnya kekurangan infrastruktur dan teknologi, infrastruktur dan dokter untuk memprediksi penyakit pada tahap awal untuk menghindari komplikasi mengurangi kematian. Pertumbuhan teknologi informasi dan telekomunikasi telah menguntungkan pasien kaya hingga miskin dengan memberikan informasi real time kepada pasien dengan biaya diagnosis dan pemantauan kesehatan pasien yang lebih rendah. Hal ini telah meningkatkan detail catatan kesehatan pasien secara dramatis. Rekam medis yang luas tersedia untuk penelitian. Industri medis menghadapi tantangan besar dalam menggunakan data medis yang sangat besar. Sejumlah besar data diubah untuk mendapatkan informasi yang berharga dan akurat dengan cepat oleh machine. Dengan demikian, Machine Learning adalah bidang yang penting. Model Machine Learning yang sangat berguna digunakan untuk menemukan pola tersembunyi dan korelasi antar fitur dalam kumpulan data sehingga dapat menentukan status penyakit jantung pasien.
# > Saat ini, tantangan utama dan hal yang menjadi urgent industri medis adalah memprediksi heart disease dengan metode yang lebih murah dan lebih andal untuk menghindari penyakit compounding effect di negara berpenghasilan rendah atau negara berkembang. Deteksi dini tidak hanya mengurangi biaya tetapi juga meningkatkan kualitas hidup.
# # **Metrics**
# Dataset ini merupakan data sekunder yang diambil dari [Kaggle](https://www.kaggle.com/datasets/johnsmith88/heart-disease-dataset). Dataset ini berasal dari tahun 1988 dan terdiri dari empat database: Cleveland, Hungaria, Swiss, dan Long Beach V. Ini berisi 76 atribut, termaksud yang akan diprediksi, tetapi semua percobaan yang dipublikasikan mengacu pada penggunaan subset dari 14 atribut tersebut. "Target" mengacu pada adanya penyakit jantung pada pasien, dengan 0 merepresentasikan tidak ada penyakit. Sedangkan 1, merepresentasikan ada penyakit. Berikut informasi detail mengenai 14 Artibut yang digunakan:
# 
# # **Goals**
# > Tujuan akhir analisis ini adalah untuk mengklasifikasikan apakah pasien-pasien terdiagnosa heart disease atau tidak dan mengetahui feature-feature yang berpengaruh signifikan terhadap status heart disease. Prediksi ini diharapkan dapat mengurangi biaya tetapi juga meningkatkan kualitas hidup
# # **Hipotesis**
# !
# import library
import pandas as pd
import numpy as np
import missingno as msno
import matplotlib.pyplot as plt
from pylab import rcParams
import seaborn as sns
import altair as alt
# splitting dataset
from sklearn.model_selection import train_test_split
# Models from Scikit-Learn
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier, plot_tree
from sklearn.svm import SVC
# Cross Validation
from sklearn.model_selection import StratifiedKFold
# Model Evaluations
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.model_selection import RandomizedSearchCV, GridSearchCV, ParameterGrid
from sklearn.metrics import (
confusion_matrix,
classification_report,
accuracy_score,
make_scorer,
)
from sklearn.metrics import roc_curve, auc
from sklearn.metrics import precision_score
from sklearn.metrics import roc_auc_score
from sklearn.metrics import recall_score
from sklearn.metrics import f1_score
from sklearn.metrics import precision_recall_curve
# learning curve
from sklearn.model_selection import learning_curve
# load data
heart = pd.read_csv("/kaggle/input/heart-disease-dataset/heart.csv")
heart.head()
# info data
heart.info()
# # **Data Preprocessing**
# Missing Value
heart.isna().sum()
msno.matrix(heart, color=(0.27, 0.52, 1.0))
plt.figure(figsize=(15, 9))
plt.show()
# Melalui visualisasi diatas, terlihat bahwa dataset ini tidak memiliki missing value.
# Check Duplicate
heart.duplicated().sum()
# Handling Duplicate
heart = heart.drop_duplicates()
# **Outlier Check**
# - Pengecekan outlier ini dilakukan dengan menggunakan formula:
# - Outlier bawah jika dantum Q3 + 1.5 x IQR.
# - Selanjutnya, penulis akan mengecek apakah terdapat ekstrem value pada atribut ini dengan menggunakan formula:
# - ekstreem value bawah jika dantum Q3 + 3 x IQR.
# Outlier
def find_outliers_IQR(df):
q1 = df.quantile(0.25)
q3 = df.quantile(0.75)
IQR = q3 - q1
outliers = df[((df < (q1 - 1.5 * IQR)) | (df > (q3 + 1.5 * IQR)))]
return outliers
num = ["age", "trestbps", "chol", "thalach", "oldpeak", "ca"]
outliers = find_outliers_IQR(heart[num])
outliers.notnull().sum()
# Ekstrem Value
def find_ekstrem_IQR(df):
q1 = df.quantile(0.25)
q3 = df.quantile(0.75)
IQR = q3 - q1
ekstrem = df[((df < (q1 - 3 * IQR)) | (df > (q3 + 3 * IQR)))]
return ekstrem
num = ["age", "trestbps", "chol", "thalach", "oldpeak", "ca"]
ekstrem = find_ekstrem_IQR(heart[num])
ekstrem.notnull().sum()
# Check Data Imbalance
(
alt.Chart(heart)
.mark_bar()
.encode(
x=alt.X("target:N", title="Heart Disease (0:tidak, 1:ya)"),
y=alt.Y("count(target):Q", title="Count of target"),
tooltip="count(target):Q",
)
.properties(height=300, width=400, title="Barplot of Heart Disease Status")
)
# Barplot diatas menunjukkan jumlah target, dengan jumlah kelas 0 (pasien yang tidak heart disease) sebanyak 138 dan jumlah kelas 1 (pasien yang heart disease) sebanyak 164. Berdasarkan barplot ini, variabel target memiliki kelas yang cukup balance.
# # **Feature Selection**
# Pemilihan feature untuk analisis selanjutnya didasarkan pada:
# 1. Untuk feature Numerik akan dilihat melalui koefisien korelasi pada masing-masing feature terhadap variabel target.
# 2. Untuk feature kategorik akan dilihat melalui p-value hasil dari uji Chi-Square.
# ## **Korelasi Variabel Numerik terhadap Variabel Target**
# **Point-biserial correlation coefficient:** This is a measure specifically designed for assessing the correlation between a binary (dichotomous) nominal variable and a numeric variable. It calculates the correlation between the binary variable (e.g., presence or absence of a characteristic) and the numeric variable (e.g., continuous or discrete variable).
from scipy.stats import pointbiserialr
numeric_vars = heart[
["age", "trestbps", "chol", "thalach", "oldpeak", "ca"]
] # Numeric independent variables
# Loop through each numeric variable and calculate point-biserial correlation coefficient
for col in numeric_vars:
correlation_coefficient, p_value = pointbiserialr(
heart["target"], numeric_vars[col]
)
print(
f"Point-Biserial Correlation Coefficient for {col}: {correlation_coefficient:.3f}"
)
# Penulis prefer tidak menyertakan feature chol, trestbps, dan age karena masing-masing feature tersebut memiliki korelasi yang rendah terhadap variabel target, yaitu secara berurutan -0.081, -0.146, -0.221.
# ## **Chi Square Test**
# The chi-square test can be used to test the association between a nominal dependent variable and a categorical independent variable, whether it is nominal or ordinal.
from sklearn.feature_selection import chi2
X = heart.drop(
["target", "age", "trestbps", "chol", "thalach", "oldpeak", "ca"], axis=1
)
y = heart["target"]
chi_scores = chi2(X, y)
p_values = pd.Series(chi_scores[1], index=X.columns)
p_values.sort_values(ascending=False, inplace=True)
p_values.plot.bar()
# Penulis akan menggunakan tingkat signifikasi 5%, maka dari itu diantara feature-feature categorik, fbs dan restecg secara masing-masing tidak berpengaruh terhadap variabel target. Maka dari itu, penulis tidak menyertakan feature ini dalam analisis selanjutnya.
heart = heart.drop(["fbs", "chol", "age", "trestbps", "restecg"], axis=1)
# # **One Hot Encoding**
# Dataset ini terdiri dari fitur numerik dan kategorik dengan tipe data nominal, maka variabel kategorik lebih cocok jika dilakukan **one hot encoding**. Variabel kategorik yang digunakan dalam analisis ini adalah sex, cp, exang, slope, dan thal. Karena sex dan exang sudah bernilai dua kategori 0 dan 1, tidak akan berpengaruh signifikan jika kedua variabel ini tidak dilakukan one hot encoding.
categorical_val = ["cp", "slope", "thal"]
heart_dmy = pd.get_dummies(heart, columns=categorical_val)
heart_dmy.head()
# # **Splitting Dataset**
X = heart_dmy.drop(["target"], axis=1)
y = heart_dmy["target"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# # **Feature Scaling**
# variabel numerik
num = ["thalach", "oldpeak", "ca"]
# Feature Scaling dengan MinMaxScaler
from sklearn.preprocessing import MinMaxScaler
sc = MinMaxScaler()
X_train[num] = sc.fit_transform(X_train[num])
X_test[num] = sc.transform(X_test[num])
# # **Model Baseline**
# Penulis akan mencoba tiga model yaitu, **Logistic Regression, Decision Tree, dan Support Vector Machine** untuk memprediksi pasien yang terdiagnosis heart disease atau tidak. Penulis tertarik untuk melakukan pemodelan klasifikasi dengan **Logistic Regression dan Decision Tree** karena model ini mudah diinterpretasikan. Selanjutnya, penulis tertarik untuk melakukan pemodelan klasifikasi dengan **Support Vector Machine** karena algoritma ini menjadi salah satu algoritma yang sudah sering digunakan untuk menganalisis dataset medis.
# **1. Logistic Regression**
# Salah satu Algoritma Klasifikasi Machine Learning yang simple dan terbaik adalah Logistic Regression(LR). LR adalah supervised learning, algoritma binary classification telah banyak digunakan di banyak aplikasi. Algoritma ini bekerja pada discrete atau binary categorical variable 0 atau 1. Sigmoid function ini digunakan sebagai cost function. Fungsi sigmoid ini memetakan nilai real yang diprediksi kedalam nilai probabilistik antara 0 dan 1.
# **Logistik Sigmoid Function:**
# $P(x) = 1/(1+\mathrm{e}^{-x})$
# Dimana, $P(x)$ adalah probabilitas fungsi estimasi dengan nilai antara 0 dan 1, $x$ adalah input kedalam fungsi probabilitas (algorithm's prediction value), dan konstanta matematika $\mathrm{e}$ adalah bilangan euler.
# **2. Decision Tree Classifier**
# Salah satu algoritma klasifikasi Machine Learning yang menggunakan serangkaian rules untuk membuat keputusan. Algoritma ini memiliki beberapa keunggulan seperti, Interpretability (hasil pemodelan mudah diinterpretasikan), No preprocessing required, Data robustness (Algoritma dapat menghandle semua tipe data dengan baik).
# Pada algoritma ini terdapat ukuran seleksi atribut. Ukuran ini disebut juga sebagai aturan pemisahan (splitting rules), karena aturan ini menentukan bagaimana decision tree "dibentuk". Terdapat beberapa aturan pemisahan dan tipe decision tree berdasarkan aturan pemisahannya yang cukup
# sering digunakan, diantaranya adalah sebagai berikut.
# 1. Information Gain, digunakan di decision tree ID3 (Quinlan, 1983).
# 2. Gain Ratio, digunakan di decision tree C4.5 (Quinlan, 1992).
# 3. Gini Index, digunakan di decision tree CART (Breiman et al, 1984).
# **3. Support Vector Machine Classifier**
# Support Vector Machine (SVM) merupakan salah satu metode dalam supervised learning yang biasanya digunakan untuk klasifikasi. Dalam pemodelan klasifikasi, SVM memiliki konsep yang lebih matang dan lebih jelas secara matematis dibandingkan dengan teknik-teknik klasifikasi lainnya. SVM juga dapat mengatasi masalah klasifikasi dan regresi dengan linear maupun non linear.
# SVM digunakan untuk mencari hyperplane terbaik dengan memaksimalkan jarak antar kelas. Hyperplane adalah sebuah fungsi yang dapat digunakan untuk pemisah antar kelas. Dalam 2-D fungsi yang digunakan untuk klasifikasi antar kelas disebut sebagai line whereas, fungsi yang digunakan untuk klasifikasi antas kelas dalam 3-D disebut plane similarly, sedangkan fungsi yang digunakan untuk klasifikasi di dalam ruang kelas dimensi yang lebih tinggi di sebut hyperplane.
# ## **1. Logistic Regression**
lr = LogisticRegression()
lr.fit(X_train, y_train)
# Prediction of the test variable
lr_y_preds = lr.predict(X_test)
# Training score
print("Training Score :", lr.score(X_train, y_train))
print("Testing Score :", lr.score(X_test, y_test))
print(classification_report(y_test, lr_y_preds))
# ## **2. Decision Tree**
clf2 = DecisionTreeClassifier(criterion="gini") # Gini Index
clf2.fit(X_train, y_train)
print("Training Score :", clf2.score(X_train, y_train))
print("Testing Score :", clf2.score(X_test, y_test))
Y_pred = clf2.predict(X_test)
print(classification_report(y_test, Y_pred))
# ## **3. Support Vector Machine**
# Pemodelan svm baik model baseline maupun hyperparameter tuning menggunakan kernel sigmoid. Untuk hyperparameter tuning dilakukan grid search ditambah dengan parameter-parameter seperti, gamma, C, degree, dan probability.
from sklearn.svm import SVC
model_sv = SVC(kernel="sigmoid")
model_sv.fit(X_train, y_train) # Train/Fit model
y_pred_sv = model_sv.predict(X_test) # get y predictions
print(classification_report(y_test, y_pred_sv)) # output accuracy
print("Training Score :", model_sv.score(X_train, y_train))
print("Testing Score :", model_sv.score(X_test, y_test))
# 
# Overfitting adalah perilaku pembelajaran mesin yang tidak diinginkan yang terjadi ketika model pembelajaran mesin memberikan prediksi akurat untuk data pelatihan tetapi tidak untuk data baru. Terjadi atau tidaknya Overfitting, salah satunya dapat dilihat dari nilai akurasi data train dan data test. Terlihat bahwa, model Decision Tree memiliki akurasi yang sangat tinggi didata train, tetapi tidak didata test. Maka dari itu, terjadi overfitting untuk pemodelan dengan Decision Tree.
# Selain itu, melalui classification report diatas, pada model baseline ini, Pemodelan Logistic Regression memiliki F1-Score paling tinggi, sebesar 0.82. Selanjutnya pemodelan dengan Decision Tree dengan F1-Score sebesar 0.79, dan pemodelan SVM dengan sigmoid kernel memiliki F1-Score terendah, sebesar 0.75.
# # **Hyperparameter Tuning - Cross Validation**
# #### **1. Hyperparameter Tuning**
# Hyperparameter Tuning dilakukan untuk mencari best parameter pada masing-masing pemodelan.
# #### **2. Cross Validation(CV)**
# Dengan Cross Validation, peneliti dapat melakukan penilaian model dengan memperkirakan kesalahan pengujian atau pemilihan model dengan memilih tingkat fleksibilitas berdasarkan kesalahan pengujian masing-masing model. Ide dasar dari CV adalah
# 1. Membagi dataset kedalam training dan validation set.
# 2. Menggunakan training set untuk fit sebuah model
# 3. Mengevaluasi model performance dengan validation set.
#
# 
# #### **3. K-fold cross-validation**
# Dengan CV k-fold, pertama-tama peneliti memilih nilai k. Kemudian, membagi kumpulan data menjadi k kumpulan. Set pertama menjadi validation set dan sisanya menjadi training set. Setelah menyesuaikan dan mengevaluasi model, pindah ke set kedua dan ulangi prosesnya. CV k-fold mengulang k kali dan rata-rata estimasi kesalahan uji k menjadi estimasi CV k-fold.
# 
#
# #### **4. Stratified KFold Cross Validation**
# Stratified k-fold cross-validation sama dengan k-fold cross-validation, tetapi Stratified k-fold cross-validation, melakukan stratified sampling, bukan random sampling.
# ## **1. Logistic Regression**
Accuracies = {} # make dictionry to save all accuracies models
# define model
lg = LogisticRegression()
# parameters
parameters = [
{
"penalty": ["l1", "l2"],
"C": np.logspace(-2, 2, 40),
"solver": ["liblinear", "saga"],
}
]
# create 10 folds
folds = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)
# define search
search = GridSearchCV(
lg, parameters, cv=folds, refit=True, verbose=2, scoring="accuracy", n_jobs=-1
)
# execute search
result = search.fit(X_train, y_train)
# summarize result
print("Best Score: %s" % result.best_score_)
print("Best Hyperparameters: %s" % result.best_params_)
# Predict how well your model is in a test dataset
Logreg = LogisticRegression(C=1.4251026703029979, penalty="l1", solver="liblinear")
Logreg.fit(X_train, y_train)
y_pred = Logreg.predict(X_test)
# Classification Report
print(classification_report(y_test, y_pred))
# ***
# **Insight**
# ***
# - Precision
# - Merupakan rasio **prediksi benar positif** dibandingkan dengan keseluruhan hasil yang **diprediksi positif**
# - Pada pemodelan ini, diperoleh presisi sebesar 0.78, artinya : Pada model ini, Persentase Pasien yang sebenarnya terkena penyakit jantung dari keseluruhan pasien yang **diprediksi terkena penyakit jantung** sebesar 78%.
# - Recall (Sensitifitas)
# - Merupakan rasio prediksi benar positif dibandingkan dengan **keseluruhan data yang benar positif**.
# - Pada pemodelan ini diperoleh recall sebesar 0.88, artinya : Pada model ini, Persentase pasien yang diprediksi terkena penyakit jantung dari keseluruhan pasien yang sebenarnya terkena penyakit jantung sebesar 88%.
# - F1 Score
# - F1 Score merupakan perbandingan rata-rata presisi dan recall yang dibobotkan. F1 Score dapat dihitung dengan formula:
# $ F1 \; Score = 2 * (Recall*Precission) / (Recall + Precission) $
# $ F1 \; Score = 2 * (0.88*0.78) / (0.88 + 0.78) = 0.83$
# - Pada pemodelan ini diperoleh F1 Score sebesar 0.83, artinya : perbandingan rata-rata presisi dan recall yang dibobotkan sebesar 83%. Dengan kata lain, kemampuan model yang seimbang untuk menangkap kasus positif (mengingat) dan akurat dengan kasus yang ditangkapnya (presisi) sebesar 0.83.
# - Accuracy
# - Merupakan rasio prediksi Benar (positif dan negatif) dengan keseluruhan data.
# - Pada pemodelan ini diperoleh akurasi sebesar 0.82, artinya : Persentase Pasien yang benar diprediksi terkena heart disease dan tidak terkena heart disease dari keseluruhan pasien sebesar 82%
#
# Confusion Matrix
cm = confusion_matrix(y_test, y_pred)
ax = plt.subplot()
sns.heatmap(cm, annot=True, fmt="g", ax=ax)
# annot=True to annotate cells, ftm='g' to disable scientific notation
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix")
ax.xaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
ax.yaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
# ***
# **Insight**
# ***
# - Dari 48 pasien yang tidak terkena heart disease, sebanyak 37 pasien diprediksi tidak terkena heart disease (TN) dan sisanya 11 pasien diprediksi terkena heart disease (FP).
# - Selanjutnya, dari 43 Pasien yang terkena heart disease, sebanyak 38 pasien diprediksi terkena heart disease (TP) dan sisanya 5 pasien diprediksi tidak terkena heart disease (FN)
# Dimana
# - TN : True Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease dan sebenarnya pasien tersebut tidak terkena heart disease)
# - FP : False Positif (Kasus dimana pasien diprediksi terkena heart disease, tetapi pada faktanya pasien tersebut tidak terkena heart disease)
# - FN : False Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease, tetapi pada faktanya pasien tersebut terkens heart disease)
# - TP : True Positif (Kasus dimana pasien diprediksi terkena heart disease dan sebenarnya pasien tersebut terkena heart disease)
# ## **2. Decision Tree**
# define model
dt = DecisionTreeClassifier(random_state=42)
# parameters
parameters = [
{
"criterion": ["gini", "entropy", "log_loss"],
"max_depth": [1, 2, 3, 4, 5],
"min_samples_split": [2, 3, 4, 5],
"min_samples_leaf": [1, 2, 3, 4, 5],
"max_features": ["sqrt", "log2"],
"random_state": [42],
}
]
# create 10 folds
folds = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)
# define search
search = GridSearchCV(dt, parameters, cv=folds, scoring="accuracy")
# execute search
result = search.fit(X_train, y_train)
# summarize result
print("Best Score: %s" % result.best_score_)
print("Best Hyperparameters: %s" % result.best_params_)
# Predict Model in Test Dataset
Dtree = DecisionTreeClassifier(
criterion="entropy",
max_depth=5,
max_features="sqrt",
min_samples_leaf=1,
min_samples_split=2,
random_state=42,
)
Dtree.fit(X_train, y_train)
y_pred_tree = Dtree.predict(X_test)
# Classification Report
print(classification_report(y_test, y_pred_tree))
# ***
# **Insight**
# ***
# - Precision
# - Merupakan rasio **prediksi benar positif** dibandingkan dengan keseluruhan hasil yang **diprediksi positif**
# - Pada pemodelan ini, diperoleh presisi sebesar 0.77, artinya : Pada model ini, Persentase Pasien yang sebenarnya terkena penyakit jantung dari keseluruhan pasien yang **diprediksi terkena penyakit jantung** sebesar 77%.
# - Recall (Sensitifitas)
# - Merupakan rasio prediksi benar positif dibandingkan dengan **keseluruhan data yang benar positif**.
# - Pada pemodelan ini diperoleh recall sebesar 0.84, artinya : Pada model ini, Persentase pasien yang diprediksi terkena penyakit jantung dari keseluruhan pasien yang sebenarnya terkena penyakit jantung sebesar 84%.
# - F1 Score
# - F1 Score merupakan perbandingan rata-rata presisi dan recall yang dibobotkan. F1 Score dapat dihitung dengan formula:
# $ F1 \; Score = 2 * (Recall*Precission) / (Recall + Precission) $
# $ F1 \; Score = 2 * (0.77*0.84) / (0.77 + 0.84) = 0.80$
# - Pada pemodelan ini diperoleh F1 Score sebesar 0.80, artinya : perbandingan rata-rata presisi dan recall yang dibobotkan sebesar 80%. Dengan kata lain, kemampuan model yang seimbang untuk menangkap kasus positif (mengingat) dan akurat dengan kasus yang ditangkapnya (presisi) sebesar 0.80.
# - Accuracy
# - Merupakan rasio prediksi Benar (positif dan negatif) dengan keseluruhan data.
# - Pada pemodelan ini diperoleh akurasi sebesar 0.80, artinya : Persentase Pasien yang benar diprediksi terkena heart disease dan tidak terkena heart disease dari keseluruhan pasien sebesar 80%
# Confusion Matrix
cm = confusion_matrix(y_test, y_pred_tree)
ax = plt.subplot()
sns.heatmap(cm, annot=True, fmt="g", ax=ax)
# annot=True to annotate cells, ftm='g' to disable scientific notation
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix")
ax.xaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
ax.yaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
# ***
# **Insight**
# ***
# - Dari 48 pasien yang tidak terkena heart disease, sebanyak 37 pasien diprediksi tidak terkena heart disease (TN) dan sisanya 11 pasien diprediksi terkena heart disease (FP).
# - Selanjutnya, dari 43 Pasien yang terkena heart disease, sebanyak 36 pasien diprediksi terkena heart disease (TP) dan sisanya 7 pasien diprediksi tidak terkena heart disease (FN)
# Dimana
# - TN : True Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease dan sebenarnya pasien tersebut tidak terkena heart disease)
# - FP : False Positif (Kasus dimana pasien diprediksi terkena heart disease, tetapi pada faktanya pasien tersebut tidak terkena heart disease)
# - FN : False Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease, tetapi pada faktanya pasien tersebut terkens heart disease)
# - TP : True Positif (Kasus dimana pasien diprediksi terkena heart disease dan sebenarnya pasien tersebut terkena heart disease)
# ## **3. Support Vector Machine**
# define model
svc = SVC()
# parameters
parameters = [
{
"kernel": ["sigmoid"],
"gamma": [0.1, 0.9, 0.06, 0.05, 0.3, 1],
"C": [0.1, 1, 2, 3, 4, 5, 6, 10, 100],
"degree": [3],
"probability": [True, False],
}
]
# create 10 folds
folds = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)
# define search
search = GridSearchCV(svc, parameters, cv=folds, scoring="accuracy")
# execute search
result = search.fit(X_train, y_train)
# summarize result
print("Best Score: %s" % result.best_score_)
print("Best Hyperparameters: %s" % result.best_params_)
# Predict Model in Test Dataset
sv = SVC(C=5, degree=3, gamma=0.06, kernel="sigmoid", probability=True)
sv.fit(X_train, y_train)
y_pred_s = sv.predict(X_test)
# Classification Report
print(classification_report(y_test, y_pred_s))
# ***
# **Insight**
# ***
# - Precision
# - Merupakan rasio **prediksi benar positif** dibandingkan dengan keseluruhan hasil yang **diprediksi positif**
# - Pada pemodelan ini, diperoleh presisi sebesar 0.74, artinya : Pada model ini, Persentase Pasien yang sebenarnya terkena penyakit jantung dari keseluruhan pasien yang **diprediksi terkena penyakit jantung** sebesar 74%.
# - Recall (Sensitifitas)
# - Merupakan rasio prediksi benar positif dibandingkan dengan **keseluruhan data yang benar positif**.
# - Pada pemodelan ini diperoleh recall sebesar 0.93, artinya : Pada model ini, Persentase pasien yang diprediksi terkena penyakit jantung dari keseluruhan pasien yang sebenarnya terkena penyakit jantung sebesar 93%.
# - F1 Score
# - F1 Score merupakan perbandingan rata-rata presisi dan recall yang dibobotkan. F1 Score dapat dihitung dengan formula:
# $ F1 \; Score = 2 * (Recall*Precission) / (Recall + Precission) $
# $ F1 \; Score = 2 * (0.93*0.74) / (0.93+0.74 ) = 0.82$
# - Pada pemodelan ini diperoleh F1 Score sebesar 0.82, artinya : perbandingan rata-rata presisi dan recall yang dibobotkan sebesar 82%. Dengan kata lain, kemampuan model yang seimbang untuk menangkap kasus positif (mengingat) dan akurat dengan kasus yang ditangkapnya (presisi) sebesar 0.82.
# - Accuracy
# - Merupakan rasio prediksi Benar (positif dan negatif) dengan keseluruhan data.
# - Pada pemodelan ini diperoleh akurasi sebesar 0.81, artinya : Persentase Pasien yang benar diprediksi terkena heart disease dan tidak terkena heart disease dari keseluruhan pasien sebesar 81%
# Confusion Matrix
cm = confusion_matrix(y_test, y_pred_s)
ax = plt.subplot()
sns.heatmap(cm, annot=True, fmt="g", ax=ax)
# annot=True to annotate cells, ftm='g' to disable scientific notation
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix")
ax.xaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
ax.yaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
# ***
# **Insight**
# ***
# - Dari 48 pasien yang tidak terkena heart disease, sebanyak 34 pasien diprediksi tidak terkena heart disease (TN) dan sisanya 14 pasien diprediksi terkena heart disease (FP).
# - Selanjutnya, dari 43 Pasien yang terkena heart disease, sebanyak 40 pasien diprediksi terkena heart disease (TP) dan sisanya 3 pasien diprediksi tidak terkena heart disease (FN)
# Dimana
# - TN : True Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease dan sebenarnya pasien tersebut tidak terkena heart disease)
# - FP : False Positif (Kasus dimana pasien diprediksi terkena heart disease, tetapi pada faktanya pasien tersebut tidak terkena heart disease)
# - FN : False Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease, tetapi pada faktanya pasien tersebut terkena heart disease)
# - TP : True Positif (Kasus dimana pasien diprediksi terkena heart disease dan sebenarnya pasien tersebut terkena heart disease)
# # Learning Curve
# **Dalam learning curve, penulis menggunakan scoring akurasi. Penulis menggunakan scoring akurasi karena data target cukup balance.**
# Learning curve dengan akurasi pada klasifikasi adalah sebuah grafik yang menunjukkan perubahan performa atau akurasi model klasifikasi seiring dengan perubahan ukuran data latih (train data) yang digunakan untuk melatih model. Grafik learning curve biasanya memiliki sumbu x yang menunjukkan ukuran data latih (misalnya, jumlah sampel atau proporsi data latih), dan sumbu y yang menunjukkan akurasi model pada data latih (training accuracy) dan akurasi model pada data uji (validation accuracy) dalam bentuk nilai atau persentase.
# Learning curve dapat memberikan informasi berharga tentang performa model klasifikasi. Beberapa interpretasi yang dapat diambil dari learning curve dengan akurasi pada klasifikasi antara lain:
# 1. Underfitting: Jika training accuracy dan validation accuracy keduanya rendah dan tidak mengalami peningkatan seiring dengan peningkatan ukuran data latih, hal ini bisa mengindikasikan bahwa model mungkin terlalu sederhana (underfitting) dan membutuhkan peningkatan kompleksitas untuk dapat melakukan generalisasi yang baik pada data uji.
# 2. Overfitting: Jika training accuracy tinggi tetapi validation accuracy rendah dan tidak mengalami peningkatan seiring dengan peningkatan ukuran data latih, hal ini bisa mengindikasikan bahwa model mungkin terlalu kompleks (overfitting) dan tidak dapat menggeneralisasi dengan baik pada data uji. Solusinya bisa dengan mengurangi kompleksitas model, menggunakan teknik regularisasi, atau memperbesar ukuran data latih.
# 3. Good fit: Jika training accuracy dan validation accuracy keduanya tinggi dan cenderung stabil seiring dengan peningkatan ukuran data latih, hal ini bisa mengindikasikan bahwa model telah mencapai performa yang baik dan dapat melakukan generalisasi yang baik pada data uji. Ukuran data latih yang lebih besar mungkin tidak diperlukan.
# import library
from sklearn.model_selection import learning_curve
# ## **1. Logistic Regression**
# Membuat learning curve dengan scoring accuracy
train_sizes, train_scores, test_scores = learning_curve(
Logreg,
X_train,
y_train,
cv=folds,
scoring="accuracy",
train_sizes=np.linspace(0.1, 1.0, 10),
)
# Menghitung rata-rata skor pada setiap ukuran data latih
train_scores_mean = np.mean(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
# Visualisasi learning curve
plt.figure(figsize=(8, 6))
plt.plot(train_sizes, train_scores_mean, "o-", color="r", label="Training Score")
plt.plot(train_sizes, test_scores_mean, "o-", color="g", label="Validation Score")
plt.xlabel("Training Set Size")
plt.ylabel("Accuracy")
plt.legend(loc="best")
plt.title("Learning Curve - Logistic Regression (Accuracy)")
plt.show()
# Melalui learning curve ini dapat dilihat bahwa **secara general** pemodelan dengan logistic regression dapat dikatakan baik (good fit). Selanjutnya, pemodelan ini jauh lebih baik jika jumlah data train diatas 112 karena memiliki training score dan cross validation score yang relatif tidak berbeda jauh. Selanjutnya pada training set size ini model juga tidak mengalami overfitting. Selain itu, dengan training set size diatas 100 cross validation score cenderung naik hingga disekitar training set size sebanyak 174 terdapat cross validation score yang nilainya lebih besar dari training score. Akurasi cross validation score yang lebih besar dari training score menunjukkan bahwa pada training set size ini akan memberikan performa model yang baik. Hal ini juga dapat mengindikasikan bahwa model mungkin tidak mengalami overfitting dan mampu melakukan generalisasi yang baik pada data yang belum pernah dilihat sebelumnya (data uji). Selanjutnya, training set size diatas 175 **hampir tidak lagi signifikan** dalam meningkatkan akurasi model. Ini bisa menjadi indikasi bahwa ukuran data latih yang sudah ada sudah cukup besar untuk model ini dan penambahan data latih lebih lanjut **berpeluang** tidak memberikan peningkatan signifikan dalam performa model.
# ## **2. Decision Tree**
# Membuat learning curve dengan scoring accuracy
train_sizes, train_scores, test_scores = learning_curve(
Dtree,
X_train,
y_train,
cv=folds,
scoring="accuracy",
train_sizes=np.linspace(0.1, 1.0, 10),
)
# Menghitung rata-rata skor pada setiap ukuran data latih
train_scores_mean = np.mean(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
# Visualisasi learning curve
plt.figure(figsize=(8, 6))
plt.plot(train_sizes, train_scores_mean, "o-", color="r", label="Training Score")
plt.plot(train_sizes, test_scores_mean, "o-", color="g", label="Validation Score")
plt.xlabel("Training Set Size")
plt.ylabel("Accuracy")
plt.legend(loc="best")
plt.title("Learning Curve - Decision Tree (Accuracy)")
plt.show()
# Melalui learning curve ini, pemodelan dengan Decision Tree **secara general cenderung mengalami overfitting**, hal ini dapat kita lihat dari akurasi dari data train yang lebih besar daripada data validasi dan gap antara training score dan cross-validation score yang cukup besar.
# ## **3. Support Vector Machine**
# Membuat learning curve dengan scoring akurasi
train_sizes, train_scores, test_scores = learning_curve(
sv,
X_train,
y_train,
cv=folds,
scoring="accuracy",
train_sizes=np.linspace(0.1, 1.0, 10),
)
# Menghitung rata-rata skor pada setiap ukuran data latih
train_scores_mean = np.mean(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
# Visualisasi learning curve
plt.figure(figsize=(8, 6))
plt.plot(train_sizes, train_scores_mean, "o-", color="r", label="Training Score")
plt.plot(train_sizes, test_scores_mean, "o-", color="g", label="Validation Score")
plt.xlabel("Training Set Size")
plt.ylabel("Accuracy")
plt.legend(loc="best")
plt.title("Learning Curve - Support Vector Machine (SVM) with Accuracy Scoring")
plt.show()
# Melalui learning curve ini, **secara general** pemodelan dengan SVM dapat dikatakan baik (good fit). Selain itu, pemodelan dengan SVM lebih baik untuk jumlah data train diatas 75 karena memiliki training score dan cross validation score yang akurasinya relatif tidak terlalu jauh. Selanjutnya pada training set size ini model juga tidak mengalami overfitting. Bahkan, **disekitar** training set size sebanyak 130 terdapat cross validation score yang nilainya lebih besar dari training score. Akurasi cross validation score yang lebih besar dari training score menunjukkan bahwa pada training set size ini akan memberikan performa model yang baik. Hal ini juga dapat mengindikasikan bahwa model mungkin tidak mengalami overfitting dan mampu melakukan generalisasi yang baik pada data yang belum pernah dilihat sebelumnya (data uji).
# # ROC Analysis
# Logistic regression
y_pred_prob_lr = Logreg.predict_proba(X_test)[:, 1]
fpr_lr, tpr_lr, thresholds_lr = roc_curve(y_test, y_pred_prob_lr)
roc_auc_lr = auc(fpr_lr, tpr_lr)
precision_lr, recall_lr, th_lr = precision_recall_curve(y_test, y_pred_prob_lr)
# Decision Tree
y_pred_prob_DT = Dtree.predict_proba(X_test)[:, 1]
fpr_DT, tpr_DT, thresholds_DT = roc_curve(y_test, y_pred_prob_DT)
roc_auc_DT = auc(fpr_DT, tpr_DT)
precision_DT, recall_DT, th_DT = precision_recall_curve(y_test, y_pred_prob_DT)
# SVM
y_pred_prob_SV = sv.predict_proba(X_test)[:, 1]
fpr_SV, tpr_SV, thresholds_SV = roc_curve(y_test, y_pred_prob_SV)
roc_auc_SV = auc(fpr_SV, tpr_SV)
precision_SV, recall_SV, th_SV = precision_recall_curve(y_test, y_pred_prob_SV)
# Plot ROC curve
plt.plot([0, 1], [0, 1], "k--")
plt.plot(fpr_lr, tpr_lr, label="Log Reg (area = %0.3f)" % roc_auc_lr)
plt.plot(fpr_DT, tpr_DT, label="Decision Tree (area = %0.3f)" % roc_auc_DT)
plt.plot(fpr_SV, tpr_SV, label="SVM Sigmoid (area = %0.3f)" % roc_auc_SV)
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("ROC curves")
plt.legend(loc="best")
plt.show()
# ***
# **Insight**
# ***
# Kurva AUC - ROC adalah pengukuran kinerja untuk masalah klasifikasi pada berbagai pengaturan ambang batas. ROC adalah kurva probabilitas dan AUC mewakili tingkat atau ukuran keterpisahan. Ini memberi tahu seberapa banyak model mampu membedakan antar kelas. Semakin tinggi AUC, semakin baik model dalam memprediksi kelas 0 sebagai 0 dan kelas 1 sebagai 1. Dengan analogi, semakin tinggi AUC, semakin baik model dalam membedakan antara pasien yang terkena heart disease dengan pasien yang tidak terkena heart disease.
# 
# Secara general, melalui ROC Curve ini, pemodelan dengan Logistic Regression dan Support Vector Machine memiliki performa model yang lebih baik dibandingkan dengan Decision Tree. Selain itu, performa model Logistic Regression dan Support Vector Machine relatif sama bagusnya.
# **Jika ditinjau dari nilai AUC:**
# - Logistic Regression : Pemodelan ini memiliki nilai AUC sebesar 0.911. Hal ini berarti model yang terbentuk memiliki 91,1% area dibawah kurva. Dengan kata lain, pemodelan ini sudah "excellent" dalam memisahkan/membedakan antara kelas 1 (Pasien yang terkena heart disease) dengan kelas 0 (pasien yang tidak terkena heart disease).
# - Decision Tree : Pemodelan ini memiliki nilai AUC sebesar 0.835. Hal ini berarti model yang terbentuk memiliki 83.5% area dibawah kurva. Dengan kata lain, pemodelan ini sudah sangat baik dalam memisahkan/membedakan antara kelas 1 (Pasien yang terkena heart disease) dengan kelas 0 (pasien yang tidak terkena heart disease).
# - Support Vector Machine : Pemodelan ini memiliki nilai AUC sebesar 0.915. Hal ini berarti model yang terbentuk memiliki 91.5% area dibawah kurva. Dengan kata lain, pemodelan ini sudah "excellent" dalam memisahkan/membedakan antara kelas 1 (Pasien yang terkena heart disease) dengan kelas 0 (pasien yang tidak terkena heart disease).
# # Best Model
# Pertama, Penentuan model terbaik didasarkan pada AUC dan F1-Score. Selanjutnya, membandingkan nilai precision, recall dan akurasi dari model klasifikasi. Kurva ROC (Receiver Operator Characteristics) dapat digunakan untuk penyelidikan lebih lanjut ke dalam model. Kinerja model dapat divisualisasikan oleh ROC Curve dan tradeoff antara TPR (True Positive Rate) dan FPR (False Positive Rate). Ini berkisar dari 0 hingga 1 dan area di bawahnya menandakan kemampuan membedakan kelas model ML. Kurva ROC mendekati satu yang lebih mampu mengklasifikasikan.
# Penilaian model klasifikasi dari masing-masing classifier akan mengacu pada confusion matrix. Confusion matrix menunjukkan prediksi dan klasifikasi terbaik melalui nilai accuracy, sensitivity, dan specificity. Ketika nilai accuracy, sensitivity, dan specificity semakin mendekati 1 (satu) maka prediksi semakin lebih baik. Hasil klasifikasi dari confusion matrix terdiri dari 4 (empat) yakni true positive, false positive, false negative, dan true negative.
# 
# Terlihat bahwa model SVM memiliki nilai AUC paling baik. Selanjutnya, model Logistic Regression memiliki nilai F1-Score paling baik. Namun, baik nilai AUC dan F1-Score antara model Logistic Regression dan SVM hampir sama. Selanjutnya akan dilibatkan metrics lain dalam penentuan model terbaik.
# 
# Terlihat bahwa Klasifikasi dengan algoritma Logistic Regression memenuhi kriteria pemilihan model terbanyak, dimana algoritma ini memiliki F1-score, precision, dan akurasi tertinggi diantara dua model klasifikasi lainnya. Selain itu, nilai AUC sebesar 0.91, artinya : pemodelan ini sudah "excellent" dalam memisahkan/membedakan antara kelas 1 (Pasien yang terkena heart disease) dengan kelas 0 (pasien yang tidak terkena heart disease). Ditambah lagi, pemodelan ini memiliki evaluasi metrics yang lebih stabil dibandingkan dengan SVM dan pemodelan dengan Logistic Regression memiliki interpretabilitas model yang mudah diinterpretasikan. Maka dari itu, penulis memilih model Logistic Regression sebagai model terbaik untuk study kasus ini.
# # Feature Importance from Best Model
# These coef's tell how much and in what way did each one of it contribute to predicting the target variable
feature_dict = dict(zip(X_train.columns, list(Logreg.coef_[0])))
feature_dict
# **Odds Ratio**
# Melalui Odds Ratio, akan memudahkan penulis untuk melihat fitur-fitur yang berperan penting dalam mendiagnosa heart disease. Dimana Odds Ratio diperoleh dari $\mathrm{e}^{\beta_i}$ dengan $\beta_i$ adalah koefisien dari masing-masing fitur.
import math
feature_importance = pd.DataFrame(X_train.columns, columns=["feature"])
feature_importance["importance"] = pow(math.e, Logreg.coef_[0])
feature_importance = feature_importance.sort_values(by=["importance"], ascending=True)
feature_importance_1 = feature_importance[feature_importance["importance"] != 1]
feature_importance_1
sns.barplot(
x=feature_importance_1["importance"], y=feature_importance_1["feature"], orient="h"
)
plt.xlabel("Importance", fontsize=16)
plt.ylabel("Feature", fontsize=16)
plt.title("Feature Importance With Odds Ratio", fontsize=16)
plt.tick_params(axis="both", which="major", labelsize=12)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/130/129130052.ipynb
|
heart-disease-dataset
|
johnsmith88
|
[{"Id": 129130052, "ScriptId": 38378976, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10032783, "CreationDate": "05/11/2023 08:13:18", "VersionNumber": 1.0, "Title": "Heart Disease Prediction_Assignment 3-4", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 702.0, "LinesInsertedFromPrevious": 702.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184910572, "KernelVersionId": 129130052, "SourceDatasetVersionId": 477177}]
|
[{"Id": 477177, "DatasetId": 216167, "DatasourceVersionId": 493143, "CreatorUserId": 3308439, "LicenseName": "Unknown", "CreationDate": "06/06/2019 15:33:55", "VersionNumber": 2.0, "Title": "Heart Disease Dataset", "Slug": "heart-disease-dataset", "Subtitle": "Public Health Dataset", "Description": "### Context\n\nThis data set dates from 1988 and consists of four databases: Cleveland, Hungary, Switzerland, and Long Beach V. It contains 76 attributes, including the predicted attribute, but all published experiments refer to using a subset of 14 of them. The \"target\" field refers to the presence of heart disease in the patient. It is integer valued 0 = no disease and 1 = disease.\n\n\n### Content\n\nAttribute Information: \n> 1. age \n> 2. sex \n> 3. chest pain type (4 values) \n> 4. resting blood pressure \n> 5. serum cholestoral in mg/dl \n> 6. fasting blood sugar > 120 mg/dl\n> 7. resting electrocardiographic results (values 0,1,2)\n> 8. maximum heart rate achieved \n> 9. exercise induced angina \n> 10. oldpeak = ST depression induced by exercise relative to rest \n> 11. the slope of the peak exercise ST segment \n> 12. number of major vessels (0-3) colored by flourosopy \n> 13. thal: 0 = normal; 1 = fixed defect; 2 = reversable defect\nThe names and social security numbers of the patients were recently removed from the database, replaced with dummy values.", "VersionNotes": "Update data", "TotalCompressedBytes": 38114.0, "TotalUncompressedBytes": 38114.0}]
|
[{"Id": 216167, "CreatorUserId": 3308439, "OwnerUserId": 3308439.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 477177.0, "CurrentDatasourceVersionId": 493143.0, "ForumId": 227259, "Type": 2, "CreationDate": "06/04/2019 02:58:45", "LastActivityDate": "06/04/2019", "TotalViews": 578180, "TotalDownloads": 97361, "TotalVotes": 747, "TotalKernels": 308}]
|
[{"Id": 3308439, "UserName": "johnsmith88", "DisplayName": "David Lapp", "RegisterDate": "06/04/2019", "PerformanceTier": 0}]
|
# # **Business Problem**
# Saat ini tantangan terbesar bagi industri medis adalah untuk menyediakan fasilitas tingkat tinggi pada infrastruktur kesehatan untuk mendiagnosa penyakit pada hari pertama dan memberikan perawatan tepat waktu untuk meningkatkan kualitas hidup melalui kualitas layanan. Sekitar 31% kematian terjadi di dunia karena penyakit jantung. Bahkan, Menurut WHO, Cardiovaskular Disease (CVD) adalah penyebab utama kematian secara global, merenggut sekitar 17.9 juta jiwa setiap tahun. Negara berkembang dan dibawahnya kekurangan infrastruktur dan teknologi, infrastruktur dan dokter untuk memprediksi penyakit pada tahap awal untuk menghindari komplikasi mengurangi kematian. Pertumbuhan teknologi informasi dan telekomunikasi telah menguntungkan pasien kaya hingga miskin dengan memberikan informasi real time kepada pasien dengan biaya diagnosis dan pemantauan kesehatan pasien yang lebih rendah. Hal ini telah meningkatkan detail catatan kesehatan pasien secara dramatis. Rekam medis yang luas tersedia untuk penelitian. Industri medis menghadapi tantangan besar dalam menggunakan data medis yang sangat besar. Sejumlah besar data diubah untuk mendapatkan informasi yang berharga dan akurat dengan cepat oleh machine. Dengan demikian, Machine Learning adalah bidang yang penting. Model Machine Learning yang sangat berguna digunakan untuk menemukan pola tersembunyi dan korelasi antar fitur dalam kumpulan data sehingga dapat menentukan status penyakit jantung pasien.
# > Saat ini, tantangan utama dan hal yang menjadi urgent industri medis adalah memprediksi heart disease dengan metode yang lebih murah dan lebih andal untuk menghindari penyakit compounding effect di negara berpenghasilan rendah atau negara berkembang. Deteksi dini tidak hanya mengurangi biaya tetapi juga meningkatkan kualitas hidup.
# # **Metrics**
# Dataset ini merupakan data sekunder yang diambil dari [Kaggle](https://www.kaggle.com/datasets/johnsmith88/heart-disease-dataset). Dataset ini berasal dari tahun 1988 dan terdiri dari empat database: Cleveland, Hungaria, Swiss, dan Long Beach V. Ini berisi 76 atribut, termaksud yang akan diprediksi, tetapi semua percobaan yang dipublikasikan mengacu pada penggunaan subset dari 14 atribut tersebut. "Target" mengacu pada adanya penyakit jantung pada pasien, dengan 0 merepresentasikan tidak ada penyakit. Sedangkan 1, merepresentasikan ada penyakit. Berikut informasi detail mengenai 14 Artibut yang digunakan:
# 
# # **Goals**
# > Tujuan akhir analisis ini adalah untuk mengklasifikasikan apakah pasien-pasien terdiagnosa heart disease atau tidak dan mengetahui feature-feature yang berpengaruh signifikan terhadap status heart disease. Prediksi ini diharapkan dapat mengurangi biaya tetapi juga meningkatkan kualitas hidup
# # **Hipotesis**
# !
# import library
import pandas as pd
import numpy as np
import missingno as msno
import matplotlib.pyplot as plt
from pylab import rcParams
import seaborn as sns
import altair as alt
# splitting dataset
from sklearn.model_selection import train_test_split
# Models from Scikit-Learn
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier, plot_tree
from sklearn.svm import SVC
# Cross Validation
from sklearn.model_selection import StratifiedKFold
# Model Evaluations
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.model_selection import RandomizedSearchCV, GridSearchCV, ParameterGrid
from sklearn.metrics import (
confusion_matrix,
classification_report,
accuracy_score,
make_scorer,
)
from sklearn.metrics import roc_curve, auc
from sklearn.metrics import precision_score
from sklearn.metrics import roc_auc_score
from sklearn.metrics import recall_score
from sklearn.metrics import f1_score
from sklearn.metrics import precision_recall_curve
# learning curve
from sklearn.model_selection import learning_curve
# load data
heart = pd.read_csv("/kaggle/input/heart-disease-dataset/heart.csv")
heart.head()
# info data
heart.info()
# # **Data Preprocessing**
# Missing Value
heart.isna().sum()
msno.matrix(heart, color=(0.27, 0.52, 1.0))
plt.figure(figsize=(15, 9))
plt.show()
# Melalui visualisasi diatas, terlihat bahwa dataset ini tidak memiliki missing value.
# Check Duplicate
heart.duplicated().sum()
# Handling Duplicate
heart = heart.drop_duplicates()
# **Outlier Check**
# - Pengecekan outlier ini dilakukan dengan menggunakan formula:
# - Outlier bawah jika dantum Q3 + 1.5 x IQR.
# - Selanjutnya, penulis akan mengecek apakah terdapat ekstrem value pada atribut ini dengan menggunakan formula:
# - ekstreem value bawah jika dantum Q3 + 3 x IQR.
# Outlier
def find_outliers_IQR(df):
q1 = df.quantile(0.25)
q3 = df.quantile(0.75)
IQR = q3 - q1
outliers = df[((df < (q1 - 1.5 * IQR)) | (df > (q3 + 1.5 * IQR)))]
return outliers
num = ["age", "trestbps", "chol", "thalach", "oldpeak", "ca"]
outliers = find_outliers_IQR(heart[num])
outliers.notnull().sum()
# Ekstrem Value
def find_ekstrem_IQR(df):
q1 = df.quantile(0.25)
q3 = df.quantile(0.75)
IQR = q3 - q1
ekstrem = df[((df < (q1 - 3 * IQR)) | (df > (q3 + 3 * IQR)))]
return ekstrem
num = ["age", "trestbps", "chol", "thalach", "oldpeak", "ca"]
ekstrem = find_ekstrem_IQR(heart[num])
ekstrem.notnull().sum()
# Check Data Imbalance
(
alt.Chart(heart)
.mark_bar()
.encode(
x=alt.X("target:N", title="Heart Disease (0:tidak, 1:ya)"),
y=alt.Y("count(target):Q", title="Count of target"),
tooltip="count(target):Q",
)
.properties(height=300, width=400, title="Barplot of Heart Disease Status")
)
# Barplot diatas menunjukkan jumlah target, dengan jumlah kelas 0 (pasien yang tidak heart disease) sebanyak 138 dan jumlah kelas 1 (pasien yang heart disease) sebanyak 164. Berdasarkan barplot ini, variabel target memiliki kelas yang cukup balance.
# # **Feature Selection**
# Pemilihan feature untuk analisis selanjutnya didasarkan pada:
# 1. Untuk feature Numerik akan dilihat melalui koefisien korelasi pada masing-masing feature terhadap variabel target.
# 2. Untuk feature kategorik akan dilihat melalui p-value hasil dari uji Chi-Square.
# ## **Korelasi Variabel Numerik terhadap Variabel Target**
# **Point-biserial correlation coefficient:** This is a measure specifically designed for assessing the correlation between a binary (dichotomous) nominal variable and a numeric variable. It calculates the correlation between the binary variable (e.g., presence or absence of a characteristic) and the numeric variable (e.g., continuous or discrete variable).
from scipy.stats import pointbiserialr
numeric_vars = heart[
["age", "trestbps", "chol", "thalach", "oldpeak", "ca"]
] # Numeric independent variables
# Loop through each numeric variable and calculate point-biserial correlation coefficient
for col in numeric_vars:
correlation_coefficient, p_value = pointbiserialr(
heart["target"], numeric_vars[col]
)
print(
f"Point-Biserial Correlation Coefficient for {col}: {correlation_coefficient:.3f}"
)
# Penulis prefer tidak menyertakan feature chol, trestbps, dan age karena masing-masing feature tersebut memiliki korelasi yang rendah terhadap variabel target, yaitu secara berurutan -0.081, -0.146, -0.221.
# ## **Chi Square Test**
# The chi-square test can be used to test the association between a nominal dependent variable and a categorical independent variable, whether it is nominal or ordinal.
from sklearn.feature_selection import chi2
X = heart.drop(
["target", "age", "trestbps", "chol", "thalach", "oldpeak", "ca"], axis=1
)
y = heart["target"]
chi_scores = chi2(X, y)
p_values = pd.Series(chi_scores[1], index=X.columns)
p_values.sort_values(ascending=False, inplace=True)
p_values.plot.bar()
# Penulis akan menggunakan tingkat signifikasi 5%, maka dari itu diantara feature-feature categorik, fbs dan restecg secara masing-masing tidak berpengaruh terhadap variabel target. Maka dari itu, penulis tidak menyertakan feature ini dalam analisis selanjutnya.
heart = heart.drop(["fbs", "chol", "age", "trestbps", "restecg"], axis=1)
# # **One Hot Encoding**
# Dataset ini terdiri dari fitur numerik dan kategorik dengan tipe data nominal, maka variabel kategorik lebih cocok jika dilakukan **one hot encoding**. Variabel kategorik yang digunakan dalam analisis ini adalah sex, cp, exang, slope, dan thal. Karena sex dan exang sudah bernilai dua kategori 0 dan 1, tidak akan berpengaruh signifikan jika kedua variabel ini tidak dilakukan one hot encoding.
categorical_val = ["cp", "slope", "thal"]
heart_dmy = pd.get_dummies(heart, columns=categorical_val)
heart_dmy.head()
# # **Splitting Dataset**
X = heart_dmy.drop(["target"], axis=1)
y = heart_dmy["target"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# # **Feature Scaling**
# variabel numerik
num = ["thalach", "oldpeak", "ca"]
# Feature Scaling dengan MinMaxScaler
from sklearn.preprocessing import MinMaxScaler
sc = MinMaxScaler()
X_train[num] = sc.fit_transform(X_train[num])
X_test[num] = sc.transform(X_test[num])
# # **Model Baseline**
# Penulis akan mencoba tiga model yaitu, **Logistic Regression, Decision Tree, dan Support Vector Machine** untuk memprediksi pasien yang terdiagnosis heart disease atau tidak. Penulis tertarik untuk melakukan pemodelan klasifikasi dengan **Logistic Regression dan Decision Tree** karena model ini mudah diinterpretasikan. Selanjutnya, penulis tertarik untuk melakukan pemodelan klasifikasi dengan **Support Vector Machine** karena algoritma ini menjadi salah satu algoritma yang sudah sering digunakan untuk menganalisis dataset medis.
# **1. Logistic Regression**
# Salah satu Algoritma Klasifikasi Machine Learning yang simple dan terbaik adalah Logistic Regression(LR). LR adalah supervised learning, algoritma binary classification telah banyak digunakan di banyak aplikasi. Algoritma ini bekerja pada discrete atau binary categorical variable 0 atau 1. Sigmoid function ini digunakan sebagai cost function. Fungsi sigmoid ini memetakan nilai real yang diprediksi kedalam nilai probabilistik antara 0 dan 1.
# **Logistik Sigmoid Function:**
# $P(x) = 1/(1+\mathrm{e}^{-x})$
# Dimana, $P(x)$ adalah probabilitas fungsi estimasi dengan nilai antara 0 dan 1, $x$ adalah input kedalam fungsi probabilitas (algorithm's prediction value), dan konstanta matematika $\mathrm{e}$ adalah bilangan euler.
# **2. Decision Tree Classifier**
# Salah satu algoritma klasifikasi Machine Learning yang menggunakan serangkaian rules untuk membuat keputusan. Algoritma ini memiliki beberapa keunggulan seperti, Interpretability (hasil pemodelan mudah diinterpretasikan), No preprocessing required, Data robustness (Algoritma dapat menghandle semua tipe data dengan baik).
# Pada algoritma ini terdapat ukuran seleksi atribut. Ukuran ini disebut juga sebagai aturan pemisahan (splitting rules), karena aturan ini menentukan bagaimana decision tree "dibentuk". Terdapat beberapa aturan pemisahan dan tipe decision tree berdasarkan aturan pemisahannya yang cukup
# sering digunakan, diantaranya adalah sebagai berikut.
# 1. Information Gain, digunakan di decision tree ID3 (Quinlan, 1983).
# 2. Gain Ratio, digunakan di decision tree C4.5 (Quinlan, 1992).
# 3. Gini Index, digunakan di decision tree CART (Breiman et al, 1984).
# **3. Support Vector Machine Classifier**
# Support Vector Machine (SVM) merupakan salah satu metode dalam supervised learning yang biasanya digunakan untuk klasifikasi. Dalam pemodelan klasifikasi, SVM memiliki konsep yang lebih matang dan lebih jelas secara matematis dibandingkan dengan teknik-teknik klasifikasi lainnya. SVM juga dapat mengatasi masalah klasifikasi dan regresi dengan linear maupun non linear.
# SVM digunakan untuk mencari hyperplane terbaik dengan memaksimalkan jarak antar kelas. Hyperplane adalah sebuah fungsi yang dapat digunakan untuk pemisah antar kelas. Dalam 2-D fungsi yang digunakan untuk klasifikasi antar kelas disebut sebagai line whereas, fungsi yang digunakan untuk klasifikasi antas kelas dalam 3-D disebut plane similarly, sedangkan fungsi yang digunakan untuk klasifikasi di dalam ruang kelas dimensi yang lebih tinggi di sebut hyperplane.
# ## **1. Logistic Regression**
lr = LogisticRegression()
lr.fit(X_train, y_train)
# Prediction of the test variable
lr_y_preds = lr.predict(X_test)
# Training score
print("Training Score :", lr.score(X_train, y_train))
print("Testing Score :", lr.score(X_test, y_test))
print(classification_report(y_test, lr_y_preds))
# ## **2. Decision Tree**
clf2 = DecisionTreeClassifier(criterion="gini") # Gini Index
clf2.fit(X_train, y_train)
print("Training Score :", clf2.score(X_train, y_train))
print("Testing Score :", clf2.score(X_test, y_test))
Y_pred = clf2.predict(X_test)
print(classification_report(y_test, Y_pred))
# ## **3. Support Vector Machine**
# Pemodelan svm baik model baseline maupun hyperparameter tuning menggunakan kernel sigmoid. Untuk hyperparameter tuning dilakukan grid search ditambah dengan parameter-parameter seperti, gamma, C, degree, dan probability.
from sklearn.svm import SVC
model_sv = SVC(kernel="sigmoid")
model_sv.fit(X_train, y_train) # Train/Fit model
y_pred_sv = model_sv.predict(X_test) # get y predictions
print(classification_report(y_test, y_pred_sv)) # output accuracy
print("Training Score :", model_sv.score(X_train, y_train))
print("Testing Score :", model_sv.score(X_test, y_test))
# 
# Overfitting adalah perilaku pembelajaran mesin yang tidak diinginkan yang terjadi ketika model pembelajaran mesin memberikan prediksi akurat untuk data pelatihan tetapi tidak untuk data baru. Terjadi atau tidaknya Overfitting, salah satunya dapat dilihat dari nilai akurasi data train dan data test. Terlihat bahwa, model Decision Tree memiliki akurasi yang sangat tinggi didata train, tetapi tidak didata test. Maka dari itu, terjadi overfitting untuk pemodelan dengan Decision Tree.
# Selain itu, melalui classification report diatas, pada model baseline ini, Pemodelan Logistic Regression memiliki F1-Score paling tinggi, sebesar 0.82. Selanjutnya pemodelan dengan Decision Tree dengan F1-Score sebesar 0.79, dan pemodelan SVM dengan sigmoid kernel memiliki F1-Score terendah, sebesar 0.75.
# # **Hyperparameter Tuning - Cross Validation**
# #### **1. Hyperparameter Tuning**
# Hyperparameter Tuning dilakukan untuk mencari best parameter pada masing-masing pemodelan.
# #### **2. Cross Validation(CV)**
# Dengan Cross Validation, peneliti dapat melakukan penilaian model dengan memperkirakan kesalahan pengujian atau pemilihan model dengan memilih tingkat fleksibilitas berdasarkan kesalahan pengujian masing-masing model. Ide dasar dari CV adalah
# 1. Membagi dataset kedalam training dan validation set.
# 2. Menggunakan training set untuk fit sebuah model
# 3. Mengevaluasi model performance dengan validation set.
#
# 
# #### **3. K-fold cross-validation**
# Dengan CV k-fold, pertama-tama peneliti memilih nilai k. Kemudian, membagi kumpulan data menjadi k kumpulan. Set pertama menjadi validation set dan sisanya menjadi training set. Setelah menyesuaikan dan mengevaluasi model, pindah ke set kedua dan ulangi prosesnya. CV k-fold mengulang k kali dan rata-rata estimasi kesalahan uji k menjadi estimasi CV k-fold.
# 
#
# #### **4. Stratified KFold Cross Validation**
# Stratified k-fold cross-validation sama dengan k-fold cross-validation, tetapi Stratified k-fold cross-validation, melakukan stratified sampling, bukan random sampling.
# ## **1. Logistic Regression**
Accuracies = {} # make dictionry to save all accuracies models
# define model
lg = LogisticRegression()
# parameters
parameters = [
{
"penalty": ["l1", "l2"],
"C": np.logspace(-2, 2, 40),
"solver": ["liblinear", "saga"],
}
]
# create 10 folds
folds = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)
# define search
search = GridSearchCV(
lg, parameters, cv=folds, refit=True, verbose=2, scoring="accuracy", n_jobs=-1
)
# execute search
result = search.fit(X_train, y_train)
# summarize result
print("Best Score: %s" % result.best_score_)
print("Best Hyperparameters: %s" % result.best_params_)
# Predict how well your model is in a test dataset
Logreg = LogisticRegression(C=1.4251026703029979, penalty="l1", solver="liblinear")
Logreg.fit(X_train, y_train)
y_pred = Logreg.predict(X_test)
# Classification Report
print(classification_report(y_test, y_pred))
# ***
# **Insight**
# ***
# - Precision
# - Merupakan rasio **prediksi benar positif** dibandingkan dengan keseluruhan hasil yang **diprediksi positif**
# - Pada pemodelan ini, diperoleh presisi sebesar 0.78, artinya : Pada model ini, Persentase Pasien yang sebenarnya terkena penyakit jantung dari keseluruhan pasien yang **diprediksi terkena penyakit jantung** sebesar 78%.
# - Recall (Sensitifitas)
# - Merupakan rasio prediksi benar positif dibandingkan dengan **keseluruhan data yang benar positif**.
# - Pada pemodelan ini diperoleh recall sebesar 0.88, artinya : Pada model ini, Persentase pasien yang diprediksi terkena penyakit jantung dari keseluruhan pasien yang sebenarnya terkena penyakit jantung sebesar 88%.
# - F1 Score
# - F1 Score merupakan perbandingan rata-rata presisi dan recall yang dibobotkan. F1 Score dapat dihitung dengan formula:
# $ F1 \; Score = 2 * (Recall*Precission) / (Recall + Precission) $
# $ F1 \; Score = 2 * (0.88*0.78) / (0.88 + 0.78) = 0.83$
# - Pada pemodelan ini diperoleh F1 Score sebesar 0.83, artinya : perbandingan rata-rata presisi dan recall yang dibobotkan sebesar 83%. Dengan kata lain, kemampuan model yang seimbang untuk menangkap kasus positif (mengingat) dan akurat dengan kasus yang ditangkapnya (presisi) sebesar 0.83.
# - Accuracy
# - Merupakan rasio prediksi Benar (positif dan negatif) dengan keseluruhan data.
# - Pada pemodelan ini diperoleh akurasi sebesar 0.82, artinya : Persentase Pasien yang benar diprediksi terkena heart disease dan tidak terkena heart disease dari keseluruhan pasien sebesar 82%
#
# Confusion Matrix
cm = confusion_matrix(y_test, y_pred)
ax = plt.subplot()
sns.heatmap(cm, annot=True, fmt="g", ax=ax)
# annot=True to annotate cells, ftm='g' to disable scientific notation
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix")
ax.xaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
ax.yaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
# ***
# **Insight**
# ***
# - Dari 48 pasien yang tidak terkena heart disease, sebanyak 37 pasien diprediksi tidak terkena heart disease (TN) dan sisanya 11 pasien diprediksi terkena heart disease (FP).
# - Selanjutnya, dari 43 Pasien yang terkena heart disease, sebanyak 38 pasien diprediksi terkena heart disease (TP) dan sisanya 5 pasien diprediksi tidak terkena heart disease (FN)
# Dimana
# - TN : True Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease dan sebenarnya pasien tersebut tidak terkena heart disease)
# - FP : False Positif (Kasus dimana pasien diprediksi terkena heart disease, tetapi pada faktanya pasien tersebut tidak terkena heart disease)
# - FN : False Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease, tetapi pada faktanya pasien tersebut terkens heart disease)
# - TP : True Positif (Kasus dimana pasien diprediksi terkena heart disease dan sebenarnya pasien tersebut terkena heart disease)
# ## **2. Decision Tree**
# define model
dt = DecisionTreeClassifier(random_state=42)
# parameters
parameters = [
{
"criterion": ["gini", "entropy", "log_loss"],
"max_depth": [1, 2, 3, 4, 5],
"min_samples_split": [2, 3, 4, 5],
"min_samples_leaf": [1, 2, 3, 4, 5],
"max_features": ["sqrt", "log2"],
"random_state": [42],
}
]
# create 10 folds
folds = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)
# define search
search = GridSearchCV(dt, parameters, cv=folds, scoring="accuracy")
# execute search
result = search.fit(X_train, y_train)
# summarize result
print("Best Score: %s" % result.best_score_)
print("Best Hyperparameters: %s" % result.best_params_)
# Predict Model in Test Dataset
Dtree = DecisionTreeClassifier(
criterion="entropy",
max_depth=5,
max_features="sqrt",
min_samples_leaf=1,
min_samples_split=2,
random_state=42,
)
Dtree.fit(X_train, y_train)
y_pred_tree = Dtree.predict(X_test)
# Classification Report
print(classification_report(y_test, y_pred_tree))
# ***
# **Insight**
# ***
# - Precision
# - Merupakan rasio **prediksi benar positif** dibandingkan dengan keseluruhan hasil yang **diprediksi positif**
# - Pada pemodelan ini, diperoleh presisi sebesar 0.77, artinya : Pada model ini, Persentase Pasien yang sebenarnya terkena penyakit jantung dari keseluruhan pasien yang **diprediksi terkena penyakit jantung** sebesar 77%.
# - Recall (Sensitifitas)
# - Merupakan rasio prediksi benar positif dibandingkan dengan **keseluruhan data yang benar positif**.
# - Pada pemodelan ini diperoleh recall sebesar 0.84, artinya : Pada model ini, Persentase pasien yang diprediksi terkena penyakit jantung dari keseluruhan pasien yang sebenarnya terkena penyakit jantung sebesar 84%.
# - F1 Score
# - F1 Score merupakan perbandingan rata-rata presisi dan recall yang dibobotkan. F1 Score dapat dihitung dengan formula:
# $ F1 \; Score = 2 * (Recall*Precission) / (Recall + Precission) $
# $ F1 \; Score = 2 * (0.77*0.84) / (0.77 + 0.84) = 0.80$
# - Pada pemodelan ini diperoleh F1 Score sebesar 0.80, artinya : perbandingan rata-rata presisi dan recall yang dibobotkan sebesar 80%. Dengan kata lain, kemampuan model yang seimbang untuk menangkap kasus positif (mengingat) dan akurat dengan kasus yang ditangkapnya (presisi) sebesar 0.80.
# - Accuracy
# - Merupakan rasio prediksi Benar (positif dan negatif) dengan keseluruhan data.
# - Pada pemodelan ini diperoleh akurasi sebesar 0.80, artinya : Persentase Pasien yang benar diprediksi terkena heart disease dan tidak terkena heart disease dari keseluruhan pasien sebesar 80%
# Confusion Matrix
cm = confusion_matrix(y_test, y_pred_tree)
ax = plt.subplot()
sns.heatmap(cm, annot=True, fmt="g", ax=ax)
# annot=True to annotate cells, ftm='g' to disable scientific notation
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix")
ax.xaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
ax.yaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
# ***
# **Insight**
# ***
# - Dari 48 pasien yang tidak terkena heart disease, sebanyak 37 pasien diprediksi tidak terkena heart disease (TN) dan sisanya 11 pasien diprediksi terkena heart disease (FP).
# - Selanjutnya, dari 43 Pasien yang terkena heart disease, sebanyak 36 pasien diprediksi terkena heart disease (TP) dan sisanya 7 pasien diprediksi tidak terkena heart disease (FN)
# Dimana
# - TN : True Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease dan sebenarnya pasien tersebut tidak terkena heart disease)
# - FP : False Positif (Kasus dimana pasien diprediksi terkena heart disease, tetapi pada faktanya pasien tersebut tidak terkena heart disease)
# - FN : False Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease, tetapi pada faktanya pasien tersebut terkens heart disease)
# - TP : True Positif (Kasus dimana pasien diprediksi terkena heart disease dan sebenarnya pasien tersebut terkena heart disease)
# ## **3. Support Vector Machine**
# define model
svc = SVC()
# parameters
parameters = [
{
"kernel": ["sigmoid"],
"gamma": [0.1, 0.9, 0.06, 0.05, 0.3, 1],
"C": [0.1, 1, 2, 3, 4, 5, 6, 10, 100],
"degree": [3],
"probability": [True, False],
}
]
# create 10 folds
folds = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)
# define search
search = GridSearchCV(svc, parameters, cv=folds, scoring="accuracy")
# execute search
result = search.fit(X_train, y_train)
# summarize result
print("Best Score: %s" % result.best_score_)
print("Best Hyperparameters: %s" % result.best_params_)
# Predict Model in Test Dataset
sv = SVC(C=5, degree=3, gamma=0.06, kernel="sigmoid", probability=True)
sv.fit(X_train, y_train)
y_pred_s = sv.predict(X_test)
# Classification Report
print(classification_report(y_test, y_pred_s))
# ***
# **Insight**
# ***
# - Precision
# - Merupakan rasio **prediksi benar positif** dibandingkan dengan keseluruhan hasil yang **diprediksi positif**
# - Pada pemodelan ini, diperoleh presisi sebesar 0.74, artinya : Pada model ini, Persentase Pasien yang sebenarnya terkena penyakit jantung dari keseluruhan pasien yang **diprediksi terkena penyakit jantung** sebesar 74%.
# - Recall (Sensitifitas)
# - Merupakan rasio prediksi benar positif dibandingkan dengan **keseluruhan data yang benar positif**.
# - Pada pemodelan ini diperoleh recall sebesar 0.93, artinya : Pada model ini, Persentase pasien yang diprediksi terkena penyakit jantung dari keseluruhan pasien yang sebenarnya terkena penyakit jantung sebesar 93%.
# - F1 Score
# - F1 Score merupakan perbandingan rata-rata presisi dan recall yang dibobotkan. F1 Score dapat dihitung dengan formula:
# $ F1 \; Score = 2 * (Recall*Precission) / (Recall + Precission) $
# $ F1 \; Score = 2 * (0.93*0.74) / (0.93+0.74 ) = 0.82$
# - Pada pemodelan ini diperoleh F1 Score sebesar 0.82, artinya : perbandingan rata-rata presisi dan recall yang dibobotkan sebesar 82%. Dengan kata lain, kemampuan model yang seimbang untuk menangkap kasus positif (mengingat) dan akurat dengan kasus yang ditangkapnya (presisi) sebesar 0.82.
# - Accuracy
# - Merupakan rasio prediksi Benar (positif dan negatif) dengan keseluruhan data.
# - Pada pemodelan ini diperoleh akurasi sebesar 0.81, artinya : Persentase Pasien yang benar diprediksi terkena heart disease dan tidak terkena heart disease dari keseluruhan pasien sebesar 81%
# Confusion Matrix
cm = confusion_matrix(y_test, y_pred_s)
ax = plt.subplot()
sns.heatmap(cm, annot=True, fmt="g", ax=ax)
# annot=True to annotate cells, ftm='g' to disable scientific notation
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix")
ax.xaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
ax.yaxis.set_ticklabels(["No Heart Disease", "Heart Disease"])
# ***
# **Insight**
# ***
# - Dari 48 pasien yang tidak terkena heart disease, sebanyak 34 pasien diprediksi tidak terkena heart disease (TN) dan sisanya 14 pasien diprediksi terkena heart disease (FP).
# - Selanjutnya, dari 43 Pasien yang terkena heart disease, sebanyak 40 pasien diprediksi terkena heart disease (TP) dan sisanya 3 pasien diprediksi tidak terkena heart disease (FN)
# Dimana
# - TN : True Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease dan sebenarnya pasien tersebut tidak terkena heart disease)
# - FP : False Positif (Kasus dimana pasien diprediksi terkena heart disease, tetapi pada faktanya pasien tersebut tidak terkena heart disease)
# - FN : False Negatif (Kasus dimana pasien diprediksi tidak terkena heart disease, tetapi pada faktanya pasien tersebut terkena heart disease)
# - TP : True Positif (Kasus dimana pasien diprediksi terkena heart disease dan sebenarnya pasien tersebut terkena heart disease)
# # Learning Curve
# **Dalam learning curve, penulis menggunakan scoring akurasi. Penulis menggunakan scoring akurasi karena data target cukup balance.**
# Learning curve dengan akurasi pada klasifikasi adalah sebuah grafik yang menunjukkan perubahan performa atau akurasi model klasifikasi seiring dengan perubahan ukuran data latih (train data) yang digunakan untuk melatih model. Grafik learning curve biasanya memiliki sumbu x yang menunjukkan ukuran data latih (misalnya, jumlah sampel atau proporsi data latih), dan sumbu y yang menunjukkan akurasi model pada data latih (training accuracy) dan akurasi model pada data uji (validation accuracy) dalam bentuk nilai atau persentase.
# Learning curve dapat memberikan informasi berharga tentang performa model klasifikasi. Beberapa interpretasi yang dapat diambil dari learning curve dengan akurasi pada klasifikasi antara lain:
# 1. Underfitting: Jika training accuracy dan validation accuracy keduanya rendah dan tidak mengalami peningkatan seiring dengan peningkatan ukuran data latih, hal ini bisa mengindikasikan bahwa model mungkin terlalu sederhana (underfitting) dan membutuhkan peningkatan kompleksitas untuk dapat melakukan generalisasi yang baik pada data uji.
# 2. Overfitting: Jika training accuracy tinggi tetapi validation accuracy rendah dan tidak mengalami peningkatan seiring dengan peningkatan ukuran data latih, hal ini bisa mengindikasikan bahwa model mungkin terlalu kompleks (overfitting) dan tidak dapat menggeneralisasi dengan baik pada data uji. Solusinya bisa dengan mengurangi kompleksitas model, menggunakan teknik regularisasi, atau memperbesar ukuran data latih.
# 3. Good fit: Jika training accuracy dan validation accuracy keduanya tinggi dan cenderung stabil seiring dengan peningkatan ukuran data latih, hal ini bisa mengindikasikan bahwa model telah mencapai performa yang baik dan dapat melakukan generalisasi yang baik pada data uji. Ukuran data latih yang lebih besar mungkin tidak diperlukan.
# import library
from sklearn.model_selection import learning_curve
# ## **1. Logistic Regression**
# Membuat learning curve dengan scoring accuracy
train_sizes, train_scores, test_scores = learning_curve(
Logreg,
X_train,
y_train,
cv=folds,
scoring="accuracy",
train_sizes=np.linspace(0.1, 1.0, 10),
)
# Menghitung rata-rata skor pada setiap ukuran data latih
train_scores_mean = np.mean(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
# Visualisasi learning curve
plt.figure(figsize=(8, 6))
plt.plot(train_sizes, train_scores_mean, "o-", color="r", label="Training Score")
plt.plot(train_sizes, test_scores_mean, "o-", color="g", label="Validation Score")
plt.xlabel("Training Set Size")
plt.ylabel("Accuracy")
plt.legend(loc="best")
plt.title("Learning Curve - Logistic Regression (Accuracy)")
plt.show()
# Melalui learning curve ini dapat dilihat bahwa **secara general** pemodelan dengan logistic regression dapat dikatakan baik (good fit). Selanjutnya, pemodelan ini jauh lebih baik jika jumlah data train diatas 112 karena memiliki training score dan cross validation score yang relatif tidak berbeda jauh. Selanjutnya pada training set size ini model juga tidak mengalami overfitting. Selain itu, dengan training set size diatas 100 cross validation score cenderung naik hingga disekitar training set size sebanyak 174 terdapat cross validation score yang nilainya lebih besar dari training score. Akurasi cross validation score yang lebih besar dari training score menunjukkan bahwa pada training set size ini akan memberikan performa model yang baik. Hal ini juga dapat mengindikasikan bahwa model mungkin tidak mengalami overfitting dan mampu melakukan generalisasi yang baik pada data yang belum pernah dilihat sebelumnya (data uji). Selanjutnya, training set size diatas 175 **hampir tidak lagi signifikan** dalam meningkatkan akurasi model. Ini bisa menjadi indikasi bahwa ukuran data latih yang sudah ada sudah cukup besar untuk model ini dan penambahan data latih lebih lanjut **berpeluang** tidak memberikan peningkatan signifikan dalam performa model.
# ## **2. Decision Tree**
# Membuat learning curve dengan scoring accuracy
train_sizes, train_scores, test_scores = learning_curve(
Dtree,
X_train,
y_train,
cv=folds,
scoring="accuracy",
train_sizes=np.linspace(0.1, 1.0, 10),
)
# Menghitung rata-rata skor pada setiap ukuran data latih
train_scores_mean = np.mean(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
# Visualisasi learning curve
plt.figure(figsize=(8, 6))
plt.plot(train_sizes, train_scores_mean, "o-", color="r", label="Training Score")
plt.plot(train_sizes, test_scores_mean, "o-", color="g", label="Validation Score")
plt.xlabel("Training Set Size")
plt.ylabel("Accuracy")
plt.legend(loc="best")
plt.title("Learning Curve - Decision Tree (Accuracy)")
plt.show()
# Melalui learning curve ini, pemodelan dengan Decision Tree **secara general cenderung mengalami overfitting**, hal ini dapat kita lihat dari akurasi dari data train yang lebih besar daripada data validasi dan gap antara training score dan cross-validation score yang cukup besar.
# ## **3. Support Vector Machine**
# Membuat learning curve dengan scoring akurasi
train_sizes, train_scores, test_scores = learning_curve(
sv,
X_train,
y_train,
cv=folds,
scoring="accuracy",
train_sizes=np.linspace(0.1, 1.0, 10),
)
# Menghitung rata-rata skor pada setiap ukuran data latih
train_scores_mean = np.mean(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
# Visualisasi learning curve
plt.figure(figsize=(8, 6))
plt.plot(train_sizes, train_scores_mean, "o-", color="r", label="Training Score")
plt.plot(train_sizes, test_scores_mean, "o-", color="g", label="Validation Score")
plt.xlabel("Training Set Size")
plt.ylabel("Accuracy")
plt.legend(loc="best")
plt.title("Learning Curve - Support Vector Machine (SVM) with Accuracy Scoring")
plt.show()
# Melalui learning curve ini, **secara general** pemodelan dengan SVM dapat dikatakan baik (good fit). Selain itu, pemodelan dengan SVM lebih baik untuk jumlah data train diatas 75 karena memiliki training score dan cross validation score yang akurasinya relatif tidak terlalu jauh. Selanjutnya pada training set size ini model juga tidak mengalami overfitting. Bahkan, **disekitar** training set size sebanyak 130 terdapat cross validation score yang nilainya lebih besar dari training score. Akurasi cross validation score yang lebih besar dari training score menunjukkan bahwa pada training set size ini akan memberikan performa model yang baik. Hal ini juga dapat mengindikasikan bahwa model mungkin tidak mengalami overfitting dan mampu melakukan generalisasi yang baik pada data yang belum pernah dilihat sebelumnya (data uji).
# # ROC Analysis
# Logistic regression
y_pred_prob_lr = Logreg.predict_proba(X_test)[:, 1]
fpr_lr, tpr_lr, thresholds_lr = roc_curve(y_test, y_pred_prob_lr)
roc_auc_lr = auc(fpr_lr, tpr_lr)
precision_lr, recall_lr, th_lr = precision_recall_curve(y_test, y_pred_prob_lr)
# Decision Tree
y_pred_prob_DT = Dtree.predict_proba(X_test)[:, 1]
fpr_DT, tpr_DT, thresholds_DT = roc_curve(y_test, y_pred_prob_DT)
roc_auc_DT = auc(fpr_DT, tpr_DT)
precision_DT, recall_DT, th_DT = precision_recall_curve(y_test, y_pred_prob_DT)
# SVM
y_pred_prob_SV = sv.predict_proba(X_test)[:, 1]
fpr_SV, tpr_SV, thresholds_SV = roc_curve(y_test, y_pred_prob_SV)
roc_auc_SV = auc(fpr_SV, tpr_SV)
precision_SV, recall_SV, th_SV = precision_recall_curve(y_test, y_pred_prob_SV)
# Plot ROC curve
plt.plot([0, 1], [0, 1], "k--")
plt.plot(fpr_lr, tpr_lr, label="Log Reg (area = %0.3f)" % roc_auc_lr)
plt.plot(fpr_DT, tpr_DT, label="Decision Tree (area = %0.3f)" % roc_auc_DT)
plt.plot(fpr_SV, tpr_SV, label="SVM Sigmoid (area = %0.3f)" % roc_auc_SV)
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("ROC curves")
plt.legend(loc="best")
plt.show()
# ***
# **Insight**
# ***
# Kurva AUC - ROC adalah pengukuran kinerja untuk masalah klasifikasi pada berbagai pengaturan ambang batas. ROC adalah kurva probabilitas dan AUC mewakili tingkat atau ukuran keterpisahan. Ini memberi tahu seberapa banyak model mampu membedakan antar kelas. Semakin tinggi AUC, semakin baik model dalam memprediksi kelas 0 sebagai 0 dan kelas 1 sebagai 1. Dengan analogi, semakin tinggi AUC, semakin baik model dalam membedakan antara pasien yang terkena heart disease dengan pasien yang tidak terkena heart disease.
# 
# Secara general, melalui ROC Curve ini, pemodelan dengan Logistic Regression dan Support Vector Machine memiliki performa model yang lebih baik dibandingkan dengan Decision Tree. Selain itu, performa model Logistic Regression dan Support Vector Machine relatif sama bagusnya.
# **Jika ditinjau dari nilai AUC:**
# - Logistic Regression : Pemodelan ini memiliki nilai AUC sebesar 0.911. Hal ini berarti model yang terbentuk memiliki 91,1% area dibawah kurva. Dengan kata lain, pemodelan ini sudah "excellent" dalam memisahkan/membedakan antara kelas 1 (Pasien yang terkena heart disease) dengan kelas 0 (pasien yang tidak terkena heart disease).
# - Decision Tree : Pemodelan ini memiliki nilai AUC sebesar 0.835. Hal ini berarti model yang terbentuk memiliki 83.5% area dibawah kurva. Dengan kata lain, pemodelan ini sudah sangat baik dalam memisahkan/membedakan antara kelas 1 (Pasien yang terkena heart disease) dengan kelas 0 (pasien yang tidak terkena heart disease).
# - Support Vector Machine : Pemodelan ini memiliki nilai AUC sebesar 0.915. Hal ini berarti model yang terbentuk memiliki 91.5% area dibawah kurva. Dengan kata lain, pemodelan ini sudah "excellent" dalam memisahkan/membedakan antara kelas 1 (Pasien yang terkena heart disease) dengan kelas 0 (pasien yang tidak terkena heart disease).
# # Best Model
# Pertama, Penentuan model terbaik didasarkan pada AUC dan F1-Score. Selanjutnya, membandingkan nilai precision, recall dan akurasi dari model klasifikasi. Kurva ROC (Receiver Operator Characteristics) dapat digunakan untuk penyelidikan lebih lanjut ke dalam model. Kinerja model dapat divisualisasikan oleh ROC Curve dan tradeoff antara TPR (True Positive Rate) dan FPR (False Positive Rate). Ini berkisar dari 0 hingga 1 dan area di bawahnya menandakan kemampuan membedakan kelas model ML. Kurva ROC mendekati satu yang lebih mampu mengklasifikasikan.
# Penilaian model klasifikasi dari masing-masing classifier akan mengacu pada confusion matrix. Confusion matrix menunjukkan prediksi dan klasifikasi terbaik melalui nilai accuracy, sensitivity, dan specificity. Ketika nilai accuracy, sensitivity, dan specificity semakin mendekati 1 (satu) maka prediksi semakin lebih baik. Hasil klasifikasi dari confusion matrix terdiri dari 4 (empat) yakni true positive, false positive, false negative, dan true negative.
# 
# Terlihat bahwa model SVM memiliki nilai AUC paling baik. Selanjutnya, model Logistic Regression memiliki nilai F1-Score paling baik. Namun, baik nilai AUC dan F1-Score antara model Logistic Regression dan SVM hampir sama. Selanjutnya akan dilibatkan metrics lain dalam penentuan model terbaik.
# 
# Terlihat bahwa Klasifikasi dengan algoritma Logistic Regression memenuhi kriteria pemilihan model terbanyak, dimana algoritma ini memiliki F1-score, precision, dan akurasi tertinggi diantara dua model klasifikasi lainnya. Selain itu, nilai AUC sebesar 0.91, artinya : pemodelan ini sudah "excellent" dalam memisahkan/membedakan antara kelas 1 (Pasien yang terkena heart disease) dengan kelas 0 (pasien yang tidak terkena heart disease). Ditambah lagi, pemodelan ini memiliki evaluasi metrics yang lebih stabil dibandingkan dengan SVM dan pemodelan dengan Logistic Regression memiliki interpretabilitas model yang mudah diinterpretasikan. Maka dari itu, penulis memilih model Logistic Regression sebagai model terbaik untuk study kasus ini.
# # Feature Importance from Best Model
# These coef's tell how much and in what way did each one of it contribute to predicting the target variable
feature_dict = dict(zip(X_train.columns, list(Logreg.coef_[0])))
feature_dict
# **Odds Ratio**
# Melalui Odds Ratio, akan memudahkan penulis untuk melihat fitur-fitur yang berperan penting dalam mendiagnosa heart disease. Dimana Odds Ratio diperoleh dari $\mathrm{e}^{\beta_i}$ dengan $\beta_i$ adalah koefisien dari masing-masing fitur.
import math
feature_importance = pd.DataFrame(X_train.columns, columns=["feature"])
feature_importance["importance"] = pow(math.e, Logreg.coef_[0])
feature_importance = feature_importance.sort_values(by=["importance"], ascending=True)
feature_importance_1 = feature_importance[feature_importance["importance"] != 1]
feature_importance_1
sns.barplot(
x=feature_importance_1["importance"], y=feature_importance_1["feature"], orient="h"
)
plt.xlabel("Importance", fontsize=16)
plt.ylabel("Feature", fontsize=16)
plt.title("Feature Importance With Odds Ratio", fontsize=16)
plt.tick_params(axis="both", which="major", labelsize=12)
|
[{"heart-disease-dataset/heart.csv": {"column_names": "[\"age\", \"sex\", \"cp\", \"trestbps\", \"chol\", \"fbs\", \"restecg\", \"thalach\", \"exang\", \"oldpeak\", \"slope\", \"ca\", \"thal\", \"target\"]", "column_data_types": "{\"age\": \"int64\", \"sex\": \"int64\", \"cp\": \"int64\", \"trestbps\": \"int64\", \"chol\": \"int64\", \"fbs\": \"int64\", \"restecg\": \"int64\", \"thalach\": \"int64\", \"exang\": \"int64\", \"oldpeak\": \"float64\", \"slope\": \"int64\", \"ca\": \"int64\", \"thal\": \"int64\", \"target\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1025 entries, 0 to 1024\nData columns (total 14 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 age 1025 non-null int64 \n 1 sex 1025 non-null int64 \n 2 cp 1025 non-null int64 \n 3 trestbps 1025 non-null int64 \n 4 chol 1025 non-null int64 \n 5 fbs 1025 non-null int64 \n 6 restecg 1025 non-null int64 \n 7 thalach 1025 non-null int64 \n 8 exang 1025 non-null int64 \n 9 oldpeak 1025 non-null float64\n 10 slope 1025 non-null int64 \n 11 ca 1025 non-null int64 \n 12 thal 1025 non-null int64 \n 13 target 1025 non-null int64 \ndtypes: float64(1), int64(13)\nmemory usage: 112.2 KB\n", "summary": "{\"age\": {\"count\": 1025.0, \"mean\": 54.43414634146342, \"std\": 9.072290233244278, \"min\": 29.0, \"25%\": 48.0, \"50%\": 56.0, \"75%\": 61.0, \"max\": 77.0}, \"sex\": {\"count\": 1025.0, \"mean\": 0.6956097560975609, \"std\": 0.4603733241196493, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"cp\": {\"count\": 1025.0, \"mean\": 0.9424390243902439, \"std\": 1.029640743645865, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 3.0}, \"trestbps\": {\"count\": 1025.0, \"mean\": 131.61170731707318, \"std\": 17.516718005376408, \"min\": 94.0, \"25%\": 120.0, \"50%\": 130.0, \"75%\": 140.0, \"max\": 200.0}, \"chol\": {\"count\": 1025.0, \"mean\": 246.0, \"std\": 51.59251020618206, \"min\": 126.0, \"25%\": 211.0, \"50%\": 240.0, \"75%\": 275.0, \"max\": 564.0}, \"fbs\": {\"count\": 1025.0, \"mean\": 0.14926829268292682, \"std\": 0.3565266897271575, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"restecg\": {\"count\": 1025.0, \"mean\": 0.5297560975609756, \"std\": 0.5278775668748921, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 2.0}, \"thalach\": {\"count\": 1025.0, \"mean\": 149.11414634146342, \"std\": 23.005723745977207, \"min\": 71.0, \"25%\": 132.0, \"50%\": 152.0, \"75%\": 166.0, \"max\": 202.0}, \"exang\": {\"count\": 1025.0, \"mean\": 0.33658536585365856, \"std\": 0.47277237600371186, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"oldpeak\": {\"count\": 1025.0, \"mean\": 1.0715121951219515, \"std\": 1.175053255150176, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.8, \"75%\": 1.8, \"max\": 6.2}, \"slope\": {\"count\": 1025.0, \"mean\": 1.3853658536585365, \"std\": 0.6177552671745918, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 2.0}, \"ca\": {\"count\": 1025.0, \"mean\": 0.7541463414634146, \"std\": 1.0307976650242823, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 4.0}, \"thal\": {\"count\": 1025.0, \"mean\": 2.32390243902439, \"std\": 0.6206602380510298, \"min\": 0.0, \"25%\": 2.0, \"50%\": 2.0, \"75%\": 3.0, \"max\": 3.0}, \"target\": {\"count\": 1025.0, \"mean\": 0.5131707317073171, \"std\": 0.5000704980788014, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}}", "examples": "{\"age\":{\"0\":52,\"1\":53,\"2\":70,\"3\":61},\"sex\":{\"0\":1,\"1\":1,\"2\":1,\"3\":1},\"cp\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"trestbps\":{\"0\":125,\"1\":140,\"2\":145,\"3\":148},\"chol\":{\"0\":212,\"1\":203,\"2\":174,\"3\":203},\"fbs\":{\"0\":0,\"1\":1,\"2\":0,\"3\":0},\"restecg\":{\"0\":1,\"1\":0,\"2\":1,\"3\":1},\"thalach\":{\"0\":168,\"1\":155,\"2\":125,\"3\":161},\"exang\":{\"0\":0,\"1\":1,\"2\":1,\"3\":0},\"oldpeak\":{\"0\":1.0,\"1\":3.1,\"2\":2.6,\"3\":0.0},\"slope\":{\"0\":2,\"1\":0,\"2\":0,\"3\":2},\"ca\":{\"0\":2,\"1\":0,\"2\":0,\"3\":1},\"thal\":{\"0\":3,\"1\":3,\"2\":3,\"3\":3},\"target\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0}}"}}]
| true | 1 |
<start_data_description><data_path>heart-disease-dataset/heart.csv:
<column_names>
['age', 'sex', 'cp', 'trestbps', 'chol', 'fbs', 'restecg', 'thalach', 'exang', 'oldpeak', 'slope', 'ca', 'thal', 'target']
<column_types>
{'age': 'int64', 'sex': 'int64', 'cp': 'int64', 'trestbps': 'int64', 'chol': 'int64', 'fbs': 'int64', 'restecg': 'int64', 'thalach': 'int64', 'exang': 'int64', 'oldpeak': 'float64', 'slope': 'int64', 'ca': 'int64', 'thal': 'int64', 'target': 'int64'}
<dataframe_Summary>
{'age': {'count': 1025.0, 'mean': 54.43414634146342, 'std': 9.072290233244278, 'min': 29.0, '25%': 48.0, '50%': 56.0, '75%': 61.0, 'max': 77.0}, 'sex': {'count': 1025.0, 'mean': 0.6956097560975609, 'std': 0.4603733241196493, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'cp': {'count': 1025.0, 'mean': 0.9424390243902439, 'std': 1.029640743645865, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 2.0, 'max': 3.0}, 'trestbps': {'count': 1025.0, 'mean': 131.61170731707318, 'std': 17.516718005376408, 'min': 94.0, '25%': 120.0, '50%': 130.0, '75%': 140.0, 'max': 200.0}, 'chol': {'count': 1025.0, 'mean': 246.0, 'std': 51.59251020618206, 'min': 126.0, '25%': 211.0, '50%': 240.0, '75%': 275.0, 'max': 564.0}, 'fbs': {'count': 1025.0, 'mean': 0.14926829268292682, 'std': 0.3565266897271575, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'restecg': {'count': 1025.0, 'mean': 0.5297560975609756, 'std': 0.5278775668748921, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 2.0}, 'thalach': {'count': 1025.0, 'mean': 149.11414634146342, 'std': 23.005723745977207, 'min': 71.0, '25%': 132.0, '50%': 152.0, '75%': 166.0, 'max': 202.0}, 'exang': {'count': 1025.0, 'mean': 0.33658536585365856, 'std': 0.47277237600371186, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'oldpeak': {'count': 1025.0, 'mean': 1.0715121951219515, 'std': 1.175053255150176, 'min': 0.0, '25%': 0.0, '50%': 0.8, '75%': 1.8, 'max': 6.2}, 'slope': {'count': 1025.0, 'mean': 1.3853658536585365, 'std': 0.6177552671745918, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 2.0}, 'ca': {'count': 1025.0, 'mean': 0.7541463414634146, 'std': 1.0307976650242823, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 4.0}, 'thal': {'count': 1025.0, 'mean': 2.32390243902439, 'std': 0.6206602380510298, 'min': 0.0, '25%': 2.0, '50%': 2.0, '75%': 3.0, 'max': 3.0}, 'target': {'count': 1025.0, 'mean': 0.5131707317073171, 'std': 0.5000704980788014, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 1025 entries, 0 to 1024
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 1025 non-null int64
1 sex 1025 non-null int64
2 cp 1025 non-null int64
3 trestbps 1025 non-null int64
4 chol 1025 non-null int64
5 fbs 1025 non-null int64
6 restecg 1025 non-null int64
7 thalach 1025 non-null int64
8 exang 1025 non-null int64
9 oldpeak 1025 non-null float64
10 slope 1025 non-null int64
11 ca 1025 non-null int64
12 thal 1025 non-null int64
13 target 1025 non-null int64
dtypes: float64(1), int64(13)
memory usage: 112.2 KB
<some_examples>
{'age': {'0': 52, '1': 53, '2': 70, '3': 61}, 'sex': {'0': 1, '1': 1, '2': 1, '3': 1}, 'cp': {'0': 0, '1': 0, '2': 0, '3': 0}, 'trestbps': {'0': 125, '1': 140, '2': 145, '3': 148}, 'chol': {'0': 212, '1': 203, '2': 174, '3': 203}, 'fbs': {'0': 0, '1': 1, '2': 0, '3': 0}, 'restecg': {'0': 1, '1': 0, '2': 1, '3': 1}, 'thalach': {'0': 168, '1': 155, '2': 125, '3': 161}, 'exang': {'0': 0, '1': 1, '2': 1, '3': 0}, 'oldpeak': {'0': 1.0, '1': 3.1, '2': 2.6, '3': 0.0}, 'slope': {'0': 2, '1': 0, '2': 0, '3': 2}, 'ca': {'0': 2, '1': 0, '2': 0, '3': 1}, 'thal': {'0': 3, '1': 3, '2': 3, '3': 3}, 'target': {'0': 0, '1': 0, '2': 0, '3': 0}}
<end_description>
| 13,659 | 0 | 14,996 | 13,659 |
129147575
|
# # Developing an exploitative alternative to A/B testing
# This is the code accompaning my [blog post](https://medium.com/@Lando-L/beyond-the-basics-reinforcement-learning-with-jax-part-ii-developing-an-exploitative-9423cb6b2fa5) on multi-arm bandits.
# ## Implementing the environment
import jax
# Numpy API with hardware acceleration and automatic differentiation
from jax import numpy as jnp
# Low level operators
from jax import lax
# API for working with pseudorandom number generators
from jax import random
# Random seed to make our experiment replicable
SEED = 42
# Number of visitors we want to simulate
NUM_VISITS = 10000
# Expected click rates for the five variants with the
CLICK_RATES = [0.042, 0.03, 0.035, 0.038, 0.045]
def visit(state, timestep, click_rates, policy_fn, update_fn):
"""
Simulates a user visit.
"""
# Unpacking the experiment state into
# the agent's parameters and the random number generator
params, rng = state
# Splitting the random number generator
next_rng, policy_rng, user_rng = random.split(rng, num=3)
# Selecting the variant to show the user, based on
# the given policy, the agent's paramters, and the current timestep
variant = policy_fn(params, timestep, policy_rng)
# Randomly simulating the user click, based on
# the variant's click rate
clicked = random.uniform(user_rng) < click_rates[variant]
# Calculating the agent's updated parameters, based on
# the current parameters, the selected variant,
# and whether or not the user clicked
next_params = update_fn(params, variant, clicked)
# Returning the updated experiment state (params and rng) and
# whether or not the user clicked
return (next_params, next_rng), clicked
# ## Implementing the policies
def action_value_init(num_variants):
"""
Returns the initial action values
"""
return {
"n": jnp.ones(num_variants, dtype=jnp.int32),
"q": jnp.ones(num_variants, dtype=jnp.float32),
}
def action_value_update(params, variant, clicked):
"""
Calculates the updated action values
"""
# Reading n and q parameters of the selected variant
n, q = params["n"][variant], params["q"][variant]
# Converting the boolean clicked variable to a float value
r = clicked.astype(jnp.float32)
return {
# Incrementing the counter of the taken action by one
"n": params["n"].at[variant].add(1),
# Incrementally updating the action-value estimate
"q": params["q"].at[variant].add((r - q) / n),
}
def epsilon_greedy_policy(params, timestep, rng, epsilon):
"""
Randomly selects either the variant with highest action-value,
or an arbitrary variant.
"""
# Selecting a random variant
def explore(q, rng):
return random.choice(rng, jnp.arange(len(q)))
# Selecting the variant with the highest action-value estimate
def exploit(q, rng):
return jnp.argmax(q)
# Splitting the random number generator
uniform_rng, choice_rng = random.split(rng)
# Deciding randomly whether to explore or to exploit
return lax.cond(
random.uniform(uniform_rng) < epsilon, explore, exploit, params["q"], choice_rng
)
def boltzmann_policy(params, timestep, rng, tau):
"""
Randomly selects a variant proportional to the current action-values
"""
return random.choice(
rng,
jnp.arange(len(params["q"])),
# Turning the action-value estimates into a probability distribution
# by applying the softmax function controlled by tau
p=jax.nn.softmax(params["q"] / tau),
)
def upper_confidence_bound_policy(params, timestep, rng, confidence):
"""
Selects the variant with highest action-value plus upper confidence bound
"""
# Read n and q parameters
n, q = params["n"], params["q"]
# Calculating each variant's upper confidence bound
# and selecting the variant with the highest value
return jnp.argmax(q + confidence * jnp.sqrt(jnp.log(timestep) / n))
def beta_init(num_variants):
"""
Returns the initial hyperparameters of the beta distribution
"""
return {
"a": jnp.ones(num_variants, dtype=jnp.int32),
"b": jnp.ones(num_variants, dtype=jnp.int32),
}
def beta_update(params, variant, clicked):
"""
Calculates the updated hyperparameters of the beta distribution
"""
# Incrementing alpha by one
def increment_alpha(a, b):
return {"a": a.at[variant].add(1), "b": b}
# Incrementing beta by one
def increment_beta(a, b):
return {"b": b.at[variant].add(1), "a": a}
# Incrementing either alpha or beta
# depending on whether or not the user clicked
return lax.cond(clicked, increment_alpha, increment_beta, params["a"], params["b"])
def thompson_policy(params, timestep, rng):
"""
Randomly sampling click rates for all variants
and selecting the variant with the highest sample
"""
return jnp.argmax(random.beta(rng, params["a"], params["b"]))
# ## Implementing the evaluation
from functools import partial
from matplotlib import pyplot as plt
def evaluate(policy_fn, init_fn, update_fn):
"""
Simulating the experiment for NUM_VISITS users
while accumulating the click history
"""
return lax.scan(
# Compiling the visit function using just-in-time (JIT) compilation
# for better performance
jax.jit(
# Partially applying the visit function by fixing
# the click_rates, policy_fn, and update_fn parameters
partial(
visit,
click_rates=jnp.array(CLICK_RATES),
policy_fn=jax.jit(policy_fn),
update_fn=jax.jit(update_fn),
)
),
# Initialising the experiment state using
# init_fn and a new PRNG key
(init_fn(len(CLICK_RATES)), random.PRNGKey(SEED)),
# Setting the number steps of the experiment
jnp.arange(1, NUM_VISITS + 1),
)
def regret(history):
"""
Calculates the regret for every action in the experiment history
"""
# Calculating regret with regard to picking the optimal (0.045) variant
def fn(acc, reward):
n, v = acc[0] + 1, acc[1] + reward
return (n, v), 0.045 - (v / n)
# Calculating regret values over entire history
_, result = lax.scan(jax.jit(fn), (jnp.array(0), jnp.array(0)), history)
return result
# Epsilon greedy policy
(epsilon_greedy_params, _), epsilon_greedy_history = evaluate(
policy_fn=partial(epsilon_greedy_policy, epsilon=0.1),
init_fn=action_value_init,
update_fn=action_value_update,
)
# Boltzmann policy
(boltzmann_params, _), boltzmann_history = evaluate(
policy_fn=partial(boltzmann_policy, tau=1.0),
init_fn=action_value_init,
update_fn=action_value_update,
)
# Upper confidence bound policy
(ucb_params, _), ucb_history = evaluate(
policy_fn=partial(upper_confidence_bound_policy, confidence=2),
init_fn=action_value_init,
update_fn=action_value_update,
)
# Thompson sampling policy
(ts_params, _), ts_history = evaluate(
policy_fn=thompson_policy, init_fn=beta_init, update_fn=beta_update
)
# Visualisation
fig, ax = plt.subplots(figsize=(16, 8))
x = jnp.arange(1, NUM_VISITS + 1)
ax.set_xlabel("Number of visits")
ax.set_ylabel("Regret")
ax.plot(
x, jnp.repeat(jnp.mean(jnp.array(CLICK_RATES)), NUM_VISITS), label="A/B Testing"
)
ax.plot(x, regret(epsilon_greedy_history), label="Espilon Greedy Policy")
ax.plot(x, regret(boltzmann_history), label="Boltzmann Policy")
ax.plot(x, regret(ucb_history), label="UCB Policy")
ax.plot(x, regret(ts_history), label="TS Policy")
plt.legend()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/147/129147575.ipynb
| null | null |
[{"Id": 129147575, "ScriptId": 37367205, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11953241, "CreationDate": "05/11/2023 10:49:52", "VersionNumber": 1.0, "Title": "Exploitative alternative to A/B testing", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 274.0, "LinesInsertedFromPrevious": 274.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
# # Developing an exploitative alternative to A/B testing
# This is the code accompaning my [blog post](https://medium.com/@Lando-L/beyond-the-basics-reinforcement-learning-with-jax-part-ii-developing-an-exploitative-9423cb6b2fa5) on multi-arm bandits.
# ## Implementing the environment
import jax
# Numpy API with hardware acceleration and automatic differentiation
from jax import numpy as jnp
# Low level operators
from jax import lax
# API for working with pseudorandom number generators
from jax import random
# Random seed to make our experiment replicable
SEED = 42
# Number of visitors we want to simulate
NUM_VISITS = 10000
# Expected click rates for the five variants with the
CLICK_RATES = [0.042, 0.03, 0.035, 0.038, 0.045]
def visit(state, timestep, click_rates, policy_fn, update_fn):
"""
Simulates a user visit.
"""
# Unpacking the experiment state into
# the agent's parameters and the random number generator
params, rng = state
# Splitting the random number generator
next_rng, policy_rng, user_rng = random.split(rng, num=3)
# Selecting the variant to show the user, based on
# the given policy, the agent's paramters, and the current timestep
variant = policy_fn(params, timestep, policy_rng)
# Randomly simulating the user click, based on
# the variant's click rate
clicked = random.uniform(user_rng) < click_rates[variant]
# Calculating the agent's updated parameters, based on
# the current parameters, the selected variant,
# and whether or not the user clicked
next_params = update_fn(params, variant, clicked)
# Returning the updated experiment state (params and rng) and
# whether or not the user clicked
return (next_params, next_rng), clicked
# ## Implementing the policies
def action_value_init(num_variants):
"""
Returns the initial action values
"""
return {
"n": jnp.ones(num_variants, dtype=jnp.int32),
"q": jnp.ones(num_variants, dtype=jnp.float32),
}
def action_value_update(params, variant, clicked):
"""
Calculates the updated action values
"""
# Reading n and q parameters of the selected variant
n, q = params["n"][variant], params["q"][variant]
# Converting the boolean clicked variable to a float value
r = clicked.astype(jnp.float32)
return {
# Incrementing the counter of the taken action by one
"n": params["n"].at[variant].add(1),
# Incrementally updating the action-value estimate
"q": params["q"].at[variant].add((r - q) / n),
}
def epsilon_greedy_policy(params, timestep, rng, epsilon):
"""
Randomly selects either the variant with highest action-value,
or an arbitrary variant.
"""
# Selecting a random variant
def explore(q, rng):
return random.choice(rng, jnp.arange(len(q)))
# Selecting the variant with the highest action-value estimate
def exploit(q, rng):
return jnp.argmax(q)
# Splitting the random number generator
uniform_rng, choice_rng = random.split(rng)
# Deciding randomly whether to explore or to exploit
return lax.cond(
random.uniform(uniform_rng) < epsilon, explore, exploit, params["q"], choice_rng
)
def boltzmann_policy(params, timestep, rng, tau):
"""
Randomly selects a variant proportional to the current action-values
"""
return random.choice(
rng,
jnp.arange(len(params["q"])),
# Turning the action-value estimates into a probability distribution
# by applying the softmax function controlled by tau
p=jax.nn.softmax(params["q"] / tau),
)
def upper_confidence_bound_policy(params, timestep, rng, confidence):
"""
Selects the variant with highest action-value plus upper confidence bound
"""
# Read n and q parameters
n, q = params["n"], params["q"]
# Calculating each variant's upper confidence bound
# and selecting the variant with the highest value
return jnp.argmax(q + confidence * jnp.sqrt(jnp.log(timestep) / n))
def beta_init(num_variants):
"""
Returns the initial hyperparameters of the beta distribution
"""
return {
"a": jnp.ones(num_variants, dtype=jnp.int32),
"b": jnp.ones(num_variants, dtype=jnp.int32),
}
def beta_update(params, variant, clicked):
"""
Calculates the updated hyperparameters of the beta distribution
"""
# Incrementing alpha by one
def increment_alpha(a, b):
return {"a": a.at[variant].add(1), "b": b}
# Incrementing beta by one
def increment_beta(a, b):
return {"b": b.at[variant].add(1), "a": a}
# Incrementing either alpha or beta
# depending on whether or not the user clicked
return lax.cond(clicked, increment_alpha, increment_beta, params["a"], params["b"])
def thompson_policy(params, timestep, rng):
"""
Randomly sampling click rates for all variants
and selecting the variant with the highest sample
"""
return jnp.argmax(random.beta(rng, params["a"], params["b"]))
# ## Implementing the evaluation
from functools import partial
from matplotlib import pyplot as plt
def evaluate(policy_fn, init_fn, update_fn):
"""
Simulating the experiment for NUM_VISITS users
while accumulating the click history
"""
return lax.scan(
# Compiling the visit function using just-in-time (JIT) compilation
# for better performance
jax.jit(
# Partially applying the visit function by fixing
# the click_rates, policy_fn, and update_fn parameters
partial(
visit,
click_rates=jnp.array(CLICK_RATES),
policy_fn=jax.jit(policy_fn),
update_fn=jax.jit(update_fn),
)
),
# Initialising the experiment state using
# init_fn and a new PRNG key
(init_fn(len(CLICK_RATES)), random.PRNGKey(SEED)),
# Setting the number steps of the experiment
jnp.arange(1, NUM_VISITS + 1),
)
def regret(history):
"""
Calculates the regret for every action in the experiment history
"""
# Calculating regret with regard to picking the optimal (0.045) variant
def fn(acc, reward):
n, v = acc[0] + 1, acc[1] + reward
return (n, v), 0.045 - (v / n)
# Calculating regret values over entire history
_, result = lax.scan(jax.jit(fn), (jnp.array(0), jnp.array(0)), history)
return result
# Epsilon greedy policy
(epsilon_greedy_params, _), epsilon_greedy_history = evaluate(
policy_fn=partial(epsilon_greedy_policy, epsilon=0.1),
init_fn=action_value_init,
update_fn=action_value_update,
)
# Boltzmann policy
(boltzmann_params, _), boltzmann_history = evaluate(
policy_fn=partial(boltzmann_policy, tau=1.0),
init_fn=action_value_init,
update_fn=action_value_update,
)
# Upper confidence bound policy
(ucb_params, _), ucb_history = evaluate(
policy_fn=partial(upper_confidence_bound_policy, confidence=2),
init_fn=action_value_init,
update_fn=action_value_update,
)
# Thompson sampling policy
(ts_params, _), ts_history = evaluate(
policy_fn=thompson_policy, init_fn=beta_init, update_fn=beta_update
)
# Visualisation
fig, ax = plt.subplots(figsize=(16, 8))
x = jnp.arange(1, NUM_VISITS + 1)
ax.set_xlabel("Number of visits")
ax.set_ylabel("Regret")
ax.plot(
x, jnp.repeat(jnp.mean(jnp.array(CLICK_RATES)), NUM_VISITS), label="A/B Testing"
)
ax.plot(x, regret(epsilon_greedy_history), label="Espilon Greedy Policy")
ax.plot(x, regret(boltzmann_history), label="Boltzmann Policy")
ax.plot(x, regret(ucb_history), label="UCB Policy")
ax.plot(x, regret(ts_history), label="TS Policy")
plt.legend()
plt.show()
| false | 0 | 2,186 | 1 | 2,186 | 2,186 |
||
129147019
|
<jupyter_start><jupyter_text>OTTO train and validation (extracted from train)
This is a version of the OTTO dataset where I created a validation set from the last week of train using the script from the organizer's repo.
I have also minimized the memory footprint (which required dividing the `ts` column by 1000 to not lose information).
**If you find this useful, I would be very grateful for an upvote 🙏 Thank you**
Kaggle dataset identifier: otto-train-and-test-data-for-local-validation
<jupyter_script># Here is a sample code for gru4rec using recbole.
# I made it based on the following code from a past competition
# https://www.kaggle.com/code/astrung/recbole-lstm-sequential-for-recomendation-tutorial
# I think you can get a better score if you change the training data or epoch size, etc. Enjoy!
# !pip install --upgrade torch
import torch
torch.__version__
import tqdm
import polars as pl
import numpy as np
import pandas as pd
import seaborn as sns
import random
import os
import h5py
import sys
import gc
from matplotlib import pyplot as plt
import pyarrow.parquet as pq
# # 1. Create atomic file
train = pl.read_parquet(
"/kaggle/input/otto-train-and-test-data-for-local-validation/test.parquet"
)
test = pl.read_parquet(
"/kaggle/input/otto-full-optimized-memory-footprint/test.parquet"
)
df = pl.concat([train, test])
# df = pl.read_parquet('../input/otto-train-and-test-data-for-local-validation/test.parquet')
df = df.sort(["session", "aid", "ts"])
df = df.with_columns((pl.col("ts") * 1e9).alias("ts"))
df = df.rename({"session": "session:token", "aid": "aid:token", "ts": "ts:float"})
df.head()
df.select([pl.col("session:token", "aid:token", "ts:float")]).write_csv(
"/kaggle/working/recbox_data/recbox_data.inter", separator="\t"
)
del df, train, test
gc.collect()
# # 3. Create dataset and train model with Recbole
# For anyone need instruction document, please check this link: https://recbole.io/docs/user_guide/usage/use_modules.html
import logging
from logging import getLogger
from recbole.config import Config
from recbole.data import create_dataset, data_preparation
from recbole.model.sequential_recommender import GRU4Rec
from recbole.trainer import Trainer
from recbole.utils import init_seed, init_logger
from recbole.utils.case_study import full_sort_topk
# 64维
MAX_ITEM = 20
parameter_dict = {
"data_path": "/kaggle/working/",
"USER_ID_FIELD": "session",
"ITEM_ID_FIELD": "aid",
"TIME_FIELD": "ts",
"user_inter_num_interval": "[5,Inf)",
"item_inter_num_interval": "[5,Inf)",
"load_col": {"inter": ["session", "aid", "ts"]},
"train_neg_sample_args": None,
"learning_rate": 0.002,
"epochs": 15,
"stopping_step": 3,
"embedding_size": 64,
"eval_batch_size": 1024,
#'train_batch_size': 1024,
# 'enable_amp':True,
"MAX_ITEM_LIST_LENGTH": MAX_ITEM,
"eval_args": {
"split": {"RS": [9, 1, 0]},
"group_by": "user",
"order": "TO",
"mode": "full",
},
}
config = Config(model="GRU4Rec", dataset="recbox_data", config_dict=parameter_dict)
# init random seed
init_seed(config["seed"], config["reproducibility"])
# logger initialization
init_logger(config)
logger = getLogger()
# Create handlers
c_handler = logging.StreamHandler()
c_handler.setLevel(logging.INFO)
logger.addHandler(c_handler)
# write config info into log
logger.info(config)
# 32维
MAX_ITEM = 20
parameter_dict = {
"data_path": "/kaggle/working/",
"USER_ID_FIELD": "session",
"ITEM_ID_FIELD": "aid",
"TIME_FIELD": "ts",
"user_inter_num_interval": "[5,Inf)",
"item_inter_num_interval": "[5,Inf)",
"load_col": {"inter": ["session", "aid", "ts"]},
"train_neg_sample_args": None,
"learning_rate": 0.002,
"epochs": 15,
"stopping_step": 3,
"embedding_size": 32,
"eval_batch_size": 1024,
#'train_batch_size': 1024,
# 'enable_amp':True,
"MAX_ITEM_LIST_LENGTH": MAX_ITEM,
"eval_args": {
"split": {"RS": [9, 1, 0]},
"group_by": "user",
"order": "TO",
"mode": "full",
},
}
config = Config(model="GRU4Rec", dataset="recbox_data", config_dict=parameter_dict)
# init random seed
init_seed(config["seed"], config["reproducibility"])
# logger initialization
init_logger(config)
logger = getLogger()
# Create handlers
c_handler = logging.StreamHandler()
c_handler.setLevel(logging.INFO)
logger.addHandler(c_handler)
# write config info into log
logger.info(config)
# 128维
MAX_ITEM = 20
parameter_dict = {
"data_path": "/kaggle/working/",
"USER_ID_FIELD": "session",
"ITEM_ID_FIELD": "aid",
"TIME_FIELD": "ts",
"user_inter_num_interval": "[5,Inf)",
"item_inter_num_interval": "[5,Inf)",
"load_col": {"inter": ["session", "aid", "ts"]},
"train_neg_sample_args": None,
"learning_rate": 0.001,
"epochs": 15,
"stopping_step": 3,
"num_layers": 1,
"embedding_size": 128,
"eval_batch_size": 1024,
#'train_batch_size': 1024,
# 'enable_amp':True,
"MAX_ITEM_LIST_LENGTH": MAX_ITEM,
"eval_args": {
"split": {"RS": [9, 1, 0]},
"group_by": "user",
"order": "TO",
"mode": "full",
},
}
config = Config(model="GRU4Rec", dataset="recbox_data", config_dict=parameter_dict)
# init random seed
init_seed(config["seed"], config["reproducibility"])
# logger initialization
init_logger(config)
logger = getLogger()
# Create handlers
c_handler = logging.StreamHandler()
c_handler.setLevel(logging.INFO)
logger.addHandler(c_handler)
# write config info into log
logger.info(config)
dataset = create_dataset(config)
logger.info(dataset)
# dataset splitting
train_data, valid_data, test_data = data_preparation(config, dataset)
# # 64维
# model = GRU4Rec(config, train_data.dataset).to(config['device'])
# logger.info(model)
# # trainer loading and initialization
# trainer = Trainer(config, model)
# # model training
# best_valid_score, best_valid_result = trainer.fit(train_data, valid_data)
# #best_valid_score, best_valid_result = trainer.fit(train_data)
# 32维
model = GRU4Rec(config, train_data.dataset).to(config["device"])
logger.info(model)
# trainer loading and initialization
trainer = Trainer(config, model)
# model training
best_valid_score, best_valid_result = trainer.fit(train_data, valid_data)
# best_valid_score, best_valid_result = trainer.fit(train_data)
# 128维
model = GRU4Rec(config, train_data.dataset).to(config["device"])
logger.info(model)
# trainer loading and initialization
trainer = Trainer(config, model)
# model training
best_valid_score, best_valid_result = trainer.fit(train_data, valid_data)
# best_valid_score, best_valid_result = trainer.fit(train_data)
# import torch
from recbole.model.abstract_recommender import GeneralRecommender
class TransformRec(GeneralRecommender):
def __init__(self, config, dataset):
super(TransformRec, self).__init__(config, dataset)
self.embedding_size = config["embedding_size"]
self.max_seq_length = config["max_seq_length"]
self.num_heads = config["num_heads"]
self.num_layers = config["num_layers"]
self.ff_num_hidden_units = config["ff_num_hidden_units"]
self.dropout = config["dropout"]
self.embedding_layer = torch.nn.Embedding(315004, self.embedding_size)
self.transformer_layer = torch.nn.TransformerEncoder(
layer=torch.nn.TransformerEncoderLayer(
d_model=self.embedding_size,
nhead=self.num_heads,
dim_feedforward=self.ff_num_hidden_units,
dropout=self.dropout,
),
num_layers=self.num_layers,
)
self.fc_layer = torch.nn.Linear(self.embedding_size, 315004)
def forward(self, user, item_seq):
# item_seq shape: [batch_size, seq_len]
input_seq = self.embedding_layer(
item_seq
) # shape: [batch_size, seq_len, embedding_size]
input_seq = input_seq.permute(
1, 0, 2
) # shape: [seq_len, batch_size, embedding_size]
output_seq = self.transformer_layer(
input_seq
) # shape: [seq_len, batch_size, embedding_size]
# 选择序列中最后一个物品做预测
output = output_seq[-1] # shape: [batch_size, embedding_size]
output = self.fc_layer(output) # shape: [batch_size, num_items]
return output
torch.__version__
config["ff_num_hidden_units"]
dataset
import gc
del trainer, train_data, valid_data, test_data
gc.collect()
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 7))
plt.plot(list(range(1, len(auc798) + 1)), auc798)
plt.scatter(list(range(1, len(auc798) + 1)), auc798)
plt.plot(list(range(1, len(auc835) + 1)), auc835)
plt.scatter(list(range(1, len(auc835) + 1)), auc835)
plt.plot(list(range(1, len(auc848) + 1)), auc848)
plt.scatter(list(range(1, len(auc848) + 1)), auc848)
plt.plot(list(range(1, len(auc827) + 1)), auc827)
plt.scatter(list(range(1, len(auc827) + 1)), auc827)
plt.xlabel("Epoch", fontsize=15)
plt.ylabel("pr auc", fontsize=15)
plt.legend(["dim 4", "", "dim 8", "", "dim 16", "", "dim 20", ""])
plt.title("pr auc", fontsize=20)
plt.savefig("pr_auc_hist.png")
# # 4. Create recommendation result from trained model
# I note document here for any one want to customize it: https://recbole.io/docs/user_guide/usage/case_study.html
# # https://qiita.com/fufufukakaka/items/e03df3a7299b2b8f99cf
# from typing import List, Tuple
# import numpy as np
# import torch
# from pydantic import BaseModel
# from recbole.data import create_dataset
# from recbole.data.dataset.sequential_dataset import SequentialDataset
# from recbole.data.interaction import Interaction
# from recbole.model.sequential_recommender.sine import SINE
# from recbole.utils import get_model, init_seed
# class ItemHistory(BaseModel):
# sequence: List[str]
# topk: int
# class RecommendedItems(BaseModel):
# score_list: List[float]
# item_list: List[str]
# def pred_user_to_item(item_history: ItemHistory):
# item_history_dict = item_history.dict()
# item_sequence = item_history_dict["sequence"]
# item_length = len(item_sequence)
# pad_length = MAX_ITEM # pre-defined by recbole
# padded_item_sequence = torch.nn.functional.pad(
# torch.tensor(dataset.token2id(dataset.iid_field, item_sequence)),
# (0, pad_length - item_length),
# "constant",
# 0,
# )
# input_interaction = Interaction(
# {
# "aid_list": padded_item_sequence.reshape(1, -1),
# "item_length": torch.tensor([item_length]),
# }
# )
# scores = model.full_sort_predict(input_interaction.to(model.device))
# scores = scores.view(-1, dataset.item_num)
# scores[:, 0] = -np.inf # pad item score -> -inf
# topk_score, topk_iid_list = torch.topk(scores, item_history_dict["topk"])
# predicted_score_list = topk_score.tolist()[0]
# predicted_item_list = dataset.id2token(
# dataset.iid_field, topk_iid_list.tolist()
# ).tolist()
# recommended_items = {
# "score_list": predicted_score_list,
# "item_list": predicted_item_list,
# }
# return recommended_items
# #test = pl.read_parquet('../input/otto-train-and-test-data-for-local-validation/test.parquet')
# test = pl.read_parquet('../input/otto-full-optimized-memory-footprint/test.parquet')
# import pandas as pd
# import numpy as np
# from collections import defaultdict
# #sample_sub = pd.read_csv('../input/otto-recommender-system//sample_submission.csv')
# session_types = ['clicks', 'carts', 'orders']
# test_session_AIDs = test.to_pandas().reset_index(drop=True).groupby('session')['aid'].apply(list)
# test_session_types = test.to_pandas().reset_index(drop=True).groupby('session')['type'].apply(list)
# del test
# gc.collect()
# labels = []
# type_weight_multipliers = {0: 1, 1: 6, 2: 3}
# for AIDs, types in zip(test_session_AIDs, test_session_types):
# if len(AIDs) >= 20:
# # if we have enough aids (over equals 20) we don't need to look for candidates! we just use the old logic
# weights=np.logspace(0.1,1,len(AIDs),base=2, endpoint=True)-1
# aids_temp=defaultdict(lambda: 0)
# for aid,w,t in zip(AIDs,weights,types):
# aids_temp[aid]+= w * type_weight_multipliers[t]
# sorted_aids=[k for k, v in sorted(aids_temp.items(), key=lambda item: -item[1])]
# labels.append(sorted_aids[:20])
# else:
# AIDs = list(dict.fromkeys(AIDs))
# item = ItemHistory(sequence=AIDs, topk=20)
# try:
# nns = [ int(v) for v in pred_user_to_item(item)['item_list']]
# except:
# nns = []
# for word in nns:
# if len(AIDs) == 20:
# break
# if int(word) not in AIDs:
# AIDs.append(word)
# labels.append(AIDs[:20])
# pred_user_to_item(item)['item_list']
# labels_as_strings = [' '.join([str(l) for l in lls]) for lls in labels]
# predictions = pd.DataFrame(data={'session_type': test_session_AIDs.index, 'labels': labels_as_strings})
# labels_as_strings = [' '.join([str(l) for l in lls]) for lls in labels]
# predictions = pd.DataFrame(data={'session_type': test_session_AIDs.index, 'labels': labels_as_strings})
# prediction_dfs = []
# for st in session_types:
# modified_predictions = predictions.copy()
# modified_predictions.session_type = modified_predictions.session_type.astype('str') + f'_{st}'
# prediction_dfs.append(modified_predictions)
# submission = pd.concat(prediction_dfs).reset_index(drop=True)
# submission.to_csv('submission.csv', index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/147/129147019.ipynb
|
otto-train-and-test-data-for-local-validation
|
radek1
|
[{"Id": 129147019, "ScriptId": 38284073, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5265145, "CreationDate": "05/11/2023 10:45:10", "VersionNumber": 2.0, "Title": "Recbole GRU4Rec sample code", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 433.0, "LinesInsertedFromPrevious": 312.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 121.0, "LinesInsertedFromFork": 312.0, "LinesDeletedFromFork": 123.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 121.0, "TotalVotes": 0}]
|
[{"Id": 184937951, "KernelVersionId": 129147019, "SourceDatasetVersionId": 4461402}, {"Id": 184937952, "KernelVersionId": 129147019, "SourceDatasetVersionId": 4474043}]
|
[{"Id": 4461402, "DatasetId": 2611514, "DatasourceVersionId": 4521320, "CreatorUserId": 83267, "LicenseName": "Unknown", "CreationDate": "11/07/2022 03:52:44", "VersionNumber": 1.0, "Title": "OTTO train and validation (extracted from train)", "Slug": "otto-train-and-test-data-for-local-validation", "Subtitle": NaN, "Description": "This is a version of the OTTO dataset where I created a validation set from the last week of train using the script from the organizer's repo.\n\nI have also minimized the memory footprint (which required dividing the `ts` column by 1000 to not lose information).\n\n**If you find this useful, I would be very grateful for an upvote \ud83d\ude4f Thank you**", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2611514, "CreatorUserId": 83267, "OwnerUserId": 83267.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4461402.0, "CurrentDatasourceVersionId": 4521320.0, "ForumId": 2642226, "Type": 2, "CreationDate": "11/07/2022 03:52:44", "LastActivityDate": "11/07/2022", "TotalViews": 9136, "TotalDownloads": 2324, "TotalVotes": 120, "TotalKernels": 48}]
|
[{"Id": 83267, "UserName": "radek1", "DisplayName": "Radek Osmulski", "RegisterDate": "02/02/2013", "PerformanceTier": 4}]
|
# Here is a sample code for gru4rec using recbole.
# I made it based on the following code from a past competition
# https://www.kaggle.com/code/astrung/recbole-lstm-sequential-for-recomendation-tutorial
# I think you can get a better score if you change the training data or epoch size, etc. Enjoy!
# !pip install --upgrade torch
import torch
torch.__version__
import tqdm
import polars as pl
import numpy as np
import pandas as pd
import seaborn as sns
import random
import os
import h5py
import sys
import gc
from matplotlib import pyplot as plt
import pyarrow.parquet as pq
# # 1. Create atomic file
train = pl.read_parquet(
"/kaggle/input/otto-train-and-test-data-for-local-validation/test.parquet"
)
test = pl.read_parquet(
"/kaggle/input/otto-full-optimized-memory-footprint/test.parquet"
)
df = pl.concat([train, test])
# df = pl.read_parquet('../input/otto-train-and-test-data-for-local-validation/test.parquet')
df = df.sort(["session", "aid", "ts"])
df = df.with_columns((pl.col("ts") * 1e9).alias("ts"))
df = df.rename({"session": "session:token", "aid": "aid:token", "ts": "ts:float"})
df.head()
df.select([pl.col("session:token", "aid:token", "ts:float")]).write_csv(
"/kaggle/working/recbox_data/recbox_data.inter", separator="\t"
)
del df, train, test
gc.collect()
# # 3. Create dataset and train model with Recbole
# For anyone need instruction document, please check this link: https://recbole.io/docs/user_guide/usage/use_modules.html
import logging
from logging import getLogger
from recbole.config import Config
from recbole.data import create_dataset, data_preparation
from recbole.model.sequential_recommender import GRU4Rec
from recbole.trainer import Trainer
from recbole.utils import init_seed, init_logger
from recbole.utils.case_study import full_sort_topk
# 64维
MAX_ITEM = 20
parameter_dict = {
"data_path": "/kaggle/working/",
"USER_ID_FIELD": "session",
"ITEM_ID_FIELD": "aid",
"TIME_FIELD": "ts",
"user_inter_num_interval": "[5,Inf)",
"item_inter_num_interval": "[5,Inf)",
"load_col": {"inter": ["session", "aid", "ts"]},
"train_neg_sample_args": None,
"learning_rate": 0.002,
"epochs": 15,
"stopping_step": 3,
"embedding_size": 64,
"eval_batch_size": 1024,
#'train_batch_size': 1024,
# 'enable_amp':True,
"MAX_ITEM_LIST_LENGTH": MAX_ITEM,
"eval_args": {
"split": {"RS": [9, 1, 0]},
"group_by": "user",
"order": "TO",
"mode": "full",
},
}
config = Config(model="GRU4Rec", dataset="recbox_data", config_dict=parameter_dict)
# init random seed
init_seed(config["seed"], config["reproducibility"])
# logger initialization
init_logger(config)
logger = getLogger()
# Create handlers
c_handler = logging.StreamHandler()
c_handler.setLevel(logging.INFO)
logger.addHandler(c_handler)
# write config info into log
logger.info(config)
# 32维
MAX_ITEM = 20
parameter_dict = {
"data_path": "/kaggle/working/",
"USER_ID_FIELD": "session",
"ITEM_ID_FIELD": "aid",
"TIME_FIELD": "ts",
"user_inter_num_interval": "[5,Inf)",
"item_inter_num_interval": "[5,Inf)",
"load_col": {"inter": ["session", "aid", "ts"]},
"train_neg_sample_args": None,
"learning_rate": 0.002,
"epochs": 15,
"stopping_step": 3,
"embedding_size": 32,
"eval_batch_size": 1024,
#'train_batch_size': 1024,
# 'enable_amp':True,
"MAX_ITEM_LIST_LENGTH": MAX_ITEM,
"eval_args": {
"split": {"RS": [9, 1, 0]},
"group_by": "user",
"order": "TO",
"mode": "full",
},
}
config = Config(model="GRU4Rec", dataset="recbox_data", config_dict=parameter_dict)
# init random seed
init_seed(config["seed"], config["reproducibility"])
# logger initialization
init_logger(config)
logger = getLogger()
# Create handlers
c_handler = logging.StreamHandler()
c_handler.setLevel(logging.INFO)
logger.addHandler(c_handler)
# write config info into log
logger.info(config)
# 128维
MAX_ITEM = 20
parameter_dict = {
"data_path": "/kaggle/working/",
"USER_ID_FIELD": "session",
"ITEM_ID_FIELD": "aid",
"TIME_FIELD": "ts",
"user_inter_num_interval": "[5,Inf)",
"item_inter_num_interval": "[5,Inf)",
"load_col": {"inter": ["session", "aid", "ts"]},
"train_neg_sample_args": None,
"learning_rate": 0.001,
"epochs": 15,
"stopping_step": 3,
"num_layers": 1,
"embedding_size": 128,
"eval_batch_size": 1024,
#'train_batch_size': 1024,
# 'enable_amp':True,
"MAX_ITEM_LIST_LENGTH": MAX_ITEM,
"eval_args": {
"split": {"RS": [9, 1, 0]},
"group_by": "user",
"order": "TO",
"mode": "full",
},
}
config = Config(model="GRU4Rec", dataset="recbox_data", config_dict=parameter_dict)
# init random seed
init_seed(config["seed"], config["reproducibility"])
# logger initialization
init_logger(config)
logger = getLogger()
# Create handlers
c_handler = logging.StreamHandler()
c_handler.setLevel(logging.INFO)
logger.addHandler(c_handler)
# write config info into log
logger.info(config)
dataset = create_dataset(config)
logger.info(dataset)
# dataset splitting
train_data, valid_data, test_data = data_preparation(config, dataset)
# # 64维
# model = GRU4Rec(config, train_data.dataset).to(config['device'])
# logger.info(model)
# # trainer loading and initialization
# trainer = Trainer(config, model)
# # model training
# best_valid_score, best_valid_result = trainer.fit(train_data, valid_data)
# #best_valid_score, best_valid_result = trainer.fit(train_data)
# 32维
model = GRU4Rec(config, train_data.dataset).to(config["device"])
logger.info(model)
# trainer loading and initialization
trainer = Trainer(config, model)
# model training
best_valid_score, best_valid_result = trainer.fit(train_data, valid_data)
# best_valid_score, best_valid_result = trainer.fit(train_data)
# 128维
model = GRU4Rec(config, train_data.dataset).to(config["device"])
logger.info(model)
# trainer loading and initialization
trainer = Trainer(config, model)
# model training
best_valid_score, best_valid_result = trainer.fit(train_data, valid_data)
# best_valid_score, best_valid_result = trainer.fit(train_data)
# import torch
from recbole.model.abstract_recommender import GeneralRecommender
class TransformRec(GeneralRecommender):
def __init__(self, config, dataset):
super(TransformRec, self).__init__(config, dataset)
self.embedding_size = config["embedding_size"]
self.max_seq_length = config["max_seq_length"]
self.num_heads = config["num_heads"]
self.num_layers = config["num_layers"]
self.ff_num_hidden_units = config["ff_num_hidden_units"]
self.dropout = config["dropout"]
self.embedding_layer = torch.nn.Embedding(315004, self.embedding_size)
self.transformer_layer = torch.nn.TransformerEncoder(
layer=torch.nn.TransformerEncoderLayer(
d_model=self.embedding_size,
nhead=self.num_heads,
dim_feedforward=self.ff_num_hidden_units,
dropout=self.dropout,
),
num_layers=self.num_layers,
)
self.fc_layer = torch.nn.Linear(self.embedding_size, 315004)
def forward(self, user, item_seq):
# item_seq shape: [batch_size, seq_len]
input_seq = self.embedding_layer(
item_seq
) # shape: [batch_size, seq_len, embedding_size]
input_seq = input_seq.permute(
1, 0, 2
) # shape: [seq_len, batch_size, embedding_size]
output_seq = self.transformer_layer(
input_seq
) # shape: [seq_len, batch_size, embedding_size]
# 选择序列中最后一个物品做预测
output = output_seq[-1] # shape: [batch_size, embedding_size]
output = self.fc_layer(output) # shape: [batch_size, num_items]
return output
torch.__version__
config["ff_num_hidden_units"]
dataset
import gc
del trainer, train_data, valid_data, test_data
gc.collect()
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 7))
plt.plot(list(range(1, len(auc798) + 1)), auc798)
plt.scatter(list(range(1, len(auc798) + 1)), auc798)
plt.plot(list(range(1, len(auc835) + 1)), auc835)
plt.scatter(list(range(1, len(auc835) + 1)), auc835)
plt.plot(list(range(1, len(auc848) + 1)), auc848)
plt.scatter(list(range(1, len(auc848) + 1)), auc848)
plt.plot(list(range(1, len(auc827) + 1)), auc827)
plt.scatter(list(range(1, len(auc827) + 1)), auc827)
plt.xlabel("Epoch", fontsize=15)
plt.ylabel("pr auc", fontsize=15)
plt.legend(["dim 4", "", "dim 8", "", "dim 16", "", "dim 20", ""])
plt.title("pr auc", fontsize=20)
plt.savefig("pr_auc_hist.png")
# # 4. Create recommendation result from trained model
# I note document here for any one want to customize it: https://recbole.io/docs/user_guide/usage/case_study.html
# # https://qiita.com/fufufukakaka/items/e03df3a7299b2b8f99cf
# from typing import List, Tuple
# import numpy as np
# import torch
# from pydantic import BaseModel
# from recbole.data import create_dataset
# from recbole.data.dataset.sequential_dataset import SequentialDataset
# from recbole.data.interaction import Interaction
# from recbole.model.sequential_recommender.sine import SINE
# from recbole.utils import get_model, init_seed
# class ItemHistory(BaseModel):
# sequence: List[str]
# topk: int
# class RecommendedItems(BaseModel):
# score_list: List[float]
# item_list: List[str]
# def pred_user_to_item(item_history: ItemHistory):
# item_history_dict = item_history.dict()
# item_sequence = item_history_dict["sequence"]
# item_length = len(item_sequence)
# pad_length = MAX_ITEM # pre-defined by recbole
# padded_item_sequence = torch.nn.functional.pad(
# torch.tensor(dataset.token2id(dataset.iid_field, item_sequence)),
# (0, pad_length - item_length),
# "constant",
# 0,
# )
# input_interaction = Interaction(
# {
# "aid_list": padded_item_sequence.reshape(1, -1),
# "item_length": torch.tensor([item_length]),
# }
# )
# scores = model.full_sort_predict(input_interaction.to(model.device))
# scores = scores.view(-1, dataset.item_num)
# scores[:, 0] = -np.inf # pad item score -> -inf
# topk_score, topk_iid_list = torch.topk(scores, item_history_dict["topk"])
# predicted_score_list = topk_score.tolist()[0]
# predicted_item_list = dataset.id2token(
# dataset.iid_field, topk_iid_list.tolist()
# ).tolist()
# recommended_items = {
# "score_list": predicted_score_list,
# "item_list": predicted_item_list,
# }
# return recommended_items
# #test = pl.read_parquet('../input/otto-train-and-test-data-for-local-validation/test.parquet')
# test = pl.read_parquet('../input/otto-full-optimized-memory-footprint/test.parquet')
# import pandas as pd
# import numpy as np
# from collections import defaultdict
# #sample_sub = pd.read_csv('../input/otto-recommender-system//sample_submission.csv')
# session_types = ['clicks', 'carts', 'orders']
# test_session_AIDs = test.to_pandas().reset_index(drop=True).groupby('session')['aid'].apply(list)
# test_session_types = test.to_pandas().reset_index(drop=True).groupby('session')['type'].apply(list)
# del test
# gc.collect()
# labels = []
# type_weight_multipliers = {0: 1, 1: 6, 2: 3}
# for AIDs, types in zip(test_session_AIDs, test_session_types):
# if len(AIDs) >= 20:
# # if we have enough aids (over equals 20) we don't need to look for candidates! we just use the old logic
# weights=np.logspace(0.1,1,len(AIDs),base=2, endpoint=True)-1
# aids_temp=defaultdict(lambda: 0)
# for aid,w,t in zip(AIDs,weights,types):
# aids_temp[aid]+= w * type_weight_multipliers[t]
# sorted_aids=[k for k, v in sorted(aids_temp.items(), key=lambda item: -item[1])]
# labels.append(sorted_aids[:20])
# else:
# AIDs = list(dict.fromkeys(AIDs))
# item = ItemHistory(sequence=AIDs, topk=20)
# try:
# nns = [ int(v) for v in pred_user_to_item(item)['item_list']]
# except:
# nns = []
# for word in nns:
# if len(AIDs) == 20:
# break
# if int(word) not in AIDs:
# AIDs.append(word)
# labels.append(AIDs[:20])
# pred_user_to_item(item)['item_list']
# labels_as_strings = [' '.join([str(l) for l in lls]) for lls in labels]
# predictions = pd.DataFrame(data={'session_type': test_session_AIDs.index, 'labels': labels_as_strings})
# labels_as_strings = [' '.join([str(l) for l in lls]) for lls in labels]
# predictions = pd.DataFrame(data={'session_type': test_session_AIDs.index, 'labels': labels_as_strings})
# prediction_dfs = []
# for st in session_types:
# modified_predictions = predictions.copy()
# modified_predictions.session_type = modified_predictions.session_type.astype('str') + f'_{st}'
# prediction_dfs.append(modified_predictions)
# submission = pd.concat(prediction_dfs).reset_index(drop=True)
# submission.to_csv('submission.csv', index=False)
| false | 0 | 4,385 | 0 | 4,515 | 4,385 |
||
129147937
|
# To check the model summary like in keras, install torchsummary
#!pip install torchsummary
# To create a progress bar to check the progress of training
# To split dataset into train, validation, and test sections in a more controlled manner
# Import libraries to deal with files
import os
# Import Image from PIL to read Image
from PIL import Image
# Import libraries for imaging viewing
import numpy as np
import matplotlib.pyplot as plt
from collections import Counter
# Import libraries for creating network and handeling data
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import Dataset, DataLoader, random_split
# Import torchvision libraries to manage images
from torchvision.io import read_image
import torchvision.transforms as transforms
# Import train_test_split to split Full Dataset
from sklearn.model_selection import train_test_split
# from torchsummary import summary
from tqdm import tqdm
class MiniDataset(Dataset):
def __init__(self, images, labels, transform=None):
# Set the list values
self.images = images
self.labels = labels
self.transform = transform
def __len__(self):
return len(self.images)
def __getitem__(self, index):
# Read the image (returns a tensor) and get the label
image = Image.open(self.images[index])
label = self.labels[index]
# Get the width and height of the image
width, height = image.size
# If the width is larger than the height
if width > height:
# Calculate the difference, and crop the image so the width is cropped
difference = width - height
image = image.crop(
((difference // 2), 0, width - (difference // 2), height)
)
# If the height is larger than the width
elif height > width:
# Calculate the difference, and crop the image so the height is cropped
difference = height - width
image = image.crop(
(0, (difference // 2), width, height - (difference // 2))
)
# If the dataset included a transform for the image, apply the transform
if self.transform:
image = self.transform(image)
# Return the image and the label
return image, label
class CreateDataset(Dataset):
def __init__(self, source_dir, splitRatio, transform=None, balanceDataset=False):
# Lists to store the important information for a dataset
images = []
labels = []
# If the source directory provided isn't a directory, return
if os.path.isdir(source_dir) == False:
print(f"{source_dir} not a valid directory")
return
# If the splitRatio is not a valid percentage, return
if sum(splitRatio) != 1:
print(f"{splitRatio} does not add up to 1")
return
# Get all the folder names of all classes from the source_dir
inner_dir = os.listdir(source_dir)
# Go through all the folders in source directory
for i in range(len(inner_dir)):
# Go through all the files within the folder
for file in os.listdir(os.path.join(source_dir, inner_dir[i])):
# Check if the file extension is a valid image extension
if file.lower().endswith(("jpg", "jpeg", "png")):
# Add the file path to images list
images.append(os.path.join(source_dir, inner_dir[i], file))
labels.append(int(inner_dir[i]))
# Split up the full Dataset into Train and Temp sections
trainImages, tempImages, trainLabels, tempLabels = train_test_split(
images, labels, train_size=splitRatio[1], random_state=13, stratify=labels
)
# Split up the Temp Dataset into Valid and Test sections
testImages, validImages, testLabels, validLabels = train_test_split(
tempImages, tempLabels, train_size=0.5, random_state=13, stratify=tempLabels
)
defaultAugmentation = transforms.Compose(
[transforms.ToTensor(), transforms.Resize((224, 224), antialias=True)]
)
# Create Three separate Datasets Objects from the Full Dataset
self.trainDataset = MiniDataset(trainImages, trainLabels, transform)
self.validDataset = MiniDataset(validImages, validLabels, defaultAugmentation)
self.testDataset = MiniDataset(testImages, testLabels, defaultAugmentation)
# If we want to balance the dataset
if balanceDataset:
# Count how many times each label occurs
countedLabels = Counter(trainLabels)
# Sort the dictionary by highest occuronce first into a list
sortedTuples = countedLabels.most_common()
# Get the maximum amount of occurunces
maximumAmount = sortedTuples[0][1]
# Go through the sortedTuples list
for i in range(len(sortedTuples)):
# Save the label and it's amounts into a variable
label = int(sortedTuples[i][0])
amount = sortedTuples[i][1]
# If the amount isn't the same as maximum amount
if amount != maximumAmount:
# Calculate the difference
difference = maximumAmount - amount
# Find the index of first occurence of the `label` in the labels list
base = trainLabels.index(label)
# Calculate the last index that would correlate to that `label` class
top = amount + base
# Set the current index to base
index = base
# While the difference is greater than 0
while difference > 0:
# Check if index reached the last value of the list, if so reset it to base
if index >= top:
index = base
# Add the label and the corresponding image to the lists
trainLabels.append(label)
trainImages.append(trainImages[index])
# Increment the index and decrement the difference
index = index + 1
difference = difference - 1
# Function to display the image with it's corresponding label
def showImage(image, label):
plt.figure()
plt.imshow(image.permute(1, 2, 0))
plt.title(str(label))
plt.xticks([])
plt.yticks([])
trainAugmentation = transforms.Compose(
[
transforms.ToTensor(),
transforms.RandomRotation(45),
transforms.RandomHorizontalFlip(0.5),
transforms.RandomVerticalFlip(0.5),
transforms.Resize((224, 224), antialias=True),
]
)
# Creating a Dataset
fullDataset = CreateDataset(
"/kaggle/input/diabetic-retinopathy-classification-dataset/annotated",
[0.70, 0.30],
transform=trainAugmentation,
balanceDataset=True,
)
# Create a DataLoader for each Dataset
trainLoader = DataLoader(
fullDataset.trainDataset,
batch_size=16,
shuffle=True,
num_workers=4,
pin_memory=True,
)
validLoader = DataLoader(
fullDataset.validDataset, batch_size=16, num_workers=4, pin_memory=True
)
testLoader = DataLoader(
fullDataset.testDataset, batch_size=1, num_workers=4, pin_memory=True
)
# Display the Image
showImage(fullDataset.trainDataset[0][0], fullDataset.trainDataset[0][1])
print(Counter(fullDataset.trainDataset.labels))
print(Counter(fullDataset.validDataset.labels))
print(Counter(fullDataset.testDataset.labels))
# Creating a base model that takes the input of size (224, 224, 3)
model = nn.Sequential(
nn.Conv2d(in_channels=3, out_channels=16, kernel_size=3, stride=1, padding=0),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(in_channels=16, out_channels=32, kernel_size=3, stride=1, padding=0),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(in_channels=32, out_channels=32, kernel_size=3, stride=1, padding=0),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(in_channels=32, out_channels=64, kernel_size=3, stride=1, padding=0),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
nn.Flatten(),
nn.Linear(in_features=9216, out_features=1024, bias=True),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(in_features=1024, out_features=6, bias=True),
)
# Set the device to Cuda if available, otherwise keep using cpu
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
# Set the model to use the cuda or cpu
model = model.to(device)
# Function to train the model on the Dataset
def train_model(model, loader, criterion, optimizer, epochs):
# Loop for number of epochs
for epoch in range(epochs):
# Go through the training and validity datasets and set model state accordingly
for phase in ["train", "val"]:
if phase == "train":
model.train()
else:
model.eval()
# Create the progress bar
progress_bar = tqdm(
enumerate(loader[phase]),
total=len(loader[phase]),
desc=f"Epoch [{epoch+1}/{epochs}]",
)
# Values to calculate the accuracy while training
running_loss = 0.0
running_corrects = 0
# Go through the images and labels in Dataset
for i, (images, labels) in progress_bar:
# Set the tensors to correspond to correct device
images = images.to(device)
labels = labels.to(device)
# Set the gradients of all optimized tensors to zero
optimizer.zero_grad()
# If training, enable the gradients and predict with images and label
with torch.set_grad_enabled(phase == "train"):
outputs = model(images)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# If in training phase, do backward propogation
if phase == "train":
loss.backward()
optimizer.step()
# Calculate loss and accuracy for current forward pass
running_loss += loss.item() * images.size(0)
running_corrects += torch.sum(preds == labels.data)
if (i + 1) % 100 == 0:
print(
f"Epoch [{epoch+1}/{epochs}], Step [{i+1}/{len(loader[phase])}], Loss: {loss.item()}"
)
# Calculate epoch loss and accuracy
epoch_loss = running_loss / len(loader[phase].dataset)
epoch_acc = running_corrects.double() / len(loader[phase].dataset)
# Print the loss and accuracy
print("{} Loss: {:.4f} Acc: {:.4f}".format(phase, epoch_loss, epoch_acc))
# Return the trained model
return model
# Runs below code with no_grad() so gradients don't change
@torch.no_grad()
# Function to evaluate the model
def evaluate_model(model, loader):
# Save the current state of the model
state = model.training
# Set the model to evaluation mode
model.eval()
# Variables to calculate loss and accuracy
running_loss = 0.0
running_corrects = 0
# Go through the dataLoader
for i, (images, labels) in enumerate(loader):
# Convert Image and Label tensors to support the device
images = images.to(device)
labels = labels.to(device)
# Get the predictions and loss of the model on the input
outputs = model(images)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# Get the loss and accuracy of the current pass
running_loss += loss.item() * images.size(0)
running_corrects += torch.sum(preds == labels.data)
# Calculate the total loss and accuracy
total_loss = running_loss / len(loader.dataset)
total_acc = running_corrects.double() / len(loader.dataset)
# For good measures, reset the state of the model we saved before evaluation
model.training = state
# Print the accuracy and Loss
print("Loss: {:.4f} Acc: {:.4f}".format(total_loss, total_acc))
loaders = {"train": trainLoader, "val": validLoader}
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(model.parameters(), lr=0.001, momentum=0.8, weight_decay=0.001)
trainedModel = train_model(model, loaders, criterion, optimizer, epochs=10)
torch.save(model.state_dict(), "/kaggle/working/savedModel.pth")
evaluate_model(trainedModel, testLoader)
# summary(model, (3, 224, 224))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/147/129147937.ipynb
| null | null |
[{"Id": 129147937, "ScriptId": 38208278, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13983771, "CreationDate": "05/11/2023 10:53:16", "VersionNumber": 2.0, "Title": "AIRS Research Project - Saksham", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 363.0, "LinesInsertedFromPrevious": 25.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 338.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# To check the model summary like in keras, install torchsummary
#!pip install torchsummary
# To create a progress bar to check the progress of training
# To split dataset into train, validation, and test sections in a more controlled manner
# Import libraries to deal with files
import os
# Import Image from PIL to read Image
from PIL import Image
# Import libraries for imaging viewing
import numpy as np
import matplotlib.pyplot as plt
from collections import Counter
# Import libraries for creating network and handeling data
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import Dataset, DataLoader, random_split
# Import torchvision libraries to manage images
from torchvision.io import read_image
import torchvision.transforms as transforms
# Import train_test_split to split Full Dataset
from sklearn.model_selection import train_test_split
# from torchsummary import summary
from tqdm import tqdm
class MiniDataset(Dataset):
def __init__(self, images, labels, transform=None):
# Set the list values
self.images = images
self.labels = labels
self.transform = transform
def __len__(self):
return len(self.images)
def __getitem__(self, index):
# Read the image (returns a tensor) and get the label
image = Image.open(self.images[index])
label = self.labels[index]
# Get the width and height of the image
width, height = image.size
# If the width is larger than the height
if width > height:
# Calculate the difference, and crop the image so the width is cropped
difference = width - height
image = image.crop(
((difference // 2), 0, width - (difference // 2), height)
)
# If the height is larger than the width
elif height > width:
# Calculate the difference, and crop the image so the height is cropped
difference = height - width
image = image.crop(
(0, (difference // 2), width, height - (difference // 2))
)
# If the dataset included a transform for the image, apply the transform
if self.transform:
image = self.transform(image)
# Return the image and the label
return image, label
class CreateDataset(Dataset):
def __init__(self, source_dir, splitRatio, transform=None, balanceDataset=False):
# Lists to store the important information for a dataset
images = []
labels = []
# If the source directory provided isn't a directory, return
if os.path.isdir(source_dir) == False:
print(f"{source_dir} not a valid directory")
return
# If the splitRatio is not a valid percentage, return
if sum(splitRatio) != 1:
print(f"{splitRatio} does not add up to 1")
return
# Get all the folder names of all classes from the source_dir
inner_dir = os.listdir(source_dir)
# Go through all the folders in source directory
for i in range(len(inner_dir)):
# Go through all the files within the folder
for file in os.listdir(os.path.join(source_dir, inner_dir[i])):
# Check if the file extension is a valid image extension
if file.lower().endswith(("jpg", "jpeg", "png")):
# Add the file path to images list
images.append(os.path.join(source_dir, inner_dir[i], file))
labels.append(int(inner_dir[i]))
# Split up the full Dataset into Train and Temp sections
trainImages, tempImages, trainLabels, tempLabels = train_test_split(
images, labels, train_size=splitRatio[1], random_state=13, stratify=labels
)
# Split up the Temp Dataset into Valid and Test sections
testImages, validImages, testLabels, validLabels = train_test_split(
tempImages, tempLabels, train_size=0.5, random_state=13, stratify=tempLabels
)
defaultAugmentation = transforms.Compose(
[transforms.ToTensor(), transforms.Resize((224, 224), antialias=True)]
)
# Create Three separate Datasets Objects from the Full Dataset
self.trainDataset = MiniDataset(trainImages, trainLabels, transform)
self.validDataset = MiniDataset(validImages, validLabels, defaultAugmentation)
self.testDataset = MiniDataset(testImages, testLabels, defaultAugmentation)
# If we want to balance the dataset
if balanceDataset:
# Count how many times each label occurs
countedLabels = Counter(trainLabels)
# Sort the dictionary by highest occuronce first into a list
sortedTuples = countedLabels.most_common()
# Get the maximum amount of occurunces
maximumAmount = sortedTuples[0][1]
# Go through the sortedTuples list
for i in range(len(sortedTuples)):
# Save the label and it's amounts into a variable
label = int(sortedTuples[i][0])
amount = sortedTuples[i][1]
# If the amount isn't the same as maximum amount
if amount != maximumAmount:
# Calculate the difference
difference = maximumAmount - amount
# Find the index of first occurence of the `label` in the labels list
base = trainLabels.index(label)
# Calculate the last index that would correlate to that `label` class
top = amount + base
# Set the current index to base
index = base
# While the difference is greater than 0
while difference > 0:
# Check if index reached the last value of the list, if so reset it to base
if index >= top:
index = base
# Add the label and the corresponding image to the lists
trainLabels.append(label)
trainImages.append(trainImages[index])
# Increment the index and decrement the difference
index = index + 1
difference = difference - 1
# Function to display the image with it's corresponding label
def showImage(image, label):
plt.figure()
plt.imshow(image.permute(1, 2, 0))
plt.title(str(label))
plt.xticks([])
plt.yticks([])
trainAugmentation = transforms.Compose(
[
transforms.ToTensor(),
transforms.RandomRotation(45),
transforms.RandomHorizontalFlip(0.5),
transforms.RandomVerticalFlip(0.5),
transforms.Resize((224, 224), antialias=True),
]
)
# Creating a Dataset
fullDataset = CreateDataset(
"/kaggle/input/diabetic-retinopathy-classification-dataset/annotated",
[0.70, 0.30],
transform=trainAugmentation,
balanceDataset=True,
)
# Create a DataLoader for each Dataset
trainLoader = DataLoader(
fullDataset.trainDataset,
batch_size=16,
shuffle=True,
num_workers=4,
pin_memory=True,
)
validLoader = DataLoader(
fullDataset.validDataset, batch_size=16, num_workers=4, pin_memory=True
)
testLoader = DataLoader(
fullDataset.testDataset, batch_size=1, num_workers=4, pin_memory=True
)
# Display the Image
showImage(fullDataset.trainDataset[0][0], fullDataset.trainDataset[0][1])
print(Counter(fullDataset.trainDataset.labels))
print(Counter(fullDataset.validDataset.labels))
print(Counter(fullDataset.testDataset.labels))
# Creating a base model that takes the input of size (224, 224, 3)
model = nn.Sequential(
nn.Conv2d(in_channels=3, out_channels=16, kernel_size=3, stride=1, padding=0),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(in_channels=16, out_channels=32, kernel_size=3, stride=1, padding=0),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(in_channels=32, out_channels=32, kernel_size=3, stride=1, padding=0),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(in_channels=32, out_channels=64, kernel_size=3, stride=1, padding=0),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
nn.Flatten(),
nn.Linear(in_features=9216, out_features=1024, bias=True),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(in_features=1024, out_features=6, bias=True),
)
# Set the device to Cuda if available, otherwise keep using cpu
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
# Set the model to use the cuda or cpu
model = model.to(device)
# Function to train the model on the Dataset
def train_model(model, loader, criterion, optimizer, epochs):
# Loop for number of epochs
for epoch in range(epochs):
# Go through the training and validity datasets and set model state accordingly
for phase in ["train", "val"]:
if phase == "train":
model.train()
else:
model.eval()
# Create the progress bar
progress_bar = tqdm(
enumerate(loader[phase]),
total=len(loader[phase]),
desc=f"Epoch [{epoch+1}/{epochs}]",
)
# Values to calculate the accuracy while training
running_loss = 0.0
running_corrects = 0
# Go through the images and labels in Dataset
for i, (images, labels) in progress_bar:
# Set the tensors to correspond to correct device
images = images.to(device)
labels = labels.to(device)
# Set the gradients of all optimized tensors to zero
optimizer.zero_grad()
# If training, enable the gradients and predict with images and label
with torch.set_grad_enabled(phase == "train"):
outputs = model(images)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# If in training phase, do backward propogation
if phase == "train":
loss.backward()
optimizer.step()
# Calculate loss and accuracy for current forward pass
running_loss += loss.item() * images.size(0)
running_corrects += torch.sum(preds == labels.data)
if (i + 1) % 100 == 0:
print(
f"Epoch [{epoch+1}/{epochs}], Step [{i+1}/{len(loader[phase])}], Loss: {loss.item()}"
)
# Calculate epoch loss and accuracy
epoch_loss = running_loss / len(loader[phase].dataset)
epoch_acc = running_corrects.double() / len(loader[phase].dataset)
# Print the loss and accuracy
print("{} Loss: {:.4f} Acc: {:.4f}".format(phase, epoch_loss, epoch_acc))
# Return the trained model
return model
# Runs below code with no_grad() so gradients don't change
@torch.no_grad()
# Function to evaluate the model
def evaluate_model(model, loader):
# Save the current state of the model
state = model.training
# Set the model to evaluation mode
model.eval()
# Variables to calculate loss and accuracy
running_loss = 0.0
running_corrects = 0
# Go through the dataLoader
for i, (images, labels) in enumerate(loader):
# Convert Image and Label tensors to support the device
images = images.to(device)
labels = labels.to(device)
# Get the predictions and loss of the model on the input
outputs = model(images)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# Get the loss and accuracy of the current pass
running_loss += loss.item() * images.size(0)
running_corrects += torch.sum(preds == labels.data)
# Calculate the total loss and accuracy
total_loss = running_loss / len(loader.dataset)
total_acc = running_corrects.double() / len(loader.dataset)
# For good measures, reset the state of the model we saved before evaluation
model.training = state
# Print the accuracy and Loss
print("Loss: {:.4f} Acc: {:.4f}".format(total_loss, total_acc))
loaders = {"train": trainLoader, "val": validLoader}
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(model.parameters(), lr=0.001, momentum=0.8, weight_decay=0.001)
trainedModel = train_model(model, loaders, criterion, optimizer, epochs=10)
torch.save(model.state_dict(), "/kaggle/working/savedModel.pth")
evaluate_model(trainedModel, testLoader)
# summary(model, (3, 224, 224))
| false | 0 | 3,080 | 0 | 3,080 | 3,080 |
||
129147023
|
<jupyter_start><jupyter_text>dataset-2-cv-model-4
The dataset has two folder for each train and test folder: happy and sad
- The size of each image is 48x48 due to the input shape of model 3
Note:
- [Training Model 4 Notebook](https://www.kaggle.com/code/ahsanfirdaus/model-4-v3-94/notebook)
- [Testing Model 4 Notebook](https://www.kaggle.com/code/ahsanfirdaus/model-4-testing-implementation/notebook)
- [Model 3 & Model 4](https://www.kaggle.com/datasets/ahsanfirdaus/ahsan-model-3-testing-implementation?select=model_4+%282%29)
Kaggle dataset identifier: emotion-dataset-2
<jupyter_script># # Import Library
from keras.models import load_model # TensorFlow is required for Keras to work
from PIL import Image, ImageOps # Install pillow instead of PIL
import numpy as np
import matplotlib.pyplot as plt
# # Disable Scientific Notation
np.set_printoptions(suppress=True)
# # Load Model
model = load_model("/kaggle/input/model-4-v3/model_4_v3.h5", compile=False)
# # Initiate Labels
class_names = ["happy", "sad"]
# # Load Image
image = Image.open(
"/kaggle/input/emotion-dataset-2/resized-48-48/test/happy/size_48_48_13694565514_c8b8614cb1_w_face.png"
).convert("L")
# The predicted image should be **happy**
# ## Before convert into gray
plt.imshow(image, cmap="gray")
# ## After convert into gray
plt.imshow(image)
# ## Image info
im_width = image.width
im_height = image.height
print("Image Width: ", im_width)
print("Image Height: ", im_height)
# Current image size is 48 x 48
# # Resizing the image
# **Just in case the image is not in 48 x 48**
size = (48, 48)
image = ImageOps.fit(image, size, Image.Resampling.LANCZOS)
# # Turn image into array
image_array = np.asarray(image)
image_array.shape
# # Normalize Image
normalized_image_array = (image_array.astype(np.float32) / 127.5) - 1
normalized_image_array.shape
# # Array Image
data = np.ndarray(shape=(1, 48, 48), dtype=np.float32)
data.shape
# # Load Image (Again)
data[0] = normalized_image_array
# # Prediction
prediction = model.predict(data)
index = np.argmax(prediction)
class_name = class_names[index]
confidence_score = prediction[0][index]
print("Class:", class_name[0:], end=" ")
print("Confidence Score:", confidence_score)
# ---
# # Load Image 2
image2 = Image.open(
"/kaggle/input/emotion-dataset-2/resized-48-48/test/sad/size_48_48_13895174901_768e9f188c_n_face.png"
).convert("L")
# ## Before convert into gray
plt.imshow(image2, cmap="gray")
# ## After convert into gray
plt.imshow(image2)
# ## Image Info
im_width = image2.width
im_height = image2.height
print("Image Width: ", im_width)
print("Image Height: ", im_height)
# # Resizing the image
size = (48, 48)
image2 = ImageOps.fit(image2, size, Image.Resampling.LANCZOS)
# # Turn image into array
image2_array = np.asarray(image2)
image2_array.shape
# # Normalize Image
normalized_image2_array = (image2_array.astype(np.float32) / 127.5) - 1
normalized_image2_array.shape
# # Array Image
data2 = np.ndarray(shape=(1, 48, 48), dtype=np.float32)
data2.shape
# # Load Image 2 (Again)
data2[0] = normalized_image2_array
# # Prediction
prediction = model.predict(data2)
index = np.argmax(prediction)
class_name = class_names[index]
confidence_score = prediction[0][index]
print("Class:", class_name[0:], end=" ")
print("Confidence Score:", confidence_score)
# ---
# # Import Library for using image url
import urllib.request
# # Initiate Image
url = "https://images.unsplash.com/photo-1494790108377-be9c29b29330?ixlib=rb-4.0.3&ixid=MnwxMjA3fDB8MHxwaG90by1wYWdlfHx8fGVufDB8fHx8&auto=format&fit=crop&w=687&q=80"
image3 = Image.open(urllib.request.urlopen(url)).convert("L")
# ## Before Convert into Gray
plt.imshow(image3, cmap="gray")
# ## After convert into gray
plt.imshow(image3)
# The expected prediction should be **happy**
# ## Image info
im_width = image3.width
im_height = image3.height
print("Image Width: ", im_width)
print("Image Height: ", im_height)
# # Resizing the image
# The resizing process is needed in order to predict using the existing model (model_4_v3)
size = (48, 48)
image3 = ImageOps.fit(image3, size, Image.Resampling.LANCZOS)
plt.imshow(image3)
# The image might looks so blury since the size of image is converted into 48 x 48
# # Turn image into array
image3_array = np.asarray(image3)
image3_array.shape
# The image is already in 48 x 48 size
# # Normalize image 3
normalized_image3_array = (image_array.astype(np.float32) / 127.5) - 1
normalized_image3_array.shape
# # Array Image 3
data3 = np.ndarray(shape=(1, 48, 48), dtype=np.float32)
data3.shape
# # Load Image 3 (Again)
data3[0] = normalized_image3_array
# # Prediction for Image 3
prediction = model.predict(data3)
index = np.argmax(prediction)
class_name = class_names[index]
confidence_score = prediction[0][index]
print("Class:", class_name[0:], end=" ")
print("Confidence Score:", confidence_score)
# ---
# # Initiate Image
url = "https://images.unsplash.com/photo-1486182706240-e597091e0cfc?ixlib=rb-4.0.3&ixid=MnwxMjA3fDB8MHxwaG90by1wYWdlfHx8fGVufDB8fHx8&auto=format&fit=crop&w=1471&q=80"
image4 = Image.open(urllib.request.urlopen(url)).convert("L")
# ## Before Convert into Gray
plt.imshow(image4, cmap="gray")
# ## After Convert into Gray
plt.imshow(image4)
# ## Image Info
im_width = image4.width
im_height = image4.height
print("Image Width: ", im_width)
print("Image Height: ", im_height)
# # Resizing the image
# The resizing process is needed in order to predict using the existing model (model_4_v3)
#
size = (48, 48)
image4 = ImageOps.fit(image4, size, Image.Resampling.LANCZOS)
plt.imshow(image4)
# The image might looks so blury since the size of image is converted into 48 x 48
# # Turn Image into array
image4_array = np.asarray(image4)
image4_array.shape
# Now, the image size is already in 48 x 48 size
# # Normalize image 4
normalized_image4_array = (image4_array.astype(np.float32) / 127.5) - 1
normalized_image4_array.shape
plt.imshow(normalized_image4_array)
# # Array Image 4
data4 = np.ndarray(shape=(1, 48, 48), dtype=np.float32)
data4.shape
# # Load Image 4 (Again)
data4[0] = normalized_image4_array
# # Prediction for Image 4
prediction = model.predict(data4)
index = np.argmax(prediction)
class_name = class_names[index]
confidence_score = prediction[0][index]
print("Class:", class_name[0:], end=" ")
print("Confidence Score:", confidence_score)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/147/129147023.ipynb
|
emotion-dataset-2
|
ahsanfirdaus
|
[{"Id": 129147023, "ScriptId": 38151823, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10264206, "CreationDate": "05/11/2023 10:45:13", "VersionNumber": 1.0, "Title": "model_4_testing_implementation", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 283.0, "LinesInsertedFromPrevious": 283.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184937955, "KernelVersionId": 129147023, "SourceDatasetVersionId": 5661223}]
|
[{"Id": 5661223, "DatasetId": 3253176, "DatasourceVersionId": 5736661, "CreatorUserId": 10264206, "LicenseName": "Unknown", "CreationDate": "05/11/2023 07:42:52", "VersionNumber": 3.0, "Title": "dataset-2-cv-model-4", "Slug": "emotion-dataset-2", "Subtitle": NaN, "Description": " The dataset has two folder for each train and test folder: happy and sad\n- The size of each image is 48x48 due to the input shape of model 3\n\nNote:\n- [Training Model 4 Notebook](https://www.kaggle.com/code/ahsanfirdaus/model-4-v3-94/notebook)\n- [Testing Model 4 Notebook](https://www.kaggle.com/code/ahsanfirdaus/model-4-testing-implementation/notebook)\n- [Model 3 & Model 4](https://www.kaggle.com/datasets/ahsanfirdaus/ahsan-model-3-testing-implementation?select=model_4+%282%29)", "VersionNotes": "Data Update 2023-05-11", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3253176, "CreatorUserId": 10264206, "OwnerUserId": 10264206.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5661223.0, "CurrentDatasourceVersionId": 5736661.0, "ForumId": 3318619, "Type": 2, "CreationDate": "05/11/2023 04:24:17", "LastActivityDate": "05/11/2023", "TotalViews": 50, "TotalDownloads": 2, "TotalVotes": 0, "TotalKernels": 2}]
|
[{"Id": 10264206, "UserName": "ahsanfirdaus", "DisplayName": "Ahsan Firdaus", "RegisterDate": "04/17/2022", "PerformanceTier": 0}]
|
# # Import Library
from keras.models import load_model # TensorFlow is required for Keras to work
from PIL import Image, ImageOps # Install pillow instead of PIL
import numpy as np
import matplotlib.pyplot as plt
# # Disable Scientific Notation
np.set_printoptions(suppress=True)
# # Load Model
model = load_model("/kaggle/input/model-4-v3/model_4_v3.h5", compile=False)
# # Initiate Labels
class_names = ["happy", "sad"]
# # Load Image
image = Image.open(
"/kaggle/input/emotion-dataset-2/resized-48-48/test/happy/size_48_48_13694565514_c8b8614cb1_w_face.png"
).convert("L")
# The predicted image should be **happy**
# ## Before convert into gray
plt.imshow(image, cmap="gray")
# ## After convert into gray
plt.imshow(image)
# ## Image info
im_width = image.width
im_height = image.height
print("Image Width: ", im_width)
print("Image Height: ", im_height)
# Current image size is 48 x 48
# # Resizing the image
# **Just in case the image is not in 48 x 48**
size = (48, 48)
image = ImageOps.fit(image, size, Image.Resampling.LANCZOS)
# # Turn image into array
image_array = np.asarray(image)
image_array.shape
# # Normalize Image
normalized_image_array = (image_array.astype(np.float32) / 127.5) - 1
normalized_image_array.shape
# # Array Image
data = np.ndarray(shape=(1, 48, 48), dtype=np.float32)
data.shape
# # Load Image (Again)
data[0] = normalized_image_array
# # Prediction
prediction = model.predict(data)
index = np.argmax(prediction)
class_name = class_names[index]
confidence_score = prediction[0][index]
print("Class:", class_name[0:], end=" ")
print("Confidence Score:", confidence_score)
# ---
# # Load Image 2
image2 = Image.open(
"/kaggle/input/emotion-dataset-2/resized-48-48/test/sad/size_48_48_13895174901_768e9f188c_n_face.png"
).convert("L")
# ## Before convert into gray
plt.imshow(image2, cmap="gray")
# ## After convert into gray
plt.imshow(image2)
# ## Image Info
im_width = image2.width
im_height = image2.height
print("Image Width: ", im_width)
print("Image Height: ", im_height)
# # Resizing the image
size = (48, 48)
image2 = ImageOps.fit(image2, size, Image.Resampling.LANCZOS)
# # Turn image into array
image2_array = np.asarray(image2)
image2_array.shape
# # Normalize Image
normalized_image2_array = (image2_array.astype(np.float32) / 127.5) - 1
normalized_image2_array.shape
# # Array Image
data2 = np.ndarray(shape=(1, 48, 48), dtype=np.float32)
data2.shape
# # Load Image 2 (Again)
data2[0] = normalized_image2_array
# # Prediction
prediction = model.predict(data2)
index = np.argmax(prediction)
class_name = class_names[index]
confidence_score = prediction[0][index]
print("Class:", class_name[0:], end=" ")
print("Confidence Score:", confidence_score)
# ---
# # Import Library for using image url
import urllib.request
# # Initiate Image
url = "https://images.unsplash.com/photo-1494790108377-be9c29b29330?ixlib=rb-4.0.3&ixid=MnwxMjA3fDB8MHxwaG90by1wYWdlfHx8fGVufDB8fHx8&auto=format&fit=crop&w=687&q=80"
image3 = Image.open(urllib.request.urlopen(url)).convert("L")
# ## Before Convert into Gray
plt.imshow(image3, cmap="gray")
# ## After convert into gray
plt.imshow(image3)
# The expected prediction should be **happy**
# ## Image info
im_width = image3.width
im_height = image3.height
print("Image Width: ", im_width)
print("Image Height: ", im_height)
# # Resizing the image
# The resizing process is needed in order to predict using the existing model (model_4_v3)
size = (48, 48)
image3 = ImageOps.fit(image3, size, Image.Resampling.LANCZOS)
plt.imshow(image3)
# The image might looks so blury since the size of image is converted into 48 x 48
# # Turn image into array
image3_array = np.asarray(image3)
image3_array.shape
# The image is already in 48 x 48 size
# # Normalize image 3
normalized_image3_array = (image_array.astype(np.float32) / 127.5) - 1
normalized_image3_array.shape
# # Array Image 3
data3 = np.ndarray(shape=(1, 48, 48), dtype=np.float32)
data3.shape
# # Load Image 3 (Again)
data3[0] = normalized_image3_array
# # Prediction for Image 3
prediction = model.predict(data3)
index = np.argmax(prediction)
class_name = class_names[index]
confidence_score = prediction[0][index]
print("Class:", class_name[0:], end=" ")
print("Confidence Score:", confidence_score)
# ---
# # Initiate Image
url = "https://images.unsplash.com/photo-1486182706240-e597091e0cfc?ixlib=rb-4.0.3&ixid=MnwxMjA3fDB8MHxwaG90by1wYWdlfHx8fGVufDB8fHx8&auto=format&fit=crop&w=1471&q=80"
image4 = Image.open(urllib.request.urlopen(url)).convert("L")
# ## Before Convert into Gray
plt.imshow(image4, cmap="gray")
# ## After Convert into Gray
plt.imshow(image4)
# ## Image Info
im_width = image4.width
im_height = image4.height
print("Image Width: ", im_width)
print("Image Height: ", im_height)
# # Resizing the image
# The resizing process is needed in order to predict using the existing model (model_4_v3)
#
size = (48, 48)
image4 = ImageOps.fit(image4, size, Image.Resampling.LANCZOS)
plt.imshow(image4)
# The image might looks so blury since the size of image is converted into 48 x 48
# # Turn Image into array
image4_array = np.asarray(image4)
image4_array.shape
# Now, the image size is already in 48 x 48 size
# # Normalize image 4
normalized_image4_array = (image4_array.astype(np.float32) / 127.5) - 1
normalized_image4_array.shape
plt.imshow(normalized_image4_array)
# # Array Image 4
data4 = np.ndarray(shape=(1, 48, 48), dtype=np.float32)
data4.shape
# # Load Image 4 (Again)
data4[0] = normalized_image4_array
# # Prediction for Image 4
prediction = model.predict(data4)
index = np.argmax(prediction)
class_name = class_names[index]
confidence_score = prediction[0][index]
print("Class:", class_name[0:], end=" ")
print("Confidence Score:", confidence_score)
| false | 0 | 2,123 | 0 | 2,320 | 2,123 |
||
129147657
|
<jupyter_start><jupyter_text>Weather Data set
Kaggle dataset identifier: weather-data-set
<jupyter_script># # Weather data set
# # Importing Libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
data = pd.read_csv("/kaggle/input/weather-data-set/Weather Data.csv")
data
data.dtypes
# # 1. Finding the Null values
data.head(2)
data.isnull().sum()
# ## Umm... there is no null values. GOOD :)
# # 2. Seeing the Wind
data.head(2)
wind = data["Wind Speed_km/h"].value_counts()
wind.to_frame()
# ## Ploting
x_values = wind.values
y_values = wind.index
plt.figure(figsize=(10, 5))
sns.barplot(x=x_values, y=y_values, palette="rainbow")
plt.xlabel("How many time of Number in wind")
plt.ylabel("Number of wind per hour")
plt.xticks(rotation=90)
# # 3. Top 10 Rel Hum_%
data.head(2)
rel_hum = data["Rel Hum_%"].value_counts().head(10)
rel_hum.to_frame()
# ## Ploting
x_values = rel_hum.values
y_values = rel_hum.index
plt.figure(figsize=(10, 5))
sns.barplot(x=x_values, y=y_values, palette="rainbow")
plt.xlabel("How many time of Number in rel hum")
plt.ylabel("Number of rel hum per hour")
plt.xticks(rotation=90)
# # 4. How many Weather are there?
data.head(2)
weather = data["Weather"].value_counts().head(10)
weather.to_frame()
# ## Ploting
x_values = weather.values
y_values = weather.index
plt.figure(figsize=(7, 5))
sns.barplot(x=x_values, y=y_values, palette="rainbow")
plt.xlabel("Number of Weather")
plt.ylabel("Name of time")
plt.xticks(rotation=90)
# # 5. Top 10 Dew Temperature in C
data.head(2)
dew = data["Dew Point Temp_C"].value_counts().head(10)
dew.to_frame()
# ## Ploting
x_values = dew.values
y_values = dew.index
plt.figure(figsize=(7, 5))
sns.countplot(x=x_values, palette="rainbow")
plt.xlabel("Number of Time")
plt.ylabel("Number of Temperature in C")
plt.xticks(rotation=90)
# # 6. Top 100 Temperature in C
data.head(2)
tem = data["Temp_C"].value_counts().head(100)
tem.to_frame()
# ## Ploting
x_values = tem.values
y_values = tem.index
plt.figure(figsize=(7, 5))
sns.countplot(y=x_values, palette="rainbow")
plt.xlabel("Number of Time")
plt.ylabel("Number of Temperature in C")
plt.xticks(rotation=90)
# # 6. Weather in Months
data.head(2)
data["Date/Time"] = pd.to_datetime(data["Date/Time"])
data.dtypes
data["Date/Time"] = data["Date/Time"].dt.month
month = data["Date/Time"].value_counts()
month.to_frame()
# ## Ploting
x_values = month.values
y_values = month.index
plt.figure(figsize=(7, 5))
sns.countplot(x=x_values, palette="rainbow")
plt.ylabel("Number of Time")
plt.xlabel("Number of Temperature in C")
plt.xticks(rotation=90)
# # 7. Visibility in Km
data.head(2)
visibility = data["Visibility_km"].value_counts()
visibility.to_frame()
# ## Ploting
x_values = visibility.values
y_values = visibility.index
plt.figure(figsize=(7, 5))
sns.barplot(x=x_values, y=y_values, palette="rainbow")
plt.xlabel("Number of Time")
plt.ylabel("Number of Temperature in C")
plt.xticks(rotation=90)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/147/129147657.ipynb
|
weather-data-set
|
learnwithsrishti
|
[{"Id": 129147657, "ScriptId": 38384785, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14633433, "CreationDate": "05/11/2023 10:50:42", "VersionNumber": 1.0, "Title": "Weather data sets", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 146.0, "LinesInsertedFromPrevious": 146.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184939092, "KernelVersionId": 129147657, "SourceDatasetVersionId": 5643355}]
|
[{"Id": 5643355, "DatasetId": 3243717, "DatasourceVersionId": 5718680, "CreatorUserId": 14633433, "LicenseName": "Unknown", "CreationDate": "05/09/2023 12:51:06", "VersionNumber": 1.0, "Title": "Weather Data set", "Slug": "weather-data-set", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3243717, "CreatorUserId": 14633433, "OwnerUserId": 14633433.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5643355.0, "CurrentDatasourceVersionId": 5718680.0, "ForumId": 3308998, "Type": 2, "CreationDate": "05/09/2023 12:51:06", "LastActivityDate": "05/09/2023", "TotalViews": 76, "TotalDownloads": 7, "TotalVotes": 1, "TotalKernels": 1}]
|
[{"Id": 14633433, "UserName": "learnwithsrishti", "DisplayName": "Learn_with_Srishti", "RegisterDate": "04/14/2023", "PerformanceTier": 0}]
|
# # Weather data set
# # Importing Libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
data = pd.read_csv("/kaggle/input/weather-data-set/Weather Data.csv")
data
data.dtypes
# # 1. Finding the Null values
data.head(2)
data.isnull().sum()
# ## Umm... there is no null values. GOOD :)
# # 2. Seeing the Wind
data.head(2)
wind = data["Wind Speed_km/h"].value_counts()
wind.to_frame()
# ## Ploting
x_values = wind.values
y_values = wind.index
plt.figure(figsize=(10, 5))
sns.barplot(x=x_values, y=y_values, palette="rainbow")
plt.xlabel("How many time of Number in wind")
plt.ylabel("Number of wind per hour")
plt.xticks(rotation=90)
# # 3. Top 10 Rel Hum_%
data.head(2)
rel_hum = data["Rel Hum_%"].value_counts().head(10)
rel_hum.to_frame()
# ## Ploting
x_values = rel_hum.values
y_values = rel_hum.index
plt.figure(figsize=(10, 5))
sns.barplot(x=x_values, y=y_values, palette="rainbow")
plt.xlabel("How many time of Number in rel hum")
plt.ylabel("Number of rel hum per hour")
plt.xticks(rotation=90)
# # 4. How many Weather are there?
data.head(2)
weather = data["Weather"].value_counts().head(10)
weather.to_frame()
# ## Ploting
x_values = weather.values
y_values = weather.index
plt.figure(figsize=(7, 5))
sns.barplot(x=x_values, y=y_values, palette="rainbow")
plt.xlabel("Number of Weather")
plt.ylabel("Name of time")
plt.xticks(rotation=90)
# # 5. Top 10 Dew Temperature in C
data.head(2)
dew = data["Dew Point Temp_C"].value_counts().head(10)
dew.to_frame()
# ## Ploting
x_values = dew.values
y_values = dew.index
plt.figure(figsize=(7, 5))
sns.countplot(x=x_values, palette="rainbow")
plt.xlabel("Number of Time")
plt.ylabel("Number of Temperature in C")
plt.xticks(rotation=90)
# # 6. Top 100 Temperature in C
data.head(2)
tem = data["Temp_C"].value_counts().head(100)
tem.to_frame()
# ## Ploting
x_values = tem.values
y_values = tem.index
plt.figure(figsize=(7, 5))
sns.countplot(y=x_values, palette="rainbow")
plt.xlabel("Number of Time")
plt.ylabel("Number of Temperature in C")
plt.xticks(rotation=90)
# # 6. Weather in Months
data.head(2)
data["Date/Time"] = pd.to_datetime(data["Date/Time"])
data.dtypes
data["Date/Time"] = data["Date/Time"].dt.month
month = data["Date/Time"].value_counts()
month.to_frame()
# ## Ploting
x_values = month.values
y_values = month.index
plt.figure(figsize=(7, 5))
sns.countplot(x=x_values, palette="rainbow")
plt.ylabel("Number of Time")
plt.xlabel("Number of Temperature in C")
plt.xticks(rotation=90)
# # 7. Visibility in Km
data.head(2)
visibility = data["Visibility_km"].value_counts()
visibility.to_frame()
# ## Ploting
x_values = visibility.values
y_values = visibility.index
plt.figure(figsize=(7, 5))
sns.barplot(x=x_values, y=y_values, palette="rainbow")
plt.xlabel("Number of Time")
plt.ylabel("Number of Temperature in C")
plt.xticks(rotation=90)
| false | 1 | 1,077 | 0 | 1,098 | 1,077 |
||
129494327
|
# # Dog Breed Identification
# ___
# **Description :**
# ___
# - You are provided with a training set and a test set of images of dogs. Each image has a filename that is its unique id. The dataset comprises 120 breeds of dogs. The goal of the competition is to create a classifier capable of determining a dog's breed from a photo. The list of breeds is as follows: 120 breed
# ___
# **Files Descriptions :**
# - `train.zip` - The training set, you are provided the breed for these dogs
# - `test.zip` - The test set, you must predict the probability of each breed for each image
# - `sample_submission.csv` - A sample submission file in the correct format
# - `labels.csv` - The breeds for the images in the train set
# ## Importing , Preprocessing Data :
# ___
# - `Read` Images
# - `Resize` Images
# - Convert Type to `tf.float32`
# - Normalize by dividing on `255`
# ___
# Basic imports
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import cv2 as cv
import os
import tensorflow as tf
import tensorflow_hub as hub
import time
import datetime
os.chdir("/kaggle/input/dog-breed-identification")
labels = pd.read_csv("labels.csv")
labels
labels.breed.value_counts().plot(kind="bar", figsize=(30, 10), title="Breeds Names")
IMG_SZ = 224
bat = 32
# read images in grey mode
train_paths = labels.id.map(lambda x: f"train//{x}.jpg")
# Minimize comp power
# read images
imgs = labels.id.map(
lambda x: tf.constant(
cv.resize(cv.imread(f"train//{x}.jpg"), (IMG_SZ, IMG_SZ)), tf.float32
)
/ 255
).to_list()
imgs = np.array(imgs)
gr_imgs = labels.id.map(
lambda x: tf.constant(
cv.resize(cv.imread(f"train//{x}.jpg", 0), (IMG_SZ, IMG_SZ)), tf.float32
)
/ 255
).to_list()
gr_imgs = np.array(gr_imgs)
plt.imshow(imgs[5])
plt.title(labels.breed[5])
plt.imshow(gr_imgs[5], cmap="gray")
plt.title(labels.breed[5])
from sklearn.preprocessing import LabelEncoder
y_train = labels.breed
unique = labels.breed.unique()
y_train = np.array([list(map(lambda y: x == y, unique)) for x in y_train])
len(unique), y_train.shape
y_train[20], labels.breed.unique()[y_train[20].argmax()], labels.breed[20]
# ## Building Models
# ___
# - MOBILENET_VS_130_224
# ____
# - VGG:
# - `16`
# - `19`
# _____
# - RESNET_V2:
# - `50`
# - `101`
# - `152`
# ____
from sklearn.model_selection import train_test_split
import tensorflow_hub as hub
from keras.layers import (
Flatten,
Dense,
Input,
MaxPooling2D,
Dropout,
GlobalMaxPool2D,
Lambda,
GlobalAveragePooling2D,
BatchNormalization,
Add,
)
from keras.models import Model, load_model
from keras.callbacks import TensorBoard, EarlyStopping, ModelCheckpoint
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import datetime
from sklearn.metrics import classification_report
IN_SHP = imgs[0].shape
OUT_SHP = len(set(labels.breed))
np.random.seed(42)
## Create callbacks
def evaluate(val=np.arange(5), mo=None, X=None, Y=None, uni=None):
"""
Function to evaluate Model and plot custome plot contains :
____________________________________________________________
- Image
- Horizontal bar plot of highest 5 predictions
- Title show if it's True or False and the right breed
____________________________________________________________
- mo : The Model
- X : Features for predictions
- Y : True Labels " Predictions "
- uni : Set of Breeds
"""
pre = mo.predict(X)
for i in val:
cols = np.array(["green", "gray", "gray", "gray", "gray"])
resu = pd.DataFrame(data=(uni, pre[i].round(4) * 100)).T.sort_values(
by=[1], ascending=False
)[:5]
plt.figure(figsize=(20, 5))
ax_1 = plt.subplot(1, 2, 1)
ax_1.margins(2, 2)
ax_1.imshow(cv.cvtColor(imgs[i], cv.COLOR_BGR2RGB), cmap="gray")
ax_2 = plt.subplot(1, 2, 2)
ax_2.margins(0.5, 0.1)
# ax_2.barh(resu[0],resu[1],color=['salmon','gray','k','k','k'])
if pre[i].argmax() == Y[i].argmax():
ax_2.barh(resu[0], resu[1], color=cols)
plt.title(
f"{uni[pre[i].argmax()]} By {np.round(pre[i][pre[i].argmax()]*100,2)}%",
color="green",
)
else:
cols[0] = "gray"
cols[resu[0] == uni[Y[i].argmax()]] = "green"
ax_2.barh(resu[0], resu[1], color=cols)
plt.title(
f"{uni[pre[i].argmax()]} By {np.round(pre[i][pre[i].argmax()]*100,2)}% , True : {uni[Y[i].argmax()]}",
color="red",
)
return pre
# ### MOBILENET_VS_130_224
# - `Try single pretrained model`
IN_SHP = imgs[0].shape
OUT_SHP = len(set(labels.breed))
MODEL_PATH = r"https://tfhub.dev/google/imagenet/mobilenet_v2_130_224/classification/5"
i = Input(IN_SHP)
x = hub.KerasLayer(MODEL_PATH)(i)
x = Dropout(0.2)(x)
x = BatchNormalization()(x)
x = Dense(1024, activation="relu")(x)
x = Dropout(0.2)(x)
x = BatchNormalization()(x)
x = Dense(512, activation="relu")(x)
x = Dropout(0.2)(x)
x = BatchNormalization()(x)
x = Dense(1024, activation="relu")(x)
x = Dense(OUT_SHP, activation="softmax")(x)
model = Model(i, x)
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.summary()
def tb_call():
loc = os.path.join("logs", datetime.datetime.now().strftime("%Y-%m-%d -- %H-%M-%S"))
return TensorBoard(loc)
tb_call_1 = tb_call()
es_call = EarlyStopping("val_accuracy", verbose=1, patience=3)
mc_call = ModelCheckpoint(
r"checkpoint\mobilenet_v2_130_224_best.h5",
monitor="val_accuracy",
save_best_only=True,
mode="max",
verbose=2,
)
EP = 50
BT_SZ = 128
data_gen = ImageDataGenerator(
horizontal_flip=True,
vertical_flip=True,
width_shift_range=0.1,
height_shift_range=0.1,
)
x_tr, x_val, y_tr, y_val = train_test_split(imgs, y_train, test_size=0.1)
data = data_gen.flow(x_tr, y_tr, batch_size=BT_SZ)
# val_data=data_gen.flow(x_val,y_val,batch_size=BT_SZ)
model.fit(
data,
validation_data=(x_val, y_val),
batch_size=BT_SZ,
epochs=EP,
callbacks=[es_call, mc_call],
)
model.evaluate(x_val, y_val)
# ### VGG19 + InceptionResNetV2 + Xception + NASNetLarge :
# ___
# - `Fearures Extraction`
from sklearn.preprocessing import PolynomialFeatures
def get_features(model, preprocessor, input_size, imgs):
"""
Function to extract features from images using fine tunned models
"""
x = Input(input_size)
x = Lambda(preprocessor)(x)
x = model(x)
fe_mo = Model(inputs=input_layer, outputs=base_model)
features = fe_mo.predict(imgs, verbose=1)
print("Feature maps shape: ", features.shape)
return features
# Load and use fine tuned models to extract features
# MobileNet
mb_fein = tf.keras.applications.mobilenet_v3.preprocess_input
mb_clf = load_model(
r"checkpoint\mobilenet_v2_130_224_best.h5",
custom_objects={"KerasLayer": hub.KerasLayer},
)
mb_fe = get_features(mb_clf, mb_fein, IN_SHP, imgs)
# ResNet
res_fein = tf.keras.applications.resnet_v2.preprocess_input
res_clf = load_model(r"checkpoint\RESNET152v2.h5")
res_fe = get_features(res_clf, res_fein, IN_SHP, imgs)
# Xception
xc_fein = tf.keras.applications.xception.preprocess_input
xc_clf = load_model(r"checkpoint\xception.h5")
xc_fe = get_features(xc_clf, xc_fein, IN_SHP, imgs)
# NASNET
nan_fein = tf.keras.applications.nasnet.preprocess_input
nan_clf = load_model(r"checkpoint\NASNET.h5")
nan_fe = get_features(nan_clf, nan_fein, IN_SHP, imgs)
# VGG
vg_fein = tf.keras.applications.vgg19.preprocess_input
vg_clf = load_model(r"checkpoint\VGG.h5")
vg_fe = get_features(vg_clf, vg_fein, IN_SHP, imgs)
ad_fe = Add()([mb_fe, res_fe, xc_fe, nan_fe, vg_fe])
ad_fe = PolynomialFeatures(3).fit_transform(ad_fe)
ad_fe.shape
# Prepare Deep net
i = Input(shape=ad_fe.shape[1])
x = Dropout(0.7)(i)
# x = BatchNormalization()(x)
x = Dense(120, activation="relu")(x)
# x = Dropout(0.7)(x)
# x = BatchNormalization()(x)
x = Dense(240, activation="relu")(x)
# x = Dropout(0.2)(x)
# x = BatchNormalization()(x)
x = Dense(120, activation="relu")(x)
x = Dense(OUT_SHP, activation="softmax")(x)
n_model = Model(i, x)
n_model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
# Training the model.
n_model.fit(ad_fe, y_train, batch_size=120, epochs=20, validation_split=0.1)
n_xtr, n_xte, n_ytr, n_yte = train_test_split(ad_fe, y_train)
ev = evaluate(
val=np.arange(20, len(n_xte), 500), mo=n_model, X=n_xte, Y=n_yte, uni=unique
)
n_model.evaluate(n_xte, n_yte)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/494/129494327.ipynb
| null | null |
[{"Id": 129494327, "ScriptId": 38505027, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12356201, "CreationDate": "05/14/2023 09:31:35", "VersionNumber": 1.0, "Title": "notebook22ee7b81cb", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 291.0, "LinesInsertedFromPrevious": 291.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
# # Dog Breed Identification
# ___
# **Description :**
# ___
# - You are provided with a training set and a test set of images of dogs. Each image has a filename that is its unique id. The dataset comprises 120 breeds of dogs. The goal of the competition is to create a classifier capable of determining a dog's breed from a photo. The list of breeds is as follows: 120 breed
# ___
# **Files Descriptions :**
# - `train.zip` - The training set, you are provided the breed for these dogs
# - `test.zip` - The test set, you must predict the probability of each breed for each image
# - `sample_submission.csv` - A sample submission file in the correct format
# - `labels.csv` - The breeds for the images in the train set
# ## Importing , Preprocessing Data :
# ___
# - `Read` Images
# - `Resize` Images
# - Convert Type to `tf.float32`
# - Normalize by dividing on `255`
# ___
# Basic imports
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import cv2 as cv
import os
import tensorflow as tf
import tensorflow_hub as hub
import time
import datetime
os.chdir("/kaggle/input/dog-breed-identification")
labels = pd.read_csv("labels.csv")
labels
labels.breed.value_counts().plot(kind="bar", figsize=(30, 10), title="Breeds Names")
IMG_SZ = 224
bat = 32
# read images in grey mode
train_paths = labels.id.map(lambda x: f"train//{x}.jpg")
# Minimize comp power
# read images
imgs = labels.id.map(
lambda x: tf.constant(
cv.resize(cv.imread(f"train//{x}.jpg"), (IMG_SZ, IMG_SZ)), tf.float32
)
/ 255
).to_list()
imgs = np.array(imgs)
gr_imgs = labels.id.map(
lambda x: tf.constant(
cv.resize(cv.imread(f"train//{x}.jpg", 0), (IMG_SZ, IMG_SZ)), tf.float32
)
/ 255
).to_list()
gr_imgs = np.array(gr_imgs)
plt.imshow(imgs[5])
plt.title(labels.breed[5])
plt.imshow(gr_imgs[5], cmap="gray")
plt.title(labels.breed[5])
from sklearn.preprocessing import LabelEncoder
y_train = labels.breed
unique = labels.breed.unique()
y_train = np.array([list(map(lambda y: x == y, unique)) for x in y_train])
len(unique), y_train.shape
y_train[20], labels.breed.unique()[y_train[20].argmax()], labels.breed[20]
# ## Building Models
# ___
# - MOBILENET_VS_130_224
# ____
# - VGG:
# - `16`
# - `19`
# _____
# - RESNET_V2:
# - `50`
# - `101`
# - `152`
# ____
from sklearn.model_selection import train_test_split
import tensorflow_hub as hub
from keras.layers import (
Flatten,
Dense,
Input,
MaxPooling2D,
Dropout,
GlobalMaxPool2D,
Lambda,
GlobalAveragePooling2D,
BatchNormalization,
Add,
)
from keras.models import Model, load_model
from keras.callbacks import TensorBoard, EarlyStopping, ModelCheckpoint
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import datetime
from sklearn.metrics import classification_report
IN_SHP = imgs[0].shape
OUT_SHP = len(set(labels.breed))
np.random.seed(42)
## Create callbacks
def evaluate(val=np.arange(5), mo=None, X=None, Y=None, uni=None):
"""
Function to evaluate Model and plot custome plot contains :
____________________________________________________________
- Image
- Horizontal bar plot of highest 5 predictions
- Title show if it's True or False and the right breed
____________________________________________________________
- mo : The Model
- X : Features for predictions
- Y : True Labels " Predictions "
- uni : Set of Breeds
"""
pre = mo.predict(X)
for i in val:
cols = np.array(["green", "gray", "gray", "gray", "gray"])
resu = pd.DataFrame(data=(uni, pre[i].round(4) * 100)).T.sort_values(
by=[1], ascending=False
)[:5]
plt.figure(figsize=(20, 5))
ax_1 = plt.subplot(1, 2, 1)
ax_1.margins(2, 2)
ax_1.imshow(cv.cvtColor(imgs[i], cv.COLOR_BGR2RGB), cmap="gray")
ax_2 = plt.subplot(1, 2, 2)
ax_2.margins(0.5, 0.1)
# ax_2.barh(resu[0],resu[1],color=['salmon','gray','k','k','k'])
if pre[i].argmax() == Y[i].argmax():
ax_2.barh(resu[0], resu[1], color=cols)
plt.title(
f"{uni[pre[i].argmax()]} By {np.round(pre[i][pre[i].argmax()]*100,2)}%",
color="green",
)
else:
cols[0] = "gray"
cols[resu[0] == uni[Y[i].argmax()]] = "green"
ax_2.barh(resu[0], resu[1], color=cols)
plt.title(
f"{uni[pre[i].argmax()]} By {np.round(pre[i][pre[i].argmax()]*100,2)}% , True : {uni[Y[i].argmax()]}",
color="red",
)
return pre
# ### MOBILENET_VS_130_224
# - `Try single pretrained model`
IN_SHP = imgs[0].shape
OUT_SHP = len(set(labels.breed))
MODEL_PATH = r"https://tfhub.dev/google/imagenet/mobilenet_v2_130_224/classification/5"
i = Input(IN_SHP)
x = hub.KerasLayer(MODEL_PATH)(i)
x = Dropout(0.2)(x)
x = BatchNormalization()(x)
x = Dense(1024, activation="relu")(x)
x = Dropout(0.2)(x)
x = BatchNormalization()(x)
x = Dense(512, activation="relu")(x)
x = Dropout(0.2)(x)
x = BatchNormalization()(x)
x = Dense(1024, activation="relu")(x)
x = Dense(OUT_SHP, activation="softmax")(x)
model = Model(i, x)
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.summary()
def tb_call():
loc = os.path.join("logs", datetime.datetime.now().strftime("%Y-%m-%d -- %H-%M-%S"))
return TensorBoard(loc)
tb_call_1 = tb_call()
es_call = EarlyStopping("val_accuracy", verbose=1, patience=3)
mc_call = ModelCheckpoint(
r"checkpoint\mobilenet_v2_130_224_best.h5",
monitor="val_accuracy",
save_best_only=True,
mode="max",
verbose=2,
)
EP = 50
BT_SZ = 128
data_gen = ImageDataGenerator(
horizontal_flip=True,
vertical_flip=True,
width_shift_range=0.1,
height_shift_range=0.1,
)
x_tr, x_val, y_tr, y_val = train_test_split(imgs, y_train, test_size=0.1)
data = data_gen.flow(x_tr, y_tr, batch_size=BT_SZ)
# val_data=data_gen.flow(x_val,y_val,batch_size=BT_SZ)
model.fit(
data,
validation_data=(x_val, y_val),
batch_size=BT_SZ,
epochs=EP,
callbacks=[es_call, mc_call],
)
model.evaluate(x_val, y_val)
# ### VGG19 + InceptionResNetV2 + Xception + NASNetLarge :
# ___
# - `Fearures Extraction`
from sklearn.preprocessing import PolynomialFeatures
def get_features(model, preprocessor, input_size, imgs):
"""
Function to extract features from images using fine tunned models
"""
x = Input(input_size)
x = Lambda(preprocessor)(x)
x = model(x)
fe_mo = Model(inputs=input_layer, outputs=base_model)
features = fe_mo.predict(imgs, verbose=1)
print("Feature maps shape: ", features.shape)
return features
# Load and use fine tuned models to extract features
# MobileNet
mb_fein = tf.keras.applications.mobilenet_v3.preprocess_input
mb_clf = load_model(
r"checkpoint\mobilenet_v2_130_224_best.h5",
custom_objects={"KerasLayer": hub.KerasLayer},
)
mb_fe = get_features(mb_clf, mb_fein, IN_SHP, imgs)
# ResNet
res_fein = tf.keras.applications.resnet_v2.preprocess_input
res_clf = load_model(r"checkpoint\RESNET152v2.h5")
res_fe = get_features(res_clf, res_fein, IN_SHP, imgs)
# Xception
xc_fein = tf.keras.applications.xception.preprocess_input
xc_clf = load_model(r"checkpoint\xception.h5")
xc_fe = get_features(xc_clf, xc_fein, IN_SHP, imgs)
# NASNET
nan_fein = tf.keras.applications.nasnet.preprocess_input
nan_clf = load_model(r"checkpoint\NASNET.h5")
nan_fe = get_features(nan_clf, nan_fein, IN_SHP, imgs)
# VGG
vg_fein = tf.keras.applications.vgg19.preprocess_input
vg_clf = load_model(r"checkpoint\VGG.h5")
vg_fe = get_features(vg_clf, vg_fein, IN_SHP, imgs)
ad_fe = Add()([mb_fe, res_fe, xc_fe, nan_fe, vg_fe])
ad_fe = PolynomialFeatures(3).fit_transform(ad_fe)
ad_fe.shape
# Prepare Deep net
i = Input(shape=ad_fe.shape[1])
x = Dropout(0.7)(i)
# x = BatchNormalization()(x)
x = Dense(120, activation="relu")(x)
# x = Dropout(0.7)(x)
# x = BatchNormalization()(x)
x = Dense(240, activation="relu")(x)
# x = Dropout(0.2)(x)
# x = BatchNormalization()(x)
x = Dense(120, activation="relu")(x)
x = Dense(OUT_SHP, activation="softmax")(x)
n_model = Model(i, x)
n_model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
# Training the model.
n_model.fit(ad_fe, y_train, batch_size=120, epochs=20, validation_split=0.1)
n_xtr, n_xte, n_ytr, n_yte = train_test_split(ad_fe, y_train)
ev = evaluate(
val=np.arange(20, len(n_xte), 500), mo=n_model, X=n_xte, Y=n_yte, uni=unique
)
n_model.evaluate(n_xte, n_yte)
| false | 0 | 3,088 | 1 | 3,088 | 3,088 |
||
129494597
|
<jupyter_start><jupyter_text>Phishing website dataset
Kaggle dataset identifier: phishing-website-dataset
<jupyter_script>import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# > *Dataset credit:> https://www.kaggle.com/datasets/akashkr/phishing-website-dataset*
# > *Description and labeling of data columns credit goes to :> https://www.kaggle.com/code/akashkr/phishing-url-eda-and-modelling.*
dataset = pd.read_csv("/kaggle/input/phishing-website-dataset/dataset.csv")
dataset.head()
# > *1 means legitimate, 0 is suspicious and -1 is phishing.*
dataset["Result"].value_counts()
dataset["Page_Rank"].value_counts()
# > *PageRank
# PageRank is a value ranging from “0” to “1”. PageRank aims to measure how important a webpage is on the Internet. The greater the PageRank value the more important the webpage. In our datasets, we find that about 95% of phishing webpages have no PageRank. Moreover, we find that the remaining 5% of phishing webpages may reach a PageRank value up to “0.2”.
# Rule: IF
# {PageRank<0.2 → Phishing
# {Otherwise → Legitimate*
dataset["Google_Index"].value_counts()
# > *Google Index
# This feature examines whether a website is in Google’s index or not. When a site is indexed by Google, it is displayed on search results (Webmaster resources, 2014). Usually, phishing webpages are merely accessible for a short period and as a result, many phishing webpages may not be found on the Google index.
# Rule: IF
# {Webpage Indexed by Google → Legitimate
# {Otherwise → Phishing*
dataset["URLURL_Length"].value_counts()
# > *Long URL to Hide the Suspicious Part
# Phishers can use long URL to hide the doubtful part in the address bar. For example: http://federmacedoadv.com.br/3f/aze/ab51e2e319e51502f416dbe46b773a5e/?cmd=_home&dispatch=11004d58f5b74f8dc1e7c2e8dd4105e811004d58f5b74f8dc1e7c2e8dd4105e8@phishing.website.html To ensure accuracy of our study, we calculated the length of URLs in the dataset and produced an average URL length. The results showed that if the length of the URL is greater than or equal 54 characters then the URL classified as phishing. By reviewing our dataset we were able to find 1220 URLs lengths equals to 54 or more which constitute 48.8% of the total dataset size. We have been able to update this feature rule by using a method based on frequency and thus improving upon its accuracy.
# RULE: IF
# {URL length<54 → feature = Legitimate
# {else if URL length≥54 and ≤75 → feature = Suspicious
# {otherwise→ feature = Phishing*
dataset["having_At_Symbol"].value_counts()
# > *URL’s having “@” Symbol
# Using “@” symbol in the URL leads the browser to ignore everything preceding the “@” symbol and the real address often follows the “@” symbol.
# RULE: IF
# {Url Having @ Symbol→ Phishing
# {Otherwise→ Legitimate*
dataset["double_slash_redirecting"].value_counts()
# > *Redirecting using “//”
# The existence of “//” within the URL path means that the user will be redirected to another website. An example of such URL’s is: “http://www.legitimate.com//http://www.phishing.com”. We examin the location where the “//” appears. We find that if the URL starts with “HTTP”, that means the “//” should appear in the sixth position. However, if the URL employs “HTTPS” then the “//” should appear in seventh position.
# RULE: IF
# {The Position of the Last Occurrence of "//\" " in the URL > 7→ Phishing
# {Otherwise→ Legitimate*
dataset["Prefix_Suffix"].value_counts()
# > *Adding Prefix or Suffix Separated by (-) to the Domain
# The dash symbol is rarely used in legitimate URLs. Phishers tend to add prefixes or suffixes separated by (-) to the domain name so that users feel that they are dealing with a legitimate webpage. For example http://www.Confirme-paypal.com/.
# RULE: IF
# {Domain Name Part Includes (-) Symbol → Phishing
# {Otherwise → Legitimate*
selected_features = [
"URLURL_Length",
"having_At_Symbol",
"double_slash_redirecting",
"Prefix_Suffix",
"Page_Rank",
"Google_Index",
"Result",
]
df = dataset[selected_features]
df.head()
new_columns = {"URLURL_Length": "URL_Length", "Prefix_Suffix": "HavingHyphen"}
df = df.rename(columns=new_columns)
df = df.drop_duplicates()
df.shape
# > *There are total 107 different combinations, this may sounds really less but keep in mind that all the features and labels has combination of only two or three unique values.*
# > *The less number will generalize more but detection time will be alot faster.*
df.isnull().sum()
df["Result"].value_counts(normalize=True)
# > *1 means legitimate, and -1 is phishing.*
X = df.drop("Result", axis=1)
y = df["Result"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=0, stratify=y
)
print(X_train.shape)
print(X_test.shape)
# # > *Logistic Regression*
# > *Aim:> Maximizing True Positive Rate leveraging False Positive Rate using ROC curve.*
# > *Nice tutorial on ROC and AUC by StatQuest with Josh Starmer
# :> https://youtu.be/4jRBRDbJemM*
# > *Reason for analysing roc on only Logistic Regression algorithm.
# 1.They are fast in predicting a new data instance.
# 2.They outputs the result of each instance in probability between 0-1(Some algorithms does'nt and this is important because we can pick and choose threshold properly)..*
# > *SVM has greater AUC(Area Under Curve) so we will go from here.*
import numpy as np
import matplotlib.pyplot as plt
from sklearn.metrics import roc_curve, auc
from sklearn.linear_model import LogisticRegression
LG = LogisticRegression()
LG.fit(X_train, y_train)
y_pred_prob = LG.predict_proba(X_train)[:, 1]
# Calculate the FPR, TPR, and thresholds
fpr, tpr, thresholds = roc_curve(y_train, y_pred_prob)
# Plot the ROC curve
plt.figure(figsize=(8, 6))
plt.plot(fpr, tpr, label="ROC curve (AUC = {:.2f})".format(auc(fpr, tpr)))
plt.plot([0, 1], [0, 1], "k--") # Diagonal line representing random guessing
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic (ROC) Curve")
plt.legend(loc="lower right")
# Display threshold values as annotations
for i, threshold in enumerate(thresholds):
plt.annotate(
"{:.2f}".format(threshold),
(fpr[i], tpr[i]),
textcoords="offset points",
xytext=(5, -10),
ha="center",
)
plt.show()
from sklearn.metrics import confusion_matrix
LG = LogisticRegression()
LG.fit(X_train, y_train)
# Predict on the test set
print("Before:>")
print("Training Accuracy: ", LG.score(X_train, y_train))
print("Testing Accuracy ", LG.score(X_test, y_test))
tn, fp, fn, tp = confusion_matrix(y_train, LG.predict(X_train)).ravel()
print("Training True Positive Rate: ", (tp) / (tp + fn))
tn, fp, fn, tp = confusion_matrix(y_test, LG.predict(X_test)).ravel()
print("Testing True Positive Rate: ", (tp) / (tp + fn))
print("After:>")
y_pred_train = LG.predict_proba(X_train)
y_pred_test = LG.predict_proba(X_test)
result_train = np.where(y_pred_train[:, 1] < 0.42, -1, 1)
result_test = np.where(y_pred_test[:, 1] < 0.42, -1, 1)
print("Training Accuracy: ", LG.score(X_train, result_train))
print("Testing Accuracy ", LG.score(X_test, result_test))
tn, fp, fn, tp = confusion_matrix(y_train, result_train).ravel()
print("Training True Positive Rate: ", (tp) / (tp + fn))
tn, fp, fn, tp = confusion_matrix(y_test, result_test).ravel()
print("Testing True Positive Rate: ", (tp) / (tp + fn))
# # > *Predicting a new URL if its a Phishing or not.*
new_url = "https://www.google.com"
df.head()
# > *1 means legitimate, 0 is suspicious and -1 is phishing.*
url_len = len(new_url)
URL_Length = -1
if url_len < 54:
URL_Length = 1
elif url_len >= 54 and url_len <= 75:
URL_Length = 0
having_At_Symbol = -1
if new_url.find("@") == -1:
having_At_Symbol = 1
print(having_At_Symbol)
try:
position = new_url.index("//")
except ValueError:
double_slash_redirecting = -1
if position + 1 > 7:
double_slash_redirecting = -1
else:
double_slash_redirecting = 1
print(double_slash_redirecting)
HavingHyphen = -1
if new_url.find("-") == -1:
HavingHyphen = 1
print(HavingHyphen)
# > *Page Rank value is received through api from https://www.domcop.com/openpagerank/documentation*
import requests
from urllib.parse import urlparse
headers = {"API-OPR": "c8g4404gswswcok8s0k4404ko4g00oo8w4ks84g4"}
requested_url = "https://www.google.com/docs/about/"
parsed_url = urlparse(requested_url)
domain = parsed_url.netloc
url = "https://openpagerank.com/api/v1.0/getPageRank?domains%5B0%5D=" + domain
request = requests.get(url, headers=headers)
result = request.json()
page_rank = result["response"][0]["page_rank_decimal"]
if page_rank < 20:
Page_Rank = -1
else:
Page_Rank = 1
print(Page_Rank)
requested_url = "http://ebrpnvvjgd.duckdns.org"
parsed_url = urlparse(requested_url)
domain = parsed_url.netloc
domain
import re
import requests
from bs4 import BeautifulSoup
Google_Index = -1
google = "https://www.google.com/search?q=site:" + domain + "&hl=en"
response = requests.get(google, cookies={"CONSENT": "YES+1"})
soup = BeautifulSoup(response.content, "html.parser")
not_indexed = re.compile("did not match any documents")
if soup(text=not_indexed):
Google_Index = -1
else:
Google_Index = 1
print(Google_Index)
df.head()
X_pred = pd.DataFrame(
{
"URL_Length": [URL_Length],
"having_At_Symbol": [having_At_Symbol],
"double_slash_redirecting": [double_slash_redirecting],
"HavingHyphen": [HavingHyphen],
"Page_Rank": [Page_Rank],
"Google_Index": [Google_Index],
}
)
X_pred.columns = X_train.columns
X_pred
LG.predict(X_pred)
# # > *Saving the model*
import pickle
# Save the model to a file using pickle
filename = "logistic_regression_model.pkl"
with open(filename, "wb") as file:
pickle.dump(LG, file)
# Load the model from the pickle file
filename = "logistic_regression_model.pkl"
with open(filename, "rb") as file:
loaded_model = pickle.load(file)
# Use the loaded model for predictions
loaded_model.predict(X_pred)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/494/129494597.ipynb
|
phishing-website-dataset
|
akashkr
|
[{"Id": 129494597, "ScriptId": 38316130, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6223792, "CreationDate": "05/14/2023 09:34:13", "VersionNumber": 4.0, "Title": "Phishing website Detection", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 289.0, "LinesInsertedFromPrevious": 101.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 188.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185608884, "KernelVersionId": 129494597, "SourceDatasetVersionId": 14849}]
|
[{"Id": 14849, "DatasetId": 10598, "DatasourceVersionId": 14849, "CreatorUserId": 1481907, "LicenseName": "CC0: Public Domain", "CreationDate": "01/22/2018 15:01:48", "VersionNumber": 2.0, "Title": "Phishing website dataset", "Slug": "phishing-website-dataset", "Subtitle": "This website lists 30 optimized features of phishing website.", "Description": NaN, "VersionNotes": "Column names and index added", "TotalCompressedBytes": 855445.0, "TotalUncompressedBytes": 855445.0}]
|
[{"Id": 10598, "CreatorUserId": 1481907, "OwnerUserId": 1481907.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 14849.0, "CurrentDatasourceVersionId": 14849.0, "ForumId": 17963, "Type": 2, "CreationDate": "01/22/2018 12:40:06", "LastActivityDate": "02/05/2018", "TotalViews": 228931, "TotalDownloads": 6972, "TotalVotes": 92, "TotalKernels": 7}]
|
[{"Id": 1481907, "UserName": "akashkr", "DisplayName": "Akash Kumar", "RegisterDate": "12/08/2017", "PerformanceTier": 2}]
|
import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# > *Dataset credit:> https://www.kaggle.com/datasets/akashkr/phishing-website-dataset*
# > *Description and labeling of data columns credit goes to :> https://www.kaggle.com/code/akashkr/phishing-url-eda-and-modelling.*
dataset = pd.read_csv("/kaggle/input/phishing-website-dataset/dataset.csv")
dataset.head()
# > *1 means legitimate, 0 is suspicious and -1 is phishing.*
dataset["Result"].value_counts()
dataset["Page_Rank"].value_counts()
# > *PageRank
# PageRank is a value ranging from “0” to “1”. PageRank aims to measure how important a webpage is on the Internet. The greater the PageRank value the more important the webpage. In our datasets, we find that about 95% of phishing webpages have no PageRank. Moreover, we find that the remaining 5% of phishing webpages may reach a PageRank value up to “0.2”.
# Rule: IF
# {PageRank<0.2 → Phishing
# {Otherwise → Legitimate*
dataset["Google_Index"].value_counts()
# > *Google Index
# This feature examines whether a website is in Google’s index or not. When a site is indexed by Google, it is displayed on search results (Webmaster resources, 2014). Usually, phishing webpages are merely accessible for a short period and as a result, many phishing webpages may not be found on the Google index.
# Rule: IF
# {Webpage Indexed by Google → Legitimate
# {Otherwise → Phishing*
dataset["URLURL_Length"].value_counts()
# > *Long URL to Hide the Suspicious Part
# Phishers can use long URL to hide the doubtful part in the address bar. For example: http://federmacedoadv.com.br/3f/aze/ab51e2e319e51502f416dbe46b773a5e/?cmd=_home&dispatch=11004d58f5b74f8dc1e7c2e8dd4105e811004d58f5b74f8dc1e7c2e8dd4105e8@phishing.website.html To ensure accuracy of our study, we calculated the length of URLs in the dataset and produced an average URL length. The results showed that if the length of the URL is greater than or equal 54 characters then the URL classified as phishing. By reviewing our dataset we were able to find 1220 URLs lengths equals to 54 or more which constitute 48.8% of the total dataset size. We have been able to update this feature rule by using a method based on frequency and thus improving upon its accuracy.
# RULE: IF
# {URL length<54 → feature = Legitimate
# {else if URL length≥54 and ≤75 → feature = Suspicious
# {otherwise→ feature = Phishing*
dataset["having_At_Symbol"].value_counts()
# > *URL’s having “@” Symbol
# Using “@” symbol in the URL leads the browser to ignore everything preceding the “@” symbol and the real address often follows the “@” symbol.
# RULE: IF
# {Url Having @ Symbol→ Phishing
# {Otherwise→ Legitimate*
dataset["double_slash_redirecting"].value_counts()
# > *Redirecting using “//”
# The existence of “//” within the URL path means that the user will be redirected to another website. An example of such URL’s is: “http://www.legitimate.com//http://www.phishing.com”. We examin the location where the “//” appears. We find that if the URL starts with “HTTP”, that means the “//” should appear in the sixth position. However, if the URL employs “HTTPS” then the “//” should appear in seventh position.
# RULE: IF
# {The Position of the Last Occurrence of "//\" " in the URL > 7→ Phishing
# {Otherwise→ Legitimate*
dataset["Prefix_Suffix"].value_counts()
# > *Adding Prefix or Suffix Separated by (-) to the Domain
# The dash symbol is rarely used in legitimate URLs. Phishers tend to add prefixes or suffixes separated by (-) to the domain name so that users feel that they are dealing with a legitimate webpage. For example http://www.Confirme-paypal.com/.
# RULE: IF
# {Domain Name Part Includes (-) Symbol → Phishing
# {Otherwise → Legitimate*
selected_features = [
"URLURL_Length",
"having_At_Symbol",
"double_slash_redirecting",
"Prefix_Suffix",
"Page_Rank",
"Google_Index",
"Result",
]
df = dataset[selected_features]
df.head()
new_columns = {"URLURL_Length": "URL_Length", "Prefix_Suffix": "HavingHyphen"}
df = df.rename(columns=new_columns)
df = df.drop_duplicates()
df.shape
# > *There are total 107 different combinations, this may sounds really less but keep in mind that all the features and labels has combination of only two or three unique values.*
# > *The less number will generalize more but detection time will be alot faster.*
df.isnull().sum()
df["Result"].value_counts(normalize=True)
# > *1 means legitimate, and -1 is phishing.*
X = df.drop("Result", axis=1)
y = df["Result"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=0, stratify=y
)
print(X_train.shape)
print(X_test.shape)
# # > *Logistic Regression*
# > *Aim:> Maximizing True Positive Rate leveraging False Positive Rate using ROC curve.*
# > *Nice tutorial on ROC and AUC by StatQuest with Josh Starmer
# :> https://youtu.be/4jRBRDbJemM*
# > *Reason for analysing roc on only Logistic Regression algorithm.
# 1.They are fast in predicting a new data instance.
# 2.They outputs the result of each instance in probability between 0-1(Some algorithms does'nt and this is important because we can pick and choose threshold properly)..*
# > *SVM has greater AUC(Area Under Curve) so we will go from here.*
import numpy as np
import matplotlib.pyplot as plt
from sklearn.metrics import roc_curve, auc
from sklearn.linear_model import LogisticRegression
LG = LogisticRegression()
LG.fit(X_train, y_train)
y_pred_prob = LG.predict_proba(X_train)[:, 1]
# Calculate the FPR, TPR, and thresholds
fpr, tpr, thresholds = roc_curve(y_train, y_pred_prob)
# Plot the ROC curve
plt.figure(figsize=(8, 6))
plt.plot(fpr, tpr, label="ROC curve (AUC = {:.2f})".format(auc(fpr, tpr)))
plt.plot([0, 1], [0, 1], "k--") # Diagonal line representing random guessing
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic (ROC) Curve")
plt.legend(loc="lower right")
# Display threshold values as annotations
for i, threshold in enumerate(thresholds):
plt.annotate(
"{:.2f}".format(threshold),
(fpr[i], tpr[i]),
textcoords="offset points",
xytext=(5, -10),
ha="center",
)
plt.show()
from sklearn.metrics import confusion_matrix
LG = LogisticRegression()
LG.fit(X_train, y_train)
# Predict on the test set
print("Before:>")
print("Training Accuracy: ", LG.score(X_train, y_train))
print("Testing Accuracy ", LG.score(X_test, y_test))
tn, fp, fn, tp = confusion_matrix(y_train, LG.predict(X_train)).ravel()
print("Training True Positive Rate: ", (tp) / (tp + fn))
tn, fp, fn, tp = confusion_matrix(y_test, LG.predict(X_test)).ravel()
print("Testing True Positive Rate: ", (tp) / (tp + fn))
print("After:>")
y_pred_train = LG.predict_proba(X_train)
y_pred_test = LG.predict_proba(X_test)
result_train = np.where(y_pred_train[:, 1] < 0.42, -1, 1)
result_test = np.where(y_pred_test[:, 1] < 0.42, -1, 1)
print("Training Accuracy: ", LG.score(X_train, result_train))
print("Testing Accuracy ", LG.score(X_test, result_test))
tn, fp, fn, tp = confusion_matrix(y_train, result_train).ravel()
print("Training True Positive Rate: ", (tp) / (tp + fn))
tn, fp, fn, tp = confusion_matrix(y_test, result_test).ravel()
print("Testing True Positive Rate: ", (tp) / (tp + fn))
# # > *Predicting a new URL if its a Phishing or not.*
new_url = "https://www.google.com"
df.head()
# > *1 means legitimate, 0 is suspicious and -1 is phishing.*
url_len = len(new_url)
URL_Length = -1
if url_len < 54:
URL_Length = 1
elif url_len >= 54 and url_len <= 75:
URL_Length = 0
having_At_Symbol = -1
if new_url.find("@") == -1:
having_At_Symbol = 1
print(having_At_Symbol)
try:
position = new_url.index("//")
except ValueError:
double_slash_redirecting = -1
if position + 1 > 7:
double_slash_redirecting = -1
else:
double_slash_redirecting = 1
print(double_slash_redirecting)
HavingHyphen = -1
if new_url.find("-") == -1:
HavingHyphen = 1
print(HavingHyphen)
# > *Page Rank value is received through api from https://www.domcop.com/openpagerank/documentation*
import requests
from urllib.parse import urlparse
headers = {"API-OPR": "c8g4404gswswcok8s0k4404ko4g00oo8w4ks84g4"}
requested_url = "https://www.google.com/docs/about/"
parsed_url = urlparse(requested_url)
domain = parsed_url.netloc
url = "https://openpagerank.com/api/v1.0/getPageRank?domains%5B0%5D=" + domain
request = requests.get(url, headers=headers)
result = request.json()
page_rank = result["response"][0]["page_rank_decimal"]
if page_rank < 20:
Page_Rank = -1
else:
Page_Rank = 1
print(Page_Rank)
requested_url = "http://ebrpnvvjgd.duckdns.org"
parsed_url = urlparse(requested_url)
domain = parsed_url.netloc
domain
import re
import requests
from bs4 import BeautifulSoup
Google_Index = -1
google = "https://www.google.com/search?q=site:" + domain + "&hl=en"
response = requests.get(google, cookies={"CONSENT": "YES+1"})
soup = BeautifulSoup(response.content, "html.parser")
not_indexed = re.compile("did not match any documents")
if soup(text=not_indexed):
Google_Index = -1
else:
Google_Index = 1
print(Google_Index)
df.head()
X_pred = pd.DataFrame(
{
"URL_Length": [URL_Length],
"having_At_Symbol": [having_At_Symbol],
"double_slash_redirecting": [double_slash_redirecting],
"HavingHyphen": [HavingHyphen],
"Page_Rank": [Page_Rank],
"Google_Index": [Google_Index],
}
)
X_pred.columns = X_train.columns
X_pred
LG.predict(X_pred)
# # > *Saving the model*
import pickle
# Save the model to a file using pickle
filename = "logistic_regression_model.pkl"
with open(filename, "wb") as file:
pickle.dump(LG, file)
# Load the model from the pickle file
filename = "logistic_regression_model.pkl"
with open(filename, "rb") as file:
loaded_model = pickle.load(file)
# Use the loaded model for predictions
loaded_model.predict(X_pred)
| false | 1 | 3,247 | 0 | 3,271 | 3,247 |
||
129494072
|
<jupyter_start><jupyter_text>IMDB Dataset of 50K Movie Reviews
IMDB dataset having 50K movie reviews for natural language processing or Text analytics.
This is a dataset for binary sentiment classification containing substantially more data than previous benchmark datasets. We provide a set of 25,000 highly polar movie reviews for training and 25,000 for testing. So, predict the number of positive and negative reviews using either classification or deep learning algorithms.
For more dataset information, please go through the following link,
http://ai.stanford.edu/~amaas/data/sentiment/
Kaggle dataset identifier: imdb-dataset-of-50k-movie-reviews
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import torch
import torch.nn as nn
import torch.nn.functional as F
from nltk.corpus import stopwords
from collections import Counter
import string
import re
import seaborn as sns
from tqdm import tqdm
import matplotlib.pyplot as plt
from torch.utils.data import TensorDataset, DataLoader
from sklearn.model_selection import train_test_split
is_cuda = torch.cuda.is_available()
if is_cuda:
device = torch.device("cuda")
else:
device = torch.device("cpu")
csv_path = "/kaggle/input/imdb-dataset-of-50k-movie-reviews/IMDB Dataset.csv"
df = pd.read_csv(csv_path)
df.tail() # Data preview
english_stopwords = set(stopwords.words("english"))
def preprocess(x, y):
# Preprocess reviews
x = x.replace({"<.*?>": ""}, regex=True) # remove html
x = x.replace({"[^A-Za-z]": " "}, regex=True) # remove non-alphabet
x = x.replace({"\s+": " "}, regex=True) # remove multiple spaces
x = x.apply(
lambda review: [w for w in review.split() if w not in english_stopwords]
) # remove stop words
x = x.apply(lambda review: [w.lower() for w in review]) # lower case
# Encode labels
y = y.replace("positive", 1)
y = y.replace("negative", 0)
return x.to_numpy(), y.to_numpy()
# Split train-valid-test dataset
x, y = preprocess(df["review"], df["sentiment"])
x_train, x_test, y_train, y_test = train_test_split(x, y, shuffle=True, test_size=10000)
x_train, x_val, y_train, y_val = train_test_split(
x_train, y_train, shuffle=True, test_size=10000
)
print(f"shape of train data is {x_train.shape}")
print(f"shape of valid data is {x_val.shape}")
print(f"shape of test data is {x_test.shape}")
# Check sentiment rate in train set
dd = pd.Series(y_train).value_counts()
sns.barplot(x=np.array(["negative", "positive"]), y=dd.values)
plt.show()
# TOKENIZATION
def createVocab(x_train):
# Create a word list
word_list = []
for sent in x_train:
for word in sent:
word_list.append(word)
corpus = Counter(word_list)
corpus_ = sorted(corpus, key=corpus.get, reverse=True)[:1000]
onehot_dict = {w: i + 1 for i, w in enumerate(corpus_)}
return onehot_dict
def tokenize(x_set, y_set, vocab):
final_set = []
for sent in x_set:
final_set.append([vocab[word] for word in sent if vocab.get(word) != None])
return np.array(final_set, dtype=object), np.array(y_set, dtype="float32")
vocab = createVocab(x_train)
x_train, y_train = tokenize(x_train, y_train, vocab)
x_val, y_val = tokenize(x_val, y_val, vocab)
x_test, y_test = tokenize(x_test, y_test, vocab)
print(f"Length of vocabulary is {len(vocab)}")
def padding_(sentences, seq_len):
features = np.zeros((len(sentences), seq_len), dtype=int)
for ii, review in enumerate(sentences):
if len(review) != 0:
features[ii, -len(review) :] = np.array(review)[:seq_len]
return features
# Padding to max length of 500
x_train_pad = padding_(x_train, 500)
x_val_pad = padding_(x_val, 500)
# create Tensor datasets
train_data = TensorDataset(torch.from_numpy(x_train_pad), torch.from_numpy(y_train))
valid_data = TensorDataset(torch.from_numpy(x_val_pad), torch.from_numpy(y_val))
# dataloaders
batch_size = 50
# make sure to SHUFFLE your data
train_loader = DataLoader(train_data, shuffle=True, batch_size=batch_size)
valid_loader = DataLoader(valid_data, shuffle=True, batch_size=batch_size)
# # obtain one batch of training data
# dataiter = iter(train_loader)
# sample_x, sample_y = dataiter.next()
# print('Sample input size: ', sample_x.size()) # batch_size, seq_length
# print('Sample input: \n', sample_x)
# print('Sample input: \n', sample_y)
class SentimentRNN(nn.Module):
def __init__(self, no_layers, vocab_size, hidden_dim, embedding_dim, drop_prob=0.5):
super(SentimentRNN, self).__init__()
self.output_dim = output_dim
self.hidden_dim = hidden_dim
self.no_layers = no_layers
self.vocab_size = vocab_size
# embedding and LSTM layers
self.embedding = nn.Embedding(vocab_size, embedding_dim)
# lstm
self.lstm = nn.LSTM(
input_size=embedding_dim,
hidden_size=self.hidden_dim,
num_layers=no_layers,
batch_first=True,
)
# dropout layer
self.dropout = nn.Dropout(drop_prob)
# linear and sigmoid layer
self.fc = nn.Linear(self.hidden_dim, output_dim)
self.sig = nn.Sigmoid()
def forward(self, x, hidden):
batch_size = x.size(0)
# embeddings and lstm_out
embeds = self.embedding(x) # shape: B x S x Feature since batch = True
# print(embeds.shape) #[50, 500, 1000]
lstm_out, hidden = self.lstm(embeds, hidden)
lstm_out = lstm_out.contiguous().view(-1, self.hidden_dim)
# dropout and fully connected layer
out = self.dropout(lstm_out)
out = self.fc(out)
# sigmoid function
sig_out = self.sig(out)
# reshape to be batch_size first
sig_out = sig_out.view(batch_size, -1)
sig_out = sig_out[:, -1] # get last batch of labels
# return last sigmoid output and hidden state
return sig_out, hidden
def init_hidden(self, batch_size):
"""Initializes hidden state"""
# Create two new tensors with sizes n_layers x batch_size x hidden_dim,
# initialized to zero, for hidden state and cell state of LSTM
h0 = torch.zeros((self.no_layers, batch_size, self.hidden_dim)).to(device)
c0 = torch.zeros((self.no_layers, batch_size, self.hidden_dim)).to(device)
hidden = (h0, c0)
return hidden
no_layers = 2
vocab_size = len(vocab) + 1 # extra 1 for padding
embedding_dim = 64
output_dim = 1
hidden_dim = 256
model = SentimentRNN(no_layers, vocab_size, hidden_dim, embedding_dim, drop_prob=0.3)
# moving to gpu
model.to(device)
print(model)
# loss and optimization functions
lr = 0.001
criterion = nn.BCELoss()
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
# function to predict accuracy
def acc(pred, label):
pred = torch.round(pred.squeeze())
return torch.sum(pred == label.squeeze()).item()
clip = 5
epochs = 7
valid_loss_min = np.Inf
# train for some number of epochs
epoch_tr_loss, epoch_vl_loss = [], []
epoch_tr_acc, epoch_vl_acc = [], []
for epoch in range(epochs):
train_losses = []
train_acc = 0.0
model.train()
# initialize hidden state
h = model.init_hidden(batch_size)
for inputs, labels in train_loader:
inputs, labels = inputs.to(device), labels.to(device)
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
h = tuple([each.data for each in h])
model.zero_grad()
output, h = model(inputs, h)
# calculate the loss and perform backprop
loss = criterion(output.squeeze(), labels.float())
loss.backward()
train_losses.append(loss.item())
# calculating accuracy
accuracy = acc(output, labels)
train_acc += accuracy
# `clip_grad_norm` helps prevent the exploding gradient problem in RNNs / LSTMs.
nn.utils.clip_grad_norm_(model.parameters(), clip)
optimizer.step()
val_h = model.init_hidden(batch_size)
val_losses = []
val_acc = 0.0
model.eval()
for inputs, labels in valid_loader:
val_h = tuple([each.data for each in val_h])
inputs, labels = inputs.to(device), labels.to(device)
output, val_h = model(inputs, val_h)
val_loss = criterion(output.squeeze(), labels.float())
val_losses.append(val_loss.item())
accuracy = acc(output, labels)
val_acc += accuracy
epoch_train_loss = np.mean(train_losses)
epoch_val_loss = np.mean(val_losses)
epoch_train_acc = train_acc / len(train_loader.dataset)
epoch_val_acc = val_acc / len(valid_loader.dataset)
epoch_tr_loss.append(epoch_train_loss)
epoch_vl_loss.append(epoch_val_loss)
epoch_tr_acc.append(epoch_train_acc)
epoch_vl_acc.append(epoch_val_acc)
print(f"Epoch {epoch+1}")
print(f"train_loss : {epoch_train_loss} val_loss : {epoch_val_loss}")
print(f"train_accuracy : {epoch_train_acc*100} val_accuracy : {epoch_val_acc*100}")
if epoch_val_loss <= valid_loss_min:
torch.save(model.state_dict(), "../working/state_dict.pt")
print(
"Validation loss decreased ({:.6f} --> {:.6f}). Saving model ...".format(
valid_loss_min, epoch_val_loss
)
)
valid_loss_min = epoch_val_loss
print(25 * "==")
fig = plt.figure(figsize=(20, 6))
plt.subplot(1, 2, 1)
plt.plot(epoch_tr_acc, label="Train Acc")
plt.plot(epoch_vl_acc, label="Validation Acc")
plt.title("Accuracy")
plt.legend()
plt.grid()
plt.subplot(1, 2, 2)
plt.plot(epoch_tr_loss, label="Train loss")
plt.plot(epoch_vl_loss, label="Validation loss")
plt.title("Loss")
plt.legend()
plt.grid()
plt.show()
x_test_pad = padding_(x_test, 500)
test_data = TensorDataset(torch.from_numpy(x_test_pad), torch.from_numpy(y_test))
test_loader = DataLoader(test_data, shuffle=True, batch_size=batch_size)
test_h = model.init_hidden(batch_size)
test_losses = []
test_acc = 0.0
model.eval()
for inputs, labels in test_loader:
test_h = tuple([each.data for each in test_h])
inputs, labels = inputs.to(device), labels.to(device)
output, test_h = model(inputs, test_h)
test_loss = criterion(output.squeeze(), labels.float())
test_losses.append(test_loss.item())
accuracy = acc(output, labels)
test_acc += accuracy
full_test_loss = np.mean(test_losses)
full_test_acc = test_acc / len(test_loader.dataset)
print(f"Accuracy on test dataset: {full_test_acc}")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/494/129494072.ipynb
|
imdb-dataset-of-50k-movie-reviews
|
lakshmi25npathi
|
[{"Id": 129494072, "ScriptId": 38497673, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10427111, "CreationDate": "05/14/2023 09:29:12", "VersionNumber": 1.0, "Title": "Sentiment Analysis on IMDB Reviews", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 323.0, "LinesInsertedFromPrevious": 323.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185607825, "KernelVersionId": 129494072, "SourceDatasetVersionId": 320111}]
|
[{"Id": 320111, "DatasetId": 134715, "DatasourceVersionId": 333307, "CreatorUserId": 2483565, "LicenseName": "Other (specified in description)", "CreationDate": "03/09/2019 06:32:21", "VersionNumber": 1.0, "Title": "IMDB Dataset of 50K Movie Reviews", "Slug": "imdb-dataset-of-50k-movie-reviews", "Subtitle": "Large Movie Review Dataset", "Description": "IMDB dataset having 50K movie reviews for natural language processing or Text analytics.\nThis is a dataset for binary sentiment classification containing substantially more data than previous benchmark datasets. We provide a set of 25,000 highly polar movie reviews for training and 25,000 for testing. So, predict the number of positive and negative reviews using either classification or deep learning algorithms.\nFor more dataset information, please go through the following link,\nhttp://ai.stanford.edu/~amaas/data/sentiment/", "VersionNotes": "Initial release", "TotalCompressedBytes": 66212309.0, "TotalUncompressedBytes": 26558952.0}]
|
[{"Id": 134715, "CreatorUserId": 2483565, "OwnerUserId": 2483565.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 320111.0, "CurrentDatasourceVersionId": 333307.0, "ForumId": 144904, "Type": 2, "CreationDate": "03/09/2019 06:32:21", "LastActivityDate": "03/09/2019", "TotalViews": 739266, "TotalDownloads": 131721, "TotalVotes": 959, "TotalKernels": 746}]
|
[{"Id": 2483565, "UserName": "lakshmi25npathi", "DisplayName": "Lakshmipathi N", "RegisterDate": "11/12/2018", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import torch
import torch.nn as nn
import torch.nn.functional as F
from nltk.corpus import stopwords
from collections import Counter
import string
import re
import seaborn as sns
from tqdm import tqdm
import matplotlib.pyplot as plt
from torch.utils.data import TensorDataset, DataLoader
from sklearn.model_selection import train_test_split
is_cuda = torch.cuda.is_available()
if is_cuda:
device = torch.device("cuda")
else:
device = torch.device("cpu")
csv_path = "/kaggle/input/imdb-dataset-of-50k-movie-reviews/IMDB Dataset.csv"
df = pd.read_csv(csv_path)
df.tail() # Data preview
english_stopwords = set(stopwords.words("english"))
def preprocess(x, y):
# Preprocess reviews
x = x.replace({"<.*?>": ""}, regex=True) # remove html
x = x.replace({"[^A-Za-z]": " "}, regex=True) # remove non-alphabet
x = x.replace({"\s+": " "}, regex=True) # remove multiple spaces
x = x.apply(
lambda review: [w for w in review.split() if w not in english_stopwords]
) # remove stop words
x = x.apply(lambda review: [w.lower() for w in review]) # lower case
# Encode labels
y = y.replace("positive", 1)
y = y.replace("negative", 0)
return x.to_numpy(), y.to_numpy()
# Split train-valid-test dataset
x, y = preprocess(df["review"], df["sentiment"])
x_train, x_test, y_train, y_test = train_test_split(x, y, shuffle=True, test_size=10000)
x_train, x_val, y_train, y_val = train_test_split(
x_train, y_train, shuffle=True, test_size=10000
)
print(f"shape of train data is {x_train.shape}")
print(f"shape of valid data is {x_val.shape}")
print(f"shape of test data is {x_test.shape}")
# Check sentiment rate in train set
dd = pd.Series(y_train).value_counts()
sns.barplot(x=np.array(["negative", "positive"]), y=dd.values)
plt.show()
# TOKENIZATION
def createVocab(x_train):
# Create a word list
word_list = []
for sent in x_train:
for word in sent:
word_list.append(word)
corpus = Counter(word_list)
corpus_ = sorted(corpus, key=corpus.get, reverse=True)[:1000]
onehot_dict = {w: i + 1 for i, w in enumerate(corpus_)}
return onehot_dict
def tokenize(x_set, y_set, vocab):
final_set = []
for sent in x_set:
final_set.append([vocab[word] for word in sent if vocab.get(word) != None])
return np.array(final_set, dtype=object), np.array(y_set, dtype="float32")
vocab = createVocab(x_train)
x_train, y_train = tokenize(x_train, y_train, vocab)
x_val, y_val = tokenize(x_val, y_val, vocab)
x_test, y_test = tokenize(x_test, y_test, vocab)
print(f"Length of vocabulary is {len(vocab)}")
def padding_(sentences, seq_len):
features = np.zeros((len(sentences), seq_len), dtype=int)
for ii, review in enumerate(sentences):
if len(review) != 0:
features[ii, -len(review) :] = np.array(review)[:seq_len]
return features
# Padding to max length of 500
x_train_pad = padding_(x_train, 500)
x_val_pad = padding_(x_val, 500)
# create Tensor datasets
train_data = TensorDataset(torch.from_numpy(x_train_pad), torch.from_numpy(y_train))
valid_data = TensorDataset(torch.from_numpy(x_val_pad), torch.from_numpy(y_val))
# dataloaders
batch_size = 50
# make sure to SHUFFLE your data
train_loader = DataLoader(train_data, shuffle=True, batch_size=batch_size)
valid_loader = DataLoader(valid_data, shuffle=True, batch_size=batch_size)
# # obtain one batch of training data
# dataiter = iter(train_loader)
# sample_x, sample_y = dataiter.next()
# print('Sample input size: ', sample_x.size()) # batch_size, seq_length
# print('Sample input: \n', sample_x)
# print('Sample input: \n', sample_y)
class SentimentRNN(nn.Module):
def __init__(self, no_layers, vocab_size, hidden_dim, embedding_dim, drop_prob=0.5):
super(SentimentRNN, self).__init__()
self.output_dim = output_dim
self.hidden_dim = hidden_dim
self.no_layers = no_layers
self.vocab_size = vocab_size
# embedding and LSTM layers
self.embedding = nn.Embedding(vocab_size, embedding_dim)
# lstm
self.lstm = nn.LSTM(
input_size=embedding_dim,
hidden_size=self.hidden_dim,
num_layers=no_layers,
batch_first=True,
)
# dropout layer
self.dropout = nn.Dropout(drop_prob)
# linear and sigmoid layer
self.fc = nn.Linear(self.hidden_dim, output_dim)
self.sig = nn.Sigmoid()
def forward(self, x, hidden):
batch_size = x.size(0)
# embeddings and lstm_out
embeds = self.embedding(x) # shape: B x S x Feature since batch = True
# print(embeds.shape) #[50, 500, 1000]
lstm_out, hidden = self.lstm(embeds, hidden)
lstm_out = lstm_out.contiguous().view(-1, self.hidden_dim)
# dropout and fully connected layer
out = self.dropout(lstm_out)
out = self.fc(out)
# sigmoid function
sig_out = self.sig(out)
# reshape to be batch_size first
sig_out = sig_out.view(batch_size, -1)
sig_out = sig_out[:, -1] # get last batch of labels
# return last sigmoid output and hidden state
return sig_out, hidden
def init_hidden(self, batch_size):
"""Initializes hidden state"""
# Create two new tensors with sizes n_layers x batch_size x hidden_dim,
# initialized to zero, for hidden state and cell state of LSTM
h0 = torch.zeros((self.no_layers, batch_size, self.hidden_dim)).to(device)
c0 = torch.zeros((self.no_layers, batch_size, self.hidden_dim)).to(device)
hidden = (h0, c0)
return hidden
no_layers = 2
vocab_size = len(vocab) + 1 # extra 1 for padding
embedding_dim = 64
output_dim = 1
hidden_dim = 256
model = SentimentRNN(no_layers, vocab_size, hidden_dim, embedding_dim, drop_prob=0.3)
# moving to gpu
model.to(device)
print(model)
# loss and optimization functions
lr = 0.001
criterion = nn.BCELoss()
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
# function to predict accuracy
def acc(pred, label):
pred = torch.round(pred.squeeze())
return torch.sum(pred == label.squeeze()).item()
clip = 5
epochs = 7
valid_loss_min = np.Inf
# train for some number of epochs
epoch_tr_loss, epoch_vl_loss = [], []
epoch_tr_acc, epoch_vl_acc = [], []
for epoch in range(epochs):
train_losses = []
train_acc = 0.0
model.train()
# initialize hidden state
h = model.init_hidden(batch_size)
for inputs, labels in train_loader:
inputs, labels = inputs.to(device), labels.to(device)
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
h = tuple([each.data for each in h])
model.zero_grad()
output, h = model(inputs, h)
# calculate the loss and perform backprop
loss = criterion(output.squeeze(), labels.float())
loss.backward()
train_losses.append(loss.item())
# calculating accuracy
accuracy = acc(output, labels)
train_acc += accuracy
# `clip_grad_norm` helps prevent the exploding gradient problem in RNNs / LSTMs.
nn.utils.clip_grad_norm_(model.parameters(), clip)
optimizer.step()
val_h = model.init_hidden(batch_size)
val_losses = []
val_acc = 0.0
model.eval()
for inputs, labels in valid_loader:
val_h = tuple([each.data for each in val_h])
inputs, labels = inputs.to(device), labels.to(device)
output, val_h = model(inputs, val_h)
val_loss = criterion(output.squeeze(), labels.float())
val_losses.append(val_loss.item())
accuracy = acc(output, labels)
val_acc += accuracy
epoch_train_loss = np.mean(train_losses)
epoch_val_loss = np.mean(val_losses)
epoch_train_acc = train_acc / len(train_loader.dataset)
epoch_val_acc = val_acc / len(valid_loader.dataset)
epoch_tr_loss.append(epoch_train_loss)
epoch_vl_loss.append(epoch_val_loss)
epoch_tr_acc.append(epoch_train_acc)
epoch_vl_acc.append(epoch_val_acc)
print(f"Epoch {epoch+1}")
print(f"train_loss : {epoch_train_loss} val_loss : {epoch_val_loss}")
print(f"train_accuracy : {epoch_train_acc*100} val_accuracy : {epoch_val_acc*100}")
if epoch_val_loss <= valid_loss_min:
torch.save(model.state_dict(), "../working/state_dict.pt")
print(
"Validation loss decreased ({:.6f} --> {:.6f}). Saving model ...".format(
valid_loss_min, epoch_val_loss
)
)
valid_loss_min = epoch_val_loss
print(25 * "==")
fig = plt.figure(figsize=(20, 6))
plt.subplot(1, 2, 1)
plt.plot(epoch_tr_acc, label="Train Acc")
plt.plot(epoch_vl_acc, label="Validation Acc")
plt.title("Accuracy")
plt.legend()
plt.grid()
plt.subplot(1, 2, 2)
plt.plot(epoch_tr_loss, label="Train loss")
plt.plot(epoch_vl_loss, label="Validation loss")
plt.title("Loss")
plt.legend()
plt.grid()
plt.show()
x_test_pad = padding_(x_test, 500)
test_data = TensorDataset(torch.from_numpy(x_test_pad), torch.from_numpy(y_test))
test_loader = DataLoader(test_data, shuffle=True, batch_size=batch_size)
test_h = model.init_hidden(batch_size)
test_losses = []
test_acc = 0.0
model.eval()
for inputs, labels in test_loader:
test_h = tuple([each.data for each in test_h])
inputs, labels = inputs.to(device), labels.to(device)
output, test_h = model(inputs, test_h)
test_loss = criterion(output.squeeze(), labels.float())
test_losses.append(test_loss.item())
accuracy = acc(output, labels)
test_acc += accuracy
full_test_loss = np.mean(test_losses)
full_test_acc = test_acc / len(test_loader.dataset)
print(f"Accuracy on test dataset: {full_test_acc}")
| false | 0 | 3,271 | 0 | 3,424 | 3,271 |
||
129494496
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
df = pd.read_csv("C:/Users/DELL/OneDrive/Desktop/KAGGLE/Zomato.csv")
df.head()
df.columns
df.info()
df.describe()
# ### things to do in data analysis
# 1.Missing values
# 2.explore numerical variables
# 3.explore categorical variables
# 4.finding relationship between features
df.isnull().sum()
df.shape
df.drop(
[
"url",
"address",
"phone",
"menu_item",
"dish_liked",
"reviews_list",
"listed_in(city)",
],
axis=1,
inplace=True,
)
df.rename(
columns={
"name": "Restaurants",
"book_table": "booking",
"listed_in(city)": "city",
"rate": "rating",
"approx_cost(for two people)": "cost",
"listed_in(type)": "types",
},
inplace=True,
)
df.head(5)
df.dropna(inplace=True)
df.info()
# ### Finding out the duplicate rows
df[df.duplicated()].count().sum()
df.drop_duplicates(inplace=True)
df.info()
# ### cleaning individual columns
mean1 = df["votes"].mean()
df["votes"].replace(np.nan, mean1, inplace=True)
mode1 = df["Restaurants"].mode().values[0]
mode2 = df["online_order"].mode().values[0]
mode3 = df["booking"].mode().values[0]
mode4 = df["rating"].mode().values[0]
mode5 = df["location"].mode().values[0]
mode6 = df["rest_type"].mode().values[0]
mode7 = df["cuisines"].mode().values[0]
mode8 = df["cost"].mode().values[0]
mode9 = df["types"].mode().values[0]
df["Restaurants"] = df["Restaurants"].replace(np.nan, mode1)
df["online_order"] = df["online_order"].replace(np.nan, mode2)
df["booking"] = df["booking"].replace(np.nan, mode3)
df["rating"] = df["rating"].replace(np.nan, mode4)
df["location"] = df["location"].replace(np.nan, mode5)
df["rest_type"] = df["rest_type"].replace(np.nan, mode6)
df["cuisines"] = df["cuisines"].replace(np.nan, mode7)
df["cost"] = df["cost"].replace(np.nan, mode8)
df["types"] = df["types"].replace(np.nan, mode9)
df.isnull().sum()
df["Restaurants"].unique()
df["online_order"].unique()
df["booking"].unique()
df["rating"].unique()
def rate(value):
if value == "NEW" or value == "-":
return int(0)
else:
value = value.split("/")
value = value[0]
return float(value)
df["rating"] = df["rating"].apply(rate)
df["rating"].head()
df["votes"].unique()
df["location"].unique()
df["rest_type"].unique()
df["cuisines"].unique()
df["cost"].unique()
def price(value):
value = str(value)
if "," in value:
value = value.replace(",", "")
return int(value)
else:
return int(value)
df["cost"] = df["cost"].apply(price)
df["cost"].head()
# ### Check if any non-null values are present after cleaning the dataset
df.info()
df.describe()
plt.figure(figsize=(8, 8))
sns.distplot(x=df["rating"], color="green")
plt.title("Distplot of Rating")
plt.tight_layout()
plt.figure(figsize=(5, 5))
sns.distplot(x=df["votes"], color="blue")
plt.title("Distplot of Votes")
plt.tight_layout()
plt.figure(figsize=(5, 5))
sns.distplot(x=df["cost"], color="yellow")
plt.title("Distplot of Cost")
plt.tight_layout()
plt.figure(figsize=(5, 3), dpi=90)
sns.countplot(x="online_order", data=df, palette="winter")
plt.title("Online delivery")
fig, axes = plt.subplots(figsize=(16, 11), nrows=1, ncols=2)
sns.barplot(
x=df["Restaurants"].value_counts()[:10].values,
y=df["rest_type"].value_counts()[:10].index,
ax=axes[0],
palette="coolwarm_r",
)
axes[0].set_title("Restaurants & rest_type")
sns.barplot(
x=df["votes"].value_counts()[:10].values,
y=df["location"].value_counts()[:10].index,
ax=axes[1],
)
axes[1].set_title("Location wise votes")
fig.tight_layout()
plt.show()
plt.figure(figsize=(12, 5))
sns.countplot(df["cost"])
plt.xticks(rotation=90)
plt.title("Cost of restaurant")
plt.show()
hotels = df["Restaurants"].value_counts().head(10)
plt.figure(figsize=(9, 5), dpi=95)
sns.barplot(y=hotels.index, x=hotels, orient="h", palette="colorblind")
plt.title("Famous restaurant chains in Banglore(no.of restaurants)")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/494/129494496.ipynb
| null | null |
[{"Id": 129494496, "ScriptId": 38505149, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13709521, "CreationDate": "05/14/2023 09:33:15", "VersionNumber": 1.0, "Title": "Zomato @ Bangalore_EDA", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 165.0, "LinesInsertedFromPrevious": 165.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
df = pd.read_csv("C:/Users/DELL/OneDrive/Desktop/KAGGLE/Zomato.csv")
df.head()
df.columns
df.info()
df.describe()
# ### things to do in data analysis
# 1.Missing values
# 2.explore numerical variables
# 3.explore categorical variables
# 4.finding relationship between features
df.isnull().sum()
df.shape
df.drop(
[
"url",
"address",
"phone",
"menu_item",
"dish_liked",
"reviews_list",
"listed_in(city)",
],
axis=1,
inplace=True,
)
df.rename(
columns={
"name": "Restaurants",
"book_table": "booking",
"listed_in(city)": "city",
"rate": "rating",
"approx_cost(for two people)": "cost",
"listed_in(type)": "types",
},
inplace=True,
)
df.head(5)
df.dropna(inplace=True)
df.info()
# ### Finding out the duplicate rows
df[df.duplicated()].count().sum()
df.drop_duplicates(inplace=True)
df.info()
# ### cleaning individual columns
mean1 = df["votes"].mean()
df["votes"].replace(np.nan, mean1, inplace=True)
mode1 = df["Restaurants"].mode().values[0]
mode2 = df["online_order"].mode().values[0]
mode3 = df["booking"].mode().values[0]
mode4 = df["rating"].mode().values[0]
mode5 = df["location"].mode().values[0]
mode6 = df["rest_type"].mode().values[0]
mode7 = df["cuisines"].mode().values[0]
mode8 = df["cost"].mode().values[0]
mode9 = df["types"].mode().values[0]
df["Restaurants"] = df["Restaurants"].replace(np.nan, mode1)
df["online_order"] = df["online_order"].replace(np.nan, mode2)
df["booking"] = df["booking"].replace(np.nan, mode3)
df["rating"] = df["rating"].replace(np.nan, mode4)
df["location"] = df["location"].replace(np.nan, mode5)
df["rest_type"] = df["rest_type"].replace(np.nan, mode6)
df["cuisines"] = df["cuisines"].replace(np.nan, mode7)
df["cost"] = df["cost"].replace(np.nan, mode8)
df["types"] = df["types"].replace(np.nan, mode9)
df.isnull().sum()
df["Restaurants"].unique()
df["online_order"].unique()
df["booking"].unique()
df["rating"].unique()
def rate(value):
if value == "NEW" or value == "-":
return int(0)
else:
value = value.split("/")
value = value[0]
return float(value)
df["rating"] = df["rating"].apply(rate)
df["rating"].head()
df["votes"].unique()
df["location"].unique()
df["rest_type"].unique()
df["cuisines"].unique()
df["cost"].unique()
def price(value):
value = str(value)
if "," in value:
value = value.replace(",", "")
return int(value)
else:
return int(value)
df["cost"] = df["cost"].apply(price)
df["cost"].head()
# ### Check if any non-null values are present after cleaning the dataset
df.info()
df.describe()
plt.figure(figsize=(8, 8))
sns.distplot(x=df["rating"], color="green")
plt.title("Distplot of Rating")
plt.tight_layout()
plt.figure(figsize=(5, 5))
sns.distplot(x=df["votes"], color="blue")
plt.title("Distplot of Votes")
plt.tight_layout()
plt.figure(figsize=(5, 5))
sns.distplot(x=df["cost"], color="yellow")
plt.title("Distplot of Cost")
plt.tight_layout()
plt.figure(figsize=(5, 3), dpi=90)
sns.countplot(x="online_order", data=df, palette="winter")
plt.title("Online delivery")
fig, axes = plt.subplots(figsize=(16, 11), nrows=1, ncols=2)
sns.barplot(
x=df["Restaurants"].value_counts()[:10].values,
y=df["rest_type"].value_counts()[:10].index,
ax=axes[0],
palette="coolwarm_r",
)
axes[0].set_title("Restaurants & rest_type")
sns.barplot(
x=df["votes"].value_counts()[:10].values,
y=df["location"].value_counts()[:10].index,
ax=axes[1],
)
axes[1].set_title("Location wise votes")
fig.tight_layout()
plt.show()
plt.figure(figsize=(12, 5))
sns.countplot(df["cost"])
plt.xticks(rotation=90)
plt.title("Cost of restaurant")
plt.show()
hotels = df["Restaurants"].value_counts().head(10)
plt.figure(figsize=(9, 5), dpi=95)
sns.barplot(y=hotels.index, x=hotels, orient="h", palette="colorblind")
plt.title("Famous restaurant chains in Banglore(no.of restaurants)")
plt.show()
| false | 0 | 1,416 | 1 | 1,416 | 1,416 |
||
129500079
|
<jupyter_start><jupyter_text>BigBasket Entire Product List (~28K datapoints)
E-commerce (electronic commerce) is the activity of electronically buying or selling of products on online services or over the Internet. E-commerce draws on technologies such as mobile commerce, electronic funds transfer, supply chain management, Internet marketing, online transaction processing, electronic data interchange (EDI), inventory management systems, and automated data collection systems. E-commerce is in turn driven by the technological advances of the semiconductor industry, and is the largest sector of the electronics industry.
Bigbasket is the largest online grocery supermarket in India. Was launched somewhere around in 2011 since then they've been expanding their business. Though some new competitors have been able to set their foot in the nation such as Blinkit etc. but BigBasket has still not loose anything - thanks to ever expanding popular base and their shift to online buying.
Kaggle dataset identifier: bigbasket-entire-product-list-28k-datapoints
<jupyter_script># # This notebook is a part of individual project for CSC532 machine learning class from SIT, KMUTT
# ### by Phichayaphak Phengphan 63130500220
# In this notebook, I will be exploring and implementing 3 different **recommendation techniques for an e-commerce website**. The goal is to learn and develop a functional recommendation system that can suggest relevant products to users based on their purchase history and browsing behavior.
# The three techniques that I will be exploring are popularity based, collaborative filtering and content-based filtering.
# * **Popularity based filtering** is a simple and effective recommendation technique that does not require any user-specific data or complex algorithms. It suggests items based on their popularity or overall usage frequency.
# * **Content-based filtering** is a technique that uses product attributes such as category, brand, and price to recommend items that are similar to what the user has previously purchased or viewed.
# * **Collaborative filtering**,on the other hand, relies on user behavior data to identify patterns and make recommendations based on similar user preferences.
# Throughout this notebook, I will provide an overview of each technique, explain how it works, and demonstrate how to implement it using Python and relevant libraries such as Pandas, NumPy, and Scikit-learn.
# ## Import basic library and data
import numpy as np
import pandas as pd
# import matplotlib.pyplot as plt
# import seaborn as sns
# import the data
df = pd.read_csv(
"/kaggle/input/bigbasket-entire-product-list-28k-datapoints/BigBasket Products.csv"
)
df.info()
# Sample of input data
df.head()
# index column is unnecessary, so we can remove it
df.drop("index", axis=1, inplace=True)
print("drop index already")
# Drop the duplicate data out, as I preview it, there are a lot of dupplicate product that has all the same name and description.
df.drop_duplicates(inplace=True, subset=["product"])
df.head()
save_df = df.copy()
# Look fo how many category and subcategory we have
print("category values")
df.category.value_counts()
print("subcategory values")
df.sub_category.value_counts()
# ## Popularity based filtering
# Popularity-based technique use the idea of recommend popular items, so I will recommend high rating items descendingPopularity-based technique use the idea of recommend popular items, so I will recommend high rating items descending
# On the website where new user visit, I will show each top 5 items of each categories. So, the user can explore what currently trending or popular.
# * Beauty & Hygiene
# * Gourmet & World Food
# * Kitchen, Garden & Pets
# * Snacks & Branded Foods
# * Foodgrains, Oil & Masala
# * Cleaning & Household
# * Beverages
# * Bakery, Cakes & Dairy
# * Baby Care
# * Fruits & Vegetables
# * Eggs, Meat & Fish
#
# Group the DataFrame by category, and sort by rating
grouped = df.sort_values(by="rating", ascending=False).groupby("category")
# Define a function to return the top 5 products from each group
def top_5(group):
return group.head(5)
# Apply the function to each group, and concatenate the results
result = grouped.apply(top_5).reset_index(drop=True)
# Display the resulting DataFrame
print(result[["category", "sub_category", "product", "rating"]])
# Now we get the result!! ready to display it on website
# ## Content based filtering
# Content-based filtering uses the characteristics or details of items that the user has searched for to suggest similar items.
# Starting by, calculate item similarity: For each item, I will calculate a similarity score between the item's characteristics. This could be done using a similarity metric such as cosine similarity.
# Generate recommendations: Once you have calculated the similarity scores between items and the user's preferences, you can generate a list of recommended items. This could involve selecting the top N items with the highest similarity scores, or using a more sophisticated algorithm such as collaborative filtering or matrix factorization.
# Evaluate the recommendations: Finally, you'll need to evaluate the quality of the recommendations. This could be done using metrics such as precision, recall, and F1 score, or by conducting user studies to measure user satisfaction with the recommendations.
# ### Recommend based on clicked product
#
import re
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.metrics.pairwise import linear_kernel, cosine_similarity
# I will drop rows that has null value so we can use the product that full of content
df = df.dropna()
df = df.reset_index(drop=True)
df[["category", "sub_category", "brand", "product", "type", "description"]]
# Do feature extraction of text data by using the TfidfVectorizer from scikit-learn. It use to transform the text data in the description column of a DataFrame into a sparse matrix of TF-IDF features. The resulting matrix has one row for each document in the DataFrame and one column for each unique word in the corpus. The stop_words parameter is set to 'english', which filters out common English words.
# TF-IDF (term frequency-inverse document frequency) is a numerical technique that assigns weights to words in a document based on how frequently they appear in the document and how rare they are across the entire corpus of documents being analyzed. The resulting scores highlight the most important and meaningful words in a document, while filtering out common and unimportant words. This technique is commonly used in natural language processing and information retrieval to preprocess and extract meaningful features from text data for further analysis.
tfidf = TfidfVectorizer(stop_words="english")
tfidf_matrix = tfidf.fit_transform(df["description"])
tfidf_matrix.shape
# calculates the cosine similarity between all pairs of documents in a corpus using the TF-IDF matrix generated from the text data. The linear_kernel function from scikit-learn is used to compute the cosine similarity between each pair of documents represented as rows in the tfidf_matrix. The resulting cosine_sim matrix is a symmetric matrix where each element represents the cosine similarity score between two documents. This technique is commonly used in natural language processing and information retrieval to find documents that are most similar to a given query document.
cosine_sim = linear_kernel(tfidf_matrix, tfidf_matrix)
cosine_sim
mapping = pd.Series(df.index, index=df["product"])
mapping
# Define recommend function that get product as parameter and return dataset of ralated products
#
def recommend_product_based_on_click(product_input):
product_index = mapping[product_input]
# get similarity values with other movies
# similarity_score is the list of index and similarity matrix
similarity_score = list(enumerate(cosine_sim[product_index]))
# sort in descending order the similarity score of movie inputted with all the other movies
similarity_score = sorted(similarity_score, key=lambda x: x[1], reverse=True)
# Get the scores of the 15 most similar movies. Ignore the first movie.
similarity_score = similarity_score[1:15]
# return movie names using the mapping series
movie_indices = [i[0] for i in similarity_score]
return df["product"].iloc[movie_indices]
# Try our function by put the 'product' as parameter
recommend_product_based_on_click("Diabetea Herbal Tea")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/500/129500079.ipynb
|
bigbasket-entire-product-list-28k-datapoints
|
surajjha101
|
[{"Id": 129500079, "ScriptId": 38325133, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5293368, "CreationDate": "05/14/2023 10:31:39", "VersionNumber": 1.0, "Title": "Recommend system for ML project", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 143.0, "LinesInsertedFromPrevious": 143.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185619948, "KernelVersionId": 129500079, "SourceDatasetVersionId": 3846442}]
|
[{"Id": 3846442, "DatasetId": 2288739, "DatasourceVersionId": 3901320, "CreatorUserId": 9590174, "LicenseName": "CC BY-NC-SA 4.0", "CreationDate": "06/22/2022 12:51:18", "VersionNumber": 1.0, "Title": "BigBasket Entire Product List (~28K datapoints)", "Slug": "bigbasket-entire-product-list-28k-datapoints", "Subtitle": "Analyzing BB Products and their performance across", "Description": "E-commerce (electronic commerce) is the activity of electronically buying or selling of products on online services or over the Internet. E-commerce draws on technologies such as mobile commerce, electronic funds transfer, supply chain management, Internet marketing, online transaction processing, electronic data interchange (EDI), inventory management systems, and automated data collection systems. E-commerce is in turn driven by the technological advances of the semiconductor industry, and is the largest sector of the electronics industry.\n\nBigbasket is the largest online grocery supermarket in India. Was launched somewhere around in 2011 since then they've been expanding their business. Though some new competitors have been able to set their foot in the nation such as Blinkit etc. but BigBasket has still not loose anything - thanks to ever expanding popular base and their shift to online buying.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2288739, "CreatorUserId": 9590174, "OwnerUserId": 9590174.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3846442.0, "CurrentDatasourceVersionId": 3901320.0, "ForumId": 2315335, "Type": 2, "CreationDate": "06/22/2022 12:51:18", "LastActivityDate": "06/22/2022", "TotalViews": 38953, "TotalDownloads": 7669, "TotalVotes": 204, "TotalKernels": 97}]
|
[{"Id": 9590174, "UserName": "surajjha101", "DisplayName": "SJ", "RegisterDate": "02/06/2022", "PerformanceTier": 4}]
|
# # This notebook is a part of individual project for CSC532 machine learning class from SIT, KMUTT
# ### by Phichayaphak Phengphan 63130500220
# In this notebook, I will be exploring and implementing 3 different **recommendation techniques for an e-commerce website**. The goal is to learn and develop a functional recommendation system that can suggest relevant products to users based on their purchase history and browsing behavior.
# The three techniques that I will be exploring are popularity based, collaborative filtering and content-based filtering.
# * **Popularity based filtering** is a simple and effective recommendation technique that does not require any user-specific data or complex algorithms. It suggests items based on their popularity or overall usage frequency.
# * **Content-based filtering** is a technique that uses product attributes such as category, brand, and price to recommend items that are similar to what the user has previously purchased or viewed.
# * **Collaborative filtering**,on the other hand, relies on user behavior data to identify patterns and make recommendations based on similar user preferences.
# Throughout this notebook, I will provide an overview of each technique, explain how it works, and demonstrate how to implement it using Python and relevant libraries such as Pandas, NumPy, and Scikit-learn.
# ## Import basic library and data
import numpy as np
import pandas as pd
# import matplotlib.pyplot as plt
# import seaborn as sns
# import the data
df = pd.read_csv(
"/kaggle/input/bigbasket-entire-product-list-28k-datapoints/BigBasket Products.csv"
)
df.info()
# Sample of input data
df.head()
# index column is unnecessary, so we can remove it
df.drop("index", axis=1, inplace=True)
print("drop index already")
# Drop the duplicate data out, as I preview it, there are a lot of dupplicate product that has all the same name and description.
df.drop_duplicates(inplace=True, subset=["product"])
df.head()
save_df = df.copy()
# Look fo how many category and subcategory we have
print("category values")
df.category.value_counts()
print("subcategory values")
df.sub_category.value_counts()
# ## Popularity based filtering
# Popularity-based technique use the idea of recommend popular items, so I will recommend high rating items descendingPopularity-based technique use the idea of recommend popular items, so I will recommend high rating items descending
# On the website where new user visit, I will show each top 5 items of each categories. So, the user can explore what currently trending or popular.
# * Beauty & Hygiene
# * Gourmet & World Food
# * Kitchen, Garden & Pets
# * Snacks & Branded Foods
# * Foodgrains, Oil & Masala
# * Cleaning & Household
# * Beverages
# * Bakery, Cakes & Dairy
# * Baby Care
# * Fruits & Vegetables
# * Eggs, Meat & Fish
#
# Group the DataFrame by category, and sort by rating
grouped = df.sort_values(by="rating", ascending=False).groupby("category")
# Define a function to return the top 5 products from each group
def top_5(group):
return group.head(5)
# Apply the function to each group, and concatenate the results
result = grouped.apply(top_5).reset_index(drop=True)
# Display the resulting DataFrame
print(result[["category", "sub_category", "product", "rating"]])
# Now we get the result!! ready to display it on website
# ## Content based filtering
# Content-based filtering uses the characteristics or details of items that the user has searched for to suggest similar items.
# Starting by, calculate item similarity: For each item, I will calculate a similarity score between the item's characteristics. This could be done using a similarity metric such as cosine similarity.
# Generate recommendations: Once you have calculated the similarity scores between items and the user's preferences, you can generate a list of recommended items. This could involve selecting the top N items with the highest similarity scores, or using a more sophisticated algorithm such as collaborative filtering or matrix factorization.
# Evaluate the recommendations: Finally, you'll need to evaluate the quality of the recommendations. This could be done using metrics such as precision, recall, and F1 score, or by conducting user studies to measure user satisfaction with the recommendations.
# ### Recommend based on clicked product
#
import re
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.metrics.pairwise import linear_kernel, cosine_similarity
# I will drop rows that has null value so we can use the product that full of content
df = df.dropna()
df = df.reset_index(drop=True)
df[["category", "sub_category", "brand", "product", "type", "description"]]
# Do feature extraction of text data by using the TfidfVectorizer from scikit-learn. It use to transform the text data in the description column of a DataFrame into a sparse matrix of TF-IDF features. The resulting matrix has one row for each document in the DataFrame and one column for each unique word in the corpus. The stop_words parameter is set to 'english', which filters out common English words.
# TF-IDF (term frequency-inverse document frequency) is a numerical technique that assigns weights to words in a document based on how frequently they appear in the document and how rare they are across the entire corpus of documents being analyzed. The resulting scores highlight the most important and meaningful words in a document, while filtering out common and unimportant words. This technique is commonly used in natural language processing and information retrieval to preprocess and extract meaningful features from text data for further analysis.
tfidf = TfidfVectorizer(stop_words="english")
tfidf_matrix = tfidf.fit_transform(df["description"])
tfidf_matrix.shape
# calculates the cosine similarity between all pairs of documents in a corpus using the TF-IDF matrix generated from the text data. The linear_kernel function from scikit-learn is used to compute the cosine similarity between each pair of documents represented as rows in the tfidf_matrix. The resulting cosine_sim matrix is a symmetric matrix where each element represents the cosine similarity score between two documents. This technique is commonly used in natural language processing and information retrieval to find documents that are most similar to a given query document.
cosine_sim = linear_kernel(tfidf_matrix, tfidf_matrix)
cosine_sim
mapping = pd.Series(df.index, index=df["product"])
mapping
# Define recommend function that get product as parameter and return dataset of ralated products
#
def recommend_product_based_on_click(product_input):
product_index = mapping[product_input]
# get similarity values with other movies
# similarity_score is the list of index and similarity matrix
similarity_score = list(enumerate(cosine_sim[product_index]))
# sort in descending order the similarity score of movie inputted with all the other movies
similarity_score = sorted(similarity_score, key=lambda x: x[1], reverse=True)
# Get the scores of the 15 most similar movies. Ignore the first movie.
similarity_score = similarity_score[1:15]
# return movie names using the mapping series
movie_indices = [i[0] for i in similarity_score]
return df["product"].iloc[movie_indices]
# Try our function by put the 'product' as parameter
recommend_product_based_on_click("Diabetea Herbal Tea")
| false | 1 | 1,769 | 0 | 2,005 | 1,769 |
||
129577062
|
import numpy as np
import pandas as pd
import librosa
import lightgbm as lgb
from glob import glob
from tqdm.auto import tqdm
from matplotlib import pyplot as plt
from sklearn.metrics import roc_auc_score
from imblearn.ensemble import BalancedRandomForestClassifier
from sklearn.linear_model import LogisticRegression
INPUT_DIR = "/kaggle/input/data-science-osaka-spring-2023/"
MOTION_DIR = INPUT_DIR + "motion/motion/"
MUSIC_DIR = INPUT_DIR + "music/music/"
df_label = pd.read_csv(INPUT_DIR + "train.csv")
# # 1. Feature Enginnering
# 基本的なアイデアは、モーションセンサーの周期性が楽曲のテンポとどの程度同期しているかを周波数解析でマッチングするということである。
# 以下で用いる関数は、下記サイトに掲載のものを元に改変(シンプル化)した。
# https://www.wizard-notes.com/entry/music-analysis/tempogram
# 音声ファイルからリズムに関する成分 (novelty function) を抽出する関数
def compute_spectral_based_novelty(y, hop_length):
Y = np.log(np.abs(librosa.stft(y=y, hop_length=hop_length)) + 1)
n_freq, n_tf = Y.shape
spectral_novelty = np.zeros(n_tf - 1)
# Compute and accumulate novelty function each frequencies
for f in range(0, n_freq):
tmp = Y[f, 1:] - Y[f, :-1]
tmp[tmp < 0.0] = 0.0
spectral_novelty += tmp
# Normalization
spectral_novelty /= np.max(spectral_novelty)
return spectral_novelty
# novelty functionからフーリエテンポグラムを得る関数
def compute_fourier_tempogram(novelty_function, sampling_rate, hop_length, n_fft):
ftg = np.abs(librosa.stft(novelty_function, hop_length=hop_length, n_fft=n_fft))
sr_minite = 60 * sampling_rate
ftg_bpms = np.linspace(0, float(sr_minite) / 2, int(1 + n_fft // 2), endpoint=True)
return ftg, ftg_bpms
# テンポの範囲を制限 (clipping) する関数
def adjust_tempograms(tg, bpms, bpm_min=-np.inf, bpm_max=np.inf):
indices = np.where(np.logical_and(bpms >= bpm_min, bpms <= bpm_max))[0]
return tg[indices], bpms[indices]
# テンポグラムの描画関数
def plot_tg(tg, bpms, ticks_base=10):
plt.figure(figsize=(5, 3))
plt.imshow(tg[::-1], aspect="auto", cmap="jet")
plt.colorbar()
plt.xlabel("time [s]")
plt.ylabel("BPM")
ticks = 2 * (len(bpms) // ticks_base)
plt.yticks(np.arange(0, len(bpms), ticks), np.round(bpms[::-1][::ticks], 3))
plt.show()
# ここでは、上述の関数がどう機能するかを理解するため、まずは0.wavを例に適用する。
sampling_rate = 100 # novelty function抽出後のサンプリングレート(ハイパラ)
bpm_resolution = 4 # BPMの分解能(音声ファイルの長さ的にこれが限界の模様)
bpm_min = 40
bpm_max = 200
y, sr = librosa.load(MUSIC_DIR + "0.wav", sr=None)
# Spectral based novelty curve
hop_length = sr // sampling_rate # novelty functionの粗さ(適用後の所望sampling_rateから逆算)
novelty_function = compute_spectral_based_novelty(y, hop_length)
hop_length = sampling_rate # テンポグラムのセル幅(ハイパラ、ここでは1秒刻みとした)
n_fft = sampling_rate * 60 // bpm_resolution # 所望のbpm_resolutionを得るための値を逆算
ftg, ftg_bpms = compute_fourier_tempogram(
novelty_function, sampling_rate, hop_length, n_fft
)
ftg, ftg_bpms = adjust_tempograms(ftg, ftg_bpms, bpm_min=bpm_min, bpm_max=bpm_max)
plot_tg(ftg, ftg_bpms)
# 時間方向は1秒刻み、BPM方向は4刻みでテンポグラムが算出された。
# 今回の音声ファイルは時間が短いため、途中でのテンポの変化は無いと考え、時間方向に集約する。
# その上で、成分の最小値を0にシフトし、成分の2乗和が1になるように規格化する。
ftg_agg = np.median(ftg, axis=1)
ftg_agg -= ftg_agg.min()
ftg_agg /= (ftg_agg**2).sum() ** 0.5
plt.figure(figsize=(5, 3))
plt.plot(ftg_bpms, ftg_agg, marker=".")
plt.xlabel("BPM")
plt.show()
# これを見ると、BPM=100, 200近辺の成分が大きく、楽曲のテンポとしてはどちらかと思われる。
# ただ、今回はテンポを一意に決める必要は無く、モーションセンサーデータに対しても同様に計算した上で、同じところにピークが来ているかどうかをマッチングすればよい。
# なお、librosaではbeat_track関数で直接テンポを推定することもできる。
tempo, beats = librosa.beat.beat_track(y=y, sr=sr)
tempo
# 楽曲のテンポはBPM=103と推定されており、テンポグラムによる分析結果と整合している。
# マッチングの際は、こちらの情報も用いることにする。
# 次に、モーションセンサーデータに対して、同様の分析を行う。
# ここでは、0.wavとペアになっている31.csvを例にとる。
df_motion = pd.read_csv(MOTION_DIR + "31.csv")
# resampleメソッドを使い、データ間隔を半ば無理やりにsampling_rateに合わせる
df_motion["index"] = (
df_motion["Time"] * sampling_rate
) # 1/sampling_rate秒毎にインクリメントされるindex
df_motion["index"] = pd.to_datetime(df_motion["index"] * 10**9) # 10**9s = 1ns
df_motion = (
df_motion.set_index("index").resample("1s").mean().interpolate()
) # sampling_rateでresample
# 音声ファイルの場合と同様
hop_length = sampling_rate
n_fft = sampling_rate * 60 // bpm_resolution
# ここではGYRO_Xを例にとる
ftg, ftg_bpms = compute_fourier_tempogram(
df_motion["GYRO_X"].values, sampling_rate, hop_length, n_fft
)
ftg, ftg_bpms = adjust_tempograms(ftg, ftg_bpms, bpm_min=bpm_min, bpm_max=bpm_max)
plot_tg(ftg, ftg_bpms)
ftg_agg = np.median(ftg, axis=1)
ftg_agg -= ftg_agg.min()
ftg_agg /= (ftg_agg**2).sum() ** 0.5
plt.figure(figsize=(5, 3))
plt.plot(ftg_bpms, ftg_agg, marker=".")
plt.xlabel("BPM")
plt.show()
# モーションセンサーについても、200がピークに来ており、音声ファイルと同期していると言えそうである。
# 全ての楽曲、モーションセンサーデータに対して、上述のプロファイルを計算する。
array_ftg_music = []
for i in tqdm(range(250)):
y, sr = librosa.load(MUSIC_DIR + f"{i}.wav", sr=None)
hop_length = sr // sampling_rate
novelty_function = compute_spectral_based_novelty(y, hop_length)
hop_length = sampling_rate
n_fft = sampling_rate * 60 // bpm_resolution
ftg, ftg_bpms = compute_fourier_tempogram(
novelty_function, sampling_rate, hop_length, n_fft
)
ftg, ftg_bpms = adjust_tempograms(ftg, ftg_bpms, bpm_min=bpm_min, bpm_max=bpm_max)
ftg = np.median(ftg, axis=1)
ftg -= ftg.min()
ftg /= (ftg**2).sum() ** 0.5
array_ftg_music.append(ftg)
sensors = df_motion.columns[1:].tolist()
sensors
n_fft = sampling_rate * 60 // bpm_resolution
hop_length = sampling_rate
array_ftg_motion = []
for i in tqdm(range(250)):
df_motion = pd.read_csv(MOTION_DIR + f"{i}.csv")
df_motion["index"] = (
df_motion["Time"] * sampling_rate
) # 1/sampling_rate秒毎にインクリメントされるindex
df_motion["index"] = pd.to_datetime(df_motion["index"] * 10**9) # 10**9s = 1ns
df_motion = (
df_motion.set_index("index").resample("1s").mean().interpolate()
) # sampling_rateでresample
array_ftg_motion.append([])
for col in sensors:
ftg, ftg_bpms = compute_fourier_tempogram(
df_motion[col].values, sampling_rate, hop_length, n_fft
)
ftg, ftg_bpms = adjust_tempograms(
ftg, ftg_bpms, bpm_min=bpm_min, bpm_max=bpm_max
)
ftg = np.median(ftg, axis=1)
ftg -= ftg.min()
ftg /= (ftg**2).sum() ** 0.5
array_ftg_motion[-1].append(ftg)
# beat_track関数によるテンポ推定も全楽曲に対して行う。
array_tempo = np.zeros(250)
for i in tqdm(range(250)):
y, sr = librosa.load(MUSIC_DIR + f"{i}.wav", sr=None)
tempo, beats = librosa.beat.beat_track(y=y, sr=sr)
array_tempo[i] = tempo
# train.csvのペアについて、両者の関係を可視化しておく。
# なお、モーションセンサーについては、最も相関の取れていたGYRO_Xを選択した。
fig, axes = plt.subplots(figsize=(12, 60), nrows=25, ncols=4)
for i, (motion_id, music_id, genre) in enumerate(
zip(df_label["ID"].values, df_label["music"].values, df_label["genre"].values)
):
ax = axes[i // 4, i % 4]
ax.plot(
ftg_bpms,
array_ftg_motion[motion_id][17],
linewidth=1,
marker=".",
label="GYRO_X",
)
ax.plot(ftg_bpms, array_ftg_music[music_id], linewidth=1, marker=".", label="music")
ax.set_title(f"ID: {motion_id}, music: {music_id} ({genre})")
ax.set_xlabel("BPM")
ax.legend()
fig.tight_layout()
plt.show()
# 両者のプロファイルに一致が見られるものと見られないものがある。
# 一致が見られないものについては、これとは別の方針を考える必要がありそう。
# 両者の一致度合いを数値化し、DataFrameを構築する。
array = []
for motion in range(250):
for music in range(250):
row = [motion, music]
tempo = array_tempo[music]
if tempo > 200:
tempo /= 2
for i, col in enumerate(sensors):
# モーションセンサーと楽曲のテンポ成分の積の最大値(ピークが同じ所に来ていれば大)
row.append((array_ftg_motion[motion][i] * array_ftg_music[music]).max())
for i, col in enumerate(sensors):
# beat_trackにより推定されたテンポに一番近い成分の値
row.append(
array_ftg_motion[motion][i][round((tempo - bpm_min) / bpm_resolution)]
)
array.append(row)
columns = ["ID", "music"]
columns += [f"ftg_similarity_{col}" for col in sensors]
columns += [f"tempo_match_{col}" for col in sensors]
df_feature = pd.DataFrame(array, columns=columns)
df_feature.to_csv("tempo_matching.csv", index=False)
df_feature
# ## 2. 特徴量毎のAUC
# 先ほど算出した各特徴量をそのまま使用したときのAUCを算出。
df = pd.merge(df_feature, df_label, how="left")
df = df.query("ID < 100")
df["target"] = 1 - df["genre"].isna()
df
for col in df.columns[2:-2]:
auc = roc_auc_score(df["target"], df[col])
print(f"{col}\t: {auc:.5f}")
# 使えそうなのは、ACCEL_X, ACCEL_Y, ACCEL_Z, GYRO_Xぐらいか。
# ftg_similarity_GYRO_Xは単独でAUC0.79を超えており、かなり有望。
def apk(actual, predicted, k=7):
if len(predicted) > k:
predicted = predicted[:k]
score = 0.0
num_hits = 0.0
for i, p in enumerate(predicted):
if p in actual and p not in predicted[:i]:
num_hits += 1.0
score += num_hits / (i + 1.0)
return score / min(len(actual), k)
def mapk(actual, predicted, k=7):
return np.mean([apk(a, p, k) for a, p in zip(actual, predicted)])
df_agg = (
df.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": list(
x.sort_values("ftg_similarity_GYRO_X", ascending=False)
.iloc[:7]["music"]
.values
)
}
)
)
.reset_index()
)
df_agg = df_agg.merge(df_label, on="ID", how="left")
df_agg
map7 = mapk(df_agg["music_y"].apply(lambda x: [x]), df_agg["music_x"], k=7)
print(f"MAP@7: {map7:.5f}")
# ftg_similarity_GYRO_Xが上位のmusicを7個選ぶだけで、MAP@7=0.13212が出た。
# これは、testも含めた全250のmusicから7個のmusicを選んでいるので、LBよりもシビアな条件である。
# この特徴量だけでsubmissionを作成してみる。
df_test = pd.read_csv("/kaggle/input/dsos2023-validation-dataset/test.csv")
df_test = df_test.merge(df_feature)
df_agg = (
df_test.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": " ".join(
x.sort_values("ftg_similarity_GYRO_X", ascending=False)
.iloc[:7]["music"]
.values.astype(str)
)
}
)
)
.reset_index()
)
df_agg.to_csv("submission_1feature.csv", index=False)
df_agg
# このsubmissionのPublic LBスコアは、0.13304であった。
# ## 3. Local Validation
# 別途作成した独自のバリデーションデータセットで評価を行う。
# まず、今回のデータは正例が極めて少ないため、容易に過学習することが予想される。
# したがって、単体のAUCの確認で有効そうだった特徴量に絞る。
features = [
f"ftg_similarity_{col}" for col in ["ACCEL_X", "ACCEL_Y", "ACCEL_Z", "GYRO_X"]
]
features += [
f"tempo_match_{col}" for col in ["ACCEL_X", "ACCEL_Y", "ACCEL_Z", "GYRO_X"]
]
# 学習データについては、負例の水増しを行うかどうかで、3パターン作成した。
# 3つそれぞれのパターンで、seedを変えて5回繰り返し評価を行う。
# まずは、多少パラメータチューニングを手で行ったLightGBM。
params = {
"objective": "binary",
"metric": "binary",
"verbosity": -1,
"learning_rate": 0.01,
"num_leaves": 2,
"feature_fraction_bynode": 0.2,
"bagging_freq": 1,
"bagging_fraction": 0.9,
}
df_test = pd.read_csv("/kaggle/input/dsos2023-validation-dataset/test.csv")
df_test = df_test.merge(df_feature)
n_seeds = 5
for train_data in ["train", "train_all", "train_all2"]:
print(train_data)
auc_mean = 0.0
pauc_mean = 0.0
map7_mean = 0.0
preds_test = np.zeros(len(df_test))
for seed in range(n_seeds):
params["seed"] = seed
list_valid = []
for i in range(5):
df_train = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/{train_data}_{i}.csv"
)
df_valid = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/valid_{i}.csv"
)
df_train = df_train.merge(df_feature)
df_valid = df_valid.merge(df_feature)
X_train = df_train.drop(["ID", "music", "target"], axis=1)[features]
X_valid = df_valid.drop(["ID", "music", "target"], axis=1)[features]
X_test = df_test.drop(["ID", "music"], axis=1)[features]
y_train = df_train["target"]
y_valid = df_valid["target"]
lgb_train = lgb.Dataset(X_train, y_train)
lgb_valid = lgb.Dataset(X_valid, y_valid)
callbacks = [lgb.early_stopping(stopping_rounds=50, verbose=False)]
model = lgb.train(
params=params,
train_set=lgb_train,
num_boost_round=10000,
valid_sets=[lgb_train, lgb_valid],
valid_names=["train", "valid"],
callbacks=callbacks,
)
preds_valid = model.predict(X_valid)
preds_test += model.predict(X_test) / 5 / n_seeds
df_valid = df_valid[["ID", "music", "target"]]
df_valid["pred"] = preds_valid
list_valid.append(df_valid)
df_pred = pd.concat(list_valid, ignore_index=True)
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": list(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values
)
}
)
)
.reset_index()
)
df_pred_agg = df_pred_agg.merge(df_label, on="ID", how="left")
auc = roc_auc_score(df_pred["target"], df_pred["pred"])
pauc = roc_auc_score(df_pred["target"], df_pred["pred"], max_fpr=0.1)
map7 = mapk(
df_pred_agg["music_y"].apply(lambda x: [x]), df_pred_agg["music_x"], k=7
)
print(f"seed:{seed}\tAUC:{auc:.5f}\tp-AUC(0.1):{pauc:.5f}\tMAP@7:{map7:.5f}")
auc_mean += auc / n_seeds
pauc_mean += pauc / n_seeds
map7_mean += map7 / n_seeds
print(f"AUC:{auc_mean:.5f}\tp-AUC(0.1):{pauc_mean:.5f}\tMAP@7:{map7_mean:.5f}")
print()
df_pred = df_test[["ID", "music"]].copy()
df_pred["music"] = df_pred["music"].astype(str)
df_pred["pred"] = preds_test
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": " ".join(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values.astype(str)
)
}
)
)
.reset_index()
)
df_pred_agg.to_csv(f"submission_lightgbm_{train_data}.csv", index=False)
# seedを変えて5回算出したが、いずれのケースでもMAP@7のブレが大きく、一方AUCは安定しているように見える。
# なお、今回のタスクでは予測の上位が評価対象となるため、partial AUCをmax_fpr=0.1で算出した。
# 負例の水増しの効果は、train_allでは明確に出ている。
# さらに水増ししたtrain_all2では、AUCは微減、p-AUCは微増である。
# MAP@7は比較的大きく向上しているが、あまり鵜吞みにしない方がよさそう。
# 次に、ベースラインnotebookで用いられていたBalancedRandomForestClassifier。
# seed以外のハイパーパラメータは変更していない。
n_seeds = 5
for train_data in ["train", "train_all", "train_all2"]:
print(train_data)
auc_mean = 0.0
pauc_mean = 0.0
map7_mean = 0.0
preds_test = np.zeros(len(df_test))
for seed in range(n_seeds):
params["seed"] = seed
list_valid = []
for i in range(5):
df_train = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/{train_data}_{i}.csv"
)
df_valid = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/valid_{i}.csv"
)
df_train = df_train.merge(df_feature)
df_valid = df_valid.merge(df_feature)
X_train = df_train.drop(["ID", "music", "target"], axis=1)[features]
X_valid = df_valid.drop(["ID", "music", "target"], axis=1)[features]
X_test = df_test.drop(["ID", "music"], axis=1)[features]
y_train = df_train["target"]
y_valid = df_valid["target"]
model = BalancedRandomForestClassifier(
n_estimators=2500,
criterion="entropy",
max_features=0.7,
min_samples_leaf=5,
random_state=seed,
n_jobs=-1,
)
model.fit(X_train, y_train)
preds_valid = model.predict_proba(X_valid)[:, 1]
preds_test += model.predict_proba(X_test)[:, 1] / 5 / 5
df_valid = df_valid[["ID", "music", "target"]]
df_valid["pred"] = preds_valid
list_valid.append(df_valid)
df_pred = pd.concat(list_valid, ignore_index=True)
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": list(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values
)
}
)
)
.reset_index()
)
df_pred_agg = df_pred_agg.merge(df_label, on="ID", how="left")
auc = roc_auc_score(df_pred["target"], df_pred["pred"])
pauc = roc_auc_score(df_pred["target"], df_pred["pred"], max_fpr=0.1)
map7 = mapk(
df_pred_agg["music_y"].apply(lambda x: [x]), df_pred_agg["music_x"], k=7
)
print(f"seed:{seed}\tAUC:{auc:.5f}\tp-AUC(0.1):{pauc:.5f}\tMAP@7:{map7:.5f}")
auc_mean += auc / n_seeds
pauc_mean += pauc / n_seeds
map7_mean += map7 / n_seeds
print(f"AUC:{auc_mean:.5f}\tp-AUC(0.1):{pauc_mean:.5f}\tMAP@7:{map7_mean:.5f}")
print()
df_pred = df_test[["ID", "music"]].copy()
df_pred["music"] = df_pred["music"].astype(str)
df_pred["pred"] = preds_test
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": " ".join(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values.astype(str)
)
}
)
)
.reset_index()
)
df_pred_agg.to_csv(f"submission_barancedRF_{train_data}.csv", index=False)
# 最後にロジスティック回帰。
n_seeds = 5
for train_data in ["train", "train_all", "train_all2"]:
print(train_data)
auc_mean = 0.0
pauc_mean = 0.0
map7_mean = 0.0
preds_test = np.zeros(len(df_test))
for seed in range(n_seeds):
params["seed"] = seed
list_valid = []
for i in range(5):
df_train = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/{train_data}_{i}.csv"
)
df_valid = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/valid_{i}.csv"
)
df_train = df_train.merge(df_feature)
df_valid = df_valid.merge(df_feature)
X_train = df_train.drop(["ID", "music", "target"], axis=1)[features]
X_valid = df_valid.drop(["ID", "music", "target"], axis=1)[features]
X_test = df_test.drop(["ID", "music"], axis=1)[features]
y_train = df_train["target"]
y_valid = df_valid["target"]
model = LogisticRegression(
tol=1e-10,
C=0.1,
class_weight="balanced",
random_state=seed,
solver="liblinear",
)
model.fit(X_train, y_train)
preds_valid = model.predict_proba(X_valid)[:, 1]
preds_test += model.predict_proba(X_test)[:, 1] / 5 / 5
df_valid = df_valid[["ID", "music", "target"]]
df_valid["pred"] = preds_valid
list_valid.append(df_valid)
df_pred = pd.concat(list_valid, ignore_index=True)
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": list(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values
)
}
)
)
.reset_index()
)
df_pred_agg = df_pred_agg.merge(df_label, on="ID", how="left")
auc = roc_auc_score(df_pred["target"], df_pred["pred"])
pauc = roc_auc_score(df_pred["target"], df_pred["pred"], max_fpr=0.1)
map7 = mapk(
df_pred_agg["music_y"].apply(lambda x: [x]), df_pred_agg["music_x"], k=7
)
print(f"seed:{seed}\tAUC:{auc:.5f}\tp-AUC(0.1):{pauc:.5f}\tMAP@7:{map7:.5f}")
auc_mean += auc / n_seeds
pauc_mean += pauc / n_seeds
map7_mean += map7 / n_seeds
print(f"AUC:{auc_mean:.5f}\tp-AUC(0.1):{pauc_mean:.5f}\tMAP@7:{map7_mean:.5f}")
print()
df_pred = df_test[["ID", "music"]].copy()
df_pred["music"] = df_pred["music"].astype(str)
df_pred["pred"] = preds_test
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": " ".join(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values.astype(str)
)
}
)
)
.reset_index()
)
df_pred_agg.to_csv(f"submission_logistic_{train_data}.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/577/129577062.ipynb
| null | null |
[{"Id": 129577062, "ScriptId": 38530642, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 169364, "CreationDate": "05/15/2023 01:52:56", "VersionNumber": 1.0, "Title": "DSOS2023 1st Place Solution", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 577.0, "LinesInsertedFromPrevious": 2.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 575.0, "LinesInsertedFromFork": 2.0, "LinesDeletedFromFork": 2.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 575.0, "TotalVotes": 26}]
| null | null | null | null |
import numpy as np
import pandas as pd
import librosa
import lightgbm as lgb
from glob import glob
from tqdm.auto import tqdm
from matplotlib import pyplot as plt
from sklearn.metrics import roc_auc_score
from imblearn.ensemble import BalancedRandomForestClassifier
from sklearn.linear_model import LogisticRegression
INPUT_DIR = "/kaggle/input/data-science-osaka-spring-2023/"
MOTION_DIR = INPUT_DIR + "motion/motion/"
MUSIC_DIR = INPUT_DIR + "music/music/"
df_label = pd.read_csv(INPUT_DIR + "train.csv")
# # 1. Feature Enginnering
# 基本的なアイデアは、モーションセンサーの周期性が楽曲のテンポとどの程度同期しているかを周波数解析でマッチングするということである。
# 以下で用いる関数は、下記サイトに掲載のものを元に改変(シンプル化)した。
# https://www.wizard-notes.com/entry/music-analysis/tempogram
# 音声ファイルからリズムに関する成分 (novelty function) を抽出する関数
def compute_spectral_based_novelty(y, hop_length):
Y = np.log(np.abs(librosa.stft(y=y, hop_length=hop_length)) + 1)
n_freq, n_tf = Y.shape
spectral_novelty = np.zeros(n_tf - 1)
# Compute and accumulate novelty function each frequencies
for f in range(0, n_freq):
tmp = Y[f, 1:] - Y[f, :-1]
tmp[tmp < 0.0] = 0.0
spectral_novelty += tmp
# Normalization
spectral_novelty /= np.max(spectral_novelty)
return spectral_novelty
# novelty functionからフーリエテンポグラムを得る関数
def compute_fourier_tempogram(novelty_function, sampling_rate, hop_length, n_fft):
ftg = np.abs(librosa.stft(novelty_function, hop_length=hop_length, n_fft=n_fft))
sr_minite = 60 * sampling_rate
ftg_bpms = np.linspace(0, float(sr_minite) / 2, int(1 + n_fft // 2), endpoint=True)
return ftg, ftg_bpms
# テンポの範囲を制限 (clipping) する関数
def adjust_tempograms(tg, bpms, bpm_min=-np.inf, bpm_max=np.inf):
indices = np.where(np.logical_and(bpms >= bpm_min, bpms <= bpm_max))[0]
return tg[indices], bpms[indices]
# テンポグラムの描画関数
def plot_tg(tg, bpms, ticks_base=10):
plt.figure(figsize=(5, 3))
plt.imshow(tg[::-1], aspect="auto", cmap="jet")
plt.colorbar()
plt.xlabel("time [s]")
plt.ylabel("BPM")
ticks = 2 * (len(bpms) // ticks_base)
plt.yticks(np.arange(0, len(bpms), ticks), np.round(bpms[::-1][::ticks], 3))
plt.show()
# ここでは、上述の関数がどう機能するかを理解するため、まずは0.wavを例に適用する。
sampling_rate = 100 # novelty function抽出後のサンプリングレート(ハイパラ)
bpm_resolution = 4 # BPMの分解能(音声ファイルの長さ的にこれが限界の模様)
bpm_min = 40
bpm_max = 200
y, sr = librosa.load(MUSIC_DIR + "0.wav", sr=None)
# Spectral based novelty curve
hop_length = sr // sampling_rate # novelty functionの粗さ(適用後の所望sampling_rateから逆算)
novelty_function = compute_spectral_based_novelty(y, hop_length)
hop_length = sampling_rate # テンポグラムのセル幅(ハイパラ、ここでは1秒刻みとした)
n_fft = sampling_rate * 60 // bpm_resolution # 所望のbpm_resolutionを得るための値を逆算
ftg, ftg_bpms = compute_fourier_tempogram(
novelty_function, sampling_rate, hop_length, n_fft
)
ftg, ftg_bpms = adjust_tempograms(ftg, ftg_bpms, bpm_min=bpm_min, bpm_max=bpm_max)
plot_tg(ftg, ftg_bpms)
# 時間方向は1秒刻み、BPM方向は4刻みでテンポグラムが算出された。
# 今回の音声ファイルは時間が短いため、途中でのテンポの変化は無いと考え、時間方向に集約する。
# その上で、成分の最小値を0にシフトし、成分の2乗和が1になるように規格化する。
ftg_agg = np.median(ftg, axis=1)
ftg_agg -= ftg_agg.min()
ftg_agg /= (ftg_agg**2).sum() ** 0.5
plt.figure(figsize=(5, 3))
plt.plot(ftg_bpms, ftg_agg, marker=".")
plt.xlabel("BPM")
plt.show()
# これを見ると、BPM=100, 200近辺の成分が大きく、楽曲のテンポとしてはどちらかと思われる。
# ただ、今回はテンポを一意に決める必要は無く、モーションセンサーデータに対しても同様に計算した上で、同じところにピークが来ているかどうかをマッチングすればよい。
# なお、librosaではbeat_track関数で直接テンポを推定することもできる。
tempo, beats = librosa.beat.beat_track(y=y, sr=sr)
tempo
# 楽曲のテンポはBPM=103と推定されており、テンポグラムによる分析結果と整合している。
# マッチングの際は、こちらの情報も用いることにする。
# 次に、モーションセンサーデータに対して、同様の分析を行う。
# ここでは、0.wavとペアになっている31.csvを例にとる。
df_motion = pd.read_csv(MOTION_DIR + "31.csv")
# resampleメソッドを使い、データ間隔を半ば無理やりにsampling_rateに合わせる
df_motion["index"] = (
df_motion["Time"] * sampling_rate
) # 1/sampling_rate秒毎にインクリメントされるindex
df_motion["index"] = pd.to_datetime(df_motion["index"] * 10**9) # 10**9s = 1ns
df_motion = (
df_motion.set_index("index").resample("1s").mean().interpolate()
) # sampling_rateでresample
# 音声ファイルの場合と同様
hop_length = sampling_rate
n_fft = sampling_rate * 60 // bpm_resolution
# ここではGYRO_Xを例にとる
ftg, ftg_bpms = compute_fourier_tempogram(
df_motion["GYRO_X"].values, sampling_rate, hop_length, n_fft
)
ftg, ftg_bpms = adjust_tempograms(ftg, ftg_bpms, bpm_min=bpm_min, bpm_max=bpm_max)
plot_tg(ftg, ftg_bpms)
ftg_agg = np.median(ftg, axis=1)
ftg_agg -= ftg_agg.min()
ftg_agg /= (ftg_agg**2).sum() ** 0.5
plt.figure(figsize=(5, 3))
plt.plot(ftg_bpms, ftg_agg, marker=".")
plt.xlabel("BPM")
plt.show()
# モーションセンサーについても、200がピークに来ており、音声ファイルと同期していると言えそうである。
# 全ての楽曲、モーションセンサーデータに対して、上述のプロファイルを計算する。
array_ftg_music = []
for i in tqdm(range(250)):
y, sr = librosa.load(MUSIC_DIR + f"{i}.wav", sr=None)
hop_length = sr // sampling_rate
novelty_function = compute_spectral_based_novelty(y, hop_length)
hop_length = sampling_rate
n_fft = sampling_rate * 60 // bpm_resolution
ftg, ftg_bpms = compute_fourier_tempogram(
novelty_function, sampling_rate, hop_length, n_fft
)
ftg, ftg_bpms = adjust_tempograms(ftg, ftg_bpms, bpm_min=bpm_min, bpm_max=bpm_max)
ftg = np.median(ftg, axis=1)
ftg -= ftg.min()
ftg /= (ftg**2).sum() ** 0.5
array_ftg_music.append(ftg)
sensors = df_motion.columns[1:].tolist()
sensors
n_fft = sampling_rate * 60 // bpm_resolution
hop_length = sampling_rate
array_ftg_motion = []
for i in tqdm(range(250)):
df_motion = pd.read_csv(MOTION_DIR + f"{i}.csv")
df_motion["index"] = (
df_motion["Time"] * sampling_rate
) # 1/sampling_rate秒毎にインクリメントされるindex
df_motion["index"] = pd.to_datetime(df_motion["index"] * 10**9) # 10**9s = 1ns
df_motion = (
df_motion.set_index("index").resample("1s").mean().interpolate()
) # sampling_rateでresample
array_ftg_motion.append([])
for col in sensors:
ftg, ftg_bpms = compute_fourier_tempogram(
df_motion[col].values, sampling_rate, hop_length, n_fft
)
ftg, ftg_bpms = adjust_tempograms(
ftg, ftg_bpms, bpm_min=bpm_min, bpm_max=bpm_max
)
ftg = np.median(ftg, axis=1)
ftg -= ftg.min()
ftg /= (ftg**2).sum() ** 0.5
array_ftg_motion[-1].append(ftg)
# beat_track関数によるテンポ推定も全楽曲に対して行う。
array_tempo = np.zeros(250)
for i in tqdm(range(250)):
y, sr = librosa.load(MUSIC_DIR + f"{i}.wav", sr=None)
tempo, beats = librosa.beat.beat_track(y=y, sr=sr)
array_tempo[i] = tempo
# train.csvのペアについて、両者の関係を可視化しておく。
# なお、モーションセンサーについては、最も相関の取れていたGYRO_Xを選択した。
fig, axes = plt.subplots(figsize=(12, 60), nrows=25, ncols=4)
for i, (motion_id, music_id, genre) in enumerate(
zip(df_label["ID"].values, df_label["music"].values, df_label["genre"].values)
):
ax = axes[i // 4, i % 4]
ax.plot(
ftg_bpms,
array_ftg_motion[motion_id][17],
linewidth=1,
marker=".",
label="GYRO_X",
)
ax.plot(ftg_bpms, array_ftg_music[music_id], linewidth=1, marker=".", label="music")
ax.set_title(f"ID: {motion_id}, music: {music_id} ({genre})")
ax.set_xlabel("BPM")
ax.legend()
fig.tight_layout()
plt.show()
# 両者のプロファイルに一致が見られるものと見られないものがある。
# 一致が見られないものについては、これとは別の方針を考える必要がありそう。
# 両者の一致度合いを数値化し、DataFrameを構築する。
array = []
for motion in range(250):
for music in range(250):
row = [motion, music]
tempo = array_tempo[music]
if tempo > 200:
tempo /= 2
for i, col in enumerate(sensors):
# モーションセンサーと楽曲のテンポ成分の積の最大値(ピークが同じ所に来ていれば大)
row.append((array_ftg_motion[motion][i] * array_ftg_music[music]).max())
for i, col in enumerate(sensors):
# beat_trackにより推定されたテンポに一番近い成分の値
row.append(
array_ftg_motion[motion][i][round((tempo - bpm_min) / bpm_resolution)]
)
array.append(row)
columns = ["ID", "music"]
columns += [f"ftg_similarity_{col}" for col in sensors]
columns += [f"tempo_match_{col}" for col in sensors]
df_feature = pd.DataFrame(array, columns=columns)
df_feature.to_csv("tempo_matching.csv", index=False)
df_feature
# ## 2. 特徴量毎のAUC
# 先ほど算出した各特徴量をそのまま使用したときのAUCを算出。
df = pd.merge(df_feature, df_label, how="left")
df = df.query("ID < 100")
df["target"] = 1 - df["genre"].isna()
df
for col in df.columns[2:-2]:
auc = roc_auc_score(df["target"], df[col])
print(f"{col}\t: {auc:.5f}")
# 使えそうなのは、ACCEL_X, ACCEL_Y, ACCEL_Z, GYRO_Xぐらいか。
# ftg_similarity_GYRO_Xは単独でAUC0.79を超えており、かなり有望。
def apk(actual, predicted, k=7):
if len(predicted) > k:
predicted = predicted[:k]
score = 0.0
num_hits = 0.0
for i, p in enumerate(predicted):
if p in actual and p not in predicted[:i]:
num_hits += 1.0
score += num_hits / (i + 1.0)
return score / min(len(actual), k)
def mapk(actual, predicted, k=7):
return np.mean([apk(a, p, k) for a, p in zip(actual, predicted)])
df_agg = (
df.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": list(
x.sort_values("ftg_similarity_GYRO_X", ascending=False)
.iloc[:7]["music"]
.values
)
}
)
)
.reset_index()
)
df_agg = df_agg.merge(df_label, on="ID", how="left")
df_agg
map7 = mapk(df_agg["music_y"].apply(lambda x: [x]), df_agg["music_x"], k=7)
print(f"MAP@7: {map7:.5f}")
# ftg_similarity_GYRO_Xが上位のmusicを7個選ぶだけで、MAP@7=0.13212が出た。
# これは、testも含めた全250のmusicから7個のmusicを選んでいるので、LBよりもシビアな条件である。
# この特徴量だけでsubmissionを作成してみる。
df_test = pd.read_csv("/kaggle/input/dsos2023-validation-dataset/test.csv")
df_test = df_test.merge(df_feature)
df_agg = (
df_test.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": " ".join(
x.sort_values("ftg_similarity_GYRO_X", ascending=False)
.iloc[:7]["music"]
.values.astype(str)
)
}
)
)
.reset_index()
)
df_agg.to_csv("submission_1feature.csv", index=False)
df_agg
# このsubmissionのPublic LBスコアは、0.13304であった。
# ## 3. Local Validation
# 別途作成した独自のバリデーションデータセットで評価を行う。
# まず、今回のデータは正例が極めて少ないため、容易に過学習することが予想される。
# したがって、単体のAUCの確認で有効そうだった特徴量に絞る。
features = [
f"ftg_similarity_{col}" for col in ["ACCEL_X", "ACCEL_Y", "ACCEL_Z", "GYRO_X"]
]
features += [
f"tempo_match_{col}" for col in ["ACCEL_X", "ACCEL_Y", "ACCEL_Z", "GYRO_X"]
]
# 学習データについては、負例の水増しを行うかどうかで、3パターン作成した。
# 3つそれぞれのパターンで、seedを変えて5回繰り返し評価を行う。
# まずは、多少パラメータチューニングを手で行ったLightGBM。
params = {
"objective": "binary",
"metric": "binary",
"verbosity": -1,
"learning_rate": 0.01,
"num_leaves": 2,
"feature_fraction_bynode": 0.2,
"bagging_freq": 1,
"bagging_fraction": 0.9,
}
df_test = pd.read_csv("/kaggle/input/dsos2023-validation-dataset/test.csv")
df_test = df_test.merge(df_feature)
n_seeds = 5
for train_data in ["train", "train_all", "train_all2"]:
print(train_data)
auc_mean = 0.0
pauc_mean = 0.0
map7_mean = 0.0
preds_test = np.zeros(len(df_test))
for seed in range(n_seeds):
params["seed"] = seed
list_valid = []
for i in range(5):
df_train = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/{train_data}_{i}.csv"
)
df_valid = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/valid_{i}.csv"
)
df_train = df_train.merge(df_feature)
df_valid = df_valid.merge(df_feature)
X_train = df_train.drop(["ID", "music", "target"], axis=1)[features]
X_valid = df_valid.drop(["ID", "music", "target"], axis=1)[features]
X_test = df_test.drop(["ID", "music"], axis=1)[features]
y_train = df_train["target"]
y_valid = df_valid["target"]
lgb_train = lgb.Dataset(X_train, y_train)
lgb_valid = lgb.Dataset(X_valid, y_valid)
callbacks = [lgb.early_stopping(stopping_rounds=50, verbose=False)]
model = lgb.train(
params=params,
train_set=lgb_train,
num_boost_round=10000,
valid_sets=[lgb_train, lgb_valid],
valid_names=["train", "valid"],
callbacks=callbacks,
)
preds_valid = model.predict(X_valid)
preds_test += model.predict(X_test) / 5 / n_seeds
df_valid = df_valid[["ID", "music", "target"]]
df_valid["pred"] = preds_valid
list_valid.append(df_valid)
df_pred = pd.concat(list_valid, ignore_index=True)
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": list(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values
)
}
)
)
.reset_index()
)
df_pred_agg = df_pred_agg.merge(df_label, on="ID", how="left")
auc = roc_auc_score(df_pred["target"], df_pred["pred"])
pauc = roc_auc_score(df_pred["target"], df_pred["pred"], max_fpr=0.1)
map7 = mapk(
df_pred_agg["music_y"].apply(lambda x: [x]), df_pred_agg["music_x"], k=7
)
print(f"seed:{seed}\tAUC:{auc:.5f}\tp-AUC(0.1):{pauc:.5f}\tMAP@7:{map7:.5f}")
auc_mean += auc / n_seeds
pauc_mean += pauc / n_seeds
map7_mean += map7 / n_seeds
print(f"AUC:{auc_mean:.5f}\tp-AUC(0.1):{pauc_mean:.5f}\tMAP@7:{map7_mean:.5f}")
print()
df_pred = df_test[["ID", "music"]].copy()
df_pred["music"] = df_pred["music"].astype(str)
df_pred["pred"] = preds_test
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": " ".join(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values.astype(str)
)
}
)
)
.reset_index()
)
df_pred_agg.to_csv(f"submission_lightgbm_{train_data}.csv", index=False)
# seedを変えて5回算出したが、いずれのケースでもMAP@7のブレが大きく、一方AUCは安定しているように見える。
# なお、今回のタスクでは予測の上位が評価対象となるため、partial AUCをmax_fpr=0.1で算出した。
# 負例の水増しの効果は、train_allでは明確に出ている。
# さらに水増ししたtrain_all2では、AUCは微減、p-AUCは微増である。
# MAP@7は比較的大きく向上しているが、あまり鵜吞みにしない方がよさそう。
# 次に、ベースラインnotebookで用いられていたBalancedRandomForestClassifier。
# seed以外のハイパーパラメータは変更していない。
n_seeds = 5
for train_data in ["train", "train_all", "train_all2"]:
print(train_data)
auc_mean = 0.0
pauc_mean = 0.0
map7_mean = 0.0
preds_test = np.zeros(len(df_test))
for seed in range(n_seeds):
params["seed"] = seed
list_valid = []
for i in range(5):
df_train = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/{train_data}_{i}.csv"
)
df_valid = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/valid_{i}.csv"
)
df_train = df_train.merge(df_feature)
df_valid = df_valid.merge(df_feature)
X_train = df_train.drop(["ID", "music", "target"], axis=1)[features]
X_valid = df_valid.drop(["ID", "music", "target"], axis=1)[features]
X_test = df_test.drop(["ID", "music"], axis=1)[features]
y_train = df_train["target"]
y_valid = df_valid["target"]
model = BalancedRandomForestClassifier(
n_estimators=2500,
criterion="entropy",
max_features=0.7,
min_samples_leaf=5,
random_state=seed,
n_jobs=-1,
)
model.fit(X_train, y_train)
preds_valid = model.predict_proba(X_valid)[:, 1]
preds_test += model.predict_proba(X_test)[:, 1] / 5 / 5
df_valid = df_valid[["ID", "music", "target"]]
df_valid["pred"] = preds_valid
list_valid.append(df_valid)
df_pred = pd.concat(list_valid, ignore_index=True)
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": list(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values
)
}
)
)
.reset_index()
)
df_pred_agg = df_pred_agg.merge(df_label, on="ID", how="left")
auc = roc_auc_score(df_pred["target"], df_pred["pred"])
pauc = roc_auc_score(df_pred["target"], df_pred["pred"], max_fpr=0.1)
map7 = mapk(
df_pred_agg["music_y"].apply(lambda x: [x]), df_pred_agg["music_x"], k=7
)
print(f"seed:{seed}\tAUC:{auc:.5f}\tp-AUC(0.1):{pauc:.5f}\tMAP@7:{map7:.5f}")
auc_mean += auc / n_seeds
pauc_mean += pauc / n_seeds
map7_mean += map7 / n_seeds
print(f"AUC:{auc_mean:.5f}\tp-AUC(0.1):{pauc_mean:.5f}\tMAP@7:{map7_mean:.5f}")
print()
df_pred = df_test[["ID", "music"]].copy()
df_pred["music"] = df_pred["music"].astype(str)
df_pred["pred"] = preds_test
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": " ".join(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values.astype(str)
)
}
)
)
.reset_index()
)
df_pred_agg.to_csv(f"submission_barancedRF_{train_data}.csv", index=False)
# 最後にロジスティック回帰。
n_seeds = 5
for train_data in ["train", "train_all", "train_all2"]:
print(train_data)
auc_mean = 0.0
pauc_mean = 0.0
map7_mean = 0.0
preds_test = np.zeros(len(df_test))
for seed in range(n_seeds):
params["seed"] = seed
list_valid = []
for i in range(5):
df_train = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/{train_data}_{i}.csv"
)
df_valid = pd.read_csv(
f"/kaggle/input/dsos2023-validation-dataset/valid_{i}.csv"
)
df_train = df_train.merge(df_feature)
df_valid = df_valid.merge(df_feature)
X_train = df_train.drop(["ID", "music", "target"], axis=1)[features]
X_valid = df_valid.drop(["ID", "music", "target"], axis=1)[features]
X_test = df_test.drop(["ID", "music"], axis=1)[features]
y_train = df_train["target"]
y_valid = df_valid["target"]
model = LogisticRegression(
tol=1e-10,
C=0.1,
class_weight="balanced",
random_state=seed,
solver="liblinear",
)
model.fit(X_train, y_train)
preds_valid = model.predict_proba(X_valid)[:, 1]
preds_test += model.predict_proba(X_test)[:, 1] / 5 / 5
df_valid = df_valid[["ID", "music", "target"]]
df_valid["pred"] = preds_valid
list_valid.append(df_valid)
df_pred = pd.concat(list_valid, ignore_index=True)
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": list(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values
)
}
)
)
.reset_index()
)
df_pred_agg = df_pred_agg.merge(df_label, on="ID", how="left")
auc = roc_auc_score(df_pred["target"], df_pred["pred"])
pauc = roc_auc_score(df_pred["target"], df_pred["pred"], max_fpr=0.1)
map7 = mapk(
df_pred_agg["music_y"].apply(lambda x: [x]), df_pred_agg["music_x"], k=7
)
print(f"seed:{seed}\tAUC:{auc:.5f}\tp-AUC(0.1):{pauc:.5f}\tMAP@7:{map7:.5f}")
auc_mean += auc / n_seeds
pauc_mean += pauc / n_seeds
map7_mean += map7 / n_seeds
print(f"AUC:{auc_mean:.5f}\tp-AUC(0.1):{pauc_mean:.5f}\tMAP@7:{map7_mean:.5f}")
print()
df_pred = df_test[["ID", "music"]].copy()
df_pred["music"] = df_pred["music"].astype(str)
df_pred["pred"] = preds_test
df_pred_agg = (
df_pred.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": " ".join(
x.sort_values("pred", ascending=False)
.iloc[:7]["music"]
.values.astype(str)
)
}
)
)
.reset_index()
)
df_pred_agg.to_csv(f"submission_logistic_{train_data}.csv", index=False)
| false | 0 | 7,926 | 26 | 7,926 | 7,926 |
||
129577861
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import zipfile
train_zip = zipfile.ZipFile(
"/kaggle/input/sberbank-russian-housing-market/train.csv.zip"
)
df = pd.read_csv(train_zip.open("train.csv"))
df.head()
# Utiliza a biblioteca para preencher valores numericos onde existe texto na base de dados
from sklearn.preprocessing import LabelEncoder
df_filled = df
for column in df_filled.columns:
if df_filled[column].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_filled[column].values))
df_filled[column] = lbl.transform(list(df_filled[column].values))
# Verifica os valores varios da base
missing = df_filled.isnull().sum() * 100 / len(df_filled)
missing_df = pd.DataFrame({"col": df_filled.columns, "missing_values": missing})
missing_df = missing_df[missing_df.missing_values != 0]
missing_df.sort_values("missing_values", inplace=True, ascending=False)
missing_df.head(10)
# Remove as colunas que possuem muitos valores vazios
missing_df = missing_df[missing_df.missing_values > 40]
cols_to_remove = missing_df.col.to_list()
df_filled.drop(cols_to_remove, inplace=True, axis=1)
# Preenche os dados vazios com os valores medios
import pandas as pd
import numpy as np
for col in df_filled.columns:
if df_filled[col].isnull().sum() > 0:
mean = df_filled[col].mean()
df_filled[col] = df_filled[col].fillna(mean)
# Transforma o timestamp em um valor numerico
df_filled["timestamp"] = df_removed.timestamp.map(
lambda t: int(time.mktime(time.strptime(t, "%Y-%m-%d")))
)
# Utiliza biblioteca para verificar a importancia das colunas (Codigo comentado para diminuit o tempo de execução final)
# from xgboost import XGBRegressor
# X = df_filled.drop('price_doc', axis=1)
# y = df_filled['price_doc']
# model = XGBRegressor(n_estimators=200, max_depth=13, random_state=987, eta=0.01)
# model.fit(X, y)
# feature_importance_df = pd.DataFrame({'col': X.columns, 'importance': model.feature_importances_})
# feature_importance_df.sort_values('importance', inplace=True, ascending=False)
# feature_importance_df.head(20)
# Seleciona as colunas desejadas para fazer a regressão
X = df_filled[
[
"timestamp",
"full_sq",
"culture_objects_top_25",
"female_f",
"life_sq",
"floor",
"max_floor",
"material",
"num_room",
"ecology",
"build_count_monolith",
"cafe_count_3000",
]
]
y = df_filled.price_doc
# Separa os dados de treino e teste
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=25
)
# Faz um tratamento dos dados para tentar melhorar o resultado final
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
# Faz o treinamento
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso, Ridge, ElasticNet
modelo = Lasso()
modelo.fit(X_train, y_train)
# Faz as metricas
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_absolute_percentage_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_log_error
import numpy as np
y_pred = modelo.predict(X_train)
mae = mean_absolute_error(y_train, y_pred)
mape = mean_absolute_percentage_error(y_train, y_pred)
rmse = mean_squared_error(y_train, y_pred) ** 0.5
r2 = r2_score(y_train, y_pred)
rmsle = np.sqrt(mean_squared_log_error(y_train, y_pred))
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("R2:", r2)
print("RMSLE:", rmsle)
print("")
test_zip = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/test.csv.zip")
df = pd.read_csv(test_zip.open("test.csv"))
df_filled = df
for column in df_filled.columns:
if df_filled[column].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_filled[column].values))
df_filled[column] = lbl.transform(list(df_filled[column].values))
for col in df_filled.columns:
if df_filled[col].isnull().sum() > 0:
mean = df_filled[col].mean()
df_filled[col] = df_filled[col].fillna(mean)
df_filled["timestamp"] = df_removed.timestamp.map(
lambda t: int(time.mktime(time.strptime(t, "%Y-%m-%d")))
)
X_test = df_filled[
[
"timestamp",
"full_sq",
"culture_objects_top_25",
"female_f",
"life_sq",
"floor",
"max_floor",
"material",
"num_room",
"ecology",
"build_count_monolith",
"cafe_count_3000",
]
]
y_pred = modelo.predict(X_test)
output = pd.DataFrame({"id": df.id, "price_doc": y_pred})
output.to_csv("/kaggle/working/submission.csv", index=False)
print("Your submission was successfully saved!")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/577/129577861.ipynb
| null | null |
[{"Id": 129577861, "ScriptId": 38172405, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14641521, "CreationDate": "05/15/2023 02:03:42", "VersionNumber": 12.0, "Title": "Regress\u00e3o Ac2", "EvaluationDate": "05/15/2023", "IsChange": false, "TotalLines": 166.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 166.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import zipfile
train_zip = zipfile.ZipFile(
"/kaggle/input/sberbank-russian-housing-market/train.csv.zip"
)
df = pd.read_csv(train_zip.open("train.csv"))
df.head()
# Utiliza a biblioteca para preencher valores numericos onde existe texto na base de dados
from sklearn.preprocessing import LabelEncoder
df_filled = df
for column in df_filled.columns:
if df_filled[column].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_filled[column].values))
df_filled[column] = lbl.transform(list(df_filled[column].values))
# Verifica os valores varios da base
missing = df_filled.isnull().sum() * 100 / len(df_filled)
missing_df = pd.DataFrame({"col": df_filled.columns, "missing_values": missing})
missing_df = missing_df[missing_df.missing_values != 0]
missing_df.sort_values("missing_values", inplace=True, ascending=False)
missing_df.head(10)
# Remove as colunas que possuem muitos valores vazios
missing_df = missing_df[missing_df.missing_values > 40]
cols_to_remove = missing_df.col.to_list()
df_filled.drop(cols_to_remove, inplace=True, axis=1)
# Preenche os dados vazios com os valores medios
import pandas as pd
import numpy as np
for col in df_filled.columns:
if df_filled[col].isnull().sum() > 0:
mean = df_filled[col].mean()
df_filled[col] = df_filled[col].fillna(mean)
# Transforma o timestamp em um valor numerico
df_filled["timestamp"] = df_removed.timestamp.map(
lambda t: int(time.mktime(time.strptime(t, "%Y-%m-%d")))
)
# Utiliza biblioteca para verificar a importancia das colunas (Codigo comentado para diminuit o tempo de execução final)
# from xgboost import XGBRegressor
# X = df_filled.drop('price_doc', axis=1)
# y = df_filled['price_doc']
# model = XGBRegressor(n_estimators=200, max_depth=13, random_state=987, eta=0.01)
# model.fit(X, y)
# feature_importance_df = pd.DataFrame({'col': X.columns, 'importance': model.feature_importances_})
# feature_importance_df.sort_values('importance', inplace=True, ascending=False)
# feature_importance_df.head(20)
# Seleciona as colunas desejadas para fazer a regressão
X = df_filled[
[
"timestamp",
"full_sq",
"culture_objects_top_25",
"female_f",
"life_sq",
"floor",
"max_floor",
"material",
"num_room",
"ecology",
"build_count_monolith",
"cafe_count_3000",
]
]
y = df_filled.price_doc
# Separa os dados de treino e teste
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=25
)
# Faz um tratamento dos dados para tentar melhorar o resultado final
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
# Faz o treinamento
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso, Ridge, ElasticNet
modelo = Lasso()
modelo.fit(X_train, y_train)
# Faz as metricas
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_absolute_percentage_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_log_error
import numpy as np
y_pred = modelo.predict(X_train)
mae = mean_absolute_error(y_train, y_pred)
mape = mean_absolute_percentage_error(y_train, y_pred)
rmse = mean_squared_error(y_train, y_pred) ** 0.5
r2 = r2_score(y_train, y_pred)
rmsle = np.sqrt(mean_squared_log_error(y_train, y_pred))
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("R2:", r2)
print("RMSLE:", rmsle)
print("")
test_zip = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/test.csv.zip")
df = pd.read_csv(test_zip.open("test.csv"))
df_filled = df
for column in df_filled.columns:
if df_filled[column].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_filled[column].values))
df_filled[column] = lbl.transform(list(df_filled[column].values))
for col in df_filled.columns:
if df_filled[col].isnull().sum() > 0:
mean = df_filled[col].mean()
df_filled[col] = df_filled[col].fillna(mean)
df_filled["timestamp"] = df_removed.timestamp.map(
lambda t: int(time.mktime(time.strptime(t, "%Y-%m-%d")))
)
X_test = df_filled[
[
"timestamp",
"full_sq",
"culture_objects_top_25",
"female_f",
"life_sq",
"floor",
"max_floor",
"material",
"num_room",
"ecology",
"build_count_monolith",
"cafe_count_3000",
]
]
y_pred = modelo.predict(X_test)
output = pd.DataFrame({"id": df.id, "price_doc": y_pred})
output.to_csv("/kaggle/working/submission.csv", index=False)
print("Your submission was successfully saved!")
| false | 0 | 1,801 | 0 | 1,801 | 1,801 |
||
129577015
|
import numpy as np
import pandas as pd
import os
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OrdinalEncoder
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
pd.set_option("display.max_columns", None)
df = pd.read_csv("/kaggle/input/playground-series-s3e13/train.csv")
df.info()
df.corr()
df.prognosis.value_counts()
enc = OrdinalEncoder()
df["prognosis"] = enc.fit_transform(df[["prognosis"]])
prognosis = df.pop("prognosis")
id = df.pop("id")
df.head()
X_train, X_test, y_train, y_test = train_test_split(df, prognosis)
clf = LogisticRegression()
clf.fit(X_train, y_train)
probs = clf.predict_proba(X_test)
indices = np.argsort(-probs, axis=1)
top_three = indices[:, :3]
print(top_three)
origin_shape = top_three.shape
top_3_names = enc.inverse_transform(top_three.reshape(-1, 1))
top_3_names = top_3_names.reshape(origin_shape)
top_3_names[0:10]
top_3_strs = np.apply_along_axis(
lambda x: np.array(" ".join(x), dtype="object"), 1, top_3_names
)
top_3_strs[:10]
clf.fit(df, prognosis)
test_df = pd.read_csv("/kaggle/input/playground-series-s3e13/test.csv")
id = test_df.pop("id")
probs_test = clf.predict_proba(test_df)
indices_test = np.argsort(-probs_test, axis=1)
top_three_test = indices_test[:, :3]
origin_shape = top_three_test.shape
top_3_names = enc.inverse_transform(top_three_test.reshape(-1, 1))
top_3_names = top_3_names.reshape(origin_shape)
top_3_strs_test = np.apply_along_axis(
lambda x: np.array(" ".join(x), dtype="object"), 1, top_3_names
)
example = pd.read_csv("/kaggle/input/playground-series-s3e13/sample_submission.csv")
example.head()
submission = pd.DataFrame({"id": id, "prognosis": top_3_strs_test})
submission.head()
submission.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/577/129577015.ipynb
| null | null |
[{"Id": 129577015, "ScriptId": 37828840, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12180553, "CreationDate": "05/15/2023 01:52:19", "VersionNumber": 1.0, "Title": "Prognosis Predictions", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 60.0, "LinesInsertedFromPrevious": 60.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
| null | null | null | null |
import numpy as np
import pandas as pd
import os
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OrdinalEncoder
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
pd.set_option("display.max_columns", None)
df = pd.read_csv("/kaggle/input/playground-series-s3e13/train.csv")
df.info()
df.corr()
df.prognosis.value_counts()
enc = OrdinalEncoder()
df["prognosis"] = enc.fit_transform(df[["prognosis"]])
prognosis = df.pop("prognosis")
id = df.pop("id")
df.head()
X_train, X_test, y_train, y_test = train_test_split(df, prognosis)
clf = LogisticRegression()
clf.fit(X_train, y_train)
probs = clf.predict_proba(X_test)
indices = np.argsort(-probs, axis=1)
top_three = indices[:, :3]
print(top_three)
origin_shape = top_three.shape
top_3_names = enc.inverse_transform(top_three.reshape(-1, 1))
top_3_names = top_3_names.reshape(origin_shape)
top_3_names[0:10]
top_3_strs = np.apply_along_axis(
lambda x: np.array(" ".join(x), dtype="object"), 1, top_3_names
)
top_3_strs[:10]
clf.fit(df, prognosis)
test_df = pd.read_csv("/kaggle/input/playground-series-s3e13/test.csv")
id = test_df.pop("id")
probs_test = clf.predict_proba(test_df)
indices_test = np.argsort(-probs_test, axis=1)
top_three_test = indices_test[:, :3]
origin_shape = top_three_test.shape
top_3_names = enc.inverse_transform(top_three_test.reshape(-1, 1))
top_3_names = top_3_names.reshape(origin_shape)
top_3_strs_test = np.apply_along_axis(
lambda x: np.array(" ".join(x), dtype="object"), 1, top_3_names
)
example = pd.read_csv("/kaggle/input/playground-series-s3e13/sample_submission.csv")
example.head()
submission = pd.DataFrame({"id": id, "prognosis": top_3_strs_test})
submission.head()
submission.to_csv("submission.csv", index=False)
| false | 0 | 691 | 2 | 691 | 691 |
||
129577205
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/train.csv.zip")
z.extractall()
df_train = pd.read_csv("/kaggle/working/train.csv")
df_train.head()
num_linhas = len(df_train)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# transforma as categorias em valores numericos
from sklearn.preprocessing import LabelEncoder
for f in df_train.columns:
if df_train[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_train[f].values))
df_train[f] = lbl.transform(list(df_train[f].values))
df_train.head()
# remove valores nulos através de uma média.
for col in df_train.columns:
if df_train[col].isnull().sum() > 0:
mean = df_train[col].mean()
df_train[col] = df_train[col].fillna(mean)
df_train.head()
# determina as colunas que serão utilizadas no treino e qual será considerada Target.
X = df_train[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
y = np.log(df_train.price_doc)
# separa o arquivo teste em treino e teste
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# torna as partes "normais"
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
# regressão ao modelo
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso, Ridge, ElasticNet
modelo = ElasticNet()
modelo.fit(X_train, y_train)
# define os coeficientes do modelo
modelo.coef_, modelo.intercept_
# expressa os indicadores de performance do modelo
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_absolute_percentage_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_log_error
import numpy as np
y_pred = modelo.predict(X_train)
mae = mean_absolute_error(y_train, y_pred)
mape = mean_absolute_percentage_error(y_train, y_pred)
rmse = mean_squared_error(y_train, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_train, y_pred))
r2 = r2_score(y_train, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
print("")
y_pred = modelo.predict(X_test)
mae = mean_absolute_error(y_test, y_pred)
mape = mean_absolute_percentage_error(y_test, y_pred)
rmse = mean_squared_error(y_test, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_test, y_pred))
r2 = r2_score(y_test, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
# upload do arquivo de Teste
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/test.csv.zip")
z.extractall()
df_test = pd.read_csv("/kaggle/working/test.csv")
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# transforma as categorias em valores numericos
from sklearn.preprocessing import LabelEncoder
for f in df_test.columns:
if df_test[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_test[f].values))
df_test[f] = lbl.transform(list(df_test[f].values))
import pandas as pd
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# remove valores nulos através de uma média.
for col in df_test.columns:
if df_test[col].isnull().sum() > 0:
mean = df_test[col].mean()
df_test[col] = df_test[col].fillna(mean)
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# determina as colunas que serão utilizadas no treino e qual será considerada Target.
X_test = df_test[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
# utiliza as colunas definidas em X_test para fazer a previsão no modelo.
y_pred = modelo.predict(X_test)
# modelos previstos recebe uma função exponencial (o modelo preve numeros logaritimos e não valores reais).
y_pred = np.exp(y_pred)
# cria uma coluna utilizado os preços previstos pelo o metodo e salva
output = pd.DataFrame({"id": df_test.id, "price_doc": y_pred})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
output.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/577/129577205.ipynb
| null | null |
[{"Id": 129577205, "ScriptId": 38530442, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14753237, "CreationDate": "05/15/2023 01:55:03", "VersionNumber": 1.0, "Title": "notebook34d5930ece", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 167.0, "LinesInsertedFromPrevious": 167.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/train.csv.zip")
z.extractall()
df_train = pd.read_csv("/kaggle/working/train.csv")
df_train.head()
num_linhas = len(df_train)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# transforma as categorias em valores numericos
from sklearn.preprocessing import LabelEncoder
for f in df_train.columns:
if df_train[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_train[f].values))
df_train[f] = lbl.transform(list(df_train[f].values))
df_train.head()
# remove valores nulos através de uma média.
for col in df_train.columns:
if df_train[col].isnull().sum() > 0:
mean = df_train[col].mean()
df_train[col] = df_train[col].fillna(mean)
df_train.head()
# determina as colunas que serão utilizadas no treino e qual será considerada Target.
X = df_train[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
y = np.log(df_train.price_doc)
# separa o arquivo teste em treino e teste
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# torna as partes "normais"
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
# regressão ao modelo
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso, Ridge, ElasticNet
modelo = ElasticNet()
modelo.fit(X_train, y_train)
# define os coeficientes do modelo
modelo.coef_, modelo.intercept_
# expressa os indicadores de performance do modelo
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_absolute_percentage_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_log_error
import numpy as np
y_pred = modelo.predict(X_train)
mae = mean_absolute_error(y_train, y_pred)
mape = mean_absolute_percentage_error(y_train, y_pred)
rmse = mean_squared_error(y_train, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_train, y_pred))
r2 = r2_score(y_train, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
print("")
y_pred = modelo.predict(X_test)
mae = mean_absolute_error(y_test, y_pred)
mape = mean_absolute_percentage_error(y_test, y_pred)
rmse = mean_squared_error(y_test, y_pred) ** 0.5
rmsle = np.sqrt(mean_squared_log_error(y_test, y_pred))
r2 = r2_score(y_test, y_pred)
print("MAE:", mae)
print("MAPE:", mape)
print("RMSE:", rmse)
print("RMSLE:", rmsle)
print("R2:", r2)
# upload do arquivo de Teste
import zipfile
z = zipfile.ZipFile("/kaggle/input/sberbank-russian-housing-market/test.csv.zip")
z.extractall()
df_test = pd.read_csv("/kaggle/working/test.csv")
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# transforma as categorias em valores numericos
from sklearn.preprocessing import LabelEncoder
for f in df_test.columns:
if df_test[f].dtype == "object":
lbl = LabelEncoder()
lbl.fit(list(df_test[f].values))
df_test[f] = lbl.transform(list(df_test[f].values))
import pandas as pd
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# remove valores nulos através de uma média.
for col in df_test.columns:
if df_test[col].isnull().sum() > 0:
mean = df_test[col].mean()
df_test[col] = df_test[col].fillna(mean)
num_linhas = len(df_test)
print("O arquivo 'dados.csv' contém", num_linhas, "linhas.")
# determina as colunas que serão utilizadas no treino e qual será considerada Target.
X_test = df_test[
[
"full_sq",
"life_sq",
"floor",
"school_km",
"ecology",
"max_floor",
"material",
"build_year",
"num_room",
]
]
# utiliza as colunas definidas em X_test para fazer a previsão no modelo.
y_pred = modelo.predict(X_test)
# modelos previstos recebe uma função exponencial (o modelo preve numeros logaritimos e não valores reais).
y_pred = np.exp(y_pred)
# cria uma coluna utilizado os preços previstos pelo o metodo e salva
output = pd.DataFrame({"id": df_test.id, "price_doc": y_pred})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
output.head()
| false | 0 | 1,733 | 0 | 1,733 | 1,733 |
||
129577706
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import json
class SFTokenizer:
def __init__(self):
json_file = "/kaggle/input/selfiestokens/SELFIES_Tokens.json"
with open(json_file, "r") as file:
self.mapping = json.load(file)
self.inverse_mapping = {v: k for k, v in self.mapping.items()}
self.sorted_keys = sorted(self.mapping.keys(), key=len, reverse=True)
def encode(self, input_string):
encoded_string = ""
cursor = 0
while cursor < len(input_string):
found = False
for key in self.sorted_keys:
if input_string[cursor:].startswith(key):
if encoded_string != "":
encoded_string += " " # add space as separator
# append encoded value
encoded_string += str(self.mapping[key])
cursor += len(key)
found = True
break
if not found:
raise ValueError(
f"Token not found in the dictionary: {input_string[cursor:]}"
)
return encoded_string
def decode(self, encoded_string):
decoded_string = ""
# split input string by space to get encoded list
encoded_list = encoded_string.split(" ")
for num in encoded_list:
# decode each token
decoded_string += self.inverse_mapping[int(num)]
return decoded_string
from pynvml import *
from huggingface_hub import notebook_login
import pandas as pd
from datasets import Dataset
def print_gpu_utilization():
nvmlInit()
handle = nvmlDeviceGetHandleByIndex(0)
info = nvmlDeviceGetMemoryInfo(handle)
print(f"GPU memory occupied: {info.used//1024**2} MB.")
def print_summary(result):
print(f"Time: {result.metrics['train_runtime']:.2f}")
print(f"Samples/second: {result.metrics['train_samples_per_second']:.2f}")
print_gpu_utilization()
# Read the CSV file using pandas
df = pd.read_csv(
"/kaggle/input/vitamincdataset300gen3kpop/May13VitCRun_300gen3kpop.csv"
)
# # Convert pandas DataFrame to Hugging Face dataset
dataset = Dataset.from_pandas(df)
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained("distilroberta-base")
# Text to tokenize
text = "[ 1 0 , 3 2 , 1 1 , 1 0 , 1 6 , 1 1"
# Tokenize the text
tokens = tokenizer.tokenize(text)
# Print the tokens
print(tokens)
SFtokenizer = SFTokenizer()
import re
# Tokenize the dataset
copy = df.copy()
copy["target"] = copy["target"].apply(SFtokenizer.encode)
copy["kids"] = copy["kids"].apply(SFtokenizer.encode)
copy["p_score"] = copy["p_score"].apply(
lambda x: re.sub(r"^\d|\.", "", str(x)).replace("", " ")[1:]
)
copy["p_score"] = copy["p_score"].apply(lambda x: re.sub(r"(?<=\d)(?=\d)", " ", str(x)))
dataset = Dataset.from_pandas(copy)
dataset = dataset.train_test_split(test_size=0.2)
import wandb
wandb.login()
# One column dataset
copy["TrainingData"] = "S " + copy["p_score"] + " K " + copy["kids"]
dataset = Dataset.from_pandas(copy)
dataset = dataset.train_test_split(test_size=0.2)
def preprocess_function(examples):
return tokenizer([" ".join(x) for x in examples["TrainingData"]])
encoded_dataset = dataset.map(
preprocess_function,
batched=True,
num_proc=4,
remove_columns=dataset["train"].column_names,
)
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
# I have saved my API token with "wandb_api" as Label.
# If you use some other Label make sure to change the same below.
wandb_api = user_secrets.get_secret("wandb_api")
wandb.login(key=wandb_api)
block_size = 128
def group_texts(examples):
# Concatenate all texts.
concatenated_examples = {k: sum(examples[k], []) for k in examples.keys()}
total_length = len(concatenated_examples[list(examples.keys())[0]])
# We drop the small remainder, we could add padding if the model supported it instead of this drop, you can
# customize this part to your needs.
if total_length >= block_size:
total_length = (total_length // block_size) * block_size
# Split by chunks of block_size.
result = {
k: [t[i : i + block_size] for i in range(0, total_length, block_size)]
for k, t in concatenated_examples.items()
}
result["labels"] = result["input_ids"].copy()
return result
lm_dataset = encoded_dataset.map(group_texts, batched=True, num_proc=4)
from transformers import DataCollatorForLanguageModeling
tokenizer.pad_token = tokenizer.eos_token
data_collator = DataCollatorForLanguageModeling(
tokenizer=tokenizer, mlm_probability=0.15
)
from transformers import AutoModelForMaskedLM
model = AutoModelForMaskedLM.from_pretrained("distilroberta-base")
import wandb
wandb.init(
project="VitCRun",
config={
"learning_rate": 2e-5,
"num_train_epochs": 10,
},
)
from transformers import TrainingArguments, Trainer
training_args = TrainingArguments(
output_dir="my_model",
evaluation_strategy="epoch",
save_strategy="epoch",
learning_rate=2e-5,
num_train_epochs=10,
weight_decay=0.01,
push_to_hub=True,
report_to="wandb", # Enables reporting to W&B.
run_name="test_run", # Name of the W&B run.
gradient_accumulation_steps=4,
gradient_checkpointing=True,
fp16=True,
)
trainer = Trainer(
model=model,
args=training_args,
train_dataset=lm_dataset["train"],
eval_dataset=lm_dataset["test"],
data_collator=data_collator,
)
trainer.train()
notebook_login()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/577/129577706.ipynb
| null | null |
[{"Id": 129577706, "ScriptId": 38521345, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6269331, "CreationDate": "05/15/2023 02:01:57", "VersionNumber": 2.0, "Title": "distilRobertA", "EvaluationDate": "05/15/2023", "IsChange": false, "TotalLines": 200.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 200.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import json
class SFTokenizer:
def __init__(self):
json_file = "/kaggle/input/selfiestokens/SELFIES_Tokens.json"
with open(json_file, "r") as file:
self.mapping = json.load(file)
self.inverse_mapping = {v: k for k, v in self.mapping.items()}
self.sorted_keys = sorted(self.mapping.keys(), key=len, reverse=True)
def encode(self, input_string):
encoded_string = ""
cursor = 0
while cursor < len(input_string):
found = False
for key in self.sorted_keys:
if input_string[cursor:].startswith(key):
if encoded_string != "":
encoded_string += " " # add space as separator
# append encoded value
encoded_string += str(self.mapping[key])
cursor += len(key)
found = True
break
if not found:
raise ValueError(
f"Token not found in the dictionary: {input_string[cursor:]}"
)
return encoded_string
def decode(self, encoded_string):
decoded_string = ""
# split input string by space to get encoded list
encoded_list = encoded_string.split(" ")
for num in encoded_list:
# decode each token
decoded_string += self.inverse_mapping[int(num)]
return decoded_string
from pynvml import *
from huggingface_hub import notebook_login
import pandas as pd
from datasets import Dataset
def print_gpu_utilization():
nvmlInit()
handle = nvmlDeviceGetHandleByIndex(0)
info = nvmlDeviceGetMemoryInfo(handle)
print(f"GPU memory occupied: {info.used//1024**2} MB.")
def print_summary(result):
print(f"Time: {result.metrics['train_runtime']:.2f}")
print(f"Samples/second: {result.metrics['train_samples_per_second']:.2f}")
print_gpu_utilization()
# Read the CSV file using pandas
df = pd.read_csv(
"/kaggle/input/vitamincdataset300gen3kpop/May13VitCRun_300gen3kpop.csv"
)
# # Convert pandas DataFrame to Hugging Face dataset
dataset = Dataset.from_pandas(df)
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained("distilroberta-base")
# Text to tokenize
text = "[ 1 0 , 3 2 , 1 1 , 1 0 , 1 6 , 1 1"
# Tokenize the text
tokens = tokenizer.tokenize(text)
# Print the tokens
print(tokens)
SFtokenizer = SFTokenizer()
import re
# Tokenize the dataset
copy = df.copy()
copy["target"] = copy["target"].apply(SFtokenizer.encode)
copy["kids"] = copy["kids"].apply(SFtokenizer.encode)
copy["p_score"] = copy["p_score"].apply(
lambda x: re.sub(r"^\d|\.", "", str(x)).replace("", " ")[1:]
)
copy["p_score"] = copy["p_score"].apply(lambda x: re.sub(r"(?<=\d)(?=\d)", " ", str(x)))
dataset = Dataset.from_pandas(copy)
dataset = dataset.train_test_split(test_size=0.2)
import wandb
wandb.login()
# One column dataset
copy["TrainingData"] = "S " + copy["p_score"] + " K " + copy["kids"]
dataset = Dataset.from_pandas(copy)
dataset = dataset.train_test_split(test_size=0.2)
def preprocess_function(examples):
return tokenizer([" ".join(x) for x in examples["TrainingData"]])
encoded_dataset = dataset.map(
preprocess_function,
batched=True,
num_proc=4,
remove_columns=dataset["train"].column_names,
)
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
# I have saved my API token with "wandb_api" as Label.
# If you use some other Label make sure to change the same below.
wandb_api = user_secrets.get_secret("wandb_api")
wandb.login(key=wandb_api)
block_size = 128
def group_texts(examples):
# Concatenate all texts.
concatenated_examples = {k: sum(examples[k], []) for k in examples.keys()}
total_length = len(concatenated_examples[list(examples.keys())[0]])
# We drop the small remainder, we could add padding if the model supported it instead of this drop, you can
# customize this part to your needs.
if total_length >= block_size:
total_length = (total_length // block_size) * block_size
# Split by chunks of block_size.
result = {
k: [t[i : i + block_size] for i in range(0, total_length, block_size)]
for k, t in concatenated_examples.items()
}
result["labels"] = result["input_ids"].copy()
return result
lm_dataset = encoded_dataset.map(group_texts, batched=True, num_proc=4)
from transformers import DataCollatorForLanguageModeling
tokenizer.pad_token = tokenizer.eos_token
data_collator = DataCollatorForLanguageModeling(
tokenizer=tokenizer, mlm_probability=0.15
)
from transformers import AutoModelForMaskedLM
model = AutoModelForMaskedLM.from_pretrained("distilroberta-base")
import wandb
wandb.init(
project="VitCRun",
config={
"learning_rate": 2e-5,
"num_train_epochs": 10,
},
)
from transformers import TrainingArguments, Trainer
training_args = TrainingArguments(
output_dir="my_model",
evaluation_strategy="epoch",
save_strategy="epoch",
learning_rate=2e-5,
num_train_epochs=10,
weight_decay=0.01,
push_to_hub=True,
report_to="wandb", # Enables reporting to W&B.
run_name="test_run", # Name of the W&B run.
gradient_accumulation_steps=4,
gradient_checkpointing=True,
fp16=True,
)
trainer = Trainer(
model=model,
args=training_args,
train_dataset=lm_dataset["train"],
eval_dataset=lm_dataset["test"],
data_collator=data_collator,
)
trainer.train()
notebook_login()
| false | 0 | 1,813 | 0 | 1,813 | 1,813 |
||
129599853
|
# # 2 Модель Б
# Загрузка данных
import pandas as pd
articles_full = pd.read_pickle("articles_full_end.pkl")
articles_full
articles_full.info()
# ## 2.1 Построение модели классификации
# ### Разбиение выборки на обучающую и тестовую
# Данные для классификации должны быть числовыми, потому что многие алгоритмы машинного обучения работают с числовыми данными. Поэтому нужно закодировать строковые данные: name_company - категориальный признак, можно закодировать LabelEncoder, text, description и activity - текстовые значения, векоризируем, rating - изменить тип
from sklearn.preprocessing import LabelEncoder
encoder = LabelEncoder()
encoded_data = encoder.fit_transform(articles_full["name_company"])
articles_full["name_code"] = encoded_data
articles_full["rating"] = articles_full["rating"].astype("float")
from tqdm.auto import tqdm
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "text"] = " ".join(articles_full["text"][i])
articles_full.at[i, "activity"] = " ".join(articles_full["activity"][i])
articles_full.at[i, "description"] = " ".join(articles_full["description"][i])
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer()
tfidf_matrix_text = vectorizer.fit_transform(articles_full["text"])
tfidf_matrix_description = vectorizer.fit_transform(articles_full["description"])
tfidf_matrix_activity = vectorizer.fit_transform(articles_full["activity"])
# На шаге 1.5 "Разведочный анализ" показана не сбалансированность данных: большое количество нулевой и третей групп номинации, это нужно учесть для грамотного разбиение выборки на обучающую и тестовую, параметр "**stratify**" отвечает за это. Он разбивает данные таким способом, что в обучающейся и тестовой выборках равное соотношений номинаций
# Выборка состоит из 1217 записей. Количество данных достаточно для проведения надлежащего обучения модели. Будет использовано 80% выборки для обучения, что позволит модели получить достаточно информации для корректной работы. Оставшиеся 20% будут использованы, для проверки работоспособности на новых, не изученных данных
from sklearn.model_selection import train_test_split
data = articles_full[["name_code", "rating", "date"]]
X = pd.concat(
[
pd.DataFrame(tfidf_matrix_text.toarray()),
data,
pd.DataFrame(tfidf_matrix_description.toarray()),
pd.DataFrame(tfidf_matrix_activity.toarray()),
],
axis=1,
)
X.columns = X.columns.astype(str)
y = articles_full["code"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, stratify=y, random_state=42
)
# На шаге 1.2 "Формирование структуры набора данных" описаны важные признаки.
# - **name_company** - строковый тип данных, не имеет пропусков, является основным признаком, определяющий компанию номинанта.
# - **text** - является одним из основных признаков так, как определяет то, о чем пишет компания.
# - **date** - не является основным признаком, может отобразить только разницу в дате публикации статей, что не сильно коррелирует с номинированными на премию
# - **description** - не является основным признаком, зачастую не несет рекламный характер, практически ни как не отражает деятельность компании и имеет много пропусков
# - **activity** - не является основным признаком, но описывает сферу деятельности компании, что поможет выявить наиболее подходящую номинацию
# - **rating** - не является основным признаком, отражает положения компании, в топе компаний, что не поможет номинировать их, много пропусков
# ### Обучение моделей
from sklearn.metrics import accuracy_score
from sklearn.metrics import f1_score
from sklearn.metrics import roc_auc_score
info_model = {"name_model": [], "accuracy": [], "F1-score": []}
# Логистическая регрессия - это статистический алгоритм, который используется для решения задач классификации:
# - Прост и быстро реализует алгоритм
# - Эффективен на больших наборах данных
# - Он не справляется с сложными нелинейными зависимостями и не учитывает взаимодействия между признаками
from sklearn.linear_model import LogisticRegression
# Создание модели логистической регрессии
model_lr = LogisticRegression()
# Обучение модели на тренировочных данных
model_lr.fit(X_train, y_train)
# Прогнозирование классов на тестовых данных
predictions_lr = model_lr.predict(X_test)
info_model["name_model"].append("LogisticRegression")
info_model["accuracy"].append(accuracy_score(y_test, predictions_lr))
info_model["F1-score"].append(f1_score(y_test, predictions_lr, average="macro"))
# Метод опорных векторов (Support Vector Machine) - широко применяется для задач классификации и регрессии:
# - Стремится найти оптимальную гиперплоскость, которая наилучшим образом разделяет данные разных классов
# - Регулируемый баланс между сложностью модели и обобщающей способностью
# - Он может быть чувствителен к выбору ядра и параметров модели
# - Вычислительно требовательным при работе с очень большими наборами данных
from sklearn.svm import SVC
# Создание модели метода опорных векторов
model_svm = SVC()
# Обучение модели на тренировочных данных
model_svm.fit(X_train, y_train)
# Прогнозирование классов на тестовых данных
predictions_svm = model_svm.predict(X_test)
info_model["name_model"].append("SVC")
info_model["accuracy"].append(accuracy_score(y_test, predictions_svm))
info_model["F1-score"].append(f1_score(y_test, predictions_svm, average="macro"))
# Случайный лес (Random Forest) - это алгоритм машинного обучения, который использует ансамбль решающих деревьев для решения задач классификации и регрессии:
# - Высокая точность
# - Устойчивость к переобучению
# - Обработка большого количества признаков
# - Возможность оценки важности признаков
# - Быстрюч))
from sklearn.ensemble import RandomForestClassifier
# Создание модели случайного леса
model_rf = RandomForestClassifier()
# Обучение модели на тренировочных данных
model_rf.fit(X_train, y_train)
# Прогнозирование классов на тестовых данных
predictions_rf = model_rf.predict(X_test)
info_model["name_model"].append("RandomForestClassifier")
info_model["accuracy"].append(accuracy_score(y_test, predictions_rf))
info_model["F1-score"].append(f1_score(y_test, predictions_rf, average="macro"))
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Случайный лес показал наиболее хорошие результаты на тестовых данных, и именно accuracy=76% и F1-score=42%. Он и будет применен в дальнейшем улучшении модели
# ## 2.2 Оптимизация модели
# ### Выделение ключевых признаков
# В пинте 1.3 "Предварительная обработка текстовых данных" в столбцах: description, activity, rating было заменено много пустых значений, вероятно это пагубно сказывается на модели, уберем их
data = articles_full[["name_code"]]
X_partial = pd.concat([pd.DataFrame(tfidf_matrix_text.toarray()), data], axis=1)
X_partial.columns = X_partial.columns.astype(str)
y = articles_full["code"]
X_train_p, X_test_p, y_train_p, y_test_p = train_test_split(
X_partial, y, test_size=0.2, stratify=y, random_state=42
)
from sklearn.ensemble import RandomForestClassifier
# Создание модели случайного леса
model_rf = RandomForestClassifier()
# Обучение модели на тренировочных данных
model_rf.fit(X_train_p, y_train_p)
# Прогнозирование классов на тестовых данных
predictions_rf = model_rf.predict(X_test_p)
info_model["name_model"].append("RandomForestClassifier_partial")
info_model["accuracy"].append(accuracy_score(y_test_p, predictions_rf))
info_model["F1-score"].append(f1_score(y_test, predictions_rf, average="macro"))
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Модель стала только хуже, вернем все назад
# ### Понижение размерности
data = articles_full[["name_code", "rating", "date"]]
X = pd.concat(
[
pd.DataFrame(tfidf_matrix_text.toarray()),
data,
pd.DataFrame(tfidf_matrix_description.toarray()),
pd.DataFrame(tfidf_matrix_activity.toarray()),
],
axis=1,
)
X.columns = X.columns.astype(str)
from sklearn.decomposition import PCA
# Создание объекта PCA
pca = PCA(n_components=2) # Указываем количество компонент
# Преобразование данных
X_transformed = pca.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(
X_transformed, y, test_size=0.2, stratify=y, random_state=42
)
# Использование преобразованных данных для обучения модели
model_rf.fit(X_train, y_train)
predictions_rf = model_rf.predict(X_test)
info_model["name_model"].append("RandomForestClassifier_PCA")
info_model["accuracy"].append(accuracy_score(y_test, predictions_rf))
info_model["F1-score"].append(f1_score(y_test, predictions_rf, average="macro"))
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Модель улучшилась, в дальнейшом ипользовать эти данные
# ### Настройки гиперпараметров модели
from sklearn.model_selection import GridSearchCV
param_grid = {
"n_estimators": [100, 200, 300],
"max_depth": [None, 5, 10],
"min_samples_split": [1, 2, 5, 10],
}
grid_search = GridSearchCV(estimator=model_rf, param_grid=param_grid, cv=5)
grid_search.fit(X_train, y_train)
# Вывод наилучших гиперпараметров
print("Наилучшие гиперпараметры:", grid_search.best_params_)
predictions_rf = grid_search.predict(X_test)
info_model["name_model"].append("GridSearchCV")
info_model["accuracy"].append(accuracy_score(y_test, predictions_rf))
info_model["F1-score"].append(f1_score(y_test, predictions_rf, average="macro"))
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Оценка производительности модели на тестовом наборе данных
accuracy = grid_search.score(X_test, y_test)
print("Точность на тестовом наборе данных:", accuracy)
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Лучшие параметры не отличаться от параметром по умолчанию, способ не работает
# ### Визуализация
from yellowbrick.classifier import ClassificationReport
# Создание экземпляра классификатора
model = RandomForestClassifier()
# Обучение модели
model.fit(X_train, y_train)
# Получение предсказаний на тестовых данных
y_pred = model.predict(X_test)
# Визуализация Classification Report
visualizer = ClassificationReport(
model_rf, classes=articles_full["nominations"].unique()
)
visualizer.score(X_test, y_test)
visualizer.show()
# Наука и техника - не определяет
# Здоровья и медицина - отлично классифицируется
# так же и игровая индустрия и киберспорт
# остальное хорошо
from yellowbrick.classifier import ConfusionMatrix
# Создание экземпляра ConfusionMatrix с моделью
visualizer = ConfusionMatrix(model_rf)
# Обучение модели и визуализация матрицы ошибок
visualizer.fit(X_train, y_train)
visualizer.score(X_test, y_test)
# Отображение визуализации
visualizer.show()
from yellowbrick.classifier import ROCAUC
# Создание экземпляра ROCAUC с моделью
visualizer = ROCAUC(model_rf)
# Обучение модели и визуализация кривой ROC-AUC
visualizer.fit(X_train, y_train)
visualizer.score(X_test, y_test)
# Отображение визуализации
visualizer.show()
# Отлично определились категории 0 и 3, не отстаёт категории 2 и 4, все же остальные категории определились плохо, так как в изначальном dataframe их было малое количество
# ### Сохранение модели
import pickle
with open("model.pkl", "wb") as f:
pickle.dump(grid_search, f)
# Загрузка сохраненной модели из файла .pkl
with open("model.pkl", "rb") as f:
model = pickle.load(f)
# Использование загруженной модели для прогнозирования
predictions = model.predict(X_test)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/599/129599853.ipynb
| null | null |
[{"Id": 129599853, "ScriptId": 38538383, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13417746, "CreationDate": "05/15/2023 06:41:12", "VersionNumber": 1.0, "Title": "Report2-KSH-djostit", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 333.0, "LinesInsertedFromPrevious": 333.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
# # 2 Модель Б
# Загрузка данных
import pandas as pd
articles_full = pd.read_pickle("articles_full_end.pkl")
articles_full
articles_full.info()
# ## 2.1 Построение модели классификации
# ### Разбиение выборки на обучающую и тестовую
# Данные для классификации должны быть числовыми, потому что многие алгоритмы машинного обучения работают с числовыми данными. Поэтому нужно закодировать строковые данные: name_company - категориальный признак, можно закодировать LabelEncoder, text, description и activity - текстовые значения, векоризируем, rating - изменить тип
from sklearn.preprocessing import LabelEncoder
encoder = LabelEncoder()
encoded_data = encoder.fit_transform(articles_full["name_company"])
articles_full["name_code"] = encoded_data
articles_full["rating"] = articles_full["rating"].astype("float")
from tqdm.auto import tqdm
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "text"] = " ".join(articles_full["text"][i])
articles_full.at[i, "activity"] = " ".join(articles_full["activity"][i])
articles_full.at[i, "description"] = " ".join(articles_full["description"][i])
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer()
tfidf_matrix_text = vectorizer.fit_transform(articles_full["text"])
tfidf_matrix_description = vectorizer.fit_transform(articles_full["description"])
tfidf_matrix_activity = vectorizer.fit_transform(articles_full["activity"])
# На шаге 1.5 "Разведочный анализ" показана не сбалансированность данных: большое количество нулевой и третей групп номинации, это нужно учесть для грамотного разбиение выборки на обучающую и тестовую, параметр "**stratify**" отвечает за это. Он разбивает данные таким способом, что в обучающейся и тестовой выборках равное соотношений номинаций
# Выборка состоит из 1217 записей. Количество данных достаточно для проведения надлежащего обучения модели. Будет использовано 80% выборки для обучения, что позволит модели получить достаточно информации для корректной работы. Оставшиеся 20% будут использованы, для проверки работоспособности на новых, не изученных данных
from sklearn.model_selection import train_test_split
data = articles_full[["name_code", "rating", "date"]]
X = pd.concat(
[
pd.DataFrame(tfidf_matrix_text.toarray()),
data,
pd.DataFrame(tfidf_matrix_description.toarray()),
pd.DataFrame(tfidf_matrix_activity.toarray()),
],
axis=1,
)
X.columns = X.columns.astype(str)
y = articles_full["code"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, stratify=y, random_state=42
)
# На шаге 1.2 "Формирование структуры набора данных" описаны важные признаки.
# - **name_company** - строковый тип данных, не имеет пропусков, является основным признаком, определяющий компанию номинанта.
# - **text** - является одним из основных признаков так, как определяет то, о чем пишет компания.
# - **date** - не является основным признаком, может отобразить только разницу в дате публикации статей, что не сильно коррелирует с номинированными на премию
# - **description** - не является основным признаком, зачастую не несет рекламный характер, практически ни как не отражает деятельность компании и имеет много пропусков
# - **activity** - не является основным признаком, но описывает сферу деятельности компании, что поможет выявить наиболее подходящую номинацию
# - **rating** - не является основным признаком, отражает положения компании, в топе компаний, что не поможет номинировать их, много пропусков
# ### Обучение моделей
from sklearn.metrics import accuracy_score
from sklearn.metrics import f1_score
from sklearn.metrics import roc_auc_score
info_model = {"name_model": [], "accuracy": [], "F1-score": []}
# Логистическая регрессия - это статистический алгоритм, который используется для решения задач классификации:
# - Прост и быстро реализует алгоритм
# - Эффективен на больших наборах данных
# - Он не справляется с сложными нелинейными зависимостями и не учитывает взаимодействия между признаками
from sklearn.linear_model import LogisticRegression
# Создание модели логистической регрессии
model_lr = LogisticRegression()
# Обучение модели на тренировочных данных
model_lr.fit(X_train, y_train)
# Прогнозирование классов на тестовых данных
predictions_lr = model_lr.predict(X_test)
info_model["name_model"].append("LogisticRegression")
info_model["accuracy"].append(accuracy_score(y_test, predictions_lr))
info_model["F1-score"].append(f1_score(y_test, predictions_lr, average="macro"))
# Метод опорных векторов (Support Vector Machine) - широко применяется для задач классификации и регрессии:
# - Стремится найти оптимальную гиперплоскость, которая наилучшим образом разделяет данные разных классов
# - Регулируемый баланс между сложностью модели и обобщающей способностью
# - Он может быть чувствителен к выбору ядра и параметров модели
# - Вычислительно требовательным при работе с очень большими наборами данных
from sklearn.svm import SVC
# Создание модели метода опорных векторов
model_svm = SVC()
# Обучение модели на тренировочных данных
model_svm.fit(X_train, y_train)
# Прогнозирование классов на тестовых данных
predictions_svm = model_svm.predict(X_test)
info_model["name_model"].append("SVC")
info_model["accuracy"].append(accuracy_score(y_test, predictions_svm))
info_model["F1-score"].append(f1_score(y_test, predictions_svm, average="macro"))
# Случайный лес (Random Forest) - это алгоритм машинного обучения, который использует ансамбль решающих деревьев для решения задач классификации и регрессии:
# - Высокая точность
# - Устойчивость к переобучению
# - Обработка большого количества признаков
# - Возможность оценки важности признаков
# - Быстрюч))
from sklearn.ensemble import RandomForestClassifier
# Создание модели случайного леса
model_rf = RandomForestClassifier()
# Обучение модели на тренировочных данных
model_rf.fit(X_train, y_train)
# Прогнозирование классов на тестовых данных
predictions_rf = model_rf.predict(X_test)
info_model["name_model"].append("RandomForestClassifier")
info_model["accuracy"].append(accuracy_score(y_test, predictions_rf))
info_model["F1-score"].append(f1_score(y_test, predictions_rf, average="macro"))
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Случайный лес показал наиболее хорошие результаты на тестовых данных, и именно accuracy=76% и F1-score=42%. Он и будет применен в дальнейшем улучшении модели
# ## 2.2 Оптимизация модели
# ### Выделение ключевых признаков
# В пинте 1.3 "Предварительная обработка текстовых данных" в столбцах: description, activity, rating было заменено много пустых значений, вероятно это пагубно сказывается на модели, уберем их
data = articles_full[["name_code"]]
X_partial = pd.concat([pd.DataFrame(tfidf_matrix_text.toarray()), data], axis=1)
X_partial.columns = X_partial.columns.astype(str)
y = articles_full["code"]
X_train_p, X_test_p, y_train_p, y_test_p = train_test_split(
X_partial, y, test_size=0.2, stratify=y, random_state=42
)
from sklearn.ensemble import RandomForestClassifier
# Создание модели случайного леса
model_rf = RandomForestClassifier()
# Обучение модели на тренировочных данных
model_rf.fit(X_train_p, y_train_p)
# Прогнозирование классов на тестовых данных
predictions_rf = model_rf.predict(X_test_p)
info_model["name_model"].append("RandomForestClassifier_partial")
info_model["accuracy"].append(accuracy_score(y_test_p, predictions_rf))
info_model["F1-score"].append(f1_score(y_test, predictions_rf, average="macro"))
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Модель стала только хуже, вернем все назад
# ### Понижение размерности
data = articles_full[["name_code", "rating", "date"]]
X = pd.concat(
[
pd.DataFrame(tfidf_matrix_text.toarray()),
data,
pd.DataFrame(tfidf_matrix_description.toarray()),
pd.DataFrame(tfidf_matrix_activity.toarray()),
],
axis=1,
)
X.columns = X.columns.astype(str)
from sklearn.decomposition import PCA
# Создание объекта PCA
pca = PCA(n_components=2) # Указываем количество компонент
# Преобразование данных
X_transformed = pca.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(
X_transformed, y, test_size=0.2, stratify=y, random_state=42
)
# Использование преобразованных данных для обучения модели
model_rf.fit(X_train, y_train)
predictions_rf = model_rf.predict(X_test)
info_model["name_model"].append("RandomForestClassifier_PCA")
info_model["accuracy"].append(accuracy_score(y_test, predictions_rf))
info_model["F1-score"].append(f1_score(y_test, predictions_rf, average="macro"))
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Модель улучшилась, в дальнейшом ипользовать эти данные
# ### Настройки гиперпараметров модели
from sklearn.model_selection import GridSearchCV
param_grid = {
"n_estimators": [100, 200, 300],
"max_depth": [None, 5, 10],
"min_samples_split": [1, 2, 5, 10],
}
grid_search = GridSearchCV(estimator=model_rf, param_grid=param_grid, cv=5)
grid_search.fit(X_train, y_train)
# Вывод наилучших гиперпараметров
print("Наилучшие гиперпараметры:", grid_search.best_params_)
predictions_rf = grid_search.predict(X_test)
info_model["name_model"].append("GridSearchCV")
info_model["accuracy"].append(accuracy_score(y_test, predictions_rf))
info_model["F1-score"].append(f1_score(y_test, predictions_rf, average="macro"))
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Оценка производительности модели на тестовом наборе данных
accuracy = grid_search.score(X_test, y_test)
print("Точность на тестовом наборе данных:", accuracy)
info_model_DF = pd.DataFrame(info_model)
info_model_DF
# Лучшие параметры не отличаться от параметром по умолчанию, способ не работает
# ### Визуализация
from yellowbrick.classifier import ClassificationReport
# Создание экземпляра классификатора
model = RandomForestClassifier()
# Обучение модели
model.fit(X_train, y_train)
# Получение предсказаний на тестовых данных
y_pred = model.predict(X_test)
# Визуализация Classification Report
visualizer = ClassificationReport(
model_rf, classes=articles_full["nominations"].unique()
)
visualizer.score(X_test, y_test)
visualizer.show()
# Наука и техника - не определяет
# Здоровья и медицина - отлично классифицируется
# так же и игровая индустрия и киберспорт
# остальное хорошо
from yellowbrick.classifier import ConfusionMatrix
# Создание экземпляра ConfusionMatrix с моделью
visualizer = ConfusionMatrix(model_rf)
# Обучение модели и визуализация матрицы ошибок
visualizer.fit(X_train, y_train)
visualizer.score(X_test, y_test)
# Отображение визуализации
visualizer.show()
from yellowbrick.classifier import ROCAUC
# Создание экземпляра ROCAUC с моделью
visualizer = ROCAUC(model_rf)
# Обучение модели и визуализация кривой ROC-AUC
visualizer.fit(X_train, y_train)
visualizer.score(X_test, y_test)
# Отображение визуализации
visualizer.show()
# Отлично определились категории 0 и 3, не отстаёт категории 2 и 4, все же остальные категории определились плохо, так как в изначальном dataframe их было малое количество
# ### Сохранение модели
import pickle
with open("model.pkl", "wb") as f:
pickle.dump(grid_search, f)
# Загрузка сохраненной модели из файла .pkl
with open("model.pkl", "rb") as f:
model = pickle.load(f)
# Использование загруженной модели для прогнозирования
predictions = model.predict(X_test)
| false | 0 | 4,259 | 1 | 4,259 | 4,259 |
||
129599742
|
<jupyter_start><jupyter_text>Titanic dataset

### Context
I took the titanic test file and the gender_submission and put them together in excel to make a csv. This is great for making charts to help you visualize. This also will help you know who died or survived. At least 70% right, but its up to you to make it 100% Thanks to the titanic beginners competitions for providing with the data. Please **Upvote** my dataset, it will mean a lot to me. Thank you!
Kaggle dataset identifier: test-file
<jupyter_code>import pandas as pd
df = pd.read_csv('test-file/tested.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 418 entries, 0 to 417
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 418 non-null int64
1 Survived 418 non-null int64
2 Pclass 418 non-null int64
3 Name 418 non-null object
4 Sex 418 non-null object
5 Age 332 non-null float64
6 SibSp 418 non-null int64
7 Parch 418 non-null int64
8 Ticket 418 non-null object
9 Fare 417 non-null float64
10 Cabin 91 non-null object
11 Embarked 418 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 39.3+ KB
<jupyter_text>Examples:
{
"PassengerId": 892,
"Survived": 0,
"Pclass": 3,
"Name": "Kelly, Mr. James",
"Sex": "male",
"Age": 34.5,
"SibSp": 0,
"Parch": 0,
"Ticket": 330911,
"Fare": 7.8292,
"Cabin": NaN,
"Embarked": "Q"
}
{
"PassengerId": 893,
"Survived": 1,
"Pclass": 3,
"Name": "Wilkes, Mrs. James (Ellen Needs)",
"Sex": "female",
"Age": 47.0,
"SibSp": 1,
"Parch": 0,
"Ticket": 363272,
"Fare": 7.0,
"Cabin": NaN,
"Embarked": "S"
}
{
"PassengerId": 894,
"Survived": 0,
"Pclass": 2,
"Name": "Myles, Mr. Thomas Francis",
"Sex": "male",
"Age": 62.0,
"SibSp": 0,
"Parch": 0,
"Ticket": 240276,
"Fare": 9.6875,
"Cabin": NaN,
"Embarked": "Q"
}
{
"PassengerId": 895,
"Survived": 0,
"Pclass": 3,
"Name": "Wirz, Mr. Albert",
"Sex": "male",
"Age": 27.0,
"SibSp": 0,
"Parch": 0,
"Ticket": 315154,
"Fare": 8.6625,
"Cabin": NaN,
"Embarked": "S"
}
<jupyter_script># # Titanic Dataset
# ## Importing Libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import math
# ## Reading dataset
titanic_df = pd.read_csv("/kaggle/input/test-file/tested.csv")
titanic_df.head(10)
print("# of passengers:", len(titanic_df.index))
# ## EDA
sns.countplot(x="survived", data=titanic_df)
sns.countplot(x="sex", data=titanic_df)
sns.countplot(x="survived", hue="sex", data=titanic_df)
titanic_df["age"].plot.hist(bins=20)
plt.title("Age distribution")
sns.countplot(x="survived", hue="pclass", data=titanic_df)
plt.ylabel("Passenger count")
plt.title("Count of Passenger survival")
titanic_df["fare"].plot.hist(bins=25, figsize=(10, 5))
sns.countplot(x="sibsp", data=titanic_df)
sns.countplot(x="parch", data=titanic_df)
# ## Data Wrangling
titanic_df.isnull().sum()
sns.heatmap(titanic_df.isnull(), yticklabels=False, cmap="flare", cbar=False)
plt.show()
## dropping categorical features irrelevant for model
titanic_df.drop(["cabin", "home.dest", "body", "boat"], axis=1, inplace=True)
titanic_df.dropna(inplace=True)
print("# of rows after dropping null values:", len(titanic_df.index))
sex = pd.get_dummies(titanic_df["sex"], drop_first=True)
sex.head()
pclass = pd.get_dummies(titanic_df["pclass"], drop_first=True)
pclass.head()
embarked = pd.get_dummies(titanic_df["embarked"], drop_first=True)
embarked.head()
titanic_df = pd.concat([titanic_df, sex, embarked, pclass], axis=1)
# freeing up kernel variable space
del sex
del embarked
del pclass
## dropping redundant features
titanic_df.drop(["pclass", "embarked", "sex", "name", "ticket"], axis=1, inplace=True)
titanic_df.head()
# ## Model Building
# ### Splitting dataset into Training and Testing data
# X representing independent variables
# Y representing dependent variable
X = titanic_df.drop("survived", axis=1)
y = titanic_df["survived"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=101
)
X_train, X_test = X_train.to_numpy(), X_test.to_numpy()
print(
f"Training dataset contains {X_train.shape[1]} columns and {X_train.shape[0]} rows."
)
print(f"Testing dataset contains {X_test.shape[1]} columns and {X_test.shape[0]} rows.")
# ### Training the model
from sklearn.linear_model import LogisticRegression
logreg_model = LogisticRegression(max_iter=1000)
logreg_model.fit(X_train, y_train)
predictions = logreg_model.predict(X_test)
# ### Checking model Performance
from sklearn.metrics import classification_report, accuracy_score
print(classification_report(y_test, predictions))
accuracy_score(y_test, predictions)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/599/129599742.ipynb
|
test-file
|
brendan45774
|
[{"Id": 129599742, "ScriptId": 38538176, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4669508, "CreationDate": "05/15/2023 06:40:19", "VersionNumber": 1.0, "Title": "titanic_data_analysis", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 109.0, "LinesInsertedFromPrevious": 109.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185832244, "KernelVersionId": 129599742, "SourceDatasetVersionId": 2879186}]
|
[{"Id": 2879186, "DatasetId": 826163, "DatasourceVersionId": 2926173, "CreatorUserId": 2681031, "LicenseName": "CC0: Public Domain", "CreationDate": "12/02/2021 16:11:42", "VersionNumber": 6.0, "Title": "Titanic dataset", "Slug": "test-file", "Subtitle": "Gender submission and test file merged", "Description": "\n\n### Context\n\nI took the titanic test file and the gender_submission and put them together in excel to make a csv. This is great for making charts to help you visualize. This also will help you know who died or survived. At least 70% right, but its up to you to make it 100% Thanks to the titanic beginners competitions for providing with the data. Please **Upvote** my dataset, it will mean a lot to me. Thank you!", "VersionNotes": "tested", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 826163, "CreatorUserId": 2681031, "OwnerUserId": 2681031.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2879186.0, "CurrentDatasourceVersionId": 2926173.0, "ForumId": 841293, "Type": 2, "CreationDate": "08/11/2020 14:08:36", "LastActivityDate": "08/11/2020", "TotalViews": 262161, "TotalDownloads": 72658, "TotalVotes": 665, "TotalKernels": 203}]
|
[{"Id": 2681031, "UserName": "brendan45774", "DisplayName": "Brenda N", "RegisterDate": "01/07/2019", "PerformanceTier": 3}]
|
# # Titanic Dataset
# ## Importing Libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import math
# ## Reading dataset
titanic_df = pd.read_csv("/kaggle/input/test-file/tested.csv")
titanic_df.head(10)
print("# of passengers:", len(titanic_df.index))
# ## EDA
sns.countplot(x="survived", data=titanic_df)
sns.countplot(x="sex", data=titanic_df)
sns.countplot(x="survived", hue="sex", data=titanic_df)
titanic_df["age"].plot.hist(bins=20)
plt.title("Age distribution")
sns.countplot(x="survived", hue="pclass", data=titanic_df)
plt.ylabel("Passenger count")
plt.title("Count of Passenger survival")
titanic_df["fare"].plot.hist(bins=25, figsize=(10, 5))
sns.countplot(x="sibsp", data=titanic_df)
sns.countplot(x="parch", data=titanic_df)
# ## Data Wrangling
titanic_df.isnull().sum()
sns.heatmap(titanic_df.isnull(), yticklabels=False, cmap="flare", cbar=False)
plt.show()
## dropping categorical features irrelevant for model
titanic_df.drop(["cabin", "home.dest", "body", "boat"], axis=1, inplace=True)
titanic_df.dropna(inplace=True)
print("# of rows after dropping null values:", len(titanic_df.index))
sex = pd.get_dummies(titanic_df["sex"], drop_first=True)
sex.head()
pclass = pd.get_dummies(titanic_df["pclass"], drop_first=True)
pclass.head()
embarked = pd.get_dummies(titanic_df["embarked"], drop_first=True)
embarked.head()
titanic_df = pd.concat([titanic_df, sex, embarked, pclass], axis=1)
# freeing up kernel variable space
del sex
del embarked
del pclass
## dropping redundant features
titanic_df.drop(["pclass", "embarked", "sex", "name", "ticket"], axis=1, inplace=True)
titanic_df.head()
# ## Model Building
# ### Splitting dataset into Training and Testing data
# X representing independent variables
# Y representing dependent variable
X = titanic_df.drop("survived", axis=1)
y = titanic_df["survived"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=101
)
X_train, X_test = X_train.to_numpy(), X_test.to_numpy()
print(
f"Training dataset contains {X_train.shape[1]} columns and {X_train.shape[0]} rows."
)
print(f"Testing dataset contains {X_test.shape[1]} columns and {X_test.shape[0]} rows.")
# ### Training the model
from sklearn.linear_model import LogisticRegression
logreg_model = LogisticRegression(max_iter=1000)
logreg_model.fit(X_train, y_train)
predictions = logreg_model.predict(X_test)
# ### Checking model Performance
from sklearn.metrics import classification_report, accuracy_score
print(classification_report(y_test, predictions))
accuracy_score(y_test, predictions)
|
[{"test-file/tested.csv": {"column_names": "[\"PassengerId\", \"Survived\", \"Pclass\", \"Name\", \"Sex\", \"Age\", \"SibSp\", \"Parch\", \"Ticket\", \"Fare\", \"Cabin\", \"Embarked\"]", "column_data_types": "{\"PassengerId\": \"int64\", \"Survived\": \"int64\", \"Pclass\": \"int64\", \"Name\": \"object\", \"Sex\": \"object\", \"Age\": \"float64\", \"SibSp\": \"int64\", \"Parch\": \"int64\", \"Ticket\": \"object\", \"Fare\": \"float64\", \"Cabin\": \"object\", \"Embarked\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 418 entries, 0 to 417\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 PassengerId 418 non-null int64 \n 1 Survived 418 non-null int64 \n 2 Pclass 418 non-null int64 \n 3 Name 418 non-null object \n 4 Sex 418 non-null object \n 5 Age 332 non-null float64\n 6 SibSp 418 non-null int64 \n 7 Parch 418 non-null int64 \n 8 Ticket 418 non-null object \n 9 Fare 417 non-null float64\n 10 Cabin 91 non-null object \n 11 Embarked 418 non-null object \ndtypes: float64(2), int64(5), object(5)\nmemory usage: 39.3+ KB\n", "summary": "{\"PassengerId\": {\"count\": 418.0, \"mean\": 1100.5, \"std\": 120.81045760473994, \"min\": 892.0, \"25%\": 996.25, \"50%\": 1100.5, \"75%\": 1204.75, \"max\": 1309.0}, \"Survived\": {\"count\": 418.0, \"mean\": 0.36363636363636365, \"std\": 0.4816221409322309, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"Pclass\": {\"count\": 418.0, \"mean\": 2.2655502392344498, \"std\": 0.8418375519640503, \"min\": 1.0, \"25%\": 1.0, \"50%\": 3.0, \"75%\": 3.0, \"max\": 3.0}, \"Age\": {\"count\": 332.0, \"mean\": 30.272590361445783, \"std\": 14.181209235624422, \"min\": 0.17, \"25%\": 21.0, \"50%\": 27.0, \"75%\": 39.0, \"max\": 76.0}, \"SibSp\": {\"count\": 418.0, \"mean\": 0.4473684210526316, \"std\": 0.8967595611217135, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 8.0}, \"Parch\": {\"count\": 418.0, \"mean\": 0.3923444976076555, \"std\": 0.9814288785371691, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 9.0}, \"Fare\": {\"count\": 417.0, \"mean\": 35.627188489208635, \"std\": 55.907576179973844, \"min\": 0.0, \"25%\": 7.8958, \"50%\": 14.4542, \"75%\": 31.5, \"max\": 512.3292}}", "examples": "{\"PassengerId\":{\"0\":892,\"1\":893,\"2\":894,\"3\":895},\"Survived\":{\"0\":0,\"1\":1,\"2\":0,\"3\":0},\"Pclass\":{\"0\":3,\"1\":3,\"2\":2,\"3\":3},\"Name\":{\"0\":\"Kelly, Mr. James\",\"1\":\"Wilkes, Mrs. James (Ellen Needs)\",\"2\":\"Myles, Mr. Thomas Francis\",\"3\":\"Wirz, Mr. Albert\"},\"Sex\":{\"0\":\"male\",\"1\":\"female\",\"2\":\"male\",\"3\":\"male\"},\"Age\":{\"0\":34.5,\"1\":47.0,\"2\":62.0,\"3\":27.0},\"SibSp\":{\"0\":0,\"1\":1,\"2\":0,\"3\":0},\"Parch\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Ticket\":{\"0\":\"330911\",\"1\":\"363272\",\"2\":\"240276\",\"3\":\"315154\"},\"Fare\":{\"0\":7.8292,\"1\":7.0,\"2\":9.6875,\"3\":8.6625},\"Cabin\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"Embarked\":{\"0\":\"Q\",\"1\":\"S\",\"2\":\"Q\",\"3\":\"S\"}}"}}]
| true | 1 |
<start_data_description><data_path>test-file/tested.csv:
<column_names>
['PassengerId', 'Survived', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp', 'Parch', 'Ticket', 'Fare', 'Cabin', 'Embarked']
<column_types>
{'PassengerId': 'int64', 'Survived': 'int64', 'Pclass': 'int64', 'Name': 'object', 'Sex': 'object', 'Age': 'float64', 'SibSp': 'int64', 'Parch': 'int64', 'Ticket': 'object', 'Fare': 'float64', 'Cabin': 'object', 'Embarked': 'object'}
<dataframe_Summary>
{'PassengerId': {'count': 418.0, 'mean': 1100.5, 'std': 120.81045760473994, 'min': 892.0, '25%': 996.25, '50%': 1100.5, '75%': 1204.75, 'max': 1309.0}, 'Survived': {'count': 418.0, 'mean': 0.36363636363636365, 'std': 0.4816221409322309, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'Pclass': {'count': 418.0, 'mean': 2.2655502392344498, 'std': 0.8418375519640503, 'min': 1.0, '25%': 1.0, '50%': 3.0, '75%': 3.0, 'max': 3.0}, 'Age': {'count': 332.0, 'mean': 30.272590361445783, 'std': 14.181209235624422, 'min': 0.17, '25%': 21.0, '50%': 27.0, '75%': 39.0, 'max': 76.0}, 'SibSp': {'count': 418.0, 'mean': 0.4473684210526316, 'std': 0.8967595611217135, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 8.0}, 'Parch': {'count': 418.0, 'mean': 0.3923444976076555, 'std': 0.9814288785371691, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 9.0}, 'Fare': {'count': 417.0, 'mean': 35.627188489208635, 'std': 55.907576179973844, 'min': 0.0, '25%': 7.8958, '50%': 14.4542, '75%': 31.5, 'max': 512.3292}}
<dataframe_info>
RangeIndex: 418 entries, 0 to 417
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 418 non-null int64
1 Survived 418 non-null int64
2 Pclass 418 non-null int64
3 Name 418 non-null object
4 Sex 418 non-null object
5 Age 332 non-null float64
6 SibSp 418 non-null int64
7 Parch 418 non-null int64
8 Ticket 418 non-null object
9 Fare 417 non-null float64
10 Cabin 91 non-null object
11 Embarked 418 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 39.3+ KB
<some_examples>
{'PassengerId': {'0': 892, '1': 893, '2': 894, '3': 895}, 'Survived': {'0': 0, '1': 1, '2': 0, '3': 0}, 'Pclass': {'0': 3, '1': 3, '2': 2, '3': 3}, 'Name': {'0': 'Kelly, Mr. James', '1': 'Wilkes, Mrs. James (Ellen Needs)', '2': 'Myles, Mr. Thomas Francis', '3': 'Wirz, Mr. Albert'}, 'Sex': {'0': 'male', '1': 'female', '2': 'male', '3': 'male'}, 'Age': {'0': 34.5, '1': 47.0, '2': 62.0, '3': 27.0}, 'SibSp': {'0': 0, '1': 1, '2': 0, '3': 0}, 'Parch': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Ticket': {'0': '330911', '1': '363272', '2': '240276', '3': '315154'}, 'Fare': {'0': 7.8292, '1': 7.0, '2': 9.6875, '3': 8.6625}, 'Cabin': {'0': None, '1': None, '2': None, '3': None}, 'Embarked': {'0': 'Q', '1': 'S', '2': 'Q', '3': 'S'}}
<end_description>
| 953 | 0 | 1,912 | 953 |
129599172
|
import torch
import torch.nn as nn
import torch.optim as optim
import torchvision.transforms as transforms
import torchvision.datasets as datasets
import matplotlib.pyplot as plt
import cv2
from torch.utils.data import Dataset
import numpy as np
import torchvision.transforms.functional as F
from torch.utils.data.dataset import random_split
import torch.nn.functional as f
from torchvision.datasets import ImageFolder
import random
import tensorflow as tf
from torchvision.datasets import DatasetFolder
from torchvision.datasets.folder import default_loader
import os
from PIL import Image
import torchvision
# Transformers
class RandomCrop(object):
def __init__(self, output_size, pad=4):
assert isinstance(output_size, (int, tuple))
assert isinstance(pad, (int, tuple))
if isinstance(output_size, int):
self.output_size = (output_size, output_size)
else:
assert len(output_size) == 2
self.output_size = output_size
self.pad = pad
def __call__(self, sample):
image, label = np.array(sample[0]), sample[1]
h, w, c = image.shape
new_h, new_w = self.output_size
crop_img = np.zeros([new_h + 2 * self.pad, new_w + 2 * self.pad, c]).astype(
np.uint8
)
resize_img = cv2.resize(image, (new_h, new_w), interpolation=cv2.INTER_CUBIC)
top = np.random.randint(0, 2 * self.pad)
left = np.random.randint(0, 2 * self.pad)
crop_img[top : top + new_h, left : left + new_w] = resize_img
crop_img = crop_img[self.pad : self.pad + new_h, self.pad : self.pad + new_w]
return [crop_img, label]
class RandomHFlip(object):
def __call__(self, sample, p=0.2):
image, label = np.array(sample[0]), sample[1]
flip = (np.random.rand() < p) * 1
flip_image = image * flip + np.fliplr(image) * (1 - flip)
return [flip_image, label]
class ToTensor(object):
def __call__(self, sample):
image, label = np.array(sample[0]), np.array(sample[1])
image = image.transpose((2, 0, 1))
return [torch.from_numpy(image), torch.from_numpy(label)]
class Normalize(object):
def __init__(self, means, stds):
assert isinstance(means, (float, tuple))
assert isinstance(stds, (float, tuple))
self.means = means
self.stds = stds
def __call__(self, sample):
image, label = sample[0], sample[1]
means_ts = torch.tensor(self.means)[:, None, None]
stds_ts = torch.tensor(self.stds)[:, None, None]
image = (image / 255.0 - means_ts) / stds_ts
return [image, label]
class RandomRotate(object):
def __init__(self, degrees):
assert isinstance(degrees, (int, float, tuple))
if isinstance(degrees, (int, float)):
self.degrees = (-degrees, degrees)
else:
assert len(degrees) == 2
self.degrees = degrees
def __call__(self, sample):
image, label = np.array(sample[0]), sample[1]
rotate = random.choice([True, False])
if rotate:
angle_choice = random.choice([True, False])
if angle_choice:
angle = self.degrees[0]
else:
angle = self.degrees[1]
image = self.rotate_image(image, angle)
return [image, label]
@staticmethod
def rotate_image(image, angle):
image = np.array(image)
h, w, _ = image.shape
center = (w / 2, h / 2)
rotation_matrix = cv2.getRotationMatrix2D(center, angle, 1.0)
cos_theta = np.abs(rotation_matrix[0, 0])
sin_theta = np.abs(rotation_matrix[0, 1])
new_w = int(h * sin_theta + w * cos_theta)
new_h = int(h * cos_theta + w * sin_theta)
rotation_matrix[0, 2] += (new_w / 2) - center[0]
rotation_matrix[1, 2] += (new_h / 2) - center[1]
rotated_image = cv2.warpAffine(
image, rotation_matrix, (new_w, new_h), flags=cv2.INTER_LINEAR
)
return rotated_image
class RandomBlur(object):
def __init__(self, kernel_sizes):
assert isinstance(kernel_sizes, (int, tuple))
if isinstance(kernel_sizes, int):
self.kernel_sizes = (kernel_sizes, kernel_sizes)
else:
assert len(kernel_sizes) == 2
self.kernel_sizes = kernel_sizes
def __call__(self, sample):
image, label = np.array(sample[0]), sample[1]
kernel_size = np.random.randint(self.kernel_sizes[0], self.kernel_sizes[1] + 1)
blurred_image = cv2.blur(image, (kernel_size, kernel_size))
return [blurred_image, label]
class ColorJitter(object):
def __init__(self, brightness=0.0):
self.brightness = brightness
def __call__(self, sample):
image, label = np.array(sample[0]), sample[1]
if self.brightness > 0:
brightness_factor = random.uniform(1, 1 + self.brightness)
image = self.adjust_brightness(image, brightness_factor)
return [image, label]
@staticmethod
def adjust_brightness(image, brightness_factor):
image = image.astype(np.float32) / 255.0
image *= brightness_factor
image = np.clip(image, 0.0, 1.0)
image = (image * 255.0).astype(np.uint8)
return image
class myDataset(Dataset):
def __init__(self, dataset, transform=None):
self.transform = transform
self.dataset = dataset
def __len__(self):
return len(self.dataset)
def __getitem__(self, idx):
image = self.dataset[idx][0]
label = self.dataset[idx][1]
if self.transform:
image = self.transform(image)
return image, label
stats = ((0.7786, 0.7310, 0.7049), (0.2234, 0.2732, 0.2731))
from typing import Sequence
class MyRotateTransform:
def __init__(self, angles: Sequence[int]):
self.angles = angles
def __call__(self, x):
angle = random.choice(self.angles)
return F.rotate(x, angle)
train_tfms = transforms.Compose(
[
MyRotateTransform([90, 90, 360]),
transforms.GaussianBlur(7, 3),
transforms.RandomHorizontalFlip(p=0.3),
transforms.ToTensor(),
transforms.Normalize(*stats),
]
)
valid_tfms = transforms.Compose(
[
MyRotateTransform([90, 90, 360]),
transforms.GaussianBlur(7, 3),
transforms.RandomHorizontalFlip(p=0.3),
transforms.ToTensor(),
transforms.Normalize(*stats),
]
)
test_tfms = transforms.Compose([transforms.ToTensor(), transforms.Normalize(*stats)])
def to_device(data, device):
if isinstance(data, (list, tuple)):
return [to_device(x, device) for x in data]
return data.to(device, non_blocking=True)
class DeviceDataLoader:
def __init__(self, dl, device):
self.dl = dl
self.device = device
def __iter__(self):
for b in self.dl:
yield to_device(b, self.device)
def __len__(self):
return len(self.dl)
def get_default_device():
if torch.cuda.is_available():
return torch.device("cuda")
else:
return torch.device("cpu")
# input_size=112*112
num_classes = 14
class FocalLoss(nn.modules.loss._WeightedLoss):
def __init__(self, weight=None, gamma=2, reduction="mean"):
super(FocalLoss, self).__init__(weight, reduction=reduction)
self.gamma = gamma
self.weight = weight
def forward(self, input, target):
ce_loss = F.cross_entropy(
input, target, reduction=self.reduction, weight=self.weight
)
pt = torch.exp(-ce_loss)
focal_loss = (1 - pt) ** self.gamma * ce_loss
if self.weight is not None:
weighted_focal_loss = self.weight * focal_loss
else:
weighted_focal_loss = focal_loss
if self.reduction == "mean":
return torch.mean(weighted_focal_loss)
elif self.reduction == "sum":
return torch.sum(weighted_focal_loss)
class SupportingClass(nn.Module):
def training_step(self, batch):
images, labels = batch
out = self(images)
loss = FocalLoss(weight=None)(out, labels)
return loss, out.detach(), labels.detach()
def accuracy(self, outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(labels))
def validation_step(self, batch):
images, labels = batch
out = self(images)
loss = FocalLoss(weight=None)(out, labels)
acc = self.accuracy(out, labels)
data = [labels, out]
return {"val_loss": loss, "val_acc": acc, "data": data}
def validation_average(self, outputs):
batch_losses = [x["val_loss"] for x in outputs]
epoch_loss = torch.stack(batch_losses).mean()
batch_accs = [x["val_acc"] for x in outputs]
epoch_acc = torch.stack(batch_accs).mean()
return {"val_loss": epoch_loss.item(), "val_acc": epoch_acc.item()}
def evaluate(self, val_loader):
outputs = [self.validation_step(batch) for batch in val_loader]
data = [output["data"] for output in outputs]
return self.validation_average(outputs), data
def print_out(self, epoch, result):
print(
"Epoch[{}], train_loss:{:.4f}, train_acc:{:.4f}, val_loss:{:.4f}, val_acc:{:.4f}".format(
epoch,
result["train_loss"],
result["train_acc"],
result["val_loss"],
result["val_acc"],
)
)
class Cnn2DModel(nn.Module):
def __init__(self):
super(Cnn2DModel, self).__init__()
self.layers = nn.Sequential(
nn.Conv2d(3, 64, kernel_size=3, stride=1, padding=1),
nn.BatchNorm2d(64),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(64, 128, kernel_size=3, stride=1, padding=1),
nn.BatchNorm2d(128),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(128, 256, kernel_size=3, stride=1, padding=1),
nn.BatchNorm2d(256),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Flatten(),
nn.Linear(256 * 28 * 28, 1024),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(1024, 14),
)
def forward(self, x):
x = self.layers(x)
return x
import torchvision.models as models
class ShallowResNetModel(nn.Module):
def __init__(self, num_classes=14):
super(ShallowResNetModel, self).__init__()
self.resnet = models.resnet18(pretrained=True)
num_features = self.resnet.fc.in_features
self.resnet.fc = nn.Linear(num_features, num_classes)
def forward(self, x):
x = self.resnet(x)
return x
def training(train_dl, model, optimizer, util):
model.train()
batch_loss = []
batch_acc = []
for batch in train_dl:
images, labels = batch
outputs = model(images)
loss = util.criterion(outputs, labels)
loss.backward()
optimizer.step()
optimizer.zero_grad()
batch_loss.append(loss.clone().detach().cpu())
batch_acc.append(util.accuracy(outputs, labels))
return torch.stack(batch_loss).mean(), torch.stack(batch_acc).mean()
def validating(val_dl, model, util):
model.eval()
batch_loss = []
batch_acc = []
for batch in val_dl:
images, labels = batch
outputs = model(images)
loss = util.criterion(outputs, labels)
batch_loss.append(loss.clone().detach().cpu())
batch_acc.append(util.accuracy(outputs, labels))
return torch.stack(batch_loss).mean(), torch.stack(batch_acc).mean()
class calculate:
def criterion(self, preds, labels):
loss = f.cross_entropy(preds, labels)
return loss
def accuracy(self, outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(preds))
def evaluate(self, val_loader):
outputs = [self.validation_step(batch) for batch in val_loader]
data = [output["data"] for output in outputs]
return self.validation_average(outputs), data
def print_all(self, epoch, result):
print(
"Epoch[{}], train_loss:{:.4f}, train_acc:{:.4f}, val_loss:{:.4f}, val_acc:{:.4f}".format(
epoch,
result["train_loss"][-1],
result["train_accu"][-1],
result["valid_loss"][-1],
result["valid_accu"][-1],
)
)
def testing(test_d, model, util):
model.eval()
batch_pred_prob = []
batch_pred_label = []
batch_label = []
with torch.no_grad():
for batch in test_d:
(
images,
labels,
) = batch
outputs = model(images)
pred_prob, pred_label = F.softmax(outputs, dim=1).max(1)
batch_pred_prob.append(pred_prob.cpu())
batch_pred_label.append(pred_label.cpu())
batch_label.append(labels.cpu())
return torch.cat(batch_label).numpy(), torch.cat(batch_pred_label).numpy()
def print_all(epoch, lr, result):
msg1 = "Epoch[{}],".format(epoch)
msg2 = "lr:{:.6f},".format(lr)
msg3 = "train_loss:{:.6f},train_acc:{:.6f},val_loss:{:.6f},val_acc:{:.6f}".format(
result["train_loss"][-1],
result["train_accu"][-1],
result["valid_loss"][-1],
result["valid_accu"][-1],
)
print(msg1 + msg2 + msg3)
def get_lr(optimizer):
for param_group in optimizer.param_groups:
return param_group["lr"]
def fit(
epochs, max_lr, model, train_dl, val_dl, weight_decay=0, opt_func=torch.optim.SGD
):
tuned_optimizer = opt_func(model.parameters(), 0.001, weight_decay=weight_decay)
sched = torch.optim.lr_scheduler.OneCycleLR(
tuned_optimizer, max_lr, epochs=epochs, steps_per_epoch=1
)
util = calculate()
lrs = []
result = {}
result["train_loss"] = []
result["train_accu"] = []
result["valid_loss"] = []
result["valid_accu"] = []
for epoch in range(epochs):
train_loss, train_accu = training(train_dl, model, tuned_optimizer, util)
valid_loss, valid_accu = validating(val_dl, model, util)
sched.step()
result["train_loss"].append(train_loss)
result["train_accu"].append(train_accu)
result["valid_loss"].append(valid_loss)
result["valid_accu"].append(valid_accu)
print_all(epoch, get_lr(tuned_optimizer), result)
return result
batch_size = 64
data_dir = "/kaggle/working/Card Classified(13) - Studend/Train Dataset"
dataset = datasets.ImageFolder(data_dir)
print("dataset size:", len(dataset))
train_size = 6000
valid_size = len(dataset) - train_size
torch.manual_seed(413) # seed
train_ds, valid_ds = random_split(dataset, [train_size, valid_size])
train_ds = myDataset(train_ds, transform=train_tfms)
valid_ds = myDataset(valid_ds, transform=valid_tfms)
train_loader = torch.utils.data.DataLoader(
train_ds, batch_size=batch_size, shuffle=True
)
val_loader = torch.utils.data.DataLoader(valid_ds, batch_size=batch_size)
device = get_default_device()
print(device)
labels = []
for idx in range(len(dataset)):
label = dataset[idx][1]
labels.append(label)
print(labels)
import os
import re
class MyTestDataset(Dataset):
def __init__(self, root_dir, transform=None):
self.root_dir = root_dir
self.transform = transform
self.image_paths = self._collect_image_paths()
def __len__(self):
return len(self.image_paths)
def __getitem__(self, idx):
img_path = self.image_paths[idx]
image = Image.open(img_path).convert("RGB")
if self.transform:
image = self.transform(image)
return image, img_path
def _collect_image_paths(self):
image_paths = []
for filename in os.listdir(self.root_dir):
if filename.endswith(".jpg") and filename.startswith("test_"):
img_path = os.path.join(self.root_dir, filename)
image_paths.append(img_path)
# Sort the image paths based on the number in the filename
image_paths = sorted(
image_paths, key=lambda x: int(re.findall(r"\d+", os.path.basename(x))[0])
)
return image_paths
test_dir = "/kaggle/working/Card Classified(13) - Studend/Test/Test Dataset"
test_ds = MyTestDataset(test_dir, transform=test_tfms)
test_loader = torch.utils.data.DataLoader(test_ds, batch_size=batch_size, shuffle=False)
train_dl = DeviceDataLoader(train_loader, device)
val_dl = DeviceDataLoader(val_loader, device)
test_dl = DeviceDataLoader(test_loader, device)
# cnt = 0
# for images, labels in test_loader:
# # Iterate over each image in the batch
# for image in images:
# # Display the image
# plt.imshow(image.permute(1, 2, 0))
# plt.show()
# break
# from google.colab import drive
# drive.mount('/content/drive')
# mean = 0.0
# std = 0.0
# for image, _ in dataset:
# image = transforms.ToTensor()(image)
# mean += torch.mean(image, dim=(1, 2))
# std += torch.std(image, dim=(1, 2))
# mean /= len(dataset)
# std /= len(dataset)
# print("Mean:", mean)
# print("Std:", std)
label_names = dataset.classes
for batch_idx, (images, labels) in enumerate(train_loader):
if batch_idx >= 1:
break
print("image size:", images.shape)
images1D = images.view(64, 3, -1)
print("imgae1D:", images1D.shape)
out = Cnn2DModel().forward(images)
print("Output1D shape:", out.shape)
for i in range(batch_size):
if i >= 20:
break
image = images[i]
label = labels[i]
label_name = label_names[label]
print(f"Batch {batch_idx}, Image {i}: Label = {label_name}")
plt.imshow(image.permute(1, 2, 0))
plt.show()
modelc = ShallowResNetModel()
modelc = to_device(modelc, device)
# print("Model info:",modelc.layers)
epoch = 45
max_lr = 0.004
weight_decay = 0.0003
optimizer = torch.optim.Adam
import time
t1 = time.perf_counter()
results1 = fit(
epoch, max_lr, modelc, train_dl, val_dl, weight_decay, opt_func=optimizer
)
t2 = time.perf_counter()
print("Time taken for training:{:.2f}s".format(t2 - t1))
# epoch = 50
# max_lr = 0.005
# weight_decay =1e-4
# optimizer =torch.optim.Adam
# import time
# t1 = time.perf_counter()
# results2 = fit(epoch,max_lr,modelc,train_dl,test_dl,weight_decay,opt_func=optimizer)
# t2 = time.perf_counter()
# print('Time taken for training:{:.2f}s'.format(t2-t1))
# confusion_matrix
from sklearn import metrics
from sklearn.metrics import multilabel_confusion_matrix
import matplotlib.pyplot as plt
def performance(model, test_d):
util = calculate()
labels, preds = testing(test_d, model, util)
cm = metrics.confusion(test_d, model, util)
accuracy = metrics.accuracy_score(labels, preds)
precision = metrics.precision_score(labels, preds, preds, average="macro")
recall = metrics.recall_score(labels, preds, preds, average="macro")
F1_score = metrics.f1_score(labels, preds, average="macro")
print("Accuracy:", accuracy)
print("precision", precision)
print("Recall", recall)
print("F1_score", F1_score)
cm_plot = metrics.ConfusionMatrixDisplay(
confusion_matrix=cm,
display_labels=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13],
)
cm_plot.plot()
plt.show()
performance(modelc, val_loader)
train_loss = results1["train_loss"]
val_loss = results1["valid_loss"]
plt.subplot(2, 1, 1)
plt.plot(train_loss, "-o", label="train_loss")
plt.plot(val_loss, "-x", label="val_loss")
plt.xlabel("epoch")
plt.ylabel("loss")
plt.legend()
plt.title("Loss vs. Number of epochs")
train_acc = results1["train_accu"]
val_acc = results1["valid_accu"]
plt.subplot(2, 1, 2)
plt.plot(train_acc, "-o", label="train_acc")
plt.plot(val_acc, "-x", label="val_acc")
plt.xlabel("epoch")
plt.ylabel("loss")
plt.legend()
plt.title("Accuracy vs. Number of epochs")
plt.tight_layout()
pred = []
modelc.eval()
with torch.no_grad():
for images, _ in test_loader:
images = images.cuda()
outputs = modelc(images)
_, test_pred = torch.max(outputs, 1)
pred.extend(test_pred.cpu().numpy().tolist())
pred = np.array(pred, dtype=np.int32)
# file_names = os.listdir("/content/Card Classified(13) - Studend/Test/Test Dataset")
# file_names.sort()
with open("p014.csv", "w") as f:
f.write("img_name,Label\n")
for i, y in enumerate(pred):
# img_name = file_names[i]
f.write("test_{}.jpg,{}\n".format(i + 1, y))
import csv
import os
# 读取CSV文件并获取文件名列表
with open("p014.csv", newline="") as csvfile:
reader = csv.reader(csvfile)
rows = list(reader)
# 获取文件名列表
filenames = rows[1:] # 从第二行开始获取文件名列表
# 提取文件名中的数字并进行排序
sorted_filenames = sorted(
filenames, key=lambda x: int(os.path.splitext(x[0])[0].split("_")[1])
)
# 将排好序的文件名写回CSV文件
with open("p014.csv", "w", newline="\n") as csvfile:
writer = csv.writer(csvfile)
writer.writerow(rows[0]) # 写回CSV文件的第一行(表头)
writer.writerows(sorted_filenames) # 写回排序后的文件名列表
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/599/129599172.ipynb
| null | null |
[{"Id": 129599172, "ScriptId": 38535614, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13311524, "CreationDate": "05/15/2023 06:35:19", "VersionNumber": 1.0, "Title": "test_v2", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 663.0, "LinesInsertedFromPrevious": 663.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import torch
import torch.nn as nn
import torch.optim as optim
import torchvision.transforms as transforms
import torchvision.datasets as datasets
import matplotlib.pyplot as plt
import cv2
from torch.utils.data import Dataset
import numpy as np
import torchvision.transforms.functional as F
from torch.utils.data.dataset import random_split
import torch.nn.functional as f
from torchvision.datasets import ImageFolder
import random
import tensorflow as tf
from torchvision.datasets import DatasetFolder
from torchvision.datasets.folder import default_loader
import os
from PIL import Image
import torchvision
# Transformers
class RandomCrop(object):
def __init__(self, output_size, pad=4):
assert isinstance(output_size, (int, tuple))
assert isinstance(pad, (int, tuple))
if isinstance(output_size, int):
self.output_size = (output_size, output_size)
else:
assert len(output_size) == 2
self.output_size = output_size
self.pad = pad
def __call__(self, sample):
image, label = np.array(sample[0]), sample[1]
h, w, c = image.shape
new_h, new_w = self.output_size
crop_img = np.zeros([new_h + 2 * self.pad, new_w + 2 * self.pad, c]).astype(
np.uint8
)
resize_img = cv2.resize(image, (new_h, new_w), interpolation=cv2.INTER_CUBIC)
top = np.random.randint(0, 2 * self.pad)
left = np.random.randint(0, 2 * self.pad)
crop_img[top : top + new_h, left : left + new_w] = resize_img
crop_img = crop_img[self.pad : self.pad + new_h, self.pad : self.pad + new_w]
return [crop_img, label]
class RandomHFlip(object):
def __call__(self, sample, p=0.2):
image, label = np.array(sample[0]), sample[1]
flip = (np.random.rand() < p) * 1
flip_image = image * flip + np.fliplr(image) * (1 - flip)
return [flip_image, label]
class ToTensor(object):
def __call__(self, sample):
image, label = np.array(sample[0]), np.array(sample[1])
image = image.transpose((2, 0, 1))
return [torch.from_numpy(image), torch.from_numpy(label)]
class Normalize(object):
def __init__(self, means, stds):
assert isinstance(means, (float, tuple))
assert isinstance(stds, (float, tuple))
self.means = means
self.stds = stds
def __call__(self, sample):
image, label = sample[0], sample[1]
means_ts = torch.tensor(self.means)[:, None, None]
stds_ts = torch.tensor(self.stds)[:, None, None]
image = (image / 255.0 - means_ts) / stds_ts
return [image, label]
class RandomRotate(object):
def __init__(self, degrees):
assert isinstance(degrees, (int, float, tuple))
if isinstance(degrees, (int, float)):
self.degrees = (-degrees, degrees)
else:
assert len(degrees) == 2
self.degrees = degrees
def __call__(self, sample):
image, label = np.array(sample[0]), sample[1]
rotate = random.choice([True, False])
if rotate:
angle_choice = random.choice([True, False])
if angle_choice:
angle = self.degrees[0]
else:
angle = self.degrees[1]
image = self.rotate_image(image, angle)
return [image, label]
@staticmethod
def rotate_image(image, angle):
image = np.array(image)
h, w, _ = image.shape
center = (w / 2, h / 2)
rotation_matrix = cv2.getRotationMatrix2D(center, angle, 1.0)
cos_theta = np.abs(rotation_matrix[0, 0])
sin_theta = np.abs(rotation_matrix[0, 1])
new_w = int(h * sin_theta + w * cos_theta)
new_h = int(h * cos_theta + w * sin_theta)
rotation_matrix[0, 2] += (new_w / 2) - center[0]
rotation_matrix[1, 2] += (new_h / 2) - center[1]
rotated_image = cv2.warpAffine(
image, rotation_matrix, (new_w, new_h), flags=cv2.INTER_LINEAR
)
return rotated_image
class RandomBlur(object):
def __init__(self, kernel_sizes):
assert isinstance(kernel_sizes, (int, tuple))
if isinstance(kernel_sizes, int):
self.kernel_sizes = (kernel_sizes, kernel_sizes)
else:
assert len(kernel_sizes) == 2
self.kernel_sizes = kernel_sizes
def __call__(self, sample):
image, label = np.array(sample[0]), sample[1]
kernel_size = np.random.randint(self.kernel_sizes[0], self.kernel_sizes[1] + 1)
blurred_image = cv2.blur(image, (kernel_size, kernel_size))
return [blurred_image, label]
class ColorJitter(object):
def __init__(self, brightness=0.0):
self.brightness = brightness
def __call__(self, sample):
image, label = np.array(sample[0]), sample[1]
if self.brightness > 0:
brightness_factor = random.uniform(1, 1 + self.brightness)
image = self.adjust_brightness(image, brightness_factor)
return [image, label]
@staticmethod
def adjust_brightness(image, brightness_factor):
image = image.astype(np.float32) / 255.0
image *= brightness_factor
image = np.clip(image, 0.0, 1.0)
image = (image * 255.0).astype(np.uint8)
return image
class myDataset(Dataset):
def __init__(self, dataset, transform=None):
self.transform = transform
self.dataset = dataset
def __len__(self):
return len(self.dataset)
def __getitem__(self, idx):
image = self.dataset[idx][0]
label = self.dataset[idx][1]
if self.transform:
image = self.transform(image)
return image, label
stats = ((0.7786, 0.7310, 0.7049), (0.2234, 0.2732, 0.2731))
from typing import Sequence
class MyRotateTransform:
def __init__(self, angles: Sequence[int]):
self.angles = angles
def __call__(self, x):
angle = random.choice(self.angles)
return F.rotate(x, angle)
train_tfms = transforms.Compose(
[
MyRotateTransform([90, 90, 360]),
transforms.GaussianBlur(7, 3),
transforms.RandomHorizontalFlip(p=0.3),
transforms.ToTensor(),
transforms.Normalize(*stats),
]
)
valid_tfms = transforms.Compose(
[
MyRotateTransform([90, 90, 360]),
transforms.GaussianBlur(7, 3),
transforms.RandomHorizontalFlip(p=0.3),
transforms.ToTensor(),
transforms.Normalize(*stats),
]
)
test_tfms = transforms.Compose([transforms.ToTensor(), transforms.Normalize(*stats)])
def to_device(data, device):
if isinstance(data, (list, tuple)):
return [to_device(x, device) for x in data]
return data.to(device, non_blocking=True)
class DeviceDataLoader:
def __init__(self, dl, device):
self.dl = dl
self.device = device
def __iter__(self):
for b in self.dl:
yield to_device(b, self.device)
def __len__(self):
return len(self.dl)
def get_default_device():
if torch.cuda.is_available():
return torch.device("cuda")
else:
return torch.device("cpu")
# input_size=112*112
num_classes = 14
class FocalLoss(nn.modules.loss._WeightedLoss):
def __init__(self, weight=None, gamma=2, reduction="mean"):
super(FocalLoss, self).__init__(weight, reduction=reduction)
self.gamma = gamma
self.weight = weight
def forward(self, input, target):
ce_loss = F.cross_entropy(
input, target, reduction=self.reduction, weight=self.weight
)
pt = torch.exp(-ce_loss)
focal_loss = (1 - pt) ** self.gamma * ce_loss
if self.weight is not None:
weighted_focal_loss = self.weight * focal_loss
else:
weighted_focal_loss = focal_loss
if self.reduction == "mean":
return torch.mean(weighted_focal_loss)
elif self.reduction == "sum":
return torch.sum(weighted_focal_loss)
class SupportingClass(nn.Module):
def training_step(self, batch):
images, labels = batch
out = self(images)
loss = FocalLoss(weight=None)(out, labels)
return loss, out.detach(), labels.detach()
def accuracy(self, outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(labels))
def validation_step(self, batch):
images, labels = batch
out = self(images)
loss = FocalLoss(weight=None)(out, labels)
acc = self.accuracy(out, labels)
data = [labels, out]
return {"val_loss": loss, "val_acc": acc, "data": data}
def validation_average(self, outputs):
batch_losses = [x["val_loss"] for x in outputs]
epoch_loss = torch.stack(batch_losses).mean()
batch_accs = [x["val_acc"] for x in outputs]
epoch_acc = torch.stack(batch_accs).mean()
return {"val_loss": epoch_loss.item(), "val_acc": epoch_acc.item()}
def evaluate(self, val_loader):
outputs = [self.validation_step(batch) for batch in val_loader]
data = [output["data"] for output in outputs]
return self.validation_average(outputs), data
def print_out(self, epoch, result):
print(
"Epoch[{}], train_loss:{:.4f}, train_acc:{:.4f}, val_loss:{:.4f}, val_acc:{:.4f}".format(
epoch,
result["train_loss"],
result["train_acc"],
result["val_loss"],
result["val_acc"],
)
)
class Cnn2DModel(nn.Module):
def __init__(self):
super(Cnn2DModel, self).__init__()
self.layers = nn.Sequential(
nn.Conv2d(3, 64, kernel_size=3, stride=1, padding=1),
nn.BatchNorm2d(64),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(64, 128, kernel_size=3, stride=1, padding=1),
nn.BatchNorm2d(128),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(128, 256, kernel_size=3, stride=1, padding=1),
nn.BatchNorm2d(256),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Flatten(),
nn.Linear(256 * 28 * 28, 1024),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(1024, 14),
)
def forward(self, x):
x = self.layers(x)
return x
import torchvision.models as models
class ShallowResNetModel(nn.Module):
def __init__(self, num_classes=14):
super(ShallowResNetModel, self).__init__()
self.resnet = models.resnet18(pretrained=True)
num_features = self.resnet.fc.in_features
self.resnet.fc = nn.Linear(num_features, num_classes)
def forward(self, x):
x = self.resnet(x)
return x
def training(train_dl, model, optimizer, util):
model.train()
batch_loss = []
batch_acc = []
for batch in train_dl:
images, labels = batch
outputs = model(images)
loss = util.criterion(outputs, labels)
loss.backward()
optimizer.step()
optimizer.zero_grad()
batch_loss.append(loss.clone().detach().cpu())
batch_acc.append(util.accuracy(outputs, labels))
return torch.stack(batch_loss).mean(), torch.stack(batch_acc).mean()
def validating(val_dl, model, util):
model.eval()
batch_loss = []
batch_acc = []
for batch in val_dl:
images, labels = batch
outputs = model(images)
loss = util.criterion(outputs, labels)
batch_loss.append(loss.clone().detach().cpu())
batch_acc.append(util.accuracy(outputs, labels))
return torch.stack(batch_loss).mean(), torch.stack(batch_acc).mean()
class calculate:
def criterion(self, preds, labels):
loss = f.cross_entropy(preds, labels)
return loss
def accuracy(self, outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(preds))
def evaluate(self, val_loader):
outputs = [self.validation_step(batch) for batch in val_loader]
data = [output["data"] for output in outputs]
return self.validation_average(outputs), data
def print_all(self, epoch, result):
print(
"Epoch[{}], train_loss:{:.4f}, train_acc:{:.4f}, val_loss:{:.4f}, val_acc:{:.4f}".format(
epoch,
result["train_loss"][-1],
result["train_accu"][-1],
result["valid_loss"][-1],
result["valid_accu"][-1],
)
)
def testing(test_d, model, util):
model.eval()
batch_pred_prob = []
batch_pred_label = []
batch_label = []
with torch.no_grad():
for batch in test_d:
(
images,
labels,
) = batch
outputs = model(images)
pred_prob, pred_label = F.softmax(outputs, dim=1).max(1)
batch_pred_prob.append(pred_prob.cpu())
batch_pred_label.append(pred_label.cpu())
batch_label.append(labels.cpu())
return torch.cat(batch_label).numpy(), torch.cat(batch_pred_label).numpy()
def print_all(epoch, lr, result):
msg1 = "Epoch[{}],".format(epoch)
msg2 = "lr:{:.6f},".format(lr)
msg3 = "train_loss:{:.6f},train_acc:{:.6f},val_loss:{:.6f},val_acc:{:.6f}".format(
result["train_loss"][-1],
result["train_accu"][-1],
result["valid_loss"][-1],
result["valid_accu"][-1],
)
print(msg1 + msg2 + msg3)
def get_lr(optimizer):
for param_group in optimizer.param_groups:
return param_group["lr"]
def fit(
epochs, max_lr, model, train_dl, val_dl, weight_decay=0, opt_func=torch.optim.SGD
):
tuned_optimizer = opt_func(model.parameters(), 0.001, weight_decay=weight_decay)
sched = torch.optim.lr_scheduler.OneCycleLR(
tuned_optimizer, max_lr, epochs=epochs, steps_per_epoch=1
)
util = calculate()
lrs = []
result = {}
result["train_loss"] = []
result["train_accu"] = []
result["valid_loss"] = []
result["valid_accu"] = []
for epoch in range(epochs):
train_loss, train_accu = training(train_dl, model, tuned_optimizer, util)
valid_loss, valid_accu = validating(val_dl, model, util)
sched.step()
result["train_loss"].append(train_loss)
result["train_accu"].append(train_accu)
result["valid_loss"].append(valid_loss)
result["valid_accu"].append(valid_accu)
print_all(epoch, get_lr(tuned_optimizer), result)
return result
batch_size = 64
data_dir = "/kaggle/working/Card Classified(13) - Studend/Train Dataset"
dataset = datasets.ImageFolder(data_dir)
print("dataset size:", len(dataset))
train_size = 6000
valid_size = len(dataset) - train_size
torch.manual_seed(413) # seed
train_ds, valid_ds = random_split(dataset, [train_size, valid_size])
train_ds = myDataset(train_ds, transform=train_tfms)
valid_ds = myDataset(valid_ds, transform=valid_tfms)
train_loader = torch.utils.data.DataLoader(
train_ds, batch_size=batch_size, shuffle=True
)
val_loader = torch.utils.data.DataLoader(valid_ds, batch_size=batch_size)
device = get_default_device()
print(device)
labels = []
for idx in range(len(dataset)):
label = dataset[idx][1]
labels.append(label)
print(labels)
import os
import re
class MyTestDataset(Dataset):
def __init__(self, root_dir, transform=None):
self.root_dir = root_dir
self.transform = transform
self.image_paths = self._collect_image_paths()
def __len__(self):
return len(self.image_paths)
def __getitem__(self, idx):
img_path = self.image_paths[idx]
image = Image.open(img_path).convert("RGB")
if self.transform:
image = self.transform(image)
return image, img_path
def _collect_image_paths(self):
image_paths = []
for filename in os.listdir(self.root_dir):
if filename.endswith(".jpg") and filename.startswith("test_"):
img_path = os.path.join(self.root_dir, filename)
image_paths.append(img_path)
# Sort the image paths based on the number in the filename
image_paths = sorted(
image_paths, key=lambda x: int(re.findall(r"\d+", os.path.basename(x))[0])
)
return image_paths
test_dir = "/kaggle/working/Card Classified(13) - Studend/Test/Test Dataset"
test_ds = MyTestDataset(test_dir, transform=test_tfms)
test_loader = torch.utils.data.DataLoader(test_ds, batch_size=batch_size, shuffle=False)
train_dl = DeviceDataLoader(train_loader, device)
val_dl = DeviceDataLoader(val_loader, device)
test_dl = DeviceDataLoader(test_loader, device)
# cnt = 0
# for images, labels in test_loader:
# # Iterate over each image in the batch
# for image in images:
# # Display the image
# plt.imshow(image.permute(1, 2, 0))
# plt.show()
# break
# from google.colab import drive
# drive.mount('/content/drive')
# mean = 0.0
# std = 0.0
# for image, _ in dataset:
# image = transforms.ToTensor()(image)
# mean += torch.mean(image, dim=(1, 2))
# std += torch.std(image, dim=(1, 2))
# mean /= len(dataset)
# std /= len(dataset)
# print("Mean:", mean)
# print("Std:", std)
label_names = dataset.classes
for batch_idx, (images, labels) in enumerate(train_loader):
if batch_idx >= 1:
break
print("image size:", images.shape)
images1D = images.view(64, 3, -1)
print("imgae1D:", images1D.shape)
out = Cnn2DModel().forward(images)
print("Output1D shape:", out.shape)
for i in range(batch_size):
if i >= 20:
break
image = images[i]
label = labels[i]
label_name = label_names[label]
print(f"Batch {batch_idx}, Image {i}: Label = {label_name}")
plt.imshow(image.permute(1, 2, 0))
plt.show()
modelc = ShallowResNetModel()
modelc = to_device(modelc, device)
# print("Model info:",modelc.layers)
epoch = 45
max_lr = 0.004
weight_decay = 0.0003
optimizer = torch.optim.Adam
import time
t1 = time.perf_counter()
results1 = fit(
epoch, max_lr, modelc, train_dl, val_dl, weight_decay, opt_func=optimizer
)
t2 = time.perf_counter()
print("Time taken for training:{:.2f}s".format(t2 - t1))
# epoch = 50
# max_lr = 0.005
# weight_decay =1e-4
# optimizer =torch.optim.Adam
# import time
# t1 = time.perf_counter()
# results2 = fit(epoch,max_lr,modelc,train_dl,test_dl,weight_decay,opt_func=optimizer)
# t2 = time.perf_counter()
# print('Time taken for training:{:.2f}s'.format(t2-t1))
# confusion_matrix
from sklearn import metrics
from sklearn.metrics import multilabel_confusion_matrix
import matplotlib.pyplot as plt
def performance(model, test_d):
util = calculate()
labels, preds = testing(test_d, model, util)
cm = metrics.confusion(test_d, model, util)
accuracy = metrics.accuracy_score(labels, preds)
precision = metrics.precision_score(labels, preds, preds, average="macro")
recall = metrics.recall_score(labels, preds, preds, average="macro")
F1_score = metrics.f1_score(labels, preds, average="macro")
print("Accuracy:", accuracy)
print("precision", precision)
print("Recall", recall)
print("F1_score", F1_score)
cm_plot = metrics.ConfusionMatrixDisplay(
confusion_matrix=cm,
display_labels=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13],
)
cm_plot.plot()
plt.show()
performance(modelc, val_loader)
train_loss = results1["train_loss"]
val_loss = results1["valid_loss"]
plt.subplot(2, 1, 1)
plt.plot(train_loss, "-o", label="train_loss")
plt.plot(val_loss, "-x", label="val_loss")
plt.xlabel("epoch")
plt.ylabel("loss")
plt.legend()
plt.title("Loss vs. Number of epochs")
train_acc = results1["train_accu"]
val_acc = results1["valid_accu"]
plt.subplot(2, 1, 2)
plt.plot(train_acc, "-o", label="train_acc")
plt.plot(val_acc, "-x", label="val_acc")
plt.xlabel("epoch")
plt.ylabel("loss")
plt.legend()
plt.title("Accuracy vs. Number of epochs")
plt.tight_layout()
pred = []
modelc.eval()
with torch.no_grad():
for images, _ in test_loader:
images = images.cuda()
outputs = modelc(images)
_, test_pred = torch.max(outputs, 1)
pred.extend(test_pred.cpu().numpy().tolist())
pred = np.array(pred, dtype=np.int32)
# file_names = os.listdir("/content/Card Classified(13) - Studend/Test/Test Dataset")
# file_names.sort()
with open("p014.csv", "w") as f:
f.write("img_name,Label\n")
for i, y in enumerate(pred):
# img_name = file_names[i]
f.write("test_{}.jpg,{}\n".format(i + 1, y))
import csv
import os
# 读取CSV文件并获取文件名列表
with open("p014.csv", newline="") as csvfile:
reader = csv.reader(csvfile)
rows = list(reader)
# 获取文件名列表
filenames = rows[1:] # 从第二行开始获取文件名列表
# 提取文件名中的数字并进行排序
sorted_filenames = sorted(
filenames, key=lambda x: int(os.path.splitext(x[0])[0].split("_")[1])
)
# 将排好序的文件名写回CSV文件
with open("p014.csv", "w", newline="\n") as csvfile:
writer = csv.writer(csvfile)
writer.writerow(rows[0]) # 写回CSV文件的第一行(表头)
writer.writerows(sorted_filenames) # 写回排序后的文件名列表
| false | 0 | 6,479 | 0 | 6,479 | 6,479 |
||
129599584
|
# # 1.1 Парсинг данных
# Подключение библиотек
import pandas as pd
import json
import docx
import requests
from bs4 import BeautifulSoup as bs
import glob
import codecs
from tqdm.auto import tqdm
import matplotlib.pyplot as plt
import seaborn as sns
# ### Парсинг docx файла
# открытие файла
condidates_docx = docx.Document("Condidates.docx")
# парсинг данных
condidates = []
text_condidates_docx = condidates_docx.paragraphs
for con in text_condidates_docx:
# проверка длинны параграфа, для избежания добавления пустых значений
if len(con.text) > 0:
condidates.append(con.text.strip().lower())
print(condidates)
print(len(condidates))
# В docx документе предоставлены 22 номинанта, удалены лишние пробелы и приведен текст в нижний регистр, для стандартизации
# ### Парсинг json файлов
all_file = glob.glob(r"Data\*.json")
print(all_file)
print(len(all_file))
norm_month = [
"января",
"февраля",
"марта",
"апреля",
"мая",
"июня",
"июля",
"августа",
"сентября",
"октября",
"ноября",
"декабря",
]
def norm_date(day, month, year=2021):
try:
day = int(day)
except:
day = 1
if day < 10:
day = "0{0}".format(day)
if month in norm_month:
month = norm_month.index(month) + 1
else:
month = 1
if month < 10:
month = "0{0}".format(month)
return int("{0}{1}{2}".format(year, month, day))
# парсинг данных компаний и статей
about_condidates_json = {
"name_company": [],
"description": [],
"activity": [],
"rating": [],
}
about_articles_json = {"name_company": [], "text": [], "date": []}
brack_articles = []
brack_read = []
are_articles = []
nan_info = []
for file in all_file:
# разбитие и срез строки для отделения имени компании
name = file.split("\\")[-1][:-5].lower()
try:
# открытие файла
with codecs.open(file, "r", "utf_8_sig") as f:
templates = json.load(f)
# считывание информации о компаниях
if templates["info"] != None:
about_condidates_json["name_company"].append(name)
about_condidates_json["description"].append(templates["info"]["about"])
about_condidates_json["activity"].append(
templates["info"]["industries"]
)
about_condidates_json["rating"].append(templates["info"]["rate"])
else:
nan_info.append(name)
if len(templates["refs"]) > 0:
are_articles.append(name)
# считывание информации о статьях
for art in templates["refs"]:
try:
about_articles_json["text"].append(art[0])
about_articles_json["date"].append(
norm_date(art[1]["day"], art[1]["month"])
)
about_articles_json["name_company"].append(name)
except:
brack_articles.append(name)
except:
brack_read.append(name)
print("Документы json, которые не считались: ", brack_read)
print("_________")
print("Нет информации о компаниях: ", nan_info)
print("_________")
print("Есть информация о кампании: ", about_condidates_json["name_company"])
print("_________")
print("У компании есть статьи: ", are_articles)
print("_________")
print("Статья не считана: ", brack_articles)
print("_________")
print("Количество статей", len(about_articles_json["name_company"]))
compani_json_df = pd.DataFrame(about_condidates_json)
compani_json_df
articles_json_df = pd.DataFrame(about_articles_json)
articles_json_df
# Все 14 json прочитаны верно, у ряда компаний отсутствует информация, есть только у двух компаний. Статьи есть не у всех компаний, всего статей 1112, есть 4 статьи, которые не считались. Информацию о компаниях и их статьях нужно дополнить с веб ресурса Habr.
# ### Парсинг веб ресурса Habr
# парсинг описание компаний
def activity_condidates(soup_new):
a = soup_new.find("a", class_="tm-company-snippet__title")
url_condidate = "https://habr.com" + a["href"]
page_condidate = requests.get(url_condidate)
soup_condidate = bs(page_condidate.text, "html.parser")
div = soup_condidate.find("div", class_="tm-company-profile__categories")
a1 = div.find_all("a")
activity = []
for i in a1:
activity.append(i.text.strip().lower())
activity = " ".join(activity)
return activity
# парсинг компаний
about_condidates_habr = {
"name_company": [],
"description": [],
"activity": [],
"rating": [],
}
for con in condidates:
con.replace(" ", "20%")
# создание ссылки
url = (
"https://habr.com/ru/search/?q="
+ con
+ "&target_type=companies&order=relevance"
)
page = requests.get(url)
soup = bs(page.text, "html.parser")
condidate_div = soup.find_all("em", class_="searched-item")
# заполнение данных
if len(condidate_div) > 0:
if condidate_div[0].text.lower() == con:
about_condidates_habr["name_company"].append(con)
description_div = soup.find("div", class_="tm-company-snippet__description")
about_condidates_habr["description"].append(description_div.text)
about_condidates_habr["activity"].append(activity_condidates(soup))
rating_span = soup.find(
"span",
class_="tm-search-companies__score-counter tm-search-companies__score-counter_rating",
)
about_condidates_habr["rating"].append(rating_span.text.split()[1])
compani_habr_df = pd.DataFrame(about_condidates_habr)
compani_habr_df
# парсинг статей
about_articles_habr = {"name_company": [], "text": [], "date": []}
for con in tqdm(condidates):
if con not in are_articles:
con = con.replace(" ", "%20")
for i in range(1, 5):
# создание ссылки
uri_page = "https://habr.com/ru/search/page{1}/?q={0}&target_type=posts&order=relevance".format(
con, i
)
page = requests.get(uri_page)
soup = bs(page.text, "html.parser")
snippetsoup_name = soup.find_all("h2", class_="tm-title tm-title_h2")
for i in snippetsoup_name:
try:
uri_article = "https://habr.com" + str(i.a.get("href"))
page_article = requests.get(uri_article)
soup_article = bs(page_article.text, "html.parser")
date = soup_article.find(
"span", class_="tm-article-datetime-published"
)
# две версии верстки, отбор данных статей
try:
note = soup_article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-1",
)
about_articles_habr["text"].append(
note.div.text.replace("\n", " ")
.replace("\r", " ")
.replace(" ", " ")
.replace("\xa0", " ")
)
about_articles_habr["name_company"].append(
con.replace("%20", " ")
)
about_articles_habr["date"].append(
date.time.get("title").split(",")[0]
)
except:
try:
note = soup_article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-2",
)
about_articles_habr["text"].append(
note.div.text.replace("\n", " ")
.replace("\r", " ")
.replace(" ", " ")
.replace("\xa0", " ")
)
about_articles_habr["name_company"].append(
con.replace("%20", " ")
)
about_articles_habr["date"].append(
date.time.get("title").split(",")[0]
)
except:
print(uri_article)
except:
print(uri_article)
print("_________")
print("Количество статей", len(about_articles_habr["name_company"]))
articles_habr_df = pd.DataFrame(about_articles_habr)
articles_habr_df
# Получены данные компании с сайта, две (skillbox, иннотех) из них совпадают с уже имеющимися. Считаю, что с сайта более актуальные данные. Получили статьи тех компаний, что не представлены в json файлах.
# ### Объединение dataframe
# конкатенация dataframe
articles = pd.concat([articles_json_df, articles_habr_df], ignore_index=True)
articles
# соединение dataframe
articles_full = articles.merge(compani_habr_df, how="left", on="name_company")
articles_full
articles_full[articles_full["name_company"] == "skillbox"]
# Получили dataframe 1686 × 6, есть пустые значения, текст не обработан.
# **Описание признаков**
# - **name_company** - компания номинант
# - **text** - тест статьи
# - **date** - дата публикации статьи
# - **description** - описание компании
# - **activity** - сфера деятельности компании
# - **rating** - рейтинг компании
FILE_NAME_ONE = "статьи.csv"
import pandas as pd
articles_full.to_csv(FILE_NAME_ONE, encoding="utf-8-sig", index=False)
# articles_full = pd.read_csv(FILE_NAME_ONE)
articles_full
# # 1.2 Формирование структуры набора данных
articles_full.info()
# - **name_company** - строковый тип данных, не имеет пропусков, является основным признаком, определяющий компанию номинанта.
# - **text** - является одним из основных признаков так, как определяет то, о чем пишет компания.
# - **date** - не является основным признаком, может отобразить только разницу в дате публикации статей, что не сильно коррелирует с номинированными на премию
# - **description** - не является основным признаком, зачастую не несет рекламный характер, практически ни как не отражает деятельность компании и имеет много пропусков
# - **activity** - не является основным признаком, но описывает сферу деятельности компании, что поможет выявить наиболее подходящую номинацию
# - **rating** - не является основным признаком, отражает положения компании, в топе компаний, что не поможет номинировать их, много пропусков
#
df = pd.DataFrame(articles_full.value_counts("name_company"))
df
# Получен dataframe 1672 × 3, новые параметры не добавлены. Все 3 признака имеют строковый тип, написаны по большой части на русском языке
for con in condidates:
if con not in articles_full["name_company"].unique():
print("У компании нет статей ни на Habr, ни в Json:", con)
# # 1.3 Предварительная обработка текстовых данных
# ### Предварительный разведочный анализ
articles_full.head(5)
# В столбцах: «description», «activity», «rating» видны пропуски
for col in articles_full.columns:
print(
f"Процент пропусков в колонке {col} = {articles_full[col].isna().sum() / articles_full.shape[0] * 100: .2f}"
)
plt.figure(figsize=(10, 6))
sns.heatmap(articles_full.isna())
articles_full.info()
articles_full.describe()
# Большая часть данных строкового типа
articles_full.name_company.unique()
# Столбец «name_company» содержит текст на русском и английском языках
articles_full["text"][0]
# Столбец «text» содержит текст на русском и английском языках, числа, знаки пунктуации, спец символы
articles_full.date.unique()
len(articles_full.date.unique())
# Столбец «date» содержит дату разных образцов: числовой и строковый
articles_full.rating.unique()
# Столбец «name_company» числовые значения и пропуски
articles_full.activity.unique()
# Столбец «activity» содержит текст на русском со знаками пунктуации и пустые значения
articles_full.description.unique()
# Столбец «description» содержит текст на русском и английском языках со знаками пунктуации и пустые значения
# Объем и разнообразие текстовых данных: Задача классификации текста для премии "Рунета" предполагает работу с большим объемом и разнообразием текстовых данных. Для обработки такого объема информации необходимо использовать эффективные методы предварительной обработки текста, чтобы извлечь значимые признаки и снизить размерность данных. Таким образом, мы можем применить методы, такие как токенизация, удаление стоп-слов, нормализация текста (лемматизация или стемминг) и векторизация слов.
# Перед тем, как набор данных можно будет использовать для создания модели машинного обучения, необходимо его подготовить. Предварительная обработка исходных данных играет важную роль для построения качественной модели.
# - name_company +
# - не трогаем +
#
#
# - text
# 1) Привести текст в нижний регистр +
# 2) Удалить спец символы +
# 3) Удалить числа +
# 3) Удалить стоп слова +
# 4) Выполнить Tокенизация
# 5) Выполнить Лемматизацию и выделение значимых частей речи
# - date
# 1) Преобразовать строки с числа +
# 2) Отсечь строки с не нужные значениями +
# - rating
# 1) Заполнить пустые значения +
# - activity
# 1) Заполнить пустые значения +
# 2) Удалить пунктуацию символы +
# 3) Выполнить Tокенизация
# 4) Выполнить Лемматизацию
# - description
# 1) Заполнить пустые значения +
# 2) Удалить пунктуацию символы +
# 3) Выполнить Tокенизация
# 4) Выполнить Лемматизацию
# Проверить результат, при необходимости выполнить дополнительные корректировки
# ### Замена пустых значений
for col in articles_full.columns:
print(
f"Процент пропусков в колонке {col} = {articles_full[col].isna().sum() / articles_full.shape[0] * 100: .2f}"
)
# **description**, **activity**, **rating** содержат много пустых значений, которые нужно заменить, для последующий классификации
# Заменим пустые значения в признаках description и activity заглушкой.
# Заглушка подобрана по принципу меньше слов и встретится встретиться в тексте
articles_full["description"].fillna("пусто", inplace=True)
articles_full["activity"].fillna("пусто", inplace=True)
# Заменим пустые значения в признаке rating заглушкой.
# Там числовой признак, заменим числом
articles_full["rating"].fillna(0, inplace=True)
for col in articles_full.columns:
print(
f"Процент пропусков в колонке {col} = {articles_full[col].isna().sum() / articles_full.shape[0] * 100: .2f}"
)
print("______")
print(
"Заменено в description:",
articles_full[articles_full["description"] == "пусто"]["description"].count(),
)
print("______")
print(
"Заменено в activity:",
articles_full[articles_full["activity"] == "пусто"]["activity"].count(),
)
print("______")
print(
"Заменено в rating:", articles_full[articles_full["rating"] == 0]["rating"].count()
)
# ### Удаление стоп-слов, пунктуации, спецсимволов.
import re
from nltk.corpus import stopwords
import nltk
nltk.download("stopwords")
nltk.download("punkt")
stopwords_ru = stopwords.words("russian")
stopwords_ru.extend(
["который", "которая", "которое", "которые", "которого", "которому"]
)
def removal_excess(text):
text = text.lower()
text = text.replace("\n", " ")
text = text.replace("\r", " ")
text = re.sub("ё", "е", text)
text = re.sub("й", "и", text)
text = re.sub(r"[^а-яa-z\s]+", "", text)
text = " ".join(
[i for i in text.split() if len(i) > 3 and i not in stopwords_ru]
) # удаление стоп слов
text = re.sub(" +", " ", text)
return text
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "text"] = removal_excess(articles_full["text"][i])
articles_full.at[i, "activity"] = removal_excess(articles_full["activity"][i])
articles_full.at[i, "description"] = removal_excess(articles_full["description"][i])
articles_full
# Удалены в "text", "description" и "activity" спец символы - пунктуация, символы переноса и табуляции, стоп слова и слова длинной менее 3 символов, лишние пробелы, что очистило данные, для дальнейшей обработки.
# ### Форматирование даты
def format_date(date):
date = str(date)
date = date.replace("-", "")
date = re.sub(" +", " ", date)
return int(date)
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "date"] = format_date(articles_full["date"][i])
articles_full["date"] = articles_full["date"].astype("int")
articles_full.info()
# ### Фильтрация по дате
articles_full = articles_full[
(articles_full["date"] > 20210000) & (articles_full["date"] < 20220000)
]
articles_full.reset_index(drop=True, inplace=True)
articles_full
articles_full.date.unique()
# ### Tокенизация
# Токенизация – это процесс разделения предложений на слова-компонентыё
# В модуле nltk.tokenize есть множество классов токенизаторов и функции nltk.word_tokenize на самом деле являются надстройкой
# TreebankWordTokenizer
# Класс используется для разделения предложений на слова и вызывается функции nltk.word_tokenize.
# Он предполагает, что текст уже был разделён на предложения и выполняет следующие шаги:
# - Разделяет стандартные сокращения, например don't -> do n't and they'll -> they 'll
# - Рассматривает большинство знаков препинания как отдельные токены
# - Отделяет запятые и одинарные кавычки, если за ними следует пробел
# - Отделяет точки в конце предложений
# word_tokenize выбран потому/ что является самым простым, разбивая текст на слова и наиболее подходит к нашей задаче.
from nltk.tokenize import word_tokenize
def tokenize(text):
return word_tokenize(text)
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "text"] = tokenize(articles_full["text"][i])
articles_full.at[i, "activity"] = tokenize(articles_full["activity"][i])
articles_full.at[i, "description"] = tokenize(articles_full["description"][i])
articles_full
# Tокенизация в "text", "description" и "activity" разбиение для дальнейшей обработки.
# ### Лемматизация и выделение значимых частей речи
# Лемматизация - основная идея в приведении слова к словарной форме.
# Выбран pymorphy2 потому что разработан российской компанией, вследствие этого как нельзя лучше лемматизирует русские текста
# Воспользуемся готовым средством маркировки из NLTK PerceptronTagger()
#
import pymorphy2
import nltk
from nltk.tag.perceptron import PerceptronTagger
nltk.download("averaged_perceptron_tagger")
morph = pymorphy2.MorphAnalyzer()
def lemmatizer(text):
return [morph.parse(i)[0].normal_form for i in text]
def translate_pos_pymorphy2(pos):
pos_dict = {
"ADJF": "Имя прилагательное (полное)",
"ADJS": "Имя прилагательное (краткое)",
"ADVB": "Наречие",
"COMP": "Сравнительная степень",
"CONJ": "Союз",
"GRND": "Деепричастие",
"INFN": "Инфинитив",
"INTJ": "Междометие",
"NOUN": "Имя существительное",
"NPRO": "Местоимение-существительное",
"NUMR": "Числительное",
"PRED": "Предикатив",
"PREP": "Предлог",
"PRTF": "Причастие (полное)",
"PRTS": "Причастие (краткое)",
"VERB": "Глагол",
}
return pos_dict.get(pos, "Неизвестно")
def perceptron(words):
words = set(words)
tagged_words = [(word, morph.parse(word)[0].tag.POS) for word in words]
tagged_words_new = []
for word, pos in tagged_words:
type_ = (word, translate_pos_pymorphy2(pos))
tagged_words_new.append(type_)
return tagged_words_new
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "text"] = lemmatizer(articles_full["text"][i])
articles_full.at[i, "activity"] = lemmatizer(articles_full["activity"][i])
articles_full.at[i, "description"] = lemmatizer(articles_full["description"][i])
per = {"part_speech": []}
for i in tqdm(range(len(articles_full))):
per["part_speech"].append(perceptron(articles_full["text"][i]))
articles_full["part_speech"] = per["part_speech"]
articles_full
# ### Проверка результатов
from sklearn.feature_extraction.text import CountVectorizer
import pandas as pd
count_vectorizer = CountVectorizer(ngram_range=(1, 3))
bag_of_words = count_vectorizer.fit_transform([" ".join(articles_full["text"][44])])
feature_names = count_vectorizer.get_feature_names_out()
uniq_and_fifa = dict(zip(feature_names, bag_of_words.toarray()[0]))
uniq_and_fifa = dict(sorted(uniq_and_fifa.items(), key=lambda item: -item[1]))
uniq_and_fifa
# Замечаны стоп слова
stopwords_ru.extend(["свой", "такой", "который", "чтоть", "наш", "сам", "этот", "ваш"])
def remove_stopwords(text):
return [i for i in text if i not in stopwords_ru]
remove_sw = {"text": []}
for i in tqdm(range(len(articles_full))):
remove_sw["text"].append(remove_stopwords(articles_full["text"][i]))
articles_full["text"] = remove_sw["text"]
count_vectorizer = CountVectorizer(ngram_range=(1, 3))
bag_of_words = count_vectorizer.fit_transform([" ".join(articles_full["text"][44])])
feature_names = count_vectorizer.get_feature_names_out()
uniq_and_fifa = dict(zip(feature_names, bag_of_words.toarray()[0]))
uniq_and_fifa = dict(sorted(uniq_and_fifa.items(), key=lambda item: -item[1]))
uniq_and_fifa
articles_full["part_speech"][0]
per = {"part_speech": []}
for i in tqdm(range(len(articles_full))):
per["part_speech"].append(perceptron(articles_full["text"][i]))
articles_full["part_speech"] = per["part_speech"]
articles_full
FILE_NAME = "статьи2.csv"
articles_full.to_csv(FILE_NAME, encoding="utf-8-sig", index=False)
articles_full.to_pickle("articles_full_obra.pkl")
# Получен dataframe c обработанным текстом, что в дальнейшем поможет нам его векторизовать, и выделенными значимыми частями речи, для статистического анализа каждой статьи
# # 1.4 Поиск ключевых слов/n-грамм. Векторизация текстов
import pandas as pd
# articles_full = pd.read_csv("статьи2.csv")
articles_full = pd.read_pickle("articles_full_obra.pkl")
articles_full
# ### Bag of words
# Mешок слов (Bag of words) выделяет вектору весь документ, и каждый элемент кодируется 1 по порядку следования слов в словаре
from sklearn.feature_extraction.text import CountVectorizer
import pandas as pd
count_vectorizer = CountVectorizer(ngram_range=(1, 3))
bag_of_words = count_vectorizer.fit_transform([" ".join(articles_full["text"][44])])
feature_names_bag = count_vectorizer.get_feature_names_out()
uniq_and_fifa_bag = dict(zip(feature_names_bag, bag_of_words.toarray()[0]))
uniq_and_fifa_bag = dict(sorted(uniq_and_fifa_bag.items(), key=lambda item: -item[1]))
uniq_and_fifa_bag
# Bag of words решает проблему размерности по одной оси. Количество строк определяется количеством документов. Однако, этот метод не учитывает важность того или иного токена, ведь одно слово может повторятся по несколько раз. В этом случае пригодится альтернативный способ, рассмотренный далее
# ### TF-IDF
# TF-IDF состоит из двух компонентов: Term Frequency (частотность слова в документе) и Inverse Document Frequency (инверсия частоты документа).
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer(ngram_range=(1, 3))
tfidf_matrix = vectorizer.fit_transform([" ".join(articles_full["text"][44])])
feature_names_tfidf = vectorizer.get_feature_names_out()
uniq_and_fifa_tfidf = dict(zip(feature_names_tfidf, tfidf_matrix.toarray()[0]))
uniq_and_fifa_tfidf = dict(
sorted(uniq_and_fifa_tfidf.items(), key=lambda item: -item[1])
)
uniq_and_fifa_tfidf
# Все вышерассмотренные NLP-методы отличаются следующими недостатками:
# не зависят от контекста — не учитывают порядок слов в предложении; обладают высокой размерностью в случае большого словаря, что может снизить производительность модели глубокого обучения (Deep Learning).
# Но имеет ряд преимуществ:
# TF-IDF учитывает не только частоту встречаемости слова в документе (TF), но и редкость слова в коллекции документов (IDF).
# ### Word Embeddings
# Word embeddings — редставляют слова в виде плотных векторов фиксированной размерности в многомерном пространстве. Векторы можно складывать, вычитать, сравнивать.
# - Эти векторные представления учитывают семантическую схожесть слов.
# - Полученные векторы обычно имеют низкую размерность (например, 100-300), поэтому они занимают меньше памяти и позволяют выявлять семантические связи между словами.
# Bag of words - дает просто подсчет частоты слова в документе, а TF-IDF важность и частоту в дробном формате, что более точно определяет значимость n-граммы, алгоритм Word Embeddings не реализован. В дальнейшем будет использоваться алгоритм TF-IDF
# ### Векторизация текст, поиск ключевых n-грамм методом TF-IDF
tfidf = {"tfidf": [], "vector": []}
for i in tqdm(range(len(articles_full["text"]))):
tfidf_matrix = vectorizer.fit_transform([" ".join(articles_full["text"][i])])
tfidf["vector"].append(tfidf_matrix)
feature_names_tfidf = vectorizer.get_feature_names_out()
uniq_and_fifa_tfidf = dict(zip(feature_names_tfidf, tfidf_matrix.toarray()[0]))
uniq_and_fifa_tfidf = dict(
sorted(uniq_and_fifa_tfidf.items(), key=lambda item: -item[1])
)
tfidf["tfidf"].append(uniq_and_fifa_tfidf)
len(tfidf["tfidf"])
articles_full["tfidf"] = tfidf["tfidf"]
articles_full["vector"] = tfidf["vector"]
articles_full
articles_full.to_pickle("articles_full_tfidf.pkl")
# Получен dataframe с 9 колонками, представлены алгоритмы векторизации/поиск ключевых n-грамм методами: Bag of words, TF-IDF
# # 1.5 Разведочный анализ
articles_full = pd.read_pickle("articles_full_tfidf.pkl")
articles_full
# ### Парсинг json
file = "Target.json"
nominations = {"name_company": [], "nominations": [], "code": []}
with codecs.open(file, "r", "utf_8_sig") as f:
templates = json.load(f)
# считывание информации о компаниях
if templates["info"] != None:
for i in range(len(templates["info"])):
nominations["name_company"].append(
templates["info"][i]["Сompany"].lower().strip()
)
nominations["nominations"].append(templates["info"][i]["Nominations"])
nominations["nominations"]
# Кодирование номинаций
from sklearn.preprocessing import LabelEncoder
encoder = LabelEncoder()
encoded_data = encoder.fit_transform(nominations["nominations"])
nominations["code"] = encoded_data
nominations_df = pd.DataFrame(nominations)
nominations_df
len(nominations_df)
# 22 компании, все есть
nominations_df.info()
articles_full = articles_full.merge(nominations_df, how="left", on="name_company")
articles_full
articles_full.info()
# ### Определений и замена пустых значений
import numpy as np
articles_full[articles_full["code"].isnull()]
articles_full["code"].fillna(0, inplace=True)
articles_full["nominations"].fillna("Здоровье и медицина", inplace=True)
articles_full.loc[
articles_full["name_company"]
== "проект по использованию технологий компьютерного зрения на базе искусственного интеллекта (ии) для анализа медицинских изображений",
"name_company",
] = "проект по использованию технологий компьютерного зрения на базе искусственного интеллекта для анализа медицинских изображений"
len(articles_full["name_company"].unique())
# Так как одна компания не представлена ни где, осталось 21 уникальная компания
articles_full.info()
import scipy.stats as stats
import seaborn as sns
from scipy import stats
import matplotlib.pyplot as plt
# ### Нормальность распределения target переменной
ax = sns.distplot(articles_full["code"])
# Признак категориальный, нормального распределения нет, категории 0 и 3 завышены, похожи на выбросы
stats.probplot(articles_full["code"], dist="norm", plot=plt)
plt.show()
# Признак категориальный, не следую нормальное распределению
ax = sns.boxplot(x=articles_full["code"])
# Ящик с усами скошен в лево, 3,4,5 кластеры малочисленно
# ### Тест Шапиро-Уилка
statistic_shapiro, p_value_shapiro = stats.shapiro(articles_full["code"])
print("Статистика теста:", statistic_shapiro)
print("p-значение:", p_value_shapiro)
# ### К-квадрат Д’Агостино
statistic_normaltest, p_value_normaltest = stats.normaltest(articles_full["code"])
print("Статистика теста:", statistic_normaltest)
print("p-значение:", p_value_normaltest)
# Математические расчеты показали, что действительно целевая переменная распределена не нормально, предположительно из-за того, что многие компании, простеленные в документе, прекратили свою деятельность, и их статей в открытом доступу оказалось очень мало
# ### Количественная оценка
# Пример категориальных данных
data = articles_full["code"]
# Построение гистограммы
data.value_counts().plot(kind="bar")
# Настройка осей и заголовка
plt.xlabel("Категория")
plt.ylabel("Частота")
plt.title("Распределение категориального признака")
# Отображение графика
plt.show()
# Пример категориальных данных
data = articles_full["code"]
# Построение круговой диаграммы
data.value_counts().plot(kind="pie")
# Настройка заголовка
plt.title("Распределение категориального признака")
# Отображение графика
plt.show()
# Количественная оценка категорий показала, что больше всего статей с номинацией 3(Наука, технологии и инновации), совсем мало статей номинацией 1(Игровая индустрия и киберспорт)
# ### Зависимость целевой переменной от месяца и дня публикации статьи
month = []
for i in tqdm(range(len(articles_full["date"]))):
month.append(float(str(articles_full["date"][i])[4:]))
articles_full["month"] = month
articles_full
sns.jointplot(x="code", y="month", data=articles_full)
# По сколько одних статей на порядок больше, чем других, нельзя сказать когда больше всего публикуется определённая номинация статей, но можно сказать, что в декабре и в июле публикуется самое большое количество статей
# ### Зависимость целевой переменной от рейтинга статьи
sns.jointplot(x="code", y="rating", data=articles_full)
# Очень много заменено пустых значений на 0. Отсутствие данных не позволяет корректно оценить корреляцию признаков
# ### Зависимость целевой переменной от n-грамм
keywords_1 = []
for i in tqdm(range(len(articles_full["tfidf"]))):
keywords_1.append(list(articles_full["tfidf"][i].keys())[0:1])
# Пример данных
topics = articles_full["code"]
keywords = keywords_1
# Создание датафрейма для визуализации
data = []
for i, topic in enumerate(topics):
for keyword in keywords[i]:
data.append([topic, keyword])
df = pd.DataFrame(data, columns=["Кластер", "Ключевое слово"])
# Построение графика
plt.figure(figsize=(8, 6))
sns.scatterplot(data=df, x="Ключевое слово", y="Кластер")
plt.xticks(rotation=45)
# Отображение графика
plt.show()
# Больше статей больше ключевых слов добавлено
nominations_df
# Для наглядности вывели номинации и их название
articles_full.to_csv("articles_full", encoding="utf-8-sig", index=False)
articles_full.to_pickle("articles_full_end.pkl")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/599/129599584.ipynb
| null | null |
[{"Id": 129599584, "ScriptId": 38538044, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13417746, "CreationDate": "05/15/2023 06:38:41", "VersionNumber": 1.0, "Title": "Report1-KSH-djostit", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 847.0, "LinesInsertedFromPrevious": 847.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # 1.1 Парсинг данных
# Подключение библиотек
import pandas as pd
import json
import docx
import requests
from bs4 import BeautifulSoup as bs
import glob
import codecs
from tqdm.auto import tqdm
import matplotlib.pyplot as plt
import seaborn as sns
# ### Парсинг docx файла
# открытие файла
condidates_docx = docx.Document("Condidates.docx")
# парсинг данных
condidates = []
text_condidates_docx = condidates_docx.paragraphs
for con in text_condidates_docx:
# проверка длинны параграфа, для избежания добавления пустых значений
if len(con.text) > 0:
condidates.append(con.text.strip().lower())
print(condidates)
print(len(condidates))
# В docx документе предоставлены 22 номинанта, удалены лишние пробелы и приведен текст в нижний регистр, для стандартизации
# ### Парсинг json файлов
all_file = glob.glob(r"Data\*.json")
print(all_file)
print(len(all_file))
norm_month = [
"января",
"февраля",
"марта",
"апреля",
"мая",
"июня",
"июля",
"августа",
"сентября",
"октября",
"ноября",
"декабря",
]
def norm_date(day, month, year=2021):
try:
day = int(day)
except:
day = 1
if day < 10:
day = "0{0}".format(day)
if month in norm_month:
month = norm_month.index(month) + 1
else:
month = 1
if month < 10:
month = "0{0}".format(month)
return int("{0}{1}{2}".format(year, month, day))
# парсинг данных компаний и статей
about_condidates_json = {
"name_company": [],
"description": [],
"activity": [],
"rating": [],
}
about_articles_json = {"name_company": [], "text": [], "date": []}
brack_articles = []
brack_read = []
are_articles = []
nan_info = []
for file in all_file:
# разбитие и срез строки для отделения имени компании
name = file.split("\\")[-1][:-5].lower()
try:
# открытие файла
with codecs.open(file, "r", "utf_8_sig") as f:
templates = json.load(f)
# считывание информации о компаниях
if templates["info"] != None:
about_condidates_json["name_company"].append(name)
about_condidates_json["description"].append(templates["info"]["about"])
about_condidates_json["activity"].append(
templates["info"]["industries"]
)
about_condidates_json["rating"].append(templates["info"]["rate"])
else:
nan_info.append(name)
if len(templates["refs"]) > 0:
are_articles.append(name)
# считывание информации о статьях
for art in templates["refs"]:
try:
about_articles_json["text"].append(art[0])
about_articles_json["date"].append(
norm_date(art[1]["day"], art[1]["month"])
)
about_articles_json["name_company"].append(name)
except:
brack_articles.append(name)
except:
brack_read.append(name)
print("Документы json, которые не считались: ", brack_read)
print("_________")
print("Нет информации о компаниях: ", nan_info)
print("_________")
print("Есть информация о кампании: ", about_condidates_json["name_company"])
print("_________")
print("У компании есть статьи: ", are_articles)
print("_________")
print("Статья не считана: ", brack_articles)
print("_________")
print("Количество статей", len(about_articles_json["name_company"]))
compani_json_df = pd.DataFrame(about_condidates_json)
compani_json_df
articles_json_df = pd.DataFrame(about_articles_json)
articles_json_df
# Все 14 json прочитаны верно, у ряда компаний отсутствует информация, есть только у двух компаний. Статьи есть не у всех компаний, всего статей 1112, есть 4 статьи, которые не считались. Информацию о компаниях и их статьях нужно дополнить с веб ресурса Habr.
# ### Парсинг веб ресурса Habr
# парсинг описание компаний
def activity_condidates(soup_new):
a = soup_new.find("a", class_="tm-company-snippet__title")
url_condidate = "https://habr.com" + a["href"]
page_condidate = requests.get(url_condidate)
soup_condidate = bs(page_condidate.text, "html.parser")
div = soup_condidate.find("div", class_="tm-company-profile__categories")
a1 = div.find_all("a")
activity = []
for i in a1:
activity.append(i.text.strip().lower())
activity = " ".join(activity)
return activity
# парсинг компаний
about_condidates_habr = {
"name_company": [],
"description": [],
"activity": [],
"rating": [],
}
for con in condidates:
con.replace(" ", "20%")
# создание ссылки
url = (
"https://habr.com/ru/search/?q="
+ con
+ "&target_type=companies&order=relevance"
)
page = requests.get(url)
soup = bs(page.text, "html.parser")
condidate_div = soup.find_all("em", class_="searched-item")
# заполнение данных
if len(condidate_div) > 0:
if condidate_div[0].text.lower() == con:
about_condidates_habr["name_company"].append(con)
description_div = soup.find("div", class_="tm-company-snippet__description")
about_condidates_habr["description"].append(description_div.text)
about_condidates_habr["activity"].append(activity_condidates(soup))
rating_span = soup.find(
"span",
class_="tm-search-companies__score-counter tm-search-companies__score-counter_rating",
)
about_condidates_habr["rating"].append(rating_span.text.split()[1])
compani_habr_df = pd.DataFrame(about_condidates_habr)
compani_habr_df
# парсинг статей
about_articles_habr = {"name_company": [], "text": [], "date": []}
for con in tqdm(condidates):
if con not in are_articles:
con = con.replace(" ", "%20")
for i in range(1, 5):
# создание ссылки
uri_page = "https://habr.com/ru/search/page{1}/?q={0}&target_type=posts&order=relevance".format(
con, i
)
page = requests.get(uri_page)
soup = bs(page.text, "html.parser")
snippetsoup_name = soup.find_all("h2", class_="tm-title tm-title_h2")
for i in snippetsoup_name:
try:
uri_article = "https://habr.com" + str(i.a.get("href"))
page_article = requests.get(uri_article)
soup_article = bs(page_article.text, "html.parser")
date = soup_article.find(
"span", class_="tm-article-datetime-published"
)
# две версии верстки, отбор данных статей
try:
note = soup_article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-1",
)
about_articles_habr["text"].append(
note.div.text.replace("\n", " ")
.replace("\r", " ")
.replace(" ", " ")
.replace("\xa0", " ")
)
about_articles_habr["name_company"].append(
con.replace("%20", " ")
)
about_articles_habr["date"].append(
date.time.get("title").split(",")[0]
)
except:
try:
note = soup_article.find(
"div",
class_="article-formatted-body article-formatted-body article-formatted-body_version-2",
)
about_articles_habr["text"].append(
note.div.text.replace("\n", " ")
.replace("\r", " ")
.replace(" ", " ")
.replace("\xa0", " ")
)
about_articles_habr["name_company"].append(
con.replace("%20", " ")
)
about_articles_habr["date"].append(
date.time.get("title").split(",")[0]
)
except:
print(uri_article)
except:
print(uri_article)
print("_________")
print("Количество статей", len(about_articles_habr["name_company"]))
articles_habr_df = pd.DataFrame(about_articles_habr)
articles_habr_df
# Получены данные компании с сайта, две (skillbox, иннотех) из них совпадают с уже имеющимися. Считаю, что с сайта более актуальные данные. Получили статьи тех компаний, что не представлены в json файлах.
# ### Объединение dataframe
# конкатенация dataframe
articles = pd.concat([articles_json_df, articles_habr_df], ignore_index=True)
articles
# соединение dataframe
articles_full = articles.merge(compani_habr_df, how="left", on="name_company")
articles_full
articles_full[articles_full["name_company"] == "skillbox"]
# Получили dataframe 1686 × 6, есть пустые значения, текст не обработан.
# **Описание признаков**
# - **name_company** - компания номинант
# - **text** - тест статьи
# - **date** - дата публикации статьи
# - **description** - описание компании
# - **activity** - сфера деятельности компании
# - **rating** - рейтинг компании
FILE_NAME_ONE = "статьи.csv"
import pandas as pd
articles_full.to_csv(FILE_NAME_ONE, encoding="utf-8-sig", index=False)
# articles_full = pd.read_csv(FILE_NAME_ONE)
articles_full
# # 1.2 Формирование структуры набора данных
articles_full.info()
# - **name_company** - строковый тип данных, не имеет пропусков, является основным признаком, определяющий компанию номинанта.
# - **text** - является одним из основных признаков так, как определяет то, о чем пишет компания.
# - **date** - не является основным признаком, может отобразить только разницу в дате публикации статей, что не сильно коррелирует с номинированными на премию
# - **description** - не является основным признаком, зачастую не несет рекламный характер, практически ни как не отражает деятельность компании и имеет много пропусков
# - **activity** - не является основным признаком, но описывает сферу деятельности компании, что поможет выявить наиболее подходящую номинацию
# - **rating** - не является основным признаком, отражает положения компании, в топе компаний, что не поможет номинировать их, много пропусков
#
df = pd.DataFrame(articles_full.value_counts("name_company"))
df
# Получен dataframe 1672 × 3, новые параметры не добавлены. Все 3 признака имеют строковый тип, написаны по большой части на русском языке
for con in condidates:
if con not in articles_full["name_company"].unique():
print("У компании нет статей ни на Habr, ни в Json:", con)
# # 1.3 Предварительная обработка текстовых данных
# ### Предварительный разведочный анализ
articles_full.head(5)
# В столбцах: «description», «activity», «rating» видны пропуски
for col in articles_full.columns:
print(
f"Процент пропусков в колонке {col} = {articles_full[col].isna().sum() / articles_full.shape[0] * 100: .2f}"
)
plt.figure(figsize=(10, 6))
sns.heatmap(articles_full.isna())
articles_full.info()
articles_full.describe()
# Большая часть данных строкового типа
articles_full.name_company.unique()
# Столбец «name_company» содержит текст на русском и английском языках
articles_full["text"][0]
# Столбец «text» содержит текст на русском и английском языках, числа, знаки пунктуации, спец символы
articles_full.date.unique()
len(articles_full.date.unique())
# Столбец «date» содержит дату разных образцов: числовой и строковый
articles_full.rating.unique()
# Столбец «name_company» числовые значения и пропуски
articles_full.activity.unique()
# Столбец «activity» содержит текст на русском со знаками пунктуации и пустые значения
articles_full.description.unique()
# Столбец «description» содержит текст на русском и английском языках со знаками пунктуации и пустые значения
# Объем и разнообразие текстовых данных: Задача классификации текста для премии "Рунета" предполагает работу с большим объемом и разнообразием текстовых данных. Для обработки такого объема информации необходимо использовать эффективные методы предварительной обработки текста, чтобы извлечь значимые признаки и снизить размерность данных. Таким образом, мы можем применить методы, такие как токенизация, удаление стоп-слов, нормализация текста (лемматизация или стемминг) и векторизация слов.
# Перед тем, как набор данных можно будет использовать для создания модели машинного обучения, необходимо его подготовить. Предварительная обработка исходных данных играет важную роль для построения качественной модели.
# - name_company +
# - не трогаем +
#
#
# - text
# 1) Привести текст в нижний регистр +
# 2) Удалить спец символы +
# 3) Удалить числа +
# 3) Удалить стоп слова +
# 4) Выполнить Tокенизация
# 5) Выполнить Лемматизацию и выделение значимых частей речи
# - date
# 1) Преобразовать строки с числа +
# 2) Отсечь строки с не нужные значениями +
# - rating
# 1) Заполнить пустые значения +
# - activity
# 1) Заполнить пустые значения +
# 2) Удалить пунктуацию символы +
# 3) Выполнить Tокенизация
# 4) Выполнить Лемматизацию
# - description
# 1) Заполнить пустые значения +
# 2) Удалить пунктуацию символы +
# 3) Выполнить Tокенизация
# 4) Выполнить Лемматизацию
# Проверить результат, при необходимости выполнить дополнительные корректировки
# ### Замена пустых значений
for col in articles_full.columns:
print(
f"Процент пропусков в колонке {col} = {articles_full[col].isna().sum() / articles_full.shape[0] * 100: .2f}"
)
# **description**, **activity**, **rating** содержат много пустых значений, которые нужно заменить, для последующий классификации
# Заменим пустые значения в признаках description и activity заглушкой.
# Заглушка подобрана по принципу меньше слов и встретится встретиться в тексте
articles_full["description"].fillna("пусто", inplace=True)
articles_full["activity"].fillna("пусто", inplace=True)
# Заменим пустые значения в признаке rating заглушкой.
# Там числовой признак, заменим числом
articles_full["rating"].fillna(0, inplace=True)
for col in articles_full.columns:
print(
f"Процент пропусков в колонке {col} = {articles_full[col].isna().sum() / articles_full.shape[0] * 100: .2f}"
)
print("______")
print(
"Заменено в description:",
articles_full[articles_full["description"] == "пусто"]["description"].count(),
)
print("______")
print(
"Заменено в activity:",
articles_full[articles_full["activity"] == "пусто"]["activity"].count(),
)
print("______")
print(
"Заменено в rating:", articles_full[articles_full["rating"] == 0]["rating"].count()
)
# ### Удаление стоп-слов, пунктуации, спецсимволов.
import re
from nltk.corpus import stopwords
import nltk
nltk.download("stopwords")
nltk.download("punkt")
stopwords_ru = stopwords.words("russian")
stopwords_ru.extend(
["который", "которая", "которое", "которые", "которого", "которому"]
)
def removal_excess(text):
text = text.lower()
text = text.replace("\n", " ")
text = text.replace("\r", " ")
text = re.sub("ё", "е", text)
text = re.sub("й", "и", text)
text = re.sub(r"[^а-яa-z\s]+", "", text)
text = " ".join(
[i for i in text.split() if len(i) > 3 and i not in stopwords_ru]
) # удаление стоп слов
text = re.sub(" +", " ", text)
return text
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "text"] = removal_excess(articles_full["text"][i])
articles_full.at[i, "activity"] = removal_excess(articles_full["activity"][i])
articles_full.at[i, "description"] = removal_excess(articles_full["description"][i])
articles_full
# Удалены в "text", "description" и "activity" спец символы - пунктуация, символы переноса и табуляции, стоп слова и слова длинной менее 3 символов, лишние пробелы, что очистило данные, для дальнейшей обработки.
# ### Форматирование даты
def format_date(date):
date = str(date)
date = date.replace("-", "")
date = re.sub(" +", " ", date)
return int(date)
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "date"] = format_date(articles_full["date"][i])
articles_full["date"] = articles_full["date"].astype("int")
articles_full.info()
# ### Фильтрация по дате
articles_full = articles_full[
(articles_full["date"] > 20210000) & (articles_full["date"] < 20220000)
]
articles_full.reset_index(drop=True, inplace=True)
articles_full
articles_full.date.unique()
# ### Tокенизация
# Токенизация – это процесс разделения предложений на слова-компонентыё
# В модуле nltk.tokenize есть множество классов токенизаторов и функции nltk.word_tokenize на самом деле являются надстройкой
# TreebankWordTokenizer
# Класс используется для разделения предложений на слова и вызывается функции nltk.word_tokenize.
# Он предполагает, что текст уже был разделён на предложения и выполняет следующие шаги:
# - Разделяет стандартные сокращения, например don't -> do n't and they'll -> they 'll
# - Рассматривает большинство знаков препинания как отдельные токены
# - Отделяет запятые и одинарные кавычки, если за ними следует пробел
# - Отделяет точки в конце предложений
# word_tokenize выбран потому/ что является самым простым, разбивая текст на слова и наиболее подходит к нашей задаче.
from nltk.tokenize import word_tokenize
def tokenize(text):
return word_tokenize(text)
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "text"] = tokenize(articles_full["text"][i])
articles_full.at[i, "activity"] = tokenize(articles_full["activity"][i])
articles_full.at[i, "description"] = tokenize(articles_full["description"][i])
articles_full
# Tокенизация в "text", "description" и "activity" разбиение для дальнейшей обработки.
# ### Лемматизация и выделение значимых частей речи
# Лемматизация - основная идея в приведении слова к словарной форме.
# Выбран pymorphy2 потому что разработан российской компанией, вследствие этого как нельзя лучше лемматизирует русские текста
# Воспользуемся готовым средством маркировки из NLTK PerceptronTagger()
#
import pymorphy2
import nltk
from nltk.tag.perceptron import PerceptronTagger
nltk.download("averaged_perceptron_tagger")
morph = pymorphy2.MorphAnalyzer()
def lemmatizer(text):
return [morph.parse(i)[0].normal_form for i in text]
def translate_pos_pymorphy2(pos):
pos_dict = {
"ADJF": "Имя прилагательное (полное)",
"ADJS": "Имя прилагательное (краткое)",
"ADVB": "Наречие",
"COMP": "Сравнительная степень",
"CONJ": "Союз",
"GRND": "Деепричастие",
"INFN": "Инфинитив",
"INTJ": "Междометие",
"NOUN": "Имя существительное",
"NPRO": "Местоимение-существительное",
"NUMR": "Числительное",
"PRED": "Предикатив",
"PREP": "Предлог",
"PRTF": "Причастие (полное)",
"PRTS": "Причастие (краткое)",
"VERB": "Глагол",
}
return pos_dict.get(pos, "Неизвестно")
def perceptron(words):
words = set(words)
tagged_words = [(word, morph.parse(word)[0].tag.POS) for word in words]
tagged_words_new = []
for word, pos in tagged_words:
type_ = (word, translate_pos_pymorphy2(pos))
tagged_words_new.append(type_)
return tagged_words_new
for i in tqdm(range(len(articles_full))):
articles_full.at[i, "text"] = lemmatizer(articles_full["text"][i])
articles_full.at[i, "activity"] = lemmatizer(articles_full["activity"][i])
articles_full.at[i, "description"] = lemmatizer(articles_full["description"][i])
per = {"part_speech": []}
for i in tqdm(range(len(articles_full))):
per["part_speech"].append(perceptron(articles_full["text"][i]))
articles_full["part_speech"] = per["part_speech"]
articles_full
# ### Проверка результатов
from sklearn.feature_extraction.text import CountVectorizer
import pandas as pd
count_vectorizer = CountVectorizer(ngram_range=(1, 3))
bag_of_words = count_vectorizer.fit_transform([" ".join(articles_full["text"][44])])
feature_names = count_vectorizer.get_feature_names_out()
uniq_and_fifa = dict(zip(feature_names, bag_of_words.toarray()[0]))
uniq_and_fifa = dict(sorted(uniq_and_fifa.items(), key=lambda item: -item[1]))
uniq_and_fifa
# Замечаны стоп слова
stopwords_ru.extend(["свой", "такой", "который", "чтоть", "наш", "сам", "этот", "ваш"])
def remove_stopwords(text):
return [i for i in text if i not in stopwords_ru]
remove_sw = {"text": []}
for i in tqdm(range(len(articles_full))):
remove_sw["text"].append(remove_stopwords(articles_full["text"][i]))
articles_full["text"] = remove_sw["text"]
count_vectorizer = CountVectorizer(ngram_range=(1, 3))
bag_of_words = count_vectorizer.fit_transform([" ".join(articles_full["text"][44])])
feature_names = count_vectorizer.get_feature_names_out()
uniq_and_fifa = dict(zip(feature_names, bag_of_words.toarray()[0]))
uniq_and_fifa = dict(sorted(uniq_and_fifa.items(), key=lambda item: -item[1]))
uniq_and_fifa
articles_full["part_speech"][0]
per = {"part_speech": []}
for i in tqdm(range(len(articles_full))):
per["part_speech"].append(perceptron(articles_full["text"][i]))
articles_full["part_speech"] = per["part_speech"]
articles_full
FILE_NAME = "статьи2.csv"
articles_full.to_csv(FILE_NAME, encoding="utf-8-sig", index=False)
articles_full.to_pickle("articles_full_obra.pkl")
# Получен dataframe c обработанным текстом, что в дальнейшем поможет нам его векторизовать, и выделенными значимыми частями речи, для статистического анализа каждой статьи
# # 1.4 Поиск ключевых слов/n-грамм. Векторизация текстов
import pandas as pd
# articles_full = pd.read_csv("статьи2.csv")
articles_full = pd.read_pickle("articles_full_obra.pkl")
articles_full
# ### Bag of words
# Mешок слов (Bag of words) выделяет вектору весь документ, и каждый элемент кодируется 1 по порядку следования слов в словаре
from sklearn.feature_extraction.text import CountVectorizer
import pandas as pd
count_vectorizer = CountVectorizer(ngram_range=(1, 3))
bag_of_words = count_vectorizer.fit_transform([" ".join(articles_full["text"][44])])
feature_names_bag = count_vectorizer.get_feature_names_out()
uniq_and_fifa_bag = dict(zip(feature_names_bag, bag_of_words.toarray()[0]))
uniq_and_fifa_bag = dict(sorted(uniq_and_fifa_bag.items(), key=lambda item: -item[1]))
uniq_and_fifa_bag
# Bag of words решает проблему размерности по одной оси. Количество строк определяется количеством документов. Однако, этот метод не учитывает важность того или иного токена, ведь одно слово может повторятся по несколько раз. В этом случае пригодится альтернативный способ, рассмотренный далее
# ### TF-IDF
# TF-IDF состоит из двух компонентов: Term Frequency (частотность слова в документе) и Inverse Document Frequency (инверсия частоты документа).
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer(ngram_range=(1, 3))
tfidf_matrix = vectorizer.fit_transform([" ".join(articles_full["text"][44])])
feature_names_tfidf = vectorizer.get_feature_names_out()
uniq_and_fifa_tfidf = dict(zip(feature_names_tfidf, tfidf_matrix.toarray()[0]))
uniq_and_fifa_tfidf = dict(
sorted(uniq_and_fifa_tfidf.items(), key=lambda item: -item[1])
)
uniq_and_fifa_tfidf
# Все вышерассмотренные NLP-методы отличаются следующими недостатками:
# не зависят от контекста — не учитывают порядок слов в предложении; обладают высокой размерностью в случае большого словаря, что может снизить производительность модели глубокого обучения (Deep Learning).
# Но имеет ряд преимуществ:
# TF-IDF учитывает не только частоту встречаемости слова в документе (TF), но и редкость слова в коллекции документов (IDF).
# ### Word Embeddings
# Word embeddings — редставляют слова в виде плотных векторов фиксированной размерности в многомерном пространстве. Векторы можно складывать, вычитать, сравнивать.
# - Эти векторные представления учитывают семантическую схожесть слов.
# - Полученные векторы обычно имеют низкую размерность (например, 100-300), поэтому они занимают меньше памяти и позволяют выявлять семантические связи между словами.
# Bag of words - дает просто подсчет частоты слова в документе, а TF-IDF важность и частоту в дробном формате, что более точно определяет значимость n-граммы, алгоритм Word Embeddings не реализован. В дальнейшем будет использоваться алгоритм TF-IDF
# ### Векторизация текст, поиск ключевых n-грамм методом TF-IDF
tfidf = {"tfidf": [], "vector": []}
for i in tqdm(range(len(articles_full["text"]))):
tfidf_matrix = vectorizer.fit_transform([" ".join(articles_full["text"][i])])
tfidf["vector"].append(tfidf_matrix)
feature_names_tfidf = vectorizer.get_feature_names_out()
uniq_and_fifa_tfidf = dict(zip(feature_names_tfidf, tfidf_matrix.toarray()[0]))
uniq_and_fifa_tfidf = dict(
sorted(uniq_and_fifa_tfidf.items(), key=lambda item: -item[1])
)
tfidf["tfidf"].append(uniq_and_fifa_tfidf)
len(tfidf["tfidf"])
articles_full["tfidf"] = tfidf["tfidf"]
articles_full["vector"] = tfidf["vector"]
articles_full
articles_full.to_pickle("articles_full_tfidf.pkl")
# Получен dataframe с 9 колонками, представлены алгоритмы векторизации/поиск ключевых n-грамм методами: Bag of words, TF-IDF
# # 1.5 Разведочный анализ
articles_full = pd.read_pickle("articles_full_tfidf.pkl")
articles_full
# ### Парсинг json
file = "Target.json"
nominations = {"name_company": [], "nominations": [], "code": []}
with codecs.open(file, "r", "utf_8_sig") as f:
templates = json.load(f)
# считывание информации о компаниях
if templates["info"] != None:
for i in range(len(templates["info"])):
nominations["name_company"].append(
templates["info"][i]["Сompany"].lower().strip()
)
nominations["nominations"].append(templates["info"][i]["Nominations"])
nominations["nominations"]
# Кодирование номинаций
from sklearn.preprocessing import LabelEncoder
encoder = LabelEncoder()
encoded_data = encoder.fit_transform(nominations["nominations"])
nominations["code"] = encoded_data
nominations_df = pd.DataFrame(nominations)
nominations_df
len(nominations_df)
# 22 компании, все есть
nominations_df.info()
articles_full = articles_full.merge(nominations_df, how="left", on="name_company")
articles_full
articles_full.info()
# ### Определений и замена пустых значений
import numpy as np
articles_full[articles_full["code"].isnull()]
articles_full["code"].fillna(0, inplace=True)
articles_full["nominations"].fillna("Здоровье и медицина", inplace=True)
articles_full.loc[
articles_full["name_company"]
== "проект по использованию технологий компьютерного зрения на базе искусственного интеллекта (ии) для анализа медицинских изображений",
"name_company",
] = "проект по использованию технологий компьютерного зрения на базе искусственного интеллекта для анализа медицинских изображений"
len(articles_full["name_company"].unique())
# Так как одна компания не представлена ни где, осталось 21 уникальная компания
articles_full.info()
import scipy.stats as stats
import seaborn as sns
from scipy import stats
import matplotlib.pyplot as plt
# ### Нормальность распределения target переменной
ax = sns.distplot(articles_full["code"])
# Признак категориальный, нормального распределения нет, категории 0 и 3 завышены, похожи на выбросы
stats.probplot(articles_full["code"], dist="norm", plot=plt)
plt.show()
# Признак категориальный, не следую нормальное распределению
ax = sns.boxplot(x=articles_full["code"])
# Ящик с усами скошен в лево, 3,4,5 кластеры малочисленно
# ### Тест Шапиро-Уилка
statistic_shapiro, p_value_shapiro = stats.shapiro(articles_full["code"])
print("Статистика теста:", statistic_shapiro)
print("p-значение:", p_value_shapiro)
# ### К-квадрат Д’Агостино
statistic_normaltest, p_value_normaltest = stats.normaltest(articles_full["code"])
print("Статистика теста:", statistic_normaltest)
print("p-значение:", p_value_normaltest)
# Математические расчеты показали, что действительно целевая переменная распределена не нормально, предположительно из-за того, что многие компании, простеленные в документе, прекратили свою деятельность, и их статей в открытом доступу оказалось очень мало
# ### Количественная оценка
# Пример категориальных данных
data = articles_full["code"]
# Построение гистограммы
data.value_counts().plot(kind="bar")
# Настройка осей и заголовка
plt.xlabel("Категория")
plt.ylabel("Частота")
plt.title("Распределение категориального признака")
# Отображение графика
plt.show()
# Пример категориальных данных
data = articles_full["code"]
# Построение круговой диаграммы
data.value_counts().plot(kind="pie")
# Настройка заголовка
plt.title("Распределение категориального признака")
# Отображение графика
plt.show()
# Количественная оценка категорий показала, что больше всего статей с номинацией 3(Наука, технологии и инновации), совсем мало статей номинацией 1(Игровая индустрия и киберспорт)
# ### Зависимость целевой переменной от месяца и дня публикации статьи
month = []
for i in tqdm(range(len(articles_full["date"]))):
month.append(float(str(articles_full["date"][i])[4:]))
articles_full["month"] = month
articles_full
sns.jointplot(x="code", y="month", data=articles_full)
# По сколько одних статей на порядок больше, чем других, нельзя сказать когда больше всего публикуется определённая номинация статей, но можно сказать, что в декабре и в июле публикуется самое большое количество статей
# ### Зависимость целевой переменной от рейтинга статьи
sns.jointplot(x="code", y="rating", data=articles_full)
# Очень много заменено пустых значений на 0. Отсутствие данных не позволяет корректно оценить корреляцию признаков
# ### Зависимость целевой переменной от n-грамм
keywords_1 = []
for i in tqdm(range(len(articles_full["tfidf"]))):
keywords_1.append(list(articles_full["tfidf"][i].keys())[0:1])
# Пример данных
topics = articles_full["code"]
keywords = keywords_1
# Создание датафрейма для визуализации
data = []
for i, topic in enumerate(topics):
for keyword in keywords[i]:
data.append([topic, keyword])
df = pd.DataFrame(data, columns=["Кластер", "Ключевое слово"])
# Построение графика
plt.figure(figsize=(8, 6))
sns.scatterplot(data=df, x="Ключевое слово", y="Кластер")
plt.xticks(rotation=45)
# Отображение графика
plt.show()
# Больше статей больше ключевых слов добавлено
nominations_df
# Для наглядности вывели номинации и их название
articles_full.to_csv("articles_full", encoding="utf-8-sig", index=False)
articles_full.to_pickle("articles_full_end.pkl")
| false | 0 | 11,158 | 0 | 11,158 | 11,158 |
||
129599461
|
<jupyter_start><jupyter_text>Contrails normalized float16 part 5
This is a optimized version of [google's contrails dataset](https://www.kaggle.com/competitions/google-research-identify-contrails-reduce-global-warming/data).
Features of this dataset:
- about 55% of the original dataset did not contain contrails which have been removed here.
- each example target pair is saved in a single npz file, instead of 10 npy files, making it load much faster with a lot less cpu usage, since number of IO operation are 10 times less.
- Examples stored in float16. This saves a lot of space. The loss of information from using float16 instead of float32 (mean absolute reconstruction error) was less than 0.019%.
- All Examples have been normalized as per channel mean and std.
Disadvantages:
- Individual human mask have been omitted.
- The examples without contrails have been omitted
- Information lost (less than 0.019%) due to the use of float16
There are 5 parts:
[part_1](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-1) [part_2](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-2) [part_3](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-3) [part_4](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-4) [part_5](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-5)
Kaggle dataset identifier: google-contrails-normalized-float16-part-5
<jupyter_script># # SuperFast Data Loading
# optimized dataset details - https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-1
# This is a optimized version of [google's contrails dataset](https://www.kaggle.com/competitions/google-research-identify-contrails-reduce-global-warming/data).
# Features of this dataset:
# - about 55% of the original dataset did not contain contrails which have been removed here.
# - each example target pair is saved in a single npz file, instead of 10 npy files, making it load much faster with a lot less cpu usage, since number of IO operation are 10 times less.
# - Examples stored in float16. This saves a lot of space. The loss of information from using float16 instead of float32 (mean absolute reconstruction error) was less than 0.019%.
# - All Examples have been normalized as per channel mean and std.
# Disadvantages:
# - Individual human mask have been omitted.
# - The examples without contrails have been omitted
# - Information lost (less than 0.019%) due to the use of float16
# There are 5 parts:
# [part_1](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-1) [part_2](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-2) [part_3](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-3) [part_4](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-4) [part_5](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-5)
import numpy as np
import os
from torch.utils.data import Dataset, DataLoader
from tqdm.auto import tqdm
# ### Get file path list for train and val
base_dir_1 = (
"/kaggle/input/google-contrails-normalized-float16-part-1/contrail_f16_part_1"
)
base_dir_2 = (
"/kaggle/input/google-contrails-normalized-float16-part-2/contrail_f16_part_2"
)
base_dir_3 = (
"/kaggle/input/google-contrails-normalized-float16-part-3/contrail_f16_part_3"
)
base_dir_4 = (
"/kaggle/input/google-contrails-normalized-float16-part-4/contrail_f16_part_4"
)
base_dir_5 = (
"/kaggle/input/google-contrails-normalized-float16-part-5/contrail_f16_part_5"
)
record_id_paths = []
record_ids_only = []
for base_dir in [base_dir_1, base_dir_2, base_dir_3, base_dir_4, base_dir_5]:
record_ids = os.listdir(base_dir)
for record_id in record_ids:
record_id_paths.append(os.path.join(base_dir, record_id))
record_ids_only.append(str(record_id)[:-4])
train_ids_non_empty = np.load(
"/kaggle/input/google-contrails-normalized-float16-part-3/train_ids_non_empty.npy"
)
val_ids_non_empty = np.load(
"/kaggle/input/google-contrails-normalized-float16-part-3/val_ids_non_empty.npy"
)
val_ids_non_empty_paths = []
train_ids_non_empty_paths = []
for record_id, path in zip(record_ids_only, record_id_paths):
if record_id in train_ids_non_empty:
train_ids_non_empty_paths.append(path)
elif record_id in val_ids_non_empty:
val_ids_non_empty_paths.append(path)
else:
print("Whay?! how?!!")
len(record_id_paths)
# The dataset return x of shape (8,9,256,256) : (time,channel,H,W)
class FastContrails(Dataset):
def __init__(self, record_id_paths):
self.record_id_paths = record_id_paths
self.length = len(self.record_id_paths)
def __getitem__(self, idx):
data = np.load(self.record_id_paths[idx])
return {"image": data["x"], "mask": data["y"]}
def __len__(self):
return self.length
train_dataset = FastContrails(train_ids_non_empty_paths)
val_dataset = FastContrails(val_ids_non_empty_paths)
train_loader = DataLoader(train_dataset, batch_size=32, num_workers=4)
val_loader = DataLoader(val_dataset, batch_size=32, num_workers=4)
for data in tqdm(train_loader):
x = data["image"]
y = data["mask"]
# print(x.shape, y.shape, x.dtype, y.dtype)
for data in tqdm(val_loader):
x = data["image"]
y = data["mask"]
# print(x.shape, y.shape, x.dtype, y.dtype)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/599/129599461.ipynb
|
google-contrails-normalized-float16-part-5
|
soumyadeepkhandual
|
[{"Id": 129599461, "ScriptId": 38523868, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10708605, "CreationDate": "05/15/2023 06:37:52", "VersionNumber": 3.0, "Title": "SuperFast DataLoading", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 91.0, "LinesInsertedFromPrevious": 34.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 57.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 7}]
|
[{"Id": 185831646, "KernelVersionId": 129599461, "SourceDatasetVersionId": 5687640}, {"Id": 185831645, "KernelVersionId": 129599461, "SourceDatasetVersionId": 5687524}, {"Id": 185831643, "KernelVersionId": 129599461, "SourceDatasetVersionId": 5685462}, {"Id": 185831642, "KernelVersionId": 129599461, "SourceDatasetVersionId": 5684997}]
|
[{"Id": 5687640, "DatasetId": 3269901, "DatasourceVersionId": 5763231, "CreatorUserId": 10708605, "LicenseName": "Unknown", "CreationDate": "05/15/2023 06:04:42", "VersionNumber": 1.0, "Title": "Contrails normalized float16 part 5", "Slug": "google-contrails-normalized-float16-part-5", "Subtitle": NaN, "Description": "This is a optimized version of [google's contrails dataset](https://www.kaggle.com/competitions/google-research-identify-contrails-reduce-global-warming/data).\n\nFeatures of this dataset:\n- about 55% of the original dataset did not contain contrails which have been removed here.\n- each example target pair is saved in a single npz file, instead of 10 npy files, making it load much faster with a lot less cpu usage, since number of IO operation are 10 times less.\n- Examples stored in float16. This saves a lot of space. The loss of information from using float16 instead of float32 (mean absolute reconstruction error) was less than 0.019%.\n- All Examples have been normalized as per channel mean and std.\n\nDisadvantages:\n- Individual human mask have been omitted.\n- The examples without contrails have been omitted\n- Information lost (less than 0.019%) due to the use of float16 \n\nThere are 5 parts:\n[part_1](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-1) [part_2](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-2) [part_3](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-3) [part_4](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-4) [part_5](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-5)", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3269901, "CreatorUserId": 10708605, "OwnerUserId": 10708605.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5687640.0, "CurrentDatasourceVersionId": 5763231.0, "ForumId": 3335531, "Type": 2, "CreationDate": "05/15/2023 06:04:42", "LastActivityDate": "05/15/2023", "TotalViews": 101, "TotalDownloads": 11, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 10708605, "UserName": "soumyadeepkhandual", "DisplayName": "Soumyadeep Khandual", "RegisterDate": "06/01/2022", "PerformanceTier": 1}]
|
# # SuperFast Data Loading
# optimized dataset details - https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-1
# This is a optimized version of [google's contrails dataset](https://www.kaggle.com/competitions/google-research-identify-contrails-reduce-global-warming/data).
# Features of this dataset:
# - about 55% of the original dataset did not contain contrails which have been removed here.
# - each example target pair is saved in a single npz file, instead of 10 npy files, making it load much faster with a lot less cpu usage, since number of IO operation are 10 times less.
# - Examples stored in float16. This saves a lot of space. The loss of information from using float16 instead of float32 (mean absolute reconstruction error) was less than 0.019%.
# - All Examples have been normalized as per channel mean and std.
# Disadvantages:
# - Individual human mask have been omitted.
# - The examples without contrails have been omitted
# - Information lost (less than 0.019%) due to the use of float16
# There are 5 parts:
# [part_1](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-1) [part_2](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-2) [part_3](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-3) [part_4](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-4) [part_5](https://www.kaggle.com/datasets/soumyadeepkhandual/google-contrails-normalized-float16-part-5)
import numpy as np
import os
from torch.utils.data import Dataset, DataLoader
from tqdm.auto import tqdm
# ### Get file path list for train and val
base_dir_1 = (
"/kaggle/input/google-contrails-normalized-float16-part-1/contrail_f16_part_1"
)
base_dir_2 = (
"/kaggle/input/google-contrails-normalized-float16-part-2/contrail_f16_part_2"
)
base_dir_3 = (
"/kaggle/input/google-contrails-normalized-float16-part-3/contrail_f16_part_3"
)
base_dir_4 = (
"/kaggle/input/google-contrails-normalized-float16-part-4/contrail_f16_part_4"
)
base_dir_5 = (
"/kaggle/input/google-contrails-normalized-float16-part-5/contrail_f16_part_5"
)
record_id_paths = []
record_ids_only = []
for base_dir in [base_dir_1, base_dir_2, base_dir_3, base_dir_4, base_dir_5]:
record_ids = os.listdir(base_dir)
for record_id in record_ids:
record_id_paths.append(os.path.join(base_dir, record_id))
record_ids_only.append(str(record_id)[:-4])
train_ids_non_empty = np.load(
"/kaggle/input/google-contrails-normalized-float16-part-3/train_ids_non_empty.npy"
)
val_ids_non_empty = np.load(
"/kaggle/input/google-contrails-normalized-float16-part-3/val_ids_non_empty.npy"
)
val_ids_non_empty_paths = []
train_ids_non_empty_paths = []
for record_id, path in zip(record_ids_only, record_id_paths):
if record_id in train_ids_non_empty:
train_ids_non_empty_paths.append(path)
elif record_id in val_ids_non_empty:
val_ids_non_empty_paths.append(path)
else:
print("Whay?! how?!!")
len(record_id_paths)
# The dataset return x of shape (8,9,256,256) : (time,channel,H,W)
class FastContrails(Dataset):
def __init__(self, record_id_paths):
self.record_id_paths = record_id_paths
self.length = len(self.record_id_paths)
def __getitem__(self, idx):
data = np.load(self.record_id_paths[idx])
return {"image": data["x"], "mask": data["y"]}
def __len__(self):
return self.length
train_dataset = FastContrails(train_ids_non_empty_paths)
val_dataset = FastContrails(val_ids_non_empty_paths)
train_loader = DataLoader(train_dataset, batch_size=32, num_workers=4)
val_loader = DataLoader(val_dataset, batch_size=32, num_workers=4)
for data in tqdm(train_loader):
x = data["image"]
y = data["mask"]
# print(x.shape, y.shape, x.dtype, y.dtype)
for data in tqdm(val_loader):
x = data["image"]
y = data["mask"]
# print(x.shape, y.shape, x.dtype, y.dtype)
| false | 0 | 1,396 | 7 | 1,869 | 1,396 |
||
129599059
|
import numpy as np
import pandas as pd
from tensorflow.keras.utils import load_img
from keras.preprocessing.image import ImageDataGenerator
from keras.utils import to_categorical
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import random
import os
# import seaborn as sns
import keras
from keras.models import Sequential
from keras.layers import (
Conv2D,
MaxPooling2D,
Dropout,
Flatten,
Dense,
Activation,
BatchNormalization,
)
# from keras.layers import Lambda # convolution layers
from keras.preprocessing.image import ImageDataGenerator
# from keras.utils.np_utils import to_categorical
train = pd.read_csv("../input/mnist-train-mini/mnist_train_mini.csv", header=None)
test = pd.read_csv("../input/mnist-test-mini/mnist_test_mini.csv", header=None)
X = train.drop([0], 1).values
y = train[0].values
# X = train[1:].values
# y = train[0].values
X = X / 255.0
X = X.reshape(-1, 28, 28, 1)
y = to_categorical(y)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1, random_state=0)
mean = np.mean(X_train)
std = np.std(X_train)
def standardize(x):
return (x - mean) / std
model = Sequential()
# model.add(Lambda(standardize,input_shape=(28,28,1)))
model.add(
Conv2D(filters=64, kernel_size=(3, 3), activation="relu", input_shape=(28, 28, 1))
)
model.add(Conv2D(filters=64, kernel_size=(3, 3), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(BatchNormalization())
model.add(Conv2D(filters=128, kernel_size=(3, 3), activation="relu"))
model.add(Conv2D(filters=128, kernel_size=(3, 3), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(BatchNormalization())
model.add(Conv2D(filters=256, kernel_size=(3, 3), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(BatchNormalization())
model.add(Flatten())
model.add(Dense(512, activation="relu"))
model.add(Dense(10, activation="softmax"))
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
model.summary()
datagen = ImageDataGenerator(
featurewise_center=False, # set input mean to 0 over the dataset
samplewise_center=False, # set each sample mean to 0
featurewise_std_normalization=False, # divide inputs by std of the dataset
samplewise_std_normalization=False, # divide each input by its std
zca_whitening=False, # apply ZCA whitening
rotation_range=15, # randomly rotate images in the range (degrees, 0 to 180)
zoom_range=0.01, # Randomly zoom image
width_shift_range=0.1, # randomly shift images horizontally (fraction of total width)
height_shift_range=0.1, # randomly shift images vertically (fraction of total height)
horizontal_flip=False, # randomly flip images
vertical_flip=False,
) # randomly flip images
# datagen.fit(X_train)
train_gen = datagen.flow(X_train, y_train, batch_size=128)
test_gen = datagen.flow(X_test, y_test, batch_size=128)
epochs = 100
batch_size = 128
train_steps = X_train.shape[0] // batch_size
valid_steps = X_test.shape[0] // batch_size
es = keras.callbacks.EarlyStopping(
monitor="val_acc", # metrics to monitor
patience=10, # how many epochs before stop
verbose=1,
mode="max", # we need the maximum accuracy.
restore_best_weights=True, #
)
rp = keras.callbacks.ReduceLROnPlateau(
monitor="val_acc",
factor=0.2,
patience=3,
verbose=1,
mode="max",
min_lr=0.00001,
)
history = model.fit(
train_gen,
epochs=epochs,
steps_per_epoch=train_steps,
validation_data=test_gen,
validation_steps=valid_steps,
callbacks=[es, rp],
)
fig, (ax2) = plt.subplots(1, 1, figsize=(12, 12))
ax2.plot(history.history["accuracy"], color="b", label="Training accuracy")
ax2.plot(history.history["val_accuracy"], color="r", label="Test accuracy")
ax2.set_xticks(np.arange(1, epochs, 1))
legend = plt.legend(loc="best", shadow=True)
plt.tight_layout()
plt.show()
y_pred = model.predict(X_test)
X_test__ = X_test.reshape(X_test.shape[0], 28, 28)
fig, axis = plt.subplots(4, 4, figsize=(12, 14))
for i, ax in enumerate(axis.flat):
ax.imshow(X_test__[i], cmap="binary")
ax.set(
title=f"Real Number is {y_test[i].argmax()}\nPredict Number is {y_pred[i].argmax()}"
)
test_x = test.drop([0], 1).values
test_x = test_x / 255.0
test_x = test_x.reshape(-1, 28, 28, 1)
# predict test data
pred = model.predict(test_x, verbose=1)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/599/129599059.ipynb
| null | null |
[{"Id": 129599059, "ScriptId": 38536308, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15001012, "CreationDate": "05/15/2023 06:34:21", "VersionNumber": 1.0, "Title": "CNN Part 2", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 137.0, "LinesInsertedFromPrevious": 137.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
from tensorflow.keras.utils import load_img
from keras.preprocessing.image import ImageDataGenerator
from keras.utils import to_categorical
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import random
import os
# import seaborn as sns
import keras
from keras.models import Sequential
from keras.layers import (
Conv2D,
MaxPooling2D,
Dropout,
Flatten,
Dense,
Activation,
BatchNormalization,
)
# from keras.layers import Lambda # convolution layers
from keras.preprocessing.image import ImageDataGenerator
# from keras.utils.np_utils import to_categorical
train = pd.read_csv("../input/mnist-train-mini/mnist_train_mini.csv", header=None)
test = pd.read_csv("../input/mnist-test-mini/mnist_test_mini.csv", header=None)
X = train.drop([0], 1).values
y = train[0].values
# X = train[1:].values
# y = train[0].values
X = X / 255.0
X = X.reshape(-1, 28, 28, 1)
y = to_categorical(y)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1, random_state=0)
mean = np.mean(X_train)
std = np.std(X_train)
def standardize(x):
return (x - mean) / std
model = Sequential()
# model.add(Lambda(standardize,input_shape=(28,28,1)))
model.add(
Conv2D(filters=64, kernel_size=(3, 3), activation="relu", input_shape=(28, 28, 1))
)
model.add(Conv2D(filters=64, kernel_size=(3, 3), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(BatchNormalization())
model.add(Conv2D(filters=128, kernel_size=(3, 3), activation="relu"))
model.add(Conv2D(filters=128, kernel_size=(3, 3), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(BatchNormalization())
model.add(Conv2D(filters=256, kernel_size=(3, 3), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(BatchNormalization())
model.add(Flatten())
model.add(Dense(512, activation="relu"))
model.add(Dense(10, activation="softmax"))
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
model.summary()
datagen = ImageDataGenerator(
featurewise_center=False, # set input mean to 0 over the dataset
samplewise_center=False, # set each sample mean to 0
featurewise_std_normalization=False, # divide inputs by std of the dataset
samplewise_std_normalization=False, # divide each input by its std
zca_whitening=False, # apply ZCA whitening
rotation_range=15, # randomly rotate images in the range (degrees, 0 to 180)
zoom_range=0.01, # Randomly zoom image
width_shift_range=0.1, # randomly shift images horizontally (fraction of total width)
height_shift_range=0.1, # randomly shift images vertically (fraction of total height)
horizontal_flip=False, # randomly flip images
vertical_flip=False,
) # randomly flip images
# datagen.fit(X_train)
train_gen = datagen.flow(X_train, y_train, batch_size=128)
test_gen = datagen.flow(X_test, y_test, batch_size=128)
epochs = 100
batch_size = 128
train_steps = X_train.shape[0] // batch_size
valid_steps = X_test.shape[0] // batch_size
es = keras.callbacks.EarlyStopping(
monitor="val_acc", # metrics to monitor
patience=10, # how many epochs before stop
verbose=1,
mode="max", # we need the maximum accuracy.
restore_best_weights=True, #
)
rp = keras.callbacks.ReduceLROnPlateau(
monitor="val_acc",
factor=0.2,
patience=3,
verbose=1,
mode="max",
min_lr=0.00001,
)
history = model.fit(
train_gen,
epochs=epochs,
steps_per_epoch=train_steps,
validation_data=test_gen,
validation_steps=valid_steps,
callbacks=[es, rp],
)
fig, (ax2) = plt.subplots(1, 1, figsize=(12, 12))
ax2.plot(history.history["accuracy"], color="b", label="Training accuracy")
ax2.plot(history.history["val_accuracy"], color="r", label="Test accuracy")
ax2.set_xticks(np.arange(1, epochs, 1))
legend = plt.legend(loc="best", shadow=True)
plt.tight_layout()
plt.show()
y_pred = model.predict(X_test)
X_test__ = X_test.reshape(X_test.shape[0], 28, 28)
fig, axis = plt.subplots(4, 4, figsize=(12, 14))
for i, ax in enumerate(axis.flat):
ax.imshow(X_test__[i], cmap="binary")
ax.set(
title=f"Real Number is {y_test[i].argmax()}\nPredict Number is {y_pred[i].argmax()}"
)
test_x = test.drop([0], 1).values
test_x = test_x / 255.0
test_x = test_x.reshape(-1, 28, 28, 1)
# predict test data
pred = model.predict(test_x, verbose=1)
| false | 0 | 1,537 | 0 | 1,537 | 1,537 |
||
129599977
|
# # 1 . Libraries
from language_tool_python import LanguageTool
# # 2 . initialize
# ### Grammar Errors
def displayGrammarErrors(matches):
if len(matches) > 0:
print("Total Grammar Errors: {}".format(len(matches)))
for match in matches:
print("\nError: {}".format(match.ruleId))
print("Message: {}".format(match.message))
print("Replacement: {}\n".format(match.replacements))
else:
print("No Grammar Errors Found!")
# # 3 . Check Grammar
def Check(text="I Love you PMC 🤠"):
tool = LanguageTool("en-US")
matches = tool.check(text)
print("Check :", text)
displayGrammarErrors(matches)
# # 4 . Fire 🔥
Check()
Check("love you.")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/599/129599977.ipynb
| null | null |
[{"Id": 129599977, "ScriptId": 38537763, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8788103, "CreationDate": "05/15/2023 06:42:15", "VersionNumber": 1.0, "Title": "Grammar-With-Pyth\u2764\ufe0fn", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 32.0, "LinesInsertedFromPrevious": 32.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 43}]
| null | null | null | null |
# # 1 . Libraries
from language_tool_python import LanguageTool
# # 2 . initialize
# ### Grammar Errors
def displayGrammarErrors(matches):
if len(matches) > 0:
print("Total Grammar Errors: {}".format(len(matches)))
for match in matches:
print("\nError: {}".format(match.ruleId))
print("Message: {}".format(match.message))
print("Replacement: {}\n".format(match.replacements))
else:
print("No Grammar Errors Found!")
# # 3 . Check Grammar
def Check(text="I Love you PMC 🤠"):
tool = LanguageTool("en-US")
matches = tool.check(text)
print("Check :", text)
displayGrammarErrors(matches)
# # 4 . Fire 🔥
Check()
Check("love you.")
| false | 0 | 200 | 43 | 200 | 200 |
||
129857996
|
<jupyter_start><jupyter_text>New Plant Diseases Dataset
**This dataset is recreated using offline augmentation from the original dataset. The original dataset can be found on [this][1] github repo. This dataset consists of about 87K rgb images of healthy and diseased crop leaves which is categorized into 38 different classes. The total dataset is divided into 80/20 ratio of training and validation set preserving the directory structure.
A new directory containing 33 test images is created later for prediction purpose.**
[1]: https://github.com/spMohanty/PlantVillage-Dataset
Kaggle dataset identifier: new-plant-diseases-dataset
<jupyter_script>import tensorflow as tf # Imports tensorflow, used for neural network building
from tensorflow.keras.applications.resnet50 import preprocess_input
from keras.preprocessing.image import ImageDataGenerator
import pandas as pd # used for data frames
from keras.callbacks import EarlyStopping
import os
# This code is for implementing my GPU
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
except ValueError:
tpu = None
gpus = tf.config.experimental.list_logical_devices("GPU")
if tpu:
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
print("Running on TPU: " + str(tpu.master()))
elif len(gpus) > 0:
strategy = tf.distribute.MirroredStrategy(gpus)
print("Running on ", len(gpus), " GPU(s)")
else:
strategy = tf.distribute.get_strategy()
print("Running on CPU")
num_classes = 38
resnet_weights_path = (
"../input/resnet50/resnet50_weights_tf_dim_ordering_tf_kernels_notop.h5"
)
my_new_model = tf.keras.Sequential(
[
tf.keras.applications.ResNet50(
include_top=False,
weights=resnet_weights_path,
input_shape=(224, 224, 3),
pooling="avg",
),
tf.keras.layers.Dense(num_classes, activation="softmax"),
]
)
my_new_model.layers[0].trainable = False
my_new_model.summary()
my_new_model.compile(
loss=tf.keras.losses.CategoricalCrossentropy(),
optimizer=tf.keras.optimizers.Adam(learning_rate=0.001),
metrics=["accuracy"],
)
image_size = 224
data_generator = ImageDataGenerator(preprocessing_function=preprocess_input)
root = "/kaggle/input/new-plant-diseases-dataset/New Plant Diseases Dataset(Augmented)/New Plant Diseases Dataset(Augmented)"
train_generator = data_generator.flow_from_directory(
root + "/train",
target_size=(image_size, image_size),
batch_size=32,
class_mode="categorical",
)
validation_generator = data_generator.flow_from_directory(
root + "/valid",
target_size=(image_size, image_size),
batch_size=32,
class_mode="categorical",
)
earlystop_callback = EarlyStopping(monitor="val_loss", verbose=2, patience=3)
history = my_new_model.fit(
train_generator,
batch_size=100,
epochs=9,
steps_per_epoch=80,
validation_data=validation_generator,
callbacks=[earlystop_callback],
)
# 100 batch size, 80 steps
my_new_model.save("PlantDiseaseModel-v1.h5")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/857/129857996.ipynb
|
new-plant-diseases-dataset
|
vipoooool
|
[{"Id": 129857996, "ScriptId": 38049151, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12174204, "CreationDate": "05/17/2023 02:38:35", "VersionNumber": 17.0, "Title": "Neural Network for Plant Disease", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 65.0, "LinesInsertedFromPrevious": 2.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 63.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186252692, "KernelVersionId": 129857996, "SourceDatasetVersionId": 182633}, {"Id": 186252693, "KernelVersionId": 129857996, "SourceDatasetVersionId": 872462}]
|
[{"Id": 182633, "DatasetId": 78313, "DatasourceVersionId": 193494, "CreatorUserId": 2009285, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "11/18/2018 07:09:16", "VersionNumber": 2.0, "Title": "New Plant Diseases Dataset", "Slug": "new-plant-diseases-dataset", "Subtitle": "Image dataset containing different healthy and unhealthy crop leaves.", "Description": "**This dataset is recreated using offline augmentation from the original dataset. The original dataset can be found on [this][1] github repo. This dataset consists of about 87K rgb images of healthy and diseased crop leaves which is categorized into 38 different classes. The total dataset is divided into 80/20 ratio of training and validation set preserving the directory structure.\nA new directory containing 33 test images is created later for prediction purpose.**\n\n\n [1]: https://github.com/spMohanty/PlantVillage-Dataset", "VersionNotes": "New Test Images", "TotalCompressedBytes": 1445887779.0, "TotalUncompressedBytes": 1445887779.0}]
|
[{"Id": 78313, "CreatorUserId": 2009285, "OwnerUserId": 2009285.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 182633.0, "CurrentDatasourceVersionId": 193494.0, "ForumId": 87652, "Type": 2, "CreationDate": "11/16/2018 12:17:57", "LastActivityDate": "11/16/2018", "TotalViews": 387678, "TotalDownloads": 47287, "TotalVotes": 766, "TotalKernels": 244}]
|
[{"Id": 2009285, "UserName": "vipoooool", "DisplayName": "Samir Bhattarai", "RegisterDate": "06/21/2018", "PerformanceTier": 0}]
|
import tensorflow as tf # Imports tensorflow, used for neural network building
from tensorflow.keras.applications.resnet50 import preprocess_input
from keras.preprocessing.image import ImageDataGenerator
import pandas as pd # used for data frames
from keras.callbacks import EarlyStopping
import os
# This code is for implementing my GPU
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
except ValueError:
tpu = None
gpus = tf.config.experimental.list_logical_devices("GPU")
if tpu:
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
print("Running on TPU: " + str(tpu.master()))
elif len(gpus) > 0:
strategy = tf.distribute.MirroredStrategy(gpus)
print("Running on ", len(gpus), " GPU(s)")
else:
strategy = tf.distribute.get_strategy()
print("Running on CPU")
num_classes = 38
resnet_weights_path = (
"../input/resnet50/resnet50_weights_tf_dim_ordering_tf_kernels_notop.h5"
)
my_new_model = tf.keras.Sequential(
[
tf.keras.applications.ResNet50(
include_top=False,
weights=resnet_weights_path,
input_shape=(224, 224, 3),
pooling="avg",
),
tf.keras.layers.Dense(num_classes, activation="softmax"),
]
)
my_new_model.layers[0].trainable = False
my_new_model.summary()
my_new_model.compile(
loss=tf.keras.losses.CategoricalCrossentropy(),
optimizer=tf.keras.optimizers.Adam(learning_rate=0.001),
metrics=["accuracy"],
)
image_size = 224
data_generator = ImageDataGenerator(preprocessing_function=preprocess_input)
root = "/kaggle/input/new-plant-diseases-dataset/New Plant Diseases Dataset(Augmented)/New Plant Diseases Dataset(Augmented)"
train_generator = data_generator.flow_from_directory(
root + "/train",
target_size=(image_size, image_size),
batch_size=32,
class_mode="categorical",
)
validation_generator = data_generator.flow_from_directory(
root + "/valid",
target_size=(image_size, image_size),
batch_size=32,
class_mode="categorical",
)
earlystop_callback = EarlyStopping(monitor="val_loss", verbose=2, patience=3)
history = my_new_model.fit(
train_generator,
batch_size=100,
epochs=9,
steps_per_epoch=80,
validation_data=validation_generator,
callbacks=[earlystop_callback],
)
# 100 batch size, 80 steps
my_new_model.save("PlantDiseaseModel-v1.h5")
| false | 0 | 753 | 0 | 904 | 753 |
||
129857138
|
<jupyter_start><jupyter_text>Intel Image Classification
### Context
This is image data of Natural Scenes around the world.
### Content
This Data contains around 25k images of size 150x150 distributed under 6 categories.
{'buildings' -> 0,
'forest' -> 1,
'glacier' -> 2,
'mountain' -> 3,
'sea' -> 4,
'street' -> 5 }
The Train, Test and Prediction data is separated in each zip files. There are around 14k images in Train, 3k in Test and 7k in Prediction.
This data was initially published on https://datahack.analyticsvidhya.com by Intel to host a Image classification Challenge.
Kaggle dataset identifier: intel-image-classification
<jupyter_script>import glob
import matplotlib
from matplotlib import pyplot as plt
import matplotlib.image as mpimg
import numpy as np
import imageio as im
from keras import models
from keras.models import Sequential
from keras.models import Model
from keras.layers import Conv2D
from keras.layers import MaxPooling2D
from keras.layers import Flatten
from keras.layers import Dense
from keras.layers import Dropout
# from keras.preprocessing import image
import keras.utils as image
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import ModelCheckpoint
import pickle
from pickle import dump
from pickle import load
def save_pickle(file, file_name):
dump(file, open(file_name, "wb"))
print("Saved: %s" % file_name)
def load_pickle(file_name):
return load(open(file_name, "rb"))
# Loading model:
# PATHS FROM MODELS
# path to the folder containing the subfolders with the training images
model_path = "/kaggle/input/models-assignment-05/my-pickle-model.pkl"
my_pickle_model = load_pickle(model_path)
# my_pickle_model = load_model(model_path)
# ### Predicting the class of unseen images
# Remembering class names ordered alphabetically
class_names = ["buildings", "forest", "glacier", "mountain", "sea", "street"]
class_labels = {class_name: i for i, class_name in enumerate(class_names)}
print(class_labels)
number_classes = len(class_names)
print(f"Images Types: {number_classes}")
img_path = "/kaggle/input/intel-image-classification/seg_test/seg_test/forest/20619.jpg"
IMAGE_SIZE = (150, 150)
img = image.load_img(img_path, target_size=IMAGE_SIZE)
img_tensor = image.img_to_array(img)
img_tensor = np.expand_dims(img_tensor, axis=0)
img_tensor /= 255.0
plt.imshow(img_tensor[0])
plt.show()
print(img_tensor.shape)
# predicting images
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
images = np.vstack([x])
# classes = my_pickle_model.predict_classes(images, batch_size=10)
predict_x = my_pickle_model.predict(images)
classes_x = np.argmax(predict_x, axis=1)
print("Predicted class is:", classes_x)
# # Visualizing intermediate activations
# ### Instantiating a model from an input tensor and a list of output tensors
# Extracts the outputs of the top 12 layers
layer_outputs = [layer.output for layer in my_pickle_model.layers[:12]]
# Creates a model that will return these outputs, given the model input
activation_model = Model(inputs=my_pickle_model.input, outputs=layer_outputs)
# Returns a list of five Numpy arrays: one array per layer activation
activations = activation_model.predict(img_tensor)
first_layer_activation = activations[0]
print(first_layer_activation.shape)
plt.matshow(first_layer_activation[0, :, :, 4], cmap="viridis")
# ### Visualizing every channel in every intermediate activation
layer_names = []
for layer in my_pickle_model.layers[:12]:
layer_names.append(
layer.name
) # Names of the layers, so you can have them as part of your plot
images_per_row = 16
for layer_name, layer_activation in zip(
layer_names, activations
): # Displays the feature maps
n_features = layer_activation.shape[-1] # Number of features in the feature map
size = layer_activation.shape[
1
] # The feature map has shape (1, size, size, n_features).
n_cols = (
n_features // images_per_row
) # Tiles the activation channels in this matrix
display_grid = np.zeros((size * n_cols, images_per_row * size))
for col in range(n_cols): # Tiles each filter into a big horizontal grid
for row in range(images_per_row):
channel_image = layer_activation[0, :, :, col * images_per_row + row]
channel_image -= (
channel_image.mean()
) # Post-processes the feature to make it visually palatable
channel_image /= channel_image.std()
channel_image *= 64
channel_image += 128
channel_image = np.clip(channel_image, 0, 255).astype("uint8")
display_grid[
col * size : (col + 1) * size, # Displays the grid
row * size : (row + 1) * size,
] = channel_image
scale = 1.0 / size
plt.figure(figsize=(scale * display_grid.shape[1], scale * display_grid.shape[0]))
plt.title(layer_name)
plt.grid(False)
plt.imshow(display_grid, aspect="auto", cmap="viridis")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/857/129857138.ipynb
|
intel-image-classification
|
puneet6060
|
[{"Id": 129857138, "ScriptId": 38621466, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10470120, "CreationDate": "05/17/2023 02:26:23", "VersionNumber": 2.0, "Title": "Visualizing Intermediate Activation Images", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 118.0, "LinesInsertedFromPrevious": 71.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 47.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186251087, "KernelVersionId": 129857138, "SourceDatasetVersionId": 269359}]
|
[{"Id": 269359, "DatasetId": 111880, "DatasourceVersionId": 281586, "CreatorUserId": 2307235, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "01/30/2019 09:22:58", "VersionNumber": 2.0, "Title": "Intel Image Classification", "Slug": "intel-image-classification", "Subtitle": "Image Scene Classification of Multiclass", "Description": "### Context\n\nThis is image data of Natural Scenes around the world. \n\n### Content\n\nThis Data contains around 25k images of size 150x150 distributed under 6 categories.\n{'buildings' -> 0, \n'forest' -> 1,\n'glacier' -> 2,\n'mountain' -> 3,\n'sea' -> 4,\n'street' -> 5 }\n\nThe Train, Test and Prediction data is separated in each zip files. There are around 14k images in Train, 3k in Test and 7k in Prediction.\nThis data was initially published on https://datahack.analyticsvidhya.com by Intel to host a Image classification Challenge.\n\n\n### Acknowledgements\n\nThanks to https://datahack.analyticsvidhya.com for the challenge and Intel for the Data\n\nPhoto by [Jan B\u00f6ttinger on Unsplash][1]\n\n### Inspiration\n\nWant to build powerful Neural network that can classify these images with more accuracy.\n\n\n [1]: https://unsplash.com/photos/27xFENkt-lc", "VersionNotes": "Added Prediction Images", "TotalCompressedBytes": 108365415.0, "TotalUncompressedBytes": 361713334.0}]
|
[{"Id": 111880, "CreatorUserId": 2307235, "OwnerUserId": 2307235.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 269359.0, "CurrentDatasourceVersionId": 281586.0, "ForumId": 121691, "Type": 2, "CreationDate": "01/29/2019 10:37:42", "LastActivityDate": "01/29/2019", "TotalViews": 441103, "TotalDownloads": 83887, "TotalVotes": 1345, "TotalKernels": 815}]
|
[{"Id": 2307235, "UserName": "puneet6060", "DisplayName": "Puneet Bansal", "RegisterDate": "10/01/2018", "PerformanceTier": 0}]
|
import glob
import matplotlib
from matplotlib import pyplot as plt
import matplotlib.image as mpimg
import numpy as np
import imageio as im
from keras import models
from keras.models import Sequential
from keras.models import Model
from keras.layers import Conv2D
from keras.layers import MaxPooling2D
from keras.layers import Flatten
from keras.layers import Dense
from keras.layers import Dropout
# from keras.preprocessing import image
import keras.utils as image
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import ModelCheckpoint
import pickle
from pickle import dump
from pickle import load
def save_pickle(file, file_name):
dump(file, open(file_name, "wb"))
print("Saved: %s" % file_name)
def load_pickle(file_name):
return load(open(file_name, "rb"))
# Loading model:
# PATHS FROM MODELS
# path to the folder containing the subfolders with the training images
model_path = "/kaggle/input/models-assignment-05/my-pickle-model.pkl"
my_pickle_model = load_pickle(model_path)
# my_pickle_model = load_model(model_path)
# ### Predicting the class of unseen images
# Remembering class names ordered alphabetically
class_names = ["buildings", "forest", "glacier", "mountain", "sea", "street"]
class_labels = {class_name: i for i, class_name in enumerate(class_names)}
print(class_labels)
number_classes = len(class_names)
print(f"Images Types: {number_classes}")
img_path = "/kaggle/input/intel-image-classification/seg_test/seg_test/forest/20619.jpg"
IMAGE_SIZE = (150, 150)
img = image.load_img(img_path, target_size=IMAGE_SIZE)
img_tensor = image.img_to_array(img)
img_tensor = np.expand_dims(img_tensor, axis=0)
img_tensor /= 255.0
plt.imshow(img_tensor[0])
plt.show()
print(img_tensor.shape)
# predicting images
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
images = np.vstack([x])
# classes = my_pickle_model.predict_classes(images, batch_size=10)
predict_x = my_pickle_model.predict(images)
classes_x = np.argmax(predict_x, axis=1)
print("Predicted class is:", classes_x)
# # Visualizing intermediate activations
# ### Instantiating a model from an input tensor and a list of output tensors
# Extracts the outputs of the top 12 layers
layer_outputs = [layer.output for layer in my_pickle_model.layers[:12]]
# Creates a model that will return these outputs, given the model input
activation_model = Model(inputs=my_pickle_model.input, outputs=layer_outputs)
# Returns a list of five Numpy arrays: one array per layer activation
activations = activation_model.predict(img_tensor)
first_layer_activation = activations[0]
print(first_layer_activation.shape)
plt.matshow(first_layer_activation[0, :, :, 4], cmap="viridis")
# ### Visualizing every channel in every intermediate activation
layer_names = []
for layer in my_pickle_model.layers[:12]:
layer_names.append(
layer.name
) # Names of the layers, so you can have them as part of your plot
images_per_row = 16
for layer_name, layer_activation in zip(
layer_names, activations
): # Displays the feature maps
n_features = layer_activation.shape[-1] # Number of features in the feature map
size = layer_activation.shape[
1
] # The feature map has shape (1, size, size, n_features).
n_cols = (
n_features // images_per_row
) # Tiles the activation channels in this matrix
display_grid = np.zeros((size * n_cols, images_per_row * size))
for col in range(n_cols): # Tiles each filter into a big horizontal grid
for row in range(images_per_row):
channel_image = layer_activation[0, :, :, col * images_per_row + row]
channel_image -= (
channel_image.mean()
) # Post-processes the feature to make it visually palatable
channel_image /= channel_image.std()
channel_image *= 64
channel_image += 128
channel_image = np.clip(channel_image, 0, 255).astype("uint8")
display_grid[
col * size : (col + 1) * size, # Displays the grid
row * size : (row + 1) * size,
] = channel_image
scale = 1.0 / size
plt.figure(figsize=(scale * display_grid.shape[1], scale * display_grid.shape[0]))
plt.title(layer_name)
plt.grid(False)
plt.imshow(display_grid, aspect="auto", cmap="viridis")
| false | 0 | 1,279 | 0 | 1,481 | 1,279 |
||
129857818
|
<jupyter_start><jupyter_text>Industrial Quality Control of Packages
**Summary:**
This dataset consists of synthetically created images of packages produced in an industrial production line in which the packaging machine occasionally produces faulty packages. One of the goals for the data scientist is to create a model that can identify damaged packages.
**Details:**
Many companies in the manufacturing industry use packaging machines to wrap their products. These machines usually work fully automatically, but from time to time, faulty packages are produced, for example because small deviations in the position of the packages cause them to be dented or bent. Therefore, in-line quality control inspection points exist across the production line in which different quality measures are taken that should ensure that faulty packages are excluded from the process before they are sent out.
This dataset consists of RGB images taken from a “virtual” production line (meaning, the images were created procedurally, see below) with two classes: damaged and intact. For each class and each package, two camera captures exist: One image taken from a camera mounted above the package, one picture taken from a camera mounted on the side of the inspection belt. Each package is identified by a unique serial number (SN) visible on the side-view of the package. This serial number is also used for naming the files ({sn}_side.png and {sn}_top.png).
The goal of this dataset is to give data scientists the possibility to work on data inspired by industrial manufacturing scenarios. Among others the following questions are interesting:
• Can you train a model that identifies damaged packages using both, top and side view?
• Can you train a model that identifies damaged packages using just the top view?
• Can you train a model that performs OCR to extract the serial numbers (some SN may be blurry due to simulated motion blur)
• Can you create a one-shot computer vision algorithms that can classify packages by just learning from one or two samples?
As mentioned above, the images were created synthetically using Blender. The code to produce these images is open-sourced and can be found at https://github.com/christian-vorhemus/procedural-3d-image-generation
Kaggle dataset identifier: industrial-quality-control-of-packages
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
import cv2
import numpy as np
# Define the root directory and the subdirectories
root_dir = "/kaggle/input/industrial-quality-control-of-packages/"
damaged_side_dir = os.path.join(root_dir, "damaged/side/")
damaged_top_dir = os.path.join(root_dir, "damaged/top/")
intact_side_dir = os.path.join(root_dir, "intact/side/")
intact_top_dir = os.path.join(root_dir, "intact/top/")
# Function to load images from a directory
def load_images(directory):
images = []
for filename in os.listdir(directory):
if filename.endswith(".jpg") or filename.endswith(".png"):
img_path = os.path.join(directory, filename)
img = cv2.imread(img_path)
if img is not None:
images.append(img)
return images
# Load the images from each category
damaged_side_images = load_images(damaged_side_dir)
damaged_top_images = load_images(damaged_top_dir)
intact_side_images = load_images(intact_side_dir)
intact_top_images = load_images(intact_top_dir)
# Create labels for the images
damaged_labels = np.ones(len(damaged_side_images) + len(damaged_top_images))
intact_labels = np.zeros(len(intact_side_images) + len(intact_top_images))
# Concatenate the images and labels
images = np.concatenate(
(damaged_side_images, damaged_top_images, intact_side_images, intact_top_images)
)
labels = np.concatenate((damaged_labels, intact_labels))
# Print the shape of the loaded data
print("Shape of images:", images.shape)
print("Shape of labels:", labels.shape)
image_data = images
image_labels = labels
# Generate an array of indices for the samples
indices = np.arange(len(image_data))
# Shuffle the indices randomly
np.random.shuffle(indices)
# Use the shuffled indices to shuffle the data and labels arrays
image_data = image_data[indices]
image_labels = image_labels[indices]
from collections import Counter
# Split the data into training, validation, and testing sets
num_samples = len(image_data)
num_train = int(num_samples * 0.7)
num_val = int(num_samples * 0.15)
num_test = num_samples - num_train - num_val
train_data = image_data[:num_train]
train_labels = image_labels[:num_train]
val_data = image_data[num_train : num_train + num_val]
val_labels = image_labels[num_train : num_train + num_val]
test_data = image_data[num_train + num_val :]
test_labels = image_labels[num_train + num_val :]
print("Number of training samples:", len(train_data))
print("Number of validation samples:", len(val_data))
print("Number of testing samples:", len(test_data))
unique_labels, label_counts = np.unique(train_labels, return_counts=True)
print("training samples:", (dict(zip(unique_labels, label_counts))))
unique_labels, label_counts = np.unique(val_labels, return_counts=True)
print("validation samples:", (dict(zip(unique_labels, label_counts))))
unique_labels, label_counts = np.unique(test_labels, return_counts=True)
print("testing samples:", (dict(zip(unique_labels, label_counts))))
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dense
cnn = Sequential()
cnn.add(Conv2D(filters=64, kernel_size=3, activation="relu", input_shape=(540, 960, 3)))
cnn.add(MaxPooling2D(pool_size=2))
cnn.add(Flatten())
cnn.add(Dense(32, activation="relu"))
cnn.add(Dense(1, activation="sigmoid"))
cnn.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
cnn.summary()
cnn.fit(
train_data,
train_labels,
batch_size=32,
epochs=10,
validation_data=(val_data, val_labels),
)
test_loss, test_accuracy = cnn.evaluate(test_data, test_labels)
print(f"Test loss: ", test_loss)
print(f"Test accuracy: ", test_accuracy)
# YOLO
import cv2
import numpy as np
from tensorflow.keras.layers import Input
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dense
# Function to load images from a directory
def load_images(directory):
images = []
for filename in os.listdir(directory):
if filename.endswith(".jpg") or filename.endswith(".png"):
img_path = os.path.join(directory, filename)
img = cv2.imread(img_path)
if img is not None:
images.append(img)
return images
# Load the images from each category
damaged_side_images = load_images(damaged_side_dir)
damaged_top_images = load_images(damaged_top_dir)
intact_side_images = load_images(intact_side_dir)
intact_top_images = load_images(intact_top_dir)
# Create labels for the images
damaged_labels = np.ones(len(damaged_side_images) + len(damaged_top_images))
intact_labels = np.zeros(len(intact_side_images) + len(intact_top_images))
# Concatenate the images and labels
images = np.concatenate(
(damaged_side_images, damaged_top_images, intact_side_images, intact_top_images)
)
labels = np.concatenate((damaged_labels, intact_labels))
# Initialize the YOLOv4 model
def load_yolov4_weights(model, weights_file):
with open(weights_file, "rb") as f:
major, minor, revision, seen, _ = np.fromfile(f, dtype=np.int32, count=5)
layers = model.layers
for i in range(len(layers)):
if layers[i].get_weights():
weight_shape = [w.shape for w in layers[i].get_weights()]
if weight_shape != []:
k = 0
for j in range(len(weight_shape)):
shape = weight_shape[j]
if len(shape) == 4:
if shape[-2] != 1:
if k < seen:
conv_weights = np.fromfile(
f, dtype=np.float32, count=np.prod(shape)
)
conv_weights = conv_weights.reshape(
shape
).transpose([2, 3, 1, 0])
layers[i].set_weights([conv_weights])
k += shape[-2]
else:
break
input_size = 608
input_layer = Input(shape=(input_size, input_size, 3))
model = YOLOv4(input_layer)
weights_file = "yolov4.weights"
load_yolov4_weights(model, weights_file)
# Define the class names
class_names = ["damage", "intact"]
# Perform object detection and classification
is_damaged = False
for image in images:
# Resize the image to the YOLOv4 input size
resized_image = cv2.resize(image, (input_size, input_size))
# Perform object detection
boxes, scores, classes, nums = model.predict(np.expand_dims(resized_image, axis=0))
# Check if 'damage' class is detected
for i in range(nums[0]):
class_index = int(classes[0][i])
class_name = class_names[class_index]
if class_name == "damage":
is_damaged = True
break
if is_damaged:
break
# Print the classification result
if is_damaged:
print("The image is damaged.")
else:
print("The image is intact.")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/857/129857818.ipynb
|
industrial-quality-control-of-packages
|
christianvorhemus
|
[{"Id": 129857818, "ScriptId": 38510466, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7912931, "CreationDate": "05/17/2023 02:36:31", "VersionNumber": 1.0, "Title": "notebookeddf8115a4", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 204.0, "LinesInsertedFromPrevious": 204.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186252385, "KernelVersionId": 129857818, "SourceDatasetVersionId": 1803333}]
|
[{"Id": 1803333, "DatasetId": 1007061, "DatasourceVersionId": 1840798, "CreatorUserId": 1136755, "LicenseName": "GPL 2", "CreationDate": "01/01/2021 16:32:34", "VersionNumber": 2.0, "Title": "Industrial Quality Control of Packages", "Slug": "industrial-quality-control-of-packages", "Subtitle": "Detect damaged packages in an in-line quality inspection scenario", "Description": "**Summary:**\n\nThis dataset consists of synthetically created images of packages produced in an industrial production line in which the packaging machine occasionally produces faulty packages. One of the goals for the data scientist is to create a model that can identify damaged packages.\n\n\n\n**Details:**\n\nMany companies in the manufacturing industry use packaging machines to wrap their products. These machines usually work fully automatically, but from time to time, faulty packages are produced, for example because small deviations in the position of the packages cause them to be dented or bent. Therefore, in-line quality control inspection points exist across the production line in which different quality measures are taken that should ensure that faulty packages are excluded from the process before they are sent out.\n\nThis dataset consists of RGB images taken from a \u201cvirtual\u201d production line (meaning, the images were created procedurally, see below) with two classes: damaged and intact. For each class and each package, two camera captures exist: One image taken from a camera mounted above the package, one picture taken from a camera mounted on the side of the inspection belt. Each package is identified by a unique serial number (SN) visible on the side-view of the package. This serial number is also used for naming the files ({sn}_side.png and {sn}_top.png). \n\nThe goal of this dataset is to give data scientists the possibility to work on data inspired by industrial manufacturing scenarios. Among others the following questions are interesting:\n\n\u2022\tCan you train a model that identifies damaged packages using both, top and side view?\n\u2022\tCan you train a model that identifies damaged packages using just the top view?\n\u2022\tCan you train a model that performs OCR to extract the serial numbers (some SN may be blurry due to simulated motion blur)\n\u2022\tCan you create a one-shot computer vision algorithms that can classify packages by just learning from one or two samples? \n\nAs mentioned above, the images were created synthetically using Blender. The code to produce these images is open-sourced and can be found at https://github.com/christian-vorhemus/procedural-3d-image-generation", "VersionNotes": "Cleaned images", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1007061, "CreatorUserId": 1136755, "OwnerUserId": 1136755.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1803333.0, "CurrentDatasourceVersionId": 1840798.0, "ForumId": 1023746, "Type": 2, "CreationDate": "11/30/2020 09:01:47", "LastActivityDate": "11/30/2020", "TotalViews": 24595, "TotalDownloads": 1274, "TotalVotes": 37, "TotalKernels": 1}]
|
[{"Id": 1136755, "UserName": "christianvorhemus", "DisplayName": "Christian Vorhemus", "RegisterDate": "06/22/2017", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
import cv2
import numpy as np
# Define the root directory and the subdirectories
root_dir = "/kaggle/input/industrial-quality-control-of-packages/"
damaged_side_dir = os.path.join(root_dir, "damaged/side/")
damaged_top_dir = os.path.join(root_dir, "damaged/top/")
intact_side_dir = os.path.join(root_dir, "intact/side/")
intact_top_dir = os.path.join(root_dir, "intact/top/")
# Function to load images from a directory
def load_images(directory):
images = []
for filename in os.listdir(directory):
if filename.endswith(".jpg") or filename.endswith(".png"):
img_path = os.path.join(directory, filename)
img = cv2.imread(img_path)
if img is not None:
images.append(img)
return images
# Load the images from each category
damaged_side_images = load_images(damaged_side_dir)
damaged_top_images = load_images(damaged_top_dir)
intact_side_images = load_images(intact_side_dir)
intact_top_images = load_images(intact_top_dir)
# Create labels for the images
damaged_labels = np.ones(len(damaged_side_images) + len(damaged_top_images))
intact_labels = np.zeros(len(intact_side_images) + len(intact_top_images))
# Concatenate the images and labels
images = np.concatenate(
(damaged_side_images, damaged_top_images, intact_side_images, intact_top_images)
)
labels = np.concatenate((damaged_labels, intact_labels))
# Print the shape of the loaded data
print("Shape of images:", images.shape)
print("Shape of labels:", labels.shape)
image_data = images
image_labels = labels
# Generate an array of indices for the samples
indices = np.arange(len(image_data))
# Shuffle the indices randomly
np.random.shuffle(indices)
# Use the shuffled indices to shuffle the data and labels arrays
image_data = image_data[indices]
image_labels = image_labels[indices]
from collections import Counter
# Split the data into training, validation, and testing sets
num_samples = len(image_data)
num_train = int(num_samples * 0.7)
num_val = int(num_samples * 0.15)
num_test = num_samples - num_train - num_val
train_data = image_data[:num_train]
train_labels = image_labels[:num_train]
val_data = image_data[num_train : num_train + num_val]
val_labels = image_labels[num_train : num_train + num_val]
test_data = image_data[num_train + num_val :]
test_labels = image_labels[num_train + num_val :]
print("Number of training samples:", len(train_data))
print("Number of validation samples:", len(val_data))
print("Number of testing samples:", len(test_data))
unique_labels, label_counts = np.unique(train_labels, return_counts=True)
print("training samples:", (dict(zip(unique_labels, label_counts))))
unique_labels, label_counts = np.unique(val_labels, return_counts=True)
print("validation samples:", (dict(zip(unique_labels, label_counts))))
unique_labels, label_counts = np.unique(test_labels, return_counts=True)
print("testing samples:", (dict(zip(unique_labels, label_counts))))
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dense
cnn = Sequential()
cnn.add(Conv2D(filters=64, kernel_size=3, activation="relu", input_shape=(540, 960, 3)))
cnn.add(MaxPooling2D(pool_size=2))
cnn.add(Flatten())
cnn.add(Dense(32, activation="relu"))
cnn.add(Dense(1, activation="sigmoid"))
cnn.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
cnn.summary()
cnn.fit(
train_data,
train_labels,
batch_size=32,
epochs=10,
validation_data=(val_data, val_labels),
)
test_loss, test_accuracy = cnn.evaluate(test_data, test_labels)
print(f"Test loss: ", test_loss)
print(f"Test accuracy: ", test_accuracy)
# YOLO
import cv2
import numpy as np
from tensorflow.keras.layers import Input
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dense
# Function to load images from a directory
def load_images(directory):
images = []
for filename in os.listdir(directory):
if filename.endswith(".jpg") or filename.endswith(".png"):
img_path = os.path.join(directory, filename)
img = cv2.imread(img_path)
if img is not None:
images.append(img)
return images
# Load the images from each category
damaged_side_images = load_images(damaged_side_dir)
damaged_top_images = load_images(damaged_top_dir)
intact_side_images = load_images(intact_side_dir)
intact_top_images = load_images(intact_top_dir)
# Create labels for the images
damaged_labels = np.ones(len(damaged_side_images) + len(damaged_top_images))
intact_labels = np.zeros(len(intact_side_images) + len(intact_top_images))
# Concatenate the images and labels
images = np.concatenate(
(damaged_side_images, damaged_top_images, intact_side_images, intact_top_images)
)
labels = np.concatenate((damaged_labels, intact_labels))
# Initialize the YOLOv4 model
def load_yolov4_weights(model, weights_file):
with open(weights_file, "rb") as f:
major, minor, revision, seen, _ = np.fromfile(f, dtype=np.int32, count=5)
layers = model.layers
for i in range(len(layers)):
if layers[i].get_weights():
weight_shape = [w.shape for w in layers[i].get_weights()]
if weight_shape != []:
k = 0
for j in range(len(weight_shape)):
shape = weight_shape[j]
if len(shape) == 4:
if shape[-2] != 1:
if k < seen:
conv_weights = np.fromfile(
f, dtype=np.float32, count=np.prod(shape)
)
conv_weights = conv_weights.reshape(
shape
).transpose([2, 3, 1, 0])
layers[i].set_weights([conv_weights])
k += shape[-2]
else:
break
input_size = 608
input_layer = Input(shape=(input_size, input_size, 3))
model = YOLOv4(input_layer)
weights_file = "yolov4.weights"
load_yolov4_weights(model, weights_file)
# Define the class names
class_names = ["damage", "intact"]
# Perform object detection and classification
is_damaged = False
for image in images:
# Resize the image to the YOLOv4 input size
resized_image = cv2.resize(image, (input_size, input_size))
# Perform object detection
boxes, scores, classes, nums = model.predict(np.expand_dims(resized_image, axis=0))
# Check if 'damage' class is detected
for i in range(nums[0]):
class_index = int(classes[0][i])
class_name = class_names[class_index]
if class_name == "damage":
is_damaged = True
break
if is_damaged:
break
# Print the classification result
if is_damaged:
print("The image is damaged.")
else:
print("The image is intact.")
| false | 0 | 2,090 | 0 | 2,700 | 2,090 |
||
129820024
|
import geopandas as gpd
import matplotlib.pyplot as plt
import contextily as cx
# read-in files
wi_co = gpd.read_file(
"https://raw.githubusercontent.com/mhaffner/data/master/wi_counties.geojson"
)
iat = gpd.read_file(
"https://raw.githubusercontent.com/mhaffner/data/master/Ice_Age_Trail.geojson"
)
# retrieve crs for wi_co
wi_co.crs
# retrieve crs for iat
iat.crs
# give better crs
wi_co_3070 = wi_co.to_crs(3070)
iat_3070 = iat.to_crs(3070)
# plot both on single ax
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
wi_co_3070.plot(ax=ax)
iat_3070.plot(ax=ax, color="gold")
# calculate length of each trail feature
# shapelength here is length of each segment
# as a new column
iat_3070["iat_length_meters"] = iat_3070.length
wi_co
trail_co = gpd.sjoin(wi_co_3070, iat_3070, how="inner", predicate="intersects")
trail_co
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
wi_co_3070.plot(ax=ax)
iat_3070.plot(ax=ax, color="gold")
trail_co.plot(ax=ax, color="violet")
# miles
trail_co["iat_length_miles"] = trail_co["iat_length_meters"] / 1609.34
trail_co
# sort descending
sorted_trail = trail_co.sort_values(by=(["iat_length_miles"]), ascending=False)
sorted_trail
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/820/129820024.ipynb
| null | null |
[{"Id": 129820024, "ScriptId": 37759535, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12217581, "CreationDate": "05/16/2023 17:44:01", "VersionNumber": 1.0, "Title": "lab_10_435", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 54.0, "LinesInsertedFromPrevious": 54.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import geopandas as gpd
import matplotlib.pyplot as plt
import contextily as cx
# read-in files
wi_co = gpd.read_file(
"https://raw.githubusercontent.com/mhaffner/data/master/wi_counties.geojson"
)
iat = gpd.read_file(
"https://raw.githubusercontent.com/mhaffner/data/master/Ice_Age_Trail.geojson"
)
# retrieve crs for wi_co
wi_co.crs
# retrieve crs for iat
iat.crs
# give better crs
wi_co_3070 = wi_co.to_crs(3070)
iat_3070 = iat.to_crs(3070)
# plot both on single ax
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
wi_co_3070.plot(ax=ax)
iat_3070.plot(ax=ax, color="gold")
# calculate length of each trail feature
# shapelength here is length of each segment
# as a new column
iat_3070["iat_length_meters"] = iat_3070.length
wi_co
trail_co = gpd.sjoin(wi_co_3070, iat_3070, how="inner", predicate="intersects")
trail_co
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
wi_co_3070.plot(ax=ax)
iat_3070.plot(ax=ax, color="gold")
trail_co.plot(ax=ax, color="violet")
# miles
trail_co["iat_length_miles"] = trail_co["iat_length_meters"] / 1609.34
trail_co
# sort descending
sorted_trail = trail_co.sort_values(by=(["iat_length_miles"]), ascending=False)
sorted_trail
| false | 0 | 492 | 0 | 492 | 492 |
||
129820124
|
<jupyter_start><jupyter_text>Customer segmentation
The data source was taken from the Kaggle challenge called Credit Card Dataset for Clustering. The sample Dataset summarizes the usage behavior of about 9000 active credit cardholders during the last six months
The file is at a customer level with 18 behavioral variables.
Following is the Data Dictionary for Credit Card dataset:
- CUSTID: Identification of Credit Cardholder (Categorical)
- BALANCE: Balance amount left in their account to make purchases
- BALANCEFREQUENCY: How frequently the Balance is updated, score between 0 and 1 (1 = frequently updated, 0 = not frequently updated)
- PURCHASES: Amount of purchases made from the account
- ONEOFFPURCHASES: Maximum purchase amount did in one-go
- INSTALLMENTSPURCHASES: Amount of purchase done in installment
- CASH ADVANCE: Cash in advance given by the user
- PURCHASESFREQUENCY: How frequently the Purchases are being made score between 0 and 1 (1 = frequently purchased, 0 = not frequently purchased)
- ONEOFFPURCHASESFREQUENCY: How frequently Purchases are happening in one-go (1 = frequently purchased, 0 = not frequently purchased)
- PURCHASESINSTALLMENTSFREQUENCY: How frequently purchases in installments are being done (1 = frequently done, 0 = not frequently done)
- CASHADVANCEFREQUENCY: How frequently the cash in advance being paid
- CASHADVANCETRX: Number of Transactions made with “Cash in Advanced”
- PURCHASESTRX: Number of purchase transactions made
- CREDIT LIMIT: Limit of Credit Card for user
- PAYMENTS: Amount of Payment done by the user
- MINIMUM_PAYMENTS: Minimum amount of payments made by the user
- PRCFULLPAYMENT: Percent of full payment paid by the user
- TENURE: Tenure of credit card service for user
Kaggle dataset identifier: customer-segmentation
<jupyter_script># This case requires to develop a customer segmentation to define marketing strategy. The sample Dataset summarizes the usage behavior of about 9000 active credit card holders during the last 6 months. The file is at a customer level with 18 behavioral variables.
# Following is the Data Dictionary for Credit Card dataset :-
# CUST_ID : Identification of Credit Card holder (Categorical)
# BALANCE : Balance amount left in their account to make purchases
# BALANCE_FREQUENCY : How frequently the Balance is updated, score between 0 and 1 (1 = frequently updated, 0 = not frequently updated)
# PURCHASES : Amount of purchases made from account
# ONEOFF_PURCHASES : Maximum purchase amount done in one-go
# INSTALLMENTS_PURCHASES : Amount of purchase done in installment
# CASH_ADVANCE : Cash in advance given by the user
# PURCHASES_FREQUENCY : How frequently the Purchases are being made, score between 0 and 1 (1 = frequently purchased, 0 = not frequently purchased)
# ONEOFFPURCHASESFREQUENCY : How frequently Purchases are happening in one-go (1 = frequently purchased, 0 = not frequently purchased)
# PURCHASESINSTALLMENTSFREQUENCY : How frequently purchases in installments are being done (1 = frequently done, 0 = not frequently done)
# CASHADVANCEFREQUENCY : How frequently the cash in advance being paid
# CASHADVANCETRX : Number of Transactions made with "Cash in Advanced"
# PURCHASES_TRX : Numbe of purchase transactions made
# CREDIT_LIMIT : Limit of Credit Card for user
# PAYMENTS : Amount of Payment done by user
# MINIMUM_PAYMENTS : Minimum amount of payments made by user
# PRCFULLPAYMENT : Percent of full payment paid by user
# TENURE : Tenure of credit card service for user
# # Import libraries
import warnings
warnings.filterwarnings("ignore")
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
from yellowbrick.cluster import KElbowVisualizer
from yellowbrick.cluster import SilhouetteVisualizer
from sklearn.cluster import DBSCAN
from sklearn.cluster import MiniBatchKMeans
from sklearn.mixture import GaussianMixture
from scipy import stats
from scipy.stats import norm, skew
from sklearn.impute import KNNImputer
from sklearn.metrics import r2_score, mean_squared_error, mean_absolute_error
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
# # Read data
data = pd.read_csv("CC GENERAL.csv")
data.head(10)
# **data information**
data.info()
# **drop ID**
data.drop("CUST_ID", axis=1, inplace=True)
# **Statistics Information from std we can observe alot of skewness in the data**
data.describe()
# **sum of missing values**
data.isnull().sum()
# **check correlation**
plt.figure(figsize=(15, 8))
sns.heatmap(data.corr(), annot=True)
list_col = [
"BALANCE",
"BALANCE_FREQUENCY",
"PURCHASES",
"INSTALLMENTS_PURCHASES",
"CASH_ADVANCE",
"CASH_ADVANCE_TRX",
]
fig, ax = plt.subplots(figsize=(16, 7))
for i, j in enumerate(list_col):
plt.subplot(2, 3, i + 1)
sns.distplot(data[j], fit=norm, kde=False, color="red")
(mu, sigma) = norm.fit(data[j])
plt.legend(
["Normal dist. ($\mu=$ {:.2f} and $\sigma=$ {:.2f} )".format(mu, sigma)],
loc="best",
)
plt.show()
sns.pairplot(
data,
x_vars=[
"PURCHASES",
"ONEOFF_PURCHASES",
"CASH_ADVANCE_TRX",
"CASH_ADVANCE_FREQUENCY",
"PURCHASES_FREQUENCY",
"PURCHASES_INSTALLMENTS_FREQUENCY",
],
y_vars=[
"PURCHASES",
"ONEOFF_PURCHASES",
"CASH_ADVANCE_TRX",
"CASH_ADVANCE_FREQUENCY",
"PURCHASES_FREQUENCY",
"PURCHASES_INSTALLMENTS_FREQUENCY",
],
diag_kind="hist",
diag_kws=dict(multiple="stack"),
)
# **fill missing values**
data["CREDIT_LIMIT"].fillna(data["CREDIT_LIMIT"].median(), inplace=True)
missing = data[data["MINIMUM_PAYMENTS"].isnull()]
missing.shape
data.dropna(inplace=True)
data.isnull().sum()
# **create a model that predict with MINIMUM_PAYMENTS to fill the missing values**
X = data.drop("MINIMUM_PAYMENTS", axis=1)
y = data["MINIMUM_PAYMENTS"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.15, random_state=0
)
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.fit_transform(X_test)
from xgboost import XGBRegressor
model = XGBRegressor(n_estimators=300)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
rmse = np.sqrt(mean_squared_error(y_test, y_pred))
rmse
mean_absolute_error(y_test, y_pred)
r2_score(y_test, y_pred)
missing.head()
y_test2 = missing.drop("MINIMUM_PAYMENTS", axis=1)
# **predict with missing values**
y_pred2 = model.predict(y_test2)
y_pred2.shape
df = pd.DataFrame(y_pred2, columns=["MINIMUM_PAYMENTS"]).reset_index(drop=True)
df.head()
y_test2 = y_test2.reset_index(drop=True)
y_test2.head()
fill_missing = pd.concat([y_test2, df], axis=1)
df = pd.concat([data, fill_missing], axis=0)
# **Clean data from missing values**
df.head()
df.shape
df.isnull().sum()
# **check Outliers**
list_col = [
"BALANCE",
"BALANCE_FREQUENCY",
"PURCHASES",
"INSTALLMENTS_PURCHASES",
"CASH_ADVANCE",
"CASH_ADVANCE_TRX",
]
fig, ax = plt.subplots(figsize=(16, 10))
for i, j in enumerate(list_col):
plt.subplot(2, 3, i + 1)
sns.boxplot(x=data["TENURE"], y=data[j], palette="viridis")
plt.show()
# **Percentage of outliers in every column**
def outlier_percent(data):
Q1 = data.quantile(0.25)
Q3 = data.quantile(0.75)
IQR = Q3 - Q1
minimum = Q1 - (1.5 * IQR)
maximum = Q3 + (1.5 * IQR)
num_outliers = np.sum((data < minimum) | (data > maximum))
num_total = data.count()
return (num_outliers / num_total) * 100
for column in df.columns:
data = df[column]
percent = str(round(outlier_percent(data), 2))
print(f'Outliers in "{column}": {percent}%')
# **Replace Outliers with np.nan**
for column in df.columns:
data = df[column]
Q1 = data.quantile(0.25)
Q3 = data.quantile(0.75)
IQR = Q3 - Q1
minimum = Q1 - (1.5 * IQR)
maximum = Q3 + (1.5 * IQR)
outliers = (data < minimum) | (data > maximum)
df[column].loc[outliers] = np.nan
df.isna().sum()
# **KNN imputer: Each sample’s missing values are imputed using the mean value from n_neighbors nearest neighbors found in the training set.**
imputer = KNNImputer()
imp_data = pd.DataFrame(imputer.fit_transform(df), columns=df.columns)
imp_data.isna().sum()
imp_data.describe()
std_imp_data = pd.DataFrame(
StandardScaler().fit_transform(imp_data), columns=imp_data.columns
)
std_imp_data.describe()
for column in std_imp_data.columns:
data = std_imp_data[column]
percent = str(round(outlier_percent(data), 2))
print(f'Outliers in "{column}": {percent}%')
pca = PCA(n_components=0.9, random_state=42)
pca.fit(std_imp_data)
PC_names = ["PC" + str(x) for x in range(1, len(pca.components_) + 1)]
pca_data = pd.DataFrame(pca.transform(std_imp_data), columns=PC_names)
fig, ax = plt.subplots(figsize=(24, 16))
plt.imshow(
pca.components_.T,
cmap="Spectral",
vmin=-1,
vmax=1,
)
plt.yticks(range(len(std_imp_data.columns)), std_imp_data.columns)
plt.xticks(range(len(pca_data.columns)), pca_data.columns)
plt.xlabel("Principal Component")
plt.ylabel("Contribution")
plt.title("Contribution of Features to Components")
plt.colorbar()
# # Kmeans Model
inertia = []
silhouette = []
for k in range(2, 20):
kmeans = KMeans(n_clusters=k, random_state=0)
y_pred = kmeans.fit_predict(pca_data)
inertia.append(kmeans.inertia_)
silhouette.append(silhouette_score(pca_data, kmeans.labels_))
plt.plot(range(2, 20), inertia, "o-")
plt.xlabel("Number of K")
plt.ylabel("inertia")
plt.title("K Elbow Method")
plt.plot(range(2, 20), silhouette, "o-")
plt.xlabel("Number of K")
plt.ylabel("silhouette Score")
model = KMeans(random_state=42)
distortion_visualizer = KElbowVisualizer(model, k=(2, 10))
distortion_visualizer.fit(pca_data)
distortion_visualizer.show()
# Instantiate the clustering model and visualizer
model = KMeans(4, random_state=42)
visualizer = SilhouetteVisualizer(model, colors="yellowbrick")
visualizer.fit(pca_data) # Fit the data to the visualizer
visualizer.show() # Finalize and render the figure
# Instantiate the clustering model and visualizer
model = KMeans(5, random_state=42)
visualizer = SilhouetteVisualizer(model, colors="yellowbrick")
visualizer.fit(pca_data) # Fit the data to the visualizer
visualizer.show() # Finalize and render the figure
km_model = KMeans(distortion_visualizer.elbow_value_, random_state=42)
labels = km_model.fit_predict(pca_data)
labels
# # MiniBatchKMeans
inertia = []
silhouette = []
for k in range(2, 20):
minibatch_kmeans = MiniBatchKMeans(n_clusters=k, random_state=42)
minibatch_kmeans.fit(pca_data)
inertia.append(minibatch_kmeans.inertia_)
silhouette.append(silhouette_score(pca_data, minibatch_kmeans.labels_))
plt.plot(
range(2, 20),
inertia,
"o-",
)
plt.xlabel("Number of K")
plt.ylabel("inertia")
plt.title("K Elbow Method")
plt.plot(range(2, 20), silhouette, "o-")
plt.xlabel("Number of K")
plt.ylabel("silhouette Score")
model = MiniBatchKMeans(random_state=42)
distortion_visualizer = KElbowVisualizer(model, k=(2, 10))
distortion_visualizer.fit(pca_data)
distortion_visualizer.show()
# Instantiate the clustering model and visualizer
model = MiniBatchKMeans(5, random_state=42)
visualizer = SilhouetteVisualizer(model, colors="yellowbrick")
visualizer.fit(pca_data) # Fit the data to the visualizer
visualizer.show() # Finalize and render the figure
# Instantiate the clustering model and visualizer
model = MiniBatchKMeans(4, random_state=42)
visualizer = SilhouetteVisualizer(model, colors="yellowbrick")
visualizer.fit(pca_data) # Fit the data to the visualizer
visualizer.show() # Finalize and render the figure
# # GaussianMixture Model
BIC = []
AIC = []
for k in range(2, 10):
gm = GaussianMixture(n_components=k, n_init=10)
gm.fit(pca_data)
BIC.append(gm.bic(pca_data))
AIC.append(gm.aic(pca_data))
plt.plot(range(2, 10), BIC, "o-")
plt.xlabel("Number of K")
plt.ylabel("Bayesian information criterion (BIC)")
plt.plot(range(2, 10), AIC, "o-")
plt.xlabel("Number of K")
plt.ylabel("Akaike information criterion")
plt.errorbar(range(2, 10), np.gradient(BIC), label="BIC")
plt.title("Gradient of BIC Scores", fontsize=20)
plt.xticks(range(2, 10))
plt.xlabel("N. of clusters")
plt.ylabel("grad(BIC)")
plt.legend()
plt.errorbar(range(2, 10), np.gradient(AIC), label="AIC")
plt.title("Gradient of AIC Scores", fontsize=20)
plt.xticks(range(2, 10))
plt.xlabel("N. of clusters")
plt.ylabel("grad(AIC)")
plt.legend()
# # BayesianGaussianMixture
from sklearn.mixture import BayesianGaussianMixture
bgm = BayesianGaussianMixture(n_components=10, n_init=10, random_state=42)
bgm.fit(pca_data)
bgm.weights_.round(3)
# # AgglomerativeClustering
from scipy.cluster.hierarchy import dendrogram
from sklearn.cluster import AgglomerativeClustering
def plot_dendrogram(model, **kwargs):
# Create linkage matrix and then plot the dendrogram
# create the counts of samples under each node
counts = np.zeros(model.children_.shape[0])
n_samples = len(model.labels_)
for i, merge in enumerate(model.children_):
current_count = 0
for child_idx in merge:
if child_idx < n_samples:
current_count += 1 # leaf node
else:
current_count += counts[child_idx - n_samples]
counts[i] = current_count
linkage_matrix = np.column_stack(
[model.children_, model.distances_, counts]
).astype(float)
# Plot the corresponding dendrogram
dendrogram(linkage_matrix, **kwargs)
# setting distance_threshold=0 ensures we compute the full tree.
model = AgglomerativeClustering(distance_threshold=0, n_clusters=None)
model = model.fit(pca_data)
plt.title("Hierarchical Clustering Dendrogram")
# plot the top three levels of the dendrogram
plot_dendrogram(model, truncate_mode="level", p=3)
plt.xlabel("Number of points in node.")
plt.show()
# # ADD Labels to the data
pca_data["Target"] = labels
pca_data.head()
imp_data["Target"] = labels
imp_data.head()
# # Clusters Analysis
imp_data["Target"].value_counts().plot.pie(
autopct="%1.0f%%", pctdistance=0.7, labeldistance=1.1
)
def colorful_scatter(data):
LABEL_COLOR_MAP = {0: "y", 1: "g", 2: "m", 3: "k"}
sns.jointplot(
data=data,
x="BALANCE",
y="PURCHASES",
hue="Target",
palette=LABEL_COLOR_MAP,
alpha=0.6,
height=10,
)
colorful_scatter(imp_data)
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="BALANCE",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "BALANCE")
plt.title("Average of BALANCE in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="BALANCE_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "BALANCE_FREQUENCY")
plt.title("Average of BALANCE_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PURCHASES",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PURCHASES")
plt.title("Average of PURCHASES in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="ONEOFF_PURCHASES",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "ONEOFF_PURCHASES")
plt.title("Average of ONEOFF_PURCHASES in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="INSTALLMENTS_PURCHASES",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "INSTALLMENTS_PURCHASES")
plt.title("Average of INSTALLMENTS_PURCHASES in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="CASH_ADVANCE",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "CASH_ADVANCE")
plt.title("Average of CASH_ADVANCE in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PURCHASES_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PURCHASES_FREQUENCY")
plt.title("Average of PURCHASES_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="ONEOFF_PURCHASES_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "ONEOFF_PURCHASES_FREQUENCY")
plt.title("Average of ONEOFF_PURCHASES_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PURCHASES_INSTALLMENTS_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PURCHASES_INSTALLMENTS_FREQUENCY")
plt.title("Average of PURCHASES_INSTALLMENTS_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="CASH_ADVANCE_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "CASH_ADVANCE_FREQUENCY")
plt.title("Average of CASH_ADVANCE_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="CASH_ADVANCE_TRX",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "CASH_ADVANCE_TRX")
plt.title("Average of CASH_ADVANCE_TRX in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PURCHASES_TRX",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PURCHASES_TRX")
plt.title("Average of PURCHASES_TRX in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="CREDIT_LIMIT",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "CREDIT_LIMIT")
plt.title("Average of CREDIT_LIMIT in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PAYMENTS",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PAYMENTS")
plt.title("Average of 'PAYMENTS' in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="MINIMUM_PAYMENTS",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "MINIMUM_PAYMENTS")
plt.title("Average of 'MINIMUM_PAYMENTS' in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PRC_FULL_PAYMENT",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PRC_FULL_PAYMENT")
plt.title("Average of PRC_FULL_PAYMENT in Clusters")
plt.show()
x, y = pca_data.iloc[:, 0], pca_data.iloc[:, 1]
colors = {0: "red", 1: "blue", 2: "green", 3: "yellow"}
names = {
0: "Cluster 0",
1: "Cluster 1",
2: "Cluster 2",
3: "Cluseer 3",
}
df = pd.DataFrame({"x": x, "y": y, "label": pca_data.iloc[:, -1]})
groups = df.groupby("label")
fig, ax = plt.subplots(figsize=(20, 13))
for name, group in groups:
ax.plot(
group.x,
group.y,
marker="o",
linestyle="",
ms=5,
color=colors[name],
label=names[name],
mec="none",
)
ax.set_aspect("auto")
ax.tick_params(axis="x", which="both", bottom="off", top="off", labelbottom="off")
ax.tick_params(axis="y", which="both", left="off", top="off", labelleft="off")
ax.legend()
ax.set_title("Customers Segmentation based on their Credit Card usage bhaviour.")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/820/129820124.ipynb
|
customer-segmentation
|
mahnazarjmand
|
[{"Id": 129820124, "ScriptId": 38609533, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8074611, "CreationDate": "05/16/2023 17:44:54", "VersionNumber": 2.0, "Title": "customer_segmentation", "EvaluationDate": "05/16/2023", "IsChange": false, "TotalLines": 576.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 576.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 186196848, "KernelVersionId": 129820124, "SourceDatasetVersionId": 4740787}]
|
[{"Id": 4740787, "DatasetId": 2743505, "DatasourceVersionId": 4803791, "CreatorUserId": 10814621, "LicenseName": "Unknown", "CreationDate": "12/18/2022 16:26:19", "VersionNumber": 1.0, "Title": "Customer segmentation", "Slug": "customer-segmentation", "Subtitle": NaN, "Description": "The data source was taken from the Kaggle challenge called Credit Card Dataset for Clustering. The sample Dataset summarizes the usage behavior of about 9000 active credit cardholders during the last six months\n\nThe file is at a customer level with 18 behavioral variables.\n\nFollowing is the Data Dictionary for Credit Card dataset:\n\n- CUSTID: Identification of Credit Cardholder (Categorical)\n- BALANCE: Balance amount left in their account to make purchases\n- BALANCEFREQUENCY: How frequently the Balance is updated, score between 0 and 1 (1 = frequently updated, 0 = not frequently updated)\n- PURCHASES: Amount of purchases made from the account\n- ONEOFFPURCHASES: Maximum purchase amount did in one-go\n- INSTALLMENTSPURCHASES: Amount of purchase done in installment\n- CASH ADVANCE: Cash in advance given by the user\n- PURCHASESFREQUENCY: How frequently the Purchases are being made score between 0 and 1 (1 = frequently purchased, 0 = not frequently purchased)\n- ONEOFFPURCHASESFREQUENCY: How frequently Purchases are happening in one-go (1 = frequently purchased, 0 = not frequently purchased)\n- PURCHASESINSTALLMENTSFREQUENCY: How frequently purchases in installments are being done (1 = frequently done, 0 = not frequently done)\n- CASHADVANCEFREQUENCY: How frequently the cash in advance being paid\n- CASHADVANCETRX: Number of Transactions made with \u201cCash in Advanced\u201d\n- PURCHASESTRX: Number of purchase transactions made\n- CREDIT LIMIT: Limit of Credit Card for user\n- PAYMENTS: Amount of Payment done by the user\n- MINIMUM_PAYMENTS: Minimum amount of payments made by the user\n- PRCFULLPAYMENT: Percent of full payment paid by the user\n- TENURE: Tenure of credit card service for user", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2743505, "CreatorUserId": 10814621, "OwnerUserId": 10814621.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4740787.0, "CurrentDatasourceVersionId": 4803791.0, "ForumId": 2776996, "Type": 2, "CreationDate": "12/18/2022 16:26:19", "LastActivityDate": "12/18/2022", "TotalViews": 1432, "TotalDownloads": 173, "TotalVotes": 35, "TotalKernels": 16}]
|
[{"Id": 10814621, "UserName": "mahnazarjmand", "DisplayName": "Mahnaz Arjmand", "RegisterDate": "06/14/2022", "PerformanceTier": 4}]
|
# This case requires to develop a customer segmentation to define marketing strategy. The sample Dataset summarizes the usage behavior of about 9000 active credit card holders during the last 6 months. The file is at a customer level with 18 behavioral variables.
# Following is the Data Dictionary for Credit Card dataset :-
# CUST_ID : Identification of Credit Card holder (Categorical)
# BALANCE : Balance amount left in their account to make purchases
# BALANCE_FREQUENCY : How frequently the Balance is updated, score between 0 and 1 (1 = frequently updated, 0 = not frequently updated)
# PURCHASES : Amount of purchases made from account
# ONEOFF_PURCHASES : Maximum purchase amount done in one-go
# INSTALLMENTS_PURCHASES : Amount of purchase done in installment
# CASH_ADVANCE : Cash in advance given by the user
# PURCHASES_FREQUENCY : How frequently the Purchases are being made, score between 0 and 1 (1 = frequently purchased, 0 = not frequently purchased)
# ONEOFFPURCHASESFREQUENCY : How frequently Purchases are happening in one-go (1 = frequently purchased, 0 = not frequently purchased)
# PURCHASESINSTALLMENTSFREQUENCY : How frequently purchases in installments are being done (1 = frequently done, 0 = not frequently done)
# CASHADVANCEFREQUENCY : How frequently the cash in advance being paid
# CASHADVANCETRX : Number of Transactions made with "Cash in Advanced"
# PURCHASES_TRX : Numbe of purchase transactions made
# CREDIT_LIMIT : Limit of Credit Card for user
# PAYMENTS : Amount of Payment done by user
# MINIMUM_PAYMENTS : Minimum amount of payments made by user
# PRCFULLPAYMENT : Percent of full payment paid by user
# TENURE : Tenure of credit card service for user
# # Import libraries
import warnings
warnings.filterwarnings("ignore")
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
from yellowbrick.cluster import KElbowVisualizer
from yellowbrick.cluster import SilhouetteVisualizer
from sklearn.cluster import DBSCAN
from sklearn.cluster import MiniBatchKMeans
from sklearn.mixture import GaussianMixture
from scipy import stats
from scipy.stats import norm, skew
from sklearn.impute import KNNImputer
from sklearn.metrics import r2_score, mean_squared_error, mean_absolute_error
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
# # Read data
data = pd.read_csv("CC GENERAL.csv")
data.head(10)
# **data information**
data.info()
# **drop ID**
data.drop("CUST_ID", axis=1, inplace=True)
# **Statistics Information from std we can observe alot of skewness in the data**
data.describe()
# **sum of missing values**
data.isnull().sum()
# **check correlation**
plt.figure(figsize=(15, 8))
sns.heatmap(data.corr(), annot=True)
list_col = [
"BALANCE",
"BALANCE_FREQUENCY",
"PURCHASES",
"INSTALLMENTS_PURCHASES",
"CASH_ADVANCE",
"CASH_ADVANCE_TRX",
]
fig, ax = plt.subplots(figsize=(16, 7))
for i, j in enumerate(list_col):
plt.subplot(2, 3, i + 1)
sns.distplot(data[j], fit=norm, kde=False, color="red")
(mu, sigma) = norm.fit(data[j])
plt.legend(
["Normal dist. ($\mu=$ {:.2f} and $\sigma=$ {:.2f} )".format(mu, sigma)],
loc="best",
)
plt.show()
sns.pairplot(
data,
x_vars=[
"PURCHASES",
"ONEOFF_PURCHASES",
"CASH_ADVANCE_TRX",
"CASH_ADVANCE_FREQUENCY",
"PURCHASES_FREQUENCY",
"PURCHASES_INSTALLMENTS_FREQUENCY",
],
y_vars=[
"PURCHASES",
"ONEOFF_PURCHASES",
"CASH_ADVANCE_TRX",
"CASH_ADVANCE_FREQUENCY",
"PURCHASES_FREQUENCY",
"PURCHASES_INSTALLMENTS_FREQUENCY",
],
diag_kind="hist",
diag_kws=dict(multiple="stack"),
)
# **fill missing values**
data["CREDIT_LIMIT"].fillna(data["CREDIT_LIMIT"].median(), inplace=True)
missing = data[data["MINIMUM_PAYMENTS"].isnull()]
missing.shape
data.dropna(inplace=True)
data.isnull().sum()
# **create a model that predict with MINIMUM_PAYMENTS to fill the missing values**
X = data.drop("MINIMUM_PAYMENTS", axis=1)
y = data["MINIMUM_PAYMENTS"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.15, random_state=0
)
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.fit_transform(X_test)
from xgboost import XGBRegressor
model = XGBRegressor(n_estimators=300)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
rmse = np.sqrt(mean_squared_error(y_test, y_pred))
rmse
mean_absolute_error(y_test, y_pred)
r2_score(y_test, y_pred)
missing.head()
y_test2 = missing.drop("MINIMUM_PAYMENTS", axis=1)
# **predict with missing values**
y_pred2 = model.predict(y_test2)
y_pred2.shape
df = pd.DataFrame(y_pred2, columns=["MINIMUM_PAYMENTS"]).reset_index(drop=True)
df.head()
y_test2 = y_test2.reset_index(drop=True)
y_test2.head()
fill_missing = pd.concat([y_test2, df], axis=1)
df = pd.concat([data, fill_missing], axis=0)
# **Clean data from missing values**
df.head()
df.shape
df.isnull().sum()
# **check Outliers**
list_col = [
"BALANCE",
"BALANCE_FREQUENCY",
"PURCHASES",
"INSTALLMENTS_PURCHASES",
"CASH_ADVANCE",
"CASH_ADVANCE_TRX",
]
fig, ax = plt.subplots(figsize=(16, 10))
for i, j in enumerate(list_col):
plt.subplot(2, 3, i + 1)
sns.boxplot(x=data["TENURE"], y=data[j], palette="viridis")
plt.show()
# **Percentage of outliers in every column**
def outlier_percent(data):
Q1 = data.quantile(0.25)
Q3 = data.quantile(0.75)
IQR = Q3 - Q1
minimum = Q1 - (1.5 * IQR)
maximum = Q3 + (1.5 * IQR)
num_outliers = np.sum((data < minimum) | (data > maximum))
num_total = data.count()
return (num_outliers / num_total) * 100
for column in df.columns:
data = df[column]
percent = str(round(outlier_percent(data), 2))
print(f'Outliers in "{column}": {percent}%')
# **Replace Outliers with np.nan**
for column in df.columns:
data = df[column]
Q1 = data.quantile(0.25)
Q3 = data.quantile(0.75)
IQR = Q3 - Q1
minimum = Q1 - (1.5 * IQR)
maximum = Q3 + (1.5 * IQR)
outliers = (data < minimum) | (data > maximum)
df[column].loc[outliers] = np.nan
df.isna().sum()
# **KNN imputer: Each sample’s missing values are imputed using the mean value from n_neighbors nearest neighbors found in the training set.**
imputer = KNNImputer()
imp_data = pd.DataFrame(imputer.fit_transform(df), columns=df.columns)
imp_data.isna().sum()
imp_data.describe()
std_imp_data = pd.DataFrame(
StandardScaler().fit_transform(imp_data), columns=imp_data.columns
)
std_imp_data.describe()
for column in std_imp_data.columns:
data = std_imp_data[column]
percent = str(round(outlier_percent(data), 2))
print(f'Outliers in "{column}": {percent}%')
pca = PCA(n_components=0.9, random_state=42)
pca.fit(std_imp_data)
PC_names = ["PC" + str(x) for x in range(1, len(pca.components_) + 1)]
pca_data = pd.DataFrame(pca.transform(std_imp_data), columns=PC_names)
fig, ax = plt.subplots(figsize=(24, 16))
plt.imshow(
pca.components_.T,
cmap="Spectral",
vmin=-1,
vmax=1,
)
plt.yticks(range(len(std_imp_data.columns)), std_imp_data.columns)
plt.xticks(range(len(pca_data.columns)), pca_data.columns)
plt.xlabel("Principal Component")
plt.ylabel("Contribution")
plt.title("Contribution of Features to Components")
plt.colorbar()
# # Kmeans Model
inertia = []
silhouette = []
for k in range(2, 20):
kmeans = KMeans(n_clusters=k, random_state=0)
y_pred = kmeans.fit_predict(pca_data)
inertia.append(kmeans.inertia_)
silhouette.append(silhouette_score(pca_data, kmeans.labels_))
plt.plot(range(2, 20), inertia, "o-")
plt.xlabel("Number of K")
plt.ylabel("inertia")
plt.title("K Elbow Method")
plt.plot(range(2, 20), silhouette, "o-")
plt.xlabel("Number of K")
plt.ylabel("silhouette Score")
model = KMeans(random_state=42)
distortion_visualizer = KElbowVisualizer(model, k=(2, 10))
distortion_visualizer.fit(pca_data)
distortion_visualizer.show()
# Instantiate the clustering model and visualizer
model = KMeans(4, random_state=42)
visualizer = SilhouetteVisualizer(model, colors="yellowbrick")
visualizer.fit(pca_data) # Fit the data to the visualizer
visualizer.show() # Finalize and render the figure
# Instantiate the clustering model and visualizer
model = KMeans(5, random_state=42)
visualizer = SilhouetteVisualizer(model, colors="yellowbrick")
visualizer.fit(pca_data) # Fit the data to the visualizer
visualizer.show() # Finalize and render the figure
km_model = KMeans(distortion_visualizer.elbow_value_, random_state=42)
labels = km_model.fit_predict(pca_data)
labels
# # MiniBatchKMeans
inertia = []
silhouette = []
for k in range(2, 20):
minibatch_kmeans = MiniBatchKMeans(n_clusters=k, random_state=42)
minibatch_kmeans.fit(pca_data)
inertia.append(minibatch_kmeans.inertia_)
silhouette.append(silhouette_score(pca_data, minibatch_kmeans.labels_))
plt.plot(
range(2, 20),
inertia,
"o-",
)
plt.xlabel("Number of K")
plt.ylabel("inertia")
plt.title("K Elbow Method")
plt.plot(range(2, 20), silhouette, "o-")
plt.xlabel("Number of K")
plt.ylabel("silhouette Score")
model = MiniBatchKMeans(random_state=42)
distortion_visualizer = KElbowVisualizer(model, k=(2, 10))
distortion_visualizer.fit(pca_data)
distortion_visualizer.show()
# Instantiate the clustering model and visualizer
model = MiniBatchKMeans(5, random_state=42)
visualizer = SilhouetteVisualizer(model, colors="yellowbrick")
visualizer.fit(pca_data) # Fit the data to the visualizer
visualizer.show() # Finalize and render the figure
# Instantiate the clustering model and visualizer
model = MiniBatchKMeans(4, random_state=42)
visualizer = SilhouetteVisualizer(model, colors="yellowbrick")
visualizer.fit(pca_data) # Fit the data to the visualizer
visualizer.show() # Finalize and render the figure
# # GaussianMixture Model
BIC = []
AIC = []
for k in range(2, 10):
gm = GaussianMixture(n_components=k, n_init=10)
gm.fit(pca_data)
BIC.append(gm.bic(pca_data))
AIC.append(gm.aic(pca_data))
plt.plot(range(2, 10), BIC, "o-")
plt.xlabel("Number of K")
plt.ylabel("Bayesian information criterion (BIC)")
plt.plot(range(2, 10), AIC, "o-")
plt.xlabel("Number of K")
plt.ylabel("Akaike information criterion")
plt.errorbar(range(2, 10), np.gradient(BIC), label="BIC")
plt.title("Gradient of BIC Scores", fontsize=20)
plt.xticks(range(2, 10))
plt.xlabel("N. of clusters")
plt.ylabel("grad(BIC)")
plt.legend()
plt.errorbar(range(2, 10), np.gradient(AIC), label="AIC")
plt.title("Gradient of AIC Scores", fontsize=20)
plt.xticks(range(2, 10))
plt.xlabel("N. of clusters")
plt.ylabel("grad(AIC)")
plt.legend()
# # BayesianGaussianMixture
from sklearn.mixture import BayesianGaussianMixture
bgm = BayesianGaussianMixture(n_components=10, n_init=10, random_state=42)
bgm.fit(pca_data)
bgm.weights_.round(3)
# # AgglomerativeClustering
from scipy.cluster.hierarchy import dendrogram
from sklearn.cluster import AgglomerativeClustering
def plot_dendrogram(model, **kwargs):
# Create linkage matrix and then plot the dendrogram
# create the counts of samples under each node
counts = np.zeros(model.children_.shape[0])
n_samples = len(model.labels_)
for i, merge in enumerate(model.children_):
current_count = 0
for child_idx in merge:
if child_idx < n_samples:
current_count += 1 # leaf node
else:
current_count += counts[child_idx - n_samples]
counts[i] = current_count
linkage_matrix = np.column_stack(
[model.children_, model.distances_, counts]
).astype(float)
# Plot the corresponding dendrogram
dendrogram(linkage_matrix, **kwargs)
# setting distance_threshold=0 ensures we compute the full tree.
model = AgglomerativeClustering(distance_threshold=0, n_clusters=None)
model = model.fit(pca_data)
plt.title("Hierarchical Clustering Dendrogram")
# plot the top three levels of the dendrogram
plot_dendrogram(model, truncate_mode="level", p=3)
plt.xlabel("Number of points in node.")
plt.show()
# # ADD Labels to the data
pca_data["Target"] = labels
pca_data.head()
imp_data["Target"] = labels
imp_data.head()
# # Clusters Analysis
imp_data["Target"].value_counts().plot.pie(
autopct="%1.0f%%", pctdistance=0.7, labeldistance=1.1
)
def colorful_scatter(data):
LABEL_COLOR_MAP = {0: "y", 1: "g", 2: "m", 3: "k"}
sns.jointplot(
data=data,
x="BALANCE",
y="PURCHASES",
hue="Target",
palette=LABEL_COLOR_MAP,
alpha=0.6,
height=10,
)
colorful_scatter(imp_data)
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="BALANCE",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "BALANCE")
plt.title("Average of BALANCE in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="BALANCE_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "BALANCE_FREQUENCY")
plt.title("Average of BALANCE_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PURCHASES",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PURCHASES")
plt.title("Average of PURCHASES in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="ONEOFF_PURCHASES",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "ONEOFF_PURCHASES")
plt.title("Average of ONEOFF_PURCHASES in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="INSTALLMENTS_PURCHASES",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "INSTALLMENTS_PURCHASES")
plt.title("Average of INSTALLMENTS_PURCHASES in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="CASH_ADVANCE",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "CASH_ADVANCE")
plt.title("Average of CASH_ADVANCE in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PURCHASES_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PURCHASES_FREQUENCY")
plt.title("Average of PURCHASES_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="ONEOFF_PURCHASES_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "ONEOFF_PURCHASES_FREQUENCY")
plt.title("Average of ONEOFF_PURCHASES_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PURCHASES_INSTALLMENTS_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PURCHASES_INSTALLMENTS_FREQUENCY")
plt.title("Average of PURCHASES_INSTALLMENTS_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="CASH_ADVANCE_FREQUENCY",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "CASH_ADVANCE_FREQUENCY")
plt.title("Average of CASH_ADVANCE_FREQUENCY in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="CASH_ADVANCE_TRX",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "CASH_ADVANCE_TRX")
plt.title("Average of CASH_ADVANCE_TRX in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PURCHASES_TRX",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PURCHASES_TRX")
plt.title("Average of PURCHASES_TRX in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="CREDIT_LIMIT",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "CREDIT_LIMIT")
plt.title("Average of CREDIT_LIMIT in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PAYMENTS",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PAYMENTS")
plt.title("Average of 'PAYMENTS' in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="MINIMUM_PAYMENTS",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "MINIMUM_PAYMENTS")
plt.title("Average of 'MINIMUM_PAYMENTS' in Clusters")
plt.show()
sns.set_theme(style="whitegrid")
g = sns.catplot(
data=imp_data,
kind="bar",
x="Target",
y="PRC_FULL_PAYMENT",
palette=["red", "green", "blue", "gray"],
)
g.despine(left=True)
g.set_axis_labels("Cluster Number", "PRC_FULL_PAYMENT")
plt.title("Average of PRC_FULL_PAYMENT in Clusters")
plt.show()
x, y = pca_data.iloc[:, 0], pca_data.iloc[:, 1]
colors = {0: "red", 1: "blue", 2: "green", 3: "yellow"}
names = {
0: "Cluster 0",
1: "Cluster 1",
2: "Cluster 2",
3: "Cluseer 3",
}
df = pd.DataFrame({"x": x, "y": y, "label": pca_data.iloc[:, -1]})
groups = df.groupby("label")
fig, ax = plt.subplots(figsize=(20, 13))
for name, group in groups:
ax.plot(
group.x,
group.y,
marker="o",
linestyle="",
ms=5,
color=colors[name],
label=names[name],
mec="none",
)
ax.set_aspect("auto")
ax.tick_params(axis="x", which="both", bottom="off", top="off", labelbottom="off")
ax.tick_params(axis="y", which="both", left="off", top="off", labelleft="off")
ax.legend()
ax.set_title("Customers Segmentation based on their Credit Card usage bhaviour.")
plt.show()
| false | 0 | 6,453 | 3 | 6,910 | 6,453 |
||
129820079
|
import geopandas as gpd
import matplotlib.pyplot as plt
import contextily as cx
##read in files
wi_co = gpd.read_file(
"https://raw.githubusercontent.com/mhaffner/data/master/wi_counties.geojson"
)
ponds = gpd.read_file(
"https://gitlab.com/mhaffner/data/-/raw/master/Classified_Trout_Spring_Ponds2.geojson"
)
ponds
# change crs
wi_co_3070 = wi_co.to_crs(3070)
ponds_3070 = ponds.to_crs(3070)
ponds_3070.crs
# quick plot on single axis
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
wi_co_3070.plot(ax=ax, color="green")
ponds_3070.plot(ax=ax, color="orange")
# spatial join
join = gpd.sjoin(wi_co_3070, ponds_3070, how="left", predicate="intersects")
join
# assign a count column, 1 per value
join["count"] = 1
join
# add number of trout ponds per county
group_join = join.groupby("NAME").sum()[["count"]]
group_join
# sort descending
group_join.sort_values(by=(["count"]), ascending=False)
join_dissolve = join.dissolve("NAME")
join_dissolve
# add column to gdf
join_dissolve["count_sum"] = group_join
join_dissolve
# graduated color map
pond_map = join_dissolve.plot(
column="count_sum", legend=True, cmap="plasma", figsize=(12, 12)
)
plt.title("Trout Ponds per WI County")
plt.xlabel("Longitude")
plt.ylabel("Latitude")
cx.add_basemap(ax=pond_map, source=cx.providers.CartoDB.Positron, crs="EPSG:3070")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/820/129820079.ipynb
| null | null |
[{"Id": 129820079, "ScriptId": 37847892, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12217581, "CreationDate": "05/16/2023 17:44:29", "VersionNumber": 1.0, "Title": "lab_10_435_part_2", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 52.0, "LinesInsertedFromPrevious": 52.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import geopandas as gpd
import matplotlib.pyplot as plt
import contextily as cx
##read in files
wi_co = gpd.read_file(
"https://raw.githubusercontent.com/mhaffner/data/master/wi_counties.geojson"
)
ponds = gpd.read_file(
"https://gitlab.com/mhaffner/data/-/raw/master/Classified_Trout_Spring_Ponds2.geojson"
)
ponds
# change crs
wi_co_3070 = wi_co.to_crs(3070)
ponds_3070 = ponds.to_crs(3070)
ponds_3070.crs
# quick plot on single axis
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
wi_co_3070.plot(ax=ax, color="green")
ponds_3070.plot(ax=ax, color="orange")
# spatial join
join = gpd.sjoin(wi_co_3070, ponds_3070, how="left", predicate="intersects")
join
# assign a count column, 1 per value
join["count"] = 1
join
# add number of trout ponds per county
group_join = join.groupby("NAME").sum()[["count"]]
group_join
# sort descending
group_join.sort_values(by=(["count"]), ascending=False)
join_dissolve = join.dissolve("NAME")
join_dissolve
# add column to gdf
join_dissolve["count_sum"] = group_join
join_dissolve
# graduated color map
pond_map = join_dissolve.plot(
column="count_sum", legend=True, cmap="plasma", figsize=(12, 12)
)
plt.title("Trout Ponds per WI County")
plt.xlabel("Longitude")
plt.ylabel("Latitude")
cx.add_basemap(ax=pond_map, source=cx.providers.CartoDB.Positron, crs="EPSG:3070")
| false | 0 | 521 | 0 | 521 | 521 |
||
129820515
|
<jupyter_start><jupyter_text>COVID-19 World Vaccination Progress
### Context
Data is collected daily from [**Our World in Data**](https://ourworldindata.org/) GitHub repository for [covid-19](https://github.com/owid/covid-19-data), merged and uploaded. Country level vaccination data is gathered and assembled in one single file. Then, this data file is merged with locations data file to include vaccination sources information. A second file, with manufacturers information, is included.
### Content
The data (country vaccinations) contains the following information:
* **Country**- this is the country for which the vaccination information is provided;
* **Country ISO Code** - ISO code for the country;
* **Date** - date for the data entry; for some of the dates we have only the daily vaccinations, for others, only the (cumulative) total;
* **Total number of vaccinations** - this is the absolute number of total immunizations in the country;
* **Total number of people vaccinated** - a person, depending on the immunization scheme, will receive one or more (typically 2) vaccines; at a certain moment, the number of vaccination might be larger than the number of people;
* **Total number of people fully vaccinated** - this is the number of people that received the entire set of immunization according to the immunization scheme (typically 2); at a certain moment in time, there might be a certain number of people that received one vaccine and another number (smaller) of people that received all vaccines in the scheme;
* **Daily vaccinations (raw)** - for a certain data entry, the number of vaccination for that date/country;
* **Daily vaccinations** - for a certain data entry, the number of vaccination for that date/country;
* **Total vaccinations per hundred** - ratio (in percent) between vaccination number and total population up to the date in the country;
* **Total number of people vaccinated per hundred** - ratio (in percent) between population immunized and total population up to the date in the country;
* **Total number of people fully vaccinated per hundred** - ratio (in percent) between population fully immunized and total population up to the date in the country;
* **Number of vaccinations per day** - number of daily vaccination for that day and country;
* **Daily vaccinations per million** - ratio (in ppm) between vaccination number and total population for the current date in the country;
* **Vaccines used in the country** - total number of vaccines used in the country (up to date);
* **Source name** - source of the information (national authority, international organization, local organization etc.);
* **Source website** - website of the source of information;
There is a second file added recently (country vaccinations by manufacturer), with the following columns:
* **Location** - country;
* **Date** - date;
* **Vaccine** - vaccine type;
* **Total number of vaccinations** - total number of vaccinations / current time and vaccine type.
Kaggle dataset identifier: covid-world-vaccination-progress
<jupyter_code>import pandas as pd
df = pd.read_csv('covid-world-vaccination-progress/country_vaccinations.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 86512 entries, 0 to 86511
Data columns (total 15 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 country 86512 non-null object
1 iso_code 86512 non-null object
2 date 86512 non-null object
3 total_vaccinations 43607 non-null float64
4 people_vaccinated 41294 non-null float64
5 people_fully_vaccinated 38802 non-null float64
6 daily_vaccinations_raw 35362 non-null float64
7 daily_vaccinations 86213 non-null float64
8 total_vaccinations_per_hundred 43607 non-null float64
9 people_vaccinated_per_hundred 41294 non-null float64
10 people_fully_vaccinated_per_hundred 38802 non-null float64
11 daily_vaccinations_per_million 86213 non-null float64
12 vaccines 86512 non-null object
13 source_name 86512 non-null object
14 source_website 86512 non-null object
dtypes: float64(9), object(6)
memory usage: 9.9+ MB
<jupyter_text>Examples:
{
"country": "Afghanistan",
"iso_code": "AFG",
"date": "2021-02-22 00:00:00",
"total_vaccinations": 0.0,
"people_vaccinated": 0.0,
"people_fully_vaccinated": NaN,
"daily_vaccinations_raw": NaN,
"daily_vaccinations": NaN,
"total_vaccinations_per_hundred": 0.0,
"people_vaccinated_per_hundred": 0.0,
"people_fully_vaccinated_per_hundred": NaN,
"daily_vaccinations_per_million": NaN,
"vaccines": "Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",
"source_name": "World Health Organization",
"source_website": "https://covid19.who.int/"
}
{
"country": "Afghanistan",
"iso_code": "AFG",
"date": "2021-02-23 00:00:00",
"total_vaccinations": NaN,
"people_vaccinated": NaN,
"people_fully_vaccinated": NaN,
"daily_vaccinations_raw": NaN,
"daily_vaccinations": 1367.0,
"total_vaccinations_per_hundred": NaN,
"people_vaccinated_per_hundred": NaN,
"people_fully_vaccinated_per_hundred": NaN,
"daily_vaccinations_per_million": 34.0,
"vaccines": "Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",
"source_name": "World Health Organization",
"source_website": "https://covid19.who.int/"
}
{
"country": "Afghanistan",
"iso_code": "AFG",
"date": "2021-02-24 00:00:00",
"total_vaccinations": NaN,
"people_vaccinated": NaN,
"people_fully_vaccinated": NaN,
"daily_vaccinations_raw": NaN,
"daily_vaccinations": 1367.0,
"total_vaccinations_per_hundred": NaN,
"people_vaccinated_per_hundred": NaN,
"people_fully_vaccinated_per_hundred": NaN,
"daily_vaccinations_per_million": 34.0,
"vaccines": "Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",
"source_name": "World Health Organization",
"source_website": "https://covid19.who.int/"
}
{
"country": "Afghanistan",
"iso_code": "AFG",
"date": "2021-02-25 00:00:00",
"total_vaccinations": NaN,
"people_vaccinated": NaN,
"people_fully_vaccinated": NaN,
"daily_vaccinations_raw": NaN,
"daily_vaccinations": 1367.0,
"total_vaccinations_per_hundred": NaN,
"people_vaccinated_per_hundred": NaN,
"people_fully_vaccinated_per_hundred": NaN,
"daily_vaccinations_per_million": 34.0,
"vaccines": "Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",
"source_name": "World Health Organization",
"source_website": "https://covid19.who.int/"
}
<jupyter_script>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv(
"/kaggle/input/covid-world-vaccination-progress/country_vaccinations.csv"
)
data.head()
data.describe()
pd.to_datetime(data.date)
data.country.value_counts()
data = data[
data.country.apply(
lambda x: x not in ["England", "Scotland", "Wales", "Northern Ireland"]
)
]
data.country.value_counts()
data.vaccines.value_counts()
df = data[["vaccines", "country"]]
df.head()
dict_ = {}
for i in df.vaccines.unique():
dict_[i] = [df["country"][j] for j in df[df["vaccines"] == i].index]
vaccines = {}
for key, value in dict_.items():
vaccines[key] = set(value)
for i, j in vaccines.items():
print(f"{i}:>>{j}")
import plotly.express as px
import plotly.offline as py
vaccine_map = px.choropleth(data, locations="iso_code", color="vaccines")
vaccine_map.update_layout(height=300, margin={"r": 0, "t": 0, "l": 0, "b": 0})
vaccine_map.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/820/129820515.ipynb
|
covid-world-vaccination-progress
|
gpreda
|
[{"Id": 129820515, "ScriptId": 38609384, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6012166, "CreationDate": "05/16/2023 17:48:41", "VersionNumber": 1.0, "Title": "Covid-19 Vaccines Analysis", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 39.0, "LinesInsertedFromPrevious": 39.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 186197380, "KernelVersionId": 129820515, "SourceDatasetVersionId": 3385976}]
|
[{"Id": 3385976, "DatasetId": 1093816, "DatasourceVersionId": 3437501, "CreatorUserId": 769452, "LicenseName": "CC0: Public Domain", "CreationDate": "03/31/2022 07:55:09", "VersionNumber": 249.0, "Title": "COVID-19 World Vaccination Progress", "Slug": "covid-world-vaccination-progress", "Subtitle": "Daily and Total Vaccination for COVID-19 in the World from Our World in Data", "Description": "### Context\n\nData is collected daily from [**Our World in Data**](https://ourworldindata.org/) GitHub repository for [covid-19](https://github.com/owid/covid-19-data), merged and uploaded. Country level vaccination data is gathered and assembled in one single file. Then, this data file is merged with locations data file to include vaccination sources information. A second file, with manufacturers information, is included.\n\n\n### Content\n\nThe data (country vaccinations) contains the following information:\n* **Country**- this is the country for which the vaccination information is provided; \n* **Country ISO Code** - ISO code for the country; \n* **Date** - date for the data entry; for some of the dates we have only the daily vaccinations, for others, only the (cumulative) total; \n* **Total number of vaccinations** - this is the absolute number of total immunizations in the country; \n* **Total number of people vaccinated** - a person, depending on the immunization scheme, will receive one or more (typically 2) vaccines; at a certain moment, the number of vaccination might be larger than the number of people; \n* **Total number of people fully vaccinated** - this is the number of people that received the entire set of immunization according to the immunization scheme (typically 2); at a certain moment in time, there might be a certain number of people that received one vaccine and another number (smaller) of people that received all vaccines in the scheme; \n* **Daily vaccinations (raw)** - for a certain data entry, the number of vaccination for that date/country; \n* **Daily vaccinations** - for a certain data entry, the number of vaccination for that date/country; \n* **Total vaccinations per hundred** - ratio (in percent) between vaccination number and total population up to the date in the country; \n* **Total number of people vaccinated per hundred** - ratio (in percent) between population immunized and total population up to the date in the country; \n* **Total number of people fully vaccinated per hundred** - ratio (in percent) between population fully immunized and total population up to the date in the country; \n* **Number of vaccinations per day** - number of daily vaccination for that day and country; \n* **Daily vaccinations per million** - ratio (in ppm) between vaccination number and total population for the current date in the country; \n* **Vaccines used in the country** - total number of vaccines used in the country (up to date); \n* **Source name** - source of the information (national authority, international organization, local organization etc.); \n* **Source website** - website of the source of information; \n\n\nThere is a second file added recently (country vaccinations by manufacturer), with the following columns:\n* **Location** - country; \n* **Date** - date; \n* **Vaccine** - vaccine type; \n* **Total number of vaccinations** - total number of vaccinations / current time and vaccine type.\n\n\n### Acknowledgements\n\nI would like to specify that I am only making available **Our World in Data** collected data about vaccinations to Kagglers. My contribution is very small, just daily collection, merge and upload of the updated version, as maintained by **Our World in Data** in their GitHub repository.\n\n### Inspiration\n\nTrack COVID-19 vaccination in the World, answer instantly to your questions: \n- Which country is using what vaccine? \n- In which country the vaccination programme is more advanced? \n- Where are vaccinated more people per day? But in terms of percent from entire population ?\n\nCombine this dataset with [COVID-19 World Testing Progress](https://www.kaggle.com/gpreda/covid19-world-testing-progress) and [COVID-19 Variants Worldwide Evolution](https://www.kaggle.com/gpreda/covid19-variants) to get more insights on the dynamics of the pandemics, as reflected in the interdependence of amount of testing performed, results of sequencing and vaccination campaigns.", "VersionNotes": "Data Update 2022/03/31", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1093816, "CreatorUserId": 769452, "OwnerUserId": 769452.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3385976.0, "CurrentDatasourceVersionId": 3437501.0, "ForumId": 1110973, "Type": 2, "CreationDate": "01/12/2021 17:01:16", "LastActivityDate": "01/12/2021", "TotalViews": 542075, "TotalDownloads": 96188, "TotalVotes": 2164, "TotalKernels": 428}]
|
[{"Id": 769452, "UserName": "gpreda", "DisplayName": "Gabriel Preda", "RegisterDate": "10/27/2016", "PerformanceTier": 4}]
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv(
"/kaggle/input/covid-world-vaccination-progress/country_vaccinations.csv"
)
data.head()
data.describe()
pd.to_datetime(data.date)
data.country.value_counts()
data = data[
data.country.apply(
lambda x: x not in ["England", "Scotland", "Wales", "Northern Ireland"]
)
]
data.country.value_counts()
data.vaccines.value_counts()
df = data[["vaccines", "country"]]
df.head()
dict_ = {}
for i in df.vaccines.unique():
dict_[i] = [df["country"][j] for j in df[df["vaccines"] == i].index]
vaccines = {}
for key, value in dict_.items():
vaccines[key] = set(value)
for i, j in vaccines.items():
print(f"{i}:>>{j}")
import plotly.express as px
import plotly.offline as py
vaccine_map = px.choropleth(data, locations="iso_code", color="vaccines")
vaccine_map.update_layout(height=300, margin={"r": 0, "t": 0, "l": 0, "b": 0})
vaccine_map.show()
|
[{"covid-world-vaccination-progress/country_vaccinations.csv": {"column_names": "[\"country\", \"iso_code\", \"date\", \"total_vaccinations\", \"people_vaccinated\", \"people_fully_vaccinated\", \"daily_vaccinations_raw\", \"daily_vaccinations\", \"total_vaccinations_per_hundred\", \"people_vaccinated_per_hundred\", \"people_fully_vaccinated_per_hundred\", \"daily_vaccinations_per_million\", \"vaccines\", \"source_name\", \"source_website\"]", "column_data_types": "{\"country\": \"object\", \"iso_code\": \"object\", \"date\": \"object\", \"total_vaccinations\": \"float64\", \"people_vaccinated\": \"float64\", \"people_fully_vaccinated\": \"float64\", \"daily_vaccinations_raw\": \"float64\", \"daily_vaccinations\": \"float64\", \"total_vaccinations_per_hundred\": \"float64\", \"people_vaccinated_per_hundred\": \"float64\", \"people_fully_vaccinated_per_hundred\": \"float64\", \"daily_vaccinations_per_million\": \"float64\", \"vaccines\": \"object\", \"source_name\": \"object\", \"source_website\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 86512 entries, 0 to 86511\nData columns (total 15 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 country 86512 non-null object \n 1 iso_code 86512 non-null object \n 2 date 86512 non-null object \n 3 total_vaccinations 43607 non-null float64\n 4 people_vaccinated 41294 non-null float64\n 5 people_fully_vaccinated 38802 non-null float64\n 6 daily_vaccinations_raw 35362 non-null float64\n 7 daily_vaccinations 86213 non-null float64\n 8 total_vaccinations_per_hundred 43607 non-null float64\n 9 people_vaccinated_per_hundred 41294 non-null float64\n 10 people_fully_vaccinated_per_hundred 38802 non-null float64\n 11 daily_vaccinations_per_million 86213 non-null float64\n 12 vaccines 86512 non-null object \n 13 source_name 86512 non-null object \n 14 source_website 86512 non-null object \ndtypes: float64(9), object(6)\nmemory usage: 9.9+ MB\n", "summary": "{\"total_vaccinations\": {\"count\": 43607.0, \"mean\": 45929644.638727725, \"std\": 224600360.18166688, \"min\": 0.0, \"25%\": 526410.0, \"50%\": 3590096.0, \"75%\": 17012303.5, \"max\": 3263129000.0}, \"people_vaccinated\": {\"count\": 41294.0, \"mean\": 17705077.78979997, \"std\": 70787311.5004759, \"min\": 0.0, \"25%\": 349464.25, \"50%\": 2187310.5, \"75%\": 9152519.75, \"max\": 1275541000.0}, \"people_fully_vaccinated\": {\"count\": 38802.0, \"mean\": 14138299.848152157, \"std\": 57139201.71915868, \"min\": 1.0, \"25%\": 243962.25, \"50%\": 1722140.5, \"75%\": 7559869.5, \"max\": 1240777000.0}, \"daily_vaccinations_raw\": {\"count\": 35362.0, \"mean\": 270599.5782478367, \"std\": 1212426.60195391, \"min\": 0.0, \"25%\": 4668.0, \"50%\": 25309.0, \"75%\": 123492.5, \"max\": 24741000.0}, \"daily_vaccinations\": {\"count\": 86213.0, \"mean\": 131305.48607518588, \"std\": 768238.7732930565, \"min\": 0.0, \"25%\": 900.0, \"50%\": 7343.0, \"75%\": 44098.0, \"max\": 22424286.0}, \"total_vaccinations_per_hundred\": {\"count\": 43607.0, \"mean\": 80.18854312381039, \"std\": 67.91357674747688, \"min\": 0.0, \"25%\": 16.05, \"50%\": 67.52, \"75%\": 132.735, \"max\": 345.37}, \"people_vaccinated_per_hundred\": {\"count\": 41294.0, \"mean\": 40.92731728580423, \"std\": 29.29075864533803, \"min\": 0.0, \"25%\": 11.37, \"50%\": 41.435, \"75%\": 67.91, \"max\": 124.76}, \"people_fully_vaccinated_per_hundred\": {\"count\": 38802.0, \"mean\": 35.52324287407866, \"std\": 28.37625180924737, \"min\": 0.0, \"25%\": 7.02, \"50%\": 31.75, \"75%\": 62.08, \"max\": 122.37}, \"daily_vaccinations_per_million\": {\"count\": 86213.0, \"mean\": 3257.049157319662, \"std\": 3934.3124401057307, \"min\": 0.0, \"25%\": 636.0, \"50%\": 2050.0, \"75%\": 4682.0, \"max\": 117497.0}}", "examples": "{\"country\":{\"0\":\"Afghanistan\",\"1\":\"Afghanistan\",\"2\":\"Afghanistan\",\"3\":\"Afghanistan\"},\"iso_code\":{\"0\":\"AFG\",\"1\":\"AFG\",\"2\":\"AFG\",\"3\":\"AFG\"},\"date\":{\"0\":\"2021-02-22\",\"1\":\"2021-02-23\",\"2\":\"2021-02-24\",\"3\":\"2021-02-25\"},\"total_vaccinations\":{\"0\":0.0,\"1\":null,\"2\":null,\"3\":null},\"people_vaccinated\":{\"0\":0.0,\"1\":null,\"2\":null,\"3\":null},\"people_fully_vaccinated\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"daily_vaccinations_raw\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"daily_vaccinations\":{\"0\":null,\"1\":1367.0,\"2\":1367.0,\"3\":1367.0},\"total_vaccinations_per_hundred\":{\"0\":0.0,\"1\":null,\"2\":null,\"3\":null},\"people_vaccinated_per_hundred\":{\"0\":0.0,\"1\":null,\"2\":null,\"3\":null},\"people_fully_vaccinated_per_hundred\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"daily_vaccinations_per_million\":{\"0\":null,\"1\":34.0,\"2\":34.0,\"3\":34.0},\"vaccines\":{\"0\":\"Johnson&Johnson, Oxford\\/AstraZeneca, Pfizer\\/BioNTech, Sinopharm\\/Beijing\",\"1\":\"Johnson&Johnson, Oxford\\/AstraZeneca, Pfizer\\/BioNTech, Sinopharm\\/Beijing\",\"2\":\"Johnson&Johnson, Oxford\\/AstraZeneca, Pfizer\\/BioNTech, Sinopharm\\/Beijing\",\"3\":\"Johnson&Johnson, Oxford\\/AstraZeneca, Pfizer\\/BioNTech, Sinopharm\\/Beijing\"},\"source_name\":{\"0\":\"World Health Organization\",\"1\":\"World Health Organization\",\"2\":\"World Health Organization\",\"3\":\"World Health Organization\"},\"source_website\":{\"0\":\"https:\\/\\/covid19.who.int\\/\",\"1\":\"https:\\/\\/covid19.who.int\\/\",\"2\":\"https:\\/\\/covid19.who.int\\/\",\"3\":\"https:\\/\\/covid19.who.int\\/\"}}"}}]
| true | 1 |
<start_data_description><data_path>covid-world-vaccination-progress/country_vaccinations.csv:
<column_names>
['country', 'iso_code', 'date', 'total_vaccinations', 'people_vaccinated', 'people_fully_vaccinated', 'daily_vaccinations_raw', 'daily_vaccinations', 'total_vaccinations_per_hundred', 'people_vaccinated_per_hundred', 'people_fully_vaccinated_per_hundred', 'daily_vaccinations_per_million', 'vaccines', 'source_name', 'source_website']
<column_types>
{'country': 'object', 'iso_code': 'object', 'date': 'object', 'total_vaccinations': 'float64', 'people_vaccinated': 'float64', 'people_fully_vaccinated': 'float64', 'daily_vaccinations_raw': 'float64', 'daily_vaccinations': 'float64', 'total_vaccinations_per_hundred': 'float64', 'people_vaccinated_per_hundred': 'float64', 'people_fully_vaccinated_per_hundred': 'float64', 'daily_vaccinations_per_million': 'float64', 'vaccines': 'object', 'source_name': 'object', 'source_website': 'object'}
<dataframe_Summary>
{'total_vaccinations': {'count': 43607.0, 'mean': 45929644.638727725, 'std': 224600360.18166688, 'min': 0.0, '25%': 526410.0, '50%': 3590096.0, '75%': 17012303.5, 'max': 3263129000.0}, 'people_vaccinated': {'count': 41294.0, 'mean': 17705077.78979997, 'std': 70787311.5004759, 'min': 0.0, '25%': 349464.25, '50%': 2187310.5, '75%': 9152519.75, 'max': 1275541000.0}, 'people_fully_vaccinated': {'count': 38802.0, 'mean': 14138299.848152157, 'std': 57139201.71915868, 'min': 1.0, '25%': 243962.25, '50%': 1722140.5, '75%': 7559869.5, 'max': 1240777000.0}, 'daily_vaccinations_raw': {'count': 35362.0, 'mean': 270599.5782478367, 'std': 1212426.60195391, 'min': 0.0, '25%': 4668.0, '50%': 25309.0, '75%': 123492.5, 'max': 24741000.0}, 'daily_vaccinations': {'count': 86213.0, 'mean': 131305.48607518588, 'std': 768238.7732930565, 'min': 0.0, '25%': 900.0, '50%': 7343.0, '75%': 44098.0, 'max': 22424286.0}, 'total_vaccinations_per_hundred': {'count': 43607.0, 'mean': 80.18854312381039, 'std': 67.91357674747688, 'min': 0.0, '25%': 16.05, '50%': 67.52, '75%': 132.735, 'max': 345.37}, 'people_vaccinated_per_hundred': {'count': 41294.0, 'mean': 40.92731728580423, 'std': 29.29075864533803, 'min': 0.0, '25%': 11.37, '50%': 41.435, '75%': 67.91, 'max': 124.76}, 'people_fully_vaccinated_per_hundred': {'count': 38802.0, 'mean': 35.52324287407866, 'std': 28.37625180924737, 'min': 0.0, '25%': 7.02, '50%': 31.75, '75%': 62.08, 'max': 122.37}, 'daily_vaccinations_per_million': {'count': 86213.0, 'mean': 3257.049157319662, 'std': 3934.3124401057307, 'min': 0.0, '25%': 636.0, '50%': 2050.0, '75%': 4682.0, 'max': 117497.0}}
<dataframe_info>
RangeIndex: 86512 entries, 0 to 86511
Data columns (total 15 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 country 86512 non-null object
1 iso_code 86512 non-null object
2 date 86512 non-null object
3 total_vaccinations 43607 non-null float64
4 people_vaccinated 41294 non-null float64
5 people_fully_vaccinated 38802 non-null float64
6 daily_vaccinations_raw 35362 non-null float64
7 daily_vaccinations 86213 non-null float64
8 total_vaccinations_per_hundred 43607 non-null float64
9 people_vaccinated_per_hundred 41294 non-null float64
10 people_fully_vaccinated_per_hundred 38802 non-null float64
11 daily_vaccinations_per_million 86213 non-null float64
12 vaccines 86512 non-null object
13 source_name 86512 non-null object
14 source_website 86512 non-null object
dtypes: float64(9), object(6)
memory usage: 9.9+ MB
<some_examples>
{'country': {'0': 'Afghanistan', '1': 'Afghanistan', '2': 'Afghanistan', '3': 'Afghanistan'}, 'iso_code': {'0': 'AFG', '1': 'AFG', '2': 'AFG', '3': 'AFG'}, 'date': {'0': '2021-02-22', '1': '2021-02-23', '2': '2021-02-24', '3': '2021-02-25'}, 'total_vaccinations': {'0': 0.0, '1': None, '2': None, '3': None}, 'people_vaccinated': {'0': 0.0, '1': None, '2': None, '3': None}, 'people_fully_vaccinated': {'0': None, '1': None, '2': None, '3': None}, 'daily_vaccinations_raw': {'0': None, '1': None, '2': None, '3': None}, 'daily_vaccinations': {'0': None, '1': 1367.0, '2': 1367.0, '3': 1367.0}, 'total_vaccinations_per_hundred': {'0': 0.0, '1': None, '2': None, '3': None}, 'people_vaccinated_per_hundred': {'0': 0.0, '1': None, '2': None, '3': None}, 'people_fully_vaccinated_per_hundred': {'0': None, '1': None, '2': None, '3': None}, 'daily_vaccinations_per_million': {'0': None, '1': 34.0, '2': 34.0, '3': 34.0}, 'vaccines': {'0': 'Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing', '1': 'Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing', '2': 'Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing', '3': 'Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing'}, 'source_name': {'0': 'World Health Organization', '1': 'World Health Organization', '2': 'World Health Organization', '3': 'World Health Organization'}, 'source_website': {'0': 'https://covid19.who.int/', '1': 'https://covid19.who.int/', '2': 'https://covid19.who.int/', '3': 'https://covid19.who.int/'}}
<end_description>
| 398 | 2 | 2,581 | 398 |
129820075
|
<jupyter_start><jupyter_text>Mental Health and Suicide Rates
### Context
Close to 800 000 people die due to suicide every year, which is one person every 40 seconds. Suicide is a global phenomenon and occurs throughout the lifespan. Effective and evidence-based interventions can be implemented at population, sub-population and individual levels to prevent suicide and suicide attempts. There are indications that for each adult who died by suicide there may have been more than 20 others attempting suicide.
Suicide is a complex issue and therefore suicide prevention efforts require coordination and collaboration among multiple sectors of society, including the health sector and other sectors such as education, labour, agriculture, business, justice, law, defense, politics, and the media. These efforts must be comprehensive and integrated as no single approach alone can make an impact on an issue as complex as suicide.
### Do leave an upvote if you found this dataset useful!
Kaggle dataset identifier: mental-health-and-suicide-rates
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/mental-health-and-suicide-rates/Crude suicide rates.csv"
)
df
import matplotlib.pyplot as plt
import seaborn as sns
# countplot = sns.countplot(data=df, x="20to29")
plot = sns.catplot(x="Sex", y=" 20to29", data=df)
# df.columns
# age_20to29 = df[df[" 20to29"]>0]
top_35_values = df.nlargest(5, " 70to79")
top_35_values
by_age = top_35_values.groupby("Country", as_index=False)
by_age_2029 = by_age[" 70to79"].mean()
by_age_2029 = by_age_2029.tail(5)
barplot = sns.barplot(x="Country", y=" 70to79", data=by_age_2029)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/820/129820075.ipynb
|
mental-health-and-suicide-rates
|
twinkle0705
|
[{"Id": 129820075, "ScriptId": 38605557, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14721492, "CreationDate": "05/16/2023 17:44:26", "VersionNumber": 1.0, "Title": "Mental Health2", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 36.0, "LinesInsertedFromPrevious": 36.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186196801, "KernelVersionId": 129820075, "SourceDatasetVersionId": 1338480}]
|
[{"Id": 1338480, "DatasetId": 748724, "DatasourceVersionId": 1370824, "CreatorUserId": 3649586, "LicenseName": "Attribution-NonCommercial-ShareAlike 3.0 IGO (CC BY-NC-SA 3.0 IGO)", "CreationDate": "07/15/2020 12:33:00", "VersionNumber": 2.0, "Title": "Mental Health and Suicide Rates", "Slug": "mental-health-and-suicide-rates", "Subtitle": "Suicide Rates of age groups in different countries along with Health Facility", "Description": "### Context\n\nClose to 800 000 people die due to suicide every year, which is one person every 40 seconds. Suicide is a global phenomenon and occurs throughout the lifespan. Effective and evidence-based interventions can be implemented at population, sub-population and individual levels to prevent suicide and suicide attempts. There are indications that for each adult who died by suicide there may have been more than 20 others attempting suicide.\n\nSuicide is a complex issue and therefore suicide prevention efforts require coordination and collaboration among multiple sectors of society, including the health sector and other sectors such as education, labour, agriculture, business, justice, law, defense, politics, and the media. These efforts must be comprehensive and integrated as no single approach alone can make an impact on an issue as complex as suicide.\n\n### Do leave an upvote if you found this dataset useful!", "VersionNotes": "updated files", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 748724, "CreatorUserId": 3649586, "OwnerUserId": 3649586.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1338480.0, "CurrentDatasourceVersionId": 1370824.0, "ForumId": 763628, "Type": 2, "CreationDate": "06/30/2020 16:51:51", "LastActivityDate": "06/30/2020", "TotalViews": 106199, "TotalDownloads": 12135, "TotalVotes": 211, "TotalKernels": 10}]
|
[{"Id": 3649586, "UserName": "twinkle0705", "DisplayName": "Twinkle Khanna", "RegisterDate": "09/01/2019", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/mental-health-and-suicide-rates/Crude suicide rates.csv"
)
df
import matplotlib.pyplot as plt
import seaborn as sns
# countplot = sns.countplot(data=df, x="20to29")
plot = sns.catplot(x="Sex", y=" 20to29", data=df)
# df.columns
# age_20to29 = df[df[" 20to29"]>0]
top_35_values = df.nlargest(5, " 70to79")
top_35_values
by_age = top_35_values.groupby("Country", as_index=False)
by_age_2029 = by_age[" 70to79"].mean()
by_age_2029 = by_age_2029.tail(5)
barplot = sns.barplot(x="Country", y=" 70to79", data=by_age_2029)
| false | 1 | 446 | 0 | 704 | 446 |
||
129820956
|
<jupyter_start><jupyter_text>NASA - Nearest Earth Objects
# Context
There is an infinite number of objects in the outer space. Some of them are closer than we think. Even though we might think that a distance of 70,000 Km can not potentially harm us, but at an astronomical scale, this is a very small distance and can disrupt many natural phenomena. These objects/asteroids can thus prove to be harmful. Hence, it is wise to know what is surrounding us and what can harm us amongst those. Thus, this dataset compiles the list of NASA certified asteroids that are classified as the nearest earth object.
# Sources
[NASA Open API](https://api.nasa.gov/)
[NEO Earth Close Approaches](https://cneos.jpl.nasa.gov/ca/)
Kaggle dataset identifier: nasa-nearest-earth-objects
<jupyter_code>import pandas as pd
df = pd.read_csv('nasa-nearest-earth-objects/neo.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 90836 entries, 0 to 90835
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 90836 non-null int64
1 name 90836 non-null object
2 est_diameter_min 90836 non-null float64
3 est_diameter_max 90836 non-null float64
4 relative_velocity 90836 non-null float64
5 miss_distance 90836 non-null float64
6 orbiting_body 90836 non-null object
7 sentry_object 90836 non-null bool
8 absolute_magnitude 90836 non-null float64
9 hazardous 90836 non-null bool
dtypes: bool(2), float64(5), int64(1), object(2)
memory usage: 5.7+ MB
<jupyter_text>Examples:
{
"id": 2162635,
"name": "162635 (2000 SS164)",
"est_diameter_min": 1.1982708007,
"est_diameter_max": 2.6794149658,
"relative_velocity": 13569.2492241812,
"miss_distance": 54839744.08284605,
"orbiting_body": "Earth",
"sentry_object": false,
"absolute_magnitude": 16.73,
"hazardous": false
}
{
"id": 2277475,
"name": "277475 (2005 WK4)",
"est_diameter_min": 0.26580000000000004,
"est_diameter_max": 0.5943468684000001,
"relative_velocity": 73588.7266634981,
"miss_distance": 61438126.52395093,
"orbiting_body": "Earth",
"sentry_object": false,
"absolute_magnitude": 20.0,
"hazardous": true
}
{
"id": 2512244,
"name": "512244 (2015 YE18)",
"est_diameter_min": 0.7220295577,
"est_diameter_max": 1.6145071727,
"relative_velocity": 114258.6921290512,
"miss_distance": 49798724.94045679,
"orbiting_body": "Earth",
"sentry_object": false,
"absolute_magnitude": 17.83,
"hazardous": false
}
{
"id": 3596030,
"name": "(2012 BV13)",
"est_diameter_min": 0.096506147,
"est_diameter_max": 0.2157943048,
"relative_velocity": 24764.3031380016,
"miss_distance": 25434972.72075825,
"orbiting_body": "Earth",
"sentry_object": false,
"absolute_magnitude": 22.2,
"hazardous": false
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## NASA - Nearest Earth Objects
# #### A cumulative data for Nearest Earth Objects by NASA
# ### CONTEXT
# There is an infinite number of objects in the outer space.
# Some of them are closer than we think. Even though we might
# think that a distance of 70,000 Km can’t potentially harm us,
# but at an astronomical scale, this is a very small distance
# and can disrupt many natural phenomena. These objects/asteroids
# can thus prove to be harmful. Hence, it is wise to know what is
# surrounding us and what can harm us amongst those. Thus, this
# dataset compiles the list of NASA certified asteroids that are c
# lassified as the nearest earth object.
# ### ABSTRACT OF THE PROBLEM:
# Sentry is a highly automated collision monitoring system that continually scans the most current asteroid catalogue for possibilities of future impact with Earth over the next 100 years. So, this concludes whether that object is hazardous for the earth are not.
#
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
neo = pd.read_csv(r"/kaggle/input/nasa-nearest-earth-objects/neo.csv")
neo.head()
# ### DATA PREPROCESSING
# #### Checking for null values
neo.isnull().sum()
# #### Checking the data types of the data set
neo.dtypes
# #### Changing the bool values into Binary 0 or 1
# #### True = 1, False = 0
neo["hazardous"] = neo["hazardous"].astype(int)
print(neo["hazardous"].head())
neo.columns
# #### Getting unique values for the each column in a dataset
unique_values = neo.nunique()
print(unique_values)
# ### Oribiting body : Earth
# ### Sentry_object : False
# #### so changing the value into 1
neo["sentry_object"] = neo["sentry_object"].astype(int)
print(neo["sentry_object"].head())
def my_func(row):
if row["orbiting_body"] == "Earth":
val = 1
else:
val = 0
return val
neo["orbiting_body"] = neo.apply(my_func, axis=1)
neo["orbiting_body"].head()
# Checking for the data types
neo.dtypes
# ### Multiple Linear Regression
# We use Multiple linear regression model to estimate the relationship
# between a quantitative dependent variable ['hazardous'] and
# independent variables
# ['est_diameter_min', 'est_diameter_max', 'relative_velocity',
# 'miss_distance', 'orbiting_body', 'sentry_object', 'absolute_magnitude']
# using a straight line.
X = neo.iloc[:, 2:9]
y = neo["hazardous"]
# train_test_split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
print("X train shape:", X_train.shape)
print("y_train shape", y_train.shape)
print("X_test shape:", X_test.shape)
print("y_test shape", y_test.shape)
# Multiple Linear Regression
from sklearn.linear_model import LinearRegression
regressor = LinearRegression()
regressor.fit(X_train, y_train)
print("Regression co-efficient:", regressor.coef_)
print()
print("Regressor intercept:", regressor.intercept_)
print()
y_pred = regressor.predict(X_test)
print("Predicted values:", y_pred)
print()
print("Actual values:", y_test.values)
sns.pairplot(
neo,
x_vars=[
"est_diameter_min",
"est_diameter_max",
"relative_velocity",
"miss_distance",
"orbiting_body",
"sentry_object",
"absolute_magnitude",
],
y_vars="hazardous",
kind="reg",
)
# The argument kind='reg' adds a linear regression line to each scatter
# plot, allowing us to visualize the relationship between the independent
# and dependent variables.
# #### Dimensionality Reduction
import statsmodels.regression.linear_model as sm
# add a column of ones as integer data type
X = np.append(arr=np.ones((90836, 1)).astype(int), values=X, axis=1)
# choose a Significance level usually 0.05, if p>0.05
# for the highest values parameter, remove that value
X_opt = X[:, [0, 3, 4, 5]]
ols = sm.OLS(endog=y, exog=X_opt).fit()
ols.summary()
sns.pairplot(
neo,
x_vars=["est_diameter_min", "est_diameter_max", "relative_velocity"],
y_vars="hazardous",
kind="reg",
)
# ### Logistic Regression
from sklearn.linear_model import LogisticRegression
logreg = LogisticRegression()
logreg.fit(X_train, y_train)
y_pred = logreg.predict(X_test)
print("Accuracy:", logreg.score(X_test, y_test))
from sklearn.metrics import confusion_matrix, roc_curve, roc_auc_score
fpr, tpr, thresholds = roc_curve(y_test, logreg.predict_proba(X_test)[:, 1])
roc_auc = roc_auc_score(y_test, y_pred)
plt.plot(fpr, tpr, label="ROC Curve( area = %0.2f)" % roc_auc)
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.legend(loc="lower right")
plt.show()
# ROC curve and AUC provided a useful way to evaluate and compare the
# performance of binary classification of the model.
# ### Decision Tree
from sklearn.tree import DecisionTreeClassifier
import graphviz
from sklearn.metrics import accuracy_score
clf = DecisionTreeClassifier()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy)
# ### Random Forest
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
print("Accuracy:", accuracy)
# ### Naive Baye's
from sklearn.naive_bayes import GaussianNB
clf = GaussianNB()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
accuracy = clf.score(X_test, y_test)
print("Accuracy:", accuracy)
from sklearn.metrics import classification_report, confusion_matrix
print("Classification Report:\n", classification_report(y_test, y_pred))
print("Confusion Matrix:\n", confusion_matrix(y_test, y_pred))
sns.heatmap(confusion_matrix(y_test, y_pred), annot=True, cmap="Blues")
# ### K- Nearest Neighbours(KNN)
from sklearn.neighbors import KNeighborsClassifier
kclassifier = KNeighborsClassifier(n_neighbors=251)
kclassifier.fit(X_train, y_train)
kpredict = kclassifier.predict(X_test)
from sklearn.metrics import confusion_matrix, classification_report
cm = confusion_matrix(y_test, kpredict)
sns.heatmap(cm, annot=True)
print(classification_report(y_test, kpredict))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/820/129820956.ipynb
|
nasa-nearest-earth-objects
|
sameepvani
|
[{"Id": 129820956, "ScriptId": 38609611, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11154128, "CreationDate": "05/16/2023 17:53:40", "VersionNumber": 1.0, "Title": "notebookaba04a48d0", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 210.0, "LinesInsertedFromPrevious": 210.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186197908, "KernelVersionId": 129820956, "SourceDatasetVersionId": 3816674}]
|
[{"Id": 3816674, "DatasetId": 2272878, "DatasourceVersionId": 3871445, "CreatorUserId": 8299318, "LicenseName": "CC0: Public Domain", "CreationDate": "06/17/2022 02:32:18", "VersionNumber": 2.0, "Title": "NASA - Nearest Earth Objects", "Slug": "nasa-nearest-earth-objects", "Subtitle": "A cumulative data for Nearest Earth Objects by NASA", "Description": "# Context\nThere is an infinite number of objects in the outer space. Some of them are closer than we think. Even though we might think that a distance of 70,000 Km can not potentially harm us, but at an astronomical scale, this is a very small distance and can disrupt many natural phenomena. These objects/asteroids can thus prove to be harmful. Hence, it is wise to know what is surrounding us and what can harm us amongst those. Thus, this dataset compiles the list of NASA certified asteroids that are classified as the nearest earth object. \n\n# Sources\n[NASA Open API](https://api.nasa.gov/)\n[NEO Earth Close Approaches](https://cneos.jpl.nasa.gov/ca/)", "VersionNotes": "Bug fix try: Same upload", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2272878, "CreatorUserId": 8299318, "OwnerUserId": 8299318.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3816674.0, "CurrentDatasourceVersionId": 3871445.0, "ForumId": 2299403, "Type": 2, "CreationDate": "06/16/2022 16:39:26", "LastActivityDate": "06/16/2022", "TotalViews": 46484, "TotalDownloads": 5646, "TotalVotes": 187, "TotalKernels": 75}]
|
[{"Id": 8299318, "UserName": "sameepvani", "DisplayName": "Sameep Vani", "RegisterDate": "09/07/2021", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## NASA - Nearest Earth Objects
# #### A cumulative data for Nearest Earth Objects by NASA
# ### CONTEXT
# There is an infinite number of objects in the outer space.
# Some of them are closer than we think. Even though we might
# think that a distance of 70,000 Km can’t potentially harm us,
# but at an astronomical scale, this is a very small distance
# and can disrupt many natural phenomena. These objects/asteroids
# can thus prove to be harmful. Hence, it is wise to know what is
# surrounding us and what can harm us amongst those. Thus, this
# dataset compiles the list of NASA certified asteroids that are c
# lassified as the nearest earth object.
# ### ABSTRACT OF THE PROBLEM:
# Sentry is a highly automated collision monitoring system that continually scans the most current asteroid catalogue for possibilities of future impact with Earth over the next 100 years. So, this concludes whether that object is hazardous for the earth are not.
#
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
neo = pd.read_csv(r"/kaggle/input/nasa-nearest-earth-objects/neo.csv")
neo.head()
# ### DATA PREPROCESSING
# #### Checking for null values
neo.isnull().sum()
# #### Checking the data types of the data set
neo.dtypes
# #### Changing the bool values into Binary 0 or 1
# #### True = 1, False = 0
neo["hazardous"] = neo["hazardous"].astype(int)
print(neo["hazardous"].head())
neo.columns
# #### Getting unique values for the each column in a dataset
unique_values = neo.nunique()
print(unique_values)
# ### Oribiting body : Earth
# ### Sentry_object : False
# #### so changing the value into 1
neo["sentry_object"] = neo["sentry_object"].astype(int)
print(neo["sentry_object"].head())
def my_func(row):
if row["orbiting_body"] == "Earth":
val = 1
else:
val = 0
return val
neo["orbiting_body"] = neo.apply(my_func, axis=1)
neo["orbiting_body"].head()
# Checking for the data types
neo.dtypes
# ### Multiple Linear Regression
# We use Multiple linear regression model to estimate the relationship
# between a quantitative dependent variable ['hazardous'] and
# independent variables
# ['est_diameter_min', 'est_diameter_max', 'relative_velocity',
# 'miss_distance', 'orbiting_body', 'sentry_object', 'absolute_magnitude']
# using a straight line.
X = neo.iloc[:, 2:9]
y = neo["hazardous"]
# train_test_split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
print("X train shape:", X_train.shape)
print("y_train shape", y_train.shape)
print("X_test shape:", X_test.shape)
print("y_test shape", y_test.shape)
# Multiple Linear Regression
from sklearn.linear_model import LinearRegression
regressor = LinearRegression()
regressor.fit(X_train, y_train)
print("Regression co-efficient:", regressor.coef_)
print()
print("Regressor intercept:", regressor.intercept_)
print()
y_pred = regressor.predict(X_test)
print("Predicted values:", y_pred)
print()
print("Actual values:", y_test.values)
sns.pairplot(
neo,
x_vars=[
"est_diameter_min",
"est_diameter_max",
"relative_velocity",
"miss_distance",
"orbiting_body",
"sentry_object",
"absolute_magnitude",
],
y_vars="hazardous",
kind="reg",
)
# The argument kind='reg' adds a linear regression line to each scatter
# plot, allowing us to visualize the relationship between the independent
# and dependent variables.
# #### Dimensionality Reduction
import statsmodels.regression.linear_model as sm
# add a column of ones as integer data type
X = np.append(arr=np.ones((90836, 1)).astype(int), values=X, axis=1)
# choose a Significance level usually 0.05, if p>0.05
# for the highest values parameter, remove that value
X_opt = X[:, [0, 3, 4, 5]]
ols = sm.OLS(endog=y, exog=X_opt).fit()
ols.summary()
sns.pairplot(
neo,
x_vars=["est_diameter_min", "est_diameter_max", "relative_velocity"],
y_vars="hazardous",
kind="reg",
)
# ### Logistic Regression
from sklearn.linear_model import LogisticRegression
logreg = LogisticRegression()
logreg.fit(X_train, y_train)
y_pred = logreg.predict(X_test)
print("Accuracy:", logreg.score(X_test, y_test))
from sklearn.metrics import confusion_matrix, roc_curve, roc_auc_score
fpr, tpr, thresholds = roc_curve(y_test, logreg.predict_proba(X_test)[:, 1])
roc_auc = roc_auc_score(y_test, y_pred)
plt.plot(fpr, tpr, label="ROC Curve( area = %0.2f)" % roc_auc)
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.legend(loc="lower right")
plt.show()
# ROC curve and AUC provided a useful way to evaluate and compare the
# performance of binary classification of the model.
# ### Decision Tree
from sklearn.tree import DecisionTreeClassifier
import graphviz
from sklearn.metrics import accuracy_score
clf = DecisionTreeClassifier()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy)
# ### Random Forest
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
print("Accuracy:", accuracy)
# ### Naive Baye's
from sklearn.naive_bayes import GaussianNB
clf = GaussianNB()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
accuracy = clf.score(X_test, y_test)
print("Accuracy:", accuracy)
from sklearn.metrics import classification_report, confusion_matrix
print("Classification Report:\n", classification_report(y_test, y_pred))
print("Confusion Matrix:\n", confusion_matrix(y_test, y_pred))
sns.heatmap(confusion_matrix(y_test, y_pred), annot=True, cmap="Blues")
# ### K- Nearest Neighbours(KNN)
from sklearn.neighbors import KNeighborsClassifier
kclassifier = KNeighborsClassifier(n_neighbors=251)
kclassifier.fit(X_train, y_train)
kpredict = kclassifier.predict(X_test)
from sklearn.metrics import confusion_matrix, classification_report
cm = confusion_matrix(y_test, kpredict)
sns.heatmap(cm, annot=True)
print(classification_report(y_test, kpredict))
|
[{"nasa-nearest-earth-objects/neo.csv": {"column_names": "[\"id\", \"name\", \"est_diameter_min\", \"est_diameter_max\", \"relative_velocity\", \"miss_distance\", \"orbiting_body\", \"sentry_object\", \"absolute_magnitude\", \"hazardous\"]", "column_data_types": "{\"id\": \"int64\", \"name\": \"object\", \"est_diameter_min\": \"float64\", \"est_diameter_max\": \"float64\", \"relative_velocity\": \"float64\", \"miss_distance\": \"float64\", \"orbiting_body\": \"object\", \"sentry_object\": \"bool\", \"absolute_magnitude\": \"float64\", \"hazardous\": \"bool\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 90836 entries, 0 to 90835\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 id 90836 non-null int64 \n 1 name 90836 non-null object \n 2 est_diameter_min 90836 non-null float64\n 3 est_diameter_max 90836 non-null float64\n 4 relative_velocity 90836 non-null float64\n 5 miss_distance 90836 non-null float64\n 6 orbiting_body 90836 non-null object \n 7 sentry_object 90836 non-null bool \n 8 absolute_magnitude 90836 non-null float64\n 9 hazardous 90836 non-null bool \ndtypes: bool(2), float64(5), int64(1), object(2)\nmemory usage: 5.7+ MB\n", "summary": "{\"id\": {\"count\": 90836.0, \"mean\": 14382878.052159937, \"std\": 20872018.35389277, \"min\": 2000433.0, \"25%\": 3448110.0, \"50%\": 3748362.0, \"75%\": 3884023.0, \"max\": 54275914.0}, \"est_diameter_min\": {\"count\": 90836.0, \"mean\": 0.12743210631522306, \"std\": 0.2985112435285911, \"min\": 0.0006089126, \"25%\": 0.0192555078, \"50%\": 0.0483676488, \"75%\": 0.1434019235, \"max\": 37.8926498379}, \"est_diameter_max\": {\"count\": 90836.0, \"mean\": 0.2849468522354066, \"std\": 0.6674914325765064, \"min\": 0.00136157, \"25%\": 0.0430566244, \"50%\": 0.1081533507, \"75%\": 0.320656449, \"max\": 84.7305408852}, \"relative_velocity\": {\"count\": 90836.0, \"mean\": 48066.918917642935, \"std\": 25293.296960671203, \"min\": 203.34643253, \"25%\": 28619.02064490995, \"50%\": 44190.117890331996, \"75%\": 62923.60463276395, \"max\": 236990.1280878666}, \"miss_distance\": {\"count\": 90836.0, \"mean\": 37066546.03042213, \"std\": 22352040.599189546, \"min\": 6745.532515957, \"25%\": 17210820.23576468, \"50%\": 37846579.263426416, \"75%\": 56548996.45139917, \"max\": 74798651.4521972}, \"absolute_magnitude\": {\"count\": 90836.0, \"mean\": 23.52710347219164, \"std\": 2.89408550693204, \"min\": 9.23, \"25%\": 21.34, \"50%\": 23.7, \"75%\": 25.7, \"max\": 33.2}}", "examples": "{\"id\":{\"0\":2162635,\"1\":2277475,\"2\":2512244,\"3\":3596030},\"name\":{\"0\":\"162635 (2000 SS164)\",\"1\":\"277475 (2005 WK4)\",\"2\":\"512244 (2015 YE18)\",\"3\":\"(2012 BV13)\"},\"est_diameter_min\":{\"0\":1.1982708007,\"1\":0.2658,\"2\":0.7220295577,\"3\":0.096506147},\"est_diameter_max\":{\"0\":2.6794149658,\"1\":0.5943468684,\"2\":1.6145071727,\"3\":0.2157943048},\"relative_velocity\":{\"0\":13569.2492241812,\"1\":73588.7266634981,\"2\":114258.6921290512,\"3\":24764.3031380016},\"miss_distance\":{\"0\":54839744.0828460529,\"1\":61438126.523950927,\"2\":49798724.9404567927,\"3\":25434972.7207582518},\"orbiting_body\":{\"0\":\"Earth\",\"1\":\"Earth\",\"2\":\"Earth\",\"3\":\"Earth\"},\"sentry_object\":{\"0\":false,\"1\":false,\"2\":false,\"3\":false},\"absolute_magnitude\":{\"0\":16.73,\"1\":20.0,\"2\":17.83,\"3\":22.2},\"hazardous\":{\"0\":false,\"1\":true,\"2\":false,\"3\":false}}"}}]
| true | 1 |
<start_data_description><data_path>nasa-nearest-earth-objects/neo.csv:
<column_names>
['id', 'name', 'est_diameter_min', 'est_diameter_max', 'relative_velocity', 'miss_distance', 'orbiting_body', 'sentry_object', 'absolute_magnitude', 'hazardous']
<column_types>
{'id': 'int64', 'name': 'object', 'est_diameter_min': 'float64', 'est_diameter_max': 'float64', 'relative_velocity': 'float64', 'miss_distance': 'float64', 'orbiting_body': 'object', 'sentry_object': 'bool', 'absolute_magnitude': 'float64', 'hazardous': 'bool'}
<dataframe_Summary>
{'id': {'count': 90836.0, 'mean': 14382878.052159937, 'std': 20872018.35389277, 'min': 2000433.0, '25%': 3448110.0, '50%': 3748362.0, '75%': 3884023.0, 'max': 54275914.0}, 'est_diameter_min': {'count': 90836.0, 'mean': 0.12743210631522306, 'std': 0.2985112435285911, 'min': 0.0006089126, '25%': 0.0192555078, '50%': 0.0483676488, '75%': 0.1434019235, 'max': 37.8926498379}, 'est_diameter_max': {'count': 90836.0, 'mean': 0.2849468522354066, 'std': 0.6674914325765064, 'min': 0.00136157, '25%': 0.0430566244, '50%': 0.1081533507, '75%': 0.320656449, 'max': 84.7305408852}, 'relative_velocity': {'count': 90836.0, 'mean': 48066.918917642935, 'std': 25293.296960671203, 'min': 203.34643253, '25%': 28619.02064490995, '50%': 44190.117890331996, '75%': 62923.60463276395, 'max': 236990.1280878666}, 'miss_distance': {'count': 90836.0, 'mean': 37066546.03042213, 'std': 22352040.599189546, 'min': 6745.532515957, '25%': 17210820.23576468, '50%': 37846579.263426416, '75%': 56548996.45139917, 'max': 74798651.4521972}, 'absolute_magnitude': {'count': 90836.0, 'mean': 23.52710347219164, 'std': 2.89408550693204, 'min': 9.23, '25%': 21.34, '50%': 23.7, '75%': 25.7, 'max': 33.2}}
<dataframe_info>
RangeIndex: 90836 entries, 0 to 90835
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 90836 non-null int64
1 name 90836 non-null object
2 est_diameter_min 90836 non-null float64
3 est_diameter_max 90836 non-null float64
4 relative_velocity 90836 non-null float64
5 miss_distance 90836 non-null float64
6 orbiting_body 90836 non-null object
7 sentry_object 90836 non-null bool
8 absolute_magnitude 90836 non-null float64
9 hazardous 90836 non-null bool
dtypes: bool(2), float64(5), int64(1), object(2)
memory usage: 5.7+ MB
<some_examples>
{'id': {'0': 2162635, '1': 2277475, '2': 2512244, '3': 3596030}, 'name': {'0': '162635 (2000 SS164)', '1': '277475 (2005 WK4)', '2': '512244 (2015 YE18)', '3': '(2012 BV13)'}, 'est_diameter_min': {'0': 1.1982708007, '1': 0.2658, '2': 0.7220295577, '3': 0.096506147}, 'est_diameter_max': {'0': 2.6794149658, '1': 0.5943468684, '2': 1.6145071727, '3': 0.2157943048}, 'relative_velocity': {'0': 13569.2492241812, '1': 73588.7266634981, '2': 114258.6921290512, '3': 24764.3031380016}, 'miss_distance': {'0': 54839744.08284605, '1': 61438126.52395093, '2': 49798724.94045679, '3': 25434972.72075825}, 'orbiting_body': {'0': 'Earth', '1': 'Earth', '2': 'Earth', '3': 'Earth'}, 'sentry_object': {'0': False, '1': False, '2': False, '3': False}, 'absolute_magnitude': {'0': 16.73, '1': 20.0, '2': 17.83, '3': 22.2}, 'hazardous': {'0': False, '1': True, '2': False, '3': False}}
<end_description>
| 2,054 | 0 | 3,258 | 2,054 |
129820179
|
<jupyter_start><jupyter_text>Personal Key Indicators of Heart Disease
# Key Indicators of Heart Disease
## 2020 annual CDC survey data of 400k adults related to their health status
### What topic does the dataset cover?
According to the [CDC](https://www.cdc.gov/heartdisease/risk_factors.htm), heart disease is one of the leading causes of death for people of most races in the US (African Americans, American Indians and Alaska Natives, and white people). About half of all Americans (47%) have at least 1 of 3 key risk factors for heart disease: high blood pressure, high cholesterol, and smoking. Other key indicator include diabetic status, obesity (high BMI), not getting enough physical activity or drinking too much alcohol. Detecting and preventing the factors that have the greatest impact on heart disease is very important in healthcare. Computational developments, in turn, allow the application of machine learning methods to detect "patterns" from the data that can predict a patient's condition.
### Where did the dataset come from and what treatments did it undergo?
Originally, the dataset come from the CDC and is a major part of the Behavioral Risk Factor Surveillance System (BRFSS), which conducts annual telephone surveys to gather data on the health status of U.S. residents. As the [CDC](https://www.cdc.gov/heartdisease/risk_factors.htm) describes: "Established in 1984 with 15 states, BRFSS now collects data in all 50 states as well as the District of Columbia and three U.S. territories. BRFSS completes more than 400,000 adult interviews each year, making it the largest continuously conducted health survey system in the world.". The most recent dataset (as of February 15, 2022) includes data from 2020. It consists of 401,958 rows and 279 columns. The vast majority of columns are questions asked to respondents about their health status, such as "Do you have serious difficulty walking or climbing stairs?" or "Have you smoked at least 100 cigarettes in your entire life? [Note: 5 packs = 100 cigarettes]". In this dataset, I noticed many different factors (questions) that directly or indirectly influence heart disease, so I decided to select the most relevant variables from it and do some cleaning so that it would be usable for machine learning projects.
### What can you do with this dataset?
As described above, the original dataset of nearly 300 variables was reduced to just about 20 variables. In addition to classical EDA, this dataset can be used to apply a range of machine learning methods, most notably classifier models (logistic regression, SVM, random forest, etc.). You should treat the variable "HeartDisease" as a binary ("Yes" - respondent had heart disease; "No" - respondent had no heart disease). But note that classes are not balanced, so the classic model application approach is not advisable. Fixing the weights/undersampling should yield significantly betters results. Based on the dataset, I constructed a logistic regression model and embedded it in an application you might be inspired by: https://share.streamlit.io/kamilpytlak/heart-condition-checker/main/app.py. Can you indicate which variables have a significant effect on the likelihood of heart disease?
Kaggle dataset identifier: personal-key-indicators-of-heart-disease
<jupyter_code>import pandas as pd
df = pd.read_csv('personal-key-indicators-of-heart-disease/heart_2020_cleaned.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 319795 entries, 0 to 319794
Data columns (total 18 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 HeartDisease 319795 non-null object
1 BMI 319795 non-null float64
2 Smoking 319795 non-null object
3 AlcoholDrinking 319795 non-null object
4 Stroke 319795 non-null object
5 PhysicalHealth 319795 non-null float64
6 MentalHealth 319795 non-null float64
7 DiffWalking 319795 non-null object
8 Sex 319795 non-null object
9 AgeCategory 319795 non-null object
10 Race 319795 non-null object
11 Diabetic 319795 non-null object
12 PhysicalActivity 319795 non-null object
13 GenHealth 319795 non-null object
14 SleepTime 319795 non-null float64
15 Asthma 319795 non-null object
16 KidneyDisease 319795 non-null object
17 SkinCancer 319795 non-null object
dtypes: float64(4), object(14)
memory usage: 43.9+ MB
<jupyter_text>Examples:
{
"HeartDisease": "No",
"BMI": 16.6,
"Smoking": "Yes",
"AlcoholDrinking": "No",
"Stroke": "No",
"PhysicalHealth": 3,
"MentalHealth": 30,
"DiffWalking": "No",
"Sex": "Female",
"AgeCategory": "55-59",
"Race": "White",
"Diabetic": "Yes",
"PhysicalActivity": "Yes",
"GenHealth": "Very good",
"SleepTime": 5,
"Asthma": "Yes",
"KidneyDisease": "No",
"SkinCancer": "Yes"
}
{
"HeartDisease": "No",
"BMI": 20.34,
"Smoking": "No",
"AlcoholDrinking": "No",
"Stroke": "Yes",
"PhysicalHealth": 0,
"MentalHealth": 0,
"DiffWalking": "No",
"Sex": "Female",
"AgeCategory": "80 or older",
"Race": "White",
"Diabetic": "No",
"PhysicalActivity": "Yes",
"GenHealth": "Very good",
"SleepTime": 7,
"Asthma": "No",
"KidneyDisease": "No",
"SkinCancer": "No"
}
{
"HeartDisease": "No",
"BMI": 26.58,
"Smoking": "Yes",
"AlcoholDrinking": "No",
"Stroke": "No",
"PhysicalHealth": 20,
"MentalHealth": 30,
"DiffWalking": "No",
"Sex": "Male",
"AgeCategory": "65-69",
"Race": "White",
"Diabetic": "Yes",
"PhysicalActivity": "Yes",
"GenHealth": "Fair",
"SleepTime": 8,
"Asthma": "Yes",
"KidneyDisease": "No",
"SkinCancer": "No"
}
{
"HeartDisease": "No",
"BMI": 24.21,
"Smoking": "No",
"AlcoholDrinking": "No",
"Stroke": "No",
"PhysicalHealth": 0,
"MentalHealth": 0,
"DiffWalking": "No",
"Sex": "Female",
"AgeCategory": "75-79",
"Race": "White",
"Diabetic": "No",
"PhysicalActivity": "No",
"GenHealth": "Good",
"SleepTime": 6,
"Asthma": "No",
"KidneyDisease": "No",
"SkinCancer": "Yes"
}
<jupyter_script># Heart disease Analysis
# Anaylzing key data from sample patients to better understand the risk factors for heart disease
import pandas as pd
import matplotlib.pyplot as plt
def import_heartdisease():
return pd.read_csv(
"/kaggle/input/personal-key-indicators-of-heart-disease/heart_2020_cleaned.csv"
)
heartdisease = import_heartdisease()
heartdisease.shape
heartdisease.describe()
# Majority of the population is classified as overwieght tettering on the edge of obese
heartdisease.notnull()
heartdisease.isnull().sum()
# # No missing data, sweet!
heartdisease.dtypes
BMI = heartdisease[["HeartDisease", "BMI"]]
BMI
BMI.drop(BMI[BMI["HeartDisease"] == "No"].index)
BMI
BMI.plot(kind="hist", bins=20)
plt.ylabel("Frequency of heart disease")
plt.xlabel("BMI")
plt.title("Number of cases of heart disease for number of BMI")
# Based on the above chart is seems the frequency of heart disease becomes more common with a BMI greater than 20
BMI6 = heartdisease[["HeartDisease", "BMI"]]
BMI6
BMI6.drop(BMI6[BMI6["BMI"] <= 20].index, inplace=True)
BMI6
BMI6["HeartDisease"].value_counts()
BMI6["HeartDisease"].value_counts().plot(
kind="bar", xlabel="numbers", ylabel="frequency"
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/820/129820179.ipynb
|
personal-key-indicators-of-heart-disease
|
kamilpytlak
|
[{"Id": 129820179, "ScriptId": 38135983, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14086290, "CreationDate": "05/16/2023 17:45:28", "VersionNumber": 8.0, "Title": "Final_Project", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 52.0, "LinesInsertedFromPrevious": 9.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 43.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186196912, "KernelVersionId": 129820179, "SourceDatasetVersionId": 3191579}]
|
[{"Id": 3191579, "DatasetId": 1936563, "DatasourceVersionId": 3241234, "CreatorUserId": 9492796, "LicenseName": "CC0: Public Domain", "CreationDate": "02/16/2022 10:18:03", "VersionNumber": 2.0, "Title": "Personal Key Indicators of Heart Disease", "Slug": "personal-key-indicators-of-heart-disease", "Subtitle": "2020 annual CDC survey data of 400k adults related to their health status", "Description": "# Key Indicators of Heart Disease\n## 2020 annual CDC survey data of 400k adults related to their health status\n\n### What topic does the dataset cover?\nAccording to the [CDC](https://www.cdc.gov/heartdisease/risk_factors.htm), heart disease is one of the leading causes of death for people of most races in the US (African Americans, American Indians and Alaska Natives, and white people). About half of all Americans (47%) have at least 1 of 3 key risk factors for heart disease: high blood pressure, high cholesterol, and smoking. Other key indicator include diabetic status, obesity (high BMI), not getting enough physical activity or drinking too much alcohol. Detecting and preventing the factors that have the greatest impact on heart disease is very important in healthcare. Computational developments, in turn, allow the application of machine learning methods to detect \"patterns\" from the data that can predict a patient's condition.\n\n### Where did the dataset come from and what treatments did it undergo?\nOriginally, the dataset come from the CDC and is a major part of the Behavioral Risk Factor Surveillance System (BRFSS), which conducts annual telephone surveys to gather data on the health status of U.S. residents. As the [CDC](https://www.cdc.gov/heartdisease/risk_factors.htm) describes: \"Established in 1984 with 15 states, BRFSS now collects data in all 50 states as well as the District of Columbia and three U.S. territories. BRFSS completes more than 400,000 adult interviews each year, making it the largest continuously conducted health survey system in the world.\". The most recent dataset (as of February 15, 2022) includes data from 2020. It consists of 401,958 rows and 279 columns. The vast majority of columns are questions asked to respondents about their health status, such as \"Do you have serious difficulty walking or climbing stairs?\" or \"Have you smoked at least 100 cigarettes in your entire life? [Note: 5 packs = 100 cigarettes]\". In this dataset, I noticed many different factors (questions) that directly or indirectly influence heart disease, so I decided to select the most relevant variables from it and do some cleaning so that it would be usable for machine learning projects.\n\n### What can you do with this dataset?\nAs described above, the original dataset of nearly 300 variables was reduced to just about 20 variables. In addition to classical EDA, this dataset can be used to apply a range of machine learning methods, most notably classifier models (logistic regression, SVM, random forest, etc.). You should treat the variable \"HeartDisease\" as a binary (\"Yes\" - respondent had heart disease; \"No\" - respondent had no heart disease). But note that classes are not balanced, so the classic model application approach is not advisable. Fixing the weights/undersampling should yield significantly betters results. Based on the dataset, I constructed a logistic regression model and embedded it in an application you might be inspired by: https://share.streamlit.io/kamilpytlak/heart-condition-checker/main/app.py. Can you indicate which variables have a significant effect on the likelihood of heart disease?", "VersionNotes": "Data Update 2022/02/16", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1936563, "CreatorUserId": 9492796, "OwnerUserId": 9492796.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3191579.0, "CurrentDatasourceVersionId": 3241234.0, "ForumId": 1960316, "Type": 2, "CreationDate": "02/15/2022 19:28:49", "LastActivityDate": "02/15/2022", "TotalViews": 320603, "TotalDownloads": 46135, "TotalVotes": 694, "TotalKernels": 186}]
|
[{"Id": 9492796, "UserName": "kamilpytlak", "DisplayName": "Kamil Pytlak", "RegisterDate": "01/25/2022", "PerformanceTier": 1}]
|
# Heart disease Analysis
# Anaylzing key data from sample patients to better understand the risk factors for heart disease
import pandas as pd
import matplotlib.pyplot as plt
def import_heartdisease():
return pd.read_csv(
"/kaggle/input/personal-key-indicators-of-heart-disease/heart_2020_cleaned.csv"
)
heartdisease = import_heartdisease()
heartdisease.shape
heartdisease.describe()
# Majority of the population is classified as overwieght tettering on the edge of obese
heartdisease.notnull()
heartdisease.isnull().sum()
# # No missing data, sweet!
heartdisease.dtypes
BMI = heartdisease[["HeartDisease", "BMI"]]
BMI
BMI.drop(BMI[BMI["HeartDisease"] == "No"].index)
BMI
BMI.plot(kind="hist", bins=20)
plt.ylabel("Frequency of heart disease")
plt.xlabel("BMI")
plt.title("Number of cases of heart disease for number of BMI")
# Based on the above chart is seems the frequency of heart disease becomes more common with a BMI greater than 20
BMI6 = heartdisease[["HeartDisease", "BMI"]]
BMI6
BMI6.drop(BMI6[BMI6["BMI"] <= 20].index, inplace=True)
BMI6
BMI6["HeartDisease"].value_counts()
BMI6["HeartDisease"].value_counts().plot(
kind="bar", xlabel="numbers", ylabel="frequency"
)
|
[{"personal-key-indicators-of-heart-disease/heart_2020_cleaned.csv": {"column_names": "[\"HeartDisease\", \"BMI\", \"Smoking\", \"AlcoholDrinking\", \"Stroke\", \"PhysicalHealth\", \"MentalHealth\", \"DiffWalking\", \"Sex\", \"AgeCategory\", \"Race\", \"Diabetic\", \"PhysicalActivity\", \"GenHealth\", \"SleepTime\", \"Asthma\", \"KidneyDisease\", \"SkinCancer\"]", "column_data_types": "{\"HeartDisease\": \"object\", \"BMI\": \"float64\", \"Smoking\": \"object\", \"AlcoholDrinking\": \"object\", \"Stroke\": \"object\", \"PhysicalHealth\": \"float64\", \"MentalHealth\": \"float64\", \"DiffWalking\": \"object\", \"Sex\": \"object\", \"AgeCategory\": \"object\", \"Race\": \"object\", \"Diabetic\": \"object\", \"PhysicalActivity\": \"object\", \"GenHealth\": \"object\", \"SleepTime\": \"float64\", \"Asthma\": \"object\", \"KidneyDisease\": \"object\", \"SkinCancer\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 319795 entries, 0 to 319794\nData columns (total 18 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 HeartDisease 319795 non-null object \n 1 BMI 319795 non-null float64\n 2 Smoking 319795 non-null object \n 3 AlcoholDrinking 319795 non-null object \n 4 Stroke 319795 non-null object \n 5 PhysicalHealth 319795 non-null float64\n 6 MentalHealth 319795 non-null float64\n 7 DiffWalking 319795 non-null object \n 8 Sex 319795 non-null object \n 9 AgeCategory 319795 non-null object \n 10 Race 319795 non-null object \n 11 Diabetic 319795 non-null object \n 12 PhysicalActivity 319795 non-null object \n 13 GenHealth 319795 non-null object \n 14 SleepTime 319795 non-null float64\n 15 Asthma 319795 non-null object \n 16 KidneyDisease 319795 non-null object \n 17 SkinCancer 319795 non-null object \ndtypes: float64(4), object(14)\nmemory usage: 43.9+ MB\n", "summary": "{\"BMI\": {\"count\": 319795.0, \"mean\": 28.325398520927465, \"std\": 6.356100200470739, \"min\": 12.02, \"25%\": 24.03, \"50%\": 27.34, \"75%\": 31.42, \"max\": 94.85}, \"PhysicalHealth\": {\"count\": 319795.0, \"mean\": 3.3717100017198516, \"std\": 7.950850182571368, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 2.0, \"max\": 30.0}, \"MentalHealth\": {\"count\": 319795.0, \"mean\": 3.898366140808956, \"std\": 7.955235218943607, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 3.0, \"max\": 30.0}, \"SleepTime\": {\"count\": 319795.0, \"mean\": 7.097074688472302, \"std\": 1.4360070609642825, \"min\": 1.0, \"25%\": 6.0, \"50%\": 7.0, \"75%\": 8.0, \"max\": 24.0}}", "examples": "{\"HeartDisease\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"BMI\":{\"0\":16.6,\"1\":20.34,\"2\":26.58,\"3\":24.21},\"Smoking\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"Yes\",\"3\":\"No\"},\"AlcoholDrinking\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"Stroke\":{\"0\":\"No\",\"1\":\"Yes\",\"2\":\"No\",\"3\":\"No\"},\"PhysicalHealth\":{\"0\":3.0,\"1\":0.0,\"2\":20.0,\"3\":0.0},\"MentalHealth\":{\"0\":30.0,\"1\":0.0,\"2\":30.0,\"3\":0.0},\"DiffWalking\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"Sex\":{\"0\":\"Female\",\"1\":\"Female\",\"2\":\"Male\",\"3\":\"Female\"},\"AgeCategory\":{\"0\":\"55-59\",\"1\":\"80 or older\",\"2\":\"65-69\",\"3\":\"75-79\"},\"Race\":{\"0\":\"White\",\"1\":\"White\",\"2\":\"White\",\"3\":\"White\"},\"Diabetic\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"Yes\",\"3\":\"No\"},\"PhysicalActivity\":{\"0\":\"Yes\",\"1\":\"Yes\",\"2\":\"Yes\",\"3\":\"No\"},\"GenHealth\":{\"0\":\"Very good\",\"1\":\"Very good\",\"2\":\"Fair\",\"3\":\"Good\"},\"SleepTime\":{\"0\":5.0,\"1\":7.0,\"2\":8.0,\"3\":6.0},\"Asthma\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"Yes\",\"3\":\"No\"},\"KidneyDisease\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"SkinCancer\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"No\",\"3\":\"Yes\"}}"}}]
| true | 1 |
<start_data_description><data_path>personal-key-indicators-of-heart-disease/heart_2020_cleaned.csv:
<column_names>
['HeartDisease', 'BMI', 'Smoking', 'AlcoholDrinking', 'Stroke', 'PhysicalHealth', 'MentalHealth', 'DiffWalking', 'Sex', 'AgeCategory', 'Race', 'Diabetic', 'PhysicalActivity', 'GenHealth', 'SleepTime', 'Asthma', 'KidneyDisease', 'SkinCancer']
<column_types>
{'HeartDisease': 'object', 'BMI': 'float64', 'Smoking': 'object', 'AlcoholDrinking': 'object', 'Stroke': 'object', 'PhysicalHealth': 'float64', 'MentalHealth': 'float64', 'DiffWalking': 'object', 'Sex': 'object', 'AgeCategory': 'object', 'Race': 'object', 'Diabetic': 'object', 'PhysicalActivity': 'object', 'GenHealth': 'object', 'SleepTime': 'float64', 'Asthma': 'object', 'KidneyDisease': 'object', 'SkinCancer': 'object'}
<dataframe_Summary>
{'BMI': {'count': 319795.0, 'mean': 28.325398520927465, 'std': 6.356100200470739, 'min': 12.02, '25%': 24.03, '50%': 27.34, '75%': 31.42, 'max': 94.85}, 'PhysicalHealth': {'count': 319795.0, 'mean': 3.3717100017198516, 'std': 7.950850182571368, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 2.0, 'max': 30.0}, 'MentalHealth': {'count': 319795.0, 'mean': 3.898366140808956, 'std': 7.955235218943607, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 3.0, 'max': 30.0}, 'SleepTime': {'count': 319795.0, 'mean': 7.097074688472302, 'std': 1.4360070609642825, 'min': 1.0, '25%': 6.0, '50%': 7.0, '75%': 8.0, 'max': 24.0}}
<dataframe_info>
RangeIndex: 319795 entries, 0 to 319794
Data columns (total 18 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 HeartDisease 319795 non-null object
1 BMI 319795 non-null float64
2 Smoking 319795 non-null object
3 AlcoholDrinking 319795 non-null object
4 Stroke 319795 non-null object
5 PhysicalHealth 319795 non-null float64
6 MentalHealth 319795 non-null float64
7 DiffWalking 319795 non-null object
8 Sex 319795 non-null object
9 AgeCategory 319795 non-null object
10 Race 319795 non-null object
11 Diabetic 319795 non-null object
12 PhysicalActivity 319795 non-null object
13 GenHealth 319795 non-null object
14 SleepTime 319795 non-null float64
15 Asthma 319795 non-null object
16 KidneyDisease 319795 non-null object
17 SkinCancer 319795 non-null object
dtypes: float64(4), object(14)
memory usage: 43.9+ MB
<some_examples>
{'HeartDisease': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'BMI': {'0': 16.6, '1': 20.34, '2': 26.58, '3': 24.21}, 'Smoking': {'0': 'Yes', '1': 'No', '2': 'Yes', '3': 'No'}, 'AlcoholDrinking': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'Stroke': {'0': 'No', '1': 'Yes', '2': 'No', '3': 'No'}, 'PhysicalHealth': {'0': 3.0, '1': 0.0, '2': 20.0, '3': 0.0}, 'MentalHealth': {'0': 30.0, '1': 0.0, '2': 30.0, '3': 0.0}, 'DiffWalking': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'Sex': {'0': 'Female', '1': 'Female', '2': 'Male', '3': 'Female'}, 'AgeCategory': {'0': '55-59', '1': '80 or older', '2': '65-69', '3': '75-79'}, 'Race': {'0': 'White', '1': 'White', '2': 'White', '3': 'White'}, 'Diabetic': {'0': 'Yes', '1': 'No', '2': 'Yes', '3': 'No'}, 'PhysicalActivity': {'0': 'Yes', '1': 'Yes', '2': 'Yes', '3': 'No'}, 'GenHealth': {'0': 'Very good', '1': 'Very good', '2': 'Fair', '3': 'Good'}, 'SleepTime': {'0': 5.0, '1': 7.0, '2': 8.0, '3': 6.0}, 'Asthma': {'0': 'Yes', '1': 'No', '2': 'Yes', '3': 'No'}, 'KidneyDisease': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'SkinCancer': {'0': 'Yes', '1': 'No', '2': 'No', '3': 'Yes'}}
<end_description>
| 395 | 0 | 2,378 | 395 |
129927236
|
<jupyter_start><jupyter_text>Flu Shot Learning
The data for this competition comes from the National 2009 H1N1 Flu Survey (NHFS).
In their own words:
> The National 2009 H1N1 Flu Survey (NHFS) was sponsored by the National Center for Immunization and Respiratory Diseases (NCIRD) and conducted jointly by NCIRD and the National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention (CDC). The NHFS was a list-assisted random-digit-dialing telephone survey of households, designed to monitor influenza immunization coverage in the 2009-10 season.
>
> The target population for the NHFS was all persons 6 months or older living in the United States at the time of the interview. Data from the NHFS were used to produce timely estimates of vaccination coverage rates for both the monovalent pH1N1 and trivalent seasonal influenza vaccines.
[National 2009 H1N1 Flu Survey Public-Use Data File Readme](ftp://ftp.cdc.gov/pub/Health_Statistics/NCHS/Datasets/nis/nhfs/nhfspuf_readme.txt)
The NHFS was conducted between October 2009 and June 2010. It was one-time survey designed specifically to monitor vaccination during the 2009-2010 flu season in response to the 2009 H1N1 pandemic. The CDC has other ongoing programs for annual phone surveys that continue to monitor seasonal flu vaccination.
## Data use restrictions
The source dataset comes with the following data use restrictions:
> The Public Health Service Act (Section 308(d)) provides that the data collected by the National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention (CDC), may be used only for the purpose of health statistical reporting and analysis.
>
> Any effort to determine the identity of any reported case is prohibited by this law.
>
> NCHS does all it can to ensure that the identity of data subjects cannot be disclosed. All direct identifiers, as well as any characteristics that might lead to identification, are omitted from the data files. Any intentional identification or disclosure of a person or establishment violates the assurances of confidentiality given to the providers of the information.
>
> Therefore, users will:
>
> 1. Use the data in these data files for statistical reporting and analysis only.
> 2. Make no use of the identity of any person or establishment discovered inadvertently and advise the Director, NCHS, of any such discovery (1 (800) 232-4636).
> 3. Not link these data files with individually identifiable data from other NCHS or non-NCHS data files.
>
> By using these data, you signify your agreement to comply with the above requirements.
[National 2009 H1N1 Flu Survey Public-Use Data File Readme](ftp://ftp.cdc.gov/pub/Health_Statistics/NCHS/Datasets/nis/nhfs/nhfspuf_readme.txt)
## Additional resources
- [U.S. National 2009 H1N1 Flu Survey (NHFS)](https://webarchive.loc.gov/all/20140511031000/http://www.cdc.gov/nchs/nis/about_nis.htm#h1n1)
- [U.S. National Immunization Surveys (NIS)](https://www.cdc.gov/vaccines/imz-managers/nis/about.html)
- [2009 H1N1 Pandemic (H1N1pdm09 virus)](https://www.cdc.gov/flu/pandemic-resources/2009-h1n1-pandemic.html), by the U.S. CDC
- [About Flu](https://www.cdc.gov/flu/about/index.html), by the U.S. CDC
- [Key Facts About Seasonal Flu Vaccine](https://www.cdc.gov/flu/prevent/keyfacts.htm), by the U.S. CDC
Data is provided courtesy of the United States [National Center for Health Statistics](https://www.cdc.gov/nchs/index.htm).
U.S. Department of Health and Human Services (DHHS). National Center for Health Statistics. The National 2009 H1N1 Flu Survey. Hyattsville, MD: Centers for Disease Control and Prevention, 2012.
Images courtesy of the [U.S. Navy](https://www.flickr.com/photos/navcent/15607260325/) and the [Fort Meade Public Affairs Office](https://www.flickr.com/photos/ftmeade/15242740638/) via Flickr under the [CC BY 2.0 license](https://creativecommons.org/licenses/by/2.0/legalcode).
The data for this competition comes from the National 2009 H1N1 Flu Survey (NHFS).
In their own words:
> The National 2009 H1N1 Flu Survey (NHFS) was sponsored by the National Center for Immunization and Respiratory Diseases (NCIRD) and conducted jointly by NCIRD and the National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention (CDC). The NHFS was a list-assisted random-digit-dialing telephone survey of households, designed to monitor influenza immunization coverage in the 2009-10 season.
>
> The target population for the NHFS was all persons 6 months or older living in the United States at the time of the interview. Data from the NHFS were used to produce timely estimates of vaccination coverage rates for both the monovalent pH1N1 and trivalent seasonal influenza vaccines.
[National 2009 H1N1 Flu Survey Public-Use Data File Readme](ftp://ftp.cdc.gov/pub/Health_Statistics/NCHS/Datasets/nis/nhfs/nhfspuf_readme.txt)
The NHFS was conducted between October 2009 and June 2010. It was one-time survey designed specifically to monitor vaccination during the 2009-2010 flu season in response to the 2009 H1N1 pandemic. The CDC has other ongoing programs for annual phone surveys that continue to monitor seasonal flu vaccination.
## Data use restrictions
The source dataset comes with the following data use restrictions:
> The Public Health Service Act (Section 308(d)) provides that the data collected by the National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention (CDC), may be used only for the purpose of health statistical reporting and analysis.
>
> Any effort to determine the identity of any reported case is prohibited by this law.
>
> NCHS does all it can to ensure that the identity of data subjects cannot be disclosed. All direct identifiers, as well as any characteristics that might lead to identification, are omitted from the data files. Any intentional identification or disclosure of a person or establishment violates the assurances of confidentiality given to the providers of the information.
>
> Therefore, users will:
>
> 1. Use the data in these data files for statistical reporting and analysis only.
> 2. Make no use of the identity of any person or establishment discovered inadvertently and advise the Director, NCHS, of any such discovery (1 (800) 232-4636).
> 3. Not link these data files with individually identifiable data from other NCHS or non-NCHS data files.
>
> By using these data, you signify your agreement to comply with the above requirements.
[National 2009 H1N1 Flu Survey Public-Use Data File Readme](ftp://ftp.cdc.gov/pub/Health_Statistics/NCHS/Datasets/nis/nhfs/nhfspuf_readme.txt)
## Additional resources
- [U.S. National 2009 H1N1 Flu Survey (NHFS)](https://webarchive.loc.gov/all/20140511031000/http://www.cdc.gov/nchs/nis/about_nis.htm#h1n1)
- [U.S. National Immunization Surveys (NIS)](https://www.cdc.gov/vaccines/imz-managers/nis/about.html)
- [2009 H1N1 Pandemic (H1N1pdm09 virus)](https://www.cdc.gov/flu/pandemic-resources/2009-h1n1-pandemic.html), by the U.S. CDC
- [About Flu](https://www.cdc.gov/flu/about/index.html), by the U.S. CDC
- [Key Facts About Seasonal Flu Vaccine](https://www.cdc.gov/flu/prevent/keyfacts.htm), by the U.S. CDC
Kaggle dataset identifier: flu-shot-learning
<jupyter_script># # **Data Analytics Competition FIND IT 2023 - Introduction**
# Swine flu, caused by the H1N1 flu virus, is a type of influenza A virus that can cause seasonal flu. Its symptoms are similar to those of seasonal flu. During the 2009-2010 flu season, the H1N1 virus caused a respiratory tract infection in humans known as swine flu, leading the WHO to declare it a pandemic. The H1N1 flu virus became one of the strains that cause seasonal flu after the pandemic ended.
# Flu vaccines are now available to protect against H1N1 flu. COVID-19 is also categorized as a seasonal flu disease and is delaying efforts to produce a vaccine for seasonal flu.
# The data set we are using represents individuals from the **2009 National H1N1 Flu Survey (NHFS)** conducted by the CDC.
# ## **Goals and Evaluation:**
# The goal of our analysis is to predict the likelihood of people getting H1N1 flu and receiving their yearly flu vaccine. We will specifically forecast two probabilities: one for the **H1N1 vaccine** and one for the **seasonal flu vaccine**.
# The performance of the model will be measured using the area under the receiver operating characteristic curve **(ROC AUC)** for each of the two target variables. The overall score will be calculated by taking the mean of these two scores. A **higher score** indicates better performance.
# **Made by BukanRISTEK**
# - Fathi Qushoyyi Ahimsa
# - Edward Salim
# - Muhammad Fakhri Robbani
# # **Load Libraries**
# Utilities
import pandas as pd
import numpy as np
pd.set_option(
"display.max_columns", None
) # Set the option to display all columns in pandas
# Google colab
from google.colab import drive
drive.mount("/content/drive") # Mount the Google Drive in Google Colab
# Data visualization
import matplotlib.pyplot as plt
import seaborn as sns
# Feature engineering
from sklearn.feature_selection import (
mutual_info_classif as mic,
) # Import mutual_info_classif for feature selection
from sklearn.impute import (
SimpleImputer,
) # Import SimpleImputer for missing value imputation
# Modeling
import optuna # Import optuna library for hyperparameter optimization
from sklearn.model_selection import KFold # Import KFold for cross-validation
from sklearn.metrics import (
roc_auc_score as ras,
) # Import roc_auc_score for evaluation metric
from sklearn.model_selection import (
cross_val_score,
) # Import cross_val_score for cross-validation
from sklearn.model_selection import GridSearchCV # Import GridSearchCV for grid search
from sklearn.model_selection import (
RandomizedSearchCV,
) # Import RandomizedSearchCV for randomized search
# Models
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.neural_network import MLPClassifier
import xgboost as xgb
import lightgbm as lgbm
import catboost as cb
# # **1. Data Wrangling**
# To kick off the analysis, we will begin with data wrangling, which encompasses a series of tasks including data cleaning, data transformation, data integration, and data reduction. Through these processes, we aim to ensure the accuracy, completeness, and analyzability of the dataset.
# ### **1.1 Load Dataset**
train_feature = pd.read_csv(
"/content/drive/MyDrive/DAC-FINDIT/training_set_features.csv",
index_col="respondent_id",
)
train_label = pd.read_csv(
"/content/drive/MyDrive/DAC-FINDIT/training_set_labels.csv",
index_col="respondent_id",
)
sample_submission = pd.read_csv(
"/content/drive/MyDrive/DAC-FINDIT/submission_format.csv", index_col="respondent_id"
)
train = train_feature.merge(
train_label, on="respondent_id"
) # Merge train_feature and train_label
test = pd.read_csv(
"/content/drive/MyDrive/DAC-FINDIT/test_set_features.csv", index_col="respondent_id"
)
train.head()
# Upon examining the given dataframe, it is evident that all the columns in the dataset are categorical in nature.
# ### **1.2 Data Inspection**
print(f"Train shape: {train.shape}") # See number of rows and columns in train dataset
print(f"Test shape: {test.shape}") # See number of rows and columns in test dataset
train.info() # Displays a summary of the dataframe
# We'll create a function to evaluate the missing value proportions in the train and test datasets. This function will facilitate the assessment of missing data and simplify future missing value checks.
def missing_values(train, test):
miss_perc_df = pd.DataFrame(
index=["train_miss", "test_miss"], # Set index of dataframe
columns=test.columns, # Set columns of dataframe
)
# Loop through the columns of the "test" dataframe and calculate the percentage of missing values for both the "train" and "test" dataframes
for i, col in enumerate(test.columns):
miss_perc_df.loc["train_miss", col] = np.round(
train.isna().sum()[i] / train[col].shape[0] * 100, 2
) # Calculate missing values in train dataset
miss_perc_df.loc["test_miss", col] = np.round(
test.isna().sum()[i] / test[col].shape[0] * 100, 2
) # Calculate missing values in test dataset
# Convert the values in the "miss_perc_df" dataframe to strings and append a "%" sign
miss_perc_df = miss_perc_df.astype(str) + "%"
return miss_perc_df
missing_values(train, test)
# From the given dataframe, we can gather the following information:
# * The columns `health_insurance`, `employment_industry`, and `employment_occupation` contain a significant number of missing values.
# * The `income_poverty` column also has a considerable amount of missing data.
# * The `doctor_recc_h1n1` and `doctor_recc_seasonal` columns share the same proportion of missing values.
# Since the columns with the dtype `float64` are already ordinal encoded, we can directly utilize them as features for our machine learning models without any additional preprocessing.
# To streamline the preprocessing of our data, we will segregate the columns into two separate variables based on their types: ordinal and nominal. This division will enable us to apply specific preprocessing techniques more efficiently to the nominal columns.
# Select columns from the 'train' dataframe that have a data type of float64 and convert them to a list.
ordinal_cols = train.select_dtypes("float64").columns.to_list()
# Select columns from the 'train' dataframe that have a data type of object (strings) and convert them to a list.
object_cols = train.select_dtypes("object").columns.to_list()
behavioral_cols = [
"behavioral_antiviral_meds",
"behavioral_avoidance",
"behavioral_face_mask",
"behavioral_wash_hands",
"behavioral_large_gatherings",
"behavioral_outside_home",
"behavioral_touch_face",
]
opinion_cols = [
"opinion_h1n1_vacc_effective",
"opinion_h1n1_risk",
"opinion_h1n1_sick_from_vacc",
"opinion_seas_vacc_effective",
"opinion_seas_risk",
"opinion_seas_sick_from_vacc",
]
# Given that all the columns in the dataset are categorical, it would be beneficial to examine the mode for each column. This is particularly important as we are likely to use the mode to impute missing values in our dataset.
# Ordinal columns
ord_summary = train.loc[:, ordinal_cols].describe()
pd.concat([ord_summary, train[ordinal_cols].mode()]).rename(index={0: "mean"})
# Nominal columns
train.loc[:, object_cols].describe()
print("Ordinal columns:", len(ordinal_cols))
print("Nominal columns:", len(object_cols))
# To compare missing values with other categories, we will create a copy of the train dataframe and fill in all the missing values with "Missing". This will allow us to perform a crosstabular analysis and compare the missing values to other categories more effectively.
nan_to_miss = train.copy()
# Replace all missing values in the 'nan_to_miss' dataframe with the string "Missing"
nan_to_miss.fillna("Missing", inplace=True)
train[train["education"].isna()].tail()
# We observe that the presence of a missing value in the `education` column is usually accompanied by missing values in the `income_poverty`, `marital_status`, `rent_or_own`, `employment_status`, `employment_industry`, and `employment_occupation` columns.
# Computes a frequency table or contingency table showing the distribution of two categorical variables in the DataFrame
pd.crosstab(nan_to_miss["doctor_recc_h1n1"], nan_to_miss["doctor_recc_seasonal"])
# It is evident that when a row in the `doctor_recc_seasonal` column contains a NaN value, the corresponding row in the `doctor_recc_h1n1` column will also be NaN. This specific missing value pattern holds significant importance and should be captured by our model.
pd.crosstab(nan_to_miss["employment_status"], nan_to_miss["employment_industry"])
pd.crosstab(nan_to_miss["employment_status"], nan_to_miss["employment_occupation"])
# An intriguing observation is that when a row in the `employment_status` column is missing or when a person is categorized as "Not in Labor Force" or "Unemployed," we can guarantee that the corresponding rows in the `employment_industry` and `employment_occupation` columns will also be missing. This pattern aligns with our expectations and makes logical sense (MNAR).
# To preserve and capture the observed pattern, we can introduce a new category (i.e. "None") in the `employment_industry` and `employment_occupation` columns specifically for cases where a person is **not in the labor force** or **unemployed**. By including this new category, we maintain the integrity of the pattern within our data, allowing our model to effectively utilize this information.
# ### **1.3 Handling Missing Values**
# To capture the missing value pattern in the dataset, we will create a new category called "Unknown" for nominal columns and assign a value of 2 as "Unknown" for ordinal columns in both the training and test data. This approach allows us to effectively handle and represent missing values in our dataset without significantly increasing the length of our code.
for df in [train, test]:
df["doctor_recc_h1n1"].fillna(2.0, inplace=True)
df["doctor_recc_seasonal"].fillna(2.0, inplace=True)
df["health_insurance"].fillna(2.0, inplace=True)
df["education"].fillna("Unknown", inplace=True)
df["marital_status"].fillna("Unknown", inplace=True)
df["rent_or_own"].fillna("Unknown", inplace=True)
df["income_poverty"].fillna("Unknown", inplace=True)
# We can safely impute missing values in the `employment_status` column with "Not in Labor Force" because when a row has a missing value in `employment_status`, the corresponding rows in `employment_industry` and `employment_occupation` will also be missing.
# This relationship is similar to the cases where the `employment_status` is "Not in Labor Force" or "Unemployed". We choose "Not in Labor Force" as the imputation value because it is the second most frequent value in the column.
for df in [train, test]:
df["employment_status"].fillna("Not in Labor Force", inplace=True)
for df in [train, test]:
for row in range(
df.index[0], df.index[-1] + 1
): # Iterate over the rows of the DataFrame.
if (df.loc[row, "employment_status"] == "Not in Labor Force") or (
df.loc[row, "employment_status"] == "Unemployed"
):
df.loc[
row, "employment_occupation"
] = "None" # Set the value of 'employment_occupation' to "None" for the current row.
df.loc[
row, "employment_industry"
] = "None" # Set the value of 'employment_industry' to "None" for the current row.
# To avoid introducing bias, we will impute the missing values in the **respondent's opinion columns** with the value 3, which represents "Don't know." This approach ensures that the missing values are treated uniformly and does not influence the analysis based on the respondents' opinions.
for df in [train, test]:
df[opinion_cols] = df[opinion_cols].fillna(3.0)
missing_values(train, test)
# For the remaining missing values in both the train and test datasets, which are **mostly** missing at random (MAR), we will fill them with the mode of each respective column. Since the dataset consists of categorical variables, using the mode as the imputation strategy helps maintain the categorical nature of the data.
imputer = SimpleImputer(
strategy="most_frequent"
) # Create an instance of SimpleImputer with 'most_frequent' strategy.
# Ordinal Columns
train[ordinal_cols] = imputer.fit_transform(
train[ordinal_cols]
) # Fill missing values in ordinal columns of 'train' DataFrame using the fitted imputer.
test[ordinal_cols] = imputer.transform(
test[ordinal_cols]
) # Fill missing values in ordinal columns of 'test' DataFrame using the fitted imputer.
# Nominal Columns
train[object_cols] = imputer.fit_transform(
train[object_cols]
) # Fill missing values in nominal columns of 'train' DataFrame using the fitted imputer.
test[object_cols] = imputer.transform(
test[object_cols]
) # Fill missing values in nominal columns of 'test' DataFrame using the fitted imputer.
missing_values(train, test)
# ### **1.4 Handling Other Issues**
# To ensure compatibility with various machine learning algorithms and statistical techniques, we will convert the columns with dtype `float64` to `int64`. This conversion will effectively transform these variables from continuous to discrete, as many algorithms require discrete variables as input. By making these variables discrete, we can leverage the full range of analytical tools and methods available for categorical data analysis.
train[ordinal_cols] = train[ordinal_cols].astype(
"int64"
) # Convert train[ordinal_cols] to integer
test[ordinal_cols] = test[ordinal_cols].astype(
"int64"
) # Convert test[ordinal_cols] to integer
print("Train duplicates:", train.duplicated().sum()) # Find duplicates value in train
print("Test duplicates:", test.duplicated().sum()) # Find duplicates value in test
# The presence of a single duplicated row is negligible and is unlikely to significantly impact the analysis. Therefore, we can safely ignore this duplicated row without compromising the integrity or accuracy of our results.
# When categorical variables are represented as ranges or intervals, such as `age_group`, assigning a single value to each category helps in reducing the loss of information and provides a more precise estimate for analysis. By assigning a midpoint, we treat the variable as if it takes on a continuous scale, albeit with some level of discretization.
# Define a dictionary to map age_group categories to numerical values
dict_age_group = {
"65+ Years": 75,
"55 - 64 Years": 60,
"45 - 54 Years": 50,
"18 - 34 Years": 26,
"35 - 44 Years": 40,
}
for df in [train, test]:
# Replace the values in the age_group column with the corresponding numerical value from dict_age_group
df["age_group"] = df["age_group"].map(dict_age_group)
# To assign a more representative midpoint to the "65+ Years" age category, an approximate value of 75 can be considered. This choice is based on global data trends indicating that the average age at which many people pass away in various countries is approximately 85 years.
# Please note that this information is sourced from a study titled "Deaths: final data for 2009" (2011, December 29) available on PubMed. It is important to mention that this specific data is used solely for the purpose of assigning a midpoint to the age group range of 65+ years. https://pubmed.ncbi.nlm.nih.gov/24974587/#:~:text=Life%20expectancy%20at%20birth%20rose,74%2C%20and%2075%2D84.
# Check inconsistency -- Ordinal columns
for col in ordinal_cols:
print(
col + ":", sorted(train[col].unique())
) # Print unique values in every columns, sorted
# Check inconsistency -- Object columns
for col in object_cols:
print(col + ":", train[col].unique()) # Print unique values in every columns
print()
# After careful manual inspection, we have determined that there are no instances of inconsistent data entries in the dataset.
# Since the dataset exclusively consists of categorical columns, there is no need for **normalization**. Categorical features do not possess a natural numerical order or scale, rendering normalization irrelevant in this context.
# # **2. Exploratory Data Analysis**
# ### **2.1 Univariate Exploration**
# We will begin by directing our attention to comprehending the distribution of categorical data, prioritizing an understanding of its characteristics without delving into its relationship with other variables.
# Select the ordinal columns (non-binary) with more than 2 unique values.
non_binary_cols = [col for col in ordinal_cols if train[col].nunique() > 2]
fig, ax = plt.subplots(
nrows=4, ncols=4, figsize=(30, 20)
) # Creating subplots for countplots with specified dimensions and figure size.
for i, col in enumerate(
non_binary_cols
): # Iterate over the non-binary ordinal columns.
bar = sns.countplot(
data=train, y=col, ax=ax[i // 4, i % 4], palette="ch:.25"
) # Create a countplot for each column and assign it to the corresponding subplot.
bar.bar_label(bar.containers[0]) # Add labels to the bars in the countplot.
# Select the ordinal columns (binary) with less than or equal to 2 unique values.
binary_cols = [col for col in ordinal_cols if train[col].nunique() <= 2]
fig, ax = plt.subplots(nrows=3, ncols=4, figsize=(30, 15))
for i, col in enumerate(binary_cols):
bar = sns.countplot(data=train, y=col, ax=ax[i // 4, i % 4], palette="coolwarm")
bar.bar_label(bar.containers[0])
# Considering that the "None" category overwhelmingly dominates all other categories in the `employment_industry` and `employment_occupation` columns, it is apparent that plotting these columns would not yield any valuable insights. Therefore, we will exclude them from our analysis.
# Countplot for Object columns
fig, ax = plt.subplots(nrows=4, ncols=3, figsize=(28, 20))
for i, col in enumerate(object_cols):
if col not in ["employment_industry", "employment_occupation"]:
bar = sns.countplot(data=train, x=col, ax=ax[i // 3, i % 3], palette="rainbow")
bar.bar_label(bar.containers[0])
# ### **2.2. Bivariate Exploration**
# Our attention now turns to comprehending the correspondence and relationship between two specific columns, particularly in relation to the target variable. The aim of this endeavor is to gain a deeper understanding of the fundamental patterns and connections inherent within our dataset.
fig, ax = plt.subplots(nrows=2, ncols=4, figsize=(25, 10))
for i, col in enumerate(behavioral_cols):
bar = sns.barplot(
data=train,
y=col,
x="chronic_med_condition",
ax=ax[i // 4, i % 4],
palette="rainbow",
)
bar.bar_label(bar.containers[0])
# The bar plot indicates a notable pattern where individuals with specific chronic medical conditions, such as asthma, diabetes, heart conditions, kidney conditions, sickle cell anemia, neurological or neuromuscular conditions, liver conditions, or weakened immune systems due to chronic illnesses or medications, tend to exhibit more proactive behavioral precautions to prevent the occurrence of flu-like illnesses.
# Create a figure with subplots to plot the barplots and violinplots.
fig, ax = plt.subplots(nrows=3, ncols=4, figsize=(25, 13))
# Health related columns
for i, col in enumerate(["health_worker", "doctor_recc_h1n1", "doctor_recc_seasonal"]):
bar = sns.barplot(
data=train, y="h1n1_vaccine", x=col, ax=ax[i, 0], palette="Paired"
)
bar.bar_label(bar.containers[0])
# Plot a violinplot for 'h1n1_vaccine' on the i-th subplot.
sns.violinplot(data=train, y="h1n1_vaccine", x=col, ax=ax[i, 1], palette="Paired")
bar = sns.barplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 2], palette="Paired"
)
bar.bar_label(bar.containers[0])
# Plot a violinplot for 'seasonal_vaccine' on the i-th subplot.
sns.violinplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 3], palette="Paired"
)
# The visualization reveals clear patterns. Healthcare workers are more likely to receive both the H1N1 flu vaccine and the seasonal flu vaccine.
# Similarly, individuals recommended the H1N1 vaccine by a doctor are highly likely to receive both the H1N1 vaccine and the seasonal vaccine. Moreover, those recommended the seasonal vaccine are also highly likely to receive both the seasonal and H1N1 vaccines. These findings highlight the influence of healthcare worker status and doctor recommendations on vaccination behavior, indicating a strong association with vaccine uptake.
# Create a figure with subplots to plot the barplots and violinplots.
fig, ax = plt.subplots(nrows=4, ncols=2, figsize=(30, 15))
# Employment related columns
# Create barplots and violinplots for 'h1n1_vaccine' for each column.
for i, col in enumerate(["employment_industry", "employment_occupation"]):
bar = sns.barplot(
data=train, y="h1n1_vaccine", x=col, ax=ax[i, 0], palette="rainbow"
)
bar.bar_label(bar.containers[0])
sns.violinplot(data=train, y="h1n1_vaccine", x=col, ax=ax[i, 1], palette="rainbow")
# Create barplots and violinplots for 'seasonal_vaccine' for each column.
for i, col in enumerate(["employment_industry", "employment_occupation"]):
bar = sns.barplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i + 2, 0], palette="rainbow"
)
bar.bar_label(bar.containers[0])
sns.violinplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i + 2, 1], palette="rainbow"
)
# The bar plot clearly indicates that individuals employed in the "haxffmxo" and "fcxhlnwr" industries have a higher likelihood of receiving both the H1N1 and seasonal vaccines. Similarly, those with employment occupations such as "dcjcmpih" and "cmhcxjea" also exhibit a higher likelihood of receiving both vaccines. These findings emphasize the positive association between specific employment industries and occupations with vaccine uptake for both H1N1 and seasonal viruses.
# Create a figure with subplots to plot the barplots and violinplots.
fig, ax = plt.subplots(nrows=4, ncols=4, figsize=(25, 17))
# Opinion related columns
for i, col in enumerate(
[
"opinion_seas_risk",
"opinion_seas_vacc_effective",
"opinion_h1n1_risk",
"opinion_h1n1_vacc_effective",
]
):
bar = sns.barplot(
data=train, y="h1n1_vaccine", x=col, ax=ax[i, 0], palette="Spectral"
)
bar.bar_label(bar.containers[0])
sns.violinplot(data=train, y="h1n1_vaccine", x=col, ax=ax[i, 1], palette="Spectral")
bar = sns.barplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 2], palette="Spectral"
)
bar.bar_label(bar.containers[0])
sns.violinplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 3], palette="Spectral"
)
# There is a clear relationship observed in the data where individuals who perceive the H1N1 vaccine and seasonal vaccines to be effective are more likely to receive both vaccines. Additionally, individuals who believe that the risk of getting sick with H1N1 flu and seasonal flu without vaccination is higher also demonstrate a higher likelihood of receiving both the H1N1 vaccine and the seasonal vaccine.
# These findings highlight the significant impact of perceived vaccine effectiveness and perceived risk of illness on the decision to receive the H1N1 and seasonal vaccines.
fig, ax = plt.subplots(nrows=5, ncols=2, figsize=(25, 21))
# Other interesting columns
for i, col in enumerate(
[
"h1n1_concern",
"chronic_med_condition",
"age_group",
"employment_status",
"household_children",
]
):
bar = sns.barplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 0], palette="coolwarm"
)
bar.bar_label(bar.containers[0])
sns.violinplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 1], palette="coolwarm"
)
# The bar plot reveals additional insights. Individuals who express concern about the H1N1 flu, have chronic medical conditions, belong to older age groups, are not in the labor force, or have zero children in their household exhibit a higher likelihood of receiving the seasonal vaccine.
# These findings suggest that individuals who fall into these categories are more inclined to prioritize their health and take proactive measures by receiving the seasonal vaccine.
# ### **Summary:**
# * People with chronic medical conditions are more likely to take preventive measures against flu-like illnesses.
# * Specific employment industries and occupations show higher vaccine uptake for both H1N1 and seasonal flu vaccines.
# * Healthcare workers have a higher likelihood of receiving both vaccines.
# * Doctor recommendations strongly influence vaccine uptake.
# * Perceived vaccine effectiveness and risk of illness are important factors.
# * Other factors like concern about H1N1, older age, not being in the labor force, and no children in the household are associated with higher seasonal flu vaccine uptake.
# # **3. Feature Engineering**
# To further enhance the predictive performance of our model, we will create additional relevant features that capture important patterns and relationships in the data. We propose the following new features:
# * `good_behavioral_count` -- represent the cumulative count of flu precaution behaviors practiced by an individual. A higher value would indicate a greater adherence to flu prevention measures.
# * `has_doc_recc` -- indicate whether a person has received a recommendation from a doctor for either the H1N1 vaccine or the seasonal flu vaccine (binary). A value of 1 represents that a doctor has recommended at least one of the vaccines, and a value of 0 indicates no doctor recommendation.
# Count-based features are beneficial for tree-based models because these models lack the inherent ability to aggregate information across multiple features simultaneously. By introducing count-based features, we allow tree models to leverage valuable insights into the occurrence or frequency of particular events or behaviors.
for df in [train, test]:
# Calculate the sum of behavioral columns and create a new column "good_behavioral_count"
df["good_behavioral_count"] = df[behavioral_cols].sum(axis=1)
for i in range(df.index[0], df.index[-1] + 1):
# Check if both "doctor_recc_h1n1" and "doctor_recc_seasonal" columns have a value of 2 (Unknown)
if df.loc[i, ["doctor_recc_h1n1", "doctor_recc_seasonal"]].isin([2]).all():
# If both columns have a value of 2, set "has_doc_recc" to 0
df.loc[i, "has_doc_recc"] = 0
else:
# If any of the columns have a value other than 2, set "has_doc_recc" to 1
df.loc[i, "has_doc_recc"] = 1
# Append the new ordinal feature to the list of ordinal columns
for new_ord_feature in ["good_behavioral_count", "has_doc_recc"]:
ordinal_cols.append(new_ord_feature)
# We will proceed with encoding the nominal columns in our dataset to ensure that categorical variables are in a suitable format for machine learning algorithms. We will use two different encoding techniques:
# * **Ordinal Encoding** -- apply to `age_group` and `education` columns in order to preserve the inherent order or hierarchy present in these columns, allowing the algorithm to understand the relative differences between categories.
# * **One-Hot Encoding** -- apply to `race`, `sex`, `marital_status`, `rent_or_own`, `employment_status`, `hhs_geo_region`, `census_msa`, `employment_industry`, `employment_occupation`, `income_poverty` in order to indicate the presence or absence of a category in the observation.
# To ensure consistency and avoid discrepancies, we will merge the test and train data before encoding the categorical variables. This merging allows us to encode the variables based on the entire dataset, eliminating any potential issues if a category is present in only one dataset. Once the encoding is complete, we will separate the merged dataset back into their respective test and train datasets.
# Get the number of rows in the train dataframe
train_rows = train.shape[0]
# Concatenate the train and test dataframes vertically
merged_df = pd.concat([train, test])
# Encode object columns using Ordinal Encoding
for col in ["age_group", "education"]:
merged_df[col], _ = merged_df[col].factorize()
# Encode object columns using One-Hot Encoding
merged_df = pd.get_dummies(
merged_df,
columns=[
"race",
"sex",
"marital_status",
"rent_or_own",
"employment_status",
"hhs_geo_region",
"census_msa",
"employment_industry",
"employment_occupation",
"income_poverty",
],
)
# Separate the merged dataframe back into train and test dataframes
train = merged_df.iloc[:train_rows]
test = merged_df.iloc[train_rows:].drop(columns=["h1n1_vaccine", "seasonal_vaccine"])
train.head()
# We will employ mutual information to measure the relationship between two columns. It quantifies how knowing one variable reduces uncertainty about the other. Higher mutual information indicates a stronger relationship, while lower values indicate weaker or no relationship. This helps confirm the relationships found during EDA.
# Mutual Information function to calculate MI scores
def make_mi_scores(X, y, discrete_features):
# Calculate Mutual Information (MI) scores between features (X) and target (y)
mi_scores = mic(X, y, discrete_features=discrete_features)
# Convert the MI scores to a Pandas Series with feature names as index and sort in descending order
mi_scores = pd.Series(mi_scores, name="MI Scores", index=X.columns).sort_values(
ascending=False
)
return mi_scores
# Create a copy of the train dataframe as X
X = train.copy()
# Separate the target variables from X and store them in y_h1n1 and y_seas
y_h1n1 = X.pop("h1n1_vaccine")
y_seas = X.pop("seasonal_vaccine")
# Identify the discrete features in X (features with dtype 'int64')
discrete_features = X.dtypes == "int64"
# H1N1 MI Score
mi_scores_h1n1 = make_mi_scores(X, y_h1n1, discrete_features)
# Convert the MI scores into a transposed DataFrame
pd.DataFrame(mi_scores_h1n1).T
# Seasonal MI Score
mi_scores_seas = make_mi_scores(X, y_seas, discrete_features)
# Convert the MI scores into a transposed DataFrame
pd.DataFrame(mi_scores_seas).T
# # **4. Modeling**
# To start modeling for the classification problem, we'll choose the best model for both the h1n1_vaccine and seasonal_vaccine targets. Then, we'll test the selected model with default parameters and evaluate its performance using the ROC AUC metric.
# By starting with the default parameters, we can establish a baseline performance for the model. This will allow us to compare and assess the effectiveness of subsequent parameter tuning and optimizations.
models = {
# "lr": LogisticRegression(random_state=42), # Error
"dtc": DecisionTreeClassifier(random_state=42),
"rfc": RandomForestClassifier(random_state=42),
"knn": KNeighborsClassifier(),
"gnb": GaussianNB(),
"svc": SVC(random_state=42),
# 'xgboost': xgb.XGBClassifier(random_state=42), # Error
# 'lightgbm': lgbm.LGBMClassifier(random_state=42), # Error
"catboost": cb.CatBoostClassifier(verbose=0, random_state=42),
"nn": MLPClassifier(random_state=42, hidden_layer_sizes=(10, 10), max_iter=500),
}
# We will utilize k-fold cross-validation to obtain a more reliable and robust estimate of our model's performance. By leveraging k-fold cross-validation, we can gain a more comprehensive understanding of our model's effectiveness and its ability to generalize to unseen data.
# Function for performing k-fold cross-validation
def kfold_cv(model, X, y):
# Define KFold cross-validation with 5 splits, random state of 42, and shuffling of data
kf = KFold(n_splits=5, random_state=42, shuffle=True)
scores = [] # List to store the evaluation scores
# Iterate over the train and validation splits
for train_idx, test_idx in kf.split(X, y):
# Split the data into training and validation sets based on the indices
X_train, X_val = X.iloc[train_idx], X.iloc[test_idx]
y_train, y_val = y.iloc[train_idx], y.iloc[test_idx]
# Fit the model on the training data
model.fit(X_train, y_train)
# Make predictions on the validation data
predictions = model.predict(X_val)
# Calculate the evaluation score (here, using RAS macro-average)
ras_score = ras(y_val, predictions, average="macro")
# Append the score to the list of scores
scores.append(ras_score)
return np.mean(
scores
) # Return the mean of the scores as the overall evaluation score
# Prepare the input features and target variables for H1N1 vaccine prediction
X_h1n1 = train.drop(["h1n1_vaccine", "seasonal_vaccine"], axis=1)
y_h1n1 = train.pop("h1n1_vaccine")
# Prepare the input features and target variables for seasonal flu vaccine prediction
X_seas = train.drop("seasonal_vaccine", axis=1)
y_seas = train.pop("seasonal_vaccine")
# H1N1 Vaccine
# Iterate over the models dictionary
for name, model in models.items():
# Perform k-fold cross-validation and obtain the evaluation score
score = kfold_cv(model, X_h1n1, y_h1n1)
# Print the model name and its corresponding score
print(f"{name}: {score}")
# Seasonal Vaccine
for name, model in models.items():
score = kfold_cv(model, X_seas, y_seas)
print(f"{name}: {score}")
# The results indicate that the CatBoost model consistently outperforms the other models, yielding higher scores. This suggests that the CatBoost model is better suited for our classification task and exhibits superior predictive performance compared to the alternative models.
# Now, we will proceed to utilize CatBoost with tuned parameters obtained from Optuna to further enhance our prediction score. By incorporating the optimized parameter settings, we aim to improve the performance of the CatBoost model and achieve an even higher prediction score.
h1n1_param = {
"iterations": 600, # Number of boosting iterations
"learning_rate": 0.02857091731949716, # Learning rate for boosting
"random_strength": 1, # Random strength for boosting
"bagging_temperature": 2, # Bagging temperature for boosting
"max_bin": 21, # Maximum number of bins for numerical features
"grow_policy": "Lossguide", # Tree growth policy
"min_data_in_leaf": 18, # Minimum number of samples required in a leaf
"max_depth": 6, # Maximum depth of trees
"l2_leaf_reg": 30.38300354991344, # L2 regularization coefficient
"one_hot_max_size": 13, # Maximum size of one-hot encoding for categorical features
"auto_class_weights": "Balanced", # Automatic class weights adjustment
"verbose": 0, # Verbosity level
}
seas_param = {
"iterations": 800, # Number of boosting iterations
"learning_rate": 0.017179768319739466, # Learning rate for boosting
"random_strength": 0, # Random strength for boosting
"bagging_temperature": 7, # Bagging temperature for boosting
"max_bin": 106, # Maximum number of bins for numerical features
"grow_policy": "Lossguide", # Tree growth policy
"min_data_in_leaf": 20, # Minimum number of samples required in a leaf
"max_depth": 4, # Maximum depth of trees
"l2_leaf_reg": 45.5914670615121, # L2 regularization coefficient
"one_hot_max_size": 8, # Maximum size of one-hot encoding for categorical features
"auto_class_weights": "Balanced", # Automatic class weights adjustment
"verbose": 0, # Verbosity level
}
# Create a CatBoostClassifier model with the H1N1 vaccine parameters and fit it to the data
model_h1n1 = cb.CatBoostClassifier(**h1n1_param)
model_h1n1.fit(X_h1n1, y_h1n1)
# Create a CatBoostClassifier model with the seasonal flu vaccine parameters and fit it to the data
model_seas = cb.CatBoostClassifier(**seas_param)
model_seas.fit(X_seas, y_seas)
score = kfold_cv(model_h1n1, X_h1n1, y_h1n1)
print(f"Catboost H1N1: {score}")
score = kfold_cv(model_seas, X_seas, y_seas)
print(f"CatBoost Seasonal: {score}")
# By incorporating the optimized parameters into CatBoost, we observe a significant increase in the prediction score. With this improved performance, we can now proceed to follow the prescribed format for the submission file in this competition.
# Create a DataFrame for the submission
submission = pd.DataFrame(
{
"respondent_id": test.index, # Use the respondent IDs from the test data
"h1n1_vaccine": model_h1n1.predict_proba(test)[
:, 1
], # Use the probability of the positive class for H1N1 vaccine
"seasonal_vaccine": model_seas.predict_proba(test)[
:, 1
], # Use the probability of the positive class for seasonal flu vaccine
}
)
# Save the submission DataFrame to a CSV file without including the index
submission.to_csv("submission.csv", index=False)
submission.head()
#
# # **Hyperparameter Tuning**
# We will utilize Optuna for hyperparameter tuning of our models, specifically focusing on the CatBoost algorithm. This technique has shown significant improvement in our ROC AUC prediction score, with an increase of over 0.05.
# By leveraging Optuna's optimization capabilities, we aim to fine-tune the hyperparameters of the CatBoost model to maximize its predictive performance and enhance the overall accuracy of our predictions.
# Function for tuning a model using k-fold cross-validation
def tune_kfold_cv(model, X, y, metric, n_folds=69):
# Create a k-fold cross-validation object with specified number of folds
skfold = KFold(n_splits=n_folds, random_state=0, shuffle=True)
# List to store the scores obtained from each fold
scores = []
# Iterate over the folds
for train_index, test_index in skfold.split(X, y):
# Split the data into training and testing sets based on the fold indices
X_train, X_test = X.iloc[train_index], X.iloc[test_index]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
# Fit the model on the training data and evaluate it on the testing data
model.fit(X_train, y_train, eval_set=[(X_test, y_test)], verbose=0)
predictions = model.predict(X_test.values)
# Calculate the score using the specified evaluation metric
score = metric(y_test, predictions, average="macro")
# Append the score to the list of scores
scores.append(score)
# Print the ROC AUC score for the current fold
print(f"ROC AUC: {score:.3f}")
# Return the list of scores
return scores
# Objective function for hyperparameter optimization using Optuna
def objective(trial, data=X_h1n1, target=y_h1n1):
# Define the hyperparameter search space using trial suggestions
params = {
"iterations": trial.suggest_int("iterations", 100, 2000, step=100),
"learning_rate": trial.suggest_float("learning_rate", 1e-5, 0.1),
"random_strength": trial.suggest_int("random_strength", 0.0, 10.0),
"bagging_temperature": trial.suggest_int("bagging_temperature", 0.0, 10.0),
"max_bin": trial.suggest_int("max_bin", 1, 255),
"grow_policy": trial.suggest_categorical(
"grow_policy", ["SymmetricTree", "Depthwise", "Lossguide"]
),
"min_data_in_leaf": trial.suggest_int("min_data_in_leaf", 1, 20),
"max_depth": trial.suggest_int("max_depth", 1, 10),
"l2_leaf_reg": trial.suggest_float("l2_leaf_reg", 1e-8, 100.0),
"one_hot_max_size": trial.suggest_int("one_hot_max_size", 2, 16),
"auto_class_weights": trial.suggest_categorical(
"auto_class_weights", ["None", "Balanced", "SqrtBalanced"]
),
"verbose": 0,
"early_stopping_rounds": 200,
}
# Create a CatBoostClassifier with the suggested hyperparameters
model = cb.CatBoostClassifier(**params)
# Perform k-fold cross-validation with the tuned model and calculate the average score
scores = tune_kfold_cv(model, X_h1n1, y_h1n1, ras, 8)
return np.sum(scores) / 8
# Create an Optuna study with the direction set to maximize
study = optuna.create_study(direction="maximize")
# Run the optimization process using the objective function defined earlier, with a specified number of trials
study.optimize(objective, n_trials=30)
# Print the number of finished trials (i.e., the number of trials that have been executed)
print("Number of finished trials:", len(study.trials))
# Print the best trial's parameters, which correspond to the set of hyperparameters that yielded the highest objective value
print("Best trial:", study.best_trial.params)
# Assign the best parameters found during optimization to a variable
parameter_tuned = study.best_trial.params
# Create a CatBoostClassifier model using the best parameters
model = cb.CatBoostClassifier(**parameter_tuned)
# Calculate the score using k-fold cross-validation with the model and the H1N1 dataset
score = kfold_cv(model, X_h1n1, y_h1n1)
# Print the score obtained from k-fold cross-validation
print(f"CatBoost H1N1: {score}")
# Define the objective function for parameter optimization
def objective(trial, data=X_seas, target=y_seas):
# Define the parameter space to search within
params = {
"iterations": trial.suggest_int("iterations", 100, 2000, step=100),
"learning_rate": trial.suggest_float("learning_rate", 1e-5, 0.1),
"random_strength": trial.suggest_int("random_strength", 0.0, 10.0),
"bagging_temperature": trial.suggest_int("bagging_temperature", 0.0, 10.0),
"max_bin": trial.suggest_int("max_bin", 1, 255),
"grow_policy": trial.suggest_categorical(
"grow_policy", ["SymmetricTree", "Depthwise", "Lossguide"]
),
"min_data_in_leaf": trial.suggest_int("min_data_in_leaf", 1, 20),
"max_depth": trial.suggest_int("max_depth", 1, 10),
"l2_leaf_reg": trial.suggest_float("l2_leaf_reg", 1e-8, 100.0),
"one_hot_max_size": trial.suggest_int("one_hot_max_size", 2, 16),
"auto_class_weights": trial.suggest_categorical(
"auto_class_weights", ["None", "Balanced", "SqrtBalanced"]
),
"verbose": 0,
"early_stopping_rounds": 200,
}
# Create a CatBoostClassifier model with the suggested parameters
model = cb.CatBoostClassifier(**params)
# Perform k-fold cross-validation and calculate scores
scores = tune_kfold_cv(model, X_seas, y_seas, ras, 8)
# Return the average of the scores
return np.sum(scores) / 8
# Create an Optuna study object for parameter optimization
study = optuna.create_study(direction="maximize")
# Perform the optimization by running the objective function for a specified number of trials
study.optimize(objective, n_trials=30)
# Print the number of finished trials
print("Number of finished trials:", len(study.trials))
# Print the best trial's parameters
print("Best trial:", study.best_trial.params)
# Retrieve the tuned parameters from the best trial of the study
parameter_tuned = study.best_trial.params
# Create a CatBoostClassifier model using the tuned parameters
model = cb.CatBoostClassifier(**parameter_tuned)
# Compute the score using k-fold cross-validation with the tuned model
score = kfold_cv(model, X_seas, y_seas)
# Print the score for the CatBoost model on the seasonal vaccine prediction
print(f"CatBoost Seas: {score}")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/927/129927236.ipynb
|
flu-shot-learning
|
soundslikedata
|
[{"Id": 129927236, "ScriptId": 38634037, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4238805, "CreationDate": "05/17/2023 13:31:56", "VersionNumber": 1.0, "Title": "DAC FINDIT 2023 - BukanRISTEK", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 719.0, "LinesInsertedFromPrevious": 719.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 4}]
|
[{"Id": 186350786, "KernelVersionId": 129927236, "SourceDatasetVersionId": 3385921}]
|
[{"Id": 3385921, "DatasetId": 2041219, "DatasourceVersionId": 3437446, "CreatorUserId": 8683201, "LicenseName": "Other (specified in description)", "CreationDate": "03/31/2022 07:46:17", "VersionNumber": 1.0, "Title": "Flu Shot Learning", "Slug": "flu-shot-learning", "Subtitle": "HOSTED BY DRIVENDATA", "Description": "\n\nThe data for this competition comes from the National 2009 H1N1 Flu Survey (NHFS).\n\nIn their own words:\n\n> The National 2009 H1N1 Flu Survey (NHFS) was sponsored by the National Center for Immunization and Respiratory Diseases (NCIRD) and conducted jointly by NCIRD and the National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention (CDC). The NHFS was a list-assisted random-digit-dialing telephone survey of households, designed to monitor influenza immunization coverage in the 2009-10 season.\n> \n> The target population for the NHFS was all persons 6 months or older living in the United States at the time of the interview. Data from the NHFS were used to produce timely estimates of vaccination coverage rates for both the monovalent pH1N1 and trivalent seasonal influenza vaccines.\n\n[National 2009 H1N1 Flu Survey Public-Use Data File Readme](ftp://ftp.cdc.gov/pub/Health_Statistics/NCHS/Datasets/nis/nhfs/nhfspuf_readme.txt)\n\n \n\nThe NHFS was conducted between October 2009 and June 2010. It was one-time survey designed specifically to monitor vaccination during the 2009-2010 flu season in response to the 2009 H1N1 pandemic. The CDC has other ongoing programs for annual phone surveys that continue to monitor seasonal flu vaccination.\n\n## Data use restrictions\n\nThe source dataset comes with the following data use restrictions:\n\n> The Public Health Service Act (Section 308(d)) provides that the data collected by the National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention (CDC), may be used only for the purpose of health statistical reporting and analysis.\n> \n> Any effort to determine the identity of any reported case is prohibited by this law.\n> \n> NCHS does all it can to ensure that the identity of data subjects cannot be disclosed. All direct identifiers, as well as any characteristics that might lead to identification, are omitted from the data files. Any intentional identification or disclosure of a person or establishment violates the assurances of confidentiality given to the providers of the information.\n> \n> Therefore, users will:\n> \n> 1. Use the data in these data files for statistical reporting and analysis only.\n> 2. Make no use of the identity of any person or establishment discovered inadvertently and advise the Director, NCHS, of any such discovery (1 (800) 232-4636).\n> 3. Not link these data files with individually identifiable data from other NCHS or non-NCHS data files.\n> \n> By using these data, you signify your agreement to comply with the above requirements.\n\n[National 2009 H1N1 Flu Survey Public-Use Data File Readme](ftp://ftp.cdc.gov/pub/Health_Statistics/NCHS/Datasets/nis/nhfs/nhfspuf_readme.txt)\n\n## Additional resources\n\n- [U.S. National 2009 H1N1 Flu Survey (NHFS)](https://webarchive.loc.gov/all/20140511031000/http://www.cdc.gov/nchs/nis/about_nis.htm#h1n1)\n- [U.S. National Immunization Surveys (NIS)](https://www.cdc.gov/vaccines/imz-managers/nis/about.html)\n- [2009 H1N1 Pandemic (H1N1pdm09 virus)](https://www.cdc.gov/flu/pandemic-resources/2009-h1n1-pandemic.html), by the U.S. CDC\n- [About Flu](https://www.cdc.gov/flu/about/index.html), by the U.S. CDC\n- [Key Facts About Seasonal Flu Vaccine](https://www.cdc.gov/flu/prevent/keyfacts.htm), by the U.S. CDC\n\nData is provided courtesy of the United States [National Center for Health Statistics](https://www.cdc.gov/nchs/index.htm).\n\nU.S. Department of Health and Human Services (DHHS). National Center for Health Statistics. The National 2009 H1N1 Flu Survey. Hyattsville, MD: Centers for Disease Control and Prevention, 2012.\n\nImages courtesy of the [U.S. Navy](https://www.flickr.com/photos/navcent/15607260325/) and the [Fort Meade Public Affairs Office](https://www.flickr.com/photos/ftmeade/15242740638/) via Flickr under the [CC BY 2.0 license](https://creativecommons.org/licenses/by/2.0/legalcode).\n\nThe data for this competition comes from the National 2009 H1N1 Flu Survey (NHFS).\n\nIn their own words:\n\n> The National 2009 H1N1 Flu Survey (NHFS) was sponsored by the National Center for Immunization and Respiratory Diseases (NCIRD) and conducted jointly by NCIRD and the National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention (CDC). The NHFS was a list-assisted random-digit-dialing telephone survey of households, designed to monitor influenza immunization coverage in the 2009-10 season.\n> \n> The target population for the NHFS was all persons 6 months or older living in the United States at the time of the interview. Data from the NHFS were used to produce timely estimates of vaccination coverage rates for both the monovalent pH1N1 and trivalent seasonal influenza vaccines.\n\n[National 2009 H1N1 Flu Survey Public-Use Data File Readme](ftp://ftp.cdc.gov/pub/Health_Statistics/NCHS/Datasets/nis/nhfs/nhfspuf_readme.txt)\n\n \n\nThe NHFS was conducted between October 2009 and June 2010. It was one-time survey designed specifically to monitor vaccination during the 2009-2010 flu season in response to the 2009 H1N1 pandemic. The CDC has other ongoing programs for annual phone surveys that continue to monitor seasonal flu vaccination.\n\n## Data use restrictions\n\nThe source dataset comes with the following data use restrictions:\n\n> The Public Health Service Act (Section 308(d)) provides that the data collected by the National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention (CDC), may be used only for the purpose of health statistical reporting and analysis.\n> \n> Any effort to determine the identity of any reported case is prohibited by this law.\n> \n> NCHS does all it can to ensure that the identity of data subjects cannot be disclosed. All direct identifiers, as well as any characteristics that might lead to identification, are omitted from the data files. Any intentional identification or disclosure of a person or establishment violates the assurances of confidentiality given to the providers of the information.\n> \n> Therefore, users will:\n> \n> 1. Use the data in these data files for statistical reporting and analysis only.\n> 2. Make no use of the identity of any person or establishment discovered inadvertently and advise the Director, NCHS, of any such discovery (1 (800) 232-4636).\n> 3. Not link these data files with individually identifiable data from other NCHS or non-NCHS data files.\n> \n> By using these data, you signify your agreement to comply with the above requirements.\n\n[National 2009 H1N1 Flu Survey Public-Use Data File Readme](ftp://ftp.cdc.gov/pub/Health_Statistics/NCHS/Datasets/nis/nhfs/nhfspuf_readme.txt)\n\n## Additional resources\n\n- [U.S. National 2009 H1N1 Flu Survey (NHFS)](https://webarchive.loc.gov/all/20140511031000/http://www.cdc.gov/nchs/nis/about_nis.htm#h1n1)\n- [U.S. National Immunization Surveys (NIS)](https://www.cdc.gov/vaccines/imz-managers/nis/about.html)\n- [2009 H1N1 Pandemic (H1N1pdm09 virus)](https://www.cdc.gov/flu/pandemic-resources/2009-h1n1-pandemic.html), by the U.S. CDC\n- [About Flu](https://www.cdc.gov/flu/about/index.html), by the U.S. CDC\n- [Key Facts About Seasonal Flu Vaccine](https://www.cdc.gov/flu/prevent/keyfacts.htm), by the U.S. CDC", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2041219, "CreatorUserId": 8683201, "OwnerUserId": 8683201.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3385921.0, "CurrentDatasourceVersionId": 3437446.0, "ForumId": 2066159, "Type": 2, "CreationDate": "03/31/2022 07:46:17", "LastActivityDate": "03/31/2022", "TotalViews": 3715, "TotalDownloads": 286, "TotalVotes": 17, "TotalKernels": 1}]
|
[{"Id": 8683201, "UserName": "soundslikedata", "DisplayName": "Vishnu Pandey", "RegisterDate": "10/24/2021", "PerformanceTier": 1}]
|
# # **Data Analytics Competition FIND IT 2023 - Introduction**
# Swine flu, caused by the H1N1 flu virus, is a type of influenza A virus that can cause seasonal flu. Its symptoms are similar to those of seasonal flu. During the 2009-2010 flu season, the H1N1 virus caused a respiratory tract infection in humans known as swine flu, leading the WHO to declare it a pandemic. The H1N1 flu virus became one of the strains that cause seasonal flu after the pandemic ended.
# Flu vaccines are now available to protect against H1N1 flu. COVID-19 is also categorized as a seasonal flu disease and is delaying efforts to produce a vaccine for seasonal flu.
# The data set we are using represents individuals from the **2009 National H1N1 Flu Survey (NHFS)** conducted by the CDC.
# ## **Goals and Evaluation:**
# The goal of our analysis is to predict the likelihood of people getting H1N1 flu and receiving their yearly flu vaccine. We will specifically forecast two probabilities: one for the **H1N1 vaccine** and one for the **seasonal flu vaccine**.
# The performance of the model will be measured using the area under the receiver operating characteristic curve **(ROC AUC)** for each of the two target variables. The overall score will be calculated by taking the mean of these two scores. A **higher score** indicates better performance.
# **Made by BukanRISTEK**
# - Fathi Qushoyyi Ahimsa
# - Edward Salim
# - Muhammad Fakhri Robbani
# # **Load Libraries**
# Utilities
import pandas as pd
import numpy as np
pd.set_option(
"display.max_columns", None
) # Set the option to display all columns in pandas
# Google colab
from google.colab import drive
drive.mount("/content/drive") # Mount the Google Drive in Google Colab
# Data visualization
import matplotlib.pyplot as plt
import seaborn as sns
# Feature engineering
from sklearn.feature_selection import (
mutual_info_classif as mic,
) # Import mutual_info_classif for feature selection
from sklearn.impute import (
SimpleImputer,
) # Import SimpleImputer for missing value imputation
# Modeling
import optuna # Import optuna library for hyperparameter optimization
from sklearn.model_selection import KFold # Import KFold for cross-validation
from sklearn.metrics import (
roc_auc_score as ras,
) # Import roc_auc_score for evaluation metric
from sklearn.model_selection import (
cross_val_score,
) # Import cross_val_score for cross-validation
from sklearn.model_selection import GridSearchCV # Import GridSearchCV for grid search
from sklearn.model_selection import (
RandomizedSearchCV,
) # Import RandomizedSearchCV for randomized search
# Models
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.neural_network import MLPClassifier
import xgboost as xgb
import lightgbm as lgbm
import catboost as cb
# # **1. Data Wrangling**
# To kick off the analysis, we will begin with data wrangling, which encompasses a series of tasks including data cleaning, data transformation, data integration, and data reduction. Through these processes, we aim to ensure the accuracy, completeness, and analyzability of the dataset.
# ### **1.1 Load Dataset**
train_feature = pd.read_csv(
"/content/drive/MyDrive/DAC-FINDIT/training_set_features.csv",
index_col="respondent_id",
)
train_label = pd.read_csv(
"/content/drive/MyDrive/DAC-FINDIT/training_set_labels.csv",
index_col="respondent_id",
)
sample_submission = pd.read_csv(
"/content/drive/MyDrive/DAC-FINDIT/submission_format.csv", index_col="respondent_id"
)
train = train_feature.merge(
train_label, on="respondent_id"
) # Merge train_feature and train_label
test = pd.read_csv(
"/content/drive/MyDrive/DAC-FINDIT/test_set_features.csv", index_col="respondent_id"
)
train.head()
# Upon examining the given dataframe, it is evident that all the columns in the dataset are categorical in nature.
# ### **1.2 Data Inspection**
print(f"Train shape: {train.shape}") # See number of rows and columns in train dataset
print(f"Test shape: {test.shape}") # See number of rows and columns in test dataset
train.info() # Displays a summary of the dataframe
# We'll create a function to evaluate the missing value proportions in the train and test datasets. This function will facilitate the assessment of missing data and simplify future missing value checks.
def missing_values(train, test):
miss_perc_df = pd.DataFrame(
index=["train_miss", "test_miss"], # Set index of dataframe
columns=test.columns, # Set columns of dataframe
)
# Loop through the columns of the "test" dataframe and calculate the percentage of missing values for both the "train" and "test" dataframes
for i, col in enumerate(test.columns):
miss_perc_df.loc["train_miss", col] = np.round(
train.isna().sum()[i] / train[col].shape[0] * 100, 2
) # Calculate missing values in train dataset
miss_perc_df.loc["test_miss", col] = np.round(
test.isna().sum()[i] / test[col].shape[0] * 100, 2
) # Calculate missing values in test dataset
# Convert the values in the "miss_perc_df" dataframe to strings and append a "%" sign
miss_perc_df = miss_perc_df.astype(str) + "%"
return miss_perc_df
missing_values(train, test)
# From the given dataframe, we can gather the following information:
# * The columns `health_insurance`, `employment_industry`, and `employment_occupation` contain a significant number of missing values.
# * The `income_poverty` column also has a considerable amount of missing data.
# * The `doctor_recc_h1n1` and `doctor_recc_seasonal` columns share the same proportion of missing values.
# Since the columns with the dtype `float64` are already ordinal encoded, we can directly utilize them as features for our machine learning models without any additional preprocessing.
# To streamline the preprocessing of our data, we will segregate the columns into two separate variables based on their types: ordinal and nominal. This division will enable us to apply specific preprocessing techniques more efficiently to the nominal columns.
# Select columns from the 'train' dataframe that have a data type of float64 and convert them to a list.
ordinal_cols = train.select_dtypes("float64").columns.to_list()
# Select columns from the 'train' dataframe that have a data type of object (strings) and convert them to a list.
object_cols = train.select_dtypes("object").columns.to_list()
behavioral_cols = [
"behavioral_antiviral_meds",
"behavioral_avoidance",
"behavioral_face_mask",
"behavioral_wash_hands",
"behavioral_large_gatherings",
"behavioral_outside_home",
"behavioral_touch_face",
]
opinion_cols = [
"opinion_h1n1_vacc_effective",
"opinion_h1n1_risk",
"opinion_h1n1_sick_from_vacc",
"opinion_seas_vacc_effective",
"opinion_seas_risk",
"opinion_seas_sick_from_vacc",
]
# Given that all the columns in the dataset are categorical, it would be beneficial to examine the mode for each column. This is particularly important as we are likely to use the mode to impute missing values in our dataset.
# Ordinal columns
ord_summary = train.loc[:, ordinal_cols].describe()
pd.concat([ord_summary, train[ordinal_cols].mode()]).rename(index={0: "mean"})
# Nominal columns
train.loc[:, object_cols].describe()
print("Ordinal columns:", len(ordinal_cols))
print("Nominal columns:", len(object_cols))
# To compare missing values with other categories, we will create a copy of the train dataframe and fill in all the missing values with "Missing". This will allow us to perform a crosstabular analysis and compare the missing values to other categories more effectively.
nan_to_miss = train.copy()
# Replace all missing values in the 'nan_to_miss' dataframe with the string "Missing"
nan_to_miss.fillna("Missing", inplace=True)
train[train["education"].isna()].tail()
# We observe that the presence of a missing value in the `education` column is usually accompanied by missing values in the `income_poverty`, `marital_status`, `rent_or_own`, `employment_status`, `employment_industry`, and `employment_occupation` columns.
# Computes a frequency table or contingency table showing the distribution of two categorical variables in the DataFrame
pd.crosstab(nan_to_miss["doctor_recc_h1n1"], nan_to_miss["doctor_recc_seasonal"])
# It is evident that when a row in the `doctor_recc_seasonal` column contains a NaN value, the corresponding row in the `doctor_recc_h1n1` column will also be NaN. This specific missing value pattern holds significant importance and should be captured by our model.
pd.crosstab(nan_to_miss["employment_status"], nan_to_miss["employment_industry"])
pd.crosstab(nan_to_miss["employment_status"], nan_to_miss["employment_occupation"])
# An intriguing observation is that when a row in the `employment_status` column is missing or when a person is categorized as "Not in Labor Force" or "Unemployed," we can guarantee that the corresponding rows in the `employment_industry` and `employment_occupation` columns will also be missing. This pattern aligns with our expectations and makes logical sense (MNAR).
# To preserve and capture the observed pattern, we can introduce a new category (i.e. "None") in the `employment_industry` and `employment_occupation` columns specifically for cases where a person is **not in the labor force** or **unemployed**. By including this new category, we maintain the integrity of the pattern within our data, allowing our model to effectively utilize this information.
# ### **1.3 Handling Missing Values**
# To capture the missing value pattern in the dataset, we will create a new category called "Unknown" for nominal columns and assign a value of 2 as "Unknown" for ordinal columns in both the training and test data. This approach allows us to effectively handle and represent missing values in our dataset without significantly increasing the length of our code.
for df in [train, test]:
df["doctor_recc_h1n1"].fillna(2.0, inplace=True)
df["doctor_recc_seasonal"].fillna(2.0, inplace=True)
df["health_insurance"].fillna(2.0, inplace=True)
df["education"].fillna("Unknown", inplace=True)
df["marital_status"].fillna("Unknown", inplace=True)
df["rent_or_own"].fillna("Unknown", inplace=True)
df["income_poverty"].fillna("Unknown", inplace=True)
# We can safely impute missing values in the `employment_status` column with "Not in Labor Force" because when a row has a missing value in `employment_status`, the corresponding rows in `employment_industry` and `employment_occupation` will also be missing.
# This relationship is similar to the cases where the `employment_status` is "Not in Labor Force" or "Unemployed". We choose "Not in Labor Force" as the imputation value because it is the second most frequent value in the column.
for df in [train, test]:
df["employment_status"].fillna("Not in Labor Force", inplace=True)
for df in [train, test]:
for row in range(
df.index[0], df.index[-1] + 1
): # Iterate over the rows of the DataFrame.
if (df.loc[row, "employment_status"] == "Not in Labor Force") or (
df.loc[row, "employment_status"] == "Unemployed"
):
df.loc[
row, "employment_occupation"
] = "None" # Set the value of 'employment_occupation' to "None" for the current row.
df.loc[
row, "employment_industry"
] = "None" # Set the value of 'employment_industry' to "None" for the current row.
# To avoid introducing bias, we will impute the missing values in the **respondent's opinion columns** with the value 3, which represents "Don't know." This approach ensures that the missing values are treated uniformly and does not influence the analysis based on the respondents' opinions.
for df in [train, test]:
df[opinion_cols] = df[opinion_cols].fillna(3.0)
missing_values(train, test)
# For the remaining missing values in both the train and test datasets, which are **mostly** missing at random (MAR), we will fill them with the mode of each respective column. Since the dataset consists of categorical variables, using the mode as the imputation strategy helps maintain the categorical nature of the data.
imputer = SimpleImputer(
strategy="most_frequent"
) # Create an instance of SimpleImputer with 'most_frequent' strategy.
# Ordinal Columns
train[ordinal_cols] = imputer.fit_transform(
train[ordinal_cols]
) # Fill missing values in ordinal columns of 'train' DataFrame using the fitted imputer.
test[ordinal_cols] = imputer.transform(
test[ordinal_cols]
) # Fill missing values in ordinal columns of 'test' DataFrame using the fitted imputer.
# Nominal Columns
train[object_cols] = imputer.fit_transform(
train[object_cols]
) # Fill missing values in nominal columns of 'train' DataFrame using the fitted imputer.
test[object_cols] = imputer.transform(
test[object_cols]
) # Fill missing values in nominal columns of 'test' DataFrame using the fitted imputer.
missing_values(train, test)
# ### **1.4 Handling Other Issues**
# To ensure compatibility with various machine learning algorithms and statistical techniques, we will convert the columns with dtype `float64` to `int64`. This conversion will effectively transform these variables from continuous to discrete, as many algorithms require discrete variables as input. By making these variables discrete, we can leverage the full range of analytical tools and methods available for categorical data analysis.
train[ordinal_cols] = train[ordinal_cols].astype(
"int64"
) # Convert train[ordinal_cols] to integer
test[ordinal_cols] = test[ordinal_cols].astype(
"int64"
) # Convert test[ordinal_cols] to integer
print("Train duplicates:", train.duplicated().sum()) # Find duplicates value in train
print("Test duplicates:", test.duplicated().sum()) # Find duplicates value in test
# The presence of a single duplicated row is negligible and is unlikely to significantly impact the analysis. Therefore, we can safely ignore this duplicated row without compromising the integrity or accuracy of our results.
# When categorical variables are represented as ranges or intervals, such as `age_group`, assigning a single value to each category helps in reducing the loss of information and provides a more precise estimate for analysis. By assigning a midpoint, we treat the variable as if it takes on a continuous scale, albeit with some level of discretization.
# Define a dictionary to map age_group categories to numerical values
dict_age_group = {
"65+ Years": 75,
"55 - 64 Years": 60,
"45 - 54 Years": 50,
"18 - 34 Years": 26,
"35 - 44 Years": 40,
}
for df in [train, test]:
# Replace the values in the age_group column with the corresponding numerical value from dict_age_group
df["age_group"] = df["age_group"].map(dict_age_group)
# To assign a more representative midpoint to the "65+ Years" age category, an approximate value of 75 can be considered. This choice is based on global data trends indicating that the average age at which many people pass away in various countries is approximately 85 years.
# Please note that this information is sourced from a study titled "Deaths: final data for 2009" (2011, December 29) available on PubMed. It is important to mention that this specific data is used solely for the purpose of assigning a midpoint to the age group range of 65+ years. https://pubmed.ncbi.nlm.nih.gov/24974587/#:~:text=Life%20expectancy%20at%20birth%20rose,74%2C%20and%2075%2D84.
# Check inconsistency -- Ordinal columns
for col in ordinal_cols:
print(
col + ":", sorted(train[col].unique())
) # Print unique values in every columns, sorted
# Check inconsistency -- Object columns
for col in object_cols:
print(col + ":", train[col].unique()) # Print unique values in every columns
print()
# After careful manual inspection, we have determined that there are no instances of inconsistent data entries in the dataset.
# Since the dataset exclusively consists of categorical columns, there is no need for **normalization**. Categorical features do not possess a natural numerical order or scale, rendering normalization irrelevant in this context.
# # **2. Exploratory Data Analysis**
# ### **2.1 Univariate Exploration**
# We will begin by directing our attention to comprehending the distribution of categorical data, prioritizing an understanding of its characteristics without delving into its relationship with other variables.
# Select the ordinal columns (non-binary) with more than 2 unique values.
non_binary_cols = [col for col in ordinal_cols if train[col].nunique() > 2]
fig, ax = plt.subplots(
nrows=4, ncols=4, figsize=(30, 20)
) # Creating subplots for countplots with specified dimensions and figure size.
for i, col in enumerate(
non_binary_cols
): # Iterate over the non-binary ordinal columns.
bar = sns.countplot(
data=train, y=col, ax=ax[i // 4, i % 4], palette="ch:.25"
) # Create a countplot for each column and assign it to the corresponding subplot.
bar.bar_label(bar.containers[0]) # Add labels to the bars in the countplot.
# Select the ordinal columns (binary) with less than or equal to 2 unique values.
binary_cols = [col for col in ordinal_cols if train[col].nunique() <= 2]
fig, ax = plt.subplots(nrows=3, ncols=4, figsize=(30, 15))
for i, col in enumerate(binary_cols):
bar = sns.countplot(data=train, y=col, ax=ax[i // 4, i % 4], palette="coolwarm")
bar.bar_label(bar.containers[0])
# Considering that the "None" category overwhelmingly dominates all other categories in the `employment_industry` and `employment_occupation` columns, it is apparent that plotting these columns would not yield any valuable insights. Therefore, we will exclude them from our analysis.
# Countplot for Object columns
fig, ax = plt.subplots(nrows=4, ncols=3, figsize=(28, 20))
for i, col in enumerate(object_cols):
if col not in ["employment_industry", "employment_occupation"]:
bar = sns.countplot(data=train, x=col, ax=ax[i // 3, i % 3], palette="rainbow")
bar.bar_label(bar.containers[0])
# ### **2.2. Bivariate Exploration**
# Our attention now turns to comprehending the correspondence and relationship between two specific columns, particularly in relation to the target variable. The aim of this endeavor is to gain a deeper understanding of the fundamental patterns and connections inherent within our dataset.
fig, ax = plt.subplots(nrows=2, ncols=4, figsize=(25, 10))
for i, col in enumerate(behavioral_cols):
bar = sns.barplot(
data=train,
y=col,
x="chronic_med_condition",
ax=ax[i // 4, i % 4],
palette="rainbow",
)
bar.bar_label(bar.containers[0])
# The bar plot indicates a notable pattern where individuals with specific chronic medical conditions, such as asthma, diabetes, heart conditions, kidney conditions, sickle cell anemia, neurological or neuromuscular conditions, liver conditions, or weakened immune systems due to chronic illnesses or medications, tend to exhibit more proactive behavioral precautions to prevent the occurrence of flu-like illnesses.
# Create a figure with subplots to plot the barplots and violinplots.
fig, ax = plt.subplots(nrows=3, ncols=4, figsize=(25, 13))
# Health related columns
for i, col in enumerate(["health_worker", "doctor_recc_h1n1", "doctor_recc_seasonal"]):
bar = sns.barplot(
data=train, y="h1n1_vaccine", x=col, ax=ax[i, 0], palette="Paired"
)
bar.bar_label(bar.containers[0])
# Plot a violinplot for 'h1n1_vaccine' on the i-th subplot.
sns.violinplot(data=train, y="h1n1_vaccine", x=col, ax=ax[i, 1], palette="Paired")
bar = sns.barplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 2], palette="Paired"
)
bar.bar_label(bar.containers[0])
# Plot a violinplot for 'seasonal_vaccine' on the i-th subplot.
sns.violinplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 3], palette="Paired"
)
# The visualization reveals clear patterns. Healthcare workers are more likely to receive both the H1N1 flu vaccine and the seasonal flu vaccine.
# Similarly, individuals recommended the H1N1 vaccine by a doctor are highly likely to receive both the H1N1 vaccine and the seasonal vaccine. Moreover, those recommended the seasonal vaccine are also highly likely to receive both the seasonal and H1N1 vaccines. These findings highlight the influence of healthcare worker status and doctor recommendations on vaccination behavior, indicating a strong association with vaccine uptake.
# Create a figure with subplots to plot the barplots and violinplots.
fig, ax = plt.subplots(nrows=4, ncols=2, figsize=(30, 15))
# Employment related columns
# Create barplots and violinplots for 'h1n1_vaccine' for each column.
for i, col in enumerate(["employment_industry", "employment_occupation"]):
bar = sns.barplot(
data=train, y="h1n1_vaccine", x=col, ax=ax[i, 0], palette="rainbow"
)
bar.bar_label(bar.containers[0])
sns.violinplot(data=train, y="h1n1_vaccine", x=col, ax=ax[i, 1], palette="rainbow")
# Create barplots and violinplots for 'seasonal_vaccine' for each column.
for i, col in enumerate(["employment_industry", "employment_occupation"]):
bar = sns.barplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i + 2, 0], palette="rainbow"
)
bar.bar_label(bar.containers[0])
sns.violinplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i + 2, 1], palette="rainbow"
)
# The bar plot clearly indicates that individuals employed in the "haxffmxo" and "fcxhlnwr" industries have a higher likelihood of receiving both the H1N1 and seasonal vaccines. Similarly, those with employment occupations such as "dcjcmpih" and "cmhcxjea" also exhibit a higher likelihood of receiving both vaccines. These findings emphasize the positive association between specific employment industries and occupations with vaccine uptake for both H1N1 and seasonal viruses.
# Create a figure with subplots to plot the barplots and violinplots.
fig, ax = plt.subplots(nrows=4, ncols=4, figsize=(25, 17))
# Opinion related columns
for i, col in enumerate(
[
"opinion_seas_risk",
"opinion_seas_vacc_effective",
"opinion_h1n1_risk",
"opinion_h1n1_vacc_effective",
]
):
bar = sns.barplot(
data=train, y="h1n1_vaccine", x=col, ax=ax[i, 0], palette="Spectral"
)
bar.bar_label(bar.containers[0])
sns.violinplot(data=train, y="h1n1_vaccine", x=col, ax=ax[i, 1], palette="Spectral")
bar = sns.barplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 2], palette="Spectral"
)
bar.bar_label(bar.containers[0])
sns.violinplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 3], palette="Spectral"
)
# There is a clear relationship observed in the data where individuals who perceive the H1N1 vaccine and seasonal vaccines to be effective are more likely to receive both vaccines. Additionally, individuals who believe that the risk of getting sick with H1N1 flu and seasonal flu without vaccination is higher also demonstrate a higher likelihood of receiving both the H1N1 vaccine and the seasonal vaccine.
# These findings highlight the significant impact of perceived vaccine effectiveness and perceived risk of illness on the decision to receive the H1N1 and seasonal vaccines.
fig, ax = plt.subplots(nrows=5, ncols=2, figsize=(25, 21))
# Other interesting columns
for i, col in enumerate(
[
"h1n1_concern",
"chronic_med_condition",
"age_group",
"employment_status",
"household_children",
]
):
bar = sns.barplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 0], palette="coolwarm"
)
bar.bar_label(bar.containers[0])
sns.violinplot(
data=train, y="seasonal_vaccine", x=col, ax=ax[i, 1], palette="coolwarm"
)
# The bar plot reveals additional insights. Individuals who express concern about the H1N1 flu, have chronic medical conditions, belong to older age groups, are not in the labor force, or have zero children in their household exhibit a higher likelihood of receiving the seasonal vaccine.
# These findings suggest that individuals who fall into these categories are more inclined to prioritize their health and take proactive measures by receiving the seasonal vaccine.
# ### **Summary:**
# * People with chronic medical conditions are more likely to take preventive measures against flu-like illnesses.
# * Specific employment industries and occupations show higher vaccine uptake for both H1N1 and seasonal flu vaccines.
# * Healthcare workers have a higher likelihood of receiving both vaccines.
# * Doctor recommendations strongly influence vaccine uptake.
# * Perceived vaccine effectiveness and risk of illness are important factors.
# * Other factors like concern about H1N1, older age, not being in the labor force, and no children in the household are associated with higher seasonal flu vaccine uptake.
# # **3. Feature Engineering**
# To further enhance the predictive performance of our model, we will create additional relevant features that capture important patterns and relationships in the data. We propose the following new features:
# * `good_behavioral_count` -- represent the cumulative count of flu precaution behaviors practiced by an individual. A higher value would indicate a greater adherence to flu prevention measures.
# * `has_doc_recc` -- indicate whether a person has received a recommendation from a doctor for either the H1N1 vaccine or the seasonal flu vaccine (binary). A value of 1 represents that a doctor has recommended at least one of the vaccines, and a value of 0 indicates no doctor recommendation.
# Count-based features are beneficial for tree-based models because these models lack the inherent ability to aggregate information across multiple features simultaneously. By introducing count-based features, we allow tree models to leverage valuable insights into the occurrence or frequency of particular events or behaviors.
for df in [train, test]:
# Calculate the sum of behavioral columns and create a new column "good_behavioral_count"
df["good_behavioral_count"] = df[behavioral_cols].sum(axis=1)
for i in range(df.index[0], df.index[-1] + 1):
# Check if both "doctor_recc_h1n1" and "doctor_recc_seasonal" columns have a value of 2 (Unknown)
if df.loc[i, ["doctor_recc_h1n1", "doctor_recc_seasonal"]].isin([2]).all():
# If both columns have a value of 2, set "has_doc_recc" to 0
df.loc[i, "has_doc_recc"] = 0
else:
# If any of the columns have a value other than 2, set "has_doc_recc" to 1
df.loc[i, "has_doc_recc"] = 1
# Append the new ordinal feature to the list of ordinal columns
for new_ord_feature in ["good_behavioral_count", "has_doc_recc"]:
ordinal_cols.append(new_ord_feature)
# We will proceed with encoding the nominal columns in our dataset to ensure that categorical variables are in a suitable format for machine learning algorithms. We will use two different encoding techniques:
# * **Ordinal Encoding** -- apply to `age_group` and `education` columns in order to preserve the inherent order or hierarchy present in these columns, allowing the algorithm to understand the relative differences between categories.
# * **One-Hot Encoding** -- apply to `race`, `sex`, `marital_status`, `rent_or_own`, `employment_status`, `hhs_geo_region`, `census_msa`, `employment_industry`, `employment_occupation`, `income_poverty` in order to indicate the presence or absence of a category in the observation.
# To ensure consistency and avoid discrepancies, we will merge the test and train data before encoding the categorical variables. This merging allows us to encode the variables based on the entire dataset, eliminating any potential issues if a category is present in only one dataset. Once the encoding is complete, we will separate the merged dataset back into their respective test and train datasets.
# Get the number of rows in the train dataframe
train_rows = train.shape[0]
# Concatenate the train and test dataframes vertically
merged_df = pd.concat([train, test])
# Encode object columns using Ordinal Encoding
for col in ["age_group", "education"]:
merged_df[col], _ = merged_df[col].factorize()
# Encode object columns using One-Hot Encoding
merged_df = pd.get_dummies(
merged_df,
columns=[
"race",
"sex",
"marital_status",
"rent_or_own",
"employment_status",
"hhs_geo_region",
"census_msa",
"employment_industry",
"employment_occupation",
"income_poverty",
],
)
# Separate the merged dataframe back into train and test dataframes
train = merged_df.iloc[:train_rows]
test = merged_df.iloc[train_rows:].drop(columns=["h1n1_vaccine", "seasonal_vaccine"])
train.head()
# We will employ mutual information to measure the relationship between two columns. It quantifies how knowing one variable reduces uncertainty about the other. Higher mutual information indicates a stronger relationship, while lower values indicate weaker or no relationship. This helps confirm the relationships found during EDA.
# Mutual Information function to calculate MI scores
def make_mi_scores(X, y, discrete_features):
# Calculate Mutual Information (MI) scores between features (X) and target (y)
mi_scores = mic(X, y, discrete_features=discrete_features)
# Convert the MI scores to a Pandas Series with feature names as index and sort in descending order
mi_scores = pd.Series(mi_scores, name="MI Scores", index=X.columns).sort_values(
ascending=False
)
return mi_scores
# Create a copy of the train dataframe as X
X = train.copy()
# Separate the target variables from X and store them in y_h1n1 and y_seas
y_h1n1 = X.pop("h1n1_vaccine")
y_seas = X.pop("seasonal_vaccine")
# Identify the discrete features in X (features with dtype 'int64')
discrete_features = X.dtypes == "int64"
# H1N1 MI Score
mi_scores_h1n1 = make_mi_scores(X, y_h1n1, discrete_features)
# Convert the MI scores into a transposed DataFrame
pd.DataFrame(mi_scores_h1n1).T
# Seasonal MI Score
mi_scores_seas = make_mi_scores(X, y_seas, discrete_features)
# Convert the MI scores into a transposed DataFrame
pd.DataFrame(mi_scores_seas).T
# # **4. Modeling**
# To start modeling for the classification problem, we'll choose the best model for both the h1n1_vaccine and seasonal_vaccine targets. Then, we'll test the selected model with default parameters and evaluate its performance using the ROC AUC metric.
# By starting with the default parameters, we can establish a baseline performance for the model. This will allow us to compare and assess the effectiveness of subsequent parameter tuning and optimizations.
models = {
# "lr": LogisticRegression(random_state=42), # Error
"dtc": DecisionTreeClassifier(random_state=42),
"rfc": RandomForestClassifier(random_state=42),
"knn": KNeighborsClassifier(),
"gnb": GaussianNB(),
"svc": SVC(random_state=42),
# 'xgboost': xgb.XGBClassifier(random_state=42), # Error
# 'lightgbm': lgbm.LGBMClassifier(random_state=42), # Error
"catboost": cb.CatBoostClassifier(verbose=0, random_state=42),
"nn": MLPClassifier(random_state=42, hidden_layer_sizes=(10, 10), max_iter=500),
}
# We will utilize k-fold cross-validation to obtain a more reliable and robust estimate of our model's performance. By leveraging k-fold cross-validation, we can gain a more comprehensive understanding of our model's effectiveness and its ability to generalize to unseen data.
# Function for performing k-fold cross-validation
def kfold_cv(model, X, y):
# Define KFold cross-validation with 5 splits, random state of 42, and shuffling of data
kf = KFold(n_splits=5, random_state=42, shuffle=True)
scores = [] # List to store the evaluation scores
# Iterate over the train and validation splits
for train_idx, test_idx in kf.split(X, y):
# Split the data into training and validation sets based on the indices
X_train, X_val = X.iloc[train_idx], X.iloc[test_idx]
y_train, y_val = y.iloc[train_idx], y.iloc[test_idx]
# Fit the model on the training data
model.fit(X_train, y_train)
# Make predictions on the validation data
predictions = model.predict(X_val)
# Calculate the evaluation score (here, using RAS macro-average)
ras_score = ras(y_val, predictions, average="macro")
# Append the score to the list of scores
scores.append(ras_score)
return np.mean(
scores
) # Return the mean of the scores as the overall evaluation score
# Prepare the input features and target variables for H1N1 vaccine prediction
X_h1n1 = train.drop(["h1n1_vaccine", "seasonal_vaccine"], axis=1)
y_h1n1 = train.pop("h1n1_vaccine")
# Prepare the input features and target variables for seasonal flu vaccine prediction
X_seas = train.drop("seasonal_vaccine", axis=1)
y_seas = train.pop("seasonal_vaccine")
# H1N1 Vaccine
# Iterate over the models dictionary
for name, model in models.items():
# Perform k-fold cross-validation and obtain the evaluation score
score = kfold_cv(model, X_h1n1, y_h1n1)
# Print the model name and its corresponding score
print(f"{name}: {score}")
# Seasonal Vaccine
for name, model in models.items():
score = kfold_cv(model, X_seas, y_seas)
print(f"{name}: {score}")
# The results indicate that the CatBoost model consistently outperforms the other models, yielding higher scores. This suggests that the CatBoost model is better suited for our classification task and exhibits superior predictive performance compared to the alternative models.
# Now, we will proceed to utilize CatBoost with tuned parameters obtained from Optuna to further enhance our prediction score. By incorporating the optimized parameter settings, we aim to improve the performance of the CatBoost model and achieve an even higher prediction score.
h1n1_param = {
"iterations": 600, # Number of boosting iterations
"learning_rate": 0.02857091731949716, # Learning rate for boosting
"random_strength": 1, # Random strength for boosting
"bagging_temperature": 2, # Bagging temperature for boosting
"max_bin": 21, # Maximum number of bins for numerical features
"grow_policy": "Lossguide", # Tree growth policy
"min_data_in_leaf": 18, # Minimum number of samples required in a leaf
"max_depth": 6, # Maximum depth of trees
"l2_leaf_reg": 30.38300354991344, # L2 regularization coefficient
"one_hot_max_size": 13, # Maximum size of one-hot encoding for categorical features
"auto_class_weights": "Balanced", # Automatic class weights adjustment
"verbose": 0, # Verbosity level
}
seas_param = {
"iterations": 800, # Number of boosting iterations
"learning_rate": 0.017179768319739466, # Learning rate for boosting
"random_strength": 0, # Random strength for boosting
"bagging_temperature": 7, # Bagging temperature for boosting
"max_bin": 106, # Maximum number of bins for numerical features
"grow_policy": "Lossguide", # Tree growth policy
"min_data_in_leaf": 20, # Minimum number of samples required in a leaf
"max_depth": 4, # Maximum depth of trees
"l2_leaf_reg": 45.5914670615121, # L2 regularization coefficient
"one_hot_max_size": 8, # Maximum size of one-hot encoding for categorical features
"auto_class_weights": "Balanced", # Automatic class weights adjustment
"verbose": 0, # Verbosity level
}
# Create a CatBoostClassifier model with the H1N1 vaccine parameters and fit it to the data
model_h1n1 = cb.CatBoostClassifier(**h1n1_param)
model_h1n1.fit(X_h1n1, y_h1n1)
# Create a CatBoostClassifier model with the seasonal flu vaccine parameters and fit it to the data
model_seas = cb.CatBoostClassifier(**seas_param)
model_seas.fit(X_seas, y_seas)
score = kfold_cv(model_h1n1, X_h1n1, y_h1n1)
print(f"Catboost H1N1: {score}")
score = kfold_cv(model_seas, X_seas, y_seas)
print(f"CatBoost Seasonal: {score}")
# By incorporating the optimized parameters into CatBoost, we observe a significant increase in the prediction score. With this improved performance, we can now proceed to follow the prescribed format for the submission file in this competition.
# Create a DataFrame for the submission
submission = pd.DataFrame(
{
"respondent_id": test.index, # Use the respondent IDs from the test data
"h1n1_vaccine": model_h1n1.predict_proba(test)[
:, 1
], # Use the probability of the positive class for H1N1 vaccine
"seasonal_vaccine": model_seas.predict_proba(test)[
:, 1
], # Use the probability of the positive class for seasonal flu vaccine
}
)
# Save the submission DataFrame to a CSV file without including the index
submission.to_csv("submission.csv", index=False)
submission.head()
#
# # **Hyperparameter Tuning**
# We will utilize Optuna for hyperparameter tuning of our models, specifically focusing on the CatBoost algorithm. This technique has shown significant improvement in our ROC AUC prediction score, with an increase of over 0.05.
# By leveraging Optuna's optimization capabilities, we aim to fine-tune the hyperparameters of the CatBoost model to maximize its predictive performance and enhance the overall accuracy of our predictions.
# Function for tuning a model using k-fold cross-validation
def tune_kfold_cv(model, X, y, metric, n_folds=69):
# Create a k-fold cross-validation object with specified number of folds
skfold = KFold(n_splits=n_folds, random_state=0, shuffle=True)
# List to store the scores obtained from each fold
scores = []
# Iterate over the folds
for train_index, test_index in skfold.split(X, y):
# Split the data into training and testing sets based on the fold indices
X_train, X_test = X.iloc[train_index], X.iloc[test_index]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
# Fit the model on the training data and evaluate it on the testing data
model.fit(X_train, y_train, eval_set=[(X_test, y_test)], verbose=0)
predictions = model.predict(X_test.values)
# Calculate the score using the specified evaluation metric
score = metric(y_test, predictions, average="macro")
# Append the score to the list of scores
scores.append(score)
# Print the ROC AUC score for the current fold
print(f"ROC AUC: {score:.3f}")
# Return the list of scores
return scores
# Objective function for hyperparameter optimization using Optuna
def objective(trial, data=X_h1n1, target=y_h1n1):
# Define the hyperparameter search space using trial suggestions
params = {
"iterations": trial.suggest_int("iterations", 100, 2000, step=100),
"learning_rate": trial.suggest_float("learning_rate", 1e-5, 0.1),
"random_strength": trial.suggest_int("random_strength", 0.0, 10.0),
"bagging_temperature": trial.suggest_int("bagging_temperature", 0.0, 10.0),
"max_bin": trial.suggest_int("max_bin", 1, 255),
"grow_policy": trial.suggest_categorical(
"grow_policy", ["SymmetricTree", "Depthwise", "Lossguide"]
),
"min_data_in_leaf": trial.suggest_int("min_data_in_leaf", 1, 20),
"max_depth": trial.suggest_int("max_depth", 1, 10),
"l2_leaf_reg": trial.suggest_float("l2_leaf_reg", 1e-8, 100.0),
"one_hot_max_size": trial.suggest_int("one_hot_max_size", 2, 16),
"auto_class_weights": trial.suggest_categorical(
"auto_class_weights", ["None", "Balanced", "SqrtBalanced"]
),
"verbose": 0,
"early_stopping_rounds": 200,
}
# Create a CatBoostClassifier with the suggested hyperparameters
model = cb.CatBoostClassifier(**params)
# Perform k-fold cross-validation with the tuned model and calculate the average score
scores = tune_kfold_cv(model, X_h1n1, y_h1n1, ras, 8)
return np.sum(scores) / 8
# Create an Optuna study with the direction set to maximize
study = optuna.create_study(direction="maximize")
# Run the optimization process using the objective function defined earlier, with a specified number of trials
study.optimize(objective, n_trials=30)
# Print the number of finished trials (i.e., the number of trials that have been executed)
print("Number of finished trials:", len(study.trials))
# Print the best trial's parameters, which correspond to the set of hyperparameters that yielded the highest objective value
print("Best trial:", study.best_trial.params)
# Assign the best parameters found during optimization to a variable
parameter_tuned = study.best_trial.params
# Create a CatBoostClassifier model using the best parameters
model = cb.CatBoostClassifier(**parameter_tuned)
# Calculate the score using k-fold cross-validation with the model and the H1N1 dataset
score = kfold_cv(model, X_h1n1, y_h1n1)
# Print the score obtained from k-fold cross-validation
print(f"CatBoost H1N1: {score}")
# Define the objective function for parameter optimization
def objective(trial, data=X_seas, target=y_seas):
# Define the parameter space to search within
params = {
"iterations": trial.suggest_int("iterations", 100, 2000, step=100),
"learning_rate": trial.suggest_float("learning_rate", 1e-5, 0.1),
"random_strength": trial.suggest_int("random_strength", 0.0, 10.0),
"bagging_temperature": trial.suggest_int("bagging_temperature", 0.0, 10.0),
"max_bin": trial.suggest_int("max_bin", 1, 255),
"grow_policy": trial.suggest_categorical(
"grow_policy", ["SymmetricTree", "Depthwise", "Lossguide"]
),
"min_data_in_leaf": trial.suggest_int("min_data_in_leaf", 1, 20),
"max_depth": trial.suggest_int("max_depth", 1, 10),
"l2_leaf_reg": trial.suggest_float("l2_leaf_reg", 1e-8, 100.0),
"one_hot_max_size": trial.suggest_int("one_hot_max_size", 2, 16),
"auto_class_weights": trial.suggest_categorical(
"auto_class_weights", ["None", "Balanced", "SqrtBalanced"]
),
"verbose": 0,
"early_stopping_rounds": 200,
}
# Create a CatBoostClassifier model with the suggested parameters
model = cb.CatBoostClassifier(**params)
# Perform k-fold cross-validation and calculate scores
scores = tune_kfold_cv(model, X_seas, y_seas, ras, 8)
# Return the average of the scores
return np.sum(scores) / 8
# Create an Optuna study object for parameter optimization
study = optuna.create_study(direction="maximize")
# Perform the optimization by running the objective function for a specified number of trials
study.optimize(objective, n_trials=30)
# Print the number of finished trials
print("Number of finished trials:", len(study.trials))
# Print the best trial's parameters
print("Best trial:", study.best_trial.params)
# Retrieve the tuned parameters from the best trial of the study
parameter_tuned = study.best_trial.params
# Create a CatBoostClassifier model using the tuned parameters
model = cb.CatBoostClassifier(**parameter_tuned)
# Compute the score using k-fold cross-validation with the tuned model
score = kfold_cv(model, X_seas, y_seas)
# Print the score for the CatBoost model on the seasonal vaccine prediction
print(f"CatBoost Seas: {score}")
| false | 0 | 12,226 | 4 | 14,553 | 12,226 |
||
129927390
|
# Importing necessary libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from alpha_vantage.timeseries import TimeSeries
from alpha_vantage.foreignexchange import ForeignExchange
import time
# Setting Seaborn style
sns.set()
# Define your API Key
key = "TZIE3ZXR18XZMI26"
# Initialize the TimeSeries class with your key
ts = TimeSeries(key)
# Get the data
data, metadata = ts.get_daily(symbol="WHEAT", outputsize="full")
# Convert the returned dict into a DataFrame
df_wheat = pd.DataFrame.from_dict(data).T
# Rename the columns for clarity
df_wheat.columns = ["open", "high", "low", "close", "volume"]
# Change the index to datetime format
df_wheat.index = pd.to_datetime(df_wheat.index)
# Sort the DataFrame by the index
df_wheat.sort_index(ascending=True, inplace=True)
# Filter data from 1990 to 03/31/2023
start_date = "1990-01-01"
end_date = "2023-03-31"
df_wheat = df_wheat.loc[start_date:end_date]
# Because of my lack of access to quality commodities data, I will use the stock price data of companies related to the agricultural industry as a proxy for commodity prices. We'll consider three companies for our analysis: Archer-Daniels-Midland Company (ADM), Bunge Limited (BG), and Deere & Company (DE). These companies are heavily involved in agricultural commodities, so their stock prices may reflect trends in the industry.
# Let's fetch the data for these three companies:
import pandas as pd
import numpy as np
import yfinance as yf
tickers = ["ADM", "BG", "DE"]
start_date = "2001-09-01"
end_date = "2023-03-31"
data = yf.download(tickers, start=start_date, end=end_date)
print(data)
import matplotlib.pyplot as plt
import seaborn as sns
# Plot the stock prices
plt.figure(figsize=(12, 8))
sns.set(style="darkgrid")
data["Adj Close"]["ADM"].plot(label="ADM")
data["Adj Close"]["BG"].plot(label="BG")
data["Adj Close"]["DE"].plot(label="DE")
plt.title("Stock Prices of ADM, BG, and DE")
plt.xlabel("Date")
plt.ylabel("Adjusted Close Price")
plt.legend()
plt.show()
# Perform time series analysis
returns = data["Adj Close"].pct_change().dropna()
mean_returns = returns.mean()
cov_matrix = returns.cov()
print("Mean Returns:")
print(mean_returns)
print("\nCovariance Matrix:")
print(cov_matrix)
correlation = returns.corr()
covariance = returns.cov()
cumulative_returns = (1 + returns).cumprod() - 1
plt.figure(figsize=(12, 8))
sns.set(style="darkgrid")
cumulative_returns["ADM"].plot(label="ADM")
cumulative_returns["BG"].plot(label="BG")
cumulative_returns["DE"].plot(label="DE")
plt.title("Cumulative Returns of ADM, BG, and DE")
plt.xlabel("Date")
plt.ylabel("Cumulative Return")
plt.legend()
plt.show()
rolling_mean = returns.rolling(window=30).mean()
rolling_std = returns.rolling(window=30).std()
plt.figure(figsize=(12, 8))
sns.set(style="darkgrid")
returns["ADM"].plot(label="ADM Returns")
rolling_mean["ADM"].plot(label="ADM Rolling Mean")
rolling_std["ADM"].plot(label="ADM Rolling Std")
plt.title("ADM Returns, Rolling Mean, and Rolling Std")
plt.xlabel("Date")
plt.ylabel("Returns")
plt.legend()
plt.show()
# As you can see above, this code calculates the rolling mean and standard deviation for a specific window size (30 in this example) and plots the stock returns, rolling mean, and rolling standard deviation for ADM.
# These are just a few examples of the historical/time series analysis you can perform. Based on the calculated metrics and visualizations, you could derive insights to guide relative value-based trading recommendations. It's important to note that trading recommendations should be based on a comprehensive analysis that takes into account additional factors such as market conditions, fundamental analysis, and risk management.
# We could also perform a regression analysis on the stock price data obtained from Yahoo Finance, we can use the statsmodels library in Python.
import statsmodels.api as sm
# Prepare the data for regression analysis
X = sm.add_constant(returns["ADM"]) # Independent variable (ADM stock returns)
Y = returns["DE"] # Dependent variable (DE stock returns)
# Fit the linear regression model
model = sm.OLS(Y, X)
results = model.fit()
# Print the regression results
print(results.summary())
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/927/129927390.ipynb
| null | null |
[{"Id": 129927390, "ScriptId": 38647261, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4454153, "CreationDate": "05/17/2023 13:32:56", "VersionNumber": 1.0, "Title": "Time Series Analysis", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 156.0, "LinesInsertedFromPrevious": 156.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# Importing necessary libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from alpha_vantage.timeseries import TimeSeries
from alpha_vantage.foreignexchange import ForeignExchange
import time
# Setting Seaborn style
sns.set()
# Define your API Key
key = "TZIE3ZXR18XZMI26"
# Initialize the TimeSeries class with your key
ts = TimeSeries(key)
# Get the data
data, metadata = ts.get_daily(symbol="WHEAT", outputsize="full")
# Convert the returned dict into a DataFrame
df_wheat = pd.DataFrame.from_dict(data).T
# Rename the columns for clarity
df_wheat.columns = ["open", "high", "low", "close", "volume"]
# Change the index to datetime format
df_wheat.index = pd.to_datetime(df_wheat.index)
# Sort the DataFrame by the index
df_wheat.sort_index(ascending=True, inplace=True)
# Filter data from 1990 to 03/31/2023
start_date = "1990-01-01"
end_date = "2023-03-31"
df_wheat = df_wheat.loc[start_date:end_date]
# Because of my lack of access to quality commodities data, I will use the stock price data of companies related to the agricultural industry as a proxy for commodity prices. We'll consider three companies for our analysis: Archer-Daniels-Midland Company (ADM), Bunge Limited (BG), and Deere & Company (DE). These companies are heavily involved in agricultural commodities, so their stock prices may reflect trends in the industry.
# Let's fetch the data for these three companies:
import pandas as pd
import numpy as np
import yfinance as yf
tickers = ["ADM", "BG", "DE"]
start_date = "2001-09-01"
end_date = "2023-03-31"
data = yf.download(tickers, start=start_date, end=end_date)
print(data)
import matplotlib.pyplot as plt
import seaborn as sns
# Plot the stock prices
plt.figure(figsize=(12, 8))
sns.set(style="darkgrid")
data["Adj Close"]["ADM"].plot(label="ADM")
data["Adj Close"]["BG"].plot(label="BG")
data["Adj Close"]["DE"].plot(label="DE")
plt.title("Stock Prices of ADM, BG, and DE")
plt.xlabel("Date")
plt.ylabel("Adjusted Close Price")
plt.legend()
plt.show()
# Perform time series analysis
returns = data["Adj Close"].pct_change().dropna()
mean_returns = returns.mean()
cov_matrix = returns.cov()
print("Mean Returns:")
print(mean_returns)
print("\nCovariance Matrix:")
print(cov_matrix)
correlation = returns.corr()
covariance = returns.cov()
cumulative_returns = (1 + returns).cumprod() - 1
plt.figure(figsize=(12, 8))
sns.set(style="darkgrid")
cumulative_returns["ADM"].plot(label="ADM")
cumulative_returns["BG"].plot(label="BG")
cumulative_returns["DE"].plot(label="DE")
plt.title("Cumulative Returns of ADM, BG, and DE")
plt.xlabel("Date")
plt.ylabel("Cumulative Return")
plt.legend()
plt.show()
rolling_mean = returns.rolling(window=30).mean()
rolling_std = returns.rolling(window=30).std()
plt.figure(figsize=(12, 8))
sns.set(style="darkgrid")
returns["ADM"].plot(label="ADM Returns")
rolling_mean["ADM"].plot(label="ADM Rolling Mean")
rolling_std["ADM"].plot(label="ADM Rolling Std")
plt.title("ADM Returns, Rolling Mean, and Rolling Std")
plt.xlabel("Date")
plt.ylabel("Returns")
plt.legend()
plt.show()
# As you can see above, this code calculates the rolling mean and standard deviation for a specific window size (30 in this example) and plots the stock returns, rolling mean, and rolling standard deviation for ADM.
# These are just a few examples of the historical/time series analysis you can perform. Based on the calculated metrics and visualizations, you could derive insights to guide relative value-based trading recommendations. It's important to note that trading recommendations should be based on a comprehensive analysis that takes into account additional factors such as market conditions, fundamental analysis, and risk management.
# We could also perform a regression analysis on the stock price data obtained from Yahoo Finance, we can use the statsmodels library in Python.
import statsmodels.api as sm
# Prepare the data for regression analysis
X = sm.add_constant(returns["ADM"]) # Independent variable (ADM stock returns)
Y = returns["DE"] # Dependent variable (DE stock returns)
# Fit the linear regression model
model = sm.OLS(Y, X)
results = model.fit()
# Print the regression results
print(results.summary())
| false | 0 | 1,261 | 0 | 1,261 | 1,261 |
||
129927565
|
<jupyter_start><jupyter_text>Stanford Dogs Dataset
### Context
The Stanford Dogs dataset contains images of 120 breeds of dogs from around the world. This dataset has been built using images and annotation from ImageNet for the task of fine-grained image categorization. It was originally collected for fine-grain image categorization, a challenging problem as certain dog breeds have near identical features or differ in colour and age.
### Content
- Number of categories: 120
- Number of images: 20,580
- Annotations: Class labels, Bounding boxes
Kaggle dataset identifier: stanford-dogs-dataset
<jupyter_script>import torchvision.datasets as datasets
import torchvision.transforms as transforms
import torch
import torchvision
import matplotlib.pyplot as plt
import numpy as np
import ssl
ssl._create_default_https_context = ssl._create_unverified_context
# # Loading the dataset
# Initializing normalizing transform for the dataset
normalize_transform = transforms.Compose(
[
transforms.RandomResizedCrop(32),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
# Downloading the "Stanford Dogs" dataset into train and test sets
train_dataset = datasets.ImageFolder(
root="/kaggle/input/stanford-dogs-dataset/images/Images/",
transform=normalize_transform,
)
test_dataset = datasets.ImageFolder(
root="/kaggle/input/stanford-dogs-dataset/images/Images/",
transform=normalize_transform,
)
# Generating data loaders from the corresponding datasets
batch_size = 128
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size=batch_size)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size=batch_size)
# Plotting some images from the dataset
plt.rcParams["figure.figsize"] = 14, 6
dataiter = iter(train_loader)
images, labels = next(dataiter)
plt.imshow(
np.transpose(
torchvision.utils.make_grid(
images[:30], normalize=True, padding=1, nrow=5
).numpy(),
(1, 2, 0),
)
)
plt.axis("off")
# # Plotting class distribution of the dataset
# Iterating over the training dataset and storing the target class for each sample
classes = []
for batch_idx, data in enumerate(train_loader, 0):
x, y = data
classes.extend(y.tolist())
# Calculating the unique classes and the respective counts
unique, counts = np.unique(classes, return_counts=True)
names = list(test_dataset.class_to_idx.keys())
print("unique : {}".format(unique))
# Plotting the unique counts and the respective counts
plt.bar(names, counts)
plt.xlabel("Target Classes")
plt.ylabel("Number of training instances")
plt.xticks([])
names
# # Implementing the CNN architecture
# ## Define the model
class CNN(torch.nn.Module):
def __init__(self):
super().__init__()
self.model = torch.nn.Sequential(
# Input = 3 x 32 x 32, Output = 32 x 32 x 32
torch.nn.Conv2d(in_channels=3, out_channels=32, kernel_size=3, padding=1),
torch.nn.ReLU(),
# Input = 32 x 32 x 32, Output = 32 x 16 x 16
torch.nn.MaxPool2d(kernel_size=2),
# Input = 32 x 16 x 16, Output = 64 x 16 x 16
torch.nn.Conv2d(in_channels=32, out_channels=64, kernel_size=3, padding=1),
torch.nn.ReLU(),
# Input = 64 x 16 x 16, Output = 64 x 8 x 8
torch.nn.MaxPool2d(kernel_size=2),
# Input = 64 x 8 x 8, Output = 64 x 8 x 8
torch.nn.Conv2d(in_channels=64, out_channels=64, kernel_size=3, padding=1),
torch.nn.ReLU(),
# Input = 64 x 8 x 8, Output = 64 x 4 x 4
torch.nn.MaxPool2d(kernel_size=2),
torch.nn.Flatten(),
torch.nn.Linear(64 * 4 * 4, 512),
torch.nn.ReLU(),
torch.nn.Linear(512, 120),
)
def forward(self, x):
return self.model(x)
# ### Defining hyper parameters
# Selecting the appropriate training device
device = "cuda" if torch.cuda.is_available() else "cpu"
model = CNN().to(device)
# Defining the model hyper parameters
num_epochs = 50
learning_rate = 0.001
weight_decay = 0.01
criterion = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(
model.parameters(), lr=learning_rate, weight_decay=weight_decay
)
# ## Train the model
train_loss_list = []
for epoch in range(num_epochs):
print(f"Epoch {epoch+1}/{num_epochs}:", end=" ")
train_loss = 0
# Iterating over the training dataset in batches
model.train()
for i, (images, labels) in enumerate(train_loader):
# Extracting images and target labels for the batch being iterated
images = images.to(device)
labels = labels.to(device)
# Calculating the model output and the cross entropy loss
outputs = model(images)
loss = criterion(outputs, labels)
# Updating weights according to calculated loss
optimizer.zero_grad()
loss.backward()
optimizer.step()
train_loss += loss.item()
# Printing loss for each epoch
train_loss_list.append(train_loss / len(train_loader))
print(f"Training loss = {train_loss_list[-1]}")
# ## Plotting the results
plt.plot(range(1, num_epochs + 1), train_loss_list)
plt.xlabel("Number of epochs")
plt.ylabel("Training loss")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/927/129927565.ipynb
|
stanford-dogs-dataset
|
jessicali9530
|
[{"Id": 129927565, "ScriptId": 37822811, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14086997, "CreationDate": "05/17/2023 13:34:24", "VersionNumber": 2.0, "Title": "lab3_ml", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 145.0, "LinesInsertedFromPrevious": 106.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 39.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186351189, "KernelVersionId": 129927565, "SourceDatasetVersionId": 791828}]
|
[{"Id": 791828, "DatasetId": 119698, "DatasourceVersionId": 813580, "CreatorUserId": 998023, "LicenseName": "Other (specified in description)", "CreationDate": "11/13/2019 06:20:35", "VersionNumber": 2.0, "Title": "Stanford Dogs Dataset", "Slug": "stanford-dogs-dataset", "Subtitle": "Over 20,000 images of 120 dog breeds", "Description": "### Context\nThe Stanford Dogs dataset contains images of 120 breeds of dogs from around the world. This dataset has been built using images and annotation from ImageNet for the task of fine-grained image categorization. It was originally collected for fine-grain image categorization, a challenging problem as certain dog breeds have near identical features or differ in colour and age.\n\n### Content\n\n- Number of categories: 120\n- Number of images: 20,580\n- Annotations: Class labels, Bounding boxes\n\n### Acknowledgements\nThe original data source is found on http://vision.stanford.edu/aditya86/ImageNetDogs/ and contains additional information on the train/test splits and baseline results.\n\nIf you use this dataset in a publication, please cite the dataset on the following papers:\n\n Aditya Khosla, Nityananda Jayadevaprakash, Bangpeng Yao and Li Fei-Fei. Novel dataset for Fine-Grained Image Categorization. First Workshop on Fine-Grained Visual Categorization (FGVC), IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2011. [pdf] [poster] [BibTex]\n\nSecondary:\n J. Deng, W. Dong, R. Socher, L.-J. Li, K. Li and L. Fei-Fei, ImageNet: A Large-Scale Hierarchical Image Database. IEEE Computer Vision and Pattern Recognition (CVPR), 2009. [pdf] [BibTex]\n\n[Banner Image from Hannah Lim on Unsplash][1]\n\n### Inspiration\n- Can you correctly identify dog breeds that have similar features, such as the basset hound and bloodhound?\n- Is this chihuahua young or old?\n\n\n [1]: https://unsplash.com/photos/U6nlG0Y5sfs", "VersionNotes": "Fix data", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 119698, "CreatorUserId": 1772071, "OwnerUserId": 1772071.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 791828.0, "CurrentDatasourceVersionId": 813580.0, "ForumId": 129616, "Type": 2, "CreationDate": "02/13/2019 05:45:25", "LastActivityDate": "02/13/2019", "TotalViews": 393260, "TotalDownloads": 39071, "TotalVotes": 928, "TotalKernels": 240}]
|
[{"Id": 1772071, "UserName": "jessicali9530", "DisplayName": "Jessica Li", "RegisterDate": "03/29/2018", "PerformanceTier": 5}]
|
import torchvision.datasets as datasets
import torchvision.transforms as transforms
import torch
import torchvision
import matplotlib.pyplot as plt
import numpy as np
import ssl
ssl._create_default_https_context = ssl._create_unverified_context
# # Loading the dataset
# Initializing normalizing transform for the dataset
normalize_transform = transforms.Compose(
[
transforms.RandomResizedCrop(32),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
# Downloading the "Stanford Dogs" dataset into train and test sets
train_dataset = datasets.ImageFolder(
root="/kaggle/input/stanford-dogs-dataset/images/Images/",
transform=normalize_transform,
)
test_dataset = datasets.ImageFolder(
root="/kaggle/input/stanford-dogs-dataset/images/Images/",
transform=normalize_transform,
)
# Generating data loaders from the corresponding datasets
batch_size = 128
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size=batch_size)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size=batch_size)
# Plotting some images from the dataset
plt.rcParams["figure.figsize"] = 14, 6
dataiter = iter(train_loader)
images, labels = next(dataiter)
plt.imshow(
np.transpose(
torchvision.utils.make_grid(
images[:30], normalize=True, padding=1, nrow=5
).numpy(),
(1, 2, 0),
)
)
plt.axis("off")
# # Plotting class distribution of the dataset
# Iterating over the training dataset and storing the target class for each sample
classes = []
for batch_idx, data in enumerate(train_loader, 0):
x, y = data
classes.extend(y.tolist())
# Calculating the unique classes and the respective counts
unique, counts = np.unique(classes, return_counts=True)
names = list(test_dataset.class_to_idx.keys())
print("unique : {}".format(unique))
# Plotting the unique counts and the respective counts
plt.bar(names, counts)
plt.xlabel("Target Classes")
plt.ylabel("Number of training instances")
plt.xticks([])
names
# # Implementing the CNN architecture
# ## Define the model
class CNN(torch.nn.Module):
def __init__(self):
super().__init__()
self.model = torch.nn.Sequential(
# Input = 3 x 32 x 32, Output = 32 x 32 x 32
torch.nn.Conv2d(in_channels=3, out_channels=32, kernel_size=3, padding=1),
torch.nn.ReLU(),
# Input = 32 x 32 x 32, Output = 32 x 16 x 16
torch.nn.MaxPool2d(kernel_size=2),
# Input = 32 x 16 x 16, Output = 64 x 16 x 16
torch.nn.Conv2d(in_channels=32, out_channels=64, kernel_size=3, padding=1),
torch.nn.ReLU(),
# Input = 64 x 16 x 16, Output = 64 x 8 x 8
torch.nn.MaxPool2d(kernel_size=2),
# Input = 64 x 8 x 8, Output = 64 x 8 x 8
torch.nn.Conv2d(in_channels=64, out_channels=64, kernel_size=3, padding=1),
torch.nn.ReLU(),
# Input = 64 x 8 x 8, Output = 64 x 4 x 4
torch.nn.MaxPool2d(kernel_size=2),
torch.nn.Flatten(),
torch.nn.Linear(64 * 4 * 4, 512),
torch.nn.ReLU(),
torch.nn.Linear(512, 120),
)
def forward(self, x):
return self.model(x)
# ### Defining hyper parameters
# Selecting the appropriate training device
device = "cuda" if torch.cuda.is_available() else "cpu"
model = CNN().to(device)
# Defining the model hyper parameters
num_epochs = 50
learning_rate = 0.001
weight_decay = 0.01
criterion = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(
model.parameters(), lr=learning_rate, weight_decay=weight_decay
)
# ## Train the model
train_loss_list = []
for epoch in range(num_epochs):
print(f"Epoch {epoch+1}/{num_epochs}:", end=" ")
train_loss = 0
# Iterating over the training dataset in batches
model.train()
for i, (images, labels) in enumerate(train_loader):
# Extracting images and target labels for the batch being iterated
images = images.to(device)
labels = labels.to(device)
# Calculating the model output and the cross entropy loss
outputs = model(images)
loss = criterion(outputs, labels)
# Updating weights according to calculated loss
optimizer.zero_grad()
loss.backward()
optimizer.step()
train_loss += loss.item()
# Printing loss for each epoch
train_loss_list.append(train_loss / len(train_loader))
print(f"Training loss = {train_loss_list[-1]}")
# ## Plotting the results
plt.plot(range(1, num_epochs + 1), train_loss_list)
plt.xlabel("Number of epochs")
plt.ylabel("Training loss")
| false | 0 | 1,405 | 0 | 1,557 | 1,405 |
||
129927517
|
<jupyter_start><jupyter_text>COVID-19 Dataset
[](https://forthebadge.com) [](https://forthebadge.com)
### Context
- A new coronavirus designated 2019-nCoV was first identified in Wuhan, the capital of China's Hubei province
- People developed pneumonia without a clear cause and for which existing vaccines or treatments were not effective.
- The virus has shown evidence of human-to-human transmission
- Transmission rate (rate of infection) appeared to escalate in mid-January 2020
- As of 30 January 2020, approximately 8,243 cases have been confirmed
### Content
> * **full_grouped.csv** - Day to day country wise no. of cases (Has County/State/Province level data)
> * **covid_19_clean_complete.csv** - Day to day country wise no. of cases (Doesn't have County/State/Province level data)
> * **country_wise_latest.csv** - Latest country level no. of cases
> * **day_wise.csv** - Day wise no. of cases (Doesn't have country level data)
> * **usa_county_wise.csv** - Day to day county level no. of cases
> * **worldometer_data.csv** - Latest data from https://www.worldometers.info/
Kaggle dataset identifier: corona-virus-report
<jupyter_code>import pandas as pd
df = pd.read_csv('corona-virus-report/covid_19_clean_complete.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 49068 entries, 0 to 49067
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Province/State 14664 non-null object
1 Country/Region 49068 non-null object
2 Lat 49068 non-null float64
3 Long 49068 non-null float64
4 Date 49068 non-null object
5 Confirmed 49068 non-null int64
6 Deaths 49068 non-null int64
7 Recovered 49068 non-null int64
8 Active 49068 non-null int64
9 WHO Region 49068 non-null object
dtypes: float64(2), int64(4), object(4)
memory usage: 3.7+ MB
<jupyter_text>Examples:
{
"Province/State": NaN,
"Country/Region": "Afghanistan",
"Lat": 33.93911,
"Long": 67.709953,
"Date": "2020-01-22 00:00:00",
"Confirmed": 0,
"Deaths": 0,
"Recovered": 0,
"Active": 0,
"WHO Region": "Eastern Mediterranean"
}
{
"Province/State": NaN,
"Country/Region": "Albania",
"Lat": 41.1533,
"Long": 20.1683,
"Date": "2020-01-22 00:00:00",
"Confirmed": 0,
"Deaths": 0,
"Recovered": 0,
"Active": 0,
"WHO Region": "Europe"
}
{
"Province/State": NaN,
"Country/Region": "Algeria",
"Lat": 28.0339,
"Long": 1.6596000000000002,
"Date": "2020-01-22 00:00:00",
"Confirmed": 0,
"Deaths": 0,
"Recovered": 0,
"Active": 0,
"WHO Region": "Africa"
}
{
"Province/State": NaN,
"Country/Region": "Andorra",
"Lat": 42.5063,
"Long": 1.5218,
"Date": "2020-01-22 00:00:00",
"Confirmed": 0,
"Deaths": 0,
"Recovered": 0,
"Active": 0,
"WHO Region": "Europe"
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/corona-virus-report/covid_19_clean_complete.csv")
df
df = df.drop(columns=["Lat", "Long", "Province/State", "WHO Region"])
df.head()
# # Top Country
import matplotlib.pyplot as plt
plt.figure(figsize=(13, 5))
c = df["Country/Region"].value_counts().head(10)
x = c.index
y = c.values
plt.bar(x, y, color=["red", "blue", "green"])
plt.show()
# #
df.head()
ndf = df.groupby("Country/Region").agg(
total_confirmed=("Confirmed", "sum"),
total_deaths=("Deaths", "sum"),
total_recovered=("Recovered", "sum"),
total_active=("Active", "sum"),
)
ndf = ndf.sort_values(by="total_confirmed", ascending=False)
ndf = ndf.head(10)
ndf
ndf = ndf.reset_index()
ndf
plt.figure(figsize=(20, 5))
original_x_axis = ndf["Country/Region"]
numerical_x_axis = np.arange(1, 11) # [1,2,3,4,5,6,7,8,9,10]
plt.xticks(numerical_x_axis, original_x_axis, rotation=45)
plt.bar(
numerical_x_axis + 0.2,
ndf["total_confirmed"],
label="Confirmed",
width=0.2,
color="blue",
)
plt.bar(
numerical_x_axis + 0.4,
ndf["total_active"],
label="Active",
width=0.2,
color="brown",
)
plt.bar(
numerical_x_axis + 0.6, ndf["total_deaths"], label="Deaths", width=0.2, color="red"
)
plt.bar(
numerical_x_axis + 0.8,
ndf["total_recovered"],
label="Recovered",
width=0.2,
color="green",
)
plt.grid()
plt.legend()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/927/129927517.ipynb
|
corona-virus-report
|
imdevskp
|
[{"Id": 129927517, "ScriptId": 38556941, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6071377, "CreationDate": "05/17/2023 13:33:58", "VersionNumber": 1.0, "Title": "notebookbdffe2329a", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 75.0, "LinesInsertedFromPrevious": 75.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 186351123, "KernelVersionId": 129927517, "SourceDatasetVersionId": 1402868}]
|
[{"Id": 1402868, "DatasetId": 494766, "DatasourceVersionId": 1435700, "CreatorUserId": 1302389, "LicenseName": "Other (specified in description)", "CreationDate": "08/07/2020 03:47:47", "VersionNumber": 166.0, "Title": "COVID-19 Dataset", "Slug": "corona-virus-report", "Subtitle": "Number of Confirmed, Death and Recovered cases every day across the globe", "Description": "[](https://forthebadge.com) [](https://forthebadge.com)\n\n### Context\n\n- A new coronavirus designated 2019-nCoV was first identified in Wuhan, the capital of China's Hubei province\n- People developed pneumonia without a clear cause and for which existing vaccines or treatments were not effective. \n- The virus has shown evidence of human-to-human transmission\n- Transmission rate (rate of infection) appeared to escalate in mid-January 2020\n- As of 30 January 2020, approximately 8,243 cases have been confirmed\n\n\n### Content\n\n> * **full_grouped.csv** - Day to day country wise no. of cases (Has County/State/Province level data) \n> * **covid_19_clean_complete.csv** - Day to day country wise no. of cases (Doesn't have County/State/Province level data) \n> * **country_wise_latest.csv** - Latest country level no. of cases \n> * **day_wise.csv** - Day wise no. of cases (Doesn't have country level data) \n> * **usa_county_wise.csv** - Day to day county level no. of cases \n> * **worldometer_data.csv** - Latest data from https://www.worldometers.info/ \n\n\n### Acknowledgements / Data Source\n\n> https://github.com/CSSEGISandData/COVID-19\n> https://www.worldometers.info/\n\n### Collection methodology\n\n> https://github.com/imdevskp/covid_19_jhu_data_web_scrap_and_cleaning\n\n### Cover Photo\n\n> Photo from National Institutes of Allergy and Infectious Diseases\n> https://www.niaid.nih.gov/news-events/novel-coronavirus-sarscov2-images\n> https://blogs.cdc.gov/publichealthmatters/2019/04/h1n1/\n\n### Similar Datasets\n\n> * COVID-19 - https://www.kaggle.com/imdevskp/corona-virus-report \n> * MERS - https://www.kaggle.com/imdevskp/mers-outbreak-dataset-20122019\n> * Ebola Western Africa 2014 Outbreak - https://www.kaggle.com/imdevskp/ebola-outbreak-20142016-complete-dataset\n> * H1N1 | Swine Flu 2009 Pandemic Dataset - https://www.kaggle.com/imdevskp/h1n1-swine-flu-2009-pandemic-dataset\n> * SARS 2003 Pandemic - https://www.kaggle.com/imdevskp/sars-outbreak-2003-complete-dataset\n> * HIV AIDS - https://www.kaggle.com/imdevskp/hiv-aids-dataset", "VersionNotes": "update", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 494766, "CreatorUserId": 1302389, "OwnerUserId": 1302389.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1402868.0, "CurrentDatasourceVersionId": 1435700.0, "ForumId": 507860, "Type": 2, "CreationDate": "01/30/2020 14:46:58", "LastActivityDate": "01/30/2020", "TotalViews": 1009073, "TotalDownloads": 271389, "TotalVotes": 2056, "TotalKernels": 642}]
|
[{"Id": 1302389, "UserName": "imdevskp", "DisplayName": "Devakumar K. P.", "RegisterDate": "09/30/2017", "PerformanceTier": 3}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/corona-virus-report/covid_19_clean_complete.csv")
df
df = df.drop(columns=["Lat", "Long", "Province/State", "WHO Region"])
df.head()
# # Top Country
import matplotlib.pyplot as plt
plt.figure(figsize=(13, 5))
c = df["Country/Region"].value_counts().head(10)
x = c.index
y = c.values
plt.bar(x, y, color=["red", "blue", "green"])
plt.show()
# #
df.head()
ndf = df.groupby("Country/Region").agg(
total_confirmed=("Confirmed", "sum"),
total_deaths=("Deaths", "sum"),
total_recovered=("Recovered", "sum"),
total_active=("Active", "sum"),
)
ndf = ndf.sort_values(by="total_confirmed", ascending=False)
ndf = ndf.head(10)
ndf
ndf = ndf.reset_index()
ndf
plt.figure(figsize=(20, 5))
original_x_axis = ndf["Country/Region"]
numerical_x_axis = np.arange(1, 11) # [1,2,3,4,5,6,7,8,9,10]
plt.xticks(numerical_x_axis, original_x_axis, rotation=45)
plt.bar(
numerical_x_axis + 0.2,
ndf["total_confirmed"],
label="Confirmed",
width=0.2,
color="blue",
)
plt.bar(
numerical_x_axis + 0.4,
ndf["total_active"],
label="Active",
width=0.2,
color="brown",
)
plt.bar(
numerical_x_axis + 0.6, ndf["total_deaths"], label="Deaths", width=0.2, color="red"
)
plt.bar(
numerical_x_axis + 0.8,
ndf["total_recovered"],
label="Recovered",
width=0.2,
color="green",
)
plt.grid()
plt.legend()
plt.show()
|
[{"corona-virus-report/covid_19_clean_complete.csv": {"column_names": "[\"Province/State\", \"Country/Region\", \"Lat\", \"Long\", \"Date\", \"Confirmed\", \"Deaths\", \"Recovered\", \"Active\", \"WHO Region\"]", "column_data_types": "{\"Province/State\": \"object\", \"Country/Region\": \"object\", \"Lat\": \"float64\", \"Long\": \"float64\", \"Date\": \"object\", \"Confirmed\": \"int64\", \"Deaths\": \"int64\", \"Recovered\": \"int64\", \"Active\": \"int64\", \"WHO Region\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 49068 entries, 0 to 49067\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Province/State 14664 non-null object \n 1 Country/Region 49068 non-null object \n 2 Lat 49068 non-null float64\n 3 Long 49068 non-null float64\n 4 Date 49068 non-null object \n 5 Confirmed 49068 non-null int64 \n 6 Deaths 49068 non-null int64 \n 7 Recovered 49068 non-null int64 \n 8 Active 49068 non-null int64 \n 9 WHO Region 49068 non-null object \ndtypes: float64(2), int64(4), object(4)\nmemory usage: 3.7+ MB\n", "summary": "{\"Lat\": {\"count\": 49068.0, \"mean\": 21.433730459770114, \"std\": 24.950319826065034, \"min\": -51.7963, \"25%\": 7.873054, \"50%\": 23.6345, \"75%\": 41.20438, \"max\": 71.7069}, \"Long\": {\"count\": 49068.0, \"mean\": 23.52823645210728, \"std\": 70.4427397445027, \"min\": -135.0, \"25%\": -15.3101, \"50%\": 21.7453, \"75%\": 80.77179699999998, \"max\": 178.065}, \"Confirmed\": {\"count\": 49068.0, \"mean\": 16884.90425531915, \"std\": 127300.20527228057, \"min\": 0.0, \"25%\": 4.0, \"50%\": 168.0, \"75%\": 1518.25, \"max\": 4290259.0}, \"Deaths\": {\"count\": 49068.0, \"mean\": 884.1791595337083, \"std\": 6313.5844105965425, \"min\": 0.0, \"25%\": 0.0, \"50%\": 2.0, \"75%\": 30.0, \"max\": 148011.0}, \"Recovered\": {\"count\": 49068.0, \"mean\": 7915.713479253282, \"std\": 54800.91873054017, \"min\": 0.0, \"25%\": 0.0, \"50%\": 29.0, \"75%\": 666.0, \"max\": 1846641.0}, \"Active\": {\"count\": 49068.0, \"mean\": 8085.01161653216, \"std\": 76258.9030255069, \"min\": -14.0, \"25%\": 0.0, \"50%\": 26.0, \"75%\": 606.0, \"max\": 2816444.0}}", "examples": "{\"Province\\/State\":{\"0\":null,\"1\":null,\"2\":null,\"3\":null},\"Country\\/Region\":{\"0\":\"Afghanistan\",\"1\":\"Albania\",\"2\":\"Algeria\",\"3\":\"Andorra\"},\"Lat\":{\"0\":33.93911,\"1\":41.1533,\"2\":28.0339,\"3\":42.5063},\"Long\":{\"0\":67.709953,\"1\":20.1683,\"2\":1.6596,\"3\":1.5218},\"Date\":{\"0\":\"2020-01-22\",\"1\":\"2020-01-22\",\"2\":\"2020-01-22\",\"3\":\"2020-01-22\"},\"Confirmed\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Deaths\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Recovered\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Active\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"WHO Region\":{\"0\":\"Eastern Mediterranean\",\"1\":\"Europe\",\"2\":\"Africa\",\"3\":\"Europe\"}}"}}]
| true | 1 |
<start_data_description><data_path>corona-virus-report/covid_19_clean_complete.csv:
<column_names>
['Province/State', 'Country/Region', 'Lat', 'Long', 'Date', 'Confirmed', 'Deaths', 'Recovered', 'Active', 'WHO Region']
<column_types>
{'Province/State': 'object', 'Country/Region': 'object', 'Lat': 'float64', 'Long': 'float64', 'Date': 'object', 'Confirmed': 'int64', 'Deaths': 'int64', 'Recovered': 'int64', 'Active': 'int64', 'WHO Region': 'object'}
<dataframe_Summary>
{'Lat': {'count': 49068.0, 'mean': 21.433730459770114, 'std': 24.950319826065034, 'min': -51.7963, '25%': 7.873054, '50%': 23.6345, '75%': 41.20438, 'max': 71.7069}, 'Long': {'count': 49068.0, 'mean': 23.52823645210728, 'std': 70.4427397445027, 'min': -135.0, '25%': -15.3101, '50%': 21.7453, '75%': 80.77179699999998, 'max': 178.065}, 'Confirmed': {'count': 49068.0, 'mean': 16884.90425531915, 'std': 127300.20527228057, 'min': 0.0, '25%': 4.0, '50%': 168.0, '75%': 1518.25, 'max': 4290259.0}, 'Deaths': {'count': 49068.0, 'mean': 884.1791595337083, 'std': 6313.5844105965425, 'min': 0.0, '25%': 0.0, '50%': 2.0, '75%': 30.0, 'max': 148011.0}, 'Recovered': {'count': 49068.0, 'mean': 7915.713479253282, 'std': 54800.91873054017, 'min': 0.0, '25%': 0.0, '50%': 29.0, '75%': 666.0, 'max': 1846641.0}, 'Active': {'count': 49068.0, 'mean': 8085.01161653216, 'std': 76258.9030255069, 'min': -14.0, '25%': 0.0, '50%': 26.0, '75%': 606.0, 'max': 2816444.0}}
<dataframe_info>
RangeIndex: 49068 entries, 0 to 49067
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Province/State 14664 non-null object
1 Country/Region 49068 non-null object
2 Lat 49068 non-null float64
3 Long 49068 non-null float64
4 Date 49068 non-null object
5 Confirmed 49068 non-null int64
6 Deaths 49068 non-null int64
7 Recovered 49068 non-null int64
8 Active 49068 non-null int64
9 WHO Region 49068 non-null object
dtypes: float64(2), int64(4), object(4)
memory usage: 3.7+ MB
<some_examples>
{'Province/State': {'0': None, '1': None, '2': None, '3': None}, 'Country/Region': {'0': 'Afghanistan', '1': 'Albania', '2': 'Algeria', '3': 'Andorra'}, 'Lat': {'0': 33.93911, '1': 41.1533, '2': 28.0339, '3': 42.5063}, 'Long': {'0': 67.709953, '1': 20.1683, '2': 1.6596, '3': 1.5218}, 'Date': {'0': '2020-01-22', '1': '2020-01-22', '2': '2020-01-22', '3': '2020-01-22'}, 'Confirmed': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Deaths': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Recovered': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Active': {'0': 0, '1': 0, '2': 0, '3': 0}, 'WHO Region': {'0': 'Eastern Mediterranean', '1': 'Europe', '2': 'Africa', '3': 'Europe'}}
<end_description>
| 729 | 1 | 1,920 | 729 |
129950473
|
<jupyter_start><jupyter_text>Synthetic Speech Commands Dataset
## Context
- We would like to have good open source speech recognition
- Commercial companies try to solve a hard problem: map arbitrary, open-ended speech to text and identify meaning
- The easier problem should be: detect a predefined sequence of sounds and map it to a predefined action.
- Lets tackle the simplest problem first: Classifying single, short words (commands)
- Audio training data is difficult to obtain.
## Approaches
- The parent project ([spoken verbs][1]) created synthetic speech datasets using text-to-speech programs. The focus there is on single-syllable verbs (commands).
- The Speech Commands dataset (by Pete Warden, see the [TensorFlow Speech Recognition Challenge][2]) asked volunteers to pronounce a small set of words: (yes, no, up, down, left, right, on, off, stop, go, and 0-9).
- This data set provides synthetic counterparts to this real world dataset.
## Open questions
One can use these two datasets in various ways. Here are some things I am interested in seeing answered:
1. What is it in an audio sample that makes it "sound similar"? Our ears can easily classify both synthetic and real speech, but for algorithms this is still hard. Extending the real dataset with the synthetic data yields a larger training sample and more diversity.
2. How well does an algorithm trained on one data set perform on the other? (transfer learning) If it works poorly, the algorithm probably has not found the key to audio similarity.
3. Are synthetic data sufficient for classifying real datasets? If this is the case, the implications are huge. You would not need to ask thousands of volunteers for hours of time. Instead, you could easily create arbitrary synthetic datasets for your target words.
A interesting challenge (idea for competition) would be to train on this data set and evaluate on the real dataset.
## Synthetic data creation
Here I describe how the synthetic audio samples were created.
Code is available at https://github.com/JohannesBuchner/spoken-command-recognition, in the "tensorflow-speech-words" folder.
1. The list of words is in "inputwords". "marvin" was changed to "marvel", because "marvin" does not have a pronounciation coding yet.
2. Pronounciations were taken from the British English Example Pronciation dictionary (BEEP, http://svr-www.eng.cam.ac.uk/comp.speech/Section1/Lexical/beep.html ). The phonemes were translated for the next step with a translation table (see compile.py for details). This creates the file "words". There are multiple pronounciations and stresses for each word.
3. A text-to-speech program (espeak) was used to pronounce these words (see generatetfspeech.sh for details). The pronounciation, stress, pitch, speed and speaker were varied. This gives >1000 clean examples for each word.
4. Noise samples were obtained. Noise samples (airport babble car exhibition restaurant street subway train) come from AURORA (https://www.ee.columbia.edu/~dpwe/sounds/noise/), and additional noise samples were synthetically created (ocean white brown pink). (see ../generatenoise.sh for details)
5. Noise and speech were mixed. The speech volume and offset were varied. The noise source, volume was also varied. See addnoise.py for details. addnoise2.py is the same, but with lower speech volume and higher noise volume. All audio files are one second (1s) long and are in wav format (16 bit, mono, 16000 Hz).
6. Finally, the data was compressed into an archive and uploaded to kaggle.
## Acknowledgements
This work built upon
- Pronounciation dictionary: BEEP: http://svr-www.eng.cam.ac.uk/comp.speech/Section1/Lexical/beep.html
- Noise samples: AURORA: https://www.ee.columbia.edu/~dpwe/sounds/noise/
- eSPEAK: http://espeak.sourceforge.net/ and mbrola voices http://www.tcts.fpms.ac.be/synthesis/mbrola/mbrcopybin.html
Please provide appropriate citations to the above when using this work.
To cite the resulting dataset, you can use:
APA-style citation: "Buchner J. Synthetic Speech Commands: A public dataset for single-word speech recognition, 2017. Available from https://www.kaggle.com/jbuchner/synthetic-speech-commands-dataset/".
BibTeX @article{speechcommands, title={Synthetic Speech Commands: A public dataset for single-word speech recognition.}, author={Buchner, Johannes}, journal={Dataset available from https://www.kaggle.com/jbuchner/synthetic-speech-commands-dataset/}, year={2017} }
Thanks to everyone trying to improve open source voice detection and speech recognition.
## Links
- https://www.kaggle.com/jbuchner/spokenverbs
- https://www.kaggle.com/c/tensorflow-speech-recognition-challenge
- https://github.com/JohannesBuchner/spoken-command-recognition/
[1]: https://www.kaggle.com/jbuchner/spokenverbs
[2]: https://www.kaggle.com/c/tensorflow-speech-recognition-challenge/data
Kaggle dataset identifier: synthetic-speech-commands-dataset
<jupyter_script># # By : 𝔄𝔥𝔪𝔢𝔡 𝔄𝔰𝔥𝔯𝔞𝔣
# dataset:https://www.kaggle.com/datasets/jbuchner/synthetic-speech-commands-dataset
# we'll build Model to use it speech recognition for thirty words
# dataset consisting of 41849 audio
# # Importing Libraries & Data
import librosa as lr
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import os
import IPython.display as ipd
import keras
from sklearn.model_selection import train_test_split
import tensorflow as tf
import pickle as pk
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
from scipy.io.wavfile import write
dataset = "/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset"
pd.DataFrame(os.listdir(dataset), columns=["Files"])
def count(path):
size = []
for file in os.listdir(path):
size.append(len(os.listdir(os.path.join(path, file))))
return pd.DataFrame(size, columns=["Number Of Sample"], index=os.listdir(path))
tr = count(dataset)
tr
plt.figure(figsize=(10, 10))
plt.pie(x="Number Of Sample", labels=os.listdir(dataset), autopct="%1.1f%%", data=tr)
plt.title("Distribution Of Data In Train", fontsize=20)
plt.show()
sns.kdeplot(tr["Number Of Sample"], color="red")
plt.title("Kdeplot For Train", fontsize=20)
plt.xlabel("Number", fontsize=20)
plt.ylabel("Count", fontsize=20)
# bed Audio
ipd.Audio(
"/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset/bed/1014.wav"
)
# bird Audio
ipd.Audio(
"/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset/bird/1019.wav"
)
# cat Audio
ipd.Audio(
"/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset/cat/1009.wav"
)
# yes Audio
ipd.Audio(
"/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset/yes/1014.wav"
)
def normalize_signal(signal):
# Calculate the peak amplitude of the signal
peak_amplitude = np.max(np.abs(signal))
# Normalize the signal by dividing it by the peak amplitude
normalized_signal = signal / peak_amplitude
return normalized_signal
def load(path):
data = []
label = []
sample = []
for file in os.listdir(path):
path_ = os.path.join(path, file)
for fil in os.listdir(path_):
data_contain, sample_rate = lr.load(os.path.join(path_, fil), sr=16000)
data_contain = normalize_signal(data_contain)
data.append(data_contain)
sample.append(sample_rate)
label.append(file)
return data, label, sample
data, label, sample = load(dataset)
df = pd.DataFrame()
df["Label"], df["sample"] = label, sample
df
# # Audio Feature Extraction
# waveform
def waveform(data, sr, label):
plt.figure(figsize=(14, 5))
lr.display.waveshow(data, sr=sr)
plt.suptitle(label)
plt.title("Waveform plot")
plt.xlabel("Time (s)")
plt.ylabel("Amplitude")
# MFCC features
def mfcc(data, sr):
mfccs = lr.feature.mfcc(y=data, sr=sr, n_mfcc=13)
return np.mean(mfccs), mfccs
def mfcc_v(mfccs, label):
plt.figure(figsize=(10, 4))
lr.display.specshow(mfccs, x_axis="time")
plt.colorbar()
plt.title("MFCC")
plt.suptitle(label)
# Mel-spectrogram
def Mel(data, sr):
mel_spec = lr.feature.melspectrogram(y=data, sr=sr)
return np.mean(mel_spec), mel_spec
def mel_v(mel_spec, label, sr):
# Convert to decibel scale
mel_spec_db = lr.power_to_db(mel_spec, ref=np.max)
# Visualize Mel-spectrogram
plt.figure(figsize=(10, 4))
lr.display.specshow(mel_spec_db, x_axis="time", y_axis="mel", sr=sr)
plt.colorbar(format="%+2.0f dB")
plt.title("Mel-spectrogram")
plt.suptitle(label)
# zero_crossing_rate
def zero_crossing(data, sr):
# Compute zero-crossing rate
zcr = lr.feature.zero_crossing_rate(data)
# Print average zero-crossing rate
avg_zcr = sum(zcr[0]) / len(zcr[0])
print("Average zero-crossing rate:", avg_zcr)
return zcr
def zero_crossing_v(zcr, label, data, sr):
time = lr.times_like(zcr)
# Create waveform plot
plt.figure(figsize=(14, 5))
lr.display.waveshow(data, sr=sr, alpha=0.5)
plt.plot(time, zcr[0], color="r")
plt.title("Zero-crossing rate")
plt.xlabel("Time (s)")
plt.ylabel("Amplitude")
plt.suptitle(label)
# waveform data[0]
waveform(data[0], sample[0], label[0])
plt.legend()
# MFCC data[0]
mfccs_mean, mfccs = mfcc(data[0], sample[0])
print("MFCCs Mean:", mfccs_mean)
print("MFCCs shape:", mfccs.shape)
mfcc_v(mfccs, label[0])
##Mel-spectrogram data[0]
mel_mean, mel = Mel(data[0], sample[0])
print("Mel Mean:", mel_mean)
print("Mel :", mel.shape)
mel_v(mel, label[0], sample[0])
# zero_crossing data[0]
zcr = zero_crossing(data[0], sample[0])
print("Zcr:", zcr.shape)
zero_crossing_v(zcr, label[0], data[0], sample[0])
# waveform data[1000]
waveform(data[1000], sample[1000], label[1000])
plt.legend()
# MFCC data[1000]
mfccs_mean, mfccs = mfcc(data[1000], sample[1000])
print("MFCCs Mean:", mfccs_mean)
print("MFCCs shape:", mfccs.shape)
mfcc_v(mfccs, label[1000])
##Mel-spectrogram data[1000]
mel_mean, mel = Mel(data[1000], sample[1000])
print("Mel Mean:", mel_mean)
print("Mel :", mel.shape)
mel_v(mel, label[1000], sample[1000])
# zero_crossing data[1000]
zcr = zero_crossing(data[1000], sample[1000])
print("Zcr:", zcr.shape)
zero_crossing_v(zcr, label[1000], data[1000], sample[1000])
code = {}
x = 0
for i in pd.unique(label):
code[i] = x
x += 1
pd.DataFrame(code.values(), columns=["Value"], index=code.keys())
def get_Name(N):
for x, y in code.items():
if y == N:
return x
for i in range(len(label)):
label[i] = code[label[i]]
pd.DataFrame(label, columns=["Labels"])
mfccs = [lr.feature.mfcc(y=d, sr=s, n_mfcc=13) for d, s in zip(data, sample)]
mfccs = np.array(mfccs)
# # Splitting Data
label = np.array(label)
X_train, X_test, y_train, y_test = train_test_split(
mfccs, label, test_size=0.1, random_state=44, shuffle=True
)
print("X_train shape is ", X_train.shape)
print("X_test shape is ", X_test.shape)
print("y_train shape is ", y_train.shape)
print("y_test shape is ", y_test.shape)
X_train = X_train.reshape((X_train.shape[0], -1))
X_test = X_test.reshape((X_test.shape[0], -1))
print("X_train shape is ", X_train.shape)
print("X_test shape is ", X_test.shape)
# # Model Building
num_class = len(pd.unique(y_train))
model = keras.Sequential()
model.add(keras.layers.Dense(512, activation="relu", input_shape=(X_train.shape[1],)))
model.add(keras.layers.Dense(128, activation="relu"))
model.add(keras.layers.Dense(64, activation="relu"))
model.add(keras.layers.Dense(num_class, activation="softmax"))
model.summary()
tf.keras.utils.plot_model(
model,
to_file="model.png",
show_shapes=True,
show_layer_names=True,
show_dtype=True,
dpi=120,
)
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
hist = model.fit(X_train, y_train, epochs=50)
loss, acc = model.evaluate(X_test, y_test)
print("Loss is :", loss)
print("ACC is :", acc)
hist_ = hist.history
pd.DataFrame(hist_)
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
plt.plot(hist_["loss"], c="r", marker="*", label="Loss")
plt.title("Overall Loss", fontsize=20)
plt.legend()
plt.subplot(1, 2, 2)
plt.plot(hist_["accuracy"], label="Accuracy")
plt.title("Overall Accuracy", fontsize=20)
plt.legend()
predict = model.predict(X_test)
predict[0]
preN = []
prename = []
for row in predict:
N = np.argmax(row)
preN.append(N)
prename.append(get_Name(N))
pd.DataFrame(prename, columns=["Predictions"])
predict = []
y_act = []
for p in range(30):
y_act.append(get_Name(y_test[p]))
predict.append(prename[p])
pd_p = pd.DataFrame(y_act, columns=["y_act"])
pd_p["predict"] = predict
pd_p
# # Model Check
plt.figure(figsize=(15, 15))
ax = plt.subplot()
CM = confusion_matrix(y_test, preN)
sns.heatmap(CM, annot=True, fmt="g", ax=ax, cbar=False, cmap="RdBu")
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix")
ax.xaxis.set_ticklabels(code.keys())
ax.yaxis.set_ticklabels(code.keys())
plt.show()
CM
ClassificationReport = classification_report(y_test, preN)
print("Classification Report is : ", ClassificationReport)
# # Save Model
model.save("/kaggle/working/speech_model.h5")
# # REAL TRIAL
code = {
"no": 0,
"two": 1,
"four": 2,
"five": 3,
"nine": 4,
"right": 5,
"off": 6,
"yes": 7,
"six": 8,
"dog": 9,
"left": 10,
"bird": 11,
"marvel": 12,
"wow": 13,
"zero": 14,
"eight": 15,
"bed": 16,
"go": 17,
"house": 18,
"tree": 19,
"seven": 20,
"on": 21,
"three": 22,
"one": 23,
"down": 24,
"stop": 25,
"up": 26,
"happy": 27,
"cat": 28,
"sheila": 29,
}
def get_Name(N):
for x, y in code.items():
if y == N:
return x
def record():
sample_rate = 16000
duration = 1
recording = sd.rec(int(duration * sample_rate), samplerate=sample_rate, channels=1)
sd.wait()
write("recording.wav", sample_rate, recording)
def predict():
data, sr = lr.load("recording.wav", sr=16000)
data = normalize_signal(data)
data = data.reshape(-1, 16000, 1)
savedmodel = keras.models.load_model("speech_model.h5")
pred = savedmodel.predict(data)
for row in pred:
N = np.argmax(row)
print(get_Name(N))
record()
ipd.Audio("recording.wav")
# waveform data
data, sr = lr.load("recording.wav", sr=16000)
data = normalize_signal(data)
waveform(data, sr, "label")
# MFCC data[0]
mfccs_mean, mfccs = mfcc(data, sr)
print("MFCCs Mean:", mfccs_mean)
print("MFCCs shape:", mfccs.shape)
mfcc_v(mfccs, "label")
mel_mean, mel = Mel(data, sr)
print("Mel Mean:", mel_mean)
print("Mel :", mel.shape)
mel_v(mel, "label", sr)
# zero_crossing data
zcr = zero_crossing(data, sr)
print("Zcr:", zcr.shape)
zero_crossing_v(zcr, "label", data, sr)
predict()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/950/129950473.ipynb
|
synthetic-speech-commands-dataset
|
jbuchner
|
[{"Id": 129950473, "ScriptId": 35674489, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10995749, "CreationDate": "05/17/2023 16:33:11", "VersionNumber": 8.0, "Title": "Speech using CNN", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 342.0, "LinesInsertedFromPrevious": 83.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 259.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186382396, "KernelVersionId": 129950473, "SourceDatasetVersionId": 39731}]
|
[{"Id": 39731, "DatasetId": 7691, "DatasourceVersionId": 41901, "CreatorUserId": 21849, "LicenseName": "CC BY-SA 4.0", "CreationDate": "06/12/2018 06:21:36", "VersionNumber": 4.0, "Title": "Synthetic Speech Commands Dataset", "Slug": "synthetic-speech-commands-dataset", "Subtitle": "Text-to-speech counterparts for the \"Speech Commands Data Set v0.01\"", "Description": "## Context\n\n - We would like to have good open source speech recognition\n - Commercial companies try to solve a hard problem: map arbitrary, open-ended speech to text and identify meaning\n - The easier problem should be: detect a predefined sequence of sounds and map it to a predefined action.\n - Lets tackle the simplest problem first: Classifying single, short words (commands)\n - Audio training data is difficult to obtain.\n\n## Approaches\n\n - The parent project ([spoken verbs][1]) created synthetic speech datasets using text-to-speech programs. The focus there is on single-syllable verbs (commands).\n - The Speech Commands dataset (by Pete Warden, see the [TensorFlow Speech Recognition Challenge][2]) asked volunteers to pronounce a small set of words: (yes, no, up, down, left, right, on, off, stop, go, and 0-9).\n - This data set provides synthetic counterparts to this real world dataset.\n\n## Open questions\n\nOne can use these two datasets in various ways. Here are some things I am interested in seeing answered:\n\n \n\n 1. What is it in an audio sample that makes it \"sound similar\"? Our ears can easily classify both synthetic and real speech, but for algorithms this is still hard. Extending the real dataset with the synthetic data yields a larger training sample and more diversity.\n 2. How well does an algorithm trained on one data set perform on the other? (transfer learning) If it works poorly, the algorithm probably has not found the key to audio similarity.\n 3. Are synthetic data sufficient for classifying real datasets? If this is the case, the implications are huge. You would not need to ask thousands of volunteers for hours of time. Instead, you could easily create arbitrary synthetic datasets for your target words.\n\nA interesting challenge (idea for competition) would be to train on this data set and evaluate on the real dataset.\n\n## Synthetic data creation\n\nHere I describe how the synthetic audio samples were created.\nCode is available at https://github.com/JohannesBuchner/spoken-command-recognition, in the \"tensorflow-speech-words\" folder.\n\n 1. The list of words is in \"inputwords\". \"marvin\" was changed to \"marvel\", because \"marvin\" does not have a pronounciation coding yet.\n 2. Pronounciations were taken from the British English Example Pronciation dictionary (BEEP, http://svr-www.eng.cam.ac.uk/comp.speech/Section1/Lexical/beep.html ). The phonemes were translated for the next step with a translation table (see compile.py for details). This creates the file \"words\". There are multiple pronounciations and stresses for each word.\n 3. A text-to-speech program (espeak) was used to pronounce these words (see generatetfspeech.sh for details). The pronounciation, stress, pitch, speed and speaker were varied. This gives >1000 clean examples for each word.\n 4. Noise samples were obtained. Noise samples (airport babble car exhibition restaurant street subway train) come from AURORA (https://www.ee.columbia.edu/~dpwe/sounds/noise/), and additional noise samples were synthetically created (ocean white brown pink). (see ../generatenoise.sh for details)\n 5. Noise and speech were mixed. The speech volume and offset were varied. The noise source, volume was also varied. See addnoise.py for details. addnoise2.py is the same, but with lower speech volume and higher noise volume. All audio files are one second (1s) long and are in wav format (16 bit, mono, 16000 Hz).\n 6. Finally, the data was compressed into an archive and uploaded to kaggle.\n\n## Acknowledgements\n\nThis work built upon\n\n - Pronounciation dictionary: BEEP: http://svr-www.eng.cam.ac.uk/comp.speech/Section1/Lexical/beep.html\n - Noise samples: AURORA: https://www.ee.columbia.edu/~dpwe/sounds/noise/\n - eSPEAK: http://espeak.sourceforge.net/ and mbrola voices http://www.tcts.fpms.ac.be/synthesis/mbrola/mbrcopybin.html\n\nPlease provide appropriate citations to the above when using this work.\n\nTo cite the resulting dataset, you can use:\n\nAPA-style citation: \"Buchner J. Synthetic Speech Commands: A public dataset for single-word speech recognition, 2017. Available from https://www.kaggle.com/jbuchner/synthetic-speech-commands-dataset/\".\n\nBibTeX @article{speechcommands, title={Synthetic Speech Commands: A public dataset for single-word speech recognition.}, author={Buchner, Johannes}, journal={Dataset available from https://www.kaggle.com/jbuchner/synthetic-speech-commands-dataset/}, year={2017} }\n\nThanks to everyone trying to improve open source voice detection and speech recognition.\n\n## Links\n\n- https://www.kaggle.com/jbuchner/spokenverbs\n- https://www.kaggle.com/c/tensorflow-speech-recognition-challenge\n- https://github.com/JohannesBuchner/spoken-command-recognition/\n\n [1]: https://www.kaggle.com/jbuchner/spokenverbs\n [2]: https://www.kaggle.com/c/tensorflow-speech-recognition-challenge/data", "VersionNotes": "Add directory layout", "TotalCompressedBytes": 2698665224.0, "TotalUncompressedBytes": 2698665224.0}]
|
[{"Id": 7691, "CreatorUserId": 1360065, "OwnerUserId": 1360065.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 39731.0, "CurrentDatasourceVersionId": 41901.0, "ForumId": 14555, "Type": 2, "CreationDate": "12/22/2017 18:43:07", "LastActivityDate": "02/05/2018", "TotalViews": 34651, "TotalDownloads": 3106, "TotalVotes": 83, "TotalKernels": 10}]
|
[{"Id": 1360065, "UserName": "jbuchner", "DisplayName": "JohannesBuchner", "RegisterDate": "10/25/2017", "PerformanceTier": 1}]
|
# # By : 𝔄𝔥𝔪𝔢𝔡 𝔄𝔰𝔥𝔯𝔞𝔣
# dataset:https://www.kaggle.com/datasets/jbuchner/synthetic-speech-commands-dataset
# we'll build Model to use it speech recognition for thirty words
# dataset consisting of 41849 audio
# # Importing Libraries & Data
import librosa as lr
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import os
import IPython.display as ipd
import keras
from sklearn.model_selection import train_test_split
import tensorflow as tf
import pickle as pk
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
from scipy.io.wavfile import write
dataset = "/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset"
pd.DataFrame(os.listdir(dataset), columns=["Files"])
def count(path):
size = []
for file in os.listdir(path):
size.append(len(os.listdir(os.path.join(path, file))))
return pd.DataFrame(size, columns=["Number Of Sample"], index=os.listdir(path))
tr = count(dataset)
tr
plt.figure(figsize=(10, 10))
plt.pie(x="Number Of Sample", labels=os.listdir(dataset), autopct="%1.1f%%", data=tr)
plt.title("Distribution Of Data In Train", fontsize=20)
plt.show()
sns.kdeplot(tr["Number Of Sample"], color="red")
plt.title("Kdeplot For Train", fontsize=20)
plt.xlabel("Number", fontsize=20)
plt.ylabel("Count", fontsize=20)
# bed Audio
ipd.Audio(
"/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset/bed/1014.wav"
)
# bird Audio
ipd.Audio(
"/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset/bird/1019.wav"
)
# cat Audio
ipd.Audio(
"/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset/cat/1009.wav"
)
# yes Audio
ipd.Audio(
"/kaggle/input/synthetic-speech-commands-dataset/augmented_dataset/augmented_dataset/yes/1014.wav"
)
def normalize_signal(signal):
# Calculate the peak amplitude of the signal
peak_amplitude = np.max(np.abs(signal))
# Normalize the signal by dividing it by the peak amplitude
normalized_signal = signal / peak_amplitude
return normalized_signal
def load(path):
data = []
label = []
sample = []
for file in os.listdir(path):
path_ = os.path.join(path, file)
for fil in os.listdir(path_):
data_contain, sample_rate = lr.load(os.path.join(path_, fil), sr=16000)
data_contain = normalize_signal(data_contain)
data.append(data_contain)
sample.append(sample_rate)
label.append(file)
return data, label, sample
data, label, sample = load(dataset)
df = pd.DataFrame()
df["Label"], df["sample"] = label, sample
df
# # Audio Feature Extraction
# waveform
def waveform(data, sr, label):
plt.figure(figsize=(14, 5))
lr.display.waveshow(data, sr=sr)
plt.suptitle(label)
plt.title("Waveform plot")
plt.xlabel("Time (s)")
plt.ylabel("Amplitude")
# MFCC features
def mfcc(data, sr):
mfccs = lr.feature.mfcc(y=data, sr=sr, n_mfcc=13)
return np.mean(mfccs), mfccs
def mfcc_v(mfccs, label):
plt.figure(figsize=(10, 4))
lr.display.specshow(mfccs, x_axis="time")
plt.colorbar()
plt.title("MFCC")
plt.suptitle(label)
# Mel-spectrogram
def Mel(data, sr):
mel_spec = lr.feature.melspectrogram(y=data, sr=sr)
return np.mean(mel_spec), mel_spec
def mel_v(mel_spec, label, sr):
# Convert to decibel scale
mel_spec_db = lr.power_to_db(mel_spec, ref=np.max)
# Visualize Mel-spectrogram
plt.figure(figsize=(10, 4))
lr.display.specshow(mel_spec_db, x_axis="time", y_axis="mel", sr=sr)
plt.colorbar(format="%+2.0f dB")
plt.title("Mel-spectrogram")
plt.suptitle(label)
# zero_crossing_rate
def zero_crossing(data, sr):
# Compute zero-crossing rate
zcr = lr.feature.zero_crossing_rate(data)
# Print average zero-crossing rate
avg_zcr = sum(zcr[0]) / len(zcr[0])
print("Average zero-crossing rate:", avg_zcr)
return zcr
def zero_crossing_v(zcr, label, data, sr):
time = lr.times_like(zcr)
# Create waveform plot
plt.figure(figsize=(14, 5))
lr.display.waveshow(data, sr=sr, alpha=0.5)
plt.plot(time, zcr[0], color="r")
plt.title("Zero-crossing rate")
plt.xlabel("Time (s)")
plt.ylabel("Amplitude")
plt.suptitle(label)
# waveform data[0]
waveform(data[0], sample[0], label[0])
plt.legend()
# MFCC data[0]
mfccs_mean, mfccs = mfcc(data[0], sample[0])
print("MFCCs Mean:", mfccs_mean)
print("MFCCs shape:", mfccs.shape)
mfcc_v(mfccs, label[0])
##Mel-spectrogram data[0]
mel_mean, mel = Mel(data[0], sample[0])
print("Mel Mean:", mel_mean)
print("Mel :", mel.shape)
mel_v(mel, label[0], sample[0])
# zero_crossing data[0]
zcr = zero_crossing(data[0], sample[0])
print("Zcr:", zcr.shape)
zero_crossing_v(zcr, label[0], data[0], sample[0])
# waveform data[1000]
waveform(data[1000], sample[1000], label[1000])
plt.legend()
# MFCC data[1000]
mfccs_mean, mfccs = mfcc(data[1000], sample[1000])
print("MFCCs Mean:", mfccs_mean)
print("MFCCs shape:", mfccs.shape)
mfcc_v(mfccs, label[1000])
##Mel-spectrogram data[1000]
mel_mean, mel = Mel(data[1000], sample[1000])
print("Mel Mean:", mel_mean)
print("Mel :", mel.shape)
mel_v(mel, label[1000], sample[1000])
# zero_crossing data[1000]
zcr = zero_crossing(data[1000], sample[1000])
print("Zcr:", zcr.shape)
zero_crossing_v(zcr, label[1000], data[1000], sample[1000])
code = {}
x = 0
for i in pd.unique(label):
code[i] = x
x += 1
pd.DataFrame(code.values(), columns=["Value"], index=code.keys())
def get_Name(N):
for x, y in code.items():
if y == N:
return x
for i in range(len(label)):
label[i] = code[label[i]]
pd.DataFrame(label, columns=["Labels"])
mfccs = [lr.feature.mfcc(y=d, sr=s, n_mfcc=13) for d, s in zip(data, sample)]
mfccs = np.array(mfccs)
# # Splitting Data
label = np.array(label)
X_train, X_test, y_train, y_test = train_test_split(
mfccs, label, test_size=0.1, random_state=44, shuffle=True
)
print("X_train shape is ", X_train.shape)
print("X_test shape is ", X_test.shape)
print("y_train shape is ", y_train.shape)
print("y_test shape is ", y_test.shape)
X_train = X_train.reshape((X_train.shape[0], -1))
X_test = X_test.reshape((X_test.shape[0], -1))
print("X_train shape is ", X_train.shape)
print("X_test shape is ", X_test.shape)
# # Model Building
num_class = len(pd.unique(y_train))
model = keras.Sequential()
model.add(keras.layers.Dense(512, activation="relu", input_shape=(X_train.shape[1],)))
model.add(keras.layers.Dense(128, activation="relu"))
model.add(keras.layers.Dense(64, activation="relu"))
model.add(keras.layers.Dense(num_class, activation="softmax"))
model.summary()
tf.keras.utils.plot_model(
model,
to_file="model.png",
show_shapes=True,
show_layer_names=True,
show_dtype=True,
dpi=120,
)
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
hist = model.fit(X_train, y_train, epochs=50)
loss, acc = model.evaluate(X_test, y_test)
print("Loss is :", loss)
print("ACC is :", acc)
hist_ = hist.history
pd.DataFrame(hist_)
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
plt.plot(hist_["loss"], c="r", marker="*", label="Loss")
plt.title("Overall Loss", fontsize=20)
plt.legend()
plt.subplot(1, 2, 2)
plt.plot(hist_["accuracy"], label="Accuracy")
plt.title("Overall Accuracy", fontsize=20)
plt.legend()
predict = model.predict(X_test)
predict[0]
preN = []
prename = []
for row in predict:
N = np.argmax(row)
preN.append(N)
prename.append(get_Name(N))
pd.DataFrame(prename, columns=["Predictions"])
predict = []
y_act = []
for p in range(30):
y_act.append(get_Name(y_test[p]))
predict.append(prename[p])
pd_p = pd.DataFrame(y_act, columns=["y_act"])
pd_p["predict"] = predict
pd_p
# # Model Check
plt.figure(figsize=(15, 15))
ax = plt.subplot()
CM = confusion_matrix(y_test, preN)
sns.heatmap(CM, annot=True, fmt="g", ax=ax, cbar=False, cmap="RdBu")
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix")
ax.xaxis.set_ticklabels(code.keys())
ax.yaxis.set_ticklabels(code.keys())
plt.show()
CM
ClassificationReport = classification_report(y_test, preN)
print("Classification Report is : ", ClassificationReport)
# # Save Model
model.save("/kaggle/working/speech_model.h5")
# # REAL TRIAL
code = {
"no": 0,
"two": 1,
"four": 2,
"five": 3,
"nine": 4,
"right": 5,
"off": 6,
"yes": 7,
"six": 8,
"dog": 9,
"left": 10,
"bird": 11,
"marvel": 12,
"wow": 13,
"zero": 14,
"eight": 15,
"bed": 16,
"go": 17,
"house": 18,
"tree": 19,
"seven": 20,
"on": 21,
"three": 22,
"one": 23,
"down": 24,
"stop": 25,
"up": 26,
"happy": 27,
"cat": 28,
"sheila": 29,
}
def get_Name(N):
for x, y in code.items():
if y == N:
return x
def record():
sample_rate = 16000
duration = 1
recording = sd.rec(int(duration * sample_rate), samplerate=sample_rate, channels=1)
sd.wait()
write("recording.wav", sample_rate, recording)
def predict():
data, sr = lr.load("recording.wav", sr=16000)
data = normalize_signal(data)
data = data.reshape(-1, 16000, 1)
savedmodel = keras.models.load_model("speech_model.h5")
pred = savedmodel.predict(data)
for row in pred:
N = np.argmax(row)
print(get_Name(N))
record()
ipd.Audio("recording.wav")
# waveform data
data, sr = lr.load("recording.wav", sr=16000)
data = normalize_signal(data)
waveform(data, sr, "label")
# MFCC data[0]
mfccs_mean, mfccs = mfcc(data, sr)
print("MFCCs Mean:", mfccs_mean)
print("MFCCs shape:", mfccs.shape)
mfcc_v(mfccs, "label")
mel_mean, mel = Mel(data, sr)
print("Mel Mean:", mel_mean)
print("Mel :", mel.shape)
mel_v(mel, "label", sr)
# zero_crossing data
zcr = zero_crossing(data, sr)
print("Zcr:", zcr.shape)
zero_crossing_v(zcr, "label", data, sr)
predict()
| false | 0 | 3,710 | 0 | 5,031 | 3,710 |
||
129470747
|
<jupyter_start><jupyter_text>Cancer Data
**570 cancer cells and 30 features to determine whether the cancer cells in our data are benign or malignant**
**Our cancer data contains 2 types of cancers: 1. benign cancer (B) and 2. malignant cancer (M).**
Kaggle dataset identifier: cancer-data
<jupyter_script>import pandas as pd
df = pd.read_csv("/kaggle/input/cancer-data/Cancer_Data.csv")
df.head()
from sklearn.model_selection import train_test_split
x = df.drop(["Unnamed: 32", "diagnosis"], axis=1)
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier()
model.fit(x_train, y_train)
y_pred = model.predict(x_test)
model.score(x_test, y_test)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/470/129470747.ipynb
|
cancer-data
|
erdemtaha
|
[{"Id": 129470747, "ScriptId": 38496359, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14115484, "CreationDate": "05/14/2023 05:35:37", "VersionNumber": 1.0, "Title": "cancer stage detection", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 23.0, "LinesInsertedFromPrevious": 23.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185558952, "KernelVersionId": 129470747, "SourceDatasetVersionId": 5212576}]
|
[{"Id": 5212576, "DatasetId": 3032092, "DatasourceVersionId": 5284991, "CreatorUserId": 2498226, "LicenseName": "Other (specified in description)", "CreationDate": "03/22/2023 07:57:00", "VersionNumber": 1.0, "Title": "Cancer Data", "Slug": "cancer-data", "Subtitle": "Benign and malignant cancer data", "Description": "**570 cancer cells and 30 features to determine whether the cancer cells in our data are benign or malignant**\n\n**Our cancer data contains 2 types of cancers: 1. benign cancer (B) and 2. malignant cancer (M).**", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3032092, "CreatorUserId": 2498226, "OwnerUserId": 2498226.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5212576.0, "CurrentDatasourceVersionId": 5284991.0, "ForumId": 3071494, "Type": 2, "CreationDate": "03/22/2023 07:57:00", "LastActivityDate": "03/22/2023", "TotalViews": 66608, "TotalDownloads": 11493, "TotalVotes": 209, "TotalKernels": 70}]
|
[{"Id": 2498226, "UserName": "erdemtaha", "DisplayName": "Erdem Taha", "RegisterDate": "11/15/2018", "PerformanceTier": 1}]
|
import pandas as pd
df = pd.read_csv("/kaggle/input/cancer-data/Cancer_Data.csv")
df.head()
from sklearn.model_selection import train_test_split
x = df.drop(["Unnamed: 32", "diagnosis"], axis=1)
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier()
model.fit(x_train, y_train)
y_pred = model.predict(x_test)
model.score(x_test, y_test)
| false | 1 | 163 | 0 | 254 | 163 |
||
129470308
|
import torch
import torch.nn as nn
import torchvision
import torchvision.transforms as transforms
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from torch.utils.data import TensorDataset, DataLoader
# device config
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# hyper parameters
input_size = 28 * 28
hidden_size = 100
num_classes = 10
num_epochs = 2
batch_size = 100
lr_rate = 0.001
trainds = pd.read_csv("/kaggle/input/digit-recognizer/train.csv")
X_train = torch.tensor(trainds.drop("label", axis=1).values, dtype=torch.float32)
y_train = torch.tensor(trainds["label"], dtype=torch.float32)
ds = TensorDataset(X_train, y_train)
ds[0]
dl = DataLoader(dataset=ds, batch_size=batch_size, shuffle=True)
examples = iter(dl)
samples, labels = next(examples)
for i in range(6):
plt.subplot(2, 3, i + 1)
plt.imshow(samples[i].reshape(28, 28), cmap="gray")
plt.show()
class MNISTNN(nn.Module):
def __init__(self, input_layer, hidden_layer, output_layer):
super(MNISTNN, self).__init__()
self.l1 = nn.Linear(input_layer, hidden_layer)
self.relu = nn.ReLU()
self.l2 = nn.Linear(hidden_layer, output_layer)
def forward(self, x):
out = self.l1(x)
out = self.relu(out)
out = self.l2(out)
# no softmax/ activation function
return out
model = MNISTNN(input_size, hidden_size, num_classes)
# **Note** Here if you observe we don't use the softmax activation function for multi class classification as later for the loss calculation we will use the cross entropy loss which performs the softmax activation itself.
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(params=model.parameters(), lr=lr_rate)
print(device)
n_total_steps = len(dl)
for epoch in range(num_epochs):
for i, (images, labels) in enumerate(dl):
images = images.reshape(-1, 28 * 28).to(device)
labels = labels.type(torch.LongTensor)
labels = labels.to(device)
model = model.to(device)
y_pred = model(images)
loss = criterion(y_pred, labels)
optimizer.zero_grad()
loss.backward()
optimizer.step()
if (i + 1) % 100 == 0:
print(
f"epoch: {epoch+1}/ {num_epochs}, step: {i+1}/{n_total_steps}, loss: {loss.item():.4f}"
)
testds = pd.read_csv("/kaggle/input/digit-recognizer/test.csv")
X_test = torch.tensor(testds.values, dtype=torch.float32)
# with torch.no_grad():
# n_correct = 0
# n_samples = 0
# for images, samples in dl:
# images = images.reshape(-1,28*28).to(device)
# labels = labels.to(device)
# output = model(images)
# _, predictions = torch.max(output,1)
# n_samples += labels.shape[0]
# n_correct += (predictions == labels).sum().item()
# accuracy = 100 * (n_correct/n_samples)
# print(f'accuracy: {accuracy:.4f}')
ImageId = []
Labels = []
for i, image in enumerate(X_test):
output = model(image.reshape(-1, 28 * 28).to(device))
_, predictions = torch.max(output, 1)
ImageId.append(i)
Labels.append(predictions.item())
# final_output = predictions.cpu().numpy()
df = pd.DataFrame({"ImageId": ImageId, "Label": Labels})
df.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/470/129470308.ipynb
| null | null |
[{"Id": 129470308, "ScriptId": 38494135, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13032715, "CreationDate": "05/14/2023 05:29:44", "VersionNumber": 3.0, "Title": "Digit Recognizer using Pytorch", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 109.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 108.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import torch
import torch.nn as nn
import torchvision
import torchvision.transforms as transforms
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from torch.utils.data import TensorDataset, DataLoader
# device config
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# hyper parameters
input_size = 28 * 28
hidden_size = 100
num_classes = 10
num_epochs = 2
batch_size = 100
lr_rate = 0.001
trainds = pd.read_csv("/kaggle/input/digit-recognizer/train.csv")
X_train = torch.tensor(trainds.drop("label", axis=1).values, dtype=torch.float32)
y_train = torch.tensor(trainds["label"], dtype=torch.float32)
ds = TensorDataset(X_train, y_train)
ds[0]
dl = DataLoader(dataset=ds, batch_size=batch_size, shuffle=True)
examples = iter(dl)
samples, labels = next(examples)
for i in range(6):
plt.subplot(2, 3, i + 1)
plt.imshow(samples[i].reshape(28, 28), cmap="gray")
plt.show()
class MNISTNN(nn.Module):
def __init__(self, input_layer, hidden_layer, output_layer):
super(MNISTNN, self).__init__()
self.l1 = nn.Linear(input_layer, hidden_layer)
self.relu = nn.ReLU()
self.l2 = nn.Linear(hidden_layer, output_layer)
def forward(self, x):
out = self.l1(x)
out = self.relu(out)
out = self.l2(out)
# no softmax/ activation function
return out
model = MNISTNN(input_size, hidden_size, num_classes)
# **Note** Here if you observe we don't use the softmax activation function for multi class classification as later for the loss calculation we will use the cross entropy loss which performs the softmax activation itself.
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(params=model.parameters(), lr=lr_rate)
print(device)
n_total_steps = len(dl)
for epoch in range(num_epochs):
for i, (images, labels) in enumerate(dl):
images = images.reshape(-1, 28 * 28).to(device)
labels = labels.type(torch.LongTensor)
labels = labels.to(device)
model = model.to(device)
y_pred = model(images)
loss = criterion(y_pred, labels)
optimizer.zero_grad()
loss.backward()
optimizer.step()
if (i + 1) % 100 == 0:
print(
f"epoch: {epoch+1}/ {num_epochs}, step: {i+1}/{n_total_steps}, loss: {loss.item():.4f}"
)
testds = pd.read_csv("/kaggle/input/digit-recognizer/test.csv")
X_test = torch.tensor(testds.values, dtype=torch.float32)
# with torch.no_grad():
# n_correct = 0
# n_samples = 0
# for images, samples in dl:
# images = images.reshape(-1,28*28).to(device)
# labels = labels.to(device)
# output = model(images)
# _, predictions = torch.max(output,1)
# n_samples += labels.shape[0]
# n_correct += (predictions == labels).sum().item()
# accuracy = 100 * (n_correct/n_samples)
# print(f'accuracy: {accuracy:.4f}')
ImageId = []
Labels = []
for i, image in enumerate(X_test):
output = model(image.reshape(-1, 28 * 28).to(device))
_, predictions = torch.max(output, 1)
ImageId.append(i)
Labels.append(predictions.item())
# final_output = predictions.cpu().numpy()
df = pd.DataFrame({"ImageId": ImageId, "Label": Labels})
df.to_csv("submission.csv", index=False)
| false | 0 | 1,057 | 0 | 1,057 | 1,057 |
||
129470071
|
<jupyter_start><jupyter_text>2021 NWSL Stats by Team
All data obtained is from the [National Womens Soccer League's Website](https://www.nwslsoccer.com/stats/teams).
The inspiration behind this dataset was to compare the NWSL stats of each team for that given year. I personally love stats and wtching soccer, so I created one myself. As I scrolled the available datasets I didn't see to many on Women's sports another reason I created the dataset.
Kaggle dataset identifier: 2021-nwsl-stats-by-team
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
nwsl_full = pd.read_csv(
"/kaggle/input/2021-nwsl-stats-by-team/2021 NWSL Stats by Team - Sheet1.csv"
)
nwsl_2018 = pd.read_csv(
"/kaggle/input/2018-nwsl-stats-by-team/2018 NWSL Stats by Team - Sheet1.csv"
)
# Number of rows?
nwsl_full.shape
# The 2021 dataset has 10 rows
# What other data is available about NWSL teams? *The 2018 season*
# #
# Could other data be gathered to extend analysis? *Unsure about this question still*
# Below is the dataset that joins the 2021 stats and the 2018 stats
nwsl_full.set_index("Team")
nwsl_years = nwsl_full.join(nwsl_2018, lsuffix="21", rsuffix="18")
nwsl_years.describe()
# Lookup Question: Which team has the most shots on goal? Is that also the team who was ranked number 1?
max_sog = nwsl_full.SOG.max()
nwsl_full.loc[nwsl_full.SOG == max_sog]
# Portland was ranked 5th but had the most shots on goal.
# Arithmetic Question: What is the average amount of fouls comitted per team?
nwsl_full.FC.describe()
# The average fouls per team was 236.1
# Could a formula be created using past season stats to predict winners for future seasons? *I don't know if I currently have the skills to answer this question*
# Are there any correlations that might not be expected like do teams that have more fouls also take more shots on goal?
# What does the distribution of various statistics look like and how do they compare to each other?
nwsl_full.describe()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/470/129470071.ipynb
|
2021-nwsl-stats-by-team
|
graciebarnes
|
[{"Id": 129470071, "ScriptId": 38482249, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14516202, "CreationDate": "05/14/2023 05:26:44", "VersionNumber": 1.0, "Title": "NWSL Data Exploration", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 57.0, "LinesInsertedFromPrevious": 57.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185557181, "KernelVersionId": 129470071, "SourceDatasetVersionId": 4059370}, {"Id": 185557182, "KernelVersionId": 129470071, "SourceDatasetVersionId": 4059726}]
|
[{"Id": 4059370, "DatasetId": 2403615, "DatasourceVersionId": 4115426, "CreatorUserId": 11245219, "LicenseName": "CC0: Public Domain", "CreationDate": "08/11/2022 18:25:05", "VersionNumber": 1.0, "Title": "2021 NWSL Stats by Team", "Slug": "2021-nwsl-stats-by-team", "Subtitle": "The statistics of the National Women's Soccer League by Team for the 2021 season", "Description": "All data obtained is from the [National Womens Soccer League's Website](https://www.nwslsoccer.com/stats/teams).\n\nThe inspiration behind this dataset was to compare the NWSL stats of each team for that given year. I personally love stats and wtching soccer, so I created one myself. As I scrolled the available datasets I didn't see to many on Women's sports another reason I created the dataset.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2403615, "CreatorUserId": 11245219, "OwnerUserId": 11245219.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4059370.0, "CurrentDatasourceVersionId": 4115426.0, "ForumId": 2430945, "Type": 2, "CreationDate": "08/11/2022 18:25:05", "LastActivityDate": "08/11/2022", "TotalViews": 276, "TotalDownloads": 32, "TotalVotes": 7, "TotalKernels": 1}]
|
[{"Id": 11245219, "UserName": "graciebarnes", "DisplayName": "Gracie Barnes", "RegisterDate": "08/08/2022", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
nwsl_full = pd.read_csv(
"/kaggle/input/2021-nwsl-stats-by-team/2021 NWSL Stats by Team - Sheet1.csv"
)
nwsl_2018 = pd.read_csv(
"/kaggle/input/2018-nwsl-stats-by-team/2018 NWSL Stats by Team - Sheet1.csv"
)
# Number of rows?
nwsl_full.shape
# The 2021 dataset has 10 rows
# What other data is available about NWSL teams? *The 2018 season*
# #
# Could other data be gathered to extend analysis? *Unsure about this question still*
# Below is the dataset that joins the 2021 stats and the 2018 stats
nwsl_full.set_index("Team")
nwsl_years = nwsl_full.join(nwsl_2018, lsuffix="21", rsuffix="18")
nwsl_years.describe()
# Lookup Question: Which team has the most shots on goal? Is that also the team who was ranked number 1?
max_sog = nwsl_full.SOG.max()
nwsl_full.loc[nwsl_full.SOG == max_sog]
# Portland was ranked 5th but had the most shots on goal.
# Arithmetic Question: What is the average amount of fouls comitted per team?
nwsl_full.FC.describe()
# The average fouls per team was 236.1
# Could a formula be created using past season stats to predict winners for future seasons? *I don't know if I currently have the skills to answer this question*
# Are there any correlations that might not be expected like do teams that have more fouls also take more shots on goal?
# What does the distribution of various statistics look like and how do they compare to each other?
nwsl_full.describe()
| false | 2 | 661 | 0 | 800 | 661 |
||
129470970
|
<jupyter_start><jupyter_text>gen-text
Kaggle dataset identifier: gen-text
<jupyter_script>import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# ## Загрузка библиотек
import pandas as pd
import matplotlib.pyplot as plt
import re
import time
import warnings
import numpy as np
from nltk.corpus import stopwords
from sklearn.decomposition import TruncatedSVD
from sklearn.preprocessing import normalize
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.manifold import TSNE
import seaborn as sns
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import confusion_matrix
from sklearn.metrics import accuracy_score, log_loss
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.linear_model import SGDClassifier
from imblearn.over_sampling import SMOTE
from collections import Counter
from scipy.sparse import hstack
from sklearn.multiclass import OneVsRestClassifier
from sklearn.svm import SVC
from sklearn.model_selection import StratifiedKFold
from collections import Counter, defaultdict
from sklearn.calibration import CalibratedClassifierCV
from sklearn.naive_bayes import MultinomialNB
from sklearn.naive_bayes import GaussianNB
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
import math
from sklearn.metrics import normalized_mutual_info_score
warnings.filterwarnings("ignore")
import six
import sys
sys.modules["sklearn.externals.six"] = six
from mlxtend.classifier import StackingClassifier
from sklearn import model_selection
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import RandomizedSearchCV
from sklearn.metrics import log_loss, make_scorer
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report
from sklearn.svm import SVC
import plotly.express as px
# ### Чтение данных, изучение, визуальное представление
data = pd.read_csv("/kaggle/input/gen-competition/training_variants")
print("Количество записей в таблице : ", data.shape[0])
print("Количество полей в таблице: ", data.shape[1])
print("Поля: ", data.columns.values)
data.sample(7)
data_text = pd.read_csv(
"/kaggle/input/gen-text/training_text", sep="\|\|", names=["ID", "text"], skiprows=1
)
print("Количество записей в таблице : ", data_text.shape[0])
print("Количество полей в таблице: ", data_text.shape[1])
print("Поля: ", data_text.columns.values)
data_text.sample(8)
# ### Предварительный анализ данных
data.describe()
data.groupby("Class").size().plot(
kind="bar",
color="maroon",
y="ID",
figsize=(4, 4),
title="Количество данных по классам",
)
d = data.groupby("Class").size()
print(d)
d.plot(
kind="pie",
y="ID",
title="Количество данных по классам",
figsize=(4, 4),
autopct="%1.1f%%",
startangle=200,
)
plt.show()
print(data.groupby("Class").size())
# Проанализируем выборку по количестве записей по каждому гену
d = data.groupby(["Gene", "Class"]).size()
d.head(50)
gene_count = data.groupby("Gene").size()
# data.groupby('Gene').size().plot(kind='bar', color = 'maroon', y='Gene' , title='Количественное соотношение записей выборки по генам')
fig = px.bar(
gene_count,
title="Количественное соотношение записей выборки по разновидности генов",
)
fig.show()
# ## Подключаем библиотеки для предварительного анализа текста
import pymorphy2
import nltk
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
a = 1
# ### Предобработка текста: удаляем лишние символы, преобразуем к нижнему регистру, разбиваем на токены - по словам, приводим в нормальную форму
def goodtext(text):
# избавляемся от ненужных символов и убираем несколько пробелов, также убираем "одиночно-стоящие" буквы, числа оставляем
text = re.sub("[^a-z0-9\n]", " ", text)
text = re.sub(r"\b[a-zA-Z]\b", " ", text)
text = re.sub("\s+", " ", text)
return text
def preprocess(text, stop_words, morph):
tokens = word_tokenize(goodtext(text.lower()))
preprocessed_text = []
for token in tokens:
lemma = morph.parse(token)[0].normal_form
if lemma not in stop_words:
preprocessed_text.append(lemma)
global a
if a % 100 == 0:
print(a)
a += 1
return preprocessed_text
# punctuation_marks = ['!', ',', '(', ')', ':', '-', '?', '.', '..', '...', '«', '»', ';', '–', '--', '*','[',']', '%','>','<', '=','’','“','”', '±','{','}','"',"'"]
stop_words = stopwords.words("english")
morph = pymorphy2.MorphAnalyzer()
data_text.head()
stop_words.append("whose")
stop_words.append("also")
# ## удаляем пустые значения из таблицы и удаляем связанные с ними данные в другой таблице
data_text.describe()
data_text = data_text.dropna()
index = [id for id in data_text["ID"]]
# удаляем записи которых нет в первой таблице
data = data.query("ID in `index`")
data_text.describe()
start_time = time.clock()
data_text["text"] = data_text.apply(
lambda row: preprocess(row["text"], stop_words, morph), axis=1
)
print(data_text["text"])
print("Time took for preprocessing the text :", time.clock() - start_time, "seconds")
result = pd.merge(data, data_text, on="ID", how="left")
result.to_pickle("result.pkl")
data_text["text"]
# ### Собираем данные в одну таблицу и сохраняемв scv-файл
result = pd.merge(data, data_text, on="ID", how="left")
# result.to_csv('result.csv', index= False)
result.head()
result.to_pickle("result.pkl")
data_new = pd.read_pickle("/kaggle/input/data-txt/result2.pkl")
# data_new['text'][0]
# ### создаём мешок слов
from collections import Counter
# ## проверим работу "Счетчика слов" на тексте первой строки - построим словарь
test_counter = Counter(data_new["text"][0])
# rint(type(data_new['text'][0]))
test_counter.most_common(100)
# ### Построим словарь слово:кол-во для текстового поля всей выборки
# Следует наделить контекстом поля "text" с нулевыми значениями
# заменяем пустые значения
data_new["text"] = data_new["text"].fillna("0")
data_new.loc[data_new["text"] == "0"].text = data_new.apply(
lambda x: [x["Gene"], x["Variation"]], axis=1
)
# data_new = data_new.dropna() # удалим пустые значения
pd.set_option("max_colwidth", 180)
pd.set_option("display.width", 600)
print("Количество записей в таблице : ", data_new.shape[0])
print("Количество полей в таблице: ", data_new.shape[1])
print("Поля: ", data_new.columns.values)
data_new.sample(9)
# Создаем словарь, упорядоченный по частоте
# В словаре будем использовать 2 специальных кода:
# Код заполнитель: 0
# Неизвестное слово: 1
# Нумерация слов в словаре начинается с 2.
# Словарь, отображающий слова в коды
word_to_index = dict()
# Словарь, отображающий коды в слова
index_to_word = dict()
all_text = Counter()
for txt in data_new["text"]:
all_text.update(txt)
print("Часть мешка слов:", all_text.most_common(20))
print("\nКоличество элементов Мешка слов:", len(all_text))
max_words = 150000
for i, w in enumerate(all_text.most_common(max_words - 2)):
word_to_index[w[0]] = i + 2
index_to_word[i + 2] = w[0]
# ### Функция для преобразования списка слов в список кодов
def text_to_sequence(txt, word_to_index):
seq = []
for word in txt:
index = word_to_index.get(word, 1) # 1 означает неизвестное слово
# Неизвестные слова не добавляем в выходную последовательность
if index != 1:
seq.append(index)
return seq
def class_(id):
return id - 1
# Преобразуем все тексты в последовательность кодов слов
data_new["digital"] = data_new.apply(
lambda row: text_to_sequence(row["text"], word_to_index), axis=1
)
data_new["Class"] = data_new.apply(lambda row: class_(row["Class"]), axis=1)
print("Количество записей в таблице : ", data_new.shape[0])
print("Количество полей в таблице: ", data_new.shape[1])
print("Поля: ", data_new.columns.values)
data_new.sample(9)
# ### разбиваем выборку на две части - данные для тестирования, данные для обучения
train, test = train_test_split(data_new, test_size=0.2)
x_train_seq = train["digital"]
y_train = train["Class"]
x_test_seq = test["digital"]
y_test = test["Class"]
def vectorize_sequences(sequences, dimension=100000):
results = np.zeros((len(sequences), dimension))
for i, sequence in enumerate(sequences):
for index in sequence:
results[i, index] += 1.0
return results
x_train = vectorize_sequences(x_train_seq, max_words)
x_test = vectorize_sequences(x_test_seq, max_words)
print(x_train)
# ### Создаем модель машинного обучения
random_state = 10000
lr = LogisticRegression(random_state=random_state, max_iter=500)
# ### Обучаем модель машинного обучения
lr.fit(x_train, y_train)
lr.score(x_test, y_test)
lr_prediction = lr.predict(x_test)
accuracy = accuracy_score(y_test, lr_prediction)
print(f"Accuracy: {accuracy}")
conf_mtrx = confusion_matrix(y_test, lr_prediction)
print(conf_mtrx)
lr2 = LogisticRegression(
C=1.0,
class_weight=None,
dual=False,
fit_intercept=True,
intercept_scaling=1,
max_iter=500,
multi_class="multinomial",
n_jobs=1,
penalty="l2",
random_state=0,
tol=0.0001,
verbose=0,
warm_start=False,
)
lr2.fit(x_train, y_train)
lr2_prediction = lr2.predict(x_test)
accuracy2 = accuracy_score(y_test, lr2_prediction)
print(f"Accuracy: {accuracy2}")
conf_mtrx2 = confusion_matrix(y_test, lr2_prediction)
print(conf_mtrx2)
# ###
from scipy.stats import loguniform
from sklearn.model_selection import RandomizedSearchCV
nb = MultinomialNB()
param = dict()
param["alpha"] = loguniform(1e-5, 1)
# parameter={'alpha':[0.00001,0.0001,0.001,0.1,1,10,100,1000]}
LogLoss = make_scorer(log_loss, greater_is_better=False, needs_proba=True)
clf = RandomizedSearchCV(nb, param, n_iter=500, scoring=LogLoss)
clf.fit(x_train, y_train)
from scipy.stats import loguniform
from sklearn.model_selection import RandomizedSearchCV
nb = MultinomialNB(alpha=0.0060888387195).fit(x_train, y_train)
prediction = nb.predict_proba(x_test)
print("Функция потерь:", log_loss(y_test, prediction))
# ### Векторизация текста на основе эмбединга TfidfVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer()
data_new["digital_1"] = vectorizer.fit_transform(data_new["text"])
# X = vectorizer.fit_transform(corpus)
# print(vectorizer.get_feature_names())
print(data_new.sample(9))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/470/129470970.ipynb
|
gen-text
|
olgapolovikova
|
[{"Id": 129470970, "ScriptId": 37109682, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8842968, "CreationDate": "05/14/2023 05:38:55", "VersionNumber": 3.0, "Title": "notebook_for_competishion_2", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 327.0, "LinesInsertedFromPrevious": 40.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 287.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185559433, "KernelVersionId": 129470970, "SourceDatasetVersionId": 5412588}, {"Id": 185559434, "KernelVersionId": 129470970, "SourceDatasetVersionId": 5653113}]
|
[{"Id": 5412588, "DatasetId": 3134615, "DatasourceVersionId": 5486476, "CreatorUserId": 8842968, "LicenseName": "Unknown", "CreationDate": "04/15/2023 09:33:45", "VersionNumber": 1.0, "Title": "gen-text", "Slug": "gen-text", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3134615, "CreatorUserId": 8842968, "OwnerUserId": 8842968.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5956622.0, "CurrentDatasourceVersionId": 6034275.0, "ForumId": 3198330, "Type": 2, "CreationDate": "04/15/2023 09:33:45", "LastActivityDate": "04/15/2023", "TotalViews": 64, "TotalDownloads": 3, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 8842968, "UserName": "olgapolovikova", "DisplayName": "Olga Polovikova", "RegisterDate": "11/10/2021", "PerformanceTier": 0}]
|
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# ## Загрузка библиотек
import pandas as pd
import matplotlib.pyplot as plt
import re
import time
import warnings
import numpy as np
from nltk.corpus import stopwords
from sklearn.decomposition import TruncatedSVD
from sklearn.preprocessing import normalize
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.manifold import TSNE
import seaborn as sns
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import confusion_matrix
from sklearn.metrics import accuracy_score, log_loss
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.linear_model import SGDClassifier
from imblearn.over_sampling import SMOTE
from collections import Counter
from scipy.sparse import hstack
from sklearn.multiclass import OneVsRestClassifier
from sklearn.svm import SVC
from sklearn.model_selection import StratifiedKFold
from collections import Counter, defaultdict
from sklearn.calibration import CalibratedClassifierCV
from sklearn.naive_bayes import MultinomialNB
from sklearn.naive_bayes import GaussianNB
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
import math
from sklearn.metrics import normalized_mutual_info_score
warnings.filterwarnings("ignore")
import six
import sys
sys.modules["sklearn.externals.six"] = six
from mlxtend.classifier import StackingClassifier
from sklearn import model_selection
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import RandomizedSearchCV
from sklearn.metrics import log_loss, make_scorer
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report
from sklearn.svm import SVC
import plotly.express as px
# ### Чтение данных, изучение, визуальное представление
data = pd.read_csv("/kaggle/input/gen-competition/training_variants")
print("Количество записей в таблице : ", data.shape[0])
print("Количество полей в таблице: ", data.shape[1])
print("Поля: ", data.columns.values)
data.sample(7)
data_text = pd.read_csv(
"/kaggle/input/gen-text/training_text", sep="\|\|", names=["ID", "text"], skiprows=1
)
print("Количество записей в таблице : ", data_text.shape[0])
print("Количество полей в таблице: ", data_text.shape[1])
print("Поля: ", data_text.columns.values)
data_text.sample(8)
# ### Предварительный анализ данных
data.describe()
data.groupby("Class").size().plot(
kind="bar",
color="maroon",
y="ID",
figsize=(4, 4),
title="Количество данных по классам",
)
d = data.groupby("Class").size()
print(d)
d.plot(
kind="pie",
y="ID",
title="Количество данных по классам",
figsize=(4, 4),
autopct="%1.1f%%",
startangle=200,
)
plt.show()
print(data.groupby("Class").size())
# Проанализируем выборку по количестве записей по каждому гену
d = data.groupby(["Gene", "Class"]).size()
d.head(50)
gene_count = data.groupby("Gene").size()
# data.groupby('Gene').size().plot(kind='bar', color = 'maroon', y='Gene' , title='Количественное соотношение записей выборки по генам')
fig = px.bar(
gene_count,
title="Количественное соотношение записей выборки по разновидности генов",
)
fig.show()
# ## Подключаем библиотеки для предварительного анализа текста
import pymorphy2
import nltk
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
a = 1
# ### Предобработка текста: удаляем лишние символы, преобразуем к нижнему регистру, разбиваем на токены - по словам, приводим в нормальную форму
def goodtext(text):
# избавляемся от ненужных символов и убираем несколько пробелов, также убираем "одиночно-стоящие" буквы, числа оставляем
text = re.sub("[^a-z0-9\n]", " ", text)
text = re.sub(r"\b[a-zA-Z]\b", " ", text)
text = re.sub("\s+", " ", text)
return text
def preprocess(text, stop_words, morph):
tokens = word_tokenize(goodtext(text.lower()))
preprocessed_text = []
for token in tokens:
lemma = morph.parse(token)[0].normal_form
if lemma not in stop_words:
preprocessed_text.append(lemma)
global a
if a % 100 == 0:
print(a)
a += 1
return preprocessed_text
# punctuation_marks = ['!', ',', '(', ')', ':', '-', '?', '.', '..', '...', '«', '»', ';', '–', '--', '*','[',']', '%','>','<', '=','’','“','”', '±','{','}','"',"'"]
stop_words = stopwords.words("english")
morph = pymorphy2.MorphAnalyzer()
data_text.head()
stop_words.append("whose")
stop_words.append("also")
# ## удаляем пустые значения из таблицы и удаляем связанные с ними данные в другой таблице
data_text.describe()
data_text = data_text.dropna()
index = [id for id in data_text["ID"]]
# удаляем записи которых нет в первой таблице
data = data.query("ID in `index`")
data_text.describe()
start_time = time.clock()
data_text["text"] = data_text.apply(
lambda row: preprocess(row["text"], stop_words, morph), axis=1
)
print(data_text["text"])
print("Time took for preprocessing the text :", time.clock() - start_time, "seconds")
result = pd.merge(data, data_text, on="ID", how="left")
result.to_pickle("result.pkl")
data_text["text"]
# ### Собираем данные в одну таблицу и сохраняемв scv-файл
result = pd.merge(data, data_text, on="ID", how="left")
# result.to_csv('result.csv', index= False)
result.head()
result.to_pickle("result.pkl")
data_new = pd.read_pickle("/kaggle/input/data-txt/result2.pkl")
# data_new['text'][0]
# ### создаём мешок слов
from collections import Counter
# ## проверим работу "Счетчика слов" на тексте первой строки - построим словарь
test_counter = Counter(data_new["text"][0])
# rint(type(data_new['text'][0]))
test_counter.most_common(100)
# ### Построим словарь слово:кол-во для текстового поля всей выборки
# Следует наделить контекстом поля "text" с нулевыми значениями
# заменяем пустые значения
data_new["text"] = data_new["text"].fillna("0")
data_new.loc[data_new["text"] == "0"].text = data_new.apply(
lambda x: [x["Gene"], x["Variation"]], axis=1
)
# data_new = data_new.dropna() # удалим пустые значения
pd.set_option("max_colwidth", 180)
pd.set_option("display.width", 600)
print("Количество записей в таблице : ", data_new.shape[0])
print("Количество полей в таблице: ", data_new.shape[1])
print("Поля: ", data_new.columns.values)
data_new.sample(9)
# Создаем словарь, упорядоченный по частоте
# В словаре будем использовать 2 специальных кода:
# Код заполнитель: 0
# Неизвестное слово: 1
# Нумерация слов в словаре начинается с 2.
# Словарь, отображающий слова в коды
word_to_index = dict()
# Словарь, отображающий коды в слова
index_to_word = dict()
all_text = Counter()
for txt in data_new["text"]:
all_text.update(txt)
print("Часть мешка слов:", all_text.most_common(20))
print("\nКоличество элементов Мешка слов:", len(all_text))
max_words = 150000
for i, w in enumerate(all_text.most_common(max_words - 2)):
word_to_index[w[0]] = i + 2
index_to_word[i + 2] = w[0]
# ### Функция для преобразования списка слов в список кодов
def text_to_sequence(txt, word_to_index):
seq = []
for word in txt:
index = word_to_index.get(word, 1) # 1 означает неизвестное слово
# Неизвестные слова не добавляем в выходную последовательность
if index != 1:
seq.append(index)
return seq
def class_(id):
return id - 1
# Преобразуем все тексты в последовательность кодов слов
data_new["digital"] = data_new.apply(
lambda row: text_to_sequence(row["text"], word_to_index), axis=1
)
data_new["Class"] = data_new.apply(lambda row: class_(row["Class"]), axis=1)
print("Количество записей в таблице : ", data_new.shape[0])
print("Количество полей в таблице: ", data_new.shape[1])
print("Поля: ", data_new.columns.values)
data_new.sample(9)
# ### разбиваем выборку на две части - данные для тестирования, данные для обучения
train, test = train_test_split(data_new, test_size=0.2)
x_train_seq = train["digital"]
y_train = train["Class"]
x_test_seq = test["digital"]
y_test = test["Class"]
def vectorize_sequences(sequences, dimension=100000):
results = np.zeros((len(sequences), dimension))
for i, sequence in enumerate(sequences):
for index in sequence:
results[i, index] += 1.0
return results
x_train = vectorize_sequences(x_train_seq, max_words)
x_test = vectorize_sequences(x_test_seq, max_words)
print(x_train)
# ### Создаем модель машинного обучения
random_state = 10000
lr = LogisticRegression(random_state=random_state, max_iter=500)
# ### Обучаем модель машинного обучения
lr.fit(x_train, y_train)
lr.score(x_test, y_test)
lr_prediction = lr.predict(x_test)
accuracy = accuracy_score(y_test, lr_prediction)
print(f"Accuracy: {accuracy}")
conf_mtrx = confusion_matrix(y_test, lr_prediction)
print(conf_mtrx)
lr2 = LogisticRegression(
C=1.0,
class_weight=None,
dual=False,
fit_intercept=True,
intercept_scaling=1,
max_iter=500,
multi_class="multinomial",
n_jobs=1,
penalty="l2",
random_state=0,
tol=0.0001,
verbose=0,
warm_start=False,
)
lr2.fit(x_train, y_train)
lr2_prediction = lr2.predict(x_test)
accuracy2 = accuracy_score(y_test, lr2_prediction)
print(f"Accuracy: {accuracy2}")
conf_mtrx2 = confusion_matrix(y_test, lr2_prediction)
print(conf_mtrx2)
# ###
from scipy.stats import loguniform
from sklearn.model_selection import RandomizedSearchCV
nb = MultinomialNB()
param = dict()
param["alpha"] = loguniform(1e-5, 1)
# parameter={'alpha':[0.00001,0.0001,0.001,0.1,1,10,100,1000]}
LogLoss = make_scorer(log_loss, greater_is_better=False, needs_proba=True)
clf = RandomizedSearchCV(nb, param, n_iter=500, scoring=LogLoss)
clf.fit(x_train, y_train)
from scipy.stats import loguniform
from sklearn.model_selection import RandomizedSearchCV
nb = MultinomialNB(alpha=0.0060888387195).fit(x_train, y_train)
prediction = nb.predict_proba(x_test)
print("Функция потерь:", log_loss(y_test, prediction))
# ### Векторизация текста на основе эмбединга TfidfVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer()
data_new["digital_1"] = vectorizer.fit_transform(data_new["text"])
# X = vectorizer.fit_transform(corpus)
# print(vectorizer.get_feature_names())
print(data_new.sample(9))
| false | 0 | 3,689 | 0 | 3,709 | 3,689 |
||
129407662
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # **BASIC OPERATIONS**
import numpy as np
lst = [1, 2, 3, 4, 5]
print(lst)
# Array (in numpy we can store multi-dimentional array)
print("1D Array")
a = np.array([1, 2, 3, 4, 5])
print(a)
print("2D Array")
b = np.array([[1, 2, 3, 4, 5], [6, 7, 8, 9, 10]])
print(b)
print("3D Array")
c = np.array([[[1, 2, 3, 4, 5], [6, 7, 8, 9, 10], [11, 12, 13, 14, 15]]])
print(c)
type(a)
# SIZE (it gives us the number of elements)
print(a.size)
print(b.size)
print(c.size)
# SHAPE (it gives us number of rows and columns)
print(a.shape)
print(b.shape)
print(c.shape)
# DATATYPES (it gives us the type of data present in array)
print(a.dtype)
print(b.dtype)
print(c.dtype)
d = np.array([[1, 2, 3.3, 4, 5.5], [6.1, 7, 8, 9.5, 10], [11, 12, 13.5, 14.7, 15]])
print(d.dtype)
# arr.transpose (it replace rows into column & column into rows)
d.transpose()
# # np.ones, np.zeros, np.empty functions
# np.empty((tuple), dtype)
np.empty((4, 4), dtype=int)
np.empty((4, 4), dtype=float)
# np.ones((rows,cols), dtype)
x = np.ones(8)
x
y = np.ones((5, 4))
y
z = np.ones((4, 3), dtype=int)
z
# np.zeros((rows, cols), dtype)
x = np.zeros(4)
x
y = np.zeros((4, 6))
y
z = np.zeros((3, 6), dtype=int)
z
z = np.ones((4, 5), dtype=str)
z
y = np.ones((3, 7), dtype=bool)
y
z = np.zeros((4, 5), dtype=str)
z
y = np.zeros((3, 7), dtype=bool)
y
# # ADVANCED FUNCTIONS (np.arange(), np.flatten(), np.ravel(), np.reshape())
# ### It use in every data analysis & when we need to give axis in Matplotlib
# np.arange(start,end,step)---- its like for loop, but here we are giving 3 parameters:-
a = np.arange(1, 20)
print(a)
##odd number
a = np.arange(1, 20, 2)
print(a)
##even number
a = np.arange(2, 20, 2)
print(a)
# Reshape
##arr.reshape((rows, cols))
a = a.reshape((3, 3))
a
b = np.arange(1, 100, 2)
b
b = b.reshape((10, 5))
b
##inverse process of reshape i,e single row & multi cols
b = b.flatten()
b
a = a.ravel()
a
# # ARRAY SLICING OPERATIONS
a = np.arange(1, 51)
a = a.reshape(10, 5)
a
##SLICING
a[0]
a[2]
a[3]
a[0, 0]
a[3, 4] # 3rd row & 4th element
a[2:5] # 2nd row to 4th row bcoz 5th will not appear
a[0:10] # print all rows & cols
a[:, 2] # (:) means all the rows & (2) means the 2nd col
a[2:5, 4] # 2:5 means 2nd row to 5th row & 4 means from the 4th col
a[:, :] # all row & cols
a[:, 2:5] # all rows & 2:5 means 2nd col to 4th col
a[:, 2:5].dtype # define the datatype
# # MATHEMATICAL OPERATIONS
# To see the Mathematical operations, we need to define 2 arrays
a = np.arange(0, 18).reshape((6, 3))
b = np.arange(20, 38).reshape((6, 3))
print(a)
print(b)
a + b # it will add vertically i.e 0+20,1+21--------
np.add(a, b) # this is the function of addition
a - b # subtract each and every element to each other
np.subtract(a, b) # this is the function of subtraction
a * b # multiply by vertically
np.multiply(a, b) # this is the function of multiply
a / b # it will give the float result
np.divide(a, b) # this is the function of dividw
# # # # #metrix multiplication start from here
a @ b
print(a.shape)
print(b.shape)
b = b.reshape((3, 6))
b
a @ b # here cross multiplication will happen
a.dot(b) # this is the function of above multiplication with @
b.max()
b.min()
b.argmax() # it will give us the index no. of the max value
np.sum(b) # sum of all elements in b
np.sum(b, axis=1) # axis = 1 means row , there it will give us sum of each & every row
np.sum(b, axis=0) # axis = 0 means cols, there it will give us sum of each & every cols
np.mean(b)
np.sqrt(b)
np.std(b)
np.log(b)
# # TRIGONOMETRIC OPERATIONS
import matplotlib.pyplot as plt
plt.style.use("dark_background")
np.pi
##Sin 90 Degree
np.sin(np.pi / 2)
##Sin 30 Degree
np.sin(np.pi / 6)
##Cos 90 Degree
np.cos(np.pi / 2)
##Cos 30 Degree
np.cos(np.pi / 6)
##Tan 90 Degree
np.tan(np.pi / 2)
np.tan(0)
# **###Using Matplotlib with Numpy:--******
x = np.arange(1, 11)
y = np.arange(10, 110, 10)
plt.figure(figsize=(6, 6))
plt.plot(x, y, "r--")
plt.show()
# **##Plotting Trignometric Curves**
##SIN CURVE
x_sin = np.arange(0, 2 * np.pi, 0.1)
y_sin = np.sin(x_sin)
print(y_sin)
plt.figure(figsize=(6, 6))
plt.plot(x_sin, y_sin)
plt.title("Sin Curve")
plt.show()
x_cos = np.arange(0, 2 * np.pi, 0.1)
y_cos = np.cos(x_cos)
print(y_cos)
plt.figure(figsize=(6, 6))
plt.plot(x_cos, y_cos)
plt.title("Cos Curve")
plt.show()
x_tan = np.arange(0, 2 * np.pi, 0.1)
y_tan = np.tan(x_tan)
print(y_tan)
plt.figure(figsize=(6, 6))
plt.plot(x_tan, y_tan)
plt.title("Tan Curve")
plt.show()
x_cot = np.arange(0, 2 * np.pi, 0.1)
y_cot = 1 / np.tan(x_cot)
print(y_cot)
plt.figure(figsize=(6, 6))
plt.plot(x_cot, y_cot)
plt.title("Cot Curve")
plt.show()
# **##SUBPLOT** (all curves should be in one plot)
plt.figure(figsize=(8, 8))
plt.subplot(2, 2, 1)
plt.plot(x_sin, y_sin, "r--")
plt.title("Sin Curve")
plt.subplot(2, 2, 2)
plt.plot(x_cos, y_cos, "b--")
plt.title("Cos Curve")
plt.subplot(2, 2, 3)
plt.plot(x_tan, y_tan, "y--")
plt.title("Tan Curve")
plt.subplot(2, 2, 4)
plt.plot(x_cot, y_cot, "g--")
plt.title("Cot Curve")
plt.show()
# # # RANDOM OPERATIONS
np.random.random(1)
np.random.random(2)
np.random.random((2, 2))
np.random.randint(1, 10)
np.random.randint(1, 10, (2, 2))
np.random.randint(1, 10, (3, 4, 5))
np.random.rand(2, 3)
np.random.randn(2, 5)
a = np.arange(1, 10)
print(a)
np.random.choice(a)
# # STRING OPERATIONS
# **#Strings with Numpy library**
s1 = "Hoshangi is my Name"
s2 = "I am the wife of Hardik"
np.char.add(s1, s2)
np.char.upper(s1)
np.char.lower(s1)
np.char.split(s2)
s3 = "Hoshangi is my \nname"
np.char.splitlines(s3)
np.char.replace(s1, "Name", "Sirname")
print(np.char.center("NUMPY COMPLETED", 80, "*"))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/407/129407662.ipynb
| null | null |
[{"Id": 129407662, "ScriptId": 38470678, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13392235, "CreationDate": "05/13/2023 14:22:56", "VersionNumber": 1.0, "Title": "Numpy practice", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 387.0, "LinesInsertedFromPrevious": 387.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # **BASIC OPERATIONS**
import numpy as np
lst = [1, 2, 3, 4, 5]
print(lst)
# Array (in numpy we can store multi-dimentional array)
print("1D Array")
a = np.array([1, 2, 3, 4, 5])
print(a)
print("2D Array")
b = np.array([[1, 2, 3, 4, 5], [6, 7, 8, 9, 10]])
print(b)
print("3D Array")
c = np.array([[[1, 2, 3, 4, 5], [6, 7, 8, 9, 10], [11, 12, 13, 14, 15]]])
print(c)
type(a)
# SIZE (it gives us the number of elements)
print(a.size)
print(b.size)
print(c.size)
# SHAPE (it gives us number of rows and columns)
print(a.shape)
print(b.shape)
print(c.shape)
# DATATYPES (it gives us the type of data present in array)
print(a.dtype)
print(b.dtype)
print(c.dtype)
d = np.array([[1, 2, 3.3, 4, 5.5], [6.1, 7, 8, 9.5, 10], [11, 12, 13.5, 14.7, 15]])
print(d.dtype)
# arr.transpose (it replace rows into column & column into rows)
d.transpose()
# # np.ones, np.zeros, np.empty functions
# np.empty((tuple), dtype)
np.empty((4, 4), dtype=int)
np.empty((4, 4), dtype=float)
# np.ones((rows,cols), dtype)
x = np.ones(8)
x
y = np.ones((5, 4))
y
z = np.ones((4, 3), dtype=int)
z
# np.zeros((rows, cols), dtype)
x = np.zeros(4)
x
y = np.zeros((4, 6))
y
z = np.zeros((3, 6), dtype=int)
z
z = np.ones((4, 5), dtype=str)
z
y = np.ones((3, 7), dtype=bool)
y
z = np.zeros((4, 5), dtype=str)
z
y = np.zeros((3, 7), dtype=bool)
y
# # ADVANCED FUNCTIONS (np.arange(), np.flatten(), np.ravel(), np.reshape())
# ### It use in every data analysis & when we need to give axis in Matplotlib
# np.arange(start,end,step)---- its like for loop, but here we are giving 3 parameters:-
a = np.arange(1, 20)
print(a)
##odd number
a = np.arange(1, 20, 2)
print(a)
##even number
a = np.arange(2, 20, 2)
print(a)
# Reshape
##arr.reshape((rows, cols))
a = a.reshape((3, 3))
a
b = np.arange(1, 100, 2)
b
b = b.reshape((10, 5))
b
##inverse process of reshape i,e single row & multi cols
b = b.flatten()
b
a = a.ravel()
a
# # ARRAY SLICING OPERATIONS
a = np.arange(1, 51)
a = a.reshape(10, 5)
a
##SLICING
a[0]
a[2]
a[3]
a[0, 0]
a[3, 4] # 3rd row & 4th element
a[2:5] # 2nd row to 4th row bcoz 5th will not appear
a[0:10] # print all rows & cols
a[:, 2] # (:) means all the rows & (2) means the 2nd col
a[2:5, 4] # 2:5 means 2nd row to 5th row & 4 means from the 4th col
a[:, :] # all row & cols
a[:, 2:5] # all rows & 2:5 means 2nd col to 4th col
a[:, 2:5].dtype # define the datatype
# # MATHEMATICAL OPERATIONS
# To see the Mathematical operations, we need to define 2 arrays
a = np.arange(0, 18).reshape((6, 3))
b = np.arange(20, 38).reshape((6, 3))
print(a)
print(b)
a + b # it will add vertically i.e 0+20,1+21--------
np.add(a, b) # this is the function of addition
a - b # subtract each and every element to each other
np.subtract(a, b) # this is the function of subtraction
a * b # multiply by vertically
np.multiply(a, b) # this is the function of multiply
a / b # it will give the float result
np.divide(a, b) # this is the function of dividw
# # # # #metrix multiplication start from here
a @ b
print(a.shape)
print(b.shape)
b = b.reshape((3, 6))
b
a @ b # here cross multiplication will happen
a.dot(b) # this is the function of above multiplication with @
b.max()
b.min()
b.argmax() # it will give us the index no. of the max value
np.sum(b) # sum of all elements in b
np.sum(b, axis=1) # axis = 1 means row , there it will give us sum of each & every row
np.sum(b, axis=0) # axis = 0 means cols, there it will give us sum of each & every cols
np.mean(b)
np.sqrt(b)
np.std(b)
np.log(b)
# # TRIGONOMETRIC OPERATIONS
import matplotlib.pyplot as plt
plt.style.use("dark_background")
np.pi
##Sin 90 Degree
np.sin(np.pi / 2)
##Sin 30 Degree
np.sin(np.pi / 6)
##Cos 90 Degree
np.cos(np.pi / 2)
##Cos 30 Degree
np.cos(np.pi / 6)
##Tan 90 Degree
np.tan(np.pi / 2)
np.tan(0)
# **###Using Matplotlib with Numpy:--******
x = np.arange(1, 11)
y = np.arange(10, 110, 10)
plt.figure(figsize=(6, 6))
plt.plot(x, y, "r--")
plt.show()
# **##Plotting Trignometric Curves**
##SIN CURVE
x_sin = np.arange(0, 2 * np.pi, 0.1)
y_sin = np.sin(x_sin)
print(y_sin)
plt.figure(figsize=(6, 6))
plt.plot(x_sin, y_sin)
plt.title("Sin Curve")
plt.show()
x_cos = np.arange(0, 2 * np.pi, 0.1)
y_cos = np.cos(x_cos)
print(y_cos)
plt.figure(figsize=(6, 6))
plt.plot(x_cos, y_cos)
plt.title("Cos Curve")
plt.show()
x_tan = np.arange(0, 2 * np.pi, 0.1)
y_tan = np.tan(x_tan)
print(y_tan)
plt.figure(figsize=(6, 6))
plt.plot(x_tan, y_tan)
plt.title("Tan Curve")
plt.show()
x_cot = np.arange(0, 2 * np.pi, 0.1)
y_cot = 1 / np.tan(x_cot)
print(y_cot)
plt.figure(figsize=(6, 6))
plt.plot(x_cot, y_cot)
plt.title("Cot Curve")
plt.show()
# **##SUBPLOT** (all curves should be in one plot)
plt.figure(figsize=(8, 8))
plt.subplot(2, 2, 1)
plt.plot(x_sin, y_sin, "r--")
plt.title("Sin Curve")
plt.subplot(2, 2, 2)
plt.plot(x_cos, y_cos, "b--")
plt.title("Cos Curve")
plt.subplot(2, 2, 3)
plt.plot(x_tan, y_tan, "y--")
plt.title("Tan Curve")
plt.subplot(2, 2, 4)
plt.plot(x_cot, y_cot, "g--")
plt.title("Cot Curve")
plt.show()
# # # RANDOM OPERATIONS
np.random.random(1)
np.random.random(2)
np.random.random((2, 2))
np.random.randint(1, 10)
np.random.randint(1, 10, (2, 2))
np.random.randint(1, 10, (3, 4, 5))
np.random.rand(2, 3)
np.random.randn(2, 5)
a = np.arange(1, 10)
print(a)
np.random.choice(a)
# # STRING OPERATIONS
# **#Strings with Numpy library**
s1 = "Hoshangi is my Name"
s2 = "I am the wife of Hardik"
np.char.add(s1, s2)
np.char.upper(s1)
np.char.lower(s1)
np.char.split(s2)
s3 = "Hoshangi is my \nname"
np.char.splitlines(s3)
np.char.replace(s1, "Name", "Sirname")
print(np.char.center("NUMPY COMPLETED", 80, "*"))
| false | 0 | 2,767 | 0 | 2,767 | 2,767 |
||
129407321
|
<jupyter_start><jupyter_text>Nike, Adidas and Converse Shoes Images
<p> <img src="https://www.googleapis.com/download/storage/v1/b/kaggle-user-content/o/inbox%2F6372737%2F2d0c8c299f63bb8a5823683346ba1ba8%2FImage2.jpg?generation=1659570752665846&alt=media"> </p>
The dataset contains 2 folders: one with the test data and the other one with train data.
The test-train-split ratio is 0.14, with the test dataset containing 114 images and the train dataset containing 711.
The images have a resolution of 240x240 pixels in RGB color model.
Both the folders contain 3 classes:
- Adidas
- Converse
- Nike
** **
### **Inspiration**
This dataset is ideal for performing multiclass classification with deep neural networks like CNNs or simpler machine learning classification models.
You can use `Tensorflow`, his high-level API `keras`, `Sklearn`, `PyTorch` or other deep/machine learning libraries to building the model from scratch or, as an alternative, fetching pretrained models as well as fine-tuning them.
It is also possible to modify the size of the images or preprocessing them using `OpenCV` , and check if the accuracy of the model improves. <br>
**Remember to upvote if you found the dataset useful :)**.
** **
### **Collection methodology**
The dataset was obtained downloading images from `Google images`.
The images with a `.webp` format were transformed into .jpg images. The obtained images were randomly shuffled and resized so that all the images had a resolution of 240x240 pixels.
Then, they were split into train and test datasets and saved.
Kaggle dataset identifier: nike-adidas-and-converse-imaged
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import tensorflow as tf
from tensorflow.keras import datasets, layers, models
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
import cv2
from skimage.io import imread
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = tf.keras.utils.image_dataset_from_directory(
"/kaggle/input/nike-adidas-and-converse-imaged/train"
)
data_iterator = data.as_numpy_iterator()
batch = data_iterator.next()
batch[1].shape
fig, ax = plt.subplots(ncols=4, figsize=(20, 20))
for idx, img in enumerate(batch[0][:4]):
ax[idx].imshow(img.astype(int))
ax[idx].title.set_text(batch[1][idx])
data = data.map(lambda x, y: (x / 255, y))
fig, ax = plt.subplots(ncols=4, figsize=(20, 20))
for idx, img in enumerate(batch[0][:4]):
ax[idx].imshow(img.astype(int))
ax[idx].title.set_text(batch[1][idx])
len(data)
train_size = int(len(data) * 0.7)
val_size = int(len(data) * 0.2)
test_size = int(len(data) * 0.1) + 1
train = data.take(train_size)
val = data.skip(train_size).take(val_size)
test = data.skip(train_size).skip(val_size).take(test_size)
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Dense, Flatten, Dropout
model = Sequential()
model.add(layers.Conv2D(32, (3, 3), activation="relu", input_shape=(256, 256, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.Flatten())
model.add(layers.Dense(64, activation="relu"))
model.add(layers.Dense(3))
logdir = "logs"
tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=logdir)
model.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
hist = model.fit(
train, epochs=20, validation_data=val, callbacks=[tensorboard_callback]
)
img = imread("/kaggle/input/airforce-test/2560px-Nike_air_Force_1_white_on_white.jpg")
plt.imshow(img)
plt.show()
img.shape
down_sized = cv2.resize(img, (256, 256), interpolation=cv2.INTER_LINEAR)
plt.imshow(down_sized)
plt.show()
yhat = model.predict(np.expand_dims(down_sized / 255, 0))
yhat
if np.argmax(yhat) == 0:
print("Predicted Class is Adidas")
if np.argmax(yhat) == 1:
print("Predicted Class is Converse")
else:
print("Predicted Class is Nike")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/407/129407321.ipynb
|
nike-adidas-and-converse-imaged
|
die9origephit
|
[{"Id": 129407321, "ScriptId": 38091728, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13949158, "CreationDate": "05/13/2023 14:20:22", "VersionNumber": 1.0, "Title": "notebook52310545e6", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 102.0, "LinesInsertedFromPrevious": 102.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185427353, "KernelVersionId": 129407321, "SourceDatasetVersionId": 4025361}]
|
[{"Id": 4025361, "DatasetId": 2385323, "DatasourceVersionId": 4081276, "CreatorUserId": 6372737, "LicenseName": "CC0: Public Domain", "CreationDate": "08/03/2022 17:13:58", "VersionNumber": 1.0, "Title": "Nike, Adidas and Converse Shoes Images", "Slug": "nike-adidas-and-converse-imaged", "Subtitle": "Nike, Adidas and Convers shoes dataset for classification", "Description": "<p> <img src=\"https://www.googleapis.com/download/storage/v1/b/kaggle-user-content/o/inbox%2F6372737%2F2d0c8c299f63bb8a5823683346ba1ba8%2FImage2.jpg?generation=1659570752665846&alt=media\"> </p>\n\n\n\n\nThe dataset contains 2 folders: one with the test data and the other one with train data. \nThe test-train-split ratio is 0.14, with the test dataset containing 114 images and the train dataset containing 711.\nThe images have a resolution of 240x240 pixels in RGB color model.\nBoth the folders contain 3 classes:\n\n- Adidas\n- Converse\n- Nike\n** **\n### **Inspiration**\n\nThis dataset is ideal for performing multiclass classification with deep neural networks like CNNs or simpler machine learning classification models.\nYou can use `Tensorflow`, his high-level API `keras`, `Sklearn`, `PyTorch` or other deep/machine learning libraries to building the model from scratch or, as an alternative, fetching pretrained models as well as fine-tuning them.\nIt is also possible to modify the size of the images or preprocessing them using `OpenCV` , and check if the accuracy of the model improves. <br>\n**Remember to upvote if you found the dataset useful :)**.\n** **\n### **Collection methodology**\n\nThe dataset was obtained downloading images from `Google images`.\n\nThe images with a `.webp` format were transformed into .jpg images. The obtained images were randomly shuffled and resized so that all the images had a resolution of 240x240 pixels.\nThen, they were split into train and test datasets and saved.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2385323, "CreatorUserId": 6372737, "OwnerUserId": 6372737.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4025361.0, "CurrentDatasourceVersionId": 4081276.0, "ForumId": 2412562, "Type": 2, "CreationDate": "08/03/2022 17:13:58", "LastActivityDate": "08/03/2022", "TotalViews": 23036, "TotalDownloads": 3217, "TotalVotes": 82, "TotalKernels": 32}]
|
[{"Id": 6372737, "UserName": "die9origephit", "DisplayName": "Iron486", "RegisterDate": "12/12/2020", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import tensorflow as tf
from tensorflow.keras import datasets, layers, models
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
import cv2
from skimage.io import imread
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = tf.keras.utils.image_dataset_from_directory(
"/kaggle/input/nike-adidas-and-converse-imaged/train"
)
data_iterator = data.as_numpy_iterator()
batch = data_iterator.next()
batch[1].shape
fig, ax = plt.subplots(ncols=4, figsize=(20, 20))
for idx, img in enumerate(batch[0][:4]):
ax[idx].imshow(img.astype(int))
ax[idx].title.set_text(batch[1][idx])
data = data.map(lambda x, y: (x / 255, y))
fig, ax = plt.subplots(ncols=4, figsize=(20, 20))
for idx, img in enumerate(batch[0][:4]):
ax[idx].imshow(img.astype(int))
ax[idx].title.set_text(batch[1][idx])
len(data)
train_size = int(len(data) * 0.7)
val_size = int(len(data) * 0.2)
test_size = int(len(data) * 0.1) + 1
train = data.take(train_size)
val = data.skip(train_size).take(val_size)
test = data.skip(train_size).skip(val_size).take(test_size)
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Dense, Flatten, Dropout
model = Sequential()
model.add(layers.Conv2D(32, (3, 3), activation="relu", input_shape=(256, 256, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.Flatten())
model.add(layers.Dense(64, activation="relu"))
model.add(layers.Dense(3))
logdir = "logs"
tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=logdir)
model.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
hist = model.fit(
train, epochs=20, validation_data=val, callbacks=[tensorboard_callback]
)
img = imread("/kaggle/input/airforce-test/2560px-Nike_air_Force_1_white_on_white.jpg")
plt.imshow(img)
plt.show()
img.shape
down_sized = cv2.resize(img, (256, 256), interpolation=cv2.INTER_LINEAR)
plt.imshow(down_sized)
plt.show()
yhat = model.predict(np.expand_dims(down_sized / 255, 0))
yhat
if np.argmax(yhat) == 0:
print("Predicted Class is Adidas")
if np.argmax(yhat) == 1:
print("Predicted Class is Converse")
else:
print("Predicted Class is Nike")
| false | 0 | 999 | 0 | 1,471 | 999 |
||
129407067
|
<jupyter_start><jupyter_text>Wild Blueberry Yield Prediction
### Context
A number of research is underway in the agricultural sector to better predict crop yield using machine learning algorithms. Many machine learning algorithms require large amounts of data in order to give useful results. One of the major challenges in training and experimenting with machine learning algorithms is the availability of training data in sufficient quality and quantity remains a limiting factor. In the paper, “Wild blueberry yield prediction using a combination of computer simulation and machine learning algorithms”, we used dataset generated by the Wild Blueberry Pollination Model, a spatially explicit simulation model validated by field observation and experimental data collected in Maine USA during the last 30 years. The blueberry yields predictive models require data that sufficiently characterize the influence of plant spatial traits, bee species composition, and weather conditions on production. In a multi-step process, we designed simulation experiments and conducted the runs on the calibrated version of the blueberry simulation model. The simulated dataset was then examined, and important features were selected to build four machine-learning-based predictive models. This simulated data provides researchers who have actual data collected from field observation and those who wants to experiment the potential of machine learning algorithms response to real data and computer simulation modelling generated data as input for crop yield prediction models.
Kaggle dataset identifier: wild-blueberry-yield-prediction
<jupyter_script>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import RepeatedStratifiedKFold, StratifiedKFold, KFold
import warnings
warnings.filterwarnings("ignore")
import lightgbm as lgb
from sklearn.metrics import mean_absolute_error
train = pd.read_csv(r"../input/playground-series-s3e14/train.csv")
test = pd.read_csv(r"../input/playground-series-s3e14/test.csv")
# train2=pd.read_csv("/kaggle/input/wild-blueberry-yield-prediction/Data in Brief/Data in Brief/WildBlueberryPollinationSimulationData.csv")
# train = pd.concat([train,train2],axis=0)
# pleaadd_suffixse refer to this notebook for feature selection :
# https://www.kaggle.com/code/iqbalsyahakbar/ps3e14-eda-fe-models-ensemble-for-starters
# features_to_exclude = ['id','MinOfUpperTRange', 'AverageOfUpperTRange', 'MaxOfLowerTRange', 'MinOfLowerTRange',
# 'AverageOfLowerTRange', 'AverageRainingDays',
# "honeybee", "bumbles", "andrena",'clonesize', 'osmia', ]
# train.drop(features_to_exclude, axis = 1, inplace = True)
# test = test_df.drop(features_to_exclude, axis = 1)
features_to_exclude = ["id"]
# features_to_exclude1 = ['id',"Row#"]
# features_to_exclude2 = ['id', ]
train.drop(features_to_exclude, axis=1, inplace=True)
test = test.drop(features_to_exclude, axis=1)
train = train.reset_index(drop=True)
# params = {"random_state":42,
# 'eval_metric': 'mae',
# 'max_depth': 3,
# 'subsample': 1,
# "max_leaves":2,
# 'learning_rate': 0.1,
# "objective" :"reg:absoluteerror",
# "n_estimators" : 50000,
# "early_stopping_rounds" : 500
# }
params = {
"random_state": 42,
"eval_metric": "mae",
"max_depth": 3,
"subsample": 1,
"max_leaves": 2,
"learning_rate": 0.3,
"objective": "reg:absoluteerror",
"n_estimators": 50000,
"early_stopping_rounds": 500,
}
X = train.copy()
y = X.pop("yield")
# total_preds = []
# Define LightGBM parameters
# Parameters by Bayesian optimization
seed = 42
folds = 5
# Initialize KFold cross-validation
kf = KFold(n_splits=folds, shuffle=True, random_state=42)
# kf = RepeatedKFold(n_repeats=3, n_splits=config['FOLDS'], random_state=config['SEED'])
# Initialize empty arrays to store validation predictions and scores
val_preds = np.zeros(len(X))
val_scores = []
prediction = np.zeros((len(test)))
import xgboost as xgb
# 'reg_alpha': 0.6469675138495181, 'reg_lambda': 0.14870311599361627,'min_child_samples': 14
from sklearn.preprocessing import RobustScaler
# Loop through each fold
predss2 = []
val_preds2 = np.zeros(len(X))
val_scores2 = []
prediction2 = np.zeros((len(test)))
for fold, (train_idx, val_idx) in enumerate(kf.split(X, y)):
X_train, y_train = X.iloc[train_idx], y.iloc[train_idx]
X_val, y_val = X.iloc[val_idx], y.iloc[val_idx]
model2 = xgb.XGBRegressor(**params)
model2.fit(
X_train,
y_train,
eval_set=[(X_train, y_train), (X_val, y_val)],
# early_stopping_rounds = 1000,
verbose=10000,
)
val_preds2[val_idx] += model2.predict(X_val)
val_score2 = mean_absolute_error(y_val, val_preds2[val_idx])
val_scores2.append(val_score2)
print(f"Fold {fold+1}: Validation score: {val_score2:.4f}")
prediction2 += model2.predict(test)
predss2.append(model2.predict(test))
# prediction += model.predict(test)
# Calculate and print average validation score
avg_val_score = np.mean(val_scores2)
print(f"Average validation score: {avg_val_score:.4f}")
prediction2 /= folds
test_df = pd.read_csv("/kaggle/input/playground-series-s3e14/sample_submission.csv")
test_df.drop(list(test_df.drop("id", axis=1)), axis=1, inplace=True)
test_df["yield"] = prediction2
test_df.to_csv("xgboost.csv", index=False)
test_df
# test_df.to_csv('submission_discussion.csv', index = False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/407/129407067.ipynb
|
wild-blueberry-yield-prediction
|
saurabhshahane
|
[{"Id": 129407067, "ScriptId": 38474350, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9494633, "CreationDate": "05/13/2023 14:18:15", "VersionNumber": 1.0, "Title": "xgboost(cv : 346, sub: 347.92)", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 130.0, "LinesInsertedFromPrevious": 130.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 9}]
|
[{"Id": 185426886, "KernelVersionId": 129407067, "SourceDatasetVersionId": 1978710}, {"Id": 185426887, "KernelVersionId": 129407067, "SourceDatasetVersionId": 2462316}]
|
[{"Id": 1978710, "DatasetId": 1182602, "DatasourceVersionId": 2017807, "CreatorUserId": 2411256, "LicenseName": "Attribution 4.0 International (CC BY 4.0)", "CreationDate": "02/26/2021 17:24:39", "VersionNumber": 1.0, "Title": "Wild Blueberry Yield Prediction", "Slug": "wild-blueberry-yield-prediction", "Subtitle": "Wild blueberry yield prediction using machine learning", "Description": "### Context\n\nA number of research is underway in the agricultural sector to better predict crop yield using machine learning algorithms. Many machine learning algorithms require large amounts of data in order to give useful results. One of the major challenges in training and experimenting with machine learning algorithms is the availability of training data in sufficient quality and quantity remains a limiting factor. In the paper, \u201cWild blueberry yield prediction using a combination of computer simulation and machine learning algorithms\u201d, we used dataset generated by the Wild Blueberry Pollination Model, a spatially explicit simulation model validated by field observation and experimental data collected in Maine USA during the last 30 years. The blueberry yields predictive models require data that sufficiently characterize the influence of plant spatial traits, bee species composition, and weather conditions on production. In a multi-step process, we designed simulation experiments and conducted the runs on the calibrated version of the blueberry simulation model. The simulated dataset was then examined, and important features were selected to build four machine-learning-based predictive models. This simulated data provides researchers who have actual data collected from field observation and those who wants to experiment the potential of machine learning algorithms response to real data and computer simulation modelling generated data as input for crop yield prediction models.\n\n\n### Acknowledgements\n\nQu, Hongchun; Obsie, Efrem; Drummond, Frank (2020), \u201cData for: Wild blueberry yield prediction using a combination of computer simulation and machine learning algorithms\u201d, Mendeley Data, V1, doi: 10.17632/p5hvjzsvn8.1", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1182602, "CreatorUserId": 2411256, "OwnerUserId": 2411256.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1978710.0, "CurrentDatasourceVersionId": 2017807.0, "ForumId": 1200381, "Type": 2, "CreationDate": "02/26/2021 17:24:39", "LastActivityDate": "02/26/2021", "TotalViews": 15727, "TotalDownloads": 1549, "TotalVotes": 35, "TotalKernels": 25}]
|
[{"Id": 2411256, "UserName": "saurabhshahane", "DisplayName": "Saurabh Shahane", "RegisterDate": "10/26/2018", "PerformanceTier": 4}]
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import RepeatedStratifiedKFold, StratifiedKFold, KFold
import warnings
warnings.filterwarnings("ignore")
import lightgbm as lgb
from sklearn.metrics import mean_absolute_error
train = pd.read_csv(r"../input/playground-series-s3e14/train.csv")
test = pd.read_csv(r"../input/playground-series-s3e14/test.csv")
# train2=pd.read_csv("/kaggle/input/wild-blueberry-yield-prediction/Data in Brief/Data in Brief/WildBlueberryPollinationSimulationData.csv")
# train = pd.concat([train,train2],axis=0)
# pleaadd_suffixse refer to this notebook for feature selection :
# https://www.kaggle.com/code/iqbalsyahakbar/ps3e14-eda-fe-models-ensemble-for-starters
# features_to_exclude = ['id','MinOfUpperTRange', 'AverageOfUpperTRange', 'MaxOfLowerTRange', 'MinOfLowerTRange',
# 'AverageOfLowerTRange', 'AverageRainingDays',
# "honeybee", "bumbles", "andrena",'clonesize', 'osmia', ]
# train.drop(features_to_exclude, axis = 1, inplace = True)
# test = test_df.drop(features_to_exclude, axis = 1)
features_to_exclude = ["id"]
# features_to_exclude1 = ['id',"Row#"]
# features_to_exclude2 = ['id', ]
train.drop(features_to_exclude, axis=1, inplace=True)
test = test.drop(features_to_exclude, axis=1)
train = train.reset_index(drop=True)
# params = {"random_state":42,
# 'eval_metric': 'mae',
# 'max_depth': 3,
# 'subsample': 1,
# "max_leaves":2,
# 'learning_rate': 0.1,
# "objective" :"reg:absoluteerror",
# "n_estimators" : 50000,
# "early_stopping_rounds" : 500
# }
params = {
"random_state": 42,
"eval_metric": "mae",
"max_depth": 3,
"subsample": 1,
"max_leaves": 2,
"learning_rate": 0.3,
"objective": "reg:absoluteerror",
"n_estimators": 50000,
"early_stopping_rounds": 500,
}
X = train.copy()
y = X.pop("yield")
# total_preds = []
# Define LightGBM parameters
# Parameters by Bayesian optimization
seed = 42
folds = 5
# Initialize KFold cross-validation
kf = KFold(n_splits=folds, shuffle=True, random_state=42)
# kf = RepeatedKFold(n_repeats=3, n_splits=config['FOLDS'], random_state=config['SEED'])
# Initialize empty arrays to store validation predictions and scores
val_preds = np.zeros(len(X))
val_scores = []
prediction = np.zeros((len(test)))
import xgboost as xgb
# 'reg_alpha': 0.6469675138495181, 'reg_lambda': 0.14870311599361627,'min_child_samples': 14
from sklearn.preprocessing import RobustScaler
# Loop through each fold
predss2 = []
val_preds2 = np.zeros(len(X))
val_scores2 = []
prediction2 = np.zeros((len(test)))
for fold, (train_idx, val_idx) in enumerate(kf.split(X, y)):
X_train, y_train = X.iloc[train_idx], y.iloc[train_idx]
X_val, y_val = X.iloc[val_idx], y.iloc[val_idx]
model2 = xgb.XGBRegressor(**params)
model2.fit(
X_train,
y_train,
eval_set=[(X_train, y_train), (X_val, y_val)],
# early_stopping_rounds = 1000,
verbose=10000,
)
val_preds2[val_idx] += model2.predict(X_val)
val_score2 = mean_absolute_error(y_val, val_preds2[val_idx])
val_scores2.append(val_score2)
print(f"Fold {fold+1}: Validation score: {val_score2:.4f}")
prediction2 += model2.predict(test)
predss2.append(model2.predict(test))
# prediction += model.predict(test)
# Calculate and print average validation score
avg_val_score = np.mean(val_scores2)
print(f"Average validation score: {avg_val_score:.4f}")
prediction2 /= folds
test_df = pd.read_csv("/kaggle/input/playground-series-s3e14/sample_submission.csv")
test_df.drop(list(test_df.drop("id", axis=1)), axis=1, inplace=True)
test_df["yield"] = prediction2
test_df.to_csv("xgboost.csv", index=False)
test_df
# test_df.to_csv('submission_discussion.csv', index = False)
| false | 3 | 1,388 | 9 | 1,688 | 1,388 |
||
129407850
|
<jupyter_start><jupyter_text>Human Stress Prediction
“subreddit – post_id – sentence_range – text-label-confidence-social_timestamp” represents the titles for Stress.csv file.
Stress detection is a challenging task, as there are so many words that can be used by people on their posts that can show whether a person is having psychological stress or not. look for datasets that you can use to train a machine learning model for stress detection.
The dataset contains data posted on subreddits related to mental health. This dataset contains various mental health problems shared by people about their life. Fortunately, this dataset is labelled as 0 and 1, where 0 indicates no stress and 1 indicates stress.
Kaggle dataset identifier: human-stress-prediction
<jupyter_script>import pandas as pd
import numpy as np
import warnings
warnings.filterwarnings("ignore")
# ## Packages
# ## Reading csv
df = pd.read_csv("/kaggle/input/human-stress-prediction/Stress.csv")
print(df.shape)
df.head()
# ## Data Exploration
df.isna().sum()
# #### No 'NA' values
df["label"].value_counts()
# #### Both label have almost equal data points
df["subreddit"].value_counts()
# #### Type of issues and mapping values to these issues
df["type_num"] = df["subreddit"].map(
dict(
zip(
pd.Series(df["subreddit"].unique()),
pd.Series([i for i in range(df["subreddit"].unique().shape[0])]),
)
)
)
# ## GENSIM 'glove-twitter-50' model
import gensim.downloader as api
glove = api.load("glove-twitter-50")
# ## SPACY for preprocess and GENSIM for vectorization
import spacy
nlp = spacy.load("en_core_web_sm")
def preprocess_and_vectorise(text):
list = []
for token in nlp(text):
if token.is_space:
continue
list.append(token.lemma_)
return glove.get_mean_vector(list)
df["vector"] = df["text"].apply(lambda text: preprocess_and_vectorise(text))
df.head()
# ## Scaling the vectors using MINMAXSCALER and Converting array of vector into DataFrame
X_vector = df["vector"].values
X_vector_stack = np.stack(X_vector)
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
X_vector_scaled = scaler.fit_transform(X_vector_stack)
X_list = []
for i in range(X_vector_scaled.shape[0]):
X_list.append((X_vector_scaled[i]).tolist())
X_vector_df = pd.DataFrame(X_list)
X_vector_df.head()
# ## Creating X and y
X = pd.concat([X_vector_df, df["type_num"], df["confidence"]], axis=1)
y = df["label"]
X.head()
# ## Train Test Split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=23, stratify=y
)
# ## SVC()
from sklearn.svm import SVC
model = SVC(C=1, kernel="poly", degree=3, gamma="scale")
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
model.score(X_test, y_test)
# ## Classification Report
from sklearn.metrics import classification_report
print("\n\n\n Classification Report :\n\n\n ", classification_report(y_test, y_pred))
# ## Visualisation Time
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
import matplotlib.pyplot as plt
import seaborn as sn
plt.figure(figsize=(4, 4), facecolor="pink", edgecolor="brown")
sn.heatmap(cm, annot=True, fmt="d")
plt.xlabel("Prediction", fontdict={"family": "fantasy", "color": "black", "size": 15})
plt.ylabel("Truth", fontdict={"family": "fantasy", "color": "black", "size": 15})
plt.title(
"Confusion Matrix", fontdict={"family": "fantasy", "color": "red", "size": 20}
)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/407/129407850.ipynb
|
human-stress-prediction
|
kreeshrajani
|
[{"Id": 129407850, "ScriptId": 37863132, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13051013, "CreationDate": "05/13/2023 14:24:35", "VersionNumber": 1.0, "Title": "GENSIM(Word Embeddings): Stress Prediction", "EvaluationDate": "05/13/2023", "IsChange": true, "TotalLines": 122.0, "LinesInsertedFromPrevious": 122.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 185428569, "KernelVersionId": 129407850, "SourceDatasetVersionId": 5100130}]
|
[{"Id": 5100130, "DatasetId": 2961947, "DatasourceVersionId": 5171271, "CreatorUserId": 9348276, "LicenseName": "Other (specified in description)", "CreationDate": "03/03/2023 13:59:49", "VersionNumber": 1.0, "Title": "Human Stress Prediction", "Slug": "human-stress-prediction", "Subtitle": "Insights from Speech-based Human Stress Dataset", "Description": "\u201csubreddit \u2013 post_id \u2013 sentence_range \u2013 text-label-confidence-social_timestamp\u201d represents the titles for Stress.csv file.\n\nStress detection is a challenging task, as there are so many words that can be used by people on their posts that can show whether a person is having psychological stress or not. look for datasets that you can use to train a machine learning model for stress detection.\n\nThe dataset contains data posted on subreddits related to mental health. This dataset contains various mental health problems shared by people about their life. Fortunately, this dataset is labelled as 0 and 1, where 0 indicates no stress and 1 indicates stress.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2961947, "CreatorUserId": 9348276, "OwnerUserId": 9348276.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5100130.0, "CurrentDatasourceVersionId": 5171271.0, "ForumId": 3000227, "Type": 2, "CreationDate": "03/03/2023 13:59:49", "LastActivityDate": "03/03/2023", "TotalViews": 25541, "TotalDownloads": 2990, "TotalVotes": 43, "TotalKernels": 20}]
|
[{"Id": 9348276, "UserName": "kreeshrajani", "DisplayName": "Kreesh Rajani", "RegisterDate": "01/08/2022", "PerformanceTier": 2}]
|
import pandas as pd
import numpy as np
import warnings
warnings.filterwarnings("ignore")
# ## Packages
# ## Reading csv
df = pd.read_csv("/kaggle/input/human-stress-prediction/Stress.csv")
print(df.shape)
df.head()
# ## Data Exploration
df.isna().sum()
# #### No 'NA' values
df["label"].value_counts()
# #### Both label have almost equal data points
df["subreddit"].value_counts()
# #### Type of issues and mapping values to these issues
df["type_num"] = df["subreddit"].map(
dict(
zip(
pd.Series(df["subreddit"].unique()),
pd.Series([i for i in range(df["subreddit"].unique().shape[0])]),
)
)
)
# ## GENSIM 'glove-twitter-50' model
import gensim.downloader as api
glove = api.load("glove-twitter-50")
# ## SPACY for preprocess and GENSIM for vectorization
import spacy
nlp = spacy.load("en_core_web_sm")
def preprocess_and_vectorise(text):
list = []
for token in nlp(text):
if token.is_space:
continue
list.append(token.lemma_)
return glove.get_mean_vector(list)
df["vector"] = df["text"].apply(lambda text: preprocess_and_vectorise(text))
df.head()
# ## Scaling the vectors using MINMAXSCALER and Converting array of vector into DataFrame
X_vector = df["vector"].values
X_vector_stack = np.stack(X_vector)
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
X_vector_scaled = scaler.fit_transform(X_vector_stack)
X_list = []
for i in range(X_vector_scaled.shape[0]):
X_list.append((X_vector_scaled[i]).tolist())
X_vector_df = pd.DataFrame(X_list)
X_vector_df.head()
# ## Creating X and y
X = pd.concat([X_vector_df, df["type_num"], df["confidence"]], axis=1)
y = df["label"]
X.head()
# ## Train Test Split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=23, stratify=y
)
# ## SVC()
from sklearn.svm import SVC
model = SVC(C=1, kernel="poly", degree=3, gamma="scale")
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
model.score(X_test, y_test)
# ## Classification Report
from sklearn.metrics import classification_report
print("\n\n\n Classification Report :\n\n\n ", classification_report(y_test, y_pred))
# ## Visualisation Time
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
import matplotlib.pyplot as plt
import seaborn as sn
plt.figure(figsize=(4, 4), facecolor="pink", edgecolor="brown")
sn.heatmap(cm, annot=True, fmt="d")
plt.xlabel("Prediction", fontdict={"family": "fantasy", "color": "black", "size": 15})
plt.ylabel("Truth", fontdict={"family": "fantasy", "color": "black", "size": 15})
plt.title(
"Confusion Matrix", fontdict={"family": "fantasy", "color": "red", "size": 20}
)
plt.show()
| false | 1 | 929 | 1 | 1,105 | 929 |
||
129593999
|
<jupyter_start><jupyter_text>bertforfastbertenglish
Kaggle dataset identifier: bertforfastbertenglish
<jupyter_script>"""
This script provides an exmaple to the fine-tuning and self-distillation
peocess of the FastBERT.
"""
import os, sys
import torch
import json
import random
import argparse
import collections
import torch.nn as nn
from uer.utils.vocab import Vocab
from uer.utils.constants import *
from uer.utils.tokenizer import *
from uer.model_builder import build_model
from uer.utils.optimizers import *
from uer.utils.config import load_hyperparam
from uer.utils.seed import set_seed
from uer.model_saver import save_model
from uer.model_loader import load_model
from uer.layers.multi_headed_attn import MultiHeadedAttention
import numpy as np
import time
from thop import profile
# import os
# assert os.environ['COLAB_TPU_ADDR']
# 日志
# import logging
# logging.basicConfig(filename= './log/fastbert_adv_01-1.log',
# format='[%(levelname)s:%(message)s]',
# level = logging.DEBUG,
# filemode='a',
# datefmt='%Y-%m-%d%I:%M:%S %p')
# print(device_lib.list_local_devices()) # 检查可用设备
# os.environ["CUDA_VISIBLE_DEVICES"] = "0"
print(torch.__version__)
print(torch.cuda.is_available())
# tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
# tf.config.experimental_connect_to_cluster(tpu)
# tf.tpu.experimental.initialize_tpu_system(tpu)
# strategy = tf.distribute.experimental.TPUStrategy(tpu)
torch.set_num_threads(1)
def normal_shannon_entropy(p, labels_num):
entropy = torch.distributions.Categorical(probs=p).entropy()
normal = -np.log(1.0 / labels_num)
return entropy / normal
class Classifier(nn.Module):
def __init__(self, args, input_size, labels_num):
super(Classifier, self).__init__()
self.input_size = input_size
self.cla_hidden_size = 128
self.cla_heads_num = 2
self.labels_num = labels_num
self.pooling = args.pooling
self.output_layer_0 = nn.Linear(input_size, self.cla_hidden_size)
self.self_atten = MultiHeadedAttention(
self.cla_hidden_size, self.cla_heads_num, args.dropout
)
self.output_layer_1 = nn.Linear(self.cla_hidden_size, self.cla_hidden_size)
self.output_layer_2 = nn.Linear(self.cla_hidden_size, labels_num)
def forward(self, hidden, mask):
hidden = torch.tanh(self.output_layer_0(hidden))
hidden = self.self_atten(hidden, hidden, hidden, mask)
if self.pooling == "mean":
hidden = torch.mean(hidden, dim=-1)
elif self.pooling == "max":
hidden = torch.max(hidden, dim=1)[0]
elif self.pooling == "last":
hidden = hidden[:, -1, :]
else:
hidden = hidden[:, 0, :]
output_1 = torch.tanh(self.output_layer_1(hidden))
logits = self.output_layer_2(output_1)
return logits
class FastBertClassifier(nn.Module):
def __init__(self, args, model):
super(FastBertClassifier, self).__init__()
self.embedding = model.embedding
self.encoder = model.encoder
self.labels_num = args.labels_num
self.classifiers = nn.ModuleList(
[
Classifier(args, args.hidden_size, self.labels_num)
for i in range(self.encoder.layers_num)
]
)
self.softmax = nn.LogSoftmax(dim=-1)
self.criterion = nn.NLLLoss()
self.soft_criterion = nn.KLDivLoss(reduction="batchmean")
self.threshold = args.speed
def forward(self, src, label, mask, fast=True):
"""
Args:
src: [batch_size x seq_length]
label: [batch_size]
mask: [batch_size x seq_length]
"""
# Embedding.
emb = self.embedding(src, mask)
# Encoder.
seq_length = emb.size(1)
mask = (mask > 0).unsqueeze(1).repeat(1, seq_length, 1).unsqueeze(1)
mask = mask.float()
mask = (1.0 - mask) * -10000.0
if self.training:
if label is not None:
# training main part of the model
hidden = emb
for i in range(self.encoder.layers_num):
hidden = self.encoder.transformer[i](hidden, mask)
logits = self.classifiers[-1](hidden, mask)
loss = self.criterion(
self.softmax(logits.view(-1, self.labels_num)), label.view(-1)
)
return loss, logits
else:
# distillate the subclassifiers
loss, hidden, hidden_list = 0, emb, []
with torch.no_grad():
for i in range(self.encoder.layers_num):
hidden = self.encoder.transformer[i](hidden, mask)
hidden_list.append(hidden)
teacher_logits = self.classifiers[-1](hidden_list[-1], mask).view(
-1, self.labels_num
)
teacher_probs = nn.functional.softmax(teacher_logits, dim=1)
loss = 0
for i in range(self.encoder.layers_num - 1):
student_logits = self.classifiers[i](hidden_list[i], mask).view(
-1, self.labels_num
)
loss += self.soft_criterion(
self.softmax(student_logits), teacher_probs
)
return loss, teacher_logits
else:
# inference
if fast:
# fast mode
hidden = emb # (batch_size, seq_len, emb_size)
batch_size = hidden.size(0)
logits = torch.zeros(
batch_size,
self.labels_num,
dtype=hidden.dtype,
device=hidden.device,
)
abs_diff_idxs = torch.arange(
0, batch_size, dtype=torch.long, device=hidden.device
)
for i in range(self.encoder.layers_num):
hidden = self.encoder.transformer[i](hidden, mask)
logits_this_layer = self.classifiers[i](
hidden, mask
) # (batch_size, labels_num)
logits[abs_diff_idxs] = logits_this_layer
# filter easy sample
abs_diff_idxs, rel_diff_idxs = self._difficult_samples_idxs(
abs_diff_idxs, logits_this_layer
)
hidden = hidden[rel_diff_idxs, :, :]
mask = mask[rel_diff_idxs, :, :]
if len(abs_diff_idxs) == 0:
break
return None, logits
else:
# normal mode
hidden = emb
for i in range(self.encoder.layers_num):
hidden = self.encoder.transformer[i](hidden, mask)
logits = self.classifiers[-1](hidden, mask)
return None, logits
def _difficult_samples_idxs(self, idxs, logits):
# logits: (batch_size, labels_num)
probs = nn.Softmax(dim=1)(logits)
entropys = normal_shannon_entropy(probs, self.labels_num)
# torch.nonzero() is very time-consuming on GPU
# Please see https://github.com/pytorch/pytorch/issues/14848
# If anyone can optimize this operation, please contact me, thank you!
rel_diff_idxs = (entropys > self.threshold).nonzero().view(-1)
abs_diff_idxs = torch.tensor(
[idxs[i] for i in rel_diff_idxs], device=logits.device
)
return abs_diff_idxs, rel_diff_idxs
class FGM:
def __init__(self, model, epsilon):
self.model = model
self.epsilon = epsilon
self.backup = {}
def attack(self, emb_name="word_embedding"):
# emb_name这个参数要换成你模型中embedding的参数名
for name, param in self.model.named_parameters():
if param.requires_grad and emb_name in name:
self.backup[name] = param.data.clone()
norm = torch.norm(param.grad)
if norm != 0:
r_at = self.epsilon * param.grad / norm
param.data.add_(r_at)
def restore(self, emb_name="word_embedding"):
# emb_name这个参数要换成你模型中embedding的参数名
for name, param in self.model.named_parameters():
if param.requires_grad and emb_name in name:
assert name in self.backup
param.data = self.backup[name]
self.backup = {}
def getargs(datapath, w_ad=True):
parser = argparse.ArgumentParser(
formatter_class=argparse.ArgumentDefaultsHelpFormatter
)
# ../input/d/datasets/googletsai/models/ccf_fastbert4_at.bin
# ../input/bertforfastbertenglish/English_uncased_base_model.bin
# Path options.
if datapath == "ci":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/citation_intent/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/citation_intent/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/citation_intent/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "ca":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert1_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert1.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/carbonsci/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/carbonsci/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/carbonsci/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "cp":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert2_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert2.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/chemprot/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/chemprot/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/chemprot/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "mag":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert3_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert3.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/mag/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/mag/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/mag/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "rct":
parser.add_argument(
"--pretrained_model_path",
default="../input/d/datasets/googletsai/models/ccf_fastbert4_at.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert4_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert4.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/rct-20k/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/rct-20k/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/rct-20k/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "sci":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert5_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert5.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/sci-cite/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/sci-cite/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/sci-cite/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
# Model options.
parser.add_argument("--batch_size", type=int, default=16, help="Batch size.")
parser.add_argument("--seq_length", type=int, default=128, help="Sequence length.")
parser.add_argument(
"--embedding", choices=["bert", "word"], default="bert", help="Emebdding type."
)
parser.add_argument(
"--encoder",
choices=[
"bert",
"lstm",
"gru",
"cnn",
"gatedcnn",
"attn",
"rcnn",
"crnn",
"gpt",
"bilstm",
],
default="bert",
help="Encoder type.",
)
parser.add_argument(
"--bidirectional", action="store_true", help="Specific to recurrent model."
)
parser.add_argument(
"--pooling",
choices=["mean", "max", "first", "last"],
default="first",
help="Pooling type.",
)
# Subword options.
parser.add_argument(
"--subword_type",
choices=["none", "char"],
default="none",
help="Subword feature type.",
)
parser.add_argument(
"--sub_vocab_path",
type=str,
default="models/sub_vocab.txt",
help="Path of the subword vocabulary file.",
)
parser.add_argument(
"--subencoder",
choices=["avg", "lstm", "gru", "cnn"],
default="avg",
help="Subencoder type.",
)
parser.add_argument(
"--sub_layers_num", type=int, default=2, help="The number of subencoder layers."
)
# Tokenizer options.
parser.add_argument(
"--tokenizer",
choices=["bert", "char", "space"],
default="bert",
help="Specify the tokenizer."
"Original Google BERT uses bert tokenizer on Chinese corpus."
"Char tokenizer segments sentences into characters."
"Space tokenizer segments sentences into words according to space.",
)
# Optimizer options.
parser.add_argument(
"--learning_rate", type=float, default=2e-5, help="Learning rate."
)
parser.add_argument("--warmup", type=float, default=0.1, help="Warm up value.")
# Training options.
parser.add_argument("--dropout", type=float, default=0.5, help="Dropout.")
parser.add_argument("--epochs_num", type=int, default=3, help="Number of epochs.")
parser.add_argument(
"--distill_epochs_num",
type=int,
default=10,
help="Number of distillation epochs.",
)
parser.add_argument(
"--report_steps", type=int, default=100, help="Specific steps to print prompt."
)
parser.add_argument("--seed", type=int, default=7, help="Random seed.")
# Evaluation options.
parser.add_argument(
"--mean_reciprocal_rank",
action="store_true",
help="Evaluation metrics for DBQA dataset.",
)
parser.add_argument(
"--fast_mode",
default=True,
dest="fast_mode",
action="store_true",
help="Whether turn on fast mode",
)
parser.add_argument(
"--speed",
type=float,
default=0.5,
help="Threshold of Uncertainty, i.e., the Speed in paper.",
)
args = parser.parse_args([])
# Load the hyperparameters from the config file.
args = load_hyperparam(args)
set_seed(args.seed)
# Count the number of labels.
labels_set = set()
columns = {}
with open(args.train_path, mode="r", encoding="utf-8") as f:
for line_id, line in enumerate(f):
try:
line = line.strip().split("\t")
if line_id == 0:
for i, column_name in enumerate(line):
columns[column_name] = i
continue
label = int(line[columns["label"]])
labels_set.add(label)
except:
pass
args.labels_num = len(labels_set)
# Load vocabulary.
vocab = Vocab()
vocab.load(args.vocab_path)
args.vocab = vocab
return args
def trainandtest(
speed,
fast_mode,
batch_size,
distill_epch_num,
epochs_num,
epsilon,
w_train,
w_ad=True,
):
# parserset();
args.speed = speed
args.fast_mode = fast_mode
args.batch_size = batch_size
args.distill_epochs_num = distill_epch_num
args.epochs_num = epochs_num
args.target = "bert"
vocab = args.vocab
# Build bert model.
# A pseudo target is added.
model = build_model(args)
# Load or initialize parameters.
if args.pretrained_model_path is not None:
# Initialize with pretrained model.
model.load_state_dict(torch.load(args.pretrained_model_path), strict=False)
else:
# Initialize with normal distribution.
for n, p in list(model.named_parameters()):
if "gamma" not in n and "beta" not in n:
p.data.normal_(0, 0.02)
# Build classification model.
model = FastBertClassifier(args, model)
# For simplicity, we use DataParallel wrapper to use multiple GPUs.
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
if torch.cuda.device_count() > 1:
print(
"{} GPUs are available. Let's use them.".format(torch.cuda.device_count())
)
model = nn.DataParallel(model)
model = model.to(device)
# Datset loader.
def batch_loader(batch_size, input_ids, label_ids, mask_ids):
instances_num = input_ids.size()[0]
for i in range(instances_num // batch_size):
input_ids_batch = input_ids[i * batch_size : (i + 1) * batch_size, :]
label_ids_batch = label_ids[i * batch_size : (i + 1) * batch_size]
mask_ids_batch = mask_ids[i * batch_size : (i + 1) * batch_size, :]
yield input_ids_batch, label_ids_batch, mask_ids_batch
if instances_num > instances_num // batch_size * batch_size:
input_ids_batch = input_ids[instances_num // batch_size * batch_size :, :]
label_ids_batch = label_ids[instances_num // batch_size * batch_size :]
mask_ids_batch = mask_ids[instances_num // batch_size * batch_size :, :]
yield input_ids_batch, label_ids_batch, mask_ids_batch
# Build tokenizer.
tokenizer = globals()[args.tokenizer.capitalize() + "Tokenizer"](args)
# Read dataset.
def read_dataset(path):
dataset = []
with open(path, mode="r", encoding="utf-8") as f:
for line_id, line in enumerate(f):
if line_id == 0:
continue
try:
line = line.strip().split("\t")
if len(line) == 2:
label = int(line[0])
text = line[1]
tokens = [vocab.get(t) for t in tokenizer.tokenize(text)]
tokens = [CLS_ID] + tokens
mask = [1] * len(tokens)
if len(tokens) > args.seq_length:
tokens = tokens[: args.seq_length]
mask = mask[: args.seq_length]
while len(tokens) < args.seq_length:
tokens.append(0)
mask.append(0)
dataset.append((tokens, label, mask))
elif len(line) == 3: # For sentence pair input.
label = int(line[columns["label"]])
text_a, text_b = (
line[columns["text_a"]],
line[columns["text_b"]],
)
tokens_a = [vocab.get(t) for t in tokenizer.tokenize(text_a)]
tokens_a = [CLS_ID] + tokens_a + [SEP_ID]
tokens_b = [vocab.get(t) for t in tokenizer.tokenize(text_b)]
tokens_b = tokens_b + [SEP_ID]
tokens = tokens_a + tokens_b
mask = [1] * len(tokens_a) + [2] * len(tokens_b)
if len(tokens) > args.seq_length:
tokens = tokens[: args.seq_length]
mask = mask[: args.seq_length]
while len(tokens) < args.seq_length:
tokens.append(0)
mask.append(0)
dataset.append((tokens, label, mask))
elif len(line) == 4: # For dbqa input.
qid = int(line[columns["qid"]])
label = int(line[columns["label"]])
text_a, text_b = (
line[columns["text_a"]],
line[columns["text_b"]],
)
tokens_a = [vocab.get(t) for t in tokenizer.tokenize(text_a)]
tokens_a = [CLS_ID] + tokens_a + [SEP_ID]
tokens_b = [vocab.get(t) for t in tokenizer.tokenize(text_b)]
tokens_b = tokens_b + [SEP_ID]
tokens = tokens_a + tokens_b
mask = [1] * len(tokens_a) + [2] * len(tokens_b)
if len(tokens) > args.seq_length:
tokens = tokens[: args.seq_length]
mask = mask[: args.seq_length]
while len(tokens) < args.seq_length:
tokens.append(0)
mask.append(0)
dataset.append((tokens, label, mask, qid))
else:
pass
except:
pass
return dataset
# Evaluation function.
def evaluate(args, is_test, fast_mode=False):
if is_test:
dataset = read_dataset(args.test_path)
else:
dataset = read_dataset(args.dev_path)
# print(dataset)
input_ids = torch.LongTensor([sample[0] for sample in dataset])
label_ids = torch.LongTensor([sample[1] for sample in dataset])
mask_ids = torch.LongTensor([sample[2] for sample in dataset])
batch_size = 32
instances_num = input_ids.size()[0]
print("The number of evaluation instances: ", instances_num)
print("Fast mode: ", fast_mode)
correct = 0
# Confusion matrix.
confusion = torch.zeros(args.labels_num, args.labels_num, dtype=torch.long)
model.eval()
if not args.mean_reciprocal_rank:
total_flops, model_params_num = 0, 0
for i, (input_ids_batch, label_ids_batch, mask_ids_batch) in enumerate(
batch_loader(batch_size, input_ids, label_ids, mask_ids)
):
input_ids_batch = input_ids_batch.to(device)
label_ids_batch = label_ids_batch.to(device)
mask_ids_batch = mask_ids_batch.to(device)
with torch.no_grad():
# Get FLOPs at this batch
inputs = (
input_ids_batch,
label_ids_batch,
mask_ids_batch,
fast_mode,
)
flops, params = profile(model, inputs, verbose=False)
total_flops += flops
model_params_num = params
# inference
loss, logits = model(
input_ids_batch, label_ids_batch, mask_ids_batch, fast=fast_mode
)
logits = nn.Softmax(dim=1)(logits)
pred = torch.argmax(logits, dim=1)
gold = label_ids_batch
for j in range(pred.size()[0]):
confusion[pred[j], gold[j]] += 1
correct += torch.sum(pred == gold).item()
print("Number of model parameters: {}".format(model_params_num))
print(
"FLOPs per sample in average: {}".format(
total_flops / float(instances_num)
)
)
if is_test:
print("Confusion matrix:")
print(confusion)
print("Report precision, recall, and f1:")
for i in range(confusion.size()[0]):
pi = (
1
if confusion[i, :].sum().item() == 0
else confusion[i, :].sum().item()
)
ri = (
1
if confusion[:, i].sum().item() == 0
else confusion[:, i].sum().item()
)
p = confusion[i, i].item() / pi
r = confusion[i, i].item() / ri
f1 = 0 if (p + r) == 0 else 2 * p * r / (p + r)
if is_test:
print("Label {}: {:.3f}, {:.3f}, {:.3f}".format(i, p, r, f1))
print(
"Acc. (Correct/Total): {:.4f} ({}/{}) ".format(
correct / len(dataset), correct, len(dataset)
)
)
return correct / len(dataset)
else:
for i, (input_ids_batch, label_ids_batch, mask_ids_batch) in enumerate(
batch_loader(batch_size, input_ids, label_ids, mask_ids)
):
input_ids_batch = input_ids_batch.to(device)
label_ids_batch = label_ids_batch.to(device)
mask_ids_batch = mask_ids_batch.to(device)
with torch.no_grad():
loss, logits = model(
input_ids_batch, label_ids_batch, mask_ids_batch
)
logits = nn.Softmax(dim=1)(logits)
if i == 0:
logits_all = logits
if i >= 1:
logits_all = torch.cat((logits_all, logits), 0)
order = -1
gold = []
for i in range(len(dataset)):
qid = dataset[i][3]
label = dataset[i][1]
if qid == order:
j += 1
if label == 1:
gold.append((qid, j))
else:
order = qid
j = 0
if label == 1:
gold.append((qid, j))
label_order = []
order = -1
for i in range(len(gold)):
if gold[i][0] == order:
templist.append(gold[i][1])
elif gold[i][0] != order:
order = gold[i][0]
if i > 0:
label_order.append(templist)
templist = []
templist.append(gold[i][1])
label_order.append(templist)
order = -1
score_list = []
for i in range(len(logits_all)):
score = float(logits_all[i][1])
qid = int(dataset[i][3])
if qid == order:
templist.append(score)
else:
order = qid
if i > 0:
score_list.append(templist)
templist = []
templist.append(score)
score_list.append(templist)
rank = []
pred = []
for i in range(len(score_list)):
if len(label_order[i]) == 1:
if label_order[i][0] < len(score_list[i]):
true_score = score_list[i][label_order[i][0]]
score_list[i].sort(reverse=True)
for j in range(len(score_list[i])):
if score_list[i][j] == true_score:
rank.append(1 / (j + 1))
else:
rank.append(0)
else:
true_rank = len(score_list[i])
for k in range(len(label_order[i])):
if label_order[i][k] < len(score_list[i]):
true_score = score_list[i][label_order[i][k]]
temp = sorted(score_list[i], reverse=True)
for j in range(len(temp)):
if temp[j] == true_score:
if j < true_rank:
true_rank = j
if true_rank < len(score_list[i]):
rank.append(1 / (true_rank + 1))
else:
rank.append(0)
MRR = sum(rank) / len(rank)
print("Mean Reciprocal Rank: {:.4f}".format(MRR))
return MRR
# if train
if w_train:
# Training phase.
print("Start training.")
trainset = read_dataset(args.train_path)
random.shuffle(trainset)
instances_num = len(trainset)
batch_size = args.batch_size
input_ids = torch.LongTensor([example[0] for example in trainset])
label_ids = torch.LongTensor([example[1] for example in trainset])
mask_ids = torch.LongTensor([example[2] for example in trainset])
train_steps = int(instances_num * args.epochs_num / batch_size) + 1
print("Batch size: ", batch_size)
print("The number of training instances:", instances_num)
param_optimizer = list(model.named_parameters())
no_decay = ["bias", "gamma", "beta"]
optimizer_grouped_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.01,
},
{
"params": [
p for n, p in param_optimizer if any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.0,
},
]
optimizer = AdamW(
optimizer_grouped_parameters, lr=args.learning_rate, correct_bias=False
)
scheduler = WarmupLinearSchedule(
optimizer, warmup_steps=train_steps * args.warmup, t_total=train_steps
)
# traning main part of model
print("Start fine-tuning the backbone of the model.")
# logging.info("Start fine-tuning the backbone of the model.")
total_loss = 0.0
result = 0.0
best_result = 0.0
# add adversial training 1
# print("add adversarial training of fine-tuning the backbone of the model.")
if w_ad:
fgm = FGM(model, epsilon)
for epoch in range(1, args.epochs_num + 1):
model.train()
for i, (input_ids_batch, label_ids_batch, mask_ids_batch) in enumerate(
batch_loader(batch_size, input_ids, label_ids, mask_ids)
):
model.zero_grad()
input_ids_batch = input_ids_batch.to(device)
label_ids_batch = label_ids_batch.to(device)
mask_ids_batch = mask_ids_batch.to(device)
loss, _ = model(
input_ids_batch, label_ids_batch, mask_ids_batch
) # training
if torch.cuda.device_count() > 1:
loss = torch.mean(loss)
total_loss += loss.item()
if (i + 1) % args.report_steps == 0:
print(
"Epoch id: {}, backbone fine-tuning steps: {}, Avg loss: {:.3f}".format(
epoch, i + 1, total_loss / args.report_steps
)
)
total_loss = 0.0
loss.backward()
# add adversial training
if w_ad:
fgm.attack() # 在embedding上添加对抗训练
loss_adv, _ = model(
input_ids_batch, label_ids_batch, mask_ids_batch
)
loss_adv.backward() # 反向传播,并在正常的grad基础上,累加对抗训练的梯度
fgm.restore() # 恢复embedding参数
optimizer.step()
scheduler.step()
result = evaluate(args, False, False)
if result > best_result:
best_result = result
save_model(model, args.output_model_path)
else:
continue
# Evaluation phase.
if args.test_path is not None:
print("Test set evaluation after bakbone fine-tuning.")
# logging.info("Test set evaluation after bakbone fine-tuning.")
model = load_model(model, args.output_model_path)
print("Test on normal model")
# logging.info("Test on normal model")
evaluate(args, True, False)
# logging.info(evaluate(args, True, False))
if args.fast_mode:
print("Test on Fast mode")
# logging.info("Test on Fast mode")
evaluate(args, True, args.fast_mode)
# logging.info(evaluate(args, True, False))
# Distillate subclassifiers
print("Start self-distillation for student-classifiers.")
# logging.info("Start self-distillation for student-classifiers.")
param_optimizer = list(model.named_parameters())
no_decay = ["bias", "gamma", "beta"]
optimizer_grouped_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.01,
},
{
"params": [
p for n, p in param_optimizer if any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.0,
},
]
optimizer = AdamW(
optimizer_grouped_parameters, lr=args.learning_rate * 10, correct_bias=False
)
scheduler = WarmupLinearSchedule(
optimizer, warmup_steps=train_steps * args.warmup, t_total=train_steps
)
model = load_model(model, args.output_model_path)
total_loss = 0.0
result = 0.0
best_result = 0.0
for epoch in range(1, args.distill_epochs_num + 1):
model.train()
for i, (input_ids_batch, label_ids_batch, mask_ids_batch) in enumerate(
batch_loader(batch_size, input_ids, label_ids, mask_ids)
):
model.zero_grad()
input_ids_batch = input_ids_batch.to(device)
label_ids_batch = label_ids_batch.to(device)
mask_ids_batch = mask_ids_batch.to(device)
loss, _ = model(input_ids_batch, None, mask_ids_batch) # distillation
if torch.cuda.device_count() > 1:
loss = torch.mean(loss)
total_loss += loss.item()
if (i + 1) % args.report_steps == 0:
print(
"Epoch id: {}, self-distillation steps: {}, Avg loss: {:.3f}".format(
epoch, i + 1, total_loss / args.report_steps
)
)
total_loss = 0.0
loss.backward()
optimizer.step()
scheduler.step()
result = evaluate(args, False, args.fast_mode)
save_model(model, args.output_model_path)
# Evaluation phase.
if args.test_path is not None:
print("Test set evaluation after self-distillation.")
# logging.info("Test set evaluation after self-distillation.")
model = load_model(model, args.output_model_path)
evaluate(args, True, args.fast_mode)
# logging.info(evaluate(args, True, args.fast_mode))
else:
if args.test_path is not None:
print("Test set evaluation after self-distillation.")
# logging.info("Test set evaluation after self-distillation.")
model = load_model(model, args.output_model_path)
evaluate(args, True, args.fast_mode)
import pandas as pd
import re
import numpy as np
import matplotlib.pyplot as plt
import random
from matplotlib import pyplot as plt
import matplotlib
from matplotlib.ticker import MultipleLocator, FormatStrFormatter
from matplotlib.font_manager import FontProperties
# font_set = FontProperties(fname=r"c:\windows\fonts\simsun.ttc", size=15)
def getpd(path, FLOPv, parav):
acc = []
FLOPs = []
parameters = []
speed = []
flat = []
consum = []
# speedup=[]
# speedup2=[]
# int p=13?
for line in open(path, "r", encoding="utf-8"):
# print(line)
if line.startswith("Acc. (Correct/Total)"):
acc.append(line.split(":")[1])
elif line.startswith("Number of model parameters"):
parameters.append(line.split(":")[1])
elif line.startswith("FLOPs per sample in average"):
FLOPs.append(line.split(":")[1])
elif line.startswith("speed"):
speed.append(line.split(":")[1])
elif line.startswith("flat"):
flat.append(line.split(":")[1])
elif line.startswith("consumed"):
consum.append(line.split(":")[1])
if len(speed) == 0:
speed = [str(i * 0.1) for i in range(1, len(acc) + 1)]
if len(flat) == 0:
flat = [str(1) for i in range(1, len(acc) + 1)]
# speedup=[FLOPs for i in range(1,len(acc)+1)]
# speedup2=[parao for i in range(1,len(acc)+1)]
# print("len of speed: "+str(len(speed)))
# print("len of acc: "+str(len(acc)))
# print("len of f: "+str(len(FLOPs)))
# print("len of parameters: "+str(len(parameters)))
# print("len of flat: "+str(len(flat)))
# print("len of consum: "+str(len(consum)))
# ,'speedup':speedup,'speedup2':speedup2
data = {
"sp": speed,
"acc": acc,
"flop": FLOPs,
"para": parameters,
"flat": flat,
"consum": consum,
}
respd = pd.DataFrame(data)
def clear_characters(text):
if "s" in text:
texts = text.split("s")
text = texts[0]
if "(" in text:
texts = text.split("(")
text = texts[0]
return re.sub("\n", "", text)
respd["sp"] = respd["sp"].apply(clear_characters).apply(float).round(1)
respd["acc"] = respd["acc"].apply(clear_characters).apply(float)
respd["flop"] = respd["flop"].apply(clear_characters).apply(float)
respd["para"] = respd["para"].apply(clear_characters).apply(float)
respd["flat"] = respd["flat"].apply(clear_characters).apply(float)
respd["consum"] = respd["consum"].apply(clear_characters).apply(float)
respd["ratio"] = respd["acc"] / respd["flop"]
respd["pratio"] = respd["acc"] / respd["para"]
respd["catio"] = respd["acc"] / respd["consum"]
respd["speedup"] = FLOPv / respd["flop"]
respd["speedup2"] = parav / respd["para"]
dira = path.split("/")[-1]
return respd, dira
# at_pd: 对抗数据
# pd: 无对抗数据
# pname :图名
def muitdraw(at_pd, pd, pname):
y_max = max(at_pd["acc"]) if max(at_pd["acc"]) > max(pd["acc"]) else max(pd["acc"])
y_min = min(at_pd["acc"]) if min(at_pd["acc"]) < min(pd["acc"]) else min(pd["acc"])
offset = (y_max - y_min) / 2
plt.figure(figsize=(4, 3), dpi=150) # figsize设置图片大小,dpi设置清晰度
plt.rcParams["font.sans-serif"] = ["SimHei"] # 用来正常显示中文标签
# plt.title('epoch与F1-score值的变化关系')
ax = plt.gca() # gca:get current axis得到当前轴
# 设置图片的右边框和上边框为不显示
ax.spines["right"].set_color("none")
ax.spines["top"].set_color("none")
ax.spines["bottom"].set_linewidth(0.5)
ax.spines["left"].set_linewidth(0.5)
plt.plot(
at_pd["speedup"],
at_pd["acc"],
c="black",
linewidth=0.5,
marker="o",
markersize=4,
label=pname + "-at",
)
plt.plot(
pd["speedup"],
pd["acc"],
c="black",
linewidth=0.5,
marker="^",
markersize=4,
label=pname,
)
# # 设置x轴的刻度
# plt.xticks(t1)
# plt.xticks(range(3,6))
plt.ylim(y_min - offset, y_max + offset)
font2 = {
"family": "Times New Roman",
"weight": "normal",
"size": 10,
}
plt.tick_params(labelsize=8)
plt.rcParams["xtick.direction"] = "in" # 将x周的刻度线方向设置向内
plt.rcParams["ytick.direction"] = "in" # 将y轴的刻度方向设置向内
plt.xlabel("speedup", font2) # ϵ
plt.ylabel("Acc", font2)
# plt.legend()
plt.legend(frameon=False, loc="upper right", fontsize="small") # 设置图例无边框,将图例放在左上角
plt.savefig(pname + ".png", bbox_inches="tight")
plt.show()
def sigdraw(pd, pname):
y_max = max(pd["acc"])
y_min = min(pd["acc"])
offset = (y_max - y_min) / 2
plt.figure(figsize=(4, 3), dpi=150) # figsize设置图片大小,dpi设置清晰度
plt.rcParams["font.sans-serif"] = ["SimHei"] # 用来正常显示中文标签
# plt.title('epoch与F1-score值的变化关系')
ax = plt.gca() # gca:get current axis得到当前轴
# 设置图片的右边框和上边框为不显示
ax.spines["right"].set_color("none")
ax.spines["top"].set_color("none")
ax.spines["bottom"].set_linewidth(0.5)
ax.spines["left"].set_linewidth(0.5)
plt.plot(
pd["speedup"],
pd["acc"],
c="black",
linewidth=0.5,
marker="^",
markersize=4,
label=pname,
)
# # 设置x轴的刻度
# plt.xticks(t1)
# plt.xticks(range(3,6))
plt.ylim(y_min - offset, y_max + offset)
font2 = {
"family": "Times New Roman",
"weight": "normal",
"size": 10,
}
plt.tick_params(labelsize=8)
plt.rcParams["xtick.direction"] = "in" # 将x周的刻度线方向设置向内
plt.rcParams["ytick.direction"] = "in" # 将y轴的刻度方向设置向内
plt.xlabel("speedup", font2) # ϵ
plt.ylabel("Acc", font2)
# plt.legend()
plt.legend(frameon=False, loc="upper right", fontsize="small") # 设置图例无边框,将图例放在左上角
plt.savefig(pname + ".png", bbox_inches="tight")
plt.show()
import sys
import os
class Logger(object):
def __init__(self, filename="log_p3.txt"):
self.terminal = sys.stdout
self.log = open(filename, "a")
def write(self, message):
self.terminal.write(message)
self.log.write(message)
self.log.flush() # 缓冲区的内容及时更新到log文件中
def flush(self):
pass
type = sys.getfilesystemencoding()
sys.stdout = Logger()
datapath = "sci"
args = getargs(datapath)
# 测试多次
print("start ad test: " + datapath)
for i in range(1, 9):
old_time = time.time()
speed = 0.1 * i
print("train num: " + str(i))
print("*** speed is " + str(speed) + " ***")
trainandtest(
speed=speed,
fast_mode=True,
batch_size=16,
distill_epch_num=5,
epochs_num=10,
epsilon=0.5,
w_train=False,
)
current_time = time.time()
print("consumed time:" + str(current_time - old_time) + "s")
sys.stdout = __console
print("back to screen!")
parav = 85200269.0
FLOPv = 10892625536.0
nres_adv, dir = getpd("../input/tmplog/log_p3.txt", FLOPv, parav)
# /kaggle/working/log_l3.txt
# nres_adv,dir=getpd('/kaggle/working/log_p3.txt',FLOPv,parav)
offset = 8
sci_pd = nres_adv[:offset]
# ci_pd = nres_adv[offset:offset*2]
sigdraw(sci_pd, "sci")
# muitdraw(ci_pd, rct_pd, 'ci')
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/593/129593999.ipynb
|
bertforfastbertenglish
|
mamainwuxi
|
[{"Id": 129593999, "ScriptId": 38534834, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6324470, "CreationDate": "05/15/2023 05:41:30", "VersionNumber": 1.0, "Title": "fastbert -scicite-citation-intent", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 1040.0, "LinesInsertedFromPrevious": 1040.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185821762, "KernelVersionId": 129593999, "SourceDatasetVersionId": 3640169}, {"Id": 185821763, "KernelVersionId": 129593999, "SourceDatasetVersionId": 3663107}, {"Id": 185821764, "KernelVersionId": 129593999, "SourceDatasetVersionId": 3717619}, {"Id": 185821765, "KernelVersionId": 129593999, "SourceDatasetVersionId": 3799657}, {"Id": 185821767, "KernelVersionId": 129593999, "SourceDatasetVersionId": 3817753}]
|
[{"Id": 3640169, "DatasetId": 2180178, "DatasourceVersionId": 3693855, "CreatorUserId": 10412325, "LicenseName": "Unknown", "CreationDate": "05/15/2022 13:32:05", "VersionNumber": 1.0, "Title": "bertforfastbertenglish", "Slug": "bertforfastbertenglish", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2180178, "CreatorUserId": 10412325, "OwnerUserId": 10412325.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3640169.0, "CurrentDatasourceVersionId": 3693855.0, "ForumId": 2206126, "Type": 2, "CreationDate": "05/15/2022 13:32:05", "LastActivityDate": "05/15/2022", "TotalViews": 198, "TotalDownloads": 1, "TotalVotes": 0, "TotalKernels": 7}]
|
[{"Id": 10412325, "UserName": "mamainwuxi", "DisplayName": "hukai_wlw_jn", "RegisterDate": "05/01/2022", "PerformanceTier": 0}]
|
"""
This script provides an exmaple to the fine-tuning and self-distillation
peocess of the FastBERT.
"""
import os, sys
import torch
import json
import random
import argparse
import collections
import torch.nn as nn
from uer.utils.vocab import Vocab
from uer.utils.constants import *
from uer.utils.tokenizer import *
from uer.model_builder import build_model
from uer.utils.optimizers import *
from uer.utils.config import load_hyperparam
from uer.utils.seed import set_seed
from uer.model_saver import save_model
from uer.model_loader import load_model
from uer.layers.multi_headed_attn import MultiHeadedAttention
import numpy as np
import time
from thop import profile
# import os
# assert os.environ['COLAB_TPU_ADDR']
# 日志
# import logging
# logging.basicConfig(filename= './log/fastbert_adv_01-1.log',
# format='[%(levelname)s:%(message)s]',
# level = logging.DEBUG,
# filemode='a',
# datefmt='%Y-%m-%d%I:%M:%S %p')
# print(device_lib.list_local_devices()) # 检查可用设备
# os.environ["CUDA_VISIBLE_DEVICES"] = "0"
print(torch.__version__)
print(torch.cuda.is_available())
# tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
# tf.config.experimental_connect_to_cluster(tpu)
# tf.tpu.experimental.initialize_tpu_system(tpu)
# strategy = tf.distribute.experimental.TPUStrategy(tpu)
torch.set_num_threads(1)
def normal_shannon_entropy(p, labels_num):
entropy = torch.distributions.Categorical(probs=p).entropy()
normal = -np.log(1.0 / labels_num)
return entropy / normal
class Classifier(nn.Module):
def __init__(self, args, input_size, labels_num):
super(Classifier, self).__init__()
self.input_size = input_size
self.cla_hidden_size = 128
self.cla_heads_num = 2
self.labels_num = labels_num
self.pooling = args.pooling
self.output_layer_0 = nn.Linear(input_size, self.cla_hidden_size)
self.self_atten = MultiHeadedAttention(
self.cla_hidden_size, self.cla_heads_num, args.dropout
)
self.output_layer_1 = nn.Linear(self.cla_hidden_size, self.cla_hidden_size)
self.output_layer_2 = nn.Linear(self.cla_hidden_size, labels_num)
def forward(self, hidden, mask):
hidden = torch.tanh(self.output_layer_0(hidden))
hidden = self.self_atten(hidden, hidden, hidden, mask)
if self.pooling == "mean":
hidden = torch.mean(hidden, dim=-1)
elif self.pooling == "max":
hidden = torch.max(hidden, dim=1)[0]
elif self.pooling == "last":
hidden = hidden[:, -1, :]
else:
hidden = hidden[:, 0, :]
output_1 = torch.tanh(self.output_layer_1(hidden))
logits = self.output_layer_2(output_1)
return logits
class FastBertClassifier(nn.Module):
def __init__(self, args, model):
super(FastBertClassifier, self).__init__()
self.embedding = model.embedding
self.encoder = model.encoder
self.labels_num = args.labels_num
self.classifiers = nn.ModuleList(
[
Classifier(args, args.hidden_size, self.labels_num)
for i in range(self.encoder.layers_num)
]
)
self.softmax = nn.LogSoftmax(dim=-1)
self.criterion = nn.NLLLoss()
self.soft_criterion = nn.KLDivLoss(reduction="batchmean")
self.threshold = args.speed
def forward(self, src, label, mask, fast=True):
"""
Args:
src: [batch_size x seq_length]
label: [batch_size]
mask: [batch_size x seq_length]
"""
# Embedding.
emb = self.embedding(src, mask)
# Encoder.
seq_length = emb.size(1)
mask = (mask > 0).unsqueeze(1).repeat(1, seq_length, 1).unsqueeze(1)
mask = mask.float()
mask = (1.0 - mask) * -10000.0
if self.training:
if label is not None:
# training main part of the model
hidden = emb
for i in range(self.encoder.layers_num):
hidden = self.encoder.transformer[i](hidden, mask)
logits = self.classifiers[-1](hidden, mask)
loss = self.criterion(
self.softmax(logits.view(-1, self.labels_num)), label.view(-1)
)
return loss, logits
else:
# distillate the subclassifiers
loss, hidden, hidden_list = 0, emb, []
with torch.no_grad():
for i in range(self.encoder.layers_num):
hidden = self.encoder.transformer[i](hidden, mask)
hidden_list.append(hidden)
teacher_logits = self.classifiers[-1](hidden_list[-1], mask).view(
-1, self.labels_num
)
teacher_probs = nn.functional.softmax(teacher_logits, dim=1)
loss = 0
for i in range(self.encoder.layers_num - 1):
student_logits = self.classifiers[i](hidden_list[i], mask).view(
-1, self.labels_num
)
loss += self.soft_criterion(
self.softmax(student_logits), teacher_probs
)
return loss, teacher_logits
else:
# inference
if fast:
# fast mode
hidden = emb # (batch_size, seq_len, emb_size)
batch_size = hidden.size(0)
logits = torch.zeros(
batch_size,
self.labels_num,
dtype=hidden.dtype,
device=hidden.device,
)
abs_diff_idxs = torch.arange(
0, batch_size, dtype=torch.long, device=hidden.device
)
for i in range(self.encoder.layers_num):
hidden = self.encoder.transformer[i](hidden, mask)
logits_this_layer = self.classifiers[i](
hidden, mask
) # (batch_size, labels_num)
logits[abs_diff_idxs] = logits_this_layer
# filter easy sample
abs_diff_idxs, rel_diff_idxs = self._difficult_samples_idxs(
abs_diff_idxs, logits_this_layer
)
hidden = hidden[rel_diff_idxs, :, :]
mask = mask[rel_diff_idxs, :, :]
if len(abs_diff_idxs) == 0:
break
return None, logits
else:
# normal mode
hidden = emb
for i in range(self.encoder.layers_num):
hidden = self.encoder.transformer[i](hidden, mask)
logits = self.classifiers[-1](hidden, mask)
return None, logits
def _difficult_samples_idxs(self, idxs, logits):
# logits: (batch_size, labels_num)
probs = nn.Softmax(dim=1)(logits)
entropys = normal_shannon_entropy(probs, self.labels_num)
# torch.nonzero() is very time-consuming on GPU
# Please see https://github.com/pytorch/pytorch/issues/14848
# If anyone can optimize this operation, please contact me, thank you!
rel_diff_idxs = (entropys > self.threshold).nonzero().view(-1)
abs_diff_idxs = torch.tensor(
[idxs[i] for i in rel_diff_idxs], device=logits.device
)
return abs_diff_idxs, rel_diff_idxs
class FGM:
def __init__(self, model, epsilon):
self.model = model
self.epsilon = epsilon
self.backup = {}
def attack(self, emb_name="word_embedding"):
# emb_name这个参数要换成你模型中embedding的参数名
for name, param in self.model.named_parameters():
if param.requires_grad and emb_name in name:
self.backup[name] = param.data.clone()
norm = torch.norm(param.grad)
if norm != 0:
r_at = self.epsilon * param.grad / norm
param.data.add_(r_at)
def restore(self, emb_name="word_embedding"):
# emb_name这个参数要换成你模型中embedding的参数名
for name, param in self.model.named_parameters():
if param.requires_grad and emb_name in name:
assert name in self.backup
param.data = self.backup[name]
self.backup = {}
def getargs(datapath, w_ad=True):
parser = argparse.ArgumentParser(
formatter_class=argparse.ArgumentDefaultsHelpFormatter
)
# ../input/d/datasets/googletsai/models/ccf_fastbert4_at.bin
# ../input/bertforfastbertenglish/English_uncased_base_model.bin
# Path options.
if datapath == "ci":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/citation_intent/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/citation_intent/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/citation_intent/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "ca":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert1_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert1.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/carbonsci/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/carbonsci/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/carbonsci/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "cp":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert2_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert2.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/chemprot/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/chemprot/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/chemprot/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "mag":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert3_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert3.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/mag/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/mag/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/mag/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "rct":
parser.add_argument(
"--pretrained_model_path",
default="../input/d/datasets/googletsai/models/ccf_fastbert4_at.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert4_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert4.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/rct-20k/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/rct-20k/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/rct-20k/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
elif datapath == "sci":
parser.add_argument(
"--pretrained_model_path",
default="../input/bertforfastbertenglish/English_uncased_base_model.bin",
type=str,
help="Path of the pretrained model.",
)
if w_ad:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert5_at.bin",
type=str,
help="Path of the output model.",
)
else:
parser.add_argument(
"--output_model_path",
default="./models/ccf_fastbert5.bin",
type=str,
help="Path of the output model.",
)
parser.add_argument(
"--vocab_path",
default="./models/google_uncased_en_vocab.txt",
type=str,
help="Path of the vocabulary file.",
)
parser.add_argument(
"--train_path",
default="../input/ipm-datasets/sci-cite/train.tsv",
type=str,
help="Path of the trainset.",
)
parser.add_argument(
"--dev_path",
default="../input/ipm-datasets/sci-cite/dev.tsv",
type=str,
help="Path of the devset.",
)
parser.add_argument(
"--test_path",
default="../input/ipm-datasets/sci-cite/test.tsv",
type=str,
help="Path of the testset.",
)
parser.add_argument(
"--config_path",
default="./models/bert_base_config.json",
type=str,
help="Path of the config file.",
)
# Model options.
parser.add_argument("--batch_size", type=int, default=16, help="Batch size.")
parser.add_argument("--seq_length", type=int, default=128, help="Sequence length.")
parser.add_argument(
"--embedding", choices=["bert", "word"], default="bert", help="Emebdding type."
)
parser.add_argument(
"--encoder",
choices=[
"bert",
"lstm",
"gru",
"cnn",
"gatedcnn",
"attn",
"rcnn",
"crnn",
"gpt",
"bilstm",
],
default="bert",
help="Encoder type.",
)
parser.add_argument(
"--bidirectional", action="store_true", help="Specific to recurrent model."
)
parser.add_argument(
"--pooling",
choices=["mean", "max", "first", "last"],
default="first",
help="Pooling type.",
)
# Subword options.
parser.add_argument(
"--subword_type",
choices=["none", "char"],
default="none",
help="Subword feature type.",
)
parser.add_argument(
"--sub_vocab_path",
type=str,
default="models/sub_vocab.txt",
help="Path of the subword vocabulary file.",
)
parser.add_argument(
"--subencoder",
choices=["avg", "lstm", "gru", "cnn"],
default="avg",
help="Subencoder type.",
)
parser.add_argument(
"--sub_layers_num", type=int, default=2, help="The number of subencoder layers."
)
# Tokenizer options.
parser.add_argument(
"--tokenizer",
choices=["bert", "char", "space"],
default="bert",
help="Specify the tokenizer."
"Original Google BERT uses bert tokenizer on Chinese corpus."
"Char tokenizer segments sentences into characters."
"Space tokenizer segments sentences into words according to space.",
)
# Optimizer options.
parser.add_argument(
"--learning_rate", type=float, default=2e-5, help="Learning rate."
)
parser.add_argument("--warmup", type=float, default=0.1, help="Warm up value.")
# Training options.
parser.add_argument("--dropout", type=float, default=0.5, help="Dropout.")
parser.add_argument("--epochs_num", type=int, default=3, help="Number of epochs.")
parser.add_argument(
"--distill_epochs_num",
type=int,
default=10,
help="Number of distillation epochs.",
)
parser.add_argument(
"--report_steps", type=int, default=100, help="Specific steps to print prompt."
)
parser.add_argument("--seed", type=int, default=7, help="Random seed.")
# Evaluation options.
parser.add_argument(
"--mean_reciprocal_rank",
action="store_true",
help="Evaluation metrics for DBQA dataset.",
)
parser.add_argument(
"--fast_mode",
default=True,
dest="fast_mode",
action="store_true",
help="Whether turn on fast mode",
)
parser.add_argument(
"--speed",
type=float,
default=0.5,
help="Threshold of Uncertainty, i.e., the Speed in paper.",
)
args = parser.parse_args([])
# Load the hyperparameters from the config file.
args = load_hyperparam(args)
set_seed(args.seed)
# Count the number of labels.
labels_set = set()
columns = {}
with open(args.train_path, mode="r", encoding="utf-8") as f:
for line_id, line in enumerate(f):
try:
line = line.strip().split("\t")
if line_id == 0:
for i, column_name in enumerate(line):
columns[column_name] = i
continue
label = int(line[columns["label"]])
labels_set.add(label)
except:
pass
args.labels_num = len(labels_set)
# Load vocabulary.
vocab = Vocab()
vocab.load(args.vocab_path)
args.vocab = vocab
return args
def trainandtest(
speed,
fast_mode,
batch_size,
distill_epch_num,
epochs_num,
epsilon,
w_train,
w_ad=True,
):
# parserset();
args.speed = speed
args.fast_mode = fast_mode
args.batch_size = batch_size
args.distill_epochs_num = distill_epch_num
args.epochs_num = epochs_num
args.target = "bert"
vocab = args.vocab
# Build bert model.
# A pseudo target is added.
model = build_model(args)
# Load or initialize parameters.
if args.pretrained_model_path is not None:
# Initialize with pretrained model.
model.load_state_dict(torch.load(args.pretrained_model_path), strict=False)
else:
# Initialize with normal distribution.
for n, p in list(model.named_parameters()):
if "gamma" not in n and "beta" not in n:
p.data.normal_(0, 0.02)
# Build classification model.
model = FastBertClassifier(args, model)
# For simplicity, we use DataParallel wrapper to use multiple GPUs.
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
if torch.cuda.device_count() > 1:
print(
"{} GPUs are available. Let's use them.".format(torch.cuda.device_count())
)
model = nn.DataParallel(model)
model = model.to(device)
# Datset loader.
def batch_loader(batch_size, input_ids, label_ids, mask_ids):
instances_num = input_ids.size()[0]
for i in range(instances_num // batch_size):
input_ids_batch = input_ids[i * batch_size : (i + 1) * batch_size, :]
label_ids_batch = label_ids[i * batch_size : (i + 1) * batch_size]
mask_ids_batch = mask_ids[i * batch_size : (i + 1) * batch_size, :]
yield input_ids_batch, label_ids_batch, mask_ids_batch
if instances_num > instances_num // batch_size * batch_size:
input_ids_batch = input_ids[instances_num // batch_size * batch_size :, :]
label_ids_batch = label_ids[instances_num // batch_size * batch_size :]
mask_ids_batch = mask_ids[instances_num // batch_size * batch_size :, :]
yield input_ids_batch, label_ids_batch, mask_ids_batch
# Build tokenizer.
tokenizer = globals()[args.tokenizer.capitalize() + "Tokenizer"](args)
# Read dataset.
def read_dataset(path):
dataset = []
with open(path, mode="r", encoding="utf-8") as f:
for line_id, line in enumerate(f):
if line_id == 0:
continue
try:
line = line.strip().split("\t")
if len(line) == 2:
label = int(line[0])
text = line[1]
tokens = [vocab.get(t) for t in tokenizer.tokenize(text)]
tokens = [CLS_ID] + tokens
mask = [1] * len(tokens)
if len(tokens) > args.seq_length:
tokens = tokens[: args.seq_length]
mask = mask[: args.seq_length]
while len(tokens) < args.seq_length:
tokens.append(0)
mask.append(0)
dataset.append((tokens, label, mask))
elif len(line) == 3: # For sentence pair input.
label = int(line[columns["label"]])
text_a, text_b = (
line[columns["text_a"]],
line[columns["text_b"]],
)
tokens_a = [vocab.get(t) for t in tokenizer.tokenize(text_a)]
tokens_a = [CLS_ID] + tokens_a + [SEP_ID]
tokens_b = [vocab.get(t) for t in tokenizer.tokenize(text_b)]
tokens_b = tokens_b + [SEP_ID]
tokens = tokens_a + tokens_b
mask = [1] * len(tokens_a) + [2] * len(tokens_b)
if len(tokens) > args.seq_length:
tokens = tokens[: args.seq_length]
mask = mask[: args.seq_length]
while len(tokens) < args.seq_length:
tokens.append(0)
mask.append(0)
dataset.append((tokens, label, mask))
elif len(line) == 4: # For dbqa input.
qid = int(line[columns["qid"]])
label = int(line[columns["label"]])
text_a, text_b = (
line[columns["text_a"]],
line[columns["text_b"]],
)
tokens_a = [vocab.get(t) for t in tokenizer.tokenize(text_a)]
tokens_a = [CLS_ID] + tokens_a + [SEP_ID]
tokens_b = [vocab.get(t) for t in tokenizer.tokenize(text_b)]
tokens_b = tokens_b + [SEP_ID]
tokens = tokens_a + tokens_b
mask = [1] * len(tokens_a) + [2] * len(tokens_b)
if len(tokens) > args.seq_length:
tokens = tokens[: args.seq_length]
mask = mask[: args.seq_length]
while len(tokens) < args.seq_length:
tokens.append(0)
mask.append(0)
dataset.append((tokens, label, mask, qid))
else:
pass
except:
pass
return dataset
# Evaluation function.
def evaluate(args, is_test, fast_mode=False):
if is_test:
dataset = read_dataset(args.test_path)
else:
dataset = read_dataset(args.dev_path)
# print(dataset)
input_ids = torch.LongTensor([sample[0] for sample in dataset])
label_ids = torch.LongTensor([sample[1] for sample in dataset])
mask_ids = torch.LongTensor([sample[2] for sample in dataset])
batch_size = 32
instances_num = input_ids.size()[0]
print("The number of evaluation instances: ", instances_num)
print("Fast mode: ", fast_mode)
correct = 0
# Confusion matrix.
confusion = torch.zeros(args.labels_num, args.labels_num, dtype=torch.long)
model.eval()
if not args.mean_reciprocal_rank:
total_flops, model_params_num = 0, 0
for i, (input_ids_batch, label_ids_batch, mask_ids_batch) in enumerate(
batch_loader(batch_size, input_ids, label_ids, mask_ids)
):
input_ids_batch = input_ids_batch.to(device)
label_ids_batch = label_ids_batch.to(device)
mask_ids_batch = mask_ids_batch.to(device)
with torch.no_grad():
# Get FLOPs at this batch
inputs = (
input_ids_batch,
label_ids_batch,
mask_ids_batch,
fast_mode,
)
flops, params = profile(model, inputs, verbose=False)
total_flops += flops
model_params_num = params
# inference
loss, logits = model(
input_ids_batch, label_ids_batch, mask_ids_batch, fast=fast_mode
)
logits = nn.Softmax(dim=1)(logits)
pred = torch.argmax(logits, dim=1)
gold = label_ids_batch
for j in range(pred.size()[0]):
confusion[pred[j], gold[j]] += 1
correct += torch.sum(pred == gold).item()
print("Number of model parameters: {}".format(model_params_num))
print(
"FLOPs per sample in average: {}".format(
total_flops / float(instances_num)
)
)
if is_test:
print("Confusion matrix:")
print(confusion)
print("Report precision, recall, and f1:")
for i in range(confusion.size()[0]):
pi = (
1
if confusion[i, :].sum().item() == 0
else confusion[i, :].sum().item()
)
ri = (
1
if confusion[:, i].sum().item() == 0
else confusion[:, i].sum().item()
)
p = confusion[i, i].item() / pi
r = confusion[i, i].item() / ri
f1 = 0 if (p + r) == 0 else 2 * p * r / (p + r)
if is_test:
print("Label {}: {:.3f}, {:.3f}, {:.3f}".format(i, p, r, f1))
print(
"Acc. (Correct/Total): {:.4f} ({}/{}) ".format(
correct / len(dataset), correct, len(dataset)
)
)
return correct / len(dataset)
else:
for i, (input_ids_batch, label_ids_batch, mask_ids_batch) in enumerate(
batch_loader(batch_size, input_ids, label_ids, mask_ids)
):
input_ids_batch = input_ids_batch.to(device)
label_ids_batch = label_ids_batch.to(device)
mask_ids_batch = mask_ids_batch.to(device)
with torch.no_grad():
loss, logits = model(
input_ids_batch, label_ids_batch, mask_ids_batch
)
logits = nn.Softmax(dim=1)(logits)
if i == 0:
logits_all = logits
if i >= 1:
logits_all = torch.cat((logits_all, logits), 0)
order = -1
gold = []
for i in range(len(dataset)):
qid = dataset[i][3]
label = dataset[i][1]
if qid == order:
j += 1
if label == 1:
gold.append((qid, j))
else:
order = qid
j = 0
if label == 1:
gold.append((qid, j))
label_order = []
order = -1
for i in range(len(gold)):
if gold[i][0] == order:
templist.append(gold[i][1])
elif gold[i][0] != order:
order = gold[i][0]
if i > 0:
label_order.append(templist)
templist = []
templist.append(gold[i][1])
label_order.append(templist)
order = -1
score_list = []
for i in range(len(logits_all)):
score = float(logits_all[i][1])
qid = int(dataset[i][3])
if qid == order:
templist.append(score)
else:
order = qid
if i > 0:
score_list.append(templist)
templist = []
templist.append(score)
score_list.append(templist)
rank = []
pred = []
for i in range(len(score_list)):
if len(label_order[i]) == 1:
if label_order[i][0] < len(score_list[i]):
true_score = score_list[i][label_order[i][0]]
score_list[i].sort(reverse=True)
for j in range(len(score_list[i])):
if score_list[i][j] == true_score:
rank.append(1 / (j + 1))
else:
rank.append(0)
else:
true_rank = len(score_list[i])
for k in range(len(label_order[i])):
if label_order[i][k] < len(score_list[i]):
true_score = score_list[i][label_order[i][k]]
temp = sorted(score_list[i], reverse=True)
for j in range(len(temp)):
if temp[j] == true_score:
if j < true_rank:
true_rank = j
if true_rank < len(score_list[i]):
rank.append(1 / (true_rank + 1))
else:
rank.append(0)
MRR = sum(rank) / len(rank)
print("Mean Reciprocal Rank: {:.4f}".format(MRR))
return MRR
# if train
if w_train:
# Training phase.
print("Start training.")
trainset = read_dataset(args.train_path)
random.shuffle(trainset)
instances_num = len(trainset)
batch_size = args.batch_size
input_ids = torch.LongTensor([example[0] for example in trainset])
label_ids = torch.LongTensor([example[1] for example in trainset])
mask_ids = torch.LongTensor([example[2] for example in trainset])
train_steps = int(instances_num * args.epochs_num / batch_size) + 1
print("Batch size: ", batch_size)
print("The number of training instances:", instances_num)
param_optimizer = list(model.named_parameters())
no_decay = ["bias", "gamma", "beta"]
optimizer_grouped_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.01,
},
{
"params": [
p for n, p in param_optimizer if any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.0,
},
]
optimizer = AdamW(
optimizer_grouped_parameters, lr=args.learning_rate, correct_bias=False
)
scheduler = WarmupLinearSchedule(
optimizer, warmup_steps=train_steps * args.warmup, t_total=train_steps
)
# traning main part of model
print("Start fine-tuning the backbone of the model.")
# logging.info("Start fine-tuning the backbone of the model.")
total_loss = 0.0
result = 0.0
best_result = 0.0
# add adversial training 1
# print("add adversarial training of fine-tuning the backbone of the model.")
if w_ad:
fgm = FGM(model, epsilon)
for epoch in range(1, args.epochs_num + 1):
model.train()
for i, (input_ids_batch, label_ids_batch, mask_ids_batch) in enumerate(
batch_loader(batch_size, input_ids, label_ids, mask_ids)
):
model.zero_grad()
input_ids_batch = input_ids_batch.to(device)
label_ids_batch = label_ids_batch.to(device)
mask_ids_batch = mask_ids_batch.to(device)
loss, _ = model(
input_ids_batch, label_ids_batch, mask_ids_batch
) # training
if torch.cuda.device_count() > 1:
loss = torch.mean(loss)
total_loss += loss.item()
if (i + 1) % args.report_steps == 0:
print(
"Epoch id: {}, backbone fine-tuning steps: {}, Avg loss: {:.3f}".format(
epoch, i + 1, total_loss / args.report_steps
)
)
total_loss = 0.0
loss.backward()
# add adversial training
if w_ad:
fgm.attack() # 在embedding上添加对抗训练
loss_adv, _ = model(
input_ids_batch, label_ids_batch, mask_ids_batch
)
loss_adv.backward() # 反向传播,并在正常的grad基础上,累加对抗训练的梯度
fgm.restore() # 恢复embedding参数
optimizer.step()
scheduler.step()
result = evaluate(args, False, False)
if result > best_result:
best_result = result
save_model(model, args.output_model_path)
else:
continue
# Evaluation phase.
if args.test_path is not None:
print("Test set evaluation after bakbone fine-tuning.")
# logging.info("Test set evaluation after bakbone fine-tuning.")
model = load_model(model, args.output_model_path)
print("Test on normal model")
# logging.info("Test on normal model")
evaluate(args, True, False)
# logging.info(evaluate(args, True, False))
if args.fast_mode:
print("Test on Fast mode")
# logging.info("Test on Fast mode")
evaluate(args, True, args.fast_mode)
# logging.info(evaluate(args, True, False))
# Distillate subclassifiers
print("Start self-distillation for student-classifiers.")
# logging.info("Start self-distillation for student-classifiers.")
param_optimizer = list(model.named_parameters())
no_decay = ["bias", "gamma", "beta"]
optimizer_grouped_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.01,
},
{
"params": [
p for n, p in param_optimizer if any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.0,
},
]
optimizer = AdamW(
optimizer_grouped_parameters, lr=args.learning_rate * 10, correct_bias=False
)
scheduler = WarmupLinearSchedule(
optimizer, warmup_steps=train_steps * args.warmup, t_total=train_steps
)
model = load_model(model, args.output_model_path)
total_loss = 0.0
result = 0.0
best_result = 0.0
for epoch in range(1, args.distill_epochs_num + 1):
model.train()
for i, (input_ids_batch, label_ids_batch, mask_ids_batch) in enumerate(
batch_loader(batch_size, input_ids, label_ids, mask_ids)
):
model.zero_grad()
input_ids_batch = input_ids_batch.to(device)
label_ids_batch = label_ids_batch.to(device)
mask_ids_batch = mask_ids_batch.to(device)
loss, _ = model(input_ids_batch, None, mask_ids_batch) # distillation
if torch.cuda.device_count() > 1:
loss = torch.mean(loss)
total_loss += loss.item()
if (i + 1) % args.report_steps == 0:
print(
"Epoch id: {}, self-distillation steps: {}, Avg loss: {:.3f}".format(
epoch, i + 1, total_loss / args.report_steps
)
)
total_loss = 0.0
loss.backward()
optimizer.step()
scheduler.step()
result = evaluate(args, False, args.fast_mode)
save_model(model, args.output_model_path)
# Evaluation phase.
if args.test_path is not None:
print("Test set evaluation after self-distillation.")
# logging.info("Test set evaluation after self-distillation.")
model = load_model(model, args.output_model_path)
evaluate(args, True, args.fast_mode)
# logging.info(evaluate(args, True, args.fast_mode))
else:
if args.test_path is not None:
print("Test set evaluation after self-distillation.")
# logging.info("Test set evaluation after self-distillation.")
model = load_model(model, args.output_model_path)
evaluate(args, True, args.fast_mode)
import pandas as pd
import re
import numpy as np
import matplotlib.pyplot as plt
import random
from matplotlib import pyplot as plt
import matplotlib
from matplotlib.ticker import MultipleLocator, FormatStrFormatter
from matplotlib.font_manager import FontProperties
# font_set = FontProperties(fname=r"c:\windows\fonts\simsun.ttc", size=15)
def getpd(path, FLOPv, parav):
acc = []
FLOPs = []
parameters = []
speed = []
flat = []
consum = []
# speedup=[]
# speedup2=[]
# int p=13?
for line in open(path, "r", encoding="utf-8"):
# print(line)
if line.startswith("Acc. (Correct/Total)"):
acc.append(line.split(":")[1])
elif line.startswith("Number of model parameters"):
parameters.append(line.split(":")[1])
elif line.startswith("FLOPs per sample in average"):
FLOPs.append(line.split(":")[1])
elif line.startswith("speed"):
speed.append(line.split(":")[1])
elif line.startswith("flat"):
flat.append(line.split(":")[1])
elif line.startswith("consumed"):
consum.append(line.split(":")[1])
if len(speed) == 0:
speed = [str(i * 0.1) for i in range(1, len(acc) + 1)]
if len(flat) == 0:
flat = [str(1) for i in range(1, len(acc) + 1)]
# speedup=[FLOPs for i in range(1,len(acc)+1)]
# speedup2=[parao for i in range(1,len(acc)+1)]
# print("len of speed: "+str(len(speed)))
# print("len of acc: "+str(len(acc)))
# print("len of f: "+str(len(FLOPs)))
# print("len of parameters: "+str(len(parameters)))
# print("len of flat: "+str(len(flat)))
# print("len of consum: "+str(len(consum)))
# ,'speedup':speedup,'speedup2':speedup2
data = {
"sp": speed,
"acc": acc,
"flop": FLOPs,
"para": parameters,
"flat": flat,
"consum": consum,
}
respd = pd.DataFrame(data)
def clear_characters(text):
if "s" in text:
texts = text.split("s")
text = texts[0]
if "(" in text:
texts = text.split("(")
text = texts[0]
return re.sub("\n", "", text)
respd["sp"] = respd["sp"].apply(clear_characters).apply(float).round(1)
respd["acc"] = respd["acc"].apply(clear_characters).apply(float)
respd["flop"] = respd["flop"].apply(clear_characters).apply(float)
respd["para"] = respd["para"].apply(clear_characters).apply(float)
respd["flat"] = respd["flat"].apply(clear_characters).apply(float)
respd["consum"] = respd["consum"].apply(clear_characters).apply(float)
respd["ratio"] = respd["acc"] / respd["flop"]
respd["pratio"] = respd["acc"] / respd["para"]
respd["catio"] = respd["acc"] / respd["consum"]
respd["speedup"] = FLOPv / respd["flop"]
respd["speedup2"] = parav / respd["para"]
dira = path.split("/")[-1]
return respd, dira
# at_pd: 对抗数据
# pd: 无对抗数据
# pname :图名
def muitdraw(at_pd, pd, pname):
y_max = max(at_pd["acc"]) if max(at_pd["acc"]) > max(pd["acc"]) else max(pd["acc"])
y_min = min(at_pd["acc"]) if min(at_pd["acc"]) < min(pd["acc"]) else min(pd["acc"])
offset = (y_max - y_min) / 2
plt.figure(figsize=(4, 3), dpi=150) # figsize设置图片大小,dpi设置清晰度
plt.rcParams["font.sans-serif"] = ["SimHei"] # 用来正常显示中文标签
# plt.title('epoch与F1-score值的变化关系')
ax = plt.gca() # gca:get current axis得到当前轴
# 设置图片的右边框和上边框为不显示
ax.spines["right"].set_color("none")
ax.spines["top"].set_color("none")
ax.spines["bottom"].set_linewidth(0.5)
ax.spines["left"].set_linewidth(0.5)
plt.plot(
at_pd["speedup"],
at_pd["acc"],
c="black",
linewidth=0.5,
marker="o",
markersize=4,
label=pname + "-at",
)
plt.plot(
pd["speedup"],
pd["acc"],
c="black",
linewidth=0.5,
marker="^",
markersize=4,
label=pname,
)
# # 设置x轴的刻度
# plt.xticks(t1)
# plt.xticks(range(3,6))
plt.ylim(y_min - offset, y_max + offset)
font2 = {
"family": "Times New Roman",
"weight": "normal",
"size": 10,
}
plt.tick_params(labelsize=8)
plt.rcParams["xtick.direction"] = "in" # 将x周的刻度线方向设置向内
plt.rcParams["ytick.direction"] = "in" # 将y轴的刻度方向设置向内
plt.xlabel("speedup", font2) # ϵ
plt.ylabel("Acc", font2)
# plt.legend()
plt.legend(frameon=False, loc="upper right", fontsize="small") # 设置图例无边框,将图例放在左上角
plt.savefig(pname + ".png", bbox_inches="tight")
plt.show()
def sigdraw(pd, pname):
y_max = max(pd["acc"])
y_min = min(pd["acc"])
offset = (y_max - y_min) / 2
plt.figure(figsize=(4, 3), dpi=150) # figsize设置图片大小,dpi设置清晰度
plt.rcParams["font.sans-serif"] = ["SimHei"] # 用来正常显示中文标签
# plt.title('epoch与F1-score值的变化关系')
ax = plt.gca() # gca:get current axis得到当前轴
# 设置图片的右边框和上边框为不显示
ax.spines["right"].set_color("none")
ax.spines["top"].set_color("none")
ax.spines["bottom"].set_linewidth(0.5)
ax.spines["left"].set_linewidth(0.5)
plt.plot(
pd["speedup"],
pd["acc"],
c="black",
linewidth=0.5,
marker="^",
markersize=4,
label=pname,
)
# # 设置x轴的刻度
# plt.xticks(t1)
# plt.xticks(range(3,6))
plt.ylim(y_min - offset, y_max + offset)
font2 = {
"family": "Times New Roman",
"weight": "normal",
"size": 10,
}
plt.tick_params(labelsize=8)
plt.rcParams["xtick.direction"] = "in" # 将x周的刻度线方向设置向内
plt.rcParams["ytick.direction"] = "in" # 将y轴的刻度方向设置向内
plt.xlabel("speedup", font2) # ϵ
plt.ylabel("Acc", font2)
# plt.legend()
plt.legend(frameon=False, loc="upper right", fontsize="small") # 设置图例无边框,将图例放在左上角
plt.savefig(pname + ".png", bbox_inches="tight")
plt.show()
import sys
import os
class Logger(object):
def __init__(self, filename="log_p3.txt"):
self.terminal = sys.stdout
self.log = open(filename, "a")
def write(self, message):
self.terminal.write(message)
self.log.write(message)
self.log.flush() # 缓冲区的内容及时更新到log文件中
def flush(self):
pass
type = sys.getfilesystemencoding()
sys.stdout = Logger()
datapath = "sci"
args = getargs(datapath)
# 测试多次
print("start ad test: " + datapath)
for i in range(1, 9):
old_time = time.time()
speed = 0.1 * i
print("train num: " + str(i))
print("*** speed is " + str(speed) + " ***")
trainandtest(
speed=speed,
fast_mode=True,
batch_size=16,
distill_epch_num=5,
epochs_num=10,
epsilon=0.5,
w_train=False,
)
current_time = time.time()
print("consumed time:" + str(current_time - old_time) + "s")
sys.stdout = __console
print("back to screen!")
parav = 85200269.0
FLOPv = 10892625536.0
nres_adv, dir = getpd("../input/tmplog/log_p3.txt", FLOPv, parav)
# /kaggle/working/log_l3.txt
# nres_adv,dir=getpd('/kaggle/working/log_p3.txt',FLOPv,parav)
offset = 8
sci_pd = nres_adv[:offset]
# ci_pd = nres_adv[offset:offset*2]
sigdraw(sci_pd, "sci")
# muitdraw(ci_pd, rct_pd, 'ci')
| false | 0 | 13,045 | 0 | 13,070 | 13,045 |
||
129593359
|
<jupyter_start><jupyter_text>Dermatology Dataset (Multi-class classification)
* The differential diagnosis of **"erythemato-squamous"** diseases is a real problem in dermatology. They all share the clinical features of erythema and scaling, with minimal differences. The disorders in this group are psoriasis, seborrheic dermatitis, lichen planus, pityriasis rosea, chronic dermatitis, and pityriasis rubra pilaris. Usually, a biopsy is necessary for the diagnosis, but unfortunately, these diseases share many histopathological features as well.
* Patients were first evaluated clinically with 12 features. Afterward, skin samples were taken for the evaluation of 22 histopathological features. The values of the histopathological features are determined by an analysis of the samples under a microscope
## Feature Value Information
In the dataset constructed for this domain, the **family history feature** has the value 1 if any of these diseases has been observed in the family, and 0 otherwise. The age feature simply represents the age of the patient.
Every other feature **clinical and histopathological** was given a degree in the range of 0 to 3. Here, 0 indicates that the feature was not present, 3 indicates the largest amount possible, and 1, 2 indicate the relative intermediate values.
## **Exploration Ideas**
* **Distribution of each attribute:** Explore the distribution of each attribute (column) in the dataset. You can use histograms or boxplots to visualize the distribution of each attribute and look for any patterns or outliers.
* **Correlation analysis**: Use correlation matrices to explore the relationship between the different attributes in the dataset. This can help identify which attributes are most closely related to each other and may be useful in predicting the class labels.
* **Missing values analysis**: Investigate the missing values in the Age attribute, which are represented with '?' in the dataset. Determine the proportion of missing values and evaluate whether imputation is needed.
* **Class distribution**: Explore the distribution of the class labels in the dataset. You can use bar plots to visualize the number of instances for each class, and determine whether the dataset is balanced or imbalanced.
* **Feature engineering**: Consider creating new features that may be useful in predicting the class labels. For example, you could create a feature that combines the presence of specific clinical attributes or histopathological attributes.
* **Outlier detection**: Explore the presence of any outliers in the dataset. Outliers can skew the distribution of the data and impact the performance of machine learning models. You can use boxplots or scatterplots to visualize the distribution of each attribute and identify any potential outliers.
Kaggle dataset identifier: dermatology-dataset-classification
<jupyter_script># # **Dermatology Dataset (Multi-class classification)**
# The aim is to determine the type of Eryhemato-Squamous Disease.
# **Font:** https://www.kaggle.com/datasets/olcaybolat1/dermatology-dataset-classification
# **Dev:** GabrielMoli (Gabriel Moraes de Oliveira)
# (obs: this is my first deep learning model)
# **Github:** GabrielMoli https://github.com/GabrielMoli
import tensorflow as tf
import keras as ke
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.model_selection import train_test_split
import sklearn.metrics as skm
from sklearn.preprocessing import LabelBinarizer
from sklearn.metrics import roc_curve, auc, roc_auc_score
import seaborn as sns
import os, random
# Importing and Spliting Datas
dermatoDataset = pd.read_csv("/content/dermatology_database_1.csv")
# I put the missing ones as 40 as an empirical average to calculate the average by the functions and then changed it with the average
dermatoDataset.age.replace("?", 36.0, inplace=True)
# y = np.asarray(dermatoDataset["class"].values.astype(float))
y = np.asarray(dermatoDataset["class"].values)
X = np.asarray(dermatoDataset.drop("class", axis=1).values.astype(float))
X_train = tf.cast(X, dtype=tf.float32)
# Y_train = tf.cast(y, dtype=tf.float32)
X2, X_test, y2, y_test = train_test_split(
X, y, test_size=0.1, random_state=20, shuffle=True, stratify=y
)
X_train, X_val, y_train, y_val = train_test_split(
X, y, test_size=0.2, random_state=20, shuffle=True, stratify=y
)
# Analysing Content:
print("treino x:", np.shape(X_train))
print("treio y:", np.shape(y_train))
pd.DataFrame(y_val).value_counts()
# Creating Model:
model = ke.Sequential()
model.add(ke.layers.Input(X_train.shape[1]))
model.add(ke.layers.Dropout(0.2))
model.add(ke.layers.BatchNormalization())
model.add(ke.layers.Dense(20, activation="relu"))
model.add(ke.layers.Dense(13, activation="relu"))
model.add(ke.layers.Dense(9, activation="relu"))
model.add(ke.layers.Dense(7, activation="softmax"))
model.summary()
# Compiling and Training
opt = tf.optimizers.Adam(learning_rate=1e-4)
model.compile(
optimizer="Adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
# defining inicialization
i = 543
os.environ["PYTHONHASHSEED"] = str(i)
tf.random.set_seed(i)
np.random.seed(i)
random.seed(i)
history = model.fit(
x=X_train, y=y_train, epochs=275, batch_size=700, validation_data=(X_val, y_val)
)
# Plotting LOSS Curve:
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
plt.plot(history.history["loss"], label="treino")
plt.plot(history.history["val_loss"], label="validacao")
plt.xlabel("epocas")
plt.ylabel("Loss")
plt.legend()
plt.subplot(1, 2, 2)
plt.plot(history.history["accuracy"], label="treino")
plt.plot(history.history["val_accuracy"], label="validacao")
plt.xlabel("epocas")
plt.ylabel("acurácia")
plt.legend()
Y_pred_prob = model.predict(X_test)
Y_pred = np.argmax(Y_pred_prob, axis=1)
cmat = skm.confusion_matrix(y_test, Y_pred)
cm_df = pd.DataFrame(cmat)
ax = plt.subplot()
sns.heatmap(cm_df, annot=True, cmap="Blues", fmt="d", cbar=False)
ax.set_xlabel("Predito")
ax.set_ylabel("Real")
ax.set_title("Confusion Matrix: test datas")
# Metrics:
acuracia = np.sum(np.diag(cmat)) * 100 / np.sum(cmat)
sensibilidade = cmat[1, 1] / (cmat[0, 1] + cmat[1, 1]) * 100
especificidade = cmat[0, 0] / (cmat[1, 0] + cmat[0, 0]) * 100
print("acuracia", acuracia, "%")
print("sensibilidad", sensibilidade, "%")
print("especificidade", especificidade, "%")
# ROC Curve:
target = [1, 2, 3, 4, 5, 6]
# set plot figure size
fig, c_ax = plt.subplots(1, 1, figsize=(12, 8))
# function for scoring roc auc score for multi-class
def multiclass_roc_auc_score(y_test, y_pred, average="macro"):
lb = LabelBinarizer()
lb.fit(y_test)
y_test = lb.transform(y_test)
y_pred = lb.transform(y_pred)
for idx, c_label in enumerate(target):
fpr, tpr, thresholds = roc_curve(y_test[:, idx].astype(int), y_pred[:, idx])
c_ax.plot(fpr, tpr, label="%s (AUC:%0.2f)" % (c_label, auc(fpr, tpr)))
c_ax.plot(fpr, fpr, "b-", label="Random Guessing")
return roc_auc_score(y_test, y_pred, average=average)
print("ROC AUC score:", multiclass_roc_auc_score(y_test, Y_pred))
c_ax.legend()
c_ax.set_xlabel("False Positive Rate")
c_ax.set_ylabel("True Positive Rate")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/593/129593359.ipynb
|
dermatology-dataset-classification
|
olcaybolat1
|
[{"Id": 129593359, "ScriptId": 38535054, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15080392, "CreationDate": "05/15/2023 05:33:17", "VersionNumber": 3.0, "Title": "Deep learning classification KERAS/TENSORFLOW", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 153.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 152.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 185820397, "KernelVersionId": 129593359, "SourceDatasetVersionId": 5642556}]
|
[{"Id": 5642556, "DatasetId": 3243099, "DatasourceVersionId": 5717877, "CreatorUserId": 10724258, "LicenseName": "Other (specified in description)", "CreationDate": "05/09/2023 11:21:39", "VersionNumber": 5.0, "Title": "Dermatology Dataset (Multi-class classification)", "Slug": "dermatology-dataset-classification", "Subtitle": "The aim is to determine the type of Eryhemato-Squamous Disease.", "Description": "* The differential diagnosis of **\"erythemato-squamous\"** diseases is a real problem in dermatology. They all share the clinical features of erythema and scaling, with minimal differences. The disorders in this group are psoriasis, seborrheic dermatitis, lichen planus, pityriasis rosea, chronic dermatitis, and pityriasis rubra pilaris. Usually, a biopsy is necessary for the diagnosis, but unfortunately, these diseases share many histopathological features as well. \n\n* Patients were first evaluated clinically with 12 features. Afterward, skin samples were taken for the evaluation of 22 histopathological features. The values of the histopathological features are determined by an analysis of the samples under a microscope\n\n## Feature Value Information \n\nIn the dataset constructed for this domain, the **family history feature** has the value 1 if any of these diseases has been observed in the family, and 0 otherwise. The age feature simply represents the age of the patient. \n\nEvery other feature **clinical and histopathological** was given a degree in the range of 0 to 3. Here, 0 indicates that the feature was not present, 3 indicates the largest amount possible, and 1, 2 indicate the relative intermediate values.\n\n## **Exploration Ideas**\n\n* **Distribution of each attribute:** Explore the distribution of each attribute (column) in the dataset. You can use histograms or boxplots to visualize the distribution of each attribute and look for any patterns or outliers.\n\n* **Correlation analysis**: Use correlation matrices to explore the relationship between the different attributes in the dataset. This can help identify which attributes are most closely related to each other and may be useful in predicting the class labels.\n\n* **Missing values analysis**: Investigate the missing values in the Age attribute, which are represented with '?' in the dataset. Determine the proportion of missing values and evaluate whether imputation is needed.\n\n* **Class distribution**: Explore the distribution of the class labels in the dataset. You can use bar plots to visualize the number of instances for each class, and determine whether the dataset is balanced or imbalanced.\n\n* **Feature engineering**: Consider creating new features that may be useful in predicting the class labels. For example, you could create a feature that combines the presence of specific clinical attributes or histopathological attributes.\n\n* **Outlier detection**: Explore the presence of any outliers in the dataset. Outliers can skew the distribution of the data and impact the performance of machine learning models. You can use boxplots or scatterplots to visualize the distribution of each attribute and identify any potential outliers.", "VersionNotes": "Data Update 2023-05-09", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3243099, "CreatorUserId": 10724258, "OwnerUserId": 10724258.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5642556.0, "CurrentDatasourceVersionId": 5717877.0, "ForumId": 3308372, "Type": 2, "CreationDate": "05/09/2023 10:30:47", "LastActivityDate": "05/09/2023", "TotalViews": 11274, "TotalDownloads": 1486, "TotalVotes": 51, "TotalKernels": 10}]
|
[{"Id": 10724258, "UserName": "olcaybolat1", "DisplayName": "olcay_bolat", "RegisterDate": "06/03/2022", "PerformanceTier": 1}]
|
# # **Dermatology Dataset (Multi-class classification)**
# The aim is to determine the type of Eryhemato-Squamous Disease.
# **Font:** https://www.kaggle.com/datasets/olcaybolat1/dermatology-dataset-classification
# **Dev:** GabrielMoli (Gabriel Moraes de Oliveira)
# (obs: this is my first deep learning model)
# **Github:** GabrielMoli https://github.com/GabrielMoli
import tensorflow as tf
import keras as ke
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.model_selection import train_test_split
import sklearn.metrics as skm
from sklearn.preprocessing import LabelBinarizer
from sklearn.metrics import roc_curve, auc, roc_auc_score
import seaborn as sns
import os, random
# Importing and Spliting Datas
dermatoDataset = pd.read_csv("/content/dermatology_database_1.csv")
# I put the missing ones as 40 as an empirical average to calculate the average by the functions and then changed it with the average
dermatoDataset.age.replace("?", 36.0, inplace=True)
# y = np.asarray(dermatoDataset["class"].values.astype(float))
y = np.asarray(dermatoDataset["class"].values)
X = np.asarray(dermatoDataset.drop("class", axis=1).values.astype(float))
X_train = tf.cast(X, dtype=tf.float32)
# Y_train = tf.cast(y, dtype=tf.float32)
X2, X_test, y2, y_test = train_test_split(
X, y, test_size=0.1, random_state=20, shuffle=True, stratify=y
)
X_train, X_val, y_train, y_val = train_test_split(
X, y, test_size=0.2, random_state=20, shuffle=True, stratify=y
)
# Analysing Content:
print("treino x:", np.shape(X_train))
print("treio y:", np.shape(y_train))
pd.DataFrame(y_val).value_counts()
# Creating Model:
model = ke.Sequential()
model.add(ke.layers.Input(X_train.shape[1]))
model.add(ke.layers.Dropout(0.2))
model.add(ke.layers.BatchNormalization())
model.add(ke.layers.Dense(20, activation="relu"))
model.add(ke.layers.Dense(13, activation="relu"))
model.add(ke.layers.Dense(9, activation="relu"))
model.add(ke.layers.Dense(7, activation="softmax"))
model.summary()
# Compiling and Training
opt = tf.optimizers.Adam(learning_rate=1e-4)
model.compile(
optimizer="Adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
# defining inicialization
i = 543
os.environ["PYTHONHASHSEED"] = str(i)
tf.random.set_seed(i)
np.random.seed(i)
random.seed(i)
history = model.fit(
x=X_train, y=y_train, epochs=275, batch_size=700, validation_data=(X_val, y_val)
)
# Plotting LOSS Curve:
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
plt.plot(history.history["loss"], label="treino")
plt.plot(history.history["val_loss"], label="validacao")
plt.xlabel("epocas")
plt.ylabel("Loss")
plt.legend()
plt.subplot(1, 2, 2)
plt.plot(history.history["accuracy"], label="treino")
plt.plot(history.history["val_accuracy"], label="validacao")
plt.xlabel("epocas")
plt.ylabel("acurácia")
plt.legend()
Y_pred_prob = model.predict(X_test)
Y_pred = np.argmax(Y_pred_prob, axis=1)
cmat = skm.confusion_matrix(y_test, Y_pred)
cm_df = pd.DataFrame(cmat)
ax = plt.subplot()
sns.heatmap(cm_df, annot=True, cmap="Blues", fmt="d", cbar=False)
ax.set_xlabel("Predito")
ax.set_ylabel("Real")
ax.set_title("Confusion Matrix: test datas")
# Metrics:
acuracia = np.sum(np.diag(cmat)) * 100 / np.sum(cmat)
sensibilidade = cmat[1, 1] / (cmat[0, 1] + cmat[1, 1]) * 100
especificidade = cmat[0, 0] / (cmat[1, 0] + cmat[0, 0]) * 100
print("acuracia", acuracia, "%")
print("sensibilidad", sensibilidade, "%")
print("especificidade", especificidade, "%")
# ROC Curve:
target = [1, 2, 3, 4, 5, 6]
# set plot figure size
fig, c_ax = plt.subplots(1, 1, figsize=(12, 8))
# function for scoring roc auc score for multi-class
def multiclass_roc_auc_score(y_test, y_pred, average="macro"):
lb = LabelBinarizer()
lb.fit(y_test)
y_test = lb.transform(y_test)
y_pred = lb.transform(y_pred)
for idx, c_label in enumerate(target):
fpr, tpr, thresholds = roc_curve(y_test[:, idx].astype(int), y_pred[:, idx])
c_ax.plot(fpr, tpr, label="%s (AUC:%0.2f)" % (c_label, auc(fpr, tpr)))
c_ax.plot(fpr, fpr, "b-", label="Random Guessing")
return roc_auc_score(y_test, y_pred, average=average)
print("ROC AUC score:", multiclass_roc_auc_score(y_test, Y_pred))
c_ax.legend()
c_ax.set_xlabel("False Positive Rate")
c_ax.set_ylabel("True Positive Rate")
plt.show()
| false | 0 | 1,596 | 3 | 2,227 | 1,596 |
||
129593862
|
# # Simple GAN Implementation
# 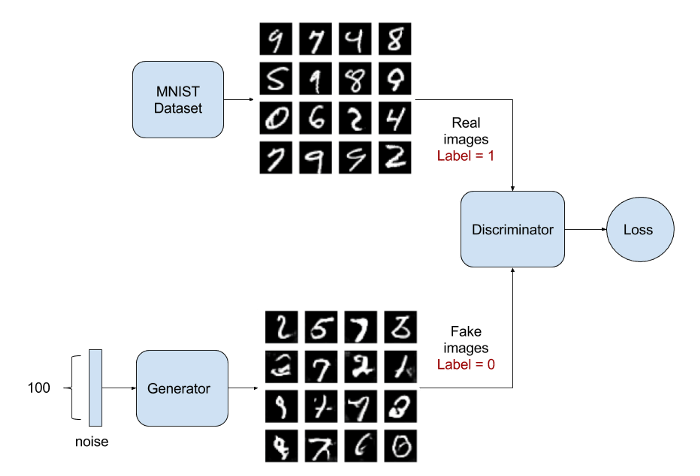
# A simple GAN implementation can be broken down into the following steps:
# Define the generator and discriminator networks. The generator network is responsible for creating new data that resembles the training data. The discriminator network is responsible for distinguishing between real and generated data.
# Initialize the weights of the generator and discriminator networks. The weights of the networks are initialized randomly.
# Train the generator and discriminator networks. The generator and discriminator networks are trained alternately. In each iteration, the generator network is trained to create data that is more likely to be classified as real by the discriminator network. The discriminator network is trained to distinguish between real and generated data more accurately.
# Continue training until the generator network is able to create data that is indistinguishable from real data.
# **1. Define the generator and discriminator networks.**
# The generator network is typically a generative network, such as a deconvolutional neural network (DCNN). The DCNN takes in a vector of random noise as input and outputs an image. The discriminator network is typically a discriminative network, such as a convolutional neural network (CNN). The CNN takes in an image as input and outputs a probability that the image is real or generated.
# **2. Initialize the weights of the generator and discriminator networks.**
# The weights of the generator and discriminator networks are initialized randomly. This is done to prevent the networks from becoming stuck in a local minimum.
# **3. Train the generator and discriminator networks.**
# The generator and discriminator networks are trained alternately. In each iteration, the generator network is trained to create data that is more likely to be classified as real by the discriminator network. The discriminator network is trained to distinguish between real and generated data more accurately.
# The training process is repeated until the generator network is able to create data that is indistinguishable from real data.
# **4. Continue training until the generator network is able to create data that is indistinguishable from real data.**
# The training process can be continued for a number of epochs. The number of epochs that are required to train a GAN depends on the complexity of the data and the complexity of the networks.
# GANs are a powerful tool for generating new data. They have been used to generate images, text, and music. GANs are still under development, but they have the potential to revolutionize the way that we create new data
# # **Objective: TO generate MNIST Images**
# # **Importing Necessary Libraries**
import tensorflow as tf
import matplotlib.pyplot as plt
# # **Loading MNIST Dataset**
# 
# (x_train, y_train), (_, _) = tf.keras.datasets.mnist.load_data() loads the MNIST dataset into NumPy arrays.
# **The MNIST dataset is a collection of handwritten digits, and it is a popular dataset for training machine learning models to recognize handwritten digits.**
# The load_data() function returns two tuples:
# x_train is a NumPy array of 60,000 training images, each of which is 28x28 pixels in size.
# y_train is a NumPy array of 60,000 labels, each of which is an integer from 0 to 9, representing the digit that is written in the image.
# x_test is a NumPy array of 10,000 testing images, each of which is 28x28 pixels in size.
# y_test is a NumPy array of 10,000 labels, each of which is an integer from 0 to 9, representing the digit that is written in the image.
# The _ in the code snippet represents the fact that the load_data() function returns two tuples, but we are only interested in the first tuple (x_train and y_train).
(x_train, y_train), (_, _) = tf.keras.datasets.mnist.load_data()
# # **Normalize The Data**
# **The MNIST dataset is a collection of handwritten digits, and each image is represented as a 28x28 matrix of pixels. The values of the pixels range from 0 to 255, where 0 represents black and 255 represents white.**
# **Normalizing the data means to scale the values of the pixels so that they all lie in the range [0, 1]. This is done by dividing each pixel value by 255.**
# Normalizing the data is important for machine learning models because it helps them to learn more effectively. When the values of the pixels are all in the same range, the model does not have to learn as much about the scale of the data. This can lead to faster training times and better accuracy
x_train = x_train / 255.0
# # **Define the generator and discriminator networks**
# **The generator network is responsible for creating new data that resembles the training data. The discriminator network is responsible for distinguishing between real and generated data.**
# The generator network is a generative network, such as a deconvolutional neural network (DCNN). The DCNN takes in a vector of random noise as input and outputs an image. The discriminator network is typically a discriminative network, such as a convolutional neural network (CNN). The CNN takes in an image as input and outputs a probability that the image is real or generated.
# The generator network is defined using the tf.keras.Sequential() class. The tf.keras.Sequential() class is a container class for a stack of layers. The layers in the generator network are defined using the tf.keras.layers module.
# The discriminator network is also defined using the tf.keras.Sequential() class. The layers in the discriminator network are defined using the tf.keras.layers module.
# The generator and discriminator networks are trained using the adversarial training algorithm. The adversarial training algorithm is an iterative algorithm that trains the generator and discriminator networks alternately. In each iteration, the generator network is trained to create data that is more likely to be classified as real by the discriminator network. The discriminator network is trained to distinguish between real and generated data more accurately.
# The adversarial training algorithm is a powerful tool for training GANs. GANs have been used to generate images, text, and music. GANs are still under development, but they have the potential to revolutionize the way that we create new data.
# Define the generator network
generator = tf.keras.Sequential(
[
tf.keras.layers.Dense(7 * 7 * 256, input_shape=(100,), activation="relu"),
tf.keras.layers.Reshape((7, 7, 256)),
tf.keras.layers.Conv2DTranspose(
128, kernel_size=3, strides=2, padding="same", activation="relu"
),
tf.keras.layers.Conv2DTranspose(
64, kernel_size=3, strides=1, padding="same", activation="relu"
),
tf.keras.layers.Conv2DTranspose(
1, kernel_size=3, strides=2, padding="same", activation="tanh"
),
]
)
# Define the discriminator network
discriminator = tf.keras.Sequential(
[
tf.keras.layers.Conv2D(
64, kernel_size=3, strides=2, padding="same", input_shape=(28, 28, 1)
),
tf.keras.layers.LeakyReLU(),
tf.keras.layers.Conv2D(128, kernel_size=3, strides=2, padding="same"),
tf.keras.layers.LeakyReLU(),
tf.keras.layers.GlobalMaxPooling2D(),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
# # **Compile the models**
# The compile() method is used to compile a Keras model. **The optimizer argument specifies the optimizer that will be used to train the model. The loss argument specifies the loss function that will be used to evaluate the model.**
# The adam optimizer is a popular optimizer for training deep learning models. The binary_crossentropy loss function is a loss function that is commonly used for classification tasks.
# Once the generator and discriminator networks are compiled, they can be trained using the adversarial training algorithm.
generator.compile(optimizer="adam", loss="binary_crossentropy")
discriminator.compile(optimizer="adam", loss="binary_crossentropy")
# # Define the GAN loss
# The gan_loss() function defines the loss function for the GAN. The loss function is a function that measures how well the model is performing. The loss function for the GAN is defined as the sum of the losses for the generator and discriminator networks.
# **The generator network is trained to minimize the loss function. The discriminator network is trained to maximize the loss function.**
# The gan_loss() function is called in the train() function to train the GAN.
# # **Calculate generator loss**
#
# **Calculates the loss for the generator network. The generator network is trained to create images that are indistinguishable from real images. The loss function for the generator network is defined as the binary cross-entropy between the discriminator's output for the generated images and a one-hot vector of ones.**
# The code snippet return generator_loss, discriminator_loss returns the loss for the generator and discriminator networks. The generator and discriminator networks are trained alternately using the adversarial training algorithm.
def gan_loss(generator, discriminator):
noise = tf.random.normal([batch_size, 100])
fake_images = generator(noise)
real_images = x_train[
tf.random.uniform([batch_size], maxval=x_train.shape[0], dtype=tf.int32)
]
# Add a channel dimension to real_images to match fake_images
real_images = tf.expand_dims(real_images, axis=-1)
# Cast real_images to double tensor
real_images = tf.cast(real_images, dtype=tf.float64)
fake_images = tf.cast(fake_images, dtype=tf.float64)
images = tf.concat([real_images, fake_images], axis=0)
targets = tf.concat([tf.ones((batch_size, 1)), tf.zeros((batch_size, 1))], axis=0)
targets = tf.random.shuffle(targets)
discriminator_loss = discriminator.train_on_batch(images, targets)
noise = tf.random.normal([batch_size, 100])
# Calculate generator loss
generator_loss = tf.reduce_mean(
tf.keras.losses.binary_crossentropy(
tf.ones((batch_size, 1)), discriminator(generator(noise))
)
)
return generator_loss, discriminator_loss
# # Train the GAN
# The epochs variable specifies the number of epochs to train the GAN for. The batch_size variable specifies the size of the batches to use when training the GAN.
# The for epoch in range(epochs): loop iterates over the epochs. In each epoch, the for i in range(0, len(x_train), batch_size): loop iterates over the batches. In each batch, the following steps are performed:
# * Generate a batch of noise.
# * Generate a batch of fake images.
# * Get a batch of real images.
# * Add a channel dimension to the real images.
# * Cast the real images to double tensor.
# * Cast the fake images to double tensor.
# * Concatenate the real and fake images.
# * Concatenate ones and zeros for the targets.
# * Shuffle the targets.
# * Train the discriminator on the concatenated images and targets.
# * Generate a batch of noise.
# * Generate a batch of fake images.
# * Train the generator on the fake images and ones.
# **Print the epoch number, the discriminator loss, and the generator loss.
# The GAN is trained for a specified number of epochs. In each epoch, the generator and discriminator networks are trained alternately. The generator network is trained to create images that are indistinguishable from real images. The discriminator network is trained to distinguish between real and generated images.**
epochs = 10
batch_size = 32
for epoch in range(epochs):
for i in range(0, len(x_train), batch_size):
generator_loss, discriminator_loss = gan_loss(generator, discriminator)
print(
f"Epoch {epoch + 1}/{epochs}, Discriminator loss: {discriminator_loss}, Generator loss: {generator_loss}"
)
# # **Generate images**
# The code snippet noise = tf.random.normal([10, 100]) generates a batch of 10 noise vectors. The noise vectors are used as input to the generator network. The generator network is a generative network that takes in a vector of random noise as input and outputs an image. The code snippet generated_images = generator(noise) generates 10 images using the generator network.
noise = tf.random.normal([10, 100])
generated_images = generator(noise)
# # **Plotting Images Using Matplot**
# The code snippet plt.figure(figsize=(10, 10)) creates a figure with a width of 10 inches and a height of 10 inches. The code snippet for i in range(10): iterates over the range of 10. In each iteration, the following steps are performed:
# **A subplot is created with the index i + 1.
# The image generated_images[i].numpy().reshape(28, 28) is displayed in the subplot.
# The axis of the subplot is turned off.
# The code snippet plt.show() displays the figure.**
# The code snippet displays 10 images generated by the generator network. The images are displayed in a 2x5 grid. The images are grayscale images with a resolution of 28x28 pixels.
plt.figure(figsize=(10, 10))
for i in range(10):
plt.subplot(2, 5, i + 1)
plt.imshow(generated_images[i].numpy().reshape(28, 28), cmap="gray")
plt.axis("off")
plt.show()
# # S**ave the model**
# he code snippet generator.save('generator.h5') saves the generator network to the file generator.h5. The file can be loaded later to use the generator network.
generator.save("generator.h5")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/593/129593862.ipynb
| null | null |
[{"Id": 129593862, "ScriptId": 38526543, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7949888, "CreationDate": "05/15/2023 05:39:54", "VersionNumber": 3.0, "Title": "notebookce97325170", "EvaluationDate": "05/15/2023", "IsChange": true, "TotalLines": 207.0, "LinesInsertedFromPrevious": 23.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 184.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Simple GAN Implementation
# 
# A simple GAN implementation can be broken down into the following steps:
# Define the generator and discriminator networks. The generator network is responsible for creating new data that resembles the training data. The discriminator network is responsible for distinguishing between real and generated data.
# Initialize the weights of the generator and discriminator networks. The weights of the networks are initialized randomly.
# Train the generator and discriminator networks. The generator and discriminator networks are trained alternately. In each iteration, the generator network is trained to create data that is more likely to be classified as real by the discriminator network. The discriminator network is trained to distinguish between real and generated data more accurately.
# Continue training until the generator network is able to create data that is indistinguishable from real data.
# **1. Define the generator and discriminator networks.**
# The generator network is typically a generative network, such as a deconvolutional neural network (DCNN). The DCNN takes in a vector of random noise as input and outputs an image. The discriminator network is typically a discriminative network, such as a convolutional neural network (CNN). The CNN takes in an image as input and outputs a probability that the image is real or generated.
# **2. Initialize the weights of the generator and discriminator networks.**
# The weights of the generator and discriminator networks are initialized randomly. This is done to prevent the networks from becoming stuck in a local minimum.
# **3. Train the generator and discriminator networks.**
# The generator and discriminator networks are trained alternately. In each iteration, the generator network is trained to create data that is more likely to be classified as real by the discriminator network. The discriminator network is trained to distinguish between real and generated data more accurately.
# The training process is repeated until the generator network is able to create data that is indistinguishable from real data.
# **4. Continue training until the generator network is able to create data that is indistinguishable from real data.**
# The training process can be continued for a number of epochs. The number of epochs that are required to train a GAN depends on the complexity of the data and the complexity of the networks.
# GANs are a powerful tool for generating new data. They have been used to generate images, text, and music. GANs are still under development, but they have the potential to revolutionize the way that we create new data
# # **Objective: TO generate MNIST Images**
# # **Importing Necessary Libraries**
import tensorflow as tf
import matplotlib.pyplot as plt
# # **Loading MNIST Dataset**
# 
# (x_train, y_train), (_, _) = tf.keras.datasets.mnist.load_data() loads the MNIST dataset into NumPy arrays.
# **The MNIST dataset is a collection of handwritten digits, and it is a popular dataset for training machine learning models to recognize handwritten digits.**
# The load_data() function returns two tuples:
# x_train is a NumPy array of 60,000 training images, each of which is 28x28 pixels in size.
# y_train is a NumPy array of 60,000 labels, each of which is an integer from 0 to 9, representing the digit that is written in the image.
# x_test is a NumPy array of 10,000 testing images, each of which is 28x28 pixels in size.
# y_test is a NumPy array of 10,000 labels, each of which is an integer from 0 to 9, representing the digit that is written in the image.
# The _ in the code snippet represents the fact that the load_data() function returns two tuples, but we are only interested in the first tuple (x_train and y_train).
(x_train, y_train), (_, _) = tf.keras.datasets.mnist.load_data()
# # **Normalize The Data**
# **The MNIST dataset is a collection of handwritten digits, and each image is represented as a 28x28 matrix of pixels. The values of the pixels range from 0 to 255, where 0 represents black and 255 represents white.**
# **Normalizing the data means to scale the values of the pixels so that they all lie in the range [0, 1]. This is done by dividing each pixel value by 255.**
# Normalizing the data is important for machine learning models because it helps them to learn more effectively. When the values of the pixels are all in the same range, the model does not have to learn as much about the scale of the data. This can lead to faster training times and better accuracy
x_train = x_train / 255.0
# # **Define the generator and discriminator networks**
# **The generator network is responsible for creating new data that resembles the training data. The discriminator network is responsible for distinguishing between real and generated data.**
# The generator network is a generative network, such as a deconvolutional neural network (DCNN). The DCNN takes in a vector of random noise as input and outputs an image. The discriminator network is typically a discriminative network, such as a convolutional neural network (CNN). The CNN takes in an image as input and outputs a probability that the image is real or generated.
# The generator network is defined using the tf.keras.Sequential() class. The tf.keras.Sequential() class is a container class for a stack of layers. The layers in the generator network are defined using the tf.keras.layers module.
# The discriminator network is also defined using the tf.keras.Sequential() class. The layers in the discriminator network are defined using the tf.keras.layers module.
# The generator and discriminator networks are trained using the adversarial training algorithm. The adversarial training algorithm is an iterative algorithm that trains the generator and discriminator networks alternately. In each iteration, the generator network is trained to create data that is more likely to be classified as real by the discriminator network. The discriminator network is trained to distinguish between real and generated data more accurately.
# The adversarial training algorithm is a powerful tool for training GANs. GANs have been used to generate images, text, and music. GANs are still under development, but they have the potential to revolutionize the way that we create new data.
# Define the generator network
generator = tf.keras.Sequential(
[
tf.keras.layers.Dense(7 * 7 * 256, input_shape=(100,), activation="relu"),
tf.keras.layers.Reshape((7, 7, 256)),
tf.keras.layers.Conv2DTranspose(
128, kernel_size=3, strides=2, padding="same", activation="relu"
),
tf.keras.layers.Conv2DTranspose(
64, kernel_size=3, strides=1, padding="same", activation="relu"
),
tf.keras.layers.Conv2DTranspose(
1, kernel_size=3, strides=2, padding="same", activation="tanh"
),
]
)
# Define the discriminator network
discriminator = tf.keras.Sequential(
[
tf.keras.layers.Conv2D(
64, kernel_size=3, strides=2, padding="same", input_shape=(28, 28, 1)
),
tf.keras.layers.LeakyReLU(),
tf.keras.layers.Conv2D(128, kernel_size=3, strides=2, padding="same"),
tf.keras.layers.LeakyReLU(),
tf.keras.layers.GlobalMaxPooling2D(),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
# # **Compile the models**
# The compile() method is used to compile a Keras model. **The optimizer argument specifies the optimizer that will be used to train the model. The loss argument specifies the loss function that will be used to evaluate the model.**
# The adam optimizer is a popular optimizer for training deep learning models. The binary_crossentropy loss function is a loss function that is commonly used for classification tasks.
# Once the generator and discriminator networks are compiled, they can be trained using the adversarial training algorithm.
generator.compile(optimizer="adam", loss="binary_crossentropy")
discriminator.compile(optimizer="adam", loss="binary_crossentropy")
# # Define the GAN loss
# The gan_loss() function defines the loss function for the GAN. The loss function is a function that measures how well the model is performing. The loss function for the GAN is defined as the sum of the losses for the generator and discriminator networks.
# **The generator network is trained to minimize the loss function. The discriminator network is trained to maximize the loss function.**
# The gan_loss() function is called in the train() function to train the GAN.
# # **Calculate generator loss**
#
# **Calculates the loss for the generator network. The generator network is trained to create images that are indistinguishable from real images. The loss function for the generator network is defined as the binary cross-entropy between the discriminator's output for the generated images and a one-hot vector of ones.**
# The code snippet return generator_loss, discriminator_loss returns the loss for the generator and discriminator networks. The generator and discriminator networks are trained alternately using the adversarial training algorithm.
def gan_loss(generator, discriminator):
noise = tf.random.normal([batch_size, 100])
fake_images = generator(noise)
real_images = x_train[
tf.random.uniform([batch_size], maxval=x_train.shape[0], dtype=tf.int32)
]
# Add a channel dimension to real_images to match fake_images
real_images = tf.expand_dims(real_images, axis=-1)
# Cast real_images to double tensor
real_images = tf.cast(real_images, dtype=tf.float64)
fake_images = tf.cast(fake_images, dtype=tf.float64)
images = tf.concat([real_images, fake_images], axis=0)
targets = tf.concat([tf.ones((batch_size, 1)), tf.zeros((batch_size, 1))], axis=0)
targets = tf.random.shuffle(targets)
discriminator_loss = discriminator.train_on_batch(images, targets)
noise = tf.random.normal([batch_size, 100])
# Calculate generator loss
generator_loss = tf.reduce_mean(
tf.keras.losses.binary_crossentropy(
tf.ones((batch_size, 1)), discriminator(generator(noise))
)
)
return generator_loss, discriminator_loss
# # Train the GAN
# The epochs variable specifies the number of epochs to train the GAN for. The batch_size variable specifies the size of the batches to use when training the GAN.
# The for epoch in range(epochs): loop iterates over the epochs. In each epoch, the for i in range(0, len(x_train), batch_size): loop iterates over the batches. In each batch, the following steps are performed:
# * Generate a batch of noise.
# * Generate a batch of fake images.
# * Get a batch of real images.
# * Add a channel dimension to the real images.
# * Cast the real images to double tensor.
# * Cast the fake images to double tensor.
# * Concatenate the real and fake images.
# * Concatenate ones and zeros for the targets.
# * Shuffle the targets.
# * Train the discriminator on the concatenated images and targets.
# * Generate a batch of noise.
# * Generate a batch of fake images.
# * Train the generator on the fake images and ones.
# **Print the epoch number, the discriminator loss, and the generator loss.
# The GAN is trained for a specified number of epochs. In each epoch, the generator and discriminator networks are trained alternately. The generator network is trained to create images that are indistinguishable from real images. The discriminator network is trained to distinguish between real and generated images.**
epochs = 10
batch_size = 32
for epoch in range(epochs):
for i in range(0, len(x_train), batch_size):
generator_loss, discriminator_loss = gan_loss(generator, discriminator)
print(
f"Epoch {epoch + 1}/{epochs}, Discriminator loss: {discriminator_loss}, Generator loss: {generator_loss}"
)
# # **Generate images**
# The code snippet noise = tf.random.normal([10, 100]) generates a batch of 10 noise vectors. The noise vectors are used as input to the generator network. The generator network is a generative network that takes in a vector of random noise as input and outputs an image. The code snippet generated_images = generator(noise) generates 10 images using the generator network.
noise = tf.random.normal([10, 100])
generated_images = generator(noise)
# # **Plotting Images Using Matplot**
# The code snippet plt.figure(figsize=(10, 10)) creates a figure with a width of 10 inches and a height of 10 inches. The code snippet for i in range(10): iterates over the range of 10. In each iteration, the following steps are performed:
# **A subplot is created with the index i + 1.
# The image generated_images[i].numpy().reshape(28, 28) is displayed in the subplot.
# The axis of the subplot is turned off.
# The code snippet plt.show() displays the figure.**
# The code snippet displays 10 images generated by the generator network. The images are displayed in a 2x5 grid. The images are grayscale images with a resolution of 28x28 pixels.
plt.figure(figsize=(10, 10))
for i in range(10):
plt.subplot(2, 5, i + 1)
plt.imshow(generated_images[i].numpy().reshape(28, 28), cmap="gray")
plt.axis("off")
plt.show()
# # S**ave the model**
# he code snippet generator.save('generator.h5') saves the generator network to the file generator.h5. The file can be loaded later to use the generator network.
generator.save("generator.h5")
| false | 0 | 3,405 | 0 | 3,405 | 3,405 |
||
129040406
|
<jupyter_start><jupyter_text>Iris Species
The Iris dataset was used in R.A. Fisher's classic 1936 paper, [The Use of Multiple Measurements in Taxonomic Problems](http://rcs.chemometrics.ru/Tutorials/classification/Fisher.pdf), and can also be found on the [UCI Machine Learning Repository][1].
It includes three iris species with 50 samples each as well as some properties about each flower. One flower species is linearly separable from the other two, but the other two are not linearly separable from each other.
The columns in this dataset are:
- Id
- SepalLengthCm
- SepalWidthCm
- PetalLengthCm
- PetalWidthCm
- Species
[](https://www.kaggle.com/benhamner/d/uciml/iris/sepal-width-vs-length)
[1]: http://archive.ics.uci.edu/ml/
Kaggle dataset identifier: iris
<jupyter_script>import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
CSV_PATH = "/kaggle/input/iris/Iris.csv"
ID = "Id"
TARGET = "Species"
TEST_SIZE = 0.3
VAL_SIZE = 0.3
SEED = 2023
# # load Data
total = pd.read_csv(CSV_PATH)
total.head()
TOTAL_LEN = len(total)
TOTAL_LEN
total[TARGET].unique()
total[TARGET].value_counts()
# # devide feature and target
y = total[TARGET]
X = total.drop([TARGET], axis=1)
# # devide trainval set and test set (total = trainval + test)
X_trainval, X_test, y_trainval, y_test = train_test_split(
X, y, test_size=TEST_SIZE, random_state=SEED, stratify=y
)
print("train+val set = ", len(X_trainval), " test set", len(X_test))
X_trainval.head()
y_trainval.value_counts()
X_test.head()
y_test.value_counts()
# # devide train set and val set (trainval = train + val)
X_train, X_val, y_train, y_val = train_test_split(
X_trainval, y_trainval, test_size=VAL_SIZE, random_state=SEED, stratify=y_trainval
)
print("train set = ", len(X_train), " val set", len(X_val))
X_train.head()
y_train.value_counts()
X_val.head()
y_val.value_counts()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/040/129040406.ipynb
|
iris
| null |
[{"Id": 129040406, "ScriptId": 38357969, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7300186, "CreationDate": "05/10/2023 13:59:20", "VersionNumber": 1.0, "Title": "[sklearn Basic]train_test_split train+val+test", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 58.0, "LinesInsertedFromPrevious": 58.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184746347, "KernelVersionId": 129040406, "SourceDatasetVersionId": 420}]
|
[{"Id": 420, "DatasetId": 19, "DatasourceVersionId": 420, "CreatorUserId": 1, "LicenseName": "CC0: Public Domain", "CreationDate": "09/27/2016 07:38:05", "VersionNumber": 2.0, "Title": "Iris Species", "Slug": "iris", "Subtitle": "Classify iris plants into three species in this classic dataset", "Description": "The Iris dataset was used in R.A. Fisher's classic 1936 paper, [The Use of Multiple Measurements in Taxonomic Problems](http://rcs.chemometrics.ru/Tutorials/classification/Fisher.pdf), and can also be found on the [UCI Machine Learning Repository][1].\n\nIt includes three iris species with 50 samples each as well as some properties about each flower. One flower species is linearly separable from the other two, but the other two are not linearly separable from each other.\n\nThe columns in this dataset are:\n\n - Id\n - SepalLengthCm\n - SepalWidthCm\n - PetalLengthCm\n - PetalWidthCm\n - Species\n\n[](https://www.kaggle.com/benhamner/d/uciml/iris/sepal-width-vs-length)\n\n\n [1]: http://archive.ics.uci.edu/ml/", "VersionNotes": "Republishing files so they're formally in our system", "TotalCompressedBytes": 15347.0, "TotalUncompressedBytes": 15347.0}]
|
[{"Id": 19, "CreatorUserId": 1, "OwnerUserId": NaN, "OwnerOrganizationId": 7.0, "CurrentDatasetVersionId": 420.0, "CurrentDatasourceVersionId": 420.0, "ForumId": 997, "Type": 2, "CreationDate": "01/12/2016 00:33:31", "LastActivityDate": "02/06/2018", "TotalViews": 1637863, "TotalDownloads": 423540, "TotalVotes": 3416, "TotalKernels": 6420}]
| null |
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
CSV_PATH = "/kaggle/input/iris/Iris.csv"
ID = "Id"
TARGET = "Species"
TEST_SIZE = 0.3
VAL_SIZE = 0.3
SEED = 2023
# # load Data
total = pd.read_csv(CSV_PATH)
total.head()
TOTAL_LEN = len(total)
TOTAL_LEN
total[TARGET].unique()
total[TARGET].value_counts()
# # devide feature and target
y = total[TARGET]
X = total.drop([TARGET], axis=1)
# # devide trainval set and test set (total = trainval + test)
X_trainval, X_test, y_trainval, y_test = train_test_split(
X, y, test_size=TEST_SIZE, random_state=SEED, stratify=y
)
print("train+val set = ", len(X_trainval), " test set", len(X_test))
X_trainval.head()
y_trainval.value_counts()
X_test.head()
y_test.value_counts()
# # devide train set and val set (trainval = train + val)
X_train, X_val, y_train, y_val = train_test_split(
X_trainval, y_trainval, test_size=VAL_SIZE, random_state=SEED, stratify=y_trainval
)
print("train set = ", len(X_train), " val set", len(X_val))
X_train.head()
y_train.value_counts()
X_val.head()
y_val.value_counts()
| false | 0 | 423 | 0 | 722 | 423 |
||
129040249
|
<jupyter_start><jupyter_text>Video Game Sales
This dataset contains a list of video games with sales greater than 100,000 copies. It was generated by a scrape of [vgchartz.com][1].
Fields include
* Rank - Ranking of overall sales
* Name - The games name
* Platform - Platform of the games release (i.e. PC,PS4, etc.)
* Year - Year of the game's release
* Genre - Genre of the game
* Publisher - Publisher of the game
* NA_Sales - Sales in North America (in millions)
* EU_Sales - Sales in Europe (in millions)
* JP_Sales - Sales in Japan (in millions)
* Other_Sales - Sales in the rest of the world (in millions)
* Global_Sales - Total worldwide sales.
The script to scrape the data is available at https://github.com/GregorUT/vgchartzScrape.
It is based on BeautifulSoup using Python.
There are 16,598 records. 2 records were dropped due to incomplete information.
[1]: http://www.vgchartz.com/
Kaggle dataset identifier: videogamesales
<jupyter_code>import pandas as pd
df = pd.read_csv('videogamesales/vgsales.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 16598 entries, 0 to 16597
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Rank 16598 non-null int64
1 Name 16598 non-null object
2 Platform 16598 non-null object
3 Year 16327 non-null float64
4 Genre 16598 non-null object
5 Publisher 16540 non-null object
6 NA_Sales 16598 non-null float64
7 EU_Sales 16598 non-null float64
8 JP_Sales 16598 non-null float64
9 Other_Sales 16598 non-null float64
10 Global_Sales 16598 non-null float64
dtypes: float64(6), int64(1), object(4)
memory usage: 1.4+ MB
<jupyter_text>Examples:
{
"Rank": 1,
"Name": "Wii Sports",
"Platform": "Wii",
"Year": 2006,
"Genre": "Sports",
"Publisher": "Nintendo",
"NA_Sales": 41.49,
"EU_Sales": 29.02,
"JP_Sales": 3.77,
"Other_Sales": 8.46,
"Global_Sales": 82.74
}
{
"Rank": 2,
"Name": "Super Mario Bros.",
"Platform": "NES",
"Year": 1985,
"Genre": "Platform",
"Publisher": "Nintendo",
"NA_Sales": 29.08,
"EU_Sales": 3.58,
"JP_Sales": 6.8100000000000005,
"Other_Sales": 0.77,
"Global_Sales": 40.24
}
{
"Rank": 3,
"Name": "Mario Kart Wii",
"Platform": "Wii",
"Year": 2008,
"Genre": "Racing",
"Publisher": "Nintendo",
"NA_Sales": 15.85,
"EU_Sales": 12.88,
"JP_Sales": 3.79,
"Other_Sales": 3.31,
"Global_Sales": 35.82
}
{
"Rank": 4,
"Name": "Wii Sports Resort",
"Platform": "Wii",
"Year": 2009,
"Genre": "Sports",
"Publisher": "Nintendo",
"NA_Sales": 15.75,
"EU_Sales": 11.01,
"JP_Sales": 3.2800000000000002,
"Other_Sales": 2.96,
"Global_Sales": 33.0
}
<jupyter_script># # **Introduction**
# Hey there fellow Data Enthusiasts! In this EDA project we are going to do Video Game Sales Analysis using a datset which is taken from Kaggle. We are going to analyze the factors which can be responsible for the better sales of Video Games. We are going to use multiple libraries which will come in handy for analysing and visualizing the dataset.
# **Ready. Set. GOOOO.**
# # **Getting Started**
# **Description about the dataset**
# This dataset contains a list of video games with sales greater than 100,000 copies.
# The dataset consists of the following columns:
# **Rank** - Ranking of overall sales.
# **Name** - The game's name.
# **Platform** - Platform of the games release (i.e. PC,PS4, etc.)
# **Year** - Year of the game's release
# **Genre** - Genre of the game
# **Publisher** - Publisher of the game
# **NA_Sales** - Sales in North America (in millions)
# **EU_Sales** - Sales in Europe (in millions)
# **JP_Sales** - Sales in Japan (in millions)
# **Other_Sales** - Sales in the rest of the world (in millions)
# **Global_Sales** - Total worldwide sales.
# **Installing and importing dependencies**
# We will be using the following libraries:
# **Pandas**: It is a Python library used for working with data sets. It has functions for analyzing, cleaning, exploring, and manipulating data.
# **Numpy**: It is a Python library used for working with arrays. It also has functions for working in domain of linear algebra, fourier transform, and matrices.
# **Matplotlib**: It is a cross-platform, data visualization and graphical plotting library (histograms, scatter plots, bar charts, etc) for Python.
#
import pandas as pd
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
# # **Data Preparation and Cleaning**
data = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv")
data.head(5)
data = pd.read_csv(
"/kaggle/input/videogamesales/vgsales.csv", index_col="Rank"
) # here we are using column Rank as index for this dataset
data.head(5)
# **Let's drop the columns we are not going to use**
data.shape
data.dtypes
# null values
data.isnull().sum()
# Handle duplicates
duplicate_rows_data = data[data.duplicated()]
print("number of duplicate rows: ", duplicate_rows_data.shape)
data.drop_duplicates()
data["Genre"].astype("category")
# # **Exploratory analysis & Visualization Matplotlib**
data.info()
data.describe()
# **Top 5 best selling Games**
data.head(5)
data["Genre"].value_counts()
plt.figure(figsize=(10, 4))
sns.countplot(
x="Genre", data=data, order=data["Genre"].value_counts().index
) # Note here we are using seaborn as sns which is an in-nuilt library in python that uses mtplotlib underneath to plot graphs.
plt.xticks(rotation="vertical")
plt.title("Genre vs. No. of Games released", fontsize=14) # Note labelling the data
plt.ylabel("No. of Games", fontsize=12) # Note labelling the y-label
plt.xlabel("Genre", fontsize=12) # Note labelling the x-label
plt.show()
Action_data = data[data["Genre"] == "Action"]
Action_data
# **Most Popular Game in North America**
JAPAN_data = data.sort_values(by=["JP_Sales", "Genre"], ascending=False)
JAPAN_data = JAPAN_data.reset_index()
JAPAN_data.drop(
["Rank", "Name", "Platform", "Year", "Publisher", "Global_Sales"], axis=1
)
JAPAN_data = JAPAN_data.groupby("Genre").sum()
JAPAN_data
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/040/129040249.ipynb
|
videogamesales
|
gregorut
|
[{"Id": 129040249, "ScriptId": 38354260, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8315101, "CreationDate": "05/10/2023 13:58:11", "VersionNumber": 1.0, "Title": "Video Game Sales Analysis : FULL EDA", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 113.0, "LinesInsertedFromPrevious": 113.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184746059, "KernelVersionId": 129040249, "SourceDatasetVersionId": 618}]
|
[{"Id": 618, "DatasetId": 284, "DatasourceVersionId": 618, "CreatorUserId": 462330, "LicenseName": "Unknown", "CreationDate": "10/26/2016 09:10:49", "VersionNumber": 2.0, "Title": "Video Game Sales", "Slug": "videogamesales", "Subtitle": "Analyze sales data from more than 16,500 games.", "Description": "This dataset contains a list of video games with sales greater than 100,000 copies. It was generated by a scrape of [vgchartz.com][1].\n\nFields include\n\n* Rank - Ranking of overall sales\n\n* Name - The games name\n\n* Platform - Platform of the games release (i.e. PC,PS4, etc.)\n\n* Year - Year of the game's release\n\n* Genre - Genre of the game\n\n* Publisher - Publisher of the game\n\n* NA_Sales - Sales in North America (in millions)\n\n* EU_Sales - Sales in Europe (in millions)\n\n* JP_Sales - Sales in Japan (in millions)\n\n* Other_Sales - Sales in the rest of the world (in millions)\n\n* Global_Sales - Total worldwide sales.\n\nThe script to scrape the data is available at https://github.com/GregorUT/vgchartzScrape.\nIt is based on BeautifulSoup using Python.\nThere are 16,598 records. 2 records were dropped due to incomplete information.\n\n\n [1]: http://www.vgchartz.com/", "VersionNotes": "Cleaned up formating", "TotalCompressedBytes": 1355781.0, "TotalUncompressedBytes": 1355781.0}]
|
[{"Id": 284, "CreatorUserId": 462330, "OwnerUserId": 462330.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 618.0, "CurrentDatasourceVersionId": 618.0, "ForumId": 1788, "Type": 2, "CreationDate": "10/26/2016 08:17:30", "LastActivityDate": "02/06/2018", "TotalViews": 1798828, "TotalDownloads": 471172, "TotalVotes": 5485, "TotalKernels": 1480}]
|
[{"Id": 462330, "UserName": "gregorut", "DisplayName": "GregorySmith", "RegisterDate": "11/09/2015", "PerformanceTier": 1}]
|
# # **Introduction**
# Hey there fellow Data Enthusiasts! In this EDA project we are going to do Video Game Sales Analysis using a datset which is taken from Kaggle. We are going to analyze the factors which can be responsible for the better sales of Video Games. We are going to use multiple libraries which will come in handy for analysing and visualizing the dataset.
# **Ready. Set. GOOOO.**
# # **Getting Started**
# **Description about the dataset**
# This dataset contains a list of video games with sales greater than 100,000 copies.
# The dataset consists of the following columns:
# **Rank** - Ranking of overall sales.
# **Name** - The game's name.
# **Platform** - Platform of the games release (i.e. PC,PS4, etc.)
# **Year** - Year of the game's release
# **Genre** - Genre of the game
# **Publisher** - Publisher of the game
# **NA_Sales** - Sales in North America (in millions)
# **EU_Sales** - Sales in Europe (in millions)
# **JP_Sales** - Sales in Japan (in millions)
# **Other_Sales** - Sales in the rest of the world (in millions)
# **Global_Sales** - Total worldwide sales.
# **Installing and importing dependencies**
# We will be using the following libraries:
# **Pandas**: It is a Python library used for working with data sets. It has functions for analyzing, cleaning, exploring, and manipulating data.
# **Numpy**: It is a Python library used for working with arrays. It also has functions for working in domain of linear algebra, fourier transform, and matrices.
# **Matplotlib**: It is a cross-platform, data visualization and graphical plotting library (histograms, scatter plots, bar charts, etc) for Python.
#
import pandas as pd
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
# # **Data Preparation and Cleaning**
data = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv")
data.head(5)
data = pd.read_csv(
"/kaggle/input/videogamesales/vgsales.csv", index_col="Rank"
) # here we are using column Rank as index for this dataset
data.head(5)
# **Let's drop the columns we are not going to use**
data.shape
data.dtypes
# null values
data.isnull().sum()
# Handle duplicates
duplicate_rows_data = data[data.duplicated()]
print("number of duplicate rows: ", duplicate_rows_data.shape)
data.drop_duplicates()
data["Genre"].astype("category")
# # **Exploratory analysis & Visualization Matplotlib**
data.info()
data.describe()
# **Top 5 best selling Games**
data.head(5)
data["Genre"].value_counts()
plt.figure(figsize=(10, 4))
sns.countplot(
x="Genre", data=data, order=data["Genre"].value_counts().index
) # Note here we are using seaborn as sns which is an in-nuilt library in python that uses mtplotlib underneath to plot graphs.
plt.xticks(rotation="vertical")
plt.title("Genre vs. No. of Games released", fontsize=14) # Note labelling the data
plt.ylabel("No. of Games", fontsize=12) # Note labelling the y-label
plt.xlabel("Genre", fontsize=12) # Note labelling the x-label
plt.show()
Action_data = data[data["Genre"] == "Action"]
Action_data
# **Most Popular Game in North America**
JAPAN_data = data.sort_values(by=["JP_Sales", "Genre"], ascending=False)
JAPAN_data = JAPAN_data.reset_index()
JAPAN_data.drop(
["Rank", "Name", "Platform", "Year", "Publisher", "Global_Sales"], axis=1
)
JAPAN_data = JAPAN_data.groupby("Genre").sum()
JAPAN_data
|
[{"videogamesales/vgsales.csv": {"column_names": "[\"Rank\", \"Name\", \"Platform\", \"Year\", \"Genre\", \"Publisher\", \"NA_Sales\", \"EU_Sales\", \"JP_Sales\", \"Other_Sales\", \"Global_Sales\"]", "column_data_types": "{\"Rank\": \"int64\", \"Name\": \"object\", \"Platform\": \"object\", \"Year\": \"float64\", \"Genre\": \"object\", \"Publisher\": \"object\", \"NA_Sales\": \"float64\", \"EU_Sales\": \"float64\", \"JP_Sales\": \"float64\", \"Other_Sales\": \"float64\", \"Global_Sales\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 16598 entries, 0 to 16597\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Rank 16598 non-null int64 \n 1 Name 16598 non-null object \n 2 Platform 16598 non-null object \n 3 Year 16327 non-null float64\n 4 Genre 16598 non-null object \n 5 Publisher 16540 non-null object \n 6 NA_Sales 16598 non-null float64\n 7 EU_Sales 16598 non-null float64\n 8 JP_Sales 16598 non-null float64\n 9 Other_Sales 16598 non-null float64\n 10 Global_Sales 16598 non-null float64\ndtypes: float64(6), int64(1), object(4)\nmemory usage: 1.4+ MB\n", "summary": "{\"Rank\": {\"count\": 16598.0, \"mean\": 8300.605253645017, \"std\": 4791.853932896403, \"min\": 1.0, \"25%\": 4151.25, \"50%\": 8300.5, \"75%\": 12449.75, \"max\": 16600.0}, \"Year\": {\"count\": 16327.0, \"mean\": 2006.4064433147546, \"std\": 5.828981114712805, \"min\": 1980.0, \"25%\": 2003.0, \"50%\": 2007.0, \"75%\": 2010.0, \"max\": 2020.0}, \"NA_Sales\": {\"count\": 16598.0, \"mean\": 0.26466742981082064, \"std\": 0.8166830292988796, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.08, \"75%\": 0.24, \"max\": 41.49}, \"EU_Sales\": {\"count\": 16598.0, \"mean\": 0.14665200626581515, \"std\": 0.5053512312869116, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.02, \"75%\": 0.11, \"max\": 29.02}, \"JP_Sales\": {\"count\": 16598.0, \"mean\": 0.077781660441017, \"std\": 0.30929064808220297, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.04, \"max\": 10.22}, \"Other_Sales\": {\"count\": 16598.0, \"mean\": 0.0480630196409206, \"std\": 0.18858840291271461, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.01, \"75%\": 0.04, \"max\": 10.57}, \"Global_Sales\": {\"count\": 16598.0, \"mean\": 0.5374406555006628, \"std\": 1.5550279355699124, \"min\": 0.01, \"25%\": 0.06, \"50%\": 0.17, \"75%\": 0.47, \"max\": 82.74}}", "examples": "{\"Rank\":{\"0\":1,\"1\":2,\"2\":3,\"3\":4},\"Name\":{\"0\":\"Wii Sports\",\"1\":\"Super Mario Bros.\",\"2\":\"Mario Kart Wii\",\"3\":\"Wii Sports Resort\"},\"Platform\":{\"0\":\"Wii\",\"1\":\"NES\",\"2\":\"Wii\",\"3\":\"Wii\"},\"Year\":{\"0\":2006.0,\"1\":1985.0,\"2\":2008.0,\"3\":2009.0},\"Genre\":{\"0\":\"Sports\",\"1\":\"Platform\",\"2\":\"Racing\",\"3\":\"Sports\"},\"Publisher\":{\"0\":\"Nintendo\",\"1\":\"Nintendo\",\"2\":\"Nintendo\",\"3\":\"Nintendo\"},\"NA_Sales\":{\"0\":41.49,\"1\":29.08,\"2\":15.85,\"3\":15.75},\"EU_Sales\":{\"0\":29.02,\"1\":3.58,\"2\":12.88,\"3\":11.01},\"JP_Sales\":{\"0\":3.77,\"1\":6.81,\"2\":3.79,\"3\":3.28},\"Other_Sales\":{\"0\":8.46,\"1\":0.77,\"2\":3.31,\"3\":2.96},\"Global_Sales\":{\"0\":82.74,\"1\":40.24,\"2\":35.82,\"3\":33.0}}"}}]
| true | 1 |
<start_data_description><data_path>videogamesales/vgsales.csv:
<column_names>
['Rank', 'Name', 'Platform', 'Year', 'Genre', 'Publisher', 'NA_Sales', 'EU_Sales', 'JP_Sales', 'Other_Sales', 'Global_Sales']
<column_types>
{'Rank': 'int64', 'Name': 'object', 'Platform': 'object', 'Year': 'float64', 'Genre': 'object', 'Publisher': 'object', 'NA_Sales': 'float64', 'EU_Sales': 'float64', 'JP_Sales': 'float64', 'Other_Sales': 'float64', 'Global_Sales': 'float64'}
<dataframe_Summary>
{'Rank': {'count': 16598.0, 'mean': 8300.605253645017, 'std': 4791.853932896403, 'min': 1.0, '25%': 4151.25, '50%': 8300.5, '75%': 12449.75, 'max': 16600.0}, 'Year': {'count': 16327.0, 'mean': 2006.4064433147546, 'std': 5.828981114712805, 'min': 1980.0, '25%': 2003.0, '50%': 2007.0, '75%': 2010.0, 'max': 2020.0}, 'NA_Sales': {'count': 16598.0, 'mean': 0.26466742981082064, 'std': 0.8166830292988796, 'min': 0.0, '25%': 0.0, '50%': 0.08, '75%': 0.24, 'max': 41.49}, 'EU_Sales': {'count': 16598.0, 'mean': 0.14665200626581515, 'std': 0.5053512312869116, 'min': 0.0, '25%': 0.0, '50%': 0.02, '75%': 0.11, 'max': 29.02}, 'JP_Sales': {'count': 16598.0, 'mean': 0.077781660441017, 'std': 0.30929064808220297, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.04, 'max': 10.22}, 'Other_Sales': {'count': 16598.0, 'mean': 0.0480630196409206, 'std': 0.18858840291271461, 'min': 0.0, '25%': 0.0, '50%': 0.01, '75%': 0.04, 'max': 10.57}, 'Global_Sales': {'count': 16598.0, 'mean': 0.5374406555006628, 'std': 1.5550279355699124, 'min': 0.01, '25%': 0.06, '50%': 0.17, '75%': 0.47, 'max': 82.74}}
<dataframe_info>
RangeIndex: 16598 entries, 0 to 16597
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Rank 16598 non-null int64
1 Name 16598 non-null object
2 Platform 16598 non-null object
3 Year 16327 non-null float64
4 Genre 16598 non-null object
5 Publisher 16540 non-null object
6 NA_Sales 16598 non-null float64
7 EU_Sales 16598 non-null float64
8 JP_Sales 16598 non-null float64
9 Other_Sales 16598 non-null float64
10 Global_Sales 16598 non-null float64
dtypes: float64(6), int64(1), object(4)
memory usage: 1.4+ MB
<some_examples>
{'Rank': {'0': 1, '1': 2, '2': 3, '3': 4}, 'Name': {'0': 'Wii Sports', '1': 'Super Mario Bros.', '2': 'Mario Kart Wii', '3': 'Wii Sports Resort'}, 'Platform': {'0': 'Wii', '1': 'NES', '2': 'Wii', '3': 'Wii'}, 'Year': {'0': 2006.0, '1': 1985.0, '2': 2008.0, '3': 2009.0}, 'Genre': {'0': 'Sports', '1': 'Platform', '2': 'Racing', '3': 'Sports'}, 'Publisher': {'0': 'Nintendo', '1': 'Nintendo', '2': 'Nintendo', '3': 'Nintendo'}, 'NA_Sales': {'0': 41.49, '1': 29.08, '2': 15.85, '3': 15.75}, 'EU_Sales': {'0': 29.02, '1': 3.58, '2': 12.88, '3': 11.01}, 'JP_Sales': {'0': 3.77, '1': 6.81, '2': 3.79, '3': 3.28}, 'Other_Sales': {'0': 8.46, '1': 0.77, '2': 3.31, '3': 2.96}, 'Global_Sales': {'0': 82.74, '1': 40.24, '2': 35.82, '3': 33.0}}
<end_description>
| 986 | 0 | 2,099 | 986 |
129040633
|
import numpy as np
from torch.autograd import Variable
from torchvision import datasets, transforms
from skimage import io
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torch.utils.data as data_utils
import torch
import matplotlib.pyplot as plt
import os
import pandas as pd
from skimage.transform import resize
from IPython.display import clear_output
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
def fetch_dataset(
path,
attrs_name="lfw_attributes.txt",
images_name="lfw-deepfunneled",
dx=80,
dy=80,
dimx=64,
dimy=64,
):
# download if not exists
if not os.path.exists(images_name):
print("images not found, donwloading...")
os.system(
"wget http://vis-www.cs.umass.edu/lfw/lfw-deepfunneled.tgz -O tmp.tgz"
)
print("extracting...")
os.system("tar xvzf tmp.tgz && rm tmp.tgz")
print("done")
assert os.path.exists(images_name)
if not os.path.exists(attrs_name):
print("attributes not found, downloading...")
os.system(
"wget http://www.cs.columbia.edu/CAVE/databases/pubfig/download/%s"
% attrs_name
)
print("done")
transform = transforms.Compose(
[
transforms.ToPILImage(),
transforms.CenterCrop((dx, dy)),
transforms.Resize((dimx, dimy)),
transforms.ToTensor(),
]
)
# read attrs
df_attrs = pd.read_csv(
os.path.join(path, attrs_name),
sep="\t",
skiprows=1,
)
df_attrs = pd.DataFrame(df_attrs.iloc[:, :-1].values, columns=df_attrs.columns[1:])
# read photos
photo_ids = []
for dirpath, dirnames, filenames in os.walk(os.path.join(path, images_name)):
for fname in filenames:
if fname.endswith(".jpg"):
fpath = os.path.join(dirpath, fname)
photo_id = fname[:-4].replace("_", " ").split()
person_id = " ".join(photo_id[:-1])
photo_number = int(photo_id[-1])
photo_ids.append(
{"person": person_id, "imagenum": photo_number, "photo_path": fpath}
)
photo_ids = pd.DataFrame(photo_ids)
# print(photo_ids)
# mass-merge
# (photos now have same order as attributes)
df = pd.merge(df_attrs, photo_ids, on=("person", "imagenum"))
assert len(df) == len(df_attrs), "lost some data when merging dataframes"
# print(df.shape)
# image preprocessing
all_photos = df["photo_path"].apply(io.imread).apply(transform)
all_photos = all_photos.values
all_attrs = df.drop(["photo_path", "person", "imagenum"], axis=1)
return all_photos, all_attrs
img_size = 64
path = os.path.abspath("")
data, attrs = fetch_dataset(path=path, dimx=img_size, dimy=img_size)
from sklearn.model_selection import train_test_split
import random
batch_size = 32
train_photos, val_photos, train_attrs, val_attrs = train_test_split(
data, attrs, train_size=0.8, shuffle=False
)
print("Training input shape: ", train_photos.shape)
data_tr = torch.utils.data.DataLoader(train_photos, batch_size=batch_size)
data_val = torch.utils.data.DataLoader(val_photos, batch_size=batch_size)
plt.figure(figsize=(18, 6))
for i in range(12):
plt.subplot(2, 6, i + 1)
plt.axis("off")
plt.imshow(data_tr.dataset[i].permute(1, 2, 0))
plt.show()
dim_code = 32
class CVAE(nn.Module):
def __init__(
self,
base_channel_size: int,
latent_dim: int,
num_classes: int,
num_input_channels: int = 3,
act_fn=nn.ReLU,
):
super().__init__()
self.dummy_param = nn.Parameter(torch.empty(0))
self.latent_dim = latent_dim
self.c_hid = base_channel_size
self.num_classes = num_classes
conv_size = int(np.exp2((np.log2(self.c_hid) - 3)))
ln_size = 2 * self.c_hid * conv_size * conv_size
self.encoder = nn.Sequential(
nn.Conv2d(
num_input_channels, self.c_hid, kernel_size=3, padding=1, stride=2
),
act_fn(),
nn.Conv2d(self.c_hid, self.c_hid, kernel_size=3, padding=1),
act_fn(),
nn.Conv2d(self.c_hid, 2 * self.c_hid, kernel_size=3, padding=1, stride=2),
act_fn(),
nn.Conv2d(2 * self.c_hid, 2 * self.c_hid, kernel_size=3, padding=1),
act_fn(),
nn.Conv2d(
2 * self.c_hid, 2 * self.c_hid, kernel_size=3, padding=1, stride=2
),
act_fn(),
)
self.flatten = nn.Flatten(start_dim=1)
self.linear_mu = nn.Sequential(nn.Linear(ln_size, latent_dim))
self.linear_logvar = nn.Sequential(nn.Linear(ln_size, latent_dim))
self.linear_decoder = nn.Sequential(
nn.Linear(latent_dim + num_classes, ln_size), act_fn()
)
self.unflatten = nn.Sequential(
nn.Unflatten(dim=1, unflattened_size=(2 * self.c_hid, conv_size, conv_size))
)
self.decoder = nn.Sequential(
nn.ConvTranspose2d(
2 * self.c_hid,
2 * self.c_hid,
kernel_size=3,
output_padding=1,
padding=1,
stride=2,
),
act_fn(),
nn.Conv2d(2 * self.c_hid, 2 * self.c_hid, kernel_size=3, padding=1),
act_fn(),
nn.ConvTranspose2d(
2 * self.c_hid,
self.c_hid,
kernel_size=3,
output_padding=1,
padding=1,
stride=2,
),
act_fn(),
nn.Conv2d(self.c_hid, self.c_hid, kernel_size=3, padding=1),
act_fn(),
nn.ConvTranspose2d(
self.c_hid,
num_input_channels,
kernel_size=3,
output_padding=1,
padding=1,
stride=2,
),
nn.Sigmoid(),
)
def encode(self, x):
x = self.encoder(x)
x = self.flatten(x)
mu = self.linear_mu(x)
logvar = self.linear_logvar(x)
return mu, logvar
def reparameterize(self, mu, logvar):
if self.training:
std = torch.exp(logvar / 2)
eps = torch.randn_like(std)
return eps * std + mu
else:
return mu
def decode(self, x):
x = self.linear_decoder(x)
x = self.unflatten(x)
x = self.decoder(x)
return x
def forward(self, x, **kwargs):
y = kwargs["labels"]
y = torch.nn.functional.one_hot(y, num_classes=self.num_classes)
mu, logvar = self.encode(x)
z = self.reparameterize(mu, logvar)
z = torch.cat([z, y], dim=1)
z = self.decode(z)
return mu, logvar, z
def sample(self, labels: list):
y = torch.tensor(labels, dtype=torch.int64).to(self.dummy_param.device)
y = torch.nn.functional.one_hot(y, num_classes=self.num_classes)
z = torch.randn(y.size()[0], 32).to(self.dummy_param.device)
z = torch.cat([z, y], dim=1)
return self.decode(z)
def loss_vae(x, mu, logsigma, reconstruction):
kl = KL_divergence(mu, logsigma)
ll = log_likelihood(x, reconstruction)
# print(f"KL_divergence:{kl} + log_likelihood:{ll}")
return kl + ll
def KL_divergence(mu, logvar):
"""
часть функции потерь, которая отвечает за "близость" латентных представлений разных людей
"""
loss = -0.5 * torch.sum(1 + logvar - mu.pow(2) - logvar.exp())
return loss
def log_likelihood(x, reconstruction):
"""
часть функции потерь, которая отвечает за качество реконструкции (как mse в обычном autoencoder)
"""
loss = nn.BCELoss(reduction="sum")
# loss = nn.MSELoss(reduction='sum')
return loss(reconstruction, x)
def loss_vae(x, mu, logsigma, reconstruction):
kl = KL_divergence(mu, logsigma)
ll = log_likelihood(x, reconstruction)
# print(f"KL_divergence:{kl} + log_likelihood:{ll}")
return kl + ll
batch_size = 32
size = 32
transform = transforms.Compose([transforms.Resize(size), transforms.ToTensor()])
# MNIST Dataset
train_dataset = datasets.MNIST(
root="./mnist_data/", transform=transform, train=True, download=True
)
test_dataset = datasets.MNIST(
root="./mnist_data/", transform=transform, train=False, download=False
)
# Data Loader (Input Pipeline)
train_loader = torch.utils.data.DataLoader(
dataset=train_dataset, batch_size=batch_size, shuffle=True
)
test_loader = torch.utils.data.DataLoader(
dataset=test_dataset, batch_size=batch_size, shuffle=False
)
criterion = loss_vae
autoencoder = CVAE(
num_input_channels=1,
base_channel_size=32,
num_classes=train_dataset.targets.unique().size()[0],
latent_dim=dim_code,
)
optimizer = torch.optim.Adam(autoencoder.parameters(), lr=1e-3)
def train(
model,
opt,
loss_fn,
epochs,
data_tr,
data_val,
scheduler=None,
device="cpu",
show=True,
show_num=3,
):
from time import time
from tqdm.autonotebook import tqdm
model = model.to(device)
X_val, Y_val = next(iter(data_val))
train_losses = []
val_losses = []
log_template = (
"Epoch {ep:03d}/{epochs:03d} train loss: {t_loss:0.4f} val loss {v_loss:0.4f}"
)
with tqdm(desc="epoch", total=epochs) as pbar_outer:
for epoch in range(epochs):
tic = time()
avg_loss = 0
model.train() # train mode
for X_batch, Y_batch in data_tr:
# data to device
X_batch = X_batch.to(device, dtype=torch.float32)
Y_batch = Y_batch.to(device)
# set parameter gradients to zero
opt.zero_grad()
# forward
mu, logvar, X_pred = model(X_batch, labels=Y_batch)
loss = loss_fn(X_batch, mu, logvar, X_pred) # forward-pass
loss.backward() # backward-pass
opt.step() # update weights
# calculate loss to show the user
avg_loss += loss / len(data_tr)
toc = time()
# show intermediate results
model.eval() # testing mode
mu, logvar, X_hat = model(
X_val.to(device, dtype=torch.float32), labels=Y_val.to(device)
)
X_hat = X_hat.detach().to("cpu") # detach and put into cpu
train_losses.append(avg_loss.item())
val_losses.append(loss_fn(X_val, mu, logvar, X_hat).item())
nums = np.random.randint(10, size=show_num)
output_nums = model.sample(nums).detach()
output_nums = output_nums.detach().to("cpu")
if scheduler:
scheduler.step()
pbar_outer.update(1)
# Visualize tools
if show:
clear_output(wait=True)
plt.clf()
for k in range(show_num):
plt.subplot(3, show_num, k + 1)
plt.imshow(X_val[k].permute(1, 2, 0))
plt.title(f"Real {Y_val[k]}")
plt.axis("off")
plt.subplot(3, show_num, k + 1 + show_num)
plt.imshow(X_hat[k].permute(1, 2, 0))
plt.title(f"Output {Y_val[k]}")
plt.axis("off")
plt.subplot(3, show_num, k + 1 + 2 * show_num)
plt.imshow(output_nums[k].permute(1, 2, 0))
plt.title(f"Class {nums[k]}")
plt.axis("off")
plt.suptitle(
log_template.format(
ep=epoch + 1,
epochs=epochs,
t_loss=train_losses[-1],
v_loss=val_losses[-1],
)
)
plt.show()
else:
tqdm.write(
log_template.format(
ep=epoch + 1,
epochs=epochs,
t_loss=train_losses[-1],
v_loss=val_losses[-1],
)
)
return train_losses, val_losses, X_hat, mu, logvar
from torch.optim import lr_scheduler
lr_scheduler = lr_scheduler.StepLR(optimizer, step_size=10, gamma=0.1)
max_epochs = 20
cvae_train_loss, cvae_val_loss, cvae_predict_img_val, cvae_mu, cvae_logvar = train(
model=autoencoder,
opt=optimizer,
loss_fn=criterion,
epochs=max_epochs,
data_tr=train_loader,
data_val=test_loader,
device=device,
scheduler=lr_scheduler,
show=True,
)
nums = range(10)
output_num = autoencoder.sample(nums).cpu().detach()
plt.figure(figsize=(8, len(nums) * 3))
for i, n in enumerate(nums):
plt.subplot(len(nums), 3, i + 1)
plt.imshow(output_num[i].permute(1, 2, 0))
plt.title(f"Output {n}")
plt.axis("off")
plt.show()
if autoencoder.training:
autoencoder.eval()
mu_val = torch.Tensor()
label_val = torch.Tensor()
for batch, label in test_loader:
mu, logvar, X_pred = autoencoder(batch.to(device), labels=label.to(device))
mu_val = torch.cat((mu_val, mu.to("cpu")), 0)
label_val = torch.cat((label_val, label.to("cpu")), dim=0)
from sklearn.manifold import TSNE
mu_val_TSNE = torch.from_numpy(
TSNE(n_components=2, init="pca", learning_rate="auto").fit_transform(
mu_val.detach()
)
)
mu_val_embedded = torch.cat((mu_val_TSNE, label_val.view(-1, 1)), dim=1)
plt.figure(figsize=(10, 10))
scatter = plt.scatter(
mu_val_embedded[:, 0],
mu_val_embedded[:, 1],
c=mu_val_embedded[:, 2],
cmap="gist_rainbow",
)
plt.legend(*scatter.legend_elements())
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/040/129040633.ipynb
| null | null |
[{"Id": 129040633, "ScriptId": 38358341, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6850515, "CreationDate": "05/10/2023 14:01:06", "VersionNumber": 1.0, "Title": "autoencoders_ CVAE", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 412.0, "LinesInsertedFromPrevious": 412.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
| null | null | null | null |
import numpy as np
from torch.autograd import Variable
from torchvision import datasets, transforms
from skimage import io
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torch.utils.data as data_utils
import torch
import matplotlib.pyplot as plt
import os
import pandas as pd
from skimage.transform import resize
from IPython.display import clear_output
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
def fetch_dataset(
path,
attrs_name="lfw_attributes.txt",
images_name="lfw-deepfunneled",
dx=80,
dy=80,
dimx=64,
dimy=64,
):
# download if not exists
if not os.path.exists(images_name):
print("images not found, donwloading...")
os.system(
"wget http://vis-www.cs.umass.edu/lfw/lfw-deepfunneled.tgz -O tmp.tgz"
)
print("extracting...")
os.system("tar xvzf tmp.tgz && rm tmp.tgz")
print("done")
assert os.path.exists(images_name)
if not os.path.exists(attrs_name):
print("attributes not found, downloading...")
os.system(
"wget http://www.cs.columbia.edu/CAVE/databases/pubfig/download/%s"
% attrs_name
)
print("done")
transform = transforms.Compose(
[
transforms.ToPILImage(),
transforms.CenterCrop((dx, dy)),
transforms.Resize((dimx, dimy)),
transforms.ToTensor(),
]
)
# read attrs
df_attrs = pd.read_csv(
os.path.join(path, attrs_name),
sep="\t",
skiprows=1,
)
df_attrs = pd.DataFrame(df_attrs.iloc[:, :-1].values, columns=df_attrs.columns[1:])
# read photos
photo_ids = []
for dirpath, dirnames, filenames in os.walk(os.path.join(path, images_name)):
for fname in filenames:
if fname.endswith(".jpg"):
fpath = os.path.join(dirpath, fname)
photo_id = fname[:-4].replace("_", " ").split()
person_id = " ".join(photo_id[:-1])
photo_number = int(photo_id[-1])
photo_ids.append(
{"person": person_id, "imagenum": photo_number, "photo_path": fpath}
)
photo_ids = pd.DataFrame(photo_ids)
# print(photo_ids)
# mass-merge
# (photos now have same order as attributes)
df = pd.merge(df_attrs, photo_ids, on=("person", "imagenum"))
assert len(df) == len(df_attrs), "lost some data when merging dataframes"
# print(df.shape)
# image preprocessing
all_photos = df["photo_path"].apply(io.imread).apply(transform)
all_photos = all_photos.values
all_attrs = df.drop(["photo_path", "person", "imagenum"], axis=1)
return all_photos, all_attrs
img_size = 64
path = os.path.abspath("")
data, attrs = fetch_dataset(path=path, dimx=img_size, dimy=img_size)
from sklearn.model_selection import train_test_split
import random
batch_size = 32
train_photos, val_photos, train_attrs, val_attrs = train_test_split(
data, attrs, train_size=0.8, shuffle=False
)
print("Training input shape: ", train_photos.shape)
data_tr = torch.utils.data.DataLoader(train_photos, batch_size=batch_size)
data_val = torch.utils.data.DataLoader(val_photos, batch_size=batch_size)
plt.figure(figsize=(18, 6))
for i in range(12):
plt.subplot(2, 6, i + 1)
plt.axis("off")
plt.imshow(data_tr.dataset[i].permute(1, 2, 0))
plt.show()
dim_code = 32
class CVAE(nn.Module):
def __init__(
self,
base_channel_size: int,
latent_dim: int,
num_classes: int,
num_input_channels: int = 3,
act_fn=nn.ReLU,
):
super().__init__()
self.dummy_param = nn.Parameter(torch.empty(0))
self.latent_dim = latent_dim
self.c_hid = base_channel_size
self.num_classes = num_classes
conv_size = int(np.exp2((np.log2(self.c_hid) - 3)))
ln_size = 2 * self.c_hid * conv_size * conv_size
self.encoder = nn.Sequential(
nn.Conv2d(
num_input_channels, self.c_hid, kernel_size=3, padding=1, stride=2
),
act_fn(),
nn.Conv2d(self.c_hid, self.c_hid, kernel_size=3, padding=1),
act_fn(),
nn.Conv2d(self.c_hid, 2 * self.c_hid, kernel_size=3, padding=1, stride=2),
act_fn(),
nn.Conv2d(2 * self.c_hid, 2 * self.c_hid, kernel_size=3, padding=1),
act_fn(),
nn.Conv2d(
2 * self.c_hid, 2 * self.c_hid, kernel_size=3, padding=1, stride=2
),
act_fn(),
)
self.flatten = nn.Flatten(start_dim=1)
self.linear_mu = nn.Sequential(nn.Linear(ln_size, latent_dim))
self.linear_logvar = nn.Sequential(nn.Linear(ln_size, latent_dim))
self.linear_decoder = nn.Sequential(
nn.Linear(latent_dim + num_classes, ln_size), act_fn()
)
self.unflatten = nn.Sequential(
nn.Unflatten(dim=1, unflattened_size=(2 * self.c_hid, conv_size, conv_size))
)
self.decoder = nn.Sequential(
nn.ConvTranspose2d(
2 * self.c_hid,
2 * self.c_hid,
kernel_size=3,
output_padding=1,
padding=1,
stride=2,
),
act_fn(),
nn.Conv2d(2 * self.c_hid, 2 * self.c_hid, kernel_size=3, padding=1),
act_fn(),
nn.ConvTranspose2d(
2 * self.c_hid,
self.c_hid,
kernel_size=3,
output_padding=1,
padding=1,
stride=2,
),
act_fn(),
nn.Conv2d(self.c_hid, self.c_hid, kernel_size=3, padding=1),
act_fn(),
nn.ConvTranspose2d(
self.c_hid,
num_input_channels,
kernel_size=3,
output_padding=1,
padding=1,
stride=2,
),
nn.Sigmoid(),
)
def encode(self, x):
x = self.encoder(x)
x = self.flatten(x)
mu = self.linear_mu(x)
logvar = self.linear_logvar(x)
return mu, logvar
def reparameterize(self, mu, logvar):
if self.training:
std = torch.exp(logvar / 2)
eps = torch.randn_like(std)
return eps * std + mu
else:
return mu
def decode(self, x):
x = self.linear_decoder(x)
x = self.unflatten(x)
x = self.decoder(x)
return x
def forward(self, x, **kwargs):
y = kwargs["labels"]
y = torch.nn.functional.one_hot(y, num_classes=self.num_classes)
mu, logvar = self.encode(x)
z = self.reparameterize(mu, logvar)
z = torch.cat([z, y], dim=1)
z = self.decode(z)
return mu, logvar, z
def sample(self, labels: list):
y = torch.tensor(labels, dtype=torch.int64).to(self.dummy_param.device)
y = torch.nn.functional.one_hot(y, num_classes=self.num_classes)
z = torch.randn(y.size()[0], 32).to(self.dummy_param.device)
z = torch.cat([z, y], dim=1)
return self.decode(z)
def loss_vae(x, mu, logsigma, reconstruction):
kl = KL_divergence(mu, logsigma)
ll = log_likelihood(x, reconstruction)
# print(f"KL_divergence:{kl} + log_likelihood:{ll}")
return kl + ll
def KL_divergence(mu, logvar):
"""
часть функции потерь, которая отвечает за "близость" латентных представлений разных людей
"""
loss = -0.5 * torch.sum(1 + logvar - mu.pow(2) - logvar.exp())
return loss
def log_likelihood(x, reconstruction):
"""
часть функции потерь, которая отвечает за качество реконструкции (как mse в обычном autoencoder)
"""
loss = nn.BCELoss(reduction="sum")
# loss = nn.MSELoss(reduction='sum')
return loss(reconstruction, x)
def loss_vae(x, mu, logsigma, reconstruction):
kl = KL_divergence(mu, logsigma)
ll = log_likelihood(x, reconstruction)
# print(f"KL_divergence:{kl} + log_likelihood:{ll}")
return kl + ll
batch_size = 32
size = 32
transform = transforms.Compose([transforms.Resize(size), transforms.ToTensor()])
# MNIST Dataset
train_dataset = datasets.MNIST(
root="./mnist_data/", transform=transform, train=True, download=True
)
test_dataset = datasets.MNIST(
root="./mnist_data/", transform=transform, train=False, download=False
)
# Data Loader (Input Pipeline)
train_loader = torch.utils.data.DataLoader(
dataset=train_dataset, batch_size=batch_size, shuffle=True
)
test_loader = torch.utils.data.DataLoader(
dataset=test_dataset, batch_size=batch_size, shuffle=False
)
criterion = loss_vae
autoencoder = CVAE(
num_input_channels=1,
base_channel_size=32,
num_classes=train_dataset.targets.unique().size()[0],
latent_dim=dim_code,
)
optimizer = torch.optim.Adam(autoencoder.parameters(), lr=1e-3)
def train(
model,
opt,
loss_fn,
epochs,
data_tr,
data_val,
scheduler=None,
device="cpu",
show=True,
show_num=3,
):
from time import time
from tqdm.autonotebook import tqdm
model = model.to(device)
X_val, Y_val = next(iter(data_val))
train_losses = []
val_losses = []
log_template = (
"Epoch {ep:03d}/{epochs:03d} train loss: {t_loss:0.4f} val loss {v_loss:0.4f}"
)
with tqdm(desc="epoch", total=epochs) as pbar_outer:
for epoch in range(epochs):
tic = time()
avg_loss = 0
model.train() # train mode
for X_batch, Y_batch in data_tr:
# data to device
X_batch = X_batch.to(device, dtype=torch.float32)
Y_batch = Y_batch.to(device)
# set parameter gradients to zero
opt.zero_grad()
# forward
mu, logvar, X_pred = model(X_batch, labels=Y_batch)
loss = loss_fn(X_batch, mu, logvar, X_pred) # forward-pass
loss.backward() # backward-pass
opt.step() # update weights
# calculate loss to show the user
avg_loss += loss / len(data_tr)
toc = time()
# show intermediate results
model.eval() # testing mode
mu, logvar, X_hat = model(
X_val.to(device, dtype=torch.float32), labels=Y_val.to(device)
)
X_hat = X_hat.detach().to("cpu") # detach and put into cpu
train_losses.append(avg_loss.item())
val_losses.append(loss_fn(X_val, mu, logvar, X_hat).item())
nums = np.random.randint(10, size=show_num)
output_nums = model.sample(nums).detach()
output_nums = output_nums.detach().to("cpu")
if scheduler:
scheduler.step()
pbar_outer.update(1)
# Visualize tools
if show:
clear_output(wait=True)
plt.clf()
for k in range(show_num):
plt.subplot(3, show_num, k + 1)
plt.imshow(X_val[k].permute(1, 2, 0))
plt.title(f"Real {Y_val[k]}")
plt.axis("off")
plt.subplot(3, show_num, k + 1 + show_num)
plt.imshow(X_hat[k].permute(1, 2, 0))
plt.title(f"Output {Y_val[k]}")
plt.axis("off")
plt.subplot(3, show_num, k + 1 + 2 * show_num)
plt.imshow(output_nums[k].permute(1, 2, 0))
plt.title(f"Class {nums[k]}")
plt.axis("off")
plt.suptitle(
log_template.format(
ep=epoch + 1,
epochs=epochs,
t_loss=train_losses[-1],
v_loss=val_losses[-1],
)
)
plt.show()
else:
tqdm.write(
log_template.format(
ep=epoch + 1,
epochs=epochs,
t_loss=train_losses[-1],
v_loss=val_losses[-1],
)
)
return train_losses, val_losses, X_hat, mu, logvar
from torch.optim import lr_scheduler
lr_scheduler = lr_scheduler.StepLR(optimizer, step_size=10, gamma=0.1)
max_epochs = 20
cvae_train_loss, cvae_val_loss, cvae_predict_img_val, cvae_mu, cvae_logvar = train(
model=autoencoder,
opt=optimizer,
loss_fn=criterion,
epochs=max_epochs,
data_tr=train_loader,
data_val=test_loader,
device=device,
scheduler=lr_scheduler,
show=True,
)
nums = range(10)
output_num = autoencoder.sample(nums).cpu().detach()
plt.figure(figsize=(8, len(nums) * 3))
for i, n in enumerate(nums):
plt.subplot(len(nums), 3, i + 1)
plt.imshow(output_num[i].permute(1, 2, 0))
plt.title(f"Output {n}")
plt.axis("off")
plt.show()
if autoencoder.training:
autoencoder.eval()
mu_val = torch.Tensor()
label_val = torch.Tensor()
for batch, label in test_loader:
mu, logvar, X_pred = autoencoder(batch.to(device), labels=label.to(device))
mu_val = torch.cat((mu_val, mu.to("cpu")), 0)
label_val = torch.cat((label_val, label.to("cpu")), dim=0)
from sklearn.manifold import TSNE
mu_val_TSNE = torch.from_numpy(
TSNE(n_components=2, init="pca", learning_rate="auto").fit_transform(
mu_val.detach()
)
)
mu_val_embedded = torch.cat((mu_val_TSNE, label_val.view(-1, 1)), dim=1)
plt.figure(figsize=(10, 10))
scatter = plt.scatter(
mu_val_embedded[:, 0],
mu_val_embedded[:, 1],
c=mu_val_embedded[:, 2],
cmap="gist_rainbow",
)
plt.legend(*scatter.legend_elements())
plt.show()
| false | 0 | 4,183 | 2 | 4,183 | 4,183 |
||
129037105
|
<jupyter_start><jupyter_text>Credit Card Lead Prediction
Credit Card Lead Prediction
Happy Customer Bank is a mid-sized private bank that deals in all kinds of banking products, like Savings accounts, Current accounts, investment products, credit products, among other offerings.
The bank also cross-sells products to its existing customers and to do so they use different kinds of communication like telecasting, e-mails, recommendations on net banking, mobile banking, etc.
In this case, the Happy Customer Bank wants to cross-sell its credit cards to its existing customers. The bank has identified a set of customers that are eligible for taking these credit cards.
Now, the bank is looking for your help in identifying customers that could show higher intent towards a recommended credit card, given:
This dataset was part of May 2021 Jobathon conducted my analytics vidhya, for more info check:https://datahack.analyticsvidhya.com/contest/job-a-thon-2/
Kaggle dataset identifier: credit-card-buyers
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## CREDIT CARD LEAD GENERATION PREDICTION
# ### About The Dataset
# * In this project or case, the bank wants to cross-sell its credit cards to its existing customers.the bank has identified a set of customers that are eligible for taking these credit cards
# * This dataset is taking from "KAGGLE"
# ### Problem Statement:
# * TO predict which individuals or groups of people are most likely to apply for a credit card in "NEAR FUTURE"
# ### Project Aim:
# * The aim of this prediction is to identify potential customers who are more likeye to be intersted in credit card.
# * In this project i used "FOUR" machine learning algorithm and check which algorithm perform well on this data.
# * The algorithms are.
# * 1.Logestic Regression
# * 2.Decision Tree
# * 3.Random Forest
# * 4.Ada boost Classifer
# ### Importing Necessary Libraries:
import pandas as pd # it's is core library for data manipulation and data analysis
import numpy as np # it's is core library for numeric and scientific computing
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# ### Loading The Dataset
df = pd.read_csv("/kaggle/input/credit-card-buyers/train data credit card.csv")
df.head()
# ### Lets check what the columns mentioned in the dataset says
# * ID=Unique identifier for a row
# * Gender = Gender of the Customer
# * Age = Age of the customer (in Years)
# * Region_code = code of the Region for the Customers
# * Occuption = Occuption Type for the customer
# * Channel_code = Acquisition Channel code for Customer
# * Vintage = Vintage for thr customer (In Months)
# * Credit_Product = if the customer has any active Credit product(home loan,Personal loan,Credit card etc.)
# * Avg Account Balance = Averge account balance for the Customer in last 12 Months
# * Is_Active = if the Customer is active in last 3 Months
# * Is_Lead(Target) = if the Customer is interested for the Credit Card
# ### Checking Dataset
# checking the size of the dataset
df.size
# *
# The size of Dataset are "2457250"
#
# checking shape of dadaset
df.shape
# *
# In this Dataset we have "245725" Rows and "10" Columns
# checking the info of the dataset
df.info()
# info() function provide a summary of the DataFrame including its column names,datatypes and number of null value in each column
# * This Dataset contained four(4) numerical colunms and six(6) are catgorical colunms.
# * And also see the "Credit_Product" column contained null values.
#
df.describe()
# describe () function provide a summary of statistical information such as mean,standard deviation,minimum,maximum and quartile value(25%,50%,75%)
# ### Data Preprocessing
# drop ID columes coz its onlye a Unique identifier for a row so it's nots much importants.
df.drop("ID", axis=1, inplace=True)
# checking the unique value of the dataset
df.nunique()
# * the unique value contained bye each colume are very small so we consider to taking this value for calculation
# checking the null or missing value of the dataset
df.isnull().sum()
# * in my data set the "Credit_Product" column contained the missing value to it's nesessary tred with them.
# #### Replacing The Null Value
# The "Credit_Product" colume is in catgorical in nature so i used mode to replace this missing value
df["Credit_Product"].fillna(df["Credit_Product"].mode()[0], inplace=True)
df.isnull().sum()
# checking the duplicate value of the dataset
df.duplicated().sum()
# #### Removing The Duplicate Values
df.drop_duplicates(inplace=True)
df.duplicated().sum()
# ### Data Visualization
# #### 1. Look For Imbalanced Data Set
# Checking value counts for 'IS_Lead' column.
df["Is_Lead"].value_counts()
plt.figure(figsize=(5, 5)) # Plotting a graph
df["Is_Lead"].value_counts().plot(
kind="pie", autopct="%1.1f%%"
) # autopct display the percent value using string formatting
plt.title("Is_Lead", size=20)
# #### Insight:
# * if we take 100 people than form this 100 peoples only 23.7% people are our customer or possible to used credit card.
#
# pair plot are used to shows relatinship between multiple varible
sns.pairplot(df)
# #### insight:
# * 1.we can see as age increase the 'vintage' and "avg_account_balance' also get increase.
# * 2.as age increase the avg account balance also get increase.
# * 3.lead value is same for age,vintage and avg account balance
# ### Checking outliers
# i used box plot to check outilers present in data.
plt.figure(figsize=(30, 10))
sns.boxplot(df["Avg_Account_Balance"])
# ### Insight:
# 1. As we can see there are outliers we can't remove them because they are considrable outliers..
# ### 2. Check whether data is normally distributed
# * plot histrograms for each numerical varible
col = ["Age", "Vintage", "Avg_Account_Balance"]
df[col].hist(bins=50, figsize=(20, 15))
plt.show()
# #### insights:
# * 1.we can seen in plots the data is not normally distrubuted, in all the plots the data is right skewed(postive skewed)
# #### Convert data into normally distrbute
# i used log transformer to normalize the data coz data is right skewed
col_to_transform = ["Age", "Vintage", "Avg_Account_Balance"]
df[col_to_transform] = df[col_to_transform].apply(lambda x: np.log(x))
# visualize the transformed data using histograms
fig, axes = plt.subplots(nrows=1, ncols=len(col_to_transform), figsize=(15, 5))
for i, col in enumerate(col_to_transform):
axes[i].hist(df[col])
axes[i].set_xlabel(f"log({col})")
axes[i].set_ylabel("Frequency")
plt.show()
# * * we can see the data is normallye distrubuted
# #### Graph shows relation between gender and is_active
plt.figure(figsize=(10, 8))
sns.countplot(x="Gender", hue="Is_Active", data=df)
# #### Insight:
# * 1. In both male and female customer the rate of inactive customer is higher as compared to active customer.
# * 2. the rate of active male customer is higher as compared to female customer.
# #### Graph shows relation between gender and Credit_product
plt.figure(figsize=(10, 8))
sns.countplot(x="Gender", hue="Credit_Product", data=df)
# #### Insight:
# * 1. In both male and female customer the rate of customer having credit_product on there credit card is higher as compared to customer not having credit_product.
# * 2. male customer having_credit product is higher as compared to female having credit product.
# #### Graph shows relation between accupation and Avg_account_Balance
plt.figure(figsize=(10, 8))
sns.barplot(data=df, x="Occupation", y="Avg_Account_Balance", ci=True)
# #### Insight:
# * 1. in the given chart the entrepreneur are having the highest number of account balance
# * 2. and also show thoes are customer works as employe are having lower number of account balance
# ### Encoding the columns
# Encoding tecnique are used to convert catgorical data into numerical,coz many algoeithm require numerical input in order to function properly
# i used one_hot encoding to perform encoding operation.
cat_col = [
"Gender",
"Occupation",
"Channel_Code",
"Credit_Product",
"Is_Active",
"Region_Code",
]
print(df[cat_col].nunique())
# i used 'get_dummies()'function
encoded_df = pd.get_dummies(df, columns=cat_col)
encoded_df
# replacing encoded data in original variable
df = encoded_df
df
# ### Checking the shape of data
df.shape
# * Now we can se that after encoding the value of number of columns is increase from (11 to 55 ) this improve dimensionality
# ### Check Correlation
correlations = df.corr()["Is_Lead"].sort_values(ascending=False)
print(correlations)
# #### from correlation we see that (occupation_salaried,Credit_product_no and Channel_code_x1) have a high negative correlation with output column so we remove this cloumns
# ### Drop the columns
df.drop("Occupation_Salaried", axis=1, inplace=True)
df.drop("Credit_Product_No", axis=1, inplace=True)
df.drop("Channel_Code_X1", axis=1, inplace=True)
df.shape
plt.figure(figsize=(40, 30))
sns.heatmap(df.corr(), annot=True)
# ### Model Building
# ##### Define The Input And Target Varible Using Train Test Split
# input varible
x = df.drop(columns=["Is_Lead"])
# output or target varible
y = df["Is_Lead"]
# Import Segregating data from scikit learn
from sklearn.model_selection import train_test_split
# split data into training and testing
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.3, random_state=100
)
x_train.shape
x_test.shape
# ### Standardization of data
# importing StandardScaler from scikit lerarn
from sklearn.preprocessing import StandardScaler, MinMaxScaler
scaler = StandardScaler()
x_train = scaler.fit_transform(x_train)
x_test = scaler.transform(x_test)
x_train
# ### 1.Logestic Regression
# Importing Logestic Regression model from scikit learn
from sklearn.linear_model import LogisticRegression, LinearRegression
logr = LogisticRegression()
logr.fit(x_train, y_train)
y_pred_logr = logr.predict(x_test)
# importing performance matrix from scikit learn
from sklearn.metrics import r2_score, accuracy_score, recall_score
print(
accuracy_score(y_test, y_pred_logr)
) # taking accuracy score for calculation coz problem statement more focus on type 1 error
print(recall_score(y_test, y_pred_logr)) # ('Flase postive')
# #### Accuracy of logestic regression is 0.778 ...(77.8%)
# used differnt tecnique to improve model performance...
# 1.Regulariation tecnique..
# i used l1 regularization
log = LogisticRegression(penalty="l1", solver="liblinear")
log.fit(x_train, y_train)
score = log.score(x_test, y_test)
print("accuracy:{:.2f}%".format(score * 100))
# * After regularization we see that we got same accuracy
# 2.Cross-Validation..
from sklearn.model_selection import cross_val_score
# 1st create logistic regression object
log = LogisticRegression(max_iter=1000)
# data devided into 5 fold k-5
scores = cross_val_score(log, x, y, cv=5)
print("Cross-validation scores:", scores)
print("mean accuracy: {:.2f}%".format(scores.mean() * 100))
# 3.GridSearcheCV tecnique..
# importing GridsearchCV from skilt learn
from sklearn.model_selection import GridSearchCV
logr.get_params()
# slecting the parameter
params = {"max_iter": [20, 100], "C": [0.05, 1.0], "penalty": ["l2"]}
logetun = GridSearchCV(logr, params, cv=20)
logetun.fit(x_train, y_train)
# slecting best parameters
logetun.best_params_
logetun.best_score_
print("accuracy:{:.2f}%".format(logetun.best_score_ * 100))
# #### final accuracy for logestic regression is =78.16
# ## 2.Decision Tree
# Importing DecisionTreeClassifier model from scikit learn
from sklearn.tree import DecisionTreeClassifier, plot_tree
# instantiate the DecisionTreeClassifier model with criterion entropy
dtc = DecisionTreeClassifier(criterion="entropy", max_depth=6, random_state=0)
# fit the model
dtc.fit(x_train, y_train)
y_pred_dtcm = dtc.predict(x_test)
# #### Check accuracy score with criterion entropy..
# * 1st check accuracy score with entropy criterion
from sklearn.metrics import accuracy_score
print(
"Model accuracy score with criterion entropy: {0:0.4f}".format(
accuracy_score(y_test, y_pred_dtcm)
)
)
# #### Check accuracy score with criterion gini index..
# * 2nd used gini index criterion
dtc = DecisionTreeClassifier(criterion="gini", max_depth=6, random_state=0)
dtc.fit(x_train, y_train)
y_pred_gini = dtc.predict(x_test)
from sklearn.metrics import accuracy_score
print(
"Model accuracy score with criterion gini index: {0:0.4f}".format(
accuracy_score(y_test, y_pred_gini)
)
)
# ##### we get same accuracy for both the cases...
# ### Visualize decision-trees
plt.figure(figsize=(12, 8))
from sklearn import tree
tree.plot_tree(dtc.fit(x_train, y_train))
# #### Used differnt tecnique to improve model performance...
# #### 1.Gridsearchcv()
dtc.get_params()
# define parameter
params = {
"max_depth": [10, 20],
"max_leaf_nodes": [250, 270],
"min_samples_split": [5, 8],
}
dtct = GridSearchCV(dtc, params, cv=10)
dtct.fit(x_train, y_train)
# selecting the best parameter
dtct.best_params_
# training accuracy
dtct.score(x_train, y_train)
# testing accuracy
dtct.score(x_test, y_test)
# ##### We can see that the training-set score and test-set score is same as above. The training-set accuracy score is 0.7959 while the test-set accuracy to be 0.790. These two values are quite comparable. So, there is no sign of overfitting...
# #### Used Confusion Metrics
# confusion matrix are used to evalute the model perforamce
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred_dtcm)
print("Confusion matrix\n\n", cm)
# visualise useing seaborn
import seaborn as sns
sns.heatmap(cm, annot=True, fmt="d")
# #### from heatmap we see that our model predicted well
# ##### Classification Report
# * Classification report is another way to evaluate the classification model performance. It displays the precision, recall, f1 and support scores for the model. I have described these terms in later.
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred_dtcm))
# #### Final accuracy for logestic regression is =79.50
# ## 3.Random forest...
# Importing Random Forest model from scikit learn
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
# instantiate the DecisionTreeClassifier model with criterion entropy
rfc = RandomForestClassifier()
# fit the model
rfc.fit(x_train, y_train)
# make a prediction
y_pred_rfc = rfc.predict(x_test)
accuracy_score(y_test, y_pred_rfc)
# ##### For the Random forest we get accuracy_score '0.77532'
# ### Using gridsearchcv() to improve model performance
# Gets the parameters
rfc.get_params()
# selecting parameters.
params = {
"max_depth": [2, 5, 10, 30],
"min_samples_split": [5, 10, 30],
"min_impurity_decrease": [1.0, 2.0],
"n_estimators": [20, 40],
"random_state": [34, 40],
}
rfct = GridSearchCV(rfc, params, cv=5)
rfct.fit(x_train, y_train)
# slecting the best paramters.
rfct.best_params_
rfct.best_score_
# ##### As we can see that accuracy decreases after the gridsearchcv so we will consider our normal accuracy
# ## 4. Ada boost classifier
# Importing Ada Boost Classifier model from scikit learn
from sklearn.ensemble import AdaBoostClassifier
ada = AdaBoostClassifier()
# fit the model
ada.fit(x_train, y_train)
# make a prediction
y_pred_ada = ada.predict(x_test)
# accuracy score
ada.score(x_test, y_test)
# ##### For the Ada boost classifier we get accuracy_score '0.77961'
# #### Using gridsearchcv() to improve model performance
# getting all parameters
ada.get_params()
# slecting the parameter
params = {"learning_rate": [1.0], "n_estimators": [200], "random_state": [10]}
adat = GridSearchCV(ada, params, cv=10)
adat.fit(x_train, y_train)
# best parameters
adat.best_params_
# final score
adat.best_score_
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/037/129037105.ipynb
|
credit-card-buyers
|
shelvigarg
|
[{"Id": 129037105, "ScriptId": 37253183, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12428991, "CreationDate": "05/10/2023 13:34:47", "VersionNumber": 1.0, "Title": "Machine Learning Project", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 633.0, "LinesInsertedFromPrevious": 633.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 7}]
|
[{"Id": 184740475, "KernelVersionId": 129037105, "SourceDatasetVersionId": 2616327}]
|
[{"Id": 2616327, "DatasetId": 1590268, "DatasourceVersionId": 2659985, "CreatorUserId": 5450951, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "09/15/2021 04:14:07", "VersionNumber": 1.0, "Title": "Credit Card Lead Prediction", "Slug": "credit-card-buyers", "Subtitle": "Predict whether a bank account holder is Potential Credit Card buyer or not!", "Description": "Credit Card Lead Prediction\n\nHappy Customer Bank is a mid-sized private bank that deals in all kinds of banking products, like Savings accounts, Current accounts, investment products, credit products, among other offerings.\n\nThe bank also cross-sells products to its existing customers and to do so they use different kinds of communication like telecasting, e-mails, recommendations on net banking, mobile banking, etc.\n\nIn this case, the Happy Customer Bank wants to cross-sell its credit cards to its existing customers. The bank has identified a set of customers that are eligible for taking these credit cards.\n\nNow, the bank is looking for your help in identifying customers that could show higher intent towards a recommended credit card, given:\n\nThis dataset was part of May 2021 Jobathon conducted my analytics vidhya, for more info check:https://datahack.analyticsvidhya.com/contest/job-a-thon-2/", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1590268, "CreatorUserId": 5450951, "OwnerUserId": 5450951.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2616327.0, "CurrentDatasourceVersionId": 2659985.0, "ForumId": 1610452, "Type": 2, "CreationDate": "09/15/2021 04:14:07", "LastActivityDate": "09/15/2021", "TotalViews": 11340, "TotalDownloads": 1374, "TotalVotes": 23, "TotalKernels": 8}]
|
[{"Id": 5450951, "UserName": "shelvigarg", "DisplayName": "Shelvi Garg", "RegisterDate": "07/11/2020", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## CREDIT CARD LEAD GENERATION PREDICTION
# ### About The Dataset
# * In this project or case, the bank wants to cross-sell its credit cards to its existing customers.the bank has identified a set of customers that are eligible for taking these credit cards
# * This dataset is taking from "KAGGLE"
# ### Problem Statement:
# * TO predict which individuals or groups of people are most likely to apply for a credit card in "NEAR FUTURE"
# ### Project Aim:
# * The aim of this prediction is to identify potential customers who are more likeye to be intersted in credit card.
# * In this project i used "FOUR" machine learning algorithm and check which algorithm perform well on this data.
# * The algorithms are.
# * 1.Logestic Regression
# * 2.Decision Tree
# * 3.Random Forest
# * 4.Ada boost Classifer
# ### Importing Necessary Libraries:
import pandas as pd # it's is core library for data manipulation and data analysis
import numpy as np # it's is core library for numeric and scientific computing
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# ### Loading The Dataset
df = pd.read_csv("/kaggle/input/credit-card-buyers/train data credit card.csv")
df.head()
# ### Lets check what the columns mentioned in the dataset says
# * ID=Unique identifier for a row
# * Gender = Gender of the Customer
# * Age = Age of the customer (in Years)
# * Region_code = code of the Region for the Customers
# * Occuption = Occuption Type for the customer
# * Channel_code = Acquisition Channel code for Customer
# * Vintage = Vintage for thr customer (In Months)
# * Credit_Product = if the customer has any active Credit product(home loan,Personal loan,Credit card etc.)
# * Avg Account Balance = Averge account balance for the Customer in last 12 Months
# * Is_Active = if the Customer is active in last 3 Months
# * Is_Lead(Target) = if the Customer is interested for the Credit Card
# ### Checking Dataset
# checking the size of the dataset
df.size
# *
# The size of Dataset are "2457250"
#
# checking shape of dadaset
df.shape
# *
# In this Dataset we have "245725" Rows and "10" Columns
# checking the info of the dataset
df.info()
# info() function provide a summary of the DataFrame including its column names,datatypes and number of null value in each column
# * This Dataset contained four(4) numerical colunms and six(6) are catgorical colunms.
# * And also see the "Credit_Product" column contained null values.
#
df.describe()
# describe () function provide a summary of statistical information such as mean,standard deviation,minimum,maximum and quartile value(25%,50%,75%)
# ### Data Preprocessing
# drop ID columes coz its onlye a Unique identifier for a row so it's nots much importants.
df.drop("ID", axis=1, inplace=True)
# checking the unique value of the dataset
df.nunique()
# * the unique value contained bye each colume are very small so we consider to taking this value for calculation
# checking the null or missing value of the dataset
df.isnull().sum()
# * in my data set the "Credit_Product" column contained the missing value to it's nesessary tred with them.
# #### Replacing The Null Value
# The "Credit_Product" colume is in catgorical in nature so i used mode to replace this missing value
df["Credit_Product"].fillna(df["Credit_Product"].mode()[0], inplace=True)
df.isnull().sum()
# checking the duplicate value of the dataset
df.duplicated().sum()
# #### Removing The Duplicate Values
df.drop_duplicates(inplace=True)
df.duplicated().sum()
# ### Data Visualization
# #### 1. Look For Imbalanced Data Set
# Checking value counts for 'IS_Lead' column.
df["Is_Lead"].value_counts()
plt.figure(figsize=(5, 5)) # Plotting a graph
df["Is_Lead"].value_counts().plot(
kind="pie", autopct="%1.1f%%"
) # autopct display the percent value using string formatting
plt.title("Is_Lead", size=20)
# #### Insight:
# * if we take 100 people than form this 100 peoples only 23.7% people are our customer or possible to used credit card.
#
# pair plot are used to shows relatinship between multiple varible
sns.pairplot(df)
# #### insight:
# * 1.we can see as age increase the 'vintage' and "avg_account_balance' also get increase.
# * 2.as age increase the avg account balance also get increase.
# * 3.lead value is same for age,vintage and avg account balance
# ### Checking outliers
# i used box plot to check outilers present in data.
plt.figure(figsize=(30, 10))
sns.boxplot(df["Avg_Account_Balance"])
# ### Insight:
# 1. As we can see there are outliers we can't remove them because they are considrable outliers..
# ### 2. Check whether data is normally distributed
# * plot histrograms for each numerical varible
col = ["Age", "Vintage", "Avg_Account_Balance"]
df[col].hist(bins=50, figsize=(20, 15))
plt.show()
# #### insights:
# * 1.we can seen in plots the data is not normally distrubuted, in all the plots the data is right skewed(postive skewed)
# #### Convert data into normally distrbute
# i used log transformer to normalize the data coz data is right skewed
col_to_transform = ["Age", "Vintage", "Avg_Account_Balance"]
df[col_to_transform] = df[col_to_transform].apply(lambda x: np.log(x))
# visualize the transformed data using histograms
fig, axes = plt.subplots(nrows=1, ncols=len(col_to_transform), figsize=(15, 5))
for i, col in enumerate(col_to_transform):
axes[i].hist(df[col])
axes[i].set_xlabel(f"log({col})")
axes[i].set_ylabel("Frequency")
plt.show()
# * * we can see the data is normallye distrubuted
# #### Graph shows relation between gender and is_active
plt.figure(figsize=(10, 8))
sns.countplot(x="Gender", hue="Is_Active", data=df)
# #### Insight:
# * 1. In both male and female customer the rate of inactive customer is higher as compared to active customer.
# * 2. the rate of active male customer is higher as compared to female customer.
# #### Graph shows relation between gender and Credit_product
plt.figure(figsize=(10, 8))
sns.countplot(x="Gender", hue="Credit_Product", data=df)
# #### Insight:
# * 1. In both male and female customer the rate of customer having credit_product on there credit card is higher as compared to customer not having credit_product.
# * 2. male customer having_credit product is higher as compared to female having credit product.
# #### Graph shows relation between accupation and Avg_account_Balance
plt.figure(figsize=(10, 8))
sns.barplot(data=df, x="Occupation", y="Avg_Account_Balance", ci=True)
# #### Insight:
# * 1. in the given chart the entrepreneur are having the highest number of account balance
# * 2. and also show thoes are customer works as employe are having lower number of account balance
# ### Encoding the columns
# Encoding tecnique are used to convert catgorical data into numerical,coz many algoeithm require numerical input in order to function properly
# i used one_hot encoding to perform encoding operation.
cat_col = [
"Gender",
"Occupation",
"Channel_Code",
"Credit_Product",
"Is_Active",
"Region_Code",
]
print(df[cat_col].nunique())
# i used 'get_dummies()'function
encoded_df = pd.get_dummies(df, columns=cat_col)
encoded_df
# replacing encoded data in original variable
df = encoded_df
df
# ### Checking the shape of data
df.shape
# * Now we can se that after encoding the value of number of columns is increase from (11 to 55 ) this improve dimensionality
# ### Check Correlation
correlations = df.corr()["Is_Lead"].sort_values(ascending=False)
print(correlations)
# #### from correlation we see that (occupation_salaried,Credit_product_no and Channel_code_x1) have a high negative correlation with output column so we remove this cloumns
# ### Drop the columns
df.drop("Occupation_Salaried", axis=1, inplace=True)
df.drop("Credit_Product_No", axis=1, inplace=True)
df.drop("Channel_Code_X1", axis=1, inplace=True)
df.shape
plt.figure(figsize=(40, 30))
sns.heatmap(df.corr(), annot=True)
# ### Model Building
# ##### Define The Input And Target Varible Using Train Test Split
# input varible
x = df.drop(columns=["Is_Lead"])
# output or target varible
y = df["Is_Lead"]
# Import Segregating data from scikit learn
from sklearn.model_selection import train_test_split
# split data into training and testing
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.3, random_state=100
)
x_train.shape
x_test.shape
# ### Standardization of data
# importing StandardScaler from scikit lerarn
from sklearn.preprocessing import StandardScaler, MinMaxScaler
scaler = StandardScaler()
x_train = scaler.fit_transform(x_train)
x_test = scaler.transform(x_test)
x_train
# ### 1.Logestic Regression
# Importing Logestic Regression model from scikit learn
from sklearn.linear_model import LogisticRegression, LinearRegression
logr = LogisticRegression()
logr.fit(x_train, y_train)
y_pred_logr = logr.predict(x_test)
# importing performance matrix from scikit learn
from sklearn.metrics import r2_score, accuracy_score, recall_score
print(
accuracy_score(y_test, y_pred_logr)
) # taking accuracy score for calculation coz problem statement more focus on type 1 error
print(recall_score(y_test, y_pred_logr)) # ('Flase postive')
# #### Accuracy of logestic regression is 0.778 ...(77.8%)
# used differnt tecnique to improve model performance...
# 1.Regulariation tecnique..
# i used l1 regularization
log = LogisticRegression(penalty="l1", solver="liblinear")
log.fit(x_train, y_train)
score = log.score(x_test, y_test)
print("accuracy:{:.2f}%".format(score * 100))
# * After regularization we see that we got same accuracy
# 2.Cross-Validation..
from sklearn.model_selection import cross_val_score
# 1st create logistic regression object
log = LogisticRegression(max_iter=1000)
# data devided into 5 fold k-5
scores = cross_val_score(log, x, y, cv=5)
print("Cross-validation scores:", scores)
print("mean accuracy: {:.2f}%".format(scores.mean() * 100))
# 3.GridSearcheCV tecnique..
# importing GridsearchCV from skilt learn
from sklearn.model_selection import GridSearchCV
logr.get_params()
# slecting the parameter
params = {"max_iter": [20, 100], "C": [0.05, 1.0], "penalty": ["l2"]}
logetun = GridSearchCV(logr, params, cv=20)
logetun.fit(x_train, y_train)
# slecting best parameters
logetun.best_params_
logetun.best_score_
print("accuracy:{:.2f}%".format(logetun.best_score_ * 100))
# #### final accuracy for logestic regression is =78.16
# ## 2.Decision Tree
# Importing DecisionTreeClassifier model from scikit learn
from sklearn.tree import DecisionTreeClassifier, plot_tree
# instantiate the DecisionTreeClassifier model with criterion entropy
dtc = DecisionTreeClassifier(criterion="entropy", max_depth=6, random_state=0)
# fit the model
dtc.fit(x_train, y_train)
y_pred_dtcm = dtc.predict(x_test)
# #### Check accuracy score with criterion entropy..
# * 1st check accuracy score with entropy criterion
from sklearn.metrics import accuracy_score
print(
"Model accuracy score with criterion entropy: {0:0.4f}".format(
accuracy_score(y_test, y_pred_dtcm)
)
)
# #### Check accuracy score with criterion gini index..
# * 2nd used gini index criterion
dtc = DecisionTreeClassifier(criterion="gini", max_depth=6, random_state=0)
dtc.fit(x_train, y_train)
y_pred_gini = dtc.predict(x_test)
from sklearn.metrics import accuracy_score
print(
"Model accuracy score with criterion gini index: {0:0.4f}".format(
accuracy_score(y_test, y_pred_gini)
)
)
# ##### we get same accuracy for both the cases...
# ### Visualize decision-trees
plt.figure(figsize=(12, 8))
from sklearn import tree
tree.plot_tree(dtc.fit(x_train, y_train))
# #### Used differnt tecnique to improve model performance...
# #### 1.Gridsearchcv()
dtc.get_params()
# define parameter
params = {
"max_depth": [10, 20],
"max_leaf_nodes": [250, 270],
"min_samples_split": [5, 8],
}
dtct = GridSearchCV(dtc, params, cv=10)
dtct.fit(x_train, y_train)
# selecting the best parameter
dtct.best_params_
# training accuracy
dtct.score(x_train, y_train)
# testing accuracy
dtct.score(x_test, y_test)
# ##### We can see that the training-set score and test-set score is same as above. The training-set accuracy score is 0.7959 while the test-set accuracy to be 0.790. These two values are quite comparable. So, there is no sign of overfitting...
# #### Used Confusion Metrics
# confusion matrix are used to evalute the model perforamce
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred_dtcm)
print("Confusion matrix\n\n", cm)
# visualise useing seaborn
import seaborn as sns
sns.heatmap(cm, annot=True, fmt="d")
# #### from heatmap we see that our model predicted well
# ##### Classification Report
# * Classification report is another way to evaluate the classification model performance. It displays the precision, recall, f1 and support scores for the model. I have described these terms in later.
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred_dtcm))
# #### Final accuracy for logestic regression is =79.50
# ## 3.Random forest...
# Importing Random Forest model from scikit learn
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
# instantiate the DecisionTreeClassifier model with criterion entropy
rfc = RandomForestClassifier()
# fit the model
rfc.fit(x_train, y_train)
# make a prediction
y_pred_rfc = rfc.predict(x_test)
accuracy_score(y_test, y_pred_rfc)
# ##### For the Random forest we get accuracy_score '0.77532'
# ### Using gridsearchcv() to improve model performance
# Gets the parameters
rfc.get_params()
# selecting parameters.
params = {
"max_depth": [2, 5, 10, 30],
"min_samples_split": [5, 10, 30],
"min_impurity_decrease": [1.0, 2.0],
"n_estimators": [20, 40],
"random_state": [34, 40],
}
rfct = GridSearchCV(rfc, params, cv=5)
rfct.fit(x_train, y_train)
# slecting the best paramters.
rfct.best_params_
rfct.best_score_
# ##### As we can see that accuracy decreases after the gridsearchcv so we will consider our normal accuracy
# ## 4. Ada boost classifier
# Importing Ada Boost Classifier model from scikit learn
from sklearn.ensemble import AdaBoostClassifier
ada = AdaBoostClassifier()
# fit the model
ada.fit(x_train, y_train)
# make a prediction
y_pred_ada = ada.predict(x_test)
# accuracy score
ada.score(x_test, y_test)
# ##### For the Ada boost classifier we get accuracy_score '0.77961'
# #### Using gridsearchcv() to improve model performance
# getting all parameters
ada.get_params()
# slecting the parameter
params = {"learning_rate": [1.0], "n_estimators": [200], "random_state": [10]}
adat = GridSearchCV(ada, params, cv=10)
adat.fit(x_train, y_train)
# best parameters
adat.best_params_
# final score
adat.best_score_
| false | 1 | 4,690 | 7 | 4,925 | 4,690 |
||
129037699
|
<jupyter_start><jupyter_text>Video Game Sales
This dataset contains a list of video games with sales greater than 100,000 copies. It was generated by a scrape of [vgchartz.com][1].
Fields include
* Rank - Ranking of overall sales
* Name - The games name
* Platform - Platform of the games release (i.e. PC,PS4, etc.)
* Year - Year of the game's release
* Genre - Genre of the game
* Publisher - Publisher of the game
* NA_Sales - Sales in North America (in millions)
* EU_Sales - Sales in Europe (in millions)
* JP_Sales - Sales in Japan (in millions)
* Other_Sales - Sales in the rest of the world (in millions)
* Global_Sales - Total worldwide sales.
The script to scrape the data is available at https://github.com/GregorUT/vgchartzScrape.
It is based on BeautifulSoup using Python.
There are 16,598 records. 2 records were dropped due to incomplete information.
[1]: http://www.vgchartz.com/
Kaggle dataset identifier: videogamesales
<jupyter_code>import pandas as pd
df = pd.read_csv('videogamesales/vgsales.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 16598 entries, 0 to 16597
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Rank 16598 non-null int64
1 Name 16598 non-null object
2 Platform 16598 non-null object
3 Year 16327 non-null float64
4 Genre 16598 non-null object
5 Publisher 16540 non-null object
6 NA_Sales 16598 non-null float64
7 EU_Sales 16598 non-null float64
8 JP_Sales 16598 non-null float64
9 Other_Sales 16598 non-null float64
10 Global_Sales 16598 non-null float64
dtypes: float64(6), int64(1), object(4)
memory usage: 1.4+ MB
<jupyter_text>Examples:
{
"Rank": 1,
"Name": "Wii Sports",
"Platform": "Wii",
"Year": 2006,
"Genre": "Sports",
"Publisher": "Nintendo",
"NA_Sales": 41.49,
"EU_Sales": 29.02,
"JP_Sales": 3.77,
"Other_Sales": 8.46,
"Global_Sales": 82.74
}
{
"Rank": 2,
"Name": "Super Mario Bros.",
"Platform": "NES",
"Year": 1985,
"Genre": "Platform",
"Publisher": "Nintendo",
"NA_Sales": 29.08,
"EU_Sales": 3.58,
"JP_Sales": 6.8100000000000005,
"Other_Sales": 0.77,
"Global_Sales": 40.24
}
{
"Rank": 3,
"Name": "Mario Kart Wii",
"Platform": "Wii",
"Year": 2008,
"Genre": "Racing",
"Publisher": "Nintendo",
"NA_Sales": 15.85,
"EU_Sales": 12.88,
"JP_Sales": 3.79,
"Other_Sales": 3.31,
"Global_Sales": 35.82
}
{
"Rank": 4,
"Name": "Wii Sports Resort",
"Platform": "Wii",
"Year": 2009,
"Genre": "Sports",
"Publisher": "Nintendo",
"NA_Sales": 15.75,
"EU_Sales": 11.01,
"JP_Sales": 3.2800000000000002,
"Other_Sales": 2.96,
"Global_Sales": 33.0
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Data Analysis with Pandas - Tala Asfan - 09.05.2023
df = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv")
df
# ### Which company is the most common video game publisher?
#
publisher = df["Publisher"].value_counts().idxmax()
publisher
# ### What’s the most common platform?
#
platform = df["Platform"].value_counts().idxmax()
platform
# ### What about the most common genre?
#
genre = df["Genre"].value_counts().idxmax()
genre
# ### What are the top 20 highest grossing games?
#
dfsorted = df.sort_values("Global_Sales", ascending=False)
top20 = dfsorted.head(20)
top20
# ### For North American video game sales, what’s the median?
# - Provide a secondary output showing ten games surrounding the median sales output.
# - Assume that games with same median value are sorted in descending order.
NAsorted = df.sort_values("NA_Sales", ascending=False)
NA_median = NAsorted["NA_Sales"].median()
surrounding_games = NAsorted.loc[
(NAsorted["NA_Sales"] >= NA_median) & (NAsorted["NA_Sales"] <= NA_median)
].head(10)
print(NA_median)
surrounding_games
# ### For the top-selling game of all time, how many standard deviations above/below the mean are its sales for North America?
# - Z = (X - µ)/σ
# - where µ denotes the mean,
# - σ denotes the standard deviation,
# - X is the value of standard deviation away from the mean.
# - ⇒ X = µ ± Zσ
df_sorted = df.sort_values("Global_Sales", ascending=False)
na_sales_top_game = df_sorted["NA_Sales"].iloc[0]
mean_na_sales = df_sorted["NA_Sales"].mean()
std_na_sales = df_sorted["NA_Sales"].std()
num_std = (na_sales_top_game - mean_na_sales) / std_na_sales
print(na_sales_top_game)
print(num_std)
# ### The Nintendo Wii seems to have outdone itself with games. How does its average number of sales compare with all of the other platforms?
grouped_by_platform = df.groupby("Platform")["Global_Sales"].mean().reset_index()
wii_data = grouped_by_platform.loc[grouped_by_platform["Platform"] == "Wii"]
mean_except_wii = grouped_by_platform.loc[grouped_by_platform["Platform"] != "Wii"][
"Global_Sales"
].mean()
if wii_data["Global_Sales"].values > mean_except_wii:
print(
"The average number of sales for the Nintendo Wii is higher than the other platforms."
)
else:
print(
"The average number of sales for the Nintendo Wii is not higher than the other platforms."
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/037/129037699.ipynb
|
videogamesales
|
gregorut
|
[{"Id": 129037699, "ScriptId": 38325180, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14997001, "CreationDate": "05/10/2023 13:39:14", "VersionNumber": 2.0, "Title": "vg-stats", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 102.0, "LinesInsertedFromPrevious": 32.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 70.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184741434, "KernelVersionId": 129037699, "SourceDatasetVersionId": 618}]
|
[{"Id": 618, "DatasetId": 284, "DatasourceVersionId": 618, "CreatorUserId": 462330, "LicenseName": "Unknown", "CreationDate": "10/26/2016 09:10:49", "VersionNumber": 2.0, "Title": "Video Game Sales", "Slug": "videogamesales", "Subtitle": "Analyze sales data from more than 16,500 games.", "Description": "This dataset contains a list of video games with sales greater than 100,000 copies. It was generated by a scrape of [vgchartz.com][1].\n\nFields include\n\n* Rank - Ranking of overall sales\n\n* Name - The games name\n\n* Platform - Platform of the games release (i.e. PC,PS4, etc.)\n\n* Year - Year of the game's release\n\n* Genre - Genre of the game\n\n* Publisher - Publisher of the game\n\n* NA_Sales - Sales in North America (in millions)\n\n* EU_Sales - Sales in Europe (in millions)\n\n* JP_Sales - Sales in Japan (in millions)\n\n* Other_Sales - Sales in the rest of the world (in millions)\n\n* Global_Sales - Total worldwide sales.\n\nThe script to scrape the data is available at https://github.com/GregorUT/vgchartzScrape.\nIt is based on BeautifulSoup using Python.\nThere are 16,598 records. 2 records were dropped due to incomplete information.\n\n\n [1]: http://www.vgchartz.com/", "VersionNotes": "Cleaned up formating", "TotalCompressedBytes": 1355781.0, "TotalUncompressedBytes": 1355781.0}]
|
[{"Id": 284, "CreatorUserId": 462330, "OwnerUserId": 462330.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 618.0, "CurrentDatasourceVersionId": 618.0, "ForumId": 1788, "Type": 2, "CreationDate": "10/26/2016 08:17:30", "LastActivityDate": "02/06/2018", "TotalViews": 1798828, "TotalDownloads": 471172, "TotalVotes": 5485, "TotalKernels": 1480}]
|
[{"Id": 462330, "UserName": "gregorut", "DisplayName": "GregorySmith", "RegisterDate": "11/09/2015", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Data Analysis with Pandas - Tala Asfan - 09.05.2023
df = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv")
df
# ### Which company is the most common video game publisher?
#
publisher = df["Publisher"].value_counts().idxmax()
publisher
# ### What’s the most common platform?
#
platform = df["Platform"].value_counts().idxmax()
platform
# ### What about the most common genre?
#
genre = df["Genre"].value_counts().idxmax()
genre
# ### What are the top 20 highest grossing games?
#
dfsorted = df.sort_values("Global_Sales", ascending=False)
top20 = dfsorted.head(20)
top20
# ### For North American video game sales, what’s the median?
# - Provide a secondary output showing ten games surrounding the median sales output.
# - Assume that games with same median value are sorted in descending order.
NAsorted = df.sort_values("NA_Sales", ascending=False)
NA_median = NAsorted["NA_Sales"].median()
surrounding_games = NAsorted.loc[
(NAsorted["NA_Sales"] >= NA_median) & (NAsorted["NA_Sales"] <= NA_median)
].head(10)
print(NA_median)
surrounding_games
# ### For the top-selling game of all time, how many standard deviations above/below the mean are its sales for North America?
# - Z = (X - µ)/σ
# - where µ denotes the mean,
# - σ denotes the standard deviation,
# - X is the value of standard deviation away from the mean.
# - ⇒ X = µ ± Zσ
df_sorted = df.sort_values("Global_Sales", ascending=False)
na_sales_top_game = df_sorted["NA_Sales"].iloc[0]
mean_na_sales = df_sorted["NA_Sales"].mean()
std_na_sales = df_sorted["NA_Sales"].std()
num_std = (na_sales_top_game - mean_na_sales) / std_na_sales
print(na_sales_top_game)
print(num_std)
# ### The Nintendo Wii seems to have outdone itself with games. How does its average number of sales compare with all of the other platforms?
grouped_by_platform = df.groupby("Platform")["Global_Sales"].mean().reset_index()
wii_data = grouped_by_platform.loc[grouped_by_platform["Platform"] == "Wii"]
mean_except_wii = grouped_by_platform.loc[grouped_by_platform["Platform"] != "Wii"][
"Global_Sales"
].mean()
if wii_data["Global_Sales"].values > mean_except_wii:
print(
"The average number of sales for the Nintendo Wii is higher than the other platforms."
)
else:
print(
"The average number of sales for the Nintendo Wii is not higher than the other platforms."
)
|
[{"videogamesales/vgsales.csv": {"column_names": "[\"Rank\", \"Name\", \"Platform\", \"Year\", \"Genre\", \"Publisher\", \"NA_Sales\", \"EU_Sales\", \"JP_Sales\", \"Other_Sales\", \"Global_Sales\"]", "column_data_types": "{\"Rank\": \"int64\", \"Name\": \"object\", \"Platform\": \"object\", \"Year\": \"float64\", \"Genre\": \"object\", \"Publisher\": \"object\", \"NA_Sales\": \"float64\", \"EU_Sales\": \"float64\", \"JP_Sales\": \"float64\", \"Other_Sales\": \"float64\", \"Global_Sales\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 16598 entries, 0 to 16597\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Rank 16598 non-null int64 \n 1 Name 16598 non-null object \n 2 Platform 16598 non-null object \n 3 Year 16327 non-null float64\n 4 Genre 16598 non-null object \n 5 Publisher 16540 non-null object \n 6 NA_Sales 16598 non-null float64\n 7 EU_Sales 16598 non-null float64\n 8 JP_Sales 16598 non-null float64\n 9 Other_Sales 16598 non-null float64\n 10 Global_Sales 16598 non-null float64\ndtypes: float64(6), int64(1), object(4)\nmemory usage: 1.4+ MB\n", "summary": "{\"Rank\": {\"count\": 16598.0, \"mean\": 8300.605253645017, \"std\": 4791.853932896403, \"min\": 1.0, \"25%\": 4151.25, \"50%\": 8300.5, \"75%\": 12449.75, \"max\": 16600.0}, \"Year\": {\"count\": 16327.0, \"mean\": 2006.4064433147546, \"std\": 5.828981114712805, \"min\": 1980.0, \"25%\": 2003.0, \"50%\": 2007.0, \"75%\": 2010.0, \"max\": 2020.0}, \"NA_Sales\": {\"count\": 16598.0, \"mean\": 0.26466742981082064, \"std\": 0.8166830292988796, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.08, \"75%\": 0.24, \"max\": 41.49}, \"EU_Sales\": {\"count\": 16598.0, \"mean\": 0.14665200626581515, \"std\": 0.5053512312869116, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.02, \"75%\": 0.11, \"max\": 29.02}, \"JP_Sales\": {\"count\": 16598.0, \"mean\": 0.077781660441017, \"std\": 0.30929064808220297, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.04, \"max\": 10.22}, \"Other_Sales\": {\"count\": 16598.0, \"mean\": 0.0480630196409206, \"std\": 0.18858840291271461, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.01, \"75%\": 0.04, \"max\": 10.57}, \"Global_Sales\": {\"count\": 16598.0, \"mean\": 0.5374406555006628, \"std\": 1.5550279355699124, \"min\": 0.01, \"25%\": 0.06, \"50%\": 0.17, \"75%\": 0.47, \"max\": 82.74}}", "examples": "{\"Rank\":{\"0\":1,\"1\":2,\"2\":3,\"3\":4},\"Name\":{\"0\":\"Wii Sports\",\"1\":\"Super Mario Bros.\",\"2\":\"Mario Kart Wii\",\"3\":\"Wii Sports Resort\"},\"Platform\":{\"0\":\"Wii\",\"1\":\"NES\",\"2\":\"Wii\",\"3\":\"Wii\"},\"Year\":{\"0\":2006.0,\"1\":1985.0,\"2\":2008.0,\"3\":2009.0},\"Genre\":{\"0\":\"Sports\",\"1\":\"Platform\",\"2\":\"Racing\",\"3\":\"Sports\"},\"Publisher\":{\"0\":\"Nintendo\",\"1\":\"Nintendo\",\"2\":\"Nintendo\",\"3\":\"Nintendo\"},\"NA_Sales\":{\"0\":41.49,\"1\":29.08,\"2\":15.85,\"3\":15.75},\"EU_Sales\":{\"0\":29.02,\"1\":3.58,\"2\":12.88,\"3\":11.01},\"JP_Sales\":{\"0\":3.77,\"1\":6.81,\"2\":3.79,\"3\":3.28},\"Other_Sales\":{\"0\":8.46,\"1\":0.77,\"2\":3.31,\"3\":2.96},\"Global_Sales\":{\"0\":82.74,\"1\":40.24,\"2\":35.82,\"3\":33.0}}"}}]
| true | 1 |
<start_data_description><data_path>videogamesales/vgsales.csv:
<column_names>
['Rank', 'Name', 'Platform', 'Year', 'Genre', 'Publisher', 'NA_Sales', 'EU_Sales', 'JP_Sales', 'Other_Sales', 'Global_Sales']
<column_types>
{'Rank': 'int64', 'Name': 'object', 'Platform': 'object', 'Year': 'float64', 'Genre': 'object', 'Publisher': 'object', 'NA_Sales': 'float64', 'EU_Sales': 'float64', 'JP_Sales': 'float64', 'Other_Sales': 'float64', 'Global_Sales': 'float64'}
<dataframe_Summary>
{'Rank': {'count': 16598.0, 'mean': 8300.605253645017, 'std': 4791.853932896403, 'min': 1.0, '25%': 4151.25, '50%': 8300.5, '75%': 12449.75, 'max': 16600.0}, 'Year': {'count': 16327.0, 'mean': 2006.4064433147546, 'std': 5.828981114712805, 'min': 1980.0, '25%': 2003.0, '50%': 2007.0, '75%': 2010.0, 'max': 2020.0}, 'NA_Sales': {'count': 16598.0, 'mean': 0.26466742981082064, 'std': 0.8166830292988796, 'min': 0.0, '25%': 0.0, '50%': 0.08, '75%': 0.24, 'max': 41.49}, 'EU_Sales': {'count': 16598.0, 'mean': 0.14665200626581515, 'std': 0.5053512312869116, 'min': 0.0, '25%': 0.0, '50%': 0.02, '75%': 0.11, 'max': 29.02}, 'JP_Sales': {'count': 16598.0, 'mean': 0.077781660441017, 'std': 0.30929064808220297, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.04, 'max': 10.22}, 'Other_Sales': {'count': 16598.0, 'mean': 0.0480630196409206, 'std': 0.18858840291271461, 'min': 0.0, '25%': 0.0, '50%': 0.01, '75%': 0.04, 'max': 10.57}, 'Global_Sales': {'count': 16598.0, 'mean': 0.5374406555006628, 'std': 1.5550279355699124, 'min': 0.01, '25%': 0.06, '50%': 0.17, '75%': 0.47, 'max': 82.74}}
<dataframe_info>
RangeIndex: 16598 entries, 0 to 16597
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Rank 16598 non-null int64
1 Name 16598 non-null object
2 Platform 16598 non-null object
3 Year 16327 non-null float64
4 Genre 16598 non-null object
5 Publisher 16540 non-null object
6 NA_Sales 16598 non-null float64
7 EU_Sales 16598 non-null float64
8 JP_Sales 16598 non-null float64
9 Other_Sales 16598 non-null float64
10 Global_Sales 16598 non-null float64
dtypes: float64(6), int64(1), object(4)
memory usage: 1.4+ MB
<some_examples>
{'Rank': {'0': 1, '1': 2, '2': 3, '3': 4}, 'Name': {'0': 'Wii Sports', '1': 'Super Mario Bros.', '2': 'Mario Kart Wii', '3': 'Wii Sports Resort'}, 'Platform': {'0': 'Wii', '1': 'NES', '2': 'Wii', '3': 'Wii'}, 'Year': {'0': 2006.0, '1': 1985.0, '2': 2008.0, '3': 2009.0}, 'Genre': {'0': 'Sports', '1': 'Platform', '2': 'Racing', '3': 'Sports'}, 'Publisher': {'0': 'Nintendo', '1': 'Nintendo', '2': 'Nintendo', '3': 'Nintendo'}, 'NA_Sales': {'0': 41.49, '1': 29.08, '2': 15.85, '3': 15.75}, 'EU_Sales': {'0': 29.02, '1': 3.58, '2': 12.88, '3': 11.01}, 'JP_Sales': {'0': 3.77, '1': 6.81, '2': 3.79, '3': 3.28}, 'Other_Sales': {'0': 8.46, '1': 0.77, '2': 3.31, '3': 2.96}, 'Global_Sales': {'0': 82.74, '1': 40.24, '2': 35.82, '3': 33.0}}
<end_description>
| 931 | 0 | 2,045 | 931 |
129547925
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
def add_date_features(df):
df["year"] = df.date.dt.year
df["month"] = df.date.dt.month
df["day_of_week"] = df.date.dt.dayofweek + 1
return df
def add_one_hot(df, category_cols):
encoder = OneHotEncoder(handle_unknown="ignore", sparse=False)
OH_cols = pd.DataFrame(encoder.fit_transform(df[category_cols]))
OH_cols.index = df.index
num_cols = df.drop(category_cols, axis=1)
OH_X = pd.concat([OH_cols, num_cols], axis=1)
return OH_X
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder
train = pd.read_csv("/kaggle/input/store-sales-time-series-forecasting/train.csv")
train_y = train.sales
train["date"] = pd.to_datetime(train.date)
train.drop(["sales"], axis=1, inplace=True)
category = train.dtypes == "object"
category_cols = list(category[category].index)
train = add_date_features(train)
train = add_one_hot(train, category_cols)
train = train.drop("date", axis=1)
train = train.drop("id", axis=1)
X_train, X_valid, y_train, y_valid = train_test_split(
train, train_y, train_size=0.8, test_size=0.2, random_state=0
)
from xgboost import XGBRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.ensemble import RandomForestRegressor
my_model_1 = XGBRegressor(random_state=0) # Your code here
my_model_1.fit(X_train, y_train)
predictions_1 = my_model_1.predict(X_valid)
mae_1 = mean_absolute_error(predictions_1, y_valid)
print("Mean Absolute Error XGB:", mae_1)
my_model_2 = RandomForestRegressor(max_leaf_nodes=100, random_state=1)
X_train_str = X_train
X_valid_str = X_valid
X_train_str.columns = X_train_str.columns.astype(str)
X_valid_str.columns = X_valid_str.columns.astype(str)
my_model_2.fit(X_train_str, y_train)
predictions_2 = my_model_2.predict(X_valid_str)
mae_2 = mean_absolute_error(predictions_2, y_valid)
print("Mean Absolute Error FR:", mae_2)
test = pd.read_csv("/kaggle/input/store-sales-time-series-forecasting/test.csv")
test["date"] = pd.to_datetime(test.date)
test.head(10)
test = add_date_features(test)
test = add_one_hot(test, category_cols)
test = test.drop("date", axis=1)
test_ids = test["id"]
test = test.drop("id", axis=1)
predictions_test = my_model_1.predict(test)
output = pd.DataFrame({"id": test_ids, "sales": predictions_test})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/547/129547925.ipynb
| null | null |
[{"Id": 129547925, "ScriptId": 38520990, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4733467, "CreationDate": "05/14/2023 18:11:31", "VersionNumber": 1.0, "Title": "APP basic example", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 85.0, "LinesInsertedFromPrevious": 85.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 4}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
def add_date_features(df):
df["year"] = df.date.dt.year
df["month"] = df.date.dt.month
df["day_of_week"] = df.date.dt.dayofweek + 1
return df
def add_one_hot(df, category_cols):
encoder = OneHotEncoder(handle_unknown="ignore", sparse=False)
OH_cols = pd.DataFrame(encoder.fit_transform(df[category_cols]))
OH_cols.index = df.index
num_cols = df.drop(category_cols, axis=1)
OH_X = pd.concat([OH_cols, num_cols], axis=1)
return OH_X
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder
train = pd.read_csv("/kaggle/input/store-sales-time-series-forecasting/train.csv")
train_y = train.sales
train["date"] = pd.to_datetime(train.date)
train.drop(["sales"], axis=1, inplace=True)
category = train.dtypes == "object"
category_cols = list(category[category].index)
train = add_date_features(train)
train = add_one_hot(train, category_cols)
train = train.drop("date", axis=1)
train = train.drop("id", axis=1)
X_train, X_valid, y_train, y_valid = train_test_split(
train, train_y, train_size=0.8, test_size=0.2, random_state=0
)
from xgboost import XGBRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.ensemble import RandomForestRegressor
my_model_1 = XGBRegressor(random_state=0) # Your code here
my_model_1.fit(X_train, y_train)
predictions_1 = my_model_1.predict(X_valid)
mae_1 = mean_absolute_error(predictions_1, y_valid)
print("Mean Absolute Error XGB:", mae_1)
my_model_2 = RandomForestRegressor(max_leaf_nodes=100, random_state=1)
X_train_str = X_train
X_valid_str = X_valid
X_train_str.columns = X_train_str.columns.astype(str)
X_valid_str.columns = X_valid_str.columns.astype(str)
my_model_2.fit(X_train_str, y_train)
predictions_2 = my_model_2.predict(X_valid_str)
mae_2 = mean_absolute_error(predictions_2, y_valid)
print("Mean Absolute Error FR:", mae_2)
test = pd.read_csv("/kaggle/input/store-sales-time-series-forecasting/test.csv")
test["date"] = pd.to_datetime(test.date)
test.head(10)
test = add_date_features(test)
test = add_one_hot(test, category_cols)
test = test.drop("date", axis=1)
test_ids = test["id"]
test = test.drop("id", axis=1)
predictions_test = my_model_1.predict(test)
output = pd.DataFrame({"id": test_ids, "sales": predictions_test})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
| false | 0 | 1,025 | 4 | 1,025 | 1,025 |
||
129547756
|
<jupyter_start><jupyter_text>12000 Data Science Jobs in India - Naukri.com
## About
**Naukri.com** is an Indian employment website operating in India and Middle East. It was founded in March 1997. Naukri was ranked No.1 by 9 independent sources, placing it way ahead of competition.Google Trends names Naukri “the most preferred job search destination in India”.
This dataset contains data of first 12,000 results obtained for '**data scientist**' job role in '**India**' from Naukri.com, on **May 18, 2022**
**Starter Notebook** : https://www.kaggle.com/code/anandhuh/data-science-jobs-in-india-eda-naukri-com
## Attribute Information
1. **Job_Role** - Job Roles
2. **Company** - Name of the Company
3. **Location** - Location of the company
4. **Job Experience** - Required Job Experience (Min. Experience-Max. Experience)
5. **Skills/Description** - Required Skills for the Job
## Source
Link : **https://www.naukri.com/**
## Other Updated Datasets
https://www.kaggle.com/anandhuh/datasets
Please appreciate the effort with an **upvote** 👍
### Thank You
Kaggle dataset identifier: data-science-jobs-in-india
<jupyter_code>import pandas as pd
df = pd.read_csv('data-science-jobs-in-india/naukri_data_science_jobs_india.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 12000 entries, 0 to 11999
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Job_Role 12000 non-null object
1 Company 12000 non-null object
2 Location 12000 non-null object
3 Job Experience 12000 non-null object
4 Skills/Description 12000 non-null object
dtypes: object(5)
memory usage: 468.9+ KB
<jupyter_text>Examples:
{
"Job_Role": "Senior Data Scientist",
"Company": "UPL",
"Location": "Bangalore/Bengaluru, Mumbai (All Areas)",
"Job Experience": "3-6",
"Skills/Description": "python, MLT, statistical modeling, machine learning, IT Skills, advanced analytics, scala, statistics"
}
{
"Job_Role": "Senior Data Scientist",
"Company": "Walmart",
"Location": "Bangalore/Bengaluru",
"Job Experience": "5-9",
"Skills/Description": "Data Science, Machine learning, Python, Azure, BiqQuery, GCP, PySpark, tensorflow"
}
{
"Job_Role": "Applied Data Scientist / ML Senior Engineer (Python / SQL)",
"Company": "SAP India Pvt.Ltd",
"Location": "Bangalore/Bengaluru",
"Job Experience": "5-10",
"Skills/Description": "Python, IT Skills, Testing, Cloud, Product Management, SAP, Cloud computing, NLP"
}
{
"Job_Role": "Data Scientist",
"Company": "UPL",
"Location": "Bangalore/Bengaluru, Mumbai (All Areas)",
"Job Experience": "1-4",
"Skills/Description": "python, machine learning, Data Science, data analysis, aws, azure"
}
<jupyter_script># **CUSTOMIZED INDEX**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
# ***Reset_index()***
a.reset_index(inplace=True)
a
a.reset_index(drop=True)
# ***Set_index() methods***
a.set_index("Company", inplace=True)
a.reset_index(drop=True)
a.reset_index(inplace=True)
a.set_index("Job_Role")
a.set_index("Job_Role", drop=False, inplace=True)
a
# ***Append Argument***
a.set_index("Company", append=True)
a.head()
a.reset_index()
a.set_index("Company")
# **ACCESS DATAFRAME**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
a.sort_index(inplace=True)
a
a.loc["senior Data Analyst"]
a.loc["Data Analyst"]
a.loc[[".NET Developer", "Data Analyst"]]
a.loc["Data Analyst":"senior Data Analyst":2]
# **ACCESS DATAFRAME-1**
a.sort_index(inplace=True, ascending=False)
a
a.iloc[0]
a.iloc[0:100:10]
a.loc["lead software engineer":"Web / Data Analyst"]
a.iloc[0:10]
# **DATA ACCESS-3**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head
a.loc["Tech Lead - Azure", "Company"]
a.loc["Tech Lead - Azure", "Location"]
a.loc["Full Stack Developer - Machine Learning", ["Company", "Location"]]
a.loc[:, ["Company", "Skills/Description"]]
a.loc["Senior Data Scientist":"Data Scientist", "Company":"Job Experience"]
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Skills/Description"],
)
a.head()
a.loc[
"python, MLT, statistical modeling, machine learning, IT Skills, advanced analytics, scala, statistics",
"Job_Role":"Location",
]
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
a.loc["Tech Lead - Azure", "Company"] = "cliqHR.com"
a.tail(3)
a.loc["Senior Data Scientist", ["Location", "Job Experience"]] = ["Bengaluru", "3-6"]
a.head()
mask = a.loc[:, "Location"].isin(["Bangalore", "Bangalore/Bengaluru"])
a[mask]
a.loc[mask, "Location"] = "Bengaluru"
a.loc[mask]
# ***INDEX RENAME***
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
a.rename(
mapper={
"Senior Data Scientist": "Senior Data Analyst",
"Data Analyst": "Data Scientist",
},
axis=0,
inplace=True,
)
a.head()
a.rename(
index={
"Senior Data Analyst": "Senior Data Scientist",
"Data Analyst": "Data Scientist",
},
inplace=True,
)
a.head()
# **COLUMN RENAMING**
import pandas as pd
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
a.rename(
columns={"Company": "Firm", "Skills/Description": "Job Requirement"}, inplace=True
)
a.columns
a
a.rename(
columns={"Firm": "Company", "Job Requirement": "Skills/Description"}, inplace=True
)
a.columns
# ***Delete Rows***
a.head()
a.drop(labels="Senior Data Scientist", inplace=True)
a.describe()
a.drop(labels="Skills/Description", axis=1, inplace=True)
a.head()
# ***Pop methods***
loc = a.pop("Location")
a.head()
# ***del KEYWORD***
del a["Company"]
a
# **RANDOM SAMPLE**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
a.sample()
a.sample(n=5)
a.sample(n=3, axis=1)
a.sample(frac=0.1)
a.sample(n=20, replace=True)
# **LARGEST VALUES**
import numpy as np
a["salary"] = np.random.randint(10000, 250000, size=12000)
a.head()
a.sort_values("salary", ascending=False).head()
a.nlargest(3, "salary")
a["salary"].nlargest(3)
a.nsmallest(3, "salary")
a["salary"].nsmallest(3)
# **WHERE() METHODS**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
mask = a["Job_Role"] == "Senior Data Scientist"
a[mask]
a.where(mask)
np.random.randint(10, 100, (4, 4))
a = pd.DataFrame(
np.random.randint(10, 100, (4, 4)),
index=["lakshmi", "akshaya", "charithran", "pooja"],
columns=["eng", "tamil", "maths", "soc"],
)
a
a < 80
a.where(a < 80, "A", inplace=True)
a
a = pd.DataFrame(
np.random.randint(10, 100, (4, 4)),
index=["lakshmi", "akshaya", "charithran", "pooja"],
columns=["eng", "tamil", "maths", "soc"],
)
a
m = a["eng"] < 75
m
a.where(m, "B")
c = lambda x: x < 80
c(a)
a.where(lambda x: x < 80, "A", inplace=True)
a
a.where(a > 35, lambda x: x + 2)
# **QUERY() METHODS**
import pandas as pd
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
a.query("Job_Role=='Data Scientist'")
a.query("`Job Experience`=='0-0'")
a.columns
a.columns = [i.replace(" ", "_") for i in a.columns]
a.columns
a.query("Job_Role=='Senior Data Scientist'and Job_Experience=='3-5'")
a.query("Location=='Chennai' and Job_Experience=='1-2'")
a.query("Job_Role in ['Senior Data Scientist']")
# **APPLY() METHODS**
s1 = pd.Series(["LAKSHMI KANTH", "JAI KUMAR", "DHONI SINGH", "MADHAN RAJ"])
s1
s1 = s1.apply(lambda x: x.replace(" ", "_"))
s1
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
a.info()
import numpy as np
a["salary"] = np.random.randint(10000, 250000, size=12000)
a
a["salary"] = a["salary"].apply(lambda x: "RS." + str(x))
a
def band(x):
if x > 200000:
return "A"
elif x > 100000:
return "B"
else:
return "c"
a["category"] = a["salary"].apply(band)
a
a["category"].value_counts()
# ***applying the transformation to a particular***
def flag(x):
loc = x["Location"]
job = x["Job_Role"]
if loc in ["Chennai"] and job == "Data Analyst":
return "Yes"
else:
return "No"
a["apply or not"] = a.apply(flag, axis=1)
a
a["apply or not"].value_counts()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/547/129547756.ipynb
|
data-science-jobs-in-india
|
anandhuh
|
[{"Id": 129547756, "ScriptId": 38510029, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13282236, "CreationDate": "05/14/2023 18:09:30", "VersionNumber": 1.0, "Title": "DATA EXTRACTION", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 307.0, "LinesInsertedFromPrevious": 307.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185719763, "KernelVersionId": 129547756, "SourceDatasetVersionId": 3860888}]
|
[{"Id": 3860888, "DatasetId": 2179348, "DatasourceVersionId": 3915815, "CreatorUserId": 6096594, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "06/25/2022 11:49:35", "VersionNumber": 3.0, "Title": "12000 Data Science Jobs in India - Naukri.com", "Slug": "data-science-jobs-in-india", "Subtitle": "Data of 12,000 Data Science Jobs in India Scraped from Naukri.com", "Description": "## About\n\n**Naukri.com** is an Indian employment website operating in India and Middle East. It was founded in March 1997. Naukri was ranked No.1 by 9 independent sources, placing it way ahead of competition.Google Trends names Naukri \u201cthe most preferred job search destination in India\u201d.\n\nThis dataset contains data of first 12,000 results obtained for '**data scientist**' job role in '**India**' from Naukri.com, on **May 18, 2022**\n\n**Starter Notebook** : https://www.kaggle.com/code/anandhuh/data-science-jobs-in-india-eda-naukri-com\n\n## Attribute Information\n\n1. **Job_Role** - Job Roles\n2. **Company** - Name of the Company\n3. **Location** - Location of the company\n4. **Job Experience** - Required Job Experience (Min. Experience-Max. Experience)\n5. **Skills/Description** - Required Skills for the Job\n\n## Source\n\nLink : **https://www.naukri.com/**\n\n## Other Updated Datasets\n\nhttps://www.kaggle.com/anandhuh/datasets\nPlease appreciate the effort with an **upvote** \ud83d\udc4d \n\n### Thank You", "VersionNotes": "Data Update 2022/06/25", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2179348, "CreatorUserId": 6096594, "OwnerUserId": 6096594.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 3860888.0, "CurrentDatasourceVersionId": 3915815.0, "ForumId": 2205291, "Type": 2, "CreationDate": "05/15/2022 07:54:06", "LastActivityDate": "05/15/2022", "TotalViews": 6391, "TotalDownloads": 942, "TotalVotes": 84, "TotalKernels": 148}]
|
[{"Id": 6096594, "UserName": "anandhuh", "DisplayName": "Anandhu H", "RegisterDate": "11/04/2020", "PerformanceTier": 4}]
|
# **CUSTOMIZED INDEX**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
# ***Reset_index()***
a.reset_index(inplace=True)
a
a.reset_index(drop=True)
# ***Set_index() methods***
a.set_index("Company", inplace=True)
a.reset_index(drop=True)
a.reset_index(inplace=True)
a.set_index("Job_Role")
a.set_index("Job_Role", drop=False, inplace=True)
a
# ***Append Argument***
a.set_index("Company", append=True)
a.head()
a.reset_index()
a.set_index("Company")
# **ACCESS DATAFRAME**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
a.sort_index(inplace=True)
a
a.loc["senior Data Analyst"]
a.loc["Data Analyst"]
a.loc[[".NET Developer", "Data Analyst"]]
a.loc["Data Analyst":"senior Data Analyst":2]
# **ACCESS DATAFRAME-1**
a.sort_index(inplace=True, ascending=False)
a
a.iloc[0]
a.iloc[0:100:10]
a.loc["lead software engineer":"Web / Data Analyst"]
a.iloc[0:10]
# **DATA ACCESS-3**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head
a.loc["Tech Lead - Azure", "Company"]
a.loc["Tech Lead - Azure", "Location"]
a.loc["Full Stack Developer - Machine Learning", ["Company", "Location"]]
a.loc[:, ["Company", "Skills/Description"]]
a.loc["Senior Data Scientist":"Data Scientist", "Company":"Job Experience"]
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Skills/Description"],
)
a.head()
a.loc[
"python, MLT, statistical modeling, machine learning, IT Skills, advanced analytics, scala, statistics",
"Job_Role":"Location",
]
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
a.loc["Tech Lead - Azure", "Company"] = "cliqHR.com"
a.tail(3)
a.loc["Senior Data Scientist", ["Location", "Job Experience"]] = ["Bengaluru", "3-6"]
a.head()
mask = a.loc[:, "Location"].isin(["Bangalore", "Bangalore/Bengaluru"])
a[mask]
a.loc[mask, "Location"] = "Bengaluru"
a.loc[mask]
# ***INDEX RENAME***
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
a.rename(
mapper={
"Senior Data Scientist": "Senior Data Analyst",
"Data Analyst": "Data Scientist",
},
axis=0,
inplace=True,
)
a.head()
a.rename(
index={
"Senior Data Analyst": "Senior Data Scientist",
"Data Analyst": "Data Scientist",
},
inplace=True,
)
a.head()
# **COLUMN RENAMING**
import pandas as pd
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv",
index_col=["Job_Role"],
)
a.head()
a.rename(
columns={"Company": "Firm", "Skills/Description": "Job Requirement"}, inplace=True
)
a.columns
a
a.rename(
columns={"Firm": "Company", "Job Requirement": "Skills/Description"}, inplace=True
)
a.columns
# ***Delete Rows***
a.head()
a.drop(labels="Senior Data Scientist", inplace=True)
a.describe()
a.drop(labels="Skills/Description", axis=1, inplace=True)
a.head()
# ***Pop methods***
loc = a.pop("Location")
a.head()
# ***del KEYWORD***
del a["Company"]
a
# **RANDOM SAMPLE**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
a.sample()
a.sample(n=5)
a.sample(n=3, axis=1)
a.sample(frac=0.1)
a.sample(n=20, replace=True)
# **LARGEST VALUES**
import numpy as np
a["salary"] = np.random.randint(10000, 250000, size=12000)
a.head()
a.sort_values("salary", ascending=False).head()
a.nlargest(3, "salary")
a["salary"].nlargest(3)
a.nsmallest(3, "salary")
a["salary"].nsmallest(3)
# **WHERE() METHODS**
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
mask = a["Job_Role"] == "Senior Data Scientist"
a[mask]
a.where(mask)
np.random.randint(10, 100, (4, 4))
a = pd.DataFrame(
np.random.randint(10, 100, (4, 4)),
index=["lakshmi", "akshaya", "charithran", "pooja"],
columns=["eng", "tamil", "maths", "soc"],
)
a
a < 80
a.where(a < 80, "A", inplace=True)
a
a = pd.DataFrame(
np.random.randint(10, 100, (4, 4)),
index=["lakshmi", "akshaya", "charithran", "pooja"],
columns=["eng", "tamil", "maths", "soc"],
)
a
m = a["eng"] < 75
m
a.where(m, "B")
c = lambda x: x < 80
c(a)
a.where(lambda x: x < 80, "A", inplace=True)
a
a.where(a > 35, lambda x: x + 2)
# **QUERY() METHODS**
import pandas as pd
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
a.query("Job_Role=='Data Scientist'")
a.query("`Job Experience`=='0-0'")
a.columns
a.columns = [i.replace(" ", "_") for i in a.columns]
a.columns
a.query("Job_Role=='Senior Data Scientist'and Job_Experience=='3-5'")
a.query("Location=='Chennai' and Job_Experience=='1-2'")
a.query("Job_Role in ['Senior Data Scientist']")
# **APPLY() METHODS**
s1 = pd.Series(["LAKSHMI KANTH", "JAI KUMAR", "DHONI SINGH", "MADHAN RAJ"])
s1
s1 = s1.apply(lambda x: x.replace(" ", "_"))
s1
a = pd.read_csv(
"/kaggle/input/data-science-jobs-in-india/naukri_data_science_jobs_india.csv"
)
a.head()
a.info()
import numpy as np
a["salary"] = np.random.randint(10000, 250000, size=12000)
a
a["salary"] = a["salary"].apply(lambda x: "RS." + str(x))
a
def band(x):
if x > 200000:
return "A"
elif x > 100000:
return "B"
else:
return "c"
a["category"] = a["salary"].apply(band)
a
a["category"].value_counts()
# ***applying the transformation to a particular***
def flag(x):
loc = x["Location"]
job = x["Job_Role"]
if loc in ["Chennai"] and job == "Data Analyst":
return "Yes"
else:
return "No"
a["apply or not"] = a.apply(flag, axis=1)
a
a["apply or not"].value_counts()
|
[{"data-science-jobs-in-india/naukri_data_science_jobs_india.csv": {"column_names": "[\"Job_Role\", \"Company\", \"Location\", \"Job Experience\", \"Skills/Description\"]", "column_data_types": "{\"Job_Role\": \"object\", \"Company\": \"object\", \"Location\": \"object\", \"Job Experience\": \"object\", \"Skills/Description\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 12000 entries, 0 to 11999\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Job_Role 12000 non-null object\n 1 Company 12000 non-null object\n 2 Location 12000 non-null object\n 3 Job Experience 12000 non-null object\n 4 Skills/Description 12000 non-null object\ndtypes: object(5)\nmemory usage: 468.9+ KB\n", "summary": "{\"Job_Role\": {\"count\": 12000, \"unique\": 6563, \"top\": \"Data Engineer\", \"freq\": 580}, \"Company\": {\"count\": 12000, \"unique\": 3507, \"top\": \"Accenture\", \"freq\": 490}, \"Location\": {\"count\": 12000, \"unique\": 822, \"top\": \"Bangalore/Bengaluru\", \"freq\": 3383}, \"Job Experience\": {\"count\": 12000, \"unique\": 143, \"top\": \"5-10\", \"freq\": 944}, \"Skills/Description\": {\"count\": 12000, \"unique\": 11356, \"top\": \"Computer science, Manager Quality Assurance, Front end, Coding, Javascript, Test planning, HTML, Application development\", \"freq\": 8}}", "examples": "{\"Job_Role\":{\"0\":\"Senior Data Scientist\",\"1\":\"Senior Data Scientist\",\"2\":\"Applied Data Scientist \\/ ML Senior Engineer (Python \\/ SQL)\",\"3\":\"Data Scientist\"},\"Company\":{\"0\":\"UPL\",\"1\":\"Walmart\",\"2\":\"SAP India Pvt.Ltd\",\"3\":\"UPL\"},\"Location\":{\"0\":\"Bangalore\\/Bengaluru, Mumbai (All Areas)\",\"1\":\"Bangalore\\/Bengaluru\",\"2\":\"Bangalore\\/Bengaluru\",\"3\":\"Bangalore\\/Bengaluru, Mumbai (All Areas)\"},\"Job Experience\":{\"0\":\"3-6\",\"1\":\"5-9\",\"2\":\"5-10\",\"3\":\"1-4\"},\"Skills\\/Description\":{\"0\":\"python, MLT, statistical modeling, machine learning, IT Skills, advanced analytics, scala, statistics\",\"1\":\"Data Science, Machine learning, Python, Azure, BiqQuery, GCP, PySpark, tensorflow\",\"2\":\"Python, IT Skills, Testing, Cloud, Product Management, SAP, Cloud computing, NLP\",\"3\":\"python, machine learning, Data Science, data analysis, aws, azure\"}}"}}]
| true | 1 |
<start_data_description><data_path>data-science-jobs-in-india/naukri_data_science_jobs_india.csv:
<column_names>
['Job_Role', 'Company', 'Location', 'Job Experience', 'Skills/Description']
<column_types>
{'Job_Role': 'object', 'Company': 'object', 'Location': 'object', 'Job Experience': 'object', 'Skills/Description': 'object'}
<dataframe_Summary>
{'Job_Role': {'count': 12000, 'unique': 6563, 'top': 'Data Engineer', 'freq': 580}, 'Company': {'count': 12000, 'unique': 3507, 'top': 'Accenture', 'freq': 490}, 'Location': {'count': 12000, 'unique': 822, 'top': 'Bangalore/Bengaluru', 'freq': 3383}, 'Job Experience': {'count': 12000, 'unique': 143, 'top': '5-10', 'freq': 944}, 'Skills/Description': {'count': 12000, 'unique': 11356, 'top': 'Computer science, Manager Quality Assurance, Front end, Coding, Javascript, Test planning, HTML, Application development', 'freq': 8}}
<dataframe_info>
RangeIndex: 12000 entries, 0 to 11999
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Job_Role 12000 non-null object
1 Company 12000 non-null object
2 Location 12000 non-null object
3 Job Experience 12000 non-null object
4 Skills/Description 12000 non-null object
dtypes: object(5)
memory usage: 468.9+ KB
<some_examples>
{'Job_Role': {'0': 'Senior Data Scientist', '1': 'Senior Data Scientist', '2': 'Applied Data Scientist / ML Senior Engineer (Python / SQL)', '3': 'Data Scientist'}, 'Company': {'0': 'UPL', '1': 'Walmart', '2': 'SAP India Pvt.Ltd', '3': 'UPL'}, 'Location': {'0': 'Bangalore/Bengaluru, Mumbai (All Areas)', '1': 'Bangalore/Bengaluru', '2': 'Bangalore/Bengaluru', '3': 'Bangalore/Bengaluru, Mumbai (All Areas)'}, 'Job Experience': {'0': '3-6', '1': '5-9', '2': '5-10', '3': '1-4'}, 'Skills/Description': {'0': 'python, MLT, statistical modeling, machine learning, IT Skills, advanced analytics, scala, statistics', '1': 'Data Science, Machine learning, Python, Azure, BiqQuery, GCP, PySpark, tensorflow', '2': 'Python, IT Skills, Testing, Cloud, Product Management, SAP, Cloud computing, NLP', '3': 'python, machine learning, Data Science, data analysis, aws, azure'}}
<end_description>
| 2,434 | 0 | 3,340 | 2,434 |
129547684
|
<jupyter_start><jupyter_text>Students Score Dataset - Linear Regression
### Context
There's a story behind every dataset and here's your opportunity to share yours.
### Content
This contains only two columns Hours and Scores. Linear regression very effective used to predict the scores based on the number of hours.
Kaggle dataset identifier: students-score-dataset-linear-regression
<jupyter_code>import pandas as pd
df = pd.read_csv('students-score-dataset-linear-regression/student_scores.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 25 entries, 0 to 24
Data columns (total 2 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Hours 25 non-null float64
1 Scores 25 non-null int64
dtypes: float64(1), int64(1)
memory usage: 528.0 bytes
<jupyter_text>Examples:
{
"Hours": 2.5,
"Scores": 21.0
}
{
"Hours": 5.1,
"Scores": 47.0
}
{
"Hours": 3.2,
"Scores": 27.0
}
{
"Hours": 8.5,
"Scores": 75.0
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df = pd.read_csv(
"/kaggle/input/students-score-dataset-linear-regression/student_scores.csv"
)
df
hours = df.Hours.values
hours
scores = df["Scores"].values
scores
from matplotlib import pyplot as plt
x = hours
y = scores
plt.scatter(x, y, color="black")
plt.xlabel("Hours")
plt.ylabel("Scores")
plt.plot
x = x.reshape(-1, 1)
x, len(x)
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, train_size=0.8, random_state=600
)
x_train, len(x_train)
plt.scatter(x_train, y_train, color="black")
plt.xlabel("Hours")
plt.ylabel("Scores")
plt.plot
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(x_train, y_train)
y_predict = lr.predict(x_test)
y_predict
lr.score(x_test, y_test) * 100
y_predict = lr.predict(x_test)
y_predict
plt.scatter(x_train, y_train, color="black")
plt.scatter(x_test, y_predict, color="red")
plt.xlabel("Hours")
plt.ylabel("Scores")
plt.plot
plt.scatter(x, y, color="black")
plt.xlabel("Hours")
plt.ylabel("Scorse")
plt.plot
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/547/129547684.ipynb
|
students-score-dataset-linear-regression
|
shubham47
|
[{"Id": 129547684, "ScriptId": 38241542, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14989886, "CreationDate": "05/14/2023 18:08:32", "VersionNumber": 2.0, "Title": "Linear Regression", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 66.0, "LinesInsertedFromPrevious": 47.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 19.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185719694, "KernelVersionId": 129547684, "SourceDatasetVersionId": 1962502}]
|
[{"Id": 1962502, "DatasetId": 1171622, "DatasourceVersionId": 2001374, "CreatorUserId": 1115324, "LicenseName": "Other (specified in description)", "CreationDate": "02/20/2021 19:25:46", "VersionNumber": 1.0, "Title": "Students Score Dataset - Linear Regression", "Slug": "students-score-dataset-linear-regression", "Subtitle": NaN, "Description": "### Context\n\nThere's a story behind every dataset and here's your opportunity to share yours.\n\n\n### Content\n\nThis contains only two columns Hours and Scores. Linear regression very effective used to predict the scores based on the number of hours.\n\n\n### Acknowledgements\n\nWe wouldn't be here without the help of others. If you owe any attributions or thanks, include them here along with any citations of past research.\n\n\n### Inspiration\n\nYour data will be in front of the world's largest data science community. What questions do you want to see answered?", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1171622, "CreatorUserId": 1115324, "OwnerUserId": 1115324.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1962502.0, "CurrentDatasourceVersionId": 2001374.0, "ForumId": 1189295, "Type": 2, "CreationDate": "02/20/2021 19:25:46", "LastActivityDate": "02/20/2021", "TotalViews": 23753, "TotalDownloads": 3418, "TotalVotes": 38, "TotalKernels": 24}]
|
[{"Id": 1115324, "UserName": "shubham47", "DisplayName": "Shubham", "RegisterDate": "06/08/2017", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df = pd.read_csv(
"/kaggle/input/students-score-dataset-linear-regression/student_scores.csv"
)
df
hours = df.Hours.values
hours
scores = df["Scores"].values
scores
from matplotlib import pyplot as plt
x = hours
y = scores
plt.scatter(x, y, color="black")
plt.xlabel("Hours")
plt.ylabel("Scores")
plt.plot
x = x.reshape(-1, 1)
x, len(x)
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, train_size=0.8, random_state=600
)
x_train, len(x_train)
plt.scatter(x_train, y_train, color="black")
plt.xlabel("Hours")
plt.ylabel("Scores")
plt.plot
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(x_train, y_train)
y_predict = lr.predict(x_test)
y_predict
lr.score(x_test, y_test) * 100
y_predict = lr.predict(x_test)
y_predict
plt.scatter(x_train, y_train, color="black")
plt.scatter(x_test, y_predict, color="red")
plt.xlabel("Hours")
plt.ylabel("Scores")
plt.plot
plt.scatter(x, y, color="black")
plt.xlabel("Hours")
plt.ylabel("Scorse")
plt.plot
|
[{"students-score-dataset-linear-regression/student_scores.csv": {"column_names": "[\"Hours\", \"Scores\"]", "column_data_types": "{\"Hours\": \"float64\", \"Scores\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 25 entries, 0 to 24\nData columns (total 2 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Hours 25 non-null float64\n 1 Scores 25 non-null int64 \ndtypes: float64(1), int64(1)\nmemory usage: 528.0 bytes\n", "summary": "{\"Hours\": {\"count\": 25.0, \"mean\": 5.012, \"std\": 2.5250940576540906, \"min\": 1.1, \"25%\": 2.7, \"50%\": 4.8, \"75%\": 7.4, \"max\": 9.2}, \"Scores\": {\"count\": 25.0, \"mean\": 51.48, \"std\": 25.28688724747802, \"min\": 17.0, \"25%\": 30.0, \"50%\": 47.0, \"75%\": 75.0, \"max\": 95.0}}", "examples": "{\"Hours\":{\"0\":2.5,\"1\":5.1,\"2\":3.2,\"3\":8.5},\"Scores\":{\"0\":21,\"1\":47,\"2\":27,\"3\":75}}"}}]
| true | 1 |
<start_data_description><data_path>students-score-dataset-linear-regression/student_scores.csv:
<column_names>
['Hours', 'Scores']
<column_types>
{'Hours': 'float64', 'Scores': 'int64'}
<dataframe_Summary>
{'Hours': {'count': 25.0, 'mean': 5.012, 'std': 2.5250940576540906, 'min': 1.1, '25%': 2.7, '50%': 4.8, '75%': 7.4, 'max': 9.2}, 'Scores': {'count': 25.0, 'mean': 51.48, 'std': 25.28688724747802, 'min': 17.0, '25%': 30.0, '50%': 47.0, '75%': 75.0, 'max': 95.0}}
<dataframe_info>
RangeIndex: 25 entries, 0 to 24
Data columns (total 2 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Hours 25 non-null float64
1 Scores 25 non-null int64
dtypes: float64(1), int64(1)
memory usage: 528.0 bytes
<some_examples>
{'Hours': {'0': 2.5, '1': 5.1, '2': 3.2, '3': 8.5}, 'Scores': {'0': 21, '1': 47, '2': 27, '3': 75}}
<end_description>
| 468 | 0 | 795 | 468 |
129547189
|
import lightgbm as lgbm
import matplotlib.pyplot as plt
import numpy as np
import optuna
import pandas as pd
import seaborn as sns
from catboost import CatBoostClassifier
from lightgbm import LGBMClassifier
from matplotlib import pyplot
from scipy import stats
from scipy.stats import rankdata
from sklearn import metrics
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.decomposition import TruncatedSVD
from sklearn.feature_selection import SelectKBest, chi2, f_classif, mutual_info_classif
from sklearn.metrics import f1_score, make_scorer
from sklearn.model_selection import (
train_test_split,
StratifiedKFold,
StratifiedShuffleSplit,
GridSearchCV,
RandomizedSearchCV,
cross_val_score,
)
from sklearn.preprocessing import StandardScaler, RobustScaler, OneHotEncoder
from xgboost import XGBClassifier
# uploading a file with a token
from google.colab import files
files.upload()
# downloading a dataset
train_df = pd.read_csv(
"classify-cancer-type-no-clustering/train_dataset.csv", index_col=0
)
train_df.shape
train_df.info() # get basic info
display(train_df.head(10)) # get the first observations
print(train_df.isna().sum()[train_df.isna().sum() != 0])
train_df["label"].unique()
# plotting the labels dist
fig, ay = plt.subplots(figsize=(20, 6))
sns.barplot(
data=train_df,
y=train_df["label"].value_counts().index,
x=train_df["label"].value_counts(),
)
# assigning features and target
features = train_df.drop(["label"], axis=1)
target = train_df["label"]
# performing rank normalization
ranks = rankdata(features, axis=0)
norm_ranks = (ranks - 1) / (np.max(ranks) - 1)
print("Normalized ranks:")
print(norm_ranks)
norm_data = pd.DataFrame(
data=norm_ranks, index=train_df.index, columns=features.columns
)
target_for_ohe = pd.DataFrame(data=target, index=features.index, columns=["label"])
target_for_ohe.info()
# encoding target for feature selection
ohe = OneHotEncoder()
target_ohe = ohe.fit_transform(target_for_ohe)
target_ohe = pd.DataFrame(
target_ohe.toarray(), index=features.index, columns=ohe.categories_
)
target_ohe.info()
# feature selection
def select_features(x, y):
fs = SelectKBest(score_func=chi2, k="all")
fs.fit(x, y)
x_fs = fs.transform(x)
return x_fs, fs
def find_best_features(fs):
feat = []
score = []
for i in range(len(norm_data.columns)):
feat.append(fs.get_feature_names_out()[i])
score.append(fs.scores_[i])
d = {"feat": feat, "score": score}
all_features = pd.DataFrame(d) # getting a df with all features and their scores
all_features = all_features.sort_values(by=["score"])
threshold = all_features["score"].quantile(0.975)
# filtering those which are of low importance
selected_features = all_features[all_features["score"] >= threshold].sort_values(
by="score", ascending=False
)["feat"]
return selected_features
target.unique()
# applying feature selection
features_fs_brain_type1, fs_brain_type1 = select_features(
norm_data, target_ohe["brain_type1"]
)
selected_brain_type1 = find_best_features(fs_brain_type1)
len(selected_brain_type1)
features_fs_brain_type2, fs_brain_type2 = select_features(
norm_data, target_ohe["brain_type2"]
)
selected_brain_type2 = find_best_features(fs_brain_type2)
len(selected_brain_type2)
features_fs_brain_type3, fs_brain_type3 = select_features(
norm_data, target_ohe["brain_type3"]
)
selected_brain_type3 = find_best_features(fs_brain_type3)
len(selected_brain_type3)
features_fs_brest_type1, fs_brest_type1 = select_features(
norm_data, target_ohe["brest_type1"]
)
selected_brest_type1 = find_best_features(fs_brest_type1)
len(selected_brest_type1)
features_fs_brest_type2, fs_brest_type2 = select_features(
norm_data, target_ohe["brest_type2"]
)
selected_brest_type2 = find_best_features(fs_brest_type2)
len(selected_brest_type2)
features_fs_brest_type3, fs_brest_type3 = select_features(
norm_data, target_ohe["brest_type3"]
)
selected_brest_type3 = find_best_features(fs_brest_type3)
len(selected_brest_type3)
features_fs_esophageal, fs_esophageal = select_features(
norm_data, target_ohe["esophageal"]
)
selected_esophageal = find_best_features(fs_esophageal)
len(selected_esophageal)
features_fs_colorectal, fs_colorectal = select_features(
norm_data, target_ohe["colorectal"]
)
selected_colorectal = find_best_features(fs_colorectal)
len(selected_colorectal)
# getting a list of all selected features
selected_feat = pd.concat(
[
selected_brain_type1,
selected_brain_type2,
selected_brain_type3,
selected_brest_type1,
selected_brest_type2,
selected_brest_type3,
selected_esophageal,
selected_colorectal,
],
join="outer",
axis=0,
)
# counting the unique ones
unique_selected_features = selected_feat.unique()
len(unique_selected_features)
# applying feature selection to a normalized df
features_after_selection = norm_data[unique_selected_features]
features_after_selection.shape
# # Create correlation matrix
# corr_matrix = features_after_selection.corr().abs()
# # Select upper triangle of correlation matrix
# upper = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(bool))
# # Find features with correlation greater than 0.95
# to_drop = [column for column in upper.columns if any(upper[column] > 0.95)]
# # Drop features
# features_after_selection.drop(to_drop, axis=1, inplace=True)
# features_after_selection = features_after_selection.reset_index().drop('index', axis=1)
# target = target.reset_index().drop('index', axis=1)
diagnoses = {
"brain_type1": 0,
"brain_type2": 1,
"brain_type3": 2,
"brest_type1": 3,
"brest_type2": 4,
"brest_type3": 5,
"esophageal": 6,
"colorectal": 7,
}
# encoding target for XGB to process
target_enc = target.copy()
target_enc = target_enc.map(diagnoses)
# tuning hyperparams
def objective(features, target, random_state=12345):
xgb = XGBClassifier(random_state=random_state)
xgb_params = {
"learning_rate": [0.05, 0.10, 0.15, 0.20, 0.25, 0.30],
"max_depth": [3, 4, 5],
"min_child_weight": [1, 3, 5, 7],
"gamma": [0.0, 0.1, 0.2, 0.3, 0.4],
"colsample_bytree": [0.3, 0.4, 0.5, 0.7],
}
# xgb_params['num_leaves'] = [2**k for k in xgb_params['max_depth']]
sss = StratifiedShuffleSplit(n_splits=3, test_size=0.5, random_state=12345)
sss.get_n_splits(features_after_selection, target_enc)
type(sss.split(features, target))
# split data
for train_index, test_index in sss.split(features, target):
X_train, X_test = features.iloc[train_index], features.iloc[test_index]
y_train, y_test = target.iloc[train_index], target.iloc[test_index]
f1 = make_scorer(f1_score, average="weighted")
xgb_grid = RandomizedSearchCV(
xgb,
param_distributions=xgb_params,
n_iter=5,
scoring=f1,
n_jobs=-1,
cv=5,
verbose=1,
)
xgb_test_scores = []
search = xgb_grid.fit(X_train, y_train)
xgb_best_params = search.best_params_
xgb_best_score = xgb_grid.best_score_
preds = search.predict(X_test)
xgb_test_scores.append(f1_score(preds, y_test, average="weighted"))
print("Лучшие параметры модели XGBoost:", xgb_best_params)
return xgb_best_params, xgb_best_score, xgb_test_scores
best_params, best_score, test_scores = objective(features_after_selection, target_enc)
best_params
# {'min_child_weight': 7,
# 'max_depth': 4,
# 'learning_rate': 0.3,
# 'gamma': 0.4,
# 'colsample_bytree': 0.5}
best_score
test_scores
# [0.9261633012702547]
# study = optuna.create_study(direction="maximize", study_name="XGB Classifier")
# func = lambda trial: objective(trial, X_train, X_test, y_train, y_test)
# study.optimize(func, n_trials=3)
# best_trial = study.best_trial
# best_trial.values
# best_trial.params
# best_params = best_trial.params
# model = XGBClassifier(**best_params)
model = XGBClassifier(**best_params, random_state=12345)
# X_train, X_test, y_train, y_test = train_test_split(features_after_selection,
# target_enc,
# test_size=0.25,
# stratify=target_enc,
# random_state=12345)
scores = cross_val_score(model, features_after_selection, target_enc, cv=5)
scores
# [0.94909091, 0.96363636, 0.92727273, 0.94525547, 0.93430657]
model.fit(features_after_selection, target_enc)
# metrics.f1_score(model.predict(X_test), y_test, average='weighted')
test_df = pd.read_csv(
"classify-cancer-type-no-clustering/test_dataset.csv", index_col=0
)
test_df.shape
test_df.head()
ranks_test = rankdata(test_df, axis=0)
norm_ranks_test = (ranks_test - 1) / (np.max(ranks_test) - 1)
print("Normalized ranks:")
print(norm_ranks)
norm_data_test = pd.DataFrame(
data=norm_ranks_test, index=test_df.index, columns=test_df.columns
)
test_df_selected = norm_data_test[unique_selected_features]
predictions = model.predict(test_df_selected)
inv_diagnosis = {v: k for k, v in diagnoses.items()}
inv_diagnosis
predictions_df = pd.DataFrame(
data=predictions, index=test_df.index, columns=["Predicted"]
)
predictions_df["Predicted"] = predictions_df["Predicted"].map(inv_diagnosis)
predictions_df.head()
predictions_df.index.name = "Id"
predictions_df.head()
predictions_df.to_csv("submission8.csv")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/547/129547189.ipynb
| null | null |
[{"Id": 129547189, "ScriptId": 38520967, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10507731, "CreationDate": "05/14/2023 18:02:39", "VersionNumber": 1.0, "Title": "notebookb6c8b87509", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 338.0, "LinesInsertedFromPrevious": 338.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import lightgbm as lgbm
import matplotlib.pyplot as plt
import numpy as np
import optuna
import pandas as pd
import seaborn as sns
from catboost import CatBoostClassifier
from lightgbm import LGBMClassifier
from matplotlib import pyplot
from scipy import stats
from scipy.stats import rankdata
from sklearn import metrics
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.decomposition import TruncatedSVD
from sklearn.feature_selection import SelectKBest, chi2, f_classif, mutual_info_classif
from sklearn.metrics import f1_score, make_scorer
from sklearn.model_selection import (
train_test_split,
StratifiedKFold,
StratifiedShuffleSplit,
GridSearchCV,
RandomizedSearchCV,
cross_val_score,
)
from sklearn.preprocessing import StandardScaler, RobustScaler, OneHotEncoder
from xgboost import XGBClassifier
# uploading a file with a token
from google.colab import files
files.upload()
# downloading a dataset
train_df = pd.read_csv(
"classify-cancer-type-no-clustering/train_dataset.csv", index_col=0
)
train_df.shape
train_df.info() # get basic info
display(train_df.head(10)) # get the first observations
print(train_df.isna().sum()[train_df.isna().sum() != 0])
train_df["label"].unique()
# plotting the labels dist
fig, ay = plt.subplots(figsize=(20, 6))
sns.barplot(
data=train_df,
y=train_df["label"].value_counts().index,
x=train_df["label"].value_counts(),
)
# assigning features and target
features = train_df.drop(["label"], axis=1)
target = train_df["label"]
# performing rank normalization
ranks = rankdata(features, axis=0)
norm_ranks = (ranks - 1) / (np.max(ranks) - 1)
print("Normalized ranks:")
print(norm_ranks)
norm_data = pd.DataFrame(
data=norm_ranks, index=train_df.index, columns=features.columns
)
target_for_ohe = pd.DataFrame(data=target, index=features.index, columns=["label"])
target_for_ohe.info()
# encoding target for feature selection
ohe = OneHotEncoder()
target_ohe = ohe.fit_transform(target_for_ohe)
target_ohe = pd.DataFrame(
target_ohe.toarray(), index=features.index, columns=ohe.categories_
)
target_ohe.info()
# feature selection
def select_features(x, y):
fs = SelectKBest(score_func=chi2, k="all")
fs.fit(x, y)
x_fs = fs.transform(x)
return x_fs, fs
def find_best_features(fs):
feat = []
score = []
for i in range(len(norm_data.columns)):
feat.append(fs.get_feature_names_out()[i])
score.append(fs.scores_[i])
d = {"feat": feat, "score": score}
all_features = pd.DataFrame(d) # getting a df with all features and their scores
all_features = all_features.sort_values(by=["score"])
threshold = all_features["score"].quantile(0.975)
# filtering those which are of low importance
selected_features = all_features[all_features["score"] >= threshold].sort_values(
by="score", ascending=False
)["feat"]
return selected_features
target.unique()
# applying feature selection
features_fs_brain_type1, fs_brain_type1 = select_features(
norm_data, target_ohe["brain_type1"]
)
selected_brain_type1 = find_best_features(fs_brain_type1)
len(selected_brain_type1)
features_fs_brain_type2, fs_brain_type2 = select_features(
norm_data, target_ohe["brain_type2"]
)
selected_brain_type2 = find_best_features(fs_brain_type2)
len(selected_brain_type2)
features_fs_brain_type3, fs_brain_type3 = select_features(
norm_data, target_ohe["brain_type3"]
)
selected_brain_type3 = find_best_features(fs_brain_type3)
len(selected_brain_type3)
features_fs_brest_type1, fs_brest_type1 = select_features(
norm_data, target_ohe["brest_type1"]
)
selected_brest_type1 = find_best_features(fs_brest_type1)
len(selected_brest_type1)
features_fs_brest_type2, fs_brest_type2 = select_features(
norm_data, target_ohe["brest_type2"]
)
selected_brest_type2 = find_best_features(fs_brest_type2)
len(selected_brest_type2)
features_fs_brest_type3, fs_brest_type3 = select_features(
norm_data, target_ohe["brest_type3"]
)
selected_brest_type3 = find_best_features(fs_brest_type3)
len(selected_brest_type3)
features_fs_esophageal, fs_esophageal = select_features(
norm_data, target_ohe["esophageal"]
)
selected_esophageal = find_best_features(fs_esophageal)
len(selected_esophageal)
features_fs_colorectal, fs_colorectal = select_features(
norm_data, target_ohe["colorectal"]
)
selected_colorectal = find_best_features(fs_colorectal)
len(selected_colorectal)
# getting a list of all selected features
selected_feat = pd.concat(
[
selected_brain_type1,
selected_brain_type2,
selected_brain_type3,
selected_brest_type1,
selected_brest_type2,
selected_brest_type3,
selected_esophageal,
selected_colorectal,
],
join="outer",
axis=0,
)
# counting the unique ones
unique_selected_features = selected_feat.unique()
len(unique_selected_features)
# applying feature selection to a normalized df
features_after_selection = norm_data[unique_selected_features]
features_after_selection.shape
# # Create correlation matrix
# corr_matrix = features_after_selection.corr().abs()
# # Select upper triangle of correlation matrix
# upper = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(bool))
# # Find features with correlation greater than 0.95
# to_drop = [column for column in upper.columns if any(upper[column] > 0.95)]
# # Drop features
# features_after_selection.drop(to_drop, axis=1, inplace=True)
# features_after_selection = features_after_selection.reset_index().drop('index', axis=1)
# target = target.reset_index().drop('index', axis=1)
diagnoses = {
"brain_type1": 0,
"brain_type2": 1,
"brain_type3": 2,
"brest_type1": 3,
"brest_type2": 4,
"brest_type3": 5,
"esophageal": 6,
"colorectal": 7,
}
# encoding target for XGB to process
target_enc = target.copy()
target_enc = target_enc.map(diagnoses)
# tuning hyperparams
def objective(features, target, random_state=12345):
xgb = XGBClassifier(random_state=random_state)
xgb_params = {
"learning_rate": [0.05, 0.10, 0.15, 0.20, 0.25, 0.30],
"max_depth": [3, 4, 5],
"min_child_weight": [1, 3, 5, 7],
"gamma": [0.0, 0.1, 0.2, 0.3, 0.4],
"colsample_bytree": [0.3, 0.4, 0.5, 0.7],
}
# xgb_params['num_leaves'] = [2**k for k in xgb_params['max_depth']]
sss = StratifiedShuffleSplit(n_splits=3, test_size=0.5, random_state=12345)
sss.get_n_splits(features_after_selection, target_enc)
type(sss.split(features, target))
# split data
for train_index, test_index in sss.split(features, target):
X_train, X_test = features.iloc[train_index], features.iloc[test_index]
y_train, y_test = target.iloc[train_index], target.iloc[test_index]
f1 = make_scorer(f1_score, average="weighted")
xgb_grid = RandomizedSearchCV(
xgb,
param_distributions=xgb_params,
n_iter=5,
scoring=f1,
n_jobs=-1,
cv=5,
verbose=1,
)
xgb_test_scores = []
search = xgb_grid.fit(X_train, y_train)
xgb_best_params = search.best_params_
xgb_best_score = xgb_grid.best_score_
preds = search.predict(X_test)
xgb_test_scores.append(f1_score(preds, y_test, average="weighted"))
print("Лучшие параметры модели XGBoost:", xgb_best_params)
return xgb_best_params, xgb_best_score, xgb_test_scores
best_params, best_score, test_scores = objective(features_after_selection, target_enc)
best_params
# {'min_child_weight': 7,
# 'max_depth': 4,
# 'learning_rate': 0.3,
# 'gamma': 0.4,
# 'colsample_bytree': 0.5}
best_score
test_scores
# [0.9261633012702547]
# study = optuna.create_study(direction="maximize", study_name="XGB Classifier")
# func = lambda trial: objective(trial, X_train, X_test, y_train, y_test)
# study.optimize(func, n_trials=3)
# best_trial = study.best_trial
# best_trial.values
# best_trial.params
# best_params = best_trial.params
# model = XGBClassifier(**best_params)
model = XGBClassifier(**best_params, random_state=12345)
# X_train, X_test, y_train, y_test = train_test_split(features_after_selection,
# target_enc,
# test_size=0.25,
# stratify=target_enc,
# random_state=12345)
scores = cross_val_score(model, features_after_selection, target_enc, cv=5)
scores
# [0.94909091, 0.96363636, 0.92727273, 0.94525547, 0.93430657]
model.fit(features_after_selection, target_enc)
# metrics.f1_score(model.predict(X_test), y_test, average='weighted')
test_df = pd.read_csv(
"classify-cancer-type-no-clustering/test_dataset.csv", index_col=0
)
test_df.shape
test_df.head()
ranks_test = rankdata(test_df, axis=0)
norm_ranks_test = (ranks_test - 1) / (np.max(ranks_test) - 1)
print("Normalized ranks:")
print(norm_ranks)
norm_data_test = pd.DataFrame(
data=norm_ranks_test, index=test_df.index, columns=test_df.columns
)
test_df_selected = norm_data_test[unique_selected_features]
predictions = model.predict(test_df_selected)
inv_diagnosis = {v: k for k, v in diagnoses.items()}
inv_diagnosis
predictions_df = pd.DataFrame(
data=predictions, index=test_df.index, columns=["Predicted"]
)
predictions_df["Predicted"] = predictions_df["Predicted"].map(inv_diagnosis)
predictions_df.head()
predictions_df.index.name = "Id"
predictions_df.head()
predictions_df.to_csv("submission8.csv")
| false | 0 | 3,235 | 0 | 3,235 | 3,235 |
||
129547672
|
<jupyter_start><jupyter_text>Bank Customer Churn
RowNumber—corresponds to the record (row) number and has no effect on the output.
CustomerId—contains random values and has no effect on customer leaving the bank.
Surname—the surname of a customer has no impact on their decision to leave the bank.
CreditScore—can have an effect on customer churn, since a customer with a higher credit score is less likely to leave the bank.
Geography—a customer’s location can affect their decision to leave the bank.
Gender—it’s interesting to explore whether gender plays a role in a customer leaving the bank.
Age—this is certainly relevant, since older customers are less likely to leave their bank than younger ones.
Tenure—refers to the number of years that the customer has been a client of the bank. Normally, older clients are more loyal and less likely to leave a bank.
Balance—also a very good indicator of customer churn, as people with a higher balance in their accounts are less likely to leave the bank compared to those with lower balances.
NumOfProducts—refers to the number of products that a customer has purchased through the bank.
HasCrCard—denotes whether or not a customer has a credit card. This column is also relevant, since people with a credit card are less likely to leave the bank.
IsActiveMember—active customers are less likely to leave the bank.
EstimatedSalary—as with balance, people with lower salaries are more likely to leave the bank compared to those with higher salaries.
Exited—whether or not the customer left the bank.
Complain—customer has complaint or not.
Satisfaction Score—Score provided by the customer for their complaint resolution.
Card Type—type of card hold by the customer.
Points Earned—the points earned by the customer for using credit card.
Acknowledgements
As we know, it is much more expensive to sign in a new client than keeping an existing one.
It is advantageous for banks to know what leads a client towards the decision to leave the company.
Churn prevention allows companies to develop loyalty programs and retention campaigns to keep as many customers as possible.
Kaggle dataset identifier: bank-customer-churn
<jupyter_script>import warnings
warnings.filterwarnings("ignore")
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
cp = sns.color_palette("pastel")
plt.style.use(plt.style.available)
# plt.style.use('darkgrid')
# plt.style.available
data = pd.read_csv("/kaggle/input/bank-customer-churn/Customer-Churn-Records.csv")
data
feat = [
"Geography",
"Gender",
"NumOfProducts",
"HasCrCard",
"IsActiveMember",
"Exited",
"Complain",
"Satisfaction Score",
"Card Type",
]
fig, axes = plt.subplots(3, 3, figsize=(15, 13), sharex=False, sharey=False)
for i, feature in enumerate(feat):
row = i // 3
column = i % 3
ax = axes[row, column]
data.groupby(feature).count()["RowNumber"].sort_values(ascending=False).plot(
kind="bar", ax=ax, color=cp[1:]
)
ax.set_title(feature, backgroundcolor="skyblue", font="Arial", fontsize=14)
ax.tick_params(axis="x", labelrotation=0)
labels = data.groupby(feature).count()["RowNumber"].sort_values(ascending=False)
ax.bar_label(ax.containers[0], labels=labels, label_type="edge")
plt.tight_layout()
plt.show()
feat = ["CreditScore", "Age", "Balance"]
fig, axes = plt.subplots(2, 2, figsize=(15, 7), sharex=False, sharey=False)
for i, feature in enumerate(feat):
row = i // 2
column = i % 2
ax = axes[row, column]
sns.histplot(data, x=feature, ax=ax, kde=True, palette="pastel")
ax.tick_params(axis="x", labelrotation=45)
ax.set_title(feature, backgroundcolor="skyblue", font="Arial", fontsize=14)
plt.tight_layout()
plt.show()
sns.boxplot(data=data, x="Balance")
data.columns
datacorr = data[
[
"CreditScore",
"Geography",
"Gender",
"Age",
"Tenure",
"Balance",
"NumOfProducts",
"HasCrCard",
"IsActiveMember",
"EstimatedSalary",
"Exited",
"Complain",
"Satisfaction Score",
"Card Type",
"Point Earned",
]
]
correlation = datacorr.corr()
correlation.style.background_gradient(cmap="coolwarm")
# datacorr
## Work in progress
## Upvote if you like
## Suggestions are welcome
## Thanks
{
column: len(datacorr[column].unique())
for column in datacorr.select_dtypes("object").columns
}.keys()
def onehot_encode(df, column):
df = df.copy()
dummies = pd.get_dummies(df[column], prefix=column)
if len(df[column].unique()) == 2:
dummies = dummies.drop(dummies.columns[0], axis=1)
df = pd.concat([df, dummies], axis=1)
df = df.drop(column, axis=1)
return df
## One Hot encode categorical features:
for column in ["Geography", "Gender", "Card Type"]:
datacorr = onehot_encode(datacorr, column=column)
X = datacorr.drop("Exited", axis=1)
y = datacorr.Exited
X
from sklearn.linear_model import LogisticRegression
from sklearn.discriminant_analysis import (
LinearDiscriminantAnalysis,
QuadraticDiscriminantAnalysis,
)
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
from catboost import CatBoostClassifier
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import accuracy_score
from sklearn.metrics import precision_score, recall_score, f1_score, roc_auc_score
models = {
"Logistic Regression": LogisticRegression(),
"Linear Discriminant Analysis": LinearDiscriminantAnalysis(),
"Quadratic Discriminant Analysis": QuadraticDiscriminantAnalysis(),
"K-Nearest Neighbors": KNeighborsClassifier(),
"Decision Tree": DecisionTreeClassifier(),
# "Random Forest": RandomForestClassifier(),
# "Gradient Boosting": GradientBoostingClassifier(),
# "XGBoost": XGBClassifier(),
# "LightGBM": LGBMClassifier(),
# "CatBoost": CatBoostClassifier(verbose=0),
# "Support Vector Machine (Linear Kernel)": SVC(kernel='linear', probability=True),
# "Support Vector Machine (RBF Kernel)": SVC(kernel='rbf', probability=True)
}
params = {
"Logistic Regression": {"C": [0.1, 1, 10]},
"Linear Discriminant Analysis": {"solver": ["svd", "lsqr"]},
"Quadratic Discriminant Analysis": {},
"K-Nearest Neighbors": {"n_neighbors": [3, 5, 7, 9]},
"Decision Tree": {"max_depth": [3, 5, 7, 9]},
"Random Forest": {"n_estimators": [50, 100, 150], "max_depth": [3, 5, 7, 9]},
"Gradient Boosting": {"learning_rate": [0.01, 0.1, 1], "max_depth": [3, 5, 7, 9]},
"XGBoost": {"learning_rate": [0.01, 0.1, 1], "max_depth": [3, 5, 7, 9]},
"LightGBM": {"learning_rate": [0.01, 0.1, 1], "max_depth": [3, 5, 7, 9]},
"CatBoost": {"learning_rate": [0.01, 0.1, 1], "max_depth": [3, 5, 7, 9]},
"Support Vector Machine (Linear Kernel)": {"C": [0.1, 1, 10]},
"Support Vector Machine (RBF Kernel)": {"C": [0.1, 1, 10], "gamma": [0.01, 0.1, 1]},
}
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
best_params = []
for name, model in models.items():
if name in params:
# print('\n')
print("Tuning hyperparameters for " + name)
param_grid = params[name]
grid_search = GridSearchCV(model, param_grid=param_grid, cv=5, n_jobs=-1)
grid_search.fit(X_train, y_train)
print("Best parameters:", grid_search.best_params_)
models[name] = grid_search.best_estimator_
# best_params[name] = grid_search.best_params_
else:
print("Skipping " + name + " as it does not have any hyperparameters to tune.")
results = []
for name, model in models.items():
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred, average="weighted")
recall = recall_score(y_test, y_pred, average="weighted")
f1 = f1_score(y_test, y_pred, average="weighted")
roc_auc = roc_auc_score(y_test, y_pred, multi_class="ovr")
results.append([name, accuracy, precision, recall, f1, roc_auc])
results_df = pd.DataFrame(
results, columns=["Model", "Accuracy", "Precision", "Recall", "F1 score", "ROC AUC"]
)
results_df = results_df.sort_values(by=["Accuracy"], ascending=False)
results_df
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/547/129547672.ipynb
|
bank-customer-churn
|
radheshyamkollipara
|
[{"Id": 129547672, "ScriptId": 38503843, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 9201197, "CreationDate": "05/14/2023 18:08:26", "VersionNumber": 2.0, "Title": "Subplots_Customer_churn", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 170.0, "LinesInsertedFromPrevious": 121.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 49.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 185719684, "KernelVersionId": 129547672, "SourceDatasetVersionId": 5550559}]
|
[{"Id": 5550559, "DatasetId": 3197960, "DatasourceVersionId": 5625285, "CreatorUserId": 14862076, "LicenseName": "Other (specified in description)", "CreationDate": "04/28/2023 16:32:01", "VersionNumber": 1.0, "Title": "Bank Customer Churn", "Slug": "bank-customer-churn", "Subtitle": "Bank Customer Data for Customer Churn", "Description": "RowNumber\u2014corresponds to the record (row) number and has no effect on the output.\nCustomerId\u2014contains random values and has no effect on customer leaving the bank.\nSurname\u2014the surname of a customer has no impact on their decision to leave the bank.\nCreditScore\u2014can have an effect on customer churn, since a customer with a higher credit score is less likely to leave the bank.\nGeography\u2014a customer\u2019s location can affect their decision to leave the bank.\nGender\u2014it\u2019s interesting to explore whether gender plays a role in a customer leaving the bank.\nAge\u2014this is certainly relevant, since older customers are less likely to leave their bank than younger ones.\nTenure\u2014refers to the number of years that the customer has been a client of the bank. Normally, older clients are more loyal and less likely to leave a bank.\nBalance\u2014also a very good indicator of customer churn, as people with a higher balance in their accounts are less likely to leave the bank compared to those with lower balances.\nNumOfProducts\u2014refers to the number of products that a customer has purchased through the bank.\nHasCrCard\u2014denotes whether or not a customer has a credit card. This column is also relevant, since people with a credit card are less likely to leave the bank.\nIsActiveMember\u2014active customers are less likely to leave the bank.\nEstimatedSalary\u2014as with balance, people with lower salaries are more likely to leave the bank compared to those with higher salaries.\nExited\u2014whether or not the customer left the bank.\nComplain\u2014customer has complaint or not.\nSatisfaction Score\u2014Score provided by the customer for their complaint resolution.\nCard Type\u2014type of card hold by the customer.\nPoints Earned\u2014the points earned by the customer for using credit card.\n\nAcknowledgements\n\nAs we know, it is much more expensive to sign in a new client than keeping an existing one.\n\nIt is advantageous for banks to know what leads a client towards the decision to leave the company.\n\nChurn prevention allows companies to develop loyalty programs and retention campaigns to keep as many customers as possible.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3197960, "CreatorUserId": 14862076, "OwnerUserId": 14862076.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5550559.0, "CurrentDatasourceVersionId": 5625285.0, "ForumId": 3262570, "Type": 2, "CreationDate": "04/28/2023 16:32:01", "LastActivityDate": "04/28/2023", "TotalViews": 39315, "TotalDownloads": 6814, "TotalVotes": 97, "TotalKernels": 52}]
|
[{"Id": 14862076, "UserName": "radheshyamkollipara", "DisplayName": "Radheshyam Kollipara", "RegisterDate": "04/28/2023", "PerformanceTier": 0}]
|
import warnings
warnings.filterwarnings("ignore")
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
cp = sns.color_palette("pastel")
plt.style.use(plt.style.available)
# plt.style.use('darkgrid')
# plt.style.available
data = pd.read_csv("/kaggle/input/bank-customer-churn/Customer-Churn-Records.csv")
data
feat = [
"Geography",
"Gender",
"NumOfProducts",
"HasCrCard",
"IsActiveMember",
"Exited",
"Complain",
"Satisfaction Score",
"Card Type",
]
fig, axes = plt.subplots(3, 3, figsize=(15, 13), sharex=False, sharey=False)
for i, feature in enumerate(feat):
row = i // 3
column = i % 3
ax = axes[row, column]
data.groupby(feature).count()["RowNumber"].sort_values(ascending=False).plot(
kind="bar", ax=ax, color=cp[1:]
)
ax.set_title(feature, backgroundcolor="skyblue", font="Arial", fontsize=14)
ax.tick_params(axis="x", labelrotation=0)
labels = data.groupby(feature).count()["RowNumber"].sort_values(ascending=False)
ax.bar_label(ax.containers[0], labels=labels, label_type="edge")
plt.tight_layout()
plt.show()
feat = ["CreditScore", "Age", "Balance"]
fig, axes = plt.subplots(2, 2, figsize=(15, 7), sharex=False, sharey=False)
for i, feature in enumerate(feat):
row = i // 2
column = i % 2
ax = axes[row, column]
sns.histplot(data, x=feature, ax=ax, kde=True, palette="pastel")
ax.tick_params(axis="x", labelrotation=45)
ax.set_title(feature, backgroundcolor="skyblue", font="Arial", fontsize=14)
plt.tight_layout()
plt.show()
sns.boxplot(data=data, x="Balance")
data.columns
datacorr = data[
[
"CreditScore",
"Geography",
"Gender",
"Age",
"Tenure",
"Balance",
"NumOfProducts",
"HasCrCard",
"IsActiveMember",
"EstimatedSalary",
"Exited",
"Complain",
"Satisfaction Score",
"Card Type",
"Point Earned",
]
]
correlation = datacorr.corr()
correlation.style.background_gradient(cmap="coolwarm")
# datacorr
## Work in progress
## Upvote if you like
## Suggestions are welcome
## Thanks
{
column: len(datacorr[column].unique())
for column in datacorr.select_dtypes("object").columns
}.keys()
def onehot_encode(df, column):
df = df.copy()
dummies = pd.get_dummies(df[column], prefix=column)
if len(df[column].unique()) == 2:
dummies = dummies.drop(dummies.columns[0], axis=1)
df = pd.concat([df, dummies], axis=1)
df = df.drop(column, axis=1)
return df
## One Hot encode categorical features:
for column in ["Geography", "Gender", "Card Type"]:
datacorr = onehot_encode(datacorr, column=column)
X = datacorr.drop("Exited", axis=1)
y = datacorr.Exited
X
from sklearn.linear_model import LogisticRegression
from sklearn.discriminant_analysis import (
LinearDiscriminantAnalysis,
QuadraticDiscriminantAnalysis,
)
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
from catboost import CatBoostClassifier
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import accuracy_score
from sklearn.metrics import precision_score, recall_score, f1_score, roc_auc_score
models = {
"Logistic Regression": LogisticRegression(),
"Linear Discriminant Analysis": LinearDiscriminantAnalysis(),
"Quadratic Discriminant Analysis": QuadraticDiscriminantAnalysis(),
"K-Nearest Neighbors": KNeighborsClassifier(),
"Decision Tree": DecisionTreeClassifier(),
# "Random Forest": RandomForestClassifier(),
# "Gradient Boosting": GradientBoostingClassifier(),
# "XGBoost": XGBClassifier(),
# "LightGBM": LGBMClassifier(),
# "CatBoost": CatBoostClassifier(verbose=0),
# "Support Vector Machine (Linear Kernel)": SVC(kernel='linear', probability=True),
# "Support Vector Machine (RBF Kernel)": SVC(kernel='rbf', probability=True)
}
params = {
"Logistic Regression": {"C": [0.1, 1, 10]},
"Linear Discriminant Analysis": {"solver": ["svd", "lsqr"]},
"Quadratic Discriminant Analysis": {},
"K-Nearest Neighbors": {"n_neighbors": [3, 5, 7, 9]},
"Decision Tree": {"max_depth": [3, 5, 7, 9]},
"Random Forest": {"n_estimators": [50, 100, 150], "max_depth": [3, 5, 7, 9]},
"Gradient Boosting": {"learning_rate": [0.01, 0.1, 1], "max_depth": [3, 5, 7, 9]},
"XGBoost": {"learning_rate": [0.01, 0.1, 1], "max_depth": [3, 5, 7, 9]},
"LightGBM": {"learning_rate": [0.01, 0.1, 1], "max_depth": [3, 5, 7, 9]},
"CatBoost": {"learning_rate": [0.01, 0.1, 1], "max_depth": [3, 5, 7, 9]},
"Support Vector Machine (Linear Kernel)": {"C": [0.1, 1, 10]},
"Support Vector Machine (RBF Kernel)": {"C": [0.1, 1, 10], "gamma": [0.01, 0.1, 1]},
}
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
best_params = []
for name, model in models.items():
if name in params:
# print('\n')
print("Tuning hyperparameters for " + name)
param_grid = params[name]
grid_search = GridSearchCV(model, param_grid=param_grid, cv=5, n_jobs=-1)
grid_search.fit(X_train, y_train)
print("Best parameters:", grid_search.best_params_)
models[name] = grid_search.best_estimator_
# best_params[name] = grid_search.best_params_
else:
print("Skipping " + name + " as it does not have any hyperparameters to tune.")
results = []
for name, model in models.items():
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred, average="weighted")
recall = recall_score(y_test, y_pred, average="weighted")
f1 = f1_score(y_test, y_pred, average="weighted")
roc_auc = roc_auc_score(y_test, y_pred, multi_class="ovr")
results.append([name, accuracy, precision, recall, f1, roc_auc])
results_df = pd.DataFrame(
results, columns=["Model", "Accuracy", "Precision", "Recall", "F1 score", "ROC AUC"]
)
results_df = results_df.sort_values(by=["Accuracy"], ascending=False)
results_df
| false | 1 | 2,073 | 3 | 2,574 | 2,073 |
||
129889353
|
# # Library
import pandas as pd
import numpy as np
from sklearn.preprocessing import OneHotEncoder, MinMaxScaler
from sklearn.linear_model import LogisticRegression
import matplotlib.pyplot as plt
import seaborn as sns
# # Load data
data_dir = "/kaggle/input/loan-status-binary-classification/"
train = pd.read_csv(data_dir + "train.csv")
test = pd.read_csv(data_dir + "test.csv")
sample_submission = pd.read_csv(data_dir + "sample_submission.csv")
# print(train.shape)
# print(train.head())
# print('=============================')
# print(test.shape)
# print(test.head())
# # Data Exploration
train.Gender.value_counts(dropna=False)
sns.countplot(x="Gender", data=train)
plt.show()
train.Term.value_counts(dropna=False)
train.Education.value_counts(dropna=False)
train.Married.value_counts(dropna=False)
# ## check null values in each column
for col in train.columns:
print(f"""[{col}] has [{train[col].isnull().sum()}] null values""")
# print(col, train[col].isnull().sum())
# ## Dealing with missing values
# There should be different ways to deal with missing values in different columns.
# For example, for columns with **numerical** values, we can fill the missing values with the **mean** of the column; For columns with **categorical** values, we can fill the missing values with the **mode** of the column.
# For all columns with missing values, numerical variables are:
# - `Term`
# For all columns with missing values, categorical variables are:
# - `Gender`
# - `Married`
# - `Self_Employed`
# - `Credit_History`
# - `Dependents`
# We then deal with missing values in each column.
# ### Dealing with categorical variables
# complete missing value in column `Gender` in train and test set with mode in train
mode_gender = train["Gender"].mode()[0]
train["Gender"].fillna(mode_gender, inplace=True)
test["Gender"].fillna(mode_gender, inplace=True)
# do the same as above for column `Married`, `self_Employed`, `Dependents` and `Credit_History`
# `Married`
mode_married = train["Married"].mode()[0]
train["Married"].fillna(mode_married, inplace=True)
test["Married"].fillna(mode_married, inplace=True)
# `Self_Employed`
mode_self_employed = train["Self_Employed"].mode()[0]
train["Self_Employed"].fillna(mode_self_employed, inplace=True)
test["Self_Employed"].fillna(mode_self_employed, inplace=True)
# `Dependents`
mode_dependents = train["Dependents"].mode()[0]
train["Dependents"].fillna(mode_dependents, inplace=True)
test["Dependents"].fillna(mode_dependents, inplace=True)
# `Credit_History`
mode_credit_history = train["Credit_History"].mode()[0]
train["Credit_History"].fillna(mode_credit_history, inplace=True)
test["Credit_History"].fillna(mode_credit_history, inplace=True)
# ### Dealing with numerical variables
# complete missing value in column `Term` in train and test set with mean in train
mean_term = train["Term"].mean()
train["Term"].fillna(mean_term, inplace=True)
test["Term"].fillna(mean_term, inplace=True)
# # Feature Engineering
# # Encoding Categorical Variables
# type('Gender')
# type('Married')
# type('Dependents')
# type('Education')
# type('Self_Employed')
# type('Credit_History')
# type('Area')
# Select the categorical variables to be encoded
cat_vars = [
"Gender",
"Married",
"Dependents",
"Education",
"Self_Employed",
"Credit_History",
"Area",
]
# Concatenate the training and testing sets
all_data = pd.concat([train, test], axis=0)
# 将train和test合起来,pd.concat能够把某一个维度上的数据框合在一起
# pd.concat( objs, axis=0, join=‘outer’, join_axes=None,ignore_index=False, keys=None, levels=None, names=None, verify_integrity=False, sort=None, copy=True,)
# objs 表示需要连接的对象,比如:[df1, df2],需要将合并的数据用综括号包围;
# axis=0 表拼接方式是上下堆叠,当axis=1表示左右拼接;一般默认是上下堆叠
# join 参数控制的是外连接还是内连接,join='outer’表示外连接,保留两个表中的所有信息;join="inner"表示内连接,拼接结果只保留两个表共有的信息;
# join_axes参数是在内连接时选择要完整保留哪个表的索引,但是这个参数在官方文档中提醒即将被弃用,所以不做详细讲解,只看一下join参数的表现吧;
# Create an instance of the one-hot encoder
enc = OneHotEncoder()
# 100 010 001 将一个变量变成三个维度,分离详细信息
# Fit the encoder on all data and transform the categorical variables
all_data_cat_encoded = enc.fit_transform(all_data[cat_vars])
# Split the encoded data back into training and testing sets
train_cat_encoded = all_data_cat_encoded[: train.shape[0]]
test_cat_encoded = all_data_cat_encoded[train.shape[0] :]
# Print the shape of the encoded training and testing sets
print(train_cat_encoded.shape)
print(test_cat_encoded.shape)
# 存储的信息更多,也更加准确
# ### Have a look at the datatype of `train_encoded` and `test_encoded`, it's tricky!
# print(type(test_cat_encoded)) it is a matrix
print(test_cat_encoded)
print(test_cat_encoded.toarray())
# 转化成可看的矩阵
# # Scaling Numerical Variables
train.columns
# select the numerical variables to be scaled
num_vars = ["Applicant_Income", "Coapplicant_Income", "Loan_Amount", "Term"]
# + \
# ['Total_Income'] # add the interaction terms
# Create an instance of the MinMaxScaler
scaler = MinMaxScaler()
# 把所有的数变换成0-1之间的数,找到最大最小投到0-1之间,(value-min)/(max-min)
# 效果:有些数值单位不一样,可能差别很大,投影以后更加准确
# Fit the scaler on the training set and transform both the training and test sets
train_num_scaled = scaler.fit_transform(train[num_vars])
test_num_scaled = scaler.transform(test[num_vars])
# 在测试集中不要fit,因为这样就导致将数据透露给模型
# 在encode里面合在一起是为了不漏掉变量
print(train_num_scaled.shape)
print(test_num_scaled.shape)
# print(type(test_num_scaled)) it is an array
print(test_num_scaled)
# # Concatenate transformed categorical featuers and numerical features
# 类别变量和数字变量的输出都是矩阵
# ## Example of using `np.hstack`
# 水平线上进行拼接
# Suppose we have two arrays `a` and `b`:
# ```python
# import numpy as np
# a = np.array([1, 2, 3])
# b = np.array([4, 5, 6])
# ```
# If we want to horizontally stack these two arrays, we can use `np.hstack`:
# ```python
# c = np.hstack((a, b))
# print(c)
# ```
# This will output:
# ```
# [1 2 3 4 5 6]
# ```
# Here, `np.hstack` takes a tuple of arrays as its argument, and stacks them horizontally (i.e., along the second axis).
import numpy as np
# concatenate the encoded categorical features and the scaled numerical features for train set
train_processed = np.hstack((train_cat_encoded.toarray(), train_num_scaled))
# concatenate the encoded categorical features and the scaled numerical features for test set
test_processed = np.hstack((test_cat_encoded.toarray(), test_num_scaled))
# # Using SVC(104 19)
from sklearn.svm import SVC
clf = SVC()
# Fit the classifier
clf.fit(train_processed, train["Status"])
# Make predictions
preds = clf.predict(test_processed)
# # Using Logistic Regression to do prediction(104 19)
# from sklearn.linear_model import LogisticRegression
# # Instantiate the classifier
# clf = LogisticRegression()
# # Fit the classifier
# clf.fit(train_processed, train['Status'])
# # Make predictions
# preds = clf.predict(test_processed)
# # Create a submission dataframe
# Create a submission dataframe
sub = pd.DataFrame({"id": test["id"], "Status": preds})
# Write the submission dataframe to a csv file
sub.to_csv("/kaggle/working/submission.csv", index=False)
result = pd.read_csv("submission.csv")
result["Status"].value_counts(dropna=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/889/129889353.ipynb
| null | null |
[{"Id": 129889353, "ScriptId": 38635053, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 15122399, "CreationDate": "05/17/2023 08:20:27", "VersionNumber": 1.0, "Title": "second hand in(SVC)", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 243.0, "LinesInsertedFromPrevious": 243.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Library
import pandas as pd
import numpy as np
from sklearn.preprocessing import OneHotEncoder, MinMaxScaler
from sklearn.linear_model import LogisticRegression
import matplotlib.pyplot as plt
import seaborn as sns
# # Load data
data_dir = "/kaggle/input/loan-status-binary-classification/"
train = pd.read_csv(data_dir + "train.csv")
test = pd.read_csv(data_dir + "test.csv")
sample_submission = pd.read_csv(data_dir + "sample_submission.csv")
# print(train.shape)
# print(train.head())
# print('=============================')
# print(test.shape)
# print(test.head())
# # Data Exploration
train.Gender.value_counts(dropna=False)
sns.countplot(x="Gender", data=train)
plt.show()
train.Term.value_counts(dropna=False)
train.Education.value_counts(dropna=False)
train.Married.value_counts(dropna=False)
# ## check null values in each column
for col in train.columns:
print(f"""[{col}] has [{train[col].isnull().sum()}] null values""")
# print(col, train[col].isnull().sum())
# ## Dealing with missing values
# There should be different ways to deal with missing values in different columns.
# For example, for columns with **numerical** values, we can fill the missing values with the **mean** of the column; For columns with **categorical** values, we can fill the missing values with the **mode** of the column.
# For all columns with missing values, numerical variables are:
# - `Term`
# For all columns with missing values, categorical variables are:
# - `Gender`
# - `Married`
# - `Self_Employed`
# - `Credit_History`
# - `Dependents`
# We then deal with missing values in each column.
# ### Dealing with categorical variables
# complete missing value in column `Gender` in train and test set with mode in train
mode_gender = train["Gender"].mode()[0]
train["Gender"].fillna(mode_gender, inplace=True)
test["Gender"].fillna(mode_gender, inplace=True)
# do the same as above for column `Married`, `self_Employed`, `Dependents` and `Credit_History`
# `Married`
mode_married = train["Married"].mode()[0]
train["Married"].fillna(mode_married, inplace=True)
test["Married"].fillna(mode_married, inplace=True)
# `Self_Employed`
mode_self_employed = train["Self_Employed"].mode()[0]
train["Self_Employed"].fillna(mode_self_employed, inplace=True)
test["Self_Employed"].fillna(mode_self_employed, inplace=True)
# `Dependents`
mode_dependents = train["Dependents"].mode()[0]
train["Dependents"].fillna(mode_dependents, inplace=True)
test["Dependents"].fillna(mode_dependents, inplace=True)
# `Credit_History`
mode_credit_history = train["Credit_History"].mode()[0]
train["Credit_History"].fillna(mode_credit_history, inplace=True)
test["Credit_History"].fillna(mode_credit_history, inplace=True)
# ### Dealing with numerical variables
# complete missing value in column `Term` in train and test set with mean in train
mean_term = train["Term"].mean()
train["Term"].fillna(mean_term, inplace=True)
test["Term"].fillna(mean_term, inplace=True)
# # Feature Engineering
# # Encoding Categorical Variables
# type('Gender')
# type('Married')
# type('Dependents')
# type('Education')
# type('Self_Employed')
# type('Credit_History')
# type('Area')
# Select the categorical variables to be encoded
cat_vars = [
"Gender",
"Married",
"Dependents",
"Education",
"Self_Employed",
"Credit_History",
"Area",
]
# Concatenate the training and testing sets
all_data = pd.concat([train, test], axis=0)
# 将train和test合起来,pd.concat能够把某一个维度上的数据框合在一起
# pd.concat( objs, axis=0, join=‘outer’, join_axes=None,ignore_index=False, keys=None, levels=None, names=None, verify_integrity=False, sort=None, copy=True,)
# objs 表示需要连接的对象,比如:[df1, df2],需要将合并的数据用综括号包围;
# axis=0 表拼接方式是上下堆叠,当axis=1表示左右拼接;一般默认是上下堆叠
# join 参数控制的是外连接还是内连接,join='outer’表示外连接,保留两个表中的所有信息;join="inner"表示内连接,拼接结果只保留两个表共有的信息;
# join_axes参数是在内连接时选择要完整保留哪个表的索引,但是这个参数在官方文档中提醒即将被弃用,所以不做详细讲解,只看一下join参数的表现吧;
# Create an instance of the one-hot encoder
enc = OneHotEncoder()
# 100 010 001 将一个变量变成三个维度,分离详细信息
# Fit the encoder on all data and transform the categorical variables
all_data_cat_encoded = enc.fit_transform(all_data[cat_vars])
# Split the encoded data back into training and testing sets
train_cat_encoded = all_data_cat_encoded[: train.shape[0]]
test_cat_encoded = all_data_cat_encoded[train.shape[0] :]
# Print the shape of the encoded training and testing sets
print(train_cat_encoded.shape)
print(test_cat_encoded.shape)
# 存储的信息更多,也更加准确
# ### Have a look at the datatype of `train_encoded` and `test_encoded`, it's tricky!
# print(type(test_cat_encoded)) it is a matrix
print(test_cat_encoded)
print(test_cat_encoded.toarray())
# 转化成可看的矩阵
# # Scaling Numerical Variables
train.columns
# select the numerical variables to be scaled
num_vars = ["Applicant_Income", "Coapplicant_Income", "Loan_Amount", "Term"]
# + \
# ['Total_Income'] # add the interaction terms
# Create an instance of the MinMaxScaler
scaler = MinMaxScaler()
# 把所有的数变换成0-1之间的数,找到最大最小投到0-1之间,(value-min)/(max-min)
# 效果:有些数值单位不一样,可能差别很大,投影以后更加准确
# Fit the scaler on the training set and transform both the training and test sets
train_num_scaled = scaler.fit_transform(train[num_vars])
test_num_scaled = scaler.transform(test[num_vars])
# 在测试集中不要fit,因为这样就导致将数据透露给模型
# 在encode里面合在一起是为了不漏掉变量
print(train_num_scaled.shape)
print(test_num_scaled.shape)
# print(type(test_num_scaled)) it is an array
print(test_num_scaled)
# # Concatenate transformed categorical featuers and numerical features
# 类别变量和数字变量的输出都是矩阵
# ## Example of using `np.hstack`
# 水平线上进行拼接
# Suppose we have two arrays `a` and `b`:
# ```python
# import numpy as np
# a = np.array([1, 2, 3])
# b = np.array([4, 5, 6])
# ```
# If we want to horizontally stack these two arrays, we can use `np.hstack`:
# ```python
# c = np.hstack((a, b))
# print(c)
# ```
# This will output:
# ```
# [1 2 3 4 5 6]
# ```
# Here, `np.hstack` takes a tuple of arrays as its argument, and stacks them horizontally (i.e., along the second axis).
import numpy as np
# concatenate the encoded categorical features and the scaled numerical features for train set
train_processed = np.hstack((train_cat_encoded.toarray(), train_num_scaled))
# concatenate the encoded categorical features and the scaled numerical features for test set
test_processed = np.hstack((test_cat_encoded.toarray(), test_num_scaled))
# # Using SVC(104 19)
from sklearn.svm import SVC
clf = SVC()
# Fit the classifier
clf.fit(train_processed, train["Status"])
# Make predictions
preds = clf.predict(test_processed)
# # Using Logistic Regression to do prediction(104 19)
# from sklearn.linear_model import LogisticRegression
# # Instantiate the classifier
# clf = LogisticRegression()
# # Fit the classifier
# clf.fit(train_processed, train['Status'])
# # Make predictions
# preds = clf.predict(test_processed)
# # Create a submission dataframe
# Create a submission dataframe
sub = pd.DataFrame({"id": test["id"], "Status": preds})
# Write the submission dataframe to a csv file
sub.to_csv("/kaggle/working/submission.csv", index=False)
result = pd.read_csv("submission.csv")
result["Status"].value_counts(dropna=False)
| false | 0 | 2,333 | 0 | 2,333 | 2,333 |
||
129889715
|
<jupyter_start><jupyter_text>Drugs A, B, C, X, Y for Decision Trees
Imagine that you are a medical researcher compiling data for a study. You have collected data about a set of patients, all of whom suffered from the same illness. During their course of treatment, each patient responded to one of 5 medications, Drug A, Drug B, Drug c, Drug x and y.
Part of your job is to build a model to find out which drug might be appropriate for a future patient with the same illness. The features of this dataset are Age, Sex, Blood Pressure, and the Cholesterol of the patients, and the target is the drug that each patient responded to.
It is a sample of multiclass classifier, and you can use the training part of the dataset to build a decision tree, and then use it to predict the class of a unknown patient, or to prescribe a drug to a new patient.
DATA Source: IBM
Kaggle dataset identifier: drugs-a-b-c-x-y-for-decision-trees
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from warnings import filterwarnings
filterwarnings("ignore")
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn import tree
from sklearn.metrics import accuracy_score
# Read Data
df = pd.read_csv("/kaggle/input/drugs-a-b-c-x-y-for-decision-trees/drug200.csv")
df.head()
df.isnull().sum()
df.size
# > Data has categorical values.Hence I used encoding to change the BP, cholestrol and Sex into Categorical data
df.info()
df.columns
# Encoding features
features_list = ["Sex", "BP", "Cholesterol", "Drug"]
for i in features_list:
print(i + " " + str(df[i].unique()))
from category_encoders import OrdinalEncoder
mapping = [{"col": "Sex", "mapping": {"F": 0, "M": 1}}]
df = OrdinalEncoder(cols=["Sex"], mapping=mapping).fit(df).transform(df)
mapping = [{"col": "Cholesterol", "mapping": {"HIGH": 1, "NORMAL": 0}}]
df = OrdinalEncoder(cols=["Cholesterol"], mapping=mapping).fit(df).transform(df)
mapping = [{"col": "BP", "mapping": {"HIGH": 2, "NORMAL": 1, "LOW": 0}}]
df = OrdinalEncoder(cols=["BP"], mapping=mapping).fit(df).transform(df)
mapping = [
{
"col": "Drug",
"mapping": {"drugA": 0, "drugB": 1, "drugC": 2, "drugX": 3, "drugY": 4},
}
]
df = OrdinalEncoder(cols=["Drug"], mapping=mapping).fit(df).transform(df)
df
df.isnull().sum()
X = df.drop("Drug", axis=1)
y = df["Drug"]
# Data splitting and training the model
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=1
)
model = DecisionTreeClassifier()
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
accuracy_score(y_test, y_pred)
from sklearn import tree
tree.plot_tree(model)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/889/129889715.ipynb
|
drugs-a-b-c-x-y-for-decision-trees
|
pablomgomez21
|
[{"Id": 129889715, "ScriptId": 38584271, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4441620, "CreationDate": "05/17/2023 08:22:41", "VersionNumber": 1.0, "Title": "DecisionTree", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 85.0, "LinesInsertedFromPrevious": 85.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186298432, "KernelVersionId": 129889715, "SourceDatasetVersionId": 2371056}]
|
[{"Id": 2371056, "DatasetId": 1432504, "DatasourceVersionId": 2412902, "CreatorUserId": 6769968, "LicenseName": "Unknown", "CreationDate": "06/26/2021 11:13:21", "VersionNumber": 1.0, "Title": "Drugs A, B, C, X, Y for Decision Trees", "Slug": "drugs-a-b-c-x-y-for-decision-trees", "Subtitle": "Practice DATASET for Decision Trees learning", "Description": "Imagine that you are a medical researcher compiling data for a study. You have collected data about a set of patients, all of whom suffered from the same illness. During their course of treatment, each patient responded to one of 5 medications, Drug A, Drug B, Drug c, Drug x and y.\n\nPart of your job is to build a model to find out which drug might be appropriate for a future patient with the same illness. The features of this dataset are Age, Sex, Blood Pressure, and the Cholesterol of the patients, and the target is the drug that each patient responded to.\n\nIt is a sample of multiclass classifier, and you can use the training part of the dataset to build a decision tree, and then use it to predict the class of a unknown patient, or to prescribe a drug to a new patient.\n\nDATA Source: IBM", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1432504, "CreatorUserId": 6769968, "OwnerUserId": 6769968.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2371056.0, "CurrentDatasourceVersionId": 2412902.0, "ForumId": 1451957, "Type": 2, "CreationDate": "06/26/2021 11:13:21", "LastActivityDate": "06/26/2021", "TotalViews": 36696, "TotalDownloads": 6216, "TotalVotes": 71, "TotalKernels": 48}]
|
[{"Id": 6769968, "UserName": "pablomgomez21", "DisplayName": "Pablo M. G\u00f3mez", "RegisterDate": "02/20/2021", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from warnings import filterwarnings
filterwarnings("ignore")
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn import tree
from sklearn.metrics import accuracy_score
# Read Data
df = pd.read_csv("/kaggle/input/drugs-a-b-c-x-y-for-decision-trees/drug200.csv")
df.head()
df.isnull().sum()
df.size
# > Data has categorical values.Hence I used encoding to change the BP, cholestrol and Sex into Categorical data
df.info()
df.columns
# Encoding features
features_list = ["Sex", "BP", "Cholesterol", "Drug"]
for i in features_list:
print(i + " " + str(df[i].unique()))
from category_encoders import OrdinalEncoder
mapping = [{"col": "Sex", "mapping": {"F": 0, "M": 1}}]
df = OrdinalEncoder(cols=["Sex"], mapping=mapping).fit(df).transform(df)
mapping = [{"col": "Cholesterol", "mapping": {"HIGH": 1, "NORMAL": 0}}]
df = OrdinalEncoder(cols=["Cholesterol"], mapping=mapping).fit(df).transform(df)
mapping = [{"col": "BP", "mapping": {"HIGH": 2, "NORMAL": 1, "LOW": 0}}]
df = OrdinalEncoder(cols=["BP"], mapping=mapping).fit(df).transform(df)
mapping = [
{
"col": "Drug",
"mapping": {"drugA": 0, "drugB": 1, "drugC": 2, "drugX": 3, "drugY": 4},
}
]
df = OrdinalEncoder(cols=["Drug"], mapping=mapping).fit(df).transform(df)
df
df.isnull().sum()
X = df.drop("Drug", axis=1)
y = df["Drug"]
# Data splitting and training the model
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=1
)
model = DecisionTreeClassifier()
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
accuracy_score(y_test, y_pred)
from sklearn import tree
tree.plot_tree(model)
| false | 1 | 794 | 0 | 1,041 | 794 |
||
129530641
|
<jupyter_start><jupyter_text>Financial Inclusion in Africa
You are asked to predict the likelihood of the person having a bank account or not (Yes = 1, No = 0), for each unique id in the test dataset . You will train your model on 70% of the data and test your model on the final 30% of the data, across four East African countries - Kenya, Rwanda, Tanzania, and Uganda.
Kaggle dataset identifier: financial-inclusion-in-africa
<jupyter_script>import pandas as pd
train = pd.read_csv("/kaggle/input/financial-inclusion-in-africa/Train.csv")
train.head()
test = pd.read_csv("/kaggle/input/financial-inclusion-in-africa/Test.csv")
test.head()
train.nunique()
train.info()
train = train.drop(["uniqueid"], axis=1)
train.head()
test = test.drop(["uniqueid"], axis=1)
test.head()
train["country"].value_counts()
train["bank_account"] = train["bank_account"].replace({"Yes": 1, "No": 0})
train.head()
train["cellphone_access"] = train["cellphone_access"].replace({"Yes": 1, "No": 0})
train.head()
test["cellphone_access"] = test["cellphone_access"].replace({"Yes": 1, "No": 0})
test.head()
train["relationship_with_head"].value_counts()
train["marital_status"].value_counts()
train["education_level"].value_counts()
train["job_type"].value_counts()
round(train["bank_account"].value_counts() * 100 / len(train), 2)
from sklearn.metrics import mean_absolute_percentage_error
from flaml import AutoML
automl = AutoML()
y = train.pop("bank_account")
X = train
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42, shuffle=True, stratify=y
)
automl.fit(X_train, y_train, task="classification", metric="roc_auc", time_budget=1800)
print("Best ML leaner:", automl.best_estimator)
print("Best hyperparmeter config:", automl.best_config)
print("Best roc_auc on validation data: {0:.4g}".format(1 - automl.best_loss))
print("Training duration of best run: {0:.4g} s".format(automl.best_config_train_time))
from sklearn.metrics import classification_report
print(classification_report(y_train, automl.predict(X_train)))
print(classification_report(y_test, automl.predict(X_test)))
y_pred = automl.predict(test)
y_pred[:5]
df = pd.DataFrame(y_pred, columns=["bank_account"])
df.head()
sol = pd.read_csv("/kaggle/input/financial-inclusion-in-africa/Test.csv")
sol = sol[["uniqueid", "country"]]
sol.head()
sol["unique_id"] = sol["uniqueid"] + " x " + sol["country"]
sol = sol[["unique_id"]]
sol.head()
sol["bank_account"] = df["bank_account"]
sol.head()
sol.to_csv("./roc_auc.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/530/129530641.ipynb
|
financial-inclusion-in-africa
|
gauravduttakiit
|
[{"Id": 129530641, "ScriptId": 38514284, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4760409, "CreationDate": "05/14/2023 15:17:06", "VersionNumber": 2.0, "Title": "Bank Account Prediction : FLAML : roc_auc", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 80.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 80.0, "LinesInsertedFromFork": 31.0, "LinesDeletedFromFork": 9.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 49.0, "TotalVotes": 2}]
|
[{"Id": 185682830, "KernelVersionId": 129530641, "SourceDatasetVersionId": 5683202}]
|
[{"Id": 5683202, "DatasetId": 3267290, "DatasourceVersionId": 5758770, "CreatorUserId": 4760409, "LicenseName": "Unknown", "CreationDate": "05/14/2023 13:56:48", "VersionNumber": 1.0, "Title": "Financial Inclusion in Africa", "Slug": "financial-inclusion-in-africa", "Subtitle": NaN, "Description": "You are asked to predict the likelihood of the person having a bank account or not (Yes = 1, No = 0), for each unique id in the test dataset . You will train your model on 70% of the data and test your model on the final 30% of the data, across four East African countries - Kenya, Rwanda, Tanzania, and Uganda.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3267290, "CreatorUserId": 4760409, "OwnerUserId": 4760409.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5683202.0, "CurrentDatasourceVersionId": 5758770.0, "ForumId": 3332906, "Type": 2, "CreationDate": "05/14/2023 13:56:48", "LastActivityDate": "05/14/2023", "TotalViews": 108, "TotalDownloads": 6, "TotalVotes": 1, "TotalKernels": 2}]
|
[{"Id": 4760409, "UserName": "gauravduttakiit", "DisplayName": "Gaurav Dutta", "RegisterDate": "03/28/2020", "PerformanceTier": 3}]
|
import pandas as pd
train = pd.read_csv("/kaggle/input/financial-inclusion-in-africa/Train.csv")
train.head()
test = pd.read_csv("/kaggle/input/financial-inclusion-in-africa/Test.csv")
test.head()
train.nunique()
train.info()
train = train.drop(["uniqueid"], axis=1)
train.head()
test = test.drop(["uniqueid"], axis=1)
test.head()
train["country"].value_counts()
train["bank_account"] = train["bank_account"].replace({"Yes": 1, "No": 0})
train.head()
train["cellphone_access"] = train["cellphone_access"].replace({"Yes": 1, "No": 0})
train.head()
test["cellphone_access"] = test["cellphone_access"].replace({"Yes": 1, "No": 0})
test.head()
train["relationship_with_head"].value_counts()
train["marital_status"].value_counts()
train["education_level"].value_counts()
train["job_type"].value_counts()
round(train["bank_account"].value_counts() * 100 / len(train), 2)
from sklearn.metrics import mean_absolute_percentage_error
from flaml import AutoML
automl = AutoML()
y = train.pop("bank_account")
X = train
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42, shuffle=True, stratify=y
)
automl.fit(X_train, y_train, task="classification", metric="roc_auc", time_budget=1800)
print("Best ML leaner:", automl.best_estimator)
print("Best hyperparmeter config:", automl.best_config)
print("Best roc_auc on validation data: {0:.4g}".format(1 - automl.best_loss))
print("Training duration of best run: {0:.4g} s".format(automl.best_config_train_time))
from sklearn.metrics import classification_report
print(classification_report(y_train, automl.predict(X_train)))
print(classification_report(y_test, automl.predict(X_test)))
y_pred = automl.predict(test)
y_pred[:5]
df = pd.DataFrame(y_pred, columns=["bank_account"])
df.head()
sol = pd.read_csv("/kaggle/input/financial-inclusion-in-africa/Test.csv")
sol = sol[["uniqueid", "country"]]
sol.head()
sol["unique_id"] = sol["uniqueid"] + " x " + sol["country"]
sol = sol[["unique_id"]]
sol.head()
sol["bank_account"] = df["bank_account"]
sol.head()
sol.to_csv("./roc_auc.csv", index=False)
| false | 2 | 747 | 2 | 866 | 747 |
||
129530321
|
import warnings
warnings.filterwarnings("ignore")
import pandas as pd
import numpy as np
import re
import string
from collections import Counter
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, confusion_matrix, roc_curve, auc
from keras_preprocessing.sequence import pad_sequences
import torch
from torch.utils.data import TensorDataset, DataLoader, RandomSampler, SequentialSampler
from transformers import XLNetTokenizer, XLNetForSequenceClassification
from pytorch_transformers import AdamW
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
from tqdm import tqdm, trange
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
df = pd.read_csv("/kaggle/input/graduation-project/kaggle-dataset.csv")
# Drop any rows with missing values in the Sentence column & Convert the Sentence column to strings
df = df.dropna(subset=["Sentence"])
df.Sentence = [str(text) for text in df.Sentence]
# Randomly sample 250K rows from the dataframe (Due to limited resources)
df = df.sample(n=250000, random_state=0)
print(f"Dataframe shape: {df.shape}")
df["Subreddit"].value_counts()
label_encoder = preprocessing.LabelEncoder()
# Encode the Subreddit column of the df dataframe & Store the label names in a list
df["Subreddit"] = label_encoder.fit_transform(df["Subreddit"])
classes = list(label_encoder.classes_)
# Print two random rows from the encoded dataframe
print(df.sample(2))
# Get sentences and labels from data
sentences = df.Sentence.values
labels = df.Subreddit.values
# Add special tokens to sentences
sentences = [str(sentence) + " [SEP] [CLS]" for sentence in sentences]
# Initialize XLNetTokenizer with pre-trained model
tokenizer = XLNetTokenizer.from_pretrained("xlnet-base-cased")
# Tokenize sentences using tokenizer
tokenized_texts = [tokenizer.tokenize(sent) for sent in sentences]
# Print tokenized version of the first sentence
print("Tokenize the first sentence:")
print(tokenized_texts[0])
MAX_LEN = 128
# Convert tokenized texts to input IDs
input_ids = [tokenizer.convert_tokens_to_ids(x) for x in tokenized_texts]
# Pad input IDs to a maximum length of MAX_LEN
input_ids = pad_sequences(
input_ids, maxlen=MAX_LEN, dtype="long", truncating="post", padding="post"
)
# Create attention masks
attention_masks = []
# Create a mask of 1s for each token followed by 0s for padding
for seq in input_ids:
seq_mask = [float(i > 0) for i in seq]
attention_masks.append(seq_mask)
# Split data into training , validation and testing sets
train_inputs, test_inputs, train_labels, test_labels = train_test_split(
input_ids, labels, random_state=56, test_size=0.2
)
train_masks, test_masks, _, _ = train_test_split(
attention_masks, input_ids, random_state=56, test_size=0.2
)
test_inputs, validation_inputs, test_labels, validation_labels = train_test_split(
train_inputs, train_labels, random_state=56, test_size=0.5
)
test_masks, validation_masks, _, _ = train_test_split(
train_masks, train_inputs, random_state=56, test_size=0.5
)
# Convert data to PyTorch tensors
train_inputs = torch.tensor(train_inputs)
train_labels = torch.tensor(train_labels)
train_masks = torch.tensor(train_masks)
validation_inputs = torch.tensor(validation_inputs)
validation_labels = torch.tensor(validation_labels)
validation_masks = torch.tensor(validation_masks)
test_inputs = torch.tensor(test_inputs)
test_labels = torch.tensor(test_labels)
test_masks = torch.tensor(test_masks)
train_inputs[0]
# Define batch size
batch_size = 32
# Create training data iterator
train_data = TensorDataset(train_inputs, train_masks, train_labels)
train_sampler = RandomSampler(train_data)
train_dataloader = DataLoader(train_data, sampler=train_sampler, batch_size=batch_size)
# Create validation data iterator
validation_data = TensorDataset(validation_inputs, validation_masks, validation_labels)
validation_sampler = SequentialSampler(validation_data)
validation_dataloader = DataLoader(
validation_data, sampler=validation_sampler, batch_size=batch_size
)
# Create validation data iterator
test_data = TensorDataset(test_inputs, test_masks, test_labels)
test_sampler = SequentialSampler(test_data)
test_dataloader = DataLoader(test_data, sampler=test_sampler, batch_size=batch_size)
# Load pre-trained XLNet model for sequence classification
model = XLNetForSequenceClassification.from_pretrained("xlnet-base-cased", num_labels=5)
# Move model to GPU for faster training
model.cuda()
# Get model parameters and group them by weight decay rate
param_optimizer = list(model.named_parameters())
no_decay = ["bias", "gamma", "beta"]
optimizer_grouped_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.01,
},
{
"params": [p for n, p in param_optimizer if any(nd in n for nd in no_decay)],
"weight_decay_rate": 0.0,
},
]
# Create AdamW optimizer with learning rate of 2e-5
optimizer = AdamW(optimizer_grouped_parameters, lr=2e-5)
# Determine device type for training (GPU or CPU)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Get name of GPU device
torch.cuda.get_device_name()
# Initialize variables
train_loss_set = []
train_acc_set = []
val_loss_set = []
val_acc_set = []
epochs = 10
best_loss = 0
early_stop_counter = 0
# Train for each epoch
for epoch in range(epochs):
print(f"Epoch {epoch+1}/{epochs}")
model.train()
nb_tr_examples, nb_tr_steps = 0, 0
# Train the data for one epoch and calculate training loss and accuracy
train_acc, train_loss = 0, 0
for step, batch in enumerate(tqdm(train_dataloader, desc=f"Training")):
batch = tuple(t.to(device) for t in batch)
b_input_ids, b_input_mask, b_labels = batch
optimizer.zero_grad()
outputs = model(
b_input_ids,
token_type_ids=None,
attention_mask=b_input_mask,
labels=b_labels,
)
loss = outputs[0]
logits = outputs[1]
loss.backward()
optimizer.step()
train_loss += loss.item()
nb_tr_examples += b_input_ids.size(0)
nb_tr_steps += 1
# Calculate training accuracy
prediction = torch.argmax(logits, dim=1)
train_acc += (prediction == b_labels).sum().item()
# Calculate training loss and accuracy for the epoch
train_loss = train_loss / nb_tr_steps
train_acc /= nb_tr_examples
train_acc_set.append(train_acc)
train_loss_set.append(train_loss)
# Calculate validation loss and accuracy
val_loss, val_acc = 0, 0
with torch.no_grad():
for batch in tqdm(validation_dataloader, desc=f"Validation "):
batch = tuple(t.to(device) for t in batch)
b_input_ids, b_input_mask, b_labels = batch
outputs = model(
b_input_ids,
token_type_ids=None,
attention_mask=b_input_mask,
labels=b_labels,
)
loss = outputs[0]
val_loss += loss.item()
prediction = torch.argmax(outputs[1], dim=1)
val_acc += (prediction == b_labels).sum().item()
val_loss /= len(validation_dataloader)
val_loss_set.append(val_loss)
val_acc /= len(validation_dataloader.dataset)
val_acc_set.append(val_acc)
# Print epoch results
print(
f"Train loss={train_loss:.4f} | Train acc={train_acc:.4f} | Val loss={val_loss:.4f} | Val acc={val_acc:.4f}"
)
# Check for early stopping
if val_loss > best_loss:
best_loss = val_loss
early_stop_counter = 0
torch.save(model.state_dict(), "best_model.pth")
else:
early_stop_counter += 1
if early_stop_counter >= 3:
print("Early stopping after {} epochs".format(epoch + 1))
break
# Load the best model
model.load_state_dict(torch.load("best_model.pth"))
# Plot loss and accuracy curves
fig, axs = plt.subplots(1, 2, figsize=(12, 6))
# Plot training and validation loss
axs[0].plot(range(1, len(train_loss_set) + 1), train_loss_set, label="Train Loss")
axs[0].plot(range(1, len(val_loss_set) + 1), val_loss_set, label="Val Loss")
axs[0].set_xlabel("Epoch")
axs[0].set_ylabel("Loss")
axs[0].set_title("Training and Validation Loss")
axs[0].legend()
# Plot training and validation accuracy
axs[1].plot(range(1, len(train_acc_set) + 1), train_acc_set, label="Train Acc")
axs[1].plot(range(1, len(val_acc_set) + 1), val_acc_set, label="Val Acc")
axs[1].set_xlabel("Epoch")
axs[1].set_ylabel("Accuracy")
axs[1].set_title("Training and Validation Accuracy")
axs[1].legend()
# Set x-axis to be numeric
for ax in axs:
ax.xaxis.set_major_locator(MaxNLocator(integer=True))
plt.show()
def evaluate(model, dataloader):
y_true = []
y_pred = []
y_pred_prob = []
model.eval()
with torch.no_grad():
for i, batch in enumerate(tqdm(dataloader, desc="Testing")):
batch = tuple(t.to(device) for t in batch)
b_input_ids, b_input_mask, b_labels = batch
outputs = model(
b_input_ids, token_type_ids=None, attention_mask=b_input_mask
)
probs = torch.nn.functional.softmax(outputs[0], dim=-1)
_, preds = torch.max(probs, dim=1)
y_pred.extend(preds.tolist())
y_pred_prob.extend(probs.tolist())
y_true.extend(b_labels.tolist())
return y_true, y_pred, y_pred_prob
# Evaluate the model on the test set and get true labels, predicted labels, and predicted probabilities
y_true, y_pred, y_pred_prob = evaluate(model, test_dataloader)
# Compute the ROC curve and AUC for each class separately
n_classes = len(label_encoder.classes_)
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
y_true_ = [1 if x == i else 0 for x in y_true]
y_pred_prob_ = [x[i] for x in y_pred_prob]
fpr[i], tpr[i], _ = roc_curve(y_true_, y_pred_prob_)
roc_auc[i] = auc(fpr[i], tpr[i])
# Plot the ROC curve for each class
labels = list(label_encoder.classes_)
plt.figure()
colors = ["blue", "red", "green", "purple", "orange", "brown"]
for i, color in zip(range(n_classes), colors):
plt.plot(
fpr[i],
tpr[i],
color=color,
lw=2,
label="ROC curve of class {0} (area = {1:0.2f})"
"".format(labels[i], roc_auc[i]),
)
plt.plot([0, 1], [0, 1], "k--", lw=2)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver operating characteristic for multiclass")
plt.legend(loc="lower right")
plt.show()
# Print classification report
print("Classification Report:")
print(classification_report(test_labels, y_pred, target_names=classes))
cm = confusion_matrix(test_labels, y_pred)
df_cm = pd.DataFrame(cm, index=labels, columns=labels)
hmap = sns.heatmap(df_cm, annot=True, fmt="d")
hmap.yaxis.set_ticklabels(hmap.yaxis.get_ticklabels(), rotation=0, ha="right")
hmap.xaxis.set_ticklabels(hmap.xaxis.get_ticklabels(), rotation=30, ha="right")
plt.title("Confusion Matrix")
plt.ylabel("True label")
plt.xlabel("Predicted label")
def tokenize_inputs(text_list, tokenizer, num_embeddings=120):
tokenized_texts = list(
map(lambda t: tokenizer.tokenize(t)[: num_embeddings - 2], text_list)
)
input_ids = [tokenizer.convert_tokens_to_ids(x) for x in tokenized_texts]
input_ids = [tokenizer.build_inputs_with_special_tokens(x) for x in input_ids]
input_ids = pad_sequences(
input_ids,
maxlen=num_embeddings,
dtype="long",
truncating="post",
padding="post",
)
return input_ids
def create_attn_masks(input_ids):
attention_masks = []
for seq in input_ids:
seq_mask = [float(i > 0) for i in seq]
attention_masks.append(seq_mask)
return attention_masks
sample = ["currently violently emotionally push away good"]
input_id = tokenize_inputs(sample, tokenizer, num_embeddings=120)
input_id = torch.tensor(input_id)
attention_masks = create_attn_masks(input_id)
attention_masks = torch.tensor(attention_masks)
outputs = model(
input_id.cuda(), token_type_ids=None, attention_mask=attention_masks.cuda()
)
prediction = torch.argmax(outputs[0], dim=1)
predicted_class = label_encoder.inverse_transform([prediction[0].item()])
print("The predicted class is:", predicted_class)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/530/129530321.ipynb
| null | null |
[{"Id": 129530321, "ScriptId": 38474318, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14630152, "CreationDate": "05/14/2023 15:14:25", "VersionNumber": 1.0, "Title": "xlent_kaggle", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 355.0, "LinesInsertedFromPrevious": 271.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 84.0, "LinesInsertedFromFork": 271.0, "LinesDeletedFromFork": 147.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 84.0, "TotalVotes": 0}]
| null | null | null | null |
import warnings
warnings.filterwarnings("ignore")
import pandas as pd
import numpy as np
import re
import string
from collections import Counter
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, confusion_matrix, roc_curve, auc
from keras_preprocessing.sequence import pad_sequences
import torch
from torch.utils.data import TensorDataset, DataLoader, RandomSampler, SequentialSampler
from transformers import XLNetTokenizer, XLNetForSequenceClassification
from pytorch_transformers import AdamW
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
from tqdm import tqdm, trange
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
df = pd.read_csv("/kaggle/input/graduation-project/kaggle-dataset.csv")
# Drop any rows with missing values in the Sentence column & Convert the Sentence column to strings
df = df.dropna(subset=["Sentence"])
df.Sentence = [str(text) for text in df.Sentence]
# Randomly sample 250K rows from the dataframe (Due to limited resources)
df = df.sample(n=250000, random_state=0)
print(f"Dataframe shape: {df.shape}")
df["Subreddit"].value_counts()
label_encoder = preprocessing.LabelEncoder()
# Encode the Subreddit column of the df dataframe & Store the label names in a list
df["Subreddit"] = label_encoder.fit_transform(df["Subreddit"])
classes = list(label_encoder.classes_)
# Print two random rows from the encoded dataframe
print(df.sample(2))
# Get sentences and labels from data
sentences = df.Sentence.values
labels = df.Subreddit.values
# Add special tokens to sentences
sentences = [str(sentence) + " [SEP] [CLS]" for sentence in sentences]
# Initialize XLNetTokenizer with pre-trained model
tokenizer = XLNetTokenizer.from_pretrained("xlnet-base-cased")
# Tokenize sentences using tokenizer
tokenized_texts = [tokenizer.tokenize(sent) for sent in sentences]
# Print tokenized version of the first sentence
print("Tokenize the first sentence:")
print(tokenized_texts[0])
MAX_LEN = 128
# Convert tokenized texts to input IDs
input_ids = [tokenizer.convert_tokens_to_ids(x) for x in tokenized_texts]
# Pad input IDs to a maximum length of MAX_LEN
input_ids = pad_sequences(
input_ids, maxlen=MAX_LEN, dtype="long", truncating="post", padding="post"
)
# Create attention masks
attention_masks = []
# Create a mask of 1s for each token followed by 0s for padding
for seq in input_ids:
seq_mask = [float(i > 0) for i in seq]
attention_masks.append(seq_mask)
# Split data into training , validation and testing sets
train_inputs, test_inputs, train_labels, test_labels = train_test_split(
input_ids, labels, random_state=56, test_size=0.2
)
train_masks, test_masks, _, _ = train_test_split(
attention_masks, input_ids, random_state=56, test_size=0.2
)
test_inputs, validation_inputs, test_labels, validation_labels = train_test_split(
train_inputs, train_labels, random_state=56, test_size=0.5
)
test_masks, validation_masks, _, _ = train_test_split(
train_masks, train_inputs, random_state=56, test_size=0.5
)
# Convert data to PyTorch tensors
train_inputs = torch.tensor(train_inputs)
train_labels = torch.tensor(train_labels)
train_masks = torch.tensor(train_masks)
validation_inputs = torch.tensor(validation_inputs)
validation_labels = torch.tensor(validation_labels)
validation_masks = torch.tensor(validation_masks)
test_inputs = torch.tensor(test_inputs)
test_labels = torch.tensor(test_labels)
test_masks = torch.tensor(test_masks)
train_inputs[0]
# Define batch size
batch_size = 32
# Create training data iterator
train_data = TensorDataset(train_inputs, train_masks, train_labels)
train_sampler = RandomSampler(train_data)
train_dataloader = DataLoader(train_data, sampler=train_sampler, batch_size=batch_size)
# Create validation data iterator
validation_data = TensorDataset(validation_inputs, validation_masks, validation_labels)
validation_sampler = SequentialSampler(validation_data)
validation_dataloader = DataLoader(
validation_data, sampler=validation_sampler, batch_size=batch_size
)
# Create validation data iterator
test_data = TensorDataset(test_inputs, test_masks, test_labels)
test_sampler = SequentialSampler(test_data)
test_dataloader = DataLoader(test_data, sampler=test_sampler, batch_size=batch_size)
# Load pre-trained XLNet model for sequence classification
model = XLNetForSequenceClassification.from_pretrained("xlnet-base-cased", num_labels=5)
# Move model to GPU for faster training
model.cuda()
# Get model parameters and group them by weight decay rate
param_optimizer = list(model.named_parameters())
no_decay = ["bias", "gamma", "beta"]
optimizer_grouped_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.01,
},
{
"params": [p for n, p in param_optimizer if any(nd in n for nd in no_decay)],
"weight_decay_rate": 0.0,
},
]
# Create AdamW optimizer with learning rate of 2e-5
optimizer = AdamW(optimizer_grouped_parameters, lr=2e-5)
# Determine device type for training (GPU or CPU)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Get name of GPU device
torch.cuda.get_device_name()
# Initialize variables
train_loss_set = []
train_acc_set = []
val_loss_set = []
val_acc_set = []
epochs = 10
best_loss = 0
early_stop_counter = 0
# Train for each epoch
for epoch in range(epochs):
print(f"Epoch {epoch+1}/{epochs}")
model.train()
nb_tr_examples, nb_tr_steps = 0, 0
# Train the data for one epoch and calculate training loss and accuracy
train_acc, train_loss = 0, 0
for step, batch in enumerate(tqdm(train_dataloader, desc=f"Training")):
batch = tuple(t.to(device) for t in batch)
b_input_ids, b_input_mask, b_labels = batch
optimizer.zero_grad()
outputs = model(
b_input_ids,
token_type_ids=None,
attention_mask=b_input_mask,
labels=b_labels,
)
loss = outputs[0]
logits = outputs[1]
loss.backward()
optimizer.step()
train_loss += loss.item()
nb_tr_examples += b_input_ids.size(0)
nb_tr_steps += 1
# Calculate training accuracy
prediction = torch.argmax(logits, dim=1)
train_acc += (prediction == b_labels).sum().item()
# Calculate training loss and accuracy for the epoch
train_loss = train_loss / nb_tr_steps
train_acc /= nb_tr_examples
train_acc_set.append(train_acc)
train_loss_set.append(train_loss)
# Calculate validation loss and accuracy
val_loss, val_acc = 0, 0
with torch.no_grad():
for batch in tqdm(validation_dataloader, desc=f"Validation "):
batch = tuple(t.to(device) for t in batch)
b_input_ids, b_input_mask, b_labels = batch
outputs = model(
b_input_ids,
token_type_ids=None,
attention_mask=b_input_mask,
labels=b_labels,
)
loss = outputs[0]
val_loss += loss.item()
prediction = torch.argmax(outputs[1], dim=1)
val_acc += (prediction == b_labels).sum().item()
val_loss /= len(validation_dataloader)
val_loss_set.append(val_loss)
val_acc /= len(validation_dataloader.dataset)
val_acc_set.append(val_acc)
# Print epoch results
print(
f"Train loss={train_loss:.4f} | Train acc={train_acc:.4f} | Val loss={val_loss:.4f} | Val acc={val_acc:.4f}"
)
# Check for early stopping
if val_loss > best_loss:
best_loss = val_loss
early_stop_counter = 0
torch.save(model.state_dict(), "best_model.pth")
else:
early_stop_counter += 1
if early_stop_counter >= 3:
print("Early stopping after {} epochs".format(epoch + 1))
break
# Load the best model
model.load_state_dict(torch.load("best_model.pth"))
# Plot loss and accuracy curves
fig, axs = plt.subplots(1, 2, figsize=(12, 6))
# Plot training and validation loss
axs[0].plot(range(1, len(train_loss_set) + 1), train_loss_set, label="Train Loss")
axs[0].plot(range(1, len(val_loss_set) + 1), val_loss_set, label="Val Loss")
axs[0].set_xlabel("Epoch")
axs[0].set_ylabel("Loss")
axs[0].set_title("Training and Validation Loss")
axs[0].legend()
# Plot training and validation accuracy
axs[1].plot(range(1, len(train_acc_set) + 1), train_acc_set, label="Train Acc")
axs[1].plot(range(1, len(val_acc_set) + 1), val_acc_set, label="Val Acc")
axs[1].set_xlabel("Epoch")
axs[1].set_ylabel("Accuracy")
axs[1].set_title("Training and Validation Accuracy")
axs[1].legend()
# Set x-axis to be numeric
for ax in axs:
ax.xaxis.set_major_locator(MaxNLocator(integer=True))
plt.show()
def evaluate(model, dataloader):
y_true = []
y_pred = []
y_pred_prob = []
model.eval()
with torch.no_grad():
for i, batch in enumerate(tqdm(dataloader, desc="Testing")):
batch = tuple(t.to(device) for t in batch)
b_input_ids, b_input_mask, b_labels = batch
outputs = model(
b_input_ids, token_type_ids=None, attention_mask=b_input_mask
)
probs = torch.nn.functional.softmax(outputs[0], dim=-1)
_, preds = torch.max(probs, dim=1)
y_pred.extend(preds.tolist())
y_pred_prob.extend(probs.tolist())
y_true.extend(b_labels.tolist())
return y_true, y_pred, y_pred_prob
# Evaluate the model on the test set and get true labels, predicted labels, and predicted probabilities
y_true, y_pred, y_pred_prob = evaluate(model, test_dataloader)
# Compute the ROC curve and AUC for each class separately
n_classes = len(label_encoder.classes_)
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
y_true_ = [1 if x == i else 0 for x in y_true]
y_pred_prob_ = [x[i] for x in y_pred_prob]
fpr[i], tpr[i], _ = roc_curve(y_true_, y_pred_prob_)
roc_auc[i] = auc(fpr[i], tpr[i])
# Plot the ROC curve for each class
labels = list(label_encoder.classes_)
plt.figure()
colors = ["blue", "red", "green", "purple", "orange", "brown"]
for i, color in zip(range(n_classes), colors):
plt.plot(
fpr[i],
tpr[i],
color=color,
lw=2,
label="ROC curve of class {0} (area = {1:0.2f})"
"".format(labels[i], roc_auc[i]),
)
plt.plot([0, 1], [0, 1], "k--", lw=2)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver operating characteristic for multiclass")
plt.legend(loc="lower right")
plt.show()
# Print classification report
print("Classification Report:")
print(classification_report(test_labels, y_pred, target_names=classes))
cm = confusion_matrix(test_labels, y_pred)
df_cm = pd.DataFrame(cm, index=labels, columns=labels)
hmap = sns.heatmap(df_cm, annot=True, fmt="d")
hmap.yaxis.set_ticklabels(hmap.yaxis.get_ticklabels(), rotation=0, ha="right")
hmap.xaxis.set_ticklabels(hmap.xaxis.get_ticklabels(), rotation=30, ha="right")
plt.title("Confusion Matrix")
plt.ylabel("True label")
plt.xlabel("Predicted label")
def tokenize_inputs(text_list, tokenizer, num_embeddings=120):
tokenized_texts = list(
map(lambda t: tokenizer.tokenize(t)[: num_embeddings - 2], text_list)
)
input_ids = [tokenizer.convert_tokens_to_ids(x) for x in tokenized_texts]
input_ids = [tokenizer.build_inputs_with_special_tokens(x) for x in input_ids]
input_ids = pad_sequences(
input_ids,
maxlen=num_embeddings,
dtype="long",
truncating="post",
padding="post",
)
return input_ids
def create_attn_masks(input_ids):
attention_masks = []
for seq in input_ids:
seq_mask = [float(i > 0) for i in seq]
attention_masks.append(seq_mask)
return attention_masks
sample = ["currently violently emotionally push away good"]
input_id = tokenize_inputs(sample, tokenizer, num_embeddings=120)
input_id = torch.tensor(input_id)
attention_masks = create_attn_masks(input_id)
attention_masks = torch.tensor(attention_masks)
outputs = model(
input_id.cuda(), token_type_ids=None, attention_mask=attention_masks.cuda()
)
prediction = torch.argmax(outputs[0], dim=1)
predicted_class = label_encoder.inverse_transform([prediction[0].item()])
print("The predicted class is:", predicted_class)
| false | 0 | 3,771 | 0 | 3,771 | 3,771 |
||
129530976
|
# ### Importing Libraries
from sklearn.datasets import make_classification # generate classification datasets
import numpy as np
import matplotlib.pyplot as plt
# ### Dataset Generation
X, y = make_classification(
n_samples=100,
n_features=2,
n_informative=2,
n_redundant=0,
n_classes=2,
n_clusters_per_class=1,
random_state=41,
hypercube=False,
class_sep=10,
)
# hypercube - if True then the clusters are kept at the vertices of a hypercube(geometrical shape), if False the clusters are put on the vertices of a random polytope.
# class_sep - greater values spread out the clusters
X.shape
# ### Data Representation
plt.figure(figsize=(8, 5))
plt.scatter(X[:, 0], X[:, 1], c=y, s=100) # c - colours, s - marker size
plt.show()
# ### Perceptron Trick Algorithm
X = np.insert(X, 0, 1, axis=1) # Adding a col of 1s to X
epochs = 1000
lr = 0.1 # learning rate
def step(z):
if z > 0:
return 1
return 0
X.shape
def perceptron(X, y):
weights = np.ones(X.shape[1])
for i in range(epochs):
j = np.random.randint(0, 100)
y_pred = step(np.dot(X[j], weights))
weights = weights + (lr * (y[j] - y_pred)) * X[j]
return weights[0], weights[1:]
intercept_, coef_ = perceptron(X, y)
intercept_
coef_
# using line equations
m = -(coef_[0] / coef_[1])
b = -(intercept_ / coef_[1])
# (x, y) values for plotting the line
x_input = np.linspace(-3, 3, 100)
y_input = (m * x_input) + b
plt.figure(figsize=(8, 5))
plt.plot(x_input, y_input)
plt.scatter(X[:, 1], X[:, 2], c=y, s=100)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/530/129530976.ipynb
| null | null |
[{"Id": 129530976, "ScriptId": 38508762, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6819371, "CreationDate": "05/14/2023 15:19:48", "VersionNumber": 1.0, "Title": "Logistic Regression Using Perceptron Trick", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 61.0, "LinesInsertedFromPrevious": 61.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ### Importing Libraries
from sklearn.datasets import make_classification # generate classification datasets
import numpy as np
import matplotlib.pyplot as plt
# ### Dataset Generation
X, y = make_classification(
n_samples=100,
n_features=2,
n_informative=2,
n_redundant=0,
n_classes=2,
n_clusters_per_class=1,
random_state=41,
hypercube=False,
class_sep=10,
)
# hypercube - if True then the clusters are kept at the vertices of a hypercube(geometrical shape), if False the clusters are put on the vertices of a random polytope.
# class_sep - greater values spread out the clusters
X.shape
# ### Data Representation
plt.figure(figsize=(8, 5))
plt.scatter(X[:, 0], X[:, 1], c=y, s=100) # c - colours, s - marker size
plt.show()
# ### Perceptron Trick Algorithm
X = np.insert(X, 0, 1, axis=1) # Adding a col of 1s to X
epochs = 1000
lr = 0.1 # learning rate
def step(z):
if z > 0:
return 1
return 0
X.shape
def perceptron(X, y):
weights = np.ones(X.shape[1])
for i in range(epochs):
j = np.random.randint(0, 100)
y_pred = step(np.dot(X[j], weights))
weights = weights + (lr * (y[j] - y_pred)) * X[j]
return weights[0], weights[1:]
intercept_, coef_ = perceptron(X, y)
intercept_
coef_
# using line equations
m = -(coef_[0] / coef_[1])
b = -(intercept_ / coef_[1])
# (x, y) values for plotting the line
x_input = np.linspace(-3, 3, 100)
y_input = (m * x_input) + b
plt.figure(figsize=(8, 5))
plt.plot(x_input, y_input)
plt.scatter(X[:, 1], X[:, 2], c=y, s=100)
plt.show()
| false | 0 | 575 | 0 | 575 | 575 |
||
129530993
|
<jupyter_start><jupyter_text>Blood Cell Detection Dataset
## Introduction
This dataset contains annotated red blood cells(RBC) and white blood cells(WBC) from peripheral blood smear taken from a light microscope.

## About Blood Cell Detection Dataset
- Images are collected from peripheral blood smear slides on a light microscope with high magnification and resolution.
- It contains 100 annotated images with labeled RBC as 2237and WBC as 103.
- Every image contains RGB channels and also is 256 pixels in both height and width.
- **images** folder contains image files as a png format. **annotations.csv** file contains both locations and labels.
- The total dataset file size is approximately 14MB.
- All PRs are welcome.
## What is Peripheral Blood Smear?
A peripheral blood smear is a thin layer of blood smeared on a glass microscope slide and then stained in such a way as to allow the various blood cells to be examined microscopically. Blood films are examined in the investigation of hematological (blood) disorders and are routinely employed to look for blood parasites, such as those of malaria and filariasis. ([Wikipedia](https://en.wikipedia.org/wiki/Blood_film))
Examination of the peripheral blood smear is an inexpensive but powerful diagnostic tool in both children and adults. In some ways it is becoming a "lost art" but it often provides rapid, reliable access to information about a variety of hematologic disorders. The smear offers a window into the functional status of the bone marrow, the factory producing all blood elements. It is particularly important when assessing cytopenic states (eg, anemia, leukopenia, thrombocytopenia). Review of the smear is an important adjunct to other clinical data; in some cases, the peripheral smear alone is sufficient to establish a diagnosis. ([UpToDate](https://www.uptodate.com/contents/evaluation-of-the-peripheral-blood-smear))
## Licence
See LICENSE for details.
Kaggle dataset identifier: blood-cell-detection-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import torch
import torchvision
print("PyTorch version:", torch.__version__)
print("Torchvision version:", torchvision.__version__)
print("CUDA is available:", torch.cuda.is_available())
import sys
def show_anns(anns):
if len(anns) == 0:
return
sorted_anns = sorted(anns, key=(lambda x: x["area"]), reverse=True)
ax = plt.gca()
ax.set_autoscale_on(False)
polygons = []
color = []
for ann in sorted_anns:
m = ann["segmentation"]
img = np.ones((m.shape[0], m.shape[1], 3))
color_mask = np.random.random((1, 3)).tolist()[0]
for i in range(3):
img[:, :, i] = color_mask[i]
ax.imshow(np.dstack((img, m * 0.35)))
import sys
sys.path.append("..")
from segment_anything import sam_model_registry, SamAutomaticMaskGenerator, SamPredictor
# sam_checkpoint = "/kaggle/input/segment-anything/pytorch/vit-b/1/model.pth"
# model_type = "vit_b"
sam_checkpoint = "/kaggle/input/segment-anything/pytorch/vit-h/1/model.pth"
model_type = "vit_h"
device = "cuda"
sam = sam_model_registry[model_type](checkpoint=sam_checkpoint)
sam.to(device=device)
# mask_generator = SamAutomaticMaskGenerator(sam)
mask_generator = SamAutomaticMaskGenerator(
model=sam,
points_per_side=10,
pred_iou_thresh=0.9,
stability_score_thresh=0.92,
crop_n_layers=1,
min_mask_region_area=100, # Requires open-cv to run post-processing
)
from glob import glob
from PIL import Image, ImageOps
import numpy as np
import torch
import matplotlib.pyplot as plt
import cv2
patch_paths = glob("/kaggle/input/blood-cell-detection-dataset/images/*")
print("No. of images = ", len(patch_paths))
# for ii in range(len(patch_paths)):
for ii in range(4):
image = cv2.imread(patch_paths[ii])
masks = mask_generator.generate(image)
print(ii, patch_paths[ii])
print("No. of detected cells = ", len(masks))
plt.figure(figsize=(8, 5))
plt.subplot(121)
plt.imshow(image)
plt.axis("off")
plt.subplot(122)
plt.imshow(image)
show_anns(masks)
plt.axis("off")
plt.show()
# for i in range(len(masks)):
for i in range(10):
x = int(masks[i]["bbox"][0])
y = int(masks[i]["bbox"][1])
a = int(masks[i]["bbox"][2])
b = int(masks[i]["bbox"][3])
cropped_im = image[y : y + b, x : x + a, :]
cropped_mask = masks[i]["segmentation"][y : y + b, x : x + a]
plt.figure(figsize=(4, 3))
plt.subplot(121)
plt.imshow(cropped_im)
plt.axis("off")
plt.subplot(122)
plt.imshow(cropped_mask)
plt.axis("off")
plt.show()
plt.figure(figsize=(4, 4))
plt.imshow(masks[i]["segmentation"])
plt.axis("off")
plt.show()
print("---")
print("image no. ", str(i))
print("Area: ", masks[i]["area"])
print("predicted_iou: ", masks[i]["predicted_iou"])
print("stability_score: ", masks[i]["stability_score"])
print("-----------------------------------------------------------------")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/530/129530993.ipynb
|
blood-cell-detection-dataset
|
draaslan
|
[{"Id": 129530993, "ScriptId": 38514382, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5121112, "CreationDate": "05/14/2023 15:19:57", "VersionNumber": 1.0, "Title": "Cell Segmentation using Segment Anything Model", "EvaluationDate": "05/14/2023", "IsChange": true, "TotalLines": 106.0, "LinesInsertedFromPrevious": 106.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 185683639, "KernelVersionId": 129530993, "SourceDatasetVersionId": 1113577}]
|
[{"Id": 1113577, "DatasetId": 624559, "DatasourceVersionId": 1143857, "CreatorUserId": 972998, "LicenseName": "Other (specified in description)", "CreationDate": "04/28/2020 01:20:35", "VersionNumber": 1.0, "Title": "Blood Cell Detection Dataset", "Slug": "blood-cell-detection-dataset", "Subtitle": "WBC & RBC detection dataset from peripheral blood smears.", "Description": "## Introduction\n\nThis dataset contains annotated red blood cells(RBC) and white blood cells(WBC) from peripheral blood smear taken from a light microscope.\n\n\n\n## About Blood Cell Detection Dataset\n\n- Images are collected from peripheral blood smear slides on a light microscope with high magnification and resolution.\n- It contains 100 annotated images with labeled RBC as 2237and WBC as 103.\n- Every image contains RGB channels and also is 256 pixels in both height and width.\n- **images** folder contains image files as a png format. **annotations.csv** file contains both locations and labels.\n- The total dataset file size is approximately 14MB.\n- All PRs are welcome.\n\n## What is Peripheral Blood Smear?\n\nA peripheral blood smear is a thin layer of blood smeared on a glass microscope slide and then stained in such a way as to allow the various blood cells to be examined microscopically. Blood films are examined in the investigation of hematological (blood) disorders and are routinely employed to look for blood parasites, such as those of malaria and filariasis. ([Wikipedia](https://en.wikipedia.org/wiki/Blood_film))\n\nExamination of the peripheral blood smear is an inexpensive but powerful diagnostic tool in both children and adults. In some ways it is becoming a \"lost art\" but it often provides rapid, reliable access to information about a variety of hematologic disorders. The smear offers a window into the functional status of the bone marrow, the factory producing all blood elements. It is particularly important when assessing cytopenic states (eg, anemia, leukopenia, thrombocytopenia). Review of the smear is an important adjunct to other clinical data; in some cases, the peripheral smear alone is sufficient to establish a diagnosis. ([UpToDate](https://www.uptodate.com/contents/evaluation-of-the-peripheral-blood-smear))\n\n## Licence\n\nSee LICENSE for details.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 624559, "CreatorUserId": 972998, "OwnerUserId": 972998.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1113577.0, "CurrentDatasourceVersionId": 1143857.0, "ForumId": 638744, "Type": 2, "CreationDate": "04/28/2020 01:20:35", "LastActivityDate": "04/28/2020", "TotalViews": 29176, "TotalDownloads": 2213, "TotalVotes": 28, "TotalKernels": 6}]
|
[{"Id": 972998, "UserName": "draaslan", "DisplayName": "Abd\u00fcssamet Aslan, MD", "RegisterDate": "03/17/2017", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import torch
import torchvision
print("PyTorch version:", torch.__version__)
print("Torchvision version:", torchvision.__version__)
print("CUDA is available:", torch.cuda.is_available())
import sys
def show_anns(anns):
if len(anns) == 0:
return
sorted_anns = sorted(anns, key=(lambda x: x["area"]), reverse=True)
ax = plt.gca()
ax.set_autoscale_on(False)
polygons = []
color = []
for ann in sorted_anns:
m = ann["segmentation"]
img = np.ones((m.shape[0], m.shape[1], 3))
color_mask = np.random.random((1, 3)).tolist()[0]
for i in range(3):
img[:, :, i] = color_mask[i]
ax.imshow(np.dstack((img, m * 0.35)))
import sys
sys.path.append("..")
from segment_anything import sam_model_registry, SamAutomaticMaskGenerator, SamPredictor
# sam_checkpoint = "/kaggle/input/segment-anything/pytorch/vit-b/1/model.pth"
# model_type = "vit_b"
sam_checkpoint = "/kaggle/input/segment-anything/pytorch/vit-h/1/model.pth"
model_type = "vit_h"
device = "cuda"
sam = sam_model_registry[model_type](checkpoint=sam_checkpoint)
sam.to(device=device)
# mask_generator = SamAutomaticMaskGenerator(sam)
mask_generator = SamAutomaticMaskGenerator(
model=sam,
points_per_side=10,
pred_iou_thresh=0.9,
stability_score_thresh=0.92,
crop_n_layers=1,
min_mask_region_area=100, # Requires open-cv to run post-processing
)
from glob import glob
from PIL import Image, ImageOps
import numpy as np
import torch
import matplotlib.pyplot as plt
import cv2
patch_paths = glob("/kaggle/input/blood-cell-detection-dataset/images/*")
print("No. of images = ", len(patch_paths))
# for ii in range(len(patch_paths)):
for ii in range(4):
image = cv2.imread(patch_paths[ii])
masks = mask_generator.generate(image)
print(ii, patch_paths[ii])
print("No. of detected cells = ", len(masks))
plt.figure(figsize=(8, 5))
plt.subplot(121)
plt.imshow(image)
plt.axis("off")
plt.subplot(122)
plt.imshow(image)
show_anns(masks)
plt.axis("off")
plt.show()
# for i in range(len(masks)):
for i in range(10):
x = int(masks[i]["bbox"][0])
y = int(masks[i]["bbox"][1])
a = int(masks[i]["bbox"][2])
b = int(masks[i]["bbox"][3])
cropped_im = image[y : y + b, x : x + a, :]
cropped_mask = masks[i]["segmentation"][y : y + b, x : x + a]
plt.figure(figsize=(4, 3))
plt.subplot(121)
plt.imshow(cropped_im)
plt.axis("off")
plt.subplot(122)
plt.imshow(cropped_mask)
plt.axis("off")
plt.show()
plt.figure(figsize=(4, 4))
plt.imshow(masks[i]["segmentation"])
plt.axis("off")
plt.show()
print("---")
print("image no. ", str(i))
print("Area: ", masks[i]["area"])
print("predicted_iou: ", masks[i]["predicted_iou"])
print("stability_score: ", masks[i]["stability_score"])
print("-----------------------------------------------------------------")
| false | 0 | 1,002 | 0 | 1,581 | 1,002 |
||
129984224
|
<jupyter_start><jupyter_text>Human Stress Prediction
“subreddit – post_id – sentence_range – text-label-confidence-social_timestamp” represents the titles for Stress.csv file.
Stress detection is a challenging task, as there are so many words that can be used by people on their posts that can show whether a person is having psychological stress or not. look for datasets that you can use to train a machine learning model for stress detection.
The dataset contains data posted on subreddits related to mental health. This dataset contains various mental health problems shared by people about their life. Fortunately, this dataset is labelled as 0 and 1, where 0 indicates no stress and 1 indicates stress.
Kaggle dataset identifier: human-stress-prediction
<jupyter_script># 0. About Human Stress
# 1. Import dependencies
import pandas as pd
import numpy as np
import re
import seaborn as sns
import collections
import matplotlib.pyplot as plt
from wordcloud import WordCloud
from unidecode import unidecode
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from nltk.stem import SnowballStemmer
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import GaussianNB, MultinomialNB, BernoulliNB
from sklearn.neural_network import MLPClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, VotingClassifier
from sklearn.svm import SVC
from sklearn.linear_model import SGDClassifier
from sklearn.metrics import accuracy_score
from sklearn.metrics import classification_report
import pickle
data = pd.read_csv("/kaggle/input/human-stress-prediction/Stress.csv")
# 2. Data ProfilinG / PreProcessinG
data.head()
# view data stats describtion
data.describe().T
# Check Null Values
data.isnull().sum()
def clean(data_frame):
# Variable to replace all characters that are not letters or whitespace
regex = re.compile("[^a-z\s]")
# Convert text to lower case
data_frame["clean_text"] = data_frame["text"].str.lower()
# Removes all characters that are not letters or spaces
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: regex.sub("", x)
)
# Removes all characters between square brackets
data_frame["clean_text"] = data_frame["clean_text"].str.replace(
"[%s]", "", regex=True
)
# Remove the accents
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: regex.sub("", unidecode(x))
)
# Remove http
data_frame["clean_text"] = data_frame["clean_text"].str.replace(
"http", "", regex=True
)
# Remove words with less than 3 characters
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: " ".join([w for w in x.split() if len(w) > 2])
)
# Tokenization
data_frame["clean_text"] = data_frame["clean_text"].apply(word_tokenize)
# Removing stopwords
stop_words = set(stopwords.words("english"))
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: [word for word in x if word not in stop_words]
)
# Word normalization
stemmer = SnowballStemmer("english")
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: [stemmer.stem(word) for word in x]
)
# Joining the words back into a single text
data_frame["clean_text"] = data_frame["clean_text"].apply(lambda x: " ".join(x))
return data_frame
def clean_text(text):
# Variable to replace all characters that are not letters or whitespace
regex = re.compile("[^a-z\s]")
# Convert text to lower case
clean_text = text.lower()
# Removes all characters that are not letters or spaces
clean_text = regex.sub("", clean_text)
# Removes all characters between square brackets
clean_text = re.sub(r"\[[^]]*\]", "", clean_text)
# Remove the accents
clean_text = unidecode(clean_text)
# Remove http
clean_text = clean_text.replace("http", "")
# Remove words with less than 3 characters
clean_text = " ".join([w for w in clean_text.split() if len(w) > 2])
# Tokenization
tokens = word_tokenize(clean_text)
# Removing stopwords
stop_words = set(stopwords.words("english"))
tokens = [word for word in tokens if word not in stop_words]
# Word normalization
stemmer = SnowballStemmer("english")
normalized_tokens = [stemmer.stem(word) for word in tokens]
# Joining the words back into a single text
cleaned_text = " ".join(normalized_tokens)
return cleaned_text
clean_data = clean(data)
clean_data.head()
plt.style.use("dark_background") # Dark Background
sns.countplot(x=clean_data["label"], palette="viridis") # Create the Countplot
plt.title("Label Distribution", fontsize=18)
# Title
# Top 10 most used words
words = []
for text in clean_data["clean_text"]:
words.extend(text.split())
word_count = collections.Counter(words)
top_words = dict(word_count.most_common(10))
plt.style.use("dark_background") # Dark Background
plt.figure(figsize=(10, 6)) # Figure Size
plt.bar(
range(len(top_words)), list(top_words.values()), align="center"
) # Create the Barplot
plt.xticks(
range(len(top_words)), list(top_words.keys())
) # Creating a y axis with words
plt.grid(alpha=0.5) # Grid Opacity
plt.title("Top 10 most used words", fontsize=18) # Grid Opacity
plt.xlabel("Words")
plt.ylabel("Frequency")
plt.show()
# Word Cloud
# Concatenating all cleaned texts into a single string
text = " ".join(caption for caption in clean_data["clean_text"])
wordcloud = WordCloud(
width=800, height=500, background_color="black", min_font_size=10
).generate(text)
plt.figure(figsize=(10, 6), facecolor=None)
plt.imshow(wordcloud)
plt.axis("off")
plt.show()
# 3. ML Model Creation and Evaluation
# Renaming label column values
clean_data["new_label"] = clean_data["label"].map({0: "No Stress", 1: "Stress"})
# Split X and Y
x = np.array(clean_data["clean_text"])
y = np.array(clean_data["new_label"])
cv = CountVectorizer() # Convert text to numerical
X = cv.fit_transform(x)
# Split Train and Test
xtrain, xtest, ytrain, ytest = train_test_split(X, y, test_size=0.20, random_state=31)
# Models
DTC = DecisionTreeClassifier()
GNB = GaussianNB()
SGD = SGDClassifier()
MLP = MLPClassifier()
MNB = MultinomialNB()
BNB = BernoulliNB()
SVM = SVC()
DTC.fit(xtrain, ytrain)
SGD.fit(xtrain, ytrain)
MLP.fit(xtrain, ytrain)
MNB.fit(xtrain, ytrain)
BNB.fit(xtrain, ytrain)
SVM.fit(xtrain, ytrain)
# Predictions
pred_dtc = DTC.predict(xtest)
pred_sgd = SGD.predict(xtest)
pred_mlp = MLP.predict(xtest)
pred_mnb = MNB.predict(xtest)
pred_bnb = BNB.predict(xtest)
pred_svm = SVM.predict(xtest)
# 4. CommunicatinG Results
# classification report of DecisionTreeClassifier
print("*" * 55)
print(f"DecisionTreeClassifier Accuracy: {accuracy_score(ytest, pred_dtc)}")
print("*" * 55)
print(classification_report(ytest, pred_dtc))
# classification report of SGDClassifier
print("*" * 55)
print(f"SGDClassifier Accuracy: {accuracy_score(ytest, pred_sgd)}")
print("*" * 55)
print(classification_report(ytest, pred_sgd))
# classification report of MLPClassifier
print("*" * 55)
print(f"MLPClassifier Accuracy: {accuracy_score(ytest, pred_mlp)}")
print("*" * 55)
print(classification_report(ytest, pred_mlp))
# classification report of MultinomialNB
print("*" * 55)
print(f"Multinomia Accuracy: {accuracy_score(ytest, pred_mnb)}")
print("*" * 55)
print(classification_report(ytest, pred_mnb))
# Classification report BernoulliNB
print("*" * 55)
print(f"Bernoulli Accuracy: {accuracy_score(ytest, pred_bnb)}")
print("*" * 55)
print(classification_report(ytest, pred_bnb))
# Classification report SVM
print("*" * 55)
print(f"SVM Accuracy: {accuracy_score(ytest, pred_svm)}")
print("*" * 55)
print(classification_report(ytest, pred_svm))
# save countvectorizer
with open("CountVectorizer.pkl", "wb") as file:
pickle.dump(cv, file)
with open("MNB_Model.pkl", "wb") as file:
pickle.dump(MNB, file)
# Load the CountVectorizer object from the pickle file
with open("/kaggle/working/CountVectorizer.pkl", "rb") as file:
vectorizer = pickle.load(file)
# Load the MNB Model object from the pickle file
with open("/kaggle/working/MNB_Model.pkl", "rb") as file:
model = pickle.load(file)
# prediction Example
xin = vectorizer.transform(input_text)
xin_array = xin.toarray()
# Predict the class for the input array
predicted_class = model.predict(xin_array)
print("Predicted class:", predicted_class[0])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/984/129984224.ipynb
|
human-stress-prediction
|
kreeshrajani
|
[{"Id": 129984224, "ScriptId": 37185572, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11603852, "CreationDate": "05/17/2023 23:39:19", "VersionNumber": 3.0, "Title": "Human Stress Prediction \ud83e\udde0", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 248.0, "LinesInsertedFromPrevious": 92.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 156.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 3}]
|
[{"Id": 186430056, "KernelVersionId": 129984224, "SourceDatasetVersionId": 5100130}]
|
[{"Id": 5100130, "DatasetId": 2961947, "DatasourceVersionId": 5171271, "CreatorUserId": 9348276, "LicenseName": "Other (specified in description)", "CreationDate": "03/03/2023 13:59:49", "VersionNumber": 1.0, "Title": "Human Stress Prediction", "Slug": "human-stress-prediction", "Subtitle": "Insights from Speech-based Human Stress Dataset", "Description": "\u201csubreddit \u2013 post_id \u2013 sentence_range \u2013 text-label-confidence-social_timestamp\u201d represents the titles for Stress.csv file.\n\nStress detection is a challenging task, as there are so many words that can be used by people on their posts that can show whether a person is having psychological stress or not. look for datasets that you can use to train a machine learning model for stress detection.\n\nThe dataset contains data posted on subreddits related to mental health. This dataset contains various mental health problems shared by people about their life. Fortunately, this dataset is labelled as 0 and 1, where 0 indicates no stress and 1 indicates stress.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2961947, "CreatorUserId": 9348276, "OwnerUserId": 9348276.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5100130.0, "CurrentDatasourceVersionId": 5171271.0, "ForumId": 3000227, "Type": 2, "CreationDate": "03/03/2023 13:59:49", "LastActivityDate": "03/03/2023", "TotalViews": 25541, "TotalDownloads": 2990, "TotalVotes": 43, "TotalKernels": 20}]
|
[{"Id": 9348276, "UserName": "kreeshrajani", "DisplayName": "Kreesh Rajani", "RegisterDate": "01/08/2022", "PerformanceTier": 2}]
|
# 0. About Human Stress
# 1. Import dependencies
import pandas as pd
import numpy as np
import re
import seaborn as sns
import collections
import matplotlib.pyplot as plt
from wordcloud import WordCloud
from unidecode import unidecode
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from nltk.stem import SnowballStemmer
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import GaussianNB, MultinomialNB, BernoulliNB
from sklearn.neural_network import MLPClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, VotingClassifier
from sklearn.svm import SVC
from sklearn.linear_model import SGDClassifier
from sklearn.metrics import accuracy_score
from sklearn.metrics import classification_report
import pickle
data = pd.read_csv("/kaggle/input/human-stress-prediction/Stress.csv")
# 2. Data ProfilinG / PreProcessinG
data.head()
# view data stats describtion
data.describe().T
# Check Null Values
data.isnull().sum()
def clean(data_frame):
# Variable to replace all characters that are not letters or whitespace
regex = re.compile("[^a-z\s]")
# Convert text to lower case
data_frame["clean_text"] = data_frame["text"].str.lower()
# Removes all characters that are not letters or spaces
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: regex.sub("", x)
)
# Removes all characters between square brackets
data_frame["clean_text"] = data_frame["clean_text"].str.replace(
"[%s]", "", regex=True
)
# Remove the accents
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: regex.sub("", unidecode(x))
)
# Remove http
data_frame["clean_text"] = data_frame["clean_text"].str.replace(
"http", "", regex=True
)
# Remove words with less than 3 characters
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: " ".join([w for w in x.split() if len(w) > 2])
)
# Tokenization
data_frame["clean_text"] = data_frame["clean_text"].apply(word_tokenize)
# Removing stopwords
stop_words = set(stopwords.words("english"))
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: [word for word in x if word not in stop_words]
)
# Word normalization
stemmer = SnowballStemmer("english")
data_frame["clean_text"] = data_frame["clean_text"].apply(
lambda x: [stemmer.stem(word) for word in x]
)
# Joining the words back into a single text
data_frame["clean_text"] = data_frame["clean_text"].apply(lambda x: " ".join(x))
return data_frame
def clean_text(text):
# Variable to replace all characters that are not letters or whitespace
regex = re.compile("[^a-z\s]")
# Convert text to lower case
clean_text = text.lower()
# Removes all characters that are not letters or spaces
clean_text = regex.sub("", clean_text)
# Removes all characters between square brackets
clean_text = re.sub(r"\[[^]]*\]", "", clean_text)
# Remove the accents
clean_text = unidecode(clean_text)
# Remove http
clean_text = clean_text.replace("http", "")
# Remove words with less than 3 characters
clean_text = " ".join([w for w in clean_text.split() if len(w) > 2])
# Tokenization
tokens = word_tokenize(clean_text)
# Removing stopwords
stop_words = set(stopwords.words("english"))
tokens = [word for word in tokens if word not in stop_words]
# Word normalization
stemmer = SnowballStemmer("english")
normalized_tokens = [stemmer.stem(word) for word in tokens]
# Joining the words back into a single text
cleaned_text = " ".join(normalized_tokens)
return cleaned_text
clean_data = clean(data)
clean_data.head()
plt.style.use("dark_background") # Dark Background
sns.countplot(x=clean_data["label"], palette="viridis") # Create the Countplot
plt.title("Label Distribution", fontsize=18)
# Title
# Top 10 most used words
words = []
for text in clean_data["clean_text"]:
words.extend(text.split())
word_count = collections.Counter(words)
top_words = dict(word_count.most_common(10))
plt.style.use("dark_background") # Dark Background
plt.figure(figsize=(10, 6)) # Figure Size
plt.bar(
range(len(top_words)), list(top_words.values()), align="center"
) # Create the Barplot
plt.xticks(
range(len(top_words)), list(top_words.keys())
) # Creating a y axis with words
plt.grid(alpha=0.5) # Grid Opacity
plt.title("Top 10 most used words", fontsize=18) # Grid Opacity
plt.xlabel("Words")
plt.ylabel("Frequency")
plt.show()
# Word Cloud
# Concatenating all cleaned texts into a single string
text = " ".join(caption for caption in clean_data["clean_text"])
wordcloud = WordCloud(
width=800, height=500, background_color="black", min_font_size=10
).generate(text)
plt.figure(figsize=(10, 6), facecolor=None)
plt.imshow(wordcloud)
plt.axis("off")
plt.show()
# 3. ML Model Creation and Evaluation
# Renaming label column values
clean_data["new_label"] = clean_data["label"].map({0: "No Stress", 1: "Stress"})
# Split X and Y
x = np.array(clean_data["clean_text"])
y = np.array(clean_data["new_label"])
cv = CountVectorizer() # Convert text to numerical
X = cv.fit_transform(x)
# Split Train and Test
xtrain, xtest, ytrain, ytest = train_test_split(X, y, test_size=0.20, random_state=31)
# Models
DTC = DecisionTreeClassifier()
GNB = GaussianNB()
SGD = SGDClassifier()
MLP = MLPClassifier()
MNB = MultinomialNB()
BNB = BernoulliNB()
SVM = SVC()
DTC.fit(xtrain, ytrain)
SGD.fit(xtrain, ytrain)
MLP.fit(xtrain, ytrain)
MNB.fit(xtrain, ytrain)
BNB.fit(xtrain, ytrain)
SVM.fit(xtrain, ytrain)
# Predictions
pred_dtc = DTC.predict(xtest)
pred_sgd = SGD.predict(xtest)
pred_mlp = MLP.predict(xtest)
pred_mnb = MNB.predict(xtest)
pred_bnb = BNB.predict(xtest)
pred_svm = SVM.predict(xtest)
# 4. CommunicatinG Results
# classification report of DecisionTreeClassifier
print("*" * 55)
print(f"DecisionTreeClassifier Accuracy: {accuracy_score(ytest, pred_dtc)}")
print("*" * 55)
print(classification_report(ytest, pred_dtc))
# classification report of SGDClassifier
print("*" * 55)
print(f"SGDClassifier Accuracy: {accuracy_score(ytest, pred_sgd)}")
print("*" * 55)
print(classification_report(ytest, pred_sgd))
# classification report of MLPClassifier
print("*" * 55)
print(f"MLPClassifier Accuracy: {accuracy_score(ytest, pred_mlp)}")
print("*" * 55)
print(classification_report(ytest, pred_mlp))
# classification report of MultinomialNB
print("*" * 55)
print(f"Multinomia Accuracy: {accuracy_score(ytest, pred_mnb)}")
print("*" * 55)
print(classification_report(ytest, pred_mnb))
# Classification report BernoulliNB
print("*" * 55)
print(f"Bernoulli Accuracy: {accuracy_score(ytest, pred_bnb)}")
print("*" * 55)
print(classification_report(ytest, pred_bnb))
# Classification report SVM
print("*" * 55)
print(f"SVM Accuracy: {accuracy_score(ytest, pred_svm)}")
print("*" * 55)
print(classification_report(ytest, pred_svm))
# save countvectorizer
with open("CountVectorizer.pkl", "wb") as file:
pickle.dump(cv, file)
with open("MNB_Model.pkl", "wb") as file:
pickle.dump(MNB, file)
# Load the CountVectorizer object from the pickle file
with open("/kaggle/working/CountVectorizer.pkl", "rb") as file:
vectorizer = pickle.load(file)
# Load the MNB Model object from the pickle file
with open("/kaggle/working/MNB_Model.pkl", "rb") as file:
model = pickle.load(file)
# prediction Example
xin = vectorizer.transform(input_text)
xin_array = xin.toarray()
# Predict the class for the input array
predicted_class = model.predict(xin_array)
print("Predicted class:", predicted_class[0])
| false | 1 | 2,434 | 3 | 2,609 | 2,434 |
||
129984568
|
<jupyter_start><jupyter_text>IBM HR Analytics Employee Attrition & Performance
Uncover the factors that lead to employee attrition and explore important questions such as ‘show me a breakdown of distance from home by job role and attrition’ or ‘compare average monthly income by education and attrition’. This is a fictional data set created by IBM data scientists.
Education
1 'Below College'
2 'College'
3 'Bachelor'
4 'Master'
5 'Doctor'
EnvironmentSatisfaction
1 'Low'
2 'Medium'
3 'High'
4 'Very High'
JobInvolvement
1 'Low'
2 'Medium'
3 'High'
4 'Very High'
JobSatisfaction
1 'Low'
2 'Medium'
3 'High'
4 'Very High'
PerformanceRating
1 'Low'
2 'Good'
3 'Excellent'
4 'Outstanding'
RelationshipSatisfaction
1 'Low'
2 'Medium'
3 'High'
4 'Very High'
WorkLifeBalance
1 'Bad'
2 'Good'
3 'Better'
4 'Best'
Kaggle dataset identifier: ibm-hr-analytics-attrition-dataset
<jupyter_script># # **The IBM Employee Attrition Dataset**
# Done by: Saja Abdalaal
# # **Background Information**
# Attrition is a problem that impacts all businesses, irrespective of geography, industry and size of the company. Employee attrition leads to significant costs for a business, including the cost of business disruption, hiring new staff and training new staff. As such, there is great business interest in understanding the drivers of, and minimizing staff attrition. Let us therefore turn to our predictive modelling capabilities and see if we can predict employee attrition on this IBM dataset.
# # **Data**
# Import basic libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# ------------------------------
# Import train and test datasets
train_url = "/kaggle/input/employee/train.csv"
test_url = "/kaggle/input/employee/test.csv"
train_df = pd.read_csv(train_url)
test_df = pd.read_csv(test_url)
train_df.head()
test_df.head()
# Concatenating both train and test data to clean, preprocess and analyse the entire dataset
df = pd.concat(
[train_df.drop("Attrition", axis=1), test_df], axis=0
) # dropped the target column
df.info()
# # **Data preprocessing and EDA**
# ## Null values
df.isnull().sum()
train_df["Attrition"].isnull().sum()
# No Null values!
# -----------------------------
# ## Categorical attributes (Handling object columns)
df.nunique()
df_cat = df.select_dtypes(include="object")
df_num = df.select_dtypes(include=["float64", "int64"])
df_cat = pd.get_dummies(df_cat, drop_first=True)
# ## Drop irrelevant columns
# Concatnating num and cat dataframes
df_new = pd.concat([df_num, df_cat], axis=1)
df_new.hist(bins=50, figsize=(50, 30))
plt.show()
# - From the above visualisation it can be observed that **EmployeeCount, and StandardHours** just include 1 value which wont make a difference in model and might act as noise so it can be dropped instead to build efficient model
#
df_new.drop(["EmployeeCount", "StandardHours"], axis=1, inplace=True)
# ## Correlation
corr = df_new.corr()
plt.figure(figsize=(50, 30))
sns.heatmap(
corr, cbar=True, fmt=".2f", annot=True, annot_kws={"size": 15}, cmap="Greens"
)
# - **Both (Department_Research & Development and Department_Sales) and (JobLevel and MonthelyIncome)** basically concludes the
# same insights so 2 of the 4 columns can be dropped to avoid noise in our model
df_new.drop(["Department_Research & Development", "JobLevel"], axis=1, inplace=True)
# # **Split Data**
train = df_new.iloc[:1058, :]
test = df_new.iloc[1059:, :]
from sklearn.model_selection import train_test_split
X = train
y = train_df["Attrition"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
# # **Model Training**
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score, classification_report
# ## Random Forest Classifier
model = RandomForestClassifier()
model.fit(X_train, y_train)
model_predictions = model.predict(X_test)
print("Accuracy: ", accuracy_score(y_test, model_predictions))
print(classification_report(y_test, model_predictions))
# ## Logistic Regression
lr = LogisticRegression().fit(X_train, y_train)
model_predictions = lr.predict(X_test)
print("Accuracy: ", accuracy_score(y_test, model_predictions))
print(classification_report(y_test, model_predictions))
# ## K-Neighbors Classifier
kn = KNeighborsClassifier().fit(X_train, y_train)
model_predictions = kn.predict(X_test)
print("Accuracy: ", accuracy_score(y_test, model_predictions))
print(classification_report(y_test, model_predictions))
# ## SVC
svc = SVC().fit(X_train, y_train)
model_predictions = svc.predict(X_test)
print("Accuracy: ", accuracy_score(y_test, model_predictions))
print(classification_report(y_test, model_predictions))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/984/129984568.ipynb
|
ibm-hr-analytics-attrition-dataset
|
pavansubhasht
|
[{"Id": 129984568, "ScriptId": 38667563, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10540686, "CreationDate": "05/17/2023 23:45:24", "VersionNumber": 1.0, "Title": "The IBM Employee Attrition", "EvaluationDate": "05/17/2023", "IsChange": true, "TotalLines": 135.0, "LinesInsertedFromPrevious": 135.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 186430511, "KernelVersionId": 129984568, "SourceDatasetVersionId": 1925}, {"Id": 186430512, "KernelVersionId": 129984568, "SourceDatasetVersionId": 1480240}]
|
[{"Id": 1925, "DatasetId": 1067, "DatasourceVersionId": 1925, "CreatorUserId": 862007, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "03/31/2017 06:55:16", "VersionNumber": 1.0, "Title": "IBM HR Analytics Employee Attrition & Performance", "Slug": "ibm-hr-analytics-attrition-dataset", "Subtitle": "Predict attrition of your valuable employees", "Description": "Uncover the factors that lead to employee attrition and explore important questions such as \u2018show me a breakdown of distance from home by job role and attrition\u2019 or \u2018compare average monthly income by education and attrition\u2019. This is a fictional data set created by IBM data scientists.\n\nEducation\n\t1 'Below College'\n\t2 'College'\n\t3 'Bachelor'\n\t4 'Master'\n\t5 'Doctor'\n\t\nEnvironmentSatisfaction\n\t1 'Low'\n\t2 'Medium'\n\t3 'High'\n\t4 'Very High'\n\t\nJobInvolvement\t\n 1 'Low'\n\t2 'Medium'\n\t3 'High'\n\t4 'Very High'\n\t\nJobSatisfaction\t\n 1 'Low'\n\t2 'Medium'\n\t3 'High'\n\t4 'Very High'\n\t\nPerformanceRating\t\n 1 'Low'\n\t2 'Good'\n\t3 'Excellent'\n\t4 'Outstanding'\n\t\nRelationshipSatisfaction\t\n 1 'Low'\n\t2 'Medium'\n\t3 'High'\n\t4 'Very High'\n\t\nWorkLifeBalance\t\n 1 'Bad'\n\t2 'Good'\n\t3 'Better'\n\t4 'Best'", "VersionNotes": "Initial release", "TotalCompressedBytes": 227977.0, "TotalUncompressedBytes": 227977.0}]
|
[{"Id": 1067, "CreatorUserId": 862007, "OwnerUserId": 862007.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1925.0, "CurrentDatasourceVersionId": 1925.0, "ForumId": 3045, "Type": 2, "CreationDate": "03/31/2017 06:55:16", "LastActivityDate": "02/06/2018", "TotalViews": 1254989, "TotalDownloads": 142350, "TotalVotes": 2254, "TotalKernels": 821}]
|
[{"Id": 862007, "UserName": "pavansubhasht", "DisplayName": "pavansubhash", "RegisterDate": "01/10/2017", "PerformanceTier": 0}]
|
# # **The IBM Employee Attrition Dataset**
# Done by: Saja Abdalaal
# # **Background Information**
# Attrition is a problem that impacts all businesses, irrespective of geography, industry and size of the company. Employee attrition leads to significant costs for a business, including the cost of business disruption, hiring new staff and training new staff. As such, there is great business interest in understanding the drivers of, and minimizing staff attrition. Let us therefore turn to our predictive modelling capabilities and see if we can predict employee attrition on this IBM dataset.
# # **Data**
# Import basic libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# ------------------------------
# Import train and test datasets
train_url = "/kaggle/input/employee/train.csv"
test_url = "/kaggle/input/employee/test.csv"
train_df = pd.read_csv(train_url)
test_df = pd.read_csv(test_url)
train_df.head()
test_df.head()
# Concatenating both train and test data to clean, preprocess and analyse the entire dataset
df = pd.concat(
[train_df.drop("Attrition", axis=1), test_df], axis=0
) # dropped the target column
df.info()
# # **Data preprocessing and EDA**
# ## Null values
df.isnull().sum()
train_df["Attrition"].isnull().sum()
# No Null values!
# -----------------------------
# ## Categorical attributes (Handling object columns)
df.nunique()
df_cat = df.select_dtypes(include="object")
df_num = df.select_dtypes(include=["float64", "int64"])
df_cat = pd.get_dummies(df_cat, drop_first=True)
# ## Drop irrelevant columns
# Concatnating num and cat dataframes
df_new = pd.concat([df_num, df_cat], axis=1)
df_new.hist(bins=50, figsize=(50, 30))
plt.show()
# - From the above visualisation it can be observed that **EmployeeCount, and StandardHours** just include 1 value which wont make a difference in model and might act as noise so it can be dropped instead to build efficient model
#
df_new.drop(["EmployeeCount", "StandardHours"], axis=1, inplace=True)
# ## Correlation
corr = df_new.corr()
plt.figure(figsize=(50, 30))
sns.heatmap(
corr, cbar=True, fmt=".2f", annot=True, annot_kws={"size": 15}, cmap="Greens"
)
# - **Both (Department_Research & Development and Department_Sales) and (JobLevel and MonthelyIncome)** basically concludes the
# same insights so 2 of the 4 columns can be dropped to avoid noise in our model
df_new.drop(["Department_Research & Development", "JobLevel"], axis=1, inplace=True)
# # **Split Data**
train = df_new.iloc[:1058, :]
test = df_new.iloc[1059:, :]
from sklearn.model_selection import train_test_split
X = train
y = train_df["Attrition"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
# # **Model Training**
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score, classification_report
# ## Random Forest Classifier
model = RandomForestClassifier()
model.fit(X_train, y_train)
model_predictions = model.predict(X_test)
print("Accuracy: ", accuracy_score(y_test, model_predictions))
print(classification_report(y_test, model_predictions))
# ## Logistic Regression
lr = LogisticRegression().fit(X_train, y_train)
model_predictions = lr.predict(X_test)
print("Accuracy: ", accuracy_score(y_test, model_predictions))
print(classification_report(y_test, model_predictions))
# ## K-Neighbors Classifier
kn = KNeighborsClassifier().fit(X_train, y_train)
model_predictions = kn.predict(X_test)
print("Accuracy: ", accuracy_score(y_test, model_predictions))
print(classification_report(y_test, model_predictions))
# ## SVC
svc = SVC().fit(X_train, y_train)
model_predictions = svc.predict(X_test)
print("Accuracy: ", accuracy_score(y_test, model_predictions))
print(classification_report(y_test, model_predictions))
| false | 0 | 1,222 | 2 | 1,510 | 1,222 |
||
129810036
|
<jupyter_start><jupyter_text>Human Faces (Object Detection)
A diverse compilation of human facial images encompassing various races, age groups, and profiles, with the aim of creating an unbiased dataset that includes coordinates of facial regions suitable for training object detection models.
Buy me a coffee: https://bmc.link/baghbidi
Kaggle dataset identifier: human-faces-object-detection
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
break
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import cv2 as cv
import matplotlib.pyplot as plt
from matplotlib import patches
import tensorflow as tf
from PIL import Image
boxes = pd.read_csv("/kaggle/input/human-faces-object-detection/faces.csv")
boxes.set_index("image_name", inplace=True)
boxes.head()
images_dir = "/kaggle/input/human-faces-object-detection/images/"
image_name = "00001722.jpg"
box_x, box_y = boxes.loc[image_name].x0, boxes.loc[image_name].y0
box_width, box_height = (
boxes.loc[image_name].x1 - box_x,
boxes.loc[image_name].y1 - box_y,
)
image = cv.imread(images_dir + image_name)[
..., ::-1
] # cv.imread returns in brg format, not rgb
plt.imshow(image)
plt.gca().add_patch(
patches.Rectangle(
(box_x, box_y), box_width, box_height, edgecolor="g", facecolor="none"
)
)
# ## All images need to be reshaped to the same shape, so are boxes
reshaped_size = 256
boxes.x0 = (boxes.x0 / boxes.width * reshaped_size).astype(int)
boxes.x1 = (boxes.x1 / boxes.width * reshaped_size).astype(int)
boxes.y0 = (boxes.y0 / boxes.height * reshaped_size).astype(int)
boxes.y1 = (boxes.y1 / boxes.height * reshaped_size).astype(int)
box_x, box_y = boxes.loc[image_name].x0, boxes.loc[image_name].y0
box_width, box_height = (
boxes.loc[image_name].x1 - box_x,
boxes.loc[image_name].y1 - box_y,
)
plt.imshow(cv.resize(image, (reshaped_size, reshaped_size)))
plt.gca().add_patch(
patches.Rectangle(
(box_x, box_y), box_width, box_height, edgecolor="g", facecolor="none"
)
)
def load_image_and_boxes(path):
# loads image and box for this image from given path
# also normalizes image and box coords
image = cv.imread(path.decode("utf-8"))[..., ::-1]
image = cv.resize(image, (reshaped_size, reshaped_size)) / 255.0
label = boxes.loc[path.decode("utf-8").split("/")[-1]].values[2:] / 255.0
return [image.astype(np.float32), label.astype(np.float32)]
images_path = [os.path.join(images_dir, f) for f in os.listdir(images_dir)]
ds = tf.data.Dataset.from_tensor_slices(images_path)
ds = ds.map(
lambda x: tf.numpy_function(load_image_and_boxes, [x], [np.float32, np.float32])
)
for i in ds.take(1):
plt.imshow(i[0])
box = i[1] * 255
plt.gca().add_patch(
patches.Rectangle(
(box[0], box[1]),
box[2] - box[0],
box[3] - box[1],
edgecolor="g",
facecolor="none",
)
)
# Create horizontal motion-blur kernel
kernel_size = 10
kernel = np.zeros((kernel_size, kernel_size, 3))
kernel[int((kernel_size - 1) / 2), :, :] = np.ones((kernel_size, 3)) / kernel_size
# Apply convolution to perform bluring
def blur_image(image):
blurred_array = np.zeros_like(image, dtype=np.float32)
for channel in range(3):
blurred_array[:, :, channel] = np.convolve(
image[:, :, channel].flatten(), kernel[:, :, channel].flatten(), mode="same"
).reshape(image.shape[:2])
return blurred_array
image = ds.take(1)
def add_gaussian_noise(image, mean, std_dev):
noisy_image = np.copy(image)
h, w, _ = noisy_image.shape
noise = np.random.normal(mean, std_dev, (h, w, 3))
noisy_image = noisy_image + noise
noisy_image = np.clip(noisy_image, 0, 255).astype(np.float32)
return noisy_image
def perform_svd(blurred_channel, blur_matrix, rank=100):
U, S, V = np.linalg.svd(blur_matrix)
U = U[:, :rank]
S = S[:rank]
V = V[:rank, :]
S_inv = np.zeros((S.shape[0], S.shape[0]))
S_inv[: S.shape[0], : S.shape[0]] = np.diag(1 / S)
reconstructed_channel = V.T @ S_inv @ U.T @ blurred_channel
return reconstructed_channel
for i in image:
x = np.array(i[0], dtype=np.float32)
blurred = blur_image(x)
A_0 = blurred[:, :, 0] @ np.linalg.inv(x[:, :, 0])
A_1 = blurred[:, :, 1] @ np.linalg.inv(x[:, :, 1])
A_2 = blurred[:, :, 2] @ np.linalg.inv(x[:, :, 2])
res = np.stack([A_0 @ x[:, :, 0], A_1 @ x[:, :, 1], A_2 @ x[:, :, 2]], 2)
res = add_gaussian_noise(res, 0.002, 0.002)
# Using svd to deblure an image
reconstructed_channel_0 = perform_svd(res[:, :, 0], A_0)
reconstructed_channel_1 = perform_svd(res[:, :, 1], A_1)
reconstructed_channel_2 = perform_svd(res[:, :, 2], A_2)
reconstructed_image = np.stack(
[reconstructed_channel_0, reconstructed_channel_1, reconstructed_channel_2], 2
)
# print(reconstructed_image.shape)
plt.imshow(reconstructed_image)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/810/129810036.ipynb
|
human-faces-object-detection
|
sbaghbidi
|
[{"Id": 129810036, "ScriptId": 38556904, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 10630531, "CreationDate": "05/16/2023 16:08:50", "VersionNumber": 4.0, "Title": "face_recognition", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 142.0, "LinesInsertedFromPrevious": 3.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 139.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186184303, "KernelVersionId": 129810036, "SourceDatasetVersionId": 5377440}]
|
[{"Id": 5377440, "DatasetId": 3119215, "DatasourceVersionId": 5451066, "CreatorUserId": 2371623, "LicenseName": "CC0: Public Domain", "CreationDate": "04/12/2023 01:38:47", "VersionNumber": 1.0, "Title": "Human Faces (Object Detection)", "Slug": "human-faces-object-detection", "Subtitle": "A curated collection of human facial images for training object detection models", "Description": "A diverse compilation of human facial images encompassing various races, age groups, and profiles, with the aim of creating an unbiased dataset that includes coordinates of facial regions suitable for training object detection models.\n\nBuy me a coffee: https://bmc.link/baghbidi", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3119215, "CreatorUserId": 2371623, "OwnerUserId": 2371623.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5377440.0, "CurrentDatasourceVersionId": 5451066.0, "ForumId": 3182683, "Type": 2, "CreationDate": "04/12/2023 01:38:47", "LastActivityDate": "04/12/2023", "TotalViews": 16459, "TotalDownloads": 2207, "TotalVotes": 59, "TotalKernels": 12}]
|
[{"Id": 2371623, "UserName": "sbaghbidi", "DisplayName": "Saeid", "RegisterDate": "10/17/2018", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
break
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import cv2 as cv
import matplotlib.pyplot as plt
from matplotlib import patches
import tensorflow as tf
from PIL import Image
boxes = pd.read_csv("/kaggle/input/human-faces-object-detection/faces.csv")
boxes.set_index("image_name", inplace=True)
boxes.head()
images_dir = "/kaggle/input/human-faces-object-detection/images/"
image_name = "00001722.jpg"
box_x, box_y = boxes.loc[image_name].x0, boxes.loc[image_name].y0
box_width, box_height = (
boxes.loc[image_name].x1 - box_x,
boxes.loc[image_name].y1 - box_y,
)
image = cv.imread(images_dir + image_name)[
..., ::-1
] # cv.imread returns in brg format, not rgb
plt.imshow(image)
plt.gca().add_patch(
patches.Rectangle(
(box_x, box_y), box_width, box_height, edgecolor="g", facecolor="none"
)
)
# ## All images need to be reshaped to the same shape, so are boxes
reshaped_size = 256
boxes.x0 = (boxes.x0 / boxes.width * reshaped_size).astype(int)
boxes.x1 = (boxes.x1 / boxes.width * reshaped_size).astype(int)
boxes.y0 = (boxes.y0 / boxes.height * reshaped_size).astype(int)
boxes.y1 = (boxes.y1 / boxes.height * reshaped_size).astype(int)
box_x, box_y = boxes.loc[image_name].x0, boxes.loc[image_name].y0
box_width, box_height = (
boxes.loc[image_name].x1 - box_x,
boxes.loc[image_name].y1 - box_y,
)
plt.imshow(cv.resize(image, (reshaped_size, reshaped_size)))
plt.gca().add_patch(
patches.Rectangle(
(box_x, box_y), box_width, box_height, edgecolor="g", facecolor="none"
)
)
def load_image_and_boxes(path):
# loads image and box for this image from given path
# also normalizes image and box coords
image = cv.imread(path.decode("utf-8"))[..., ::-1]
image = cv.resize(image, (reshaped_size, reshaped_size)) / 255.0
label = boxes.loc[path.decode("utf-8").split("/")[-1]].values[2:] / 255.0
return [image.astype(np.float32), label.astype(np.float32)]
images_path = [os.path.join(images_dir, f) for f in os.listdir(images_dir)]
ds = tf.data.Dataset.from_tensor_slices(images_path)
ds = ds.map(
lambda x: tf.numpy_function(load_image_and_boxes, [x], [np.float32, np.float32])
)
for i in ds.take(1):
plt.imshow(i[0])
box = i[1] * 255
plt.gca().add_patch(
patches.Rectangle(
(box[0], box[1]),
box[2] - box[0],
box[3] - box[1],
edgecolor="g",
facecolor="none",
)
)
# Create horizontal motion-blur kernel
kernel_size = 10
kernel = np.zeros((kernel_size, kernel_size, 3))
kernel[int((kernel_size - 1) / 2), :, :] = np.ones((kernel_size, 3)) / kernel_size
# Apply convolution to perform bluring
def blur_image(image):
blurred_array = np.zeros_like(image, dtype=np.float32)
for channel in range(3):
blurred_array[:, :, channel] = np.convolve(
image[:, :, channel].flatten(), kernel[:, :, channel].flatten(), mode="same"
).reshape(image.shape[:2])
return blurred_array
image = ds.take(1)
def add_gaussian_noise(image, mean, std_dev):
noisy_image = np.copy(image)
h, w, _ = noisy_image.shape
noise = np.random.normal(mean, std_dev, (h, w, 3))
noisy_image = noisy_image + noise
noisy_image = np.clip(noisy_image, 0, 255).astype(np.float32)
return noisy_image
def perform_svd(blurred_channel, blur_matrix, rank=100):
U, S, V = np.linalg.svd(blur_matrix)
U = U[:, :rank]
S = S[:rank]
V = V[:rank, :]
S_inv = np.zeros((S.shape[0], S.shape[0]))
S_inv[: S.shape[0], : S.shape[0]] = np.diag(1 / S)
reconstructed_channel = V.T @ S_inv @ U.T @ blurred_channel
return reconstructed_channel
for i in image:
x = np.array(i[0], dtype=np.float32)
blurred = blur_image(x)
A_0 = blurred[:, :, 0] @ np.linalg.inv(x[:, :, 0])
A_1 = blurred[:, :, 1] @ np.linalg.inv(x[:, :, 1])
A_2 = blurred[:, :, 2] @ np.linalg.inv(x[:, :, 2])
res = np.stack([A_0 @ x[:, :, 0], A_1 @ x[:, :, 1], A_2 @ x[:, :, 2]], 2)
res = add_gaussian_noise(res, 0.002, 0.002)
# Using svd to deblure an image
reconstructed_channel_0 = perform_svd(res[:, :, 0], A_0)
reconstructed_channel_1 = perform_svd(res[:, :, 1], A_1)
reconstructed_channel_2 = perform_svd(res[:, :, 2], A_2)
reconstructed_image = np.stack(
[reconstructed_channel_0, reconstructed_channel_1, reconstructed_channel_2], 2
)
# print(reconstructed_image.shape)
plt.imshow(reconstructed_image)
| false | 1 | 1,808 | 0 | 1,900 | 1,808 |
||
129810349
|
# ### Импортируем библиотеки для работы.
import pandas as pd
import numpy as np
from sklearn.pipeline import Pipeline
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn.feature_extraction.text import CountVectorizer
import matplotlib.pyplot as plt
pd.set_option("display.max_columns", None)
# ## 2.0 Построение, обучение и оптимизация модели
# ### Загрузим данные из предыдущей сессии
df = pd.read_csv("result_habr.csv", encoding="utf-8-sig")
# ### Просмотрим данные в нашем наборе.
df.head()
# Видим ненужный столбец "Unnamed: 0"
# Удалим столбец "Unnamed: 0".
df = df.drop(columns="Unnamed: 0")
# Просмотрим информацию о наборе данных
df.info()
# Видим пустые значения
# Узнаем подробности
df.isnull().sum()
# В наборе данных каким-то образом появились пустые значения, удалим их.
df = df[df["TextArticle"].notnull()]
df = df[df["Lemmatization"].notnull()]
df = df[df["Target"].notnull()]
df.isnull().sum()
# Можно продолжить работу
# ## 2.1 Построение модели классификации
# Оставим названия существующих номинаций.
targets = df["Target"].unique()
print(targets)
# Прежде чем раделять на выборки удалим ненужные поля.
df = df.drop(
columns=[
"Name",
"Raiting",
"Field",
"DataPublish",
"TextArticle",
"Vector",
"Target",
]
)
# Просмотрим, что получилось.
df.head()
# ### 2.1.1 Разделение выборки на обучающую и тестовую
from sklearn.model_selection import train_test_split
# Разобъём набор данных по принципу 1/3. Обоснование приведенно ниже
# Обоснование разбиения набора данных: разбиение набора данных на обучающую и тестирующую выборки будут произведенны на основании официального сайта функций машинного обучение - scikit-learn. Для этого обратимся к официальному сайту библиотеки scikit-learn
# https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html
# На примере данном на офиациальном сайте можно увидеть, что там также используют разбиение 1/3. Из данной информации мы можем понять, что данное разбиение будет наиболее оптимальным.
# 1/3 - это значит, что тестовая выборка будет иметь 0.33 данных
X_train, X_test, y_train, y_test = train_test_split(
df["Lemmatization"], df["Target2"], test_size=0.33, random_state=42, shuffle=True
)
# ### 2.1.2 Метод ближайших соседей | K-nearest Neighbors (KNN)
# Алгоритм является черезвычайно легким в использовании, но при этом может выполнять и сложные функции.
# KNN — это непараметрический алгоритм обучения, что означает, что он ничего не предполагает о базовых данных. Это чрезвычайно полезная функция, поскольку большинство реальных данных на самом деле не следуют никаким теоретическим предположениям
# Данные причины были основными для выбора данного алгоритма
# Импортируем библиотеку
from sklearn.neighbors import KNeighborsClassifier
# Создадим объект типа PipeLine для дальнейшей работы
# Выберем стандартное количество соседей в виде 3-х
knn = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("knn", KNeighborsClassifier(n_neighbors=3)),
]
)
print(knn)
# Обучение модели
knn.fit(X_train, y_train)
y_pred = knn.predict(X_test)
# Для вычисление точности используем метрику f1
# Метрика F1 score является хорошим выбором для оценки качества моделей классификации, особенно если классы несбалансированы. Она учитывает как точность (precision), так и полноту (recall) модели, что позволяет более полно оценивать ее работу.
from sklearn.metrics import f1_score
print("f1 %s" % f1_score(y_pred, y_test, average="micro"))
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred, target_names=targets))
# ### 2.1.3 Метод Наивный Байесовский | The Naive Bayes Algorithm
# Алгоритм Naive Bayes прост в реализации и не требует большого количества времени для обучения.
# Алгоритм Naive Bayes обрабатывает данные быстро, что делает его решение многих задач в реальном времени.
from sklearn.naive_bayes import MultinomialNB
nb = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("clf", MultinomialNB()),
]
)
# Обучение модели
nb.fit(X_train, y_train)
y_pred = nb.predict(X_test)
print("f1 %s" % f1_score(y_pred, y_test, average="micro"))
print(classification_report(y_test, y_pred, target_names=targets))
# ### 2.1.4 Метод Классификатор дерева решений | DecisionTreeClassifier
# Деревья решений устойчивы к выбросам и шуму в данных, а также деревья решений могут обрабатывать большие объемы данных с высокой скоростью.
from sklearn.tree import DecisionTreeClassifier
dtc = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("dtc", DecisionTreeClassifier()),
]
)
# Обучение модели
dtc.fit(X_train, y_train)
y_pred = dtc.predict(X_test)
print("f1 %s" % f1_score(y_pred, y_test, average="micro"))
print(classification_report(y_test, y_pred, target_names=targets))
from yellowbrick.classifier import ROCAUC
visualizer = ROCAUC(dtc)
visualizer.fit(X_train, y_train)
visualizer.score(X_test, y_test)
visualizer.show()
from yellowbrick.classifier import PrecisionRecallCurve
vizual_1 = PrecisionRecallCurve(dtc)
vizual_1.fit(X_train, y_train)
vizual_1.score(X_test, y_test)
vizual_1.show()
# ## Вывод о проделанной работе:
# Разбиение выборки на 1/3 и обучение моделей различными методами классификации такими как __K-nearest Neighbors__, __Naive Bayes Algorithm__, __DecisionTreeClassifier__ и __Logistic Regression__.
# Оценив точность модели с помощью метрики __f1_score__, можно увидеть, что метод классификации __DecisionTreeClassifier__ имеет наибольшую точность, а именно 0.792.
# ## 2.2 Оптимизация модели
# ### 2.2.1 Настройка гиперпараметров
# Проведем поиск лучшего значения для параметра max_depth
best_depth = 1
best_f1_accuracy = 0
for i in range(1, 25):
dtcc = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("dtc", DecisionTreeClassifier(max_depth=i)),
]
)
dtcc.fit(X_train, y_train)
y_pred = dtcc.predict(X_test)
accuracy = f1_score(y_pred, y_test, average="micro")
print("f1 %s" % accuracy)
if accuracy > best_f1_accuracy:
best_f1_accuracy = accuracy
best_depth = i
# Просмотрим лучшее значение
print(best_depth)
# Просмотрим точность при этом значении
print(best_f1_accuracy)
# ### 2.2.2 Обучим модель на лучшем параметре
dtc = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("dtc", DecisionTreeClassifier(max_depth=best_depth)),
]
)
dtc.fit(X_train, y_train)
y_pred = dtc.predict(X_test)
accuracy = f1_score(y_pred, y_test, average="micro")
print("f1 %s" % accuracy)
# ### 2.2.3 Кривые валидации и обучения
# Построим кривую на оптимизированной модели.
visualizer = ROCAUC(dtc)
visualizer.fit(X_train, y_train)
visualizer.score(X_test, y_test)
visualizer.show()
# Проверим верность предугадывания модели, выберем любое число
dtc.predict([df["Lemmatization"][500]])
# Давайте сравним предугаданное от реального
df["Target2"][500]
# Как можно увидеть модель правильно определила номинацию для 500 статьи.
# ### 2.2.4 Сохранение модели для использования в будущем.
# Импортируем библиотеку
import joblib
# Сохранение модели
joblib.dump(dtc, "model.pkl")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/810/129810349.ipynb
| null | null |
[{"Id": 129810349, "ScriptId": 38606443, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13417746, "CreationDate": "05/16/2023 16:11:23", "VersionNumber": 1.0, "Title": "Report2-SHA-djostit", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 270.0, "LinesInsertedFromPrevious": 270.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
# ### Импортируем библиотеки для работы.
import pandas as pd
import numpy as np
from sklearn.pipeline import Pipeline
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn.feature_extraction.text import CountVectorizer
import matplotlib.pyplot as plt
pd.set_option("display.max_columns", None)
# ## 2.0 Построение, обучение и оптимизация модели
# ### Загрузим данные из предыдущей сессии
df = pd.read_csv("result_habr.csv", encoding="utf-8-sig")
# ### Просмотрим данные в нашем наборе.
df.head()
# Видим ненужный столбец "Unnamed: 0"
# Удалим столбец "Unnamed: 0".
df = df.drop(columns="Unnamed: 0")
# Просмотрим информацию о наборе данных
df.info()
# Видим пустые значения
# Узнаем подробности
df.isnull().sum()
# В наборе данных каким-то образом появились пустые значения, удалим их.
df = df[df["TextArticle"].notnull()]
df = df[df["Lemmatization"].notnull()]
df = df[df["Target"].notnull()]
df.isnull().sum()
# Можно продолжить работу
# ## 2.1 Построение модели классификации
# Оставим названия существующих номинаций.
targets = df["Target"].unique()
print(targets)
# Прежде чем раделять на выборки удалим ненужные поля.
df = df.drop(
columns=[
"Name",
"Raiting",
"Field",
"DataPublish",
"TextArticle",
"Vector",
"Target",
]
)
# Просмотрим, что получилось.
df.head()
# ### 2.1.1 Разделение выборки на обучающую и тестовую
from sklearn.model_selection import train_test_split
# Разобъём набор данных по принципу 1/3. Обоснование приведенно ниже
# Обоснование разбиения набора данных: разбиение набора данных на обучающую и тестирующую выборки будут произведенны на основании официального сайта функций машинного обучение - scikit-learn. Для этого обратимся к официальному сайту библиотеки scikit-learn
# https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html
# На примере данном на офиациальном сайте можно увидеть, что там также используют разбиение 1/3. Из данной информации мы можем понять, что данное разбиение будет наиболее оптимальным.
# 1/3 - это значит, что тестовая выборка будет иметь 0.33 данных
X_train, X_test, y_train, y_test = train_test_split(
df["Lemmatization"], df["Target2"], test_size=0.33, random_state=42, shuffle=True
)
# ### 2.1.2 Метод ближайших соседей | K-nearest Neighbors (KNN)
# Алгоритм является черезвычайно легким в использовании, но при этом может выполнять и сложные функции.
# KNN — это непараметрический алгоритм обучения, что означает, что он ничего не предполагает о базовых данных. Это чрезвычайно полезная функция, поскольку большинство реальных данных на самом деле не следуют никаким теоретическим предположениям
# Данные причины были основными для выбора данного алгоритма
# Импортируем библиотеку
from sklearn.neighbors import KNeighborsClassifier
# Создадим объект типа PipeLine для дальнейшей работы
# Выберем стандартное количество соседей в виде 3-х
knn = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("knn", KNeighborsClassifier(n_neighbors=3)),
]
)
print(knn)
# Обучение модели
knn.fit(X_train, y_train)
y_pred = knn.predict(X_test)
# Для вычисление точности используем метрику f1
# Метрика F1 score является хорошим выбором для оценки качества моделей классификации, особенно если классы несбалансированы. Она учитывает как точность (precision), так и полноту (recall) модели, что позволяет более полно оценивать ее работу.
from sklearn.metrics import f1_score
print("f1 %s" % f1_score(y_pred, y_test, average="micro"))
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred, target_names=targets))
# ### 2.1.3 Метод Наивный Байесовский | The Naive Bayes Algorithm
# Алгоритм Naive Bayes прост в реализации и не требует большого количества времени для обучения.
# Алгоритм Naive Bayes обрабатывает данные быстро, что делает его решение многих задач в реальном времени.
from sklearn.naive_bayes import MultinomialNB
nb = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("clf", MultinomialNB()),
]
)
# Обучение модели
nb.fit(X_train, y_train)
y_pred = nb.predict(X_test)
print("f1 %s" % f1_score(y_pred, y_test, average="micro"))
print(classification_report(y_test, y_pred, target_names=targets))
# ### 2.1.4 Метод Классификатор дерева решений | DecisionTreeClassifier
# Деревья решений устойчивы к выбросам и шуму в данных, а также деревья решений могут обрабатывать большие объемы данных с высокой скоростью.
from sklearn.tree import DecisionTreeClassifier
dtc = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("dtc", DecisionTreeClassifier()),
]
)
# Обучение модели
dtc.fit(X_train, y_train)
y_pred = dtc.predict(X_test)
print("f1 %s" % f1_score(y_pred, y_test, average="micro"))
print(classification_report(y_test, y_pred, target_names=targets))
from yellowbrick.classifier import ROCAUC
visualizer = ROCAUC(dtc)
visualizer.fit(X_train, y_train)
visualizer.score(X_test, y_test)
visualizer.show()
from yellowbrick.classifier import PrecisionRecallCurve
vizual_1 = PrecisionRecallCurve(dtc)
vizual_1.fit(X_train, y_train)
vizual_1.score(X_test, y_test)
vizual_1.show()
# ## Вывод о проделанной работе:
# Разбиение выборки на 1/3 и обучение моделей различными методами классификации такими как __K-nearest Neighbors__, __Naive Bayes Algorithm__, __DecisionTreeClassifier__ и __Logistic Regression__.
# Оценив точность модели с помощью метрики __f1_score__, можно увидеть, что метод классификации __DecisionTreeClassifier__ имеет наибольшую точность, а именно 0.792.
# ## 2.2 Оптимизация модели
# ### 2.2.1 Настройка гиперпараметров
# Проведем поиск лучшего значения для параметра max_depth
best_depth = 1
best_f1_accuracy = 0
for i in range(1, 25):
dtcc = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("dtc", DecisionTreeClassifier(max_depth=i)),
]
)
dtcc.fit(X_train, y_train)
y_pred = dtcc.predict(X_test)
accuracy = f1_score(y_pred, y_test, average="micro")
print("f1 %s" % accuracy)
if accuracy > best_f1_accuracy:
best_f1_accuracy = accuracy
best_depth = i
# Просмотрим лучшее значение
print(best_depth)
# Просмотрим точность при этом значении
print(best_f1_accuracy)
# ### 2.2.2 Обучим модель на лучшем параметре
dtc = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("dtc", DecisionTreeClassifier(max_depth=best_depth)),
]
)
dtc.fit(X_train, y_train)
y_pred = dtc.predict(X_test)
accuracy = f1_score(y_pred, y_test, average="micro")
print("f1 %s" % accuracy)
# ### 2.2.3 Кривые валидации и обучения
# Построим кривую на оптимизированной модели.
visualizer = ROCAUC(dtc)
visualizer.fit(X_train, y_train)
visualizer.score(X_test, y_test)
visualizer.show()
# Проверим верность предугадывания модели, выберем любое число
dtc.predict([df["Lemmatization"][500]])
# Давайте сравним предугаданное от реального
df["Target2"][500]
# Как можно увидеть модель правильно определила номинацию для 500 статьи.
# ### 2.2.4 Сохранение модели для использования в будущем.
# Импортируем библиотеку
import joblib
# Сохранение модели
joblib.dump(dtc, "model.pkl")
| false | 0 | 2,851 | 1 | 2,851 | 2,851 |
||
129810327
|
<jupyter_start><jupyter_text>Iris.csv
Kaggle dataset identifier: iriscsv
<jupyter_script># # Ex - 09 22MCA1049 PRADEEP KUMAR
# # RANDOM FOREST CLASSIFIER
# 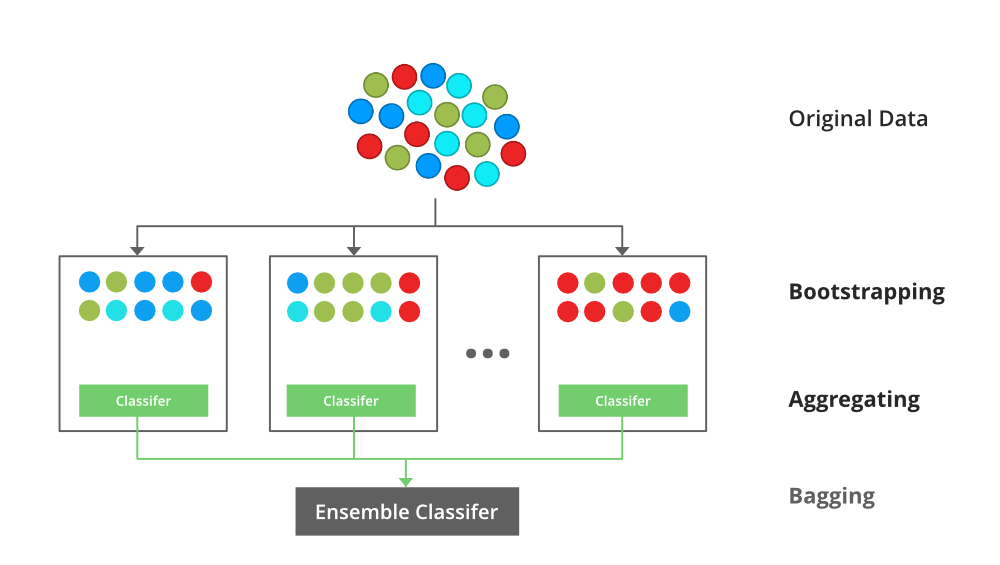
# ## 1. Load the dataset (iris.csv).
import pandas as pd
iris_df = pd.read_csv("/kaggle/input/iriscsv/Iris.csv")
iris_df.shape
iris_df.tail()
# ## 2. Load the dataset (Churnprediction.csv).
churn_df = pd.read_csv(
"/kaggle/input/telecom-customer-churn-by-maven-analytics/telecom_customer_churn.csv"
)
churn_df.head()
# ## 3. Drop columns that are not required for classification of Churn Risk.
iris_df = iris_df.drop(["Id"], axis=1)
churn_df = churn_df.drop(["Customer ID"], axis=1)
# ## 4. If require perform data preprocessing.
# There is no need for data preprocessing in this case.
# ## 5. Split dataset into test and train (20:80).
from sklearn.model_selection import train_test_split
# Separate the features from the target variable
X = iris_df.drop("Species", axis=1)
y = iris_df["Species"]
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
X_churn = churn_df.drop("Churn Category", axis=1)
y_churn = churn_df["Churn Category"]
X_train_churn, X_test_churn, y_train_churn, y_test_churn = train_test_split(
X_churn, y_churn, test_size=0.2
)
# ## 6. Build any three classification models for identifying Churn Risk.
# Logistic Regression
from sklearn.linear_model import LogisticRegression
logistic_regression = LogisticRegression(max_iter=1000)
logistic_regression.fit(X_train, y_train)
# Decision Tree
from sklearn.tree import DecisionTreeClassifier
decision_tree = DecisionTreeClassifier()
decision_tree.fit(X_train, y_train)
# Random Forest
from sklearn.ensemble import RandomForestClassifier
random_forest = RandomForestClassifier()
random_forest.fit(X_train, y_train)
# ## 7. Build Voting ensemble classifier on the training dataset.
from sklearn.ensemble import VotingClassifier
voting_classifier = VotingClassifier(
estimators=[
("lr", logistic_regression),
("dt", decision_tree),
("rf", random_forest),
],
voting="hard",
)
voting_classifier.fit(X_train, y_train)
# ## 8. Build Bagging ensemble classifier on the training dataset.
import warnings
# Ignore the FutureWarning for the base_estimator parameter
warnings.filterwarnings("ignore", category=FutureWarning)
from sklearn.ensemble import BaggingClassifier
bagging_classifier = BaggingClassifier(
base_estimator=logistic_regression, n_estimators=100
)
bagging_classifier.fit(X_train, y_train)
# ## 9. Build Boosting ensemble classifier on the training dataset.
from sklearn.ensemble import AdaBoostClassifier
adaboost_classifier = AdaBoostClassifier(
base_estimator=logistic_regression, n_estimators=100
)
adaboost_classifier.fit(X_train, y_train)
# ## 10. Fit the models designed from step-5 to step-8 on the test dataset.
logistic_regression.score(X_test, y_test)
decision_tree.score(X_test, y_test)
random_forest.score(X_test, y_test)
voting_classifier.score(X_test, y_test)
bagging_classifier.score(X_test, y_test)
adaboost_classifier.score(X_test, y_test)
# ## 11. Evaluate the designed models from step-5 to step-8 with appropriate classification metrics.
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
accuracy_score(y_test, logistic_regression.predict(X_test))
import pickle
# Assuming you have a trained model object named 'model'
# Save the model to a file using pickle
with open("model.pkl", "wb") as file:
pickle.dump(adaboost_classifier, file)
import pickle
# Load the model from the pickle file
with open("model.pkl", "rb") as file:
adaboost_classifier = pickle.load(file)
# Assuming you have loaded the model from the pickle file
# And you have new data stored in a variable named 'X_new'
# Make predictions using the loaded model
predictions = adaboost_classifier.predict([[5.1, 3.5, 1.4, 0.2]])
predictions
import pandas as pd
# Define the new data
new_data = {
"sepal_length": [5.1, 6.2, 7.3],
"sepal_width": [3.5, 2.8, 3.0],
"petal_length": [1.4, 4.8, 6.3],
"petal_width": [0.2, 1.8, 2.5],
"species": ["Setosa", "Versicolor", "Virginica"],
}
# Create a DataFrame from the new data
df = pd.DataFrame(new_data)
# Save the DataFrame to a CSV file
df.to_csv("new_data.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/810/129810327.ipynb
|
iriscsv
|
saurabh00007
|
[{"Id": 129810327, "ScriptId": 38583553, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13322048, "CreationDate": "05/16/2023 16:11:07", "VersionNumber": 4.0, "Title": "ML Ex. 9 22MCA1049 PRADEEP KUMAR", "EvaluationDate": "05/16/2023", "IsChange": false, "TotalLines": 154.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 154.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 186184622, "KernelVersionId": 129810327, "SourceDatasetVersionId": 6570}, {"Id": 186184623, "KernelVersionId": 129810327, "SourceDatasetVersionId": 23404}, {"Id": 186184624, "KernelVersionId": 129810327, "SourceDatasetVersionId": 3907949}]
|
[{"Id": 6570, "DatasetId": 4247, "DatasourceVersionId": 6570, "CreatorUserId": 1301025, "LicenseName": "CC0: Public Domain", "CreationDate": "11/09/2017 07:34:35", "VersionNumber": 1.0, "Title": "Iris.csv", "Slug": "iriscsv", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 5107.0, "TotalUncompressedBytes": 5107.0}]
|
[{"Id": 4247, "CreatorUserId": 1301025, "OwnerUserId": 1301025.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 6570.0, "CurrentDatasourceVersionId": 6570.0, "ForumId": 9871, "Type": 2, "CreationDate": "11/09/2017 07:34:35", "LastActivityDate": "02/05/2018", "TotalViews": 274115, "TotalDownloads": 76238, "TotalVotes": 136, "TotalKernels": 90}]
|
[{"Id": 1301025, "UserName": "saurabh00007", "DisplayName": "saurabh singh", "RegisterDate": "09/29/2017", "PerformanceTier": 0}]
|
# # Ex - 09 22MCA1049 PRADEEP KUMAR
# # RANDOM FOREST CLASSIFIER
# 
# ## 1. Load the dataset (iris.csv).
import pandas as pd
iris_df = pd.read_csv("/kaggle/input/iriscsv/Iris.csv")
iris_df.shape
iris_df.tail()
# ## 2. Load the dataset (Churnprediction.csv).
churn_df = pd.read_csv(
"/kaggle/input/telecom-customer-churn-by-maven-analytics/telecom_customer_churn.csv"
)
churn_df.head()
# ## 3. Drop columns that are not required for classification of Churn Risk.
iris_df = iris_df.drop(["Id"], axis=1)
churn_df = churn_df.drop(["Customer ID"], axis=1)
# ## 4. If require perform data preprocessing.
# There is no need for data preprocessing in this case.
# ## 5. Split dataset into test and train (20:80).
from sklearn.model_selection import train_test_split
# Separate the features from the target variable
X = iris_df.drop("Species", axis=1)
y = iris_df["Species"]
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
X_churn = churn_df.drop("Churn Category", axis=1)
y_churn = churn_df["Churn Category"]
X_train_churn, X_test_churn, y_train_churn, y_test_churn = train_test_split(
X_churn, y_churn, test_size=0.2
)
# ## 6. Build any three classification models for identifying Churn Risk.
# Logistic Regression
from sklearn.linear_model import LogisticRegression
logistic_regression = LogisticRegression(max_iter=1000)
logistic_regression.fit(X_train, y_train)
# Decision Tree
from sklearn.tree import DecisionTreeClassifier
decision_tree = DecisionTreeClassifier()
decision_tree.fit(X_train, y_train)
# Random Forest
from sklearn.ensemble import RandomForestClassifier
random_forest = RandomForestClassifier()
random_forest.fit(X_train, y_train)
# ## 7. Build Voting ensemble classifier on the training dataset.
from sklearn.ensemble import VotingClassifier
voting_classifier = VotingClassifier(
estimators=[
("lr", logistic_regression),
("dt", decision_tree),
("rf", random_forest),
],
voting="hard",
)
voting_classifier.fit(X_train, y_train)
# ## 8. Build Bagging ensemble classifier on the training dataset.
import warnings
# Ignore the FutureWarning for the base_estimator parameter
warnings.filterwarnings("ignore", category=FutureWarning)
from sklearn.ensemble import BaggingClassifier
bagging_classifier = BaggingClassifier(
base_estimator=logistic_regression, n_estimators=100
)
bagging_classifier.fit(X_train, y_train)
# ## 9. Build Boosting ensemble classifier on the training dataset.
from sklearn.ensemble import AdaBoostClassifier
adaboost_classifier = AdaBoostClassifier(
base_estimator=logistic_regression, n_estimators=100
)
adaboost_classifier.fit(X_train, y_train)
# ## 10. Fit the models designed from step-5 to step-8 on the test dataset.
logistic_regression.score(X_test, y_test)
decision_tree.score(X_test, y_test)
random_forest.score(X_test, y_test)
voting_classifier.score(X_test, y_test)
bagging_classifier.score(X_test, y_test)
adaboost_classifier.score(X_test, y_test)
# ## 11. Evaluate the designed models from step-5 to step-8 with appropriate classification metrics.
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
accuracy_score(y_test, logistic_regression.predict(X_test))
import pickle
# Assuming you have a trained model object named 'model'
# Save the model to a file using pickle
with open("model.pkl", "wb") as file:
pickle.dump(adaboost_classifier, file)
import pickle
# Load the model from the pickle file
with open("model.pkl", "rb") as file:
adaboost_classifier = pickle.load(file)
# Assuming you have loaded the model from the pickle file
# And you have new data stored in a variable named 'X_new'
# Make predictions using the loaded model
predictions = adaboost_classifier.predict([[5.1, 3.5, 1.4, 0.2]])
predictions
import pandas as pd
# Define the new data
new_data = {
"sepal_length": [5.1, 6.2, 7.3],
"sepal_width": [3.5, 2.8, 3.0],
"petal_length": [1.4, 4.8, 6.3],
"petal_width": [0.2, 1.8, 2.5],
"species": ["Setosa", "Versicolor", "Virginica"],
}
# Create a DataFrame from the new data
df = pd.DataFrame(new_data)
# Save the DataFrame to a CSV file
df.to_csv("new_data.csv", index=False)
| false | 2 | 1,431 | 0 | 1,451 | 1,431 |
||
129810107
|
# ## Gradient descent
# Gradient descent is an iterative optimization algorithm used to minimize a cost function by adjusting the model parameters in the direction of steepest descent of the gradient. The goal is to find the optimal values of the model parameters that result in the minimum value of the cost function. The gradient is calculated by computing the partial derivatives of the cost function with respect to each of the model parameters. The learning rate determines the size of the step taken in the direction of the gradient, and the process is repeated until convergence is achieved or a maximum number of iterations is reached. Gradient descent is widely used in machine learning and deep learning for optimizing models such as linear regression, logistic regression, and neural networks.
# 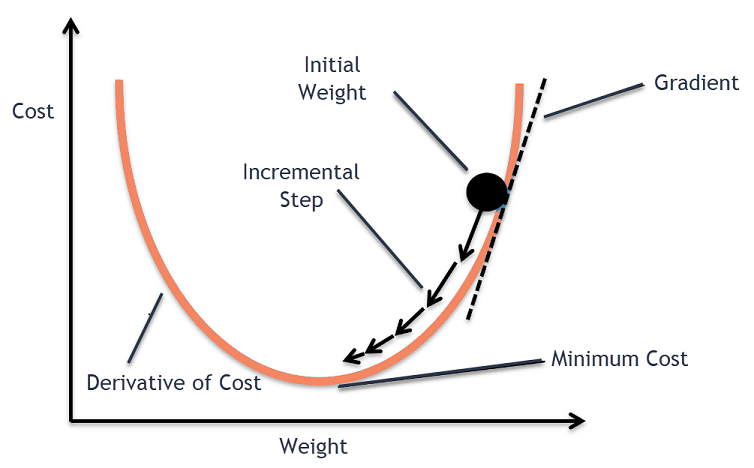
# The mathematical formulation of gradient descent involves computing the gradient of the cost function with respect to the parameters of the model, and using this gradient to update the parameters in the direction of steepest descent.
# Let's consider a cost function J(θ) which is a function of the parameters θ. The goal of gradient descent is to find the values of θ that minimize the cost function. We start with an initial guess for θ, say θ_0. At each iteration of the algorithm, we update θ using the following update rule:
# **θ = θ - α * ∇J(θ)**
# where
# - **α** is the learning rate
# - **∇J(θ)** is the gradient of the cost function with respect to θ.
# **What is learning rate?**
# Learning rate (also referred to as step size or the alpha) is the size of the steps that are taken to reach the minimum. This is typically a small value, and it is evaluated and updated based on the behavior of the cost function. High learning rates result in larger steps but risks overshooting the minimum. Conversely, a low learning rate has small step sizes. While it has the advantage of more precision, the number of iterations compromises overall efficiency as this takes more time and computations to reach the minimum.
# **What is cost function?**
# The cost (or loss) function measures the difference, or error, between actual y and predicted y at its current position. This improves the machine learning model's efficacy by providing feedback to the model so that it can adjust the parameters to minimize the error and find the local or global minimum. It continuously iterates, moving along the direction of steepest descent (or the negative gradient) until the cost function is close to or at zero. At this point, the model will stop learning. Additionally, while the terms, cost function and loss function, are considered synonymous, there is a slight difference between them. It’s worth noting that a loss function refers to the error of one training example, while a cost function calculates the average error across an entire training set.
# 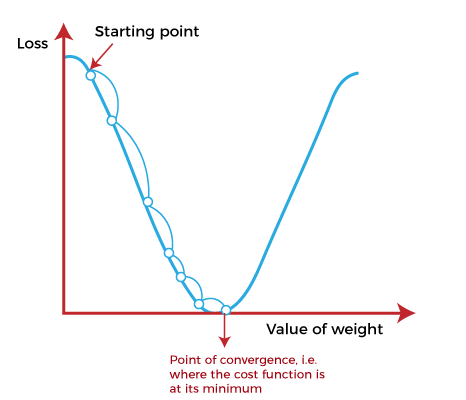
# **How does Gradient Descent work?**
# The gradient descent algorithm’s purpose is to minimise a given function (say cost function). It executes two phases iteratively to attain this goal:
# - Calculate the function’s gradient (slope) and first order derivative at that point.
# - Make a step (move) in the opposite direction of the gradient, increasing the slope by alpha times the gradient at that point from the current position.
# 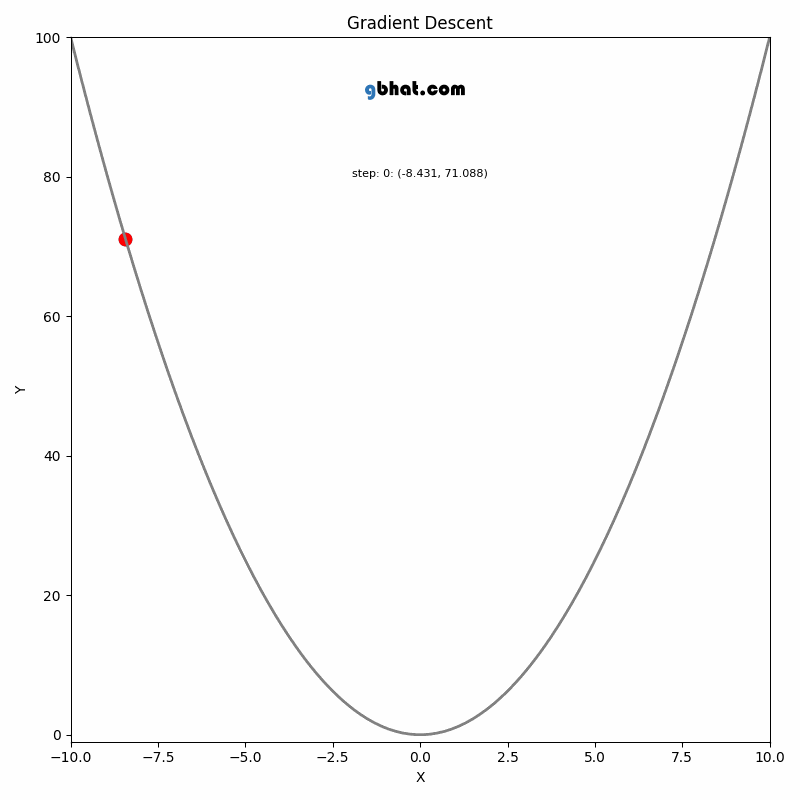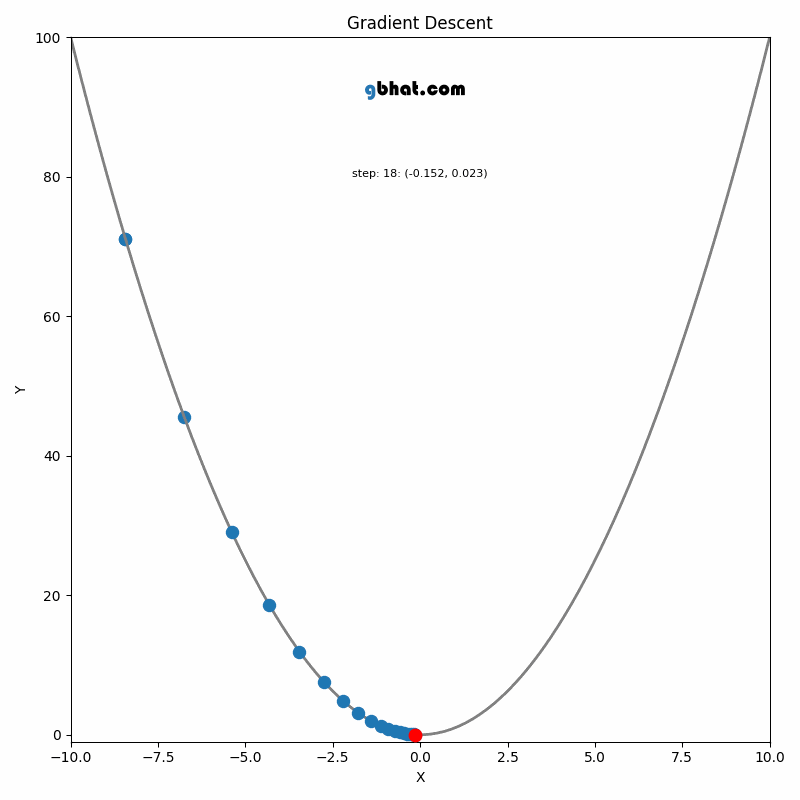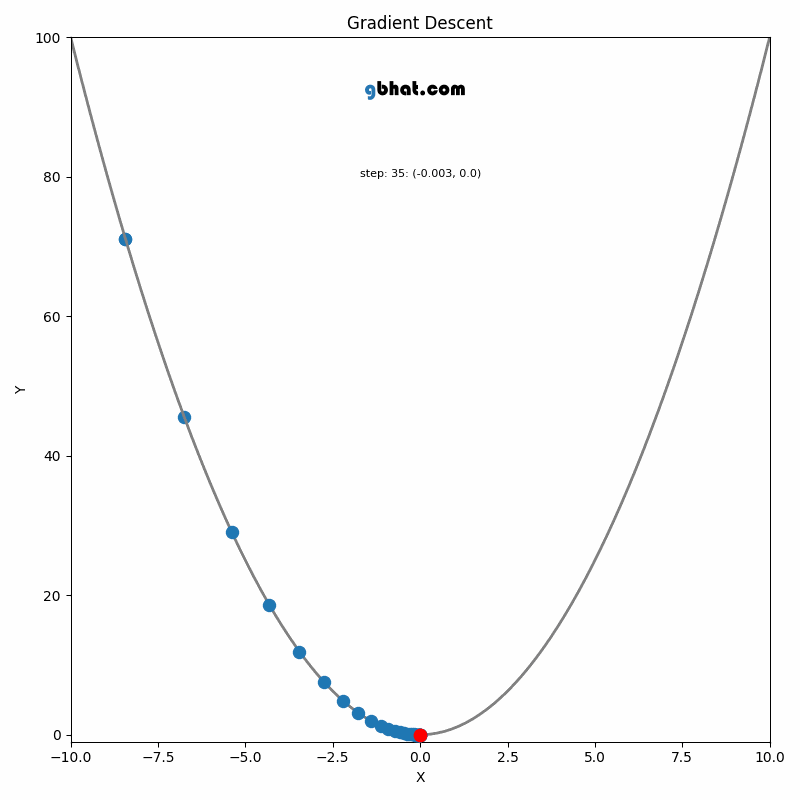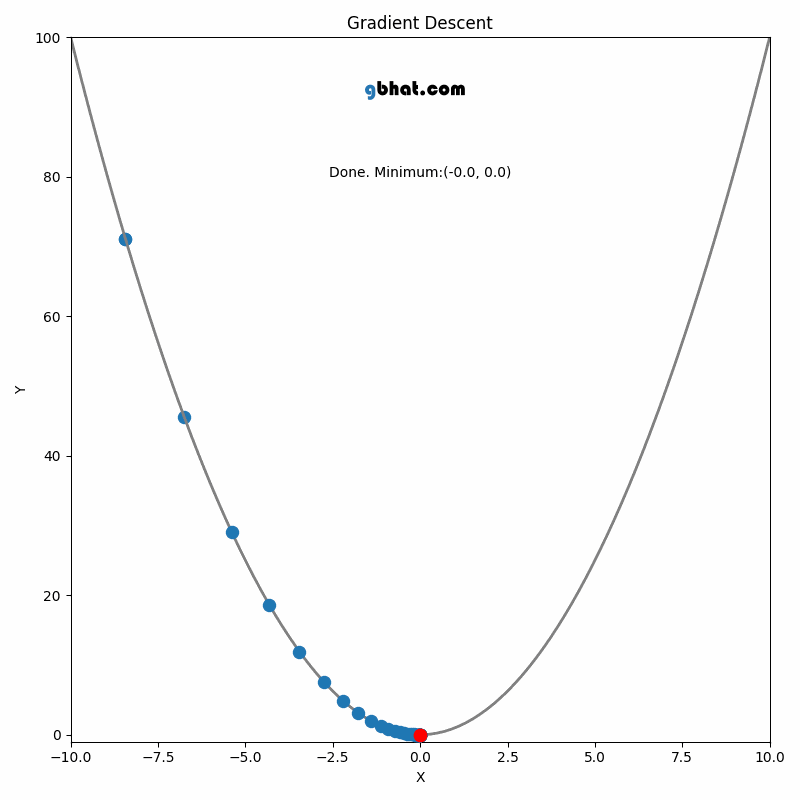
# **Lets see the Gradient Descent from scratch below:**
# Import Libraries
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
def compute_cost(X, y, theta):
"""
Compute the cost (mean squared error) for a given set of parameters.
Args:
X (ndarray): Input feature matrix
y (ndarray): Target values
theta (ndarray): Model parameters
Returns:
float: Cost value
"""
m = len(y)
predictions = X.dot(theta)
cost = (1 / (2 * m)) * np.sum((predictions - y) ** 2)
return cost
def gradient_descent(X, y, theta, learning_rate, num_iterations):
"""
Perform gradient descent to optimize model parameters.
Args:
X (ndarray): Input feature matrix
y (ndarray): Target values
theta (ndarray): Initial model parameters
learning_rate (float): Learning rate for gradient descent
num_iterations (int): Number of iterations for gradient descent
Returns:
ndarray: Optimized model parameters
list: Cost history during optimization
"""
m = len(y)
cost_history = []
for iteration in range(num_iterations):
# Compute gradients
gradients = 1 / m * X.T.dot(X.dot(theta) - y)
# Update parameters
theta = theta - learning_rate * gradients
# Compute cost
cost = compute_cost(X, y, theta)
cost_history.append(cost)
return theta, cost_history
# Generate random data
np.random.seed(0)
X = 2 * np.random.rand(100, 1)
y = 4 + 3 * X + np.random.randn(100, 1)
# Add bias term to X
X_b = np.c_[np.ones((100, 1)), X]
# Set learning rate and number of iterations
learning_rate = 0.01
num_iterations = 500
# Initialize theta
theta = np.random.randn(2, 1)
# Perform gradient descent
theta_optimized, cost_history = gradient_descent(
X_b, y, theta, learning_rate, num_iterations
)
# Plot the cost history over iterations
plt.plot(range(num_iterations), cost_history)
plt.title("Gradient Descent Optimization")
plt.xlabel("Iteration")
plt.ylabel("Cost")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/810/129810107.ipynb
| null | null |
[{"Id": 129810107, "ScriptId": 38104244, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7900156, "CreationDate": "05/16/2023 16:09:29", "VersionNumber": 1.0, "Title": "Gradient Descent", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 149.0, "LinesInsertedFromPrevious": 149.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 11}]
| null | null | null | null |
# ## Gradient descent
# Gradient descent is an iterative optimization algorithm used to minimize a cost function by adjusting the model parameters in the direction of steepest descent of the gradient. The goal is to find the optimal values of the model parameters that result in the minimum value of the cost function. The gradient is calculated by computing the partial derivatives of the cost function with respect to each of the model parameters. The learning rate determines the size of the step taken in the direction of the gradient, and the process is repeated until convergence is achieved or a maximum number of iterations is reached. Gradient descent is widely used in machine learning and deep learning for optimizing models such as linear regression, logistic regression, and neural networks.
# 
# The mathematical formulation of gradient descent involves computing the gradient of the cost function with respect to the parameters of the model, and using this gradient to update the parameters in the direction of steepest descent.
# Let's consider a cost function J(θ) which is a function of the parameters θ. The goal of gradient descent is to find the values of θ that minimize the cost function. We start with an initial guess for θ, say θ_0. At each iteration of the algorithm, we update θ using the following update rule:
# **θ = θ - α * ∇J(θ)**
# where
# - **α** is the learning rate
# - **∇J(θ)** is the gradient of the cost function with respect to θ.
# **What is learning rate?**
# Learning rate (also referred to as step size or the alpha) is the size of the steps that are taken to reach the minimum. This is typically a small value, and it is evaluated and updated based on the behavior of the cost function. High learning rates result in larger steps but risks overshooting the minimum. Conversely, a low learning rate has small step sizes. While it has the advantage of more precision, the number of iterations compromises overall efficiency as this takes more time and computations to reach the minimum.
# **What is cost function?**
# The cost (or loss) function measures the difference, or error, between actual y and predicted y at its current position. This improves the machine learning model's efficacy by providing feedback to the model so that it can adjust the parameters to minimize the error and find the local or global minimum. It continuously iterates, moving along the direction of steepest descent (or the negative gradient) until the cost function is close to or at zero. At this point, the model will stop learning. Additionally, while the terms, cost function and loss function, are considered synonymous, there is a slight difference between them. It’s worth noting that a loss function refers to the error of one training example, while a cost function calculates the average error across an entire training set.
# 
# **How does Gradient Descent work?**
# The gradient descent algorithm’s purpose is to minimise a given function (say cost function). It executes two phases iteratively to attain this goal:
# - Calculate the function’s gradient (slope) and first order derivative at that point.
# - Make a step (move) in the opposite direction of the gradient, increasing the slope by alpha times the gradient at that point from the current position.
# 
# **Lets see the Gradient Descent from scratch below:**
# Import Libraries
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
def compute_cost(X, y, theta):
"""
Compute the cost (mean squared error) for a given set of parameters.
Args:
X (ndarray): Input feature matrix
y (ndarray): Target values
theta (ndarray): Model parameters
Returns:
float: Cost value
"""
m = len(y)
predictions = X.dot(theta)
cost = (1 / (2 * m)) * np.sum((predictions - y) ** 2)
return cost
def gradient_descent(X, y, theta, learning_rate, num_iterations):
"""
Perform gradient descent to optimize model parameters.
Args:
X (ndarray): Input feature matrix
y (ndarray): Target values
theta (ndarray): Initial model parameters
learning_rate (float): Learning rate for gradient descent
num_iterations (int): Number of iterations for gradient descent
Returns:
ndarray: Optimized model parameters
list: Cost history during optimization
"""
m = len(y)
cost_history = []
for iteration in range(num_iterations):
# Compute gradients
gradients = 1 / m * X.T.dot(X.dot(theta) - y)
# Update parameters
theta = theta - learning_rate * gradients
# Compute cost
cost = compute_cost(X, y, theta)
cost_history.append(cost)
return theta, cost_history
# Generate random data
np.random.seed(0)
X = 2 * np.random.rand(100, 1)
y = 4 + 3 * X + np.random.randn(100, 1)
# Add bias term to X
X_b = np.c_[np.ones((100, 1)), X]
# Set learning rate and number of iterations
learning_rate = 0.01
num_iterations = 500
# Initialize theta
theta = np.random.randn(2, 1)
# Perform gradient descent
theta_optimized, cost_history = gradient_descent(
X_b, y, theta, learning_rate, num_iterations
)
# Plot the cost history over iterations
plt.plot(range(num_iterations), cost_history)
plt.title("Gradient Descent Optimization")
plt.xlabel("Iteration")
plt.ylabel("Cost")
plt.show()
| false | 0 | 1,384 | 11 | 1,384 | 1,384 |
||
129810807
|
# pip install fitz
# pip install PyMuPDF
import pandas as pd
import numpy as np
import fitz
import io
import json
import codecs
import glob
# # JSON
list_111 = {
"Id": [],
"district_name": [],
"widget": [],
"key": [],
"payloadID": [],
"status": [],
"title": [],
"content_type": [],
"body": [],
"response_performers": [],
}
# Метод для считывания json файла
def read_in_utf8(file):
with codecs.open(file, "r", encoding="utf_8_sig") as f: # открытие файла
return json.load(f)
list_json = glob.glob("dada/*.json")
list_json
for file in list_json:
load_file = read_in_utf8(file)
id1 = load_file["id"]
dist = load_file["district_name"]
for j in range(0, len(load_file["feed"])):
list_111["Id"].append(id1)
list_111["district_name"].append(dist)
list_111["widget"].append(load_file["feed"][j]["widget"])
list_111["key"].append(load_file["feed"][j]["key"])
list_111["payloadID"].append(load_file["feed"][j]["payload"]["id"])
try:
list_111["status"].append(load_file["feed"][j]["payload"]["status"])
except:
list_111["status"].append("NaN")
list_111["title"].append(load_file["feed"][j]["meta"]["title"])
list_111["content_type"].append(load_file["feed"][j]["content_type"])
try:
list_111["body"].append(load_file["feed"][j]["payload"]["body"])
except:
list_111["body"].append("NaN")
try:
list_111["response_performers"].append(
load_file["feed"][j]["payload"]["response_performers"]["name"]
)
except:
list_111["response_performers"].append("NaN")
df = pd.DataFrame.from_dict(data=list_111, orient="index")
df = df.transpose()
df.head(10)
# # PDF
# загрузка в list пути всех нужных pdf файлов
all_pdf = glob.glob(r"C:\Users\HONOR\Downloads\Aboba\*.pdf")
len(all_pdf)
all_pdf
pdf_document = r"C:\Users\HONOR\Downloads\Aboba\Cocoapods,_Carthage,_SPM_как_выбрать_менеджер_зависимостей_в_iOS.pdf"
doc = fitz.open(pdf_document)
print("Исходный документ: ", doc)
print("\nКоличество страниц: %i\n\n------------------\n\n" % doc.page_count)
print(doc.metadata)
for current_page in range(len(doc)):
page = doc.load_page(current_page)
page_text = page.get_text("text")
print("Стр. ", current_page + 1, "\n")
print(page_text)
pdf_document = (
r"C:\Users\HONOR\Downloads\Aboba\imgonline-com-ua-site2pdfB28eKGLy6EjA.pdf"
)
doc = fitz.open(pdf_document)
print("Исходный документ: ", doc)
text = []
for current_page in range(len(doc)):
page = doc.load_page(current_page)
page_text = page.get_text("text")
text.append(page_text)
print(page_text)
pdf_document = r"C:\Users\HONOR\Downloads\Aboba\Быстрое начало работы с Gitlab CICD.pdf"
doc = fitz.open(pdf_document)
print("Исходный документ: ", doc)
text = []
for current_page in range(len(doc)):
page = doc.load_page(current_page)
page_text = page.get_text("text")
text.append(page_text)
print(page_text)
# Можно было наверное использовать метод написанный ниже
text = "".join(text).replace("\n", " ")
# text
def extract_text_from_pdf(pdf):
doc = fitz.open(pdf)
text = []
for current_page in range(len(doc)):
page = doc.load_page(current_page)
page_text = page.get_text("text")
text.append(page_text)
return text
def pars_pdf(text):
text2 = text.split("\n")
print(text2)
if "Рейтинг" in text2:
num = text2.index("Рейтинг")
raiting = text2[num - 1]
name = text2[num + 1]
activity = text2[num + 2]
for i in text2:
if "назад" in i:
result = len(i.split())
if result > 3:
result_txt = i.split()
num = result_txt.index("назад")
date_publish = (
result_txt[num - 2]
+ " "
+ result_txt[num - 1]
+ " "
+ result_txt[num]
)
else:
date_publish = i
break
else:
date_publish = ""
print(num, raiting, name, activity, date_publish)
else:
raiting = 0
name = ""
activity = ""
for i in text2:
if "назад" in i:
date_publish = i
break
else:
date_publish = ""
print(num, raiting, name, activity, date_publish)
return name, raiting, activity, date_publish
x = extract_text_from_pdf(
r"C:\Users\HONOR\Downloads\Aboba\imgonline-com-ua-site2pdfB28eKGLy6EjA.pdf"
)
pars_pdf(x[0])
# Перебор текста из pdf файла, парсинг
Brak = []
NameCompany = []
Description = []
Raiting = []
DataPublish = []
Activity = []
TextArticle = []
for pdf in all_pdf:
try:
x = extract_text_from_pdf(pdf)
print(pdf.split("\\")[-1])
N, R, A, D = pars_pdf(x[0])
T = "".join(x).replace("\n", " ")
NameCompany.append(N)
Raiting.append(R)
DataPublish.append(D)
Activity.append(A)
T = "".join(T).replace(A, "")
T = "".join(T).replace(D, "")
text = T.split(" ")
text = text[5:]
T = " ".join(text)
TextArticle.append(T)
except:
Brak.append(pdf)
# T
Brak
# Создание DataFrame
df = pd.DataFrame(
{
"NameCompany": [],
"Description": [],
"Raiting": [],
"DataPublish": [],
"Activity": [],
"TextArticle": [],
}
)
df
print(
len(NameCompany),
len(Description),
len(Raiting),
len(DataPublish),
len(Activity),
len(TextArticle),
)
NameCompany
df["NameCompany"] = NameCompany
df["Raiting"] = Raiting
df["DataPublish"] = DataPublish
df["Activity"] = Activity
df["TextArticle"] = TextArticle
df
import pandas
df.to_csv(r"C:\Users\HONOR\Downloads\Aboba\dataset.csv")
df2 = pd.read_csv(r"C:\Users\HONOR\Downloads\Aboba\dataset.csv")
df2.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/810/129810807.ipynb
| null | null |
[{"Id": 129810807, "ScriptId": 38606699, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13417746, "CreationDate": "05/16/2023 16:15:02", "VersionNumber": 1.0, "Title": "PDF-SHA-djostit", "EvaluationDate": "05/16/2023", "IsChange": true, "TotalLines": 215.0, "LinesInsertedFromPrevious": 215.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
# pip install fitz
# pip install PyMuPDF
import pandas as pd
import numpy as np
import fitz
import io
import json
import codecs
import glob
# # JSON
list_111 = {
"Id": [],
"district_name": [],
"widget": [],
"key": [],
"payloadID": [],
"status": [],
"title": [],
"content_type": [],
"body": [],
"response_performers": [],
}
# Метод для считывания json файла
def read_in_utf8(file):
with codecs.open(file, "r", encoding="utf_8_sig") as f: # открытие файла
return json.load(f)
list_json = glob.glob("dada/*.json")
list_json
for file in list_json:
load_file = read_in_utf8(file)
id1 = load_file["id"]
dist = load_file["district_name"]
for j in range(0, len(load_file["feed"])):
list_111["Id"].append(id1)
list_111["district_name"].append(dist)
list_111["widget"].append(load_file["feed"][j]["widget"])
list_111["key"].append(load_file["feed"][j]["key"])
list_111["payloadID"].append(load_file["feed"][j]["payload"]["id"])
try:
list_111["status"].append(load_file["feed"][j]["payload"]["status"])
except:
list_111["status"].append("NaN")
list_111["title"].append(load_file["feed"][j]["meta"]["title"])
list_111["content_type"].append(load_file["feed"][j]["content_type"])
try:
list_111["body"].append(load_file["feed"][j]["payload"]["body"])
except:
list_111["body"].append("NaN")
try:
list_111["response_performers"].append(
load_file["feed"][j]["payload"]["response_performers"]["name"]
)
except:
list_111["response_performers"].append("NaN")
df = pd.DataFrame.from_dict(data=list_111, orient="index")
df = df.transpose()
df.head(10)
# # PDF
# загрузка в list пути всех нужных pdf файлов
all_pdf = glob.glob(r"C:\Users\HONOR\Downloads\Aboba\*.pdf")
len(all_pdf)
all_pdf
pdf_document = r"C:\Users\HONOR\Downloads\Aboba\Cocoapods,_Carthage,_SPM_как_выбрать_менеджер_зависимостей_в_iOS.pdf"
doc = fitz.open(pdf_document)
print("Исходный документ: ", doc)
print("\nКоличество страниц: %i\n\n------------------\n\n" % doc.page_count)
print(doc.metadata)
for current_page in range(len(doc)):
page = doc.load_page(current_page)
page_text = page.get_text("text")
print("Стр. ", current_page + 1, "\n")
print(page_text)
pdf_document = (
r"C:\Users\HONOR\Downloads\Aboba\imgonline-com-ua-site2pdfB28eKGLy6EjA.pdf"
)
doc = fitz.open(pdf_document)
print("Исходный документ: ", doc)
text = []
for current_page in range(len(doc)):
page = doc.load_page(current_page)
page_text = page.get_text("text")
text.append(page_text)
print(page_text)
pdf_document = r"C:\Users\HONOR\Downloads\Aboba\Быстрое начало работы с Gitlab CICD.pdf"
doc = fitz.open(pdf_document)
print("Исходный документ: ", doc)
text = []
for current_page in range(len(doc)):
page = doc.load_page(current_page)
page_text = page.get_text("text")
text.append(page_text)
print(page_text)
# Можно было наверное использовать метод написанный ниже
text = "".join(text).replace("\n", " ")
# text
def extract_text_from_pdf(pdf):
doc = fitz.open(pdf)
text = []
for current_page in range(len(doc)):
page = doc.load_page(current_page)
page_text = page.get_text("text")
text.append(page_text)
return text
def pars_pdf(text):
text2 = text.split("\n")
print(text2)
if "Рейтинг" in text2:
num = text2.index("Рейтинг")
raiting = text2[num - 1]
name = text2[num + 1]
activity = text2[num + 2]
for i in text2:
if "назад" in i:
result = len(i.split())
if result > 3:
result_txt = i.split()
num = result_txt.index("назад")
date_publish = (
result_txt[num - 2]
+ " "
+ result_txt[num - 1]
+ " "
+ result_txt[num]
)
else:
date_publish = i
break
else:
date_publish = ""
print(num, raiting, name, activity, date_publish)
else:
raiting = 0
name = ""
activity = ""
for i in text2:
if "назад" in i:
date_publish = i
break
else:
date_publish = ""
print(num, raiting, name, activity, date_publish)
return name, raiting, activity, date_publish
x = extract_text_from_pdf(
r"C:\Users\HONOR\Downloads\Aboba\imgonline-com-ua-site2pdfB28eKGLy6EjA.pdf"
)
pars_pdf(x[0])
# Перебор текста из pdf файла, парсинг
Brak = []
NameCompany = []
Description = []
Raiting = []
DataPublish = []
Activity = []
TextArticle = []
for pdf in all_pdf:
try:
x = extract_text_from_pdf(pdf)
print(pdf.split("\\")[-1])
N, R, A, D = pars_pdf(x[0])
T = "".join(x).replace("\n", " ")
NameCompany.append(N)
Raiting.append(R)
DataPublish.append(D)
Activity.append(A)
T = "".join(T).replace(A, "")
T = "".join(T).replace(D, "")
text = T.split(" ")
text = text[5:]
T = " ".join(text)
TextArticle.append(T)
except:
Brak.append(pdf)
# T
Brak
# Создание DataFrame
df = pd.DataFrame(
{
"NameCompany": [],
"Description": [],
"Raiting": [],
"DataPublish": [],
"Activity": [],
"TextArticle": [],
}
)
df
print(
len(NameCompany),
len(Description),
len(Raiting),
len(DataPublish),
len(Activity),
len(TextArticle),
)
NameCompany
df["NameCompany"] = NameCompany
df["Raiting"] = Raiting
df["DataPublish"] = DataPublish
df["Activity"] = Activity
df["TextArticle"] = TextArticle
df
import pandas
df.to_csv(r"C:\Users\HONOR\Downloads\Aboba\dataset.csv")
df2 = pd.read_csv(r"C:\Users\HONOR\Downloads\Aboba\dataset.csv")
df2.head()
| false | 0 | 2,002 | 1 | 2,002 | 2,002 |
||
129177244
|
import torch
# encoder representations of four different words
word_1 = torch.tensor([1.0, 0.0, 0.0])
word_2 = torch.tensor([0.0, 1.0, 0.0])
word_3 = torch.tensor([1.0, 1.0, 0.0])
word_4 = torch.tensor([0.0, 0.0, 1.0])
# Stacking the word embeddings into a single array
input_embeddings = torch.stack([word_1, word_2, word_3, word_4], dim=0)
input_embeddings
# Generating the weight matrices
torch.manual_seed(42)
W_Q = torch.randint(0, 2, (3, 3)).type(torch.float)
W_K = torch.randint(0, 2, (3, 3)).type(torch.float)
W_V = torch.randint(0, 2, (3, 3)).type(torch.float)
print("W_Q:")
print(W_Q)
print("\n")
print("W_K:")
print(W_K)
print("\n")
print("W_V:")
print(W_V)
print("\n")
# Generating the queries, keys and values
Q = torch.matmul(input_embeddings, W_Q)
K = torch.matmul(input_embeddings, W_K)
V = torch.matmul(input_embeddings, W_V)
print("Q:")
print(Q)
print("\n")
print("K:")
print(K)
print("\n")
print("V:")
print(V)
print("\n")
# Scoring the query vectors againsts all the key vectors
attention_scores = torch.matmul(Q, K.transpose(1, 0)) / K.shape[1] ** 5
print("Scores:")
print(attention_scores)
# Computing the weights by a softmax operation
attention_weights = torch.softmax(attention_scores, axis=1)
attention_weights
# Computing the attention matrix
attention_matrix = torch.matmul(attention_weights, V)
attention_matrix
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/177/129177244.ipynb
| null | null |
[{"Id": 129177244, "ScriptId": 37959359, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6877706, "CreationDate": "05/11/2023 14:58:33", "VersionNumber": 1.0, "Title": "simple_transformer_attention_by_pyTorch", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 64.0, "LinesInsertedFromPrevious": 64.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import torch
# encoder representations of four different words
word_1 = torch.tensor([1.0, 0.0, 0.0])
word_2 = torch.tensor([0.0, 1.0, 0.0])
word_3 = torch.tensor([1.0, 1.0, 0.0])
word_4 = torch.tensor([0.0, 0.0, 1.0])
# Stacking the word embeddings into a single array
input_embeddings = torch.stack([word_1, word_2, word_3, word_4], dim=0)
input_embeddings
# Generating the weight matrices
torch.manual_seed(42)
W_Q = torch.randint(0, 2, (3, 3)).type(torch.float)
W_K = torch.randint(0, 2, (3, 3)).type(torch.float)
W_V = torch.randint(0, 2, (3, 3)).type(torch.float)
print("W_Q:")
print(W_Q)
print("\n")
print("W_K:")
print(W_K)
print("\n")
print("W_V:")
print(W_V)
print("\n")
# Generating the queries, keys and values
Q = torch.matmul(input_embeddings, W_Q)
K = torch.matmul(input_embeddings, W_K)
V = torch.matmul(input_embeddings, W_V)
print("Q:")
print(Q)
print("\n")
print("K:")
print(K)
print("\n")
print("V:")
print(V)
print("\n")
# Scoring the query vectors againsts all the key vectors
attention_scores = torch.matmul(Q, K.transpose(1, 0)) / K.shape[1] ** 5
print("Scores:")
print(attention_scores)
# Computing the weights by a softmax operation
attention_weights = torch.softmax(attention_scores, axis=1)
attention_weights
# Computing the attention matrix
attention_matrix = torch.matmul(attention_weights, V)
attention_matrix
| false | 0 | 510 | 0 | 510 | 510 |
||
129177441
|
import tensorflow as tf
from tensorflow import convert_to_tensor, string
from tensorflow.keras.layers import TextVectorization, Embedding, Layer
from tensorflow.data import Dataset
import numpy as np
import matplotlib.pyplot as plt
# **Text Vectorization Layer**
output_sequence_length = 5
vocab_size = 10
sentences = [["I am a robot"], ["you too robot"]]
sentence_data = Dataset.from_tensor_slices(sentences)
# Create the TextVectorization layer
vectorize_layer = TextVectorization(
output_sequence_length=output_sequence_length, max_tokens=vocab_size
)
# Train the layer to create a dictionary
vectorize_layer.adapt(sentence_data)
# Convert all sentences to tensors
word_tensors = convert_to_tensor(sentences, dtype=tf.string)
# Use the word tensors to get vectorized phrases
vectorized_words = vectorize_layer(word_tensors)
sentence_data
vectorized_words
word_tensors
print("Vocabulary:", vectorize_layer.get_vocabulary())
print("Vectorized words:", vectorized_words)
# ## **The Embedding layer**
# **The Word Embedding**
output_length = 6
word_embedding_layer = Embedding(vocab_size, output_length)
embedded_word = word_embedding_layer(vectorized_words)
print(embedded_word)
word_embedding_layer
# **The Position Embedding**
position_embedding_layer = Embedding(output_sequence_length, output_length)
position_indices = tf.range(output_sequence_length)
embedded_indices = position_embedding_layer(position_indices)
print(embedded_indices)
position_embedding_layer
# **The output of positional Encoding layer in transformers**
final_output_embedding = embedded_word + embedded_indices
print("Final Output:", final_output_embedding)
# ## **Subclassing the Keras Embedding Layer**
class PositionEmbeddingLayer(Layer):
def __init__(self, output_seq_length, vocab_length, output_length, **kwargs):
super(PositionEmbeddingLayer, self).__init__(**kwargs)
self.word_embedding_layer = Embedding(
input_dim=vocab_length, output_dim=output_length
)
self.position_embedding_layer = Embedding(
input_dim=output_seq_length, output_dim=output_length
)
def call(self, inputs):
embedded_words = self.word_embedding_layer(inputs)
position_indices = tf.range(tf.shape(inputs)[-1])
embedded_indices = self.position_embedding_layer(position_indices)
return embedded_words + embedded_indices
my_embedding_layer = PositionEmbeddingLayer(
output_sequence_length, vocab_size, output_length
)
embedded_layer_output = my_embedding_layer(vectorized_words)
print("Output from my_embedded_layer", embedded_layer_output)
def get_position_encoding(seq_len, d, n=10000):
P = np.zeros((seq_len, d))
for k in range(seq_len):
for i in np.arange(int(d / 2)):
denominator = np.power(n, 2 * i / d)
P[k, 2 * i] = np.sin(k / denominator)
P[k, 2 * i + 1] = np.cos(k / denominator)
return P
get_position_encoding(10, 6)
# ## **Positional Encoding in Transformers: Attention is All you need**
class PositionEmbeddingFixedWeights(Layer):
def __init__(self, sequence_length, vocab_size, output_dim, **kwargs):
super(PositionEmbeddingFixedWeights, self).__init__(**kwargs)
word_embedding_matrix = self.get_position_encoding(vocab_size, output_dim)
position_embedding_matrix = self.get_position_encoding(
sequence_length, output_dim
)
self.word_embedding_layer = Embedding(
input_dim=vocab_size,
output_dim=output_dim,
weights=[word_embedding_matrix],
trainable=False,
)
self.position_embedding_layer = Embedding(
input_dim=sequence_length,
output_dim=output_dim,
weights=[position_embedding_matrix],
trainable=False,
)
def get_position_encoding(self, seq_len, d, n=10000):
P = np.zeros((seq_len, d))
for k in range(seq_len):
for i in np.arange(int(d / 2)):
denominator = np.power(n, 2 * i / d)
P[k, 2 * i] = np.sin(k / denominator)
P[k, 2 * i + 1] = np.cos(k / denominator)
return P
def call(self, inputs):
position_indices = tf.range(tf.shape(inputs)[-1])
embedded_indices = self.position_embedding_layer(position_indices)
embedded_words = self.word_embedding_layer(inputs)
return embedded_words + embedded_indices
attnisallyouneed = PositionEmbeddingFixedWeights(
output_sequence_length, vocab_size, output_length
)
attnisallyouneed_output = attnisallyouneed(vectorized_words)
print("Attention is all you need Output:", attnisallyouneed_output)
# ## **Visualizing the Final Embedding**
technical_phrase = (
"to understand machine learning algorithms you need"
+ " to understand concepts such as gradient of a function "
+ "Hessians of a matrix and optimization etc"
)
wise_quote = (
"patrick henry said give me liberty or give me death "
+ "when he addressed the second virginia convention in march"
)
total_vocabulary = 200
sequence_length = 20
final_output_length = 50
phrase_vectorization_layer = TextVectorization(
output_sequence_length=sequence_length, max_tokens=total_vocabulary
)
# Learn the dictionary
phrase_vectorization_layer.adapt([technical_phrase, wise_quote])
# Convert all sentences to tensors
phrase_tensors = convert_to_tensor([technical_phrase, wise_quote], dtype=tf.string)
# Use the word tensor to get vectorized phrases
vectorized_phrases = phrase_vectorization_layer(phrase_tensors)
# Initialize Random Weight Embedding Layer
random_weight_embedding_layer = PositionEmbeddingLayer(
sequence_length, total_vocabulary, final_output_length
)
# Random Embedding
random_embedding = random_weight_embedding_layer(vectorized_phrases)
# Initialize Fixed Weight Embedding Layer
fixed_weight_embedding_layer = PositionEmbeddingFixedWeights(
sequence_length, total_vocabulary, final_output_length
)
# Fixed Embedding
fixed_embedding = fixed_weight_embedding_layer(vectorized_phrases)
# **Random Embedding Plot for Both the Phrases**
fig = plt.figure(figsize=(15, 5))
title = ["Tech Phrase", "Wise Phrase"]
for i in range(2):
ax = plt.subplot(1, 2, 1 + i)
matrix = tf.reshape(
random_embedding[i, :, :], (sequence_length, final_output_length)
)
cax = ax.matshow(matrix)
plt.gcf().colorbar(cax)
plt.title(title[i], y=1.2)
fig.suptitle("Random Embedding")
plt.show()
# **Fixed weight embedding for both phrases**
fig = plt.figure(figsize=(15, 5))
title = ["Tech Phrase", "Wise Phrase"]
for i in range(2):
ax = plt.subplot(1, 2, 1 + i)
matrix = tf.reshape(
fixed_embedding[i, :, :], (sequence_length, final_output_length)
)
cax = ax.matshow(matrix)
plt.gcf().colorbar(cax)
plt.title(title[i], y=1.2)
fig.suptitle("Fixed Weight Embedding from Attention is All You Need")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/177/129177441.ipynb
| null | null |
[{"Id": 129177441, "ScriptId": 38167031, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6877706, "CreationDate": "05/11/2023 15:00:22", "VersionNumber": 1.0, "Title": "Transformer_positional_encoding", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 181.0, "LinesInsertedFromPrevious": 181.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import tensorflow as tf
from tensorflow import convert_to_tensor, string
from tensorflow.keras.layers import TextVectorization, Embedding, Layer
from tensorflow.data import Dataset
import numpy as np
import matplotlib.pyplot as plt
# **Text Vectorization Layer**
output_sequence_length = 5
vocab_size = 10
sentences = [["I am a robot"], ["you too robot"]]
sentence_data = Dataset.from_tensor_slices(sentences)
# Create the TextVectorization layer
vectorize_layer = TextVectorization(
output_sequence_length=output_sequence_length, max_tokens=vocab_size
)
# Train the layer to create a dictionary
vectorize_layer.adapt(sentence_data)
# Convert all sentences to tensors
word_tensors = convert_to_tensor(sentences, dtype=tf.string)
# Use the word tensors to get vectorized phrases
vectorized_words = vectorize_layer(word_tensors)
sentence_data
vectorized_words
word_tensors
print("Vocabulary:", vectorize_layer.get_vocabulary())
print("Vectorized words:", vectorized_words)
# ## **The Embedding layer**
# **The Word Embedding**
output_length = 6
word_embedding_layer = Embedding(vocab_size, output_length)
embedded_word = word_embedding_layer(vectorized_words)
print(embedded_word)
word_embedding_layer
# **The Position Embedding**
position_embedding_layer = Embedding(output_sequence_length, output_length)
position_indices = tf.range(output_sequence_length)
embedded_indices = position_embedding_layer(position_indices)
print(embedded_indices)
position_embedding_layer
# **The output of positional Encoding layer in transformers**
final_output_embedding = embedded_word + embedded_indices
print("Final Output:", final_output_embedding)
# ## **Subclassing the Keras Embedding Layer**
class PositionEmbeddingLayer(Layer):
def __init__(self, output_seq_length, vocab_length, output_length, **kwargs):
super(PositionEmbeddingLayer, self).__init__(**kwargs)
self.word_embedding_layer = Embedding(
input_dim=vocab_length, output_dim=output_length
)
self.position_embedding_layer = Embedding(
input_dim=output_seq_length, output_dim=output_length
)
def call(self, inputs):
embedded_words = self.word_embedding_layer(inputs)
position_indices = tf.range(tf.shape(inputs)[-1])
embedded_indices = self.position_embedding_layer(position_indices)
return embedded_words + embedded_indices
my_embedding_layer = PositionEmbeddingLayer(
output_sequence_length, vocab_size, output_length
)
embedded_layer_output = my_embedding_layer(vectorized_words)
print("Output from my_embedded_layer", embedded_layer_output)
def get_position_encoding(seq_len, d, n=10000):
P = np.zeros((seq_len, d))
for k in range(seq_len):
for i in np.arange(int(d / 2)):
denominator = np.power(n, 2 * i / d)
P[k, 2 * i] = np.sin(k / denominator)
P[k, 2 * i + 1] = np.cos(k / denominator)
return P
get_position_encoding(10, 6)
# ## **Positional Encoding in Transformers: Attention is All you need**
class PositionEmbeddingFixedWeights(Layer):
def __init__(self, sequence_length, vocab_size, output_dim, **kwargs):
super(PositionEmbeddingFixedWeights, self).__init__(**kwargs)
word_embedding_matrix = self.get_position_encoding(vocab_size, output_dim)
position_embedding_matrix = self.get_position_encoding(
sequence_length, output_dim
)
self.word_embedding_layer = Embedding(
input_dim=vocab_size,
output_dim=output_dim,
weights=[word_embedding_matrix],
trainable=False,
)
self.position_embedding_layer = Embedding(
input_dim=sequence_length,
output_dim=output_dim,
weights=[position_embedding_matrix],
trainable=False,
)
def get_position_encoding(self, seq_len, d, n=10000):
P = np.zeros((seq_len, d))
for k in range(seq_len):
for i in np.arange(int(d / 2)):
denominator = np.power(n, 2 * i / d)
P[k, 2 * i] = np.sin(k / denominator)
P[k, 2 * i + 1] = np.cos(k / denominator)
return P
def call(self, inputs):
position_indices = tf.range(tf.shape(inputs)[-1])
embedded_indices = self.position_embedding_layer(position_indices)
embedded_words = self.word_embedding_layer(inputs)
return embedded_words + embedded_indices
attnisallyouneed = PositionEmbeddingFixedWeights(
output_sequence_length, vocab_size, output_length
)
attnisallyouneed_output = attnisallyouneed(vectorized_words)
print("Attention is all you need Output:", attnisallyouneed_output)
# ## **Visualizing the Final Embedding**
technical_phrase = (
"to understand machine learning algorithms you need"
+ " to understand concepts such as gradient of a function "
+ "Hessians of a matrix and optimization etc"
)
wise_quote = (
"patrick henry said give me liberty or give me death "
+ "when he addressed the second virginia convention in march"
)
total_vocabulary = 200
sequence_length = 20
final_output_length = 50
phrase_vectorization_layer = TextVectorization(
output_sequence_length=sequence_length, max_tokens=total_vocabulary
)
# Learn the dictionary
phrase_vectorization_layer.adapt([technical_phrase, wise_quote])
# Convert all sentences to tensors
phrase_tensors = convert_to_tensor([technical_phrase, wise_quote], dtype=tf.string)
# Use the word tensor to get vectorized phrases
vectorized_phrases = phrase_vectorization_layer(phrase_tensors)
# Initialize Random Weight Embedding Layer
random_weight_embedding_layer = PositionEmbeddingLayer(
sequence_length, total_vocabulary, final_output_length
)
# Random Embedding
random_embedding = random_weight_embedding_layer(vectorized_phrases)
# Initialize Fixed Weight Embedding Layer
fixed_weight_embedding_layer = PositionEmbeddingFixedWeights(
sequence_length, total_vocabulary, final_output_length
)
# Fixed Embedding
fixed_embedding = fixed_weight_embedding_layer(vectorized_phrases)
# **Random Embedding Plot for Both the Phrases**
fig = plt.figure(figsize=(15, 5))
title = ["Tech Phrase", "Wise Phrase"]
for i in range(2):
ax = plt.subplot(1, 2, 1 + i)
matrix = tf.reshape(
random_embedding[i, :, :], (sequence_length, final_output_length)
)
cax = ax.matshow(matrix)
plt.gcf().colorbar(cax)
plt.title(title[i], y=1.2)
fig.suptitle("Random Embedding")
plt.show()
# **Fixed weight embedding for both phrases**
fig = plt.figure(figsize=(15, 5))
title = ["Tech Phrase", "Wise Phrase"]
for i in range(2):
ax = plt.subplot(1, 2, 1 + i)
matrix = tf.reshape(
fixed_embedding[i, :, :], (sequence_length, final_output_length)
)
cax = ax.matshow(matrix)
plt.gcf().colorbar(cax)
plt.title(title[i], y=1.2)
fig.suptitle("Fixed Weight Embedding from Attention is All You Need")
plt.show()
| false | 0 | 1,974 | 0 | 1,974 | 1,974 |
||
129177805
|
# 
# ## **Step 1: Represent the Input**
import numpy as np
from scipy.special import softmax
print("Step 1: Input : 3 inputs, d_model = 4")
x = np.array(
[
[1.0, 0.0, 1.0, 0.0], # Input 1
[0.0, 2.0, 0.0, 2.0], # input 2
[1.0, 1.0, 1.0, 1.0],
]
) # input 3
print(x)
# **The output shows that we have 3 vectors of d_model = 4**
# ## **Step 2 : Initialize the weight metrices**
# * q_w to train the queries
# * k_w to train the keys
# * v_w to train the values
print("step 2 : weights 3 dimentions x d_model = 4")
print("\n w_query:\n")
w_query = np.array([[1, 0, 1], [1, 0, 0], [0, 0, 1], [0, 1, 1]])
print(w_query)
print("\n w_key:\n")
w_key = np.array([[0, 0, 1], [1, 1, 0], [0, 1, 0], [1, 1, 0]])
print(w_key)
print("\n w_value:\n")
w_value = np.array([[0, 2, 0], [0, 3, 0], [1, 0, 3], [1, 1, 0]])
print(w_value)
# ## **Step 3 : Matrix multiplication to obtain Q, K, V**
# * x dimention : 3 * 4
# * w_query dimention : 4 * 3
# * Q dimention : 3 * 3
print("Step 3: Matrix Multiplication to obtain Q,K,V")
print("Query: x * w_query")
Q = np.matmul(x, w_query)
print(Q)
print("Key : x * w_key")
K = np.matmul(x, w_key)
print(K)
print("Value: x * w_value")
V = np.matmul(x, w_value)
print(V)
# ## **Step 4 : Scaled Attention Scores**
# 
# * Q dimention : 3 * 3
# * K dimention : 3 * 3
# * attention_scores dimention = 3 * 3
print("Step 4 : Scaled Attention Scores")
k_d = 1 # square_root of k_d=3 rouded down to 1 for this example
attention_scores = (Q @ K.transpose()) / k_d
print(attention_scores)
# ## **Step 5: Scaled softmax attention scores for each other**
print("Step 5 : Scaled softmax attention_scores for each vactor:")
attention_scores[0] = softmax(attention_scores[0])
attention_scores[1] = softmax(attention_scores[1])
attention_scores[2] = softmax(attention_scores[2])
print(attention_scores[0])
print(attention_scores[1])
print(attention_scores[2])
# ## **Step 6: The final attention representations**
attention_scores[0].reshape(-1, 1)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/177/129177805.ipynb
| null | null |
[{"Id": 129177805, "ScriptId": 38321976, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6877706, "CreationDate": "05/11/2023 15:03:26", "VersionNumber": 1.0, "Title": "Denis_Rothman_Book_multi_head_attention", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 89.0, "LinesInsertedFromPrevious": 89.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
| null | null | null | null |
# 
# ## **Step 1: Represent the Input**
import numpy as np
from scipy.special import softmax
print("Step 1: Input : 3 inputs, d_model = 4")
x = np.array(
[
[1.0, 0.0, 1.0, 0.0], # Input 1
[0.0, 2.0, 0.0, 2.0], # input 2
[1.0, 1.0, 1.0, 1.0],
]
) # input 3
print(x)
# **The output shows that we have 3 vectors of d_model = 4**
# ## **Step 2 : Initialize the weight metrices**
# * q_w to train the queries
# * k_w to train the keys
# * v_w to train the values
print("step 2 : weights 3 dimentions x d_model = 4")
print("\n w_query:\n")
w_query = np.array([[1, 0, 1], [1, 0, 0], [0, 0, 1], [0, 1, 1]])
print(w_query)
print("\n w_key:\n")
w_key = np.array([[0, 0, 1], [1, 1, 0], [0, 1, 0], [1, 1, 0]])
print(w_key)
print("\n w_value:\n")
w_value = np.array([[0, 2, 0], [0, 3, 0], [1, 0, 3], [1, 1, 0]])
print(w_value)
# ## **Step 3 : Matrix multiplication to obtain Q, K, V**
# * x dimention : 3 * 4
# * w_query dimention : 4 * 3
# * Q dimention : 3 * 3
print("Step 3: Matrix Multiplication to obtain Q,K,V")
print("Query: x * w_query")
Q = np.matmul(x, w_query)
print(Q)
print("Key : x * w_key")
K = np.matmul(x, w_key)
print(K)
print("Value: x * w_value")
V = np.matmul(x, w_value)
print(V)
# ## **Step 4 : Scaled Attention Scores**
# 
# * Q dimention : 3 * 3
# * K dimention : 3 * 3
# * attention_scores dimention = 3 * 3
print("Step 4 : Scaled Attention Scores")
k_d = 1 # square_root of k_d=3 rouded down to 1 for this example
attention_scores = (Q @ K.transpose()) / k_d
print(attention_scores)
# ## **Step 5: Scaled softmax attention scores for each other**
print("Step 5 : Scaled softmax attention_scores for each vactor:")
attention_scores[0] = softmax(attention_scores[0])
attention_scores[1] = softmax(attention_scores[1])
attention_scores[2] = softmax(attention_scores[2])
print(attention_scores[0])
print(attention_scores[1])
print(attention_scores[2])
# ## **Step 6: The final attention representations**
attention_scores[0].reshape(-1, 1)
| false | 0 | 1,038 | 2 | 1,038 | 1,038 |
||
129177071
|
<jupyter_start><jupyter_text>Most Drugs
Kaggle dataset identifier: most-drugs
<jupyter_script>import pandas as pd
df = pd.read_csv(
"/kaggle/input/most-drugs/All Drugs.csv", on_bad_lines="skip"
) # upload all compounds CSV
df = pd.read_csv(
"/kaggle/input/most-drugs/All Drugs.csv", on_bad_lines="skip", sep=";"
) # Print the name of the header in the file
print(df.columns)
df_approved = df[df["Max Phase"] == 4] # Approved drugs from FDA
print(df_approved)
df_pahse3 = df[df["Max Phase"] == 3] # phase 3 drugs
print(df_pahse3)
df_pahse2 = df[df["Max Phase"] == 2] # Phase 2
print(df_pahse2)
df_pahse1 = df[df["Max Phase"] == 1] # phase1
print(df_pahse1)
df_early_phase = df[df["Max Phase"] == 0.5] # early_phase
print(df_early_phase)
df_priclinical_1 = df[df["Max Phase"].isnull()] # priclinical 1
print(df_priclinical_1)
df_priclinical_2 = df[df["Max Phase"] == -1] # priclinical 2
print(df_priclinical_2)
df["Max Phase"].fillna(-1, inplace=True) # combine all priclinical together
grouped = df.groupby("Max Phase")[
"Max Phase"
].value_counts() # Total number of each pahse, -1 priclinical, 0.5 early phase, 1,2 and 3 are phase, 4 approved
print(grouped)
# Group the rows by the Type column and count the number of rows for each group
grouped = df.groupby("Type")["Type"].count()
# Print the result for each group
print("Number of rows for Antibody type: ", grouped["Antibody"])
print("Number of rows for Cell type: ", grouped["Cell"])
print("Number of rows for Enzyme type: ", grouped["Enzyme"])
print("Number of rows for Gene type: ", grouped["Gene"])
print("Number of rows for Oligonucleotide type: ", grouped["Oligonucleotide"])
print("Number of rows for Oligosaccharide type: ", grouped["Oligosaccharide"])
print("Number of rows for Protein type: ", grouped["Protein"])
print("Number of rows for Small molecule type: ", grouped["Small molecule"])
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/177/129177071.ipynb
|
most-drugs
|
ahmedelmaamounamin
|
[{"Id": 129177071, "ScriptId": 36974853, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14449681, "CreationDate": "05/11/2023 14:57:07", "VersionNumber": 10.0, "Title": "Drugs", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 46.0, "LinesInsertedFromPrevious": 40.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 6.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184993388, "KernelVersionId": 129177071, "SourceDatasetVersionId": 5340272}]
|
[{"Id": 5340272, "DatasetId": 3101102, "DatasourceVersionId": 5413629, "CreatorUserId": 14449681, "LicenseName": "Unknown", "CreationDate": "04/07/2023 15:39:34", "VersionNumber": 1.0, "Title": "Most Drugs", "Slug": "most-drugs", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3101102, "CreatorUserId": 14449681, "OwnerUserId": 14449681.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5340272.0, "CurrentDatasourceVersionId": 5413629.0, "ForumId": 3164313, "Type": 2, "CreationDate": "04/07/2023 15:39:34", "LastActivityDate": "04/07/2023", "TotalViews": 99, "TotalDownloads": 2, "TotalVotes": 0, "TotalKernels": 1}]
|
[{"Id": 14449681, "UserName": "ahmedelmaamounamin", "DisplayName": "Ahmed Elmaamounamin", "RegisterDate": "04/01/2023", "PerformanceTier": 0}]
|
import pandas as pd
df = pd.read_csv(
"/kaggle/input/most-drugs/All Drugs.csv", on_bad_lines="skip"
) # upload all compounds CSV
df = pd.read_csv(
"/kaggle/input/most-drugs/All Drugs.csv", on_bad_lines="skip", sep=";"
) # Print the name of the header in the file
print(df.columns)
df_approved = df[df["Max Phase"] == 4] # Approved drugs from FDA
print(df_approved)
df_pahse3 = df[df["Max Phase"] == 3] # phase 3 drugs
print(df_pahse3)
df_pahse2 = df[df["Max Phase"] == 2] # Phase 2
print(df_pahse2)
df_pahse1 = df[df["Max Phase"] == 1] # phase1
print(df_pahse1)
df_early_phase = df[df["Max Phase"] == 0.5] # early_phase
print(df_early_phase)
df_priclinical_1 = df[df["Max Phase"].isnull()] # priclinical 1
print(df_priclinical_1)
df_priclinical_2 = df[df["Max Phase"] == -1] # priclinical 2
print(df_priclinical_2)
df["Max Phase"].fillna(-1, inplace=True) # combine all priclinical together
grouped = df.groupby("Max Phase")[
"Max Phase"
].value_counts() # Total number of each pahse, -1 priclinical, 0.5 early phase, 1,2 and 3 are phase, 4 approved
print(grouped)
# Group the rows by the Type column and count the number of rows for each group
grouped = df.groupby("Type")["Type"].count()
# Print the result for each group
print("Number of rows for Antibody type: ", grouped["Antibody"])
print("Number of rows for Cell type: ", grouped["Cell"])
print("Number of rows for Enzyme type: ", grouped["Enzyme"])
print("Number of rows for Gene type: ", grouped["Gene"])
print("Number of rows for Oligonucleotide type: ", grouped["Oligonucleotide"])
print("Number of rows for Oligosaccharide type: ", grouped["Oligosaccharide"])
print("Number of rows for Protein type: ", grouped["Protein"])
print("Number of rows for Small molecule type: ", grouped["Small molecule"])
| false | 1 | 627 | 0 | 649 | 627 |
||
129177614
|
from tensorflow import matmul, math, reshape, shape, transpose, cast, float32
from tensorflow.keras.layers import Dense, Layer
from keras.backend import softmax
from numpy import random
# **Implement the scaled-dot product attention**
class DotProductAttention(Layer):
def __init__(self, **kwargs):
super(DotProductAttention, self).__init__(**kwargs)
def call(self, queries, keys, values, d_k, mask=None):
# Scoring the queries againsts the keys after transposing the latter, and scaling
scores = matmul(queries, keys, transpose_b=True) / math.sqrt(cast(d_k, float32))
# Apply mask to the attention scores
if mask is not None:
scores += -1e9 * mask
# Computing the weights by a softmax operation
weigths = softmax(scores)
# Computing the attention by a weighted sum of the value vectors
return matmul(weigths, values)
# **Implementing Multi-Head Attention**
class MultiHeadAttention(Layer):
def __init__(self, h, d_k, d_v, d_model, **kwargs):
super(MultiHeadAttention, self).__init__(**kwargs)
self.attention = DotProductAttention() # scaled dot product attention
self.heads = h # Number of attention heads to use
self.d_k = d_k # Dimentionality of the linearly projected queries and keys
self.d_v = d_v # Dimentionality of the linearly projected values
self.d_model = d_model # Dimentionality of the model
self.W_q = Dense(d_k) # Learned projected matrix for queries
self.W_k = Dense(d_k) # Learned projected matrix for keys
self.W_v = Dense(d_v) # Learned projected matrix for values
self.W_o = Dense(d_model) # Learned projection matrix for multi-head output
def reshape_tensor(self, x, heads, flag):
if flag:
# Tensor shape after reshaping and transposing: (batch_size, heads, seq_lenght, -1)
x = reshape(x, shape=(shape(x)[0], shape(x)[1], heads, -1))
x = transpose(x, perm=(0, 2, 1, 3))
else:
x = transpose(x, perm=(0, 2, 1, 3))
x = reshape(x, shape=(shape(x)[0], shape(x)[1], self.d_k))
return x
def call(self, queries, keys, values, mask=None):
# Rearrange the queires to be able to compute all heads in parallel
q_reshaped = self.reshape_tensor(self.W_q(queries), self.heads, True)
# Resulting tensor shape (batch_size, number of heads, sequence length, depth)
# Rearrange the keys to be able to compute all heads in parallel
k_reshaped = self.reshape_tensor(self.W_k(keys), self.heads, True)
# Resulting tensor shape (batch_size, number of heads, sequence length, depth)
# Rearrange the queires to be able to compute all heads in parallel
v_reshaped = self.reshape_tensor(self.W_v(values), self.heads, True)
# Resulting tensor shape (batch_size, number of heads, sequence length, depth)
# Compute the multi-head attention output using the reshaped queries, keys and values
o_reshaped = self.attention(q_reshaped, k_reshaped, v_reshaped, self.d_k, mask)
# Resulting tensor shape (batch_size, input_seq_length, -1)
# Rearrange back the output into concatenated form
output = self.reshape_tensor(o_reshaped, self.heads, False)
# Resulting tensor shape (batch_size, input_seq_length, d_v)
# Apply one final linear projection to the output to generate the multi-head attention
# Resulting tensor shape: (batch_size, input_seq_length, d_model)
return self.W_o(output)
# **Testing out the code**
h = 8 # Number of self-attention heads
d_k = 64 # Dimentionality of the linearly projected queries and keys
d_v = 64 # Dimentionality of the linearly projected values
d_model = 512 # Dimentionality of the model sub-layer outputs
batch_size = 64 # Batch size from the training process
input_seq_length = 5 # Maximum length of the input sequence
queries = random.random((batch_size, input_seq_length, d_k))
keys = random.random((batch_size, input_seq_length, d_k))
values = random.random((batch_size, input_seq_length, d_v))
multihead_attention = MultiHeadAttention(h, d_k, d_v, d_model)
print(multihead_attention(queries, keys, values))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/177/129177614.ipynb
| null | null |
[{"Id": 129177614, "ScriptId": 37960847, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6877706, "CreationDate": "05/11/2023 15:01:51", "VersionNumber": 1.0, "Title": "transformer_multi_head_attention", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 97.0, "LinesInsertedFromPrevious": 97.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
from tensorflow import matmul, math, reshape, shape, transpose, cast, float32
from tensorflow.keras.layers import Dense, Layer
from keras.backend import softmax
from numpy import random
# **Implement the scaled-dot product attention**
class DotProductAttention(Layer):
def __init__(self, **kwargs):
super(DotProductAttention, self).__init__(**kwargs)
def call(self, queries, keys, values, d_k, mask=None):
# Scoring the queries againsts the keys after transposing the latter, and scaling
scores = matmul(queries, keys, transpose_b=True) / math.sqrt(cast(d_k, float32))
# Apply mask to the attention scores
if mask is not None:
scores += -1e9 * mask
# Computing the weights by a softmax operation
weigths = softmax(scores)
# Computing the attention by a weighted sum of the value vectors
return matmul(weigths, values)
# **Implementing Multi-Head Attention**
class MultiHeadAttention(Layer):
def __init__(self, h, d_k, d_v, d_model, **kwargs):
super(MultiHeadAttention, self).__init__(**kwargs)
self.attention = DotProductAttention() # scaled dot product attention
self.heads = h # Number of attention heads to use
self.d_k = d_k # Dimentionality of the linearly projected queries and keys
self.d_v = d_v # Dimentionality of the linearly projected values
self.d_model = d_model # Dimentionality of the model
self.W_q = Dense(d_k) # Learned projected matrix for queries
self.W_k = Dense(d_k) # Learned projected matrix for keys
self.W_v = Dense(d_v) # Learned projected matrix for values
self.W_o = Dense(d_model) # Learned projection matrix for multi-head output
def reshape_tensor(self, x, heads, flag):
if flag:
# Tensor shape after reshaping and transposing: (batch_size, heads, seq_lenght, -1)
x = reshape(x, shape=(shape(x)[0], shape(x)[1], heads, -1))
x = transpose(x, perm=(0, 2, 1, 3))
else:
x = transpose(x, perm=(0, 2, 1, 3))
x = reshape(x, shape=(shape(x)[0], shape(x)[1], self.d_k))
return x
def call(self, queries, keys, values, mask=None):
# Rearrange the queires to be able to compute all heads in parallel
q_reshaped = self.reshape_tensor(self.W_q(queries), self.heads, True)
# Resulting tensor shape (batch_size, number of heads, sequence length, depth)
# Rearrange the keys to be able to compute all heads in parallel
k_reshaped = self.reshape_tensor(self.W_k(keys), self.heads, True)
# Resulting tensor shape (batch_size, number of heads, sequence length, depth)
# Rearrange the queires to be able to compute all heads in parallel
v_reshaped = self.reshape_tensor(self.W_v(values), self.heads, True)
# Resulting tensor shape (batch_size, number of heads, sequence length, depth)
# Compute the multi-head attention output using the reshaped queries, keys and values
o_reshaped = self.attention(q_reshaped, k_reshaped, v_reshaped, self.d_k, mask)
# Resulting tensor shape (batch_size, input_seq_length, -1)
# Rearrange back the output into concatenated form
output = self.reshape_tensor(o_reshaped, self.heads, False)
# Resulting tensor shape (batch_size, input_seq_length, d_v)
# Apply one final linear projection to the output to generate the multi-head attention
# Resulting tensor shape: (batch_size, input_seq_length, d_model)
return self.W_o(output)
# **Testing out the code**
h = 8 # Number of self-attention heads
d_k = 64 # Dimentionality of the linearly projected queries and keys
d_v = 64 # Dimentionality of the linearly projected values
d_model = 512 # Dimentionality of the model sub-layer outputs
batch_size = 64 # Batch size from the training process
input_seq_length = 5 # Maximum length of the input sequence
queries = random.random((batch_size, input_seq_length, d_k))
keys = random.random((batch_size, input_seq_length, d_k))
values = random.random((batch_size, input_seq_length, d_v))
multihead_attention = MultiHeadAttention(h, d_k, d_v, d_model)
print(multihead_attention(queries, keys, values))
| false | 0 | 1,203 | 0 | 1,203 | 1,203 |
||
129177343
|
# import the required libraries
from tensorflow import matmul, math, cast, float32
from tensorflow.keras.layers import Layer
from keras.backend import softmax
# **Implementing the scaled-Dot product attention**
class DotProductAttention(Layer):
def __init__(self, **kwargs):
super(DotProductAttention, self).__init__(**kwargs)
def call(self, queries, keys, values, d_k, mask=None):
# Scoring the queries againsts keys after transposing the latter, and scaling
scores = matmul(queries, keys, transpose_b=True) / math.sqrt(cast(d_k, float32))
# Apply mask to the attention scores
if mask is not None:
scores += -1e9 * mask
# Computing the weights by a softmax operation
weights = softmax(scores)
# Computing the attention by a weighted sum of the value vectors
return matmul(weights, values)
# **Testing out the code**
from numpy import random
input_seq_length = 5 # Maximum length of the input sequence
d_k = 64 # Dimentionality of the linearly projected queries and keys
d_v = 64 # Dimentionality of the linearly projected values
batch_size = 64 # Batch size from training process
queries = random.random((batch_size, input_seq_length, d_k))
keys = random.random((batch_size, input_seq_length, d_k))
values = random.random((batch_size, input_seq_length, d_v))
values
attention = DotProductAttention()
print(attention(queries, keys, values, d_k))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/177/129177343.ipynb
| null | null |
[{"Id": 129177343, "ScriptId": 37967696, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6877706, "CreationDate": "05/11/2023 14:59:31", "VersionNumber": 1.0, "Title": "transformer_dot_product_attention", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 43.0, "LinesInsertedFromPrevious": 43.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# import the required libraries
from tensorflow import matmul, math, cast, float32
from tensorflow.keras.layers import Layer
from keras.backend import softmax
# **Implementing the scaled-Dot product attention**
class DotProductAttention(Layer):
def __init__(self, **kwargs):
super(DotProductAttention, self).__init__(**kwargs)
def call(self, queries, keys, values, d_k, mask=None):
# Scoring the queries againsts keys after transposing the latter, and scaling
scores = matmul(queries, keys, transpose_b=True) / math.sqrt(cast(d_k, float32))
# Apply mask to the attention scores
if mask is not None:
scores += -1e9 * mask
# Computing the weights by a softmax operation
weights = softmax(scores)
# Computing the attention by a weighted sum of the value vectors
return matmul(weights, values)
# **Testing out the code**
from numpy import random
input_seq_length = 5 # Maximum length of the input sequence
d_k = 64 # Dimentionality of the linearly projected queries and keys
d_v = 64 # Dimentionality of the linearly projected values
batch_size = 64 # Batch size from training process
queries = random.random((batch_size, input_seq_length, d_k))
keys = random.random((batch_size, input_seq_length, d_k))
values = random.random((batch_size, input_seq_length, d_v))
values
attention = DotProductAttention()
print(attention(queries, keys, values, d_k))
| false | 0 | 391 | 0 | 391 | 391 |
||
129094603
|
<jupyter_start><jupyter_text>HR Competency Scores for Screening
##### Context
Recruitment and candidate selection play a critical role in determining the success of an organization. An effective initial screening process can significantly improve the quality of the hiring pool and increase the chances of finding the right candidate for any given role. This dataset focuses on both behavioral and functional competency scores, which are essential aspects of a candidate's potential fit and contribution to the organization.
##### Sources
The data in this dataset has been collected from an anonymous company's internal HR department and published in a normalized form. The dataset combines the scores from two key assessments:
1. Functional competency test: Utilized to evaluate a candidate's hard skills and domain knowledge.
2. HR behavior test: An assessment tool focused on evaluating soft or behavior skills, crucial for teamwork and adaptability within an organization.
##### Young Researchers' Contribution
We were approached by a group of young researchers interested in the explainable AI (XAI) problem. They aimed to analyze HR data to understand why specific candidates were called for interviews while others were not. With their valuable input and help in preprocessing the data, we have made this dataset available for the wider research community.
##### Inspiration
The inspiration behind sharing this dataset was the growing need for insights into the hiring process and the importance of selecting candidates who possess a balance of functional and behavioral competencies. With the added value of XAI research, we hope to encourage researchers and data scientists to analyze the initial screening process, build models to optimize candidate selection, explain their decisions, and uncover new insights that can enhance recruitment strategies.
The dataset can be employed for a wide range of applications, including:
1. Identifying the most significant factors in determining a candidate's eligibility for an interview.
2. Developing machine learning models to predict and explain the likelihood of a candidate being called for an interview.
3. Analyzing the balance between functional competencies and behavioral skills required for a good fit in the organization.
4. Investigating the impact of different skill combinations on the overall competency scores.
We hope this dataset inspires researchers to explore new dimensions of the hiring process and contribute to building better and more transparent recruitment strategies.
Kaggle dataset identifier: hr-competency-scores-for-screening
<jupyter_script># # Classifier Comparison
# * **Problem Statement** : HR competency score for screening potential candidates
# * **Solution/aim** : First comparing 10 different SK-Learn ML models for their accuracy and then making a prediction on the test-data
# standard libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.simplefilter("ignore")
# sklearn libraries
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, log_loss
# sklearn Models
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC, LinearSVC, NuSVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import (
RandomForestClassifier,
AdaBoostClassifier,
GradientBoostingClassifier,
)
from sklearn.naive_bayes import GaussianNB
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
# ## Data Load & Preparation
#
# load
FILE_PATH = "/kaggle/input/hr-competency-scores-for-screening/dataset.csv"
df = pd.read_csv(FILE_PATH)
# remove NULLs
df.dropna(inplace=True)
df.reset_index(drop=True, inplace=True)
# view
df.head()
# feature engineering
x, y = df.drop("call_for_interview", axis=1), df[["call_for_interview"]]
# scaling the data
ss = StandardScaler()
x = ss.fit_transform(x)
# split the data
X_train, X_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42, stratify=y
)
# ## Sklearn Classifier Comparison
# Looping through all the standard out-of-the box classifiers and printing the accuracy. Obviously, these will perform much better after tuning their hyperparameters, but thats the story for another time.
# loop de loop
classifiers = [
KNeighborsClassifier(3),
SVC(kernel="rbf", C=0.025, probability=True),
NuSVC(probability=True),
DecisionTreeClassifier(),
RandomForestClassifier(),
AdaBoostClassifier(),
GradientBoostingClassifier(),
GaussianNB(),
LinearDiscriminantAnalysis(),
QuadraticDiscriminantAnalysis(),
]
# Logging for Visual Comparison
log_cols = ["Classifier", "Accuracy", "Log Loss"]
log = pd.DataFrame(columns=log_cols)
for clf in classifiers:
clf.fit(X_train, y_train)
name = clf.__class__.__name__
print("=" * 40)
print(name)
print("****Results****")
train_predictions = clf.predict(X_test)
acc = accuracy_score(y_test, train_predictions)
print("Accuracy: {:.2%}".format(acc))
train_predictions = clf.predict_proba(X_test)
ll = log_loss(y_test, train_predictions)
print("Log Loss: {:.3}".format(ll))
log_entry = pd.DataFrame([[name, acc * 100, ll]], columns=log_cols)
log = log.append(log_entry)
print("=" * 40)
# plot the Accuracy & Log Loss
fig, ax = plt.subplots(nrows=1, ncols=2, sharey=True, figsize=(20, 8))
plt.subplot(1, 2, 1)
sns.set_color_codes("muted")
c = sns.barplot(x="Accuracy", y="Classifier", data=log, color="b")
c.set(ylabel=None)
c.bar_label(c.containers[0], padding=-30, fmt="%.2f")
plt.xlabel("Accuracy %")
plt.title("Classifier Accuracy")
plt.subplot(1, 2, 2)
sns.set_color_codes("muted")
l = sns.barplot(
x="Log Loss",
y="Classifier",
data=log,
color="g",
)
l.set(ylabel=None)
l.bar_label(l.containers[0], padding=-25, fmt="%.2f")
plt.xlabel("Log Loss")
plt.title("Classifier Log Loss")
fig.supylabel("Classifier")
plt.show()
# **Some Observations:**
# * Decision Tree Classifier has the highest loss
# * Almost all of them have a 90% + accuracy
# * Gradient Boosting has the highest accuracy of 96.67& with loss of 0.17
# * Quadratic Discriminant has accuracy of 95% (little less than Gradient Booster) and loss of 0.16 (less than Gradient Booster)
# ### My Choice of Classifier for making prediction = Quadratic Discriminant Analysis
# **Note: Due to low volume of data & incredibly high accuracy, I will not attempt to fine tune this classifier**
# instantiate the classifier - once again !
qda = QuadraticDiscriminantAnalysis()
# train the model on training data
qda.fit(X_train, y_train)
# make predictions on the test-data
pred = qda.predict(X_test)
# add new prediction column to the test-data
y_test["prediction"] = pred.tolist()
# view
y_test.head(10)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/094/129094603.ipynb
|
hr-competency-scores-for-screening
|
muhammadjawwadismail
|
[{"Id": 129094603, "ScriptId": 38375068, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3828601, "CreationDate": "05/11/2023 01:18:38", "VersionNumber": 1.0, "Title": "notebooke900980fe7", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 147.0, "LinesInsertedFromPrevious": 147.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184846242, "KernelVersionId": 129094603, "SourceDatasetVersionId": 5658852}]
|
[{"Id": 5658852, "DatasetId": 3252282, "DatasourceVersionId": 5734269, "CreatorUserId": 4429155, "LicenseName": "CC BY-SA 4.0", "CreationDate": "05/10/2023 21:38:25", "VersionNumber": 1.0, "Title": "HR Competency Scores for Screening", "Slug": "hr-competency-scores-for-screening", "Subtitle": "Anonymized HR Data for Evaluating Candidate Screening Processes", "Description": "##### Context\n\nRecruitment and candidate selection play a critical role in determining the success of an organization. An effective initial screening process can significantly improve the quality of the hiring pool and increase the chances of finding the right candidate for any given role. This dataset focuses on both behavioral and functional competency scores, which are essential aspects of a candidate's potential fit and contribution to the organization.\n\n##### Sources\n\nThe data in this dataset has been collected from an anonymous company's internal HR department and published in a normalized form. The dataset combines the scores from two key assessments:\n\n1. Functional competency test: Utilized to evaluate a candidate's hard skills and domain knowledge.\n2. HR behavior test: An assessment tool focused on evaluating soft or behavior skills, crucial for teamwork and adaptability within an organization.\n\n##### Young Researchers' Contribution\n\nWe were approached by a group of young researchers interested in the explainable AI (XAI) problem. They aimed to analyze HR data to understand why specific candidates were called for interviews while others were not. With their valuable input and help in preprocessing the data, we have made this dataset available for the wider research community.\n\n##### Inspiration\n\nThe inspiration behind sharing this dataset was the growing need for insights into the hiring process and the importance of selecting candidates who possess a balance of functional and behavioral competencies. With the added value of XAI research, we hope to encourage researchers and data scientists to analyze the initial screening process, build models to optimize candidate selection, explain their decisions, and uncover new insights that can enhance recruitment strategies.\n\nThe dataset can be employed for a wide range of applications, including:\n\n1. Identifying the most significant factors in determining a candidate's eligibility for an interview.\n2. Developing machine learning models to predict and explain the likelihood of a candidate being called for an interview.\n3. Analyzing the balance between functional competencies and behavioral skills required for a good fit in the organization.\n4. Investigating the impact of different skill combinations on the overall competency scores.\n\nWe hope this dataset inspires researchers to explore new dimensions of the hiring process and contribute to building better and more transparent recruitment strategies.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3252282, "CreatorUserId": 4429155, "OwnerUserId": 4429155.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5658852.0, "CurrentDatasourceVersionId": 5734269.0, "ForumId": 3317708, "Type": 2, "CreationDate": "05/10/2023 21:38:25", "LastActivityDate": "05/10/2023", "TotalViews": 5601, "TotalDownloads": 897, "TotalVotes": 37, "TotalKernels": 12}]
|
[{"Id": 4429155, "UserName": "muhammadjawwadismail", "DisplayName": "Muhammad Jawad", "RegisterDate": "02/03/2020", "PerformanceTier": 0}]
|
# # Classifier Comparison
# * **Problem Statement** : HR competency score for screening potential candidates
# * **Solution/aim** : First comparing 10 different SK-Learn ML models for their accuracy and then making a prediction on the test-data
# standard libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.simplefilter("ignore")
# sklearn libraries
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, log_loss
# sklearn Models
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC, LinearSVC, NuSVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import (
RandomForestClassifier,
AdaBoostClassifier,
GradientBoostingClassifier,
)
from sklearn.naive_bayes import GaussianNB
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
# ## Data Load & Preparation
#
# load
FILE_PATH = "/kaggle/input/hr-competency-scores-for-screening/dataset.csv"
df = pd.read_csv(FILE_PATH)
# remove NULLs
df.dropna(inplace=True)
df.reset_index(drop=True, inplace=True)
# view
df.head()
# feature engineering
x, y = df.drop("call_for_interview", axis=1), df[["call_for_interview"]]
# scaling the data
ss = StandardScaler()
x = ss.fit_transform(x)
# split the data
X_train, X_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42, stratify=y
)
# ## Sklearn Classifier Comparison
# Looping through all the standard out-of-the box classifiers and printing the accuracy. Obviously, these will perform much better after tuning their hyperparameters, but thats the story for another time.
# loop de loop
classifiers = [
KNeighborsClassifier(3),
SVC(kernel="rbf", C=0.025, probability=True),
NuSVC(probability=True),
DecisionTreeClassifier(),
RandomForestClassifier(),
AdaBoostClassifier(),
GradientBoostingClassifier(),
GaussianNB(),
LinearDiscriminantAnalysis(),
QuadraticDiscriminantAnalysis(),
]
# Logging for Visual Comparison
log_cols = ["Classifier", "Accuracy", "Log Loss"]
log = pd.DataFrame(columns=log_cols)
for clf in classifiers:
clf.fit(X_train, y_train)
name = clf.__class__.__name__
print("=" * 40)
print(name)
print("****Results****")
train_predictions = clf.predict(X_test)
acc = accuracy_score(y_test, train_predictions)
print("Accuracy: {:.2%}".format(acc))
train_predictions = clf.predict_proba(X_test)
ll = log_loss(y_test, train_predictions)
print("Log Loss: {:.3}".format(ll))
log_entry = pd.DataFrame([[name, acc * 100, ll]], columns=log_cols)
log = log.append(log_entry)
print("=" * 40)
# plot the Accuracy & Log Loss
fig, ax = plt.subplots(nrows=1, ncols=2, sharey=True, figsize=(20, 8))
plt.subplot(1, 2, 1)
sns.set_color_codes("muted")
c = sns.barplot(x="Accuracy", y="Classifier", data=log, color="b")
c.set(ylabel=None)
c.bar_label(c.containers[0], padding=-30, fmt="%.2f")
plt.xlabel("Accuracy %")
plt.title("Classifier Accuracy")
plt.subplot(1, 2, 2)
sns.set_color_codes("muted")
l = sns.barplot(
x="Log Loss",
y="Classifier",
data=log,
color="g",
)
l.set(ylabel=None)
l.bar_label(l.containers[0], padding=-25, fmt="%.2f")
plt.xlabel("Log Loss")
plt.title("Classifier Log Loss")
fig.supylabel("Classifier")
plt.show()
# **Some Observations:**
# * Decision Tree Classifier has the highest loss
# * Almost all of them have a 90% + accuracy
# * Gradient Boosting has the highest accuracy of 96.67& with loss of 0.17
# * Quadratic Discriminant has accuracy of 95% (little less than Gradient Booster) and loss of 0.16 (less than Gradient Booster)
# ### My Choice of Classifier for making prediction = Quadratic Discriminant Analysis
# **Note: Due to low volume of data & incredibly high accuracy, I will not attempt to fine tune this classifier**
# instantiate the classifier - once again !
qda = QuadraticDiscriminantAnalysis()
# train the model on training data
qda.fit(X_train, y_train)
# make predictions on the test-data
pred = qda.predict(X_test)
# add new prediction column to the test-data
y_test["prediction"] = pred.tolist()
# view
y_test.head(10)
| false | 0 | 1,295 | 0 | 1,822 | 1,295 |
||
129094203
|
# import libraries
from random import randint, seed
import pandas as pd
import numpy as np
import plotly.express as px
seed(10)
my_data = pd.read_csv("/kaggle/input/titanic/Titanic.tsv", sep="\t")
my_data.info()
my_data.head(11)
# Checking Outliers
my_data.columns
# Handling duplicates
my_data.drop_duplicates(keep=False, inplace=True)
my_data.shape
# Handling missing data, NaNs, Blanks (missing values)
my_data.isna().sum()
my_data = my_data.dropna() # delete rows with missing data
my_data.isna().sum()
my_data.shape
# Wrong/improper values
my_data["Age"].value_counts()
# Convert incorrectly formatted age values to float
my_data["Age"] = my_data["Age"].apply(
lambda x: float(x.replace(".", "")) if isinstance(x, str) and "." in x else x
)
my_data["Age"].value_counts()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/094/129094203.ipynb
| null | null |
[{"Id": 129094203, "ScriptId": 38204470, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14627695, "CreationDate": "05/11/2023 01:12:26", "VersionNumber": 8.0, "Title": "Data Cleaning and Preparation", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 43.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 42.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# import libraries
from random import randint, seed
import pandas as pd
import numpy as np
import plotly.express as px
seed(10)
my_data = pd.read_csv("/kaggle/input/titanic/Titanic.tsv", sep="\t")
my_data.info()
my_data.head(11)
# Checking Outliers
my_data.columns
# Handling duplicates
my_data.drop_duplicates(keep=False, inplace=True)
my_data.shape
# Handling missing data, NaNs, Blanks (missing values)
my_data.isna().sum()
my_data = my_data.dropna() # delete rows with missing data
my_data.isna().sum()
my_data.shape
# Wrong/improper values
my_data["Age"].value_counts()
# Convert incorrectly formatted age values to float
my_data["Age"] = my_data["Age"].apply(
lambda x: float(x.replace(".", "")) if isinstance(x, str) and "." in x else x
)
my_data["Age"].value_counts()
| false | 0 | 264 | 0 | 264 | 264 |
||
129094639
|
import numpy as np
x = np.array([-3, -2, -1, 1, 2, 3])
print("Sigmoid ", 1 / (1 + np.exp(-z)))
print("Tanh ", np.tanh(x))
print("ReLU ", np.maximum(x, 0))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/094/129094639.ipynb
| null | null |
[{"Id": 129094639, "ScriptId": 38376704, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1848770, "CreationDate": "05/11/2023 01:19:09", "VersionNumber": 1.0, "Title": "1.2.1 Activation Functions", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 8.0, "LinesInsertedFromPrevious": 8.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
x = np.array([-3, -2, -1, 1, 2, 3])
print("Sigmoid ", 1 / (1 + np.exp(-z)))
print("Tanh ", np.tanh(x))
print("ReLU ", np.maximum(x, 0))
| false | 0 | 74 | 0 | 74 | 74 |
||
129100065
|
<jupyter_start><jupyter_text>Mall_Customers
Kaggle dataset identifier: mall-customers
<jupyter_code>import pandas as pd
df = pd.read_csv('mall-customers/Mall_Customers.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 200 entries, 0 to 199
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 CustomerID 200 non-null int64
1 Genre 200 non-null object
2 Age 200 non-null int64
3 Annual Income (k$) 200 non-null int64
4 Spending Score (1-100) 200 non-null int64
dtypes: int64(4), object(1)
memory usage: 7.9+ KB
<jupyter_text>Examples:
{
"CustomerID": 1,
"Genre": "Male",
"Age": 19,
"Annual Income (k$)": 15,
"Spending Score (1-100)": 39
}
{
"CustomerID": 2,
"Genre": "Male",
"Age": 21,
"Annual Income (k$)": 15,
"Spending Score (1-100)": 81
}
{
"CustomerID": 3,
"Genre": "Female",
"Age": 20,
"Annual Income (k$)": 16,
"Spending Score (1-100)": 6
}
{
"CustomerID": 4,
"Genre": "Female",
"Age": 23,
"Annual Income (k$)": 16,
"Spending Score (1-100)": 77
}
<jupyter_script># Install a pip package in the current Jupyter kernel
import sys
# from pycaret.utils import enable_colab
# enable_colab()
import pandas as pd
dataset = pd.read_csv("/kaggle/input/mall-customers/Mall_Customers.csv")
dataset.head()
dataset.shape
data = dataset.sample(frac=0.95, random_state=786)
data_unseen = dataset.drop(data.index)
data.reset_index(drop=True, inplace=True)
data_unseen.reset_index(drop=True, inplace=True)
print("Data for Modeling: " + str(data.shape))
print("Unseen Data For Predictions: " + str(data_unseen.shape))
from pycaret.clustering import *
exp_clu101 = setup(data, normalize=True, ignore_features=["CustomerID"], session_id=123)
plot_model(kmeans, "elbow")
kmeans = create_model("kmeans", num_clusters=6)
print(kmeans)
kmean_results = assign_model(kmeans)
kmean_results.head()
plot_model(kmeans)
models()
print(Agglo)
Agglo = create_model("hclust", num_clusters=3)
kmean_results_2 = assign_model(Agglo)
kmean_results_2.head()
plot_model(Agglo)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/100/129100065.ipynb
|
mall-customers
|
shwetabh123
|
[{"Id": 129100065, "ScriptId": 38377779, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8292053, "CreationDate": "05/11/2023 02:39:14", "VersionNumber": 4.0, "Title": "Calvin - Mall_Customers - Clustering with PyCaret", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 51.0, "LinesInsertedFromPrevious": 3.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 48.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184856748, "KernelVersionId": 129100065, "SourceDatasetVersionId": 10938}]
|
[{"Id": 10938, "DatasetId": 7721, "DatasourceVersionId": 10938, "CreatorUserId": 1508014, "LicenseName": "CC0: Public Domain", "CreationDate": "12/23/2017 06:12:40", "VersionNumber": 1.0, "Title": "Mall_Customers", "Slug": "mall-customers", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 4286.0, "TotalUncompressedBytes": 4286.0}]
|
[{"Id": 7721, "CreatorUserId": 1508014, "OwnerUserId": 1508014.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 10938.0, "CurrentDatasourceVersionId": 10938.0, "ForumId": 14591, "Type": 2, "CreationDate": "12/23/2017 06:12:40", "LastActivityDate": "02/01/2018", "TotalViews": 246607, "TotalDownloads": 43908, "TotalVotes": 192, "TotalKernels": 140}]
|
[{"Id": 1508014, "UserName": "shwetabh123", "DisplayName": "shwetabh123", "RegisterDate": "12/20/2017", "PerformanceTier": 1}]
|
# Install a pip package in the current Jupyter kernel
import sys
# from pycaret.utils import enable_colab
# enable_colab()
import pandas as pd
dataset = pd.read_csv("/kaggle/input/mall-customers/Mall_Customers.csv")
dataset.head()
dataset.shape
data = dataset.sample(frac=0.95, random_state=786)
data_unseen = dataset.drop(data.index)
data.reset_index(drop=True, inplace=True)
data_unseen.reset_index(drop=True, inplace=True)
print("Data for Modeling: " + str(data.shape))
print("Unseen Data For Predictions: " + str(data_unseen.shape))
from pycaret.clustering import *
exp_clu101 = setup(data, normalize=True, ignore_features=["CustomerID"], session_id=123)
plot_model(kmeans, "elbow")
kmeans = create_model("kmeans", num_clusters=6)
print(kmeans)
kmean_results = assign_model(kmeans)
kmean_results.head()
plot_model(kmeans)
models()
print(Agglo)
Agglo = create_model("hclust", num_clusters=3)
kmean_results_2 = assign_model(Agglo)
kmean_results_2.head()
plot_model(Agglo)
|
[{"mall-customers/Mall_Customers.csv": {"column_names": "[\"CustomerID\", \"Genre\", \"Age\", \"Annual Income (k$)\", \"Spending Score (1-100)\"]", "column_data_types": "{\"CustomerID\": \"int64\", \"Genre\": \"object\", \"Age\": \"int64\", \"Annual Income (k$)\": \"int64\", \"Spending Score (1-100)\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 200 entries, 0 to 199\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 CustomerID 200 non-null int64 \n 1 Genre 200 non-null object\n 2 Age 200 non-null int64 \n 3 Annual Income (k$) 200 non-null int64 \n 4 Spending Score (1-100) 200 non-null int64 \ndtypes: int64(4), object(1)\nmemory usage: 7.9+ KB\n", "summary": "{\"CustomerID\": {\"count\": 200.0, \"mean\": 100.5, \"std\": 57.879184513951124, \"min\": 1.0, \"25%\": 50.75, \"50%\": 100.5, \"75%\": 150.25, \"max\": 200.0}, \"Age\": {\"count\": 200.0, \"mean\": 38.85, \"std\": 13.96900733155888, \"min\": 18.0, \"25%\": 28.75, \"50%\": 36.0, \"75%\": 49.0, \"max\": 70.0}, \"Annual Income (k$)\": {\"count\": 200.0, \"mean\": 60.56, \"std\": 26.264721165271244, \"min\": 15.0, \"25%\": 41.5, \"50%\": 61.5, \"75%\": 78.0, \"max\": 137.0}, \"Spending Score (1-100)\": {\"count\": 200.0, \"mean\": 50.2, \"std\": 25.823521668370173, \"min\": 1.0, \"25%\": 34.75, \"50%\": 50.0, \"75%\": 73.0, \"max\": 99.0}}", "examples": "{\"CustomerID\":{\"0\":1,\"1\":2,\"2\":3,\"3\":4},\"Genre\":{\"0\":\"Male\",\"1\":\"Male\",\"2\":\"Female\",\"3\":\"Female\"},\"Age\":{\"0\":19,\"1\":21,\"2\":20,\"3\":23},\"Annual Income (k$)\":{\"0\":15,\"1\":15,\"2\":16,\"3\":16},\"Spending Score (1-100)\":{\"0\":39,\"1\":81,\"2\":6,\"3\":77}}"}}]
| true | 1 |
<start_data_description><data_path>mall-customers/Mall_Customers.csv:
<column_names>
['CustomerID', 'Genre', 'Age', 'Annual Income (k$)', 'Spending Score (1-100)']
<column_types>
{'CustomerID': 'int64', 'Genre': 'object', 'Age': 'int64', 'Annual Income (k$)': 'int64', 'Spending Score (1-100)': 'int64'}
<dataframe_Summary>
{'CustomerID': {'count': 200.0, 'mean': 100.5, 'std': 57.879184513951124, 'min': 1.0, '25%': 50.75, '50%': 100.5, '75%': 150.25, 'max': 200.0}, 'Age': {'count': 200.0, 'mean': 38.85, 'std': 13.96900733155888, 'min': 18.0, '25%': 28.75, '50%': 36.0, '75%': 49.0, 'max': 70.0}, 'Annual Income (k$)': {'count': 200.0, 'mean': 60.56, 'std': 26.264721165271244, 'min': 15.0, '25%': 41.5, '50%': 61.5, '75%': 78.0, 'max': 137.0}, 'Spending Score (1-100)': {'count': 200.0, 'mean': 50.2, 'std': 25.823521668370173, 'min': 1.0, '25%': 34.75, '50%': 50.0, '75%': 73.0, 'max': 99.0}}
<dataframe_info>
RangeIndex: 200 entries, 0 to 199
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 CustomerID 200 non-null int64
1 Genre 200 non-null object
2 Age 200 non-null int64
3 Annual Income (k$) 200 non-null int64
4 Spending Score (1-100) 200 non-null int64
dtypes: int64(4), object(1)
memory usage: 7.9+ KB
<some_examples>
{'CustomerID': {'0': 1, '1': 2, '2': 3, '3': 4}, 'Genre': {'0': 'Male', '1': 'Male', '2': 'Female', '3': 'Female'}, 'Age': {'0': 19, '1': 21, '2': 20, '3': 23}, 'Annual Income (k$)': {'0': 15, '1': 15, '2': 16, '3': 16}, 'Spending Score (1-100)': {'0': 39, '1': 81, '2': 6, '3': 77}}
<end_description>
| 344 | 0 | 798 | 344 |
129100433
|
<jupyter_start><jupyter_text>Covid_19_india dataset
Kaggle dataset identifier: covid-19-india-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
from plotly.subplots import make_subplots
from datetime import datetime
covid_df = pd.read_csv("/kaggle/input/covid-19-india-dataset/covid_19_india.csv")
covid_df.head(10)
covid_df.info()
covid_df.describe()
covid_df.drop(
["Sno", "Time", "ConfirmedIndianNational", "ConfirmedForeignNational"],
inplace=True,
axis=1,
)
covid_df
# no of active cases
covid_df["Active_Cases"] = covid_df["Confirmed"] - (
covid_df["Cured"] + covid_df["Deaths"]
)
covid_df.tail()
# to convert to pivot table
statewise = pd.pivot_table(
covid_df,
values=["Confirmed", "Deaths", "Cured"],
index="State/UnionTerritory",
aggfunc=max,
)
statewise["Recovery Rate"] = statewise["Cured"] * 100 / statewise["Confirmed"]
statewise["Mortality Rate"] = statewise["Deaths"] * 100 / statewise["Confirmed"]
statewise = statewise.sort_values(by="Confirmed", ascending=False)
statewise.style.background_gradient(cmap="cubehelix")
top_10_active_cases = (
covid_df.groupby("State/UnionTerritory")
.max()[["Active_Cases", "Date"]]
.sort_values(by="Active_Cases", ascending=False)
.reset_index()
)
fig = plt.figure(figsize=(16, 9))
plt.title("Top 10 states with most active cases in India", size=25)
ax = sns.barplot(
data=top_10_active_cases.iloc[:10],
y="Active_Cases",
x="State/UnionTerritory",
linewidth=2,
edgecolor="red",
)
top_10_active_cases = (
covid_df.groupby(by="State/UnionTerritory")
.max()[["Active_Cases", "Date"]]
.sort_values(by="Active_Cases", ascending=False)
.reset_index()
)
fig = plt.figure(figsize=(18, 5))
plt.title("Top 10 states with most active cases in India", size=25)
ax = sns.barplot(
data=top_10_active_cases.iloc[:10],
y="Active_Cases",
x="State/UnionTerritory",
linewidth=2,
edgecolor="red",
)
plt.xlabel("States")
plt.ylabel("Total Active Cases")
plt.show()
# top states with highest deaths reported
top_10_deaths = (
covid_df.groupby(by="State/UnionTerritory")
.max()[["Deaths", "Date"]]
.sort_values(by="Deaths", ascending=False)
.reset_index()
)
fig = plt.figure(figsize=(18, 5))
plt.title("Top 10 states with most deaths in India", size=25)
ax = sns.barplot(
data=top_10_deaths.iloc[:12],
y="Deaths",
x="State/UnionTerritory",
linewidth=2,
edgecolor="black",
)
plt.xlabel("States")
plt.ylabel("Total Death Cases")
plt.show()
# #growth trend
# fig = plt.figure(figsize=(12,6))
# ax= sns.lineplot(data=covid_df[covid_df['State/UnionTerritory'].isin(['Maharashtra','Karnataka','Kerala','Tamil Nadu','Uttar Pradesh']),x='Date',y='Active_Cases',hue='State/UnionTerritory'])
# ax.set_title("Top 5 Affected States in India",size=16)
# covid_df['Date'] = covid_df['Date'].dt.strftime('%Y-%m-%d')
covid_df["Date"] = covid_df["Date"].dt.strftime("%m/%Y")
import seaborn as sns
fig = plt.figure(figsize=(12, 6))
states = ["Maharashtra", "Karnataka", "Kerala", "Tamil Nadu", "Uttar Pradesh"]
covid_df_states = covid_df[covid_df["State/UnionTerritory"].isin(states)]
ax = sns.lineplot(
data=covid_df_states, x="Date", y="Active_Cases", hue="State/UnionTerritory"
)
ax.set_title("Top 5 Affected States in India", size=16)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/100/129100433.ipynb
|
covid-19-india-dataset
|
lingutlaaswini
|
[{"Id": 129100433, "ScriptId": 38378211, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13174409, "CreationDate": "05/11/2023 02:44:06", "VersionNumber": 1.0, "Title": "notebook3ee58f19bd", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 108.0, "LinesInsertedFromPrevious": 108.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184857224, "KernelVersionId": 129100433, "SourceDatasetVersionId": 1281858}]
|
[{"Id": 1281858, "DatasetId": 739770, "DatasourceVersionId": 1313915, "CreatorUserId": 5134929, "LicenseName": "Unknown", "CreationDate": "06/26/2020 12:21:57", "VersionNumber": 1.0, "Title": "Covid_19_india dataset", "Slug": "covid-19-india-dataset", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 739770, "CreatorUserId": 5134929, "OwnerUserId": 5134929.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1281858.0, "CurrentDatasourceVersionId": 1313915.0, "ForumId": 754649, "Type": 2, "CreationDate": "06/26/2020 12:21:57", "LastActivityDate": "06/26/2020", "TotalViews": 2337, "TotalDownloads": 380, "TotalVotes": 8, "TotalKernels": 3}]
|
[{"Id": 5134929, "UserName": "lingutlaaswini", "DisplayName": "lingutla aswini", "RegisterDate": "05/21/2020", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
from plotly.subplots import make_subplots
from datetime import datetime
covid_df = pd.read_csv("/kaggle/input/covid-19-india-dataset/covid_19_india.csv")
covid_df.head(10)
covid_df.info()
covid_df.describe()
covid_df.drop(
["Sno", "Time", "ConfirmedIndianNational", "ConfirmedForeignNational"],
inplace=True,
axis=1,
)
covid_df
# no of active cases
covid_df["Active_Cases"] = covid_df["Confirmed"] - (
covid_df["Cured"] + covid_df["Deaths"]
)
covid_df.tail()
# to convert to pivot table
statewise = pd.pivot_table(
covid_df,
values=["Confirmed", "Deaths", "Cured"],
index="State/UnionTerritory",
aggfunc=max,
)
statewise["Recovery Rate"] = statewise["Cured"] * 100 / statewise["Confirmed"]
statewise["Mortality Rate"] = statewise["Deaths"] * 100 / statewise["Confirmed"]
statewise = statewise.sort_values(by="Confirmed", ascending=False)
statewise.style.background_gradient(cmap="cubehelix")
top_10_active_cases = (
covid_df.groupby("State/UnionTerritory")
.max()[["Active_Cases", "Date"]]
.sort_values(by="Active_Cases", ascending=False)
.reset_index()
)
fig = plt.figure(figsize=(16, 9))
plt.title("Top 10 states with most active cases in India", size=25)
ax = sns.barplot(
data=top_10_active_cases.iloc[:10],
y="Active_Cases",
x="State/UnionTerritory",
linewidth=2,
edgecolor="red",
)
top_10_active_cases = (
covid_df.groupby(by="State/UnionTerritory")
.max()[["Active_Cases", "Date"]]
.sort_values(by="Active_Cases", ascending=False)
.reset_index()
)
fig = plt.figure(figsize=(18, 5))
plt.title("Top 10 states with most active cases in India", size=25)
ax = sns.barplot(
data=top_10_active_cases.iloc[:10],
y="Active_Cases",
x="State/UnionTerritory",
linewidth=2,
edgecolor="red",
)
plt.xlabel("States")
plt.ylabel("Total Active Cases")
plt.show()
# top states with highest deaths reported
top_10_deaths = (
covid_df.groupby(by="State/UnionTerritory")
.max()[["Deaths", "Date"]]
.sort_values(by="Deaths", ascending=False)
.reset_index()
)
fig = plt.figure(figsize=(18, 5))
plt.title("Top 10 states with most deaths in India", size=25)
ax = sns.barplot(
data=top_10_deaths.iloc[:12],
y="Deaths",
x="State/UnionTerritory",
linewidth=2,
edgecolor="black",
)
plt.xlabel("States")
plt.ylabel("Total Death Cases")
plt.show()
# #growth trend
# fig = plt.figure(figsize=(12,6))
# ax= sns.lineplot(data=covid_df[covid_df['State/UnionTerritory'].isin(['Maharashtra','Karnataka','Kerala','Tamil Nadu','Uttar Pradesh']),x='Date',y='Active_Cases',hue='State/UnionTerritory'])
# ax.set_title("Top 5 Affected States in India",size=16)
# covid_df['Date'] = covid_df['Date'].dt.strftime('%Y-%m-%d')
covid_df["Date"] = covid_df["Date"].dt.strftime("%m/%Y")
import seaborn as sns
fig = plt.figure(figsize=(12, 6))
states = ["Maharashtra", "Karnataka", "Kerala", "Tamil Nadu", "Uttar Pradesh"]
covid_df_states = covid_df[covid_df["State/UnionTerritory"].isin(states)]
ax = sns.lineplot(
data=covid_df_states, x="Date", y="Active_Cases", hue="State/UnionTerritory"
)
ax.set_title("Top 5 Affected States in India", size=16)
| false | 1 | 1,335 | 0 | 1,368 | 1,335 |
||
129199421
|
# RAW DATA SITE NEEDED TO LOAD PARAM DATA, AND CONSTRUCT THE GFN2-XTB CALCULATOR
# Sources:
# https://github.com/grimme-lab/xtb/blob/main/param_gfn2-xtb.txt
# https://github.com/grimme-lab/xtb
# https://zenodo.org/record/4486287#.ZE28g3bMKUl
# https://xtb-python.readthedocs.io/en/latest/
# https://xtb-python.readthedocs.io/_/downloads/en/stable/pdf/
# http://rdkit.org/docs/source/rdkit.Chem.rdchem.html?highlight=getatoms
# https://www.rdkit.org/docs/source/rdkit.Chem.rdForceFieldHelpers.html
# https://www.scm.com/wp-content/uploads/pdfs/2014/UFF.pdf
# https://stackoverflow.com/questions/71915443/rdkit-coordinates-for-atoms-in-a-molecule
import numpy as np
from scipy import constants
import random as rand
from rdkit import Chem
from rdkit.Chem import AllChem
from xtb.interface import Calculator, Molecule, Param
from xtb.utils import get_method
from xtb.utils import get_solvent, Solvent
from xtb.libxtb import VERBOSITY_MUTED
# Unit conversions
Hartree_to_kcal_mol = 627.5
Bohr_to_angstrom = constants.physical_constants["Bohr radius"][0] * 1.0e10
# List of reference states for each element.
ref_states = {
"H": "[H][H]",
"C": "[C]#[C]",
"N": "N#N",
"O": "O=O",
"F": "FF",
"P": "P#P",
"S": "S=S",
"Cl": "ClCl",
}
def RDK_FP(mol):
"""
Takes a molecule and returns its RDK footprint.
"""
fp = AllChem.RDKFingerprint(mol, fpSize=512)
fp_arr = np.array(list(fp), dtype=int)
return fp_arr
def aq_structure(mol, element_num_list):
"""
Optimizes the aqueous-phase structure of a molecule and returns the 3D-coordinates and energy of the structure.
Also calculates and returns the energy of the optimized structure with no solvent.
"""
AllChem.EmbedMolecule(mol, randomSeed=-1)
AllChem.UFFOptimizeMolecule(
mol
) # first performs a UFF optimziation to get an initial guess.
xyz = mol.GetConformer().GetPositions()
calc = Calculator(get_method("GFN2-xTB"), np.array(element_num_list), xyz)
calc.set_verbosity(VERBOSITY_MUTED)
calc.set_solvent(Solvent.h2o)
step_range = rand.choice([30, 35, 40, 45, 50, 55, 60])
for step_i in range(
step_range
): # Iteratively optimizing the molecule using the calculated gradients.
calc.update(xyz / Bohr_to_angstrom)
result = calc.singlepoint()
E_aq = result.get_energy() * Hartree_to_kcal_mol
Eformation_grad = result.get_gradient() / Bohr_to_angstrom
xyz -= 0.05 * Eformation_grad # gradient descent with a step size of 0.05.
if (
np.linalg.norm(Eformation_grad, 2) < 1e-5
): # stop the optimization when gradient is close to zero.
break
calc.set_solvent() # remove the solvent to get the energy of the molecule in the gas phase.
result_gas = calc.singlepoint()
E_gas = result_gas.get_energy() * Hartree_to_kcal_mol
return xyz, E_aq, E_gas
def ref_energy(element_list):
"""
Given a list of elements in a molecule, returns the total energy of the molecule based on the reference states of the elements.
The molecule's reference state energy is the sum of the energies of each of the atoms in the molecule.
"""
E_ref = 0
for element in element_list:
mol = Chem.MolFromSmiles(
ref_states[element]
) # gets the reference state diatomic molecule for the element.
atoms = mol.GetAtoms()
element_num_list = [atom.GetAtomicNum() for atom in atoms]
energy = (
aq_structure(mol, element_num_list)[1] / 2
) # for every element in the molecule, calculate the aqueous phase energy of the diatomic molecule it comes from.
E_ref += energy # the total energy of the reference state.
return E_ref
def smiles_to_properties(smiles, n_calculations=10):
"""
Given a SMILES string representing a molecule, returns the following properties of the molecule:
the 3D coordinates of the lowest energy structure of the molecule in the aqueous phase, the energy of formation, the hydration energy, and the molecular fingerprint.
By default, the function runs 10 tests for the given molecule, but that can be changed by the user. The more tests run, the longer the calculations take.
"""
# first converts the smiles string into a list of its elements and a list of the atomic numbers of the elements.
mol = Chem.MolFromSmiles(smiles)
mol = Chem.AddHs(mol)
atoms = mol.GetAtoms()
element_list = [atom.GetSymbol() for atom in atoms]
element_num_list = [atom.GetAtomicNum() for atom in atoms]
Eformation_Aq_values = (
[]
) # keeps track of the calculated formation energies of the molecule in the aqueous phase for each calculation.
hydration_energy_values = (
[]
) # # keeps track of the calculated hydration energies of the molecule for each calculation.
for i in range(n_calculations):
xyz, E_aq, E_gas = aq_structure(
mol, element_num_list
) # optimize the molecule and get its coordinates, energy in aq phase, and energy in has phase
Eref = ref_energy(element_list) # obtain the reference energy of the molecule
Eformation = E_aq - Eref
Ehydration = E_aq - E_gas
hydration_energy_values.append(Ehydration)
Eformation_Aq_values.append(Eformation)
Eformation_Aq_mean = np.round(np.mean(Eformation_Aq_values), 3)
Eformation_Aq_std = np.round(np.std(Eformation_Aq_values), 3)
hydration_energy_mean = np.round(np.mean(hydration_energy_values), 3)
hydration_energy_std = np.round(np.std(hydration_energy_values), 3)
xyz = np.round(xyz, 3)
fp = RDK_FP(mol)
return (
(element_list, xyz),
f"Formation energy vs reference: {Eformation_Aq_mean} ± {Eformation_Aq_std} kcal/mol",
f"Hydration energy: {hydration_energy_mean} ± {hydration_energy_std} kcal/mol",
fp,
)
smiles_to_properties("CC")
"""
SMILE strings of molecules tested:
'CCCCC(=O)OC' #methyl hexanoate
'CCCCO' #butan-1-ol
'C1CCC(CC1)N' #cyclohexanamine
'C1cc(cc1)OC=O' #phentyl formate
'CCC(=O)OC' #methyl propanoate
'C1CC(CC(C1)Cl)Cl' #1,3-dichlorobenzene
'COC(=O)CC#N' #methyl 2-cyanoacetate
'CCOCC' #ethoxyethane
'CCCCC[N+](=O)[O-]' #1-nitropentane
'CCCCCCCC(=O)C' #nonan-2-one
'CC1CCNCC1' 4-#methylpyridine
'CCCCS' #butane-1-thiol
'CC1CCCC(C1C)O' #2,3-dimehtylphenol
'CO' #methanol
'CS(=O)(=O)C' #methylsulfonylmethane
'C1(C(C(C1(F)F)(F)F)(F)F)(F)F' #octafluorocyclobutane
'CCCS' #propanethiol
'CC#N' #acetonitrile
'CSSC' #methyldisulfanylmethane
'C1CCC(C1)O' #cylopentanol
"""
# TESTING CODE - ATOMIC DISTANCES
import math
import numpy as np
element_list, xyz = smiles_to_properties("CC")[0]
def distances(element_list, xyz):
n = len(element_list)
dists = np.zeros((n, n))
for i in range(n):
for j in range(i, n):
dist = math.sqrt(sum((xyz[i] - xyz[j]) ** 2))
dists[i, j] = dist
dists[j, i] = dist
return dists
dists = distances(element_list, xyz)
print(dists)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/199/129199421.ipynb
| null | null |
[{"Id": 129199421, "ScriptId": 38406212, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13344059, "CreationDate": "05/11/2023 18:48:23", "VersionNumber": 3.0, "Title": "Che352 Final Project", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 167.0, "LinesInsertedFromPrevious": 4.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 163.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# RAW DATA SITE NEEDED TO LOAD PARAM DATA, AND CONSTRUCT THE GFN2-XTB CALCULATOR
# Sources:
# https://github.com/grimme-lab/xtb/blob/main/param_gfn2-xtb.txt
# https://github.com/grimme-lab/xtb
# https://zenodo.org/record/4486287#.ZE28g3bMKUl
# https://xtb-python.readthedocs.io/en/latest/
# https://xtb-python.readthedocs.io/_/downloads/en/stable/pdf/
# http://rdkit.org/docs/source/rdkit.Chem.rdchem.html?highlight=getatoms
# https://www.rdkit.org/docs/source/rdkit.Chem.rdForceFieldHelpers.html
# https://www.scm.com/wp-content/uploads/pdfs/2014/UFF.pdf
# https://stackoverflow.com/questions/71915443/rdkit-coordinates-for-atoms-in-a-molecule
import numpy as np
from scipy import constants
import random as rand
from rdkit import Chem
from rdkit.Chem import AllChem
from xtb.interface import Calculator, Molecule, Param
from xtb.utils import get_method
from xtb.utils import get_solvent, Solvent
from xtb.libxtb import VERBOSITY_MUTED
# Unit conversions
Hartree_to_kcal_mol = 627.5
Bohr_to_angstrom = constants.physical_constants["Bohr radius"][0] * 1.0e10
# List of reference states for each element.
ref_states = {
"H": "[H][H]",
"C": "[C]#[C]",
"N": "N#N",
"O": "O=O",
"F": "FF",
"P": "P#P",
"S": "S=S",
"Cl": "ClCl",
}
def RDK_FP(mol):
"""
Takes a molecule and returns its RDK footprint.
"""
fp = AllChem.RDKFingerprint(mol, fpSize=512)
fp_arr = np.array(list(fp), dtype=int)
return fp_arr
def aq_structure(mol, element_num_list):
"""
Optimizes the aqueous-phase structure of a molecule and returns the 3D-coordinates and energy of the structure.
Also calculates and returns the energy of the optimized structure with no solvent.
"""
AllChem.EmbedMolecule(mol, randomSeed=-1)
AllChem.UFFOptimizeMolecule(
mol
) # first performs a UFF optimziation to get an initial guess.
xyz = mol.GetConformer().GetPositions()
calc = Calculator(get_method("GFN2-xTB"), np.array(element_num_list), xyz)
calc.set_verbosity(VERBOSITY_MUTED)
calc.set_solvent(Solvent.h2o)
step_range = rand.choice([30, 35, 40, 45, 50, 55, 60])
for step_i in range(
step_range
): # Iteratively optimizing the molecule using the calculated gradients.
calc.update(xyz / Bohr_to_angstrom)
result = calc.singlepoint()
E_aq = result.get_energy() * Hartree_to_kcal_mol
Eformation_grad = result.get_gradient() / Bohr_to_angstrom
xyz -= 0.05 * Eformation_grad # gradient descent with a step size of 0.05.
if (
np.linalg.norm(Eformation_grad, 2) < 1e-5
): # stop the optimization when gradient is close to zero.
break
calc.set_solvent() # remove the solvent to get the energy of the molecule in the gas phase.
result_gas = calc.singlepoint()
E_gas = result_gas.get_energy() * Hartree_to_kcal_mol
return xyz, E_aq, E_gas
def ref_energy(element_list):
"""
Given a list of elements in a molecule, returns the total energy of the molecule based on the reference states of the elements.
The molecule's reference state energy is the sum of the energies of each of the atoms in the molecule.
"""
E_ref = 0
for element in element_list:
mol = Chem.MolFromSmiles(
ref_states[element]
) # gets the reference state diatomic molecule for the element.
atoms = mol.GetAtoms()
element_num_list = [atom.GetAtomicNum() for atom in atoms]
energy = (
aq_structure(mol, element_num_list)[1] / 2
) # for every element in the molecule, calculate the aqueous phase energy of the diatomic molecule it comes from.
E_ref += energy # the total energy of the reference state.
return E_ref
def smiles_to_properties(smiles, n_calculations=10):
"""
Given a SMILES string representing a molecule, returns the following properties of the molecule:
the 3D coordinates of the lowest energy structure of the molecule in the aqueous phase, the energy of formation, the hydration energy, and the molecular fingerprint.
By default, the function runs 10 tests for the given molecule, but that can be changed by the user. The more tests run, the longer the calculations take.
"""
# first converts the smiles string into a list of its elements and a list of the atomic numbers of the elements.
mol = Chem.MolFromSmiles(smiles)
mol = Chem.AddHs(mol)
atoms = mol.GetAtoms()
element_list = [atom.GetSymbol() for atom in atoms]
element_num_list = [atom.GetAtomicNum() for atom in atoms]
Eformation_Aq_values = (
[]
) # keeps track of the calculated formation energies of the molecule in the aqueous phase for each calculation.
hydration_energy_values = (
[]
) # # keeps track of the calculated hydration energies of the molecule for each calculation.
for i in range(n_calculations):
xyz, E_aq, E_gas = aq_structure(
mol, element_num_list
) # optimize the molecule and get its coordinates, energy in aq phase, and energy in has phase
Eref = ref_energy(element_list) # obtain the reference energy of the molecule
Eformation = E_aq - Eref
Ehydration = E_aq - E_gas
hydration_energy_values.append(Ehydration)
Eformation_Aq_values.append(Eformation)
Eformation_Aq_mean = np.round(np.mean(Eformation_Aq_values), 3)
Eformation_Aq_std = np.round(np.std(Eformation_Aq_values), 3)
hydration_energy_mean = np.round(np.mean(hydration_energy_values), 3)
hydration_energy_std = np.round(np.std(hydration_energy_values), 3)
xyz = np.round(xyz, 3)
fp = RDK_FP(mol)
return (
(element_list, xyz),
f"Formation energy vs reference: {Eformation_Aq_mean} ± {Eformation_Aq_std} kcal/mol",
f"Hydration energy: {hydration_energy_mean} ± {hydration_energy_std} kcal/mol",
fp,
)
smiles_to_properties("CC")
"""
SMILE strings of molecules tested:
'CCCCC(=O)OC' #methyl hexanoate
'CCCCO' #butan-1-ol
'C1CCC(CC1)N' #cyclohexanamine
'C1cc(cc1)OC=O' #phentyl formate
'CCC(=O)OC' #methyl propanoate
'C1CC(CC(C1)Cl)Cl' #1,3-dichlorobenzene
'COC(=O)CC#N' #methyl 2-cyanoacetate
'CCOCC' #ethoxyethane
'CCCCC[N+](=O)[O-]' #1-nitropentane
'CCCCCCCC(=O)C' #nonan-2-one
'CC1CCNCC1' 4-#methylpyridine
'CCCCS' #butane-1-thiol
'CC1CCCC(C1C)O' #2,3-dimehtylphenol
'CO' #methanol
'CS(=O)(=O)C' #methylsulfonylmethane
'C1(C(C(C1(F)F)(F)F)(F)F)(F)F' #octafluorocyclobutane
'CCCS' #propanethiol
'CC#N' #acetonitrile
'CSSC' #methyldisulfanylmethane
'C1CCC(C1)O' #cylopentanol
"""
# TESTING CODE - ATOMIC DISTANCES
import math
import numpy as np
element_list, xyz = smiles_to_properties("CC")[0]
def distances(element_list, xyz):
n = len(element_list)
dists = np.zeros((n, n))
for i in range(n):
for j in range(i, n):
dist = math.sqrt(sum((xyz[i] - xyz[j]) ** 2))
dists[i, j] = dist
dists[j, i] = dist
return dists
dists = distances(element_list, xyz)
print(dists)
| false | 0 | 2,344 | 0 | 2,344 | 2,344 |
||
129199981
|
<jupyter_start><jupyter_text>Choclate_Sales_project
The dataset provided contains information about chocolate sales made by different salespeople in various regions. It includes details such as the name of the salesperson, the geographic location of the sale, the product sold, the amount generated from the sale, the number of units sold, the cost per unit, the total cost, the profit earned, and the profit percentage.
The data shows that sales vary greatly across different regions, with the highest sales generated in Canada by Carla Molina, who sold Drinking Coco for $9,632 and earned a profit of $7,769, which is an impressive profit margin of 81%. On the other hand, Curtice Advani from the UK had a negative profit margin of -159%, indicating that the cost of selling White Choc was higher than the amount generated from sales.
The dataset also provides insights into the most popular products sold, with Choco Coated Almonds being the most popular product sold in the US
Overall, the dataset provides valuable insights into chocolate sales, highlighting the importance of factors such as product popularity, regional variations, and profit margins in determining the success of sales.
Format : CSV
Dataset Contains:
Sales Person
Geography
Product
Amount
Units
cost per unit
Cost
Profit
profit %
Kaggle dataset identifier: choclate-sales-project
<jupyter_script>import pandas as pd
import numpy as np
from numpy import random
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
df1 = pd.read_csv(
r"/kaggle/input/choclate-sales-project/choclate protfolio project - 11.csv"
)
# **Analayzing the data**
df1
pd.options.display.max_rows = 100
df1.head()
df1.tail()
df1.shape
df1["Sales Person"].describe()
df1["Profit"].describe()
df1.info()
df1.columns
df1.describe().T
df1.isnull().sum()
df1.duplicated()
# ****Cleaning*****
# To fill up the missing values
df2 = df1.fillna("")
df2.head(10)
# use the count function to find out how many difference countries are inside the (Geography) with the index top 5
df2["Geography"].value_counts()[:5]
# # we can drop all the empty Values
# Drop all Empty values
df2.dropna(inplace=True)
# Check How many empty values you have inside the dataset
df2.isnull().sum()
# Joining 2 columns
df3 = df2["Sales Person"] + " From " + df2["Geography"]
df3.head(10)
# selecting the saller by indexing
df2["Sales Person"][:2]
# # Checking the Relationships
df2.corr()
# **We can Change the colors as well**
sns.heatmap(df2.corr(), cmap="coolwarm", annot=True)
df2.pivot_table(
index="Sales Person", columns="Geography", values="Amount", aggfunc=np.sum
)
# # VESUALIZATION
df2["Geography"].value_counts().plot(kind="barh", color="hotpink").grid(axis="x")
plt.title("The Geography", color="hotpink")
plt.figure(figsize=(5, 3))
plt.show()
df2["Sales Person"].value_counts().plot(kind="barh", color="black").grid(axis="x")
plt.title("Sales Person")
plt.figure(figsize=(5, 3))
plt.show()
df2["Amount"].value_counts()[:10].plot(kind="pie", startangle=90)
plt.title("Sales Amount")
df2["Profit"].value_counts()[:15].plot(kind="barh", color="r")
plt.title("Sales Profit")
plt.show()
df2["Product"].value_counts()[:15].plot(kind="barh", color="black")
plt.title("Products")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/199/129199981.ipynb
|
choclate-sales-project
|
prajwal6362venom
|
[{"Id": 129199981, "ScriptId": 38269910, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 11228334, "CreationDate": "05/11/2023 18:55:35", "VersionNumber": 1.0, "Title": "chocolate", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 93.0, "LinesInsertedFromPrevious": 93.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 185033243, "KernelVersionId": 129199981, "SourceDatasetVersionId": 5469358}]
|
[{"Id": 5469358, "DatasetId": 3159078, "DatasourceVersionId": 5543568, "CreatorUserId": 14233780, "LicenseName": "Unknown", "CreationDate": "04/20/2023 15:19:13", "VersionNumber": 1.0, "Title": "Choclate_Sales_project", "Slug": "choclate-sales-project", "Subtitle": "Sales of types of chocolate in US", "Description": "The dataset provided contains information about chocolate sales made by different salespeople in various regions. It includes details such as the name of the salesperson, the geographic location of the sale, the product sold, the amount generated from the sale, the number of units sold, the cost per unit, the total cost, the profit earned, and the profit percentage.\n\nThe data shows that sales vary greatly across different regions, with the highest sales generated in Canada by Carla Molina, who sold Drinking Coco for $9,632 and earned a profit of $7,769, which is an impressive profit margin of 81%. On the other hand, Curtice Advani from the UK had a negative profit margin of -159%, indicating that the cost of selling White Choc was higher than the amount generated from sales.\n\nThe dataset also provides insights into the most popular products sold, with Choco Coated Almonds being the most popular product sold in the US\n\nOverall, the dataset provides valuable insights into chocolate sales, highlighting the importance of factors such as product popularity, regional variations, and profit margins in determining the success of sales.\n \nFormat : CSV\n\nDataset Contains:\nSales Person\t\nGeography\t\nProduct\t\nAmount\t\nUnits\t\ncost per unit\t\nCost\t\nProfit\t\nprofit %", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3159078, "CreatorUserId": 14233780, "OwnerUserId": 14233780.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5469358.0, "CurrentDatasourceVersionId": 5543568.0, "ForumId": 3223132, "Type": 2, "CreationDate": "04/20/2023 15:19:13", "LastActivityDate": "04/20/2023", "TotalViews": 6399, "TotalDownloads": 1383, "TotalVotes": 29, "TotalKernels": 9}]
|
[{"Id": 14233780, "UserName": "prajwal6362venom", "DisplayName": "Prajwal N", "RegisterDate": "03/19/2023", "PerformanceTier": 0}]
|
import pandas as pd
import numpy as np
from numpy import random
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
df1 = pd.read_csv(
r"/kaggle/input/choclate-sales-project/choclate protfolio project - 11.csv"
)
# **Analayzing the data**
df1
pd.options.display.max_rows = 100
df1.head()
df1.tail()
df1.shape
df1["Sales Person"].describe()
df1["Profit"].describe()
df1.info()
df1.columns
df1.describe().T
df1.isnull().sum()
df1.duplicated()
# ****Cleaning*****
# To fill up the missing values
df2 = df1.fillna("")
df2.head(10)
# use the count function to find out how many difference countries are inside the (Geography) with the index top 5
df2["Geography"].value_counts()[:5]
# # we can drop all the empty Values
# Drop all Empty values
df2.dropna(inplace=True)
# Check How many empty values you have inside the dataset
df2.isnull().sum()
# Joining 2 columns
df3 = df2["Sales Person"] + " From " + df2["Geography"]
df3.head(10)
# selecting the saller by indexing
df2["Sales Person"][:2]
# # Checking the Relationships
df2.corr()
# **We can Change the colors as well**
sns.heatmap(df2.corr(), cmap="coolwarm", annot=True)
df2.pivot_table(
index="Sales Person", columns="Geography", values="Amount", aggfunc=np.sum
)
# # VESUALIZATION
df2["Geography"].value_counts().plot(kind="barh", color="hotpink").grid(axis="x")
plt.title("The Geography", color="hotpink")
plt.figure(figsize=(5, 3))
plt.show()
df2["Sales Person"].value_counts().plot(kind="barh", color="black").grid(axis="x")
plt.title("Sales Person")
plt.figure(figsize=(5, 3))
plt.show()
df2["Amount"].value_counts()[:10].plot(kind="pie", startangle=90)
plt.title("Sales Amount")
df2["Profit"].value_counts()[:15].plot(kind="barh", color="r")
plt.title("Sales Profit")
plt.show()
df2["Product"].value_counts()[:15].plot(kind="barh", color="black")
plt.title("Products")
plt.show()
| false | 1 | 663 | 2 | 989 | 663 |
||
129199262
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Gerando Listas com números aleatórios
# Para gerar uma lista com 100 números aleatórios desordenados em Python, podemos utilizar o módulo random. O código abaixo irá gerar uma lista com números aleatórios entre 1 e 100:
import random
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
# Esse código utiliza a função randint da biblioteca random para gerar números inteiros aleatórios entre 0 e 1000, e cria uma lista com 100 desses números.
# Agora vamos ver exemplos de algoritmos de ordenação em Python:
# ## Bubble Sort
# O Bubble Sort é um dos algoritmos mais simples de ordenação. Ele percorre a lista diversas vezes, comparando elementos adjacentes e realizando a troca se necessário, até que a lista esteja completamente ordenada.
# O algoritmo funciona da seguinte maneira: o primeiro elemento é comparado com o segundo, se o primeiro for maior do que o segundo, a troca é feita. Em seguida, o segundo elemento é comparado com o terceiro, e assim por diante. Esse processo é repetido até o penúltimo elemento da lista. Depois disso, a lista é percorrida novamente, comparando o primeiro com o segundo, o segundo com o terceiro, e assim por diante, até que a lista esteja completamente ordenada.
# Esse algoritmo recebe esse nome porque os maiores elementos “sobem” para o final da lista, como se fossem bolhas de ar em um líquido. O Bubble Sort é muito fácil de entender e implementar, mas pode ser muito ineficiente para listas grandes, já que tem complexidade O(n²) no pior caso.
# O código começa definindo uma função chamada bubble_sort que recebe uma lista como argumento. Em seguida, a função usa um loop for para percorrer a lista n vezes, onde n é o tamanho da lista. Dentro desse loop for, temos outro loop for que percorre a lista da posição 0 até a posição n-i-2 (onde i é o índice do loop externo).
# Dentro desse segundo loop, temos uma estrutura if que verifica se o elemento atual é maior do que o próximo elemento. Se for, então os elementos são trocados de posição usando uma tupla. Essa tupla realiza a troca de valores entre as duas posições da lista.
# O código finaliza com a definição da lista, que é criada com 100 números aleatórios entre 0 e 1000. Em seguida, a função bubble_sort é chamada passando essa lista como argumento. Por fim, a lista é impressa na tela, agora ordenada pelo Bubble Sort.
def bubble_sort(lista):
n = len(lista)
for i in range(n):
for j in range(0, n - i - 1):
if lista[j] > lista[j + 1]:
lista[j], lista[j + 1] = lista[j + 1], lista[j]
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
bubble_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Selection Sort
# O Selection Sort é um algoritmo de ordenação que busca o menor valor em uma lista e o coloca na primeira posição, depois busca o segundo menor valor e o coloca na segunda posição, e assim por diante, até que toda a lista esteja ordenada.
# O algoritmo funciona da seguinte forma: ele percorre toda a lista e seleciona o menor elemento, em seguida, troca o elemento selecionado com o primeiro elemento não ordenado da lista. Depois, o algoritmo percorre novamente a lista, desta vez ignorando o primeiro elemento, e seleciona novamente o menor elemento, trocando-o com o segundo elemento não ordenado da lista. O algoritmo continua este processo até que a lista esteja completamente ordenada.
# A complexidade do Selection Sort é O(n^2), o que significa que seu desempenho não é tão bom quanto outros algoritmos como o Quick Sort e o Merge Sort, mas é um algoritmo simples de entender e implementar, sendo adequado para listas de tamanho pequeno ou médio.
# Exemplo de código:
def selection_sort(lista):
n = len(lista)
for i in range(n):
min_idx = i
for j in range(i + 1, n):
if lista[j] < lista[min_idx]:
min_idx = j
lista[i], lista[min_idx] = lista[min_idx], lista[i]
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
selection_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Insertion Sort
# O método de ordenação por inserção, ou insertion sort em inglês, é um algoritmo que percorre uma lista de elementos do começo ao fim, comparando cada elemento com os anteriores e trocando-os de lugar, caso estejam fora de ordem.
# O funcionamento do algoritmo é bastante simples. Ele começa percorrendo a lista a partir do segundo elemento, e comparando-o com o elemento anterior. Caso este seja maior do que o elemento anterior, ele é movido para a posição correta na lista, ou seja, para uma posição anterior.
# O processo é repetido para cada elemento subsequente, até que a lista esteja completamente ordenada. Para isso, o algoritmo mantém uma variável que representa a posição em que o elemento que está sendo analisado deverá ser inserido, chamada de chave.
# O insertion sort é considerado um algoritmo simples e eficiente, com complexidade O(n^2) no pior caso, e O(n) no melhor caso (quando a lista já está ordenada). Além disso, ele é um algoritmo in-place, ou seja, não utiliza memória extra para ordenar os elementos, o que o torna útil em situações em que o espaço de memória é limitado.
# No código apresentado, o algoritmo é implementado na função insertion_sort, que recebe uma lista como parâmetro e utiliza duas variáveis, n e i, para percorrer a lista. A variável chave é utilizada para armazenar o valor do elemento que está sendo analisado, e a variável j é utilizada para percorrer a lista da posição anterior à chave até o início da lista, comparando cada elemento com a chave e movendo-os para a posição correta na lista.
# Por fim, o código gera uma lista aleatória de 100 elementos, imprime a lista original, chama a função insertion_sort para ordenar a lista e imprime a lista ordenada.
# Exemplo de código:
def insertion_sort(lista):
n = len(lista)
for i in range(1, n):
chave = lista[i]
j = i - 1
while j >= 0 and chave < lista[j]:
lista[j + 1] = lista[j]
j -= 1
lista[j + 1] = chave
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
insertion_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Merge Sort
# O código implementa o algoritmo de ordenação Merge Sort, que é um algoritmo de ordenação recursivo e baseado em comparações. Ele divide a lista não ordenada em metades iguais, ordena cada metade separadamente e, em seguida, mescla as metades ordenadas para produzir a lista final ordenada.
# O processo começa com a verificação do comprimento da lista, se for maior que 1, o algoritmo divide a lista em duas partes, a esquerda e a direita. Em seguida, o algoritmo chama a si mesmo para ordenar cada uma das partes de forma recursiva.
# Após a chamada recursiva, as partes são mescladas em ordem crescente. Esse processo de mesclagem ocorre em três loops enquanto as duas partes ainda possuem elementos a serem comparados. No primeiro loop, os elementos de ambas as partes são comparados e o menor é colocado na posição correta na lista final. Nos outros dois loops, os elementos restantes são adicionados à lista final.
# O processo se repete recursivamente até que toda a lista esteja ordenada em ordem crescente.
# Por fim, o código imprime a lista original e a lista ordenada.
# Exemplo de código:
def merge_sort(lista):
if len(lista) > 1:
meio = len(lista) // 2
esquerda = lista[:meio]
direita = lista[meio:]
merge_sort(esquerda)
merge_sort(direita)
i = j = k = 0
while i < len(esquerda) and j < len(direita):
if esquerda[i] < direita[j]:
lista[k] = esquerda[i]
i += 1
else:
lista[k] = direita[j]
j += 1
k += 1
while i < len(esquerda):
lista[k] = esquerda[i]
i += 1
k += 1
while j < len(direita):
lista[k] = direita[j]
j += 1
k += 1
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
merge_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Heap Sort
# Heap Sort é um algoritmo de ordenação que utiliza a estrutura de dados heap para organizar os elementos em ordem crescente ou decrescente. O heap é uma árvore binária completa em que todos os seus nós são menores ou maiores que seus filhos. A raiz da árvore é o elemento que deve ser removido ou adicionado para manter a ordem.
# O Heap Sort funciona da seguinte maneira: primeiro, ele cria uma árvore heap com os elementos da lista desordenada. Em seguida, remove o elemento raiz (o maior ou menor, dependendo da ordem escolhida) e o coloca na última posição da lista. O processo é repetido com a árvore heap restante até que todos os elementos tenham sido removidos e adicionados à lista em ordem.
# A complexidade de tempo do Heap Sort é O(n log n) em todos os casos, o que significa que ele é muito eficiente em grandes conjuntos de dados.
# Exemplo de código Python que ordena uma lista aleatória usando o Heap Sort:
import random
def heapify(lista, n, i):
maior = i
esquerda = 2 * i + 1
direita = 2 * i + 2
if esquerda < n and lista[i] < lista[esquerda]:
maior = esquerda
if direita < n and lista[maior] < lista[direita]:
maior = direita
if maior != i:
lista[i], lista[maior] = lista[maior], lista[i]
heapify(lista, n, maior)
def heap_sort(lista):
n = len(lista)
for i in range(n // 2 - 1, -1, -1):
heapify(lista, n, i)
for i in range(n - 1, 0, -1):
lista[i], lista[0] = lista[0], lista[i]
heapify(lista, i, 0)
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
heap_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Shell Sort
# O Shell Sort é um algoritmo de ordenação que utiliza uma sequência de lacunas crescentes para dividir a lista em sub-listas e ordená-las usando o método de inserção. Ele foi proposto pelo matemático Donald Shell em 1959 como uma melhoria do algoritmo de inserção.
# A ideia básica por trás do Shell Sort é que, ao invés de comparar elementos adjacentes como no algoritmo de inserção, ele compara elementos que estão a uma distância fixa de lacunas chamadas de incrementos. Esses incrementos são escolhidos de forma a diminuir gradualmente o tamanho da lacuna, até que se chegue no valor 1, momento em que o algoritmo se transforma em um simples algoritmo de inserção.
# O algoritmo de Shell Sort é implementado através de um loop externo que percorre todos os incrementos possíveis. Dentro desse loop, há um loop interno que percorre a sub-lista que começa no incremento atual até o final da lista. Dentro desse loop interno, o algoritmo realiza comparações e trocas de elementos adjacentes.
# O Shell Sort pode ser implementado de várias maneiras diferentes, dependendo da sequência de incrementos escolhida. Uma das sequências mais populares é a sequência de incrementos de Knuth, que é dada por h = 1, 4, 13, 40, 121, 364, ... (3h + 1). Outra sequência comum é a sequência de Sedgewick, que é dada por h = 1, 5, 19, 41, 109, 209, 505, 929, 2161, ...
# O tempo de execução do Shell Sort depende da sequência de incrementos escolhida. Em média, o tempo de execução é O(n log n), mas em pior caso pode chegar a O(n²). Apesar disso, o Shell Sort é geralmente mais rápido do que outros algoritmos de ordenação com tempo de execução O(n²) como o Bubble Sort e o Insertion Sort.
# Exemplo de código em Python que implementa o algoritmo de Shell Sort:
def shell_sort(lista):
n = len(lista)
h = 1
while h < n // 3:
h = 3 * h + 1
while h >= 1:
for i in range(h, n):
j = i
while j >= h and lista[j] < lista[j - h]:
lista[j], lista[j - h] = lista[j - h], lista[j]
j -= h
h //= 3
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
shell_sort(lista)
print("Lista ordenada:")
print(lista)
# Neste exemplo de código, utilizamos a sequência de incrementos de Knuth e o método de inserção para ordenar as sub-listas. No primeiro loop while, definimos o valor inicial do incremento como sendo a maior potência de 3 menor do que o tamanho da lista. No segundo loop while, iteramos sobre todos os incrementos de h até o valor 1, realizando as comparações e trocas de elementos necessárias em cada sub-lista.
# ## O Quick Sort com Mediana de Três
# O Quick Sort com Mediana de Três é uma variação do algoritmo de ordenação Quick Sort que utiliza a mediana de três elementos para escolher o pivô e evitar o pior caso. O pior caso ocorre quando a lista está quase ordenada e o pivô é sempre escolhido como o primeiro ou o último elemento, fazendo com que o algoritmo tenha tempo de execução O(n²).
# A ideia por trás da Mediana de Três é escolher três elementos da lista: o primeiro, o último e o elemento do meio, e utilizar o valor do elemento mediano como pivô. Isso faz com que o pivô seja escolhido de forma mais adequada, pois é menos provável que o pivô escolhido seja o menor ou o maior elemento da lista. Além disso, a escolha do pivô baseado em três elementos em vez de apenas um também torna o algoritmo menos vulnerável a casos extremos.
# O algoritmo de Quick Sort com Mediana de Três tem tempo de execução médio O(n log n) e pior caso O(n²). O tempo de execução médio é alcançado na maioria dos casos, já que a escolha do pivô baseado em três elementos faz com que o algoritmo seja mais eficiente na maioria dos casos.
# Segue abaixo um exemplo de código em Python para o Quick Sort com Mediana de Três:
import random
def quick_sort_mediana(lista):
if len(lista) <= 1:
return lista
# escolhe a mediana de três elementos como pivô
primeiro = lista[0]
ultimo = lista[-1]
meio = lista[len(lista) // 2]
if primeiro > meio:
primeiro, meio = meio, primeiro
if primeiro > ultimo:
primeiro, ultimo = ultimo, primeiro
if meio > ultimo:
meio, ultimo = ultimo, meio
pivot = meio
# particiona a lista
esquerda = []
direita = []
iguais = []
for elemento in lista:
if elemento < pivot:
esquerda.append(elemento)
elif elemento > pivot:
direita.append(elemento)
else:
iguais.append(elemento)
# ordena as sublistas
esquerda = quick_sort_mediana(esquerda)
direita = quick_sort_mediana(direita)
# junta as sublistas
return esquerda + iguais + direita
# exemplo de uso
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
lista_ordenada = quick_sort_mediana(lista)
print("Lista ordenada:")
print(lista_ordenada)
# Nesse exemplo, a mediana de três elementos é escolhida a partir dos índices 0, len(lista)//2 e -1. Em seguida, a lista é particionada em três sub-listas: uma com elementos menores que o pivô, outra com elementos maiores que o pivô e outra com elementos iguais ao pivô. As sublistas menores e maiores são ordenadas recursivamente e, por fim, as sublistas são concatenadas para formar a lista ordenada.
# ## O Counting Sort
# O Counting Sort é um algoritmo de ordenação que utiliza uma contagem dos elementos para ordená-los. É um algoritmo eficiente para listas com um intervalo limitado de valores, pois a complexidade do algoritmo depende do tamanho desse intervalo e não do número de elementos na lista.
# O algoritmo funciona criando um vetor auxiliar com tamanho igual ao maior valor da lista, e inicializando todas as suas posições com zero. Em seguida, ele percorre a lista original e conta quantas vezes cada elemento aparece, incrementando o valor na posição correspondente do vetor auxiliar. Depois disso, o algoritmo percorre o vetor auxiliar e gera a lista ordenada, colocando cada elemento na lista o número de vezes que ele apareceu no vetor auxiliar.
# O Counting Sort tem tempo de execução O(n + k), onde n é o número de elementos na lista e k é o maior valor da lista. Isso significa que o algoritmo é muito rápido quando k é relativamente pequeno, mas pode se tornar muito lento quando k é muito grande.
# Segue abaixo um exemplo de implementação do Counting Sort em Python:
def counting_sort(lista):
max_value = max(lista)
counts = [0] * (max_value + 1)
for num in lista:
counts[num] += 1
sorted_list = []
for i in range(len(counts)):
sorted_list.extend([i] * counts[i])
return sorted_list
# Nesse exemplo, a função counting_sort recebe a lista a ser ordenada como parâmetro. A primeira linha do método determina o maior valor da lista. Em seguida, é criado um vetor counts com tamanho igual a max_value + 1 e inicializado com zeros. O algoritmo percorre a lista original, incrementando o valor na posição correspondente do vetor counts para cada elemento. Depois disso, o algoritmo percorre o vetor counts e gera a lista ordenada, colocando cada elemento na lista o número de vezes que ele apareceu no vetor counts.
# Segue abaixo um exemplo de uso do Counting Sort:
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
lista_ordenada = counting_sort(lista)
print("Lista ordenada:")
print(lista_ordenada)
# ## Radix Sort
# O Radix Sort é um algoritmo de ordenação que trabalha com a posição dos dígitos dos números na lista. Ele ordena os elementos por dígitos, começando pelo dígito menos significativo e indo até o mais significativo. Para isso, ele utiliza uma lista de baldes, cada um correspondendo a um dígito (0 a 9).
# O processo de ordenação do Radix Sort começa com a separação dos elementos em baldes de acordo com o valor do dígito menos significativo. Em seguida, os elementos são reunidos em uma nova lista, na ordem em que foram colocados nos baldes. Esse processo é repetido para cada dígito, até que todos os dígitos tenham sido considerados. Ao final, a lista estará ordenada.
# Uma vantagem do Radix Sort é que ele é capaz de ordenar números com diferentes quantidades de dígitos de maneira eficiente, pois ele não compara diretamente os números, mas sim os seus dígitos. No entanto, ele só é eficiente quando o maior valor da lista não é muito grande, pois o número de baldes a serem criados pode se tornar muito grande.
# Segue abaixo um exemplo de código em Python que utiliza o Radix Sort para ordenar uma lista aleatória de números inteiros:
def radix_sort(lista):
# Encontra o maior valor da lista para saber o número de dígitos
max_valor = max(lista)
num_digitos = len(str(max_valor))
# Itera sobre os dígitos, começando pelo menos significativo
for digito in range(num_digitos):
# Cria a lista de baldes
baldes = [[] for _ in range(10)]
# Coloca cada elemento em um balde de acordo com o dígito atual
for valor in lista:
digito_valor = (valor // (10**digito)) % 10
baldes[digito_valor].append(valor)
# Reúne os elementos dos baldes em uma nova lista
lista = [valor for balde in baldes for valor in balde]
return lista
# Exemplo de uso
import random
lista = [random.randint(0, 1000) for _ in range(100)]
print("Lista original:")
print(lista)
lista_ordenada = radix_sort(lista)
print("Lista ordenada:")
print(lista_ordenada)
# Neste exemplo, a função radix_sort recebe uma lista como parâmetro e retorna a lista ordenada pelo Radix Sort. A cada iteração do loop externo, o algoritmo ordena os elementos de acordo com um dígito diferente, começando pelo menos significativo. O loop interno coloca cada elemento em um balde de acordo com o dígito atual, e o loop externo reúne os elementos dos baldes em uma nova lista. No final das iterações, a função retorna a lista ordenada.
# ## O Bucket Sort
# O Bucket Sort é um algoritmo de ordenação que é especialmente útil para lidar com listas que possuem uma distribuição uniforme de elementos. A ideia básica do Bucket Sort é dividir a lista original em um número fixo de "baldes" ou intervalos, colocando cada elemento da lista em seu respectivo balde. Em seguida, cada balde é ordenado individualmente, e os elementos de todos os baldes são concatenados em uma única lista ordenada.
# O primeiro passo do Bucket Sort é determinar o número de baldes que serão usados. Isso pode ser feito de diversas formas, mas uma abordagem comum é escolher o número de baldes como o número de elementos na lista original. Em seguida, cada elemento da lista é colocado em seu respectivo balde. Isso pode ser feito usando uma função de mapeamento que associa cada elemento da lista a um dos baldes.
# Uma vez que todos os elementos da lista foram distribuídos entre os baldes, cada balde é ordenado individualmente. Isso pode ser feito usando qualquer algoritmo de ordenação, mas o mais comum é usar recursivamente o próprio Bucket Sort. Em seguida, os elementos de todos os baldes são concatenados em uma única lista ordenada.
# O tempo de execução do Bucket Sort depende da distribuição dos elementos na lista original. Em geral, se a distribuição dos elementos for uniforme, o Bucket Sort é muito eficiente, com tempo de execução O(n + k), onde n é o número de elementos na lista e k é o número de baldes. No entanto, se a distribuição dos elementos for muito desigual, o Bucket Sort pode ser muito ineficiente, com tempo de execução O(n²).
# Aqui está um exemplo de código em Python que implementa o Bucket Sort:
import random
def bucket_sort(lista):
# determina o número de baldes
n = len(lista)
num_baldes = n
# cria os baldes
baldes = []
for i in range(num_baldes):
baldes.append([])
# distribui os elementos entre os baldes
for i in range(n):
j = int(lista[i] * num_baldes)
baldes[j].append(lista[i])
# ordena cada balde e concatena os elementos em uma lista ordenada
lista_ordenada = []
for i in range(num_baldes):
baldes[i].sort()
lista_ordenada += baldes[i]
return lista_ordenada
# exemplo de uso
lista = [random.random() for i in range(100)]
print("Lista original:")
print(lista)
lista_ordenada = bucket_sort(lista)
print("Lista ordenada:")
print(lista_ordenada)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/199/129199262.ipynb
| null | null |
[{"Id": 129199262, "ScriptId": 38409162, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 12114345, "CreationDate": "05/11/2023 18:46:07", "VersionNumber": 1.0, "Title": "M\u00e9todos de Ordena\u00e7\u00e3o em Python", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 466.0, "LinesInsertedFromPrevious": 466.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Gerando Listas com números aleatórios
# Para gerar uma lista com 100 números aleatórios desordenados em Python, podemos utilizar o módulo random. O código abaixo irá gerar uma lista com números aleatórios entre 1 e 100:
import random
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
# Esse código utiliza a função randint da biblioteca random para gerar números inteiros aleatórios entre 0 e 1000, e cria uma lista com 100 desses números.
# Agora vamos ver exemplos de algoritmos de ordenação em Python:
# ## Bubble Sort
# O Bubble Sort é um dos algoritmos mais simples de ordenação. Ele percorre a lista diversas vezes, comparando elementos adjacentes e realizando a troca se necessário, até que a lista esteja completamente ordenada.
# O algoritmo funciona da seguinte maneira: o primeiro elemento é comparado com o segundo, se o primeiro for maior do que o segundo, a troca é feita. Em seguida, o segundo elemento é comparado com o terceiro, e assim por diante. Esse processo é repetido até o penúltimo elemento da lista. Depois disso, a lista é percorrida novamente, comparando o primeiro com o segundo, o segundo com o terceiro, e assim por diante, até que a lista esteja completamente ordenada.
# Esse algoritmo recebe esse nome porque os maiores elementos “sobem” para o final da lista, como se fossem bolhas de ar em um líquido. O Bubble Sort é muito fácil de entender e implementar, mas pode ser muito ineficiente para listas grandes, já que tem complexidade O(n²) no pior caso.
# O código começa definindo uma função chamada bubble_sort que recebe uma lista como argumento. Em seguida, a função usa um loop for para percorrer a lista n vezes, onde n é o tamanho da lista. Dentro desse loop for, temos outro loop for que percorre a lista da posição 0 até a posição n-i-2 (onde i é o índice do loop externo).
# Dentro desse segundo loop, temos uma estrutura if que verifica se o elemento atual é maior do que o próximo elemento. Se for, então os elementos são trocados de posição usando uma tupla. Essa tupla realiza a troca de valores entre as duas posições da lista.
# O código finaliza com a definição da lista, que é criada com 100 números aleatórios entre 0 e 1000. Em seguida, a função bubble_sort é chamada passando essa lista como argumento. Por fim, a lista é impressa na tela, agora ordenada pelo Bubble Sort.
def bubble_sort(lista):
n = len(lista)
for i in range(n):
for j in range(0, n - i - 1):
if lista[j] > lista[j + 1]:
lista[j], lista[j + 1] = lista[j + 1], lista[j]
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
bubble_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Selection Sort
# O Selection Sort é um algoritmo de ordenação que busca o menor valor em uma lista e o coloca na primeira posição, depois busca o segundo menor valor e o coloca na segunda posição, e assim por diante, até que toda a lista esteja ordenada.
# O algoritmo funciona da seguinte forma: ele percorre toda a lista e seleciona o menor elemento, em seguida, troca o elemento selecionado com o primeiro elemento não ordenado da lista. Depois, o algoritmo percorre novamente a lista, desta vez ignorando o primeiro elemento, e seleciona novamente o menor elemento, trocando-o com o segundo elemento não ordenado da lista. O algoritmo continua este processo até que a lista esteja completamente ordenada.
# A complexidade do Selection Sort é O(n^2), o que significa que seu desempenho não é tão bom quanto outros algoritmos como o Quick Sort e o Merge Sort, mas é um algoritmo simples de entender e implementar, sendo adequado para listas de tamanho pequeno ou médio.
# Exemplo de código:
def selection_sort(lista):
n = len(lista)
for i in range(n):
min_idx = i
for j in range(i + 1, n):
if lista[j] < lista[min_idx]:
min_idx = j
lista[i], lista[min_idx] = lista[min_idx], lista[i]
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
selection_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Insertion Sort
# O método de ordenação por inserção, ou insertion sort em inglês, é um algoritmo que percorre uma lista de elementos do começo ao fim, comparando cada elemento com os anteriores e trocando-os de lugar, caso estejam fora de ordem.
# O funcionamento do algoritmo é bastante simples. Ele começa percorrendo a lista a partir do segundo elemento, e comparando-o com o elemento anterior. Caso este seja maior do que o elemento anterior, ele é movido para a posição correta na lista, ou seja, para uma posição anterior.
# O processo é repetido para cada elemento subsequente, até que a lista esteja completamente ordenada. Para isso, o algoritmo mantém uma variável que representa a posição em que o elemento que está sendo analisado deverá ser inserido, chamada de chave.
# O insertion sort é considerado um algoritmo simples e eficiente, com complexidade O(n^2) no pior caso, e O(n) no melhor caso (quando a lista já está ordenada). Além disso, ele é um algoritmo in-place, ou seja, não utiliza memória extra para ordenar os elementos, o que o torna útil em situações em que o espaço de memória é limitado.
# No código apresentado, o algoritmo é implementado na função insertion_sort, que recebe uma lista como parâmetro e utiliza duas variáveis, n e i, para percorrer a lista. A variável chave é utilizada para armazenar o valor do elemento que está sendo analisado, e a variável j é utilizada para percorrer a lista da posição anterior à chave até o início da lista, comparando cada elemento com a chave e movendo-os para a posição correta na lista.
# Por fim, o código gera uma lista aleatória de 100 elementos, imprime a lista original, chama a função insertion_sort para ordenar a lista e imprime a lista ordenada.
# Exemplo de código:
def insertion_sort(lista):
n = len(lista)
for i in range(1, n):
chave = lista[i]
j = i - 1
while j >= 0 and chave < lista[j]:
lista[j + 1] = lista[j]
j -= 1
lista[j + 1] = chave
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
insertion_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Merge Sort
# O código implementa o algoritmo de ordenação Merge Sort, que é um algoritmo de ordenação recursivo e baseado em comparações. Ele divide a lista não ordenada em metades iguais, ordena cada metade separadamente e, em seguida, mescla as metades ordenadas para produzir a lista final ordenada.
# O processo começa com a verificação do comprimento da lista, se for maior que 1, o algoritmo divide a lista em duas partes, a esquerda e a direita. Em seguida, o algoritmo chama a si mesmo para ordenar cada uma das partes de forma recursiva.
# Após a chamada recursiva, as partes são mescladas em ordem crescente. Esse processo de mesclagem ocorre em três loops enquanto as duas partes ainda possuem elementos a serem comparados. No primeiro loop, os elementos de ambas as partes são comparados e o menor é colocado na posição correta na lista final. Nos outros dois loops, os elementos restantes são adicionados à lista final.
# O processo se repete recursivamente até que toda a lista esteja ordenada em ordem crescente.
# Por fim, o código imprime a lista original e a lista ordenada.
# Exemplo de código:
def merge_sort(lista):
if len(lista) > 1:
meio = len(lista) // 2
esquerda = lista[:meio]
direita = lista[meio:]
merge_sort(esquerda)
merge_sort(direita)
i = j = k = 0
while i < len(esquerda) and j < len(direita):
if esquerda[i] < direita[j]:
lista[k] = esquerda[i]
i += 1
else:
lista[k] = direita[j]
j += 1
k += 1
while i < len(esquerda):
lista[k] = esquerda[i]
i += 1
k += 1
while j < len(direita):
lista[k] = direita[j]
j += 1
k += 1
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
merge_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Heap Sort
# Heap Sort é um algoritmo de ordenação que utiliza a estrutura de dados heap para organizar os elementos em ordem crescente ou decrescente. O heap é uma árvore binária completa em que todos os seus nós são menores ou maiores que seus filhos. A raiz da árvore é o elemento que deve ser removido ou adicionado para manter a ordem.
# O Heap Sort funciona da seguinte maneira: primeiro, ele cria uma árvore heap com os elementos da lista desordenada. Em seguida, remove o elemento raiz (o maior ou menor, dependendo da ordem escolhida) e o coloca na última posição da lista. O processo é repetido com a árvore heap restante até que todos os elementos tenham sido removidos e adicionados à lista em ordem.
# A complexidade de tempo do Heap Sort é O(n log n) em todos os casos, o que significa que ele é muito eficiente em grandes conjuntos de dados.
# Exemplo de código Python que ordena uma lista aleatória usando o Heap Sort:
import random
def heapify(lista, n, i):
maior = i
esquerda = 2 * i + 1
direita = 2 * i + 2
if esquerda < n and lista[i] < lista[esquerda]:
maior = esquerda
if direita < n and lista[maior] < lista[direita]:
maior = direita
if maior != i:
lista[i], lista[maior] = lista[maior], lista[i]
heapify(lista, n, maior)
def heap_sort(lista):
n = len(lista)
for i in range(n // 2 - 1, -1, -1):
heapify(lista, n, i)
for i in range(n - 1, 0, -1):
lista[i], lista[0] = lista[0], lista[i]
heapify(lista, i, 0)
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
heap_sort(lista)
print("Lista ordenada:")
print(lista)
# ## Shell Sort
# O Shell Sort é um algoritmo de ordenação que utiliza uma sequência de lacunas crescentes para dividir a lista em sub-listas e ordená-las usando o método de inserção. Ele foi proposto pelo matemático Donald Shell em 1959 como uma melhoria do algoritmo de inserção.
# A ideia básica por trás do Shell Sort é que, ao invés de comparar elementos adjacentes como no algoritmo de inserção, ele compara elementos que estão a uma distância fixa de lacunas chamadas de incrementos. Esses incrementos são escolhidos de forma a diminuir gradualmente o tamanho da lacuna, até que se chegue no valor 1, momento em que o algoritmo se transforma em um simples algoritmo de inserção.
# O algoritmo de Shell Sort é implementado através de um loop externo que percorre todos os incrementos possíveis. Dentro desse loop, há um loop interno que percorre a sub-lista que começa no incremento atual até o final da lista. Dentro desse loop interno, o algoritmo realiza comparações e trocas de elementos adjacentes.
# O Shell Sort pode ser implementado de várias maneiras diferentes, dependendo da sequência de incrementos escolhida. Uma das sequências mais populares é a sequência de incrementos de Knuth, que é dada por h = 1, 4, 13, 40, 121, 364, ... (3h + 1). Outra sequência comum é a sequência de Sedgewick, que é dada por h = 1, 5, 19, 41, 109, 209, 505, 929, 2161, ...
# O tempo de execução do Shell Sort depende da sequência de incrementos escolhida. Em média, o tempo de execução é O(n log n), mas em pior caso pode chegar a O(n²). Apesar disso, o Shell Sort é geralmente mais rápido do que outros algoritmos de ordenação com tempo de execução O(n²) como o Bubble Sort e o Insertion Sort.
# Exemplo de código em Python que implementa o algoritmo de Shell Sort:
def shell_sort(lista):
n = len(lista)
h = 1
while h < n // 3:
h = 3 * h + 1
while h >= 1:
for i in range(h, n):
j = i
while j >= h and lista[j] < lista[j - h]:
lista[j], lista[j - h] = lista[j - h], lista[j]
j -= h
h //= 3
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
shell_sort(lista)
print("Lista ordenada:")
print(lista)
# Neste exemplo de código, utilizamos a sequência de incrementos de Knuth e o método de inserção para ordenar as sub-listas. No primeiro loop while, definimos o valor inicial do incremento como sendo a maior potência de 3 menor do que o tamanho da lista. No segundo loop while, iteramos sobre todos os incrementos de h até o valor 1, realizando as comparações e trocas de elementos necessárias em cada sub-lista.
# ## O Quick Sort com Mediana de Três
# O Quick Sort com Mediana de Três é uma variação do algoritmo de ordenação Quick Sort que utiliza a mediana de três elementos para escolher o pivô e evitar o pior caso. O pior caso ocorre quando a lista está quase ordenada e o pivô é sempre escolhido como o primeiro ou o último elemento, fazendo com que o algoritmo tenha tempo de execução O(n²).
# A ideia por trás da Mediana de Três é escolher três elementos da lista: o primeiro, o último e o elemento do meio, e utilizar o valor do elemento mediano como pivô. Isso faz com que o pivô seja escolhido de forma mais adequada, pois é menos provável que o pivô escolhido seja o menor ou o maior elemento da lista. Além disso, a escolha do pivô baseado em três elementos em vez de apenas um também torna o algoritmo menos vulnerável a casos extremos.
# O algoritmo de Quick Sort com Mediana de Três tem tempo de execução médio O(n log n) e pior caso O(n²). O tempo de execução médio é alcançado na maioria dos casos, já que a escolha do pivô baseado em três elementos faz com que o algoritmo seja mais eficiente na maioria dos casos.
# Segue abaixo um exemplo de código em Python para o Quick Sort com Mediana de Três:
import random
def quick_sort_mediana(lista):
if len(lista) <= 1:
return lista
# escolhe a mediana de três elementos como pivô
primeiro = lista[0]
ultimo = lista[-1]
meio = lista[len(lista) // 2]
if primeiro > meio:
primeiro, meio = meio, primeiro
if primeiro > ultimo:
primeiro, ultimo = ultimo, primeiro
if meio > ultimo:
meio, ultimo = ultimo, meio
pivot = meio
# particiona a lista
esquerda = []
direita = []
iguais = []
for elemento in lista:
if elemento < pivot:
esquerda.append(elemento)
elif elemento > pivot:
direita.append(elemento)
else:
iguais.append(elemento)
# ordena as sublistas
esquerda = quick_sort_mediana(esquerda)
direita = quick_sort_mediana(direita)
# junta as sublistas
return esquerda + iguais + direita
# exemplo de uso
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
lista_ordenada = quick_sort_mediana(lista)
print("Lista ordenada:")
print(lista_ordenada)
# Nesse exemplo, a mediana de três elementos é escolhida a partir dos índices 0, len(lista)//2 e -1. Em seguida, a lista é particionada em três sub-listas: uma com elementos menores que o pivô, outra com elementos maiores que o pivô e outra com elementos iguais ao pivô. As sublistas menores e maiores são ordenadas recursivamente e, por fim, as sublistas são concatenadas para formar a lista ordenada.
# ## O Counting Sort
# O Counting Sort é um algoritmo de ordenação que utiliza uma contagem dos elementos para ordená-los. É um algoritmo eficiente para listas com um intervalo limitado de valores, pois a complexidade do algoritmo depende do tamanho desse intervalo e não do número de elementos na lista.
# O algoritmo funciona criando um vetor auxiliar com tamanho igual ao maior valor da lista, e inicializando todas as suas posições com zero. Em seguida, ele percorre a lista original e conta quantas vezes cada elemento aparece, incrementando o valor na posição correspondente do vetor auxiliar. Depois disso, o algoritmo percorre o vetor auxiliar e gera a lista ordenada, colocando cada elemento na lista o número de vezes que ele apareceu no vetor auxiliar.
# O Counting Sort tem tempo de execução O(n + k), onde n é o número de elementos na lista e k é o maior valor da lista. Isso significa que o algoritmo é muito rápido quando k é relativamente pequeno, mas pode se tornar muito lento quando k é muito grande.
# Segue abaixo um exemplo de implementação do Counting Sort em Python:
def counting_sort(lista):
max_value = max(lista)
counts = [0] * (max_value + 1)
for num in lista:
counts[num] += 1
sorted_list = []
for i in range(len(counts)):
sorted_list.extend([i] * counts[i])
return sorted_list
# Nesse exemplo, a função counting_sort recebe a lista a ser ordenada como parâmetro. A primeira linha do método determina o maior valor da lista. Em seguida, é criado um vetor counts com tamanho igual a max_value + 1 e inicializado com zeros. O algoritmo percorre a lista original, incrementando o valor na posição correspondente do vetor counts para cada elemento. Depois disso, o algoritmo percorre o vetor counts e gera a lista ordenada, colocando cada elemento na lista o número de vezes que ele apareceu no vetor counts.
# Segue abaixo um exemplo de uso do Counting Sort:
lista = [random.randint(0, 1000) for i in range(100)]
print("Lista original:")
print(lista)
lista_ordenada = counting_sort(lista)
print("Lista ordenada:")
print(lista_ordenada)
# ## Radix Sort
# O Radix Sort é um algoritmo de ordenação que trabalha com a posição dos dígitos dos números na lista. Ele ordena os elementos por dígitos, começando pelo dígito menos significativo e indo até o mais significativo. Para isso, ele utiliza uma lista de baldes, cada um correspondendo a um dígito (0 a 9).
# O processo de ordenação do Radix Sort começa com a separação dos elementos em baldes de acordo com o valor do dígito menos significativo. Em seguida, os elementos são reunidos em uma nova lista, na ordem em que foram colocados nos baldes. Esse processo é repetido para cada dígito, até que todos os dígitos tenham sido considerados. Ao final, a lista estará ordenada.
# Uma vantagem do Radix Sort é que ele é capaz de ordenar números com diferentes quantidades de dígitos de maneira eficiente, pois ele não compara diretamente os números, mas sim os seus dígitos. No entanto, ele só é eficiente quando o maior valor da lista não é muito grande, pois o número de baldes a serem criados pode se tornar muito grande.
# Segue abaixo um exemplo de código em Python que utiliza o Radix Sort para ordenar uma lista aleatória de números inteiros:
def radix_sort(lista):
# Encontra o maior valor da lista para saber o número de dígitos
max_valor = max(lista)
num_digitos = len(str(max_valor))
# Itera sobre os dígitos, começando pelo menos significativo
for digito in range(num_digitos):
# Cria a lista de baldes
baldes = [[] for _ in range(10)]
# Coloca cada elemento em um balde de acordo com o dígito atual
for valor in lista:
digito_valor = (valor // (10**digito)) % 10
baldes[digito_valor].append(valor)
# Reúne os elementos dos baldes em uma nova lista
lista = [valor for balde in baldes for valor in balde]
return lista
# Exemplo de uso
import random
lista = [random.randint(0, 1000) for _ in range(100)]
print("Lista original:")
print(lista)
lista_ordenada = radix_sort(lista)
print("Lista ordenada:")
print(lista_ordenada)
# Neste exemplo, a função radix_sort recebe uma lista como parâmetro e retorna a lista ordenada pelo Radix Sort. A cada iteração do loop externo, o algoritmo ordena os elementos de acordo com um dígito diferente, começando pelo menos significativo. O loop interno coloca cada elemento em um balde de acordo com o dígito atual, e o loop externo reúne os elementos dos baldes em uma nova lista. No final das iterações, a função retorna a lista ordenada.
# ## O Bucket Sort
# O Bucket Sort é um algoritmo de ordenação que é especialmente útil para lidar com listas que possuem uma distribuição uniforme de elementos. A ideia básica do Bucket Sort é dividir a lista original em um número fixo de "baldes" ou intervalos, colocando cada elemento da lista em seu respectivo balde. Em seguida, cada balde é ordenado individualmente, e os elementos de todos os baldes são concatenados em uma única lista ordenada.
# O primeiro passo do Bucket Sort é determinar o número de baldes que serão usados. Isso pode ser feito de diversas formas, mas uma abordagem comum é escolher o número de baldes como o número de elementos na lista original. Em seguida, cada elemento da lista é colocado em seu respectivo balde. Isso pode ser feito usando uma função de mapeamento que associa cada elemento da lista a um dos baldes.
# Uma vez que todos os elementos da lista foram distribuídos entre os baldes, cada balde é ordenado individualmente. Isso pode ser feito usando qualquer algoritmo de ordenação, mas o mais comum é usar recursivamente o próprio Bucket Sort. Em seguida, os elementos de todos os baldes são concatenados em uma única lista ordenada.
# O tempo de execução do Bucket Sort depende da distribuição dos elementos na lista original. Em geral, se a distribuição dos elementos for uniforme, o Bucket Sort é muito eficiente, com tempo de execução O(n + k), onde n é o número de elementos na lista e k é o número de baldes. No entanto, se a distribuição dos elementos for muito desigual, o Bucket Sort pode ser muito ineficiente, com tempo de execução O(n²).
# Aqui está um exemplo de código em Python que implementa o Bucket Sort:
import random
def bucket_sort(lista):
# determina o número de baldes
n = len(lista)
num_baldes = n
# cria os baldes
baldes = []
for i in range(num_baldes):
baldes.append([])
# distribui os elementos entre os baldes
for i in range(n):
j = int(lista[i] * num_baldes)
baldes[j].append(lista[i])
# ordena cada balde e concatena os elementos em uma lista ordenada
lista_ordenada = []
for i in range(num_baldes):
baldes[i].sort()
lista_ordenada += baldes[i]
return lista_ordenada
# exemplo de uso
lista = [random.random() for i in range(100)]
print("Lista original:")
print(lista)
lista_ordenada = bucket_sort(lista)
print("Lista ordenada:")
print(lista_ordenada)
| false | 0 | 6,986 | 0 | 6,986 | 6,986 |
||
129007065
|
<jupyter_start><jupyter_text>GoogleNews-vectors-negative300 ( word2vec )
## word2vec
This repository hosts the word2vec pre-trained Google News corpus (3 billion running words) word vector model (3 million 300-dimension English word vectors).
Kaggle dataset identifier: googlenewsvectors
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.neighbors import NearestNeighbors
from gensim.models import KeyedVectors
import numpy as np
import ast
import pandas as pd
# Veri setini yükleme
data_path = (
"/kaggle/input/2-recommended-reads-conversion-of-data-to-num/vectorizedData.csv"
)
data = pd.read_csv(data_path)
data = data.drop_duplicates(subset=["booktitle", "authorname"], keep="first")
data["word2vec"] = data["word2vec"].apply(lambda x: x.strip("[]"))
data["word2vec"] = data["word2vec"].apply(lambda x: x.split())
data["word2vec"] = data["word2vec"].apply(lambda x: [float(y) for y in x])
# Google News Binary modelinin yolu
model_path = "/kaggle/input/googlenewsvectors/GoogleNews-vectors-negative300.bin"
# Google News Binary modelini yükleme
word_vectors = KeyedVectors.load_word2vec_format(model_path, binary=True)
# Kullanıcının girdiği kitap betimlemesi
user_description = "a space adventure with friends"
user_vector = np.mean(
[
word_vectors[word]
for word in user_description.split()
if word in word_vectors.key_to_index
],
axis=0,
)
nn_model = NearestNeighbors(n_neighbors=10, metric="cosine")
X = np.array(data["word2vec"].tolist())
nn_model.fit(X)
distances, indices = nn_model.kneighbors([user_vector])
recommended_books = data.iloc[indices[0]][["booktitle", "authorname"]].values.tolist()
# Yakınlık oranlarını hesaplama ve yüzde olarak gösterme
print("Kullanıcının girdiği kitaba benzer kitaplar:")
for i, book in enumerate(recommended_books):
similarity = 1 - distances[0][i]
similarity_percent = round(similarity * 100, 2)
print(f"{book[0]} by {book[1]} ({similarity_percent}%)")
import matplotlib.pyplot as plt
books = [book[0] for book in recommended_books]
similarity_percents = [round((1 - distance) * 100, 2) for distance in distances[0]]
fig, ax = plt.subplots(figsize=(8, 6))
ax.barh(books, similarity_percents, align="center", color="skyblue")
ax.set_xlabel("Benzerlik Oranı (%)")
ax.set_title("Kullanıcının Girdiği Kitaba Benzer Kitaplar")
plt.subplots_adjust(left=0.3)
for i, v in enumerate(similarity_percents):
ax.text(v + 2, i - 0.15, str(v), color="blue", fontweight="bold")
plt.gca().invert_yaxis()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/007/129007065.ipynb
|
googlenewsvectors
|
adarshsng
|
[{"Id": 129007065, "ScriptId": 38342277, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4842847, "CreationDate": "05/10/2023 09:11:34", "VersionNumber": 1.0, "Title": "#5 recommended reads - KNN", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 77.0, "LinesInsertedFromPrevious": 77.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184684386, "KernelVersionId": 129007065, "SourceDatasetVersionId": 2307650}]
|
[{"Id": 2307650, "DatasetId": 1391881, "DatasourceVersionId": 2349008, "CreatorUserId": 3974712, "LicenseName": "Unknown", "CreationDate": "06/06/2021 09:26:00", "VersionNumber": 1.0, "Title": "GoogleNews-vectors-negative300 ( word2vec )", "Slug": "googlenewsvectors", "Subtitle": "This repository hosts the word2vec pre-trained Google News corpus - word2vec", "Description": "## word2vec\n\nThis repository hosts the word2vec pre-trained Google News corpus (3 billion running words) word vector model (3 million 300-dimension English word vectors).", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1391881, "CreatorUserId": 3974712, "OwnerUserId": 3974712.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2307650.0, "CurrentDatasourceVersionId": 2349008.0, "ForumId": 1411117, "Type": 2, "CreationDate": "06/06/2021 09:26:00", "LastActivityDate": "06/06/2021", "TotalViews": 3273, "TotalDownloads": 216, "TotalVotes": 16, "TotalKernels": 12}]
|
[{"Id": 3974712, "UserName": "adarshsng", "DisplayName": "KA-KA-shi", "RegisterDate": "11/04/2019", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.neighbors import NearestNeighbors
from gensim.models import KeyedVectors
import numpy as np
import ast
import pandas as pd
# Veri setini yükleme
data_path = (
"/kaggle/input/2-recommended-reads-conversion-of-data-to-num/vectorizedData.csv"
)
data = pd.read_csv(data_path)
data = data.drop_duplicates(subset=["booktitle", "authorname"], keep="first")
data["word2vec"] = data["word2vec"].apply(lambda x: x.strip("[]"))
data["word2vec"] = data["word2vec"].apply(lambda x: x.split())
data["word2vec"] = data["word2vec"].apply(lambda x: [float(y) for y in x])
# Google News Binary modelinin yolu
model_path = "/kaggle/input/googlenewsvectors/GoogleNews-vectors-negative300.bin"
# Google News Binary modelini yükleme
word_vectors = KeyedVectors.load_word2vec_format(model_path, binary=True)
# Kullanıcının girdiği kitap betimlemesi
user_description = "a space adventure with friends"
user_vector = np.mean(
[
word_vectors[word]
for word in user_description.split()
if word in word_vectors.key_to_index
],
axis=0,
)
nn_model = NearestNeighbors(n_neighbors=10, metric="cosine")
X = np.array(data["word2vec"].tolist())
nn_model.fit(X)
distances, indices = nn_model.kneighbors([user_vector])
recommended_books = data.iloc[indices[0]][["booktitle", "authorname"]].values.tolist()
# Yakınlık oranlarını hesaplama ve yüzde olarak gösterme
print("Kullanıcının girdiği kitaba benzer kitaplar:")
for i, book in enumerate(recommended_books):
similarity = 1 - distances[0][i]
similarity_percent = round(similarity * 100, 2)
print(f"{book[0]} by {book[1]} ({similarity_percent}%)")
import matplotlib.pyplot as plt
books = [book[0] for book in recommended_books]
similarity_percents = [round((1 - distance) * 100, 2) for distance in distances[0]]
fig, ax = plt.subplots(figsize=(8, 6))
ax.barh(books, similarity_percents, align="center", color="skyblue")
ax.set_xlabel("Benzerlik Oranı (%)")
ax.set_title("Kullanıcının Girdiği Kitaba Benzer Kitaplar")
plt.subplots_adjust(left=0.3)
for i, v in enumerate(similarity_percents):
ax.text(v + 2, i - 0.15, str(v), color="blue", fontweight="bold")
plt.gca().invert_yaxis()
plt.show()
| false | 0 | 913 | 0 | 990 | 913 |
||
129007116
|
<jupyter_start><jupyter_text>Salary Dataset - Simple linear regression
## Dataset Description
Salary Dataset in CSV for Simple linear regression. It has also been used in Machine Learning A to Z course of my series.
## Columns
- #
- YearsExperience
- Salary
Kaggle dataset identifier: salary-dataset-simple-linear-regression
<jupyter_code>import pandas as pd
df = pd.read_csv('salary-dataset-simple-linear-regression/Salary_dataset.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 30 entries, 0 to 29
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 30 non-null int64
1 YearsExperience 30 non-null float64
2 Salary 30 non-null float64
dtypes: float64(2), int64(1)
memory usage: 848.0 bytes
<jupyter_text>Examples:
{
"Unnamed: 0": 0.0,
"YearsExperience": 1.2,
"Salary": 39344.0
}
{
"Unnamed: 0": 1.0,
"YearsExperience": 1.4,
"Salary": 46206.0
}
{
"Unnamed: 0": 2.0,
"YearsExperience": 1.6,
"Salary": 37732.0
}
{
"Unnamed: 0": 3.0,
"YearsExperience": 2.1,
"Salary": 43526.0
}
<jupyter_script># import Libraries
# !pip install matplotlib
# need if you haven't install matplotlib
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import math
# # Problem Understanding
# **Hi,I learning ML from coursrea**
# **Nowdays,people want to know if they have how many experience , how many they can be paided**
# **let's find out with this data set**
# **In here, i will use simple linear Regression which one of the easiest algorithm**
# **it fit the data with stright line.**
# 
# # Data Understanding
# load data
data = pd.read_csv(
"/kaggle/input/salary-dataset-simple-linear-regression/Salary_dataset.csv"
)
data.head() # show the head of data
data.tail() # show the tail of data
# **Checking the data**
# There are alot of method to check data :
# data.info() #show basic informations of data
# data.shape() #show colums and rows of data
# data.describe() # show mini and max value and other value of data in each feature
#
print(data.info()) # Basic informations of data
# By seeing it there are 3 colums and Non-Null value
# **"non-null" usually refers to a value that is not null or missing. A null value represents the absence of a value or a missing value, and it can cause errors in data analysis or processing. Therefore, it is important to identify and handle null values appropriately in data. A non-null value is a valid value that can be used in data analysis or processing.**
# **shape**
print("data shape :", data.shape)
# From this we can see there are 3 colums and 30 rows but we only need two colums we don't need unnamed.
# for your model,
# YearExperience will be x .
# Salary will be y .
# before splitting the data, we need to check NaN values and duplicate values that can distrube your model.
# **checking null value**
# checking null value
data.isnull().sum()
data.isnull().sum().to_frame().rename(columns={0: "Total No. of Missing Values"})
# more code but more easier to understand
# if there is any NaN value use this code
# data.dropna(inplace= True)
# 1. data.isnull() returns a DataFrame with the same shape as the original data DataFrame, but with boolean values. Each element in the returned DataFrame is True if the corresponding element in the original DataFrame is NaN, and False otherwise.
# 2. .sum() method is then applied to the resulting DataFrame to count the number of True values in each column. Since True is treated as 1 and False is treated as 0 in Python, the sum of all True values in each column gives us the total number of NaN values in that column.
# 3. .to_frame() method is applied to convert the resulting Series object to a DataFrame.
# 4. .rename(columns={0:"Total No. of Missing Values"}) method is applied to rename the column in the resulting DataFrame to "Total No. of Missing Values".
# **checking Duplicate values**
# checking Duplicate values
print("Duplicate Values =", data.duplicated().sum())
# duplicated value is repeated values in the data. It can effect the model.
# **Now it's time to split the data into *feature(x)* and *target(y)***.
# **We use x to predict y**
# **in real world, there are *x_train,y_train* to fit the model and *x_test,y_tes*t to test the model after fitting**
# # Split data into features (x) and target (y)
x = data.YearsExperience
y = data.Salary
print("x_train data is")
x
print("y_train data is")
y
# **visualization data plot**
# visualization data is easier to understand and give more informations about data
# checking data with vis
plt.title("Salary Data")
plt.scatter(x, y, marker="x", c="r")
plt.xlabel("Years of Experience")
plt.ylabel("Salary (per year)")
plt.show()
w = 9909
b = 21641.1797
print("w :", w)
print("b :", b)
# Try other value of w and b
# # **Building linear regression function for model**
# we have 30 x, so we will predict 30 y. to make it easier let's use for loop
def linearRegression(x, w, b):
pre_y = []
m = len(x)
for i in range(m):
f_wb = w * x[i] + b
pre_y.append(f_wb)
return pre_y
y_pred = linearRegression(x, w, b)
# Plot our model prediction
plt.plot(x, y_pred, c="b", label="Our Prediction")
# Plot the data points
plt.scatter(x, y, marker="x", c="r", label="Actual Values")
# Set the title
plt.title("Housing Prices")
# Set the y-axis label
plt.ylabel("Price (in 1000s of dollars)")
# Set the x-axis label
plt.xlabel("Size (1000 sqft)")
# to show box for label
plt.legend()
plt.show()
# # **Squared Error Cost Function**
# **Now let see how much our model great, by seeing difference between y_pred and y (which is called error)**
# **There are still a lot of cost functions!**
# **In here, I will use Squared error cost function!**
def SquaredErrorCost(y_pred, y):
totalCost = 0
m = len(y_pred)
for i in range(m):
cost = (y_pred[i] - y[i]) ** 2
totalCost += cost
totalCost /= 2 * m
return totalCost
# Function to calculate the cost for gradient descent
# instand of y_pred, it use w , b
def compute_cost(x, y, w, b):
m = x.shape[0]
cost = 0
for i in range(m):
f_wb = w * x[i] + b
cost = cost + (f_wb - y[i]) ** 2
total_cost = 1 / (2 * m) * cost
return total_cost
# assuming y, y_pred and cost are already defined
data = {"y": y, "y_pred": y_pred, "error": abs(y_pred - y)}
df = pd.DataFrame(data)
# print the data frame
print(df)
cost = SquaredErrorCost(y_pred, y)
print(" Squared Error Cost :", cost)
print(f" Squared Error Cost : {cost:10}")
print("Squared Error Cost: {:.5e}".format(cost))
# # **Grandient Descent**
# **The algorithm that can find best w,b if you use it correctly**
def compute_gradient(x, y, w, b): # to compute derivate
m = len(x)
dj_dw = w
dj_db = b
for i in range(m):
f_wb = w * x[i] + b
dj_dw_i = (f_wb - y[i]) * x[i]
dj_db_i = f_wb - y[i]
dj_dw += dj_dw_i
dj_db += dj_db_i
dj_dw /= m
dj_db /= m
return dj_dw, dj_db
def gradient_descent(
x, y, w_in, b_in, alpha, num_iters, cost_function, gradient_function
):
"""
Performs gradient descent to fit w,b. Updates w,b by taking
num_iters gradient steps with learning rate alpha
Args:
x (ndarray (m,)) : Data, m examples
y (ndarray (m,)) : target values
w_in,b_in (scalar): initial values of model parameters
alpha (float): Learning rate
num_iters (int): number of iterations to run gradient descent
cost_function: function to call to produce cost
gradient_function: function to call to produce gradient
Returns:
w (scalar): Updated value of parameter after running gradient descent
b (scalar): Updated value of parameter after running gradient descent
J_history (List): History of cost values
p_history (list): History of parameters [w,b]
"""
# An array to store cost J and w's at each iteration primarily for graphing later
J_history = []
p_history = []
b = b_in
w = w_in
for i in range(num_iters):
# Calculate the gradient and update the parameters using gradient_function
dj_dw, dj_db = gradient_function(x, y, w, b)
# Update Parameters using equation (3) above
b = b - alpha * dj_db
w = w - alpha * dj_dw
# Save cost J at each iteration
if i < 100000: # prevent resource exhaustion
J_history.append(cost_function(x, y, w, b))
p_history.append([w, b])
# Print cost every at intervals 10 times or as many iterations if < 10
if i % math.ceil(num_iters / 10) == 0:
print(
f"Iteration {i:4}: Cost {J_history[-1]:0.2e} ",
f"dj_dw: {dj_dw: 0.3e}, dj_db: {dj_db: 0.3e} ",
f"w: {w: 0.3e}, b:{b: 0.5e}",
)
return w, b, J_history, p_history # return w and J,w history for graphing
# Now let's check our gradient descent.
# w_init and b_init will be 0 , you know according above 0 is pretty bad.
# let's use gradient descent to find the best values of w and b
# initialize parameters
w_init = 0
b_init = 0
# some gradient descent settings
iterations = 10000
tmp_alpha = 1.0e-2
# run gradient descent
w_final, b_final, J_hist, p_hist = gradient_descent(
x, y, w_init, b_init, tmp_alpha, iterations, compute_cost, compute_gradient
)
print(f"(w,b) found by gradient descent: ({w_final:8.4f},{b_final:8.4f})")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/007/129007116.ipynb
|
salary-dataset-simple-linear-regression
|
abhishek14398
|
[{"Id": 129007116, "ScriptId": 37398256, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 14091700, "CreationDate": "05/10/2023 09:12:02", "VersionNumber": 3.0, "Title": "simple linear regression", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 300.0, "LinesInsertedFromPrevious": 138.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 162.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 184684459, "KernelVersionId": 129007116, "SourceDatasetVersionId": 4832081}]
|
[{"Id": 4832081, "DatasetId": 2799910, "DatasourceVersionId": 4895851, "CreatorUserId": 3259703, "LicenseName": "CC0: Public Domain", "CreationDate": "01/10/2023 03:55:40", "VersionNumber": 1.0, "Title": "Salary Dataset - Simple linear regression", "Slug": "salary-dataset-simple-linear-regression", "Subtitle": "Simple Linear Regression Dataset, used in Machine Learning A - Z", "Description": "## Dataset Description\nSalary Dataset in CSV for Simple linear regression. It has also been used in Machine Learning A to Z course of my series.\n\n## Columns\n- #\n- YearsExperience\n- Salary", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2799910, "CreatorUserId": 3259703, "OwnerUserId": 3259703.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 4832081.0, "CurrentDatasourceVersionId": 4895851.0, "ForumId": 2834222, "Type": 2, "CreationDate": "01/10/2023 03:55:40", "LastActivityDate": "01/10/2023", "TotalViews": 65295, "TotalDownloads": 13051, "TotalVotes": 139, "TotalKernels": 93}]
|
[{"Id": 3259703, "UserName": "abhishek14398", "DisplayName": "Allena Venkata Sai Aby", "RegisterDate": "05/22/2019", "PerformanceTier": 2}]
|
# import Libraries
# !pip install matplotlib
# need if you haven't install matplotlib
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import math
# # Problem Understanding
# **Hi,I learning ML from coursrea**
# **Nowdays,people want to know if they have how many experience , how many they can be paided**
# **let's find out with this data set**
# **In here, i will use simple linear Regression which one of the easiest algorithm**
# **it fit the data with stright line.**
# 
# # Data Understanding
# load data
data = pd.read_csv(
"/kaggle/input/salary-dataset-simple-linear-regression/Salary_dataset.csv"
)
data.head() # show the head of data
data.tail() # show the tail of data
# **Checking the data**
# There are alot of method to check data :
# data.info() #show basic informations of data
# data.shape() #show colums and rows of data
# data.describe() # show mini and max value and other value of data in each feature
#
print(data.info()) # Basic informations of data
# By seeing it there are 3 colums and Non-Null value
# **"non-null" usually refers to a value that is not null or missing. A null value represents the absence of a value or a missing value, and it can cause errors in data analysis or processing. Therefore, it is important to identify and handle null values appropriately in data. A non-null value is a valid value that can be used in data analysis or processing.**
# **shape**
print("data shape :", data.shape)
# From this we can see there are 3 colums and 30 rows but we only need two colums we don't need unnamed.
# for your model,
# YearExperience will be x .
# Salary will be y .
# before splitting the data, we need to check NaN values and duplicate values that can distrube your model.
# **checking null value**
# checking null value
data.isnull().sum()
data.isnull().sum().to_frame().rename(columns={0: "Total No. of Missing Values"})
# more code but more easier to understand
# if there is any NaN value use this code
# data.dropna(inplace= True)
# 1. data.isnull() returns a DataFrame with the same shape as the original data DataFrame, but with boolean values. Each element in the returned DataFrame is True if the corresponding element in the original DataFrame is NaN, and False otherwise.
# 2. .sum() method is then applied to the resulting DataFrame to count the number of True values in each column. Since True is treated as 1 and False is treated as 0 in Python, the sum of all True values in each column gives us the total number of NaN values in that column.
# 3. .to_frame() method is applied to convert the resulting Series object to a DataFrame.
# 4. .rename(columns={0:"Total No. of Missing Values"}) method is applied to rename the column in the resulting DataFrame to "Total No. of Missing Values".
# **checking Duplicate values**
# checking Duplicate values
print("Duplicate Values =", data.duplicated().sum())
# duplicated value is repeated values in the data. It can effect the model.
# **Now it's time to split the data into *feature(x)* and *target(y)***.
# **We use x to predict y**
# **in real world, there are *x_train,y_train* to fit the model and *x_test,y_tes*t to test the model after fitting**
# # Split data into features (x) and target (y)
x = data.YearsExperience
y = data.Salary
print("x_train data is")
x
print("y_train data is")
y
# **visualization data plot**
# visualization data is easier to understand and give more informations about data
# checking data with vis
plt.title("Salary Data")
plt.scatter(x, y, marker="x", c="r")
plt.xlabel("Years of Experience")
plt.ylabel("Salary (per year)")
plt.show()
w = 9909
b = 21641.1797
print("w :", w)
print("b :", b)
# Try other value of w and b
# # **Building linear regression function for model**
# we have 30 x, so we will predict 30 y. to make it easier let's use for loop
def linearRegression(x, w, b):
pre_y = []
m = len(x)
for i in range(m):
f_wb = w * x[i] + b
pre_y.append(f_wb)
return pre_y
y_pred = linearRegression(x, w, b)
# Plot our model prediction
plt.plot(x, y_pred, c="b", label="Our Prediction")
# Plot the data points
plt.scatter(x, y, marker="x", c="r", label="Actual Values")
# Set the title
plt.title("Housing Prices")
# Set the y-axis label
plt.ylabel("Price (in 1000s of dollars)")
# Set the x-axis label
plt.xlabel("Size (1000 sqft)")
# to show box for label
plt.legend()
plt.show()
# # **Squared Error Cost Function**
# **Now let see how much our model great, by seeing difference between y_pred and y (which is called error)**
# **There are still a lot of cost functions!**
# **In here, I will use Squared error cost function!**
def SquaredErrorCost(y_pred, y):
totalCost = 0
m = len(y_pred)
for i in range(m):
cost = (y_pred[i] - y[i]) ** 2
totalCost += cost
totalCost /= 2 * m
return totalCost
# Function to calculate the cost for gradient descent
# instand of y_pred, it use w , b
def compute_cost(x, y, w, b):
m = x.shape[0]
cost = 0
for i in range(m):
f_wb = w * x[i] + b
cost = cost + (f_wb - y[i]) ** 2
total_cost = 1 / (2 * m) * cost
return total_cost
# assuming y, y_pred and cost are already defined
data = {"y": y, "y_pred": y_pred, "error": abs(y_pred - y)}
df = pd.DataFrame(data)
# print the data frame
print(df)
cost = SquaredErrorCost(y_pred, y)
print(" Squared Error Cost :", cost)
print(f" Squared Error Cost : {cost:10}")
print("Squared Error Cost: {:.5e}".format(cost))
# # **Grandient Descent**
# **The algorithm that can find best w,b if you use it correctly**
def compute_gradient(x, y, w, b): # to compute derivate
m = len(x)
dj_dw = w
dj_db = b
for i in range(m):
f_wb = w * x[i] + b
dj_dw_i = (f_wb - y[i]) * x[i]
dj_db_i = f_wb - y[i]
dj_dw += dj_dw_i
dj_db += dj_db_i
dj_dw /= m
dj_db /= m
return dj_dw, dj_db
def gradient_descent(
x, y, w_in, b_in, alpha, num_iters, cost_function, gradient_function
):
"""
Performs gradient descent to fit w,b. Updates w,b by taking
num_iters gradient steps with learning rate alpha
Args:
x (ndarray (m,)) : Data, m examples
y (ndarray (m,)) : target values
w_in,b_in (scalar): initial values of model parameters
alpha (float): Learning rate
num_iters (int): number of iterations to run gradient descent
cost_function: function to call to produce cost
gradient_function: function to call to produce gradient
Returns:
w (scalar): Updated value of parameter after running gradient descent
b (scalar): Updated value of parameter after running gradient descent
J_history (List): History of cost values
p_history (list): History of parameters [w,b]
"""
# An array to store cost J and w's at each iteration primarily for graphing later
J_history = []
p_history = []
b = b_in
w = w_in
for i in range(num_iters):
# Calculate the gradient and update the parameters using gradient_function
dj_dw, dj_db = gradient_function(x, y, w, b)
# Update Parameters using equation (3) above
b = b - alpha * dj_db
w = w - alpha * dj_dw
# Save cost J at each iteration
if i < 100000: # prevent resource exhaustion
J_history.append(cost_function(x, y, w, b))
p_history.append([w, b])
# Print cost every at intervals 10 times or as many iterations if < 10
if i % math.ceil(num_iters / 10) == 0:
print(
f"Iteration {i:4}: Cost {J_history[-1]:0.2e} ",
f"dj_dw: {dj_dw: 0.3e}, dj_db: {dj_db: 0.3e} ",
f"w: {w: 0.3e}, b:{b: 0.5e}",
)
return w, b, J_history, p_history # return w and J,w history for graphing
# Now let's check our gradient descent.
# w_init and b_init will be 0 , you know according above 0 is pretty bad.
# let's use gradient descent to find the best values of w and b
# initialize parameters
w_init = 0
b_init = 0
# some gradient descent settings
iterations = 10000
tmp_alpha = 1.0e-2
# run gradient descent
w_final, b_final, J_hist, p_hist = gradient_descent(
x, y, w_init, b_init, tmp_alpha, iterations, compute_cost, compute_gradient
)
print(f"(w,b) found by gradient descent: ({w_final:8.4f},{b_final:8.4f})")
|
[{"salary-dataset-simple-linear-regression/Salary_dataset.csv": {"column_names": "[\"Unnamed: 0\", \"YearsExperience\", \"Salary\"]", "column_data_types": "{\"Unnamed: 0\": \"int64\", \"YearsExperience\": \"float64\", \"Salary\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 30 entries, 0 to 29\nData columns (total 3 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 30 non-null int64 \n 1 YearsExperience 30 non-null float64\n 2 Salary 30 non-null float64\ndtypes: float64(2), int64(1)\nmemory usage: 848.0 bytes\n", "summary": "{\"Unnamed: 0\": {\"count\": 30.0, \"mean\": 14.5, \"std\": 8.803408430829505, \"min\": 0.0, \"25%\": 7.25, \"50%\": 14.5, \"75%\": 21.75, \"max\": 29.0}, \"YearsExperience\": {\"count\": 30.0, \"mean\": 5.413333333333332, \"std\": 2.8378881576627184, \"min\": 1.2000000000000002, \"25%\": 3.3000000000000003, \"50%\": 4.8, \"75%\": 7.8, \"max\": 10.6}, \"Salary\": {\"count\": 30.0, \"mean\": 76004.0, \"std\": 27414.4297845823, \"min\": 37732.0, \"25%\": 56721.75, \"50%\": 65238.0, \"75%\": 100545.75, \"max\": 122392.0}}", "examples": "{\"Unnamed: 0\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"YearsExperience\":{\"0\":1.2,\"1\":1.4,\"2\":1.6,\"3\":2.1},\"Salary\":{\"0\":39344.0,\"1\":46206.0,\"2\":37732.0,\"3\":43526.0}}"}}]
| true | 1 |
<start_data_description><data_path>salary-dataset-simple-linear-regression/Salary_dataset.csv:
<column_names>
['Unnamed: 0', 'YearsExperience', 'Salary']
<column_types>
{'Unnamed: 0': 'int64', 'YearsExperience': 'float64', 'Salary': 'float64'}
<dataframe_Summary>
{'Unnamed: 0': {'count': 30.0, 'mean': 14.5, 'std': 8.803408430829505, 'min': 0.0, '25%': 7.25, '50%': 14.5, '75%': 21.75, 'max': 29.0}, 'YearsExperience': {'count': 30.0, 'mean': 5.413333333333332, 'std': 2.8378881576627184, 'min': 1.2000000000000002, '25%': 3.3000000000000003, '50%': 4.8, '75%': 7.8, 'max': 10.6}, 'Salary': {'count': 30.0, 'mean': 76004.0, 'std': 27414.4297845823, 'min': 37732.0, '25%': 56721.75, '50%': 65238.0, '75%': 100545.75, 'max': 122392.0}}
<dataframe_info>
RangeIndex: 30 entries, 0 to 29
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 30 non-null int64
1 YearsExperience 30 non-null float64
2 Salary 30 non-null float64
dtypes: float64(2), int64(1)
memory usage: 848.0 bytes
<some_examples>
{'Unnamed: 0': {'0': 0, '1': 1, '2': 2, '3': 3}, 'YearsExperience': {'0': 1.2, '1': 1.4, '2': 1.6, '3': 2.1}, 'Salary': {'0': 39344.0, '1': 46206.0, '2': 37732.0, '3': 43526.0}}
<end_description>
| 2,568 | 1 | 2,968 | 2,568 |
129007856
|
<jupyter_start><jupyter_text>Flicktime
Kaggle dataset identifier: flicktime
<jupyter_script>import numpy as np
import pandas as pd
data = pd.read_csv("/kaggle/input/flicktime/rating.csv")
import pandas as pd
import numpy as np
import tensorflow as tf
from tensorflow.keras.layers import Input, Embedding, Dot, Flatten, Dense, Dropout
from tensorflow.keras.models import Model
# Load data
# data = pd.read_csv('ratings.csv')
# Create user-item matrix
n_users = data.userId.nunique()
n_movies = data.movieId.nunique()
user_ids = data.userId.astype("category").cat.codes.values
movie_ids = data.movieId.astype("category").cat.codes.values
ratings = data.rating.values
user_item_matrix = np.zeros((n_users, n_movies))
for i in range(len(ratings)):
user_item_matrix[user_ids[i], movie_ids[i]] = ratings[i]
# Define model
embedding_size = 64
user_input = Input(shape=(1,))
user_embed = Embedding(n_users, embedding_size)(user_input)
user_embed = Flatten()(user_embed)
movie_input = Input(shape=(1,))
movie_embed = Embedding(n_movies, embedding_size)(movie_input)
movie_embed = Flatten()(movie_embed)
dot_product = Dot(axes=1)([user_embed, movie_embed])
dense1 = Dense(64, activation="relu")(dot_product)
dropout1 = Dropout(0.2)(dense1)
dense2 = Dense(32, activation="relu")(dropout1)
dropout2 = Dropout(0.2)(dense2)
output = Dense(1)(dropout2)
model = Model(inputs=[user_input, movie_input], outputs=output)
model.compile(loss="mse", optimizer="adam")
# Train model
history = model.fit(
[user_ids, movie_ids], ratings, batch_size=128, epochs=5, validation_split=0.2
)
del data
import gc
gc.collect()
import ctypes
libc = ctypes.CDLL("libc.so.6") # clearing cache
libc.malloc_trim(0)
# Make predictions
user_ids_test = np.random.choice(n_users, 10)
movie_ids_test = np.random.choice(n_movies, 10)
predictions = model.predict([user_ids_test, movie_ids_test]).flatten()
print(predictions)
import matplotlib.pyplot as plt
# Plot the training and validation loss over epochs
plt.plot(history.history["loss"], label="train_loss")
plt.plot(history.history["val_loss"], label="val_loss")
plt.title("Model Learning Curve")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
plt.show()
import pickle
pickle.dump(model, open("model_rating.pkl", "wb"))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/007/129007856.ipynb
|
flicktime
|
jy2040
|
[{"Id": 129007856, "ScriptId": 38345333, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1591561, "CreationDate": "05/10/2023 09:18:14", "VersionNumber": 1.0, "Title": "flicktime_nn_rating_1", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 86.0, "LinesInsertedFromPrevious": 86.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184685527, "KernelVersionId": 129007856, "SourceDatasetVersionId": 5647522}]
|
[{"Id": 5647522, "DatasetId": 3232464, "DatasourceVersionId": 5722880, "CreatorUserId": 1591561, "LicenseName": "Unknown", "CreationDate": "05/09/2023 23:01:06", "VersionNumber": 5.0, "Title": "Flicktime", "Slug": "flicktime", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Data Update 2023-05-09", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 3232464, "CreatorUserId": 1591561, "OwnerUserId": 1591561.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5658766.0, "CurrentDatasourceVersionId": 5734182.0, "ForumId": 3297613, "Type": 2, "CreationDate": "05/07/2023 01:12:33", "LastActivityDate": "05/07/2023", "TotalViews": 65, "TotalDownloads": 4, "TotalVotes": 0, "TotalKernels": 4}]
|
[{"Id": 1591561, "UserName": "jy2040", "DisplayName": "Jay Bharadva", "RegisterDate": "01/29/2018", "PerformanceTier": 1}]
|
import numpy as np
import pandas as pd
data = pd.read_csv("/kaggle/input/flicktime/rating.csv")
import pandas as pd
import numpy as np
import tensorflow as tf
from tensorflow.keras.layers import Input, Embedding, Dot, Flatten, Dense, Dropout
from tensorflow.keras.models import Model
# Load data
# data = pd.read_csv('ratings.csv')
# Create user-item matrix
n_users = data.userId.nunique()
n_movies = data.movieId.nunique()
user_ids = data.userId.astype("category").cat.codes.values
movie_ids = data.movieId.astype("category").cat.codes.values
ratings = data.rating.values
user_item_matrix = np.zeros((n_users, n_movies))
for i in range(len(ratings)):
user_item_matrix[user_ids[i], movie_ids[i]] = ratings[i]
# Define model
embedding_size = 64
user_input = Input(shape=(1,))
user_embed = Embedding(n_users, embedding_size)(user_input)
user_embed = Flatten()(user_embed)
movie_input = Input(shape=(1,))
movie_embed = Embedding(n_movies, embedding_size)(movie_input)
movie_embed = Flatten()(movie_embed)
dot_product = Dot(axes=1)([user_embed, movie_embed])
dense1 = Dense(64, activation="relu")(dot_product)
dropout1 = Dropout(0.2)(dense1)
dense2 = Dense(32, activation="relu")(dropout1)
dropout2 = Dropout(0.2)(dense2)
output = Dense(1)(dropout2)
model = Model(inputs=[user_input, movie_input], outputs=output)
model.compile(loss="mse", optimizer="adam")
# Train model
history = model.fit(
[user_ids, movie_ids], ratings, batch_size=128, epochs=5, validation_split=0.2
)
del data
import gc
gc.collect()
import ctypes
libc = ctypes.CDLL("libc.so.6") # clearing cache
libc.malloc_trim(0)
# Make predictions
user_ids_test = np.random.choice(n_users, 10)
movie_ids_test = np.random.choice(n_movies, 10)
predictions = model.predict([user_ids_test, movie_ids_test]).flatten()
print(predictions)
import matplotlib.pyplot as plt
# Plot the training and validation loss over epochs
plt.plot(history.history["loss"], label="train_loss")
plt.plot(history.history["val_loss"], label="val_loss")
plt.title("Model Learning Curve")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
plt.show()
import pickle
pickle.dump(model, open("model_rating.pkl", "wb"))
| false | 1 | 717 | 0 | 737 | 717 |
||
129007439
|
<jupyter_start><jupyter_text>Mall Customer Segmentation Data
### Context
This data set is created only for the learning purpose of the customer segmentation concepts , also known as market basket analysis . I will demonstrate this by using unsupervised ML technique (KMeans Clustering Algorithm) in the simplest form.
### Content
You are owing a supermarket mall and through membership cards , you have some basic data about your customers like Customer ID, age, gender, annual income and spending score.
Spending Score is something you assign to the customer based on your defined parameters like customer behavior and purchasing data.
**Problem Statement**
You own the mall and want to understand the customers like who can be easily converge [Target Customers] so that the sense can be given to marketing team and plan the strategy accordingly.
Kaggle dataset identifier: customer-segmentation-tutorial-in-python
<jupyter_code>import pandas as pd
df = pd.read_csv('customer-segmentation-tutorial-in-python/Mall_Customers.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 200 entries, 0 to 199
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 CustomerID 200 non-null int64
1 Gender 200 non-null object
2 Age 200 non-null int64
3 Annual Income (k$) 200 non-null int64
4 Spending Score (1-100) 200 non-null int64
dtypes: int64(4), object(1)
memory usage: 7.9+ KB
<jupyter_text>Examples:
{
"CustomerID": 1,
"Gender": "Male",
"Age": 19,
"Annual Income (k$)": 15,
"Spending Score (1-100)": 39
}
{
"CustomerID": 2,
"Gender": "Male",
"Age": 21,
"Annual Income (k$)": 15,
"Spending Score (1-100)": 81
}
{
"CustomerID": 3,
"Gender": "Female",
"Age": 20,
"Annual Income (k$)": 16,
"Spending Score (1-100)": 6
}
{
"CustomerID": 4,
"Gender": "Female",
"Age": 23,
"Annual Income (k$)": 16,
"Spending Score (1-100)": 77
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd
# data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import warnings
warnings.filterwarnings("ignore")
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/customer-segmentation-tutorial-in-python/Mall_Customers.csv"
)
df
df.info()
df.isna().sum()
df.describe()
plt.figure(figsize=(12, 6))
sns.scatterplot(
data=df,
x=df["Annual Income (k$)"],
y=df["Spending Score (1-100)"],
hue=df["Gender"],
)
df_copy = df.copy()
df_copy
df1 = df.drop(["CustomerID", "Gender"], axis=1)
df1
df2 = df["Gender"]
df2
# K Means Clustering
from sklearn.cluster import KMeans
import plotly.express as px
fig = px.scatter(df, x="Annual Income (k$)", y="Spending Score (1-100)", color="Gender")
fig.show()
x = df1["Annual Income (k$)"]
y = df1["Spending Score (1-100)"]
plt.scatter(x, y)
kmeans = KMeans(n_clusters=4, max_iter=1000)
kmeans.fit(df1)
kmeans.cluster_centers_
clusters = kmeans.predict(df1)
# clusters
clusters[50:60]
x = df1["Annual Income (k$)"]
y = df1["Spending Score (1-100)"]
plt.scatter(x, y, c=clusters)
fig = px.scatter(
df1, x="Annual Income (k$)", y="Spending Score (1-100)", color=clusters
)
fig.show()
# plt.figure(figsize=(40,20))
# plt.subplot(321)
plt.figure(figsize=(20, 9))
x = df1["Annual Income (k$)"]
y = df1["Spending Score (1-100)"]
plt.subplot(321)
plt.title("K-means Predictions")
sns.scatterplot(data=df, x=x, y=y, hue=clusters)
plt.subplot(322)
plt.title("Actual Clusters")
sns.scatterplot(data=df, x=x, y=y, hue=df2)
x = df1["Age"]
y = df1["Spending Score (1-100)"]
# plt.figure(figsize=(10, 4))
plt.subplot(323)
plt.title("K-means Predictions")
sns.scatterplot(data=df, x=x, y=y, hue=clusters)
plt.subplot(324)
plt.title("Actual Clusters")
sns.scatterplot(data=df, x=x, y=y, hue=df2)
x = df1["Age"]
y = df1["Annual Income (k$)"]
# plt.figure(figsize=(10, 4))
plt.subplot(325)
plt.title("K-means Predictions")
sns.scatterplot(data=df, x=x, y=y, hue=clusters)
plt.subplot(326)
plt.title("Actual Clusters")
sns.scatterplot(data=df, x=x, y=y, hue=df2)
plt.tight_layout()
kmeans.inertia_
sse = {}
for k in range(1, 10):
kmeans = KMeans(n_clusters=k, max_iter=1000).fit(df1)
sse[k] = kmeans.inertia_
sse
plt.plot(list(sse.keys()), list(sse.values()))
plt.xlabel("Number of cluster")
plt.ylabel("SSE")
# k=4
model = KMeans(n_clusters=4, max_iter=1000)
model.fit(df1)
model.predict([[31, 17, 40]])
from sklearn.metrics import silhouette_score
score = silhouette_score(df1, model.labels_)
score
for n_cluster in range(2, 11):
kmeans = KMeans(n_clusters=n_cluster).fit(df1)
label = kmeans.labels_
sil_coeff = silhouette_score(df1, label, metric="euclidean")
print(
"For n_clusters={}, The Silhouette Coefficient is {}".format(
n_cluster, sil_coeff
)
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/007/129007439.ipynb
|
customer-segmentation-tutorial-in-python
|
vjchoudhary7
|
[{"Id": 129007439, "ScriptId": 38345802, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 13693366, "CreationDate": "05/10/2023 09:14:56", "VersionNumber": 1.0, "Title": "mall_customer_k-means", "EvaluationDate": "05/10/2023", "IsChange": true, "TotalLines": 150.0, "LinesInsertedFromPrevious": 150.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 184684921, "KernelVersionId": 129007439, "SourceDatasetVersionId": 74935}]
|
[{"Id": 74935, "DatasetId": 42674, "DatasourceVersionId": 77392, "CreatorUserId": 1790645, "LicenseName": "Other (specified in description)", "CreationDate": "08/11/2018 07:23:02", "VersionNumber": 1.0, "Title": "Mall Customer Segmentation Data", "Slug": "customer-segmentation-tutorial-in-python", "Subtitle": "Market Basket Analysis", "Description": "### Context\n\nThis data set is created only for the learning purpose of the customer segmentation concepts , also known as market basket analysis . I will demonstrate this by using unsupervised ML technique (KMeans Clustering Algorithm) in the simplest form. \n\n\n### Content\n\nYou are owing a supermarket mall and through membership cards , you have some basic data about your customers like Customer ID, age, gender, annual income and spending score. \nSpending Score is something you assign to the customer based on your defined parameters like customer behavior and purchasing data. \n\n**Problem Statement**\nYou own the mall and want to understand the customers like who can be easily converge [Target Customers] so that the sense can be given to marketing team and plan the strategy accordingly. \n\n\n### Acknowledgements\n\nFrom Udemy's Machine Learning A-Z course.\n\nI am new to Data science field and want to share my knowledge to others\n\nhttps://github.com/SteffiPeTaffy/machineLearningAZ/blob/master/Machine%20Learning%20A-Z%20Template%20Folder/Part%204%20-%20Clustering/Section%2025%20-%20Hierarchical%20Clustering/Mall_Customers.csv\n\n### Inspiration\n\nBy the end of this case study , you would be able to answer below questions. \n1- How to achieve customer segmentation using machine learning algorithm (KMeans Clustering) in Python in simplest way.\n2- Who are your target customers with whom you can start marketing strategy [easy to converse]\n3- How the marketing strategy works in real world", "VersionNotes": "Initial release", "TotalCompressedBytes": 3981.0, "TotalUncompressedBytes": 3981.0}]
|
[{"Id": 42674, "CreatorUserId": 1790645, "OwnerUserId": 1790645.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 74935.0, "CurrentDatasourceVersionId": 77392.0, "ForumId": 51177, "Type": 2, "CreationDate": "08/11/2018 07:23:02", "LastActivityDate": "08/11/2018", "TotalViews": 710472, "TotalDownloads": 132442, "TotalVotes": 1487, "TotalKernels": 1044}]
|
[{"Id": 1790645, "UserName": "vjchoudhary7", "DisplayName": "Vijay Choudhary", "RegisterDate": "04/05/2018", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd
# data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import warnings
warnings.filterwarnings("ignore")
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/customer-segmentation-tutorial-in-python/Mall_Customers.csv"
)
df
df.info()
df.isna().sum()
df.describe()
plt.figure(figsize=(12, 6))
sns.scatterplot(
data=df,
x=df["Annual Income (k$)"],
y=df["Spending Score (1-100)"],
hue=df["Gender"],
)
df_copy = df.copy()
df_copy
df1 = df.drop(["CustomerID", "Gender"], axis=1)
df1
df2 = df["Gender"]
df2
# K Means Clustering
from sklearn.cluster import KMeans
import plotly.express as px
fig = px.scatter(df, x="Annual Income (k$)", y="Spending Score (1-100)", color="Gender")
fig.show()
x = df1["Annual Income (k$)"]
y = df1["Spending Score (1-100)"]
plt.scatter(x, y)
kmeans = KMeans(n_clusters=4, max_iter=1000)
kmeans.fit(df1)
kmeans.cluster_centers_
clusters = kmeans.predict(df1)
# clusters
clusters[50:60]
x = df1["Annual Income (k$)"]
y = df1["Spending Score (1-100)"]
plt.scatter(x, y, c=clusters)
fig = px.scatter(
df1, x="Annual Income (k$)", y="Spending Score (1-100)", color=clusters
)
fig.show()
# plt.figure(figsize=(40,20))
# plt.subplot(321)
plt.figure(figsize=(20, 9))
x = df1["Annual Income (k$)"]
y = df1["Spending Score (1-100)"]
plt.subplot(321)
plt.title("K-means Predictions")
sns.scatterplot(data=df, x=x, y=y, hue=clusters)
plt.subplot(322)
plt.title("Actual Clusters")
sns.scatterplot(data=df, x=x, y=y, hue=df2)
x = df1["Age"]
y = df1["Spending Score (1-100)"]
# plt.figure(figsize=(10, 4))
plt.subplot(323)
plt.title("K-means Predictions")
sns.scatterplot(data=df, x=x, y=y, hue=clusters)
plt.subplot(324)
plt.title("Actual Clusters")
sns.scatterplot(data=df, x=x, y=y, hue=df2)
x = df1["Age"]
y = df1["Annual Income (k$)"]
# plt.figure(figsize=(10, 4))
plt.subplot(325)
plt.title("K-means Predictions")
sns.scatterplot(data=df, x=x, y=y, hue=clusters)
plt.subplot(326)
plt.title("Actual Clusters")
sns.scatterplot(data=df, x=x, y=y, hue=df2)
plt.tight_layout()
kmeans.inertia_
sse = {}
for k in range(1, 10):
kmeans = KMeans(n_clusters=k, max_iter=1000).fit(df1)
sse[k] = kmeans.inertia_
sse
plt.plot(list(sse.keys()), list(sse.values()))
plt.xlabel("Number of cluster")
plt.ylabel("SSE")
# k=4
model = KMeans(n_clusters=4, max_iter=1000)
model.fit(df1)
model.predict([[31, 17, 40]])
from sklearn.metrics import silhouette_score
score = silhouette_score(df1, model.labels_)
score
for n_cluster in range(2, 11):
kmeans = KMeans(n_clusters=n_cluster).fit(df1)
label = kmeans.labels_
sil_coeff = silhouette_score(df1, label, metric="euclidean")
print(
"For n_clusters={}, The Silhouette Coefficient is {}".format(
n_cluster, sil_coeff
)
)
|
[{"customer-segmentation-tutorial-in-python/Mall_Customers.csv": {"column_names": "[\"CustomerID\", \"Gender\", \"Age\", \"Annual Income (k$)\", \"Spending Score (1-100)\"]", "column_data_types": "{\"CustomerID\": \"int64\", \"Gender\": \"object\", \"Age\": \"int64\", \"Annual Income (k$)\": \"int64\", \"Spending Score (1-100)\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 200 entries, 0 to 199\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 CustomerID 200 non-null int64 \n 1 Gender 200 non-null object\n 2 Age 200 non-null int64 \n 3 Annual Income (k$) 200 non-null int64 \n 4 Spending Score (1-100) 200 non-null int64 \ndtypes: int64(4), object(1)\nmemory usage: 7.9+ KB\n", "summary": "{\"CustomerID\": {\"count\": 200.0, \"mean\": 100.5, \"std\": 57.879184513951124, \"min\": 1.0, \"25%\": 50.75, \"50%\": 100.5, \"75%\": 150.25, \"max\": 200.0}, \"Age\": {\"count\": 200.0, \"mean\": 38.85, \"std\": 13.96900733155888, \"min\": 18.0, \"25%\": 28.75, \"50%\": 36.0, \"75%\": 49.0, \"max\": 70.0}, \"Annual Income (k$)\": {\"count\": 200.0, \"mean\": 60.56, \"std\": 26.264721165271244, \"min\": 15.0, \"25%\": 41.5, \"50%\": 61.5, \"75%\": 78.0, \"max\": 137.0}, \"Spending Score (1-100)\": {\"count\": 200.0, \"mean\": 50.2, \"std\": 25.823521668370173, \"min\": 1.0, \"25%\": 34.75, \"50%\": 50.0, \"75%\": 73.0, \"max\": 99.0}}", "examples": "{\"CustomerID\":{\"0\":1,\"1\":2,\"2\":3,\"3\":4},\"Gender\":{\"0\":\"Male\",\"1\":\"Male\",\"2\":\"Female\",\"3\":\"Female\"},\"Age\":{\"0\":19,\"1\":21,\"2\":20,\"3\":23},\"Annual Income (k$)\":{\"0\":15,\"1\":15,\"2\":16,\"3\":16},\"Spending Score (1-100)\":{\"0\":39,\"1\":81,\"2\":6,\"3\":77}}"}}]
| true | 1 |
<start_data_description><data_path>customer-segmentation-tutorial-in-python/Mall_Customers.csv:
<column_names>
['CustomerID', 'Gender', 'Age', 'Annual Income (k$)', 'Spending Score (1-100)']
<column_types>
{'CustomerID': 'int64', 'Gender': 'object', 'Age': 'int64', 'Annual Income (k$)': 'int64', 'Spending Score (1-100)': 'int64'}
<dataframe_Summary>
{'CustomerID': {'count': 200.0, 'mean': 100.5, 'std': 57.879184513951124, 'min': 1.0, '25%': 50.75, '50%': 100.5, '75%': 150.25, 'max': 200.0}, 'Age': {'count': 200.0, 'mean': 38.85, 'std': 13.96900733155888, 'min': 18.0, '25%': 28.75, '50%': 36.0, '75%': 49.0, 'max': 70.0}, 'Annual Income (k$)': {'count': 200.0, 'mean': 60.56, 'std': 26.264721165271244, 'min': 15.0, '25%': 41.5, '50%': 61.5, '75%': 78.0, 'max': 137.0}, 'Spending Score (1-100)': {'count': 200.0, 'mean': 50.2, 'std': 25.823521668370173, 'min': 1.0, '25%': 34.75, '50%': 50.0, '75%': 73.0, 'max': 99.0}}
<dataframe_info>
RangeIndex: 200 entries, 0 to 199
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 CustomerID 200 non-null int64
1 Gender 200 non-null object
2 Age 200 non-null int64
3 Annual Income (k$) 200 non-null int64
4 Spending Score (1-100) 200 non-null int64
dtypes: int64(4), object(1)
memory usage: 7.9+ KB
<some_examples>
{'CustomerID': {'0': 1, '1': 2, '2': 3, '3': 4}, 'Gender': {'0': 'Male', '1': 'Male', '2': 'Female', '3': 'Female'}, 'Age': {'0': 19, '1': 21, '2': 20, '3': 23}, 'Annual Income (k$)': {'0': 15, '1': 15, '2': 16, '3': 16}, 'Spending Score (1-100)': {'0': 39, '1': 81, '2': 6, '3': 77}}
<end_description>
| 1,294 | 0 | 1,926 | 1,294 |
129193870
|
<jupyter_start><jupyter_text>Pins Face Recognition
### Content
This images has been collected from Pinterest and cropped. There are 105 celebrities and 17534 faces.

### Usage
If you are looking for a face recognition project, i highly recommend [my project that i shared on GitHub](https://github.com/aangfanboy/liyana).
Kaggle dataset identifier: pins-face-recognition
<jupyter_script># # Face Image Segment Anything Automatic Mask
# https://github.com/facebookresearch/segment-anything
# https://arxiv.org/abs/2304.02643
import os
import cv2
import random
from segment_anything import sam_model_registry
from segment_anything import SamAutomaticMaskGenerator
from segment_anything import SamPredictor
import torch
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
# # get mask image
def show_anns(anns, axes=None):
if len(anns) == 0:
return
if axes:
ax = axes
else:
ax = plt.gca()
ax.set_autoscale_on(False)
sorted_anns = sorted(anns, key=(lambda x: x["area"]), reverse=True)
polygons = []
color = []
for ann in sorted_anns:
m = ann["segmentation"]
img = np.ones((m.shape[0], m.shape[1], 3))
color_mask = np.random.random((1, 3)).tolist()[0]
for i in range(3):
img[:, :, i] = color_mask[i]
ax.imshow(np.dstack((img, m * 1.0))) # m*0.5
img2 = np.dstack((img, m * 1.0)) ### mask image
return img2 ### get mask image
sam_checkpoint = "/kaggle/input/segment-anything-models/sam_vit_h_4b8939.pth"
model_type = "vit_h" #
device = "cpu" # cpu,cuda
sam = sam_model_registry[model_type](checkpoint=sam_checkpoint)
sam.to(device=device)
mask_generator1 = SamAutomaticMaskGenerator(sam)
# # Generate Automatic Mask
path0 = "/kaggle/input/pins-face-recognition/105_classes_pins_dataset/pins_Mark Zuckerberg/Mark Zuckerberg106_1992.jpg"
image = cv2.imread(path0)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
masks1 = mask_generator1.generate(image)
fig, axs = plt.subplots(1, 3, figsize=(12, 4))
axs[0].imshow(image)
axs[2].imshow(image)
show_anns(masks1, axs[1])
show_anns(masks1, axs[2])
axs[0].axis("off")
axs[1].axis("off")
axs[2].axis("off")
plt.show()
print(image.shape)
bgw = np.ones(image.shape) * 255
print(len(masks1))
print(masks1[0].keys())
print(bgw.shape)
# # The sliced mask image
# show slices of masks1
mks = list("aaa")
for i in range(3):
plt.figure(figsize=(3, 3))
plt.imshow(bgw)
mks[i] = show_anns([masks1[i]])
plt.title(f"mask{i}")
plt.axis("off")
plt.show()
# mask0
plt.figure(figsize=(3, 3))
plt.imshow(mks[0])
plt.axis("off")
plt.show()
# mask2
plt.figure(figsize=(3, 3))
plt.imshow(mks[2])
plt.axis("off")
plt.show()
# mask0+mask2
mask = cv2.add(mks[0], mks[2])
plt.figure(figsize=(3, 3))
plt.imshow(mask)
plt.axis("off")
plt.show()
# # Synthesized Positive Mask
print(mask.flatten().min(), mask.flatten().max())
plt.hist(mask.flatten() * 255, bins=50, range=[0, 256], alpha=0.5, color="r")
plt.xlabel("Pixel value")
plt.ylabel("Frequency")
plt.show()
max_value = 255
threshold_type = cv2.THRESH_BINARY
_, positive_img = cv2.threshold(mask * 255, 250, max_value, threshold_type)
negative_img0 = np.logical_not(positive_img)
# reverse_img cannot be shown
negative_img = negative_img0.astype(np.uint8) * 255
# positive mask
plt.figure(figsize=(3, 3))
plt.imshow(positive_img)
plt.axis("off")
plt.show()
print(positive_img.shape)
# negative mask
plt.figure(figsize=(3, 3))
plt.imshow(negative_img)
plt.axis("off")
plt.show()
print(negative_img.shape)
plt.hist(negative_img.flatten(), bins=50, range=[0, 256], alpha=0.5, color="r")
plt.xlabel("Pixel value")
plt.ylabel("Frequency")
plt.show()
# # negative mask of 4 channels
print("original image")
plt.figure(figsize=(3, 3))
plt.imshow(image)
plt.axis("off")
plt.show()
print("negative mask")
plt.figure(figsize=(3, 3))
plt.imshow(negative_img)
plt.axis("off")
plt.show()
print(negative_img.shape)
# # image with positive mask of 3 channels
four_channel_image = negative_img
alpha_channel = four_channel_image[:, :, 3]
print(alpha_channel.shape)
plt.figure(figsize=(3, 3))
plt.imshow(alpha_channel)
plt.axis("off")
plt.show()
alpha_channel = cv2.cvtColor(alpha_channel, cv2.COLOR_GRAY2BGR)
alpha_channel = alpha_channel.astype(float) / 255.0
print("positive mask")
plt.figure(figsize=(3, 3))
plt.imshow(alpha_channel)
plt.axis("off")
plt.show()
masked_image = cv2.multiply(alpha_channel, image.astype(float)).astype(np.uint8)
print("image with positive mask")
plt.figure(figsize=(3, 3))
plt.imshow(masked_image)
plt.axis("off")
plt.show()
# # image with negative mask of 3 channels
alpha_channel2 = four_channel_image[:, :, 3]
alpha_channel3 = np.logical_not(alpha_channel2).astype(np.uint8)
print(alpha_channel3.shape)
plt.figure(figsize=(3, 3))
plt.imshow(alpha_channel3)
plt.axis("off")
plt.show()
alpha_channel3 = cv2.cvtColor(alpha_channel3, cv2.COLOR_GRAY2BGR)
alpha_channel3 = alpha_channel3.astype(float)
print("negative mask")
plt.figure(figsize=(3, 3))
plt.imshow(alpha_channel3)
plt.axis("off")
plt.show()
masked_image3 = cv2.multiply(alpha_channel3, image.astype(float))
masked_image3 = masked_image3.astype(np.uint8)
print("image with negative emask")
plt.figure(figsize=(3, 3))
plt.imshow(masked_image3)
plt.axis("off")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0129/193/129193870.ipynb
|
pins-face-recognition
|
hereisburak
|
[{"Id": 129193870, "ScriptId": 38313647, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2648923, "CreationDate": "05/11/2023 17:43:53", "VersionNumber": 38.0, "Title": "Face Image Segment Anything Automatic Mask", "EvaluationDate": "05/11/2023", "IsChange": true, "TotalLines": 216.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 215.0, "LinesInsertedFromFork": 144.0, "LinesDeletedFromFork": 70.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 72.0, "TotalVotes": 1}]
|
[{"Id": 185023640, "KernelVersionId": 129193870, "SourceDatasetVersionId": 992580}, {"Id": 185023641, "KernelVersionId": 129193870, "SourceDatasetVersionId": 5334514}]
|
[{"Id": 992580, "DatasetId": 543939, "DatasourceVersionId": 1021153, "CreatorUserId": 4625385, "LicenseName": "CC0: Public Domain", "CreationDate": "03/07/2020 11:57:06", "VersionNumber": 1.0, "Title": "Pins Face Recognition", "Slug": "pins-face-recognition", "Subtitle": "Facial Recognition Dataset collected from Pinterest", "Description": "### Content\n\nThis images has been collected from Pinterest and cropped. There are 105 celebrities and 17534 faces.\n\n\n\n### Usage\n\nIf you are looking for a face recognition project, i highly recommend [my project that i shared on GitHub](https://github.com/aangfanboy/liyana).", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 543939, "CreatorUserId": 4625385, "OwnerUserId": 4625385.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 992580.0, "CurrentDatasourceVersionId": 1021153.0, "ForumId": 557490, "Type": 2, "CreationDate": "03/07/2020 11:57:06", "LastActivityDate": "03/07/2020", "TotalViews": 48629, "TotalDownloads": 6330, "TotalVotes": 87, "TotalKernels": 35}]
|
[{"Id": 4625385, "UserName": "hereisburak", "DisplayName": "Burak", "RegisterDate": "03/07/2020", "PerformanceTier": 0}]
|
# # Face Image Segment Anything Automatic Mask
# https://github.com/facebookresearch/segment-anything
# https://arxiv.org/abs/2304.02643
import os
import cv2
import random
from segment_anything import sam_model_registry
from segment_anything import SamAutomaticMaskGenerator
from segment_anything import SamPredictor
import torch
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
# # get mask image
def show_anns(anns, axes=None):
if len(anns) == 0:
return
if axes:
ax = axes
else:
ax = plt.gca()
ax.set_autoscale_on(False)
sorted_anns = sorted(anns, key=(lambda x: x["area"]), reverse=True)
polygons = []
color = []
for ann in sorted_anns:
m = ann["segmentation"]
img = np.ones((m.shape[0], m.shape[1], 3))
color_mask = np.random.random((1, 3)).tolist()[0]
for i in range(3):
img[:, :, i] = color_mask[i]
ax.imshow(np.dstack((img, m * 1.0))) # m*0.5
img2 = np.dstack((img, m * 1.0)) ### mask image
return img2 ### get mask image
sam_checkpoint = "/kaggle/input/segment-anything-models/sam_vit_h_4b8939.pth"
model_type = "vit_h" #
device = "cpu" # cpu,cuda
sam = sam_model_registry[model_type](checkpoint=sam_checkpoint)
sam.to(device=device)
mask_generator1 = SamAutomaticMaskGenerator(sam)
# # Generate Automatic Mask
path0 = "/kaggle/input/pins-face-recognition/105_classes_pins_dataset/pins_Mark Zuckerberg/Mark Zuckerberg106_1992.jpg"
image = cv2.imread(path0)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
masks1 = mask_generator1.generate(image)
fig, axs = plt.subplots(1, 3, figsize=(12, 4))
axs[0].imshow(image)
axs[2].imshow(image)
show_anns(masks1, axs[1])
show_anns(masks1, axs[2])
axs[0].axis("off")
axs[1].axis("off")
axs[2].axis("off")
plt.show()
print(image.shape)
bgw = np.ones(image.shape) * 255
print(len(masks1))
print(masks1[0].keys())
print(bgw.shape)
# # The sliced mask image
# show slices of masks1
mks = list("aaa")
for i in range(3):
plt.figure(figsize=(3, 3))
plt.imshow(bgw)
mks[i] = show_anns([masks1[i]])
plt.title(f"mask{i}")
plt.axis("off")
plt.show()
# mask0
plt.figure(figsize=(3, 3))
plt.imshow(mks[0])
plt.axis("off")
plt.show()
# mask2
plt.figure(figsize=(3, 3))
plt.imshow(mks[2])
plt.axis("off")
plt.show()
# mask0+mask2
mask = cv2.add(mks[0], mks[2])
plt.figure(figsize=(3, 3))
plt.imshow(mask)
plt.axis("off")
plt.show()
# # Synthesized Positive Mask
print(mask.flatten().min(), mask.flatten().max())
plt.hist(mask.flatten() * 255, bins=50, range=[0, 256], alpha=0.5, color="r")
plt.xlabel("Pixel value")
plt.ylabel("Frequency")
plt.show()
max_value = 255
threshold_type = cv2.THRESH_BINARY
_, positive_img = cv2.threshold(mask * 255, 250, max_value, threshold_type)
negative_img0 = np.logical_not(positive_img)
# reverse_img cannot be shown
negative_img = negative_img0.astype(np.uint8) * 255
# positive mask
plt.figure(figsize=(3, 3))
plt.imshow(positive_img)
plt.axis("off")
plt.show()
print(positive_img.shape)
# negative mask
plt.figure(figsize=(3, 3))
plt.imshow(negative_img)
plt.axis("off")
plt.show()
print(negative_img.shape)
plt.hist(negative_img.flatten(), bins=50, range=[0, 256], alpha=0.5, color="r")
plt.xlabel("Pixel value")
plt.ylabel("Frequency")
plt.show()
# # negative mask of 4 channels
print("original image")
plt.figure(figsize=(3, 3))
plt.imshow(image)
plt.axis("off")
plt.show()
print("negative mask")
plt.figure(figsize=(3, 3))
plt.imshow(negative_img)
plt.axis("off")
plt.show()
print(negative_img.shape)
# # image with positive mask of 3 channels
four_channel_image = negative_img
alpha_channel = four_channel_image[:, :, 3]
print(alpha_channel.shape)
plt.figure(figsize=(3, 3))
plt.imshow(alpha_channel)
plt.axis("off")
plt.show()
alpha_channel = cv2.cvtColor(alpha_channel, cv2.COLOR_GRAY2BGR)
alpha_channel = alpha_channel.astype(float) / 255.0
print("positive mask")
plt.figure(figsize=(3, 3))
plt.imshow(alpha_channel)
plt.axis("off")
plt.show()
masked_image = cv2.multiply(alpha_channel, image.astype(float)).astype(np.uint8)
print("image with positive mask")
plt.figure(figsize=(3, 3))
plt.imshow(masked_image)
plt.axis("off")
plt.show()
# # image with negative mask of 3 channels
alpha_channel2 = four_channel_image[:, :, 3]
alpha_channel3 = np.logical_not(alpha_channel2).astype(np.uint8)
print(alpha_channel3.shape)
plt.figure(figsize=(3, 3))
plt.imshow(alpha_channel3)
plt.axis("off")
plt.show()
alpha_channel3 = cv2.cvtColor(alpha_channel3, cv2.COLOR_GRAY2BGR)
alpha_channel3 = alpha_channel3.astype(float)
print("negative mask")
plt.figure(figsize=(3, 3))
plt.imshow(alpha_channel3)
plt.axis("off")
plt.show()
masked_image3 = cv2.multiply(alpha_channel3, image.astype(float))
masked_image3 = masked_image3.astype(np.uint8)
print("image with negative emask")
plt.figure(figsize=(3, 3))
plt.imshow(masked_image3)
plt.axis("off")
plt.show()
| false | 0 | 1,834 | 1 | 1,971 | 1,834 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.