file_id
stringlengths 5
9
| content
stringlengths 100
5.25M
| local_path
stringlengths 66
70
| kaggle_dataset_name
stringlengths 3
50
⌀ | kaggle_dataset_owner
stringlengths 3
20
⌀ | kversion
stringlengths 497
763
⌀ | kversion_datasetsources
stringlengths 71
5.46k
⌀ | dataset_versions
stringlengths 338
235k
⌀ | datasets
stringlengths 334
371
⌀ | users
stringlengths 111
264
⌀ | script
stringlengths 100
5.25M
| df_info
stringlengths 0
4.87M
| has_data_info
bool 2
classes | nb_filenames
int64 0
370
| retreived_data_description
stringlengths 0
4.44M
| script_nb_tokens
int64 25
663k
| upvotes
int64 0
1.65k
| tokens_description
int64 25
663k
| tokens_script
int64 25
663k
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
69492655
|
<jupyter_start><jupyter_text>Titanic
### Context
This Data was originally taken from [Titanic: Machine Learning from Disaster][1] .But its better refined and cleaned & some features have been self engineered typically for logistic regression . If you use this data for other models and benefit from it , I would be happy to receive your comments and improvements.
### Content
There are two files namely:-
**train_data.csv** :- Typically a data set of 792x16 . The **survived** column is your target variable (The output you want to predict).The **parch & sibsb** columns from the original data set has been replaced with a single column called **Family size**.
All Categorical data like **Embarked , pclass** have been re-encoded using the one hot encoding method .
Additionally, 4 more columns have been added , re-engineered from the **Name column** to **Title_1 to Title_4** signifying males & females depending on whether they were married or not .(Mr , Mrs ,Master,Miss). An additional analysis to see if Married or in other words people with social responsibilities had more survival instincts/or not & is the trend similar for both genders.
All missing values have been filled with a median of the column values . All real valued data columns have been normalized.
**test_data.csv** :- A data of 100x16 , for testing your model , The arrangement of **test_data** exactly matches the **train_data**
I am open to feedbacks & suggesstions
[1]: https://www.kaggle.com/c/titanic/data
Kaggle dataset identifier: titanic
<jupyter_code>import pandas as pd
df = pd.read_csv('titanic/test_data.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 100 entries, 0 to 99
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 100 non-null int64
1 PassengerId 100 non-null int64
2 Survived 100 non-null int64
3 Sex 100 non-null int64
4 Age 100 non-null float64
5 Fare 100 non-null float64
6 Pclass_1 100 non-null int64
7 Pclass_2 100 non-null int64
8 Pclass_3 100 non-null int64
9 Family_size 100 non-null float64
10 Title_1 100 non-null int64
11 Title_2 100 non-null int64
12 Title_3 100 non-null int64
13 Title_4 100 non-null int64
14 Emb_1 100 non-null int64
15 Emb_2 100 non-null int64
16 Emb_3 100 non-null int64
dtypes: float64(3), int64(14)
memory usage: 13.4 KB
<jupyter_text>Examples:
{
"Unnamed: 0": 791.0,
"PassengerId": 792.0,
"Survived": 0.0,
"Sex": 1.0,
"Age": 0.2,
"Fare": 0.050748620200000004,
"Pclass_1": 0.0,
"Pclass_2": 1.0,
"Pclass_3": 0.0,
"Family_size": 0.0,
"Title_1": 1.0,
"Title_2": 0.0,
"Title_3": 0.0,
"Title_4": 0.0,
"Emb_1": 0.0,
"Emb_2": 0.0,
"Emb_3": 1.0
}
{
"Unnamed: 0": 792.0,
"PassengerId": 793.0,
"Survived": 0.0,
"Sex": 0.0,
"Age": 0.35000000000000003,
"Fare": 0.1357525591,
"Pclass_1": 0.0,
"Pclass_2": 0.0,
"Pclass_3": 1.0,
"Family_size": 1.0,
"Title_1": 0.0,
"Title_2": 0.0,
"Title_3": 0.0,
"Title_4": 1.0,
"Emb_1": 0.0,
"Emb_2": 0.0,
"Emb_3": 1.0
}
{
"Unnamed: 0": 793.0,
"PassengerId": 794.0,
"Survived": 0.0,
"Sex": 1.0,
"Age": 0.35000000000000003,
"Fare": 0.0599142114,
"Pclass_1": 1.0,
"Pclass_2": 0.0,
"Pclass_3": 0.0,
"Family_size": 0.0,
"Title_1": 1.0,
"Title_2": 0.0,
"Title_3": 0.0,
"Title_4": 0.0,
"Emb_1": 1.0,
"Emb_2": 0.0,
"Emb_3": 0.0
}
{
"Unnamed: 0": 794.0,
"PassengerId": 795.0,
"Survived": 0.0,
"Sex": 1.0,
"Age": 0.3125,
"Fare": 0.015411575200000001,
"Pclass_1": 0.0,
"Pclass_2": 0.0,
"Pclass_3": 1.0,
"Family_size": 0.0,
"Title_1": 1.0,
"Title_2": 0.0,
"Title_3": 0.0,
"Title_4": 0.0,
"Emb_1": 0.0,
"Emb_2": 0.0,
"Emb_3": 1.0
}
<jupyter_code>import pandas as pd
df = pd.read_csv('titanic/train_data.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 792 entries, 0 to 791
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 792 non-null int64
1 PassengerId 792 non-null int64
2 Survived 792 non-null int64
3 Sex 792 non-null int64
4 Age 792 non-null float64
5 Fare 792 non-null float64
6 Pclass_1 792 non-null int64
7 Pclass_2 792 non-null int64
8 Pclass_3 792 non-null int64
9 Family_size 792 non-null float64
10 Title_1 792 non-null int64
11 Title_2 792 non-null int64
12 Title_3 792 non-null int64
13 Title_4 792 non-null int64
14 Emb_1 792 non-null int64
15 Emb_2 792 non-null int64
16 Emb_3 792 non-null int64
dtypes: float64(3), int64(14)
memory usage: 105.3 KB
<jupyter_text>Examples:
{
"Unnamed: 0": 0.0,
"PassengerId": 1.0,
"Survived": 0.0,
"Sex": 1.0,
"Age": 0.275,
"Fare": 0.014151057600000001,
"Pclass_1": 0.0,
"Pclass_2": 0.0,
"Pclass_3": 1.0,
"Family_size": 0.1,
"Title_1": 1.0,
"Title_2": 0.0,
"Title_3": 0.0,
"Title_4": 0.0,
"Emb_1": 0.0,
"Emb_2": 0.0,
"Emb_3": 1.0
}
{
"Unnamed: 0": 1.0,
"PassengerId": 2.0,
"Survived": 1.0,
"Sex": 0.0,
"Age": 0.47500000000000003,
"Fare": 0.13913573540000002,
"Pclass_1": 1.0,
"Pclass_2": 0.0,
"Pclass_3": 0.0,
"Family_size": 0.1,
"Title_1": 1.0,
"Title_2": 0.0,
"Title_3": 0.0,
"Title_4": 0.0,
"Emb_1": 1.0,
"Emb_2": 0.0,
"Emb_3": 0.0
}
{
"Unnamed: 0": 2.0,
"PassengerId": 3.0,
"Survived": 1.0,
"Sex": 0.0,
"Age": 0.325,
"Fare": 0.0154685698,
"Pclass_1": 0.0,
"Pclass_2": 0.0,
"Pclass_3": 1.0,
"Family_size": 0.0,
"Title_1": 0.0,
"Title_2": 0.0,
"Title_3": 0.0,
"Title_4": 1.0,
"Emb_1": 0.0,
"Emb_2": 0.0,
"Emb_3": 1.0
}
{
"Unnamed: 0": 3.0,
"PassengerId": 4.0,
"Survived": 1.0,
"Sex": 0.0,
"Age": 0.4375,
"Fare": 0.10364429750000001,
"Pclass_1": 1.0,
"Pclass_2": 0.0,
"Pclass_3": 0.0,
"Family_size": 0.1,
"Title_1": 1.0,
"Title_2": 0.0,
"Title_3": 0.0,
"Title_4": 0.0,
"Emb_1": 0.0,
"Emb_2": 0.0,
"Emb_3": 1.0
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import mean_squared_error
from sklearn.metrics import accuracy_score
from sklearn.metrics import roc_auc_score
import matplotlib.pyplot as pt
import seaborn as sns
from sklearn.model_selection import train_test_split
pd.set_option("display.max_columns", None)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Importing data
input_ads = pd.read_csv("../input/titanic/train_data.csv")
input_ads.drop(
columns=["Unnamed: 0", "Title_1", "Title_2", "Title_3", "Title_4"], inplace=True
) # Dropping un-necessary columns
# -----------------------------------------------------------------
print(input_ads.shape)
input_ads.head()
# # Null Check
pd.DataFrame(input_ads.isnull().sum()).T
# # Description of the data
input_ads.describe()
# # Description of target variable
# Total survived vs not-survived split in the training data
input_ads["Survived"].value_counts()
# # Manipulation of data into train-test
target = "Survived" # To predict
# --------------------------------------------------------------------------------
# Splitting into X & Y datasets (supervised training)
X = input_ads[[cols for cols in list(input_ads.columns) if target not in cols]]
y = input_ads[target]
# --------------------------------------------------------------------------------
# Since test data is already placed in the input folder separately, we will just import it
test_ads = pd.read_csv("../input/titanic/test_data.csv")
test_ads.drop(
columns=["Unnamed: 0", "Title_1", "Title_2", "Title_3", "Title_4"], inplace=True
) # Dropping un-necessary columns
# Splitting into X & Y datasets (supervised training)
X_test = test_ads[[cols for cols in list(test_ads.columns) if target not in cols]]
y_test = test_ads[target]
print("Train % of total data:", 100 * X.shape[0] / (X.shape[0] + X_test.shape[0]))
# --------------------------------------------------------------------------------
# Manipulation of datasets for convenience and consistency
X_arr = np.array(X)
X_test_arr = np.array(X_test)
y_arr = np.array(y).reshape(X_arr.shape[0], 1)
y_test_arr = np.array(y_test).reshape(X_test_arr.shape[0], 1)
# --------------------------------------------------------------------------------
# Basic Summary
print(X_arr.shape)
print(X_test_arr.shape)
print(y_arr.shape)
# # Standard scaling the x-data
from sklearn.preprocessing import StandardScaler
# ----------------------------------------------------------
scaler = StandardScaler()
X_arr = scaler.fit_transform(X_arr)
X_test_arr = scaler.transform(X_test_arr)
# ----------------------------------------------------------
X_arr[0:3]
# # Artificial Neural Network (ANN) from Scratch
# ## UDFs for activation, initialization, layer_propagation
# All popular activation functions
def activation_fn(z, type_):
# print('Activation : ',type_)
if type_ == "linear":
activated_arr = z
elif type_ == "sigmoid":
activated_arr = 1 / (1 + np.exp(-z))
elif type_ == "relu":
activated_arr = np.maximum(np.zeros(z.shape), z)
elif type_ == "tanh":
activated_arr = (np.exp(z) - np.exp(-z)) / (np.exp(z) + np.exp(-z))
elif type_ == "leaky_relu":
activated_arr = np.maximum(0.01 * z, z)
elif type_ == "softmax":
exp_ = np.exp(z)
exp_sum = np.sum(exp_)
activated_arr = exp_ / exp_sum
return activated_arr
# ----------------------------------------------------------------------------------------------------------------------------
# Initialization of params
def generate_param_grid(a_prev, n_hidden, hidden_size_list):
parameters = {}
features = a_prev.shape[0] # Total features
n_examples = a_prev.shape[1]
for n_hidden_idx in range(1, n_hidden + 1):
n_hidden_nodes = hidden_size_list[n_hidden_idx] # Should start from 0
# print('#------------ Layer :',n_hidden_idx,'---- Size :',n_hidden_nodes,'---- Prev features :',features,'------#')
parameters["w" + str(n_hidden_idx)] = (
np.random.rand(n_hidden_nodes, features) * 0.1
) # Xavier Initialization
parameters["b" + str(n_hidden_idx)] = np.zeros((n_hidden_nodes, 1)) * 0.1
features = n_hidden_nodes
return parameters # Return randomly initiated params
# ---------------------------------------------------------------------------------------------------------------------------
# Propagation between z and activation
def layer_propagation(a_prev, w, b, activation):
# print(a_prev.shape)
# print(w.shape)
# print(b.shape)
z_ = np.dot(w, a_prev) + b
a = activation_fn(z=z_, type_=activation)
return z_, a
# ## UDF for forward propagation
def forward_propagation(
params_dict, data_x, data_y, n_hidden, hidden_size_list, activation_list
):
cache = {"a0": data_x.T}
a = data_x.T.copy()
for layer_idx in range(1, n_hidden + 1):
# print('#---------- Layer :',layer_idx,'-- No of Nodes :',hidden_size_list[layer_idx])
# nodes = hidden_size_list[layer_idx]
activation_ = activation_list[layer_idx]
w_ = params_dict["w" + str(layer_idx)]
b_ = params_dict["b" + str(layer_idx)]
z, a = layer_propagation(a_prev=a, w=w_, b=b_, activation=activation_)
cache["z" + str(layer_idx)] = z
cache["a" + str(layer_idx)] = a
return cache, a
# ## UDF for cost calculation, gradient calculation & back-propagation
# Calculation of the total cost incurred by the model
def cost_calculation(activation_list, y_true, y_pred):
if activation_list[-1] == "sigmoid":
# print('sig')
m = y_true.shape[1]
cost = (-1 / m) * np.sum(
(y_true * np.log(y_pred)) + ((1 - y_true) * np.log(1 - y_pred))
)
elif activation_list[-1] == "linear":
m = y_true.shape[1]
cost = (1 / m) * np.sum(np.square(y_true - y_pred))
##-------------------->> Softmax to be added <<----------------------
return cost
# Gradient of the activation functions wrt corresponding z
# --------------------------------------------------------------------------------------------
# Gradient for each activation type
def grad_fn_dz(activation, a):
if activation == "linear":
grad = 1
elif activation == "sigmoid":
grad = a * (1 - a)
elif activation == "tanh":
grad = np.square(1 - a)
elif activation == "relu":
grad = np.where(a >= 0, 1, 0)
elif activation == "leaky_relu":
grad = np.where(a >= 0, 1, 0.01)
##-------------------->> Softmax to be added <<----------------------
return grad
# --------------------------------------------------------------------------------------------
# UDF for gradient of loss function wrt last layer
def dL_last_layer(activation_list, y_true, y_pred):
if activation_list[-1] == "sigmoid":
# print('Last Layer y true shape :',y_true.shape)
# print('Last Layer y pred shape :',y_pred.shape)
grad_final_layer = -((y_true / y_pred) - ((1 - y_true) / (1 - y_pred)))
# print('Last Layer gradient shape :',grad_final_layer.shape)
elif activation_list[-1] == "linear":
grad_final_layer = -2 * (y_true - y_pred) # Check the sign
return grad_final_layer
# --------------------------------------------------------------------------------------------
# Back=Propagation
def back_propagation(
cache,
params_dict,
data_x,
data_y,
n_hidden,
hidden_size_list,
activation_list,
y_pred,
):
grads_cache = {}
# db_cache = {}
da = dL_last_layer(activation_list=activation_list, y_true=data_y.T, y_pred=y_pred)
# print('Final da shape :',da.shape)
m = data_y.shape[0] # Data in the batches
# print('dm in backprop :',m)
for layer_idx in list(reversed(range(1, n_hidden + 1))):
# print('# -------- Layer :',layer_idx,'-------- Size :',hidden_size_list[layer_idx],'--------#')
activation_ = activation_list[layer_idx]
a = cache["a" + str(layer_idx)]
a_prev = cache["a" + str(layer_idx - 1)]
w = params_dict["w" + str(layer_idx)]
# print('Shape of a:',a.shape)
# print('Shape of a_prev:',a_prev.shape)
# print('SHape of w:',w.shape)
# z =
dz = da * (grad_fn_dz(activation=activation_, a=a))
# print('dz shape :',dz.shape)
dw = (1 / m) * np.dot(dz, a_prev.T)
# print('dw shape :',dw.shape)
grads_cache["dw" + str(layer_idx)] = dw
db = (1 / m) * np.sum(dz, axis=1, keepdims=True)
# print('db shape :',db.shape)
grads_cache["db" + str(layer_idx)] = db
da = np.dot(w.T, dz)
# print('da shape :',da.shape)
return grads_cache
# ## UDF for updating weights through gradient descent
def update_weights(params, grads_cache, alpha, n_hidden):
for layer_idx in list(reversed(range(1, n_hidden + 1))):
# print('#---- layer :',layer_idx,'----#')
dw = grads_cache["dw" + str(layer_idx)]
db = grads_cache["db" + str(layer_idx)]
# print('dw shape :',dw.shape)
# print('db shape :',db.shape)
# print('w shape :',params['w'+str(layer_idx)].shape)
params["w" + str(layer_idx)] -= alpha * dw
params["b" + str(layer_idx)] -= alpha * db
return params
# ## UDF for predictions
def prediction(params, test_x, n_hidden, hidden_size_list, activation_list, threshold):
# -----------------------------------------------------------------
# Forward Propagation on trained weights
cache, y_pred = forward_propagation(
params_dict=params,
data_x=test_x,
data_y=None,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
)
# print(cache)
preds = np.where(y_pred > threshold, 1, 0).astype(float)
return cache, np.round(y_pred, 4), preds
# ## Simple Gradient Descent
def ANN_train_gd(
data_x, data_y, alpha, n_iters, n_hidden, hidden_size_list, activation_list
):
cost_history = [] # Record of cost through epochs
# -------------------------------------------------------------------------
# Initialization of params
params_dict = generate_param_grid(
a_prev=data_x.T, n_hidden=n_hidden, hidden_size_list=hidden_size_list
)
print("#----------------- Initial params ------------------#")
print(params_dict)
initial_params_abcd = params_dict.copy()
# -------------------------------------------------------------------------------------------
cache_tray = []
for epoch in range(n_iters):
if (epoch > 0) & (epoch % 1000 == 0):
print(
"#----------------------------------- Epoch :",
epoch,
"--------------------------------------#",
)
print("cost :", cost)
# -------------------------------------------------------------------------
# Forward Propagation
cache, y_pred = forward_propagation(
params_dict=params_dict,
data_x=data_x,
data_y=data_y,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
)
# print(np.max(y_pred))
cache_tray.append(cache)
# -------------------------------------------------------------------------
# Cost calculation
cost = cost_calculation(
activation_list=activation_list, y_true=data_y.T, y_pred=y_pred
)
cost_history.append(cost)
# print('cost :',cost)
# -------------------------------------------------------------------------
# Back Propagation
grads_cache_ = back_propagation(
cache=cache,
params_dict=params_dict,
data_x=data_x,
data_y=data_y,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
y_pred=y_pred,
)
# ------------------------------------------------------------------------
# Updating weights
params_dict = update_weights(
params=params_dict, grads_cache=grads_cache_, alpha=alpha, n_hidden=n_hidden
)
return (
initial_params_abcd,
params_dict,
grads_cache_,
cost_history,
y_pred,
cache_tray,
)
def ANN_train_gd(
data_x_overall,
data_y_overall,
batch_size,
alpha,
n_iters,
n_hidden,
hidden_size_list,
activation_list,
):
print("Total training rows :", data_x_overall.shape[0])
# ----------------------------------------------------------------------------------------
# Creating x-y batches according to the provided batch_size
n_batches = data_x_overall.shape[0] // batch_size
print("Total Batches to create in each epoch/iter :", n_batches)
batches_x = np.array_split(data_x_overall, n_batches)
print("Total Batches of X:", len(batches_x))
batches_y = np.array_split(data_y_overall, n_batches)
print("Total Batches of y:", len(batches_y))
# -------------------------------------------------------------------------------------------
cost_history = [] # Record of cost through epochs
# -------------------------------------------------------------------------------------------
# Initialization of params
params_dict = generate_param_grid(
a_prev=data_x_overall.T, n_hidden=n_hidden, hidden_size_list=hidden_size_list
)
print("#----------------- Initial params ------------------#")
print(params_dict)
initial_params_abcd = params_dict.copy()
# -------------------------------------------------------------------------------------------
cache_tray = []
for epoch in range(n_iters):
if (epoch > 0) & (epoch % 1000 == 0):
print(
"#----------------------------------- Epoch :",
epoch,
"--------------------------------------#",
)
print("cost :", cost)
for j in range(len(batches_x)): # For each batch created for each epoch/iter
# -------------------------------------------------------------------------
# For each batch of data
data_x = batches_x[j]
data_y = batches_y[j]
# -------------------------------------------------------------------------
# Forward Propagation
cache, y_pred = forward_propagation(
params_dict=params_dict,
data_x=data_x,
data_y=data_y,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
)
# print(np.max(y_pred))
# cache_tray.append(cache)
# -------------------------------------------------------------------------
# Cost calculation
cost = cost_calculation(
activation_list=activation_list, y_true=data_y.T, y_pred=y_pred
)
# cost_history.append(cost)
# print('cost :',cost)
# -------------------------------------------------------------------------
# Back Propagation
grads_cache_ = back_propagation(
cache=cache,
params_dict=params_dict,
data_x=data_x,
data_y=data_y,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
y_pred=y_pred,
)
# ------------------------------------------------------------------------
# Updating weights
params_dict = update_weights(
params=params_dict,
grads_cache=grads_cache_,
alpha=alpha,
n_hidden=n_hidden,
)
cost_history.append(cost) # Appending cost after each epoch
return (
initial_params_abcd,
params_dict,
grads_cache_,
cost_history,
y_pred,
cache_tray,
)
# Defining hyper-parameters for ANN
# --------------------------------------------------------------------------------------------------------------------------
n_hidden = 2 # No of hidden layers
alpha = 0.003 # Learning_rate
n_iters = 3001 # Total epochs
hidden_size_list = [0, 3, 1] # first element will be 0 and not counted in hidden layers
activation_list = [
0,
"relu",
"sigmoid",
] # first element will be 0 and not counted in hidden layers
batch_size = 25 # Batch wise gradient descent
# --------------------------------------------------------------------------------------------------------------------------
(
initial_params_train,
params_dict_train,
grads,
cost_history_train,
y_pred_train,
cache_tray,
) = ANN_train_gd(
data_x_overall=X_arr,
data_y_overall=y_arr,
batch_size=batch_size,
alpha=alpha,
n_iters=n_iters,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
)
# # Cost-Epoch plot for the manual ANN training
# Cost plot over epochs (1 value at end of each epoch) - over the last batch
ax = sns.lineplot(x=list(range(n_iters)), y=cost_history_train)
ax.set(xlabel="epochs", ylabel="cost", title="Cost vs epoch plot for Manual ANN")
# # Predict on the test data
cache, preds_proba, manual_preds = prediction(
params=params_dict_train,
test_x=X_test_arr,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
threshold=0.5,
)
# -------------------------------------------------------------------------------------------
print("Shape of prediction array :", preds_proba.shape)
print("Unique predictions :", np.unique(manual_preds))
print("Unique of predict proba :", np.unique(preds_proba), "\n")
print("#--------------------- Evaluation ----------------------#")
# Evaluation of the predictions
print("ROC AUC of test set :", roc_auc_score(y_test_arr.ravel(), manual_preds.ravel()))
print(
"Accuracy of test set :", accuracy_score(y_test_arr.ravel(), manual_preds.ravel())
)
# # Benchmarking with Keras functional API
# ## Importing necessary libraries
import tensorflow as tf
import keras
import tensorflow.keras.models
import tensorflow.keras.layers as tfl
from tensorflow.keras import Input
from tensorflow.keras import Model
from sklearn.preprocessing import StandardScaler
from keras.layers import BatchNormalization
# ## Defining the model with same specifications as manual
def ANN_keras(x):
input_ = tfl.Input(shape=(x.shape[1],))
x = tfl.Dense(3, activation="relu", name="Dense_3")(input_) # Layer 1
preds = tfl.Dense(1, activation="sigmoid", name="pred")(x) # Output layer
model = Model(input_, preds, name="ANN_keras")
model.compile(
loss="binary_crossentropy",
optimizer=tf.keras.optimizers.Adam(learning_rate=alpha),
)
return model
model = ANN_keras(X_arr)
model.summary()
# ## Training the model
history = model.fit(
X_arr,
y_arr,
epochs=101,
batch_size=X_arr.shape[0],
validation_data=(X_test_arr, y_test_arr),
verbose=0,
)
history = model.fit(
X_arr,
y_arr,
epochs=101,
batch_size=25,
validation_data=(X_test_arr, y_test_arr),
verbose=0,
)
# ## Predicting through keras model
keras_pred = model.predict(X_test_arr)
keras_pred = np.where(keras_pred > 0.5, 1, 0)
# print(np.unique(keras_pred))
print("#--------------------- Evaluation ----------------------#")
# Evaluation of the predictions
print("ROC AUC of test set :", roc_auc_score(y_test_arr.ravel(), keras_pred.ravel()))
print("Accuracy of test set :", accuracy_score(y_test_arr.ravel(), keras_pred.ravel()))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492655.ipynb
|
titanic
|
azeembootwala
|
[{"Id": 69492655, "ScriptId": 18938006, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4811800, "CreationDate": "07/31/2021 17:03:30", "VersionNumber": 3.0, "Title": "Artificial Neural Network from scratch (L Layer)", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 575.0, "LinesInsertedFromPrevious": 219.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 356.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92729106, "KernelVersionId": 69492655, "SourceDatasetVersionId": 2445}]
|
[{"Id": 2445, "DatasetId": 1355, "DatasourceVersionId": 2445, "CreatorUserId": 1059540, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "06/05/2017 12:14:37", "VersionNumber": 1.0, "Title": "Titanic", "Slug": "titanic", "Subtitle": "For Binary logistic regression", "Description": "### Context \nThis Data was originally taken from [Titanic: Machine Learning from Disaster][1] .But its better refined and cleaned & some features have been self engineered typically for logistic regression . If you use this data for other models and benefit from it , I would be happy to receive your comments and improvements. \n\n\n### Content\nThere are two files namely:- \n**train_data.csv** :- Typically a data set of 792x16 . The **survived** column is your target variable (The output you want to predict).The **parch & sibsb** columns from the original data set has been replaced with a single column called **Family size**. \n\nAll Categorical data like **Embarked , pclass** have been re-encoded using the one hot encoding method . \n\nAdditionally, 4 more columns have been added , re-engineered from the **Name column** to **Title_1 to Title_4** signifying males & females depending on whether they were married or not .(Mr , Mrs ,Master,Miss). An additional analysis to see if Married or in other words people with social responsibilities had more survival instincts/or not & is the trend similar for both genders. \n\nAll missing values have been filled with a median of the column values . All real valued data columns have been normalized. \n\n**test_data.csv** :- A data of 100x16 , for testing your model , The arrangement of **test_data** exactly matches the **train_data** \n\nI am open to feedbacks & suggesstions \n\n\n [1]: https://www.kaggle.com/c/titanic/data", "VersionNotes": "Initial release", "TotalCompressedBytes": 55456.0, "TotalUncompressedBytes": 55456.0}]
|
[{"Id": 1355, "CreatorUserId": 1059540, "OwnerUserId": 1059540.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2445.0, "CurrentDatasourceVersionId": 2445.0, "ForumId": 3937, "Type": 2, "CreationDate": "06/05/2017 12:14:37", "LastActivityDate": "02/05/2018", "TotalViews": 77731, "TotalDownloads": 17264, "TotalVotes": 166, "TotalKernels": 87}]
|
[{"Id": 1059540, "UserName": "azeembootwala", "DisplayName": "Azeem Bootwala", "RegisterDate": "05/05/2017", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import mean_squared_error
from sklearn.metrics import accuracy_score
from sklearn.metrics import roc_auc_score
import matplotlib.pyplot as pt
import seaborn as sns
from sklearn.model_selection import train_test_split
pd.set_option("display.max_columns", None)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Importing data
input_ads = pd.read_csv("../input/titanic/train_data.csv")
input_ads.drop(
columns=["Unnamed: 0", "Title_1", "Title_2", "Title_3", "Title_4"], inplace=True
) # Dropping un-necessary columns
# -----------------------------------------------------------------
print(input_ads.shape)
input_ads.head()
# # Null Check
pd.DataFrame(input_ads.isnull().sum()).T
# # Description of the data
input_ads.describe()
# # Description of target variable
# Total survived vs not-survived split in the training data
input_ads["Survived"].value_counts()
# # Manipulation of data into train-test
target = "Survived" # To predict
# --------------------------------------------------------------------------------
# Splitting into X & Y datasets (supervised training)
X = input_ads[[cols for cols in list(input_ads.columns) if target not in cols]]
y = input_ads[target]
# --------------------------------------------------------------------------------
# Since test data is already placed in the input folder separately, we will just import it
test_ads = pd.read_csv("../input/titanic/test_data.csv")
test_ads.drop(
columns=["Unnamed: 0", "Title_1", "Title_2", "Title_3", "Title_4"], inplace=True
) # Dropping un-necessary columns
# Splitting into X & Y datasets (supervised training)
X_test = test_ads[[cols for cols in list(test_ads.columns) if target not in cols]]
y_test = test_ads[target]
print("Train % of total data:", 100 * X.shape[0] / (X.shape[0] + X_test.shape[0]))
# --------------------------------------------------------------------------------
# Manipulation of datasets for convenience and consistency
X_arr = np.array(X)
X_test_arr = np.array(X_test)
y_arr = np.array(y).reshape(X_arr.shape[0], 1)
y_test_arr = np.array(y_test).reshape(X_test_arr.shape[0], 1)
# --------------------------------------------------------------------------------
# Basic Summary
print(X_arr.shape)
print(X_test_arr.shape)
print(y_arr.shape)
# # Standard scaling the x-data
from sklearn.preprocessing import StandardScaler
# ----------------------------------------------------------
scaler = StandardScaler()
X_arr = scaler.fit_transform(X_arr)
X_test_arr = scaler.transform(X_test_arr)
# ----------------------------------------------------------
X_arr[0:3]
# # Artificial Neural Network (ANN) from Scratch
# ## UDFs for activation, initialization, layer_propagation
# All popular activation functions
def activation_fn(z, type_):
# print('Activation : ',type_)
if type_ == "linear":
activated_arr = z
elif type_ == "sigmoid":
activated_arr = 1 / (1 + np.exp(-z))
elif type_ == "relu":
activated_arr = np.maximum(np.zeros(z.shape), z)
elif type_ == "tanh":
activated_arr = (np.exp(z) - np.exp(-z)) / (np.exp(z) + np.exp(-z))
elif type_ == "leaky_relu":
activated_arr = np.maximum(0.01 * z, z)
elif type_ == "softmax":
exp_ = np.exp(z)
exp_sum = np.sum(exp_)
activated_arr = exp_ / exp_sum
return activated_arr
# ----------------------------------------------------------------------------------------------------------------------------
# Initialization of params
def generate_param_grid(a_prev, n_hidden, hidden_size_list):
parameters = {}
features = a_prev.shape[0] # Total features
n_examples = a_prev.shape[1]
for n_hidden_idx in range(1, n_hidden + 1):
n_hidden_nodes = hidden_size_list[n_hidden_idx] # Should start from 0
# print('#------------ Layer :',n_hidden_idx,'---- Size :',n_hidden_nodes,'---- Prev features :',features,'------#')
parameters["w" + str(n_hidden_idx)] = (
np.random.rand(n_hidden_nodes, features) * 0.1
) # Xavier Initialization
parameters["b" + str(n_hidden_idx)] = np.zeros((n_hidden_nodes, 1)) * 0.1
features = n_hidden_nodes
return parameters # Return randomly initiated params
# ---------------------------------------------------------------------------------------------------------------------------
# Propagation between z and activation
def layer_propagation(a_prev, w, b, activation):
# print(a_prev.shape)
# print(w.shape)
# print(b.shape)
z_ = np.dot(w, a_prev) + b
a = activation_fn(z=z_, type_=activation)
return z_, a
# ## UDF for forward propagation
def forward_propagation(
params_dict, data_x, data_y, n_hidden, hidden_size_list, activation_list
):
cache = {"a0": data_x.T}
a = data_x.T.copy()
for layer_idx in range(1, n_hidden + 1):
# print('#---------- Layer :',layer_idx,'-- No of Nodes :',hidden_size_list[layer_idx])
# nodes = hidden_size_list[layer_idx]
activation_ = activation_list[layer_idx]
w_ = params_dict["w" + str(layer_idx)]
b_ = params_dict["b" + str(layer_idx)]
z, a = layer_propagation(a_prev=a, w=w_, b=b_, activation=activation_)
cache["z" + str(layer_idx)] = z
cache["a" + str(layer_idx)] = a
return cache, a
# ## UDF for cost calculation, gradient calculation & back-propagation
# Calculation of the total cost incurred by the model
def cost_calculation(activation_list, y_true, y_pred):
if activation_list[-1] == "sigmoid":
# print('sig')
m = y_true.shape[1]
cost = (-1 / m) * np.sum(
(y_true * np.log(y_pred)) + ((1 - y_true) * np.log(1 - y_pred))
)
elif activation_list[-1] == "linear":
m = y_true.shape[1]
cost = (1 / m) * np.sum(np.square(y_true - y_pred))
##-------------------->> Softmax to be added <<----------------------
return cost
# Gradient of the activation functions wrt corresponding z
# --------------------------------------------------------------------------------------------
# Gradient for each activation type
def grad_fn_dz(activation, a):
if activation == "linear":
grad = 1
elif activation == "sigmoid":
grad = a * (1 - a)
elif activation == "tanh":
grad = np.square(1 - a)
elif activation == "relu":
grad = np.where(a >= 0, 1, 0)
elif activation == "leaky_relu":
grad = np.where(a >= 0, 1, 0.01)
##-------------------->> Softmax to be added <<----------------------
return grad
# --------------------------------------------------------------------------------------------
# UDF for gradient of loss function wrt last layer
def dL_last_layer(activation_list, y_true, y_pred):
if activation_list[-1] == "sigmoid":
# print('Last Layer y true shape :',y_true.shape)
# print('Last Layer y pred shape :',y_pred.shape)
grad_final_layer = -((y_true / y_pred) - ((1 - y_true) / (1 - y_pred)))
# print('Last Layer gradient shape :',grad_final_layer.shape)
elif activation_list[-1] == "linear":
grad_final_layer = -2 * (y_true - y_pred) # Check the sign
return grad_final_layer
# --------------------------------------------------------------------------------------------
# Back=Propagation
def back_propagation(
cache,
params_dict,
data_x,
data_y,
n_hidden,
hidden_size_list,
activation_list,
y_pred,
):
grads_cache = {}
# db_cache = {}
da = dL_last_layer(activation_list=activation_list, y_true=data_y.T, y_pred=y_pred)
# print('Final da shape :',da.shape)
m = data_y.shape[0] # Data in the batches
# print('dm in backprop :',m)
for layer_idx in list(reversed(range(1, n_hidden + 1))):
# print('# -------- Layer :',layer_idx,'-------- Size :',hidden_size_list[layer_idx],'--------#')
activation_ = activation_list[layer_idx]
a = cache["a" + str(layer_idx)]
a_prev = cache["a" + str(layer_idx - 1)]
w = params_dict["w" + str(layer_idx)]
# print('Shape of a:',a.shape)
# print('Shape of a_prev:',a_prev.shape)
# print('SHape of w:',w.shape)
# z =
dz = da * (grad_fn_dz(activation=activation_, a=a))
# print('dz shape :',dz.shape)
dw = (1 / m) * np.dot(dz, a_prev.T)
# print('dw shape :',dw.shape)
grads_cache["dw" + str(layer_idx)] = dw
db = (1 / m) * np.sum(dz, axis=1, keepdims=True)
# print('db shape :',db.shape)
grads_cache["db" + str(layer_idx)] = db
da = np.dot(w.T, dz)
# print('da shape :',da.shape)
return grads_cache
# ## UDF for updating weights through gradient descent
def update_weights(params, grads_cache, alpha, n_hidden):
for layer_idx in list(reversed(range(1, n_hidden + 1))):
# print('#---- layer :',layer_idx,'----#')
dw = grads_cache["dw" + str(layer_idx)]
db = grads_cache["db" + str(layer_idx)]
# print('dw shape :',dw.shape)
# print('db shape :',db.shape)
# print('w shape :',params['w'+str(layer_idx)].shape)
params["w" + str(layer_idx)] -= alpha * dw
params["b" + str(layer_idx)] -= alpha * db
return params
# ## UDF for predictions
def prediction(params, test_x, n_hidden, hidden_size_list, activation_list, threshold):
# -----------------------------------------------------------------
# Forward Propagation on trained weights
cache, y_pred = forward_propagation(
params_dict=params,
data_x=test_x,
data_y=None,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
)
# print(cache)
preds = np.where(y_pred > threshold, 1, 0).astype(float)
return cache, np.round(y_pred, 4), preds
# ## Simple Gradient Descent
def ANN_train_gd(
data_x, data_y, alpha, n_iters, n_hidden, hidden_size_list, activation_list
):
cost_history = [] # Record of cost through epochs
# -------------------------------------------------------------------------
# Initialization of params
params_dict = generate_param_grid(
a_prev=data_x.T, n_hidden=n_hidden, hidden_size_list=hidden_size_list
)
print("#----------------- Initial params ------------------#")
print(params_dict)
initial_params_abcd = params_dict.copy()
# -------------------------------------------------------------------------------------------
cache_tray = []
for epoch in range(n_iters):
if (epoch > 0) & (epoch % 1000 == 0):
print(
"#----------------------------------- Epoch :",
epoch,
"--------------------------------------#",
)
print("cost :", cost)
# -------------------------------------------------------------------------
# Forward Propagation
cache, y_pred = forward_propagation(
params_dict=params_dict,
data_x=data_x,
data_y=data_y,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
)
# print(np.max(y_pred))
cache_tray.append(cache)
# -------------------------------------------------------------------------
# Cost calculation
cost = cost_calculation(
activation_list=activation_list, y_true=data_y.T, y_pred=y_pred
)
cost_history.append(cost)
# print('cost :',cost)
# -------------------------------------------------------------------------
# Back Propagation
grads_cache_ = back_propagation(
cache=cache,
params_dict=params_dict,
data_x=data_x,
data_y=data_y,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
y_pred=y_pred,
)
# ------------------------------------------------------------------------
# Updating weights
params_dict = update_weights(
params=params_dict, grads_cache=grads_cache_, alpha=alpha, n_hidden=n_hidden
)
return (
initial_params_abcd,
params_dict,
grads_cache_,
cost_history,
y_pred,
cache_tray,
)
def ANN_train_gd(
data_x_overall,
data_y_overall,
batch_size,
alpha,
n_iters,
n_hidden,
hidden_size_list,
activation_list,
):
print("Total training rows :", data_x_overall.shape[0])
# ----------------------------------------------------------------------------------------
# Creating x-y batches according to the provided batch_size
n_batches = data_x_overall.shape[0] // batch_size
print("Total Batches to create in each epoch/iter :", n_batches)
batches_x = np.array_split(data_x_overall, n_batches)
print("Total Batches of X:", len(batches_x))
batches_y = np.array_split(data_y_overall, n_batches)
print("Total Batches of y:", len(batches_y))
# -------------------------------------------------------------------------------------------
cost_history = [] # Record of cost through epochs
# -------------------------------------------------------------------------------------------
# Initialization of params
params_dict = generate_param_grid(
a_prev=data_x_overall.T, n_hidden=n_hidden, hidden_size_list=hidden_size_list
)
print("#----------------- Initial params ------------------#")
print(params_dict)
initial_params_abcd = params_dict.copy()
# -------------------------------------------------------------------------------------------
cache_tray = []
for epoch in range(n_iters):
if (epoch > 0) & (epoch % 1000 == 0):
print(
"#----------------------------------- Epoch :",
epoch,
"--------------------------------------#",
)
print("cost :", cost)
for j in range(len(batches_x)): # For each batch created for each epoch/iter
# -------------------------------------------------------------------------
# For each batch of data
data_x = batches_x[j]
data_y = batches_y[j]
# -------------------------------------------------------------------------
# Forward Propagation
cache, y_pred = forward_propagation(
params_dict=params_dict,
data_x=data_x,
data_y=data_y,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
)
# print(np.max(y_pred))
# cache_tray.append(cache)
# -------------------------------------------------------------------------
# Cost calculation
cost = cost_calculation(
activation_list=activation_list, y_true=data_y.T, y_pred=y_pred
)
# cost_history.append(cost)
# print('cost :',cost)
# -------------------------------------------------------------------------
# Back Propagation
grads_cache_ = back_propagation(
cache=cache,
params_dict=params_dict,
data_x=data_x,
data_y=data_y,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
y_pred=y_pred,
)
# ------------------------------------------------------------------------
# Updating weights
params_dict = update_weights(
params=params_dict,
grads_cache=grads_cache_,
alpha=alpha,
n_hidden=n_hidden,
)
cost_history.append(cost) # Appending cost after each epoch
return (
initial_params_abcd,
params_dict,
grads_cache_,
cost_history,
y_pred,
cache_tray,
)
# Defining hyper-parameters for ANN
# --------------------------------------------------------------------------------------------------------------------------
n_hidden = 2 # No of hidden layers
alpha = 0.003 # Learning_rate
n_iters = 3001 # Total epochs
hidden_size_list = [0, 3, 1] # first element will be 0 and not counted in hidden layers
activation_list = [
0,
"relu",
"sigmoid",
] # first element will be 0 and not counted in hidden layers
batch_size = 25 # Batch wise gradient descent
# --------------------------------------------------------------------------------------------------------------------------
(
initial_params_train,
params_dict_train,
grads,
cost_history_train,
y_pred_train,
cache_tray,
) = ANN_train_gd(
data_x_overall=X_arr,
data_y_overall=y_arr,
batch_size=batch_size,
alpha=alpha,
n_iters=n_iters,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
)
# # Cost-Epoch plot for the manual ANN training
# Cost plot over epochs (1 value at end of each epoch) - over the last batch
ax = sns.lineplot(x=list(range(n_iters)), y=cost_history_train)
ax.set(xlabel="epochs", ylabel="cost", title="Cost vs epoch plot for Manual ANN")
# # Predict on the test data
cache, preds_proba, manual_preds = prediction(
params=params_dict_train,
test_x=X_test_arr,
n_hidden=n_hidden,
hidden_size_list=hidden_size_list,
activation_list=activation_list,
threshold=0.5,
)
# -------------------------------------------------------------------------------------------
print("Shape of prediction array :", preds_proba.shape)
print("Unique predictions :", np.unique(manual_preds))
print("Unique of predict proba :", np.unique(preds_proba), "\n")
print("#--------------------- Evaluation ----------------------#")
# Evaluation of the predictions
print("ROC AUC of test set :", roc_auc_score(y_test_arr.ravel(), manual_preds.ravel()))
print(
"Accuracy of test set :", accuracy_score(y_test_arr.ravel(), manual_preds.ravel())
)
# # Benchmarking with Keras functional API
# ## Importing necessary libraries
import tensorflow as tf
import keras
import tensorflow.keras.models
import tensorflow.keras.layers as tfl
from tensorflow.keras import Input
from tensorflow.keras import Model
from sklearn.preprocessing import StandardScaler
from keras.layers import BatchNormalization
# ## Defining the model with same specifications as manual
def ANN_keras(x):
input_ = tfl.Input(shape=(x.shape[1],))
x = tfl.Dense(3, activation="relu", name="Dense_3")(input_) # Layer 1
preds = tfl.Dense(1, activation="sigmoid", name="pred")(x) # Output layer
model = Model(input_, preds, name="ANN_keras")
model.compile(
loss="binary_crossentropy",
optimizer=tf.keras.optimizers.Adam(learning_rate=alpha),
)
return model
model = ANN_keras(X_arr)
model.summary()
# ## Training the model
history = model.fit(
X_arr,
y_arr,
epochs=101,
batch_size=X_arr.shape[0],
validation_data=(X_test_arr, y_test_arr),
verbose=0,
)
history = model.fit(
X_arr,
y_arr,
epochs=101,
batch_size=25,
validation_data=(X_test_arr, y_test_arr),
verbose=0,
)
# ## Predicting through keras model
keras_pred = model.predict(X_test_arr)
keras_pred = np.where(keras_pred > 0.5, 1, 0)
# print(np.unique(keras_pred))
print("#--------------------- Evaluation ----------------------#")
# Evaluation of the predictions
print("ROC AUC of test set :", roc_auc_score(y_test_arr.ravel(), keras_pred.ravel()))
print("Accuracy of test set :", accuracy_score(y_test_arr.ravel(), keras_pred.ravel()))
|
[{"titanic/test_data.csv": {"column_names": "[\"Unnamed: 0\", \"PassengerId\", \"Survived\", \"Sex\", \"Age\", \"Fare\", \"Pclass_1\", \"Pclass_2\", \"Pclass_3\", \"Family_size\", \"Title_1\", \"Title_2\", \"Title_3\", \"Title_4\", \"Emb_1\", \"Emb_2\", \"Emb_3\"]", "column_data_types": "{\"Unnamed: 0\": \"int64\", \"PassengerId\": \"int64\", \"Survived\": \"int64\", \"Sex\": \"int64\", \"Age\": \"float64\", \"Fare\": \"float64\", \"Pclass_1\": \"int64\", \"Pclass_2\": \"int64\", \"Pclass_3\": \"int64\", \"Family_size\": \"float64\", \"Title_1\": \"int64\", \"Title_2\": \"int64\", \"Title_3\": \"int64\", \"Title_4\": \"int64\", \"Emb_1\": \"int64\", \"Emb_2\": \"int64\", \"Emb_3\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 100 entries, 0 to 99\nData columns (total 17 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 100 non-null int64 \n 1 PassengerId 100 non-null int64 \n 2 Survived 100 non-null int64 \n 3 Sex 100 non-null int64 \n 4 Age 100 non-null float64\n 5 Fare 100 non-null float64\n 6 Pclass_1 100 non-null int64 \n 7 Pclass_2 100 non-null int64 \n 8 Pclass_3 100 non-null int64 \n 9 Family_size 100 non-null float64\n 10 Title_1 100 non-null int64 \n 11 Title_2 100 non-null int64 \n 12 Title_3 100 non-null int64 \n 13 Title_4 100 non-null int64 \n 14 Emb_1 100 non-null int64 \n 15 Emb_2 100 non-null int64 \n 16 Emb_3 100 non-null int64 \ndtypes: float64(3), int64(14)\nmemory usage: 13.4 KB\n", "summary": "{\"Unnamed: 0\": {\"count\": 100.0, \"mean\": 840.5, \"std\": 29.011491975882016, \"min\": 791.0, \"25%\": 815.75, \"50%\": 840.5, \"75%\": 865.25, \"max\": 890.0}, \"PassengerId\": {\"count\": 100.0, \"mean\": 841.5, \"std\": 29.011491975882016, \"min\": 792.0, \"25%\": 816.75, \"50%\": 841.5, \"75%\": 866.25, \"max\": 891.0}, \"Survived\": {\"count\": 100.0, \"mean\": 0.36, \"std\": 0.4824181513244218, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"Sex\": {\"count\": 100.0, \"mean\": 0.65, \"std\": 0.4793724854411023, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"Age\": {\"count\": 100.0, \"mean\": 0.35565624999999995, \"std\": 0.16118783389306676, \"min\": 0.0052499999999999, \"25%\": 0.27187500000000003, \"50%\": 0.35, \"75%\": 0.4265625, \"max\": 0.925}, \"Fare\": {\"count\": 100.0, \"mean\": 0.04833325916227294, \"std\": 0.05342856309679655, \"min\": 0.0, \"25%\": 0.0154115752137492, \"50%\": 0.0253743101115454, \"75%\": 0.0585561002574126, \"max\": 0.3217983671436256}, \"Pclass_1\": {\"count\": 100.0, \"mean\": 0.23, \"std\": 0.4229525846816507, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Pclass_2\": {\"count\": 100.0, \"mean\": 0.2, \"std\": 0.40201512610368484, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Pclass_3\": {\"count\": 100.0, \"mean\": 0.57, \"std\": 0.497569851956243, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"Family_size\": {\"count\": 100.0, \"mean\": 0.10400000000000002, \"std\": 0.2078849720786994, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.1, \"max\": 1.0}, \"Title_1\": {\"count\": 100.0, \"mean\": 0.77, \"std\": 0.4229525846816507, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"Title_2\": {\"count\": 100.0, \"mean\": 0.01, \"std\": 0.09999999999999999, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Title_3\": {\"count\": 100.0, \"mean\": 0.08, \"std\": 0.27265992434429076, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Title_4\": {\"count\": 100.0, \"mean\": 0.14, \"std\": 0.348735088019777, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Emb_1\": {\"count\": 100.0, \"mean\": 0.21, \"std\": 0.40936018074033237, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Emb_2\": {\"count\": 100.0, \"mean\": 0.04, \"std\": 0.19694638556693236, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Emb_3\": {\"count\": 100.0, \"mean\": 0.74, \"std\": 0.44084400227680803, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}}", "examples": "{\"Unnamed: 0\":{\"0\":791,\"1\":792,\"2\":793,\"3\":794},\"PassengerId\":{\"0\":792,\"1\":793,\"2\":794,\"3\":795},\"Survived\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Sex\":{\"0\":1,\"1\":0,\"2\":1,\"3\":1},\"Age\":{\"0\":0.2,\"1\":0.35,\"2\":0.35,\"3\":0.3125},\"Fare\":{\"0\":0.0507486202,\"1\":0.1357525591,\"2\":0.0599142114,\"3\":0.0154115752},\"Pclass_1\":{\"0\":0,\"1\":0,\"2\":1,\"3\":0},\"Pclass_2\":{\"0\":1,\"1\":0,\"2\":0,\"3\":0},\"Pclass_3\":{\"0\":0,\"1\":1,\"2\":0,\"3\":1},\"Family_size\":{\"0\":0.0,\"1\":1.0,\"2\":0.0,\"3\":0.0},\"Title_1\":{\"0\":1,\"1\":0,\"2\":1,\"3\":1},\"Title_2\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Title_3\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Title_4\":{\"0\":0,\"1\":1,\"2\":0,\"3\":0},\"Emb_1\":{\"0\":0,\"1\":0,\"2\":1,\"3\":0},\"Emb_2\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Emb_3\":{\"0\":1,\"1\":1,\"2\":0,\"3\":1}}"}}, {"titanic/train_data.csv": {"column_names": "[\"Unnamed: 0\", \"PassengerId\", \"Survived\", \"Sex\", \"Age\", \"Fare\", \"Pclass_1\", \"Pclass_2\", \"Pclass_3\", \"Family_size\", \"Title_1\", \"Title_2\", \"Title_3\", \"Title_4\", \"Emb_1\", \"Emb_2\", \"Emb_3\"]", "column_data_types": "{\"Unnamed: 0\": \"int64\", \"PassengerId\": \"int64\", \"Survived\": \"int64\", \"Sex\": \"int64\", \"Age\": \"float64\", \"Fare\": \"float64\", \"Pclass_1\": \"int64\", \"Pclass_2\": \"int64\", \"Pclass_3\": \"int64\", \"Family_size\": \"float64\", \"Title_1\": \"int64\", \"Title_2\": \"int64\", \"Title_3\": \"int64\", \"Title_4\": \"int64\", \"Emb_1\": \"int64\", \"Emb_2\": \"int64\", \"Emb_3\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 792 entries, 0 to 791\nData columns (total 17 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 792 non-null int64 \n 1 PassengerId 792 non-null int64 \n 2 Survived 792 non-null int64 \n 3 Sex 792 non-null int64 \n 4 Age 792 non-null float64\n 5 Fare 792 non-null float64\n 6 Pclass_1 792 non-null int64 \n 7 Pclass_2 792 non-null int64 \n 8 Pclass_3 792 non-null int64 \n 9 Family_size 792 non-null float64\n 10 Title_1 792 non-null int64 \n 11 Title_2 792 non-null int64 \n 12 Title_3 792 non-null int64 \n 13 Title_4 792 non-null int64 \n 14 Emb_1 792 non-null int64 \n 15 Emb_2 792 non-null int64 \n 16 Emb_3 792 non-null int64 \ndtypes: float64(3), int64(14)\nmemory usage: 105.3 KB\n", "summary": "{\"Unnamed: 0\": {\"count\": 792.0, \"mean\": 395.5, \"std\": 228.77499863402906, \"min\": 0.0, \"25%\": 197.75, \"50%\": 395.5, \"75%\": 593.25, \"max\": 791.0}, \"PassengerId\": {\"count\": 792.0, \"mean\": 396.5, \"std\": 228.77499863402906, \"min\": 1.0, \"25%\": 198.75, \"50%\": 396.5, \"75%\": 594.25, \"max\": 792.0}, \"Survived\": {\"count\": 792.0, \"mean\": 0.38636363636363635, \"std\": 0.4872232622710165, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"Sex\": {\"count\": 792.0, \"mean\": 0.6477272727272727, \"std\": 0.4779802495415942, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"Age\": {\"count\": 792.0, \"mean\": 0.36824368686868686, \"std\": 0.162993712818546, \"min\": 0.008375, \"25%\": 0.275, \"50%\": 0.35, \"75%\": 0.4375, \"max\": 1.0}, \"Fare\": {\"count\": 792.0, \"mean\": 0.06467712344746579, \"std\": 0.10098746729808539, \"min\": 0.0, \"25%\": 0.0154685698179998, \"50%\": 0.028302115124416, \"75%\": 0.0610447345183526, \"max\": 1.0}, \"Pclass_1\": {\"count\": 792.0, \"mean\": 0.24368686868686867, \"std\": 0.4295772101125523, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Pclass_2\": {\"count\": 792.0, \"mean\": 0.20833333333333334, \"std\": 0.40637306071557894, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Pclass_3\": {\"count\": 792.0, \"mean\": 0.547979797979798, \"std\": 0.4980071126944576, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"Family_size\": {\"count\": 792.0, \"mean\": 0.08863636363636362, \"std\": 0.1544851048700475, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.1, \"max\": 1.0}, \"Title_1\": {\"count\": 792.0, \"mean\": 0.7449494949494949, \"std\": 0.4361650454554044, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"Title_2\": {\"count\": 792.0, \"mean\": 0.005050505050505051, \"std\": 0.07093201085612565, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Title_3\": {\"count\": 792.0, \"mean\": 0.04040404040404041, \"std\": 0.19702936277118982, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Title_4\": {\"count\": 792.0, \"mean\": 0.20959595959595959, \"std\": 0.40727746237872897, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Emb_1\": {\"count\": 792.0, \"mean\": 0.1856060606060606, \"std\": 0.38903411965925394, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Emb_2\": {\"count\": 792.0, \"mean\": 0.09217171717171717, \"std\": 0.2894509922654781, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"Emb_3\": {\"count\": 792.0, \"mean\": 0.7209595959595959, \"std\": 0.44881086134701287, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}}", "examples": "{\"Unnamed: 0\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"PassengerId\":{\"0\":1,\"1\":2,\"2\":3,\"3\":4},\"Survived\":{\"0\":0,\"1\":1,\"2\":1,\"3\":1},\"Sex\":{\"0\":1,\"1\":0,\"2\":0,\"3\":0},\"Age\":{\"0\":0.275,\"1\":0.475,\"2\":0.325,\"3\":0.4375},\"Fare\":{\"0\":0.0141510576,\"1\":0.1391357354,\"2\":0.0154685698,\"3\":0.1036442975},\"Pclass_1\":{\"0\":0,\"1\":1,\"2\":0,\"3\":1},\"Pclass_2\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Pclass_3\":{\"0\":1,\"1\":0,\"2\":1,\"3\":0},\"Family_size\":{\"0\":0.1,\"1\":0.1,\"2\":0.0,\"3\":0.1},\"Title_1\":{\"0\":1,\"1\":1,\"2\":0,\"3\":1},\"Title_2\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Title_3\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Title_4\":{\"0\":0,\"1\":0,\"2\":1,\"3\":0},\"Emb_1\":{\"0\":0,\"1\":1,\"2\":0,\"3\":0},\"Emb_2\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Emb_3\":{\"0\":1,\"1\":0,\"2\":1,\"3\":1}}"}}]
| true | 2 |
<start_data_description><data_path>titanic/test_data.csv:
<column_names>
['Unnamed: 0', 'PassengerId', 'Survived', 'Sex', 'Age', 'Fare', 'Pclass_1', 'Pclass_2', 'Pclass_3', 'Family_size', 'Title_1', 'Title_2', 'Title_3', 'Title_4', 'Emb_1', 'Emb_2', 'Emb_3']
<column_types>
{'Unnamed: 0': 'int64', 'PassengerId': 'int64', 'Survived': 'int64', 'Sex': 'int64', 'Age': 'float64', 'Fare': 'float64', 'Pclass_1': 'int64', 'Pclass_2': 'int64', 'Pclass_3': 'int64', 'Family_size': 'float64', 'Title_1': 'int64', 'Title_2': 'int64', 'Title_3': 'int64', 'Title_4': 'int64', 'Emb_1': 'int64', 'Emb_2': 'int64', 'Emb_3': 'int64'}
<dataframe_Summary>
{'Unnamed: 0': {'count': 100.0, 'mean': 840.5, 'std': 29.011491975882016, 'min': 791.0, '25%': 815.75, '50%': 840.5, '75%': 865.25, 'max': 890.0}, 'PassengerId': {'count': 100.0, 'mean': 841.5, 'std': 29.011491975882016, 'min': 792.0, '25%': 816.75, '50%': 841.5, '75%': 866.25, 'max': 891.0}, 'Survived': {'count': 100.0, 'mean': 0.36, 'std': 0.4824181513244218, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'Sex': {'count': 100.0, 'mean': 0.65, 'std': 0.4793724854411023, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'Age': {'count': 100.0, 'mean': 0.35565624999999995, 'std': 0.16118783389306676, 'min': 0.0052499999999999, '25%': 0.27187500000000003, '50%': 0.35, '75%': 0.4265625, 'max': 0.925}, 'Fare': {'count': 100.0, 'mean': 0.04833325916227294, 'std': 0.05342856309679655, 'min': 0.0, '25%': 0.0154115752137492, '50%': 0.0253743101115454, '75%': 0.0585561002574126, 'max': 0.3217983671436256}, 'Pclass_1': {'count': 100.0, 'mean': 0.23, 'std': 0.4229525846816507, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Pclass_2': {'count': 100.0, 'mean': 0.2, 'std': 0.40201512610368484, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Pclass_3': {'count': 100.0, 'mean': 0.57, 'std': 0.497569851956243, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'Family_size': {'count': 100.0, 'mean': 0.10400000000000002, 'std': 0.2078849720786994, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.1, 'max': 1.0}, 'Title_1': {'count': 100.0, 'mean': 0.77, 'std': 0.4229525846816507, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'Title_2': {'count': 100.0, 'mean': 0.01, 'std': 0.09999999999999999, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Title_3': {'count': 100.0, 'mean': 0.08, 'std': 0.27265992434429076, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Title_4': {'count': 100.0, 'mean': 0.14, 'std': 0.348735088019777, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Emb_1': {'count': 100.0, 'mean': 0.21, 'std': 0.40936018074033237, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Emb_2': {'count': 100.0, 'mean': 0.04, 'std': 0.19694638556693236, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Emb_3': {'count': 100.0, 'mean': 0.74, 'std': 0.44084400227680803, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 100 entries, 0 to 99
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 100 non-null int64
1 PassengerId 100 non-null int64
2 Survived 100 non-null int64
3 Sex 100 non-null int64
4 Age 100 non-null float64
5 Fare 100 non-null float64
6 Pclass_1 100 non-null int64
7 Pclass_2 100 non-null int64
8 Pclass_3 100 non-null int64
9 Family_size 100 non-null float64
10 Title_1 100 non-null int64
11 Title_2 100 non-null int64
12 Title_3 100 non-null int64
13 Title_4 100 non-null int64
14 Emb_1 100 non-null int64
15 Emb_2 100 non-null int64
16 Emb_3 100 non-null int64
dtypes: float64(3), int64(14)
memory usage: 13.4 KB
<some_examples>
{'Unnamed: 0': {'0': 791, '1': 792, '2': 793, '3': 794}, 'PassengerId': {'0': 792, '1': 793, '2': 794, '3': 795}, 'Survived': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Sex': {'0': 1, '1': 0, '2': 1, '3': 1}, 'Age': {'0': 0.2, '1': 0.35, '2': 0.35, '3': 0.3125}, 'Fare': {'0': 0.0507486202, '1': 0.1357525591, '2': 0.0599142114, '3': 0.0154115752}, 'Pclass_1': {'0': 0, '1': 0, '2': 1, '3': 0}, 'Pclass_2': {'0': 1, '1': 0, '2': 0, '3': 0}, 'Pclass_3': {'0': 0, '1': 1, '2': 0, '3': 1}, 'Family_size': {'0': 0.0, '1': 1.0, '2': 0.0, '3': 0.0}, 'Title_1': {'0': 1, '1': 0, '2': 1, '3': 1}, 'Title_2': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Title_3': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Title_4': {'0': 0, '1': 1, '2': 0, '3': 0}, 'Emb_1': {'0': 0, '1': 0, '2': 1, '3': 0}, 'Emb_2': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Emb_3': {'0': 1, '1': 1, '2': 0, '3': 1}}
<end_description>
<start_data_description><data_path>titanic/train_data.csv:
<column_names>
['Unnamed: 0', 'PassengerId', 'Survived', 'Sex', 'Age', 'Fare', 'Pclass_1', 'Pclass_2', 'Pclass_3', 'Family_size', 'Title_1', 'Title_2', 'Title_3', 'Title_4', 'Emb_1', 'Emb_2', 'Emb_3']
<column_types>
{'Unnamed: 0': 'int64', 'PassengerId': 'int64', 'Survived': 'int64', 'Sex': 'int64', 'Age': 'float64', 'Fare': 'float64', 'Pclass_1': 'int64', 'Pclass_2': 'int64', 'Pclass_3': 'int64', 'Family_size': 'float64', 'Title_1': 'int64', 'Title_2': 'int64', 'Title_3': 'int64', 'Title_4': 'int64', 'Emb_1': 'int64', 'Emb_2': 'int64', 'Emb_3': 'int64'}
<dataframe_Summary>
{'Unnamed: 0': {'count': 792.0, 'mean': 395.5, 'std': 228.77499863402906, 'min': 0.0, '25%': 197.75, '50%': 395.5, '75%': 593.25, 'max': 791.0}, 'PassengerId': {'count': 792.0, 'mean': 396.5, 'std': 228.77499863402906, 'min': 1.0, '25%': 198.75, '50%': 396.5, '75%': 594.25, 'max': 792.0}, 'Survived': {'count': 792.0, 'mean': 0.38636363636363635, 'std': 0.4872232622710165, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'Sex': {'count': 792.0, 'mean': 0.6477272727272727, 'std': 0.4779802495415942, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'Age': {'count': 792.0, 'mean': 0.36824368686868686, 'std': 0.162993712818546, 'min': 0.008375, '25%': 0.275, '50%': 0.35, '75%': 0.4375, 'max': 1.0}, 'Fare': {'count': 792.0, 'mean': 0.06467712344746579, 'std': 0.10098746729808539, 'min': 0.0, '25%': 0.0154685698179998, '50%': 0.028302115124416, '75%': 0.0610447345183526, 'max': 1.0}, 'Pclass_1': {'count': 792.0, 'mean': 0.24368686868686867, 'std': 0.4295772101125523, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Pclass_2': {'count': 792.0, 'mean': 0.20833333333333334, 'std': 0.40637306071557894, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Pclass_3': {'count': 792.0, 'mean': 0.547979797979798, 'std': 0.4980071126944576, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'Family_size': {'count': 792.0, 'mean': 0.08863636363636362, 'std': 0.1544851048700475, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.1, 'max': 1.0}, 'Title_1': {'count': 792.0, 'mean': 0.7449494949494949, 'std': 0.4361650454554044, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'Title_2': {'count': 792.0, 'mean': 0.005050505050505051, 'std': 0.07093201085612565, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Title_3': {'count': 792.0, 'mean': 0.04040404040404041, 'std': 0.19702936277118982, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Title_4': {'count': 792.0, 'mean': 0.20959595959595959, 'std': 0.40727746237872897, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Emb_1': {'count': 792.0, 'mean': 0.1856060606060606, 'std': 0.38903411965925394, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Emb_2': {'count': 792.0, 'mean': 0.09217171717171717, 'std': 0.2894509922654781, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'Emb_3': {'count': 792.0, 'mean': 0.7209595959595959, 'std': 0.44881086134701287, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 792 entries, 0 to 791
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 792 non-null int64
1 PassengerId 792 non-null int64
2 Survived 792 non-null int64
3 Sex 792 non-null int64
4 Age 792 non-null float64
5 Fare 792 non-null float64
6 Pclass_1 792 non-null int64
7 Pclass_2 792 non-null int64
8 Pclass_3 792 non-null int64
9 Family_size 792 non-null float64
10 Title_1 792 non-null int64
11 Title_2 792 non-null int64
12 Title_3 792 non-null int64
13 Title_4 792 non-null int64
14 Emb_1 792 non-null int64
15 Emb_2 792 non-null int64
16 Emb_3 792 non-null int64
dtypes: float64(3), int64(14)
memory usage: 105.3 KB
<some_examples>
{'Unnamed: 0': {'0': 0, '1': 1, '2': 2, '3': 3}, 'PassengerId': {'0': 1, '1': 2, '2': 3, '3': 4}, 'Survived': {'0': 0, '1': 1, '2': 1, '3': 1}, 'Sex': {'0': 1, '1': 0, '2': 0, '3': 0}, 'Age': {'0': 0.275, '1': 0.475, '2': 0.325, '3': 0.4375}, 'Fare': {'0': 0.0141510576, '1': 0.1391357354, '2': 0.0154685698, '3': 0.1036442975}, 'Pclass_1': {'0': 0, '1': 1, '2': 0, '3': 1}, 'Pclass_2': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Pclass_3': {'0': 1, '1': 0, '2': 1, '3': 0}, 'Family_size': {'0': 0.1, '1': 0.1, '2': 0.0, '3': 0.1}, 'Title_1': {'0': 1, '1': 1, '2': 0, '3': 1}, 'Title_2': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Title_3': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Title_4': {'0': 0, '1': 0, '2': 1, '3': 0}, 'Emb_1': {'0': 0, '1': 1, '2': 0, '3': 0}, 'Emb_2': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Emb_3': {'0': 1, '1': 0, '2': 1, '3': 1}}
<end_description>
| 5,420 | 0 | 8,390 | 5,420 |
69492463
|
<jupyter_start><jupyter_text>AMZ preprocessed csv
Kaggle dataset identifier: amz-preprocessed-csv
<jupyter_code>import pandas as pd
df = pd.read_csv('amz-preprocessed-csv/amz_df.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 3013799 entries, 0 to 3013798
Data columns (total 7 columns):
# Column Dtype
--- ------ -----
0 index int64
1 TITLE object
2 DESCRIPTION object
3 BULLET_POINTS object
4 BRAND object
5 BROWSE_NODE_ID object
6 PRODUCT_ID object
dtypes: int64(1), object(6)
memory usage: 161.0+ MB
<jupyter_text>Examples:
{
"index": 0,
"TITLE": "pete cat bedtime blues doll inch",
"DESCRIPTION": "pete cat coolest popular cat town new pete cat bedtime blues doll merrymakers rocks striped pj red slippers one sleepy cat ready cuddle measures inches tall safe ages removable clothing surface wash new",
"BULLET_POINTS": "pete cat bedtime blues plush doll based popular pete cat books james dean super cuddly ready naptime bedtime safe ages perfect ages measures inches",
"BRAND": "merrymakers",
"BROWSE_NODE_ID": 0,
"PRODUCT_ID": "error"
}
{
"index": 1,
"TITLE": "new yorker nyhm refrigerator magnet x",
"DESCRIPTION": "new yorker handsome cello wrapped hard magnet measures inch width inch height highlight one many beautiful new yorker covers full color cat tea cup new yorker cover artist gurbuz dogan eksioglu",
"BULLET_POINTS": "cat tea cup new yorker cover artist gurbuz dogan eksioglu handsome cello wrapped hard magnet ideal home office gift new yorker magazine lover highlight one many beautiful new yorker covers full color rigid magnet measures inch width inch height",
"BRAND": "new yorker",
"BROWSE_NODE_ID": 1,
"PRODUCT_ID": "error"
}
{
"index": 2,
"TITLE": "ultimate self sufficiency handbook complete guide baking crafts gardening preserving harvest raising animals",
"DESCRIPTION": "error",
"BULLET_POINTS": "skyhorse publishing",
"BRAND": "imusti",
"BROWSE_NODE_ID": 2,
"PRODUCT_ID": "error"
}
{
"index": 3,
"TITLE": "amway nutrilite kids chewable iron tablets",
"DESCRIPTION": "error",
"BULLET_POINTS": "nutrilite kids chewable iron tablets quantity tablets",
"BRAND": "amway",
"BROWSE_NODE_ID": 3,
"PRODUCT_ID": "error"
}
<jupyter_script>import numpy as np
import pandas as pd
from tqdm import tqdm
tqdm.pandas()
import missingno as msno
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow as tf
from tensorflow.keras import optimizers
import tensorflow_hub as hub
from tensorflow.keras.callbacks import ReduceLROnPlateau, ModelCheckpoint, EarlyStopping
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
from nltk import word_tokenize, RegexpTokenizer, TweetTokenizer, PorterStemmer
from re import sub
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.model_selection import train_test_split
import tensorflow as tf
from sklearn.model_selection import train_test_split
import datetime
from pytz import timezone
df_train = pd.read_csv(
"../input/amz-preprocessed-csv/amz_df.csv", escapechar="\\", quoting=3
)
df_mini = df_train[:2903024]
df_mini.BROWSE_NODE_ID = pd.to_numeric(df_mini.BROWSE_NODE_ID, errors="coerce")
# df_test= pd.read_csv('../input/amz-data/dataset/test.csv', escapechar = "\\", quoting = 3)
# df= pd.concat([df_train, df_test])
# df.reset_index(inplace=True)
# print(df_train.shape, df_test.shape, df.shape)
df_mini.BROWSE_NODE_ID.unique().shape
plt.hist(df_mini.BROWSE_NODE_ID.values, 200)
plt.show()
def reject_outliers(data, m=0.1):
return data[abs(data - np.mean(data)) < m * np.std(data)]
node = reject_outliers(df_mini.BROWSE_NODE_ID.values)
plt.hist(node, 200)
plt.show()
node.shape
sns.boxplot(df_mini.BROWSE_NODE_ID.values)
plt.show()
sns.boxplot(node)
plt.show()
df_mini.loc[df_mini["BROWSE_NODE_ID"] <= 1500]
df_mini.shape
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492463.ipynb
|
amz-preprocessed-csv
|
akhileshdkapse
|
[{"Id": 69492463, "ScriptId": 18975443, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4684168, "CreationDate": "07/31/2021 17:00:26", "VersionNumber": 1.0, "Title": "BROWSE_NODE_ID EDA", "EvaluationDate": NaN, "IsChange": true, "TotalLines": 61.0, "LinesInsertedFromPrevious": 61.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92728608, "KernelVersionId": 69492463, "SourceDatasetVersionId": 2483867}]
|
[{"Id": 2483867, "DatasetId": 1503468, "DatasourceVersionId": 2526419, "CreatorUserId": 4684168, "LicenseName": "Unknown", "CreationDate": "07/31/2021 12:07:02", "VersionNumber": 1.0, "Title": "AMZ preprocessed csv", "Slug": "amz-preprocessed-csv", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1503468, "CreatorUserId": 4684168, "OwnerUserId": 4684168.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2486211.0, "CurrentDatasourceVersionId": 2528772.0, "ForumId": 1523207, "Type": 2, "CreationDate": "07/31/2021 12:07:02", "LastActivityDate": "07/31/2021", "TotalViews": 1426, "TotalDownloads": 11, "TotalVotes": 1, "TotalKernels": 4}]
|
[{"Id": 4684168, "UserName": "akhileshdkapse", "DisplayName": "Akhilesh D. Kapse", "RegisterDate": "03/17/2020", "PerformanceTier": 2}]
|
import numpy as np
import pandas as pd
from tqdm import tqdm
tqdm.pandas()
import missingno as msno
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow as tf
from tensorflow.keras import optimizers
import tensorflow_hub as hub
from tensorflow.keras.callbacks import ReduceLROnPlateau, ModelCheckpoint, EarlyStopping
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
from nltk import word_tokenize, RegexpTokenizer, TweetTokenizer, PorterStemmer
from re import sub
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.model_selection import train_test_split
import tensorflow as tf
from sklearn.model_selection import train_test_split
import datetime
from pytz import timezone
df_train = pd.read_csv(
"../input/amz-preprocessed-csv/amz_df.csv", escapechar="\\", quoting=3
)
df_mini = df_train[:2903024]
df_mini.BROWSE_NODE_ID = pd.to_numeric(df_mini.BROWSE_NODE_ID, errors="coerce")
# df_test= pd.read_csv('../input/amz-data/dataset/test.csv', escapechar = "\\", quoting = 3)
# df= pd.concat([df_train, df_test])
# df.reset_index(inplace=True)
# print(df_train.shape, df_test.shape, df.shape)
df_mini.BROWSE_NODE_ID.unique().shape
plt.hist(df_mini.BROWSE_NODE_ID.values, 200)
plt.show()
def reject_outliers(data, m=0.1):
return data[abs(data - np.mean(data)) < m * np.std(data)]
node = reject_outliers(df_mini.BROWSE_NODE_ID.values)
plt.hist(node, 200)
plt.show()
node.shape
sns.boxplot(df_mini.BROWSE_NODE_ID.values)
plt.show()
sns.boxplot(node)
plt.show()
df_mini.loc[df_mini["BROWSE_NODE_ID"] <= 1500]
df_mini.shape
|
[{"amz-preprocessed-csv/amz_df.csv": {"column_names": "[\"index\", \"TITLE\", \"DESCRIPTION\", \"BULLET_POINTS\", \"BRAND\", \"BROWSE_NODE_ID\", \"PRODUCT_ID\"]", "column_data_types": "{\"index\": \"int64\", \"TITLE\": \"object\", \"DESCRIPTION\": \"object\", \"BULLET_POINTS\": \"object\", \"BRAND\": \"object\", \"BROWSE_NODE_ID\": \"object\", \"PRODUCT_ID\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 3013799 entries, 0 to 3013798\nData columns (total 7 columns):\n # Column Dtype \n--- ------ ----- \n 0 index int64 \n 1 TITLE object\n 2 DESCRIPTION object\n 3 BULLET_POINTS object\n 4 BRAND object\n 5 BROWSE_NODE_ID object\n 6 PRODUCT_ID object\ndtypes: int64(1), object(6)\nmemory usage: 161.0+ MB\n", "summary": "{\"index\": {\"count\": 3013799.0, \"mean\": 1400195.6386942195, \"std\": 863440.7957458469, \"min\": 0.0, \"25%\": 642674.5, \"50%\": 1396124.0, \"75%\": 2149573.5, \"max\": 2903023.0}}", "examples": "{\"index\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"TITLE\":{\"0\":\"pete cat bedtime blues doll inch\",\"1\":\"new yorker nyhm refrigerator magnet x\",\"2\":\"ultimate self sufficiency handbook complete guide baking crafts gardening preserving harvest raising animals\",\"3\":\"amway nutrilite kids chewable iron tablets\"},\"DESCRIPTION\":{\"0\":\"pete cat coolest popular cat town new pete cat bedtime blues doll merrymakers rocks striped pj red slippers one sleepy cat ready cuddle measures inches tall safe ages removable clothing surface wash new\",\"1\":\"new yorker handsome cello wrapped hard magnet measures inch width inch height highlight one many beautiful new yorker covers full color cat tea cup new yorker cover artist gurbuz dogan eksioglu\",\"2\":\"error\",\"3\":\"error\"},\"BULLET_POINTS\":{\"0\":\"pete cat bedtime blues plush doll based popular pete cat books james dean super cuddly ready naptime bedtime safe ages perfect ages measures inches\",\"1\":\"cat tea cup new yorker cover artist gurbuz dogan eksioglu handsome cello wrapped hard magnet ideal home office gift new yorker magazine lover highlight one many beautiful new yorker covers full color rigid magnet measures inch width inch height\",\"2\":\"skyhorse publishing\",\"3\":\"nutrilite kids chewable iron tablets quantity tablets\"},\"BRAND\":{\"0\":\"merrymakers\",\"1\":\"new yorker\",\"2\":\"imusti\",\"3\":\"amway\"},\"BROWSE_NODE_ID\":{\"0\":0.0,\"1\":1.0,\"2\":2.0,\"3\":3.0},\"PRODUCT_ID\":{\"0\":\"error\",\"1\":\"error\",\"2\":\"error\",\"3\":\"error\"}}"}}]
| true | 1 |
<start_data_description><data_path>amz-preprocessed-csv/amz_df.csv:
<column_names>
['index', 'TITLE', 'DESCRIPTION', 'BULLET_POINTS', 'BRAND', 'BROWSE_NODE_ID', 'PRODUCT_ID']
<column_types>
{'index': 'int64', 'TITLE': 'object', 'DESCRIPTION': 'object', 'BULLET_POINTS': 'object', 'BRAND': 'object', 'BROWSE_NODE_ID': 'object', 'PRODUCT_ID': 'object'}
<dataframe_Summary>
{'index': {'count': 3013799.0, 'mean': 1400195.6386942195, 'std': 863440.7957458469, 'min': 0.0, '25%': 642674.5, '50%': 1396124.0, '75%': 2149573.5, 'max': 2903023.0}}
<dataframe_info>
RangeIndex: 3013799 entries, 0 to 3013798
Data columns (total 7 columns):
# Column Dtype
--- ------ -----
0 index int64
1 TITLE object
2 DESCRIPTION object
3 BULLET_POINTS object
4 BRAND object
5 BROWSE_NODE_ID object
6 PRODUCT_ID object
dtypes: int64(1), object(6)
memory usage: 161.0+ MB
<some_examples>
{'index': {'0': 0, '1': 1, '2': 2, '3': 3}, 'TITLE': {'0': 'pete cat bedtime blues doll inch', '1': 'new yorker nyhm refrigerator magnet x', '2': 'ultimate self sufficiency handbook complete guide baking crafts gardening preserving harvest raising animals', '3': 'amway nutrilite kids chewable iron tablets'}, 'DESCRIPTION': {'0': 'pete cat coolest popular cat town new pete cat bedtime blues doll merrymakers rocks striped pj red slippers one sleepy cat ready cuddle measures inches tall safe ages removable clothing surface wash new', '1': 'new yorker handsome cello wrapped hard magnet measures inch width inch height highlight one many beautiful new yorker covers full color cat tea cup new yorker cover artist gurbuz dogan eksioglu', '2': 'error', '3': 'error'}, 'BULLET_POINTS': {'0': 'pete cat bedtime blues plush doll based popular pete cat books james dean super cuddly ready naptime bedtime safe ages perfect ages measures inches', '1': 'cat tea cup new yorker cover artist gurbuz dogan eksioglu handsome cello wrapped hard magnet ideal home office gift new yorker magazine lover highlight one many beautiful new yorker covers full color rigid magnet measures inch width inch height', '2': 'skyhorse publishing', '3': 'nutrilite kids chewable iron tablets quantity tablets'}, 'BRAND': {'0': 'merrymakers', '1': 'new yorker', '2': 'imusti', '3': 'amway'}, 'BROWSE_NODE_ID': {'0': 0.0, '1': 1.0, '2': 2.0, '3': 3.0}, 'PRODUCT_ID': {'0': 'error', '1': 'error', '2': 'error', '3': 'error'}}
<end_description>
| 543 | 0 | 1,295 | 543 |
69492248
|
<jupyter_start><jupyter_text>Data Science - Facens - Exercício 1
<h3 style="text-align: center">Visualização de Dados - Especialização em Ciência de Dados - FACENS</h3>
Instruções:
1. Clonar o notebook-template: https://www.kaggle.com/matheusmota/template-exerc-cio-1
2. Enviar o link do notebook no portal da disciplina.
Observação: Caso prefira não deixar o notebook público, mantenha-o privado, compartilhe com o professor e faça a submissão do link no portal. Apenas o link será aceito.
Kaggle dataset identifier: dataviz-facens-20182-aula-1-exerccio-2
<jupyter_code>import pandas as pd
df = pd.read_csv('dataviz-facens-20182-aula-1-exerccio-2/BR_eleitorado_2016_municipio.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 5568 entries, 0 to 5567
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 cod_municipio_tse 5568 non-null int64
1 uf 5568 non-null object
2 nome_municipio 5568 non-null object
3 total_eleitores 5568 non-null int64
4 f_16 5568 non-null int64
5 f_17 5568 non-null int64
6 f_18_20 5568 non-null int64
7 f_21_24 5568 non-null int64
8 f_25_34 5568 non-null int64
9 f_35_44 5568 non-null int64
10 f_45_59 5568 non-null int64
11 f_60_69 5568 non-null int64
12 f_70_79 5568 non-null int64
13 f_sup_79 5568 non-null int64
14 gen_feminino 5568 non-null int64
15 gen_masculino 5568 non-null int64
16 gen_nao_informado 5568 non-null int64
dtypes: int64(15), object(2)
memory usage: 739.6+ KB
<jupyter_text>Examples:
{
"cod_municipio_tse": 16691,
"uf": "RN",
"nome_municipio": "ESP\u00cdRITO SANTO",
"total_eleitores": 6435,
"f_16": 120,
"f_17": 155,
"f_18_20": 492,
"f_21_24": 627,
"f_25_34": 1482,
"f_35_44": 1233,
"f_45_59": 1363,
"f_60_69": 579,
"f_70_79": 322,
"f_sup_79": 62,
"gen_feminino": 3353,
"gen_masculino": 3082,
"gen_nao_informado": 0
}
{
"cod_municipio_tse": 58033,
"uf": "RJ",
"nome_municipio": "ARARUAMA",
"total_eleitores": 92990,
"f_16": 193,
"f_17": 524,
"f_18_20": 4584,
"f_21_24": 7704,
"f_25_34": 18136,
"f_35_44": 17749,
"f_45_59": 23552,
"f_60_69": 11185,
"f_70_79": 5988,
"f_sup_79": 3371,
"gen_feminino": 49435,
"gen_masculino": 43407,
"gen_nao_informado": 148
}
{
"cod_municipio_tse": 58386,
"uf": "RJ",
"nome_municipio": "MACUCO",
"total_eleitores": 7113,
"f_16": 88,
"f_17": 123,
"f_18_20": 419,
"f_21_24": 544,
"f_25_34": 1533,
"f_35_44": 1461,
"f_45_59": 1646,
"f_60_69": 776,
"f_70_79": 356,
"f_sup_79": 166,
"gen_feminino": 3641,
"gen_masculino": 3461,
"gen_nao_informado": 11
}
{
"cod_municipio_tse": 6130,
"uf": "AP",
"nome_municipio": "LARANJAL DO JARI",
"total_eleitores": 27718,
"f_16": 509,
"f_17": 707,
"f_18_20": 2710,
"f_21_24": 3314,
"f_25_34": 7041,
"f_35_44": 5643,
"f_45_59": 5343,
"f_60_69": 1572,
"f_70_79": 660,
"f_sup_79": 219,
"gen_feminino": 13583,
"gen_masculino": 14135,
"gen_nao_informado": 0
}
<jupyter_script>#
#
# Especialização em Ciência de Dados - FACENS
# Visualização de Dados
# # Exercício 1 - Variáveis
# (valendo nota)
# * **Data de entrega:** Verifique o prazo na página da disciplina
# * **Professor:** Matheus Mota
# * **Aluno:** Alison Carlos Santos
# * **RA:** 202064
# ## Questão Única
# **Enunciado:** Este notebook está associado ao *Kaggle Dataset* chamado "Exercício 1". Este *Kaggle Dataset* possui dois arquivos em formato CSV (anv.csv e BR_eleitorado_2016_municipio ). Escolha um dos datasets disponíveis e já conhecidos, a seu critério. Uma vez definido o csv, escolha no mínimo 7 e no máximo 12 variáveis (colunas) que você avalia como sendo relevantes. Para cada uma das suas variáveis escolhidas, forneça:
# ### Item A - Classificação das variáveis
# Classifique todas as variáveis escolhidas, e construa um dataframe com sua resposta.
# Exemplo:
# **Importando as libs utilizadas neste notebook**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
# **Verificando o dataset.**
df = pd.read_csv(
"/kaggle/input/dataviz-facens-20182-aula-1-exerccio-2/BR_eleitorado_2016_municipio.csv",
delimiter=",",
)
df.sample(10)
# **Selecionando as váriaveis**
df = df[
[
"uf",
"nome_municipio",
"total_eleitores",
"f_25_34",
"gen_feminino",
"gen_masculino",
"gen_nao_informado",
]
]
df.sample(5)
variable_classification = [
["uf", "Qualitativa Nominal"],
["nome_municipio", "Qualitativa Nominal"],
["total_eleitores", "Quantitativa Discreta"],
["f_25_34", "Quantitativa Discreta"],
["gen_feminino", "Quantitativa Discreta"],
["gen_masculino", "Quantitativa Discreta"],
["gen_nao_informado", "Quantitativa Discreta"],
]
variable_classification = pd.DataFrame(
variable_classification, columns=["Variável", "Classificação"]
)
variable_classification
# ### Item B - Tabela de frequência
# Construa uma tabela de frequência para cada uma das **variáveis qualitativas** que você escolheu (caso não tenha escolhido nenhuma, deixe esta questão em branco). Uma dica: a função *value_counts()* do Pandas pode ser muito útil. =)
# # **Frequência das Unidades Federativas no Dataset**
df["uf"].value_counts()
# # **Frequência dos Município no Dataset**
df["nome_municipio"].value_counts()
# ### Item C - Representação Gráfica
# Para cada uma das variáveis, produza um ou mais gráficos, usando matplotlib ou Pandas plot, que descreva seu comportamento / característica. Lembre-se que o(s) gráfico(s) precisa(m) ser compatível(eis) com a classificação da variável.
uf = df.groupby("uf")["total_eleitores"].sum().sort_values(ascending=False)
# Parametros do gráfico
fig, ax = plt.subplots(figsize=(20, 10))
ax.set_ylabel("Total de eleitores (em milhões)", fontsize="15")
ax.set_xlabel("UF", fontsize="15")
plt.title(
"Quantidade de eleitores por UF",
fontweight="bold",
color="black",
fontsize="20",
horizontalalignment="center",
)
plt.bar(uf.index, uf.values, color="orange")
df.sample(5)
nome_municipio = (
df.groupby("nome_municipio")[["gen_feminino", "gen_masculino"]]
.sum()
.sort_values(by="nome_municipio", ascending=True)
)
sorocaba = nome_municipio.loc["SOROCABA"]
plt.title(
"Distribuição de eleitores por gênero no município de Sorocaba",
fontweight="bold",
color="black",
fontsize="15",
horizontalalignment="center",
)
plt.pie(sorocaba, labels=["Feminino", "Masculino"], autopct="%1.2f%%")
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492248.ipynb
|
dataviz-facens-20182-aula-1-exerccio-2
|
matheusmota
|
[{"Id": 69492248, "ScriptId": 18973569, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6118182, "CreationDate": "07/31/2021 16:57:18", "VersionNumber": 5.0, "Title": "Exerc\u00edcio 1 - Alison Carlos Santos - 202064", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 96.0, "LinesInsertedFromPrevious": 15.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 81.0, "LinesInsertedFromFork": 60.0, "LinesDeletedFromFork": 13.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 36.0, "TotalVotes": 0}]
|
[{"Id": 92727945, "KernelVersionId": 69492248, "SourceDatasetVersionId": 2456609}]
|
[{"Id": 2456609, "DatasetId": 212085, "DatasourceVersionId": 2499007, "CreatorUserId": 2201395, "LicenseName": "Unknown", "CreationDate": "07/24/2021 00:23:56", "VersionNumber": 3.0, "Title": "Data Science - Facens - Exerc\u00edcio 1", "Slug": "dataviz-facens-20182-aula-1-exerccio-2", "Subtitle": "Classifica\u00e7\u00e3o de Vari\u00e1veis e visualiza\u00e7\u00f5es b\u00e1sicas de vari\u00e1veis", "Description": "<h3 style=\"text-align: center\">Visualiza\u00e7\u00e3o de Dados - Especializa\u00e7\u00e3o em Ci\u00eancia de Dados - FACENS</h3>\nInstru\u00e7\u00f5es:\n1. Clonar o notebook-template: https://www.kaggle.com/matheusmota/template-exerc-cio-1\n2. Enviar o link do notebook no portal da disciplina.\n\nObserva\u00e7\u00e3o: Caso prefira n\u00e3o deixar o notebook p\u00fablico, mantenha-o privado, compartilhe com o professor e fa\u00e7a a submiss\u00e3o do link no portal. Apenas o link ser\u00e1 aceito.", "VersionNotes": "Novo dataset", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 212085, "CreatorUserId": 2201395, "OwnerUserId": 2201395.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2456609.0, "CurrentDatasourceVersionId": 2499007.0, "ForumId": 223155, "Type": 2, "CreationDate": "05/31/2019 21:14:53", "LastActivityDate": "05/31/2019", "TotalViews": 3901, "TotalDownloads": 115, "TotalVotes": 7, "TotalKernels": 70}]
|
[{"Id": 2201395, "UserName": "matheusmota", "DisplayName": "Matheus Mota", "RegisterDate": "09/01/2018", "PerformanceTier": 0}]
|
#
#
# Especialização em Ciência de Dados - FACENS
# Visualização de Dados
# # Exercício 1 - Variáveis
# (valendo nota)
# * **Data de entrega:** Verifique o prazo na página da disciplina
# * **Professor:** Matheus Mota
# * **Aluno:** Alison Carlos Santos
# * **RA:** 202064
# ## Questão Única
# **Enunciado:** Este notebook está associado ao *Kaggle Dataset* chamado "Exercício 1". Este *Kaggle Dataset* possui dois arquivos em formato CSV (anv.csv e BR_eleitorado_2016_municipio ). Escolha um dos datasets disponíveis e já conhecidos, a seu critério. Uma vez definido o csv, escolha no mínimo 7 e no máximo 12 variáveis (colunas) que você avalia como sendo relevantes. Para cada uma das suas variáveis escolhidas, forneça:
# ### Item A - Classificação das variáveis
# Classifique todas as variáveis escolhidas, e construa um dataframe com sua resposta.
# Exemplo:
# **Importando as libs utilizadas neste notebook**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
# **Verificando o dataset.**
df = pd.read_csv(
"/kaggle/input/dataviz-facens-20182-aula-1-exerccio-2/BR_eleitorado_2016_municipio.csv",
delimiter=",",
)
df.sample(10)
# **Selecionando as váriaveis**
df = df[
[
"uf",
"nome_municipio",
"total_eleitores",
"f_25_34",
"gen_feminino",
"gen_masculino",
"gen_nao_informado",
]
]
df.sample(5)
variable_classification = [
["uf", "Qualitativa Nominal"],
["nome_municipio", "Qualitativa Nominal"],
["total_eleitores", "Quantitativa Discreta"],
["f_25_34", "Quantitativa Discreta"],
["gen_feminino", "Quantitativa Discreta"],
["gen_masculino", "Quantitativa Discreta"],
["gen_nao_informado", "Quantitativa Discreta"],
]
variable_classification = pd.DataFrame(
variable_classification, columns=["Variável", "Classificação"]
)
variable_classification
# ### Item B - Tabela de frequência
# Construa uma tabela de frequência para cada uma das **variáveis qualitativas** que você escolheu (caso não tenha escolhido nenhuma, deixe esta questão em branco). Uma dica: a função *value_counts()* do Pandas pode ser muito útil. =)
# # **Frequência das Unidades Federativas no Dataset**
df["uf"].value_counts()
# # **Frequência dos Município no Dataset**
df["nome_municipio"].value_counts()
# ### Item C - Representação Gráfica
# Para cada uma das variáveis, produza um ou mais gráficos, usando matplotlib ou Pandas plot, que descreva seu comportamento / característica. Lembre-se que o(s) gráfico(s) precisa(m) ser compatível(eis) com a classificação da variável.
uf = df.groupby("uf")["total_eleitores"].sum().sort_values(ascending=False)
# Parametros do gráfico
fig, ax = plt.subplots(figsize=(20, 10))
ax.set_ylabel("Total de eleitores (em milhões)", fontsize="15")
ax.set_xlabel("UF", fontsize="15")
plt.title(
"Quantidade de eleitores por UF",
fontweight="bold",
color="black",
fontsize="20",
horizontalalignment="center",
)
plt.bar(uf.index, uf.values, color="orange")
df.sample(5)
nome_municipio = (
df.groupby("nome_municipio")[["gen_feminino", "gen_masculino"]]
.sum()
.sort_values(by="nome_municipio", ascending=True)
)
sorocaba = nome_municipio.loc["SOROCABA"]
plt.title(
"Distribuição de eleitores por gênero no município de Sorocaba",
fontweight="bold",
color="black",
fontsize="15",
horizontalalignment="center",
)
plt.pie(sorocaba, labels=["Feminino", "Masculino"], autopct="%1.2f%%")
plt.show()
|
[{"dataviz-facens-20182-aula-1-exerccio-2/BR_eleitorado_2016_municipio.csv": {"column_names": "[\"cod_municipio_tse\", \"uf\", \"nome_municipio\", \"total_eleitores\", \"f_16\", \"f_17\", \"f_18_20\", \"f_21_24\", \"f_25_34\", \"f_35_44\", \"f_45_59\", \"f_60_69\", \"f_70_79\", \"f_sup_79\", \"gen_feminino\", \"gen_masculino\", \"gen_nao_informado\"]", "column_data_types": "{\"cod_municipio_tse\": \"int64\", \"uf\": \"object\", \"nome_municipio\": \"object\", \"total_eleitores\": \"int64\", \"f_16\": \"int64\", \"f_17\": \"int64\", \"f_18_20\": \"int64\", \"f_21_24\": \"int64\", \"f_25_34\": \"int64\", \"f_35_44\": \"int64\", \"f_45_59\": \"int64\", \"f_60_69\": \"int64\", \"f_70_79\": \"int64\", \"f_sup_79\": \"int64\", \"gen_feminino\": \"int64\", \"gen_masculino\": \"int64\", \"gen_nao_informado\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 5568 entries, 0 to 5567\nData columns (total 17 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 cod_municipio_tse 5568 non-null int64 \n 1 uf 5568 non-null object\n 2 nome_municipio 5568 non-null object\n 3 total_eleitores 5568 non-null int64 \n 4 f_16 5568 non-null int64 \n 5 f_17 5568 non-null int64 \n 6 f_18_20 5568 non-null int64 \n 7 f_21_24 5568 non-null int64 \n 8 f_25_34 5568 non-null int64 \n 9 f_35_44 5568 non-null int64 \n 10 f_45_59 5568 non-null int64 \n 11 f_60_69 5568 non-null int64 \n 12 f_70_79 5568 non-null int64 \n 13 f_sup_79 5568 non-null int64 \n 14 gen_feminino 5568 non-null int64 \n 15 gen_masculino 5568 non-null int64 \n 16 gen_nao_informado 5568 non-null int64 \ndtypes: int64(15), object(2)\nmemory usage: 739.6+ KB\n", "summary": "{\"cod_municipio_tse\": {\"count\": 5568.0, \"mean\": 51465.073096264365, \"std\": 29372.607384834984, \"min\": 19.0, \"25%\": 24248.0, \"50%\": 51241.5, \"75%\": 79202.5, \"max\": 99074.0}, \"total_eleitores\": {\"count\": 5568.0, \"mean\": 25878.037356321838, \"std\": 154825.6362859667, \"min\": 954.0, \"25%\": 4620.0, \"50%\": 8831.5, \"75%\": 18254.25, \"max\": 8886324.0}, \"f_16\": {\"count\": 5568.0, \"mean\": 149.664691091954, \"std\": 258.21273028083925, \"min\": 2.0, \"25%\": 49.0, \"50%\": 90.0, \"75%\": 177.0, \"max\": 11402.0}, \"f_17\": {\"count\": 5568.0, \"mean\": 265.40714798850576, \"std\": 783.6650434366487, \"min\": 8.0, \"25%\": 72.0, \"50%\": 136.0, \"75%\": 272.0, \"max\": 40888.0}, \"f_18_20\": {\"count\": 5568.0, \"mean\": 1510.9288793103449, \"std\": 7733.040439804812, \"min\": 33.0, \"25%\": 277.0, \"50%\": 550.0, \"75%\": 1159.75, \"max\": 439186.0}, \"f_21_24\": {\"count\": 5568.0, \"mean\": 2253.5463362068967, \"std\": 12048.388015821112, \"min\": 63.0, \"25%\": 391.75, \"50%\": 785.5, \"75%\": 1670.5, \"max\": 677646.0}, \"f_25_34\": {\"count\": 5568.0, \"mean\": 5707.740481321839, \"std\": 32431.034529935587, \"min\": 156.0, \"25%\": 971.0, \"50%\": 1924.0, \"75%\": 4068.0, \"max\": 1866143.0}, \"f_35_44\": {\"count\": 5568.0, \"mean\": 5191.82507183908, \"std\": 31791.957161891016, \"min\": 167.0, \"25%\": 854.0, \"50%\": 1659.5, \"75%\": 3552.25, \"max\": 1849841.0}, \"f_45_59\": {\"count\": 5568.0, \"mean\": 6164.22665229885, \"std\": 38688.819190422364, \"min\": 231.0, \"25%\": 1066.0, \"50%\": 1986.5, \"75%\": 4115.75, \"max\": 2226691.0}, \"f_60_69\": {\"count\": 5568.0, \"mean\": 2595.044540229885, \"std\": 16947.823715650822, \"min\": 96.0, \"25%\": 467.0, \"50%\": 870.0, \"75%\": 1763.5, \"max\": 955342.0}, \"f_70_79\": {\"count\": 5568.0, \"mean\": 1310.7106681034484, \"std\": 8599.37348724144, \"min\": 40.0, \"25%\": 259.0, \"50%\": 487.0, \"75%\": 978.0, \"max\": 482908.0}, \"f_sup_79\": {\"count\": 5568.0, \"mean\": 728.2374281609195, \"std\": 6375.566777203154, \"min\": 7.0, \"25%\": 107.0, \"50%\": 232.0, \"75%\": 533.0, \"max\": 336149.0}, \"gen_feminino\": {\"count\": 5568.0, \"mean\": 13510.426724137931, \"std\": 83594.37051569676, \"min\": 436.0, \"25%\": 2272.25, \"50%\": 4412.0, \"75%\": 9312.0, \"max\": 4783972.0}, \"gen_masculino\": {\"count\": 5568.0, \"mean\": 12350.508979885057, \"std\": 71050.02536999786, \"min\": 513.0, \"25%\": 2344.75, \"50%\": 4407.0, \"75%\": 8980.75, \"max\": 4089081.0}, \"gen_nao_informado\": {\"count\": 5568.0, \"mean\": 17.101652298850574, \"std\": 205.70460842832563, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 7.0, \"max\": 13271.0}}", "examples": "{\"cod_municipio_tse\":{\"0\":16691,\"1\":58033,\"2\":58386,\"3\":6130},\"uf\":{\"0\":\"RN\",\"1\":\"RJ\",\"2\":\"RJ\",\"3\":\"AP\"},\"nome_municipio\":{\"0\":\"ESP\\u00cdRITO SANTO\",\"1\":\"ARARUAMA\",\"2\":\"MACUCO\",\"3\":\"LARANJAL DO JARI\"},\"total_eleitores\":{\"0\":6435,\"1\":92990,\"2\":7113,\"3\":27718},\"f_16\":{\"0\":120,\"1\":193,\"2\":88,\"3\":509},\"f_17\":{\"0\":155,\"1\":524,\"2\":123,\"3\":707},\"f_18_20\":{\"0\":492,\"1\":4584,\"2\":419,\"3\":2710},\"f_21_24\":{\"0\":627,\"1\":7704,\"2\":544,\"3\":3314},\"f_25_34\":{\"0\":1482,\"1\":18136,\"2\":1533,\"3\":7041},\"f_35_44\":{\"0\":1233,\"1\":17749,\"2\":1461,\"3\":5643},\"f_45_59\":{\"0\":1363,\"1\":23552,\"2\":1646,\"3\":5343},\"f_60_69\":{\"0\":579,\"1\":11185,\"2\":776,\"3\":1572},\"f_70_79\":{\"0\":322,\"1\":5988,\"2\":356,\"3\":660},\"f_sup_79\":{\"0\":62,\"1\":3371,\"2\":166,\"3\":219},\"gen_feminino\":{\"0\":3353,\"1\":49435,\"2\":3641,\"3\":13583},\"gen_masculino\":{\"0\":3082,\"1\":43407,\"2\":3461,\"3\":14135},\"gen_nao_informado\":{\"0\":0,\"1\":148,\"2\":11,\"3\":0}}"}}]
| true | 1 |
<start_data_description><data_path>dataviz-facens-20182-aula-1-exerccio-2/BR_eleitorado_2016_municipio.csv:
<column_names>
['cod_municipio_tse', 'uf', 'nome_municipio', 'total_eleitores', 'f_16', 'f_17', 'f_18_20', 'f_21_24', 'f_25_34', 'f_35_44', 'f_45_59', 'f_60_69', 'f_70_79', 'f_sup_79', 'gen_feminino', 'gen_masculino', 'gen_nao_informado']
<column_types>
{'cod_municipio_tse': 'int64', 'uf': 'object', 'nome_municipio': 'object', 'total_eleitores': 'int64', 'f_16': 'int64', 'f_17': 'int64', 'f_18_20': 'int64', 'f_21_24': 'int64', 'f_25_34': 'int64', 'f_35_44': 'int64', 'f_45_59': 'int64', 'f_60_69': 'int64', 'f_70_79': 'int64', 'f_sup_79': 'int64', 'gen_feminino': 'int64', 'gen_masculino': 'int64', 'gen_nao_informado': 'int64'}
<dataframe_Summary>
{'cod_municipio_tse': {'count': 5568.0, 'mean': 51465.073096264365, 'std': 29372.607384834984, 'min': 19.0, '25%': 24248.0, '50%': 51241.5, '75%': 79202.5, 'max': 99074.0}, 'total_eleitores': {'count': 5568.0, 'mean': 25878.037356321838, 'std': 154825.6362859667, 'min': 954.0, '25%': 4620.0, '50%': 8831.5, '75%': 18254.25, 'max': 8886324.0}, 'f_16': {'count': 5568.0, 'mean': 149.664691091954, 'std': 258.21273028083925, 'min': 2.0, '25%': 49.0, '50%': 90.0, '75%': 177.0, 'max': 11402.0}, 'f_17': {'count': 5568.0, 'mean': 265.40714798850576, 'std': 783.6650434366487, 'min': 8.0, '25%': 72.0, '50%': 136.0, '75%': 272.0, 'max': 40888.0}, 'f_18_20': {'count': 5568.0, 'mean': 1510.9288793103449, 'std': 7733.040439804812, 'min': 33.0, '25%': 277.0, '50%': 550.0, '75%': 1159.75, 'max': 439186.0}, 'f_21_24': {'count': 5568.0, 'mean': 2253.5463362068967, 'std': 12048.388015821112, 'min': 63.0, '25%': 391.75, '50%': 785.5, '75%': 1670.5, 'max': 677646.0}, 'f_25_34': {'count': 5568.0, 'mean': 5707.740481321839, 'std': 32431.034529935587, 'min': 156.0, '25%': 971.0, '50%': 1924.0, '75%': 4068.0, 'max': 1866143.0}, 'f_35_44': {'count': 5568.0, 'mean': 5191.82507183908, 'std': 31791.957161891016, 'min': 167.0, '25%': 854.0, '50%': 1659.5, '75%': 3552.25, 'max': 1849841.0}, 'f_45_59': {'count': 5568.0, 'mean': 6164.22665229885, 'std': 38688.819190422364, 'min': 231.0, '25%': 1066.0, '50%': 1986.5, '75%': 4115.75, 'max': 2226691.0}, 'f_60_69': {'count': 5568.0, 'mean': 2595.044540229885, 'std': 16947.823715650822, 'min': 96.0, '25%': 467.0, '50%': 870.0, '75%': 1763.5, 'max': 955342.0}, 'f_70_79': {'count': 5568.0, 'mean': 1310.7106681034484, 'std': 8599.37348724144, 'min': 40.0, '25%': 259.0, '50%': 487.0, '75%': 978.0, 'max': 482908.0}, 'f_sup_79': {'count': 5568.0, 'mean': 728.2374281609195, 'std': 6375.566777203154, 'min': 7.0, '25%': 107.0, '50%': 232.0, '75%': 533.0, 'max': 336149.0}, 'gen_feminino': {'count': 5568.0, 'mean': 13510.426724137931, 'std': 83594.37051569676, 'min': 436.0, '25%': 2272.25, '50%': 4412.0, '75%': 9312.0, 'max': 4783972.0}, 'gen_masculino': {'count': 5568.0, 'mean': 12350.508979885057, 'std': 71050.02536999786, 'min': 513.0, '25%': 2344.75, '50%': 4407.0, '75%': 8980.75, 'max': 4089081.0}, 'gen_nao_informado': {'count': 5568.0, 'mean': 17.101652298850574, 'std': 205.70460842832563, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 7.0, 'max': 13271.0}}
<dataframe_info>
RangeIndex: 5568 entries, 0 to 5567
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 cod_municipio_tse 5568 non-null int64
1 uf 5568 non-null object
2 nome_municipio 5568 non-null object
3 total_eleitores 5568 non-null int64
4 f_16 5568 non-null int64
5 f_17 5568 non-null int64
6 f_18_20 5568 non-null int64
7 f_21_24 5568 non-null int64
8 f_25_34 5568 non-null int64
9 f_35_44 5568 non-null int64
10 f_45_59 5568 non-null int64
11 f_60_69 5568 non-null int64
12 f_70_79 5568 non-null int64
13 f_sup_79 5568 non-null int64
14 gen_feminino 5568 non-null int64
15 gen_masculino 5568 non-null int64
16 gen_nao_informado 5568 non-null int64
dtypes: int64(15), object(2)
memory usage: 739.6+ KB
<some_examples>
{'cod_municipio_tse': {'0': 16691, '1': 58033, '2': 58386, '3': 6130}, 'uf': {'0': 'RN', '1': 'RJ', '2': 'RJ', '3': 'AP'}, 'nome_municipio': {'0': 'ESPÍRITO SANTO', '1': 'ARARUAMA', '2': 'MACUCO', '3': 'LARANJAL DO JARI'}, 'total_eleitores': {'0': 6435, '1': 92990, '2': 7113, '3': 27718}, 'f_16': {'0': 120, '1': 193, '2': 88, '3': 509}, 'f_17': {'0': 155, '1': 524, '2': 123, '3': 707}, 'f_18_20': {'0': 492, '1': 4584, '2': 419, '3': 2710}, 'f_21_24': {'0': 627, '1': 7704, '2': 544, '3': 3314}, 'f_25_34': {'0': 1482, '1': 18136, '2': 1533, '3': 7041}, 'f_35_44': {'0': 1233, '1': 17749, '2': 1461, '3': 5643}, 'f_45_59': {'0': 1363, '1': 23552, '2': 1646, '3': 5343}, 'f_60_69': {'0': 579, '1': 11185, '2': 776, '3': 1572}, 'f_70_79': {'0': 322, '1': 5988, '2': 356, '3': 660}, 'f_sup_79': {'0': 62, '1': 3371, '2': 166, '3': 219}, 'gen_feminino': {'0': 3353, '1': 49435, '2': 3641, '3': 13583}, 'gen_masculino': {'0': 3082, '1': 43407, '2': 3461, '3': 14135}, 'gen_nao_informado': {'0': 0, '1': 148, '2': 11, '3': 0}}
<end_description>
| 1,180 | 0 | 2,887 | 1,180 |
69492590
|
<jupyter_start><jupyter_text>Telco Customer Churn
### Context
"Predict behavior to retain customers. You can analyze all relevant customer data and develop focused customer retention programs." [IBM Sample Data Sets]
### Content
Each row represents a customer, each column contains customer’s attributes described on the column Metadata.
**The data set includes information about:**
+ Customers who left within the last month – the column is called Churn
+ Services that each customer has signed up for – phone, multiple lines, internet, online security, online backup, device protection, tech support, and streaming TV and movies
+ Customer account information – how long they’ve been a customer, contract, payment method, paperless billing, monthly charges, and total charges
+ Demographic info about customers – gender, age range, and if they have partners and dependents
### Inspiration
To explore this type of models and learn more about the subject.
**New version from IBM:**
https://community.ibm.com/community/user/businessanalytics/blogs/steven-macko/2019/07/11/telco-customer-churn-1113
Kaggle dataset identifier: telco-customer-churn
<jupyter_code>import pandas as pd
df = pd.read_csv('telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 7043 entries, 0 to 7042
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 customerID 7043 non-null object
1 gender 7043 non-null object
2 SeniorCitizen 7043 non-null int64
3 Partner 7043 non-null object
4 Dependents 7043 non-null object
5 tenure 7043 non-null int64
6 PhoneService 7043 non-null object
7 MultipleLines 7043 non-null object
8 InternetService 7043 non-null object
9 OnlineSecurity 7043 non-null object
10 OnlineBackup 7043 non-null object
11 DeviceProtection 7043 non-null object
12 TechSupport 7043 non-null object
13 StreamingTV 7043 non-null object
14 StreamingMovies 7043 non-null object
15 Contract 7043 non-null object
16 PaperlessBilling 7043 non-null object
17 PaymentMethod 7043 non-null object
18 MonthlyCharges 7043 non-null float64
19 TotalCharges 7043 non-null object
20 Churn 7043 non-null object
dtypes: float64(1), int64(2), object(18)
memory usage: 1.1+ MB
<jupyter_text>Examples:
{
"customerID": "7590-VHVEG",
"gender": "Female",
"SeniorCitizen": 0,
"Partner": "Yes",
"Dependents": "No",
"tenure": 1,
"PhoneService": "No",
"MultipleLines": "No phone service",
"InternetService": "DSL",
"OnlineSecurity": "No",
"OnlineBackup": "Yes",
"DeviceProtection": "No",
"TechSupport": "No",
"StreamingTV": "No",
"StreamingMovies": "No",
"Contract": "Month-to-month",
"PaperlessBilling": "Yes",
"PaymentMethod": "Electronic check",
"MonthlyCharges": 29.85,
"TotalCharges": 29.85,
"...": "and 1 more columns"
}
{
"customerID": "5575-GNVDE",
"gender": "Male",
"SeniorCitizen": 0,
"Partner": "No",
"Dependents": "No",
"tenure": 34,
"PhoneService": "Yes",
"MultipleLines": "No",
"InternetService": "DSL",
"OnlineSecurity": "Yes",
"OnlineBackup": "No",
"DeviceProtection": "Yes",
"TechSupport": "No",
"StreamingTV": "No",
"StreamingMovies": "No",
"Contract": "One year",
"PaperlessBilling": "No",
"PaymentMethod": "Mailed check",
"MonthlyCharges": 56.95,
"TotalCharges": 1889.5,
"...": "and 1 more columns"
}
{
"customerID": "3668-QPYBK",
"gender": "Male",
"SeniorCitizen": 0,
"Partner": "No",
"Dependents": "No",
"tenure": 2,
"PhoneService": "Yes",
"MultipleLines": "No",
"InternetService": "DSL",
"OnlineSecurity": "Yes",
"OnlineBackup": "Yes",
"DeviceProtection": "No",
"TechSupport": "No",
"StreamingTV": "No",
"StreamingMovies": "No",
"Contract": "Month-to-month",
"PaperlessBilling": "Yes",
"PaymentMethod": "Mailed check",
"MonthlyCharges": 53.85,
"TotalCharges": 108.15,
"...": "and 1 more columns"
}
{
"customerID": "7795-CFOCW",
"gender": "Male",
"SeniorCitizen": 0,
"Partner": "No",
"Dependents": "No",
"tenure": 45,
"PhoneService": "No",
"MultipleLines": "No phone service",
"InternetService": "DSL",
"OnlineSecurity": "Yes",
"OnlineBackup": "No",
"DeviceProtection": "Yes",
"TechSupport": "Yes",
"StreamingTV": "No",
"StreamingMovies": "No",
"Contract": "One year",
"PaperlessBilling": "No",
"PaymentMethod": "Bank transfer (automatic)",
"MonthlyCharges": 42.3,
"TotalCharges": 1840.75,
"...": "and 1 more columns"
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Installation of required libraries
import missingno as msno
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.ensemble import (
RandomForestClassifier,
GradientBoostingClassifier,
VotingClassifier,
AdaBoostClassifier,
)
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import cross_validate, GridSearchCV
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.preprocessing import StandardScaler, LabelEncoder
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
from sklearn.ensemble import GradientBoostingClassifier
from catboost import CatBoostClassifier
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.metrics import accuracy_score
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score, GridSearchCV
import warnings
warnings.filterwarnings("ignore", category=DeprecationWarning)
warnings.filterwarnings("ignore", category=FutureWarning)
warnings.filterwarnings("ignore", category=UserWarning)
# 1. Introduction
# Customer churn is defined as when customers or subscribers discontinue doing business with a firm or service.
# Customers in the telecom industry can choose from a variety of service providers and actively switch from one to the next. The telecommunications business has an annual churn rate of 15-25 percent in this highly competitive market.
# Individualized customer retention is tough because most firms have a large number of customers and can't afford to devote much time to each of them. The costs would be too great, outweighing the additional revenue. However, if a corporation could forecast which customers are likely to leave ahead of time, it could focus customer retention efforts only on these "high risk" clients. The ultimate goal is to expand its coverage area and retrieve more customers loyalty. The core to succeed in this market lies in the customer itself.
# Customer churn is a critical metric because it is much less expensive to retain existing customers than it is to acquire new customers.
# To reduce customer churn, telecom companies need to predict which customers are at high risk of churn.
# To detect early signs of potential churn, one must first develop a holistic view of the customers and their interactions across numerous channels, including store/branch visits, product purchase histories, customer service calls, Web-based transactions, and social media interactions, to mention a few.
# As a result, by addressing churn, these businesses may not only preserve their market position, but also grow and thrive. More customers they have in their network, the lower the cost of initiation and the larger the profit. As a result, the company's key focus for success is reducing client attrition and implementing effective retention strategy.
# ### Data Description
# * Context
# * "Predict behavior to retain customers. You can analyze all relevant customer data and develop focused customer retention programs."
# * Content
# * Each row represents a customer, each column contains customer’s attributes described on the column Metadata.
# * The data set includes information about:
# * Customers who left within the last month – the column is called Churn
# * Services that each customer has signed up for – phone, multiple lines, internet, online security, online backup, device protection, tech support, and streaming TV and movies
# * Customer account information – how long they’ve been a customer, contract, payment method, paperless billing, monthly charges, and total charges
# * Demographic info about customers – gender, age range, and if they have partners and dependents
# * 1. Exploratory Data Analysis
# * 2. Data Preprocessing & Feature Engineering
# * 3. Modeling
# * 4. Feature Selection
# * 5. Hyperparameter Optimization with Selected Features
# * 6. Submitting the results
# * Customer ID
# * Gender - Whether the customer is a male or a female
# * SeniorCitizen - Whether the customer is a senior citizen or not (1, 0)
# * Partner - Whether the customer has a partner or not (Yes, No)
# * Dependents - Whether the customer has dependents or not (Yes, No)
# * tenure - Number of months the customer has stayed with the company
# * PhoneService - Whether the customer has a phone service or not (Yes, No)
# * MultipleLines - Whether the customer has multiple lines or not (Yes, No, No phone service)
# * InternetService - Customer’s internet service provider (DSL, Fiber optic, No)
# * OnlineSecurity - Whether the customer has online security or not (Yes, No, No internet service)
# * OnlineBackup - Whether the customer has online backup or not (Yes, No, No internet service)
# * DeviceProtection - Whether the customer has device protection or not (Yes, No, No internet service)
# * TechSupport - Whether the customer has tech support or not (Yes, No, No internet service)
# * StreamingTV - Whether the customer has streaming TV or not (Yes, No, No internet service)
# * StreamingMovies Whether the customer has streaming movies or not (Yes, No, No internet service)
# * Contract - The contract term of the customer (Month-to-month, One year, Two year)
# * PaperlessBilling - Whether the customer has paperless billing or not (Yes, No)
# * PaymentMethod - The customer’s payment method (Electronic check, Mailed check, Bank transfer (automatic), Credit card (automatic))
# * MonthlyCharges - The amount charged to the customer monthly
# * TotalCharges - The total amount charged to the customer
# * Churn - Whether the customer churned or not (Yes or No)
#
#### Functions of the project.
def grab_col_names(dataframe, cat_th=10, car_th=20):
"""
Veri setindeki kategorik, numerik ve kategorik fakat kardinal değişkenlerin isimlerini verir.
Not: Kategorik değişkenlerin içerisine numerik görünümlü kategorik değişkenler de dahildir.
Parameters
------
dataframe: dataframe
Değişken isimleri alınmak istenilen dataframe
cat_th: int, optional
numerik fakat kategorik olan değişkenler için sınıf eşik değeri
car_th: int, optinal
kategorik fakat kardinal değişkenler için sınıf eşik değeri
Returns
------
cat_cols: list
Kategorik değişken listesi
num_cols: list
Numerik değişken listesi
cat_but_car: list
Kategorik görünümlü kardinal değişken listesi
Examples
------
import seaborn as sns
df = sns.load_dataset("iris")
print(grab_col_names(df))
Notes
------
cat_cols + num_cols + cat_but_car = toplam değişken sayısı
num_but_cat cat_cols'un içerisinde.
Return olan 3 liste toplamı toplam değişken sayısına eşittir: cat_cols + num_cols + cat_but_car = değişken sayısı
"""
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
def cat_summary(dataframe, col_name, target, plot=False):
print(f"#######{col_name.upper()}############")
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
),
end="\n\n\n",
)
print(
pd.DataFrame({"TARGET_MEAN": dataframe.groupby(col_name)[target].mean()}),
end="\n\n\n",
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
print("##################END######################", end="\n\n\n")
def num_summary(dataframe, numerical_col, target, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T, end="\n\n\n")
print(dataframe.groupby(target).agg({numerical_col: "mean"}), end="\n\n\n")
if plot:
dataframe[numerical_col].hist(bins=20)
plt.xlabel(numerical_col)
plt.title(numerical_col)
plt.show()
def target_summary_with_cat(dataframe, target, categorical_col):
print(
pd.DataFrame(
{"TARGET_MEAN": dataframe.groupby(categorical_col)[target].mean()}
),
end="\n\n\n",
)
def target_summary_with_num(dataframe, target, numerical_col):
print(dataframe.groupby(target).agg({numerical_col: "mean"}), end="\n\n\n")
def rare_analyser(dataframe, target, cat_cols):
for col in cat_cols:
print(col, ":", len(dataframe[col].value_counts()))
print(
pd.DataFrame(
{
"COUNT": dataframe[col].value_counts(),
"RATIO": dataframe[col].value_counts() / len(dataframe),
"TARGET_MEAN": dataframe.groupby(col)[target].mean(),
}
).sort_values(by="COUNT"),
end="\n\n\n",
)
def rare_encoder(dataframe, rare_perc):
temp_df = dataframe.copy()
rare_columns = [
col
for col in temp_df.columns
if temp_df[col].dtypes == "O"
and (temp_df[col].value_counts() / len(temp_df) < rare_perc).any(axis=None)
]
for var in rare_columns:
tmp = temp_df[var].value_counts() / len(temp_df)
rare_labels = tmp[tmp < rare_perc].index
temp_df[var] = np.where(temp_df[var].isin(rare_labels), "Rare", temp_df[var])
return temp_df
def one_hot_encoder(dataframe, categorical_cols, drop_first=False):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
def missing_values_table(dataframe, na_name=False):
"""eksik değerlerin sayını ve oranını na_name= True olursa kolon isimleirinide verir."""
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
def check_outlier(dataframe, col_name, q1=0.05, q3=0.95):
"""
aykırı değer var mı yok sonucunu döner
"""
low_limit, up_limit = outlier_thresholds(dataframe, col_name, q1=q1, q3=q3)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
def outlier_thresholds(dataframe, col_name, q1=0.25, q3=0.75):
# üst v alt limitleri hesaplayıp döndürür.
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def replace_with_thresholds(dataframe, variable, q1=0.25, q3=0.75):
"""
outliarları alt ve üst limite dönüştürüp baskılar.
"""
low_limit, up_limit = outlier_thresholds(dataframe, variable, q1=q1, q3=q3)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
######################################
# Exploratory Data Analysis
######################################
# reading dataset
df_ = pd.read_csv("../input/telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv")
df = df_.copy()
df.head()
# Feature information
print("##################### Shape #####################")
print(df.shape)
print("##################### Types #####################")
print(df.dtypes)
print("##################### Head #####################")
print(df.head())
print("##################### Tail #####################")
print(df.tail())
print("##################### NA #####################")
print(df.isnull().sum())
print("##################### INFO #####################")
print(df.info())
print("##################### NumUnique #####################")
print(df.nunique())
print("##################### Describe #####################")
print(df.describe())
# # Açıklamalar;
# * Veri hiç eksik gözlem olmadığı görülüyor ama yaptığım ilerideki işlemler ile TotalChaerges değişkeni 10 tane eksik gözleme sahip. Burada eksikliği rassalıktan kaynaklanıp kaynaklanmadığı araştırmalıyız.
# * Tenure değeri 0 olanlar dikkat çekiyor incelenmesi gerekir.
# * Değişkenlerin büyük bir bölümü kategorik değişkenlerden oluşuyor.
# * SeniorCitizen değikenini numerik tipte kategorik değere dönüştüreceğim.
# yazdığım fonksiyon ile değişkenleri tiplerine göre listelere ayırıyorum
cat_cols, num_cols, cat_but_car = grab_col_names(df)
# 'TotalCharges' veri tipini float yapacağım.
cat_but_car
# Kategorik değişkenler
cat_cols
# numerical columns
num_cols
# "TotalCharges" değişkenini numerik formata dönüştürdüm
df["TotalCharges"] = pd.to_numeric(df["TotalCharges"], errors="coerce")
num_cols.append("TotalCharges")
# Churn analizine bir etkisi olmadığı için "customerID" değişkenini veri setinden çıkarıyorum.
df.drop("customerID", axis=1, inplace=True)
# Data setimizi biraz dengesiz olarak görünüyor burada veri seti dengelenebilir veya daha fazla veri ile analiz
# yapılabilir. şimdilik bu durumu göz ardı ediyorum.
df["Churn"].value_counts()
# yukarda da tenure değeri sıfır olanların araştırmamız gerektiğinden bahsetmiştim
# burda anlaşılacağı üzere TotalCharges değişkenindeki na değerler rassal değil veri setine yeni gelen müşteriler
# anlaşılıyor. burda bu na değerlerini ilgili gözlem biriminin MonthlyCharges değeri ile dolduracağım çünkü henüz ödemeye
# başladıkları için bu değer ile doldurmak mantıklı.
df[df["TotalCharges"].isnull()]
# dediğim gibi indexlerde tutuyor.
df[df["TotalCharges"].isnull()].index
df[df["tenure"] == 0].index
# yukarda değidiğim gibi tenure lar 0 olduğu için ilk aylık ödemeleri ile totalcharges değişkenini doldurdum.
df.loc[df["tenure"] == 0, "TotalCharges"] = df.loc[df["tenure"] == 0, "MonthlyCharges"]
# na olan gözlem birimleri dolduruldu.
df.loc[df["tenure"] == 0, "TotalCharges"]
# SeniorCitizen değişkeninin tipini object yapıyorum.
df["SeniorCitizen"].astype(object)
# Hedef değişkenimiz olan Churn değişkeninindeki Yes değerlerini 1 No değerlerini 0 olarak dönüştüyorum hem makine
# öğrenmesi bölümü hemde hedef değişkene göre değişkenlerin analizi için buna ihtiyacım var.
# Ayrıca Churn ün değişken tipini integer yapıyorum.(analiz için)
df["Churn"] = df["Churn"].apply(lambda x: 1 if x == "Yes" else 0)
df["Churn"].astype(int)
for col in cat_cols:
# print(df.groupby(col).agg({"Churn":["mean", "count"]}))
# burda kategorik değişkenleri analiz etmek için tanımladığım cat_summary fonksiyonunu çağırıyorum.
cat_summary(df, col, "Churn", plot=True)
# summurazing numerical variables
for col in num_cols:
target_summary_with_num(df, "Churn", col)
# ### Açıklamalar;
# * Gender - değişkenindeki farklılığın Churn olup olamamaya herhangi bir etkisi bulunmamaktadır.
# * SeniorCitizen - yaşlı müşterilerin daha fazla churn oldukları gözlemlenmektedir. Bunun nedeni araştırılmalıdır. Müşteriler artık telekom hizmetlerini kullanamadıkları için yani firma dışı nedenlerden mi churnm oluyolar.
# * Partner - Partnerı olan müşteriler daha az churn olma eğilimindeler burada firma partnerlara yönelik kampanyalar yaparak partner şeklinde müşteri kazanıp müşterilerin churn olmasını azaltabilir.
# * Dependents - bakmakla yükümlü oldukları insanlar olan kişiler olmayanlara %50 daha az churn oluyorlar.
# * Tenure - Müşteri tenure(şirket yaşı) artıkça churn olma oranı düşüyor. churn olanlar genelde tenure düşük olan müşteriler churn olma eğiliminde firma başlangıçta bu müşterilerin churn olmasını engellemek için belirli aralıklarla hatırlatma veya avantajlı bir kampanyalar ile daha uzun sözleşmeler yapmalıdır.
# * Phoneservice - değişkenindeki farklılığın Churn olup olamamaya herhangi bir etkisi bulunmamaktadır.
# * MultipleLines - değişkenindeki farklılığın Churn olup olamamaya belirgin bir etkisi bulunmamaktadır.
# * InternetService - burda fiber obtik servisi olan müşterilerin churn olma eğiliminin yüksek olduğu görülüyor. Bu müşterilerin teknolojiyi ve trendleri yakından taqkip ettiklerini ve daha fazla duyarlı olduklarını düşünüyorum bu müşterilere hem yeni teknolojiler ile ilgili bilgi akışı sağlanmalı ve bu müşterileri elde tutmak için farklı yan hizmetler verilebilir mesela bir teknoloji dergisi aboneliği veya trend olan bir site veya uygulamadan ücretsiz yararlanma gibi farklı alternatifler ile firma bağlılığı kazandırılmalıdır. Bu durum aynı zamanda Telco firmasının teknolojiyi çok rekabetin olduğu bu piyasada iyi bir şekilde takip etmesi gerektiğine de işaret ediyor.
# * Onlinesecurity - online güvenliği olmayan müşterilerin yüksek churn olma eğiliminde oldukları görülmüştür. Bu hizmetin günümüz internet ortamının güvensizliği de tanıtılarak ve gerekirse bu hizmetin uygun sağlanması için çeşitli kampanyalar düzenlerek müşterilerek bu ürünün satılması müşteri kaybını önlemede büyük destek sağlayacaktır.
# * OnlineBackup - Online backup da online güvenlik ile aynı özelliği gösteriyor ve benzer önlem ve staretejiler bu değişken içinde yapılabilir.
# * DeviceProtection - bu değişken Onlinesecurity ve OnlineBackup ile benzer şekilde etkiye sahip. Bu üç ürünü veya en az ikisini çapraz satış yaparak müşterilerin churn olmasına engel olmada büyük başarı elde edilebilir.
# * TechSupport - teknolojik desteği olmayan müşterilerin churn olma oranları yüksek, burda firma müşterilere bir şekilde ulaşarak hem yerinde hemde online teknolojik destek olmalıdır.
# * StreamingTV - internet servisi olmayan müşterilere hariç değişkenindeki farklılığın Churn olup olamamaya herhangi bir etkisi bulunmamaktadır.
# * StreamingMovies - internet servisi olmayan müşterilere hariç değişkenindeki farklılığın Churn olup olamamaya herhangi bir etkisi bulunmamaktadır.
# * Contract - kontrat süreleri artıkça churn oranları düşmektedir. Firma uzun kontratlarda rakabetçi ve kısa süreli kontratlara göre uygun tarifeler vererek müşteri bağlılığını artırmalıdır.
# * PaperlessBilling - kağıt fatura alan müşterilerin churn oranı daha düşük görünmekte olup firma kağıt ve posta maliyetinin yüksek olduğu düşünülürse kağıt fatura almayan müşterilerin bilgilendirmelerini daha etkili bir şekilde yapabilir.
# * PaymentMethod - electronik check ödemesi olan müşterilerin churn oranı yüksek ve özellikle kredi kartı otomatik ödeme talimatları churn oranları düşüktür müşterileri bu kanala itmek müşterinin fiyat duyarlılığını bir nebze azaltacaktır hesaba verilen otomatik ödemeden ziyade çünkü hesap hareketleri kredi kartı ekstresinden daha çok kontrol edilir.
# * MonthlyCharges - aylık sabit ödemeleri yüksek olan müşterilerin churn oranları yüksek burda aylık sabit ödemenin düşük tutulup müşteriye çapraz satış yapılarak gelir artışı sağlamak churn olmayı azaltabilir çünkü totalcharges değişkenin yüksek olması churn olmaya etkisi az olanlara göre düşük.
#
# Feature information
print("##################### Shape #####################")
print(df.shape)
print("##################### Types #####################")
print(df.dtypes)
print("##################### Head #####################")
print(df.head())
print("##################### Tail #####################")
print(df.tail())
print("##################### NA #####################")
print(df.isnull().sum())
print("##################### INFO #####################")
print(df.info())
print("##################### NumUnique #####################")
print(df.nunique())
print("##################### Describe #####################")
print(df.describe())
# * According to these values and in the figure ,people will be tend to churn if
# 1. their contract type is month to month
# 2. there is no online security
# 3. there is no technical support to the customer
# 4. internet service is fiber optic
# 5. payment method is electronic check
# 6. there is no online backup
# 7. there is no device protection
# 8. there is monthly charges
# 9. there is paperless billing
# * According to these valuues,people will not churn if
# 1. there is higher tenure
# 2. there is two years period contract
# 3. there is no internet service
# 4. there is no streaming TV
# rare değişken bulunmadığı görülmüştür.
rare_analyser(df, "Churn", cat_cols)
cat_cols = [col for col in cat_cols if col != "Churn"]
scaler = StandardScaler()
df[num_cols] = scaler.fit_transform(df[num_cols])
# Here I use my custom one_hot_encoder() function ,n order to transform categorical columns into dummy numbers
df = one_hot_encoder(df, cat_cols, drop_first=True)
df.head()
def base_models(X, y, scoring="roc_auc"):
print("Base Models....")
classifiers = [
("LR", LogisticRegression()),
("KNN", KNeighborsClassifier()),
("SVC", SVC()),
("CART", DecisionTreeClassifier()),
("RF", RandomForestClassifier()),
("Adaboost", AdaBoostClassifier()),
("GBM", GradientBoostingClassifier()),
("XGBoost", XGBClassifier()),
("LightGBM", LGBMClassifier()),
# ('CatBoost', CatBoostClassifier(verbose=False))
]
for name, classifier in classifiers:
cv_results = cross_validate(classifier, X, y, cv=3, scoring=scoring)
print(f"{scoring}: {round(cv_results['test_score'].mean(), 4)} ({name}) ")
X = df.drop(["Churn"], axis=1)
y = df["Churn"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.20)
df.head()
base_models(X_train, y_train, scoring="roc_auc")
base_models(X_test, y_test, scoring="roc_auc")
######################################################
# Automated Hyperparameter Optimization
######################################################
lgr_params = {
"solver": ["newton-cg", "liblinear"],
"penalty": ["l2"],
"C": [100, 1, 0.01],
}
gbm_params = {
"learning_rate": [0.1, 0.01, 1],
"max_depth": [1, 2, 4],
"n_estimators": [100, 1000, 2000],
"subsample": [0.5, 0.75, 1],
}
knn_params = {"n_neighbors": range(2, 50)}
cart_params = {"max_depth": range(1, 20), "min_samples_split": range(2, 30)}
rf_params = {
"max_depth": [8, 15, None],
"max_features": [5, 7, "auto"],
"min_samples_split": [15, 20],
"n_estimators": [200, 300],
}
xgboost_params = {
"learning_rate": [0.1, 0.01],
"max_depth": [5, 8],
"n_estimators": [100, 200],
"colsample_bytree": [0.5, 1],
}
lightgbm_params = {
"learning_rate": [0.01, 0.1, 0.001],
"n_estimators": [300, 500, 1500],
"colsample_bytree": [0.7, 1],
}
classifiers = [
("KNN", KNeighborsClassifier(), knn_params),
("CART", DecisionTreeClassifier(), cart_params),
("RF", RandomForestClassifier(), rf_params),
# ('XGBoost', XGBClassifier(), xgboost_params),
("LightGBM", LGBMClassifier(), lightgbm_params),
("GBM", GradientBoostingClassifier(), gbm_params),
("LR", LogisticRegression(), lgr_params),
]
def hyperparameter_optimization(X, y, cv=3, scoring="roc_auc"):
print("Hyperparameter Optimization....")
best_models = {}
for name, classifier, params in classifiers:
print(f"########## {name} ##########", end="\n\n")
print("########## before ##########", end="\n\n")
cv_results = cross_validate(
classifier, X, y, cv=3, scoring=["accuracy", "f1", "roc_auc"]
)
print(f"Accuracy: {cv_results['test_accuracy'].mean()}")
print(f"F1Score: {cv_results['test_f1'].mean()}")
print(f"ROC_AUC: {cv_results['test_roc_auc'].mean()}", end="\n\n")
print("########## after ##########", end="\n\n")
gs_best = GridSearchCV(classifier, params, cv=cv, n_jobs=-1, verbose=False).fit(
X, y
)
final_model = classifier.set_params(**gs_best.best_params_)
cv_results = cross_validate(final_model, X, y, cv=cv, scoring=scoring)
print(f"Accuracy: {cv_results['test_accuracy'].mean()}")
print(f"F1Score: {cv_results['test_f1'].mean()}")
print(f"ROC_AUC: {cv_results['test_roc_auc'].mean()}")
print(f"{name} best params: {gs_best.best_params_}", end="\n\n")
best_models[name] = final_model
return best_models
best_models = hyperparameter_optimization(
X_train, y_train, cv=3, scoring=["accuracy", "f1", "roc_auc"]
)
# the function for calculate the tet scores
def test_scores(best_models, X_test, y_test):
for i, j in best_models.items():
print(i)
j.fit(X_train, y_train)
predictions = j.predict(X_test)
print(confusion_matrix(y_test, predictions), end="\n\n")
print(classification_report(y_test, predictions), end="\n\n")
print(accuracy_score(y_test, predictions), end="\n\n")
best_models
from sklearn.metrics import (
classification_report,
confusion_matrix,
accuracy_score,
roc_auc_score,
)
test_scores(best_models, X_test, y_test)
######################################################
# Stacking & Ensemble Learning
######################################################
def voting_classifier(best_models, X, y):
print("Voting Classifier...")
voting_clf = VotingClassifier(
estimators=[
("GBM", best_models["GBM"]),
("LR", best_models["LR"]),
("LightGBM", best_models["LightGBM"]),
],
voting="soft",
).fit(X, y)
cv_results = cross_validate(
voting_clf, X, y, cv=3, scoring=["accuracy", "f1", "roc_auc"]
)
print(f"Accuracy: {cv_results['test_accuracy'].mean()}")
print(f"F1Score: {cv_results['test_f1'].mean()}")
print(f"ROC_AUC: {cv_results['test_roc_auc'].mean()}")
return voting_clf
voting_clf = voting_classifier(best_models, X, y)
voting_clf
voting_clf.predict(X_test)
voting_clf.fit(X_train, y_train)
predictions = voting_clf.predict(X_test)
print(confusion_matrix(y_test, predictions), end="\n\n")
print(classification_report(y_test, predictions), end="\n\n")
print(accuracy_score(y_test, predictions), end="\n\n")
#######################################
# Feature Selection
#######################################
def plot_importance(model, features, num=len(X), save=False):
feature_imp = pd.DataFrame(
{"Value": model.feature_importances_, "Feature": features.columns}
)
plt.figure(figsize=(30, 30))
sns.set(font_scale=1)
sns.barplot(
x="Value",
y="Feature",
data=feature_imp.sort_values(by="Value", ascending=False)[0:num],
)
plt.title("Features")
plt.tight_layout()
plt.show()
if save:
plt.savefig("importances.png")
return
features = pd.DataFrame(
{"Feature": X.columns, "Value": j.feature_importances_}
).sort_values(by="Value", ascending=False)
plot_importance(j, features)
features = pd.DataFrame(
{"Feature": X.columns, "Value": LGBMClassifier().feature_importances_}
).sort_values(by="Value", ascending=False)
plot_importance(voting_clf, features)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492590.ipynb
|
telco-customer-churn
|
blastchar
|
[{"Id": 69492590, "ScriptId": 18923150, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6385159, "CreationDate": "07/31/2021 17:02:21", "VersionNumber": 2.0, "Title": "Telco Customer Churn Analysis", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 608.0, "LinesInsertedFromPrevious": 213.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 395.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92728922, "KernelVersionId": 69492590, "SourceDatasetVersionId": 18858}]
|
[{"Id": 18858, "DatasetId": 13996, "DatasourceVersionId": 18858, "CreatorUserId": 1574575, "LicenseName": "Data files \u00a9 Original Authors", "CreationDate": "02/23/2018 18:20:00", "VersionNumber": 1.0, "Title": "Telco Customer Churn", "Slug": "telco-customer-churn", "Subtitle": "Focused customer retention programs", "Description": "### Context\n\n\"Predict behavior to retain customers. You can analyze all relevant customer data and develop focused customer retention programs.\" [IBM Sample Data Sets]\n\n### Content\n\nEach row represents a customer, each column contains customer\u2019s attributes described on the column Metadata.\n\n**The data set includes information about:**\n\n+ Customers who left within the last month \u2013 the column is called Churn\n+ Services that each customer has signed up for \u2013 phone, multiple lines, internet, online security, online backup, device protection, tech support, and streaming TV and movies\n+ Customer account information \u2013 how long they\u2019ve been a customer, contract, payment method, paperless billing, monthly charges, and total charges\n+ Demographic info about customers \u2013 gender, age range, and if they have partners and dependents\n\n### Inspiration\n\nTo explore this type of models and learn more about the subject.\n\n**New version from IBM:**\nhttps://community.ibm.com/community/user/businessanalytics/blogs/steven-macko/2019/07/11/telco-customer-churn-1113", "VersionNotes": "Initial release", "TotalCompressedBytes": 977501.0, "TotalUncompressedBytes": 977501.0}]
|
[{"Id": 13996, "CreatorUserId": 1574575, "OwnerUserId": 1574575.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 18858.0, "CurrentDatasourceVersionId": 18858.0, "ForumId": 21551, "Type": 2, "CreationDate": "02/23/2018 18:20:00", "LastActivityDate": "02/23/2018", "TotalViews": 1765427, "TotalDownloads": 190987, "TotalVotes": 2480, "TotalKernels": 1184}]
|
[{"Id": 1574575, "UserName": "blastchar", "DisplayName": "BlastChar", "RegisterDate": "01/23/2018", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Installation of required libraries
import missingno as msno
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.ensemble import (
RandomForestClassifier,
GradientBoostingClassifier,
VotingClassifier,
AdaBoostClassifier,
)
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import cross_validate, GridSearchCV
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.preprocessing import StandardScaler, LabelEncoder
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
from sklearn.ensemble import GradientBoostingClassifier
from catboost import CatBoostClassifier
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.metrics import accuracy_score
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score, GridSearchCV
import warnings
warnings.filterwarnings("ignore", category=DeprecationWarning)
warnings.filterwarnings("ignore", category=FutureWarning)
warnings.filterwarnings("ignore", category=UserWarning)
# 1. Introduction
# Customer churn is defined as when customers or subscribers discontinue doing business with a firm or service.
# Customers in the telecom industry can choose from a variety of service providers and actively switch from one to the next. The telecommunications business has an annual churn rate of 15-25 percent in this highly competitive market.
# Individualized customer retention is tough because most firms have a large number of customers and can't afford to devote much time to each of them. The costs would be too great, outweighing the additional revenue. However, if a corporation could forecast which customers are likely to leave ahead of time, it could focus customer retention efforts only on these "high risk" clients. The ultimate goal is to expand its coverage area and retrieve more customers loyalty. The core to succeed in this market lies in the customer itself.
# Customer churn is a critical metric because it is much less expensive to retain existing customers than it is to acquire new customers.
# To reduce customer churn, telecom companies need to predict which customers are at high risk of churn.
# To detect early signs of potential churn, one must first develop a holistic view of the customers and their interactions across numerous channels, including store/branch visits, product purchase histories, customer service calls, Web-based transactions, and social media interactions, to mention a few.
# As a result, by addressing churn, these businesses may not only preserve their market position, but also grow and thrive. More customers they have in their network, the lower the cost of initiation and the larger the profit. As a result, the company's key focus for success is reducing client attrition and implementing effective retention strategy.
# ### Data Description
# * Context
# * "Predict behavior to retain customers. You can analyze all relevant customer data and develop focused customer retention programs."
# * Content
# * Each row represents a customer, each column contains customer’s attributes described on the column Metadata.
# * The data set includes information about:
# * Customers who left within the last month – the column is called Churn
# * Services that each customer has signed up for – phone, multiple lines, internet, online security, online backup, device protection, tech support, and streaming TV and movies
# * Customer account information – how long they’ve been a customer, contract, payment method, paperless billing, monthly charges, and total charges
# * Demographic info about customers – gender, age range, and if they have partners and dependents
# * 1. Exploratory Data Analysis
# * 2. Data Preprocessing & Feature Engineering
# * 3. Modeling
# * 4. Feature Selection
# * 5. Hyperparameter Optimization with Selected Features
# * 6. Submitting the results
# * Customer ID
# * Gender - Whether the customer is a male or a female
# * SeniorCitizen - Whether the customer is a senior citizen or not (1, 0)
# * Partner - Whether the customer has a partner or not (Yes, No)
# * Dependents - Whether the customer has dependents or not (Yes, No)
# * tenure - Number of months the customer has stayed with the company
# * PhoneService - Whether the customer has a phone service or not (Yes, No)
# * MultipleLines - Whether the customer has multiple lines or not (Yes, No, No phone service)
# * InternetService - Customer’s internet service provider (DSL, Fiber optic, No)
# * OnlineSecurity - Whether the customer has online security or not (Yes, No, No internet service)
# * OnlineBackup - Whether the customer has online backup or not (Yes, No, No internet service)
# * DeviceProtection - Whether the customer has device protection or not (Yes, No, No internet service)
# * TechSupport - Whether the customer has tech support or not (Yes, No, No internet service)
# * StreamingTV - Whether the customer has streaming TV or not (Yes, No, No internet service)
# * StreamingMovies Whether the customer has streaming movies or not (Yes, No, No internet service)
# * Contract - The contract term of the customer (Month-to-month, One year, Two year)
# * PaperlessBilling - Whether the customer has paperless billing or not (Yes, No)
# * PaymentMethod - The customer’s payment method (Electronic check, Mailed check, Bank transfer (automatic), Credit card (automatic))
# * MonthlyCharges - The amount charged to the customer monthly
# * TotalCharges - The total amount charged to the customer
# * Churn - Whether the customer churned or not (Yes or No)
#
#### Functions of the project.
def grab_col_names(dataframe, cat_th=10, car_th=20):
"""
Veri setindeki kategorik, numerik ve kategorik fakat kardinal değişkenlerin isimlerini verir.
Not: Kategorik değişkenlerin içerisine numerik görünümlü kategorik değişkenler de dahildir.
Parameters
------
dataframe: dataframe
Değişken isimleri alınmak istenilen dataframe
cat_th: int, optional
numerik fakat kategorik olan değişkenler için sınıf eşik değeri
car_th: int, optinal
kategorik fakat kardinal değişkenler için sınıf eşik değeri
Returns
------
cat_cols: list
Kategorik değişken listesi
num_cols: list
Numerik değişken listesi
cat_but_car: list
Kategorik görünümlü kardinal değişken listesi
Examples
------
import seaborn as sns
df = sns.load_dataset("iris")
print(grab_col_names(df))
Notes
------
cat_cols + num_cols + cat_but_car = toplam değişken sayısı
num_but_cat cat_cols'un içerisinde.
Return olan 3 liste toplamı toplam değişken sayısına eşittir: cat_cols + num_cols + cat_but_car = değişken sayısı
"""
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
def cat_summary(dataframe, col_name, target, plot=False):
print(f"#######{col_name.upper()}############")
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
),
end="\n\n\n",
)
print(
pd.DataFrame({"TARGET_MEAN": dataframe.groupby(col_name)[target].mean()}),
end="\n\n\n",
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
print("##################END######################", end="\n\n\n")
def num_summary(dataframe, numerical_col, target, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T, end="\n\n\n")
print(dataframe.groupby(target).agg({numerical_col: "mean"}), end="\n\n\n")
if plot:
dataframe[numerical_col].hist(bins=20)
plt.xlabel(numerical_col)
plt.title(numerical_col)
plt.show()
def target_summary_with_cat(dataframe, target, categorical_col):
print(
pd.DataFrame(
{"TARGET_MEAN": dataframe.groupby(categorical_col)[target].mean()}
),
end="\n\n\n",
)
def target_summary_with_num(dataframe, target, numerical_col):
print(dataframe.groupby(target).agg({numerical_col: "mean"}), end="\n\n\n")
def rare_analyser(dataframe, target, cat_cols):
for col in cat_cols:
print(col, ":", len(dataframe[col].value_counts()))
print(
pd.DataFrame(
{
"COUNT": dataframe[col].value_counts(),
"RATIO": dataframe[col].value_counts() / len(dataframe),
"TARGET_MEAN": dataframe.groupby(col)[target].mean(),
}
).sort_values(by="COUNT"),
end="\n\n\n",
)
def rare_encoder(dataframe, rare_perc):
temp_df = dataframe.copy()
rare_columns = [
col
for col in temp_df.columns
if temp_df[col].dtypes == "O"
and (temp_df[col].value_counts() / len(temp_df) < rare_perc).any(axis=None)
]
for var in rare_columns:
tmp = temp_df[var].value_counts() / len(temp_df)
rare_labels = tmp[tmp < rare_perc].index
temp_df[var] = np.where(temp_df[var].isin(rare_labels), "Rare", temp_df[var])
return temp_df
def one_hot_encoder(dataframe, categorical_cols, drop_first=False):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
def missing_values_table(dataframe, na_name=False):
"""eksik değerlerin sayını ve oranını na_name= True olursa kolon isimleirinide verir."""
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
def check_outlier(dataframe, col_name, q1=0.05, q3=0.95):
"""
aykırı değer var mı yok sonucunu döner
"""
low_limit, up_limit = outlier_thresholds(dataframe, col_name, q1=q1, q3=q3)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
def outlier_thresholds(dataframe, col_name, q1=0.25, q3=0.75):
# üst v alt limitleri hesaplayıp döndürür.
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def replace_with_thresholds(dataframe, variable, q1=0.25, q3=0.75):
"""
outliarları alt ve üst limite dönüştürüp baskılar.
"""
low_limit, up_limit = outlier_thresholds(dataframe, variable, q1=q1, q3=q3)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
######################################
# Exploratory Data Analysis
######################################
# reading dataset
df_ = pd.read_csv("../input/telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv")
df = df_.copy()
df.head()
# Feature information
print("##################### Shape #####################")
print(df.shape)
print("##################### Types #####################")
print(df.dtypes)
print("##################### Head #####################")
print(df.head())
print("##################### Tail #####################")
print(df.tail())
print("##################### NA #####################")
print(df.isnull().sum())
print("##################### INFO #####################")
print(df.info())
print("##################### NumUnique #####################")
print(df.nunique())
print("##################### Describe #####################")
print(df.describe())
# # Açıklamalar;
# * Veri hiç eksik gözlem olmadığı görülüyor ama yaptığım ilerideki işlemler ile TotalChaerges değişkeni 10 tane eksik gözleme sahip. Burada eksikliği rassalıktan kaynaklanıp kaynaklanmadığı araştırmalıyız.
# * Tenure değeri 0 olanlar dikkat çekiyor incelenmesi gerekir.
# * Değişkenlerin büyük bir bölümü kategorik değişkenlerden oluşuyor.
# * SeniorCitizen değikenini numerik tipte kategorik değere dönüştüreceğim.
# yazdığım fonksiyon ile değişkenleri tiplerine göre listelere ayırıyorum
cat_cols, num_cols, cat_but_car = grab_col_names(df)
# 'TotalCharges' veri tipini float yapacağım.
cat_but_car
# Kategorik değişkenler
cat_cols
# numerical columns
num_cols
# "TotalCharges" değişkenini numerik formata dönüştürdüm
df["TotalCharges"] = pd.to_numeric(df["TotalCharges"], errors="coerce")
num_cols.append("TotalCharges")
# Churn analizine bir etkisi olmadığı için "customerID" değişkenini veri setinden çıkarıyorum.
df.drop("customerID", axis=1, inplace=True)
# Data setimizi biraz dengesiz olarak görünüyor burada veri seti dengelenebilir veya daha fazla veri ile analiz
# yapılabilir. şimdilik bu durumu göz ardı ediyorum.
df["Churn"].value_counts()
# yukarda da tenure değeri sıfır olanların araştırmamız gerektiğinden bahsetmiştim
# burda anlaşılacağı üzere TotalCharges değişkenindeki na değerler rassal değil veri setine yeni gelen müşteriler
# anlaşılıyor. burda bu na değerlerini ilgili gözlem biriminin MonthlyCharges değeri ile dolduracağım çünkü henüz ödemeye
# başladıkları için bu değer ile doldurmak mantıklı.
df[df["TotalCharges"].isnull()]
# dediğim gibi indexlerde tutuyor.
df[df["TotalCharges"].isnull()].index
df[df["tenure"] == 0].index
# yukarda değidiğim gibi tenure lar 0 olduğu için ilk aylık ödemeleri ile totalcharges değişkenini doldurdum.
df.loc[df["tenure"] == 0, "TotalCharges"] = df.loc[df["tenure"] == 0, "MonthlyCharges"]
# na olan gözlem birimleri dolduruldu.
df.loc[df["tenure"] == 0, "TotalCharges"]
# SeniorCitizen değişkeninin tipini object yapıyorum.
df["SeniorCitizen"].astype(object)
# Hedef değişkenimiz olan Churn değişkeninindeki Yes değerlerini 1 No değerlerini 0 olarak dönüştüyorum hem makine
# öğrenmesi bölümü hemde hedef değişkene göre değişkenlerin analizi için buna ihtiyacım var.
# Ayrıca Churn ün değişken tipini integer yapıyorum.(analiz için)
df["Churn"] = df["Churn"].apply(lambda x: 1 if x == "Yes" else 0)
df["Churn"].astype(int)
for col in cat_cols:
# print(df.groupby(col).agg({"Churn":["mean", "count"]}))
# burda kategorik değişkenleri analiz etmek için tanımladığım cat_summary fonksiyonunu çağırıyorum.
cat_summary(df, col, "Churn", plot=True)
# summurazing numerical variables
for col in num_cols:
target_summary_with_num(df, "Churn", col)
# ### Açıklamalar;
# * Gender - değişkenindeki farklılığın Churn olup olamamaya herhangi bir etkisi bulunmamaktadır.
# * SeniorCitizen - yaşlı müşterilerin daha fazla churn oldukları gözlemlenmektedir. Bunun nedeni araştırılmalıdır. Müşteriler artık telekom hizmetlerini kullanamadıkları için yani firma dışı nedenlerden mi churnm oluyolar.
# * Partner - Partnerı olan müşteriler daha az churn olma eğilimindeler burada firma partnerlara yönelik kampanyalar yaparak partner şeklinde müşteri kazanıp müşterilerin churn olmasını azaltabilir.
# * Dependents - bakmakla yükümlü oldukları insanlar olan kişiler olmayanlara %50 daha az churn oluyorlar.
# * Tenure - Müşteri tenure(şirket yaşı) artıkça churn olma oranı düşüyor. churn olanlar genelde tenure düşük olan müşteriler churn olma eğiliminde firma başlangıçta bu müşterilerin churn olmasını engellemek için belirli aralıklarla hatırlatma veya avantajlı bir kampanyalar ile daha uzun sözleşmeler yapmalıdır.
# * Phoneservice - değişkenindeki farklılığın Churn olup olamamaya herhangi bir etkisi bulunmamaktadır.
# * MultipleLines - değişkenindeki farklılığın Churn olup olamamaya belirgin bir etkisi bulunmamaktadır.
# * InternetService - burda fiber obtik servisi olan müşterilerin churn olma eğiliminin yüksek olduğu görülüyor. Bu müşterilerin teknolojiyi ve trendleri yakından taqkip ettiklerini ve daha fazla duyarlı olduklarını düşünüyorum bu müşterilere hem yeni teknolojiler ile ilgili bilgi akışı sağlanmalı ve bu müşterileri elde tutmak için farklı yan hizmetler verilebilir mesela bir teknoloji dergisi aboneliği veya trend olan bir site veya uygulamadan ücretsiz yararlanma gibi farklı alternatifler ile firma bağlılığı kazandırılmalıdır. Bu durum aynı zamanda Telco firmasının teknolojiyi çok rekabetin olduğu bu piyasada iyi bir şekilde takip etmesi gerektiğine de işaret ediyor.
# * Onlinesecurity - online güvenliği olmayan müşterilerin yüksek churn olma eğiliminde oldukları görülmüştür. Bu hizmetin günümüz internet ortamının güvensizliği de tanıtılarak ve gerekirse bu hizmetin uygun sağlanması için çeşitli kampanyalar düzenlerek müşterilerek bu ürünün satılması müşteri kaybını önlemede büyük destek sağlayacaktır.
# * OnlineBackup - Online backup da online güvenlik ile aynı özelliği gösteriyor ve benzer önlem ve staretejiler bu değişken içinde yapılabilir.
# * DeviceProtection - bu değişken Onlinesecurity ve OnlineBackup ile benzer şekilde etkiye sahip. Bu üç ürünü veya en az ikisini çapraz satış yaparak müşterilerin churn olmasına engel olmada büyük başarı elde edilebilir.
# * TechSupport - teknolojik desteği olmayan müşterilerin churn olma oranları yüksek, burda firma müşterilere bir şekilde ulaşarak hem yerinde hemde online teknolojik destek olmalıdır.
# * StreamingTV - internet servisi olmayan müşterilere hariç değişkenindeki farklılığın Churn olup olamamaya herhangi bir etkisi bulunmamaktadır.
# * StreamingMovies - internet servisi olmayan müşterilere hariç değişkenindeki farklılığın Churn olup olamamaya herhangi bir etkisi bulunmamaktadır.
# * Contract - kontrat süreleri artıkça churn oranları düşmektedir. Firma uzun kontratlarda rakabetçi ve kısa süreli kontratlara göre uygun tarifeler vererek müşteri bağlılığını artırmalıdır.
# * PaperlessBilling - kağıt fatura alan müşterilerin churn oranı daha düşük görünmekte olup firma kağıt ve posta maliyetinin yüksek olduğu düşünülürse kağıt fatura almayan müşterilerin bilgilendirmelerini daha etkili bir şekilde yapabilir.
# * PaymentMethod - electronik check ödemesi olan müşterilerin churn oranı yüksek ve özellikle kredi kartı otomatik ödeme talimatları churn oranları düşüktür müşterileri bu kanala itmek müşterinin fiyat duyarlılığını bir nebze azaltacaktır hesaba verilen otomatik ödemeden ziyade çünkü hesap hareketleri kredi kartı ekstresinden daha çok kontrol edilir.
# * MonthlyCharges - aylık sabit ödemeleri yüksek olan müşterilerin churn oranları yüksek burda aylık sabit ödemenin düşük tutulup müşteriye çapraz satış yapılarak gelir artışı sağlamak churn olmayı azaltabilir çünkü totalcharges değişkenin yüksek olması churn olmaya etkisi az olanlara göre düşük.
#
# Feature information
print("##################### Shape #####################")
print(df.shape)
print("##################### Types #####################")
print(df.dtypes)
print("##################### Head #####################")
print(df.head())
print("##################### Tail #####################")
print(df.tail())
print("##################### NA #####################")
print(df.isnull().sum())
print("##################### INFO #####################")
print(df.info())
print("##################### NumUnique #####################")
print(df.nunique())
print("##################### Describe #####################")
print(df.describe())
# * According to these values and in the figure ,people will be tend to churn if
# 1. their contract type is month to month
# 2. there is no online security
# 3. there is no technical support to the customer
# 4. internet service is fiber optic
# 5. payment method is electronic check
# 6. there is no online backup
# 7. there is no device protection
# 8. there is monthly charges
# 9. there is paperless billing
# * According to these valuues,people will not churn if
# 1. there is higher tenure
# 2. there is two years period contract
# 3. there is no internet service
# 4. there is no streaming TV
# rare değişken bulunmadığı görülmüştür.
rare_analyser(df, "Churn", cat_cols)
cat_cols = [col for col in cat_cols if col != "Churn"]
scaler = StandardScaler()
df[num_cols] = scaler.fit_transform(df[num_cols])
# Here I use my custom one_hot_encoder() function ,n order to transform categorical columns into dummy numbers
df = one_hot_encoder(df, cat_cols, drop_first=True)
df.head()
def base_models(X, y, scoring="roc_auc"):
print("Base Models....")
classifiers = [
("LR", LogisticRegression()),
("KNN", KNeighborsClassifier()),
("SVC", SVC()),
("CART", DecisionTreeClassifier()),
("RF", RandomForestClassifier()),
("Adaboost", AdaBoostClassifier()),
("GBM", GradientBoostingClassifier()),
("XGBoost", XGBClassifier()),
("LightGBM", LGBMClassifier()),
# ('CatBoost', CatBoostClassifier(verbose=False))
]
for name, classifier in classifiers:
cv_results = cross_validate(classifier, X, y, cv=3, scoring=scoring)
print(f"{scoring}: {round(cv_results['test_score'].mean(), 4)} ({name}) ")
X = df.drop(["Churn"], axis=1)
y = df["Churn"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.20)
df.head()
base_models(X_train, y_train, scoring="roc_auc")
base_models(X_test, y_test, scoring="roc_auc")
######################################################
# Automated Hyperparameter Optimization
######################################################
lgr_params = {
"solver": ["newton-cg", "liblinear"],
"penalty": ["l2"],
"C": [100, 1, 0.01],
}
gbm_params = {
"learning_rate": [0.1, 0.01, 1],
"max_depth": [1, 2, 4],
"n_estimators": [100, 1000, 2000],
"subsample": [0.5, 0.75, 1],
}
knn_params = {"n_neighbors": range(2, 50)}
cart_params = {"max_depth": range(1, 20), "min_samples_split": range(2, 30)}
rf_params = {
"max_depth": [8, 15, None],
"max_features": [5, 7, "auto"],
"min_samples_split": [15, 20],
"n_estimators": [200, 300],
}
xgboost_params = {
"learning_rate": [0.1, 0.01],
"max_depth": [5, 8],
"n_estimators": [100, 200],
"colsample_bytree": [0.5, 1],
}
lightgbm_params = {
"learning_rate": [0.01, 0.1, 0.001],
"n_estimators": [300, 500, 1500],
"colsample_bytree": [0.7, 1],
}
classifiers = [
("KNN", KNeighborsClassifier(), knn_params),
("CART", DecisionTreeClassifier(), cart_params),
("RF", RandomForestClassifier(), rf_params),
# ('XGBoost', XGBClassifier(), xgboost_params),
("LightGBM", LGBMClassifier(), lightgbm_params),
("GBM", GradientBoostingClassifier(), gbm_params),
("LR", LogisticRegression(), lgr_params),
]
def hyperparameter_optimization(X, y, cv=3, scoring="roc_auc"):
print("Hyperparameter Optimization....")
best_models = {}
for name, classifier, params in classifiers:
print(f"########## {name} ##########", end="\n\n")
print("########## before ##########", end="\n\n")
cv_results = cross_validate(
classifier, X, y, cv=3, scoring=["accuracy", "f1", "roc_auc"]
)
print(f"Accuracy: {cv_results['test_accuracy'].mean()}")
print(f"F1Score: {cv_results['test_f1'].mean()}")
print(f"ROC_AUC: {cv_results['test_roc_auc'].mean()}", end="\n\n")
print("########## after ##########", end="\n\n")
gs_best = GridSearchCV(classifier, params, cv=cv, n_jobs=-1, verbose=False).fit(
X, y
)
final_model = classifier.set_params(**gs_best.best_params_)
cv_results = cross_validate(final_model, X, y, cv=cv, scoring=scoring)
print(f"Accuracy: {cv_results['test_accuracy'].mean()}")
print(f"F1Score: {cv_results['test_f1'].mean()}")
print(f"ROC_AUC: {cv_results['test_roc_auc'].mean()}")
print(f"{name} best params: {gs_best.best_params_}", end="\n\n")
best_models[name] = final_model
return best_models
best_models = hyperparameter_optimization(
X_train, y_train, cv=3, scoring=["accuracy", "f1", "roc_auc"]
)
# the function for calculate the tet scores
def test_scores(best_models, X_test, y_test):
for i, j in best_models.items():
print(i)
j.fit(X_train, y_train)
predictions = j.predict(X_test)
print(confusion_matrix(y_test, predictions), end="\n\n")
print(classification_report(y_test, predictions), end="\n\n")
print(accuracy_score(y_test, predictions), end="\n\n")
best_models
from sklearn.metrics import (
classification_report,
confusion_matrix,
accuracy_score,
roc_auc_score,
)
test_scores(best_models, X_test, y_test)
######################################################
# Stacking & Ensemble Learning
######################################################
def voting_classifier(best_models, X, y):
print("Voting Classifier...")
voting_clf = VotingClassifier(
estimators=[
("GBM", best_models["GBM"]),
("LR", best_models["LR"]),
("LightGBM", best_models["LightGBM"]),
],
voting="soft",
).fit(X, y)
cv_results = cross_validate(
voting_clf, X, y, cv=3, scoring=["accuracy", "f1", "roc_auc"]
)
print(f"Accuracy: {cv_results['test_accuracy'].mean()}")
print(f"F1Score: {cv_results['test_f1'].mean()}")
print(f"ROC_AUC: {cv_results['test_roc_auc'].mean()}")
return voting_clf
voting_clf = voting_classifier(best_models, X, y)
voting_clf
voting_clf.predict(X_test)
voting_clf.fit(X_train, y_train)
predictions = voting_clf.predict(X_test)
print(confusion_matrix(y_test, predictions), end="\n\n")
print(classification_report(y_test, predictions), end="\n\n")
print(accuracy_score(y_test, predictions), end="\n\n")
#######################################
# Feature Selection
#######################################
def plot_importance(model, features, num=len(X), save=False):
feature_imp = pd.DataFrame(
{"Value": model.feature_importances_, "Feature": features.columns}
)
plt.figure(figsize=(30, 30))
sns.set(font_scale=1)
sns.barplot(
x="Value",
y="Feature",
data=feature_imp.sort_values(by="Value", ascending=False)[0:num],
)
plt.title("Features")
plt.tight_layout()
plt.show()
if save:
plt.savefig("importances.png")
return
features = pd.DataFrame(
{"Feature": X.columns, "Value": j.feature_importances_}
).sort_values(by="Value", ascending=False)
plot_importance(j, features)
features = pd.DataFrame(
{"Feature": X.columns, "Value": LGBMClassifier().feature_importances_}
).sort_values(by="Value", ascending=False)
plot_importance(voting_clf, features)
|
[{"telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv": {"column_names": "[\"customerID\", \"gender\", \"SeniorCitizen\", \"Partner\", \"Dependents\", \"tenure\", \"PhoneService\", \"MultipleLines\", \"InternetService\", \"OnlineSecurity\", \"OnlineBackup\", \"DeviceProtection\", \"TechSupport\", \"StreamingTV\", \"StreamingMovies\", \"Contract\", \"PaperlessBilling\", \"PaymentMethod\", \"MonthlyCharges\", \"TotalCharges\", \"Churn\"]", "column_data_types": "{\"customerID\": \"object\", \"gender\": \"object\", \"SeniorCitizen\": \"int64\", \"Partner\": \"object\", \"Dependents\": \"object\", \"tenure\": \"int64\", \"PhoneService\": \"object\", \"MultipleLines\": \"object\", \"InternetService\": \"object\", \"OnlineSecurity\": \"object\", \"OnlineBackup\": \"object\", \"DeviceProtection\": \"object\", \"TechSupport\": \"object\", \"StreamingTV\": \"object\", \"StreamingMovies\": \"object\", \"Contract\": \"object\", \"PaperlessBilling\": \"object\", \"PaymentMethod\": \"object\", \"MonthlyCharges\": \"float64\", \"TotalCharges\": \"object\", \"Churn\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 7043 entries, 0 to 7042\nData columns (total 21 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 customerID 7043 non-null object \n 1 gender 7043 non-null object \n 2 SeniorCitizen 7043 non-null int64 \n 3 Partner 7043 non-null object \n 4 Dependents 7043 non-null object \n 5 tenure 7043 non-null int64 \n 6 PhoneService 7043 non-null object \n 7 MultipleLines 7043 non-null object \n 8 InternetService 7043 non-null object \n 9 OnlineSecurity 7043 non-null object \n 10 OnlineBackup 7043 non-null object \n 11 DeviceProtection 7043 non-null object \n 12 TechSupport 7043 non-null object \n 13 StreamingTV 7043 non-null object \n 14 StreamingMovies 7043 non-null object \n 15 Contract 7043 non-null object \n 16 PaperlessBilling 7043 non-null object \n 17 PaymentMethod 7043 non-null object \n 18 MonthlyCharges 7043 non-null float64\n 19 TotalCharges 7043 non-null object \n 20 Churn 7043 non-null object \ndtypes: float64(1), int64(2), object(18)\nmemory usage: 1.1+ MB\n", "summary": "{\"SeniorCitizen\": {\"count\": 7043.0, \"mean\": 0.1621468124378816, \"std\": 0.3686116056100131, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"tenure\": {\"count\": 7043.0, \"mean\": 32.37114865824223, \"std\": 24.55948102309446, \"min\": 0.0, \"25%\": 9.0, \"50%\": 29.0, \"75%\": 55.0, \"max\": 72.0}, \"MonthlyCharges\": {\"count\": 7043.0, \"mean\": 64.76169246059918, \"std\": 30.090047097678493, \"min\": 18.25, \"25%\": 35.5, \"50%\": 70.35, \"75%\": 89.85, \"max\": 118.75}}", "examples": "{\"customerID\":{\"0\":\"7590-VHVEG\",\"1\":\"5575-GNVDE\",\"2\":\"3668-QPYBK\",\"3\":\"7795-CFOCW\"},\"gender\":{\"0\":\"Female\",\"1\":\"Male\",\"2\":\"Male\",\"3\":\"Male\"},\"SeniorCitizen\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Partner\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"Dependents\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"tenure\":{\"0\":1,\"1\":34,\"2\":2,\"3\":45},\"PhoneService\":{\"0\":\"No\",\"1\":\"Yes\",\"2\":\"Yes\",\"3\":\"No\"},\"MultipleLines\":{\"0\":\"No phone service\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No phone service\"},\"InternetService\":{\"0\":\"DSL\",\"1\":\"DSL\",\"2\":\"DSL\",\"3\":\"DSL\"},\"OnlineSecurity\":{\"0\":\"No\",\"1\":\"Yes\",\"2\":\"Yes\",\"3\":\"Yes\"},\"OnlineBackup\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"Yes\",\"3\":\"No\"},\"DeviceProtection\":{\"0\":\"No\",\"1\":\"Yes\",\"2\":\"No\",\"3\":\"Yes\"},\"TechSupport\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"Yes\"},\"StreamingTV\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"StreamingMovies\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"No\",\"3\":\"No\"},\"Contract\":{\"0\":\"Month-to-month\",\"1\":\"One year\",\"2\":\"Month-to-month\",\"3\":\"One year\"},\"PaperlessBilling\":{\"0\":\"Yes\",\"1\":\"No\",\"2\":\"Yes\",\"3\":\"No\"},\"PaymentMethod\":{\"0\":\"Electronic check\",\"1\":\"Mailed check\",\"2\":\"Mailed check\",\"3\":\"Bank transfer (automatic)\"},\"MonthlyCharges\":{\"0\":29.85,\"1\":56.95,\"2\":53.85,\"3\":42.3},\"TotalCharges\":{\"0\":\"29.85\",\"1\":\"1889.5\",\"2\":\"108.15\",\"3\":\"1840.75\"},\"Churn\":{\"0\":\"No\",\"1\":\"No\",\"2\":\"Yes\",\"3\":\"No\"}}"}}]
| true | 1 |
<start_data_description><data_path>telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv:
<column_names>
['customerID', 'gender', 'SeniorCitizen', 'Partner', 'Dependents', 'tenure', 'PhoneService', 'MultipleLines', 'InternetService', 'OnlineSecurity', 'OnlineBackup', 'DeviceProtection', 'TechSupport', 'StreamingTV', 'StreamingMovies', 'Contract', 'PaperlessBilling', 'PaymentMethod', 'MonthlyCharges', 'TotalCharges', 'Churn']
<column_types>
{'customerID': 'object', 'gender': 'object', 'SeniorCitizen': 'int64', 'Partner': 'object', 'Dependents': 'object', 'tenure': 'int64', 'PhoneService': 'object', 'MultipleLines': 'object', 'InternetService': 'object', 'OnlineSecurity': 'object', 'OnlineBackup': 'object', 'DeviceProtection': 'object', 'TechSupport': 'object', 'StreamingTV': 'object', 'StreamingMovies': 'object', 'Contract': 'object', 'PaperlessBilling': 'object', 'PaymentMethod': 'object', 'MonthlyCharges': 'float64', 'TotalCharges': 'object', 'Churn': 'object'}
<dataframe_Summary>
{'SeniorCitizen': {'count': 7043.0, 'mean': 0.1621468124378816, 'std': 0.3686116056100131, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'tenure': {'count': 7043.0, 'mean': 32.37114865824223, 'std': 24.55948102309446, 'min': 0.0, '25%': 9.0, '50%': 29.0, '75%': 55.0, 'max': 72.0}, 'MonthlyCharges': {'count': 7043.0, 'mean': 64.76169246059918, 'std': 30.090047097678493, 'min': 18.25, '25%': 35.5, '50%': 70.35, '75%': 89.85, 'max': 118.75}}
<dataframe_info>
RangeIndex: 7043 entries, 0 to 7042
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 customerID 7043 non-null object
1 gender 7043 non-null object
2 SeniorCitizen 7043 non-null int64
3 Partner 7043 non-null object
4 Dependents 7043 non-null object
5 tenure 7043 non-null int64
6 PhoneService 7043 non-null object
7 MultipleLines 7043 non-null object
8 InternetService 7043 non-null object
9 OnlineSecurity 7043 non-null object
10 OnlineBackup 7043 non-null object
11 DeviceProtection 7043 non-null object
12 TechSupport 7043 non-null object
13 StreamingTV 7043 non-null object
14 StreamingMovies 7043 non-null object
15 Contract 7043 non-null object
16 PaperlessBilling 7043 non-null object
17 PaymentMethod 7043 non-null object
18 MonthlyCharges 7043 non-null float64
19 TotalCharges 7043 non-null object
20 Churn 7043 non-null object
dtypes: float64(1), int64(2), object(18)
memory usage: 1.1+ MB
<some_examples>
{'customerID': {'0': '7590-VHVEG', '1': '5575-GNVDE', '2': '3668-QPYBK', '3': '7795-CFOCW'}, 'gender': {'0': 'Female', '1': 'Male', '2': 'Male', '3': 'Male'}, 'SeniorCitizen': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Partner': {'0': 'Yes', '1': 'No', '2': 'No', '3': 'No'}, 'Dependents': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'tenure': {'0': 1, '1': 34, '2': 2, '3': 45}, 'PhoneService': {'0': 'No', '1': 'Yes', '2': 'Yes', '3': 'No'}, 'MultipleLines': {'0': 'No phone service', '1': 'No', '2': 'No', '3': 'No phone service'}, 'InternetService': {'0': 'DSL', '1': 'DSL', '2': 'DSL', '3': 'DSL'}, 'OnlineSecurity': {'0': 'No', '1': 'Yes', '2': 'Yes', '3': 'Yes'}, 'OnlineBackup': {'0': 'Yes', '1': 'No', '2': 'Yes', '3': 'No'}, 'DeviceProtection': {'0': 'No', '1': 'Yes', '2': 'No', '3': 'Yes'}, 'TechSupport': {'0': 'No', '1': 'No', '2': 'No', '3': 'Yes'}, 'StreamingTV': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'StreamingMovies': {'0': 'No', '1': 'No', '2': 'No', '3': 'No'}, 'Contract': {'0': 'Month-to-month', '1': 'One year', '2': 'Month-to-month', '3': 'One year'}, 'PaperlessBilling': {'0': 'Yes', '1': 'No', '2': 'Yes', '3': 'No'}, 'PaymentMethod': {'0': 'Electronic check', '1': 'Mailed check', '2': 'Mailed check', '3': 'Bank transfer (automatic)'}, 'MonthlyCharges': {'0': 29.85, '1': 56.95, '2': 53.85, '3': 42.3}, 'TotalCharges': {'0': '29.85', '1': '1889.5', '2': '108.15', '3': '1840.75'}, 'Churn': {'0': 'No', '1': 'No', '2': 'Yes', '3': 'No'}}
<end_description>
| 8,924 | 0 | 10,472 | 8,924 |
69492089
|
<jupyter_start><jupyter_text>Volcanic Eruptions in the Holocene Period
# Content
The Smithsonian Institution's Global Volcanism Program (GVP) documents Earth's volcanoes and their eruptive history over the past 10,000 years. The GVP reports on current eruptions from around the world and maintains a database repository on active volcanoes and their eruptions. The GVP is housed in the Department of Mineral Sciences, part of the National Museum of Natural History, on the National Mall in Washington, D.C.
The GVP database includes the names, locations, types, and features of more than 1,500 volcanoes with eruptions during the Holocene period (approximately the last 10,000 years) or exhibiting current unrest.
Kaggle dataset identifier: volcanic-eruptions
<jupyter_script># ### All days of the challange:
# * [Day 1: Handling missing values](https://www.kaggle.com/rtatman/data-cleaning-challenge-handling-missing-values)
# * [Day 2: Scaling and normalization](https://www.kaggle.com/rtatman/data-cleaning-challenge-scale-and-normalize-data)
# * [Day 3: Parsing dates](https://www.kaggle.com/rtatman/data-cleaning-challenge-parsing-dates/)
# * [Day 4: Character encodings](https://www.kaggle.com/rtatman/data-cleaning-challenge-character-encodings/)
# * [Day 5: Inconsistent Data Entry](https://www.kaggle.com/rtatman/data-cleaning-challenge-inconsistent-data-entry/)
# ___
# Welcome to day 3 of the 5-Day Data Challenge! Today, we're going to work with dates. To get started, click the blue "Fork Notebook" button in the upper, right hand corner. This will create a private copy of this notebook that you can edit and play with. Once you're finished with the exercises, you can choose to make your notebook public to share with others. :)
# > **Your turn!** As we work through this notebook, you'll see some notebook cells (a block of either code or text) that has "Your Turn!" written in it. These are exercises for you to do to help cement your understanding of the concepts we're talking about. Once you've written the code to answer a specific question, you can run the code by clicking inside the cell (box with code in it) with the code you want to run and then hit CTRL + ENTER (CMD + ENTER on a Mac). You can also click in a cell and then click on the right "play" arrow to the left of the code. If you want to run all the code in your notebook, you can use the double, "fast forward" arrows at the bottom of the notebook editor.
# Here's what we're going to do today:
# * [Get our environment set up](#Get-our-environment-set-up)
# * [Check the data type of our date column](#Check-the-data-type-of-our-date-column)
# * [Convert our date columns to datetime](#Convert-our-date-columns-to-datetime)
# * [Select just the day of the month from our column](#Select-just-the-day-of-the-month-from-our-column)
# * [Plot the day of the month to check the date parsing](#Plot-the-day-of-the-month-to-the-date-parsing)
# Let's get started!
# # Get our environment set up
# ________
# The first thing we'll need to do is load in the libraries and datasets we'll be using. For today, we'll be working with two datasets: one containing information on earthquakes that occured between 1965 and 2016, and another that contains information on landslides that occured between 2007 and 2016.
# > **Important!** Make sure you run this cell yourself or the rest of your code won't work!
# modules we'll use
import pandas as pd
import numpy as np
import seaborn as sns
import datetime
# read in our data
earthquakes = pd.read_csv("../input/earthquake-database/database.csv")
landslides = pd.read_csv("../input/landslide-events/catalog.csv")
volcanos = pd.read_csv("../input/volcanic-eruptions/database.csv")
# set seed for reproducibility
np.random.seed(0)
earthquakes.head(3)
# Now we're ready to look at some dates! (If you like, you can take this opportunity to take a look at some of the data.)
# # Check the data type of our date column
# ___
# For this part of the challenge, I'll be working with the `date` column from the `landslides` dataframe. The very first thing I'm going to do is take a peek at the first few rows to make sure it actually looks like it contains dates.
# print the first few rows of the date column
print(landslides["date"].head())
# Yep, those are dates! But just because I, a human, can tell that these are dates doesn't mean that Python knows that they're dates. Notice that the at the bottom of the output of `head()`, you can see that it says that the data type of this column is "object".
# > Pandas uses the "object" dtype for storing various types of data types, but most often when you see a column with the dtype "object" it will have strings in it.
# If you check the pandas dtype documentation [here](http://pandas.pydata.org/pandas-docs/stable/basics.html#dtypes), you'll notice that there's also a specific `datetime64` dtypes. Because the dtype of our column is `object` rather than `datetime64`, we can tell that Python doesn't know that this column contains dates.
# We can also look at just the dtype of your column without printing the first few rows if we like:
# check the data type of our date column
landslides["date"].dtype
# You may have to check the [numpy documentation](https://docs.scipy.org/doc/numpy-1.12.0/reference/generated/numpy.dtype.kind.html#numpy.dtype.kind) to match the letter code to the dtype of the object. "O" is the code for "object", so we can see that these two methods give us the same information.
volcanos.head(3)
# Your turn! Check the data type of the Date column in the earthquakes dataframe
# (note the capital 'D' in date!)
earthquakes["Date"].dtype
# # Convert our date columns to datetime
# ___
# Now that we know that our date column isn't being recognized as a date, it's time to convert it so that it *is* recognized as a date. This is called "parsing dates" because we're taking in a string and identifying its component parts.
# We can pandas what the format of our dates are with a guide called as ["strftime directive", which you can find more information on at this link](http://strftime.org/). The basic idea is that you need to point out which parts of the date are where and what punctuation is between them. There are [lots of possible parts of a date](http://strftime.org/), but the most common are `%d` for day, `%m` for month, `%y` for a two-digit year and `%Y` for a four digit year.
# Some examples:
# * 1/17/07 has the format "%m/%d/%y"
# * 17-1-2007 has the format "%d-%m-%Y"
#
# Looking back up at the head of the `date` column in the landslides dataset, we can see that it's in the format "month/day/two-digit year", so we can use the same syntax as the first example to parse in our dates:
# create a new column, date_parsed, with the parsed dates
landslides["date_parsed"] = pd.to_datetime(landslides["date"], format="%m/%d/%y")
# Now when I check the first few rows of the new column, I can see that the dtype is `datetime64`. I can also see that my dates have been slightly rearranged so that they fit the default order datetime objects (year-month-day).
# print the first few rows
landslides["date_parsed"].head()
# Now that our dates are parsed correctly, we can interact with them in useful ways.
# ___
# * **What if I run into an error with multiple date formats?** While we're specifying the date format here, sometimes you'll run into an error when there are multiple date formats in a single column. If that happens, you have have pandas try to infer what the right date format should be. You can do that like so:
# `landslides['date_parsed'] = pd.to_datetime(landslides['Date'], infer_datetime_format=True)`
# * **Why don't you always use `infer_datetime_format = True?`** There are two big reasons not to always have pandas guess the time format. The first is that pandas won't always been able to figure out the correct date format, especially if someone has gotten creative with data entry. The second is that it's much slower than specifying the exact format of the dates.
# ____
# Your turn! Create a new column, date_parsed, in the earthquakes
# dataset that has correctly parsed dates in it. (Don't forget to
# double-check that the dtype is correct!)
earthquakes["date_parsed"] = pd.to_datetime(
earthquakes["Date"], infer_datetime_format=True
)
earthquakes["date_parsed"].head()
# # Select just the day of the month from our column
# ___
# "Ok, Rachael," you may be saying at this point, "This messing around with data types is fine, I guess, but what's the *point*?" To answer your question, let's try to get information on the day of the month that a landslide occured on from the original "date" column, which has an "object" dtype:
# try to get the day of the month from the date column
day_of_month_landslides = landslides["date"].dt.day
# We got an error! The important part to look at here is the part at the very end that says `AttributeError: Can only use .dt accessor with datetimelike values`. We're getting this error because the dt.day() function doesn't know how to deal with a column with the dtype "object". Even though our dataframe has dates in it, because they haven't been parsed we can't interact with them in a useful way.
# Luckily, we have a column that we parsed earlier , and that lets us get the day of the month out no problem:
# get the day of the month from the date_parsed column
day_of_month_landslides = landslides["date_parsed"].dt.day
# Your turn! get the day of the month from the date_parsed column
day_of_month_earthqueks = earthquakes["date_parsed"].dt.day
day_of_month_earthqueks
# # Plot the day of the month to check the date parsing
# ___
# One of the biggest dangers in parsing dates is mixing up the months and days. The to_datetime() function does have very helpful error messages, but it doesn't hurt to double-check that the days of the month we've extracted make sense.
# To do this, let's plot a histogram of the days of the month. We expect it to have values between 1 and 31 and, since there's no reason to suppose the landslides are more common on some days of the month than others, a relatively even distribution. (With a dip on 31 because not all months have 31 days.) Let's see if that's the case:
# remove na's
day_of_month_landslides = day_of_month_landslides.dropna()
# plot the day of the month
sns.distplot(day_of_month_landslides, kde=False, bins=31)
# Yep, it looks like we did parse our dates correctly & this graph makes good sense to me. Why don't you take a turn checking the dates you parsed earlier?
# Your turn! Plot the days of the month from your
# earthquake dataset and make sure they make sense.
# remove na's
day_of_month_earthqueks = day_of_month_earthqueks.dropna()
# plot the day of the month
sns.distplot(day_of_month_earthqueks, kde=False, bins=31)
# And that's it for today! If you have any questions, be sure to post them in the comments below or [on the forums](https://www.kaggle.com/questions-and-answers).
# Remember that your notebook is private by default, and in order to share it with other people or ask for help with it, you'll need to make it public. First, you'll need to save a version of your notebook that shows your current work by hitting the "Commit & Run" button. (Your work is saved automatically, but versioning your work lets you go back and look at what it was like at the point you saved it. It also lets you share a nice compiled notebook instead of just the raw code.) Then, once your notebook is finished running, you can go to the Settings tab in the panel to the left (you may have to expand it by hitting the [<] button next to the "Commit & Run" button) and setting the "Visibility" dropdown to "Public".
# # More practice!
# ___
# If you're interested in graphing time series, [check out this Learn tutorial](https://www.kaggle.com/residentmario/time-series-plotting-optional).
# You can also look into passing columns that you know have dates in them the `parse_dates` argument in `read_csv`. (The documention [is here](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html).) Do note that this method can be very slow, but depending on your needs it may sometimes be handy to use.
# For an extra challenge, you can try try parsing the column `Last Known Eruption` from the `volcanos` dataframe. This column contains a mixture of text ("Unknown") and years both before the common era (BCE, also known as BC) and in the common era (CE, also known as AD).
volcanos["Last Known Eruption"].sample(5)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492089.ipynb
|
volcanic-eruptions
| null |
[{"Id": 69492089, "ScriptId": 18975494, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2018936, "CreationDate": "07/31/2021 16:55:08", "VersionNumber": 1.0, "Title": "Data Cleaning Challenge: Parsing Dates", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 171.0, "LinesInsertedFromPrevious": 15.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 156.0, "LinesInsertedFromFork": 15.0, "LinesDeletedFromFork": 2.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 156.0, "TotalVotes": 0}]
|
[{"Id": 92727417, "KernelVersionId": 69492089, "SourceDatasetVersionId": 1325}, {"Id": 92727416, "KernelVersionId": 69492089, "SourceDatasetVersionId": 1296}, {"Id": 92727418, "KernelVersionId": 69492089, "SourceDatasetVersionId": 1360}]
|
[{"Id": 1325, "DatasetId": 705, "DatasourceVersionId": 1325, "CreatorUserId": 797864, "LicenseName": "CC0: Public Domain", "CreationDate": "01/23/2017 23:00:21", "VersionNumber": 1.0, "Title": "Volcanic Eruptions in the Holocene Period", "Slug": "volcanic-eruptions", "Subtitle": "Name, location, and type of volcanoes active in the past 10,000 years", "Description": "# Content\n\nThe Smithsonian Institution's Global Volcanism Program (GVP) documents Earth's volcanoes and their eruptive history over the past 10,000 years. The GVP reports on current eruptions from around the world and maintains a database repository on active volcanoes and their eruptions. The GVP is housed in the Department of Mineral Sciences, part of the National Museum of Natural History, on the National Mall in Washington, D.C.\n\nThe GVP database includes the names, locations, types, and features of more than 1,500 volcanoes with eruptions during the Holocene period (approximately the last 10,000 years) or exhibiting current unrest.", "VersionNotes": "Initial release", "TotalCompressedBytes": 259712.0, "TotalUncompressedBytes": 259712.0}]
|
[{"Id": 705, "CreatorUserId": 797864, "OwnerUserId": NaN, "OwnerOrganizationId": 389.0, "CurrentDatasetVersionId": 1325.0, "CurrentDatasourceVersionId": 1325.0, "ForumId": 2439, "Type": 2, "CreationDate": "01/23/2017 23:00:21", "LastActivityDate": "02/04/2018", "TotalViews": 34397, "TotalDownloads": 7042, "TotalVotes": 135, "TotalKernels": 827}]
| null |
# ### All days of the challange:
# * [Day 1: Handling missing values](https://www.kaggle.com/rtatman/data-cleaning-challenge-handling-missing-values)
# * [Day 2: Scaling and normalization](https://www.kaggle.com/rtatman/data-cleaning-challenge-scale-and-normalize-data)
# * [Day 3: Parsing dates](https://www.kaggle.com/rtatman/data-cleaning-challenge-parsing-dates/)
# * [Day 4: Character encodings](https://www.kaggle.com/rtatman/data-cleaning-challenge-character-encodings/)
# * [Day 5: Inconsistent Data Entry](https://www.kaggle.com/rtatman/data-cleaning-challenge-inconsistent-data-entry/)
# ___
# Welcome to day 3 of the 5-Day Data Challenge! Today, we're going to work with dates. To get started, click the blue "Fork Notebook" button in the upper, right hand corner. This will create a private copy of this notebook that you can edit and play with. Once you're finished with the exercises, you can choose to make your notebook public to share with others. :)
# > **Your turn!** As we work through this notebook, you'll see some notebook cells (a block of either code or text) that has "Your Turn!" written in it. These are exercises for you to do to help cement your understanding of the concepts we're talking about. Once you've written the code to answer a specific question, you can run the code by clicking inside the cell (box with code in it) with the code you want to run and then hit CTRL + ENTER (CMD + ENTER on a Mac). You can also click in a cell and then click on the right "play" arrow to the left of the code. If you want to run all the code in your notebook, you can use the double, "fast forward" arrows at the bottom of the notebook editor.
# Here's what we're going to do today:
# * [Get our environment set up](#Get-our-environment-set-up)
# * [Check the data type of our date column](#Check-the-data-type-of-our-date-column)
# * [Convert our date columns to datetime](#Convert-our-date-columns-to-datetime)
# * [Select just the day of the month from our column](#Select-just-the-day-of-the-month-from-our-column)
# * [Plot the day of the month to check the date parsing](#Plot-the-day-of-the-month-to-the-date-parsing)
# Let's get started!
# # Get our environment set up
# ________
# The first thing we'll need to do is load in the libraries and datasets we'll be using. For today, we'll be working with two datasets: one containing information on earthquakes that occured between 1965 and 2016, and another that contains information on landslides that occured between 2007 and 2016.
# > **Important!** Make sure you run this cell yourself or the rest of your code won't work!
# modules we'll use
import pandas as pd
import numpy as np
import seaborn as sns
import datetime
# read in our data
earthquakes = pd.read_csv("../input/earthquake-database/database.csv")
landslides = pd.read_csv("../input/landslide-events/catalog.csv")
volcanos = pd.read_csv("../input/volcanic-eruptions/database.csv")
# set seed for reproducibility
np.random.seed(0)
earthquakes.head(3)
# Now we're ready to look at some dates! (If you like, you can take this opportunity to take a look at some of the data.)
# # Check the data type of our date column
# ___
# For this part of the challenge, I'll be working with the `date` column from the `landslides` dataframe. The very first thing I'm going to do is take a peek at the first few rows to make sure it actually looks like it contains dates.
# print the first few rows of the date column
print(landslides["date"].head())
# Yep, those are dates! But just because I, a human, can tell that these are dates doesn't mean that Python knows that they're dates. Notice that the at the bottom of the output of `head()`, you can see that it says that the data type of this column is "object".
# > Pandas uses the "object" dtype for storing various types of data types, but most often when you see a column with the dtype "object" it will have strings in it.
# If you check the pandas dtype documentation [here](http://pandas.pydata.org/pandas-docs/stable/basics.html#dtypes), you'll notice that there's also a specific `datetime64` dtypes. Because the dtype of our column is `object` rather than `datetime64`, we can tell that Python doesn't know that this column contains dates.
# We can also look at just the dtype of your column without printing the first few rows if we like:
# check the data type of our date column
landslides["date"].dtype
# You may have to check the [numpy documentation](https://docs.scipy.org/doc/numpy-1.12.0/reference/generated/numpy.dtype.kind.html#numpy.dtype.kind) to match the letter code to the dtype of the object. "O" is the code for "object", so we can see that these two methods give us the same information.
volcanos.head(3)
# Your turn! Check the data type of the Date column in the earthquakes dataframe
# (note the capital 'D' in date!)
earthquakes["Date"].dtype
# # Convert our date columns to datetime
# ___
# Now that we know that our date column isn't being recognized as a date, it's time to convert it so that it *is* recognized as a date. This is called "parsing dates" because we're taking in a string and identifying its component parts.
# We can pandas what the format of our dates are with a guide called as ["strftime directive", which you can find more information on at this link](http://strftime.org/). The basic idea is that you need to point out which parts of the date are where and what punctuation is between them. There are [lots of possible parts of a date](http://strftime.org/), but the most common are `%d` for day, `%m` for month, `%y` for a two-digit year and `%Y` for a four digit year.
# Some examples:
# * 1/17/07 has the format "%m/%d/%y"
# * 17-1-2007 has the format "%d-%m-%Y"
#
# Looking back up at the head of the `date` column in the landslides dataset, we can see that it's in the format "month/day/two-digit year", so we can use the same syntax as the first example to parse in our dates:
# create a new column, date_parsed, with the parsed dates
landslides["date_parsed"] = pd.to_datetime(landslides["date"], format="%m/%d/%y")
# Now when I check the first few rows of the new column, I can see that the dtype is `datetime64`. I can also see that my dates have been slightly rearranged so that they fit the default order datetime objects (year-month-day).
# print the first few rows
landslides["date_parsed"].head()
# Now that our dates are parsed correctly, we can interact with them in useful ways.
# ___
# * **What if I run into an error with multiple date formats?** While we're specifying the date format here, sometimes you'll run into an error when there are multiple date formats in a single column. If that happens, you have have pandas try to infer what the right date format should be. You can do that like so:
# `landslides['date_parsed'] = pd.to_datetime(landslides['Date'], infer_datetime_format=True)`
# * **Why don't you always use `infer_datetime_format = True?`** There are two big reasons not to always have pandas guess the time format. The first is that pandas won't always been able to figure out the correct date format, especially if someone has gotten creative with data entry. The second is that it's much slower than specifying the exact format of the dates.
# ____
# Your turn! Create a new column, date_parsed, in the earthquakes
# dataset that has correctly parsed dates in it. (Don't forget to
# double-check that the dtype is correct!)
earthquakes["date_parsed"] = pd.to_datetime(
earthquakes["Date"], infer_datetime_format=True
)
earthquakes["date_parsed"].head()
# # Select just the day of the month from our column
# ___
# "Ok, Rachael," you may be saying at this point, "This messing around with data types is fine, I guess, but what's the *point*?" To answer your question, let's try to get information on the day of the month that a landslide occured on from the original "date" column, which has an "object" dtype:
# try to get the day of the month from the date column
day_of_month_landslides = landslides["date"].dt.day
# We got an error! The important part to look at here is the part at the very end that says `AttributeError: Can only use .dt accessor with datetimelike values`. We're getting this error because the dt.day() function doesn't know how to deal with a column with the dtype "object". Even though our dataframe has dates in it, because they haven't been parsed we can't interact with them in a useful way.
# Luckily, we have a column that we parsed earlier , and that lets us get the day of the month out no problem:
# get the day of the month from the date_parsed column
day_of_month_landslides = landslides["date_parsed"].dt.day
# Your turn! get the day of the month from the date_parsed column
day_of_month_earthqueks = earthquakes["date_parsed"].dt.day
day_of_month_earthqueks
# # Plot the day of the month to check the date parsing
# ___
# One of the biggest dangers in parsing dates is mixing up the months and days. The to_datetime() function does have very helpful error messages, but it doesn't hurt to double-check that the days of the month we've extracted make sense.
# To do this, let's plot a histogram of the days of the month. We expect it to have values between 1 and 31 and, since there's no reason to suppose the landslides are more common on some days of the month than others, a relatively even distribution. (With a dip on 31 because not all months have 31 days.) Let's see if that's the case:
# remove na's
day_of_month_landslides = day_of_month_landslides.dropna()
# plot the day of the month
sns.distplot(day_of_month_landslides, kde=False, bins=31)
# Yep, it looks like we did parse our dates correctly & this graph makes good sense to me. Why don't you take a turn checking the dates you parsed earlier?
# Your turn! Plot the days of the month from your
# earthquake dataset and make sure they make sense.
# remove na's
day_of_month_earthqueks = day_of_month_earthqueks.dropna()
# plot the day of the month
sns.distplot(day_of_month_earthqueks, kde=False, bins=31)
# And that's it for today! If you have any questions, be sure to post them in the comments below or [on the forums](https://www.kaggle.com/questions-and-answers).
# Remember that your notebook is private by default, and in order to share it with other people or ask for help with it, you'll need to make it public. First, you'll need to save a version of your notebook that shows your current work by hitting the "Commit & Run" button. (Your work is saved automatically, but versioning your work lets you go back and look at what it was like at the point you saved it. It also lets you share a nice compiled notebook instead of just the raw code.) Then, once your notebook is finished running, you can go to the Settings tab in the panel to the left (you may have to expand it by hitting the [<] button next to the "Commit & Run" button) and setting the "Visibility" dropdown to "Public".
# # More practice!
# ___
# If you're interested in graphing time series, [check out this Learn tutorial](https://www.kaggle.com/residentmario/time-series-plotting-optional).
# You can also look into passing columns that you know have dates in them the `parse_dates` argument in `read_csv`. (The documention [is here](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html).) Do note that this method can be very slow, but depending on your needs it may sometimes be handy to use.
# For an extra challenge, you can try try parsing the column `Last Known Eruption` from the `volcanos` dataframe. This column contains a mixture of text ("Unknown") and years both before the common era (BCE, also known as BC) and in the common era (CE, also known as AD).
volcanos["Last Known Eruption"].sample(5)
| false | 0 | 3,171 | 0 | 3,376 | 3,171 |
||
69492055
|
<jupyter_start><jupyter_text>RoBERTa | Transformers | Pytorch
Created using the following kernel: https://www.kaggle.com/maroberti/transformers-model-downloader-pytorch-tf2-0
Kaggle dataset identifier: roberta-transformers-pytorch
<jupyter_script># ### ver17
# * robertabase
# * roberta large
# * roberta large meanpooling
# * roberta base embedding svm+ridge
# * roberta large embedding svm+ridge
# のブレンド
# * robertabase + roberta + roberta large meanpooling のstacking
# # Define
import os
import math
import random
import time
import glob
import re
import gc
gc.enable()
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import pickle
from tqdm import tqdm
import gc
from sklearn.model_selection import KFold, StratifiedKFold, train_test_split
from sklearn.metrics import mean_squared_error as mse
from sklearn.linear_model import LinearRegression, Ridge
from sklearn.svm import SVR
import warnings
warnings.filterwarnings("ignore")
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold
import torch
import torch.nn as nn
# from torch.utils.data import Dataset
# from torch.utils.data import DataLoader
from torch.utils.data import Dataset, SequentialSampler, DataLoader
import transformers
# from transformers import AdamW
# from transformers import AutoTokenizer
# from transformers import AutoModel
# from transformers import AutoConfig
from transformers import get_cosine_schedule_with_warmup
from transformers import (
AutoConfig,
AutoModel,
AutoTokenizer,
AdamW,
get_linear_schedule_with_warmup,
logging,
)
import gc
gc.enable()
import tensorflow as tf
from tensorflow.keras.layers import (
Input,
LSTM,
Bidirectional,
Embedding,
Dense,
Conv1D,
Dropout,
MaxPool1D,
MaxPooling1D,
GlobalAveragePooling2D,
GlobalAveragePooling1D,
GlobalMaxPooling1D,
concatenate,
Flatten,
)
from tensorflow.keras.metrics import RootMeanSquaredError
from tensorflow.keras.models import Model, load_model, save_model, model_from_json
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import (
ReduceLROnPlateau,
ModelCheckpoint,
EarlyStopping,
LearningRateScheduler,
)
from tensorflow.keras.utils import plot_model
from tensorflow.keras import backend as K
from transformers import (
TFBertModel,
BertTokenizerFast,
BertTokenizer,
RobertaTokenizerFast,
TFRobertaModel,
RobertaConfig,
TFAutoModel,
AutoTokenizer,
)
train_df = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_df = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
submission_df = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
# # roberta large embedding SVM & Ridge CV: 0.4822 LB:0.471
# https://www.kaggle.com/iamnishipy/submit-my-roberta-large-svm?scriptVersionId=69275054
# NUM_FOLDS = 5
# NUM_EPOCHS = 3
# BATCH_SIZE = 8
# MAX_LEN = 248
# EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1., 1)]
ROBERTA_PATH = "../input/roberta-large-20210712191259-mlm/clrp_roberta_large/"
TOKENIZER_PATH = "../input/roberta-large-20210712191259-mlm/clrp_roberta_large/"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
class Model(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
# https://towardsdatascience.com/attention-based-deep-multiple-instance-learning-1bb3df857e24
# 768: node fully connected layer
# 512: node attention layer
# self.attention = nn.Sequential(
# nn.Linear(768, 512),
# nn.Tanh(),
# nn.Linear(512, 1),
# nn.Softmax(dim=1)
# )
# self.regressor = nn.Sequential(
# nn.Linear(768, 1)
# )
# 768 -> 1024
# 512 -> 768
self.attention = nn.Sequential(
nn.Linear(1024, 768), nn.Tanh(), nn.Linear(768, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(1024, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
train_data = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
# test_data = pd.read_csv('../input/commonlitreadabilityprize/test.csv')
# sample = pd.read_csv('../input/commonlitreadabilityprize/sample_submission.csv')
num_bins = int(np.floor(1 + np.log2(len(train_data))))
train_data.loc[:, "bins"] = pd.cut(train_data["target"], bins=num_bins, labels=False)
target = train_data["target"].to_numpy()
bins = train_data.bins.to_numpy()
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": 8,
"max_len": 248,
"nfolds": 10,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
def get_embeddings(df, path, plot_losses=True, verbose=True):
# cuda使えたら使う構文
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
# tokenizer = AutoTokenizer.from_pretrained('../input/clrp-roberta-base/clrp_roberta_base')
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
# 以下でpredictionsを抽出するために使った構文を使ってembeddingsをreturnしている.
# SVMの手法とは、embeddingsの意味は?
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
gc.collect
return np.array(embeddings)
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": 8,
"max_len": 248,
"nfolds": 10,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
def get_preds_svm(X, y, X_test, bins=bins, nfolds=10, C=10, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
preds_ridge = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(
n_splits=config["nfolds"], shuffle=True, random_state=config["seed"]
)
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
model = SVR(C=5, kernel=kernel, gamma="auto")
model_ridge = Ridge(alpha=10)
# print(train_idx)
# print(valid_idx)
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
model_ridge.fit(X_train, y_train)
prediction = model.predict(X_valid)
pred_ridge = model_ridge.predict(X_valid)
# score = rmse_score(prediction,y_valid)
score = rmse_score(y_valid, prediction)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
preds_ridge += model_ridge.predict(X_test)
print("mean rmse", np.mean(scores))
return (np.array(preds) / nfolds + np.array(preds_ridge) / nfolds) / 2
# ## Inference
preds = []
for i in range(5):
train_embeddings = get_embeddings(
train_data, f"../input/roberta-large-20210720020531-sch/model_{i+1}.pth"
)
test_embeddings = get_embeddings(
test_df, f"../input/roberta-large-20210720020531-sch/model_{i+1}.pth"
)
svm_pred = get_preds_svm(train_embeddings, target, test_embeddings)
preds.append(svm_pred)
del train_embeddings, test_embeddings, svm_pred
gc.collect()
large_svmridge_pred = (preds[0] + preds[1] + preds[2] + preds[3] + preds[4]) / 5
del tokenizer, train_data, num_bins, target, bins
gc.collect
# test_embeddings1 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_1.pth')
# train_embedding = get_embeddings(train_df,'../input/roberta-large-20210720020531-sch/model_1.pth')
# test_embeddings2 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_2.pth')
# test_embeddings3 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_3.pth')
# test_embeddings4 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_4.pth')
# test_embeddings5 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_5.pth')
# train_embeddings1 = np.loadtxt('../input/robertalargeembedding/train1.csv', delimiter=',')
# train_embeddings2 = np.loadtxt('../input/robertalargeembedding/train2.csv', delimiter=',')
# train_embeddings3 = np.loadtxt('../input/robertalargeembedding/train3.csv', delimiter=',')
# train_embeddings4 = np.loadtxt('../input/robertalargeembedding/train4.csv', delimiter=',')
# train_embeddings5 = np.loadtxt('../input/robertalargeembedding/train5.csv', delimiter=',')
# # roberta base embedding SVM & Ridge CV: 0.4787 LB:0.465
# https://www.kaggle.com/iamnishipy/submit-my-roberta-base-svm?scriptVersionId=69168038
NUM_FOLDS = 5
NUM_EPOCHS = 3
BATCH_SIZE = 16
MAX_LEN = 248
EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1.0, 1)]
ROBERTA_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
TOKENIZER_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
train_data = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_data = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
sample = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
num_bins = int(np.floor(1 + np.log2(len(train_data))))
train_data.loc[:, "bins"] = pd.cut(train_data["target"], bins=num_bins, labels=False)
target = train_data["target"].to_numpy()
bins = train_data.bins.to_numpy()
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": BATCH_SIZE,
"max_len": MAX_LEN,
"nfolds": 10,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
class Model(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(768, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
def get_embeddings(df, path, plot_losses=True, verbose=True):
# cuda使えたら使う構文
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
# tokenizer = AutoTokenizer.from_pretrained('../input/clrp-roberta-base/clrp_roberta_base')
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
# 以下でpredictionsを抽出するために使った構文を使ってembeddingsをreturnしている.
# SVMの手法とは、embeddingsの意味は?
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
return np.array(embeddings)
def get_preds_svm(X, y, X_test, bins=bins, nfolds=10, C=10, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
preds_ridge = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(
n_splits=config["nfolds"], shuffle=True, random_state=config["seed"]
)
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
model = SVR(C=5, kernel=kernel, gamma="auto")
model_ridge = Ridge(alpha=10)
# print(train_idx)
# print(valid_idx)
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
model_ridge.fit(X_train, y_train)
prediction = model.predict(X_valid)
pred_ridge = model_ridge.predict(X_valid)
# score = rmse_score(prediction,y_valid)
score = rmse_score(y_valid, prediction)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
preds_ridge += model_ridge.predict(X_test)
print("mean rmse", np.mean(scores))
return (np.array(preds) / nfolds + np.array(preds_ridge) / nfolds) / 2
# ## Inference
###
# preds = []
# for i in range(5):
# train_embeddings = get_embeddings(train_data, f'../input/roberta-base-20210711202147-sche/model_{i+1}.pth')
# test_embeddings = get_embeddings(test_data, f'../input/roberta-base-20210711202147-sche/model_{i+1}.pth')
# svm_pred = get_preds_svm(train_embeddings, target, test_embeddings)
# preds.append(svm_pred)
# del train_embeddings,test_embeddings,svm_pred
# gc.collect()
# roberta_svmridge_pred = (preds[0] + preds[1] + preds[2] + preds[3] + preds[4])/5
# roberta_svmridge_pred
# del tokenizer,train_data,num_bins,target,bins
# gc.collect
# #train/testでembeddingsを取得している
# train_embeddings1 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_1.pth')
# test_embeddings1 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_1.pth')
# train_embeddings2 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_2.pth')
# test_embeddings2 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_2.pth')
# train_embeddings3 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_3.pth')
# test_embeddings3 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_3.pth')
# train_embeddings4 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_4.pth')
# test_embeddings4 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_4.pth')
# train_embeddings5 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_5.pth')
# test_embeddings5 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_5.pth')
# preds1 = get_preds_svm(train_embeddings1,target,test_embeddings1)
# preds2 = get_preds_svm(train_embeddings2,target,test_embeddings2)
# preds3 = get_preds_svm(train_embeddings3,target,test_embeddings3)
# preds4 = get_preds_svm(train_embeddings4,target,test_embeddings4)
# preds5 = get_preds_svm(train_embeddings5,target,test_embeddings5)
# roberta_svmridge_pred = (preds1 + preds2 + preds3 + preds4 + preds5)/5
# del preds1,preds2,preds3,preds4,preds5
# del train_embeddings1,train_embeddings1,train_embeddings1,train_embeddings1,train_embeddings1
# del test_embeddings1,test_embeddings2,test_embeddings3,test_embeddings4,test_embeddings5
# # pytorch_t5_large_svm
# https://www.kaggle.com/gilfernandes/commonlit-pytorch-t5-large-svm
import yaml, gc
from pathlib import Path
from transformers import T5EncoderModel
class CommonLitModel(nn.Module):
def __init__(self):
super(CommonLitModel, self).__init__()
config = AutoConfig.from_pretrained(cfg.MODEL_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.transformer_model = T5EncoderModel.from_pretrained(
cfg.MODEL_PATH, config=config
)
self.attention = AttentionHead(config.hidden_size, 512, 1)
self.regressor = nn.Linear(config.hidden_size, 1)
def forward(self, input_ids, attention_mask):
last_layer_hidden_states = self.transformer_model(
input_ids=input_ids, attention_mask=attention_mask
)["last_hidden_state"]
weights = self.attention(last_layer_hidden_states)
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
return self.regressor(context_vector), context_vector
BASE_PATH = Path("/kaggle/input/commonlit-t5-large")
DATA_PATH = Path("/kaggle/input/commonlitreadabilityprize/")
assert DATA_PATH.exists()
MODELS_PATH = Path(BASE_PATH / "best_models")
assert MODELS_PATH.exists()
class Config:
NUM_FOLDS = 6
NUM_EPOCHS = 3
BATCH_SIZE = 16
MAX_LEN = 248
MODEL_PATH = BASE_PATH / "lm"
# TOKENIZER_PATH = str(MODELS_PATH/'roberta-base-0')
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
SEED = 1000
NUM_WORKERS = 2
MODEL_FOLDER = MODELS_PATH
model_name = "t5-large"
svm_kernels = ["rbf"]
svm_c = 5
cfg = Config()
train_df["normalized_target"] = (
train_df["target"] - train_df["target"].mean()
) / train_df["target"].std()
model_path = MODELS_PATH
assert model_path.exists()
from transformers import T5EncoderModel
class AttentionHead(nn.Module):
def __init__(self, in_features, hidden_dim, num_targets):
super().__init__()
self.in_features = in_features
self.hidden_layer = nn.Linear(in_features, hidden_dim)
self.final_layer = nn.Linear(hidden_dim, num_targets)
self.out_features = hidden_dim
def forward(self, features):
att = torch.tanh(self.hidden_layer(features))
score = self.final_layer(att)
attention_weights = torch.softmax(score, dim=1)
return attention_weights
class CommonLitModel(nn.Module):
def __init__(self):
super(CommonLitModel, self).__init__()
config = AutoConfig.from_pretrained(cfg.MODEL_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.transformer_model = T5EncoderModel.from_pretrained(
cfg.MODEL_PATH, config=config
)
self.attention = AttentionHead(config.hidden_size, 512, 1)
self.regressor = nn.Linear(config.hidden_size, 1)
def forward(self, input_ids, attention_mask):
last_layer_hidden_states = self.transformer_model(
input_ids=input_ids, attention_mask=attention_mask
)["last_hidden_state"]
weights = self.attention(last_layer_hidden_states)
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
return self.regressor(context_vector), context_vector
def load_model(i):
inference_model = CommonLitModel()
inference_model = inference_model.cuda()
inference_model.load_state_dict(
torch.load(str(model_path / f"{i + 1}_pytorch_model.bin"))
)
inference_model.eval()
return inference_model
def convert_to_list(t):
return t.flatten().long()
class CommonLitDataset(nn.Module):
def __init__(self, text, test_id, tokenizer, max_len=128):
self.excerpt = text
self.test_id = test_id
self.max_len = max_len
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=self.max_len,
padding="max_length",
truncation=True,
)
return {
"input_ids": convert_to_list(encode["input_ids"]),
"attention_mask": convert_to_list(encode["attention_mask"]),
"id": self.test_id[idx],
}
def __len__(self):
return len(self.excerpt)
def create_dl(df, tokenizer):
text = df["excerpt"].values
ids = df["id"].values
ds = CommonLitDataset(text, ids, tokenizer, max_len=cfg.MAX_LEN)
return DataLoader(
ds,
batch_size=cfg.BATCH_SIZE,
shuffle=False,
num_workers=1,
pin_memory=True,
drop_last=False,
)
def get_cls_embeddings(dl, transformer_model):
cls_embeddings = []
with torch.no_grad():
for input_features in tqdm(dl, total=len(dl)):
_, context_vector = transformer_model(
input_features["input_ids"].cuda(),
input_features["attention_mask"].cuda(),
)
# cls_embeddings.extend(output['last_hidden_state'][:,0,:].detach().cpu().numpy())
embedding_out = context_vector.detach().cpu().numpy()
cls_embeddings.extend(embedding_out)
return np.array(cls_embeddings)
def rmse_score(X, y):
return np.sqrt(mean_squared_error(X, y))
def calc_mean(scores):
return np.mean(np.array(scores), axis=0)
from transformers import T5Tokenizer
tokenizers = []
for i in range(1, cfg.NUM_FOLDS):
tokenizer_path = MODELS_PATH / f"tokenizer-{i}"
print(tokenizer_path)
assert Path(tokenizer_path).exists()
tokenizer = T5Tokenizer.from_pretrained(str(tokenizer_path))
tokenizers.append(tokenizer)
num_bins = int(np.ceil(np.log2(len(train_df))))
train_df["bins"] = pd.cut(train_df["target"], bins=num_bins, labels=False)
bins = train_df["bins"].values
train_target = train_df["normalized_target"].values
each_mean_score = []
each_fold_score = []
c = 10
al = 10
# for c in C:
final_scores_svm = []
final_scores_ridge = []
final_rmse = []
for j, tokenizer in enumerate(tokenizers):
print("Model", j)
test_dl = create_dl(test_df, tokenizer)
train_dl = create_dl(train_df, tokenizer)
transformer_model = load_model(j)
transformer_model.cuda()
X = get_cls_embeddings(train_dl, transformer_model)
y = train_target
X_test = get_cls_embeddings(test_dl, transformer_model)
kfold = StratifiedKFold(n_splits=cfg.NUM_FOLDS)
scores = []
scores_svm = []
scores_ridge = []
rmse_scores_svm = []
rmse_scores_ridge = []
rmse_scores = []
for kernel in cfg.svm_kernels:
print("Kernel", kernel)
kernel_scores_svm = []
kernel_scores_ridge = []
kernel_scores = []
kernel_rmse_scores = []
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
print("Fold", k, train_idx.shape, valid_idx.shape)
model_svm = SVR(C=c, kernel=kernel, gamma="auto")
model_ridge = Ridge(alpha=al)
# model = SVR(C=cfg.svm_c, kernel=kernel, gamma='auto')
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model_svm.fit(X_train, y_train)
model_ridge.fit(X_train, y_train)
pred_svm = model_svm.predict(X_valid)
pred_ridge = model_ridge.predict(X_valid)
prediction = (pred_svm + pred_ridge) / 2
kernel_rmse_scores.append(rmse_score(prediction, y_valid))
print("rmse_score", kernel_rmse_scores[k])
kernel_scores_svm.append(model_svm.predict(X_test))
kernel_scores_ridge.append(model_ridge.predict(X_test))
score_mean = (
np.array(kernel_scores_svm) + np.array(kernel_scores_ridge)
) / 2
kernel_scores.append(score_mean)
# scores.append(calc_mean(kernel_scores))
scores_svm.append(calc_mean(kernel_scores_svm))
scores_ridge.append(calc_mean(kernel_scores_ridge))
rmse_scores.append(calc_mean(kernel_rmse_scores))
final_scores_svm.append(calc_mean(scores_svm))
final_scores_ridge.append(calc_mean(scores_ridge))
# final_rmse.append(calc_mean(rmse_scores))
final_rmse.append(calc_mean(rmse_scores))
del transformer_model
torch.cuda.empty_cache()
del tokenizer
gc.collect()
print("FINAL RMSE score", np.mean(np.array(final_rmse)))
each_mean_score.append(np.mean(np.array(final_rmse)))
each_fold_score.append(final_rmse)
print("BEST SCORE : ", each_mean_score)
print("FOLD SCORE : ", each_fold_score)
final_scores = (np.array(final_scores_svm) + np.array(final_scores_ridge)) / 2
final_scores2 = final_scores * train_df["target"].std() + train_df["target"].mean()
target_mean = train_df["target"].mean()
t5_embedding_pred = calc_mean(final_scores2).flatten()
t5_embedding_pred
# del final_scores,train_target,tokenizers,model_svm,model_ridge,X_train,X_valid,y_valid
# gc.collect
# # roberta large pretrain svm&ridge LB:0.468(使えないのでこのモデルは使わん)
# cpptake Note:https://www.kaggle.com/takeshikobayashi/clrp-roberta-svm-roberta-large/edit/run/69118503
# 元Note:https://www.kaggle.com/tutty1992/clrp-roberta-svm-roberta-large
# # train_data = pd.read_csv('../input/commonlitreadabilityprize/train.csv')
# # test_data = pd.read_csv('../input/commonlitreadabilityprize/test.csv')
# # sample = pd.read_csv('../input/commonlitreadabilityprize/sample_submission.csv')
# num_bins = int(np.floor(1 + np.log2(len(train_df))))
# train_df.loc[:,'bins'] = pd.cut(train_df['target'],bins=num_bins,labels=False)
# target = train_df['target'].to_numpy()
# bins = train_df.bins.to_numpy()
# def rmse_score(y_true,y_pred):
# return np.sqrt(mean_squared_error(y_true,y_pred))
# config = {
# 'batch_size':128,
# 'max_len':256,
# 'nfolds':10,
# 'seed':42,
# }
# def seed_everything(seed=42):
# random.seed(seed)
# os.environ['PYTHONASSEED'] = str(seed)
# np.random.seed(seed)
# torch.manual_seed(seed)
# torch.cuda.manual_seed(seed)
# torch.backends.cudnn.deterministic = True
# torch.backends.cudnn.benchmark = True
# seed_everything(seed=config['seed'])
# from sklearn.ensemble import RandomForestRegressor as RFR
# class CLRPDataset(Dataset):
# def __init__(self,df,tokenizer):
# self.excerpt = df['excerpt'].to_numpy()
# self.tokenizer = tokenizer
# def __getitem__(self,idx):
# encode = self.tokenizer(self.excerpt[idx],return_tensors='pt',
# max_length=config['max_len'],
# padding='max_length',truncation=True)
# return encode
# def __len__(self):
# return len(self.excerpt)
# class AttentionHead(nn.Module):
# def __init__(self, in_features, hidden_dim, num_targets):
# super().__init__()
# self.in_features = in_features
# self.middle_features = hidden_dim
# self.W = nn.Linear(in_features, hidden_dim)
# self.V = nn.Linear(hidden_dim, 1)
# self.out_features = hidden_dim
# def forward(self, features):
# att = torch.tanh(self.W(features))
# score = self.V(att)
# attention_weights = torch.softmax(score, dim=1)
# context_vector = attention_weights * features
# context_vector = torch.sum(context_vector, dim=1)
# return context_vector
# class Model(nn.Module):
# def __init__(self):
# super(Model,self).__init__()
# self.roberta = AutoModel.from_pretrained('../input/clrp-pytorch-roberta-pretrain-roberta-large/clrp_roberta_large/')
# #changed attentionHead Dimension from 768 to 1024 by changing model from roberta-base to roberta-large
# self.head = AttentionHead(1024,1024,1)
# self.dropout = nn.Dropout(0.1)
# self.linear = nn.Linear(self.head.out_features,1)
# def forward(self,**xb):
# x = self.roberta(**xb)[0]
# x = self.head(x)
# return x
# def get_embeddings(df,path,plot_losses=True, verbose=True):
# device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# print(f"{device} is used")
# model = Model()
# model.load_state_dict(torch.load(path))
# model.to(device)
# model.eval()
# tokenizer = AutoTokenizer.from_pretrained('../input/clrp-pytorch-roberta-pretrain-roberta-large/clrp_roberta_large/')
# ds = CLRPDataset(df,tokenizer)
# dl = DataLoader(ds,
# batch_size = config["batch_size"],
# shuffle=False,
# num_workers = 4,
# pin_memory=True,
# drop_last=False
# )
# embeddings = list()
# with torch.no_grad():
# for i, inputs in tqdm(enumerate(dl)):
# inputs = {key:val.reshape(val.shape[0],-1).to(device) for key,val in inputs.items()}
# outputs = model(**inputs)
# outputs = outputs.detach().cpu().numpy()
# embeddings.extend(outputs)
# return np.array(embeddings)
# def get_preds_svm_ridge(X,y,X_test,bins=bins,nfolds=10,C=10,kernel='rbf'):
# scores = list()
# preds = np.zeros((X_test.shape[0]))
# preds_ridge = np.zeros((X_test.shape[0]))
# kfold = StratifiedKFold(n_splits=config['nfolds'],shuffle=True,random_state=config['seed'])
# for k, (train_idx,valid_idx) in enumerate(kfold.split(X,bins)):
# model = SVR(C=5,kernel=kernel,gamma='auto')
# model_ridge = Ridge(alpha=10)
# X_train,y_train = X[train_idx], y[train_idx]
# X_valid,y_valid = X[valid_idx], y[valid_idx]
# model.fit(X_train,y_train)
# model_ridge.fit(X_train, y_train)
# prediction = model.predict(X_valid)
# pred_ridge = model_ridge.predict(X_valid)
# score = rmse_score(prediction,y_valid)
# print(f'Fold {k} , rmse score: {score}')
# scores.append(score)
# preds += model.predict(X_test)
# preds_ridge += model_ridge.predict(X_test)
# print("mean rmse",np.mean(scores))
# return (np.array(preds)/nfolds + np.array(preds_ridge)/nfolds)/2
# ## embedding パート
# embedding重すぎたため、特徴量のみdataset化して保存して使ってる
# # np.loadtxt('data/src/sample.csv', delimiter=',')
# train_embeddings1 = np.loadtxt('../input/roberta-embedding-dataset/train1.csv', delimiter=',')
# train_embeddings2 = np.loadtxt('../input/roberta-embedding-dataset/train2.csv', delimiter=',')
# train_embeddings3 = np.loadtxt('../input/roberta-embedding-dataset/train3.csv', delimiter=',')
# train_embeddings4 = np.loadtxt('../input/roberta-embedding-dataset/train4.csv', delimiter=',')
# train_embeddings5 = np.loadtxt('../input/roberta-embedding-dataset/train5.csv', delimiter=',')
# test_embeddings1 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model0/model0.bin')
# test_embeddings2 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model1/model1.bin')
# test_embeddings3 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model2/model2.bin')
# test_embeddings4 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model3/model3.bin')
# test_embeddings5 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model4/model4.bin')
# ## pred SVM & Ridge
# from sklearn.linear_model import Ridge
# embedding_preds1 = get_preds_svm_ridge(train_embeddings1,target,test_embeddings1)
# embedding_preds2 = get_preds_svm_ridge(train_embeddings2,target,test_embeddings2)
# embedding_preds3 = get_preds_svm_ridge(train_embeddings3,target,test_embeddings3)
# embedding_preds4 = get_preds_svm_ridge(train_embeddings4,target,test_embeddings4)
# embedding_preds5 = get_preds_svm_ridge(train_embeddings5,target,test_embeddings5)
# svm_ridge_preds = (embedding_preds1 + embedding_preds2 + embedding_preds3 + embedding_preds4 + embedding_preds5)/5
# svm_ridge_preds
# del train_embeddings1,test_embeddings1
# del train_embeddings2,test_embeddings2
# del train_embeddings3,test_embeddings3
# del train_embeddings4,test_embeddings4
# del train_embeddings5,test_embeddings5
# gc.collect
# # nishipy roberta base SVM&Ridge
# NUM_FOLDS = 5
# NUM_EPOCHS = 3
# BATCH_SIZE = 16
# MAX_LEN = 248
# EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1., 1)]
# ROBERTA_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
# TOKENIZER_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
# DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
# tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
# config = {
# 'batch_size':128,
# 'max_len':256,
# 'nfolds':10,
# 'seed':42,
# }
# def seed_everything(seed=42):
# random.seed(seed)
# os.environ['PYTHONASSEED'] = str(seed)
# np.random.seed(seed)
# torch.manual_seed(seed)
# torch.cuda.manual_seed(seed)
# torch.backends.cudnn.deterministic = True
# torch.backends.cudnn.benchmark = True
# seed_everything(seed=config['seed'])
# class CLRPDataset(Dataset):
# def __init__(self,df,tokenizer):
# self.excerpt = df['excerpt'].to_numpy()
# self.tokenizer = tokenizer
# def __getitem__(self,idx):
# encode = self.tokenizer(self.excerpt[idx],return_tensors='pt',
# max_length=config['max_len'],
# padding='max_length',truncation=True)
# return encode
# def __len__(self):
# return len(self.excerpt)
# class Model(nn.Module):
# def __init__(self):
# super().__init__()
# config = AutoConfig.from_pretrained(ROBERTA_PATH)
# config.update({"output_hidden_states":True,
# "hidden_dropout_prob": 0.0,
# "layer_norm_eps": 1e-7})
# self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
# self.attention = nn.Sequential(
# nn.Linear(768, 512),
# nn.Tanh(),
# nn.Linear(512, 1),
# nn.Softmax(dim=1)
# )
# self.regressor = nn.Sequential(
# nn.Linear(768, 1)
# )
# def forward(self, input_ids, attention_mask):
# roberta_output = self.roberta(input_ids=input_ids,
# attention_mask=attention_mask)
# # There are a total of 13 layers of hidden states.
# # 1 for the embedding layer, and 12 for the 12 Roberta layers.
# # We take the hidden states from the last Roberta layer.
# last_layer_hidden_states = roberta_output.hidden_states[-1]
# # The number of cells is MAX_LEN.
# # The size of the hidden state of each cell is 768 (for roberta-base).
# # In order to condense hidden states of all cells to a context vector,
# # we compute a weighted average of the hidden states of all cells.
# # We compute the weight of each cell, using the attention neural network.
# weights = self.attention(last_layer_hidden_states)
# # weights.shape is BATCH_SIZE x MAX_LEN x 1
# # last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# # Now we compute context_vector as the weighted average.
# # context_vector.shape is BATCH_SIZE x 768
# context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# # Now we reduce the context vector to the prediction score.
# return self.regressor(context_vector)
# def predict(model, data_loader):
# """Returns an np.array with predictions of the |model| on |data_loader|"""
# model.eval()
# result = np.zeros(len(data_loader.dataset))
# index = 0
# with torch.no_grad():
# for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
# input_ids = input_ids.to(DEVICE)
# attention_mask = attention_mask.to(DEVICE)
# pred = model(input_ids, attention_mask)
# result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
# index += pred.shape[0]
# return result
# def rmse_score(y_true,y_pred):
# return np.sqrt(mean_squared_error(y_true,y_pred))
# def seed_everything(seed=42):
# random.seed(seed)
# os.environ['PYTHONASSEED'] = str(seed)
# np.random.seed(seed)
# torch.manual_seed(seed)
# torch.cuda.manual_seed(seed)
# torch.backends.cudnn.deterministic = True
# torch.backends.cudnn.benchmark = True
# def get_embeddings(df,path,plot_losses=True, verbose=True):
# #cuda使えたら使う構文
# device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# print(f"{device} is used")
# model = Model()
# model.load_state_dict(torch.load(path))
# model.to(device)
# model.eval()
# #tokenizer = AutoTokenizer.from_pretrained('../input/clrp-roberta-base/clrp_roberta_base')
# tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
# ds = CLRPDataset(df, tokenizer)
# dl = DataLoader(ds,
# batch_size = config["batch_size"],
# shuffle=False,
# num_workers = 4,
# pin_memory=True,
# drop_last=False
# )
# #以下でpredictionsを抽出するために使った構文を使ってembeddingsをreturnしている.
# #SVMの手法とは、embeddingsの意味は?
# embeddings = list()
# with torch.no_grad():
# for i, inputs in tqdm(enumerate(dl)):
# inputs = {key:val.reshape(val.shape[0],-1).to(device) for key,val in inputs.items()}
# outputs = model(**inputs)
# outputs = outputs.detach().cpu().numpy()
# embeddings.extend(outputs)
# return np.array(embeddings)
# def get_preds_svm(X,y,X_test,bins=bins,nfolds=10,C=10,kernel='rbf'):
# scores = list()
# preds = np.zeros((X_test.shape[0]))
# preds_ridge = np.zeros((X_test.shape[0]))
# kfold = StratifiedKFold(n_splits=config['nfolds'],shuffle=True,random_state=config['seed'])
# for k, (train_idx,valid_idx) in enumerate(kfold.split(X,bins)):
# model = SVR(C=5,kernel=kernel,gamma='auto')
# model_ridge = Ridge(alpha=10)
# #print(train_idx)
# #print(valid_idx)
# X_train,y_train = X[train_idx], y[train_idx]
# X_valid,y_valid = X[valid_idx], y[valid_idx]
# model.fit(X_train,y_train)
# model_ridge.fit(X_train, y_train)
# prediction = model.predict(X_valid)
# pred_ridge = model_ridge.predict(X_valid)
# #score = rmse_score(prediction,y_valid)
# score = rmse_score(y_valid, prediction)
# print(f'Fold {k} , rmse score: {score}')
# scores.append(score)
# preds += model.predict(X_test)
# preds_ridge += model_ridge.predict(X_test)
# print("mean rmse",np.mean(scores))
# return (np.array(preds)/nfolds + np.array(preds_ridge)/nfolds)/2
# num_bins = int(np.floor(1 + np.log2(len(train_df))))
# train_df.loc[:,'bins'] = pd.cut(train_df['target'],bins=num_bins,labels=False)
# target = train_df['target'].to_numpy()
# bins = train_df.bins.to_numpy()
# config = {
# 'batch_size': BATCH_SIZE,
# 'max_len': MAX_LEN,
# 'nfolds':10,
# 'seed':42,
# }
# seed_everything(seed=config['seed'])
# # np.loadtxt('data/src/sample.csv', delimiter=',')
# # train_embeddings1 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train1.csv', delimiter=',')
# # train_embeddings2 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train2.csv', delimiter=',')
# # train_embeddings3 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train3.csv', delimiter=',')
# # train_embeddings4 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train4.csv', delimiter=',')
# # train_embeddings5 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train5.csv', delimiter=',')
# train_embeddings1 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_1.pth')
# train_embeddings2 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_2.pth')
# train_embeddings3 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_3.pth')
# train_embeddings4 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_4.pth')
# train_embeddings5 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_5.pth')
# test_embeddings1 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_1.pth')
# test_embeddings2 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_2.pth')
# test_embeddings3 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_3.pth')
# test_embeddings4 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_4.pth')
# test_embeddings5 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_5.pth')
# ## 予測
# svm_ridge_preds1 = get_preds_svm(train_embeddings1,target,test_embeddings1)
# svm_ridge_preds2 = get_preds_svm(train_embeddings2,target,test_embeddings2)
# svm_ridge_preds3 = get_preds_svm(train_embeddings3,target,test_embeddings3)
# svm_ridge_preds4 = get_preds_svm(train_embeddings4,target,test_embeddings4)
# svm_ridge_preds5 = get_preds_svm(train_embeddings5,target,test_embeddings5)
# roberta_svm_ridge_preds = (svm_ridge_preds1 + svm_ridge_preds2 + svm_ridge_preds3 + svm_ridge_preds4 + svm_ridge_preds5)/5
# roberta_svm_ridge_preds
# del train_embeddings1,test_embeddings1
# del train_embeddings2,test_embeddings2
# del train_embeddings3,test_embeddings3
# del train_embeddings4,test_embeddings4
# del train_embeddings5,test_embeddings5
# gc.collect
# # prepare stacking
train_df = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
### nishipyに合わせてtarget and std = 0を消す
train_df.drop(
train_df[(train_df.target == 0) & (train_df.standard_error == 0)].index,
inplace=True,
)
train_df.reset_index(drop=True, inplace=True)
roberta_df = pd.read_csv("../input/nishipyroberta-dataset/nishipy_roberta_stacking.csv")
robertalarge_df = pd.read_csv(
"../input/nishipy-robertalarge-dataset/nishipy_robertalarge_stacking.csv"
)
meanpooling_df = pd.read_csv("../input/meanpoolingdataset/meanpooling_stackingdata.csv")
# robertamean_df = pd.read_csv('../input/meanpoolingrobertabasedataset/meanpooling_roberta_stacking.csv')
# ## 外れ値削除
# robertamean_df = robertamean_df[robertamean_df['id'] != '436ce79fe'].reset_index()
roberta_pred = roberta_df["meanpooling pred"]
robertalarge_pred = robertalarge_df["meanpooling pred"]
meanpooling_pred = meanpooling_df["meanpooling pred"]
# robertamean_pred = robertamean_df['meanpooling roberta pred']
stacking_df = train_df.drop(
["url_legal", "license", "excerpt", "standard_error"], axis=1
)
# stacking_df[['roberta_pred','robertalarge_pred','meanpooling_pred']] =
stacking_df["roberta_pred"] = roberta_pred
stacking_df["robertalarge_pred"] = robertalarge_pred
stacking_df["meanpooling_pred"] = meanpooling_pred
# stacking_df['robertamean_pred'] = robertamean_pred
## 行数が違っていたため、
# stacking_df = stacking_df.merge(robertamean_df,on='id',how='inner').drop(['excerpt','target_y'],axis = 1).rename(columns={'target_x': 'target','meanpooling roberta pred': 'robertamean_pred'})
# stacking_df.head()
import seaborn as sns
sns.distplot(stacking_df["roberta_pred"], bins=20, color="red")
sns.distplot(stacking_df["robertalarge_pred"], bins=20, color="green")
sns.distplot(stacking_df["meanpooling_pred"], bins=20, color="yellow")
# sns.distplot(stacking_df['robertamean_pred'], bins=20, color='blue')
# sns.displot()
import numpy as np
from sklearn.metrics import mean_squared_error
print(
"nishipy roberta base RMSE is : ",
np.sqrt(mean_squared_error(stacking_df["target"], stacking_df["roberta_pred"])),
)
print(
"nishipy roberta large RMSE is : ",
np.sqrt(
mean_squared_error(stacking_df["target"], stacking_df["robertalarge_pred"])
),
)
print(
"cpptake roberta large RMSE is : ",
np.sqrt(mean_squared_error(stacking_df["target"], stacking_df["meanpooling_pred"])),
)
# print('cpptake roberta base RMSE is : ',np.sqrt(mean_squared_error(stacking_df['target'], stacking_df['robertamean_pred'])))
# # RoBERTa Base LB:0.466
NUM_FOLDS = 5
NUM_EPOCHS = 3
BATCH_SIZE = 16
MAX_LEN = 248
EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1.0, 1)]
ROBERTA_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
TOKENIZER_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
class LitDataset(Dataset):
def __init__(self, df, inference_only=False):
super().__init__()
self.df = df
self.inference_only = inference_only
self.text = df.excerpt.tolist()
# self.text = [text.replace("\n", " ") for text in self.text]
if not self.inference_only:
self.target = torch.tensor(df.target.values, dtype=torch.float32)
self.encoded = tokenizer.batch_encode_plus(
self.text,
padding="max_length",
max_length=MAX_LEN,
truncation=True,
return_attention_mask=True,
)
def __len__(self):
return len(self.df)
def __getitem__(self, index):
input_ids = torch.tensor(self.encoded["input_ids"][index])
attention_mask = torch.tensor(self.encoded["attention_mask"][index])
if self.inference_only:
return (input_ids, attention_mask)
else:
target = self.target[index]
return (input_ids, attention_mask, target)
class LitModel(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(768, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
all_predictions = np.zeros((5, len(test_df)))
test_dataset = LitDataset(test_df, inference_only=True)
test_loader = DataLoader(
test_dataset, batch_size=BATCH_SIZE, drop_last=False, shuffle=False, num_workers=2
)
for index in range(5):
# CHANGEME
model_path = f"../input/roberta-base-20210711202147-sche/model_{index + 1}.pth"
print(f"\nUsing {model_path}")
model = LitModel()
model.load_state_dict(torch.load(model_path))
model.to(DEVICE)
all_predictions[index] = predict(model, test_loader)
del model
gc.collect()
### nishipy_roberta にNumpy形式で結果を保存
nishipy_roberta = all_predictions.mean(axis=0)
# nishipy_roberta.target = predictions
# ## release memory
del all_predictions, test_dataset, test_loader, tokenizer
gc.collect()
# # RoBERTa Large LB:0.468
NUM_FOLDS = 5
NUM_EPOCHS = 3
BATCH_SIZE = 8
MAX_LEN = 248
EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1.0, 1)]
ROBERTA_PATH = "../input/roberta-large-20210712191259-mlm/clrp_roberta_large"
TOKENIZER_PATH = "../input/roberta-large-20210712191259-mlm/clrp_roberta_large"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
# nishipy_robertalarge = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
class LitDataset(Dataset):
def __init__(self, df, inference_only=False):
super().__init__()
self.df = df
self.inference_only = inference_only
self.text = df.excerpt.tolist()
# self.text = [text.replace("\n", " ") for text in self.text]
if not self.inference_only:
self.target = torch.tensor(df.target.values, dtype=torch.float32)
self.encoded = tokenizer.batch_encode_plus(
self.text,
padding="max_length",
max_length=MAX_LEN,
truncation=True,
return_attention_mask=True,
)
def __len__(self):
return len(self.df)
def __getitem__(self, index):
input_ids = torch.tensor(self.encoded["input_ids"][index])
attention_mask = torch.tensor(self.encoded["attention_mask"][index])
if self.inference_only:
return (input_ids, attention_mask)
else:
target = self.target[index]
return (input_ids, attention_mask, target)
class LitModel(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
# https://towardsdatascience.com/attention-based-deep-multiple-instance-learning-1bb3df857e24
# 768: node fully connected layer
# 512: node attention layer
# self.attention = nn.Sequential(
# nn.Linear(768, 512),
# nn.Tanh(),
# nn.Linear(512, 1),
# nn.Softmax(dim=1)
# )
# self.regressor = nn.Sequential(
# nn.Linear(768, 1)
# )
# 768 -> 1024
# 512 -> 768
self.attention = nn.Sequential(
nn.Linear(1024, 768), nn.Tanh(), nn.Linear(768, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(1024, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
all_predictions = np.zeros((5, len(test_df)))
test_dataset = LitDataset(test_df, inference_only=True)
test_loader = DataLoader(
test_dataset, batch_size=BATCH_SIZE, drop_last=False, shuffle=False, num_workers=2
)
for index in range(5):
# CHANGEME
model_path = f"../input/roberta-large-20210720020531-sch/model_{index + 1}.pth"
print(f"\nUsing {model_path}")
model = LitModel()
model.load_state_dict(torch.load(model_path))
model.to(DEVICE)
all_predictions[index] = predict(model, test_loader)
del model
gc.collect()
## nishipy_robertalargeにNumpy形式で保存
nishipy_robertalarge = all_predictions.mean(axis=0)
# predictions = all_predictions.mean(axis=0)
# nishipy_robertalarge.target = predictions
# # nishipy_robertalarge
# nishipy_robertalarge
# ## release memory
del all_predictions, test_dataset, test_loader, tokenizer
gc.collect()
# # Robertalarge meanpooling LB:0.468
# 参照:https://www.kaggle.com/jcesquiveld/roberta-large-5-fold-single-model-meanpooling
# ## import
SEED = 42
HIDDEN_SIZE = 1024
MAX_LEN = 300
INPUT_DIR = "../input/commonlitreadabilityprize"
# BASELINE_DIR = '../input/commonlit-readability-models/'
BASELINE_DIR = "../input/robertalargemeanpooling"
MODEL_DIR = "../input/roberta-transformers-pytorch/roberta-large"
TOKENIZER = AutoTokenizer.from_pretrained(MODEL_DIR)
DEVICE = torch.device("cuda" if torch.cuda.is_available() else "cpu")
BATCH_SIZE = 8
# robertalarge_meanpool = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
# test = pd.read_csv('../input/commonlitreadabilityprize/test.csv')
# test.head()
def seed_everything(seed):
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(SEED)
class CLRPDataset(Dataset):
def __init__(self, texts, tokenizer):
self.texts = texts
self.tokenizer = tokenizer
def __len__(self):
return len(self.texts)
def __getitem__(self, idx):
encode = self.tokenizer(
self.texts[idx],
padding="max_length",
max_length=MAX_LEN,
truncation=True,
add_special_tokens=True,
return_attention_mask=True,
return_tensors="pt",
)
return encode
class MeanPoolingModel(nn.Module):
def __init__(self, model_name):
super().__init__()
config = AutoConfig.from_pretrained(model_name)
self.model = AutoModel.from_pretrained(model_name, config=config)
self.linear = nn.Linear(HIDDEN_SIZE, 1)
self.loss = nn.MSELoss()
def forward(self, input_ids, attention_mask, labels=None):
outputs = self.model(input_ids, attention_mask)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
logits = self.linear(mean_embeddings)
preds = logits.squeeze(-1).squeeze(-1)
if labels is not None:
loss = self.loss(preds.view(-1).float(), labels.view(-1).float())
return loss
else:
return preds
def predict(df, model):
ds = CLRPDataset(df.excerpt.tolist(), TOKENIZER)
dl = DataLoader(ds, batch_size=BATCH_SIZE, shuffle=False, pin_memory=False)
model.to(DEVICE)
model.eval()
model.zero_grad()
predictions = []
for batch in tqdm(dl):
inputs = {
key: val.reshape(val.shape[0], -1).to(DEVICE) for key, val in batch.items()
}
outputs = model(**inputs)
predictions.extend(outputs.detach().cpu().numpy().ravel())
return predictions
# ## Inference
#
fold_predictions = []
# modelpath = glob.glob(BASELINE_DIR)
modelpath = glob.glob(BASELINE_DIR + "/*")
# 余計なデータを削除
modelpath.remove("../input/robertalargemeanpooling/experiment-1.csv")
# for path in glob.glob(BASELINE_DIR + '/*.ckpt'):
for path in modelpath:
print(path)
model = MeanPoolingModel(MODEL_DIR)
model.load_state_dict(torch.load(path))
# fold = int(re.match(r'.*_f(\d)_.*', path).group(1))
# print(f'*** fold {fold} ***')
# y_pred = predict(test, model)
y_pred = predict(test_df, model)
fold_predictions.append(y_pred)
# Free memory
del model
gc.collect()
robertalarge_meanpool = np.mean(fold_predictions, axis=0)
del fold_predictions, modelpath, y_pred, TOKENIZER
gc.collect()
# # Roberta base mean pooling LB:0.476
# test_df = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
# submission_df = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
config = {
"lr": 2e-5,
"wd": 0.01,
"batch_size": 16,
"valid_step": 10,
"max_len": 256,
"epochs": 3,
"nfolds": 5,
"seed": 42,
"model_path": "../input/clrp-roberta-base/clrp_roberta_base",
}
# for i in range(config['nfolds']):
# os.makedirs(f'model{i}',exist_ok=True)
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
### mean pooling
class Model(nn.Module):
def __init__(self, path):
super(Model, self).__init__()
self.config = AutoConfig.from_pretrained(path)
self.config.update({"output_hidden_states": True, "hidden_dropout_prob": 0.0})
self.roberta = AutoModel.from_pretrained(path, config=self.config)
# self.linear = nn.Linear(self.config.hidden_size*4, 1, 1)
self.linear = nn.Linear(self.config.hidden_size, 1)
self.layer_norm = nn.LayerNorm(self.config.hidden_size)
def forward(self, **xb):
attention_mask = xb["attention_mask"]
outputs = self.roberta(**xb)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
norm_mean_embeddings = self.layer_norm(mean_embeddings)
logits = self.linear(norm_mean_embeddings)
preds = logits.squeeze(-1).squeeze(-1)
return preds.view(-1).float()
def get_prediction(df, path, model_path, device="cuda"):
model = Model(model_path)
model.load_state_dict(torch.load(path, map_location=device))
model.to(device)
model.eval()
tokenizer = AutoTokenizer.from_pretrained(model_path)
test_ds = CLRPDataset(df, tokenizer)
test_dl = DataLoader(
test_ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
)
predictions = list()
for i, (inputs) in tqdm(enumerate(test_dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device) for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.cpu().detach().numpy().ravel().tolist()
predictions.extend(outputs)
torch.cuda.empty_cache()
del model, tokenizer
gc.collect
return np.array(predictions)
# all_predictions = np.zeros((5, len(test_df)))
# for fold in range(5):
# #CHANGEME
# path = f"../input/robertabasemeanpooling/model{fold}/model{fold}.bin"
# model_path = '../input/clrp-roberta-base/clrp_roberta_base'
# print(f"\nUsing {path} ")
# # model = Model('../input/clrp-roberta-base/clrp_roberta_base')
# # model.load_state_dict(torch.load(model_path))
# # model.to(DEVICE)
# pred = get_prediction(test_df,path,model_path)
# all_predictions[fold] = pred
# gc.collect()
# robertabase_meanpool = all_predictions.mean(axis=0)
# robertabase_meanpool
# del all_predictions,pred
# # Stacking 実装
submission_df = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
submission_df = submission_df.drop("target", axis=1)
submission_df["roberta_pred"] = nishipy_roberta
submission_df["robertalarge_pred"] = nishipy_robertalarge
submission_df["large_meanpooling_pred"] = robertalarge_meanpool
# submission_df['roberta_meanpooling_pred'] = robertabase_meanpool
submission_df
# ## 学習
y_train = stacking_df.target
X_train = stacking_df.drop(["id", "target"], axis=1)
X_test = submission_df.drop("id", axis=1)
# y_test = submission_df.shape[0]
y_train.shape
X_train.shape
#### データサイズあってないから無理やり合わせるけど非常に怪しい
# X_train2 = X_train.dropna()
# y_train2 = y_train.iloc[:2833]
from sklearn.linear_model import LinearRegression
stacking_model = SVR(kernel="linear", gamma="auto") #'linear'
# stacking_model = LinearRegression()
stacking_model.fit(X_train, y_train)
stacking_pred = stacking_model.predict(X_test)
stacking_pred
# # Emsemble
submission_df = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
predictions = pd.DataFrame()
# predictions = y_test * 0.6 + svm_ridge_preds * 0.2 + roberta_svm_ridge_preds * 0.2
## ver10 t5_large_svm利用
# predictions = y_test * 0.6 + svm_ridge_pred * 0.2 + roberta_svm_ridge_preds * 0.2
# ## ver12 roberta meanpooling , stacking + blending + t5_large_svm + nishipy roberta svm利用
# blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool + robertabase_meanpool)/3
# predictions = stacking_pred * 0.3 + blending_pred * 0.3 + svm_ridge_pred * 0.2 + roberta_svm_ridge_preds * 0.2
## ver14 エラーの原因調査 SVMモデルを削除して Roberta meanが悪いのか切り分け
# blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool + robertabase_meanpool)/4
# predictions = stacking_pred * 0.3 + blending_pred * 0.3 + large_svmridge_pred * 0.2 + roberta_svmridge_pred * 0.2
# ## ver17
# blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool)/3
# predictions = stacking_pred * 0.3 + blending_pred * 0.3 + large_svmridge_pred * 0.2 + roberta_svmridge_pred * 0.2
# ## ver17
# blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool)/3
# predictions = stacking_pred * 0.3 + blending_pred * 0.3 + ((large_svmridge_pred + roberta_svmridge_pred + t5_embedding_pred)/3) * 0.4
## ver21
blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool) / 3
predictions = (
stacking_pred * 0.3
+ blending_pred * 0.3
+ ((large_svmridge_pred + t5_embedding_pred) / 3) * 0.4
)
submission_df.target = predictions
print(submission_df)
submission_df.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492055.ipynb
|
roberta-transformers-pytorch
|
maroberti
|
[{"Id": 69492055, "ScriptId": 18851026, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4717884, "CreationDate": "07/31/2021 16:54:39", "VersionNumber": 21.0, "Title": "Submit my roberta stacking & ensemble", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 2028.0, "LinesInsertedFromPrevious": 23.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 2005.0, "LinesInsertedFromFork": 1262.0, "LinesDeletedFromFork": 512.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 766.0, "TotalVotes": 0}]
|
[{"Id": 92727346, "KernelVersionId": 69492055, "SourceDatasetVersionId": 906797}, {"Id": 92727343, "KernelVersionId": 69492055, "SourceDatasetVersionId": 2379259}, {"Id": 92727344, "KernelVersionId": 69492055, "SourceDatasetVersionId": 2236772}, {"Id": 92727345, "KernelVersionId": 69492055, "SourceDatasetVersionId": 2219267}]
|
[{"Id": 906797, "DatasetId": 486306, "DatasourceVersionId": 933877, "CreatorUserId": 2802303, "LicenseName": "Unknown", "CreationDate": "01/22/2020 16:41:47", "VersionNumber": 1.0, "Title": "RoBERTa | Transformers | Pytorch", "Slug": "roberta-transformers-pytorch", "Subtitle": "'roberta-base', 'roberta-large', 'distilroberta-base', 'roberta-large-mnli'", "Description": "Created using the following kernel: https://www.kaggle.com/maroberti/transformers-model-downloader-pytorch-tf2-0", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 486306, "CreatorUserId": 2802303, "OwnerUserId": 2802303.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 906797.0, "CurrentDatasourceVersionId": 933877.0, "ForumId": 499342, "Type": 2, "CreationDate": "01/22/2020 16:41:47", "LastActivityDate": "01/22/2020", "TotalViews": 5186, "TotalDownloads": 803, "TotalVotes": 41, "TotalKernels": 98}]
|
[{"Id": 2802303, "UserName": "maroberti", "DisplayName": "Maximilien Roberti", "RegisterDate": "02/11/2019", "PerformanceTier": 1}]
|
# ### ver17
# * robertabase
# * roberta large
# * roberta large meanpooling
# * roberta base embedding svm+ridge
# * roberta large embedding svm+ridge
# のブレンド
# * robertabase + roberta + roberta large meanpooling のstacking
# # Define
import os
import math
import random
import time
import glob
import re
import gc
gc.enable()
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import pickle
from tqdm import tqdm
import gc
from sklearn.model_selection import KFold, StratifiedKFold, train_test_split
from sklearn.metrics import mean_squared_error as mse
from sklearn.linear_model import LinearRegression, Ridge
from sklearn.svm import SVR
import warnings
warnings.filterwarnings("ignore")
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold
import torch
import torch.nn as nn
# from torch.utils.data import Dataset
# from torch.utils.data import DataLoader
from torch.utils.data import Dataset, SequentialSampler, DataLoader
import transformers
# from transformers import AdamW
# from transformers import AutoTokenizer
# from transformers import AutoModel
# from transformers import AutoConfig
from transformers import get_cosine_schedule_with_warmup
from transformers import (
AutoConfig,
AutoModel,
AutoTokenizer,
AdamW,
get_linear_schedule_with_warmup,
logging,
)
import gc
gc.enable()
import tensorflow as tf
from tensorflow.keras.layers import (
Input,
LSTM,
Bidirectional,
Embedding,
Dense,
Conv1D,
Dropout,
MaxPool1D,
MaxPooling1D,
GlobalAveragePooling2D,
GlobalAveragePooling1D,
GlobalMaxPooling1D,
concatenate,
Flatten,
)
from tensorflow.keras.metrics import RootMeanSquaredError
from tensorflow.keras.models import Model, load_model, save_model, model_from_json
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import (
ReduceLROnPlateau,
ModelCheckpoint,
EarlyStopping,
LearningRateScheduler,
)
from tensorflow.keras.utils import plot_model
from tensorflow.keras import backend as K
from transformers import (
TFBertModel,
BertTokenizerFast,
BertTokenizer,
RobertaTokenizerFast,
TFRobertaModel,
RobertaConfig,
TFAutoModel,
AutoTokenizer,
)
train_df = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_df = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
submission_df = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
# # roberta large embedding SVM & Ridge CV: 0.4822 LB:0.471
# https://www.kaggle.com/iamnishipy/submit-my-roberta-large-svm?scriptVersionId=69275054
# NUM_FOLDS = 5
# NUM_EPOCHS = 3
# BATCH_SIZE = 8
# MAX_LEN = 248
# EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1., 1)]
ROBERTA_PATH = "../input/roberta-large-20210712191259-mlm/clrp_roberta_large/"
TOKENIZER_PATH = "../input/roberta-large-20210712191259-mlm/clrp_roberta_large/"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
class Model(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
# https://towardsdatascience.com/attention-based-deep-multiple-instance-learning-1bb3df857e24
# 768: node fully connected layer
# 512: node attention layer
# self.attention = nn.Sequential(
# nn.Linear(768, 512),
# nn.Tanh(),
# nn.Linear(512, 1),
# nn.Softmax(dim=1)
# )
# self.regressor = nn.Sequential(
# nn.Linear(768, 1)
# )
# 768 -> 1024
# 512 -> 768
self.attention = nn.Sequential(
nn.Linear(1024, 768), nn.Tanh(), nn.Linear(768, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(1024, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
train_data = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
# test_data = pd.read_csv('../input/commonlitreadabilityprize/test.csv')
# sample = pd.read_csv('../input/commonlitreadabilityprize/sample_submission.csv')
num_bins = int(np.floor(1 + np.log2(len(train_data))))
train_data.loc[:, "bins"] = pd.cut(train_data["target"], bins=num_bins, labels=False)
target = train_data["target"].to_numpy()
bins = train_data.bins.to_numpy()
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": 8,
"max_len": 248,
"nfolds": 10,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
def get_embeddings(df, path, plot_losses=True, verbose=True):
# cuda使えたら使う構文
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
# tokenizer = AutoTokenizer.from_pretrained('../input/clrp-roberta-base/clrp_roberta_base')
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
# 以下でpredictionsを抽出するために使った構文を使ってembeddingsをreturnしている.
# SVMの手法とは、embeddingsの意味は?
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
gc.collect
return np.array(embeddings)
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": 8,
"max_len": 248,
"nfolds": 10,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
def get_preds_svm(X, y, X_test, bins=bins, nfolds=10, C=10, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
preds_ridge = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(
n_splits=config["nfolds"], shuffle=True, random_state=config["seed"]
)
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
model = SVR(C=5, kernel=kernel, gamma="auto")
model_ridge = Ridge(alpha=10)
# print(train_idx)
# print(valid_idx)
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
model_ridge.fit(X_train, y_train)
prediction = model.predict(X_valid)
pred_ridge = model_ridge.predict(X_valid)
# score = rmse_score(prediction,y_valid)
score = rmse_score(y_valid, prediction)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
preds_ridge += model_ridge.predict(X_test)
print("mean rmse", np.mean(scores))
return (np.array(preds) / nfolds + np.array(preds_ridge) / nfolds) / 2
# ## Inference
preds = []
for i in range(5):
train_embeddings = get_embeddings(
train_data, f"../input/roberta-large-20210720020531-sch/model_{i+1}.pth"
)
test_embeddings = get_embeddings(
test_df, f"../input/roberta-large-20210720020531-sch/model_{i+1}.pth"
)
svm_pred = get_preds_svm(train_embeddings, target, test_embeddings)
preds.append(svm_pred)
del train_embeddings, test_embeddings, svm_pred
gc.collect()
large_svmridge_pred = (preds[0] + preds[1] + preds[2] + preds[3] + preds[4]) / 5
del tokenizer, train_data, num_bins, target, bins
gc.collect
# test_embeddings1 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_1.pth')
# train_embedding = get_embeddings(train_df,'../input/roberta-large-20210720020531-sch/model_1.pth')
# test_embeddings2 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_2.pth')
# test_embeddings3 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_3.pth')
# test_embeddings4 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_4.pth')
# test_embeddings5 = get_embeddings(test_df,'../input/roberta-large-20210720020531-sch/model_5.pth')
# train_embeddings1 = np.loadtxt('../input/robertalargeembedding/train1.csv', delimiter=',')
# train_embeddings2 = np.loadtxt('../input/robertalargeembedding/train2.csv', delimiter=',')
# train_embeddings3 = np.loadtxt('../input/robertalargeembedding/train3.csv', delimiter=',')
# train_embeddings4 = np.loadtxt('../input/robertalargeembedding/train4.csv', delimiter=',')
# train_embeddings5 = np.loadtxt('../input/robertalargeembedding/train5.csv', delimiter=',')
# # roberta base embedding SVM & Ridge CV: 0.4787 LB:0.465
# https://www.kaggle.com/iamnishipy/submit-my-roberta-base-svm?scriptVersionId=69168038
NUM_FOLDS = 5
NUM_EPOCHS = 3
BATCH_SIZE = 16
MAX_LEN = 248
EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1.0, 1)]
ROBERTA_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
TOKENIZER_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
train_data = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_data = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
sample = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
num_bins = int(np.floor(1 + np.log2(len(train_data))))
train_data.loc[:, "bins"] = pd.cut(train_data["target"], bins=num_bins, labels=False)
target = train_data["target"].to_numpy()
bins = train_data.bins.to_numpy()
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": BATCH_SIZE,
"max_len": MAX_LEN,
"nfolds": 10,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
class Model(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(768, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
def get_embeddings(df, path, plot_losses=True, verbose=True):
# cuda使えたら使う構文
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
# tokenizer = AutoTokenizer.from_pretrained('../input/clrp-roberta-base/clrp_roberta_base')
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
# 以下でpredictionsを抽出するために使った構文を使ってembeddingsをreturnしている.
# SVMの手法とは、embeddingsの意味は?
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
return np.array(embeddings)
def get_preds_svm(X, y, X_test, bins=bins, nfolds=10, C=10, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
preds_ridge = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(
n_splits=config["nfolds"], shuffle=True, random_state=config["seed"]
)
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
model = SVR(C=5, kernel=kernel, gamma="auto")
model_ridge = Ridge(alpha=10)
# print(train_idx)
# print(valid_idx)
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
model_ridge.fit(X_train, y_train)
prediction = model.predict(X_valid)
pred_ridge = model_ridge.predict(X_valid)
# score = rmse_score(prediction,y_valid)
score = rmse_score(y_valid, prediction)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
preds_ridge += model_ridge.predict(X_test)
print("mean rmse", np.mean(scores))
return (np.array(preds) / nfolds + np.array(preds_ridge) / nfolds) / 2
# ## Inference
###
# preds = []
# for i in range(5):
# train_embeddings = get_embeddings(train_data, f'../input/roberta-base-20210711202147-sche/model_{i+1}.pth')
# test_embeddings = get_embeddings(test_data, f'../input/roberta-base-20210711202147-sche/model_{i+1}.pth')
# svm_pred = get_preds_svm(train_embeddings, target, test_embeddings)
# preds.append(svm_pred)
# del train_embeddings,test_embeddings,svm_pred
# gc.collect()
# roberta_svmridge_pred = (preds[0] + preds[1] + preds[2] + preds[3] + preds[4])/5
# roberta_svmridge_pred
# del tokenizer,train_data,num_bins,target,bins
# gc.collect
# #train/testでembeddingsを取得している
# train_embeddings1 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_1.pth')
# test_embeddings1 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_1.pth')
# train_embeddings2 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_2.pth')
# test_embeddings2 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_2.pth')
# train_embeddings3 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_3.pth')
# test_embeddings3 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_3.pth')
# train_embeddings4 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_4.pth')
# test_embeddings4 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_4.pth')
# train_embeddings5 = get_embeddings(train_data,'../input/roberta-base-20210711202147-sche/model_5.pth')
# test_embeddings5 = get_embeddings(test_data,'../input/roberta-base-20210711202147-sche/model_5.pth')
# preds1 = get_preds_svm(train_embeddings1,target,test_embeddings1)
# preds2 = get_preds_svm(train_embeddings2,target,test_embeddings2)
# preds3 = get_preds_svm(train_embeddings3,target,test_embeddings3)
# preds4 = get_preds_svm(train_embeddings4,target,test_embeddings4)
# preds5 = get_preds_svm(train_embeddings5,target,test_embeddings5)
# roberta_svmridge_pred = (preds1 + preds2 + preds3 + preds4 + preds5)/5
# del preds1,preds2,preds3,preds4,preds5
# del train_embeddings1,train_embeddings1,train_embeddings1,train_embeddings1,train_embeddings1
# del test_embeddings1,test_embeddings2,test_embeddings3,test_embeddings4,test_embeddings5
# # pytorch_t5_large_svm
# https://www.kaggle.com/gilfernandes/commonlit-pytorch-t5-large-svm
import yaml, gc
from pathlib import Path
from transformers import T5EncoderModel
class CommonLitModel(nn.Module):
def __init__(self):
super(CommonLitModel, self).__init__()
config = AutoConfig.from_pretrained(cfg.MODEL_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.transformer_model = T5EncoderModel.from_pretrained(
cfg.MODEL_PATH, config=config
)
self.attention = AttentionHead(config.hidden_size, 512, 1)
self.regressor = nn.Linear(config.hidden_size, 1)
def forward(self, input_ids, attention_mask):
last_layer_hidden_states = self.transformer_model(
input_ids=input_ids, attention_mask=attention_mask
)["last_hidden_state"]
weights = self.attention(last_layer_hidden_states)
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
return self.regressor(context_vector), context_vector
BASE_PATH = Path("/kaggle/input/commonlit-t5-large")
DATA_PATH = Path("/kaggle/input/commonlitreadabilityprize/")
assert DATA_PATH.exists()
MODELS_PATH = Path(BASE_PATH / "best_models")
assert MODELS_PATH.exists()
class Config:
NUM_FOLDS = 6
NUM_EPOCHS = 3
BATCH_SIZE = 16
MAX_LEN = 248
MODEL_PATH = BASE_PATH / "lm"
# TOKENIZER_PATH = str(MODELS_PATH/'roberta-base-0')
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
SEED = 1000
NUM_WORKERS = 2
MODEL_FOLDER = MODELS_PATH
model_name = "t5-large"
svm_kernels = ["rbf"]
svm_c = 5
cfg = Config()
train_df["normalized_target"] = (
train_df["target"] - train_df["target"].mean()
) / train_df["target"].std()
model_path = MODELS_PATH
assert model_path.exists()
from transformers import T5EncoderModel
class AttentionHead(nn.Module):
def __init__(self, in_features, hidden_dim, num_targets):
super().__init__()
self.in_features = in_features
self.hidden_layer = nn.Linear(in_features, hidden_dim)
self.final_layer = nn.Linear(hidden_dim, num_targets)
self.out_features = hidden_dim
def forward(self, features):
att = torch.tanh(self.hidden_layer(features))
score = self.final_layer(att)
attention_weights = torch.softmax(score, dim=1)
return attention_weights
class CommonLitModel(nn.Module):
def __init__(self):
super(CommonLitModel, self).__init__()
config = AutoConfig.from_pretrained(cfg.MODEL_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.transformer_model = T5EncoderModel.from_pretrained(
cfg.MODEL_PATH, config=config
)
self.attention = AttentionHead(config.hidden_size, 512, 1)
self.regressor = nn.Linear(config.hidden_size, 1)
def forward(self, input_ids, attention_mask):
last_layer_hidden_states = self.transformer_model(
input_ids=input_ids, attention_mask=attention_mask
)["last_hidden_state"]
weights = self.attention(last_layer_hidden_states)
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
return self.regressor(context_vector), context_vector
def load_model(i):
inference_model = CommonLitModel()
inference_model = inference_model.cuda()
inference_model.load_state_dict(
torch.load(str(model_path / f"{i + 1}_pytorch_model.bin"))
)
inference_model.eval()
return inference_model
def convert_to_list(t):
return t.flatten().long()
class CommonLitDataset(nn.Module):
def __init__(self, text, test_id, tokenizer, max_len=128):
self.excerpt = text
self.test_id = test_id
self.max_len = max_len
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=self.max_len,
padding="max_length",
truncation=True,
)
return {
"input_ids": convert_to_list(encode["input_ids"]),
"attention_mask": convert_to_list(encode["attention_mask"]),
"id": self.test_id[idx],
}
def __len__(self):
return len(self.excerpt)
def create_dl(df, tokenizer):
text = df["excerpt"].values
ids = df["id"].values
ds = CommonLitDataset(text, ids, tokenizer, max_len=cfg.MAX_LEN)
return DataLoader(
ds,
batch_size=cfg.BATCH_SIZE,
shuffle=False,
num_workers=1,
pin_memory=True,
drop_last=False,
)
def get_cls_embeddings(dl, transformer_model):
cls_embeddings = []
with torch.no_grad():
for input_features in tqdm(dl, total=len(dl)):
_, context_vector = transformer_model(
input_features["input_ids"].cuda(),
input_features["attention_mask"].cuda(),
)
# cls_embeddings.extend(output['last_hidden_state'][:,0,:].detach().cpu().numpy())
embedding_out = context_vector.detach().cpu().numpy()
cls_embeddings.extend(embedding_out)
return np.array(cls_embeddings)
def rmse_score(X, y):
return np.sqrt(mean_squared_error(X, y))
def calc_mean(scores):
return np.mean(np.array(scores), axis=0)
from transformers import T5Tokenizer
tokenizers = []
for i in range(1, cfg.NUM_FOLDS):
tokenizer_path = MODELS_PATH / f"tokenizer-{i}"
print(tokenizer_path)
assert Path(tokenizer_path).exists()
tokenizer = T5Tokenizer.from_pretrained(str(tokenizer_path))
tokenizers.append(tokenizer)
num_bins = int(np.ceil(np.log2(len(train_df))))
train_df["bins"] = pd.cut(train_df["target"], bins=num_bins, labels=False)
bins = train_df["bins"].values
train_target = train_df["normalized_target"].values
each_mean_score = []
each_fold_score = []
c = 10
al = 10
# for c in C:
final_scores_svm = []
final_scores_ridge = []
final_rmse = []
for j, tokenizer in enumerate(tokenizers):
print("Model", j)
test_dl = create_dl(test_df, tokenizer)
train_dl = create_dl(train_df, tokenizer)
transformer_model = load_model(j)
transformer_model.cuda()
X = get_cls_embeddings(train_dl, transformer_model)
y = train_target
X_test = get_cls_embeddings(test_dl, transformer_model)
kfold = StratifiedKFold(n_splits=cfg.NUM_FOLDS)
scores = []
scores_svm = []
scores_ridge = []
rmse_scores_svm = []
rmse_scores_ridge = []
rmse_scores = []
for kernel in cfg.svm_kernels:
print("Kernel", kernel)
kernel_scores_svm = []
kernel_scores_ridge = []
kernel_scores = []
kernel_rmse_scores = []
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
print("Fold", k, train_idx.shape, valid_idx.shape)
model_svm = SVR(C=c, kernel=kernel, gamma="auto")
model_ridge = Ridge(alpha=al)
# model = SVR(C=cfg.svm_c, kernel=kernel, gamma='auto')
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model_svm.fit(X_train, y_train)
model_ridge.fit(X_train, y_train)
pred_svm = model_svm.predict(X_valid)
pred_ridge = model_ridge.predict(X_valid)
prediction = (pred_svm + pred_ridge) / 2
kernel_rmse_scores.append(rmse_score(prediction, y_valid))
print("rmse_score", kernel_rmse_scores[k])
kernel_scores_svm.append(model_svm.predict(X_test))
kernel_scores_ridge.append(model_ridge.predict(X_test))
score_mean = (
np.array(kernel_scores_svm) + np.array(kernel_scores_ridge)
) / 2
kernel_scores.append(score_mean)
# scores.append(calc_mean(kernel_scores))
scores_svm.append(calc_mean(kernel_scores_svm))
scores_ridge.append(calc_mean(kernel_scores_ridge))
rmse_scores.append(calc_mean(kernel_rmse_scores))
final_scores_svm.append(calc_mean(scores_svm))
final_scores_ridge.append(calc_mean(scores_ridge))
# final_rmse.append(calc_mean(rmse_scores))
final_rmse.append(calc_mean(rmse_scores))
del transformer_model
torch.cuda.empty_cache()
del tokenizer
gc.collect()
print("FINAL RMSE score", np.mean(np.array(final_rmse)))
each_mean_score.append(np.mean(np.array(final_rmse)))
each_fold_score.append(final_rmse)
print("BEST SCORE : ", each_mean_score)
print("FOLD SCORE : ", each_fold_score)
final_scores = (np.array(final_scores_svm) + np.array(final_scores_ridge)) / 2
final_scores2 = final_scores * train_df["target"].std() + train_df["target"].mean()
target_mean = train_df["target"].mean()
t5_embedding_pred = calc_mean(final_scores2).flatten()
t5_embedding_pred
# del final_scores,train_target,tokenizers,model_svm,model_ridge,X_train,X_valid,y_valid
# gc.collect
# # roberta large pretrain svm&ridge LB:0.468(使えないのでこのモデルは使わん)
# cpptake Note:https://www.kaggle.com/takeshikobayashi/clrp-roberta-svm-roberta-large/edit/run/69118503
# 元Note:https://www.kaggle.com/tutty1992/clrp-roberta-svm-roberta-large
# # train_data = pd.read_csv('../input/commonlitreadabilityprize/train.csv')
# # test_data = pd.read_csv('../input/commonlitreadabilityprize/test.csv')
# # sample = pd.read_csv('../input/commonlitreadabilityprize/sample_submission.csv')
# num_bins = int(np.floor(1 + np.log2(len(train_df))))
# train_df.loc[:,'bins'] = pd.cut(train_df['target'],bins=num_bins,labels=False)
# target = train_df['target'].to_numpy()
# bins = train_df.bins.to_numpy()
# def rmse_score(y_true,y_pred):
# return np.sqrt(mean_squared_error(y_true,y_pred))
# config = {
# 'batch_size':128,
# 'max_len':256,
# 'nfolds':10,
# 'seed':42,
# }
# def seed_everything(seed=42):
# random.seed(seed)
# os.environ['PYTHONASSEED'] = str(seed)
# np.random.seed(seed)
# torch.manual_seed(seed)
# torch.cuda.manual_seed(seed)
# torch.backends.cudnn.deterministic = True
# torch.backends.cudnn.benchmark = True
# seed_everything(seed=config['seed'])
# from sklearn.ensemble import RandomForestRegressor as RFR
# class CLRPDataset(Dataset):
# def __init__(self,df,tokenizer):
# self.excerpt = df['excerpt'].to_numpy()
# self.tokenizer = tokenizer
# def __getitem__(self,idx):
# encode = self.tokenizer(self.excerpt[idx],return_tensors='pt',
# max_length=config['max_len'],
# padding='max_length',truncation=True)
# return encode
# def __len__(self):
# return len(self.excerpt)
# class AttentionHead(nn.Module):
# def __init__(self, in_features, hidden_dim, num_targets):
# super().__init__()
# self.in_features = in_features
# self.middle_features = hidden_dim
# self.W = nn.Linear(in_features, hidden_dim)
# self.V = nn.Linear(hidden_dim, 1)
# self.out_features = hidden_dim
# def forward(self, features):
# att = torch.tanh(self.W(features))
# score = self.V(att)
# attention_weights = torch.softmax(score, dim=1)
# context_vector = attention_weights * features
# context_vector = torch.sum(context_vector, dim=1)
# return context_vector
# class Model(nn.Module):
# def __init__(self):
# super(Model,self).__init__()
# self.roberta = AutoModel.from_pretrained('../input/clrp-pytorch-roberta-pretrain-roberta-large/clrp_roberta_large/')
# #changed attentionHead Dimension from 768 to 1024 by changing model from roberta-base to roberta-large
# self.head = AttentionHead(1024,1024,1)
# self.dropout = nn.Dropout(0.1)
# self.linear = nn.Linear(self.head.out_features,1)
# def forward(self,**xb):
# x = self.roberta(**xb)[0]
# x = self.head(x)
# return x
# def get_embeddings(df,path,plot_losses=True, verbose=True):
# device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# print(f"{device} is used")
# model = Model()
# model.load_state_dict(torch.load(path))
# model.to(device)
# model.eval()
# tokenizer = AutoTokenizer.from_pretrained('../input/clrp-pytorch-roberta-pretrain-roberta-large/clrp_roberta_large/')
# ds = CLRPDataset(df,tokenizer)
# dl = DataLoader(ds,
# batch_size = config["batch_size"],
# shuffle=False,
# num_workers = 4,
# pin_memory=True,
# drop_last=False
# )
# embeddings = list()
# with torch.no_grad():
# for i, inputs in tqdm(enumerate(dl)):
# inputs = {key:val.reshape(val.shape[0],-1).to(device) for key,val in inputs.items()}
# outputs = model(**inputs)
# outputs = outputs.detach().cpu().numpy()
# embeddings.extend(outputs)
# return np.array(embeddings)
# def get_preds_svm_ridge(X,y,X_test,bins=bins,nfolds=10,C=10,kernel='rbf'):
# scores = list()
# preds = np.zeros((X_test.shape[0]))
# preds_ridge = np.zeros((X_test.shape[0]))
# kfold = StratifiedKFold(n_splits=config['nfolds'],shuffle=True,random_state=config['seed'])
# for k, (train_idx,valid_idx) in enumerate(kfold.split(X,bins)):
# model = SVR(C=5,kernel=kernel,gamma='auto')
# model_ridge = Ridge(alpha=10)
# X_train,y_train = X[train_idx], y[train_idx]
# X_valid,y_valid = X[valid_idx], y[valid_idx]
# model.fit(X_train,y_train)
# model_ridge.fit(X_train, y_train)
# prediction = model.predict(X_valid)
# pred_ridge = model_ridge.predict(X_valid)
# score = rmse_score(prediction,y_valid)
# print(f'Fold {k} , rmse score: {score}')
# scores.append(score)
# preds += model.predict(X_test)
# preds_ridge += model_ridge.predict(X_test)
# print("mean rmse",np.mean(scores))
# return (np.array(preds)/nfolds + np.array(preds_ridge)/nfolds)/2
# ## embedding パート
# embedding重すぎたため、特徴量のみdataset化して保存して使ってる
# # np.loadtxt('data/src/sample.csv', delimiter=',')
# train_embeddings1 = np.loadtxt('../input/roberta-embedding-dataset/train1.csv', delimiter=',')
# train_embeddings2 = np.loadtxt('../input/roberta-embedding-dataset/train2.csv', delimiter=',')
# train_embeddings3 = np.loadtxt('../input/roberta-embedding-dataset/train3.csv', delimiter=',')
# train_embeddings4 = np.loadtxt('../input/roberta-embedding-dataset/train4.csv', delimiter=',')
# train_embeddings5 = np.loadtxt('../input/roberta-embedding-dataset/train5.csv', delimiter=',')
# test_embeddings1 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model0/model0.bin')
# test_embeddings2 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model1/model1.bin')
# test_embeddings3 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model2/model2.bin')
# test_embeddings4 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model3/model3.bin')
# test_embeddings5 = get_embeddings(test_df,'../input/clrp-pytorch-roberta-finetune-roberta-large/model4/model4.bin')
# ## pred SVM & Ridge
# from sklearn.linear_model import Ridge
# embedding_preds1 = get_preds_svm_ridge(train_embeddings1,target,test_embeddings1)
# embedding_preds2 = get_preds_svm_ridge(train_embeddings2,target,test_embeddings2)
# embedding_preds3 = get_preds_svm_ridge(train_embeddings3,target,test_embeddings3)
# embedding_preds4 = get_preds_svm_ridge(train_embeddings4,target,test_embeddings4)
# embedding_preds5 = get_preds_svm_ridge(train_embeddings5,target,test_embeddings5)
# svm_ridge_preds = (embedding_preds1 + embedding_preds2 + embedding_preds3 + embedding_preds4 + embedding_preds5)/5
# svm_ridge_preds
# del train_embeddings1,test_embeddings1
# del train_embeddings2,test_embeddings2
# del train_embeddings3,test_embeddings3
# del train_embeddings4,test_embeddings4
# del train_embeddings5,test_embeddings5
# gc.collect
# # nishipy roberta base SVM&Ridge
# NUM_FOLDS = 5
# NUM_EPOCHS = 3
# BATCH_SIZE = 16
# MAX_LEN = 248
# EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1., 1)]
# ROBERTA_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
# TOKENIZER_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
# DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
# tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
# config = {
# 'batch_size':128,
# 'max_len':256,
# 'nfolds':10,
# 'seed':42,
# }
# def seed_everything(seed=42):
# random.seed(seed)
# os.environ['PYTHONASSEED'] = str(seed)
# np.random.seed(seed)
# torch.manual_seed(seed)
# torch.cuda.manual_seed(seed)
# torch.backends.cudnn.deterministic = True
# torch.backends.cudnn.benchmark = True
# seed_everything(seed=config['seed'])
# class CLRPDataset(Dataset):
# def __init__(self,df,tokenizer):
# self.excerpt = df['excerpt'].to_numpy()
# self.tokenizer = tokenizer
# def __getitem__(self,idx):
# encode = self.tokenizer(self.excerpt[idx],return_tensors='pt',
# max_length=config['max_len'],
# padding='max_length',truncation=True)
# return encode
# def __len__(self):
# return len(self.excerpt)
# class Model(nn.Module):
# def __init__(self):
# super().__init__()
# config = AutoConfig.from_pretrained(ROBERTA_PATH)
# config.update({"output_hidden_states":True,
# "hidden_dropout_prob": 0.0,
# "layer_norm_eps": 1e-7})
# self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
# self.attention = nn.Sequential(
# nn.Linear(768, 512),
# nn.Tanh(),
# nn.Linear(512, 1),
# nn.Softmax(dim=1)
# )
# self.regressor = nn.Sequential(
# nn.Linear(768, 1)
# )
# def forward(self, input_ids, attention_mask):
# roberta_output = self.roberta(input_ids=input_ids,
# attention_mask=attention_mask)
# # There are a total of 13 layers of hidden states.
# # 1 for the embedding layer, and 12 for the 12 Roberta layers.
# # We take the hidden states from the last Roberta layer.
# last_layer_hidden_states = roberta_output.hidden_states[-1]
# # The number of cells is MAX_LEN.
# # The size of the hidden state of each cell is 768 (for roberta-base).
# # In order to condense hidden states of all cells to a context vector,
# # we compute a weighted average of the hidden states of all cells.
# # We compute the weight of each cell, using the attention neural network.
# weights = self.attention(last_layer_hidden_states)
# # weights.shape is BATCH_SIZE x MAX_LEN x 1
# # last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# # Now we compute context_vector as the weighted average.
# # context_vector.shape is BATCH_SIZE x 768
# context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# # Now we reduce the context vector to the prediction score.
# return self.regressor(context_vector)
# def predict(model, data_loader):
# """Returns an np.array with predictions of the |model| on |data_loader|"""
# model.eval()
# result = np.zeros(len(data_loader.dataset))
# index = 0
# with torch.no_grad():
# for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
# input_ids = input_ids.to(DEVICE)
# attention_mask = attention_mask.to(DEVICE)
# pred = model(input_ids, attention_mask)
# result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
# index += pred.shape[0]
# return result
# def rmse_score(y_true,y_pred):
# return np.sqrt(mean_squared_error(y_true,y_pred))
# def seed_everything(seed=42):
# random.seed(seed)
# os.environ['PYTHONASSEED'] = str(seed)
# np.random.seed(seed)
# torch.manual_seed(seed)
# torch.cuda.manual_seed(seed)
# torch.backends.cudnn.deterministic = True
# torch.backends.cudnn.benchmark = True
# def get_embeddings(df,path,plot_losses=True, verbose=True):
# #cuda使えたら使う構文
# device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# print(f"{device} is used")
# model = Model()
# model.load_state_dict(torch.load(path))
# model.to(device)
# model.eval()
# #tokenizer = AutoTokenizer.from_pretrained('../input/clrp-roberta-base/clrp_roberta_base')
# tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
# ds = CLRPDataset(df, tokenizer)
# dl = DataLoader(ds,
# batch_size = config["batch_size"],
# shuffle=False,
# num_workers = 4,
# pin_memory=True,
# drop_last=False
# )
# #以下でpredictionsを抽出するために使った構文を使ってembeddingsをreturnしている.
# #SVMの手法とは、embeddingsの意味は?
# embeddings = list()
# with torch.no_grad():
# for i, inputs in tqdm(enumerate(dl)):
# inputs = {key:val.reshape(val.shape[0],-1).to(device) for key,val in inputs.items()}
# outputs = model(**inputs)
# outputs = outputs.detach().cpu().numpy()
# embeddings.extend(outputs)
# return np.array(embeddings)
# def get_preds_svm(X,y,X_test,bins=bins,nfolds=10,C=10,kernel='rbf'):
# scores = list()
# preds = np.zeros((X_test.shape[0]))
# preds_ridge = np.zeros((X_test.shape[0]))
# kfold = StratifiedKFold(n_splits=config['nfolds'],shuffle=True,random_state=config['seed'])
# for k, (train_idx,valid_idx) in enumerate(kfold.split(X,bins)):
# model = SVR(C=5,kernel=kernel,gamma='auto')
# model_ridge = Ridge(alpha=10)
# #print(train_idx)
# #print(valid_idx)
# X_train,y_train = X[train_idx], y[train_idx]
# X_valid,y_valid = X[valid_idx], y[valid_idx]
# model.fit(X_train,y_train)
# model_ridge.fit(X_train, y_train)
# prediction = model.predict(X_valid)
# pred_ridge = model_ridge.predict(X_valid)
# #score = rmse_score(prediction,y_valid)
# score = rmse_score(y_valid, prediction)
# print(f'Fold {k} , rmse score: {score}')
# scores.append(score)
# preds += model.predict(X_test)
# preds_ridge += model_ridge.predict(X_test)
# print("mean rmse",np.mean(scores))
# return (np.array(preds)/nfolds + np.array(preds_ridge)/nfolds)/2
# num_bins = int(np.floor(1 + np.log2(len(train_df))))
# train_df.loc[:,'bins'] = pd.cut(train_df['target'],bins=num_bins,labels=False)
# target = train_df['target'].to_numpy()
# bins = train_df.bins.to_numpy()
# config = {
# 'batch_size': BATCH_SIZE,
# 'max_len': MAX_LEN,
# 'nfolds':10,
# 'seed':42,
# }
# seed_everything(seed=config['seed'])
# # np.loadtxt('data/src/sample.csv', delimiter=',')
# # train_embeddings1 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train1.csv', delimiter=',')
# # train_embeddings2 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train2.csv', delimiter=',')
# # train_embeddings3 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train3.csv', delimiter=',')
# # train_embeddings4 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train4.csv', delimiter=',')
# # train_embeddings5 = np.loadtxt('../input/nishipy-robertabase-embedding-dataset/train5.csv', delimiter=',')
# train_embeddings1 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_1.pth')
# train_embeddings2 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_2.pth')
# train_embeddings3 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_3.pth')
# train_embeddings4 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_4.pth')
# train_embeddings5 = get_embeddings(train_df,'../input/roberta-base-20210711202147-sche/model_5.pth')
# test_embeddings1 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_1.pth')
# test_embeddings2 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_2.pth')
# test_embeddings3 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_3.pth')
# test_embeddings4 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_4.pth')
# test_embeddings5 = get_embeddings(test_df,'../input/roberta-base-20210711202147-sche/model_5.pth')
# ## 予測
# svm_ridge_preds1 = get_preds_svm(train_embeddings1,target,test_embeddings1)
# svm_ridge_preds2 = get_preds_svm(train_embeddings2,target,test_embeddings2)
# svm_ridge_preds3 = get_preds_svm(train_embeddings3,target,test_embeddings3)
# svm_ridge_preds4 = get_preds_svm(train_embeddings4,target,test_embeddings4)
# svm_ridge_preds5 = get_preds_svm(train_embeddings5,target,test_embeddings5)
# roberta_svm_ridge_preds = (svm_ridge_preds1 + svm_ridge_preds2 + svm_ridge_preds3 + svm_ridge_preds4 + svm_ridge_preds5)/5
# roberta_svm_ridge_preds
# del train_embeddings1,test_embeddings1
# del train_embeddings2,test_embeddings2
# del train_embeddings3,test_embeddings3
# del train_embeddings4,test_embeddings4
# del train_embeddings5,test_embeddings5
# gc.collect
# # prepare stacking
train_df = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
### nishipyに合わせてtarget and std = 0を消す
train_df.drop(
train_df[(train_df.target == 0) & (train_df.standard_error == 0)].index,
inplace=True,
)
train_df.reset_index(drop=True, inplace=True)
roberta_df = pd.read_csv("../input/nishipyroberta-dataset/nishipy_roberta_stacking.csv")
robertalarge_df = pd.read_csv(
"../input/nishipy-robertalarge-dataset/nishipy_robertalarge_stacking.csv"
)
meanpooling_df = pd.read_csv("../input/meanpoolingdataset/meanpooling_stackingdata.csv")
# robertamean_df = pd.read_csv('../input/meanpoolingrobertabasedataset/meanpooling_roberta_stacking.csv')
# ## 外れ値削除
# robertamean_df = robertamean_df[robertamean_df['id'] != '436ce79fe'].reset_index()
roberta_pred = roberta_df["meanpooling pred"]
robertalarge_pred = robertalarge_df["meanpooling pred"]
meanpooling_pred = meanpooling_df["meanpooling pred"]
# robertamean_pred = robertamean_df['meanpooling roberta pred']
stacking_df = train_df.drop(
["url_legal", "license", "excerpt", "standard_error"], axis=1
)
# stacking_df[['roberta_pred','robertalarge_pred','meanpooling_pred']] =
stacking_df["roberta_pred"] = roberta_pred
stacking_df["robertalarge_pred"] = robertalarge_pred
stacking_df["meanpooling_pred"] = meanpooling_pred
# stacking_df['robertamean_pred'] = robertamean_pred
## 行数が違っていたため、
# stacking_df = stacking_df.merge(robertamean_df,on='id',how='inner').drop(['excerpt','target_y'],axis = 1).rename(columns={'target_x': 'target','meanpooling roberta pred': 'robertamean_pred'})
# stacking_df.head()
import seaborn as sns
sns.distplot(stacking_df["roberta_pred"], bins=20, color="red")
sns.distplot(stacking_df["robertalarge_pred"], bins=20, color="green")
sns.distplot(stacking_df["meanpooling_pred"], bins=20, color="yellow")
# sns.distplot(stacking_df['robertamean_pred'], bins=20, color='blue')
# sns.displot()
import numpy as np
from sklearn.metrics import mean_squared_error
print(
"nishipy roberta base RMSE is : ",
np.sqrt(mean_squared_error(stacking_df["target"], stacking_df["roberta_pred"])),
)
print(
"nishipy roberta large RMSE is : ",
np.sqrt(
mean_squared_error(stacking_df["target"], stacking_df["robertalarge_pred"])
),
)
print(
"cpptake roberta large RMSE is : ",
np.sqrt(mean_squared_error(stacking_df["target"], stacking_df["meanpooling_pred"])),
)
# print('cpptake roberta base RMSE is : ',np.sqrt(mean_squared_error(stacking_df['target'], stacking_df['robertamean_pred'])))
# # RoBERTa Base LB:0.466
NUM_FOLDS = 5
NUM_EPOCHS = 3
BATCH_SIZE = 16
MAX_LEN = 248
EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1.0, 1)]
ROBERTA_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
TOKENIZER_PATH = "../input/clrp-roberta-base/clrp_roberta_base/"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
class LitDataset(Dataset):
def __init__(self, df, inference_only=False):
super().__init__()
self.df = df
self.inference_only = inference_only
self.text = df.excerpt.tolist()
# self.text = [text.replace("\n", " ") for text in self.text]
if not self.inference_only:
self.target = torch.tensor(df.target.values, dtype=torch.float32)
self.encoded = tokenizer.batch_encode_plus(
self.text,
padding="max_length",
max_length=MAX_LEN,
truncation=True,
return_attention_mask=True,
)
def __len__(self):
return len(self.df)
def __getitem__(self, index):
input_ids = torch.tensor(self.encoded["input_ids"][index])
attention_mask = torch.tensor(self.encoded["attention_mask"][index])
if self.inference_only:
return (input_ids, attention_mask)
else:
target = self.target[index]
return (input_ids, attention_mask, target)
class LitModel(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(768, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
all_predictions = np.zeros((5, len(test_df)))
test_dataset = LitDataset(test_df, inference_only=True)
test_loader = DataLoader(
test_dataset, batch_size=BATCH_SIZE, drop_last=False, shuffle=False, num_workers=2
)
for index in range(5):
# CHANGEME
model_path = f"../input/roberta-base-20210711202147-sche/model_{index + 1}.pth"
print(f"\nUsing {model_path}")
model = LitModel()
model.load_state_dict(torch.load(model_path))
model.to(DEVICE)
all_predictions[index] = predict(model, test_loader)
del model
gc.collect()
### nishipy_roberta にNumpy形式で結果を保存
nishipy_roberta = all_predictions.mean(axis=0)
# nishipy_roberta.target = predictions
# ## release memory
del all_predictions, test_dataset, test_loader, tokenizer
gc.collect()
# # RoBERTa Large LB:0.468
NUM_FOLDS = 5
NUM_EPOCHS = 3
BATCH_SIZE = 8
MAX_LEN = 248
EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1.0, 1)]
ROBERTA_PATH = "../input/roberta-large-20210712191259-mlm/clrp_roberta_large"
TOKENIZER_PATH = "../input/roberta-large-20210712191259-mlm/clrp_roberta_large"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
# nishipy_robertalarge = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
class LitDataset(Dataset):
def __init__(self, df, inference_only=False):
super().__init__()
self.df = df
self.inference_only = inference_only
self.text = df.excerpt.tolist()
# self.text = [text.replace("\n", " ") for text in self.text]
if not self.inference_only:
self.target = torch.tensor(df.target.values, dtype=torch.float32)
self.encoded = tokenizer.batch_encode_plus(
self.text,
padding="max_length",
max_length=MAX_LEN,
truncation=True,
return_attention_mask=True,
)
def __len__(self):
return len(self.df)
def __getitem__(self, index):
input_ids = torch.tensor(self.encoded["input_ids"][index])
attention_mask = torch.tensor(self.encoded["attention_mask"][index])
if self.inference_only:
return (input_ids, attention_mask)
else:
target = self.target[index]
return (input_ids, attention_mask, target)
class LitModel(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
# https://towardsdatascience.com/attention-based-deep-multiple-instance-learning-1bb3df857e24
# 768: node fully connected layer
# 512: node attention layer
# self.attention = nn.Sequential(
# nn.Linear(768, 512),
# nn.Tanh(),
# nn.Linear(512, 1),
# nn.Softmax(dim=1)
# )
# self.regressor = nn.Sequential(
# nn.Linear(768, 1)
# )
# 768 -> 1024
# 512 -> 768
self.attention = nn.Sequential(
nn.Linear(1024, 768), nn.Tanh(), nn.Linear(768, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(1024, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
all_predictions = np.zeros((5, len(test_df)))
test_dataset = LitDataset(test_df, inference_only=True)
test_loader = DataLoader(
test_dataset, batch_size=BATCH_SIZE, drop_last=False, shuffle=False, num_workers=2
)
for index in range(5):
# CHANGEME
model_path = f"../input/roberta-large-20210720020531-sch/model_{index + 1}.pth"
print(f"\nUsing {model_path}")
model = LitModel()
model.load_state_dict(torch.load(model_path))
model.to(DEVICE)
all_predictions[index] = predict(model, test_loader)
del model
gc.collect()
## nishipy_robertalargeにNumpy形式で保存
nishipy_robertalarge = all_predictions.mean(axis=0)
# predictions = all_predictions.mean(axis=0)
# nishipy_robertalarge.target = predictions
# # nishipy_robertalarge
# nishipy_robertalarge
# ## release memory
del all_predictions, test_dataset, test_loader, tokenizer
gc.collect()
# # Robertalarge meanpooling LB:0.468
# 参照:https://www.kaggle.com/jcesquiveld/roberta-large-5-fold-single-model-meanpooling
# ## import
SEED = 42
HIDDEN_SIZE = 1024
MAX_LEN = 300
INPUT_DIR = "../input/commonlitreadabilityprize"
# BASELINE_DIR = '../input/commonlit-readability-models/'
BASELINE_DIR = "../input/robertalargemeanpooling"
MODEL_DIR = "../input/roberta-transformers-pytorch/roberta-large"
TOKENIZER = AutoTokenizer.from_pretrained(MODEL_DIR)
DEVICE = torch.device("cuda" if torch.cuda.is_available() else "cpu")
BATCH_SIZE = 8
# robertalarge_meanpool = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
# test = pd.read_csv('../input/commonlitreadabilityprize/test.csv')
# test.head()
def seed_everything(seed):
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(SEED)
class CLRPDataset(Dataset):
def __init__(self, texts, tokenizer):
self.texts = texts
self.tokenizer = tokenizer
def __len__(self):
return len(self.texts)
def __getitem__(self, idx):
encode = self.tokenizer(
self.texts[idx],
padding="max_length",
max_length=MAX_LEN,
truncation=True,
add_special_tokens=True,
return_attention_mask=True,
return_tensors="pt",
)
return encode
class MeanPoolingModel(nn.Module):
def __init__(self, model_name):
super().__init__()
config = AutoConfig.from_pretrained(model_name)
self.model = AutoModel.from_pretrained(model_name, config=config)
self.linear = nn.Linear(HIDDEN_SIZE, 1)
self.loss = nn.MSELoss()
def forward(self, input_ids, attention_mask, labels=None):
outputs = self.model(input_ids, attention_mask)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
logits = self.linear(mean_embeddings)
preds = logits.squeeze(-1).squeeze(-1)
if labels is not None:
loss = self.loss(preds.view(-1).float(), labels.view(-1).float())
return loss
else:
return preds
def predict(df, model):
ds = CLRPDataset(df.excerpt.tolist(), TOKENIZER)
dl = DataLoader(ds, batch_size=BATCH_SIZE, shuffle=False, pin_memory=False)
model.to(DEVICE)
model.eval()
model.zero_grad()
predictions = []
for batch in tqdm(dl):
inputs = {
key: val.reshape(val.shape[0], -1).to(DEVICE) for key, val in batch.items()
}
outputs = model(**inputs)
predictions.extend(outputs.detach().cpu().numpy().ravel())
return predictions
# ## Inference
#
fold_predictions = []
# modelpath = glob.glob(BASELINE_DIR)
modelpath = glob.glob(BASELINE_DIR + "/*")
# 余計なデータを削除
modelpath.remove("../input/robertalargemeanpooling/experiment-1.csv")
# for path in glob.glob(BASELINE_DIR + '/*.ckpt'):
for path in modelpath:
print(path)
model = MeanPoolingModel(MODEL_DIR)
model.load_state_dict(torch.load(path))
# fold = int(re.match(r'.*_f(\d)_.*', path).group(1))
# print(f'*** fold {fold} ***')
# y_pred = predict(test, model)
y_pred = predict(test_df, model)
fold_predictions.append(y_pred)
# Free memory
del model
gc.collect()
robertalarge_meanpool = np.mean(fold_predictions, axis=0)
del fold_predictions, modelpath, y_pred, TOKENIZER
gc.collect()
# # Roberta base mean pooling LB:0.476
# test_df = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
# submission_df = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
config = {
"lr": 2e-5,
"wd": 0.01,
"batch_size": 16,
"valid_step": 10,
"max_len": 256,
"epochs": 3,
"nfolds": 5,
"seed": 42,
"model_path": "../input/clrp-roberta-base/clrp_roberta_base",
}
# for i in range(config['nfolds']):
# os.makedirs(f'model{i}',exist_ok=True)
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
### mean pooling
class Model(nn.Module):
def __init__(self, path):
super(Model, self).__init__()
self.config = AutoConfig.from_pretrained(path)
self.config.update({"output_hidden_states": True, "hidden_dropout_prob": 0.0})
self.roberta = AutoModel.from_pretrained(path, config=self.config)
# self.linear = nn.Linear(self.config.hidden_size*4, 1, 1)
self.linear = nn.Linear(self.config.hidden_size, 1)
self.layer_norm = nn.LayerNorm(self.config.hidden_size)
def forward(self, **xb):
attention_mask = xb["attention_mask"]
outputs = self.roberta(**xb)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
norm_mean_embeddings = self.layer_norm(mean_embeddings)
logits = self.linear(norm_mean_embeddings)
preds = logits.squeeze(-1).squeeze(-1)
return preds.view(-1).float()
def get_prediction(df, path, model_path, device="cuda"):
model = Model(model_path)
model.load_state_dict(torch.load(path, map_location=device))
model.to(device)
model.eval()
tokenizer = AutoTokenizer.from_pretrained(model_path)
test_ds = CLRPDataset(df, tokenizer)
test_dl = DataLoader(
test_ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
)
predictions = list()
for i, (inputs) in tqdm(enumerate(test_dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device) for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.cpu().detach().numpy().ravel().tolist()
predictions.extend(outputs)
torch.cuda.empty_cache()
del model, tokenizer
gc.collect
return np.array(predictions)
# all_predictions = np.zeros((5, len(test_df)))
# for fold in range(5):
# #CHANGEME
# path = f"../input/robertabasemeanpooling/model{fold}/model{fold}.bin"
# model_path = '../input/clrp-roberta-base/clrp_roberta_base'
# print(f"\nUsing {path} ")
# # model = Model('../input/clrp-roberta-base/clrp_roberta_base')
# # model.load_state_dict(torch.load(model_path))
# # model.to(DEVICE)
# pred = get_prediction(test_df,path,model_path)
# all_predictions[fold] = pred
# gc.collect()
# robertabase_meanpool = all_predictions.mean(axis=0)
# robertabase_meanpool
# del all_predictions,pred
# # Stacking 実装
submission_df = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
submission_df = submission_df.drop("target", axis=1)
submission_df["roberta_pred"] = nishipy_roberta
submission_df["robertalarge_pred"] = nishipy_robertalarge
submission_df["large_meanpooling_pred"] = robertalarge_meanpool
# submission_df['roberta_meanpooling_pred'] = robertabase_meanpool
submission_df
# ## 学習
y_train = stacking_df.target
X_train = stacking_df.drop(["id", "target"], axis=1)
X_test = submission_df.drop("id", axis=1)
# y_test = submission_df.shape[0]
y_train.shape
X_train.shape
#### データサイズあってないから無理やり合わせるけど非常に怪しい
# X_train2 = X_train.dropna()
# y_train2 = y_train.iloc[:2833]
from sklearn.linear_model import LinearRegression
stacking_model = SVR(kernel="linear", gamma="auto") #'linear'
# stacking_model = LinearRegression()
stacking_model.fit(X_train, y_train)
stacking_pred = stacking_model.predict(X_test)
stacking_pred
# # Emsemble
submission_df = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
predictions = pd.DataFrame()
# predictions = y_test * 0.6 + svm_ridge_preds * 0.2 + roberta_svm_ridge_preds * 0.2
## ver10 t5_large_svm利用
# predictions = y_test * 0.6 + svm_ridge_pred * 0.2 + roberta_svm_ridge_preds * 0.2
# ## ver12 roberta meanpooling , stacking + blending + t5_large_svm + nishipy roberta svm利用
# blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool + robertabase_meanpool)/3
# predictions = stacking_pred * 0.3 + blending_pred * 0.3 + svm_ridge_pred * 0.2 + roberta_svm_ridge_preds * 0.2
## ver14 エラーの原因調査 SVMモデルを削除して Roberta meanが悪いのか切り分け
# blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool + robertabase_meanpool)/4
# predictions = stacking_pred * 0.3 + blending_pred * 0.3 + large_svmridge_pred * 0.2 + roberta_svmridge_pred * 0.2
# ## ver17
# blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool)/3
# predictions = stacking_pred * 0.3 + blending_pred * 0.3 + large_svmridge_pred * 0.2 + roberta_svmridge_pred * 0.2
# ## ver17
# blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool)/3
# predictions = stacking_pred * 0.3 + blending_pred * 0.3 + ((large_svmridge_pred + roberta_svmridge_pred + t5_embedding_pred)/3) * 0.4
## ver21
blending_pred = (nishipy_roberta + nishipy_robertalarge + robertalarge_meanpool) / 3
predictions = (
stacking_pred * 0.3
+ blending_pred * 0.3
+ ((large_svmridge_pred + t5_embedding_pred) / 3) * 0.4
)
submission_df.target = predictions
print(submission_df)
submission_df.to_csv("submission.csv", index=False)
| false | 6 | 22,667 | 0 | 22,729 | 22,667 |
||
69492266
|
<jupyter_start><jupyter_text>Heart Attack Analysis & Prediction Dataset
## Hone your analytical and ML skills by participating in tasks of my other dataset's. Given below.
[Data Science Job Posting on Glassdoor](https://www.kaggle.com/rashikrahmanpritom/data-science-job-posting-on-glassdoor)
[Groceries dataset for Market Basket Analysis(MBA)](https://www.kaggle.com/rashikrahmanpritom/groceries-dataset-for-market-basket-analysismba)
[Dataset for Facial recognition using ML approach](https://www.kaggle.com/rashikrahmanpritom/dataset-for-facial-recognition-using-ml-approach)
[Covid_w/wo_Pneumonia Chest Xray](https://www.kaggle.com/rashikrahmanpritom/covid-wwo-pneumonia-chest-xray)
[Disney Movies 1937-2016 Gross Income](https://www.kaggle.com/rashikrahmanpritom/disney-movies-19372016-total-gross)
[Bollywood Movie data from 2000 to 2019](https://www.kaggle.com/rashikrahmanpritom/bollywood-movie-data-from-2000-to-2019)
[17.7K English song data from 2008-2017](https://www.kaggle.com/rashikrahmanpritom/177k-english-song-data-from-20082017)
## About this dataset
- Age : Age of the patient
- Sex : Sex of the patient
- exang: exercise induced angina (1 = yes; 0 = no)
- ca: number of major vessels (0-3)
- cp : Chest Pain type chest pain type
- Value 1: typical angina
- Value 2: atypical angina
- Value 3: non-anginal pain
- Value 4: asymptomatic
- trtbps : resting blood pressure (in mm Hg)
- chol : cholestoral in mg/dl fetched via BMI sensor
- fbs : (fasting blood sugar > 120 mg/dl) (1 = true; 0 = false)
- rest_ecg : resting electrocardiographic results
- Value 0: normal
- Value 1: having ST-T wave abnormality (T wave inversions and/or ST elevation or depression of > 0.05 mV)
- Value 2: showing probable or definite left ventricular hypertrophy by Estes' criteria
- thalach : maximum heart rate achieved
- target : 0= less chance of heart attack 1= more chance of heart attack
n
Kaggle dataset identifier: heart-attack-analysis-prediction-dataset
<jupyter_code>import pandas as pd
df = pd.read_csv('heart-attack-analysis-prediction-dataset/heart.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 303 entries, 0 to 302
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 303 non-null int64
1 sex 303 non-null int64
2 cp 303 non-null int64
3 trtbps 303 non-null int64
4 chol 303 non-null int64
5 fbs 303 non-null int64
6 restecg 303 non-null int64
7 thalachh 303 non-null int64
8 exng 303 non-null int64
9 oldpeak 303 non-null float64
10 slp 303 non-null int64
11 caa 303 non-null int64
12 thall 303 non-null int64
13 output 303 non-null int64
dtypes: float64(1), int64(13)
memory usage: 33.3 KB
<jupyter_text>Examples:
{
"age": 63.0,
"sex": 1.0,
"cp": 3.0,
"trtbps": 145.0,
"chol": 233.0,
"fbs": 1.0,
"restecg": 0.0,
"thalachh": 150.0,
"exng": 0.0,
"oldpeak": 2.3,
"slp": 0.0,
"caa": 0.0,
"thall": 1.0,
"output": 1.0
}
{
"age": 37.0,
"sex": 1.0,
"cp": 2.0,
"trtbps": 130.0,
"chol": 250.0,
"fbs": 0.0,
"restecg": 1.0,
"thalachh": 187.0,
"exng": 0.0,
"oldpeak": 3.5,
"slp": 0.0,
"caa": 0.0,
"thall": 2.0,
"output": 1.0
}
{
"age": 41.0,
"sex": 0.0,
"cp": 1.0,
"trtbps": 130.0,
"chol": 204.0,
"fbs": 0.0,
"restecg": 0.0,
"thalachh": 172.0,
"exng": 0.0,
"oldpeak": 1.4,
"slp": 2.0,
"caa": 0.0,
"thall": 2.0,
"output": 1.0
}
{
"age": 56.0,
"sex": 1.0,
"cp": 1.0,
"trtbps": 120.0,
"chol": 236.0,
"fbs": 0.0,
"restecg": 1.0,
"thalachh": 178.0,
"exng": 0.0,
"oldpeak": 0.8,
"slp": 2.0,
"caa": 0.0,
"thall": 2.0,
"output": 1.0
}
<jupyter_script>import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
classification_report,
recall_score,
precision_score,
f1_score,
roc_auc_score,
roc_curve,
)
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.neural_network import MLPClassifier
import keras
from keras.models import Sequential
from keras.layers import Dense
df = pd.read_csv("../input/heart-attack-analysis-prediction-dataset/heart.csv")
display(df.head(10))
print("\n\n")
print(df.shape)
features_num = ["age", "trtbps", "chol", "thalachh", "oldpeak"]
features_cat = ["sex", "exng", "caa", "cp", "fbs", "restecg", "slp", "thall"]
scaler = StandardScaler()
ohe = OneHotEncoder(sparse=False)
scaled_columns = scaler.fit_transform(df[features_num])
encoded_columns = ohe.fit_transform(df[features_cat])
X = np.concatenate([scaled_columns, encoded_columns], axis=1)
y = df["output"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=4
)
print(df["output"]) # access data frame specific column in this way OR
print(df.output) # access data frame this way
print(df.output.value_counts())
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492266.ipynb
|
heart-attack-analysis-prediction-dataset
|
rashikrahmanpritom
|
[{"Id": 69492266, "ScriptId": 18975210, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8019645, "CreationDate": "07/31/2021 16:57:34", "VersionNumber": 1.0, "Title": "Heart Failure Precition Project", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 43.0, "LinesInsertedFromPrevious": 43.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92727992, "KernelVersionId": 69492266, "SourceDatasetVersionId": 2047221}]
|
[{"Id": 2047221, "DatasetId": 1226038, "DatasourceVersionId": 2087216, "CreatorUserId": 4730101, "LicenseName": "CC0: Public Domain", "CreationDate": "03/22/2021 11:40:59", "VersionNumber": 2.0, "Title": "Heart Attack Analysis & Prediction Dataset", "Slug": "heart-attack-analysis-prediction-dataset", "Subtitle": "A dataset for heart attack classification", "Description": "## Hone your analytical and ML skills by participating in tasks of my other dataset's. Given below.\n\n\n[Data Science Job Posting on Glassdoor](https://www.kaggle.com/rashikrahmanpritom/data-science-job-posting-on-glassdoor)\n\n[Groceries dataset for Market Basket Analysis(MBA)](https://www.kaggle.com/rashikrahmanpritom/groceries-dataset-for-market-basket-analysismba)\n\n[Dataset for Facial recognition using ML approach](https://www.kaggle.com/rashikrahmanpritom/dataset-for-facial-recognition-using-ml-approach)\n\n[Covid_w/wo_Pneumonia Chest Xray](https://www.kaggle.com/rashikrahmanpritom/covid-wwo-pneumonia-chest-xray)\n\n[Disney Movies 1937-2016 Gross Income](https://www.kaggle.com/rashikrahmanpritom/disney-movies-19372016-total-gross)\n\n[Bollywood Movie data from 2000 to 2019](https://www.kaggle.com/rashikrahmanpritom/bollywood-movie-data-from-2000-to-2019)\n\n[17.7K English song data from 2008-2017](https://www.kaggle.com/rashikrahmanpritom/177k-english-song-data-from-20082017)\n\n## About this dataset\n\n- Age : Age of the patient\n\n- Sex : Sex of the patient\n\n- exang: exercise induced angina (1 = yes; 0 = no)\n\n- ca: number of major vessels (0-3)\n\n- cp : Chest Pain type chest pain type\n - Value 1: typical angina\n - Value 2: atypical angina\n - Value 3: non-anginal pain\n - Value 4: asymptomatic\n \n- trtbps : resting blood pressure (in mm Hg)\n- chol : cholestoral in mg/dl fetched via BMI sensor\n- fbs : (fasting blood sugar > 120 mg/dl) (1 = true; 0 = false)\n- rest_ecg : resting electrocardiographic results\n - Value 0: normal\n - Value 1: having ST-T wave abnormality (T wave inversions and/or ST elevation or depression of > 0.05 mV)\n - Value 2: showing probable or definite left ventricular hypertrophy by Estes' criteria\n \n- thalach : maximum heart rate achieved\n- target : 0= less chance of heart attack 1= more chance of heart attack\n\nn", "VersionNotes": "heart csv update", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1226038, "CreatorUserId": 4730101, "OwnerUserId": 4730101.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2047221.0, "CurrentDatasourceVersionId": 2087216.0, "ForumId": 1244179, "Type": 2, "CreationDate": "03/22/2021 08:19:12", "LastActivityDate": "03/22/2021", "TotalViews": 870835, "TotalDownloads": 138216, "TotalVotes": 3197, "TotalKernels": 1050}]
|
[{"Id": 4730101, "UserName": "rashikrahmanpritom", "DisplayName": "Rashik Rahman", "RegisterDate": "03/24/2020", "PerformanceTier": 3}]
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
classification_report,
recall_score,
precision_score,
f1_score,
roc_auc_score,
roc_curve,
)
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.neural_network import MLPClassifier
import keras
from keras.models import Sequential
from keras.layers import Dense
df = pd.read_csv("../input/heart-attack-analysis-prediction-dataset/heart.csv")
display(df.head(10))
print("\n\n")
print(df.shape)
features_num = ["age", "trtbps", "chol", "thalachh", "oldpeak"]
features_cat = ["sex", "exng", "caa", "cp", "fbs", "restecg", "slp", "thall"]
scaler = StandardScaler()
ohe = OneHotEncoder(sparse=False)
scaled_columns = scaler.fit_transform(df[features_num])
encoded_columns = ohe.fit_transform(df[features_cat])
X = np.concatenate([scaled_columns, encoded_columns], axis=1)
y = df["output"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=4
)
print(df["output"]) # access data frame specific column in this way OR
print(df.output) # access data frame this way
print(df.output.value_counts())
|
[{"heart-attack-analysis-prediction-dataset/heart.csv": {"column_names": "[\"age\", \"sex\", \"cp\", \"trtbps\", \"chol\", \"fbs\", \"restecg\", \"thalachh\", \"exng\", \"oldpeak\", \"slp\", \"caa\", \"thall\", \"output\"]", "column_data_types": "{\"age\": \"int64\", \"sex\": \"int64\", \"cp\": \"int64\", \"trtbps\": \"int64\", \"chol\": \"int64\", \"fbs\": \"int64\", \"restecg\": \"int64\", \"thalachh\": \"int64\", \"exng\": \"int64\", \"oldpeak\": \"float64\", \"slp\": \"int64\", \"caa\": \"int64\", \"thall\": \"int64\", \"output\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 303 entries, 0 to 302\nData columns (total 14 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 age 303 non-null int64 \n 1 sex 303 non-null int64 \n 2 cp 303 non-null int64 \n 3 trtbps 303 non-null int64 \n 4 chol 303 non-null int64 \n 5 fbs 303 non-null int64 \n 6 restecg 303 non-null int64 \n 7 thalachh 303 non-null int64 \n 8 exng 303 non-null int64 \n 9 oldpeak 303 non-null float64\n 10 slp 303 non-null int64 \n 11 caa 303 non-null int64 \n 12 thall 303 non-null int64 \n 13 output 303 non-null int64 \ndtypes: float64(1), int64(13)\nmemory usage: 33.3 KB\n", "summary": "{\"age\": {\"count\": 303.0, \"mean\": 54.366336633663366, \"std\": 9.082100989837857, \"min\": 29.0, \"25%\": 47.5, \"50%\": 55.0, \"75%\": 61.0, \"max\": 77.0}, \"sex\": {\"count\": 303.0, \"mean\": 0.6831683168316832, \"std\": 0.46601082333962385, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"cp\": {\"count\": 303.0, \"mean\": 0.966996699669967, \"std\": 1.0320524894832985, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 3.0}, \"trtbps\": {\"count\": 303.0, \"mean\": 131.62376237623764, \"std\": 17.5381428135171, \"min\": 94.0, \"25%\": 120.0, \"50%\": 130.0, \"75%\": 140.0, \"max\": 200.0}, \"chol\": {\"count\": 303.0, \"mean\": 246.26402640264027, \"std\": 51.83075098793003, \"min\": 126.0, \"25%\": 211.0, \"50%\": 240.0, \"75%\": 274.5, \"max\": 564.0}, \"fbs\": {\"count\": 303.0, \"mean\": 0.1485148514851485, \"std\": 0.35619787492797644, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"restecg\": {\"count\": 303.0, \"mean\": 0.528052805280528, \"std\": 0.525859596359298, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 2.0}, \"thalachh\": {\"count\": 303.0, \"mean\": 149.64686468646866, \"std\": 22.905161114914094, \"min\": 71.0, \"25%\": 133.5, \"50%\": 153.0, \"75%\": 166.0, \"max\": 202.0}, \"exng\": {\"count\": 303.0, \"mean\": 0.32673267326732675, \"std\": 0.4697944645223165, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"oldpeak\": {\"count\": 303.0, \"mean\": 1.0396039603960396, \"std\": 1.1610750220686348, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.8, \"75%\": 1.6, \"max\": 6.2}, \"slp\": {\"count\": 303.0, \"mean\": 1.3993399339933994, \"std\": 0.6162261453459619, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 2.0}, \"caa\": {\"count\": 303.0, \"mean\": 0.7293729372937293, \"std\": 1.022606364969327, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 4.0}, \"thall\": {\"count\": 303.0, \"mean\": 2.3135313531353137, \"std\": 0.6122765072781409, \"min\": 0.0, \"25%\": 2.0, \"50%\": 2.0, \"75%\": 3.0, \"max\": 3.0}, \"output\": {\"count\": 303.0, \"mean\": 0.5445544554455446, \"std\": 0.4988347841643913, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}}", "examples": "{\"age\":{\"0\":63,\"1\":37,\"2\":41,\"3\":56},\"sex\":{\"0\":1,\"1\":1,\"2\":0,\"3\":1},\"cp\":{\"0\":3,\"1\":2,\"2\":1,\"3\":1},\"trtbps\":{\"0\":145,\"1\":130,\"2\":130,\"3\":120},\"chol\":{\"0\":233,\"1\":250,\"2\":204,\"3\":236},\"fbs\":{\"0\":1,\"1\":0,\"2\":0,\"3\":0},\"restecg\":{\"0\":0,\"1\":1,\"2\":0,\"3\":1},\"thalachh\":{\"0\":150,\"1\":187,\"2\":172,\"3\":178},\"exng\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"oldpeak\":{\"0\":2.3,\"1\":3.5,\"2\":1.4,\"3\":0.8},\"slp\":{\"0\":0,\"1\":0,\"2\":2,\"3\":2},\"caa\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"thall\":{\"0\":1,\"1\":2,\"2\":2,\"3\":2},\"output\":{\"0\":1,\"1\":1,\"2\":1,\"3\":1}}"}}]
| true | 1 |
<start_data_description><data_path>heart-attack-analysis-prediction-dataset/heart.csv:
<column_names>
['age', 'sex', 'cp', 'trtbps', 'chol', 'fbs', 'restecg', 'thalachh', 'exng', 'oldpeak', 'slp', 'caa', 'thall', 'output']
<column_types>
{'age': 'int64', 'sex': 'int64', 'cp': 'int64', 'trtbps': 'int64', 'chol': 'int64', 'fbs': 'int64', 'restecg': 'int64', 'thalachh': 'int64', 'exng': 'int64', 'oldpeak': 'float64', 'slp': 'int64', 'caa': 'int64', 'thall': 'int64', 'output': 'int64'}
<dataframe_Summary>
{'age': {'count': 303.0, 'mean': 54.366336633663366, 'std': 9.082100989837857, 'min': 29.0, '25%': 47.5, '50%': 55.0, '75%': 61.0, 'max': 77.0}, 'sex': {'count': 303.0, 'mean': 0.6831683168316832, 'std': 0.46601082333962385, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'cp': {'count': 303.0, 'mean': 0.966996699669967, 'std': 1.0320524894832985, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 2.0, 'max': 3.0}, 'trtbps': {'count': 303.0, 'mean': 131.62376237623764, 'std': 17.5381428135171, 'min': 94.0, '25%': 120.0, '50%': 130.0, '75%': 140.0, 'max': 200.0}, 'chol': {'count': 303.0, 'mean': 246.26402640264027, 'std': 51.83075098793003, 'min': 126.0, '25%': 211.0, '50%': 240.0, '75%': 274.5, 'max': 564.0}, 'fbs': {'count': 303.0, 'mean': 0.1485148514851485, 'std': 0.35619787492797644, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'restecg': {'count': 303.0, 'mean': 0.528052805280528, 'std': 0.525859596359298, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 2.0}, 'thalachh': {'count': 303.0, 'mean': 149.64686468646866, 'std': 22.905161114914094, 'min': 71.0, '25%': 133.5, '50%': 153.0, '75%': 166.0, 'max': 202.0}, 'exng': {'count': 303.0, 'mean': 0.32673267326732675, 'std': 0.4697944645223165, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'oldpeak': {'count': 303.0, 'mean': 1.0396039603960396, 'std': 1.1610750220686348, 'min': 0.0, '25%': 0.0, '50%': 0.8, '75%': 1.6, 'max': 6.2}, 'slp': {'count': 303.0, 'mean': 1.3993399339933994, 'std': 0.6162261453459619, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 2.0}, 'caa': {'count': 303.0, 'mean': 0.7293729372937293, 'std': 1.022606364969327, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 4.0}, 'thall': {'count': 303.0, 'mean': 2.3135313531353137, 'std': 0.6122765072781409, 'min': 0.0, '25%': 2.0, '50%': 2.0, '75%': 3.0, 'max': 3.0}, 'output': {'count': 303.0, 'mean': 0.5445544554455446, 'std': 0.4988347841643913, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 303 entries, 0 to 302
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 303 non-null int64
1 sex 303 non-null int64
2 cp 303 non-null int64
3 trtbps 303 non-null int64
4 chol 303 non-null int64
5 fbs 303 non-null int64
6 restecg 303 non-null int64
7 thalachh 303 non-null int64
8 exng 303 non-null int64
9 oldpeak 303 non-null float64
10 slp 303 non-null int64
11 caa 303 non-null int64
12 thall 303 non-null int64
13 output 303 non-null int64
dtypes: float64(1), int64(13)
memory usage: 33.3 KB
<some_examples>
{'age': {'0': 63, '1': 37, '2': 41, '3': 56}, 'sex': {'0': 1, '1': 1, '2': 0, '3': 1}, 'cp': {'0': 3, '1': 2, '2': 1, '3': 1}, 'trtbps': {'0': 145, '1': 130, '2': 130, '3': 120}, 'chol': {'0': 233, '1': 250, '2': 204, '3': 236}, 'fbs': {'0': 1, '1': 0, '2': 0, '3': 0}, 'restecg': {'0': 0, '1': 1, '2': 0, '3': 1}, 'thalachh': {'0': 150, '1': 187, '2': 172, '3': 178}, 'exng': {'0': 0, '1': 0, '2': 0, '3': 0}, 'oldpeak': {'0': 2.3, '1': 3.5, '2': 1.4, '3': 0.8}, 'slp': {'0': 0, '1': 0, '2': 2, '3': 2}, 'caa': {'0': 0, '1': 0, '2': 0, '3': 0}, 'thall': {'0': 1, '1': 2, '2': 2, '3': 2}, 'output': {'0': 1, '1': 1, '2': 1, '3': 1}}
<end_description>
| 464 | 0 | 2,159 | 464 |
69492116
|
<jupyter_start><jupyter_text>Chest X-Ray Images (Pneumonia)
### Context
http://www.cell.com/cell/fulltext/S0092-8674(18)30154-5

Figure S6. Illustrative Examples of Chest X-Rays in Patients with Pneumonia, Related to Figure 6
The normal chest X-ray (left panel) depicts clear lungs without any areas of abnormal opacification in the image. Bacterial pneumonia (middle) typically exhibits a focal lobar consolidation, in this case in the right upper lobe (white arrows), whereas viral pneumonia (right) manifests with a more diffuse ‘‘interstitial’’ pattern in both lungs.
http://www.cell.com/cell/fulltext/S0092-8674(18)30154-5
### Content
The dataset is organized into 3 folders (train, test, val) and contains subfolders for each image category (Pneumonia/Normal). There are 5,863 X-Ray images (JPEG) and 2 categories (Pneumonia/Normal).
Chest X-ray images (anterior-posterior) were selected from retrospective cohorts of pediatric patients of one to five years old from Guangzhou Women and Children’s Medical Center, Guangzhou. All chest X-ray imaging was performed as part of patients’ routine clinical care.
For the analysis of chest x-ray images, all chest radiographs were initially screened for quality control by removing all low quality or unreadable scans. The diagnoses for the images were then graded by two expert physicians before being cleared for training the AI system. In order to account for any grading errors, the evaluation set was also checked by a third expert.
Kaggle dataset identifier: chest-xray-pneumonia
<jupyter_script># ## **Pneumonia Detection from X-Ray Images**
# ### Data Augmenatation and Transfer Learning
# ### The Data
# 
# - The dataset is organized into 3 folders (train, test, val) and contains subfolders for each image category (Pneumonia/Normal). There are 5,863 X-Ray images (JPEG) and 2 categories (Pneumonia/Normal).
# **Dependencies**
import os
import numpy as np
import tensorflow as tf
from tensorflow import keras
from keras.preprocessing.image import ImageDataGenerator as imgen
from keras.models import Model
from keras.layers import Dropout, Dense, Input
from keras.callbacks import (
EarlyStopping,
ModelCheckpoint,
LearningRateScheduler,
ReduceLROnPlateau,
CSVLogger,
)
from keras.optimizers import Adam
from keras.optimizers.schedules import ExponentialDecay
from keras.applications.mobilenet_v2 import MobileNetV2
from keras.applications.mobilenet_v2 import preprocess_input
from tqdm.notebook import tqdm
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
# ## Data Preparation and Augmentation
IMAGE_SIZE = 150
BATCH_SIZE = 32
traingen = imgen(
rescale=1 / 255,
zoom_range=0.2,
width_shift_range=0.2,
height_shift_range=0.2,
fill_mode="nearest",
# preprocessing_function = preprocess_input
)
testgen = imgen(
rescale=1 / 255,
)
# preprocessing_function = preprocess_input)
train_ds = traingen.flow_from_directory(
"../input/chest-xray-pneumonia/chest_xray/train",
target_size=(IMAGE_SIZE, IMAGE_SIZE),
seed=123,
batch_size=BATCH_SIZE,
class_mode="binary",
)
val_ds = testgen.flow_from_directory(
"../input/chest-xray-pneumonia/chest_xray/val",
target_size=(IMAGE_SIZE, IMAGE_SIZE),
seed=123,
batch_size=16,
class_mode="binary",
)
test_ds = testgen.flow_from_directory(
"../input/chest-xray-pneumonia/chest_xray/test",
target_size=(IMAGE_SIZE, IMAGE_SIZE),
seed=123,
batch_size=BATCH_SIZE,
shuffle=False,
class_mode="binary",
)
class_names = train_ds.class_indices
class_names = list(class_names.keys())
class_names
# ## EDA
# **Distribution of classes.**
class_dist = train_ds.classes
sns.countplot(x=class_dist)
# - **Unbalanced.**
# **Images/Labels.**
images, labels = next(val_ds)
def plotImages(image, label):
plt.figure(figsize=[22, 14])
for i in range(16):
plt.subplot(4, 4, i + 1)
plt.imshow(image[i])
plt.title(f"Class : {class_names[np.argmax(label[i])]}")
plt.axis("off")
plt.show()
plotImages(images, labels)
# ## **Defining out Model.**
# get the pre-trained model
base_model = ResNet50V2(include_top=False, weights="imagenet", pooling="avg")
base_model.trainable = False
# **MODEL**
image_input = Input(shape=(IMAGE_SIZE, IMAGE_SIZE, 3))
x = base_model(image_input, training=False)
x = Dense(512, activation="relu")(x)
x = Dropout(0.3)(x)
x = Dense(128, activation="relu")(x)
image_output = Dense(1, activation="sigmoid")(x)
model = Model(image_input, image_output)
model.summary()
# ## Loss, Optimizer, metrics and Checkpoints
# callbacks
def scheduler(epoch, lr):
if epoch < 10:
return lr
else:
return lr * tf.math.exp(-0.1)
my_callbacks = [
EarlyStopping(monitor="val_loss", patience=3),
ModelCheckpoint("Pneumonia_model.h5", verbose=1, save_best_only=True),
LearningRateScheduler(scheduler),
CSVLogger("training.log"),
]
# compile
METRICS = ["accuracy", tf.keras.metrics.AUC(name="AUC")]
model.compile(optimizer=Adam(), loss="binary_crossentropy", metrics=METRICS)
# before training
model.evaluate(test_ds)
# ## **Training**
history = model.fit(
train_ds, epochs=20, validation_data=test_ds, callbacks=my_callbacks
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492116.ipynb
|
chest-xray-pneumonia
|
paultimothymooney
|
[{"Id": 69492116, "ScriptId": 18925637, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6681037, "CreationDate": "07/31/2021 16:55:37", "VersionNumber": 1.0, "Title": "Pneumonia Detection", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 158.0, "LinesInsertedFromPrevious": 158.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92727527, "KernelVersionId": 69492116, "SourceDatasetVersionId": 23812}]
|
[{"Id": 23812, "DatasetId": 17810, "DatasourceVersionId": 23851, "CreatorUserId": 1314380, "LicenseName": "Other (specified in description)", "CreationDate": "03/24/2018 19:41:59", "VersionNumber": 2.0, "Title": "Chest X-Ray Images (Pneumonia)", "Slug": "chest-xray-pneumonia", "Subtitle": "5,863 images, 2 categories", "Description": "### Context\n\nhttp://www.cell.com/cell/fulltext/S0092-8674(18)30154-5\n\n\n\nFigure S6. Illustrative Examples of Chest X-Rays in Patients with Pneumonia, Related to Figure 6\nThe normal chest X-ray (left panel) depicts clear lungs without any areas of abnormal opacification in the image. Bacterial pneumonia (middle) typically exhibits a focal lobar consolidation, in this case in the right upper lobe (white arrows), whereas viral pneumonia (right) manifests with a more diffuse \u2018\u2018interstitial\u2019\u2019 pattern in both lungs.\nhttp://www.cell.com/cell/fulltext/S0092-8674(18)30154-5\n\n### Content\n\nThe dataset is organized into 3 folders (train, test, val) and contains subfolders for each image category (Pneumonia/Normal). There are 5,863 X-Ray images (JPEG) and 2 categories (Pneumonia/Normal). \n\nChest X-ray images (anterior-posterior) were selected from retrospective cohorts of pediatric patients of one to five years old from Guangzhou Women and Children\u2019s Medical Center, Guangzhou. All chest X-ray imaging was performed as part of patients\u2019 routine clinical care. \n\nFor the analysis of chest x-ray images, all chest radiographs were initially screened for quality control by removing all low quality or unreadable scans. The diagnoses for the images were then graded by two expert physicians before being cleared for training the AI system. In order to account for any grading errors, the evaluation set was also checked by a third expert.\n\n### Acknowledgements\n\nData: https://data.mendeley.com/datasets/rscbjbr9sj/2\n\nLicense: [CC BY 4.0][1]\n\nCitation: http://www.cell.com/cell/fulltext/S0092-8674(18)30154-5\n\n![enter image description here][2]\n\n\n### Inspiration\n\nAutomated methods to detect and classify human diseases from medical images.\n\n\n [1]: https://creativecommons.org/licenses/by/4.0/\n [2]: https://i.imgur.com/8AUJkin.png", "VersionNotes": "train/test/val", "TotalCompressedBytes": 1237249419.0, "TotalUncompressedBytes": 1237249419.0}]
|
[{"Id": 17810, "CreatorUserId": 1314380, "OwnerUserId": 1314380.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 23812.0, "CurrentDatasourceVersionId": 23851.0, "ForumId": 25540, "Type": 2, "CreationDate": "03/22/2018 05:42:41", "LastActivityDate": "03/22/2018", "TotalViews": 2063138, "TotalDownloads": 237932, "TotalVotes": 5834, "TotalKernels": 2058}]
|
[{"Id": 1314380, "UserName": "paultimothymooney", "DisplayName": "Paul Mooney", "RegisterDate": "10/05/2017", "PerformanceTier": 5}]
|
# ## **Pneumonia Detection from X-Ray Images**
# ### Data Augmenatation and Transfer Learning
# ### The Data
# 
# - The dataset is organized into 3 folders (train, test, val) and contains subfolders for each image category (Pneumonia/Normal). There are 5,863 X-Ray images (JPEG) and 2 categories (Pneumonia/Normal).
# **Dependencies**
import os
import numpy as np
import tensorflow as tf
from tensorflow import keras
from keras.preprocessing.image import ImageDataGenerator as imgen
from keras.models import Model
from keras.layers import Dropout, Dense, Input
from keras.callbacks import (
EarlyStopping,
ModelCheckpoint,
LearningRateScheduler,
ReduceLROnPlateau,
CSVLogger,
)
from keras.optimizers import Adam
from keras.optimizers.schedules import ExponentialDecay
from keras.applications.mobilenet_v2 import MobileNetV2
from keras.applications.mobilenet_v2 import preprocess_input
from tqdm.notebook import tqdm
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
# ## Data Preparation and Augmentation
IMAGE_SIZE = 150
BATCH_SIZE = 32
traingen = imgen(
rescale=1 / 255,
zoom_range=0.2,
width_shift_range=0.2,
height_shift_range=0.2,
fill_mode="nearest",
# preprocessing_function = preprocess_input
)
testgen = imgen(
rescale=1 / 255,
)
# preprocessing_function = preprocess_input)
train_ds = traingen.flow_from_directory(
"../input/chest-xray-pneumonia/chest_xray/train",
target_size=(IMAGE_SIZE, IMAGE_SIZE),
seed=123,
batch_size=BATCH_SIZE,
class_mode="binary",
)
val_ds = testgen.flow_from_directory(
"../input/chest-xray-pneumonia/chest_xray/val",
target_size=(IMAGE_SIZE, IMAGE_SIZE),
seed=123,
batch_size=16,
class_mode="binary",
)
test_ds = testgen.flow_from_directory(
"../input/chest-xray-pneumonia/chest_xray/test",
target_size=(IMAGE_SIZE, IMAGE_SIZE),
seed=123,
batch_size=BATCH_SIZE,
shuffle=False,
class_mode="binary",
)
class_names = train_ds.class_indices
class_names = list(class_names.keys())
class_names
# ## EDA
# **Distribution of classes.**
class_dist = train_ds.classes
sns.countplot(x=class_dist)
# - **Unbalanced.**
# **Images/Labels.**
images, labels = next(val_ds)
def plotImages(image, label):
plt.figure(figsize=[22, 14])
for i in range(16):
plt.subplot(4, 4, i + 1)
plt.imshow(image[i])
plt.title(f"Class : {class_names[np.argmax(label[i])]}")
plt.axis("off")
plt.show()
plotImages(images, labels)
# ## **Defining out Model.**
# get the pre-trained model
base_model = ResNet50V2(include_top=False, weights="imagenet", pooling="avg")
base_model.trainable = False
# **MODEL**
image_input = Input(shape=(IMAGE_SIZE, IMAGE_SIZE, 3))
x = base_model(image_input, training=False)
x = Dense(512, activation="relu")(x)
x = Dropout(0.3)(x)
x = Dense(128, activation="relu")(x)
image_output = Dense(1, activation="sigmoid")(x)
model = Model(image_input, image_output)
model.summary()
# ## Loss, Optimizer, metrics and Checkpoints
# callbacks
def scheduler(epoch, lr):
if epoch < 10:
return lr
else:
return lr * tf.math.exp(-0.1)
my_callbacks = [
EarlyStopping(monitor="val_loss", patience=3),
ModelCheckpoint("Pneumonia_model.h5", verbose=1, save_best_only=True),
LearningRateScheduler(scheduler),
CSVLogger("training.log"),
]
# compile
METRICS = ["accuracy", tf.keras.metrics.AUC(name="AUC")]
model.compile(optimizer=Adam(), loss="binary_crossentropy", metrics=METRICS)
# before training
model.evaluate(test_ds)
# ## **Training**
history = model.fit(
train_ds, epochs=20, validation_data=test_ds, callbacks=my_callbacks
)
| false | 0 | 1,219 | 0 | 1,695 | 1,219 |
||
69492937
|
# # Loading the data and exploring its shape and values
# # 1. библиотеки
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.utils import resample
import itertools
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import normalize
from sklearn.metrics import classification_report
from sklearn.metrics import roc_auc_score
from sklearn.metrics import balanced_accuracy_score
from sklearn.metrics import f1_score
import tensorflow as tf
import tensorflow_addons as tfa
from tensorflow import keras
from tensorflow.keras.layers import (
Dense,
Conv1D,
MaxPool1D,
Flatten,
Dropout,
InputLayer,
LSTM,
GRU,
BatchNormalization,
Bidirectional,
Concatenate,
)
from tensorflow.keras.models import Model
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
from tensorflow.keras.preprocessing import sequence
from tensorflow.keras.optimizers import SGD, RMSprop
from tensorflow.keras.utils import to_categorical
# # 2. вспомогательные функции
# ## 2.1 класс детекторов для нахождения пик
# класс детекторов дл нахождения пик
import numpy as np
import pywt
try:
import pathlib
except ImportError:
import pathlib2 as pathlib
import scipy.signal as signal
class Detectors:
"""ECG heartbeat detection algorithms
General useage instructions:
r_peaks = detectors.the_detector(ecg_in_samples)
The argument ecg_in_samples is a single channel ECG in volt
at the given sample rate.
"""
def __init__(self, sampling_frequency):
"""
The constructor takes the sampling rate in Hz of the ECG data.
"""
self.fs = sampling_frequency
# this is set to a positive value for benchmarking
self.engzee_fake_delay = 0
def hamilton_detector(self, unfiltered_ecg):
"""
P.S. Hamilton,
Open Source ECG Analysis Software Documentation, E.P.Limited, 2002.
"""
f1 = 8 / self.fs
f2 = 16 / self.fs
b, a = signal.butter(1, [f1 * 2, f2 * 2], btype="bandpass")
filtered_ecg = signal.lfilter(b, a, unfiltered_ecg)
diff = abs(np.diff(filtered_ecg))
b = np.ones(int(0.08 * self.fs))
b = b / int(0.08 * self.fs)
a = [1]
ma = signal.lfilter(b, a, diff)
ma[0 : len(b) * 2] = 0
n_pks = []
n_pks_ave = 0.0
s_pks = []
s_pks_ave = 0.0
QRS = [0]
RR = []
RR_ave = 0.0
th = 0.0
i = 0
idx = []
peaks = []
for i in range(len(ma)):
if i > 0 and i < len(ma) - 1:
if ma[i - 1] < ma[i] and ma[i + 1] < ma[i]:
peak = i
peaks.append(i)
if ma[peak] > th and (peak - QRS[-1]) > 0.3 * self.fs:
QRS.append(peak)
idx.append(i)
s_pks.append(ma[peak])
if len(n_pks) > 8:
s_pks.pop(0)
s_pks_ave = np.mean(s_pks)
if RR_ave != 0.0:
if QRS[-1] - QRS[-2] > 1.5 * RR_ave:
missed_peaks = peaks[idx[-2] + 1 : idx[-1]]
for missed_peak in missed_peaks:
if (
missed_peak - peaks[idx[-2]]
> int(0.360 * self.fs)
and ma[missed_peak] > 0.5 * th
):
QRS.append(missed_peak)
QRS.sort()
break
if len(QRS) > 2:
RR.append(QRS[-1] - QRS[-2])
if len(RR) > 8:
RR.pop(0)
RR_ave = int(np.mean(RR))
else:
n_pks.append(ma[peak])
if len(n_pks) > 8:
n_pks.pop(0)
n_pks_ave = np.mean(n_pks)
th = n_pks_ave + 0.45 * (s_pks_ave - n_pks_ave)
i += 1
QRS.pop(0)
return QRS
def christov_detector(self, unfiltered_ecg):
"""
Ivaylo I. Christov,
Real time electrocardiogram QRS detection using combined
adaptive threshold, BioMedical Engineering OnLine 2004,
vol. 3:28, 2004.
"""
total_taps = 0
b = np.ones(int(0.02 * self.fs))
b = b / int(0.02 * self.fs)
total_taps += len(b)
a = [1]
MA1 = signal.lfilter(b, a, unfiltered_ecg)
b = np.ones(int(0.028 * self.fs))
b = b / int(0.028 * self.fs)
total_taps += len(b)
a = [1]
MA2 = signal.lfilter(b, a, MA1)
Y = []
for i in range(1, len(MA2) - 1):
diff = abs(MA2[i + 1] - MA2[i - 1])
Y.append(diff)
b = np.ones(int(0.040 * self.fs))
b = b / int(0.040 * self.fs)
total_taps += len(b)
a = [1]
MA3 = signal.lfilter(b, a, Y)
MA3[0:total_taps] = 0
ms50 = int(0.05 * self.fs)
ms200 = int(0.2 * self.fs)
ms1200 = int(1.2 * self.fs)
ms350 = int(0.35 * self.fs)
M = 0
newM5 = 0
M_list = []
MM = []
M_slope = np.linspace(1.0, 0.6, ms1200 - ms200)
F = 0
F_list = []
R = 0
RR = []
Rm = 0
R_list = []
MFR = 0
MFR_list = []
QRS = []
for i in range(len(MA3)):
# M
if i < 5 * self.fs:
M = 0.6 * np.max(MA3[: i + 1])
MM.append(M)
if len(MM) > 5:
MM.pop(0)
elif QRS and i < QRS[-1] + ms200:
newM5 = 0.6 * np.max(MA3[QRS[-1] : i])
if newM5 > 1.5 * MM[-1]:
newM5 = 1.1 * MM[-1]
elif QRS and i == QRS[-1] + ms200:
if newM5 == 0:
newM5 = MM[-1]
MM.append(newM5)
if len(MM) > 5:
MM.pop(0)
M = np.mean(MM)
elif QRS and i > QRS[-1] + ms200 and i < QRS[-1] + ms1200:
M = np.mean(MM) * M_slope[i - (QRS[-1] + ms200)]
elif QRS and i > QRS[-1] + ms1200:
M = 0.6 * np.mean(MM)
# F
if i > ms350:
F_section = MA3[i - ms350 : i]
max_latest = np.max(F_section[-ms50:])
max_earliest = np.max(F_section[:ms50])
F = F + ((max_latest - max_earliest) / 150.0)
# R
if QRS and i < QRS[-1] + int((2.0 / 3.0 * Rm)):
R = 0
elif QRS and i > QRS[-1] + int((2.0 / 3.0 * Rm)) and i < QRS[-1] + Rm:
dec = (M - np.mean(MM)) / 1.4
R = 0 + dec
MFR = M + F + R
M_list.append(M)
F_list.append(F)
R_list.append(R)
MFR_list.append(MFR)
if not QRS and MA3[i] > MFR:
QRS.append(i)
elif QRS and i > QRS[-1] + ms200 and MA3[i] > MFR:
QRS.append(i)
if len(QRS) > 2:
RR.append(QRS[-1] - QRS[-2])
if len(RR) > 5:
RR.pop(0)
Rm = int(np.mean(RR))
QRS.pop(0)
return QRS
def engzee_detector(self, unfiltered_ecg):
"""
C. Zeelenberg, A single scan algorithm for QRS detection and
feature extraction, IEEE Comp. in Cardiology, vol. 6,
pp. 37-42, 1979 with modifications A. Lourenco, H. Silva,
P. Leite, R. Lourenco and A. Fred, “Real Time
Electrocardiogram Segmentation for Finger Based ECG
Biometrics”, BIOSIGNALS 2012, pp. 49-54, 2012.
"""
f1 = 48 / self.fs
f2 = 52 / self.fs
b, a = signal.butter(4, [f1 * 2, f2 * 2], btype="bandstop")
filtered_ecg = signal.lfilter(b, a, unfiltered_ecg)
diff = np.zeros(len(filtered_ecg))
for i in range(4, len(diff)):
diff[i] = filtered_ecg[i] - filtered_ecg[i - 4]
ci = [1, 4, 6, 4, 1]
low_pass = signal.lfilter(ci, 1, diff)
low_pass[: int(0.2 * self.fs)] = 0
ms200 = int(0.2 * self.fs)
ms1200 = int(1.2 * self.fs)
ms160 = int(0.16 * self.fs)
neg_threshold = int(0.01 * self.fs)
M = 0
M_list = []
neg_m = []
MM = []
M_slope = np.linspace(1.0, 0.6, ms1200 - ms200)
QRS = []
r_peaks = []
counter = 0
thi_list = []
thi = False
thf_list = []
thf = False
for i in range(len(low_pass)):
# M
if i < 5 * self.fs:
M = 0.6 * np.max(low_pass[: i + 1])
MM.append(M)
if len(MM) > 5:
MM.pop(0)
elif QRS and i < QRS[-1] + ms200:
newM5 = 0.6 * np.max(low_pass[QRS[-1] : i])
if newM5 > 1.5 * MM[-1]:
newM5 = 1.1 * MM[-1]
elif QRS and i == QRS[-1] + ms200:
MM.append(newM5)
if len(MM) > 5:
MM.pop(0)
M = np.mean(MM)
elif QRS and i > QRS[-1] + ms200 and i < QRS[-1] + ms1200:
M = np.mean(MM) * M_slope[i - (QRS[-1] + ms200)]
elif QRS and i > QRS[-1] + ms1200:
M = 0.6 * np.mean(MM)
M_list.append(M)
neg_m.append(-M)
if not QRS and low_pass[i] > M:
QRS.append(i)
thi_list.append(i)
thi = True
elif QRS and i > QRS[-1] + ms200 and low_pass[i] > M:
QRS.append(i)
thi_list.append(i)
thi = True
if thi and i < thi_list[-1] + ms160:
if low_pass[i] < -M and low_pass[i - 1] > -M:
# thf_list.append(i)
thf = True
if thf and low_pass[i] < -M:
thf_list.append(i)
counter += 1
elif low_pass[i] > -M and thf:
counter = 0
thi = False
thf = False
elif thi and i > thi_list[-1] + ms160:
counter = 0
thi = False
thf = False
if counter > neg_threshold:
unfiltered_section = unfiltered_ecg[
thi_list[-1] - int(0.01 * self.fs) : i
]
r_peaks.append(
self.engzee_fake_delay
+ np.argmax(unfiltered_section)
+ thi_list[-1]
- int(0.01 * self.fs)
)
counter = 0
thi = False
thf = False
return r_peaks
def matched_filter_detector(self, unfiltered_ecg, template_file=""):
"""
FIR matched filter using template of QRS complex.
Template provided for 250Hz and 360Hz. Optionally provide your
own template file where every line has one sample.
Uses the Pan and Tompkins thresholding method.
"""
current_dir = pathlib.Path(__file__).resolve()
if len(template_file) > 1:
template = np.loadtxt(template_file)
else:
if self.fs == 250:
template_dir = current_dir.parent / "templates" / "template_250hz.csv"
template = np.loadtxt(template_dir)
elif self.fs == 360:
template_dir = current_dir.parent / "templates" / "template_360hz.csv"
template = np.loadtxt(template_dir)
else:
print("\n!!No template for this frequency!!\n")
return False
f0 = 0.1 / self.fs
f1 = 48 / self.fs
b, a = signal.butter(4, [f0 * 2, f1 * 2], btype="bandpass")
prefiltered_ecg = signal.lfilter(b, a, unfiltered_ecg)
matched_coeffs = template[::-1] # time reversing template
detection = signal.lfilter(
matched_coeffs, 1, prefiltered_ecg
) # matched filter FIR filtering
squared = detection * detection # squaring matched filter output
squared[: len(template)] = 0
squared_peaks = panPeakDetect(squared, self.fs)
return squared_peaks
def swt_detector(self, unfiltered_ecg):
"""
Stationary Wavelet Transform
based on Vignesh Kalidas and Lakshman Tamil.
Real-time QRS detector using Stationary Wavelet Transform
for Automated ECG Analysis.
In: 2017 IEEE 17th International Conference on
Bioinformatics and Bioengineering (BIBE).
Uses the Pan and Tompkins thresolding.
"""
swt_level = 3
padding = -1
for i in range(1000):
if (len(unfiltered_ecg) + i) % 2**swt_level == 0:
padding = i
break
if padding > 0:
unfiltered_ecg = np.pad(unfiltered_ecg, (0, padding), "edge")
elif padding == -1:
print("Padding greater than 1000 required\n")
swt_ecg = pywt.swt(unfiltered_ecg, "db3", level=swt_level)
swt_ecg = np.array(swt_ecg)
swt_ecg = swt_ecg[0, 1, :]
squared = swt_ecg * swt_ecg
f1 = 0.01 / self.fs
f2 = 10 / self.fs
b, a = signal.butter(3, [f1 * 2, f2 * 2], btype="bandpass")
filtered_squared = signal.lfilter(b, a, squared)
filt_peaks = panPeakDetect(filtered_squared, self.fs)
return filt_peaks
def pan_tompkins_detector(self, unfiltered_ecg):
"""
Jiapu Pan and Willis J. Tompkins.
A Real-Time QRS Detection Algorithm.
In: IEEE Transactions on Biomedical Engineering
BME-32.3 (1985), pp. 230–236.
"""
f1 = 5 / self.fs
f2 = 15 / self.fs
b, a = signal.butter(1, [f1 * 2, f2 * 2], btype="bandpass")
filtered_ecg = signal.lfilter(b, a, unfiltered_ecg)
diff = np.diff(filtered_ecg)
squared = diff * diff
N = int(0.12 * self.fs)
mwa = MWA(squared, N)
mwa[: int(0.2 * self.fs)] = 0
mwa_peaks = panPeakDetect(mwa, self.fs)
return mwa_peaks
def two_average_detector(self, unfiltered_ecg):
"""
Elgendi, Mohamed & Jonkman,
Mirjam & De Boer, Friso. (2010).
Frequency Bands Effects on QRS Detection.
The 3rd International Conference on Bio-inspired Systems
and Signal Processing (BIOSIGNALS2010). 428-431.
"""
f1 = 8 / self.fs
f2 = 20 / self.fs
b, a = signal.butter(2, [f1 * 2, f2 * 2], btype="bandpass")
filtered_ecg = signal.lfilter(b, a, unfiltered_ecg)
window1 = int(0.12 * self.fs)
mwa_qrs = MWA(abs(filtered_ecg), window1)
window2 = int(0.6 * self.fs)
mwa_beat = MWA(abs(filtered_ecg), window2)
blocks = np.zeros(len(unfiltered_ecg))
block_height = np.max(filtered_ecg)
for i in range(len(mwa_qrs)):
if mwa_qrs[i] > mwa_beat[i]:
blocks[i] = block_height
else:
blocks[i] = 0
QRS = []
for i in range(1, len(blocks)):
if blocks[i - 1] == 0 and blocks[i] == block_height:
start = i
elif blocks[i - 1] == block_height and blocks[i] == 0:
end = i - 1
if end - start > int(0.08 * self.fs):
detection = np.argmax(filtered_ecg[start : end + 1]) + start
if QRS:
if detection - QRS[-1] > int(0.3 * self.fs):
QRS.append(detection)
else:
QRS.append(detection)
return QRS
def MWA(input_array, window_size):
mwa = np.zeros(len(input_array))
for i in range(len(input_array)):
if i < window_size:
section = input_array[0:i]
else:
section = input_array[i - window_size : i]
if i != 0:
mwa[i] = np.mean(section)
else:
mwa[i] = input_array[i]
return mwa
def normalise(input_array):
output_array = (input_array - np.min(input_array)) / (
np.max(input_array) - np.min(input_array)
)
return output_array
def panPeakDetect(detection, fs):
min_distance = int(0.25 * fs)
signal_peaks = [0]
noise_peaks = []
SPKI = 0.0
NPKI = 0.0
threshold_I1 = 0.0
threshold_I2 = 0.0
RR_missed = 0
index = 0
indexes = []
missed_peaks = []
peaks = []
for i in range(len(detection)):
if i > 0 and i < len(detection) - 1:
if detection[i - 1] < detection[i] and detection[i + 1] < detection[i]:
peak = i
peaks.append(i)
if (
detection[peak] > threshold_I1
and (peak - signal_peaks[-1]) > 0.3 * fs
):
signal_peaks.append(peak)
indexes.append(index)
SPKI = 0.125 * detection[signal_peaks[-1]] + 0.875 * SPKI
if RR_missed != 0:
if signal_peaks[-1] - signal_peaks[-2] > RR_missed:
missed_section_peaks = peaks[indexes[-2] + 1 : indexes[-1]]
missed_section_peaks2 = []
for missed_peak in missed_section_peaks:
if (
missed_peak - signal_peaks[-2] > min_distance
and signal_peaks[-1] - missed_peak > min_distance
and detection[missed_peak] > threshold_I2
):
missed_section_peaks2.append(missed_peak)
if len(missed_section_peaks2) > 0:
missed_peak = missed_section_peaks2[
np.argmax(detection[missed_section_peaks2])
]
missed_peaks.append(missed_peak)
signal_peaks.append(signal_peaks[-1])
signal_peaks[-2] = missed_peak
else:
noise_peaks.append(peak)
NPKI = 0.125 * detection[noise_peaks[-1]] + 0.875 * NPKI
threshold_I1 = NPKI + 0.25 * (SPKI - NPKI)
threshold_I2 = 0.5 * threshold_I1
if len(signal_peaks) > 8:
RR = np.diff(signal_peaks[-9:])
RR_ave = int(np.mean(RR))
RR_missed = int(1.66 * RR_ave)
index = index + 1
signal_peaks.pop(0)
return signal_peaks
# ## 2.2 функция выранного детектора
def detect(unfiltered_ecg, fs=250):
detectors = Detectors(fs)
r_peaks = []
# r_peaks_two_average = detectors.two_average_detector(unfiltered_ecg)
# r_peaks = detectors.matched_filter_detector(unfiltered_ecg,"templates/template_250hz.csv")
# r_peaks_swt = detectors.swt_detector(unfiltered_ecg)
# r_peaks_engzee = detectors.engzee_detector(unfiltered_ecg)
# r_peaks_christ = detectors.christov_detector(unfiltered_ecg)
# r_peaks_ham = detectors.hamilton_detector(unfiltered_ecg)
r_peaks = detectors.pan_tompkins_detector(unfiltered_ecg)
# r_peaks.append(sum(r_peaks_two_average, r_peaks_swt, r_peaks_engzee, \
# r_peaks_christ, r_peaks_ham, r_peaks_pan_tom)/(r_peaks_two_average, \
# r_peaks_swt, r_peaks_engzee, \
# r_peaks_christ, r_peaks_ham, r_peaks_pan_tom).count())
return r_peaks
def visualize(unfiltered_ecg, r_peaks):
plt.figure()
plt.plot(unfiltered_ecg)
plt.plot(r_peaks, unfiltered_ecg[r_peaks], "ro")
plt.title("Detected R-peaks")
plt.show()
# # 3. Загрузка подготовленных датасетов
data_test = pd.read_csv("../input/test-ecg/new_test_df.csv")
data_train = pd.read_csv("../input/train-ecg/new_train_df.csv")
data_valid = pd.read_csv("../input/valid-ecg/new_valid_df.csv")
# # 4. Выбор таргета
# ## 4.1 Таргет для модели - 1
# таргеты будем выбирать по очереди
y_variables = data_train.columns.tolist()[-5:]
# ## 4.2 Таргет для модели - 2 по конкретному ЭКГ
find_y(df_with_peaks, get_unfiltered_ecg(df, ecg_id, False))
# ## 5.1 Датафрейм с конкретным ЭКГ по указанным датасету и номеру ecg_id
# функция, возвращающая фрейм только по указанному экг
def get_unfiltered_ecg(df, ecg_id, cut_target_data=True):
ecg_unique = df[df["ecg_id"] == ecg_id]
return ecg_unique[ecg_unique.columns[1:-5]] if cut_target_data else ecg_unique
# ## 5.2 Формируем датасет с пиками поканально по указанным датасету и ЭКГ
# функция, возвращающая словарь из пиков к конкретному экг
def get_peaks(df, ecg_id):
list_of_peaks = []
num_channels = []
ecg_unique = get_unfiltered_ecg(df, ecg_id)
ecg_data = ecg_unique.to_numpy()
# подавать несколько каналов (пики попеременно)
for num_channel in range(ecg_data.shape[1]):
unfiltered_ecg = ecg_data[:, num_channel]
r_peaks = detect(unfiltered_ecg, 100)
# visualize(unfiltered_ecg, r_peaks)
list_of_peaks.append(r_peaks)
num_channels.append(f"channel-{num_channel}")
dict_peaks = dict(list(zip(num_channels, list_of_peaks)))
# dict_peaks_list[ecg_id_data] = dict_peaks
# peaks_data = pd.DataFrame(dict_peaks_list).T
return dict_peaks
# функция, возвращающая фрейм из пиков для дальнейшего слияния с общими данными по этому экг
def pad_nan_peaks(dict_peaks, pad_value=9999):
len_max = 0
for channel, list_peaks in dict_peaks.items():
if len_max < len(list_peaks):
len_max = len(list_peaks)
for channel, list_peaks in dict_peaks.items():
dict_peaks[channel] = np.pad(
list_peaks,
(0, len_max - len(list_peaks)),
"constant",
constant_values=pad_value,
)
peaks_df = pd.DataFrame(dict_peaks)
return peaks_df
# ## 5.3 Формируем датасет с неотфильтрованными данными и пиками поканально по указанным датасету и ЭКГ
def merge_peaks_to_data(df, ecg_id):
ecg_unique = get_unfiltered_ecg(df, ecg_id)
peaks_df = pad_nan_peaks(get_peaks(df, ecg_id))
df_with_peaks = pd.concat([ecg_unique, peaks_df], ignore_index=True)
df_with_peaks["ecg_id"] = ecg_id
return df_with_peaks
# если y с пиками и надо определить длину таргета
def find_y(df_with_peaks, start_df):
ecg_id = df_with_peaks["ecg_id"][0]
y_value = start_df["sinus_rythm"].tolist()[0]
df_with_peaks["target"] = y_value
return df_with_peaks["target"]
find_y(
merge_peaks_to_data(data_train[:2000], 1),
get_unfiltered_ecg(data_train[:2000], 1, False),
)
df_with_peaks = merge_peaks_to_data(data_valid, 17)
start_df = get_unfiltered_ecg(data_valid, 17, False)
# y_valid_ptb = find_y(df_with_peaks, get_unfiltered_ecg(data_valid, 17, False))
ecg_id = df_with_peaks["ecg_id"][0]
start_df["sinus_rythm"].tolist()[0]
# df_with_peaks['target'] = y_value
def X_Y_valid(data_valid):
y_valid_list = []
x_valid_list = []
for ecg_id in data_valid["ecg_id"].unique():
df_with_peaks = merge_peaks_to_data(data_valid, ecg_id)
y_valid_ptb = find_y(
df_with_peaks, get_unfiltered_ecg(data_valid, ecg_id, False)
)
valid_ptb = normalize(df_with_peaks, axis=0, norm="max")
x_valid_ptb = valid_ptb.reshape(len(valid_ptb), valid_ptb.shape[1], 1)
y_valid_list.append(y_valid_ptb)
x_valid_list.append(x_valid_ptb)
y_valid_ptb = list(itertools.chain(*y_valid_list))
x_valid_ptb = list(itertools.chain(*x_valid_list))
return y_valid_list, x_valid_list
y_valid_list, x_valid_list = X_Y_valid(data_valid)
def df_X_y_from_ecg_id(data_train):
for ecg_id_data in data_train["ecg_id"].unique():
df_with_peaks = merge_peaks_to_data(ecg_id_data)
y_train_ptb = to_categorical(
find_y(df_with_peaks, get_unfiltered_ecg(ecg_id_data, False))
)
train_ptb = normalize(df_with_peaks, axis=0, norm="max")
x_train_ptb = train_ptb.reshape(len(train_ptb), train_ptb.shape[1], 1)
input_shape_x = (x_train_ptb.shape[1], 1)
history_conv1d_ptb = history_model(
x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb
)
model_conv1d_ptb.load_weights("conv1d_ptb.h5")
# TODO: надо сделать цикл для fit из 14000 отдельных датасетов экг+пики,
# и мы получим в саммери окончательный вес, затем этот вес мы отдельной
# строчкой запускаем в предтренировку и в отдельной строчкой оцениваем тестовые данные
def history_model(x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb):
history_conv1d_ptb = model_conv1d_ptb.fit(
x_train_ptb,
y_train_ptb,
epochs=10,
batch_size=32,
steps_per_epoch=1000,
validation_data=(x_valid_ptb, y_valid_ptb),
validation_steps=500,
callbacks=[checkpoint_cb, earlystop_cb],
)
return history_conv1d_ptb
def df_with_peaks(df):
labels = []
for ecg_id in df["ecg_id"].unique():
if ecg_id == min(df["ecg_id"].unique()):
df_with_peaks = merge_peaks_to_data(df, ecg_id)
label = np.pad(
[], (0, df_with_peaks.shape[0]), "constant", constant_values=ecg_id
)
labels.append(label)
else:
df_with_peaks = pd.concat([df_with_peaks, merge_peaks_to_data(df, ecg_id)])
label = np.pad(
[],
(0, merge_peaks_to_data(df, ecg_id).shape[0]),
"constant",
constant_values=ecg_id,
)
labels.append(label)
return df_with_peaks
df_with_peaks(data_train[:2000])
# тренировочная, тестовая и валидационная выборки X
X_cols = []
for col in data_train.columns.tolist():
if "channel" in col:
X_cols.append(col)
train_ptb = data_train[X_cols]
test_ptb = data_test[X_cols]
valid_ptb = data_valid[X_cols]
# тренировочная выборка y - синусовый ритм
out_train_ptb = data_train[y_variables[0]]
out_test_ptb = data_test[y_variables[0]]
out_valid_ptb = data_valid[y_variables[0]]
print("Traing dataset size: ", train_ptb.shape)
print("Validation dataset size: ", valid_ptb.shape)
print("Test dataset size: ", test_ptb.shape)
# Normalizing the training, validation & test data
train_ptb = normalize(train_ptb, axis=0, norm="max")
valid_ptb = normalize(valid_ptb, axis=0, norm="max")
test_ptb = normalize(test_ptb, axis=0, norm="max")
# Reshaping the dataframe into a 3-D Numpy array (batch, Time Period, Value)
x_train_ptb = train_ptb.reshape(len(train_ptb), train_ptb.shape[1], 1)
x_valid_ptb = valid_ptb.reshape(len(valid_ptb), valid_ptb.shape[1], 1)
x_test_ptb = test_ptb.reshape(len(test_ptb), test_ptb.shape[1], 1)
# Converting the output into a categorical array
y_train_ptb = to_categorical(out_train_ptb)
y_valid_ptb = to_categorical(out_valid_ptb)
y_test_ptb = to_categorical(out_test_ptb)
print("Traing dataset size: ", x_train_ptb.shape, " -- Y size: ", y_train_ptb.shape)
print("Validation dataset size: ", x_valid_ptb.shape, " -- Y size: ", y_valid_ptb.shape)
print("Test dataset size: ", x_test_ptb.shape, " -- Y size: ", y_test_ptb.shape)
# ## Defining Conv1D model for PTB
# Creating a model based on a series of Conv1D layers that are connected to another series of full connected dense layers
tf.keras.backend.clear_session()
# Function to build Convolutional 1D Networks
# def build_conv1d_model_old (input_shape=(x_train_ptb.shape[1],1)):
# model = keras.models.Sequential()
# model.add(Conv1D(32,7, padding='same', input_shape=input_shape))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Conv1D(64,7, padding='same'))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Conv1D(128,7, padding='same', input_shape=input_shape))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Conv1D(256,7, padding='same'))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Conv1D(512,7, padding='same', input_shape=input_shape))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Flatten())
# model.add(Dense(512, activation='relu'))
# model.add(Dropout(0.5))
# model.add(Dense(256, activation='relu'))
# model.add(Dropout(0.5))
# model.add(Dense(128, activation='relu'))
# model.add(Dense(64, activation='relu'))
# model.add(Dense(32, activation='relu'))
# model.add(Dense(2, activation="softmax"))
# model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=[tfa.metrics.F1Score(2,"micro")])
# return model
def build_conv1d_model(input_shape):
model = keras.models.Sequential()
model.add(Conv1D(32, 7, padding="same", input_shape=input_shape))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Conv1D(64, 7, padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Conv1D(128, 7, padding="same", input_shape=input_shape))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Conv1D(256, 7, padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Conv1D(512, 7, padding="same", input_shape=input_shape))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Flatten())
model.add(Dense(512, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(256, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(128, activation="relu"))
model.add(Dense(64, activation="relu"))
model.add(Dense(32, activation="relu"))
model.add(Dense(2, activation="softmax"))
model.compile(
optimizer="adam",
loss="categorical_crossentropy",
metrics=[tfa.metrics.F1Score(2, "micro")],
)
return model
# checkpoint_cb = ModelCheckpoint("conv1d_ptb.h5", save_best_only=True)
# earlystop_cb = EarlyStopping(patience=5, restore_best_weights=True)
# model_conv1d_ptb= build_conv1d_model(input_shape=(x_train_ptb.shape[1], x_train_ptb.shape[2]))
# model_conv1d_ptb.summary()
def summary_model(x_train_ptb):
checkpoint_cb = ModelCheckpoint("conv1d_ptb.h5", save_best_only=True)
earlystop_cb = EarlyStopping(patience=5, restore_best_weights=True)
model_conv1d_ptb = build_conv1d_model(
input_shape=(x_train_ptb.shape[1], x_train_ptb.shape[2])
)
return model_conv1d_ptb.summary()
def history_model(x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb):
history_conv1d_ptb = model_conv1d_ptb.fit(
x_train_ptb,
y_train_ptb,
epochs=5,
batch_size=32,
steps_per_epoch=1000,
validation_data=(x_valid_ptb, y_valid_ptb),
validation_steps=500,
callbacks=[checkpoint_cb, earlystop_cb],
)
return history_conv1d_ptb
# for i in list(range(2)):
# history = model.fit(training_set_1,epochs=1)
# history = model.fit(training_set_2,epochs=1)
history_conv1d_ptb = model_conv1d_ptb.fit(
x_train_ptb,
y_train_ptb,
epochs=10,
batch_size=32,
steps_per_epoch=1000,
# class_weight=class_weight,
validation_data=(x_valid_ptb, y_valid_ptb),
validation_steps=500,
callbacks=[checkpoint_cb, earlystop_cb],
)
# history_conv1d_ptb = model_conv1d_ptb.fit(x_train_ptb[100000:200000], y_train_ptb[100000:200000],
# epochs=1, batch_size=32,
# # class_weight=class_weight,
# validation_data=(x_valid_ptb[100000:200000], y_valid_ptb[100000:200000]),
# callbacks=[checkpoint_cb, earlystop_cb])
# history_conv1d_ptb = model_conv1d_ptb.fit(x_train_ptb, y_train_ptb, epochs=40, batch_size=32,
# # class_weight=class_weight,
# validation_data=(x_valid_ptb, y_valid_ptb),
# callbacks=[checkpoint_cb, earlystop_cb])
def evaluate_model(
x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb, x_test_ptb, y_test_ptb
):
model_conv1d_ptb = history_model(x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb)
model_conv1d_ptb.evaluate(x_test_ptb, y_test_ptb)
model_conv1d_ptb.load_weights("conv1d_ptb.h5")
model_conv1d_ptb.evaluate(x_test_ptb, y_test_ptb)
# Calculating the predictions based on the highest probability class
conv1d_pred_proba_ptb = model_conv1d_ptb.predict(x_test_ptb)
conv1d_pred_ptb = np.argmax(conv1d_pred_proba_ptb, axis=1)
print(
classification_report(
out_test_ptb,
conv1d_pred_ptb > 0.5,
target_names=[PTB_Outcome[i] for i in PTB_Outcome],
)
)
print(roc_auc_score(conv1d_pred_proba_ptb, out_test_ptb))
print(balanced_accuracy_score(conv1d_pred_proba_ptb, out_test_ptb))
print(f1_score(conv1d_pred_proba_ptb, out_test_ptb))
# Plotting the training and validatoin results
plt.figure(figsize=(25, 12))
plt.plot(
history_conv1d_ptb.epoch,
history_conv1d_ptb.history["loss"],
color="r",
label="Train loss",
)
plt.plot(
history_conv1d_ptb.epoch,
history_conv1d_ptb.history["val_loss"],
color="b",
label="Val loss",
linestyle="--",
)
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.plot(
history_conv1d_ptb.epoch,
history_conv1d_ptb.history["f1_score"],
color="g",
label="Train F1",
)
plt.plot(
history_conv1d_ptb.epoch,
history_conv1d_ptb.history["val_f1_score"],
color="c",
label="Val F1",
linestyle="--",
)
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
plt.show()
# ## Defining Conv1D Residual model for PTB
# Creating a model based on a series of Conv1D layers with 2 residual blocks that are connected to another series of full connected dense layers
def build_conv1d_res_model(input_shape=(x_train_ptb.shape[1], 1)):
model = keras.models.Sequential()
input_ = tf.keras.layers.Input(shape=(input_shape))
conv1_1 = Conv1D(64, 7, padding="same", input_shape=input_shape)(input_)
conv1_1 = BatchNormalization()(conv1_1)
conv1_1 = tf.keras.layers.ReLU()(conv1_1)
conv1_2 = Conv1D(64, 7, padding="same")(conv1_1)
conv1_2 = BatchNormalization()(conv1_2)
conv1_2 = tf.keras.layers.ReLU()(conv1_2)
conv1_3 = Conv1D(64, 7, padding="same")(conv1_2)
conv1_3 = BatchNormalization()(conv1_3)
conv1_3 = tf.keras.layers.ReLU()(conv1_3)
concat_1 = Concatenate()([conv1_1, conv1_3])
max_1 = MaxPool1D(5, padding="same")(concat_1)
conv1_4 = Conv1D(128, 7, padding="same")(max_1)
conv1_4 = BatchNormalization()(conv1_4)
conv1_4 = tf.keras.layers.ReLU()(conv1_4)
conv1_5 = Conv1D(128, 7, padding="same", input_shape=input_shape)(conv1_4)
conv1_5 = BatchNormalization()(conv1_5)
conv1_5 = tf.keras.layers.ReLU()(conv1_5)
conv1_6 = Conv1D(128, 7, padding="same", input_shape=input_shape)(conv1_5)
conv1_6 = BatchNormalization()(conv1_6)
conv1_6 = tf.keras.layers.ReLU()(conv1_6)
concat_2 = Concatenate()([conv1_4, conv1_6])
max_2 = MaxPool1D(5, padding="same")(concat_2)
flat = Flatten()(max_2)
dense_1 = Dense(512, activation="relu")(flat)
drop_1 = Dropout(0.5)(dense_1)
dense_2 = Dense(256, activation="relu")(drop_1)
drop_2 = Dropout(0.5)(dense_2)
dense_3 = Dense(128, activation="relu")(drop_2)
dense_4 = Dense(64, activation="relu")(dense_3)
dense_5 = Dense(32, activation="relu")(dense_4)
dense_6 = Dense(2, activation="softmax")(dense_5)
model = Model(inputs=input_, outputs=dense_6)
model.compile(
optimizer="adam",
loss="categorical_crossentropy",
metrics=[tfa.metrics.F1Score(2, "micro")],
)
return model
checkpoint_cb = ModelCheckpoint("conv1d_res_ptb.h5", save_best_only=True)
earlystop_cb = EarlyStopping(patience=5, restore_best_weights=True)
inp_shape = (x_train_ptb.shape[1], x_train_ptb.shape[2])
model_conv1d_res_ptb = build_conv1d_res_model(
input_shape=(x_train_ptb.shape[1], x_train_ptb.shape[2])
)
# model_conv1d_res_ptb.build(inp_shape)
history_conv1d_res_ptb = model_conv1d_res_ptb.fit(
x_train_ptb,
y_train_ptb,
epochs=10,
batch_size=32,
steps_per_epoch=1000,
# class_weight=class_weight,
validation_data=(x_valid_ptb, y_valid_ptb),
validation_steps=500,
callbacks=[checkpoint_cb, earlystop_cb],
)
# history_conv1d_res_ptb = model_conv1d_res_ptb.fit(x_train_ptb, y_train_ptb, epochs=40, batch_size=32,
# class_weight=class_weight, validation_data=(x_valid_ptb, y_valid_ptb),
# callbacks=[checkpoint_cb, earlystop_cb])
model_conv1d_res_ptb.load_weights("conv1d_res_ptb.h5")
model_conv1d_res_ptb.evaluate(x_test_ptb, y_test_ptb)
# Calculating the predictions based on the highest probability class
conv1d_res_pred_proba_ptb = model_conv1d_res_ptb.predict(x_test_ptb)
conv1d_res_pred_ptb = np.argmax(conv1d_res_pred_proba_ptb, axis=1)
print(
classification_report(
out_test_ptb,
conv1d_res_pred_ptb > 0.5,
target_names=[PTB_Outcome[i] for i in PTB_Outcome],
)
)
print(roc_auc_score(conv1d_res_pred_ptb, out_test_ptb))
print(balanced_accuracy_score(conv1d_res_pred_ptb, out_test_ptb))
print(f1_score(conv1d_res_pred_ptb, out_test_ptb))
# Plotting the training and validatoin results
plt.figure(figsize=(25, 12))
plt.plot(
history_conv1d_res_ptb.epoch,
history_conv1d_res_ptb.history["loss"],
color="r",
label="Train loss",
)
plt.plot(
history_conv1d_res_ptb.epoch,
history_conv1d_res_ptb.history["val_loss"],
color="b",
label="Val loss",
linestyle="--",
)
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.plot(
history_conv1d_res_ptb.epoch,
history_conv1d_res_ptb.history["f1_score"],
color="g",
label="Train F1",
)
plt.plot(
history_conv1d_res_ptb.epoch,
history_conv1d_res_ptb.history["val_f1_score"],
color="c",
label="Val F1",
linestyle="--",
)
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/492/69492937.ipynb
| null | null |
[{"Id": 69492937, "ScriptId": 18596145, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4125499, "CreationDate": "07/31/2021 17:07:43", "VersionNumber": 21.0, "Title": "ECG Heartbeat Categorization (Neural Network)", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 1140.0, "LinesInsertedFromPrevious": 51.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1089.0, "LinesInsertedFromFork": 932.0, "LinesDeletedFromFork": 351.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 208.0, "TotalVotes": 0}]
| null | null | null | null |
# # Loading the data and exploring its shape and values
# # 1. библиотеки
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.utils import resample
import itertools
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import normalize
from sklearn.metrics import classification_report
from sklearn.metrics import roc_auc_score
from sklearn.metrics import balanced_accuracy_score
from sklearn.metrics import f1_score
import tensorflow as tf
import tensorflow_addons as tfa
from tensorflow import keras
from tensorflow.keras.layers import (
Dense,
Conv1D,
MaxPool1D,
Flatten,
Dropout,
InputLayer,
LSTM,
GRU,
BatchNormalization,
Bidirectional,
Concatenate,
)
from tensorflow.keras.models import Model
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
from tensorflow.keras.preprocessing import sequence
from tensorflow.keras.optimizers import SGD, RMSprop
from tensorflow.keras.utils import to_categorical
# # 2. вспомогательные функции
# ## 2.1 класс детекторов для нахождения пик
# класс детекторов дл нахождения пик
import numpy as np
import pywt
try:
import pathlib
except ImportError:
import pathlib2 as pathlib
import scipy.signal as signal
class Detectors:
"""ECG heartbeat detection algorithms
General useage instructions:
r_peaks = detectors.the_detector(ecg_in_samples)
The argument ecg_in_samples is a single channel ECG in volt
at the given sample rate.
"""
def __init__(self, sampling_frequency):
"""
The constructor takes the sampling rate in Hz of the ECG data.
"""
self.fs = sampling_frequency
# this is set to a positive value for benchmarking
self.engzee_fake_delay = 0
def hamilton_detector(self, unfiltered_ecg):
"""
P.S. Hamilton,
Open Source ECG Analysis Software Documentation, E.P.Limited, 2002.
"""
f1 = 8 / self.fs
f2 = 16 / self.fs
b, a = signal.butter(1, [f1 * 2, f2 * 2], btype="bandpass")
filtered_ecg = signal.lfilter(b, a, unfiltered_ecg)
diff = abs(np.diff(filtered_ecg))
b = np.ones(int(0.08 * self.fs))
b = b / int(0.08 * self.fs)
a = [1]
ma = signal.lfilter(b, a, diff)
ma[0 : len(b) * 2] = 0
n_pks = []
n_pks_ave = 0.0
s_pks = []
s_pks_ave = 0.0
QRS = [0]
RR = []
RR_ave = 0.0
th = 0.0
i = 0
idx = []
peaks = []
for i in range(len(ma)):
if i > 0 and i < len(ma) - 1:
if ma[i - 1] < ma[i] and ma[i + 1] < ma[i]:
peak = i
peaks.append(i)
if ma[peak] > th and (peak - QRS[-1]) > 0.3 * self.fs:
QRS.append(peak)
idx.append(i)
s_pks.append(ma[peak])
if len(n_pks) > 8:
s_pks.pop(0)
s_pks_ave = np.mean(s_pks)
if RR_ave != 0.0:
if QRS[-1] - QRS[-2] > 1.5 * RR_ave:
missed_peaks = peaks[idx[-2] + 1 : idx[-1]]
for missed_peak in missed_peaks:
if (
missed_peak - peaks[idx[-2]]
> int(0.360 * self.fs)
and ma[missed_peak] > 0.5 * th
):
QRS.append(missed_peak)
QRS.sort()
break
if len(QRS) > 2:
RR.append(QRS[-1] - QRS[-2])
if len(RR) > 8:
RR.pop(0)
RR_ave = int(np.mean(RR))
else:
n_pks.append(ma[peak])
if len(n_pks) > 8:
n_pks.pop(0)
n_pks_ave = np.mean(n_pks)
th = n_pks_ave + 0.45 * (s_pks_ave - n_pks_ave)
i += 1
QRS.pop(0)
return QRS
def christov_detector(self, unfiltered_ecg):
"""
Ivaylo I. Christov,
Real time electrocardiogram QRS detection using combined
adaptive threshold, BioMedical Engineering OnLine 2004,
vol. 3:28, 2004.
"""
total_taps = 0
b = np.ones(int(0.02 * self.fs))
b = b / int(0.02 * self.fs)
total_taps += len(b)
a = [1]
MA1 = signal.lfilter(b, a, unfiltered_ecg)
b = np.ones(int(0.028 * self.fs))
b = b / int(0.028 * self.fs)
total_taps += len(b)
a = [1]
MA2 = signal.lfilter(b, a, MA1)
Y = []
for i in range(1, len(MA2) - 1):
diff = abs(MA2[i + 1] - MA2[i - 1])
Y.append(diff)
b = np.ones(int(0.040 * self.fs))
b = b / int(0.040 * self.fs)
total_taps += len(b)
a = [1]
MA3 = signal.lfilter(b, a, Y)
MA3[0:total_taps] = 0
ms50 = int(0.05 * self.fs)
ms200 = int(0.2 * self.fs)
ms1200 = int(1.2 * self.fs)
ms350 = int(0.35 * self.fs)
M = 0
newM5 = 0
M_list = []
MM = []
M_slope = np.linspace(1.0, 0.6, ms1200 - ms200)
F = 0
F_list = []
R = 0
RR = []
Rm = 0
R_list = []
MFR = 0
MFR_list = []
QRS = []
for i in range(len(MA3)):
# M
if i < 5 * self.fs:
M = 0.6 * np.max(MA3[: i + 1])
MM.append(M)
if len(MM) > 5:
MM.pop(0)
elif QRS and i < QRS[-1] + ms200:
newM5 = 0.6 * np.max(MA3[QRS[-1] : i])
if newM5 > 1.5 * MM[-1]:
newM5 = 1.1 * MM[-1]
elif QRS and i == QRS[-1] + ms200:
if newM5 == 0:
newM5 = MM[-1]
MM.append(newM5)
if len(MM) > 5:
MM.pop(0)
M = np.mean(MM)
elif QRS and i > QRS[-1] + ms200 and i < QRS[-1] + ms1200:
M = np.mean(MM) * M_slope[i - (QRS[-1] + ms200)]
elif QRS and i > QRS[-1] + ms1200:
M = 0.6 * np.mean(MM)
# F
if i > ms350:
F_section = MA3[i - ms350 : i]
max_latest = np.max(F_section[-ms50:])
max_earliest = np.max(F_section[:ms50])
F = F + ((max_latest - max_earliest) / 150.0)
# R
if QRS and i < QRS[-1] + int((2.0 / 3.0 * Rm)):
R = 0
elif QRS and i > QRS[-1] + int((2.0 / 3.0 * Rm)) and i < QRS[-1] + Rm:
dec = (M - np.mean(MM)) / 1.4
R = 0 + dec
MFR = M + F + R
M_list.append(M)
F_list.append(F)
R_list.append(R)
MFR_list.append(MFR)
if not QRS and MA3[i] > MFR:
QRS.append(i)
elif QRS and i > QRS[-1] + ms200 and MA3[i] > MFR:
QRS.append(i)
if len(QRS) > 2:
RR.append(QRS[-1] - QRS[-2])
if len(RR) > 5:
RR.pop(0)
Rm = int(np.mean(RR))
QRS.pop(0)
return QRS
def engzee_detector(self, unfiltered_ecg):
"""
C. Zeelenberg, A single scan algorithm for QRS detection and
feature extraction, IEEE Comp. in Cardiology, vol. 6,
pp. 37-42, 1979 with modifications A. Lourenco, H. Silva,
P. Leite, R. Lourenco and A. Fred, “Real Time
Electrocardiogram Segmentation for Finger Based ECG
Biometrics”, BIOSIGNALS 2012, pp. 49-54, 2012.
"""
f1 = 48 / self.fs
f2 = 52 / self.fs
b, a = signal.butter(4, [f1 * 2, f2 * 2], btype="bandstop")
filtered_ecg = signal.lfilter(b, a, unfiltered_ecg)
diff = np.zeros(len(filtered_ecg))
for i in range(4, len(diff)):
diff[i] = filtered_ecg[i] - filtered_ecg[i - 4]
ci = [1, 4, 6, 4, 1]
low_pass = signal.lfilter(ci, 1, diff)
low_pass[: int(0.2 * self.fs)] = 0
ms200 = int(0.2 * self.fs)
ms1200 = int(1.2 * self.fs)
ms160 = int(0.16 * self.fs)
neg_threshold = int(0.01 * self.fs)
M = 0
M_list = []
neg_m = []
MM = []
M_slope = np.linspace(1.0, 0.6, ms1200 - ms200)
QRS = []
r_peaks = []
counter = 0
thi_list = []
thi = False
thf_list = []
thf = False
for i in range(len(low_pass)):
# M
if i < 5 * self.fs:
M = 0.6 * np.max(low_pass[: i + 1])
MM.append(M)
if len(MM) > 5:
MM.pop(0)
elif QRS and i < QRS[-1] + ms200:
newM5 = 0.6 * np.max(low_pass[QRS[-1] : i])
if newM5 > 1.5 * MM[-1]:
newM5 = 1.1 * MM[-1]
elif QRS and i == QRS[-1] + ms200:
MM.append(newM5)
if len(MM) > 5:
MM.pop(0)
M = np.mean(MM)
elif QRS and i > QRS[-1] + ms200 and i < QRS[-1] + ms1200:
M = np.mean(MM) * M_slope[i - (QRS[-1] + ms200)]
elif QRS and i > QRS[-1] + ms1200:
M = 0.6 * np.mean(MM)
M_list.append(M)
neg_m.append(-M)
if not QRS and low_pass[i] > M:
QRS.append(i)
thi_list.append(i)
thi = True
elif QRS and i > QRS[-1] + ms200 and low_pass[i] > M:
QRS.append(i)
thi_list.append(i)
thi = True
if thi and i < thi_list[-1] + ms160:
if low_pass[i] < -M and low_pass[i - 1] > -M:
# thf_list.append(i)
thf = True
if thf and low_pass[i] < -M:
thf_list.append(i)
counter += 1
elif low_pass[i] > -M and thf:
counter = 0
thi = False
thf = False
elif thi and i > thi_list[-1] + ms160:
counter = 0
thi = False
thf = False
if counter > neg_threshold:
unfiltered_section = unfiltered_ecg[
thi_list[-1] - int(0.01 * self.fs) : i
]
r_peaks.append(
self.engzee_fake_delay
+ np.argmax(unfiltered_section)
+ thi_list[-1]
- int(0.01 * self.fs)
)
counter = 0
thi = False
thf = False
return r_peaks
def matched_filter_detector(self, unfiltered_ecg, template_file=""):
"""
FIR matched filter using template of QRS complex.
Template provided for 250Hz and 360Hz. Optionally provide your
own template file where every line has one sample.
Uses the Pan and Tompkins thresholding method.
"""
current_dir = pathlib.Path(__file__).resolve()
if len(template_file) > 1:
template = np.loadtxt(template_file)
else:
if self.fs == 250:
template_dir = current_dir.parent / "templates" / "template_250hz.csv"
template = np.loadtxt(template_dir)
elif self.fs == 360:
template_dir = current_dir.parent / "templates" / "template_360hz.csv"
template = np.loadtxt(template_dir)
else:
print("\n!!No template for this frequency!!\n")
return False
f0 = 0.1 / self.fs
f1 = 48 / self.fs
b, a = signal.butter(4, [f0 * 2, f1 * 2], btype="bandpass")
prefiltered_ecg = signal.lfilter(b, a, unfiltered_ecg)
matched_coeffs = template[::-1] # time reversing template
detection = signal.lfilter(
matched_coeffs, 1, prefiltered_ecg
) # matched filter FIR filtering
squared = detection * detection # squaring matched filter output
squared[: len(template)] = 0
squared_peaks = panPeakDetect(squared, self.fs)
return squared_peaks
def swt_detector(self, unfiltered_ecg):
"""
Stationary Wavelet Transform
based on Vignesh Kalidas and Lakshman Tamil.
Real-time QRS detector using Stationary Wavelet Transform
for Automated ECG Analysis.
In: 2017 IEEE 17th International Conference on
Bioinformatics and Bioengineering (BIBE).
Uses the Pan and Tompkins thresolding.
"""
swt_level = 3
padding = -1
for i in range(1000):
if (len(unfiltered_ecg) + i) % 2**swt_level == 0:
padding = i
break
if padding > 0:
unfiltered_ecg = np.pad(unfiltered_ecg, (0, padding), "edge")
elif padding == -1:
print("Padding greater than 1000 required\n")
swt_ecg = pywt.swt(unfiltered_ecg, "db3", level=swt_level)
swt_ecg = np.array(swt_ecg)
swt_ecg = swt_ecg[0, 1, :]
squared = swt_ecg * swt_ecg
f1 = 0.01 / self.fs
f2 = 10 / self.fs
b, a = signal.butter(3, [f1 * 2, f2 * 2], btype="bandpass")
filtered_squared = signal.lfilter(b, a, squared)
filt_peaks = panPeakDetect(filtered_squared, self.fs)
return filt_peaks
def pan_tompkins_detector(self, unfiltered_ecg):
"""
Jiapu Pan and Willis J. Tompkins.
A Real-Time QRS Detection Algorithm.
In: IEEE Transactions on Biomedical Engineering
BME-32.3 (1985), pp. 230–236.
"""
f1 = 5 / self.fs
f2 = 15 / self.fs
b, a = signal.butter(1, [f1 * 2, f2 * 2], btype="bandpass")
filtered_ecg = signal.lfilter(b, a, unfiltered_ecg)
diff = np.diff(filtered_ecg)
squared = diff * diff
N = int(0.12 * self.fs)
mwa = MWA(squared, N)
mwa[: int(0.2 * self.fs)] = 0
mwa_peaks = panPeakDetect(mwa, self.fs)
return mwa_peaks
def two_average_detector(self, unfiltered_ecg):
"""
Elgendi, Mohamed & Jonkman,
Mirjam & De Boer, Friso. (2010).
Frequency Bands Effects on QRS Detection.
The 3rd International Conference on Bio-inspired Systems
and Signal Processing (BIOSIGNALS2010). 428-431.
"""
f1 = 8 / self.fs
f2 = 20 / self.fs
b, a = signal.butter(2, [f1 * 2, f2 * 2], btype="bandpass")
filtered_ecg = signal.lfilter(b, a, unfiltered_ecg)
window1 = int(0.12 * self.fs)
mwa_qrs = MWA(abs(filtered_ecg), window1)
window2 = int(0.6 * self.fs)
mwa_beat = MWA(abs(filtered_ecg), window2)
blocks = np.zeros(len(unfiltered_ecg))
block_height = np.max(filtered_ecg)
for i in range(len(mwa_qrs)):
if mwa_qrs[i] > mwa_beat[i]:
blocks[i] = block_height
else:
blocks[i] = 0
QRS = []
for i in range(1, len(blocks)):
if blocks[i - 1] == 0 and blocks[i] == block_height:
start = i
elif blocks[i - 1] == block_height and blocks[i] == 0:
end = i - 1
if end - start > int(0.08 * self.fs):
detection = np.argmax(filtered_ecg[start : end + 1]) + start
if QRS:
if detection - QRS[-1] > int(0.3 * self.fs):
QRS.append(detection)
else:
QRS.append(detection)
return QRS
def MWA(input_array, window_size):
mwa = np.zeros(len(input_array))
for i in range(len(input_array)):
if i < window_size:
section = input_array[0:i]
else:
section = input_array[i - window_size : i]
if i != 0:
mwa[i] = np.mean(section)
else:
mwa[i] = input_array[i]
return mwa
def normalise(input_array):
output_array = (input_array - np.min(input_array)) / (
np.max(input_array) - np.min(input_array)
)
return output_array
def panPeakDetect(detection, fs):
min_distance = int(0.25 * fs)
signal_peaks = [0]
noise_peaks = []
SPKI = 0.0
NPKI = 0.0
threshold_I1 = 0.0
threshold_I2 = 0.0
RR_missed = 0
index = 0
indexes = []
missed_peaks = []
peaks = []
for i in range(len(detection)):
if i > 0 and i < len(detection) - 1:
if detection[i - 1] < detection[i] and detection[i + 1] < detection[i]:
peak = i
peaks.append(i)
if (
detection[peak] > threshold_I1
and (peak - signal_peaks[-1]) > 0.3 * fs
):
signal_peaks.append(peak)
indexes.append(index)
SPKI = 0.125 * detection[signal_peaks[-1]] + 0.875 * SPKI
if RR_missed != 0:
if signal_peaks[-1] - signal_peaks[-2] > RR_missed:
missed_section_peaks = peaks[indexes[-2] + 1 : indexes[-1]]
missed_section_peaks2 = []
for missed_peak in missed_section_peaks:
if (
missed_peak - signal_peaks[-2] > min_distance
and signal_peaks[-1] - missed_peak > min_distance
and detection[missed_peak] > threshold_I2
):
missed_section_peaks2.append(missed_peak)
if len(missed_section_peaks2) > 0:
missed_peak = missed_section_peaks2[
np.argmax(detection[missed_section_peaks2])
]
missed_peaks.append(missed_peak)
signal_peaks.append(signal_peaks[-1])
signal_peaks[-2] = missed_peak
else:
noise_peaks.append(peak)
NPKI = 0.125 * detection[noise_peaks[-1]] + 0.875 * NPKI
threshold_I1 = NPKI + 0.25 * (SPKI - NPKI)
threshold_I2 = 0.5 * threshold_I1
if len(signal_peaks) > 8:
RR = np.diff(signal_peaks[-9:])
RR_ave = int(np.mean(RR))
RR_missed = int(1.66 * RR_ave)
index = index + 1
signal_peaks.pop(0)
return signal_peaks
# ## 2.2 функция выранного детектора
def detect(unfiltered_ecg, fs=250):
detectors = Detectors(fs)
r_peaks = []
# r_peaks_two_average = detectors.two_average_detector(unfiltered_ecg)
# r_peaks = detectors.matched_filter_detector(unfiltered_ecg,"templates/template_250hz.csv")
# r_peaks_swt = detectors.swt_detector(unfiltered_ecg)
# r_peaks_engzee = detectors.engzee_detector(unfiltered_ecg)
# r_peaks_christ = detectors.christov_detector(unfiltered_ecg)
# r_peaks_ham = detectors.hamilton_detector(unfiltered_ecg)
r_peaks = detectors.pan_tompkins_detector(unfiltered_ecg)
# r_peaks.append(sum(r_peaks_two_average, r_peaks_swt, r_peaks_engzee, \
# r_peaks_christ, r_peaks_ham, r_peaks_pan_tom)/(r_peaks_two_average, \
# r_peaks_swt, r_peaks_engzee, \
# r_peaks_christ, r_peaks_ham, r_peaks_pan_tom).count())
return r_peaks
def visualize(unfiltered_ecg, r_peaks):
plt.figure()
plt.plot(unfiltered_ecg)
plt.plot(r_peaks, unfiltered_ecg[r_peaks], "ro")
plt.title("Detected R-peaks")
plt.show()
# # 3. Загрузка подготовленных датасетов
data_test = pd.read_csv("../input/test-ecg/new_test_df.csv")
data_train = pd.read_csv("../input/train-ecg/new_train_df.csv")
data_valid = pd.read_csv("../input/valid-ecg/new_valid_df.csv")
# # 4. Выбор таргета
# ## 4.1 Таргет для модели - 1
# таргеты будем выбирать по очереди
y_variables = data_train.columns.tolist()[-5:]
# ## 4.2 Таргет для модели - 2 по конкретному ЭКГ
find_y(df_with_peaks, get_unfiltered_ecg(df, ecg_id, False))
# ## 5.1 Датафрейм с конкретным ЭКГ по указанным датасету и номеру ecg_id
# функция, возвращающая фрейм только по указанному экг
def get_unfiltered_ecg(df, ecg_id, cut_target_data=True):
ecg_unique = df[df["ecg_id"] == ecg_id]
return ecg_unique[ecg_unique.columns[1:-5]] if cut_target_data else ecg_unique
# ## 5.2 Формируем датасет с пиками поканально по указанным датасету и ЭКГ
# функция, возвращающая словарь из пиков к конкретному экг
def get_peaks(df, ecg_id):
list_of_peaks = []
num_channels = []
ecg_unique = get_unfiltered_ecg(df, ecg_id)
ecg_data = ecg_unique.to_numpy()
# подавать несколько каналов (пики попеременно)
for num_channel in range(ecg_data.shape[1]):
unfiltered_ecg = ecg_data[:, num_channel]
r_peaks = detect(unfiltered_ecg, 100)
# visualize(unfiltered_ecg, r_peaks)
list_of_peaks.append(r_peaks)
num_channels.append(f"channel-{num_channel}")
dict_peaks = dict(list(zip(num_channels, list_of_peaks)))
# dict_peaks_list[ecg_id_data] = dict_peaks
# peaks_data = pd.DataFrame(dict_peaks_list).T
return dict_peaks
# функция, возвращающая фрейм из пиков для дальнейшего слияния с общими данными по этому экг
def pad_nan_peaks(dict_peaks, pad_value=9999):
len_max = 0
for channel, list_peaks in dict_peaks.items():
if len_max < len(list_peaks):
len_max = len(list_peaks)
for channel, list_peaks in dict_peaks.items():
dict_peaks[channel] = np.pad(
list_peaks,
(0, len_max - len(list_peaks)),
"constant",
constant_values=pad_value,
)
peaks_df = pd.DataFrame(dict_peaks)
return peaks_df
# ## 5.3 Формируем датасет с неотфильтрованными данными и пиками поканально по указанным датасету и ЭКГ
def merge_peaks_to_data(df, ecg_id):
ecg_unique = get_unfiltered_ecg(df, ecg_id)
peaks_df = pad_nan_peaks(get_peaks(df, ecg_id))
df_with_peaks = pd.concat([ecg_unique, peaks_df], ignore_index=True)
df_with_peaks["ecg_id"] = ecg_id
return df_with_peaks
# если y с пиками и надо определить длину таргета
def find_y(df_with_peaks, start_df):
ecg_id = df_with_peaks["ecg_id"][0]
y_value = start_df["sinus_rythm"].tolist()[0]
df_with_peaks["target"] = y_value
return df_with_peaks["target"]
find_y(
merge_peaks_to_data(data_train[:2000], 1),
get_unfiltered_ecg(data_train[:2000], 1, False),
)
df_with_peaks = merge_peaks_to_data(data_valid, 17)
start_df = get_unfiltered_ecg(data_valid, 17, False)
# y_valid_ptb = find_y(df_with_peaks, get_unfiltered_ecg(data_valid, 17, False))
ecg_id = df_with_peaks["ecg_id"][0]
start_df["sinus_rythm"].tolist()[0]
# df_with_peaks['target'] = y_value
def X_Y_valid(data_valid):
y_valid_list = []
x_valid_list = []
for ecg_id in data_valid["ecg_id"].unique():
df_with_peaks = merge_peaks_to_data(data_valid, ecg_id)
y_valid_ptb = find_y(
df_with_peaks, get_unfiltered_ecg(data_valid, ecg_id, False)
)
valid_ptb = normalize(df_with_peaks, axis=0, norm="max")
x_valid_ptb = valid_ptb.reshape(len(valid_ptb), valid_ptb.shape[1], 1)
y_valid_list.append(y_valid_ptb)
x_valid_list.append(x_valid_ptb)
y_valid_ptb = list(itertools.chain(*y_valid_list))
x_valid_ptb = list(itertools.chain(*x_valid_list))
return y_valid_list, x_valid_list
y_valid_list, x_valid_list = X_Y_valid(data_valid)
def df_X_y_from_ecg_id(data_train):
for ecg_id_data in data_train["ecg_id"].unique():
df_with_peaks = merge_peaks_to_data(ecg_id_data)
y_train_ptb = to_categorical(
find_y(df_with_peaks, get_unfiltered_ecg(ecg_id_data, False))
)
train_ptb = normalize(df_with_peaks, axis=0, norm="max")
x_train_ptb = train_ptb.reshape(len(train_ptb), train_ptb.shape[1], 1)
input_shape_x = (x_train_ptb.shape[1], 1)
history_conv1d_ptb = history_model(
x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb
)
model_conv1d_ptb.load_weights("conv1d_ptb.h5")
# TODO: надо сделать цикл для fit из 14000 отдельных датасетов экг+пики,
# и мы получим в саммери окончательный вес, затем этот вес мы отдельной
# строчкой запускаем в предтренировку и в отдельной строчкой оцениваем тестовые данные
def history_model(x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb):
history_conv1d_ptb = model_conv1d_ptb.fit(
x_train_ptb,
y_train_ptb,
epochs=10,
batch_size=32,
steps_per_epoch=1000,
validation_data=(x_valid_ptb, y_valid_ptb),
validation_steps=500,
callbacks=[checkpoint_cb, earlystop_cb],
)
return history_conv1d_ptb
def df_with_peaks(df):
labels = []
for ecg_id in df["ecg_id"].unique():
if ecg_id == min(df["ecg_id"].unique()):
df_with_peaks = merge_peaks_to_data(df, ecg_id)
label = np.pad(
[], (0, df_with_peaks.shape[0]), "constant", constant_values=ecg_id
)
labels.append(label)
else:
df_with_peaks = pd.concat([df_with_peaks, merge_peaks_to_data(df, ecg_id)])
label = np.pad(
[],
(0, merge_peaks_to_data(df, ecg_id).shape[0]),
"constant",
constant_values=ecg_id,
)
labels.append(label)
return df_with_peaks
df_with_peaks(data_train[:2000])
# тренировочная, тестовая и валидационная выборки X
X_cols = []
for col in data_train.columns.tolist():
if "channel" in col:
X_cols.append(col)
train_ptb = data_train[X_cols]
test_ptb = data_test[X_cols]
valid_ptb = data_valid[X_cols]
# тренировочная выборка y - синусовый ритм
out_train_ptb = data_train[y_variables[0]]
out_test_ptb = data_test[y_variables[0]]
out_valid_ptb = data_valid[y_variables[0]]
print("Traing dataset size: ", train_ptb.shape)
print("Validation dataset size: ", valid_ptb.shape)
print("Test dataset size: ", test_ptb.shape)
# Normalizing the training, validation & test data
train_ptb = normalize(train_ptb, axis=0, norm="max")
valid_ptb = normalize(valid_ptb, axis=0, norm="max")
test_ptb = normalize(test_ptb, axis=0, norm="max")
# Reshaping the dataframe into a 3-D Numpy array (batch, Time Period, Value)
x_train_ptb = train_ptb.reshape(len(train_ptb), train_ptb.shape[1], 1)
x_valid_ptb = valid_ptb.reshape(len(valid_ptb), valid_ptb.shape[1], 1)
x_test_ptb = test_ptb.reshape(len(test_ptb), test_ptb.shape[1], 1)
# Converting the output into a categorical array
y_train_ptb = to_categorical(out_train_ptb)
y_valid_ptb = to_categorical(out_valid_ptb)
y_test_ptb = to_categorical(out_test_ptb)
print("Traing dataset size: ", x_train_ptb.shape, " -- Y size: ", y_train_ptb.shape)
print("Validation dataset size: ", x_valid_ptb.shape, " -- Y size: ", y_valid_ptb.shape)
print("Test dataset size: ", x_test_ptb.shape, " -- Y size: ", y_test_ptb.shape)
# ## Defining Conv1D model for PTB
# Creating a model based on a series of Conv1D layers that are connected to another series of full connected dense layers
tf.keras.backend.clear_session()
# Function to build Convolutional 1D Networks
# def build_conv1d_model_old (input_shape=(x_train_ptb.shape[1],1)):
# model = keras.models.Sequential()
# model.add(Conv1D(32,7, padding='same', input_shape=input_shape))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Conv1D(64,7, padding='same'))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Conv1D(128,7, padding='same', input_shape=input_shape))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Conv1D(256,7, padding='same'))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Conv1D(512,7, padding='same', input_shape=input_shape))
# model.add(BatchNormalization())
# model.add(tf.keras.layers.ReLU())
# model.add(MaxPool1D(5,padding='same'))
# model.add(Flatten())
# model.add(Dense(512, activation='relu'))
# model.add(Dropout(0.5))
# model.add(Dense(256, activation='relu'))
# model.add(Dropout(0.5))
# model.add(Dense(128, activation='relu'))
# model.add(Dense(64, activation='relu'))
# model.add(Dense(32, activation='relu'))
# model.add(Dense(2, activation="softmax"))
# model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=[tfa.metrics.F1Score(2,"micro")])
# return model
def build_conv1d_model(input_shape):
model = keras.models.Sequential()
model.add(Conv1D(32, 7, padding="same", input_shape=input_shape))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Conv1D(64, 7, padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Conv1D(128, 7, padding="same", input_shape=input_shape))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Conv1D(256, 7, padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Conv1D(512, 7, padding="same", input_shape=input_shape))
model.add(BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(MaxPool1D(5, padding="same"))
model.add(Flatten())
model.add(Dense(512, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(256, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(128, activation="relu"))
model.add(Dense(64, activation="relu"))
model.add(Dense(32, activation="relu"))
model.add(Dense(2, activation="softmax"))
model.compile(
optimizer="adam",
loss="categorical_crossentropy",
metrics=[tfa.metrics.F1Score(2, "micro")],
)
return model
# checkpoint_cb = ModelCheckpoint("conv1d_ptb.h5", save_best_only=True)
# earlystop_cb = EarlyStopping(patience=5, restore_best_weights=True)
# model_conv1d_ptb= build_conv1d_model(input_shape=(x_train_ptb.shape[1], x_train_ptb.shape[2]))
# model_conv1d_ptb.summary()
def summary_model(x_train_ptb):
checkpoint_cb = ModelCheckpoint("conv1d_ptb.h5", save_best_only=True)
earlystop_cb = EarlyStopping(patience=5, restore_best_weights=True)
model_conv1d_ptb = build_conv1d_model(
input_shape=(x_train_ptb.shape[1], x_train_ptb.shape[2])
)
return model_conv1d_ptb.summary()
def history_model(x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb):
history_conv1d_ptb = model_conv1d_ptb.fit(
x_train_ptb,
y_train_ptb,
epochs=5,
batch_size=32,
steps_per_epoch=1000,
validation_data=(x_valid_ptb, y_valid_ptb),
validation_steps=500,
callbacks=[checkpoint_cb, earlystop_cb],
)
return history_conv1d_ptb
# for i in list(range(2)):
# history = model.fit(training_set_1,epochs=1)
# history = model.fit(training_set_2,epochs=1)
history_conv1d_ptb = model_conv1d_ptb.fit(
x_train_ptb,
y_train_ptb,
epochs=10,
batch_size=32,
steps_per_epoch=1000,
# class_weight=class_weight,
validation_data=(x_valid_ptb, y_valid_ptb),
validation_steps=500,
callbacks=[checkpoint_cb, earlystop_cb],
)
# history_conv1d_ptb = model_conv1d_ptb.fit(x_train_ptb[100000:200000], y_train_ptb[100000:200000],
# epochs=1, batch_size=32,
# # class_weight=class_weight,
# validation_data=(x_valid_ptb[100000:200000], y_valid_ptb[100000:200000]),
# callbacks=[checkpoint_cb, earlystop_cb])
# history_conv1d_ptb = model_conv1d_ptb.fit(x_train_ptb, y_train_ptb, epochs=40, batch_size=32,
# # class_weight=class_weight,
# validation_data=(x_valid_ptb, y_valid_ptb),
# callbacks=[checkpoint_cb, earlystop_cb])
def evaluate_model(
x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb, x_test_ptb, y_test_ptb
):
model_conv1d_ptb = history_model(x_train_ptb, y_train_ptb, x_valid_ptb, y_valid_ptb)
model_conv1d_ptb.evaluate(x_test_ptb, y_test_ptb)
model_conv1d_ptb.load_weights("conv1d_ptb.h5")
model_conv1d_ptb.evaluate(x_test_ptb, y_test_ptb)
# Calculating the predictions based on the highest probability class
conv1d_pred_proba_ptb = model_conv1d_ptb.predict(x_test_ptb)
conv1d_pred_ptb = np.argmax(conv1d_pred_proba_ptb, axis=1)
print(
classification_report(
out_test_ptb,
conv1d_pred_ptb > 0.5,
target_names=[PTB_Outcome[i] for i in PTB_Outcome],
)
)
print(roc_auc_score(conv1d_pred_proba_ptb, out_test_ptb))
print(balanced_accuracy_score(conv1d_pred_proba_ptb, out_test_ptb))
print(f1_score(conv1d_pred_proba_ptb, out_test_ptb))
# Plotting the training and validatoin results
plt.figure(figsize=(25, 12))
plt.plot(
history_conv1d_ptb.epoch,
history_conv1d_ptb.history["loss"],
color="r",
label="Train loss",
)
plt.plot(
history_conv1d_ptb.epoch,
history_conv1d_ptb.history["val_loss"],
color="b",
label="Val loss",
linestyle="--",
)
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.plot(
history_conv1d_ptb.epoch,
history_conv1d_ptb.history["f1_score"],
color="g",
label="Train F1",
)
plt.plot(
history_conv1d_ptb.epoch,
history_conv1d_ptb.history["val_f1_score"],
color="c",
label="Val F1",
linestyle="--",
)
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
plt.show()
# ## Defining Conv1D Residual model for PTB
# Creating a model based on a series of Conv1D layers with 2 residual blocks that are connected to another series of full connected dense layers
def build_conv1d_res_model(input_shape=(x_train_ptb.shape[1], 1)):
model = keras.models.Sequential()
input_ = tf.keras.layers.Input(shape=(input_shape))
conv1_1 = Conv1D(64, 7, padding="same", input_shape=input_shape)(input_)
conv1_1 = BatchNormalization()(conv1_1)
conv1_1 = tf.keras.layers.ReLU()(conv1_1)
conv1_2 = Conv1D(64, 7, padding="same")(conv1_1)
conv1_2 = BatchNormalization()(conv1_2)
conv1_2 = tf.keras.layers.ReLU()(conv1_2)
conv1_3 = Conv1D(64, 7, padding="same")(conv1_2)
conv1_3 = BatchNormalization()(conv1_3)
conv1_3 = tf.keras.layers.ReLU()(conv1_3)
concat_1 = Concatenate()([conv1_1, conv1_3])
max_1 = MaxPool1D(5, padding="same")(concat_1)
conv1_4 = Conv1D(128, 7, padding="same")(max_1)
conv1_4 = BatchNormalization()(conv1_4)
conv1_4 = tf.keras.layers.ReLU()(conv1_4)
conv1_5 = Conv1D(128, 7, padding="same", input_shape=input_shape)(conv1_4)
conv1_5 = BatchNormalization()(conv1_5)
conv1_5 = tf.keras.layers.ReLU()(conv1_5)
conv1_6 = Conv1D(128, 7, padding="same", input_shape=input_shape)(conv1_5)
conv1_6 = BatchNormalization()(conv1_6)
conv1_6 = tf.keras.layers.ReLU()(conv1_6)
concat_2 = Concatenate()([conv1_4, conv1_6])
max_2 = MaxPool1D(5, padding="same")(concat_2)
flat = Flatten()(max_2)
dense_1 = Dense(512, activation="relu")(flat)
drop_1 = Dropout(0.5)(dense_1)
dense_2 = Dense(256, activation="relu")(drop_1)
drop_2 = Dropout(0.5)(dense_2)
dense_3 = Dense(128, activation="relu")(drop_2)
dense_4 = Dense(64, activation="relu")(dense_3)
dense_5 = Dense(32, activation="relu")(dense_4)
dense_6 = Dense(2, activation="softmax")(dense_5)
model = Model(inputs=input_, outputs=dense_6)
model.compile(
optimizer="adam",
loss="categorical_crossentropy",
metrics=[tfa.metrics.F1Score(2, "micro")],
)
return model
checkpoint_cb = ModelCheckpoint("conv1d_res_ptb.h5", save_best_only=True)
earlystop_cb = EarlyStopping(patience=5, restore_best_weights=True)
inp_shape = (x_train_ptb.shape[1], x_train_ptb.shape[2])
model_conv1d_res_ptb = build_conv1d_res_model(
input_shape=(x_train_ptb.shape[1], x_train_ptb.shape[2])
)
# model_conv1d_res_ptb.build(inp_shape)
history_conv1d_res_ptb = model_conv1d_res_ptb.fit(
x_train_ptb,
y_train_ptb,
epochs=10,
batch_size=32,
steps_per_epoch=1000,
# class_weight=class_weight,
validation_data=(x_valid_ptb, y_valid_ptb),
validation_steps=500,
callbacks=[checkpoint_cb, earlystop_cb],
)
# history_conv1d_res_ptb = model_conv1d_res_ptb.fit(x_train_ptb, y_train_ptb, epochs=40, batch_size=32,
# class_weight=class_weight, validation_data=(x_valid_ptb, y_valid_ptb),
# callbacks=[checkpoint_cb, earlystop_cb])
model_conv1d_res_ptb.load_weights("conv1d_res_ptb.h5")
model_conv1d_res_ptb.evaluate(x_test_ptb, y_test_ptb)
# Calculating the predictions based on the highest probability class
conv1d_res_pred_proba_ptb = model_conv1d_res_ptb.predict(x_test_ptb)
conv1d_res_pred_ptb = np.argmax(conv1d_res_pred_proba_ptb, axis=1)
print(
classification_report(
out_test_ptb,
conv1d_res_pred_ptb > 0.5,
target_names=[PTB_Outcome[i] for i in PTB_Outcome],
)
)
print(roc_auc_score(conv1d_res_pred_ptb, out_test_ptb))
print(balanced_accuracy_score(conv1d_res_pred_ptb, out_test_ptb))
print(f1_score(conv1d_res_pred_ptb, out_test_ptb))
# Plotting the training and validatoin results
plt.figure(figsize=(25, 12))
plt.plot(
history_conv1d_res_ptb.epoch,
history_conv1d_res_ptb.history["loss"],
color="r",
label="Train loss",
)
plt.plot(
history_conv1d_res_ptb.epoch,
history_conv1d_res_ptb.history["val_loss"],
color="b",
label="Val loss",
linestyle="--",
)
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.plot(
history_conv1d_res_ptb.epoch,
history_conv1d_res_ptb.history["f1_score"],
color="g",
label="Train F1",
)
plt.plot(
history_conv1d_res_ptb.epoch,
history_conv1d_res_ptb.history["val_f1_score"],
color="c",
label="Val F1",
linestyle="--",
)
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
plt.show()
| false | 0 | 13,359 | 0 | 13,359 | 13,359 |
||
69506296
|
<jupyter_start><jupyter_text>Germany Cars Dataset
### Context
AutoScout24 is one of the largest Europe's car market for new and used cars. We've collected car data from 2011 to 2021.
### Content
It shows basic fields like make, model, mileage, horse power, etc...
Data was collected and scraped automatically using the tool we've been building at [ZenRows](https://www.zenrows.com/blog/collecting-data-to-map-housing-prices?utm_source=kaggle&utm_medium=dataset&utm_campaign=cars_germany).
Kaggle dataset identifier: cars-germany
<jupyter_code>import pandas as pd
df = pd.read_csv('cars-germany/autoscout24-germany-dataset.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 46405 entries, 0 to 46404
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 mileage 46405 non-null int64
1 make 46405 non-null object
2 model 46262 non-null object
3 fuel 46405 non-null object
4 gear 46223 non-null object
5 offerType 46405 non-null object
6 price 46405 non-null int64
7 hp 46376 non-null float64
8 year 46405 non-null int64
dtypes: float64(1), int64(3), object(5)
memory usage: 3.2+ MB
<jupyter_text>Examples:
{
"mileage": 235000,
"make": "BMW",
"model": "316",
"fuel": "Diesel",
"gear": "Manual",
"offerType": "Used",
"price": 6800,
"hp": 116,
"year": 2011
}
{
"mileage": 92800,
"make": "Volkswagen",
"model": "Golf",
"fuel": "Gasoline",
"gear": "Manual",
"offerType": "Used",
"price": 6877,
"hp": 122,
"year": 2011
}
{
"mileage": 149300,
"make": "SEAT",
"model": "Exeo",
"fuel": "Gasoline",
"gear": "Manual",
"offerType": "Used",
"price": 6900,
"hp": 160,
"year": 2011
}
{
"mileage": 96200,
"make": "Renault",
"model": "Megane",
"fuel": "Gasoline",
"gear": "Manual",
"offerType": "Used",
"price": 6950,
"hp": 110,
"year": 2011
}
<jupyter_script># # Can we predict the price of a car based on its properties?
# Can we predict the price of a car based on the dataset we have through a linear regression?
# A somewhat classical example of a linear regression, I will be working on a dataset where some properties of more than 45k cars to predict their price.
# So, we should start as usual with importing the relevant libraries.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# Then, we are opening our dataset, again as usual.
df = pd.read_csv("../input/cars-germany/autoscout24-germany-dataset.csv")
pd.options.display.max_rows = 1000
df
# ## Feature Engineering
# ### Outliers
# Let's detect, if any, outliers and get rid of them.
df.corr()["price"].sort_values()
# As seen the most correlated property is 'hp' vis-à-vis the price.
# Let's plot the data, to visually see the outliers, again if any.
sns.scatterplot(x="hp", y="price", data=df)
sns.scatterplot(x="year", y="price", data=df)
# So, we do have a few outliers, as visually seen from the graphs. We will remove them from our dataset.
df[(df["price"] > 600000)]
# These three cars are outliers due to their prices.
drop_ind = df[(df["price"] > 600000)].index
df = df.drop(drop_ind, axis=0)
# Let's see the current version of our dataset.
sns.scatterplot(x="hp", y="price", data=df)
# The data is dispersed especially after the 600 hp, but the current version is better to work on.
df.info()
# We have some null values in our dataset. We need to either get rid of them, or fill them with some rational values.
df.isnull().sum()
# This is a better representation of our null data.
# Let's see if these null values are meaningful.
100 * df.isnull().sum() / len(df)
# 1)As seen from the table, there are 3 features containing null values. We are more interested in 'make', 'mileage', 'hp' and 'year'.
# 2)In order to have as many data as possible, I will try to fill these values rather than simply removing them.
#
df["model"] = df["model"].fillna("None")
df["gear"] = df["gear"].fillna("None")
100 * df.isnull().sum() / len(df)
# In order not to remove the rows where 'hp' data is missing, I will adopt a very unorthodox approach and fill these data with the average values of 'hp', which we will later on see that it is '132'.
df["hp"].mean()
df["hp"] = df["hp"].fillna(132)
100 * df.isnull().sum() / len(df)
# ## Creating the Dummy Variables
my_object_df = df.select_dtypes(include="object")
my_numeric_df = df.select_dtypes(exclude="object")
my_object_df
df_objects_dummies = pd.get_dummies(my_object_df, drop_first=True)
df_objects_dummies
# So we have created dummy variables, instead of having them as strings.
final_df = pd.concat([my_numeric_df, df_objects_dummies], axis=1)
final_df
# We are concatenating the dummy variables with the numeric columns.
final_df.info()
# Let's see the final version of our dataframe.
# ## Creating our Features (X) and Target (y)
X = final_df.drop("price", axis=1)
y = final_df["price"]
# ## Creating our Training and Test Sets
from sklearn.model_selection import train_test_split
# At this stage, we are certainly importing train_test_split module from sklearn.
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, shuffle=True)
# ## Scaling our X Features
from sklearn.preprocessing import StandardScaler
# For this we are going to need StandScaler module from sklearn library.
scaler = StandardScaler()
scaled_X_train = scaler.fit_transform(X_train)
scaled_X_test = scaler.transform(X_test)
# ## Creating our Linear Regression
from sklearn.linear_model import LinearRegression
# We are importing LinearRegression module in order to create and run our linear regression.
reg = LinearRegression()
reg.fit(X_train, y_train)
reg.score(X_train, y_train)
# Our regression score is around 0.924. Does not seem so bad I guess.
reg.fit(X_test, y_test)
reg.score(X_test, y_test)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506296.ipynb
|
cars-germany
|
ander289386
|
[{"Id": 69506296, "ScriptId": 18979330, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6778319, "CreationDate": "07/31/2021 21:15:02", "VersionNumber": 2.0, "Title": "German_Cars_Dataset_Linear_Regression", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 133.0, "LinesInsertedFromPrevious": 128.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 5.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 92761523, "KernelVersionId": 69506296, "SourceDatasetVersionId": 2318097}]
|
[{"Id": 2318097, "DatasetId": 1398432, "DatasourceVersionId": 2359558, "CreatorUserId": 289386, "LicenseName": "CC0: Public Domain", "CreationDate": "06/09/2021 16:14:27", "VersionNumber": 3.0, "Title": "Germany Cars Dataset", "Slug": "cars-germany", "Subtitle": "Dataset scraped from AutoScout24 with information about new and used cars.", "Description": "### Context\n\nAutoScout24 is one of the largest Europe's car market for new and used cars. We've collected car data from 2011 to 2021.\n\n\n### Content\n\nIt shows basic fields like make, model, mileage, horse power, etc...\n\nData was collected and scraped automatically using the tool we've been building at [ZenRows](https://www.zenrows.com/blog/collecting-data-to-map-housing-prices?utm_source=kaggle&utm_medium=dataset&utm_campaign=cars_germany).", "VersionNotes": "More data", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1398432, "CreatorUserId": 289386, "OwnerUserId": 289386.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2318097.0, "CurrentDatasourceVersionId": 2359558.0, "ForumId": 1417705, "Type": 2, "CreationDate": "06/09/2021 10:47:15", "LastActivityDate": "06/09/2021", "TotalViews": 24206, "TotalDownloads": 3296, "TotalVotes": 40, "TotalKernels": 17}]
|
[{"Id": 289386, "UserName": "ander289386", "DisplayName": "Ander", "RegisterDate": "02/03/2015", "PerformanceTier": 0}]
|
# # Can we predict the price of a car based on its properties?
# Can we predict the price of a car based on the dataset we have through a linear regression?
# A somewhat classical example of a linear regression, I will be working on a dataset where some properties of more than 45k cars to predict their price.
# So, we should start as usual with importing the relevant libraries.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# Then, we are opening our dataset, again as usual.
df = pd.read_csv("../input/cars-germany/autoscout24-germany-dataset.csv")
pd.options.display.max_rows = 1000
df
# ## Feature Engineering
# ### Outliers
# Let's detect, if any, outliers and get rid of them.
df.corr()["price"].sort_values()
# As seen the most correlated property is 'hp' vis-à-vis the price.
# Let's plot the data, to visually see the outliers, again if any.
sns.scatterplot(x="hp", y="price", data=df)
sns.scatterplot(x="year", y="price", data=df)
# So, we do have a few outliers, as visually seen from the graphs. We will remove them from our dataset.
df[(df["price"] > 600000)]
# These three cars are outliers due to their prices.
drop_ind = df[(df["price"] > 600000)].index
df = df.drop(drop_ind, axis=0)
# Let's see the current version of our dataset.
sns.scatterplot(x="hp", y="price", data=df)
# The data is dispersed especially after the 600 hp, but the current version is better to work on.
df.info()
# We have some null values in our dataset. We need to either get rid of them, or fill them with some rational values.
df.isnull().sum()
# This is a better representation of our null data.
# Let's see if these null values are meaningful.
100 * df.isnull().sum() / len(df)
# 1)As seen from the table, there are 3 features containing null values. We are more interested in 'make', 'mileage', 'hp' and 'year'.
# 2)In order to have as many data as possible, I will try to fill these values rather than simply removing them.
#
df["model"] = df["model"].fillna("None")
df["gear"] = df["gear"].fillna("None")
100 * df.isnull().sum() / len(df)
# In order not to remove the rows where 'hp' data is missing, I will adopt a very unorthodox approach and fill these data with the average values of 'hp', which we will later on see that it is '132'.
df["hp"].mean()
df["hp"] = df["hp"].fillna(132)
100 * df.isnull().sum() / len(df)
# ## Creating the Dummy Variables
my_object_df = df.select_dtypes(include="object")
my_numeric_df = df.select_dtypes(exclude="object")
my_object_df
df_objects_dummies = pd.get_dummies(my_object_df, drop_first=True)
df_objects_dummies
# So we have created dummy variables, instead of having them as strings.
final_df = pd.concat([my_numeric_df, df_objects_dummies], axis=1)
final_df
# We are concatenating the dummy variables with the numeric columns.
final_df.info()
# Let's see the final version of our dataframe.
# ## Creating our Features (X) and Target (y)
X = final_df.drop("price", axis=1)
y = final_df["price"]
# ## Creating our Training and Test Sets
from sklearn.model_selection import train_test_split
# At this stage, we are certainly importing train_test_split module from sklearn.
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, shuffle=True)
# ## Scaling our X Features
from sklearn.preprocessing import StandardScaler
# For this we are going to need StandScaler module from sklearn library.
scaler = StandardScaler()
scaled_X_train = scaler.fit_transform(X_train)
scaled_X_test = scaler.transform(X_test)
# ## Creating our Linear Regression
from sklearn.linear_model import LinearRegression
# We are importing LinearRegression module in order to create and run our linear regression.
reg = LinearRegression()
reg.fit(X_train, y_train)
reg.score(X_train, y_train)
# Our regression score is around 0.924. Does not seem so bad I guess.
reg.fit(X_test, y_test)
reg.score(X_test, y_test)
|
[{"cars-germany/autoscout24-germany-dataset.csv": {"column_names": "[\"mileage\", \"make\", \"model\", \"fuel\", \"gear\", \"offerType\", \"price\", \"hp\", \"year\"]", "column_data_types": "{\"mileage\": \"int64\", \"make\": \"object\", \"model\": \"object\", \"fuel\": \"object\", \"gear\": \"object\", \"offerType\": \"object\", \"price\": \"int64\", \"hp\": \"float64\", \"year\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 46405 entries, 0 to 46404\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 mileage 46405 non-null int64 \n 1 make 46405 non-null object \n 2 model 46262 non-null object \n 3 fuel 46405 non-null object \n 4 gear 46223 non-null object \n 5 offerType 46405 non-null object \n 6 price 46405 non-null int64 \n 7 hp 46376 non-null float64\n 8 year 46405 non-null int64 \ndtypes: float64(1), int64(3), object(5)\nmemory usage: 3.2+ MB\n", "summary": "{\"mileage\": {\"count\": 46405.0, \"mean\": 71177.86410947096, \"std\": 62625.30845633686, \"min\": 0.0, \"25%\": 19800.0, \"50%\": 60000.0, \"75%\": 105000.0, \"max\": 1111111.0}, \"price\": {\"count\": 46405.0, \"mean\": 16572.33722659196, \"std\": 19304.6959239994, \"min\": 1100.0, \"25%\": 7490.0, \"50%\": 10999.0, \"75%\": 19490.0, \"max\": 1199900.0}, \"hp\": {\"count\": 46376.0, \"mean\": 132.99098671726756, \"std\": 75.44928433298794, \"min\": 1.0, \"25%\": 86.0, \"50%\": 116.0, \"75%\": 150.0, \"max\": 850.0}, \"year\": {\"count\": 46405.0, \"mean\": 2016.0129511906046, \"std\": 3.15521392122357, \"min\": 2011.0, \"25%\": 2013.0, \"50%\": 2016.0, \"75%\": 2019.0, \"max\": 2021.0}}", "examples": "{\"mileage\":{\"0\":235000,\"1\":92800,\"2\":149300,\"3\":96200},\"make\":{\"0\":\"BMW\",\"1\":\"Volkswagen\",\"2\":\"SEAT\",\"3\":\"Renault\"},\"model\":{\"0\":\"316\",\"1\":\"Golf\",\"2\":\"Exeo\",\"3\":\"Megane\"},\"fuel\":{\"0\":\"Diesel\",\"1\":\"Gasoline\",\"2\":\"Gasoline\",\"3\":\"Gasoline\"},\"gear\":{\"0\":\"Manual\",\"1\":\"Manual\",\"2\":\"Manual\",\"3\":\"Manual\"},\"offerType\":{\"0\":\"Used\",\"1\":\"Used\",\"2\":\"Used\",\"3\":\"Used\"},\"price\":{\"0\":6800,\"1\":6877,\"2\":6900,\"3\":6950},\"hp\":{\"0\":116.0,\"1\":122.0,\"2\":160.0,\"3\":110.0},\"year\":{\"0\":2011,\"1\":2011,\"2\":2011,\"3\":2011}}"}}]
| true | 1 |
<start_data_description><data_path>cars-germany/autoscout24-germany-dataset.csv:
<column_names>
['mileage', 'make', 'model', 'fuel', 'gear', 'offerType', 'price', 'hp', 'year']
<column_types>
{'mileage': 'int64', 'make': 'object', 'model': 'object', 'fuel': 'object', 'gear': 'object', 'offerType': 'object', 'price': 'int64', 'hp': 'float64', 'year': 'int64'}
<dataframe_Summary>
{'mileage': {'count': 46405.0, 'mean': 71177.86410947096, 'std': 62625.30845633686, 'min': 0.0, '25%': 19800.0, '50%': 60000.0, '75%': 105000.0, 'max': 1111111.0}, 'price': {'count': 46405.0, 'mean': 16572.33722659196, 'std': 19304.6959239994, 'min': 1100.0, '25%': 7490.0, '50%': 10999.0, '75%': 19490.0, 'max': 1199900.0}, 'hp': {'count': 46376.0, 'mean': 132.99098671726756, 'std': 75.44928433298794, 'min': 1.0, '25%': 86.0, '50%': 116.0, '75%': 150.0, 'max': 850.0}, 'year': {'count': 46405.0, 'mean': 2016.0129511906046, 'std': 3.15521392122357, 'min': 2011.0, '25%': 2013.0, '50%': 2016.0, '75%': 2019.0, 'max': 2021.0}}
<dataframe_info>
RangeIndex: 46405 entries, 0 to 46404
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 mileage 46405 non-null int64
1 make 46405 non-null object
2 model 46262 non-null object
3 fuel 46405 non-null object
4 gear 46223 non-null object
5 offerType 46405 non-null object
6 price 46405 non-null int64
7 hp 46376 non-null float64
8 year 46405 non-null int64
dtypes: float64(1), int64(3), object(5)
memory usage: 3.2+ MB
<some_examples>
{'mileage': {'0': 235000, '1': 92800, '2': 149300, '3': 96200}, 'make': {'0': 'BMW', '1': 'Volkswagen', '2': 'SEAT', '3': 'Renault'}, 'model': {'0': '316', '1': 'Golf', '2': 'Exeo', '3': 'Megane'}, 'fuel': {'0': 'Diesel', '1': 'Gasoline', '2': 'Gasoline', '3': 'Gasoline'}, 'gear': {'0': 'Manual', '1': 'Manual', '2': 'Manual', '3': 'Manual'}, 'offerType': {'0': 'Used', '1': 'Used', '2': 'Used', '3': 'Used'}, 'price': {'0': 6800, '1': 6877, '2': 6900, '3': 6950}, 'hp': {'0': 116.0, '1': 122.0, '2': 160.0, '3': 110.0}, 'year': {'0': 2011, '1': 2011, '2': 2011, '3': 2011}}
<end_description>
| 1,205 | 2 | 1,988 | 1,205 |
69506612
|
<jupyter_start><jupyter_text>Face Mask Detection
### Context
Having seen multiple datasets related to face mask detection on Kaggle, one dataset which stood out contained 3 classes (with mask, without a mask, and wearing mask incorrectly), unfortunately, the dataset was highly imbalanced and uncleaned. So to improve this dataset, images had to be augmented in such a way that each class has an equal distribution of images and removing noisy images which could be considered as outliers. Thus this dataset that I've created is a combination of an existing dataset that has been cleaned and equally distributed across each class.
### Content
The dataset contains 3 folders labeled as to which class they belong to. the 3 classes are "with_mask", "withou_mask", and "mask_weared_incorrect". Each folder holds 2994 images of people that belong to such a labeled class.
Kaggle dataset identifier: face-mask-detection
<jupyter_script>import numpy as np
import tensorflow as tf
from tensorflow import keras
# First we will split images to train/test data. And we don't want to apply augmentation to test data. Because of that we will manually split data and will give to ImageDataGenerator
import splitfolders
splitfolders.ratio(
"../input/face-mask-detection/Dataset", output="./", seed=1337, ratio=(0.9, 0.1)
)
train_data_generator = keras.preprocessing.image.ImageDataGenerator(
horizontal_flip=True,
vertical_flip=True,
zoom_range=0.1,
shear_range=0.1,
width_shift_range=0.2,
height_shift_range=0.2,
rotation_range=90,
)
test_data_generator = keras.preprocessing.image.ImageDataGenerator()
train_data = train_data_generator.flow_from_directory(
"./train", target_size=(128, 128), batch_size=1, shuffle=True
)
test_data = test_data_generator.flow_from_directory(
"./val", target_size=(128, 128), batch_size=1, shuffle=True
)
labels = train_data.class_indices
labels
# ImageDataGenerator does not really assign values to an array, it just hold pointers. Because of that every learning step CPU perform reading operations. This very slows learning speed. We will store data in numpy array type.
def get_array_from_datagen(train_generator):
x = []
y = []
train_generator.reset()
for i in range(train_generator.__len__()):
a, b = train_generator.next()
x.append(a)
y.append(b)
x = np.array(x, dtype=np.float32)
y = np.array(y, dtype=np.float32)
print(x.shape)
print(y.shape)
return x, y
X_train, y_train = get_array_from_datagen(train_data)
X_test, y_test = get_array_from_datagen(test_data)
X_train = X_train.reshape(-1, 128, 128, 3)
X_test = X_test.reshape(-1, 128, 128, 3)
y_train = y_train.reshape(-1, 3)
y_test = y_test.reshape(-1, 3)
import gc
from keras import Sequential
from keras.layers import (
Conv2D,
MaxPooling2D,
Flatten,
Dense,
BatchNormalization,
Dropout,
)
from keras.optimizers import SGD, RMSprop, Adam, Adadelta, Adagrad, Adamax, Nadam, Ftrl
input_shape = (128, 128, 3)
class_num = len(labels)
def cnn1():
return Sequential(
[
Conv2D(
64,
kernel_size=(3, 3),
activation="relu",
padding="same",
input_shape=input_shape,
),
BatchNormalization(),
MaxPooling2D(pool_size=(2, 2), strides=2),
Dropout(0.3),
Conv2D(64, kernel_size=(5, 5), activation="relu"),
MaxPooling2D(pool_size=(2, 2), strides=2),
Dropout(0.3),
Flatten(),
Dense(512, activation="relu"),
Dropout(0.3),
Dense(64, activation="relu"),
Dense(class_num, activation="softmax"),
]
)
def cnn2():
return Sequential(
[
Conv2D(64, kernel_size=(3, 3), activation="relu", input_shape=input_shape),
BatchNormalization(),
MaxPooling2D(pool_size=(2, 2), strides=2),
Dropout(0.3),
Conv2D(64, kernel_size=(3, 3), activation="relu"),
Dropout(0.3),
MaxPooling2D(pool_size=(2, 2), strides=2),
Conv2D(64, kernel_size=(3, 3), activation="relu"),
Dropout(0.3),
MaxPooling2D(pool_size=(2, 2), strides=2),
Flatten(),
Dense(128, activation="relu"),
Dense(class_num, activation="softmax"),
]
)
def cnn3():
return Sequential(
[
Conv2D(
128,
kernel_size=(3, 3),
activation="relu",
padding="same",
input_shape=input_shape,
),
BatchNormalization(),
Conv2D(128, kernel_size=(3, 3), activation="relu", padding="same"),
Dropout(0.3),
MaxPooling2D(pool_size=(2, 2), strides=2),
Conv2D(128, kernel_size=(3, 3), activation="relu"),
Dropout(0.3),
Conv2D(128, kernel_size=(3, 3), activation="relu"),
Dropout(0.3),
MaxPooling2D(pool_size=(2, 2), strides=2),
Flatten(),
Dense(128, activation="relu"),
Dense(class_num, activation="softmax"),
]
)
learning_rate_reduction = keras.callbacks.ReduceLROnPlateau(
monitor="val_accuracy", factor=0.5, patience=3, verbose=0, min_lr=0.00001
)
early_stopping = keras.callbacks.EarlyStopping(patience=5, verbose=1)
import matplotlib.pyplot as plt
def plot_history(history):
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# summarize history for loss
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
model = cnn1()
model.compile(optimizer="Adam", loss=["categorical_crossentropy"], metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
epochs=25,
validation_data=(X_test, y_test),
verbose=2,
callbacks=[learning_rate_reduction, early_stopping],
)
gc.collect()
plot_history(history)
model = cnn2()
model.compile(optimizer="Adam", loss=["categorical_crossentropy"], metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
epochs=25,
validation_data=(X_test, y_test),
verbose=2,
callbacks=[learning_rate_reduction, early_stopping],
)
gc.collect()
plot_history(history)
model = cnn3()
model.compile(optimizer="Adam", loss=["categorical_crossentropy"], metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
epochs=25,
validation_data=(X_test, y_test),
verbose=2,
callbacks=[learning_rate_reduction, early_stopping],
)
gc.collect()
plot_history(history)
base_model = tf.keras.applications.EfficientNetB0(include_top=False)
model = keras.Sequential(
[
base_model,
keras.layers.GlobalAveragePooling2D(),
keras.layers.Dropout(0.5),
keras.layers.Dense(3, activation="softmax"),
]
)
model.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
validation_data=(X_test, y_test),
epochs=25,
callbacks=[learning_rate_reduction, early_stopping],
)
plot_history(history)
base_model = tf.keras.applications.ResNet50(include_top=False)
model = keras.Sequential(
[
base_model,
keras.layers.GlobalAveragePooling2D(),
keras.layers.Dropout(0.5),
keras.layers.Dense(3, activation="softmax"),
]
)
model.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
validation_data=(X_test, y_test),
epochs=25,
callbacks=[learning_rate_reduction, early_stopping],
)
plot_history(history)
# Accuricies :
# CNN -> 0.8933
# ResNet50 -> 0.9833
# EfficientNet -> 0.9956
# Best Model is EfficientNet, we will check recall, precision and f1-score values
base_model = tf.keras.applications.EfficientNetB0(include_top=False)
model = keras.Sequential(
[
base_model,
keras.layers.GlobalAveragePooling2D(),
keras.layers.Dropout(0.5),
keras.layers.Dense(3, activation="softmax"),
]
)
model.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.fit(
X_train,
y_train,
validation_data=(X_test, y_test),
epochs=25,
callbacks=[learning_rate_reduction, early_stopping],
verbose=0,
)
from sklearn.metrics import classification_report
print(
classification_report(y_test.argmax(axis=1), model.predict(X_test).argmax(axis=1))
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506612.ipynb
|
face-mask-detection
|
vijaykumar1799
|
[{"Id": 69506612, "ScriptId": 18976993, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6399716, "CreationDate": "07/31/2021 21:22:29", "VersionNumber": 1.0, "Title": "Mask Detection - CNN, ResNet50, EfficientNet", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 231.0, "LinesInsertedFromPrevious": 231.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 92762457, "KernelVersionId": 69506612, "SourceDatasetVersionId": 2249954}]
|
[{"Id": 2249954, "DatasetId": 1353321, "DatasourceVersionId": 2290881, "CreatorUserId": 3174210, "LicenseName": "CC0: Public Domain", "CreationDate": "05/19/2021 15:24:14", "VersionNumber": 1.0, "Title": "Face Mask Detection", "Slug": "face-mask-detection", "Subtitle": "Building a face mask classifier", "Description": "### Context\n\nHaving seen multiple datasets related to face mask detection on Kaggle, one dataset which stood out contained 3 classes (with mask, without a mask, and wearing mask incorrectly), unfortunately, the dataset was highly imbalanced and uncleaned. So to improve this dataset, images had to be augmented in such a way that each class has an equal distribution of images and removing noisy images which could be considered as outliers. Thus this dataset that I've created is a combination of an existing dataset that has been cleaned and equally distributed across each class.\n\n\n### Content\n\nThe dataset contains 3 folders labeled as to which class they belong to. the 3 classes are \"with_mask\", \"withou_mask\", and \"mask_weared_incorrect\". Each folder holds 2994 images of people that belong to such a labeled class.\n\n### Acknowledgements\n\nI would like to acknowledge the usage of those 2 datasets:\n- https://www.kaggle.com/ashishjangra27/face-mask-12k-images-dataset\n- https://www.kaggle.com/andrewmvd/face-mask-detection\n\nUsing the above-mentioned datasets, data was manually extracted from both datasets in such a way that the new dataset is equally distributed and contains good quality samples without noise.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1353321, "CreatorUserId": 3174210, "OwnerUserId": 3174210.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2249954.0, "CurrentDatasourceVersionId": 2290881.0, "ForumId": 1372371, "Type": 2, "CreationDate": "05/19/2021 15:24:14", "LastActivityDate": "05/19/2021", "TotalViews": 34574, "TotalDownloads": 5120, "TotalVotes": 70, "TotalKernels": 21}]
|
[{"Id": 3174210, "UserName": "vijaykumar1799", "DisplayName": "vijay kumar", "RegisterDate": "05/03/2019", "PerformanceTier": 1}]
|
import numpy as np
import tensorflow as tf
from tensorflow import keras
# First we will split images to train/test data. And we don't want to apply augmentation to test data. Because of that we will manually split data and will give to ImageDataGenerator
import splitfolders
splitfolders.ratio(
"../input/face-mask-detection/Dataset", output="./", seed=1337, ratio=(0.9, 0.1)
)
train_data_generator = keras.preprocessing.image.ImageDataGenerator(
horizontal_flip=True,
vertical_flip=True,
zoom_range=0.1,
shear_range=0.1,
width_shift_range=0.2,
height_shift_range=0.2,
rotation_range=90,
)
test_data_generator = keras.preprocessing.image.ImageDataGenerator()
train_data = train_data_generator.flow_from_directory(
"./train", target_size=(128, 128), batch_size=1, shuffle=True
)
test_data = test_data_generator.flow_from_directory(
"./val", target_size=(128, 128), batch_size=1, shuffle=True
)
labels = train_data.class_indices
labels
# ImageDataGenerator does not really assign values to an array, it just hold pointers. Because of that every learning step CPU perform reading operations. This very slows learning speed. We will store data in numpy array type.
def get_array_from_datagen(train_generator):
x = []
y = []
train_generator.reset()
for i in range(train_generator.__len__()):
a, b = train_generator.next()
x.append(a)
y.append(b)
x = np.array(x, dtype=np.float32)
y = np.array(y, dtype=np.float32)
print(x.shape)
print(y.shape)
return x, y
X_train, y_train = get_array_from_datagen(train_data)
X_test, y_test = get_array_from_datagen(test_data)
X_train = X_train.reshape(-1, 128, 128, 3)
X_test = X_test.reshape(-1, 128, 128, 3)
y_train = y_train.reshape(-1, 3)
y_test = y_test.reshape(-1, 3)
import gc
from keras import Sequential
from keras.layers import (
Conv2D,
MaxPooling2D,
Flatten,
Dense,
BatchNormalization,
Dropout,
)
from keras.optimizers import SGD, RMSprop, Adam, Adadelta, Adagrad, Adamax, Nadam, Ftrl
input_shape = (128, 128, 3)
class_num = len(labels)
def cnn1():
return Sequential(
[
Conv2D(
64,
kernel_size=(3, 3),
activation="relu",
padding="same",
input_shape=input_shape,
),
BatchNormalization(),
MaxPooling2D(pool_size=(2, 2), strides=2),
Dropout(0.3),
Conv2D(64, kernel_size=(5, 5), activation="relu"),
MaxPooling2D(pool_size=(2, 2), strides=2),
Dropout(0.3),
Flatten(),
Dense(512, activation="relu"),
Dropout(0.3),
Dense(64, activation="relu"),
Dense(class_num, activation="softmax"),
]
)
def cnn2():
return Sequential(
[
Conv2D(64, kernel_size=(3, 3), activation="relu", input_shape=input_shape),
BatchNormalization(),
MaxPooling2D(pool_size=(2, 2), strides=2),
Dropout(0.3),
Conv2D(64, kernel_size=(3, 3), activation="relu"),
Dropout(0.3),
MaxPooling2D(pool_size=(2, 2), strides=2),
Conv2D(64, kernel_size=(3, 3), activation="relu"),
Dropout(0.3),
MaxPooling2D(pool_size=(2, 2), strides=2),
Flatten(),
Dense(128, activation="relu"),
Dense(class_num, activation="softmax"),
]
)
def cnn3():
return Sequential(
[
Conv2D(
128,
kernel_size=(3, 3),
activation="relu",
padding="same",
input_shape=input_shape,
),
BatchNormalization(),
Conv2D(128, kernel_size=(3, 3), activation="relu", padding="same"),
Dropout(0.3),
MaxPooling2D(pool_size=(2, 2), strides=2),
Conv2D(128, kernel_size=(3, 3), activation="relu"),
Dropout(0.3),
Conv2D(128, kernel_size=(3, 3), activation="relu"),
Dropout(0.3),
MaxPooling2D(pool_size=(2, 2), strides=2),
Flatten(),
Dense(128, activation="relu"),
Dense(class_num, activation="softmax"),
]
)
learning_rate_reduction = keras.callbacks.ReduceLROnPlateau(
monitor="val_accuracy", factor=0.5, patience=3, verbose=0, min_lr=0.00001
)
early_stopping = keras.callbacks.EarlyStopping(patience=5, verbose=1)
import matplotlib.pyplot as plt
def plot_history(history):
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# summarize history for loss
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
model = cnn1()
model.compile(optimizer="Adam", loss=["categorical_crossentropy"], metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
epochs=25,
validation_data=(X_test, y_test),
verbose=2,
callbacks=[learning_rate_reduction, early_stopping],
)
gc.collect()
plot_history(history)
model = cnn2()
model.compile(optimizer="Adam", loss=["categorical_crossentropy"], metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
epochs=25,
validation_data=(X_test, y_test),
verbose=2,
callbacks=[learning_rate_reduction, early_stopping],
)
gc.collect()
plot_history(history)
model = cnn3()
model.compile(optimizer="Adam", loss=["categorical_crossentropy"], metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
epochs=25,
validation_data=(X_test, y_test),
verbose=2,
callbacks=[learning_rate_reduction, early_stopping],
)
gc.collect()
plot_history(history)
base_model = tf.keras.applications.EfficientNetB0(include_top=False)
model = keras.Sequential(
[
base_model,
keras.layers.GlobalAveragePooling2D(),
keras.layers.Dropout(0.5),
keras.layers.Dense(3, activation="softmax"),
]
)
model.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
validation_data=(X_test, y_test),
epochs=25,
callbacks=[learning_rate_reduction, early_stopping],
)
plot_history(history)
base_model = tf.keras.applications.ResNet50(include_top=False)
model = keras.Sequential(
[
base_model,
keras.layers.GlobalAveragePooling2D(),
keras.layers.Dropout(0.5),
keras.layers.Dense(3, activation="softmax"),
]
)
model.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
history = model.fit(
X_train,
y_train,
validation_data=(X_test, y_test),
epochs=25,
callbacks=[learning_rate_reduction, early_stopping],
)
plot_history(history)
# Accuricies :
# CNN -> 0.8933
# ResNet50 -> 0.9833
# EfficientNet -> 0.9956
# Best Model is EfficientNet, we will check recall, precision and f1-score values
base_model = tf.keras.applications.EfficientNetB0(include_top=False)
model = keras.Sequential(
[
base_model,
keras.layers.GlobalAveragePooling2D(),
keras.layers.Dropout(0.5),
keras.layers.Dense(3, activation="softmax"),
]
)
model.compile(optimizer="Adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.fit(
X_train,
y_train,
validation_data=(X_test, y_test),
epochs=25,
callbacks=[learning_rate_reduction, early_stopping],
verbose=0,
)
from sklearn.metrics import classification_report
print(
classification_report(y_test.argmax(axis=1), model.predict(X_test).argmax(axis=1))
)
| false | 0 | 2,384 | 1 | 2,589 | 2,384 |
||
69506894
|
# # Operators - Τελεστές
#
print(3 + 2)
print(3 - 2)
print(3 * 2)
print(3 / 2)
print(4 % 2)
print(3 > 2)
print(3 < 2)
print(3 == 2)
print(3 >= 2)
print(3 <= 2)
print(3 != 2)
num = 1 + 2.1
print(num)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506894.ipynb
| null | null |
[{"Id": 69506894, "ScriptId": 17898323, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7721280, "CreationDate": "07/31/2021 21:29:24", "VersionNumber": 1.0, "Title": "\u039c\u03ac\u03b8\u03b7\u03bc\u03b1 1", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 55.0, "LinesInsertedFromPrevious": 55.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Operators - Τελεστές
#
print(3 + 2)
print(3 - 2)
print(3 * 2)
print(3 / 2)
print(4 % 2)
print(3 > 2)
print(3 < 2)
print(3 == 2)
print(3 >= 2)
print(3 <= 2)
print(3 != 2)
num = 1 + 2.1
print(num)
| false | 0 | 119 | 0 | 119 | 119 |
||
69506263
|
import os
import gc
import numpy as np
import pandas as pd
from scipy.stats import kurtosis
from sklearn.metrics import roc_auc_score
from sklearn.preprocessing import MinMaxScaler
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LogisticRegression
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
from sklearn.model_selection import train_test_split, cross_val_score, StratifiedKFold
import xgboost as xgb
from xgboost import XGBClassifier
warnings.simplefilter(action="ignore", category=FutureWarning)
DATA_DIRECTORY = "../input/home-credit-default-risk"
df_train = pd.read_csv(os.path.join(DATA_DIRECTORY, "application_train.csv"))
df_test = pd.read_csv(os.path.join(DATA_DIRECTORY, "application_test.csv"))
df = df_train.append(df_test)
del df_train, df_test
gc.collect()
df = df[df["AMT_INCOME_TOTAL"] < 20000000]
df = df[df["CODE_GENDER"] != "XNA"]
df["DAYS_EMPLOYED"].replace(365243, np.nan, inplace=True)
df["DAYS_LAST_PHONE_CHANGE"].replace(0, np.nan, inplace=True)
def get_age_group(days_birth):
age_years = -days_birth / 365
if age_years < 27:
return 1
elif age_years < 40:
return 2
elif age_years < 50:
return 3
elif age_years < 65:
return 4
elif age_years < 99:
return 5
else:
return 0
docs = [f for f in df.columns if "FLAG_DOC" in f]
df["DOCUMENT_COUNT"] = df[docs].sum(axis=1)
df["NEW_DOC_KURT"] = df[docs].kurtosis(axis=1)
df["AGE_RANGE"] = df["DAYS_BIRTH"].apply(lambda x: get_age_group(x))
df["EXT_SOURCES_PROD"] = df["EXT_SOURCE_1"] * df["EXT_SOURCE_2"] * df["EXT_SOURCE_3"]
df["EXT_SOURCES_WEIGHTED"] = (
df.EXT_SOURCE_1 * 2 + df.EXT_SOURCE_2 * 1 + df.EXT_SOURCE_3 * 3
)
np.warnings.filterwarnings("ignore", r"All-NaN (slice|axis) encountered")
for function_name in ["min", "max", "mean", "nanmedian", "var"]:
feature_name = "EXT_SOURCES_{}".format(function_name.upper())
df[feature_name] = eval("np.{}".format(function_name))(
df[["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3"]], axis=1
)
df["CREDIT_TO_ANNUITY_RATIO"] = df["AMT_CREDIT"] / df["AMT_ANNUITY"]
df["CREDIT_TO_GOODS_RATIO"] = df["AMT_CREDIT"] / df["AMT_GOODS_PRICE"]
df["ANNUITY_TO_INCOME_RATIO"] = df["AMT_ANNUITY"] / df["AMT_INCOME_TOTAL"]
df["CREDIT_TO_INCOME_RATIO"] = df["AMT_CREDIT"] / df["AMT_INCOME_TOTAL"]
df["INCOME_TO_EMPLOYED_RATIO"] = df["AMT_INCOME_TOTAL"] / df["DAYS_EMPLOYED"]
df["INCOME_TO_BIRTH_RATIO"] = df["AMT_INCOME_TOTAL"] / df["DAYS_BIRTH"]
df["EMPLOYED_TO_BIRTH_RATIO"] = df["DAYS_EMPLOYED"] / df["DAYS_BIRTH"]
df["ID_TO_BIRTH_RATIO"] = df["DAYS_ID_PUBLISH"] / df["DAYS_BIRTH"]
df["CAR_TO_BIRTH_RATIO"] = df["OWN_CAR_AGE"] / df["DAYS_BIRTH"]
df["CAR_TO_EMPLOYED_RATIO"] = df["OWN_CAR_AGE"] / df["DAYS_EMPLOYED"]
df["PHONE_TO_BIRTH_RATIO"] = df["DAYS_LAST_PHONE_CHANGE"] / df["DAYS_BIRTH"]
def do_mean(df, group_cols, counted, agg_name):
gp = (
df[group_cols + [counted]]
.groupby(group_cols)[counted]
.mean()
.reset_index()
.rename(columns={counted: agg_name})
)
df = df.merge(gp, on=group_cols, how="left")
del gp
gc.collect()
return df
def do_median(df, group_cols, counted, agg_name):
gp = (
df[group_cols + [counted]]
.groupby(group_cols)[counted]
.median()
.reset_index()
.rename(columns={counted: agg_name})
)
df = df.merge(gp, on=group_cols, how="left")
del gp
gc.collect()
return df
def do_std(df, group_cols, counted, agg_name):
gp = (
df[group_cols + [counted]]
.groupby(group_cols)[counted]
.std()
.reset_index()
.rename(columns={counted: agg_name})
)
df = df.merge(gp, on=group_cols, how="left")
del gp
gc.collect()
return df
def do_sum(df, group_cols, counted, agg_name):
gp = (
df[group_cols + [counted]]
.groupby(group_cols)[counted]
.sum()
.reset_index()
.rename(columns={counted: agg_name})
)
df = df.merge(gp, on=group_cols, how="left")
del gp
gc.collect()
return df
group = [
"ORGANIZATION_TYPE",
"NAME_EDUCATION_TYPE",
"OCCUPATION_TYPE",
"AGE_RANGE",
"CODE_GENDER",
]
df = do_median(df, group, "EXT_SOURCES_MEAN", "GROUP_EXT_SOURCES_MEDIAN")
df = do_std(df, group, "EXT_SOURCES_MEAN", "GROUP_EXT_SOURCES_STD")
df = do_mean(df, group, "AMT_INCOME_TOTAL", "GROUP_INCOME_MEAN")
df = do_std(df, group, "AMT_INCOME_TOTAL", "GROUP_INCOME_STD")
df = do_mean(df, group, "CREDIT_TO_ANNUITY_RATIO", "GROUP_CREDIT_TO_ANNUITY_MEAN")
df = do_std(df, group, "CREDIT_TO_ANNUITY_RATIO", "GROUP_CREDIT_TO_ANNUITY_STD")
df = do_mean(df, group, "AMT_CREDIT", "GROUP_CREDIT_MEAN")
df = do_mean(df, group, "AMT_ANNUITY", "GROUP_ANNUITY_MEAN")
df = do_std(df, group, "AMT_ANNUITY", "GROUP_ANNUITY_STD")
def label_encoder(df, categorical_columns=None):
if not categorical_columns:
categorical_columns = [col for col in df.columns if df[col].dtype == "object"]
for col in categorical_columns:
df[col], uniques = pd.factorize(df[col])
return df, categorical_columns
def drop_application_columns(df):
drop_list = [
"CNT_CHILDREN",
"CNT_FAM_MEMBERS",
"HOUR_APPR_PROCESS_START",
"FLAG_EMP_PHONE",
"FLAG_MOBIL",
"FLAG_CONT_MOBILE",
"FLAG_EMAIL",
"FLAG_PHONE",
"FLAG_OWN_REALTY",
"REG_REGION_NOT_LIVE_REGION",
"REG_REGION_NOT_WORK_REGION",
"REG_CITY_NOT_WORK_CITY",
"OBS_30_CNT_SOCIAL_CIRCLE",
"OBS_60_CNT_SOCIAL_CIRCLE",
"AMT_REQ_CREDIT_BUREAU_DAY",
"AMT_REQ_CREDIT_BUREAU_MON",
"AMT_REQ_CREDIT_BUREAU_YEAR",
"COMMONAREA_MODE",
"NONLIVINGAREA_MODE",
"ELEVATORS_MODE",
"NONLIVINGAREA_AVG",
"FLOORSMIN_MEDI",
"LANDAREA_MODE",
"NONLIVINGAREA_MEDI",
"LIVINGAPARTMENTS_MODE",
"FLOORSMIN_AVG",
"LANDAREA_AVG",
"FLOORSMIN_MODE",
"LANDAREA_MEDI",
"COMMONAREA_MEDI",
"YEARS_BUILD_AVG",
"COMMONAREA_AVG",
"BASEMENTAREA_AVG",
"BASEMENTAREA_MODE",
"NONLIVINGAPARTMENTS_MEDI",
"BASEMENTAREA_MEDI",
"LIVINGAPARTMENTS_AVG",
"ELEVATORS_AVG",
"YEARS_BUILD_MEDI",
"ENTRANCES_MODE",
"NONLIVINGAPARTMENTS_MODE",
"LIVINGAREA_MODE",
"LIVINGAPARTMENTS_MEDI",
"YEARS_BUILD_MODE",
"YEARS_BEGINEXPLUATATION_AVG",
"ELEVATORS_MEDI",
"LIVINGAREA_MEDI",
"YEARS_BEGINEXPLUATATION_MODE",
"NONLIVINGAPARTMENTS_AVG",
"HOUSETYPE_MODE",
"FONDKAPREMONT_MODE",
"EMERGENCYSTATE_MODE",
]
for doc_num in [2, 4, 5, 6, 7, 9, 10, 11, 12, 13, 14, 15, 16, 17, 19, 20, 21]:
drop_list.append("FLAG_DOCUMENT_{}".format(doc_num))
df.drop(drop_list, axis=1, inplace=True)
return df
df, le_encoded_cols = label_encoder(df, None)
df = drop_application_columns(df)
df = pd.get_dummies(df)
bureau = pd.read_csv(os.path.join(DATA_DIRECTORY, "bureau.csv"))
bureau["CREDIT_DURATION"] = -bureau["DAYS_CREDIT"] + bureau["DAYS_CREDIT_ENDDATE"]
bureau["ENDDATE_DIF"] = bureau["DAYS_CREDIT_ENDDATE"] - bureau["DAYS_ENDDATE_FACT"]
bureau["DEBT_PERCENTAGE"] = bureau["AMT_CREDIT_SUM"] / bureau["AMT_CREDIT_SUM_DEBT"]
bureau["DEBT_CREDIT_DIFF"] = bureau["AMT_CREDIT_SUM"] - bureau["AMT_CREDIT_SUM_DEBT"]
bureau["CREDIT_TO_ANNUITY_RATIO"] = bureau["AMT_CREDIT_SUM"] / bureau["AMT_ANNUITY"]
def one_hot_encoder(df, categorical_columns=None, nan_as_category=True):
original_columns = list(df.columns)
if not categorical_columns:
categorical_columns = [col for col in df.columns if df[col].dtype == "object"]
df = pd.get_dummies(df, columns=categorical_columns, dummy_na=nan_as_category)
categorical_columns = [c for c in df.columns if c not in original_columns]
return df, categorical_columns
def group(df_to_agg, prefix, aggregations, aggregate_by="SK_ID_CURR"):
agg_df = df_to_agg.groupby(aggregate_by).agg(aggregations)
agg_df.columns = pd.Index(
["{}{}_{}".format(prefix, e[0], e[1].upper()) for e in agg_df.columns.tolist()]
)
return agg_df.reset_index()
def group_and_merge(
df_to_agg, df_to_merge, prefix, aggregations, aggregate_by="SK_ID_CURR"
):
agg_df = group(df_to_agg, prefix, aggregations, aggregate_by=aggregate_by)
return df_to_merge.merge(agg_df, how="left", on=aggregate_by)
def get_bureau_balance(path, num_rows=None):
bb = pd.read_csv(os.path.join(path, "bureau_balance.csv"))
bb, categorical_cols = one_hot_encoder(bb, nan_as_category=False)
# Calculate rate for each category with decay
bb_processed = bb.groupby("SK_ID_BUREAU")[categorical_cols].mean().reset_index()
# Min, Max, Count and mean duration of payments (months)
agg = {"MONTHS_BALANCE": ["min", "max", "mean", "size"]}
bb_processed = group_and_merge(bb, bb_processed, "", agg, "SK_ID_BUREAU")
del bb
gc.collect()
return bb_processed
bureau, categorical_cols = one_hot_encoder(bureau, nan_as_category=False)
bureau = bureau.merge(get_bureau_balance(DATA_DIRECTORY), how="left", on="SK_ID_BUREAU")
bureau["STATUS_12345"] = 0
for i in range(1, 6):
bureau["STATUS_12345"] += bureau["STATUS_{}".format(i)]
features = [
"AMT_CREDIT_MAX_OVERDUE",
"AMT_CREDIT_SUM_OVERDUE",
"AMT_CREDIT_SUM",
"AMT_CREDIT_SUM_DEBT",
"DEBT_PERCENTAGE",
"DEBT_CREDIT_DIFF",
"STATUS_0",
"STATUS_12345",
]
agg_length = bureau.groupby("MONTHS_BALANCE_SIZE")[features].mean().reset_index()
agg_length.rename({feat: "LL_" + feat for feat in features}, axis=1, inplace=True)
bureau = bureau.merge(agg_length, how="left", on="MONTHS_BALANCE_SIZE")
del agg_length
gc.collect()
BUREAU_AGG = {
"SK_ID_BUREAU": ["nunique"],
"DAYS_CREDIT": ["min", "max", "mean"],
"DAYS_CREDIT_ENDDATE": ["min", "max"],
"AMT_CREDIT_MAX_OVERDUE": ["max", "mean"],
"AMT_CREDIT_SUM": ["max", "mean", "sum"],
"AMT_CREDIT_SUM_DEBT": ["max", "mean", "sum"],
"AMT_CREDIT_SUM_OVERDUE": ["max", "mean", "sum"],
"AMT_ANNUITY": ["mean"],
"DEBT_CREDIT_DIFF": ["mean", "sum"],
"MONTHS_BALANCE_MEAN": ["mean", "var"],
"MONTHS_BALANCE_SIZE": ["mean", "sum"],
"STATUS_0": ["mean"],
"STATUS_1": ["mean"],
"STATUS_12345": ["mean"],
"STATUS_C": ["mean"],
"STATUS_X": ["mean"],
"CREDIT_ACTIVE_Active": ["mean"],
"CREDIT_ACTIVE_Closed": ["mean"],
"CREDIT_ACTIVE_Sold": ["mean"],
"CREDIT_TYPE_Consumer credit": ["mean"],
"CREDIT_TYPE_Credit card": ["mean"],
"CREDIT_TYPE_Car loan": ["mean"],
"CREDIT_TYPE_Mortgage": ["mean"],
"CREDIT_TYPE_Microloan": ["mean"],
"LL_AMT_CREDIT_SUM_OVERDUE": ["mean"],
"LL_DEBT_CREDIT_DIFF": ["mean"],
"LL_STATUS_12345": ["mean"],
}
BUREAU_ACTIVE_AGG = {
"DAYS_CREDIT": ["max", "mean"],
"DAYS_CREDIT_ENDDATE": ["min", "max"],
"AMT_CREDIT_MAX_OVERDUE": ["max", "mean"],
"AMT_CREDIT_SUM": ["max", "sum"],
"AMT_CREDIT_SUM_DEBT": ["mean", "sum"],
"AMT_CREDIT_SUM_OVERDUE": ["max", "mean"],
"DAYS_CREDIT_UPDATE": ["min", "mean"],
"DEBT_PERCENTAGE": ["mean"],
"DEBT_CREDIT_DIFF": ["mean"],
"CREDIT_TO_ANNUITY_RATIO": ["mean"],
"MONTHS_BALANCE_MEAN": ["mean", "var"],
"MONTHS_BALANCE_SIZE": ["mean", "sum"],
}
BUREAU_CLOSED_AGG = {
"DAYS_CREDIT": ["max", "var"],
"DAYS_CREDIT_ENDDATE": ["max"],
"AMT_CREDIT_MAX_OVERDUE": ["max", "mean"],
"AMT_CREDIT_SUM_OVERDUE": ["mean"],
"AMT_CREDIT_SUM": ["max", "mean", "sum"],
"AMT_CREDIT_SUM_DEBT": ["max", "sum"],
"DAYS_CREDIT_UPDATE": ["max"],
"ENDDATE_DIF": ["mean"],
"STATUS_12345": ["mean"],
}
BUREAU_LOAN_TYPE_AGG = {
"DAYS_CREDIT": ["mean", "max"],
"AMT_CREDIT_MAX_OVERDUE": ["mean", "max"],
"AMT_CREDIT_SUM": ["mean", "max"],
"AMT_CREDIT_SUM_DEBT": ["mean", "max"],
"DEBT_PERCENTAGE": ["mean"],
"DEBT_CREDIT_DIFF": ["mean"],
"DAYS_CREDIT_ENDDATE": ["max"],
}
BUREAU_TIME_AGG = {
"AMT_CREDIT_MAX_OVERDUE": ["max", "mean"],
"AMT_CREDIT_SUM_OVERDUE": ["mean"],
"AMT_CREDIT_SUM": ["max", "sum"],
"AMT_CREDIT_SUM_DEBT": ["mean", "sum"],
"DEBT_PERCENTAGE": ["mean"],
"DEBT_CREDIT_DIFF": ["mean"],
"STATUS_0": ["mean"],
"STATUS_12345": ["mean"],
}
agg_bureau = group(bureau, "BUREAU_", BUREAU_AGG)
active = bureau[bureau["CREDIT_ACTIVE_Active"] == 1]
agg_bureau = group_and_merge(active, agg_bureau, "BUREAU_ACTIVE_", BUREAU_ACTIVE_AGG)
closed = bureau[bureau["CREDIT_ACTIVE_Closed"] == 1]
agg_bureau = group_and_merge(closed, agg_bureau, "BUREAU_CLOSED_", BUREAU_CLOSED_AGG)
del active, closed
gc.collect()
for credit_type in [
"Consumer credit",
"Credit card",
"Mortgage",
"Car loan",
"Microloan",
]:
type_df = bureau[bureau["CREDIT_TYPE_" + credit_type] == 1]
prefix = "BUREAU_" + credit_type.split(" ")[0].upper() + "_"
agg_bureau = group_and_merge(type_df, agg_bureau, prefix, BUREAU_LOAN_TYPE_AGG)
del type_df
gc.collect()
for time_frame in [6, 12]:
prefix = "BUREAU_LAST{}M_".format(time_frame)
time_frame_df = bureau[bureau["DAYS_CREDIT"] >= -30 * time_frame]
agg_bureau = group_and_merge(time_frame_df, agg_bureau, prefix, BUREAU_TIME_AGG)
del time_frame_df
gc.collect()
sort_bureau = bureau.sort_values(by=["DAYS_CREDIT"])
gr = sort_bureau.groupby("SK_ID_CURR")["AMT_CREDIT_MAX_OVERDUE"].last().reset_index()
gr.rename({"AMT_CREDIT_MAX_OVERDUE": "BUREAU_LAST_LOAN_MAX_OVERDUE"}, inplace=True)
agg_bureau = agg_bureau.merge(gr, on="SK_ID_CURR", how="left")
agg_bureau["BUREAU_DEBT_OVER_CREDIT"] = (
agg_bureau["BUREAU_AMT_CREDIT_SUM_DEBT_SUM"]
/ agg_bureau["BUREAU_AMT_CREDIT_SUM_SUM"]
)
agg_bureau["BUREAU_ACTIVE_DEBT_OVER_CREDIT"] = (
agg_bureau["BUREAU_ACTIVE_AMT_CREDIT_SUM_DEBT_SUM"]
/ agg_bureau["BUREAU_ACTIVE_AMT_CREDIT_SUM_SUM"]
)
df = pd.merge(df, agg_bureau, on="SK_ID_CURR", how="left")
del agg_bureau, bureau
gc.collect()
prev = pd.read_csv(os.path.join(DATA_DIRECTORY, "previous_application.csv"))
pay = pd.read_csv(os.path.join(DATA_DIRECTORY, "installments_payments.csv"))
PREVIOUS_AGG = {
"SK_ID_PREV": ["nunique"],
"AMT_ANNUITY": ["min", "max", "mean"],
"AMT_DOWN_PAYMENT": ["max", "mean"],
"HOUR_APPR_PROCESS_START": ["min", "max", "mean"],
"RATE_DOWN_PAYMENT": ["max", "mean"],
"DAYS_DECISION": ["min", "max", "mean"],
"CNT_PAYMENT": ["max", "mean"],
"DAYS_TERMINATION": ["max"],
# Engineered features
"CREDIT_TO_ANNUITY_RATIO": ["mean", "max"],
"APPLICATION_CREDIT_DIFF": ["min", "max", "mean"],
"APPLICATION_CREDIT_RATIO": ["min", "max", "mean", "var"],
"DOWN_PAYMENT_TO_CREDIT": ["mean"],
}
PREVIOUS_ACTIVE_AGG = {
"SK_ID_PREV": ["nunique"],
"SIMPLE_INTERESTS": ["mean"],
"AMT_ANNUITY": ["max", "sum"],
"AMT_APPLICATION": ["max", "mean"],
"AMT_CREDIT": ["sum"],
"AMT_DOWN_PAYMENT": ["max", "mean"],
"DAYS_DECISION": ["min", "mean"],
"CNT_PAYMENT": ["mean", "sum"],
"DAYS_LAST_DUE_1ST_VERSION": ["min", "max", "mean"],
# Engineered features
"AMT_PAYMENT": ["sum"],
"INSTALMENT_PAYMENT_DIFF": ["mean", "max"],
"REMAINING_DEBT": ["max", "mean", "sum"],
"REPAYMENT_RATIO": ["mean"],
}
PREVIOUS_LATE_PAYMENTS_AGG = {
"DAYS_DECISION": ["min", "max", "mean"],
"DAYS_LAST_DUE_1ST_VERSION": ["min", "max", "mean"],
# Engineered features
"APPLICATION_CREDIT_DIFF": ["min"],
"NAME_CONTRACT_TYPE_Consumer loans": ["mean"],
"NAME_CONTRACT_TYPE_Cash loans": ["mean"],
"NAME_CONTRACT_TYPE_Revolving loans": ["mean"],
}
PREVIOUS_LOAN_TYPE_AGG = {
"AMT_CREDIT": ["sum"],
"AMT_ANNUITY": ["mean", "max"],
"SIMPLE_INTERESTS": ["min", "mean", "max", "var"],
"APPLICATION_CREDIT_DIFF": ["min", "var"],
"APPLICATION_CREDIT_RATIO": ["min", "max", "mean"],
"DAYS_DECISION": ["max"],
"DAYS_LAST_DUE_1ST_VERSION": ["max", "mean"],
"CNT_PAYMENT": ["mean"],
}
PREVIOUS_TIME_AGG = {
"AMT_CREDIT": ["sum"],
"AMT_ANNUITY": ["mean", "max"],
"SIMPLE_INTERESTS": ["mean", "max"],
"DAYS_DECISION": ["min", "mean"],
"DAYS_LAST_DUE_1ST_VERSION": ["min", "max", "mean"],
# Engineered features
"APPLICATION_CREDIT_DIFF": ["min"],
"APPLICATION_CREDIT_RATIO": ["min", "max", "mean"],
"NAME_CONTRACT_TYPE_Consumer loans": ["mean"],
"NAME_CONTRACT_TYPE_Cash loans": ["mean"],
"NAME_CONTRACT_TYPE_Revolving loans": ["mean"],
}
PREVIOUS_APPROVED_AGG = {
"SK_ID_PREV": ["nunique"],
"AMT_ANNUITY": ["min", "max", "mean"],
"AMT_CREDIT": ["min", "max", "mean"],
"AMT_DOWN_PAYMENT": ["max"],
"AMT_GOODS_PRICE": ["max"],
"HOUR_APPR_PROCESS_START": ["min", "max"],
"DAYS_DECISION": ["min", "mean"],
"CNT_PAYMENT": ["max", "mean"],
"DAYS_TERMINATION": ["mean"],
# Engineered features
"CREDIT_TO_ANNUITY_RATIO": ["mean", "max"],
"APPLICATION_CREDIT_DIFF": ["max"],
"APPLICATION_CREDIT_RATIO": ["min", "max", "mean"],
# The following features are only for approved applications
"DAYS_FIRST_DRAWING": ["max", "mean"],
"DAYS_FIRST_DUE": ["min", "mean"],
"DAYS_LAST_DUE_1ST_VERSION": ["min", "max", "mean"],
"DAYS_LAST_DUE": ["max", "mean"],
"DAYS_LAST_DUE_DIFF": ["min", "max", "mean"],
"SIMPLE_INTERESTS": ["min", "max", "mean"],
}
PREVIOUS_REFUSED_AGG = {
"AMT_APPLICATION": ["max", "mean"],
"AMT_CREDIT": ["min", "max"],
"DAYS_DECISION": ["min", "max", "mean"],
"CNT_PAYMENT": ["max", "mean"],
# Engineered features
"APPLICATION_CREDIT_DIFF": ["min", "max", "mean", "var"],
"APPLICATION_CREDIT_RATIO": ["min", "mean"],
"NAME_CONTRACT_TYPE_Consumer loans": ["mean"],
"NAME_CONTRACT_TYPE_Cash loans": ["mean"],
"NAME_CONTRACT_TYPE_Revolving loans": ["mean"],
}
ohe_columns = [
"NAME_CONTRACT_STATUS",
"NAME_CONTRACT_TYPE",
"CHANNEL_TYPE",
"NAME_TYPE_SUITE",
"NAME_YIELD_GROUP",
"PRODUCT_COMBINATION",
"NAME_PRODUCT_TYPE",
"NAME_CLIENT_TYPE",
]
prev, categorical_cols = one_hot_encoder(prev, ohe_columns, nan_as_category=False)
prev["APPLICATION_CREDIT_DIFF"] = prev["AMT_APPLICATION"] - prev["AMT_CREDIT"]
prev["APPLICATION_CREDIT_RATIO"] = prev["AMT_APPLICATION"] / prev["AMT_CREDIT"]
prev["CREDIT_TO_ANNUITY_RATIO"] = prev["AMT_CREDIT"] / prev["AMT_ANNUITY"]
prev["DOWN_PAYMENT_TO_CREDIT"] = prev["AMT_DOWN_PAYMENT"] / prev["AMT_CREDIT"]
total_payment = prev["AMT_ANNUITY"] * prev["CNT_PAYMENT"]
prev["SIMPLE_INTERESTS"] = (total_payment / prev["AMT_CREDIT"] - 1) / prev[
"CNT_PAYMENT"
]
approved = prev[prev["NAME_CONTRACT_STATUS_Approved"] == 1]
active_df = approved[approved["DAYS_LAST_DUE"] == 365243]
active_pay = pay[pay["SK_ID_PREV"].isin(active_df["SK_ID_PREV"])]
active_pay_agg = active_pay.groupby("SK_ID_PREV")[
["AMT_INSTALMENT", "AMT_PAYMENT"]
].sum()
active_pay_agg.reset_index(inplace=True)
active_pay_agg["INSTALMENT_PAYMENT_DIFF"] = (
active_pay_agg["AMT_INSTALMENT"] - active_pay_agg["AMT_PAYMENT"]
)
active_df = active_df.merge(active_pay_agg, on="SK_ID_PREV", how="left")
active_df["REMAINING_DEBT"] = active_df["AMT_CREDIT"] - active_df["AMT_PAYMENT"]
active_df["REPAYMENT_RATIO"] = active_df["AMT_PAYMENT"] / active_df["AMT_CREDIT"]
active_agg_df = group(active_df, "PREV_ACTIVE_", PREVIOUS_ACTIVE_AGG)
active_agg_df["TOTAL_REPAYMENT_RATIO"] = (
active_agg_df["PREV_ACTIVE_AMT_PAYMENT_SUM"]
/ active_agg_df["PREV_ACTIVE_AMT_CREDIT_SUM"]
)
del active_pay, active_pay_agg, active_df
gc.collect()
prev["DAYS_FIRST_DRAWING"].replace(365243, np.nan, inplace=True)
prev["DAYS_FIRST_DUE"].replace(365243, np.nan, inplace=True)
prev["DAYS_LAST_DUE_1ST_VERSION"].replace(365243, np.nan, inplace=True)
prev["DAYS_LAST_DUE"].replace(365243, np.nan, inplace=True)
prev["DAYS_TERMINATION"].replace(365243, np.nan, inplace=True)
prev["DAYS_LAST_DUE_DIFF"] = prev["DAYS_LAST_DUE_1ST_VERSION"] - prev["DAYS_LAST_DUE"]
approved["DAYS_LAST_DUE_DIFF"] = (
approved["DAYS_LAST_DUE_1ST_VERSION"] - approved["DAYS_LAST_DUE"]
)
categorical_agg = {key: ["mean"] for key in categorical_cols}
agg_prev = group(prev, "PREV_", {**PREVIOUS_AGG, **categorical_agg})
agg_prev = agg_prev.merge(active_agg_df, how="left", on="SK_ID_CURR")
del active_agg_df
gc.collect()
agg_prev = group_and_merge(approved, agg_prev, "APPROVED_", PREVIOUS_APPROVED_AGG)
refused = prev[prev["NAME_CONTRACT_STATUS_Refused"] == 1]
agg_prev = group_and_merge(refused, agg_prev, "REFUSED_", PREVIOUS_REFUSED_AGG)
del approved, refused
gc.collect()
for loan_type in ["Consumer loans", "Cash loans"]:
type_df = prev[prev["NAME_CONTRACT_TYPE_{}".format(loan_type)] == 1]
prefix = "PREV_" + loan_type.split(" ")[0] + "_"
agg_prev = group_and_merge(type_df, agg_prev, prefix, PREVIOUS_LOAN_TYPE_AGG)
del type_df
gc.collect()
pay["LATE_PAYMENT"] = pay["DAYS_ENTRY_PAYMENT"] - pay["DAYS_INSTALMENT"]
pay["LATE_PAYMENT"] = pay["LATE_PAYMENT"].apply(lambda x: 1 if x > 0 else 0)
dpd_id = pay[pay["LATE_PAYMENT"] > 0]["SK_ID_PREV"].unique()
agg_dpd = group_and_merge(
prev[prev["SK_ID_PREV"].isin(dpd_id)],
agg_prev,
"PREV_LATE_",
PREVIOUS_LATE_PAYMENTS_AGG,
)
del agg_dpd, dpd_id
gc.collect()
for time_frame in [12, 24]:
time_frame_df = prev[prev["DAYS_DECISION"] >= -30 * time_frame]
prefix = "PREV_LAST{}M_".format(time_frame)
agg_prev = group_and_merge(time_frame_df, agg_prev, prefix, PREVIOUS_TIME_AGG)
del time_frame_df
gc.collect()
del prev
gc.collect()
df = pd.merge(df, agg_prev, on="SK_ID_CURR", how="left")
train = df[df["TARGET"].notnull()]
test = df[df["TARGET"].isnull()]
del df
del agg_prev
gc.collect()
labels = train["TARGET"]
test_lebels = test["TARGET"]
train = train.drop(columns=["TARGET"])
test = test.drop(columns=["TARGET"])
feature = list(train.columns)
train.replace([np.inf, -np.inf], np.nan, inplace=True)
test.replace([np.inf, -np.inf], np.nan, inplace=True)
test_df = test.copy()
train_df = train.copy()
train_df["TARGET"] = labels
test_df["TARGET"] = test_lebels
imputer = SimpleImputer(strategy="median")
imputer.fit(train)
imputer.fit(test)
train1 = imputer.transform(train)
test1 = imputer.transform(test)
del train
del test
gc.collect()
scaler = MinMaxScaler(feature_range=(0, 1))
scaler.fit(train1)
scaler.fit(test1)
train = scaler.transform(train1)
test = scaler.transform(test1)
del train1
del test1
gc.collect()
# Model Class to be used for different ML algorithms
class ClassifierModel(object):
def __init__(self, clf, params=None):
self.clf = clf(**params)
def train(self, x_train, y_train):
self.clf.fit(x_train, y_train)
def fit(self, x, y):
return self.clf.fit(x, y)
def feature_importances(self, x, y):
return self.clf.fit(x, y).feature_importances_
def predict(self, x):
return self.clf.predict(x)
def trainModel(model, x_train, y_train, x_test, n_folds, seed):
cv = KFold(n_splits=n_folds, random_state=seed)
scores = cross_val_score(
model.clf, x_train, y_train, scoring="accuracy", cv=cv, n_jobs=-1
)
y_pred = cross_val_predict(model.clf, x_train, y_train, cv=cv, n_jobs=-1)
return scores, y_pred
# # **Ensabmle Model-2 (GB+logistic+SVM)**
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import cross_val_predict
gb_params = {"n_estimators": 100, "max_depth": 1}
gb_model = ClassifierModel(clf=GradientBoostingClassifier, params=gb_params)
gb_scores, gb_train_pred = trainModel(gb_model, train, labels, test, 5, None)
gb_scores
lg_params = {"penalty": "l1", "solver": "saga", "tol": 0.1}
lg_model = ClassifierModel(clf=LogisticRegression, params=lg_params)
lg_scores, lg_train_pred = trainModel(lg_model, train, labels, test, 5, None)
lg_scores
acc_pred_train = pd.DataFrame(
{"GradientBoosting": gb_scores.ravel(), "Logistic": lg_scores.ravel()}
)
acc_pred_train.head()
x_train = np.column_stack((gb_train_pred, lg_train_pred))
def trainStackModel(x_train, y_train, x_test, n_folds, seed):
cv = KFold(n_splits=n_folds, random_state=seed)
svm = SVC().fit(x_train, y_train)
scores = cross_val_score(svm, x_train, y_train, scoring="accuracy", cv=cv)
return scores
stackModel_scores = trainStackModel(x_train, labels, test, 5, None)
acc_pred_train["stackingModel"] = stackModel_scores
acc_pred_train
predictions = lg_model.predict_proba(test)[:, 1]
submit = test_df[["SK_ID_CURR"]]
submit["TARGET"] = predictions
submit.to_csv("ensamble_2.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506263.ipynb
| null | null |
[{"Id": 69506263, "ScriptId": 18805376, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7966267, "CreationDate": "07/31/2021 21:14:28", "VersionNumber": 8.0, "Title": "CSE499", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 598.0, "LinesInsertedFromPrevious": 3.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 595.0, "LinesInsertedFromFork": 97.0, "LinesDeletedFromFork": 283.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 501.0, "TotalVotes": 0}]
| null | null | null | null |
import os
import gc
import numpy as np
import pandas as pd
from scipy.stats import kurtosis
from sklearn.metrics import roc_auc_score
from sklearn.preprocessing import MinMaxScaler
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LogisticRegression
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
from sklearn.model_selection import train_test_split, cross_val_score, StratifiedKFold
import xgboost as xgb
from xgboost import XGBClassifier
warnings.simplefilter(action="ignore", category=FutureWarning)
DATA_DIRECTORY = "../input/home-credit-default-risk"
df_train = pd.read_csv(os.path.join(DATA_DIRECTORY, "application_train.csv"))
df_test = pd.read_csv(os.path.join(DATA_DIRECTORY, "application_test.csv"))
df = df_train.append(df_test)
del df_train, df_test
gc.collect()
df = df[df["AMT_INCOME_TOTAL"] < 20000000]
df = df[df["CODE_GENDER"] != "XNA"]
df["DAYS_EMPLOYED"].replace(365243, np.nan, inplace=True)
df["DAYS_LAST_PHONE_CHANGE"].replace(0, np.nan, inplace=True)
def get_age_group(days_birth):
age_years = -days_birth / 365
if age_years < 27:
return 1
elif age_years < 40:
return 2
elif age_years < 50:
return 3
elif age_years < 65:
return 4
elif age_years < 99:
return 5
else:
return 0
docs = [f for f in df.columns if "FLAG_DOC" in f]
df["DOCUMENT_COUNT"] = df[docs].sum(axis=1)
df["NEW_DOC_KURT"] = df[docs].kurtosis(axis=1)
df["AGE_RANGE"] = df["DAYS_BIRTH"].apply(lambda x: get_age_group(x))
df["EXT_SOURCES_PROD"] = df["EXT_SOURCE_1"] * df["EXT_SOURCE_2"] * df["EXT_SOURCE_3"]
df["EXT_SOURCES_WEIGHTED"] = (
df.EXT_SOURCE_1 * 2 + df.EXT_SOURCE_2 * 1 + df.EXT_SOURCE_3 * 3
)
np.warnings.filterwarnings("ignore", r"All-NaN (slice|axis) encountered")
for function_name in ["min", "max", "mean", "nanmedian", "var"]:
feature_name = "EXT_SOURCES_{}".format(function_name.upper())
df[feature_name] = eval("np.{}".format(function_name))(
df[["EXT_SOURCE_1", "EXT_SOURCE_2", "EXT_SOURCE_3"]], axis=1
)
df["CREDIT_TO_ANNUITY_RATIO"] = df["AMT_CREDIT"] / df["AMT_ANNUITY"]
df["CREDIT_TO_GOODS_RATIO"] = df["AMT_CREDIT"] / df["AMT_GOODS_PRICE"]
df["ANNUITY_TO_INCOME_RATIO"] = df["AMT_ANNUITY"] / df["AMT_INCOME_TOTAL"]
df["CREDIT_TO_INCOME_RATIO"] = df["AMT_CREDIT"] / df["AMT_INCOME_TOTAL"]
df["INCOME_TO_EMPLOYED_RATIO"] = df["AMT_INCOME_TOTAL"] / df["DAYS_EMPLOYED"]
df["INCOME_TO_BIRTH_RATIO"] = df["AMT_INCOME_TOTAL"] / df["DAYS_BIRTH"]
df["EMPLOYED_TO_BIRTH_RATIO"] = df["DAYS_EMPLOYED"] / df["DAYS_BIRTH"]
df["ID_TO_BIRTH_RATIO"] = df["DAYS_ID_PUBLISH"] / df["DAYS_BIRTH"]
df["CAR_TO_BIRTH_RATIO"] = df["OWN_CAR_AGE"] / df["DAYS_BIRTH"]
df["CAR_TO_EMPLOYED_RATIO"] = df["OWN_CAR_AGE"] / df["DAYS_EMPLOYED"]
df["PHONE_TO_BIRTH_RATIO"] = df["DAYS_LAST_PHONE_CHANGE"] / df["DAYS_BIRTH"]
def do_mean(df, group_cols, counted, agg_name):
gp = (
df[group_cols + [counted]]
.groupby(group_cols)[counted]
.mean()
.reset_index()
.rename(columns={counted: agg_name})
)
df = df.merge(gp, on=group_cols, how="left")
del gp
gc.collect()
return df
def do_median(df, group_cols, counted, agg_name):
gp = (
df[group_cols + [counted]]
.groupby(group_cols)[counted]
.median()
.reset_index()
.rename(columns={counted: agg_name})
)
df = df.merge(gp, on=group_cols, how="left")
del gp
gc.collect()
return df
def do_std(df, group_cols, counted, agg_name):
gp = (
df[group_cols + [counted]]
.groupby(group_cols)[counted]
.std()
.reset_index()
.rename(columns={counted: agg_name})
)
df = df.merge(gp, on=group_cols, how="left")
del gp
gc.collect()
return df
def do_sum(df, group_cols, counted, agg_name):
gp = (
df[group_cols + [counted]]
.groupby(group_cols)[counted]
.sum()
.reset_index()
.rename(columns={counted: agg_name})
)
df = df.merge(gp, on=group_cols, how="left")
del gp
gc.collect()
return df
group = [
"ORGANIZATION_TYPE",
"NAME_EDUCATION_TYPE",
"OCCUPATION_TYPE",
"AGE_RANGE",
"CODE_GENDER",
]
df = do_median(df, group, "EXT_SOURCES_MEAN", "GROUP_EXT_SOURCES_MEDIAN")
df = do_std(df, group, "EXT_SOURCES_MEAN", "GROUP_EXT_SOURCES_STD")
df = do_mean(df, group, "AMT_INCOME_TOTAL", "GROUP_INCOME_MEAN")
df = do_std(df, group, "AMT_INCOME_TOTAL", "GROUP_INCOME_STD")
df = do_mean(df, group, "CREDIT_TO_ANNUITY_RATIO", "GROUP_CREDIT_TO_ANNUITY_MEAN")
df = do_std(df, group, "CREDIT_TO_ANNUITY_RATIO", "GROUP_CREDIT_TO_ANNUITY_STD")
df = do_mean(df, group, "AMT_CREDIT", "GROUP_CREDIT_MEAN")
df = do_mean(df, group, "AMT_ANNUITY", "GROUP_ANNUITY_MEAN")
df = do_std(df, group, "AMT_ANNUITY", "GROUP_ANNUITY_STD")
def label_encoder(df, categorical_columns=None):
if not categorical_columns:
categorical_columns = [col for col in df.columns if df[col].dtype == "object"]
for col in categorical_columns:
df[col], uniques = pd.factorize(df[col])
return df, categorical_columns
def drop_application_columns(df):
drop_list = [
"CNT_CHILDREN",
"CNT_FAM_MEMBERS",
"HOUR_APPR_PROCESS_START",
"FLAG_EMP_PHONE",
"FLAG_MOBIL",
"FLAG_CONT_MOBILE",
"FLAG_EMAIL",
"FLAG_PHONE",
"FLAG_OWN_REALTY",
"REG_REGION_NOT_LIVE_REGION",
"REG_REGION_NOT_WORK_REGION",
"REG_CITY_NOT_WORK_CITY",
"OBS_30_CNT_SOCIAL_CIRCLE",
"OBS_60_CNT_SOCIAL_CIRCLE",
"AMT_REQ_CREDIT_BUREAU_DAY",
"AMT_REQ_CREDIT_BUREAU_MON",
"AMT_REQ_CREDIT_BUREAU_YEAR",
"COMMONAREA_MODE",
"NONLIVINGAREA_MODE",
"ELEVATORS_MODE",
"NONLIVINGAREA_AVG",
"FLOORSMIN_MEDI",
"LANDAREA_MODE",
"NONLIVINGAREA_MEDI",
"LIVINGAPARTMENTS_MODE",
"FLOORSMIN_AVG",
"LANDAREA_AVG",
"FLOORSMIN_MODE",
"LANDAREA_MEDI",
"COMMONAREA_MEDI",
"YEARS_BUILD_AVG",
"COMMONAREA_AVG",
"BASEMENTAREA_AVG",
"BASEMENTAREA_MODE",
"NONLIVINGAPARTMENTS_MEDI",
"BASEMENTAREA_MEDI",
"LIVINGAPARTMENTS_AVG",
"ELEVATORS_AVG",
"YEARS_BUILD_MEDI",
"ENTRANCES_MODE",
"NONLIVINGAPARTMENTS_MODE",
"LIVINGAREA_MODE",
"LIVINGAPARTMENTS_MEDI",
"YEARS_BUILD_MODE",
"YEARS_BEGINEXPLUATATION_AVG",
"ELEVATORS_MEDI",
"LIVINGAREA_MEDI",
"YEARS_BEGINEXPLUATATION_MODE",
"NONLIVINGAPARTMENTS_AVG",
"HOUSETYPE_MODE",
"FONDKAPREMONT_MODE",
"EMERGENCYSTATE_MODE",
]
for doc_num in [2, 4, 5, 6, 7, 9, 10, 11, 12, 13, 14, 15, 16, 17, 19, 20, 21]:
drop_list.append("FLAG_DOCUMENT_{}".format(doc_num))
df.drop(drop_list, axis=1, inplace=True)
return df
df, le_encoded_cols = label_encoder(df, None)
df = drop_application_columns(df)
df = pd.get_dummies(df)
bureau = pd.read_csv(os.path.join(DATA_DIRECTORY, "bureau.csv"))
bureau["CREDIT_DURATION"] = -bureau["DAYS_CREDIT"] + bureau["DAYS_CREDIT_ENDDATE"]
bureau["ENDDATE_DIF"] = bureau["DAYS_CREDIT_ENDDATE"] - bureau["DAYS_ENDDATE_FACT"]
bureau["DEBT_PERCENTAGE"] = bureau["AMT_CREDIT_SUM"] / bureau["AMT_CREDIT_SUM_DEBT"]
bureau["DEBT_CREDIT_DIFF"] = bureau["AMT_CREDIT_SUM"] - bureau["AMT_CREDIT_SUM_DEBT"]
bureau["CREDIT_TO_ANNUITY_RATIO"] = bureau["AMT_CREDIT_SUM"] / bureau["AMT_ANNUITY"]
def one_hot_encoder(df, categorical_columns=None, nan_as_category=True):
original_columns = list(df.columns)
if not categorical_columns:
categorical_columns = [col for col in df.columns if df[col].dtype == "object"]
df = pd.get_dummies(df, columns=categorical_columns, dummy_na=nan_as_category)
categorical_columns = [c for c in df.columns if c not in original_columns]
return df, categorical_columns
def group(df_to_agg, prefix, aggregations, aggregate_by="SK_ID_CURR"):
agg_df = df_to_agg.groupby(aggregate_by).agg(aggregations)
agg_df.columns = pd.Index(
["{}{}_{}".format(prefix, e[0], e[1].upper()) for e in agg_df.columns.tolist()]
)
return agg_df.reset_index()
def group_and_merge(
df_to_agg, df_to_merge, prefix, aggregations, aggregate_by="SK_ID_CURR"
):
agg_df = group(df_to_agg, prefix, aggregations, aggregate_by=aggregate_by)
return df_to_merge.merge(agg_df, how="left", on=aggregate_by)
def get_bureau_balance(path, num_rows=None):
bb = pd.read_csv(os.path.join(path, "bureau_balance.csv"))
bb, categorical_cols = one_hot_encoder(bb, nan_as_category=False)
# Calculate rate for each category with decay
bb_processed = bb.groupby("SK_ID_BUREAU")[categorical_cols].mean().reset_index()
# Min, Max, Count and mean duration of payments (months)
agg = {"MONTHS_BALANCE": ["min", "max", "mean", "size"]}
bb_processed = group_and_merge(bb, bb_processed, "", agg, "SK_ID_BUREAU")
del bb
gc.collect()
return bb_processed
bureau, categorical_cols = one_hot_encoder(bureau, nan_as_category=False)
bureau = bureau.merge(get_bureau_balance(DATA_DIRECTORY), how="left", on="SK_ID_BUREAU")
bureau["STATUS_12345"] = 0
for i in range(1, 6):
bureau["STATUS_12345"] += bureau["STATUS_{}".format(i)]
features = [
"AMT_CREDIT_MAX_OVERDUE",
"AMT_CREDIT_SUM_OVERDUE",
"AMT_CREDIT_SUM",
"AMT_CREDIT_SUM_DEBT",
"DEBT_PERCENTAGE",
"DEBT_CREDIT_DIFF",
"STATUS_0",
"STATUS_12345",
]
agg_length = bureau.groupby("MONTHS_BALANCE_SIZE")[features].mean().reset_index()
agg_length.rename({feat: "LL_" + feat for feat in features}, axis=1, inplace=True)
bureau = bureau.merge(agg_length, how="left", on="MONTHS_BALANCE_SIZE")
del agg_length
gc.collect()
BUREAU_AGG = {
"SK_ID_BUREAU": ["nunique"],
"DAYS_CREDIT": ["min", "max", "mean"],
"DAYS_CREDIT_ENDDATE": ["min", "max"],
"AMT_CREDIT_MAX_OVERDUE": ["max", "mean"],
"AMT_CREDIT_SUM": ["max", "mean", "sum"],
"AMT_CREDIT_SUM_DEBT": ["max", "mean", "sum"],
"AMT_CREDIT_SUM_OVERDUE": ["max", "mean", "sum"],
"AMT_ANNUITY": ["mean"],
"DEBT_CREDIT_DIFF": ["mean", "sum"],
"MONTHS_BALANCE_MEAN": ["mean", "var"],
"MONTHS_BALANCE_SIZE": ["mean", "sum"],
"STATUS_0": ["mean"],
"STATUS_1": ["mean"],
"STATUS_12345": ["mean"],
"STATUS_C": ["mean"],
"STATUS_X": ["mean"],
"CREDIT_ACTIVE_Active": ["mean"],
"CREDIT_ACTIVE_Closed": ["mean"],
"CREDIT_ACTIVE_Sold": ["mean"],
"CREDIT_TYPE_Consumer credit": ["mean"],
"CREDIT_TYPE_Credit card": ["mean"],
"CREDIT_TYPE_Car loan": ["mean"],
"CREDIT_TYPE_Mortgage": ["mean"],
"CREDIT_TYPE_Microloan": ["mean"],
"LL_AMT_CREDIT_SUM_OVERDUE": ["mean"],
"LL_DEBT_CREDIT_DIFF": ["mean"],
"LL_STATUS_12345": ["mean"],
}
BUREAU_ACTIVE_AGG = {
"DAYS_CREDIT": ["max", "mean"],
"DAYS_CREDIT_ENDDATE": ["min", "max"],
"AMT_CREDIT_MAX_OVERDUE": ["max", "mean"],
"AMT_CREDIT_SUM": ["max", "sum"],
"AMT_CREDIT_SUM_DEBT": ["mean", "sum"],
"AMT_CREDIT_SUM_OVERDUE": ["max", "mean"],
"DAYS_CREDIT_UPDATE": ["min", "mean"],
"DEBT_PERCENTAGE": ["mean"],
"DEBT_CREDIT_DIFF": ["mean"],
"CREDIT_TO_ANNUITY_RATIO": ["mean"],
"MONTHS_BALANCE_MEAN": ["mean", "var"],
"MONTHS_BALANCE_SIZE": ["mean", "sum"],
}
BUREAU_CLOSED_AGG = {
"DAYS_CREDIT": ["max", "var"],
"DAYS_CREDIT_ENDDATE": ["max"],
"AMT_CREDIT_MAX_OVERDUE": ["max", "mean"],
"AMT_CREDIT_SUM_OVERDUE": ["mean"],
"AMT_CREDIT_SUM": ["max", "mean", "sum"],
"AMT_CREDIT_SUM_DEBT": ["max", "sum"],
"DAYS_CREDIT_UPDATE": ["max"],
"ENDDATE_DIF": ["mean"],
"STATUS_12345": ["mean"],
}
BUREAU_LOAN_TYPE_AGG = {
"DAYS_CREDIT": ["mean", "max"],
"AMT_CREDIT_MAX_OVERDUE": ["mean", "max"],
"AMT_CREDIT_SUM": ["mean", "max"],
"AMT_CREDIT_SUM_DEBT": ["mean", "max"],
"DEBT_PERCENTAGE": ["mean"],
"DEBT_CREDIT_DIFF": ["mean"],
"DAYS_CREDIT_ENDDATE": ["max"],
}
BUREAU_TIME_AGG = {
"AMT_CREDIT_MAX_OVERDUE": ["max", "mean"],
"AMT_CREDIT_SUM_OVERDUE": ["mean"],
"AMT_CREDIT_SUM": ["max", "sum"],
"AMT_CREDIT_SUM_DEBT": ["mean", "sum"],
"DEBT_PERCENTAGE": ["mean"],
"DEBT_CREDIT_DIFF": ["mean"],
"STATUS_0": ["mean"],
"STATUS_12345": ["mean"],
}
agg_bureau = group(bureau, "BUREAU_", BUREAU_AGG)
active = bureau[bureau["CREDIT_ACTIVE_Active"] == 1]
agg_bureau = group_and_merge(active, agg_bureau, "BUREAU_ACTIVE_", BUREAU_ACTIVE_AGG)
closed = bureau[bureau["CREDIT_ACTIVE_Closed"] == 1]
agg_bureau = group_and_merge(closed, agg_bureau, "BUREAU_CLOSED_", BUREAU_CLOSED_AGG)
del active, closed
gc.collect()
for credit_type in [
"Consumer credit",
"Credit card",
"Mortgage",
"Car loan",
"Microloan",
]:
type_df = bureau[bureau["CREDIT_TYPE_" + credit_type] == 1]
prefix = "BUREAU_" + credit_type.split(" ")[0].upper() + "_"
agg_bureau = group_and_merge(type_df, agg_bureau, prefix, BUREAU_LOAN_TYPE_AGG)
del type_df
gc.collect()
for time_frame in [6, 12]:
prefix = "BUREAU_LAST{}M_".format(time_frame)
time_frame_df = bureau[bureau["DAYS_CREDIT"] >= -30 * time_frame]
agg_bureau = group_and_merge(time_frame_df, agg_bureau, prefix, BUREAU_TIME_AGG)
del time_frame_df
gc.collect()
sort_bureau = bureau.sort_values(by=["DAYS_CREDIT"])
gr = sort_bureau.groupby("SK_ID_CURR")["AMT_CREDIT_MAX_OVERDUE"].last().reset_index()
gr.rename({"AMT_CREDIT_MAX_OVERDUE": "BUREAU_LAST_LOAN_MAX_OVERDUE"}, inplace=True)
agg_bureau = agg_bureau.merge(gr, on="SK_ID_CURR", how="left")
agg_bureau["BUREAU_DEBT_OVER_CREDIT"] = (
agg_bureau["BUREAU_AMT_CREDIT_SUM_DEBT_SUM"]
/ agg_bureau["BUREAU_AMT_CREDIT_SUM_SUM"]
)
agg_bureau["BUREAU_ACTIVE_DEBT_OVER_CREDIT"] = (
agg_bureau["BUREAU_ACTIVE_AMT_CREDIT_SUM_DEBT_SUM"]
/ agg_bureau["BUREAU_ACTIVE_AMT_CREDIT_SUM_SUM"]
)
df = pd.merge(df, agg_bureau, on="SK_ID_CURR", how="left")
del agg_bureau, bureau
gc.collect()
prev = pd.read_csv(os.path.join(DATA_DIRECTORY, "previous_application.csv"))
pay = pd.read_csv(os.path.join(DATA_DIRECTORY, "installments_payments.csv"))
PREVIOUS_AGG = {
"SK_ID_PREV": ["nunique"],
"AMT_ANNUITY": ["min", "max", "mean"],
"AMT_DOWN_PAYMENT": ["max", "mean"],
"HOUR_APPR_PROCESS_START": ["min", "max", "mean"],
"RATE_DOWN_PAYMENT": ["max", "mean"],
"DAYS_DECISION": ["min", "max", "mean"],
"CNT_PAYMENT": ["max", "mean"],
"DAYS_TERMINATION": ["max"],
# Engineered features
"CREDIT_TO_ANNUITY_RATIO": ["mean", "max"],
"APPLICATION_CREDIT_DIFF": ["min", "max", "mean"],
"APPLICATION_CREDIT_RATIO": ["min", "max", "mean", "var"],
"DOWN_PAYMENT_TO_CREDIT": ["mean"],
}
PREVIOUS_ACTIVE_AGG = {
"SK_ID_PREV": ["nunique"],
"SIMPLE_INTERESTS": ["mean"],
"AMT_ANNUITY": ["max", "sum"],
"AMT_APPLICATION": ["max", "mean"],
"AMT_CREDIT": ["sum"],
"AMT_DOWN_PAYMENT": ["max", "mean"],
"DAYS_DECISION": ["min", "mean"],
"CNT_PAYMENT": ["mean", "sum"],
"DAYS_LAST_DUE_1ST_VERSION": ["min", "max", "mean"],
# Engineered features
"AMT_PAYMENT": ["sum"],
"INSTALMENT_PAYMENT_DIFF": ["mean", "max"],
"REMAINING_DEBT": ["max", "mean", "sum"],
"REPAYMENT_RATIO": ["mean"],
}
PREVIOUS_LATE_PAYMENTS_AGG = {
"DAYS_DECISION": ["min", "max", "mean"],
"DAYS_LAST_DUE_1ST_VERSION": ["min", "max", "mean"],
# Engineered features
"APPLICATION_CREDIT_DIFF": ["min"],
"NAME_CONTRACT_TYPE_Consumer loans": ["mean"],
"NAME_CONTRACT_TYPE_Cash loans": ["mean"],
"NAME_CONTRACT_TYPE_Revolving loans": ["mean"],
}
PREVIOUS_LOAN_TYPE_AGG = {
"AMT_CREDIT": ["sum"],
"AMT_ANNUITY": ["mean", "max"],
"SIMPLE_INTERESTS": ["min", "mean", "max", "var"],
"APPLICATION_CREDIT_DIFF": ["min", "var"],
"APPLICATION_CREDIT_RATIO": ["min", "max", "mean"],
"DAYS_DECISION": ["max"],
"DAYS_LAST_DUE_1ST_VERSION": ["max", "mean"],
"CNT_PAYMENT": ["mean"],
}
PREVIOUS_TIME_AGG = {
"AMT_CREDIT": ["sum"],
"AMT_ANNUITY": ["mean", "max"],
"SIMPLE_INTERESTS": ["mean", "max"],
"DAYS_DECISION": ["min", "mean"],
"DAYS_LAST_DUE_1ST_VERSION": ["min", "max", "mean"],
# Engineered features
"APPLICATION_CREDIT_DIFF": ["min"],
"APPLICATION_CREDIT_RATIO": ["min", "max", "mean"],
"NAME_CONTRACT_TYPE_Consumer loans": ["mean"],
"NAME_CONTRACT_TYPE_Cash loans": ["mean"],
"NAME_CONTRACT_TYPE_Revolving loans": ["mean"],
}
PREVIOUS_APPROVED_AGG = {
"SK_ID_PREV": ["nunique"],
"AMT_ANNUITY": ["min", "max", "mean"],
"AMT_CREDIT": ["min", "max", "mean"],
"AMT_DOWN_PAYMENT": ["max"],
"AMT_GOODS_PRICE": ["max"],
"HOUR_APPR_PROCESS_START": ["min", "max"],
"DAYS_DECISION": ["min", "mean"],
"CNT_PAYMENT": ["max", "mean"],
"DAYS_TERMINATION": ["mean"],
# Engineered features
"CREDIT_TO_ANNUITY_RATIO": ["mean", "max"],
"APPLICATION_CREDIT_DIFF": ["max"],
"APPLICATION_CREDIT_RATIO": ["min", "max", "mean"],
# The following features are only for approved applications
"DAYS_FIRST_DRAWING": ["max", "mean"],
"DAYS_FIRST_DUE": ["min", "mean"],
"DAYS_LAST_DUE_1ST_VERSION": ["min", "max", "mean"],
"DAYS_LAST_DUE": ["max", "mean"],
"DAYS_LAST_DUE_DIFF": ["min", "max", "mean"],
"SIMPLE_INTERESTS": ["min", "max", "mean"],
}
PREVIOUS_REFUSED_AGG = {
"AMT_APPLICATION": ["max", "mean"],
"AMT_CREDIT": ["min", "max"],
"DAYS_DECISION": ["min", "max", "mean"],
"CNT_PAYMENT": ["max", "mean"],
# Engineered features
"APPLICATION_CREDIT_DIFF": ["min", "max", "mean", "var"],
"APPLICATION_CREDIT_RATIO": ["min", "mean"],
"NAME_CONTRACT_TYPE_Consumer loans": ["mean"],
"NAME_CONTRACT_TYPE_Cash loans": ["mean"],
"NAME_CONTRACT_TYPE_Revolving loans": ["mean"],
}
ohe_columns = [
"NAME_CONTRACT_STATUS",
"NAME_CONTRACT_TYPE",
"CHANNEL_TYPE",
"NAME_TYPE_SUITE",
"NAME_YIELD_GROUP",
"PRODUCT_COMBINATION",
"NAME_PRODUCT_TYPE",
"NAME_CLIENT_TYPE",
]
prev, categorical_cols = one_hot_encoder(prev, ohe_columns, nan_as_category=False)
prev["APPLICATION_CREDIT_DIFF"] = prev["AMT_APPLICATION"] - prev["AMT_CREDIT"]
prev["APPLICATION_CREDIT_RATIO"] = prev["AMT_APPLICATION"] / prev["AMT_CREDIT"]
prev["CREDIT_TO_ANNUITY_RATIO"] = prev["AMT_CREDIT"] / prev["AMT_ANNUITY"]
prev["DOWN_PAYMENT_TO_CREDIT"] = prev["AMT_DOWN_PAYMENT"] / prev["AMT_CREDIT"]
total_payment = prev["AMT_ANNUITY"] * prev["CNT_PAYMENT"]
prev["SIMPLE_INTERESTS"] = (total_payment / prev["AMT_CREDIT"] - 1) / prev[
"CNT_PAYMENT"
]
approved = prev[prev["NAME_CONTRACT_STATUS_Approved"] == 1]
active_df = approved[approved["DAYS_LAST_DUE"] == 365243]
active_pay = pay[pay["SK_ID_PREV"].isin(active_df["SK_ID_PREV"])]
active_pay_agg = active_pay.groupby("SK_ID_PREV")[
["AMT_INSTALMENT", "AMT_PAYMENT"]
].sum()
active_pay_agg.reset_index(inplace=True)
active_pay_agg["INSTALMENT_PAYMENT_DIFF"] = (
active_pay_agg["AMT_INSTALMENT"] - active_pay_agg["AMT_PAYMENT"]
)
active_df = active_df.merge(active_pay_agg, on="SK_ID_PREV", how="left")
active_df["REMAINING_DEBT"] = active_df["AMT_CREDIT"] - active_df["AMT_PAYMENT"]
active_df["REPAYMENT_RATIO"] = active_df["AMT_PAYMENT"] / active_df["AMT_CREDIT"]
active_agg_df = group(active_df, "PREV_ACTIVE_", PREVIOUS_ACTIVE_AGG)
active_agg_df["TOTAL_REPAYMENT_RATIO"] = (
active_agg_df["PREV_ACTIVE_AMT_PAYMENT_SUM"]
/ active_agg_df["PREV_ACTIVE_AMT_CREDIT_SUM"]
)
del active_pay, active_pay_agg, active_df
gc.collect()
prev["DAYS_FIRST_DRAWING"].replace(365243, np.nan, inplace=True)
prev["DAYS_FIRST_DUE"].replace(365243, np.nan, inplace=True)
prev["DAYS_LAST_DUE_1ST_VERSION"].replace(365243, np.nan, inplace=True)
prev["DAYS_LAST_DUE"].replace(365243, np.nan, inplace=True)
prev["DAYS_TERMINATION"].replace(365243, np.nan, inplace=True)
prev["DAYS_LAST_DUE_DIFF"] = prev["DAYS_LAST_DUE_1ST_VERSION"] - prev["DAYS_LAST_DUE"]
approved["DAYS_LAST_DUE_DIFF"] = (
approved["DAYS_LAST_DUE_1ST_VERSION"] - approved["DAYS_LAST_DUE"]
)
categorical_agg = {key: ["mean"] for key in categorical_cols}
agg_prev = group(prev, "PREV_", {**PREVIOUS_AGG, **categorical_agg})
agg_prev = agg_prev.merge(active_agg_df, how="left", on="SK_ID_CURR")
del active_agg_df
gc.collect()
agg_prev = group_and_merge(approved, agg_prev, "APPROVED_", PREVIOUS_APPROVED_AGG)
refused = prev[prev["NAME_CONTRACT_STATUS_Refused"] == 1]
agg_prev = group_and_merge(refused, agg_prev, "REFUSED_", PREVIOUS_REFUSED_AGG)
del approved, refused
gc.collect()
for loan_type in ["Consumer loans", "Cash loans"]:
type_df = prev[prev["NAME_CONTRACT_TYPE_{}".format(loan_type)] == 1]
prefix = "PREV_" + loan_type.split(" ")[0] + "_"
agg_prev = group_and_merge(type_df, agg_prev, prefix, PREVIOUS_LOAN_TYPE_AGG)
del type_df
gc.collect()
pay["LATE_PAYMENT"] = pay["DAYS_ENTRY_PAYMENT"] - pay["DAYS_INSTALMENT"]
pay["LATE_PAYMENT"] = pay["LATE_PAYMENT"].apply(lambda x: 1 if x > 0 else 0)
dpd_id = pay[pay["LATE_PAYMENT"] > 0]["SK_ID_PREV"].unique()
agg_dpd = group_and_merge(
prev[prev["SK_ID_PREV"].isin(dpd_id)],
agg_prev,
"PREV_LATE_",
PREVIOUS_LATE_PAYMENTS_AGG,
)
del agg_dpd, dpd_id
gc.collect()
for time_frame in [12, 24]:
time_frame_df = prev[prev["DAYS_DECISION"] >= -30 * time_frame]
prefix = "PREV_LAST{}M_".format(time_frame)
agg_prev = group_and_merge(time_frame_df, agg_prev, prefix, PREVIOUS_TIME_AGG)
del time_frame_df
gc.collect()
del prev
gc.collect()
df = pd.merge(df, agg_prev, on="SK_ID_CURR", how="left")
train = df[df["TARGET"].notnull()]
test = df[df["TARGET"].isnull()]
del df
del agg_prev
gc.collect()
labels = train["TARGET"]
test_lebels = test["TARGET"]
train = train.drop(columns=["TARGET"])
test = test.drop(columns=["TARGET"])
feature = list(train.columns)
train.replace([np.inf, -np.inf], np.nan, inplace=True)
test.replace([np.inf, -np.inf], np.nan, inplace=True)
test_df = test.copy()
train_df = train.copy()
train_df["TARGET"] = labels
test_df["TARGET"] = test_lebels
imputer = SimpleImputer(strategy="median")
imputer.fit(train)
imputer.fit(test)
train1 = imputer.transform(train)
test1 = imputer.transform(test)
del train
del test
gc.collect()
scaler = MinMaxScaler(feature_range=(0, 1))
scaler.fit(train1)
scaler.fit(test1)
train = scaler.transform(train1)
test = scaler.transform(test1)
del train1
del test1
gc.collect()
# Model Class to be used for different ML algorithms
class ClassifierModel(object):
def __init__(self, clf, params=None):
self.clf = clf(**params)
def train(self, x_train, y_train):
self.clf.fit(x_train, y_train)
def fit(self, x, y):
return self.clf.fit(x, y)
def feature_importances(self, x, y):
return self.clf.fit(x, y).feature_importances_
def predict(self, x):
return self.clf.predict(x)
def trainModel(model, x_train, y_train, x_test, n_folds, seed):
cv = KFold(n_splits=n_folds, random_state=seed)
scores = cross_val_score(
model.clf, x_train, y_train, scoring="accuracy", cv=cv, n_jobs=-1
)
y_pred = cross_val_predict(model.clf, x_train, y_train, cv=cv, n_jobs=-1)
return scores, y_pred
# # **Ensabmle Model-2 (GB+logistic+SVM)**
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import cross_val_predict
gb_params = {"n_estimators": 100, "max_depth": 1}
gb_model = ClassifierModel(clf=GradientBoostingClassifier, params=gb_params)
gb_scores, gb_train_pred = trainModel(gb_model, train, labels, test, 5, None)
gb_scores
lg_params = {"penalty": "l1", "solver": "saga", "tol": 0.1}
lg_model = ClassifierModel(clf=LogisticRegression, params=lg_params)
lg_scores, lg_train_pred = trainModel(lg_model, train, labels, test, 5, None)
lg_scores
acc_pred_train = pd.DataFrame(
{"GradientBoosting": gb_scores.ravel(), "Logistic": lg_scores.ravel()}
)
acc_pred_train.head()
x_train = np.column_stack((gb_train_pred, lg_train_pred))
def trainStackModel(x_train, y_train, x_test, n_folds, seed):
cv = KFold(n_splits=n_folds, random_state=seed)
svm = SVC().fit(x_train, y_train)
scores = cross_val_score(svm, x_train, y_train, scoring="accuracy", cv=cv)
return scores
stackModel_scores = trainStackModel(x_train, labels, test, 5, None)
acc_pred_train["stackingModel"] = stackModel_scores
acc_pred_train
predictions = lg_model.predict_proba(test)[:, 1]
submit = test_df[["SK_ID_CURR"]]
submit["TARGET"] = predictions
submit.to_csv("ensamble_2.csv", index=False)
| false | 0 | 9,123 | 0 | 9,123 | 9,123 |
||
69506025
|
<jupyter_start><jupyter_text>Simple/Normal Wikipedia Sections
Kaggle dataset identifier: simplenormal-wikipedia-sections
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from catboost import CatBoostClassifier, Pool
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import StratifiedKFold
FOLDS = 5
import pickle
import gzip
def pickle_save(object, filename, gzipping=True, protocol=4):
"""Save an object to a compressed disk file.
Works well with huge objects.
"""
open_f = gzip.GzipFile if gzipping else open
with open_f(filename, "wb") as f:
pickle.dump(object, f, protocol)
wp = pd.read_pickle(
"../input/simplenormal-wikipedia-sections/wikipedia_sections.pkl.xz"
)
data = wp[["text", "label"]].copy()
del wp
X_train, X_test, y_train, y_test = train_test_split(
data[[x for x in data.columns if x != "label"]],
data["label"],
test_size=0.3,
random_state=42,
)
def fit_model_classifier(train_pool, test_pool, **kwargs):
model = CatBoostClassifier(
task_type="GPU",
iterations=5000,
eval_metric="AUC",
od_type="Iter",
od_wait=500,
l2_leaf_reg=10,
bootstrap_type="Bernoulli",
subsample=0.7,
learning_rate=0.2,
**kwargs,
)
return model.fit(
train_pool, eval_set=test_pool, verbose=100, plot=False, use_best_model=True
)
def get_oof_classifier(n_folds, x_train, y, x_test, text_features, seeds):
ntrain = x_train.shape[0]
ntest = x_test.shape[0]
tpo = {
"tokenizers": [
{
"tokenizer_id": "Sense",
"separator_type": "BySense",
"lowercasing": "True",
"token_types": ["Word", "Number"],
}
],
"dictionaries": [
{
"dictionary_id": "Word",
"token_level_type": "Word",
"occurrence_lower_bound": "500",
"max_dictionary_size": "10000",
},
{
"dictionary_id": "Bigram",
"token_level_type": "Word",
"gram_order": "2",
"occurrence_lower_bound": "100",
"max_dictionary_size": "1000",
},
{
"dictionary_id": "Trigram",
"token_level_type": "Word",
"gram_order": "3",
"occurrence_lower_bound": "100",
"max_dictionary_size": "1000",
},
],
"feature_processing": {
"0": [
{
"tokenizers_names": ["Sense"],
"dictionaries_names": ["Word"],
"feature_calcers": ["BoW", "BM25"],
},
{
"tokenizers_names": ["Sense"],
"dictionaries_names": ["Bigram", "Trigram"],
"feature_calcers": ["BoW"],
},
]
},
}
oof_train = np.zeros((len(seeds), ntrain))
oof_test = np.zeros((ntest))
oof_test_skf = np.empty((len(seeds), n_folds, ntest))
test_pool = Pool(data=x_test, text_features=text_features)
models = {}
for iseed, seed in enumerate(seeds):
kf = StratifiedKFold(n_splits=n_folds, shuffle=True, random_state=seed)
for i, (tr_i, t_i) in enumerate(kf.split(x_train, y)):
print(f"\nSeed {seed}, Fold {i}")
x_tr = x_train.iloc[tr_i, :]
y_tr = y[tr_i]
x_te = x_train.iloc[t_i, :]
y_te = y[t_i]
train_pool = Pool(data=x_tr, label=y_tr, text_features=text_features)
valid_pool = Pool(data=x_te, label=y_te, text_features=text_features)
model = fit_model_classifier(
train_pool, valid_pool, random_seed=seed, text_processing=tpo
)
oof_train[iseed, t_i] = model.predict_proba(valid_pool)[:, 1]
oof_test_skf[iseed, i, :] = model.predict_proba(test_pool)[:, 1]
models[(seed, i)] = model
oof_test[:] = oof_test_skf.mean(axis=1).mean(axis=0)
oof_train = oof_train.mean(axis=0)
return oof_train, oof_test, models
oof_train, oof_test, models = get_oof_classifier(
n_folds=FOLDS,
x_train=X_train[0:100000],
y=y_train[0:100000].values,
x_test=X_test,
text_features=["text"],
seeds=[0, 42, 888],
)
roc_auc_score(y_test, oof_test)
pickle_save(models, "models.zpkl")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506025.ipynb
|
simplenormal-wikipedia-sections
|
markwijkhuizen
|
[{"Id": 69506025, "ScriptId": 18978731, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 526714, "CreationDate": "07/31/2021 21:08:40", "VersionNumber": 1.0, "Title": "notebookcb392fab86", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 160.0, "LinesInsertedFromPrevious": 160.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92760770, "KernelVersionId": 69506025, "SourceDatasetVersionId": 2452800}]
|
[{"Id": 2452800, "DatasetId": 1455125, "DatasourceVersionId": 2495170, "CreatorUserId": 4433335, "LicenseName": "CC BY-SA 3.0", "CreationDate": "07/22/2021 19:12:39", "VersionNumber": 8.0, "Title": "Simple/Normal Wikipedia Sections", "Slug": "simplenormal-wikipedia-sections", "Subtitle": "Simple and Normal Wikipedia Sections", "Description": NaN, "VersionNotes": "Added pretrained RoBERTa weights", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1455125, "CreatorUserId": 4433335, "OwnerUserId": 4433335.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2452800.0, "CurrentDatasourceVersionId": 2495170.0, "ForumId": 1474692, "Type": 2, "CreationDate": "07/08/2021 08:21:17", "LastActivityDate": "07/08/2021", "TotalViews": 2305, "TotalDownloads": 46, "TotalVotes": 5, "TotalKernels": 4}]
|
[{"Id": 4433335, "UserName": "markwijkhuizen", "DisplayName": "Mark Wijkhuizen", "RegisterDate": "02/04/2020", "PerformanceTier": 3}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from catboost import CatBoostClassifier, Pool
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import StratifiedKFold
FOLDS = 5
import pickle
import gzip
def pickle_save(object, filename, gzipping=True, protocol=4):
"""Save an object to a compressed disk file.
Works well with huge objects.
"""
open_f = gzip.GzipFile if gzipping else open
with open_f(filename, "wb") as f:
pickle.dump(object, f, protocol)
wp = pd.read_pickle(
"../input/simplenormal-wikipedia-sections/wikipedia_sections.pkl.xz"
)
data = wp[["text", "label"]].copy()
del wp
X_train, X_test, y_train, y_test = train_test_split(
data[[x for x in data.columns if x != "label"]],
data["label"],
test_size=0.3,
random_state=42,
)
def fit_model_classifier(train_pool, test_pool, **kwargs):
model = CatBoostClassifier(
task_type="GPU",
iterations=5000,
eval_metric="AUC",
od_type="Iter",
od_wait=500,
l2_leaf_reg=10,
bootstrap_type="Bernoulli",
subsample=0.7,
learning_rate=0.2,
**kwargs,
)
return model.fit(
train_pool, eval_set=test_pool, verbose=100, plot=False, use_best_model=True
)
def get_oof_classifier(n_folds, x_train, y, x_test, text_features, seeds):
ntrain = x_train.shape[0]
ntest = x_test.shape[0]
tpo = {
"tokenizers": [
{
"tokenizer_id": "Sense",
"separator_type": "BySense",
"lowercasing": "True",
"token_types": ["Word", "Number"],
}
],
"dictionaries": [
{
"dictionary_id": "Word",
"token_level_type": "Word",
"occurrence_lower_bound": "500",
"max_dictionary_size": "10000",
},
{
"dictionary_id": "Bigram",
"token_level_type": "Word",
"gram_order": "2",
"occurrence_lower_bound": "100",
"max_dictionary_size": "1000",
},
{
"dictionary_id": "Trigram",
"token_level_type": "Word",
"gram_order": "3",
"occurrence_lower_bound": "100",
"max_dictionary_size": "1000",
},
],
"feature_processing": {
"0": [
{
"tokenizers_names": ["Sense"],
"dictionaries_names": ["Word"],
"feature_calcers": ["BoW", "BM25"],
},
{
"tokenizers_names": ["Sense"],
"dictionaries_names": ["Bigram", "Trigram"],
"feature_calcers": ["BoW"],
},
]
},
}
oof_train = np.zeros((len(seeds), ntrain))
oof_test = np.zeros((ntest))
oof_test_skf = np.empty((len(seeds), n_folds, ntest))
test_pool = Pool(data=x_test, text_features=text_features)
models = {}
for iseed, seed in enumerate(seeds):
kf = StratifiedKFold(n_splits=n_folds, shuffle=True, random_state=seed)
for i, (tr_i, t_i) in enumerate(kf.split(x_train, y)):
print(f"\nSeed {seed}, Fold {i}")
x_tr = x_train.iloc[tr_i, :]
y_tr = y[tr_i]
x_te = x_train.iloc[t_i, :]
y_te = y[t_i]
train_pool = Pool(data=x_tr, label=y_tr, text_features=text_features)
valid_pool = Pool(data=x_te, label=y_te, text_features=text_features)
model = fit_model_classifier(
train_pool, valid_pool, random_seed=seed, text_processing=tpo
)
oof_train[iseed, t_i] = model.predict_proba(valid_pool)[:, 1]
oof_test_skf[iseed, i, :] = model.predict_proba(test_pool)[:, 1]
models[(seed, i)] = model
oof_test[:] = oof_test_skf.mean(axis=1).mean(axis=0)
oof_train = oof_train.mean(axis=0)
return oof_train, oof_test, models
oof_train, oof_test, models = get_oof_classifier(
n_folds=FOLDS,
x_train=X_train[0:100000],
y=y_train[0:100000].values,
x_test=X_test,
text_features=["text"],
seeds=[0, 42, 888],
)
roc_auc_score(y_test, oof_test)
pickle_save(models, "models.zpkl")
| false | 0 | 1,523 | 0 | 1,551 | 1,523 |
||
69506999
|
<jupyter_start><jupyter_text>Segmentation Full Body TikTok Dancing Dataset
# Segmentation Full Body TikTok Dancing
Dataset includes 2615 images of a segmented dancing people.
Videos of people dancing from [TikTok](https://www.tiktok.com/) were dowloaded and cut into frames. On each frame, all the dancing people were selected in Photoshop.
# Get the Dataset
This is just an example of the data.
If you need access to the entire dataset, contact us via [[email protected]](mailto:[email protected]) or leave a request on **https://trainingdata.pro/data-market**
# Image

# Content
There are 3 folders in the dataset:
- collages - collages of original images and images with segmentation
- images - original images
- masks - segmentation masks for the original images
**[TrainingData](https://trainingdata.pro)** provides high-quality data annotation tailored to your needs.
Kaggle dataset identifier: segmentation-full-body-tiktok-dancing-dataset
<jupyter_script>#!pip install cloud-tpu-client==0.10 https://storage.googleapis.com/tpu-pytorch/wheels/torch_xla-1.9-cp37-cp37m-linux_x86_64.whl
import tqdm
print(tqdm.__version__)
assert tqdm.__version__ >= "4.47.0", "tqdm version should be >=4.47.0"
import albumentations as A
assert A.__version__ >= "1.0.0", "albumentations version should be >=1.0.0"
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
from glob import glob
from sklearn.model_selection import train_test_split
path = "../input/segmentation-full-body-tiktok-dancing-dataset/segmentation_full_body_tik_tok_2615_img/"
images = sorted(glob(path + "images/*.png"))
segs = sorted(glob(path + "masks/*.png"))
data_dicts = [
{"image": image_name, "label": label_name}
for image_name, label_name in zip(images, segs)
]
print(len(data_dicts))
train_files, val_files = train_test_split(data_dicts, test_size=0.2, random_state=21)
len(train_files), len(val_files)
import cv2
image = cv2.imread(train_files[0]["image"])
mask = cv2.imread(train_files[0]["label"], 0)
print(image.shape)
print(mask.max(), mask.min())
fig, ax = plt.subplots(1, 2)
ax[0].imshow(image)
ax[1].imshow(mask, cmap="gray")
from pytorch_lightning import seed_everything, LightningModule, Trainer
from pytorch_lightning.callbacks import (
EarlyStopping,
ModelCheckpoint,
LearningRateMonitor,
)
from torch.utils.data import DataLoader, Dataset
from pytorch_lightning.loggers import TensorBoardLogger
from torch.optim.lr_scheduler import ReduceLROnPlateau, CosineAnnealingWarmRestarts
import torch.nn as nn
import torch
import torchvision
from torch.nn import functional as F
import albumentations as A
from albumentations.pytorch import ToTensorV2
aug = A.Compose(
[
A.Resize(512, 512),
A.HorizontalFlip(p=0.5),
A.VerticalFlip(p=0.1),
A.Rotate(10, border_mode=cv2.BORDER_CONSTANT, value=0, mask_value=0),
A.GaussNoise(p=0.1),
A.GlassBlur(p=0.1),
A.GridDropout(p=0.1),
A.Equalize(),
A.CoarseDropout(p=0.1),
A.Normalize(mean=(0), std=(1)),
ToTensorV2(p=1.0),
],
p=1.0,
)
class DataReader(Dataset):
def __init__(self, data, transform=None):
super(DataReader, self).__init__()
self.data = data
self.transform = transform
def __len__(self):
return len(self.data)
def __getitem__(self, index):
image_path = self.data[index]["image"]
mask_path = self.data[index]["label"]
image = cv2.imread(image_path)
mask = cv2.imread(mask_path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
mask = cv2.cvtColor(mask, cv2.COLOR_BGR2GRAY)
if self.transform:
transformed = self.transform(image=image, mask=mask)
image = transformed["image"]
mask = transformed["mask"]
mask = np.expand_dims(mask, 0) / 255
return image, mask
ds = DataReader(data=data_dicts, transform=aug)
loader = DataLoader(ds, batch_size=8, shuffle=True, num_workers=4)
batch = next(iter(loader))
print(batch[0].shape, batch[1].shape)
print(batch[1].max(), batch[1].min())
plt.figure()
grid_img = torchvision.utils.make_grid(batch[0], 4, 4)
plt.imshow(grid_img.permute(1, 2, 0))
plt.title("batch of images")
plt.figure()
grid_img = torchvision.utils.make_grid(batch[1], 4, 4)
plt.imshow(grid_img.permute(1, 2, 0) * 255)
plt.title("batch of masks")
from einops import rearrange
def dice_coef_multilabel(mask_pred, mask_gt):
def compute_dice_coefficient(mask_pred, mask_gt, smooth=0.0001):
"""Compute soerensen-dice coefficient.
compute the soerensen-dice coefficient between the ground truth mask `mask_gt`
and the predicted mask `mask_pred`.
Args:
mask_gt: 4-dim Numpy array of type bool. The ground truth mask. [B, 1, H, W]
mask_pred: 4-dim Numpy array of type bool. The predicted mask. [B, C, H, W]
Returns:
the dice coeffcient as float. If both masks are empty, the result is NaN
"""
volume_sum = mask_gt.sum() + mask_pred.sum()
volume_intersect = (mask_gt * mask_pred).sum()
return (2 * volume_intersect + smooth) / (volume_sum + smooth)
dice = 0
n_pred_ch = mask_pred.shape[1]
mask_pred = torch.softmax(mask_pred, 1)
mask_gt = F.one_hot(mask_gt.long(), num_classes=n_pred_ch) # create one hot vector
mask_gt = rearrange(
mask_gt, "d0 d1 d2 d3 d4 -> d0 (d1 d4) d2 d3 "
) # reshape one hot vector
for ind in range(1, n_pred_ch):
dice += compute_dice_coefficient(mask_gt[:, ind, :, :], mask_pred[:, ind, :, :])
return dice / n_pred_ch # taking average
from einops import rearrange
def dice_loss(
pred,
true,
softmax=True,
sigmoid=False,
one_hot=True,
background=True,
smooth=0.0001,
):
"""
pred: predicted values without applying any activation at the end
shape (B,C,H,W) example: (4, 59, 512, 512)
true: ground truth shape (B,1,H,W) example: (4, 1, 512, 512)
softmax: for multiclass
sigmoid: for binaryclass
one_hot: convert true values to one hot encoded
background: calculate background
"""
n_pred_ch = pred.shape[1]
if softmax:
assert n_pred_ch != 1, "single channel found"
pred = torch.softmax(pred, 1)
if sigmoid:
pred = torch.sigmoid(pred, 1)
if one_hot:
assert n_pred_ch != 1, "single channel found"
true = F.one_hot(true.long(), num_classes=n_pred_ch)
true = rearrange(true, "d0 d1 d2 d3 d4 -> d0 (d1 d4) d2 d3 ")
if background is False:
assert one_hot != True, "apply one hot encode "
true = true[:, 1:]
pred = pred[:, 1:]
reduce_axis = torch.arange(
1, len(true.shape)
).tolist() # reducing only spatial dimensions (not batch nor channels)
intersection = torch.sum(true * pred, dim=reduce_axis)
denominator = torch.sum(true, dim=reduce_axis) + torch.sum(pred, dim=reduce_axis)
dice = (2.0 * intersection + smooth) / (denominator + smooth)
return 1.0 - torch.mean(dice) # the batch and channel average
def comboloss(
logits,
true,
softmax=True,
sigmoid=False,
one_hot=True,
background=True,
smooth=1e-5,
epsilon=1e-7,
alpha=0.5,
beta=0.5,
):
"""
https://arxiv.org/pdf/1805.02798.pdf
https://github.com/asgsaeid/ComboLoss/blob/master/combo_loss.py
Combo Loss is weightage average of dice loss and modified
crossenropy loss to handle the class-imbalance problem, i.e.,
segmenting a small foreground from a large context/background,
while at the same time controlling the trade-off between F P and F N.
"""
n_pred_ch = logits.shape[1]
if softmax:
assert n_pred_ch != 1, "single channel found"
pred = torch.softmax(logits, 1)
if sigmoid:
pred = torch.sigmoid(logits, 1)
if one_hot:
assert n_pred_ch != 1, "single channel found"
true = F.one_hot(true.long(), num_classes=n_pred_ch)
true = rearrange(true, "d0 d1 d2 d3 d4 -> d0 (d1 d4) d2 d3 ")
if background is False:
assert one_hot != True, "apply one hot encode "
true = true[:, 1:]
pred = pred[:, 1:]
reduce_axis = torch.arange(
1, len(true.shape)
).tolist() # reducing only spatial dimensions (not batch nor channels)
intersection = torch.sum(true * pred, dim=reduce_axis)
denominator = torch.sum(true, dim=reduce_axis) + torch.sum(pred, dim=reduce_axis)
dice = (2.0 * intersection + smooth) / (denominator + smooth)
pred = torch.clamp(pred, epsilon, 1.0 - epsilon)
out = beta * (
(true - torch.log(pred)) + ((1 - beta) * (1.0 - true) * torch.log(1.0 - pred))
)
weighted_ce = -torch.mean(out, dim=reduce_axis)
dice = torch.mean(dice)
weighted_ce = torch.mean(weighted_ce)
# print((alpha * (1-dice)),weighted_ce)
combo = (alpha * weighted_ce) + (alpha * (1 - dice))
return combo
from einops import rearrange
def focal_dice_loss(pred, true, softmax=True, alpha=0.5, gamma=2):
"""
pred: predicted values without applying any activation at the end
shape (B,C,H,W) example: (4, 59, 512, 512)
true: ground truth shape (B,1,H,W) example: (4, 1, 512, 512)
"""
n_pred_ch = pred.shape[1]
if softmax:
assert n_pred_ch != 1, "single channel found"
pred = torch.softmax(pred, 1)
celoss = F.cross_entropy(pred, torch.squeeze(true, dim=1).long(), reduction="none")
celoss = torch.exp(-celoss)
focal_loss = alpha * (1 - celoss) ** gamma * celoss
focal_loss = torch.mean(focal_loss)
diceloss = dice_loss(
pred, true, softmax=False
) # softmax false, beacuase already applied
return 0.5 * focal_loss + 0.5 * diceloss
import segmentation_models_pytorch as smp
from torchmetrics.functional import dice_score
class OurModel(LightningModule):
def __init__(self):
super(OurModel, self).__init__()
# architecute
self.layer = smp.Unet(
encoder_name="efficientnet-b7", # choose encoder, e.g. mobilenet_v2 or efficientnet-b7
encoder_weights="imagenet", # use `imagenet` pre-trained weights for encoder initialization
in_channels=3, # model input channels (1 for gray-scale images, 3 for RGB, etc.)
classes=2, # model output channels (number of classes in your dataset)
# activation='softmax2d' # could be None for logits or 'softmax2d' for multiclass segmentation
)
# parameters
self.lr = 1e-3
self.batch_size = 6
self.numworker = 4
# self.criterion= smp.losses.DiceLoss(mode='multiclass')
def forward(self, x):
return self.layer(x)
def configure_optimizers(self):
opt = torch.optim.AdamW(self.parameters(), lr=self.lr, weight_decay=1e-5)
scheduler = CosineAnnealingWarmRestarts(
opt, T_0=10, T_mult=1, eta_min=1e-5, last_epoch=-1
)
return {"optimizer": opt, "lr_scheduler": scheduler}
# return opt
def train_dataloader(self):
ds = DataReader(data=train_files, transform=aug)
loader = DataLoader(
ds, batch_size=self.batch_size, shuffle=True, num_workers=self.numworker
)
return loader
def training_step(self, batch, batch_idx):
image, segment = batch[0], batch[1]
out = self(image)
loss = focal_dice_loss(out, segment)
dice = dice_coef_multilabel(out, segment)
self.log("train_loss", loss, on_step=True, on_epoch=True, prog_bar=True)
self.log("train_dice", dice, on_step=True, on_epoch=True, prog_bar=True)
return loss
def val_dataloader(self):
ds = DataReader(data=val_files, transform=aug)
loader = DataLoader(
ds, batch_size=self.batch_size, shuffle=False, num_workers=self.numworker
)
return loader
def validation_step(self, batch, batch_idx):
image, segment = batch[0], batch[1]
out = self(image)
loss = focal_dice_loss(out, segment)
dice = dice_coef_multilabel(out, segment)
self.log("val_dice", dice, on_step=True, on_epoch=True, prog_bar=True)
self.log("val_loss", loss, on_step=True, on_epoch=True, prog_bar=True)
return loss
model = OurModel()
lr_monitor = LearningRateMonitor(logging_interval="epoch")
logger = TensorBoardLogger("unet", name="logs")
checkpoint_callback = ModelCheckpoint(
monitor="val_loss", dirpath="unet", filename="{epoch}-{val_loss:.2f}-{val_dice:.2f}"
)
trainer = Trainer(
max_epochs=15,
auto_lr_find=True,
# tpu_cores=8,precision=16,
gpus=-1,
precision=16,
logger=logger,
accumulate_grad_batches=4,
stochastic_weight_avg=True,
progress_bar_refresh_rate=120,
# auto_scale_batch_size='binsearch',
# resume_from_checkpoint='',
callbacks=[checkpoint_callback, lr_monitor],
)
# lr_finder = trainer.tuner.lr_find(model)
# fig = lr_finder.plot(suggest=True)
# fig.show()
# new_lr = lr_finder.suggestion()
# model.hparams.lr = new_lr
# %%time
# trainer.tune(model)
trainer.fit(model)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506999.ipynb
|
segmentation-full-body-tiktok-dancing-dataset
|
tapakah68
|
[{"Id": 69506999, "ScriptId": 18978916, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1836841, "CreationDate": "07/31/2021 21:32:06", "VersionNumber": 1.0, "Title": "PL", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 325.0, "LinesInsertedFromPrevious": 325.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92763714, "KernelVersionId": 69506999, "SourceDatasetVersionId": 1932509}]
|
[{"Id": 1932509, "DatasetId": 1152755, "DatasourceVersionId": 1971110, "CreatorUserId": 618942, "LicenseName": "Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0)", "CreationDate": "02/11/2021 16:47:40", "VersionNumber": 1.0, "Title": "Segmentation Full Body TikTok Dancing Dataset", "Slug": "segmentation-full-body-tiktok-dancing-dataset", "Subtitle": "2615 images of a segmented dancing people", "Description": "# Segmentation Full Body TikTok Dancing\n\nDataset includes 2615 images of a segmented dancing people.\nVideos of people dancing from [TikTok](https://www.tiktok.com/) were dowloaded and cut into frames. On each frame, all the dancing people were selected in Photoshop.\n\n# Get the Dataset\nThis is just an example of the data. \nIf you need access to the entire dataset, contact us via [[email protected]](mailto:[email protected]) or leave a request on **https://trainingdata.pro/data-market**\n\n# Image\n\n\n\n# Content\nThere are 3 folders in the dataset:\n- collages - collages of original images and images with segmentation\n- images - original images\n- masks - segmentation masks for the original images\n\n**[TrainingData](https://trainingdata.pro)** provides high-quality data annotation tailored to your needs.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1152755, "CreatorUserId": 618942, "OwnerUserId": 618942.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5506926.0, "CurrentDatasourceVersionId": 5581346.0, "ForumId": 1170311, "Type": 2, "CreationDate": "02/11/2021 16:47:40", "LastActivityDate": "02/11/2021", "TotalViews": 10107, "TotalDownloads": 890, "TotalVotes": 29, "TotalKernels": 5}]
|
[{"Id": 618942, "UserName": "tapakah68", "DisplayName": "KUCEV ROMAN", "RegisterDate": "05/20/2016", "PerformanceTier": 2}]
|
#!pip install cloud-tpu-client==0.10 https://storage.googleapis.com/tpu-pytorch/wheels/torch_xla-1.9-cp37-cp37m-linux_x86_64.whl
import tqdm
print(tqdm.__version__)
assert tqdm.__version__ >= "4.47.0", "tqdm version should be >=4.47.0"
import albumentations as A
assert A.__version__ >= "1.0.0", "albumentations version should be >=1.0.0"
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
from glob import glob
from sklearn.model_selection import train_test_split
path = "../input/segmentation-full-body-tiktok-dancing-dataset/segmentation_full_body_tik_tok_2615_img/"
images = sorted(glob(path + "images/*.png"))
segs = sorted(glob(path + "masks/*.png"))
data_dicts = [
{"image": image_name, "label": label_name}
for image_name, label_name in zip(images, segs)
]
print(len(data_dicts))
train_files, val_files = train_test_split(data_dicts, test_size=0.2, random_state=21)
len(train_files), len(val_files)
import cv2
image = cv2.imread(train_files[0]["image"])
mask = cv2.imread(train_files[0]["label"], 0)
print(image.shape)
print(mask.max(), mask.min())
fig, ax = plt.subplots(1, 2)
ax[0].imshow(image)
ax[1].imshow(mask, cmap="gray")
from pytorch_lightning import seed_everything, LightningModule, Trainer
from pytorch_lightning.callbacks import (
EarlyStopping,
ModelCheckpoint,
LearningRateMonitor,
)
from torch.utils.data import DataLoader, Dataset
from pytorch_lightning.loggers import TensorBoardLogger
from torch.optim.lr_scheduler import ReduceLROnPlateau, CosineAnnealingWarmRestarts
import torch.nn as nn
import torch
import torchvision
from torch.nn import functional as F
import albumentations as A
from albumentations.pytorch import ToTensorV2
aug = A.Compose(
[
A.Resize(512, 512),
A.HorizontalFlip(p=0.5),
A.VerticalFlip(p=0.1),
A.Rotate(10, border_mode=cv2.BORDER_CONSTANT, value=0, mask_value=0),
A.GaussNoise(p=0.1),
A.GlassBlur(p=0.1),
A.GridDropout(p=0.1),
A.Equalize(),
A.CoarseDropout(p=0.1),
A.Normalize(mean=(0), std=(1)),
ToTensorV2(p=1.0),
],
p=1.0,
)
class DataReader(Dataset):
def __init__(self, data, transform=None):
super(DataReader, self).__init__()
self.data = data
self.transform = transform
def __len__(self):
return len(self.data)
def __getitem__(self, index):
image_path = self.data[index]["image"]
mask_path = self.data[index]["label"]
image = cv2.imread(image_path)
mask = cv2.imread(mask_path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
mask = cv2.cvtColor(mask, cv2.COLOR_BGR2GRAY)
if self.transform:
transformed = self.transform(image=image, mask=mask)
image = transformed["image"]
mask = transformed["mask"]
mask = np.expand_dims(mask, 0) / 255
return image, mask
ds = DataReader(data=data_dicts, transform=aug)
loader = DataLoader(ds, batch_size=8, shuffle=True, num_workers=4)
batch = next(iter(loader))
print(batch[0].shape, batch[1].shape)
print(batch[1].max(), batch[1].min())
plt.figure()
grid_img = torchvision.utils.make_grid(batch[0], 4, 4)
plt.imshow(grid_img.permute(1, 2, 0))
plt.title("batch of images")
plt.figure()
grid_img = torchvision.utils.make_grid(batch[1], 4, 4)
plt.imshow(grid_img.permute(1, 2, 0) * 255)
plt.title("batch of masks")
from einops import rearrange
def dice_coef_multilabel(mask_pred, mask_gt):
def compute_dice_coefficient(mask_pred, mask_gt, smooth=0.0001):
"""Compute soerensen-dice coefficient.
compute the soerensen-dice coefficient between the ground truth mask `mask_gt`
and the predicted mask `mask_pred`.
Args:
mask_gt: 4-dim Numpy array of type bool. The ground truth mask. [B, 1, H, W]
mask_pred: 4-dim Numpy array of type bool. The predicted mask. [B, C, H, W]
Returns:
the dice coeffcient as float. If both masks are empty, the result is NaN
"""
volume_sum = mask_gt.sum() + mask_pred.sum()
volume_intersect = (mask_gt * mask_pred).sum()
return (2 * volume_intersect + smooth) / (volume_sum + smooth)
dice = 0
n_pred_ch = mask_pred.shape[1]
mask_pred = torch.softmax(mask_pred, 1)
mask_gt = F.one_hot(mask_gt.long(), num_classes=n_pred_ch) # create one hot vector
mask_gt = rearrange(
mask_gt, "d0 d1 d2 d3 d4 -> d0 (d1 d4) d2 d3 "
) # reshape one hot vector
for ind in range(1, n_pred_ch):
dice += compute_dice_coefficient(mask_gt[:, ind, :, :], mask_pred[:, ind, :, :])
return dice / n_pred_ch # taking average
from einops import rearrange
def dice_loss(
pred,
true,
softmax=True,
sigmoid=False,
one_hot=True,
background=True,
smooth=0.0001,
):
"""
pred: predicted values without applying any activation at the end
shape (B,C,H,W) example: (4, 59, 512, 512)
true: ground truth shape (B,1,H,W) example: (4, 1, 512, 512)
softmax: for multiclass
sigmoid: for binaryclass
one_hot: convert true values to one hot encoded
background: calculate background
"""
n_pred_ch = pred.shape[1]
if softmax:
assert n_pred_ch != 1, "single channel found"
pred = torch.softmax(pred, 1)
if sigmoid:
pred = torch.sigmoid(pred, 1)
if one_hot:
assert n_pred_ch != 1, "single channel found"
true = F.one_hot(true.long(), num_classes=n_pred_ch)
true = rearrange(true, "d0 d1 d2 d3 d4 -> d0 (d1 d4) d2 d3 ")
if background is False:
assert one_hot != True, "apply one hot encode "
true = true[:, 1:]
pred = pred[:, 1:]
reduce_axis = torch.arange(
1, len(true.shape)
).tolist() # reducing only spatial dimensions (not batch nor channels)
intersection = torch.sum(true * pred, dim=reduce_axis)
denominator = torch.sum(true, dim=reduce_axis) + torch.sum(pred, dim=reduce_axis)
dice = (2.0 * intersection + smooth) / (denominator + smooth)
return 1.0 - torch.mean(dice) # the batch and channel average
def comboloss(
logits,
true,
softmax=True,
sigmoid=False,
one_hot=True,
background=True,
smooth=1e-5,
epsilon=1e-7,
alpha=0.5,
beta=0.5,
):
"""
https://arxiv.org/pdf/1805.02798.pdf
https://github.com/asgsaeid/ComboLoss/blob/master/combo_loss.py
Combo Loss is weightage average of dice loss and modified
crossenropy loss to handle the class-imbalance problem, i.e.,
segmenting a small foreground from a large context/background,
while at the same time controlling the trade-off between F P and F N.
"""
n_pred_ch = logits.shape[1]
if softmax:
assert n_pred_ch != 1, "single channel found"
pred = torch.softmax(logits, 1)
if sigmoid:
pred = torch.sigmoid(logits, 1)
if one_hot:
assert n_pred_ch != 1, "single channel found"
true = F.one_hot(true.long(), num_classes=n_pred_ch)
true = rearrange(true, "d0 d1 d2 d3 d4 -> d0 (d1 d4) d2 d3 ")
if background is False:
assert one_hot != True, "apply one hot encode "
true = true[:, 1:]
pred = pred[:, 1:]
reduce_axis = torch.arange(
1, len(true.shape)
).tolist() # reducing only spatial dimensions (not batch nor channels)
intersection = torch.sum(true * pred, dim=reduce_axis)
denominator = torch.sum(true, dim=reduce_axis) + torch.sum(pred, dim=reduce_axis)
dice = (2.0 * intersection + smooth) / (denominator + smooth)
pred = torch.clamp(pred, epsilon, 1.0 - epsilon)
out = beta * (
(true - torch.log(pred)) + ((1 - beta) * (1.0 - true) * torch.log(1.0 - pred))
)
weighted_ce = -torch.mean(out, dim=reduce_axis)
dice = torch.mean(dice)
weighted_ce = torch.mean(weighted_ce)
# print((alpha * (1-dice)),weighted_ce)
combo = (alpha * weighted_ce) + (alpha * (1 - dice))
return combo
from einops import rearrange
def focal_dice_loss(pred, true, softmax=True, alpha=0.5, gamma=2):
"""
pred: predicted values without applying any activation at the end
shape (B,C,H,W) example: (4, 59, 512, 512)
true: ground truth shape (B,1,H,W) example: (4, 1, 512, 512)
"""
n_pred_ch = pred.shape[1]
if softmax:
assert n_pred_ch != 1, "single channel found"
pred = torch.softmax(pred, 1)
celoss = F.cross_entropy(pred, torch.squeeze(true, dim=1).long(), reduction="none")
celoss = torch.exp(-celoss)
focal_loss = alpha * (1 - celoss) ** gamma * celoss
focal_loss = torch.mean(focal_loss)
diceloss = dice_loss(
pred, true, softmax=False
) # softmax false, beacuase already applied
return 0.5 * focal_loss + 0.5 * diceloss
import segmentation_models_pytorch as smp
from torchmetrics.functional import dice_score
class OurModel(LightningModule):
def __init__(self):
super(OurModel, self).__init__()
# architecute
self.layer = smp.Unet(
encoder_name="efficientnet-b7", # choose encoder, e.g. mobilenet_v2 or efficientnet-b7
encoder_weights="imagenet", # use `imagenet` pre-trained weights for encoder initialization
in_channels=3, # model input channels (1 for gray-scale images, 3 for RGB, etc.)
classes=2, # model output channels (number of classes in your dataset)
# activation='softmax2d' # could be None for logits or 'softmax2d' for multiclass segmentation
)
# parameters
self.lr = 1e-3
self.batch_size = 6
self.numworker = 4
# self.criterion= smp.losses.DiceLoss(mode='multiclass')
def forward(self, x):
return self.layer(x)
def configure_optimizers(self):
opt = torch.optim.AdamW(self.parameters(), lr=self.lr, weight_decay=1e-5)
scheduler = CosineAnnealingWarmRestarts(
opt, T_0=10, T_mult=1, eta_min=1e-5, last_epoch=-1
)
return {"optimizer": opt, "lr_scheduler": scheduler}
# return opt
def train_dataloader(self):
ds = DataReader(data=train_files, transform=aug)
loader = DataLoader(
ds, batch_size=self.batch_size, shuffle=True, num_workers=self.numworker
)
return loader
def training_step(self, batch, batch_idx):
image, segment = batch[0], batch[1]
out = self(image)
loss = focal_dice_loss(out, segment)
dice = dice_coef_multilabel(out, segment)
self.log("train_loss", loss, on_step=True, on_epoch=True, prog_bar=True)
self.log("train_dice", dice, on_step=True, on_epoch=True, prog_bar=True)
return loss
def val_dataloader(self):
ds = DataReader(data=val_files, transform=aug)
loader = DataLoader(
ds, batch_size=self.batch_size, shuffle=False, num_workers=self.numworker
)
return loader
def validation_step(self, batch, batch_idx):
image, segment = batch[0], batch[1]
out = self(image)
loss = focal_dice_loss(out, segment)
dice = dice_coef_multilabel(out, segment)
self.log("val_dice", dice, on_step=True, on_epoch=True, prog_bar=True)
self.log("val_loss", loss, on_step=True, on_epoch=True, prog_bar=True)
return loss
model = OurModel()
lr_monitor = LearningRateMonitor(logging_interval="epoch")
logger = TensorBoardLogger("unet", name="logs")
checkpoint_callback = ModelCheckpoint(
monitor="val_loss", dirpath="unet", filename="{epoch}-{val_loss:.2f}-{val_dice:.2f}"
)
trainer = Trainer(
max_epochs=15,
auto_lr_find=True,
# tpu_cores=8,precision=16,
gpus=-1,
precision=16,
logger=logger,
accumulate_grad_batches=4,
stochastic_weight_avg=True,
progress_bar_refresh_rate=120,
# auto_scale_batch_size='binsearch',
# resume_from_checkpoint='',
callbacks=[checkpoint_callback, lr_monitor],
)
# lr_finder = trainer.tuner.lr_find(model)
# fig = lr_finder.plot(suggest=True)
# fig.show()
# new_lr = lr_finder.suggestion()
# model.hparams.lr = new_lr
# %%time
# trainer.tune(model)
trainer.fit(model)
| false | 0 | 3,890 | 0 | 4,250 | 3,890 |
||
69506408
|
<jupyter_start><jupyter_text>roberta-model-objects
Kaggle dataset identifier: robertamodelobjects
<jupyter_script># # Settings
# CONTROLS
MODEL_PREFIX = "V01"
MODEL_NUMBER = MODEL_PREFIX[-2:]
MODEL_NAME = "roberta" # options include 'xlm' or 'distilbert' or 'roberta'
ON_KAGGLE = True
MAX_SEQ_LEN = 200
TRAIN_SPLIT_RATIO = 0.2
# for the current setup NUM_FOLDS * NUM_EPOCHS = 135 = 3 * 3 * 3 * 5 with per epoch per fold time = 1.33 seconds
NUM_EPOCHS = [32]
NUM_FOLDS = 5
MIN_LR = 1e-7
MAX_LR = 5e-3
CLR_STEP_SIZE = 2
DROPOUT = 0.3
if ON_KAGGLE:
BATCH_SIZE = 12
PREDICT_BATCH_SIZE = 128
else:
BATCH_SIZE = 16
PREDICT_BATCH_SIZE = 256
RETRAIN_TRANSFORMER = True
USE_LOGITS = True
USE_MINMAXSCALING = False
RUN_ON_SAMPLE = 0
print(
*[
"RE_TRFMR",
"T" if RETRAIN_TRANSFORMER else "F",
"LOGITS",
"T" if USE_LOGITS else "F",
"MMS",
"T" if USE_MINMAXSCALING else "F",
],
sep="-",
)
if ON_KAGGLE:
RESULTS_DIR = "../working/"
DATA_DIR = "../input/commonlitreadabilityprize/"
if MODEL_NAME == "roberta":
MODEL_DIR = "../input/robertamodelobjects/"
else:
MODEL_DIR = "../input/tf-distilbert-base-multilingual-cased/"
else:
PATH = ".."
RESULTS_DIR = os.path.join(PATH, "results")
DATA_DIR = os.path.join(PATH, "data")
if MODEL_NAME == "xlm":
MODEL_DIR = os.path.join(PATH, "models", "robertamodelobjects")
else:
MODEL_DIR = os.path.join(PATH, "models", "distilbert-base-multilingual-cased")
# # Libraries
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn.model_selection import StratifiedKFold, train_test_split, KFold
from sklearn.preprocessing import MinMaxScaler
from sklearn.utils import class_weight
import pickle, os, sys, re, json, gc
from time import time, ctime
from pprint import pprint
from collections import Counter
import tensorflow as tf
from tensorflow.keras import Model, Input
from tensorflow.keras.layers import (
Conv1D,
Conv2D,
LSTM,
Embedding,
Dense,
concatenate,
MaxPooling2D,
Softmax,
Flatten,
)
from tensorflow.keras.layers import (
BatchNormalization,
Dropout,
Reshape,
Activation,
Bidirectional,
TimeDistributed,
)
from tensorflow.keras.layers import RepeatVector, Multiply, Layer, LeakyReLU, Subtract
from tensorflow.keras.activations import softmax
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras import initializers, regularizers, constraints
from tensorflow.keras.callbacks import *
from tensorflow.keras.callbacks import CSVLogger, ModelCheckpoint
import tensorflow.keras.backend as K
from tensorflow.keras.losses import SparseCategoricalCrossentropy
from tensorflow.keras.models import save_model, load_model
from tensorflow.keras.utils import to_categorical
from tensorflow.data import Dataset
import tokenizers, transformers
from transformers import *
import tensorflow_addons as tfa
from tensorflow_addons.optimizers import TriangularCyclicalLearningRate
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
import os, sys, pickle
from time import time, ctime
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.model_selection import cross_val_score, KFold
from sklearn.metrics import mean_squared_error
import xgboost as xgb
from hyperopt import hp, tpe
from hyperopt.fmin import fmin
from hyperopt import STATUS_OK
from hyperopt import Trials
from functools import partial
seeded_value = 12345
pd.set_option("display.max_colwidth", None)
np.random.seed(seeded_value)
tf.random.set_seed(seeded_value)
def sigmoid(X):
return 1 / (1 + np.exp(-X))
print(ctime(time()))
print([tf.__version__, transformers.__version__, tokenizers.__version__])
# Limiting GPU memory growth
# By default, TensorFlow maps nearly all of the GPU memory of all GPUs (subject to
# CUDA_VISIBLE_DEVICES) visible to the process. This is done to more efficiently use the relatively precious GPU memory resources on the devices by reducing memory fragmentation. To limit TensorFlow to a specific set of GPUs we use the tf.config.experimental.set_visible_devices method.
print(tf.config.experimental.list_logical_devices("CPU"))
print(tf.config.experimental.list_logical_devices("GPU"))
print(tf.config.experimental.list_physical_devices("CPU"))
print(tf.config.experimental.list_physical_devices("GPU"))
physical_devices = tf.config.list_physical_devices("GPU")
try:
tf.config.experimental.set_memory_growth(physical_devices[0], True)
except:
# Invalid device or cannot modify virtual devices once initialized.
pass
# # Import Data
data = pd.read_csv(DATA_DIR + "train.csv")
test = pd.read_csv(DATA_DIR + "test.csv")
print(data.columns, test.columns)
if RUN_ON_SAMPLE:
data = data.sample(RUN_ON_SAMPLE)
data = data.reset_index(drop=True)
data.columns = ["id", "url_legal", "license", "excerpt", "target", "standard_error"]
test.columns = ["id", "url_legal", "license", "excerpt"]
REQ_COLS = ["id", "url_legal", "license", "excerpt", "target"]
data["excerpt"] = data["excerpt"].astype(str)
test["excerpt"] = test["excerpt"].astype(str)
data.shape
data.sample(2)
print(dict(data["target"].describe()))
target_histogram = plt.hist(data["target"], bins=100)
excerpt_histogram = plt.hist(
data["excerpt"].apply(lambda x: len(x.split(" "))), bins=100
)
# # Tokenizer, Config & Model Initialization
# 1. https://arxiv.org/pdf/1911.02116.pdf
# 2. https://huggingface.co/transformers/model_doc/xlmroberta.html
MODEL_DIR = "../input/robertamodelobjects/"
if MODEL_NAME == "xlm":
model_tokenizer = transformers.XLMRobertaTokenizer.from_pretrained(MODEL_DIR)
elif MODEL_NAME == "roberta":
model_tokenizer = transformers.RobertaTokenizer.from_pretrained(MODEL_DIR)
else:
model_tokenizer = transformers.DistilBertTokenizer.from_pretrained(MODEL_DIR)
with open(MODEL_DIR + "special_tokens_map.json") as f:
special_tokens = json.load(f)
model_tokenizer.add_special_tokens(special_tokens)
VOCAB_SIZE = model_tokenizer.vocab_size
print(VOCAB_SIZE)
# # Tokenization
def find_max_len():
X_tokens_ = []
for t in data.excerpt.tolist():
encoded_text_ = model_tokenizer.encode_plus(
t, padding="do_not_pad", truncation="do_not_truncate"
)
X_tokens_.append(encoded_text_["input_ids"])
return max([len(el) for el in X_tokens_])
MAX_SEQ_LEN = find_max_len()
def preprocess_data(data, MAX_SEQ_LEN):
X_tokens, X_att = [], []
for t in data.excerpt.tolist():
encoded_text = model_tokenizer.encode_plus(
t, padding="max_length", truncation=True, max_length=MAX_SEQ_LEN
)
X_tokens.append(encoded_text["input_ids"])
X_att.append(encoded_text["attention_mask"])
X_tokens, X_att = X_tokens, X_att
Y = data["target"].tolist()
X_tokens, X_att, Y = (
np.array(X_tokens, dtype=np.int32),
np.array(X_att, dtype=np.int32),
np.array(Y, dtype=np.float32),
)
return X_tokens, X_att, Y
X_tokens, X_att, Y = preprocess_data(data, MAX_SEQ_LEN)
print(
"\n \t Sample\n",
len(X_tokens),
"\t: X_tokens ",
"\n",
len(X_att),
"\t: X_att ",
"\n",
len(Y),
"\t: Y ",
"\n",
)
X_tokens_test, X_att_test = [], []
for t in test.excerpt.tolist():
encoded_text = model_tokenizer.encode_plus(
t, padding="max_length", truncation=True, max_length=MAX_SEQ_LEN
)
X_tokens_test.append(encoded_text["input_ids"])
X_att_test.append(encoded_text["attention_mask"])
X_tokens_test, X_att_test = np.array(X_tokens_test, dtype=np.int32), np.array(
X_att_test, dtype=np.int32
)
print(
"\n",
len(X_tokens_test),
"\t: X_tokens_test ",
"\n",
len(X_att_test),
"\t: X_att_test ",
"\n",
)
# # Model Specifications
def build_model():
input_sequences = Input((MAX_SEQ_LEN), dtype=tf.int32, name="words")
input_att_flags = Input((MAX_SEQ_LEN), dtype=tf.int32, name="att_flags")
MODEL_DIR = "../input/robertamodelobjects/"
if MODEL_NAME == "xlm":
config = transformers.XLMRobertaConfig.from_pretrained(MODEL_DIR)
model = transformers.TFXLMRobertaModel.from_pretrained(MODEL_DIR, config=config)
x = model({"inputs": input_sequences, "attention_mask": input_att_flags})
elif MODEL_NAME == "roberta":
config = transformers.RobertaConfig.from_pretrained(MODEL_DIR)
model = transformers.TFRobertaModel.from_pretrained(MODEL_DIR, config=config)
x = model({"inputs": input_sequences, "attention_mask": input_att_flags})
else:
config = transformers.DistilBertConfig.from_pretrained(MODEL_DIR)
model = transformers.TFDistilBertModel.from_pretrained(MODEL_DIR, config=config)
x = model({"inputs": input_sequences, "attention_mask": input_att_flags})
model_ = Model([input_att_flags, input_sequences], x)
return model_
model = build_model()
X_ = model.predict(
x={"att_flags": X_att, "words": X_tokens}, batch_size=PREDICT_BATCH_SIZE
)
X_test_ = model.predict(
x={"att_flags": X_att_test, "words": X_tokens_test}, batch_size=PREDICT_BATCH_SIZE
)
X_.keys(), X_["last_hidden_state"].shape
X, y, X_test = (
X_["last_hidden_state"].mean(axis=1),
data.target.values,
X_test_["last_hidden_state"].mean(axis=1),
)
X.shape, y.shape, X_test.shape
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, test_size=0.33, random_state=42
)
for i in [X_train, X_valid, y_train, y_valid]:
print(i.shape)
from sklearn.neighbors import KNeighborsRegressor
knn_model = KNeighborsRegressor(
n_neighbors=30,
weights="uniform",
algorithm="auto",
leaf_size=30,
p=2,
metric="manhattan",
metric_params=None,
n_jobs=-1,
)
knn_model.fit(X_train, y_train)
np.sqrt(mean_squared_error(y_valid, knn_model.predict(X_valid)))
from sklearn.cluster import KMeans
sse = {}
for k in range(2, 100):
kmeans = KMeans(n_clusters=k, max_iter=1000).fit(X)
sse[k] = kmeans.inertia_
plt.figure()
plt.plot(list(sse.keys()), list(sse.values()))
plt.xlabel("Number of cluster")
plt.ylabel("SSE")
plt.show()
Kmeans_model = KMeans(n_clusters=500, random_state=np.random.randint(0, 100_000_000))
Kmeans_model.fit(X_train)
Kmeans_model.labels_
lookup_matrix_train = pd.DataFrame(
{"targets": y_train, "cluster_memberships": Kmeans_model.labels_}
)
lookup_matrix_train.groupby("cluster_memberships")[["targets"]].describe()
lookup_dict = (
lookup_matrix_train.groupby("cluster_memberships", as_index=True)[["targets"]]
.mean()
.to_dict("dict")["targets"]
)
pred_valid = np.array(list(map(lookup_dict.get, Kmeans_model.predict(X_valid))))
np.sqrt(mean_squared_error(y_valid, pred_valid))
seeded_value = 12345
NUM_FOLDS = 3
NUM_EVALS = 40
NUM_TREES = 100
def objective(params, X, y):
params = {
"max_depth": int(params["max_depth"]),
"gamma": params["gamma"],
"colsample_bytree": params["colsample_bytree"],
"learning_rate": params["learning_rate"],
}
model = xgb.XGBRegressor(
n_estimators=NUM_TREES,
verbosity=1,
objective="reg:squarederror",
random_state=seeded_value,
n_jobs=2,
# tree_method= 'gpu_hist',
**params,
)
score = cross_val_score(
estimator=model,
X=X,
y=y,
scoring="neg_mean_squared_error",
cv=KFold(
n_splits=NUM_FOLDS,
shuffle=True,
random_state=np.random.randint(0, 10_000_000),
),
).mean()
print("\tRMSE Score {:.3f} params {}".format(-1.0 * score, params))
del model
return {"loss": -1.0 * score, "params": params, "status": STATUS_OK}
def hpt(X, y):
bayes_trials = Trials()
space = {
"max_depth": hp.quniform("max_depth", 2, 8, 1),
"colsample_bytree": hp.uniform("colsample_bytree", 0.3, 1.0),
"gamma": hp.uniform("gamma", 0.0, 0.5),
"learning_rate": hp.loguniform("learning_rate", np.log(0.001), np.log(0.5)),
}
partial_objective = partial(objective, X=X, y=y)
best = fmin(
fn=partial_objective,
space=space,
algo=tpe.suggest,
max_evals=NUM_EVALS,
trials=bayes_trials,
)
print("\tbest: ", best)
optimal_params = {
"max_depth": int(best["max_depth"]),
"gamma": best["gamma"],
"colsample_bytree": best["colsample_bytree"],
"learning_rate": best["learning_rate"],
}
print("\toptimal_params", optimal_params)
return optimal_params
def retrain_after_hpt(optimal_params, X, y):
tuned_model = xgb.XGBRegressor(
**{
"n_estimators": NUM_TREES,
"verbosity": 1,
"objective": "reg:squarederror",
"random_state": seeded_value,
"n_jobs": 12,
# 'tree_method': 'gpu_hist',
**optimal_params,
}
)
tuned_model.fit(X=X, y=y)
return tuned_model
def evaluate(y_train, pred_train, y_valid, pred_valid, fold_num, prefix):
trains = pd.DataFrame({"target": y_train, "preds": pred_train})
valids = pd.DataFrame({"target": y_valid, "preds": pred_valid})
trains.to_csv(f"{prefix}-train-preds-{fold_num}.csv", index=False)
valids.to_csv(f"{prefix}-valid-preds-{fold_num}.csv", index=False)
print("Training Correlations: ", "\n\t\t\t", trains[["target", "preds"]].corr())
print(
"Training np.sign: ",
"\n\t\t\t",
np.sign(trains["target"] - trains["preds"]).value_counts(),
)
print(
"Training RMSE: ",
"\n\t\t\t",
np.sqrt(mean_squared_error(trains["target"], trains["preds"])),
)
print("Training distribution stats: ", "\n\t\t\t", valids.describe())
print("Validation Correlations: ", "\n\t\t\t", valids[["target", "preds"]].corr())
print(
"Validation np.sign: ",
"\n\t\t\t",
np.sign(valids["target"] - valids["preds"]).value_counts(),
)
print(
"Validation RMSE: ",
"\n\t\t\t",
np.sqrt(mean_squared_error(valids["target"], valids["preds"])),
)
print("Validation distribution stats: ", "\n\t\t\t", trains.describe())
def apply_minmax_(y_ref, pred_y):
min_ = y_ref.min()
max_ = y_ref.max()
pred_min_ = pred_y.min()
pred_max_ = pred_y.max()
pred_norm = (pred_y - pred_min_) / (pred_max_ - pred_min_)
y_pred_adj = (pred_norm * (max_ - min_)) + min_
return y_pred_adj
def apply_norm_(y_ref, pred_y):
mean_ = y_ref.mean()
std_ = y_ref.std()
pred_mean_ = pred_y.mean()
pred_std_ = pred_y.std()
pred_norm = (pred_y - pred_mean_) / pred_std_
y_pred_adj = pred_norm * std_ + mean_
return y_pred_adj
ENABLE_POSTPROCESSING = True
kf = KFold(n_splits=NUM_FOLDS, shuffle=True, random_state=seeded_value)
for fold_num, (t_i, v_i) in enumerate(kf.split(X)):
print("=" * 15, f" START: {str(fold_num)} ", "=" * 15)
if fold_num == 0:
predictions = {}
predictions_norm = {}
predictions_minmax = {}
X_train, y_train = X[t_i], y[t_i]
X_valid, y_valid = X[v_i], y[v_i]
optimal_params = hpt(X_train, y_train)
eval_model = retrain_after_hpt(optimal_params, X_train, y_train)
eval_model.save_model(
f"eval_model_xgb_{fold_num}.txt"
) # loaded_model = xgb.XGBRegressor(); loaded_model.load_model("path-to-file")
pred_train = eval_model.predict(X_train)
pred_valid = eval_model.predict(X_valid)
print("\tTrain RMSE:", mean_squared_error(y_train, pred_train, squared=False))
print("\tValid RMSE:", mean_squared_error(y_valid, pred_valid, squared=False))
evaluate(y_train, pred_train, y_valid, pred_valid, fold_num, "original")
if ENABLE_POSTPROCESSING:
print(
"=" * 25,
"After Normalizing Predictions on Training and Validation Datasets",
"=" * 25,
)
evaluate(
y_train,
apply_norm_(y_train, pred_train),
y_valid,
apply_norm_(y_train, pred_valid),
fold_num,
"norm",
)
print(
"=" * 25,
"After MinMaxScaling Predictions on Training and Validation Datasets",
"=" * 25,
)
evaluate(
y_train,
apply_minmax_(y_train, pred_train),
y_valid,
apply_minmax_(y_train, pred_valid),
fold_num,
"minmax",
)
model = retrain_after_hpt(optimal_params, X, y)
model.save_model(
f"model_xgb_{fold_num}.txt"
) # loaded_model = xgb.XGBRegressor(); loaded_model.load_model("path-to-file")
pred_test = model.predict(X_test)
predictions["F_" + str(fold_num)] = pred_test
if ENABLE_POSTPROCESSING:
predictions_norm["F_" + str(fold_num)] = apply_norm_(y, pred_test)
predictions_minmax["F_" + str(fold_num)] = apply_minmax_(y, pred_test)
print("=" * 15, " COMPLETE ", "=" * 15)
final_preds_to_submit = (
predictions_minmax # predictions, predictions_norm, predictions_minmax
)
test_results = pd.DataFrame(
{k: v.flatten() for k, v in final_preds_to_submit.items()},
columns=["F_" + str(num) for num in range(NUM_FOLDS)],
).values
test_results = test_results.mean(axis=1)
test["target"] = test_results
test[["id", "target"]].to_csv("submission.csv", index=False)
test[["id", "target"]].head()
print(ctime(time()))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506408.ipynb
|
robertamodelobjects
|
deepaksadulla
|
[{"Id": 69506408, "ScriptId": 18961894, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 406530, "CreationDate": "07/31/2021 21:17:24", "VersionNumber": 11.0, "Title": "roberta inference with CV pairwise kmeans", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 528.0, "LinesInsertedFromPrevious": 60.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 468.0, "LinesInsertedFromFork": 209.0, "LinesDeletedFromFork": 85.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 319.0, "TotalVotes": 0}]
|
[{"Id": 92761778, "KernelVersionId": 69506408, "SourceDatasetVersionId": 1150198}]
|
[{"Id": 1150198, "DatasetId": 649649, "DatasourceVersionId": 1180913, "CreatorUserId": 406530, "LicenseName": "Unknown", "CreationDate": "05/12/2020 10:24:53", "VersionNumber": 1.0, "Title": "roberta-model-objects", "Slug": "robertamodelobjects", "Subtitle": "Roberta Transformer Models & Tokenizer Objects", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 649649, "CreatorUserId": 406530, "OwnerUserId": 406530.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1150198.0, "CurrentDatasourceVersionId": 1180913.0, "ForumId": 663998, "Type": 2, "CreationDate": "05/12/2020 10:24:53", "LastActivityDate": "05/12/2020", "TotalViews": 1743, "TotalDownloads": 5, "TotalVotes": 1, "TotalKernels": 1}]
|
[{"Id": 406530, "UserName": "deepaksadulla", "DisplayName": "DeepakSadulla", "RegisterDate": "08/26/2015", "PerformanceTier": 1}]
|
# # Settings
# CONTROLS
MODEL_PREFIX = "V01"
MODEL_NUMBER = MODEL_PREFIX[-2:]
MODEL_NAME = "roberta" # options include 'xlm' or 'distilbert' or 'roberta'
ON_KAGGLE = True
MAX_SEQ_LEN = 200
TRAIN_SPLIT_RATIO = 0.2
# for the current setup NUM_FOLDS * NUM_EPOCHS = 135 = 3 * 3 * 3 * 5 with per epoch per fold time = 1.33 seconds
NUM_EPOCHS = [32]
NUM_FOLDS = 5
MIN_LR = 1e-7
MAX_LR = 5e-3
CLR_STEP_SIZE = 2
DROPOUT = 0.3
if ON_KAGGLE:
BATCH_SIZE = 12
PREDICT_BATCH_SIZE = 128
else:
BATCH_SIZE = 16
PREDICT_BATCH_SIZE = 256
RETRAIN_TRANSFORMER = True
USE_LOGITS = True
USE_MINMAXSCALING = False
RUN_ON_SAMPLE = 0
print(
*[
"RE_TRFMR",
"T" if RETRAIN_TRANSFORMER else "F",
"LOGITS",
"T" if USE_LOGITS else "F",
"MMS",
"T" if USE_MINMAXSCALING else "F",
],
sep="-",
)
if ON_KAGGLE:
RESULTS_DIR = "../working/"
DATA_DIR = "../input/commonlitreadabilityprize/"
if MODEL_NAME == "roberta":
MODEL_DIR = "../input/robertamodelobjects/"
else:
MODEL_DIR = "../input/tf-distilbert-base-multilingual-cased/"
else:
PATH = ".."
RESULTS_DIR = os.path.join(PATH, "results")
DATA_DIR = os.path.join(PATH, "data")
if MODEL_NAME == "xlm":
MODEL_DIR = os.path.join(PATH, "models", "robertamodelobjects")
else:
MODEL_DIR = os.path.join(PATH, "models", "distilbert-base-multilingual-cased")
# # Libraries
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn.model_selection import StratifiedKFold, train_test_split, KFold
from sklearn.preprocessing import MinMaxScaler
from sklearn.utils import class_weight
import pickle, os, sys, re, json, gc
from time import time, ctime
from pprint import pprint
from collections import Counter
import tensorflow as tf
from tensorflow.keras import Model, Input
from tensorflow.keras.layers import (
Conv1D,
Conv2D,
LSTM,
Embedding,
Dense,
concatenate,
MaxPooling2D,
Softmax,
Flatten,
)
from tensorflow.keras.layers import (
BatchNormalization,
Dropout,
Reshape,
Activation,
Bidirectional,
TimeDistributed,
)
from tensorflow.keras.layers import RepeatVector, Multiply, Layer, LeakyReLU, Subtract
from tensorflow.keras.activations import softmax
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras import initializers, regularizers, constraints
from tensorflow.keras.callbacks import *
from tensorflow.keras.callbacks import CSVLogger, ModelCheckpoint
import tensorflow.keras.backend as K
from tensorflow.keras.losses import SparseCategoricalCrossentropy
from tensorflow.keras.models import save_model, load_model
from tensorflow.keras.utils import to_categorical
from tensorflow.data import Dataset
import tokenizers, transformers
from transformers import *
import tensorflow_addons as tfa
from tensorflow_addons.optimizers import TriangularCyclicalLearningRate
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
import os, sys, pickle
from time import time, ctime
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.model_selection import cross_val_score, KFold
from sklearn.metrics import mean_squared_error
import xgboost as xgb
from hyperopt import hp, tpe
from hyperopt.fmin import fmin
from hyperopt import STATUS_OK
from hyperopt import Trials
from functools import partial
seeded_value = 12345
pd.set_option("display.max_colwidth", None)
np.random.seed(seeded_value)
tf.random.set_seed(seeded_value)
def sigmoid(X):
return 1 / (1 + np.exp(-X))
print(ctime(time()))
print([tf.__version__, transformers.__version__, tokenizers.__version__])
# Limiting GPU memory growth
# By default, TensorFlow maps nearly all of the GPU memory of all GPUs (subject to
# CUDA_VISIBLE_DEVICES) visible to the process. This is done to more efficiently use the relatively precious GPU memory resources on the devices by reducing memory fragmentation. To limit TensorFlow to a specific set of GPUs we use the tf.config.experimental.set_visible_devices method.
print(tf.config.experimental.list_logical_devices("CPU"))
print(tf.config.experimental.list_logical_devices("GPU"))
print(tf.config.experimental.list_physical_devices("CPU"))
print(tf.config.experimental.list_physical_devices("GPU"))
physical_devices = tf.config.list_physical_devices("GPU")
try:
tf.config.experimental.set_memory_growth(physical_devices[0], True)
except:
# Invalid device or cannot modify virtual devices once initialized.
pass
# # Import Data
data = pd.read_csv(DATA_DIR + "train.csv")
test = pd.read_csv(DATA_DIR + "test.csv")
print(data.columns, test.columns)
if RUN_ON_SAMPLE:
data = data.sample(RUN_ON_SAMPLE)
data = data.reset_index(drop=True)
data.columns = ["id", "url_legal", "license", "excerpt", "target", "standard_error"]
test.columns = ["id", "url_legal", "license", "excerpt"]
REQ_COLS = ["id", "url_legal", "license", "excerpt", "target"]
data["excerpt"] = data["excerpt"].astype(str)
test["excerpt"] = test["excerpt"].astype(str)
data.shape
data.sample(2)
print(dict(data["target"].describe()))
target_histogram = plt.hist(data["target"], bins=100)
excerpt_histogram = plt.hist(
data["excerpt"].apply(lambda x: len(x.split(" "))), bins=100
)
# # Tokenizer, Config & Model Initialization
# 1. https://arxiv.org/pdf/1911.02116.pdf
# 2. https://huggingface.co/transformers/model_doc/xlmroberta.html
MODEL_DIR = "../input/robertamodelobjects/"
if MODEL_NAME == "xlm":
model_tokenizer = transformers.XLMRobertaTokenizer.from_pretrained(MODEL_DIR)
elif MODEL_NAME == "roberta":
model_tokenizer = transformers.RobertaTokenizer.from_pretrained(MODEL_DIR)
else:
model_tokenizer = transformers.DistilBertTokenizer.from_pretrained(MODEL_DIR)
with open(MODEL_DIR + "special_tokens_map.json") as f:
special_tokens = json.load(f)
model_tokenizer.add_special_tokens(special_tokens)
VOCAB_SIZE = model_tokenizer.vocab_size
print(VOCAB_SIZE)
# # Tokenization
def find_max_len():
X_tokens_ = []
for t in data.excerpt.tolist():
encoded_text_ = model_tokenizer.encode_plus(
t, padding="do_not_pad", truncation="do_not_truncate"
)
X_tokens_.append(encoded_text_["input_ids"])
return max([len(el) for el in X_tokens_])
MAX_SEQ_LEN = find_max_len()
def preprocess_data(data, MAX_SEQ_LEN):
X_tokens, X_att = [], []
for t in data.excerpt.tolist():
encoded_text = model_tokenizer.encode_plus(
t, padding="max_length", truncation=True, max_length=MAX_SEQ_LEN
)
X_tokens.append(encoded_text["input_ids"])
X_att.append(encoded_text["attention_mask"])
X_tokens, X_att = X_tokens, X_att
Y = data["target"].tolist()
X_tokens, X_att, Y = (
np.array(X_tokens, dtype=np.int32),
np.array(X_att, dtype=np.int32),
np.array(Y, dtype=np.float32),
)
return X_tokens, X_att, Y
X_tokens, X_att, Y = preprocess_data(data, MAX_SEQ_LEN)
print(
"\n \t Sample\n",
len(X_tokens),
"\t: X_tokens ",
"\n",
len(X_att),
"\t: X_att ",
"\n",
len(Y),
"\t: Y ",
"\n",
)
X_tokens_test, X_att_test = [], []
for t in test.excerpt.tolist():
encoded_text = model_tokenizer.encode_plus(
t, padding="max_length", truncation=True, max_length=MAX_SEQ_LEN
)
X_tokens_test.append(encoded_text["input_ids"])
X_att_test.append(encoded_text["attention_mask"])
X_tokens_test, X_att_test = np.array(X_tokens_test, dtype=np.int32), np.array(
X_att_test, dtype=np.int32
)
print(
"\n",
len(X_tokens_test),
"\t: X_tokens_test ",
"\n",
len(X_att_test),
"\t: X_att_test ",
"\n",
)
# # Model Specifications
def build_model():
input_sequences = Input((MAX_SEQ_LEN), dtype=tf.int32, name="words")
input_att_flags = Input((MAX_SEQ_LEN), dtype=tf.int32, name="att_flags")
MODEL_DIR = "../input/robertamodelobjects/"
if MODEL_NAME == "xlm":
config = transformers.XLMRobertaConfig.from_pretrained(MODEL_DIR)
model = transformers.TFXLMRobertaModel.from_pretrained(MODEL_DIR, config=config)
x = model({"inputs": input_sequences, "attention_mask": input_att_flags})
elif MODEL_NAME == "roberta":
config = transformers.RobertaConfig.from_pretrained(MODEL_DIR)
model = transformers.TFRobertaModel.from_pretrained(MODEL_DIR, config=config)
x = model({"inputs": input_sequences, "attention_mask": input_att_flags})
else:
config = transformers.DistilBertConfig.from_pretrained(MODEL_DIR)
model = transformers.TFDistilBertModel.from_pretrained(MODEL_DIR, config=config)
x = model({"inputs": input_sequences, "attention_mask": input_att_flags})
model_ = Model([input_att_flags, input_sequences], x)
return model_
model = build_model()
X_ = model.predict(
x={"att_flags": X_att, "words": X_tokens}, batch_size=PREDICT_BATCH_SIZE
)
X_test_ = model.predict(
x={"att_flags": X_att_test, "words": X_tokens_test}, batch_size=PREDICT_BATCH_SIZE
)
X_.keys(), X_["last_hidden_state"].shape
X, y, X_test = (
X_["last_hidden_state"].mean(axis=1),
data.target.values,
X_test_["last_hidden_state"].mean(axis=1),
)
X.shape, y.shape, X_test.shape
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, test_size=0.33, random_state=42
)
for i in [X_train, X_valid, y_train, y_valid]:
print(i.shape)
from sklearn.neighbors import KNeighborsRegressor
knn_model = KNeighborsRegressor(
n_neighbors=30,
weights="uniform",
algorithm="auto",
leaf_size=30,
p=2,
metric="manhattan",
metric_params=None,
n_jobs=-1,
)
knn_model.fit(X_train, y_train)
np.sqrt(mean_squared_error(y_valid, knn_model.predict(X_valid)))
from sklearn.cluster import KMeans
sse = {}
for k in range(2, 100):
kmeans = KMeans(n_clusters=k, max_iter=1000).fit(X)
sse[k] = kmeans.inertia_
plt.figure()
plt.plot(list(sse.keys()), list(sse.values()))
plt.xlabel("Number of cluster")
plt.ylabel("SSE")
plt.show()
Kmeans_model = KMeans(n_clusters=500, random_state=np.random.randint(0, 100_000_000))
Kmeans_model.fit(X_train)
Kmeans_model.labels_
lookup_matrix_train = pd.DataFrame(
{"targets": y_train, "cluster_memberships": Kmeans_model.labels_}
)
lookup_matrix_train.groupby("cluster_memberships")[["targets"]].describe()
lookup_dict = (
lookup_matrix_train.groupby("cluster_memberships", as_index=True)[["targets"]]
.mean()
.to_dict("dict")["targets"]
)
pred_valid = np.array(list(map(lookup_dict.get, Kmeans_model.predict(X_valid))))
np.sqrt(mean_squared_error(y_valid, pred_valid))
seeded_value = 12345
NUM_FOLDS = 3
NUM_EVALS = 40
NUM_TREES = 100
def objective(params, X, y):
params = {
"max_depth": int(params["max_depth"]),
"gamma": params["gamma"],
"colsample_bytree": params["colsample_bytree"],
"learning_rate": params["learning_rate"],
}
model = xgb.XGBRegressor(
n_estimators=NUM_TREES,
verbosity=1,
objective="reg:squarederror",
random_state=seeded_value,
n_jobs=2,
# tree_method= 'gpu_hist',
**params,
)
score = cross_val_score(
estimator=model,
X=X,
y=y,
scoring="neg_mean_squared_error",
cv=KFold(
n_splits=NUM_FOLDS,
shuffle=True,
random_state=np.random.randint(0, 10_000_000),
),
).mean()
print("\tRMSE Score {:.3f} params {}".format(-1.0 * score, params))
del model
return {"loss": -1.0 * score, "params": params, "status": STATUS_OK}
def hpt(X, y):
bayes_trials = Trials()
space = {
"max_depth": hp.quniform("max_depth", 2, 8, 1),
"colsample_bytree": hp.uniform("colsample_bytree", 0.3, 1.0),
"gamma": hp.uniform("gamma", 0.0, 0.5),
"learning_rate": hp.loguniform("learning_rate", np.log(0.001), np.log(0.5)),
}
partial_objective = partial(objective, X=X, y=y)
best = fmin(
fn=partial_objective,
space=space,
algo=tpe.suggest,
max_evals=NUM_EVALS,
trials=bayes_trials,
)
print("\tbest: ", best)
optimal_params = {
"max_depth": int(best["max_depth"]),
"gamma": best["gamma"],
"colsample_bytree": best["colsample_bytree"],
"learning_rate": best["learning_rate"],
}
print("\toptimal_params", optimal_params)
return optimal_params
def retrain_after_hpt(optimal_params, X, y):
tuned_model = xgb.XGBRegressor(
**{
"n_estimators": NUM_TREES,
"verbosity": 1,
"objective": "reg:squarederror",
"random_state": seeded_value,
"n_jobs": 12,
# 'tree_method': 'gpu_hist',
**optimal_params,
}
)
tuned_model.fit(X=X, y=y)
return tuned_model
def evaluate(y_train, pred_train, y_valid, pred_valid, fold_num, prefix):
trains = pd.DataFrame({"target": y_train, "preds": pred_train})
valids = pd.DataFrame({"target": y_valid, "preds": pred_valid})
trains.to_csv(f"{prefix}-train-preds-{fold_num}.csv", index=False)
valids.to_csv(f"{prefix}-valid-preds-{fold_num}.csv", index=False)
print("Training Correlations: ", "\n\t\t\t", trains[["target", "preds"]].corr())
print(
"Training np.sign: ",
"\n\t\t\t",
np.sign(trains["target"] - trains["preds"]).value_counts(),
)
print(
"Training RMSE: ",
"\n\t\t\t",
np.sqrt(mean_squared_error(trains["target"], trains["preds"])),
)
print("Training distribution stats: ", "\n\t\t\t", valids.describe())
print("Validation Correlations: ", "\n\t\t\t", valids[["target", "preds"]].corr())
print(
"Validation np.sign: ",
"\n\t\t\t",
np.sign(valids["target"] - valids["preds"]).value_counts(),
)
print(
"Validation RMSE: ",
"\n\t\t\t",
np.sqrt(mean_squared_error(valids["target"], valids["preds"])),
)
print("Validation distribution stats: ", "\n\t\t\t", trains.describe())
def apply_minmax_(y_ref, pred_y):
min_ = y_ref.min()
max_ = y_ref.max()
pred_min_ = pred_y.min()
pred_max_ = pred_y.max()
pred_norm = (pred_y - pred_min_) / (pred_max_ - pred_min_)
y_pred_adj = (pred_norm * (max_ - min_)) + min_
return y_pred_adj
def apply_norm_(y_ref, pred_y):
mean_ = y_ref.mean()
std_ = y_ref.std()
pred_mean_ = pred_y.mean()
pred_std_ = pred_y.std()
pred_norm = (pred_y - pred_mean_) / pred_std_
y_pred_adj = pred_norm * std_ + mean_
return y_pred_adj
ENABLE_POSTPROCESSING = True
kf = KFold(n_splits=NUM_FOLDS, shuffle=True, random_state=seeded_value)
for fold_num, (t_i, v_i) in enumerate(kf.split(X)):
print("=" * 15, f" START: {str(fold_num)} ", "=" * 15)
if fold_num == 0:
predictions = {}
predictions_norm = {}
predictions_minmax = {}
X_train, y_train = X[t_i], y[t_i]
X_valid, y_valid = X[v_i], y[v_i]
optimal_params = hpt(X_train, y_train)
eval_model = retrain_after_hpt(optimal_params, X_train, y_train)
eval_model.save_model(
f"eval_model_xgb_{fold_num}.txt"
) # loaded_model = xgb.XGBRegressor(); loaded_model.load_model("path-to-file")
pred_train = eval_model.predict(X_train)
pred_valid = eval_model.predict(X_valid)
print("\tTrain RMSE:", mean_squared_error(y_train, pred_train, squared=False))
print("\tValid RMSE:", mean_squared_error(y_valid, pred_valid, squared=False))
evaluate(y_train, pred_train, y_valid, pred_valid, fold_num, "original")
if ENABLE_POSTPROCESSING:
print(
"=" * 25,
"After Normalizing Predictions on Training and Validation Datasets",
"=" * 25,
)
evaluate(
y_train,
apply_norm_(y_train, pred_train),
y_valid,
apply_norm_(y_train, pred_valid),
fold_num,
"norm",
)
print(
"=" * 25,
"After MinMaxScaling Predictions on Training and Validation Datasets",
"=" * 25,
)
evaluate(
y_train,
apply_minmax_(y_train, pred_train),
y_valid,
apply_minmax_(y_train, pred_valid),
fold_num,
"minmax",
)
model = retrain_after_hpt(optimal_params, X, y)
model.save_model(
f"model_xgb_{fold_num}.txt"
) # loaded_model = xgb.XGBRegressor(); loaded_model.load_model("path-to-file")
pred_test = model.predict(X_test)
predictions["F_" + str(fold_num)] = pred_test
if ENABLE_POSTPROCESSING:
predictions_norm["F_" + str(fold_num)] = apply_norm_(y, pred_test)
predictions_minmax["F_" + str(fold_num)] = apply_minmax_(y, pred_test)
print("=" * 15, " COMPLETE ", "=" * 15)
final_preds_to_submit = (
predictions_minmax # predictions, predictions_norm, predictions_minmax
)
test_results = pd.DataFrame(
{k: v.flatten() for k, v in final_preds_to_submit.items()},
columns=["F_" + str(num) for num in range(NUM_FOLDS)],
).values
test_results = test_results.mean(axis=1)
test["target"] = test_results
test[["id", "target"]].to_csv("submission.csv", index=False)
test[["id", "target"]].head()
print(ctime(time()))
| false | 0 | 5,659 | 0 | 5,685 | 5,659 |
||
69506743
|
#
# **Synopsis :**
# Titanic, in full Royal Mail Ship (RMS) Titanic, British luxury passenger liner that sank on April 14–15, 1912, during its maiden voyage, en route to New York City from Southampton, England, killing about 1,500 passengers and ship personnel. One of the most famous tragedies in modern history, it inspired numerous stories, several films, and a musical and has been the subject of much scholarship and scientific speculation.
# **Data :**
# There are tow datasets one dataset is titled `train.csv` and the other is titled `test.csv`.
# Train.csv will contain the details of a subset of the passengers on board (891 to be exact) and importantly, will reveal whether they survived or not, also known as the “ground truth”.
# The `test.csv` dataset contains similar information but does not disclose the “ground truth” for each passenger. It’s your job to predict these outcomes.
# **Goal :**
# Knowing from a training set of samples listing passengers who survived or did not survive the Titanic disaster, can our model determine based on a given test dataset not containing the survival information, if these passengers in the test dataset survived or not.
# ### Required Libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import math
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import confusion_matrix
from sklearn.metrics import accuracy_score
# ## Dataset
train = pd.read_csv("../input/titanic/train.csv")
test = pd.read_csv("../input/titanic/test.csv")
data = [train, test]
# ### let‘s talk about train dataset!
train.head()
train.tail()
train.shape
train.info()
# Drop useless columns
train = train.drop(["Cabin", "Ticket", "Name", "PassengerId"], axis=1)
# **"NAN" values in the taining dataset**
# number of "NAN" values
train.isnull().sum()
## Dealing with mising values ##
freq = train.Embarked.dropna().mode()
print(freq, "\n")
train["Embarked"] = train["Embarked"].fillna(
freq[0]
) # fill "NAN" values with the most frequent value
mean = train["Age"].dropna().mean()
train["Age"] = train["Age"].fillna(round(mean))
print(round(mean))
# **Converting categorical feature to numeric**
train["Sex"].replace("female", 0, inplace=True)
train["Sex"].replace("male", 1, inplace=True)
train["Embarked"].replace("S", 0, inplace=True)
train["Embarked"].replace("C", 1, inplace=True)
train["Embarked"].replace("Q", 2, inplace=True)
print(
train.isnull().sum(),
train.shape,
train.head(),
train.describe().T,
sep=" \n *********** ************* *********** \n ",
)
# ### Data Analysis & Visualization
sns.set(rc={"figure.figsize": (13, 13)})
ax = sns.heatmap(train.corr(), annot=True)
cols = ["Pclass", "Sex", "SibSp", "Parch", "Embarked"]
for col in cols:
print(
train[[col, "Survived"]]
.groupby([col], as_index=False)
.mean()
.sort_values(by="Survived", ascending=False),
end=" \n ******** ******* ********* \n ",
)
fig, axes = plt.subplots(5, 1, figsize=(6, 12))
axes = axes.flatten()
for ax, catplot in zip(axes, train[cols]):
_ = sns.countplot(
x=catplot, data=train, ax=ax, hue=train["Survived"], palette="OrRd"
)
_.legend(loc="upper right")
plt.tight_layout()
plt.show()
_ = sns.FacetGrid(train, col="Survived")
_.map(plt.hist, "Age", bins=15)
_ = sns.FacetGrid(train, col="Pclass")
_.map(plt.hist, "Age", bins=15)
_ = sns.FacetGrid(train, col="Sex")
_.map(plt.hist, "Age", bins=15)
fig, ax = plt.subplots(1, 3, figsize=(20, 8))
sns.histplot(
train[train["Pclass"] == 1].Fare, ax=ax[0], kde=True, stat="density", linewidth=0
)
ax[0].set_title("Fares in Pclass 1")
sns.histplot(
train[train["Pclass"] == 2].Fare, ax=ax[1], kde=True, stat="density", linewidth=0
)
ax[1].set_title("Fares in Pclass 2")
sns.histplot(
train[train["Pclass"] == 3].Fare, ax=ax[2], kde=True, stat="density", linewidth=0
)
ax[2].set_title("Fares in Pclass 3")
plt.show()
# ### let‘s talk about test dataset!
test.head()
print(
test.shape,
test.info(),
test.isnull().sum(),
sep=" \n *********** ************* ************ \n",
)
# Drop useless columns
test = test.drop(["Cabin", "Ticket", "Name", "PassengerId"], axis=1)
## Dealing with mising values ##
freq = test.Fare.dropna().mode()
print(freq, "\n")
test["Fare"] = test["Fare"].fillna(
freq[0]
) # fill "NAN" values with the most frequent value
mean = test["Age"].dropna().mean()
test["Age"] = test["Age"].fillna(round(mean))
print(round(mean))
test["Sex"].replace("female", 0, inplace=True)
test["Sex"].replace("male", 1, inplace=True)
test["Embarked"].replace("S", 0, inplace=True)
test["Embarked"].replace("C", 1, inplace=True)
test["Embarked"].replace("Q", 2, inplace=True)
test.head()
# ## Machine learning model
# ### Training and Predictions
x_test = test
x_train = train.drop("Survived", axis=1)
y_train = train["Survived"]
log = LogisticRegression(solver="liblinear")
log.fit(x_train, y_train)
y_pred = log.predict(x_test)
y_pred[:20]
# ### Evaluating
y_test = pd.read_csv("../input/titanic/gender_submission.csv")
y_test = np.array(y_test["Survived"])
accuracy_score(y_test, y_pred)
from sklearn import metrics
print("Mean Absolute Error:", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error:", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error:", np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506743.ipynb
| null | null |
[{"Id": 69506743, "ScriptId": 18977672, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5578861, "CreationDate": "07/31/2021 21:25:46", "VersionNumber": 1.0, "Title": "Titanic is here !", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 173.0, "LinesInsertedFromPrevious": 173.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
#
# **Synopsis :**
# Titanic, in full Royal Mail Ship (RMS) Titanic, British luxury passenger liner that sank on April 14–15, 1912, during its maiden voyage, en route to New York City from Southampton, England, killing about 1,500 passengers and ship personnel. One of the most famous tragedies in modern history, it inspired numerous stories, several films, and a musical and has been the subject of much scholarship and scientific speculation.
# **Data :**
# There are tow datasets one dataset is titled `train.csv` and the other is titled `test.csv`.
# Train.csv will contain the details of a subset of the passengers on board (891 to be exact) and importantly, will reveal whether they survived or not, also known as the “ground truth”.
# The `test.csv` dataset contains similar information but does not disclose the “ground truth” for each passenger. It’s your job to predict these outcomes.
# **Goal :**
# Knowing from a training set of samples listing passengers who survived or did not survive the Titanic disaster, can our model determine based on a given test dataset not containing the survival information, if these passengers in the test dataset survived or not.
# ### Required Libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import math
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import confusion_matrix
from sklearn.metrics import accuracy_score
# ## Dataset
train = pd.read_csv("../input/titanic/train.csv")
test = pd.read_csv("../input/titanic/test.csv")
data = [train, test]
# ### let‘s talk about train dataset!
train.head()
train.tail()
train.shape
train.info()
# Drop useless columns
train = train.drop(["Cabin", "Ticket", "Name", "PassengerId"], axis=1)
# **"NAN" values in the taining dataset**
# number of "NAN" values
train.isnull().sum()
## Dealing with mising values ##
freq = train.Embarked.dropna().mode()
print(freq, "\n")
train["Embarked"] = train["Embarked"].fillna(
freq[0]
) # fill "NAN" values with the most frequent value
mean = train["Age"].dropna().mean()
train["Age"] = train["Age"].fillna(round(mean))
print(round(mean))
# **Converting categorical feature to numeric**
train["Sex"].replace("female", 0, inplace=True)
train["Sex"].replace("male", 1, inplace=True)
train["Embarked"].replace("S", 0, inplace=True)
train["Embarked"].replace("C", 1, inplace=True)
train["Embarked"].replace("Q", 2, inplace=True)
print(
train.isnull().sum(),
train.shape,
train.head(),
train.describe().T,
sep=" \n *********** ************* *********** \n ",
)
# ### Data Analysis & Visualization
sns.set(rc={"figure.figsize": (13, 13)})
ax = sns.heatmap(train.corr(), annot=True)
cols = ["Pclass", "Sex", "SibSp", "Parch", "Embarked"]
for col in cols:
print(
train[[col, "Survived"]]
.groupby([col], as_index=False)
.mean()
.sort_values(by="Survived", ascending=False),
end=" \n ******** ******* ********* \n ",
)
fig, axes = plt.subplots(5, 1, figsize=(6, 12))
axes = axes.flatten()
for ax, catplot in zip(axes, train[cols]):
_ = sns.countplot(
x=catplot, data=train, ax=ax, hue=train["Survived"], palette="OrRd"
)
_.legend(loc="upper right")
plt.tight_layout()
plt.show()
_ = sns.FacetGrid(train, col="Survived")
_.map(plt.hist, "Age", bins=15)
_ = sns.FacetGrid(train, col="Pclass")
_.map(plt.hist, "Age", bins=15)
_ = sns.FacetGrid(train, col="Sex")
_.map(plt.hist, "Age", bins=15)
fig, ax = plt.subplots(1, 3, figsize=(20, 8))
sns.histplot(
train[train["Pclass"] == 1].Fare, ax=ax[0], kde=True, stat="density", linewidth=0
)
ax[0].set_title("Fares in Pclass 1")
sns.histplot(
train[train["Pclass"] == 2].Fare, ax=ax[1], kde=True, stat="density", linewidth=0
)
ax[1].set_title("Fares in Pclass 2")
sns.histplot(
train[train["Pclass"] == 3].Fare, ax=ax[2], kde=True, stat="density", linewidth=0
)
ax[2].set_title("Fares in Pclass 3")
plt.show()
# ### let‘s talk about test dataset!
test.head()
print(
test.shape,
test.info(),
test.isnull().sum(),
sep=" \n *********** ************* ************ \n",
)
# Drop useless columns
test = test.drop(["Cabin", "Ticket", "Name", "PassengerId"], axis=1)
## Dealing with mising values ##
freq = test.Fare.dropna().mode()
print(freq, "\n")
test["Fare"] = test["Fare"].fillna(
freq[0]
) # fill "NAN" values with the most frequent value
mean = test["Age"].dropna().mean()
test["Age"] = test["Age"].fillna(round(mean))
print(round(mean))
test["Sex"].replace("female", 0, inplace=True)
test["Sex"].replace("male", 1, inplace=True)
test["Embarked"].replace("S", 0, inplace=True)
test["Embarked"].replace("C", 1, inplace=True)
test["Embarked"].replace("Q", 2, inplace=True)
test.head()
# ## Machine learning model
# ### Training and Predictions
x_test = test
x_train = train.drop("Survived", axis=1)
y_train = train["Survived"]
log = LogisticRegression(solver="liblinear")
log.fit(x_train, y_train)
y_pred = log.predict(x_test)
y_pred[:20]
# ### Evaluating
y_test = pd.read_csv("../input/titanic/gender_submission.csv")
y_test = np.array(y_test["Survived"])
accuracy_score(y_test, y_pred)
from sklearn import metrics
print("Mean Absolute Error:", metrics.mean_absolute_error(y_test, y_pred))
print("Mean Squared Error:", metrics.mean_squared_error(y_test, y_pred))
print("Root Mean Squared Error:", np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
| false | 0 | 1,805 | 0 | 1,805 | 1,805 |
||
69506385
|
#
#
#
import numpy as np
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
from tensorflow import (
function,
GradientTape,
sqrt,
abs,
reduce_mean,
ones_like,
zeros_like,
convert_to_tensor,
float32,
)
from tensorflow import data as tfdata
from tensorflow import config as tfconfig
from tensorflow import nn
from tensorflow.keras import Model, Sequential, Input
from tensorflow.keras.layers import GRU, LSTM, Dense
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.losses import BinaryCrossentropy, MeanSquaredError
import numpy as np
from tqdm import tqdm, trange
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
#
# Dataset
# ¶
#
#
df = pd.read_csv("../input/yahootoapple20152021/yahoo-to-apple-map-2015-2021.csv")
# df = df.set_index('Date').sort_index()
del df["Unnamed: 0"]
del df["Date"]
df.head()
#
# Parameters
# ¶
#
#
seq_len = 24
n_seq = 6
hidden_dim = 24
gamma = 1
noise_dim = 32
dim = 128
batch_size = 128
log_step = 100
learning_rate = 5e-4
train_steps = 5000
gan_args = batch_size, learning_rate, noise_dim, 24, 2, (0, 1), dim
#
# Preprocess
# ¶
#
#
def preprocess(data, seq_len):
ori_data = data[::-1]
scaler = MinMaxScaler().fit(ori_data)
ori_data = scaler.transform(ori_data)
temp_data = []
for i in range(0, len(ori_data) - seq_len):
_x = ori_data[i : i + seq_len]
temp_data.append(_x)
idx = np.random.permutation(len(temp_data))
data = []
for i in range(len(temp_data)):
data.append(temp_data[idx[i]])
return data
stock_data = preprocess(df.values, seq_len)
#
# Modules
# ¶
#
#
def net(model, n_layers, hidden_units, output_units, net_type="GRU"):
if net_type == "GRU":
for i in range(n_layers):
model.add(
GRU(units=hidden_units, return_sequences=True, name=f"GRU_{i + 1}")
)
else:
for i in range(n_layers):
model.add(
LSTM(units=hidden_units, return_sequences=True, name=f"LSTM_{i + 1}")
)
model.add(Dense(units=output_units, activation="sigmoid", name="OUT"))
return model
class Generator(Model):
def __init__(self, hidden_dim, net_type="GRU"):
self.hidden_dim = hidden_dim
self.net_type = net_type
def build(self, input_shape):
model = Sequential(name="Generator")
model = net(
model,
n_layers=3,
hidden_units=self.hidden_dim,
output_units=self.hidden_dim,
net_type=self.net_type,
)
return model
class Discriminator(Model):
def __init__(self, hidden_dim, net_type="GRU"):
self.hidden_dim = hidden_dim
self.net_type = net_type
def build(self, input_shape):
model = Sequential(name="Discriminator")
model = net(
model,
n_layers=3,
hidden_units=self.hidden_dim,
output_units=1,
net_type=self.net_type,
)
return model
class Recovery(Model):
def __init__(self, hidden_dim, n_seq):
self.hidden_dim = hidden_dim
self.n_seq = n_seq
return
def build(self, input_shape):
recovery = Sequential(name="Recovery")
recovery = net(
recovery, n_layers=3, hidden_units=self.hidden_dim, output_units=self.n_seq
)
return recovery
class Embedder(Model):
def __init__(self, hidden_dim):
self.hidden_dim = hidden_dim
return
def build(self, input_shape):
embedder = Sequential(name="Embedder")
embedder = net(
embedder,
n_layers=3,
hidden_units=self.hidden_dim,
output_units=self.hidden_dim,
)
return embedder
class Supervisor(Model):
def __init__(self, hidden_dim):
self.hidden_dim = hidden_dim
def build(self, input_shape):
model = Sequential(name="Supervisor")
model = net(
model,
n_layers=2,
hidden_units=self.hidden_dim,
output_units=self.hidden_dim,
)
return model
#
# Definitions
# ¶
#
#
class TimeGAN:
def __init__(self, model_parameters, hidden_dim, seq_len, n_seq, gamma):
self.seq_len = seq_len
(
self.batch_size,
self.lr,
self.beta_1,
self.beta_2,
self.noise_dim,
self.data_dim,
self.layers_dim,
) = model_parameters
self.n_seq = n_seq
self.hidden_dim = hidden_dim
self.gamma = gamma
self.define_gan()
def define_gan(self):
self.generator_aux = Generator(self.hidden_dim).build(
input_shape=(self.seq_len, self.n_seq)
)
self.supervisor = Supervisor(self.hidden_dim).build(
input_shape=(self.hidden_dim, self.hidden_dim)
)
self.discriminator = Discriminator(self.hidden_dim).build(
input_shape=(self.hidden_dim, self.hidden_dim)
)
self.recovery = Recovery(self.hidden_dim, self.n_seq).build(
input_shape=(self.hidden_dim, self.hidden_dim)
)
self.embedder = Embedder(self.hidden_dim).build(
input_shape=(self.seq_len, self.n_seq)
)
X = Input(
shape=[self.seq_len, self.n_seq],
batch_size=self.batch_size,
name="RealData",
)
Z = Input(
shape=[self.seq_len, self.n_seq],
batch_size=self.batch_size,
name="RandomNoise",
)
# AutoEncoder
H = self.embedder(X)
X_tilde = self.recovery(H)
self.autoencoder = Model(inputs=X, outputs=X_tilde)
# Adversarial Supervise Architecture
E_Hat = self.generator_aux(Z)
H_hat = self.supervisor(E_Hat)
Y_fake = self.discriminator(H_hat)
self.adversarial_supervised = Model(
inputs=Z, outputs=Y_fake, name="AdversarialSupervised"
)
# Adversarial architecture in latent space
Y_fake_e = self.discriminator(E_Hat)
self.adversarial_embedded = Model(
inputs=Z, outputs=Y_fake_e, name="AdversarialEmbedded"
)
# Synthetic data generation
X_hat = self.recovery(H_hat)
self.generator = Model(inputs=Z, outputs=X_hat, name="FinalGenerator")
# Final discriminator model
Y_real = self.discriminator(H)
self.discriminator_model = Model(
inputs=X, outputs=Y_real, name="RealDiscriminator"
)
# Loss functions
self._mse = MeanSquaredError()
self._bce = BinaryCrossentropy()
#
# Training Modules
# ¶
#
#
class TimeGAN(TimeGAN):
def __init__(self, model_parameters, hidden_dim, seq_len, n_seq, gamma):
super().__init__(model_parameters, hidden_dim, seq_len, n_seq, gamma)
@function
def train_autoencoder(self, x, opt):
with GradientTape() as tape:
x_tilde = self.autoencoder(x)
embedding_loss_t0 = self._mse(x, x_tilde)
e_loss_0 = 10 * sqrt(embedding_loss_t0)
var_list = self.embedder.trainable_variables + self.recovery.trainable_variables
gradients = tape.gradient(e_loss_0, var_list)
opt.apply_gradients(zip(gradients, var_list))
return sqrt(embedding_loss_t0)
@function
def train_supervisor(self, x, opt):
with GradientTape() as tape:
h = self.embedder(x)
h_hat_supervised = self.supervisor(h)
g_loss_s = self._mse(h[:, 1:, :], h_hat_supervised[:, 1:, :])
var_list = (
self.supervisor.trainable_variables + self.generator.trainable_variables
)
gradients = tape.gradient(g_loss_s, var_list)
apply_grads = [
(grad, var) for (grad, var) in zip(gradients, var_list) if grad is not None
]
opt.apply_gradients(apply_grads)
return g_loss_s
@function
def train_embedder(self, x, opt):
with GradientTape() as tape:
h = self.embedder(x)
h_hat_supervised = self.supervisor(h)
generator_loss_supervised = self._mse(
h[:, 1:, :], h_hat_supervised[:, 1:, :]
)
x_tilde = self.autoencoder(x)
embedding_loss_t0 = self._mse(x, x_tilde)
e_loss = 10 * sqrt(embedding_loss_t0) + 0.1 * generator_loss_supervised
var_list = self.embedder.trainable_variables + self.recovery.trainable_variables
gradients = tape.gradient(e_loss, var_list)
opt.apply_gradients(zip(gradients, var_list))
return sqrt(embedding_loss_t0)
def discriminator_loss(self, x, z):
y_real = self.discriminator_model(x)
discriminator_loss_real = self._bce(y_true=ones_like(y_real), y_pred=y_real)
y_fake = self.adversarial_supervised(z)
discriminator_loss_fake = self._bce(y_true=zeros_like(y_fake), y_pred=y_fake)
y_fake_e = self.adversarial_embedded(z)
discriminator_loss_fake_e = self._bce(
y_true=zeros_like(y_fake_e), y_pred=y_fake_e
)
return (
discriminator_loss_real
+ discriminator_loss_fake
+ self.gamma * discriminator_loss_fake_e
)
@staticmethod
def calc_generator_moments_loss(y_true, y_pred):
y_true_mean, y_true_var = nn.moments(x=y_true, axes=[0])
y_pred_mean, y_pred_var = nn.moments(x=y_pred, axes=[0])
g_loss_mean = reduce_mean(abs(y_true_mean - y_pred_mean))
g_loss_var = reduce_mean(abs(sqrt(y_true_var + 1e-6) - sqrt(y_pred_var + 1e-6)))
return g_loss_mean + g_loss_var
@function
def train_generator(self, x, z, opt):
with GradientTape() as tape:
y_fake = self.adversarial_supervised(z)
generator_loss_unsupervised = self._bce(
y_true=ones_like(y_fake), y_pred=y_fake
)
y_fake_e = self.adversarial_embedded(z)
generator_loss_unsupervised_e = self._bce(
y_true=ones_like(y_fake_e), y_pred=y_fake_e
)
h = self.embedder(x)
h_hat_supervised = self.supervisor(h)
generator_loss_supervised = self._mse(
h[:, 1:, :], h_hat_supervised[:, 1:, :]
)
x_hat = self.generator(z)
generator_moment_loss = self.calc_generator_moments_loss(x, x_hat)
generator_loss = (
generator_loss_unsupervised
+ generator_loss_unsupervised_e
+ 100 * sqrt(generator_loss_supervised)
+ 100 * generator_moment_loss
)
var_list = (
self.generator_aux.trainable_variables + self.supervisor.trainable_variables
)
gradients = tape.gradient(generator_loss, var_list)
opt.apply_gradients(zip(gradients, var_list))
return (
generator_loss_unsupervised,
generator_loss_supervised,
generator_moment_loss,
)
@function
def train_discriminator(self, x, z, opt):
with GradientTape() as tape:
discriminator_loss = self.discriminator_loss(x, z)
var_list = self.discriminator.trainable_variables
gradients = tape.gradient(discriminator_loss, var_list)
opt.apply_gradients(zip(gradients, var_list))
return discriminator_loss
def get_batch_data(self, data, n_windows):
data = convert_to_tensor(data, dtype=float32)
return iter(
tfdata.Dataset.from_tensor_slices(data)
.shuffle(buffer_size=n_windows)
.batch(self.batch_size)
.repeat()
)
def _generate_noise(self):
while True:
yield np.random.uniform(low=0, high=1, size=(self.seq_len, self.n_seq))
def get_batch_noise(self):
return iter(
tfdata.Dataset.from_generator(self._generate_noise, output_types=float32)
.batch(self.batch_size)
.repeat()
)
def sample(self, n_samples):
steps = n_samples // self.batch_size + 1
data = []
for _ in trange(steps, desc="Synthetic data generation"):
Z_ = next(self.get_batch_noise())
records = self.generator(Z_)
data.append(records)
return np.array(np.vstack(data))
synth = TimeGAN(
model_parameters=gan_args, hidden_dim=24, seq_len=seq_len, n_seq=n_seq, gamma=1
)
#
# Training
# ¶
#
#
autoencoder_opt = Adam(learning_rate=learning_rate)
for _ in tqdm(range(train_steps), desc="Emddeding network training"):
X_ = next(synth.get_batch_data(stock_data, n_windows=len(stock_data)))
step_e_loss_t0 = synth.train_autoencoder(X_, autoencoder_opt)
supervisor_opt = Adam(learning_rate=learning_rate)
for _ in tqdm(range(train_steps), desc="Supervised network training"):
X_ = next(synth.get_batch_data(stock_data, n_windows=len(stock_data)))
step_g_loss_s = synth.train_supervisor(X_, supervisor_opt)
generator_opt = Adam(learning_rate=learning_rate)
embedder_opt = Adam(learning_rate=learning_rate)
discriminator_opt = Adam(learning_rate=learning_rate)
step_g_loss_u = step_g_loss_s = step_g_loss_v = step_e_loss_t0 = step_d_loss = 0
for _ in tqdm(range(train_steps), desc="Joint networks training"):
# Train the generator (k times as often as the discriminator)
# Here k=2
for _ in range(2):
X_ = next(synth.get_batch_data(stock_data, n_windows=len(stock_data)))
Z_ = next(synth.get_batch_noise())
# Train the generator
step_g_loss_u, step_g_loss_s, step_g_loss_v = synth.train_generator(
X_, Z_, generator_opt
)
# Train the embedder
step_e_loss_t0 = synth.train_embedder(X_, embedder_opt)
X_ = next(synth.get_batch_data(stock_data, n_windows=len(stock_data)))
Z_ = next(synth.get_batch_noise())
step_d_loss = synth.discriminator_loss(X_, Z_)
if step_d_loss > 0.15:
step_d_loss = synth.train_discriminator(X_, Z_, discriminator_opt)
#
# Analyzing
# ¶
#
#
sample_size = 250
idx = np.random.permutation(len(stock_data))[:sample_size]
real_sample = np.asarray(stock_data)[idx]
synth_data = synth.sample(len(stock_data))
synthetic_sample = np.asarray(synth_data)[idx]
# for the purpose of comparision we need the data to be 2-Dimensional. For that reason we are going to use only two componentes for both the PCA and TSNE.
synth_data_reduced = real_sample.reshape(-1, seq_len)
stock_data_reduced = np.asarray(synthetic_sample).reshape(-1, seq_len)
n_components = 2
pca = PCA(n_components=n_components)
tsne = TSNE(n_components=n_components, n_iter=300)
# The fit of the methods must be done only using the real sequential data
pca.fit(stock_data_reduced)
pca_real = pd.DataFrame(pca.transform(stock_data_reduced))
pca_synth = pd.DataFrame(pca.transform(synth_data_reduced))
data_reduced = np.concatenate((stock_data_reduced, synth_data_reduced), axis=0)
tsne_results = pd.DataFrame(tsne.fit_transform(data_reduced))
fig = plt.figure(constrained_layout=True, figsize=(20, 10))
spec = gridspec.GridSpec(ncols=2, nrows=1, figure=fig)
# TSNE scatter plot
ax = fig.add_subplot(spec[0, 0])
ax.set_title("PCA results", fontsize=20, color="red", pad=10)
# PCA scatter plot
plt.scatter(
pca_real.iloc[:, 0].values,
pca_real.iloc[:, 1].values,
c="black",
alpha=0.2,
label="Original",
)
plt.scatter(
pca_synth.iloc[:, 0], pca_synth.iloc[:, 1], c="red", alpha=0.2, label="Synthetic"
)
ax.legend()
ax2 = fig.add_subplot(spec[0, 1])
ax2.set_title("TSNE results", fontsize=20, color="red", pad=10)
plt.scatter(
tsne_results.iloc[:sample_size, 0].values,
tsne_results.iloc[:sample_size, 1].values,
c="black",
alpha=0.2,
label="Original",
)
plt.scatter(
tsne_results.iloc[sample_size:, 0],
tsne_results.iloc[sample_size:, 1],
c="red",
alpha=0.2,
label="Synthetic",
)
ax2.legend()
fig.suptitle(
"Validating synthetic vs real data diversity and distributions",
fontsize=16,
color="grey",
)
# Reshaping the data
cols = ["Open", "High", "Low", "Close", "Adj Close", "Volume"]
# Plotting some generated samples. Both Synthetic and Original data are still standartized with values between [0,1]
fig, axes = plt.subplots(nrows=3, ncols=2, figsize=(15, 10))
axes = axes.flatten()
time = list(range(1, 25))
obs = np.random.randint(len(stock_data))
for j, col in enumerate(cols):
df = pd.DataFrame(
{"Real": stock_data[obs][:, j], "Synthetic": synth_data[obs][:, j]}
)
df.plot(ax=axes[j], title=col, secondary_y="Synthetic data", style=["-", "--"])
fig.tight_layout()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506385.ipynb
| null | null |
[{"Id": 69506385, "ScriptId": 18975064, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2969707, "CreationDate": "07/31/2021 21:17:03", "VersionNumber": 1.0, "Title": "GANSTOCKAPPLEYAHOO2015", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 512.0, "LinesInsertedFromPrevious": 512.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 13}]
| null | null | null | null |
#
#
#
import numpy as np
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
from tensorflow import (
function,
GradientTape,
sqrt,
abs,
reduce_mean,
ones_like,
zeros_like,
convert_to_tensor,
float32,
)
from tensorflow import data as tfdata
from tensorflow import config as tfconfig
from tensorflow import nn
from tensorflow.keras import Model, Sequential, Input
from tensorflow.keras.layers import GRU, LSTM, Dense
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.losses import BinaryCrossentropy, MeanSquaredError
import numpy as np
from tqdm import tqdm, trange
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
#
# Dataset
# ¶
#
#
df = pd.read_csv("../input/yahootoapple20152021/yahoo-to-apple-map-2015-2021.csv")
# df = df.set_index('Date').sort_index()
del df["Unnamed: 0"]
del df["Date"]
df.head()
#
# Parameters
# ¶
#
#
seq_len = 24
n_seq = 6
hidden_dim = 24
gamma = 1
noise_dim = 32
dim = 128
batch_size = 128
log_step = 100
learning_rate = 5e-4
train_steps = 5000
gan_args = batch_size, learning_rate, noise_dim, 24, 2, (0, 1), dim
#
# Preprocess
# ¶
#
#
def preprocess(data, seq_len):
ori_data = data[::-1]
scaler = MinMaxScaler().fit(ori_data)
ori_data = scaler.transform(ori_data)
temp_data = []
for i in range(0, len(ori_data) - seq_len):
_x = ori_data[i : i + seq_len]
temp_data.append(_x)
idx = np.random.permutation(len(temp_data))
data = []
for i in range(len(temp_data)):
data.append(temp_data[idx[i]])
return data
stock_data = preprocess(df.values, seq_len)
#
# Modules
# ¶
#
#
def net(model, n_layers, hidden_units, output_units, net_type="GRU"):
if net_type == "GRU":
for i in range(n_layers):
model.add(
GRU(units=hidden_units, return_sequences=True, name=f"GRU_{i + 1}")
)
else:
for i in range(n_layers):
model.add(
LSTM(units=hidden_units, return_sequences=True, name=f"LSTM_{i + 1}")
)
model.add(Dense(units=output_units, activation="sigmoid", name="OUT"))
return model
class Generator(Model):
def __init__(self, hidden_dim, net_type="GRU"):
self.hidden_dim = hidden_dim
self.net_type = net_type
def build(self, input_shape):
model = Sequential(name="Generator")
model = net(
model,
n_layers=3,
hidden_units=self.hidden_dim,
output_units=self.hidden_dim,
net_type=self.net_type,
)
return model
class Discriminator(Model):
def __init__(self, hidden_dim, net_type="GRU"):
self.hidden_dim = hidden_dim
self.net_type = net_type
def build(self, input_shape):
model = Sequential(name="Discriminator")
model = net(
model,
n_layers=3,
hidden_units=self.hidden_dim,
output_units=1,
net_type=self.net_type,
)
return model
class Recovery(Model):
def __init__(self, hidden_dim, n_seq):
self.hidden_dim = hidden_dim
self.n_seq = n_seq
return
def build(self, input_shape):
recovery = Sequential(name="Recovery")
recovery = net(
recovery, n_layers=3, hidden_units=self.hidden_dim, output_units=self.n_seq
)
return recovery
class Embedder(Model):
def __init__(self, hidden_dim):
self.hidden_dim = hidden_dim
return
def build(self, input_shape):
embedder = Sequential(name="Embedder")
embedder = net(
embedder,
n_layers=3,
hidden_units=self.hidden_dim,
output_units=self.hidden_dim,
)
return embedder
class Supervisor(Model):
def __init__(self, hidden_dim):
self.hidden_dim = hidden_dim
def build(self, input_shape):
model = Sequential(name="Supervisor")
model = net(
model,
n_layers=2,
hidden_units=self.hidden_dim,
output_units=self.hidden_dim,
)
return model
#
# Definitions
# ¶
#
#
class TimeGAN:
def __init__(self, model_parameters, hidden_dim, seq_len, n_seq, gamma):
self.seq_len = seq_len
(
self.batch_size,
self.lr,
self.beta_1,
self.beta_2,
self.noise_dim,
self.data_dim,
self.layers_dim,
) = model_parameters
self.n_seq = n_seq
self.hidden_dim = hidden_dim
self.gamma = gamma
self.define_gan()
def define_gan(self):
self.generator_aux = Generator(self.hidden_dim).build(
input_shape=(self.seq_len, self.n_seq)
)
self.supervisor = Supervisor(self.hidden_dim).build(
input_shape=(self.hidden_dim, self.hidden_dim)
)
self.discriminator = Discriminator(self.hidden_dim).build(
input_shape=(self.hidden_dim, self.hidden_dim)
)
self.recovery = Recovery(self.hidden_dim, self.n_seq).build(
input_shape=(self.hidden_dim, self.hidden_dim)
)
self.embedder = Embedder(self.hidden_dim).build(
input_shape=(self.seq_len, self.n_seq)
)
X = Input(
shape=[self.seq_len, self.n_seq],
batch_size=self.batch_size,
name="RealData",
)
Z = Input(
shape=[self.seq_len, self.n_seq],
batch_size=self.batch_size,
name="RandomNoise",
)
# AutoEncoder
H = self.embedder(X)
X_tilde = self.recovery(H)
self.autoencoder = Model(inputs=X, outputs=X_tilde)
# Adversarial Supervise Architecture
E_Hat = self.generator_aux(Z)
H_hat = self.supervisor(E_Hat)
Y_fake = self.discriminator(H_hat)
self.adversarial_supervised = Model(
inputs=Z, outputs=Y_fake, name="AdversarialSupervised"
)
# Adversarial architecture in latent space
Y_fake_e = self.discriminator(E_Hat)
self.adversarial_embedded = Model(
inputs=Z, outputs=Y_fake_e, name="AdversarialEmbedded"
)
# Synthetic data generation
X_hat = self.recovery(H_hat)
self.generator = Model(inputs=Z, outputs=X_hat, name="FinalGenerator")
# Final discriminator model
Y_real = self.discriminator(H)
self.discriminator_model = Model(
inputs=X, outputs=Y_real, name="RealDiscriminator"
)
# Loss functions
self._mse = MeanSquaredError()
self._bce = BinaryCrossentropy()
#
# Training Modules
# ¶
#
#
class TimeGAN(TimeGAN):
def __init__(self, model_parameters, hidden_dim, seq_len, n_seq, gamma):
super().__init__(model_parameters, hidden_dim, seq_len, n_seq, gamma)
@function
def train_autoencoder(self, x, opt):
with GradientTape() as tape:
x_tilde = self.autoencoder(x)
embedding_loss_t0 = self._mse(x, x_tilde)
e_loss_0 = 10 * sqrt(embedding_loss_t0)
var_list = self.embedder.trainable_variables + self.recovery.trainable_variables
gradients = tape.gradient(e_loss_0, var_list)
opt.apply_gradients(zip(gradients, var_list))
return sqrt(embedding_loss_t0)
@function
def train_supervisor(self, x, opt):
with GradientTape() as tape:
h = self.embedder(x)
h_hat_supervised = self.supervisor(h)
g_loss_s = self._mse(h[:, 1:, :], h_hat_supervised[:, 1:, :])
var_list = (
self.supervisor.trainable_variables + self.generator.trainable_variables
)
gradients = tape.gradient(g_loss_s, var_list)
apply_grads = [
(grad, var) for (grad, var) in zip(gradients, var_list) if grad is not None
]
opt.apply_gradients(apply_grads)
return g_loss_s
@function
def train_embedder(self, x, opt):
with GradientTape() as tape:
h = self.embedder(x)
h_hat_supervised = self.supervisor(h)
generator_loss_supervised = self._mse(
h[:, 1:, :], h_hat_supervised[:, 1:, :]
)
x_tilde = self.autoencoder(x)
embedding_loss_t0 = self._mse(x, x_tilde)
e_loss = 10 * sqrt(embedding_loss_t0) + 0.1 * generator_loss_supervised
var_list = self.embedder.trainable_variables + self.recovery.trainable_variables
gradients = tape.gradient(e_loss, var_list)
opt.apply_gradients(zip(gradients, var_list))
return sqrt(embedding_loss_t0)
def discriminator_loss(self, x, z):
y_real = self.discriminator_model(x)
discriminator_loss_real = self._bce(y_true=ones_like(y_real), y_pred=y_real)
y_fake = self.adversarial_supervised(z)
discriminator_loss_fake = self._bce(y_true=zeros_like(y_fake), y_pred=y_fake)
y_fake_e = self.adversarial_embedded(z)
discriminator_loss_fake_e = self._bce(
y_true=zeros_like(y_fake_e), y_pred=y_fake_e
)
return (
discriminator_loss_real
+ discriminator_loss_fake
+ self.gamma * discriminator_loss_fake_e
)
@staticmethod
def calc_generator_moments_loss(y_true, y_pred):
y_true_mean, y_true_var = nn.moments(x=y_true, axes=[0])
y_pred_mean, y_pred_var = nn.moments(x=y_pred, axes=[0])
g_loss_mean = reduce_mean(abs(y_true_mean - y_pred_mean))
g_loss_var = reduce_mean(abs(sqrt(y_true_var + 1e-6) - sqrt(y_pred_var + 1e-6)))
return g_loss_mean + g_loss_var
@function
def train_generator(self, x, z, opt):
with GradientTape() as tape:
y_fake = self.adversarial_supervised(z)
generator_loss_unsupervised = self._bce(
y_true=ones_like(y_fake), y_pred=y_fake
)
y_fake_e = self.adversarial_embedded(z)
generator_loss_unsupervised_e = self._bce(
y_true=ones_like(y_fake_e), y_pred=y_fake_e
)
h = self.embedder(x)
h_hat_supervised = self.supervisor(h)
generator_loss_supervised = self._mse(
h[:, 1:, :], h_hat_supervised[:, 1:, :]
)
x_hat = self.generator(z)
generator_moment_loss = self.calc_generator_moments_loss(x, x_hat)
generator_loss = (
generator_loss_unsupervised
+ generator_loss_unsupervised_e
+ 100 * sqrt(generator_loss_supervised)
+ 100 * generator_moment_loss
)
var_list = (
self.generator_aux.trainable_variables + self.supervisor.trainable_variables
)
gradients = tape.gradient(generator_loss, var_list)
opt.apply_gradients(zip(gradients, var_list))
return (
generator_loss_unsupervised,
generator_loss_supervised,
generator_moment_loss,
)
@function
def train_discriminator(self, x, z, opt):
with GradientTape() as tape:
discriminator_loss = self.discriminator_loss(x, z)
var_list = self.discriminator.trainable_variables
gradients = tape.gradient(discriminator_loss, var_list)
opt.apply_gradients(zip(gradients, var_list))
return discriminator_loss
def get_batch_data(self, data, n_windows):
data = convert_to_tensor(data, dtype=float32)
return iter(
tfdata.Dataset.from_tensor_slices(data)
.shuffle(buffer_size=n_windows)
.batch(self.batch_size)
.repeat()
)
def _generate_noise(self):
while True:
yield np.random.uniform(low=0, high=1, size=(self.seq_len, self.n_seq))
def get_batch_noise(self):
return iter(
tfdata.Dataset.from_generator(self._generate_noise, output_types=float32)
.batch(self.batch_size)
.repeat()
)
def sample(self, n_samples):
steps = n_samples // self.batch_size + 1
data = []
for _ in trange(steps, desc="Synthetic data generation"):
Z_ = next(self.get_batch_noise())
records = self.generator(Z_)
data.append(records)
return np.array(np.vstack(data))
synth = TimeGAN(
model_parameters=gan_args, hidden_dim=24, seq_len=seq_len, n_seq=n_seq, gamma=1
)
#
# Training
# ¶
#
#
autoencoder_opt = Adam(learning_rate=learning_rate)
for _ in tqdm(range(train_steps), desc="Emddeding network training"):
X_ = next(synth.get_batch_data(stock_data, n_windows=len(stock_data)))
step_e_loss_t0 = synth.train_autoencoder(X_, autoencoder_opt)
supervisor_opt = Adam(learning_rate=learning_rate)
for _ in tqdm(range(train_steps), desc="Supervised network training"):
X_ = next(synth.get_batch_data(stock_data, n_windows=len(stock_data)))
step_g_loss_s = synth.train_supervisor(X_, supervisor_opt)
generator_opt = Adam(learning_rate=learning_rate)
embedder_opt = Adam(learning_rate=learning_rate)
discriminator_opt = Adam(learning_rate=learning_rate)
step_g_loss_u = step_g_loss_s = step_g_loss_v = step_e_loss_t0 = step_d_loss = 0
for _ in tqdm(range(train_steps), desc="Joint networks training"):
# Train the generator (k times as often as the discriminator)
# Here k=2
for _ in range(2):
X_ = next(synth.get_batch_data(stock_data, n_windows=len(stock_data)))
Z_ = next(synth.get_batch_noise())
# Train the generator
step_g_loss_u, step_g_loss_s, step_g_loss_v = synth.train_generator(
X_, Z_, generator_opt
)
# Train the embedder
step_e_loss_t0 = synth.train_embedder(X_, embedder_opt)
X_ = next(synth.get_batch_data(stock_data, n_windows=len(stock_data)))
Z_ = next(synth.get_batch_noise())
step_d_loss = synth.discriminator_loss(X_, Z_)
if step_d_loss > 0.15:
step_d_loss = synth.train_discriminator(X_, Z_, discriminator_opt)
#
# Analyzing
# ¶
#
#
sample_size = 250
idx = np.random.permutation(len(stock_data))[:sample_size]
real_sample = np.asarray(stock_data)[idx]
synth_data = synth.sample(len(stock_data))
synthetic_sample = np.asarray(synth_data)[idx]
# for the purpose of comparision we need the data to be 2-Dimensional. For that reason we are going to use only two componentes for both the PCA and TSNE.
synth_data_reduced = real_sample.reshape(-1, seq_len)
stock_data_reduced = np.asarray(synthetic_sample).reshape(-1, seq_len)
n_components = 2
pca = PCA(n_components=n_components)
tsne = TSNE(n_components=n_components, n_iter=300)
# The fit of the methods must be done only using the real sequential data
pca.fit(stock_data_reduced)
pca_real = pd.DataFrame(pca.transform(stock_data_reduced))
pca_synth = pd.DataFrame(pca.transform(synth_data_reduced))
data_reduced = np.concatenate((stock_data_reduced, synth_data_reduced), axis=0)
tsne_results = pd.DataFrame(tsne.fit_transform(data_reduced))
fig = plt.figure(constrained_layout=True, figsize=(20, 10))
spec = gridspec.GridSpec(ncols=2, nrows=1, figure=fig)
# TSNE scatter plot
ax = fig.add_subplot(spec[0, 0])
ax.set_title("PCA results", fontsize=20, color="red", pad=10)
# PCA scatter plot
plt.scatter(
pca_real.iloc[:, 0].values,
pca_real.iloc[:, 1].values,
c="black",
alpha=0.2,
label="Original",
)
plt.scatter(
pca_synth.iloc[:, 0], pca_synth.iloc[:, 1], c="red", alpha=0.2, label="Synthetic"
)
ax.legend()
ax2 = fig.add_subplot(spec[0, 1])
ax2.set_title("TSNE results", fontsize=20, color="red", pad=10)
plt.scatter(
tsne_results.iloc[:sample_size, 0].values,
tsne_results.iloc[:sample_size, 1].values,
c="black",
alpha=0.2,
label="Original",
)
plt.scatter(
tsne_results.iloc[sample_size:, 0],
tsne_results.iloc[sample_size:, 1],
c="red",
alpha=0.2,
label="Synthetic",
)
ax2.legend()
fig.suptitle(
"Validating synthetic vs real data diversity and distributions",
fontsize=16,
color="grey",
)
# Reshaping the data
cols = ["Open", "High", "Low", "Close", "Adj Close", "Volume"]
# Plotting some generated samples. Both Synthetic and Original data are still standartized with values between [0,1]
fig, axes = plt.subplots(nrows=3, ncols=2, figsize=(15, 10))
axes = axes.flatten()
time = list(range(1, 25))
obs = np.random.randint(len(stock_data))
for j, col in enumerate(cols):
df = pd.DataFrame(
{"Real": stock_data[obs][:, j], "Synthetic": synth_data[obs][:, j]}
)
df.plot(ax=axes[j], title=col, secondary_y="Synthetic data", style=["-", "--"])
fig.tight_layout()
| false | 0 | 5,131 | 13 | 5,131 | 5,131 |
||
69506131
|
<jupyter_start><jupyter_text>Auto-mpg dataset
### Context
The data is technical spec of cars. The dataset is downloaded from UCI Machine Learning Repository
### Content
1. Title: Auto-Mpg Data
2. Sources:
(a) Origin: This dataset was taken from the StatLib library which is
maintained at Carnegie Mellon University. The dataset was
used in the 1983 American Statistical Association Exposition.
(c) Date: July 7, 1993
3. Past Usage:
- See 2b (above)
- Quinlan,R. (1993). Combining Instance-Based and Model-Based Learning.
In Proceedings on the Tenth International Conference of Machine
Learning, 236-243, University of Massachusetts, Amherst. Morgan
Kaufmann.
4. Relevant Information:
This dataset is a slightly modified version of the dataset provided in
the StatLib library. In line with the use by Ross Quinlan (1993) in
predicting the attribute "mpg", 8 of the original instances were removed
because they had unknown values for the "mpg" attribute. The original
dataset is available in the file "auto-mpg.data-original".
"The data concerns city-cycle fuel consumption in miles per gallon,
to be predicted in terms of 3 multivalued discrete and 5 continuous
attributes." (Quinlan, 1993)
5. Number of Instances: 398
6. Number of Attributes: 9 including the class attribute
7. Attribute Information:
1. mpg: continuous
2. cylinders: multi-valued discrete
3. displacement: continuous
4. horsepower: continuous
5. weight: continuous
6. acceleration: continuous
7. model year: multi-valued discrete
8. origin: multi-valued discrete
9. car name: string (unique for each instance)
8. Missing Attribute Values: horsepower has 6 missing values
Kaggle dataset identifier: autompg-dataset
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.linear_model import LinearRegression
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
auto = pd.read_csv("../input/autompg-dataset/auto-mpg.csv")
auto.head()
auto.shape
# Car Name cannot be used in predicting the dependent variable (mpg), so I drop it:
auto = auto.drop("car name", axis=1)
auto.head()
# I replace the categorical values with the actual categories:
auto["origin"] = auto["origin"].replace({1: "america", 2: "europe", 3: "asia"})
auto.head()
# Given the origin categories, I can now create dummy variables:
auto = pd.get_dummies(auto, columns=["origin"])
auto.head()
auto.describe()
auto.dtypes
# horsepower is represented as an object when we needed a numerical field
hp_is_digit = pd.DataFrame(auto.horsepower.str.isdigit())
auto[hp_is_digit["horsepower"] == False]
auto = auto.replace("?", np.nan)
auto[hp_is_digit["horsepower"] == False]
auto.median()
# Replace the missing values with the median value
median_filler = lambda x: x.fillna(x.median())
auto = auto.apply(median_filler, axis=0)
auto["horsepower"] = auto["horsepower"].astype("float64")
auto_attr = auto.iloc[:, 0:7]
sns.pairplot(auto_attr, diag_kind="kde")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/506/69506131.ipynb
|
autompg-dataset
| null |
[{"Id": 69506131, "ScriptId": 18979029, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4714677, "CreationDate": "07/31/2021 21:11:34", "VersionNumber": 3.0, "Title": "auto mpg", "EvaluationDate": "07/31/2021", "IsChange": true, "TotalLines": 68.0, "LinesInsertedFromPrevious": 23.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 45.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 92761042, "KernelVersionId": 69506131, "SourceDatasetVersionId": 2704}]
|
[{"Id": 2704, "DatasetId": 1489, "DatasourceVersionId": 2704, "CreatorUserId": 693660, "LicenseName": "CC0: Public Domain", "CreationDate": "07/02/2017 05:25:54", "VersionNumber": 3.0, "Title": "Auto-mpg dataset", "Slug": "autompg-dataset", "Subtitle": "Mileage per gallon performances of various cars", "Description": "### Context\n\nThe data is technical spec of cars. The dataset is downloaded from UCI Machine Learning Repository\n\n\n### Content\n\n1. Title: Auto-Mpg Data\n\n2. Sources:\n (a) Origin: This dataset was taken from the StatLib library which is\n maintained at Carnegie Mellon University. The dataset was \n used in the 1983 American Statistical Association Exposition.\n (c) Date: July 7, 1993\n\n3. Past Usage:\n - See 2b (above)\n - Quinlan,R. (1993). Combining Instance-Based and Model-Based Learning.\n In Proceedings on the Tenth International Conference of Machine \n Learning, 236-243, University of Massachusetts, Amherst. Morgan\n Kaufmann.\n\n4. Relevant Information:\n\n This dataset is a slightly modified version of the dataset provided in\n the StatLib library. In line with the use by Ross Quinlan (1993) in\n predicting the attribute \"mpg\", 8 of the original instances were removed \n because they had unknown values for the \"mpg\" attribute. The original \n dataset is available in the file \"auto-mpg.data-original\".\n\n \"The data concerns city-cycle fuel consumption in miles per gallon,\n to be predicted in terms of 3 multivalued discrete and 5 continuous\n attributes.\" (Quinlan, 1993)\n\n5. Number of Instances: 398\n\n6. Number of Attributes: 9 including the class attribute\n\n7. Attribute Information:\n\n 1. mpg: continuous\n 2. cylinders: multi-valued discrete\n 3. displacement: continuous\n 4. horsepower: continuous\n 5. weight: continuous\n 6. acceleration: continuous\n 7. model year: multi-valued discrete\n 8. origin: multi-valued discrete\n 9. car name: string (unique for each instance)\n\n8. Missing Attribute Values: horsepower has 6 missing values\n\n### Acknowledgements\n\nDataset: UCI Machine Learning Repository \nData link : https://archive.ics.uci.edu/ml/datasets/auto+mpg\n\n\n### Inspiration\n\nI have used this dataset for practicing my exploratory analysis skills.", "VersionNotes": "Auto-mpg.csv with header", "TotalCompressedBytes": 18131.0, "TotalUncompressedBytes": 18131.0}]
|
[{"Id": 1489, "CreatorUserId": 693660, "OwnerUserId": NaN, "OwnerOrganizationId": 7.0, "CurrentDatasetVersionId": 2704.0, "CurrentDatasourceVersionId": 2704.0, "ForumId": 4406, "Type": 2, "CreationDate": "06/28/2017 10:09:21", "LastActivityDate": "02/05/2018", "TotalViews": 274788, "TotalDownloads": 40258, "TotalVotes": 275, "TotalKernels": 260}]
| null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.linear_model import LinearRegression
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
auto = pd.read_csv("../input/autompg-dataset/auto-mpg.csv")
auto.head()
auto.shape
# Car Name cannot be used in predicting the dependent variable (mpg), so I drop it:
auto = auto.drop("car name", axis=1)
auto.head()
# I replace the categorical values with the actual categories:
auto["origin"] = auto["origin"].replace({1: "america", 2: "europe", 3: "asia"})
auto.head()
# Given the origin categories, I can now create dummy variables:
auto = pd.get_dummies(auto, columns=["origin"])
auto.head()
auto.describe()
auto.dtypes
# horsepower is represented as an object when we needed a numerical field
hp_is_digit = pd.DataFrame(auto.horsepower.str.isdigit())
auto[hp_is_digit["horsepower"] == False]
auto = auto.replace("?", np.nan)
auto[hp_is_digit["horsepower"] == False]
auto.median()
# Replace the missing values with the median value
median_filler = lambda x: x.fillna(x.median())
auto = auto.apply(median_filler, axis=0)
auto["horsepower"] = auto["horsepower"].astype("float64")
auto_attr = auto.iloc[:, 0:7]
sns.pairplot(auto_attr, diag_kind="kde")
| false | 0 | 569 | 1 | 1,064 | 569 |
||
69571331
|
<jupyter_start><jupyter_text>UEFA Euro Cup 2020
<img src="https://upload.wikimedia.org/wikipedia/commons/thumb/a/ab/UEFA_Euro_2020_logo.svg/1200px-UEFA_Euro_2020_logo.svg.png" style="width: 400px">
### UEFA 2020!
This dataset has all information related to the 2020 UEFA European Football Championship in the main stage.
Please feel free to share and use the dataset.
### Columns
| Column | Type | Description | Example |
| --- | --- | --- | --- |
| stage | string |Competition Fase/Stage. | "Final", "Semi-finals", and etc. |
| date | string | When the match occurred. | "11.07.2021" |
| pens | bool | If the match ends with penalties or normal time. | "True" or "False"|
| pens\_home\_score| int or bool| In case of penalties, the team home scores. | "1", "2", and etc... or "False"|
| pens\_away\_score | int or bool| In case of penalties, the team away scores. | "1", "2", and etc... or "False"|
| team\_name\_home | string | The team home name. | "England" |
| team\_name\_away | string | The team away name. | "Italy" |
| team\_home\_score | int| The team home's scores. | "1", "2", and etc...|
| team\_away\_score | int| The team away's scores. | "1", "2", and etc... |
| possession_home | string| Ball possession for the team home. | "10%", "20%", and etc... |
| possession_away | string| Ball possession for the team away. | "10%", "20%", and etc...|
| total\_shots\_home| int| Total shots for the team home. | "5", "8", and etc... |
| total\_shots\_away | int| Total shots for the team away. | "5", "8", and etc... |
| shots\_on\_target\_home | int| How many total shots were on target for the team home?| "5", "8", and etc... |
| shots\_on\_target\_away | int| How many total shots were on target for the team away?| "5", "8", and etc... |
| duels\_won\_home | int| Win possession of the ball against other team's player (for home).| "40%", "60%", and etc...|
| duels\_won\_away | int| Win possession of the ball against other team's player (for away). | "40%", "60%", and etc...|
| events_list | list:json| All events happened during the match: Eg: Goals, Cards, Penalty and etc. | [{'event_team': 'away', 'event_time': " 2' ", 'event_type': 'Goal', 'action_player_1': ' Luke Shaw ', 'action_player_2': ' Kieran Trippier '},...]|
| lineup_home| list:json| The lineup for the team home. | [{'Player_Name': 'Insigne', 'Player_Number': '10'},...]|
| lineup_away | list:json| The lineup for the team away. | [{'Player_Name': 'Kane', 'Player_Number': '9'},...]|
### Inspiration
The inspiration for creating this dataset is to analyze the performance of teams during the competition and relate them to the bet on other platforms around the world
### Source
All data were taken from [One Football](https://onefootball.com/en/competition/uefa-euro-2020-20/results) platform.
The images were taken from [Wikipedia](https://en.wikipedia.org/wiki/UEFA_Euro_2020).
Kaggle dataset identifier: euro-cup-2020
<jupyter_code>import pandas as pd
df = pd.read_csv('euro-cup-2020/eurocup_2020_results.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 51 entries, 0 to 50
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 stage 51 non-null object
1 date 51 non-null object
2 pens 51 non-null bool
3 pens_home_score 51 non-null object
4 pens_away_score 51 non-null object
5 team_name_home 51 non-null object
6 team_name_away 51 non-null object
7 team_home_score 51 non-null int64
8 team_away_score 51 non-null int64
9 possession_home 51 non-null object
10 possession_away 51 non-null object
11 total_shots_home 51 non-null int64
12 total_shots_away 51 non-null int64
13 shots_on_target_home 51 non-null int64
14 shots_on_target_away 51 non-null int64
15 duels_won_home 51 non-null object
16 duels_won_away 51 non-null object
17 events_list 51 non-null object
18 lineup_home 51 non-null object
19 lineup_away 51 non-null object
dtypes: bool(1), int64(6), object(13)
memory usage: 7.7+ KB
<jupyter_text>Examples:
{
"stage": " Final ",
"date": "2021-11-07 00:00:00",
"pens": true,
"pens_home_score": "3",
"pens_away_score": "2",
"team_name_home": " Italy ",
"team_name_away": " England ",
"team_home_score": 1,
"team_away_score": 1,
"possession_home": "66%",
"possession_away": " 34% ",
"total_shots_home": 19,
"total_shots_away": 6,
"shots_on_target_home": 6,
"shots_on_target_away": 2,
"duels_won_home": "53%",
"duels_won_away": " 47% ",
"events_list": "[{'event_team': 'away', 'event_time': \" 2' \", 'event_type': 'Goal', 'action_player_1': ' Luke Shaw ', 'action_player_2': ' Kieran Trippier '}, {'event_team': 'home', 'event_time': \" 47' \", 'event_type': 'Yellow card', 'action_player_1': ' Nicolo Barella '}, {'event_team': 'ho...(truncated)",
"lineup_home": "[{'Player_Name': 'Insigne', 'Player_Number': '10'}, {'Player_Name': 'Immobile', 'Player_Number': '17'}, {'Player_Name': 'Chiesa', 'Player_Number': '14'}, {'Player_Name': 'Verratti', 'Player_Number': '6'}, {'Player_Name': 'Jorginho', 'Player_Number': '8'}, {'Player_Name': 'Barella...(truncated)",
"lineup_away": "[{'Player_Name': 'Kane', 'Player_Number': '9'}, {'Player_Name': 'Mount', 'Player_Number': '19'}, {'Player_Name': 'Sterling', 'Player_Number': '10'}, {'Player_Name': 'Shaw', 'Player_Number': '3'}, {'Player_Name': 'Rice', 'Player_Number': '4'}, {'Player_Name': 'Phillips', 'Player_N...(truncated)",
}
{
"stage": " Semi-finals ",
"date": "2021-07-07 00:00:00",
"pens": false,
"pens_home_score": "False",
"pens_away_score": "False",
"team_name_home": " England ",
"team_name_away": " Denmark ",
"team_home_score": 2,
"team_away_score": 1,
"possession_home": "59%",
"possession_away": " 41% ",
"total_shots_home": 20,
"total_shots_away": 6,
"shots_on_target_home": 10,
"shots_on_target_away": 3,
"duels_won_home": "50%",
"duels_won_away": " 50% ",
"events_list": "[{'event_team': 'away', 'event_time': \" 30' \", 'event_type': 'Goal', 'action_player_1': ' Mikkel Damsgaard '}, {'event_team': 'home', 'event_time': \" 39' \", 'event_type': 'Own goal', 'action_player_1': ' Simon Kjaer ', 'action_player_2': ' Own goal '}, {'event_team': 'home', ...(truncated)",
"lineup_home": "[{'Player_Name': 'Kane', 'Player_Number': '9'}, {'Player_Name': 'Sterling', 'Player_Number': '10'}, {'Player_Name': 'Mount', 'Player_Number': '19'}, {'Player_Name': 'Saka', 'Player_Number': '25'}, {'Player_Name': 'Rice', 'Player_Number': '4'}, {'Player_Name': 'Phillips', 'Player_...(truncated)",
"lineup_away": "[{'Player_Name': 'Krogh Damsgaard', 'Player_Number': '14'}, {'Player_Name': 'Dolberg', 'Player_Number': '12'}, {'Player_Name': 'Braithwaite', 'Player_Number': '9'}, {'Player_Name': 'M\u00e6hle Pedersen', 'Player_Number': '5'}, {'Player_Name': 'Delaney', 'Player_Number': '8'}, {'P...(truncated)",
}
{
"stage": " Semi-finals ",
"date": "2021-06-07 00:00:00",
"pens": true,
"pens_home_score": "4",
"pens_away_score": "2",
"team_name_home": " Italy ",
"team_name_away": " Spain ",
"team_home_score": 1,
"team_away_score": 1,
"possession_home": "29%",
"possession_away": " 71% ",
"total_shots_home": 7,
"total_shots_away": 16,
"shots_on_target_home": 4,
"shots_on_target_away": 5,
"duels_won_home": "49%",
"duels_won_away": " 51% ",
"events_list": "[{'event_team': 'away', 'event_time': \" 51' \", 'event_type': 'Yellow card', 'action_player_1': ' Sergio Busquets '}, {'event_team': 'home', 'event_time': \" 60' \", 'event_type': 'Goal', 'action_player_1': ' Federico Chiesa ', 'action_player_2': ' Ciro Immobile '}, {'event_team...(truncated)",
"lineup_home": "[{'Player_Name': 'Insigne', 'Player_Number': '10'}, {'Player_Name': 'Immobile', 'Player_Number': '17'}, {'Player_Name': 'Chiesa', 'Player_Number': '14'}, {'Player_Name': 'Verratti', 'Player_Number': '6'}, {'Player_Name': 'Jorginho', 'Player_Number': '8'}, {'Player_Name': 'Barella...(truncated)",
"lineup_away": "[{'Player_Name': 'Torres', 'Player_Number': '11'}, {'Player_Name': 'Olmo Carvajal', 'Player_Number': '19'}, {'Player_Name': 'Oyarzabal', 'Player_Number': '21'}, {'Player_Name': 'Gonz\u00e1lez L\u00f3pez', 'Player_Number': '26'}, {'Player_Name': 'Busquets', 'Player_Number': '5'}, ...(truncated)",
}
{
"stage": " Quarter-finals ",
"date": "2021-03-07 00:00:00",
"pens": false,
"pens_home_score": "False",
"pens_away_score": "False",
"team_name_home": " Ukraine ",
"team_name_away": " England ",
"team_home_score": 0,
"team_away_score": 4,
"possession_home": "48%",
"possession_away": " 52% ",
"total_shots_home": 7,
"total_shots_away": 10,
"shots_on_target_home": 2,
"shots_on_target_away": 6,
"duels_won_home": "42%",
"duels_won_away": " 59% ",
"events_list": "[{'event_team': 'away', 'event_time': \" 4' \", 'event_type': 'Goal', 'action_player_1': ' Harry Kane ', 'action_player_2': ' Raheem Sterling '}, {'event_team': 'home', 'event_time': \" 35' \", 'event_type': 'Substitution', 'action_player_1': ' Viktor Tsygankov ', 'action_player_...(truncated)",
"lineup_home": "[{'Player_Name': 'Yaremchuk', 'Player_Number': '9'}, {'Player_Name': 'Yarmolenko', 'Player_Number': '7'}, {'Player_Name': 'Mykolenko', 'Player_Number': '16'}, {'Player_Name': 'Zinchenko', 'Player_Number': '17'}, {'Player_Name': 'Sydorchuk', 'Player_Number': '5'}, {'Player_Name': ...(truncated)",
"lineup_away": "[{'Player_Name': 'Kane', 'Player_Number': '9'}, {'Player_Name': 'Sterling', 'Player_Number': '10'}, {'Player_Name': 'Mount', 'Player_Number': '19'}, {'Player_Name': 'Sancho', 'Player_Number': '17'}, {'Player_Name': 'Rice', 'Player_Number': '4'}, {'Player_Name': 'Phillips', 'Playe...(truncated)",
}
<jupyter_script># # This is an analysis of the 2020 UEFA European Football Championship.
# **Importing Libraries**
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df = pd.read_csv("../input/euro-cup-2020/eurocup_2020_results.csv")
df.head()
matches = len(df["stage"])
print(f"A total of {matches} matches were played")
# **Converting penalty columns from object to int**
df["pens_home_score"].replace({"False": 0}, inplace=True)
df["pens_away_score"].replace({"False": 0}, inplace=True)
df.astype({"pens_home_score": "int64", "pens_away_score": "int64"}).dtypes
# **Analyzing Penalty Matches**
pen = df[df["pens"] == True]
print(len(pen), "matches went to a penalty shootout")
home_pen = pen[pen["pens_home_score"] > pen["pens_away_score"]]
away_pen = pen[pen["pens_home_score"] < pen["pens_away_score"]]
home_pen_win = home_pen["team_name_home"]
away_pen_win = away_pen["team_name_away"]
home_pen_lose = home_pen["team_name_away"]
away_pen_lose = away_pen["team_name_home"]
print(home_pen_win, "\n", home_pen_lose)
print("\n\n")
print(away_pen_win, "\n", away_pen_lose)
# As we can see Italy won both the penalty shootouts:
# one against Spain and one against England (Final Match)
# Spain won against Switzerland in the quater-finals and Switzerland won against France in round of 16
# # Analysing Other Matches
# **We need to convert possession and duels won into int64**
df["possession_home"] = df["possession_home"].str.rstrip("%").astype("float") / 100.0
df["possession_away"] = df["possession_away"].str.strip(" %").astype("float") / 100.0
df["duels_won_home"] = df["duels_won_home"].str.strip(" %").astype("float") / 100.0
df["duels_won_away"] = df["duels_won_away"].str.strip(" %").astype("float") / 100.0
df.head(3)
home_win = df[df["team_home_score"] > df["team_away_score"]]
away_win = df[df["team_home_score"] < df["team_away_score"]]
print("Teams that won as home: \n", home_win["team_name_home"].value_counts())
print("\nTeams that won as away: \n", away_win["team_name_away"].value_counts())
print("\nTotal games won by home teams: ", len(home_win))
print("\nTotal games won by away teams: ", len(away_win))
# **Which teams had more possession and more duels (Win possession of the ball against other team's player)**
# In the data below, home teams are the winner
home_win[
[
"team_name_home",
"team_name_away",
"possession_home",
"possession_away",
"duels_won_home",
"duels_won_away",
]
]
# In the data below, away teams are the winner
away_win[
[
"team_name_home",
"team_name_away",
"possession_home",
"possession_away",
"duels_won_home",
"duels_won_away",
]
]
# **Possession for home teams that won**
sns.catplot(
x="team_name_home",
y="possession_home",
jitter=False,
data=home_win,
height=5,
aspect=2,
)
# **Possession for away teams that won**
sns.catplot(
x="team_name_away",
y="possession_away",
jitter=False,
data=away_win,
height=5,
aspect=3,
)
# **Shots by winning home teams**
sns.catplot(
x="team_name_home",
y="total_shots_home",
hue="shots_on_target_home",
jitter=False,
data=home_win,
height=5,
aspect=3,
)
# **Shots by winning away teams**
sns.catplot(
x="team_name_away",
y="total_shots_away",
hue="shots_on_target_away",
jitter=False,
data=away_win,
height=5,
aspect=3,
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/571/69571331.ipynb
|
euro-cup-2020
|
mcarujo
|
[{"Id": 69571331, "ScriptId": 18679592, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7151095, "CreationDate": "08/01/2021 16:55:14", "VersionNumber": 1.0, "Title": "Euro_2020_EDA", "EvaluationDate": "08/01/2021", "IsChange": true, "TotalLines": 99.0, "LinesInsertedFromPrevious": 99.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 7}]
|
[{"Id": 92918543, "KernelVersionId": 69571331, "SourceDatasetVersionId": 2438592}]
|
[{"Id": 2438592, "DatasetId": 1475678, "DatasourceVersionId": 2480859, "CreatorUserId": 3565979, "LicenseName": "CC0: Public Domain", "CreationDate": "07/18/2021 18:03:38", "VersionNumber": 1.0, "Title": "UEFA Euro Cup 2020", "Slug": "euro-cup-2020", "Subtitle": "All match results from The UEFA Euro Cup 2020 (2021 due to Covid-19 pandemic).", "Description": "<img src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/a/ab/UEFA_Euro_2020_logo.svg/1200px-UEFA_Euro_2020_logo.svg.png\" style=\"width: 400px\">\n### UEFA 2020!\nThis dataset has all information related to the 2020 UEFA European Football Championship in the main stage.\nPlease feel free to share and use the dataset.\n\n\n### Columns\n| Column | Type | Description | Example |\n| --- | --- | --- | --- |\n| stage | string |Competition Fase/Stage. | \"Final\", \"Semi-finals\", and etc. |\n| date | string | When the match occurred. | \"11.07.2021\" |\n| pens | bool | If the match ends with penalties or normal time. | \"True\" or \"False\"|\n| pens\\_home\\_score| int or bool| In case of penalties, the team home scores. | \"1\", \"2\", and etc... or \"False\"|\n| pens\\_away\\_score\t| int or bool| In case of penalties, the team away scores. | \"1\", \"2\", and etc... or \"False\"|\n| team\\_name\\_home | string | The team home name. | \"England\" |\n| team\\_name\\_away | string | The team away name. | \"Italy\" |\n| team\\_home\\_score | int| The team home's scores. | \"1\", \"2\", and etc...|\n| team\\_away\\_score | int| The team away's scores. | \"1\", \"2\", and etc... |\n| possession_home | string| Ball possession for the team home. | \"10%\", \"20%\", and etc... |\n| possession_away | string| Ball possession for the team away. | \"10%\", \"20%\", and etc...|\n| total\\_shots\\_home| int| Total shots for the team home. | \"5\", \"8\", and etc... |\n| total\\_shots\\_away | int| Total shots for the team away. | \"5\", \"8\", and etc... |\n| shots\\_on\\_target\\_home | int| How many total shots were on target for the team home?| \"5\", \"8\", and etc... |\n| shots\\_on\\_target\\_away | int| How many total shots were on target for the team away?| \"5\", \"8\", and etc... |\n| duels\\_won\\_home | int| Win possession of the ball against other team's player (for home).| \"40%\", \"60%\", and etc...|\n| duels\\_won\\_away | int| Win possession of the ball against other team's player (for away). | \"40%\", \"60%\", and etc...|\n| events_list | list:json| All events happened during the match: Eg: Goals, Cards, Penalty and etc. | [{'event_team': 'away', 'event_time': \" 2' \", 'event_type': 'Goal', 'action_player_1': ' Luke Shaw ', 'action_player_2': ' Kieran Trippier '},...]|\n| lineup_home| list:json| The lineup for the team home. | [{'Player_Name': 'Insigne', 'Player_Number': '10'},...]|\n| lineup_away | list:json| The lineup for the team away. | [{'Player_Name': 'Kane', 'Player_Number': '9'},...]|\n\n\n### Inspiration\n\nThe inspiration for creating this dataset is to analyze the performance of teams during the competition and relate them to the bet on other platforms around the world\n\n\n### Source\nAll data were taken from [One Football](https://onefootball.com/en/competition/uefa-euro-2020-20/results) platform.\nThe images were taken from [Wikipedia](https://en.wikipedia.org/wiki/UEFA_Euro_2020).", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1475678, "CreatorUserId": 3565979, "OwnerUserId": 3565979.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2438592.0, "CurrentDatasourceVersionId": 2480859.0, "ForumId": 1495333, "Type": 2, "CreationDate": "07/18/2021 18:03:38", "LastActivityDate": "07/18/2021", "TotalViews": 8605, "TotalDownloads": 836, "TotalVotes": 30, "TotalKernels": 1}]
|
[{"Id": 3565979, "UserName": "mcarujo", "DisplayName": "Marco Carujo", "RegisterDate": "08/10/2019", "PerformanceTier": 2}]
|
# # This is an analysis of the 2020 UEFA European Football Championship.
# **Importing Libraries**
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df = pd.read_csv("../input/euro-cup-2020/eurocup_2020_results.csv")
df.head()
matches = len(df["stage"])
print(f"A total of {matches} matches were played")
# **Converting penalty columns from object to int**
df["pens_home_score"].replace({"False": 0}, inplace=True)
df["pens_away_score"].replace({"False": 0}, inplace=True)
df.astype({"pens_home_score": "int64", "pens_away_score": "int64"}).dtypes
# **Analyzing Penalty Matches**
pen = df[df["pens"] == True]
print(len(pen), "matches went to a penalty shootout")
home_pen = pen[pen["pens_home_score"] > pen["pens_away_score"]]
away_pen = pen[pen["pens_home_score"] < pen["pens_away_score"]]
home_pen_win = home_pen["team_name_home"]
away_pen_win = away_pen["team_name_away"]
home_pen_lose = home_pen["team_name_away"]
away_pen_lose = away_pen["team_name_home"]
print(home_pen_win, "\n", home_pen_lose)
print("\n\n")
print(away_pen_win, "\n", away_pen_lose)
# As we can see Italy won both the penalty shootouts:
# one against Spain and one against England (Final Match)
# Spain won against Switzerland in the quater-finals and Switzerland won against France in round of 16
# # Analysing Other Matches
# **We need to convert possession and duels won into int64**
df["possession_home"] = df["possession_home"].str.rstrip("%").astype("float") / 100.0
df["possession_away"] = df["possession_away"].str.strip(" %").astype("float") / 100.0
df["duels_won_home"] = df["duels_won_home"].str.strip(" %").astype("float") / 100.0
df["duels_won_away"] = df["duels_won_away"].str.strip(" %").astype("float") / 100.0
df.head(3)
home_win = df[df["team_home_score"] > df["team_away_score"]]
away_win = df[df["team_home_score"] < df["team_away_score"]]
print("Teams that won as home: \n", home_win["team_name_home"].value_counts())
print("\nTeams that won as away: \n", away_win["team_name_away"].value_counts())
print("\nTotal games won by home teams: ", len(home_win))
print("\nTotal games won by away teams: ", len(away_win))
# **Which teams had more possession and more duels (Win possession of the ball against other team's player)**
# In the data below, home teams are the winner
home_win[
[
"team_name_home",
"team_name_away",
"possession_home",
"possession_away",
"duels_won_home",
"duels_won_away",
]
]
# In the data below, away teams are the winner
away_win[
[
"team_name_home",
"team_name_away",
"possession_home",
"possession_away",
"duels_won_home",
"duels_won_away",
]
]
# **Possession for home teams that won**
sns.catplot(
x="team_name_home",
y="possession_home",
jitter=False,
data=home_win,
height=5,
aspect=2,
)
# **Possession for away teams that won**
sns.catplot(
x="team_name_away",
y="possession_away",
jitter=False,
data=away_win,
height=5,
aspect=3,
)
# **Shots by winning home teams**
sns.catplot(
x="team_name_home",
y="total_shots_home",
hue="shots_on_target_home",
jitter=False,
data=home_win,
height=5,
aspect=3,
)
# **Shots by winning away teams**
sns.catplot(
x="team_name_away",
y="total_shots_away",
hue="shots_on_target_away",
jitter=False,
data=away_win,
height=5,
aspect=3,
)
|
[{"euro-cup-2020/eurocup_2020_results.csv": {"column_names": "[\"stage\", \"date\", \"pens\", \"pens_home_score\", \"pens_away_score\", \"team_name_home\", \"team_name_away\", \"team_home_score\", \"team_away_score\", \"possession_home\", \"possession_away\", \"total_shots_home\", \"total_shots_away\", \"shots_on_target_home\", \"shots_on_target_away\", \"duels_won_home\", \"duels_won_away\", \"events_list\", \"lineup_home\", \"lineup_away\"]", "column_data_types": "{\"stage\": \"object\", \"date\": \"object\", \"pens\": \"bool\", \"pens_home_score\": \"object\", \"pens_away_score\": \"object\", \"team_name_home\": \"object\", \"team_name_away\": \"object\", \"team_home_score\": \"int64\", \"team_away_score\": \"int64\", \"possession_home\": \"object\", \"possession_away\": \"object\", \"total_shots_home\": \"int64\", \"total_shots_away\": \"int64\", \"shots_on_target_home\": \"int64\", \"shots_on_target_away\": \"int64\", \"duels_won_home\": \"object\", \"duels_won_away\": \"object\", \"events_list\": \"object\", \"lineup_home\": \"object\", \"lineup_away\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 51 entries, 0 to 50\nData columns (total 20 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 stage 51 non-null object\n 1 date 51 non-null object\n 2 pens 51 non-null bool \n 3 pens_home_score 51 non-null object\n 4 pens_away_score 51 non-null object\n 5 team_name_home 51 non-null object\n 6 team_name_away 51 non-null object\n 7 team_home_score 51 non-null int64 \n 8 team_away_score 51 non-null int64 \n 9 possession_home 51 non-null object\n 10 possession_away 51 non-null object\n 11 total_shots_home 51 non-null int64 \n 12 total_shots_away 51 non-null int64 \n 13 shots_on_target_home 51 non-null int64 \n 14 shots_on_target_away 51 non-null int64 \n 15 duels_won_home 51 non-null object\n 16 duels_won_away 51 non-null object\n 17 events_list 51 non-null object\n 18 lineup_home 51 non-null object\n 19 lineup_away 51 non-null object\ndtypes: bool(1), int64(6), object(13)\nmemory usage: 7.7+ KB\n", "summary": "{\"team_home_score\": {\"count\": 51.0, \"mean\": 1.196078431372549, \"std\": 1.0773970084075277, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 3.0}, \"team_away_score\": {\"count\": 51.0, \"mean\": 1.588235294117647, \"std\": 1.3442688806668894, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 2.0, \"max\": 5.0}, \"total_shots_home\": {\"count\": 51.0, \"mean\": 12.137254901960784, \"std\": 6.1903783659583755, \"min\": 3.0, \"25%\": 7.0, \"50%\": 11.0, \"75%\": 17.0, \"max\": 27.0}, \"total_shots_away\": {\"count\": 51.0, \"mean\": 12.235294117647058, \"std\": 5.788223338103386, \"min\": 1.0, \"25%\": 8.5, \"50%\": 11.0, \"75%\": 16.0, \"max\": 28.0}, \"shots_on_target_home\": {\"count\": 51.0, \"mean\": 3.803921568627451, \"std\": 2.545738461375302, \"min\": 0.0, \"25%\": 2.0, \"50%\": 4.0, \"75%\": 6.0, \"max\": 10.0}, \"shots_on_target_away\": {\"count\": 51.0, \"mean\": 4.352941176470588, \"std\": 2.681965916351397, \"min\": 0.0, \"25%\": 2.0, \"50%\": 4.0, \"75%\": 6.5, \"max\": 10.0}}", "examples": "{\"stage\":{\"0\":\" Final \",\"1\":\" Semi-finals \",\"2\":\" Semi-finals \",\"3\":\" Quarter-finals \"},\"date\":{\"0\":\"11.07.2021\",\"1\":\" 07.07.2021 \",\"2\":\"06.07.2021\",\"3\":\" 03.07.2021 \"},\"pens\":{\"0\":true,\"1\":false,\"2\":true,\"3\":false},\"pens_home_score\":{\"0\":\"3\",\"1\":\"False\",\"2\":\"4\",\"3\":\"False\"},\"pens_away_score\":{\"0\":\"2\",\"1\":\"False\",\"2\":\"2\",\"3\":\"False\"},\"team_name_home\":{\"0\":\" Italy \",\"1\":\" England \",\"2\":\" Italy \",\"3\":\" Ukraine \"},\"team_name_away\":{\"0\":\" England \",\"1\":\" Denmark \",\"2\":\" Spain \",\"3\":\" England \"},\"team_home_score\":{\"0\":1,\"1\":2,\"2\":1,\"3\":0},\"team_away_score\":{\"0\":1,\"1\":1,\"2\":1,\"3\":4},\"possession_home\":{\"0\":\"66%\",\"1\":\"59%\",\"2\":\"29%\",\"3\":\"48%\"},\"possession_away\":{\"0\":\" 34% \",\"1\":\" 41% \",\"2\":\" 71% \",\"3\":\" 52% \"},\"total_shots_home\":{\"0\":19,\"1\":20,\"2\":7,\"3\":7},\"total_shots_away\":{\"0\":6,\"1\":6,\"2\":16,\"3\":10},\"shots_on_target_home\":{\"0\":6,\"1\":10,\"2\":4,\"3\":2},\"shots_on_target_away\":{\"0\":2,\"1\":3,\"2\":5,\"3\":6},\"duels_won_home\":{\"0\":\"53%\",\"1\":\"50%\",\"2\":\"49%\",\"3\":\"42%\"},\"duels_won_away\":{\"0\":\" 47% \",\"1\":\" 50% \",\"2\":\" 51% \",\"3\":\" 59% \"},\"events_list\":{\"0\":\"[{'event_team': 'away', 'event_time': \\\" 2' \\\", 'event_type': 'Goal', 'action_player_1': ' Luke Shaw ', 'action_player_2': ' Kieran Trippier '}, {'event_team': 'home', 'event_time': \\\" 47' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Nicolo Barella '}, {'event_team': 'home', 'event_time': \\\" 54' \\\", 'event_type': 'Substitution', 'action_player_1': ' Bryan Cristante ', 'action_player_2': ' Nicolo Barella '}, {'event_team': 'home', 'event_time': \\\" 55' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Leonardo Bonucci '}, {'event_team': 'home', 'event_time': \\\" 55' \\\", 'event_type': 'Substitution', 'action_player_1': ' Domenico Berardi ', 'action_player_2': ' Ciro Immobile '}, {'event_team': 'home', 'event_time': \\\" 67' \\\", 'event_type': 'Goal', 'action_player_1': ' Leonardo Bonucci '}, {'event_team': 'away', 'event_time': \\\" 70' \\\", 'event_type': 'Substitution', 'action_player_1': ' Bukayo Saka ', 'action_player_2': ' Kieran Trippier '}, {'event_team': 'away', 'event_time': \\\" 74' \\\", 'event_type': 'Substitution', 'action_player_1': ' Jordan Henderson ', 'action_player_2': ' Declan Rice '}, {'event_team': 'home', 'event_time': \\\" 84' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Lorenzo Insigne '}, {'event_team': 'home', 'event_time': \\\" 86' \\\", 'event_type': 'Substitution', 'action_player_1': ' Federico Bernardeschi ', 'action_player_2': ' Federico Chiesa '}, {'event_team': 'home', 'event_time': \\\" 96' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Giorgio Chiellini '}, {'event_team': 'home', 'event_time': \\\" 90' \\\", 'event_type': 'Substitution', 'action_player_1': ' Andrea Belotti ', 'action_player_2': ' Lorenzo Insigne '}, {'event_team': 'home', 'event_time': \\\" 96' \\\", 'event_type': 'Substitution', 'action_player_1': ' Manuel Locatelli ', 'action_player_2': ' Marco Verratti '}, {'event_team': 'away', 'event_time': \\\" 99' \\\", 'event_type': 'Substitution', 'action_player_1': ' Jack Grealish ', 'action_player_2': ' Mason Mount '}, {'event_team': 'away', 'event_time': \\\" 106' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Harry Maguire '}, {'event_team': 'home', 'event_time': \\\" 114' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Jorginho '}, {'event_team': 'home', 'event_time': \\\" 118' \\\", 'event_type': 'Substitution', 'action_player_1': ' Alessandro Florenzi ', 'action_player_2': ' Emerson '}, {'event_team': 'away', 'event_time': \\\" 120' \\\", 'event_type': 'Substitution', 'action_player_1': ' Marcus Rashford ', 'action_player_2': ' Jordan Henderson '}, {'event_team': 'away', 'event_time': \\\" 120' \\\", 'event_type': 'Substitution', 'action_player_1': ' Jadon Sancho ', 'action_player_2': ' Kyle Walker '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Domenico Berardi '}, {'event_team': 'away', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Harry Kane '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Missed', 'event_player': ' Andrea Belotti '}, {'event_team': 'away', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Harry Maguire '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Leonardo Bonucci '}, {'event_team': 'away', 'event_time': False, 'event_type': 'PK', 'event_result': 'Missed', 'event_player': ' Marcus Rashford '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Federico Bernardeschi '}, {'event_team': 'away', 'event_time': False, 'event_type': 'PK', 'event_result': 'Missed', 'event_player': ' Jadon Sancho '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Missed', 'event_player': ' Jorginho '}, {'event_team': 'away', 'event_time': False, 'event_type': 'PK', 'event_result': 'Missed', 'event_player': ' Bukayo Saka '}]\",\"1\":\"[{'event_team': 'away', 'event_time': \\\" 30' \\\", 'event_type': 'Goal', 'action_player_1': ' Mikkel Damsgaard '}, {'event_team': 'home', 'event_time': \\\" 39' \\\", 'event_type': 'Own goal', 'action_player_1': ' Simon Kjaer ', 'action_player_2': ' Own goal '}, {'event_team': 'home', 'event_time': \\\" 49' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Harry Maguire '}, {'event_team': 'away', 'event_time': \\\" 67' \\\", 'event_type': 'Substitution', 'action_player_1': ' Daniel Wass ', 'action_player_2': ' Jens Stryger Larsen '}, {'event_team': 'away', 'event_time': \\\" 67' \\\", 'event_type': 'Substitution', 'action_player_1': ' Yussuf Poulsen ', 'action_player_2': ' Mikkel Damsgaard '}, {'event_team': 'away', 'event_time': \\\" 67' \\\", 'event_type': 'Substitution', 'action_player_1': ' Christian N\\u00f8rgaard ', 'action_player_2': ' Kasper Dolberg '}, {'event_team': 'home', 'event_time': \\\" 69' \\\", 'event_type': 'Substitution', 'action_player_1': ' Jack Grealish ', 'action_player_2': ' Bukayo Saka '}, {'event_team': 'away', 'event_time': \\\" 72' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Daniel Wass '}, {'event_team': 'away', 'event_time': \\\" 79' \\\", 'event_type': 'Substitution', 'action_player_1': ' Joachim Andersen ', 'action_player_2': ' Andreas Christensen '}, {'event_team': 'away', 'event_time': \\\" 88' \\\", 'event_type': 'Substitution', 'action_player_1': ' Mathias Jensen ', 'action_player_2': ' Thomas Delaney '}, {'event_team': 'home', 'event_time': \\\" 95' \\\", 'event_type': 'Substitution', 'action_player_1': ' Jordan Henderson ', 'action_player_2': ' Declan Rice '}, {'event_team': 'home', 'event_time': \\\" 95' \\\", 'event_type': 'Substitution', 'action_player_1': ' Philip Foden ', 'action_player_2': ' Mason Mount '}, {'event_team': 'home', 'event_time': \\\" 104' \\\", 'event_type': 'Goal', 'action_player_1': ' Harry Kane '}, {'event_team': 'away', 'event_time': \\\" 105' \\\", 'event_type': 'Substitution', 'action_player_1': ' Jonas Wind ', 'action_player_2': ' Jannik Vestergaard '}, {'event_team': 'home', 'event_time': \\\" 105' \\\", 'event_type': 'Substitution', 'action_player_1': ' Kieran Trippier ', 'action_player_2': ' Jack Grealish '}]\",\"2\":\"[{'event_team': 'away', 'event_time': \\\" 51' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Sergio Busquets '}, {'event_team': 'home', 'event_time': \\\" 60' \\\", 'event_type': 'Goal', 'action_player_1': ' Federico Chiesa ', 'action_player_2': ' Ciro Immobile '}, {'event_team': 'home', 'event_time': \\\" 61' \\\", 'event_type': 'Substitution', 'action_player_1': ' Domenico Berardi ', 'action_player_2': ' Ciro Immobile '}, {'event_team': 'away', 'event_time': \\\" 62' \\\", 'event_type': 'Substitution', 'action_player_1': ' Alvaro Morata ', 'action_player_2': ' Ferran Torres '}, {'event_team': 'away', 'event_time': \\\" 70' \\\", 'event_type': 'Substitution', 'action_player_1': ' Gerard Moreno ', 'action_player_2': ' Mikel Oyarzabal '}, {'event_team': 'away', 'event_time': \\\" 70' \\\", 'event_type': 'Substitution', 'action_player_1': ' Rodri ', 'action_player_2': ' Koke '}, {'event_team': 'home', 'event_time': \\\" 74' \\\", 'event_type': 'Substitution', 'action_player_1': ' Matteo Pessina ', 'action_player_2': ' Marco Verratti '}, {'event_team': 'home', 'event_time': \\\" 74' \\\", 'event_type': 'Substitution', 'action_player_1': ' Rafael Tol\\u00f3i ', 'action_player_2': ' Emerson '}, {'event_team': 'away', 'event_time': \\\" 80' \\\", 'event_type': 'Goal', 'action_player_1': ' Alvaro Morata ', 'action_player_2': ' Daniel Olmo '}, {'event_team': 'home', 'event_time': \\\" 85' \\\", 'event_type': 'Substitution', 'action_player_1': ' Manuel Locatelli ', 'action_player_2': ' Nicolo Barella '}, {'event_team': 'away', 'event_time': \\\" 85' \\\", 'event_type': 'Substitution', 'action_player_1': ' Marcos Llorente ', 'action_player_2': ' Cesar Azpilicueta '}, {'event_team': 'home', 'event_time': \\\" 85' \\\", 'event_type': 'Substitution', 'action_player_1': ' Andrea Belotti ', 'action_player_2': ' Lorenzo Insigne '}, {'event_team': 'home', 'event_time': \\\" 97' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Rafael Tol\\u00f3i '}, {'event_team': 'away', 'event_time': \\\" 105' \\\", 'event_type': 'Substitution', 'action_player_1': ' Thiago Alcantara ', 'action_player_2': ' Sergio Busquets '}, {'event_team': 'home', 'event_time': \\\" 107' \\\", 'event_type': 'Substitution', 'action_player_1': ' Federico Bernardeschi ', 'action_player_2': ' Federico Chiesa '}, {'event_team': 'away', 'event_time': \\\" 109' \\\", 'event_type': 'Substitution', 'action_player_1': ' Pau Torres ', 'action_player_2': ' Eric Garc\\u00eda '}, {'event_team': 'home', 'event_time': \\\" 118' \\\", 'event_type': 'Yellow card', 'action_player_1': ' Leonardo Bonucci '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Missed', 'event_player': ' Manuel Locatelli '}, {'event_team': 'away', 'event_time': False, 'event_type': 'PK', 'event_result': 'Missed', 'event_player': ' Daniel Olmo '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Andrea Belotti '}, {'event_team': 'away', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Gerard Moreno '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Leonardo Bonucci '}, {'event_team': 'away', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Thiago Alcantara '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Federico Bernardeschi '}, {'event_team': 'away', 'event_time': False, 'event_type': 'PK', 'event_result': 'Missed', 'event_player': ' Alvaro Morata '}, {'event_team': 'home', 'event_time': False, 'event_type': 'PK', 'event_result': 'Goal', 'event_player': ' Jorginho '}]\",\"3\":\"[{'event_team': 'away', 'event_time': \\\" 4' \\\", 'event_type': 'Goal', 'action_player_1': ' Harry Kane ', 'action_player_2': ' Raheem Sterling '}, {'event_team': 'home', 'event_time': \\\" 35' \\\", 'event_type': 'Substitution', 'action_player_1': ' Viktor Tsygankov ', 'action_player_2': ' Serhii Kryvtsov '}, {'event_team': 'away', 'event_time': \\\" 46' \\\", 'event_type': 'Goal', 'action_player_1': ' Harry Maguire ', 'action_player_2': ' Luke Shaw '}, {'event_team': 'away', 'event_time': \\\" 50' \\\", 'event_type': 'Goal', 'action_player_1': ' Harry Kane ', 'action_player_2': ' Luke Shaw '}, {'event_team': 'away', 'event_time': \\\" 57' \\\", 'event_type': 'Substitution', 'action_player_1': ' Jordan Henderson ', 'action_player_2': ' Declan Rice '}, {'event_team': 'away', 'event_time': \\\" 63' \\\", 'event_type': 'Goal', 'action_player_1': ' Jordan Henderson ', 'action_player_2': ' Mason Mount '}, {'event_team': 'home', 'event_time': \\\" 64' \\\", 'event_type': 'Substitution', 'action_player_1': ' Yevhen Makarenko ', 'action_player_2': ' Serhiy Sydorchuk '}, {'event_team': 'away', 'event_time': \\\" 65' \\\", 'event_type': 'Substitution', 'action_player_1': ' Kieran Trippier ', 'action_player_2': ' Luke Shaw '}, {'event_team': 'away', 'event_time': \\\" 65' \\\", 'event_type': 'Substitution', 'action_player_1': ' Marcus Rashford ', 'action_player_2': ' Raheem Sterling '}, {'event_team': 'away', 'event_time': \\\" 65' \\\", 'event_type': 'Substitution', 'action_player_1': ' Jude Bellingham ', 'action_player_2': ' Kalvin Phillips '}, {'event_team': 'away', 'event_time': \\\" 73' \\\", 'event_type': 'Substitution', 'action_player_1': ' Dominic Calvert-Lewin ', 'action_player_2': ' Harry Kane '}]\"},\"lineup_home\":{\"0\":\"[{'Player_Name': 'Insigne', 'Player_Number': '10'}, {'Player_Name': 'Immobile', 'Player_Number': '17'}, {'Player_Name': 'Chiesa', 'Player_Number': '14'}, {'Player_Name': 'Verratti', 'Player_Number': '6'}, {'Player_Name': 'Jorginho', 'Player_Number': '8'}, {'Player_Name': 'Barella', 'Player_Number': '18'}, {'Player_Name': 'Emerson', 'Player_Number': '13'}, {'Player_Name': 'Chiellini', 'Player_Number': '3'}, {'Player_Name': 'Bonucci', 'Player_Number': '19'}, {'Player_Name': 'Di Lorenzo', 'Player_Number': '2'}, {'Player_Name': 'Donnarumma', 'Player_Number': '21'}]\",\"1\":\"[{'Player_Name': 'Kane', 'Player_Number': '9'}, {'Player_Name': 'Sterling', 'Player_Number': '10'}, {'Player_Name': 'Mount', 'Player_Number': '19'}, {'Player_Name': 'Saka', 'Player_Number': '25'}, {'Player_Name': 'Rice', 'Player_Number': '4'}, {'Player_Name': 'Phillips', 'Player_Number': '14'}, {'Player_Name': 'Shaw', 'Player_Number': '3'}, {'Player_Name': 'Maguire', 'Player_Number': '6'}, {'Player_Name': 'Stones', 'Player_Number': '5'}, {'Player_Name': 'Walker', 'Player_Number': '2'}, {'Player_Name': 'Pickford', 'Player_Number': '1'}]\",\"2\":\"[{'Player_Name': 'Insigne', 'Player_Number': '10'}, {'Player_Name': 'Immobile', 'Player_Number': '17'}, {'Player_Name': 'Chiesa', 'Player_Number': '14'}, {'Player_Name': 'Verratti', 'Player_Number': '6'}, {'Player_Name': 'Jorginho', 'Player_Number': '8'}, {'Player_Name': 'Barella', 'Player_Number': '18'}, {'Player_Name': 'Emerson', 'Player_Number': '13'}, {'Player_Name': 'Chiellini', 'Player_Number': '3'}, {'Player_Name': 'Bonucci', 'Player_Number': '19'}, {'Player_Name': 'Di Lorenzo', 'Player_Number': '2'}, {'Player_Name': 'Donnarumma', 'Player_Number': '21'}]\",\"3\":\"[{'Player_Name': 'Yaremchuk', 'Player_Number': '9'}, {'Player_Name': 'Yarmolenko', 'Player_Number': '7'}, {'Player_Name': 'Mykolenko', 'Player_Number': '16'}, {'Player_Name': 'Zinchenko', 'Player_Number': '17'}, {'Player_Name': 'Sydorchuk', 'Player_Number': '5'}, {'Player_Name': 'Shaparenko', 'Player_Number': '10'}, {'Player_Name': 'Karavaev', 'Player_Number': '21'}, {'Player_Name': 'Matvienko', 'Player_Number': '22'}, {'Player_Name': 'Kryvtsov', 'Player_Number': '4'}, {'Player_Name': 'Zabarnyi', 'Player_Number': '13'}, {'Player_Name': 'Bushchan', 'Player_Number': '1'}]\"},\"lineup_away\":{\"0\":\"[{'Player_Name': 'Kane', 'Player_Number': '9'}, {'Player_Name': 'Mount', 'Player_Number': '19'}, {'Player_Name': 'Sterling', 'Player_Number': '10'}, {'Player_Name': 'Shaw', 'Player_Number': '3'}, {'Player_Name': 'Rice', 'Player_Number': '4'}, {'Player_Name': 'Phillips', 'Player_Number': '14'}, {'Player_Name': 'Trippier', 'Player_Number': '12'}, {'Player_Name': 'Maguire', 'Player_Number': '6'}, {'Player_Name': 'Stones', 'Player_Number': '5'}, {'Player_Name': 'Walker', 'Player_Number': '2'}, {'Player_Name': 'Pickford', 'Player_Number': '1'}]\",\"1\":\"[{'Player_Name': 'Krogh Damsgaard', 'Player_Number': '14'}, {'Player_Name': 'Dolberg', 'Player_Number': '12'}, {'Player_Name': 'Braithwaite', 'Player_Number': '9'}, {'Player_Name': 'M\\u00e6hle Pedersen', 'Player_Number': '5'}, {'Player_Name': 'Delaney', 'Player_Number': '8'}, {'Player_Name': 'H\\u00f8jbjerg', 'Player_Number': '23'}, {'Player_Name': 'Stryger Larsen', 'Player_Number': '17'}, {'Player_Name': 'Vestergaard', 'Player_Number': '3'}, {'Player_Name': 'Kjaer', 'Player_Number': '4'}, {'Player_Name': 'Christensen', 'Player_Number': '6'}, {'Player_Name': 'Schmeichel', 'Player_Number': '1'}]\",\"2\":\"[{'Player_Name': 'Torres', 'Player_Number': '11'}, {'Player_Name': 'Olmo Carvajal', 'Player_Number': '19'}, {'Player_Name': 'Oyarzabal', 'Player_Number': '21'}, {'Player_Name': 'Gonz\\u00e1lez L\\u00f3pez', 'Player_Number': '26'}, {'Player_Name': 'Busquets', 'Player_Number': '5'}, {'Player_Name': 'Koke', 'Player_Number': '8'}, {'Player_Name': 'Alba', 'Player_Number': '18'}, {'Player_Name': 'Laporte', 'Player_Number': '24'}, {'Player_Name': 'Garc\\u00eda Martret', 'Player_Number': '12'}, {'Player_Name': 'Azpilicueta', 'Player_Number': '2'}, {'Player_Name': 'Simon', 'Player_Number': '23'}]\",\"3\":\"[{'Player_Name': 'Kane', 'Player_Number': '9'}, {'Player_Name': 'Sterling', 'Player_Number': '10'}, {'Player_Name': 'Mount', 'Player_Number': '19'}, {'Player_Name': 'Sancho', 'Player_Number': '17'}, {'Player_Name': 'Rice', 'Player_Number': '4'}, {'Player_Name': 'Phillips', 'Player_Number': '14'}, {'Player_Name': 'Shaw', 'Player_Number': '3'}, {'Player_Name': 'Maguire', 'Player_Number': '6'}, {'Player_Name': 'Stones', 'Player_Number': '5'}, {'Player_Name': 'Walker', 'Player_Number': '2'}, {'Player_Name': 'Pickford', 'Player_Number': '1'}]\"}}"}}]
| true | 1 |
<start_data_description><data_path>euro-cup-2020/eurocup_2020_results.csv:
<column_names>
['stage', 'date', 'pens', 'pens_home_score', 'pens_away_score', 'team_name_home', 'team_name_away', 'team_home_score', 'team_away_score', 'possession_home', 'possession_away', 'total_shots_home', 'total_shots_away', 'shots_on_target_home', 'shots_on_target_away', 'duels_won_home', 'duels_won_away', 'events_list', 'lineup_home', 'lineup_away']
<column_types>
{'stage': 'object', 'date': 'object', 'pens': 'bool', 'pens_home_score': 'object', 'pens_away_score': 'object', 'team_name_home': 'object', 'team_name_away': 'object', 'team_home_score': 'int64', 'team_away_score': 'int64', 'possession_home': 'object', 'possession_away': 'object', 'total_shots_home': 'int64', 'total_shots_away': 'int64', 'shots_on_target_home': 'int64', 'shots_on_target_away': 'int64', 'duels_won_home': 'object', 'duels_won_away': 'object', 'events_list': 'object', 'lineup_home': 'object', 'lineup_away': 'object'}
<dataframe_Summary>
{'team_home_score': {'count': 51.0, 'mean': 1.196078431372549, 'std': 1.0773970084075277, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 2.0, 'max': 3.0}, 'team_away_score': {'count': 51.0, 'mean': 1.588235294117647, 'std': 1.3442688806668894, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 2.0, 'max': 5.0}, 'total_shots_home': {'count': 51.0, 'mean': 12.137254901960784, 'std': 6.1903783659583755, 'min': 3.0, '25%': 7.0, '50%': 11.0, '75%': 17.0, 'max': 27.0}, 'total_shots_away': {'count': 51.0, 'mean': 12.235294117647058, 'std': 5.788223338103386, 'min': 1.0, '25%': 8.5, '50%': 11.0, '75%': 16.0, 'max': 28.0}, 'shots_on_target_home': {'count': 51.0, 'mean': 3.803921568627451, 'std': 2.545738461375302, 'min': 0.0, '25%': 2.0, '50%': 4.0, '75%': 6.0, 'max': 10.0}, 'shots_on_target_away': {'count': 51.0, 'mean': 4.352941176470588, 'std': 2.681965916351397, 'min': 0.0, '25%': 2.0, '50%': 4.0, '75%': 6.5, 'max': 10.0}}
<dataframe_info>
RangeIndex: 51 entries, 0 to 50
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 stage 51 non-null object
1 date 51 non-null object
2 pens 51 non-null bool
3 pens_home_score 51 non-null object
4 pens_away_score 51 non-null object
5 team_name_home 51 non-null object
6 team_name_away 51 non-null object
7 team_home_score 51 non-null int64
8 team_away_score 51 non-null int64
9 possession_home 51 non-null object
10 possession_away 51 non-null object
11 total_shots_home 51 non-null int64
12 total_shots_away 51 non-null int64
13 shots_on_target_home 51 non-null int64
14 shots_on_target_away 51 non-null int64
15 duels_won_home 51 non-null object
16 duels_won_away 51 non-null object
17 events_list 51 non-null object
18 lineup_home 51 non-null object
19 lineup_away 51 non-null object
dtypes: bool(1), int64(6), object(13)
memory usage: 7.7+ KB
<some_examples>
{'stage': {'0': ' Final ', '1': ' Semi-finals ', '2': ' Semi-finals ', '3': ' Quarter-finals '}, 'date': {'0': '11.07.2021', '1': ' 07.07.2021 ', '2': '06.07.2021', '3': ' 03.07.2021 '}, 'pens': {'0': True, '1': False, '2': True, '3': False}, 'pens_home_score': {'0': '3', '1': 'False', '2': '4', '3': 'False'}, 'pens_away_score': {'0': '2', '1': 'False', '2': '2', '3': 'False'}, 'team_name_home': {'0': ' Italy ', '1': ' England ', '2': ' Italy ', '3': ' Ukraine '}, 'team_name_away': {'0': ' England ', '1': ' Denmark ', '2': ' Spain ', '3': ' England '}, 'team_home_score': {'0': 1, '1': 2, '2': 1, '3': 0}, 'team_away_score': {'0': 1, '1': 1, '2': 1, '3': 4}, 'possession_home': {'0': '66%', '1': '59%', '2': '29%', '3': '48%'}, 'possession_away': {'0': ' 34% ', '1': ' 41% ', '2': ' 71% ', '3': ' 52% '}, 'total_shots_home': {'0': 19, '1': 20, '2': 7, '3': 7}, 'total_shots_away': {'0': 6, '1': 6, '2': 16, '3': 10}, 'shots_on_target_home': {'0': 6, '1': 10, '2': 4, '3': 2}, 'shots_on_target_away': {'0': 2, '1': 3, '2': 5, '3': 6}, 'duels_won_home': {'0': '53%', '1': '50%', '2': '49%', '3': '42%'}, 'duels_won_away': {'0': ' 47% ', '1': ' 50% ', '2': ' 51% ', '3': ' 59% '}, 'events_list': {'0': '[{\'event_team\': \'away\', \'event_time\': " 2\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Luke Shaw \', \'action_player_2\': \' Kieran Trippier \'}, {\'event_team\': \'home\', \'event_time\': " 47\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Nicolo Barella \'}, {\'event_team\': \'home\', \'event_time\': " 54\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Bryan Cristante \', \'action_player_2\': \' Nicolo Barella \'}, {\'event_team\': \'home\', \'event_time\': " 55\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Leonardo Bonucci \'}, {\'event_team\': \'home\', \'event_time\': " 55\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Domenico Berardi \', \'action_player_2\': \' Ciro Immobile \'}, {\'event_team\': \'home\', \'event_time\': " 67\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Leonardo Bonucci \'}, {\'event_team\': \'away\', \'event_time\': " 70\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Bukayo Saka \', \'action_player_2\': \' Kieran Trippier \'}, {\'event_team\': \'away\', \'event_time\': " 74\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Jordan Henderson \', \'action_player_2\': \' Declan Rice \'}, {\'event_team\': \'home\', \'event_time\': " 84\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Lorenzo Insigne \'}, {\'event_team\': \'home\', \'event_time\': " 86\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Federico Bernardeschi \', \'action_player_2\': \' Federico Chiesa \'}, {\'event_team\': \'home\', \'event_time\': " 96\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Giorgio Chiellini \'}, {\'event_team\': \'home\', \'event_time\': " 90\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Andrea Belotti \', \'action_player_2\': \' Lorenzo Insigne \'}, {\'event_team\': \'home\', \'event_time\': " 96\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Manuel Locatelli \', \'action_player_2\': \' Marco Verratti \'}, {\'event_team\': \'away\', \'event_time\': " 99\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Jack Grealish \', \'action_player_2\': \' Mason Mount \'}, {\'event_team\': \'away\', \'event_time\': " 106\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Harry Maguire \'}, {\'event_team\': \'home\', \'event_time\': " 114\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Jorginho \'}, {\'event_team\': \'home\', \'event_time\': " 118\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Alessandro Florenzi \', \'action_player_2\': \' Emerson \'}, {\'event_team\': \'away\', \'event_time\': " 120\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Marcus Rashford \', \'action_player_2\': \' Jordan Henderson \'}, {\'event_team\': \'away\', \'event_time\': " 120\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Jadon Sancho \', \'action_player_2\': \' Kyle Walker \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Domenico Berardi \'}, {\'event_team\': \'away\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Harry Kane \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Missed\', \'event_player\': \' Andrea Belotti \'}, {\'event_team\': \'away\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Harry Maguire \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Leonardo Bonucci \'}, {\'event_team\': \'away\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Missed\', \'event_player\': \' Marcus Rashford \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Federico Bernardeschi \'}, {\'event_team\': \'away\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Missed\', \'event_player\': \' Jadon Sancho \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Missed\', \'event_player\': \' Jorginho \'}, {\'event_team\': \'away\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Missed\', \'event_player\': \' Bukayo Saka \'}]', '1': '[{\'event_team\': \'away\', \'event_time\': " 30\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Mikkel Damsgaard \'}, {\'event_team\': \'home\', \'event_time\': " 39\' ", \'event_type\': \'Own goal\', \'action_player_1\': \' Simon Kjaer \', \'action_player_2\': \' Own goal \'}, {\'event_team\': \'home\', \'event_time\': " 49\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Harry Maguire \'}, {\'event_team\': \'away\', \'event_time\': " 67\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Daniel Wass \', \'action_player_2\': \' Jens Stryger Larsen \'}, {\'event_team\': \'away\', \'event_time\': " 67\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Yussuf Poulsen \', \'action_player_2\': \' Mikkel Damsgaard \'}, {\'event_team\': \'away\', \'event_time\': " 67\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Christian Nørgaard \', \'action_player_2\': \' Kasper Dolberg \'}, {\'event_team\': \'home\', \'event_time\': " 69\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Jack Grealish \', \'action_player_2\': \' Bukayo Saka \'}, {\'event_team\': \'away\', \'event_time\': " 72\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Daniel Wass \'}, {\'event_team\': \'away\', \'event_time\': " 79\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Joachim Andersen \', \'action_player_2\': \' Andreas Christensen \'}, {\'event_team\': \'away\', \'event_time\': " 88\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Mathias Jensen \', \'action_player_2\': \' Thomas Delaney \'}, {\'event_team\': \'home\', \'event_time\': " 95\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Jordan Henderson \', \'action_player_2\': \' Declan Rice \'}, {\'event_team\': \'home\', \'event_time\': " 95\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Philip Foden \', \'action_player_2\': \' Mason Mount \'}, {\'event_team\': \'home\', \'event_time\': " 104\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Harry Kane \'}, {\'event_team\': \'away\', \'event_time\': " 105\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Jonas Wind \', \'action_player_2\': \' Jannik Vestergaard \'}, {\'event_team\': \'home\', \'event_time\': " 105\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Kieran Trippier \', \'action_player_2\': \' Jack Grealish \'}]', '2': '[{\'event_team\': \'away\', \'event_time\': " 51\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Sergio Busquets \'}, {\'event_team\': \'home\', \'event_time\': " 60\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Federico Chiesa \', \'action_player_2\': \' Ciro Immobile \'}, {\'event_team\': \'home\', \'event_time\': " 61\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Domenico Berardi \', \'action_player_2\': \' Ciro Immobile \'}, {\'event_team\': \'away\', \'event_time\': " 62\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Alvaro Morata \', \'action_player_2\': \' Ferran Torres \'}, {\'event_team\': \'away\', \'event_time\': " 70\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Gerard Moreno \', \'action_player_2\': \' Mikel Oyarzabal \'}, {\'event_team\': \'away\', \'event_time\': " 70\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Rodri \', \'action_player_2\': \' Koke \'}, {\'event_team\': \'home\', \'event_time\': " 74\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Matteo Pessina \', \'action_player_2\': \' Marco Verratti \'}, {\'event_team\': \'home\', \'event_time\': " 74\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Rafael Tolói \', \'action_player_2\': \' Emerson \'}, {\'event_team\': \'away\', \'event_time\': " 80\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Alvaro Morata \', \'action_player_2\': \' Daniel Olmo \'}, {\'event_team\': \'home\', \'event_time\': " 85\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Manuel Locatelli \', \'action_player_2\': \' Nicolo Barella \'}, {\'event_team\': \'away\', \'event_time\': " 85\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Marcos Llorente \', \'action_player_2\': \' Cesar Azpilicueta \'}, {\'event_team\': \'home\', \'event_time\': " 85\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Andrea Belotti \', \'action_player_2\': \' Lorenzo Insigne \'}, {\'event_team\': \'home\', \'event_time\': " 97\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Rafael Tolói \'}, {\'event_team\': \'away\', \'event_time\': " 105\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Thiago Alcantara \', \'action_player_2\': \' Sergio Busquets \'}, {\'event_team\': \'home\', \'event_time\': " 107\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Federico Bernardeschi \', \'action_player_2\': \' Federico Chiesa \'}, {\'event_team\': \'away\', \'event_time\': " 109\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Pau Torres \', \'action_player_2\': \' Eric García \'}, {\'event_team\': \'home\', \'event_time\': " 118\' ", \'event_type\': \'Yellow card\', \'action_player_1\': \' Leonardo Bonucci \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Missed\', \'event_player\': \' Manuel Locatelli \'}, {\'event_team\': \'away\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Missed\', \'event_player\': \' Daniel Olmo \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Andrea Belotti \'}, {\'event_team\': \'away\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Gerard Moreno \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Leonardo Bonucci \'}, {\'event_team\': \'away\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Thiago Alcantara \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Federico Bernardeschi \'}, {\'event_team\': \'away\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Missed\', \'event_player\': \' Alvaro Morata \'}, {\'event_team\': \'home\', \'event_time\': False, \'event_type\': \'PK\', \'event_result\': \'Goal\', \'event_player\': \' Jorginho \'}]', '3': '[{\'event_team\': \'away\', \'event_time\': " 4\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Harry Kane \', \'action_player_2\': \' Raheem Sterling \'}, {\'event_team\': \'home\', \'event_time\': " 35\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Viktor Tsygankov \', \'action_player_2\': \' Serhii Kryvtsov \'}, {\'event_team\': \'away\', \'event_time\': " 46\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Harry Maguire \', \'action_player_2\': \' Luke Shaw \'}, {\'event_team\': \'away\', \'event_time\': " 50\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Harry Kane \', \'action_player_2\': \' Luke Shaw \'}, {\'event_team\': \'away\', \'event_time\': " 57\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Jordan Henderson \', \'action_player_2\': \' Declan Rice \'}, {\'event_team\': \'away\', \'event_time\': " 63\' ", \'event_type\': \'Goal\', \'action_player_1\': \' Jordan Henderson \', \'action_player_2\': \' Mason Mount \'}, {\'event_team\': \'home\', \'event_time\': " 64\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Yevhen Makarenko \', \'action_player_2\': \' Serhiy Sydorchuk \'}, {\'event_team\': \'away\', \'event_time\': " 65\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Kieran Trippier \', \'action_player_2\': \' Luke Shaw \'}, {\'event_team\': \'away\', \'event_time\': " 65\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Marcus Rashford \', \'action_player_2\': \' Raheem Sterling \'}, {\'event_team\': \'away\', \'event_time\': " 65\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Jude Bellingham \', \'action_player_2\': \' Kalvin Phillips \'}, {\'event_team\': \'away\', \'event_time\': " 73\' ", \'event_type\': \'Substitution\', \'action_player_1\': \' Dominic Calvert-Lewin \', \'action_player_2\': \' Harry Kane \'}]'}, 'lineup_home': {'0': "[{'Player_Name': 'Insigne', 'Player_Number': '10'}, {'Player_Name': 'Immobile', 'Player_Number': '17'}, {'Player_Name': 'Chiesa', 'Player_Number': '14'}, {'Player_Name': 'Verratti', 'Player_Number': '6'}, {'Player_Name': 'Jorginho', 'Player_Number': '8'}, {'Player_Name': 'Barella', 'Player_Number': '18'}, {'Player_Name': 'Emerson', 'Player_Number': '13'}, {'Player_Name': 'Chiellini', 'Player_Number': '3'}, {'Player_Name': 'Bonucci', 'Player_Number': '19'}, {'Player_Name': 'Di Lorenzo', 'Player_Number': '2'}, {'Player_Name': 'Donnarumma', 'Player_Number': '21'}]", '1': "[{'Player_Name': 'Kane', 'Player_Number': '9'}, {'Player_Name': 'Sterling', 'Player_Number': '10'}, {'Player_Name': 'Mount', 'Player_Number': '19'}, {'Player_Name': 'Saka', 'Player_Number': '25'}, {'Player_Name': 'Rice', 'Player_Number': '4'}, {'Player_Name': 'Phillips', 'Player_Number': '14'}, {'Player_Name': 'Shaw', 'Player_Number': '3'}, {'Player_Name': 'Maguire', 'Player_Number': '6'}, {'Player_Name': 'Stones', 'Player_Number': '5'}, {'Player_Name': 'Walker', 'Player_Number': '2'}, {'Player_Name': 'Pickford', 'Player_Number': '1'}]", '2': "[{'Player_Name': 'Insigne', 'Player_Number': '10'}, {'Player_Name': 'Immobile', 'Player_Number': '17'}, {'Player_Name': 'Chiesa', 'Player_Number': '14'}, {'Player_Name': 'Verratti', 'Player_Number': '6'}, {'Player_Name': 'Jorginho', 'Player_Number': '8'}, {'Player_Name': 'Barella', 'Player_Number': '18'}, {'Player_Name': 'Emerson', 'Player_Number': '13'}, {'Player_Name': 'Chiellini', 'Player_Number': '3'}, {'Player_Name': 'Bonucci', 'Player_Number': '19'}, {'Player_Name': 'Di Lorenzo', 'Player_Number': '2'}, {'Player_Name': 'Donnarumma', 'Player_Number': '21'}]", '3': "[{'Player_Name': 'Yaremchuk', 'Player_Number': '9'}, {'Player_Name': 'Yarmolenko', 'Player_Number': '7'}, {'Player_Name': 'Mykolenko', 'Player_Number': '16'}, {'Player_Name': 'Zinchenko', 'Player_Number': '17'}, {'Player_Name': 'Sydorchuk', 'Player_Number': '5'}, {'Player_Name': 'Shaparenko', 'Player_Number': '10'}, {'Player_Name': 'Karavaev', 'Player_Number': '21'}, {'Player_Name': 'Matvienko', 'Player_Number': '22'}, {'Player_Name': 'Kryvtsov', 'Player_Number': '4'}, {'Player_Name': 'Zabarnyi', 'Player_Number': '13'}, {'Player_Name': 'Bushchan', 'Player_Number': '1'}]"}, 'lineup_away': {'0': "[{'Player_Name': 'Kane', 'Player_Number': '9'}, {'Player_Name': 'Mount', 'Player_Number': '19'}, {'Player_Name': 'Sterling', 'Player_Number': '10'}, {'Player_Name': 'Shaw', 'Player_Number': '3'}, {'Player_Name': 'Rice', 'Player_Number': '4'}, {'Player_Name': 'Phillips', 'Player_Number': '14'}, {'Player_Name': 'Trippier', 'Player_Number': '12'}, {'Player_Name': 'Maguire', 'Player_Number': '6'}, {'Player_Name': 'Stones', 'Player_Number': '5'}, {'Player_Name': 'Walker', 'Player_Number': '2'}, {'Player_Name': 'Pickford', 'Player_Number': '1'}]", '1': "[{'Player_Name': 'Krogh Damsgaard', 'Player_Number': '14'}, {'Player_Name': 'Dolberg', 'Player_Number': '12'}, {'Player_Name': 'Braithwaite', 'Player_Number': '9'}, {'Player_Name': 'Mæhle Pedersen', 'Player_Number': '5'}, {'Player_Name': 'Delaney', 'Player_Number': '8'}, {'Player_Name': 'Højbjerg', 'Player_Number': '23'}, {'Player_Name': 'Stryger Larsen', 'Player_Number': '17'}, {'Player_Name': 'Vestergaard', 'Player_Number': '3'}, {'Player_Name': 'Kjaer', 'Player_Number': '4'}, {'Player_Name': 'Christensen', 'Player_Number': '6'}, {'Player_Name': 'Schmeichel', 'Player_Number': '1'}]", '2': "[{'Player_Name': 'Torres', 'Player_Number': '11'}, {'Player_Name': 'Olmo Carvajal', 'Player_Number': '19'}, {'Player_Name': 'Oyarzabal', 'Player_Number': '21'}, {'Player_Name': 'González López', 'Player_Number': '26'}, {'Player_Name': 'Busquets', 'Player_Number': '5'}, {'Player_Name': 'Koke', 'Player_Number': '8'}, {'Player_Name': 'Alba', 'Player_Number': '18'}, {'Player_Name': 'Laporte', 'Player_Number': '24'}, {'Player_Name': 'García Martret', 'Player_Number': '12'}, {'Player_Name': 'Azpilicueta', 'Player_Number': '2'}, {'Player_Name': 'Simon', 'Player_Number': '23'}]", '3': "[{'Player_Name': 'Kane', 'Player_Number': '9'}, {'Player_Name': 'Sterling', 'Player_Number': '10'}, {'Player_Name': 'Mount', 'Player_Number': '19'}, {'Player_Name': 'Sancho', 'Player_Number': '17'}, {'Player_Name': 'Rice', 'Player_Number': '4'}, {'Player_Name': 'Phillips', 'Player_Number': '14'}, {'Player_Name': 'Shaw', 'Player_Number': '3'}, {'Player_Name': 'Maguire', 'Player_Number': '6'}, {'Player_Name': 'Stones', 'Player_Number': '5'}, {'Player_Name': 'Walker', 'Player_Number': '2'}, {'Player_Name': 'Pickford', 'Player_Number': '1'}]"}}
<end_description>
| 1,242 | 7 | 4,968 | 1,242 |
69571670
|
# # Loss Given Default Analysis [TPS August]
# 
#
# Photo by
# Konstantin Evdokimov
# on
# Unsplash. All images are by author unless specified otherwise.
#
# # 1. Problem definition
# In this month's TPS competition, we are tasked to predict the amount of money a bank or a financial institution might lose if a loan goes into default.
# Before we start the EDA, let's make sure we are all on the same page on some of the key terms of the problem definition:
# 1. What is loan default?
# - Default is a failure to repay a debt/loan on time. It can occur when a borrower fails to make timely payments on loans such as mortgage, bank loans, car leases, etc.
# 2. What is a loss given default (LGD)?
# - LGD is the amount of money a bank or financial institution might lose if a loan goes into default. Calculating and predicting LGD can be complex and involve many factors.
# As you will see in just a bit, the dataset for the competition has over 100 features and the target `loss` is (I think) LGD. For more information on these terms, check out [this](https://www.kaggle.com/c/tabular-playground-series-aug-2021/discussion/256337) discussion thread.
# The metric used in this competition is Root Mean Squared Error, a regression metric:
# 
# # 2. Setup
import warnings
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import plotly.express as px
import plotly.graph_objects as go
import seaborn as sns
from matplotlib import rcParams
warnings.filterwarnings("ignore")
# Global plot configs
rcParams["axes.spines.top"] = False
rcParams["axes.spines.right"] = False
rcParams["font.family"] = "monospace"
# Pandas global settings
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
pd.options.display.float_format = "{:.5f}".format
# Import data
train_df = pd.read_csv(
"../input/tabular-playground-series-aug-2021/train.csv", index_col="id"
)
test_df = pd.read_csv(
"../input/tabular-playground-series-aug-2021/test.csv", index_col="id"
)
sub = pd.read_csv("../input/tabular-playground-series-aug-2021/sample_submission.csv")
# Colors
dark_red = "#b20710"
black = "#221f1f"
green = "#009473"
# # 3. Overview of the datasets
# Both training and test sets have 100 features, excluding the ID column. The target is given as `loss` and has a discrete distribution.
# Some other observations:
# - Training and test data contain **250k and 150k** observations, respectively
# - There are **no missing values** in both sets
# - All features either have `float64` or `int64` type
# Here are the first few rows of train and test datasets:
train_df.head()
test_df.head()
# # 4. Analyzing the distributions of the features
# Let's start by looking at the high-level summary of both datasets:
stat_summary_train = (
train_df.describe().drop("loss", axis=1).T[["mean", "std", "min", "50%", "max"]]
)
stat_summary_test = test_df.describe().T[["mean", "std", "min", "50%", "max"]]
stat_summary_train.sample(10)
# From a random sample of the summary, we can see that features have rather different scales. As a single-metric overview, we will choose and categorize features based on their mean:
# Bin the mean into categories
bins = [-np.inf, 100, 10000, np.inf]
labels = ["Below 100", "Between 100-10000", "Above 10000"]
stat_summary_train["mean_cats_train"] = pd.cut(
stat_summary_train["mean"], bins=bins, labels=labels
)
# Group by mean categories
grouped_train = stat_summary_train.value_counts("mean_cats_train").sort_values(
ascending=False
)
cmap = [dark_red] * 4
fig, ax = plt.subplots(figsize=(12, 6))
# Plot the bar
bar = grouped_train.plot(kind="bar", ax=ax, color=cmap)
# Display title
fig.text(
0.5,
1,
"Features Grouped by Mean Categories in Train/Test Sets",
fontfamily="monospace",
size="20",
ha="center",
)
# Remove unnecessary elements
ax.yaxis.set_visible(False)
ax.set_xlabel("")
for s in ["top", "left", "right"]:
ax.spines[s].set_visible(False)
# Annotate above the bars
for patch in ax.patches:
text = f"{patch.get_height():.0f}"
x = patch.get_x() + patch.get_width() / 2
y = patch.get_height() + 5
ax.text(x, y, text, ha="center", va="center", fontsize=20)
# Format xticklabels
plt.setp(ax.get_xmajorticklabels(), rotation=0, fontsize="large")
ax.spines["bottom"].set_linewidth(1.5)
fig.text(
0.6,
0.5,
"""
The majority of features in
boths sets have rather low mean.
However, we can observe features
with larger scales. This suggests
we do feature scaling when we get
to preprocessing.
""",
bbox=dict(boxstyle="round", fc="#009473"),
fontsize="large",
)
# Now, let's look at the distributions of both sets as a whole:
fig = plt.figure(figsize=(15, 60))
gs = fig.add_gridspec(20, 5)
gs.update(wspace=0.1, hspace=0.4)
# Add 100 subplots for all features
temp = 0
for row in range(0, 20):
for col in range(0, 5):
locals()[f"ax_{temp}"] = fig.add_subplot(gs[row, col])
locals()[f"ax_{temp}"].tick_params(axis="y", left=False)
locals()[f"ax_{temp}"].get_yaxis().set_visible(False)
locals()[f"ax_{temp}"].set_axisbelow(True)
for s in ["top", "right", "left"]:
locals()[f"ax_{temp}"].spines[s].set_visible(False)
temp += 1
# General texts
fig.suptitle(
"Distribution of Features in Train and Test Sets",
y=0.9,
fontsize=25,
fontweight="bold",
fontfamily="monospace",
)
# Fill subplots with KDEplots of both train and test set features
temp = 0
for feature in test_df.columns.to_list():
for df, color in zip([train_df, test_df], [dark_red, green]):
sns.kdeplot(
df[feature],
shade=True,
color=color,
linewidth=1.5,
alpha=0.7,
zorder=3,
legend=False,
ax=locals()[f"ax_{temp}"],
)
locals()[f"ax_{temp}"].grid(
which="major", axis="x", zorder=0, color="gray", linestyle=":", dashes=(1, 5)
)
locals()[f"ax_{temp}"].set_xlabel(feature)
temp += 1
plt.show()
# Key observations:
# - Train and test sets have roughly the same distributions in terms of features.
# - Many features have or almost have **normal distributions**
# - Some features are **bimodal**
# - Some features are even **trimodal**
# - Most features have **skewed distributions**.
# We need to think about how to make all these normally distributed if we decide to use non-tree based models.
# > Checking the correlation revealed no significant relationships between features (most were between -0.3 and 0.3).
# Next, there are 5 features that are discrete. Let's check their cardinality to see any of them may be categorical:
discrete_cols = [col for col in test_df.columns if test_df[col].dtype == "int64"]
cardinality = train_df[discrete_cols].nunique().sort_values(ascending=False)
colors = [dark_red] * 5
fig, ax = plt.subplots(figsize=(12, 6))
cardinality.plot(kind="bar", color=colors)
# Display title
fig.text(
0.5,
1,
"Cardinality of Discrete Features",
fontfamily="monospace",
size="20",
ha="center",
)
# Remove unnecessary elements
ax.yaxis.set_visible(False)
ax.set_xlabel("")
for s in ["top", "left", "right"]:
ax.spines[s].set_visible(False)
# Annotate above the bars
for patch in ax.patches:
text = f"{patch.get_height():.0f}"
x = patch.get_x() + patch.get_width() / 2
y = patch.get_height() + 10000
ax.text(x, y, text, ha="center", va="center", fontsize=20)
# Format xticklabels
plt.setp(ax.get_xmajorticklabels(), rotation=0, fontsize="large")
ax.spines["bottom"].set_linewidth(1.5)
fig.text(
0.4,
0.5,
"""
These discrete features
all have very high cardinality,
meaning it isn't a good idea to
treat them as categorical.
""",
bbox=dict(boxstyle="round", fc="#009473"),
fontsize="large",
)
# # 5. Analyzing the target
# Let's look at the distribution of the target:
fig, ax = plt.subplots(figsize=(8, 4))
sns.kdeplot(
train_df["loss"],
color=dark_red,
shade=True,
ax=ax,
)
ax.set(xlabel="Target - loss")
plt.title(
"Distribution of the Target",
ha="center",
fontfamily="monospace",
fontsize="large",
fontweight="bold",
size=20,
)
fig.text(
0.4,
0.5,
"""
The target has a skewed distribution.
""",
bbox=dict(boxstyle="round", fc="#009473"),
fontsize="medium",
)
ax.yaxis.set_visible(False)
ax.spines["left"].set_visible(False)
# # 6. XGBoost Baseline
# Since the dataset has high dimensionality, we will apply Principal Component Analysis as a base reduction method. We will pass 0.95 to `n_components` so that PCA will find the minimum number of features we need to keep to preserve at least 95% variance of the dataset.
# We will perform all of the steps inside Sklearn pipelines ending with a baseline XGBoost regressor:
from sklearn.model_selection import KFold, cross_validate, train_test_split
from xgboost import XGBRegressor
# Log transform all features and the target
X, y = train_df.drop("loss", axis=1), train_df[["loss"]]
reg = XGBRegressor(
objective="reg:squarederror",
tree_method="gpu_hist",
)
# Validation set to be used inside early_stopping
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, test_size=0.1, random_state=1121218
)
# Set up `fit_params` for XGBoost
eval_set = [(X_valid, y_valid)]
fit_params = {
"eval_set": eval_set,
"eval_metric": "rmse",
"early_stopping_rounds": 100,
"verbose": False,
}
_ = reg.fit(X_train, y_train, **fit_params)
preds = reg.predict(test_df)
submission = pd.DataFrame({"id": test_df.index, "loss": preds})
submission.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/571/69571670.ipynb
| null | null |
[{"Id": 69571670, "ScriptId": 18988978, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4686011, "CreationDate": "08/01/2021 17:00:02", "VersionNumber": 5.0, "Title": "Relevant EDA + XGBoost CV Baseline", "EvaluationDate": "08/01/2021", "IsChange": true, "TotalLines": 330.0, "LinesInsertedFromPrevious": 5.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 325.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 12}]
| null | null | null | null |
# # Loss Given Default Analysis [TPS August]
# 
#
# Photo by
# Konstantin Evdokimov
# on
# Unsplash. All images are by author unless specified otherwise.
#
# # 1. Problem definition
# In this month's TPS competition, we are tasked to predict the amount of money a bank or a financial institution might lose if a loan goes into default.
# Before we start the EDA, let's make sure we are all on the same page on some of the key terms of the problem definition:
# 1. What is loan default?
# - Default is a failure to repay a debt/loan on time. It can occur when a borrower fails to make timely payments on loans such as mortgage, bank loans, car leases, etc.
# 2. What is a loss given default (LGD)?
# - LGD is the amount of money a bank or financial institution might lose if a loan goes into default. Calculating and predicting LGD can be complex and involve many factors.
# As you will see in just a bit, the dataset for the competition has over 100 features and the target `loss` is (I think) LGD. For more information on these terms, check out [this](https://www.kaggle.com/c/tabular-playground-series-aug-2021/discussion/256337) discussion thread.
# The metric used in this competition is Root Mean Squared Error, a regression metric:
# 
# # 2. Setup
import warnings
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import plotly.express as px
import plotly.graph_objects as go
import seaborn as sns
from matplotlib import rcParams
warnings.filterwarnings("ignore")
# Global plot configs
rcParams["axes.spines.top"] = False
rcParams["axes.spines.right"] = False
rcParams["font.family"] = "monospace"
# Pandas global settings
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
pd.options.display.float_format = "{:.5f}".format
# Import data
train_df = pd.read_csv(
"../input/tabular-playground-series-aug-2021/train.csv", index_col="id"
)
test_df = pd.read_csv(
"../input/tabular-playground-series-aug-2021/test.csv", index_col="id"
)
sub = pd.read_csv("../input/tabular-playground-series-aug-2021/sample_submission.csv")
# Colors
dark_red = "#b20710"
black = "#221f1f"
green = "#009473"
# # 3. Overview of the datasets
# Both training and test sets have 100 features, excluding the ID column. The target is given as `loss` and has a discrete distribution.
# Some other observations:
# - Training and test data contain **250k and 150k** observations, respectively
# - There are **no missing values** in both sets
# - All features either have `float64` or `int64` type
# Here are the first few rows of train and test datasets:
train_df.head()
test_df.head()
# # 4. Analyzing the distributions of the features
# Let's start by looking at the high-level summary of both datasets:
stat_summary_train = (
train_df.describe().drop("loss", axis=1).T[["mean", "std", "min", "50%", "max"]]
)
stat_summary_test = test_df.describe().T[["mean", "std", "min", "50%", "max"]]
stat_summary_train.sample(10)
# From a random sample of the summary, we can see that features have rather different scales. As a single-metric overview, we will choose and categorize features based on their mean:
# Bin the mean into categories
bins = [-np.inf, 100, 10000, np.inf]
labels = ["Below 100", "Between 100-10000", "Above 10000"]
stat_summary_train["mean_cats_train"] = pd.cut(
stat_summary_train["mean"], bins=bins, labels=labels
)
# Group by mean categories
grouped_train = stat_summary_train.value_counts("mean_cats_train").sort_values(
ascending=False
)
cmap = [dark_red] * 4
fig, ax = plt.subplots(figsize=(12, 6))
# Plot the bar
bar = grouped_train.plot(kind="bar", ax=ax, color=cmap)
# Display title
fig.text(
0.5,
1,
"Features Grouped by Mean Categories in Train/Test Sets",
fontfamily="monospace",
size="20",
ha="center",
)
# Remove unnecessary elements
ax.yaxis.set_visible(False)
ax.set_xlabel("")
for s in ["top", "left", "right"]:
ax.spines[s].set_visible(False)
# Annotate above the bars
for patch in ax.patches:
text = f"{patch.get_height():.0f}"
x = patch.get_x() + patch.get_width() / 2
y = patch.get_height() + 5
ax.text(x, y, text, ha="center", va="center", fontsize=20)
# Format xticklabels
plt.setp(ax.get_xmajorticklabels(), rotation=0, fontsize="large")
ax.spines["bottom"].set_linewidth(1.5)
fig.text(
0.6,
0.5,
"""
The majority of features in
boths sets have rather low mean.
However, we can observe features
with larger scales. This suggests
we do feature scaling when we get
to preprocessing.
""",
bbox=dict(boxstyle="round", fc="#009473"),
fontsize="large",
)
# Now, let's look at the distributions of both sets as a whole:
fig = plt.figure(figsize=(15, 60))
gs = fig.add_gridspec(20, 5)
gs.update(wspace=0.1, hspace=0.4)
# Add 100 subplots for all features
temp = 0
for row in range(0, 20):
for col in range(0, 5):
locals()[f"ax_{temp}"] = fig.add_subplot(gs[row, col])
locals()[f"ax_{temp}"].tick_params(axis="y", left=False)
locals()[f"ax_{temp}"].get_yaxis().set_visible(False)
locals()[f"ax_{temp}"].set_axisbelow(True)
for s in ["top", "right", "left"]:
locals()[f"ax_{temp}"].spines[s].set_visible(False)
temp += 1
# General texts
fig.suptitle(
"Distribution of Features in Train and Test Sets",
y=0.9,
fontsize=25,
fontweight="bold",
fontfamily="monospace",
)
# Fill subplots with KDEplots of both train and test set features
temp = 0
for feature in test_df.columns.to_list():
for df, color in zip([train_df, test_df], [dark_red, green]):
sns.kdeplot(
df[feature],
shade=True,
color=color,
linewidth=1.5,
alpha=0.7,
zorder=3,
legend=False,
ax=locals()[f"ax_{temp}"],
)
locals()[f"ax_{temp}"].grid(
which="major", axis="x", zorder=0, color="gray", linestyle=":", dashes=(1, 5)
)
locals()[f"ax_{temp}"].set_xlabel(feature)
temp += 1
plt.show()
# Key observations:
# - Train and test sets have roughly the same distributions in terms of features.
# - Many features have or almost have **normal distributions**
# - Some features are **bimodal**
# - Some features are even **trimodal**
# - Most features have **skewed distributions**.
# We need to think about how to make all these normally distributed if we decide to use non-tree based models.
# > Checking the correlation revealed no significant relationships between features (most were between -0.3 and 0.3).
# Next, there are 5 features that are discrete. Let's check their cardinality to see any of them may be categorical:
discrete_cols = [col for col in test_df.columns if test_df[col].dtype == "int64"]
cardinality = train_df[discrete_cols].nunique().sort_values(ascending=False)
colors = [dark_red] * 5
fig, ax = plt.subplots(figsize=(12, 6))
cardinality.plot(kind="bar", color=colors)
# Display title
fig.text(
0.5,
1,
"Cardinality of Discrete Features",
fontfamily="monospace",
size="20",
ha="center",
)
# Remove unnecessary elements
ax.yaxis.set_visible(False)
ax.set_xlabel("")
for s in ["top", "left", "right"]:
ax.spines[s].set_visible(False)
# Annotate above the bars
for patch in ax.patches:
text = f"{patch.get_height():.0f}"
x = patch.get_x() + patch.get_width() / 2
y = patch.get_height() + 10000
ax.text(x, y, text, ha="center", va="center", fontsize=20)
# Format xticklabels
plt.setp(ax.get_xmajorticklabels(), rotation=0, fontsize="large")
ax.spines["bottom"].set_linewidth(1.5)
fig.text(
0.4,
0.5,
"""
These discrete features
all have very high cardinality,
meaning it isn't a good idea to
treat them as categorical.
""",
bbox=dict(boxstyle="round", fc="#009473"),
fontsize="large",
)
# # 5. Analyzing the target
# Let's look at the distribution of the target:
fig, ax = plt.subplots(figsize=(8, 4))
sns.kdeplot(
train_df["loss"],
color=dark_red,
shade=True,
ax=ax,
)
ax.set(xlabel="Target - loss")
plt.title(
"Distribution of the Target",
ha="center",
fontfamily="monospace",
fontsize="large",
fontweight="bold",
size=20,
)
fig.text(
0.4,
0.5,
"""
The target has a skewed distribution.
""",
bbox=dict(boxstyle="round", fc="#009473"),
fontsize="medium",
)
ax.yaxis.set_visible(False)
ax.spines["left"].set_visible(False)
# # 6. XGBoost Baseline
# Since the dataset has high dimensionality, we will apply Principal Component Analysis as a base reduction method. We will pass 0.95 to `n_components` so that PCA will find the minimum number of features we need to keep to preserve at least 95% variance of the dataset.
# We will perform all of the steps inside Sklearn pipelines ending with a baseline XGBoost regressor:
from sklearn.model_selection import KFold, cross_validate, train_test_split
from xgboost import XGBRegressor
# Log transform all features and the target
X, y = train_df.drop("loss", axis=1), train_df[["loss"]]
reg = XGBRegressor(
objective="reg:squarederror",
tree_method="gpu_hist",
)
# Validation set to be used inside early_stopping
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, test_size=0.1, random_state=1121218
)
# Set up `fit_params` for XGBoost
eval_set = [(X_valid, y_valid)]
fit_params = {
"eval_set": eval_set,
"eval_metric": "rmse",
"early_stopping_rounds": 100,
"verbose": False,
}
_ = reg.fit(X_train, y_train, **fit_params)
preds = reg.predict(test_df)
submission = pd.DataFrame({"id": test_df.index, "loss": preds})
submission.to_csv("submission.csv", index=False)
| false | 0 | 3,209 | 12 | 3,209 | 3,209 |
||
69571457
|
<jupyter_start><jupyter_text>Wav2Lip-preprocessed
Kaggle dataset identifier: wav2lippreprocessed
<jupyter_script># ### 之前代码有问题,以最新版为准
# # 任务
# 本次实践涉及到对Wav2Lip模型的,以及相关代码实现。总体上分为以下几个部分:
# 1. 环境的配置
# 2. 数据集准备及预处理
# 3. 模型的训练
# 4. 模型的推理
# ## Wav2Lip
# **[Wav2Lip](https://arxiv.org/pdf/2008.10010.pdf)** 是一种基于对抗生成网络的由语音驱动的人脸说话视频生成模型。如下图所示,Wav2Lip的网络模型总体上分成三块:生成器、判别器和一个预训练好的Lip-Sync Expert组成。网络的输入有2个:任意的一段视频和一段语音,输出为一段唇音同步的视频。生成器是基于encoder-decoder的网络结构,分别利用2个encoder: speech encoder, identity encoder去对输入的语音和视频人脸进行编码,并将二者的编码结果进行拼接,送入到 face decoder 中进行解码得到输出的视频帧。判别器Visual Quality Discriminator对生成结果的质量进行规范,提高生成视频的清晰度。为了更好的保证生成结果的唇音同步性,Wav2Lip引入了一个预预训练的唇音同步判别模型 Pre-trained Lip-sync Expert,作为衡量生成结果的唇音同步性的额外损失。
# ### Lip-Sync Expert
# Lip-sync Expert基于 **[SyncNet](https://www.robots.ox.ac.uk/~vgg/publications/2016/Chung16a/)**,是一种用来判别语音和视频是否同步的网络模型。如下图所示,SyncNet的输入也是两种:语音特征MFCC和嘴唇的视频帧,利用两个基于卷积神经网络的Encoder分别对输入的语音和视频帧进行降纬和特征提取,将二者的特征都映射到同一个纬度空间中去,最后利用contrastive loss对唇音同步性进行衡量,结果的值越大代表越不同步,结果值越小则代表越同步。在Wav2Lip模型中,进一步改进了SyncNet的网络结构:网络更深;加入了残差网络结构;输入的语音特征被替换成了mel-spectrogram特征。
# ## 1. 环境的配置
# - `建议准备一台有显卡的linux系统电脑,或者可以选择使用第三方云服务器(Google Colab)`
# - `Python 3.6 或者更高版本`
# - ffmpeg: `sudo apt-get install ffmpeg`
# - 必要的python包的安装,所需要的库名称都已经包含在`requirements.txt`文件中,可以使用 `pip install -r requirements.txt`一次性安装.
# - 在本实验中利用到了人脸检测的相关技术,需要下载人脸检测预训练模型:Face detection [pre-trained model](https://www.adrianbulat.com/downloads/python-fan/s3fd-619a316812.pth) 并移动到 `face_detection/detection/sfd/s3fd.pth`文件夹下.
# !pip install -r requirements.txt
# ## 2. 数据集的准备及预处理
# **LRS2 数据集的下载**
# 实验所需要的数据集下载地址为:LRS2 dataset,下载该数据集需要获得BBC的许可,需要发送申请邮件以获取下载密钥,具体操作详见网页中的指示。下载完成后对数据集进行解压到本目录的`mvlrs_v1/`文件夹下,并将LRS2中的文件列表文件`train.txt, val.txt, test.txt` 移动到`filelists/`文件夹下,最终得到的数据集目录结构如下所示。
# ```
# data_root (mvlrs_v1)
# ├── main, pretrain (我们只使用main文件夹下的数据)
# | ├── 文件夹列表
# | │ ├── 5位以.mp4结尾的视频ID
# ```
# **数据集预处理**
# 数据集中大多数视频都是包含人的半身或者全身的画面,而我们的模型只需要人脸这一小部分。所以在预处理阶段,我们要对每一个视频进行分帧操作,提取视频的每一帧,之后使用`face detection`工具包对人脸位置进行定位并裁减,只保留人脸的图片帧。同时,我们也需要将每一个视频中的语音分离出来。
# !python preprocess.py --data_root "./mvlrs_v1/main" --preprocessed_root "./lrs2_preprocessed"
# 预处理后的`lrs2_preprocessed/`文件夹下的目录结构如下
# ```
# preprocessed_root (lrs2_preprocessed)
# ├── 文件夹列表
# | ├── 五位的视频ID
# | │ ├── *.jpg
# | │ ├── audio.wav
# ```
# ## 3. 模型训练
# 模型的训练主要分为两个部分:
# 1. Lip-Sync Expert Discriminator的训练。这里提供官方的预训练模型 [weight](https://iiitaphyd-my.sharepoint.com/:u:/g/personal/radrabha_m_research_iiit_ac_in/EQRvmiZg-HRAjvI6zqN9eTEBP74KefynCwPWVmF57l-AYA?e=ZRPHKP)
# 2. Wav2Lip 模型的训练。
# ### 3.1 预训练Lip-Sync Expert
# #### 1. 网络的搭建
# 上面我们已经介绍了SyncNet的基本网络结构,主要有一系列的(Conv+BatchNorm+Relu)组成,这里我们对其进行了一些改进,加入了残差结构。为了方便之后的使用,我们对(Conv+BatchNorm+Relu)以及残差模块进行了封装。
import torch
from torch import nn
from torch.nn import functional as F
class Conv2d(nn.Module):
def __init__(
self, cin, cout, kernel_size, stride, padding, residual=False, *args, **kwargs
):
super().__init__(*args, **kwargs)
########TODO######################
# 按下面的网络结构要求,补全代码
# self.conv_block: Sequential结构,Conv2d+BatchNorm
# self.act: relu激活函数
self.conv_block = nn.Sequential(
nn.Conv2d(cin, cout, kernel_size, stride, padding), nn.BatchNorm2d(cout)
)
self.act = nn.ReLU()
self.residual = residual
def forward(self, x):
out = self.conv_block(x)
if self.residual:
out += x
return self.act(out)
# SyncNet的主要包含两个部分:Face_encoder和Audio_encoder。每一个部分都由多个Conv2d模块组成,通过指定卷积核的大小实现对输入的下采样和特征提取
import torch
from torch import nn
from torch.nn import functional as F
class SyncNet_color(nn.Module):
def __init__(self):
super(SyncNet_color, self).__init__()
################TODO###################
# 根据上面提供的网络结构图,补全下面卷积网络的参数
self.face_encoder = nn.Sequential(
Conv2d(15, 32, kernel_size=(7, 7), stride=1, padding=3),
Conv2d(32, 64, kernel_size=5, stride=(1, 2), padding=1),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 128, kernel_size=3, stride=2, padding=1),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 256, kernel_size=3, stride=2, padding=1),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 512, kernel_size=3, stride=2, padding=1),
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(512, 512, kernel_size=3, stride=2, padding=1),
Conv2d(512, 512, kernel_size=3, stride=1, padding=0),
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
)
self.audio_encoder = nn.Sequential(
Conv2d(1, 32, kernel_size=3, stride=1, padding=1),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 64, kernel_size=3, stride=(3, 1), padding=1),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 128, kernel_size=3, stride=3, padding=1),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 256, kernel_size=3, stride=(3, 2), padding=1),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 512, kernel_size=3, stride=1, padding=0),
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
)
def forward(
self, audio_sequences, face_sequences
): # audio_sequences := (B, dim, T)
#########################TODO#######################
# 正向传播
face_embedding = self.face_encoder(face_sequences)
audio_embedding = self.audio_encoder(audio_sequences)
audio_embedding = audio_embedding.view(audio_embedding.size(0), -1)
face_embedding = face_embedding.view(face_embedding.size(0), -1)
audio_embedding = F.normalize(audio_embedding, p=2, dim=1)
face_embedding = F.normalize(face_embedding, p=2, dim=1)
return audio_embedding, face_embedding
from os.path import dirname, join, basename, isfile
from tqdm import tqdm
from models import SyncNet_color as SyncNet
import audio
import torch
from torch import nn
from torch import optim
import torch.backends.cudnn as cudnn
from torch.utils import data as data_utils
import numpy as np
from glob import glob
import os, random, cv2, argparse
from hparams import hparams, get_image_list
# #### 2.数据集的定义
global_step = 0 # 起始的step
global_epoch = 0 # 起始的epoch
use_cuda = torch.cuda.is_available() # 训练的设备 cpu or gpu
print("use_cuda: {}".format(use_cuda))
syncnet_T = 5 ## 每次选取200ms的视频片段进行训练,视频的fps为25,所以200ms对应的帧数为:25*0.2=5帧
syncnet_mel_step_size = 16 # 200ms对应的声音的mel-spectrogram特征的长度为16.
data_root = "/kaggle/input/wav2lippreprocessed/lrs2_preprocessed" # 数据集的位置
class Dataset(object):
def __init__(self, split):
self.all_videos = get_image_list(data_root, split)
def get_frame_id(self, frame):
return int(basename(frame).split(".")[0])
def get_window(self, start_frame):
start_id = self.get_frame_id(start_frame)
vidname = dirname(start_frame)
window_fnames = []
for frame_id in range(start_id, start_id + syncnet_T):
frame = join(vidname, "{}.jpg".format(frame_id))
if not isfile(frame):
return None
window_fnames.append(frame)
return window_fnames
def crop_audio_window(self, spec, start_frame):
# num_frames = (T x hop_size * fps) / sample_rate
start_frame_num = self.get_frame_id(start_frame)
start_idx = int(80.0 * (start_frame_num / float(hparams.fps)))
end_idx = start_idx + syncnet_mel_step_size
return spec[start_idx:end_idx, :]
def __len__(self):
return len(self.all_videos)
def __getitem__(self, idx):
"""
return: x,mel,y
x: 五张嘴唇图片
mel:对应的语音的mel spectrogram
t:同步or不同步
"""
while 1:
idx = random.randint(0, len(self.all_videos) - 1)
vidname = self.all_videos[idx]
img_names = list(glob(join(vidname, "*.jpg")))
if len(img_names) <= 3 * syncnet_T:
continue
img_name = random.choice(img_names)
wrong_img_name = random.choice(img_names)
while wrong_img_name == img_name:
wrong_img_name = random.choice(img_names)
# 随机决定是产生负样本还是正样本
if random.choice([True, False]):
y = torch.ones(1).float()
chosen = img_name
else:
y = torch.zeros(1).float()
chosen = wrong_img_name
window_fnames = self.get_window(chosen)
if window_fnames is None:
continue
window = []
all_read = True
for fname in window_fnames:
img = cv2.imread(fname)
if img is None:
all_read = False
break
try:
img = cv2.resize(img, (hparams.img_size, hparams.img_size))
except Exception as e:
all_read = False
break
window.append(img)
if not all_read:
continue
try:
wavpath = join(vidname, "audio.wav")
wav = audio.load_wav(wavpath, hparams.sample_rate)
orig_mel = audio.melspectrogram(wav).T
except Exception as e:
continue
mel = self.crop_audio_window(orig_mel.copy(), img_name)
if mel.shape[0] != syncnet_mel_step_size:
continue
# H x W x 3 * T
x = np.concatenate(window, axis=2) / 255.0
x = x.transpose(2, 0, 1)
x = x[:, x.shape[1] // 2 :]
x = torch.FloatTensor(x)
mel = torch.FloatTensor(mel.T).unsqueeze(0)
return x, mel, y
ds = Dataset("train")
x, mel, t = ds[0]
print(x.shape)
print(mel.shape)
print(t.shape)
import matplotlib.pyplot as plt
plt.imshow(mel[0].numpy())
plt.imshow(x[:3, :, :].transpose(0, 2).numpy())
# #### 3.训练
# 使用cosine_loss 作为损失函数
# 损失函数的定义
logloss = nn.BCELoss() # 交叉熵损失
def cosine_loss(a, v, y): # 余弦相似度损失
"""
a: audio_encoder的输出
v: video face_encoder的输出
y: 是否同步的真实值
"""
d = nn.functional.cosine_similarity(a, v)
loss = logloss(d.unsqueeze(1), y)
return loss
def train(
device,
model,
train_data_loader,
test_data_loader,
optimizer,
checkpoint_dir=None,
checkpoint_interval=None,
nepochs=None,
):
global global_step, global_epoch
resumed_step = global_step
while global_epoch < nepochs:
running_loss = 0.0
prog_bar = tqdm(enumerate(train_data_loader))
for step, (x, mel, y) in prog_bar:
model.train()
optimizer.zero_grad()
#####TODO###########
####################
# 补全模型的训练
x = x.to(device)
mel = mel.to(device)
a, v = model(mel, x)
y = y.to(device)
loss = cosine_loss(a, v, y)
loss.backward()
optimizer.step()
global_step += 1
cur_session_steps = global_step - resumed_step
running_loss += loss.item()
if global_step == 1 or global_step % checkpoint_interval == 0:
save_checkpoint(
model, optimizer, global_step, checkpoint_dir, global_epoch
)
if global_step % hparams.syncnet_eval_interval == 0:
with torch.no_grad():
eval_model(
test_data_loader, global_step, device, model, checkpoint_dir
)
prog_bar.set_description(
"Epoch: {} Loss: {}".format(global_epoch, running_loss / (step + 1))
)
global_epoch += 1
def eval_model(test_data_loader, global_step, device, model, checkpoint_dir):
# 在测试集上进行评估
eval_steps = 1400
print("Evaluating for {} steps".format(eval_steps))
losses = []
while 1:
for step, (x, mel, y) in enumerate(test_data_loader):
model.eval()
# Transform data to CUDA device
x = x.to(device)
mel = mel.to(device)
a, v = model(mel, x)
y = y.to(device)
loss = cosine_loss(a, v, y)
losses.append(loss.item())
if step > eval_steps:
break
averaged_loss = sum(losses) / len(losses)
print(averaged_loss)
return
latest_checkpoint_path = ""
def save_checkpoint(model, optimizer, step, checkpoint_dir, epoch):
# 保存训练的结果 checkpoint
global latest_checkpoint_path
checkpoint_path = join(
checkpoint_dir, "checkpoint_step{:09d}.pth".format(global_step)
)
optimizer_state = optimizer.state_dict() if hparams.save_optimizer_state else None
torch.save(
{
"state_dict": model.state_dict(),
"optimizer": optimizer_state,
"global_step": step,
"global_epoch": epoch,
},
checkpoint_path,
)
latest_checkpoint_path = checkpoint_path
print("Saved checkpoint:", checkpoint_path)
def _load(checkpoint_path):
if use_cuda:
checkpoint = torch.load(checkpoint_path)
else:
checkpoint = torch.load(
checkpoint_path, map_location=lambda storage, loc: storage
)
return checkpoint
def load_checkpoint(path, model, optimizer, reset_optimizer=False):
# 读取指定checkpoint的保存信息
global global_step
global global_epoch
print("Load checkpoint from: {}".format(path))
checkpoint = _load(path)
model.load_state_dict(checkpoint["state_dict"])
if not reset_optimizer:
optimizer_state = checkpoint["optimizer"]
if optimizer_state is not None:
print("Load optimizer state from {}".format(path))
optimizer.load_state_dict(checkpoint["optimizer"])
global_step = checkpoint["global_step"]
global_epoch = checkpoint["global_epoch"]
return model
# **下面开始训练,最终的Loss参考值为0.20左右,此时模型能达到较好的判别效果**
checkpoint_dir = "/kaggle/working/expert_checkpoints/" # 指定存储 checkpoint的位置
checkpoint_path = (
"/kaggle/input/wav2lip24epoch/expert_checkpoints/checkpoint_step000060000.pth"
)
# 指定加载checkpoint的路径,第一次训练时不需要,后续如果想从某个checkpoint恢复训练,可指定。
if not os.path.exists(checkpoint_dir):
os.mkdir(checkpoint_dir)
# Dataset and Dataloader setup
train_dataset = Dataset("train")
test_dataset = Dataset("val")
############TODO#########
#####Train Dataloader and Test Dataloader
#### 具体的bacthsize等参数,参考 hparams.py文件
train_data_loader = data_utils.DataLoader(
train_dataset,
batch_size=hparams.batch_size,
shuffle=True,
num_workers=hparams.num_workers,
)
test_data_loader = data_utils.DataLoader(
test_dataset, batch_size=hparams.batch_size, num_workers=8
)
device = torch.device("cuda" if use_cuda else "cpu")
# Model
#####定义 SynNet模型,并加载到指定的device上
model = SyncNet().to(device)
print(
"total trainable params {}".format(
sum(p.numel() for p in model.parameters() if p.requires_grad)
)
)
####定义优化器,使用adam,lr参考hparams.py文件
optimizer = optim.Adam([p for p in model.parameters() if p.requires_grad], lr=1e-6)
if checkpoint_path is not None:
load_checkpoint(checkpoint_path, model, optimizer, reset_optimizer=True)
train(
device,
model,
train_data_loader,
test_data_loader,
optimizer,
checkpoint_dir=checkpoint_dir,
checkpoint_interval=hparams.syncnet_checkpoint_interval,
nepochs=48,
)
# ### 3.2 训练Wav2Lip
# 预训练模型 [weight](https://iiitaphyd-my.sharepoint.com/:u:/g/personal/radrabha_m_research_iiit_ac_in/EdjI7bZlgApMqsVoEUUXpLsBxqXbn5z8VTmoxp55YNDcIA?e=n9ljGW)
# #### 1. 模型的定义
# wav2lip模型的生成器首先对输入进行下采样,然后再经过上采样恢复成原来的大小。为了方便,我们对其中重复利用到的模块进行了封装。
class nonorm_Conv2d(nn.Module): # 不需要进行 norm的卷积
def __init__(
self, cin, cout, kernel_size, stride, padding, residual=False, *args, **kwargs
):
super().__init__(*args, **kwargs)
self.conv_block = nn.Sequential(
nn.Conv2d(cin, cout, kernel_size, stride, padding),
)
self.act = nn.LeakyReLU(0.01, inplace=True)
def forward(self, x):
out = self.conv_block(x)
return self.act(out)
class Conv2dTranspose(nn.Module): # 逆卷积,上采样
def __init__(
self, cin, cout, kernel_size, stride, padding, output_padding=0, *args, **kwargs
):
super().__init__(*args, **kwargs)
############TODO###########
## 完成self.conv_block: 一个逆卷积和batchnorm组成的 Sequential结构
self.conv_block = nn.Sequential(
nn.ConvTranspose2d(cin, cout, kernel_size, stride, padding, output_padding),
nn.BatchNorm2d(cout),
)
self.act = nn.ReLU()
def forward(self, x):
out = self.conv_block(x)
return self.act(out)
# **生成器**
# 由两个encoder: face_encoder和 audio_encoder, 一个decoder:face_decoder组成。face encoder 和 audio encoder 分别对输入的人脸和语音特征进行降维,得到(1,1,512)的特征,并将二者进行拼接送入到 face decoder中去进行上采样,最终得到和输入一样大小的人脸图像。
#####################TODO############################
# 根据下面打印的网络模型图,补全网络的参数
class Wav2Lip(nn.Module):
def __init__(self):
super(Wav2Lip, self).__init__()
self.face_encoder_blocks = nn.ModuleList(
[
nn.Sequential(
Conv2d(6, 16, kernel_size=7, stride=1, padding=3)
), # 96,96
nn.Sequential(
Conv2d(16, 32, kernel_size=3, stride=2, padding=1), # 48,48
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(32, 64, kernel_size=3, stride=2, padding=1), # 24,24
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(64, 128, kernel_size=3, stride=2, padding=1), # 12,12
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(128, 256, kernel_size=3, stride=2, padding=1), # 6,6
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(256, 512, kernel_size=3, stride=2, padding=1), # 3,3
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(512, 512, kernel_size=3, stride=1, padding=0), # 1, 1
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
),
]
)
self.audio_encoder = nn.Sequential(
Conv2d(1, 32, kernel_size=3, stride=1, padding=1),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 64, kernel_size=3, stride=(3, 1), padding=1),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 128, kernel_size=3, stride=3, padding=1),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 256, kernel_size=3, stride=(3, 2), padding=1),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 512, kernel_size=3, stride=1, padding=0),
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
)
self.face_decoder_blocks = nn.ModuleList(
[
nn.Sequential(
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
),
nn.Sequential(
Conv2dTranspose(
1024, 512, kernel_size=3, stride=1, padding=0
), # 3,3
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2dTranspose(
1024, 512, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
), # 6, 6
nn.Sequential(
Conv2dTranspose(
768, 384, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(384, 384, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(384, 384, kernel_size=3, stride=1, padding=1, residual=True),
), # 12, 12
nn.Sequential(
Conv2dTranspose(
512, 256, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
), # 24, 24
nn.Sequential(
Conv2dTranspose(
320, 128, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
), # 48, 48
nn.Sequential(
Conv2dTranspose(
160, 64, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
),
]
) # 96,96
self.output_block = nn.Sequential(
Conv2d(80, 32, kernel_size=3, stride=1, padding=1),
nn.Conv2d(32, 3, kernel_size=1, stride=1, padding=0),
nn.Sigmoid(),
)
def forward(self, audio_sequences, face_sequences):
# audio_sequences = (B, T, 1, 80, 16)
B = audio_sequences.size(0)
input_dim_size = len(face_sequences.size())
if input_dim_size > 4:
audio_sequences = torch.cat(
[audio_sequences[:, i] for i in range(audio_sequences.size(1))], dim=0
)
face_sequences = torch.cat(
[face_sequences[:, :, i] for i in range(face_sequences.size(2))], dim=0
)
audio_embedding = self.audio_encoder(audio_sequences) # B, 512, 1, 1
feats = []
x = face_sequences
for f in self.face_encoder_blocks:
x = f(x)
feats.append(x)
x = audio_embedding
for f in self.face_decoder_blocks:
x = f(x)
try:
x = torch.cat((x, feats[-1]), dim=1)
except Exception as e:
print(x.size())
print(feats[-1].size())
raise e
feats.pop()
x = self.output_block(x)
if input_dim_size > 4:
x = torch.split(x, B, dim=0) # [(B, C, H, W)]
outputs = torch.stack(x, dim=2) # (B, C, T, H, W)
else:
outputs = x
return outputs
# **判别器**
# 判别器也是由一系列的卷积神经网络组成,输入一张人脸图片,利用face encoder对其进行降维到512维。
###########TODO##################
####补全判别器模型
class Wav2Lip_disc_qual(nn.Module):
def __init__(self):
super(Wav2Lip_disc_qual, self).__init__()
self.face_encoder_blocks = nn.ModuleList(
[
nn.Sequential(
nonorm_Conv2d(3, 32, kernel_size=7, stride=1, padding=3)
), # 48,96
nn.Sequential(
nonorm_Conv2d(
32, 64, kernel_size=5, stride=(1, 2), padding=2
), # 48,48
nonorm_Conv2d(64, 64, kernel_size=5, stride=1, padding=2),
),
nn.Sequential(
nonorm_Conv2d(64, 128, kernel_size=5, stride=2, padding=2), # 24,24
nonorm_Conv2d(128, 128, kernel_size=5, stride=1, padding=2),
),
nn.Sequential(
nonorm_Conv2d(
128, 256, kernel_size=5, stride=2, padding=2
), # 12,12
nonorm_Conv2d(256, 256, kernel_size=5, stride=1, padding=2),
),
nn.Sequential(
nonorm_Conv2d(256, 512, kernel_size=3, stride=2, padding=1), # 6,6
nonorm_Conv2d(512, 512, kernel_size=3, stride=1, padding=1),
),
nn.Sequential(
nonorm_Conv2d(512, 512, kernel_size=3, stride=2, padding=1), # 3,3
nonorm_Conv2d(512, 512, kernel_size=3, stride=1, padding=1),
),
nn.Sequential(
nonorm_Conv2d(512, 512, kernel_size=3, stride=1, padding=0), # 1, 1
nonorm_Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
),
]
)
self.binary_pred = nn.Sequential(
nn.Conv2d(512, 1, kernel_size=1, stride=1, padding=0), nn.Sigmoid()
)
self.label_noise = 0.0
def get_lower_half(self, face_sequences):
return face_sequences[:, :, face_sequences.size(2) // 2 :]
def to_2d(self, face_sequences):
B = face_sequences.size(0)
face_sequences = torch.cat(
[face_sequences[:, :, i] for i in range(face_sequences.size(2))], dim=0
)
return face_sequences
def perceptual_forward(self, false_face_sequences):
false_face_sequences = self.to_2d(false_face_sequences)
false_face_sequences = self.get_lower_half(false_face_sequences)
false_feats = false_face_sequences
for f in self.face_encoder_blocks:
false_feats = f(false_feats)
false_pred_loss = F.binary_cross_entropy(
self.binary_pred(false_feats).view(len(false_feats), -1),
torch.ones((len(false_feats), 1)).cuda(),
)
return false_pred_loss
def forward(self, face_sequences):
face_sequences = self.to_2d(face_sequences)
face_sequences = self.get_lower_half(face_sequences)
x = face_sequences
for f in self.face_encoder_blocks:
x = f(x)
return self.binary_pred(x).view(len(x), -1)
# #### 2. 数据集的定义
# 在训练时,会用到4个数据:
# 1. x:输入的图片
# 2. indiv_mels: 每一张图片所对应语音的mel-spectrogram特征
# 3. mel: 所有帧对应的200ms的语音mel-spectrogram,用于SyncNet进行唇音同步损失的计算
# 4. y:真实的与语音对应的,唇音同步的图片。
#
global_step = 0
global_epoch = 0
use_cuda = torch.cuda.is_available()
print("use_cuda: {}".format(use_cuda))
syncnet_T = 5
syncnet_mel_step_size = 16
class Dataset(object):
def __init__(self, split):
self.all_videos = get_image_list(data_root, split)
def get_frame_id(self, frame):
return int(basename(frame).split(".")[0])
def get_window(self, start_frame):
start_id = self.get_frame_id(start_frame)
vidname = dirname(start_frame)
window_fnames = []
for frame_id in range(start_id, start_id + syncnet_T):
frame = join(vidname, "{}.jpg".format(frame_id))
if not isfile(frame):
return None
window_fnames.append(frame)
return window_fnames
def read_window(self, window_fnames):
if window_fnames is None:
return None
window = []
for fname in window_fnames:
img = cv2.imread(fname)
if img is None:
return None
try:
img = cv2.resize(img, (hparams.img_size, hparams.img_size))
except Exception as e:
return None
window.append(img)
return window
def crop_audio_window(self, spec, start_frame):
if type(start_frame) == int:
start_frame_num = start_frame
else:
start_frame_num = self.get_frame_id(
start_frame
) # 0-indexing ---> 1-indexing
start_idx = int(80.0 * (start_frame_num / float(hparams.fps)))
end_idx = start_idx + syncnet_mel_step_size
return spec[start_idx:end_idx, :]
def get_segmented_mels(self, spec, start_frame):
mels = []
assert syncnet_T == 5
start_frame_num = (
self.get_frame_id(start_frame) + 1
) # 0-indexing ---> 1-indexing
if start_frame_num - 2 < 0:
return None
for i in range(start_frame_num, start_frame_num + syncnet_T):
m = self.crop_audio_window(spec, i - 2)
if m.shape[0] != syncnet_mel_step_size:
return None
mels.append(m.T)
mels = np.asarray(mels)
return mels
def prepare_window(self, window):
# 3 x T x H x W
x = np.asarray(window) / 255.0
x = np.transpose(x, (3, 0, 1, 2))
return x
def __len__(self):
return len(self.all_videos)
def __getitem__(self, idx):
while 1:
idx = random.randint(0, len(self.all_videos) - 1) # 随机选择一个视频id
vidname = self.all_videos[idx]
img_names = list(glob(join(vidname, "*.jpg")))
if len(img_names) <= 3 * syncnet_T:
continue
img_name = random.choice(img_names)
wrong_img_name = random.choice(img_names) # 随机选择帧
while wrong_img_name == img_name:
wrong_img_name = random.choice(img_names)
window_fnames = self.get_window(img_name)
wrong_window_fnames = self.get_window(wrong_img_name)
if window_fnames is None or wrong_window_fnames is None:
continue
window = self.read_window(window_fnames)
if window is None:
continue
wrong_window = self.read_window(wrong_window_fnames)
if wrong_window is None:
continue
try:
# 读取音频
wavpath = join(vidname, "audio.wav")
wav = audio.load_wav(wavpath, hparams.sample_rate)
# 提取完整mel-spectrogram
orig_mel = audio.melspectrogram(wav).T
except Exception as e:
continue
# 分割 mel-spectrogram
mel = self.crop_audio_window(orig_mel.copy(), img_name)
if mel.shape[0] != syncnet_mel_step_size:
continue
indiv_mels = self.get_segmented_mels(orig_mel.copy(), img_name)
if indiv_mels is None:
continue
window = self.prepare_window(window)
y = window.copy()
window[:, :, window.shape[2] // 2 :] = 0.0
wrong_window = self.prepare_window(wrong_window)
x = np.concatenate([window, wrong_window], axis=0)
x = torch.FloatTensor(x)
mel = torch.FloatTensor(mel.T).unsqueeze(0)
indiv_mels = torch.FloatTensor(indiv_mels).unsqueeze(1)
y = torch.FloatTensor(y)
return x, indiv_mels, mel, y
ds = Dataset("train")
x, indiv_mels, mel, y = ds[0]
print(x.shape)
print(indiv_mels.shape)
print(mel.shape)
print(y.shape)
# #### 3. 训练
# bce 交叉墒loss
logloss = nn.BCELoss()
def cosine_loss(a, v, y):
d = nn.functional.cosine_similarity(a, v)
loss = logloss(d.unsqueeze(1), y)
return loss
device = torch.device("cuda" if use_cuda else "cpu")
syncnet = SyncNet().to(device) # 定义syncnet 模型
for p in syncnet.parameters():
p.requires_grad = False
#####L1 loss
recon_loss = nn.L1Loss()
def get_sync_loss(mel, g):
g = g[:, :, :, g.size(3) // 2 :]
g = torch.cat([g[:, :, i] for i in range(syncnet_T)], dim=1)
# B, 3 * T, H//2, W
a, v = syncnet(mel, g)
y = torch.ones(g.size(0), 1).float().to(device)
return cosine_loss(a, v, y)
def train(
device,
model,
disc,
train_data_loader,
test_data_loader,
optimizer,
disc_optimizer,
checkpoint_dir=None,
checkpoint_interval=None,
nepochs=None,
):
global global_step, global_epoch
resumed_step = global_step
while global_epoch < nepochs:
print("Starting Epoch: {}".format(global_epoch))
running_sync_loss, running_l1_loss, disc_loss, running_perceptual_loss = (
0.0,
0.0,
0.0,
0.0,
)
running_disc_real_loss, running_disc_fake_loss = 0.0, 0.0
prog_bar = tqdm(enumerate(train_data_loader))
for step, (x, indiv_mels, mel, gt) in prog_bar:
disc.train()
model.train()
x = x.to(device)
mel = mel.to(device)
indiv_mels = indiv_mels.to(device)
gt = gt.to(device)
### Train generator now. Remove ALL grads.
# 训练生成器
optimizer.zero_grad()
disc_optimizer.zero_grad()
g = model(indiv_mels, x) # 得到生成的结果
if hparams.syncnet_wt > 0.0:
sync_loss = get_sync_loss(mel, g) # 从预训练的expert 模型中获得唇音同步的损失
else:
sync_loss = 0.0
if hparams.disc_wt > 0.0:
perceptual_loss = disc.perceptual_forward(g) # 判别器的感知损失
else:
perceptual_loss = 0.0
l1loss = recon_loss(g, gt) # l1 loss,重建损失
# 最终的损失函数
loss = (
hparams.syncnet_wt * sync_loss
+ hparams.disc_wt * perceptual_loss
+ (1.0 - hparams.syncnet_wt - hparams.disc_wt) * l1loss
)
loss.backward()
optimizer.step()
### Remove all gradients before Training disc
# 训练判别器
disc_optimizer.zero_grad()
pred = disc(gt)
disc_real_loss = F.binary_cross_entropy(
pred, torch.ones((len(pred), 1)).to(device)
)
disc_real_loss.backward()
pred = disc(g.detach())
disc_fake_loss = F.binary_cross_entropy(
pred, torch.zeros((len(pred), 1)).to(device)
)
disc_fake_loss.backward()
disc_optimizer.step()
running_disc_real_loss += disc_real_loss.item()
running_disc_fake_loss += disc_fake_loss.item()
# Logs
global_step += 1
cur_session_steps = global_step - resumed_step
running_l1_loss += l1loss.item()
if hparams.syncnet_wt > 0.0:
running_sync_loss += sync_loss.item()
else:
running_sync_loss += 0.0
if hparams.disc_wt > 0.0:
running_perceptual_loss += perceptual_loss.item()
else:
running_perceptual_loss += 0.0
if global_step == 1 or global_step % checkpoint_interval == 0:
save_checkpoint(
model, optimizer, global_step, checkpoint_dir, global_epoch
)
save_checkpoint(
disc,
disc_optimizer,
global_step,
checkpoint_dir,
global_epoch,
prefix="disc_",
)
if global_step % hparams.eval_interval == 0:
with torch.no_grad():
average_sync_loss = eval_model(
test_data_loader, global_step, device, model, disc
)
if average_sync_loss < 0.75:
hparams.set_hparam("syncnet_wt", 0.03)
prog_bar.set_description(
"L1: {}, Sync: {}, Percep: {} | Fake: {}, Real: {}".format(
running_l1_loss / (step + 1),
running_sync_loss / (step + 1),
running_perceptual_loss / (step + 1),
running_disc_fake_loss / (step + 1),
running_disc_real_loss / (step + 1),
)
)
global_epoch += 1
def eval_model(test_data_loader, global_step, device, model, disc):
eval_steps = 300
print("Evaluating for {} steps".format(eval_steps))
(
running_sync_loss,
running_l1_loss,
running_disc_real_loss,
running_disc_fake_loss,
running_perceptual_loss,
) = ([], [], [], [], [])
while 1:
for step, (x, indiv_mels, mel, gt) in enumerate((test_data_loader)):
model.eval()
disc.eval()
x = x.to(device)
mel = mel.to(device)
indiv_mels = indiv_mels.to(device)
gt = gt.to(device)
pred = disc(gt)
disc_real_loss = F.binary_cross_entropy(
pred, torch.ones((len(pred), 1)).to(device)
)
g = model(indiv_mels, x)
pred = disc(g)
disc_fake_loss = F.binary_cross_entropy(
pred, torch.zeros((len(pred), 1)).to(device)
)
running_disc_real_loss.append(disc_real_loss.item())
running_disc_fake_loss.append(disc_fake_loss.item())
sync_loss = get_sync_loss(mel, g)
if hparams.disc_wt > 0.0:
perceptual_loss = disc.perceptual_forward(g)
else:
perceptual_loss = 0.0
l1loss = recon_loss(g, gt)
loss = (
hparams.syncnet_wt * sync_loss
+ hparams.disc_wt * perceptual_loss
+ (1.0 - hparams.syncnet_wt - hparams.disc_wt) * l1loss
)
running_l1_loss.append(l1loss.item())
running_sync_loss.append(sync_loss.item())
if hparams.disc_wt > 0.0:
running_perceptual_loss.append(perceptual_loss.item())
else:
running_perceptual_loss.append(0.0)
if step > eval_steps:
break
print(
"L1: {}, Sync: {}, Percep: {} | Fake: {}, Real: {}".format(
sum(running_l1_loss) / len(running_l1_loss),
sum(running_sync_loss) / len(running_sync_loss),
sum(running_perceptual_loss) / len(running_perceptual_loss),
sum(running_disc_fake_loss) / len(running_disc_fake_loss),
sum(running_disc_real_loss) / len(running_disc_real_loss),
)
)
return sum(running_sync_loss) / len(running_sync_loss)
def save_checkpoint(model, optimizer, step, checkpoint_dir, epoch, prefix=""):
checkpoint_path = join(
checkpoint_dir, "{}checkpoint_step{:09d}.pth".format(prefix, global_step)
)
optimizer_state = optimizer.state_dict() if hparams.save_optimizer_state else None
torch.save(
{
"state_dict": model.state_dict(),
"optimizer": optimizer_state,
"global_step": step,
"global_epoch": epoch,
},
checkpoint_path,
)
print("Saved checkpoint:", checkpoint_path)
def _load(checkpoint_path):
if use_cuda:
checkpoint = torch.load(checkpoint_path)
else:
checkpoint = torch.load(
checkpoint_path, map_location=lambda storage, loc: storage
)
return checkpoint
def load_checkpoint(
path, model, optimizer, reset_optimizer=False, overwrite_global_states=True
):
global global_step
global global_epoch
print("Load checkpoint from: {}".format(path))
checkpoint = _load(path)
s = checkpoint["state_dict"]
new_s = {}
for k, v in s.items():
new_s[k.replace("module.", "")] = v
model.load_state_dict(new_s)
if not reset_optimizer:
optimizer_state = checkpoint["optimizer"]
if optimizer_state is not None:
print("Load optimizer state from {}".format(path))
optimizer.load_state_dict(checkpoint["optimizer"])
if overwrite_global_states:
global_step = checkpoint["global_step"]
global_epoch = checkpoint["global_epoch"]
return model
checkpoint_dir = "/kaggle/working/wav2lip_checkpoints" # checkpoint 存储的位置
# Dataset and Dataloader setup
train_dataset = Dataset("train")
test_dataset = Dataset("val")
train_data_loader = data_utils.DataLoader(
train_dataset,
batch_size=hparams.batch_size,
shuffle=True,
num_workers=hparams.num_workers,
)
test_data_loader = data_utils.DataLoader(
test_dataset, batch_size=hparams.batch_size, num_workers=4
)
device = torch.device("cuda" if use_cuda else "cpu")
# Model
model = Wav2Lip().to(device) ####### 生成器模型
disc = Wav2Lip_disc_qual().to(device) ####### 判别器模型
print(
"total trainable params {}".format(
sum(p.numel() for p in model.parameters() if p.requires_grad)
)
)
print(
"total DISC trainable params {}".format(
sum(p.numel() for p in disc.parameters() if p.requires_grad)
)
)
optimizer = optim.Adam(
[p for p in model.parameters() if p.requires_grad],
lr=hparams.initial_learning_rate,
betas=(0.5, 0.999),
) #####adam优化器,betas=[0.5,0.999]
disc_optimizer = optim.Adam(
[p for p in disc.parameters() if p.requires_grad],
lr=hparams.disc_initial_learning_rate,
betas=(0.5, 0.999),
) #####adam优化器,betas=[0.5,0.999]
# 继续训练的生成器的checkpoint位置
# checkpoint_path=""
# load_checkpoint(checkpoint_path, model, optimizer, reset_optimizer=False)
# 继续训练的判别器的checkpoint位置
# disc_checkpoint_path=""
# load_checkpoint(disc_checkpoint_path, disc, disc_optimizer,
# reset_optimizer=False, overwrite_global_states=False)
# syncnet的checkpoint位置,我们将使用此模型计算生成的帧和语音的唇音同步损失
syncnet_checkpoint_path = latest_checkpoint_path
# syncnet_checkpoint_path="/kaggle/working/expert_checkpoints/checkpoint_step000000001.pth"
load_checkpoint(
syncnet_checkpoint_path,
syncnet,
None,
reset_optimizer=True,
overwrite_global_states=False,
)
if not os.path.exists(checkpoint_dir):
os.mkdir(checkpoint_dir)
# Train!
train(
device,
model,
disc,
train_data_loader,
test_data_loader,
optimizer,
disc_optimizer,
checkpoint_dir=checkpoint_dir,
checkpoint_interval=hparams.checkpoint_interval,
nepochs=hparams.nepochs,
)
# #### 4. 命令行训练
# 上面是按步骤训练的过程,在`hq_wav2lip_train.py`文件中已经把上述的过程进行了封装,你可以通过以下的命令直接进行训练
# !python wav2lip_train.py --data_root lrs2_preprocessed/ --checkpoint_dir <folder_to_save_checkpoints> --syncnet_checkpoint_path <path_to_expert_disc_checkpoint>
# ### 4. 模型的推理
# 当模型训练完毕后,我们只使用生成器的网络模型部分作为我们的推理模型。模型的输入由一段包含人脸的参照视频和一段语音组成。
# 在这里我们可以直接使用官方提供给我们的预训练模型[weight](https://iiitaphyd-my.sharepoint.com/:u:/g/personal/radrabha_m_research_iiit_ac_in/EdjI7bZlgApMqsVoEUUXpLsBxqXbn5z8VTmoxp55YNDcIA?e=n9ljGW)下载该模型并放入到指定文件夹下,供之后的推理使用。
# 模型的推理过程主要分为以下几个步骤:
# 1. 输入数据的预处理,包含人脸抠图,视频分帧,提取mel-spectrogram特征等操作。
# 2. 利用网络模型生成唇音同步的视频帧。
# 3. 将生成的视频帧准换成视频,并和输入的语音结合,形成最终的输出视频。
#
from os import listdir, path
import numpy as np
import scipy, cv2, os, sys, argparse, audio
import json, subprocess, random, string
from tqdm import tqdm
from glob import glob
import torch, face_detection
from models import Wav2Lip
import platform
import audio
checkpoint_path = "/kaggle/working/wav2lip_checkpoints/checkpoint_step000000001.pth" # 生成器的checkpoint位置
face = "input_video.mp4" # 参照视频的文件位置, *.mp4
speech = "input_audio.wav" # 输入语音的位置,*.wav
resize_factor = 1 # 对输入的视频进行下采样的倍率
crop = [0, -1, 0, -1] # 是否对视频帧进行裁剪,处理视频中有多张人脸时有用
fps = 25 # 视频的帧率
static = False # 是否只使用固定的一帧作为视频的生成参照
if not os.path.isfile(face):
raise ValueError("--face argument must be a valid path to video/image file")
else: # 若输入的是视频格式
video_stream = cv2.VideoCapture(face) # 读取视频
fps = video_stream.get(cv2.CAP_PROP_FPS) # 读取 fps
print("Reading video frames...")
full_frames = []
# 提取所有的帧
while 1:
still_reading, frame = video_stream.read()
if not still_reading:
video_stream.release()
break
if resize_factor > 1: # 进行下采样,降低分辨率
frame = cv2.resize(
frame,
(frame.shape[1] // resize_factor, frame.shape[0] // resize_factor),
)
y1, y2, x1, x2 = crop # 裁剪
if x2 == -1:
x2 = frame.shape[1]
if y2 == -1:
y2 = frame.shape[0]
frame = frame[y1:y2, x1:x2]
full_frames.append(frame)
print("Number of frames available for inference: " + str(len(full_frames)))
# 检查输入的音频是否为 .wav格式的,若不是则进行转换
if not speech.endswith(".wav"):
print("Extracting raw audio...")
command = "ffmpeg -y -i {} -strict -2 {}".format(speech, "temp/temp.wav")
subprocess.call(command, shell=True)
speech = "temp/temp.wav"
wav = audio.load_wav(speech, 16000) # 保证采样率为16000
mel = audio.melspectrogram(wav)
print(mel.shape)
wav2lip_batch_size = 128 # 推理时输入到网络的batchsize
mel_step_size = 16
# 提取语音的mel谱
mel_chunks = []
mel_idx_multiplier = 80.0 / fps
i = 0
while 1:
start_idx = int(i * mel_idx_multiplier)
if start_idx + mel_step_size > len(mel[0]):
mel_chunks.append(mel[:, len(mel[0]) - mel_step_size :])
break
mel_chunks.append(mel[:, start_idx : start_idx + mel_step_size])
i += 1
print("Length of mel chunks: {}".format(len(mel_chunks)))
full_frames = full_frames[: len(mel_chunks)]
batch_size = wav2lip_batch_size
img_size = 96 # 默认的输入图片大小
pads = [0, 20, 0, 0] # 填充的长度,保证下巴也在抠图的范围之内
nosmooth = False
face_det_batch_size = 16
def get_smoothened_boxes(boxes, T):
for i in range(len(boxes)):
if i + T > len(boxes):
window = boxes[len(boxes) - T :]
else:
window = boxes[i : i + T]
boxes[i] = np.mean(window, axis=0)
return boxes
# 人脸检测函数
def face_detect(images):
detector = face_detection.FaceAlignment(
face_detection.LandmarksType._2D, flip_input=False, device=device
)
batch_size = face_det_batch_size
while 1:
predictions = []
try:
for i in tqdm(range(0, len(images), batch_size)):
predictions.extend(
detector.get_detections_for_batch(
np.array(images[i : i + batch_size])
)
)
except RuntimeError:
if batch_size == 1:
raise RuntimeError(
"Image too big to run face detection on GPU. Please use the --resize_factor argument"
)
batch_size //= 2
print("Recovering from OOM error; New batch size: {}".format(batch_size))
continue
break
results = []
pady1, pady2, padx1, padx2 = pads
for rect, image in zip(predictions, images):
if rect is None:
cv2.imwrite(
"temp/faulty_frame.jpg", image
) # check this frame where the face was not detected.
raise ValueError(
"Face not detected! Ensure the video contains a face in all the frames."
)
y1 = max(0, rect[1] - pady1)
y2 = min(image.shape[0], rect[3] + pady2)
x1 = max(0, rect[0] - padx1)
x2 = min(image.shape[1], rect[2] + padx2)
results.append([x1, y1, x2, y2])
boxes = np.array(results)
if not nosmooth:
boxes = get_smoothened_boxes(boxes, T=5)
results = [
[image[y1:y2, x1:x2], (y1, y2, x1, x2)]
for image, (x1, y1, x2, y2) in zip(images, boxes)
]
del detector
return results
box = [-1, -1, -1, -1]
def datagen(frames, mels):
img_batch, mel_batch, frame_batch, coords_batch = [], [], [], []
if box[0] == -1: # 如果未指定 特定的人脸边界的话
if not static: # 是否使用视频的第一帧作为参考
face_det_results = face_detect(frames) # BGR2RGB for CNN face detection
else:
face_det_results = face_detect([frames[0]])
else:
print("Using the specified bounding box instead of face detection...")
y1, y2, x1, x2 = box
face_det_results = [
[f[y1:y2, x1:x2], (y1, y2, x1, x2)] for f in frames
] # 裁剪出人脸结果
for i, m in enumerate(mels):
idx = 0 if static else i % len(frames)
frame_to_save = frames[idx].copy()
face, coords = face_det_results[idx].copy()
face = cv2.resize(face, (img_size, img_size)) # 重采样到指定大小
img_batch.append(face)
mel_batch.append(m)
frame_batch.append(frame_to_save)
coords_batch.append(coords)
if len(img_batch) >= wav2lip_batch_size:
img_batch, mel_batch = np.asarray(img_batch), np.asarray(mel_batch)
img_masked = img_batch.copy()
img_masked[:, img_size // 2 :] = 0
img_batch = np.concatenate((img_masked, img_batch), axis=3) / 255.0
mel_batch = np.reshape(
mel_batch, [len(mel_batch), mel_batch.shape[1], mel_batch.shape[2], 1]
)
yield img_batch, mel_batch, frame_batch, coords_batch
img_batch, mel_batch, frame_batch, coords_batch = [], [], [], []
if len(img_batch) > 0:
img_batch, mel_batch = np.asarray(img_batch), np.asarray(mel_batch)
img_masked = img_batch.copy()
img_masked[:, img_size // 2 :] = 0
img_batch = np.concatenate((img_masked, img_batch), axis=3) / 255.0
mel_batch = np.reshape(
mel_batch, [len(mel_batch), mel_batch.shape[1], mel_batch.shape[2], 1]
)
yield img_batch, mel_batch, frame_batch, coords_batch
mel_step_size = 16
device = "cuda" if torch.cuda.is_available() else "cpu"
print("Using {} for inference.".format(device))
# 加载模型
def _load(checkpoint_path):
if device == "cuda":
checkpoint = torch.load(checkpoint_path)
else:
checkpoint = torch.load(
checkpoint_path, map_location=lambda storage, loc: storage
)
return checkpoint
def load_model(path):
model = Wav2Lip()
print("Load checkpoint from: {}".format(path))
checkpoint = _load(path)
s = checkpoint["state_dict"]
new_s = {}
for k, v in s.items():
new_s[k.replace("module.", "")] = v
model.load_state_dict(new_s)
model = model.to(device)
return model.eval()
os.mkdir("/kaggle/working/temp/")
full_frames = full_frames[: len(mel_chunks)]
batch_size = wav2lip_batch_size
gen = datagen(full_frames.copy(), mel_chunks) # 进行人脸的裁剪与拼接,6通道
for i, (img_batch, mel_batch, frames, coords) in enumerate(
tqdm(gen, total=int(np.ceil(float(len(mel_chunks)) / batch_size)))
):
# 加载模型
if i == 0:
model = load_model(checkpoint_path)
print("Model loaded")
frame_h, frame_w = full_frames[0].shape[:-1]
# 暂存临时视频
out = cv2.VideoWriter(
"/kaggle/working/temp/result_without_audio.avi",
cv2.VideoWriter_fourcc(*"DIVX"),
fps,
(frame_w, frame_h),
)
img_batch = torch.FloatTensor(np.transpose(img_batch, (0, 3, 1, 2))).to(device)
mel_batch = torch.FloatTensor(np.transpose(mel_batch, (0, 3, 1, 2))).to(device)
##### 将 img_batch, mel_batch送入模型得到pred
##############TODO##############
with torch.no_grad():
pred = model(mel_batch, img_batch)
pred = pred.cpu().numpy().transpose(0, 2, 3, 1) * 255.0
for p, f, c in zip(pred, frames, coords):
y1, y2, x1, x2 = c
p = cv2.resize(p.astype(np.uint8), (x2 - x1, y2 - y1))
f[y1:y2, x1:x2] = p
out.write(f)
out.release()
# 将生成的视频与语音合并
outfile = "/kaggle/working/result.mp4" # 最终输出结果到该文件夹下
command = "ffmpeg -y -i {} -i {} -strict -2 -q:v 1 {}".format(
speech, "/kaggle/working/temp/result_without_audio.mp4", outfile
)
subprocess.call(command, shell=platform.system() != "Windows")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/571/69571457.ipynb
|
wav2lippreprocessed
|
yinxj24
|
[{"Id": 69571457, "ScriptId": 18799135, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6641168, "CreationDate": "08/01/2021 16:57:03", "VersionNumber": 18.0, "Title": "wav2lip_train", "EvaluationDate": "08/01/2021", "IsChange": true, "TotalLines": 1398.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1397.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92918863, "KernelVersionId": 69571457, "SourceDatasetVersionId": 2456654}, {"Id": 92918862, "KernelVersionId": 69571457, "SourceDatasetVersionId": 2446822}, {"Id": 92918864, "KernelVersionId": 69571457, "SourceDatasetVersionId": 2488763}]
|
[{"Id": 2456654, "DatasetId": 1486947, "DatasourceVersionId": 2499052, "CreatorUserId": 6641168, "LicenseName": "Unknown", "CreationDate": "07/24/2021 00:55:03", "VersionNumber": 1.0, "Title": "Wav2Lip-preprocessed", "Slug": "wav2lippreprocessed", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1486947, "CreatorUserId": 6641168, "OwnerUserId": 6641168.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2456654.0, "CurrentDatasourceVersionId": 2499052.0, "ForumId": 1506638, "Type": 2, "CreationDate": "07/24/2021 00:55:03", "LastActivityDate": "07/24/2021", "TotalViews": 2816, "TotalDownloads": 381, "TotalVotes": 8, "TotalKernels": 1}]
|
[{"Id": 6641168, "UserName": "yinxj24", "DisplayName": "YinXJ24", "RegisterDate": "02/02/2021", "PerformanceTier": 1}]
|
# ### 之前代码有问题,以最新版为准
# # 任务
# 本次实践涉及到对Wav2Lip模型的,以及相关代码实现。总体上分为以下几个部分:
# 1. 环境的配置
# 2. 数据集准备及预处理
# 3. 模型的训练
# 4. 模型的推理
# ## Wav2Lip
# **[Wav2Lip](https://arxiv.org/pdf/2008.10010.pdf)** 是一种基于对抗生成网络的由语音驱动的人脸说话视频生成模型。如下图所示,Wav2Lip的网络模型总体上分成三块:生成器、判别器和一个预训练好的Lip-Sync Expert组成。网络的输入有2个:任意的一段视频和一段语音,输出为一段唇音同步的视频。生成器是基于encoder-decoder的网络结构,分别利用2个encoder: speech encoder, identity encoder去对输入的语音和视频人脸进行编码,并将二者的编码结果进行拼接,送入到 face decoder 中进行解码得到输出的视频帧。判别器Visual Quality Discriminator对生成结果的质量进行规范,提高生成视频的清晰度。为了更好的保证生成结果的唇音同步性,Wav2Lip引入了一个预预训练的唇音同步判别模型 Pre-trained Lip-sync Expert,作为衡量生成结果的唇音同步性的额外损失。
# ### Lip-Sync Expert
# Lip-sync Expert基于 **[SyncNet](https://www.robots.ox.ac.uk/~vgg/publications/2016/Chung16a/)**,是一种用来判别语音和视频是否同步的网络模型。如下图所示,SyncNet的输入也是两种:语音特征MFCC和嘴唇的视频帧,利用两个基于卷积神经网络的Encoder分别对输入的语音和视频帧进行降纬和特征提取,将二者的特征都映射到同一个纬度空间中去,最后利用contrastive loss对唇音同步性进行衡量,结果的值越大代表越不同步,结果值越小则代表越同步。在Wav2Lip模型中,进一步改进了SyncNet的网络结构:网络更深;加入了残差网络结构;输入的语音特征被替换成了mel-spectrogram特征。
# ## 1. 环境的配置
# - `建议准备一台有显卡的linux系统电脑,或者可以选择使用第三方云服务器(Google Colab)`
# - `Python 3.6 或者更高版本`
# - ffmpeg: `sudo apt-get install ffmpeg`
# - 必要的python包的安装,所需要的库名称都已经包含在`requirements.txt`文件中,可以使用 `pip install -r requirements.txt`一次性安装.
# - 在本实验中利用到了人脸检测的相关技术,需要下载人脸检测预训练模型:Face detection [pre-trained model](https://www.adrianbulat.com/downloads/python-fan/s3fd-619a316812.pth) 并移动到 `face_detection/detection/sfd/s3fd.pth`文件夹下.
# !pip install -r requirements.txt
# ## 2. 数据集的准备及预处理
# **LRS2 数据集的下载**
# 实验所需要的数据集下载地址为:LRS2 dataset,下载该数据集需要获得BBC的许可,需要发送申请邮件以获取下载密钥,具体操作详见网页中的指示。下载完成后对数据集进行解压到本目录的`mvlrs_v1/`文件夹下,并将LRS2中的文件列表文件`train.txt, val.txt, test.txt` 移动到`filelists/`文件夹下,最终得到的数据集目录结构如下所示。
# ```
# data_root (mvlrs_v1)
# ├── main, pretrain (我们只使用main文件夹下的数据)
# | ├── 文件夹列表
# | │ ├── 5位以.mp4结尾的视频ID
# ```
# **数据集预处理**
# 数据集中大多数视频都是包含人的半身或者全身的画面,而我们的模型只需要人脸这一小部分。所以在预处理阶段,我们要对每一个视频进行分帧操作,提取视频的每一帧,之后使用`face detection`工具包对人脸位置进行定位并裁减,只保留人脸的图片帧。同时,我们也需要将每一个视频中的语音分离出来。
# !python preprocess.py --data_root "./mvlrs_v1/main" --preprocessed_root "./lrs2_preprocessed"
# 预处理后的`lrs2_preprocessed/`文件夹下的目录结构如下
# ```
# preprocessed_root (lrs2_preprocessed)
# ├── 文件夹列表
# | ├── 五位的视频ID
# | │ ├── *.jpg
# | │ ├── audio.wav
# ```
# ## 3. 模型训练
# 模型的训练主要分为两个部分:
# 1. Lip-Sync Expert Discriminator的训练。这里提供官方的预训练模型 [weight](https://iiitaphyd-my.sharepoint.com/:u:/g/personal/radrabha_m_research_iiit_ac_in/EQRvmiZg-HRAjvI6zqN9eTEBP74KefynCwPWVmF57l-AYA?e=ZRPHKP)
# 2. Wav2Lip 模型的训练。
# ### 3.1 预训练Lip-Sync Expert
# #### 1. 网络的搭建
# 上面我们已经介绍了SyncNet的基本网络结构,主要有一系列的(Conv+BatchNorm+Relu)组成,这里我们对其进行了一些改进,加入了残差结构。为了方便之后的使用,我们对(Conv+BatchNorm+Relu)以及残差模块进行了封装。
import torch
from torch import nn
from torch.nn import functional as F
class Conv2d(nn.Module):
def __init__(
self, cin, cout, kernel_size, stride, padding, residual=False, *args, **kwargs
):
super().__init__(*args, **kwargs)
########TODO######################
# 按下面的网络结构要求,补全代码
# self.conv_block: Sequential结构,Conv2d+BatchNorm
# self.act: relu激活函数
self.conv_block = nn.Sequential(
nn.Conv2d(cin, cout, kernel_size, stride, padding), nn.BatchNorm2d(cout)
)
self.act = nn.ReLU()
self.residual = residual
def forward(self, x):
out = self.conv_block(x)
if self.residual:
out += x
return self.act(out)
# SyncNet的主要包含两个部分:Face_encoder和Audio_encoder。每一个部分都由多个Conv2d模块组成,通过指定卷积核的大小实现对输入的下采样和特征提取
import torch
from torch import nn
from torch.nn import functional as F
class SyncNet_color(nn.Module):
def __init__(self):
super(SyncNet_color, self).__init__()
################TODO###################
# 根据上面提供的网络结构图,补全下面卷积网络的参数
self.face_encoder = nn.Sequential(
Conv2d(15, 32, kernel_size=(7, 7), stride=1, padding=3),
Conv2d(32, 64, kernel_size=5, stride=(1, 2), padding=1),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 128, kernel_size=3, stride=2, padding=1),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 256, kernel_size=3, stride=2, padding=1),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 512, kernel_size=3, stride=2, padding=1),
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(512, 512, kernel_size=3, stride=2, padding=1),
Conv2d(512, 512, kernel_size=3, stride=1, padding=0),
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
)
self.audio_encoder = nn.Sequential(
Conv2d(1, 32, kernel_size=3, stride=1, padding=1),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 64, kernel_size=3, stride=(3, 1), padding=1),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 128, kernel_size=3, stride=3, padding=1),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 256, kernel_size=3, stride=(3, 2), padding=1),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 512, kernel_size=3, stride=1, padding=0),
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
)
def forward(
self, audio_sequences, face_sequences
): # audio_sequences := (B, dim, T)
#########################TODO#######################
# 正向传播
face_embedding = self.face_encoder(face_sequences)
audio_embedding = self.audio_encoder(audio_sequences)
audio_embedding = audio_embedding.view(audio_embedding.size(0), -1)
face_embedding = face_embedding.view(face_embedding.size(0), -1)
audio_embedding = F.normalize(audio_embedding, p=2, dim=1)
face_embedding = F.normalize(face_embedding, p=2, dim=1)
return audio_embedding, face_embedding
from os.path import dirname, join, basename, isfile
from tqdm import tqdm
from models import SyncNet_color as SyncNet
import audio
import torch
from torch import nn
from torch import optim
import torch.backends.cudnn as cudnn
from torch.utils import data as data_utils
import numpy as np
from glob import glob
import os, random, cv2, argparse
from hparams import hparams, get_image_list
# #### 2.数据集的定义
global_step = 0 # 起始的step
global_epoch = 0 # 起始的epoch
use_cuda = torch.cuda.is_available() # 训练的设备 cpu or gpu
print("use_cuda: {}".format(use_cuda))
syncnet_T = 5 ## 每次选取200ms的视频片段进行训练,视频的fps为25,所以200ms对应的帧数为:25*0.2=5帧
syncnet_mel_step_size = 16 # 200ms对应的声音的mel-spectrogram特征的长度为16.
data_root = "/kaggle/input/wav2lippreprocessed/lrs2_preprocessed" # 数据集的位置
class Dataset(object):
def __init__(self, split):
self.all_videos = get_image_list(data_root, split)
def get_frame_id(self, frame):
return int(basename(frame).split(".")[0])
def get_window(self, start_frame):
start_id = self.get_frame_id(start_frame)
vidname = dirname(start_frame)
window_fnames = []
for frame_id in range(start_id, start_id + syncnet_T):
frame = join(vidname, "{}.jpg".format(frame_id))
if not isfile(frame):
return None
window_fnames.append(frame)
return window_fnames
def crop_audio_window(self, spec, start_frame):
# num_frames = (T x hop_size * fps) / sample_rate
start_frame_num = self.get_frame_id(start_frame)
start_idx = int(80.0 * (start_frame_num / float(hparams.fps)))
end_idx = start_idx + syncnet_mel_step_size
return spec[start_idx:end_idx, :]
def __len__(self):
return len(self.all_videos)
def __getitem__(self, idx):
"""
return: x,mel,y
x: 五张嘴唇图片
mel:对应的语音的mel spectrogram
t:同步or不同步
"""
while 1:
idx = random.randint(0, len(self.all_videos) - 1)
vidname = self.all_videos[idx]
img_names = list(glob(join(vidname, "*.jpg")))
if len(img_names) <= 3 * syncnet_T:
continue
img_name = random.choice(img_names)
wrong_img_name = random.choice(img_names)
while wrong_img_name == img_name:
wrong_img_name = random.choice(img_names)
# 随机决定是产生负样本还是正样本
if random.choice([True, False]):
y = torch.ones(1).float()
chosen = img_name
else:
y = torch.zeros(1).float()
chosen = wrong_img_name
window_fnames = self.get_window(chosen)
if window_fnames is None:
continue
window = []
all_read = True
for fname in window_fnames:
img = cv2.imread(fname)
if img is None:
all_read = False
break
try:
img = cv2.resize(img, (hparams.img_size, hparams.img_size))
except Exception as e:
all_read = False
break
window.append(img)
if not all_read:
continue
try:
wavpath = join(vidname, "audio.wav")
wav = audio.load_wav(wavpath, hparams.sample_rate)
orig_mel = audio.melspectrogram(wav).T
except Exception as e:
continue
mel = self.crop_audio_window(orig_mel.copy(), img_name)
if mel.shape[0] != syncnet_mel_step_size:
continue
# H x W x 3 * T
x = np.concatenate(window, axis=2) / 255.0
x = x.transpose(2, 0, 1)
x = x[:, x.shape[1] // 2 :]
x = torch.FloatTensor(x)
mel = torch.FloatTensor(mel.T).unsqueeze(0)
return x, mel, y
ds = Dataset("train")
x, mel, t = ds[0]
print(x.shape)
print(mel.shape)
print(t.shape)
import matplotlib.pyplot as plt
plt.imshow(mel[0].numpy())
plt.imshow(x[:3, :, :].transpose(0, 2).numpy())
# #### 3.训练
# 使用cosine_loss 作为损失函数
# 损失函数的定义
logloss = nn.BCELoss() # 交叉熵损失
def cosine_loss(a, v, y): # 余弦相似度损失
"""
a: audio_encoder的输出
v: video face_encoder的输出
y: 是否同步的真实值
"""
d = nn.functional.cosine_similarity(a, v)
loss = logloss(d.unsqueeze(1), y)
return loss
def train(
device,
model,
train_data_loader,
test_data_loader,
optimizer,
checkpoint_dir=None,
checkpoint_interval=None,
nepochs=None,
):
global global_step, global_epoch
resumed_step = global_step
while global_epoch < nepochs:
running_loss = 0.0
prog_bar = tqdm(enumerate(train_data_loader))
for step, (x, mel, y) in prog_bar:
model.train()
optimizer.zero_grad()
#####TODO###########
####################
# 补全模型的训练
x = x.to(device)
mel = mel.to(device)
a, v = model(mel, x)
y = y.to(device)
loss = cosine_loss(a, v, y)
loss.backward()
optimizer.step()
global_step += 1
cur_session_steps = global_step - resumed_step
running_loss += loss.item()
if global_step == 1 or global_step % checkpoint_interval == 0:
save_checkpoint(
model, optimizer, global_step, checkpoint_dir, global_epoch
)
if global_step % hparams.syncnet_eval_interval == 0:
with torch.no_grad():
eval_model(
test_data_loader, global_step, device, model, checkpoint_dir
)
prog_bar.set_description(
"Epoch: {} Loss: {}".format(global_epoch, running_loss / (step + 1))
)
global_epoch += 1
def eval_model(test_data_loader, global_step, device, model, checkpoint_dir):
# 在测试集上进行评估
eval_steps = 1400
print("Evaluating for {} steps".format(eval_steps))
losses = []
while 1:
for step, (x, mel, y) in enumerate(test_data_loader):
model.eval()
# Transform data to CUDA device
x = x.to(device)
mel = mel.to(device)
a, v = model(mel, x)
y = y.to(device)
loss = cosine_loss(a, v, y)
losses.append(loss.item())
if step > eval_steps:
break
averaged_loss = sum(losses) / len(losses)
print(averaged_loss)
return
latest_checkpoint_path = ""
def save_checkpoint(model, optimizer, step, checkpoint_dir, epoch):
# 保存训练的结果 checkpoint
global latest_checkpoint_path
checkpoint_path = join(
checkpoint_dir, "checkpoint_step{:09d}.pth".format(global_step)
)
optimizer_state = optimizer.state_dict() if hparams.save_optimizer_state else None
torch.save(
{
"state_dict": model.state_dict(),
"optimizer": optimizer_state,
"global_step": step,
"global_epoch": epoch,
},
checkpoint_path,
)
latest_checkpoint_path = checkpoint_path
print("Saved checkpoint:", checkpoint_path)
def _load(checkpoint_path):
if use_cuda:
checkpoint = torch.load(checkpoint_path)
else:
checkpoint = torch.load(
checkpoint_path, map_location=lambda storage, loc: storage
)
return checkpoint
def load_checkpoint(path, model, optimizer, reset_optimizer=False):
# 读取指定checkpoint的保存信息
global global_step
global global_epoch
print("Load checkpoint from: {}".format(path))
checkpoint = _load(path)
model.load_state_dict(checkpoint["state_dict"])
if not reset_optimizer:
optimizer_state = checkpoint["optimizer"]
if optimizer_state is not None:
print("Load optimizer state from {}".format(path))
optimizer.load_state_dict(checkpoint["optimizer"])
global_step = checkpoint["global_step"]
global_epoch = checkpoint["global_epoch"]
return model
# **下面开始训练,最终的Loss参考值为0.20左右,此时模型能达到较好的判别效果**
checkpoint_dir = "/kaggle/working/expert_checkpoints/" # 指定存储 checkpoint的位置
checkpoint_path = (
"/kaggle/input/wav2lip24epoch/expert_checkpoints/checkpoint_step000060000.pth"
)
# 指定加载checkpoint的路径,第一次训练时不需要,后续如果想从某个checkpoint恢复训练,可指定。
if not os.path.exists(checkpoint_dir):
os.mkdir(checkpoint_dir)
# Dataset and Dataloader setup
train_dataset = Dataset("train")
test_dataset = Dataset("val")
############TODO#########
#####Train Dataloader and Test Dataloader
#### 具体的bacthsize等参数,参考 hparams.py文件
train_data_loader = data_utils.DataLoader(
train_dataset,
batch_size=hparams.batch_size,
shuffle=True,
num_workers=hparams.num_workers,
)
test_data_loader = data_utils.DataLoader(
test_dataset, batch_size=hparams.batch_size, num_workers=8
)
device = torch.device("cuda" if use_cuda else "cpu")
# Model
#####定义 SynNet模型,并加载到指定的device上
model = SyncNet().to(device)
print(
"total trainable params {}".format(
sum(p.numel() for p in model.parameters() if p.requires_grad)
)
)
####定义优化器,使用adam,lr参考hparams.py文件
optimizer = optim.Adam([p for p in model.parameters() if p.requires_grad], lr=1e-6)
if checkpoint_path is not None:
load_checkpoint(checkpoint_path, model, optimizer, reset_optimizer=True)
train(
device,
model,
train_data_loader,
test_data_loader,
optimizer,
checkpoint_dir=checkpoint_dir,
checkpoint_interval=hparams.syncnet_checkpoint_interval,
nepochs=48,
)
# ### 3.2 训练Wav2Lip
# 预训练模型 [weight](https://iiitaphyd-my.sharepoint.com/:u:/g/personal/radrabha_m_research_iiit_ac_in/EdjI7bZlgApMqsVoEUUXpLsBxqXbn5z8VTmoxp55YNDcIA?e=n9ljGW)
# #### 1. 模型的定义
# wav2lip模型的生成器首先对输入进行下采样,然后再经过上采样恢复成原来的大小。为了方便,我们对其中重复利用到的模块进行了封装。
class nonorm_Conv2d(nn.Module): # 不需要进行 norm的卷积
def __init__(
self, cin, cout, kernel_size, stride, padding, residual=False, *args, **kwargs
):
super().__init__(*args, **kwargs)
self.conv_block = nn.Sequential(
nn.Conv2d(cin, cout, kernel_size, stride, padding),
)
self.act = nn.LeakyReLU(0.01, inplace=True)
def forward(self, x):
out = self.conv_block(x)
return self.act(out)
class Conv2dTranspose(nn.Module): # 逆卷积,上采样
def __init__(
self, cin, cout, kernel_size, stride, padding, output_padding=0, *args, **kwargs
):
super().__init__(*args, **kwargs)
############TODO###########
## 完成self.conv_block: 一个逆卷积和batchnorm组成的 Sequential结构
self.conv_block = nn.Sequential(
nn.ConvTranspose2d(cin, cout, kernel_size, stride, padding, output_padding),
nn.BatchNorm2d(cout),
)
self.act = nn.ReLU()
def forward(self, x):
out = self.conv_block(x)
return self.act(out)
# **生成器**
# 由两个encoder: face_encoder和 audio_encoder, 一个decoder:face_decoder组成。face encoder 和 audio encoder 分别对输入的人脸和语音特征进行降维,得到(1,1,512)的特征,并将二者进行拼接送入到 face decoder中去进行上采样,最终得到和输入一样大小的人脸图像。
#####################TODO############################
# 根据下面打印的网络模型图,补全网络的参数
class Wav2Lip(nn.Module):
def __init__(self):
super(Wav2Lip, self).__init__()
self.face_encoder_blocks = nn.ModuleList(
[
nn.Sequential(
Conv2d(6, 16, kernel_size=7, stride=1, padding=3)
), # 96,96
nn.Sequential(
Conv2d(16, 32, kernel_size=3, stride=2, padding=1), # 48,48
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(32, 64, kernel_size=3, stride=2, padding=1), # 24,24
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(64, 128, kernel_size=3, stride=2, padding=1), # 12,12
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(128, 256, kernel_size=3, stride=2, padding=1), # 6,6
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(256, 512, kernel_size=3, stride=2, padding=1), # 3,3
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2d(512, 512, kernel_size=3, stride=1, padding=0), # 1, 1
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
),
]
)
self.audio_encoder = nn.Sequential(
Conv2d(1, 32, kernel_size=3, stride=1, padding=1),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 32, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(32, 64, kernel_size=3, stride=(3, 1), padding=1),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 128, kernel_size=3, stride=3, padding=1),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 256, kernel_size=3, stride=(3, 2), padding=1),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 512, kernel_size=3, stride=1, padding=0),
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
)
self.face_decoder_blocks = nn.ModuleList(
[
nn.Sequential(
Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
),
nn.Sequential(
Conv2dTranspose(
1024, 512, kernel_size=3, stride=1, padding=0
), # 3,3
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
),
nn.Sequential(
Conv2dTranspose(
1024, 512, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(512, 512, kernel_size=3, stride=1, padding=1, residual=True),
), # 6, 6
nn.Sequential(
Conv2dTranspose(
768, 384, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(384, 384, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(384, 384, kernel_size=3, stride=1, padding=1, residual=True),
), # 12, 12
nn.Sequential(
Conv2dTranspose(
512, 256, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(256, 256, kernel_size=3, stride=1, padding=1, residual=True),
), # 24, 24
nn.Sequential(
Conv2dTranspose(
320, 128, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(128, 128, kernel_size=3, stride=1, padding=1, residual=True),
), # 48, 48
nn.Sequential(
Conv2dTranspose(
160, 64, kernel_size=3, stride=2, padding=1, output_padding=1
),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
Conv2d(64, 64, kernel_size=3, stride=1, padding=1, residual=True),
),
]
) # 96,96
self.output_block = nn.Sequential(
Conv2d(80, 32, kernel_size=3, stride=1, padding=1),
nn.Conv2d(32, 3, kernel_size=1, stride=1, padding=0),
nn.Sigmoid(),
)
def forward(self, audio_sequences, face_sequences):
# audio_sequences = (B, T, 1, 80, 16)
B = audio_sequences.size(0)
input_dim_size = len(face_sequences.size())
if input_dim_size > 4:
audio_sequences = torch.cat(
[audio_sequences[:, i] for i in range(audio_sequences.size(1))], dim=0
)
face_sequences = torch.cat(
[face_sequences[:, :, i] for i in range(face_sequences.size(2))], dim=0
)
audio_embedding = self.audio_encoder(audio_sequences) # B, 512, 1, 1
feats = []
x = face_sequences
for f in self.face_encoder_blocks:
x = f(x)
feats.append(x)
x = audio_embedding
for f in self.face_decoder_blocks:
x = f(x)
try:
x = torch.cat((x, feats[-1]), dim=1)
except Exception as e:
print(x.size())
print(feats[-1].size())
raise e
feats.pop()
x = self.output_block(x)
if input_dim_size > 4:
x = torch.split(x, B, dim=0) # [(B, C, H, W)]
outputs = torch.stack(x, dim=2) # (B, C, T, H, W)
else:
outputs = x
return outputs
# **判别器**
# 判别器也是由一系列的卷积神经网络组成,输入一张人脸图片,利用face encoder对其进行降维到512维。
###########TODO##################
####补全判别器模型
class Wav2Lip_disc_qual(nn.Module):
def __init__(self):
super(Wav2Lip_disc_qual, self).__init__()
self.face_encoder_blocks = nn.ModuleList(
[
nn.Sequential(
nonorm_Conv2d(3, 32, kernel_size=7, stride=1, padding=3)
), # 48,96
nn.Sequential(
nonorm_Conv2d(
32, 64, kernel_size=5, stride=(1, 2), padding=2
), # 48,48
nonorm_Conv2d(64, 64, kernel_size=5, stride=1, padding=2),
),
nn.Sequential(
nonorm_Conv2d(64, 128, kernel_size=5, stride=2, padding=2), # 24,24
nonorm_Conv2d(128, 128, kernel_size=5, stride=1, padding=2),
),
nn.Sequential(
nonorm_Conv2d(
128, 256, kernel_size=5, stride=2, padding=2
), # 12,12
nonorm_Conv2d(256, 256, kernel_size=5, stride=1, padding=2),
),
nn.Sequential(
nonorm_Conv2d(256, 512, kernel_size=3, stride=2, padding=1), # 6,6
nonorm_Conv2d(512, 512, kernel_size=3, stride=1, padding=1),
),
nn.Sequential(
nonorm_Conv2d(512, 512, kernel_size=3, stride=2, padding=1), # 3,3
nonorm_Conv2d(512, 512, kernel_size=3, stride=1, padding=1),
),
nn.Sequential(
nonorm_Conv2d(512, 512, kernel_size=3, stride=1, padding=0), # 1, 1
nonorm_Conv2d(512, 512, kernel_size=1, stride=1, padding=0),
),
]
)
self.binary_pred = nn.Sequential(
nn.Conv2d(512, 1, kernel_size=1, stride=1, padding=0), nn.Sigmoid()
)
self.label_noise = 0.0
def get_lower_half(self, face_sequences):
return face_sequences[:, :, face_sequences.size(2) // 2 :]
def to_2d(self, face_sequences):
B = face_sequences.size(0)
face_sequences = torch.cat(
[face_sequences[:, :, i] for i in range(face_sequences.size(2))], dim=0
)
return face_sequences
def perceptual_forward(self, false_face_sequences):
false_face_sequences = self.to_2d(false_face_sequences)
false_face_sequences = self.get_lower_half(false_face_sequences)
false_feats = false_face_sequences
for f in self.face_encoder_blocks:
false_feats = f(false_feats)
false_pred_loss = F.binary_cross_entropy(
self.binary_pred(false_feats).view(len(false_feats), -1),
torch.ones((len(false_feats), 1)).cuda(),
)
return false_pred_loss
def forward(self, face_sequences):
face_sequences = self.to_2d(face_sequences)
face_sequences = self.get_lower_half(face_sequences)
x = face_sequences
for f in self.face_encoder_blocks:
x = f(x)
return self.binary_pred(x).view(len(x), -1)
# #### 2. 数据集的定义
# 在训练时,会用到4个数据:
# 1. x:输入的图片
# 2. indiv_mels: 每一张图片所对应语音的mel-spectrogram特征
# 3. mel: 所有帧对应的200ms的语音mel-spectrogram,用于SyncNet进行唇音同步损失的计算
# 4. y:真实的与语音对应的,唇音同步的图片。
#
global_step = 0
global_epoch = 0
use_cuda = torch.cuda.is_available()
print("use_cuda: {}".format(use_cuda))
syncnet_T = 5
syncnet_mel_step_size = 16
class Dataset(object):
def __init__(self, split):
self.all_videos = get_image_list(data_root, split)
def get_frame_id(self, frame):
return int(basename(frame).split(".")[0])
def get_window(self, start_frame):
start_id = self.get_frame_id(start_frame)
vidname = dirname(start_frame)
window_fnames = []
for frame_id in range(start_id, start_id + syncnet_T):
frame = join(vidname, "{}.jpg".format(frame_id))
if not isfile(frame):
return None
window_fnames.append(frame)
return window_fnames
def read_window(self, window_fnames):
if window_fnames is None:
return None
window = []
for fname in window_fnames:
img = cv2.imread(fname)
if img is None:
return None
try:
img = cv2.resize(img, (hparams.img_size, hparams.img_size))
except Exception as e:
return None
window.append(img)
return window
def crop_audio_window(self, spec, start_frame):
if type(start_frame) == int:
start_frame_num = start_frame
else:
start_frame_num = self.get_frame_id(
start_frame
) # 0-indexing ---> 1-indexing
start_idx = int(80.0 * (start_frame_num / float(hparams.fps)))
end_idx = start_idx + syncnet_mel_step_size
return spec[start_idx:end_idx, :]
def get_segmented_mels(self, spec, start_frame):
mels = []
assert syncnet_T == 5
start_frame_num = (
self.get_frame_id(start_frame) + 1
) # 0-indexing ---> 1-indexing
if start_frame_num - 2 < 0:
return None
for i in range(start_frame_num, start_frame_num + syncnet_T):
m = self.crop_audio_window(spec, i - 2)
if m.shape[0] != syncnet_mel_step_size:
return None
mels.append(m.T)
mels = np.asarray(mels)
return mels
def prepare_window(self, window):
# 3 x T x H x W
x = np.asarray(window) / 255.0
x = np.transpose(x, (3, 0, 1, 2))
return x
def __len__(self):
return len(self.all_videos)
def __getitem__(self, idx):
while 1:
idx = random.randint(0, len(self.all_videos) - 1) # 随机选择一个视频id
vidname = self.all_videos[idx]
img_names = list(glob(join(vidname, "*.jpg")))
if len(img_names) <= 3 * syncnet_T:
continue
img_name = random.choice(img_names)
wrong_img_name = random.choice(img_names) # 随机选择帧
while wrong_img_name == img_name:
wrong_img_name = random.choice(img_names)
window_fnames = self.get_window(img_name)
wrong_window_fnames = self.get_window(wrong_img_name)
if window_fnames is None or wrong_window_fnames is None:
continue
window = self.read_window(window_fnames)
if window is None:
continue
wrong_window = self.read_window(wrong_window_fnames)
if wrong_window is None:
continue
try:
# 读取音频
wavpath = join(vidname, "audio.wav")
wav = audio.load_wav(wavpath, hparams.sample_rate)
# 提取完整mel-spectrogram
orig_mel = audio.melspectrogram(wav).T
except Exception as e:
continue
# 分割 mel-spectrogram
mel = self.crop_audio_window(orig_mel.copy(), img_name)
if mel.shape[0] != syncnet_mel_step_size:
continue
indiv_mels = self.get_segmented_mels(orig_mel.copy(), img_name)
if indiv_mels is None:
continue
window = self.prepare_window(window)
y = window.copy()
window[:, :, window.shape[2] // 2 :] = 0.0
wrong_window = self.prepare_window(wrong_window)
x = np.concatenate([window, wrong_window], axis=0)
x = torch.FloatTensor(x)
mel = torch.FloatTensor(mel.T).unsqueeze(0)
indiv_mels = torch.FloatTensor(indiv_mels).unsqueeze(1)
y = torch.FloatTensor(y)
return x, indiv_mels, mel, y
ds = Dataset("train")
x, indiv_mels, mel, y = ds[0]
print(x.shape)
print(indiv_mels.shape)
print(mel.shape)
print(y.shape)
# #### 3. 训练
# bce 交叉墒loss
logloss = nn.BCELoss()
def cosine_loss(a, v, y):
d = nn.functional.cosine_similarity(a, v)
loss = logloss(d.unsqueeze(1), y)
return loss
device = torch.device("cuda" if use_cuda else "cpu")
syncnet = SyncNet().to(device) # 定义syncnet 模型
for p in syncnet.parameters():
p.requires_grad = False
#####L1 loss
recon_loss = nn.L1Loss()
def get_sync_loss(mel, g):
g = g[:, :, :, g.size(3) // 2 :]
g = torch.cat([g[:, :, i] for i in range(syncnet_T)], dim=1)
# B, 3 * T, H//2, W
a, v = syncnet(mel, g)
y = torch.ones(g.size(0), 1).float().to(device)
return cosine_loss(a, v, y)
def train(
device,
model,
disc,
train_data_loader,
test_data_loader,
optimizer,
disc_optimizer,
checkpoint_dir=None,
checkpoint_interval=None,
nepochs=None,
):
global global_step, global_epoch
resumed_step = global_step
while global_epoch < nepochs:
print("Starting Epoch: {}".format(global_epoch))
running_sync_loss, running_l1_loss, disc_loss, running_perceptual_loss = (
0.0,
0.0,
0.0,
0.0,
)
running_disc_real_loss, running_disc_fake_loss = 0.0, 0.0
prog_bar = tqdm(enumerate(train_data_loader))
for step, (x, indiv_mels, mel, gt) in prog_bar:
disc.train()
model.train()
x = x.to(device)
mel = mel.to(device)
indiv_mels = indiv_mels.to(device)
gt = gt.to(device)
### Train generator now. Remove ALL grads.
# 训练生成器
optimizer.zero_grad()
disc_optimizer.zero_grad()
g = model(indiv_mels, x) # 得到生成的结果
if hparams.syncnet_wt > 0.0:
sync_loss = get_sync_loss(mel, g) # 从预训练的expert 模型中获得唇音同步的损失
else:
sync_loss = 0.0
if hparams.disc_wt > 0.0:
perceptual_loss = disc.perceptual_forward(g) # 判别器的感知损失
else:
perceptual_loss = 0.0
l1loss = recon_loss(g, gt) # l1 loss,重建损失
# 最终的损失函数
loss = (
hparams.syncnet_wt * sync_loss
+ hparams.disc_wt * perceptual_loss
+ (1.0 - hparams.syncnet_wt - hparams.disc_wt) * l1loss
)
loss.backward()
optimizer.step()
### Remove all gradients before Training disc
# 训练判别器
disc_optimizer.zero_grad()
pred = disc(gt)
disc_real_loss = F.binary_cross_entropy(
pred, torch.ones((len(pred), 1)).to(device)
)
disc_real_loss.backward()
pred = disc(g.detach())
disc_fake_loss = F.binary_cross_entropy(
pred, torch.zeros((len(pred), 1)).to(device)
)
disc_fake_loss.backward()
disc_optimizer.step()
running_disc_real_loss += disc_real_loss.item()
running_disc_fake_loss += disc_fake_loss.item()
# Logs
global_step += 1
cur_session_steps = global_step - resumed_step
running_l1_loss += l1loss.item()
if hparams.syncnet_wt > 0.0:
running_sync_loss += sync_loss.item()
else:
running_sync_loss += 0.0
if hparams.disc_wt > 0.0:
running_perceptual_loss += perceptual_loss.item()
else:
running_perceptual_loss += 0.0
if global_step == 1 or global_step % checkpoint_interval == 0:
save_checkpoint(
model, optimizer, global_step, checkpoint_dir, global_epoch
)
save_checkpoint(
disc,
disc_optimizer,
global_step,
checkpoint_dir,
global_epoch,
prefix="disc_",
)
if global_step % hparams.eval_interval == 0:
with torch.no_grad():
average_sync_loss = eval_model(
test_data_loader, global_step, device, model, disc
)
if average_sync_loss < 0.75:
hparams.set_hparam("syncnet_wt", 0.03)
prog_bar.set_description(
"L1: {}, Sync: {}, Percep: {} | Fake: {}, Real: {}".format(
running_l1_loss / (step + 1),
running_sync_loss / (step + 1),
running_perceptual_loss / (step + 1),
running_disc_fake_loss / (step + 1),
running_disc_real_loss / (step + 1),
)
)
global_epoch += 1
def eval_model(test_data_loader, global_step, device, model, disc):
eval_steps = 300
print("Evaluating for {} steps".format(eval_steps))
(
running_sync_loss,
running_l1_loss,
running_disc_real_loss,
running_disc_fake_loss,
running_perceptual_loss,
) = ([], [], [], [], [])
while 1:
for step, (x, indiv_mels, mel, gt) in enumerate((test_data_loader)):
model.eval()
disc.eval()
x = x.to(device)
mel = mel.to(device)
indiv_mels = indiv_mels.to(device)
gt = gt.to(device)
pred = disc(gt)
disc_real_loss = F.binary_cross_entropy(
pred, torch.ones((len(pred), 1)).to(device)
)
g = model(indiv_mels, x)
pred = disc(g)
disc_fake_loss = F.binary_cross_entropy(
pred, torch.zeros((len(pred), 1)).to(device)
)
running_disc_real_loss.append(disc_real_loss.item())
running_disc_fake_loss.append(disc_fake_loss.item())
sync_loss = get_sync_loss(mel, g)
if hparams.disc_wt > 0.0:
perceptual_loss = disc.perceptual_forward(g)
else:
perceptual_loss = 0.0
l1loss = recon_loss(g, gt)
loss = (
hparams.syncnet_wt * sync_loss
+ hparams.disc_wt * perceptual_loss
+ (1.0 - hparams.syncnet_wt - hparams.disc_wt) * l1loss
)
running_l1_loss.append(l1loss.item())
running_sync_loss.append(sync_loss.item())
if hparams.disc_wt > 0.0:
running_perceptual_loss.append(perceptual_loss.item())
else:
running_perceptual_loss.append(0.0)
if step > eval_steps:
break
print(
"L1: {}, Sync: {}, Percep: {} | Fake: {}, Real: {}".format(
sum(running_l1_loss) / len(running_l1_loss),
sum(running_sync_loss) / len(running_sync_loss),
sum(running_perceptual_loss) / len(running_perceptual_loss),
sum(running_disc_fake_loss) / len(running_disc_fake_loss),
sum(running_disc_real_loss) / len(running_disc_real_loss),
)
)
return sum(running_sync_loss) / len(running_sync_loss)
def save_checkpoint(model, optimizer, step, checkpoint_dir, epoch, prefix=""):
checkpoint_path = join(
checkpoint_dir, "{}checkpoint_step{:09d}.pth".format(prefix, global_step)
)
optimizer_state = optimizer.state_dict() if hparams.save_optimizer_state else None
torch.save(
{
"state_dict": model.state_dict(),
"optimizer": optimizer_state,
"global_step": step,
"global_epoch": epoch,
},
checkpoint_path,
)
print("Saved checkpoint:", checkpoint_path)
def _load(checkpoint_path):
if use_cuda:
checkpoint = torch.load(checkpoint_path)
else:
checkpoint = torch.load(
checkpoint_path, map_location=lambda storage, loc: storage
)
return checkpoint
def load_checkpoint(
path, model, optimizer, reset_optimizer=False, overwrite_global_states=True
):
global global_step
global global_epoch
print("Load checkpoint from: {}".format(path))
checkpoint = _load(path)
s = checkpoint["state_dict"]
new_s = {}
for k, v in s.items():
new_s[k.replace("module.", "")] = v
model.load_state_dict(new_s)
if not reset_optimizer:
optimizer_state = checkpoint["optimizer"]
if optimizer_state is not None:
print("Load optimizer state from {}".format(path))
optimizer.load_state_dict(checkpoint["optimizer"])
if overwrite_global_states:
global_step = checkpoint["global_step"]
global_epoch = checkpoint["global_epoch"]
return model
checkpoint_dir = "/kaggle/working/wav2lip_checkpoints" # checkpoint 存储的位置
# Dataset and Dataloader setup
train_dataset = Dataset("train")
test_dataset = Dataset("val")
train_data_loader = data_utils.DataLoader(
train_dataset,
batch_size=hparams.batch_size,
shuffle=True,
num_workers=hparams.num_workers,
)
test_data_loader = data_utils.DataLoader(
test_dataset, batch_size=hparams.batch_size, num_workers=4
)
device = torch.device("cuda" if use_cuda else "cpu")
# Model
model = Wav2Lip().to(device) ####### 生成器模型
disc = Wav2Lip_disc_qual().to(device) ####### 判别器模型
print(
"total trainable params {}".format(
sum(p.numel() for p in model.parameters() if p.requires_grad)
)
)
print(
"total DISC trainable params {}".format(
sum(p.numel() for p in disc.parameters() if p.requires_grad)
)
)
optimizer = optim.Adam(
[p for p in model.parameters() if p.requires_grad],
lr=hparams.initial_learning_rate,
betas=(0.5, 0.999),
) #####adam优化器,betas=[0.5,0.999]
disc_optimizer = optim.Adam(
[p for p in disc.parameters() if p.requires_grad],
lr=hparams.disc_initial_learning_rate,
betas=(0.5, 0.999),
) #####adam优化器,betas=[0.5,0.999]
# 继续训练的生成器的checkpoint位置
# checkpoint_path=""
# load_checkpoint(checkpoint_path, model, optimizer, reset_optimizer=False)
# 继续训练的判别器的checkpoint位置
# disc_checkpoint_path=""
# load_checkpoint(disc_checkpoint_path, disc, disc_optimizer,
# reset_optimizer=False, overwrite_global_states=False)
# syncnet的checkpoint位置,我们将使用此模型计算生成的帧和语音的唇音同步损失
syncnet_checkpoint_path = latest_checkpoint_path
# syncnet_checkpoint_path="/kaggle/working/expert_checkpoints/checkpoint_step000000001.pth"
load_checkpoint(
syncnet_checkpoint_path,
syncnet,
None,
reset_optimizer=True,
overwrite_global_states=False,
)
if not os.path.exists(checkpoint_dir):
os.mkdir(checkpoint_dir)
# Train!
train(
device,
model,
disc,
train_data_loader,
test_data_loader,
optimizer,
disc_optimizer,
checkpoint_dir=checkpoint_dir,
checkpoint_interval=hparams.checkpoint_interval,
nepochs=hparams.nepochs,
)
# #### 4. 命令行训练
# 上面是按步骤训练的过程,在`hq_wav2lip_train.py`文件中已经把上述的过程进行了封装,你可以通过以下的命令直接进行训练
# !python wav2lip_train.py --data_root lrs2_preprocessed/ --checkpoint_dir <folder_to_save_checkpoints> --syncnet_checkpoint_path <path_to_expert_disc_checkpoint>
# ### 4. 模型的推理
# 当模型训练完毕后,我们只使用生成器的网络模型部分作为我们的推理模型。模型的输入由一段包含人脸的参照视频和一段语音组成。
# 在这里我们可以直接使用官方提供给我们的预训练模型[weight](https://iiitaphyd-my.sharepoint.com/:u:/g/personal/radrabha_m_research_iiit_ac_in/EdjI7bZlgApMqsVoEUUXpLsBxqXbn5z8VTmoxp55YNDcIA?e=n9ljGW)下载该模型并放入到指定文件夹下,供之后的推理使用。
# 模型的推理过程主要分为以下几个步骤:
# 1. 输入数据的预处理,包含人脸抠图,视频分帧,提取mel-spectrogram特征等操作。
# 2. 利用网络模型生成唇音同步的视频帧。
# 3. 将生成的视频帧准换成视频,并和输入的语音结合,形成最终的输出视频。
#
from os import listdir, path
import numpy as np
import scipy, cv2, os, sys, argparse, audio
import json, subprocess, random, string
from tqdm import tqdm
from glob import glob
import torch, face_detection
from models import Wav2Lip
import platform
import audio
checkpoint_path = "/kaggle/working/wav2lip_checkpoints/checkpoint_step000000001.pth" # 生成器的checkpoint位置
face = "input_video.mp4" # 参照视频的文件位置, *.mp4
speech = "input_audio.wav" # 输入语音的位置,*.wav
resize_factor = 1 # 对输入的视频进行下采样的倍率
crop = [0, -1, 0, -1] # 是否对视频帧进行裁剪,处理视频中有多张人脸时有用
fps = 25 # 视频的帧率
static = False # 是否只使用固定的一帧作为视频的生成参照
if not os.path.isfile(face):
raise ValueError("--face argument must be a valid path to video/image file")
else: # 若输入的是视频格式
video_stream = cv2.VideoCapture(face) # 读取视频
fps = video_stream.get(cv2.CAP_PROP_FPS) # 读取 fps
print("Reading video frames...")
full_frames = []
# 提取所有的帧
while 1:
still_reading, frame = video_stream.read()
if not still_reading:
video_stream.release()
break
if resize_factor > 1: # 进行下采样,降低分辨率
frame = cv2.resize(
frame,
(frame.shape[1] // resize_factor, frame.shape[0] // resize_factor),
)
y1, y2, x1, x2 = crop # 裁剪
if x2 == -1:
x2 = frame.shape[1]
if y2 == -1:
y2 = frame.shape[0]
frame = frame[y1:y2, x1:x2]
full_frames.append(frame)
print("Number of frames available for inference: " + str(len(full_frames)))
# 检查输入的音频是否为 .wav格式的,若不是则进行转换
if not speech.endswith(".wav"):
print("Extracting raw audio...")
command = "ffmpeg -y -i {} -strict -2 {}".format(speech, "temp/temp.wav")
subprocess.call(command, shell=True)
speech = "temp/temp.wav"
wav = audio.load_wav(speech, 16000) # 保证采样率为16000
mel = audio.melspectrogram(wav)
print(mel.shape)
wav2lip_batch_size = 128 # 推理时输入到网络的batchsize
mel_step_size = 16
# 提取语音的mel谱
mel_chunks = []
mel_idx_multiplier = 80.0 / fps
i = 0
while 1:
start_idx = int(i * mel_idx_multiplier)
if start_idx + mel_step_size > len(mel[0]):
mel_chunks.append(mel[:, len(mel[0]) - mel_step_size :])
break
mel_chunks.append(mel[:, start_idx : start_idx + mel_step_size])
i += 1
print("Length of mel chunks: {}".format(len(mel_chunks)))
full_frames = full_frames[: len(mel_chunks)]
batch_size = wav2lip_batch_size
img_size = 96 # 默认的输入图片大小
pads = [0, 20, 0, 0] # 填充的长度,保证下巴也在抠图的范围之内
nosmooth = False
face_det_batch_size = 16
def get_smoothened_boxes(boxes, T):
for i in range(len(boxes)):
if i + T > len(boxes):
window = boxes[len(boxes) - T :]
else:
window = boxes[i : i + T]
boxes[i] = np.mean(window, axis=0)
return boxes
# 人脸检测函数
def face_detect(images):
detector = face_detection.FaceAlignment(
face_detection.LandmarksType._2D, flip_input=False, device=device
)
batch_size = face_det_batch_size
while 1:
predictions = []
try:
for i in tqdm(range(0, len(images), batch_size)):
predictions.extend(
detector.get_detections_for_batch(
np.array(images[i : i + batch_size])
)
)
except RuntimeError:
if batch_size == 1:
raise RuntimeError(
"Image too big to run face detection on GPU. Please use the --resize_factor argument"
)
batch_size //= 2
print("Recovering from OOM error; New batch size: {}".format(batch_size))
continue
break
results = []
pady1, pady2, padx1, padx2 = pads
for rect, image in zip(predictions, images):
if rect is None:
cv2.imwrite(
"temp/faulty_frame.jpg", image
) # check this frame where the face was not detected.
raise ValueError(
"Face not detected! Ensure the video contains a face in all the frames."
)
y1 = max(0, rect[1] - pady1)
y2 = min(image.shape[0], rect[3] + pady2)
x1 = max(0, rect[0] - padx1)
x2 = min(image.shape[1], rect[2] + padx2)
results.append([x1, y1, x2, y2])
boxes = np.array(results)
if not nosmooth:
boxes = get_smoothened_boxes(boxes, T=5)
results = [
[image[y1:y2, x1:x2], (y1, y2, x1, x2)]
for image, (x1, y1, x2, y2) in zip(images, boxes)
]
del detector
return results
box = [-1, -1, -1, -1]
def datagen(frames, mels):
img_batch, mel_batch, frame_batch, coords_batch = [], [], [], []
if box[0] == -1: # 如果未指定 特定的人脸边界的话
if not static: # 是否使用视频的第一帧作为参考
face_det_results = face_detect(frames) # BGR2RGB for CNN face detection
else:
face_det_results = face_detect([frames[0]])
else:
print("Using the specified bounding box instead of face detection...")
y1, y2, x1, x2 = box
face_det_results = [
[f[y1:y2, x1:x2], (y1, y2, x1, x2)] for f in frames
] # 裁剪出人脸结果
for i, m in enumerate(mels):
idx = 0 if static else i % len(frames)
frame_to_save = frames[idx].copy()
face, coords = face_det_results[idx].copy()
face = cv2.resize(face, (img_size, img_size)) # 重采样到指定大小
img_batch.append(face)
mel_batch.append(m)
frame_batch.append(frame_to_save)
coords_batch.append(coords)
if len(img_batch) >= wav2lip_batch_size:
img_batch, mel_batch = np.asarray(img_batch), np.asarray(mel_batch)
img_masked = img_batch.copy()
img_masked[:, img_size // 2 :] = 0
img_batch = np.concatenate((img_masked, img_batch), axis=3) / 255.0
mel_batch = np.reshape(
mel_batch, [len(mel_batch), mel_batch.shape[1], mel_batch.shape[2], 1]
)
yield img_batch, mel_batch, frame_batch, coords_batch
img_batch, mel_batch, frame_batch, coords_batch = [], [], [], []
if len(img_batch) > 0:
img_batch, mel_batch = np.asarray(img_batch), np.asarray(mel_batch)
img_masked = img_batch.copy()
img_masked[:, img_size // 2 :] = 0
img_batch = np.concatenate((img_masked, img_batch), axis=3) / 255.0
mel_batch = np.reshape(
mel_batch, [len(mel_batch), mel_batch.shape[1], mel_batch.shape[2], 1]
)
yield img_batch, mel_batch, frame_batch, coords_batch
mel_step_size = 16
device = "cuda" if torch.cuda.is_available() else "cpu"
print("Using {} for inference.".format(device))
# 加载模型
def _load(checkpoint_path):
if device == "cuda":
checkpoint = torch.load(checkpoint_path)
else:
checkpoint = torch.load(
checkpoint_path, map_location=lambda storage, loc: storage
)
return checkpoint
def load_model(path):
model = Wav2Lip()
print("Load checkpoint from: {}".format(path))
checkpoint = _load(path)
s = checkpoint["state_dict"]
new_s = {}
for k, v in s.items():
new_s[k.replace("module.", "")] = v
model.load_state_dict(new_s)
model = model.to(device)
return model.eval()
os.mkdir("/kaggle/working/temp/")
full_frames = full_frames[: len(mel_chunks)]
batch_size = wav2lip_batch_size
gen = datagen(full_frames.copy(), mel_chunks) # 进行人脸的裁剪与拼接,6通道
for i, (img_batch, mel_batch, frames, coords) in enumerate(
tqdm(gen, total=int(np.ceil(float(len(mel_chunks)) / batch_size)))
):
# 加载模型
if i == 0:
model = load_model(checkpoint_path)
print("Model loaded")
frame_h, frame_w = full_frames[0].shape[:-1]
# 暂存临时视频
out = cv2.VideoWriter(
"/kaggle/working/temp/result_without_audio.avi",
cv2.VideoWriter_fourcc(*"DIVX"),
fps,
(frame_w, frame_h),
)
img_batch = torch.FloatTensor(np.transpose(img_batch, (0, 3, 1, 2))).to(device)
mel_batch = torch.FloatTensor(np.transpose(mel_batch, (0, 3, 1, 2))).to(device)
##### 将 img_batch, mel_batch送入模型得到pred
##############TODO##############
with torch.no_grad():
pred = model(mel_batch, img_batch)
pred = pred.cpu().numpy().transpose(0, 2, 3, 1) * 255.0
for p, f, c in zip(pred, frames, coords):
y1, y2, x1, x2 = c
p = cv2.resize(p.astype(np.uint8), (x2 - x1, y2 - y1))
f[y1:y2, x1:x2] = p
out.write(f)
out.release()
# 将生成的视频与语音合并
outfile = "/kaggle/working/result.mp4" # 最终输出结果到该文件夹下
command = "ffmpeg -y -i {} -i {} -strict -2 -q:v 1 {}".format(
speech, "/kaggle/working/temp/result_without_audio.mp4", outfile
)
subprocess.call(command, shell=platform.system() != "Windows")
| false | 0 | 17,569 | 0 | 17,596 | 17,569 |
||
69571578
|
<jupyter_start><jupyter_text>Melbourne Housing Market
##Update 06/08/2018 - Well it finally happened, Melbourne housing has cooled off. So here's your challenge; 1) when did it exactly happen? , 2) Could you see it slowing down? What were the variables that showed the slowing down (was it overall price, amount sold vs unsold, change in more rentals sold and less housing, changes in which CouncilArea or Region, more houses sold in distances further away from Melbourne CBD and less closer)? 3) Could you have predicted it (I'm not sure how you would do this, but I'm sure you magicians have a way that would make me think we should burn you for being a witch) 4) Should I hold off even longer in buying a two bedroom apartment in Northcote? <-- This is the real reason for me in publishing this dataset :)
#### Update 22/05/2018 - Will continue with a smaller subset of the data (not as many columns). The web scraping was taking some time and also may potentially cause problems. Will continue to post the data.
#### Update 28/11/2017 - Last few weeks clearance levels starting to decrease (I may just be seeing a pattern I want to see.. maybe I'm just evil). Anyway, can any of you magicians make any sense of it?
Melbourne is currently experiencing a housing bubble (some experts say it may burst soon). Maybe someone can find a trend or give a prediction? Which suburbs are the best to buy in? Which ones are value for money? Where's the expensive side of town? And more importantly where should I buy a 2 bedroom unit?
## Content & Acknowledgements
This data was scraped from publicly available results posted every week from Domain.com.au, I've cleaned it as best I can, now it's up to you to make data analysis magic. The dataset includes Address, Type of Real estate, Suburb, Method of Selling, Rooms, Price, Real Estate Agent, Date of Sale and distance from C.B.D.
....Now with extra data including including property size, land size and council area, you may need to change your code!
## Some Key Details
**Suburb**: Suburb
**Address**: Address
**Rooms**: Number of rooms
**Price**: Price in Australian dollars
**Method**:
S - property sold;
SP - property sold prior;
PI - property passed in;
PN - sold prior not disclosed;
SN - sold not disclosed;
NB - no bid;
VB - vendor bid;
W - withdrawn prior to auction;
SA - sold after auction;
SS - sold after auction price not disclosed.
N/A - price or highest bid not available.
**Type**:
br - bedroom(s);
h - house,cottage,villa, semi,terrace;
u - unit, duplex;
t - townhouse;
dev site - development site;
o res - other residential.
**SellerG**: Real Estate Agent
**Date**: Date sold
**Distance**: Distance from CBD in Kilometres
**Regionname**: General Region (West, North West, North, North east ...etc)
**Propertycount**: Number of properties that exist in the suburb.
**Bedroom2** : Scraped # of Bedrooms (from different source)
**Bathroom**: Number of Bathrooms
**Car**: Number of carspots
**Landsize**: Land Size in Metres
**BuildingArea**: Building Size in Metres
**YearBuilt**: Year the house was built
**CouncilArea**: Governing council for the area
Lattitude: Self explanitory
Longtitude: Self explanitory
Kaggle dataset identifier: melbourne-housing-market
<jupyter_code>import pandas as pd
df = pd.read_csv('melbourne-housing-market/MELBOURNE_HOUSE_PRICES_LESS.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 63023 entries, 0 to 63022
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Suburb 63023 non-null object
1 Address 63023 non-null object
2 Rooms 63023 non-null int64
3 Type 63023 non-null object
4 Price 48433 non-null float64
5 Method 63023 non-null object
6 SellerG 63023 non-null object
7 Date 63023 non-null object
8 Postcode 63023 non-null int64
9 Regionname 63023 non-null object
10 Propertycount 63023 non-null int64
11 Distance 63023 non-null float64
12 CouncilArea 63023 non-null object
dtypes: float64(2), int64(3), object(8)
memory usage: 6.3+ MB
<jupyter_text>Examples:
{
"Suburb": "Abbotsford",
"Address": "49 Lithgow St",
"Rooms": 3,
"Type": "h",
"Price": 1490000,
"Method": "S",
"SellerG": "Jellis",
"Date": "2017-01-04 00:00:00",
"Postcode": 3067,
"Regionname": "Northern Metropolitan",
"Propertycount": 4019,
"Distance": 3.0,
"CouncilArea": "Yarra City Council"
}
{
"Suburb": "Abbotsford",
"Address": "59A Turner St",
"Rooms": 3,
"Type": "h",
"Price": 1220000,
"Method": "S",
"SellerG": "Marshall",
"Date": "2017-01-04 00:00:00",
"Postcode": 3067,
"Regionname": "Northern Metropolitan",
"Propertycount": 4019,
"Distance": 3.0,
"CouncilArea": "Yarra City Council"
}
{
"Suburb": "Abbotsford",
"Address": "119B Yarra St",
"Rooms": 3,
"Type": "h",
"Price": 1420000,
"Method": "S",
"SellerG": "Nelson",
"Date": "2017-01-04 00:00:00",
"Postcode": 3067,
"Regionname": "Northern Metropolitan",
"Propertycount": 4019,
"Distance": 3.0,
"CouncilArea": "Yarra City Council"
}
{
"Suburb": "Aberfeldie",
"Address": "68 Vida St",
"Rooms": 3,
"Type": "h",
"Price": 1515000,
"Method": "S",
"SellerG": "Barry",
"Date": "2017-01-04 00:00:00",
"Postcode": 3040,
"Regionname": "Western Metropolitan",
"Propertycount": 1543,
"Distance": 7.5,
"CouncilArea": "Moonee Valley City Council"
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import sklearn
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv(
"/kaggle/input/melbourne-housing-market/MELBOURNE_HOUSE_PRICES_LESS.csv"
)
print(data.columns)
data.head()
data.drop(
columns={
"Suburb",
"Address",
"Type",
"Method",
"SellerG",
"Date",
"Postcode",
"Regionname",
"Propertycount",
"Distance",
"CouncilArea",
},
inplace=True,
)
data.head()
# ## scale()
# Standardize a dataset along any axis.
# Center to the mean and component wise scale to unit variance.
# Assume **D** to be the data points.
# The scale function does this math :
# # $\frac{D_{i} - \bar{D} }{\sigma{(D)}}$
# $Where,$
# $\bar{D} : Mean $
# $\sigma{(D)} : Standard Deviation $
from sklearn.preprocessing import scale
print(data["Price"][:5])
print("\n Scaled Data:")
scale(data["Price"][:5])
X = data["Price"][:5]
(X - np.mean(X)) / (np.std(X))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/571/69571578.ipynb
|
melbourne-housing-market
|
anthonypino
|
[{"Id": 69571578, "ScriptId": 18903726, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6409095, "CreationDate": "08/01/2021 16:58:48", "VersionNumber": 2.0, "Title": "Understanding Feature Scaling with scikit-learn", "EvaluationDate": "08/01/2021", "IsChange": true, "TotalLines": 41.0, "LinesInsertedFromPrevious": 14.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 27.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 92919281, "KernelVersionId": 69571578, "SourceDatasetVersionId": 126865}]
|
[{"Id": 126865, "DatasetId": 423, "DatasourceVersionId": 136629, "CreatorUserId": 649776, "LicenseName": "CC BY-NC-SA 4.0", "CreationDate": "10/14/2018 23:33:37", "VersionNumber": 27.0, "Title": "Melbourne Housing Market", "Slug": "melbourne-housing-market", "Subtitle": "Melbourne housing clearance data from Jan 2016", "Description": "##Update 06/08/2018 - Well it finally happened, Melbourne housing has cooled off. So here's your challenge; 1) when did it exactly happen? , 2) Could you see it slowing down? What were the variables that showed the slowing down (was it overall price, amount sold vs unsold, change in more rentals sold and less housing, changes in which CouncilArea or Region, more houses sold in distances further away from Melbourne CBD and less closer)? 3) Could you have predicted it (I'm not sure how you would do this, but I'm sure you magicians have a way that would make me think we should burn you for being a witch) 4) Should I hold off even longer in buying a two bedroom apartment in Northcote? <-- This is the real reason for me in publishing this dataset :)\n\n#### Update 22/05/2018 - Will continue with a smaller subset of the data (not as many columns). The web scraping was taking some time and also may potentially cause problems. Will continue to post the data.\n\n\n#### Update 28/11/2017 - Last few weeks clearance levels starting to decrease (I may just be seeing a pattern I want to see.. maybe I'm just evil). Anyway, can any of you magicians make any sense of it?\n\n\nMelbourne is currently experiencing a housing bubble (some experts say it may burst soon). Maybe someone can find a trend or give a prediction? Which suburbs are the best to buy in? Which ones are value for money? Where's the expensive side of town? And more importantly where should I buy a 2 bedroom unit?\n\n## Content & Acknowledgements\n\nThis data was scraped from publicly available results posted every week from Domain.com.au, I've cleaned it as best I can, now it's up to you to make data analysis magic. The dataset includes Address, Type of Real estate, Suburb, Method of Selling, Rooms, Price, Real Estate Agent, Date of Sale and distance from C.B.D.\n\n....Now with extra data including including property size, land size and council area, you may need to change your code!\n\n## Some Key Details\n\n**Suburb**: Suburb\n\n**Address**: Address\n\n**Rooms**: Number of rooms\n\n**Price**: Price in Australian dollars\n\n**Method**: \n S - property sold; \n SP - property sold prior; \n PI - property passed in; \n PN - sold prior not disclosed; \n SN - sold not disclosed; \n NB - no bid; \n VB - vendor bid; \n W - withdrawn prior to auction; \n SA - sold after auction; \n SS - sold after auction price not disclosed. \n N/A - price or highest bid not available. \n\n**Type**:\n br - bedroom(s); \n h - house,cottage,villa, semi,terrace; \n u - unit, duplex;\n t - townhouse; \n dev site - development site; \n o res - other residential.\n\n**SellerG**: Real Estate Agent\n\n**Date**: Date sold\n\n**Distance**: Distance from CBD in Kilometres\n\n**Regionname**: General Region (West, North West, North, North east ...etc) \n\n**Propertycount**: Number of properties that exist in the suburb.\n\n**Bedroom2** : Scraped # of Bedrooms (from different source)\n\n**Bathroom**: Number of Bathrooms\t\n\n**Car**: Number of carspots\t\n\n**Landsize**: Land Size in Metres\n\n**BuildingArea**: Building Size\tin Metres\n\n**YearBuilt**: Year the house was built\t\n\n**CouncilArea**: Governing council for the area\t\n\nLattitude: Self explanitory\t\n\nLongtitude: Self explanitory", "VersionNotes": "Latest data 15/10/2018", "TotalCompressedBytes": 12314654.0, "TotalUncompressedBytes": 2366402.0}]
|
[{"Id": 423, "CreatorUserId": 649776, "OwnerUserId": 649776.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 126865.0, "CurrentDatasourceVersionId": 136629.0, "ForumId": 2026, "Type": 2, "CreationDate": "11/23/2016 02:12:52", "LastActivityDate": "02/06/2018", "TotalViews": 303523, "TotalDownloads": 49493, "TotalVotes": 680, "TotalKernels": 304}]
|
[{"Id": 649776, "UserName": "anthonypino", "DisplayName": "Tony Pino", "RegisterDate": "06/27/2016", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import sklearn
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv(
"/kaggle/input/melbourne-housing-market/MELBOURNE_HOUSE_PRICES_LESS.csv"
)
print(data.columns)
data.head()
data.drop(
columns={
"Suburb",
"Address",
"Type",
"Method",
"SellerG",
"Date",
"Postcode",
"Regionname",
"Propertycount",
"Distance",
"CouncilArea",
},
inplace=True,
)
data.head()
# ## scale()
# Standardize a dataset along any axis.
# Center to the mean and component wise scale to unit variance.
# Assume **D** to be the data points.
# The scale function does this math :
# # $\frac{D_{i} - \bar{D} }{\sigma{(D)}}$
# $Where,$
# $\bar{D} : Mean $
# $\sigma{(D)} : Standard Deviation $
from sklearn.preprocessing import scale
print(data["Price"][:5])
print("\n Scaled Data:")
scale(data["Price"][:5])
X = data["Price"][:5]
(X - np.mean(X)) / (np.std(X))
|
[{"melbourne-housing-market/MELBOURNE_HOUSE_PRICES_LESS.csv": {"column_names": "[\"Suburb\", \"Address\", \"Rooms\", \"Type\", \"Price\", \"Method\", \"SellerG\", \"Date\", \"Postcode\", \"Regionname\", \"Propertycount\", \"Distance\", \"CouncilArea\"]", "column_data_types": "{\"Suburb\": \"object\", \"Address\": \"object\", \"Rooms\": \"int64\", \"Type\": \"object\", \"Price\": \"float64\", \"Method\": \"object\", \"SellerG\": \"object\", \"Date\": \"object\", \"Postcode\": \"int64\", \"Regionname\": \"object\", \"Propertycount\": \"int64\", \"Distance\": \"float64\", \"CouncilArea\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 63023 entries, 0 to 63022\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Suburb 63023 non-null object \n 1 Address 63023 non-null object \n 2 Rooms 63023 non-null int64 \n 3 Type 63023 non-null object \n 4 Price 48433 non-null float64\n 5 Method 63023 non-null object \n 6 SellerG 63023 non-null object \n 7 Date 63023 non-null object \n 8 Postcode 63023 non-null int64 \n 9 Regionname 63023 non-null object \n 10 Propertycount 63023 non-null int64 \n 11 Distance 63023 non-null float64\n 12 CouncilArea 63023 non-null object \ndtypes: float64(2), int64(3), object(8)\nmemory usage: 6.3+ MB\n", "summary": "{\"Rooms\": {\"count\": 63023.0, \"mean\": 3.110594544848706, \"std\": 0.9575513092737409, \"min\": 1.0, \"25%\": 3.0, \"50%\": 3.0, \"75%\": 4.0, \"max\": 31.0}, \"Price\": {\"count\": 48433.0, \"mean\": 997898.2414882415, \"std\": 593498.9190372769, \"min\": 85000.0, \"25%\": 620000.0, \"50%\": 830000.0, \"75%\": 1220000.0, \"max\": 11200000.0}, \"Postcode\": {\"count\": 63023.0, \"mean\": 3125.6738968313157, \"std\": 125.62687746089352, \"min\": 3000.0, \"25%\": 3056.0, \"50%\": 3107.0, \"75%\": 3163.0, \"max\": 3980.0}, \"Propertycount\": {\"count\": 63023.0, \"mean\": 7617.728130999793, \"std\": 4424.423167331061, \"min\": 39.0, \"25%\": 4380.0, \"50%\": 6795.0, \"75%\": 10412.0, \"max\": 21650.0}, \"Distance\": {\"count\": 63023.0, \"mean\": 12.684829348015805, \"std\": 7.592015369125735, \"min\": 0.0, \"25%\": 7.0, \"50%\": 11.4, \"75%\": 16.7, \"max\": 64.1}}", "examples": "{\"Suburb\":{\"0\":\"Abbotsford\",\"1\":\"Abbotsford\",\"2\":\"Abbotsford\",\"3\":\"Aberfeldie\"},\"Address\":{\"0\":\"49 Lithgow St\",\"1\":\"59A Turner St\",\"2\":\"119B Yarra St\",\"3\":\"68 Vida St\"},\"Rooms\":{\"0\":3,\"1\":3,\"2\":3,\"3\":3},\"Type\":{\"0\":\"h\",\"1\":\"h\",\"2\":\"h\",\"3\":\"h\"},\"Price\":{\"0\":1490000.0,\"1\":1220000.0,\"2\":1420000.0,\"3\":1515000.0},\"Method\":{\"0\":\"S\",\"1\":\"S\",\"2\":\"S\",\"3\":\"S\"},\"SellerG\":{\"0\":\"Jellis\",\"1\":\"Marshall\",\"2\":\"Nelson\",\"3\":\"Barry\"},\"Date\":{\"0\":\"1\\/04\\/2017\",\"1\":\"1\\/04\\/2017\",\"2\":\"1\\/04\\/2017\",\"3\":\"1\\/04\\/2017\"},\"Postcode\":{\"0\":3067,\"1\":3067,\"2\":3067,\"3\":3040},\"Regionname\":{\"0\":\"Northern Metropolitan\",\"1\":\"Northern Metropolitan\",\"2\":\"Northern Metropolitan\",\"3\":\"Western Metropolitan\"},\"Propertycount\":{\"0\":4019,\"1\":4019,\"2\":4019,\"3\":1543},\"Distance\":{\"0\":3.0,\"1\":3.0,\"2\":3.0,\"3\":7.5},\"CouncilArea\":{\"0\":\"Yarra City Council\",\"1\":\"Yarra City Council\",\"2\":\"Yarra City Council\",\"3\":\"Moonee Valley City Council\"}}"}}]
| true | 1 |
<start_data_description><data_path>melbourne-housing-market/MELBOURNE_HOUSE_PRICES_LESS.csv:
<column_names>
['Suburb', 'Address', 'Rooms', 'Type', 'Price', 'Method', 'SellerG', 'Date', 'Postcode', 'Regionname', 'Propertycount', 'Distance', 'CouncilArea']
<column_types>
{'Suburb': 'object', 'Address': 'object', 'Rooms': 'int64', 'Type': 'object', 'Price': 'float64', 'Method': 'object', 'SellerG': 'object', 'Date': 'object', 'Postcode': 'int64', 'Regionname': 'object', 'Propertycount': 'int64', 'Distance': 'float64', 'CouncilArea': 'object'}
<dataframe_Summary>
{'Rooms': {'count': 63023.0, 'mean': 3.110594544848706, 'std': 0.9575513092737409, 'min': 1.0, '25%': 3.0, '50%': 3.0, '75%': 4.0, 'max': 31.0}, 'Price': {'count': 48433.0, 'mean': 997898.2414882415, 'std': 593498.9190372769, 'min': 85000.0, '25%': 620000.0, '50%': 830000.0, '75%': 1220000.0, 'max': 11200000.0}, 'Postcode': {'count': 63023.0, 'mean': 3125.6738968313157, 'std': 125.62687746089352, 'min': 3000.0, '25%': 3056.0, '50%': 3107.0, '75%': 3163.0, 'max': 3980.0}, 'Propertycount': {'count': 63023.0, 'mean': 7617.728130999793, 'std': 4424.423167331061, 'min': 39.0, '25%': 4380.0, '50%': 6795.0, '75%': 10412.0, 'max': 21650.0}, 'Distance': {'count': 63023.0, 'mean': 12.684829348015805, 'std': 7.592015369125735, 'min': 0.0, '25%': 7.0, '50%': 11.4, '75%': 16.7, 'max': 64.1}}
<dataframe_info>
RangeIndex: 63023 entries, 0 to 63022
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Suburb 63023 non-null object
1 Address 63023 non-null object
2 Rooms 63023 non-null int64
3 Type 63023 non-null object
4 Price 48433 non-null float64
5 Method 63023 non-null object
6 SellerG 63023 non-null object
7 Date 63023 non-null object
8 Postcode 63023 non-null int64
9 Regionname 63023 non-null object
10 Propertycount 63023 non-null int64
11 Distance 63023 non-null float64
12 CouncilArea 63023 non-null object
dtypes: float64(2), int64(3), object(8)
memory usage: 6.3+ MB
<some_examples>
{'Suburb': {'0': 'Abbotsford', '1': 'Abbotsford', '2': 'Abbotsford', '3': 'Aberfeldie'}, 'Address': {'0': '49 Lithgow St', '1': '59A Turner St', '2': '119B Yarra St', '3': '68 Vida St'}, 'Rooms': {'0': 3, '1': 3, '2': 3, '3': 3}, 'Type': {'0': 'h', '1': 'h', '2': 'h', '3': 'h'}, 'Price': {'0': 1490000.0, '1': 1220000.0, '2': 1420000.0, '3': 1515000.0}, 'Method': {'0': 'S', '1': 'S', '2': 'S', '3': 'S'}, 'SellerG': {'0': 'Jellis', '1': 'Marshall', '2': 'Nelson', '3': 'Barry'}, 'Date': {'0': '1/04/2017', '1': '1/04/2017', '2': '1/04/2017', '3': '1/04/2017'}, 'Postcode': {'0': 3067, '1': 3067, '2': 3067, '3': 3040}, 'Regionname': {'0': 'Northern Metropolitan', '1': 'Northern Metropolitan', '2': 'Northern Metropolitan', '3': 'Western Metropolitan'}, 'Propertycount': {'0': 4019, '1': 4019, '2': 4019, '3': 1543}, 'Distance': {'0': 3.0, '1': 3.0, '2': 3.0, '3': 7.5}, 'CouncilArea': {'0': 'Yarra City Council', '1': 'Yarra City Council', '2': 'Yarra City Council', '3': 'Moonee Valley City Council'}}
<end_description>
| 370 | 1 | 2,246 | 370 |
69571250
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# import libraries
import gzip
import numpy as np
import pandas as pd
import string
import matplotlib.pyplot as plt
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn import tree
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Ridge
from sklearn.metrics import (
accuracy_score,
classification_report,
confusion_matrix,
plot_confusion_matrix,
)
from sklearn.model_selection import cross_val_score
from sklearn.ensemble import BaggingClassifier
from sklearn.ensemble import AdaBoostRegressor
from sklearn.ensemble import RandomForestRegressor
from xgboost import XGBRegressor
train = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
# define X, y and test
X = train["excerpt"]
y = train["target"]
test_text = test["excerpt"]
# lower the text
X = X.str.lower()
test_text = test_text.str.lower()
def remove_punctuations(text):
for punctuation in string.punctuation:
text = text.replace(punctuation, "")
return text
X = X.apply(remove_punctuations)
test_text = test_text.apply(remove_punctuations)
# apply Porter Stemmer
from nltk.stem import PorterStemmer
ps = PorterStemmer()
def stem_sentences(sentence):
tokens = sentence.split()
stemmed_tokens = [ps.stem(token) for token in tokens]
return " ".join(stemmed_tokens)
X = X.apply(stem_sentences)
test_text = test_text.apply(stem_sentences)
# lemmatize the text
from nltk.stem import WordNetLemmatizer
import nltk
# nltk.download('wordnet')
lemmatizer = WordNetLemmatizer()
def lemmatize_sentences(sentence):
tokens = sentence.split()
stemmed_tokens = [lemmatizer.lemmatize(token) for token in tokens]
return " ".join(stemmed_tokens)
X = X.apply(lemmatize_sentences)
test_text = test_text.apply(lemmatize_sentences)
import nltk
# nltk.download('stopwords')
from nltk.corpus import stopwords
", ".join(stopwords.words("english"))
STOPWORDS = set(stopwords.words("english"))
def remove_stopwords(text):
"""custom function to remove the stopwords"""
return " ".join([word for word in str(text).split() if word not in STOPWORDS])
X = X.apply(lambda text: remove_stopwords(text))
test_text = test_text.apply(lambda text: remove_stopwords(text))
from collections import Counter
cnt = Counter()
for text in X.tolist() + test_text.tolist():
for word in text.split():
cnt[word] += 1
most_common_words = [x[0] for x in cnt.most_common(100)]
def remove_most_common_words(text):
return " ".join(
[word for word in str(text).split() if word not in most_common_words]
)
X = X.apply(lambda text: remove_most_common_words(text))
test_text = test_text.apply(lambda text: remove_most_common_words(text))
X_new = []
for index, value in X.items():
X_new.append(value)
y_new = []
for index, value in y.items():
y_new.append(value)
real_test = []
for index, value in test_text.items():
real_test.append(value)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_new, y_new, test_size=0.20)
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.metrics import accuracy_score
vectorizer = CountVectorizer(max_features=5000, ngram_range=(1, 2))
m = RandomForestRegressor()
m.fit(vectorizer.fit_transform(X_new + real_test)[: len(X_new)], y_new)
# predict
pred_test = m.predict(vectorizer.fit_transform(X_new + real_test)[len(X_new) :])
pred_test_list = [i for i in pred_test]
pred_test_list
submission = pd.DataFrame({"id": test["id"], "target": pred_test_list})
submission.to_csv("/kaggle/working/submission.csv", index=False)
submission.head(7)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/571/69571250.ipynb
| null | null |
[{"Id": 69571250, "ScriptId": 18807639, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7914795, "CreationDate": "08/01/2021 16:54:03", "VersionNumber": 8.0, "Title": "Group 4", "EvaluationDate": "08/01/2021", "IsChange": true, "TotalLines": 199.0, "LinesInsertedFromPrevious": 11.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 188.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# import libraries
import gzip
import numpy as np
import pandas as pd
import string
import matplotlib.pyplot as plt
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn import tree
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Ridge
from sklearn.metrics import (
accuracy_score,
classification_report,
confusion_matrix,
plot_confusion_matrix,
)
from sklearn.model_selection import cross_val_score
from sklearn.ensemble import BaggingClassifier
from sklearn.ensemble import AdaBoostRegressor
from sklearn.ensemble import RandomForestRegressor
from xgboost import XGBRegressor
train = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
# define X, y and test
X = train["excerpt"]
y = train["target"]
test_text = test["excerpt"]
# lower the text
X = X.str.lower()
test_text = test_text.str.lower()
def remove_punctuations(text):
for punctuation in string.punctuation:
text = text.replace(punctuation, "")
return text
X = X.apply(remove_punctuations)
test_text = test_text.apply(remove_punctuations)
# apply Porter Stemmer
from nltk.stem import PorterStemmer
ps = PorterStemmer()
def stem_sentences(sentence):
tokens = sentence.split()
stemmed_tokens = [ps.stem(token) for token in tokens]
return " ".join(stemmed_tokens)
X = X.apply(stem_sentences)
test_text = test_text.apply(stem_sentences)
# lemmatize the text
from nltk.stem import WordNetLemmatizer
import nltk
# nltk.download('wordnet')
lemmatizer = WordNetLemmatizer()
def lemmatize_sentences(sentence):
tokens = sentence.split()
stemmed_tokens = [lemmatizer.lemmatize(token) for token in tokens]
return " ".join(stemmed_tokens)
X = X.apply(lemmatize_sentences)
test_text = test_text.apply(lemmatize_sentences)
import nltk
# nltk.download('stopwords')
from nltk.corpus import stopwords
", ".join(stopwords.words("english"))
STOPWORDS = set(stopwords.words("english"))
def remove_stopwords(text):
"""custom function to remove the stopwords"""
return " ".join([word for word in str(text).split() if word not in STOPWORDS])
X = X.apply(lambda text: remove_stopwords(text))
test_text = test_text.apply(lambda text: remove_stopwords(text))
from collections import Counter
cnt = Counter()
for text in X.tolist() + test_text.tolist():
for word in text.split():
cnt[word] += 1
most_common_words = [x[0] for x in cnt.most_common(100)]
def remove_most_common_words(text):
return " ".join(
[word for word in str(text).split() if word not in most_common_words]
)
X = X.apply(lambda text: remove_most_common_words(text))
test_text = test_text.apply(lambda text: remove_most_common_words(text))
X_new = []
for index, value in X.items():
X_new.append(value)
y_new = []
for index, value in y.items():
y_new.append(value)
real_test = []
for index, value in test_text.items():
real_test.append(value)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_new, y_new, test_size=0.20)
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.metrics import accuracy_score
vectorizer = CountVectorizer(max_features=5000, ngram_range=(1, 2))
m = RandomForestRegressor()
m.fit(vectorizer.fit_transform(X_new + real_test)[: len(X_new)], y_new)
# predict
pred_test = m.predict(vectorizer.fit_transform(X_new + real_test)[len(X_new) :])
pred_test_list = [i for i in pred_test]
pred_test_list
submission = pd.DataFrame({"id": test["id"], "target": pred_test_list})
submission.to_csv("/kaggle/working/submission.csv", index=False)
submission.head(7)
| false | 0 | 1,374 | 0 | 1,374 | 1,374 |
||
69571806
|
<jupyter_start><jupyter_text>Iris Species
The Iris dataset was used in R.A. Fisher's classic 1936 paper, [The Use of Multiple Measurements in Taxonomic Problems](http://rcs.chemometrics.ru/Tutorials/classification/Fisher.pdf), and can also be found on the [UCI Machine Learning Repository][1].
It includes three iris species with 50 samples each as well as some properties about each flower. One flower species is linearly separable from the other two, but the other two are not linearly separable from each other.
The columns in this dataset are:
- Id
- SepalLengthCm
- SepalWidthCm
- PetalLengthCm
- PetalWidthCm
- Species
[](https://www.kaggle.com/benhamner/d/uciml/iris/sepal-width-vs-length)
[1]: http://archive.ics.uci.edu/ml/
Kaggle dataset identifier: iris
<jupyter_script># The data set includes three iris species with 50 samples each as well as some properties about each flower.
# In this notebook I will explain How to effectively use logistic regression to solve classification problems. I will try to explain each and every step in a concise and clear manner. we will go through the following, step by step:
# - [Reading and understanding the data](#read)
# - [Data visualization and explanatory data analysis](#visual)
# - [Feature engineering: Data prep for the model](#prepare)
# - [Model building](#build)
# - [Model evaluation](#eval1)
# - [Model optimization: hyper parameter tuning](#hyper)
# - [Model re-evaluation](#eval2)
# ### **Reading and understanding the data**
# import the necessary liberaries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # visuals
import seaborn as sns # advanced visuals
import warnings # ignore warnings
warnings.filterwarnings("ignore")
# read the data
df = pd.read_csv("../input/iris/Iris.csv")
# display the first 5 rows
df.head()
# main characteristics of the dataframe
df.info()
# The dataframe has 150 non-null values. It has 6 variables, all of them are in the right data type.
# the first variable "Id" seems to be redundant and unnecessary for the our analysis, we can drop it and keep the rest of variables.
# drop Id, axis = 1: tells python to drop the entire column
# Do not run this cell more than once
df = df.drop("Id", axis=1)
df.head()
# summary statistics
df.describe()
# From the summary statistics we can notice that Sepal leafs are wider and longer than Petal lefs, this can be clearly demonstrated in the following image:
# 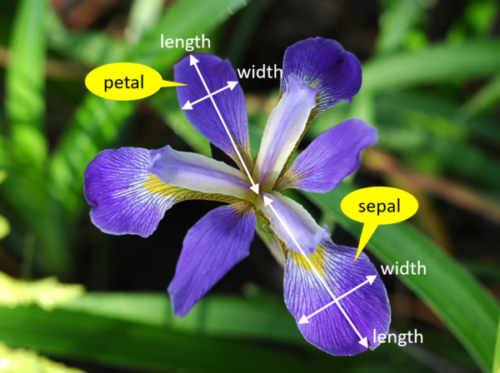
# How many species in our dataframe?
# is the data balanced?
df["Species"].value_counts()
# The data is clean and balanced with exactly the same number of flowers per species: 50 flowers. but why do we care about the balance between number of observations per class?
# Imbalanced classifications pose a challenge for predictive modeling as most of the machine learning algorithms used for classification were designed around the assumption of an equal number of examples for each class.
# For example, an imbalanced multiclass classification problem may have 80 percent examples in the first class, 18 percent in the second class, and 2 percent in a third class.
# The minority class is harder to predict because there are few examples of this class, by definition. This means it is more challenging for a model to learn the characteristics of examples from this class, and to differentiate examples from this class from the majority class (or classes).
# This is a problem because typically, the minority class is more important and therefore the problem is more sensitive to classification errors for the minority class than the majority class.
# More detailed explanation can be found [here](https://machinelearningmastery.com/what-is-imbalanced-classification/).
# ### **Data visualization and explanatory data analysis**
# This section focuses on how to produce and analyze charts that meets the best practices in both academia and industry. we will try to meet the following criteria in each graph:
# 1. **Chose the right graph that suits the variable type:** to display the distribution of categorical variables we might opt for count or bar plot. As for continuous variables we might go with a histogram. If we wan to study the distribution of a continuous variable per each calss of other categorical variable we can use a box plots or a kde plot with hue parameter... etc.
# 2. **Maximize Dagt-Ink Ration:** it equals to the ink used to display the data devided by the total ink used in the graph. Try not to use so many colors without a good reason for that. Aviod using backround colors, or borders or any other unnecessary decorations.
# 3. **Use clear well written Titles, labels, and tick marks.**
fig, axes = plt.subplots(2, 2, figsize=(10, 5), dpi=100)
fig.suptitle(
"Distribution of (sepal length, sepal width, petal length, petal width) per Species"
)
# Distribution of sepal length per Species
sns.kdeplot(
ax=axes[0, 0], data=df, x="SepalLengthCm", hue="Species", alpha=0.5, shade=True
)
axes[0, 0].set_xlabel("Sepal Length CM")
axes[0, 0].get_legend().remove()
# Distribution of sepal width per Species
sns.kdeplot(
ax=axes[0, 1], data=df, x="SepalWidthCm", hue="Species", alpha=0.5, shade=True
)
axes[0, 1].set_xlabel("Sepal width CM")
axes[0, 1].get_legend().remove()
# Distribution of petal length per Species
sns.kdeplot(
ax=axes[1, 0], data=df, x="PetalLengthCm", hue="Species", alpha=0.5, shade=True
)
axes[1, 0].set_xlabel("Petal Length CM")
axes[1, 0].get_legend().remove()
# Distribution of petal width per Species
sns.kdeplot(
ax=axes[1, 1], data=df, x="PetalWidthCm", hue="Species", alpha=0.5, shade=True
)
axes[1, 1].set_xlabel("Petal Width CM")
plt.tight_layout()
# **Main conclusions from the graph:**
# 1. Setosa is easily separable from the other species, this means that the model will be able to classify it accurately.
# 2. Petal length and width is expected to be better predictors of Species than Sepal lenght and width.
# Both conclusions can be demonstrated in the following picture where Setosa is clearly different from other sepcies especially when it comes to its petal leefs, it has a very small sepal width and length comapred to other species.
# 
# Scatter plot od petal length vs petal width
plt.figure(figsize=(7, 3), dpi=100)
sns.scatterplot(data=df, x="PetalLengthCm", y="PetalWidthCm", hue="Species")
plt.title("Species clusters based on Sepal length and width")
plt.xlabel("Petal Length Cm")
plt.ylabel("Petal Width Cm")
plt.legend(bbox_to_anchor=(1.05, 1), loc="upper left")
plt.show()
# Scatter plot od sepal length vs petal width
plt.figure(figsize=(7, 3), dpi=100)
sns.scatterplot(data=df, x="SepalLengthCm", y="SepalWidthCm", hue="Species")
plt.title("Species clusters based on Sepal length and width")
plt.xlabel("Sepal Length Cm")
plt.ylabel("Sepal Width Cm")
plt.legend(bbox_to_anchor=(1.05, 1), loc="upper left")
plt.show()
# box plots
fig, axes = plt.subplots(2, 2, figsize=(10, 5), dpi=100)
# Mean Sepal Length
sns.boxplot(ax=axes[0, 0], data=df, x="Species", y="SepalLengthCm")
axes[0, 0].set_xlabel(None)
axes[0, 0].set_ylabel(None)
axes[0, 0].set_title("Mean Sepal Length")
# Mean Sepal Width
sns.boxplot(ax=axes[0, 1], data=df, x="Species", y="SepalWidthCm")
axes[0, 1].set_xlabel(None)
axes[0, 1].set_ylabel(None)
axes[0, 1].set_title("Mean Sepal Width")
# Mean Petal Length
sns.boxplot(ax=axes[1, 0], data=df, x="Species", y="PetalLengthCm")
axes[1, 0].set_xlabel(None)
axes[1, 0].set_ylabel(None)
axes[1, 0].set_title("Mean Petal Length")
# Mean Petal Width
sns.boxplot(ax=axes[1, 1], data=df, x="Species", y="PetalWidthCm")
axes[1, 1].set_xlabel(None)
axes[1, 1].set_ylabel(None)
axes[1, 1].set_title("Mean Petal Width")
plt.tight_layout()
plt.subplots_adjust(hspace=0.5)
# Scatter and box plots confirmed the aforementioned conclusion, setosa is easily separable based on petal length and width.
# Correlation map
plt.figure(figsize=(8, 4), dpi=100)
sns.heatmap(df.corr(), annot=True, cmap="viridis", vmin=-1, vmax=1)
plt.title("Correlation map between variables")
# plt.xticks(rotation = 90)
plt.show()
# [Correlation is](https://www.jmp.com/en_in/statistics-knowledge-portal/what-is-correlation.html) a statistical measure that expresses the extent to which two variables are linearly related (meaning they change together at a constant rate). It's a common tool for describing simple relationships without making a statement about cause and effect.
# Correlation coefficient ranges between -1 (perfect negative correlation) and 1 (perfect positive correlation). As you can notice, there is a strong positive correlation between petal width and length on one hand and sepal length on the other hand.
# ### **Feature engineering: Data prep for the model**
# In this section we will make sure that the data is well prepared for training the model. We will:
# 1. Seprate the dependent variable from the independent ones.
# 2. Perform a train test split
# 3. Scale the data (feature scaling).
# 1. Seprate the dependent variable from the independent ones.
X = df.drop("Species", axis=1)
y = df["Species"]
# **Why train test split ?** we need to split the data into two parts:
# 1. Training part, we will use it to train the model.
# 2. Test part: this is unseen data (the model has never seen it before), we will use it the test the real performance of the model.
# **Why we need to test on unseen data?** why we do not simply train the model on the whole data and then reuse some of it for evaluation? because this will be like giving the student the answers before entring the exam, the model will be very familiar with the evaluation data because he has seen them before and he will get a full mark. In order for the test to be real, the model has to be evaluated on unseen data.
# 2. Perform a train test split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=101
)
# **Why feature scaling?**
# Real Life Datasets have many features with a wide range of values like for example let’s consider the house price prediction dataset. It will have many features like no. of. bedrooms, square feet area of the house, etc
# As you can guess, the no. of bedrooms will vary between 1 and 5, but the square feet area will range from 500-2000. This is a huge difference in the range of both features.
# Many machine learning algorithms that are using Euclidean distance as a metric to calculate the similarities will fail to give a reasonable recognition to the smaller feature, in this case, the number of bedrooms, which in the real case can turn out to be an actually important metric.
# 
# To aviod this problem we need to scale the features so that they all have the same scale, i.e the same range of values. We can normalize all features so that have values between (-1, 1) or standardize them to have values between (0, 1).
# The important thing to note here is that feature scaling does not affect the relative importance of features, scaled features will still have the same orginal information and importance relative to each other, this can be clearly demonstated from the image below: despite feature scaling they are still strawberry and apple, they did not lose their meaning.
# 
# 3. Feature scaling
from sklearn.preprocessing import StandardScaler # import the scaler
scaler = StandardScaler() # initiate it
Scaled_X_train = scaler.fit_transform(
X_train
) # fit the parameters and use it to trannsform the traning data
Scaled_X_test = scaler.transform(X_test) # transform the test data
# Have you noticed that we used .fit_transform() with the traning data and only used .transform() with the test data? we did it to aviod data leakage. Read more about it [from here](https://machinelearningmastery.com/data-preparation-without-data-leakage/)
# ### **Model building**
# We will use logestic regression, but the same methodology can be applied to any other classifier
# Logestic Regression
from sklearn.linear_model import LogisticRegression # import the classifier
log_model = LogisticRegression() # initiate it
log_model.fit(Scaled_X_train, y_train) # fit the model to the training data
# ### **Model evaluation**
# First we will make predictions using the model on the test data, and then evaluate its performance using the following metrics:
# 1. **Confusion matrix:** A summary of prediction results on a classification problem. The number of correct and incorrect predictions are summarized with count values and broken down by each class. Here is an example of a confusion matrix:
# 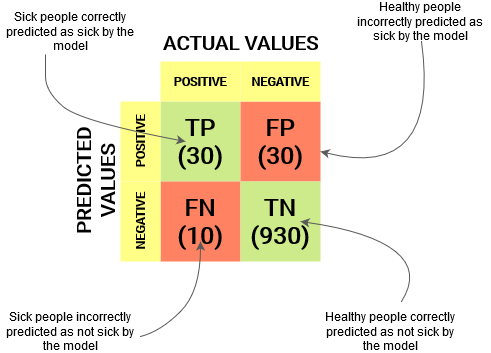
# 2. **Accuracy score:** the fraction of predictions our model got right (number of correct predictions devided by total number of predictions).
# 3. **Classification report:** used to measure the quality of predictions from a classification algorithm. How many predictions are True and how many are False. The report shows the main classification metrics precision, recall and f1-score on a per-class basis. **Precision:** What percent of your predictions were correct? - **Recall:** What percent of the positive cases did you catch? - **F1 score:** What percent of positive predictions were correct?.
# For more info, click [here](https://scikit-learn.org/stable/modules/model_evaluation.html#model-evaluation) and [here](https://muthu.co/understanding-the-classification-report-in-sklearn/).
# creating predictions
y_pred = log_model.predict(Scaled_X_test)
# import evaluation metrics
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
plot_confusion_matrix,
classification_report,
)
# create the confusion matrix
confusion_matrix(y_test, y_pred)
# plot the confusion matrix
fig, ax = plt.subplots(dpi=120)
plot_confusion_matrix(log_model, Scaled_X_test, y_test, ax=ax)
# measure the accuracy of our model
acc_score = accuracy_score(y_test, y_pred)
round(acc_score, 2)
# generate the classification report
print(
classification_report(y_test, y_pred)
) # Hint: try it without using the print() method
# As we expected before, the model did a perfect job predicting Setosa. It only misclassified one observation as versicolor, where in fact it is virginica. However, the model performance is near perfect and we could not have done better than that.
# ### **Model optimization: hyper parameter tuning**
# Hyperparameter tuning [is](https://neptune.ai/blog/hyperparameter-tuning-in-python-a-complete-guide-2020) the process of determining the right combination of parameters that allows us to maximize model performance. We will try different values for each parameter and choose the ones that give us the best predictions.
# import GridSearchCV
from sklearn.model_selection import GridSearchCV
# set the range of paprameters
penalty = ["l1", "l2", "elasticnet"]
C = np.logspace(0, 20, 50)
solver = ["newton-cg", "lbfgs", "liblinear", "sag", "saga"]
multi_class = ["ovr", "multinomial"]
l1_ratio = np.linspace(0, 1, 20)
# build the parameter grid
param_grid = {
"penalty": penalty,
"C": C,
"solver": solver,
"multi_class": multi_class,
"l1_ratio": l1_ratio,
}
# initiate and fit the Grid Search Model
grid_model = GridSearchCV(log_model, param_grid=param_grid)
grid_model.fit(Scaled_X_train, y_train)
# best parameters
grid_model.best_params_
# ### **Model re-evaluation**
# We will evaluate the optimized version of our model and see if it does better than the base model
# creating predictions
y_pred = grid_model.predict(Scaled_X_test)
# plot the confusion matrix
fig, ax = plt.subplots(dpi=120)
plot_confusion_matrix(grid_model, Scaled_X_test, y_test, ax=ax)
# measure the accuracy of our model
acc_score = accuracy_score(y_test, y_pred)
round(acc_score, 2)
# generate the classification report
print(
classification_report(y_test, y_pred)
) # Hint: try it without using the print() method
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/571/69571806.ipynb
|
iris
| null |
[{"Id": 69571806, "ScriptId": 18956937, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6541719, "CreationDate": "08/01/2021 17:02:01", "VersionNumber": 9.0, "Title": "IRIS: Complete Classification Tutorial", "EvaluationDate": "08/01/2021", "IsChange": true, "TotalLines": 300.0, "LinesInsertedFromPrevious": 82.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 218.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 21}]
|
[{"Id": 92919897, "KernelVersionId": 69571806, "SourceDatasetVersionId": 420}]
|
[{"Id": 420, "DatasetId": 19, "DatasourceVersionId": 420, "CreatorUserId": 1, "LicenseName": "CC0: Public Domain", "CreationDate": "09/27/2016 07:38:05", "VersionNumber": 2.0, "Title": "Iris Species", "Slug": "iris", "Subtitle": "Classify iris plants into three species in this classic dataset", "Description": "The Iris dataset was used in R.A. Fisher's classic 1936 paper, [The Use of Multiple Measurements in Taxonomic Problems](http://rcs.chemometrics.ru/Tutorials/classification/Fisher.pdf), and can also be found on the [UCI Machine Learning Repository][1].\n\nIt includes three iris species with 50 samples each as well as some properties about each flower. One flower species is linearly separable from the other two, but the other two are not linearly separable from each other.\n\nThe columns in this dataset are:\n\n - Id\n - SepalLengthCm\n - SepalWidthCm\n - PetalLengthCm\n - PetalWidthCm\n - Species\n\n[](https://www.kaggle.com/benhamner/d/uciml/iris/sepal-width-vs-length)\n\n\n [1]: http://archive.ics.uci.edu/ml/", "VersionNotes": "Republishing files so they're formally in our system", "TotalCompressedBytes": 15347.0, "TotalUncompressedBytes": 15347.0}]
|
[{"Id": 19, "CreatorUserId": 1, "OwnerUserId": NaN, "OwnerOrganizationId": 7.0, "CurrentDatasetVersionId": 420.0, "CurrentDatasourceVersionId": 420.0, "ForumId": 997, "Type": 2, "CreationDate": "01/12/2016 00:33:31", "LastActivityDate": "02/06/2018", "TotalViews": 1637863, "TotalDownloads": 423540, "TotalVotes": 3416, "TotalKernels": 6420}]
| null |
# The data set includes three iris species with 50 samples each as well as some properties about each flower.
# In this notebook I will explain How to effectively use logistic regression to solve classification problems. I will try to explain each and every step in a concise and clear manner. we will go through the following, step by step:
# - [Reading and understanding the data](#read)
# - [Data visualization and explanatory data analysis](#visual)
# - [Feature engineering: Data prep for the model](#prepare)
# - [Model building](#build)
# - [Model evaluation](#eval1)
# - [Model optimization: hyper parameter tuning](#hyper)
# - [Model re-evaluation](#eval2)
# ### **Reading and understanding the data**
# import the necessary liberaries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # visuals
import seaborn as sns # advanced visuals
import warnings # ignore warnings
warnings.filterwarnings("ignore")
# read the data
df = pd.read_csv("../input/iris/Iris.csv")
# display the first 5 rows
df.head()
# main characteristics of the dataframe
df.info()
# The dataframe has 150 non-null values. It has 6 variables, all of them are in the right data type.
# the first variable "Id" seems to be redundant and unnecessary for the our analysis, we can drop it and keep the rest of variables.
# drop Id, axis = 1: tells python to drop the entire column
# Do not run this cell more than once
df = df.drop("Id", axis=1)
df.head()
# summary statistics
df.describe()
# From the summary statistics we can notice that Sepal leafs are wider and longer than Petal lefs, this can be clearly demonstrated in the following image:
# 
# How many species in our dataframe?
# is the data balanced?
df["Species"].value_counts()
# The data is clean and balanced with exactly the same number of flowers per species: 50 flowers. but why do we care about the balance between number of observations per class?
# Imbalanced classifications pose a challenge for predictive modeling as most of the machine learning algorithms used for classification were designed around the assumption of an equal number of examples for each class.
# For example, an imbalanced multiclass classification problem may have 80 percent examples in the first class, 18 percent in the second class, and 2 percent in a third class.
# The minority class is harder to predict because there are few examples of this class, by definition. This means it is more challenging for a model to learn the characteristics of examples from this class, and to differentiate examples from this class from the majority class (or classes).
# This is a problem because typically, the minority class is more important and therefore the problem is more sensitive to classification errors for the minority class than the majority class.
# More detailed explanation can be found [here](https://machinelearningmastery.com/what-is-imbalanced-classification/).
# ### **Data visualization and explanatory data analysis**
# This section focuses on how to produce and analyze charts that meets the best practices in both academia and industry. we will try to meet the following criteria in each graph:
# 1. **Chose the right graph that suits the variable type:** to display the distribution of categorical variables we might opt for count or bar plot. As for continuous variables we might go with a histogram. If we wan to study the distribution of a continuous variable per each calss of other categorical variable we can use a box plots or a kde plot with hue parameter... etc.
# 2. **Maximize Dagt-Ink Ration:** it equals to the ink used to display the data devided by the total ink used in the graph. Try not to use so many colors without a good reason for that. Aviod using backround colors, or borders or any other unnecessary decorations.
# 3. **Use clear well written Titles, labels, and tick marks.**
fig, axes = plt.subplots(2, 2, figsize=(10, 5), dpi=100)
fig.suptitle(
"Distribution of (sepal length, sepal width, petal length, petal width) per Species"
)
# Distribution of sepal length per Species
sns.kdeplot(
ax=axes[0, 0], data=df, x="SepalLengthCm", hue="Species", alpha=0.5, shade=True
)
axes[0, 0].set_xlabel("Sepal Length CM")
axes[0, 0].get_legend().remove()
# Distribution of sepal width per Species
sns.kdeplot(
ax=axes[0, 1], data=df, x="SepalWidthCm", hue="Species", alpha=0.5, shade=True
)
axes[0, 1].set_xlabel("Sepal width CM")
axes[0, 1].get_legend().remove()
# Distribution of petal length per Species
sns.kdeplot(
ax=axes[1, 0], data=df, x="PetalLengthCm", hue="Species", alpha=0.5, shade=True
)
axes[1, 0].set_xlabel("Petal Length CM")
axes[1, 0].get_legend().remove()
# Distribution of petal width per Species
sns.kdeplot(
ax=axes[1, 1], data=df, x="PetalWidthCm", hue="Species", alpha=0.5, shade=True
)
axes[1, 1].set_xlabel("Petal Width CM")
plt.tight_layout()
# **Main conclusions from the graph:**
# 1. Setosa is easily separable from the other species, this means that the model will be able to classify it accurately.
# 2. Petal length and width is expected to be better predictors of Species than Sepal lenght and width.
# Both conclusions can be demonstrated in the following picture where Setosa is clearly different from other sepcies especially when it comes to its petal leefs, it has a very small sepal width and length comapred to other species.
# 
# Scatter plot od petal length vs petal width
plt.figure(figsize=(7, 3), dpi=100)
sns.scatterplot(data=df, x="PetalLengthCm", y="PetalWidthCm", hue="Species")
plt.title("Species clusters based on Sepal length and width")
plt.xlabel("Petal Length Cm")
plt.ylabel("Petal Width Cm")
plt.legend(bbox_to_anchor=(1.05, 1), loc="upper left")
plt.show()
# Scatter plot od sepal length vs petal width
plt.figure(figsize=(7, 3), dpi=100)
sns.scatterplot(data=df, x="SepalLengthCm", y="SepalWidthCm", hue="Species")
plt.title("Species clusters based on Sepal length and width")
plt.xlabel("Sepal Length Cm")
plt.ylabel("Sepal Width Cm")
plt.legend(bbox_to_anchor=(1.05, 1), loc="upper left")
plt.show()
# box plots
fig, axes = plt.subplots(2, 2, figsize=(10, 5), dpi=100)
# Mean Sepal Length
sns.boxplot(ax=axes[0, 0], data=df, x="Species", y="SepalLengthCm")
axes[0, 0].set_xlabel(None)
axes[0, 0].set_ylabel(None)
axes[0, 0].set_title("Mean Sepal Length")
# Mean Sepal Width
sns.boxplot(ax=axes[0, 1], data=df, x="Species", y="SepalWidthCm")
axes[0, 1].set_xlabel(None)
axes[0, 1].set_ylabel(None)
axes[0, 1].set_title("Mean Sepal Width")
# Mean Petal Length
sns.boxplot(ax=axes[1, 0], data=df, x="Species", y="PetalLengthCm")
axes[1, 0].set_xlabel(None)
axes[1, 0].set_ylabel(None)
axes[1, 0].set_title("Mean Petal Length")
# Mean Petal Width
sns.boxplot(ax=axes[1, 1], data=df, x="Species", y="PetalWidthCm")
axes[1, 1].set_xlabel(None)
axes[1, 1].set_ylabel(None)
axes[1, 1].set_title("Mean Petal Width")
plt.tight_layout()
plt.subplots_adjust(hspace=0.5)
# Scatter and box plots confirmed the aforementioned conclusion, setosa is easily separable based on petal length and width.
# Correlation map
plt.figure(figsize=(8, 4), dpi=100)
sns.heatmap(df.corr(), annot=True, cmap="viridis", vmin=-1, vmax=1)
plt.title("Correlation map between variables")
# plt.xticks(rotation = 90)
plt.show()
# [Correlation is](https://www.jmp.com/en_in/statistics-knowledge-portal/what-is-correlation.html) a statistical measure that expresses the extent to which two variables are linearly related (meaning they change together at a constant rate). It's a common tool for describing simple relationships without making a statement about cause and effect.
# Correlation coefficient ranges between -1 (perfect negative correlation) and 1 (perfect positive correlation). As you can notice, there is a strong positive correlation between petal width and length on one hand and sepal length on the other hand.
# ### **Feature engineering: Data prep for the model**
# In this section we will make sure that the data is well prepared for training the model. We will:
# 1. Seprate the dependent variable from the independent ones.
# 2. Perform a train test split
# 3. Scale the data (feature scaling).
# 1. Seprate the dependent variable from the independent ones.
X = df.drop("Species", axis=1)
y = df["Species"]
# **Why train test split ?** we need to split the data into two parts:
# 1. Training part, we will use it to train the model.
# 2. Test part: this is unseen data (the model has never seen it before), we will use it the test the real performance of the model.
# **Why we need to test on unseen data?** why we do not simply train the model on the whole data and then reuse some of it for evaluation? because this will be like giving the student the answers before entring the exam, the model will be very familiar with the evaluation data because he has seen them before and he will get a full mark. In order for the test to be real, the model has to be evaluated on unseen data.
# 2. Perform a train test split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=101
)
# **Why feature scaling?**
# Real Life Datasets have many features with a wide range of values like for example let’s consider the house price prediction dataset. It will have many features like no. of. bedrooms, square feet area of the house, etc
# As you can guess, the no. of bedrooms will vary between 1 and 5, but the square feet area will range from 500-2000. This is a huge difference in the range of both features.
# Many machine learning algorithms that are using Euclidean distance as a metric to calculate the similarities will fail to give a reasonable recognition to the smaller feature, in this case, the number of bedrooms, which in the real case can turn out to be an actually important metric.
# 
# To aviod this problem we need to scale the features so that they all have the same scale, i.e the same range of values. We can normalize all features so that have values between (-1, 1) or standardize them to have values between (0, 1).
# The important thing to note here is that feature scaling does not affect the relative importance of features, scaled features will still have the same orginal information and importance relative to each other, this can be clearly demonstated from the image below: despite feature scaling they are still strawberry and apple, they did not lose their meaning.
# 
# 3. Feature scaling
from sklearn.preprocessing import StandardScaler # import the scaler
scaler = StandardScaler() # initiate it
Scaled_X_train = scaler.fit_transform(
X_train
) # fit the parameters and use it to trannsform the traning data
Scaled_X_test = scaler.transform(X_test) # transform the test data
# Have you noticed that we used .fit_transform() with the traning data and only used .transform() with the test data? we did it to aviod data leakage. Read more about it [from here](https://machinelearningmastery.com/data-preparation-without-data-leakage/)
# ### **Model building**
# We will use logestic regression, but the same methodology can be applied to any other classifier
# Logestic Regression
from sklearn.linear_model import LogisticRegression # import the classifier
log_model = LogisticRegression() # initiate it
log_model.fit(Scaled_X_train, y_train) # fit the model to the training data
# ### **Model evaluation**
# First we will make predictions using the model on the test data, and then evaluate its performance using the following metrics:
# 1. **Confusion matrix:** A summary of prediction results on a classification problem. The number of correct and incorrect predictions are summarized with count values and broken down by each class. Here is an example of a confusion matrix:
# 
# 2. **Accuracy score:** the fraction of predictions our model got right (number of correct predictions devided by total number of predictions).
# 3. **Classification report:** used to measure the quality of predictions from a classification algorithm. How many predictions are True and how many are False. The report shows the main classification metrics precision, recall and f1-score on a per-class basis. **Precision:** What percent of your predictions were correct? - **Recall:** What percent of the positive cases did you catch? - **F1 score:** What percent of positive predictions were correct?.
# For more info, click [here](https://scikit-learn.org/stable/modules/model_evaluation.html#model-evaluation) and [here](https://muthu.co/understanding-the-classification-report-in-sklearn/).
# creating predictions
y_pred = log_model.predict(Scaled_X_test)
# import evaluation metrics
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
plot_confusion_matrix,
classification_report,
)
# create the confusion matrix
confusion_matrix(y_test, y_pred)
# plot the confusion matrix
fig, ax = plt.subplots(dpi=120)
plot_confusion_matrix(log_model, Scaled_X_test, y_test, ax=ax)
# measure the accuracy of our model
acc_score = accuracy_score(y_test, y_pred)
round(acc_score, 2)
# generate the classification report
print(
classification_report(y_test, y_pred)
) # Hint: try it without using the print() method
# As we expected before, the model did a perfect job predicting Setosa. It only misclassified one observation as versicolor, where in fact it is virginica. However, the model performance is near perfect and we could not have done better than that.
# ### **Model optimization: hyper parameter tuning**
# Hyperparameter tuning [is](https://neptune.ai/blog/hyperparameter-tuning-in-python-a-complete-guide-2020) the process of determining the right combination of parameters that allows us to maximize model performance. We will try different values for each parameter and choose the ones that give us the best predictions.
# import GridSearchCV
from sklearn.model_selection import GridSearchCV
# set the range of paprameters
penalty = ["l1", "l2", "elasticnet"]
C = np.logspace(0, 20, 50)
solver = ["newton-cg", "lbfgs", "liblinear", "sag", "saga"]
multi_class = ["ovr", "multinomial"]
l1_ratio = np.linspace(0, 1, 20)
# build the parameter grid
param_grid = {
"penalty": penalty,
"C": C,
"solver": solver,
"multi_class": multi_class,
"l1_ratio": l1_ratio,
}
# initiate and fit the Grid Search Model
grid_model = GridSearchCV(log_model, param_grid=param_grid)
grid_model.fit(Scaled_X_train, y_train)
# best parameters
grid_model.best_params_
# ### **Model re-evaluation**
# We will evaluate the optimized version of our model and see if it does better than the base model
# creating predictions
y_pred = grid_model.predict(Scaled_X_test)
# plot the confusion matrix
fig, ax = plt.subplots(dpi=120)
plot_confusion_matrix(grid_model, Scaled_X_test, y_test, ax=ax)
# measure the accuracy of our model
acc_score = accuracy_score(y_test, y_pred)
round(acc_score, 2)
# generate the classification report
print(
classification_report(y_test, y_pred)
) # Hint: try it without using the print() method
| false | 0 | 4,408 | 21 | 4,706 | 4,408 |
||
69571684
|
<jupyter_start><jupyter_text>Sunspots
#Context
Sunspots are temporary phenomena on the Sun's photosphere that appear as spots darker than the surrounding areas. They are regions of reduced surface temperature caused by concentrations of magnetic field flux that inhibit convection. Sunspots usually appear in pairs of opposite magnetic polarity. Their number varies according to the approximately 11-year solar cycle.
Source: https://en.wikipedia.org/wiki/Sunspot
#Content :
Monthly Mean Total Sunspot Number, from 1749/01/01 to 2017/08/31
#Acknowledgements :
SIDC and Quandl.
Database from SIDC - Solar Influences Data Analysis Center - the solar physics research department of the Royal Observatory of Belgium. [SIDC website][1]
[1]: http://sidc.oma.be/
Kaggle dataset identifier: sunspots
<jupyter_code>import pandas as pd
df = pd.read_csv('sunspots/Sunspots.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 3265 entries, 0 to 3264
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 3265 non-null int64
1 Date 3265 non-null object
2 Monthly Mean Total Sunspot Number 3265 non-null float64
dtypes: float64(1), int64(1), object(1)
memory usage: 76.6+ KB
<jupyter_text>Examples:
{
"Unnamed: 0": 0,
"Date": "1749-01-31 00:00:00",
"Monthly Mean Total Sunspot Number": 96.7
}
{
"Unnamed: 0": 1,
"Date": "1749-02-28 00:00:00",
"Monthly Mean Total Sunspot Number": 104.3
}
{
"Unnamed: 0": 2,
"Date": "1749-03-31 00:00:00",
"Monthly Mean Total Sunspot Number": 116.7
}
{
"Unnamed: 0": 3,
"Date": "1749-04-30 00:00:00",
"Monthly Mean Total Sunspot Number": 92.8
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import tensorflow as tf
import csv
import numpy as np
import matplotlib.pyplot as plt
def plot_series(time, series, format="-", start=0, end=None):
plt.plot(time[start:end], series[start:end], format)
plt.xlabel("Time")
plt.ylabel("Value")
plt.grid(True)
import pandas as pd
df = pd.read_csv("/kaggle/input/sunspots/Sunspots.csv")
df.head()
time_step = []
temps = []
with open("/kaggle/input/sunspots/Sunspots.csv") as csvfile:
reader = csv.reader(csvfile, delimiter=",")
next(reader)
step = 0
for row in reader:
temps.append(float(row[2]))
time_step.append(step)
step = step + 1
series = np.array(temps)
time = np.array(time_step)
plt.figure(figsize=(10, 6))
plot_series(time, series)
split_time = 2500
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
window_size = 30
batch_size = 32
shuffle_buffer_size = 1000
def windowed_dataset(series, window_size, batch_size, shuffle_buffer):
series = tf.expand_dims(series, axis=-1)
ds = tf.data.Dataset.from_tensor_slices(series)
ds = ds.window(window_size + 1, shift=1, drop_remainder=True)
ds = ds.flat_map(lambda w: w.batch(window_size + 1))
ds = ds.shuffle(shuffle_buffer)
ds = ds.map(lambda w: (w[:-1], w[1:]))
return ds.batch(batch_size).prefetch(1)
def model_forecast(model, series, window_size):
ds = tf.data.Dataset.from_tensor_slices(series)
ds = ds.window(window_size, shift=1, drop_remainder=True)
ds = ds.flat_map(lambda w: w.batch(window_size))
ds = ds.batch(32).prefetch(1)
forecast = model.predict(ds)
return forecast
tf.keras.backend.clear_session()
tf.random.set_seed(51)
np.random.seed(51)
window_size = 64
batch_size = 256
train_set = windowed_dataset(x_train, window_size, batch_size, shuffle_buffer_size)
print(train_set)
print(x_train.shape)
model = tf.keras.models.Sequential(
[
tf.keras.layers.Conv1D(
filters=32,
kernel_size=5,
strides=1,
padding="causal",
activation="relu",
input_shape=[None, 1],
),
tf.keras.layers.LSTM(64, return_sequences=True),
tf.keras.layers.LSTM(64, return_sequences=True),
tf.keras.layers.Dense(30, activation="relu"),
tf.keras.layers.Dense(10, activation="relu"),
tf.keras.layers.Dense(1),
tf.keras.layers.Lambda(lambda x: x * 400),
]
)
lr_schedule = tf.keras.callbacks.LearningRateScheduler(
lambda epoch: 1e-8 * 10 ** (epoch / 20)
)
optimizer = tf.keras.optimizers.SGD(learning_rate=1e-8, momentum=0.9)
model.compile(loss=tf.keras.losses.Huber(), optimizer=optimizer, metrics=["mae"])
history = model.fit(train_set, epochs=100, callbacks=[lr_schedule])
plt.semilogx(history.history["lr"], history.history["loss"])
plt.axis([1e-8, 1e-4, 0, 60])
tf.keras.backend.clear_session()
tf.random.set_seed(51)
np.random.seed(51)
train_set = windowed_dataset(
x_train, window_size=60, batch_size=100, shuffle_buffer=shuffle_buffer_size
)
model = tf.keras.models.Sequential(
[
tf.keras.layers.Conv1D(
filters=60,
kernel_size=5,
strides=1,
padding="causal",
activation="relu",
input_shape=[None, 1],
),
tf.keras.layers.LSTM(60, return_sequences=True),
tf.keras.layers.LSTM(60, return_sequences=True),
tf.keras.layers.Dense(30, activation="relu"),
tf.keras.layers.Dense(10, activation="relu"),
tf.keras.layers.Dense(1),
tf.keras.layers.Lambda(lambda x: x * 400),
]
)
optimizer = tf.keras.optimizers.SGD(lr=1e-5, momentum=0.9)
model.compile(loss=tf.keras.losses.Huber(), optimizer=optimizer, metrics=["mae"])
history = model.fit(train_set, epochs=150)
rnn_forecast = model_forecast(model, series[..., np.newaxis], window_size)
rnn_forecast = rnn_forecast[split_time - window_size : -1, -1, 0]
# original data (blue) vs forecasted values (orange)
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plot_series(time_valid, rnn_forecast)
tf.keras.metrics.mean_absolute_error(x_valid, rnn_forecast).numpy()
print(rnn_forecast)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/571/69571684.ipynb
|
sunspots
|
robervalt
|
[{"Id": 69571684, "ScriptId": 18998341, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 581435, "CreationDate": "08/01/2021 17:00:13", "VersionNumber": 1.0, "Title": "Sunspots forecasting using Tensorflow LSTMs", "EvaluationDate": "08/01/2021", "IsChange": true, "TotalLines": 145.0, "LinesInsertedFromPrevious": 145.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 92919670, "KernelVersionId": 69571684, "SourceDatasetVersionId": 1932014}]
|
[{"Id": 1932014, "DatasetId": 2418, "DatasourceVersionId": 1970611, "CreatorUserId": 958985, "LicenseName": "CC0: Public Domain", "CreationDate": "02/11/2021 13:53:52", "VersionNumber": 3.0, "Title": "Sunspots", "Slug": "sunspots", "Subtitle": "Monthly Mean Total Sunspot Number - form 1749 to july 2018", "Description": "#Context\n\nSunspots are temporary phenomena on the Sun's photosphere that appear as spots darker than the surrounding areas. They are regions of reduced surface temperature caused by concentrations of magnetic field flux that inhibit convection. Sunspots usually appear in pairs of opposite magnetic polarity. Their number varies according to the approximately 11-year solar cycle.\n\nSource: https://en.wikipedia.org/wiki/Sunspot\n\n#Content :\n\nMonthly Mean Total Sunspot Number, from 1749/01/01 to 2017/08/31\n\n#Acknowledgements :\n\nSIDC and Quandl.\n\nDatabase from SIDC - Solar Influences Data Analysis Center - the solar physics research department of the Royal Observatory of Belgium. [SIDC website][1]\n\n [1]: http://sidc.oma.be/", "VersionNotes": "Up to 2021-01-31", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 2418, "CreatorUserId": 958985, "OwnerUserId": 958985.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1932014.0, "CurrentDatasourceVersionId": 1970611.0, "ForumId": 6460, "Type": 2, "CreationDate": "09/11/2017 14:44:50", "LastActivityDate": "02/06/2018", "TotalViews": 61533, "TotalDownloads": 8993, "TotalVotes": 143, "TotalKernels": 74}]
|
[{"Id": 958985, "UserName": "robervalt", "DisplayName": "Especuloide", "RegisterDate": "03/10/2017", "PerformanceTier": 0}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import tensorflow as tf
import csv
import numpy as np
import matplotlib.pyplot as plt
def plot_series(time, series, format="-", start=0, end=None):
plt.plot(time[start:end], series[start:end], format)
plt.xlabel("Time")
plt.ylabel("Value")
plt.grid(True)
import pandas as pd
df = pd.read_csv("/kaggle/input/sunspots/Sunspots.csv")
df.head()
time_step = []
temps = []
with open("/kaggle/input/sunspots/Sunspots.csv") as csvfile:
reader = csv.reader(csvfile, delimiter=",")
next(reader)
step = 0
for row in reader:
temps.append(float(row[2]))
time_step.append(step)
step = step + 1
series = np.array(temps)
time = np.array(time_step)
plt.figure(figsize=(10, 6))
plot_series(time, series)
split_time = 2500
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
window_size = 30
batch_size = 32
shuffle_buffer_size = 1000
def windowed_dataset(series, window_size, batch_size, shuffle_buffer):
series = tf.expand_dims(series, axis=-1)
ds = tf.data.Dataset.from_tensor_slices(series)
ds = ds.window(window_size + 1, shift=1, drop_remainder=True)
ds = ds.flat_map(lambda w: w.batch(window_size + 1))
ds = ds.shuffle(shuffle_buffer)
ds = ds.map(lambda w: (w[:-1], w[1:]))
return ds.batch(batch_size).prefetch(1)
def model_forecast(model, series, window_size):
ds = tf.data.Dataset.from_tensor_slices(series)
ds = ds.window(window_size, shift=1, drop_remainder=True)
ds = ds.flat_map(lambda w: w.batch(window_size))
ds = ds.batch(32).prefetch(1)
forecast = model.predict(ds)
return forecast
tf.keras.backend.clear_session()
tf.random.set_seed(51)
np.random.seed(51)
window_size = 64
batch_size = 256
train_set = windowed_dataset(x_train, window_size, batch_size, shuffle_buffer_size)
print(train_set)
print(x_train.shape)
model = tf.keras.models.Sequential(
[
tf.keras.layers.Conv1D(
filters=32,
kernel_size=5,
strides=1,
padding="causal",
activation="relu",
input_shape=[None, 1],
),
tf.keras.layers.LSTM(64, return_sequences=True),
tf.keras.layers.LSTM(64, return_sequences=True),
tf.keras.layers.Dense(30, activation="relu"),
tf.keras.layers.Dense(10, activation="relu"),
tf.keras.layers.Dense(1),
tf.keras.layers.Lambda(lambda x: x * 400),
]
)
lr_schedule = tf.keras.callbacks.LearningRateScheduler(
lambda epoch: 1e-8 * 10 ** (epoch / 20)
)
optimizer = tf.keras.optimizers.SGD(learning_rate=1e-8, momentum=0.9)
model.compile(loss=tf.keras.losses.Huber(), optimizer=optimizer, metrics=["mae"])
history = model.fit(train_set, epochs=100, callbacks=[lr_schedule])
plt.semilogx(history.history["lr"], history.history["loss"])
plt.axis([1e-8, 1e-4, 0, 60])
tf.keras.backend.clear_session()
tf.random.set_seed(51)
np.random.seed(51)
train_set = windowed_dataset(
x_train, window_size=60, batch_size=100, shuffle_buffer=shuffle_buffer_size
)
model = tf.keras.models.Sequential(
[
tf.keras.layers.Conv1D(
filters=60,
kernel_size=5,
strides=1,
padding="causal",
activation="relu",
input_shape=[None, 1],
),
tf.keras.layers.LSTM(60, return_sequences=True),
tf.keras.layers.LSTM(60, return_sequences=True),
tf.keras.layers.Dense(30, activation="relu"),
tf.keras.layers.Dense(10, activation="relu"),
tf.keras.layers.Dense(1),
tf.keras.layers.Lambda(lambda x: x * 400),
]
)
optimizer = tf.keras.optimizers.SGD(lr=1e-5, momentum=0.9)
model.compile(loss=tf.keras.losses.Huber(), optimizer=optimizer, metrics=["mae"])
history = model.fit(train_set, epochs=150)
rnn_forecast = model_forecast(model, series[..., np.newaxis], window_size)
rnn_forecast = rnn_forecast[split_time - window_size : -1, -1, 0]
# original data (blue) vs forecasted values (orange)
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plot_series(time_valid, rnn_forecast)
tf.keras.metrics.mean_absolute_error(x_valid, rnn_forecast).numpy()
print(rnn_forecast)
|
[{"sunspots/Sunspots.csv": {"column_names": "[\"Unnamed: 0\", \"Date\", \"Monthly Mean Total Sunspot Number\"]", "column_data_types": "{\"Unnamed: 0\": \"int64\", \"Date\": \"object\", \"Monthly Mean Total Sunspot Number\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 3265 entries, 0 to 3264\nData columns (total 3 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Unnamed: 0 3265 non-null int64 \n 1 Date 3265 non-null object \n 2 Monthly Mean Total Sunspot Number 3265 non-null float64\ndtypes: float64(1), int64(1), object(1)\nmemory usage: 76.6+ KB\n", "summary": "{\"Unnamed: 0\": {\"count\": 3265.0, \"mean\": 1632.0, \"std\": 942.6686409691725, \"min\": 0.0, \"25%\": 816.0, \"50%\": 1632.0, \"75%\": 2448.0, \"max\": 3264.0}, \"Monthly Mean Total Sunspot Number\": {\"count\": 3265.0, \"mean\": 81.77877488514548, \"std\": 67.88927651806058, \"min\": 0.0, \"25%\": 23.9, \"50%\": 67.2, \"75%\": 122.5, \"max\": 398.2}}", "examples": "{\"Unnamed: 0\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"Date\":{\"0\":\"1749-01-31\",\"1\":\"1749-02-28\",\"2\":\"1749-03-31\",\"3\":\"1749-04-30\"},\"Monthly Mean Total Sunspot Number\":{\"0\":96.7,\"1\":104.3,\"2\":116.7,\"3\":92.8}}"}}]
| true | 1 |
<start_data_description><data_path>sunspots/Sunspots.csv:
<column_names>
['Unnamed: 0', 'Date', 'Monthly Mean Total Sunspot Number']
<column_types>
{'Unnamed: 0': 'int64', 'Date': 'object', 'Monthly Mean Total Sunspot Number': 'float64'}
<dataframe_Summary>
{'Unnamed: 0': {'count': 3265.0, 'mean': 1632.0, 'std': 942.6686409691725, 'min': 0.0, '25%': 816.0, '50%': 1632.0, '75%': 2448.0, 'max': 3264.0}, 'Monthly Mean Total Sunspot Number': {'count': 3265.0, 'mean': 81.77877488514548, 'std': 67.88927651806058, 'min': 0.0, '25%': 23.9, '50%': 67.2, '75%': 122.5, 'max': 398.2}}
<dataframe_info>
RangeIndex: 3265 entries, 0 to 3264
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Unnamed: 0 3265 non-null int64
1 Date 3265 non-null object
2 Monthly Mean Total Sunspot Number 3265 non-null float64
dtypes: float64(1), int64(1), object(1)
memory usage: 76.6+ KB
<some_examples>
{'Unnamed: 0': {'0': 0, '1': 1, '2': 2, '3': 3}, 'Date': {'0': '1749-01-31', '1': '1749-02-28', '2': '1749-03-31', '3': '1749-04-30'}, 'Monthly Mean Total Sunspot Number': {'0': 96.7, '1': 104.3, '2': 116.7, '3': 92.8}}
<end_description>
| 1,616 | 1 | 2,239 | 1,616 |
69571152
|
# Problem Statement
# The dataset is used for this competition is synthetic, but based on a real dataset and generated using a CTGAN. The original dataset deals with calculating the loss associated with a loan defaults. Although the features are anonymized, they have properties relating to real-world features.
# Import
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Read
train = pd.read_csv("/kaggle/input/tabular-playground-series-aug-2021/train.csv")
train
test = pd.read_csv("/kaggle/input/tabular-playground-series-aug-2021/test.csv")
test
submission = pd.read_csv(
"/kaggle/input/tabular-playground-series-aug-2021/sample_submission.csv"
)
submission
# Analyse target
sns.displot(train["loss"])
train["loss"].describe()
target = train.loss
train.drop(["loss"], axis=1, inplace=True)
train
# Combine
combi = train.append(test)
combi
combi.drop(["id"], axis=1, inplace=True)
combi
# Check for null values
combi.isnull().sum().sum()
# Normalise
combi = (combi - combi.min()) / (combi.max() - combi.min())
combi.shape
# Define X and y
length = len(train)
y = target.ravel()
X = combi[:length]
X_test = combi[length:]
y.shape, X.shape, X_test.shape
# SelectKBest
from sklearn.feature_selection import SelectPercentile, f_regression
selector = SelectPercentile(f_regression, percentile=10)
X = selector.fit_transform(X, y)
X_test = selector.transform(X_test)
y.shape, X.shape, X_test.shape
# Split
from sklearn.model_selection import train_test_split
X_train, X_val, y_train, y_val = train_test_split(
X, y, test_size=0.1, random_state=1, shuffle=True
)
X_train.shape, X_val.shape, y_train.shape, y_val.shape, X_test.shape
# Select Model
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingRegressor
model = HistGradientBoostingRegressor(max_iter=20000, random_state=1).fit(
X_train, y_train
)
print(model.score(X_train, y_train))
# Predict on validation set
y_pred = model.predict(X_val)
print(model.score(X_val, y_val))
# Evaluate
from sklearn import metrics
print("Mean Absolute Error:", metrics.mean_absolute_error(y_val, y_pred))
print("Mean Squared Error:", metrics.mean_squared_error(y_val, y_pred))
print("Root Mean Squared Error:", np.sqrt(metrics.mean_squared_error(y_val, y_pred)))
# Compare
compare = pd.DataFrame({"actual": y_val, "predicted": y_pred})
print(compare)
# Graph
plt.figure(figsize=(10, 10))
plt.scatter(y_val, y_pred, c="crimson")
plt.yscale("log")
plt.xscale("log")
p1 = max(max(y_pred), max(y_val))
p2 = min(min(y_pred), min(y_val))
plt.plot([p1, p2], [p1, p2], "b-")
plt.xlabel("Actual Values", fontsize=15)
plt.ylabel("Predictions", fontsize=15)
plt.axis("equal")
plt.show()
# Predict on test set
prediction = model.predict(X_test)
prediction[prediction < 0] = 0
prediction.shape
# Prepare submission
submission.loss = prediction
submission
submission.to_csv("submission.csv", index=False)
submission = pd.read_csv("submission.csv")
submission
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/571/69571152.ipynb
| null | null |
[{"Id": 69571152, "ScriptId": 18998740, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5286865, "CreationDate": "08/01/2021 16:52:38", "VersionNumber": 1.0, "Title": "Aug 21 - SelectPercentile", "EvaluationDate": "08/01/2021", "IsChange": true, "TotalLines": 137.0, "LinesInsertedFromPrevious": 2.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 135.0, "LinesInsertedFromFork": 2.0, "LinesDeletedFromFork": 2.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 135.0, "TotalVotes": 0}]
| null | null | null | null |
# Problem Statement
# The dataset is used for this competition is synthetic, but based on a real dataset and generated using a CTGAN. The original dataset deals with calculating the loss associated with a loan defaults. Although the features are anonymized, they have properties relating to real-world features.
# Import
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Read
train = pd.read_csv("/kaggle/input/tabular-playground-series-aug-2021/train.csv")
train
test = pd.read_csv("/kaggle/input/tabular-playground-series-aug-2021/test.csv")
test
submission = pd.read_csv(
"/kaggle/input/tabular-playground-series-aug-2021/sample_submission.csv"
)
submission
# Analyse target
sns.displot(train["loss"])
train["loss"].describe()
target = train.loss
train.drop(["loss"], axis=1, inplace=True)
train
# Combine
combi = train.append(test)
combi
combi.drop(["id"], axis=1, inplace=True)
combi
# Check for null values
combi.isnull().sum().sum()
# Normalise
combi = (combi - combi.min()) / (combi.max() - combi.min())
combi.shape
# Define X and y
length = len(train)
y = target.ravel()
X = combi[:length]
X_test = combi[length:]
y.shape, X.shape, X_test.shape
# SelectKBest
from sklearn.feature_selection import SelectPercentile, f_regression
selector = SelectPercentile(f_regression, percentile=10)
X = selector.fit_transform(X, y)
X_test = selector.transform(X_test)
y.shape, X.shape, X_test.shape
# Split
from sklearn.model_selection import train_test_split
X_train, X_val, y_train, y_val = train_test_split(
X, y, test_size=0.1, random_state=1, shuffle=True
)
X_train.shape, X_val.shape, y_train.shape, y_val.shape, X_test.shape
# Select Model
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingRegressor
model = HistGradientBoostingRegressor(max_iter=20000, random_state=1).fit(
X_train, y_train
)
print(model.score(X_train, y_train))
# Predict on validation set
y_pred = model.predict(X_val)
print(model.score(X_val, y_val))
# Evaluate
from sklearn import metrics
print("Mean Absolute Error:", metrics.mean_absolute_error(y_val, y_pred))
print("Mean Squared Error:", metrics.mean_squared_error(y_val, y_pred))
print("Root Mean Squared Error:", np.sqrt(metrics.mean_squared_error(y_val, y_pred)))
# Compare
compare = pd.DataFrame({"actual": y_val, "predicted": y_pred})
print(compare)
# Graph
plt.figure(figsize=(10, 10))
plt.scatter(y_val, y_pred, c="crimson")
plt.yscale("log")
plt.xscale("log")
p1 = max(max(y_pred), max(y_val))
p2 = min(min(y_pred), min(y_val))
plt.plot([p1, p2], [p1, p2], "b-")
plt.xlabel("Actual Values", fontsize=15)
plt.ylabel("Predictions", fontsize=15)
plt.axis("equal")
plt.show()
# Predict on test set
prediction = model.predict(X_test)
prediction[prediction < 0] = 0
prediction.shape
# Prepare submission
submission.loss = prediction
submission
submission.to_csv("submission.csv", index=False)
submission = pd.read_csv("submission.csv")
submission
| false | 0 | 1,084 | 0 | 1,084 | 1,084 |
||
69136561
|
# ## You're here!
# Welcome to your first competition in the [ITI's AI Pro training program](https://ai.iti.gov.eg/epita/ai-engineer/)! We hope you enjoy and learn as much as we did prepairing this competition.
# ## Introduction
# In the competition, it's required to predict the `Severity` of a car crash given info about the crash, e.g., location.
# This is the getting started notebook. Things are kept simple so that it's easier to understand the steps and modify it.
# Feel free to `Fork` this notebook and share it with your modifications **OR** use it to create your submissions.
# ### Prerequisites
# You should know how to use python and a little bit of Machine Learning. You can apply the techniques you learned in the training program and submit the new solutions!
# ### Checklist
# You can participate in this competition the way you perefer. However, I recommend following these steps if this is your first time joining a competition on Kaggle.
# * Fork this notebook and run the cells in order.
# * Submit this solution.
# * Make changes to the data processing step as you see fit.
# * Submit the new solutions.
# *You can submit up to 5 submissions per day. You can select only one of the submission you make to be considered in the final ranking.*
# Don't hesitate to leave a comment or contact me if you have any question!
# ## Import the libraries
# We'll use `pandas` to load and manipulate the data. Other libraries will be imported in the relevant sections.
from datetime import datetime
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.preprocessing import LabelEncoder
import os
import xml.etree.ElementTree as et
labelencoder = LabelEncoder()
# ## Exploratory Data Analysis
# In this step, one should load the data and analyze it. However, I'll load the data and do minimal analysis. You are encouraged to do thorough analysis!
# Let's load the data using `pandas` and have a look at the generated `DataFrame`.
dataset_path = "/kaggle/input/car-crashes-severity-prediction/"
df = pd.read_csv(os.path.join(dataset_path, "train.csv"))
print("The shape of the dataset is {}.\n\n".format(df.shape))
df.head()
# We've got 6407 examples in the dataset with 14 featues, 1 ID, and the `Severity` of the crash.
# By looking at the features and a sample from the data, the features look of numerical and catogerical types. What about some descriptive statistics?
df.drop(columns="ID").describe()
# The output shows desciptive statistics for the numerical features, `Lat`, `Lng`, `Distance(mi)`, and `Severity`. I'll use the numerical features to demonstrate how to train the model and make submissions. **However you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# ### Remove columns with now effect
df.drop(columns=["Bump", "Roundabout"], inplace=True)
# #### Format date data in the dataframe and extract the hour to match with weather data
df["timestamp"] = pd.to_datetime(df["timestamp"])
df["date_time"] = pd.to_datetime(df["timestamp"].dt.strftime("%Y-%m-%d %H:00:00"))
df.info()
# #### convert bool values to numeric int values for cleaning
df["Crossing"] = df["Crossing"].astype(int)
df["Give_Way"] = df["Give_Way"].astype(int)
df["Junction"] = df["Junction"].astype(int)
df["No_Exit"] = df["No_Exit"].astype(int)
df["Railway"] = df["Railway"].astype(int)
df["Stop"] = df["Stop"].astype(int)
df["Amenity"] = df["Amenity"].astype(int)
# #### encode side data as numeric
df["Side"] = labelencoder.fit_transform(df["Side"])
df.head()
# ### Import Holiday Data
xtree = et.parse(os.path.join(dataset_path, "holidays.xml"))
xroot = xtree.getroot()
df_cols = ["date", "description"]
rows = []
for node in xroot:
date = node.find("date").text if node is not None else None
description = node.find("description").text if node is not None else None
rows.append({"date": date, "description": description})
holidays_df = pd.DataFrame(rows)
holidays_df.head()
# #### format date to match with the train data
holidays_df["date"] = pd.to_datetime(holidays_df["date"])
# #### Check if the day of the accident was holiday or not
df["is_holiday"] = df["timestamp"].dt.date.isin(holidays_df["date"].dt.date).astype(int)
# ### Import Weather Data
weather_df = pd.read_csv(os.path.join(dataset_path, "weather-sfcsv.csv"))
weather_df.head()
weather_df.describe()
weather_df["date_time"] = pd.to_datetime(weather_df[["Year", "Month", "Day", "Hour"]])
weather_df.dropna(subset=["Weather_Condition"], inplace=True)
weather_df["Weather_Condition"] = labelencoder.fit_transform(
weather_df["Weather_Condition"]
)
weather_df["Visibility(mi)"].fillna(weather_df["Visibility(mi)"].mean(), inplace=True)
weather_df["Wind_Speed(mph)"].fillna(weather_df["Wind_Speed(mph)"].mean(), inplace=True)
weather_df["Wind_Speed(mph)"].replace(
0, weather_df["Wind_Speed(mph)"].median(), inplace=True
)
weather_df["Humidity(%)"].fillna(weather_df["Humidity(%)"].mean(), inplace=True)
weather_df["Temperature(F)"].fillna(weather_df["Temperature(F)"].mean(), inplace=True)
weather_df.describe()
weather_df.drop_duplicates(subset=["date_time"], inplace=True, keep="last")
weather_df.drop(
columns=[
"Wind_Chill(F)",
"Precipitation(in)",
"Selected",
"Year",
"Month",
"Day",
"Hour",
],
inplace=True,
)
weather_df.head()
df_merged = pd.merge(df, weather_df, how="left", on=["date_time"])
df_merged.dropna(inplace=True)
df_merged.info()
df_merged.drop(columns=["ID"]).describe()
df_merged.head()
corr_matrix = df_merged.drop(columns=["ID"]).corr()
corr_matrix["Severity"].sort_values(ascending=False)
df_merged.drop(columns=["date_time", "timestamp"], inplace=True)
# ## Data Splitting
# Now it's time to split the dataset for the training step. Typically the dataset is split into 3 subsets, namely, the training, validation and test sets. In our case, the test set is already predefined. So we'll split the "training" set into training and validation sets with 0.8:0.2 ratio.
# *Note: a good way to generate reproducible results is to set the seed to the algorithms that depends on randomization. This is done with the argument `random_state` in the following command*
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
df_merged, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(columns=["ID", "Severity"])
y_train = train_df["Severity"]
X_val = val_df.drop(columns=["ID", "Severity"])
y_val = val_df["Severity"]
# As pointed out eariler, I'll use the numerical features to train the classifier. **However, you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# This cell is used to select the numerical features. IT SHOULD BE REMOVED AS YOU DO YOUR WORK.
X_train = X_train[
["Lat", "Lng", "Distance(mi)", "Temperature(F)", "Humidity(%)", "Wind_Speed(mph)"]
]
X_val = X_val[
["Lat", "Lng", "Distance(mi)", "Temperature(F)", "Humidity(%)", "Wind_Speed(mph)"]
]
# ## Model Training
# Let's train a model with the data! We'll train a Random Forest Classifier to demonstrate the process of making submissions.
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
# Now let's test our classifier on the validation dataset and see the accuracy.
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# Well. That's a good start, right? A classifier that predicts all examples' `Severity` as 2 will get around 0.63. You should get better score as you add more features and do better data preprocessing.
# ## Submission File Generation
# We have built a model and we'd like to submit our predictions on the test set! In order to do that, we'll load the test set, predict the class and save the submission file.
# First, we'll load the data.
test_df = pd.read_csv(os.path.join(dataset_path, "test.csv"))
test_df.head()
# Note that the test set has the same features and doesn't have the `Severity` column.
# At this stage one must **NOT** forget to apply the same processing done on the training set on the features of the test set.
# Now we'll add `Severity` column to the test `DataFrame` and add the values of the predicted class to it.
# **I'll select the numerical features here as I did in the training set. DO NOT forget to change this step as you change the preprocessing of the training data.**
test_df.drop(columns=["Bump", "Roundabout"], inplace=True)
test_df["timestamp"] = pd.to_datetime(test_df["timestamp"])
test_df["date_time"] = pd.to_datetime(
test_df["timestamp"].dt.strftime("%Y-%m-%d %H:00:00")
)
test_df["Crossing"] = test_df["Crossing"].astype("int")
test_df["Give_Way"] = test_df["Give_Way"].astype("int")
test_df["Junction"] = test_df["Junction"].astype("int")
test_df["No_Exit"] = test_df["No_Exit"].astype("int")
test_df["Railway"] = test_df["Railway"].astype("int")
test_df["Stop"] = test_df["Stop"].astype("int")
test_df["Amenity"] = test_df["Amenity"].astype("int")
test_df["Side"] = labelencoder.fit_transform(test_df["Side"])
test_df["is_holiday"] = (
test_df["timestamp"].dt.date.isin(holidays_df["date"].dt.date).astype(int)
)
test_df_merged = pd.merge(test_df, weather_df, how="left", on=["date_time"])
test_df_merged.dropna(inplace=True)
test_df_merged.drop(columns=["date_time", "timestamp"], inplace=True)
test_df_merged.info()
test_df_merged.head()
X_test = test_df_merged.drop(columns=["ID"])
# You should update/remove the next line once you change the features used for training
X_test = X_test[
["Lat", "Lng", "Distance(mi)", "Temperature(F)", "Humidity(%)", "Wind_Speed(mph)"]
]
y_test_predicted = classifier.predict(X_test)
test_df["Severity"] = y_test_predicted
test_df.head()
# Now we're ready to generate the submission file. The submission file needs the columns `ID` and `Severity` only.
test_df[["ID", "Severity"]].to_csv("/kaggle/working/submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/136/69136561.ipynb
| null | null |
[{"Id": 69136561, "ScriptId": 18860525, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1388367, "CreationDate": "07/27/2021 09:22:45", "VersionNumber": 3.0, "Title": "Fork of Getting Started - Car Crashes' Severity 2", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 269.0, "LinesInsertedFromPrevious": 21.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 248.0, "LinesInsertedFromFork": 24.0, "LinesDeletedFromFork": 16.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 245.0, "TotalVotes": 0}]
| null | null | null | null |
# ## You're here!
# Welcome to your first competition in the [ITI's AI Pro training program](https://ai.iti.gov.eg/epita/ai-engineer/)! We hope you enjoy and learn as much as we did prepairing this competition.
# ## Introduction
# In the competition, it's required to predict the `Severity` of a car crash given info about the crash, e.g., location.
# This is the getting started notebook. Things are kept simple so that it's easier to understand the steps and modify it.
# Feel free to `Fork` this notebook and share it with your modifications **OR** use it to create your submissions.
# ### Prerequisites
# You should know how to use python and a little bit of Machine Learning. You can apply the techniques you learned in the training program and submit the new solutions!
# ### Checklist
# You can participate in this competition the way you perefer. However, I recommend following these steps if this is your first time joining a competition on Kaggle.
# * Fork this notebook and run the cells in order.
# * Submit this solution.
# * Make changes to the data processing step as you see fit.
# * Submit the new solutions.
# *You can submit up to 5 submissions per day. You can select only one of the submission you make to be considered in the final ranking.*
# Don't hesitate to leave a comment or contact me if you have any question!
# ## Import the libraries
# We'll use `pandas` to load and manipulate the data. Other libraries will be imported in the relevant sections.
from datetime import datetime
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.preprocessing import LabelEncoder
import os
import xml.etree.ElementTree as et
labelencoder = LabelEncoder()
# ## Exploratory Data Analysis
# In this step, one should load the data and analyze it. However, I'll load the data and do minimal analysis. You are encouraged to do thorough analysis!
# Let's load the data using `pandas` and have a look at the generated `DataFrame`.
dataset_path = "/kaggle/input/car-crashes-severity-prediction/"
df = pd.read_csv(os.path.join(dataset_path, "train.csv"))
print("The shape of the dataset is {}.\n\n".format(df.shape))
df.head()
# We've got 6407 examples in the dataset with 14 featues, 1 ID, and the `Severity` of the crash.
# By looking at the features and a sample from the data, the features look of numerical and catogerical types. What about some descriptive statistics?
df.drop(columns="ID").describe()
# The output shows desciptive statistics for the numerical features, `Lat`, `Lng`, `Distance(mi)`, and `Severity`. I'll use the numerical features to demonstrate how to train the model and make submissions. **However you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# ### Remove columns with now effect
df.drop(columns=["Bump", "Roundabout"], inplace=True)
# #### Format date data in the dataframe and extract the hour to match with weather data
df["timestamp"] = pd.to_datetime(df["timestamp"])
df["date_time"] = pd.to_datetime(df["timestamp"].dt.strftime("%Y-%m-%d %H:00:00"))
df.info()
# #### convert bool values to numeric int values for cleaning
df["Crossing"] = df["Crossing"].astype(int)
df["Give_Way"] = df["Give_Way"].astype(int)
df["Junction"] = df["Junction"].astype(int)
df["No_Exit"] = df["No_Exit"].astype(int)
df["Railway"] = df["Railway"].astype(int)
df["Stop"] = df["Stop"].astype(int)
df["Amenity"] = df["Amenity"].astype(int)
# #### encode side data as numeric
df["Side"] = labelencoder.fit_transform(df["Side"])
df.head()
# ### Import Holiday Data
xtree = et.parse(os.path.join(dataset_path, "holidays.xml"))
xroot = xtree.getroot()
df_cols = ["date", "description"]
rows = []
for node in xroot:
date = node.find("date").text if node is not None else None
description = node.find("description").text if node is not None else None
rows.append({"date": date, "description": description})
holidays_df = pd.DataFrame(rows)
holidays_df.head()
# #### format date to match with the train data
holidays_df["date"] = pd.to_datetime(holidays_df["date"])
# #### Check if the day of the accident was holiday or not
df["is_holiday"] = df["timestamp"].dt.date.isin(holidays_df["date"].dt.date).astype(int)
# ### Import Weather Data
weather_df = pd.read_csv(os.path.join(dataset_path, "weather-sfcsv.csv"))
weather_df.head()
weather_df.describe()
weather_df["date_time"] = pd.to_datetime(weather_df[["Year", "Month", "Day", "Hour"]])
weather_df.dropna(subset=["Weather_Condition"], inplace=True)
weather_df["Weather_Condition"] = labelencoder.fit_transform(
weather_df["Weather_Condition"]
)
weather_df["Visibility(mi)"].fillna(weather_df["Visibility(mi)"].mean(), inplace=True)
weather_df["Wind_Speed(mph)"].fillna(weather_df["Wind_Speed(mph)"].mean(), inplace=True)
weather_df["Wind_Speed(mph)"].replace(
0, weather_df["Wind_Speed(mph)"].median(), inplace=True
)
weather_df["Humidity(%)"].fillna(weather_df["Humidity(%)"].mean(), inplace=True)
weather_df["Temperature(F)"].fillna(weather_df["Temperature(F)"].mean(), inplace=True)
weather_df.describe()
weather_df.drop_duplicates(subset=["date_time"], inplace=True, keep="last")
weather_df.drop(
columns=[
"Wind_Chill(F)",
"Precipitation(in)",
"Selected",
"Year",
"Month",
"Day",
"Hour",
],
inplace=True,
)
weather_df.head()
df_merged = pd.merge(df, weather_df, how="left", on=["date_time"])
df_merged.dropna(inplace=True)
df_merged.info()
df_merged.drop(columns=["ID"]).describe()
df_merged.head()
corr_matrix = df_merged.drop(columns=["ID"]).corr()
corr_matrix["Severity"].sort_values(ascending=False)
df_merged.drop(columns=["date_time", "timestamp"], inplace=True)
# ## Data Splitting
# Now it's time to split the dataset for the training step. Typically the dataset is split into 3 subsets, namely, the training, validation and test sets. In our case, the test set is already predefined. So we'll split the "training" set into training and validation sets with 0.8:0.2 ratio.
# *Note: a good way to generate reproducible results is to set the seed to the algorithms that depends on randomization. This is done with the argument `random_state` in the following command*
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
df_merged, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(columns=["ID", "Severity"])
y_train = train_df["Severity"]
X_val = val_df.drop(columns=["ID", "Severity"])
y_val = val_df["Severity"]
# As pointed out eariler, I'll use the numerical features to train the classifier. **However, you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# This cell is used to select the numerical features. IT SHOULD BE REMOVED AS YOU DO YOUR WORK.
X_train = X_train[
["Lat", "Lng", "Distance(mi)", "Temperature(F)", "Humidity(%)", "Wind_Speed(mph)"]
]
X_val = X_val[
["Lat", "Lng", "Distance(mi)", "Temperature(F)", "Humidity(%)", "Wind_Speed(mph)"]
]
# ## Model Training
# Let's train a model with the data! We'll train a Random Forest Classifier to demonstrate the process of making submissions.
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
# Now let's test our classifier on the validation dataset and see the accuracy.
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# Well. That's a good start, right? A classifier that predicts all examples' `Severity` as 2 will get around 0.63. You should get better score as you add more features and do better data preprocessing.
# ## Submission File Generation
# We have built a model and we'd like to submit our predictions on the test set! In order to do that, we'll load the test set, predict the class and save the submission file.
# First, we'll load the data.
test_df = pd.read_csv(os.path.join(dataset_path, "test.csv"))
test_df.head()
# Note that the test set has the same features and doesn't have the `Severity` column.
# At this stage one must **NOT** forget to apply the same processing done on the training set on the features of the test set.
# Now we'll add `Severity` column to the test `DataFrame` and add the values of the predicted class to it.
# **I'll select the numerical features here as I did in the training set. DO NOT forget to change this step as you change the preprocessing of the training data.**
test_df.drop(columns=["Bump", "Roundabout"], inplace=True)
test_df["timestamp"] = pd.to_datetime(test_df["timestamp"])
test_df["date_time"] = pd.to_datetime(
test_df["timestamp"].dt.strftime("%Y-%m-%d %H:00:00")
)
test_df["Crossing"] = test_df["Crossing"].astype("int")
test_df["Give_Way"] = test_df["Give_Way"].astype("int")
test_df["Junction"] = test_df["Junction"].astype("int")
test_df["No_Exit"] = test_df["No_Exit"].astype("int")
test_df["Railway"] = test_df["Railway"].astype("int")
test_df["Stop"] = test_df["Stop"].astype("int")
test_df["Amenity"] = test_df["Amenity"].astype("int")
test_df["Side"] = labelencoder.fit_transform(test_df["Side"])
test_df["is_holiday"] = (
test_df["timestamp"].dt.date.isin(holidays_df["date"].dt.date).astype(int)
)
test_df_merged = pd.merge(test_df, weather_df, how="left", on=["date_time"])
test_df_merged.dropna(inplace=True)
test_df_merged.drop(columns=["date_time", "timestamp"], inplace=True)
test_df_merged.info()
test_df_merged.head()
X_test = test_df_merged.drop(columns=["ID"])
# You should update/remove the next line once you change the features used for training
X_test = X_test[
["Lat", "Lng", "Distance(mi)", "Temperature(F)", "Humidity(%)", "Wind_Speed(mph)"]
]
y_test_predicted = classifier.predict(X_test)
test_df["Severity"] = y_test_predicted
test_df.head()
# Now we're ready to generate the submission file. The submission file needs the columns `ID` and `Severity` only.
test_df[["ID", "Severity"]].to_csv("/kaggle/working/submission.csv", index=False)
| false | 0 | 2,978 | 0 | 2,978 | 2,978 |
||
69136479
|
import os
import time
import numpy as np
from scipy.stats import norm
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import torch
import torch.nn as nn
from torch.distributions.normal import Normal
from torch.distributions.kl import kl_divergence
from transformers import (
AutoTokenizer,
AutoModel,
AutoConfig,
AdamW,
get_linear_schedule_with_warmup,
get_cosine_schedule_with_warmup,
)
def seed_everything(s):
np.random.seed(s)
torch.manual_seed(s)
torch.cuda.manual_seed(s)
torch.cuda.manual_seed_all(s)
seed_everything(42)
# https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3
#
class CONFIG:
env = "prod" # Set test for Testing.
checkpoint = "roberta-base"
pretrain_path = "../input/clrp-roberta-base-pretrain/clrp_roberta_base"
tokenizer = AutoTokenizer.from_pretrained(checkpoint)
base_config = AutoConfig.from_pretrained(checkpoint)
hidden_size = base_config.hidden_size
pad_token_id = tokenizer.pad_token_id
max_seq_len = tokenizer.model_max_length
FREEZE_LAYERS_START = 0
TRAIN_MAX_ITERS = 350 # Roughly 3 epochs
TRAIN_WARMUP_STEPS = 204
TRAIN_SAMPLES_PER_BATCH = 20
batch_size = 20
folds = 5
bins = 9
train_sample_bins = 10
learning_rate = 2e-5
weight_decay = 0.01
optimizer = "AdamW"
epochs = 8
clip_gradient_norm = 1.0
eval_every = 10
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
if CONFIG.env == "test":
CONFIG.TRAIN_SAMPLES_PER_BATCH = 4
CONFIG.TRAIN_MAX_ITERS = 3
CONFIG.epochs = 5
CONFIG.eval_every = 1
print("Device:", CONFIG.device)
def get_qunatile_boundaries(df, bins=10):
df = df.copy()
qs = []
for i in np.arange(1 / bins, 1.1, 1 / bins):
q = train_df.target.quantile(i)
qs.append(q)
return qs
def get_quantile(target, qs):
for i, q in enumerate(qs):
if target <= q:
return i
def get_bin_ranges(df):
df = df.copy()
bin_ranges = []
min_target = train_df.target.min()
max_target = train_df.target.max()
min_std = train_df[train_df.target == min_target].standard_error.min()
max_std = train_df[train_df.target == max_target].standard_error.max()
min_val = min_target - min_std
max_val = max_target + max_std
bin_values = np.arange(min_val, max_val, 0.5)
start_bin = (-1e9, bin_values[0])
end_bin = (bin_values[-1], 1e9)
bin_ranges.append(start_bin)
for i in range(1, len(bin_values)):
bin_ranges.append((bin_values[i - 1], bin_values[i]))
bin_ranges.append(end_bin)
return bin_ranges
def get_bin_distribution(row, bin_ranges):
mu = row.target
scale = 0.2
bins = []
for bin_range in bin_ranges:
s = bin_range[0]
e = bin_range[1]
cdf1 = norm.cdf(s, mu, scale)
cdf2 = norm.cdf(e, mu, scale)
cdf = cdf2 - cdf1
bins.append(cdf)
return bins
train_df = pd.read_csv("../input/commonlit-kfold-dataset/fold_train.csv")
bin_ranges = get_bin_ranges(train_df)
# Update Bins in the configuration
CONFIG.bins = len(bin_ranges)
print(bin_ranges)
train_qs = get_qunatile_boundaries(train_df, CONFIG.train_sample_bins)
train_df["q"] = train_df.target.apply(get_quantile, args=(train_qs,))
train_df["ybin"] = train_df.apply(get_bin_distribution, args=(bin_ranges,), axis=1)
train_df.head()
# # Datasets
class CommonLitDataset(torch.utils.data.Dataset):
def __init__(self, df, phase="train"):
self.excerpts = df.excerpt.values
self.targets = df.target.values
self.standard_errors = df.standard_error.values
self.ybin = df.ybin.values
self.tokenizer = CONFIG.tokenizer
self.pad_token_id = CONFIG.pad_token_id
self.max_seq_len = CONFIG.max_seq_len
def get_tokenized_features(self, excerpt):
inputs = self.tokenizer(excerpt, truncation=True)
input_ids = inputs["input_ids"]
attention_mask = inputs["attention_mask"]
input_len = len(input_ids)
pad_len = self.max_seq_len - input_len
input_ids += [self.pad_token_id] * pad_len
attention_mask += [0] * pad_len
return {
"seq_len": input_len,
"input_ids": input_ids,
"attention_mask": attention_mask,
}
def __getitem__(self, idx):
excerpt = self.excerpts[idx]
target = self.targets[idx]
ybin = self.ybin[idx]
sigma = self.standard_errors[idx]
features = self.get_tokenized_features(excerpt)
return {
"seq_len": features["seq_len"],
"input_ids": torch.tensor(features["input_ids"], dtype=torch.long),
"attention_mask": torch.tensor(
features["attention_mask"], dtype=torch.long
),
"yreg": torch.tensor(target, dtype=torch.float32),
"ybin": torch.tensor(ybin, dtype=torch.float32),
"sigmas": torch.tensor(sigma, dtype=torch.float32),
}
def __len__(self):
return len(self.targets)
# # Train Data Sampler
class TrainDataSampler:
def __init__(self, batch_size, df):
self.qmap = {}
self.batch_size = batch_size
self.batch_fraction = 1.0
self.df = df.copy()
self.tokenizer = CONFIG.tokenizer
self.pad_token_id = CONFIG.pad_token_id
self.max_seq_len = CONFIG.max_seq_len
for i in range(CONFIG.train_sample_bins):
ids = self.df[self.df.q == i].id.values
np.random.shuffle(ids)
self.qmap[i] = ids
def get_tokenized_features(self, excerpt):
inputs = self.tokenizer(excerpt, truncation=True)
input_ids = inputs["input_ids"]
attention_mask = inputs["attention_mask"]
input_len = len(input_ids)
pad_len = self.max_seq_len - input_len
input_ids += [self.pad_token_id] * pad_len
attention_mask += [0] * pad_len
return {
"seq_len": input_len,
"input_ids": input_ids,
"attention_mask": attention_mask,
}
def get_mbs(self):
sentences = []
yreg = []
ybin = []
sigmas = []
for i in range(CONFIG.train_sample_bins):
if i not in self.qmap:
continue
yids = self.qmap[i][-2:]
sentences += list(self.df[self.df.id.isin(yids)].excerpt.values)
yreg += list(self.df[self.df.id.isin(yids)].target.values)
ybin += list(self.df[self.df.id.isin(yids)].ybin.values)
sigmas += list(self.df[self.df.id.isin(yids)].standard_error.values)
self.qmap[i] = self.qmap[i][:-2]
if len(self.qmap[i]) == 0:
self.qmap.pop(i)
num_samples = len(yreg)
self.batch_fraction = len(yreg) / self.batch_size
features = {
"seq_len": [],
"input_ids": [],
"attention_mask": [],
"yreg": [],
"ybin": [],
"sigmas": [],
}
for i, sentence in enumerate(sentences):
data = self.get_tokenized_features(sentence)
seq_len = data["seq_len"]
input_ids = data["input_ids"]
attention_mask = data["attention_mask"]
features["seq_len"].append(seq_len)
features["input_ids"].append(input_ids)
features["attention_mask"].append(attention_mask)
features["yreg"].append(yreg[i] + np.random.uniform(-0.1, 0.1))
features["ybin"].append(ybin[i])
features["sigmas"].append(sigmas[i])
features["seq_len"] = torch.tensor(features["seq_len"], dtype=torch.long)
features["input_ids"] = torch.tensor(features["input_ids"], dtype=torch.long)
features["attention_mask"] = torch.tensor(
features["attention_mask"], dtype=torch.long
)
features["yreg"] = torch.tensor(features["yreg"], dtype=torch.float32)
features["ybin"] = torch.tensor(features["ybin"], dtype=torch.float32)
features["sigmas"] = torch.tensor(features["sigmas"], dtype=torch.float32)
return features
def __iter__(self):
while len(self.qmap) > 0:
mbs = self.get_mbs()
if self.batch_fraction < 0.5:
break
yield mbs
def __next__(self):
for i in range(10):
yield i
# # Model
def freeze_roberta_layers(roberta):
max_freeze_layer = CONFIG.FREEZE_LAYERS_START
for n, p in roberta.named_parameters():
if ("embedding" in n) or (
"layer" in n and int(n.split(".")[2]) <= max_freeze_layer
):
p.requires_grad = False
class TextRegressor(nn.Module):
def __init__(self):
super(TextRegressor, self).__init__()
self.linear = nn.Linear(CONFIG.hidden_size, 1024)
self.layer_norm = nn.LayerNorm(1024)
self.relu = nn.ReLU()
self.dropout = nn.Dropout(0.5)
self.regressor = nn.Linear(1024, 1)
nn.init.uniform_(self.linear.weight, -0.025, 0.025)
nn.init.uniform_(self.regressor.weight, -0.02, 0.02)
def forward(self, x):
x = self.linear(x)
x = self.layer_norm(x)
x = self.relu(x)
x = self.dropout(x)
x = self.regressor(x)
return x
class BinEstimator(nn.Module):
def __init__(self):
super(BinEstimator, self).__init__()
self.linear = nn.Linear(CONFIG.hidden_size, 1024)
self.layer_norm = nn.LayerNorm(1024)
self.relu = nn.ReLU()
self.dropout = nn.Dropout(0.5)
self.logits = nn.Linear(1024, CONFIG.bins)
nn.init.uniform_(self.linear.weight, -0.025, 0.025)
nn.init.uniform_(self.logits.weight, -0.02, 0.02)
def forward(self, x):
x = self.linear(x)
x = self.layer_norm(x)
x = self.relu(x)
x = self.dropout(x)
x = self.logits(x)
x = torch.softmax(x, dim=1)
return x
class AttentionHead(nn.Module):
def __init__(self):
super(AttentionHead, self).__init__()
self.W = nn.Linear(CONFIG.hidden_size, CONFIG.hidden_size)
self.V = nn.Linear(CONFIG.hidden_size, 1)
def forward(self, x):
attn = torch.tanh(self.W(x))
score = self.V(attn)
attention_weights = torch.softmax(score, dim=1)
context_vector = attention_weights * x
context_vector = torch.sum(context_vector, dim=1)
return context_vector
class CommonLitModel(nn.Module):
def __init__(self):
super(CommonLitModel, self).__init__()
self.roberta = AutoModel.from_pretrained(CONFIG.pretrain_path)
self.attention_head = AttentionHead()
self.dropout = nn.Dropout(0.25)
self.layer_norm = nn.LayerNorm(CONFIG.hidden_size)
self.regressor = TextRegressor()
self.bin_estimator = BinEstimator()
freeze_roberta_layers(self.roberta)
def forward(self, input_ids, attention_mask, output_hidden_states=False):
roberta_output = self.roberta(
input_ids, attention_mask=attention_mask, output_hidden_states=True
)
last_hidden_state = roberta_output.last_hidden_state
cls_pool = roberta_output.pooler_output
attention_pool = self.attention_head(last_hidden_state)
x_pool = (cls_pool + attention_pool) / 2
x_pool = self.dropout(x_pool)
x_pool = self.layer_norm(x_pool)
yhat_reg = self.regressor(x_pool)
yhat_bin = self.bin_estimator(x_pool)
return yhat_reg, yhat_bin
# # Custom Loss
class CustomLoss:
def __init__(self):
self.criterion = nn.MSELoss()
self.kl_divergence = nn.KLDivLoss(reduction="batchmean", log_target=True)
def get_reg_loss(self, y, yreg, phase):
if phase == "val":
# y=y.numpy().reshape(-1, 1)
# yreg=yreg.numpy().reshape(-1, 1)
# y=qt.inverse_transform(y)[:, 0]
# yreg=qt.inverse_transform(yreg)[:, 0]
# ydiff=(np.abs(y - yreg)**2).mean()
# reg_loss=torch.tensor(ydiff)
reg_loss = self.criterion(yreg, y)
else:
ydiff = torch.abs(yreg - y)
alpha1 = 0.1
alpha2 = 0.5
alpha3 = 0.4
ydiff1 = ydiff[ydiff <= 0.1]
ydiff2 = ydiff[(ydiff > 0.1) & (ydiff <= 0.5)]
ydiff3 = ydiff[(ydiff > 0.5)]
reg_loss = torch.tensor(0.0, device=CONFIG.device)
if len(ydiff1) > 0:
reg_loss += alpha1 * (ydiff1**2).mean()
if len(ydiff2) > 0:
reg_loss += alpha2 * (ydiff2**2).mean()
if len(ydiff3) > 0:
reg_loss += alpha3 * (ydiff3**2).mean()
# reg_loss=(ydiff**2).mean()
# reg_loss=self.criterion(yreg, y)
return reg_loss
def get_bin_loss(self, ybin, yhat_bin, phase):
if phase == "val":
ybin = ybin.view(-1, CONFIG.bins)
yhat_bin = yhat_bin.view(-1, CONFIG.bins)
yerr = torch.abs(ybin - yhat_bin)
yerr = yerr.sum(dim=1)
loss = yerr.mean()
return loss
def get_bin_cross_entropy_loss(self, ybin, yhat_bin, phase):
if phase == "val":
ybin = ybin.view(-1, CONFIG.bins)
yhat_bin = yhat_bin.view(-1, CONFIG.bins)
loss = torch.tensor(0.0)
loss = torch.zeros(ybin.shape)
for i in range(ybin.shape[1]):
loss[:, i] = (-ybin[:, i]) * torch.log(yhat_bin[:, i] + 1e-9)
loss = loss.sum(dim=1).mean()
return loss
def get_distribution_loss(self, y_mus, y_sigmas, yhat_mus):
P = Normal(y_mus, y_sigmas)
Q = Normal(yhat_mus, y_sigmas)
loss = (kl_divergence(P, Q) + kl_divergence(Q, P)) / 2
loss = loss.mean()
loss = loss.to(CONFIG.device)
return loss
def get_consistency_kl_div_loss(self, ybin, yhat_bin, phase):
if phase == "val":
ybin = ybin.view(-1, 1 + CONFIG.bins)
yhat_bin = yhat_bin.view(-1, 1 + CONFIG.bins)
loss1 = self.kl_divergence(ybin, yhat_bin)
loss2 = self.kl_divergence(yhat_bin, ybin)
loss = (loss1 + loss2) / 2
return loss
def get_consistency_loss(self, sigmas, yhat_mus, yhat_bin, phase):
num_samples = yhat_mus.size(0)
yreg_dist = torch.zeros(num_samples, 1 + CONFIG.bins)
yreg_dist[:, 0] = Normal(yhat_mus, sigmas).cdf(qs_bins[0])
for i in range(1, CONFIG.bins):
yreg_dist[:, i] = Normal(yhat_mus, sigmas).cdf(qs_bins[i]) - Normal(
yhat_mus, sigmas
).cdf(qs_bins[i - 1])
yreg_dist[:, CONFIG.bins] = 1 - Normal(yhat_mus, sigmas).cdf(
qs_bins[CONFIG.bins - 1]
)
if phase == "train":
yreg_dist = yreg_dist.to(CONFIG.device)
loss = self.get_bin_loss(yreg_dist, yhat_bin, phase)
# loss=self.get_consistency_kl_div_loss(yreg_dist, yhat_bin, phase)
return loss
def get_loss(self, inputs, phase):
yreg = inputs["yreg"]
ybin = inputs["ybin"]
yhat_reg = inputs["yhat_reg"]
yhat_bin = inputs["yhat_bin"]
reg_loss = self.get_reg_loss(yreg, yhat_reg, phase)
bin_loss = self.get_bin_cross_entropy_loss(ybin, yhat_bin, phase)
# bin_loss=self.get_bin_loss(ybin, yhat_bin, phase)
# distribution_loss=self.get_distribution_loss(y_mus, y_sigmas, yhat_mus)
# consistency_loss=self.get_consistency_loss(y_sigmas, yhat_mus, yhat_bins, phase)
# yreg_diff=torch.abs(y_mus-yhat_mus)
# loss=(bin_loss/2)+(mu_loss+distribution_loss)/2+(consistency_loss/2)
loss = (reg_loss + bin_loss) / 2
return {"loss": loss, "reg_loss": reg_loss.item(), "bin_loss": bin_loss.item()}
# # Evaluator
class CustomEvaluator:
def __init__(self, val_dataloader):
self.val_dataloader = val_dataloader
self.criterion = nn.MSELoss()
self.custom_loss = CustomLoss()
def evaluate(self, model):
model.eval()
all_yreg = []
all_ybin = []
all_yhatreg = []
all_yhatbin = []
for batch in self.val_dataloader:
batch_max_seq_len = torch.max(batch["seq_len"])
input_ids = batch["input_ids"][:, :batch_max_seq_len].to(CONFIG.device)
attention_mask = batch["attention_mask"][:, :batch_max_seq_len].to(
CONFIG.device
)
yreg = batch["yreg"].view(-1)
ybin = batch["ybin"].view(-1)
all_yreg += yreg.tolist()
all_ybin += ybin.tolist()
with torch.no_grad():
yhat_reg, yhat_bin = model(input_ids, attention_mask)
yhat_reg = yhat_reg.view(-1).detach().cpu()
yhat_bin = yhat_bin.view(-1).detach().cpu()
all_yhatreg += yhat_reg.tolist()
all_yhatbin += yhat_bin.tolist()
all_yreg = torch.tensor(all_yreg, dtype=torch.float32)
all_ybin = torch.tensor(all_ybin, dtype=torch.float32)
all_yhatreg = torch.tensor(all_yhatreg, dtype=torch.float32)
all_yhatbin = torch.tensor(all_yhatbin, dtype=torch.float32)
ydiff = torch.abs(all_yhatreg - all_yreg).numpy()
print("ydiff Variance:", np.std(ydiff))
print("Quantiles---")
print(np.quantile(ydiff, 0.7))
print(np.quantile(ydiff, 0.8))
print(np.quantile(ydiff, 0.9))
print(np.quantile(ydiff, 0.95))
model_losses = self.custom_loss.get_loss(
{
"yreg": all_yreg,
"ybin": all_ybin,
"yhat_reg": all_yhatreg,
"yhat_bin": all_yhatbin,
},
"val",
)
return model_losses
# # Trainer
def get_optimizer_params(model):
optimizer_parameters = [
{
"params": [
p
for n, p in model.named_parameters()
if (p.requires_grad and ("roberta" in n) and ("LayerNorm" not in n))
],
},
{
"params": [
p
for n, p in model.named_parameters()
if (p.requires_grad and ("roberta" in n) and "LayerNorm" in n)
],
"weight_decay": 0,
},
{
"params": [
p
for n, p in model.named_parameters()
if (p.requires_grad and "roberta" not in n)
],
"lr": 1e-3,
},
]
return optimizer_parameters
class Trainer:
def __init__(self, model, fold_train_df, val_dataloader):
self.df = fold_train_df.copy()
self.val_dataloader = val_dataloader
self.model = model
self.optimizer = AdamW(
get_optimizer_params(model),
lr=CONFIG.learning_rate,
weight_decay=CONFIG.weight_decay,
)
self.schedular = torch.optim.lr_scheduler.OneCycleLR(
self.optimizer,
max_lr=CONFIG.learning_rate,
total_steps=CONFIG.TRAIN_MAX_ITERS,
)
# CONFIG.TRAIN_WARMUP_STEPS,
# CONFIG.TRAIN_MAX_ITERS)
self.custom_loss = CustomLoss()
self.custom_evaluator = CustomEvaluator(val_dataloader)
self.train_loss = []
self.train_reg_loss = []
self.train_bin_loss = []
self.val_loss = []
self.val_reg_loss = []
self.val_bin_loss = []
self.iter_count = 0
self.best_iter = 0
self.best_reg_iter = 0
self.best_bin_iter = 0
self.best_loss = None
self.best_reg_loss = None
self.best_bin_loss = None
def checkpoint(self, model_losses):
val_loss = model_losses["loss"].item()
val_reg_loss = model_losses["reg_loss"]
val_bin_loss = model_losses["bin_loss"]
if (self.best_loss is None) or (self.best_loss > val_loss):
self.best_loss = val_loss
self.best_iter = self.iter_count
torch.save(self.model, "best_model.pt")
if (self.best_reg_loss is None) or (self.best_reg_loss > val_reg_loss):
self.best_reg_loss = val_reg_loss
self.best_reg_iter = self.iter_count
torch.save(self.model, "best_reg_model.pt")
if (self.best_bin_loss is None) or (self.best_bin_loss > val_bin_loss):
self.best_bin_loss = val_bin_loss
self.best_bin_iter = self.iter_count
torch.save(self.model, "best_bin_model.pt")
print("===" * 10)
print(
"Iter:{} | BestIter:{} | Best Reg Iter:{} | Best Bin Iter:{}".format(
self.iter_count, self.best_iter, self.best_reg_iter, self.best_bin_iter
)
)
print("Training Losses:")
print(
"Total: {:.3f} | Reg Loss:{:.3f} | Bin Loss:{:.3f}".format(
self.train_loss[-1], self.train_reg_loss[-1], self.train_bin_loss[-1]
)
)
print()
print("Val Losses")
print(
"Total: {:.3f} | Reg Loss:{:.3f} | Bin Loss:{:.3f}".format(
val_loss, val_reg_loss, val_bin_loss
)
)
def train_ops(self, inputs):
self.optimizer.zero_grad()
model_losses = self.custom_loss.get_loss(inputs, "train")
model_losses["loss"].backward()
self.optimizer.step()
self.schedular.step()
return model_losses
def train_epoch(self):
t1 = time.time()
self.model.train()
for batch in TrainDataSampler(CONFIG.TRAIN_SAMPLES_PER_BATCH, self.df):
self.iter_count += 1
if self.iter_count > CONFIG.TRAIN_MAX_ITERS:
break
batch_seq_lens = batch["seq_len"]
batch_max_seq_len = torch.max(batch["seq_len"])
input_ids = batch["input_ids"][:, :batch_max_seq_len].to(CONFIG.device)
attention_mask = batch["attention_mask"][:, :batch_max_seq_len].to(
CONFIG.device
)
yreg = batch["yreg"].to(CONFIG.device)
ybin = batch["ybin"].to(CONFIG.device)
yhat_reg, yhat_bin = self.model(input_ids, attention_mask)
yhat_reg = yhat_reg.view(-1)
model_losses = self.train_ops(
{"yreg": yreg, "ybin": ybin, "yhat_reg": yhat_reg, "yhat_bin": yhat_bin}
)
self.train_loss.append(model_losses["loss"].item())
self.train_reg_loss.append(model_losses["reg_loss"])
self.train_bin_loss.append(model_losses["bin_loss"])
if self.iter_count % CONFIG.eval_every == 0:
model_losses = self.custom_evaluator.evaluate(model)
self.val_loss.append(model_losses["loss"].item())
self.val_reg_loss.append(model_losses["reg_loss"])
self.val_bin_loss.append(model_losses["bin_loss"])
self.checkpoint(model_losses)
def train(self):
while True:
if self.iter_count > CONFIG.TRAIN_MAX_ITERS:
break
self.train_epoch()
for k in range(CONFIG.folds):
print("===" * 10)
print()
print("Fold: ==> ", k + 1)
fold_train_df = train_df[train_df.kfold != k].copy()
fold_val_df = train_df[train_df.kfold == k].copy()
if CONFIG.env == "test":
fold_train_df = fold_train_df.head(4)
fold_val_df = fold_val_df.head(4)
val_dataset = CommonLitDataset(fold_val_df)
val_dataloader = torch.utils.data.DataLoader(
val_dataset,
batch_size=CONFIG.batch_size,
shuffle=False,
pin_memory=True,
drop_last=False,
)
model = CommonLitModel()
model = model.to(CONFIG.device)
trainer = Trainer(model, fold_train_df, val_dataloader)
trainer.train()
best_model = torch.load("best_model.pt")
best_reg_model = torch.load("best_reg_model.pt")
best_bin_model = torch.load("best_bin_model.pt")
torch.save(best_model, "best_model{}.pt".format(k + 1))
torch.save(best_reg_model, "best_reg_model{}.pt".format(k + 1))
torch.save(best_bin_model, "best_bin_model{}.pt".format(k + 1))
print("Best Iteration:", trainer.best_iter)
print("Best Reg Iteration:", trainer.best_reg_iter)
print("Best Bin Iteration:", trainer.best_bin_iter)
print("Best Loss:{}".format(trainer.best_loss))
print("Best Reg Loss:{}".format(trainer.best_reg_loss))
print("Best Bin Loss:{}".format(trainer.best_bin_loss))
plt.plot(trainer.train_loss)
plt.plot(trainer.train_reg_loss)
plt.plot(trainer.train_bin_loss)
plt.plot(trainer.val_loss)
plt.plot(trainer.val_reg_loss)
plt.plot(trainer.val_bin_loss)
# # Inference
train_df = pd.read_csv("../input/commonlit-kfold-dataset/fold_train.csv")
train_df["q"] = train_df.target.apply(get_quantile, args=(train_qs,))
train_df["ybin"] = train_df.apply(get_bin_distribution, args=(bin_ranges,), axis=1)
if CONFIG.env == "test":
train_df = train_df.head(5)
batch_size = 32
test_dataset = CommonLitDataset(train_df)
test_dataloader = torch.utils.data.DataLoader(
test_dataset, batch_size=batch_size, shuffle=False, pin_memory=True, drop_last=False
)
print(len(test_dataloader))
models = [
torch.load("./best_reg_model1.pt"),
torch.load("./best_reg_model2.pt"),
torch.load("./best_reg_model3.pt"),
torch.load("./best_reg_model4.pt"),
torch.load("./best_reg_model5.pt"),
]
ypreds = []
ypred_bins = [] # np.zeros(len(train_df), CONFIG.bins)
for batch in test_dataloader:
batch_seq_lens = batch["seq_len"]
batch_max_seq_len = torch.max(batch["seq_len"])
input_ids = batch["input_ids"][:, :batch_max_seq_len].to(CONFIG.device)
attention_mask = batch["attention_mask"][:, :batch_max_seq_len].to(CONFIG.device)
batch_size = input_ids.size(0)
with torch.no_grad():
batch_yhat = np.zeros(batch_size)
batch_yhat_bin = np.zeros((batch_size, CONFIG.bins))
for model in models:
model.eval()
yhat, yhat_bin = model(input_ids, attention_mask)
yhat = yhat.view(-1).detach().cpu()
yhat_bin = yhat_bin.detach().cpu().numpy()
batch_yhat += yhat.numpy()
batch_yhat_bin += yhat_bin
batch_yhat /= len(models)
batch_yhat_bin /= len(models)
ypreds += batch_yhat.tolist()
ypred_bins += batch_yhat_bin.tolist()
train_df["yhat_target"] = ypreds
train_df["yhat_bins"] = ypred_bins
train_df.head()
train_df.to_csv("Train Inference.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/136/69136479.ipynb
| null | null |
[{"Id": 69136479, "ScriptId": 18822308, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 964619, "CreationDate": "07/27/2021 09:21:43", "VersionNumber": 41.0, "Title": "clrp-roberta task finetune-2", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 833.0, "LinesInsertedFromPrevious": 6.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 827.0, "LinesInsertedFromFork": 360.0, "LinesDeletedFromFork": 83.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 473.0, "TotalVotes": 0}]
| null | null | null | null |
import os
import time
import numpy as np
from scipy.stats import norm
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import torch
import torch.nn as nn
from torch.distributions.normal import Normal
from torch.distributions.kl import kl_divergence
from transformers import (
AutoTokenizer,
AutoModel,
AutoConfig,
AdamW,
get_linear_schedule_with_warmup,
get_cosine_schedule_with_warmup,
)
def seed_everything(s):
np.random.seed(s)
torch.manual_seed(s)
torch.cuda.manual_seed(s)
torch.cuda.manual_seed_all(s)
seed_everything(42)
# https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3
#
class CONFIG:
env = "prod" # Set test for Testing.
checkpoint = "roberta-base"
pretrain_path = "../input/clrp-roberta-base-pretrain/clrp_roberta_base"
tokenizer = AutoTokenizer.from_pretrained(checkpoint)
base_config = AutoConfig.from_pretrained(checkpoint)
hidden_size = base_config.hidden_size
pad_token_id = tokenizer.pad_token_id
max_seq_len = tokenizer.model_max_length
FREEZE_LAYERS_START = 0
TRAIN_MAX_ITERS = 350 # Roughly 3 epochs
TRAIN_WARMUP_STEPS = 204
TRAIN_SAMPLES_PER_BATCH = 20
batch_size = 20
folds = 5
bins = 9
train_sample_bins = 10
learning_rate = 2e-5
weight_decay = 0.01
optimizer = "AdamW"
epochs = 8
clip_gradient_norm = 1.0
eval_every = 10
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
if CONFIG.env == "test":
CONFIG.TRAIN_SAMPLES_PER_BATCH = 4
CONFIG.TRAIN_MAX_ITERS = 3
CONFIG.epochs = 5
CONFIG.eval_every = 1
print("Device:", CONFIG.device)
def get_qunatile_boundaries(df, bins=10):
df = df.copy()
qs = []
for i in np.arange(1 / bins, 1.1, 1 / bins):
q = train_df.target.quantile(i)
qs.append(q)
return qs
def get_quantile(target, qs):
for i, q in enumerate(qs):
if target <= q:
return i
def get_bin_ranges(df):
df = df.copy()
bin_ranges = []
min_target = train_df.target.min()
max_target = train_df.target.max()
min_std = train_df[train_df.target == min_target].standard_error.min()
max_std = train_df[train_df.target == max_target].standard_error.max()
min_val = min_target - min_std
max_val = max_target + max_std
bin_values = np.arange(min_val, max_val, 0.5)
start_bin = (-1e9, bin_values[0])
end_bin = (bin_values[-1], 1e9)
bin_ranges.append(start_bin)
for i in range(1, len(bin_values)):
bin_ranges.append((bin_values[i - 1], bin_values[i]))
bin_ranges.append(end_bin)
return bin_ranges
def get_bin_distribution(row, bin_ranges):
mu = row.target
scale = 0.2
bins = []
for bin_range in bin_ranges:
s = bin_range[0]
e = bin_range[1]
cdf1 = norm.cdf(s, mu, scale)
cdf2 = norm.cdf(e, mu, scale)
cdf = cdf2 - cdf1
bins.append(cdf)
return bins
train_df = pd.read_csv("../input/commonlit-kfold-dataset/fold_train.csv")
bin_ranges = get_bin_ranges(train_df)
# Update Bins in the configuration
CONFIG.bins = len(bin_ranges)
print(bin_ranges)
train_qs = get_qunatile_boundaries(train_df, CONFIG.train_sample_bins)
train_df["q"] = train_df.target.apply(get_quantile, args=(train_qs,))
train_df["ybin"] = train_df.apply(get_bin_distribution, args=(bin_ranges,), axis=1)
train_df.head()
# # Datasets
class CommonLitDataset(torch.utils.data.Dataset):
def __init__(self, df, phase="train"):
self.excerpts = df.excerpt.values
self.targets = df.target.values
self.standard_errors = df.standard_error.values
self.ybin = df.ybin.values
self.tokenizer = CONFIG.tokenizer
self.pad_token_id = CONFIG.pad_token_id
self.max_seq_len = CONFIG.max_seq_len
def get_tokenized_features(self, excerpt):
inputs = self.tokenizer(excerpt, truncation=True)
input_ids = inputs["input_ids"]
attention_mask = inputs["attention_mask"]
input_len = len(input_ids)
pad_len = self.max_seq_len - input_len
input_ids += [self.pad_token_id] * pad_len
attention_mask += [0] * pad_len
return {
"seq_len": input_len,
"input_ids": input_ids,
"attention_mask": attention_mask,
}
def __getitem__(self, idx):
excerpt = self.excerpts[idx]
target = self.targets[idx]
ybin = self.ybin[idx]
sigma = self.standard_errors[idx]
features = self.get_tokenized_features(excerpt)
return {
"seq_len": features["seq_len"],
"input_ids": torch.tensor(features["input_ids"], dtype=torch.long),
"attention_mask": torch.tensor(
features["attention_mask"], dtype=torch.long
),
"yreg": torch.tensor(target, dtype=torch.float32),
"ybin": torch.tensor(ybin, dtype=torch.float32),
"sigmas": torch.tensor(sigma, dtype=torch.float32),
}
def __len__(self):
return len(self.targets)
# # Train Data Sampler
class TrainDataSampler:
def __init__(self, batch_size, df):
self.qmap = {}
self.batch_size = batch_size
self.batch_fraction = 1.0
self.df = df.copy()
self.tokenizer = CONFIG.tokenizer
self.pad_token_id = CONFIG.pad_token_id
self.max_seq_len = CONFIG.max_seq_len
for i in range(CONFIG.train_sample_bins):
ids = self.df[self.df.q == i].id.values
np.random.shuffle(ids)
self.qmap[i] = ids
def get_tokenized_features(self, excerpt):
inputs = self.tokenizer(excerpt, truncation=True)
input_ids = inputs["input_ids"]
attention_mask = inputs["attention_mask"]
input_len = len(input_ids)
pad_len = self.max_seq_len - input_len
input_ids += [self.pad_token_id] * pad_len
attention_mask += [0] * pad_len
return {
"seq_len": input_len,
"input_ids": input_ids,
"attention_mask": attention_mask,
}
def get_mbs(self):
sentences = []
yreg = []
ybin = []
sigmas = []
for i in range(CONFIG.train_sample_bins):
if i not in self.qmap:
continue
yids = self.qmap[i][-2:]
sentences += list(self.df[self.df.id.isin(yids)].excerpt.values)
yreg += list(self.df[self.df.id.isin(yids)].target.values)
ybin += list(self.df[self.df.id.isin(yids)].ybin.values)
sigmas += list(self.df[self.df.id.isin(yids)].standard_error.values)
self.qmap[i] = self.qmap[i][:-2]
if len(self.qmap[i]) == 0:
self.qmap.pop(i)
num_samples = len(yreg)
self.batch_fraction = len(yreg) / self.batch_size
features = {
"seq_len": [],
"input_ids": [],
"attention_mask": [],
"yreg": [],
"ybin": [],
"sigmas": [],
}
for i, sentence in enumerate(sentences):
data = self.get_tokenized_features(sentence)
seq_len = data["seq_len"]
input_ids = data["input_ids"]
attention_mask = data["attention_mask"]
features["seq_len"].append(seq_len)
features["input_ids"].append(input_ids)
features["attention_mask"].append(attention_mask)
features["yreg"].append(yreg[i] + np.random.uniform(-0.1, 0.1))
features["ybin"].append(ybin[i])
features["sigmas"].append(sigmas[i])
features["seq_len"] = torch.tensor(features["seq_len"], dtype=torch.long)
features["input_ids"] = torch.tensor(features["input_ids"], dtype=torch.long)
features["attention_mask"] = torch.tensor(
features["attention_mask"], dtype=torch.long
)
features["yreg"] = torch.tensor(features["yreg"], dtype=torch.float32)
features["ybin"] = torch.tensor(features["ybin"], dtype=torch.float32)
features["sigmas"] = torch.tensor(features["sigmas"], dtype=torch.float32)
return features
def __iter__(self):
while len(self.qmap) > 0:
mbs = self.get_mbs()
if self.batch_fraction < 0.5:
break
yield mbs
def __next__(self):
for i in range(10):
yield i
# # Model
def freeze_roberta_layers(roberta):
max_freeze_layer = CONFIG.FREEZE_LAYERS_START
for n, p in roberta.named_parameters():
if ("embedding" in n) or (
"layer" in n and int(n.split(".")[2]) <= max_freeze_layer
):
p.requires_grad = False
class TextRegressor(nn.Module):
def __init__(self):
super(TextRegressor, self).__init__()
self.linear = nn.Linear(CONFIG.hidden_size, 1024)
self.layer_norm = nn.LayerNorm(1024)
self.relu = nn.ReLU()
self.dropout = nn.Dropout(0.5)
self.regressor = nn.Linear(1024, 1)
nn.init.uniform_(self.linear.weight, -0.025, 0.025)
nn.init.uniform_(self.regressor.weight, -0.02, 0.02)
def forward(self, x):
x = self.linear(x)
x = self.layer_norm(x)
x = self.relu(x)
x = self.dropout(x)
x = self.regressor(x)
return x
class BinEstimator(nn.Module):
def __init__(self):
super(BinEstimator, self).__init__()
self.linear = nn.Linear(CONFIG.hidden_size, 1024)
self.layer_norm = nn.LayerNorm(1024)
self.relu = nn.ReLU()
self.dropout = nn.Dropout(0.5)
self.logits = nn.Linear(1024, CONFIG.bins)
nn.init.uniform_(self.linear.weight, -0.025, 0.025)
nn.init.uniform_(self.logits.weight, -0.02, 0.02)
def forward(self, x):
x = self.linear(x)
x = self.layer_norm(x)
x = self.relu(x)
x = self.dropout(x)
x = self.logits(x)
x = torch.softmax(x, dim=1)
return x
class AttentionHead(nn.Module):
def __init__(self):
super(AttentionHead, self).__init__()
self.W = nn.Linear(CONFIG.hidden_size, CONFIG.hidden_size)
self.V = nn.Linear(CONFIG.hidden_size, 1)
def forward(self, x):
attn = torch.tanh(self.W(x))
score = self.V(attn)
attention_weights = torch.softmax(score, dim=1)
context_vector = attention_weights * x
context_vector = torch.sum(context_vector, dim=1)
return context_vector
class CommonLitModel(nn.Module):
def __init__(self):
super(CommonLitModel, self).__init__()
self.roberta = AutoModel.from_pretrained(CONFIG.pretrain_path)
self.attention_head = AttentionHead()
self.dropout = nn.Dropout(0.25)
self.layer_norm = nn.LayerNorm(CONFIG.hidden_size)
self.regressor = TextRegressor()
self.bin_estimator = BinEstimator()
freeze_roberta_layers(self.roberta)
def forward(self, input_ids, attention_mask, output_hidden_states=False):
roberta_output = self.roberta(
input_ids, attention_mask=attention_mask, output_hidden_states=True
)
last_hidden_state = roberta_output.last_hidden_state
cls_pool = roberta_output.pooler_output
attention_pool = self.attention_head(last_hidden_state)
x_pool = (cls_pool + attention_pool) / 2
x_pool = self.dropout(x_pool)
x_pool = self.layer_norm(x_pool)
yhat_reg = self.regressor(x_pool)
yhat_bin = self.bin_estimator(x_pool)
return yhat_reg, yhat_bin
# # Custom Loss
class CustomLoss:
def __init__(self):
self.criterion = nn.MSELoss()
self.kl_divergence = nn.KLDivLoss(reduction="batchmean", log_target=True)
def get_reg_loss(self, y, yreg, phase):
if phase == "val":
# y=y.numpy().reshape(-1, 1)
# yreg=yreg.numpy().reshape(-1, 1)
# y=qt.inverse_transform(y)[:, 0]
# yreg=qt.inverse_transform(yreg)[:, 0]
# ydiff=(np.abs(y - yreg)**2).mean()
# reg_loss=torch.tensor(ydiff)
reg_loss = self.criterion(yreg, y)
else:
ydiff = torch.abs(yreg - y)
alpha1 = 0.1
alpha2 = 0.5
alpha3 = 0.4
ydiff1 = ydiff[ydiff <= 0.1]
ydiff2 = ydiff[(ydiff > 0.1) & (ydiff <= 0.5)]
ydiff3 = ydiff[(ydiff > 0.5)]
reg_loss = torch.tensor(0.0, device=CONFIG.device)
if len(ydiff1) > 0:
reg_loss += alpha1 * (ydiff1**2).mean()
if len(ydiff2) > 0:
reg_loss += alpha2 * (ydiff2**2).mean()
if len(ydiff3) > 0:
reg_loss += alpha3 * (ydiff3**2).mean()
# reg_loss=(ydiff**2).mean()
# reg_loss=self.criterion(yreg, y)
return reg_loss
def get_bin_loss(self, ybin, yhat_bin, phase):
if phase == "val":
ybin = ybin.view(-1, CONFIG.bins)
yhat_bin = yhat_bin.view(-1, CONFIG.bins)
yerr = torch.abs(ybin - yhat_bin)
yerr = yerr.sum(dim=1)
loss = yerr.mean()
return loss
def get_bin_cross_entropy_loss(self, ybin, yhat_bin, phase):
if phase == "val":
ybin = ybin.view(-1, CONFIG.bins)
yhat_bin = yhat_bin.view(-1, CONFIG.bins)
loss = torch.tensor(0.0)
loss = torch.zeros(ybin.shape)
for i in range(ybin.shape[1]):
loss[:, i] = (-ybin[:, i]) * torch.log(yhat_bin[:, i] + 1e-9)
loss = loss.sum(dim=1).mean()
return loss
def get_distribution_loss(self, y_mus, y_sigmas, yhat_mus):
P = Normal(y_mus, y_sigmas)
Q = Normal(yhat_mus, y_sigmas)
loss = (kl_divergence(P, Q) + kl_divergence(Q, P)) / 2
loss = loss.mean()
loss = loss.to(CONFIG.device)
return loss
def get_consistency_kl_div_loss(self, ybin, yhat_bin, phase):
if phase == "val":
ybin = ybin.view(-1, 1 + CONFIG.bins)
yhat_bin = yhat_bin.view(-1, 1 + CONFIG.bins)
loss1 = self.kl_divergence(ybin, yhat_bin)
loss2 = self.kl_divergence(yhat_bin, ybin)
loss = (loss1 + loss2) / 2
return loss
def get_consistency_loss(self, sigmas, yhat_mus, yhat_bin, phase):
num_samples = yhat_mus.size(0)
yreg_dist = torch.zeros(num_samples, 1 + CONFIG.bins)
yreg_dist[:, 0] = Normal(yhat_mus, sigmas).cdf(qs_bins[0])
for i in range(1, CONFIG.bins):
yreg_dist[:, i] = Normal(yhat_mus, sigmas).cdf(qs_bins[i]) - Normal(
yhat_mus, sigmas
).cdf(qs_bins[i - 1])
yreg_dist[:, CONFIG.bins] = 1 - Normal(yhat_mus, sigmas).cdf(
qs_bins[CONFIG.bins - 1]
)
if phase == "train":
yreg_dist = yreg_dist.to(CONFIG.device)
loss = self.get_bin_loss(yreg_dist, yhat_bin, phase)
# loss=self.get_consistency_kl_div_loss(yreg_dist, yhat_bin, phase)
return loss
def get_loss(self, inputs, phase):
yreg = inputs["yreg"]
ybin = inputs["ybin"]
yhat_reg = inputs["yhat_reg"]
yhat_bin = inputs["yhat_bin"]
reg_loss = self.get_reg_loss(yreg, yhat_reg, phase)
bin_loss = self.get_bin_cross_entropy_loss(ybin, yhat_bin, phase)
# bin_loss=self.get_bin_loss(ybin, yhat_bin, phase)
# distribution_loss=self.get_distribution_loss(y_mus, y_sigmas, yhat_mus)
# consistency_loss=self.get_consistency_loss(y_sigmas, yhat_mus, yhat_bins, phase)
# yreg_diff=torch.abs(y_mus-yhat_mus)
# loss=(bin_loss/2)+(mu_loss+distribution_loss)/2+(consistency_loss/2)
loss = (reg_loss + bin_loss) / 2
return {"loss": loss, "reg_loss": reg_loss.item(), "bin_loss": bin_loss.item()}
# # Evaluator
class CustomEvaluator:
def __init__(self, val_dataloader):
self.val_dataloader = val_dataloader
self.criterion = nn.MSELoss()
self.custom_loss = CustomLoss()
def evaluate(self, model):
model.eval()
all_yreg = []
all_ybin = []
all_yhatreg = []
all_yhatbin = []
for batch in self.val_dataloader:
batch_max_seq_len = torch.max(batch["seq_len"])
input_ids = batch["input_ids"][:, :batch_max_seq_len].to(CONFIG.device)
attention_mask = batch["attention_mask"][:, :batch_max_seq_len].to(
CONFIG.device
)
yreg = batch["yreg"].view(-1)
ybin = batch["ybin"].view(-1)
all_yreg += yreg.tolist()
all_ybin += ybin.tolist()
with torch.no_grad():
yhat_reg, yhat_bin = model(input_ids, attention_mask)
yhat_reg = yhat_reg.view(-1).detach().cpu()
yhat_bin = yhat_bin.view(-1).detach().cpu()
all_yhatreg += yhat_reg.tolist()
all_yhatbin += yhat_bin.tolist()
all_yreg = torch.tensor(all_yreg, dtype=torch.float32)
all_ybin = torch.tensor(all_ybin, dtype=torch.float32)
all_yhatreg = torch.tensor(all_yhatreg, dtype=torch.float32)
all_yhatbin = torch.tensor(all_yhatbin, dtype=torch.float32)
ydiff = torch.abs(all_yhatreg - all_yreg).numpy()
print("ydiff Variance:", np.std(ydiff))
print("Quantiles---")
print(np.quantile(ydiff, 0.7))
print(np.quantile(ydiff, 0.8))
print(np.quantile(ydiff, 0.9))
print(np.quantile(ydiff, 0.95))
model_losses = self.custom_loss.get_loss(
{
"yreg": all_yreg,
"ybin": all_ybin,
"yhat_reg": all_yhatreg,
"yhat_bin": all_yhatbin,
},
"val",
)
return model_losses
# # Trainer
def get_optimizer_params(model):
optimizer_parameters = [
{
"params": [
p
for n, p in model.named_parameters()
if (p.requires_grad and ("roberta" in n) and ("LayerNorm" not in n))
],
},
{
"params": [
p
for n, p in model.named_parameters()
if (p.requires_grad and ("roberta" in n) and "LayerNorm" in n)
],
"weight_decay": 0,
},
{
"params": [
p
for n, p in model.named_parameters()
if (p.requires_grad and "roberta" not in n)
],
"lr": 1e-3,
},
]
return optimizer_parameters
class Trainer:
def __init__(self, model, fold_train_df, val_dataloader):
self.df = fold_train_df.copy()
self.val_dataloader = val_dataloader
self.model = model
self.optimizer = AdamW(
get_optimizer_params(model),
lr=CONFIG.learning_rate,
weight_decay=CONFIG.weight_decay,
)
self.schedular = torch.optim.lr_scheduler.OneCycleLR(
self.optimizer,
max_lr=CONFIG.learning_rate,
total_steps=CONFIG.TRAIN_MAX_ITERS,
)
# CONFIG.TRAIN_WARMUP_STEPS,
# CONFIG.TRAIN_MAX_ITERS)
self.custom_loss = CustomLoss()
self.custom_evaluator = CustomEvaluator(val_dataloader)
self.train_loss = []
self.train_reg_loss = []
self.train_bin_loss = []
self.val_loss = []
self.val_reg_loss = []
self.val_bin_loss = []
self.iter_count = 0
self.best_iter = 0
self.best_reg_iter = 0
self.best_bin_iter = 0
self.best_loss = None
self.best_reg_loss = None
self.best_bin_loss = None
def checkpoint(self, model_losses):
val_loss = model_losses["loss"].item()
val_reg_loss = model_losses["reg_loss"]
val_bin_loss = model_losses["bin_loss"]
if (self.best_loss is None) or (self.best_loss > val_loss):
self.best_loss = val_loss
self.best_iter = self.iter_count
torch.save(self.model, "best_model.pt")
if (self.best_reg_loss is None) or (self.best_reg_loss > val_reg_loss):
self.best_reg_loss = val_reg_loss
self.best_reg_iter = self.iter_count
torch.save(self.model, "best_reg_model.pt")
if (self.best_bin_loss is None) or (self.best_bin_loss > val_bin_loss):
self.best_bin_loss = val_bin_loss
self.best_bin_iter = self.iter_count
torch.save(self.model, "best_bin_model.pt")
print("===" * 10)
print(
"Iter:{} | BestIter:{} | Best Reg Iter:{} | Best Bin Iter:{}".format(
self.iter_count, self.best_iter, self.best_reg_iter, self.best_bin_iter
)
)
print("Training Losses:")
print(
"Total: {:.3f} | Reg Loss:{:.3f} | Bin Loss:{:.3f}".format(
self.train_loss[-1], self.train_reg_loss[-1], self.train_bin_loss[-1]
)
)
print()
print("Val Losses")
print(
"Total: {:.3f} | Reg Loss:{:.3f} | Bin Loss:{:.3f}".format(
val_loss, val_reg_loss, val_bin_loss
)
)
def train_ops(self, inputs):
self.optimizer.zero_grad()
model_losses = self.custom_loss.get_loss(inputs, "train")
model_losses["loss"].backward()
self.optimizer.step()
self.schedular.step()
return model_losses
def train_epoch(self):
t1 = time.time()
self.model.train()
for batch in TrainDataSampler(CONFIG.TRAIN_SAMPLES_PER_BATCH, self.df):
self.iter_count += 1
if self.iter_count > CONFIG.TRAIN_MAX_ITERS:
break
batch_seq_lens = batch["seq_len"]
batch_max_seq_len = torch.max(batch["seq_len"])
input_ids = batch["input_ids"][:, :batch_max_seq_len].to(CONFIG.device)
attention_mask = batch["attention_mask"][:, :batch_max_seq_len].to(
CONFIG.device
)
yreg = batch["yreg"].to(CONFIG.device)
ybin = batch["ybin"].to(CONFIG.device)
yhat_reg, yhat_bin = self.model(input_ids, attention_mask)
yhat_reg = yhat_reg.view(-1)
model_losses = self.train_ops(
{"yreg": yreg, "ybin": ybin, "yhat_reg": yhat_reg, "yhat_bin": yhat_bin}
)
self.train_loss.append(model_losses["loss"].item())
self.train_reg_loss.append(model_losses["reg_loss"])
self.train_bin_loss.append(model_losses["bin_loss"])
if self.iter_count % CONFIG.eval_every == 0:
model_losses = self.custom_evaluator.evaluate(model)
self.val_loss.append(model_losses["loss"].item())
self.val_reg_loss.append(model_losses["reg_loss"])
self.val_bin_loss.append(model_losses["bin_loss"])
self.checkpoint(model_losses)
def train(self):
while True:
if self.iter_count > CONFIG.TRAIN_MAX_ITERS:
break
self.train_epoch()
for k in range(CONFIG.folds):
print("===" * 10)
print()
print("Fold: ==> ", k + 1)
fold_train_df = train_df[train_df.kfold != k].copy()
fold_val_df = train_df[train_df.kfold == k].copy()
if CONFIG.env == "test":
fold_train_df = fold_train_df.head(4)
fold_val_df = fold_val_df.head(4)
val_dataset = CommonLitDataset(fold_val_df)
val_dataloader = torch.utils.data.DataLoader(
val_dataset,
batch_size=CONFIG.batch_size,
shuffle=False,
pin_memory=True,
drop_last=False,
)
model = CommonLitModel()
model = model.to(CONFIG.device)
trainer = Trainer(model, fold_train_df, val_dataloader)
trainer.train()
best_model = torch.load("best_model.pt")
best_reg_model = torch.load("best_reg_model.pt")
best_bin_model = torch.load("best_bin_model.pt")
torch.save(best_model, "best_model{}.pt".format(k + 1))
torch.save(best_reg_model, "best_reg_model{}.pt".format(k + 1))
torch.save(best_bin_model, "best_bin_model{}.pt".format(k + 1))
print("Best Iteration:", trainer.best_iter)
print("Best Reg Iteration:", trainer.best_reg_iter)
print("Best Bin Iteration:", trainer.best_bin_iter)
print("Best Loss:{}".format(trainer.best_loss))
print("Best Reg Loss:{}".format(trainer.best_reg_loss))
print("Best Bin Loss:{}".format(trainer.best_bin_loss))
plt.plot(trainer.train_loss)
plt.plot(trainer.train_reg_loss)
plt.plot(trainer.train_bin_loss)
plt.plot(trainer.val_loss)
plt.plot(trainer.val_reg_loss)
plt.plot(trainer.val_bin_loss)
# # Inference
train_df = pd.read_csv("../input/commonlit-kfold-dataset/fold_train.csv")
train_df["q"] = train_df.target.apply(get_quantile, args=(train_qs,))
train_df["ybin"] = train_df.apply(get_bin_distribution, args=(bin_ranges,), axis=1)
if CONFIG.env == "test":
train_df = train_df.head(5)
batch_size = 32
test_dataset = CommonLitDataset(train_df)
test_dataloader = torch.utils.data.DataLoader(
test_dataset, batch_size=batch_size, shuffle=False, pin_memory=True, drop_last=False
)
print(len(test_dataloader))
models = [
torch.load("./best_reg_model1.pt"),
torch.load("./best_reg_model2.pt"),
torch.load("./best_reg_model3.pt"),
torch.load("./best_reg_model4.pt"),
torch.load("./best_reg_model5.pt"),
]
ypreds = []
ypred_bins = [] # np.zeros(len(train_df), CONFIG.bins)
for batch in test_dataloader:
batch_seq_lens = batch["seq_len"]
batch_max_seq_len = torch.max(batch["seq_len"])
input_ids = batch["input_ids"][:, :batch_max_seq_len].to(CONFIG.device)
attention_mask = batch["attention_mask"][:, :batch_max_seq_len].to(CONFIG.device)
batch_size = input_ids.size(0)
with torch.no_grad():
batch_yhat = np.zeros(batch_size)
batch_yhat_bin = np.zeros((batch_size, CONFIG.bins))
for model in models:
model.eval()
yhat, yhat_bin = model(input_ids, attention_mask)
yhat = yhat.view(-1).detach().cpu()
yhat_bin = yhat_bin.detach().cpu().numpy()
batch_yhat += yhat.numpy()
batch_yhat_bin += yhat_bin
batch_yhat /= len(models)
batch_yhat_bin /= len(models)
ypreds += batch_yhat.tolist()
ypred_bins += batch_yhat_bin.tolist()
train_df["yhat_target"] = ypreds
train_df["yhat_bins"] = ypred_bins
train_df.head()
train_df.to_csv("Train Inference.csv", index=False)
| false | 0 | 8,287 | 0 | 8,287 | 8,287 |
||
69136143
|
<jupyter_start><jupyter_text>Titanic Dataset
### Context
Titanic Shipwreck
### Content
Predicting the survival rate of titanic shipwreck
Kaggle dataset identifier: output
<jupyter_script># # Titanic Top 4% with ensemble modeling
# * **1 소개**
# * **2 데이터 로드 및 체크**
# * 2.1 데이터 로드
# * 2.2 이상치 감지
# * 2.3 학습 및 테스트 데이터 세트 조인
# * 2.4 널 및 결측치
# * **3 피처 분석**
# * 3.1 수치형 값
# * 3.2 범주형 값
# * **4 결측치 처리**
# * 4.1 Age
# * **5 피처 엔지니어링**
# * 5.1 Name/Title
# * 5.2 Family Size
# * 5.3 Cabin
# * 5.4 Ticket
# * **6 모델링**
# * 6.1 단순 ML모델
# * 6.1.1 교차 검증 모델
# * 6.1.2 베스트모델에 대한 하이터 파라미터 튜닝
# * 6.1.3 학습곡선 폴롯
# * 6.1.4 트리기반의 분류기에 대한 피처 중요도
# * 6.2 앙상블 모델링
# * 6.2.1 모델 조합
# * 6.3 예측
# * 6.3.1 예측 및 결과 예측
#
# ## 1. 소개
# 캐글에서 세번째로 타이타닉 관련 데이터셋을 가지고 다음과 같은 방법으로 진행하려고 한다.
# No matter what you're going through, there's a light at the end of the tunnel.
# It may seem hard to get to it, but you can do it.
# Just keep working towards to it and you will find the positive side of things.
# [Demi Lovato]
# * **피처 분석**
# * **피처 엔지니어링**
# * **모델링**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from collections import Counter
from sklearn.ensemble import (
RandomForestClassifier,
AdaBoostClassifier,
GradientBoostingClassifier,
ExtraTreesClassifier,
VotingClassifier,
)
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis # LDA
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.svm import SVC
from sklearn.model_selection import (
GridSearchCV,
cross_val_score,
StratifiedKFold,
learning_curve,
)
sns.set(style="white", context="notebook", palette="deep", font_scale=2)
plt.style.use("seaborn")
# ## 2.데이터 로드 및 데이터 확인
# ### 2.1 데이터 로드
def load_dataset():
train = pd.read_csv("../input/titanic/train.csv")
test = pd.read_csv("../input/titanic/test.csv")
IDTEST = test["PassengerId"] # 결과물제출시 PassengerId
# Shape및 데이터보기
display(train.shape, test.shape)
display(train.head(n=2), display(test.tail(n=2)))
return train, test, IDTEST
# 수행
train, test, IDTEST = load_dataset()
# ### 2.2 이상치 감지
def detect_outliers(df, features, n):
"""
df - DataFrame
features - feature
n - 이상치값 필터링에 사용되는 변수값
"""
outlier_index = []
for col in features:
# 25%
Q1 = np.percentile(df[col], 25)
Q3 = np.percentile(df[col], 75)
# IQR(Interquartile range)
IQR = Q3 - Q1
# Outline_step
outlier_step = 1.5 * IQR
outlier_df = df[(df[col] < Q1 - outlier_step) | (df[col] > Q3 + outlier_step)]
display("이상치 : \n", outlier_df, outlier_df.shape)
outlier_list_col = outlier_df.index
outlier_index.extend(outlier_list_col)
outlier_index = Counter(outlier_index)
multiple_outliers = list(k for k, v in outlier_index.items() if v > n)
return multiple_outliers
# 이상치탐지
multiple_outliers = detect_outliers(train, ["Age", "SibSp", "Parch", "Fare"], 2)
# 이상치(Outlier)는 데이터의 통계치를 왜곡시키기 때문에 때론, 아웃라이어를 통계치에 제외시키기도 한다.
# 1977년도 Turjey, JW가 발표한 Turkey Method에 방법에 의해 아웃라이어를 찾기위해 IQR에 outlier step만큼 더하거나 뺀 값이 이상치이다.
def remove_outliers(df, outlier_pos):
"""
이상치의 데이터프레임에서 인덱스를 매개변수로 받아
해당 데이터를 삭제
"""
display("이상치 삭제전 : ", df.shape)
# show he outlier rows
display(df.iloc[outlier_pos])
# Drop outlier
df = df.drop(outlier_pos, inplace=False, axis=0, errors="ignore").reset_index(
drop=True
)
# 삭제후
display("이상치 삭제 후 ", df.shape)
return df
# Remove outlier
# 10개의 아웃라이어
train = remove_outliers(train, multiple_outliers)
# 10개의 이상치 데이터가 발견되었으면 , 28번, 89번 그리고 342번의 승객의 요금이 비정상적으로 높음.
# 나머지 7개 로우는 매우 높은 SibSp값을 갖고 있다.
# ### 2.3 학습 데이터셋트와 테스트 데이터세트 조인
def get_join_dataset(train, test):
"""
학습 및 테스트 데이터셋을 병합한 데이터셋
"""
display(train.shape, test.shape)
dataset = pd.concat(objs=[train, test], axis=0).reset_index(drop=True)
display("\n 병합된 데이터셋 ", dataset.shape)
display("\n Dataset Info ", dataset.info())
return dataset
# 수행
dataset = get_join_dataset(train, test)
# ### 2.4 널값과 결측치 확인
def check_null_missing_val(df, trainT=False):
"""
널값과 결측치 확인
trainT 파라미터는 True인 경우는 학습데이터 확인용
"""
if trainT:
# Fill empty and NaNs values with NaN
df = df.fillna(np.nan)
# Check for Null values
display(df.isnull().sum())
# 데이터 타입
display(df.dtypes, type(df.dtypes), df.dtypes.to_list())
print("\n")
display(df.info())
print("\n Technical Statistics")
display(df.describe())
print("\n Top 5")
display(df.head(n=5))
return df
dataset = check_null_missing_val(dataset)
# train데이터셋 확인
train = check_null_missing_val(train, trainT=True)
# Age및 Cabin피처는 결측치 처리를 위해서 중요한 역할을 할 수 있다.
# ## 3. 피처 분석
# ### 3.1 숫자형 값
int_cols = train.dtypes[train.dtypes != "object"]
int_cols.drop(index=["PassengerId", "Pclass"], inplace=True)
def extract_num_feature(df, objType=None):
"""
데이터프레임을 입력받아 데이터타입을 필터링하는 해당
피처만 리스트타입으로 리턴
리턴값은 pd.Series
"""
int_cols = df.dtypes[df.dtypes != objType]
int_cols.drop(index=["PassengerId", "Pclass"], inplace=True)
return int_cols
int_cols = extract_num_feature(train, "object")
int_cols.index
# Correlation matrix between numerical values (SibSp Parch Age and Fare values) and Survived
g = sns.heatmap(train[int_cols.index].corr(), annot=True, fmt=".2f", cmap="coolwarm_r")
# Fare피처가 생존률과는 의미있는 상관관계를 가지고있습니다.
# 그렇다고 해서 다른 피처들이 전혀 필요없다는 의미는 아닙니다. 기존 피처를 이용해 새롭게 파생변수를 생성해보면 상관관계를 새롭게 도출되는 경우도 있습니다. 그런면에서 다른 피처들은 또다른 의미를 갖습니다.
# "호랑이는 죽어서 가죽을 남기고 사람은 죽어서 이름을 남긴다."
# 피처의 소멸 자체도 의미있다는 말씀이지요.
# #### SibSP
# * [seaborn - factorplot](https://www.kite.com/python/docs/seaborn.factorplot)
# * [seaborn - style 사용자 정의](http://hleecaster.com/python-seaborn-set-style-and-context/)
g = sns.factorplot(
x="SibSp", y="Survived", data=train, kind="bar", size=6, palette="muted"
)
g.set_xlabels("SibSp")
g.despine(left=True)
g.set_ylabels("Survival Probability")
# 동반가족이 2명보다 많은 경우는 생존률이 많이 낮아짐을 알수 있다.
# 피처 엔지니어링을 통해서 새로운 피처를 추가적으로 도출이 가능하다.
# #### Parch
g = sns.factorplot(x="Parch", y="Survived", data=train, palette="summer_r", kind="bar")
g.despine(left=True)
g.set_xlabels("Parch")
g.set_ylabels("Survival Possibility")
# 사이즈가 작은 이동이 생존률이 훨씬 좋은 것을 알수있다.
# 동반가족이 3인인 경우, 생존율의 표준편차가 심한 것을 알수 있다.
# #### Age
# 히스토그램과 kde plot이 같이.
g = sns.FacetGrid(train, col="Survived")
g = g.map(sns.distplot, "Age")
# 연령대를 구분할 필요가 있어보임.연령에 대한 사망과 생존의 차이는 크게 두드러지지는 않지만, 연령대가 뒤로 갈수록 사망과 생존의 구별은 확연히 드러날것으로 보임
cond_0 = (train["Survived"] == 0) & (train["Age"].notnull()) # Not Survived
cond_1 = (train["Survived"] == 1) & (train["Age"].notnull()) # Survived
g = sns.kdeplot(train["Age"][cond_0], color="red", shade=True)
g = sns.kdeplot(train["Age"][cond_1], color="Blue", shade=True, ax=g)
g.set_xlabel("Age")
g.set_ylabel("Frequency")
g = g.legend(["Not Survived", "Survived"])
# When we superimpose the two densities , we cleary see a peak correponsing (between 0 and 5) to babies and very young childrens.
# #### Fare
def show_dist_plot(df, col: str):
"""
해당 데이터프레임의 피처에 분포 및 kde plot을 시각화
"""
g = sns.distplot(
df[col], color="b", label="Skewness : {:.2f}".format(df[col].skew())
)
g.set_title("Skewness for {0}".format(col))
g = g.legend(loc="best")
num_cols = int_cols[1:].index.values
# 학습 및 테스트 데이터셋 조인한 데이터셋
dataset[num_cols].isnull().sum()
# Fill Fare missing values with the median value
dataset["Fare"] = dataset["Fare"].fillna(dataset["Fare"].median())
# Recheck
dataset["Fare"].isnull().sum()
# * Check the skewness of Fare feature
# before logarithm
show_dist_plot(dataset, "Fare")
# As we can see, Fare distribution is very skewed. This can lead to overweigth very high values in the model, even if it is scaled.
# 위의 그래프처럼 , Fare band가 매우 편향되어있음을 알수 있다. 이런 데이터를 그냥 모델에 학습시키면 Min/Max Scaler혹은 Standard Scaler로 스케일링이 되어 있다 하더락도 과적합되기 마련이다.
# 그러므로 이런 편향된 데이터의 경우엔, 로그변환을 적용해야 한다.
# np.log1p변환과 np.log()변환을 같이 한다면 이때의 편향정도는?
# What if we do both np.log1p and np.log ?
# * Logarithm transformation
dataset["Fare"] = dataset["Fare"].apply(lambda x: np.log1p(x))
# Apply log to Fare to reduce skewness distribution
dataset["Fare"] = dataset["Fare"].map(lambda i: np.log(i) if i > 0 else 0)
# After Logarithm
show_dist_plot(dataset, "Fare")
# 로그변환 후에 편향정도가 상당히 감소했음을 알 수 있다
# ### 3.2 범주형 피처의 값
def show_factor_plot(df, xCol=None, yCol=None, col=None, kind="bar", hue=None):
"""
피처별 생존률 비교
"""
g = sns.factorplot(
x=xCol, y=yCol, hue=hue, col=col, kind=kind, palette="muted", size=6, data=df
)
sns.despine(left=True)
g.set_ylabels(yCol)
# #### Sex
# * bar plot으로 성별 생존률
# * Sex로 Groupby하여 Survived의 mean()값
g = sns.barplot(x="Sex", y="Survived", data=train)
g = g.set_ylabel("Survivor Probability")
train[["Sex", "Survived"]].groupby("Sex").mean()
# 사고가 났을때 100명중에 20명정도는 남자는 살고, 여자는 75명 이상은 산다고 나타난다
# 성별 피처는 생존률을 예측할때 정말 중요한 피처라고 할 수 있다
# 1997년 타이타닉 영화가 나왔을때 영화대사 중에 이런 구절이 나온다.
# : "여자와 아이들 먼저 구조해".
# 세월호 침몰 사고때는 어떠했는가?
# #### Pclass
# Explore Pclass vs Survived
show_factor_plot(train, "Pclass", "Survived")
# Explore Pclass vs Survived by Sex
show_factor_plot(train, "Pclass", "Survived", hue="Sex")
# * 좌석별 여성 및 남성 비교
dataset[(dataset["Pclass"] == 3)].groupby("Sex")["Pclass"].count()
# 승객들의 생존률은 3등급 객실이라고 해서 다르지 않다. 1등급 객실은 2등급, 3등급 객실보다는 생존률이 높다.
# #### Embarked
dataset["Embarked"].isnull().sum()
# 사우스햄튼이 가장 승객이 많다.
dataset["Embarked"].value_counts()
# Fill Embarked nan values of dataset set with 'S' most frequent value
dataset["Embarked"] = dataset["Embarked"].fillna("S")
# Recheck
dataset["Embarked"].isnull().sum()
# Explore Embarked vs Survived
show_factor_plot(train, xCol="Embarked", yCol="Survived")
# 프랑스 노르망디 해안에 위치한 체르보르그항에서 탑승한 승객들의 생존률이 다른 사우스햄튼 혹은 퀸즈타운보다 훨씬 생존률이 좋음을 알 수 있다.
# 그렇다면, 1등급 객실에 묵은 승객들의 비율이 다른 경유지 항구 - 체르보르그, 사우스햄튼, 퀸즈타운에서 온 승객들보다 더 많을 것이다.
# 경유지항구(Embarked) v.s Pclass(객실등급)
# Explore Pclass vs Embarked
show_factor_plot(train, xCol="Pclass", col="Embarked", kind="count")
# 사우스햄튼과 퀸즈타운의 승객들의 대부분은 3등급 객실에 분포되어 있으며, 추론은 이 사망자의 대부분은 퀸즈타운과 사우스햄튼 탑승객 일 것이다.
# 반면 체르보르그 탑승객들은 1등급이 가장 많다.
# 1등급 객실의 생존률이 높은 이유는?
# -> 사회적 영향도 인가, 아니면 구조가 용이한 여객선의 구조적 문제인가?
# ## 4. 결측치 채우기
# ### 4.1 Age
# 앞에서 보듯히, Age피처는 256개의 결측치를 포함하고있다.
# Age가 속한 연령대에 따라 생존률이 극명하게 갈리는 부분이 있기 때문에 중요한 피처라고 할 수 있으며, 이를 위해서라도 결측치를 대치할 부분이 필요하다.
# 이를 위해서는 Sex, Parch, Pclass 그리고 SibSp를 이용해 새로운 피처를 생성하는 문제를 생각해보자
# Explore Age vs Sex, Parch , Pclass and SibSP
g = sns.factorplot(y="Age", x="Sex", data=dataset, kind="box")
g = sns.factorplot(y="Age", x="Sex", hue="Pclass", data=dataset, kind="box")
g = sns.factorplot(y="Age", x="Parch", data=dataset, kind="box")
g = sns.factorplot(y="Age", x="SibSp", data=dataset, kind="box")
# 성별이 연령대를 예측할 수 있는 결정인자는 아니다.
# 그러나 1등급 객실에 있는 승객의 나이가 다른 2등급 객실, 3등급 객실의 승객들보다 연령대가 높다는 점은 주목 할 만하다.
# 부모 및 아이들을 동반한 경우가 자식,배우자를 동반한 경우보다 연령대가 높다는 것을 알 수 있다.
# convert Sex into categoricl value 0 for male and 1 for female
dataset["Sex"] = dataset["Sex"].map({"male": 0, "female": 1})
# 위의 방법과 동일
dataset["Sex"] = dataset["Sex"].apply(lambda x: 0 if x == "male" else 1)
# 앞에서 한번 np.log1p()로 로그변환 한 데이터를 다시 밑에것 np.logp()를 수행하면
# 히트맵에서 이상하게 보임
df = dataset[["Sex", "Age", "SibSp", "Parch", "Pclass"]]
g = sns.heatmap(df.corr(), cmap="BrBG", annot=True)
# 상관관계 맵은 Parch 피처를 제외한 factorplot관측치를 확인합니다.
# Age피처가 Sex랑은 관련성이 떨어지고, Pclass, Parch 그리고 SibSp와 음의 상관관계를 가지고있습니다.
# Age피처가 부모 및 아이들의 수에 따라 증가하는 경향은 있지만, 전반적으로는 음의 상관관계입니다.
# 그래서 Age 결측치를 대치하기 위해 SibSp, Parch 그리고 Pclass를 사용하기로 합니다.(결측치 대치는 median)
def fill_missing_value_age(df, idx_nan_age: list):
"""
Age가 널인것들을 리스트 파라미터로 전달해 중앙값으로 대치함.
Filling missing value of Age
Fill Age with the median age of similar rows according to Pclass, Parch and SibSp
"""
for i in idx_nan_age:
age_med = df["Age"].median()
age_pred = df["Age"][
(
(df["SibSp"] == df.iloc[i]["SibSp"])
& (df["Parch"] == df.iloc[i]["Parch"])
& (df["Pclass"] == df.iloc[i]["Pclass"])
)
].median()
if not np.isnan(age_pred):
df["Age"].iloc[i] = age_pred
else:
df["Age"].iloc[i] = age_med
return df
index_NaN_age = list(dataset["Age"].isnull().index)
fill_missing_value_age(dataset, index_NaN_age)
# * [box v.s violin] (https://junklee.tistory.com/9)
show_factor_plot(train, xCol="Survived", yCol="Age", kind="box")
show_factor_plot(train, xCol="Survived", yCol="Age", kind="violin")
# 사망자와 생존자의 Age의 median값도 크게 차이가 없다.
# violin plot을 보면 나이가 어린 영역대의 생존률이 상당히 높음을 알 수 있다.
# ## 5. 피처 엔지니어링
def feature_engineering(df, titleOpt=False, familyOpt=False):
"""
1.Title에 대한 정제작업
"""
if titleOpt:
df["Title"].replace(
titleList,
[
"Mr",
"Miss",
"Mrs",
"Master",
"Mr",
"Other",
"Other",
"Other",
"Miss",
"Miss",
"Mr",
"Other",
"Mr",
"Miss",
"Other",
"Other",
"Mrs",
"Mr",
],
inplace=True,
)
df["Title"].replace(
["Mr", "Mrs", "Miss", "Master", "Other"], [0, 1, 2, 3, 4], inplace=True
)
df["Title"].astype(int)
# Drop Name variable
df.drop(labels=["Name"], axis=0, inplace=True, errors="ignore")
if familyOpt:
df["FamilySize"] = df["SibSp"] + df["Parch"] + 1
# 피처 추가 후 기존 컬럼 삭제
df.drop(labels=["SibSp", "Parch"], axis=0, inplace=True, errors="ignore")
display(df.head(n=2))
display(df.tail(n=2))
return df
# ### 5.1 Name/Title
display(dataset["Name"].head())
display(dataset["Name"].tail())
# The Name feature contains information on passenger's title.
# Since some passenger with distingused title may be preferred during the evacuation, it is interesting to add them to the model.
def get_TitleFromName(df, feature):
"""
이름에서 존칭 분리
"""
dataset_title = [i.split(",")[1].split(".")[0] for i in df[feature]]
display(dataset_title[1:10])
df["Title"] = pd.Series(dataset_title)
display(df.head(n=2))
return df
# Get Title from Name
dataset = get_TitleFromName(dataset, "Name")
g = sns.countplot(x="Title", data=dataset)
g = plt.setp(g.get_xticklabels(), rotation=45)
# There is 17 titles in the dataset, most of them are very rare and we can group them in 4 categories.
titleList = dataset["Title"].value_counts().index.values.tolist()
dataset = feature_engineering(dataset, titleOpt=True)
g = sns.countplot(dataset["Title"])
g = g.set_xticklabels(["Mr", "Mrs", "Miss", "Master", "Other"])
g = sns.factorplot(x="Title", y="Survived", data=dataset, kind="bar")
g = g.set_xticklabels(["Mr", "Mrs", "Miss", "Master", "Other"])
g = g.set_ylabels("Survival probability for Title")
# "여자와 아이먼저 구조"
# "Other" 타이틀이 승선객의 Title구성을 볼때 생존률이 높다.
# ### 5.2 Family size
# * 가족수(FamilySize)피처 추가 - SibSp(동반가족(자손 + 배우자)) , Parch(부모님 동반된 아이들 포함)
# Create a family size descriptor from SibSp and Parch
dataset = feature_engineering(dataset, familyOpt=True)
g = sns.factorplot(x="FamilySize", y="Survived", data=dataset)
g = g.set_ylabels("Survival Probability for FamilySize")
# The family size seems to play an important role, survival probability is worst for large families.
# Additionally, i decided to created 4 categories of family size.
dataset["FamilySize"].value_counts()
dataset["Single"] = dataset["FamilySize"].map(lambda s: 1 if s == 1 else 0)
dataset["SmallF"] = dataset["FamilySize"].map(lambda s: 1 if s == 2 else 0)
dataset["MedF"] = dataset["FamilySize"].map(lambda s: 1 if 3 <= s <= 4 else 0)
dataset["LargeF"] = dataset["FamilySize"].map(lambda s: 1 if s >= 5 else 0)
def show_factorplot(df, cols):
"""
데이터셋과 컬럼에 대한 정보를 받아서
Factotplot으로 피처별 생존률을 보여줌
"""
for i in range(len(cols)):
g = sns.factorplot(x=cols[i], y="Survived", data=dataset, kind="bar")
g = g.set_ylabels("Survival Probabilit for {0}".format(cols[i]))
# 피처별 생존률
cols = ["Single", "SmallF", "MedF", "LargeF"]
show_factorplot(dataset, cols)
# 2인, 3인, 4인정도의 인원으로 가족끼리 탑승한 경우 생존률이 1인 혹은 5인 이상의 가족 및 친지끼리 승선한 경우보다 생존률이 좋다는 점은 기억하자.
# 혼자보다 둘이, 둘보다는 세명이서 여행가라. 그러면 사고당해서 돌아오지 못할 확률이 줄어든다.
# convert to indicator values Title and Embarked
dataset = pd.get_dummies(dataset, columns=["Title"])
dataset = pd.get_dummies(dataset, columns=["Embarked"], prefix="Em")
dataset.head()
# At this stage, we have 22 features.
# ### 5.3 Cabin
dataset["Cabin"].head()
dataset["Cabin"].describe()
dataset["Cabin"].isnull().sum()
# The Cabin feature column contains 292 values and 1007 missing values.
# I supposed that passengers without a cabin have a missing value displayed instead of the cabin number.
dataset["Cabin"][dataset["Cabin"].notnull()].head()
# Replace the Cabin number by the type of cabin 'X' if not
dataset["Cabin"] = pd.Series(
[i[0] if not pd.isnull(i) else "X" for i in dataset["Cabin"]]
)
# The first letter of the cabin indicates the Desk, i choosed to keep this information only, since it indicates the probable location of the passenger in the Titanic.
g = sns.countplot(dataset["Cabin"], order=["A", "B", "C", "D", "E", "F", "G", "T", "X"])
g = sns.factorplot(
y="Survived",
x="Cabin",
data=dataset,
kind="bar",
order=["A", "B", "C", "D", "E", "F", "G", "T", "X"],
)
g = g.set_ylabels("Survival Probability")
# Because of the low number of passenger that have a cabin, survival probabilities have an important standard deviation and we can't distinguish between survival probability of passengers in the different desks.
# But we can see that passengers with a cabin have generally more chance to survive than passengers without (X).
# It is particularly true for cabin B, C, D, E and F.
dataset = pd.get_dummies(dataset, columns=["Cabin"], prefix="Cabin")
# ### 5.4 Ticket
dataset["Ticket"].head()
# It could mean that tickets sharing the same prefixes could be booked for cabins placed together. It could therefore lead to the actual placement of the cabins within the ship.
# Tickets with same prefixes may have a similar class and survival.
# So i decided to replace the Ticket feature column by the ticket prefixe. Which may be more informative.
## Treat Ticket by extracting the ticket prefix. When there is no prefix it returns X.
Ticket = []
for i in list(dataset.Ticket):
if not i.isdigit():
Ticket.append(
i.replace(".", "").replace("/", "").strip().split(" ")[0]
) # Take prefix
else:
Ticket.append("X")
dataset["Ticket"] = Ticket
dataset["Ticket"].head()
dataset = pd.get_dummies(dataset, columns=["Ticket"], prefix="T")
# Create categorical values for Pclass
dataset["Pclass"] = dataset["Pclass"].astype("category")
dataset = pd.get_dummies(dataset, columns=["Pclass"], prefix="Pc")
# Drop useless variables
dataset.drop(labels=["PassengerId"], axis=1, inplace=True)
dataset.head()
# ## 6. MODELING
## Separate train dataset and test dataset
train = dataset[:train_len]
test = dataset[train_len:]
test.drop(labels=["Survived"], axis=1, inplace=True)
## Separate train features and label
train["Survived"] = train["Survived"].astype(int)
Y_train = train["Survived"]
X_train = train.drop(labels=["Survived"], axis=1)
# ### 6.1 Simple modeling
# #### 6.1.1 Cross validate models
# I compared 10 popular classifiers and evaluate the mean accuracy of each of them by a stratified kfold cross validation procedure.
# * SVC
# * Decision Tree
# * AdaBoost
# * Random Forest
# * Extra Trees
# * Gradient Boosting
# * Multiple layer perceprton (neural network)
# * KNN
# * Logistic regression
# * Linear Discriminant Analysis
# Cross validate model with Kfold stratified cross val
kfold = StratifiedKFold(n_splits=10)
# Modeling step Test differents algorithms
random_state = 2
classifiers = []
classifiers.append(SVC(random_state=random_state))
classifiers.append(DecisionTreeClassifier(random_state=random_state))
classifiers.append(
AdaBoostClassifier(
DecisionTreeClassifier(random_state=random_state),
random_state=random_state,
learning_rate=0.1,
)
)
classifiers.append(RandomForestClassifier(random_state=random_state))
classifiers.append(ExtraTreesClassifier(random_state=random_state))
classifiers.append(GradientBoostingClassifier(random_state=random_state))
classifiers.append(MLPClassifier(random_state=random_state))
classifiers.append(KNeighborsClassifier())
classifiers.append(LogisticRegression(random_state=random_state))
classifiers.append(LinearDiscriminantAnalysis())
cv_results = []
for classifier in classifiers:
cv_results.append(
cross_val_score(
classifier, X_train, y=Y_train, scoring="accuracy", cv=kfold, n_jobs=4
)
)
cv_means = []
cv_std = []
for cv_result in cv_results:
cv_means.append(cv_result.mean())
cv_std.append(cv_result.std())
cv_res = pd.DataFrame(
{
"CrossValMeans": cv_means,
"CrossValerrors": cv_std,
"Algorithm": [
"SVC",
"DecisionTree",
"AdaBoost",
"RandomForest",
"ExtraTrees",
"GradientBoosting",
"MultipleLayerPerceptron",
"KNeighboors",
"LogisticRegression",
"LinearDiscriminantAnalysis",
],
}
)
g = sns.barplot(
"CrossValMeans",
"Algorithm",
data=cv_res,
palette="Set3",
orient="h",
**{"xerr": cv_std}
)
g.set_xlabel("Mean Accuracy")
g = g.set_title("Cross validation scores")
# I decided to choose the SVC, AdaBoost, RandomForest , ExtraTrees and the GradientBoosting classifiers for the ensemble modeling.
# #### 6.1.2 Hyperparameter tunning for best models
# I performed a grid search optimization for AdaBoost, ExtraTrees , RandomForest, GradientBoosting and SVC classifiers.
# I set the "n_jobs" parameter to 4 since i have 4 cpu . The computation time is clearly reduced.
# But be carefull, this step can take a long time, i took me 15 min in total on 4 cpu.
### META MODELING WITH ADABOOST, RF, EXTRATREES and GRADIENTBOOSTING
# Adaboost
DTC = DecisionTreeClassifier()
adaDTC = AdaBoostClassifier(DTC, random_state=7)
ada_param_grid = {
"base_estimator__criterion": ["gini", "entropy"],
"base_estimator__splitter": ["best", "random"],
"algorithm": ["SAMME", "SAMME.R"],
"n_estimators": [1, 2],
"learning_rate": [0.0001, 0.001, 0.01, 0.1, 0.2, 0.3, 1.5],
}
gsadaDTC = GridSearchCV(
adaDTC, param_grid=ada_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsadaDTC.fit(X_train, Y_train)
ada_best = gsadaDTC.best_estimator_
gsadaDTC.best_score_
# ExtraTrees
ExtC = ExtraTreesClassifier()
## Search grid for optimal parameters
ex_param_grid = {
"max_depth": [None],
"max_features": [1, 3, 10],
"min_samples_split": [2, 3, 10],
"min_samples_leaf": [1, 3, 10],
"bootstrap": [False],
"n_estimators": [100, 300],
"criterion": ["gini"],
}
gsExtC = GridSearchCV(
ExtC, param_grid=ex_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsExtC.fit(X_train, Y_train)
ExtC_best = gsExtC.best_estimator_
# Best score
gsExtC.best_score_
# RFC Parameters tunning
RFC = RandomForestClassifier()
## Search grid for optimal parameters
rf_param_grid = {
"max_depth": [None],
"max_features": [1, 3, 10],
"min_samples_split": [2, 3, 10],
"min_samples_leaf": [1, 3, 10],
"bootstrap": [False],
"n_estimators": [100, 300],
"criterion": ["gini"],
}
gsRFC = GridSearchCV(
RFC, param_grid=rf_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsRFC.fit(X_train, Y_train)
RFC_best = gsRFC.best_estimator_
# Best score
gsRFC.best_score_
# Gradient boosting tunning
GBC = GradientBoostingClassifier()
gb_param_grid = {
"loss": ["deviance"],
"n_estimators": [100, 200, 300],
"learning_rate": [0.1, 0.05, 0.01],
"max_depth": [4, 8],
"min_samples_leaf": [100, 150],
"max_features": [0.3, 0.1],
}
gsGBC = GridSearchCV(
GBC, param_grid=gb_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsGBC.fit(X_train, Y_train)
GBC_best = gsGBC.best_estimator_
# Best score
gsGBC.best_score_
### SVC classifier
SVMC = SVC(probability=True)
svc_param_grid = {
"kernel": ["rbf"],
"gamma": [0.001, 0.01, 0.1, 1],
"C": [1, 10, 50, 100, 200, 300, 1000],
}
gsSVMC = GridSearchCV(
SVMC, param_grid=svc_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsSVMC.fit(X_train, Y_train)
SVMC_best = gsSVMC.best_estimator_
# Best score
gsSVMC.best_score_
# #### 6.1.3 Plot learning curves
# Learning curves are a good way to see the overfitting effect on the training set and the effect of the training size on the accuracy.
def plot_learning_curve(
estimator,
title,
X,
y,
ylim=None,
cv=None,
n_jobs=-1,
train_sizes=np.linspace(0.1, 1.0, 5),
):
"""Generate a simple plot of the test and training learning curve"""
plt.figure()
plt.title(title)
if ylim is not None:
plt.ylim(*ylim)
plt.xlabel("Training examples")
plt.ylabel("Score")
train_sizes, train_scores, test_scores = learning_curve(
estimator, X, y, cv=cv, n_jobs=n_jobs, train_sizes=train_sizes
)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
plt.grid()
plt.fill_between(
train_sizes,
train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std,
alpha=0.1,
color="r",
)
plt.fill_between(
train_sizes,
test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std,
alpha=0.1,
color="g",
)
plt.plot(train_sizes, train_scores_mean, "o-", color="r", label="Training score")
plt.plot(
train_sizes, test_scores_mean, "o-", color="g", label="Cross-validation score"
)
plt.legend(loc="best")
return plt
g = plot_learning_curve(
gsRFC.best_estimator_, "RF mearning curves", X_train, Y_train, cv=kfold
)
g = plot_learning_curve(
gsExtC.best_estimator_, "ExtraTrees learning curves", X_train, Y_train, cv=kfold
)
g = plot_learning_curve(
gsSVMC.best_estimator_, "SVC learning curves", X_train, Y_train, cv=kfold
)
g = plot_learning_curve(
gsadaDTC.best_estimator_, "AdaBoost learning curves", X_train, Y_train, cv=kfold
)
g = plot_learning_curve(
gsGBC.best_estimator_,
"GradientBoosting learning curves",
X_train,
Y_train,
cv=kfold,
)
# GradientBoosting and Adaboost classifiers tend to overfit the training set. According to the growing cross-validation curves GradientBoosting and Adaboost could perform better with more training examples.
# SVC and ExtraTrees classifiers seem to better generalize the prediction since the training and cross-validation curves are close together.
# #### 6.1.4 Feature importance of tree based classifiers
# In order to see the most informative features for the prediction of passengers survival, i displayed the feature importance for the 4 tree based classifiers.
nrows = ncols = 2
fig, axes = plt.subplots(nrows=nrows, ncols=ncols, sharex="all", figsize=(15, 15))
names_classifiers = [
("AdaBoosting", ada_best),
("ExtraTrees", ExtC_best),
("RandomForest", RFC_best),
("GradientBoosting", GBC_best),
]
nclassifier = 0
for row in range(nrows):
for col in range(ncols):
name = names_classifiers[nclassifier][0]
classifier = names_classifiers[nclassifier][1]
indices = np.argsort(classifier.feature_importances_)[::-1][:40]
g = sns.barplot(
y=X_train.columns[indices][:40],
x=classifier.feature_importances_[indices][:40],
orient="h",
ax=axes[row][col],
)
g.set_xlabel("Relative importance", fontsize=12)
g.set_ylabel("Features", fontsize=12)
g.tick_params(labelsize=9)
g.set_title(name + " feature importance")
nclassifier += 1
# I plot the feature importance for the 4 tree based classifiers (Adaboost, ExtraTrees, RandomForest and GradientBoosting).
# We note that the four classifiers have different top features according to the relative importance. It means that their predictions are not based on the same features. Nevertheless, they share some common important features for the classification , for example 'Fare', 'Title_2', 'Age' and 'Sex'.
# Title_2 which indicates the Mrs/Mlle/Mme/Miss/Ms category is highly correlated with Sex.
# We can say that:
# - Pc_1, Pc_2, Pc_3 and Fare refer to the general social standing of passengers.
# - Sex and Title_2 (Mrs/Mlle/Mme/Miss/Ms) and Title_3 (Mr) refer to the gender.
# - Age and Title_1 (Master) refer to the age of passengers.
# - Fsize, LargeF, MedF, Single refer to the size of the passenger family.
# **According to the feature importance of this 4 classifiers, the prediction of the survival seems to be more associated with the Age, the Sex, the family size and the social standing of the passengers more than the location in the boat.**
test_Survived_RFC = pd.Series(RFC_best.predict(test), name="RFC")
test_Survived_ExtC = pd.Series(ExtC_best.predict(test), name="ExtC")
test_Survived_SVMC = pd.Series(SVMC_best.predict(test), name="SVC")
test_Survived_AdaC = pd.Series(ada_best.predict(test), name="Ada")
test_Survived_GBC = pd.Series(GBC_best.predict(test), name="GBC")
# Concatenate all classifier results
ensemble_results = pd.concat(
[
test_Survived_RFC,
test_Survived_ExtC,
test_Survived_AdaC,
test_Survived_GBC,
test_Survived_SVMC,
],
axis=1,
)
g = sns.heatmap(ensemble_results.corr(), annot=True)
# The prediction seems to be quite similar for the 5 classifiers except when Adaboost is compared to the others classifiers.
# The 5 classifiers give more or less the same prediction but there is some differences. Theses differences between the 5 classifier predictions are sufficient to consider an ensembling vote.
# ### 6.2 Ensemble modeling
# #### 6.2.1 Combining models
# I choosed a voting classifier to combine the predictions coming from the 5 classifiers.
# I preferred to pass the argument "soft" to the voting parameter to take into account the probability of each vote.
votingC = VotingClassifier(
estimators=[
("rfc", RFC_best),
("extc", ExtC_best),
("svc", SVMC_best),
("adac", ada_best),
("gbc", GBC_best),
],
voting="soft",
n_jobs=4,
)
votingC = votingC.fit(X_train, Y_train)
# ### 6.3 Prediction
# #### 6.3.1 Predict and Submit results
test_Survived = pd.Series(votingC.predict(test), name="Survived")
results = pd.concat([IDtest, test_Survived], axis=1)
results.to_csv("ensemble_python_voting.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/136/69136143.ipynb
|
output
|
shravanbangera
|
[{"Id": 69136143, "ScriptId": 18764016, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5052699, "CreationDate": "07/27/2021 09:16:45", "VersionNumber": 3.0, "Title": "Titanic Data analysis using ensemble modeling", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 943.0, "LinesInsertedFromPrevious": 179.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 764.0, "LinesInsertedFromFork": 454.0, "LinesDeletedFromFork": 327.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 489.0, "TotalVotes": 0}]
|
[{"Id": 91954390, "KernelVersionId": 69136143, "SourceDatasetVersionId": 1514005}]
|
[{"Id": 1514005, "DatasetId": 892201, "DatasourceVersionId": 1548266, "CreatorUserId": 4332881, "LicenseName": "Other (specified in description)", "CreationDate": "09/25/2020 05:28:42", "VersionNumber": 1.0, "Title": "Titanic Dataset", "Slug": "output", "Subtitle": "Titanic Shipwreck Dataset", "Description": "### Context\n\nTitanic Shipwreck\n\n\n### Content\n\nPredicting the survival rate of titanic shipwreck\n\n\n### Acknowledgements\n\nKaggle\n\n\n### Inspiration\n\nHow to avoid such a disaster in future?", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 892201, "CreatorUserId": 4332881, "OwnerUserId": 4332881.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1514005.0, "CurrentDatasourceVersionId": 1548266.0, "ForumId": 907776, "Type": 2, "CreationDate": "09/25/2020 05:28:42", "LastActivityDate": "09/25/2020", "TotalViews": 3132, "TotalDownloads": 219, "TotalVotes": 12, "TotalKernels": 1}]
|
[{"Id": 4332881, "UserName": "shravanbangera", "DisplayName": "Shravan Bangera", "RegisterDate": "01/15/2020", "PerformanceTier": 1}]
|
# # Titanic Top 4% with ensemble modeling
# * **1 소개**
# * **2 데이터 로드 및 체크**
# * 2.1 데이터 로드
# * 2.2 이상치 감지
# * 2.3 학습 및 테스트 데이터 세트 조인
# * 2.4 널 및 결측치
# * **3 피처 분석**
# * 3.1 수치형 값
# * 3.2 범주형 값
# * **4 결측치 처리**
# * 4.1 Age
# * **5 피처 엔지니어링**
# * 5.1 Name/Title
# * 5.2 Family Size
# * 5.3 Cabin
# * 5.4 Ticket
# * **6 모델링**
# * 6.1 단순 ML모델
# * 6.1.1 교차 검증 모델
# * 6.1.2 베스트모델에 대한 하이터 파라미터 튜닝
# * 6.1.3 학습곡선 폴롯
# * 6.1.4 트리기반의 분류기에 대한 피처 중요도
# * 6.2 앙상블 모델링
# * 6.2.1 모델 조합
# * 6.3 예측
# * 6.3.1 예측 및 결과 예측
#
# ## 1. 소개
# 캐글에서 세번째로 타이타닉 관련 데이터셋을 가지고 다음과 같은 방법으로 진행하려고 한다.
# No matter what you're going through, there's a light at the end of the tunnel.
# It may seem hard to get to it, but you can do it.
# Just keep working towards to it and you will find the positive side of things.
# [Demi Lovato]
# * **피처 분석**
# * **피처 엔지니어링**
# * **모델링**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from collections import Counter
from sklearn.ensemble import (
RandomForestClassifier,
AdaBoostClassifier,
GradientBoostingClassifier,
ExtraTreesClassifier,
VotingClassifier,
)
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis # LDA
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.svm import SVC
from sklearn.model_selection import (
GridSearchCV,
cross_val_score,
StratifiedKFold,
learning_curve,
)
sns.set(style="white", context="notebook", palette="deep", font_scale=2)
plt.style.use("seaborn")
# ## 2.데이터 로드 및 데이터 확인
# ### 2.1 데이터 로드
def load_dataset():
train = pd.read_csv("../input/titanic/train.csv")
test = pd.read_csv("../input/titanic/test.csv")
IDTEST = test["PassengerId"] # 결과물제출시 PassengerId
# Shape및 데이터보기
display(train.shape, test.shape)
display(train.head(n=2), display(test.tail(n=2)))
return train, test, IDTEST
# 수행
train, test, IDTEST = load_dataset()
# ### 2.2 이상치 감지
def detect_outliers(df, features, n):
"""
df - DataFrame
features - feature
n - 이상치값 필터링에 사용되는 변수값
"""
outlier_index = []
for col in features:
# 25%
Q1 = np.percentile(df[col], 25)
Q3 = np.percentile(df[col], 75)
# IQR(Interquartile range)
IQR = Q3 - Q1
# Outline_step
outlier_step = 1.5 * IQR
outlier_df = df[(df[col] < Q1 - outlier_step) | (df[col] > Q3 + outlier_step)]
display("이상치 : \n", outlier_df, outlier_df.shape)
outlier_list_col = outlier_df.index
outlier_index.extend(outlier_list_col)
outlier_index = Counter(outlier_index)
multiple_outliers = list(k for k, v in outlier_index.items() if v > n)
return multiple_outliers
# 이상치탐지
multiple_outliers = detect_outliers(train, ["Age", "SibSp", "Parch", "Fare"], 2)
# 이상치(Outlier)는 데이터의 통계치를 왜곡시키기 때문에 때론, 아웃라이어를 통계치에 제외시키기도 한다.
# 1977년도 Turjey, JW가 발표한 Turkey Method에 방법에 의해 아웃라이어를 찾기위해 IQR에 outlier step만큼 더하거나 뺀 값이 이상치이다.
def remove_outliers(df, outlier_pos):
"""
이상치의 데이터프레임에서 인덱스를 매개변수로 받아
해당 데이터를 삭제
"""
display("이상치 삭제전 : ", df.shape)
# show he outlier rows
display(df.iloc[outlier_pos])
# Drop outlier
df = df.drop(outlier_pos, inplace=False, axis=0, errors="ignore").reset_index(
drop=True
)
# 삭제후
display("이상치 삭제 후 ", df.shape)
return df
# Remove outlier
# 10개의 아웃라이어
train = remove_outliers(train, multiple_outliers)
# 10개의 이상치 데이터가 발견되었으면 , 28번, 89번 그리고 342번의 승객의 요금이 비정상적으로 높음.
# 나머지 7개 로우는 매우 높은 SibSp값을 갖고 있다.
# ### 2.3 학습 데이터셋트와 테스트 데이터세트 조인
def get_join_dataset(train, test):
"""
학습 및 테스트 데이터셋을 병합한 데이터셋
"""
display(train.shape, test.shape)
dataset = pd.concat(objs=[train, test], axis=0).reset_index(drop=True)
display("\n 병합된 데이터셋 ", dataset.shape)
display("\n Dataset Info ", dataset.info())
return dataset
# 수행
dataset = get_join_dataset(train, test)
# ### 2.4 널값과 결측치 확인
def check_null_missing_val(df, trainT=False):
"""
널값과 결측치 확인
trainT 파라미터는 True인 경우는 학습데이터 확인용
"""
if trainT:
# Fill empty and NaNs values with NaN
df = df.fillna(np.nan)
# Check for Null values
display(df.isnull().sum())
# 데이터 타입
display(df.dtypes, type(df.dtypes), df.dtypes.to_list())
print("\n")
display(df.info())
print("\n Technical Statistics")
display(df.describe())
print("\n Top 5")
display(df.head(n=5))
return df
dataset = check_null_missing_val(dataset)
# train데이터셋 확인
train = check_null_missing_val(train, trainT=True)
# Age및 Cabin피처는 결측치 처리를 위해서 중요한 역할을 할 수 있다.
# ## 3. 피처 분석
# ### 3.1 숫자형 값
int_cols = train.dtypes[train.dtypes != "object"]
int_cols.drop(index=["PassengerId", "Pclass"], inplace=True)
def extract_num_feature(df, objType=None):
"""
데이터프레임을 입력받아 데이터타입을 필터링하는 해당
피처만 리스트타입으로 리턴
리턴값은 pd.Series
"""
int_cols = df.dtypes[df.dtypes != objType]
int_cols.drop(index=["PassengerId", "Pclass"], inplace=True)
return int_cols
int_cols = extract_num_feature(train, "object")
int_cols.index
# Correlation matrix between numerical values (SibSp Parch Age and Fare values) and Survived
g = sns.heatmap(train[int_cols.index].corr(), annot=True, fmt=".2f", cmap="coolwarm_r")
# Fare피처가 생존률과는 의미있는 상관관계를 가지고있습니다.
# 그렇다고 해서 다른 피처들이 전혀 필요없다는 의미는 아닙니다. 기존 피처를 이용해 새롭게 파생변수를 생성해보면 상관관계를 새롭게 도출되는 경우도 있습니다. 그런면에서 다른 피처들은 또다른 의미를 갖습니다.
# "호랑이는 죽어서 가죽을 남기고 사람은 죽어서 이름을 남긴다."
# 피처의 소멸 자체도 의미있다는 말씀이지요.
# #### SibSP
# * [seaborn - factorplot](https://www.kite.com/python/docs/seaborn.factorplot)
# * [seaborn - style 사용자 정의](http://hleecaster.com/python-seaborn-set-style-and-context/)
g = sns.factorplot(
x="SibSp", y="Survived", data=train, kind="bar", size=6, palette="muted"
)
g.set_xlabels("SibSp")
g.despine(left=True)
g.set_ylabels("Survival Probability")
# 동반가족이 2명보다 많은 경우는 생존률이 많이 낮아짐을 알수 있다.
# 피처 엔지니어링을 통해서 새로운 피처를 추가적으로 도출이 가능하다.
# #### Parch
g = sns.factorplot(x="Parch", y="Survived", data=train, palette="summer_r", kind="bar")
g.despine(left=True)
g.set_xlabels("Parch")
g.set_ylabels("Survival Possibility")
# 사이즈가 작은 이동이 생존률이 훨씬 좋은 것을 알수있다.
# 동반가족이 3인인 경우, 생존율의 표준편차가 심한 것을 알수 있다.
# #### Age
# 히스토그램과 kde plot이 같이.
g = sns.FacetGrid(train, col="Survived")
g = g.map(sns.distplot, "Age")
# 연령대를 구분할 필요가 있어보임.연령에 대한 사망과 생존의 차이는 크게 두드러지지는 않지만, 연령대가 뒤로 갈수록 사망과 생존의 구별은 확연히 드러날것으로 보임
cond_0 = (train["Survived"] == 0) & (train["Age"].notnull()) # Not Survived
cond_1 = (train["Survived"] == 1) & (train["Age"].notnull()) # Survived
g = sns.kdeplot(train["Age"][cond_0], color="red", shade=True)
g = sns.kdeplot(train["Age"][cond_1], color="Blue", shade=True, ax=g)
g.set_xlabel("Age")
g.set_ylabel("Frequency")
g = g.legend(["Not Survived", "Survived"])
# When we superimpose the two densities , we cleary see a peak correponsing (between 0 and 5) to babies and very young childrens.
# #### Fare
def show_dist_plot(df, col: str):
"""
해당 데이터프레임의 피처에 분포 및 kde plot을 시각화
"""
g = sns.distplot(
df[col], color="b", label="Skewness : {:.2f}".format(df[col].skew())
)
g.set_title("Skewness for {0}".format(col))
g = g.legend(loc="best")
num_cols = int_cols[1:].index.values
# 학습 및 테스트 데이터셋 조인한 데이터셋
dataset[num_cols].isnull().sum()
# Fill Fare missing values with the median value
dataset["Fare"] = dataset["Fare"].fillna(dataset["Fare"].median())
# Recheck
dataset["Fare"].isnull().sum()
# * Check the skewness of Fare feature
# before logarithm
show_dist_plot(dataset, "Fare")
# As we can see, Fare distribution is very skewed. This can lead to overweigth very high values in the model, even if it is scaled.
# 위의 그래프처럼 , Fare band가 매우 편향되어있음을 알수 있다. 이런 데이터를 그냥 모델에 학습시키면 Min/Max Scaler혹은 Standard Scaler로 스케일링이 되어 있다 하더락도 과적합되기 마련이다.
# 그러므로 이런 편향된 데이터의 경우엔, 로그변환을 적용해야 한다.
# np.log1p변환과 np.log()변환을 같이 한다면 이때의 편향정도는?
# What if we do both np.log1p and np.log ?
# * Logarithm transformation
dataset["Fare"] = dataset["Fare"].apply(lambda x: np.log1p(x))
# Apply log to Fare to reduce skewness distribution
dataset["Fare"] = dataset["Fare"].map(lambda i: np.log(i) if i > 0 else 0)
# After Logarithm
show_dist_plot(dataset, "Fare")
# 로그변환 후에 편향정도가 상당히 감소했음을 알 수 있다
# ### 3.2 범주형 피처의 값
def show_factor_plot(df, xCol=None, yCol=None, col=None, kind="bar", hue=None):
"""
피처별 생존률 비교
"""
g = sns.factorplot(
x=xCol, y=yCol, hue=hue, col=col, kind=kind, palette="muted", size=6, data=df
)
sns.despine(left=True)
g.set_ylabels(yCol)
# #### Sex
# * bar plot으로 성별 생존률
# * Sex로 Groupby하여 Survived의 mean()값
g = sns.barplot(x="Sex", y="Survived", data=train)
g = g.set_ylabel("Survivor Probability")
train[["Sex", "Survived"]].groupby("Sex").mean()
# 사고가 났을때 100명중에 20명정도는 남자는 살고, 여자는 75명 이상은 산다고 나타난다
# 성별 피처는 생존률을 예측할때 정말 중요한 피처라고 할 수 있다
# 1997년 타이타닉 영화가 나왔을때 영화대사 중에 이런 구절이 나온다.
# : "여자와 아이들 먼저 구조해".
# 세월호 침몰 사고때는 어떠했는가?
# #### Pclass
# Explore Pclass vs Survived
show_factor_plot(train, "Pclass", "Survived")
# Explore Pclass vs Survived by Sex
show_factor_plot(train, "Pclass", "Survived", hue="Sex")
# * 좌석별 여성 및 남성 비교
dataset[(dataset["Pclass"] == 3)].groupby("Sex")["Pclass"].count()
# 승객들의 생존률은 3등급 객실이라고 해서 다르지 않다. 1등급 객실은 2등급, 3등급 객실보다는 생존률이 높다.
# #### Embarked
dataset["Embarked"].isnull().sum()
# 사우스햄튼이 가장 승객이 많다.
dataset["Embarked"].value_counts()
# Fill Embarked nan values of dataset set with 'S' most frequent value
dataset["Embarked"] = dataset["Embarked"].fillna("S")
# Recheck
dataset["Embarked"].isnull().sum()
# Explore Embarked vs Survived
show_factor_plot(train, xCol="Embarked", yCol="Survived")
# 프랑스 노르망디 해안에 위치한 체르보르그항에서 탑승한 승객들의 생존률이 다른 사우스햄튼 혹은 퀸즈타운보다 훨씬 생존률이 좋음을 알 수 있다.
# 그렇다면, 1등급 객실에 묵은 승객들의 비율이 다른 경유지 항구 - 체르보르그, 사우스햄튼, 퀸즈타운에서 온 승객들보다 더 많을 것이다.
# 경유지항구(Embarked) v.s Pclass(객실등급)
# Explore Pclass vs Embarked
show_factor_plot(train, xCol="Pclass", col="Embarked", kind="count")
# 사우스햄튼과 퀸즈타운의 승객들의 대부분은 3등급 객실에 분포되어 있으며, 추론은 이 사망자의 대부분은 퀸즈타운과 사우스햄튼 탑승객 일 것이다.
# 반면 체르보르그 탑승객들은 1등급이 가장 많다.
# 1등급 객실의 생존률이 높은 이유는?
# -> 사회적 영향도 인가, 아니면 구조가 용이한 여객선의 구조적 문제인가?
# ## 4. 결측치 채우기
# ### 4.1 Age
# 앞에서 보듯히, Age피처는 256개의 결측치를 포함하고있다.
# Age가 속한 연령대에 따라 생존률이 극명하게 갈리는 부분이 있기 때문에 중요한 피처라고 할 수 있으며, 이를 위해서라도 결측치를 대치할 부분이 필요하다.
# 이를 위해서는 Sex, Parch, Pclass 그리고 SibSp를 이용해 새로운 피처를 생성하는 문제를 생각해보자
# Explore Age vs Sex, Parch , Pclass and SibSP
g = sns.factorplot(y="Age", x="Sex", data=dataset, kind="box")
g = sns.factorplot(y="Age", x="Sex", hue="Pclass", data=dataset, kind="box")
g = sns.factorplot(y="Age", x="Parch", data=dataset, kind="box")
g = sns.factorplot(y="Age", x="SibSp", data=dataset, kind="box")
# 성별이 연령대를 예측할 수 있는 결정인자는 아니다.
# 그러나 1등급 객실에 있는 승객의 나이가 다른 2등급 객실, 3등급 객실의 승객들보다 연령대가 높다는 점은 주목 할 만하다.
# 부모 및 아이들을 동반한 경우가 자식,배우자를 동반한 경우보다 연령대가 높다는 것을 알 수 있다.
# convert Sex into categoricl value 0 for male and 1 for female
dataset["Sex"] = dataset["Sex"].map({"male": 0, "female": 1})
# 위의 방법과 동일
dataset["Sex"] = dataset["Sex"].apply(lambda x: 0 if x == "male" else 1)
# 앞에서 한번 np.log1p()로 로그변환 한 데이터를 다시 밑에것 np.logp()를 수행하면
# 히트맵에서 이상하게 보임
df = dataset[["Sex", "Age", "SibSp", "Parch", "Pclass"]]
g = sns.heatmap(df.corr(), cmap="BrBG", annot=True)
# 상관관계 맵은 Parch 피처를 제외한 factorplot관측치를 확인합니다.
# Age피처가 Sex랑은 관련성이 떨어지고, Pclass, Parch 그리고 SibSp와 음의 상관관계를 가지고있습니다.
# Age피처가 부모 및 아이들의 수에 따라 증가하는 경향은 있지만, 전반적으로는 음의 상관관계입니다.
# 그래서 Age 결측치를 대치하기 위해 SibSp, Parch 그리고 Pclass를 사용하기로 합니다.(결측치 대치는 median)
def fill_missing_value_age(df, idx_nan_age: list):
"""
Age가 널인것들을 리스트 파라미터로 전달해 중앙값으로 대치함.
Filling missing value of Age
Fill Age with the median age of similar rows according to Pclass, Parch and SibSp
"""
for i in idx_nan_age:
age_med = df["Age"].median()
age_pred = df["Age"][
(
(df["SibSp"] == df.iloc[i]["SibSp"])
& (df["Parch"] == df.iloc[i]["Parch"])
& (df["Pclass"] == df.iloc[i]["Pclass"])
)
].median()
if not np.isnan(age_pred):
df["Age"].iloc[i] = age_pred
else:
df["Age"].iloc[i] = age_med
return df
index_NaN_age = list(dataset["Age"].isnull().index)
fill_missing_value_age(dataset, index_NaN_age)
# * [box v.s violin] (https://junklee.tistory.com/9)
show_factor_plot(train, xCol="Survived", yCol="Age", kind="box")
show_factor_plot(train, xCol="Survived", yCol="Age", kind="violin")
# 사망자와 생존자의 Age의 median값도 크게 차이가 없다.
# violin plot을 보면 나이가 어린 영역대의 생존률이 상당히 높음을 알 수 있다.
# ## 5. 피처 엔지니어링
def feature_engineering(df, titleOpt=False, familyOpt=False):
"""
1.Title에 대한 정제작업
"""
if titleOpt:
df["Title"].replace(
titleList,
[
"Mr",
"Miss",
"Mrs",
"Master",
"Mr",
"Other",
"Other",
"Other",
"Miss",
"Miss",
"Mr",
"Other",
"Mr",
"Miss",
"Other",
"Other",
"Mrs",
"Mr",
],
inplace=True,
)
df["Title"].replace(
["Mr", "Mrs", "Miss", "Master", "Other"], [0, 1, 2, 3, 4], inplace=True
)
df["Title"].astype(int)
# Drop Name variable
df.drop(labels=["Name"], axis=0, inplace=True, errors="ignore")
if familyOpt:
df["FamilySize"] = df["SibSp"] + df["Parch"] + 1
# 피처 추가 후 기존 컬럼 삭제
df.drop(labels=["SibSp", "Parch"], axis=0, inplace=True, errors="ignore")
display(df.head(n=2))
display(df.tail(n=2))
return df
# ### 5.1 Name/Title
display(dataset["Name"].head())
display(dataset["Name"].tail())
# The Name feature contains information on passenger's title.
# Since some passenger with distingused title may be preferred during the evacuation, it is interesting to add them to the model.
def get_TitleFromName(df, feature):
"""
이름에서 존칭 분리
"""
dataset_title = [i.split(",")[1].split(".")[0] for i in df[feature]]
display(dataset_title[1:10])
df["Title"] = pd.Series(dataset_title)
display(df.head(n=2))
return df
# Get Title from Name
dataset = get_TitleFromName(dataset, "Name")
g = sns.countplot(x="Title", data=dataset)
g = plt.setp(g.get_xticklabels(), rotation=45)
# There is 17 titles in the dataset, most of them are very rare and we can group them in 4 categories.
titleList = dataset["Title"].value_counts().index.values.tolist()
dataset = feature_engineering(dataset, titleOpt=True)
g = sns.countplot(dataset["Title"])
g = g.set_xticklabels(["Mr", "Mrs", "Miss", "Master", "Other"])
g = sns.factorplot(x="Title", y="Survived", data=dataset, kind="bar")
g = g.set_xticklabels(["Mr", "Mrs", "Miss", "Master", "Other"])
g = g.set_ylabels("Survival probability for Title")
# "여자와 아이먼저 구조"
# "Other" 타이틀이 승선객의 Title구성을 볼때 생존률이 높다.
# ### 5.2 Family size
# * 가족수(FamilySize)피처 추가 - SibSp(동반가족(자손 + 배우자)) , Parch(부모님 동반된 아이들 포함)
# Create a family size descriptor from SibSp and Parch
dataset = feature_engineering(dataset, familyOpt=True)
g = sns.factorplot(x="FamilySize", y="Survived", data=dataset)
g = g.set_ylabels("Survival Probability for FamilySize")
# The family size seems to play an important role, survival probability is worst for large families.
# Additionally, i decided to created 4 categories of family size.
dataset["FamilySize"].value_counts()
dataset["Single"] = dataset["FamilySize"].map(lambda s: 1 if s == 1 else 0)
dataset["SmallF"] = dataset["FamilySize"].map(lambda s: 1 if s == 2 else 0)
dataset["MedF"] = dataset["FamilySize"].map(lambda s: 1 if 3 <= s <= 4 else 0)
dataset["LargeF"] = dataset["FamilySize"].map(lambda s: 1 if s >= 5 else 0)
def show_factorplot(df, cols):
"""
데이터셋과 컬럼에 대한 정보를 받아서
Factotplot으로 피처별 생존률을 보여줌
"""
for i in range(len(cols)):
g = sns.factorplot(x=cols[i], y="Survived", data=dataset, kind="bar")
g = g.set_ylabels("Survival Probabilit for {0}".format(cols[i]))
# 피처별 생존률
cols = ["Single", "SmallF", "MedF", "LargeF"]
show_factorplot(dataset, cols)
# 2인, 3인, 4인정도의 인원으로 가족끼리 탑승한 경우 생존률이 1인 혹은 5인 이상의 가족 및 친지끼리 승선한 경우보다 생존률이 좋다는 점은 기억하자.
# 혼자보다 둘이, 둘보다는 세명이서 여행가라. 그러면 사고당해서 돌아오지 못할 확률이 줄어든다.
# convert to indicator values Title and Embarked
dataset = pd.get_dummies(dataset, columns=["Title"])
dataset = pd.get_dummies(dataset, columns=["Embarked"], prefix="Em")
dataset.head()
# At this stage, we have 22 features.
# ### 5.3 Cabin
dataset["Cabin"].head()
dataset["Cabin"].describe()
dataset["Cabin"].isnull().sum()
# The Cabin feature column contains 292 values and 1007 missing values.
# I supposed that passengers without a cabin have a missing value displayed instead of the cabin number.
dataset["Cabin"][dataset["Cabin"].notnull()].head()
# Replace the Cabin number by the type of cabin 'X' if not
dataset["Cabin"] = pd.Series(
[i[0] if not pd.isnull(i) else "X" for i in dataset["Cabin"]]
)
# The first letter of the cabin indicates the Desk, i choosed to keep this information only, since it indicates the probable location of the passenger in the Titanic.
g = sns.countplot(dataset["Cabin"], order=["A", "B", "C", "D", "E", "F", "G", "T", "X"])
g = sns.factorplot(
y="Survived",
x="Cabin",
data=dataset,
kind="bar",
order=["A", "B", "C", "D", "E", "F", "G", "T", "X"],
)
g = g.set_ylabels("Survival Probability")
# Because of the low number of passenger that have a cabin, survival probabilities have an important standard deviation and we can't distinguish between survival probability of passengers in the different desks.
# But we can see that passengers with a cabin have generally more chance to survive than passengers without (X).
# It is particularly true for cabin B, C, D, E and F.
dataset = pd.get_dummies(dataset, columns=["Cabin"], prefix="Cabin")
# ### 5.4 Ticket
dataset["Ticket"].head()
# It could mean that tickets sharing the same prefixes could be booked for cabins placed together. It could therefore lead to the actual placement of the cabins within the ship.
# Tickets with same prefixes may have a similar class and survival.
# So i decided to replace the Ticket feature column by the ticket prefixe. Which may be more informative.
## Treat Ticket by extracting the ticket prefix. When there is no prefix it returns X.
Ticket = []
for i in list(dataset.Ticket):
if not i.isdigit():
Ticket.append(
i.replace(".", "").replace("/", "").strip().split(" ")[0]
) # Take prefix
else:
Ticket.append("X")
dataset["Ticket"] = Ticket
dataset["Ticket"].head()
dataset = pd.get_dummies(dataset, columns=["Ticket"], prefix="T")
# Create categorical values for Pclass
dataset["Pclass"] = dataset["Pclass"].astype("category")
dataset = pd.get_dummies(dataset, columns=["Pclass"], prefix="Pc")
# Drop useless variables
dataset.drop(labels=["PassengerId"], axis=1, inplace=True)
dataset.head()
# ## 6. MODELING
## Separate train dataset and test dataset
train = dataset[:train_len]
test = dataset[train_len:]
test.drop(labels=["Survived"], axis=1, inplace=True)
## Separate train features and label
train["Survived"] = train["Survived"].astype(int)
Y_train = train["Survived"]
X_train = train.drop(labels=["Survived"], axis=1)
# ### 6.1 Simple modeling
# #### 6.1.1 Cross validate models
# I compared 10 popular classifiers and evaluate the mean accuracy of each of them by a stratified kfold cross validation procedure.
# * SVC
# * Decision Tree
# * AdaBoost
# * Random Forest
# * Extra Trees
# * Gradient Boosting
# * Multiple layer perceprton (neural network)
# * KNN
# * Logistic regression
# * Linear Discriminant Analysis
# Cross validate model with Kfold stratified cross val
kfold = StratifiedKFold(n_splits=10)
# Modeling step Test differents algorithms
random_state = 2
classifiers = []
classifiers.append(SVC(random_state=random_state))
classifiers.append(DecisionTreeClassifier(random_state=random_state))
classifiers.append(
AdaBoostClassifier(
DecisionTreeClassifier(random_state=random_state),
random_state=random_state,
learning_rate=0.1,
)
)
classifiers.append(RandomForestClassifier(random_state=random_state))
classifiers.append(ExtraTreesClassifier(random_state=random_state))
classifiers.append(GradientBoostingClassifier(random_state=random_state))
classifiers.append(MLPClassifier(random_state=random_state))
classifiers.append(KNeighborsClassifier())
classifiers.append(LogisticRegression(random_state=random_state))
classifiers.append(LinearDiscriminantAnalysis())
cv_results = []
for classifier in classifiers:
cv_results.append(
cross_val_score(
classifier, X_train, y=Y_train, scoring="accuracy", cv=kfold, n_jobs=4
)
)
cv_means = []
cv_std = []
for cv_result in cv_results:
cv_means.append(cv_result.mean())
cv_std.append(cv_result.std())
cv_res = pd.DataFrame(
{
"CrossValMeans": cv_means,
"CrossValerrors": cv_std,
"Algorithm": [
"SVC",
"DecisionTree",
"AdaBoost",
"RandomForest",
"ExtraTrees",
"GradientBoosting",
"MultipleLayerPerceptron",
"KNeighboors",
"LogisticRegression",
"LinearDiscriminantAnalysis",
],
}
)
g = sns.barplot(
"CrossValMeans",
"Algorithm",
data=cv_res,
palette="Set3",
orient="h",
**{"xerr": cv_std}
)
g.set_xlabel("Mean Accuracy")
g = g.set_title("Cross validation scores")
# I decided to choose the SVC, AdaBoost, RandomForest , ExtraTrees and the GradientBoosting classifiers for the ensemble modeling.
# #### 6.1.2 Hyperparameter tunning for best models
# I performed a grid search optimization for AdaBoost, ExtraTrees , RandomForest, GradientBoosting and SVC classifiers.
# I set the "n_jobs" parameter to 4 since i have 4 cpu . The computation time is clearly reduced.
# But be carefull, this step can take a long time, i took me 15 min in total on 4 cpu.
### META MODELING WITH ADABOOST, RF, EXTRATREES and GRADIENTBOOSTING
# Adaboost
DTC = DecisionTreeClassifier()
adaDTC = AdaBoostClassifier(DTC, random_state=7)
ada_param_grid = {
"base_estimator__criterion": ["gini", "entropy"],
"base_estimator__splitter": ["best", "random"],
"algorithm": ["SAMME", "SAMME.R"],
"n_estimators": [1, 2],
"learning_rate": [0.0001, 0.001, 0.01, 0.1, 0.2, 0.3, 1.5],
}
gsadaDTC = GridSearchCV(
adaDTC, param_grid=ada_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsadaDTC.fit(X_train, Y_train)
ada_best = gsadaDTC.best_estimator_
gsadaDTC.best_score_
# ExtraTrees
ExtC = ExtraTreesClassifier()
## Search grid for optimal parameters
ex_param_grid = {
"max_depth": [None],
"max_features": [1, 3, 10],
"min_samples_split": [2, 3, 10],
"min_samples_leaf": [1, 3, 10],
"bootstrap": [False],
"n_estimators": [100, 300],
"criterion": ["gini"],
}
gsExtC = GridSearchCV(
ExtC, param_grid=ex_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsExtC.fit(X_train, Y_train)
ExtC_best = gsExtC.best_estimator_
# Best score
gsExtC.best_score_
# RFC Parameters tunning
RFC = RandomForestClassifier()
## Search grid for optimal parameters
rf_param_grid = {
"max_depth": [None],
"max_features": [1, 3, 10],
"min_samples_split": [2, 3, 10],
"min_samples_leaf": [1, 3, 10],
"bootstrap": [False],
"n_estimators": [100, 300],
"criterion": ["gini"],
}
gsRFC = GridSearchCV(
RFC, param_grid=rf_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsRFC.fit(X_train, Y_train)
RFC_best = gsRFC.best_estimator_
# Best score
gsRFC.best_score_
# Gradient boosting tunning
GBC = GradientBoostingClassifier()
gb_param_grid = {
"loss": ["deviance"],
"n_estimators": [100, 200, 300],
"learning_rate": [0.1, 0.05, 0.01],
"max_depth": [4, 8],
"min_samples_leaf": [100, 150],
"max_features": [0.3, 0.1],
}
gsGBC = GridSearchCV(
GBC, param_grid=gb_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsGBC.fit(X_train, Y_train)
GBC_best = gsGBC.best_estimator_
# Best score
gsGBC.best_score_
### SVC classifier
SVMC = SVC(probability=True)
svc_param_grid = {
"kernel": ["rbf"],
"gamma": [0.001, 0.01, 0.1, 1],
"C": [1, 10, 50, 100, 200, 300, 1000],
}
gsSVMC = GridSearchCV(
SVMC, param_grid=svc_param_grid, cv=kfold, scoring="accuracy", n_jobs=4, verbose=1
)
gsSVMC.fit(X_train, Y_train)
SVMC_best = gsSVMC.best_estimator_
# Best score
gsSVMC.best_score_
# #### 6.1.3 Plot learning curves
# Learning curves are a good way to see the overfitting effect on the training set and the effect of the training size on the accuracy.
def plot_learning_curve(
estimator,
title,
X,
y,
ylim=None,
cv=None,
n_jobs=-1,
train_sizes=np.linspace(0.1, 1.0, 5),
):
"""Generate a simple plot of the test and training learning curve"""
plt.figure()
plt.title(title)
if ylim is not None:
plt.ylim(*ylim)
plt.xlabel("Training examples")
plt.ylabel("Score")
train_sizes, train_scores, test_scores = learning_curve(
estimator, X, y, cv=cv, n_jobs=n_jobs, train_sizes=train_sizes
)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
plt.grid()
plt.fill_between(
train_sizes,
train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std,
alpha=0.1,
color="r",
)
plt.fill_between(
train_sizes,
test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std,
alpha=0.1,
color="g",
)
plt.plot(train_sizes, train_scores_mean, "o-", color="r", label="Training score")
plt.plot(
train_sizes, test_scores_mean, "o-", color="g", label="Cross-validation score"
)
plt.legend(loc="best")
return plt
g = plot_learning_curve(
gsRFC.best_estimator_, "RF mearning curves", X_train, Y_train, cv=kfold
)
g = plot_learning_curve(
gsExtC.best_estimator_, "ExtraTrees learning curves", X_train, Y_train, cv=kfold
)
g = plot_learning_curve(
gsSVMC.best_estimator_, "SVC learning curves", X_train, Y_train, cv=kfold
)
g = plot_learning_curve(
gsadaDTC.best_estimator_, "AdaBoost learning curves", X_train, Y_train, cv=kfold
)
g = plot_learning_curve(
gsGBC.best_estimator_,
"GradientBoosting learning curves",
X_train,
Y_train,
cv=kfold,
)
# GradientBoosting and Adaboost classifiers tend to overfit the training set. According to the growing cross-validation curves GradientBoosting and Adaboost could perform better with more training examples.
# SVC and ExtraTrees classifiers seem to better generalize the prediction since the training and cross-validation curves are close together.
# #### 6.1.4 Feature importance of tree based classifiers
# In order to see the most informative features for the prediction of passengers survival, i displayed the feature importance for the 4 tree based classifiers.
nrows = ncols = 2
fig, axes = plt.subplots(nrows=nrows, ncols=ncols, sharex="all", figsize=(15, 15))
names_classifiers = [
("AdaBoosting", ada_best),
("ExtraTrees", ExtC_best),
("RandomForest", RFC_best),
("GradientBoosting", GBC_best),
]
nclassifier = 0
for row in range(nrows):
for col in range(ncols):
name = names_classifiers[nclassifier][0]
classifier = names_classifiers[nclassifier][1]
indices = np.argsort(classifier.feature_importances_)[::-1][:40]
g = sns.barplot(
y=X_train.columns[indices][:40],
x=classifier.feature_importances_[indices][:40],
orient="h",
ax=axes[row][col],
)
g.set_xlabel("Relative importance", fontsize=12)
g.set_ylabel("Features", fontsize=12)
g.tick_params(labelsize=9)
g.set_title(name + " feature importance")
nclassifier += 1
# I plot the feature importance for the 4 tree based classifiers (Adaboost, ExtraTrees, RandomForest and GradientBoosting).
# We note that the four classifiers have different top features according to the relative importance. It means that their predictions are not based on the same features. Nevertheless, they share some common important features for the classification , for example 'Fare', 'Title_2', 'Age' and 'Sex'.
# Title_2 which indicates the Mrs/Mlle/Mme/Miss/Ms category is highly correlated with Sex.
# We can say that:
# - Pc_1, Pc_2, Pc_3 and Fare refer to the general social standing of passengers.
# - Sex and Title_2 (Mrs/Mlle/Mme/Miss/Ms) and Title_3 (Mr) refer to the gender.
# - Age and Title_1 (Master) refer to the age of passengers.
# - Fsize, LargeF, MedF, Single refer to the size of the passenger family.
# **According to the feature importance of this 4 classifiers, the prediction of the survival seems to be more associated with the Age, the Sex, the family size and the social standing of the passengers more than the location in the boat.**
test_Survived_RFC = pd.Series(RFC_best.predict(test), name="RFC")
test_Survived_ExtC = pd.Series(ExtC_best.predict(test), name="ExtC")
test_Survived_SVMC = pd.Series(SVMC_best.predict(test), name="SVC")
test_Survived_AdaC = pd.Series(ada_best.predict(test), name="Ada")
test_Survived_GBC = pd.Series(GBC_best.predict(test), name="GBC")
# Concatenate all classifier results
ensemble_results = pd.concat(
[
test_Survived_RFC,
test_Survived_ExtC,
test_Survived_AdaC,
test_Survived_GBC,
test_Survived_SVMC,
],
axis=1,
)
g = sns.heatmap(ensemble_results.corr(), annot=True)
# The prediction seems to be quite similar for the 5 classifiers except when Adaboost is compared to the others classifiers.
# The 5 classifiers give more or less the same prediction but there is some differences. Theses differences between the 5 classifier predictions are sufficient to consider an ensembling vote.
# ### 6.2 Ensemble modeling
# #### 6.2.1 Combining models
# I choosed a voting classifier to combine the predictions coming from the 5 classifiers.
# I preferred to pass the argument "soft" to the voting parameter to take into account the probability of each vote.
votingC = VotingClassifier(
estimators=[
("rfc", RFC_best),
("extc", ExtC_best),
("svc", SVMC_best),
("adac", ada_best),
("gbc", GBC_best),
],
voting="soft",
n_jobs=4,
)
votingC = votingC.fit(X_train, Y_train)
# ### 6.3 Prediction
# #### 6.3.1 Predict and Submit results
test_Survived = pd.Series(votingC.predict(test), name="Survived")
results = pd.concat([IDtest, test_Survived], axis=1)
results.to_csv("ensemble_python_voting.csv", index=False)
| false | 2 | 11,799 | 0 | 11,849 | 11,799 |
||
69136035
|
<jupyter_start><jupyter_text>Worldwide deaths by country/risk factors
### Context
This dataset shows the total annual deaths due to each risk factor, by country. The data is downloaded from WHO website.
### Details
Each record provides details for a particular country/year. Total annual deaths are provided for each risk factor, with the column name of risk factor.
Kaggle dataset identifier: worldwide-deaths-by-risk-factors
<jupyter_code>import pandas as pd
df = pd.read_csv('worldwide-deaths-by-risk-factors/number-of-deaths-by-risk-factor.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 6468 entries, 0 to 6467
Data columns (total 31 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Entity 6468 non-null object
1 Year 6468 non-null int64
2 Unsafe water source 6468 non-null float64
3 Unsafe sanitation 6468 non-null float64
4 No access to handwashing facility 6468 non-null float64
5 Household air pollution from solid fuels 6468 non-null float64
6 Non-exclusive breastfeeding 6468 non-null float64
7 Discontinued breastfeeding 6468 non-null float64
8 Child wasting 6468 non-null float64
9 Child stunting 6468 non-null float64
10 Low birth weight for gestation 6468 non-null float64
11 Secondhand smoke 6468 non-null float64
12 Alcohol use 6468 non-null float64
13 Drug use 6468 non-null float64
14 Diet low in fruits 6468 non-null float64
15 Diet low in vegetables 6468 non-null float64
16 Unsafe sex 6468 non-null float64
17 Low physical activity 6468 non-null float64
18 High fasting plasma glucose 6468 non-null float64
19 High total cholesterol 1561 non-null float64
20 High body-mass index 6468 non-null float64
21 High systolic blood pressure 6468 non-null float64
22 Smoking 6468 non-null float64
23 Iron deficiency 6468 non-null float64
24 Vitamin A deficiency 6468 non-null float64
25 Low bone mineral density 6468 non-null float64
26 Air pollution 6468 non-null float64
27 Outdoor air pollution 6467 non-null float64
28 Diet high in sodium 6468 non-null float64
29 Diet low in whole grains 6468 non-null float64
30 Diet low in nuts and seeds 6468 non-null float64
dtypes: float64(29), int64(1), object(1)
memory usage: 1.5+ MB
<jupyter_text>Examples:
{
"Entity": "Afghanistan",
"Year": 1990,
"Unsafe water source": 7554.049543,
"Unsafe sanitation": 5887.747628,
"No access to handwashing facility": 5412.314513,
"Household air pollution from solid fuels": 22388.49723,
"Non-exclusive breastfeeding": 3221.138842,
"Discontinued breastfeeding": 156.0975526,
"Child wasting": 22778.84925,
"Child stunting": 10408.43885,
"Low birth weight for gestation": 12168.56463,
"Secondhand smoke": 4234.808095,
"Alcohol use": 356.5293069,
"Drug use": 208.3254296,
"Diet low in fruits": 8538.964137,
"Diet low in vegetables": 7678.717644,
"Unsafe sex": 387.1675823,
"Low physical activity": 4221.303183,
"High fasting plasma glucose": 21610.06616,
"High total cholesterol": 9505.531962,
"...": "and 11 more columns"
}
{
"Entity": "Afghanistan",
"Year": 1991,
"Unsafe water source": 7359.676749,
"Unsafe sanitation": 5732.77016,
"No access to handwashing facility": 5287.891103,
"Household air pollution from solid fuels": 22128.75821,
"Non-exclusive breastfeeding": 3150.559597,
"Discontinued breastfeeding": 151.5398506,
"Child wasting": 22292.69111,
"Child stunting": 10271.97643,
"Low birth weight for gestation": 12360.63537,
"Secondhand smoke": 4219.597324,
"Alcohol use": 320.5984612,
"Drug use": 217.7696908,
"Diet low in fruits": 8642.847151,
"Diet low in vegetables": 7789.773033,
"Unsafe sex": 394.4482851,
"Low physical activity": 4252.630379,
"High fasting plasma glucose": 21824.93804,
"High total cholesterol": NaN,
"...": "and 11 more columns"
}
{
"Entity": "Afghanistan",
"Year": 1992,
"Unsafe water source": 7650.437822,
"Unsafe sanitation": 5954.804987,
"No access to handwashing facility": 5506.657363,
"Household air pollution from solid fuels": 22873.76879,
"Non-exclusive breastfeeding": 3331.349048,
"Discontinued breastfeeding": 156.6091937,
"Child wasting": 23102.19794,
"Child stunting": 10618.87978,
"Low birth weight for gestation": 13459.59372,
"Secondhand smoke": 4371.907968,
"Alcohol use": 293.2570162,
"Drug use": 247.8332513,
"Diet low in fruits": 8961.526496,
"Diet low in vegetables": 8083.234634,
"Unsafe sex": 422.4533018,
"Low physical activity": 4347.330897,
"High fasting plasma glucose": 22418.69881,
"High total cholesterol": NaN,
"...": "and 11 more columns"
}
{
"Entity": "Afghanistan",
"Year": 1993,
"Unsafe water source": 10270.73138,
"Unsafe sanitation": 7986.736613,
"No access to handwashing facility": 7104.620351,
"Household air pollution from solid fuels": 25599.75628,
"Non-exclusive breastfeeding": 4477.0061,
"Discontinued breastfeeding": 206.8344513,
"Child wasting": 27902.66996,
"Child stunting": 12260.09384,
"Low birth weight for gestation": 18458.42913,
"Secondhand smoke": 4863.558517,
"Alcohol use": 278.1297583,
"Drug use": 285.0361812,
"Diet low in fruits": 9377.118485,
"Diet low in vegetables": 8452.242405,
"Unsafe sex": 448.3283172,
"Low physical activity": 4465.13767,
"High fasting plasma glucose": 23140.51117,
"High total cholesterol": NaN,
"...": "and 11 more columns"
}
<jupyter_script># # Cluster model for world deaths
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
data = pd.read_csv(
"../input/worldwide-deaths-by-risk-factors/number-of-deaths-by-risk-factor.csv"
)
data.head()
data.info()
# There is a feature (High total cholesterol) that has a lot of missing data. I will drop the feature since it would the percentage of missing data is more than 70%.
data.drop("High total cholesterol", axis=1, inplace=True)
# ### Other than countries listed under the 'Entity' column, there are also come country groupings provided. They are:
# * North America,
# * Latin America and Caribbean,
# * Central Europe,
# * Eastern Europe,
# * Western Europe,
# * North Africa and Middle East,
# * Central Sub-Saharan Africa,
# * Eastern Sub-Saharan Africa,
# * Western Sub-Saharan Africa,
# * Southern Sub-Saharan Africa,
# * Central Asia,
# * East Asia,
# * Southeast Asia,
# * South Asia,
# * Australasia,
# * High-income, and
# * High-income Asia Pacific
# This should cover all the countries provided in the data.
#
# There is also 'World' data provided.
#
# Other than by countries, there are also data grouped by Socio-demographic Index (SDI).
# SDI is a summary measure that identifies where countries or other geographic areas sit on the spectrum of development. Expressed on a scale of 0 to 1, SDI is a composite average of the rankings of the incomes per capita, average educational attainment, and fertility rates of all areas in the GBD study.
# A list can be found here http://ghdx.healthdata.org/record/ihme-data/gbd-2019-socio-demographic-index-sdi-1950-2019. There is also 2 other categories for high income countries (High-income and High-income Asia Pacific).
# ### Removing the groupings to analyse the data by countries.
remove = [
"North America",
"Latin America and Caribbean",
"Central Europe",
"Eastern Europe",
"Western Europe",
"North Africa and Middle East",
"Central Sub-Saharan Africa",
"Eastern Sub-Saharan Africa",
"Western Sub-Saharan Africa",
"Southern Sub-Saharan Africa",
"Central Asia",
"East Asia",
"Southeast Asia",
"South Asia",
"Australasia",
"Central Europe, Eastern Europe, and Central Asia",
"Sub-Saharan Africa",
"Southeast Asia, East Asia, and Oceania",
"Southern Latin America",
"Central Latin America",
"Tropical Latin America",
"High SDI",
"High-middle SDI",
"Middle SDI",
"Low-middle SDI",
"Low SDI",
"High-income",
"High-income Asia Pacific",
"World",
]
country_df = data[~data["Entity"].isin(remove)]
country_df.head()
# The data will be grouped by the 'year' feature and the mean value will be used.
grouped_country_df = country_df.groupby("Entity").mean()
total_deaths = (
grouped_country_df.drop("Year", axis=1)
.sum()
.transpose()
.sort_values(ascending=False)
)
plt.figure(figsize=(10, 8))
sns.barplot(y=total_deaths.index, x=total_deaths.values, orient="h")
plt.xticks(rotation=90)
# ### Let's visualize the correlation for just a few selected features.
sns.heatmap(
grouped_country_df[
[
"High fasting plasma glucose",
"High body-mass index",
"High systolic blood pressure",
"Air pollution",
"Smoking",
]
].corr(),
annot=True,
)
# The correlation values are high for all 4 features.
# What about the effects of diet?
sns.pairplot(
grouped_country_df[
[
"Diet high in sodium",
"Diet low in whole grains",
"Diet low in nuts and seeds",
"Diet low in fruits",
"Diet low in vegetables",
]
],
kind="reg",
diag_kind="kde",
)
# There are positive correlations for 4 of the feature (Diet low in whole grains, Diet low in nuts and seeds, Diet low in fruits and Diet low in vegetables) and the values shown are high as well.
# Features with correlation value above 95% will be identified and dropped for cluster modelling.
# *Note: I have also tried to perform clustering without removing any feature to check if this would impact the outcome but I'm happy to inform that the results were the same. However, having a large number of features will reduce the interpretability of the model. *
corr_matrix = grouped_country_df.corr().abs()
upper = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(np.bool_))
high_corr_col = [column for column in upper.columns if any(upper[column] > 0.95)]
country_feat = grouped_country_df.drop(high_corr_col, axis=1)
country_feat.head()
# Total features are now reduced to 9 (not including Year which will be dropped) from the initial 28. K-Means clustering will be used and Sum of Squared Distances will be calculated to determine the cluster number.
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import KMeans
X = country_feat.drop("Year", axis=1)
scaler = StandardScaler()
scaled_X = scaler.fit_transform(X)
ssd = []
for k in range(2, 16):
model = KMeans(n_clusters=k)
model.fit(scaled_X)
ssd.append(model.inertia_)
# The elbow method will be used to determine the k value.
plt.figure(figsize=(10, 6))
plt.plot(range(2, 16), ssd, "o--")
plt.xlabel("k values")
plt.ylabel("Sum of Squared Distances")
# Selected n_clusters=6.
model = KMeans(n_clusters=6, random_state=0)
cluster_labels = model.fit_predict(scaled_X)
X["Cluster"] = cluster_labels
cluster_corr = X.corr()["Cluster"].sort_values()
# Which feature did K-Means regarded as most important?
plt.figure(figsize=(10, 4))
sns.barplot(x=cluster_corr[:-1].index, y=cluster_corr[:-1].values)
plt.title("Feature importance determined by K-Means", fontsize=16)
plt.xlabel("Risk factors")
plt.ylabel("Correlation")
plt.xticks(rotation=90)
# Let's see how K-Means clustered the countries using choropleth map.
iso = pd.read_csv("../input/country-code/country_code.csv", index_col="Country")
iso_code = iso["3let"].to_dict()
X["ISO_Code"] = X.index.map(iso_code)
import plotly.express as px
import plotly.offline as pyo
pyo.init_notebook_mode()
fig = px.choropleth(
X,
locations="ISO_Code",
color="Cluster",
hover_name=X.index,
color_continuous_scale="Rainbow",
)
fig.show()
# Was there a reason why China and India/Myanmar were having their own cluster? What about US being grouped with Russia and why was Canada was not grouped with US?
sel_countries = ["United States", "Canada", "China", "India", "Russia", "Australia"]
filt_countries = country_feat.loc[sel_countries].groupby("Entity").sum()
filt_countries = filt_countries.drop("Year", axis=1).groupby("Entity").sum().transpose()
filt_countries = filt_countries.reset_index()
filt_countries = pd.melt(
filt_countries, "index", var_name="country", value_name="value"
)
plt.figure(figsize=(12, 8))
sns.lineplot(x="index", y="value", hue="country", data=filt_countries, palette="Dark2")
plt.legend(loc=(1.05, 0.5))
plt.title("Total Deaths by Risk Factors for Selected Countries", fontsize=16)
plt.xlabel("Risk factors")
plt.ylabel("Total")
plt.xticks(rotation=90)
# In an attempt to get some insights, the chart above was plotted to see if there were any information that can be gained to explain how K-Means clustered the countries. The chart provided a clear pattern on why US and Russia were clustered together and also why China and India are in their own cluster. Canada and Australia can also be seen to have similar pattern and was probably why they were clustered together.
# Let's explore further and look at the data grouped by Socio-demographic Index (SDI).
# ## Data grouped by Socio-demographic Index (SDI)
sdi_list = ["High SDI", "High-middle SDI", "Middle SDI", "Low SDI", "Low-middle SDI"]
sdi = data[data["Entity"].isin(sdi_list)]
import matplotlib.lines as mlines
sdi1 = (
sdi[(sdi["Year"] > 2001) & (sdi["Year"] < 2011)]
.groupby("Entity")
.mean()
.sum(axis=1)
/ 1000000
)
sdi2 = (
sdi[(sdi["Year"] > 2011) & (sdi["Year"] < 2017)]
.groupby("Entity")
.mean()
.sum(axis=1)
/ 1000000
)
left_label = [
str(c) + ", " + str(round(y, 2)) + "mil" for c, y in zip(sdi1.index, sdi1.values)
]
right_label = [
str(c) + ", " + str(round(y, 2)) + "mil" for c, y in zip(sdi2.index, sdi2.values)
]
klass = [
"red" if (y1 - y2) < 0 else "green" for y1, y2 in zip(sdi1.values, sdi2.values)
]
def newline(p1, p2, color="black"):
ax = plt.gca()
l = mlines.Line2D(
[p1[0], p2[0]],
[p1[1], p2[1]],
color="red" if p1[1] - p2[1] > 0 else "green",
marker="o",
markersize=6,
)
ax.add_line(l)
return l
fig, ax = plt.subplots(figsize=(14, 14))
ax.vlines(
x=1, ymin=8, ymax=16, color="black", alpha=0.7, linewidth=1, linestyles="dotted"
)
ax.vlines(
x=3, ymin=8, ymax=16, color="black", alpha=0.7, linewidth=1, linestyles="dotted"
)
ax.scatter(y=sdi1.values, x=np.repeat(1, sdi1.shape[0]), s=10, color="black", alpha=0.7)
ax.scatter(y=sdi2.values, x=np.repeat(3, sdi2.shape[0]), s=10, color="black", alpha=0.7)
for p1, p2, c in zip(sdi1.values, sdi2.values, sdi2.index):
newline([1, p1], [3, p2])
ax.text(
1 - 0.05,
p1,
c + ", " + str(round(p1, 2)) + "mil",
horizontalalignment="right",
verticalalignment="center",
fontdict={"size": 12},
)
ax.text(
3 + 0.05,
p2,
c + ", " + str(round(p2, 2)) + "mil",
horizontalalignment="left",
verticalalignment="center",
fontdict={"size": 12},
)
ax.text(
1 - 0.05,
17,
"2001-2010",
horizontalalignment="right",
verticalalignment="center",
fontdict={"size": 18, "weight": 700},
)
ax.text(
3 + 0.05,
17,
"2011-2017",
horizontalalignment="left",
verticalalignment="center",
fontdict={"size": 18, "weight": 700},
)
ax.set_title("Slopechart: Comparing Total Deaths by SDI", fontdict={"size": 22})
ax.set(xlim=(0, 4), ylim=(8, 18), ylabel="Average Total Deaths (million)")
ax.set_xticks([1, 3])
ax.set_xticklabels(["", ""])
plt.yticks(np.arange(8, 18, 2), fontsize=12)
plt.gca().spines["top"].set_alpha(0.0)
plt.gca().spines["bottom"].set_alpha(0.0)
plt.gca().spines["right"].set_alpha(0.0)
plt.gca().spines["left"].set_alpha(0.0)
plt.show()
# It is observed that countries with Low SDIs has seen a decreased in their death rates while other SDIs had an increase. Countries grouped as Middle SDI had the biggest increase in death count. Let's explore the Low SDI and Middle SDI to get some insights.
low_sdi = sdi[sdi["Entity"] == "Low SDI"]
low_2001_2010 = (
low_sdi[(low_sdi["Year"] > 2001) & (low_sdi["Year"] < 2011)]
.groupby("Entity")
.mean()
)
low_2011_2017 = (
low_sdi[(low_sdi["Year"] > 2011) & (low_sdi["Year"] < 2017)]
.groupby("Entity")
.mean()
)
plt.figure(figsize=(10, 8))
diff = (low_2011_2017 - low_2001_2010).drop("Year", axis=1).transpose()
sns.barplot(x=diff.index, y=diff["Low SDI"], data=diff)
plt.xlabel("Risk factors")
plt.ylabel("Total")
plt.xticks(rotation=90)
# Improvements to sanitation as well as safer sex activities seemed to have improve thesurvivability of people in low SDI countries.
mid_sdi = sdi[sdi["Entity"] == "Middle SDI"]
mid_2001_2010 = (
mid_sdi[(mid_sdi["Year"] > 2001) & (mid_sdi["Year"] < 2011)]
.groupby("Entity")
.mean()
)
mid_2011_2017 = (
mid_sdi[(mid_sdi["Year"] > 2011) & (mid_sdi["Year"] < 2017)]
.groupby("Entity")
.mean()
)
plt.figure(figsize=(10, 8))
diff = (mid_2011_2017 - mid_2001_2010).drop("Year", axis=1).transpose()
sns.barplot(x=diff.index, y=diff["Middle SDI"], data=diff)
plt.xlabel("Risk factors")
plt.ylabel("Total")
plt.xticks(rotation=90)
# Higher purchasing power as well as access to modern conveniences have certainly increased the death count for middle SDI countires as risk factors related to diet as well as lifestyle have increased significantly.
# Moving on to explore more from the data for the SDIs.
sdi_deaths = sdi.drop("Year", axis=1).groupby("Entity").sum().transpose()
sdi_deaths = sdi_deaths.reset_index()
sdi_melt = pd.melt(sdi_deaths, "index", var_name="count", value_name="value")
plt.figure(figsize=(12, 8))
sns.lineplot(x="index", y="value", hue="count", data=sdi_melt, palette="Dark2")
plt.legend(loc=(1.05, 0.5))
plt.title("Total Deaths by Risk Factors for SDIs", fontsize=16)
plt.xlabel("Risk factors")
plt.ylabel("Total")
plt.xticks(rotation=90)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/136/69136035.ipynb
|
worldwide-deaths-by-risk-factors
|
varpit94
|
[{"Id": 69136035, "ScriptId": 18864840, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7227680, "CreationDate": "07/27/2021 09:15:17", "VersionNumber": 4.0, "Title": "Cluster model for world deaths", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 282.0, "LinesInsertedFromPrevious": 8.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 274.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 91954206, "KernelVersionId": 69136035, "SourceDatasetVersionId": 2403074}]
|
[{"Id": 2403074, "DatasetId": 1453305, "DatasourceVersionId": 2445120, "CreatorUserId": 1130103, "LicenseName": "Other (specified in description)", "CreationDate": "07/07/2021 11:06:58", "VersionNumber": 1.0, "Title": "Worldwide deaths by country/risk factors", "Slug": "worldwide-deaths-by-risk-factors", "Subtitle": "WHO data - deaths by country and risk factors", "Description": "### Context\n\nThis dataset shows the total annual deaths due to each risk factor, by country. The data is downloaded from WHO website.\n\n### Details\n\nEach record provides details for a particular country/year. Total annual deaths are provided for each risk factor, with the column name of risk factor.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1453305, "CreatorUserId": 1130103, "OwnerUserId": 1130103.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2403074.0, "CurrentDatasourceVersionId": 2445120.0, "ForumId": 1472864, "Type": 2, "CreationDate": "07/07/2021 11:06:58", "LastActivityDate": "07/07/2021", "TotalViews": 10990, "TotalDownloads": 1609, "TotalVotes": 32, "TotalKernels": 5}]
|
[{"Id": 1130103, "UserName": "varpit94", "DisplayName": "Arpit Verma", "RegisterDate": "06/17/2017", "PerformanceTier": 2}]
|
# # Cluster model for world deaths
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
data = pd.read_csv(
"../input/worldwide-deaths-by-risk-factors/number-of-deaths-by-risk-factor.csv"
)
data.head()
data.info()
# There is a feature (High total cholesterol) that has a lot of missing data. I will drop the feature since it would the percentage of missing data is more than 70%.
data.drop("High total cholesterol", axis=1, inplace=True)
# ### Other than countries listed under the 'Entity' column, there are also come country groupings provided. They are:
# * North America,
# * Latin America and Caribbean,
# * Central Europe,
# * Eastern Europe,
# * Western Europe,
# * North Africa and Middle East,
# * Central Sub-Saharan Africa,
# * Eastern Sub-Saharan Africa,
# * Western Sub-Saharan Africa,
# * Southern Sub-Saharan Africa,
# * Central Asia,
# * East Asia,
# * Southeast Asia,
# * South Asia,
# * Australasia,
# * High-income, and
# * High-income Asia Pacific
# This should cover all the countries provided in the data.
#
# There is also 'World' data provided.
#
# Other than by countries, there are also data grouped by Socio-demographic Index (SDI).
# SDI is a summary measure that identifies where countries or other geographic areas sit on the spectrum of development. Expressed on a scale of 0 to 1, SDI is a composite average of the rankings of the incomes per capita, average educational attainment, and fertility rates of all areas in the GBD study.
# A list can be found here http://ghdx.healthdata.org/record/ihme-data/gbd-2019-socio-demographic-index-sdi-1950-2019. There is also 2 other categories for high income countries (High-income and High-income Asia Pacific).
# ### Removing the groupings to analyse the data by countries.
remove = [
"North America",
"Latin America and Caribbean",
"Central Europe",
"Eastern Europe",
"Western Europe",
"North Africa and Middle East",
"Central Sub-Saharan Africa",
"Eastern Sub-Saharan Africa",
"Western Sub-Saharan Africa",
"Southern Sub-Saharan Africa",
"Central Asia",
"East Asia",
"Southeast Asia",
"South Asia",
"Australasia",
"Central Europe, Eastern Europe, and Central Asia",
"Sub-Saharan Africa",
"Southeast Asia, East Asia, and Oceania",
"Southern Latin America",
"Central Latin America",
"Tropical Latin America",
"High SDI",
"High-middle SDI",
"Middle SDI",
"Low-middle SDI",
"Low SDI",
"High-income",
"High-income Asia Pacific",
"World",
]
country_df = data[~data["Entity"].isin(remove)]
country_df.head()
# The data will be grouped by the 'year' feature and the mean value will be used.
grouped_country_df = country_df.groupby("Entity").mean()
total_deaths = (
grouped_country_df.drop("Year", axis=1)
.sum()
.transpose()
.sort_values(ascending=False)
)
plt.figure(figsize=(10, 8))
sns.barplot(y=total_deaths.index, x=total_deaths.values, orient="h")
plt.xticks(rotation=90)
# ### Let's visualize the correlation for just a few selected features.
sns.heatmap(
grouped_country_df[
[
"High fasting plasma glucose",
"High body-mass index",
"High systolic blood pressure",
"Air pollution",
"Smoking",
]
].corr(),
annot=True,
)
# The correlation values are high for all 4 features.
# What about the effects of diet?
sns.pairplot(
grouped_country_df[
[
"Diet high in sodium",
"Diet low in whole grains",
"Diet low in nuts and seeds",
"Diet low in fruits",
"Diet low in vegetables",
]
],
kind="reg",
diag_kind="kde",
)
# There are positive correlations for 4 of the feature (Diet low in whole grains, Diet low in nuts and seeds, Diet low in fruits and Diet low in vegetables) and the values shown are high as well.
# Features with correlation value above 95% will be identified and dropped for cluster modelling.
# *Note: I have also tried to perform clustering without removing any feature to check if this would impact the outcome but I'm happy to inform that the results were the same. However, having a large number of features will reduce the interpretability of the model. *
corr_matrix = grouped_country_df.corr().abs()
upper = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(np.bool_))
high_corr_col = [column for column in upper.columns if any(upper[column] > 0.95)]
country_feat = grouped_country_df.drop(high_corr_col, axis=1)
country_feat.head()
# Total features are now reduced to 9 (not including Year which will be dropped) from the initial 28. K-Means clustering will be used and Sum of Squared Distances will be calculated to determine the cluster number.
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import KMeans
X = country_feat.drop("Year", axis=1)
scaler = StandardScaler()
scaled_X = scaler.fit_transform(X)
ssd = []
for k in range(2, 16):
model = KMeans(n_clusters=k)
model.fit(scaled_X)
ssd.append(model.inertia_)
# The elbow method will be used to determine the k value.
plt.figure(figsize=(10, 6))
plt.plot(range(2, 16), ssd, "o--")
plt.xlabel("k values")
plt.ylabel("Sum of Squared Distances")
# Selected n_clusters=6.
model = KMeans(n_clusters=6, random_state=0)
cluster_labels = model.fit_predict(scaled_X)
X["Cluster"] = cluster_labels
cluster_corr = X.corr()["Cluster"].sort_values()
# Which feature did K-Means regarded as most important?
plt.figure(figsize=(10, 4))
sns.barplot(x=cluster_corr[:-1].index, y=cluster_corr[:-1].values)
plt.title("Feature importance determined by K-Means", fontsize=16)
plt.xlabel("Risk factors")
plt.ylabel("Correlation")
plt.xticks(rotation=90)
# Let's see how K-Means clustered the countries using choropleth map.
iso = pd.read_csv("../input/country-code/country_code.csv", index_col="Country")
iso_code = iso["3let"].to_dict()
X["ISO_Code"] = X.index.map(iso_code)
import plotly.express as px
import plotly.offline as pyo
pyo.init_notebook_mode()
fig = px.choropleth(
X,
locations="ISO_Code",
color="Cluster",
hover_name=X.index,
color_continuous_scale="Rainbow",
)
fig.show()
# Was there a reason why China and India/Myanmar were having their own cluster? What about US being grouped with Russia and why was Canada was not grouped with US?
sel_countries = ["United States", "Canada", "China", "India", "Russia", "Australia"]
filt_countries = country_feat.loc[sel_countries].groupby("Entity").sum()
filt_countries = filt_countries.drop("Year", axis=1).groupby("Entity").sum().transpose()
filt_countries = filt_countries.reset_index()
filt_countries = pd.melt(
filt_countries, "index", var_name="country", value_name="value"
)
plt.figure(figsize=(12, 8))
sns.lineplot(x="index", y="value", hue="country", data=filt_countries, palette="Dark2")
plt.legend(loc=(1.05, 0.5))
plt.title("Total Deaths by Risk Factors for Selected Countries", fontsize=16)
plt.xlabel("Risk factors")
plt.ylabel("Total")
plt.xticks(rotation=90)
# In an attempt to get some insights, the chart above was plotted to see if there were any information that can be gained to explain how K-Means clustered the countries. The chart provided a clear pattern on why US and Russia were clustered together and also why China and India are in their own cluster. Canada and Australia can also be seen to have similar pattern and was probably why they were clustered together.
# Let's explore further and look at the data grouped by Socio-demographic Index (SDI).
# ## Data grouped by Socio-demographic Index (SDI)
sdi_list = ["High SDI", "High-middle SDI", "Middle SDI", "Low SDI", "Low-middle SDI"]
sdi = data[data["Entity"].isin(sdi_list)]
import matplotlib.lines as mlines
sdi1 = (
sdi[(sdi["Year"] > 2001) & (sdi["Year"] < 2011)]
.groupby("Entity")
.mean()
.sum(axis=1)
/ 1000000
)
sdi2 = (
sdi[(sdi["Year"] > 2011) & (sdi["Year"] < 2017)]
.groupby("Entity")
.mean()
.sum(axis=1)
/ 1000000
)
left_label = [
str(c) + ", " + str(round(y, 2)) + "mil" for c, y in zip(sdi1.index, sdi1.values)
]
right_label = [
str(c) + ", " + str(round(y, 2)) + "mil" for c, y in zip(sdi2.index, sdi2.values)
]
klass = [
"red" if (y1 - y2) < 0 else "green" for y1, y2 in zip(sdi1.values, sdi2.values)
]
def newline(p1, p2, color="black"):
ax = plt.gca()
l = mlines.Line2D(
[p1[0], p2[0]],
[p1[1], p2[1]],
color="red" if p1[1] - p2[1] > 0 else "green",
marker="o",
markersize=6,
)
ax.add_line(l)
return l
fig, ax = plt.subplots(figsize=(14, 14))
ax.vlines(
x=1, ymin=8, ymax=16, color="black", alpha=0.7, linewidth=1, linestyles="dotted"
)
ax.vlines(
x=3, ymin=8, ymax=16, color="black", alpha=0.7, linewidth=1, linestyles="dotted"
)
ax.scatter(y=sdi1.values, x=np.repeat(1, sdi1.shape[0]), s=10, color="black", alpha=0.7)
ax.scatter(y=sdi2.values, x=np.repeat(3, sdi2.shape[0]), s=10, color="black", alpha=0.7)
for p1, p2, c in zip(sdi1.values, sdi2.values, sdi2.index):
newline([1, p1], [3, p2])
ax.text(
1 - 0.05,
p1,
c + ", " + str(round(p1, 2)) + "mil",
horizontalalignment="right",
verticalalignment="center",
fontdict={"size": 12},
)
ax.text(
3 + 0.05,
p2,
c + ", " + str(round(p2, 2)) + "mil",
horizontalalignment="left",
verticalalignment="center",
fontdict={"size": 12},
)
ax.text(
1 - 0.05,
17,
"2001-2010",
horizontalalignment="right",
verticalalignment="center",
fontdict={"size": 18, "weight": 700},
)
ax.text(
3 + 0.05,
17,
"2011-2017",
horizontalalignment="left",
verticalalignment="center",
fontdict={"size": 18, "weight": 700},
)
ax.set_title("Slopechart: Comparing Total Deaths by SDI", fontdict={"size": 22})
ax.set(xlim=(0, 4), ylim=(8, 18), ylabel="Average Total Deaths (million)")
ax.set_xticks([1, 3])
ax.set_xticklabels(["", ""])
plt.yticks(np.arange(8, 18, 2), fontsize=12)
plt.gca().spines["top"].set_alpha(0.0)
plt.gca().spines["bottom"].set_alpha(0.0)
plt.gca().spines["right"].set_alpha(0.0)
plt.gca().spines["left"].set_alpha(0.0)
plt.show()
# It is observed that countries with Low SDIs has seen a decreased in their death rates while other SDIs had an increase. Countries grouped as Middle SDI had the biggest increase in death count. Let's explore the Low SDI and Middle SDI to get some insights.
low_sdi = sdi[sdi["Entity"] == "Low SDI"]
low_2001_2010 = (
low_sdi[(low_sdi["Year"] > 2001) & (low_sdi["Year"] < 2011)]
.groupby("Entity")
.mean()
)
low_2011_2017 = (
low_sdi[(low_sdi["Year"] > 2011) & (low_sdi["Year"] < 2017)]
.groupby("Entity")
.mean()
)
plt.figure(figsize=(10, 8))
diff = (low_2011_2017 - low_2001_2010).drop("Year", axis=1).transpose()
sns.barplot(x=diff.index, y=diff["Low SDI"], data=diff)
plt.xlabel("Risk factors")
plt.ylabel("Total")
plt.xticks(rotation=90)
# Improvements to sanitation as well as safer sex activities seemed to have improve thesurvivability of people in low SDI countries.
mid_sdi = sdi[sdi["Entity"] == "Middle SDI"]
mid_2001_2010 = (
mid_sdi[(mid_sdi["Year"] > 2001) & (mid_sdi["Year"] < 2011)]
.groupby("Entity")
.mean()
)
mid_2011_2017 = (
mid_sdi[(mid_sdi["Year"] > 2011) & (mid_sdi["Year"] < 2017)]
.groupby("Entity")
.mean()
)
plt.figure(figsize=(10, 8))
diff = (mid_2011_2017 - mid_2001_2010).drop("Year", axis=1).transpose()
sns.barplot(x=diff.index, y=diff["Middle SDI"], data=diff)
plt.xlabel("Risk factors")
plt.ylabel("Total")
plt.xticks(rotation=90)
# Higher purchasing power as well as access to modern conveniences have certainly increased the death count for middle SDI countires as risk factors related to diet as well as lifestyle have increased significantly.
# Moving on to explore more from the data for the SDIs.
sdi_deaths = sdi.drop("Year", axis=1).groupby("Entity").sum().transpose()
sdi_deaths = sdi_deaths.reset_index()
sdi_melt = pd.melt(sdi_deaths, "index", var_name="count", value_name="value")
plt.figure(figsize=(12, 8))
sns.lineplot(x="index", y="value", hue="count", data=sdi_melt, palette="Dark2")
plt.legend(loc=(1.05, 0.5))
plt.title("Total Deaths by Risk Factors for SDIs", fontsize=16)
plt.xlabel("Risk factors")
plt.ylabel("Total")
plt.xticks(rotation=90)
|
[{"worldwide-deaths-by-risk-factors/number-of-deaths-by-risk-factor.csv": {"column_names": "[\"Entity\", \"Year\", \"Unsafe water source\", \"Unsafe sanitation\", \"No access to handwashing facility\", \"Household air pollution from solid fuels\", \"Non-exclusive breastfeeding\", \"Discontinued breastfeeding\", \"Child wasting\", \"Child stunting\", \"Low birth weight for gestation\", \"Secondhand smoke\", \"Alcohol use\", \"Drug use\", \"Diet low in fruits\", \"Diet low in vegetables\", \"Unsafe sex\", \"Low physical activity\", \"High fasting plasma glucose\", \"High total cholesterol\", \"High body-mass index\", \"High systolic blood pressure\", \"Smoking\", \"Iron deficiency\", \"Vitamin A deficiency\", \"Low bone mineral density\", \"Air pollution\", \"Outdoor air pollution\", \"Diet high in sodium\", \"Diet low in whole grains\", \"Diet low in nuts and seeds\"]", "column_data_types": "{\"Entity\": \"object\", \"Year\": \"int64\", \"Unsafe water source\": \"float64\", \"Unsafe sanitation\": \"float64\", \"No access to handwashing facility\": \"float64\", \"Household air pollution from solid fuels\": \"float64\", \"Non-exclusive breastfeeding\": \"float64\", \"Discontinued breastfeeding\": \"float64\", \"Child wasting\": \"float64\", \"Child stunting\": \"float64\", \"Low birth weight for gestation\": \"float64\", \"Secondhand smoke\": \"float64\", \"Alcohol use\": \"float64\", \"Drug use\": \"float64\", \"Diet low in fruits\": \"float64\", \"Diet low in vegetables\": \"float64\", \"Unsafe sex\": \"float64\", \"Low physical activity\": \"float64\", \"High fasting plasma glucose\": \"float64\", \"High total cholesterol\": \"float64\", \"High body-mass index\": \"float64\", \"High systolic blood pressure\": \"float64\", \"Smoking\": \"float64\", \"Iron deficiency\": \"float64\", \"Vitamin A deficiency\": \"float64\", \"Low bone mineral density\": \"float64\", \"Air pollution\": \"float64\", \"Outdoor air pollution\": \"float64\", \"Diet high in sodium\": \"float64\", \"Diet low in whole grains\": \"float64\", \"Diet low in nuts and seeds\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 6468 entries, 0 to 6467\nData columns (total 31 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Entity 6468 non-null object \n 1 Year 6468 non-null int64 \n 2 Unsafe water source 6468 non-null float64\n 3 Unsafe sanitation 6468 non-null float64\n 4 No access to handwashing facility 6468 non-null float64\n 5 Household air pollution from solid fuels 6468 non-null float64\n 6 Non-exclusive breastfeeding 6468 non-null float64\n 7 Discontinued breastfeeding 6468 non-null float64\n 8 Child wasting 6468 non-null float64\n 9 Child stunting 6468 non-null float64\n 10 Low birth weight for gestation 6468 non-null float64\n 11 Secondhand smoke 6468 non-null float64\n 12 Alcohol use 6468 non-null float64\n 13 Drug use 6468 non-null float64\n 14 Diet low in fruits 6468 non-null float64\n 15 Diet low in vegetables 6468 non-null float64\n 16 Unsafe sex 6468 non-null float64\n 17 Low physical activity 6468 non-null float64\n 18 High fasting plasma glucose 6468 non-null float64\n 19 High total cholesterol 1561 non-null float64\n 20 High body-mass index 6468 non-null float64\n 21 High systolic blood pressure 6468 non-null float64\n 22 Smoking 6468 non-null float64\n 23 Iron deficiency 6468 non-null float64\n 24 Vitamin A deficiency 6468 non-null float64\n 25 Low bone mineral density 6468 non-null float64\n 26 Air pollution 6468 non-null float64\n 27 Outdoor air pollution 6467 non-null float64\n 28 Diet high in sodium 6468 non-null float64\n 29 Diet low in whole grains 6468 non-null float64\n 30 Diet low in nuts and seeds 6468 non-null float64\ndtypes: float64(29), int64(1), object(1)\nmemory usage: 1.5+ MB\n", "summary": "{\"Year\": {\"count\": 6468.0, \"mean\": 2003.5, \"std\": 8.078371722451168, \"min\": 1990.0, \"25%\": 1996.75, \"50%\": 2003.5, \"75%\": 2010.25, \"max\": 2017.0}, \"Unsafe water source\": {\"count\": 6468.0, \"mean\": 31566.317806638537, \"std\": 152773.11646656765, \"min\": 0.008650193, \"25%\": 10.19665036, \"50%\": 279.031692, \"75%\": 5301.718018, \"max\": 2111659.077}, \"Unsafe sanitation\": {\"count\": 6468.0, \"mean\": 23374.36214076977, \"std\": 114493.0394140417, \"min\": 0.006495981, \"25%\": 4.6038451275, \"50%\": 160.19653649999998, \"75%\": 3832.3443890000003, \"max\": 1638021.199}, \"No access to handwashing facility\": {\"count\": 6468.0, \"mean\": 18933.050499989586, \"std\": 89810.37269100736, \"min\": 0.077913565, \"25%\": 16.8848687575, \"50%\": 252.49909835, \"75%\": 3811.44195125, \"max\": 1239519.421}, \"Household air pollution from solid fuels\": {\"count\": 6468.0, \"mean\": 43084.20690054527, \"std\": 187734.46452151827, \"min\": 0.020585329, \"25%\": 87.59782796500001, \"50%\": 1091.6711525, \"75%\": 9161.964472, \"max\": 2708904.82}, \"Non-exclusive breastfeeding\": {\"count\": 6468.0, \"mean\": 6231.427631749459, \"std\": 28517.846341299508, \"min\": 0.003816093, \"25%\": 4.63325407175, \"50%\": 102.428307, \"75%\": 1367.82727725, \"max\": 514102.3516}, \"Discontinued breastfeeding\": {\"count\": 6468.0, \"mean\": 409.1104231753814, \"std\": 1874.9894308596104, \"min\": 0.000519522, \"25%\": 0.26436632225, \"50%\": 6.6193269305, \"75%\": 78.2794455525, \"max\": 34850.39553}, \"Child wasting\": {\"count\": 6468.0, \"mean\": 43446.43282756805, \"std\": 202236.71052564908, \"min\": 0.101712676, \"25%\": 41.372448105, \"50%\": 730.34623745, \"75%\": 10234.538905, \"max\": 3365308.624}, \"Child stunting\": {\"count\": 6468.0, \"mean\": 11767.717972154967, \"std\": 58248.9147748556, \"min\": 0.001400828, \"25%\": 1.86371691675, \"50%\": 77.87361939499999, \"75%\": 1971.596324, \"max\": 1001277.449}, \"Low birth weight for gestation\": {\"count\": 6468.0, \"mean\": 30948.006623301553, \"std\": 134294.63265703767, \"min\": 0.326638355, \"25%\": 144.562750075, \"50%\": 1220.7169525, \"75%\": 8708.1455665, \"max\": 1976612.538}, \"Secondhand smoke\": {\"count\": 6468.0, \"mean\": 24282.250535878797, \"std\": 100256.18319294348, \"min\": 2.890665314, \"25%\": 278.067695175, \"50%\": 1196.2279005, \"75%\": 5963.6664095, \"max\": 1260994.206}, \"Alcohol use\": {\"count\": 6468.0, \"mean\": 50203.341290893026, \"std\": 195822.60820165742, \"min\": -2315.344758, \"25%\": 363.9521945, \"50%\": 2803.3219055, \"75%\": 12891.2682225, \"max\": 2842854.196}, \"Drug use\": {\"count\": 6468.0, \"mean\": 8890.242149539004, \"std\": 35415.11558940809, \"min\": 1.240062072, \"25%\": 92.909932325, \"50%\": 408.58629075, \"75%\": 2170.843581, \"max\": 585348.1802}, \"Diet low in fruits\": {\"count\": 6468.0, \"mean\": 45452.64274801748, \"std\": 183428.5650741543, \"min\": 1.578807212, \"25%\": 536.0436977749999, \"50%\": 2452.8859515000004, \"75%\": 10521.8222775, \"max\": 2423447.368}, \"Diet low in vegetables\": {\"count\": 6468.0, \"mean\": 28742.012911678834, \"std\": 111659.95288202437, \"min\": 0.776437994, \"25%\": 412.9828087, \"50%\": 1837.7530864999999, \"75%\": 7612.29874275, \"max\": 1462367.411}, \"Unsafe sex\": {\"count\": 6468.0, \"mean\": 26764.450109834786, \"std\": 121709.06324131391, \"min\": 1.021821742, \"25%\": 136.08295802499998, \"50%\": 831.8222561499999, \"75%\": 5948.95286725, \"max\": 1771140.671}, \"Low physical activity\": {\"count\": 6468.0, \"mean\": 21141.486434150796, \"std\": 82215.98589640381, \"min\": 2.416704627, \"25%\": 261.559164275, \"50%\": 1189.4123725, \"75%\": 5694.74391075, \"max\": 1263051.288}, \"High fasting plasma glucose\": {\"count\": 6468.0, \"mean\": 99555.71464861435, \"std\": 384033.01630385185, \"min\": 21.04263206, \"25%\": 2034.71416725, \"50%\": 7820.164595, \"75%\": 34704.7861075, \"max\": 6526028.193}, \"High total cholesterol\": {\"count\": 1561.0, \"mean\": 51628.248059538506, \"std\": 267299.8614611761, \"min\": 9.527324419, \"25%\": 838.89415, \"50%\": 4004.747922, \"75%\": 17422.50858, \"max\": 4392505.382}, \"High body-mass index\": {\"count\": 6468.0, \"mean\": 68685.287814527, \"std\": 268134.06582041923, \"min\": 19.99820791, \"25%\": 1141.44276975, \"50%\": 4739.652490500001, \"75%\": 21601.1724, \"max\": 4724346.293}, \"High systolic blood pressure\": {\"count\": 6468.0, \"mean\": 174383.18589704882, \"std\": 680991.5457603022, \"min\": 21.02607099, \"25%\": 2665.31336725, \"50%\": 10993.308535, \"75%\": 47322.843085, \"max\": 10440818.48}, \"Smoking\": {\"count\": 6468.0, \"mean\": 133548.3482099975, \"std\": 529931.5037142257, \"min\": 11.70747783, \"25%\": 1292.925608, \"50%\": 5935.789171, \"75%\": 31638.099524999998, \"max\": 7099111.294}, \"Iron deficiency\": {\"count\": 6468.0, \"mean\": 1878.745700797305, \"std\": 9011.891579629453, \"min\": 0.005498606, \"25%\": 2.25620886325, \"50%\": 31.990665645, \"75%\": 421.3835854, \"max\": 125242.9483}, \"Vitamin A deficiency\": {\"count\": 6468.0, \"mean\": 11908.622026943482, \"std\": 58801.64861146738, \"min\": 0.003464789, \"25%\": 1.89638615925, \"50%\": 70.490244695, \"75%\": 2081.94672175, \"max\": 986994.9962}, \"Low bone mineral density\": {\"count\": 6468.0, \"mean\": 4579.055653594658, \"std\": 18884.513383680638, \"min\": 0.381231765, \"25%\": 40.6026580675, \"50%\": 246.7507558, \"75%\": 1096.10389075, \"max\": 327314.2626}, \"Air pollution\": {\"count\": 6468.0, \"mean\": 95735.50609881866, \"std\": 390933.5348036437, \"min\": 8.524592707, \"25%\": 1076.83675575, \"50%\": 6125.098028, \"75%\": 22727.359675, \"max\": 4895475.998}, \"Outdoor air pollution\": {\"count\": 6467.0, \"mean\": 55573.12727539817, \"std\": 229803.8133202807, \"min\": 4.83, \"25%\": 553.7049999999999, \"50%\": 2242.02, \"75%\": 12821.5, \"max\": 3408877.62}, \"Diet high in sodium\": {\"count\": 6468.0, \"mean\": 54240.674046736654, \"std\": 243437.33318232978, \"min\": 2.673822838, \"25%\": 355.63730814999997, \"50%\": 1945.6382509999999, \"75%\": 9691.37605825, \"max\": 3196514.265}, \"Diet low in whole grains\": {\"count\": 6468.0, \"mean\": 53348.812853284246, \"std\": 209715.31219065792, \"min\": 9.317591906, \"25%\": 798.734878525, \"50%\": 3504.309221, \"75%\": 14463.69073, \"max\": 3065588.513}, \"Diet low in nuts and seeds\": {\"count\": 6468.0, \"mean\": 34967.03952926335, \"std\": 135943.1920663187, \"min\": 5.18878769, \"25%\": 553.348463625, \"50%\": 2279.1572859999997, \"75%\": 10038.799640000001, \"max\": 2062521.709}}", "examples": "{\"Entity\":{\"0\":\"Afghanistan\",\"1\":\"Afghanistan\",\"2\":\"Afghanistan\",\"3\":\"Afghanistan\"},\"Year\":{\"0\":1990,\"1\":1991,\"2\":1992,\"3\":1993},\"Unsafe water source\":{\"0\":7554.049543,\"1\":7359.676749,\"2\":7650.437822,\"3\":10270.73138},\"Unsafe sanitation\":{\"0\":5887.747628,\"1\":5732.77016,\"2\":5954.804987,\"3\":7986.736613},\"No access to handwashing facility\":{\"0\":5412.314513,\"1\":5287.891103,\"2\":5506.657363,\"3\":7104.620351},\"Household air pollution from solid fuels\":{\"0\":22388.49723,\"1\":22128.75821,\"2\":22873.76879,\"3\":25599.75628},\"Non-exclusive breastfeeding\":{\"0\":3221.138842,\"1\":3150.559597,\"2\":3331.349048,\"3\":4477.0061},\"Discontinued breastfeeding\":{\"0\":156.0975526,\"1\":151.5398506,\"2\":156.6091937,\"3\":206.8344513},\"Child wasting\":{\"0\":22778.84925,\"1\":22292.69111,\"2\":23102.19794,\"3\":27902.66996},\"Child stunting\":{\"0\":10408.43885,\"1\":10271.97643,\"2\":10618.87978,\"3\":12260.09384},\"Low birth weight for gestation\":{\"0\":12168.56463,\"1\":12360.63537,\"2\":13459.59372,\"3\":18458.42913},\"Secondhand smoke\":{\"0\":4234.808095,\"1\":4219.597324,\"2\":4371.907968,\"3\":4863.558517},\"Alcohol use\":{\"0\":356.5293069,\"1\":320.5984612,\"2\":293.2570162,\"3\":278.1297583},\"Drug use\":{\"0\":208.3254296,\"1\":217.7696908,\"2\":247.8332513,\"3\":285.0361812},\"Diet low in fruits\":{\"0\":8538.964137,\"1\":8642.847151,\"2\":8961.526496,\"3\":9377.118485},\"Diet low in vegetables\":{\"0\":7678.717644,\"1\":7789.773033,\"2\":8083.234634,\"3\":8452.242405},\"Unsafe sex\":{\"0\":387.1675823,\"1\":394.4482851,\"2\":422.4533018,\"3\":448.3283172},\"Low physical activity\":{\"0\":4221.303183,\"1\":4252.630379,\"2\":4347.330897,\"3\":4465.13767},\"High fasting plasma glucose\":{\"0\":21610.06616,\"1\":21824.93804,\"2\":22418.69881,\"3\":23140.51117},\"High total cholesterol\":{\"0\":9505.531962,\"1\":null,\"2\":null,\"3\":null},\"High body-mass index\":{\"0\":7701.58128,\"1\":7747.774903,\"2\":7991.018971,\"3\":8281.564408},\"High systolic blood pressure\":{\"0\":28183.98335,\"1\":28435.39751,\"2\":29173.6112,\"3\":30074.76091},\"Smoking\":{\"0\":6393.667372,\"1\":6429.25332,\"2\":6561.054957,\"3\":6731.97256},\"Iron deficiency\":{\"0\":726.4312942,\"1\":739.2457995,\"2\":873.4853406,\"3\":1040.047422},\"Vitamin A deficiency\":{\"0\":9344.131952,\"1\":9330.182378,\"2\":9769.844533,\"3\":11433.76949},\"Low bone mineral density\":{\"0\":374.8440561,\"1\":379.8542372,\"2\":388.1304337,\"3\":405.5779314},\"Air pollution\":{\"0\":26598.00673,\"1\":26379.53222,\"2\":27263.12791,\"3\":30495.5615},\"Outdoor air pollution\":{\"0\":4383.83,\"1\":4426.36,\"2\":4568.91,\"3\":5080.29},\"Diet high in sodium\":{\"0\":2737.197934,\"1\":2741.184956,\"2\":2798.560245,\"3\":2853.301679},\"Diet low in whole grains\":{\"0\":11381.37735,\"1\":11487.83239,\"2\":11866.23557,\"3\":12335.96168},\"Diet low in nuts and seeds\":{\"0\":7299.86733,\"1\":7386.764303,\"2\":7640.628526,\"3\":7968.311853}}"}}]
| true | 2 |
<start_data_description><data_path>worldwide-deaths-by-risk-factors/number-of-deaths-by-risk-factor.csv:
<column_names>
['Entity', 'Year', 'Unsafe water source', 'Unsafe sanitation', 'No access to handwashing facility', 'Household air pollution from solid fuels', 'Non-exclusive breastfeeding', 'Discontinued breastfeeding', 'Child wasting', 'Child stunting', 'Low birth weight for gestation', 'Secondhand smoke', 'Alcohol use', 'Drug use', 'Diet low in fruits', 'Diet low in vegetables', 'Unsafe sex', 'Low physical activity', 'High fasting plasma glucose', 'High total cholesterol', 'High body-mass index', 'High systolic blood pressure', 'Smoking', 'Iron deficiency', 'Vitamin A deficiency', 'Low bone mineral density', 'Air pollution', 'Outdoor air pollution', 'Diet high in sodium', 'Diet low in whole grains', 'Diet low in nuts and seeds']
<column_types>
{'Entity': 'object', 'Year': 'int64', 'Unsafe water source': 'float64', 'Unsafe sanitation': 'float64', 'No access to handwashing facility': 'float64', 'Household air pollution from solid fuels': 'float64', 'Non-exclusive breastfeeding': 'float64', 'Discontinued breastfeeding': 'float64', 'Child wasting': 'float64', 'Child stunting': 'float64', 'Low birth weight for gestation': 'float64', 'Secondhand smoke': 'float64', 'Alcohol use': 'float64', 'Drug use': 'float64', 'Diet low in fruits': 'float64', 'Diet low in vegetables': 'float64', 'Unsafe sex': 'float64', 'Low physical activity': 'float64', 'High fasting plasma glucose': 'float64', 'High total cholesterol': 'float64', 'High body-mass index': 'float64', 'High systolic blood pressure': 'float64', 'Smoking': 'float64', 'Iron deficiency': 'float64', 'Vitamin A deficiency': 'float64', 'Low bone mineral density': 'float64', 'Air pollution': 'float64', 'Outdoor air pollution': 'float64', 'Diet high in sodium': 'float64', 'Diet low in whole grains': 'float64', 'Diet low in nuts and seeds': 'float64'}
<dataframe_Summary>
{'Year': {'count': 6468.0, 'mean': 2003.5, 'std': 8.078371722451168, 'min': 1990.0, '25%': 1996.75, '50%': 2003.5, '75%': 2010.25, 'max': 2017.0}, 'Unsafe water source': {'count': 6468.0, 'mean': 31566.317806638537, 'std': 152773.11646656765, 'min': 0.008650193, '25%': 10.19665036, '50%': 279.031692, '75%': 5301.718018, 'max': 2111659.077}, 'Unsafe sanitation': {'count': 6468.0, 'mean': 23374.36214076977, 'std': 114493.0394140417, 'min': 0.006495981, '25%': 4.6038451275, '50%': 160.19653649999998, '75%': 3832.3443890000003, 'max': 1638021.199}, 'No access to handwashing facility': {'count': 6468.0, 'mean': 18933.050499989586, 'std': 89810.37269100736, 'min': 0.077913565, '25%': 16.8848687575, '50%': 252.49909835, '75%': 3811.44195125, 'max': 1239519.421}, 'Household air pollution from solid fuels': {'count': 6468.0, 'mean': 43084.20690054527, 'std': 187734.46452151827, 'min': 0.020585329, '25%': 87.59782796500001, '50%': 1091.6711525, '75%': 9161.964472, 'max': 2708904.82}, 'Non-exclusive breastfeeding': {'count': 6468.0, 'mean': 6231.427631749459, 'std': 28517.846341299508, 'min': 0.003816093, '25%': 4.63325407175, '50%': 102.428307, '75%': 1367.82727725, 'max': 514102.3516}, 'Discontinued breastfeeding': {'count': 6468.0, 'mean': 409.1104231753814, 'std': 1874.9894308596104, 'min': 0.000519522, '25%': 0.26436632225, '50%': 6.6193269305, '75%': 78.2794455525, 'max': 34850.39553}, 'Child wasting': {'count': 6468.0, 'mean': 43446.43282756805, 'std': 202236.71052564908, 'min': 0.101712676, '25%': 41.372448105, '50%': 730.34623745, '75%': 10234.538905, 'max': 3365308.624}, 'Child stunting': {'count': 6468.0, 'mean': 11767.717972154967, 'std': 58248.9147748556, 'min': 0.001400828, '25%': 1.86371691675, '50%': 77.87361939499999, '75%': 1971.596324, 'max': 1001277.449}, 'Low birth weight for gestation': {'count': 6468.0, 'mean': 30948.006623301553, 'std': 134294.63265703767, 'min': 0.326638355, '25%': 144.562750075, '50%': 1220.7169525, '75%': 8708.1455665, 'max': 1976612.538}, 'Secondhand smoke': {'count': 6468.0, 'mean': 24282.250535878797, 'std': 100256.18319294348, 'min': 2.890665314, '25%': 278.067695175, '50%': 1196.2279005, '75%': 5963.6664095, 'max': 1260994.206}, 'Alcohol use': {'count': 6468.0, 'mean': 50203.341290893026, 'std': 195822.60820165742, 'min': -2315.344758, '25%': 363.9521945, '50%': 2803.3219055, '75%': 12891.2682225, 'max': 2842854.196}, 'Drug use': {'count': 6468.0, 'mean': 8890.242149539004, 'std': 35415.11558940809, 'min': 1.240062072, '25%': 92.909932325, '50%': 408.58629075, '75%': 2170.843581, 'max': 585348.1802}, 'Diet low in fruits': {'count': 6468.0, 'mean': 45452.64274801748, 'std': 183428.5650741543, 'min': 1.578807212, '25%': 536.0436977749999, '50%': 2452.8859515000004, '75%': 10521.8222775, 'max': 2423447.368}, 'Diet low in vegetables': {'count': 6468.0, 'mean': 28742.012911678834, 'std': 111659.95288202437, 'min': 0.776437994, '25%': 412.9828087, '50%': 1837.7530864999999, '75%': 7612.29874275, 'max': 1462367.411}, 'Unsafe sex': {'count': 6468.0, 'mean': 26764.450109834786, 'std': 121709.06324131391, 'min': 1.021821742, '25%': 136.08295802499998, '50%': 831.8222561499999, '75%': 5948.95286725, 'max': 1771140.671}, 'Low physical activity': {'count': 6468.0, 'mean': 21141.486434150796, 'std': 82215.98589640381, 'min': 2.416704627, '25%': 261.559164275, '50%': 1189.4123725, '75%': 5694.74391075, 'max': 1263051.288}, 'High fasting plasma glucose': {'count': 6468.0, 'mean': 99555.71464861435, 'std': 384033.01630385185, 'min': 21.04263206, '25%': 2034.71416725, '50%': 7820.164595, '75%': 34704.7861075, 'max': 6526028.193}, 'High total cholesterol': {'count': 1561.0, 'mean': 51628.248059538506, 'std': 267299.8614611761, 'min': 9.527324419, '25%': 838.89415, '50%': 4004.747922, '75%': 17422.50858, 'max': 4392505.382}, 'High body-mass index': {'count': 6468.0, 'mean': 68685.287814527, 'std': 268134.06582041923, 'min': 19.99820791, '25%': 1141.44276975, '50%': 4739.652490500001, '75%': 21601.1724, 'max': 4724346.293}, 'High systolic blood pressure': {'count': 6468.0, 'mean': 174383.18589704882, 'std': 680991.5457603022, 'min': 21.02607099, '25%': 2665.31336725, '50%': 10993.308535, '75%': 47322.843085, 'max': 10440818.48}, 'Smoking': {'count': 6468.0, 'mean': 133548.3482099975, 'std': 529931.5037142257, 'min': 11.70747783, '25%': 1292.925608, '50%': 5935.789171, '75%': 31638.099524999998, 'max': 7099111.294}, 'Iron deficiency': {'count': 6468.0, 'mean': 1878.745700797305, 'std': 9011.891579629453, 'min': 0.005498606, '25%': 2.25620886325, '50%': 31.990665645, '75%': 421.3835854, 'max': 125242.9483}, 'Vitamin A deficiency': {'count': 6468.0, 'mean': 11908.622026943482, 'std': 58801.64861146738, 'min': 0.003464789, '25%': 1.89638615925, '50%': 70.490244695, '75%': 2081.94672175, 'max': 986994.9962}, 'Low bone mineral density': {'count': 6468.0, 'mean': 4579.055653594658, 'std': 18884.513383680638, 'min': 0.381231765, '25%': 40.6026580675, '50%': 246.7507558, '75%': 1096.10389075, 'max': 327314.2626}, 'Air pollution': {'count': 6468.0, 'mean': 95735.50609881866, 'std': 390933.5348036437, 'min': 8.524592707, '25%': 1076.83675575, '50%': 6125.098028, '75%': 22727.359675, 'max': 4895475.998}, 'Outdoor air pollution': {'count': 6467.0, 'mean': 55573.12727539817, 'std': 229803.8133202807, 'min': 4.83, '25%': 553.7049999999999, '50%': 2242.02, '75%': 12821.5, 'max': 3408877.62}, 'Diet high in sodium': {'count': 6468.0, 'mean': 54240.674046736654, 'std': 243437.33318232978, 'min': 2.673822838, '25%': 355.63730814999997, '50%': 1945.6382509999999, '75%': 9691.37605825, 'max': 3196514.265}, 'Diet low in whole grains': {'count': 6468.0, 'mean': 53348.812853284246, 'std': 209715.31219065792, 'min': 9.317591906, '25%': 798.734878525, '50%': 3504.309221, '75%': 14463.69073, 'max': 3065588.513}, 'Diet low in nuts and seeds': {'count': 6468.0, 'mean': 34967.03952926335, 'std': 135943.1920663187, 'min': 5.18878769, '25%': 553.348463625, '50%': 2279.1572859999997, '75%': 10038.799640000001, 'max': 2062521.709}}
<dataframe_info>
RangeIndex: 6468 entries, 0 to 6467
Data columns (total 31 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Entity 6468 non-null object
1 Year 6468 non-null int64
2 Unsafe water source 6468 non-null float64
3 Unsafe sanitation 6468 non-null float64
4 No access to handwashing facility 6468 non-null float64
5 Household air pollution from solid fuels 6468 non-null float64
6 Non-exclusive breastfeeding 6468 non-null float64
7 Discontinued breastfeeding 6468 non-null float64
8 Child wasting 6468 non-null float64
9 Child stunting 6468 non-null float64
10 Low birth weight for gestation 6468 non-null float64
11 Secondhand smoke 6468 non-null float64
12 Alcohol use 6468 non-null float64
13 Drug use 6468 non-null float64
14 Diet low in fruits 6468 non-null float64
15 Diet low in vegetables 6468 non-null float64
16 Unsafe sex 6468 non-null float64
17 Low physical activity 6468 non-null float64
18 High fasting plasma glucose 6468 non-null float64
19 High total cholesterol 1561 non-null float64
20 High body-mass index 6468 non-null float64
21 High systolic blood pressure 6468 non-null float64
22 Smoking 6468 non-null float64
23 Iron deficiency 6468 non-null float64
24 Vitamin A deficiency 6468 non-null float64
25 Low bone mineral density 6468 non-null float64
26 Air pollution 6468 non-null float64
27 Outdoor air pollution 6467 non-null float64
28 Diet high in sodium 6468 non-null float64
29 Diet low in whole grains 6468 non-null float64
30 Diet low in nuts and seeds 6468 non-null float64
dtypes: float64(29), int64(1), object(1)
memory usage: 1.5+ MB
<some_examples>
{'Entity': {'0': 'Afghanistan', '1': 'Afghanistan', '2': 'Afghanistan', '3': 'Afghanistan'}, 'Year': {'0': 1990, '1': 1991, '2': 1992, '3': 1993}, 'Unsafe water source': {'0': 7554.049543, '1': 7359.676749, '2': 7650.437822, '3': 10270.73138}, 'Unsafe sanitation': {'0': 5887.747628, '1': 5732.77016, '2': 5954.804987, '3': 7986.736613}, 'No access to handwashing facility': {'0': 5412.314513, '1': 5287.891103, '2': 5506.657363, '3': 7104.620351}, 'Household air pollution from solid fuels': {'0': 22388.49723, '1': 22128.75821, '2': 22873.76879, '3': 25599.75628}, 'Non-exclusive breastfeeding': {'0': 3221.138842, '1': 3150.559597, '2': 3331.349048, '3': 4477.0061}, 'Discontinued breastfeeding': {'0': 156.0975526, '1': 151.5398506, '2': 156.6091937, '3': 206.8344513}, 'Child wasting': {'0': 22778.84925, '1': 22292.69111, '2': 23102.19794, '3': 27902.66996}, 'Child stunting': {'0': 10408.43885, '1': 10271.97643, '2': 10618.87978, '3': 12260.09384}, 'Low birth weight for gestation': {'0': 12168.56463, '1': 12360.63537, '2': 13459.59372, '3': 18458.42913}, 'Secondhand smoke': {'0': 4234.808095, '1': 4219.597324, '2': 4371.907968, '3': 4863.558517}, 'Alcohol use': {'0': 356.5293069, '1': 320.5984612, '2': 293.2570162, '3': 278.1297583}, 'Drug use': {'0': 208.3254296, '1': 217.7696908, '2': 247.8332513, '3': 285.0361812}, 'Diet low in fruits': {'0': 8538.964137, '1': 8642.847151, '2': 8961.526496, '3': 9377.118485}, 'Diet low in vegetables': {'0': 7678.717644, '1': 7789.773033, '2': 8083.234634, '3': 8452.242405}, 'Unsafe sex': {'0': 387.1675823, '1': 394.4482851, '2': 422.4533018, '3': 448.3283172}, 'Low physical activity': {'0': 4221.303183, '1': 4252.630379, '2': 4347.330897, '3': 4465.13767}, 'High fasting plasma glucose': {'0': 21610.06616, '1': 21824.93804, '2': 22418.69881, '3': 23140.51117}, 'High total cholesterol': {'0': 9505.531962, '1': None, '2': None, '3': None}, 'High body-mass index': {'0': 7701.58128, '1': 7747.774903, '2': 7991.018971, '3': 8281.564408}, 'High systolic blood pressure': {'0': 28183.98335, '1': 28435.39751, '2': 29173.6112, '3': 30074.76091}, 'Smoking': {'0': 6393.667372, '1': 6429.25332, '2': 6561.054957, '3': 6731.97256}, 'Iron deficiency': {'0': 726.4312942, '1': 739.2457995, '2': 873.4853406, '3': 1040.047422}, 'Vitamin A deficiency': {'0': 9344.131952, '1': 9330.182378, '2': 9769.844533, '3': 11433.76949}, 'Low bone mineral density': {'0': 374.8440561, '1': 379.8542372, '2': 388.1304337, '3': 405.5779314}, 'Air pollution': {'0': 26598.00673, '1': 26379.53222, '2': 27263.12791, '3': 30495.5615}, 'Outdoor air pollution': {'0': 4383.83, '1': 4426.36, '2': 4568.91, '3': 5080.29}, 'Diet high in sodium': {'0': 2737.197934, '1': 2741.184956, '2': 2798.560245, '3': 2853.301679}, 'Diet low in whole grains': {'0': 11381.37735, '1': 11487.83239, '2': 11866.23557, '3': 12335.96168}, 'Diet low in nuts and seeds': {'0': 7299.86733, '1': 7386.764303, '2': 7640.628526, '3': 7968.311853}}
<end_description>
| 4,242 | 0 | 6,747 | 4,242 |
69136148
|
import pandas as pd
import os
dataset_path = "/kaggle/input/car-crashes-severity-prediction/"
# Factorization function:
def factorize(df, col_name, index):
data, unique = pd.factorize(df[col_name])
df.drop(columns=[col_name], inplace=True)
df.insert(index, col_name, data)
data = pd.read_csv(os.path.join(dataset_path, "train.csv"))
print("The shape of the dataset is {}.\n\n".format(data.shape))
data.head()
# Statistical discription of the data:
data.drop(columns="ID").describe()
# Change "timestamp" datatype to "datetime" and split it to columns:
def split_datetime(df):
df["timestamp"] = pd.to_datetime(
df["timestamp"], format="%Y-%m-%d %H:%M:%S", errors="coerce"
)
df["Year"] = df["timestamp"].dt.year
df["Month"] = df["timestamp"].dt.month
df["Day"] = df["timestamp"].dt.day
df["Hour"] = df["timestamp"].dt.hour
df.drop(columns="timestamp", inplace=True)
return df
data = split_datetime(data)
data.head()
# Read the weather data:
weather = pd.read_csv(os.path.join(dataset_path, "weather-sfcsv.csv"))
# Drop duplicate times/dates:
weather = weather.drop_duplicates(["Year", "Day", "Month", "Hour"])
print("The shape of the weather dataset is {}.\n\n".format(weather.shape))
weather.head()
# Clean weather data:
weather["Wind_Speed(mph)"].fillna(weather["Wind_Speed(mph)"].mean(), inplace=True)
weather = weather.drop(columns=["Wind_Chill(F)", "Precipitation(in)", "Selected"])
# Merge training data and weather data on "Year","Month","Day","Hour":
data_merged = data.merge(
weather,
"left",
left_on=["Year", "Month", "Day", "Hour"],
right_on=["Year", "Month", "Day", "Hour"],
)
# Drop the columns and clean the data:
data_merged = data_merged.drop(columns=["Month", "Day", "Hour", "Bump", "Roundabout"])
data_merged = data_merged.dropna()
data_merged = data_merged.drop_duplicates()
# Check the statistical discription of the prepared data:
data_merged.drop(columns="ID").describe()
columns = [
("Crossing", 5),
("Give_Way", 6),
("Junction", 7),
("No_Exit", 8),
("Railway", 9),
("Stop", 11),
("Amenity", 12),
("Side", 13),
("Weather_Condition", 15),
]
for col_name, index in columns:
factorize(data_merged, col_name, index)
data_merged.describe()
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
data_merged, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(
columns=["ID", "Severity", "Give_Way", "No_Exit", "Railway", "Amenity"]
)
y_train = train_df["Severity"]
X_val = val_df.drop(
columns=["ID", "Severity", "Give_Way", "No_Exit", "Railway", "Amenity"]
)
y_val = val_df["Severity"]
X_train.head()
# Use Random forest Regressor to show features importance:
from sklearn.ensemble import RandomForestRegressor
import numpy as np
import matplotlib.pyplot as plt
model = RandomForestRegressor(random_state=1, max_depth=10)
sss = pd.get_dummies(data_merged.drop(columns="Severity"))
model.fit(sss, data_merged.Severity)
features = sss.columns
importances = model.feature_importances_
indices = np.argsort(importances) # top 10 features
plt.title("Feature Importances")
plt.barh(range(len(indices)), importances[indices], color="b", align="center")
plt.yticks(range(len(indices)), [features[i] for i in indices])
plt.xlabel("Relative Importance")
plt.show()
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# ## Test data:
test_df = pd.read_csv(os.path.join(dataset_path, "test.csv"))
test_df.head()
# Change "timestamp" datatype to "datetime":
test_df = split_datetime(test_df)
test_df
# Merge training data and weather data on "Year","Month","Day","Hour":
test_df_merged = test_df.merge(
weather,
"left",
left_on=["Year", "Month", "Day", "Hour"],
right_on=["Year", "Month", "Day", "Hour"],
)
# Drop the columns:
test_df_merged = test_df_merged.drop(columns=["Month", "Day", "Hour"])
# Check the statistical discription of the prepared data
test_df_merged = test_df_merged.dropna()
test_df_merged = test_df_merged.drop_duplicates()
test_df_merged.drop(columns="ID").describe()
columns = [
("Bump", 3),
("Crossing", 5),
("Give_Way", 6),
("Junction", 7),
("No_Exit", 8),
("Railway", 9),
("Roundabout", 10),
("Stop", 11),
("Amenity", 12),
("Side", 13),
("Weather_Condition", 15),
]
for col_name, index in columns:
factorize(test_df_merged, col_name, index)
test_df_merged.head()
X_test = test_df_merged.drop(
columns=["ID", "Bump", "Roundabout", "Give_Way", "No_Exit", "Railway", "Amenity"]
)
y_test_predicted = classifier.predict(X_test)
test_df_merged["Severity"] = y_test_predicted
test_df_merged.head()
test_df_merged[["ID", "Severity"]].to_csv("/kaggle/working/submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/136/69136148.ipynb
| null | null |
[{"Id": 69136148, "ScriptId": 18855615, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7633490, "CreationDate": "07/27/2021 09:16:48", "VersionNumber": 7.0, "Title": "ITI AI Programming Challenge", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 144.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 143.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import os
dataset_path = "/kaggle/input/car-crashes-severity-prediction/"
# Factorization function:
def factorize(df, col_name, index):
data, unique = pd.factorize(df[col_name])
df.drop(columns=[col_name], inplace=True)
df.insert(index, col_name, data)
data = pd.read_csv(os.path.join(dataset_path, "train.csv"))
print("The shape of the dataset is {}.\n\n".format(data.shape))
data.head()
# Statistical discription of the data:
data.drop(columns="ID").describe()
# Change "timestamp" datatype to "datetime" and split it to columns:
def split_datetime(df):
df["timestamp"] = pd.to_datetime(
df["timestamp"], format="%Y-%m-%d %H:%M:%S", errors="coerce"
)
df["Year"] = df["timestamp"].dt.year
df["Month"] = df["timestamp"].dt.month
df["Day"] = df["timestamp"].dt.day
df["Hour"] = df["timestamp"].dt.hour
df.drop(columns="timestamp", inplace=True)
return df
data = split_datetime(data)
data.head()
# Read the weather data:
weather = pd.read_csv(os.path.join(dataset_path, "weather-sfcsv.csv"))
# Drop duplicate times/dates:
weather = weather.drop_duplicates(["Year", "Day", "Month", "Hour"])
print("The shape of the weather dataset is {}.\n\n".format(weather.shape))
weather.head()
# Clean weather data:
weather["Wind_Speed(mph)"].fillna(weather["Wind_Speed(mph)"].mean(), inplace=True)
weather = weather.drop(columns=["Wind_Chill(F)", "Precipitation(in)", "Selected"])
# Merge training data and weather data on "Year","Month","Day","Hour":
data_merged = data.merge(
weather,
"left",
left_on=["Year", "Month", "Day", "Hour"],
right_on=["Year", "Month", "Day", "Hour"],
)
# Drop the columns and clean the data:
data_merged = data_merged.drop(columns=["Month", "Day", "Hour", "Bump", "Roundabout"])
data_merged = data_merged.dropna()
data_merged = data_merged.drop_duplicates()
# Check the statistical discription of the prepared data:
data_merged.drop(columns="ID").describe()
columns = [
("Crossing", 5),
("Give_Way", 6),
("Junction", 7),
("No_Exit", 8),
("Railway", 9),
("Stop", 11),
("Amenity", 12),
("Side", 13),
("Weather_Condition", 15),
]
for col_name, index in columns:
factorize(data_merged, col_name, index)
data_merged.describe()
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
data_merged, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(
columns=["ID", "Severity", "Give_Way", "No_Exit", "Railway", "Amenity"]
)
y_train = train_df["Severity"]
X_val = val_df.drop(
columns=["ID", "Severity", "Give_Way", "No_Exit", "Railway", "Amenity"]
)
y_val = val_df["Severity"]
X_train.head()
# Use Random forest Regressor to show features importance:
from sklearn.ensemble import RandomForestRegressor
import numpy as np
import matplotlib.pyplot as plt
model = RandomForestRegressor(random_state=1, max_depth=10)
sss = pd.get_dummies(data_merged.drop(columns="Severity"))
model.fit(sss, data_merged.Severity)
features = sss.columns
importances = model.feature_importances_
indices = np.argsort(importances) # top 10 features
plt.title("Feature Importances")
plt.barh(range(len(indices)), importances[indices], color="b", align="center")
plt.yticks(range(len(indices)), [features[i] for i in indices])
plt.xlabel("Relative Importance")
plt.show()
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# ## Test data:
test_df = pd.read_csv(os.path.join(dataset_path, "test.csv"))
test_df.head()
# Change "timestamp" datatype to "datetime":
test_df = split_datetime(test_df)
test_df
# Merge training data and weather data on "Year","Month","Day","Hour":
test_df_merged = test_df.merge(
weather,
"left",
left_on=["Year", "Month", "Day", "Hour"],
right_on=["Year", "Month", "Day", "Hour"],
)
# Drop the columns:
test_df_merged = test_df_merged.drop(columns=["Month", "Day", "Hour"])
# Check the statistical discription of the prepared data
test_df_merged = test_df_merged.dropna()
test_df_merged = test_df_merged.drop_duplicates()
test_df_merged.drop(columns="ID").describe()
columns = [
("Bump", 3),
("Crossing", 5),
("Give_Way", 6),
("Junction", 7),
("No_Exit", 8),
("Railway", 9),
("Roundabout", 10),
("Stop", 11),
("Amenity", 12),
("Side", 13),
("Weather_Condition", 15),
]
for col_name, index in columns:
factorize(test_df_merged, col_name, index)
test_df_merged.head()
X_test = test_df_merged.drop(
columns=["ID", "Bump", "Roundabout", "Give_Way", "No_Exit", "Railway", "Amenity"]
)
y_test_predicted = classifier.predict(X_test)
test_df_merged["Severity"] = y_test_predicted
test_df_merged.head()
test_df_merged[["ID", "Severity"]].to_csv("/kaggle/working/submission.csv", index=False)
| false | 0 | 1,665 | 0 | 1,665 | 1,665 |
||
69136736
|
<jupyter_start><jupyter_text>scRNA-seq study of Neuron Development
Remark: for cell cycle analysis - see paper https://arxiv.org/abs/2208.05229
"Computational challenges of cell cycle analysis using single cell transcriptomics" Alexander Chervov, Andrei Zinovyev
## Data and Context
Data - results of single cell RNA sequencing, i.e. rows - correspond to cells, columns to genes (csv file is vice versa).
value of the matrix shows how strong is "expression" of the corresponding gene in the corresponding cell.
https://en.wikipedia.org/wiki/Single-cell_transcriptomics
Particular data from: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76381
There are original TXT files and reconversion to *.h5ad format which is more easy to work with.
There are several subdatasets human/mouse/different cell types.
Paper:
24( S),25-Epoxycholesterol and cholesterol 24S-hydroxylase ( CYP46A1) overexpression promote midbrain dopaminergic neurogenesis in vivo
https://pubmed.ncbi.nlm.nih.gov/30655290/
Abstract: The liver X receptors Lxrα/NR1H3 and Lxrβ/NR1H2 are ligand-dependent nuclear receptors critical for midbrain dopaminergic (mDA) neuron development. We found previously that 24(S),25-epoxycholesterol (24,25-EC), the most potent and abundant Lxr ligand in the developing mouse midbrain, promotes mDA neurogenesis in vitro In this study, we demonstrate that 24,25-EC promotes mDA neurogenesis in an Lxr-dependent manner in the developing mouse midbrain in vivo and also prevents toxicity induced by the Lxr inhibitor geranylgeranyl pyrophosphate. Furthermore, using MS, we show that overexpression of human cholesterol 24S-hydroxylase (CYP46A1) increases the levels of both 24(S)-hydroxycholesterol (24-HC) and 24,25-EC in the developing midbrain, resulting in a specific increase in mDA neurogenesis in vitro and in vivo, but has no effect on oculomotor or red nucleus neurogenesis. 24-HC, unlike 24,25-EC, did not affect in vitro neurogenesis, indicating that the neurogenic effect of 24,25-EC on mDA neurons is specific. Combined, our results indicate that increased levels of 24,25-EC in vivo, by intracerebroventricular delivery in WT mice or by overexpression of its biosynthetic enzyme CYP46A1, specifically promote mDA neurogenesis. We propose that increasing the levels of 24,25-EC in vivo may be a useful strategy to combat the loss of mDA neurons in Parkinson's disease.
## Inspiration
Single cell RNA sequencing is important technology in modern biology,
see e.g.
"Eleven grand challenges in single-cell data science"
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-1926-6
Also see review :
Nature. P. Kharchenko: "The triumphs and limitations of computational methods for scRNA-seq"
https://www.nature.com/articles/s41592-021-01171-x
Kaggle dataset identifier: scrnaseq-study-of-neuron-development
<jupyter_script># # What is about ?
# Cell cycle - look with splitting by subsets of data.
# Versions:
# 1 str_data_inf = ' GSE76381_ESMoleculeCounts '
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import scanpy as sc
import anndata
import scipy
import time
t0start = time.time()
import pandas as pd
import numpy as np
import os
import sys
import matplotlib.pyplot as plt
import matplotlib as mpl
mpl.rcParams["figure.dpi"] = 70
plt.style.use("dark_background")
import seaborn as sns
from sklearn.decomposition import PCA
# # Open file
# Similar to https://github.com/theislab/scanpy/issues/882
def read_mtx_by_prefix(path_and_fn_prefix):
adata = sc.read(path_and_fn_prefix + "matrix.mtx").T
genes = pd.read_csv(path_and_fn_prefix + "genes.tsv", header=None, sep="\t")
genes = genes.set_index(1)
genes.index.name = "Gene"
genes.columns = ["Ensemble"]
adata.var = genes
barcodes = pd.read_csv(
path_and_fn_prefix + "barcodes.tsv", header=None, sep="\t", index_col=0
)
barcodes.index.name = "barcodes"
adata.obs = barcodes
return adata
if 1:
str_data_inf = " GSE76381_iPSMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_iPSMoleculeCounts.h5ad"
str_data_inf = " GSE76381_MouseEmbryoMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_MouseEmbryoMoleculeCounts.h5ad"
str_data_inf = " GSE76381_MouseAdultDAMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_MouseAdultDAMoleculeCounts.h5ad"
str_data_inf = " GSE76381_EmbryoMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_EmbryoMoleculeCounts.h5ad"
str_data_inf = " GSE76381_ESMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_ESMoleculeCounts.h5ad"
adata_orig = sc.read(path_and_fn_prefix)
else:
str_data_inf = " GSM3267241_U5_all "
path_and_fn_prefix = "/kaggle/input/scrnaseq-neuroepithelialderived-cells-and-glioma/GSM3267241_U5_all_"
adata_orig = read_mtx_by_prefix(path_and_fn_prefix)
genes_upper = [
t.upper() for t in adata_orig.var.index
] # Simple conversion mouse genes to human
v = adata_orig.var.copy()
v.index = genes_upper
adata_orig.var = v.copy()
list_save_original_obs_columns = list(adata_orig.obs.columns)
adata = adata_orig.copy()
adata_orig
# # Preliminary functions and preprocessing functions
def _smooth_adata_by_pooling(
adata, X_embed, n_neighbours=10, copy=False, output_mode=1
):
# adata_pooled = adata.copy if copy else adata
nbrs = NearestNeighbors(n_neighbors=n_neighbours).fit(X_embed)
if output_mode > 0:
print("done: nbrs = NearestNeighbors(n_neighbors=n_neighbours).fit(X_embed) ")
distances, indices = nbrs.kneighbors(X_embed)
if output_mode > 0:
print("done: distances, indices = nbrs.kneighbors(X_embed) ")
nd_array_X = _get_nd_array(adata.X)
if output_mode > 0:
print("done: nd_array_X = _get_nd_array(adata.X)")
adata.X = _smooth_matrix_by_pooling(nd_array_X, indices)
if output_mode > 0:
print(
"done: adata.X = _smooth_matrix_by_pooling(_get_nd_array(adata.X), indices) "
)
if "matrix" in adata.layers:
adata.layers["matrix"] = _smooth_matrix_by_pooling(
_get_nd_array(adata.layers["matrix"]), indices
)
if "spliced" in adata.layers:
adata.layers["spliced"] = _smooth_matrix_by_pooling(
_get_nd_array(adata.layers["spliced"]), indices
)
if "unspliced" in adata.layers:
adata.layers["unspliced"] = _smooth_matrix_by_pooling(
_get_nd_array(adata.layers["unspliced"]), indices
)
adata.uns["scycle"] = True
def _smooth_matrix_by_pooling(matrix, indices):
matrix_pooled = matrix.copy()
t0 = time.time()
# print(indices.shape, matrix.shape, type(matrix) )
for i in range(len(indices)):
# print(i,len(indices), time.time()-t0 )
matrix_pooled[i, :] = np.mean(matrix[indices[i], :], axis=0)
return matrix_pooled
def _get_nd_array(arr):
x = None
if str(type(arr)):
x = arr
else:
print("start: x = arr.toarray()")
x = arr.toarray()
print("finished: x = arr.toarray()")
return x
def calc_scores(anndata, signature_dict):
matrix = anndata.to_df().to_numpy()
scores_dic = {}
for key in signature_dict:
names = np.array(signature_dict[key])
inds = np.where(np.isin(anndata.var_names, names))[0]
matrix_sel = matrix[:, inds]
scores = np.mean(matrix_sel, axis=1)
scores_dic[key] = scores
return scores_dic
def find_nonproliferative_cells(
adata,
estimation_fraction_nonproliferating_cells=0.3,
number_of_nodes=30,
max_number_of_iterations=20,
number_of_sigmas=3.0,
Mu=1.0,
):
all_markers = list(adata.uns["S-phase_genes"]) + list(adata.uns["G2-M_genes"])
Xccm = adata[:, list(set(all_markers) & set(adata.var_names))].X
cc_score = np.array(list(np.mean(Xccm, axis=1)))
ind_sorted_prolif = np.argsort(cc_score)
ind_nonprolif = ind_sorted_prolif[
0 : int(len(adata) * estimation_fraction_nonproliferating_cells)
]
adata.obs["proliferating"] = np.empty(len(adata)).astype(np.int)
adata.obs["proliferating"][:] = 1
adata.obs["proliferating"][ind_nonprolif] = 0
sc.pl.scatter(adata, x="S-phase", y="G2-M", color="proliferating")
fraction_nonprolif_old = estimation_fraction_nonproliferating_cells
for i in range(max_number_of_iterations):
X_elpigraph_training = (
adata.obs[["S-phase", "G2-M"]].to_numpy().astype(np.float64)
)
u = X_elpigraph_training.copy()
ind_prolif = np.where(np.array(adata.obs["proliferating"]) == 1)[0]
X_elpigraph_training = X_elpigraph_training[ind_prolif, :]
egr = elpigraph.computeElasticPrincipalCircle(
X_elpigraph_training,
number_of_nodes,
Mu=Mu,
drawPCAView=False,
verbose=False,
)
partition, dists = elpigraph.src.core.PartitionData(
X=u,
NodePositions=egr[0]["NodePositions"],
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(u**2, axis=1, keepdims=1),
)
mndist = np.mean(dists[ind_prolif])
# plt.hist(dists,bins=100)
# plt.show()
# plt.hist(dists[ind_prolif],bins=100)
# plt.show()
intervaldist = np.std(dists[ind_prolif]) * number_of_sigmas
tt1 = np.array(1 - adata.obs["proliferating"])
tt2 = np.zeros(len(tt1)).astype(np.int)
for k, d in enumerate(dists):
if d > mndist + intervaldist:
tt2[k] = 1
nonprolif_new = np.array(tt1) * np.array(tt2)
adata.obs["proliferating"] = 1 - nonprolif_new
# adata.obs['dists'] = dists
# sc.pl.scatter(adata,x='S-phase',y='G2-M',color='dists')
fraction_nonprolif = 1 - np.sum(adata.obs["proliferating"]) / len(adata)
print(
"\n\n===========\nIteration",
i,
"Fraction of non-proliferating cells:",
fraction_nonprolif,
"\n==============\n\n\n",
)
if np.abs(fraction_nonprolif - fraction_nonprolif_old) < 0.01:
break
fraction_nonprolif_old = fraction_nonprolif
def _compute_principal_circle(adata, number_of_nodes=30, n_components=30):
# driving_feature = np.array(adata_orig[adata.uns['ind_samples']].obs['total_counts'])
# driving_feature_weight = 1.0
# egr, partition, X, Xp = _compute_principal_circle(adata,number_of_nodes=40,
# driving_feature=driving_feature,driving_feature_weight=0.1)
X = adata.X
X_prolif = adata.X[np.where(adata.obs["proliferating"] == 1)[0], :]
# driving_feature = driving_feature[adata.obs[np.where(adata.obs['proliferating']==1)[0]]]
mn_prolif = np.mean(X_prolif, axis=0)
pca = PCA(n_components=n_components)
u = pca.fit_transform(X_prolif)
v = pca.components_.T
# X_pca = adata.obsm['X_pca'].astype(np.float64)
X_elpigraph_training = u.astype(np.float64)
# std = np.std(X_elpigraph_training)
# driving_feature = stats.zscore(driving_feature)*std*driving_feature_weight
# driving_feature = driving_feature.reshape(-1,1)
# X_elpigraph_training = np.concatenate([X_elpigraph_training,driving_feature],axis=1)
# print('std=',std)
# print('std df=',np.std(driving_feature))
# print(X_elpigraph_training.shape)
egr = elpigraph.computeElasticPrincipalCircle(
X_elpigraph_training, number_of_nodes, Mu=0.2
)
partition, dists = elpigraph.src.core.PartitionData(
X=X_elpigraph_training,
NodePositions=egr[0]["NodePositions"],
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(X_elpigraph_training**2, axis=1, keepdims=1),
)
Xp = (X - mn_prolif) @ v
# number_of_driving_features = driving_feature.shape[1]
# node_positions_reduced = egr[0]['NodePositions'][:,:-number_of_driving_features]
nodep = egr[0]["NodePositions"]
partition, dists = elpigraph.src.core.PartitionData(
X=Xp,
NodePositions=nodep,
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(Xp**2, axis=1, keepdims=1),
)
# nodep = egr[0]['NodePositions'][:,:-number_of_driving_features]
# edges = egr[0]['Edges'][0]
# X_elpigraph_training = X_elpigraph_training[:,:-number_of_driving_features]
# egr[0]['NodePositions'] = nodep
return egr, partition, X_elpigraph_training, Xp
def _compute_principal_curve(
adata, number_of_nodes=30, Mu=0.3, Lambda=0.001, n_components=30
):
X = adata.X
X_prolif = adata.X[np.where(adata.obs["proliferating"] == 1)[0], :]
mn_prolif = np.mean(X_prolif, axis=0)
pca = PCA(n_components=n_components)
u = pca.fit_transform(X_prolif)
v = pca.components_.T
# X_pca = adata.obsm['X_pca'].astype(np.float64)
adata.varm["pc_components_elpigraph"] = v
adata.varm["mean_point_elpigraph"] = mn_prolif
X_elpigraph_training = u.astype(np.float64)
egr, starting_node = compute_principal_curve_from_circle(
X_elpigraph_training,
n_nodes=number_of_nodes,
produceTree=False,
Mu=Mu,
Lambda=Lambda,
)
partition_, dists = elpigraph.src.core.PartitionData(
X=X_elpigraph_training,
NodePositions=egr[0]["NodePositions"],
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(X_elpigraph_training**2, axis=1, keepdims=1),
)
Xp = (X - mn_prolif) @ v
partition_, dists = elpigraph.src.core.PartitionData(
X=Xp,
NodePositions=egr[0]["NodePositions"],
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(Xp**2, axis=1, keepdims=1),
)
nodep = egr[0]["NodePositions"]
edges = egr[0]["Edges"][0]
return egr, partition_, starting_node, X_elpigraph_training, Xp
def compute_principal_curve_from_circle(
X, n_nodes=30, Mu=0.1, Lambda=0.01, produceTree=False
):
egr = elpigraph.computeElasticPrincipalCircle(X, int(n_nodes / 2), Mu=Mu)
nodep = egr[0]["NodePositions"]
edges = egr[0]["Edges"][0]
partition_, dists = elpigraph.src.core.PartitionData(
X=X,
NodePositions=nodep,
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(X**2, axis=1, keepdims=1),
)
node_sizes = np.array(
[len(np.where(partition_ == i)[0]) for i in range(nodep.shape[0])]
)
node_min = np.argmin(node_sizes)
edges_open = edges.copy()
k = 0
starting_node = -1
while k < edges_open.shape[0]:
e = edges_open[k, :]
if (e[0] == node_min) | (e[1] == node_min):
edges_open = np.delete(edges_open, k, axis=0)
if e[0] == node_min:
starting_node = e[1]
if e[1] == node_min:
starting_node = e[0]
else:
k = k + 1
nodep_open = np.delete(nodep, node_min, axis=0)
if starting_node > node_min:
starting_node = starting_node - 1
for e in edges_open:
if e[0] > node_min:
e[0] = e[0] - 1
if e[1] > node_min:
e[1] = e[1] - 1
if produceTree:
egrl = elpigraph.computeElasticPrincipalTree(
X,
n_nodes,
InitNodePositions=nodep_open,
InitEdges=edges_open,
Lambda=Lambda,
Mu=Mu,
alpha=0.01,
FinalEnergy="Penalized",
)
else:
egrl = elpigraph.computeElasticPrincipalCurve(
X,
n_nodes,
InitNodePositions=nodep_open,
InitEdges=edges_open,
Lambda=Lambda,
Mu=Mu,
alpha=0.01,
FinalEnergy="Penalized",
)
return egrl, starting_node
def subtract_cell_cycle_trajectory(X, partition):
points = range(X.shape[0])
r2scores = []
X1 = X[points, :]
X_ro = np.zeros((X.shape[0], X.shape[1]))
partition_points = partition[points]
inds = {}
for k in range(len(partition_points)):
j = partition_points[k][0]
if not j in inds:
inds[j] = [k]
else:
inds[j].append(k)
XT = X1.T
for j in range(X1.shape[0]):
k = partition_points[j][0]
ind = np.array(inds[k])
X_ro[j, :] = (XT[:, j] - np.mean(XT[:, ind], axis=1)).T
residue_matrix = X1 - X_ro
residues_var = np.var(residue_matrix, axis=0)
vrs = np.var(X1, axis=0)
r2scores = residues_var / vrs
return X_ro, residue_matrix, r2scores
list_genes2include_mandotory = [
"E2F1",
"FOXM1",
"ESRRB",
"NR5A2",
"HIST1H4C",
"FOXO3",
"EZH1",
"ANLN",
]
list_genes2include_mandotory += ["PCNA", "TOP2A"]
list_genes2include_mandotory += [
"WEE1",
"WEE2",
] # Inhibitor of CDK1 https://en.wikipedia.org/wiki/Wee1
list_genes2include_mandotory += [
"CDK1",
"UBE2C",
"TOP2A",
"HIST1H4E",
"HIST1H4C",
] # Hsiao2020GenomeResearch 5 genes 'HIST1H4E' = H4C5,'HIST1H4C'=H4C3
list_genes2include_mandotory += [
"MYBL2",
"TP53",
"KLF4",
"CDKN1A",
"CDKN2A",
"RB1",
"MDM2",
"MDM4",
"ZBTB20",
"GAPDH",
"EFNA5",
"LINC00511",
]
# MYBL2 https://en.wikipedia.org/wiki/MYBL2 - transcription factor
# The protein encoded by this gene, a member of the MYB family of transcription factor genes, is a nuclear protein involved in cell cycle progression. The encoded protein is phosphorylated by cyclin A/cyclin-dependent kinase 2 during the S-phase of the cell cycle and possesses both activator and repressor activities.
# https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6788831/
# Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb Christin F Down 1, Julie Millour, Eric W-F Lam, Roger J Watson
# https://pubmed.ncbi.nlm.nih.gov/23555285/
# Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB -
list_genes2include_mandotory += (
["CCNE1", "CCNE2", "CDK2"]
+ ["CCNB1", "CCNB2", "CCNB3"]
+ ["CCNA1", "CCNA2", "CCNF", "CDK1"]
+ ["CCND1", "CCND2", "CCND3"]
+ ["CDK4", "CDK6"]
)
# 'CCNA1','CCNA2', 'CCNE1', 'CCNE2',]
list_genes2include_mandotory += [
"CDK11A", # All of the form: CDKnumber* - 27
"CDK14",
"CDK17",
"CDK13",
"CDK5RAP1",
"CDK16",
"CDK6",
"CDK5RAP3",
"CDK2AP1",
"CDK18",
"CDK2",
"CDK8",
"CDK7",
"CDK4",
"CDK9",
"CDK5RAP2",
"CDK15",
"CDK19",
"CDK20",
"CDK5",
"CDK12",
"CDK2AP2",
"CDK1",
"CDK5R2",
"CDK5R1",
"CDK10",
"CDK11B",
]
list_genes2include_mandotory += [
"CDKN3", # 10 CDKN* - A cyclin-dependent kinase inhibitor protein is a protein which inhibits the enzyme cyclin-dependent kinase (CDK).
"CDKN1B",
"CDKN2C",
"CDKN1A",
"CDKN2D",
"CDKN1C",
"CDKN2A",
"CDKN2AIP",
"CDKN2AIPNL",
"CDKN2B-AS1",
]
# p16 CDKN2A Cyclin-dependent kinase 4, Cyclin-dependent kinase 6
# p15 CDKN2B Cyclin-dependent kinase 4
# p18 CDKN2C Cyclin-dependent kinase 4, Cyclin-dependent kinase 6
# p19 CDKN2D Cyclin-dependent kinase 4, Cyclin-dependent kinase 6
# p21 / WAF1 CDKN1A[2] Cyclin E1/Cyclin-dependent kinase 2
# p27 CDKN1B Cyclin D3/Cyclin-dependent kinase 4, Cyclin E1/Cyclin-dependent kinase 2
# p57 CDKN1C Cyclin E1/Cyclin-dependent kinase 2
# https://en.wikipedia.org/wiki/Cyclin-dependent_kinase_inhibitor_protein
list_genes2include_mandotory += [
"CDKL3", # All starts with CDK - 43
"CDKL5",
"CDK11A",
"CDK14",
"CDK17",
"CDK13",
"CDKL1",
"CDKN3",
"CDK5RAP1",
"CDK16",
"CDK6",
"CDK5RAP3",
"CDKN1B",
"CDK2AP1",
"CDK18",
"CDKN2C",
"CDK2",
"CDKN1A",
"CDKN2D",
"CDKN1C",
"CDK8",
"CDK7",
"CDK4",
"CDK9",
"CDK5RAP2",
"CDK15",
"CDKL2",
"CDKAL1",
"CDKN2A",
"CDK19",
"CDK20",
"CDK5",
"CDK12",
"CDK2AP2",
"CDKN2AIP",
"CDK1",
"CDK5R2",
"CDK5R1",
"CDK10",
"CDKL4",
"CDKN2AIPNL",
"CDKN2B-AS1",
"CDK11B",
]
list_genes2include_mandotory += [
"CDC27", # "cdc" in its name refers to "cell division cycle"
"CDC42",
"CDC14A",
"CDC14B",
"CDC45",
"CDC6",
"CDC23",
"CDC5L",
"CDC7",
"CDC34",
"CDC25B",
"CDC37",
"CDC37L1",
"CDCA3",
"CDC20",
"CDC42EP1",
"CDC16",
"CDC73",
"CDCA8",
"CDC42BPA",
"CDCA7",
"CDCA5",
"CDC42EP2",
"CDC123",
"CDC25C",
"CDC42SE2",
"CDC42EP3",
"CDCP1",
"CDC25A",
"CDC20B",
"CDCA7L",
"CDC42EP5",
"CDC40",
"CDCA4",
"CDC42BPG",
"CDC26",
"CDC42EP4",
"CDCA2",
"CDC42SE1",
"CDC42BPB",
"CDC42-IT1",
"CDC20P1",
"CDC42P6",
"CDC27P2",
"CDC42P5",
"CDC37L1-AS1",
"CDC27P3",
]
# Cdc25 is a dual-specificity phosphatase first isolated from the yeast Schizosaccharomyces pombe as a cell cycle defective mutant.[2] As with other cell cycle proteins or genes such as Cdc2 and Cdc4, the "cdc" in its name refers to "cell division cycle". Dual-specificity phosphatases are considered a sub-class of protein tyrosine phosphatases. By removing inhibitory phosphate residues from target cyclin-dependent kinases (Cdks),[3] Cdc25 proteins control entry into and progression through various phases of the cell cycle, including mitosis and S ("Synthesis") phase.
list_genes2include_mandotory += [
"E2F2", # 10 genes E2F transc. factors
"E2F1",
"E2F3",
"E2F8",
"E2F5",
"E2F7",
"E2F6",
"E2F3P2",
"E2F4",
"E2F3-IT1",
]
list_genes2include_mandotory += ["GATA1", "FOXA1", "BRD4", "KMT2A"]
# 'GATA1', 'FOXA1', 'BRD4', 'KMT2A' - transcription factors are retained in mitotic chromosomes (from paper "Trans landscape of the human cell cycle" )
list_histone_genes_from_wiki = [
"H1F0",
"H1FNT",
"H1FOO",
"H1FX",
"HIST1H1A",
"HIST1H1B",
"HIST1H1C",
"HIST1H1D",
"HIST1H1E",
"HIST1H1T",
"H2AFB1",
"H2AFB2",
"H2AFB3",
"H2AFJ",
"H2AFV",
"H2AFX",
"H2AFY",
"H2AFY2",
"H2AFZ",
"HIST1H2AA",
"HIST1H2AB",
"HIST1H2AC",
"HIST1H2AD",
"HIST1H2AE",
"HIST1H2AG",
"HIST1H2AI",
"HIST1H2AJ",
"HIST1H2AK",
"HIST1H2AL",
"HIST1H2AM",
"HIST2H2AA3",
"HIST2H2AC",
"H2BFM",
"H2BFS",
"H2BFWT",
"HIST1H2BA",
"HIST1H2BB",
"HIST1H2BC",
"HIST1H2BD",
"HIST1H2BE",
"HIST1H2BF",
"HIST1H2BG",
"HIST1H2BH",
"HIST1H2BI",
"HIST1H2BJ",
"HIST1H2BK",
"HIST1H2BL",
"HIST1H2BM",
"HIST1H2BN",
"HIST1H2BO",
"HIST2H2BE",
"HIST1H3A",
"HIST1H3B",
"HIST1H3C",
"HIST1H3D",
"HIST1H3E",
"HIST1H3F",
"HIST1H3G",
"HIST1H3H",
"HIST1H3I",
"HIST1H3J",
"HIST2H3C",
"HIST3H3",
"HIST1H4A",
"HIST1H4B",
"HIST1H4C",
"HIST1H4D",
"HIST1H4E",
"HIST1H4F",
"HIST1H4G",
"HIST1H4H",
"HIST1H4I",
"HIST1H4J",
"HIST1H4K",
"HIST1H4L",
"HIST4H4",
]
list_genes2include_mandotory += list_histone_genes_from_wiki
S_phase_genes_Tirosh = [
"MCM5",
"PCNA",
"TYMS",
"FEN1",
"MCM2",
"MCM4",
"RRM1",
"UNG",
"GINS2",
"MCM6",
"CDCA7",
"DTL",
"PRIM1",
"UHRF1",
"MLF1IP",
"HELLS",
"RFC2",
"RPA2",
"NASP",
"RAD51AP1",
"GMNN",
"WDR76",
"SLBP",
"CCNE2",
"UBR7",
"POLD3",
"MSH2",
"ATAD2",
"RAD51",
"RRM2",
"CDC45",
"CDC6",
"EXO1",
"TIPIN",
"DSCC1",
"BLM",
"CASP8AP2",
"USP1",
"CLSPN",
"POLA1",
"CHAF1B",
"BRIP1",
"E2F8",
]
G2_M_genes_Tirosh = [
"HMGB2",
"CDK1",
"NUSAP1",
"UBE2C",
"BIRC5",
"TPX2",
"TOP2A",
"NDC80",
"CKS2",
"NUF2",
"CKS1B",
"MKI67",
"TMPO",
"CENPF",
"TACC3",
"FAM64A",
"SMC4",
"CCNB2",
"CKAP2L",
"CKAP2",
"AURKB",
"BUB1",
"KIF11",
"ANP32E",
"TUBB4B",
"GTSE1",
"KIF20B",
"HJURP",
"CDCA3",
"HN1",
"CDC20",
"TTK",
"CDC25C",
"KIF2C",
"RANGAP1",
"NCAPD2",
"DLGAP5",
"CDCA2",
"CDCA8",
"ECT2",
"KIF23",
"HMMR",
"AURKA",
"PSRC1",
"ANLN",
"LBR",
"CKAP5",
"CENPE",
"CTCF",
"NEK2",
"G2E3",
"GAS2L3",
"CBX5",
"CENPA",
]
from sklearn.neighbors import NearestNeighbors
def preproc(adata_orig, output_mode=1):
adata = adata_orig.copy()
from sklearn.neighbors import NearestNeighbors
import scipy
# First standard preprocessing
# Then "pooling" - fighting with zeros
# Params:
n_top_genes_to_keep = 10000
threshold_pct_counts_mt = 40
min_count = 500 # Threshold on min total_counts
max_count = np.inf # 10000# Threshold on max total_counts
if output_mode > 0:
print(adata_orig.X.sum())
# ################################################################################################
# Preprocessing first step: filter CELLs by counts and level of MT-percent
# thresholds are set visually looking on violin plots
# Examples:
# threshold_pct_counts_mt = 20 - 40 min_count = 500 - 1000; max_count = 10000 - 12000;
# These thresholds depends on cell line dataset
if output_mode > 0:
print(adata)
adata.var["mt"] = adata.var_names.str.startswith(
"MT-"
) # annotate the group of mitochondrial genes as 'mt'
# sv.pp.remove_duplicate_cells(adata)
sc.pp.calculate_qc_metrics(
adata, qc_vars=["mt"], percent_top=None, log1p=False, inplace=True
)
if output_mode > 0:
sc.pl.violin(
adata,
["n_genes_by_counts", "total_counts", "pct_counts_mt"],
jitter=1.9,
multi_panel=True,
)
median_count = np.median(adata.obs["total_counts"])
if output_mode > 0:
print("Median total counts =", median_count)
print("min(n_genes_by_counts):", adata.obs["n_genes_by_counts"].min())
print("min(total_counts):", adata.obs["total_counts"].min())
# min_count = np.max((median_count/2,5000))
if output_mode > 0:
print("Thresholds: min_count=", min_count, "max_count=", max_count)
print(
"Look at total_counts va MT-percent, expect some linear dependence - but does not happen: "
)
sc.pl.scatter(adata, x="total_counts", y="pct_counts_mt")
inds1 = np.where(
(adata.obs["total_counts"] > min_count)
& (adata.obs["total_counts"] < max_count)
)
inds2 = np.where(adata.obs["pct_counts_mt"] < threshold_pct_counts_mt)
if output_mode > 0:
print(len(inds1[0]), "samples pass the count filter")
print(len(inds2[0]), " samples pass the mt filter")
ind_samples = np.intersect1d(inds1[0], inds2[0])
if output_mode > 0:
print("Samples selected", len(ind_samples))
adata.uns["ind_samples"] = ind_samples
# Here we cut cells. Filtering out those with counts too low or too big
adata = adata[ind_samples, :]
# ################################################################################################
# Preprocessing second step:
# 1) normalization to some value, i.e. median of the total counts
# 2) taking logs
# 3) keeping only higly variable genes
sc.pp.normalize_total(adata, target_sum=np.median(adata.obs["total_counts"]))
sc.pp.log1p(adata)
try:
sc.pp.highly_variable_genes(
adata, n_top_genes=np.min([X.shape[1], n_top_genes_to_keep]), n_bins=20
)
ind_genes = np.where(adata.var["highly_variable"])[0]
except:
ind_genes = np.arange(
adata.X.shape[1]
) # np.where(adata.var['highly_variable'])[0]
ind_genes2 = np.where(adata.var.index.isin(list_genes2include_mandotory))[0]
ind_genes = list(set(ind_genes) | set(ind_genes2))
adata = adata[:, ind_genes]
if output_mode > 0:
print("Violin plots after filtering cells and genes")
sc.pl.violin(
adata,
["n_genes_by_counts", "total_counts", "pct_counts_mt"],
jitter=1.9,
multi_panel=True,
)
# ################################################################################################
# Preprocessing third step:
# Mainly: Create non-sparse adata (otherwise next step - pooling works extremely slow)
# 1) calculate PCA for adata previously calculated adata (which is cutted, normed, logged) - and keep it for use in pooling
# 2) create new non-sparse adata with cells and genes, obtained by filters on previous steps
# print('Check - we should see ints, otherwise data has been corrupted: ')
# print(adata_orig.X[:4,:5].toarray())
sc.tl.pca(adata, n_comps=30)
X_pca = adata.obsm["X_pca"]
X_pca1 = X_pca.copy()
adata_orig[ind_samples, ind_genes].var.shape
if scipy.sparse.issparse(adata.X):
XX2 = adata_orig[ind_samples, ind_genes].X.toarray()
else:
XX2 = adata_orig[ind_samples, ind_genes].X.copy()
adata = anndata.AnnData(
XX2,
obs=adata_orig[ind_samples, ind_genes].obs.copy(),
var=adata_orig[ind_samples, ind_genes].var.copy(),
) #
if output_mode > 0:
print("New non-sparse adata: ", adata)
# sv.pp.remove_duplicate_cells(adata)
adata.var["mt"] = adata.var_names.str.startswith(
"MT-"
) # annotate the group of mitochondrial genes as 'mt'
sc.pp.calculate_qc_metrics(
adata, qc_vars=["mt"], percent_top=None, log1p=False, inplace=True
)
if output_mode > 0:
print("Violin plots for new adata")
sc.pl.violin(
adata,
["n_genes_by_counts", "total_counts", "pct_counts_mt"],
jitter=1.9,
multi_panel=True,
)
# ################################################################################################
# Preprocessing 4-th step: may take quite long - 10 minutes for 40K celss
# Mainly: pooling - we are making pooling on original counts, but calculate KNN graph using PCA obtained from normed/log data
# (that is why we forget adata obtained on the steps 1-2, and will do norm-log again here )
# 1) pooling
# 2) quality metrics calculations
# 3) normalization
# 4) log
t0 = time.time()
_smooth_adata_by_pooling(adata, X_pca, n_neighbours=20, output_mode=output_mode)
if output_mode > 0:
print(time.time() - t0, "seconds passed")
sc.pp.calculate_qc_metrics(adata, percent_top=None, log1p=False, inplace=True)
sc.pp.normalize_total(adata, target_sum=np.median(adata.obs["total_counts"]))
sc.pp.log1p(adata)
# adata = adata[:,ind_genes]
adata.uns["ind_genes"] = ind_genes
sc.tl.pca(adata, n_comps=30)
X_pca = adata.obsm["X_pca"]
return adata
# adata = preproc(adata_orig, output_mode = 0)
# adata
# # Main Loop
import warnings
warnings.filterwarnings("ignore")
adata = preproc(adata_orig, output_mode=0)
adata.obs["Timepoint"].unique()[::-1]
n_x_subplots = 1
t00 = time.time()
print("Start processing. ")
# print( series_GSE_and_cell_count[mm] )
df_stat = pd.DataFrame()
ix4df_stat = 0
c = 0
if 1:
mask = np.ones(adata.shape[0]).astype(bool)
ix4df_stat += 1
# print()
# print(GSE)
t0 = time.time()
list_genes_upper = [t.upper() for t in adata.var.index]
I = np.where(pd.Series(list_genes_upper).isin(S_phase_genes_Tirosh))[0]
v1 = adata[mask].X[:, I].mean(axis=1)
I = np.where(pd.Series(list_genes_upper).isin(G2_M_genes_Tirosh))[0]
v2 = adata[mask].X[:, I].mean(axis=1)
if c % n_x_subplots == 0:
fig = plt.figure(figsize=(25, 10))
c = 0
c += 1
fig.add_subplot(1, n_x_subplots, c)
color_by = adata.obs["Timepoint"]
sns.scatterplot(x=v1, y=v2, hue=color_by) # adata.obs['pct_counts_mt'])
corr_phases = np.round(np.corrcoef(v1, v2)[0, 1], 2)
plt.title(str_data_inf, fontsize=17)
if c == n_x_subplots:
plt.show()
# print( np.round(time.time() - t0,1 ) , 'seconds passed ', np.round(time.time() - t00,1 ) , 'seconds passed total')
print(
np.round(time.time() - t00, 1),
np.round((time.time() - t00) / 60, 1),
"total seconds,minutes passed",
)
plt.show()
df_stat
n_x_subplots = 2
t00 = time.time()
print("Start processing. ")
# print( series_GSE_and_cell_count[mm] )
df_stat = pd.DataFrame()
ix4df_stat = 0
c = 0
for uv in [
"day_0",
"day_12",
"day_17",
"day_35",
]: # adata.obs['Timepoint'].unique()[::-1]:
mask = adata.obs["Timepoint"] == uv
ix4df_stat += 1
# print()
# print(GSE)
t0 = time.time()
list_genes_upper = [t.upper() for t in adata.var.index]
I = np.where(pd.Series(list_genes_upper).isin(S_phase_genes_Tirosh))[0]
v1 = adata[mask].X[:, I].mean(axis=1)
I = np.where(pd.Series(list_genes_upper).isin(G2_M_genes_Tirosh))[0]
v2 = adata[mask].X[:, I].mean(axis=1)
if c % n_x_subplots == 0:
fig = plt.figure(figsize=(25, 10))
c = 0
c += 1
fig.add_subplot(1, n_x_subplots, c)
# sns.scatterplot(x = v1, y = v2 , hue = adata.obs['pct_counts_mt'])
I = np.where("CCNE2" == np.array(list_genes_upper))[0][0]
color_by = pd.Series(adata[mask].X[:, I] > np.median(adata.X[:, I]))
color_by.name = "CCNE2"
if len(np.unique(color_by)) == 2:
sns.scatterplot(
x=v1, y=v2, hue=color_by, palette=["green", "red"]
) # obs['pct_counts_mt'])
else:
sns.scatterplot(
x=v1, y=v2
) # , hue = color_by, palette = ['red','green'] )# obs['pct_counts_mt'])
corr_phases = np.round(np.corrcoef(v1, v2)[0, 1], 2)
df_stat.loc[ix4df_stat, "SubType"] = uv
df_stat.loc[ix4df_stat, "n_cells"] = mask.sum()
df_stat.loc[ix4df_stat, "Phase Correlation"] = corr_phases
df_stat.loc[ix4df_stat, "Info"] = str_data_inf
plt.title(
uv
+ " "
+ str_data_inf
+ " Corr "
+ str(corr_phases)
+ " n_cells "
+ str(mask.sum()),
fontsize=17,
)
plt.title(
uv + " Corr " + str(corr_phases) + " n_cells " + str(mask.sum()), fontsize=17
)
if c == n_x_subplots:
plt.show()
# print( np.round(time.time() - t0,1 ) , 'seconds passed ', np.round(time.time() - t00,1 ) , 'seconds passed total')
print(
np.round(time.time() - t00, 1),
np.round((time.time() - t00) / 60, 1),
"total seconds,minutes passed",
)
plt.show()
df_stat
print(
np.round(time.time() - t0start, 1),
np.round((time.time() - t0start) / 60, 1),
np.round((time.time() - t0start) / 3600, 1),
"total seconds/minutes/hours passed",
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/136/69136736.ipynb
|
scrnaseq-study-of-neuron-development
|
alexandervc
|
[{"Id": 69136736, "ScriptId": 18870863, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2262596, "CreationDate": "07/27/2021 09:25:21", "VersionNumber": 1.0, "Title": "cell cycle briefly with subsets Neuron development", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 683.0, "LinesInsertedFromPrevious": 683.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 91955415, "KernelVersionId": 69136736, "SourceDatasetVersionId": 2465622}]
|
[{"Id": 2465622, "DatasetId": 1492357, "DatasourceVersionId": 2508068, "CreatorUserId": 2262596, "LicenseName": "Unknown", "CreationDate": "07/26/2021 15:54:45", "VersionNumber": 3.0, "Title": "scRNA-seq study of Neuron Development", "Slug": "scrnaseq-study-of-neuron-development", "Subtitle": "GSE76381 Midbrain and Dopaminergic Neuron Development in Mouse,Human,Stem Cells", "Description": "Remark: for cell cycle analysis - see paper https://arxiv.org/abs/2208.05229\n\"Computational challenges of cell cycle analysis using single cell transcriptomics\" Alexander Chervov, Andrei Zinovyev\n\n## Data and Context\n\nData - results of single cell RNA sequencing, i.e. rows - correspond to cells, columns to genes (csv file is vice versa).\nvalue of the matrix shows how strong is \"expression\" of the corresponding gene in the corresponding cell.\nhttps://en.wikipedia.org/wiki/Single-cell_transcriptomics\n\nParticular data from: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76381\nThere are original TXT files and reconversion to *.h5ad format which is more easy to work with.\nThere are several subdatasets human/mouse/different cell types. \n\nPaper: \n24( S),25-Epoxycholesterol and cholesterol 24S-hydroxylase ( CYP46A1) overexpression promote midbrain dopaminergic neurogenesis in vivo \n\nhttps://pubmed.ncbi.nlm.nih.gov/30655290/\n\nAbstract: The liver X receptors Lxr\u03b1/NR1H3 and Lxr\u03b2/NR1H2 are ligand-dependent nuclear receptors critical for midbrain dopaminergic (mDA) neuron development. We found previously that 24(S),25-epoxycholesterol (24,25-EC), the most potent and abundant Lxr ligand in the developing mouse midbrain, promotes mDA neurogenesis in vitro In this study, we demonstrate that 24,25-EC promotes mDA neurogenesis in an Lxr-dependent manner in the developing mouse midbrain in vivo and also prevents toxicity induced by the Lxr inhibitor geranylgeranyl pyrophosphate. Furthermore, using MS, we show that overexpression of human cholesterol 24S-hydroxylase (CYP46A1) increases the levels of both 24(S)-hydroxycholesterol (24-HC) and 24,25-EC in the developing midbrain, resulting in a specific increase in mDA neurogenesis in vitro and in vivo, but has no effect on oculomotor or red nucleus neurogenesis. 24-HC, unlike 24,25-EC, did not affect in vitro neurogenesis, indicating that the neurogenic effect of 24,25-EC on mDA neurons is specific. Combined, our results indicate that increased levels of 24,25-EC in vivo, by intracerebroventricular delivery in WT mice or by overexpression of its biosynthetic enzyme CYP46A1, specifically promote mDA neurogenesis. We propose that increasing the levels of 24,25-EC in vivo may be a useful strategy to combat the loss of mDA neurons in Parkinson's disease.\n\n\n## Inspiration\n\nSingle cell RNA sequencing is important technology in modern biology,\nsee e.g.\n\"Eleven grand challenges in single-cell data science\"\nhttps://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-1926-6\n\nAlso see review :\nNature. P. Kharchenko: \"The triumphs and limitations of computational methods for scRNA-seq\"\nhttps://www.nature.com/articles/s41592-021-01171-x", "VersionNotes": "h5ad converted files fully added", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1492357, "CreatorUserId": 2262596, "OwnerUserId": 2262596.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2465622.0, "CurrentDatasourceVersionId": 2508068.0, "ForumId": 1512069, "Type": 2, "CreationDate": "07/26/2021 14:46:56", "LastActivityDate": "07/26/2021", "TotalViews": 679, "TotalDownloads": 12, "TotalVotes": 5, "TotalKernels": 4}]
|
[{"Id": 2262596, "UserName": "alexandervc", "DisplayName": "Alexander Chervov", "RegisterDate": "09/18/2018", "PerformanceTier": 3}]
|
# # What is about ?
# Cell cycle - look with splitting by subsets of data.
# Versions:
# 1 str_data_inf = ' GSE76381_ESMoleculeCounts '
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import scanpy as sc
import anndata
import scipy
import time
t0start = time.time()
import pandas as pd
import numpy as np
import os
import sys
import matplotlib.pyplot as plt
import matplotlib as mpl
mpl.rcParams["figure.dpi"] = 70
plt.style.use("dark_background")
import seaborn as sns
from sklearn.decomposition import PCA
# # Open file
# Similar to https://github.com/theislab/scanpy/issues/882
def read_mtx_by_prefix(path_and_fn_prefix):
adata = sc.read(path_and_fn_prefix + "matrix.mtx").T
genes = pd.read_csv(path_and_fn_prefix + "genes.tsv", header=None, sep="\t")
genes = genes.set_index(1)
genes.index.name = "Gene"
genes.columns = ["Ensemble"]
adata.var = genes
barcodes = pd.read_csv(
path_and_fn_prefix + "barcodes.tsv", header=None, sep="\t", index_col=0
)
barcodes.index.name = "barcodes"
adata.obs = barcodes
return adata
if 1:
str_data_inf = " GSE76381_iPSMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_iPSMoleculeCounts.h5ad"
str_data_inf = " GSE76381_MouseEmbryoMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_MouseEmbryoMoleculeCounts.h5ad"
str_data_inf = " GSE76381_MouseAdultDAMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_MouseAdultDAMoleculeCounts.h5ad"
str_data_inf = " GSE76381_EmbryoMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_EmbryoMoleculeCounts.h5ad"
str_data_inf = " GSE76381_ESMoleculeCounts "
path_and_fn_prefix = "/kaggle/input/scrnaseq-study-of-neuron-development/GSE76381_ESMoleculeCounts.h5ad"
adata_orig = sc.read(path_and_fn_prefix)
else:
str_data_inf = " GSM3267241_U5_all "
path_and_fn_prefix = "/kaggle/input/scrnaseq-neuroepithelialderived-cells-and-glioma/GSM3267241_U5_all_"
adata_orig = read_mtx_by_prefix(path_and_fn_prefix)
genes_upper = [
t.upper() for t in adata_orig.var.index
] # Simple conversion mouse genes to human
v = adata_orig.var.copy()
v.index = genes_upper
adata_orig.var = v.copy()
list_save_original_obs_columns = list(adata_orig.obs.columns)
adata = adata_orig.copy()
adata_orig
# # Preliminary functions and preprocessing functions
def _smooth_adata_by_pooling(
adata, X_embed, n_neighbours=10, copy=False, output_mode=1
):
# adata_pooled = adata.copy if copy else adata
nbrs = NearestNeighbors(n_neighbors=n_neighbours).fit(X_embed)
if output_mode > 0:
print("done: nbrs = NearestNeighbors(n_neighbors=n_neighbours).fit(X_embed) ")
distances, indices = nbrs.kneighbors(X_embed)
if output_mode > 0:
print("done: distances, indices = nbrs.kneighbors(X_embed) ")
nd_array_X = _get_nd_array(adata.X)
if output_mode > 0:
print("done: nd_array_X = _get_nd_array(adata.X)")
adata.X = _smooth_matrix_by_pooling(nd_array_X, indices)
if output_mode > 0:
print(
"done: adata.X = _smooth_matrix_by_pooling(_get_nd_array(adata.X), indices) "
)
if "matrix" in adata.layers:
adata.layers["matrix"] = _smooth_matrix_by_pooling(
_get_nd_array(adata.layers["matrix"]), indices
)
if "spliced" in adata.layers:
adata.layers["spliced"] = _smooth_matrix_by_pooling(
_get_nd_array(adata.layers["spliced"]), indices
)
if "unspliced" in adata.layers:
adata.layers["unspliced"] = _smooth_matrix_by_pooling(
_get_nd_array(adata.layers["unspliced"]), indices
)
adata.uns["scycle"] = True
def _smooth_matrix_by_pooling(matrix, indices):
matrix_pooled = matrix.copy()
t0 = time.time()
# print(indices.shape, matrix.shape, type(matrix) )
for i in range(len(indices)):
# print(i,len(indices), time.time()-t0 )
matrix_pooled[i, :] = np.mean(matrix[indices[i], :], axis=0)
return matrix_pooled
def _get_nd_array(arr):
x = None
if str(type(arr)):
x = arr
else:
print("start: x = arr.toarray()")
x = arr.toarray()
print("finished: x = arr.toarray()")
return x
def calc_scores(anndata, signature_dict):
matrix = anndata.to_df().to_numpy()
scores_dic = {}
for key in signature_dict:
names = np.array(signature_dict[key])
inds = np.where(np.isin(anndata.var_names, names))[0]
matrix_sel = matrix[:, inds]
scores = np.mean(matrix_sel, axis=1)
scores_dic[key] = scores
return scores_dic
def find_nonproliferative_cells(
adata,
estimation_fraction_nonproliferating_cells=0.3,
number_of_nodes=30,
max_number_of_iterations=20,
number_of_sigmas=3.0,
Mu=1.0,
):
all_markers = list(adata.uns["S-phase_genes"]) + list(adata.uns["G2-M_genes"])
Xccm = adata[:, list(set(all_markers) & set(adata.var_names))].X
cc_score = np.array(list(np.mean(Xccm, axis=1)))
ind_sorted_prolif = np.argsort(cc_score)
ind_nonprolif = ind_sorted_prolif[
0 : int(len(adata) * estimation_fraction_nonproliferating_cells)
]
adata.obs["proliferating"] = np.empty(len(adata)).astype(np.int)
adata.obs["proliferating"][:] = 1
adata.obs["proliferating"][ind_nonprolif] = 0
sc.pl.scatter(adata, x="S-phase", y="G2-M", color="proliferating")
fraction_nonprolif_old = estimation_fraction_nonproliferating_cells
for i in range(max_number_of_iterations):
X_elpigraph_training = (
adata.obs[["S-phase", "G2-M"]].to_numpy().astype(np.float64)
)
u = X_elpigraph_training.copy()
ind_prolif = np.where(np.array(adata.obs["proliferating"]) == 1)[0]
X_elpigraph_training = X_elpigraph_training[ind_prolif, :]
egr = elpigraph.computeElasticPrincipalCircle(
X_elpigraph_training,
number_of_nodes,
Mu=Mu,
drawPCAView=False,
verbose=False,
)
partition, dists = elpigraph.src.core.PartitionData(
X=u,
NodePositions=egr[0]["NodePositions"],
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(u**2, axis=1, keepdims=1),
)
mndist = np.mean(dists[ind_prolif])
# plt.hist(dists,bins=100)
# plt.show()
# plt.hist(dists[ind_prolif],bins=100)
# plt.show()
intervaldist = np.std(dists[ind_prolif]) * number_of_sigmas
tt1 = np.array(1 - adata.obs["proliferating"])
tt2 = np.zeros(len(tt1)).astype(np.int)
for k, d in enumerate(dists):
if d > mndist + intervaldist:
tt2[k] = 1
nonprolif_new = np.array(tt1) * np.array(tt2)
adata.obs["proliferating"] = 1 - nonprolif_new
# adata.obs['dists'] = dists
# sc.pl.scatter(adata,x='S-phase',y='G2-M',color='dists')
fraction_nonprolif = 1 - np.sum(adata.obs["proliferating"]) / len(adata)
print(
"\n\n===========\nIteration",
i,
"Fraction of non-proliferating cells:",
fraction_nonprolif,
"\n==============\n\n\n",
)
if np.abs(fraction_nonprolif - fraction_nonprolif_old) < 0.01:
break
fraction_nonprolif_old = fraction_nonprolif
def _compute_principal_circle(adata, number_of_nodes=30, n_components=30):
# driving_feature = np.array(adata_orig[adata.uns['ind_samples']].obs['total_counts'])
# driving_feature_weight = 1.0
# egr, partition, X, Xp = _compute_principal_circle(adata,number_of_nodes=40,
# driving_feature=driving_feature,driving_feature_weight=0.1)
X = adata.X
X_prolif = adata.X[np.where(adata.obs["proliferating"] == 1)[0], :]
# driving_feature = driving_feature[adata.obs[np.where(adata.obs['proliferating']==1)[0]]]
mn_prolif = np.mean(X_prolif, axis=0)
pca = PCA(n_components=n_components)
u = pca.fit_transform(X_prolif)
v = pca.components_.T
# X_pca = adata.obsm['X_pca'].astype(np.float64)
X_elpigraph_training = u.astype(np.float64)
# std = np.std(X_elpigraph_training)
# driving_feature = stats.zscore(driving_feature)*std*driving_feature_weight
# driving_feature = driving_feature.reshape(-1,1)
# X_elpigraph_training = np.concatenate([X_elpigraph_training,driving_feature],axis=1)
# print('std=',std)
# print('std df=',np.std(driving_feature))
# print(X_elpigraph_training.shape)
egr = elpigraph.computeElasticPrincipalCircle(
X_elpigraph_training, number_of_nodes, Mu=0.2
)
partition, dists = elpigraph.src.core.PartitionData(
X=X_elpigraph_training,
NodePositions=egr[0]["NodePositions"],
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(X_elpigraph_training**2, axis=1, keepdims=1),
)
Xp = (X - mn_prolif) @ v
# number_of_driving_features = driving_feature.shape[1]
# node_positions_reduced = egr[0]['NodePositions'][:,:-number_of_driving_features]
nodep = egr[0]["NodePositions"]
partition, dists = elpigraph.src.core.PartitionData(
X=Xp,
NodePositions=nodep,
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(Xp**2, axis=1, keepdims=1),
)
# nodep = egr[0]['NodePositions'][:,:-number_of_driving_features]
# edges = egr[0]['Edges'][0]
# X_elpigraph_training = X_elpigraph_training[:,:-number_of_driving_features]
# egr[0]['NodePositions'] = nodep
return egr, partition, X_elpigraph_training, Xp
def _compute_principal_curve(
adata, number_of_nodes=30, Mu=0.3, Lambda=0.001, n_components=30
):
X = adata.X
X_prolif = adata.X[np.where(adata.obs["proliferating"] == 1)[0], :]
mn_prolif = np.mean(X_prolif, axis=0)
pca = PCA(n_components=n_components)
u = pca.fit_transform(X_prolif)
v = pca.components_.T
# X_pca = adata.obsm['X_pca'].astype(np.float64)
adata.varm["pc_components_elpigraph"] = v
adata.varm["mean_point_elpigraph"] = mn_prolif
X_elpigraph_training = u.astype(np.float64)
egr, starting_node = compute_principal_curve_from_circle(
X_elpigraph_training,
n_nodes=number_of_nodes,
produceTree=False,
Mu=Mu,
Lambda=Lambda,
)
partition_, dists = elpigraph.src.core.PartitionData(
X=X_elpigraph_training,
NodePositions=egr[0]["NodePositions"],
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(X_elpigraph_training**2, axis=1, keepdims=1),
)
Xp = (X - mn_prolif) @ v
partition_, dists = elpigraph.src.core.PartitionData(
X=Xp,
NodePositions=egr[0]["NodePositions"],
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(Xp**2, axis=1, keepdims=1),
)
nodep = egr[0]["NodePositions"]
edges = egr[0]["Edges"][0]
return egr, partition_, starting_node, X_elpigraph_training, Xp
def compute_principal_curve_from_circle(
X, n_nodes=30, Mu=0.1, Lambda=0.01, produceTree=False
):
egr = elpigraph.computeElasticPrincipalCircle(X, int(n_nodes / 2), Mu=Mu)
nodep = egr[0]["NodePositions"]
edges = egr[0]["Edges"][0]
partition_, dists = elpigraph.src.core.PartitionData(
X=X,
NodePositions=nodep,
MaxBlockSize=100000000,
TrimmingRadius=np.inf,
SquaredX=np.sum(X**2, axis=1, keepdims=1),
)
node_sizes = np.array(
[len(np.where(partition_ == i)[0]) for i in range(nodep.shape[0])]
)
node_min = np.argmin(node_sizes)
edges_open = edges.copy()
k = 0
starting_node = -1
while k < edges_open.shape[0]:
e = edges_open[k, :]
if (e[0] == node_min) | (e[1] == node_min):
edges_open = np.delete(edges_open, k, axis=0)
if e[0] == node_min:
starting_node = e[1]
if e[1] == node_min:
starting_node = e[0]
else:
k = k + 1
nodep_open = np.delete(nodep, node_min, axis=0)
if starting_node > node_min:
starting_node = starting_node - 1
for e in edges_open:
if e[0] > node_min:
e[0] = e[0] - 1
if e[1] > node_min:
e[1] = e[1] - 1
if produceTree:
egrl = elpigraph.computeElasticPrincipalTree(
X,
n_nodes,
InitNodePositions=nodep_open,
InitEdges=edges_open,
Lambda=Lambda,
Mu=Mu,
alpha=0.01,
FinalEnergy="Penalized",
)
else:
egrl = elpigraph.computeElasticPrincipalCurve(
X,
n_nodes,
InitNodePositions=nodep_open,
InitEdges=edges_open,
Lambda=Lambda,
Mu=Mu,
alpha=0.01,
FinalEnergy="Penalized",
)
return egrl, starting_node
def subtract_cell_cycle_trajectory(X, partition):
points = range(X.shape[0])
r2scores = []
X1 = X[points, :]
X_ro = np.zeros((X.shape[0], X.shape[1]))
partition_points = partition[points]
inds = {}
for k in range(len(partition_points)):
j = partition_points[k][0]
if not j in inds:
inds[j] = [k]
else:
inds[j].append(k)
XT = X1.T
for j in range(X1.shape[0]):
k = partition_points[j][0]
ind = np.array(inds[k])
X_ro[j, :] = (XT[:, j] - np.mean(XT[:, ind], axis=1)).T
residue_matrix = X1 - X_ro
residues_var = np.var(residue_matrix, axis=0)
vrs = np.var(X1, axis=0)
r2scores = residues_var / vrs
return X_ro, residue_matrix, r2scores
list_genes2include_mandotory = [
"E2F1",
"FOXM1",
"ESRRB",
"NR5A2",
"HIST1H4C",
"FOXO3",
"EZH1",
"ANLN",
]
list_genes2include_mandotory += ["PCNA", "TOP2A"]
list_genes2include_mandotory += [
"WEE1",
"WEE2",
] # Inhibitor of CDK1 https://en.wikipedia.org/wiki/Wee1
list_genes2include_mandotory += [
"CDK1",
"UBE2C",
"TOP2A",
"HIST1H4E",
"HIST1H4C",
] # Hsiao2020GenomeResearch 5 genes 'HIST1H4E' = H4C5,'HIST1H4C'=H4C3
list_genes2include_mandotory += [
"MYBL2",
"TP53",
"KLF4",
"CDKN1A",
"CDKN2A",
"RB1",
"MDM2",
"MDM4",
"ZBTB20",
"GAPDH",
"EFNA5",
"LINC00511",
]
# MYBL2 https://en.wikipedia.org/wiki/MYBL2 - transcription factor
# The protein encoded by this gene, a member of the MYB family of transcription factor genes, is a nuclear protein involved in cell cycle progression. The encoded protein is phosphorylated by cyclin A/cyclin-dependent kinase 2 during the S-phase of the cell cycle and possesses both activator and repressor activities.
# https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6788831/
# Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb Christin F Down 1, Julie Millour, Eric W-F Lam, Roger J Watson
# https://pubmed.ncbi.nlm.nih.gov/23555285/
# Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB -
list_genes2include_mandotory += (
["CCNE1", "CCNE2", "CDK2"]
+ ["CCNB1", "CCNB2", "CCNB3"]
+ ["CCNA1", "CCNA2", "CCNF", "CDK1"]
+ ["CCND1", "CCND2", "CCND3"]
+ ["CDK4", "CDK6"]
)
# 'CCNA1','CCNA2', 'CCNE1', 'CCNE2',]
list_genes2include_mandotory += [
"CDK11A", # All of the form: CDKnumber* - 27
"CDK14",
"CDK17",
"CDK13",
"CDK5RAP1",
"CDK16",
"CDK6",
"CDK5RAP3",
"CDK2AP1",
"CDK18",
"CDK2",
"CDK8",
"CDK7",
"CDK4",
"CDK9",
"CDK5RAP2",
"CDK15",
"CDK19",
"CDK20",
"CDK5",
"CDK12",
"CDK2AP2",
"CDK1",
"CDK5R2",
"CDK5R1",
"CDK10",
"CDK11B",
]
list_genes2include_mandotory += [
"CDKN3", # 10 CDKN* - A cyclin-dependent kinase inhibitor protein is a protein which inhibits the enzyme cyclin-dependent kinase (CDK).
"CDKN1B",
"CDKN2C",
"CDKN1A",
"CDKN2D",
"CDKN1C",
"CDKN2A",
"CDKN2AIP",
"CDKN2AIPNL",
"CDKN2B-AS1",
]
# p16 CDKN2A Cyclin-dependent kinase 4, Cyclin-dependent kinase 6
# p15 CDKN2B Cyclin-dependent kinase 4
# p18 CDKN2C Cyclin-dependent kinase 4, Cyclin-dependent kinase 6
# p19 CDKN2D Cyclin-dependent kinase 4, Cyclin-dependent kinase 6
# p21 / WAF1 CDKN1A[2] Cyclin E1/Cyclin-dependent kinase 2
# p27 CDKN1B Cyclin D3/Cyclin-dependent kinase 4, Cyclin E1/Cyclin-dependent kinase 2
# p57 CDKN1C Cyclin E1/Cyclin-dependent kinase 2
# https://en.wikipedia.org/wiki/Cyclin-dependent_kinase_inhibitor_protein
list_genes2include_mandotory += [
"CDKL3", # All starts with CDK - 43
"CDKL5",
"CDK11A",
"CDK14",
"CDK17",
"CDK13",
"CDKL1",
"CDKN3",
"CDK5RAP1",
"CDK16",
"CDK6",
"CDK5RAP3",
"CDKN1B",
"CDK2AP1",
"CDK18",
"CDKN2C",
"CDK2",
"CDKN1A",
"CDKN2D",
"CDKN1C",
"CDK8",
"CDK7",
"CDK4",
"CDK9",
"CDK5RAP2",
"CDK15",
"CDKL2",
"CDKAL1",
"CDKN2A",
"CDK19",
"CDK20",
"CDK5",
"CDK12",
"CDK2AP2",
"CDKN2AIP",
"CDK1",
"CDK5R2",
"CDK5R1",
"CDK10",
"CDKL4",
"CDKN2AIPNL",
"CDKN2B-AS1",
"CDK11B",
]
list_genes2include_mandotory += [
"CDC27", # "cdc" in its name refers to "cell division cycle"
"CDC42",
"CDC14A",
"CDC14B",
"CDC45",
"CDC6",
"CDC23",
"CDC5L",
"CDC7",
"CDC34",
"CDC25B",
"CDC37",
"CDC37L1",
"CDCA3",
"CDC20",
"CDC42EP1",
"CDC16",
"CDC73",
"CDCA8",
"CDC42BPA",
"CDCA7",
"CDCA5",
"CDC42EP2",
"CDC123",
"CDC25C",
"CDC42SE2",
"CDC42EP3",
"CDCP1",
"CDC25A",
"CDC20B",
"CDCA7L",
"CDC42EP5",
"CDC40",
"CDCA4",
"CDC42BPG",
"CDC26",
"CDC42EP4",
"CDCA2",
"CDC42SE1",
"CDC42BPB",
"CDC42-IT1",
"CDC20P1",
"CDC42P6",
"CDC27P2",
"CDC42P5",
"CDC37L1-AS1",
"CDC27P3",
]
# Cdc25 is a dual-specificity phosphatase first isolated from the yeast Schizosaccharomyces pombe as a cell cycle defective mutant.[2] As with other cell cycle proteins or genes such as Cdc2 and Cdc4, the "cdc" in its name refers to "cell division cycle". Dual-specificity phosphatases are considered a sub-class of protein tyrosine phosphatases. By removing inhibitory phosphate residues from target cyclin-dependent kinases (Cdks),[3] Cdc25 proteins control entry into and progression through various phases of the cell cycle, including mitosis and S ("Synthesis") phase.
list_genes2include_mandotory += [
"E2F2", # 10 genes E2F transc. factors
"E2F1",
"E2F3",
"E2F8",
"E2F5",
"E2F7",
"E2F6",
"E2F3P2",
"E2F4",
"E2F3-IT1",
]
list_genes2include_mandotory += ["GATA1", "FOXA1", "BRD4", "KMT2A"]
# 'GATA1', 'FOXA1', 'BRD4', 'KMT2A' - transcription factors are retained in mitotic chromosomes (from paper "Trans landscape of the human cell cycle" )
list_histone_genes_from_wiki = [
"H1F0",
"H1FNT",
"H1FOO",
"H1FX",
"HIST1H1A",
"HIST1H1B",
"HIST1H1C",
"HIST1H1D",
"HIST1H1E",
"HIST1H1T",
"H2AFB1",
"H2AFB2",
"H2AFB3",
"H2AFJ",
"H2AFV",
"H2AFX",
"H2AFY",
"H2AFY2",
"H2AFZ",
"HIST1H2AA",
"HIST1H2AB",
"HIST1H2AC",
"HIST1H2AD",
"HIST1H2AE",
"HIST1H2AG",
"HIST1H2AI",
"HIST1H2AJ",
"HIST1H2AK",
"HIST1H2AL",
"HIST1H2AM",
"HIST2H2AA3",
"HIST2H2AC",
"H2BFM",
"H2BFS",
"H2BFWT",
"HIST1H2BA",
"HIST1H2BB",
"HIST1H2BC",
"HIST1H2BD",
"HIST1H2BE",
"HIST1H2BF",
"HIST1H2BG",
"HIST1H2BH",
"HIST1H2BI",
"HIST1H2BJ",
"HIST1H2BK",
"HIST1H2BL",
"HIST1H2BM",
"HIST1H2BN",
"HIST1H2BO",
"HIST2H2BE",
"HIST1H3A",
"HIST1H3B",
"HIST1H3C",
"HIST1H3D",
"HIST1H3E",
"HIST1H3F",
"HIST1H3G",
"HIST1H3H",
"HIST1H3I",
"HIST1H3J",
"HIST2H3C",
"HIST3H3",
"HIST1H4A",
"HIST1H4B",
"HIST1H4C",
"HIST1H4D",
"HIST1H4E",
"HIST1H4F",
"HIST1H4G",
"HIST1H4H",
"HIST1H4I",
"HIST1H4J",
"HIST1H4K",
"HIST1H4L",
"HIST4H4",
]
list_genes2include_mandotory += list_histone_genes_from_wiki
S_phase_genes_Tirosh = [
"MCM5",
"PCNA",
"TYMS",
"FEN1",
"MCM2",
"MCM4",
"RRM1",
"UNG",
"GINS2",
"MCM6",
"CDCA7",
"DTL",
"PRIM1",
"UHRF1",
"MLF1IP",
"HELLS",
"RFC2",
"RPA2",
"NASP",
"RAD51AP1",
"GMNN",
"WDR76",
"SLBP",
"CCNE2",
"UBR7",
"POLD3",
"MSH2",
"ATAD2",
"RAD51",
"RRM2",
"CDC45",
"CDC6",
"EXO1",
"TIPIN",
"DSCC1",
"BLM",
"CASP8AP2",
"USP1",
"CLSPN",
"POLA1",
"CHAF1B",
"BRIP1",
"E2F8",
]
G2_M_genes_Tirosh = [
"HMGB2",
"CDK1",
"NUSAP1",
"UBE2C",
"BIRC5",
"TPX2",
"TOP2A",
"NDC80",
"CKS2",
"NUF2",
"CKS1B",
"MKI67",
"TMPO",
"CENPF",
"TACC3",
"FAM64A",
"SMC4",
"CCNB2",
"CKAP2L",
"CKAP2",
"AURKB",
"BUB1",
"KIF11",
"ANP32E",
"TUBB4B",
"GTSE1",
"KIF20B",
"HJURP",
"CDCA3",
"HN1",
"CDC20",
"TTK",
"CDC25C",
"KIF2C",
"RANGAP1",
"NCAPD2",
"DLGAP5",
"CDCA2",
"CDCA8",
"ECT2",
"KIF23",
"HMMR",
"AURKA",
"PSRC1",
"ANLN",
"LBR",
"CKAP5",
"CENPE",
"CTCF",
"NEK2",
"G2E3",
"GAS2L3",
"CBX5",
"CENPA",
]
from sklearn.neighbors import NearestNeighbors
def preproc(adata_orig, output_mode=1):
adata = adata_orig.copy()
from sklearn.neighbors import NearestNeighbors
import scipy
# First standard preprocessing
# Then "pooling" - fighting with zeros
# Params:
n_top_genes_to_keep = 10000
threshold_pct_counts_mt = 40
min_count = 500 # Threshold on min total_counts
max_count = np.inf # 10000# Threshold on max total_counts
if output_mode > 0:
print(adata_orig.X.sum())
# ################################################################################################
# Preprocessing first step: filter CELLs by counts and level of MT-percent
# thresholds are set visually looking on violin plots
# Examples:
# threshold_pct_counts_mt = 20 - 40 min_count = 500 - 1000; max_count = 10000 - 12000;
# These thresholds depends on cell line dataset
if output_mode > 0:
print(adata)
adata.var["mt"] = adata.var_names.str.startswith(
"MT-"
) # annotate the group of mitochondrial genes as 'mt'
# sv.pp.remove_duplicate_cells(adata)
sc.pp.calculate_qc_metrics(
adata, qc_vars=["mt"], percent_top=None, log1p=False, inplace=True
)
if output_mode > 0:
sc.pl.violin(
adata,
["n_genes_by_counts", "total_counts", "pct_counts_mt"],
jitter=1.9,
multi_panel=True,
)
median_count = np.median(adata.obs["total_counts"])
if output_mode > 0:
print("Median total counts =", median_count)
print("min(n_genes_by_counts):", adata.obs["n_genes_by_counts"].min())
print("min(total_counts):", adata.obs["total_counts"].min())
# min_count = np.max((median_count/2,5000))
if output_mode > 0:
print("Thresholds: min_count=", min_count, "max_count=", max_count)
print(
"Look at total_counts va MT-percent, expect some linear dependence - but does not happen: "
)
sc.pl.scatter(adata, x="total_counts", y="pct_counts_mt")
inds1 = np.where(
(adata.obs["total_counts"] > min_count)
& (adata.obs["total_counts"] < max_count)
)
inds2 = np.where(adata.obs["pct_counts_mt"] < threshold_pct_counts_mt)
if output_mode > 0:
print(len(inds1[0]), "samples pass the count filter")
print(len(inds2[0]), " samples pass the mt filter")
ind_samples = np.intersect1d(inds1[0], inds2[0])
if output_mode > 0:
print("Samples selected", len(ind_samples))
adata.uns["ind_samples"] = ind_samples
# Here we cut cells. Filtering out those with counts too low or too big
adata = adata[ind_samples, :]
# ################################################################################################
# Preprocessing second step:
# 1) normalization to some value, i.e. median of the total counts
# 2) taking logs
# 3) keeping only higly variable genes
sc.pp.normalize_total(adata, target_sum=np.median(adata.obs["total_counts"]))
sc.pp.log1p(adata)
try:
sc.pp.highly_variable_genes(
adata, n_top_genes=np.min([X.shape[1], n_top_genes_to_keep]), n_bins=20
)
ind_genes = np.where(adata.var["highly_variable"])[0]
except:
ind_genes = np.arange(
adata.X.shape[1]
) # np.where(adata.var['highly_variable'])[0]
ind_genes2 = np.where(adata.var.index.isin(list_genes2include_mandotory))[0]
ind_genes = list(set(ind_genes) | set(ind_genes2))
adata = adata[:, ind_genes]
if output_mode > 0:
print("Violin plots after filtering cells and genes")
sc.pl.violin(
adata,
["n_genes_by_counts", "total_counts", "pct_counts_mt"],
jitter=1.9,
multi_panel=True,
)
# ################################################################################################
# Preprocessing third step:
# Mainly: Create non-sparse adata (otherwise next step - pooling works extremely slow)
# 1) calculate PCA for adata previously calculated adata (which is cutted, normed, logged) - and keep it for use in pooling
# 2) create new non-sparse adata with cells and genes, obtained by filters on previous steps
# print('Check - we should see ints, otherwise data has been corrupted: ')
# print(adata_orig.X[:4,:5].toarray())
sc.tl.pca(adata, n_comps=30)
X_pca = adata.obsm["X_pca"]
X_pca1 = X_pca.copy()
adata_orig[ind_samples, ind_genes].var.shape
if scipy.sparse.issparse(adata.X):
XX2 = adata_orig[ind_samples, ind_genes].X.toarray()
else:
XX2 = adata_orig[ind_samples, ind_genes].X.copy()
adata = anndata.AnnData(
XX2,
obs=adata_orig[ind_samples, ind_genes].obs.copy(),
var=adata_orig[ind_samples, ind_genes].var.copy(),
) #
if output_mode > 0:
print("New non-sparse adata: ", adata)
# sv.pp.remove_duplicate_cells(adata)
adata.var["mt"] = adata.var_names.str.startswith(
"MT-"
) # annotate the group of mitochondrial genes as 'mt'
sc.pp.calculate_qc_metrics(
adata, qc_vars=["mt"], percent_top=None, log1p=False, inplace=True
)
if output_mode > 0:
print("Violin plots for new adata")
sc.pl.violin(
adata,
["n_genes_by_counts", "total_counts", "pct_counts_mt"],
jitter=1.9,
multi_panel=True,
)
# ################################################################################################
# Preprocessing 4-th step: may take quite long - 10 minutes for 40K celss
# Mainly: pooling - we are making pooling on original counts, but calculate KNN graph using PCA obtained from normed/log data
# (that is why we forget adata obtained on the steps 1-2, and will do norm-log again here )
# 1) pooling
# 2) quality metrics calculations
# 3) normalization
# 4) log
t0 = time.time()
_smooth_adata_by_pooling(adata, X_pca, n_neighbours=20, output_mode=output_mode)
if output_mode > 0:
print(time.time() - t0, "seconds passed")
sc.pp.calculate_qc_metrics(adata, percent_top=None, log1p=False, inplace=True)
sc.pp.normalize_total(adata, target_sum=np.median(adata.obs["total_counts"]))
sc.pp.log1p(adata)
# adata = adata[:,ind_genes]
adata.uns["ind_genes"] = ind_genes
sc.tl.pca(adata, n_comps=30)
X_pca = adata.obsm["X_pca"]
return adata
# adata = preproc(adata_orig, output_mode = 0)
# adata
# # Main Loop
import warnings
warnings.filterwarnings("ignore")
adata = preproc(adata_orig, output_mode=0)
adata.obs["Timepoint"].unique()[::-1]
n_x_subplots = 1
t00 = time.time()
print("Start processing. ")
# print( series_GSE_and_cell_count[mm] )
df_stat = pd.DataFrame()
ix4df_stat = 0
c = 0
if 1:
mask = np.ones(adata.shape[0]).astype(bool)
ix4df_stat += 1
# print()
# print(GSE)
t0 = time.time()
list_genes_upper = [t.upper() for t in adata.var.index]
I = np.where(pd.Series(list_genes_upper).isin(S_phase_genes_Tirosh))[0]
v1 = adata[mask].X[:, I].mean(axis=1)
I = np.where(pd.Series(list_genes_upper).isin(G2_M_genes_Tirosh))[0]
v2 = adata[mask].X[:, I].mean(axis=1)
if c % n_x_subplots == 0:
fig = plt.figure(figsize=(25, 10))
c = 0
c += 1
fig.add_subplot(1, n_x_subplots, c)
color_by = adata.obs["Timepoint"]
sns.scatterplot(x=v1, y=v2, hue=color_by) # adata.obs['pct_counts_mt'])
corr_phases = np.round(np.corrcoef(v1, v2)[0, 1], 2)
plt.title(str_data_inf, fontsize=17)
if c == n_x_subplots:
plt.show()
# print( np.round(time.time() - t0,1 ) , 'seconds passed ', np.round(time.time() - t00,1 ) , 'seconds passed total')
print(
np.round(time.time() - t00, 1),
np.round((time.time() - t00) / 60, 1),
"total seconds,minutes passed",
)
plt.show()
df_stat
n_x_subplots = 2
t00 = time.time()
print("Start processing. ")
# print( series_GSE_and_cell_count[mm] )
df_stat = pd.DataFrame()
ix4df_stat = 0
c = 0
for uv in [
"day_0",
"day_12",
"day_17",
"day_35",
]: # adata.obs['Timepoint'].unique()[::-1]:
mask = adata.obs["Timepoint"] == uv
ix4df_stat += 1
# print()
# print(GSE)
t0 = time.time()
list_genes_upper = [t.upper() for t in adata.var.index]
I = np.where(pd.Series(list_genes_upper).isin(S_phase_genes_Tirosh))[0]
v1 = adata[mask].X[:, I].mean(axis=1)
I = np.where(pd.Series(list_genes_upper).isin(G2_M_genes_Tirosh))[0]
v2 = adata[mask].X[:, I].mean(axis=1)
if c % n_x_subplots == 0:
fig = plt.figure(figsize=(25, 10))
c = 0
c += 1
fig.add_subplot(1, n_x_subplots, c)
# sns.scatterplot(x = v1, y = v2 , hue = adata.obs['pct_counts_mt'])
I = np.where("CCNE2" == np.array(list_genes_upper))[0][0]
color_by = pd.Series(adata[mask].X[:, I] > np.median(adata.X[:, I]))
color_by.name = "CCNE2"
if len(np.unique(color_by)) == 2:
sns.scatterplot(
x=v1, y=v2, hue=color_by, palette=["green", "red"]
) # obs['pct_counts_mt'])
else:
sns.scatterplot(
x=v1, y=v2
) # , hue = color_by, palette = ['red','green'] )# obs['pct_counts_mt'])
corr_phases = np.round(np.corrcoef(v1, v2)[0, 1], 2)
df_stat.loc[ix4df_stat, "SubType"] = uv
df_stat.loc[ix4df_stat, "n_cells"] = mask.sum()
df_stat.loc[ix4df_stat, "Phase Correlation"] = corr_phases
df_stat.loc[ix4df_stat, "Info"] = str_data_inf
plt.title(
uv
+ " "
+ str_data_inf
+ " Corr "
+ str(corr_phases)
+ " n_cells "
+ str(mask.sum()),
fontsize=17,
)
plt.title(
uv + " Corr " + str(corr_phases) + " n_cells " + str(mask.sum()), fontsize=17
)
if c == n_x_subplots:
plt.show()
# print( np.round(time.time() - t0,1 ) , 'seconds passed ', np.round(time.time() - t00,1 ) , 'seconds passed total')
print(
np.round(time.time() - t00, 1),
np.round((time.time() - t00) / 60, 1),
"total seconds,minutes passed",
)
plt.show()
df_stat
print(
np.round(time.time() - t0start, 1),
np.round((time.time() - t0start) / 60, 1),
np.round((time.time() - t0start) / 3600, 1),
"total seconds/minutes/hours passed",
)
| false | 0 | 11,962 | 0 | 12,865 | 11,962 |
||
69136981
|
import json
import requests
import time
import pandas as pd
import numpy as np
import plotly.graph_objects as go
import plotly.express as px
import matplotlib.pyplot as plt
from plotly.subplots import make_subplots
from datetime import date, datetime
from fbprophet import Prophet
from sklearn.metrics import mean_absolute_error
# %%
# GET DATA
url1 = "https://raw.githubusercontent.com/CITF-Malaysia/citf-public/main/vaccination/vax_state.csv"
url2 = "https://raw.githubusercontent.com/CITF-Malaysia/citf-public/main/vaccination/vax_malaysia.csv"
url3 = "https://raw.githubusercontent.com/CITF-Malaysia/citf-public/main/static/population.csv"
url4 = "https://query.data.world/s/64itw4xd4l43sq7n6fwgagvsqcojjd"
url5 = "https://query.data.world/s/4gtzoe6nkuueyikifwpsiko5rbqzxl"
res1 = requests.get(url1, allow_redirects=True)
with open("vax_state.csv", "wb") as file:
file.write(res1.content)
vax_state_df = pd.read_csv("vax_state.csv")
res2 = requests.get(url2, allow_redirects=True)
with open("vax_malaysia.csv", "wb") as file:
file.write(res2.content)
vax_malaysia_df = pd.read_csv("vax_malaysia.csv")
res3 = requests.get(url3, allow_redirects=True)
with open("population.csv", "wb") as file:
file.write(res3.content)
population = pd.read_csv("population.csv")
res4 = requests.get(url4, allow_redirects=True)
with open("covid-19_my_state.csv", "wb") as file:
file.write(res4.content)
cases_state_df = pd.read_csv("covid-19_my_state.csv")
res5 = requests.get(url5, allow_redirects=True)
with open("covid-19_my.csv", "wb") as file:
file.write(res5.content)
cases_malaysia_df = pd.read_csv("covid-19_my.csv")
# %%
# DATA OPERATIONS
# Convert date format
vax_malaysia_df["date"] = pd.to_datetime(vax_malaysia_df["date"])
vax_state_df["date"] = pd.to_datetime(vax_state_df["date"])
cases_malaysia_df["date"] = pd.to_datetime(cases_malaysia_df["date"])
cases_state_df["date"] = pd.to_datetime(cases_state_df["date"])
population_states = pd.DataFrame(population).drop(["idxs", "pop_18", "pop_60"], axis=1)[
1:
]
population_malaysia = pd.DataFrame(population).drop(
["idxs", "pop_18", "pop_60"], axis=1
)[:1]
# %%
# Format state and columns in cases_state_df
rename_dict = {
"JOHOR": "Johor",
"KEDAH": "Kedah",
"KELANTAN": "Kelantan",
"MELAKA": "Melaka",
"NEGERI SEMBILAN": "Negeri Sembilan",
"PAHANG": "Pahang",
"PULAU PINANG": "Pulau Pinang",
"PERAK": "Perak",
"PERLIS": "Perlis",
"SELANGOR": "Selangor",
"TERENGGANU": "Terengganu",
"SABAH": "Sabah",
"SARAWAK": "Sarawak",
"WP KUALA LUMPUR": "W.P. Kuala Lumpur",
"WP LABUAN": "W.P. Labuan",
"WP PUTRAJAYA": "W.P. Putrajaya",
}
cases_state_df["state"] = cases_state_df["state"].replace(rename_dict)
# %%
# Merge population into DataFrame
vax_state_df = pd.merge(population_states, vax_state_df, on="state")
cases_state_df = pd.merge(population_states, cases_state_df, on="state")
# Calculate percentage using population
vax_state_df["percentage_fully_vaccinated"] = (
vax_state_df["dose2_cumul"] / vax_state_df["pop"] * 100
)
vax_malaysia_df["percentage_fully_vaccinated"] = (
vax_malaysia_df["dose2_cumul"] / sum(vax_state_df["pop"].unique()) * 100
)
# %%
# Key analytics
total_first_dose_malaysia = vax_malaysia_df["dose1_daily"].sum()
total_second_dose_malaysia = vax_malaysia_df["dose2_daily"].sum()
pct_fully_vax_malaysia = (
total_second_dose_malaysia / population_malaysia["pop"].sum() * 100
)
daily_new_cases = cases_malaysia_df["new_cases"].iloc[-1]
daily_deaths = cases_malaysia_df["new_deaths"].iloc[-1]
cumul_deaths = cases_malaysia_df["total_deaths"].iloc[-1]
# %%
# Group by state
daily_vax1 = vax_malaysia_df["dose1_daily"].iloc[-1]
daily_vax2 = vax_malaysia_df["dose2_daily"].iloc[-1]
daily_vaxsum = daily_vax1 + daily_vax2
# %%
pred_df = pd.DataFrame()
pred_df["date"] = vax_malaysia_df["date"]
pred_df["total_daily"] = vax_malaysia_df["total_daily"]
pred_df.columns = ["ds", "y"]
pred_df.dtypes
# %%
# train_test_split
test = pred_df[-5:]
train = pred_df[:-5]
# %%
model = Prophet()
model.fit(train)
# %%
future = model.make_future_dataframe(periods=5)
future.plot()
future
forecast = model.predict(future)
forecast[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail(5)
model.plot(forecast)
model.plot_components(forecast)
y_true = test["y"].values
y_pred = forecast["yhat"][-5:].values
mae = mean_absolute_error(y_true, y_pred)
print("MAE: %.3f" % mae)
plt.plot(y_true, label="Actual")
plt.plot(y_pred, label="Predicted")
plt.legend()
plt.show()
future_long = model.make_future_dataframe(periods=200)
forecast_long = model.predict(future_long)
forecast_long[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail(200)
model.plot(forecast_long)
model.plot_components(forecast_long)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/136/69136981.ipynb
| null | null |
[{"Id": 69136981, "ScriptId": 18752859, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7423408, "CreationDate": "07/27/2021 09:28:26", "VersionNumber": 1.0, "Title": "Malaysia Vaccination Forecast Using FBProphet", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 160.0, "LinesInsertedFromPrevious": 160.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import json
import requests
import time
import pandas as pd
import numpy as np
import plotly.graph_objects as go
import plotly.express as px
import matplotlib.pyplot as plt
from plotly.subplots import make_subplots
from datetime import date, datetime
from fbprophet import Prophet
from sklearn.metrics import mean_absolute_error
# %%
# GET DATA
url1 = "https://raw.githubusercontent.com/CITF-Malaysia/citf-public/main/vaccination/vax_state.csv"
url2 = "https://raw.githubusercontent.com/CITF-Malaysia/citf-public/main/vaccination/vax_malaysia.csv"
url3 = "https://raw.githubusercontent.com/CITF-Malaysia/citf-public/main/static/population.csv"
url4 = "https://query.data.world/s/64itw4xd4l43sq7n6fwgagvsqcojjd"
url5 = "https://query.data.world/s/4gtzoe6nkuueyikifwpsiko5rbqzxl"
res1 = requests.get(url1, allow_redirects=True)
with open("vax_state.csv", "wb") as file:
file.write(res1.content)
vax_state_df = pd.read_csv("vax_state.csv")
res2 = requests.get(url2, allow_redirects=True)
with open("vax_malaysia.csv", "wb") as file:
file.write(res2.content)
vax_malaysia_df = pd.read_csv("vax_malaysia.csv")
res3 = requests.get(url3, allow_redirects=True)
with open("population.csv", "wb") as file:
file.write(res3.content)
population = pd.read_csv("population.csv")
res4 = requests.get(url4, allow_redirects=True)
with open("covid-19_my_state.csv", "wb") as file:
file.write(res4.content)
cases_state_df = pd.read_csv("covid-19_my_state.csv")
res5 = requests.get(url5, allow_redirects=True)
with open("covid-19_my.csv", "wb") as file:
file.write(res5.content)
cases_malaysia_df = pd.read_csv("covid-19_my.csv")
# %%
# DATA OPERATIONS
# Convert date format
vax_malaysia_df["date"] = pd.to_datetime(vax_malaysia_df["date"])
vax_state_df["date"] = pd.to_datetime(vax_state_df["date"])
cases_malaysia_df["date"] = pd.to_datetime(cases_malaysia_df["date"])
cases_state_df["date"] = pd.to_datetime(cases_state_df["date"])
population_states = pd.DataFrame(population).drop(["idxs", "pop_18", "pop_60"], axis=1)[
1:
]
population_malaysia = pd.DataFrame(population).drop(
["idxs", "pop_18", "pop_60"], axis=1
)[:1]
# %%
# Format state and columns in cases_state_df
rename_dict = {
"JOHOR": "Johor",
"KEDAH": "Kedah",
"KELANTAN": "Kelantan",
"MELAKA": "Melaka",
"NEGERI SEMBILAN": "Negeri Sembilan",
"PAHANG": "Pahang",
"PULAU PINANG": "Pulau Pinang",
"PERAK": "Perak",
"PERLIS": "Perlis",
"SELANGOR": "Selangor",
"TERENGGANU": "Terengganu",
"SABAH": "Sabah",
"SARAWAK": "Sarawak",
"WP KUALA LUMPUR": "W.P. Kuala Lumpur",
"WP LABUAN": "W.P. Labuan",
"WP PUTRAJAYA": "W.P. Putrajaya",
}
cases_state_df["state"] = cases_state_df["state"].replace(rename_dict)
# %%
# Merge population into DataFrame
vax_state_df = pd.merge(population_states, vax_state_df, on="state")
cases_state_df = pd.merge(population_states, cases_state_df, on="state")
# Calculate percentage using population
vax_state_df["percentage_fully_vaccinated"] = (
vax_state_df["dose2_cumul"] / vax_state_df["pop"] * 100
)
vax_malaysia_df["percentage_fully_vaccinated"] = (
vax_malaysia_df["dose2_cumul"] / sum(vax_state_df["pop"].unique()) * 100
)
# %%
# Key analytics
total_first_dose_malaysia = vax_malaysia_df["dose1_daily"].sum()
total_second_dose_malaysia = vax_malaysia_df["dose2_daily"].sum()
pct_fully_vax_malaysia = (
total_second_dose_malaysia / population_malaysia["pop"].sum() * 100
)
daily_new_cases = cases_malaysia_df["new_cases"].iloc[-1]
daily_deaths = cases_malaysia_df["new_deaths"].iloc[-1]
cumul_deaths = cases_malaysia_df["total_deaths"].iloc[-1]
# %%
# Group by state
daily_vax1 = vax_malaysia_df["dose1_daily"].iloc[-1]
daily_vax2 = vax_malaysia_df["dose2_daily"].iloc[-1]
daily_vaxsum = daily_vax1 + daily_vax2
# %%
pred_df = pd.DataFrame()
pred_df["date"] = vax_malaysia_df["date"]
pred_df["total_daily"] = vax_malaysia_df["total_daily"]
pred_df.columns = ["ds", "y"]
pred_df.dtypes
# %%
# train_test_split
test = pred_df[-5:]
train = pred_df[:-5]
# %%
model = Prophet()
model.fit(train)
# %%
future = model.make_future_dataframe(periods=5)
future.plot()
future
forecast = model.predict(future)
forecast[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail(5)
model.plot(forecast)
model.plot_components(forecast)
y_true = test["y"].values
y_pred = forecast["yhat"][-5:].values
mae = mean_absolute_error(y_true, y_pred)
print("MAE: %.3f" % mae)
plt.plot(y_true, label="Actual")
plt.plot(y_pred, label="Predicted")
plt.legend()
plt.show()
future_long = model.make_future_dataframe(periods=200)
forecast_long = model.predict(future_long)
forecast_long[["ds", "yhat", "yhat_lower", "yhat_upper"]].tail(200)
model.plot(forecast_long)
model.plot_components(forecast_long)
| false | 0 | 1,847 | 0 | 1,847 | 1,847 |
||
69136658
|
<jupyter_start><jupyter_text>Rain drop size distribution Dataset
### Context
Rain drop size distribution measurements from a tropical site, Pune, India (JJAS, 2013-2015).
Kaggle dataset identifier: rain-drop-size-distribution
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # READINF FILES AND MAKING CSV
#
import glob
path = r"/kaggle/input/rain-drop-size-distribution/Disdrometer_RainDSD_Data/"
all_files = glob.glob(path + "/*.txt")
li = []
all_data_df = None
first = True
for filename in all_files:
if first == True:
first = False
df = pd.read_csv(filename, delimiter="\t")
all_data_df = df.copy()
print(df.columns)
if first == False:
df = pd.read_csv(filename, delimiter="\t")
all_data_df = pd.concat([all_data_df, df])
# print(df.columns)
# print(len(df))
len(all_data_df)
all_data_df["YYYY-MM-DD"] = pd.to_datetime(all_data_df["YYYY-MM-DD"])
all_data_df = all_data_df.set_index(
"YYYY-MM-DD"
) # ""] = pd.to_datetime(all_data_df['YYYY-MM-DD'])
all_data_df.isna().any()
# ## PLOTTING AVERAGE RI [mm/h]
# daily
all_data_df.resample("D").mean()["RI [mm/h]"].plot(figsize=(20, 10))
# MONTHLY
all_data_df.resample("M").mean()["RI [mm/h]"].plot(figsize=(20, 10))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/136/69136658.ipynb
|
rain-drop-size-distribution
|
saurabhshahane
|
[{"Id": 69136658, "ScriptId": 18870794, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4370733, "CreationDate": "07/27/2021 09:24:19", "VersionNumber": 1.0, "Title": "Explanatory Data Analysis", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 73.0, "LinesInsertedFromPrevious": 73.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 91955283, "KernelVersionId": 69136658, "SourceDatasetVersionId": 2462537}]
|
[{"Id": 2462537, "DatasetId": 1466632, "DatasourceVersionId": 2504964, "CreatorUserId": 2411256, "LicenseName": "Attribution 4.0 International (CC BY 4.0)", "CreationDate": "07/25/2021 19:41:24", "VersionNumber": 4.0, "Title": "Rain drop size distribution Dataset", "Slug": "rain-drop-size-distribution", "Subtitle": "Rain drop size distribution Dataset", "Description": "### Context\n\nRain drop size distribution measurements from a tropical site, Pune, India (JJAS, 2013-2015).\n\n### Acknowledgements\n\nKanawade, Vijay (2020), \u201cRain drop size distribution\u201d, Mendeley Data, V2, doi: 10.17632/n3j6x74nb3.2", "VersionNotes": "Automatic Update 2021-07-25", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1466632, "CreatorUserId": 2411256, "OwnerUserId": 2411256.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2643725.0, "CurrentDatasourceVersionId": 2687702.0, "ForumId": 1486254, "Type": 2, "CreationDate": "07/14/2021 06:13:06", "LastActivityDate": "07/14/2021", "TotalViews": 2687, "TotalDownloads": 33, "TotalVotes": 10, "TotalKernels": 1}]
|
[{"Id": 2411256, "UserName": "saurabhshahane", "DisplayName": "Saurabh Shahane", "RegisterDate": "10/26/2018", "PerformanceTier": 4}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # READINF FILES AND MAKING CSV
#
import glob
path = r"/kaggle/input/rain-drop-size-distribution/Disdrometer_RainDSD_Data/"
all_files = glob.glob(path + "/*.txt")
li = []
all_data_df = None
first = True
for filename in all_files:
if first == True:
first = False
df = pd.read_csv(filename, delimiter="\t")
all_data_df = df.copy()
print(df.columns)
if first == False:
df = pd.read_csv(filename, delimiter="\t")
all_data_df = pd.concat([all_data_df, df])
# print(df.columns)
# print(len(df))
len(all_data_df)
all_data_df["YYYY-MM-DD"] = pd.to_datetime(all_data_df["YYYY-MM-DD"])
all_data_df = all_data_df.set_index(
"YYYY-MM-DD"
) # ""] = pd.to_datetime(all_data_df['YYYY-MM-DD'])
all_data_df.isna().any()
# ## PLOTTING AVERAGE RI [mm/h]
# daily
all_data_df.resample("D").mean()["RI [mm/h]"].plot(figsize=(20, 10))
# MONTHLY
all_data_df.resample("M").mean()["RI [mm/h]"].plot(figsize=(20, 10))
| false | 0 | 558 | 2 | 621 | 558 |
||
69141940
|
# ## You're here!
# Welcome to your first competition in the [ITI's AI Pro training program](https://ai.iti.gov.eg/epita/ai-engineer/)! We hope you enjoy and learn as much as we did prepairing this competition.
# ## Introduction
# In the competition, it's required to predict the `Severity` of a car crash given info about the crash, e.g., location.
# This is the getting started notebook. Things are kept simple so that it's easier to understand the steps and modify it.
# Feel free to `Fork` this notebook and share it with your modifications **OR** use it to create your submissions.
# ### Prerequisites
# You should know how to use python and a little bit of Machine Learning. You can apply the techniques you learned in the training program and submit the new solutions!
# ### Checklist
# You can participate in this competition the way you perefer. However, I recommend following these steps if this is your first time joining a competition on Kaggle.
# * Fork this notebook and run the cells in order.
# * Submit this solution.
# * Make changes to the data processing step as you see fit.
# * Submit the new solutions.
# *You can submit up to 5 submissions per day. You can select only one of the submission you make to be considered in the final ranking.*
# Don't hesitate to leave a comment or contact me if you have any question!
# ## Import the libraries
# We'll use `pandas` to load and manipulate the data. Other libraries will be imported in the relevant sections.
# read_xml available only in pandas >= 1.3.0
# !pip install -Iv pandas==1.3.1
# the notebook requires restarting afterwards
# import os
# os._exit(00)
import pandas as pd
from datetime import datetime
import os
pd.__version__
# ## Exploratory Data Analysis
# In this step, one should load the data and analyze it. However, I'll load the data and do minimal analysis. You are encouraged to do thorough analysis!
# Let's load the data using `pandas` and have a look at the generated `DataFrame`.
dataset_path = "/kaggle/input/car-crashes-severity-prediction/"
# dataset_path = "./"
df = pd.read_csv(os.path.join(dataset_path, "train.csv"), index_col=None)
print("The shape of the dataset is {}.\n\n".format(df.shape))
# remove rows with any null values
df = df.dropna()
# remove duplicate rows
df = df.drop_duplicates()
df.head()
# converting timestamp column type to become datetime
df["timestamp"] = pd.to_datetime(df["timestamp"])
# convert side to be numeric
df["Side"] = df["Side"] == "R"
# FIXME
import matplotlib.pyplot as plt
# FIXME
plt.plot(df["Severity"], df["Distance(mi)"], marker=".", linestyle="none")
# plt.hist(df["Severity"], bins=4)
plt.xlabel("Severity")
plt.ylabel("Distance(mi)")
plt.show()
corr = df.corr()
corr
# dropping sparse features w/ no correlation
df = df.drop(columns=["Bump", "Roundabout"])
corr = df.corr()
corr
# pd.get_dummies(df[["Crossing", "Give_Way", "Junction", "No_Exit", "Railway", "Stop", "Amenity"]])
# We've got 6407 examples in the dataset with 14 featues, 1 ID, and the `Severity` of the crash.
# By looking at the features and a sample from the data, the features look of numerical and catogerical types. What about some descriptive statistics?
df.drop(columns="ID").describe()
# read weather file
df_weather = pd.read_csv(
os.path.join(dataset_path, "weather-sfcsv.csv"), index_col=None
)
df_weather.head(15)
df_weather["date"] = (
df_weather["Year"].astype(str)
+ "-"
+ df_weather["Month"].astype(str)
+ "-"
+ df_weather["Day"].astype(str)
+ " "
+ df_weather["Hour"].astype(str)
+ ":00:00"
)
df_weather["date"] = pd.to_datetime(df_weather["date"])
# df_weather = df_weather.dropna()
# df_weather = df_weather.drop_duplicates()
modes = {}
for col in df_weather.drop(
columns=["Year", "Hour", "Day", "Month", "Weather_Condition", "Selected"]
).columns:
# modes[col] = df_weather[col].mode()[0]
df_weather[col] = df_weather[col].fillna(df_weather[col].median())
# df_weather["Temperature(F)"].mode()[0]
# print(modes["Wind_Chill(F)"])
# df_weather["Wind_Chill(F)"].fillna(modes["Wind_Chill(F)"])
# df_weather["Wind_Chill(F)"].head()
# print(df_weather.shape)
# df_weather.dropna()
# df_weather.drop_duplicates()
# print(df_weather.shape)
# better naming convention
# df.columns = [
# "id", "lat", "lng", "bump", "distance_mile", "is_crossing",
# "is_give_way", "is_junction", "is_no_exit", "is_railway",
# "is_roundabout", "is_stop", "is_amenity", "side", "severity",
# "timestamp", "date"
# ]
# df_weather.columns = [
# "year", "day", "month", "hour", "weather_condition",
# "wind_chill_fahr", "perceptation_inch", "temperature_fahr",
# "humidity_percent", "wind_spd_mi_per_hr", "visibility_miles",
# "selected", "date"
# ]
df_weather_averaged = (
df_weather.groupby("date")
.mean()
.drop(columns=["Year", "Day", "Month", "Hour"])
.dropna()
.drop_duplicates()
)
timestamp = df["timestamp"].apply(
lambda ts: f"{ts.year}-{ts.month}-{ts.day} {ts.hour}:00:00"
)
timestamp = pd.to_datetime(timestamp)
df["date"] = timestamp
final_df = pd.merge(df, df_weather_averaged, how="inner", on="date").drop(
columns=["date"]
)
# FIXME illogical action to fill na with 0
# final_df = pd.merge(df, df_weather_averaged, how="left", on="date").drop(
# columns=["date"]
# ).fillna(0)
final_df.shape
corr = final_df.drop(columns=["ID"]).corr()
corr
# drop features w/ no correlation
# final_df = final_df.drop(columns=["No_Exit"])
final_df.corr()
df_holidays = pd.read_xml(os.path.join(dataset_path, "holidays.xml"), xpath="/root/*")
# df_holidays = pd.read_csv(os.path.join("../input/holidays/", "holidays.csv"))
# ../input/holidays/holidays.csv
df_holidays["date"] = pd.to_datetime(df_holidays["date"])
df_holidays.head()
to_date = lambda date_time: date_time.date()
dates_holidays = df_holidays["date"].apply(to_date)
final_df["is_holiday"] = final_df["timestamp"].apply(to_date).isin(dates_holidays)
final_df.head()
# FIXME
df = final_df.drop(columns=["timestamp"])
df.corr()
# The output shows desciptive statistics for the numerical features, `Lat`, `Lng`, `Distance(mi)`, and `Severity`. I'll use the numerical features to demonstrate how to train the model and make submissions. **However you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# ## Data Splitting
# Now it's time to split the dataset for the training step. Typically the dataset is split into 3 subsets, namely, the training, validation and test sets. In our case, the test set is already predefined. So we'll split the "training" set into training and validation sets with 0.8:0.2 ratio.
# *Note: a good way to generate reproducible results is to set the seed to the algorithms that depends on randomization. This is done with the argument `random_state` in the following command*
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
df, test_size=0.2, random_state=42, stratify=df["Amenity"]
) # Try adding `stratify` here
X_train = train_df.drop(columns=["ID", "Severity"])
y_train = train_df["Severity"]
X_val = val_df.drop(columns=["ID", "Severity"])
y_val = val_df["Severity"]
# As pointed out eariler, I'll use the numerical features to train the classifier. **However, you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# Right=True , Left= False
X_train["Side"] = X_train["Side"] == "R"
# X_train["Side"].head()
X_val["Side"] = X_val["Side"] == "R"
# ## Model Training
# Let's train a model with the data! We'll train a Random Forest Classifier to demonstrate the process of making submissions.
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
# Now let's test our classifier on the validation dataset and see the accuracy.
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# Well. That's a good start, right? A classifier that predicts all examples' `Severity` as 2 will get around 0.63. You should get better score as you add more features and do better data preprocessing.
help(train_test_split)
# ## Submission File Generation
# We have built a model and we'd like to submit our predictions on the test set! In order to do that, we'll load the test set, predict the class and save the submission file.
# First, we'll load the data.
test_df = pd.read_csv(os.path.join(dataset_path, "test.csv"))
test_df.head()
# Note that the test set has the same features and doesn't have the `Severity` column.
# At this stage one must **NOT** forget to apply the same processing done on the training set on the features of the test set.
# Now we'll add `Severity` column to the test `DataFrame` and add the values of the predicted class to it.
# **I'll select the numerical features here as I did in the training set. DO NOT forget to change this step as you change the preprocessing of the training data.**
def create_date_series(data_frame: pd.DataFrame) -> pd.Series:
"""
Args:
data_frame:
a dataframe with columns:
Return:
a series:
"""
date = data_frame["timestamp"].apply(
lambda ts: f"{ts.year}-{ts.month}-{ts.day} {ts.hour}:00:00"
)
return pd.to_datetime(date)
def get_holiday_series(data_frame: pd.DataFrame) -> pd.Series:
"""
Args:
data_frame:
a dataframe with columns:
Return:
a series:
"""
return data_frame["timestamp"].apply(to_date).isin(dates_holidays)
def prepare_set(data_frame: pd.DataFrame) -> pd.DataFrame:
"""
Args:
data_frame:
a dataframe with columns:
Return:
a dataframe with columns:
"""
data_frame = data_frame.dropna().drop_duplicates()
# df["timestamp"] = pd.to_datetime(df["timestamp"]).dropna().drop_duplicates()
data_frame["timestamp"] = pd.to_datetime(data_frame["timestamp"])
data_frame["date"] = create_date_series(data_frame)
data_frame["is_holiday"] = get_holiday_series(data_frame)
data_frame = pd.merge(data_frame, df_weather_averaged, on="date", how="left")
data_frame = (
# FIXME that no longer matches the original stream, due to the illogical action mentioned above
# df.dropna()
# .drop_duplicates()
# .drop(columns=["ID", "Bump", "Roundabout", "timestamp", "date", "No_Exit"])
data_frame.drop(columns=["ID", "Bump", "Roundabout", "timestamp", "date"])
)
data_frame["Side"] = data_frame["Side"] == "R"
for col in df_weather_averaged.columns:
data_frame[col].fillna(data_frame[col].median(), inplace=True)
return data_frame
# test_df.dropna().drop_duplicates().shape
test_df["timestamp"].isna().any()
X_test = test_df.drop(columns=["ID"])
# You should update/remove the next line once you change the features used for training
# X_test = X_test[['Lat', 'Lng', 'Distance(mi)']]
# X_test = X_test[['Lat', 'Lng', 'Distance(mi)', "Side"]]
# X_test["Side"] = (X_test["Side"] == "R")
X_test = prepare_set(test_df)
y_test_predicted = classifier.predict(X_test)
test_df["Severity"] = y_test_predicted
test_df.head()
# Now we're ready to generate the submission file. The submission file needs the columns `ID` and `Severity` only.
test_df[["ID", "Severity"]].to_csv("/kaggle/working/submission.csv", index=False)
# test_df[["ID", "Severity"]].to_csv("./submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/141/69141940.ipynb
| null | null |
[{"Id": 69141940, "ScriptId": 18861623, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6654473, "CreationDate": "07/27/2021 10:38:18", "VersionNumber": 3.0, "Title": "Getting Started - Car Crashes' Severity Pre 2c5b26", "EvaluationDate": "07/27/2021", "IsChange": false, "TotalLines": 359.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 359.0, "LinesInsertedFromFork": 223.0, "LinesDeletedFromFork": 94.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 136.0, "TotalVotes": 0}]
| null | null | null | null |
# ## You're here!
# Welcome to your first competition in the [ITI's AI Pro training program](https://ai.iti.gov.eg/epita/ai-engineer/)! We hope you enjoy and learn as much as we did prepairing this competition.
# ## Introduction
# In the competition, it's required to predict the `Severity` of a car crash given info about the crash, e.g., location.
# This is the getting started notebook. Things are kept simple so that it's easier to understand the steps and modify it.
# Feel free to `Fork` this notebook and share it with your modifications **OR** use it to create your submissions.
# ### Prerequisites
# You should know how to use python and a little bit of Machine Learning. You can apply the techniques you learned in the training program and submit the new solutions!
# ### Checklist
# You can participate in this competition the way you perefer. However, I recommend following these steps if this is your first time joining a competition on Kaggle.
# * Fork this notebook and run the cells in order.
# * Submit this solution.
# * Make changes to the data processing step as you see fit.
# * Submit the new solutions.
# *You can submit up to 5 submissions per day. You can select only one of the submission you make to be considered in the final ranking.*
# Don't hesitate to leave a comment or contact me if you have any question!
# ## Import the libraries
# We'll use `pandas` to load and manipulate the data. Other libraries will be imported in the relevant sections.
# read_xml available only in pandas >= 1.3.0
# !pip install -Iv pandas==1.3.1
# the notebook requires restarting afterwards
# import os
# os._exit(00)
import pandas as pd
from datetime import datetime
import os
pd.__version__
# ## Exploratory Data Analysis
# In this step, one should load the data and analyze it. However, I'll load the data and do minimal analysis. You are encouraged to do thorough analysis!
# Let's load the data using `pandas` and have a look at the generated `DataFrame`.
dataset_path = "/kaggle/input/car-crashes-severity-prediction/"
# dataset_path = "./"
df = pd.read_csv(os.path.join(dataset_path, "train.csv"), index_col=None)
print("The shape of the dataset is {}.\n\n".format(df.shape))
# remove rows with any null values
df = df.dropna()
# remove duplicate rows
df = df.drop_duplicates()
df.head()
# converting timestamp column type to become datetime
df["timestamp"] = pd.to_datetime(df["timestamp"])
# convert side to be numeric
df["Side"] = df["Side"] == "R"
# FIXME
import matplotlib.pyplot as plt
# FIXME
plt.plot(df["Severity"], df["Distance(mi)"], marker=".", linestyle="none")
# plt.hist(df["Severity"], bins=4)
plt.xlabel("Severity")
plt.ylabel("Distance(mi)")
plt.show()
corr = df.corr()
corr
# dropping sparse features w/ no correlation
df = df.drop(columns=["Bump", "Roundabout"])
corr = df.corr()
corr
# pd.get_dummies(df[["Crossing", "Give_Way", "Junction", "No_Exit", "Railway", "Stop", "Amenity"]])
# We've got 6407 examples in the dataset with 14 featues, 1 ID, and the `Severity` of the crash.
# By looking at the features and a sample from the data, the features look of numerical and catogerical types. What about some descriptive statistics?
df.drop(columns="ID").describe()
# read weather file
df_weather = pd.read_csv(
os.path.join(dataset_path, "weather-sfcsv.csv"), index_col=None
)
df_weather.head(15)
df_weather["date"] = (
df_weather["Year"].astype(str)
+ "-"
+ df_weather["Month"].astype(str)
+ "-"
+ df_weather["Day"].astype(str)
+ " "
+ df_weather["Hour"].astype(str)
+ ":00:00"
)
df_weather["date"] = pd.to_datetime(df_weather["date"])
# df_weather = df_weather.dropna()
# df_weather = df_weather.drop_duplicates()
modes = {}
for col in df_weather.drop(
columns=["Year", "Hour", "Day", "Month", "Weather_Condition", "Selected"]
).columns:
# modes[col] = df_weather[col].mode()[0]
df_weather[col] = df_weather[col].fillna(df_weather[col].median())
# df_weather["Temperature(F)"].mode()[0]
# print(modes["Wind_Chill(F)"])
# df_weather["Wind_Chill(F)"].fillna(modes["Wind_Chill(F)"])
# df_weather["Wind_Chill(F)"].head()
# print(df_weather.shape)
# df_weather.dropna()
# df_weather.drop_duplicates()
# print(df_weather.shape)
# better naming convention
# df.columns = [
# "id", "lat", "lng", "bump", "distance_mile", "is_crossing",
# "is_give_way", "is_junction", "is_no_exit", "is_railway",
# "is_roundabout", "is_stop", "is_amenity", "side", "severity",
# "timestamp", "date"
# ]
# df_weather.columns = [
# "year", "day", "month", "hour", "weather_condition",
# "wind_chill_fahr", "perceptation_inch", "temperature_fahr",
# "humidity_percent", "wind_spd_mi_per_hr", "visibility_miles",
# "selected", "date"
# ]
df_weather_averaged = (
df_weather.groupby("date")
.mean()
.drop(columns=["Year", "Day", "Month", "Hour"])
.dropna()
.drop_duplicates()
)
timestamp = df["timestamp"].apply(
lambda ts: f"{ts.year}-{ts.month}-{ts.day} {ts.hour}:00:00"
)
timestamp = pd.to_datetime(timestamp)
df["date"] = timestamp
final_df = pd.merge(df, df_weather_averaged, how="inner", on="date").drop(
columns=["date"]
)
# FIXME illogical action to fill na with 0
# final_df = pd.merge(df, df_weather_averaged, how="left", on="date").drop(
# columns=["date"]
# ).fillna(0)
final_df.shape
corr = final_df.drop(columns=["ID"]).corr()
corr
# drop features w/ no correlation
# final_df = final_df.drop(columns=["No_Exit"])
final_df.corr()
df_holidays = pd.read_xml(os.path.join(dataset_path, "holidays.xml"), xpath="/root/*")
# df_holidays = pd.read_csv(os.path.join("../input/holidays/", "holidays.csv"))
# ../input/holidays/holidays.csv
df_holidays["date"] = pd.to_datetime(df_holidays["date"])
df_holidays.head()
to_date = lambda date_time: date_time.date()
dates_holidays = df_holidays["date"].apply(to_date)
final_df["is_holiday"] = final_df["timestamp"].apply(to_date).isin(dates_holidays)
final_df.head()
# FIXME
df = final_df.drop(columns=["timestamp"])
df.corr()
# The output shows desciptive statistics for the numerical features, `Lat`, `Lng`, `Distance(mi)`, and `Severity`. I'll use the numerical features to demonstrate how to train the model and make submissions. **However you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# ## Data Splitting
# Now it's time to split the dataset for the training step. Typically the dataset is split into 3 subsets, namely, the training, validation and test sets. In our case, the test set is already predefined. So we'll split the "training" set into training and validation sets with 0.8:0.2 ratio.
# *Note: a good way to generate reproducible results is to set the seed to the algorithms that depends on randomization. This is done with the argument `random_state` in the following command*
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
df, test_size=0.2, random_state=42, stratify=df["Amenity"]
) # Try adding `stratify` here
X_train = train_df.drop(columns=["ID", "Severity"])
y_train = train_df["Severity"]
X_val = val_df.drop(columns=["ID", "Severity"])
y_val = val_df["Severity"]
# As pointed out eariler, I'll use the numerical features to train the classifier. **However, you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# Right=True , Left= False
X_train["Side"] = X_train["Side"] == "R"
# X_train["Side"].head()
X_val["Side"] = X_val["Side"] == "R"
# ## Model Training
# Let's train a model with the data! We'll train a Random Forest Classifier to demonstrate the process of making submissions.
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
# Now let's test our classifier on the validation dataset and see the accuracy.
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# Well. That's a good start, right? A classifier that predicts all examples' `Severity` as 2 will get around 0.63. You should get better score as you add more features and do better data preprocessing.
help(train_test_split)
# ## Submission File Generation
# We have built a model and we'd like to submit our predictions on the test set! In order to do that, we'll load the test set, predict the class and save the submission file.
# First, we'll load the data.
test_df = pd.read_csv(os.path.join(dataset_path, "test.csv"))
test_df.head()
# Note that the test set has the same features and doesn't have the `Severity` column.
# At this stage one must **NOT** forget to apply the same processing done on the training set on the features of the test set.
# Now we'll add `Severity` column to the test `DataFrame` and add the values of the predicted class to it.
# **I'll select the numerical features here as I did in the training set. DO NOT forget to change this step as you change the preprocessing of the training data.**
def create_date_series(data_frame: pd.DataFrame) -> pd.Series:
"""
Args:
data_frame:
a dataframe with columns:
Return:
a series:
"""
date = data_frame["timestamp"].apply(
lambda ts: f"{ts.year}-{ts.month}-{ts.day} {ts.hour}:00:00"
)
return pd.to_datetime(date)
def get_holiday_series(data_frame: pd.DataFrame) -> pd.Series:
"""
Args:
data_frame:
a dataframe with columns:
Return:
a series:
"""
return data_frame["timestamp"].apply(to_date).isin(dates_holidays)
def prepare_set(data_frame: pd.DataFrame) -> pd.DataFrame:
"""
Args:
data_frame:
a dataframe with columns:
Return:
a dataframe with columns:
"""
data_frame = data_frame.dropna().drop_duplicates()
# df["timestamp"] = pd.to_datetime(df["timestamp"]).dropna().drop_duplicates()
data_frame["timestamp"] = pd.to_datetime(data_frame["timestamp"])
data_frame["date"] = create_date_series(data_frame)
data_frame["is_holiday"] = get_holiday_series(data_frame)
data_frame = pd.merge(data_frame, df_weather_averaged, on="date", how="left")
data_frame = (
# FIXME that no longer matches the original stream, due to the illogical action mentioned above
# df.dropna()
# .drop_duplicates()
# .drop(columns=["ID", "Bump", "Roundabout", "timestamp", "date", "No_Exit"])
data_frame.drop(columns=["ID", "Bump", "Roundabout", "timestamp", "date"])
)
data_frame["Side"] = data_frame["Side"] == "R"
for col in df_weather_averaged.columns:
data_frame[col].fillna(data_frame[col].median(), inplace=True)
return data_frame
# test_df.dropna().drop_duplicates().shape
test_df["timestamp"].isna().any()
X_test = test_df.drop(columns=["ID"])
# You should update/remove the next line once you change the features used for training
# X_test = X_test[['Lat', 'Lng', 'Distance(mi)']]
# X_test = X_test[['Lat', 'Lng', 'Distance(mi)', "Side"]]
# X_test["Side"] = (X_test["Side"] == "R")
X_test = prepare_set(test_df)
y_test_predicted = classifier.predict(X_test)
test_df["Severity"] = y_test_predicted
test_df.head()
# Now we're ready to generate the submission file. The submission file needs the columns `ID` and `Severity` only.
test_df[["ID", "Severity"]].to_csv("/kaggle/working/submission.csv", index=False)
# test_df[["ID", "Severity"]].to_csv("./submission.csv", index=False)
| false | 0 | 3,468 | 0 | 3,468 | 3,468 |
||
69141863
|
# **This notebook is an exercise in the [Introduction to Machine Learning](https://www.kaggle.com/learn/intro-to-machine-learning) course. You can reference the tutorial at [this link](https://www.kaggle.com/alexisbcook/machine-learning-competitions).**
# ---
# # Introduction
# In this exercise, you will create and submit predictions for a Kaggle competition. You can then improve your model (e.g. by adding features) to apply what you've learned and move up the leaderboard.
# Begin by running the code cell below to set up code checking and the filepaths for the dataset.
# Set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.machine_learning.ex7 import *
# Set up filepaths
import os
if not os.path.exists("../input/train.csv"):
os.symlink("../input/home-data-for-ml-course/train.csv", "../input/train.csv")
os.symlink("../input/home-data-for-ml-course/test.csv", "../input/test.csv")
# Here's some of the code you've written so far. Start by running it again.
# Import helpful libraries
import pandas as pd
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
# Load the data, and separate the target
iowa_file_path = "../input/train.csv"
home_data = pd.read_csv(iowa_file_path)
y = home_data.SalePrice
# Create X (After completing the exercise, you can return to modify this line!)
features = [
"LotArea",
"YearBuilt",
"1stFlrSF",
"2ndFlrSF",
"FullBath",
"BedroomAbvGr",
"TotRmsAbvGrd",
]
# Select columns corresponding to features, and preview the data
X = home_data[features]
X.head()
# Split into validation and training data
train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)
# Define a random forest model
rf_model = RandomForestRegressor(random_state=1)
rf_model.fit(train_X, train_y)
rf_val_predictions = rf_model.predict(val_X)
rf_val_mae = mean_absolute_error(rf_val_predictions, val_y)
print("Validation MAE for Random Forest Model: {:,.0f}".format(rf_val_mae))
# # Train a model for the competition
# The code cell above trains a Random Forest model on **`train_X`** and **`train_y`**.
# Use the code cell below to build a Random Forest model and train it on all of **`X`** and **`y`**.
# To improve accuracy, create a new Random Forest model which you will train on all training data
rf_model_on_full_data = RandomForestRegressor(random_state=1)
# fit rf_model_on_full_data on all data from the training data
rf_model_on_full_data.fit(X, y)
# Now, read the file of "test" data, and apply your model to make predictions.
# path to file you will use for predictions
test_data_path = "../input/test.csv"
# read test data file using pandas
test_data = pd.read_csv(test_data_path)
# create test_X which comes from test_data but includes only the columns you used for prediction.
# The list of columns is stored in a variable called features
test_X = test_data[features]
# make predictions which we will submit.
test_preds = rf_model_on_full_data.predict(test_X)
# Before submitting, run a check to make sure your `test_preds` have the right format.
# Check your answer (To get credit for completing the exercise, you must get a "Correct" result!)
step_1.check()
# step_1.solution()
# # Generate a submission
# Run the code cell below to generate a CSV file with your predictions that you can use to submit to the competition.
# Run the code to save predictions in the format used for competition scoring
output = pd.DataFrame({"Id": test_data.Id, "SalePrice": test_preds})
output.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/141/69141863.ipynb
| null | null |
[{"Id": 69141863, "ScriptId": 18872882, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5929773, "CreationDate": "07/27/2021 10:37:18", "VersionNumber": 1.0, "Title": "Exercise: Machine Learning Competitions", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 175.0, "LinesInsertedFromPrevious": 5.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 170.0, "LinesInsertedFromFork": 5.0, "LinesDeletedFromFork": 5.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 170.0, "TotalVotes": 0}]
| null | null | null | null |
# **This notebook is an exercise in the [Introduction to Machine Learning](https://www.kaggle.com/learn/intro-to-machine-learning) course. You can reference the tutorial at [this link](https://www.kaggle.com/alexisbcook/machine-learning-competitions).**
# ---
# # Introduction
# In this exercise, you will create and submit predictions for a Kaggle competition. You can then improve your model (e.g. by adding features) to apply what you've learned and move up the leaderboard.
# Begin by running the code cell below to set up code checking and the filepaths for the dataset.
# Set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.machine_learning.ex7 import *
# Set up filepaths
import os
if not os.path.exists("../input/train.csv"):
os.symlink("../input/home-data-for-ml-course/train.csv", "../input/train.csv")
os.symlink("../input/home-data-for-ml-course/test.csv", "../input/test.csv")
# Here's some of the code you've written so far. Start by running it again.
# Import helpful libraries
import pandas as pd
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
# Load the data, and separate the target
iowa_file_path = "../input/train.csv"
home_data = pd.read_csv(iowa_file_path)
y = home_data.SalePrice
# Create X (After completing the exercise, you can return to modify this line!)
features = [
"LotArea",
"YearBuilt",
"1stFlrSF",
"2ndFlrSF",
"FullBath",
"BedroomAbvGr",
"TotRmsAbvGrd",
]
# Select columns corresponding to features, and preview the data
X = home_data[features]
X.head()
# Split into validation and training data
train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)
# Define a random forest model
rf_model = RandomForestRegressor(random_state=1)
rf_model.fit(train_X, train_y)
rf_val_predictions = rf_model.predict(val_X)
rf_val_mae = mean_absolute_error(rf_val_predictions, val_y)
print("Validation MAE for Random Forest Model: {:,.0f}".format(rf_val_mae))
# # Train a model for the competition
# The code cell above trains a Random Forest model on **`train_X`** and **`train_y`**.
# Use the code cell below to build a Random Forest model and train it on all of **`X`** and **`y`**.
# To improve accuracy, create a new Random Forest model which you will train on all training data
rf_model_on_full_data = RandomForestRegressor(random_state=1)
# fit rf_model_on_full_data on all data from the training data
rf_model_on_full_data.fit(X, y)
# Now, read the file of "test" data, and apply your model to make predictions.
# path to file you will use for predictions
test_data_path = "../input/test.csv"
# read test data file using pandas
test_data = pd.read_csv(test_data_path)
# create test_X which comes from test_data but includes only the columns you used for prediction.
# The list of columns is stored in a variable called features
test_X = test_data[features]
# make predictions which we will submit.
test_preds = rf_model_on_full_data.predict(test_X)
# Before submitting, run a check to make sure your `test_preds` have the right format.
# Check your answer (To get credit for completing the exercise, you must get a "Correct" result!)
step_1.check()
# step_1.solution()
# # Generate a submission
# Run the code cell below to generate a CSV file with your predictions that you can use to submit to the competition.
# Run the code to save predictions in the format used for competition scoring
output = pd.DataFrame({"Id": test_data.Id, "SalePrice": test_preds})
output.to_csv("submission.csv", index=False)
| false | 0 | 1,057 | 0 | 1,057 | 1,057 |
||
69141484
|
<jupyter_start><jupyter_text>Lyme Disease Rashes in TFRecords
Kaggle dataset identifier: lyme-disease-rashes-in-tfrecords
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
###for dirname, _, filenames in os.walk('/kaggle/input'):
### for filename in filenames:
### print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Initialize Environment
# !pip install -q efficientnet >> /dev/null
import pandas as pd, numpy as np
from kaggle_datasets import KaggleDatasets
import tensorflow as tf, re, math
import tensorflow.keras.backend as K
# import efficientnet.tfkeras as efn
from sklearn.model_selection import KFold
from sklearn.metrics import roc_auc_score
import matplotlib.pyplot as plt
import cv2
# ## Configuration
# In order to be a proper cross validation with a meaningful overall CV score (aligned with LB score), **you need to choose the same** `IMG_SIZES`, `INC2019`, `INC2018`, and `EFF_NETS` **for each fold**. If your goal is to just run lots of experiments, then you can choose to have a different experiment in each fold. Then each fold is like a holdout validation experiment. When you find a configuration you like, you can use that configuration for all folds.
# * DEVICE - is GPU or TPU
# * SEED - a different seed produces a different triple stratified kfold split.
# * FOLDS - number of folds. Best set to 3, 5, or 15 but can be any number between 2 and 15
# * IMG_SIZES - is a Python list of length FOLDS. These are the image sizes to use each fold
# * INC2019 - This includes the new half of the 2019 competition data. The second half of the 2019 data is the comp data from 2018 plus 2017
# * INC2018 - This includes the second half of the 2019 competition data which is the comp data from 2018 plus 2017
# * BATCH_SIZES - is a list of length FOLDS. These are batch sizes for each fold. For maximum speed, it is best to use the largest batch size your GPU or TPU allows.
# * EPOCHS - is a list of length FOLDS. These are maximum epochs. Note that each fold, the best epoch model is saved and used. So if epochs is too large, it won't matter.
# * EFF_NETS - is a list of length FOLDS. These are the EfficientNets to use each fold. The number refers to the B. So a number of `0` refers to EfficientNetB0, and `1` refers to EfficientNetB1, etc.
# * WGTS - this should be `1/FOLDS` for each fold. This is the weight when ensembling the folds to predict the test set. If you want a weird ensemble, you can use different weights.
# * TTA - test time augmentation. Each test image is randomly augmented and predicted TTA times and the average prediction is used. TTA is also applied to OOF during validation.
DEVICE = "TPU" # or "GPU" #
# USE DIFFERENT SEED FOR DIFFERENT STRATIFIED KFOLD
SEED = 42
# NUMBER OF FOLDS. USE 3, 5, OR 15
FOLDS = 3
# WHICH IMAGE SIZES TO LOAD EACH FOLD
# CHOOSE 128, 192, 256, 384, 512, 768, 1024, 1536
# IMG_SIZES = [384,384,384,384,384]
# IMG_SIZES = [128,128,128,128,128]
SIZE = 128
IMG_SIZES = [SIZE] * FOLDS
# INCLUDE OLD COMP DATA? YES=1 NO=0
# TOKEN = 0
# INC2019 = [TOKEN,TOKEN,TOKEN,TOKEN,TOKEN]
# INC2018 = [TOKEN,TOKEN,TOKEN,TOKEN,TOKEN]
# BATCH SIZE AND EPOCHS
# Original batch size
# BATCH_SIZES = [32]*FOLDS
# YG
# Experimental batch sizes for EfficientNetB7
# BAD
# BATCH_SIZES = [4]*FOLDS # for 768 - VERY LONG - stops after 2 folds only, ~4 hours per fold
# BATCH_SIZES = [8]*FOLDS # for 768 - VERY LONG - 3 hours per fold
# BATCH_SIZES = [12]*FOLDS # for 768 - BAD, not enough resources
# GOOD
# BATCH_SIZES = [32]*FOLDS # for 384 - GOOD - ~1 hour per fold
# BATCH_SIZES = [16]*FOLDS # for 512
# GPU: Experimental batch sizes for EfficientNetB0
# BATCH_SIZES = [2]*FOLDS # SIZE = 1024
# Experimental batch sizes for EfficientNetB1
BATCH = 4
BATCH_SIZES = [BATCH] * FOLDS # SIZE = 768
# YG
# NASNetLarge
# BAD
# BATCH_SIZES = [4]*FOLDS # for 768 - VERY LONG - ~3.5-4 hours per fold
# BATCH_SIZES = [8]*FOLDS # for 768 - BAD - not enough resources
# GOOD
# BATCH_SIZES = [16]*FOLDS # for 384 - GOOD - ~1 hour per fold
# BATCH_SIZES = [8]*FOLDS # for 512
# EPOCHS = [12]*FOLDS
EPOCH = 32
EPOCHS = [EPOCH] * FOLDS
# EfficientNet
# WHICH EFFICIENTNET B? TO USE
model_name = "MobileNetV2"
# MODEL = 7 # EfficientNetB7
MODEL = "MobileNetV2" # EfficientNetB3
# MODEL = 0 # EfficientNetB0
EFF_NETS = [MODEL] * FOLDS
SPECIFIC_SIZE = SIZE
# MobileNetV2
# MODEL = 'MobileNetV2'
# SPECIFIC_SIZE = SIZE
# model_name = 'NASNetMobile'
# MODEL = 'NASNetMobile'
# SPECIFIC_SIZE = SIZE # 224 only at the moment!
# model_name = 'NASNetLarge'
# MODEL = 'NASNetLarge'
# SPECIFIC_SIZE = SIZE # 331 only at the moment!
# WEIGHTS FOR FOLD MODELS WHEN PREDICTING TEST
WGTS = [1 / FOLDS] * FOLDS
# TEST TIME AUGMENTATION STEPS
TTA = 11
# make the weights and biases of the base model non-trainable
# by "freezing" each layer of the BASE network
TRAINABLE = False
AUGMENTATION = True
AUG_LOSSLESS = False
# Time estimation - 1 epoch
# Model = EfficientNetB0
# MODEL = 0
########################################################
# Image Size
# SIZE = 1536
# BATCH = 1
# Fold Times:
# FOLDS = 3
# 1it [03:29, 209.24s/it]
# 2it [06:58, 209.25s/it]
# 3it [10:25, 208.61s/it]
# CPU times: user 2min 11s, sys: 13.6 s, total: 2min 25s
# Wall time: 10min 25s
# + 5 min for testing
########################################################
# Image Size
# SIZE = 1024
# BATCH = 8
# CPU times: user 2min 18s, sys: 13.8 s, total: 2min 32s
# Wall time: 6min 57s
########################################################
########################################################
# Image Size
# SIZE = 768
# BATCH = 16
# CPU times: user 2min 15s, sys: 12.9 s, total: 2min 28s
# Wall time: 6min 20s
# Time estimation - 1 epoch
# Model = EfficientNetB7
# MODEL = 7
######################################################## MIN
# Image Size
# SIZE = 128
# BATCH = 32
# CPU times: user 7min 4s, sys: 46.5 s, total: 7min 50s
# Wall time: 13min 25s
######################################################## MAX
# Image Size
# SIZE = 1024
# BATCH = 1
# CPU times: user 7min 19s, sys: 47.8 s, total: 8min 6s
# Wall time: 18min 27s
########################################################
# Image Size
# SIZE = 768
# BATCH = 4
# CPU times: user 7min 27s, sys: 49.2 s, total: 8min 16s
# Wall time: 17min 40s
########################################################
# Image Size
# SIZE = 512
# BATCH = 16
# CPU times: user 7min 11s, sys: 47.5 s, total: 7min 58s
# Wall time: 15min 50s
########################################################
# Image Size
# SIZE = 256
# BATCH = 16
# CPU times: user 4min 43s, sys: 40.4 s, total: 5min 24s
# Wall time: 7min 9s
########################################################
# TRAINABLE = False
########################################################
# Image Size
# SIZE = 256
# BATCH = 16
# CPU times: user 4min 51s, sys: 41.1 s, total: 5min 32s
# Wall time: 7min 12s
########################################################
# TRAINABLE = False
# AUGMENTATION = False
########################################################
# Image Size
# SIZE = 256
# BATCH = 16
# CPU times: user 4min 35s, sys: 38.1 s, total: 5min 13s
# Wall time: 6min 56s
if DEVICE == "TPU":
print("connecting to TPU...")
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
print("Running on TPU ", tpu.master())
except ValueError:
print("Could not connect to TPU")
tpu = None
if tpu:
try:
print("initializing TPU ...")
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
print("TPU initialized")
except _:
print("failed to initialize TPU")
else:
DEVICE = "GPU"
if DEVICE != "TPU":
print("Using default strategy for CPU and single GPU")
strategy = tf.distribute.get_strategy()
if DEVICE == "GPU":
print(
"Num GPUs Available: ", len(tf.config.experimental.list_physical_devices("GPU"))
)
AUTO = tf.data.experimental.AUTOTUNE
REPLICAS = strategy.num_replicas_in_sync
print(f"REPLICAS: {REPLICAS}")
# # Step 1: Preprocess
# Preprocess has already been done and saved to TFRecords. Here we choose which size to load. We can use either 128x128, 192x192, 256x256, 384x384, 512x512, 768x768 by changing the `IMG_SIZES` variable in the preceeding code section. These TFRecords are discussed [here][1]. The advantage of using different input sizes is discussed [here][2]
# [1]: https://www.kaggle.com/c/siim-isic-melanoma-classification/discussion/155579
# [2]: https://www.kaggle.com/c/siim-isic-melanoma-classification/discussion/160147
print(KaggleDatasets().get_gcs_path("lyme-disease-rashes-in-tfrecords"))
GCS_PATH = [None] * FOLDS
# GCS_PATH2 = [None]*FOLDS
for i, k in enumerate(IMG_SIZES):
# GCS_PATH[i] = KaggleDatasets().get_gcs_path('melanoma-%ix%i'%(k,k))
# GCS_PATH2[i] = KaggleDatasets().get_gcs_path('isic2019-%ix%i'%(k,k))
GCS_PATH[i] = KaggleDatasets().get_gcs_path("lyme-disease-rashes-in-tfrecords")
files_train = np.sort(
np.array(tf.io.gfile.glob(GCS_PATH[0] + "/train_%d_*.tfrec" % SIZE))
)
files_test = np.sort(
np.array(tf.io.gfile.glob(GCS_PATH[0] + "/test_%d_*.tfrec" % SIZE))
)
files_train
files_test
# # Step 2: Data Augmentation
# This notebook uses rotation, sheer, zoom, shift augmentation first shown in this notebook [here][1] and successfully used in Melanoma comp by AgentAuers [here][2]. This notebook also uses horizontal flip, hue, saturation, contrast, brightness augmentation similar to last years winner and also similar to AgentAuers' notebook.
# Additionally we can decide to use external data by changing the variables `INC2019` and `INC2018` in the preceeding code section. These variables respectively indicate whether to load last year 2019 data and/or year 2018 + 2017 data. These datasets are discussed [here][3]
# Consider experimenting with different augmenation and/or external data. The code to load TFRecords is taken from AgentAuers' notebook [here][2]. Thank you AgentAuers, this is great work.
# [1]: https://www.kaggle.com/cdeotte/rotation-augmentation-gpu-tpu-0-96
# [2]: https://www.kaggle.com/agentauers/incredible-tpus-finetune-effnetb0-b6-at-once
# [3]: https://www.kaggle.com/c/siim-isic-melanoma-classification/discussion/164910
ROT_ = 180.0
SHR_ = 2.0
HZOOM_ = 8.0
WZOOM_ = 8.0
HSHIFT_ = 8.0
WSHIFT_ = 8.0
def get_mat(rotation, shear, height_zoom, width_zoom, height_shift, width_shift):
# returns 3x3 transformmatrix which transforms indicies
# CONVERT DEGREES TO RADIANS
rotation = math.pi * rotation / 180.0
shear = math.pi * shear / 180.0
def get_3x3_mat(lst):
return tf.reshape(tf.concat([lst], axis=0), [3, 3])
# ROTATION MATRIX
c1 = tf.math.cos(rotation)
s1 = tf.math.sin(rotation)
one = tf.constant([1], dtype="float32")
zero = tf.constant([0], dtype="float32")
rotation_matrix = get_3x3_mat([c1, s1, zero, -s1, c1, zero, zero, zero, one])
# SHEAR MATRIX
c2 = tf.math.cos(shear)
s2 = tf.math.sin(shear)
shear_matrix = get_3x3_mat([one, s2, zero, zero, c2, zero, zero, zero, one])
# ZOOM MATRIX
zoom_matrix = get_3x3_mat(
[one / height_zoom, zero, zero, zero, one / width_zoom, zero, zero, zero, one]
)
# SHIFT MATRIX
shift_matrix = get_3x3_mat(
[one, zero, height_shift, zero, one, width_shift, zero, zero, one]
)
return K.dot(K.dot(rotation_matrix, shear_matrix), K.dot(zoom_matrix, shift_matrix))
def transform(image, DIM=256):
# input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]
# output - image randomly rotated, sheared, zoomed, and shifted
XDIM = DIM % 2 # fix for size 331
rot = ROT_ * tf.random.normal([1], dtype="float32")
shr = SHR_ * tf.random.normal([1], dtype="float32")
h_zoom = 1.0 + tf.random.normal([1], dtype="float32") / HZOOM_
w_zoom = 1.0 + tf.random.normal([1], dtype="float32") / WZOOM_
h_shift = HSHIFT_ * tf.random.normal([1], dtype="float32")
w_shift = WSHIFT_ * tf.random.normal([1], dtype="float32")
# GET TRANSFORMATION MATRIX
m = get_mat(rot, shr, h_zoom, w_zoom, h_shift, w_shift)
# LIST DESTINATION PIXEL INDICES
x = tf.repeat(tf.range(DIM // 2, -DIM // 2, -1), DIM)
y = tf.tile(tf.range(-DIM // 2, DIM // 2), [DIM])
z = tf.ones([DIM * DIM], dtype="int32")
idx = tf.stack([x, y, z])
# ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS
idx2 = K.dot(m, tf.cast(idx, dtype="float32"))
idx2 = K.cast(idx2, dtype="int32")
idx2 = K.clip(idx2, -DIM // 2 + XDIM + 1, DIM // 2)
# FIND ORIGIN PIXEL VALUES
idx3 = tf.stack([DIM // 2 - idx2[0,], DIM // 2 - 1 + idx2[1,]])
d = tf.gather_nd(image, tf.transpose(idx3))
return tf.reshape(d, [DIM, DIM, 3])
def read_labeled_tfrecord(example):
tfrec_format = {
"image": tf.io.FixedLenFeature([], tf.string),
"image_name": tf.io.FixedLenFeature([], tf.string),
#'patient_id' : tf.io.FixedLenFeature([], tf.int64),
#'sex' : tf.io.FixedLenFeature([], tf.int64),
#'age_approx' : tf.io.FixedLenFeature([], tf.int64),
#'anatom_site_general_challenge': tf.io.FixedLenFeature([], tf.int64),
#'diagnosis' : tf.io.FixedLenFeature([], tf.int64),
"target": tf.io.FixedLenFeature([], tf.int64),
}
example = tf.io.parse_single_example(example, tfrec_format)
return example["image"], example["target"]
def read_unlabeled_tfrecord(example, return_image_name):
tfrec_format = {
"image": tf.io.FixedLenFeature([], tf.string),
"image_name": tf.io.FixedLenFeature([], tf.string),
}
example = tf.io.parse_single_example(example, tfrec_format)
return example["image"], example["image_name"] if return_image_name else 0
def prepare_image(img, augment=True, aug_lossless=True, dim=256):
img = tf.image.decode_jpeg(img, channels=3)
img = tf.cast(img, tf.float32) / 255.0
if augment:
if aug_lossless:
img = transform(img, DIM=dim)
img = tf.image.random_flip_left_right(img)
img = tf.image.random_flip_up_down(img)
img = tf.image.rot90(
img, tf.random.uniform(shape=[], minval=0, maxval=3, dtype=tf.int32)
)
else:
img = transform(img, DIM=dim)
img = tf.image.random_flip_left_right(img)
img = tf.image.random_flip_up_down(img)
img = tf.image.rot90(
img, tf.random.uniform(shape=[], minval=0, maxval=3, dtype=tf.int32)
)
img = tf.image.random_hue(img, 0.01)
img = tf.image.random_saturation(img, 0.7, 1.3)
img = tf.image.random_contrast(img, 0.8, 1.2)
img = tf.image.random_brightness(img, 0.1)
img = tf.image.random_jpeg_quality(img, 75, 95)
# YG - just for adaptation to NASNet ... and other families
# else:
# if(MODEL == 'NASNetMobile' or MODEL == 'NASNetLarge'):
# img = transform(img,DIM=dim)
# img = cv2.resize(img, (dim, dim),interpolation = cv2.INTER_CUBIC)
img = tf.reshape(img, [dim, dim, 3])
# img = tf.image.resize(img, [224, 224, 3])
return img
def count_data_items(filenames):
n = [
int(re.compile(r"-([0-9]*)\.").search(filename).group(1))
for filename in filenames
]
return np.sum(n)
def get_dataset(
files,
augment=False,
aug_lossless=True,
shuffle=False,
repeat=False,
labeled=True,
return_image_names=True,
batch_size=16,
dim=256,
):
ds = tf.data.TFRecordDataset(files, num_parallel_reads=AUTO)
ds = ds.cache()
if repeat:
ds = ds.repeat()
if shuffle:
ds = ds.shuffle(1024 * 8)
opt = tf.data.Options()
opt.experimental_deterministic = False
ds = ds.with_options(opt)
if labeled:
ds = ds.map(read_labeled_tfrecord, num_parallel_calls=AUTO)
else:
ds = ds.map(
lambda example: read_unlabeled_tfrecord(example, return_image_names),
num_parallel_calls=AUTO,
)
ds = ds.map(
lambda img, imgname_or_label: (
prepare_image(img, augment=augment, aug_lossless=aug_lossless, dim=dim),
imgname_or_label,
),
num_parallel_calls=AUTO,
)
ds = ds.batch(batch_size * REPLICAS)
ds = ds.prefetch(AUTO)
return ds
# # Step 3: Build Model
# This is a common model architecute. Consider experimenting with different backbones, custom heads, losses, and optimizers. Also consider inputing meta features into your CNN.
# YG
# These models HAVE PASSED the initial test!
from tensorflow.keras.applications import MobileNetV2
from tensorflow.keras.applications import NASNetMobile
from tensorflow.keras.applications import NASNetLarge
from tensorflow.keras.applications import DenseNet121
from tensorflow.keras.applications import EfficientNetB0
dim = SPECIFIC_SIZE
# if(model_name =='EfficientNet'):
# These should be checked:
# from tensorflow.keras.applications import InceptionV3
# model_name = 'InceptionV3'
# from tensorflow.keras.applications import InceptionResNetV2
# model_name = 'InceptionResNetV2'
# from tensorflow.keras.applications import MobileNetV2
# model_name = 'MobileNetV2'
# from tensorflow.keras.applications import ResNet101
# model_name = 'ResNet101'
# from tensorflow.keras.applications import ResNet101V2
# model_name = 'ResNet101V2'
# from tensorflow.keras.applications import VGG16
# model_name = 'VGG16'
# from tensorflow.keras.applications import Xception
# model_name = 'Xception'
def build_model(model_name=model_name, dim=SPECIFIC_SIZE, trainable=False):
inp = tf.keras.layers.Input(shape=(dim, dim, 3))
if model_name == "EfficientNet":
# EFNS = [efn.EfficientNetB0, efn.EfficientNetB1, efn.EfficientNetB2, efn.EfficientNetB3,
# efn.EfficientNetB4, efn.EfficientNetB5, efn.EfficientNetB6, efn.EfficientNetB7]
# base = EFNS[MODEL](input_shape=(dim,dim,3),weights='imagenet',include_top=False)
if MODEL == 0:
base = EfficientNetB0(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
model_name = "EfficientNetB" + str(MODEL)
else:
print(
"ERROR: This code is prepared for EfficientNetB, version"
+ str(MODEL)
+ ", BUT other version is used!"
)
if model_name == "MobileNetV2":
base = MobileNetV2(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
if model_name == "NASNetMobile":
base = NASNetMobile(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
if model_name == "NASNetLarge":
base = NASNetLarge(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
if model_name == "DenseNet121":
base = DenseNet121(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
# make the weights and biases of the base model non-trainable
# by "freezing" each layer of the BASE network
for layer in base.layers:
layer.trainable = trainable
x = base(inp)
x = tf.keras.layers.GlobalAveragePooling2D()(x)
x = tf.keras.layers.Dense(1, activation="sigmoid")(x)
model = tf.keras.Model(inputs=inp, outputs=x)
opt = tf.keras.optimizers.Adam(learning_rate=0.001)
loss = tf.keras.losses.BinaryCrossentropy(label_smoothing=0.05)
model.compile(optimizer=opt, loss=loss, metrics=["AUC"])
return model
# # Step 4: Train Schedule
# This is a common train schedule for transfer learning. The learning rate starts near zero, then increases to a maximum, then decays over time. Consider changing the schedule and/or learning rates. Note how the learning rate max is larger with larger batches sizes. This is a good practice to follow.
def get_lr_callback(batch_size=8):
lr_start = 0.000005
lr_max = 0.00000125 * REPLICAS * batch_size
lr_min = 0.000001
lr_ramp_ep = 5
lr_sus_ep = 0
lr_decay = 0.8
def lrfn(epoch):
if epoch < lr_ramp_ep:
lr = (lr_max - lr_start) / lr_ramp_ep * epoch + lr_start
elif epoch < lr_ramp_ep + lr_sus_ep:
lr = lr_max
else:
lr = (lr_max - lr_min) * lr_decay ** (
epoch - lr_ramp_ep - lr_sus_ep
) + lr_min
return lr
lr_callback = tf.keras.callbacks.LearningRateScheduler(lrfn, verbose=False)
return lr_callback
# ## Train Model
# Our model will be trained for the number of FOLDS and EPOCHS you chose in the configuration above. Each fold the model with lowest validation loss will be saved and used to predict OOF and test. Adjust the variables `VERBOSE` and `DISPLOY_PLOT` below to determine what output you want displayed. The variable `VERBOSE=1 or 2` will display the training and validation loss and auc for each epoch as text. The variable `DISPLAY_PLOT` shows this information as a plot.
from tqdm import tqdm
# USE VERBOSE=0 for silent, VERBOSE=1 for interactive, VERBOSE=2 for commit
VERBOSE = 1
DISPLAY_PLOT = True
skf = KFold(n_splits=FOLDS, shuffle=True, random_state=SEED)
oof_pred = []
oof_tar = []
oof_val = []
oof_names = []
oof_folds = []
preds = np.zeros((count_data_items(files_test), 1))
for fold, (idxT, idxV) in tqdm(enumerate(skf.split(np.arange(14)))):
# DISPLAY FOLD INFO
if DEVICE == "TPU":
if tpu:
tf.tpu.experimental.initialize_tpu_system(tpu)
print("#" * 25)
print("#### FOLD", fold + 1)
# YG
# print('#### Image Size %i with EfficientNet B%i and batch_size %i'%
# (IMG_SIZES[fold],EFF_NETS[fold],BATCH_SIZES[fold]*REPLICAS))
print(
"#### Image Size %i with %s and batch_size %i"
% (IMG_SIZES[fold], model_name, BATCH_SIZES[fold] * REPLICAS)
)
# CREATE TRAIN AND VALIDATION SUBSETS
# files_train = tf.io.gfile.glob([GCS_PATH[fold] + '/train%.2i*.tfrec'%x for x in idxT])
print("SIZE:", SIZE)
filename_core = GCS_PATH[fold] + "/train_%i_" % (SIZE)
files_train = tf.io.gfile.glob([filename_core + "%.2i*.tfrec" % x for x in idxT])
# print([GCS_PATH[fold]])
# print([GCS_PATH[fold] + '/train_128_%.2i*.tfrec'%x for x in idxT])
print("Files for TRAIN:")
print(files_train)
# if INC2019[fold]:
# files_train += tf.io.gfile.glob([GCS_PATH2[fold] + '/train%.2i*.tfrec'%x for x in idxT*2+1])
# print('#### Using 2019 external data')
# if INC2018[fold]:
# files_train += tf.io.gfile.glob([GCS_PATH2[fold] + '/train%.2i*.tfrec'%x for x in idxT*2])
# print('#### Using 2018+2017 external data')
np.random.shuffle(files_train)
print("#" * 25)
filename_core = GCS_PATH[fold] + "/train_%i_" % (SIZE)
files_valid = tf.io.gfile.glob([filename_core + "%.2i*.tfrec" % x for x in idxV])
print("Files for VALIDATION:")
print(files_valid)
files_test = np.sort(
np.array(tf.io.gfile.glob(GCS_PATH[fold] + "/test_%i_*.tfrec" % (SIZE)))
)
print("Files for TEST:")
print(files_test)
# BUILD MODEL
K.clear_session()
with strategy.scope():
model = build_model(
model_name=model_name, dim=SPECIFIC_SIZE, trainable=TRAINABLE
)
model.summary() # YG
# SAVE BEST MODEL EACH FOLD
sv = tf.keras.callbacks.ModelCheckpoint(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-%i.h5" % fold,
monitor="val_loss",
verbose=0,
save_best_only=True,
save_weights_only=True,
mode="min",
save_freq="epoch",
)
# TRAIN
print("Training...")
history = model.fit(
get_dataset(
files_train,
augment=True,
aug_lossless=AUG_LOSSLESS,
shuffle=True,
repeat=True,
# dim=IMG_SIZES[fold],
dim=SPECIFIC_SIZE,
batch_size=BATCH_SIZES[fold],
),
epochs=EPOCHS[fold],
callbacks=[sv, get_lr_callback(BATCH_SIZES[fold])],
steps_per_epoch=count_data_items(files_train) / BATCH_SIZES[fold] // REPLICAS,
validation_data=get_dataset(
files_valid,
augment=False,
shuffle=False,
repeat=False,
# dim=IMG_SIZES[fold]
dim=SPECIFIC_SIZE,
), # class_weight = {0:1,1:2},
verbose=VERBOSE,
)
print("Loading best model...")
model.load_weights(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-%i.h5" % fold
)
# PREDICT OOF USING TTA
print("Predicting OOF with TTA...")
ds_valid = get_dataset(
files_valid,
labeled=False,
return_image_names=False,
augment=AUGMENTATION,
aug_lossless=AUG_LOSSLESS,
repeat=True,
shuffle=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
batch_size=BATCH_SIZES[fold] * 4,
)
ct_valid = count_data_items(files_valid)
STEPS = TTA * ct_valid / BATCH_SIZES[fold] / 4 / REPLICAS
pred = model.predict(ds_valid, steps=STEPS, verbose=VERBOSE)[: TTA * ct_valid,]
oof_pred.append(np.mean(pred.reshape((ct_valid, TTA), order="F"), axis=1))
# oof_pred.append(model.predict(get_dataset(files_valid,dim=IMG_SIZES[fold]),verbose=1))
# GET OOF TARGETS AND NAMES
ds_valid = get_dataset(
files_valid,
augment=False,
aug_lossless=False,
repeat=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
labeled=True,
return_image_names=True,
)
oof_tar.append(
np.array([target.numpy() for img, target in iter(ds_valid.unbatch())])
)
oof_folds.append(np.ones_like(oof_tar[-1], dtype="int8") * fold)
ds = get_dataset(
files_valid,
augment=False,
aug_lossless=False,
repeat=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
labeled=False,
return_image_names=True,
)
oof_names.append(
np.array(
[img_name.numpy().decode("utf-8") for img, img_name in iter(ds.unbatch())]
)
)
# PREDICT TEST USING TTA
print("Predicting Test with TTA...")
ds_test = get_dataset(
files_test,
labeled=False,
return_image_names=False,
augment=True,
aug_lossless=AUG_LOSSLESS,
repeat=True,
shuffle=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
batch_size=BATCH_SIZES[fold] * 4,
)
ct_test = count_data_items(files_test)
STEPS = TTA * ct_test / BATCH_SIZES[fold] / 4 / REPLICAS
pred = model.predict(ds_test, steps=STEPS, verbose=VERBOSE)[: TTA * ct_test,]
preds[:, 0] += np.mean(pred.reshape((ct_test, TTA), order="F"), axis=1) * WGTS[fold]
# REPORT RESULTS
### BUG - YG
list_length = min(len(oof_tar[-1]), len(oof_pred[-1]))
auc = roc_auc_score(oof_tar[-1][:list_length], oof_pred[-1][:list_length])
oof_val.append(np.max(history.history["val_auc"]))
print(
"#### FOLD %i OOF AUC without TTA = %.3f, with TTA = %.3f"
% (fold + 1, oof_val[-1], auc)
)
# PLOT TRAINING
if DISPLAY_PLOT:
plt.figure(figsize=(15, 5))
plt.plot(
np.arange(EPOCHS[fold]),
history.history["auc"],
"-o",
label="Train AUC",
color="#ff7f0e",
)
plt.plot(
np.arange(EPOCHS[fold]),
history.history["val_auc"],
"-o",
label="Val AUC",
color="#1f77b4",
)
x = np.argmax(history.history["val_auc"])
y = np.max(history.history["val_auc"])
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="#1f77b4")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.2f" % y, size=14)
plt.ylabel("AUC", size=14)
plt.xlabel("Epoch", size=14)
plt.legend(loc=2)
plt2 = plt.gca().twinx()
plt2.plot(
np.arange(EPOCHS[fold]),
history.history["loss"],
"-o",
label="Train Loss",
color="#2ca02c",
)
plt2.plot(
np.arange(EPOCHS[fold]),
history.history["val_loss"],
"-o",
label="Val Loss",
color="#d62728",
)
x = np.argmin(history.history["val_loss"])
y = np.min(history.history["val_loss"])
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="#d62728")
plt.text(x - 0.03 * xdist, y + 0.05 * ydist, "min loss", size=14)
plt.ylabel("Loss", size=14)
# YG
# plt.title('FOLD %i - Image Size %i, EfficientNet B%i, inc2019=%i, inc2018=%i'%
# (fold+1,IMG_SIZES[fold],EFF_NETS[fold],INC2019[fold],INC2018[fold]),size=18)
# plt.title('FOLD %i - Image Size %i, %s, inc2019=%i, inc2018=%i'%
# (fold+1,IMG_SIZES[fold],model_name,INC2019[fold],INC2018[fold]),size=18)
plt.title(
"FOLD %i - Image Size %i, %s, Device=%s, TTA=%i"
% (fold + 1, IMG_SIZES[fold], model_name, DEVICE, TTA),
size=18,
)
plt.legend(loc=3)
plt.savefig(
"AUC_"
+ DEVICE
+ "_model"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_fold"
+ str(fold)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# ## Calculate OOF AUC
# The OOF (out of fold) predictions are saved to disk. If you wish to ensemble multiple models, use the OOF to determine what are the best weights to blend your models with. Choose weights that maximize OOF CV score when used to blend OOF. Then use those same weights to blend your test predictions.
# COMPUTE OVERALL OOF AUC
oof = np.concatenate(oof_pred)
true = np.concatenate(oof_tar)
names = np.concatenate(oof_names)
folds = np.concatenate(oof_folds)
### BUG - YG
list_length = min(len(true), len(oof))
auc = roc_auc_score(true[:list_length], oof[:list_length])
print("Overall OOF AUC with TTA = %.3f" % auc)
# SAVE OOF TO DISK
df_oof = pd.DataFrame(
dict(
image_name=names, target=true[:list_length], pred=oof[:list_length], fold=folds
)
)
df_oof.to_csv(DEVICE + "_" + str(MODEL) + "_oof.csv", index=False)
df_oof.tail()
# # Step 5: Post process
# ## Measure prediction times, AUC, model file size
# ### With TTA
import time
import statistics
VERBOSE = 1
def AUC_time(print_verbose=False, augment=True, aug_lossless=True, TTA=1):
predict_time_list = []
AUC_list = []
oof_pred = []
oof_tar = []
for fold, (idxT, idxV) in enumerate(skf.split(np.arange(15))):
if print_verbose:
print("Fold %i. Loading the current fold model..." % fold)
model.load_weights(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-%i.h5" % fold
)
# model.load_weights(DEVICE + '_fold-%i.h5'%fold)
# PREDICT with TTA # augment=True, aug_lossless = True,
ds_valid = get_dataset(
files_valid,
labeled=False,
return_image_names=False,
augment=True,
aug_lossless=True,
repeat=True,
shuffle=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
batch_size=BATCH_SIZES[fold] * 4,
)
ct_valid = count_data_items(files_valid)
STEPS = TTA * ct_valid / BATCH_SIZES[fold] / 4 / REPLICAS
if print_verbose:
print("Predicting Test with TTA_LL for %i files..." % (ct_valid))
# Start timer
ts_eval = time.time()
pred = model.predict(ds_valid, steps=STEPS, verbose=VERBOSE)[: TTA * ct_valid,]
# End timer
te_eval = time.time()
test_time = (te_eval - ts_eval) / ct_valid
predict_time_list.append(test_time)
if print_verbose:
print("Fold %i, test_time=%.6f seconds." % (fold, test_time))
# Add predictions to list
oof_pred.append(np.mean(pred.reshape((ct_valid, TTA), order="F"), axis=1))
# GET OOF TARGETS AND NAMES
ds_valid = get_dataset(
files_valid,
augment=False,
aug_lossless=True,
repeat=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
labeled=True,
return_image_names=True,
)
# Add targets to list
oof_tar.append(
np.array([target.numpy() for img, target in iter(ds_valid.unbatch())])
)
# Calculate AUC
auc = roc_auc_score(oof_tar[-1], oof_pred[-1])
# Add AUC to list
AUC_list.append(auc)
if print_verbose:
print("Fold %i, AUC=%.6f" % (fold, auc))
# Calculate AUC mean value to list
AUC_mean = statistics.mean(AUC_list) # mean
# Calculate AUC standard deviation value to list
AUC_std = statistics.stdev(AUC_list) # standard devition
print("#### TTA = %d" % TTA)
print("#### OOF AUC with TTA_LL: mean = %.6f, stdev = %.6f." % (AUC_mean, AUC_std))
# Add AUC mean value to list
predict_time_mean = statistics.mean(predict_time_list) # mean
# Add AUC standard deviation value to list
predict_time_std = statistics.stdev(predict_time_list) # standard devition
print(
"#### Time without TTA_LL: mean = %.6f, stdev = %.6f seconds."
% (predict_time_mean, predict_time_std)
)
return AUC_mean, AUC_std, predict_time_mean, predict_time_std
# #### LOSSLESS TTA - withTTA_LL
# _withTTA_LL_
VERBOSE = 0
TTA_list = [1, 2, 4, 8, 16, 32, 64, 128]
withTTA_LL_AUC_mean_list = []
withTTA_LL_AUC_std_list = []
withTTA_LL_time_mean_list = []
withTTA_LL_time_std_list = []
for i in tqdm(TTA_list):
AUC_mean, AUC_std, predict_time_mean, predict_time_std = AUC_time(
print_verbose=False, augment=True, aug_lossless=True, TTA=i
)
withTTA_LL_AUC_mean_list.append(AUC_mean)
withTTA_LL_AUC_std_list.append(AUC_std)
withTTA_LL_time_mean_list.append(predict_time_mean)
withTTA_LL_time_std_list.append(predict_time_std)
# PLOT: AUC - withTTA_LL
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
np.log2(TTA_list),
withTTA_LL_AUC_mean_list,
yerr=withTTA_LL_AUC_std_list,
fmt="-o",
label="AUC mean",
color="#1f77b4",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmax(withTTA_LL_AUC_mean_list)
y = np.max(withTTA_LL_AUC_mean_list)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.3f" % y, size=14, color="red")
plt.ylabel("AUC", size=14)
plt.xlabel("log2(TTA steps)", size=14)
plt.title(
"withTTA_LL, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
# plt.legend(loc='best')
plt.savefig(
"AUC_"
+ "withTTA_LL"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# PLOT: PREDICTION TIME - withTTA_LL
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
TTA_list,
withTTA_LL_time_mean_list,
yerr=withTTA_LL_time_std_list,
fmt="-o",
label="lossless",
color="#1f77b4",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmin(withTTA_LL_time_mean_list)
y = np.min(withTTA_LL_time_mean_list)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "min time\n%.3f" % y, size=14, color="red")
plt.ylabel("Prediction Time", size=14)
plt.xlabel("TTA steps", size=14)
plt.title(
"withTTA_LL, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
# plt.legend(loc='best')
plt.savefig(
"TIME_"
+ "withTTA_LL"
+ "_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# #### FULL TTA - withTTA
# withTTA_
VERBOSE = 0
TTA_list = [1, 2, 4, 8, 16, 32, 64, 128]
withTTA_AUC_mean_list = []
withTTA_AUC_std_list = []
withTTA_time_mean_list = []
withTTA_time_std_list = []
for i in tqdm(TTA_list):
AUC_mean, AUC_std, predict_time_mean, predict_time_std = AUC_time(
print_verbose=False, augment=True, aug_lossless=False, TTA=i
)
withTTA_AUC_mean_list.append(AUC_mean)
withTTA_AUC_std_list.append(AUC_std)
withTTA_time_mean_list.append(predict_time_mean)
withTTA_time_std_list.append(predict_time_std)
# PLOT: AUC - withTTA_LL
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
np.log2(TTA_list),
withTTA_AUC_mean_list,
yerr=withTTA_AUC_std_list,
fmt="-o",
label="full",
color="#1f77b4",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmax(withTTA_AUC_mean_list)
y = np.max(withTTA_AUC_mean_list)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.3f" % y, size=14, color="red")
plt.ylabel("AUC", size=14)
plt.xlabel("log2(TTA steps)", size=14)
plt.title(
"withTTA_full, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
# plt.legend(loc='best')
plt.savefig(
"AUC_"
+ "withTTA_full"
+ "_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# PLOT: PREDICTION TIME - withTTA_LL
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
TTA_list,
withTTA_time_mean_list,
yerr=withTTA_time_std_list,
fmt="-o",
label="full",
color="#1f77b4",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmin(withTTA_LL_time_mean_list)
y = np.min(withTTA_LL_time_mean_list)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "min time\n%.3f" % y, size=14, color="red")
plt.ylabel("Prediction Time", size=14)
plt.xlabel("TTA steps", size=14)
plt.title(
"withTTA, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
# plt.legend(loc='best')
plt.savefig(
"TIME_"
+ "withTTA_full"
+ "_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# #### Both TTAs on the same plot - AUC and TIME
# PLOT: AUC - withTTA_LL - lossless
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
np.log2(TTA_list),
withTTA_LL_AUC_mean_list,
yerr=withTTA_LL_AUC_std_list,
fmt="-o",
label="lossless",
color="blue",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmax(withTTA_LL_AUC_mean_list)
y = np.max(withTTA_LL_AUC_mean_list)
withTTA_LL_AUC_mean_max = np.max(withTTA_LL_AUC_mean_list)
withTTA_LL_AUC_std_max = withTTA_LL_AUC_std_list[np.argmax(withTTA_LL_AUC_mean_list)]
withTTA_LL_AUC_mean_TTA = np.argmax(withTTA_LL_AUC_mean_list)
print(
"i=%d," % withTTA_LL_AUC_mean_TTA,
"withTTA_LL_AUC_max=%.3f" % withTTA_LL_AUC_mean_max,
"(+-%.3f)" % withTTA_LL_AUC_std_max,
)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.3f" % y, size=14, color="blue")
# PLOT: AUC - withTTA - full
plt.errorbar(
np.log2(TTA_list),
withTTA_AUC_mean_list,
yerr=withTTA_AUC_std_list,
fmt="-o",
label="full",
color="green",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmax(withTTA_AUC_mean_list)
y = np.max(withTTA_AUC_mean_list)
withTTA_AUC_mean_max = np.max(withTTA_AUC_mean_list)
withTTA_AUC_std_max = withTTA_AUC_std_list[np.argmax(withTTA_AUC_mean_list)]
withTTA_AUC_mean_TTA = np.argmax(withTTA_AUC_mean_list)
print(
"i=%d," % withTTA_AUC_mean_TTA,
"withTTA_AUC_max=%.3f" % withTTA_AUC_mean_max,
"(+-%.3f)" % withTTA_AUC_std_max,
)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(
x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.3f" % y, size=14, color="green"
)
plt.ylabel("AUC", size=14)
plt.xlabel("log2(TTA steps)", size=14)
plt.title(
"Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
plt.legend(loc="best")
plt.savefig(
"AUC_both_TTA_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# PLOT: PREDICTION TIME - withTTA_LL - lossless
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
TTA_list,
withTTA_LL_time_mean_list,
yerr=withTTA_LL_time_std_list,
fmt="-o",
label="lossless",
color="blue",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmin(withTTA_LL_time_mean_list)
y = np.min(withTTA_LL_time_mean_list)
withTTA_LL_time_mean_max = withTTA_LL_time_mean_list[
np.argmax(withTTA_LL_AUC_mean_list)
]
withTTA_LL_time_std_max = withTTA_LL_time_std_list[np.argmax(withTTA_LL_AUC_mean_list)]
withTTA_LL_time_mean_TTA = np.argmax(withTTA_LL_AUC_mean_list)
print(
"i=%d," % withTTA_LL_time_mean_TTA,
"withTTA_LL_time_mean_max=%.3f" % withTTA_LL_time_mean_max,
"(+-%.3f)" % withTTA_LL_time_std_max,
)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(
x - 0.03 * xdist, y - 0.13 * ydist, "min time\n%.3f" % y, size=14, color="blue"
)
# PLOT: PREDICTION TIME - withTTA - full
plt.errorbar(
TTA_list,
withTTA_time_mean_list,
yerr=withTTA_time_std_list,
fmt="-o",
label="full",
color="green",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmin(withTTA_time_mean_list)
y = np.min(withTTA_time_mean_list)
withTTA_time_mean_max = withTTA_time_mean_list[np.argmax(withTTA_AUC_mean_list)]
withTTA_time_std_max = withTTA_time_std_list[np.argmax(withTTA_AUC_mean_list)]
withTTA_time_mean_TTA = np.argmax(withTTA_AUC_mean_list)
print(
"i=%d," % withTTA_time_mean_TTA,
"withTTA_time_mean_max=%.3f" % withTTA_time_mean_max,
"(+-%.3f)" % withTTA_time_std_max,
)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(
x - 0.03 * xdist, y + 0.1 * ydist, "min time\n%.3f" % y, size=14, color="green"
)
plt.ylabel("Prediction Time", size=14)
plt.xlabel("TTA steps", size=14)
plt.title(
"withTTA_LL, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
plt.legend(loc="best")
plt.savefig(
"TIME_both_TTA_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# ### Without TTA
TTA = 1
predict_woTTA_time_list = []
AUC_woTTA_list = []
oof_pred = []
oof_tar = []
for fold, (idxT, idxV) in enumerate(skf.split(np.arange(15))):
print("Fold %i. Loading the current fold model..." % fold)
model.load_weights(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-%i.h5" % fold
)
# model.load_weights(DEVICE + '_fold-%i.h5'%fold)
# PREDICT without TTA # augment=False
ds_valid = get_dataset(
files_valid,
labeled=False,
return_image_names=False,
augment=False,
repeat=True,
shuffle=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
batch_size=BATCH_SIZES[fold] * 4,
)
ct_valid = count_data_items(files_valid)
STEPS = TTA * ct_valid / BATCH_SIZES[fold] / 4 / REPLICAS
print("Predicting Test without TTA for %i files..." % (ct_valid))
# Start timer
ts_eval = time.time()
pred = model.predict(ds_valid, steps=STEPS, verbose=VERBOSE)[: TTA * ct_valid,]
# End timer
te_eval = time.time()
test_time = (te_eval - ts_eval) / ct_valid
predict_woTTA_time_list.append(test_time)
print("Fold %i, test_time=%.6f seconds." % (fold, test_time))
# Add predictions to list
oof_pred.append(np.mean(pred.reshape((ct_valid, TTA), order="F"), axis=1))
# GET OOF TARGETS AND NAMES
ds_valid = get_dataset(
files_valid,
augment=False,
repeat=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
labeled=True,
return_image_names=True,
)
# Add targets to list
oof_tar.append(
np.array([target.numpy() for img, target in iter(ds_valid.unbatch())])
)
# Calculate AUC
auc = roc_auc_score(oof_tar[-1], oof_pred[-1])
# Add AUC to list
AUC_woTTA_list.append(auc)
print("Fold %i, AUC=%.6f" % (fold, auc))
# Calculate AUC mean value to list
AUC_woTTA_mean = statistics.mean(AUC_woTTA_list) # mean
# Calculate AUC standard deviation value to list
AUC_woTTA_std = statistics.stdev(AUC_woTTA_list) # standard devition
print(
"#### OOF AUC without TTA: mean = %.6f, stdev = %.6f."
% (AUC_woTTA_mean, AUC_woTTA_std)
)
# Add AUC mean value to list
predict_woTTA_time_mean = statistics.mean(predict_woTTA_time_list) # mean
# Add AUC standard deviation value to list
predict_woTTA_time_std = statistics.stdev(predict_woTTA_time_list) # standard devition
print(
"#### Time without TTA: mean = %.6f, stdev = %.6f seconds."
% (predict_woTTA_time_mean, predict_woTTA_time_std)
)
# ### Measure model file size
import os
model_size = os.path.getsize(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-0.h5"
) # >> 20
print(str(model_size) + " Bytes")
# ### Save to file
results = pd.DataFrame(
data=[
[
MODEL,
model_size,
TRAINABLE,
AUGMENTATION,
AUG_LOSSLESS,
EPOCHS[0],
SIZE,
withTTA_LL_time_mean_max,
withTTA_LL_time_std_max,
withTTA_LL_AUC_mean_max,
withTTA_LL_AUC_std_max,
withTTA_time_mean_max,
withTTA_time_std_max,
withTTA_AUC_mean_max,
withTTA_AUC_std_max,
predict_woTTA_time_mean,
predict_woTTA_time_std,
AUC_woTTA_mean,
AUC_woTTA_std,
]
],
columns=[
"model",
"model_size",
"trainable",
"augmentation",
"aug_lossless",
"epochs",
"image_size",
"TTA_LL_time_mean",
"TTA_LL_time_std",
"TTA_LL_AUC_mean",
"TTA_LL_AUC_std",
"TTA_time_mean",
"TTA_time_std",
"TTA_AUC_mean",
"TTA_AUC_std",
"woTTA_time_mean",
"woTTA_time_std",
"woTTA_AUC_mean",
"woTTA_AUC_std",
],
)
experiment_title = (
DEVICE
+ "_"
+ str(MODEL)
+ "_s"
+ str(SIZE)
+ "_ep"
+ str(EPOCHS[0])
+ "_train"
+ str(TRAINABLE)
+ "_aug"
+ str(AUGMENTATION)
+ "_loss"
+ str(AUG_LOSSLESS)
+ "_time_AUC"
)
results.to_csv(experiment_title + ".csv", index=False)
results.head()
from zipfile import ZipFile
from os.path import basename
# Zip the files from given directory that matches the filter
def zipFilesInDir(dirName, zipFileName, filter):
# create a ZipFile object
with ZipFile(zipFileName, "w") as zipObj:
# Iterate over all the files in directory
for folderName, subfolders, filenames in os.walk(dirName):
for filename in filenames:
if filter(filename):
# create complete filepath of file in directory
filePath = os.path.join(folderName, filename)
# Add file to zip
zipObj.write(filePath, basename(filePath))
print("*** Create a zip archive of *.png *.ipynb *.csv files form a directory ***")
zipFilesInDir(".", experiment_title + "_csv_.zip", lambda name: "csv" in name)
zipFilesInDir(".", experiment_title + "_ipynb_.zip", lambda name: "ipynb" in name)
zipFilesInDir(".", experiment_title + "_png_.zip", lambda name: "png" in name)
zipFilesInDir(".", experiment_title + "_all.zip", lambda name: "_.zip" in name)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/141/69141484.ipynb
|
lyme-disease-rashes-in-tfrecords
|
yoctoman
|
[{"Id": 69141484, "ScriptId": 18783452, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2209763, "CreationDate": "07/27/2021 10:31:22", "VersionNumber": 2.0, "Title": "Dirty Lyme Dataset - EfficientNetB0-Keras - TPU", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 1135.0, "LinesInsertedFromPrevious": 5.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1130.0, "LinesInsertedFromFork": 5.0, "LinesDeletedFromFork": 5.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 1130.0, "TotalVotes": 0}]
|
[{"Id": 91963730, "KernelVersionId": 69141484, "SourceDatasetVersionId": 2339169}]
|
[{"Id": 2339169, "DatasetId": 1411076, "DatasourceVersionId": 2380799, "CreatorUserId": 88001, "LicenseName": "Unknown", "CreationDate": "06/16/2021 11:14:16", "VersionNumber": 5.0, "Title": "Lyme Disease Rashes in TFRecords", "Slug": "lyme-disease-rashes-in-tfrecords", "Subtitle": NaN, "Description": NaN, "VersionNotes": "15 TFRecords for TRAIN/TEST", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1411076, "CreatorUserId": 88001, "OwnerUserId": 88001.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2339169.0, "CurrentDatasourceVersionId": 2380799.0, "ForumId": 1430423, "Type": 2, "CreationDate": "06/15/2021 21:20:14", "LastActivityDate": "06/15/2021", "TotalViews": 1601, "TotalDownloads": 22, "TotalVotes": 1, "TotalKernels": 2}]
|
[{"Id": 88001, "UserName": "yoctoman", "DisplayName": "yoctoman", "RegisterDate": "03/02/2013", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
###for dirname, _, filenames in os.walk('/kaggle/input'):
### for filename in filenames:
### print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Initialize Environment
# !pip install -q efficientnet >> /dev/null
import pandas as pd, numpy as np
from kaggle_datasets import KaggleDatasets
import tensorflow as tf, re, math
import tensorflow.keras.backend as K
# import efficientnet.tfkeras as efn
from sklearn.model_selection import KFold
from sklearn.metrics import roc_auc_score
import matplotlib.pyplot as plt
import cv2
# ## Configuration
# In order to be a proper cross validation with a meaningful overall CV score (aligned with LB score), **you need to choose the same** `IMG_SIZES`, `INC2019`, `INC2018`, and `EFF_NETS` **for each fold**. If your goal is to just run lots of experiments, then you can choose to have a different experiment in each fold. Then each fold is like a holdout validation experiment. When you find a configuration you like, you can use that configuration for all folds.
# * DEVICE - is GPU or TPU
# * SEED - a different seed produces a different triple stratified kfold split.
# * FOLDS - number of folds. Best set to 3, 5, or 15 but can be any number between 2 and 15
# * IMG_SIZES - is a Python list of length FOLDS. These are the image sizes to use each fold
# * INC2019 - This includes the new half of the 2019 competition data. The second half of the 2019 data is the comp data from 2018 plus 2017
# * INC2018 - This includes the second half of the 2019 competition data which is the comp data from 2018 plus 2017
# * BATCH_SIZES - is a list of length FOLDS. These are batch sizes for each fold. For maximum speed, it is best to use the largest batch size your GPU or TPU allows.
# * EPOCHS - is a list of length FOLDS. These are maximum epochs. Note that each fold, the best epoch model is saved and used. So if epochs is too large, it won't matter.
# * EFF_NETS - is a list of length FOLDS. These are the EfficientNets to use each fold. The number refers to the B. So a number of `0` refers to EfficientNetB0, and `1` refers to EfficientNetB1, etc.
# * WGTS - this should be `1/FOLDS` for each fold. This is the weight when ensembling the folds to predict the test set. If you want a weird ensemble, you can use different weights.
# * TTA - test time augmentation. Each test image is randomly augmented and predicted TTA times and the average prediction is used. TTA is also applied to OOF during validation.
DEVICE = "TPU" # or "GPU" #
# USE DIFFERENT SEED FOR DIFFERENT STRATIFIED KFOLD
SEED = 42
# NUMBER OF FOLDS. USE 3, 5, OR 15
FOLDS = 3
# WHICH IMAGE SIZES TO LOAD EACH FOLD
# CHOOSE 128, 192, 256, 384, 512, 768, 1024, 1536
# IMG_SIZES = [384,384,384,384,384]
# IMG_SIZES = [128,128,128,128,128]
SIZE = 128
IMG_SIZES = [SIZE] * FOLDS
# INCLUDE OLD COMP DATA? YES=1 NO=0
# TOKEN = 0
# INC2019 = [TOKEN,TOKEN,TOKEN,TOKEN,TOKEN]
# INC2018 = [TOKEN,TOKEN,TOKEN,TOKEN,TOKEN]
# BATCH SIZE AND EPOCHS
# Original batch size
# BATCH_SIZES = [32]*FOLDS
# YG
# Experimental batch sizes for EfficientNetB7
# BAD
# BATCH_SIZES = [4]*FOLDS # for 768 - VERY LONG - stops after 2 folds only, ~4 hours per fold
# BATCH_SIZES = [8]*FOLDS # for 768 - VERY LONG - 3 hours per fold
# BATCH_SIZES = [12]*FOLDS # for 768 - BAD, not enough resources
# GOOD
# BATCH_SIZES = [32]*FOLDS # for 384 - GOOD - ~1 hour per fold
# BATCH_SIZES = [16]*FOLDS # for 512
# GPU: Experimental batch sizes for EfficientNetB0
# BATCH_SIZES = [2]*FOLDS # SIZE = 1024
# Experimental batch sizes for EfficientNetB1
BATCH = 4
BATCH_SIZES = [BATCH] * FOLDS # SIZE = 768
# YG
# NASNetLarge
# BAD
# BATCH_SIZES = [4]*FOLDS # for 768 - VERY LONG - ~3.5-4 hours per fold
# BATCH_SIZES = [8]*FOLDS # for 768 - BAD - not enough resources
# GOOD
# BATCH_SIZES = [16]*FOLDS # for 384 - GOOD - ~1 hour per fold
# BATCH_SIZES = [8]*FOLDS # for 512
# EPOCHS = [12]*FOLDS
EPOCH = 32
EPOCHS = [EPOCH] * FOLDS
# EfficientNet
# WHICH EFFICIENTNET B? TO USE
model_name = "MobileNetV2"
# MODEL = 7 # EfficientNetB7
MODEL = "MobileNetV2" # EfficientNetB3
# MODEL = 0 # EfficientNetB0
EFF_NETS = [MODEL] * FOLDS
SPECIFIC_SIZE = SIZE
# MobileNetV2
# MODEL = 'MobileNetV2'
# SPECIFIC_SIZE = SIZE
# model_name = 'NASNetMobile'
# MODEL = 'NASNetMobile'
# SPECIFIC_SIZE = SIZE # 224 only at the moment!
# model_name = 'NASNetLarge'
# MODEL = 'NASNetLarge'
# SPECIFIC_SIZE = SIZE # 331 only at the moment!
# WEIGHTS FOR FOLD MODELS WHEN PREDICTING TEST
WGTS = [1 / FOLDS] * FOLDS
# TEST TIME AUGMENTATION STEPS
TTA = 11
# make the weights and biases of the base model non-trainable
# by "freezing" each layer of the BASE network
TRAINABLE = False
AUGMENTATION = True
AUG_LOSSLESS = False
# Time estimation - 1 epoch
# Model = EfficientNetB0
# MODEL = 0
########################################################
# Image Size
# SIZE = 1536
# BATCH = 1
# Fold Times:
# FOLDS = 3
# 1it [03:29, 209.24s/it]
# 2it [06:58, 209.25s/it]
# 3it [10:25, 208.61s/it]
# CPU times: user 2min 11s, sys: 13.6 s, total: 2min 25s
# Wall time: 10min 25s
# + 5 min for testing
########################################################
# Image Size
# SIZE = 1024
# BATCH = 8
# CPU times: user 2min 18s, sys: 13.8 s, total: 2min 32s
# Wall time: 6min 57s
########################################################
########################################################
# Image Size
# SIZE = 768
# BATCH = 16
# CPU times: user 2min 15s, sys: 12.9 s, total: 2min 28s
# Wall time: 6min 20s
# Time estimation - 1 epoch
# Model = EfficientNetB7
# MODEL = 7
######################################################## MIN
# Image Size
# SIZE = 128
# BATCH = 32
# CPU times: user 7min 4s, sys: 46.5 s, total: 7min 50s
# Wall time: 13min 25s
######################################################## MAX
# Image Size
# SIZE = 1024
# BATCH = 1
# CPU times: user 7min 19s, sys: 47.8 s, total: 8min 6s
# Wall time: 18min 27s
########################################################
# Image Size
# SIZE = 768
# BATCH = 4
# CPU times: user 7min 27s, sys: 49.2 s, total: 8min 16s
# Wall time: 17min 40s
########################################################
# Image Size
# SIZE = 512
# BATCH = 16
# CPU times: user 7min 11s, sys: 47.5 s, total: 7min 58s
# Wall time: 15min 50s
########################################################
# Image Size
# SIZE = 256
# BATCH = 16
# CPU times: user 4min 43s, sys: 40.4 s, total: 5min 24s
# Wall time: 7min 9s
########################################################
# TRAINABLE = False
########################################################
# Image Size
# SIZE = 256
# BATCH = 16
# CPU times: user 4min 51s, sys: 41.1 s, total: 5min 32s
# Wall time: 7min 12s
########################################################
# TRAINABLE = False
# AUGMENTATION = False
########################################################
# Image Size
# SIZE = 256
# BATCH = 16
# CPU times: user 4min 35s, sys: 38.1 s, total: 5min 13s
# Wall time: 6min 56s
if DEVICE == "TPU":
print("connecting to TPU...")
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
print("Running on TPU ", tpu.master())
except ValueError:
print("Could not connect to TPU")
tpu = None
if tpu:
try:
print("initializing TPU ...")
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
print("TPU initialized")
except _:
print("failed to initialize TPU")
else:
DEVICE = "GPU"
if DEVICE != "TPU":
print("Using default strategy for CPU and single GPU")
strategy = tf.distribute.get_strategy()
if DEVICE == "GPU":
print(
"Num GPUs Available: ", len(tf.config.experimental.list_physical_devices("GPU"))
)
AUTO = tf.data.experimental.AUTOTUNE
REPLICAS = strategy.num_replicas_in_sync
print(f"REPLICAS: {REPLICAS}")
# # Step 1: Preprocess
# Preprocess has already been done and saved to TFRecords. Here we choose which size to load. We can use either 128x128, 192x192, 256x256, 384x384, 512x512, 768x768 by changing the `IMG_SIZES` variable in the preceeding code section. These TFRecords are discussed [here][1]. The advantage of using different input sizes is discussed [here][2]
# [1]: https://www.kaggle.com/c/siim-isic-melanoma-classification/discussion/155579
# [2]: https://www.kaggle.com/c/siim-isic-melanoma-classification/discussion/160147
print(KaggleDatasets().get_gcs_path("lyme-disease-rashes-in-tfrecords"))
GCS_PATH = [None] * FOLDS
# GCS_PATH2 = [None]*FOLDS
for i, k in enumerate(IMG_SIZES):
# GCS_PATH[i] = KaggleDatasets().get_gcs_path('melanoma-%ix%i'%(k,k))
# GCS_PATH2[i] = KaggleDatasets().get_gcs_path('isic2019-%ix%i'%(k,k))
GCS_PATH[i] = KaggleDatasets().get_gcs_path("lyme-disease-rashes-in-tfrecords")
files_train = np.sort(
np.array(tf.io.gfile.glob(GCS_PATH[0] + "/train_%d_*.tfrec" % SIZE))
)
files_test = np.sort(
np.array(tf.io.gfile.glob(GCS_PATH[0] + "/test_%d_*.tfrec" % SIZE))
)
files_train
files_test
# # Step 2: Data Augmentation
# This notebook uses rotation, sheer, zoom, shift augmentation first shown in this notebook [here][1] and successfully used in Melanoma comp by AgentAuers [here][2]. This notebook also uses horizontal flip, hue, saturation, contrast, brightness augmentation similar to last years winner and also similar to AgentAuers' notebook.
# Additionally we can decide to use external data by changing the variables `INC2019` and `INC2018` in the preceeding code section. These variables respectively indicate whether to load last year 2019 data and/or year 2018 + 2017 data. These datasets are discussed [here][3]
# Consider experimenting with different augmenation and/or external data. The code to load TFRecords is taken from AgentAuers' notebook [here][2]. Thank you AgentAuers, this is great work.
# [1]: https://www.kaggle.com/cdeotte/rotation-augmentation-gpu-tpu-0-96
# [2]: https://www.kaggle.com/agentauers/incredible-tpus-finetune-effnetb0-b6-at-once
# [3]: https://www.kaggle.com/c/siim-isic-melanoma-classification/discussion/164910
ROT_ = 180.0
SHR_ = 2.0
HZOOM_ = 8.0
WZOOM_ = 8.0
HSHIFT_ = 8.0
WSHIFT_ = 8.0
def get_mat(rotation, shear, height_zoom, width_zoom, height_shift, width_shift):
# returns 3x3 transformmatrix which transforms indicies
# CONVERT DEGREES TO RADIANS
rotation = math.pi * rotation / 180.0
shear = math.pi * shear / 180.0
def get_3x3_mat(lst):
return tf.reshape(tf.concat([lst], axis=0), [3, 3])
# ROTATION MATRIX
c1 = tf.math.cos(rotation)
s1 = tf.math.sin(rotation)
one = tf.constant([1], dtype="float32")
zero = tf.constant([0], dtype="float32")
rotation_matrix = get_3x3_mat([c1, s1, zero, -s1, c1, zero, zero, zero, one])
# SHEAR MATRIX
c2 = tf.math.cos(shear)
s2 = tf.math.sin(shear)
shear_matrix = get_3x3_mat([one, s2, zero, zero, c2, zero, zero, zero, one])
# ZOOM MATRIX
zoom_matrix = get_3x3_mat(
[one / height_zoom, zero, zero, zero, one / width_zoom, zero, zero, zero, one]
)
# SHIFT MATRIX
shift_matrix = get_3x3_mat(
[one, zero, height_shift, zero, one, width_shift, zero, zero, one]
)
return K.dot(K.dot(rotation_matrix, shear_matrix), K.dot(zoom_matrix, shift_matrix))
def transform(image, DIM=256):
# input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]
# output - image randomly rotated, sheared, zoomed, and shifted
XDIM = DIM % 2 # fix for size 331
rot = ROT_ * tf.random.normal([1], dtype="float32")
shr = SHR_ * tf.random.normal([1], dtype="float32")
h_zoom = 1.0 + tf.random.normal([1], dtype="float32") / HZOOM_
w_zoom = 1.0 + tf.random.normal([1], dtype="float32") / WZOOM_
h_shift = HSHIFT_ * tf.random.normal([1], dtype="float32")
w_shift = WSHIFT_ * tf.random.normal([1], dtype="float32")
# GET TRANSFORMATION MATRIX
m = get_mat(rot, shr, h_zoom, w_zoom, h_shift, w_shift)
# LIST DESTINATION PIXEL INDICES
x = tf.repeat(tf.range(DIM // 2, -DIM // 2, -1), DIM)
y = tf.tile(tf.range(-DIM // 2, DIM // 2), [DIM])
z = tf.ones([DIM * DIM], dtype="int32")
idx = tf.stack([x, y, z])
# ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS
idx2 = K.dot(m, tf.cast(idx, dtype="float32"))
idx2 = K.cast(idx2, dtype="int32")
idx2 = K.clip(idx2, -DIM // 2 + XDIM + 1, DIM // 2)
# FIND ORIGIN PIXEL VALUES
idx3 = tf.stack([DIM // 2 - idx2[0,], DIM // 2 - 1 + idx2[1,]])
d = tf.gather_nd(image, tf.transpose(idx3))
return tf.reshape(d, [DIM, DIM, 3])
def read_labeled_tfrecord(example):
tfrec_format = {
"image": tf.io.FixedLenFeature([], tf.string),
"image_name": tf.io.FixedLenFeature([], tf.string),
#'patient_id' : tf.io.FixedLenFeature([], tf.int64),
#'sex' : tf.io.FixedLenFeature([], tf.int64),
#'age_approx' : tf.io.FixedLenFeature([], tf.int64),
#'anatom_site_general_challenge': tf.io.FixedLenFeature([], tf.int64),
#'diagnosis' : tf.io.FixedLenFeature([], tf.int64),
"target": tf.io.FixedLenFeature([], tf.int64),
}
example = tf.io.parse_single_example(example, tfrec_format)
return example["image"], example["target"]
def read_unlabeled_tfrecord(example, return_image_name):
tfrec_format = {
"image": tf.io.FixedLenFeature([], tf.string),
"image_name": tf.io.FixedLenFeature([], tf.string),
}
example = tf.io.parse_single_example(example, tfrec_format)
return example["image"], example["image_name"] if return_image_name else 0
def prepare_image(img, augment=True, aug_lossless=True, dim=256):
img = tf.image.decode_jpeg(img, channels=3)
img = tf.cast(img, tf.float32) / 255.0
if augment:
if aug_lossless:
img = transform(img, DIM=dim)
img = tf.image.random_flip_left_right(img)
img = tf.image.random_flip_up_down(img)
img = tf.image.rot90(
img, tf.random.uniform(shape=[], minval=0, maxval=3, dtype=tf.int32)
)
else:
img = transform(img, DIM=dim)
img = tf.image.random_flip_left_right(img)
img = tf.image.random_flip_up_down(img)
img = tf.image.rot90(
img, tf.random.uniform(shape=[], minval=0, maxval=3, dtype=tf.int32)
)
img = tf.image.random_hue(img, 0.01)
img = tf.image.random_saturation(img, 0.7, 1.3)
img = tf.image.random_contrast(img, 0.8, 1.2)
img = tf.image.random_brightness(img, 0.1)
img = tf.image.random_jpeg_quality(img, 75, 95)
# YG - just for adaptation to NASNet ... and other families
# else:
# if(MODEL == 'NASNetMobile' or MODEL == 'NASNetLarge'):
# img = transform(img,DIM=dim)
# img = cv2.resize(img, (dim, dim),interpolation = cv2.INTER_CUBIC)
img = tf.reshape(img, [dim, dim, 3])
# img = tf.image.resize(img, [224, 224, 3])
return img
def count_data_items(filenames):
n = [
int(re.compile(r"-([0-9]*)\.").search(filename).group(1))
for filename in filenames
]
return np.sum(n)
def get_dataset(
files,
augment=False,
aug_lossless=True,
shuffle=False,
repeat=False,
labeled=True,
return_image_names=True,
batch_size=16,
dim=256,
):
ds = tf.data.TFRecordDataset(files, num_parallel_reads=AUTO)
ds = ds.cache()
if repeat:
ds = ds.repeat()
if shuffle:
ds = ds.shuffle(1024 * 8)
opt = tf.data.Options()
opt.experimental_deterministic = False
ds = ds.with_options(opt)
if labeled:
ds = ds.map(read_labeled_tfrecord, num_parallel_calls=AUTO)
else:
ds = ds.map(
lambda example: read_unlabeled_tfrecord(example, return_image_names),
num_parallel_calls=AUTO,
)
ds = ds.map(
lambda img, imgname_or_label: (
prepare_image(img, augment=augment, aug_lossless=aug_lossless, dim=dim),
imgname_or_label,
),
num_parallel_calls=AUTO,
)
ds = ds.batch(batch_size * REPLICAS)
ds = ds.prefetch(AUTO)
return ds
# # Step 3: Build Model
# This is a common model architecute. Consider experimenting with different backbones, custom heads, losses, and optimizers. Also consider inputing meta features into your CNN.
# YG
# These models HAVE PASSED the initial test!
from tensorflow.keras.applications import MobileNetV2
from tensorflow.keras.applications import NASNetMobile
from tensorflow.keras.applications import NASNetLarge
from tensorflow.keras.applications import DenseNet121
from tensorflow.keras.applications import EfficientNetB0
dim = SPECIFIC_SIZE
# if(model_name =='EfficientNet'):
# These should be checked:
# from tensorflow.keras.applications import InceptionV3
# model_name = 'InceptionV3'
# from tensorflow.keras.applications import InceptionResNetV2
# model_name = 'InceptionResNetV2'
# from tensorflow.keras.applications import MobileNetV2
# model_name = 'MobileNetV2'
# from tensorflow.keras.applications import ResNet101
# model_name = 'ResNet101'
# from tensorflow.keras.applications import ResNet101V2
# model_name = 'ResNet101V2'
# from tensorflow.keras.applications import VGG16
# model_name = 'VGG16'
# from tensorflow.keras.applications import Xception
# model_name = 'Xception'
def build_model(model_name=model_name, dim=SPECIFIC_SIZE, trainable=False):
inp = tf.keras.layers.Input(shape=(dim, dim, 3))
if model_name == "EfficientNet":
# EFNS = [efn.EfficientNetB0, efn.EfficientNetB1, efn.EfficientNetB2, efn.EfficientNetB3,
# efn.EfficientNetB4, efn.EfficientNetB5, efn.EfficientNetB6, efn.EfficientNetB7]
# base = EFNS[MODEL](input_shape=(dim,dim,3),weights='imagenet',include_top=False)
if MODEL == 0:
base = EfficientNetB0(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
model_name = "EfficientNetB" + str(MODEL)
else:
print(
"ERROR: This code is prepared for EfficientNetB, version"
+ str(MODEL)
+ ", BUT other version is used!"
)
if model_name == "MobileNetV2":
base = MobileNetV2(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
if model_name == "NASNetMobile":
base = NASNetMobile(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
if model_name == "NASNetLarge":
base = NASNetLarge(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
if model_name == "DenseNet121":
base = DenseNet121(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
# make the weights and biases of the base model non-trainable
# by "freezing" each layer of the BASE network
for layer in base.layers:
layer.trainable = trainable
x = base(inp)
x = tf.keras.layers.GlobalAveragePooling2D()(x)
x = tf.keras.layers.Dense(1, activation="sigmoid")(x)
model = tf.keras.Model(inputs=inp, outputs=x)
opt = tf.keras.optimizers.Adam(learning_rate=0.001)
loss = tf.keras.losses.BinaryCrossentropy(label_smoothing=0.05)
model.compile(optimizer=opt, loss=loss, metrics=["AUC"])
return model
# # Step 4: Train Schedule
# This is a common train schedule for transfer learning. The learning rate starts near zero, then increases to a maximum, then decays over time. Consider changing the schedule and/or learning rates. Note how the learning rate max is larger with larger batches sizes. This is a good practice to follow.
def get_lr_callback(batch_size=8):
lr_start = 0.000005
lr_max = 0.00000125 * REPLICAS * batch_size
lr_min = 0.000001
lr_ramp_ep = 5
lr_sus_ep = 0
lr_decay = 0.8
def lrfn(epoch):
if epoch < lr_ramp_ep:
lr = (lr_max - lr_start) / lr_ramp_ep * epoch + lr_start
elif epoch < lr_ramp_ep + lr_sus_ep:
lr = lr_max
else:
lr = (lr_max - lr_min) * lr_decay ** (
epoch - lr_ramp_ep - lr_sus_ep
) + lr_min
return lr
lr_callback = tf.keras.callbacks.LearningRateScheduler(lrfn, verbose=False)
return lr_callback
# ## Train Model
# Our model will be trained for the number of FOLDS and EPOCHS you chose in the configuration above. Each fold the model with lowest validation loss will be saved and used to predict OOF and test. Adjust the variables `VERBOSE` and `DISPLOY_PLOT` below to determine what output you want displayed. The variable `VERBOSE=1 or 2` will display the training and validation loss and auc for each epoch as text. The variable `DISPLAY_PLOT` shows this information as a plot.
from tqdm import tqdm
# USE VERBOSE=0 for silent, VERBOSE=1 for interactive, VERBOSE=2 for commit
VERBOSE = 1
DISPLAY_PLOT = True
skf = KFold(n_splits=FOLDS, shuffle=True, random_state=SEED)
oof_pred = []
oof_tar = []
oof_val = []
oof_names = []
oof_folds = []
preds = np.zeros((count_data_items(files_test), 1))
for fold, (idxT, idxV) in tqdm(enumerate(skf.split(np.arange(14)))):
# DISPLAY FOLD INFO
if DEVICE == "TPU":
if tpu:
tf.tpu.experimental.initialize_tpu_system(tpu)
print("#" * 25)
print("#### FOLD", fold + 1)
# YG
# print('#### Image Size %i with EfficientNet B%i and batch_size %i'%
# (IMG_SIZES[fold],EFF_NETS[fold],BATCH_SIZES[fold]*REPLICAS))
print(
"#### Image Size %i with %s and batch_size %i"
% (IMG_SIZES[fold], model_name, BATCH_SIZES[fold] * REPLICAS)
)
# CREATE TRAIN AND VALIDATION SUBSETS
# files_train = tf.io.gfile.glob([GCS_PATH[fold] + '/train%.2i*.tfrec'%x for x in idxT])
print("SIZE:", SIZE)
filename_core = GCS_PATH[fold] + "/train_%i_" % (SIZE)
files_train = tf.io.gfile.glob([filename_core + "%.2i*.tfrec" % x for x in idxT])
# print([GCS_PATH[fold]])
# print([GCS_PATH[fold] + '/train_128_%.2i*.tfrec'%x for x in idxT])
print("Files for TRAIN:")
print(files_train)
# if INC2019[fold]:
# files_train += tf.io.gfile.glob([GCS_PATH2[fold] + '/train%.2i*.tfrec'%x for x in idxT*2+1])
# print('#### Using 2019 external data')
# if INC2018[fold]:
# files_train += tf.io.gfile.glob([GCS_PATH2[fold] + '/train%.2i*.tfrec'%x for x in idxT*2])
# print('#### Using 2018+2017 external data')
np.random.shuffle(files_train)
print("#" * 25)
filename_core = GCS_PATH[fold] + "/train_%i_" % (SIZE)
files_valid = tf.io.gfile.glob([filename_core + "%.2i*.tfrec" % x for x in idxV])
print("Files for VALIDATION:")
print(files_valid)
files_test = np.sort(
np.array(tf.io.gfile.glob(GCS_PATH[fold] + "/test_%i_*.tfrec" % (SIZE)))
)
print("Files for TEST:")
print(files_test)
# BUILD MODEL
K.clear_session()
with strategy.scope():
model = build_model(
model_name=model_name, dim=SPECIFIC_SIZE, trainable=TRAINABLE
)
model.summary() # YG
# SAVE BEST MODEL EACH FOLD
sv = tf.keras.callbacks.ModelCheckpoint(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-%i.h5" % fold,
monitor="val_loss",
verbose=0,
save_best_only=True,
save_weights_only=True,
mode="min",
save_freq="epoch",
)
# TRAIN
print("Training...")
history = model.fit(
get_dataset(
files_train,
augment=True,
aug_lossless=AUG_LOSSLESS,
shuffle=True,
repeat=True,
# dim=IMG_SIZES[fold],
dim=SPECIFIC_SIZE,
batch_size=BATCH_SIZES[fold],
),
epochs=EPOCHS[fold],
callbacks=[sv, get_lr_callback(BATCH_SIZES[fold])],
steps_per_epoch=count_data_items(files_train) / BATCH_SIZES[fold] // REPLICAS,
validation_data=get_dataset(
files_valid,
augment=False,
shuffle=False,
repeat=False,
# dim=IMG_SIZES[fold]
dim=SPECIFIC_SIZE,
), # class_weight = {0:1,1:2},
verbose=VERBOSE,
)
print("Loading best model...")
model.load_weights(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-%i.h5" % fold
)
# PREDICT OOF USING TTA
print("Predicting OOF with TTA...")
ds_valid = get_dataset(
files_valid,
labeled=False,
return_image_names=False,
augment=AUGMENTATION,
aug_lossless=AUG_LOSSLESS,
repeat=True,
shuffle=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
batch_size=BATCH_SIZES[fold] * 4,
)
ct_valid = count_data_items(files_valid)
STEPS = TTA * ct_valid / BATCH_SIZES[fold] / 4 / REPLICAS
pred = model.predict(ds_valid, steps=STEPS, verbose=VERBOSE)[: TTA * ct_valid,]
oof_pred.append(np.mean(pred.reshape((ct_valid, TTA), order="F"), axis=1))
# oof_pred.append(model.predict(get_dataset(files_valid,dim=IMG_SIZES[fold]),verbose=1))
# GET OOF TARGETS AND NAMES
ds_valid = get_dataset(
files_valid,
augment=False,
aug_lossless=False,
repeat=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
labeled=True,
return_image_names=True,
)
oof_tar.append(
np.array([target.numpy() for img, target in iter(ds_valid.unbatch())])
)
oof_folds.append(np.ones_like(oof_tar[-1], dtype="int8") * fold)
ds = get_dataset(
files_valid,
augment=False,
aug_lossless=False,
repeat=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
labeled=False,
return_image_names=True,
)
oof_names.append(
np.array(
[img_name.numpy().decode("utf-8") for img, img_name in iter(ds.unbatch())]
)
)
# PREDICT TEST USING TTA
print("Predicting Test with TTA...")
ds_test = get_dataset(
files_test,
labeled=False,
return_image_names=False,
augment=True,
aug_lossless=AUG_LOSSLESS,
repeat=True,
shuffle=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
batch_size=BATCH_SIZES[fold] * 4,
)
ct_test = count_data_items(files_test)
STEPS = TTA * ct_test / BATCH_SIZES[fold] / 4 / REPLICAS
pred = model.predict(ds_test, steps=STEPS, verbose=VERBOSE)[: TTA * ct_test,]
preds[:, 0] += np.mean(pred.reshape((ct_test, TTA), order="F"), axis=1) * WGTS[fold]
# REPORT RESULTS
### BUG - YG
list_length = min(len(oof_tar[-1]), len(oof_pred[-1]))
auc = roc_auc_score(oof_tar[-1][:list_length], oof_pred[-1][:list_length])
oof_val.append(np.max(history.history["val_auc"]))
print(
"#### FOLD %i OOF AUC without TTA = %.3f, with TTA = %.3f"
% (fold + 1, oof_val[-1], auc)
)
# PLOT TRAINING
if DISPLAY_PLOT:
plt.figure(figsize=(15, 5))
plt.plot(
np.arange(EPOCHS[fold]),
history.history["auc"],
"-o",
label="Train AUC",
color="#ff7f0e",
)
plt.plot(
np.arange(EPOCHS[fold]),
history.history["val_auc"],
"-o",
label="Val AUC",
color="#1f77b4",
)
x = np.argmax(history.history["val_auc"])
y = np.max(history.history["val_auc"])
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="#1f77b4")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.2f" % y, size=14)
plt.ylabel("AUC", size=14)
plt.xlabel("Epoch", size=14)
plt.legend(loc=2)
plt2 = plt.gca().twinx()
plt2.plot(
np.arange(EPOCHS[fold]),
history.history["loss"],
"-o",
label="Train Loss",
color="#2ca02c",
)
plt2.plot(
np.arange(EPOCHS[fold]),
history.history["val_loss"],
"-o",
label="Val Loss",
color="#d62728",
)
x = np.argmin(history.history["val_loss"])
y = np.min(history.history["val_loss"])
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="#d62728")
plt.text(x - 0.03 * xdist, y + 0.05 * ydist, "min loss", size=14)
plt.ylabel("Loss", size=14)
# YG
# plt.title('FOLD %i - Image Size %i, EfficientNet B%i, inc2019=%i, inc2018=%i'%
# (fold+1,IMG_SIZES[fold],EFF_NETS[fold],INC2019[fold],INC2018[fold]),size=18)
# plt.title('FOLD %i - Image Size %i, %s, inc2019=%i, inc2018=%i'%
# (fold+1,IMG_SIZES[fold],model_name,INC2019[fold],INC2018[fold]),size=18)
plt.title(
"FOLD %i - Image Size %i, %s, Device=%s, TTA=%i"
% (fold + 1, IMG_SIZES[fold], model_name, DEVICE, TTA),
size=18,
)
plt.legend(loc=3)
plt.savefig(
"AUC_"
+ DEVICE
+ "_model"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_fold"
+ str(fold)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# ## Calculate OOF AUC
# The OOF (out of fold) predictions are saved to disk. If you wish to ensemble multiple models, use the OOF to determine what are the best weights to blend your models with. Choose weights that maximize OOF CV score when used to blend OOF. Then use those same weights to blend your test predictions.
# COMPUTE OVERALL OOF AUC
oof = np.concatenate(oof_pred)
true = np.concatenate(oof_tar)
names = np.concatenate(oof_names)
folds = np.concatenate(oof_folds)
### BUG - YG
list_length = min(len(true), len(oof))
auc = roc_auc_score(true[:list_length], oof[:list_length])
print("Overall OOF AUC with TTA = %.3f" % auc)
# SAVE OOF TO DISK
df_oof = pd.DataFrame(
dict(
image_name=names, target=true[:list_length], pred=oof[:list_length], fold=folds
)
)
df_oof.to_csv(DEVICE + "_" + str(MODEL) + "_oof.csv", index=False)
df_oof.tail()
# # Step 5: Post process
# ## Measure prediction times, AUC, model file size
# ### With TTA
import time
import statistics
VERBOSE = 1
def AUC_time(print_verbose=False, augment=True, aug_lossless=True, TTA=1):
predict_time_list = []
AUC_list = []
oof_pred = []
oof_tar = []
for fold, (idxT, idxV) in enumerate(skf.split(np.arange(15))):
if print_verbose:
print("Fold %i. Loading the current fold model..." % fold)
model.load_weights(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-%i.h5" % fold
)
# model.load_weights(DEVICE + '_fold-%i.h5'%fold)
# PREDICT with TTA # augment=True, aug_lossless = True,
ds_valid = get_dataset(
files_valid,
labeled=False,
return_image_names=False,
augment=True,
aug_lossless=True,
repeat=True,
shuffle=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
batch_size=BATCH_SIZES[fold] * 4,
)
ct_valid = count_data_items(files_valid)
STEPS = TTA * ct_valid / BATCH_SIZES[fold] / 4 / REPLICAS
if print_verbose:
print("Predicting Test with TTA_LL for %i files..." % (ct_valid))
# Start timer
ts_eval = time.time()
pred = model.predict(ds_valid, steps=STEPS, verbose=VERBOSE)[: TTA * ct_valid,]
# End timer
te_eval = time.time()
test_time = (te_eval - ts_eval) / ct_valid
predict_time_list.append(test_time)
if print_verbose:
print("Fold %i, test_time=%.6f seconds." % (fold, test_time))
# Add predictions to list
oof_pred.append(np.mean(pred.reshape((ct_valid, TTA), order="F"), axis=1))
# GET OOF TARGETS AND NAMES
ds_valid = get_dataset(
files_valid,
augment=False,
aug_lossless=True,
repeat=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
labeled=True,
return_image_names=True,
)
# Add targets to list
oof_tar.append(
np.array([target.numpy() for img, target in iter(ds_valid.unbatch())])
)
# Calculate AUC
auc = roc_auc_score(oof_tar[-1], oof_pred[-1])
# Add AUC to list
AUC_list.append(auc)
if print_verbose:
print("Fold %i, AUC=%.6f" % (fold, auc))
# Calculate AUC mean value to list
AUC_mean = statistics.mean(AUC_list) # mean
# Calculate AUC standard deviation value to list
AUC_std = statistics.stdev(AUC_list) # standard devition
print("#### TTA = %d" % TTA)
print("#### OOF AUC with TTA_LL: mean = %.6f, stdev = %.6f." % (AUC_mean, AUC_std))
# Add AUC mean value to list
predict_time_mean = statistics.mean(predict_time_list) # mean
# Add AUC standard deviation value to list
predict_time_std = statistics.stdev(predict_time_list) # standard devition
print(
"#### Time without TTA_LL: mean = %.6f, stdev = %.6f seconds."
% (predict_time_mean, predict_time_std)
)
return AUC_mean, AUC_std, predict_time_mean, predict_time_std
# #### LOSSLESS TTA - withTTA_LL
# _withTTA_LL_
VERBOSE = 0
TTA_list = [1, 2, 4, 8, 16, 32, 64, 128]
withTTA_LL_AUC_mean_list = []
withTTA_LL_AUC_std_list = []
withTTA_LL_time_mean_list = []
withTTA_LL_time_std_list = []
for i in tqdm(TTA_list):
AUC_mean, AUC_std, predict_time_mean, predict_time_std = AUC_time(
print_verbose=False, augment=True, aug_lossless=True, TTA=i
)
withTTA_LL_AUC_mean_list.append(AUC_mean)
withTTA_LL_AUC_std_list.append(AUC_std)
withTTA_LL_time_mean_list.append(predict_time_mean)
withTTA_LL_time_std_list.append(predict_time_std)
# PLOT: AUC - withTTA_LL
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
np.log2(TTA_list),
withTTA_LL_AUC_mean_list,
yerr=withTTA_LL_AUC_std_list,
fmt="-o",
label="AUC mean",
color="#1f77b4",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmax(withTTA_LL_AUC_mean_list)
y = np.max(withTTA_LL_AUC_mean_list)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.3f" % y, size=14, color="red")
plt.ylabel("AUC", size=14)
plt.xlabel("log2(TTA steps)", size=14)
plt.title(
"withTTA_LL, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
# plt.legend(loc='best')
plt.savefig(
"AUC_"
+ "withTTA_LL"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# PLOT: PREDICTION TIME - withTTA_LL
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
TTA_list,
withTTA_LL_time_mean_list,
yerr=withTTA_LL_time_std_list,
fmt="-o",
label="lossless",
color="#1f77b4",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmin(withTTA_LL_time_mean_list)
y = np.min(withTTA_LL_time_mean_list)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "min time\n%.3f" % y, size=14, color="red")
plt.ylabel("Prediction Time", size=14)
plt.xlabel("TTA steps", size=14)
plt.title(
"withTTA_LL, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
# plt.legend(loc='best')
plt.savefig(
"TIME_"
+ "withTTA_LL"
+ "_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# #### FULL TTA - withTTA
# withTTA_
VERBOSE = 0
TTA_list = [1, 2, 4, 8, 16, 32, 64, 128]
withTTA_AUC_mean_list = []
withTTA_AUC_std_list = []
withTTA_time_mean_list = []
withTTA_time_std_list = []
for i in tqdm(TTA_list):
AUC_mean, AUC_std, predict_time_mean, predict_time_std = AUC_time(
print_verbose=False, augment=True, aug_lossless=False, TTA=i
)
withTTA_AUC_mean_list.append(AUC_mean)
withTTA_AUC_std_list.append(AUC_std)
withTTA_time_mean_list.append(predict_time_mean)
withTTA_time_std_list.append(predict_time_std)
# PLOT: AUC - withTTA_LL
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
np.log2(TTA_list),
withTTA_AUC_mean_list,
yerr=withTTA_AUC_std_list,
fmt="-o",
label="full",
color="#1f77b4",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmax(withTTA_AUC_mean_list)
y = np.max(withTTA_AUC_mean_list)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.3f" % y, size=14, color="red")
plt.ylabel("AUC", size=14)
plt.xlabel("log2(TTA steps)", size=14)
plt.title(
"withTTA_full, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
# plt.legend(loc='best')
plt.savefig(
"AUC_"
+ "withTTA_full"
+ "_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# PLOT: PREDICTION TIME - withTTA_LL
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
TTA_list,
withTTA_time_mean_list,
yerr=withTTA_time_std_list,
fmt="-o",
label="full",
color="#1f77b4",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmin(withTTA_LL_time_mean_list)
y = np.min(withTTA_LL_time_mean_list)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "min time\n%.3f" % y, size=14, color="red")
plt.ylabel("Prediction Time", size=14)
plt.xlabel("TTA steps", size=14)
plt.title(
"withTTA, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
# plt.legend(loc='best')
plt.savefig(
"TIME_"
+ "withTTA_full"
+ "_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# #### Both TTAs on the same plot - AUC and TIME
# PLOT: AUC - withTTA_LL - lossless
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
np.log2(TTA_list),
withTTA_LL_AUC_mean_list,
yerr=withTTA_LL_AUC_std_list,
fmt="-o",
label="lossless",
color="blue",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmax(withTTA_LL_AUC_mean_list)
y = np.max(withTTA_LL_AUC_mean_list)
withTTA_LL_AUC_mean_max = np.max(withTTA_LL_AUC_mean_list)
withTTA_LL_AUC_std_max = withTTA_LL_AUC_std_list[np.argmax(withTTA_LL_AUC_mean_list)]
withTTA_LL_AUC_mean_TTA = np.argmax(withTTA_LL_AUC_mean_list)
print(
"i=%d," % withTTA_LL_AUC_mean_TTA,
"withTTA_LL_AUC_max=%.3f" % withTTA_LL_AUC_mean_max,
"(+-%.3f)" % withTTA_LL_AUC_std_max,
)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.3f" % y, size=14, color="blue")
# PLOT: AUC - withTTA - full
plt.errorbar(
np.log2(TTA_list),
withTTA_AUC_mean_list,
yerr=withTTA_AUC_std_list,
fmt="-o",
label="full",
color="green",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmax(withTTA_AUC_mean_list)
y = np.max(withTTA_AUC_mean_list)
withTTA_AUC_mean_max = np.max(withTTA_AUC_mean_list)
withTTA_AUC_std_max = withTTA_AUC_std_list[np.argmax(withTTA_AUC_mean_list)]
withTTA_AUC_mean_TTA = np.argmax(withTTA_AUC_mean_list)
print(
"i=%d," % withTTA_AUC_mean_TTA,
"withTTA_AUC_max=%.3f" % withTTA_AUC_mean_max,
"(+-%.3f)" % withTTA_AUC_std_max,
)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(
x - 0.03 * xdist, y - 0.13 * ydist, "max auc\n%.3f" % y, size=14, color="green"
)
plt.ylabel("AUC", size=14)
plt.xlabel("log2(TTA steps)", size=14)
plt.title(
"Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
plt.legend(loc="best")
plt.savefig(
"AUC_both_TTA_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# PLOT: PREDICTION TIME - withTTA_LL - lossless
plt.style.use("seaborn-whitegrid")
plt.figure(figsize=(10, 5))
# plt.plot(TTA_list,withTTA_LL_AUC_mean_list,'-o',label='AUC mean',color='#ff7f0e')
plt.errorbar(
TTA_list,
withTTA_LL_time_mean_list,
yerr=withTTA_LL_time_std_list,
fmt="-o",
label="lossless",
color="blue",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmin(withTTA_LL_time_mean_list)
y = np.min(withTTA_LL_time_mean_list)
withTTA_LL_time_mean_max = withTTA_LL_time_mean_list[
np.argmax(withTTA_LL_AUC_mean_list)
]
withTTA_LL_time_std_max = withTTA_LL_time_std_list[np.argmax(withTTA_LL_AUC_mean_list)]
withTTA_LL_time_mean_TTA = np.argmax(withTTA_LL_AUC_mean_list)
print(
"i=%d," % withTTA_LL_time_mean_TTA,
"withTTA_LL_time_mean_max=%.3f" % withTTA_LL_time_mean_max,
"(+-%.3f)" % withTTA_LL_time_std_max,
)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(
x - 0.03 * xdist, y - 0.13 * ydist, "min time\n%.3f" % y, size=14, color="blue"
)
# PLOT: PREDICTION TIME - withTTA - full
plt.errorbar(
TTA_list,
withTTA_time_mean_list,
yerr=withTTA_time_std_list,
fmt="-o",
label="full",
color="green",
)
# plt.plot(TTA_list,withTTA_LL_AUC_std_list,'-o',label='AUC std',color='#1f77b4')
x = np.argmin(withTTA_time_mean_list)
y = np.min(withTTA_time_mean_list)
withTTA_time_mean_max = withTTA_time_mean_list[np.argmax(withTTA_AUC_mean_list)]
withTTA_time_std_max = withTTA_time_std_list[np.argmax(withTTA_AUC_mean_list)]
withTTA_time_mean_TTA = np.argmax(withTTA_AUC_mean_list)
print(
"i=%d," % withTTA_time_mean_TTA,
"withTTA_time_mean_max=%.3f" % withTTA_time_mean_max,
"(+-%.3f)" % withTTA_time_std_max,
)
xdist = plt.xlim()[1] - plt.xlim()[0]
ydist = plt.ylim()[1] - plt.ylim()[0]
plt.scatter(x, y, s=200, color="red")
plt.text(
x - 0.03 * xdist, y + 0.1 * ydist, "min time\n%.3f" % y, size=14, color="green"
)
plt.ylabel("Prediction Time", size=14)
plt.xlabel("TTA steps", size=14)
plt.title(
"withTTA_LL, Image Size=%i, %s, Device=%s, Epochs=%i"
% (IMG_SIZES[fold], model_name, DEVICE, EPOCH),
size=14,
)
plt.legend(loc="best")
plt.savefig(
"TIME_both_TTA_"
+ DEVICE
+ "_"
+ str(MODEL)
+ "_"
+ str(SIZE)
+ "_ep"
+ str(EPOCH)
+ ".png",
bbox_inches="tight",
dpi=300,
)
plt.show()
# ### Without TTA
TTA = 1
predict_woTTA_time_list = []
AUC_woTTA_list = []
oof_pred = []
oof_tar = []
for fold, (idxT, idxV) in enumerate(skf.split(np.arange(15))):
print("Fold %i. Loading the current fold model..." % fold)
model.load_weights(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-%i.h5" % fold
)
# model.load_weights(DEVICE + '_fold-%i.h5'%fold)
# PREDICT without TTA # augment=False
ds_valid = get_dataset(
files_valid,
labeled=False,
return_image_names=False,
augment=False,
repeat=True,
shuffle=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
batch_size=BATCH_SIZES[fold] * 4,
)
ct_valid = count_data_items(files_valid)
STEPS = TTA * ct_valid / BATCH_SIZES[fold] / 4 / REPLICAS
print("Predicting Test without TTA for %i files..." % (ct_valid))
# Start timer
ts_eval = time.time()
pred = model.predict(ds_valid, steps=STEPS, verbose=VERBOSE)[: TTA * ct_valid,]
# End timer
te_eval = time.time()
test_time = (te_eval - ts_eval) / ct_valid
predict_woTTA_time_list.append(test_time)
print("Fold %i, test_time=%.6f seconds." % (fold, test_time))
# Add predictions to list
oof_pred.append(np.mean(pred.reshape((ct_valid, TTA), order="F"), axis=1))
# GET OOF TARGETS AND NAMES
ds_valid = get_dataset(
files_valid,
augment=False,
repeat=False,
dim=SPECIFIC_SIZE,
# dim=IMG_SIZES[fold],
labeled=True,
return_image_names=True,
)
# Add targets to list
oof_tar.append(
np.array([target.numpy() for img, target in iter(ds_valid.unbatch())])
)
# Calculate AUC
auc = roc_auc_score(oof_tar[-1], oof_pred[-1])
# Add AUC to list
AUC_woTTA_list.append(auc)
print("Fold %i, AUC=%.6f" % (fold, auc))
# Calculate AUC mean value to list
AUC_woTTA_mean = statistics.mean(AUC_woTTA_list) # mean
# Calculate AUC standard deviation value to list
AUC_woTTA_std = statistics.stdev(AUC_woTTA_list) # standard devition
print(
"#### OOF AUC without TTA: mean = %.6f, stdev = %.6f."
% (AUC_woTTA_mean, AUC_woTTA_std)
)
# Add AUC mean value to list
predict_woTTA_time_mean = statistics.mean(predict_woTTA_time_list) # mean
# Add AUC standard deviation value to list
predict_woTTA_time_std = statistics.stdev(predict_woTTA_time_list) # standard devition
print(
"#### Time without TTA: mean = %.6f, stdev = %.6f seconds."
% (predict_woTTA_time_mean, predict_woTTA_time_std)
)
# ### Measure model file size
import os
model_size = os.path.getsize(
DEVICE + "_" + str(MODEL) + "_" + str(SIZE) + "_fold-0.h5"
) # >> 20
print(str(model_size) + " Bytes")
# ### Save to file
results = pd.DataFrame(
data=[
[
MODEL,
model_size,
TRAINABLE,
AUGMENTATION,
AUG_LOSSLESS,
EPOCHS[0],
SIZE,
withTTA_LL_time_mean_max,
withTTA_LL_time_std_max,
withTTA_LL_AUC_mean_max,
withTTA_LL_AUC_std_max,
withTTA_time_mean_max,
withTTA_time_std_max,
withTTA_AUC_mean_max,
withTTA_AUC_std_max,
predict_woTTA_time_mean,
predict_woTTA_time_std,
AUC_woTTA_mean,
AUC_woTTA_std,
]
],
columns=[
"model",
"model_size",
"trainable",
"augmentation",
"aug_lossless",
"epochs",
"image_size",
"TTA_LL_time_mean",
"TTA_LL_time_std",
"TTA_LL_AUC_mean",
"TTA_LL_AUC_std",
"TTA_time_mean",
"TTA_time_std",
"TTA_AUC_mean",
"TTA_AUC_std",
"woTTA_time_mean",
"woTTA_time_std",
"woTTA_AUC_mean",
"woTTA_AUC_std",
],
)
experiment_title = (
DEVICE
+ "_"
+ str(MODEL)
+ "_s"
+ str(SIZE)
+ "_ep"
+ str(EPOCHS[0])
+ "_train"
+ str(TRAINABLE)
+ "_aug"
+ str(AUGMENTATION)
+ "_loss"
+ str(AUG_LOSSLESS)
+ "_time_AUC"
)
results.to_csv(experiment_title + ".csv", index=False)
results.head()
from zipfile import ZipFile
from os.path import basename
# Zip the files from given directory that matches the filter
def zipFilesInDir(dirName, zipFileName, filter):
# create a ZipFile object
with ZipFile(zipFileName, "w") as zipObj:
# Iterate over all the files in directory
for folderName, subfolders, filenames in os.walk(dirName):
for filename in filenames:
if filter(filename):
# create complete filepath of file in directory
filePath = os.path.join(folderName, filename)
# Add file to zip
zipObj.write(filePath, basename(filePath))
print("*** Create a zip archive of *.png *.ipynb *.csv files form a directory ***")
zipFilesInDir(".", experiment_title + "_csv_.zip", lambda name: "csv" in name)
zipFilesInDir(".", experiment_title + "_ipynb_.zip", lambda name: "ipynb" in name)
zipFilesInDir(".", experiment_title + "_png_.zip", lambda name: "png" in name)
zipFilesInDir(".", experiment_title + "_all.zip", lambda name: "_.zip" in name)
| false | 0 | 17,696 | 0 | 17,732 | 17,696 |
||
69141765
|
# # Test B
# **Test B**
# description
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from tensorflow import keras
# Read the data
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
# pull data into target (y) and predictors (X)
train_y = train.SalePrice
predictor_cols = ["LotArea", "OverallQual", "YearBuilt", "TotRmsAbvGrd"]
# Create training predictors data
train_X = train[predictor_cols]
my_model = keras.models.Sequential(
[
keras.layers.Dense(200, activation="relu", input_shape=[4]),
keras.layers.Dense(100, activation="relu"),
keras.layers.Dense(50, activation="relu"),
keras.layers.Dense(25, activation="relu"),
keras.layers.Dense(1),
]
)
my_model.compile(loss="mean_squared_error", optimizer="adam")
history = my_model.fit(train_X, train_y, epochs=300, validation_split=0.2)
pd.DataFrame(history.history).plot()
plt.grid(True)
plt.gca().set_ylim([0, 6000000000])
plt.show()
# Read the test data
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
# Treat the test data in the same way as training data. In this case, pull same columns.
test_X = test[predictor_cols]
# Use the model to make predictions
predicted_prices = my_model.predict(test_X)
# We will look at the predicted prices to ensure we have something sensible.
print(predicted_prices)
test_id = test.Id
test_id_nparray = test.Id.to_numpy()
my_submission = pd.DataFrame(
{"Id": test_id_nparray, "SalePrice": predicted_prices.ravel()}
)
# you could use any filename. We choose submission here
my_submission.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/141/69141765.ipynb
| null | null |
[{"Id": 69141765, "ScriptId": 18780948, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7949355, "CreationDate": "07/27/2021 10:35:31", "VersionNumber": 10.0, "Title": "Test B", "EvaluationDate": "07/27/2021", "IsChange": false, "TotalLines": 53.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 53.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Test B
# **Test B**
# description
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from tensorflow import keras
# Read the data
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
# pull data into target (y) and predictors (X)
train_y = train.SalePrice
predictor_cols = ["LotArea", "OverallQual", "YearBuilt", "TotRmsAbvGrd"]
# Create training predictors data
train_X = train[predictor_cols]
my_model = keras.models.Sequential(
[
keras.layers.Dense(200, activation="relu", input_shape=[4]),
keras.layers.Dense(100, activation="relu"),
keras.layers.Dense(50, activation="relu"),
keras.layers.Dense(25, activation="relu"),
keras.layers.Dense(1),
]
)
my_model.compile(loss="mean_squared_error", optimizer="adam")
history = my_model.fit(train_X, train_y, epochs=300, validation_split=0.2)
pd.DataFrame(history.history).plot()
plt.grid(True)
plt.gca().set_ylim([0, 6000000000])
plt.show()
# Read the test data
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
# Treat the test data in the same way as training data. In this case, pull same columns.
test_X = test[predictor_cols]
# Use the model to make predictions
predicted_prices = my_model.predict(test_X)
# We will look at the predicted prices to ensure we have something sensible.
print(predicted_prices)
test_id = test.Id
test_id_nparray = test.Id.to_numpy()
my_submission = pd.DataFrame(
{"Id": test_id_nparray, "SalePrice": predicted_prices.ravel()}
)
# you could use any filename. We choose submission here
my_submission.to_csv("submission.csv", index=False)
| false | 0 | 522 | 0 | 522 | 522 |
||
69141954
|
# # **EDA**
import pandas as pd
import numpy as np
from matplotlib import pyplot as pt
df_gendersub = pd.read_csv("../input/titanic/gender_submission.csv")
df_train = pd.read_csv("../input/titanic/train.csv")
df_test = pd.read_csv("../input/titanic/test.csv")
print(df_gendersub["Survived"].value_counts())
print(df_train.columns)
df_train.isnull().sum()
df_train["Age"].fillna(df_train["Age"].mean(), inplace=True)
df_train["Age"].isnull().sum()
df_train["Embarked"]
features = ["PassengerId", "Pclass", "Age", "SibSp", "Parch", "Fare"]
X = df_train[features]
y = df_train["Survived"]
print(X[0:5])
print(y[0:5])
import seaborn as sns
f, ax = pt.subplots(1, 2, figsize=(18, 8))
df_train["Survived"].value_counts().plot.pie(autopct="%1.1f%%", ax=ax[0])
ax[0].set_title("Servived")
sns.countplot("Survived", data=df_train, ax=ax[1])
ax[1].set_title("Survived")
pt.show()
# Name,Embarked --catorical
# PClass --Ordinal
# Age -continuous
print(df_train.groupby(["Sex", "Survived"])["Survived"].count())
f, ax = pt.subplots(1, 2, figsize=(12, 6))
df_train[["Sex", "Survived"]].groupby(["Sex"]).mean().plot.bar(ax=ax[0])
sns.countplot("Sex", data=df_train, ax=ax[1], hue="Survived")
plt.show()
print(pd.crosstab(df_train.Pclass, df_train.Survived, margins=True))
f, ax = pt.subplots(1, 2, figsize=(12, 6))
print(df_train["Pclass"].value_counts())
df_train["Pclass"].value_counts().plot.bar(ax=ax[0])
ax[0].set_ylabel("count")
ax[0].set_ylabel("Pclass")
sns.countplot("Pclass", hue="Survived", ax=ax[1], data=df_train)
print(pd.crosstab([df_train.Age, df_train.Pclass], df_train.Survived, margins=True))
sns.violinplot("Pclass", "Age", hue="Survived", data=df_train, split=True)
# # Data Pre-processing/enginnering
print("MIN: ", df_train["Age"].min())
print("MAX: ", df_train["Age"].max())
print("MEAN: ", df_train["Age"].mean())
df_train["AgeGroup"] = 0
df_train.loc[df_train["Age"] <= 16, "AgeGroup"] = 0
df_train.loc[((df_train["Age"] > 16) & (df_train["Age"] <= 32)), "AgeGroup"] = 1
df_train.loc[((df_train["Age"] > 32) & (df_train["Age"] <= 48)), "AgeGroup"] = 2
df_train.loc[((df_train["Age"] > 48) & (df_train["Age"] <= 64)), "AgeGroup"] = 3
df_train.loc[((df_train["Age"] > 64) & (df_train["Age"] <= 80)), "AgeGroup"] = 4
df_train["Embarked"].fillna("S", inplace=True)
df_train["Embarked"].isnull().sum()
df_train["Fare_Range"] = pd.qcut(df_train["Fare"], 4)
df_train.groupby(["Fare_Range"])["Survived"].mean().to_frame()
df_train["Fare_cat"] = 0
df_train.loc[df_train["Fare"] <= 7.91, "Fare_cat"] = 0
df_train.loc[(df_train["Fare"] > 7.91) & (df_train["Fare"] <= 14.454), "Fare_cat"] = 1
df_train.loc[(df_train["Fare"] > 14.454) & (df_train["Fare"] <= 31), "Fare_cat"] = 2
df_train.loc[(df_train["Fare"] > 31) & (df_train["Fare"] <= 513), "Fare_cat"] = 3
df_train["Sex"].replace(["male", "female"], [0, 1], inplace=True)
df_train["Embarked"].replace(["S", "C", "Q"], [0, 1, 2], inplace=True)
sns.heatmap(
df_train[
[
"Survived",
"Pclass",
"Sex",
"SibSp",
"Parch",
"Embarked",
"AgeGroup",
"Fare_cat",
]
].corr(),
annot=True,
)
#
# # Prediction
from sklearn.model_selection import train_test_split
features = ["Pclass", "Sex", "SibSp", "Parch", "Embarked", "AgeGroup", "Fare_cat"]
X = df_train[features]
y = df_train.Survived
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0, stratify=y
)
print(X_train.shape)
print(y_train.shape)
print(X_test.shape)
print(y_test.shape)
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error as mse
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
print(mse(y_test, y_pred, squared=False))
print(lr.score(X_test, y_test))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/141/69141954.ipynb
| null | null |
[{"Id": 69141954, "ScriptId": 18854123, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7221706, "CreationDate": "07/27/2021 10:38:27", "VersionNumber": 1.0, "Title": "notebook6e2eb3b032", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 121.0, "LinesInsertedFromPrevious": 121.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # **EDA**
import pandas as pd
import numpy as np
from matplotlib import pyplot as pt
df_gendersub = pd.read_csv("../input/titanic/gender_submission.csv")
df_train = pd.read_csv("../input/titanic/train.csv")
df_test = pd.read_csv("../input/titanic/test.csv")
print(df_gendersub["Survived"].value_counts())
print(df_train.columns)
df_train.isnull().sum()
df_train["Age"].fillna(df_train["Age"].mean(), inplace=True)
df_train["Age"].isnull().sum()
df_train["Embarked"]
features = ["PassengerId", "Pclass", "Age", "SibSp", "Parch", "Fare"]
X = df_train[features]
y = df_train["Survived"]
print(X[0:5])
print(y[0:5])
import seaborn as sns
f, ax = pt.subplots(1, 2, figsize=(18, 8))
df_train["Survived"].value_counts().plot.pie(autopct="%1.1f%%", ax=ax[0])
ax[0].set_title("Servived")
sns.countplot("Survived", data=df_train, ax=ax[1])
ax[1].set_title("Survived")
pt.show()
# Name,Embarked --catorical
# PClass --Ordinal
# Age -continuous
print(df_train.groupby(["Sex", "Survived"])["Survived"].count())
f, ax = pt.subplots(1, 2, figsize=(12, 6))
df_train[["Sex", "Survived"]].groupby(["Sex"]).mean().plot.bar(ax=ax[0])
sns.countplot("Sex", data=df_train, ax=ax[1], hue="Survived")
plt.show()
print(pd.crosstab(df_train.Pclass, df_train.Survived, margins=True))
f, ax = pt.subplots(1, 2, figsize=(12, 6))
print(df_train["Pclass"].value_counts())
df_train["Pclass"].value_counts().plot.bar(ax=ax[0])
ax[0].set_ylabel("count")
ax[0].set_ylabel("Pclass")
sns.countplot("Pclass", hue="Survived", ax=ax[1], data=df_train)
print(pd.crosstab([df_train.Age, df_train.Pclass], df_train.Survived, margins=True))
sns.violinplot("Pclass", "Age", hue="Survived", data=df_train, split=True)
# # Data Pre-processing/enginnering
print("MIN: ", df_train["Age"].min())
print("MAX: ", df_train["Age"].max())
print("MEAN: ", df_train["Age"].mean())
df_train["AgeGroup"] = 0
df_train.loc[df_train["Age"] <= 16, "AgeGroup"] = 0
df_train.loc[((df_train["Age"] > 16) & (df_train["Age"] <= 32)), "AgeGroup"] = 1
df_train.loc[((df_train["Age"] > 32) & (df_train["Age"] <= 48)), "AgeGroup"] = 2
df_train.loc[((df_train["Age"] > 48) & (df_train["Age"] <= 64)), "AgeGroup"] = 3
df_train.loc[((df_train["Age"] > 64) & (df_train["Age"] <= 80)), "AgeGroup"] = 4
df_train["Embarked"].fillna("S", inplace=True)
df_train["Embarked"].isnull().sum()
df_train["Fare_Range"] = pd.qcut(df_train["Fare"], 4)
df_train.groupby(["Fare_Range"])["Survived"].mean().to_frame()
df_train["Fare_cat"] = 0
df_train.loc[df_train["Fare"] <= 7.91, "Fare_cat"] = 0
df_train.loc[(df_train["Fare"] > 7.91) & (df_train["Fare"] <= 14.454), "Fare_cat"] = 1
df_train.loc[(df_train["Fare"] > 14.454) & (df_train["Fare"] <= 31), "Fare_cat"] = 2
df_train.loc[(df_train["Fare"] > 31) & (df_train["Fare"] <= 513), "Fare_cat"] = 3
df_train["Sex"].replace(["male", "female"], [0, 1], inplace=True)
df_train["Embarked"].replace(["S", "C", "Q"], [0, 1, 2], inplace=True)
sns.heatmap(
df_train[
[
"Survived",
"Pclass",
"Sex",
"SibSp",
"Parch",
"Embarked",
"AgeGroup",
"Fare_cat",
]
].corr(),
annot=True,
)
#
# # Prediction
from sklearn.model_selection import train_test_split
features = ["Pclass", "Sex", "SibSp", "Parch", "Embarked", "AgeGroup", "Fare_cat"]
X = df_train[features]
y = df_train.Survived
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0, stratify=y
)
print(X_train.shape)
print(y_train.shape)
print(X_test.shape)
print(y_test.shape)
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error as mse
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
print(mse(y_test, y_pred, squared=False))
print(lr.score(X_test, y_test))
| false | 0 | 1,565 | 0 | 1,565 | 1,565 |
||
69141250
|
# **In this notebook,I try to analysis house price dataset and predict the house prices by regression methods.**
# **I start with introdusing the dataset,then I do data analysis and house price prediction step by step.**
# # **Import the dataset:**
# **This dataset have 1460 rows and 81 columns.The SalePrice is the target variable that I am trying to predict.At first I should import all necessary libraries and then read and import the dataset:**
import numpy as np # Import all necessary libraries
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
with open(
"../input/house-prices-advanced-regression-techniques/data_description.txt"
) as f:
print(f.read())
train.head()
# Cleaning the Id columns because it doesnt have important information.
train = train.drop("Id", axis=1)
train.info()
# ### **Target Value**
fig = plt.figure(figsize=(8, 4), dpi=100) # Draw histogram for target variable
sns.distplot(
train["SalePrice"], hist_kws=dict(edgecolor="w", linewidth=1), bins=25, color="r"
)
plt.title("Sales data distribution")
# **As we see the most amount of prices are between 100000 to 300000.This SalePrice distribution is not very normal so I use log for that to make more normal distribution.**
train["saleprice"] = np.log1p(train["SalePrice"]) # Use log function in numpy
fig = plt.figure(figsize=(8, 4), dpi=100)
sns.distplot(
train["saleprice"], hist_kws=dict(edgecolor="w", linewidth=1), bins=25, color="r"
)
plt.title("Sales data distribution")
# ### **Checking Missing Data**
# **Before I continue,I want to introduce the missing value briefly.The real-world data often has a lot of missing values. The cause of missing values can be data corruption or failure to record data.
# The handling of missing data is very important during the preprocessing of the dataset as many machine learning algorithms do not support missing values.**
# **Now I want to identify the rows with the most number of missing values and drop or transform them.**
# Let's check if the data set has any missing values.
100 * (train.isnull().sum() / len(train))
def missing_values_percent(train): # we can use this function in all dataframes.
nan_percent = 100 * (train.isnull().sum() / len(train))
nan_percent = nan_percent[nan_percent > 0].sort_values()
return nan_percent
nan_percent = missing_values_percent(train)
nan_percent
# Drawing barplot for these missing values.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
# determine the missing values that their percentages are between 0 and 5.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
plt.ylim(0, 5)
# ## **In this step we want to make a decision about our missing data**
train[train["Electrical"].isnull()]
# can delet this row
train = train.drop(labels=1379, axis=0)
nan_percent = missing_values_percent(
train
) # see Electrical has been droped from missing data.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
plt.ylim(0, 5)
train["MasVnrType"] = train["MasVnrType"].fillna("None")
train["MasVnrArea"] = train["MasVnrArea"].fillna(0)
nan_percent = missing_values_percent(
train
) # see MasVnrType and MasVnrArea have been droped.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
plt.ylim(0, 5)
bsmt_str_cols = ["BsmtQual", "BsmtCond", "BsmtFinType1", "BsmtExposure", "BsmtFinType2"]
train[bsmt_str_cols] = train[bsmt_str_cols].fillna("None")
gar_str_cols = ["GarageType", "GarageFinish"] # transform the object to None
train[gar_str_cols] = train[gar_str_cols].fillna("None")
gar_num_cols = [
"GarageYrBlt",
"GarageQual",
"GarageCond",
] # transform the int or float to 0
train[gar_num_cols] = train[gar_num_cols].fillna(0)
nan_percent = missing_values_percent(
train
) # see MasVnrType and MasVnrArea have been droped.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
train["FireplaceQu"] = train["FireplaceQu"].fillna("None")
# **From the below boxplot we understand that any neighborhood have specefic LotFrontage distribution.So we can replace the average of Lotfrontage base on each Neighborhood with the missing data.**
plt.figure(figsize=(8, 12))
sns.boxplot(data=train, x="LotFrontage", y="Neighborhood")
train.groupby("Neighborhood")[
"LotFrontage"
].mean() # average of Lotfrontage base on each Neighborhood
# replace the average of Lotfrontage base on each Neighborhood with the missing data
train["LotFrontage"] = train.groupby("Neighborhood")["LotFrontage"].transform(
lambda val: val.fillna(val.mean())
)
nan_percent = missing_values_percent(
train
) # see MasVnrType and MasVnrArea have been droped.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
# **As we see,we have only 4 features that have missing data.As the percentage of missing datas in these features are above 80%,the best way is to drop them from the dataframe.**
train = train.drop(["Fence", "Alley", "MiscFeature", "PoolQC"], axis=1)
train.isnull().sum() # we have no missing data
# ## Now,we have no missing data.
# ### **Categorical data**
# **In this step,I want to encoding str to int.**
train.info()
# At first,I tranform MSSubClass to str:
train["MSSubClass"].apply(str)
train.select_dtypes(include="object") # seperate the data that are object
# Divide dataframe to 2 parts(num and str)
train_num = train.select_dtypes(exclude="object")
train_obj = train.select_dtypes(include="object")
train_num.info()
train_obj.info()
train_obj = pd.get_dummies(
train_obj, drop_first=True
) # use one-hot encoding to transform str to int and float
train_obj.shape
Final_train = pd.concat([train_num, train_obj], axis=1)
Final_train.head()
# ### **LinearRegression**
# **Linear regression is probably one of the most important and widely used regression techniques. It’s among the simplest regression methods. One of its main advantages is the ease of interpreting results.
# Linear regression performs the task to predict a dependent variable value (y) based on a given independent variable (x). So, this regression technique finds out a linear relationship between x (input) and y(output). Hence, the name is Linear Regression.**
# **In this step we want to predict our target valuable (saleprice) based on our features.**
# **After import the data,data overview and exploratory data analysis which are shown,we should determine the feature and lable.**
# Determine the feature and lable
X = Final_train.drop("SalePrice", axis=1)
y = Final_train["SalePrice"]
# Split the dataset to train and test
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.30, random_state=101
)
# train the model
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X_train, y_train)
# coeficient matrix
pd.DataFrame(model.coef_, X.columns, columns=["coeficient"])
# predicting test data
y_pred = model.predict(X_test)
# evaluating the model
from sklearn import metrics
MAD = metrics.mean_absolute_error(y_test, y_pred)
MSE = metrics.mean_squared_error(y_test, y_pred)
RMSE = np.sqrt(MSE)
pd.DataFrame(
data=[MAD, MSE, RMSE], index=["MAD", "MSE", "RMSE"], columns=["LinearRegression"]
)
# # **Residuals**
# **For linear regression it is a good idea to evaluate residuals(y- ̂y)**
#
test_residuals = (
y_test - y_pred
) # the residuals should be random and close to normal distribution.
sns.scatterplot(x=y_test, y=y_pred, color="r")
plt.xlabel("Y-Test")
plt.ylabel("Y-Pred")
sns.scatterplot(
x=y_test, y=test_residuals, color="r"
) # test residuals should not show a clear pattern.
plt.axhline(y=0, color="b", ls="--")
plt.xlabel("Y-Test")
plt.ylabel("residuals")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/141/69141250.ipynb
| null | null |
[{"Id": 69141250, "ScriptId": 18715584, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7807419, "CreationDate": "07/27/2021 10:27:46", "VersionNumber": 12.0, "Title": "Advance house price Step by Step", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 213.0, "LinesInsertedFromPrevious": 16.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 197.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# **In this notebook,I try to analysis house price dataset and predict the house prices by regression methods.**
# **I start with introdusing the dataset,then I do data analysis and house price prediction step by step.**
# # **Import the dataset:**
# **This dataset have 1460 rows and 81 columns.The SalePrice is the target variable that I am trying to predict.At first I should import all necessary libraries and then read and import the dataset:**
import numpy as np # Import all necessary libraries
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
with open(
"../input/house-prices-advanced-regression-techniques/data_description.txt"
) as f:
print(f.read())
train.head()
# Cleaning the Id columns because it doesnt have important information.
train = train.drop("Id", axis=1)
train.info()
# ### **Target Value**
fig = plt.figure(figsize=(8, 4), dpi=100) # Draw histogram for target variable
sns.distplot(
train["SalePrice"], hist_kws=dict(edgecolor="w", linewidth=1), bins=25, color="r"
)
plt.title("Sales data distribution")
# **As we see the most amount of prices are between 100000 to 300000.This SalePrice distribution is not very normal so I use log for that to make more normal distribution.**
train["saleprice"] = np.log1p(train["SalePrice"]) # Use log function in numpy
fig = plt.figure(figsize=(8, 4), dpi=100)
sns.distplot(
train["saleprice"], hist_kws=dict(edgecolor="w", linewidth=1), bins=25, color="r"
)
plt.title("Sales data distribution")
# ### **Checking Missing Data**
# **Before I continue,I want to introduce the missing value briefly.The real-world data often has a lot of missing values. The cause of missing values can be data corruption or failure to record data.
# The handling of missing data is very important during the preprocessing of the dataset as many machine learning algorithms do not support missing values.**
# **Now I want to identify the rows with the most number of missing values and drop or transform them.**
# Let's check if the data set has any missing values.
100 * (train.isnull().sum() / len(train))
def missing_values_percent(train): # we can use this function in all dataframes.
nan_percent = 100 * (train.isnull().sum() / len(train))
nan_percent = nan_percent[nan_percent > 0].sort_values()
return nan_percent
nan_percent = missing_values_percent(train)
nan_percent
# Drawing barplot for these missing values.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
# determine the missing values that their percentages are between 0 and 5.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
plt.ylim(0, 5)
# ## **In this step we want to make a decision about our missing data**
train[train["Electrical"].isnull()]
# can delet this row
train = train.drop(labels=1379, axis=0)
nan_percent = missing_values_percent(
train
) # see Electrical has been droped from missing data.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
plt.ylim(0, 5)
train["MasVnrType"] = train["MasVnrType"].fillna("None")
train["MasVnrArea"] = train["MasVnrArea"].fillna(0)
nan_percent = missing_values_percent(
train
) # see MasVnrType and MasVnrArea have been droped.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
plt.ylim(0, 5)
bsmt_str_cols = ["BsmtQual", "BsmtCond", "BsmtFinType1", "BsmtExposure", "BsmtFinType2"]
train[bsmt_str_cols] = train[bsmt_str_cols].fillna("None")
gar_str_cols = ["GarageType", "GarageFinish"] # transform the object to None
train[gar_str_cols] = train[gar_str_cols].fillna("None")
gar_num_cols = [
"GarageYrBlt",
"GarageQual",
"GarageCond",
] # transform the int or float to 0
train[gar_num_cols] = train[gar_num_cols].fillna(0)
nan_percent = missing_values_percent(
train
) # see MasVnrType and MasVnrArea have been droped.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
train["FireplaceQu"] = train["FireplaceQu"].fillna("None")
# **From the below boxplot we understand that any neighborhood have specefic LotFrontage distribution.So we can replace the average of Lotfrontage base on each Neighborhood with the missing data.**
plt.figure(figsize=(8, 12))
sns.boxplot(data=train, x="LotFrontage", y="Neighborhood")
train.groupby("Neighborhood")[
"LotFrontage"
].mean() # average of Lotfrontage base on each Neighborhood
# replace the average of Lotfrontage base on each Neighborhood with the missing data
train["LotFrontage"] = train.groupby("Neighborhood")["LotFrontage"].transform(
lambda val: val.fillna(val.mean())
)
nan_percent = missing_values_percent(
train
) # see MasVnrType and MasVnrArea have been droped.
plt.figure(figsize=(12, 6))
sns.barplot(x=nan_percent.index, y=nan_percent)
plt.xticks(rotation=45)
# **As we see,we have only 4 features that have missing data.As the percentage of missing datas in these features are above 80%,the best way is to drop them from the dataframe.**
train = train.drop(["Fence", "Alley", "MiscFeature", "PoolQC"], axis=1)
train.isnull().sum() # we have no missing data
# ## Now,we have no missing data.
# ### **Categorical data**
# **In this step,I want to encoding str to int.**
train.info()
# At first,I tranform MSSubClass to str:
train["MSSubClass"].apply(str)
train.select_dtypes(include="object") # seperate the data that are object
# Divide dataframe to 2 parts(num and str)
train_num = train.select_dtypes(exclude="object")
train_obj = train.select_dtypes(include="object")
train_num.info()
train_obj.info()
train_obj = pd.get_dummies(
train_obj, drop_first=True
) # use one-hot encoding to transform str to int and float
train_obj.shape
Final_train = pd.concat([train_num, train_obj], axis=1)
Final_train.head()
# ### **LinearRegression**
# **Linear regression is probably one of the most important and widely used regression techniques. It’s among the simplest regression methods. One of its main advantages is the ease of interpreting results.
# Linear regression performs the task to predict a dependent variable value (y) based on a given independent variable (x). So, this regression technique finds out a linear relationship between x (input) and y(output). Hence, the name is Linear Regression.**
# **In this step we want to predict our target valuable (saleprice) based on our features.**
# **After import the data,data overview and exploratory data analysis which are shown,we should determine the feature and lable.**
# Determine the feature and lable
X = Final_train.drop("SalePrice", axis=1)
y = Final_train["SalePrice"]
# Split the dataset to train and test
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.30, random_state=101
)
# train the model
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X_train, y_train)
# coeficient matrix
pd.DataFrame(model.coef_, X.columns, columns=["coeficient"])
# predicting test data
y_pred = model.predict(X_test)
# evaluating the model
from sklearn import metrics
MAD = metrics.mean_absolute_error(y_test, y_pred)
MSE = metrics.mean_squared_error(y_test, y_pred)
RMSE = np.sqrt(MSE)
pd.DataFrame(
data=[MAD, MSE, RMSE], index=["MAD", "MSE", "RMSE"], columns=["LinearRegression"]
)
# # **Residuals**
# **For linear regression it is a good idea to evaluate residuals(y- ̂y)**
#
test_residuals = (
y_test - y_pred
) # the residuals should be random and close to normal distribution.
sns.scatterplot(x=y_test, y=y_pred, color="r")
plt.xlabel("Y-Test")
plt.ylabel("Y-Pred")
sns.scatterplot(
x=y_test, y=test_residuals, color="r"
) # test residuals should not show a clear pattern.
plt.axhline(y=0, color="b", ls="--")
plt.xlabel("Y-Test")
plt.ylabel("residuals")
| false | 0 | 2,482 | 0 | 2,482 | 2,482 |
||
69141185
|
# # Reading Files
import pandas as pd
import matplotlib.pyplot as plt
plt.style.use("ggplot")
import re
import seaborn as sns
import os
import numpy as np
import pandas as pd
df = pd.read_csv("../input/car-crashes-severity-prediction/train.csv")
holidays = pd.read_csv("../input/holidayscsv/holidays.csv")
weather = pd.read_csv("../input/car-crashes-severity-prediction/weather-sfcsv.csv")
test = pd.read_csv("../input/car-crashes-severity-prediction/test.csv")
print("The shape of the dataset is {}.\n\n".format(df.shape))
test.Severity
# # Convert type of timestamp Column
# Convert object to date time
df["timestamp"] = pd.to_datetime(df["timestamp"])
# Create date column in df dataframe
df["date"] = [d.date() for d in df["timestamp"]]
df["date"] = pd.to_datetime(df["date"])
# Create date column in weather dataframe
weather["date"] = (
weather["Year"].astype(str)
+ "-"
+ weather["Month"].astype(str)
+ "-"
+ weather["Day"].astype(str)
)
weather["date"] = pd.to_datetime(weather["date"])
# Temporary weather dataframe
weather_1 = weather.drop(["Year", "Day", "Month"], axis=1)
# Take a look on sorted weather
sortred_weather = weather_1.sort_values(by=["date"])
weather = weather.drop_duplicates(subset=["Hour", "date"])
df.info()
import datetime as dt
weather_cp = weather.copy()
df_cp = df.copy()
df_cp["year"] = df_cp.date.dt.year
df_cp["month"] = df_cp.date.dt.month
df_cp["Day"] = df_cp.date.dt.day
df_cp["weekday"] = df_cp[["date"]].apply(
lambda x: dt.datetime.strftime(x["date"], "%A"), axis=1
)
def hr_func(ts):
return ts.hour
df_cp["Hour"] = df_cp["timestamp"].apply(hr_func)
# ========================================
holidays_cp = holidays.copy()
holidays_cp["date"] = pd.to_datetime(holidays_cp["date"])
# ========================================
df_cp1 = pd.merge(df_cp, weather_cp, on=["date", "Hour"], how="left")
df_cp2 = pd.merge(df_cp1, holidays_cp, on=["date"], how="left")
cleand_df_cp = df_cp2.drop(
[
"Junction",
"Year",
"Month",
"Day_y",
"date",
"ID",
"Wind_Chill(F)",
"Bump",
"Give_Way",
"No_Exit",
"Roundabout",
"timestamp",
],
axis=1,
)
# cleand_df_cp.info()
cleand_df_cp["Temperature(F)"] = cleand_df_cp["Temperature(F)"].fillna(
cleand_df_cp["Temperature(F)"].mean()
)
cleand_df_cp["Visibility(mi)"] = cleand_df_cp["Visibility(mi)"].fillna(
cleand_df_cp["Visibility(mi)"].mean()
)
cleand_df_cp["Humidity(%)"] = cleand_df_cp["Humidity(%)"].fillna(
cleand_df_cp["Humidity(%)"].mean()
)
cleand_df_cp["Weather_Condition"] = cleand_df_cp["Weather_Condition"].fillna(
"Plarty Cloudy"
)
cleand_df_cp["Wind_Speed(mph)"] = cleand_df_cp["Wind_Speed(mph)"].fillna(
cleand_df_cp["Wind_Speed(mph)"].mean()
)
cleand_df_cp["Precipitation(in)"] = cleand_df_cp["Precipitation(in)"].fillna(
cleand_df_cp["Precipitation(in)"].mean()
)
cleand_df_cp["description"] = cleand_df_cp["description"].fillna(0)
cleand_df_cp.isnull().sum().sort_values(ascending=False).head(5)
cleand_df_cp["H_day"] = cleand_df_cp["description"].copy()
bool_df1 = cleand_df_cp.select_dtypes(include="bool")
# bool_df1.columns
for i in range(len(cleand_df_cp["description"])):
if cleand_df_cp["description"][i] != 0:
cleand_df_cp["H_day"][i] = 1
for i in range(len(cleand_df_cp["weekday"])):
if cleand_df_cp["weekday"][i] == "Sunday":
cleand_df_cp["H_day"][i] = 1
elif cleand_df_cp["weekday"][i] == "Saturday":
cleand_df_cp["H_day"][i] = 1
else:
cleand_df_cp["H_day"][i] = 0
for i in range(len(cleand_df_cp["Hour"])):
if (cleand_df_cp["Hour"][i] > 18) or (cleand_df_cp["Hour"][i] <= 6):
cleand_df_cp["Hour"][i] = 1
else:
cleand_df_cp["Hour"][i] = 0
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for i in bool_df1.columns:
cleand_df_cp[i] = le.fit_transform(cleand_df_cp[i])
cleand_df_cp["Selected"] = le.fit_transform(cleand_df_cp["Selected"])
cleand_df_cp["Weather_Condition"] = le.fit_transform(cleand_df_cp["Weather_Condition"])
cleand_df_cp["Side"] = le.fit_transform(cleand_df_cp["Side"])
# obj_col1 = ["Side"]
# for i in obj_col1:
# cleand_df_cp = pd.get_dummies(cleand_df_cp, prefix=[i], columns=[i])
# ====================================================================
# from sklearn.preprocessing import MinMaxScaler
# # Create the scaler
# mms = MinMaxScaler()
# # create a copy of wine
# wine_copy_4 = cleand_df_cp.copy()
# wine_copy_5 = wine_copy_4.drop(columns=['Severity'])
# # Scaled data
# wine_copy_5_scaled = mms.fit_transform(wine_copy_5)
# #create a dataframe
# wine_copy_5_scaled_df = pd.DataFrame(wine_copy_5_scaled, columns=wine_copy_5.columns)
# ====================================================================
# from sklearn.preprocessing import StandardScaler
# # Create the scaler
# ss = StandardScaler()
# cleand_df_cp = ss.fit_transform(cleand_df_cp)
# #create a dataframe
# cleand_df_cp = pd.DataFrame(cleand_df_cp, columns=[])
# =====================================================================
# from category_encoders import BinaryEncoder
# bin_enc = BinaryEncoder(cols=obj_col)
# # fit and trasform the data
# out_enc = bin_enc.fit_transform(cleand_df_cp[obj_col])
# # concatenate the original to the converted values
# cleand_df_cp = pd.concat([cleand_df_cp, out_enc], axis=1)
# cleand_df_cp.drop_duplicates(subset=['Hour', 'month','year','Day_x'])
# wine_copy_4['Severity'].var()
# wine_copy_5_scaled_df['Severity']=wine_copy_4['Severity']
cleand_df_cp = cleand_df_cp.drop(
["description", "weekday", "Day_x", "year", "Precipitation(in)", "Humidity(%)"],
axis=1,
)
cleand_df_cp.Hour
# wine_copy_5_scaled_df.var().sort_values()
# m=np.absolute(np.log(np.absolute(cleand_df_cp)))
# m.var()
cleand_df_cp
# define a function to convert time to day or night
def day_night(x):
if (x > 18) or (x <= 6):
return "Night"
elif (x > 6) or (x <= 18):
return "Morning"
# applay day night function on weather data
sortred_weather["day_night"] = sortred_weather["Hour"].apply(day_night)
# sortred_weather
# Create day night column in df dataframe
def hr_func(ts):
return ts.hour
df["h"] = df["timestamp"].apply(hr_func)
df["day_night"] = df["h"].apply(day_night)
# Create column of date and hour
sortred_weather["date_hour"] = sortred_weather["date"].astype(str) + sortred_weather[
"day_night"
].astype(str)
sortred_weather = sortred_weather.drop_duplicates("date_hour")
sortred_weather.info()
holidays["date"] = pd.to_datetime(holidays["date"])
holidays.head()
# # Merge weather and holidays data with main dataframe
# Merge weather data with main dataframe
df_joined = pd.merge(df, sortred_weather, on=["date", "day_night"], how="left")
df_joined = pd.merge(df_joined, holidays, on=["date"], how="left")
df_joined.info()
df_joined[["h", "day_night", "Hour", "date_hour"]]
# # take a look about null data
# df_joined.info()
df_joined.isnull().sum().sort_values(ascending=False)
# # Change data types and drop dummy column
# we will drop ID column later now we noticed that **1- Wind_Chill(F)** have lots of nulls (3138).** 2- Bump** all of its data is just false. **3- Give_Way** also is just all **false** except 3 data sets. **4- All other dummy column also as "No_Exit","Roundabout",..etc**
# cleand_df
df_joined["year"] = df_joined.date.dt.year
df_joined["month"] = df_joined.date.dt.month
df_joined["day"] = df_joined.date.dt.day
cleand_df = df_joined.drop(
[
"date",
"ID",
"Wind_Chill(F)",
"Bump",
"Give_Way",
"date_hour",
"Hour",
"No_Exit",
"Roundabout",
"day_night",
"timestamp",
],
axis=1,
)
cleand_df.info()
# ["Crossing",]
# # Replace null with suitable values
cleand_df.isnull().sum().sort_values(ascending=False).head(10)
cleand_df["Precipitation(in)"].describe()
cleand_df["Temperature(F)"] = cleand_df["Temperature(F)"].fillna(
cleand_df["Temperature(F)"].mean()
)
cleand_df["Visibility(mi)"] = cleand_df["Visibility(mi)"].fillna(
cleand_df["Visibility(mi)"].mean()
)
cleand_df["Humidity(%)"] = cleand_df["Humidity(%)"].fillna(
cleand_df["Humidity(%)"].mean()
)
cleand_df["Weather_Condition"] = cleand_df["Weather_Condition"].fillna("Partly Cloudy")
cleand_df["Wind_Speed(mph)"] = cleand_df["Wind_Speed(mph)"].fillna(
cleand_df["Wind_Speed(mph)"].mean()
)
cleand_df["Precipitation(in)"] = cleand_df["Precipitation(in)"].fillna(
cleand_df["Precipitation(in)"].mean()
)
cleand_df["description"] = cleand_df["description"].fillna("None")
# # There is no missing values now!
cleand_df.isnull().sum().sort_values(ascending=False).head(5)
# # Now we want to change categorical columns data to numerical columns
bool_df = cleand_df.select_dtypes(include="bool")
bool_df.columns
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for i in bool_df.columns:
cleand_df[i] = le.fit_transform(cleand_df[i])
cleand_df["Selected"] = le.fit_transform(cleand_df["Selected"])
obj_col = ["description", "Weather_Condition", "Side"]
for i in obj_col:
cleand_df = pd.get_dummies(cleand_df, prefix=[i], columns=[i])
# from category_encoders import BinaryEncoder
# bin_enc = BinaryEncoder(cols=obj_col)
# # fit and trasform the data
# out_enc = bin_enc.fit_transform(cleand_df_cp[obj_col])
# # concatenate the original to the converted values
# cleand_df_cp = pd.concat([cleand_df_cp, out_enc], axis=1)
# ## You're here!
# Welcome to your first competition in the [ITI's AI Pro training program](https://ai.iti.gov.eg/epita/ai-engineer/)! We hope you enjoy and learn as much as we did prepairing this competition.
# ## Introduction
# In the competition, it's required to predict the `Severity` of a car crash given info about the crash, e.g., location.
# This is the getting started notebook. Things are kept simple so that it's easier to understand the steps and modify it.
# Feel free to `Fork` this notebook and share it with your modifications **OR** use it to create your submissions.
# ### Prerequisites
# You should know how to use python and a little bit of Machine Learning. You can apply the techniques you learned in the training program and submit the new solutions!
# ### Checklist
# You can participate in this competition the way you perefer. However, I recommend following these steps if this is your first time joining a competition on Kaggle.
# * Fork this notebook and run the cells in order.
# * Submit this solution.
# * Make changes to the data processing step as you see fit.
# * Submit the new solutions.
# *You can submit up to 5 submissions per day. You can select only one of the submission you make to be considered in the final ranking.*
# Don't hesitate to leave a comment or contact me if you have any question!
# ## Import the libraries
# We'll use `pandas` to load and manipulate the data. Other libraries will be imported in the relevant sections.
import pandas as pd
import os
# ## Exploratory Data Analysis
# In this step, one should load the data and analyze it. However, I'll load the data and do minimal analysis. You are encouraged to do thorough analysis!
# Let's load the data using `pandas` and have a look at the generated `DataFrame`.
# dataset_path = '/kaggle/input/car-crashes-severity-prediction/'
# df = pd.read_csv(os.path.join(dataset_path, 'train.csv'))
# print("The shape of the dataset is {}.\n\n".format(df.shape))
# df.head()
# We've got 6407 examples in the dataset with 14 featues, 1 ID, and the `Severity` of the crash.
# By looking at the features and a sample from the data, the features look of numerical and catogerical types. What about some descriptive statistics?
# The output shows desciptive statistics for the numerical features, `Lat`, `Lng`, `Distance(mi)`, and `Severity`. I'll use the numerical features to demonstrate how to train the model and make submissions. **However you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# ## Data Splitting
# Now it's time to split the dataset for the training step. Typically the dataset is split into 3 subsets, namely, the training, validation and test sets. In our case, the test set is already predefined. So we'll split the "training" set into training and validation sets with 0.8:0.2 ratio.
# *Note: a good way to generate reproducible results is to set the seed to the algorithms that depends on randomization. This is done with the argument `random_state` in the following command*
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
cleand_df_cp, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(columns=["Severity"])
y_train = train_df["Severity"]
X_val = val_df.drop(columns=["Severity"])
y_val = val_df["Severity"]
# As pointed out eariler, I'll use the numerical features to train the classifier. **However, you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# # This cell is used to select the numerical features. IT SHOULD BE REMOVED AS YOU DO YOUR WORK.
# X_train = X_train[['Lat', 'Lng', 'Distance(mi)']]
# X_val = X_val[['Lat', 'Lng', 'Distance(mi)']]
# ## Model Training
# Let's train a model with the data! We'll train a Random Forest Classifier to demonstrate the process of making submissions.
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
# Now let's test our classifier on the validation dataset and see the accuracy.
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# Well. That's a good start, right? A classifier that predicts all examples' `Severity` as 2 will get around 0.63. You should get better score as you add more features and do better data preprocessing.
# ## Submission File Generation
# We have built a model and we'd like to submit our predictions on the test set! In order to do that, we'll load the test set, predict the class and save the submission file.
# First, we'll load the data.
# test_df = pd.read_csv(os.path.join(dataset_path, 'test.csv'))
# test_df.head()
test
import datetime as dt
# Convert object to date time
test["timestamp"] = pd.to_datetime(test["timestamp"])
# Create date column in df dataframe
test["date"] = [d.date() for d in test["timestamp"]]
test["date"] = pd.to_datetime(test["date"])
weather_ts = weather.copy()
test["year"] = test.date.dt.year
test["month"] = test.date.dt.month
test["Day"] = test.date.dt.day
test["weekday"] = test[["date"]].apply(
lambda x: dt.datetime.strftime(x["date"], "%A"), axis=1
)
def hr_func(ts):
return ts.hour
test["Hour"] = test["timestamp"].apply(hr_func)
# ========================================
holidays_ts = holidays.copy()
holidays_ts["date"] = pd.to_datetime(holidays_ts["date"])
# ========================================
test1 = pd.merge(test, weather_ts, on=["date", "Hour"], how="left")
test2 = pd.merge(test1, holidays_ts, on=["date"], how="left")
test2_cp = test2.drop(
[
"Junction",
"Year",
"Month",
"Day_y",
"date",
"ID",
"Wind_Chill(F)",
"Bump",
"Give_Way",
"No_Exit",
"Roundabout",
"timestamp",
],
axis=1,
)
# cleand_df_cp.info()
test2_cp["Temperature(F)"] = test2_cp["Temperature(F)"].fillna(
test2_cp["Temperature(F)"].mean()
)
test2_cp["Visibility(mi)"] = test2_cp["Visibility(mi)"].fillna(
test2_cp["Visibility(mi)"].mean()
)
test2_cp["Humidity(%)"] = test2_cp["Humidity(%)"].fillna(test2_cp["Humidity(%)"].mean())
test2_cp["Weather_Condition"] = test2_cp["Weather_Condition"].fillna("Plarty Cloudy")
test2_cp["Wind_Speed(mph)"] = test2_cp["Wind_Speed(mph)"].fillna(
test2_cp["Wind_Speed(mph)"].mean()
)
test2_cp["Precipitation(in)"] = test2_cp["Precipitation(in)"].fillna(
test2_cp["Precipitation(in)"].mean()
)
test2_cp["description"] = test2_cp["description"].fillna(0)
test2_cp.isnull().sum().sort_values(ascending=False).head(5)
test2_cp["H_day"] = test2_cp["description"].copy()
bool_d = test2_cp.select_dtypes(include="bool")
# bool_df1.columns
for i in range(len(test2_cp["description"])):
if test2_cp["description"][i] != 0:
test2_cp["H_day"][i] = 1
for i in range(len(test2_cp["weekday"])):
if test2_cp["weekday"][i] == "Sunday":
test2_cp["H_day"][i] = 1
elif test2_cp["weekday"][i] == "Saturday":
test2_cp["H_day"][i] = 1
else:
test2_cp["H_day"][i] = 0
for i in range(len(test2_cp["Hour"])):
if (test2_cp["Hour"][i] > 18) or (test2_cp["Hour"][i] <= 6):
test2_cp["Hour"][i] = 1
else:
test2_cp["Hour"][i] = 0
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for i in bool_d.columns:
test2_cp[i] = le.fit_transform(test2_cp[i])
test2_cp["Selected"] = le.fit_transform(test2_cp["Selected"])
test2_cp["Weather_Condition"] = le.fit_transform(test2_cp["Weather_Condition"])
test2_cp["Side"] = le.fit_transform(test2_cp["Side"])
test2_cp = test2_cp.drop(
["description", "weekday", "Day_x", "year", "Precipitation(in)", "Humidity(%)"],
axis=1,
)
# Note that the test set has the same features and doesn't have the `Severity` column.
# At this stage one must **NOT** forget to apply the same processing done on the training set on the features of the test set.
# Now we'll add `Severity` column to the test `DataFrame` and add the values of the predicted class to it.
# **I'll select the numerical features here as I did in the training set. DO NOT forget to change this step as you change the preprocessing of the training data.**
X_test = test2_cp
# # You should update/remove the next line once you change the features used for training
# X_test = X_test[['Lat', 'Lng', 'Distance(mi)']]
y_test_predicted = classifier.predict(X_test)
test2_cp["Severity"] = y_test_predicted
test2_cp.head()
# Now we're ready to generate the submission file. The submission file needs the columns `ID` and `Severity` only.
test2_cp[["ID", "Severity"]].to_csv("/kaggle/working/submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/141/69141185.ipynb
| null | null |
[{"Id": 69141185, "ScriptId": 18846191, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7771740, "CreationDate": "07/27/2021 10:26:50", "VersionNumber": 2.0, "Title": "Getting Started - Car Crashes' Severity Prediction", "EvaluationDate": "07/27/2021", "IsChange": false, "TotalLines": 490.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 490.0, "LinesInsertedFromFork": 374.0, "LinesDeletedFromFork": 26.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 116.0, "TotalVotes": 0}]
| null | null | null | null |
# # Reading Files
import pandas as pd
import matplotlib.pyplot as plt
plt.style.use("ggplot")
import re
import seaborn as sns
import os
import numpy as np
import pandas as pd
df = pd.read_csv("../input/car-crashes-severity-prediction/train.csv")
holidays = pd.read_csv("../input/holidayscsv/holidays.csv")
weather = pd.read_csv("../input/car-crashes-severity-prediction/weather-sfcsv.csv")
test = pd.read_csv("../input/car-crashes-severity-prediction/test.csv")
print("The shape of the dataset is {}.\n\n".format(df.shape))
test.Severity
# # Convert type of timestamp Column
# Convert object to date time
df["timestamp"] = pd.to_datetime(df["timestamp"])
# Create date column in df dataframe
df["date"] = [d.date() for d in df["timestamp"]]
df["date"] = pd.to_datetime(df["date"])
# Create date column in weather dataframe
weather["date"] = (
weather["Year"].astype(str)
+ "-"
+ weather["Month"].astype(str)
+ "-"
+ weather["Day"].astype(str)
)
weather["date"] = pd.to_datetime(weather["date"])
# Temporary weather dataframe
weather_1 = weather.drop(["Year", "Day", "Month"], axis=1)
# Take a look on sorted weather
sortred_weather = weather_1.sort_values(by=["date"])
weather = weather.drop_duplicates(subset=["Hour", "date"])
df.info()
import datetime as dt
weather_cp = weather.copy()
df_cp = df.copy()
df_cp["year"] = df_cp.date.dt.year
df_cp["month"] = df_cp.date.dt.month
df_cp["Day"] = df_cp.date.dt.day
df_cp["weekday"] = df_cp[["date"]].apply(
lambda x: dt.datetime.strftime(x["date"], "%A"), axis=1
)
def hr_func(ts):
return ts.hour
df_cp["Hour"] = df_cp["timestamp"].apply(hr_func)
# ========================================
holidays_cp = holidays.copy()
holidays_cp["date"] = pd.to_datetime(holidays_cp["date"])
# ========================================
df_cp1 = pd.merge(df_cp, weather_cp, on=["date", "Hour"], how="left")
df_cp2 = pd.merge(df_cp1, holidays_cp, on=["date"], how="left")
cleand_df_cp = df_cp2.drop(
[
"Junction",
"Year",
"Month",
"Day_y",
"date",
"ID",
"Wind_Chill(F)",
"Bump",
"Give_Way",
"No_Exit",
"Roundabout",
"timestamp",
],
axis=1,
)
# cleand_df_cp.info()
cleand_df_cp["Temperature(F)"] = cleand_df_cp["Temperature(F)"].fillna(
cleand_df_cp["Temperature(F)"].mean()
)
cleand_df_cp["Visibility(mi)"] = cleand_df_cp["Visibility(mi)"].fillna(
cleand_df_cp["Visibility(mi)"].mean()
)
cleand_df_cp["Humidity(%)"] = cleand_df_cp["Humidity(%)"].fillna(
cleand_df_cp["Humidity(%)"].mean()
)
cleand_df_cp["Weather_Condition"] = cleand_df_cp["Weather_Condition"].fillna(
"Plarty Cloudy"
)
cleand_df_cp["Wind_Speed(mph)"] = cleand_df_cp["Wind_Speed(mph)"].fillna(
cleand_df_cp["Wind_Speed(mph)"].mean()
)
cleand_df_cp["Precipitation(in)"] = cleand_df_cp["Precipitation(in)"].fillna(
cleand_df_cp["Precipitation(in)"].mean()
)
cleand_df_cp["description"] = cleand_df_cp["description"].fillna(0)
cleand_df_cp.isnull().sum().sort_values(ascending=False).head(5)
cleand_df_cp["H_day"] = cleand_df_cp["description"].copy()
bool_df1 = cleand_df_cp.select_dtypes(include="bool")
# bool_df1.columns
for i in range(len(cleand_df_cp["description"])):
if cleand_df_cp["description"][i] != 0:
cleand_df_cp["H_day"][i] = 1
for i in range(len(cleand_df_cp["weekday"])):
if cleand_df_cp["weekday"][i] == "Sunday":
cleand_df_cp["H_day"][i] = 1
elif cleand_df_cp["weekday"][i] == "Saturday":
cleand_df_cp["H_day"][i] = 1
else:
cleand_df_cp["H_day"][i] = 0
for i in range(len(cleand_df_cp["Hour"])):
if (cleand_df_cp["Hour"][i] > 18) or (cleand_df_cp["Hour"][i] <= 6):
cleand_df_cp["Hour"][i] = 1
else:
cleand_df_cp["Hour"][i] = 0
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for i in bool_df1.columns:
cleand_df_cp[i] = le.fit_transform(cleand_df_cp[i])
cleand_df_cp["Selected"] = le.fit_transform(cleand_df_cp["Selected"])
cleand_df_cp["Weather_Condition"] = le.fit_transform(cleand_df_cp["Weather_Condition"])
cleand_df_cp["Side"] = le.fit_transform(cleand_df_cp["Side"])
# obj_col1 = ["Side"]
# for i in obj_col1:
# cleand_df_cp = pd.get_dummies(cleand_df_cp, prefix=[i], columns=[i])
# ====================================================================
# from sklearn.preprocessing import MinMaxScaler
# # Create the scaler
# mms = MinMaxScaler()
# # create a copy of wine
# wine_copy_4 = cleand_df_cp.copy()
# wine_copy_5 = wine_copy_4.drop(columns=['Severity'])
# # Scaled data
# wine_copy_5_scaled = mms.fit_transform(wine_copy_5)
# #create a dataframe
# wine_copy_5_scaled_df = pd.DataFrame(wine_copy_5_scaled, columns=wine_copy_5.columns)
# ====================================================================
# from sklearn.preprocessing import StandardScaler
# # Create the scaler
# ss = StandardScaler()
# cleand_df_cp = ss.fit_transform(cleand_df_cp)
# #create a dataframe
# cleand_df_cp = pd.DataFrame(cleand_df_cp, columns=[])
# =====================================================================
# from category_encoders import BinaryEncoder
# bin_enc = BinaryEncoder(cols=obj_col)
# # fit and trasform the data
# out_enc = bin_enc.fit_transform(cleand_df_cp[obj_col])
# # concatenate the original to the converted values
# cleand_df_cp = pd.concat([cleand_df_cp, out_enc], axis=1)
# cleand_df_cp.drop_duplicates(subset=['Hour', 'month','year','Day_x'])
# wine_copy_4['Severity'].var()
# wine_copy_5_scaled_df['Severity']=wine_copy_4['Severity']
cleand_df_cp = cleand_df_cp.drop(
["description", "weekday", "Day_x", "year", "Precipitation(in)", "Humidity(%)"],
axis=1,
)
cleand_df_cp.Hour
# wine_copy_5_scaled_df.var().sort_values()
# m=np.absolute(np.log(np.absolute(cleand_df_cp)))
# m.var()
cleand_df_cp
# define a function to convert time to day or night
def day_night(x):
if (x > 18) or (x <= 6):
return "Night"
elif (x > 6) or (x <= 18):
return "Morning"
# applay day night function on weather data
sortred_weather["day_night"] = sortred_weather["Hour"].apply(day_night)
# sortred_weather
# Create day night column in df dataframe
def hr_func(ts):
return ts.hour
df["h"] = df["timestamp"].apply(hr_func)
df["day_night"] = df["h"].apply(day_night)
# Create column of date and hour
sortred_weather["date_hour"] = sortred_weather["date"].astype(str) + sortred_weather[
"day_night"
].astype(str)
sortred_weather = sortred_weather.drop_duplicates("date_hour")
sortred_weather.info()
holidays["date"] = pd.to_datetime(holidays["date"])
holidays.head()
# # Merge weather and holidays data with main dataframe
# Merge weather data with main dataframe
df_joined = pd.merge(df, sortred_weather, on=["date", "day_night"], how="left")
df_joined = pd.merge(df_joined, holidays, on=["date"], how="left")
df_joined.info()
df_joined[["h", "day_night", "Hour", "date_hour"]]
# # take a look about null data
# df_joined.info()
df_joined.isnull().sum().sort_values(ascending=False)
# # Change data types and drop dummy column
# we will drop ID column later now we noticed that **1- Wind_Chill(F)** have lots of nulls (3138).** 2- Bump** all of its data is just false. **3- Give_Way** also is just all **false** except 3 data sets. **4- All other dummy column also as "No_Exit","Roundabout",..etc**
# cleand_df
df_joined["year"] = df_joined.date.dt.year
df_joined["month"] = df_joined.date.dt.month
df_joined["day"] = df_joined.date.dt.day
cleand_df = df_joined.drop(
[
"date",
"ID",
"Wind_Chill(F)",
"Bump",
"Give_Way",
"date_hour",
"Hour",
"No_Exit",
"Roundabout",
"day_night",
"timestamp",
],
axis=1,
)
cleand_df.info()
# ["Crossing",]
# # Replace null with suitable values
cleand_df.isnull().sum().sort_values(ascending=False).head(10)
cleand_df["Precipitation(in)"].describe()
cleand_df["Temperature(F)"] = cleand_df["Temperature(F)"].fillna(
cleand_df["Temperature(F)"].mean()
)
cleand_df["Visibility(mi)"] = cleand_df["Visibility(mi)"].fillna(
cleand_df["Visibility(mi)"].mean()
)
cleand_df["Humidity(%)"] = cleand_df["Humidity(%)"].fillna(
cleand_df["Humidity(%)"].mean()
)
cleand_df["Weather_Condition"] = cleand_df["Weather_Condition"].fillna("Partly Cloudy")
cleand_df["Wind_Speed(mph)"] = cleand_df["Wind_Speed(mph)"].fillna(
cleand_df["Wind_Speed(mph)"].mean()
)
cleand_df["Precipitation(in)"] = cleand_df["Precipitation(in)"].fillna(
cleand_df["Precipitation(in)"].mean()
)
cleand_df["description"] = cleand_df["description"].fillna("None")
# # There is no missing values now!
cleand_df.isnull().sum().sort_values(ascending=False).head(5)
# # Now we want to change categorical columns data to numerical columns
bool_df = cleand_df.select_dtypes(include="bool")
bool_df.columns
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for i in bool_df.columns:
cleand_df[i] = le.fit_transform(cleand_df[i])
cleand_df["Selected"] = le.fit_transform(cleand_df["Selected"])
obj_col = ["description", "Weather_Condition", "Side"]
for i in obj_col:
cleand_df = pd.get_dummies(cleand_df, prefix=[i], columns=[i])
# from category_encoders import BinaryEncoder
# bin_enc = BinaryEncoder(cols=obj_col)
# # fit and trasform the data
# out_enc = bin_enc.fit_transform(cleand_df_cp[obj_col])
# # concatenate the original to the converted values
# cleand_df_cp = pd.concat([cleand_df_cp, out_enc], axis=1)
# ## You're here!
# Welcome to your first competition in the [ITI's AI Pro training program](https://ai.iti.gov.eg/epita/ai-engineer/)! We hope you enjoy and learn as much as we did prepairing this competition.
# ## Introduction
# In the competition, it's required to predict the `Severity` of a car crash given info about the crash, e.g., location.
# This is the getting started notebook. Things are kept simple so that it's easier to understand the steps and modify it.
# Feel free to `Fork` this notebook and share it with your modifications **OR** use it to create your submissions.
# ### Prerequisites
# You should know how to use python and a little bit of Machine Learning. You can apply the techniques you learned in the training program and submit the new solutions!
# ### Checklist
# You can participate in this competition the way you perefer. However, I recommend following these steps if this is your first time joining a competition on Kaggle.
# * Fork this notebook and run the cells in order.
# * Submit this solution.
# * Make changes to the data processing step as you see fit.
# * Submit the new solutions.
# *You can submit up to 5 submissions per day. You can select only one of the submission you make to be considered in the final ranking.*
# Don't hesitate to leave a comment or contact me if you have any question!
# ## Import the libraries
# We'll use `pandas` to load and manipulate the data. Other libraries will be imported in the relevant sections.
import pandas as pd
import os
# ## Exploratory Data Analysis
# In this step, one should load the data and analyze it. However, I'll load the data and do minimal analysis. You are encouraged to do thorough analysis!
# Let's load the data using `pandas` and have a look at the generated `DataFrame`.
# dataset_path = '/kaggle/input/car-crashes-severity-prediction/'
# df = pd.read_csv(os.path.join(dataset_path, 'train.csv'))
# print("The shape of the dataset is {}.\n\n".format(df.shape))
# df.head()
# We've got 6407 examples in the dataset with 14 featues, 1 ID, and the `Severity` of the crash.
# By looking at the features and a sample from the data, the features look of numerical and catogerical types. What about some descriptive statistics?
# The output shows desciptive statistics for the numerical features, `Lat`, `Lng`, `Distance(mi)`, and `Severity`. I'll use the numerical features to demonstrate how to train the model and make submissions. **However you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# ## Data Splitting
# Now it's time to split the dataset for the training step. Typically the dataset is split into 3 subsets, namely, the training, validation and test sets. In our case, the test set is already predefined. So we'll split the "training" set into training and validation sets with 0.8:0.2 ratio.
# *Note: a good way to generate reproducible results is to set the seed to the algorithms that depends on randomization. This is done with the argument `random_state` in the following command*
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
cleand_df_cp, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(columns=["Severity"])
y_train = train_df["Severity"]
X_val = val_df.drop(columns=["Severity"])
y_val = val_df["Severity"]
# As pointed out eariler, I'll use the numerical features to train the classifier. **However, you shouldn't use the numerical features only to make the final submission if you want to make it to the top of the leaderboard.**
# # This cell is used to select the numerical features. IT SHOULD BE REMOVED AS YOU DO YOUR WORK.
# X_train = X_train[['Lat', 'Lng', 'Distance(mi)']]
# X_val = X_val[['Lat', 'Lng', 'Distance(mi)']]
# ## Model Training
# Let's train a model with the data! We'll train a Random Forest Classifier to demonstrate the process of making submissions.
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
# Now let's test our classifier on the validation dataset and see the accuracy.
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# Well. That's a good start, right? A classifier that predicts all examples' `Severity` as 2 will get around 0.63. You should get better score as you add more features and do better data preprocessing.
# ## Submission File Generation
# We have built a model and we'd like to submit our predictions on the test set! In order to do that, we'll load the test set, predict the class and save the submission file.
# First, we'll load the data.
# test_df = pd.read_csv(os.path.join(dataset_path, 'test.csv'))
# test_df.head()
test
import datetime as dt
# Convert object to date time
test["timestamp"] = pd.to_datetime(test["timestamp"])
# Create date column in df dataframe
test["date"] = [d.date() for d in test["timestamp"]]
test["date"] = pd.to_datetime(test["date"])
weather_ts = weather.copy()
test["year"] = test.date.dt.year
test["month"] = test.date.dt.month
test["Day"] = test.date.dt.day
test["weekday"] = test[["date"]].apply(
lambda x: dt.datetime.strftime(x["date"], "%A"), axis=1
)
def hr_func(ts):
return ts.hour
test["Hour"] = test["timestamp"].apply(hr_func)
# ========================================
holidays_ts = holidays.copy()
holidays_ts["date"] = pd.to_datetime(holidays_ts["date"])
# ========================================
test1 = pd.merge(test, weather_ts, on=["date", "Hour"], how="left")
test2 = pd.merge(test1, holidays_ts, on=["date"], how="left")
test2_cp = test2.drop(
[
"Junction",
"Year",
"Month",
"Day_y",
"date",
"ID",
"Wind_Chill(F)",
"Bump",
"Give_Way",
"No_Exit",
"Roundabout",
"timestamp",
],
axis=1,
)
# cleand_df_cp.info()
test2_cp["Temperature(F)"] = test2_cp["Temperature(F)"].fillna(
test2_cp["Temperature(F)"].mean()
)
test2_cp["Visibility(mi)"] = test2_cp["Visibility(mi)"].fillna(
test2_cp["Visibility(mi)"].mean()
)
test2_cp["Humidity(%)"] = test2_cp["Humidity(%)"].fillna(test2_cp["Humidity(%)"].mean())
test2_cp["Weather_Condition"] = test2_cp["Weather_Condition"].fillna("Plarty Cloudy")
test2_cp["Wind_Speed(mph)"] = test2_cp["Wind_Speed(mph)"].fillna(
test2_cp["Wind_Speed(mph)"].mean()
)
test2_cp["Precipitation(in)"] = test2_cp["Precipitation(in)"].fillna(
test2_cp["Precipitation(in)"].mean()
)
test2_cp["description"] = test2_cp["description"].fillna(0)
test2_cp.isnull().sum().sort_values(ascending=False).head(5)
test2_cp["H_day"] = test2_cp["description"].copy()
bool_d = test2_cp.select_dtypes(include="bool")
# bool_df1.columns
for i in range(len(test2_cp["description"])):
if test2_cp["description"][i] != 0:
test2_cp["H_day"][i] = 1
for i in range(len(test2_cp["weekday"])):
if test2_cp["weekday"][i] == "Sunday":
test2_cp["H_day"][i] = 1
elif test2_cp["weekday"][i] == "Saturday":
test2_cp["H_day"][i] = 1
else:
test2_cp["H_day"][i] = 0
for i in range(len(test2_cp["Hour"])):
if (test2_cp["Hour"][i] > 18) or (test2_cp["Hour"][i] <= 6):
test2_cp["Hour"][i] = 1
else:
test2_cp["Hour"][i] = 0
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for i in bool_d.columns:
test2_cp[i] = le.fit_transform(test2_cp[i])
test2_cp["Selected"] = le.fit_transform(test2_cp["Selected"])
test2_cp["Weather_Condition"] = le.fit_transform(test2_cp["Weather_Condition"])
test2_cp["Side"] = le.fit_transform(test2_cp["Side"])
test2_cp = test2_cp.drop(
["description", "weekday", "Day_x", "year", "Precipitation(in)", "Humidity(%)"],
axis=1,
)
# Note that the test set has the same features and doesn't have the `Severity` column.
# At this stage one must **NOT** forget to apply the same processing done on the training set on the features of the test set.
# Now we'll add `Severity` column to the test `DataFrame` and add the values of the predicted class to it.
# **I'll select the numerical features here as I did in the training set. DO NOT forget to change this step as you change the preprocessing of the training data.**
X_test = test2_cp
# # You should update/remove the next line once you change the features used for training
# X_test = X_test[['Lat', 'Lng', 'Distance(mi)']]
y_test_predicted = classifier.predict(X_test)
test2_cp["Severity"] = y_test_predicted
test2_cp.head()
# Now we're ready to generate the submission file. The submission file needs the columns `ID` and `Severity` only.
test2_cp[["ID", "Severity"]].to_csv("/kaggle/working/submission.csv", index=False)
| false | 0 | 5,889 | 0 | 5,889 | 5,889 |
||
69141621
|
import pandas as pd
import numpy as np
import xml.etree.ElementTree as ET
from sklearn import preprocessing
import os
# convert XML into pandas DF #
root = ET.parse("/kaggle/input/car-crashes-severity-prediction/holidays.xml").getroot()
tags = {"tags": []}
for elem in root:
tag = {}
tag["date"] = elem[0].text
# tag["des"] = elem[1].text
tag["ones"] = 1
tags["tags"].append(tag)
df_holidays = pd.DataFrame(tags["tags"])
# df_holidays['date'] = pd.to_datetime(df_holidays['date']).dt.date
df_holidays.head()
df_holidays.info()
df_weather = pd.read_csv(
"/kaggle/input/car-crashes-severity-prediction/weather-sfcsv.csv"
)
df_weather.drop_duplicates(
subset=["Year", "Month", "Day", "Hour"],
keep="first",
inplace=True,
ignore_index=True,
)
df_weather["date"] = pd.to_datetime(
(df_weather.Year * 10000 + df_weather.Month * 100 + df_weather.Day).apply(str),
format="%Y%m%d",
).dt.date
df_weather.fillna(df_weather.mean(), inplace=True)
df_weather.head()
# convert weather conditon into categories #
t = df_weather[["Weather_Condition"]].dropna()
lencod = preprocessing.LabelEncoder()
t = t.apply(lencod.fit_transform)
df_weather["nu_wc"] = t
t.head(10)
df_weather.info()
df_train = pd.read_csv("/kaggle/input/car-crashes-severity-prediction/train.csv")
df_train.head()
# transform catogry data #
numerical_train = df_train[
[
"Bump",
"Crossing",
"Give_Way",
"Junction",
"No_Exit",
"Railway",
"Roundabout",
"Stop",
"Amenity",
"Side",
]
].dropna()
le = preprocessing.LabelEncoder()
numerical_train = numerical_train.apply(le.fit_transform)
df_train[
[
"Bump",
"Crossing",
"Give_Way",
"Junction",
"No_Exit",
"Railway",
"Roundabout",
"Stop",
"Amenity",
"Side",
]
] = numerical_train[
[
"Bump",
"Crossing",
"Give_Way",
"Junction",
"No_Exit",
"Railway",
"Roundabout",
"Stop",
"Amenity",
"Side",
]
]
df_train.info()
## Format train datat set date colum for merge with weather data set ##
df_train["date"] = pd.to_datetime(df_train["timestamp"]).dt.date
df_train["Hour"] = pd.to_datetime(df_train["timestamp"]).dt.hour
# dftrain=pd.read_csv('train.csv')
df_test = pd.read_csv("/kaggle/input/car-crashes-severity-prediction/test.csv")
df_test.head()
# format test df timestamp date #
df_test["date"] = pd.to_datetime(df_train["timestamp"]).dt.date
df_test["Hour"] = pd.to_datetime(df_train["timestamp"]).dt.hour
df_holiday_train = pd.merge(df_train, df_holidays, on="date", how="left").fillna(0)
df_holiday_test = pd.merge(df_test, df_holidays, on="date", how="left").fillna(0)
# merging data Frames #
df_train_final = pd.merge(
df_holiday_train,
df_weather,
how="left",
left_on=["date", "Hour"],
right_on=["date", "Hour"],
)
df_test_final = pd.merge(
df_holiday_test,
df_weather,
how="left",
left_on=["date", "Hour"],
right_on=["date", "Hour"],
)
# remove duplicates #
df_train_final.drop_duplicates(subset=["ID"], inplace=True, ignore_index=True)
df_test_final.drop_duplicates(subset=["ID"], inplace=True, ignore_index=True)
df_test_final["Visibility(mi)"].fillna(
df_test_final["Visibility(mi)"].mean(), inplace=True
)
df_train_final["Visibility(mi)"].fillna(
df_train_final["Visibility(mi)"].mean(), inplace=True
)
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
df_train_final, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(columns=["ID", "Severity"])
y_train = train_df["Severity"]
X_val = val_df.drop(columns=["ID", "Severity"])
y_val = val_df["Severity"]
X_train = X_train[
[
"Lat",
"Lng",
"Distance(mi)",
"Temperature(F)",
"Amenity",
"Crossing",
"Junction",
"Visibility(mi)",
]
]
X_val = X_val[
[
"Lat",
"Lng",
"Distance(mi)",
"Temperature(F)",
"Amenity",
"Crossing",
"Junction",
"Visibility(mi)",
]
]
X_test = df_test_final[
[
"Lat",
"Lng",
"Distance(mi)",
"Temperature(F)",
"Amenity",
"Crossing",
"Junction",
"Visibility(mi)",
]
]
y_train.size
X_train.info()
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# predict for x_test
y_prdicted = classifier.predict(X_test)
y_prdicted[:-20]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/141/69141621.ipynb
| null | null |
[{"Id": 69141621, "ScriptId": 18869319, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6363989, "CreationDate": "07/27/2021 10:33:29", "VersionNumber": 2.0, "Title": "notebookbe61bd6539", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 143.0, "LinesInsertedFromPrevious": 15.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 128.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import numpy as np
import xml.etree.ElementTree as ET
from sklearn import preprocessing
import os
# convert XML into pandas DF #
root = ET.parse("/kaggle/input/car-crashes-severity-prediction/holidays.xml").getroot()
tags = {"tags": []}
for elem in root:
tag = {}
tag["date"] = elem[0].text
# tag["des"] = elem[1].text
tag["ones"] = 1
tags["tags"].append(tag)
df_holidays = pd.DataFrame(tags["tags"])
# df_holidays['date'] = pd.to_datetime(df_holidays['date']).dt.date
df_holidays.head()
df_holidays.info()
df_weather = pd.read_csv(
"/kaggle/input/car-crashes-severity-prediction/weather-sfcsv.csv"
)
df_weather.drop_duplicates(
subset=["Year", "Month", "Day", "Hour"],
keep="first",
inplace=True,
ignore_index=True,
)
df_weather["date"] = pd.to_datetime(
(df_weather.Year * 10000 + df_weather.Month * 100 + df_weather.Day).apply(str),
format="%Y%m%d",
).dt.date
df_weather.fillna(df_weather.mean(), inplace=True)
df_weather.head()
# convert weather conditon into categories #
t = df_weather[["Weather_Condition"]].dropna()
lencod = preprocessing.LabelEncoder()
t = t.apply(lencod.fit_transform)
df_weather["nu_wc"] = t
t.head(10)
df_weather.info()
df_train = pd.read_csv("/kaggle/input/car-crashes-severity-prediction/train.csv")
df_train.head()
# transform catogry data #
numerical_train = df_train[
[
"Bump",
"Crossing",
"Give_Way",
"Junction",
"No_Exit",
"Railway",
"Roundabout",
"Stop",
"Amenity",
"Side",
]
].dropna()
le = preprocessing.LabelEncoder()
numerical_train = numerical_train.apply(le.fit_transform)
df_train[
[
"Bump",
"Crossing",
"Give_Way",
"Junction",
"No_Exit",
"Railway",
"Roundabout",
"Stop",
"Amenity",
"Side",
]
] = numerical_train[
[
"Bump",
"Crossing",
"Give_Way",
"Junction",
"No_Exit",
"Railway",
"Roundabout",
"Stop",
"Amenity",
"Side",
]
]
df_train.info()
## Format train datat set date colum for merge with weather data set ##
df_train["date"] = pd.to_datetime(df_train["timestamp"]).dt.date
df_train["Hour"] = pd.to_datetime(df_train["timestamp"]).dt.hour
# dftrain=pd.read_csv('train.csv')
df_test = pd.read_csv("/kaggle/input/car-crashes-severity-prediction/test.csv")
df_test.head()
# format test df timestamp date #
df_test["date"] = pd.to_datetime(df_train["timestamp"]).dt.date
df_test["Hour"] = pd.to_datetime(df_train["timestamp"]).dt.hour
df_holiday_train = pd.merge(df_train, df_holidays, on="date", how="left").fillna(0)
df_holiday_test = pd.merge(df_test, df_holidays, on="date", how="left").fillna(0)
# merging data Frames #
df_train_final = pd.merge(
df_holiday_train,
df_weather,
how="left",
left_on=["date", "Hour"],
right_on=["date", "Hour"],
)
df_test_final = pd.merge(
df_holiday_test,
df_weather,
how="left",
left_on=["date", "Hour"],
right_on=["date", "Hour"],
)
# remove duplicates #
df_train_final.drop_duplicates(subset=["ID"], inplace=True, ignore_index=True)
df_test_final.drop_duplicates(subset=["ID"], inplace=True, ignore_index=True)
df_test_final["Visibility(mi)"].fillna(
df_test_final["Visibility(mi)"].mean(), inplace=True
)
df_train_final["Visibility(mi)"].fillna(
df_train_final["Visibility(mi)"].mean(), inplace=True
)
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(
df_train_final, test_size=0.2, random_state=42
) # Try adding `stratify` here
X_train = train_df.drop(columns=["ID", "Severity"])
y_train = train_df["Severity"]
X_val = val_df.drop(columns=["ID", "Severity"])
y_val = val_df["Severity"]
X_train = X_train[
[
"Lat",
"Lng",
"Distance(mi)",
"Temperature(F)",
"Amenity",
"Crossing",
"Junction",
"Visibility(mi)",
]
]
X_val = X_val[
[
"Lat",
"Lng",
"Distance(mi)",
"Temperature(F)",
"Amenity",
"Crossing",
"Junction",
"Visibility(mi)",
]
]
X_test = df_test_final[
[
"Lat",
"Lng",
"Distance(mi)",
"Temperature(F)",
"Amenity",
"Crossing",
"Junction",
"Visibility(mi)",
]
]
y_train.size
X_train.info()
from sklearn.ensemble import RandomForestClassifier
# Create an instance of the classifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
# Train the classifier
classifier = classifier.fit(X_train, y_train)
print(
"The accuracy of the classifier on the validation set is ",
(classifier.score(X_val, y_val)),
)
# predict for x_test
y_prdicted = classifier.predict(X_test)
y_prdicted[:-20]
| false | 0 | 1,591 | 0 | 1,591 | 1,591 |
||
69141338
|
import numpy as np
import pandas as pd
import os
import warnings
import matplotlib.pyplot as plt
from sklearn.feature_selection import mutual_info_classif
from sklearn.ensemble import RandomForestClassifier
# read and combine train and test sets
train_data = pd.read_csv("../input/titanic/train.csv")
test_data = pd.read_csv("../input/titanic/test.csv")
# combine train/test sets to process together
complete_data = [train_data, test_data]
# look at the no.of missing values in the features of train set
train_missing_values = train_data.isnull().sum()
train_missing_values[0:]
# look at the no.of missing values in the features of test set
test_missing_values = test_data.isnull().sum()
test_missing_values[0:]
# visualize data distribution in Age Column
train_data["Age"].hist(bins=5)
# visualize data distribution in Fare Column
train_data["Fare"].hist(bins=20)
# getting mutual info for features to get an idea
def make_mi_scores(X, y, discrete_features):
mi_scores = mutual_info_classif(X, y, discrete_features=discrete_features)
mi_scores = pd.Series(mi_scores, name="MI Scores", index=X.columns)
mi_scores = mi_scores.sort_values(ascending=False)
return mi_scores
train_copy = train_data.copy()
target = train_copy.pop("Survived")
for colname in train_copy.select_dtypes(["object", "float"]):
train_copy[colname], _ = train_copy[colname].factorize()
discrete_features = train_copy.dtypes == int
mi_scores = make_mi_scores(train_copy, target, discrete_features)
mi_scores
def handleBasicMissingValues(complete_data):
for data in complete_data:
# fill missing values of Fare
data["Fare"].fillna(method="bfill", axis=0, inplace=True)
# dropping Cabin column due to many missing values
data.drop(["Cabin"], axis=1, inplace=True)
return complete_data
def fillAgeMissingValues(complete_data):
for data in complete_data:
# fill missing values of Age Column using grouped mean based on Title & Parch
miss_child = data[(data["Title"].str.contains("Miss")) & (data["Parch"] != 0)][
"Age"
].mean()
miss_young = data[(data["Title"].str.contains("Miss")) & (data["Parch"] == 0)][
"Age"
].mean()
mr = data[(data["Title"].str.contains("Mr"))]["Age"].mean()
mrs = data[(data["Title"].str.contains("Mrs"))]["Age"].mean()
master = data[(data["Title"].str.contains("Master"))]["Age"].mean()
data.loc[
(data["Age"].isna()) & (data["Title"].str.contains("Mrs")), "Age"
] = mrs
data.loc[(data["Age"].isna()) & (data["Title"].str.contains("Mr")), "Age"] = mr
data.loc[
(data["Age"].isna()) & (data["Title"].str.contains("Master")), "Age"
] = master
data.loc[
(data["Age"].isna())
& (data["Title"].str.contains("Miss"))
& (data["Parch"] != 0),
"Age",
] = miss_child
data.loc[
(data["Age"].isna())
& (data["Title"].str.contains("Miss"))
& (data["Parch"] == 0),
"Age",
] = miss_young
# fill any other missing value with population mean
data["Age"].fillna(value=data["Age"].mean(), axis=0, inplace=True)
return complete_data
def createNewFeatures(complete_data):
for data in complete_data:
# create column Title using Name
name_comma_seperated = data["Name"].str.split(",", n=1, expand=True)
title_extracted = name_comma_seperated[1].str.split(".", n=1, expand=True)
data["Title"] = title_extracted[0]
# create LogFare using Fare
data["LogFare"] = data.Fare.apply(np.log1p)
# create FamilyMembers using Parch & SibSp
data["FamilyMembers"] = data.Parch + data.SibSp
# create column TicketSuffix using Ticket, to extract number part
ticket_space_seperated = data["Ticket"].str.rsplit(" ", n=1)
ticket_suffixes = []
for item in ticket_space_seperated:
if len(item) == 1:
if item[0] != "LINE":
ticket_suffixes.append(int(item[0]))
else:
# assign 1000 for LINE since no number part is there
ticket_suffixes.append(1000)
else:
ticket_suffixes.append(int(item[1]))
data["TicketSuffix"] = ticket_suffixes
return complete_data
def binningData(complete_data):
ageBins = [0, 16, 32, 48, 64, 80]
ageLabels = [1, 2, 3, 4, 5]
fareBins = [0, 1, 2, 4, 7]
fareLabels = [1, 2, 3, 4]
ticketBins = [0, 1000, 10000, 20000, 30000, 100000, 200000, 300000, 400000, 4000000]
ticketLabels = [1, 2, 3, 4, 5, 6, 7, 8, 9]
for data in complete_data:
# binning Age
data["AgeGroup"] = pd.cut(
data["Age"], bins=ageBins, labels=ageLabels, right=True
)
# binning Fare
data["FareGroup"] = pd.cut(
data["LogFare"], bins=fareBins, labels=fareLabels, right=True
)
# binning TicketSuffix
data["TicketGroup"] = pd.cut(
data["TicketSuffix"], bins=ticketBins, labels=ticketLabels, right=True
)
return complete_data
complete_data = handleBasicMissingValues(complete_data)
complete_data = createNewFeatures(complete_data)
complete_data = fillAgeMissingValues(complete_data)
complete_data = binningData(complete_data)
train_data = complete_data[0]
test_data = complete_data[1]
y = train_data["Survived"]
features = [
"AgeGroup",
"Pclass",
"Sex",
"FamilyMembers",
"Parch",
"TicketGroup",
"FareGroup",
]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = RandomForestClassifier(n_estimators=100, max_depth=6, random_state=2)
model.fit(X, y)
predictions = model.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/141/69141338.ipynb
| null | null |
[{"Id": 69141338, "ScriptId": 18797438, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7890445, "CreationDate": "07/27/2021 10:29:00", "VersionNumber": 21.0, "Title": "Titanic Competition - UOM_170504N", "EvaluationDate": "07/27/2021", "IsChange": true, "TotalLines": 155.0, "LinesInsertedFromPrevious": 7.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 148.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import os
import warnings
import matplotlib.pyplot as plt
from sklearn.feature_selection import mutual_info_classif
from sklearn.ensemble import RandomForestClassifier
# read and combine train and test sets
train_data = pd.read_csv("../input/titanic/train.csv")
test_data = pd.read_csv("../input/titanic/test.csv")
# combine train/test sets to process together
complete_data = [train_data, test_data]
# look at the no.of missing values in the features of train set
train_missing_values = train_data.isnull().sum()
train_missing_values[0:]
# look at the no.of missing values in the features of test set
test_missing_values = test_data.isnull().sum()
test_missing_values[0:]
# visualize data distribution in Age Column
train_data["Age"].hist(bins=5)
# visualize data distribution in Fare Column
train_data["Fare"].hist(bins=20)
# getting mutual info for features to get an idea
def make_mi_scores(X, y, discrete_features):
mi_scores = mutual_info_classif(X, y, discrete_features=discrete_features)
mi_scores = pd.Series(mi_scores, name="MI Scores", index=X.columns)
mi_scores = mi_scores.sort_values(ascending=False)
return mi_scores
train_copy = train_data.copy()
target = train_copy.pop("Survived")
for colname in train_copy.select_dtypes(["object", "float"]):
train_copy[colname], _ = train_copy[colname].factorize()
discrete_features = train_copy.dtypes == int
mi_scores = make_mi_scores(train_copy, target, discrete_features)
mi_scores
def handleBasicMissingValues(complete_data):
for data in complete_data:
# fill missing values of Fare
data["Fare"].fillna(method="bfill", axis=0, inplace=True)
# dropping Cabin column due to many missing values
data.drop(["Cabin"], axis=1, inplace=True)
return complete_data
def fillAgeMissingValues(complete_data):
for data in complete_data:
# fill missing values of Age Column using grouped mean based on Title & Parch
miss_child = data[(data["Title"].str.contains("Miss")) & (data["Parch"] != 0)][
"Age"
].mean()
miss_young = data[(data["Title"].str.contains("Miss")) & (data["Parch"] == 0)][
"Age"
].mean()
mr = data[(data["Title"].str.contains("Mr"))]["Age"].mean()
mrs = data[(data["Title"].str.contains("Mrs"))]["Age"].mean()
master = data[(data["Title"].str.contains("Master"))]["Age"].mean()
data.loc[
(data["Age"].isna()) & (data["Title"].str.contains("Mrs")), "Age"
] = mrs
data.loc[(data["Age"].isna()) & (data["Title"].str.contains("Mr")), "Age"] = mr
data.loc[
(data["Age"].isna()) & (data["Title"].str.contains("Master")), "Age"
] = master
data.loc[
(data["Age"].isna())
& (data["Title"].str.contains("Miss"))
& (data["Parch"] != 0),
"Age",
] = miss_child
data.loc[
(data["Age"].isna())
& (data["Title"].str.contains("Miss"))
& (data["Parch"] == 0),
"Age",
] = miss_young
# fill any other missing value with population mean
data["Age"].fillna(value=data["Age"].mean(), axis=0, inplace=True)
return complete_data
def createNewFeatures(complete_data):
for data in complete_data:
# create column Title using Name
name_comma_seperated = data["Name"].str.split(",", n=1, expand=True)
title_extracted = name_comma_seperated[1].str.split(".", n=1, expand=True)
data["Title"] = title_extracted[0]
# create LogFare using Fare
data["LogFare"] = data.Fare.apply(np.log1p)
# create FamilyMembers using Parch & SibSp
data["FamilyMembers"] = data.Parch + data.SibSp
# create column TicketSuffix using Ticket, to extract number part
ticket_space_seperated = data["Ticket"].str.rsplit(" ", n=1)
ticket_suffixes = []
for item in ticket_space_seperated:
if len(item) == 1:
if item[0] != "LINE":
ticket_suffixes.append(int(item[0]))
else:
# assign 1000 for LINE since no number part is there
ticket_suffixes.append(1000)
else:
ticket_suffixes.append(int(item[1]))
data["TicketSuffix"] = ticket_suffixes
return complete_data
def binningData(complete_data):
ageBins = [0, 16, 32, 48, 64, 80]
ageLabels = [1, 2, 3, 4, 5]
fareBins = [0, 1, 2, 4, 7]
fareLabels = [1, 2, 3, 4]
ticketBins = [0, 1000, 10000, 20000, 30000, 100000, 200000, 300000, 400000, 4000000]
ticketLabels = [1, 2, 3, 4, 5, 6, 7, 8, 9]
for data in complete_data:
# binning Age
data["AgeGroup"] = pd.cut(
data["Age"], bins=ageBins, labels=ageLabels, right=True
)
# binning Fare
data["FareGroup"] = pd.cut(
data["LogFare"], bins=fareBins, labels=fareLabels, right=True
)
# binning TicketSuffix
data["TicketGroup"] = pd.cut(
data["TicketSuffix"], bins=ticketBins, labels=ticketLabels, right=True
)
return complete_data
complete_data = handleBasicMissingValues(complete_data)
complete_data = createNewFeatures(complete_data)
complete_data = fillAgeMissingValues(complete_data)
complete_data = binningData(complete_data)
train_data = complete_data[0]
test_data = complete_data[1]
y = train_data["Survived"]
features = [
"AgeGroup",
"Pclass",
"Sex",
"FamilyMembers",
"Parch",
"TicketGroup",
"FareGroup",
]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = RandomForestClassifier(n_estimators=100, max_depth=6, random_state=2)
model.fit(X, y)
predictions = model.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
| false | 0 | 1,833 | 0 | 1,833 | 1,833 |
||
69708893
|
<jupyter_start><jupyter_text>Credit Card Fraud Detection
Context
---------
It is important that credit card companies are able to recognize fraudulent credit card transactions so that customers are not charged for items that they did not purchase.
Content
---------
The dataset contains transactions made by credit cards in September 2013 by European cardholders.
This dataset presents transactions that occurred in two days, where we have 492 frauds out of 284,807 transactions. The dataset is highly unbalanced, the positive class (frauds) account for 0.172% of all transactions.
It contains only numerical input variables which are the result of a PCA transformation. Unfortunately, due to confidentiality issues, we cannot provide the original features and more background information about the data. Features V1, V2, ... V28 are the principal components obtained with PCA, the only features which have not been transformed with PCA are 'Time' and 'Amount'. Feature 'Time' contains the seconds elapsed between each transaction and the first transaction in the dataset. The feature 'Amount' is the transaction Amount, this feature can be used for example-dependant cost-sensitive learning. Feature 'Class' is the response variable and it takes value 1 in case of fraud and 0 otherwise.
Given the class imbalance ratio, we recommend measuring the accuracy using the Area Under the Precision-Recall Curve (AUPRC). Confusion matrix accuracy is not meaningful for unbalanced classification.
Update (03/05/2021)
---------
A simulator for transaction data has been released as part of the practical handbook on Machine Learning for Credit Card Fraud Detection - https://fraud-detection-handbook.github.io/fraud-detection-handbook/Chapter_3_GettingStarted/SimulatedDataset.html. We invite all practitioners interested in fraud detection datasets to also check out this data simulator, and the methodologies for credit card fraud detection presented in the book.
Acknowledgements
---------
The dataset has been collected and analysed during a research collaboration of Worldline and the Machine Learning Group (http://mlg.ulb.ac.be) of ULB (Université Libre de Bruxelles) on big data mining and fraud detection.
More details on current and past projects on related topics are available on [https://www.researchgate.net/project/Fraud-detection-5][1] and the page of the [DefeatFraud][2] project
Please cite the following works:
Andrea Dal Pozzolo, Olivier Caelen, Reid A. Johnson and Gianluca Bontempi. [Calibrating Probability with Undersampling for Unbalanced Classification.][3] In Symposium on Computational Intelligence and Data Mining (CIDM), IEEE, 2015
Dal Pozzolo, Andrea; Caelen, Olivier; Le Borgne, Yann-Ael; Waterschoot, Serge; Bontempi, Gianluca. [Learned lessons in credit card fraud detection from a practitioner perspective][4], Expert systems with applications,41,10,4915-4928,2014, Pergamon
Dal Pozzolo, Andrea; Boracchi, Giacomo; Caelen, Olivier; Alippi, Cesare; Bontempi, Gianluca. [Credit card fraud detection: a realistic modeling and a novel learning strategy,][5] IEEE transactions on neural networks and learning systems,29,8,3784-3797,2018,IEEE
Dal Pozzolo, Andrea [Adaptive Machine learning for credit card fraud detection][6] ULB MLG PhD thesis (supervised by G. Bontempi)
Carcillo, Fabrizio; Dal Pozzolo, Andrea; Le Borgne, Yann-Aël; Caelen, Olivier; Mazzer, Yannis; Bontempi, Gianluca. [Scarff: a scalable framework for streaming credit card fraud detection with Spark][7], Information fusion,41, 182-194,2018,Elsevier
Carcillo, Fabrizio; Le Borgne, Yann-Aël; Caelen, Olivier; Bontempi, Gianluca. [Streaming active learning strategies for real-life credit card fraud detection: assessment and visualization,][8] International Journal of Data Science and Analytics, 5,4,285-300,2018,Springer International Publishing
Bertrand Lebichot, Yann-Aël Le Borgne, Liyun He, Frederic Oblé, Gianluca Bontempi [Deep-Learning Domain Adaptation Techniques for Credit Cards Fraud Detection](https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection), INNSBDDL 2019: Recent Advances in Big Data and Deep Learning, pp 78-88, 2019
Fabrizio Carcillo, Yann-Aël Le Borgne, Olivier Caelen, Frederic Oblé, Gianluca Bontempi [Combining Unsupervised and Supervised Learning in Credit Card Fraud Detection ](https://www.researchgate.net/publication/333143698_Combining_Unsupervised_and_Supervised_Learning_in_Credit_Card_Fraud_Detection) Information Sciences, 2019
Yann-Aël Le Borgne, Gianluca Bontempi [Reproducible machine Learning for Credit Card Fraud Detection - Practical Handbook ](https://www.researchgate.net/publication/351283764_Machine_Learning_for_Credit_Card_Fraud_Detection_-_Practical_Handbook)
Bertrand Lebichot, Gianmarco Paldino, Wissam Siblini, Liyun He, Frederic Oblé, Gianluca Bontempi [Incremental learning strategies for credit cards fraud detection](https://www.researchgate.net/publication/352275169_Incremental_learning_strategies_for_credit_cards_fraud_detection), IInternational Journal of Data Science and Analytics
[1]: https://www.researchgate.net/project/Fraud-detection-5
[2]: https://mlg.ulb.ac.be/wordpress/portfolio_page/defeatfraud-assessment-and-validation-of-deep-feature-engineering-and-learning-solutions-for-fraud-detection/
[3]: https://www.researchgate.net/publication/283349138_Calibrating_Probability_with_Undersampling_for_Unbalanced_Classification
[4]: https://www.researchgate.net/publication/260837261_Learned_lessons_in_credit_card_fraud_detection_from_a_practitioner_perspective
[5]: https://www.researchgate.net/publication/319867396_Credit_Card_Fraud_Detection_A_Realistic_Modeling_and_a_Novel_Learning_Strategy
[6]: http://di.ulb.ac.be/map/adalpozz/pdf/Dalpozzolo2015PhD.pdf
[7]: https://www.researchgate.net/publication/319616537_SCARFF_a_Scalable_Framework_for_Streaming_Credit_Card_Fraud_Detection_with_Spark
[8]: https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection
Kaggle dataset identifier: creditcardfraud
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from keras.layers import Input, Dense
from keras.models import Model, Sequential
from keras import regularizers
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report, accuracy_score
from sklearn.manifold import TSNE
from sklearn import preprocessing
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
sns.set(style="whitegrid")
np.random.seed(203)
data = pd.read_csv("../input/creditcardfraud/creditcard.csv")
data["Time"] = data["Time"].apply(lambda x: x / 3600 % 24)
data.head()
# We have an unlabeled dataset.
# examine the dataset, we see a imbalanced dataset in which only 492 are fraud transactions.
print(data.Class.value_counts())
# We want to randomly select 1000 rows of normal transactions, combine that 1000 rows with the fraud data.
non_fraud = data[data["Class"] == 0].sample(1000)
fraud = data[data["Class"] == 1]
# sample(frac=1): returns each row in random order; reset_index(drop=True) drops index and replaces index with increasing integer.
df = non_fraud.append(fraud).sample(frac=1).reset_index(drop=True)
# X is an Numpy Array of all the features, and Y is an Numpy Array of all actual outputs
X = df.drop(["Class"], axis=1).values
Y = df["Class"].values
# Array X has a shape of [1492,30], meaning that it has 1492 data points, each data point can be viewed as a 30-dimensional vector.
# Here we set input_layer to 30 units(30-dimensional vector)
input_layer = Input(shape=(X.shape[1],))
# Dense is fully connected layer/全连接层. Dense(units, activation = activation_function, activity_regularizier = regularizers.l1/l2)
# sigmoid activation: [0,1], tanh activation: [-1,1]; relu activation
encoded = Dense(100, activation="tanh", activity_regularizer=regularizers.l1(10e-5))(
input_layer
)
encoded = Dense(50, activation="relu")(encoded)
## decoding part
decoded = Dense(50, activation="tanh")(encoded)
decoded = Dense(100, activation="tanh")(decoded)
## output layer
output_layer = Dense(X.shape[1], activation="relu")(decoded)
# 生产神经网络, 损失函数为mean squared error, 优化算法采用adadelta(是一种相比batch gradient descent先进的优化算法)
autoencoder = Model(input_layer, output_layer)
autoencoder.compile(optimizer="adadelta", loss="mse")
x = data.drop(["Class"], axis=1)
y = data["Class"].values
# We want to preproceess our data for appropriate fitting of our autoencoder neural network. We use MinMaxScaler() and apply to all x columns here.
x_scale = preprocessing.MinMaxScaler().fit_transform(x.values)
# Now, slice table x into normal transactions and fraud transactions
x_norm, x_fraud = x_scale[y == 0], x_scale[y == 1]
autoencoder.fit(
x_norm[0:2000],
x_norm[0:2000],
batch_size=256,
epochs=10,
shuffle=True,
validation_split=0.20,
)
hidden_representation = Sequential()
hidden_representation.add(autoencoder.layers[0])
hidden_representation.add(autoencoder.layers[1])
hidden_representation.add(autoencoder.layers[2])
norm_hid_rep = hidden_representation.predict(x_norm[:3000])
fraud_hid_rep = hidden_representation.predict(x_fraud)
rep_x = np.append(norm_hid_rep, fraud_hid_rep, axis=0)
y_n = np.zeros(norm_hid_rep.shape[0])
y_f = np.ones(fraud_hid_rep.shape[0])
rep_y = np.append(y_n, y_f)
train_x, val_x, train_y, val_y = train_test_split(rep_x, rep_y, test_size=0.25)
clf = LogisticRegression(solver="lbfgs").fit(train_x, train_y)
pred_y = clf.predict(val_x)
print("")
print("Classification Report: ")
print(classification_report(val_y, pred_y))
print("")
print("Accuracy Score: ", accuracy_score(val_y, pred_y))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/708/69708893.ipynb
|
creditcardfraud
| null |
[{"Id": 69708893, "ScriptId": 19039250, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8003286, "CreationDate": "08/02/2021 21:48:49", "VersionNumber": 2.0, "Title": "Credit Card Fraud Detection with autoencoder nn", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 108.0, "LinesInsertedFromPrevious": 7.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 101.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93180890, "KernelVersionId": 69708893, "SourceDatasetVersionId": 23498}]
|
[{"Id": 23498, "DatasetId": 310, "DatasourceVersionId": 23502, "CreatorUserId": 998023, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "03/23/2018 01:17:27", "VersionNumber": 3.0, "Title": "Credit Card Fraud Detection", "Slug": "creditcardfraud", "Subtitle": "Anonymized credit card transactions labeled as fraudulent or genuine", "Description": "Context\n---------\n\nIt is important that credit card companies are able to recognize fraudulent credit card transactions so that customers are not charged for items that they did not purchase.\n\nContent\n---------\n\nThe dataset contains transactions made by credit cards in September 2013 by European cardholders. \nThis dataset presents transactions that occurred in two days, where we have 492 frauds out of 284,807 transactions. The dataset is highly unbalanced, the positive class (frauds) account for 0.172% of all transactions.\n\nIt contains only numerical input variables which are the result of a PCA transformation. Unfortunately, due to confidentiality issues, we cannot provide the original features and more background information about the data. Features V1, V2, ... V28 are the principal components obtained with PCA, the only features which have not been transformed with PCA are 'Time' and 'Amount'. Feature 'Time' contains the seconds elapsed between each transaction and the first transaction in the dataset. The feature 'Amount' is the transaction Amount, this feature can be used for example-dependant cost-sensitive learning. Feature 'Class' is the response variable and it takes value 1 in case of fraud and 0 otherwise. \n\nGiven the class imbalance ratio, we recommend measuring the accuracy using the Area Under the Precision-Recall Curve (AUPRC). Confusion matrix accuracy is not meaningful for unbalanced classification.\n\nUpdate (03/05/2021)\n---------\n\nA simulator for transaction data has been released as part of the practical handbook on Machine Learning for Credit Card Fraud Detection - https://fraud-detection-handbook.github.io/fraud-detection-handbook/Chapter_3_GettingStarted/SimulatedDataset.html. We invite all practitioners interested in fraud detection datasets to also check out this data simulator, and the methodologies for credit card fraud detection presented in the book.\n\nAcknowledgements\n---------\n\nThe dataset has been collected and analysed during a research collaboration of Worldline and the Machine Learning Group (http://mlg.ulb.ac.be) of ULB (Universit\u00e9 Libre de Bruxelles) on big data mining and fraud detection.\nMore details on current and past projects on related topics are available on [https://www.researchgate.net/project/Fraud-detection-5][1] and the page of the [DefeatFraud][2] project\n\nPlease cite the following works: \n\nAndrea Dal Pozzolo, Olivier Caelen, Reid A. Johnson and Gianluca Bontempi. [Calibrating Probability with Undersampling for Unbalanced Classification.][3] In Symposium on Computational Intelligence and Data Mining (CIDM), IEEE, 2015\n\nDal Pozzolo, Andrea; Caelen, Olivier; Le Borgne, Yann-Ael; Waterschoot, Serge; Bontempi, Gianluca. [Learned lessons in credit card fraud detection from a practitioner perspective][4], Expert systems with applications,41,10,4915-4928,2014, Pergamon\n\nDal Pozzolo, Andrea; Boracchi, Giacomo; Caelen, Olivier; Alippi, Cesare; Bontempi, Gianluca. [Credit card fraud detection: a realistic modeling and a novel learning strategy,][5] IEEE transactions on neural networks and learning systems,29,8,3784-3797,2018,IEEE\n\nDal Pozzolo, Andrea [Adaptive Machine learning for credit card fraud detection][6] ULB MLG PhD thesis (supervised by G. Bontempi)\n\nCarcillo, Fabrizio; Dal Pozzolo, Andrea; Le Borgne, Yann-A\u00ebl; Caelen, Olivier; Mazzer, Yannis; Bontempi, Gianluca. [Scarff: a scalable framework for streaming credit card fraud detection with Spark][7], Information fusion,41, 182-194,2018,Elsevier\n\nCarcillo, Fabrizio; Le Borgne, Yann-A\u00ebl; Caelen, Olivier; Bontempi, Gianluca. [Streaming active learning strategies for real-life credit card fraud detection: assessment and visualization,][8] International Journal of Data Science and Analytics, 5,4,285-300,2018,Springer International Publishing\n\nBertrand Lebichot, Yann-A\u00ebl Le Borgne, Liyun He, Frederic Obl\u00e9, Gianluca Bontempi [Deep-Learning Domain Adaptation Techniques for Credit Cards Fraud Detection](https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection), INNSBDDL 2019: Recent Advances in Big Data and Deep Learning, pp 78-88, 2019\n\nFabrizio Carcillo, Yann-A\u00ebl Le Borgne, Olivier Caelen, Frederic Obl\u00e9, Gianluca Bontempi [Combining Unsupervised and Supervised Learning in Credit Card Fraud Detection ](https://www.researchgate.net/publication/333143698_Combining_Unsupervised_and_Supervised_Learning_in_Credit_Card_Fraud_Detection) Information Sciences, 2019\n\nYann-A\u00ebl Le Borgne, Gianluca Bontempi [Reproducible machine Learning for Credit Card Fraud Detection - Practical Handbook ](https://www.researchgate.net/publication/351283764_Machine_Learning_for_Credit_Card_Fraud_Detection_-_Practical_Handbook) \n\nBertrand Lebichot, Gianmarco Paldino, Wissam Siblini, Liyun He, Frederic Obl\u00e9, Gianluca Bontempi [Incremental learning strategies for credit cards fraud detection](https://www.researchgate.net/publication/352275169_Incremental_learning_strategies_for_credit_cards_fraud_detection), IInternational Journal of Data Science and Analytics\n\n [1]: https://www.researchgate.net/project/Fraud-detection-5\n [2]: https://mlg.ulb.ac.be/wordpress/portfolio_page/defeatfraud-assessment-and-validation-of-deep-feature-engineering-and-learning-solutions-for-fraud-detection/\n [3]: https://www.researchgate.net/publication/283349138_Calibrating_Probability_with_Undersampling_for_Unbalanced_Classification\n [4]: https://www.researchgate.net/publication/260837261_Learned_lessons_in_credit_card_fraud_detection_from_a_practitioner_perspective\n [5]: https://www.researchgate.net/publication/319867396_Credit_Card_Fraud_Detection_A_Realistic_Modeling_and_a_Novel_Learning_Strategy\n [6]: http://di.ulb.ac.be/map/adalpozz/pdf/Dalpozzolo2015PhD.pdf\n [7]: https://www.researchgate.net/publication/319616537_SCARFF_a_Scalable_Framework_for_Streaming_Credit_Card_Fraud_Detection_with_Spark\n \n[8]: https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection", "VersionNotes": "Fixed preview", "TotalCompressedBytes": 150828752.0, "TotalUncompressedBytes": 69155632.0}]
|
[{"Id": 310, "CreatorUserId": 14069, "OwnerUserId": NaN, "OwnerOrganizationId": 1160.0, "CurrentDatasetVersionId": 23498.0, "CurrentDatasourceVersionId": 23502.0, "ForumId": 1838, "Type": 2, "CreationDate": "11/03/2016 13:21:36", "LastActivityDate": "02/06/2018", "TotalViews": 10310781, "TotalDownloads": 564249, "TotalVotes": 10432, "TotalKernels": 4266}]
| null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from keras.layers import Input, Dense
from keras.models import Model, Sequential
from keras import regularizers
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report, accuracy_score
from sklearn.manifold import TSNE
from sklearn import preprocessing
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
sns.set(style="whitegrid")
np.random.seed(203)
data = pd.read_csv("../input/creditcardfraud/creditcard.csv")
data["Time"] = data["Time"].apply(lambda x: x / 3600 % 24)
data.head()
# We have an unlabeled dataset.
# examine the dataset, we see a imbalanced dataset in which only 492 are fraud transactions.
print(data.Class.value_counts())
# We want to randomly select 1000 rows of normal transactions, combine that 1000 rows with the fraud data.
non_fraud = data[data["Class"] == 0].sample(1000)
fraud = data[data["Class"] == 1]
# sample(frac=1): returns each row in random order; reset_index(drop=True) drops index and replaces index with increasing integer.
df = non_fraud.append(fraud).sample(frac=1).reset_index(drop=True)
# X is an Numpy Array of all the features, and Y is an Numpy Array of all actual outputs
X = df.drop(["Class"], axis=1).values
Y = df["Class"].values
# Array X has a shape of [1492,30], meaning that it has 1492 data points, each data point can be viewed as a 30-dimensional vector.
# Here we set input_layer to 30 units(30-dimensional vector)
input_layer = Input(shape=(X.shape[1],))
# Dense is fully connected layer/全连接层. Dense(units, activation = activation_function, activity_regularizier = regularizers.l1/l2)
# sigmoid activation: [0,1], tanh activation: [-1,1]; relu activation
encoded = Dense(100, activation="tanh", activity_regularizer=regularizers.l1(10e-5))(
input_layer
)
encoded = Dense(50, activation="relu")(encoded)
## decoding part
decoded = Dense(50, activation="tanh")(encoded)
decoded = Dense(100, activation="tanh")(decoded)
## output layer
output_layer = Dense(X.shape[1], activation="relu")(decoded)
# 生产神经网络, 损失函数为mean squared error, 优化算法采用adadelta(是一种相比batch gradient descent先进的优化算法)
autoencoder = Model(input_layer, output_layer)
autoencoder.compile(optimizer="adadelta", loss="mse")
x = data.drop(["Class"], axis=1)
y = data["Class"].values
# We want to preproceess our data for appropriate fitting of our autoencoder neural network. We use MinMaxScaler() and apply to all x columns here.
x_scale = preprocessing.MinMaxScaler().fit_transform(x.values)
# Now, slice table x into normal transactions and fraud transactions
x_norm, x_fraud = x_scale[y == 0], x_scale[y == 1]
autoencoder.fit(
x_norm[0:2000],
x_norm[0:2000],
batch_size=256,
epochs=10,
shuffle=True,
validation_split=0.20,
)
hidden_representation = Sequential()
hidden_representation.add(autoencoder.layers[0])
hidden_representation.add(autoencoder.layers[1])
hidden_representation.add(autoencoder.layers[2])
norm_hid_rep = hidden_representation.predict(x_norm[:3000])
fraud_hid_rep = hidden_representation.predict(x_fraud)
rep_x = np.append(norm_hid_rep, fraud_hid_rep, axis=0)
y_n = np.zeros(norm_hid_rep.shape[0])
y_f = np.ones(fraud_hid_rep.shape[0])
rep_y = np.append(y_n, y_f)
train_x, val_x, train_y, val_y = train_test_split(rep_x, rep_y, test_size=0.25)
clf = LogisticRegression(solver="lbfgs").fit(train_x, train_y)
pred_y = clf.predict(val_x)
print("")
print("Classification Report: ")
print(classification_report(val_y, pred_y))
print("")
print("Accuracy Score: ", accuracy_score(val_y, pred_y))
| false | 0 | 1,356 | 0 | 3,230 | 1,356 |
||
69708287
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import make_column_transformer
train_data.isna().sum()
train_data = train_data.loc[
train_data.Embarked.notna(), ["Survived", "Pclass", "Sex", "Embarked"]
]
X_train = train_data.loc[:, ["Pclass", "Sex", "Embarked"]]
y_train = train_data.Survived
X_train.shape
y_train.shape
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import LogisticRegression
column_trans = make_column_transformer(
(OneHotEncoder(), ["Sex", "Embarked"]), remainder="passthrough"
)
logreg = LogisticRegression(solver="lbfgs")
pipe = make_pipeline(column_trans, logreg)
from sklearn.model_selection import cross_val_score
cross_val_score(pipe, X_train, y_train, cv=10, scoring="accuracy").mean()
pipe.fit(X_train, y_train)
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
X_test = test_data.loc[:, ["Pclass", "Sex", "Embarked"]]
# y_test = test_data.Survived
test_data.head()
X_test.head()
test_data.isna().sum()
y_pred = pipe.predict(X_test)
submissions = pd.read_csv("/kaggle/input/titanic/gender_submission.csv")
submissions.shape
y_test = submissions.Survived
from sklearn.metrics import accuracy_score
accuracy_score(y_test, y_pred)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/708/69708287.ipynb
| null | null |
[{"Id": 69708287, "ScriptId": 19046737, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7949479, "CreationDate": "08/02/2021 21:43:47", "VersionNumber": 1.0, "Title": "notebookceacef8b95", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 70.0, "LinesInsertedFromPrevious": 70.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import make_column_transformer
train_data.isna().sum()
train_data = train_data.loc[
train_data.Embarked.notna(), ["Survived", "Pclass", "Sex", "Embarked"]
]
X_train = train_data.loc[:, ["Pclass", "Sex", "Embarked"]]
y_train = train_data.Survived
X_train.shape
y_train.shape
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import LogisticRegression
column_trans = make_column_transformer(
(OneHotEncoder(), ["Sex", "Embarked"]), remainder="passthrough"
)
logreg = LogisticRegression(solver="lbfgs")
pipe = make_pipeline(column_trans, logreg)
from sklearn.model_selection import cross_val_score
cross_val_score(pipe, X_train, y_train, cv=10, scoring="accuracy").mean()
pipe.fit(X_train, y_train)
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
X_test = test_data.loc[:, ["Pclass", "Sex", "Embarked"]]
# y_test = test_data.Survived
test_data.head()
X_test.head()
test_data.isna().sum()
y_pred = pipe.predict(X_test)
submissions = pd.read_csv("/kaggle/input/titanic/gender_submission.csv")
submissions.shape
y_test = submissions.Survived
from sklearn.metrics import accuracy_score
accuracy_score(y_test, y_pred)
| false | 0 | 636 | 0 | 636 | 636 |
||
69708883
|
<jupyter_start><jupyter_text>roberta-large
Kaggle dataset identifier: robertalarge
<jupyter_script>import transformers
from transformers import (
AutoTokenizer,
AutoModel,
AutoConfig,
AutoModelForMaskedLM,
Trainer,
TrainingArguments,
DataCollatorForLanguageModeling,
RobertaForSequenceClassification,
AdamW,
get_linear_schedule_with_warmup,
)
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import torch
import torch.nn as nn
from torch.utils.data import (
dataset,
TensorDataset,
DataLoader,
RandomSampler,
SequentialSampler,
random_split,
)
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error
from typing import Dict
from transformers.tokenization_utils import PreTrainedTokenizer
from tqdm.notebook import tqdm
import os
import random
import time
import datetime
import warnings
# Set verbosity to the error level
transformers.logging.set_verbosity_error()
WARMUP_STEPS = 0
LEARNING_RATE = 5e-5
WEIGHT_DECAY = 0
EVAL_STEPS = 200
TRAIN_BATCH_SIZE = 16
VALID_BATCH_SIZE = 16
EPOCHS = 5
ROBERTA_MODEL = "../input/roberta-base"
TRAINING_FILE = "../input/commonlitreadabilityprize/train.csv"
TEST_FILE = "../input/commonlitreadabilityprize/test.csv"
SAMPLE_FILE = "../input/commonlitreadabilityprize/sample_submission.csv"
MODEL_PATH = "Models/clrp_roberta-pretrained"
TOKENIZER = transformers.AutoTokenizer.from_pretrained(
ROBERTA_MODEL, do_lower_case=True
)
MODEL = transformers.AutoModelForMaskedLM.from_pretrained(ROBERTA_MODEL)
class LineByLineTextDataset(dataset.Dataset):
def __init__(self, data, tokenizer: PreTrainedTokenizer, block_size: int):
data = data["excerpt"]
lines = [line for line in data if (len(line) > 0 and not line.isspace())]
batch_encoding = tokenizer(
lines, add_special_tokens=True, truncation=True, max_length=block_size
)
self.examples = batch_encoding["input_ids"]
self.examples = [
{"input_ids": torch.tensor(e, dtype=torch.long)} for e in self.examples
]
def __len__(self):
return len(self.examples)
def __getitem__(self, i) -> Dict[str, torch.tensor]:
return self.examples[i]
train_data = pd.read_csv(TRAINING_FILE)
test_data = pd.read_csv(TEST_FILE)
train_dataset = LineByLineTextDataset(train_data, tokenizer=TOKENIZER, block_size=256)
valid_dataset = LineByLineTextDataset(train_data, tokenizer=TOKENIZER, block_size=256)
test_dataset = LineByLineTextDataset(test_data, tokenizer=TOKENIZER, block_size=256)
data_collator = DataCollatorForLanguageModeling(
tokenizer=TOKENIZER, mlm=True, mlm_probability=0.15
)
training_args = TrainingArguments(
output_dir="./results",
overwrite_output_dir=True,
num_train_epochs=EPOCHS,
per_device_train_batch_size=TRAIN_BATCH_SIZE,
per_device_eval_batch_size=VALID_BATCH_SIZE,
evaluation_strategy="steps",
save_total_limit=2,
eval_steps=EVAL_STEPS,
metric_for_best_model="eval_loss",
greater_is_better=False,
load_best_model_at_end=True,
prediction_loss_only=True,
warmup_steps=WARMUP_STEPS,
weight_decay=WEIGHT_DECAY,
report_to="none",
)
trainer = Trainer(
model=MODEL,
args=training_args,
data_collator=data_collator,
train_dataset=train_dataset,
eval_dataset=valid_dataset,
)
trainer.train()
trainer.save_model(MODEL_PATH)
del trainer
torch.cuda.empty_cache()
# ## Finetuning class
# #### Define model architecture
class Model(nn.Module):
def __init__(self, path):
super().__init__()
self.config = AutoConfig.from_pretrained(path)
self.config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = RobertaForSequenceClassification.from_pretrained(
path, config=self.config
)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.dropout = nn.Dropout(0.1)
self.linear = nn.Linear(self.config.hidden_size, 1)
def forward(self, input_ids, attention_mask, token_type_ids):
x = self.roberta(
input_ids=input_ids,
attention_mask=attention_mask,
token_type_ids=token_type_ids,
)
weights = self.attention(x.hidden_states[-1])
x = torch.sum(weights * x.hidden_states[-1], dim=1)
x = self.dropout(x)
x = self.linear(x)
return x
# #### Define Finetuning class
class Finetune:
def __init__(
self, sentences, labels, model, base_path, seed=42, batch_size=32, epochs=4
):
self.model = model
self.tokenizer = transformers.AutoTokenizer.from_pretrained(
base_path, do_lower_case=True
)
self.device = torch.device("cuda")
self.sentences = sentences
self.labels = labels
self.batch_size = batch_size
self.epochs = epochs
self.seed = seed
def load_data(self, max_len):
input_ids = []
attention_masks = []
token_type_ids = []
for sent in self.sentences:
encoded_dict = self.tokenizer.encode_plus(
sent,
add_special_tokens=True,
max_length=max_len,
padding="max_length",
return_attention_mask=True,
return_token_type_ids=True,
return_tensors="pt",
truncation=True,
)
input_ids.append(encoded_dict["input_ids"])
attention_masks.append(encoded_dict["attention_mask"])
token_type_ids.append(encoded_dict["token_type_ids"])
input_ids = torch.cat(input_ids, dim=0)
attention_masks = torch.cat(attention_masks, dim=0)
token_type_ids = torch.cat(token_type_ids, dim=0)
labels = torch.tensor(self.labels, dtype=torch.float)
dataset = TensorDataset(input_ids, attention_masks, token_type_ids, labels)
train_size = int(0.9 * len(dataset))
val_size = len(dataset) - train_size
train_dataset, val_dataset = random_split(dataset, [train_size, val_size])
train_dataloader = DataLoader(
train_dataset,
sampler=RandomSampler(train_dataset),
batch_size=self.batch_size,
)
validation_dataloader = DataLoader(
val_dataset,
sampler=SequentialSampler(val_dataset),
batch_size=self.batch_size,
)
return train_dataloader, validation_dataloader
def optimizer(self, train_dataloader, lr=2e-5, eps=1e-8, wd=1e-2):
total_steps = len(train_dataloader) * self.epochs
param_optimizer = list(self.model.named_parameters())
no_decay = ["bias", "LayerNorm.weight"]
optimizer_grouped_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.1,
},
{
"params": [
p for n, p in param_optimizer if any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.0,
},
]
optimizer = AdamW(optimizer_grouped_parameters, lr=lr, eps=eps, weight_decay=wd)
scheduler = get_linear_schedule_with_warmup(
optimizer, num_warmup_steps=0, num_training_steps=total_steps
)
return optimizer, scheduler
def train_and_evaluate(
self, train_dataloader, validation_dataloader, optimizer, scheduler
):
self.model.cuda()
random.seed(self.seed)
np.random.seed(self.seed)
torch.manual_seed(self.seed)
torch.cuda.manual_seed_all(self.seed)
training_stats = []
total_t0 = time.time()
for epoch_i in tqdm(
range(0, self.epochs), leave=False, bar_format="Epochs {l_bar}{bar}{r_bar}"
):
t0 = time.time()
total_train_loss = 0
self.model.train()
for step, batch in enumerate(
tqdm(
train_dataloader,
leave=False,
bar_format="Steps {l_bar}{bar}{r_bar}",
)
):
b_input_ids = batch[0].to(self.device)
b_input_mask = batch[1].to(self.device)
b_input_token_type_ids = batch[2].to(self.device)
b_labels = batch[3].to(self.device)
self.model.zero_grad()
optimizer.zero_grad()
result = self.model(
b_input_ids,
attention_mask=b_input_mask,
token_type_ids=b_input_token_type_ids,
)
loss = torch.sqrt(nn.MSELoss()(result.flatten(), b_labels.view(-1)))
total_train_loss += loss.item()
loss.backward()
torch.nn.utils.clip_grad_norm_(self.model.parameters(), 1.0)
optimizer.step()
scheduler.step()
avg_train_loss = total_train_loss / len(train_dataloader)
training_time = str(
datetime.timedelta(seconds=int(round((time.time() - t0))))
)
t0 = time.time()
self.model.eval()
total_eval_score = 0
total_eval_loss = 0
nb_eval_steps = 0
for batch in validation_dataloader:
b_input_ids = batch[0].to(self.device)
b_input_mask = batch[1].to(self.device)
b_input_token_type_ids = batch[2].to(self.device)
b_labels = batch[3].to(self.device)
with torch.no_grad():
result = self.model(
b_input_ids,
attention_mask=b_input_mask,
token_type_ids=b_input_token_type_ids,
)
loss = torch.sqrt(nn.MSELoss()(result.flatten(), b_labels.view(-1)))
total_eval_loss += loss.item()
logits = result.detach().cpu().numpy()
avg_val_loss = total_eval_loss / len(validation_dataloader)
validation_time = str(
datetime.timedelta(seconds=int(round((time.time() - t0))))
)
training_stats.append(
{
"epoch": epoch_i + 1,
"Training Loss": avg_train_loss,
"Valid. Loss": avg_val_loss,
"Training Time": training_time,
"Validation Time": validation_time,
}
)
return self.model, training_stats
# #### Train
sentences = train_data.excerpt.values
labels = train_data.target.values
epochs = 4
lr = 3e-5
folds = 5
max_length = 192
for fold in tqdm(range(folds), leave=False, bar_format="Folds {l_bar}{bar}{r_bar}"):
fold = fold + 1
model = Model(MODEL_PATH)
finetune = Finetune(sentences, labels, model, ROBERTA_MODEL, epochs=epochs)
train_dataloader, validation_dataloader = finetune.load_data(max_length)
optimizer, scheduler = finetune.optimizer(train_dataloader, lr=lr)
model, training_stats = finetune.train_and_evaluate(
train_dataloader, validation_dataloader, optimizer, scheduler
)
if fold == 1:
df = pd.DataFrame(data=training_stats)
df["Fold"] = fold
else:
df1 = pd.DataFrame(data=training_stats)
df1["Fold"] = fold
df = df1.append(df)
output_dir = "./Models/"
if not os.path.exists(output_dir):
os.makedirs(output_dir)
torch.save(model.state_dict(), f"./Models/model{fold}.bin")
del model
# Display floats with two decimal places.
pd.set_option("precision", 2)
# Use the 'epoch' as the row index.
df_stats = df.set_index("Fold")
# A hack to force the column headers to wrap.
# df = df.style.set_table_styles([dict(selector="th",props=[('max-width', '70px')])])
# Display the table.
df_stats
del finetune
torch.cuda.empty_cache()
# import matplotlib.pyplot as plt
# import seaborn as sns
# # Use plot styling from seaborn.
# sns.set(style='darkgrid')
# # Increase the plot size and font size.
# sns.set(font_scale=1.5)
# plt.rcParams["figure.figsize"] = (12,6)
# # Plot the learning curve.
# plt.plot(df_stats['Training Loss'], 'b-o', label="Training")
# plt.plot(df_stats['Valid. Loss'], 'g-o', label="Validation")
# # Label the plot.
# plt.title("Training & Validation Loss")
# plt.xlabel("Epoch")
# plt.ylabel("Loss")
# plt.legend()
# plt.xticks([1, 2, 3, 4])
# plt.show()
# ### Inference
def base_model(base_path, num_labels=1):
config = AutoConfig.from_pretrained(base_path)
config.update({"num_labels": num_labels})
model = Model(base_path)
return model
class Inference:
def __init__(self, sentences, model_path, base_path, seed=42, batch_size=16):
self.model = model
self.tokenizer = transformers.AutoTokenizer.from_pretrained(
base_path, do_lower_case=True
)
self.device = torch.device("cuda")
self.sentences = sentences
self.batch_size = batch_size
def load_data(self, max_len):
input_ids = []
attention_masks = []
token_type_ids = []
for sent in self.sentences:
encoded_dict = self.tokenizer.encode_plus(
sent,
add_special_tokens=True,
max_length=max_len,
padding="max_length",
return_attention_mask=True,
return_token_type_ids=True,
return_tensors="pt",
truncation=True,
)
input_ids.append(encoded_dict["input_ids"])
attention_masks.append(encoded_dict["attention_mask"])
token_type_ids.append(encoded_dict["token_type_ids"])
input_ids = torch.cat(input_ids, dim=0)
attention_masks = torch.cat(attention_masks, dim=0)
token_type_ids = torch.cat(token_type_ids, dim=0)
test_dataset = TensorDataset(input_ids, attention_masks, token_type_ids)
test_dataloader = DataLoader(
test_dataset,
sampler=SequentialSampler(test_dataset),
batch_size=self.batch_size,
pin_memory=False,
drop_last=False,
num_workers=0,
)
return test_dataloader
def predict(self, test_dataloader):
self.model.to(self.device)
self.model.eval()
result = np.zeros(len(test_dataloader.dataset))
index = 0
with torch.no_grad():
for batch_num, batch_data in enumerate(test_dataloader):
input_ids, attention_mask, token_type_ids = (
batch_data[0],
batch_data[1],
batch_data[2],
)
input_ids, attention_mask, token_type_ids = (
input_ids.cuda(),
attention_mask.cuda(),
token_type_ids.cuda(),
)
pred = model(input_ids, attention_mask, token_type_ids)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
all_predictions = np.zeros((folds, len(test_data)))
sentences = test_data.excerpt.values
submission_df = pd.read_csv(SAMPLE_FILE)
for fold in range(folds):
model_path = f"./Models/model{fold+1}.bin"
model = base_model(ROBERTA_MODEL)
model.load_state_dict(torch.load(model_path))
inference = Inference(sentences, model, ROBERTA_MODEL)
test_dataloader = inference.load_data(144)
result = inference.predict(test_dataloader)
all_predictions[fold] = result
del model
predictions = all_predictions.mean(axis=0)
submission_df.target = predictions
submission_df
submission_df.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/708/69708883.ipynb
|
robertalarge
|
marshal02
|
[{"Id": 69708883, "ScriptId": 18886486, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5797807, "CreationDate": "08/02/2021 21:48:43", "VersionNumber": 18.0, "Title": "CLRP - kauvinlucas", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 419.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 418.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93180888, "KernelVersionId": 69708883, "SourceDatasetVersionId": 1212718}, {"Id": 93180887, "KernelVersionId": 69708883, "SourceDatasetVersionId": 1042664}]
|
[{"Id": 1212718, "DatasetId": 692034, "DatasourceVersionId": 1244142, "CreatorUserId": 1896350, "LicenseName": "CC0: Public Domain", "CreationDate": "06/04/2020 06:01:42", "VersionNumber": 2.0, "Title": "roberta-large", "Slug": "robertalarge", "Subtitle": NaN, "Description": NaN, "VersionNotes": "roberta-large", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 692034, "CreatorUserId": 1896350, "OwnerUserId": 1896350.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1212718.0, "CurrentDatasourceVersionId": 1244142.0, "ForumId": 706673, "Type": 2, "CreationDate": "06/04/2020 00:33:26", "LastActivityDate": "06/04/2020", "TotalViews": 4465, "TotalDownloads": 1801, "TotalVotes": 37, "TotalKernels": 88}]
|
[{"Id": 1896350, "UserName": "marshal02", "DisplayName": "Marshal Baskey", "RegisterDate": "05/09/2018", "PerformanceTier": 1}]
|
import transformers
from transformers import (
AutoTokenizer,
AutoModel,
AutoConfig,
AutoModelForMaskedLM,
Trainer,
TrainingArguments,
DataCollatorForLanguageModeling,
RobertaForSequenceClassification,
AdamW,
get_linear_schedule_with_warmup,
)
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import torch
import torch.nn as nn
from torch.utils.data import (
dataset,
TensorDataset,
DataLoader,
RandomSampler,
SequentialSampler,
random_split,
)
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error
from typing import Dict
from transformers.tokenization_utils import PreTrainedTokenizer
from tqdm.notebook import tqdm
import os
import random
import time
import datetime
import warnings
# Set verbosity to the error level
transformers.logging.set_verbosity_error()
WARMUP_STEPS = 0
LEARNING_RATE = 5e-5
WEIGHT_DECAY = 0
EVAL_STEPS = 200
TRAIN_BATCH_SIZE = 16
VALID_BATCH_SIZE = 16
EPOCHS = 5
ROBERTA_MODEL = "../input/roberta-base"
TRAINING_FILE = "../input/commonlitreadabilityprize/train.csv"
TEST_FILE = "../input/commonlitreadabilityprize/test.csv"
SAMPLE_FILE = "../input/commonlitreadabilityprize/sample_submission.csv"
MODEL_PATH = "Models/clrp_roberta-pretrained"
TOKENIZER = transformers.AutoTokenizer.from_pretrained(
ROBERTA_MODEL, do_lower_case=True
)
MODEL = transformers.AutoModelForMaskedLM.from_pretrained(ROBERTA_MODEL)
class LineByLineTextDataset(dataset.Dataset):
def __init__(self, data, tokenizer: PreTrainedTokenizer, block_size: int):
data = data["excerpt"]
lines = [line for line in data if (len(line) > 0 and not line.isspace())]
batch_encoding = tokenizer(
lines, add_special_tokens=True, truncation=True, max_length=block_size
)
self.examples = batch_encoding["input_ids"]
self.examples = [
{"input_ids": torch.tensor(e, dtype=torch.long)} for e in self.examples
]
def __len__(self):
return len(self.examples)
def __getitem__(self, i) -> Dict[str, torch.tensor]:
return self.examples[i]
train_data = pd.read_csv(TRAINING_FILE)
test_data = pd.read_csv(TEST_FILE)
train_dataset = LineByLineTextDataset(train_data, tokenizer=TOKENIZER, block_size=256)
valid_dataset = LineByLineTextDataset(train_data, tokenizer=TOKENIZER, block_size=256)
test_dataset = LineByLineTextDataset(test_data, tokenizer=TOKENIZER, block_size=256)
data_collator = DataCollatorForLanguageModeling(
tokenizer=TOKENIZER, mlm=True, mlm_probability=0.15
)
training_args = TrainingArguments(
output_dir="./results",
overwrite_output_dir=True,
num_train_epochs=EPOCHS,
per_device_train_batch_size=TRAIN_BATCH_SIZE,
per_device_eval_batch_size=VALID_BATCH_SIZE,
evaluation_strategy="steps",
save_total_limit=2,
eval_steps=EVAL_STEPS,
metric_for_best_model="eval_loss",
greater_is_better=False,
load_best_model_at_end=True,
prediction_loss_only=True,
warmup_steps=WARMUP_STEPS,
weight_decay=WEIGHT_DECAY,
report_to="none",
)
trainer = Trainer(
model=MODEL,
args=training_args,
data_collator=data_collator,
train_dataset=train_dataset,
eval_dataset=valid_dataset,
)
trainer.train()
trainer.save_model(MODEL_PATH)
del trainer
torch.cuda.empty_cache()
# ## Finetuning class
# #### Define model architecture
class Model(nn.Module):
def __init__(self, path):
super().__init__()
self.config = AutoConfig.from_pretrained(path)
self.config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = RobertaForSequenceClassification.from_pretrained(
path, config=self.config
)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.dropout = nn.Dropout(0.1)
self.linear = nn.Linear(self.config.hidden_size, 1)
def forward(self, input_ids, attention_mask, token_type_ids):
x = self.roberta(
input_ids=input_ids,
attention_mask=attention_mask,
token_type_ids=token_type_ids,
)
weights = self.attention(x.hidden_states[-1])
x = torch.sum(weights * x.hidden_states[-1], dim=1)
x = self.dropout(x)
x = self.linear(x)
return x
# #### Define Finetuning class
class Finetune:
def __init__(
self, sentences, labels, model, base_path, seed=42, batch_size=32, epochs=4
):
self.model = model
self.tokenizer = transformers.AutoTokenizer.from_pretrained(
base_path, do_lower_case=True
)
self.device = torch.device("cuda")
self.sentences = sentences
self.labels = labels
self.batch_size = batch_size
self.epochs = epochs
self.seed = seed
def load_data(self, max_len):
input_ids = []
attention_masks = []
token_type_ids = []
for sent in self.sentences:
encoded_dict = self.tokenizer.encode_plus(
sent,
add_special_tokens=True,
max_length=max_len,
padding="max_length",
return_attention_mask=True,
return_token_type_ids=True,
return_tensors="pt",
truncation=True,
)
input_ids.append(encoded_dict["input_ids"])
attention_masks.append(encoded_dict["attention_mask"])
token_type_ids.append(encoded_dict["token_type_ids"])
input_ids = torch.cat(input_ids, dim=0)
attention_masks = torch.cat(attention_masks, dim=0)
token_type_ids = torch.cat(token_type_ids, dim=0)
labels = torch.tensor(self.labels, dtype=torch.float)
dataset = TensorDataset(input_ids, attention_masks, token_type_ids, labels)
train_size = int(0.9 * len(dataset))
val_size = len(dataset) - train_size
train_dataset, val_dataset = random_split(dataset, [train_size, val_size])
train_dataloader = DataLoader(
train_dataset,
sampler=RandomSampler(train_dataset),
batch_size=self.batch_size,
)
validation_dataloader = DataLoader(
val_dataset,
sampler=SequentialSampler(val_dataset),
batch_size=self.batch_size,
)
return train_dataloader, validation_dataloader
def optimizer(self, train_dataloader, lr=2e-5, eps=1e-8, wd=1e-2):
total_steps = len(train_dataloader) * self.epochs
param_optimizer = list(self.model.named_parameters())
no_decay = ["bias", "LayerNorm.weight"]
optimizer_grouped_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.1,
},
{
"params": [
p for n, p in param_optimizer if any(nd in n for nd in no_decay)
],
"weight_decay_rate": 0.0,
},
]
optimizer = AdamW(optimizer_grouped_parameters, lr=lr, eps=eps, weight_decay=wd)
scheduler = get_linear_schedule_with_warmup(
optimizer, num_warmup_steps=0, num_training_steps=total_steps
)
return optimizer, scheduler
def train_and_evaluate(
self, train_dataloader, validation_dataloader, optimizer, scheduler
):
self.model.cuda()
random.seed(self.seed)
np.random.seed(self.seed)
torch.manual_seed(self.seed)
torch.cuda.manual_seed_all(self.seed)
training_stats = []
total_t0 = time.time()
for epoch_i in tqdm(
range(0, self.epochs), leave=False, bar_format="Epochs {l_bar}{bar}{r_bar}"
):
t0 = time.time()
total_train_loss = 0
self.model.train()
for step, batch in enumerate(
tqdm(
train_dataloader,
leave=False,
bar_format="Steps {l_bar}{bar}{r_bar}",
)
):
b_input_ids = batch[0].to(self.device)
b_input_mask = batch[1].to(self.device)
b_input_token_type_ids = batch[2].to(self.device)
b_labels = batch[3].to(self.device)
self.model.zero_grad()
optimizer.zero_grad()
result = self.model(
b_input_ids,
attention_mask=b_input_mask,
token_type_ids=b_input_token_type_ids,
)
loss = torch.sqrt(nn.MSELoss()(result.flatten(), b_labels.view(-1)))
total_train_loss += loss.item()
loss.backward()
torch.nn.utils.clip_grad_norm_(self.model.parameters(), 1.0)
optimizer.step()
scheduler.step()
avg_train_loss = total_train_loss / len(train_dataloader)
training_time = str(
datetime.timedelta(seconds=int(round((time.time() - t0))))
)
t0 = time.time()
self.model.eval()
total_eval_score = 0
total_eval_loss = 0
nb_eval_steps = 0
for batch in validation_dataloader:
b_input_ids = batch[0].to(self.device)
b_input_mask = batch[1].to(self.device)
b_input_token_type_ids = batch[2].to(self.device)
b_labels = batch[3].to(self.device)
with torch.no_grad():
result = self.model(
b_input_ids,
attention_mask=b_input_mask,
token_type_ids=b_input_token_type_ids,
)
loss = torch.sqrt(nn.MSELoss()(result.flatten(), b_labels.view(-1)))
total_eval_loss += loss.item()
logits = result.detach().cpu().numpy()
avg_val_loss = total_eval_loss / len(validation_dataloader)
validation_time = str(
datetime.timedelta(seconds=int(round((time.time() - t0))))
)
training_stats.append(
{
"epoch": epoch_i + 1,
"Training Loss": avg_train_loss,
"Valid. Loss": avg_val_loss,
"Training Time": training_time,
"Validation Time": validation_time,
}
)
return self.model, training_stats
# #### Train
sentences = train_data.excerpt.values
labels = train_data.target.values
epochs = 4
lr = 3e-5
folds = 5
max_length = 192
for fold in tqdm(range(folds), leave=False, bar_format="Folds {l_bar}{bar}{r_bar}"):
fold = fold + 1
model = Model(MODEL_PATH)
finetune = Finetune(sentences, labels, model, ROBERTA_MODEL, epochs=epochs)
train_dataloader, validation_dataloader = finetune.load_data(max_length)
optimizer, scheduler = finetune.optimizer(train_dataloader, lr=lr)
model, training_stats = finetune.train_and_evaluate(
train_dataloader, validation_dataloader, optimizer, scheduler
)
if fold == 1:
df = pd.DataFrame(data=training_stats)
df["Fold"] = fold
else:
df1 = pd.DataFrame(data=training_stats)
df1["Fold"] = fold
df = df1.append(df)
output_dir = "./Models/"
if not os.path.exists(output_dir):
os.makedirs(output_dir)
torch.save(model.state_dict(), f"./Models/model{fold}.bin")
del model
# Display floats with two decimal places.
pd.set_option("precision", 2)
# Use the 'epoch' as the row index.
df_stats = df.set_index("Fold")
# A hack to force the column headers to wrap.
# df = df.style.set_table_styles([dict(selector="th",props=[('max-width', '70px')])])
# Display the table.
df_stats
del finetune
torch.cuda.empty_cache()
# import matplotlib.pyplot as plt
# import seaborn as sns
# # Use plot styling from seaborn.
# sns.set(style='darkgrid')
# # Increase the plot size and font size.
# sns.set(font_scale=1.5)
# plt.rcParams["figure.figsize"] = (12,6)
# # Plot the learning curve.
# plt.plot(df_stats['Training Loss'], 'b-o', label="Training")
# plt.plot(df_stats['Valid. Loss'], 'g-o', label="Validation")
# # Label the plot.
# plt.title("Training & Validation Loss")
# plt.xlabel("Epoch")
# plt.ylabel("Loss")
# plt.legend()
# plt.xticks([1, 2, 3, 4])
# plt.show()
# ### Inference
def base_model(base_path, num_labels=1):
config = AutoConfig.from_pretrained(base_path)
config.update({"num_labels": num_labels})
model = Model(base_path)
return model
class Inference:
def __init__(self, sentences, model_path, base_path, seed=42, batch_size=16):
self.model = model
self.tokenizer = transformers.AutoTokenizer.from_pretrained(
base_path, do_lower_case=True
)
self.device = torch.device("cuda")
self.sentences = sentences
self.batch_size = batch_size
def load_data(self, max_len):
input_ids = []
attention_masks = []
token_type_ids = []
for sent in self.sentences:
encoded_dict = self.tokenizer.encode_plus(
sent,
add_special_tokens=True,
max_length=max_len,
padding="max_length",
return_attention_mask=True,
return_token_type_ids=True,
return_tensors="pt",
truncation=True,
)
input_ids.append(encoded_dict["input_ids"])
attention_masks.append(encoded_dict["attention_mask"])
token_type_ids.append(encoded_dict["token_type_ids"])
input_ids = torch.cat(input_ids, dim=0)
attention_masks = torch.cat(attention_masks, dim=0)
token_type_ids = torch.cat(token_type_ids, dim=0)
test_dataset = TensorDataset(input_ids, attention_masks, token_type_ids)
test_dataloader = DataLoader(
test_dataset,
sampler=SequentialSampler(test_dataset),
batch_size=self.batch_size,
pin_memory=False,
drop_last=False,
num_workers=0,
)
return test_dataloader
def predict(self, test_dataloader):
self.model.to(self.device)
self.model.eval()
result = np.zeros(len(test_dataloader.dataset))
index = 0
with torch.no_grad():
for batch_num, batch_data in enumerate(test_dataloader):
input_ids, attention_mask, token_type_ids = (
batch_data[0],
batch_data[1],
batch_data[2],
)
input_ids, attention_mask, token_type_ids = (
input_ids.cuda(),
attention_mask.cuda(),
token_type_ids.cuda(),
)
pred = model(input_ids, attention_mask, token_type_ids)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
all_predictions = np.zeros((folds, len(test_data)))
sentences = test_data.excerpt.values
submission_df = pd.read_csv(SAMPLE_FILE)
for fold in range(folds):
model_path = f"./Models/model{fold+1}.bin"
model = base_model(ROBERTA_MODEL)
model.load_state_dict(torch.load(model_path))
inference = Inference(sentences, model, ROBERTA_MODEL)
test_dataloader = inference.load_data(144)
result = inference.predict(test_dataloader)
all_predictions[fold] = result
del model
predictions = all_predictions.mean(axis=0)
submission_df.target = predictions
submission_df
submission_df.to_csv("submission.csv", index=False)
| false | 0 | 4,376 | 0 | 4,398 | 4,376 |
||
69708674
|
<jupyter_start><jupyter_text>CommonLit Roberta Model Set
Kaggle dataset identifier: commonlit-roberta-0467
<jupyter_script># # Model 1
import os
import math
import random
import time
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
from torch.utils.data import Dataset
from torch.utils.data import DataLoader
from transformers import AutoTokenizer
from transformers import AutoModel
from transformers import AutoConfig
from sklearn.model_selection import KFold
from sklearn.svm import SVR
import gc
gc.enable()
BATCH_SIZE = 32
MAX_LEN = 248
EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1.0, 1)]
ROBERTA_PATH = "../input/huggingface-roberta/roberta-base"
TOKENIZER_PATH = "../input/huggingface-roberta/roberta-base"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
test_df = pd.read_csv("/kaggle/input/commonlitreadabilityprize/test.csv")
submission_df = pd.read_csv(
"/kaggle/input/commonlitreadabilityprize/sample_submission.csv"
)
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
# # Dataset
class LitDataset(Dataset):
def __init__(self, df, inference_only=False):
super().__init__()
self.df = df
self.inference_only = inference_only
self.text = df.excerpt.tolist()
# self.text = [text.replace("\n", " ") for text in self.text]
if not self.inference_only:
self.target = torch.tensor(df.target.values, dtype=torch.float32)
self.encoded = tokenizer.batch_encode_plus(
self.text,
padding="max_length",
max_length=MAX_LEN,
truncation=True,
return_attention_mask=True,
)
def __len__(self):
return len(self.df)
def __getitem__(self, index):
input_ids = torch.tensor(self.encoded["input_ids"][index])
attention_mask = torch.tensor(self.encoded["attention_mask"][index])
if self.inference_only:
return (input_ids, attention_mask)
else:
target = self.target[index]
return (input_ids, attention_mask, target)
# # Model
# The model is inspired by the one from [Maunish](https://www.kaggle.com/maunish/clrp-roberta-svm).
class LitModel(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(768, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
# # Inference
test_dataset = LitDataset(test_df, inference_only=True)
NUM_MODELS = 5
all_predictions = np.zeros((NUM_MODELS, len(test_df)))
test_dataset = LitDataset(test_df, inference_only=True)
test_loader = DataLoader(
test_dataset, batch_size=BATCH_SIZE, drop_last=False, shuffle=False, num_workers=2
)
for model_index in range(NUM_MODELS):
model_path = f"../input/commonlit-roberta-0467/model_{model_index + 1}.pth"
print(f"\nUsing {model_path}")
model = LitModel()
model.load_state_dict(torch.load(model_path, map_location=DEVICE))
model.to(DEVICE)
all_predictions[model_index] = predict(model, test_loader)
del model
gc.collect()
model1_predictions = all_predictions.mean(axis=0)
# # Model 2
# Imported from [https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3](https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3)
import os
import gc
import sys
import cv2
import math
import time
import tqdm
import random
import numpy as np
import pandas as pd
import seaborn as sns
from tqdm import tqdm
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
from sklearn.svm import SVR
from sklearn.linear_model import Ridge
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold, StratifiedKFold
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.optim import Adam, lr_scheduler
from torch.utils.data import Dataset, DataLoader
from transformers import (
AutoModel,
AutoTokenizer,
AutoConfig,
AutoModelForSequenceClassification,
)
import plotly.express as px
import plotly.graph_objs as go
import plotly.figure_factory as ff
from colorama import Fore, Back, Style
y_ = Fore.YELLOW
r_ = Fore.RED
g_ = Fore.GREEN
b_ = Fore.BLUE
m_ = Fore.MAGENTA
c_ = Fore.CYAN
sr_ = Style.RESET_ALL
train_data = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_data = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
sample = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
num_bins = int(np.floor(1 + np.log2(len(train_data))))
train_data.loc[:, "bins"] = pd.cut(train_data["target"], bins=num_bins, labels=False)
target = train_data["target"].to_numpy()
bins = train_data.bins.to_numpy()
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": 8,
"max_len": 256,
"nfolds": 5,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
class Model(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(
"../input/huggingface-roberta/roberta-large"
)
self.model = AutoModel.from_pretrained(
"../input/huggingface-roberta/roberta-large", config=config
)
self.drop_out1 = nn.Dropout(0)
self.drop_out2 = nn.Dropout(0.1)
self.layer_norm = nn.LayerNorm(1024)
self.layer_norm1 = nn.LayerNorm(1024)
self.l1 = nn.Linear(1024, 512)
self.l2 = nn.Linear(512, 1)
self._init_weights(self.layer_norm1)
self._init_weights(self.l1)
self._init_weights(self.l2)
def _init_weights(self, module):
if isinstance(module, nn.Linear):
module.weight.data.normal_(mean=0.0, std=0.02)
if module.bias is not None:
module.bias.data.zero_()
elif isinstance(module, nn.Embedding):
module.weight.data.normal_(mean=0.0, std=0.02)
if module.padding_idx is not None:
module.weight.data[module.padding_idx].zero_()
elif isinstance(module, nn.LayerNorm):
module.bias.data.zero_()
module.weight.data.fill_(1.0)
def forward(self, input_ids, attention_mask, labels=None):
outputs = self.model(input_ids, attention_mask)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
out = self.layer_norm(mean_embeddings)
return out
def get_embeddings(df, path, plot_losses=True, verbose=True):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
tokenizer = AutoTokenizer.from_pretrained(
"../input/huggingface-roberta/roberta-large"
)
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
return np.array(embeddings)
def get_preds_svm(X, y, X_test, RidgeReg=0, bins=bins, nfolds=10, C=8, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(n_splits=10, shuffle=True, random_state=config["seed"])
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
if RidgeReg:
print("ridge...")
model = Ridge(alpha=80.0)
else:
model = SVR(C=C, kernel=kernel, gamma="auto")
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
prediction = model.predict(X_valid)
score = rmse_score(prediction, y_valid)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
print("mean rmse", np.mean(scores))
return np.array(preds) / nfolds
train_embeddings0 = get_embeddings(train_data, "../input/version0/model0/model0.bin")
test_embeddings0 = get_embeddings(test_data, "../input/version0/model0/model0.bin")
svm_preds0 = get_preds_svm(train_embeddings0, target, test_embeddings0)
ridge_preds0 = get_preds_svm(train_embeddings0, target, test_embeddings0, RidgeReg=1)
del train_embeddings0, test_embeddings0
gc.collect()
train_embeddings1 = get_embeddings(train_data, "../input/version0/model1/model1.bin")
test_embeddings1 = get_embeddings(test_data, "../input/version0/model1/model1.bin")
svm_preds1 = get_preds_svm(train_embeddings1, target, test_embeddings1)
ridge_preds1 = get_preds_svm(train_embeddings1, target, test_embeddings1, RidgeReg=1)
del train_embeddings1, test_embeddings1
gc.collect()
train_embeddings2 = get_embeddings(train_data, "../input/version0/model2/model2.bin")
test_embeddings2 = get_embeddings(test_data, "../input/version0/model2/model2.bin")
svm_preds2 = get_preds_svm(train_embeddings2, target, test_embeddings2)
ridge_preds2 = get_preds_svm(train_embeddings2, target, test_embeddings2, RidgeReg=1)
del train_embeddings2, test_embeddings2
gc.collect()
train_embeddings3 = get_embeddings(train_data, "../input/version0/model3/model3.bin")
test_embeddings3 = get_embeddings(test_data, "../input/version0/model3/model3.bin")
svm_preds3 = get_preds_svm(train_embeddings3, target, test_embeddings3)
ridge_preds3 = get_preds_svm(train_embeddings3, target, test_embeddings3, RidgeReg=1)
del train_embeddings3, test_embeddings3
gc.collect()
train_embeddings4 = get_embeddings(train_data, "../input/version0/model4/model4.bin")
test_embeddings4 = get_embeddings(test_data, "../input/version0/model4/model4.bin")
svm_preds4 = get_preds_svm(train_embeddings4, target, test_embeddings4)
ridge_preds4 = get_preds_svm(train_embeddings4, target, test_embeddings4, RidgeReg=1)
del train_embeddings4, test_embeddings4
gc.collect()
class Model2(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(
"../input/huggingface-roberta/roberta-large"
)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.model = AutoModel.from_pretrained(
"../input/huggingface-roberta/roberta-large", config=config
)
self.drop_out1 = nn.Dropout(0)
self.drop_out2 = nn.Dropout(0.1)
self.layer_norm = nn.LayerNorm(1024)
self.layer_norm1 = nn.LayerNorm(1024)
self.l1 = nn.Linear(1024, 512)
self.l2 = nn.Linear(512, 1)
self._init_weights(self.layer_norm1)
self._init_weights(self.l1)
self._init_weights(self.l2)
def _init_weights(self, module):
if isinstance(module, nn.Linear):
module.weight.data.normal_(mean=0.0, std=0.02)
if module.bias is not None:
module.bias.data.zero_()
elif isinstance(module, nn.Embedding):
module.weight.data.normal_(mean=0.0, std=0.02)
if module.padding_idx is not None:
module.weight.data[module.padding_idx].zero_()
elif isinstance(module, nn.LayerNorm):
module.bias.data.zero_()
module.weight.data.fill_(1.0)
def forward(self, input_ids, attention_mask, labels=None):
outputs = self.model(input_ids, attention_mask)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
out = self.layer_norm(mean_embeddings)
return out
def get_embeddings(df, path, plot_losses=True, verbose=True):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model2()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
tokenizer = AutoTokenizer.from_pretrained(
"../input/huggingface-roberta/roberta-large"
)
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
return np.array(embeddings)
def get_preds_svm(X, y, X_test, RidgeReg=0, bins=bins, nfolds=10, C=10, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(n_splits=10, shuffle=True, random_state=config["seed"])
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
if RidgeReg:
print("ridge...")
model = Ridge(alpha=80.0)
else:
model = SVR(C=C, kernel=kernel, gamma="auto")
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
prediction = model.predict(X_valid)
score = rmse_score(prediction, y_valid)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
print("mean rmse", np.mean(scores))
return np.array(preds) / nfolds
train_embeddings0 = get_embeddings(train_data, "../input/nepoch2/model0/model0.bin")
test_embeddings0 = get_embeddings(test_data, "../input/nepoch2/model0/model0.bin")
svm0 = get_preds_svm(train_embeddings0, target, test_embeddings0)
del train_embeddings0, test_embeddings0
gc.collect()
train_embeddings1 = get_embeddings(train_data, "../input/nepoch2/model1/model1.bin")
test_embeddings1 = get_embeddings(test_data, "../input/nepoch2/model1/model1.bin")
svm1 = get_preds_svm(train_embeddings1, target, test_embeddings1)
del train_embeddings1, test_embeddings1
gc.collect()
train_embeddings2 = get_embeddings(train_data, "../input/nepoch2/model2/model2.bin")
test_embeddings2 = get_embeddings(test_data, "../input/nepoch2/model2/model2.bin")
svm2 = get_preds_svm(train_embeddings2, target, test_embeddings2)
del train_embeddings2, test_embeddings2
gc.collect()
train_embeddings0 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model0/model0.bin"
)
test_embeddings0 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model0/model0.bin"
)
svmpreds0 = get_preds_svm(train_embeddings0, target, test_embeddings0)
del train_embeddings0, test_embeddings0
gc.collect()
train_embeddings1 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model1/model1.bin"
)
test_embeddings1 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model1/model1.bin"
)
svmpreds1 = get_preds_svm(train_embeddings1, target, test_embeddings1)
del train_embeddings1, test_embeddings1
gc.collect()
train_embeddings2 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model2/model2.bin"
)
test_embeddings2 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model2/model2.bin"
)
svmpreds2 = get_preds_svm(train_embeddings2, target, test_embeddings2)
del train_embeddings2, test_embeddings2
gc.collect()
train_embeddings3 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model3/model3.bin"
)
test_embeddings3 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model3/model3.bin"
)
svmpreds3 = get_preds_svm(train_embeddings3, target, test_embeddings3)
del train_embeddings3, test_embeddings3
gc.collect()
train_embeddings4 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model4/model4.bin"
)
test_embeddings4 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model4/model4.bin"
)
svmpreds4 = get_preds_svm(train_embeddings4, target, test_embeddings4)
del train_embeddings4, test_embeddings4
gc.collect()
svmpreds = (svmpreds1 + svmpreds2 + svmpreds4 + svmpreds0 + svmpreds3) / 5
svms = (svm_preds2 + svmpreds + svm_preds4 + svm_preds0) / 4
svm = (svm1 + svm2 + svm0) / 3
svm_preds = svm * 0.3 + svms * 0.7
predictions = model1_predictions * 0.3 + svm_preds * 0.7
submission_df.target = predictions
print(submission_df)
submission_df.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/708/69708674.ipynb
|
commonlit-roberta-0467
|
andretugan
|
[{"Id": 69708674, "ScriptId": 19041626, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6985000, "CreationDate": "08/02/2021 21:46:58", "VersionNumber": 2.0, "Title": "Fork of Fork of commonlit-two-models", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 573.0, "LinesInsertedFromPrevious": 44.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 529.0, "LinesInsertedFromFork": 168.0, "LinesDeletedFromFork": 3.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 405.0, "TotalVotes": 0}]
|
[{"Id": 93180625, "KernelVersionId": 69708674, "SourceDatasetVersionId": 2383189}, {"Id": 93180624, "KernelVersionId": 69708674, "SourceDatasetVersionId": 2200673}]
|
[{"Id": 2383189, "DatasetId": 1440491, "DatasourceVersionId": 2425105, "CreatorUserId": 6027330, "LicenseName": "Unknown", "CreationDate": "06/30/2021 15:03:46", "VersionNumber": 1.0, "Title": "CommonLit Roberta Model Set", "Slug": "commonlit-roberta-0467", "Subtitle": "5 Roberta models, mean of the predictions gets the score of 0.467", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1440491, "CreatorUserId": 6027330, "OwnerUserId": 6027330.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2383189.0, "CurrentDatasourceVersionId": 2425105.0, "ForumId": 1459974, "Type": 2, "CreationDate": "06/30/2021 15:03:46", "LastActivityDate": "06/30/2021", "TotalViews": 1480, "TotalDownloads": 113, "TotalVotes": 12, "TotalKernels": 32}]
|
[{"Id": 6027330, "UserName": "andretugan", "DisplayName": "Andrey Tuganov", "RegisterDate": "10/24/2020", "PerformanceTier": 1}]
|
# # Model 1
import os
import math
import random
import time
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
from torch.utils.data import Dataset
from torch.utils.data import DataLoader
from transformers import AutoTokenizer
from transformers import AutoModel
from transformers import AutoConfig
from sklearn.model_selection import KFold
from sklearn.svm import SVR
import gc
gc.enable()
BATCH_SIZE = 32
MAX_LEN = 248
EVAL_SCHEDULE = [(0.50, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1.0, 1)]
ROBERTA_PATH = "../input/huggingface-roberta/roberta-base"
TOKENIZER_PATH = "../input/huggingface-roberta/roberta-base"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
test_df = pd.read_csv("/kaggle/input/commonlitreadabilityprize/test.csv")
submission_df = pd.read_csv(
"/kaggle/input/commonlitreadabilityprize/sample_submission.csv"
)
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
# # Dataset
class LitDataset(Dataset):
def __init__(self, df, inference_only=False):
super().__init__()
self.df = df
self.inference_only = inference_only
self.text = df.excerpt.tolist()
# self.text = [text.replace("\n", " ") for text in self.text]
if not self.inference_only:
self.target = torch.tensor(df.target.values, dtype=torch.float32)
self.encoded = tokenizer.batch_encode_plus(
self.text,
padding="max_length",
max_length=MAX_LEN,
truncation=True,
return_attention_mask=True,
)
def __len__(self):
return len(self.df)
def __getitem__(self, index):
input_ids = torch.tensor(self.encoded["input_ids"][index])
attention_mask = torch.tensor(self.encoded["attention_mask"][index])
if self.inference_only:
return (input_ids, attention_mask)
else:
target = self.target[index]
return (input_ids, attention_mask, target)
# # Model
# The model is inspired by the one from [Maunish](https://www.kaggle.com/maunish/clrp-roberta-svm).
class LitModel(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(768, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
# # Inference
test_dataset = LitDataset(test_df, inference_only=True)
NUM_MODELS = 5
all_predictions = np.zeros((NUM_MODELS, len(test_df)))
test_dataset = LitDataset(test_df, inference_only=True)
test_loader = DataLoader(
test_dataset, batch_size=BATCH_SIZE, drop_last=False, shuffle=False, num_workers=2
)
for model_index in range(NUM_MODELS):
model_path = f"../input/commonlit-roberta-0467/model_{model_index + 1}.pth"
print(f"\nUsing {model_path}")
model = LitModel()
model.load_state_dict(torch.load(model_path, map_location=DEVICE))
model.to(DEVICE)
all_predictions[model_index] = predict(model, test_loader)
del model
gc.collect()
model1_predictions = all_predictions.mean(axis=0)
# # Model 2
# Imported from [https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3](https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3)
import os
import gc
import sys
import cv2
import math
import time
import tqdm
import random
import numpy as np
import pandas as pd
import seaborn as sns
from tqdm import tqdm
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
from sklearn.svm import SVR
from sklearn.linear_model import Ridge
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold, StratifiedKFold
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.optim import Adam, lr_scheduler
from torch.utils.data import Dataset, DataLoader
from transformers import (
AutoModel,
AutoTokenizer,
AutoConfig,
AutoModelForSequenceClassification,
)
import plotly.express as px
import plotly.graph_objs as go
import plotly.figure_factory as ff
from colorama import Fore, Back, Style
y_ = Fore.YELLOW
r_ = Fore.RED
g_ = Fore.GREEN
b_ = Fore.BLUE
m_ = Fore.MAGENTA
c_ = Fore.CYAN
sr_ = Style.RESET_ALL
train_data = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_data = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
sample = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
num_bins = int(np.floor(1 + np.log2(len(train_data))))
train_data.loc[:, "bins"] = pd.cut(train_data["target"], bins=num_bins, labels=False)
target = train_data["target"].to_numpy()
bins = train_data.bins.to_numpy()
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": 8,
"max_len": 256,
"nfolds": 5,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
class Model(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(
"../input/huggingface-roberta/roberta-large"
)
self.model = AutoModel.from_pretrained(
"../input/huggingface-roberta/roberta-large", config=config
)
self.drop_out1 = nn.Dropout(0)
self.drop_out2 = nn.Dropout(0.1)
self.layer_norm = nn.LayerNorm(1024)
self.layer_norm1 = nn.LayerNorm(1024)
self.l1 = nn.Linear(1024, 512)
self.l2 = nn.Linear(512, 1)
self._init_weights(self.layer_norm1)
self._init_weights(self.l1)
self._init_weights(self.l2)
def _init_weights(self, module):
if isinstance(module, nn.Linear):
module.weight.data.normal_(mean=0.0, std=0.02)
if module.bias is not None:
module.bias.data.zero_()
elif isinstance(module, nn.Embedding):
module.weight.data.normal_(mean=0.0, std=0.02)
if module.padding_idx is not None:
module.weight.data[module.padding_idx].zero_()
elif isinstance(module, nn.LayerNorm):
module.bias.data.zero_()
module.weight.data.fill_(1.0)
def forward(self, input_ids, attention_mask, labels=None):
outputs = self.model(input_ids, attention_mask)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
out = self.layer_norm(mean_embeddings)
return out
def get_embeddings(df, path, plot_losses=True, verbose=True):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
tokenizer = AutoTokenizer.from_pretrained(
"../input/huggingface-roberta/roberta-large"
)
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
return np.array(embeddings)
def get_preds_svm(X, y, X_test, RidgeReg=0, bins=bins, nfolds=10, C=8, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(n_splits=10, shuffle=True, random_state=config["seed"])
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
if RidgeReg:
print("ridge...")
model = Ridge(alpha=80.0)
else:
model = SVR(C=C, kernel=kernel, gamma="auto")
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
prediction = model.predict(X_valid)
score = rmse_score(prediction, y_valid)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
print("mean rmse", np.mean(scores))
return np.array(preds) / nfolds
train_embeddings0 = get_embeddings(train_data, "../input/version0/model0/model0.bin")
test_embeddings0 = get_embeddings(test_data, "../input/version0/model0/model0.bin")
svm_preds0 = get_preds_svm(train_embeddings0, target, test_embeddings0)
ridge_preds0 = get_preds_svm(train_embeddings0, target, test_embeddings0, RidgeReg=1)
del train_embeddings0, test_embeddings0
gc.collect()
train_embeddings1 = get_embeddings(train_data, "../input/version0/model1/model1.bin")
test_embeddings1 = get_embeddings(test_data, "../input/version0/model1/model1.bin")
svm_preds1 = get_preds_svm(train_embeddings1, target, test_embeddings1)
ridge_preds1 = get_preds_svm(train_embeddings1, target, test_embeddings1, RidgeReg=1)
del train_embeddings1, test_embeddings1
gc.collect()
train_embeddings2 = get_embeddings(train_data, "../input/version0/model2/model2.bin")
test_embeddings2 = get_embeddings(test_data, "../input/version0/model2/model2.bin")
svm_preds2 = get_preds_svm(train_embeddings2, target, test_embeddings2)
ridge_preds2 = get_preds_svm(train_embeddings2, target, test_embeddings2, RidgeReg=1)
del train_embeddings2, test_embeddings2
gc.collect()
train_embeddings3 = get_embeddings(train_data, "../input/version0/model3/model3.bin")
test_embeddings3 = get_embeddings(test_data, "../input/version0/model3/model3.bin")
svm_preds3 = get_preds_svm(train_embeddings3, target, test_embeddings3)
ridge_preds3 = get_preds_svm(train_embeddings3, target, test_embeddings3, RidgeReg=1)
del train_embeddings3, test_embeddings3
gc.collect()
train_embeddings4 = get_embeddings(train_data, "../input/version0/model4/model4.bin")
test_embeddings4 = get_embeddings(test_data, "../input/version0/model4/model4.bin")
svm_preds4 = get_preds_svm(train_embeddings4, target, test_embeddings4)
ridge_preds4 = get_preds_svm(train_embeddings4, target, test_embeddings4, RidgeReg=1)
del train_embeddings4, test_embeddings4
gc.collect()
class Model2(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(
"../input/huggingface-roberta/roberta-large"
)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.0,
"layer_norm_eps": 1e-7,
}
)
self.model = AutoModel.from_pretrained(
"../input/huggingface-roberta/roberta-large", config=config
)
self.drop_out1 = nn.Dropout(0)
self.drop_out2 = nn.Dropout(0.1)
self.layer_norm = nn.LayerNorm(1024)
self.layer_norm1 = nn.LayerNorm(1024)
self.l1 = nn.Linear(1024, 512)
self.l2 = nn.Linear(512, 1)
self._init_weights(self.layer_norm1)
self._init_weights(self.l1)
self._init_weights(self.l2)
def _init_weights(self, module):
if isinstance(module, nn.Linear):
module.weight.data.normal_(mean=0.0, std=0.02)
if module.bias is not None:
module.bias.data.zero_()
elif isinstance(module, nn.Embedding):
module.weight.data.normal_(mean=0.0, std=0.02)
if module.padding_idx is not None:
module.weight.data[module.padding_idx].zero_()
elif isinstance(module, nn.LayerNorm):
module.bias.data.zero_()
module.weight.data.fill_(1.0)
def forward(self, input_ids, attention_mask, labels=None):
outputs = self.model(input_ids, attention_mask)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
out = self.layer_norm(mean_embeddings)
return out
def get_embeddings(df, path, plot_losses=True, verbose=True):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model2()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
tokenizer = AutoTokenizer.from_pretrained(
"../input/huggingface-roberta/roberta-large"
)
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
return np.array(embeddings)
def get_preds_svm(X, y, X_test, RidgeReg=0, bins=bins, nfolds=10, C=10, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(n_splits=10, shuffle=True, random_state=config["seed"])
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
if RidgeReg:
print("ridge...")
model = Ridge(alpha=80.0)
else:
model = SVR(C=C, kernel=kernel, gamma="auto")
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
prediction = model.predict(X_valid)
score = rmse_score(prediction, y_valid)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
print("mean rmse", np.mean(scores))
return np.array(preds) / nfolds
train_embeddings0 = get_embeddings(train_data, "../input/nepoch2/model0/model0.bin")
test_embeddings0 = get_embeddings(test_data, "../input/nepoch2/model0/model0.bin")
svm0 = get_preds_svm(train_embeddings0, target, test_embeddings0)
del train_embeddings0, test_embeddings0
gc.collect()
train_embeddings1 = get_embeddings(train_data, "../input/nepoch2/model1/model1.bin")
test_embeddings1 = get_embeddings(test_data, "../input/nepoch2/model1/model1.bin")
svm1 = get_preds_svm(train_embeddings1, target, test_embeddings1)
del train_embeddings1, test_embeddings1
gc.collect()
train_embeddings2 = get_embeddings(train_data, "../input/nepoch2/model2/model2.bin")
test_embeddings2 = get_embeddings(test_data, "../input/nepoch2/model2/model2.bin")
svm2 = get_preds_svm(train_embeddings2, target, test_embeddings2)
del train_embeddings2, test_embeddings2
gc.collect()
train_embeddings0 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model0/model0.bin"
)
test_embeddings0 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model0/model0.bin"
)
svmpreds0 = get_preds_svm(train_embeddings0, target, test_embeddings0)
del train_embeddings0, test_embeddings0
gc.collect()
train_embeddings1 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model1/model1.bin"
)
test_embeddings1 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model1/model1.bin"
)
svmpreds1 = get_preds_svm(train_embeddings1, target, test_embeddings1)
del train_embeddings1, test_embeddings1
gc.collect()
train_embeddings2 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model2/model2.bin"
)
test_embeddings2 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model2/model2.bin"
)
svmpreds2 = get_preds_svm(train_embeddings2, target, test_embeddings2)
del train_embeddings2, test_embeddings2
gc.collect()
train_embeddings3 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model3/model3.bin"
)
test_embeddings3 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model3/model3.bin"
)
svmpreds3 = get_preds_svm(train_embeddings3, target, test_embeddings3)
del train_embeddings3, test_embeddings3
gc.collect()
train_embeddings4 = get_embeddings(
train_data, "../input/fork-of-fork-of-nepoch2/model4/model4.bin"
)
test_embeddings4 = get_embeddings(
test_data, "../input/fork-of-fork-of-nepoch2/model4/model4.bin"
)
svmpreds4 = get_preds_svm(train_embeddings4, target, test_embeddings4)
del train_embeddings4, test_embeddings4
gc.collect()
svmpreds = (svmpreds1 + svmpreds2 + svmpreds4 + svmpreds0 + svmpreds3) / 5
svms = (svm_preds2 + svmpreds + svm_preds4 + svm_preds0) / 4
svm = (svm1 + svm2 + svm0) / 3
svm_preds = svm * 0.3 + svms * 0.7
predictions = model1_predictions * 0.3 + svm_preds * 0.7
submission_df.target = predictions
print(submission_df)
submission_df.to_csv("submission.csv", index=False)
| false | 3 | 6,279 | 0 | 6,308 | 6,279 |
||
69708453
|
<jupyter_start><jupyter_text>Used cars from mercadolibre
Kaggle dataset identifier: used-cars-from-mercadolibre
<jupyter_code>import pandas as pd
df = pd.read_csv('used-cars-from-mercadolibre/cars.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 10000 entries, 0 to 9999
Data columns (total 16 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 10000 non-null int64
1 price 10000 non-null int64
2 used 10000 non-null object
3 engine_displacement 10000 non-null float64
4 vehicle_year 10000 non-null int64
5 brand 10000 non-null object
6 model 10000 non-null object
7 engine 8656 non-null object
8 doors 10000 non-null int64
9 traction_control 5765 non-null object
10 power 10000 non-null float64
11 fuel_type 10000 non-null object
12 km 10000 non-null float64
13 transmission 8847 non-null object
14 trim 10000 non-null object
15 permalink 10000 non-null object
dtypes: float64(3), int64(4), object(9)
memory usage: 1.2+ MB
<jupyter_text>Examples:
{
"id": 0,
"price": 12500,
"used": "Usado",
"engine_displacement": 1400,
"vehicle_year": 2015,
"brand": "Chevrolet",
"model": "Prisma",
"engine": 1.4,
"doors": 4,
"traction_control": null,
"power": -1,
"fuel_type": "Nafta",
"km": 57778,
"transmission": "Manual",
"trim": "LTZ",
"permalink": "https://auto.mercadolibre.com.uy/MLU-480867981-chevrolet-prisma-ltz-2015-gris-oscuro-4-puertas-_JM"
}
{
"id": 1,
"price": 18290,
"used": "Usado",
"engine_displacement": 1998,
"vehicle_year": 2011,
"brand": "Volkswagen",
"model": "Tiguan",
"engine": 2.0,
"doors": 5,
"traction_control": "4x4",
"power": 180,
"fuel_type": "Nafta",
"km": 157699,
"transmission": "Autom\u00e1tica",
"trim": "2.0 TURBO FSI 4X4 EXTRA FULL AUTOM\u00c1TICA",
"permalink": "https://auto.mercadolibre.com.uy/MLU-480808591-volkswagen-tiguan-20-tfsi-4x4-ex-full-aut-ale-_JM"
}
{
"id": 2,
"price": 21490,
"used": "Nuevo",
"engine_displacement": 1598,
"vehicle_year": 2022,
"brand": "Nissan",
"model": "Versa",
"engine": 1.6,
"doors": 4,
"traction_control": "Delantera",
"power": 106,
"fuel_type": "Nafta",
"km": 0,
"transmission": "Manual",
"trim": "SENSE EXTRA FULL",
"permalink": "https://auto.mercadolibre.com.uy/MLU-480791804-nissan-new-versa-sense-extra-full-0km-permuta-financia-_JM"
}
{
"id": 3,
"price": 13290,
"used": "Usado",
"engine_displacement": 1598,
"vehicle_year": 2018,
"brand": "Renault",
"model": "Sandero",
"engine": 1.6,
"doors": 5,
"traction_control": "Delantera",
"power": 105,
"fuel_type": "Nafta",
"km": 31310,
"transmission": "Manual",
"trim": "PRIVILEGE 1.6 EXTRA FULL",
"permalink": "https://auto.mercadolibre.com.uy/MLU-480719884-renault-sandero-privilege-extra-full-permuta-financia-_JM"
}
<jupyter_script>import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
import math
df = pd.read_csv("/kaggle/input/used-cars-from-mercadolibre/cars.csv")
# me quedo solo con los usados
df = df[df.used == "Usado"]
df.drop(columns="used", inplace=True)
def encode_and_bind(original_dataframe, feature_to_encode):
dummies = pd.get_dummies(original_dataframe[[feature_to_encode]])
res = pd.concat([original_dataframe, dummies], axis=1)
return res
# modelos mas comunes
most_common_models = (
df[["model", "id"]]
.groupby("model")
.count()
.reset_index()
.sort_values(by="id", ascending=False)
)
# me quedo con los autos de los 100 modelos mas frecuentes, no me interesa entrenar con aquellos que ponen cualquier fruta en modelo
df = df[df.model.isin(most_common_models.head(100).model.values)]
df.info()
df_human = df.copy()
# entreno con columnas engine_displacement, vehicle_year, model (one hot), doors, km, transm (one hot) -> price
df = df[["price", "engine_displacement", "vehicle_year", "doors", "km", "model"]]
df = encode_and_bind(df, "model")
df.drop(columns="model", inplace=True)
def split_vals(a, n):
return a[:n], a[n:]
def rmse(x, y):
return math.sqrt(((x - y) ** 2).mean())
def print_score(m):
res = [
rmse(m.predict(X_train), y_train),
rmse(m.predict(X_valid), y_valid),
m.score(X_train, y_train),
m.score(X_valid, y_valid),
]
if hasattr(m, "oob_score_"):
res.append(m.oob_score_)
print(res)
df_raw_train, df_raw_test = train_test_split(df)
n_valid = 100
n_train = len(df_raw_train) - n_valid
X_train, X_valid = split_vals(df_raw_train.drop("price", axis=1), n_train)
y_train, y_valid = split_vals(df_raw_train["price"], n_train)
X_test = df_raw_test
m = RandomForestRegressor(n_jobs=1, oob_score=True)
m.fit(X_train, y_train)
print_score(m)
def feat_importance(m, df_train):
importance = m.feature_importances_
importance = pd.DataFrame(
importance, index=df_train.columns, columns=["Importance"]
)
return importance.sort_values(by=["Importance"], ascending=False)
importance = feat_importance(m, X_train)
# importance[:]
df_human["predicted"] = m.predict(df.drop(columns="price"))
df_human["diff"] = df_human.apply(
lambda row: abs(row["price"] - row["predicted"]), axis=1
)
df_human[["brand", "model", "doors", "km", "price", "predicted", "diff"]].sort_values(
by="diff", ascending=True
).head(20)
df_human[["diff"]].describe()
df2 = df_human
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/708/69708453.ipynb
|
used-cars-from-mercadolibre
|
juanblanco32312
|
[{"Id": 69708453, "ScriptId": 19048252, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2429757, "CreationDate": "08/02/2021 21:45:09", "VersionNumber": 1.0, "Title": "notebook8f8fe6ecf5", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 77.0, "LinesInsertedFromPrevious": 77.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 93180448, "KernelVersionId": 69708453, "SourceDatasetVersionId": 2488610}]
|
[{"Id": 2488610, "DatasetId": 1506410, "DatasourceVersionId": 2531177, "CreatorUserId": 2429757, "LicenseName": "Unknown", "CreationDate": "08/01/2021 15:56:18", "VersionNumber": 1.0, "Title": "Used cars from mercadolibre", "Slug": "used-cars-from-mercadolibre", "Subtitle": "10k cars for sale from Uruguay using mercadolibre.com API", "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1506410, "CreatorUserId": 2429757, "OwnerUserId": 2429757.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2488610.0, "CurrentDatasourceVersionId": 2531177.0, "ForumId": 1526156, "Type": 2, "CreationDate": "08/01/2021 15:56:18", "LastActivityDate": "08/01/2021", "TotalViews": 2127, "TotalDownloads": 107, "TotalVotes": 3, "TotalKernels": 1}]
|
[{"Id": 2429757, "UserName": "juanblanco32312", "DisplayName": "Juan Blanco", "RegisterDate": "10/30/2018", "PerformanceTier": 0}]
|
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
import math
df = pd.read_csv("/kaggle/input/used-cars-from-mercadolibre/cars.csv")
# me quedo solo con los usados
df = df[df.used == "Usado"]
df.drop(columns="used", inplace=True)
def encode_and_bind(original_dataframe, feature_to_encode):
dummies = pd.get_dummies(original_dataframe[[feature_to_encode]])
res = pd.concat([original_dataframe, dummies], axis=1)
return res
# modelos mas comunes
most_common_models = (
df[["model", "id"]]
.groupby("model")
.count()
.reset_index()
.sort_values(by="id", ascending=False)
)
# me quedo con los autos de los 100 modelos mas frecuentes, no me interesa entrenar con aquellos que ponen cualquier fruta en modelo
df = df[df.model.isin(most_common_models.head(100).model.values)]
df.info()
df_human = df.copy()
# entreno con columnas engine_displacement, vehicle_year, model (one hot), doors, km, transm (one hot) -> price
df = df[["price", "engine_displacement", "vehicle_year", "doors", "km", "model"]]
df = encode_and_bind(df, "model")
df.drop(columns="model", inplace=True)
def split_vals(a, n):
return a[:n], a[n:]
def rmse(x, y):
return math.sqrt(((x - y) ** 2).mean())
def print_score(m):
res = [
rmse(m.predict(X_train), y_train),
rmse(m.predict(X_valid), y_valid),
m.score(X_train, y_train),
m.score(X_valid, y_valid),
]
if hasattr(m, "oob_score_"):
res.append(m.oob_score_)
print(res)
df_raw_train, df_raw_test = train_test_split(df)
n_valid = 100
n_train = len(df_raw_train) - n_valid
X_train, X_valid = split_vals(df_raw_train.drop("price", axis=1), n_train)
y_train, y_valid = split_vals(df_raw_train["price"], n_train)
X_test = df_raw_test
m = RandomForestRegressor(n_jobs=1, oob_score=True)
m.fit(X_train, y_train)
print_score(m)
def feat_importance(m, df_train):
importance = m.feature_importances_
importance = pd.DataFrame(
importance, index=df_train.columns, columns=["Importance"]
)
return importance.sort_values(by=["Importance"], ascending=False)
importance = feat_importance(m, X_train)
# importance[:]
df_human["predicted"] = m.predict(df.drop(columns="price"))
df_human["diff"] = df_human.apply(
lambda row: abs(row["price"] - row["predicted"]), axis=1
)
df_human[["brand", "model", "doors", "km", "price", "predicted", "diff"]].sort_values(
by="diff", ascending=True
).head(20)
df_human[["diff"]].describe()
df2 = df_human
|
[{"used-cars-from-mercadolibre/cars.csv": {"column_names": "[\"id\", \"price\", \"used\", \"engine_displacement\", \"vehicle_year\", \"brand\", \"model\", \"engine\", \"doors\", \"traction_control\", \"power\", \"fuel_type\", \"km\", \"transmission\", \"trim\", \"permalink\"]", "column_data_types": "{\"id\": \"int64\", \"price\": \"int64\", \"used\": \"object\", \"engine_displacement\": \"float64\", \"vehicle_year\": \"int64\", \"brand\": \"object\", \"model\": \"object\", \"engine\": \"object\", \"doors\": \"int64\", \"traction_control\": \"object\", \"power\": \"float64\", \"fuel_type\": \"object\", \"km\": \"float64\", \"transmission\": \"object\", \"trim\": \"object\", \"permalink\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 10000 entries, 0 to 9999\nData columns (total 16 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 id 10000 non-null int64 \n 1 price 10000 non-null int64 \n 2 used 10000 non-null object \n 3 engine_displacement 10000 non-null float64\n 4 vehicle_year 10000 non-null int64 \n 5 brand 10000 non-null object \n 6 model 10000 non-null object \n 7 engine 8656 non-null object \n 8 doors 10000 non-null int64 \n 9 traction_control 5765 non-null object \n 10 power 10000 non-null float64\n 11 fuel_type 10000 non-null object \n 12 km 10000 non-null float64\n 13 transmission 8847 non-null object \n 14 trim 10000 non-null object \n 15 permalink 10000 non-null object \ndtypes: float64(3), int64(4), object(9)\nmemory usage: 1.2+ MB\n", "summary": "{\"id\": {\"count\": 10000.0, \"mean\": 4999.5, \"std\": 2886.8956799071675, \"min\": 0.0, \"25%\": 2499.75, \"50%\": 4999.5, \"75%\": 7499.25, \"max\": 9999.0}, \"price\": {\"count\": 10000.0, \"mean\": 20296.5324, \"std\": 27389.407650898494, \"min\": 1.0, \"25%\": 9990.0, \"50%\": 14500.0, \"75%\": 23000.0, \"max\": 1111111.0}, \"engine_displacement\": {\"count\": 10000.0, \"mean\": 998.9946199999999, \"std\": 951.7719614170768, \"min\": -1.0, \"25%\": -1.0, \"50%\": 1200.0, \"75%\": 1600.0, \"max\": 14000.0}, \"vehicle_year\": {\"count\": 10000.0, \"mean\": 2014.5316, \"std\": 7.025164534277711, \"min\": 1931.0, \"25%\": 2012.0, \"50%\": 2016.0, \"75%\": 2020.0, \"max\": 2022.0}, \"doors\": {\"count\": 10000.0, \"mean\": 16.3115, \"std\": 1199.9572002350708, \"min\": 2.0, \"25%\": 4.0, \"50%\": 5.0, \"75%\": 5.0, \"max\": 120000.0}, \"power\": {\"count\": 10000.0, \"mean\": 66.48161, \"std\": 80.24594024560054, \"min\": -1.0, \"25%\": -1.0, \"50%\": 68.0, \"75%\": 110.0, \"max\": 3000.0}, \"km\": {\"count\": 10000.0, \"mean\": 80887.58855849999, \"std\": 82686.8121218442, \"min\": -1.0, \"25%\": 0.0, \"50%\": 75000.0, \"75%\": 123343.5, \"max\": 999999.0}}", "examples": "{\"id\":{\"0\":0,\"1\":1,\"2\":2,\"3\":3},\"price\":{\"0\":12500,\"1\":18290,\"2\":21490,\"3\":13290},\"used\":{\"0\":\"Usado\",\"1\":\"Usado\",\"2\":\"Nuevo\",\"3\":\"Usado\"},\"engine_displacement\":{\"0\":1400.0,\"1\":1998.0,\"2\":1598.0,\"3\":1598.0},\"vehicle_year\":{\"0\":2015,\"1\":2011,\"2\":2022,\"3\":2018},\"brand\":{\"0\":\"Chevrolet\",\"1\":\"Volkswagen\",\"2\":\"Nissan\",\"3\":\"Renault\"},\"model\":{\"0\":\"Prisma\",\"1\":\"Tiguan\",\"2\":\"Versa\",\"3\":\"Sandero\"},\"engine\":{\"0\":\"1.4\",\"1\":\"2.0\",\"2\":\"1.6\",\"3\":\"1.6\"},\"doors\":{\"0\":4,\"1\":5,\"2\":4,\"3\":5},\"traction_control\":{\"0\":null,\"1\":\"4x4\",\"2\":\"Delantera\",\"3\":\"Delantera\"},\"power\":{\"0\":-1.0,\"1\":180.0,\"2\":106.0,\"3\":105.0},\"fuel_type\":{\"0\":\"Nafta\",\"1\":\"Nafta\",\"2\":\"Nafta\",\"3\":\"Nafta\"},\"km\":{\"0\":57778.0,\"1\":157699.0,\"2\":0.0,\"3\":31310.0},\"transmission\":{\"0\":\"Manual\",\"1\":\"Autom\\u00e1tica\",\"2\":\"Manual\",\"3\":\"Manual\"},\"trim\":{\"0\":\"LTZ\",\"1\":\"2.0 TURBO FSI 4X4 EXTRA FULL AUTOM\\u00c1TICA\",\"2\":\"SENSE EXTRA FULL\",\"3\":\"PRIVILEGE 1.6 EXTRA FULL\"},\"permalink\":{\"0\":\"https:\\/\\/auto.mercadolibre.com.uy\\/MLU-480867981-chevrolet-prisma-ltz-2015-gris-oscuro-4-puertas-_JM\",\"1\":\"https:\\/\\/auto.mercadolibre.com.uy\\/MLU-480808591-volkswagen-tiguan-20-tfsi-4x4-ex-full-aut-ale-_JM\",\"2\":\"https:\\/\\/auto.mercadolibre.com.uy\\/MLU-480791804-nissan-new-versa-sense-extra-full-0km-permuta-financia-_JM\",\"3\":\"https:\\/\\/auto.mercadolibre.com.uy\\/MLU-480719884-renault-sandero-privilege-extra-full-permuta-financia-_JM\"}}"}}]
| true | 1 |
<start_data_description><data_path>used-cars-from-mercadolibre/cars.csv:
<column_names>
['id', 'price', 'used', 'engine_displacement', 'vehicle_year', 'brand', 'model', 'engine', 'doors', 'traction_control', 'power', 'fuel_type', 'km', 'transmission', 'trim', 'permalink']
<column_types>
{'id': 'int64', 'price': 'int64', 'used': 'object', 'engine_displacement': 'float64', 'vehicle_year': 'int64', 'brand': 'object', 'model': 'object', 'engine': 'object', 'doors': 'int64', 'traction_control': 'object', 'power': 'float64', 'fuel_type': 'object', 'km': 'float64', 'transmission': 'object', 'trim': 'object', 'permalink': 'object'}
<dataframe_Summary>
{'id': {'count': 10000.0, 'mean': 4999.5, 'std': 2886.8956799071675, 'min': 0.0, '25%': 2499.75, '50%': 4999.5, '75%': 7499.25, 'max': 9999.0}, 'price': {'count': 10000.0, 'mean': 20296.5324, 'std': 27389.407650898494, 'min': 1.0, '25%': 9990.0, '50%': 14500.0, '75%': 23000.0, 'max': 1111111.0}, 'engine_displacement': {'count': 10000.0, 'mean': 998.9946199999999, 'std': 951.7719614170768, 'min': -1.0, '25%': -1.0, '50%': 1200.0, '75%': 1600.0, 'max': 14000.0}, 'vehicle_year': {'count': 10000.0, 'mean': 2014.5316, 'std': 7.025164534277711, 'min': 1931.0, '25%': 2012.0, '50%': 2016.0, '75%': 2020.0, 'max': 2022.0}, 'doors': {'count': 10000.0, 'mean': 16.3115, 'std': 1199.9572002350708, 'min': 2.0, '25%': 4.0, '50%': 5.0, '75%': 5.0, 'max': 120000.0}, 'power': {'count': 10000.0, 'mean': 66.48161, 'std': 80.24594024560054, 'min': -1.0, '25%': -1.0, '50%': 68.0, '75%': 110.0, 'max': 3000.0}, 'km': {'count': 10000.0, 'mean': 80887.58855849999, 'std': 82686.8121218442, 'min': -1.0, '25%': 0.0, '50%': 75000.0, '75%': 123343.5, 'max': 999999.0}}
<dataframe_info>
RangeIndex: 10000 entries, 0 to 9999
Data columns (total 16 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 10000 non-null int64
1 price 10000 non-null int64
2 used 10000 non-null object
3 engine_displacement 10000 non-null float64
4 vehicle_year 10000 non-null int64
5 brand 10000 non-null object
6 model 10000 non-null object
7 engine 8656 non-null object
8 doors 10000 non-null int64
9 traction_control 5765 non-null object
10 power 10000 non-null float64
11 fuel_type 10000 non-null object
12 km 10000 non-null float64
13 transmission 8847 non-null object
14 trim 10000 non-null object
15 permalink 10000 non-null object
dtypes: float64(3), int64(4), object(9)
memory usage: 1.2+ MB
<some_examples>
{'id': {'0': 0, '1': 1, '2': 2, '3': 3}, 'price': {'0': 12500, '1': 18290, '2': 21490, '3': 13290}, 'used': {'0': 'Usado', '1': 'Usado', '2': 'Nuevo', '3': 'Usado'}, 'engine_displacement': {'0': 1400.0, '1': 1998.0, '2': 1598.0, '3': 1598.0}, 'vehicle_year': {'0': 2015, '1': 2011, '2': 2022, '3': 2018}, 'brand': {'0': 'Chevrolet', '1': 'Volkswagen', '2': 'Nissan', '3': 'Renault'}, 'model': {'0': 'Prisma', '1': 'Tiguan', '2': 'Versa', '3': 'Sandero'}, 'engine': {'0': '1.4', '1': '2.0', '2': '1.6', '3': '1.6'}, 'doors': {'0': 4, '1': 5, '2': 4, '3': 5}, 'traction_control': {'0': None, '1': '4x4', '2': 'Delantera', '3': 'Delantera'}, 'power': {'0': -1.0, '1': 180.0, '2': 106.0, '3': 105.0}, 'fuel_type': {'0': 'Nafta', '1': 'Nafta', '2': 'Nafta', '3': 'Nafta'}, 'km': {'0': 57778.0, '1': 157699.0, '2': 0.0, '3': 31310.0}, 'transmission': {'0': 'Manual', '1': 'Automática', '2': 'Manual', '3': 'Manual'}, 'trim': {'0': 'LTZ', '1': '2.0 TURBO FSI 4X4 EXTRA FULL AUTOMÁTICA', '2': 'SENSE EXTRA FULL', '3': 'PRIVILEGE 1.6 EXTRA FULL'}, 'permalink': {'0': 'https://auto.mercadolibre.com.uy/MLU-480867981-chevrolet-prisma-ltz-2015-gris-oscuro-4-puertas-_JM', '1': 'https://auto.mercadolibre.com.uy/MLU-480808591-volkswagen-tiguan-20-tfsi-4x4-ex-full-aut-ale-_JM', '2': 'https://auto.mercadolibre.com.uy/MLU-480791804-nissan-new-versa-sense-extra-full-0km-permuta-financia-_JM', '3': 'https://auto.mercadolibre.com.uy/MLU-480719884-renault-sandero-privilege-extra-full-permuta-financia-_JM'}}
<end_description>
| 903 | 1 | 2,197 | 903 |
69708302
|
# ### Goal for This notebook:
# 1. Feature Engineernig
# 2. Clean Data
# 3. Feature Extraction
# 4. Visualise Data
# ### Load data and liberary
# * Here we load basic liberary
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib as mpl
import plotly.express as px
### so that u dont have warnings
from warnings import filterwarnings
import plotly.graph_objs as go
from plotly.offline import iplot
filterwarnings("ignore")
# Custom colors
class color:
S = "\033[1m" + "\033[93m"
E = "\033[0m"
my_colors = ["#E7C84B", "#4EE4EA", "#4EA9EA", "#242179", "#AB51E9", "#E051E9"]
print(color.S + "Notebook Color Scheme:" + color.E)
sns.palplot(sns.color_palette(my_colors))
# Set Style
sns.set_style("white")
mpl.rcParams["xtick.labelsize"] = 16
mpl.rcParams["ytick.labelsize"] = 16
mpl.rcParams["axes.spines.left"] = False
mpl.rcParams["axes.spines.right"] = False
mpl.rcParams["axes.spines.top"] = False
plt.rcParams.update({"font.size": 17})
# #### Read in the training data
# Read in the training data
train_df = pd.read_csv("../input/tabular-playground-series-aug-2021/train.csv")
# Print some useful information
print(
color.S + "Train Data has:" + color.E,
"{:,}".format(train.shape[0]),
"observations.",
"\n" + color.S + "Number of Missing Values:" + color.E,
train.isna().sum()[0],
"\n" + "\n" + color.S + "Head of Training Data:" + color.E,
)
train.head(5)
# ### Read in the Test data
test_df = pd.read_csv("../input/tabular-playground-series-aug-2021/test.csv")
# Print some useful information
print(
color.S + "Test Data has:" + color.E,
"{:,}".format(test_df.shape[0]),
"observations.",
"\n" + color.S + "Number of Missing Values:" + color.E,
test_df.isna().sum()[0],
"\n" + "\n" + color.S + "Head of Training Data:" + color.E,
)
test_df.head(5)
# ### Work with Train Data
train_df.head()
train_df.tail()
train_df.shape
train_df.info()
train_df.dtypes
feature_na = [
feature for feature in train_df.columns if train_df[feature].isnull().sum() > 0
]
feature_na
# #### getting all NAN features
# % of missing values
for feature in feature_na:
print(
"{} has {} % missing values".format(
feature, np.round(trian_df[feature].isnull().sum() / len(train_df) * 100, 4)
)
)
train_df["loss"].unique()
# ### Showing Basics Statistics
# Now that you’ve seen what data types are in your dataset, it’s time to get an overview of the values each column contains. You can do this with .describe():
#
train_df.describe()
train_df.max()
train_df.describe().T.style.bar(subset=["mean"], color="red").background_gradient(
subset=["std"], cmap="Reds"
).background_gradient(subset=["50%"], cmap="coolwarm")
# * This data haven't any decimal value all value float
# Skewness of the distribution
print(train_df.skew())
# ### Data Interaction
# •Correlation
# * Correlation tells relation between two attributes.
# * Correlation requires continous data. Hence, ignore categorical data
# * Calculates pearson co-efficient for all combinations
#
data_corr = train_df.corr()
data_corr
sns.scatterplot(x="f1", y="f2", hue="loss", data=train_df)
sns.scatterplot(x=train_df["f1"], y=train_df["loss"])
sns.regplot(x=train_df["f1"], y=train_df["loss"])
# ### calculate avg f1 and loss
train_df.groupby("f1")["loss"].mean().nlargest(20).plot.bar()
# ##### Loss distribution
sns.set_style(style="whitegrid")
sns.distplot(train_df["loss"])
plt.figure(figsize=(10, 7))
chains = train_df["loss"].value_counts()[0:20]
sns.barplot(x=chains, y=chains.index, palette="deep")
plt.title("Most loss ")
plt.xlabel("Number of outlets")
x = train_df["f1"].value_counts()
labels = ["accepted", "not accepted"]
fig = px.scatter_3d(train_df.iloc[:500], x="f0", y="f1", z="f2", color="loss")
fig.show()
plt.figure(figsize=(20, 12))
train_df["loss"].value_counts().nlargest(20).plot.bar(color="red")
plt.gcf().autofmt_xdate()
# #### How many types ofLosses we have?
train_df["loss"].isna().sum()
len(train_df["loss"].unique())
trace1 = go.Bar(
x=train_df.groupby("f1")["f2"].max().nlargest(12).index,
y=train_df.groupby("f3")["f4"].max().nlargest(12),
name="loss",
)
iplot([trace1])
fig = px.box(train_df.head(500), x="f1", y="loss")
fig.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/708/69708302.ipynb
| null | null |
[{"Id": 69708302, "ScriptId": 18994630, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6470511, "CreationDate": "08/02/2021 21:43:55", "VersionNumber": 2.0, "Title": "EDA For Tabular Playground Series - Aug 2021", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 162.0, "LinesInsertedFromPrevious": 87.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 75.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ### Goal for This notebook:
# 1. Feature Engineernig
# 2. Clean Data
# 3. Feature Extraction
# 4. Visualise Data
# ### Load data and liberary
# * Here we load basic liberary
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib as mpl
import plotly.express as px
### so that u dont have warnings
from warnings import filterwarnings
import plotly.graph_objs as go
from plotly.offline import iplot
filterwarnings("ignore")
# Custom colors
class color:
S = "\033[1m" + "\033[93m"
E = "\033[0m"
my_colors = ["#E7C84B", "#4EE4EA", "#4EA9EA", "#242179", "#AB51E9", "#E051E9"]
print(color.S + "Notebook Color Scheme:" + color.E)
sns.palplot(sns.color_palette(my_colors))
# Set Style
sns.set_style("white")
mpl.rcParams["xtick.labelsize"] = 16
mpl.rcParams["ytick.labelsize"] = 16
mpl.rcParams["axes.spines.left"] = False
mpl.rcParams["axes.spines.right"] = False
mpl.rcParams["axes.spines.top"] = False
plt.rcParams.update({"font.size": 17})
# #### Read in the training data
# Read in the training data
train_df = pd.read_csv("../input/tabular-playground-series-aug-2021/train.csv")
# Print some useful information
print(
color.S + "Train Data has:" + color.E,
"{:,}".format(train.shape[0]),
"observations.",
"\n" + color.S + "Number of Missing Values:" + color.E,
train.isna().sum()[0],
"\n" + "\n" + color.S + "Head of Training Data:" + color.E,
)
train.head(5)
# ### Read in the Test data
test_df = pd.read_csv("../input/tabular-playground-series-aug-2021/test.csv")
# Print some useful information
print(
color.S + "Test Data has:" + color.E,
"{:,}".format(test_df.shape[0]),
"observations.",
"\n" + color.S + "Number of Missing Values:" + color.E,
test_df.isna().sum()[0],
"\n" + "\n" + color.S + "Head of Training Data:" + color.E,
)
test_df.head(5)
# ### Work with Train Data
train_df.head()
train_df.tail()
train_df.shape
train_df.info()
train_df.dtypes
feature_na = [
feature for feature in train_df.columns if train_df[feature].isnull().sum() > 0
]
feature_na
# #### getting all NAN features
# % of missing values
for feature in feature_na:
print(
"{} has {} % missing values".format(
feature, np.round(trian_df[feature].isnull().sum() / len(train_df) * 100, 4)
)
)
train_df["loss"].unique()
# ### Showing Basics Statistics
# Now that you’ve seen what data types are in your dataset, it’s time to get an overview of the values each column contains. You can do this with .describe():
#
train_df.describe()
train_df.max()
train_df.describe().T.style.bar(subset=["mean"], color="red").background_gradient(
subset=["std"], cmap="Reds"
).background_gradient(subset=["50%"], cmap="coolwarm")
# * This data haven't any decimal value all value float
# Skewness of the distribution
print(train_df.skew())
# ### Data Interaction
# •Correlation
# * Correlation tells relation between two attributes.
# * Correlation requires continous data. Hence, ignore categorical data
# * Calculates pearson co-efficient for all combinations
#
data_corr = train_df.corr()
data_corr
sns.scatterplot(x="f1", y="f2", hue="loss", data=train_df)
sns.scatterplot(x=train_df["f1"], y=train_df["loss"])
sns.regplot(x=train_df["f1"], y=train_df["loss"])
# ### calculate avg f1 and loss
train_df.groupby("f1")["loss"].mean().nlargest(20).plot.bar()
# ##### Loss distribution
sns.set_style(style="whitegrid")
sns.distplot(train_df["loss"])
plt.figure(figsize=(10, 7))
chains = train_df["loss"].value_counts()[0:20]
sns.barplot(x=chains, y=chains.index, palette="deep")
plt.title("Most loss ")
plt.xlabel("Number of outlets")
x = train_df["f1"].value_counts()
labels = ["accepted", "not accepted"]
fig = px.scatter_3d(train_df.iloc[:500], x="f0", y="f1", z="f2", color="loss")
fig.show()
plt.figure(figsize=(20, 12))
train_df["loss"].value_counts().nlargest(20).plot.bar(color="red")
plt.gcf().autofmt_xdate()
# #### How many types ofLosses we have?
train_df["loss"].isna().sum()
len(train_df["loss"].unique())
trace1 = go.Bar(
x=train_df.groupby("f1")["f2"].max().nlargest(12).index,
y=train_df.groupby("f3")["f4"].max().nlargest(12),
name="loss",
)
iplot([trace1])
fig = px.box(train_df.head(500), x="f1", y="loss")
fig.show()
| false | 0 | 1,497 | 0 | 1,497 | 1,497 |
||
69672309
|
import numpy as np
import pandas as pd
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
import plotly.express as px
import seaborn as sns
from matplotlib import rcParams
rcParams["axes.spines.top"] = False
rcParams["axes.spines.right"] = False
from xgboost import XGBRegressor
train_raw = pd.read_csv(
"../input/tabular-playground-series-aug-2021/train.csv", index_col="id"
)
test_raw = pd.read_csv(
"../input/tabular-playground-series-aug-2021/test.csv", index_col="id"
)
print("Train shape:", train_raw.shape)
print("Test shape:", test_raw.shape)
# combine train and test for pre processing
# save original end/start indices for re-splitting train/test later
train_end_idx = 249999
test_start_idx = 250000
all_raw = pd.concat([train_raw, test_raw])
all_raw.head(3)
# check for missing values
print("Train data null count:", train_raw.isnull().sum().sum())
print("Test data null count:", test_raw.isnull().sum().sum())
# check feature datatypes
train_raw.dtypes.unique()
# see summary statistics for features
all_raw.drop("loss", axis=1).describe().T.sample(10)
# examine distribution for features
fig, axs = plt.subplots(20, 5, figsize=(12, 40))
plt.suptitle("Feature Distributions")
for i, feat in enumerate(all_raw.loc[:, :"f99"]):
sns.histplot(all_raw[feat], kde=False, ax=axs.flat[i])
axs.flat[i].axes.get_yaxis().set_visible(False)
axs.flat[i].spines["left"].set_visible(False)
plt.tight_layout()
plt.show()
sns.kdeplot(train_raw["loss"], shade=True)
plt.show()
# set up train test data
X_train = train_raw.copy()
y_train = X_train.pop("loss")
X_test = test_raw.copy()
X_train.head()
# baseline model
xgb_regressor = XGBRegressor(tree_method="approx", max_bin=100)
xgb_regressor.fit(X_train, y_train)
predictions = xgb_regressor.predict(X_test)
output = pd.DataFrame({"id": X_test.index, "loss": predictions})
output.to_csv("my_submission.csv", index=False)
print("Submission saved.")
sns.kdeplot(output["loss"], shade=True)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/672/69672309.ipynb
| null | null |
[{"Id": 69672309, "ScriptId": 18994649, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6743276, "CreationDate": "08/02/2021 17:16:52", "VersionNumber": 3.0, "Title": "TPS - Aug 2021", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 73.0, "LinesInsertedFromPrevious": 32.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 41.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
import plotly.express as px
import seaborn as sns
from matplotlib import rcParams
rcParams["axes.spines.top"] = False
rcParams["axes.spines.right"] = False
from xgboost import XGBRegressor
train_raw = pd.read_csv(
"../input/tabular-playground-series-aug-2021/train.csv", index_col="id"
)
test_raw = pd.read_csv(
"../input/tabular-playground-series-aug-2021/test.csv", index_col="id"
)
print("Train shape:", train_raw.shape)
print("Test shape:", test_raw.shape)
# combine train and test for pre processing
# save original end/start indices for re-splitting train/test later
train_end_idx = 249999
test_start_idx = 250000
all_raw = pd.concat([train_raw, test_raw])
all_raw.head(3)
# check for missing values
print("Train data null count:", train_raw.isnull().sum().sum())
print("Test data null count:", test_raw.isnull().sum().sum())
# check feature datatypes
train_raw.dtypes.unique()
# see summary statistics for features
all_raw.drop("loss", axis=1).describe().T.sample(10)
# examine distribution for features
fig, axs = plt.subplots(20, 5, figsize=(12, 40))
plt.suptitle("Feature Distributions")
for i, feat in enumerate(all_raw.loc[:, :"f99"]):
sns.histplot(all_raw[feat], kde=False, ax=axs.flat[i])
axs.flat[i].axes.get_yaxis().set_visible(False)
axs.flat[i].spines["left"].set_visible(False)
plt.tight_layout()
plt.show()
sns.kdeplot(train_raw["loss"], shade=True)
plt.show()
# set up train test data
X_train = train_raw.copy()
y_train = X_train.pop("loss")
X_test = test_raw.copy()
X_train.head()
# baseline model
xgb_regressor = XGBRegressor(tree_method="approx", max_bin=100)
xgb_regressor.fit(X_train, y_train)
predictions = xgb_regressor.predict(X_test)
output = pd.DataFrame({"id": X_test.index, "loss": predictions})
output.to_csv("my_submission.csv", index=False)
print("Submission saved.")
sns.kdeplot(output["loss"], shade=True)
plt.show()
| false | 0 | 690 | 0 | 690 | 690 |
||
69672521
|
# **Importing Libraries**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import os
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
# **Loading Dataset **
os.listdir("../input/titanic/")
train_data = pd.read_csv("../input/titanic/train.csv")
test_data = pd.read_csv("../input/titanic/test.csv")
subm = pd.read_csv("../input/titanic/gender_submission.csv")
train_data.head()
train_data.describe()
train_data.dtypes
# **Dealing with missing values **
train_data.isnull().sum().sort_values(ascending=False)
train_data["Cabin"] = train_data["Cabin"].fillna(train_data["Cabin"].mode()[0])
train_data["Age"] = train_data["Age"].fillna(train_data["Age"].mean())
train_data["Embarked"] = train_data["Embarked"].fillna(train_data["Embarked"].mode()[0])
test_data.isnull().sum().sort_values(ascending=False)
test_data["Cabin"] = test_data["Cabin"].fillna(test_data["Cabin"].mode()[0])
test_data["Age"] = test_data["Age"].fillna(test_data["Age"].mean())
test_data["Fare"] = test_data["Fare"].fillna(test_data["Fare"].mean())
# **Plotting Graphs**
sns.barplot(x="Sex", y="Survived", data=train_data)
# Above shows that survival rate of female is high
sns.barplot(x="Pclass", y="Survived", hue="Sex", data=train_data)
# Above shows that survival rate of Pclass 1 is high
# Feature Engineering
train_data.drop(["PassengerId"], axis=1, inplace=True)
test_data.drop(["PassengerId"], axis=1, inplace=True)
data = [train_data, test_data]
# Dividing Age into groups
for dataset in data:
dataset["Age"] = dataset.Age.astype(int)
dataset.loc[(dataset["Age"] <= 11), "Age"] = 0
dataset.loc[(dataset["Age"] > 11) & (dataset["Age"] <= 18), "Age"] = 1
dataset.loc[(dataset["Age"] > 18) & (dataset["Age"] <= 22), "Age"] = 2
dataset.loc[(dataset["Age"] > 22) & (dataset["Age"] <= 27), "Age"] = 3
dataset.loc[(dataset["Age"] > 27) & (dataset["Age"] <= 34), "Age"] = 4
dataset.loc[(dataset["Age"] > 34) & (dataset["Age"] <= 40), "Age"] = 5
dataset.loc[(dataset["Age"] > 40) & (dataset["Age"] <= 60), "Age"] = 6
dataset.loc[(dataset["Age"] > 60), "Age"] = 7
train_data.Age.value_counts()
for dataset in data:
dataset["Age_Pclass"] = dataset["Age"] * dataset["Pclass"]
for dataset in data:
dataset["Family_Size"] = dataset["SibSp"] + dataset["Parch"]
dataset["Travelling_Alone"] = dataset["Family_Size"].apply(
lambda x: 0 if x > 0 else 1
)
# yes or 1 if family_size=0 else no or 1
train_data["Travelling_Alone"].dtype
train_data["Travelling_Alone"].value_counts()
train_data["Fare"].dtype
for dataset in data:
dataset.loc[dataset["Fare"] <= 7.91, "Fare"] = 0
dataset.loc[(dataset["Fare"] > 7.91) & (dataset["Fare"] <= 14.454), "Fare"] = 1
dataset.loc[(dataset["Fare"] > 14.454) & (dataset["Fare"] <= 31), "Fare"] = 2
dataset.loc[(dataset["Fare"] > 31) & (dataset["Fare"] <= 99), "Fare"] = 3
dataset.loc[(dataset["Fare"] > 99) & (dataset["Fare"] <= 250), "Fare"] = 4
dataset.loc[dataset["Fare"] > 250, "Fare"] = 5
dataset["Fare"] = dataset["Fare"].astype(int)
# Fare per person
for dataset in data:
dataset["Fare_per_person"] = dataset["Fare"] / (dataset["Family_Size"] + 1)
dataset["Fare_per_person"] = dataset["Fare_per_person"].astype(int)
train_data.Fare_per_person.value_counts()
dataset["Fare_per_person"].dtype
# As we will be using Fare per person so Fare can be dropped and Ticket have around 75 %
train_data.drop(["Fare"], axis=1, inplace=True)
test_data.drop(["Fare"], axis=1, inplace=True)
train_data.head()
for dataset in data:
dataset["Title"] = dataset["Name"].apply(
lambda x: x.split(",")[1].split(".")[0].strip()
)
dataset["Title"] = dataset["Title"].astype(str)
dataset["Title"] = dataset["Title"].map(
{
"Mr": "Mr",
"Miss": "Miss",
"Mrs": "Mrs",
"Master": "Rare",
"Dr": "Rare",
"Rev": "Rare",
"Col": "Rare",
"Major": "Rare",
"Ms": "Miss",
"Mlle": "Miss",
"the Countess": "Rare",
"Capt": "Rare",
"Dona": "Rare",
"Lady": "Rare",
"Don": "Rare",
"Jonkheer": "Mr",
"Sir": "Mr",
"Mme": "Mrs",
}
)
dataset.drop(["Name", "Ticket"], axis=1, inplace=True)
train_data.Cabin.dtype
for dataset in data:
dataset["Cabin"] = dataset["Cabin"].astype(str)
dataset["Cabin"] = dataset["Cabin"].str[0]
train_data.head()
test_data.head()
# ***Creating dummies for categorical data***
train_data = pd.get_dummies(train_data, drop_first=True)
test_data = pd.get_dummies(test_data, drop_first=True)
train_data.drop(["Cabin_T"], axis=1, inplace=True)
# Splitting Dataset
x = train_data.loc[:, "Pclass":"Title_Rare"]
y = train_data.loc[:, "Survived"]
print("x shape:", x.shape)
print("y shape:", y.shape)
# Splitting in training and testing set
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42
)
print("x_train shape:", x_train.shape)
print("y_train shape:", y_train.shape)
print("x_test shape:", x_test.shape)
print("y_test shape:", y_test.shape)
# scaling numerical features
scaler = MinMaxScaler()
x_train = scaler.fit_transform(x_train)
# x_test=scaler.transform(x_test)
# **Random forest Algorithim**
random_forest = RandomForestClassifier(n_estimators=100, max_depth=5, random_state=1)
random_forest.fit(x_train, y_train)
Y_prediction = random_forest.predict(x_test)
random_forest.score(x_train, y_train)
acc_random_forest = round(random_forest.score(x_train, y_train) * 100, 2)
acc_random_forest
# **Logistic Regression**
logreg = LogisticRegression()
logreg.fit(x_train, y_train)
Y_pred = logreg.predict(x_test)
acc_log = round(logreg.score(x_train, y_train) * 100, 2)
acc_log
# **KNN**
knn = KNeighborsClassifier(n_neighbors=3)
knn.fit(x_train, y_train)
Y_pred = knn.predict(x_test)
acc_knn = round(knn.score(x_train, y_train) * 100, 2)
acc_knn
# SVC
linear_svc = LinearSVC()
linear_svc.fit(x_train, y_train)
Y_pred = linear_svc.predict(x_test)
acc_linear_svc = round(linear_svc.score(x_train, y_train) * 100, 2)
acc_linear_svc
# **Decision Tree**
decision_tree = DecisionTreeClassifier()
decision_tree.fit(x_train, y_train)
Y_pred = decision_tree.predict(x_test)
acc_decision_tree = round(decision_tree.score(x_train, y_train) * 100, 2)
acc_decision_tree
# decission tree n random forest is showing good score
from sklearn.model_selection import cross_val_predict
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test, Y_prediction)
print(classification_report(y_test, Y_prediction))
test_data = scaler.transform(test_data)
prediction_final = random_forest.predict(test_data)
subm
del subm["Survived"]
subm["Survived"] = prediction_final
subm.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/672/69672521.ipynb
| null | null |
[{"Id": 69672521, "ScriptId": 17546293, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2985778, "CreationDate": "08/02/2021 17:18:31", "VersionNumber": 2.0, "Title": "Survival Prediction", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 254.0, "LinesInsertedFromPrevious": 4.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 250.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# **Importing Libraries**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import os
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
# **Loading Dataset **
os.listdir("../input/titanic/")
train_data = pd.read_csv("../input/titanic/train.csv")
test_data = pd.read_csv("../input/titanic/test.csv")
subm = pd.read_csv("../input/titanic/gender_submission.csv")
train_data.head()
train_data.describe()
train_data.dtypes
# **Dealing with missing values **
train_data.isnull().sum().sort_values(ascending=False)
train_data["Cabin"] = train_data["Cabin"].fillna(train_data["Cabin"].mode()[0])
train_data["Age"] = train_data["Age"].fillna(train_data["Age"].mean())
train_data["Embarked"] = train_data["Embarked"].fillna(train_data["Embarked"].mode()[0])
test_data.isnull().sum().sort_values(ascending=False)
test_data["Cabin"] = test_data["Cabin"].fillna(test_data["Cabin"].mode()[0])
test_data["Age"] = test_data["Age"].fillna(test_data["Age"].mean())
test_data["Fare"] = test_data["Fare"].fillna(test_data["Fare"].mean())
# **Plotting Graphs**
sns.barplot(x="Sex", y="Survived", data=train_data)
# Above shows that survival rate of female is high
sns.barplot(x="Pclass", y="Survived", hue="Sex", data=train_data)
# Above shows that survival rate of Pclass 1 is high
# Feature Engineering
train_data.drop(["PassengerId"], axis=1, inplace=True)
test_data.drop(["PassengerId"], axis=1, inplace=True)
data = [train_data, test_data]
# Dividing Age into groups
for dataset in data:
dataset["Age"] = dataset.Age.astype(int)
dataset.loc[(dataset["Age"] <= 11), "Age"] = 0
dataset.loc[(dataset["Age"] > 11) & (dataset["Age"] <= 18), "Age"] = 1
dataset.loc[(dataset["Age"] > 18) & (dataset["Age"] <= 22), "Age"] = 2
dataset.loc[(dataset["Age"] > 22) & (dataset["Age"] <= 27), "Age"] = 3
dataset.loc[(dataset["Age"] > 27) & (dataset["Age"] <= 34), "Age"] = 4
dataset.loc[(dataset["Age"] > 34) & (dataset["Age"] <= 40), "Age"] = 5
dataset.loc[(dataset["Age"] > 40) & (dataset["Age"] <= 60), "Age"] = 6
dataset.loc[(dataset["Age"] > 60), "Age"] = 7
train_data.Age.value_counts()
for dataset in data:
dataset["Age_Pclass"] = dataset["Age"] * dataset["Pclass"]
for dataset in data:
dataset["Family_Size"] = dataset["SibSp"] + dataset["Parch"]
dataset["Travelling_Alone"] = dataset["Family_Size"].apply(
lambda x: 0 if x > 0 else 1
)
# yes or 1 if family_size=0 else no or 1
train_data["Travelling_Alone"].dtype
train_data["Travelling_Alone"].value_counts()
train_data["Fare"].dtype
for dataset in data:
dataset.loc[dataset["Fare"] <= 7.91, "Fare"] = 0
dataset.loc[(dataset["Fare"] > 7.91) & (dataset["Fare"] <= 14.454), "Fare"] = 1
dataset.loc[(dataset["Fare"] > 14.454) & (dataset["Fare"] <= 31), "Fare"] = 2
dataset.loc[(dataset["Fare"] > 31) & (dataset["Fare"] <= 99), "Fare"] = 3
dataset.loc[(dataset["Fare"] > 99) & (dataset["Fare"] <= 250), "Fare"] = 4
dataset.loc[dataset["Fare"] > 250, "Fare"] = 5
dataset["Fare"] = dataset["Fare"].astype(int)
# Fare per person
for dataset in data:
dataset["Fare_per_person"] = dataset["Fare"] / (dataset["Family_Size"] + 1)
dataset["Fare_per_person"] = dataset["Fare_per_person"].astype(int)
train_data.Fare_per_person.value_counts()
dataset["Fare_per_person"].dtype
# As we will be using Fare per person so Fare can be dropped and Ticket have around 75 %
train_data.drop(["Fare"], axis=1, inplace=True)
test_data.drop(["Fare"], axis=1, inplace=True)
train_data.head()
for dataset in data:
dataset["Title"] = dataset["Name"].apply(
lambda x: x.split(",")[1].split(".")[0].strip()
)
dataset["Title"] = dataset["Title"].astype(str)
dataset["Title"] = dataset["Title"].map(
{
"Mr": "Mr",
"Miss": "Miss",
"Mrs": "Mrs",
"Master": "Rare",
"Dr": "Rare",
"Rev": "Rare",
"Col": "Rare",
"Major": "Rare",
"Ms": "Miss",
"Mlle": "Miss",
"the Countess": "Rare",
"Capt": "Rare",
"Dona": "Rare",
"Lady": "Rare",
"Don": "Rare",
"Jonkheer": "Mr",
"Sir": "Mr",
"Mme": "Mrs",
}
)
dataset.drop(["Name", "Ticket"], axis=1, inplace=True)
train_data.Cabin.dtype
for dataset in data:
dataset["Cabin"] = dataset["Cabin"].astype(str)
dataset["Cabin"] = dataset["Cabin"].str[0]
train_data.head()
test_data.head()
# ***Creating dummies for categorical data***
train_data = pd.get_dummies(train_data, drop_first=True)
test_data = pd.get_dummies(test_data, drop_first=True)
train_data.drop(["Cabin_T"], axis=1, inplace=True)
# Splitting Dataset
x = train_data.loc[:, "Pclass":"Title_Rare"]
y = train_data.loc[:, "Survived"]
print("x shape:", x.shape)
print("y shape:", y.shape)
# Splitting in training and testing set
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42
)
print("x_train shape:", x_train.shape)
print("y_train shape:", y_train.shape)
print("x_test shape:", x_test.shape)
print("y_test shape:", y_test.shape)
# scaling numerical features
scaler = MinMaxScaler()
x_train = scaler.fit_transform(x_train)
# x_test=scaler.transform(x_test)
# **Random forest Algorithim**
random_forest = RandomForestClassifier(n_estimators=100, max_depth=5, random_state=1)
random_forest.fit(x_train, y_train)
Y_prediction = random_forest.predict(x_test)
random_forest.score(x_train, y_train)
acc_random_forest = round(random_forest.score(x_train, y_train) * 100, 2)
acc_random_forest
# **Logistic Regression**
logreg = LogisticRegression()
logreg.fit(x_train, y_train)
Y_pred = logreg.predict(x_test)
acc_log = round(logreg.score(x_train, y_train) * 100, 2)
acc_log
# **KNN**
knn = KNeighborsClassifier(n_neighbors=3)
knn.fit(x_train, y_train)
Y_pred = knn.predict(x_test)
acc_knn = round(knn.score(x_train, y_train) * 100, 2)
acc_knn
# SVC
linear_svc = LinearSVC()
linear_svc.fit(x_train, y_train)
Y_pred = linear_svc.predict(x_test)
acc_linear_svc = round(linear_svc.score(x_train, y_train) * 100, 2)
acc_linear_svc
# **Decision Tree**
decision_tree = DecisionTreeClassifier()
decision_tree.fit(x_train, y_train)
Y_pred = decision_tree.predict(x_test)
acc_decision_tree = round(decision_tree.score(x_train, y_train) * 100, 2)
acc_decision_tree
# decission tree n random forest is showing good score
from sklearn.model_selection import cross_val_predict
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test, Y_prediction)
print(classification_report(y_test, Y_prediction))
test_data = scaler.transform(test_data)
prediction_final = random_forest.predict(test_data)
subm
del subm["Survived"]
subm["Survived"] = prediction_final
subm.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
| false | 0 | 2,484 | 0 | 2,484 | 2,484 |
||
69672345
|
# # **Simple explanation of how to handle categorical variables**
# We refer below competition for tutorial,
# https://www.kaggle.com/c/home-data-for-ml-course
# Cover below course topic,
# https://www.kaggle.com/alexisbcook/categorical-variables
# # Lets collect data for train and test
import pandas as pd
from sklearn.model_selection import train_test_split
# LodeStar has addded the option below
pd.set_option("display.max_columns", 100)
# Read the data
X_train_full = pd.read_csv(
"../input/home-data-for-ml-course/train.csv", index_col="Id"
) # Train data contain 1460 rows
X_test_full = pd.read_csv(
"../input/home-data-for-ml-course/test.csv", index_col="Id"
) # Test data contain 1459 rows
# print(X_train_full.head()) # Return all column with first 5 values
# print("X_train_full data row and col:", X_test_full.shape) # (1460, 80)
# X_train_full data contain SalePrice, while X_test_full not contain SalePrice which need to predict
# Remove rows with missing target, separate target from predictors
X_train_full.dropna(axis=0, subset=["SalePrice"], inplace=True)
# print(X_train_full.shape) # (1460, 80) i.e no value missing
Y_train_full = X_train_full.SalePrice
# print(Y_train_real.shape) # (1460,) i.e only target data, single column
X_train_full.drop(["SalePrice"], axis=1, inplace=True)
# print(X_train_full.shape) # (1460, 79) i.e only train data
# To keep things simple, we'll drop columns with missing values
cols_with_missing = [
col for col in X_train_full.columns if X_train_full[col].isnull().any()
]
# print(len(cols_with_missing)) # Return 19, as 19 column contain missing values
X_train_full.drop(cols_with_missing, axis=1, inplace=True)
X_test_full.drop(
cols_with_missing, axis=1, inplace=True
) # Also removing from test data to test with model
# Break off validation set from training data
X_train, X_valid, y_train, y_valid = train_test_split(
X_train_full, Y_train_full, train_size=0.8, test_size=0.2, random_state=0
)
# print(X_train.shape) # (1168, 60)
# X_train.head() # return all numerical as well as categorical columns
# # Create common function to get Mean absolute error
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
# function for comparing different approaches
def score_dataset(X_train, X_valid, y_train, y_valid):
model = RandomForestRegressor(n_estimators=100, random_state=0)
model.fit(X_train, y_train)
preds = model.predict(X_valid)
return mean_absolute_error(y_valid, preds)
# # Approach 1 : Drop column with categorical data
# First approach drop column with categorical data
drop_X_train = X_train.select_dtypes(exclude=["object"])
drop_X_valid = X_valid.select_dtypes(exclude=["object"])
print("MAE from Approach 1 (Drop categorical variables):")
print(score_dataset(drop_X_train, drop_X_valid, y_train, y_valid)) # 17837.82570776256
# # Approach 2 : Ordinal encoding
# First lets investigatge one column from dataset which is contain categorical data, column: Condition2
print(
"Unique values in 'Condition2' column in training data:",
X_train["Condition2"].unique(),
)
print(
"\nUnique values in 'Condition2' column in validation data:",
X_valid["Condition2"].unique(),
)
# > Fitting an ordinal encoder to a column in the training data creates a corresponding integer-valued label for each unique value that appears in the training data. In the case that the validation data contains values that don't also appear in the training data, the encoder will throw an error, because these values won't have an integer assigned to them. Notice that the 'Condition2' column in the validation data contains the values 'RRAn' and 'RRNn', but these don't appear in the training data -- thus, if we try to use an ordinal encoder with scikit-learn, the code will throw an error.
# This is a common problem that you'll encounter with real-world data, and there are many approaches to fixing this issue. For instance, you can write a custom ordinal encoder to deal with new categories. The simplest approach, however, is to drop the problematic categorical columns.
# Run the code cell below to save the problematic columns to a Python list **bad_label_cols**. Likewise, columns that can be safely ordinal encoded are stored in **good_label_cols**.
# Fetch all categorical columns
object_cols = [col for col in X_train.columns if X_train[col].dtype == "object"]
# Columns that can be safely ordinal encoded
good_label_cols = [
col for col in object_cols if set(X_valid[col]).issubset(set(X_train[col]))
]
# Problematic columns that will be dropped from the dataset
bad_label_cols = list(set(object_cols) - set(good_label_cols))
print("Categorical columns that should be ordinal encoded:", good_label_cols)
print("\n")
print("Categorical columns that should be dropped from the dataset:", bad_label_cols)
from sklearn.preprocessing import OrdinalEncoder
# Drop categorical columns that will not be encoded as treated as bad columns
label_X_train = X_train.drop(bad_label_cols, axis=1)
label_X_valid = X_valid.drop(bad_label_cols, axis=1)
# Apply ordinal encoder
ordinal_encoder = OrdinalEncoder()
label_X_train[good_label_cols] = ordinal_encoder.fit_transform(X_train[good_label_cols])
label_X_valid[good_label_cols] = ordinal_encoder.transform(X_valid[good_label_cols])
# print(label_X_train.shape) # (1168, 57) as 3 columns (bad_label_cols) are dropped
# print(X_train.head)
# print("==================================================================================")
# print(label_X_train.head)
# Uncomment above print values to see difference as how categorical columns are changed to numeric values
print("MAE from Approach 2 (Ordinal Encoding):")
print(
score_dataset(label_X_train, label_X_valid, y_train, y_valid)
) # 17098.01649543379
# # Approach 3 : One-hot encoding
# Get number of unique entries in each column with categorical data
object_nunique = list(map(lambda col: X_train[col].nunique(), object_cols))
d = dict(zip(object_cols, object_nunique))
# Print number of unique entries by column, in ascending order
sorted(d.items(), key=lambda x: x[1])
# Street have two type of unique values, where Neighborhood has 25 types of unique values
# Now lets drop high cardinality (grater than 10) columns as it will generate lots of columns, which is not feasible
# Columns that will be one-hot encoded
low_cardinality_cols = [col for col in object_cols if X_train[col].nunique() < 10]
# Columns that will be dropped from the dataset
high_cardinality_cols = list(set(object_cols) - set(low_cardinality_cols))
print("Categorical columns that will be one-hot encoded:", low_cardinality_cols)
print(
"\nCategorical columns that will be dropped from the dataset:",
high_cardinality_cols,
)
from sklearn.preprocessing import OneHotEncoder
# Apply one-hot encoder to each column with categorical data
OH_encoder = OneHotEncoder(handle_unknown="ignore", sparse=False)
OH_cols_train = pd.DataFrame(OH_encoder.fit_transform(X_train[low_cardinality_cols]))
OH_cols_valid = pd.DataFrame(OH_encoder.transform(X_valid[low_cardinality_cols]))
# One-hot encoding removed index; put it back
OH_cols_train.index = X_train.index
OH_cols_valid.index = X_valid.index
# Remove categorical columns (will replace with one-hot encoding)
num_X_train = X_train.drop(object_cols, axis=1)
num_X_valid = X_valid.drop(object_cols, axis=1)
# Add one-hot encoded columns to numerical features
OH_X_train = pd.concat([num_X_train, OH_cols_train], axis=1)
OH_X_valid = pd.concat([num_X_valid, OH_cols_valid], axis=1)
print("MAE from Approach 3 (One-Hot Encoding):")
print(score_dataset(OH_X_train, OH_X_valid, y_train, y_valid))
# # As seen above, Approach 2 : Ordinal encoding giving best result, as lowest MAE 17098.02
# # **So lets predict by test data set and submit to competition**
# Fetch all categorical columns
object_cols = [col for col in X_train.columns if X_train[col].dtype == "object"]
# Columns that can be safely ordinal encoded
good_label_cols = [
col for col in object_cols if set(X_test_full[col]).issubset(set(X_train[col]))
]
# Problematic columns that will be dropped from the dataset
bad_label_cols = list(set(object_cols) - set(good_label_cols))
from sklearn.preprocessing import OrdinalEncoder
# Drop categorical columns that will not be encoded as treated as bad columns
label_X_train = X_train.drop(bad_label_cols, axis=1)
label_X_test = X_test_full.drop(bad_label_cols, axis=1)
# Apply ordinal encoder
ordinal_encoder = OrdinalEncoder()
label_X_train[good_label_cols] = ordinal_encoder.fit_transform(X_train[good_label_cols])
label_X_test[good_label_cols] = ordinal_encoder.transform(X_test_full[good_label_cols])
# Impute Nan with mean value
from sklearn.impute import SimpleImputer
my_imputer = SimpleImputer(strategy="mean")
label_X_train = pd.DataFrame(my_imputer.fit_transform(label_X_train))
label_X_test = pd.DataFrame(my_imputer.transform(label_X_test))
model1 = RandomForestRegressor(n_estimators=100, random_state=0)
model1.fit(label_X_train, y_train)
preds = model1.predict(label_X_test)
# Save test predictions to file
output = pd.DataFrame({"Id": X_test_full.index, "SalePrice": preds})
output.to_csv(
"home-data-for-ml-course-handle-categorical-variables-submission.csv", index=False
)
# Download file and submit, Its gave me MAE: 16619.07644 for real sumbited result at, https://www.kaggle.com/c/home-data-for-ml-course
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/672/69672345.ipynb
| null | null |
[{"Id": 69672345, "ScriptId": 18881132, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6534192, "CreationDate": "08/02/2021 17:17:08", "VersionNumber": 5.0, "Title": "Simple practical explanation handle categorical V", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 206.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 205.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # **Simple explanation of how to handle categorical variables**
# We refer below competition for tutorial,
# https://www.kaggle.com/c/home-data-for-ml-course
# Cover below course topic,
# https://www.kaggle.com/alexisbcook/categorical-variables
# # Lets collect data for train and test
import pandas as pd
from sklearn.model_selection import train_test_split
# LodeStar has addded the option below
pd.set_option("display.max_columns", 100)
# Read the data
X_train_full = pd.read_csv(
"../input/home-data-for-ml-course/train.csv", index_col="Id"
) # Train data contain 1460 rows
X_test_full = pd.read_csv(
"../input/home-data-for-ml-course/test.csv", index_col="Id"
) # Test data contain 1459 rows
# print(X_train_full.head()) # Return all column with first 5 values
# print("X_train_full data row and col:", X_test_full.shape) # (1460, 80)
# X_train_full data contain SalePrice, while X_test_full not contain SalePrice which need to predict
# Remove rows with missing target, separate target from predictors
X_train_full.dropna(axis=0, subset=["SalePrice"], inplace=True)
# print(X_train_full.shape) # (1460, 80) i.e no value missing
Y_train_full = X_train_full.SalePrice
# print(Y_train_real.shape) # (1460,) i.e only target data, single column
X_train_full.drop(["SalePrice"], axis=1, inplace=True)
# print(X_train_full.shape) # (1460, 79) i.e only train data
# To keep things simple, we'll drop columns with missing values
cols_with_missing = [
col for col in X_train_full.columns if X_train_full[col].isnull().any()
]
# print(len(cols_with_missing)) # Return 19, as 19 column contain missing values
X_train_full.drop(cols_with_missing, axis=1, inplace=True)
X_test_full.drop(
cols_with_missing, axis=1, inplace=True
) # Also removing from test data to test with model
# Break off validation set from training data
X_train, X_valid, y_train, y_valid = train_test_split(
X_train_full, Y_train_full, train_size=0.8, test_size=0.2, random_state=0
)
# print(X_train.shape) # (1168, 60)
# X_train.head() # return all numerical as well as categorical columns
# # Create common function to get Mean absolute error
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
# function for comparing different approaches
def score_dataset(X_train, X_valid, y_train, y_valid):
model = RandomForestRegressor(n_estimators=100, random_state=0)
model.fit(X_train, y_train)
preds = model.predict(X_valid)
return mean_absolute_error(y_valid, preds)
# # Approach 1 : Drop column with categorical data
# First approach drop column with categorical data
drop_X_train = X_train.select_dtypes(exclude=["object"])
drop_X_valid = X_valid.select_dtypes(exclude=["object"])
print("MAE from Approach 1 (Drop categorical variables):")
print(score_dataset(drop_X_train, drop_X_valid, y_train, y_valid)) # 17837.82570776256
# # Approach 2 : Ordinal encoding
# First lets investigatge one column from dataset which is contain categorical data, column: Condition2
print(
"Unique values in 'Condition2' column in training data:",
X_train["Condition2"].unique(),
)
print(
"\nUnique values in 'Condition2' column in validation data:",
X_valid["Condition2"].unique(),
)
# > Fitting an ordinal encoder to a column in the training data creates a corresponding integer-valued label for each unique value that appears in the training data. In the case that the validation data contains values that don't also appear in the training data, the encoder will throw an error, because these values won't have an integer assigned to them. Notice that the 'Condition2' column in the validation data contains the values 'RRAn' and 'RRNn', but these don't appear in the training data -- thus, if we try to use an ordinal encoder with scikit-learn, the code will throw an error.
# This is a common problem that you'll encounter with real-world data, and there are many approaches to fixing this issue. For instance, you can write a custom ordinal encoder to deal with new categories. The simplest approach, however, is to drop the problematic categorical columns.
# Run the code cell below to save the problematic columns to a Python list **bad_label_cols**. Likewise, columns that can be safely ordinal encoded are stored in **good_label_cols**.
# Fetch all categorical columns
object_cols = [col for col in X_train.columns if X_train[col].dtype == "object"]
# Columns that can be safely ordinal encoded
good_label_cols = [
col for col in object_cols if set(X_valid[col]).issubset(set(X_train[col]))
]
# Problematic columns that will be dropped from the dataset
bad_label_cols = list(set(object_cols) - set(good_label_cols))
print("Categorical columns that should be ordinal encoded:", good_label_cols)
print("\n")
print("Categorical columns that should be dropped from the dataset:", bad_label_cols)
from sklearn.preprocessing import OrdinalEncoder
# Drop categorical columns that will not be encoded as treated as bad columns
label_X_train = X_train.drop(bad_label_cols, axis=1)
label_X_valid = X_valid.drop(bad_label_cols, axis=1)
# Apply ordinal encoder
ordinal_encoder = OrdinalEncoder()
label_X_train[good_label_cols] = ordinal_encoder.fit_transform(X_train[good_label_cols])
label_X_valid[good_label_cols] = ordinal_encoder.transform(X_valid[good_label_cols])
# print(label_X_train.shape) # (1168, 57) as 3 columns (bad_label_cols) are dropped
# print(X_train.head)
# print("==================================================================================")
# print(label_X_train.head)
# Uncomment above print values to see difference as how categorical columns are changed to numeric values
print("MAE from Approach 2 (Ordinal Encoding):")
print(
score_dataset(label_X_train, label_X_valid, y_train, y_valid)
) # 17098.01649543379
# # Approach 3 : One-hot encoding
# Get number of unique entries in each column with categorical data
object_nunique = list(map(lambda col: X_train[col].nunique(), object_cols))
d = dict(zip(object_cols, object_nunique))
# Print number of unique entries by column, in ascending order
sorted(d.items(), key=lambda x: x[1])
# Street have two type of unique values, where Neighborhood has 25 types of unique values
# Now lets drop high cardinality (grater than 10) columns as it will generate lots of columns, which is not feasible
# Columns that will be one-hot encoded
low_cardinality_cols = [col for col in object_cols if X_train[col].nunique() < 10]
# Columns that will be dropped from the dataset
high_cardinality_cols = list(set(object_cols) - set(low_cardinality_cols))
print("Categorical columns that will be one-hot encoded:", low_cardinality_cols)
print(
"\nCategorical columns that will be dropped from the dataset:",
high_cardinality_cols,
)
from sklearn.preprocessing import OneHotEncoder
# Apply one-hot encoder to each column with categorical data
OH_encoder = OneHotEncoder(handle_unknown="ignore", sparse=False)
OH_cols_train = pd.DataFrame(OH_encoder.fit_transform(X_train[low_cardinality_cols]))
OH_cols_valid = pd.DataFrame(OH_encoder.transform(X_valid[low_cardinality_cols]))
# One-hot encoding removed index; put it back
OH_cols_train.index = X_train.index
OH_cols_valid.index = X_valid.index
# Remove categorical columns (will replace with one-hot encoding)
num_X_train = X_train.drop(object_cols, axis=1)
num_X_valid = X_valid.drop(object_cols, axis=1)
# Add one-hot encoded columns to numerical features
OH_X_train = pd.concat([num_X_train, OH_cols_train], axis=1)
OH_X_valid = pd.concat([num_X_valid, OH_cols_valid], axis=1)
print("MAE from Approach 3 (One-Hot Encoding):")
print(score_dataset(OH_X_train, OH_X_valid, y_train, y_valid))
# # As seen above, Approach 2 : Ordinal encoding giving best result, as lowest MAE 17098.02
# # **So lets predict by test data set and submit to competition**
# Fetch all categorical columns
object_cols = [col for col in X_train.columns if X_train[col].dtype == "object"]
# Columns that can be safely ordinal encoded
good_label_cols = [
col for col in object_cols if set(X_test_full[col]).issubset(set(X_train[col]))
]
# Problematic columns that will be dropped from the dataset
bad_label_cols = list(set(object_cols) - set(good_label_cols))
from sklearn.preprocessing import OrdinalEncoder
# Drop categorical columns that will not be encoded as treated as bad columns
label_X_train = X_train.drop(bad_label_cols, axis=1)
label_X_test = X_test_full.drop(bad_label_cols, axis=1)
# Apply ordinal encoder
ordinal_encoder = OrdinalEncoder()
label_X_train[good_label_cols] = ordinal_encoder.fit_transform(X_train[good_label_cols])
label_X_test[good_label_cols] = ordinal_encoder.transform(X_test_full[good_label_cols])
# Impute Nan with mean value
from sklearn.impute import SimpleImputer
my_imputer = SimpleImputer(strategy="mean")
label_X_train = pd.DataFrame(my_imputer.fit_transform(label_X_train))
label_X_test = pd.DataFrame(my_imputer.transform(label_X_test))
model1 = RandomForestRegressor(n_estimators=100, random_state=0)
model1.fit(label_X_train, y_train)
preds = model1.predict(label_X_test)
# Save test predictions to file
output = pd.DataFrame({"Id": X_test_full.index, "SalePrice": preds})
output.to_csv(
"home-data-for-ml-course-handle-categorical-variables-submission.csv", index=False
)
# Download file and submit, Its gave me MAE: 16619.07644 for real sumbited result at, https://www.kaggle.com/c/home-data-for-ml-course
| false | 0 | 2,877 | 0 | 2,877 | 2,877 |
||
69672809
|
# # Drug features clustering according to mechanisms of action
# On 2021.8.2, now I'm in a hospital because my father is sick due to cerebral infarction by sudden thrombus after about one month since the 2nd pfizer vaccination. Furthermore, by the COVID-19 pandemic situation, my father and me have been isolated from the out world. My labtop display was broken when I tested my MoA(mechanisms of action) prediction model and drug feature clustering according to mechanisms of action, so my old mother bought a new monitor and brought me the monitor because I couldn't go out. I'm writing this with the sub monitor.
# Last year, I participated in the Mechanisms of Action contest, and that time, I developed an autoencoder and additive angular margin based MoA prediction model approaching this problem as clusering, not multi-classification. Of course, I devleoped an autoencoder based model as multi-classification. I thought the clustering model will be superior to the multi-classification model because the clustering model predicted mechanisms of action more accurately in practical mechanisms of action plot graphs than the multi-classification model, but rather the multi-classification model was superior to the clustering model in the viewpoint of the metric used that time.
# By the way, MoA clustering for the multi-clasification is more random than the clustering model, and in the clustering model, MoA clustering is structured and all drugs have almost all mechanisms of action. So that time, if it is valid, I thought the mRNA vaccine can have almost all known mechanisms of action. But I needed verification.
# Vaccination is mandatory, but our family and friend can die or have serious side effects due to unstable vaccines. Maybe, I hoped fortune for vaccination to my family. But fortune is only a fortune word to me.
# After I faced my father's accident, I searched for relevant papers, and I got it.
# Refer to https://www.medrxiv.org/content/10.1101/2021.04.30.21256383v1.
# This paper shows the vaccine induced immune thrombotic thrombocytopenia side effect for the BNT162b2, ChAdOx1 vaccines. So I understood my father accident's reason.
# Moreover, I'm confident that the MoA clusering model is valid.
# For mRNA vaccines, the vaccination can be carried out to children <= 13. It can be terrible in the human history, because mRNA vaccines' stability are almost unknown.
# Refer to https://coronavirus.quora.com/?__ni__=0&__nsrc__=4&__snid3__=24261290897&__tiids__=32992518
# I still think that the mRNA vaccine is only a solution to cope with fastly changing variants and mutants. But their stability is also mandatory as like that vaccination is mandatory, and this solution must be developed very fastly as the vaccination effect development.
# Via MoA prediction, we can identify side effects of vaccines and develop more stable vaccines. It is mandatory.
# ## Data analysis
import pandas as pd
import seaborn as sns
import os, sys
from scipy.special import comb
from itertools import combinations
import json
from tqdm import tqdm
import pdb
import plotly.express as px
raw_data_path = "/kaggle/input/lish-moa"
# ### Convert categorical values into categorical indexes
input_df = pd.read_csv(os.path.join(raw_data_path, "train_features.csv"))
input_df.info()
input_df.columns
len(input_df.sig_id), len(input_df.sig_id.unique())
input_df.cp_type.unique(), input_df.cp_time.unique(), input_df.cp_dose.unique()
input_df.cp_time.value_counts()
res = input_df.cp_type.astype("category")
res
len(res.cat.categories)
res = res.cat.rename_categories(range(len(res.cat.categories)))
res
res2 = res.map(lambda x: int(x))
res2
type(res2.iloc[0])
input_df.cp_type
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_type
cp_v_index_series = input_df.cp_type[input_df.cp_type == 0]
cp_v_index_series
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
len(input_df.cp_time.cat.categories), len(input_df.cp_dose.cat.categories)
input_df[["cp_type", "cp_time", "cp_dose"]].head(3)
res = input_df.isna().any()
res.any()
res = input_df.iloc[0:64]
res2 = res["cp_time"].values
type(res2)
res2.to_numpy()
g_feature_names = ["g-" + str(v) for v in range(772)]
res2 = res[g_feature_names].values
res2.shape
res2
res2 = res["sig_id"].values
type(res2), res2.shape
res2
# ### Gene expression and cell viability
input_vals = input_df.values
input_vals.shape
input_vals[0]
input_vals_p = input_vals[:, 4:]
input_vals_p.shape
input_vals_p = input_vals_p.astype("float32")
input_vals_p.shape, input_vals_p.dtype
input_vals_p[0, ...]
ge_val = input_vals_p[:, 0:772]
ct_val = input_vals_p[:, 772:]
ge_val.shape, ct_val.shape
ge_val_mean = ge_val.mean(axis=-1)
ge_val_std = ge_val.std(axis=-1)
ge_val_mean.shape
figure(figsize=(20, 20))
plot(ge_val_mean, label="mean")
plot(ge_val_std, label="std")
legend()
grid()
ct_val_mean = ct_val.mean(axis=-1)
ct_val_std = ct_val.std(axis=-1)
figure(figsize=(20, 20))
plot(ct_val_mean, label="mean")
plot(ct_val_std, label="std")
legend()
grid()
# ### Target feature
target_scored_df = pd.read_csv(os.path.join(raw_data_path, "train_targets_scored.csv"))
target_scored_df.info()
target_scored_df.columns
moa_names = list(target_scored_df.columns)[1:]
moa_names
target_scored_df.sum(axis=1)
res = target_scored_df[input_df.cp_type == 0]
res
res.sum(axis=1)
target_scored_df
target_nonscored_df = pd.read_csv(
os.path.join(raw_data_path, "train_targets_nonscored.csv")
)
target_nonscored_df.info()
target_nonscored_df.columns
target_df = pd.concat([target_scored_df, target_nonscored_df.iloc[:, 1:]], axis=1)
len(target_df.columns)
target_df
target_df.columns
res = target_nonscored_df.sum()
res
target_scored_df.iloc[:, 1:2].info()
type(target_scored_df.iloc[:, 1:2])
target_scored_df.iloc[:, 1:2].iloc[:, 0].value_counts()
for i in range(1, len(target_scored_df.columns)):
print(i, target_scored_df.iloc[:, i : (i + 1)].iloc[:, 0].value_counts())
target_scored_df.columns[1:]
len(target_scored_df.columns[1:])
# ## MoA prediction model
"""Keras unsupervised setup module.
"""
from setuptools import setup, find_packages
from os import path
from io import open
here = path.abspath(path.dirname(__file__))
# Get the long description from the README file
with open(path.join(here, "README.md"), encoding="utf-8") as f:
long_description = f.read()
setup(
name="keras-unsupervised", # Required
version="1.1.3.dev1", # Required
description="Keras based unsupervised learning framework.", # Optional
long_description=long_description, # Optional
long_description_content_type="text/markdown", # Optional (see note above)
url="https://github.com/tonandr/keras_unsupervised", # Optional
author="Inwoo Chung", # Optional
author_email="[email protected]", # Optional
classifiers=[ # Optional
# How mature is this project? Common values are
# 3 - Alpha
# 4 - Beta
# 5 - Production/Stable
"Development Status :: 4 - Beta",
# Indicate who your project is intended for
"Intended Audience :: Developers",
"Intended Audience :: Science/Research",
#'Software Development :: Libraries',
# Pick your license as you wish
"License :: OSI Approved :: BSD License",
# Specify the Python versions you support here. In particular, ensure
# that you indicate whether you support Python 2, Python 3 or both.
# These classifiers are *not* checked by 'pip install'. See instead
# 'python_requires' below.
"Programming Language :: Python :: 3",
"Programming Language :: Python :: 3.7" "Programming Language :: Python :: 3.8",
],
# This field adds keywords for your project which will appear on the
# project page. What does your project relate to?
#
# Note that this is a string of words separated by whitespace, not a list.
keywords="keras deepleaning unsupervised semisupervised restricted-botlzmann-machine deep-belief-network autoencoder generative-adversarial-networks", # Optional
# You can just specify package directories manually here if your project is
# simple. Or you can use find_packages().
#
# Alternatively, if you just want to distribute a single Python file, use
# the `py_modules` argument instead as follows, which will expect a file
# called `my_module.py` to exist:
#
# py_modules=["my_module"],
#
packages=find_packages(
exclude=["analysis", "docs", "docs_mkdocs", "resource"]
), # Required
# This field lists other packages that your project depends on to run.
# Any package you put here will be installed by pip when your project is
# installed, so they must be valid existing projects.
#
# For an analysis of "install_requires" vs pip's requirements files see:
# https://packaging.python.org/en/latest/requirements.html
install_requires=[
"tensorflow-probability==0.11",
"pandas",
"scikit-image",
"matplotlib",
"opencv-contrib-python",
], # Optional
# Specify which Python versions you support. In contrast to the
# 'Programming Language' classifiers above, 'pip install' will check this
# and refuse to install the project if the version does not match. If you
# do not support Python 2, you can simplify this to '>=3.5' or similar, see
# https://packaging.python.org/guides/distributing-packages-using-setuptools/#python-requires
python_requires="<=3.8",
# List additional groups of dependencies here (e.g. development
# dependencies). Users will be able to install these using the "extras"
# syntax, for example:
#
# $ pip install sampleproject[dev]
#
# Similar to `install_requires` above, these must be valid existing
# projects.
# extras_require={ # Optional
# 'dev': ['check-manifest'],
# 'test': ['coverage'],
# },
# If there are data files included in your packages that need to be
# installed, specify them here.
#
# If using Python 2.6 or earlier, then these have to be included in
# MANIFEST.in as well.
# package_data={ # Optional
# 'sample': ['package_data.dat'],
# },
# Although 'package_data' is the preferred approach, in some case you may
# need to place data files outside of your packages. See:
# http://docs.python.org/3.4/distutils/setupscript.html#installing-additional-files
#
# In this case, 'data_file' will be installed into '<sys.prefix>/my_data'
# data_files=[('my_data', ['data/data_file'])], # Optional
# To provide executable scripts, use entry points in preference to the
# "scripts" keyword. Entry points provide cross-platform support and allow
# `pip` to create the appropriate form of executable for the target
# platform.
#
# For example, the following would provide a command called `sample` which
# executes the function `main` from this package when invoked:
# entry_points={ # Optional
# 'console_scripts': [
# 'sample=sample:main',
# ],
# },
# List additional URLs that are relevant to your project as a dict.
#
# This field corresponds to the "Project-URL" metadata fields:
# https://packaging.python.org/specifications/core-metadata/#project-url-multiple-use
#
# Examples listed include a pattern for specifying where the package tracks
# issues, where the source is hosted, where to say thanks to the package
# maintainers, and where to support the project financially. The key is
# what's used to render the link text on PyPI.
project_urls={ # Optional
"Bug Reports": "https://github.com/tonandr/keras_unsupervised/issues",
"Source": "https://github.com/tonandr/keras_unsupervised/",
},
)
import sys
sys.path.append("/kaggle/working/keras_unsupervised")
"""
Created on Oct 8, 2020
@author: Inwoo Chung ([email protected])
"""
import os
import time
import json
import random
from random import shuffle
import ctypes
# ctypes.WinDLL('cudart64_110.dll') #?
import numpy as np
import pandas as pd
from tqdm import tqdm
from sklearn.model_selection import StratifiedKFold
import tensorflow as tf
import tensorflow.keras as keras
from tensorflow.keras import backend as K
from tensorflow.keras.losses import Loss
from tensorflow.keras.metrics import Metric
from tensorflow.keras.models import Model, load_model
from tensorflow.keras.layers import Input, Conv1D, Dense, Concatenate, Dropout
from tensorflow.keras.layers import (
LSTM,
Bidirectional,
BatchNormalization,
LayerNormalization,
)
from tensorflow.keras.layers import Embedding, Layer
from tensorflow.keras import optimizers
from tensorflow.keras.callbacks import (
TensorBoard,
ReduceLROnPlateau,
LearningRateScheduler,
ModelCheckpoint,
EarlyStopping,
)
from tensorflow.keras.constraints import UnitNorm
from tensorflow.keras.initializers import RandomUniform, TruncatedNormal
from tensorflow.keras import regularizers
from ku.composite_layer import DenseBatchNormalization
from ku.backprop import (
make_decoder_from_encoder,
make_autoencoder_from_encoder,
make_autoencoder_with_sym_sc,
)
# os.environ["CUDA_DEVICE_ORDER"] = 'PCI_BUS_ID'
# os.environ["CUDA_VISIBLE_DEVICES"] = '-1'
# Constants.
DEBUG = True
MODE_TRAIN = 0
MODE_VAL = 1
CV_TYPE_TRAIN_VAL_SPLIT = "train_val_split"
CV_TYPE_K_FOLD = "k_fold"
DATASET_TYPE_PLAIN = "plain"
DATASET_TYPE_BALANCED = "balanced"
LOSS_TYPE_MULTI_LABEL = "multi_label"
LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN = "additive_angular_margin"
epsilon = 1e-7
class MoALoss(Loss):
def __init__(
self,
W,
m=0.5,
ls=0.2,
scale=64.0,
loss_type=LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN,
name="MoA_loss",
):
super(MoALoss, self).__init__(name=name)
self.W = W
self.m = m
self.ls = ls
self.scale = scale
self.loss_type = loss_type
# @tf.function
def call(self, y_true, y_pred):
y_true = tf.cast(y_true, dtype=tf.float32)
pos_mask = y_true
neg_mask = 1.0 - y_true
# Label smoothing.
y_true = pos_mask * y_true * (1.0 - self.ls / 2.0) + neg_mask * (
y_true + self.ls / 2.0
)
"""
pos_log_loss = pos_mask * self.W[:, :, 0] * tf.sqrt(tf.square(y_true - y_pred))
pos_log_loss_mean = tf.reduce_mean(pos_log_loss, axis=0) #?
pos_loss = 1.0 * tf.reduce_mean(pos_log_loss_mean, axis=0)
neg_log_loss = neg_mask * self.W[:, :, 1] * tf.sqrt(tf.square(y_true - y_pred))
neg_log_loss_mean = tf.reduce_mean(neg_log_loss, axis=0) #?
neg_loss = 1.0 * tf.reduce_mean(neg_log_loss_mean, axis=0)
loss = pos_loss + neg_loss
"""
"""
loss = tf.reduce_mean(tf.sqrt(tf.square(y_true - y_pred)))
loss = tf.losses.binary_crossentropy(y_true, y_pred)
log_loss_mean = tf.reduce_mean(log_loss, axis=0) #?
loss = tf.reduce_mean(log_loss_mean, axis=0)
"""
if self.loss_type == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
A = y_pred
e_AM_A = tf.math.exp(self.scale * tf.math.cos(tf.math.acos(A) + self.m))
# d = A.shape[-1] #?
S = tf.tile(tf.reduce_sum(tf.math.exp(A), axis=1, keepdims=True), (1, 206))
S_p = S - tf.math.exp(A) + e_AM_A
P = e_AM_A / (S_p + epsilon)
# P = tf.clip_by_value(P, clip_value_min=epsilon, clip_value_max=(1.0 - epsilon))
# log_loss_1 = -1.0 * self.W[:, :, 0] * y_true * tf.math.log(P)
log_loss_1 = -1.0 * y_true * tf.math.log(P)
log_loss_2 = tf.reduce_sum(log_loss_1, axis=1)
loss = tf.reduce_mean(log_loss_2, axis=0)
elif self.loss_type == LOSS_TYPE_MULTI_LABEL:
y_pred = tf.sigmoid(y_pred)
y_pred = tf.maximum(tf.minimum(y_pred, 1.0 - 1e-15), 1e-15)
log_loss = -1.0 * (
y_true * tf.math.log(y_pred + epsilon)
+ (1.0 - y_true) * tf.math.log(1.0 - y_pred + epsilon)
)
log_loss_mean = tf.reduce_mean(log_loss, axis=0) # ?
loss = tf.reduce_mean(log_loss_mean, axis=0)
else:
raise ValueError("loss type is not valid.")
# tf.print(A, e_AM_A, S, S_p, P, log_loss_1, log_loss_2, loss)
return loss
def get_config(self):
"""Get configuration."""
config = {
"W": self.W,
"m": self.m,
"ls": self.ls,
"scale": self.scale,
"loss_type": self.loss_type,
}
base_config = super(MoALoss, self).get_config()
return dict(list(base_config.items()) + list(config.items()))
class MoAMetric(Metric):
def __init__(self, sn_t=2.45, name="MoA_metric", **kwargs):
super(MoAMetric, self).__init__(name=name, **kwargs)
self.sn_t = sn_t
self.total_loss = self.add_weight(name="total_loss", initializer="zeros")
self.count = self.add_weight(name="count", initializer="zeros")
def update_state(self, y_true, y_pred, sample_weight=None):
E = tf.reduce_mean(tf.math.exp(y_pred), axis=1, keepdims=True)
E_2 = tf.reduce_mean(tf.square(tf.math.exp(y_pred)), axis=1, keepdims=True)
S = tf.sqrt(E_2 - tf.square(E))
e_A = (tf.exp(y_pred) - E) / (S + epsilon)
e_A_p = tf.where(tf.math.greater(e_A, self.sn_t), self.sn_t, 0.0)
p_hat = e_A_p / (tf.reduce_sum(e_A_p, axis=1, keepdims=True) + epsilon)
y_pred = tf.maximum(tf.minimum(p_hat, 1.0 - 1e-15), 1e-15)
y_true = tf.cast(y_true, dtype=tf.float32)
log_loss = -1.0 * (
y_true * tf.math.log(y_pred + epsilon)
+ (1.0 - y_true) * tf.math.log(1.0 - y_pred + epsilon)
)
log_loss_mean = tf.reduce_mean(log_loss, axis=0) # ?
loss = tf.reduce_mean(log_loss_mean, axis=0)
self.total_loss.assign_add(loss)
self.count.assign_add(tf.constant(1.0))
def result(self):
return tf.math.divide_no_nan(self.total_loss, self.count)
def reset_states(self):
self.total_loss.assign(0.0)
self.count.assign(0.0)
def get_config(self):
"""Get configuration."""
config = {"sn_t": self.sn_t}
base_config = super(MoAMetric, self).get_config()
return dict(list(base_config.items()) + list(config.items()))
class _MoAPredictor(Layer):
def __init__(self, conf, **kwargs):
super(_MoAPredictor, self).__init__(**kwargs)
# Initialize.
self.conf = conf
self.hps = self.conf["hps"]
self.nn_arch = self.conf["nn_arch"]
# Design layers.
# First layers.
self.embed_treatment_type_0 = Embedding(
self.nn_arch["num_treatment_type"], self.nn_arch["d_input_feature"]
)
self.dense_treatment_type_0 = Dense(
self.nn_arch["d_input_feature"], activation="relu"
)
self.layer_normalization_0_1 = LayerNormalization()
self.layer_normalization_0_2 = LayerNormalization()
self.layer_normalization_0_3 = LayerNormalization()
# Autoencoder for gene expression profile.
input_gene_exp_1 = Input(shape=(self.nn_arch["d_gene_exp"],))
d_geps = [int(self.nn_arch["d_gep_init"] / np.power(2, v)) for v in range(4)]
dense_1_1 = Dense(
d_geps[0],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_1_1 = BatchNormalization()
dropout_1_1 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_1_1 = DenseBatchNormalization(
dense_1_1, batch_normalization_1_1, dropout=dropout_1_1
)
dense_1_2 = Dense(
d_geps[1],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_1_2 = BatchNormalization()
dropout_1_2 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_1_2 = DenseBatchNormalization(
dense_1_2, batch_normalization_1_2, dropout=dropout_1_2
)
dense_1_3 = Dense(
d_geps[2],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_1_3 = BatchNormalization()
dropout_1_3 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_1_3 = DenseBatchNormalization(
dense_1_3, batch_normalization_1_3, dropout=dropout_1_3
)
dense_1_4 = Dense(
d_geps[3],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_1_4 = BatchNormalization()
dropout_1_4 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_1_4 = DenseBatchNormalization(
dense_1_4, batch_normalization_1_4, dropout=dropout_1_4
)
self.encoder_gene_exp_1 = keras.Sequential(
[
input_gene_exp_1,
dense_batch_normalization_1_1,
dense_batch_normalization_1_2,
dense_batch_normalization_1_3,
dense_batch_normalization_1_4,
]
)
self.decoder_gene_exp_1 = make_decoder_from_encoder(self.encoder_gene_exp_1)
self.dropout_1 = Dropout(self.nn_arch["dropout_rate"])
# Autoencoder for cell type.
input_gene_exp_2 = Input(shape=(self.nn_arch["d_cell_type"],))
d_cvs = [int(self.nn_arch["d_cv_init"] / np.power(2, v)) for v in range(3)]
dense_2_1 = Dense(
d_cvs[0],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_2_1 = BatchNormalization()
dropout_2_1 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_2_1 = DenseBatchNormalization(
dense_2_1, batch_normalization_2_1, dropout=dropout_2_1
)
dense_2_2 = Dense(
d_cvs[1],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_2_2 = BatchNormalization()
dropout_2_2 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_2_2 = DenseBatchNormalization(
dense_2_2, batch_normalization_2_2, dropout=dropout_2_2
)
dense_2_3 = Dense(
d_cvs[2],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_2_3 = BatchNormalization()
dropout_2_3 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_2_3 = DenseBatchNormalization(
dense_2_3, batch_normalization_2_3, dropout=dropout_2_3
)
self.encoder_cell_type_2 = keras.Sequential(
[
input_gene_exp_2,
dense_batch_normalization_2_1,
dense_batch_normalization_2_2,
dense_batch_normalization_2_3,
]
)
self.decoder_cell_type_2 = make_decoder_from_encoder(self.encoder_cell_type_2)
self.dropout_2 = Dropout(self.nn_arch["dropout_rate"])
# Skip-connection autoencoder layer.
self.sc_aes = []
self.dropout_3 = Dropout(self.nn_arch["dropout_rate"])
for i in range(self.nn_arch["num_sc_ae"]):
input_sk_ae_3 = Input(shape=(self.nn_arch["d_hidden"],))
d_ae_init = d_geps[-1] + d_cvs[-1] + self.nn_arch["d_input_feature"]
d_aes = [d_ae_init, int(d_ae_init * 2), int(d_ae_init * 2), d_ae_init]
dense_3_1 = Dense(
d_aes[0],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_3_1 = BatchNormalization()
dropout_3_1 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_3_1 = DenseBatchNormalization(
dense_3_1, batch_normalization_3_1, dropout=dropout_3_1
)
dense_3_2 = Dense(
d_aes[1],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_3_2 = BatchNormalization()
dropout_3_2 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_3_2 = DenseBatchNormalization(
dense_3_2, batch_normalization_3_2, dropout=dropout_3_2
)
dense_3_3 = Dense(
d_aes[2],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_3_3 = BatchNormalization()
dropout_3_3 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_3_3 = DenseBatchNormalization(
dense_3_3, batch_normalization_3_3, dropout=dropout_3_3
)
dense_3_4 = Dense(
d_aes[3],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_3_4 = BatchNormalization()
dropout_3_4 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_3_4 = DenseBatchNormalization(
dense_3_4, batch_normalization_3_4, dropout=dropout_3_4
)
sc_encoder_3 = keras.Sequential(
[
input_sk_ae_3,
dense_batch_normalization_3_1,
dense_batch_normalization_3_2,
dense_batch_normalization_3_3,
dense_batch_normalization_3_4,
]
)
sc_autoencoder_3 = make_autoencoder_from_encoder(sc_encoder_3)
self.sc_aes.append(make_autoencoder_with_sym_sc(sc_autoencoder_3))
# Final layers.
d_fs = [int(self.nn_arch["d_f_init"] / np.power(2, v)) for v in range(3)]
self.dense_4_1 = Dense(
d_fs[0],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
self.dense_4_2 = Dense(
d_fs[1],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
self.dense_4_3 = Dense(
d_fs[2],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
self.dropout_4_3 = Dropout(self.nn_arch["dropout_rate"])
if self.conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
self.dense_4_4 = Dense(
self.nn_arch["num_moa_annotation"],
activation="linear",
kernel_initializer=TruncatedNormal(),
kernel_constraint=None,
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
use_bias=False,
) # ?
elif self.conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
self.dense_4_4 = Dense(
self.nn_arch["num_moa_annotation"],
activation="linear",
kernel_initializer=TruncatedNormal(),
kernel_constraint=UnitNorm(),
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
use_bias=False,
) # ?
else:
raise ValueError("loss type is not valid.")
def call(self, inputs):
t = inputs[0]
g = inputs[1]
c = inputs[2]
# First layers.
t = self.embed_treatment_type_0(t)
t = tf.reshape(t, (-1, self.nn_arch["d_input_feature"]))
t = self.dense_treatment_type_0(t)
t = self.layer_normalization_0_1(t)
g = self.layer_normalization_0_2(g)
c = self.layer_normalization_0_3(c)
# Gene expression.
g_e = self.encoder_gene_exp_1(g)
x_g = self.decoder_gene_exp_1(g_e)
x_g = tf.expand_dims(x_g, axis=-1)
x_g = tf.squeeze(x_g, axis=-1)
x_g = self.dropout_1(x_g)
# Cell type.
c_e = self.encoder_cell_type_2(c)
x_c = self.decoder_cell_type_2(c_e)
x_c = self.dropout_2(x_c)
# Skip-connection autoencoder and final layers.
x = tf.concat([t, g_e, c_e], axis=-1)
for i in range(self.nn_arch["num_sc_ae"]):
x = self.sc_aes[i](x)
x = self.dropout_3(x)
# Final layers.
x = self.dense_4_1(x)
x = self.dense_4_2(x)
x = self.dense_4_3(x)
x = self.dropout_4_3(x)
# Normalize x.
if self.conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
x1 = self.dense_4_4(x)
elif self.conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
x = x / tf.sqrt(tf.reduce_sum(tf.square(x), axis=1, keepdims=True))
x1 = self.dense_4_4(x)
else:
raise ValueError("loss type is not valid.")
outputs = [x_g, x_c, x1]
return outputs
def get_config(self):
"""Get configuration."""
config = {"conf": self.conf}
base_config = super(_MoAPredictor, self).get_config()
return dict(list(base_config.items()) + list(config.items()))
class MoAPredictor(object):
"""MoA predictor."""
# Constants.
MODEL_PATH = "MoA_predictor"
OUTPUT_FILE_NAME = "submission.csv"
EVALUATION_FILE_NAME = "eval.csv"
def __init__(self, conf):
"""
Parameters
----------
conf: Dictionary
Configuration dictionary.
"""
# Initialize.
self.conf = conf
self.raw_data_path = self.conf["raw_data_path"]
self.hps = self.conf["hps"]
self.nn_arch = self.conf["nn_arch"]
self.model_loading = self.conf["model_loading"]
# Create weight for classification imbalance.
W = self._create_W()
# with strategy.scope():
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
if self.model_loading:
self.model = load_model(
self.MODEL_PATH + ".h5",
custom_objects={
"MoALoss": MoALoss,
"MoAMetric": MoAMetric,
"_MoAPredictor": _MoAPredictor,
},
compile=False,
)
# self.model = load_model(self.MODEL_PATH, compile=False)
opt = optimizers.Adam(
lr=self.hps["lr"],
beta_1=self.hps["beta_1"],
beta_2=self.hps["beta_2"],
decay=self.hps["decay"],
)
self.model.compile(
optimizer=opt,
loss=[
"mse",
"mse",
MoALoss(
W,
self.nn_arch["additive_margin"],
self.hps["ls"],
self.nn_arch["scale"],
loss_type=self.conf["loss_type"],
),
],
loss_weights=self.hps["loss_weights"],
metrics=[["mse"], ["mse"], [MoAMetric(self.hps["sn_t"])]],
run_eagerly=False,
)
else:
# Design the MoA prediction model.
# Input.
input_t = Input(shape=(self.nn_arch["d_treatment_type"],))
input_g = Input(shape=(self.nn_arch["d_gene_exp"],))
input_c = Input(shape=(self.nn_arch["d_cell_type"],))
outputs = _MoAPredictor(self.conf, name="moap")(
[input_t, input_g, input_c]
)
opt = optimizers.Adam(
lr=self.hps["lr"],
beta_1=self.hps["beta_1"],
beta_2=self.hps["beta_2"],
decay=self.hps["decay"],
)
self.model = Model(inputs=[input_t, input_g, input_c], outputs=outputs)
self.model.compile(
optimizer=opt,
loss=[
"mse",
"mse",
MoALoss(
W,
self.nn_arch["additive_margin"],
self.hps["ls"],
self.nn_arch["scale"],
loss_type=self.conf["loss_type"],
),
],
loss_weights=self.hps["loss_weights"],
metrics=[["mse"], ["mse"], [MoAMetric(self.hps["sn_t"])]],
run_eagerly=False,
)
self.model.summary()
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
self.k_fold_models = []
if self.model_loading:
opt = optimizers.Adam(
lr=self.hps["lr"],
beta_1=self.hps["beta_1"],
beta_2=self.hps["beta_2"],
decay=self.hps["decay"],
)
# load models for K-fold.
for i in range(self.nn_arch["k_fold"]):
self.k_fold_models.append(
load_model(
self.MODEL_PATH + "_" + str(i) + ".h5",
custom_objects={
"MoALoss": MoALoss,
"MoAMetric": MoAMetric,
"_MoAPredictor": _MoAPredictor,
},
compile=False,
)
)
self.k_fold_models[i].compile(
optimizer=opt,
loss=[
"mse",
"mse",
MoALoss(
W,
self.nn_arch["additive_margin"],
self.hps["ls"],
self.nn_arch["scale"],
loss_type=self.conf["loss_type"],
),
],
loss_weights=self.hps["loss_weights"],
metrics=[["mse"], ["mse"], [MoAMetric(self.hps["sn_t"])]],
run_eagerly=False,
)
else:
# Create models for K-fold.
for i in range(self.nn_arch["k_fold"]):
# Design the MoA prediction model.
# Input.
input_t = Input(shape=(self.nn_arch["d_treatment_type"],))
input_g = Input(shape=(self.nn_arch["d_gene_exp"],))
input_c = Input(shape=(self.nn_arch["d_cell_type"],))
outputs = _MoAPredictor(self.conf, name="moap")(
[input_t, input_g, input_c]
)
opt = optimizers.Adam(
lr=self.hps["lr"],
beta_1=self.hps["beta_1"],
beta_2=self.hps["beta_2"],
decay=self.hps["decay"],
)
model = Model(inputs=[input_t, input_g, input_c], outputs=outputs)
model.compile(
optimizer=opt,
loss=[
"mse",
"mse",
MoALoss(
W,
self.nn_arch["additive_margin"],
self.hps["ls"],
self.nn_arch["scale"],
loss_type=self.conf["loss_type"],
),
],
loss_weights=self.hps["loss_weights"],
metrics=[["mse"], ["mse"], [MoAMetric(self.hps["sn_t"])]],
run_eagerly=False,
)
model.summary()
self.k_fold_models.append(model)
else:
raise ValueError("cv_type is not valid.")
# Create dataset.
self._create_dataset()
def _create_dataset(self):
input_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_features.csv")
) # .iloc[:1024]
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
# Remove samples of ctl_vehicle.
valid_indexes = input_df.cp_type == 1
input_df = input_df[valid_indexes]
input_df = input_df.reset_index(drop=True)
target_scored_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_targets_scored.csv")
) # .iloc[:1024]
target_scored_df = target_scored_df[valid_indexes]
target_scored_df = target_scored_df.reset_index(drop=True)
del target_scored_df["sig_id"]
target_scored_df.columns = range(len(target_scored_df.columns))
n_target_samples = target_scored_df.sum().values
if self.conf["data_aug"]:
genes = [col for col in input_df.columns if col.startswith("g-")]
cells = [col for col in input_df.columns if col.startswith("c-")]
features = genes + cells
targets = [col for col in target_scored_df if col != "sig_id"]
aug_trains = []
aug_targets = []
for t in [0, 1, 2]:
for d in [0, 1]:
for _ in range(3):
train1 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
]
target1 = target_scored_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
]
ctl1 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
].sample(train1.shape[0], replace=True)
ctl2 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
].sample(train1.shape[0], replace=True)
train1[genes + cells] = (
train1[genes + cells].values
+ ctl1[genes + cells].values
- ctl2[genes + cells].values
)
aug_trains.append(train1)
aug_targets.append(target1)
input_df = pd.concat(aug_trains).reset_index(drop=True)
target_scored_df = pd.concat(aug_targets).reset_index(drop=True)
g_feature_names = ["g-" + str(v) for v in range(self.nn_arch["d_gene_exp"])]
c_feature_names = ["c-" + str(v) for v in range(self.nn_arch["d_cell_type"])]
moa_names = [v for v in range(self.nn_arch["num_moa_annotation"])]
def get_series_from_input(idxes):
idxes = idxes.numpy() # ?
series = input_df.iloc[idxes]
# Treatment.
if isinstance(idxes, np.int32) != True:
cp_time = series["cp_time"].values.to_numpy()
cp_dose = series["cp_dose"].values.to_numpy()
else:
cp_time = np.asarray(series["cp_time"])
cp_dose = np.asarray(series["cp_dose"])
treatment_type = cp_time * 2 + cp_dose
# Gene expression.
gene_exps = series[g_feature_names].values
# Cell viability.
cell_vs = series[c_feature_names].values
return treatment_type, gene_exps, cell_vs
def make_input_target_features(idxes):
treatment_type, gene_exps, cell_vs = tf.py_function(
get_series_from_input,
inp=[idxes],
Tout=[tf.int64, tf.float64, tf.float64],
)
MoA_values = tf.py_function(
get_series_from_target, inp=[idxes], Tout=tf.int32
)
return (
(treatment_type, gene_exps, cell_vs),
(gene_exps, cell_vs, MoA_values),
)
def make_input_features(idx):
treatment_type, gene_exps, cell_vs = tf.py_function(
get_series_from_input,
inp=[idx],
Tout=[tf.int64, tf.float64, tf.float64],
)
return treatment_type, gene_exps, cell_vs
def make_a_target_features(idx):
treatment_type, gene_exps, cell_vs = tf.py_function(
get_series_from_input,
inp=[idx],
Tout=[tf.int64, tf.float64, tf.float64],
)
return gene_exps, cell_vs
def get_series_from_target(idxes):
idxes = idxes.numpy()
series = target_scored_df.iloc[idxes]
# MoA annotations' values.
MoA_values = series[moa_names].values
return MoA_values
def make_target_features(idx):
MoA_values = tf.py_function(
get_series_from_target, inp=[idx], Tout=tf.int32
)
return MoA_values
def divide_inputs(input1, input2):
return input1[0], input1[1], input2
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
if self.conf["dataset_type"] == DATASET_TYPE_PLAIN:
train_val_index = np.arange(len(input_df))
# np.random.shuffle(train_val_index)
num_val = int(self.conf["val_ratio"] * len(input_df))
num_tr = len(input_df) - num_val
train_index = train_val_index[:num_tr]
val_index = train_val_index[num_tr:]
self.train_index = train_index
self.val_index = val_index
# Training dataset.
input_dataset = tf.data.Dataset.from_tensor_slices(train_index)
input_dataset = input_dataset.map(make_input_features)
a_target_dataset = tf.data.Dataset.from_tensor_slices(train_index)
a_target_dataset = a_target_dataset.map(make_a_target_features)
target_dataset = tf.data.Dataset.from_tensor_slices(train_index)
target_dataset = target_dataset.map(make_target_features)
f_target_dataset = tf.data.Dataset.zip(
(a_target_dataset, target_dataset)
).map(divide_inputs)
# Inputs and targets.
tr_dataset = tf.data.Dataset.zip((input_dataset, f_target_dataset))
tr_dataset = (
tr_dataset.shuffle(
buffer_size=self.hps["batch_size"] * 5,
reshuffle_each_iteration=True,
)
.repeat()
.batch(self.hps["batch_size"])
)
self.step = len(train_index) // self.hps["batch_size"]
# Validation dataset.
input_dataset = tf.data.Dataset.from_tensor_slices(val_index)
input_dataset = input_dataset.map(make_input_features)
a_target_dataset = tf.data.Dataset.from_tensor_slices(val_index)
a_target_dataset = a_target_dataset.map(make_a_target_features)
target_dataset = tf.data.Dataset.from_tensor_slices(val_index)
target_dataset = target_dataset.map(make_target_features)
f_target_dataset = tf.data.Dataset.zip(
(a_target_dataset, target_dataset)
).map(divide_inputs)
# Inputs and targets.
val_dataset = tf.data.Dataset.zip((input_dataset, f_target_dataset))
val_dataset = val_dataset.batch(self.hps["batch_size"])
self.trval_dataset = (tr_dataset, val_dataset)
elif self.conf["dataset_type"] == DATASET_TYPE_BALANCED:
MoA_p_sets = []
val_index = []
for col in target_scored_df.columns:
s = target_scored_df.iloc[:, col]
s = s[s == 1]
s = list(s.index)
# shuffle(s)
n_val = int(n_target_samples[col] * self.conf["val_ratio"])
if n_val != 0:
tr_set = s[: int(-1.0 * n_val)]
val_set = s[int(-1.0 * n_val) :]
MoA_p_sets.append(tr_set)
val_index += val_set
else:
MoA_p_sets.append(s)
df = target_scored_df.sum(axis=1)
df = df[df == 0]
no_MoA_p_set = list(df.index)
# shuffle(no_MoA_p_set)
val_index += no_MoA_p_set[
int(-1.0 * len(no_MoA_p_set) * self.conf["val_ratio"]) :
]
MoA_p_sets.append(
no_MoA_p_set[
: int(-1.0 * len(no_MoA_p_set) * self.conf["val_ratio"])
]
)
idxes = []
for i in range(self.hps["rep"]):
for col in range(len(target_scored_df.columns) + 1):
if len(MoA_p_sets[col]) >= (i + 1):
idx = MoA_p_sets[col][i]
else:
idx = np.random.choice(
MoA_p_sets[col], size=1, replace=True
)[0]
idxes.append(idx)
train_index = idxes
self.train_index = train_index
self.val_index = val_index
# Training dataset.
tr_dataset = tf.data.Dataset.from_tensor_slices(train_index)
# Inputs and targets.
tr_dataset = (
tr_dataset.shuffle(
buffer_size=self.hps["batch_size"] * 5,
reshuffle_each_iteration=True,
)
.repeat()
.batch(self.hps["batch_size"])
.map(
make_input_target_features,
num_parallel_calls=tf.data.experimental.AUTOTUNE,
)
)
self.step = len(train_index) // self.hps["batch_size"]
# Validation dataset.
val_dataset = tf.data.Dataset.from_tensor_slices(val_index)
# Inputs and targets.
val_dataset = val_dataset.batch(self.hps["batch_size"]).map(
make_input_target_features,
num_parallel_calls=tf.data.experimental.AUTOTUNE,
)
# Save datasets.
# tf.data.experimental.save(tr_dataset, './tr_dataset')
# tf.data.experimental.save(val_dataset, './val_dataset')
self.trval_dataset = (tr_dataset, val_dataset)
else:
raise ValueError("dataset type is not valid.")
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
stratified_kfold = StratifiedKFold(n_splits=self.nn_arch["k_fold"])
# group_kfold = GroupKFold(n_splits=self.nn_arch['k_fold'])
self.k_fold_trval_datasets = []
for train_index, val_index in stratified_kfold.split(
input_df, input_df.cp_type
):
# Training dataset.
input_dataset = tf.data.Dataset.from_tensor_slices(train_index)
input_dataset = input_dataset.map(make_input_features)
a_target_dataset = tf.data.Dataset.from_tensor_slices(train_index)
a_target_dataset = a_target_dataset.map(make_a_target_features)
target_dataset = tf.data.Dataset.from_tensor_slices(train_index)
target_dataset = target_dataset.map(make_target_features)
f_target_dataset = tf.data.Dataset.zip(
(a_target_dataset, target_dataset)
).map(divide_inputs)
# Inputs and targets.
tr_dataset = tf.data.Dataset.zip((input_dataset, f_target_dataset))
tr_dataset = (
tr_dataset.shuffle(
buffer_size=self.hps["batch_size"] * 5,
reshuffle_each_iteration=True,
)
.repeat()
.batch(self.hps["batch_size"])
)
self.step = len(train_index) // self.hps["batch_size"]
# Validation dataset.
input_dataset = tf.data.Dataset.from_tensor_slices(val_index)
input_dataset = input_dataset.map(make_input_features)
a_target_dataset = tf.data.Dataset.from_tensor_slices(val_index)
a_target_dataset = a_target_dataset.map(make_a_target_features)
target_dataset = tf.data.Dataset.from_tensor_slices(val_index)
target_dataset = target_dataset.map(make_target_features)
f_target_dataset = tf.data.Dataset.zip(
(a_target_dataset, target_dataset)
).map(divide_inputs)
# Inputs and targets.
val_dataset = tf.data.Dataset.zip((input_dataset, f_target_dataset))
val_dataset = val_dataset.batch(self.hps["batch_size"])
self.k_fold_trval_datasets.append((tr_dataset, val_dataset))
else:
raise ValueError("cv_type is not valid.")
def _create_W(self):
target_scored_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_targets_scored.csv")
)
del target_scored_df["sig_id"]
weights = []
for c in target_scored_df.columns:
s = target_scored_df[c]
s = s.value_counts()
s = s / s.sum()
weights.append(s.values)
weight = np.expand_dims(np.array(weights), axis=0)
return weight
def train(self):
"""Train."""
reduce_lr = ReduceLROnPlateau(
monitor="val_loss",
factor=self.hps["reduce_lr_factor"],
patience=3,
min_lr=1.0e-8,
verbose=1,
)
tensorboard = TensorBoard(
histogram_freq=1, write_graph=True, write_images=True, update_freq="epoch"
)
earlystopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, mode="auto"
)
"""
def schedule_lr(e_i):
self.hps['lr'] = self.hps['reduce_lr_factor'] * self.hps['lr']
return self.hps['lr']
lr_scheduler = LearningRateScheduler(schedule_lr, verbose=1)
"""
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
model_check_point = ModelCheckpoint(
self.MODEL_PATH + ".h5",
monitor="val_loss",
verbose=1,
save_best_only=True,
)
hist = self.model.fit(
self.trval_dataset[0],
steps_per_epoch=self.step,
epochs=self.hps["epochs"],
verbose=1,
max_queue_size=80,
workers=4,
use_multiprocessing=False,
callbacks=[
model_check_point,
earlystopping,
], # , reduce_lr] #, tensorboard]
validation_data=self.trval_dataset[1],
validation_freq=1,
shuffle=True,
)
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
for i in range(self.nn_arch["k_fold"]):
model_check_point = ModelCheckpoint(
self.MODEL_PATH + "_" + str(i) + ".h5",
monitor="loss",
verbose=1,
save_best_only=True,
)
hist = self.k_fold_models[i].fit(
self.k_fold_trval_datasets[i][0],
steps_per_epoch=self.step,
epochs=self.hps["epochs"],
verbose=1,
max_queue_size=80,
workers=4,
use_multiprocessing=False,
callbacks=[
model_check_point,
earlystopping,
], # reduce_lr] #, tensorboard]
validation_data=self.k_fold_trval_datasets[i][1],
validation_freq=1,
shuffle=True,
)
else:
raise ValueError("cv_type is not valid.")
# print('Save the model.')
# self.model.save(self.MODEL_PATH, save_format='h5')
# self.model.save(self.MODEL_PATH, save_format='tf')
return hist
def evaluate(self):
"""Evaluate."""
assert self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT
input_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_features.csv")
) # .iloc[:1024]
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
# Remove samples of ctl_vehicle.
valid_indexes = input_df.cp_type == 1 # ?
target_scored_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_targets_scored.csv")
) # .iloc[:1024]
target_scored_df = target_scored_df.loc[self.val_index]
MoA_annots = target_scored_df.columns[1:]
def make_input_features(inputs):
# Treatment.
cp_time = inputs["cp_time"]
cp_dose = inputs["cp_dose"]
treatment_type = cp_time * 2 + cp_dose
# Gene expression.
gene_exps = [
inputs["g-" + str(v)] for v in range(self.nn_arch["d_gene_exp"])
]
gene_exps = tf.stack(gene_exps, axis=0)
# Cell viability.
cell_vs = [
inputs["c-" + str(v)] for v in range(self.nn_arch["d_cell_type"])
]
cell_vs = tf.stack(cell_vs, axis=0)
return (tf.expand_dims(treatment_type, axis=-1), gene_exps, cell_vs)
# Validation dataset.
val_dataset = tf.data.Dataset.from_tensor_slices(
input_df.loc[self.val_index].to_dict("list")
)
val_dataset = val_dataset.map(make_input_features)
val_iter = val_dataset.as_numpy_iterator()
# Predict MoAs.
sig_id_list = []
MoAs = [[] for _ in range(len(MoA_annots))]
for i, d in tqdm(enumerate(val_iter)):
t, g, c = d
id = target_scored_df["sig_id"].iloc[i]
t = np.expand_dims(t, axis=0)
g = np.expand_dims(g, axis=0)
c = np.expand_dims(c, axis=0)
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
_, _, result = self.model.layers[-1](
[t, g, c]
) # self.model.predict([t, g, c])
result = np.squeeze(result, axis=0)
# result = np.exp(result) / (np.sum(np.exp(result), axis=0) + epsilon)
for i, MoA in enumerate(result):
MoAs[i].append(MoA)
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
# Conduct ensemble prediction.
result_list = []
for i in range(self.nn_arch["k_fold"]):
_, _, result = self.k_fold_models[i].predict([t, g, c])
result = np.squeeze(result, axis=0)
# result = np.exp(result) / (np.sum(np.exp(result), axis=0) + epsilon)
result_list.append(result)
result_mean = np.asarray(result_list).mean(axis=0)
for i, MoA in enumerate(result_mean):
MoAs[i].append(MoA)
else:
raise ValueError("cv_type is not valid.")
sig_id_list.append(id)
# Save the result.
result_dict = {"sig_id": sig_id_list}
for i, MoA_annot in enumerate(MoA_annots):
result_dict[MoA_annot] = MoAs[i]
submission_df = pd.DataFrame(result_dict)
submission_df.to_csv(self.OUTPUT_FILE_NAME, index=False)
target_scored_df.to_csv("gt.csv", index=False)
def test(self):
"""Test."""
# Create the test dataset.
input_df = pd.read_csv(os.path.join(self.raw_data_path, "test_features.csv"))
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
# Remove samples of ctl_vehicle.
valid_indexes = input_df.cp_type == 1 # ?
target_scored_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_targets_scored.csv")
)
MoA_annots = target_scored_df.columns[1:]
def make_input_features(inputs):
id_ = inputs["sig_id"]
cp_type = inputs["cp_type"]
# Treatment.
cp_time = inputs["cp_time"]
cp_dose = inputs["cp_dose"]
treatment_type = cp_time * 2 + cp_dose
# Gene expression.
gene_exps = [
inputs["g-" + str(v)] for v in range(self.nn_arch["d_gene_exp"])
]
gene_exps = tf.stack(gene_exps, axis=0)
# Cell viability.
cell_vs = [
inputs["c-" + str(v)] for v in range(self.nn_arch["d_cell_type"])
]
cell_vs = tf.stack(cell_vs, axis=0)
return (
id_,
cp_type,
tf.expand_dims(treatment_type, axis=-1),
gene_exps,
cell_vs,
)
test_dataset = tf.data.Dataset.from_tensor_slices(input_df.to_dict("list"))
test_dataset = test_dataset.map(make_input_features)
test_iter = test_dataset.as_numpy_iterator()
# Predict MoAs.
sig_id_list = []
MoAs = [[] for _ in range(len(MoA_annots))]
def cal_prob(logit):
a = logit
a = (a + 1.0) / 2.0
a = tf.where(tf.math.greater(a, self.hps["sn_t"]), a, 0.0)
a = self.hps["m1"] * a + self.hps["m2"]
p_h = tf.sigmoid(a).numpy()
return p_h
def cal_prob_2(logit):
y_pred = logit
E = tf.reduce_mean(tf.math.exp(y_pred), axis=-1, keepdims=True)
E_2 = tf.reduce_mean(tf.square(tf.math.exp(y_pred)), axis=-1, keepdims=True)
S = tf.sqrt(E_2 - tf.square(E))
e_A = (tf.exp(y_pred) - E) / (S + epsilon)
e_A_p = tf.where(
tf.math.greater(e_A, self.hps["sn_t"]), self.hps["sn_t"], 0.0
)
p_h = e_A_p / (tf.reduce_sum(e_A_p, axis=-1, keepdims=True) + epsilon)
return p_h.numpy()
def cal_prob_3(logit):
A = logit
A = (A + 1.0) / 2.0
E = tf.reduce_mean(A, axis=-1, keepdims=True)
E_2 = tf.reduce_mean(tf.square(A), axis=-1, keepdims=True)
S = tf.sqrt(E_2 - tf.square(E))
# S_N = tf.abs(A - E) / (S + epsilon)
S_N = (A - E) / (S + epsilon)
# S_N = tf.where(tf.math.greater(S_N, self.hps['sn_t']), S_N, 0.0)
A_p = self.hps["m1"] * S_N + self.hps["m2"]
# P_h = tf.clip_by_value(A_p / 10.0, clip_value_min=0.0, clip_value_max=1.0)
P_h = tf.sigmoid(A_p)
return P_h.numpy()
def cal_prob_4(logit):
a = logit
p_h = tf.sigmoid(a).numpy()
return p_h
for id_, cp_type, t, g, c in tqdm(test_iter):
id_ = id_.decode("utf8") # ?
t = np.expand_dims(t, axis=0)
g = np.expand_dims(g, axis=0)
c = np.expand_dims(c, axis=0)
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
# _, _, result = self.model.layers[-1]([t, g, c]) #self.model.predict([t, g, c])
_, _, result = self.model.predict([t, g, c])
result = np.squeeze(result, axis=0)
if cp_type == 1:
if self.conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
result = cal_prob_4(result)
elif self.conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
result = cal_prob_3(result)
else:
raise ValueError("loss type is not valid.")
else:
result = np.zeros((len(result)))
for i, MoA in enumerate(result):
MoAs[i].append(MoA)
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
# Conduct ensemble prediction.
result_list = []
for i in range(self.nn_arch["k_fold"]):
_, _, result = self.k_fold_models[i].predict([t, g, c])
result = np.squeeze(result, axis=0)
if cp_type == 1:
if self.conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
result = cal_prob_4(result)
elif (
self.conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN
):
result = cal_prob_3(result)
else:
raise ValueError("loss type is not valid.")
else:
result = np.zeros((len(result)))
result_list.append(result)
result_mean = np.asarray(result_list).mean(axis=0)
for i, MoA in enumerate(result_mean):
MoAs[i].append(MoA)
else:
raise ValueError("cv_type is not valid.")
sig_id_list.append(id_)
# Save the result.
result_dict = {"sig_id": sig_id_list}
for i, MoA_annot in enumerate(MoA_annots):
result_dict[MoA_annot] = MoAs[i]
submission_df = pd.DataFrame(result_dict)
submission_df.to_csv(self.OUTPUT_FILE_NAME, index=False)
import time
seed = int(time.time())
# seed = 1606208227
print(f"Seed:{seed}")
np.random.seed(seed)
tf.random.set_seed(seed)
# ## Training
{
"mode": "train",
"raw_data_path": "/kaggle/input/lish-moa",
"model_loading": false,
"multi_gpu": false,
"num_gpus": 4,
"cv_type": "train_val_split",
"dataset_type": "balanced",
"val_ratio": 0.1,
"loss_type": "additive_angular_margin",
"data_aug": false,
"hps": {
"lr": 0.001,
"beta_1": 0.999,
"beta_2": 0.999,
"decay": 0.0,
"epochs": 258,
"batch_size": 512,
"reduce_lr_factor": 0.96,
"ls": 8.281e-5,
"loss_weights": [1.0, 0.1, 100.0],
"rep": 1600,
"sn_t": 1.6,
"m1": 0.8081512,
"m2": 0.011438734,
"weight_decay": 0.000858,
},
"nn_arch": {
"k_fold": 5,
"d_treatment_type": 1,
"num_treatment_type": 6,
"d_input_feature": 8,
"d_gene_exp": 772,
"d_cell_type": 100,
"d_gep_init": 1024,
"d_cv_init": 128,
"num_sc_ae": 0,
"d_f_init": 512,
"num_moa_annotation": 206,
"d_out": 772,
"dropout_rate": 0.2,
"similarity_type": "diff_abs",
"additive_margin": 0.02,
"scale": 1.0,
},
}
with open("MoA_pred_conf.json", "r") as f:
conf = json.load(f)
# Train.
model = MoAPredictor(conf)
ts = time.time()
hist = model.train()
te = time.time()
print("Elasped time: {0:f}s".format(te - ts))
# ### MoA clustering analysis
# #### Center analysis
moap = model.model.get_layer("moap")
W = moap.dense_4_4.weights[0]
W.shape
W = W.numpy()
W = W.T
W.shape
from sklearn.manifold import TSNE
W_e = TSNE(n_components=2).fit_transform(W)
W_e.shape
colors = np.arange(len(W_e))
figure(figsize=(20, 20))
scatter(W_e[:, 0], W_e[:, 1], c=colors, cmap=cm.prism, marker="^", s=100)
# for i, c in enumerate(colors):
# annotate(str(c), (tsne_embed_features[i, 0], tsne_embed_features[i, 1]))
grid()
# #### Clustering map for training and validation data
from matplotlib.patches import Rectangle
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
def plot_colortable(colors, title, sort_colors=True, emptycols=0):
cell_width = 212
cell_height = 22
swatch_width = 48
margin = 12
topmargin = 40
# Sort colors by hue, saturation, value and name.
if sort_colors is True:
by_hsv = sorted(
(tuple(mcolors.rgb_to_hsv(mcolors.to_rgb(color))), name)
for name, color in colors.items()
)
names = [name for hsv, name in by_hsv]
else:
names = list(colors)
n = len(names)
ncols = 4 - emptycols
nrows = n // ncols + int(n % ncols > 0)
width = cell_width * 4 + 2 * margin
height = cell_height * nrows + margin + topmargin
dpi = 72
fig, ax = plt.subplots(figsize=(width / dpi, height / dpi), dpi=dpi)
fig.subplots_adjust(
margin / width,
margin / height,
(width - margin) / width,
(height - topmargin) / height,
)
ax.set_xlim(0, cell_width * 4)
ax.set_ylim(cell_height * (nrows - 0.5), -cell_height / 2.0)
ax.yaxis.set_visible(False)
ax.xaxis.set_visible(False)
ax.set_axis_off()
ax.set_title(title, fontsize=24, loc="left", pad=10)
for i, name in enumerate(names):
row = i % nrows
col = i // nrows
y = row * cell_height
swatch_start_x = cell_width * col
text_pos_x = cell_width * col + swatch_width + 7
ax.text(
text_pos_x,
y,
name,
fontsize=8,
horizontalalignment="left",
verticalalignment="center",
)
ax.add_patch(
Rectangle(
xy=(swatch_start_x, y - 9),
width=swatch_width,
height=18,
facecolor=colors[name],
edgecolor="0.7",
)
)
return fig
len(mcolors.XKCD_COLORS)
colors = mcolors.XKCD_COLORS
color_items = colors.items()
color_items = list(color_items)
cls_colors = [color_items[i][1] for i in range(0, len(color_items), 4)]
cls_colors = cls_colors[:207]
moa_names = ["none"] + moa_names
moa_name_colors = dict((name, color) for name, color in zip(moa_names, cls_colors))
# #### Clustering for training data
input_df = pd.read_csv(os.path.join(raw_data_path, "train_features.csv"))
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
# Remove samples of ctl_vehicle.
valid_indexes = input_df.cp_type == 1
input_df = input_df[valid_indexes]
input_df = input_df.reset_index(drop=True)
target_scored_df = pd.read_csv(os.path.join(raw_data_path, "train_targets_scored.csv"))
target_scored_df = target_scored_df[valid_indexes]
target_scored_df = target_scored_df.reset_index(drop=True)
del target_scored_df["sig_id"]
target_scored_df.columns = range(len(target_scored_df.columns))
n_target_samples = target_scored_df.sum().values
if model.conf["data_aug"]:
genes = [col for col in input_df.columns if col.startswith("g-")]
cells = [col for col in input_df.columns if col.startswith("c-")]
features = genes + cells
targets = [col for col in target_scored_df if col != "sig_id"]
aug_trains = []
aug_targets = []
for t in [0, 1, 2]:
for d in [0, 1]:
for _ in range(3):
train1 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
]
target1 = target_scored_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
]
ctl1 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
].sample(train1.shape[0], replace=True)
ctl2 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
].sample(train1.shape[0], replace=True)
train1[genes + cells] = (
train1[genes + cells].values
+ ctl1[genes + cells].values
- ctl2[genes + cells].values
)
aug_trains.append(train1)
aug_targets.append(target1)
input_df = pd.concat(aug_trains).reset_index(drop=True)
target_scored_df = pd.concat(aug_targets).reset_index(drop=True)
g_feature_names = ["g-" + str(v) for v in range(model.nn_arch["d_gene_exp"])]
c_feature_names = ["c-" + str(v) for v in range(model.nn_arch["d_cell_type"])]
moa_names = [v for v in range(model.nn_arch["num_moa_annotation"])]
def get_series_from_input(idxes):
idxes = idxes.numpy()
df = input_df.iloc[idxes]
# Treatment.
cp_time = df["cp_time"]
cp_dose = df["cp_dose"]
treatment_type = cp_time * 2 + cp_dose
# Gene expression.
gene_exps = df[g_feature_names].values
# Cell viability.
cell_vs = df[c_feature_names].values
return treatment_type, gene_exps, cell_vs
def make_input_features(idx):
treatment_type, gene_exps, cell_vs = tf.py_function(
get_series_from_input, inp=[idx], Tout=[tf.int32, tf.float64, tf.float64]
)
return treatment_type, gene_exps, cell_vs
MoA_p_sets = []
train_index = []
val_index = []
for col in target_scored_df.columns:
s = target_scored_df.iloc[:, col]
s = s[s == 1]
s = list(s.index)
n_val = int(n_target_samples[col] * model.conf["val_ratio"])
if n_val != 0:
tr_set = s[: int(-1.0 * n_val)]
val_set = s[int(-1.0 * n_val) :]
MoA_p_sets.append(tr_set)
train_index += tr_set
val_index += val_set
else:
MoA_p_sets.append(s)
train_index += s
# Training dataset.
tr_dataset = tf.data.Dataset.from_tensor_slices(train_index)
tr_dataset = tr_dataset.map(
make_input_features
) # , num_parallel_calls=tf.data.experimental.AUTOTUNE)
tr_iter = tr_dataset.as_numpy_iterator()
# Validation dataset.
val_dataset = tf.data.Dataset.from_tensor_slices(val_index)
val_dataset = val_dataset.map(
make_input_features
) # , num_parallel_calls=tf.data.experimental.AUTOTUNE)
val_iter = val_dataset.as_numpy_iterator()
tr_iter = tr_dataset.as_numpy_iterator()
moap = model.model.get_layer("moap")
embed_feature_dicts = []
target_df = target_scored_df.loc[train_index]
for i, d in tqdm(enumerate(tr_iter)):
t, g, c = d
t = np.expand_dims(t, axis=0)
g = np.expand_dims(g, axis=0)
c = np.expand_dims(c, axis=0)
# First layers.
t = moap.embed_treatment_type_0(t)
t = tf.reshape(t, (-1, model.nn_arch["d_input_feature"]))
t = moap.dense_treatment_type_0(t)
t = moap.layer_normalization_0_1(t)
g = moap.layer_normalization_0_2(g)
c = moap.layer_normalization_0_3(c)
# Gene expression.
g_e = moap.encoder_gene_exp_1(g)
x_g = moap.decoder_gene_exp_1(g_e)
x_g = tf.expand_dims(x_g, axis=-1)
x_g = tf.squeeze(x_g, axis=-1)
# Cell type.
c_e = moap.encoder_cell_type_2(c)
x_c = moap.decoder_cell_type_2(c_e)
x_c = moap.dropout_2(x_c)
# Skip-connection autoencoder and final layers.
x = tf.concat([t, g_e, c_e], axis=-1)
for k in range(model.nn_arch["num_sc_ae"]):
x = moap.sc_aes[k](x)
# Final layers.
x = moap.dense_4_1(x)
x = moap.dense_4_2(x)
x = moap.dense_4_3(x)
# Normalize x.
if conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
x1 = moap.dense_4_4(x)
elif conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
x = x / tf.sqrt(tf.reduce_sum(tf.square(x), axis=1, keepdims=True))
x1 = moap.dense_4_4(x)
else:
raise ValueError("loss type is not valid.")
# Get embed_feature_dict.
embed_feature_dict = {}
embed_feature_dict["sig_id"] = -1
embed_feature_dict["embed_feature"] = x.numpy().ravel()
series = target_df.iloc[i]
df = series[series == 1].to_frame()
embed_feature_dict["MoA_classes"] = list(df.index)
embed_feature_dicts.append(embed_feature_dict)
embed_features = np.array([v["embed_feature"] for v in embed_feature_dicts])
tsne_embed_features = TSNE(n_components=2).fit_transform(embed_features)
classes = [v["MoA_classes"] for v in embed_feature_dicts]
for i in tqdm(range(len(classes))):
if len(classes[i]) == 0:
classes[i] = [0]
colors = [v[0] for v in classes]
figure(figsize=(20, 20))
x = []
y = []
c = []
for i in range(len(tsne_embed_features)):
for v in classes[i]:
x.append(tsne_embed_features[i, 0])
y.append(tsne_embed_features[i, 1])
c.append(cls_colors[v])
scatter(x, y, c=c, alpha=1.0)
title("Drug features clustering for training data")
grid()
moa_name_colors_fig = plot_colortable(moa_name_colors, "MoA Colors")
# #### Clustering for validation data
val_iter = val_dataset.as_numpy_iterator()
moap = model.model.get_layer("moap")
embed_feature_dicts = []
target_df = target_scored_df.loc[val_index]
for i, d in tqdm(enumerate(val_iter)):
t, g, c = d
id_ = input_df["sig_id"].iloc[i]
t = np.expand_dims(t, axis=0)
g = np.expand_dims(g, axis=0)
c = np.expand_dims(c, axis=0)
# First layers.
t = moap.embed_treatment_type_0(t)
t = tf.reshape(t, (-1, model.nn_arch["d_input_feature"]))
t = moap.dense_treatment_type_0(t)
t = moap.layer_normalization_0_1(t)
g = moap.layer_normalization_0_2(g)
c = moap.layer_normalization_0_3(c)
# Gene expression.
g_e = moap.encoder_gene_exp_1(g)
x_g = moap.decoder_gene_exp_1(g_e)
x_g = tf.expand_dims(x_g, axis=-1)
x_g = tf.squeeze(x_g, axis=-1)
# Cell type.
c_e = moap.encoder_cell_type_2(c)
x_c = moap.decoder_cell_type_2(c_e)
x_c = moap.dropout_2(x_c)
# Skip-connection autoencoder and final layers.
x = tf.concat([t, g_e, c_e], axis=-1)
for i in range(model.nn_arch["num_sc_ae"]):
x = moap.sc_aes[i](x)
# Final layers.
x = moap.dense_4_1(x)
x = moap.dense_4_2(x)
x = moap.dense_4_3(x)
# Normalize x.
if conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
x1 = moap.dense_4_4(x)
elif conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
x = x / tf.sqrt(tf.reduce_sum(tf.square(x), axis=1, keepdims=True))
x1 = moap.dense_4_4(x)
else:
raise ValueError("loss type is not valid.")
# Get embed_feature_dict.
embed_feature_dict = {}
embed_feature_dict["sig_id"] = id_
embed_feature_dict["embed_feature"] = x.numpy().ravel()
series = target_df.iloc[i]
df = series[series == 1].to_frame()
embed_feature_dict["MoA_classes"] = list(df.index)
embed_feature_dicts.append(embed_feature_dict)
embed_features = np.array([v["embed_feature"] for v in embed_feature_dicts])
tsne_embed_features = TSNE(n_components=2).fit_transform(embed_features)
classes = [v["MoA_classes"] for v in embed_feature_dicts]
for i in tqdm(range(len(classes))):
if len(classes[i]) == 0:
classes[i] = [0]
colors = [v[0] for v in classes]
figure(figsize=(20, 20))
x = []
y = []
c = []
for i in range(len(tsne_embed_features)):
for v in classes[i]:
x.append(tsne_embed_features[i, 0])
y.append(tsne_embed_features[i, 1])
c.append(cls_colors[v])
scatter(x, y, c=c, alpha=1.0)
title("Drug features clustering for validation data")
grid()
moa_name_colors_fig = plot_colortable(moa_name_colors, "MoA Colors")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/672/69672809.ipynb
| null | null |
[{"Id": 69672809, "ScriptId": 12161444, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2956435, "CreationDate": "08/02/2021 17:20:49", "VersionNumber": 12.0, "Title": "Drug features clustering according to MoA", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 1916.0, "LinesInsertedFromPrevious": 753.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1163.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Drug features clustering according to mechanisms of action
# On 2021.8.2, now I'm in a hospital because my father is sick due to cerebral infarction by sudden thrombus after about one month since the 2nd pfizer vaccination. Furthermore, by the COVID-19 pandemic situation, my father and me have been isolated from the out world. My labtop display was broken when I tested my MoA(mechanisms of action) prediction model and drug feature clustering according to mechanisms of action, so my old mother bought a new monitor and brought me the monitor because I couldn't go out. I'm writing this with the sub monitor.
# Last year, I participated in the Mechanisms of Action contest, and that time, I developed an autoencoder and additive angular margin based MoA prediction model approaching this problem as clusering, not multi-classification. Of course, I devleoped an autoencoder based model as multi-classification. I thought the clustering model will be superior to the multi-classification model because the clustering model predicted mechanisms of action more accurately in practical mechanisms of action plot graphs than the multi-classification model, but rather the multi-classification model was superior to the clustering model in the viewpoint of the metric used that time.
# By the way, MoA clustering for the multi-clasification is more random than the clustering model, and in the clustering model, MoA clustering is structured and all drugs have almost all mechanisms of action. So that time, if it is valid, I thought the mRNA vaccine can have almost all known mechanisms of action. But I needed verification.
# Vaccination is mandatory, but our family and friend can die or have serious side effects due to unstable vaccines. Maybe, I hoped fortune for vaccination to my family. But fortune is only a fortune word to me.
# After I faced my father's accident, I searched for relevant papers, and I got it.
# Refer to https://www.medrxiv.org/content/10.1101/2021.04.30.21256383v1.
# This paper shows the vaccine induced immune thrombotic thrombocytopenia side effect for the BNT162b2, ChAdOx1 vaccines. So I understood my father accident's reason.
# Moreover, I'm confident that the MoA clusering model is valid.
# For mRNA vaccines, the vaccination can be carried out to children <= 13. It can be terrible in the human history, because mRNA vaccines' stability are almost unknown.
# Refer to https://coronavirus.quora.com/?__ni__=0&__nsrc__=4&__snid3__=24261290897&__tiids__=32992518
# I still think that the mRNA vaccine is only a solution to cope with fastly changing variants and mutants. But their stability is also mandatory as like that vaccination is mandatory, and this solution must be developed very fastly as the vaccination effect development.
# Via MoA prediction, we can identify side effects of vaccines and develop more stable vaccines. It is mandatory.
# ## Data analysis
import pandas as pd
import seaborn as sns
import os, sys
from scipy.special import comb
from itertools import combinations
import json
from tqdm import tqdm
import pdb
import plotly.express as px
raw_data_path = "/kaggle/input/lish-moa"
# ### Convert categorical values into categorical indexes
input_df = pd.read_csv(os.path.join(raw_data_path, "train_features.csv"))
input_df.info()
input_df.columns
len(input_df.sig_id), len(input_df.sig_id.unique())
input_df.cp_type.unique(), input_df.cp_time.unique(), input_df.cp_dose.unique()
input_df.cp_time.value_counts()
res = input_df.cp_type.astype("category")
res
len(res.cat.categories)
res = res.cat.rename_categories(range(len(res.cat.categories)))
res
res2 = res.map(lambda x: int(x))
res2
type(res2.iloc[0])
input_df.cp_type
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_type
cp_v_index_series = input_df.cp_type[input_df.cp_type == 0]
cp_v_index_series
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
len(input_df.cp_time.cat.categories), len(input_df.cp_dose.cat.categories)
input_df[["cp_type", "cp_time", "cp_dose"]].head(3)
res = input_df.isna().any()
res.any()
res = input_df.iloc[0:64]
res2 = res["cp_time"].values
type(res2)
res2.to_numpy()
g_feature_names = ["g-" + str(v) for v in range(772)]
res2 = res[g_feature_names].values
res2.shape
res2
res2 = res["sig_id"].values
type(res2), res2.shape
res2
# ### Gene expression and cell viability
input_vals = input_df.values
input_vals.shape
input_vals[0]
input_vals_p = input_vals[:, 4:]
input_vals_p.shape
input_vals_p = input_vals_p.astype("float32")
input_vals_p.shape, input_vals_p.dtype
input_vals_p[0, ...]
ge_val = input_vals_p[:, 0:772]
ct_val = input_vals_p[:, 772:]
ge_val.shape, ct_val.shape
ge_val_mean = ge_val.mean(axis=-1)
ge_val_std = ge_val.std(axis=-1)
ge_val_mean.shape
figure(figsize=(20, 20))
plot(ge_val_mean, label="mean")
plot(ge_val_std, label="std")
legend()
grid()
ct_val_mean = ct_val.mean(axis=-1)
ct_val_std = ct_val.std(axis=-1)
figure(figsize=(20, 20))
plot(ct_val_mean, label="mean")
plot(ct_val_std, label="std")
legend()
grid()
# ### Target feature
target_scored_df = pd.read_csv(os.path.join(raw_data_path, "train_targets_scored.csv"))
target_scored_df.info()
target_scored_df.columns
moa_names = list(target_scored_df.columns)[1:]
moa_names
target_scored_df.sum(axis=1)
res = target_scored_df[input_df.cp_type == 0]
res
res.sum(axis=1)
target_scored_df
target_nonscored_df = pd.read_csv(
os.path.join(raw_data_path, "train_targets_nonscored.csv")
)
target_nonscored_df.info()
target_nonscored_df.columns
target_df = pd.concat([target_scored_df, target_nonscored_df.iloc[:, 1:]], axis=1)
len(target_df.columns)
target_df
target_df.columns
res = target_nonscored_df.sum()
res
target_scored_df.iloc[:, 1:2].info()
type(target_scored_df.iloc[:, 1:2])
target_scored_df.iloc[:, 1:2].iloc[:, 0].value_counts()
for i in range(1, len(target_scored_df.columns)):
print(i, target_scored_df.iloc[:, i : (i + 1)].iloc[:, 0].value_counts())
target_scored_df.columns[1:]
len(target_scored_df.columns[1:])
# ## MoA prediction model
"""Keras unsupervised setup module.
"""
from setuptools import setup, find_packages
from os import path
from io import open
here = path.abspath(path.dirname(__file__))
# Get the long description from the README file
with open(path.join(here, "README.md"), encoding="utf-8") as f:
long_description = f.read()
setup(
name="keras-unsupervised", # Required
version="1.1.3.dev1", # Required
description="Keras based unsupervised learning framework.", # Optional
long_description=long_description, # Optional
long_description_content_type="text/markdown", # Optional (see note above)
url="https://github.com/tonandr/keras_unsupervised", # Optional
author="Inwoo Chung", # Optional
author_email="[email protected]", # Optional
classifiers=[ # Optional
# How mature is this project? Common values are
# 3 - Alpha
# 4 - Beta
# 5 - Production/Stable
"Development Status :: 4 - Beta",
# Indicate who your project is intended for
"Intended Audience :: Developers",
"Intended Audience :: Science/Research",
#'Software Development :: Libraries',
# Pick your license as you wish
"License :: OSI Approved :: BSD License",
# Specify the Python versions you support here. In particular, ensure
# that you indicate whether you support Python 2, Python 3 or both.
# These classifiers are *not* checked by 'pip install'. See instead
# 'python_requires' below.
"Programming Language :: Python :: 3",
"Programming Language :: Python :: 3.7" "Programming Language :: Python :: 3.8",
],
# This field adds keywords for your project which will appear on the
# project page. What does your project relate to?
#
# Note that this is a string of words separated by whitespace, not a list.
keywords="keras deepleaning unsupervised semisupervised restricted-botlzmann-machine deep-belief-network autoencoder generative-adversarial-networks", # Optional
# You can just specify package directories manually here if your project is
# simple. Or you can use find_packages().
#
# Alternatively, if you just want to distribute a single Python file, use
# the `py_modules` argument instead as follows, which will expect a file
# called `my_module.py` to exist:
#
# py_modules=["my_module"],
#
packages=find_packages(
exclude=["analysis", "docs", "docs_mkdocs", "resource"]
), # Required
# This field lists other packages that your project depends on to run.
# Any package you put here will be installed by pip when your project is
# installed, so they must be valid existing projects.
#
# For an analysis of "install_requires" vs pip's requirements files see:
# https://packaging.python.org/en/latest/requirements.html
install_requires=[
"tensorflow-probability==0.11",
"pandas",
"scikit-image",
"matplotlib",
"opencv-contrib-python",
], # Optional
# Specify which Python versions you support. In contrast to the
# 'Programming Language' classifiers above, 'pip install' will check this
# and refuse to install the project if the version does not match. If you
# do not support Python 2, you can simplify this to '>=3.5' or similar, see
# https://packaging.python.org/guides/distributing-packages-using-setuptools/#python-requires
python_requires="<=3.8",
# List additional groups of dependencies here (e.g. development
# dependencies). Users will be able to install these using the "extras"
# syntax, for example:
#
# $ pip install sampleproject[dev]
#
# Similar to `install_requires` above, these must be valid existing
# projects.
# extras_require={ # Optional
# 'dev': ['check-manifest'],
# 'test': ['coverage'],
# },
# If there are data files included in your packages that need to be
# installed, specify them here.
#
# If using Python 2.6 or earlier, then these have to be included in
# MANIFEST.in as well.
# package_data={ # Optional
# 'sample': ['package_data.dat'],
# },
# Although 'package_data' is the preferred approach, in some case you may
# need to place data files outside of your packages. See:
# http://docs.python.org/3.4/distutils/setupscript.html#installing-additional-files
#
# In this case, 'data_file' will be installed into '<sys.prefix>/my_data'
# data_files=[('my_data', ['data/data_file'])], # Optional
# To provide executable scripts, use entry points in preference to the
# "scripts" keyword. Entry points provide cross-platform support and allow
# `pip` to create the appropriate form of executable for the target
# platform.
#
# For example, the following would provide a command called `sample` which
# executes the function `main` from this package when invoked:
# entry_points={ # Optional
# 'console_scripts': [
# 'sample=sample:main',
# ],
# },
# List additional URLs that are relevant to your project as a dict.
#
# This field corresponds to the "Project-URL" metadata fields:
# https://packaging.python.org/specifications/core-metadata/#project-url-multiple-use
#
# Examples listed include a pattern for specifying where the package tracks
# issues, where the source is hosted, where to say thanks to the package
# maintainers, and where to support the project financially. The key is
# what's used to render the link text on PyPI.
project_urls={ # Optional
"Bug Reports": "https://github.com/tonandr/keras_unsupervised/issues",
"Source": "https://github.com/tonandr/keras_unsupervised/",
},
)
import sys
sys.path.append("/kaggle/working/keras_unsupervised")
"""
Created on Oct 8, 2020
@author: Inwoo Chung ([email protected])
"""
import os
import time
import json
import random
from random import shuffle
import ctypes
# ctypes.WinDLL('cudart64_110.dll') #?
import numpy as np
import pandas as pd
from tqdm import tqdm
from sklearn.model_selection import StratifiedKFold
import tensorflow as tf
import tensorflow.keras as keras
from tensorflow.keras import backend as K
from tensorflow.keras.losses import Loss
from tensorflow.keras.metrics import Metric
from tensorflow.keras.models import Model, load_model
from tensorflow.keras.layers import Input, Conv1D, Dense, Concatenate, Dropout
from tensorflow.keras.layers import (
LSTM,
Bidirectional,
BatchNormalization,
LayerNormalization,
)
from tensorflow.keras.layers import Embedding, Layer
from tensorflow.keras import optimizers
from tensorflow.keras.callbacks import (
TensorBoard,
ReduceLROnPlateau,
LearningRateScheduler,
ModelCheckpoint,
EarlyStopping,
)
from tensorflow.keras.constraints import UnitNorm
from tensorflow.keras.initializers import RandomUniform, TruncatedNormal
from tensorflow.keras import regularizers
from ku.composite_layer import DenseBatchNormalization
from ku.backprop import (
make_decoder_from_encoder,
make_autoencoder_from_encoder,
make_autoencoder_with_sym_sc,
)
# os.environ["CUDA_DEVICE_ORDER"] = 'PCI_BUS_ID'
# os.environ["CUDA_VISIBLE_DEVICES"] = '-1'
# Constants.
DEBUG = True
MODE_TRAIN = 0
MODE_VAL = 1
CV_TYPE_TRAIN_VAL_SPLIT = "train_val_split"
CV_TYPE_K_FOLD = "k_fold"
DATASET_TYPE_PLAIN = "plain"
DATASET_TYPE_BALANCED = "balanced"
LOSS_TYPE_MULTI_LABEL = "multi_label"
LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN = "additive_angular_margin"
epsilon = 1e-7
class MoALoss(Loss):
def __init__(
self,
W,
m=0.5,
ls=0.2,
scale=64.0,
loss_type=LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN,
name="MoA_loss",
):
super(MoALoss, self).__init__(name=name)
self.W = W
self.m = m
self.ls = ls
self.scale = scale
self.loss_type = loss_type
# @tf.function
def call(self, y_true, y_pred):
y_true = tf.cast(y_true, dtype=tf.float32)
pos_mask = y_true
neg_mask = 1.0 - y_true
# Label smoothing.
y_true = pos_mask * y_true * (1.0 - self.ls / 2.0) + neg_mask * (
y_true + self.ls / 2.0
)
"""
pos_log_loss = pos_mask * self.W[:, :, 0] * tf.sqrt(tf.square(y_true - y_pred))
pos_log_loss_mean = tf.reduce_mean(pos_log_loss, axis=0) #?
pos_loss = 1.0 * tf.reduce_mean(pos_log_loss_mean, axis=0)
neg_log_loss = neg_mask * self.W[:, :, 1] * tf.sqrt(tf.square(y_true - y_pred))
neg_log_loss_mean = tf.reduce_mean(neg_log_loss, axis=0) #?
neg_loss = 1.0 * tf.reduce_mean(neg_log_loss_mean, axis=0)
loss = pos_loss + neg_loss
"""
"""
loss = tf.reduce_mean(tf.sqrt(tf.square(y_true - y_pred)))
loss = tf.losses.binary_crossentropy(y_true, y_pred)
log_loss_mean = tf.reduce_mean(log_loss, axis=0) #?
loss = tf.reduce_mean(log_loss_mean, axis=0)
"""
if self.loss_type == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
A = y_pred
e_AM_A = tf.math.exp(self.scale * tf.math.cos(tf.math.acos(A) + self.m))
# d = A.shape[-1] #?
S = tf.tile(tf.reduce_sum(tf.math.exp(A), axis=1, keepdims=True), (1, 206))
S_p = S - tf.math.exp(A) + e_AM_A
P = e_AM_A / (S_p + epsilon)
# P = tf.clip_by_value(P, clip_value_min=epsilon, clip_value_max=(1.0 - epsilon))
# log_loss_1 = -1.0 * self.W[:, :, 0] * y_true * tf.math.log(P)
log_loss_1 = -1.0 * y_true * tf.math.log(P)
log_loss_2 = tf.reduce_sum(log_loss_1, axis=1)
loss = tf.reduce_mean(log_loss_2, axis=0)
elif self.loss_type == LOSS_TYPE_MULTI_LABEL:
y_pred = tf.sigmoid(y_pred)
y_pred = tf.maximum(tf.minimum(y_pred, 1.0 - 1e-15), 1e-15)
log_loss = -1.0 * (
y_true * tf.math.log(y_pred + epsilon)
+ (1.0 - y_true) * tf.math.log(1.0 - y_pred + epsilon)
)
log_loss_mean = tf.reduce_mean(log_loss, axis=0) # ?
loss = tf.reduce_mean(log_loss_mean, axis=0)
else:
raise ValueError("loss type is not valid.")
# tf.print(A, e_AM_A, S, S_p, P, log_loss_1, log_loss_2, loss)
return loss
def get_config(self):
"""Get configuration."""
config = {
"W": self.W,
"m": self.m,
"ls": self.ls,
"scale": self.scale,
"loss_type": self.loss_type,
}
base_config = super(MoALoss, self).get_config()
return dict(list(base_config.items()) + list(config.items()))
class MoAMetric(Metric):
def __init__(self, sn_t=2.45, name="MoA_metric", **kwargs):
super(MoAMetric, self).__init__(name=name, **kwargs)
self.sn_t = sn_t
self.total_loss = self.add_weight(name="total_loss", initializer="zeros")
self.count = self.add_weight(name="count", initializer="zeros")
def update_state(self, y_true, y_pred, sample_weight=None):
E = tf.reduce_mean(tf.math.exp(y_pred), axis=1, keepdims=True)
E_2 = tf.reduce_mean(tf.square(tf.math.exp(y_pred)), axis=1, keepdims=True)
S = tf.sqrt(E_2 - tf.square(E))
e_A = (tf.exp(y_pred) - E) / (S + epsilon)
e_A_p = tf.where(tf.math.greater(e_A, self.sn_t), self.sn_t, 0.0)
p_hat = e_A_p / (tf.reduce_sum(e_A_p, axis=1, keepdims=True) + epsilon)
y_pred = tf.maximum(tf.minimum(p_hat, 1.0 - 1e-15), 1e-15)
y_true = tf.cast(y_true, dtype=tf.float32)
log_loss = -1.0 * (
y_true * tf.math.log(y_pred + epsilon)
+ (1.0 - y_true) * tf.math.log(1.0 - y_pred + epsilon)
)
log_loss_mean = tf.reduce_mean(log_loss, axis=0) # ?
loss = tf.reduce_mean(log_loss_mean, axis=0)
self.total_loss.assign_add(loss)
self.count.assign_add(tf.constant(1.0))
def result(self):
return tf.math.divide_no_nan(self.total_loss, self.count)
def reset_states(self):
self.total_loss.assign(0.0)
self.count.assign(0.0)
def get_config(self):
"""Get configuration."""
config = {"sn_t": self.sn_t}
base_config = super(MoAMetric, self).get_config()
return dict(list(base_config.items()) + list(config.items()))
class _MoAPredictor(Layer):
def __init__(self, conf, **kwargs):
super(_MoAPredictor, self).__init__(**kwargs)
# Initialize.
self.conf = conf
self.hps = self.conf["hps"]
self.nn_arch = self.conf["nn_arch"]
# Design layers.
# First layers.
self.embed_treatment_type_0 = Embedding(
self.nn_arch["num_treatment_type"], self.nn_arch["d_input_feature"]
)
self.dense_treatment_type_0 = Dense(
self.nn_arch["d_input_feature"], activation="relu"
)
self.layer_normalization_0_1 = LayerNormalization()
self.layer_normalization_0_2 = LayerNormalization()
self.layer_normalization_0_3 = LayerNormalization()
# Autoencoder for gene expression profile.
input_gene_exp_1 = Input(shape=(self.nn_arch["d_gene_exp"],))
d_geps = [int(self.nn_arch["d_gep_init"] / np.power(2, v)) for v in range(4)]
dense_1_1 = Dense(
d_geps[0],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_1_1 = BatchNormalization()
dropout_1_1 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_1_1 = DenseBatchNormalization(
dense_1_1, batch_normalization_1_1, dropout=dropout_1_1
)
dense_1_2 = Dense(
d_geps[1],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_1_2 = BatchNormalization()
dropout_1_2 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_1_2 = DenseBatchNormalization(
dense_1_2, batch_normalization_1_2, dropout=dropout_1_2
)
dense_1_3 = Dense(
d_geps[2],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_1_3 = BatchNormalization()
dropout_1_3 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_1_3 = DenseBatchNormalization(
dense_1_3, batch_normalization_1_3, dropout=dropout_1_3
)
dense_1_4 = Dense(
d_geps[3],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_1_4 = BatchNormalization()
dropout_1_4 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_1_4 = DenseBatchNormalization(
dense_1_4, batch_normalization_1_4, dropout=dropout_1_4
)
self.encoder_gene_exp_1 = keras.Sequential(
[
input_gene_exp_1,
dense_batch_normalization_1_1,
dense_batch_normalization_1_2,
dense_batch_normalization_1_3,
dense_batch_normalization_1_4,
]
)
self.decoder_gene_exp_1 = make_decoder_from_encoder(self.encoder_gene_exp_1)
self.dropout_1 = Dropout(self.nn_arch["dropout_rate"])
# Autoencoder for cell type.
input_gene_exp_2 = Input(shape=(self.nn_arch["d_cell_type"],))
d_cvs = [int(self.nn_arch["d_cv_init"] / np.power(2, v)) for v in range(3)]
dense_2_1 = Dense(
d_cvs[0],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_2_1 = BatchNormalization()
dropout_2_1 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_2_1 = DenseBatchNormalization(
dense_2_1, batch_normalization_2_1, dropout=dropout_2_1
)
dense_2_2 = Dense(
d_cvs[1],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_2_2 = BatchNormalization()
dropout_2_2 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_2_2 = DenseBatchNormalization(
dense_2_2, batch_normalization_2_2, dropout=dropout_2_2
)
dense_2_3 = Dense(
d_cvs[2],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_2_3 = BatchNormalization()
dropout_2_3 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_2_3 = DenseBatchNormalization(
dense_2_3, batch_normalization_2_3, dropout=dropout_2_3
)
self.encoder_cell_type_2 = keras.Sequential(
[
input_gene_exp_2,
dense_batch_normalization_2_1,
dense_batch_normalization_2_2,
dense_batch_normalization_2_3,
]
)
self.decoder_cell_type_2 = make_decoder_from_encoder(self.encoder_cell_type_2)
self.dropout_2 = Dropout(self.nn_arch["dropout_rate"])
# Skip-connection autoencoder layer.
self.sc_aes = []
self.dropout_3 = Dropout(self.nn_arch["dropout_rate"])
for i in range(self.nn_arch["num_sc_ae"]):
input_sk_ae_3 = Input(shape=(self.nn_arch["d_hidden"],))
d_ae_init = d_geps[-1] + d_cvs[-1] + self.nn_arch["d_input_feature"]
d_aes = [d_ae_init, int(d_ae_init * 2), int(d_ae_init * 2), d_ae_init]
dense_3_1 = Dense(
d_aes[0],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_3_1 = BatchNormalization()
dropout_3_1 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_3_1 = DenseBatchNormalization(
dense_3_1, batch_normalization_3_1, dropout=dropout_3_1
)
dense_3_2 = Dense(
d_aes[1],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_3_2 = BatchNormalization()
dropout_3_2 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_3_2 = DenseBatchNormalization(
dense_3_2, batch_normalization_3_2, dropout=dropout_3_2
)
dense_3_3 = Dense(
d_aes[2],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_3_3 = BatchNormalization()
dropout_3_3 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_3_3 = DenseBatchNormalization(
dense_3_3, batch_normalization_3_3, dropout=dropout_3_3
)
dense_3_4 = Dense(
d_aes[3],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
batch_normalization_3_4 = BatchNormalization()
dropout_3_4 = None # Dropout(self.nn_arch['dropout_rate'])
dense_batch_normalization_3_4 = DenseBatchNormalization(
dense_3_4, batch_normalization_3_4, dropout=dropout_3_4
)
sc_encoder_3 = keras.Sequential(
[
input_sk_ae_3,
dense_batch_normalization_3_1,
dense_batch_normalization_3_2,
dense_batch_normalization_3_3,
dense_batch_normalization_3_4,
]
)
sc_autoencoder_3 = make_autoencoder_from_encoder(sc_encoder_3)
self.sc_aes.append(make_autoencoder_with_sym_sc(sc_autoencoder_3))
# Final layers.
d_fs = [int(self.nn_arch["d_f_init"] / np.power(2, v)) for v in range(3)]
self.dense_4_1 = Dense(
d_fs[0],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
self.dense_4_2 = Dense(
d_fs[1],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
self.dense_4_3 = Dense(
d_fs[2],
activation="swish",
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
)
self.dropout_4_3 = Dropout(self.nn_arch["dropout_rate"])
if self.conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
self.dense_4_4 = Dense(
self.nn_arch["num_moa_annotation"],
activation="linear",
kernel_initializer=TruncatedNormal(),
kernel_constraint=None,
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
use_bias=False,
) # ?
elif self.conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
self.dense_4_4 = Dense(
self.nn_arch["num_moa_annotation"],
activation="linear",
kernel_initializer=TruncatedNormal(),
kernel_constraint=UnitNorm(),
kernel_regularizer=regularizers.l2(self.hps["weight_decay"]),
use_bias=False,
) # ?
else:
raise ValueError("loss type is not valid.")
def call(self, inputs):
t = inputs[0]
g = inputs[1]
c = inputs[2]
# First layers.
t = self.embed_treatment_type_0(t)
t = tf.reshape(t, (-1, self.nn_arch["d_input_feature"]))
t = self.dense_treatment_type_0(t)
t = self.layer_normalization_0_1(t)
g = self.layer_normalization_0_2(g)
c = self.layer_normalization_0_3(c)
# Gene expression.
g_e = self.encoder_gene_exp_1(g)
x_g = self.decoder_gene_exp_1(g_e)
x_g = tf.expand_dims(x_g, axis=-1)
x_g = tf.squeeze(x_g, axis=-1)
x_g = self.dropout_1(x_g)
# Cell type.
c_e = self.encoder_cell_type_2(c)
x_c = self.decoder_cell_type_2(c_e)
x_c = self.dropout_2(x_c)
# Skip-connection autoencoder and final layers.
x = tf.concat([t, g_e, c_e], axis=-1)
for i in range(self.nn_arch["num_sc_ae"]):
x = self.sc_aes[i](x)
x = self.dropout_3(x)
# Final layers.
x = self.dense_4_1(x)
x = self.dense_4_2(x)
x = self.dense_4_3(x)
x = self.dropout_4_3(x)
# Normalize x.
if self.conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
x1 = self.dense_4_4(x)
elif self.conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
x = x / tf.sqrt(tf.reduce_sum(tf.square(x), axis=1, keepdims=True))
x1 = self.dense_4_4(x)
else:
raise ValueError("loss type is not valid.")
outputs = [x_g, x_c, x1]
return outputs
def get_config(self):
"""Get configuration."""
config = {"conf": self.conf}
base_config = super(_MoAPredictor, self).get_config()
return dict(list(base_config.items()) + list(config.items()))
class MoAPredictor(object):
"""MoA predictor."""
# Constants.
MODEL_PATH = "MoA_predictor"
OUTPUT_FILE_NAME = "submission.csv"
EVALUATION_FILE_NAME = "eval.csv"
def __init__(self, conf):
"""
Parameters
----------
conf: Dictionary
Configuration dictionary.
"""
# Initialize.
self.conf = conf
self.raw_data_path = self.conf["raw_data_path"]
self.hps = self.conf["hps"]
self.nn_arch = self.conf["nn_arch"]
self.model_loading = self.conf["model_loading"]
# Create weight for classification imbalance.
W = self._create_W()
# with strategy.scope():
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
if self.model_loading:
self.model = load_model(
self.MODEL_PATH + ".h5",
custom_objects={
"MoALoss": MoALoss,
"MoAMetric": MoAMetric,
"_MoAPredictor": _MoAPredictor,
},
compile=False,
)
# self.model = load_model(self.MODEL_PATH, compile=False)
opt = optimizers.Adam(
lr=self.hps["lr"],
beta_1=self.hps["beta_1"],
beta_2=self.hps["beta_2"],
decay=self.hps["decay"],
)
self.model.compile(
optimizer=opt,
loss=[
"mse",
"mse",
MoALoss(
W,
self.nn_arch["additive_margin"],
self.hps["ls"],
self.nn_arch["scale"],
loss_type=self.conf["loss_type"],
),
],
loss_weights=self.hps["loss_weights"],
metrics=[["mse"], ["mse"], [MoAMetric(self.hps["sn_t"])]],
run_eagerly=False,
)
else:
# Design the MoA prediction model.
# Input.
input_t = Input(shape=(self.nn_arch["d_treatment_type"],))
input_g = Input(shape=(self.nn_arch["d_gene_exp"],))
input_c = Input(shape=(self.nn_arch["d_cell_type"],))
outputs = _MoAPredictor(self.conf, name="moap")(
[input_t, input_g, input_c]
)
opt = optimizers.Adam(
lr=self.hps["lr"],
beta_1=self.hps["beta_1"],
beta_2=self.hps["beta_2"],
decay=self.hps["decay"],
)
self.model = Model(inputs=[input_t, input_g, input_c], outputs=outputs)
self.model.compile(
optimizer=opt,
loss=[
"mse",
"mse",
MoALoss(
W,
self.nn_arch["additive_margin"],
self.hps["ls"],
self.nn_arch["scale"],
loss_type=self.conf["loss_type"],
),
],
loss_weights=self.hps["loss_weights"],
metrics=[["mse"], ["mse"], [MoAMetric(self.hps["sn_t"])]],
run_eagerly=False,
)
self.model.summary()
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
self.k_fold_models = []
if self.model_loading:
opt = optimizers.Adam(
lr=self.hps["lr"],
beta_1=self.hps["beta_1"],
beta_2=self.hps["beta_2"],
decay=self.hps["decay"],
)
# load models for K-fold.
for i in range(self.nn_arch["k_fold"]):
self.k_fold_models.append(
load_model(
self.MODEL_PATH + "_" + str(i) + ".h5",
custom_objects={
"MoALoss": MoALoss,
"MoAMetric": MoAMetric,
"_MoAPredictor": _MoAPredictor,
},
compile=False,
)
)
self.k_fold_models[i].compile(
optimizer=opt,
loss=[
"mse",
"mse",
MoALoss(
W,
self.nn_arch["additive_margin"],
self.hps["ls"],
self.nn_arch["scale"],
loss_type=self.conf["loss_type"],
),
],
loss_weights=self.hps["loss_weights"],
metrics=[["mse"], ["mse"], [MoAMetric(self.hps["sn_t"])]],
run_eagerly=False,
)
else:
# Create models for K-fold.
for i in range(self.nn_arch["k_fold"]):
# Design the MoA prediction model.
# Input.
input_t = Input(shape=(self.nn_arch["d_treatment_type"],))
input_g = Input(shape=(self.nn_arch["d_gene_exp"],))
input_c = Input(shape=(self.nn_arch["d_cell_type"],))
outputs = _MoAPredictor(self.conf, name="moap")(
[input_t, input_g, input_c]
)
opt = optimizers.Adam(
lr=self.hps["lr"],
beta_1=self.hps["beta_1"],
beta_2=self.hps["beta_2"],
decay=self.hps["decay"],
)
model = Model(inputs=[input_t, input_g, input_c], outputs=outputs)
model.compile(
optimizer=opt,
loss=[
"mse",
"mse",
MoALoss(
W,
self.nn_arch["additive_margin"],
self.hps["ls"],
self.nn_arch["scale"],
loss_type=self.conf["loss_type"],
),
],
loss_weights=self.hps["loss_weights"],
metrics=[["mse"], ["mse"], [MoAMetric(self.hps["sn_t"])]],
run_eagerly=False,
)
model.summary()
self.k_fold_models.append(model)
else:
raise ValueError("cv_type is not valid.")
# Create dataset.
self._create_dataset()
def _create_dataset(self):
input_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_features.csv")
) # .iloc[:1024]
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
# Remove samples of ctl_vehicle.
valid_indexes = input_df.cp_type == 1
input_df = input_df[valid_indexes]
input_df = input_df.reset_index(drop=True)
target_scored_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_targets_scored.csv")
) # .iloc[:1024]
target_scored_df = target_scored_df[valid_indexes]
target_scored_df = target_scored_df.reset_index(drop=True)
del target_scored_df["sig_id"]
target_scored_df.columns = range(len(target_scored_df.columns))
n_target_samples = target_scored_df.sum().values
if self.conf["data_aug"]:
genes = [col for col in input_df.columns if col.startswith("g-")]
cells = [col for col in input_df.columns if col.startswith("c-")]
features = genes + cells
targets = [col for col in target_scored_df if col != "sig_id"]
aug_trains = []
aug_targets = []
for t in [0, 1, 2]:
for d in [0, 1]:
for _ in range(3):
train1 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
]
target1 = target_scored_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
]
ctl1 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
].sample(train1.shape[0], replace=True)
ctl2 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
].sample(train1.shape[0], replace=True)
train1[genes + cells] = (
train1[genes + cells].values
+ ctl1[genes + cells].values
- ctl2[genes + cells].values
)
aug_trains.append(train1)
aug_targets.append(target1)
input_df = pd.concat(aug_trains).reset_index(drop=True)
target_scored_df = pd.concat(aug_targets).reset_index(drop=True)
g_feature_names = ["g-" + str(v) for v in range(self.nn_arch["d_gene_exp"])]
c_feature_names = ["c-" + str(v) for v in range(self.nn_arch["d_cell_type"])]
moa_names = [v for v in range(self.nn_arch["num_moa_annotation"])]
def get_series_from_input(idxes):
idxes = idxes.numpy() # ?
series = input_df.iloc[idxes]
# Treatment.
if isinstance(idxes, np.int32) != True:
cp_time = series["cp_time"].values.to_numpy()
cp_dose = series["cp_dose"].values.to_numpy()
else:
cp_time = np.asarray(series["cp_time"])
cp_dose = np.asarray(series["cp_dose"])
treatment_type = cp_time * 2 + cp_dose
# Gene expression.
gene_exps = series[g_feature_names].values
# Cell viability.
cell_vs = series[c_feature_names].values
return treatment_type, gene_exps, cell_vs
def make_input_target_features(idxes):
treatment_type, gene_exps, cell_vs = tf.py_function(
get_series_from_input,
inp=[idxes],
Tout=[tf.int64, tf.float64, tf.float64],
)
MoA_values = tf.py_function(
get_series_from_target, inp=[idxes], Tout=tf.int32
)
return (
(treatment_type, gene_exps, cell_vs),
(gene_exps, cell_vs, MoA_values),
)
def make_input_features(idx):
treatment_type, gene_exps, cell_vs = tf.py_function(
get_series_from_input,
inp=[idx],
Tout=[tf.int64, tf.float64, tf.float64],
)
return treatment_type, gene_exps, cell_vs
def make_a_target_features(idx):
treatment_type, gene_exps, cell_vs = tf.py_function(
get_series_from_input,
inp=[idx],
Tout=[tf.int64, tf.float64, tf.float64],
)
return gene_exps, cell_vs
def get_series_from_target(idxes):
idxes = idxes.numpy()
series = target_scored_df.iloc[idxes]
# MoA annotations' values.
MoA_values = series[moa_names].values
return MoA_values
def make_target_features(idx):
MoA_values = tf.py_function(
get_series_from_target, inp=[idx], Tout=tf.int32
)
return MoA_values
def divide_inputs(input1, input2):
return input1[0], input1[1], input2
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
if self.conf["dataset_type"] == DATASET_TYPE_PLAIN:
train_val_index = np.arange(len(input_df))
# np.random.shuffle(train_val_index)
num_val = int(self.conf["val_ratio"] * len(input_df))
num_tr = len(input_df) - num_val
train_index = train_val_index[:num_tr]
val_index = train_val_index[num_tr:]
self.train_index = train_index
self.val_index = val_index
# Training dataset.
input_dataset = tf.data.Dataset.from_tensor_slices(train_index)
input_dataset = input_dataset.map(make_input_features)
a_target_dataset = tf.data.Dataset.from_tensor_slices(train_index)
a_target_dataset = a_target_dataset.map(make_a_target_features)
target_dataset = tf.data.Dataset.from_tensor_slices(train_index)
target_dataset = target_dataset.map(make_target_features)
f_target_dataset = tf.data.Dataset.zip(
(a_target_dataset, target_dataset)
).map(divide_inputs)
# Inputs and targets.
tr_dataset = tf.data.Dataset.zip((input_dataset, f_target_dataset))
tr_dataset = (
tr_dataset.shuffle(
buffer_size=self.hps["batch_size"] * 5,
reshuffle_each_iteration=True,
)
.repeat()
.batch(self.hps["batch_size"])
)
self.step = len(train_index) // self.hps["batch_size"]
# Validation dataset.
input_dataset = tf.data.Dataset.from_tensor_slices(val_index)
input_dataset = input_dataset.map(make_input_features)
a_target_dataset = tf.data.Dataset.from_tensor_slices(val_index)
a_target_dataset = a_target_dataset.map(make_a_target_features)
target_dataset = tf.data.Dataset.from_tensor_slices(val_index)
target_dataset = target_dataset.map(make_target_features)
f_target_dataset = tf.data.Dataset.zip(
(a_target_dataset, target_dataset)
).map(divide_inputs)
# Inputs and targets.
val_dataset = tf.data.Dataset.zip((input_dataset, f_target_dataset))
val_dataset = val_dataset.batch(self.hps["batch_size"])
self.trval_dataset = (tr_dataset, val_dataset)
elif self.conf["dataset_type"] == DATASET_TYPE_BALANCED:
MoA_p_sets = []
val_index = []
for col in target_scored_df.columns:
s = target_scored_df.iloc[:, col]
s = s[s == 1]
s = list(s.index)
# shuffle(s)
n_val = int(n_target_samples[col] * self.conf["val_ratio"])
if n_val != 0:
tr_set = s[: int(-1.0 * n_val)]
val_set = s[int(-1.0 * n_val) :]
MoA_p_sets.append(tr_set)
val_index += val_set
else:
MoA_p_sets.append(s)
df = target_scored_df.sum(axis=1)
df = df[df == 0]
no_MoA_p_set = list(df.index)
# shuffle(no_MoA_p_set)
val_index += no_MoA_p_set[
int(-1.0 * len(no_MoA_p_set) * self.conf["val_ratio"]) :
]
MoA_p_sets.append(
no_MoA_p_set[
: int(-1.0 * len(no_MoA_p_set) * self.conf["val_ratio"])
]
)
idxes = []
for i in range(self.hps["rep"]):
for col in range(len(target_scored_df.columns) + 1):
if len(MoA_p_sets[col]) >= (i + 1):
idx = MoA_p_sets[col][i]
else:
idx = np.random.choice(
MoA_p_sets[col], size=1, replace=True
)[0]
idxes.append(idx)
train_index = idxes
self.train_index = train_index
self.val_index = val_index
# Training dataset.
tr_dataset = tf.data.Dataset.from_tensor_slices(train_index)
# Inputs and targets.
tr_dataset = (
tr_dataset.shuffle(
buffer_size=self.hps["batch_size"] * 5,
reshuffle_each_iteration=True,
)
.repeat()
.batch(self.hps["batch_size"])
.map(
make_input_target_features,
num_parallel_calls=tf.data.experimental.AUTOTUNE,
)
)
self.step = len(train_index) // self.hps["batch_size"]
# Validation dataset.
val_dataset = tf.data.Dataset.from_tensor_slices(val_index)
# Inputs and targets.
val_dataset = val_dataset.batch(self.hps["batch_size"]).map(
make_input_target_features,
num_parallel_calls=tf.data.experimental.AUTOTUNE,
)
# Save datasets.
# tf.data.experimental.save(tr_dataset, './tr_dataset')
# tf.data.experimental.save(val_dataset, './val_dataset')
self.trval_dataset = (tr_dataset, val_dataset)
else:
raise ValueError("dataset type is not valid.")
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
stratified_kfold = StratifiedKFold(n_splits=self.nn_arch["k_fold"])
# group_kfold = GroupKFold(n_splits=self.nn_arch['k_fold'])
self.k_fold_trval_datasets = []
for train_index, val_index in stratified_kfold.split(
input_df, input_df.cp_type
):
# Training dataset.
input_dataset = tf.data.Dataset.from_tensor_slices(train_index)
input_dataset = input_dataset.map(make_input_features)
a_target_dataset = tf.data.Dataset.from_tensor_slices(train_index)
a_target_dataset = a_target_dataset.map(make_a_target_features)
target_dataset = tf.data.Dataset.from_tensor_slices(train_index)
target_dataset = target_dataset.map(make_target_features)
f_target_dataset = tf.data.Dataset.zip(
(a_target_dataset, target_dataset)
).map(divide_inputs)
# Inputs and targets.
tr_dataset = tf.data.Dataset.zip((input_dataset, f_target_dataset))
tr_dataset = (
tr_dataset.shuffle(
buffer_size=self.hps["batch_size"] * 5,
reshuffle_each_iteration=True,
)
.repeat()
.batch(self.hps["batch_size"])
)
self.step = len(train_index) // self.hps["batch_size"]
# Validation dataset.
input_dataset = tf.data.Dataset.from_tensor_slices(val_index)
input_dataset = input_dataset.map(make_input_features)
a_target_dataset = tf.data.Dataset.from_tensor_slices(val_index)
a_target_dataset = a_target_dataset.map(make_a_target_features)
target_dataset = tf.data.Dataset.from_tensor_slices(val_index)
target_dataset = target_dataset.map(make_target_features)
f_target_dataset = tf.data.Dataset.zip(
(a_target_dataset, target_dataset)
).map(divide_inputs)
# Inputs and targets.
val_dataset = tf.data.Dataset.zip((input_dataset, f_target_dataset))
val_dataset = val_dataset.batch(self.hps["batch_size"])
self.k_fold_trval_datasets.append((tr_dataset, val_dataset))
else:
raise ValueError("cv_type is not valid.")
def _create_W(self):
target_scored_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_targets_scored.csv")
)
del target_scored_df["sig_id"]
weights = []
for c in target_scored_df.columns:
s = target_scored_df[c]
s = s.value_counts()
s = s / s.sum()
weights.append(s.values)
weight = np.expand_dims(np.array(weights), axis=0)
return weight
def train(self):
"""Train."""
reduce_lr = ReduceLROnPlateau(
monitor="val_loss",
factor=self.hps["reduce_lr_factor"],
patience=3,
min_lr=1.0e-8,
verbose=1,
)
tensorboard = TensorBoard(
histogram_freq=1, write_graph=True, write_images=True, update_freq="epoch"
)
earlystopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=5, verbose=1, mode="auto"
)
"""
def schedule_lr(e_i):
self.hps['lr'] = self.hps['reduce_lr_factor'] * self.hps['lr']
return self.hps['lr']
lr_scheduler = LearningRateScheduler(schedule_lr, verbose=1)
"""
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
model_check_point = ModelCheckpoint(
self.MODEL_PATH + ".h5",
monitor="val_loss",
verbose=1,
save_best_only=True,
)
hist = self.model.fit(
self.trval_dataset[0],
steps_per_epoch=self.step,
epochs=self.hps["epochs"],
verbose=1,
max_queue_size=80,
workers=4,
use_multiprocessing=False,
callbacks=[
model_check_point,
earlystopping,
], # , reduce_lr] #, tensorboard]
validation_data=self.trval_dataset[1],
validation_freq=1,
shuffle=True,
)
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
for i in range(self.nn_arch["k_fold"]):
model_check_point = ModelCheckpoint(
self.MODEL_PATH + "_" + str(i) + ".h5",
monitor="loss",
verbose=1,
save_best_only=True,
)
hist = self.k_fold_models[i].fit(
self.k_fold_trval_datasets[i][0],
steps_per_epoch=self.step,
epochs=self.hps["epochs"],
verbose=1,
max_queue_size=80,
workers=4,
use_multiprocessing=False,
callbacks=[
model_check_point,
earlystopping,
], # reduce_lr] #, tensorboard]
validation_data=self.k_fold_trval_datasets[i][1],
validation_freq=1,
shuffle=True,
)
else:
raise ValueError("cv_type is not valid.")
# print('Save the model.')
# self.model.save(self.MODEL_PATH, save_format='h5')
# self.model.save(self.MODEL_PATH, save_format='tf')
return hist
def evaluate(self):
"""Evaluate."""
assert self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT
input_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_features.csv")
) # .iloc[:1024]
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
# Remove samples of ctl_vehicle.
valid_indexes = input_df.cp_type == 1 # ?
target_scored_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_targets_scored.csv")
) # .iloc[:1024]
target_scored_df = target_scored_df.loc[self.val_index]
MoA_annots = target_scored_df.columns[1:]
def make_input_features(inputs):
# Treatment.
cp_time = inputs["cp_time"]
cp_dose = inputs["cp_dose"]
treatment_type = cp_time * 2 + cp_dose
# Gene expression.
gene_exps = [
inputs["g-" + str(v)] for v in range(self.nn_arch["d_gene_exp"])
]
gene_exps = tf.stack(gene_exps, axis=0)
# Cell viability.
cell_vs = [
inputs["c-" + str(v)] for v in range(self.nn_arch["d_cell_type"])
]
cell_vs = tf.stack(cell_vs, axis=0)
return (tf.expand_dims(treatment_type, axis=-1), gene_exps, cell_vs)
# Validation dataset.
val_dataset = tf.data.Dataset.from_tensor_slices(
input_df.loc[self.val_index].to_dict("list")
)
val_dataset = val_dataset.map(make_input_features)
val_iter = val_dataset.as_numpy_iterator()
# Predict MoAs.
sig_id_list = []
MoAs = [[] for _ in range(len(MoA_annots))]
for i, d in tqdm(enumerate(val_iter)):
t, g, c = d
id = target_scored_df["sig_id"].iloc[i]
t = np.expand_dims(t, axis=0)
g = np.expand_dims(g, axis=0)
c = np.expand_dims(c, axis=0)
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
_, _, result = self.model.layers[-1](
[t, g, c]
) # self.model.predict([t, g, c])
result = np.squeeze(result, axis=0)
# result = np.exp(result) / (np.sum(np.exp(result), axis=0) + epsilon)
for i, MoA in enumerate(result):
MoAs[i].append(MoA)
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
# Conduct ensemble prediction.
result_list = []
for i in range(self.nn_arch["k_fold"]):
_, _, result = self.k_fold_models[i].predict([t, g, c])
result = np.squeeze(result, axis=0)
# result = np.exp(result) / (np.sum(np.exp(result), axis=0) + epsilon)
result_list.append(result)
result_mean = np.asarray(result_list).mean(axis=0)
for i, MoA in enumerate(result_mean):
MoAs[i].append(MoA)
else:
raise ValueError("cv_type is not valid.")
sig_id_list.append(id)
# Save the result.
result_dict = {"sig_id": sig_id_list}
for i, MoA_annot in enumerate(MoA_annots):
result_dict[MoA_annot] = MoAs[i]
submission_df = pd.DataFrame(result_dict)
submission_df.to_csv(self.OUTPUT_FILE_NAME, index=False)
target_scored_df.to_csv("gt.csv", index=False)
def test(self):
"""Test."""
# Create the test dataset.
input_df = pd.read_csv(os.path.join(self.raw_data_path, "test_features.csv"))
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
# Remove samples of ctl_vehicle.
valid_indexes = input_df.cp_type == 1 # ?
target_scored_df = pd.read_csv(
os.path.join(self.raw_data_path, "train_targets_scored.csv")
)
MoA_annots = target_scored_df.columns[1:]
def make_input_features(inputs):
id_ = inputs["sig_id"]
cp_type = inputs["cp_type"]
# Treatment.
cp_time = inputs["cp_time"]
cp_dose = inputs["cp_dose"]
treatment_type = cp_time * 2 + cp_dose
# Gene expression.
gene_exps = [
inputs["g-" + str(v)] for v in range(self.nn_arch["d_gene_exp"])
]
gene_exps = tf.stack(gene_exps, axis=0)
# Cell viability.
cell_vs = [
inputs["c-" + str(v)] for v in range(self.nn_arch["d_cell_type"])
]
cell_vs = tf.stack(cell_vs, axis=0)
return (
id_,
cp_type,
tf.expand_dims(treatment_type, axis=-1),
gene_exps,
cell_vs,
)
test_dataset = tf.data.Dataset.from_tensor_slices(input_df.to_dict("list"))
test_dataset = test_dataset.map(make_input_features)
test_iter = test_dataset.as_numpy_iterator()
# Predict MoAs.
sig_id_list = []
MoAs = [[] for _ in range(len(MoA_annots))]
def cal_prob(logit):
a = logit
a = (a + 1.0) / 2.0
a = tf.where(tf.math.greater(a, self.hps["sn_t"]), a, 0.0)
a = self.hps["m1"] * a + self.hps["m2"]
p_h = tf.sigmoid(a).numpy()
return p_h
def cal_prob_2(logit):
y_pred = logit
E = tf.reduce_mean(tf.math.exp(y_pred), axis=-1, keepdims=True)
E_2 = tf.reduce_mean(tf.square(tf.math.exp(y_pred)), axis=-1, keepdims=True)
S = tf.sqrt(E_2 - tf.square(E))
e_A = (tf.exp(y_pred) - E) / (S + epsilon)
e_A_p = tf.where(
tf.math.greater(e_A, self.hps["sn_t"]), self.hps["sn_t"], 0.0
)
p_h = e_A_p / (tf.reduce_sum(e_A_p, axis=-1, keepdims=True) + epsilon)
return p_h.numpy()
def cal_prob_3(logit):
A = logit
A = (A + 1.0) / 2.0
E = tf.reduce_mean(A, axis=-1, keepdims=True)
E_2 = tf.reduce_mean(tf.square(A), axis=-1, keepdims=True)
S = tf.sqrt(E_2 - tf.square(E))
# S_N = tf.abs(A - E) / (S + epsilon)
S_N = (A - E) / (S + epsilon)
# S_N = tf.where(tf.math.greater(S_N, self.hps['sn_t']), S_N, 0.0)
A_p = self.hps["m1"] * S_N + self.hps["m2"]
# P_h = tf.clip_by_value(A_p / 10.0, clip_value_min=0.0, clip_value_max=1.0)
P_h = tf.sigmoid(A_p)
return P_h.numpy()
def cal_prob_4(logit):
a = logit
p_h = tf.sigmoid(a).numpy()
return p_h
for id_, cp_type, t, g, c in tqdm(test_iter):
id_ = id_.decode("utf8") # ?
t = np.expand_dims(t, axis=0)
g = np.expand_dims(g, axis=0)
c = np.expand_dims(c, axis=0)
if self.conf["cv_type"] == CV_TYPE_TRAIN_VAL_SPLIT:
# _, _, result = self.model.layers[-1]([t, g, c]) #self.model.predict([t, g, c])
_, _, result = self.model.predict([t, g, c])
result = np.squeeze(result, axis=0)
if cp_type == 1:
if self.conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
result = cal_prob_4(result)
elif self.conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
result = cal_prob_3(result)
else:
raise ValueError("loss type is not valid.")
else:
result = np.zeros((len(result)))
for i, MoA in enumerate(result):
MoAs[i].append(MoA)
elif self.conf["cv_type"] == CV_TYPE_K_FOLD:
# Conduct ensemble prediction.
result_list = []
for i in range(self.nn_arch["k_fold"]):
_, _, result = self.k_fold_models[i].predict([t, g, c])
result = np.squeeze(result, axis=0)
if cp_type == 1:
if self.conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
result = cal_prob_4(result)
elif (
self.conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN
):
result = cal_prob_3(result)
else:
raise ValueError("loss type is not valid.")
else:
result = np.zeros((len(result)))
result_list.append(result)
result_mean = np.asarray(result_list).mean(axis=0)
for i, MoA in enumerate(result_mean):
MoAs[i].append(MoA)
else:
raise ValueError("cv_type is not valid.")
sig_id_list.append(id_)
# Save the result.
result_dict = {"sig_id": sig_id_list}
for i, MoA_annot in enumerate(MoA_annots):
result_dict[MoA_annot] = MoAs[i]
submission_df = pd.DataFrame(result_dict)
submission_df.to_csv(self.OUTPUT_FILE_NAME, index=False)
import time
seed = int(time.time())
# seed = 1606208227
print(f"Seed:{seed}")
np.random.seed(seed)
tf.random.set_seed(seed)
# ## Training
{
"mode": "train",
"raw_data_path": "/kaggle/input/lish-moa",
"model_loading": false,
"multi_gpu": false,
"num_gpus": 4,
"cv_type": "train_val_split",
"dataset_type": "balanced",
"val_ratio": 0.1,
"loss_type": "additive_angular_margin",
"data_aug": false,
"hps": {
"lr": 0.001,
"beta_1": 0.999,
"beta_2": 0.999,
"decay": 0.0,
"epochs": 258,
"batch_size": 512,
"reduce_lr_factor": 0.96,
"ls": 8.281e-5,
"loss_weights": [1.0, 0.1, 100.0],
"rep": 1600,
"sn_t": 1.6,
"m1": 0.8081512,
"m2": 0.011438734,
"weight_decay": 0.000858,
},
"nn_arch": {
"k_fold": 5,
"d_treatment_type": 1,
"num_treatment_type": 6,
"d_input_feature": 8,
"d_gene_exp": 772,
"d_cell_type": 100,
"d_gep_init": 1024,
"d_cv_init": 128,
"num_sc_ae": 0,
"d_f_init": 512,
"num_moa_annotation": 206,
"d_out": 772,
"dropout_rate": 0.2,
"similarity_type": "diff_abs",
"additive_margin": 0.02,
"scale": 1.0,
},
}
with open("MoA_pred_conf.json", "r") as f:
conf = json.load(f)
# Train.
model = MoAPredictor(conf)
ts = time.time()
hist = model.train()
te = time.time()
print("Elasped time: {0:f}s".format(te - ts))
# ### MoA clustering analysis
# #### Center analysis
moap = model.model.get_layer("moap")
W = moap.dense_4_4.weights[0]
W.shape
W = W.numpy()
W = W.T
W.shape
from sklearn.manifold import TSNE
W_e = TSNE(n_components=2).fit_transform(W)
W_e.shape
colors = np.arange(len(W_e))
figure(figsize=(20, 20))
scatter(W_e[:, 0], W_e[:, 1], c=colors, cmap=cm.prism, marker="^", s=100)
# for i, c in enumerate(colors):
# annotate(str(c), (tsne_embed_features[i, 0], tsne_embed_features[i, 1]))
grid()
# #### Clustering map for training and validation data
from matplotlib.patches import Rectangle
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
def plot_colortable(colors, title, sort_colors=True, emptycols=0):
cell_width = 212
cell_height = 22
swatch_width = 48
margin = 12
topmargin = 40
# Sort colors by hue, saturation, value and name.
if sort_colors is True:
by_hsv = sorted(
(tuple(mcolors.rgb_to_hsv(mcolors.to_rgb(color))), name)
for name, color in colors.items()
)
names = [name for hsv, name in by_hsv]
else:
names = list(colors)
n = len(names)
ncols = 4 - emptycols
nrows = n // ncols + int(n % ncols > 0)
width = cell_width * 4 + 2 * margin
height = cell_height * nrows + margin + topmargin
dpi = 72
fig, ax = plt.subplots(figsize=(width / dpi, height / dpi), dpi=dpi)
fig.subplots_adjust(
margin / width,
margin / height,
(width - margin) / width,
(height - topmargin) / height,
)
ax.set_xlim(0, cell_width * 4)
ax.set_ylim(cell_height * (nrows - 0.5), -cell_height / 2.0)
ax.yaxis.set_visible(False)
ax.xaxis.set_visible(False)
ax.set_axis_off()
ax.set_title(title, fontsize=24, loc="left", pad=10)
for i, name in enumerate(names):
row = i % nrows
col = i // nrows
y = row * cell_height
swatch_start_x = cell_width * col
text_pos_x = cell_width * col + swatch_width + 7
ax.text(
text_pos_x,
y,
name,
fontsize=8,
horizontalalignment="left",
verticalalignment="center",
)
ax.add_patch(
Rectangle(
xy=(swatch_start_x, y - 9),
width=swatch_width,
height=18,
facecolor=colors[name],
edgecolor="0.7",
)
)
return fig
len(mcolors.XKCD_COLORS)
colors = mcolors.XKCD_COLORS
color_items = colors.items()
color_items = list(color_items)
cls_colors = [color_items[i][1] for i in range(0, len(color_items), 4)]
cls_colors = cls_colors[:207]
moa_names = ["none"] + moa_names
moa_name_colors = dict((name, color) for name, color in zip(moa_names, cls_colors))
# #### Clustering for training data
input_df = pd.read_csv(os.path.join(raw_data_path, "train_features.csv"))
input_df.cp_type = input_df.cp_type.astype("category")
input_df.cp_type = input_df.cp_type.cat.rename_categories(
range(len(input_df.cp_type.cat.categories))
)
input_df.cp_time = input_df.cp_time.astype("category")
input_df.cp_time = input_df.cp_time.cat.rename_categories(
range(len(input_df.cp_time.cat.categories))
)
input_df.cp_dose = input_df.cp_dose.astype("category")
input_df.cp_dose = input_df.cp_dose.cat.rename_categories(
range(len(input_df.cp_dose.cat.categories))
)
# Remove samples of ctl_vehicle.
valid_indexes = input_df.cp_type == 1
input_df = input_df[valid_indexes]
input_df = input_df.reset_index(drop=True)
target_scored_df = pd.read_csv(os.path.join(raw_data_path, "train_targets_scored.csv"))
target_scored_df = target_scored_df[valid_indexes]
target_scored_df = target_scored_df.reset_index(drop=True)
del target_scored_df["sig_id"]
target_scored_df.columns = range(len(target_scored_df.columns))
n_target_samples = target_scored_df.sum().values
if model.conf["data_aug"]:
genes = [col for col in input_df.columns if col.startswith("g-")]
cells = [col for col in input_df.columns if col.startswith("c-")]
features = genes + cells
targets = [col for col in target_scored_df if col != "sig_id"]
aug_trains = []
aug_targets = []
for t in [0, 1, 2]:
for d in [0, 1]:
for _ in range(3):
train1 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
]
target1 = target_scored_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
]
ctl1 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
].sample(train1.shape[0], replace=True)
ctl2 = input_df.loc[
(input_df["cp_time"] == t) & (input_df["cp_dose"] == d)
].sample(train1.shape[0], replace=True)
train1[genes + cells] = (
train1[genes + cells].values
+ ctl1[genes + cells].values
- ctl2[genes + cells].values
)
aug_trains.append(train1)
aug_targets.append(target1)
input_df = pd.concat(aug_trains).reset_index(drop=True)
target_scored_df = pd.concat(aug_targets).reset_index(drop=True)
g_feature_names = ["g-" + str(v) for v in range(model.nn_arch["d_gene_exp"])]
c_feature_names = ["c-" + str(v) for v in range(model.nn_arch["d_cell_type"])]
moa_names = [v for v in range(model.nn_arch["num_moa_annotation"])]
def get_series_from_input(idxes):
idxes = idxes.numpy()
df = input_df.iloc[idxes]
# Treatment.
cp_time = df["cp_time"]
cp_dose = df["cp_dose"]
treatment_type = cp_time * 2 + cp_dose
# Gene expression.
gene_exps = df[g_feature_names].values
# Cell viability.
cell_vs = df[c_feature_names].values
return treatment_type, gene_exps, cell_vs
def make_input_features(idx):
treatment_type, gene_exps, cell_vs = tf.py_function(
get_series_from_input, inp=[idx], Tout=[tf.int32, tf.float64, tf.float64]
)
return treatment_type, gene_exps, cell_vs
MoA_p_sets = []
train_index = []
val_index = []
for col in target_scored_df.columns:
s = target_scored_df.iloc[:, col]
s = s[s == 1]
s = list(s.index)
n_val = int(n_target_samples[col] * model.conf["val_ratio"])
if n_val != 0:
tr_set = s[: int(-1.0 * n_val)]
val_set = s[int(-1.0 * n_val) :]
MoA_p_sets.append(tr_set)
train_index += tr_set
val_index += val_set
else:
MoA_p_sets.append(s)
train_index += s
# Training dataset.
tr_dataset = tf.data.Dataset.from_tensor_slices(train_index)
tr_dataset = tr_dataset.map(
make_input_features
) # , num_parallel_calls=tf.data.experimental.AUTOTUNE)
tr_iter = tr_dataset.as_numpy_iterator()
# Validation dataset.
val_dataset = tf.data.Dataset.from_tensor_slices(val_index)
val_dataset = val_dataset.map(
make_input_features
) # , num_parallel_calls=tf.data.experimental.AUTOTUNE)
val_iter = val_dataset.as_numpy_iterator()
tr_iter = tr_dataset.as_numpy_iterator()
moap = model.model.get_layer("moap")
embed_feature_dicts = []
target_df = target_scored_df.loc[train_index]
for i, d in tqdm(enumerate(tr_iter)):
t, g, c = d
t = np.expand_dims(t, axis=0)
g = np.expand_dims(g, axis=0)
c = np.expand_dims(c, axis=0)
# First layers.
t = moap.embed_treatment_type_0(t)
t = tf.reshape(t, (-1, model.nn_arch["d_input_feature"]))
t = moap.dense_treatment_type_0(t)
t = moap.layer_normalization_0_1(t)
g = moap.layer_normalization_0_2(g)
c = moap.layer_normalization_0_3(c)
# Gene expression.
g_e = moap.encoder_gene_exp_1(g)
x_g = moap.decoder_gene_exp_1(g_e)
x_g = tf.expand_dims(x_g, axis=-1)
x_g = tf.squeeze(x_g, axis=-1)
# Cell type.
c_e = moap.encoder_cell_type_2(c)
x_c = moap.decoder_cell_type_2(c_e)
x_c = moap.dropout_2(x_c)
# Skip-connection autoencoder and final layers.
x = tf.concat([t, g_e, c_e], axis=-1)
for k in range(model.nn_arch["num_sc_ae"]):
x = moap.sc_aes[k](x)
# Final layers.
x = moap.dense_4_1(x)
x = moap.dense_4_2(x)
x = moap.dense_4_3(x)
# Normalize x.
if conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
x1 = moap.dense_4_4(x)
elif conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
x = x / tf.sqrt(tf.reduce_sum(tf.square(x), axis=1, keepdims=True))
x1 = moap.dense_4_4(x)
else:
raise ValueError("loss type is not valid.")
# Get embed_feature_dict.
embed_feature_dict = {}
embed_feature_dict["sig_id"] = -1
embed_feature_dict["embed_feature"] = x.numpy().ravel()
series = target_df.iloc[i]
df = series[series == 1].to_frame()
embed_feature_dict["MoA_classes"] = list(df.index)
embed_feature_dicts.append(embed_feature_dict)
embed_features = np.array([v["embed_feature"] for v in embed_feature_dicts])
tsne_embed_features = TSNE(n_components=2).fit_transform(embed_features)
classes = [v["MoA_classes"] for v in embed_feature_dicts]
for i in tqdm(range(len(classes))):
if len(classes[i]) == 0:
classes[i] = [0]
colors = [v[0] for v in classes]
figure(figsize=(20, 20))
x = []
y = []
c = []
for i in range(len(tsne_embed_features)):
for v in classes[i]:
x.append(tsne_embed_features[i, 0])
y.append(tsne_embed_features[i, 1])
c.append(cls_colors[v])
scatter(x, y, c=c, alpha=1.0)
title("Drug features clustering for training data")
grid()
moa_name_colors_fig = plot_colortable(moa_name_colors, "MoA Colors")
# #### Clustering for validation data
val_iter = val_dataset.as_numpy_iterator()
moap = model.model.get_layer("moap")
embed_feature_dicts = []
target_df = target_scored_df.loc[val_index]
for i, d in tqdm(enumerate(val_iter)):
t, g, c = d
id_ = input_df["sig_id"].iloc[i]
t = np.expand_dims(t, axis=0)
g = np.expand_dims(g, axis=0)
c = np.expand_dims(c, axis=0)
# First layers.
t = moap.embed_treatment_type_0(t)
t = tf.reshape(t, (-1, model.nn_arch["d_input_feature"]))
t = moap.dense_treatment_type_0(t)
t = moap.layer_normalization_0_1(t)
g = moap.layer_normalization_0_2(g)
c = moap.layer_normalization_0_3(c)
# Gene expression.
g_e = moap.encoder_gene_exp_1(g)
x_g = moap.decoder_gene_exp_1(g_e)
x_g = tf.expand_dims(x_g, axis=-1)
x_g = tf.squeeze(x_g, axis=-1)
# Cell type.
c_e = moap.encoder_cell_type_2(c)
x_c = moap.decoder_cell_type_2(c_e)
x_c = moap.dropout_2(x_c)
# Skip-connection autoencoder and final layers.
x = tf.concat([t, g_e, c_e], axis=-1)
for i in range(model.nn_arch["num_sc_ae"]):
x = moap.sc_aes[i](x)
# Final layers.
x = moap.dense_4_1(x)
x = moap.dense_4_2(x)
x = moap.dense_4_3(x)
# Normalize x.
if conf["loss_type"] == LOSS_TYPE_MULTI_LABEL:
x1 = moap.dense_4_4(x)
elif conf["loss_type"] == LOSS_TYPE_ADDITIVE_ANGULAR_MARGIN:
x = x / tf.sqrt(tf.reduce_sum(tf.square(x), axis=1, keepdims=True))
x1 = moap.dense_4_4(x)
else:
raise ValueError("loss type is not valid.")
# Get embed_feature_dict.
embed_feature_dict = {}
embed_feature_dict["sig_id"] = id_
embed_feature_dict["embed_feature"] = x.numpy().ravel()
series = target_df.iloc[i]
df = series[series == 1].to_frame()
embed_feature_dict["MoA_classes"] = list(df.index)
embed_feature_dicts.append(embed_feature_dict)
embed_features = np.array([v["embed_feature"] for v in embed_feature_dicts])
tsne_embed_features = TSNE(n_components=2).fit_transform(embed_features)
classes = [v["MoA_classes"] for v in embed_feature_dicts]
for i in tqdm(range(len(classes))):
if len(classes[i]) == 0:
classes[i] = [0]
colors = [v[0] for v in classes]
figure(figsize=(20, 20))
x = []
y = []
c = []
for i in range(len(tsne_embed_features)):
for v in classes[i]:
x.append(tsne_embed_features[i, 0])
y.append(tsne_embed_features[i, 1])
c.append(cls_colors[v])
scatter(x, y, c=c, alpha=1.0)
title("Drug features clustering for validation data")
grid()
moa_name_colors_fig = plot_colortable(moa_name_colors, "MoA Colors")
| false | 0 | 23,264 | 0 | 23,264 | 23,264 |
||
69605251
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
print(dirname, len(filenames))
# You can write up to 20GB to the current directory /kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
import torchvision
from torch.utils.data import TensorDataset, DataLoader
from torch.optim import lr_scheduler
from torchvision import datasets, models, transforms
device = "cuda" if torch.cuda.is_available() else "cpu"
print("Using device", device)
from cv2 import cv2
from PIL import Image
import matplotlib.pyplot as plt
import time
import copy
import random
from tqdm import tqdm
random_seed = 42
torch.manual_seed(random_seed)
torch.cuda.manual_seed(random_seed)
torch.cuda.manual_seed_all(random_seed) # if use multi-GPU
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
np.random.seed(random_seed)
random.seed(random_seed)
train_path = "/kaggle/input/aptos2019-blindness-detection/train_images/"
test_path = "/kaggle/input/aptos2019-blindness-detection/test_images/"
train_data = pd.read_csv("../input/aptos2019-blindness-detection/train.csv")
test_data = pd.read_csv("../input/aptos2019-blindness-detection/test.csv")
print(train_data.shape)
print(train_data.head(10))
print(test_data.shape)
print(test_data.head(10))
train_data["diagnosis"].value_counts().plot(kind="pie")
# # grayscale
def crop_image1(img, tol=7):
# img is image data
# tol is tolerance
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
def crop_image(img, tol=7):
if img.ndim == 2:
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
elif img.ndim == 3:
h, w, _ = img.shape
# print(h,w)
img1 = cv2.resize(crop_image1(img[:, :, 0]), (w, h))
img2 = cv2.resize(crop_image1(img[:, :, 1]), (w, h))
img3 = cv2.resize(crop_image1(img[:, :, 2]), (w, h))
# print(img1.shape,img2.shape,img3.shape)
img[:, :, 0] = img1
img[:, :, 1] = img2
img[:, :, 2] = img3
return img
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_data["diagnosis"].unique()):
for i, (idx, row) in enumerate(
train_data.loc[train_data["diagnosis"] == class_id].sample(5).iterrows()
):
ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[])
img = cv2.imread(train_path + "/" + row["id_code"] + ".png")
img = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
img = crop_image(img)
img = cv2.resize(img, (300, 300))
ax.imshow(img, cmap="gray")
ax.set_title("Label: %d-%d-%s" % (class_id, idx, row["id_code"]))
# # ben color
def load_ben_color(path, sigmaX=10):
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = crop_image(image)
image = cv2.resize(image, (300, 300))
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128)
return image
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_data["diagnosis"].unique()):
for i, (idx, row) in enumerate(
train_data.loc[train_data["diagnosis"] == class_id].sample(5).iterrows()
):
ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[])
path = (
f"../input/aptos2019-blindness-detection/train_images/{row['id_code']}.png"
)
image = load_ben_color(path, sigmaX=30)
plt.imshow(image)
ax.set_title("%d-%d-%s" % (class_id, idx, row["id_code"]))
def crop_image1(img, tol=7):
# img is image data
# tol is tolerance
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
def crop_image(img, tol=7):
if img.ndim == 2:
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
elif img.ndim == 3:
h, w, _ = img.shape
# print(h,w)
img1 = cv2.resize(crop_image1(img[:, :, 0]), (w, h))
img2 = cv2.resize(crop_image1(img[:, :, 1]), (w, h))
img3 = cv2.resize(crop_image1(img[:, :, 2]), (w, h))
# print(img1.shape,img2.shape,img3.shape)
img[:, :, 0] = img1
img[:, :, 1] = img2
img[:, :, 2] = img3
return img
def circle_crop(path):
img = cv2.imread(path)
img = crop_image(img)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(image, sigmaX=10):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128)
image = cv2.addWeighted(image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128)
return image
def change_gray_color(image, sigmaX=10):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128)
image = cv2.addWeighted(image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128)
return image
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_data["diagnosis"].unique()):
for i, (idx, row) in enumerate(
train_data.loc[train_data["diagnosis"] == class_id].sample(5).iterrows()
):
ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[])
path = (
f"../input/aptos2019-blindness-detection/train_images/{row['id_code']}.png"
)
image = circle_crop(path)
# image = change_gray_color(image, 40)
# image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = change_ben_color(image, 30)
image = cv2.resize(image, (300, 300))
plt.imshow(image, cmap="gray")
ax.set_title("%d-%d-%s" % (class_id, idx, row["id_code"]))
train_data["id_code"], val_set, train_data["diagnosis"], val_label = train_test_split(
train_data["id_code"], train_data["diagnosis"], test_size=0.9, random_state=42
)
train_data = train_data.dropna()
train_data = train_data.reset_index(drop=True)
train_data.head(10)
# # augmentation 1
# # transforms
transform = torchvision.transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(), # 0.5
transforms.RandomRotation(10),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
# # 1. 색 변환 x
from PIL import Image
class Dataset1:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset1(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
x, y = train_dataset.__getitem__(0)
print(x.shape)
print(y.shape)
images, labels = next(iter(dataloader))
images.shape, labels.shape
model_ft = models.resnet18(pretrained=True)
params = list(model_ft.parameters())
len(params), params[0].size()
num_features = model_ft.fc.in_features
model_ft.fc = nn.Linear(num_features, 1)
model_ft = model_ft.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model_ft.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model_ft.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model_ft(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model_ft.state_dict(), "model1.bin")
# # 2. change ben color
from PIL import Image
class Dataset2:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(self, image, sigmaX=30):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def change_gray_color(self, image, sigmaX=40):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
img = self.change_ben_color(img)
# img = self.change_gray_color(img)
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset2(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model2 = models.resnet18(pretrained=True)
num_features = model2.fc.in_features
model2.fc = nn.Linear(num_features, 1)
model2 = model2.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model2.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model2.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model2(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model2.state_dict(), "model2.bin")
# # 3. change gray color
from PIL import Image
class Dataset3:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(self, image, sigmaX=30):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def change_gray_color(self, image, sigmaX=40):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
# img = self.change_ben_color(img)
img = self.change_gray_color(img)
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset3(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model3 = models.resnet18(pretrained=True)
num_features = model3.fc.in_features
model3.fc = nn.Linear(num_features, 1)
model3 = model3.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model3.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model3.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model3(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model3.state_dict(), "model3.bin")
# # augmentation 2
# # 1. albumentation
import albumentations
import albumentations.pytorch
"""
transform = torchvision.transforms.Compose([
transforms.Resize((224,224)),
transforms.RandomHorizontalFlip(), #0.5
transforms.RandomRotation(10),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])
])
"""
albumentations_transform = albumentations.Compose(
[
albumentations.Resize(224, 224),
albumentations.HorizontalFlip(), # Same with transforms.RandomHorizontalFlip()
albumentations.RandomRotate90(),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset4:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(self, image, sigmaX=30):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def change_gray_color(self, image, sigmaX=40):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
# img = self.change_ben_color(img)
# img = self.change_gray_color(img)
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset4(train_data, train_path, albumentations_transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
images, labels = next(iter(dataloader))
print(images.shape, labels.shape)
params = list(model4.parameters())
len(params), params[0].size()
model4 = models.resnet18(pretrained=True)
num_features = model4.fc.in_features
model4.fc = nn.Linear(num_features, 1)
model4 = model4.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model4.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model4.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model4(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model4.state_dict(), "model4.bin")
# # 2. albumentation One of
# 1.ResizedRandomCrop
albumentations_transform_oneof = albumentations.Compose(
[
albumentations.Resize(256, 256),
albumentations.RandomCrop(224, 224),
albumentations.OneOf(
[
albumentations.HorizontalFlip(p=1),
albumentations.RandomRotate90(p=1),
albumentations.VerticalFlip(p=1),
],
p=1,
),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset5:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(self, image, sigmaX=30):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def change_gray_color(self, image, sigmaX=40):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
# img = self.change_ben_color(img)
# img = self.change_gray_color(img)
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset5(train_data, train_path, albumentations_transform_oneof)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model5 = models.resnet18(pretrained=True)
num_features = model5.fc.in_features
model5.fc = nn.Linear(num_features, 1)
model5 = model5.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model5.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model5.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model5(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model5.state_dict(), "model5.bin")
# 2. just resize
albumentations_transform_resize = albumentations.Compose(
[
albumentations.Resize(224, 224),
albumentations.OneOf(
[
albumentations.HorizontalFlip(p=1),
albumentations.RandomRotate90(p=1),
albumentations.VerticalFlip(p=1),
]
),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset6:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset6(train_data, train_path, albumentations_transform_resize)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model6 = models.resnet18(pretrained=True)
num_features = model6.fc.in_features
model6.fc = nn.Linear(num_features, 1)
model6 = model6.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model6.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model6.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model6(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model6.state_dict(), "model6.bin")
# 3. p =1
albumentations_transform_ = albumentations.Compose(
[
albumentations.Resize(224, 224),
albumentations.OneOf(
[
albumentations.HorizontalFlip(p=1),
albumentations.RandomRotate90(p=1),
albumentations.VerticalFlip(p=1),
],
p=1,
),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset7:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset7(train_data, train_path, albumentations_transform_)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model7 = models.resnet18(pretrained=True)
num_features = model7.fc.in_features
model7.fc = nn.Linear(num_features, 1)
model7 = model7.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model7.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model7.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model7(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model7.state_dict(), "model7.bin")
# 4. not augmented
albumentations_transform__ = albumentations.Compose(
[
albumentations.Resize(224, 224),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset8:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset8(train_data, train_path, albumentations_transform__)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model8 = models.resnet18(pretrained=True)
num_features = model8.fc.in_features
model8.fc = nn.Linear(num_features, 1)
model8 = model8.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model8.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model8.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model8(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model8.state_dict(), "model8.bin")
# # augmentation 3
# # randaugment
# - batch를 추출할 때마다 여러 Augmentation 옵션들 중에서 random하게 추출해서 적용
# - 전체 transform 중에 몇 개씩 뽑을 지(N)와 Augmentation의 강도를 어느 정도로 줄지(M)이 hyper parameter
from randaugment import RandAugment
transform = torchvision.transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(), # 0.5
RandAugment(),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
class Dataset9:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset9(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model9 = models.resnet18(pretrained=True)
num_features = model9.fc.in_features
model9.fc = nn.Linear(num_features, 1)
model9 = model9.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model9.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model9.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model9(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model9.state_dict(), "model9.bin")
# # augmentation 4
# # uniform augmentation
# - search 없이 random하게 augmentation을 확률적으로 적용
#
from UniformAugment import UniformAugment
transform = transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
# Add UniformAugment with num_ops hyperparameter (num_ops=2 is optimal)
transform.transforms.insert(0, UniformAugment())
class Dataset10:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset10(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model10 = models.resnet18(pretrained=True)
num_features = model10.fc.in_features
model10.fc = nn.Linear(num_features, 1)
model10 = model10.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model10.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model10.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model10(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model10.state_dict(), "model10.bin")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605251.ipynb
| null | null |
[{"Id": 69605251, "ScriptId": 18902370, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7933389, "CreationDate": "08/02/2021 05:34:44", "VersionNumber": 6.0, "Title": "cmd - Blindness Detection", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 1135.0, "LinesInsertedFromPrevious": 11.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1124.0, "LinesInsertedFromFork": 1018.0, "LinesDeletedFromFork": 23.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 117.0, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
print(dirname, len(filenames))
# You can write up to 20GB to the current directory /kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
import torchvision
from torch.utils.data import TensorDataset, DataLoader
from torch.optim import lr_scheduler
from torchvision import datasets, models, transforms
device = "cuda" if torch.cuda.is_available() else "cpu"
print("Using device", device)
from cv2 import cv2
from PIL import Image
import matplotlib.pyplot as plt
import time
import copy
import random
from tqdm import tqdm
random_seed = 42
torch.manual_seed(random_seed)
torch.cuda.manual_seed(random_seed)
torch.cuda.manual_seed_all(random_seed) # if use multi-GPU
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
np.random.seed(random_seed)
random.seed(random_seed)
train_path = "/kaggle/input/aptos2019-blindness-detection/train_images/"
test_path = "/kaggle/input/aptos2019-blindness-detection/test_images/"
train_data = pd.read_csv("../input/aptos2019-blindness-detection/train.csv")
test_data = pd.read_csv("../input/aptos2019-blindness-detection/test.csv")
print(train_data.shape)
print(train_data.head(10))
print(test_data.shape)
print(test_data.head(10))
train_data["diagnosis"].value_counts().plot(kind="pie")
# # grayscale
def crop_image1(img, tol=7):
# img is image data
# tol is tolerance
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
def crop_image(img, tol=7):
if img.ndim == 2:
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
elif img.ndim == 3:
h, w, _ = img.shape
# print(h,w)
img1 = cv2.resize(crop_image1(img[:, :, 0]), (w, h))
img2 = cv2.resize(crop_image1(img[:, :, 1]), (w, h))
img3 = cv2.resize(crop_image1(img[:, :, 2]), (w, h))
# print(img1.shape,img2.shape,img3.shape)
img[:, :, 0] = img1
img[:, :, 1] = img2
img[:, :, 2] = img3
return img
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_data["diagnosis"].unique()):
for i, (idx, row) in enumerate(
train_data.loc[train_data["diagnosis"] == class_id].sample(5).iterrows()
):
ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[])
img = cv2.imread(train_path + "/" + row["id_code"] + ".png")
img = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
img = crop_image(img)
img = cv2.resize(img, (300, 300))
ax.imshow(img, cmap="gray")
ax.set_title("Label: %d-%d-%s" % (class_id, idx, row["id_code"]))
# # ben color
def load_ben_color(path, sigmaX=10):
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = crop_image(image)
image = cv2.resize(image, (300, 300))
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128)
return image
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_data["diagnosis"].unique()):
for i, (idx, row) in enumerate(
train_data.loc[train_data["diagnosis"] == class_id].sample(5).iterrows()
):
ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[])
path = (
f"../input/aptos2019-blindness-detection/train_images/{row['id_code']}.png"
)
image = load_ben_color(path, sigmaX=30)
plt.imshow(image)
ax.set_title("%d-%d-%s" % (class_id, idx, row["id_code"]))
def crop_image1(img, tol=7):
# img is image data
# tol is tolerance
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
def crop_image(img, tol=7):
if img.ndim == 2:
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
elif img.ndim == 3:
h, w, _ = img.shape
# print(h,w)
img1 = cv2.resize(crop_image1(img[:, :, 0]), (w, h))
img2 = cv2.resize(crop_image1(img[:, :, 1]), (w, h))
img3 = cv2.resize(crop_image1(img[:, :, 2]), (w, h))
# print(img1.shape,img2.shape,img3.shape)
img[:, :, 0] = img1
img[:, :, 1] = img2
img[:, :, 2] = img3
return img
def circle_crop(path):
img = cv2.imread(path)
img = crop_image(img)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(image, sigmaX=10):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128)
image = cv2.addWeighted(image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128)
return image
def change_gray_color(image, sigmaX=10):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128)
image = cv2.addWeighted(image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128)
return image
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_data["diagnosis"].unique()):
for i, (idx, row) in enumerate(
train_data.loc[train_data["diagnosis"] == class_id].sample(5).iterrows()
):
ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[])
path = (
f"../input/aptos2019-blindness-detection/train_images/{row['id_code']}.png"
)
image = circle_crop(path)
# image = change_gray_color(image, 40)
# image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = change_ben_color(image, 30)
image = cv2.resize(image, (300, 300))
plt.imshow(image, cmap="gray")
ax.set_title("%d-%d-%s" % (class_id, idx, row["id_code"]))
train_data["id_code"], val_set, train_data["diagnosis"], val_label = train_test_split(
train_data["id_code"], train_data["diagnosis"], test_size=0.9, random_state=42
)
train_data = train_data.dropna()
train_data = train_data.reset_index(drop=True)
train_data.head(10)
# # augmentation 1
# # transforms
transform = torchvision.transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(), # 0.5
transforms.RandomRotation(10),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
# # 1. 색 변환 x
from PIL import Image
class Dataset1:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset1(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
x, y = train_dataset.__getitem__(0)
print(x.shape)
print(y.shape)
images, labels = next(iter(dataloader))
images.shape, labels.shape
model_ft = models.resnet18(pretrained=True)
params = list(model_ft.parameters())
len(params), params[0].size()
num_features = model_ft.fc.in_features
model_ft.fc = nn.Linear(num_features, 1)
model_ft = model_ft.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model_ft.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model_ft.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model_ft(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model_ft.state_dict(), "model1.bin")
# # 2. change ben color
from PIL import Image
class Dataset2:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(self, image, sigmaX=30):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def change_gray_color(self, image, sigmaX=40):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
img = self.change_ben_color(img)
# img = self.change_gray_color(img)
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset2(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model2 = models.resnet18(pretrained=True)
num_features = model2.fc.in_features
model2.fc = nn.Linear(num_features, 1)
model2 = model2.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model2.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model2.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model2(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model2.state_dict(), "model2.bin")
# # 3. change gray color
from PIL import Image
class Dataset3:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(self, image, sigmaX=30):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def change_gray_color(self, image, sigmaX=40):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
# img = self.change_ben_color(img)
img = self.change_gray_color(img)
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset3(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model3 = models.resnet18(pretrained=True)
num_features = model3.fc.in_features
model3.fc = nn.Linear(num_features, 1)
model3 = model3.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model3.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model3.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model3(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model3.state_dict(), "model3.bin")
# # augmentation 2
# # 1. albumentation
import albumentations
import albumentations.pytorch
"""
transform = torchvision.transforms.Compose([
transforms.Resize((224,224)),
transforms.RandomHorizontalFlip(), #0.5
transforms.RandomRotation(10),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])
])
"""
albumentations_transform = albumentations.Compose(
[
albumentations.Resize(224, 224),
albumentations.HorizontalFlip(), # Same with transforms.RandomHorizontalFlip()
albumentations.RandomRotate90(),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset4:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(self, image, sigmaX=30):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def change_gray_color(self, image, sigmaX=40):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
# img = self.change_ben_color(img)
# img = self.change_gray_color(img)
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset4(train_data, train_path, albumentations_transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
images, labels = next(iter(dataloader))
print(images.shape, labels.shape)
params = list(model4.parameters())
len(params), params[0].size()
model4 = models.resnet18(pretrained=True)
num_features = model4.fc.in_features
model4.fc = nn.Linear(num_features, 1)
model4 = model4.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model4.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model4.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model4(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model4.state_dict(), "model4.bin")
# # 2. albumentation One of
# 1.ResizedRandomCrop
albumentations_transform_oneof = albumentations.Compose(
[
albumentations.Resize(256, 256),
albumentations.RandomCrop(224, 224),
albumentations.OneOf(
[
albumentations.HorizontalFlip(p=1),
albumentations.RandomRotate90(p=1),
albumentations.VerticalFlip(p=1),
],
p=1,
),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset5:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def change_ben_color(self, image, sigmaX=30):
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def change_gray_color(self, image, sigmaX=40):
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.addWeighted(
image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128
)
image = cv2.addWeighted(
image, -4, cv2.GaussianBlur(image, (0, 0), sigmaX), 4, 128
)
return image
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
# img = self.change_ben_color(img)
# img = self.change_gray_color(img)
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset5(train_data, train_path, albumentations_transform_oneof)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model5 = models.resnet18(pretrained=True)
num_features = model5.fc.in_features
model5.fc = nn.Linear(num_features, 1)
model5 = model5.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model5.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model5.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model5(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model5.state_dict(), "model5.bin")
# 2. just resize
albumentations_transform_resize = albumentations.Compose(
[
albumentations.Resize(224, 224),
albumentations.OneOf(
[
albumentations.HorizontalFlip(p=1),
albumentations.RandomRotate90(p=1),
albumentations.VerticalFlip(p=1),
]
),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset6:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset6(train_data, train_path, albumentations_transform_resize)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model6 = models.resnet18(pretrained=True)
num_features = model6.fc.in_features
model6.fc = nn.Linear(num_features, 1)
model6 = model6.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model6.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model6.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model6(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model6.state_dict(), "model6.bin")
# 3. p =1
albumentations_transform_ = albumentations.Compose(
[
albumentations.Resize(224, 224),
albumentations.OneOf(
[
albumentations.HorizontalFlip(p=1),
albumentations.RandomRotate90(p=1),
albumentations.VerticalFlip(p=1),
],
p=1,
),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset7:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset7(train_data, train_path, albumentations_transform_)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model7 = models.resnet18(pretrained=True)
num_features = model7.fc.in_features
model7.fc = nn.Linear(num_features, 1)
model7 = model7.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model7.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model7.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model7(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model7.state_dict(), "model7.bin")
# 4. not augmented
albumentations_transform__ = albumentations.Compose(
[
albumentations.Resize(224, 224),
albumentations.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
albumentations.pytorch.transforms.ToTensorV2(),
]
)
class Dataset8:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
x = self.transform(image=img)["image"] # dictionary
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset8(train_data, train_path, albumentations_transform__)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model8 = models.resnet18(pretrained=True)
num_features = model8.fc.in_features
model8.fc = nn.Linear(num_features, 1)
model8 = model8.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model8.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model8.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model8(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model8.state_dict(), "model8.bin")
# # augmentation 3
# # randaugment
# - batch를 추출할 때마다 여러 Augmentation 옵션들 중에서 random하게 추출해서 적용
# - 전체 transform 중에 몇 개씩 뽑을 지(N)와 Augmentation의 강도를 어느 정도로 줄지(M)이 hyper parameter
from randaugment import RandAugment
transform = torchvision.transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(), # 0.5
RandAugment(),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
class Dataset9:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset9(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model9 = models.resnet18(pretrained=True)
num_features = model9.fc.in_features
model9.fc = nn.Linear(num_features, 1)
model9 = model9.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model9.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model9.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model9(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model9.state_dict(), "model9.bin")
# # augmentation 4
# # uniform augmentation
# - search 없이 random하게 augmentation을 확률적으로 적용
#
from UniformAugment import UniformAugment
transform = transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
# Add UniformAugment with num_ops hyperparameter (num_ops=2 is optimal)
transform.transforms.insert(0, UniformAugment())
class Dataset10:
def __init__(self, data, root, transform):
self.files = list(root + data["id_code"] + ".png")
self.targets = data["diagnosis"]
self.transform = transform
def __len__(self):
return len(self.files)
def circle_crop(self, path):
img = cv2.imread(path)
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def __getitem__(self, idx):
img = self.circle_crop(self.files[idx])
img = Image.fromarray(img).convert("RGB")
x = self.transform(img)
y = torch.tensor(self.targets[idx]).unsqueeze(0).float()
return x, y
train_dataset = Dataset10(train_data, train_path, transform)
dataloader = DataLoader(train_dataset, batch_size=64, shuffle=True)
model10 = models.resnet18(pretrained=True)
num_features = model10.fc.in_features
model10.fc = nn.Linear(num_features, 1)
model10 = model10.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(lr=1e-4, params=model10.parameters())
scheduler = lr_scheduler.StepLR(optimizer, step_size=10)
since = time.time()
criterion = torch.nn.MSELoss()
num_epochs = 15
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch, num_epochs - 1))
print("-" * 10)
model10.train()
running_loss = 0.0
tk0 = tqdm(dataloader, total=int(len(dataloader)))
counter = 0
for bi, (d, t) in enumerate(tk0):
inputs = d
labels = t
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.float)
optimizer.zero_grad()
with torch.set_grad_enabled(True):
outputs = model10(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
counter += 1
tk0.set_postfix(loss=(running_loss / (counter * dataloader.batch_size)))
epoch_loss = running_loss / len(dataloader)
print("Training Loss: {:.4f}".format(epoch_loss))
scheduler.step()
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_elapsed // 60, time_elapsed % 60)
)
torch.save(model10.state_dict(), "model10.bin")
| false | 0 | 12,746 | 0 | 12,746 | 12,746 |
||
69605184
|
<jupyter_start><jupyter_text>COVID-19 data from John Hopkins University
This is a daily updating version of [COVID-19 Data Repository](https://github.com/CSSEGISandData/COVID-19) by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). The data updates every day at 6am UTC, which updates just after the raw JHU data typically updates.
I'm making it available in both a raw form (files with the prefix RAW) and convenient form (files prefixed with CONVENIENT).
The data covers:
- confirmed cases and deaths on a country level
- confirmed cases and deaths by US county
- some metadata that's available in the raw JHU data
The RAW version is exactly as it's distributed in the original dataset.
The CONVENIENT version is aiming to be easier to analyze. The data is organized by column rather than by row. The metadata is stripped out into a separate file. And it converted to daily change rather than cumulative totals.
If you find any issues in the data, then you can share them in this [discussion thread](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/discussion/195628). I will attempt to address the most upvoted issues.
If you have any requests for changing or enriching this data, please add them on this [discussion thread](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/discussion/195984). Again, I will attempt to address the most upvoted requests.
I have a notebook that updates just after each data dump updates, giving a brief overview of the [latest data](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/notebooks). It's also a useful reference if you want to see how to read the CONVENIENT data into a pandas DataFrame.
Kaggle dataset identifier: covid19-data-from-john-hopkins-university
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
pd.set_option("max_rows", 100)
# # Negative values issue
# The RAW files show the cumulative number of cases. For the CONVENIENT files, I take am looking at the difference. But there are quite a lot of days with negative case counts and negative death counts. This issue is more severe for the us than global files. Please comment on this notebook if you have ideas for how to address this issue.
df_dict = {}
table_list = [
"global_confirmed_cases",
"global_deaths",
"us_confirmed_cases",
"us_deaths",
]
print("Number of negative values:")
for table in table_list:
df_dict[table] = df_convenient_global_cases = pd.read_csv(
f"/kaggle/input/covid19-data-from-john-hopkins-university/CONVENIENT_{table}.csv",
header=[0, 1],
index_col=0,
)
print(
f'{table.replace("_"," ")}: {(df_dict[table] < 0).sum().sum()} out of {df_dict[table].count().sum()}'
)
# ## Negative values are most common for unassigned counties
# However unassigned counties only make up ~11% of the problem.
print((df_dict["us_confirmed_cases"] < 0).sum().sort_values(ascending=False)[:10])
print(
f"{ (df_dict['us_confirmed_cases'].xs('Unassigned', level=1,axis=1, drop_level=False)<0).sum().sum() } out of {(df_dict['us_confirmed_cases']<0).sum().sum()} are in Unassigned counties"
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605184.ipynb
|
covid19-data-from-john-hopkins-university
|
antgoldbloom
|
[{"Id": 69605184, "ScriptId": 12687157, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 368, "CreationDate": "08/02/2021 05:33:27", "VersionNumber": 271.0, "Title": "ISSUE: negative values in CONVENIENT files", "EvaluationDate": "08/02/2021", "IsChange": false, "TotalLines": 30.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 30.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93000588, "KernelVersionId": 69605184, "SourceDatasetVersionId": 2490132}]
|
[{"Id": 2490132, "DatasetId": 951970, "DatasourceVersionId": 2532702, "CreatorUserId": 368, "LicenseName": "Attribution 4.0 International (CC BY 4.0)", "CreationDate": "08/02/2021 05:03:46", "VersionNumber": 290.0, "Title": "COVID-19 data from John Hopkins University", "Slug": "covid19-data-from-john-hopkins-university", "Subtitle": "Updated daily at 6am UTC in both raw and convenient form", "Description": "This is a daily updating version of [COVID-19 Data Repository](https://github.com/CSSEGISandData/COVID-19) by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). The data updates every day at 6am UTC, which updates just after the raw JHU data typically updates. \n\nI'm making it available in both a raw form (files with the prefix RAW) and convenient form (files prefixed with CONVENIENT). \n\nThe data covers:\n- confirmed cases and deaths on a country level \n- confirmed cases and deaths by US county\n- some metadata that's available in the raw JHU data\n\nThe RAW version is exactly as it's distributed in the original dataset.\n\nThe CONVENIENT version is aiming to be easier to analyze. The data is organized by column rather than by row. The metadata is stripped out into a separate file. And it converted to daily change rather than cumulative totals. \n\nIf you find any issues in the data, then you can share them in this [discussion thread](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/discussion/195628). I will attempt to address the most upvoted issues.\n\nIf you have any requests for changing or enriching this data, please add them on this [discussion thread](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/discussion/195984). Again, I will attempt to address the most upvoted requests. \n\nI have a notebook that updates just after each data dump updates, giving a brief overview of the [latest data](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/notebooks). It's also a useful reference if you want to see how to read the CONVENIENT data into a pandas DataFrame.", "VersionNotes": "2021-08-02 05:03:29 automated update", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 951970, "CreatorUserId": 368, "OwnerUserId": 368.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5651576.0, "CurrentDatasourceVersionId": 5726942.0, "ForumId": 968155, "Type": 2, "CreationDate": "11/02/2020 14:29:16", "LastActivityDate": "11/02/2020", "TotalViews": 119682, "TotalDownloads": 19634, "TotalVotes": 391, "TotalKernels": 60}]
|
[{"Id": 368, "UserName": "antgoldbloom", "DisplayName": "Anthony Goldbloom", "RegisterDate": "01/20/2010", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
pd.set_option("max_rows", 100)
# # Negative values issue
# The RAW files show the cumulative number of cases. For the CONVENIENT files, I take am looking at the difference. But there are quite a lot of days with negative case counts and negative death counts. This issue is more severe for the us than global files. Please comment on this notebook if you have ideas for how to address this issue.
df_dict = {}
table_list = [
"global_confirmed_cases",
"global_deaths",
"us_confirmed_cases",
"us_deaths",
]
print("Number of negative values:")
for table in table_list:
df_dict[table] = df_convenient_global_cases = pd.read_csv(
f"/kaggle/input/covid19-data-from-john-hopkins-university/CONVENIENT_{table}.csv",
header=[0, 1],
index_col=0,
)
print(
f'{table.replace("_"," ")}: {(df_dict[table] < 0).sum().sum()} out of {df_dict[table].count().sum()}'
)
# ## Negative values are most common for unassigned counties
# However unassigned counties only make up ~11% of the problem.
print((df_dict["us_confirmed_cases"] < 0).sum().sort_values(ascending=False)[:10])
print(
f"{ (df_dict['us_confirmed_cases'].xs('Unassigned', level=1,axis=1, drop_level=False)<0).sum().sum() } out of {(df_dict['us_confirmed_cases']<0).sum().sum()} are in Unassigned counties"
)
| false | 0 | 443 | 0 | 937 | 443 |
||
69605229
|
# # Predicting House Prices XGBoost + GBM Models
# **Bugra Sebati E.** - **July 2021**
# ## Introduction
# For this competiton, we are given a data set of 1460 homes, each with a few dozen features of types: float, integer, and categorical. We are tasked with building a regression model to estimate a home's sale price. Total number of attributes equals 81, of which 36 is quantitative, 43 categorical + Id and SalePrice.
# **What you can find on this notebook?**
# * Understanding the data
# * Exploratory Data Analysis
# * Data Preprocessing
# * PCA Trial
# * GBM and XGBoost Models
# * Submission
# 
# ### Lets meet variables
# * **SalePrice** : The property's sale price in dollars. This is target variable for predict
# * **MSSubClass**: The building class
# * **MSZoning**: The general zoning classification
# * **LotFrontage**: Linear feet of street connected to property
# * **LotArea**: Lot size in square feet
# * **Street**: Type of road access
# * **Alley**: Type of alley access
# * **LotShape**: General shape of property
# * **LandContour**: Flatness of the property
# * **Utilities**: Type of utilities available
# * **LotConfig**: Lot configuration
# * **LandSlope**: Slope of property
# * **Neighborhood**: Physical locations within Ames city limits
# * **Condition1**: Proximity to main road or railroad
# * **Condition2**: Proximity to main road or railroad (if a second is present)
# * **BldgType**: Type of dwelling
# * **HouseStyle**: Style of dwelling
# * **OverallQual**: Overall material and finish quality
# * **OverallCond**: Overall condition rating
# * **YearBuilt**: Original construction date
# * **YearRemodAdd**: Remodel date
# * **RoofStyle**: Type of roof
# * **RoofMatl**: Roof material
# * **Exterior1st**: Exterior covering on house
# * **Exterior2nd**: Exterior covering on house (if more than one material)
# * **MasVnrType**: Masonry veneer type
# * **MasVnrArea**: Masonry veneer area in square feet
# * **ExterQual**: Exterior material quality
# * **ExterCond**: Present condition of the material on the exterior
# * **Foundation**: Type of foundation
# * **BsmtQual**: Height of the basement
# * **BsmtCond**: General condition of the basement
# * **BsmtExposure**: Walkout or garden level basement walls
# * **BsmtFinType1**: Quality of basement finished area
# * **BsmtFinSF1**: Type 1 finished square feet
# * **BsmtFinType2**: Quality of second finished area (if present)
# * **BsmtFinSF2**: Type 2 finished square feet
# * **BsmtUnfSF**: Unfinished square feet of basement area
# * **TotalBsmtSF**: Total square feet of basement area
# * **Heating**: Type of heating
# * **HeatingQC**: Heating quality and condition
# * **CentralAir**: Central air conditioning
# * **Electrical**: Electrical system
# * **1stFlrSF**: First Floor square feet
# * **2ndFlrSF**: Second floor square feet
# * **LowQualFinSF**: Low quality finished square feet (all floors)
# * **GrLivArea**: Above grade (ground) living area square feet
# * **BsmtFullBath**: Basement full bathrooms
# * **BsmtHalfBath**: Basement half bathrooms
# * **FullBath**: Full bathrooms above grade
# * **HalfBath**: Half baths above grade
# * **Bedroom**: Number of bedrooms above basement level
# * **Kitchen**: Number of kitchens
# * **KitchenQual**: Kitchen quality
# * **TotRmsAbvGrd**: Total rooms above grade (does not include bathrooms)
# * **Functional**: Home functionality rating
# * **Fireplaces**: Number of fireplaces
# * **FireplaceQu**: Fireplace quality
# * **GarageType**: Garage location
# * **GarageYrBlt**: Year garage was built
# * **GarageFinish**: Interior finish of the garage
# * **GarageCars**: Size of garage in car capacity
# * **GarageArea**: Size of garage in square feet
# * **GarageQual**: Garage quality
# * **GarageCond**: Garage condition
# * **PavedDrive**: Paved driveway
# * **WoodDeckSF**: Wood deck area in square feet
# * **OpenPorchSF**: Open porch area in square feet
# * **EnclosedPorch**: Enclosed porch area in square feet
# * **3SsnPorch**: Three season porch area in square feet
# * **ScreenPorch**: Screen porch area in square feet
# * **PoolArea**: Pool area in square feet
# * **PoolQC**: Pool quality
# * **Fence**: Fence quality
# * **MiscFeature**: Miscellaneous feature not covered in other categories
# * **MiscVal**: Value of miscellaneous feature
# * **MoSold**: Month Sold
# * **YrSold**: Year Sold
# * **SaleType**: Type of sale
# * **SaleCondition**: Condition of sale
# #### Since we learn variables, we can start now...
# If you like this notebook,dont forget to upvote :) **Thanks !**
#### IMPORT LIBRARIES
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import StandardScaler
from scipy.stats import skew
from scipy.special import boxcox1p
from sklearn.decomposition import PCA
from sklearn.model_selection import KFold, cross_val_score
from sklearn.metrics import mean_squared_error
import xgboost as xgb
from sklearn.ensemble import GradientBoostingRegressor
import warnings
warnings.filterwarnings("ignore")
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
traindf = train.copy()
testdf = test.copy()
train.head()
test.head()
#### I like colors :-9
trainshape = ("Train Data:", train.shape[0], "obs, and", train.shape[1], "features")
print("\033[95m {}\033[00m".format(trainshape))
testshape = ("Test Data:", test.shape[0], "obs, and", test.shape[1], "features")
print("\033[95m {}\033[00m".format(testshape))
# save id
train_id = train["Id"]
test_id = test["Id"]
# drop id
train.drop("Id", axis=1, inplace=True)
test.drop("Id", axis=1, inplace=True)
train.describe().T
# Focus Target Variable
sns.distplot(train["SalePrice"], color="g", bins=60, hist_kws={"alpha": 0.4})
# As we can see at the above, the target variable SalePrice is not distributed normally.
# This can reduce the performance of the ML regression models because some of them assume normal distribution.
# Therfore we need to log transform.
sns.distplot(np.log1p(train["SalePrice"]), color="g", bins=60, hist_kws={"alpha": 0.4})
# It looks like better :)
# Now, let's look at the best 8 correlation with heatmap.
corrmatrix = train.corr()
plt.figure(figsize=(10, 6))
columnss = corrmatrix.nlargest(8, "SalePrice")["SalePrice"].index
cm = np.corrcoef(train[columnss].values.T)
sns.set(font_scale=1.1)
hm = sns.heatmap(
cm,
cbar=True,
annot=True,
square=True,
cmap="RdPu",
fmt=".2f",
annot_kws={"size": 10},
yticklabels=columnss.values,
xticklabels=columnss.values,
)
plt.show()
# Now let's look at the distribution of the variable with the 3 highest correlations.
f, ax = plt.subplots(figsize=(10, 7))
sns.boxplot(x="OverallQual", y="SalePrice", data=train)
sns.jointplot(x=train["GrLivArea"], y=train["SalePrice"], kind="reg")
sns.boxplot(x=train["GarageCars"], y=train["SalePrice"])
# #### - **Outliers**
# Can you see two points at the bottom right on GrLivArea. Yes ! It's outliers !
# Car garages result in less Sale Price? That doesn't make much sense.
# We need to remove outliers.
train = train.drop(
train[(train["GrLivArea"] > 4000) & (train["SalePrice"] < 200000)].index
).reset_index(drop=True)
train = train.drop(
train[(train["GarageCars"] > 3) & (train["SalePrice"] < 300000)].index
).reset_index(drop=True)
# It should look better.
sns.jointplot(x=train["GrLivArea"], y=train["SalePrice"], kind="reg")
sns.boxplot(x=train["GarageCars"], y=train["SalePrice"])
# They Look succesfull.
# Now, we need to concanete train and test data for some cleaning operations.
df = pd.concat((train, test)).reset_index(drop=True)
df.drop(["SalePrice"], axis=1, inplace=True)
df.shape
#### Focus missing values
df.isna().sum().nlargest(35)
sns.set_style("whitegrid")
f, ax = plt.subplots(figsize=(12, 6))
miss = round(df.isnull().mean() * 100, 2)
miss = miss[miss > 0]
miss.sort_values(inplace=True)
miss.plot.bar(color="g")
ax.set(title="Percent missing data by variables")
# As can be seen, there are many missing observations in the data.
# #### - **Filling missing values**
# For a few columns there is lots of NaN entries.
# However, reading the data description we find this is not missing data:
# For PoolQC, NaN is not missing data but means no pool, likewise for Fence, FireplaceQu etc.
# Now, lets filling NA values :)
some_miss_columns = [
"PoolQC",
"MiscFeature",
"Alley",
"Fence",
"FireplaceQu",
"GarageType",
"GarageFinish",
"GarageQual",
"GarageCond",
"BsmtQual",
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinType2",
"MasVnrType",
"MSSubClass",
]
for i in some_miss_columns:
df[i].fillna("None", inplace=True)
df["Functional"] = df["Functional"].fillna("Typ")
some_miss_columns2 = [
"MSZoning",
"BsmtFullBath",
"BsmtHalfBath",
"Utilities",
"MSZoning",
"Electrical",
"KitchenQual",
"SaleType",
"Exterior1st",
"Exterior2nd",
"MasVnrArea",
]
for i in some_miss_columns2:
df[i].fillna(df[i].mode()[0], inplace=True)
some_miss_columns3 = [
"GarageYrBlt",
"GarageArea",
"GarageCars",
"BsmtFinSF1",
"BsmtFinSF2",
"BsmtUnfSF",
"TotalBsmtSF",
]
for i in some_miss_columns3:
df[i] = df[i].fillna(0)
df["LotFrontage"] = df.groupby("Neighborhood")["LotFrontage"].transform(
lambda x: x.fillna(x.median())
)
# We've filled out all the missing data.
# Let's control.
df.isna().sum().nlargest(3)
# We should transform for some variables.
Nm = ["MSSubClass", "MoSold", "YrSold"]
for col in Nm:
df[col] = df[col].astype(str)
# #### - **Label Encoder**
# Convert this kind of categorical text data into model-understandable numerical data, we use the Label Encoder class.
lbe = LabelEncoder()
encodecolumns = (
"FireplaceQu",
"BsmtQual",
"BsmtCond",
"ExterQual",
"ExterCond",
"HeatingQC",
"GarageQual",
"GarageCond",
"PoolQC",
"KitchenQual",
"BsmtFinType1",
"BsmtFinType2",
"Functional",
"Fence",
"BsmtExposure",
"GarageFinish",
"LandSlope",
"LotShape",
"PavedDrive",
"Street",
"Alley",
"CentralAir",
"MSSubClass",
"OverallCond",
"YrSold",
"MoSold",
)
for i in encodecolumns:
lbe.fit(list(df[i].values))
df[i] = lbe.transform(list(df[i].values))
# #### - **Log Transform for SalePrice**
# We must apply logarithmic transformation to our target variable.Because ML models work better with normal distribution.
train["SalePrice"] = np.log1p(train["SalePrice"])
y = train.SalePrice.values
y[:5]
# #### - **Fixing "Skewed" features**
# We need to fix all of the skewed data to be more normal so that our models will be more accurate when making predictions.
numeric = df.dtypes[df.dtypes != "object"].index
skewed_var = df[numeric].apply(lambda x: skew(x.dropna())).sort_values(ascending=False)
skewness = pd.DataFrame({"Skewed Features": skewed_var})
skewness.head()
# Now we will apply box cox transformation to these skewed values. So what is box cox transformation?
# #### - **Box Cox Transformation**
# A Box Cox transformation is a transformation of a non-normal dependent variables into a normal shape. Normality is an important assumption for many statistical techniques; if your data isn’t normal, applying a Box-Cox means that you are able to run a broader number of tests.
#
# References : Box, G. E. P. and Cox, D. R. (1964). An analysis of transformations.
# Lets do it.
skewness = skewness[abs(skewness) > 0.75]
skewed_var2 = skewness.index
for i in skewed_var2:
df[i] = boxcox1p(df[i], 0.15)
df[i] += 1
# #### - **Dummy Variables**
# Next step is dummy variables !
# In statistics and econometrics, particularly in regression analysis, a dummy variable is one that takes only the value 0 or 1 to indicate the absence or presence of some categorical effect that may be expected to shift the outcome.
df = pd.get_dummies(df)
df.head()
X_train = df[: train.shape[0]]
X_test = df[train.shape[0] :]
# Now, we are ready to ML, but i want to try PCA. So what is the PCA ?
# #### **PCA (Principal component analysis)**
# PCA is used in exploratory data analysis and for making predictive models. It is commonly used for dimensionality reduction by projecting each data point onto only the first few principal components to obtain lower-dimensional data while preserving as much of the data's variation as possible. The first principal component can equivalently be defined as a direction that maximizes the variance of the projected data. Lets try it.
# **Note** : You need to **standardize** the data before using PCA.
dff = df.copy()
##df_standardize = StandardScaler().fit_transform(dff)
##I didn't standardize it again because the data is already close to the standard.
pca = PCA()
pca_fit = pca.fit_transform(dff)
pca = PCA().fit(dff)
plt.plot(np.cumsum(pca.explained_variance_ratio_))
# With about 30 variables, we can explain 90% of the variance in the dataset.How do we do that ?
pca = PCA(n_components=30)
pca_fit = pca.fit_transform(dff)
pca_df = pd.DataFrame(data=pca_fit)
pca_df.head()
# I didn't have much experience with PCA , so I just wanted to try it. Your positive and negative opinions are important to me :)
# Now, we will predict models ! Firstly start Cross-validation with k-folds
n_folds = 5
def rmsle_cv(model):
kf = KFold(n_folds, shuffle=True, random_state=42).get_n_splits(X_train.values)
rmse = np.sqrt(
-cross_val_score(
model, X_train.values, y, scoring="neg_mean_squared_error", cv=kf
)
)
return rmse
model_xgb = xgb.XGBRegressor(
colsample_bytree=0.2,
gamma=0.0,
learning_rate=0.05,
max_depth=6,
min_child_weight=1.5,
n_estimators=7200,
reg_alpha=0.9,
reg_lambda=0.6,
subsample=0.2,
seed=42,
random_state=7,
)
model_gbm = GradientBoostingRegressor(
n_estimators=3000,
learning_rate=0.05,
max_depth=4,
max_features="sqrt",
min_samples_leaf=15,
min_samples_split=10,
loss="huber",
random_state=5,
)
# Checking performance of base models by evaluating the cross-validation RMSLE error.
score = rmsle_cv(model_xgb)
print("XGBoost score: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
score = rmsle_cv(model_gbm)
print("Gradient Boosting score: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
## we need this func
def rmsle(y, y_pred):
return np.sqrt(mean_squared_error(y, y_pred))
# #### - **XGBoost**
model_xgb.fit(X_train, y)
xgb_train_pred = model_xgb.predict(X_train)
xgb_pred = np.expm1(model_xgb.predict(X_test))
print(rmsle(y, xgb_train_pred))
xgb_pred[:5]
# #### - **GBM (Gradient Boosting Machines)**
model_gbm.fit(X_train, y)
gbm_train_pred = model_gbm.predict(X_train)
gbm_pred = np.expm1(model_gbm.predict(X_test.values))
print(rmsle(y, gbm_train_pred))
gbm_pred[:5]
# #### - **SUBMISSION**
trybest = (0.5 * xgb_pred) + (0.5 * gbm_pred)
submission = pd.DataFrame({"Id": test_id, "SalePrice": trybest})
submission.head(5)
submission.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605229.ipynb
| null | null |
[{"Id": 69605229, "ScriptId": 18654334, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7037244, "CreationDate": "08/02/2021 05:34:15", "VersionNumber": 2.0, "Title": "Predicting House Prices XGBoost + GBM Models", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 417.0, "LinesInsertedFromPrevious": 81.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 336.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 13}]
| null | null | null | null |
# # Predicting House Prices XGBoost + GBM Models
# **Bugra Sebati E.** - **July 2021**
# ## Introduction
# For this competiton, we are given a data set of 1460 homes, each with a few dozen features of types: float, integer, and categorical. We are tasked with building a regression model to estimate a home's sale price. Total number of attributes equals 81, of which 36 is quantitative, 43 categorical + Id and SalePrice.
# **What you can find on this notebook?**
# * Understanding the data
# * Exploratory Data Analysis
# * Data Preprocessing
# * PCA Trial
# * GBM and XGBoost Models
# * Submission
# 
# ### Lets meet variables
# * **SalePrice** : The property's sale price in dollars. This is target variable for predict
# * **MSSubClass**: The building class
# * **MSZoning**: The general zoning classification
# * **LotFrontage**: Linear feet of street connected to property
# * **LotArea**: Lot size in square feet
# * **Street**: Type of road access
# * **Alley**: Type of alley access
# * **LotShape**: General shape of property
# * **LandContour**: Flatness of the property
# * **Utilities**: Type of utilities available
# * **LotConfig**: Lot configuration
# * **LandSlope**: Slope of property
# * **Neighborhood**: Physical locations within Ames city limits
# * **Condition1**: Proximity to main road or railroad
# * **Condition2**: Proximity to main road or railroad (if a second is present)
# * **BldgType**: Type of dwelling
# * **HouseStyle**: Style of dwelling
# * **OverallQual**: Overall material and finish quality
# * **OverallCond**: Overall condition rating
# * **YearBuilt**: Original construction date
# * **YearRemodAdd**: Remodel date
# * **RoofStyle**: Type of roof
# * **RoofMatl**: Roof material
# * **Exterior1st**: Exterior covering on house
# * **Exterior2nd**: Exterior covering on house (if more than one material)
# * **MasVnrType**: Masonry veneer type
# * **MasVnrArea**: Masonry veneer area in square feet
# * **ExterQual**: Exterior material quality
# * **ExterCond**: Present condition of the material on the exterior
# * **Foundation**: Type of foundation
# * **BsmtQual**: Height of the basement
# * **BsmtCond**: General condition of the basement
# * **BsmtExposure**: Walkout or garden level basement walls
# * **BsmtFinType1**: Quality of basement finished area
# * **BsmtFinSF1**: Type 1 finished square feet
# * **BsmtFinType2**: Quality of second finished area (if present)
# * **BsmtFinSF2**: Type 2 finished square feet
# * **BsmtUnfSF**: Unfinished square feet of basement area
# * **TotalBsmtSF**: Total square feet of basement area
# * **Heating**: Type of heating
# * **HeatingQC**: Heating quality and condition
# * **CentralAir**: Central air conditioning
# * **Electrical**: Electrical system
# * **1stFlrSF**: First Floor square feet
# * **2ndFlrSF**: Second floor square feet
# * **LowQualFinSF**: Low quality finished square feet (all floors)
# * **GrLivArea**: Above grade (ground) living area square feet
# * **BsmtFullBath**: Basement full bathrooms
# * **BsmtHalfBath**: Basement half bathrooms
# * **FullBath**: Full bathrooms above grade
# * **HalfBath**: Half baths above grade
# * **Bedroom**: Number of bedrooms above basement level
# * **Kitchen**: Number of kitchens
# * **KitchenQual**: Kitchen quality
# * **TotRmsAbvGrd**: Total rooms above grade (does not include bathrooms)
# * **Functional**: Home functionality rating
# * **Fireplaces**: Number of fireplaces
# * **FireplaceQu**: Fireplace quality
# * **GarageType**: Garage location
# * **GarageYrBlt**: Year garage was built
# * **GarageFinish**: Interior finish of the garage
# * **GarageCars**: Size of garage in car capacity
# * **GarageArea**: Size of garage in square feet
# * **GarageQual**: Garage quality
# * **GarageCond**: Garage condition
# * **PavedDrive**: Paved driveway
# * **WoodDeckSF**: Wood deck area in square feet
# * **OpenPorchSF**: Open porch area in square feet
# * **EnclosedPorch**: Enclosed porch area in square feet
# * **3SsnPorch**: Three season porch area in square feet
# * **ScreenPorch**: Screen porch area in square feet
# * **PoolArea**: Pool area in square feet
# * **PoolQC**: Pool quality
# * **Fence**: Fence quality
# * **MiscFeature**: Miscellaneous feature not covered in other categories
# * **MiscVal**: Value of miscellaneous feature
# * **MoSold**: Month Sold
# * **YrSold**: Year Sold
# * **SaleType**: Type of sale
# * **SaleCondition**: Condition of sale
# #### Since we learn variables, we can start now...
# If you like this notebook,dont forget to upvote :) **Thanks !**
#### IMPORT LIBRARIES
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import StandardScaler
from scipy.stats import skew
from scipy.special import boxcox1p
from sklearn.decomposition import PCA
from sklearn.model_selection import KFold, cross_val_score
from sklearn.metrics import mean_squared_error
import xgboost as xgb
from sklearn.ensemble import GradientBoostingRegressor
import warnings
warnings.filterwarnings("ignore")
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
traindf = train.copy()
testdf = test.copy()
train.head()
test.head()
#### I like colors :-9
trainshape = ("Train Data:", train.shape[0], "obs, and", train.shape[1], "features")
print("\033[95m {}\033[00m".format(trainshape))
testshape = ("Test Data:", test.shape[0], "obs, and", test.shape[1], "features")
print("\033[95m {}\033[00m".format(testshape))
# save id
train_id = train["Id"]
test_id = test["Id"]
# drop id
train.drop("Id", axis=1, inplace=True)
test.drop("Id", axis=1, inplace=True)
train.describe().T
# Focus Target Variable
sns.distplot(train["SalePrice"], color="g", bins=60, hist_kws={"alpha": 0.4})
# As we can see at the above, the target variable SalePrice is not distributed normally.
# This can reduce the performance of the ML regression models because some of them assume normal distribution.
# Therfore we need to log transform.
sns.distplot(np.log1p(train["SalePrice"]), color="g", bins=60, hist_kws={"alpha": 0.4})
# It looks like better :)
# Now, let's look at the best 8 correlation with heatmap.
corrmatrix = train.corr()
plt.figure(figsize=(10, 6))
columnss = corrmatrix.nlargest(8, "SalePrice")["SalePrice"].index
cm = np.corrcoef(train[columnss].values.T)
sns.set(font_scale=1.1)
hm = sns.heatmap(
cm,
cbar=True,
annot=True,
square=True,
cmap="RdPu",
fmt=".2f",
annot_kws={"size": 10},
yticklabels=columnss.values,
xticklabels=columnss.values,
)
plt.show()
# Now let's look at the distribution of the variable with the 3 highest correlations.
f, ax = plt.subplots(figsize=(10, 7))
sns.boxplot(x="OverallQual", y="SalePrice", data=train)
sns.jointplot(x=train["GrLivArea"], y=train["SalePrice"], kind="reg")
sns.boxplot(x=train["GarageCars"], y=train["SalePrice"])
# #### - **Outliers**
# Can you see two points at the bottom right on GrLivArea. Yes ! It's outliers !
# Car garages result in less Sale Price? That doesn't make much sense.
# We need to remove outliers.
train = train.drop(
train[(train["GrLivArea"] > 4000) & (train["SalePrice"] < 200000)].index
).reset_index(drop=True)
train = train.drop(
train[(train["GarageCars"] > 3) & (train["SalePrice"] < 300000)].index
).reset_index(drop=True)
# It should look better.
sns.jointplot(x=train["GrLivArea"], y=train["SalePrice"], kind="reg")
sns.boxplot(x=train["GarageCars"], y=train["SalePrice"])
# They Look succesfull.
# Now, we need to concanete train and test data for some cleaning operations.
df = pd.concat((train, test)).reset_index(drop=True)
df.drop(["SalePrice"], axis=1, inplace=True)
df.shape
#### Focus missing values
df.isna().sum().nlargest(35)
sns.set_style("whitegrid")
f, ax = plt.subplots(figsize=(12, 6))
miss = round(df.isnull().mean() * 100, 2)
miss = miss[miss > 0]
miss.sort_values(inplace=True)
miss.plot.bar(color="g")
ax.set(title="Percent missing data by variables")
# As can be seen, there are many missing observations in the data.
# #### - **Filling missing values**
# For a few columns there is lots of NaN entries.
# However, reading the data description we find this is not missing data:
# For PoolQC, NaN is not missing data but means no pool, likewise for Fence, FireplaceQu etc.
# Now, lets filling NA values :)
some_miss_columns = [
"PoolQC",
"MiscFeature",
"Alley",
"Fence",
"FireplaceQu",
"GarageType",
"GarageFinish",
"GarageQual",
"GarageCond",
"BsmtQual",
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinType2",
"MasVnrType",
"MSSubClass",
]
for i in some_miss_columns:
df[i].fillna("None", inplace=True)
df["Functional"] = df["Functional"].fillna("Typ")
some_miss_columns2 = [
"MSZoning",
"BsmtFullBath",
"BsmtHalfBath",
"Utilities",
"MSZoning",
"Electrical",
"KitchenQual",
"SaleType",
"Exterior1st",
"Exterior2nd",
"MasVnrArea",
]
for i in some_miss_columns2:
df[i].fillna(df[i].mode()[0], inplace=True)
some_miss_columns3 = [
"GarageYrBlt",
"GarageArea",
"GarageCars",
"BsmtFinSF1",
"BsmtFinSF2",
"BsmtUnfSF",
"TotalBsmtSF",
]
for i in some_miss_columns3:
df[i] = df[i].fillna(0)
df["LotFrontage"] = df.groupby("Neighborhood")["LotFrontage"].transform(
lambda x: x.fillna(x.median())
)
# We've filled out all the missing data.
# Let's control.
df.isna().sum().nlargest(3)
# We should transform for some variables.
Nm = ["MSSubClass", "MoSold", "YrSold"]
for col in Nm:
df[col] = df[col].astype(str)
# #### - **Label Encoder**
# Convert this kind of categorical text data into model-understandable numerical data, we use the Label Encoder class.
lbe = LabelEncoder()
encodecolumns = (
"FireplaceQu",
"BsmtQual",
"BsmtCond",
"ExterQual",
"ExterCond",
"HeatingQC",
"GarageQual",
"GarageCond",
"PoolQC",
"KitchenQual",
"BsmtFinType1",
"BsmtFinType2",
"Functional",
"Fence",
"BsmtExposure",
"GarageFinish",
"LandSlope",
"LotShape",
"PavedDrive",
"Street",
"Alley",
"CentralAir",
"MSSubClass",
"OverallCond",
"YrSold",
"MoSold",
)
for i in encodecolumns:
lbe.fit(list(df[i].values))
df[i] = lbe.transform(list(df[i].values))
# #### - **Log Transform for SalePrice**
# We must apply logarithmic transformation to our target variable.Because ML models work better with normal distribution.
train["SalePrice"] = np.log1p(train["SalePrice"])
y = train.SalePrice.values
y[:5]
# #### - **Fixing "Skewed" features**
# We need to fix all of the skewed data to be more normal so that our models will be more accurate when making predictions.
numeric = df.dtypes[df.dtypes != "object"].index
skewed_var = df[numeric].apply(lambda x: skew(x.dropna())).sort_values(ascending=False)
skewness = pd.DataFrame({"Skewed Features": skewed_var})
skewness.head()
# Now we will apply box cox transformation to these skewed values. So what is box cox transformation?
# #### - **Box Cox Transformation**
# A Box Cox transformation is a transformation of a non-normal dependent variables into a normal shape. Normality is an important assumption for many statistical techniques; if your data isn’t normal, applying a Box-Cox means that you are able to run a broader number of tests.
#
# References : Box, G. E. P. and Cox, D. R. (1964). An analysis of transformations.
# Lets do it.
skewness = skewness[abs(skewness) > 0.75]
skewed_var2 = skewness.index
for i in skewed_var2:
df[i] = boxcox1p(df[i], 0.15)
df[i] += 1
# #### - **Dummy Variables**
# Next step is dummy variables !
# In statistics and econometrics, particularly in regression analysis, a dummy variable is one that takes only the value 0 or 1 to indicate the absence or presence of some categorical effect that may be expected to shift the outcome.
df = pd.get_dummies(df)
df.head()
X_train = df[: train.shape[0]]
X_test = df[train.shape[0] :]
# Now, we are ready to ML, but i want to try PCA. So what is the PCA ?
# #### **PCA (Principal component analysis)**
# PCA is used in exploratory data analysis and for making predictive models. It is commonly used for dimensionality reduction by projecting each data point onto only the first few principal components to obtain lower-dimensional data while preserving as much of the data's variation as possible. The first principal component can equivalently be defined as a direction that maximizes the variance of the projected data. Lets try it.
# **Note** : You need to **standardize** the data before using PCA.
dff = df.copy()
##df_standardize = StandardScaler().fit_transform(dff)
##I didn't standardize it again because the data is already close to the standard.
pca = PCA()
pca_fit = pca.fit_transform(dff)
pca = PCA().fit(dff)
plt.plot(np.cumsum(pca.explained_variance_ratio_))
# With about 30 variables, we can explain 90% of the variance in the dataset.How do we do that ?
pca = PCA(n_components=30)
pca_fit = pca.fit_transform(dff)
pca_df = pd.DataFrame(data=pca_fit)
pca_df.head()
# I didn't have much experience with PCA , so I just wanted to try it. Your positive and negative opinions are important to me :)
# Now, we will predict models ! Firstly start Cross-validation with k-folds
n_folds = 5
def rmsle_cv(model):
kf = KFold(n_folds, shuffle=True, random_state=42).get_n_splits(X_train.values)
rmse = np.sqrt(
-cross_val_score(
model, X_train.values, y, scoring="neg_mean_squared_error", cv=kf
)
)
return rmse
model_xgb = xgb.XGBRegressor(
colsample_bytree=0.2,
gamma=0.0,
learning_rate=0.05,
max_depth=6,
min_child_weight=1.5,
n_estimators=7200,
reg_alpha=0.9,
reg_lambda=0.6,
subsample=0.2,
seed=42,
random_state=7,
)
model_gbm = GradientBoostingRegressor(
n_estimators=3000,
learning_rate=0.05,
max_depth=4,
max_features="sqrt",
min_samples_leaf=15,
min_samples_split=10,
loss="huber",
random_state=5,
)
# Checking performance of base models by evaluating the cross-validation RMSLE error.
score = rmsle_cv(model_xgb)
print("XGBoost score: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
score = rmsle_cv(model_gbm)
print("Gradient Boosting score: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
## we need this func
def rmsle(y, y_pred):
return np.sqrt(mean_squared_error(y, y_pred))
# #### - **XGBoost**
model_xgb.fit(X_train, y)
xgb_train_pred = model_xgb.predict(X_train)
xgb_pred = np.expm1(model_xgb.predict(X_test))
print(rmsle(y, xgb_train_pred))
xgb_pred[:5]
# #### - **GBM (Gradient Boosting Machines)**
model_gbm.fit(X_train, y)
gbm_train_pred = model_gbm.predict(X_train)
gbm_pred = np.expm1(model_gbm.predict(X_test.values))
print(rmsle(y, gbm_train_pred))
gbm_pred[:5]
# #### - **SUBMISSION**
trybest = (0.5 * xgb_pred) + (0.5 * gbm_pred)
submission = pd.DataFrame({"Id": test_id, "SalePrice": trybest})
submission.head(5)
submission.to_csv("submission.csv", index=False)
| false | 0 | 4,884 | 13 | 4,884 | 4,884 |
||
69605607
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import sys, matplotlib, IPython, sklearn, random, time, warnings
import scipy as sp
from IPython import display
from subprocess import check_output
print("Python version: {}".format(sys.version))
print("pandas version: {}".format(pd.__version__))
print("NumPy version: {}".format(np.__version__))
print("matplotlib version: {}".format(matplotlib.__version__))
print("SciPy version: {}".format(sp.__version__))
print("IPython version: {}".format(IPython.__version__))
print("scikit-learn version: {}".format(sklearn.__version__))
warnings.filterwarnings("ignore")
print("-" * 25)
print(check_output(["ls", "../input/titanic/"]).decode("utf8"))
# model algorithems
from sklearn import (
svm,
tree,
linear_model,
neighbors,
naive_bayes,
ensemble,
discriminant_analysis,
gaussian_process,
)
from xgboost import XGBClassifier
# model helpers
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
from sklearn import feature_selection
from sklearn import model_selection
from sklearn import metrics
# visualization
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.pylab as pylab
import seaborn as sns
from pandas.plotting import scatter_matrix
# show plot in Jupyter Notebook
mpl.style.use("ggplot")
sns.set_style("white")
pylab.rcParams["figure.figsize"] = 12, 8
data_raw = pd.read_csv("../input/titanic/train.csv")
data_test = pd.read_csv("../input/titanic/test.csv")
data1 = data_raw.copy(deep=True)
data_cleaner = [data1, data_test]
# print(data_raw.info())
# data_raw.head()
# data_raw.tail()
print("sex classcifications:", set(data_raw["Sex"]))
print("Age Range: {}~{}".format(min(data_raw["Age"]), max(data_raw["Age"])))
print("Fare Range: {}~{}".format(min(data_raw["Fare"]), max(data_raw["Fare"])))
data_raw.sample(5)
print("Train columns with null values:\n", data1.isnull().sum())
print("-" * 10)
print("Test columns with null values:\n", data_test.isnull().sum())
print("-" * 10)
data_raw.describe(include="all")
for dataset in data_cleaner:
dataset["Age"].fillna(dataset["Age"].median(), inplace=True)
dataset["Embarked"].fillna(dataset["Embarked"].mode()[0], inplace=True)
dataset["Fare"].fillna(dataset["Fare"].median(), inplace=True)
drop_column = ["PassengerId", "Cabin", "Ticket"]
data1.drop(drop_column, axis=1, inplace=True)
print("Train columns with null values:\n", data1.isnull().sum())
print("-" * 10)
print("Test columns with null values:\n", data_test.isnull().sum())
print("-" * 10)
for dataset in data_cleaner:
dataset["FamilySize"] = dataset["SibSp"] + dataset["Parch"] + 1
dataset["IsAlone"] = 1
dataset["IsAlone"].loc[dataset["FamilySize"] > 1] = 0
dataset["Title"] = (
dataset["Name"].str.split(", ", expand=True)[1].str.split(".", expand=True)[0]
)
dataset["FareBin"] = pd.qcut(dataset["Fare"], 4)
dataset["AgeBin"] = pd.cut(dataset["Age"].astype(int), 5)
data1["Title"].value_counts()
stat_min = 10
# create a true/false series with title name as index
title_names = data1["Title"].value_counts() < stat_min
data1["Title"] = data1["Title"].apply(
lambda x: "Misc" if title_names.loc[x] == True else x
)
print(data1["Title"].value_counts())
print("-" * 10)
# preview data
data1.info()
data_test.info()
data1.sample(10)
label = LabelEncoder()
for dataset in data_cleaner:
dataset["Sex_Code"] = label.fit_transform(dataset["Sex"])
dataset["Embarked_Code"] = label.fit_transform(dataset["Embarked"])
dataset["Title_Code"] = label.fit_transform(dataset["Title"])
dataset["AgeBin_Code"] = label.fit_transform(dataset["AgeBin"])
dataset["FareBin_Code"] = label.fit_transform(dataset["FareBin"])
data1.sample(5)
Target = ["Survived"]
## feature selection
# feature names for charts
data1_x = [
"Sex",
"Pclass",
"Embarked",
"Title",
"SibSp",
"Parch",
"Age",
"Fare",
"FamilySize",
"IsAlone",
]
# feature names for codes
data1_x_calc = [
"Sex_Code",
"Pclass",
"Embarked_Code",
"Title_Code",
"SibSp",
"Parch",
"Age",
"Fare",
]
data1_xy = Target + data1_x
print("Original X Y: ", data1_xy, "\n")
# features w/ bins
data1_x_bin = [
"Sex_Code",
"Pclass",
"Embarked_Code",
"Title_Code",
"FamilySize",
"AgeBin_Code",
"FareBin_Code",
]
data1_xy_bin = Target + data1_x_bin
print("Bin X Y: ", data1_xy_bin, "\n")
# dummy features
data1_dummy = pd.get_dummies(data1[data1_x])
data1_x_dummy = data1_dummy.columns.tolist()
data1_xy_dummy = Target + data1_x_dummy
print("Dummy X Y: ", data1_xy_dummy, "\n")
data1_dummy.head()
print("Train columns with null values: \n", data1.isnull().sum())
print("-" * 10)
print(data1.info())
print("-" * 10)
print("Test columns with null values: \n", data_test.isnull().sum())
print("-" * 10)
print(data_test.info())
print("-" * 10)
data_raw.describe(include="all")
train1_x, crossVal1_x, train1_y, crossVal1_y = model_selection.train_test_split(
data1[data1_x_calc], data1[Target], random_state=0
)
(
train1_x_bin,
crossVal1_x_bin,
train1_y_bin,
crossVal1_y_bin,
) = model_selection.train_test_split(data1[data1_x_bin], data1[Target], random_state=0)
(
train1_x_dummy,
crossVal1_x_dummy,
train1_y_dummy,
crossVal1_y_dummy,
) = model_selection.train_test_split(
data1_dummy[data1_x_dummy], data1[Target], random_state=0
)
print("Data1 Shape: {}".format(data1.shape))
print("Train1 Shape: {}".format(train1_x.shape))
print("crossVal1 Shape: {}".format(crossVal1_x.shape))
train1_x_bin.head()
for x in data1_x:
# don't do it on float values, otherwise too many groups
if data1[x].dtype != "float64":
print("Survival correlation by:", x)
print(data1[[x, Target[0]]].groupby(x, as_index=False).mean())
print("-" * 10, "\n")
print(pd.crosstab(data1["Title"], data1[Target[0]]))
plt.figure(figsize=[16, 12])
plt.subplot(231)
# plt.grid(True,axis='y')
plt.boxplot(x=data1["Fare"], showmeans=True, meanline=True)
plt.title("Fare Boxplot")
plt.ylabel("Fare($)")
plt.subplot(232)
plt.boxplot(data1["Age"], showmeans=True, meanline=True)
plt.title("Age Boxplot")
plt.ylabel("Age (Years)")
plt.subplot(233)
plt.boxplot(data1["FamilySize"], showmeans=True, meanline=True)
plt.title("Family Size Boxplot")
plt.ylabel("Family Size (#)")
plt.subplot(234)
plt.hist(
x=[data1[data1["Survived"] == 1]["Fare"], data1[data1["Survived"] == 0]["Fare"]],
stacked=False,
color=["g", "r"],
label=["Survived", "Dead"],
)
plt.title("Fare Histogram by Survival")
plt.xlabel("Fare ($)")
plt.ylabel("# of Passengers")
plt.legend()
plt.subplot(235)
plt.hist(
x=[data1[data1["Survived"] == 1]["Age"], data1[data1["Survived"] == 0]["Age"]],
stacked=False,
color=["g", "r"],
label=["Survived", "Dead"],
)
plt.title("Age Histogram by Survival")
plt.xlabel("Age (years)")
plt.ylabel("# of Passengers")
plt.legend()
plt.subplot(236)
plt.hist(
x=[
data1[data1["Survived"] == 1]["FamilySize"],
data1[data1["Survived"] == 0]["FamilySize"],
],
bins=np.arange(data1["FamilySize"].min() - 0.5, data1["FamilySize"].max() + 0.5),
stacked=False,
color=["g", "r"],
label=["Survived", "Dead"],
)
plt.title("FamilySize Histogram by Survival")
plt.xlabel("Family Size (#)")
plt.ylabel("# of Passengers")
plt.legend()
fig, axs = plt.subplots(2, 3, figsize=(16, 12))
sns.barplot(x="Embarked", y="Survived", data=data1, ax=axs[0, 0])
sns.barplot(x="Pclass", y="Survived", data=data1, ax=axs[0, 1])
sns.barplot(x="IsAlone", y="Survived", order=[1, 0], data=data1, ax=axs[0, 2])
sns.pointplot(x="FareBin", y="Survived", data=data1, ax=axs[1, 0])
sns.pointplot(x="AgeBin", y="Survived", data=data1, ax=axs[1, 1])
sns.pointplot(x="FamilySize", y="Survived", data=data1, ax=axs[1, 2])
fig, (axis1, axis2, axis3) = plt.subplots(1, 3, figsize=(16, 8))
sns.boxplot(x="Pclass", y="Fare", hue="Survived", data=data1, ax=axis1)
axis1.set_title("Pclass vs Fare Survival Comparison")
sns.violinplot(x="Pclass", y="Age", hue="Survived", data=data1, split=True, ax=axis2)
axis2.set_title("Pclass vs Age Survival Comparison")
sns.boxplot(x="Pclass", y="FamilySize", hue="Survived", data=data1, ax=axis3)
axis3.set_title("Pclass vs FamilySize Survival Comparison")
fig, qaxis = plt.subplots(2, 3, figsize=(14, 12))
sns.barplot(x="Sex", y="Survived", hue="Embarked", data=data1, ax=qaxis[0, 0])
qaxis[0, 0].set_title("Sex vs Embarked Survival Comparison")
sns.barplot(x="Sex", y="Survived", hue="Pclass", data=data1, ax=qaxis[0, 1])
qaxis[0, 1].set_title("Sex vs Pclass Survival Comparison")
sns.barplot(x="Sex", y="Survived", hue="IsAlone", data=data1, ax=qaxis[0, 2])
qaxis[0, 2].set_title("Sex vs IsAlone Survival Comparison")
sns.barplot(x="Embarked", y="Survived", hue="Sex", data=data1, ax=qaxis[1, 0])
qaxis[1, 0].set_title("Embarked vs Sex Survival Comparison")
sns.barplot(x="Pclass", y="Survived", hue="Sex", data=data1, ax=qaxis[1, 1])
qaxis[1, 1].set_title("Pclass vs Sex Survival Comparison")
sns.barplot(x="IsAlone", y="Survived", hue="Sex", data=data1, ax=qaxis[1, 2])
qaxis[1, 2].set_title("IsAlone vs Sex Survival Comparison")
fig, (maxis1, maxis2) = plt.subplots(1, 2, figsize=(16, 8))
sns.pointplot(
x="FamilySize",
y="Survived",
hue="Sex",
data=data1,
palette={"male": "blue", "female": "pink"},
markers=["*", "o"],
linestyles=["-", "--"],
ax=maxis1,
)
sns.pointplot(
x="AgeBin",
y="Survived",
hue="Sex",
data=data1,
palette={"male": "blue", "female": "pink"},
markers=["*", "o"],
linestyles=["-", "--"],
ax=maxis2,
)
# use facetgrid in seaborn for multi-plot
e = sns.FacetGrid(data1, col="Embarked")
e.map(sns.pointplot, "Pclass", "Survived", "Sex", ci=95.0, palette="deep")
e.add_legend()
a = sns.FacetGrid(data1, hue="Survived", aspect=3)
# kde - kernal density estimation
a.map(sns.kdeplot, "Age", shade=True)
a.set(xlim=(0, data1["Age"].max()))
a.add_legend()
h = sns.FacetGrid(data1, row="Sex", col="Pclass", hue="Survived")
# alpha is transparency
h.map(plt.hist, "Age", alpha=0.5)
h.add_legend()
## pair plots of the entire dataset
pp = sns.pairplot(
data1,
hue="Survived",
palette="deep",
size=1.2,
diag_kind="kde",
diag_kws=dict(shade=True),
plot_kws=dict(s=10),
)
pp.set(xticklabels=[])
# correlation heatmap of dataset
def correlation_heatmap(df):
_, ax = plt.subplots(figsize=(14, 12))
colormap = sns.diverging_palette(220, 10, as_cmap=True)
_ = sns.heatmap(
df.corr(),
cmap=colormap,
square=True,
cbar_kws={"shrink": 0.9},
ax=ax,
annot=True,
linewidths=0.1,
vmax=1.0,
linecolor="white",
annot_kws={"fontsize": 12},
)
plt.title("Pearson Correlation of Features", y=1.05, size=15)
correlation_heatmap(data1)
# ML algo selection
MLA = [
## Ensemble Methods
ensemble.AdaBoostClassifier(),
ensemble.BaggingClassifier(),
ensemble.ExtraTreesClassifier(),
ensemble.GradientBoostingClassifier(),
ensemble.RandomForestClassifier(),
## Gaussian Processes
gaussian_process.GaussianProcessClassifier(),
## GLM
linear_model.LogisticRegressionCV(),
linear_model.PassiveAggressiveClassifier(),
linear_model.RidgeClassifierCV(),
linear_model.SGDClassifier(),
linear_model.Perceptron(),
## Naives Bayes
naive_bayes.BernoulliNB(),
naive_bayes.GaussianNB(),
## Nearest Neighbor
neighbors.KNeighborsClassifier(),
## SVM
svm.SVC(probability=True),
svm.NuSVC(probability=True),
svm.LinearSVC(),
## Trees
tree.DecisionTreeClassifier(),
tree.ExtraTreeClassifier(),
## Discriminant Analysis
discriminant_analysis.LinearDiscriminantAnalysis(),
discriminant_analysis.QuadraticDiscriminantAnalysis(),
# xgboost: https://xgboost.readthedocs.io/en/latest/
XGBClassifier(),
]
# split dataset
cv_split = model_selection.ShuffleSplit(
n_splits=10, test_size=0.3, train_size=0.6, random_state=0
)
# create table to compare MLA metrics
MLA_columns = [
"MLA Name",
"MLA Parameters",
"MLA Train Accuracy Mean",
"MLA Test Accuracy Mean",
"MLA Test Accuracy 3*STD",
"MLA Time",
]
MLA_compare = pd.DataFrame(columns=MLA_columns)
MLA_predict = data1[Target]
# loop through the MLA list
row_index = 0
for alg in MLA:
MLA_name = alg.__class__.__name__
MLA_compare.loc[row_index, "MLA Name"] = MLA_name
MLA_compare.loc[row_index, "MLA Parameters"] = str(alg.get_params())
cv_results = model_selection.cross_validate(
alg, data1[data1_x_bin], data1[Target], cv=cv_split, return_train_score=True
)
# if row_index == 0:
# print(cv_results)
MLA_compare.loc[row_index, "MLA Time"] = cv_results["fit_time"].mean()
MLA_compare.loc[row_index, "MLA Train Accuracy Mean"] = cv_results[
"train_score"
].mean()
MLA_compare.loc[row_index, "MLA Test Accuracy Mean"] = cv_results[
"test_score"
].mean()
MLA_compare.loc[row_index, "MLA Test Accuracy 3*STD"] = (
cv_results["test_score"].std() * 3
)
## save MLA predictions for later useage
alg.fit(data1[data1_x_bin], data1[Target])
MLA_predict[MLA_name] = alg.predict(data1[data1_x_bin])
row_index += 1
MLA_compare.sort_values(by=["MLA Test Accuracy Mean"], ascending=False, inplace=True)
MLA_compare
sns.barplot(x="MLA Test Accuracy Mean", y="MLA Name", data=MLA_compare, color="m")
# prettify using pyplot: https://matplotlib.org/api/pyplot_api.html
plt.title("Machine Learning Algorithm Accuracy Score \n")
plt.xlabel("Accuracy Score (%)")
plt.ylabel("Algorithm")
# coin flip model with random 1/survived 0/died
# iterate over dataFrame rows as (index, Series) pairs: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.iterrows.html
data1["Random_Predict"] = 0 # assume prediction wrong
for index, row in data1.iterrows():
# random number generator: https://docs.python.org/2/library/random.html
if random.random() > 0.5: # Random float x, 0.0 <= x < 1.0
data1["Random_Predict"][index] = 1 # predict survived/1
else:
data1["Random_Predict"][index] = 0 # predict died/0
# score random guess of survival. Use shortcut 1 = Right Guess and 0 = Wrong Guess
# the mean of the column will then equal the accuracy
data1["Random_Score"] = 0 # assume prediction wrong
data1.loc[
(data1["Survived"] == data1["Random_Predict"]), "Random_Score"
] = 1 # set to 1 for correct prediction
print("Coin Flip Model Accuracy: {:.2f}%".format(data1["Random_Score"].mean() * 100))
# we can also use scikit's accuracy_score function to save us a few lines of code
print(
"Coin Flip Model Accuracy w/SciKit: {:.2f}%".format(
metrics.accuracy_score(data1["Survived"], data1["Random_Predict"]) * 100
)
)
# group by or pivot table
pivot_female = (
data1[data1.Sex == "female"]
.groupby(["Sex", "Pclass", "Embarked", "FareBin"])["Survived"]
.mean()
)
print("Survival Decision Tree w/Female Node: \n", pivot_female)
pivot_male = data1[data1.Sex == "male"].groupby(["Sex", "Title"])["Survived"].mean()
print("\n\nSurvival Decision Tree w/Male Node: \n", pivot_male)
# handmade data model using brain power (and Microsoft Excel Pivot Tables for quick calculations)
def mytree(df):
# initialize table to store predictions
Model = pd.DataFrame(data={"Predict": []})
male_title = ["Master"] # survived titles
for index, row in df.iterrows():
# Question 1: Were you on the Titanic; majority died
Model.loc[index, "Predict"] = 0
# Question 2: Are you female; majority survived
if df.loc[index, "Sex"] == "female":
Model.loc[index, "Predict"] = 1
# Question 3A Female - Class and Question 4 Embarked gain minimum information
# Question 5B Female - FareBin; set anything less than .5 in female node decision tree back to 0
if (
(df.loc[index, "Sex"] == "female")
& (df.loc[index, "Pclass"] == 3)
& (df.loc[index, "Embarked"] == "S")
& (df.loc[index, "Fare"] > 8)
):
Model.loc[index, "Predict"] = 0
# Question 3B Male: Title; set anything greater than .5 to 1 for majority survived
if (df.loc[index, "Sex"] == "male") & (df.loc[index, "Title"] in male_title):
Model.loc[index, "Predict"] = 1
return Model
# model data
Tree_Predict = mytree(data1)
# print matrix
print(
"Decision Tree Model Accuracy/Precision Score: {:.2f}%\n".format(
metrics.accuracy_score(data1["Survived"], Tree_Predict) * 100
)
)
# Accuracy Summary Report
print(metrics.classification_report(data1["Survived"], Tree_Predict))
import itertools
def my_plot_confusion_mtrix(
cm, classes, normalize=False, title="Confusion matrix", cmap=plt.cm.Blues
):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print("Confusion matrix, without normalization")
print(cm)
plt.imshow(cm, interpolation="nearest", cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes)
plt.yticks(tick_marks, classes)
fmt = ".2f" if normalize else "d"
thresh = cm.max() / 2.0
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(
j,
i,
format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black",
)
plt.tight_layout()
plt.ylabel("True label")
plt.xlabel("Predicted label")
# Compute confusion matrix
cnf_matrix = metrics.confusion_matrix(data1["Survived"], Tree_Predict)
np.set_printoptions(precision=2)
class_names = ["Dead", "Survived"]
# Plot non-normalized confusion matrix
plt.figure()
my_plot_confusion_mtrix(
cnf_matrix, class_names, False, "Confusion matrix, without normalization"
)
# Plot normalized confusion matrix
plt.figure()
my_plot_confusion_mtrix(cnf_matrix, class_names, True, "Normalized confusion matrix")
# base model
dtree = tree.DecisionTreeClassifier(random_state=0)
base_results = model_selection.cross_validate(
dtree, data1[data1_x_bin], data1[Target], cv=cv_split, return_train_score=True
)
dtree.fit(data1[data1_x_bin], data1[Target])
print("BEFORE DT Parameters: ", dtree.get_params())
print(
"BEFORE DT Training w/bin score mean: {:.2f}".format(
base_results["train_score"].mean() * 100
)
)
print(
"BEFORE DT Test w/bin score mean: {:.2f}".format(
base_results["test_score"].mean() * 100
)
)
print(
"BEFORE DT Test w/bin score 3*std: +/- {:.2f}".format(
base_results["test_score"].std() * 100 * 3
)
)
# print("BEFORE DT Test w/bin set score min: {:.2f}". format(base_results['test_score'].min()*100))
print("-" * 10)
param_grid = {
"criterion": [
"gini",
"entropy",
], # scoring methodology; two supported formulas for calculating information gain - default is gini
# 'splitter': ['best', 'random'], #splitting methodology; two supported strategies - default is best
"max_depth": [2, 4, 6, 8, 10, None], # max depth tree can grow; default is none
# 'min_samples_split': [2,5,10,.03,.05], #minimum subset size BEFORE new split (fraction is % of total); default is 2
# 'min_samples_leaf': [1,5,10,.03,.05], #minimum subset size AFTER new split split (fraction is % of total); default is 1
# 'max_features': [None, 'auto'], #max features to consider when performing split; default none or all
"random_state": [
0
], # seed or control random number generator: https://www.quora.com/What-is-seed-in-random-number-generation
}
# choose best model with grid_search
tune_model = model_selection.GridSearchCV(
tree.DecisionTreeClassifier(),
param_grid=param_grid,
scoring="roc_auc",
cv=cv_split,
return_train_score=True,
)
tune_model.fit(data1[data1_x_bin], data1[Target])
# print(tune_model.cv_results_.keys())
# print(tune_model.cv_results_['params'])
print("AFTER DT Parameters: ", tune_model.best_params_)
# print(tune_model.cv_results_['mean_train_score'])
print(
"AFTER DT Training w/bin score mean: {:.2f}".format(
tune_model.cv_results_["mean_train_score"][tune_model.best_index_] * 100
)
)
# print(tune_model.cv_results_['mean_test_score'])
print(
"AFTER DT Test w/bin score mean: {:.2f}".format(
tune_model.cv_results_["mean_test_score"][tune_model.best_index_] * 100
)
)
print(
"AFTER DT Test w/bin score 3*std: +/- {:.2f}".format(
tune_model.cv_results_["std_test_score"][tune_model.best_index_] * 100 * 3
)
)
print("-" * 10)
# base model
print("BEFORE DT RFE Training Shape Old: ", data1[data1_x_bin].shape)
print("BEFORE DT RFE Training Columns Old: ", data1[data1_x_bin].columns.values)
print(
"BEFORE DT RFE Training w/bin score mean: {:.2f}".format(
base_results["train_score"].mean() * 100
)
)
print(
"BEFORE DT RFE Test w/bin score mean: {:.2f}".format(
base_results["test_score"].mean() * 100
)
)
print(
"BEFORE DT RFE Test w/bin score 3*std: +/- {:.2f}".format(
base_results["test_score"].std() * 100 * 3
)
)
print("-" * 10)
# feature selection
dtree_rfe = feature_selection.RFECV(dtree, step=1, scoring="accuracy", cv=cv_split)
dtree_rfe.fit(data1[data1_x_bin], data1[Target])
# transform x&y to reduced features and fit new model
X_rfe = data1[data1_x_bin].columns.values[dtree_rfe.get_support()]
rfe_results = model_selection.cross_validate(
dtree, data1[X_rfe], data1[Target], cv=cv_split, return_train_score=True
)
print("AFTER DT RFE Training Shape New: ", data1[X_rfe].shape)
print("AFTER DT RFE Training Columns New: ", X_rfe)
print(
"AFTER DT RFE Training w/bin score mean: {:.2f}".format(
rfe_results["train_score"].mean() * 100
)
)
print(
"AFTER DT RFE Test w/bin score mean: {:.2f}".format(
rfe_results["test_score"].mean() * 100
)
)
print(
"AFTER DT RFE Test w/bin score 3*std: +/- {:.2f}".format(
rfe_results["test_score"].std() * 100 * 3
)
)
print("-" * 10)
# tune rfe model
rfe_tune_model = model_selection.GridSearchCV(
tree.DecisionTreeClassifier(),
param_grid=param_grid,
scoring="roc_auc",
cv=cv_split,
return_train_score=True,
)
rfe_tune_model.fit(data1[X_rfe], data1[Target])
# print(rfe_tune_model.cv_results_.keys())
# print(rfe_tune_model.cv_results_['params'])
print("AFTER DT RFE Tuned Parameters: ", rfe_tune_model.best_params_)
# print(rfe_tune_model.cv_results_['mean_train_score'])
print(
"AFTER DT RFE Tuned Training w/bin score mean: {:.2f}".format(
rfe_tune_model.cv_results_["mean_train_score"][tune_model.best_index_] * 100
)
)
# print(rfe_tune_model.cv_results_['mean_test_score'])
print(
"AFTER DT RFE Tuned Test w/bin score mean: {:.2f}".format(
rfe_tune_model.cv_results_["mean_test_score"][tune_model.best_index_] * 100
)
)
print(
"AFTER DT RFE Tuned Test w/bin score 3*std: +/- {:.2f}".format(
rfe_tune_model.cv_results_["std_test_score"][tune_model.best_index_] * 100 * 3
)
)
print("-" * 10)
# Graph MLA version of Decision Tree
import graphviz
dot_data = tree.export_graphviz(
dtree,
out_file=None,
feature_names=data1_x_bin,
class_names=True,
filled=True,
rounded=True,
)
graph = graphviz.Source(dot_data)
graph
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605607.ipynb
| null | null |
[{"Id": 69605607, "ScriptId": 18914721, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7954964, "CreationDate": "08/02/2021 05:41:43", "VersionNumber": 3.0, "Title": "Practice:Titanic_Survival", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 539.0, "LinesInsertedFromPrevious": 261.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 278.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import sys, matplotlib, IPython, sklearn, random, time, warnings
import scipy as sp
from IPython import display
from subprocess import check_output
print("Python version: {}".format(sys.version))
print("pandas version: {}".format(pd.__version__))
print("NumPy version: {}".format(np.__version__))
print("matplotlib version: {}".format(matplotlib.__version__))
print("SciPy version: {}".format(sp.__version__))
print("IPython version: {}".format(IPython.__version__))
print("scikit-learn version: {}".format(sklearn.__version__))
warnings.filterwarnings("ignore")
print("-" * 25)
print(check_output(["ls", "../input/titanic/"]).decode("utf8"))
# model algorithems
from sklearn import (
svm,
tree,
linear_model,
neighbors,
naive_bayes,
ensemble,
discriminant_analysis,
gaussian_process,
)
from xgboost import XGBClassifier
# model helpers
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
from sklearn import feature_selection
from sklearn import model_selection
from sklearn import metrics
# visualization
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.pylab as pylab
import seaborn as sns
from pandas.plotting import scatter_matrix
# show plot in Jupyter Notebook
mpl.style.use("ggplot")
sns.set_style("white")
pylab.rcParams["figure.figsize"] = 12, 8
data_raw = pd.read_csv("../input/titanic/train.csv")
data_test = pd.read_csv("../input/titanic/test.csv")
data1 = data_raw.copy(deep=True)
data_cleaner = [data1, data_test]
# print(data_raw.info())
# data_raw.head()
# data_raw.tail()
print("sex classcifications:", set(data_raw["Sex"]))
print("Age Range: {}~{}".format(min(data_raw["Age"]), max(data_raw["Age"])))
print("Fare Range: {}~{}".format(min(data_raw["Fare"]), max(data_raw["Fare"])))
data_raw.sample(5)
print("Train columns with null values:\n", data1.isnull().sum())
print("-" * 10)
print("Test columns with null values:\n", data_test.isnull().sum())
print("-" * 10)
data_raw.describe(include="all")
for dataset in data_cleaner:
dataset["Age"].fillna(dataset["Age"].median(), inplace=True)
dataset["Embarked"].fillna(dataset["Embarked"].mode()[0], inplace=True)
dataset["Fare"].fillna(dataset["Fare"].median(), inplace=True)
drop_column = ["PassengerId", "Cabin", "Ticket"]
data1.drop(drop_column, axis=1, inplace=True)
print("Train columns with null values:\n", data1.isnull().sum())
print("-" * 10)
print("Test columns with null values:\n", data_test.isnull().sum())
print("-" * 10)
for dataset in data_cleaner:
dataset["FamilySize"] = dataset["SibSp"] + dataset["Parch"] + 1
dataset["IsAlone"] = 1
dataset["IsAlone"].loc[dataset["FamilySize"] > 1] = 0
dataset["Title"] = (
dataset["Name"].str.split(", ", expand=True)[1].str.split(".", expand=True)[0]
)
dataset["FareBin"] = pd.qcut(dataset["Fare"], 4)
dataset["AgeBin"] = pd.cut(dataset["Age"].astype(int), 5)
data1["Title"].value_counts()
stat_min = 10
# create a true/false series with title name as index
title_names = data1["Title"].value_counts() < stat_min
data1["Title"] = data1["Title"].apply(
lambda x: "Misc" if title_names.loc[x] == True else x
)
print(data1["Title"].value_counts())
print("-" * 10)
# preview data
data1.info()
data_test.info()
data1.sample(10)
label = LabelEncoder()
for dataset in data_cleaner:
dataset["Sex_Code"] = label.fit_transform(dataset["Sex"])
dataset["Embarked_Code"] = label.fit_transform(dataset["Embarked"])
dataset["Title_Code"] = label.fit_transform(dataset["Title"])
dataset["AgeBin_Code"] = label.fit_transform(dataset["AgeBin"])
dataset["FareBin_Code"] = label.fit_transform(dataset["FareBin"])
data1.sample(5)
Target = ["Survived"]
## feature selection
# feature names for charts
data1_x = [
"Sex",
"Pclass",
"Embarked",
"Title",
"SibSp",
"Parch",
"Age",
"Fare",
"FamilySize",
"IsAlone",
]
# feature names for codes
data1_x_calc = [
"Sex_Code",
"Pclass",
"Embarked_Code",
"Title_Code",
"SibSp",
"Parch",
"Age",
"Fare",
]
data1_xy = Target + data1_x
print("Original X Y: ", data1_xy, "\n")
# features w/ bins
data1_x_bin = [
"Sex_Code",
"Pclass",
"Embarked_Code",
"Title_Code",
"FamilySize",
"AgeBin_Code",
"FareBin_Code",
]
data1_xy_bin = Target + data1_x_bin
print("Bin X Y: ", data1_xy_bin, "\n")
# dummy features
data1_dummy = pd.get_dummies(data1[data1_x])
data1_x_dummy = data1_dummy.columns.tolist()
data1_xy_dummy = Target + data1_x_dummy
print("Dummy X Y: ", data1_xy_dummy, "\n")
data1_dummy.head()
print("Train columns with null values: \n", data1.isnull().sum())
print("-" * 10)
print(data1.info())
print("-" * 10)
print("Test columns with null values: \n", data_test.isnull().sum())
print("-" * 10)
print(data_test.info())
print("-" * 10)
data_raw.describe(include="all")
train1_x, crossVal1_x, train1_y, crossVal1_y = model_selection.train_test_split(
data1[data1_x_calc], data1[Target], random_state=0
)
(
train1_x_bin,
crossVal1_x_bin,
train1_y_bin,
crossVal1_y_bin,
) = model_selection.train_test_split(data1[data1_x_bin], data1[Target], random_state=0)
(
train1_x_dummy,
crossVal1_x_dummy,
train1_y_dummy,
crossVal1_y_dummy,
) = model_selection.train_test_split(
data1_dummy[data1_x_dummy], data1[Target], random_state=0
)
print("Data1 Shape: {}".format(data1.shape))
print("Train1 Shape: {}".format(train1_x.shape))
print("crossVal1 Shape: {}".format(crossVal1_x.shape))
train1_x_bin.head()
for x in data1_x:
# don't do it on float values, otherwise too many groups
if data1[x].dtype != "float64":
print("Survival correlation by:", x)
print(data1[[x, Target[0]]].groupby(x, as_index=False).mean())
print("-" * 10, "\n")
print(pd.crosstab(data1["Title"], data1[Target[0]]))
plt.figure(figsize=[16, 12])
plt.subplot(231)
# plt.grid(True,axis='y')
plt.boxplot(x=data1["Fare"], showmeans=True, meanline=True)
plt.title("Fare Boxplot")
plt.ylabel("Fare($)")
plt.subplot(232)
plt.boxplot(data1["Age"], showmeans=True, meanline=True)
plt.title("Age Boxplot")
plt.ylabel("Age (Years)")
plt.subplot(233)
plt.boxplot(data1["FamilySize"], showmeans=True, meanline=True)
plt.title("Family Size Boxplot")
plt.ylabel("Family Size (#)")
plt.subplot(234)
plt.hist(
x=[data1[data1["Survived"] == 1]["Fare"], data1[data1["Survived"] == 0]["Fare"]],
stacked=False,
color=["g", "r"],
label=["Survived", "Dead"],
)
plt.title("Fare Histogram by Survival")
plt.xlabel("Fare ($)")
plt.ylabel("# of Passengers")
plt.legend()
plt.subplot(235)
plt.hist(
x=[data1[data1["Survived"] == 1]["Age"], data1[data1["Survived"] == 0]["Age"]],
stacked=False,
color=["g", "r"],
label=["Survived", "Dead"],
)
plt.title("Age Histogram by Survival")
plt.xlabel("Age (years)")
plt.ylabel("# of Passengers")
plt.legend()
plt.subplot(236)
plt.hist(
x=[
data1[data1["Survived"] == 1]["FamilySize"],
data1[data1["Survived"] == 0]["FamilySize"],
],
bins=np.arange(data1["FamilySize"].min() - 0.5, data1["FamilySize"].max() + 0.5),
stacked=False,
color=["g", "r"],
label=["Survived", "Dead"],
)
plt.title("FamilySize Histogram by Survival")
plt.xlabel("Family Size (#)")
plt.ylabel("# of Passengers")
plt.legend()
fig, axs = plt.subplots(2, 3, figsize=(16, 12))
sns.barplot(x="Embarked", y="Survived", data=data1, ax=axs[0, 0])
sns.barplot(x="Pclass", y="Survived", data=data1, ax=axs[0, 1])
sns.barplot(x="IsAlone", y="Survived", order=[1, 0], data=data1, ax=axs[0, 2])
sns.pointplot(x="FareBin", y="Survived", data=data1, ax=axs[1, 0])
sns.pointplot(x="AgeBin", y="Survived", data=data1, ax=axs[1, 1])
sns.pointplot(x="FamilySize", y="Survived", data=data1, ax=axs[1, 2])
fig, (axis1, axis2, axis3) = plt.subplots(1, 3, figsize=(16, 8))
sns.boxplot(x="Pclass", y="Fare", hue="Survived", data=data1, ax=axis1)
axis1.set_title("Pclass vs Fare Survival Comparison")
sns.violinplot(x="Pclass", y="Age", hue="Survived", data=data1, split=True, ax=axis2)
axis2.set_title("Pclass vs Age Survival Comparison")
sns.boxplot(x="Pclass", y="FamilySize", hue="Survived", data=data1, ax=axis3)
axis3.set_title("Pclass vs FamilySize Survival Comparison")
fig, qaxis = plt.subplots(2, 3, figsize=(14, 12))
sns.barplot(x="Sex", y="Survived", hue="Embarked", data=data1, ax=qaxis[0, 0])
qaxis[0, 0].set_title("Sex vs Embarked Survival Comparison")
sns.barplot(x="Sex", y="Survived", hue="Pclass", data=data1, ax=qaxis[0, 1])
qaxis[0, 1].set_title("Sex vs Pclass Survival Comparison")
sns.barplot(x="Sex", y="Survived", hue="IsAlone", data=data1, ax=qaxis[0, 2])
qaxis[0, 2].set_title("Sex vs IsAlone Survival Comparison")
sns.barplot(x="Embarked", y="Survived", hue="Sex", data=data1, ax=qaxis[1, 0])
qaxis[1, 0].set_title("Embarked vs Sex Survival Comparison")
sns.barplot(x="Pclass", y="Survived", hue="Sex", data=data1, ax=qaxis[1, 1])
qaxis[1, 1].set_title("Pclass vs Sex Survival Comparison")
sns.barplot(x="IsAlone", y="Survived", hue="Sex", data=data1, ax=qaxis[1, 2])
qaxis[1, 2].set_title("IsAlone vs Sex Survival Comparison")
fig, (maxis1, maxis2) = plt.subplots(1, 2, figsize=(16, 8))
sns.pointplot(
x="FamilySize",
y="Survived",
hue="Sex",
data=data1,
palette={"male": "blue", "female": "pink"},
markers=["*", "o"],
linestyles=["-", "--"],
ax=maxis1,
)
sns.pointplot(
x="AgeBin",
y="Survived",
hue="Sex",
data=data1,
palette={"male": "blue", "female": "pink"},
markers=["*", "o"],
linestyles=["-", "--"],
ax=maxis2,
)
# use facetgrid in seaborn for multi-plot
e = sns.FacetGrid(data1, col="Embarked")
e.map(sns.pointplot, "Pclass", "Survived", "Sex", ci=95.0, palette="deep")
e.add_legend()
a = sns.FacetGrid(data1, hue="Survived", aspect=3)
# kde - kernal density estimation
a.map(sns.kdeplot, "Age", shade=True)
a.set(xlim=(0, data1["Age"].max()))
a.add_legend()
h = sns.FacetGrid(data1, row="Sex", col="Pclass", hue="Survived")
# alpha is transparency
h.map(plt.hist, "Age", alpha=0.5)
h.add_legend()
## pair plots of the entire dataset
pp = sns.pairplot(
data1,
hue="Survived",
palette="deep",
size=1.2,
diag_kind="kde",
diag_kws=dict(shade=True),
plot_kws=dict(s=10),
)
pp.set(xticklabels=[])
# correlation heatmap of dataset
def correlation_heatmap(df):
_, ax = plt.subplots(figsize=(14, 12))
colormap = sns.diverging_palette(220, 10, as_cmap=True)
_ = sns.heatmap(
df.corr(),
cmap=colormap,
square=True,
cbar_kws={"shrink": 0.9},
ax=ax,
annot=True,
linewidths=0.1,
vmax=1.0,
linecolor="white",
annot_kws={"fontsize": 12},
)
plt.title("Pearson Correlation of Features", y=1.05, size=15)
correlation_heatmap(data1)
# ML algo selection
MLA = [
## Ensemble Methods
ensemble.AdaBoostClassifier(),
ensemble.BaggingClassifier(),
ensemble.ExtraTreesClassifier(),
ensemble.GradientBoostingClassifier(),
ensemble.RandomForestClassifier(),
## Gaussian Processes
gaussian_process.GaussianProcessClassifier(),
## GLM
linear_model.LogisticRegressionCV(),
linear_model.PassiveAggressiveClassifier(),
linear_model.RidgeClassifierCV(),
linear_model.SGDClassifier(),
linear_model.Perceptron(),
## Naives Bayes
naive_bayes.BernoulliNB(),
naive_bayes.GaussianNB(),
## Nearest Neighbor
neighbors.KNeighborsClassifier(),
## SVM
svm.SVC(probability=True),
svm.NuSVC(probability=True),
svm.LinearSVC(),
## Trees
tree.DecisionTreeClassifier(),
tree.ExtraTreeClassifier(),
## Discriminant Analysis
discriminant_analysis.LinearDiscriminantAnalysis(),
discriminant_analysis.QuadraticDiscriminantAnalysis(),
# xgboost: https://xgboost.readthedocs.io/en/latest/
XGBClassifier(),
]
# split dataset
cv_split = model_selection.ShuffleSplit(
n_splits=10, test_size=0.3, train_size=0.6, random_state=0
)
# create table to compare MLA metrics
MLA_columns = [
"MLA Name",
"MLA Parameters",
"MLA Train Accuracy Mean",
"MLA Test Accuracy Mean",
"MLA Test Accuracy 3*STD",
"MLA Time",
]
MLA_compare = pd.DataFrame(columns=MLA_columns)
MLA_predict = data1[Target]
# loop through the MLA list
row_index = 0
for alg in MLA:
MLA_name = alg.__class__.__name__
MLA_compare.loc[row_index, "MLA Name"] = MLA_name
MLA_compare.loc[row_index, "MLA Parameters"] = str(alg.get_params())
cv_results = model_selection.cross_validate(
alg, data1[data1_x_bin], data1[Target], cv=cv_split, return_train_score=True
)
# if row_index == 0:
# print(cv_results)
MLA_compare.loc[row_index, "MLA Time"] = cv_results["fit_time"].mean()
MLA_compare.loc[row_index, "MLA Train Accuracy Mean"] = cv_results[
"train_score"
].mean()
MLA_compare.loc[row_index, "MLA Test Accuracy Mean"] = cv_results[
"test_score"
].mean()
MLA_compare.loc[row_index, "MLA Test Accuracy 3*STD"] = (
cv_results["test_score"].std() * 3
)
## save MLA predictions for later useage
alg.fit(data1[data1_x_bin], data1[Target])
MLA_predict[MLA_name] = alg.predict(data1[data1_x_bin])
row_index += 1
MLA_compare.sort_values(by=["MLA Test Accuracy Mean"], ascending=False, inplace=True)
MLA_compare
sns.barplot(x="MLA Test Accuracy Mean", y="MLA Name", data=MLA_compare, color="m")
# prettify using pyplot: https://matplotlib.org/api/pyplot_api.html
plt.title("Machine Learning Algorithm Accuracy Score \n")
plt.xlabel("Accuracy Score (%)")
plt.ylabel("Algorithm")
# coin flip model with random 1/survived 0/died
# iterate over dataFrame rows as (index, Series) pairs: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.iterrows.html
data1["Random_Predict"] = 0 # assume prediction wrong
for index, row in data1.iterrows():
# random number generator: https://docs.python.org/2/library/random.html
if random.random() > 0.5: # Random float x, 0.0 <= x < 1.0
data1["Random_Predict"][index] = 1 # predict survived/1
else:
data1["Random_Predict"][index] = 0 # predict died/0
# score random guess of survival. Use shortcut 1 = Right Guess and 0 = Wrong Guess
# the mean of the column will then equal the accuracy
data1["Random_Score"] = 0 # assume prediction wrong
data1.loc[
(data1["Survived"] == data1["Random_Predict"]), "Random_Score"
] = 1 # set to 1 for correct prediction
print("Coin Flip Model Accuracy: {:.2f}%".format(data1["Random_Score"].mean() * 100))
# we can also use scikit's accuracy_score function to save us a few lines of code
print(
"Coin Flip Model Accuracy w/SciKit: {:.2f}%".format(
metrics.accuracy_score(data1["Survived"], data1["Random_Predict"]) * 100
)
)
# group by or pivot table
pivot_female = (
data1[data1.Sex == "female"]
.groupby(["Sex", "Pclass", "Embarked", "FareBin"])["Survived"]
.mean()
)
print("Survival Decision Tree w/Female Node: \n", pivot_female)
pivot_male = data1[data1.Sex == "male"].groupby(["Sex", "Title"])["Survived"].mean()
print("\n\nSurvival Decision Tree w/Male Node: \n", pivot_male)
# handmade data model using brain power (and Microsoft Excel Pivot Tables for quick calculations)
def mytree(df):
# initialize table to store predictions
Model = pd.DataFrame(data={"Predict": []})
male_title = ["Master"] # survived titles
for index, row in df.iterrows():
# Question 1: Were you on the Titanic; majority died
Model.loc[index, "Predict"] = 0
# Question 2: Are you female; majority survived
if df.loc[index, "Sex"] == "female":
Model.loc[index, "Predict"] = 1
# Question 3A Female - Class and Question 4 Embarked gain minimum information
# Question 5B Female - FareBin; set anything less than .5 in female node decision tree back to 0
if (
(df.loc[index, "Sex"] == "female")
& (df.loc[index, "Pclass"] == 3)
& (df.loc[index, "Embarked"] == "S")
& (df.loc[index, "Fare"] > 8)
):
Model.loc[index, "Predict"] = 0
# Question 3B Male: Title; set anything greater than .5 to 1 for majority survived
if (df.loc[index, "Sex"] == "male") & (df.loc[index, "Title"] in male_title):
Model.loc[index, "Predict"] = 1
return Model
# model data
Tree_Predict = mytree(data1)
# print matrix
print(
"Decision Tree Model Accuracy/Precision Score: {:.2f}%\n".format(
metrics.accuracy_score(data1["Survived"], Tree_Predict) * 100
)
)
# Accuracy Summary Report
print(metrics.classification_report(data1["Survived"], Tree_Predict))
import itertools
def my_plot_confusion_mtrix(
cm, classes, normalize=False, title="Confusion matrix", cmap=plt.cm.Blues
):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print("Confusion matrix, without normalization")
print(cm)
plt.imshow(cm, interpolation="nearest", cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes)
plt.yticks(tick_marks, classes)
fmt = ".2f" if normalize else "d"
thresh = cm.max() / 2.0
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(
j,
i,
format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black",
)
plt.tight_layout()
plt.ylabel("True label")
plt.xlabel("Predicted label")
# Compute confusion matrix
cnf_matrix = metrics.confusion_matrix(data1["Survived"], Tree_Predict)
np.set_printoptions(precision=2)
class_names = ["Dead", "Survived"]
# Plot non-normalized confusion matrix
plt.figure()
my_plot_confusion_mtrix(
cnf_matrix, class_names, False, "Confusion matrix, without normalization"
)
# Plot normalized confusion matrix
plt.figure()
my_plot_confusion_mtrix(cnf_matrix, class_names, True, "Normalized confusion matrix")
# base model
dtree = tree.DecisionTreeClassifier(random_state=0)
base_results = model_selection.cross_validate(
dtree, data1[data1_x_bin], data1[Target], cv=cv_split, return_train_score=True
)
dtree.fit(data1[data1_x_bin], data1[Target])
print("BEFORE DT Parameters: ", dtree.get_params())
print(
"BEFORE DT Training w/bin score mean: {:.2f}".format(
base_results["train_score"].mean() * 100
)
)
print(
"BEFORE DT Test w/bin score mean: {:.2f}".format(
base_results["test_score"].mean() * 100
)
)
print(
"BEFORE DT Test w/bin score 3*std: +/- {:.2f}".format(
base_results["test_score"].std() * 100 * 3
)
)
# print("BEFORE DT Test w/bin set score min: {:.2f}". format(base_results['test_score'].min()*100))
print("-" * 10)
param_grid = {
"criterion": [
"gini",
"entropy",
], # scoring methodology; two supported formulas for calculating information gain - default is gini
# 'splitter': ['best', 'random'], #splitting methodology; two supported strategies - default is best
"max_depth": [2, 4, 6, 8, 10, None], # max depth tree can grow; default is none
# 'min_samples_split': [2,5,10,.03,.05], #minimum subset size BEFORE new split (fraction is % of total); default is 2
# 'min_samples_leaf': [1,5,10,.03,.05], #minimum subset size AFTER new split split (fraction is % of total); default is 1
# 'max_features': [None, 'auto'], #max features to consider when performing split; default none or all
"random_state": [
0
], # seed or control random number generator: https://www.quora.com/What-is-seed-in-random-number-generation
}
# choose best model with grid_search
tune_model = model_selection.GridSearchCV(
tree.DecisionTreeClassifier(),
param_grid=param_grid,
scoring="roc_auc",
cv=cv_split,
return_train_score=True,
)
tune_model.fit(data1[data1_x_bin], data1[Target])
# print(tune_model.cv_results_.keys())
# print(tune_model.cv_results_['params'])
print("AFTER DT Parameters: ", tune_model.best_params_)
# print(tune_model.cv_results_['mean_train_score'])
print(
"AFTER DT Training w/bin score mean: {:.2f}".format(
tune_model.cv_results_["mean_train_score"][tune_model.best_index_] * 100
)
)
# print(tune_model.cv_results_['mean_test_score'])
print(
"AFTER DT Test w/bin score mean: {:.2f}".format(
tune_model.cv_results_["mean_test_score"][tune_model.best_index_] * 100
)
)
print(
"AFTER DT Test w/bin score 3*std: +/- {:.2f}".format(
tune_model.cv_results_["std_test_score"][tune_model.best_index_] * 100 * 3
)
)
print("-" * 10)
# base model
print("BEFORE DT RFE Training Shape Old: ", data1[data1_x_bin].shape)
print("BEFORE DT RFE Training Columns Old: ", data1[data1_x_bin].columns.values)
print(
"BEFORE DT RFE Training w/bin score mean: {:.2f}".format(
base_results["train_score"].mean() * 100
)
)
print(
"BEFORE DT RFE Test w/bin score mean: {:.2f}".format(
base_results["test_score"].mean() * 100
)
)
print(
"BEFORE DT RFE Test w/bin score 3*std: +/- {:.2f}".format(
base_results["test_score"].std() * 100 * 3
)
)
print("-" * 10)
# feature selection
dtree_rfe = feature_selection.RFECV(dtree, step=1, scoring="accuracy", cv=cv_split)
dtree_rfe.fit(data1[data1_x_bin], data1[Target])
# transform x&y to reduced features and fit new model
X_rfe = data1[data1_x_bin].columns.values[dtree_rfe.get_support()]
rfe_results = model_selection.cross_validate(
dtree, data1[X_rfe], data1[Target], cv=cv_split, return_train_score=True
)
print("AFTER DT RFE Training Shape New: ", data1[X_rfe].shape)
print("AFTER DT RFE Training Columns New: ", X_rfe)
print(
"AFTER DT RFE Training w/bin score mean: {:.2f}".format(
rfe_results["train_score"].mean() * 100
)
)
print(
"AFTER DT RFE Test w/bin score mean: {:.2f}".format(
rfe_results["test_score"].mean() * 100
)
)
print(
"AFTER DT RFE Test w/bin score 3*std: +/- {:.2f}".format(
rfe_results["test_score"].std() * 100 * 3
)
)
print("-" * 10)
# tune rfe model
rfe_tune_model = model_selection.GridSearchCV(
tree.DecisionTreeClassifier(),
param_grid=param_grid,
scoring="roc_auc",
cv=cv_split,
return_train_score=True,
)
rfe_tune_model.fit(data1[X_rfe], data1[Target])
# print(rfe_tune_model.cv_results_.keys())
# print(rfe_tune_model.cv_results_['params'])
print("AFTER DT RFE Tuned Parameters: ", rfe_tune_model.best_params_)
# print(rfe_tune_model.cv_results_['mean_train_score'])
print(
"AFTER DT RFE Tuned Training w/bin score mean: {:.2f}".format(
rfe_tune_model.cv_results_["mean_train_score"][tune_model.best_index_] * 100
)
)
# print(rfe_tune_model.cv_results_['mean_test_score'])
print(
"AFTER DT RFE Tuned Test w/bin score mean: {:.2f}".format(
rfe_tune_model.cv_results_["mean_test_score"][tune_model.best_index_] * 100
)
)
print(
"AFTER DT RFE Tuned Test w/bin score 3*std: +/- {:.2f}".format(
rfe_tune_model.cv_results_["std_test_score"][tune_model.best_index_] * 100 * 3
)
)
print("-" * 10)
# Graph MLA version of Decision Tree
import graphviz
dot_data = tree.export_graphviz(
dtree,
out_file=None,
feature_names=data1_x_bin,
class_names=True,
filled=True,
rounded=True,
)
graph = graphviz.Source(dot_data)
graph
| false | 0 | 8,245 | 0 | 8,245 | 8,245 |
||
69605479
|
<jupyter_start><jupyter_text>Credit Card Fraud Detection
Context
---------
It is important that credit card companies are able to recognize fraudulent credit card transactions so that customers are not charged for items that they did not purchase.
Content
---------
The dataset contains transactions made by credit cards in September 2013 by European cardholders.
This dataset presents transactions that occurred in two days, where we have 492 frauds out of 284,807 transactions. The dataset is highly unbalanced, the positive class (frauds) account for 0.172% of all transactions.
It contains only numerical input variables which are the result of a PCA transformation. Unfortunately, due to confidentiality issues, we cannot provide the original features and more background information about the data. Features V1, V2, ... V28 are the principal components obtained with PCA, the only features which have not been transformed with PCA are 'Time' and 'Amount'. Feature 'Time' contains the seconds elapsed between each transaction and the first transaction in the dataset. The feature 'Amount' is the transaction Amount, this feature can be used for example-dependant cost-sensitive learning. Feature 'Class' is the response variable and it takes value 1 in case of fraud and 0 otherwise.
Given the class imbalance ratio, we recommend measuring the accuracy using the Area Under the Precision-Recall Curve (AUPRC). Confusion matrix accuracy is not meaningful for unbalanced classification.
Update (03/05/2021)
---------
A simulator for transaction data has been released as part of the practical handbook on Machine Learning for Credit Card Fraud Detection - https://fraud-detection-handbook.github.io/fraud-detection-handbook/Chapter_3_GettingStarted/SimulatedDataset.html. We invite all practitioners interested in fraud detection datasets to also check out this data simulator, and the methodologies for credit card fraud detection presented in the book.
Acknowledgements
---------
The dataset has been collected and analysed during a research collaboration of Worldline and the Machine Learning Group (http://mlg.ulb.ac.be) of ULB (Université Libre de Bruxelles) on big data mining and fraud detection.
More details on current and past projects on related topics are available on [https://www.researchgate.net/project/Fraud-detection-5][1] and the page of the [DefeatFraud][2] project
Please cite the following works:
Andrea Dal Pozzolo, Olivier Caelen, Reid A. Johnson and Gianluca Bontempi. [Calibrating Probability with Undersampling for Unbalanced Classification.][3] In Symposium on Computational Intelligence and Data Mining (CIDM), IEEE, 2015
Dal Pozzolo, Andrea; Caelen, Olivier; Le Borgne, Yann-Ael; Waterschoot, Serge; Bontempi, Gianluca. [Learned lessons in credit card fraud detection from a practitioner perspective][4], Expert systems with applications,41,10,4915-4928,2014, Pergamon
Dal Pozzolo, Andrea; Boracchi, Giacomo; Caelen, Olivier; Alippi, Cesare; Bontempi, Gianluca. [Credit card fraud detection: a realistic modeling and a novel learning strategy,][5] IEEE transactions on neural networks and learning systems,29,8,3784-3797,2018,IEEE
Dal Pozzolo, Andrea [Adaptive Machine learning for credit card fraud detection][6] ULB MLG PhD thesis (supervised by G. Bontempi)
Carcillo, Fabrizio; Dal Pozzolo, Andrea; Le Borgne, Yann-Aël; Caelen, Olivier; Mazzer, Yannis; Bontempi, Gianluca. [Scarff: a scalable framework for streaming credit card fraud detection with Spark][7], Information fusion,41, 182-194,2018,Elsevier
Carcillo, Fabrizio; Le Borgne, Yann-Aël; Caelen, Olivier; Bontempi, Gianluca. [Streaming active learning strategies for real-life credit card fraud detection: assessment and visualization,][8] International Journal of Data Science and Analytics, 5,4,285-300,2018,Springer International Publishing
Bertrand Lebichot, Yann-Aël Le Borgne, Liyun He, Frederic Oblé, Gianluca Bontempi [Deep-Learning Domain Adaptation Techniques for Credit Cards Fraud Detection](https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection), INNSBDDL 2019: Recent Advances in Big Data and Deep Learning, pp 78-88, 2019
Fabrizio Carcillo, Yann-Aël Le Borgne, Olivier Caelen, Frederic Oblé, Gianluca Bontempi [Combining Unsupervised and Supervised Learning in Credit Card Fraud Detection ](https://www.researchgate.net/publication/333143698_Combining_Unsupervised_and_Supervised_Learning_in_Credit_Card_Fraud_Detection) Information Sciences, 2019
Yann-Aël Le Borgne, Gianluca Bontempi [Reproducible machine Learning for Credit Card Fraud Detection - Practical Handbook ](https://www.researchgate.net/publication/351283764_Machine_Learning_for_Credit_Card_Fraud_Detection_-_Practical_Handbook)
Bertrand Lebichot, Gianmarco Paldino, Wissam Siblini, Liyun He, Frederic Oblé, Gianluca Bontempi [Incremental learning strategies for credit cards fraud detection](https://www.researchgate.net/publication/352275169_Incremental_learning_strategies_for_credit_cards_fraud_detection), IInternational Journal of Data Science and Analytics
[1]: https://www.researchgate.net/project/Fraud-detection-5
[2]: https://mlg.ulb.ac.be/wordpress/portfolio_page/defeatfraud-assessment-and-validation-of-deep-feature-engineering-and-learning-solutions-for-fraud-detection/
[3]: https://www.researchgate.net/publication/283349138_Calibrating_Probability_with_Undersampling_for_Unbalanced_Classification
[4]: https://www.researchgate.net/publication/260837261_Learned_lessons_in_credit_card_fraud_detection_from_a_practitioner_perspective
[5]: https://www.researchgate.net/publication/319867396_Credit_Card_Fraud_Detection_A_Realistic_Modeling_and_a_Novel_Learning_Strategy
[6]: http://di.ulb.ac.be/map/adalpozz/pdf/Dalpozzolo2015PhD.pdf
[7]: https://www.researchgate.net/publication/319616537_SCARFF_a_Scalable_Framework_for_Streaming_Credit_Card_Fraud_Detection_with_Spark
[8]: https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection
Kaggle dataset identifier: creditcardfraud
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from matplotlib import gridspec
import matplotlib.pyplot as plt
import seaborn as sns
from pylab import rcParams
rcParams["figure.figsize"] = 15, 10
from imblearn.over_sampling import SMOTE
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import RobustScaler
from sklearn.manifold import TSNE
from sklearn.linear_model import LogisticRegression
import xgboost as xgb
from sklearn.metrics import (
confusion_matrix,
classification_report,
accuracy_score,
precision_recall_curve,
roc_auc_score,
)
import eli5
from eli5.sklearn import PermutationImportance
from pdpbox import pdp, get_dataset, info_plots
import shap
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("../input/creditcardfraud/creditcard.csv")
df.head()
df.describe()
# The Class column in looking highly skewed. We will see how much imbalance there really is.
df.isna().sum()
# No Null values, lesser work to do!
plt.subplot(121)
plt.pie(
x=df.groupby(["Class"]).Class.count().to_list(),
labels=["Left", "Right"],
autopct="%1.2f%%",
explode=(0, 0.2),
)
plt.subplot(122)
sns.countplot(data=df, x="Class")
fraudnt, fraud = df.Class.value_counts()
plt.text(0, fraudnt // 2, fraudnt, fontsize=20, horizontalalignment="center")
plt.text(1, fraud // 2, fraud, fontsize=20, horizontalalignment="center")
# Using such an imbalanced dataset for machine learning can lead to very poor performance in real life. We will see some ways that we can mitigate this issue later in this notebook.
features = df.iloc[:, 0:29].columns
plt.figure(figsize=(15, 29 * 4))
gs = gridspec.GridSpec(29, 1)
for i, c in enumerate(df[features]):
ax = plt.subplot(gs[i])
sns.distplot(df[c][df.Class == 1], bins=50, label="Fraud")
sns.distplot(df[c][df.Class == 0], bins=50, label="Valid")
ax.legend()
ax.set_xlabel("")
ax.set_title("histogram of feature: " + str(c))
plt.show()
# bla bla
plt.subplot(211)
sns.distplot(df.Amount.values, kde=True)
plt.subplot(212)
sns.histplot(df.Time.values, kde=True)
# The amount variable is highly left skewed as most people take part in low volume banking transactions but the time variable has visible seasonality indicating peak times and breaks that are part of a human's daily routine.
plt.subplot(221)
sns.distplot(df[df.Class == 0].Amount.values, kde=True)
plt.subplot(222)
sns.histplot(df[df.Class == 1].Amount.values, kde=True)
plt.subplot(223)
sns.histplot(df[df.Class == 0].Time.values, kde=True)
plt.subplot(224)
sns.histplot(df[df.Class == 1].Time.values, kde=True)
# Not only do fraudulent transactions cash out larger sums of money from banks, they also fail to follow the seasonal nature of the time as well.
dft = df[["Amount", "Class"]].copy()
dft["Digits"] = dft.Amount.astype(str).str[:1].astype(int)
plt.subplot(211)
sns.countplot(dft[dft.Class == 0].Digits)
plt.subplot(212)
sns.countplot(dft[dft.Class == 1].Digits)
# > Benford’s Law, also known as the Law of First Digits or the Phenomenon of Significant Digits, is the finding that the first digits (or numerals to be exact) of the numbers found in series of records of the most varied sources do not display a uniform distribution, but rather are arranged in such a way that the digit “1” is the most frequent, followed by “2”, “3”, and so in a successively decreasing manner down to “9”.
# We can see that the fraudulent transactions fail to follow this law, hence we can infer that something is wrong with those transactions.
rob_scaler = RobustScaler()
# Normalizing the dataframe
df["scaled_amount"] = rob_scaler.fit_transform(df["Amount"].values.reshape(-1, 1))
df["scaled_time"] = rob_scaler.fit_transform(df["Time"].values.reshape(-1, 1))
df.drop(["Time", "Amount"], axis=1, inplace=True)
sns.heatmap(df.corr(), cmap="coolwarm_r", vmin=-1, vmax=1, center=0)
# Looks like no feature is correlated, but this is the result of the highly imbalanced nature of the dataset, taking a subset of the dataset will give us a clearer picture.
fraud_df = df.loc[df["Class"] == 1]
non_fraud_df = df.loc[df["Class"] == 0][:492]
sub_df = (
pd.concat([fraud_df, non_fraud_df])
.sample(frac=1, random_state=420)
.reset_index(drop=True)
)
sns.heatmap(sub_df.corr(), cmap="coolwarm_r", vmin=-1, vmax=1, center=0)
# Making a subset to plot points clearly
X_sub = sub_df.copy().drop("Class", axis=1)
y_sub = sub_df.copy()["Class"]
X_reduced_tsne = TSNE(n_components=2, random_state=42).fit_transform(X_sub.values)
plt.scatter(
X_reduced_tsne[:, 0],
X_reduced_tsne[:, 1],
c=(y_sub == 0),
cmap="coolwarm_r",
label="No Fraud",
)
plt.scatter(
X_reduced_tsne[:, 0],
X_reduced_tsne[:, 1],
c=(y_sub == 1),
cmap="coolwarm_r",
label="Fraud",
)
# t-SNE helps us visualize the data points in a lower dimension that can be understood by us. Here, we can observe that these two classes can be separated by using a simple straight line with little to no overlapping, hence Logistic regression can work well on this dataset.
# # Data Preprocessing
# To mitigate the imbalance in the dataset, we can either use a subset of the non fraud cases to match the number of fraud cases, i.e. undersampling. Alternatively we can increase the number of fraud cases using SMOTE, i.e. oversampling.
X = df.copy().drop("Class", axis=1)
y = df.copy().Class
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, stratify=y, random_state=42
)
X_sub_train, X_sub_test, y_sub_train, y_sub_test = train_test_split(
X_sub, y_sub, test_size=0.2, stratify=y_sub, random_state=42
)
sm = SMOTE(random_state=666)
# New training dataset with smote applied to it
X_train_s, y_train_s = sm.fit_resample(X_train, y_train.ravel())
plt.subplot(121)
plt.pie(
x=[len(y_train_s) - sum(y_train_s), sum(y_train_s)],
labels=["Left", "Right"],
autopct="%1.2f%%",
explode=(0, 0.01),
)
plt.subplot(122)
sns.countplot(x=y_train_s)
fraudnt_s, fraud_s = len(y_train_s) - sum(y_train_s), sum(y_train_s)
plt.text(0, fraudnt_s // 2, fraudnt_s, fontsize=20, horizontalalignment="center")
plt.text(1, fraud_s // 2, fraud_s, fontsize=20, horizontalalignment="center")
# Perfectly balanced!
# # Machine Learning
def model_eval(y_actual, predicted):
print(
classification_report(y_actual, predicted, target_names=["Not Fraud", "Fraud"])
)
sns.heatmap(
data=confusion_matrix(y_test, predicted),
annot=True,
cmap="coolwarm_r",
center=0,
)
# ## Logistic Regression on original dataset
lr_clf = LogisticRegression(max_iter=1000)
lr_clf.fit(X_train, y_train)
prediction_lr_clf = lr_clf.predict(X_test)
model_eval(y_test, prediction_lr_clf)
# The accuracy maybe high but the recall is sub-par at best. We could only catch 62% of the frauds.
# ## Logistic Regression on sub-sample
lr_clf_sub = LogisticRegression(max_iter=1000)
lr_clf_sub.fit(X_sub_train, y_sub_train)
prediction_lr_clf_sub = lr_clf.predict(X_test)
print(
classification_report(
y_test, prediction_lr_clf_sub, target_names=["Not Fraud", "Fraud"]
)
)
sns.heatmap(
data=confusion_matrix(y_test, prediction_lr_clf_sub),
annot=True,
cmap="coolwarm_r",
center=0,
)
# Using a subset gave us some minor improvement in recall but it still isn't enough, lets see what oversampling does to the model.
# ## Logistic Regression with SMOTE
lr_clf_smote = LogisticRegression(max_iter=1000)
lr_clf_smote.fit(X_train_s, y_train_s)
prediction_lr_clf_smote = lr_clf_smote.predict(X_test)
model_eval(y_test, prediction_lr_clf_smote)
# The recall in this model is excellent! 92% of the frauds are caught, but the precision takes a hit. A lot of normal transactions are flagged, this means many unhappy customers.
# ## XGBoost with scale_pos_weight
# XGBoost has a parameter which gives higher priority to the minority class making the model fairly unbiased as the weights for both the categories remains the same.
scale_val = fraudnt // fraud # inverse of the ratio of number of each class present
print(f"The scale value is {scale_val}")
xgb_clf = xgb.XGBClassifier(
n_estimators=100,
verbosity=1,
scale_pos_weight=scale_val,
max_depth=5,
gamma=0.2,
colsamplebytree=0.8,
regalpha=0.1,
subsample=0.2,
learning_rate=0.3,
)
xgb_clf.fit(X_train, y_train)
prediction_xgb_clf = xgb_clf.predict(X_test)
model_eval(y_test, prediction_xgb_clf)
# Even though the recall value is lower, there is higher precision here giving us a better F1 score, making XGBoost a better all round model.
xgb.plot_importance(xgb_clf)
def prc_with_model(X_test, y_test, model):
y_prob = eval(model).predict_proba(X_test)[:, 1]
precision, recall, thresholds = precision_recall_curve(y_test, y_prob)
plt.plot(precision, recall, label=model)
def auc_score(y_test, model):
print(
f"AUC score for {model} is: ",
roc_auc_score(y_test, eval(f"prediction_{model}")),
)
models = ["lr_clf", "lr_clf_sub", "lr_clf_smote", "xgb_clf"]
for model in models:
prc_with_model(X_test, y_test, model)
auc_score(y_test, model)
plt.legend()
# Even though Logistic Regression has a higher AUC than XGBoost, I will be choosing XGBoost for further analysis as it is compatible with the libraries being used for machine learning explainibility.
# # Feature Overview
perm = PermutationImportance(xgb_clf, random_state=1).fit(X_test, y_test)
eli5.show_weights(perm, feature_names=X_test.columns.tolist())
pdp_dist = pdp.pdp_isolate(
model=xgb_clf, dataset=X_test, model_features=X_test.columns, feature="scaled_time"
)
pdp.pdp_plot(pdp_dist, "scaled_time")
plt.show()
features_to_plot = ["scaled_time", "scaled_amount"]
inter1 = pdp.pdp_interact(
model=xgb_clf,
dataset=X_test,
model_features=X_test.columns,
features=features_to_plot,
)
pdp.pdp_interact_plot(
pdp_interact_out=inter1, feature_names=features_to_plot, plot_type="contour"
)
plt.show()
data_for_prediction = X.iloc[X_test[y_test == 1].index[1]]
explainer = shap.TreeExplainer(xgb_clf)
data_for_prediction_array = data_for_prediction.values.reshape(1, -1)
shap_values = explainer.shap_values(data_for_prediction_array)
print(xgb_clf.predict_proba(data_for_prediction_array))
shap.initjs()
shap.force_plot(explainer.expected_value, shap_values, data_for_prediction_array)
data_for_prediction = X_test.iloc[5]
explainer = shap.TreeExplainer(xgb_clf)
data_for_prediction_array = data_for_prediction.values.reshape(1, -1)
shap_values = explainer.shap_values(data_for_prediction_array)
print(xgb_clf.predict_proba(data_for_prediction_array))
shap.initjs()
shap.force_plot(explainer.expected_value, shap_values, data_for_prediction_array)
explainer = shap.TreeExplainer(xgb_clf)
shap_values = explainer.shap_values(X_test)
shap.summary_plot(shap_values, X_test)
explainer = shap.TreeExplainer(xgb_clf)
shap_values = explainer.shap_values(X)
shap.dependence_plot(
"scaled_time",
shap_values,
X,
interaction_index="scaled_amount",
x_jitter=1,
dot_size=10,
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605479.ipynb
|
creditcardfraud
| null |
[{"Id": 69605479, "ScriptId": 18782262, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5850776, "CreationDate": "08/02/2021 05:39:15", "VersionNumber": 1.0, "Title": "Credit Card Fraud", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 278.0, "LinesInsertedFromPrevious": 278.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93001345, "KernelVersionId": 69605479, "SourceDatasetVersionId": 23498}]
|
[{"Id": 23498, "DatasetId": 310, "DatasourceVersionId": 23502, "CreatorUserId": 998023, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "03/23/2018 01:17:27", "VersionNumber": 3.0, "Title": "Credit Card Fraud Detection", "Slug": "creditcardfraud", "Subtitle": "Anonymized credit card transactions labeled as fraudulent or genuine", "Description": "Context\n---------\n\nIt is important that credit card companies are able to recognize fraudulent credit card transactions so that customers are not charged for items that they did not purchase.\n\nContent\n---------\n\nThe dataset contains transactions made by credit cards in September 2013 by European cardholders. \nThis dataset presents transactions that occurred in two days, where we have 492 frauds out of 284,807 transactions. The dataset is highly unbalanced, the positive class (frauds) account for 0.172% of all transactions.\n\nIt contains only numerical input variables which are the result of a PCA transformation. Unfortunately, due to confidentiality issues, we cannot provide the original features and more background information about the data. Features V1, V2, ... V28 are the principal components obtained with PCA, the only features which have not been transformed with PCA are 'Time' and 'Amount'. Feature 'Time' contains the seconds elapsed between each transaction and the first transaction in the dataset. The feature 'Amount' is the transaction Amount, this feature can be used for example-dependant cost-sensitive learning. Feature 'Class' is the response variable and it takes value 1 in case of fraud and 0 otherwise. \n\nGiven the class imbalance ratio, we recommend measuring the accuracy using the Area Under the Precision-Recall Curve (AUPRC). Confusion matrix accuracy is not meaningful for unbalanced classification.\n\nUpdate (03/05/2021)\n---------\n\nA simulator for transaction data has been released as part of the practical handbook on Machine Learning for Credit Card Fraud Detection - https://fraud-detection-handbook.github.io/fraud-detection-handbook/Chapter_3_GettingStarted/SimulatedDataset.html. We invite all practitioners interested in fraud detection datasets to also check out this data simulator, and the methodologies for credit card fraud detection presented in the book.\n\nAcknowledgements\n---------\n\nThe dataset has been collected and analysed during a research collaboration of Worldline and the Machine Learning Group (http://mlg.ulb.ac.be) of ULB (Universit\u00e9 Libre de Bruxelles) on big data mining and fraud detection.\nMore details on current and past projects on related topics are available on [https://www.researchgate.net/project/Fraud-detection-5][1] and the page of the [DefeatFraud][2] project\n\nPlease cite the following works: \n\nAndrea Dal Pozzolo, Olivier Caelen, Reid A. Johnson and Gianluca Bontempi. [Calibrating Probability with Undersampling for Unbalanced Classification.][3] In Symposium on Computational Intelligence and Data Mining (CIDM), IEEE, 2015\n\nDal Pozzolo, Andrea; Caelen, Olivier; Le Borgne, Yann-Ael; Waterschoot, Serge; Bontempi, Gianluca. [Learned lessons in credit card fraud detection from a practitioner perspective][4], Expert systems with applications,41,10,4915-4928,2014, Pergamon\n\nDal Pozzolo, Andrea; Boracchi, Giacomo; Caelen, Olivier; Alippi, Cesare; Bontempi, Gianluca. [Credit card fraud detection: a realistic modeling and a novel learning strategy,][5] IEEE transactions on neural networks and learning systems,29,8,3784-3797,2018,IEEE\n\nDal Pozzolo, Andrea [Adaptive Machine learning for credit card fraud detection][6] ULB MLG PhD thesis (supervised by G. Bontempi)\n\nCarcillo, Fabrizio; Dal Pozzolo, Andrea; Le Borgne, Yann-A\u00ebl; Caelen, Olivier; Mazzer, Yannis; Bontempi, Gianluca. [Scarff: a scalable framework for streaming credit card fraud detection with Spark][7], Information fusion,41, 182-194,2018,Elsevier\n\nCarcillo, Fabrizio; Le Borgne, Yann-A\u00ebl; Caelen, Olivier; Bontempi, Gianluca. [Streaming active learning strategies for real-life credit card fraud detection: assessment and visualization,][8] International Journal of Data Science and Analytics, 5,4,285-300,2018,Springer International Publishing\n\nBertrand Lebichot, Yann-A\u00ebl Le Borgne, Liyun He, Frederic Obl\u00e9, Gianluca Bontempi [Deep-Learning Domain Adaptation Techniques for Credit Cards Fraud Detection](https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection), INNSBDDL 2019: Recent Advances in Big Data and Deep Learning, pp 78-88, 2019\n\nFabrizio Carcillo, Yann-A\u00ebl Le Borgne, Olivier Caelen, Frederic Obl\u00e9, Gianluca Bontempi [Combining Unsupervised and Supervised Learning in Credit Card Fraud Detection ](https://www.researchgate.net/publication/333143698_Combining_Unsupervised_and_Supervised_Learning_in_Credit_Card_Fraud_Detection) Information Sciences, 2019\n\nYann-A\u00ebl Le Borgne, Gianluca Bontempi [Reproducible machine Learning for Credit Card Fraud Detection - Practical Handbook ](https://www.researchgate.net/publication/351283764_Machine_Learning_for_Credit_Card_Fraud_Detection_-_Practical_Handbook) \n\nBertrand Lebichot, Gianmarco Paldino, Wissam Siblini, Liyun He, Frederic Obl\u00e9, Gianluca Bontempi [Incremental learning strategies for credit cards fraud detection](https://www.researchgate.net/publication/352275169_Incremental_learning_strategies_for_credit_cards_fraud_detection), IInternational Journal of Data Science and Analytics\n\n [1]: https://www.researchgate.net/project/Fraud-detection-5\n [2]: https://mlg.ulb.ac.be/wordpress/portfolio_page/defeatfraud-assessment-and-validation-of-deep-feature-engineering-and-learning-solutions-for-fraud-detection/\n [3]: https://www.researchgate.net/publication/283349138_Calibrating_Probability_with_Undersampling_for_Unbalanced_Classification\n [4]: https://www.researchgate.net/publication/260837261_Learned_lessons_in_credit_card_fraud_detection_from_a_practitioner_perspective\n [5]: https://www.researchgate.net/publication/319867396_Credit_Card_Fraud_Detection_A_Realistic_Modeling_and_a_Novel_Learning_Strategy\n [6]: http://di.ulb.ac.be/map/adalpozz/pdf/Dalpozzolo2015PhD.pdf\n [7]: https://www.researchgate.net/publication/319616537_SCARFF_a_Scalable_Framework_for_Streaming_Credit_Card_Fraud_Detection_with_Spark\n \n[8]: https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection", "VersionNotes": "Fixed preview", "TotalCompressedBytes": 150828752.0, "TotalUncompressedBytes": 69155632.0}]
|
[{"Id": 310, "CreatorUserId": 14069, "OwnerUserId": NaN, "OwnerOrganizationId": 1160.0, "CurrentDatasetVersionId": 23498.0, "CurrentDatasourceVersionId": 23502.0, "ForumId": 1838, "Type": 2, "CreationDate": "11/03/2016 13:21:36", "LastActivityDate": "02/06/2018", "TotalViews": 10310781, "TotalDownloads": 564249, "TotalVotes": 10432, "TotalKernels": 4266}]
| null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from matplotlib import gridspec
import matplotlib.pyplot as plt
import seaborn as sns
from pylab import rcParams
rcParams["figure.figsize"] = 15, 10
from imblearn.over_sampling import SMOTE
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import RobustScaler
from sklearn.manifold import TSNE
from sklearn.linear_model import LogisticRegression
import xgboost as xgb
from sklearn.metrics import (
confusion_matrix,
classification_report,
accuracy_score,
precision_recall_curve,
roc_auc_score,
)
import eli5
from eli5.sklearn import PermutationImportance
from pdpbox import pdp, get_dataset, info_plots
import shap
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("../input/creditcardfraud/creditcard.csv")
df.head()
df.describe()
# The Class column in looking highly skewed. We will see how much imbalance there really is.
df.isna().sum()
# No Null values, lesser work to do!
plt.subplot(121)
plt.pie(
x=df.groupby(["Class"]).Class.count().to_list(),
labels=["Left", "Right"],
autopct="%1.2f%%",
explode=(0, 0.2),
)
plt.subplot(122)
sns.countplot(data=df, x="Class")
fraudnt, fraud = df.Class.value_counts()
plt.text(0, fraudnt // 2, fraudnt, fontsize=20, horizontalalignment="center")
plt.text(1, fraud // 2, fraud, fontsize=20, horizontalalignment="center")
# Using such an imbalanced dataset for machine learning can lead to very poor performance in real life. We will see some ways that we can mitigate this issue later in this notebook.
features = df.iloc[:, 0:29].columns
plt.figure(figsize=(15, 29 * 4))
gs = gridspec.GridSpec(29, 1)
for i, c in enumerate(df[features]):
ax = plt.subplot(gs[i])
sns.distplot(df[c][df.Class == 1], bins=50, label="Fraud")
sns.distplot(df[c][df.Class == 0], bins=50, label="Valid")
ax.legend()
ax.set_xlabel("")
ax.set_title("histogram of feature: " + str(c))
plt.show()
# bla bla
plt.subplot(211)
sns.distplot(df.Amount.values, kde=True)
plt.subplot(212)
sns.histplot(df.Time.values, kde=True)
# The amount variable is highly left skewed as most people take part in low volume banking transactions but the time variable has visible seasonality indicating peak times and breaks that are part of a human's daily routine.
plt.subplot(221)
sns.distplot(df[df.Class == 0].Amount.values, kde=True)
plt.subplot(222)
sns.histplot(df[df.Class == 1].Amount.values, kde=True)
plt.subplot(223)
sns.histplot(df[df.Class == 0].Time.values, kde=True)
plt.subplot(224)
sns.histplot(df[df.Class == 1].Time.values, kde=True)
# Not only do fraudulent transactions cash out larger sums of money from banks, they also fail to follow the seasonal nature of the time as well.
dft = df[["Amount", "Class"]].copy()
dft["Digits"] = dft.Amount.astype(str).str[:1].astype(int)
plt.subplot(211)
sns.countplot(dft[dft.Class == 0].Digits)
plt.subplot(212)
sns.countplot(dft[dft.Class == 1].Digits)
# > Benford’s Law, also known as the Law of First Digits or the Phenomenon of Significant Digits, is the finding that the first digits (or numerals to be exact) of the numbers found in series of records of the most varied sources do not display a uniform distribution, but rather are arranged in such a way that the digit “1” is the most frequent, followed by “2”, “3”, and so in a successively decreasing manner down to “9”.
# We can see that the fraudulent transactions fail to follow this law, hence we can infer that something is wrong with those transactions.
rob_scaler = RobustScaler()
# Normalizing the dataframe
df["scaled_amount"] = rob_scaler.fit_transform(df["Amount"].values.reshape(-1, 1))
df["scaled_time"] = rob_scaler.fit_transform(df["Time"].values.reshape(-1, 1))
df.drop(["Time", "Amount"], axis=1, inplace=True)
sns.heatmap(df.corr(), cmap="coolwarm_r", vmin=-1, vmax=1, center=0)
# Looks like no feature is correlated, but this is the result of the highly imbalanced nature of the dataset, taking a subset of the dataset will give us a clearer picture.
fraud_df = df.loc[df["Class"] == 1]
non_fraud_df = df.loc[df["Class"] == 0][:492]
sub_df = (
pd.concat([fraud_df, non_fraud_df])
.sample(frac=1, random_state=420)
.reset_index(drop=True)
)
sns.heatmap(sub_df.corr(), cmap="coolwarm_r", vmin=-1, vmax=1, center=0)
# Making a subset to plot points clearly
X_sub = sub_df.copy().drop("Class", axis=1)
y_sub = sub_df.copy()["Class"]
X_reduced_tsne = TSNE(n_components=2, random_state=42).fit_transform(X_sub.values)
plt.scatter(
X_reduced_tsne[:, 0],
X_reduced_tsne[:, 1],
c=(y_sub == 0),
cmap="coolwarm_r",
label="No Fraud",
)
plt.scatter(
X_reduced_tsne[:, 0],
X_reduced_tsne[:, 1],
c=(y_sub == 1),
cmap="coolwarm_r",
label="Fraud",
)
# t-SNE helps us visualize the data points in a lower dimension that can be understood by us. Here, we can observe that these two classes can be separated by using a simple straight line with little to no overlapping, hence Logistic regression can work well on this dataset.
# # Data Preprocessing
# To mitigate the imbalance in the dataset, we can either use a subset of the non fraud cases to match the number of fraud cases, i.e. undersampling. Alternatively we can increase the number of fraud cases using SMOTE, i.e. oversampling.
X = df.copy().drop("Class", axis=1)
y = df.copy().Class
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, stratify=y, random_state=42
)
X_sub_train, X_sub_test, y_sub_train, y_sub_test = train_test_split(
X_sub, y_sub, test_size=0.2, stratify=y_sub, random_state=42
)
sm = SMOTE(random_state=666)
# New training dataset with smote applied to it
X_train_s, y_train_s = sm.fit_resample(X_train, y_train.ravel())
plt.subplot(121)
plt.pie(
x=[len(y_train_s) - sum(y_train_s), sum(y_train_s)],
labels=["Left", "Right"],
autopct="%1.2f%%",
explode=(0, 0.01),
)
plt.subplot(122)
sns.countplot(x=y_train_s)
fraudnt_s, fraud_s = len(y_train_s) - sum(y_train_s), sum(y_train_s)
plt.text(0, fraudnt_s // 2, fraudnt_s, fontsize=20, horizontalalignment="center")
plt.text(1, fraud_s // 2, fraud_s, fontsize=20, horizontalalignment="center")
# Perfectly balanced!
# # Machine Learning
def model_eval(y_actual, predicted):
print(
classification_report(y_actual, predicted, target_names=["Not Fraud", "Fraud"])
)
sns.heatmap(
data=confusion_matrix(y_test, predicted),
annot=True,
cmap="coolwarm_r",
center=0,
)
# ## Logistic Regression on original dataset
lr_clf = LogisticRegression(max_iter=1000)
lr_clf.fit(X_train, y_train)
prediction_lr_clf = lr_clf.predict(X_test)
model_eval(y_test, prediction_lr_clf)
# The accuracy maybe high but the recall is sub-par at best. We could only catch 62% of the frauds.
# ## Logistic Regression on sub-sample
lr_clf_sub = LogisticRegression(max_iter=1000)
lr_clf_sub.fit(X_sub_train, y_sub_train)
prediction_lr_clf_sub = lr_clf.predict(X_test)
print(
classification_report(
y_test, prediction_lr_clf_sub, target_names=["Not Fraud", "Fraud"]
)
)
sns.heatmap(
data=confusion_matrix(y_test, prediction_lr_clf_sub),
annot=True,
cmap="coolwarm_r",
center=0,
)
# Using a subset gave us some minor improvement in recall but it still isn't enough, lets see what oversampling does to the model.
# ## Logistic Regression with SMOTE
lr_clf_smote = LogisticRegression(max_iter=1000)
lr_clf_smote.fit(X_train_s, y_train_s)
prediction_lr_clf_smote = lr_clf_smote.predict(X_test)
model_eval(y_test, prediction_lr_clf_smote)
# The recall in this model is excellent! 92% of the frauds are caught, but the precision takes a hit. A lot of normal transactions are flagged, this means many unhappy customers.
# ## XGBoost with scale_pos_weight
# XGBoost has a parameter which gives higher priority to the minority class making the model fairly unbiased as the weights for both the categories remains the same.
scale_val = fraudnt // fraud # inverse of the ratio of number of each class present
print(f"The scale value is {scale_val}")
xgb_clf = xgb.XGBClassifier(
n_estimators=100,
verbosity=1,
scale_pos_weight=scale_val,
max_depth=5,
gamma=0.2,
colsamplebytree=0.8,
regalpha=0.1,
subsample=0.2,
learning_rate=0.3,
)
xgb_clf.fit(X_train, y_train)
prediction_xgb_clf = xgb_clf.predict(X_test)
model_eval(y_test, prediction_xgb_clf)
# Even though the recall value is lower, there is higher precision here giving us a better F1 score, making XGBoost a better all round model.
xgb.plot_importance(xgb_clf)
def prc_with_model(X_test, y_test, model):
y_prob = eval(model).predict_proba(X_test)[:, 1]
precision, recall, thresholds = precision_recall_curve(y_test, y_prob)
plt.plot(precision, recall, label=model)
def auc_score(y_test, model):
print(
f"AUC score for {model} is: ",
roc_auc_score(y_test, eval(f"prediction_{model}")),
)
models = ["lr_clf", "lr_clf_sub", "lr_clf_smote", "xgb_clf"]
for model in models:
prc_with_model(X_test, y_test, model)
auc_score(y_test, model)
plt.legend()
# Even though Logistic Regression has a higher AUC than XGBoost, I will be choosing XGBoost for further analysis as it is compatible with the libraries being used for machine learning explainibility.
# # Feature Overview
perm = PermutationImportance(xgb_clf, random_state=1).fit(X_test, y_test)
eli5.show_weights(perm, feature_names=X_test.columns.tolist())
pdp_dist = pdp.pdp_isolate(
model=xgb_clf, dataset=X_test, model_features=X_test.columns, feature="scaled_time"
)
pdp.pdp_plot(pdp_dist, "scaled_time")
plt.show()
features_to_plot = ["scaled_time", "scaled_amount"]
inter1 = pdp.pdp_interact(
model=xgb_clf,
dataset=X_test,
model_features=X_test.columns,
features=features_to_plot,
)
pdp.pdp_interact_plot(
pdp_interact_out=inter1, feature_names=features_to_plot, plot_type="contour"
)
plt.show()
data_for_prediction = X.iloc[X_test[y_test == 1].index[1]]
explainer = shap.TreeExplainer(xgb_clf)
data_for_prediction_array = data_for_prediction.values.reshape(1, -1)
shap_values = explainer.shap_values(data_for_prediction_array)
print(xgb_clf.predict_proba(data_for_prediction_array))
shap.initjs()
shap.force_plot(explainer.expected_value, shap_values, data_for_prediction_array)
data_for_prediction = X_test.iloc[5]
explainer = shap.TreeExplainer(xgb_clf)
data_for_prediction_array = data_for_prediction.values.reshape(1, -1)
shap_values = explainer.shap_values(data_for_prediction_array)
print(xgb_clf.predict_proba(data_for_prediction_array))
shap.initjs()
shap.force_plot(explainer.expected_value, shap_values, data_for_prediction_array)
explainer = shap.TreeExplainer(xgb_clf)
shap_values = explainer.shap_values(X_test)
shap.summary_plot(shap_values, X_test)
explainer = shap.TreeExplainer(xgb_clf)
shap_values = explainer.shap_values(X)
shap.dependence_plot(
"scaled_time",
shap_values,
X,
interaction_index="scaled_amount",
x_jitter=1,
dot_size=10,
)
| false | 0 | 3,959 | 0 | 5,833 | 3,959 |
||
69605112
|
<jupyter_start><jupyter_text>CommonLit RoBERTa Large II
Kaggle dataset identifier: commonlit-roberta-large-ii
<jupyter_script># # Overview
# This notebook combines four models.
# # Model 1
# The model is inspired by the one from [Maunish](https://www.kaggle.com/maunish/clrp-roberta-svm).
import os
import math
import random
import time
import numpy as np
import pandas as pd
from tqdm import tqdm
import torch
import torch.nn as nn
from torch.utils.data import Dataset
from torch.utils.data import DataLoader
from transformers import AutoTokenizer
from transformers import AutoModel
from transformers import AutoConfig
from sklearn.model_selection import KFold
from sklearn.svm import SVR
import gc
gc.enable()
BATCH_SIZE = 32
MAX_LEN = 248
EVAL_SCHEDULE = [(0.5, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1, 1)]
ROBERTA_PATH = "/kaggle/input/roberta-base"
TOKENIZER_PATH = "/kaggle/input/roberta-base"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
DEVICE
test_df = pd.read_csv("/kaggle/input/commonlitreadabilityprize/test.csv")
submission_df = pd.read_csv(
"/kaggle/input/commonlitreadabilityprize/sample_submission.csv"
)
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
class LitDataset(Dataset):
def __init__(self, df, inference_only=False):
super().__init__()
self.df = df
self.inference_only = inference_only
self.text = df.excerpt.tolist()
# self.text = [text.replace("\n", " ") for text in self.text]
if not self.inference_only:
self.target = torch.tensor(df.target.values, dtype=torch.float32)
self.encoded = tokenizer.batch_encode_plus(
self.text,
padding="max_length",
max_length=MAX_LEN,
truncation=True,
return_attention_mask=True,
)
def __len__(self):
return len(self.df)
def __getitem__(self, index):
input_ids = torch.tensor(self.encoded["input_ids"][index])
attention_mask = torch.tensor(self.encoded["attention_mask"][index])
if self.inference_only:
return (input_ids, attention_mask)
else:
target = self.target[index]
return (input_ids, attention_mask, target)
class LitModel(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.5,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(768, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
NUM_MODELS = 5
all_predictions = np.zeros((NUM_MODELS, len(test_df)))
test_dataset = LitDataset(test_df, inference_only=True)
test_loader = DataLoader(
test_dataset, batch_size=BATCH_SIZE, drop_last=False, shuffle=False, num_workers=2
)
for model_index in tqdm(range(NUM_MODELS)):
model_path = f"../input/commonlit-roberta-0467/model_{model_index + 1}.pth"
print(f"\nUsing {model_path}")
model = LitModel()
model.load_state_dict(torch.load(model_path, map_location=DEVICE))
model.to(DEVICE)
all_predictions[model_index] = predict(model, test_loader)
del model
gc.collect()
model1_predictions = all_predictions.mean(axis=0)
# # Model 2
# Inspired from [https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3](https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3)
test = test_df
from glob import glob
import os
import matplotlib.pyplot as plt
import json
from collections import defaultdict
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.optim.optimizer import Optimizer
import torch.optim.lr_scheduler as lr_scheduler
from torch.utils.data import Dataset, DataLoader, SequentialSampler, RandomSampler
from transformers import RobertaConfig
from transformers import (
get_cosine_schedule_with_warmup,
get_cosine_with_hard_restarts_schedule_with_warmup,
)
from transformers import RobertaTokenizer
from transformers import RobertaModel
from IPython.display import clear_output
def convert_examples_to_features(data, tokenizer, max_len, is_test=False):
data = data.replace("\n", "")
tok = tokenizer.encode_plus(
data,
max_length=max_len,
truncation=True,
return_attention_mask=True,
return_token_type_ids=True,
)
curr_sent = {}
padding_length = max_len - len(tok["input_ids"])
curr_sent["input_ids"] = tok["input_ids"] + ([0] * padding_length)
curr_sent["token_type_ids"] = tok["token_type_ids"] + ([0] * padding_length)
curr_sent["attention_mask"] = tok["attention_mask"] + ([0] * padding_length)
return curr_sent
class DatasetRetriever(Dataset):
def __init__(self, data, tokenizer, max_len, is_test=False):
self.data = data
self.excerpts = self.data.excerpt.values.tolist()
self.tokenizer = tokenizer
self.is_test = is_test
self.max_len = max_len
def __len__(self):
return len(self.data)
def __getitem__(self, item):
if not self.is_test:
excerpt, label = self.excerpts[item], self.targets[item]
features = convert_examples_to_features(
excerpt, self.tokenizer, self.max_len, self.is_test
)
return {
"input_ids": torch.tensor(features["input_ids"], dtype=torch.long),
"token_type_ids": torch.tensor(
features["token_type_ids"], dtype=torch.long
),
"attention_mask": torch.tensor(
features["attention_mask"], dtype=torch.long
),
"label": torch.tensor(label, dtype=torch.double),
}
else:
excerpt = self.excerpts[item]
features = convert_examples_to_features(
excerpt, self.tokenizer, self.max_len, self.is_test
)
return {
"input_ids": torch.tensor(features["input_ids"], dtype=torch.long),
"token_type_ids": torch.tensor(
features["token_type_ids"], dtype=torch.long
),
"attention_mask": torch.tensor(
features["attention_mask"], dtype=torch.long
),
}
class CommonLitModel(nn.Module):
def __init__(
self, model_name, config, multisample_dropout=True, output_hidden_states=False
):
super(CommonLitModel, self).__init__()
self.config = config
self.roberta = RobertaModel.from_pretrained(
model_name, output_hidden_states=output_hidden_states
)
self.layer_norm = nn.LayerNorm(config.hidden_size)
if multisample_dropout:
self.dropouts = nn.ModuleList([nn.Dropout(0.5) for _ in range(5)])
else:
self.dropouts = nn.ModuleList([nn.Dropout(0.3)])
self.regressor = nn.Linear(config.hidden_size, 1)
self._init_weights(self.layer_norm)
self._init_weights(self.regressor)
def _init_weights(self, module):
if isinstance(module, nn.Linear):
module.weight.data.normal_(mean=0.0, std=self.config.initializer_range)
if module.bias is not None:
module.bias.data.zero_()
elif isinstance(module, nn.Embedding):
module.weight.data.normal_(mean=0.0, std=self.config.initializer_range)
if module.padding_idx is not None:
module.weight.data[module.padding_idx].zero_()
elif isinstance(module, nn.LayerNorm):
module.bias.data.zero_()
module.weight.data.fill_(1.0)
def forward(
self, input_ids=None, attention_mask=None, token_type_ids=None, labels=None
):
outputs = self.roberta(
input_ids,
attention_mask=attention_mask,
token_type_ids=token_type_ids,
)
sequence_output = outputs[1]
sequence_output = self.layer_norm(sequence_output)
# multi-sample dropout
for i, dropout in enumerate(self.dropouts):
if i == 0:
logits = self.regressor(dropout(sequence_output))
else:
logits += self.regressor(dropout(sequence_output))
logits /= len(self.dropouts)
# calculate loss
loss = None
if labels is not None:
loss_fn = torch.nn.MSELoss()
logits = logits.view(-1).to(labels.dtype)
loss = torch.sqrt(loss_fn(logits, labels.view(-1)))
output = (logits,) + outputs[1:]
return ((loss,) + output) if loss is not None else output
def make_model(model_name, num_labels=1):
tokenizer = RobertaTokenizer.from_pretrained(model_name)
config = RobertaConfig.from_pretrained(model_name)
config.update({"num_labels": num_labels})
model = CommonLitModel(model_name, config=config)
return model, tokenizer
def make_loader(
data,
tokenizer,
max_len,
batch_size,
):
test_dataset = DatasetRetriever(data, tokenizer, max_len, is_test=True)
test_sampler = SequentialSampler(test_dataset)
test_loader = DataLoader(
test_dataset,
batch_size=batch_size // 2,
sampler=test_sampler,
pin_memory=False,
drop_last=False,
num_workers=0,
)
return test_loader
class Evaluator:
def __init__(self, model, scalar=None):
self.model = model
self.scalar = scalar
def evaluate(self, data_loader, tokenizer):
preds = []
self.model.eval()
total_loss = 0
with torch.no_grad():
for batch_idx, batch_data in enumerate(data_loader):
input_ids, attention_mask, token_type_ids = (
batch_data["input_ids"],
batch_data["attention_mask"],
batch_data["token_type_ids"],
)
input_ids, attention_mask, token_type_ids = (
input_ids.cuda(),
attention_mask.cuda(),
token_type_ids.cuda(),
)
if self.scalar is not None:
with torch.cuda.amp.autocast():
outputs = self.model(
input_ids=input_ids,
attention_mask=attention_mask,
token_type_ids=token_type_ids,
)
else:
outputs = self.model(
input_ids=input_ids,
attention_mask=attention_mask,
token_type_ids=token_type_ids,
)
logits = outputs[0].detach().cpu().numpy().squeeze().tolist()
preds += logits
return preds
def config(fold, model_name, load_model_path):
torch.manual_seed(2021)
torch.cuda.manual_seed(2021)
torch.cuda.manual_seed_all(2021)
max_len = 250
batch_size = 8
model, tokenizer = make_model(model_name=model_name, num_labels=1)
model.load_state_dict(torch.load(f"{load_model_path}/model{fold}.bin"))
test_loader = make_loader(test, tokenizer, max_len=max_len, batch_size=batch_size)
if torch.cuda.device_count() >= 1:
print(
"Model pushed to {} GPU(s), type {}.".format(
torch.cuda.device_count(), torch.cuda.get_device_name(0)
)
)
model = model.cuda()
else:
raise ValueError("CPU training is not supported")
# scaler = torch.cuda.amp.GradScaler()
scaler = None
return (model, tokenizer, test_loader, scaler)
def run(fold=0, model_name=None, load_model_path=None):
model, tokenizer, test_loader, scaler = config(fold, model_name, load_model_path)
import time
evaluator = Evaluator(model, scaler)
test_time_list = []
torch.cuda.synchronize()
tic1 = time.time()
preds = evaluator.evaluate(test_loader, tokenizer)
torch.cuda.synchronize()
tic2 = time.time()
test_time_list.append(tic2 - tic1)
del model, tokenizer, test_loader, scaler
gc.collect()
torch.cuda.empty_cache()
return preds
pred_df1 = pd.DataFrame()
pred_df2 = pd.DataFrame()
pred_df3 = pd.DataFrame()
for fold in tqdm(range(5)):
pred_df1[f"fold{fold}"] = run(
fold % 5, "../input/roberta-base/", "../input/commonlit-roberta-base-i/"
)
pred_df2[f"fold{fold+5}"] = run(
fold % 5, "../input/robertalarge/", "../input/roberta-large-itptfit/"
)
pred_df3[f"fold{fold+10}"] = run(
fold % 5, "../input/robertalarge/", "../input/commonlit-roberta-large-ii/"
)
pred_df1 = np.array(pred_df1)
pred_df2 = np.array(pred_df2)
pred_df3 = np.array(pred_df3)
model2_predictions = (
(pred_df2.mean(axis=1) * 0.5)
+ (pred_df1.mean(axis=1) * 0.3)
+ (pred_df3.mean(axis=1) * 0.2)
)
# # Model 3
# Inspired from: https://www.kaggle.com/ragnar123/commonlit-readability-roberta-tf-inference
import re
import os
import numpy as np
import pandas as pd
import random
import math
import tensorflow as tf
import logging
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold
from tensorflow.keras import backend as K
from transformers import RobertaTokenizer, TFRobertaModel
from kaggle_datasets import KaggleDatasets
tf.get_logger().setLevel(logging.ERROR)
from kaggle_datasets import KaggleDatasets
# Configurations
# Number of folds for training
FOLDS = 5
# Max length
MAX_LEN = 250
# Get the trained model we want to use
MODEL = "../input/tfroberta-base"
# Let's load our model tokenizer
tokenizer = RobertaTokenizer.from_pretrained(MODEL)
# This function tokenize the text according to a transformers model tokenizer
def regular_encode(texts, tokenizer, maxlen=MAX_LEN):
enc_di = tokenizer.batch_encode_plus(
texts,
padding="max_length",
truncation=True,
max_length=maxlen,
)
return np.array(enc_di["input_ids"])
# This function encode our training sentences
def encode_texts(x_test, MAX_LEN):
x_test = regular_encode(x_test.tolist(), tokenizer, maxlen=MAX_LEN)
return x_test
# Function to build our model
def build_roberta_base_model(max_len=MAX_LEN):
transformer = TFRobertaModel.from_pretrained(MODEL)
input_word_ids = tf.keras.layers.Input(
shape=(max_len,), dtype=tf.int32, name="input_word_ids"
)
sequence_output = transformer(input_word_ids)[0]
# We only need the cls_token, resulting in a 2d array
cls_token = sequence_output[:, 0, :]
output = tf.keras.layers.Dense(1, activation="linear", dtype="float32")(cls_token)
model = tf.keras.models.Model(inputs=[input_word_ids], outputs=[output])
return model
# Function for inference
def roberta_base_inference1():
# Read our test data
df = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
# Get text features
x_test = df["excerpt"]
# Encode our text with Roberta tokenizer
x_test = encode_texts(x_test, MAX_LEN)
# Initiate an empty vector to store prediction
predictions = np.zeros(len(df))
# Predict with the 5 models (5 folds training)
for i in range(FOLDS):
print("\n")
print("-" * 50)
print(f"Predicting with model {i + 1}")
# Build model
model = build_roberta_base_model(max_len=MAX_LEN)
# Load pretrained weights
model.load_weights(
f"../input/epochs-100-lr-4e5-seed-123/Roberta_Base_123_{i + 1}.h5"
)
# Predict
fold_predictions = model.predict(x_test).reshape(-1)
# Add fold prediction to the global predictions
predictions += fold_predictions / FOLDS
return predictions
model3_predictions = roberta_base_inference1()
# ## Model 4
# Inspired from: https://www.kaggle.com/jcesquiveld/best-transformer-representations
import os
import numpy as np
import pandas as pd
import random
from transformers import (
AutoConfig,
AutoModel,
AutoTokenizer,
AdamW,
get_linear_schedule_with_warmup,
logging,
)
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import (
Dataset,
TensorDataset,
SequentialSampler,
RandomSampler,
DataLoader,
)
from tqdm.notebook import tqdm
import gc
gc.enable()
from IPython.display import clear_output
from sklearn.model_selection import StratifiedKFold
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("whitegrid")
logging.set_verbosity_error()
INPUT_DIR = "../input/commonlitreadabilityprize"
MODEL_DIR = "../input/roberta-transformers-pytorch/roberta-large"
CHECKPOINT_DIR1 = "../input/clrp-mean-pooling/"
CHECKPOINT_DIR2 = "../input/clrp-mean-pooling-seeds-17-43/"
DEVICE = torch.device("cuda" if torch.cuda.is_available() else "cpu")
MAX_LENGTH = 300
TEST_BATCH_SIZE = 1
HIDDEN_SIZE = 1024
NUM_FOLDS = 5
SEEDS = [113, 71, 17, 43]
test = pd.read_csv(os.path.join(INPUT_DIR, "test.csv"))
class MeanPoolingModel(nn.Module):
def __init__(self, model_name):
super().__init__()
config = AutoConfig.from_pretrained(model_name)
self.model = AutoModel.from_pretrained(model_name, config=config)
self.linear = nn.Linear(HIDDEN_SIZE, 1)
self.loss = nn.MSELoss()
def forward(self, input_ids, attention_mask, labels=None):
outputs = self.model(input_ids, attention_mask)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
logits = self.linear(mean_embeddings)
preds = logits.squeeze(-1).squeeze(-1)
if labels is not None:
loss = self.loss(preds.view(-1).float(), labels.view(-1).float())
return loss
else:
return preds
def get_test_loader(data):
x_test = data.excerpt.tolist()
tokenizer = AutoTokenizer.from_pretrained(MODEL_DIR)
encoded_test = tokenizer.batch_encode_plus(
x_test,
add_special_tokens=True,
return_attention_mask=True,
padding="max_length",
truncation=True,
max_length=MAX_LENGTH,
return_tensors="pt",
)
dataset_test = TensorDataset(
encoded_test["input_ids"], encoded_test["attention_mask"]
)
dataloader_test = DataLoader(
dataset_test,
sampler=SequentialSampler(dataset_test),
batch_size=TEST_BATCH_SIZE,
)
return dataloader_test
test_dataloader = get_test_loader(test)
all_predictions = []
for seed in SEEDS:
fold_predictions = []
for fold in tqdm(range(NUM_FOLDS)):
model_path = f"model_{seed + 1}_{fold + 1}.pth"
print(f"\nUsing {model_path}")
if seed in [113, 71]:
model_path = CHECKPOINT_DIR1 + f"model_{seed + 1}_{fold + 1}.pth"
else:
model_path = CHECKPOINT_DIR2 + f"model_{seed + 1}_{fold + 1}.pth"
model = MeanPoolingModel(MODEL_DIR)
model.load_state_dict(torch.load(model_path))
model.to(DEVICE)
model.eval()
predictions = []
for batch in test_dataloader:
batch = tuple(b.to(DEVICE) for b in batch)
inputs = {
"input_ids": batch[0],
"attention_mask": batch[1],
"labels": None,
}
preds = model(**inputs).item()
predictions.append(preds)
del model
gc.collect()
fold_predictions.append(predictions)
all_predictions.append(np.mean(fold_predictions, axis=0).tolist())
model4_predictions = np.mean(all_predictions, axis=0)
# # Model 5
import os
import gc
import sys
import cv2
import math
import time
import tqdm
import random
import numpy as np
import pandas as pd
import seaborn as sns
from tqdm import tqdm
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
from sklearn.svm import SVR
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold, StratifiedKFold
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.optim import Adam, lr_scheduler
from torch.utils.data import Dataset, DataLoader
from transformers import AutoModel, AutoTokenizer, AutoModelForSequenceClassification
import plotly.express as px
import plotly.graph_objs as go
import plotly.figure_factory as ff
from colorama import Fore, Back, Style
y_ = Fore.YELLOW
r_ = Fore.RED
g_ = Fore.GREEN
b_ = Fore.BLUE
m_ = Fore.MAGENTA
c_ = Fore.CYAN
sr_ = Style.RESET_ALL
train_data = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_data = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
sample = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
num_bins = int(np.floor(1 + np.log2(len(train_data))))
train_data.loc[:, "bins"] = pd.cut(train_data["target"], bins=num_bins, labels=False)
target = train_data["target"].to_numpy()
bins = train_data.bins.to_numpy()
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": 128,
"max_len": 256,
"nfolds": 5,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
class AttentionHead(nn.Module):
def __init__(self, in_features, hidden_dim, num_targets):
super().__init__()
self.in_features = in_features
self.middle_features = hidden_dim
self.W = nn.Linear(in_features, hidden_dim)
self.V = nn.Linear(hidden_dim, 1)
self.out_features = hidden_dim
def forward(self, features):
att = torch.tanh(self.W(features))
score = self.V(att)
attention_weights = torch.softmax(score, dim=1)
context_vector = attention_weights * features
context_vector = torch.sum(context_vector, dim=1)
return context_vector
class Model(nn.Module):
def __init__(self):
super(Model, self).__init__()
self.roberta = AutoModel.from_pretrained("../input/roberta-base")
self.head = AttentionHead(768, 768, 1)
self.dropout = nn.Dropout(0.1)
self.linear = nn.Linear(self.head.out_features, 1)
def forward(self, **xb):
x = self.roberta(**xb)[0]
x = self.head(x)
return x
def get_embeddings(df, path, plot_losses=True, verbose=True):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
tokenizer = AutoTokenizer.from_pretrained("../input/roberta-base")
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
return np.array(embeddings)
train_embeddings1 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model0/model0.bin"
)
test_embeddings1 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model0/model0.bin"
)
train_embeddings2 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model1/model1.bin"
)
test_embeddings2 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model1/model1.bin"
)
train_embeddings3 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model2/model2.bin"
)
test_embeddings3 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model2/model2.bin"
)
train_embeddings4 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model3/model3.bin"
)
test_embeddings4 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model3/model3.bin"
)
train_embeddings5 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model4/model4.bin"
)
test_embeddings5 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model4/model4.bin"
)
def get_preds_svm(X, y, X_test, bins=bins, nfolds=5, C=10, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(
n_splits=config["nfolds"], shuffle=True, random_state=config["seed"]
)
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
model = SVR(C=C, kernel=kernel, gamma="auto")
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
prediction = model.predict(X_valid)
score = rmse_score(prediction, y_valid)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
print("mean rmse", np.mean(scores))
return np.array(preds) / nfolds
svm_preds1 = get_preds_svm(train_embeddings1, target, test_embeddings1)
svm_preds2 = get_preds_svm(train_embeddings2, target, test_embeddings2)
svm_preds3 = get_preds_svm(train_embeddings3, target, test_embeddings3)
svm_preds4 = get_preds_svm(train_embeddings4, target, test_embeddings4)
svm_preds5 = get_preds_svm(train_embeddings5, target, test_embeddings5)
del train_embeddings1, test_embeddings1
gc.collect()
del train_embeddings2, test_embeddings2
gc.collect()
del train_embeddings3, test_embeddings3
gc.collect()
del train_embeddings4, test_embeddings4
gc.collect()
del train_embeddings5, test_embeddings5
gc.collect()
model5_predictions = (
svm_preds1 + svm_preds2 + svm_preds3 + svm_preds4 + svm_preds5
) / 5
# # Model 6
import os
from pathlib import Path
in_folder_path = Path("../input/clrp-finetune-dataset")
scripts_dir = Path(in_folder_path / "scripts")
os.chdir(scripts_dir)
exec(Path("imports.py").read_text())
exec(Path("config.py").read_text())
exec(Path("dataset.py").read_text())
exec(Path("model.py").read_text())
os.chdir("/kaggle/working")
test_df = pd.read_csv("/kaggle/input/commonlitreadabilityprize/test.csv")
tokenizer = torch.load("../input/tokenizers/roberta-tokenizer.pt")
models_folder_path = Path(in_folder_path / "models")
models_preds = []
n_models = 5
for model_num in range(n_models):
print(f"Inference#{model_num+1}/{n_models}")
test_ds = CLRPDataset(
data=test_df, tokenizer=tokenizer, max_len=Config.max_len, is_test=True
)
test_sampler = SequentialSampler(test_ds)
test_dataloader = DataLoader(
test_ds, sampler=test_sampler, batch_size=Config.batch_size
)
model = torch.load(models_folder_path / f"best_model_{model_num}.pt").to(
Config.device
)
all_preds = []
model.eval()
for step, batch in enumerate(test_dataloader):
sent_id, mask = batch["input_ids"].to(Config.device), batch[
"attention_mask"
].to(Config.device)
with torch.no_grad():
preds = model(sent_id, mask)
all_preds += preds.flatten().cpu().tolist()
models_preds.append(all_preds)
models_preds = np.array(models_preds)
model6_predictions = models_preds.mean(axis=0)
predictions = (
model1_predictions
+ model2_predictions
+ model3_predictions
+ model4_predictions
+ model5_predictions
+ model6_predictions
) / 6
predictions
submission_df.target = predictions
submission_df
submission_df.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605112.ipynb
|
commonlit-roberta-large-ii
|
rhtsingh
|
[{"Id": 69605112, "ScriptId": 18350823, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5233552, "CreationDate": "08/02/2021 05:32:11", "VersionNumber": 36.0, "Title": "commonlit-ensemble-6x-inference", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 974.0, "LinesInsertedFromPrevious": 13.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 961.0, "LinesInsertedFromFork": 530.0, "LinesDeletedFromFork": 35.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 444.0, "TotalVotes": 0}]
|
[{"Id": 93000459, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2279832}, {"Id": 93000465, "KernelVersionId": 69605112, "SourceDatasetVersionId": 906797}, {"Id": 93000455, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2397500}, {"Id": 93000467, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2488129}, {"Id": 93000460, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2263042}, {"Id": 93000454, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2417441}, {"Id": 93000466, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2475122}, {"Id": 93000452, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2441389}, {"Id": 93000456, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2383189}, {"Id": 93000451, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2451917}, {"Id": 93000453, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2418326}, {"Id": 93000462, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2210416}, {"Id": 93000463, "KernelVersionId": 69605112, "SourceDatasetVersionId": 1212718}, {"Id": 93000461, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2219267}, {"Id": 93000457, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2344115}, {"Id": 93000464, "KernelVersionId": 69605112, "SourceDatasetVersionId": 1042664}, {"Id": 93000458, "KernelVersionId": 69605112, "SourceDatasetVersionId": 2279836}]
|
[{"Id": 2279832, "DatasetId": 1373228, "DatasourceVersionId": 2320965, "CreatorUserId": 1314864, "LicenseName": "Unknown", "CreationDate": "05/28/2021 12:53:00", "VersionNumber": 1.0, "Title": "CommonLit RoBERTa Large II", "Slug": "commonlit-roberta-large-ii", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1373228, "CreatorUserId": 1314864, "OwnerUserId": 1314864.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2279832.0, "CurrentDatasourceVersionId": 2320965.0, "ForumId": 1392380, "Type": 2, "CreationDate": "05/28/2021 12:53:00", "LastActivityDate": "05/28/2021", "TotalViews": 1663, "TotalDownloads": 163, "TotalVotes": 21, "TotalKernels": 31}]
|
[{"Id": 1314864, "UserName": "rhtsingh", "DisplayName": "Rohit Singh", "RegisterDate": "10/05/2017", "PerformanceTier": 4}]
|
# # Overview
# This notebook combines four models.
# # Model 1
# The model is inspired by the one from [Maunish](https://www.kaggle.com/maunish/clrp-roberta-svm).
import os
import math
import random
import time
import numpy as np
import pandas as pd
from tqdm import tqdm
import torch
import torch.nn as nn
from torch.utils.data import Dataset
from torch.utils.data import DataLoader
from transformers import AutoTokenizer
from transformers import AutoModel
from transformers import AutoConfig
from sklearn.model_selection import KFold
from sklearn.svm import SVR
import gc
gc.enable()
BATCH_SIZE = 32
MAX_LEN = 248
EVAL_SCHEDULE = [(0.5, 16), (0.49, 8), (0.48, 4), (0.47, 2), (-1, 1)]
ROBERTA_PATH = "/kaggle/input/roberta-base"
TOKENIZER_PATH = "/kaggle/input/roberta-base"
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
DEVICE
test_df = pd.read_csv("/kaggle/input/commonlitreadabilityprize/test.csv")
submission_df = pd.read_csv(
"/kaggle/input/commonlitreadabilityprize/sample_submission.csv"
)
tokenizer = AutoTokenizer.from_pretrained(TOKENIZER_PATH)
class LitDataset(Dataset):
def __init__(self, df, inference_only=False):
super().__init__()
self.df = df
self.inference_only = inference_only
self.text = df.excerpt.tolist()
# self.text = [text.replace("\n", " ") for text in self.text]
if not self.inference_only:
self.target = torch.tensor(df.target.values, dtype=torch.float32)
self.encoded = tokenizer.batch_encode_plus(
self.text,
padding="max_length",
max_length=MAX_LEN,
truncation=True,
return_attention_mask=True,
)
def __len__(self):
return len(self.df)
def __getitem__(self, index):
input_ids = torch.tensor(self.encoded["input_ids"][index])
attention_mask = torch.tensor(self.encoded["attention_mask"][index])
if self.inference_only:
return (input_ids, attention_mask)
else:
target = self.target[index]
return (input_ids, attention_mask, target)
class LitModel(nn.Module):
def __init__(self):
super().__init__()
config = AutoConfig.from_pretrained(ROBERTA_PATH)
config.update(
{
"output_hidden_states": True,
"hidden_dropout_prob": 0.5,
"layer_norm_eps": 1e-7,
}
)
self.roberta = AutoModel.from_pretrained(ROBERTA_PATH, config=config)
self.attention = nn.Sequential(
nn.Linear(768, 512), nn.Tanh(), nn.Linear(512, 1), nn.Softmax(dim=1)
)
self.regressor = nn.Sequential(nn.Linear(768, 1))
def forward(self, input_ids, attention_mask):
roberta_output = self.roberta(
input_ids=input_ids, attention_mask=attention_mask
)
# There are a total of 13 layers of hidden states.
# 1 for the embedding layer, and 12 for the 12 Roberta layers.
# We take the hidden states from the last Roberta layer.
last_layer_hidden_states = roberta_output.hidden_states[-1]
# The number of cells is MAX_LEN.
# The size of the hidden state of each cell is 768 (for roberta-base).
# In order to condense hidden states of all cells to a context vector,
# we compute a weighted average of the hidden states of all cells.
# We compute the weight of each cell, using the attention neural network.
weights = self.attention(last_layer_hidden_states)
# weights.shape is BATCH_SIZE x MAX_LEN x 1
# last_layer_hidden_states.shape is BATCH_SIZE x MAX_LEN x 768
# Now we compute context_vector as the weighted average.
# context_vector.shape is BATCH_SIZE x 768
context_vector = torch.sum(weights * last_layer_hidden_states, dim=1)
# Now we reduce the context vector to the prediction score.
return self.regressor(context_vector)
def predict(model, data_loader):
"""Returns an np.array with predictions of the |model| on |data_loader|"""
model.eval()
result = np.zeros(len(data_loader.dataset))
index = 0
with torch.no_grad():
for batch_num, (input_ids, attention_mask) in enumerate(data_loader):
input_ids = input_ids.to(DEVICE)
attention_mask = attention_mask.to(DEVICE)
pred = model(input_ids, attention_mask)
result[index : index + pred.shape[0]] = pred.flatten().to("cpu")
index += pred.shape[0]
return result
NUM_MODELS = 5
all_predictions = np.zeros((NUM_MODELS, len(test_df)))
test_dataset = LitDataset(test_df, inference_only=True)
test_loader = DataLoader(
test_dataset, batch_size=BATCH_SIZE, drop_last=False, shuffle=False, num_workers=2
)
for model_index in tqdm(range(NUM_MODELS)):
model_path = f"../input/commonlit-roberta-0467/model_{model_index + 1}.pth"
print(f"\nUsing {model_path}")
model = LitModel()
model.load_state_dict(torch.load(model_path, map_location=DEVICE))
model.to(DEVICE)
all_predictions[model_index] = predict(model, test_loader)
del model
gc.collect()
model1_predictions = all_predictions.mean(axis=0)
# # Model 2
# Inspired from [https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3](https://www.kaggle.com/rhtsingh/commonlit-readability-prize-roberta-torch-infer-3)
test = test_df
from glob import glob
import os
import matplotlib.pyplot as plt
import json
from collections import defaultdict
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.optim.optimizer import Optimizer
import torch.optim.lr_scheduler as lr_scheduler
from torch.utils.data import Dataset, DataLoader, SequentialSampler, RandomSampler
from transformers import RobertaConfig
from transformers import (
get_cosine_schedule_with_warmup,
get_cosine_with_hard_restarts_schedule_with_warmup,
)
from transformers import RobertaTokenizer
from transformers import RobertaModel
from IPython.display import clear_output
def convert_examples_to_features(data, tokenizer, max_len, is_test=False):
data = data.replace("\n", "")
tok = tokenizer.encode_plus(
data,
max_length=max_len,
truncation=True,
return_attention_mask=True,
return_token_type_ids=True,
)
curr_sent = {}
padding_length = max_len - len(tok["input_ids"])
curr_sent["input_ids"] = tok["input_ids"] + ([0] * padding_length)
curr_sent["token_type_ids"] = tok["token_type_ids"] + ([0] * padding_length)
curr_sent["attention_mask"] = tok["attention_mask"] + ([0] * padding_length)
return curr_sent
class DatasetRetriever(Dataset):
def __init__(self, data, tokenizer, max_len, is_test=False):
self.data = data
self.excerpts = self.data.excerpt.values.tolist()
self.tokenizer = tokenizer
self.is_test = is_test
self.max_len = max_len
def __len__(self):
return len(self.data)
def __getitem__(self, item):
if not self.is_test:
excerpt, label = self.excerpts[item], self.targets[item]
features = convert_examples_to_features(
excerpt, self.tokenizer, self.max_len, self.is_test
)
return {
"input_ids": torch.tensor(features["input_ids"], dtype=torch.long),
"token_type_ids": torch.tensor(
features["token_type_ids"], dtype=torch.long
),
"attention_mask": torch.tensor(
features["attention_mask"], dtype=torch.long
),
"label": torch.tensor(label, dtype=torch.double),
}
else:
excerpt = self.excerpts[item]
features = convert_examples_to_features(
excerpt, self.tokenizer, self.max_len, self.is_test
)
return {
"input_ids": torch.tensor(features["input_ids"], dtype=torch.long),
"token_type_ids": torch.tensor(
features["token_type_ids"], dtype=torch.long
),
"attention_mask": torch.tensor(
features["attention_mask"], dtype=torch.long
),
}
class CommonLitModel(nn.Module):
def __init__(
self, model_name, config, multisample_dropout=True, output_hidden_states=False
):
super(CommonLitModel, self).__init__()
self.config = config
self.roberta = RobertaModel.from_pretrained(
model_name, output_hidden_states=output_hidden_states
)
self.layer_norm = nn.LayerNorm(config.hidden_size)
if multisample_dropout:
self.dropouts = nn.ModuleList([nn.Dropout(0.5) for _ in range(5)])
else:
self.dropouts = nn.ModuleList([nn.Dropout(0.3)])
self.regressor = nn.Linear(config.hidden_size, 1)
self._init_weights(self.layer_norm)
self._init_weights(self.regressor)
def _init_weights(self, module):
if isinstance(module, nn.Linear):
module.weight.data.normal_(mean=0.0, std=self.config.initializer_range)
if module.bias is not None:
module.bias.data.zero_()
elif isinstance(module, nn.Embedding):
module.weight.data.normal_(mean=0.0, std=self.config.initializer_range)
if module.padding_idx is not None:
module.weight.data[module.padding_idx].zero_()
elif isinstance(module, nn.LayerNorm):
module.bias.data.zero_()
module.weight.data.fill_(1.0)
def forward(
self, input_ids=None, attention_mask=None, token_type_ids=None, labels=None
):
outputs = self.roberta(
input_ids,
attention_mask=attention_mask,
token_type_ids=token_type_ids,
)
sequence_output = outputs[1]
sequence_output = self.layer_norm(sequence_output)
# multi-sample dropout
for i, dropout in enumerate(self.dropouts):
if i == 0:
logits = self.regressor(dropout(sequence_output))
else:
logits += self.regressor(dropout(sequence_output))
logits /= len(self.dropouts)
# calculate loss
loss = None
if labels is not None:
loss_fn = torch.nn.MSELoss()
logits = logits.view(-1).to(labels.dtype)
loss = torch.sqrt(loss_fn(logits, labels.view(-1)))
output = (logits,) + outputs[1:]
return ((loss,) + output) if loss is not None else output
def make_model(model_name, num_labels=1):
tokenizer = RobertaTokenizer.from_pretrained(model_name)
config = RobertaConfig.from_pretrained(model_name)
config.update({"num_labels": num_labels})
model = CommonLitModel(model_name, config=config)
return model, tokenizer
def make_loader(
data,
tokenizer,
max_len,
batch_size,
):
test_dataset = DatasetRetriever(data, tokenizer, max_len, is_test=True)
test_sampler = SequentialSampler(test_dataset)
test_loader = DataLoader(
test_dataset,
batch_size=batch_size // 2,
sampler=test_sampler,
pin_memory=False,
drop_last=False,
num_workers=0,
)
return test_loader
class Evaluator:
def __init__(self, model, scalar=None):
self.model = model
self.scalar = scalar
def evaluate(self, data_loader, tokenizer):
preds = []
self.model.eval()
total_loss = 0
with torch.no_grad():
for batch_idx, batch_data in enumerate(data_loader):
input_ids, attention_mask, token_type_ids = (
batch_data["input_ids"],
batch_data["attention_mask"],
batch_data["token_type_ids"],
)
input_ids, attention_mask, token_type_ids = (
input_ids.cuda(),
attention_mask.cuda(),
token_type_ids.cuda(),
)
if self.scalar is not None:
with torch.cuda.amp.autocast():
outputs = self.model(
input_ids=input_ids,
attention_mask=attention_mask,
token_type_ids=token_type_ids,
)
else:
outputs = self.model(
input_ids=input_ids,
attention_mask=attention_mask,
token_type_ids=token_type_ids,
)
logits = outputs[0].detach().cpu().numpy().squeeze().tolist()
preds += logits
return preds
def config(fold, model_name, load_model_path):
torch.manual_seed(2021)
torch.cuda.manual_seed(2021)
torch.cuda.manual_seed_all(2021)
max_len = 250
batch_size = 8
model, tokenizer = make_model(model_name=model_name, num_labels=1)
model.load_state_dict(torch.load(f"{load_model_path}/model{fold}.bin"))
test_loader = make_loader(test, tokenizer, max_len=max_len, batch_size=batch_size)
if torch.cuda.device_count() >= 1:
print(
"Model pushed to {} GPU(s), type {}.".format(
torch.cuda.device_count(), torch.cuda.get_device_name(0)
)
)
model = model.cuda()
else:
raise ValueError("CPU training is not supported")
# scaler = torch.cuda.amp.GradScaler()
scaler = None
return (model, tokenizer, test_loader, scaler)
def run(fold=0, model_name=None, load_model_path=None):
model, tokenizer, test_loader, scaler = config(fold, model_name, load_model_path)
import time
evaluator = Evaluator(model, scaler)
test_time_list = []
torch.cuda.synchronize()
tic1 = time.time()
preds = evaluator.evaluate(test_loader, tokenizer)
torch.cuda.synchronize()
tic2 = time.time()
test_time_list.append(tic2 - tic1)
del model, tokenizer, test_loader, scaler
gc.collect()
torch.cuda.empty_cache()
return preds
pred_df1 = pd.DataFrame()
pred_df2 = pd.DataFrame()
pred_df3 = pd.DataFrame()
for fold in tqdm(range(5)):
pred_df1[f"fold{fold}"] = run(
fold % 5, "../input/roberta-base/", "../input/commonlit-roberta-base-i/"
)
pred_df2[f"fold{fold+5}"] = run(
fold % 5, "../input/robertalarge/", "../input/roberta-large-itptfit/"
)
pred_df3[f"fold{fold+10}"] = run(
fold % 5, "../input/robertalarge/", "../input/commonlit-roberta-large-ii/"
)
pred_df1 = np.array(pred_df1)
pred_df2 = np.array(pred_df2)
pred_df3 = np.array(pred_df3)
model2_predictions = (
(pred_df2.mean(axis=1) * 0.5)
+ (pred_df1.mean(axis=1) * 0.3)
+ (pred_df3.mean(axis=1) * 0.2)
)
# # Model 3
# Inspired from: https://www.kaggle.com/ragnar123/commonlit-readability-roberta-tf-inference
import re
import os
import numpy as np
import pandas as pd
import random
import math
import tensorflow as tf
import logging
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold
from tensorflow.keras import backend as K
from transformers import RobertaTokenizer, TFRobertaModel
from kaggle_datasets import KaggleDatasets
tf.get_logger().setLevel(logging.ERROR)
from kaggle_datasets import KaggleDatasets
# Configurations
# Number of folds for training
FOLDS = 5
# Max length
MAX_LEN = 250
# Get the trained model we want to use
MODEL = "../input/tfroberta-base"
# Let's load our model tokenizer
tokenizer = RobertaTokenizer.from_pretrained(MODEL)
# This function tokenize the text according to a transformers model tokenizer
def regular_encode(texts, tokenizer, maxlen=MAX_LEN):
enc_di = tokenizer.batch_encode_plus(
texts,
padding="max_length",
truncation=True,
max_length=maxlen,
)
return np.array(enc_di["input_ids"])
# This function encode our training sentences
def encode_texts(x_test, MAX_LEN):
x_test = regular_encode(x_test.tolist(), tokenizer, maxlen=MAX_LEN)
return x_test
# Function to build our model
def build_roberta_base_model(max_len=MAX_LEN):
transformer = TFRobertaModel.from_pretrained(MODEL)
input_word_ids = tf.keras.layers.Input(
shape=(max_len,), dtype=tf.int32, name="input_word_ids"
)
sequence_output = transformer(input_word_ids)[0]
# We only need the cls_token, resulting in a 2d array
cls_token = sequence_output[:, 0, :]
output = tf.keras.layers.Dense(1, activation="linear", dtype="float32")(cls_token)
model = tf.keras.models.Model(inputs=[input_word_ids], outputs=[output])
return model
# Function for inference
def roberta_base_inference1():
# Read our test data
df = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
# Get text features
x_test = df["excerpt"]
# Encode our text with Roberta tokenizer
x_test = encode_texts(x_test, MAX_LEN)
# Initiate an empty vector to store prediction
predictions = np.zeros(len(df))
# Predict with the 5 models (5 folds training)
for i in range(FOLDS):
print("\n")
print("-" * 50)
print(f"Predicting with model {i + 1}")
# Build model
model = build_roberta_base_model(max_len=MAX_LEN)
# Load pretrained weights
model.load_weights(
f"../input/epochs-100-lr-4e5-seed-123/Roberta_Base_123_{i + 1}.h5"
)
# Predict
fold_predictions = model.predict(x_test).reshape(-1)
# Add fold prediction to the global predictions
predictions += fold_predictions / FOLDS
return predictions
model3_predictions = roberta_base_inference1()
# ## Model 4
# Inspired from: https://www.kaggle.com/jcesquiveld/best-transformer-representations
import os
import numpy as np
import pandas as pd
import random
from transformers import (
AutoConfig,
AutoModel,
AutoTokenizer,
AdamW,
get_linear_schedule_with_warmup,
logging,
)
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import (
Dataset,
TensorDataset,
SequentialSampler,
RandomSampler,
DataLoader,
)
from tqdm.notebook import tqdm
import gc
gc.enable()
from IPython.display import clear_output
from sklearn.model_selection import StratifiedKFold
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("whitegrid")
logging.set_verbosity_error()
INPUT_DIR = "../input/commonlitreadabilityprize"
MODEL_DIR = "../input/roberta-transformers-pytorch/roberta-large"
CHECKPOINT_DIR1 = "../input/clrp-mean-pooling/"
CHECKPOINT_DIR2 = "../input/clrp-mean-pooling-seeds-17-43/"
DEVICE = torch.device("cuda" if torch.cuda.is_available() else "cpu")
MAX_LENGTH = 300
TEST_BATCH_SIZE = 1
HIDDEN_SIZE = 1024
NUM_FOLDS = 5
SEEDS = [113, 71, 17, 43]
test = pd.read_csv(os.path.join(INPUT_DIR, "test.csv"))
class MeanPoolingModel(nn.Module):
def __init__(self, model_name):
super().__init__()
config = AutoConfig.from_pretrained(model_name)
self.model = AutoModel.from_pretrained(model_name, config=config)
self.linear = nn.Linear(HIDDEN_SIZE, 1)
self.loss = nn.MSELoss()
def forward(self, input_ids, attention_mask, labels=None):
outputs = self.model(input_ids, attention_mask)
last_hidden_state = outputs[0]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(last_hidden_state.size()).float()
)
sum_embeddings = torch.sum(last_hidden_state * input_mask_expanded, 1)
sum_mask = input_mask_expanded.sum(1)
sum_mask = torch.clamp(sum_mask, min=1e-9)
mean_embeddings = sum_embeddings / sum_mask
logits = self.linear(mean_embeddings)
preds = logits.squeeze(-1).squeeze(-1)
if labels is not None:
loss = self.loss(preds.view(-1).float(), labels.view(-1).float())
return loss
else:
return preds
def get_test_loader(data):
x_test = data.excerpt.tolist()
tokenizer = AutoTokenizer.from_pretrained(MODEL_DIR)
encoded_test = tokenizer.batch_encode_plus(
x_test,
add_special_tokens=True,
return_attention_mask=True,
padding="max_length",
truncation=True,
max_length=MAX_LENGTH,
return_tensors="pt",
)
dataset_test = TensorDataset(
encoded_test["input_ids"], encoded_test["attention_mask"]
)
dataloader_test = DataLoader(
dataset_test,
sampler=SequentialSampler(dataset_test),
batch_size=TEST_BATCH_SIZE,
)
return dataloader_test
test_dataloader = get_test_loader(test)
all_predictions = []
for seed in SEEDS:
fold_predictions = []
for fold in tqdm(range(NUM_FOLDS)):
model_path = f"model_{seed + 1}_{fold + 1}.pth"
print(f"\nUsing {model_path}")
if seed in [113, 71]:
model_path = CHECKPOINT_DIR1 + f"model_{seed + 1}_{fold + 1}.pth"
else:
model_path = CHECKPOINT_DIR2 + f"model_{seed + 1}_{fold + 1}.pth"
model = MeanPoolingModel(MODEL_DIR)
model.load_state_dict(torch.load(model_path))
model.to(DEVICE)
model.eval()
predictions = []
for batch in test_dataloader:
batch = tuple(b.to(DEVICE) for b in batch)
inputs = {
"input_ids": batch[0],
"attention_mask": batch[1],
"labels": None,
}
preds = model(**inputs).item()
predictions.append(preds)
del model
gc.collect()
fold_predictions.append(predictions)
all_predictions.append(np.mean(fold_predictions, axis=0).tolist())
model4_predictions = np.mean(all_predictions, axis=0)
# # Model 5
import os
import gc
import sys
import cv2
import math
import time
import tqdm
import random
import numpy as np
import pandas as pd
import seaborn as sns
from tqdm import tqdm
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
from sklearn.svm import SVR
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold, StratifiedKFold
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.optim import Adam, lr_scheduler
from torch.utils.data import Dataset, DataLoader
from transformers import AutoModel, AutoTokenizer, AutoModelForSequenceClassification
import plotly.express as px
import plotly.graph_objs as go
import plotly.figure_factory as ff
from colorama import Fore, Back, Style
y_ = Fore.YELLOW
r_ = Fore.RED
g_ = Fore.GREEN
b_ = Fore.BLUE
m_ = Fore.MAGENTA
c_ = Fore.CYAN
sr_ = Style.RESET_ALL
train_data = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_data = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
sample = pd.read_csv("../input/commonlitreadabilityprize/sample_submission.csv")
num_bins = int(np.floor(1 + np.log2(len(train_data))))
train_data.loc[:, "bins"] = pd.cut(train_data["target"], bins=num_bins, labels=False)
target = train_data["target"].to_numpy()
bins = train_data.bins.to_numpy()
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
config = {
"batch_size": 128,
"max_len": 256,
"nfolds": 5,
"seed": 42,
}
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONASSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(seed=config["seed"])
class CLRPDataset(Dataset):
def __init__(self, df, tokenizer):
self.excerpt = df["excerpt"].to_numpy()
self.tokenizer = tokenizer
def __getitem__(self, idx):
encode = self.tokenizer(
self.excerpt[idx],
return_tensors="pt",
max_length=config["max_len"],
padding="max_length",
truncation=True,
)
return encode
def __len__(self):
return len(self.excerpt)
class AttentionHead(nn.Module):
def __init__(self, in_features, hidden_dim, num_targets):
super().__init__()
self.in_features = in_features
self.middle_features = hidden_dim
self.W = nn.Linear(in_features, hidden_dim)
self.V = nn.Linear(hidden_dim, 1)
self.out_features = hidden_dim
def forward(self, features):
att = torch.tanh(self.W(features))
score = self.V(att)
attention_weights = torch.softmax(score, dim=1)
context_vector = attention_weights * features
context_vector = torch.sum(context_vector, dim=1)
return context_vector
class Model(nn.Module):
def __init__(self):
super(Model, self).__init__()
self.roberta = AutoModel.from_pretrained("../input/roberta-base")
self.head = AttentionHead(768, 768, 1)
self.dropout = nn.Dropout(0.1)
self.linear = nn.Linear(self.head.out_features, 1)
def forward(self, **xb):
x = self.roberta(**xb)[0]
x = self.head(x)
return x
def get_embeddings(df, path, plot_losses=True, verbose=True):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
model = Model()
model.load_state_dict(torch.load(path))
model.to(device)
model.eval()
tokenizer = AutoTokenizer.from_pretrained("../input/roberta-base")
ds = CLRPDataset(df, tokenizer)
dl = DataLoader(
ds,
batch_size=config["batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
drop_last=False,
)
embeddings = list()
with torch.no_grad():
for i, inputs in tqdm(enumerate(dl)):
inputs = {
key: val.reshape(val.shape[0], -1).to(device)
for key, val in inputs.items()
}
outputs = model(**inputs)
outputs = outputs.detach().cpu().numpy()
embeddings.extend(outputs)
return np.array(embeddings)
train_embeddings1 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model0/model0.bin"
)
test_embeddings1 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model0/model0.bin"
)
train_embeddings2 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model1/model1.bin"
)
test_embeddings2 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model1/model1.bin"
)
train_embeddings3 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model2/model2.bin"
)
test_embeddings3 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model2/model2.bin"
)
train_embeddings4 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model3/model3.bin"
)
test_embeddings4 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model3/model3.bin"
)
train_embeddings5 = get_embeddings(
train_data, "../input/roberta-svm-finetune/model4/model4.bin"
)
test_embeddings5 = get_embeddings(
test_data, "../input/roberta-svm-finetune/model4/model4.bin"
)
def get_preds_svm(X, y, X_test, bins=bins, nfolds=5, C=10, kernel="rbf"):
scores = list()
preds = np.zeros((X_test.shape[0]))
kfold = StratifiedKFold(
n_splits=config["nfolds"], shuffle=True, random_state=config["seed"]
)
for k, (train_idx, valid_idx) in enumerate(kfold.split(X, bins)):
model = SVR(C=C, kernel=kernel, gamma="auto")
X_train, y_train = X[train_idx], y[train_idx]
X_valid, y_valid = X[valid_idx], y[valid_idx]
model.fit(X_train, y_train)
prediction = model.predict(X_valid)
score = rmse_score(prediction, y_valid)
print(f"Fold {k} , rmse score: {score}")
scores.append(score)
preds += model.predict(X_test)
print("mean rmse", np.mean(scores))
return np.array(preds) / nfolds
svm_preds1 = get_preds_svm(train_embeddings1, target, test_embeddings1)
svm_preds2 = get_preds_svm(train_embeddings2, target, test_embeddings2)
svm_preds3 = get_preds_svm(train_embeddings3, target, test_embeddings3)
svm_preds4 = get_preds_svm(train_embeddings4, target, test_embeddings4)
svm_preds5 = get_preds_svm(train_embeddings5, target, test_embeddings5)
del train_embeddings1, test_embeddings1
gc.collect()
del train_embeddings2, test_embeddings2
gc.collect()
del train_embeddings3, test_embeddings3
gc.collect()
del train_embeddings4, test_embeddings4
gc.collect()
del train_embeddings5, test_embeddings5
gc.collect()
model5_predictions = (
svm_preds1 + svm_preds2 + svm_preds3 + svm_preds4 + svm_preds5
) / 5
# # Model 6
import os
from pathlib import Path
in_folder_path = Path("../input/clrp-finetune-dataset")
scripts_dir = Path(in_folder_path / "scripts")
os.chdir(scripts_dir)
exec(Path("imports.py").read_text())
exec(Path("config.py").read_text())
exec(Path("dataset.py").read_text())
exec(Path("model.py").read_text())
os.chdir("/kaggle/working")
test_df = pd.read_csv("/kaggle/input/commonlitreadabilityprize/test.csv")
tokenizer = torch.load("../input/tokenizers/roberta-tokenizer.pt")
models_folder_path = Path(in_folder_path / "models")
models_preds = []
n_models = 5
for model_num in range(n_models):
print(f"Inference#{model_num+1}/{n_models}")
test_ds = CLRPDataset(
data=test_df, tokenizer=tokenizer, max_len=Config.max_len, is_test=True
)
test_sampler = SequentialSampler(test_ds)
test_dataloader = DataLoader(
test_ds, sampler=test_sampler, batch_size=Config.batch_size
)
model = torch.load(models_folder_path / f"best_model_{model_num}.pt").to(
Config.device
)
all_preds = []
model.eval()
for step, batch in enumerate(test_dataloader):
sent_id, mask = batch["input_ids"].to(Config.device), batch[
"attention_mask"
].to(Config.device)
with torch.no_grad():
preds = model(sent_id, mask)
all_preds += preds.flatten().cpu().tolist()
models_preds.append(all_preds)
models_preds = np.array(models_preds)
model6_predictions = models_preds.mean(axis=0)
predictions = (
model1_predictions
+ model2_predictions
+ model3_predictions
+ model4_predictions
+ model5_predictions
+ model6_predictions
) / 6
predictions
submission_df.target = predictions
submission_df
submission_df.to_csv("submission.csv", index=False)
| false | 3 | 8,883 | 0 | 8,912 | 8,883 |
||
69605181
|
<jupyter_start><jupyter_text>COVID-19 data from John Hopkins University
This is a daily updating version of [COVID-19 Data Repository](https://github.com/CSSEGISandData/COVID-19) by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). The data updates every day at 6am UTC, which updates just after the raw JHU data typically updates.
I'm making it available in both a raw form (files with the prefix RAW) and convenient form (files prefixed with CONVENIENT).
The data covers:
- confirmed cases and deaths on a country level
- confirmed cases and deaths by US county
- some metadata that's available in the raw JHU data
The RAW version is exactly as it's distributed in the original dataset.
The CONVENIENT version is aiming to be easier to analyze. The data is organized by column rather than by row. The metadata is stripped out into a separate file. And it converted to daily change rather than cumulative totals.
If you find any issues in the data, then you can share them in this [discussion thread](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/discussion/195628). I will attempt to address the most upvoted issues.
If you have any requests for changing or enriching this data, please add them on this [discussion thread](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/discussion/195984). Again, I will attempt to address the most upvoted requests.
I have a notebook that updates just after each data dump updates, giving a brief overview of the [latest data](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/notebooks). It's also a useful reference if you want to see how to read the CONVENIENT data into a pandas DataFrame.
Kaggle dataset identifier: covid19-data-from-john-hopkins-university
<jupyter_code>import pandas as pd
df = pd.read_csv('covid19-data-from-john-hopkins-university/CONVENIENT_us_metadata.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 3342 entries, 0 to 3341
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Province_State 3342 non-null object
1 Admin2 3336 non-null object
2 Population 3342 non-null int64
3 Lat 3342 non-null float64
4 Long 3342 non-null float64
dtypes: float64(2), int64(1), object(2)
memory usage: 130.7+ KB
<jupyter_text>Examples:
{
"Province_State": "Alabama",
"Admin2": "Autauga",
"Population": 55869,
"Lat": 32.53952745,
"Long": -86.64408227
}
{
"Province_State": "Alabama",
"Admin2": "Baldwin",
"Population": 223234,
"Lat": 30.72774991,
"Long": -87.72207058
}
{
"Province_State": "Alabama",
"Admin2": "Barbour",
"Population": 24686,
"Lat": 31.868263,
"Long": -85.3871286
}
{
"Province_State": "Alabama",
"Admin2": "Bibb",
"Population": 22394,
"Lat": 32.99642064,
"Long": -87.1251146
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
df_dict = {}
table_list = [
"global_confirmed_cases",
"global_deaths",
"us_confirmed_cases",
"us_deaths",
]
for table in table_list:
df_dict[table] = df_convenient_global_cases = pd.read_csv(
f"/kaggle/input/covid19-data-from-john-hopkins-university/CONVENIENT_{table}.csv",
header=[0, 1],
index_col=0,
)
# ## Confirmed cases: Top 10 US counties - last 7 days
most_recent_date = df_dict["us_confirmed_cases"].index[
len(df_dict["us_confirmed_cases"]) - 1
]
df_dict["us_confirmed_cases"].tail(7).sum().sort_values(ascending=False)[0:30]
# ## Confirmed cases per 10K people for counties with >=10K people
df_county_metadata = pd.read_csv(
"../input/covid19-data-from-john-hopkins-university/CONVENIENT_us_metadata.csv",
index_col=[0, 1],
)
counties_to_consider = df_county_metadata[
(df_county_metadata["Population"].index.isin(df_dict["us_confirmed_cases"].columns))
& (df_county_metadata["Population"] > 10000)
].index
(
df_dict["us_confirmed_cases"].tail(7).sum()[counties_to_consider]
/ df_county_metadata["Population"][counties_to_consider]
).sort_values(ascending=False)[:30] * 10000
# ## Deaths: Top 10 US counties
# Sum last 7 days
most_recent_date = df_dict["us_deaths"].index[len(df_dict["us_deaths"]) - 1]
df_dict["us_deaths"].tail(7).sum().sort_values(ascending=False)[0:30]
# ## Confirmed cases: Top 10 countries
# Sum last 7 days
most_recent_date = df_dict["global_confirmed_cases"].index[
len(df_dict["global_confirmed_cases"]) - 1
]
df_dict["global_confirmed_cases"].T.groupby(level=0).sum().T.tail(7).sum().sort_values(
ascending=False
)[0:30]
# ## Deaths: Top 10 countries
# Sum last 7 days
most_recent_date = df_dict["global_deaths"].index[len(df_dict["global_deaths"]) - 1]
df_dict["global_deaths"].T.groupby(level=0).sum().T.tail(7).sum().sort_values(
ascending=False
)[0:30]
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605181.ipynb
|
covid19-data-from-john-hopkins-university
|
antgoldbloom
|
[{"Id": 69605181, "ScriptId": 12728489, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 368, "CreationDate": "08/02/2021 05:33:23", "VersionNumber": 274.0, "Title": "Quick look at latest data\u00a0(auto-updates daily)", "EvaluationDate": "08/02/2021", "IsChange": false, "TotalLines": 41.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 41.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93000586, "KernelVersionId": 69605181, "SourceDatasetVersionId": 2490132}]
|
[{"Id": 2490132, "DatasetId": 951970, "DatasourceVersionId": 2532702, "CreatorUserId": 368, "LicenseName": "Attribution 4.0 International (CC BY 4.0)", "CreationDate": "08/02/2021 05:03:46", "VersionNumber": 290.0, "Title": "COVID-19 data from John Hopkins University", "Slug": "covid19-data-from-john-hopkins-university", "Subtitle": "Updated daily at 6am UTC in both raw and convenient form", "Description": "This is a daily updating version of [COVID-19 Data Repository](https://github.com/CSSEGISandData/COVID-19) by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). The data updates every day at 6am UTC, which updates just after the raw JHU data typically updates. \n\nI'm making it available in both a raw form (files with the prefix RAW) and convenient form (files prefixed with CONVENIENT). \n\nThe data covers:\n- confirmed cases and deaths on a country level \n- confirmed cases and deaths by US county\n- some metadata that's available in the raw JHU data\n\nThe RAW version is exactly as it's distributed in the original dataset.\n\nThe CONVENIENT version is aiming to be easier to analyze. The data is organized by column rather than by row. The metadata is stripped out into a separate file. And it converted to daily change rather than cumulative totals. \n\nIf you find any issues in the data, then you can share them in this [discussion thread](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/discussion/195628). I will attempt to address the most upvoted issues.\n\nIf you have any requests for changing or enriching this data, please add them on this [discussion thread](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/discussion/195984). Again, I will attempt to address the most upvoted requests. \n\nI have a notebook that updates just after each data dump updates, giving a brief overview of the [latest data](https://www.kaggle.com/antgoldbloom/covid19-data-from-john-hopkins-university/notebooks). It's also a useful reference if you want to see how to read the CONVENIENT data into a pandas DataFrame.", "VersionNotes": "2021-08-02 05:03:29 automated update", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 951970, "CreatorUserId": 368, "OwnerUserId": 368.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 5651576.0, "CurrentDatasourceVersionId": 5726942.0, "ForumId": 968155, "Type": 2, "CreationDate": "11/02/2020 14:29:16", "LastActivityDate": "11/02/2020", "TotalViews": 119682, "TotalDownloads": 19634, "TotalVotes": 391, "TotalKernels": 60}]
|
[{"Id": 368, "UserName": "antgoldbloom", "DisplayName": "Anthony Goldbloom", "RegisterDate": "01/20/2010", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
df_dict = {}
table_list = [
"global_confirmed_cases",
"global_deaths",
"us_confirmed_cases",
"us_deaths",
]
for table in table_list:
df_dict[table] = df_convenient_global_cases = pd.read_csv(
f"/kaggle/input/covid19-data-from-john-hopkins-university/CONVENIENT_{table}.csv",
header=[0, 1],
index_col=0,
)
# ## Confirmed cases: Top 10 US counties - last 7 days
most_recent_date = df_dict["us_confirmed_cases"].index[
len(df_dict["us_confirmed_cases"]) - 1
]
df_dict["us_confirmed_cases"].tail(7).sum().sort_values(ascending=False)[0:30]
# ## Confirmed cases per 10K people for counties with >=10K people
df_county_metadata = pd.read_csv(
"../input/covid19-data-from-john-hopkins-university/CONVENIENT_us_metadata.csv",
index_col=[0, 1],
)
counties_to_consider = df_county_metadata[
(df_county_metadata["Population"].index.isin(df_dict["us_confirmed_cases"].columns))
& (df_county_metadata["Population"] > 10000)
].index
(
df_dict["us_confirmed_cases"].tail(7).sum()[counties_to_consider]
/ df_county_metadata["Population"][counties_to_consider]
).sort_values(ascending=False)[:30] * 10000
# ## Deaths: Top 10 US counties
# Sum last 7 days
most_recent_date = df_dict["us_deaths"].index[len(df_dict["us_deaths"]) - 1]
df_dict["us_deaths"].tail(7).sum().sort_values(ascending=False)[0:30]
# ## Confirmed cases: Top 10 countries
# Sum last 7 days
most_recent_date = df_dict["global_confirmed_cases"].index[
len(df_dict["global_confirmed_cases"]) - 1
]
df_dict["global_confirmed_cases"].T.groupby(level=0).sum().T.tail(7).sum().sort_values(
ascending=False
)[0:30]
# ## Deaths: Top 10 countries
# Sum last 7 days
most_recent_date = df_dict["global_deaths"].index[len(df_dict["global_deaths"]) - 1]
df_dict["global_deaths"].T.groupby(level=0).sum().T.tail(7).sum().sort_values(
ascending=False
)[0:30]
|
[{"covid19-data-from-john-hopkins-university/CONVENIENT_us_metadata.csv": {"column_names": "[\"Province_State\", \"Admin2\", \"Population\", \"Lat\", \"Long\"]", "column_data_types": "{\"Province_State\": \"object\", \"Admin2\": \"object\", \"Population\": \"int64\", \"Lat\": \"float64\", \"Long\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 3342 entries, 0 to 3341\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Province_State 3342 non-null object \n 1 Admin2 3336 non-null object \n 2 Population 3342 non-null int64 \n 3 Lat 3342 non-null float64\n 4 Long 3342 non-null float64\ndtypes: float64(2), int64(1), object(2)\nmemory usage: 130.7+ KB\n", "summary": "{\"Population\": {\"count\": 3342.0, \"mean\": 99603.57181328545, \"std\": 324166.10491497914, \"min\": 0.0, \"25%\": 9917.25, \"50%\": 24891.5, \"75%\": 64975.25, \"max\": 10039107.0}, \"Lat\": {\"count\": 3342.0, \"mean\": 36.7216172437672, \"std\": 9.079322310859308, \"min\": -14.271, \"25%\": 33.89680287, \"50%\": 38.005609559999996, \"75%\": 41.5792554875, \"max\": 69.31479216}, \"Long\": {\"count\": 3342.0, \"mean\": -88.64204524277079, \"std\": 21.77628672289758, \"min\": -174.1596, \"25%\": -97.80359504500001, \"50%\": -89.48886474, \"75%\": -82.31339778, \"max\": 145.6739}}", "examples": "{\"Province_State\":{\"0\":\"Alabama\",\"1\":\"Alabama\",\"2\":\"Alabama\",\"3\":\"Alabama\"},\"Admin2\":{\"0\":\"Autauga\",\"1\":\"Baldwin\",\"2\":\"Barbour\",\"3\":\"Bibb\"},\"Population\":{\"0\":55869,\"1\":223234,\"2\":24686,\"3\":22394},\"Lat\":{\"0\":32.53952745,\"1\":30.72774991,\"2\":31.868263,\"3\":32.99642064},\"Long\":{\"0\":-86.64408227,\"1\":-87.72207058,\"2\":-85.3871286,\"3\":-87.1251146}}"}}]
| true | 1 |
<start_data_description><data_path>covid19-data-from-john-hopkins-university/CONVENIENT_us_metadata.csv:
<column_names>
['Province_State', 'Admin2', 'Population', 'Lat', 'Long']
<column_types>
{'Province_State': 'object', 'Admin2': 'object', 'Population': 'int64', 'Lat': 'float64', 'Long': 'float64'}
<dataframe_Summary>
{'Population': {'count': 3342.0, 'mean': 99603.57181328545, 'std': 324166.10491497914, 'min': 0.0, '25%': 9917.25, '50%': 24891.5, '75%': 64975.25, 'max': 10039107.0}, 'Lat': {'count': 3342.0, 'mean': 36.7216172437672, 'std': 9.079322310859308, 'min': -14.271, '25%': 33.89680287, '50%': 38.005609559999996, '75%': 41.5792554875, 'max': 69.31479216}, 'Long': {'count': 3342.0, 'mean': -88.64204524277079, 'std': 21.77628672289758, 'min': -174.1596, '25%': -97.80359504500001, '50%': -89.48886474, '75%': -82.31339778, 'max': 145.6739}}
<dataframe_info>
RangeIndex: 3342 entries, 0 to 3341
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Province_State 3342 non-null object
1 Admin2 3336 non-null object
2 Population 3342 non-null int64
3 Lat 3342 non-null float64
4 Long 3342 non-null float64
dtypes: float64(2), int64(1), object(2)
memory usage: 130.7+ KB
<some_examples>
{'Province_State': {'0': 'Alabama', '1': 'Alabama', '2': 'Alabama', '3': 'Alabama'}, 'Admin2': {'0': 'Autauga', '1': 'Baldwin', '2': 'Barbour', '3': 'Bibb'}, 'Population': {'0': 55869, '1': 223234, '2': 24686, '3': 22394}, 'Lat': {'0': 32.53952745, '1': 30.72774991, '2': 31.868263, '3': 32.99642064}, 'Long': {'0': -86.64408227, '1': -87.72207058, '2': -85.3871286, '3': -87.1251146}}
<end_description>
| 724 | 0 | 1,715 | 724 |
69605615
|
# ## Summary
# This notebook aim to create Models to make predictions on Titanic - Machine Learning from Disaster dataset. And I will try to answer following questions:
# - Which Model can achieve a better performance? In order to answer this question, I use different kinds of Algorithms:
# - Deep Neural Network
# - Deep and Wide Neural Network with TensorFlow DenseFeatures layer
# - Logistic Regression
# - KNN
# - Decision Tree Classifier
# - Gradient Boosting Classifier¶
# - Random Forest Classifier
#
# - How to preprocess the data to get the optimal results? I will do EDA and try to add more features based on existing dataset. I will use statistic methods to search for features that's correlated to survival.
# ## Import Packages
import pandas as pd
import numpy as np
import sklearn
import seaborn as sns
import tensorflow as tf
from sklearn.ensemble import GradientBoostingClassifier, RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn import model_selection
from sklearn.cluster import KMeans
from sklearn.metrics import accuracy_score
# ## Common Functions
# **Save results**
def save_results(Survived, test, path):
submission = pd.DataFrame(
{"PassengerId": test["PassengerId"], "Survived": Survived}
)
submission.to_csv(path, index=False)
# **Evaluate Model, save and keep tract of best results**
sklearn.metrics.f1_score
def evaulate_and_save(
model,
validation_features,
validation_targets,
test_features,
save_path,
best_score,
best_path,
):
y_pred = model.predict(validation_features)
if y_pred.dtype != int:
if y_pred.shape[-1] == 2:
y_pred = np.argmax(y_pred, axis=-1)
if y_pred.shape[-1] == 1:
y_pred = np.array(y_pred > 0.5, dtype=int)
y_pred = y_pred.reshape(-1)
score = sklearn.metrics.accuracy_score(validation_targets, y_pred)
f1 = sklearn.metrics.f1_score(validation_targets, y_pred)
print("Accuracy Score:", score)
print("Classification Report:")
print(sklearn.metrics.classification_report(validation_targets, y_pred))
Survived = model.predict(test_features)
if Survived.dtype != int:
if Survived.shape[-1] == 2:
Survived = np.argmax(Survived, axis=-1)
if Survived.shape[-1] == 1:
Survived = np.array(Survived > 0.5, dtype=int)
Survived = Survived.reshape(-1)
save_results(Survived, test, save_path)
if f1 > best_score:
best_score = f1
best_path = path
return best_score, best_path
# ## Import datasets
train = pd.read_csv("/kaggle/input/titanic/train.csv")
test = pd.read_csv("/kaggle/input/titanic/test.csv")
train.head()
test.head()
# ## Data Wrangling and Preprocessing
# As we can see Age, Cabin and Fare information contains missing values, so we need to apply Missing Value Imputation to them.
train.isnull().sum()
test.isnull().sum()
train["Cabin"] = train["Cabin"].replace(np.NAN, train["Cabin"].mode()[0])
test["Cabin"] = test["Cabin"].replace(np.NAN, test["Cabin"].mode()[0])
train["Embarked"] = train["Embarked"].replace(np.NAN, train["Embarked"].mode()[0])
train["Age"] = train["Age"].replace(np.NAN, train["Age"].mean())
test["Age"] = test["Age"].replace(np.NAN, test["Age"].mean())
test["Fare"] = test["Fare"].replace(np.NAN, test["Fare"].mean())
# Let's see the Cabin labels, there are so many of them. But I make an assmptionn that the First Alphabet matters, it indicates the location and class of the passengers so it has an impact to survive.
cabin_labels = sorted(set(list(train["Cabin"].unique()) + list(test["Cabin"].unique())))
print(cabin_labels[:30])
train["Cabin_type"] = train["Cabin"].apply(lambda cabin: cabin[0])
test["Cabin_type"] = test["Cabin"].apply(lambda cabin: cabin[0])
# ## Handle Categorical Features
categorical_features = ["Sex", "Cabin_type", "Embarked"]
categorical_label_dictionary = dict()
for feature in categorical_features:
unique_labels = sorted(
set(list(train[feature].unique()) + list(test[feature].unique()))
)
for data in [train, test]:
categorical_label_dictionary[feature] = unique_labels
data[feature + "_value"] = data[feature].apply(
lambda item: unique_labels.index(item)
)
# Let's see after we preprocess, what does the data look like?
train.head(30)
# ## Exploratory Data Analysis
# ### Basic Statistic infos
train.info()
train.describe()
# ### What's the factor to survive?
# As we can see it's related to Gender, PClass, Status, Fare Cabin and Embarked.
train.corr()["Survived"].sort_values(ascending=False)
related_features = list(train.corr()[train.corr()["Survived"].abs() > 0.05].index)
related_features.remove("Survived")
print(related_features)
# ## Survive Rate
# **Overall Survive Rate**
train.Survived.mean()
# **Survive Rate across different genders**
# As we can see women are more likely to survive.
train.groupby("Sex").Survived.mean()
# ### Preprocess Data
train_test = pd.concat([train, test])[related_features]
train_test.head()
for feature in ["Sex", "Cabin_type", "Embarked"]:
items = pd.get_dummies(train_test[feature + "_value"])
labels = categorical_label_dictionary[feature]
items.columns = [feature + "_" + labels[column] for column in list(items.columns)]
train_test[items.columns] = items
train_test.pop(feature + "_value")
train_test.head()
train_features = train_test.iloc[0 : len(train)]
test_features = train_test.iloc[len(train) :]
train_features.head()
test_features.head()
# ### Train Validation Split
(
train_features,
validation_features,
train_targets,
validation_targets,
) = model_selection.train_test_split(
train_features, train["Survived"], test_size=0.2, random_state=88
)
print(
train_features.shape,
validation_features.shape,
train_targets.shape,
validation_targets.shape,
)
# ## Model Development and Evaluation
# I will try differnt Models and use results from best Model.
best_score = 0
best_path = ""
# ### Using Deep Neural Network
model = tf.keras.Sequential(
[
tf.keras.layers.Input(shape=(train_features.shape[1])),
tf.keras.layers.Dense(32, activation="relu"),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dense(32, activation="relu"),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
early_stop = tf.keras.callbacks.EarlyStopping(monitor="val_accuracy", patience=20)
history = model.fit(
train_features,
train_targets,
epochs=400,
validation_data=(validation_features, validation_targets),
callbacks=[early_stop],
verbose=0,
)
pd.DataFrame(history.history).plot()
best_score, best_path = evaulate_and_save(
model,
validation_features,
validation_targets,
test_features,
"submission_dnn.csv",
best_score,
best_path,
)
# ### Using Deep and Wide Model
from tensorflow import feature_column
categorical_feature_names = [
"Pclass",
"Sex_value",
"Embarked_value",
"Cabin_type_value",
]
numerical_feature_names = ["Age", "Fare"]
categorical_features = [
feature_column.indicator_column(
feature_column.categorical_column_with_vocabulary_list(
key, sorted(list(train[key].unique()))
)
)
for key in categorical_feature_names
]
numerical_features = [
feature_column.numeric_column(key) for key in numerical_feature_names
]
input_dictionary = dict()
inputs = dict()
for item in numerical_features:
inputs[item.key] = tf.keras.layers.Input(name=item.key, shape=())
for item in categorical_features:
inputs[item.categorical_column.key] = tf.keras.layers.Input(
name=item.categorical_column.key, shape=(), dtype="int32"
)
def features_and_labels(row_data):
label = row_data.pop("Survived")
features = row_data
return features, label
def create_dataset(pattern, epochs=1, batch_size=32, mode="eval"):
dataset = tf.data.experimental.make_csv_dataset(pattern, batch_size)
dataset = dataset.map(features_and_labels)
if mode == "train":
dataset = dataset.shuffle(buffer_size=128).repeat(epochs)
dataset = dataset.prefetch(1)
return dataset
def create_test_dataset(pattern, batch_size=32):
dataset = tf.data.experimental.make_csv_dataset(pattern, batch_size)
dataset = dataset.map(lambda features: features)
dataset = dataset.prefetch(1)
return dataset
from sklearn.model_selection import train_test_split
train_data, val_data = train_test_split(
train[categorical_feature_names + numerical_feature_names + ["Survived"]],
test_size=0.2,
random_state=np.random.randint(0, 1000),
)
train_data.to_csv("train_data.csv", index=False)
val_data.to_csv("val_data.csv", index=False)
test[categorical_feature_names + numerical_feature_names].to_csv(
"test_data.csv", index=False
)
batch_size = 32
train_dataset = create_dataset("train_data.csv", batch_size=batch_size, mode="train")
val_dataset = create_dataset(
"val_data.csv", batch_size=val_data.shape[0], mode="eval"
).take(1)
test_dataset = create_test_dataset("test_data.csv", batch_size=test.shape[0]).take(1)
def build_deep_and_wide_model():
deep = tf.keras.layers.DenseFeatures(numerical_features, name="deep")(inputs)
deep = tf.keras.layers.Dense(32, activation="relu")(deep)
deep = tf.keras.layers.Dropout(0.3)(deep)
deep = tf.keras.layers.Dense(32, activation="relu")(deep)
deep = tf.keras.layers.Dropout(0.3)(deep)
deep = tf.keras.layers.Dense(32, activation="relu")(deep)
deep = tf.keras.layers.Dropout(0.3)(deep)
deep = tf.keras.layers.Dense(32, activation="relu")(deep)
deep = tf.keras.layers.Dropout(0.3)(deep)
wide = tf.keras.layers.DenseFeatures(categorical_features, name="wide")(inputs)
wide = tf.keras.layers.Dense(64, activation="relu")(wide)
combined = tf.keras.layers.concatenate(inputs=[deep, wide], name="combined")
output = tf.keras.layers.Dense(2, activation="softmax")(combined)
model = tf.keras.Model(inputs=list(inputs.values()), outputs=output)
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
return model
deep_and_wide_model = build_deep_and_wide_model()
tf.keras.utils.plot_model(deep_and_wide_model, show_shapes=False, rankdir="LR")
epochs = 400
early_stop = tf.keras.callbacks.EarlyStopping(patience=10)
steps_per_epoch = train_data.shape[0] // batch_size
history = deep_and_wide_model.fit(
train_dataset,
steps_per_epoch=steps_per_epoch,
validation_data=val_dataset,
epochs=epochs,
callbacks=[early_stop],
verbose=0,
)
pd.DataFrame(history.history).plot()
y_pred = np.argmax(deep_and_wide_model.predict(val_dataset), axis=-1).reshape(-1)
score = accuracy_score(val_data["Survived"], y_pred)
print("Accuracy score:", score)
print(sklearn.metrics.classification_report(val_data["Survived"], y_pred))
Survived = np.argmax(deep_and_wide_model.predict(test_dataset), axis=-1).reshape(-1)
print(Survived.shape)
path = "submission_deep_and_wide_model.csv"
save_results(Survived, test, path)
if score > best_score:
best_score = score
best_path = path
# ### Using Logistic Regression
logitistc_related_columns = list(
train.corr()[train.corr()["Survived"].abs() > 0.2].index
)
logitistc_related_columns.remove("Survived")
logitistc_related_columns
from sklearn.linear_model import LogisticRegression
best_logit = None
best_solver = ""
best_logit_score = 0
logit_train_features, logit_val_features = train_test_split(
train[logitistc_related_columns + ["Survived"]], test_size=0.2, random_state=48
)
logit_train_targets = logit_train_features.pop("Survived")
logit_val_targets = logit_val_features.pop("Survived")
for solver in ["newton-cg", "lbfgs", "liblinear"]:
logit = LogisticRegression(solver=solver)
logit.fit(logit_train_features, logit_train_targets)
score = logit.score(logit_val_features, logit_val_targets)
if score > best_logit_score:
best_solver = solver
best_logit_score = score
best_logit = logit
print("Best Solver:", best_solver, "Score:", best_logit_score)
best_score, best_path = evaulate_and_save(
best_logit,
logit_val_features,
logit_val_targets,
test[logitistc_related_columns],
"submission_logit.csv",
best_score,
best_path,
)
# ### Using KNN
best_algorithm = ""
best_knn_score = 0
best_knn = None
for algorithm in ["ball_tree", "kd_tree", "brute"]:
knn = KNeighborsClassifier(2, algorithm=algorithm)
knn.fit(train_features, train_targets)
score = knn.score(validation_features, validation_targets)
if score > best_knn_score:
best_knn_score = score
best_knn = knn
print("Best KNN Score: ", best_knn_score, "Model:", best_knn)
best_score, best_path = evaulate_and_save(
best_knn,
validation_features,
validation_targets,
test_features,
"submission_knn.csv",
best_score,
best_path,
)
# ### Using Decision Tree Classifier
from sklearn.tree import DecisionTreeClassifier
best_tree = None
best_tree_score = 0
for max_depth in range(2, 15):
tree = sklearn.tree.DecisionTreeClassifier(max_depth=5)
tree.fit(train_features, train_targets)
score = tree.score(validation_features, validation_targets)
if score > best_tree_score:
best_tree_score = score
best_tree = tree
print("Best Decision Tree Score: ", best_tree_score, "Model:", best_tree)
best_score, best_path = evaulate_and_save(
best_tree,
validation_features,
validation_targets,
test_features,
"submission_tree.csv",
best_score,
best_path,
)
# ### Using Gradient Boosting Classifier
best_gbc_score = 0
best_depth = 3
best_n_estimators = 3
best_gbc_model = None
for depth in range(3, 15):
gbc = GradientBoostingClassifier(
n_estimators=3, learning_rate=0.1, max_depth=4, random_state=1345
)
gbc.fit(train_features, train_targets)
score = gbc.score(validation_features, validation_targets)
if score > best_gbc_score:
best_depth = depth
best_gbc_score = score
best_gbc_model = gbc
for n_estimators in range(3, 15):
gbc = GradientBoostingClassifier(
n_estimators=n_estimators,
learning_rate=0.1,
max_depth=best_depth,
random_state=1345,
)
gbc.fit(train_features, train_targets)
score = gbc.score(validation_features, validation_targets)
if score > best_gbc_score:
best_n_estimators = n_estimators
best_gbc_score = score
best_gbc_model = gbc
print(
"Best Gradient Boosting Classifier Score:",
best_gbc_score,
" Model:",
best_gbc_model,
)
best_score, best_path = evaulate_and_save(
best_gbc_model,
validation_features,
validation_targets,
test_features,
"submission_gbc.csv",
best_score,
best_path,
)
# ### Using Random Forest Classifier
best_forest = None
best_max_depth = 4
best_n_estimators = 3
best_forest_score = 0
print("Find best number of estimators")
for n_estimators in list(range(3, 40, 2)):
forest = RandomForestClassifier(
n_estimators=n_estimators, max_depth=best_max_depth, random_state=84
)
forest.fit(train_features, train_targets)
score = forest.score(validation_features, validation_targets)
print("Score: ", score)
if score > best_forest_score:
best_n_estimators = n_estimators
best_forest_score = score
best_forest = forest
print("Best Number of Estimator:", best_n_estimators)
for max_depth in range(4, 15):
forest = RandomForestClassifier(
n_estimators=best_n_estimators, max_depth=max_depth, random_state=886
)
forest.fit(train_features, train_targets)
score = forest.score(validation_features, validation_targets)
print("Score: ", score)
if score > best_forest_score:
best_max_depth = max_depth
best_forest_score = best_score
best_forest = forest
print("Best Max Depth:", best_max_depth, "\nBest score:", best_forest_score)
best_score, best_path = evaulate_and_save(
best_forest,
validation_features,
validation_targets,
test_features,
"submission_forest.csv",
best_score,
best_path,
)
kmeans = KMeans(n_clusters=2, n_init=100, max_iter=300)
kmeans.fit(train_features, train_targets)
best_score, best_path = evaulate_and_save(
kmeans,
validation_features,
validation_targets,
test_features,
"submission_kmeans.csv",
best_score,
best_path,
)
# ## Submit best Model
print("Best path:", best_path)
print("Best Score", best_score)
submission = pd.read_csv(best_path)
print(submission.head(10))
submission.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605615.ipynb
| null | null |
[{"Id": 69605615, "ScriptId": 18984265, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4562457, "CreationDate": "08/02/2021 05:41:57", "VersionNumber": 14.0, "Title": "Classification with SKLearn and Tensorflow", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 464.0, "LinesInsertedFromPrevious": 64.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 400.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ## Summary
# This notebook aim to create Models to make predictions on Titanic - Machine Learning from Disaster dataset. And I will try to answer following questions:
# - Which Model can achieve a better performance? In order to answer this question, I use different kinds of Algorithms:
# - Deep Neural Network
# - Deep and Wide Neural Network with TensorFlow DenseFeatures layer
# - Logistic Regression
# - KNN
# - Decision Tree Classifier
# - Gradient Boosting Classifier¶
# - Random Forest Classifier
#
# - How to preprocess the data to get the optimal results? I will do EDA and try to add more features based on existing dataset. I will use statistic methods to search for features that's correlated to survival.
# ## Import Packages
import pandas as pd
import numpy as np
import sklearn
import seaborn as sns
import tensorflow as tf
from sklearn.ensemble import GradientBoostingClassifier, RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn import model_selection
from sklearn.cluster import KMeans
from sklearn.metrics import accuracy_score
# ## Common Functions
# **Save results**
def save_results(Survived, test, path):
submission = pd.DataFrame(
{"PassengerId": test["PassengerId"], "Survived": Survived}
)
submission.to_csv(path, index=False)
# **Evaluate Model, save and keep tract of best results**
sklearn.metrics.f1_score
def evaulate_and_save(
model,
validation_features,
validation_targets,
test_features,
save_path,
best_score,
best_path,
):
y_pred = model.predict(validation_features)
if y_pred.dtype != int:
if y_pred.shape[-1] == 2:
y_pred = np.argmax(y_pred, axis=-1)
if y_pred.shape[-1] == 1:
y_pred = np.array(y_pred > 0.5, dtype=int)
y_pred = y_pred.reshape(-1)
score = sklearn.metrics.accuracy_score(validation_targets, y_pred)
f1 = sklearn.metrics.f1_score(validation_targets, y_pred)
print("Accuracy Score:", score)
print("Classification Report:")
print(sklearn.metrics.classification_report(validation_targets, y_pred))
Survived = model.predict(test_features)
if Survived.dtype != int:
if Survived.shape[-1] == 2:
Survived = np.argmax(Survived, axis=-1)
if Survived.shape[-1] == 1:
Survived = np.array(Survived > 0.5, dtype=int)
Survived = Survived.reshape(-1)
save_results(Survived, test, save_path)
if f1 > best_score:
best_score = f1
best_path = path
return best_score, best_path
# ## Import datasets
train = pd.read_csv("/kaggle/input/titanic/train.csv")
test = pd.read_csv("/kaggle/input/titanic/test.csv")
train.head()
test.head()
# ## Data Wrangling and Preprocessing
# As we can see Age, Cabin and Fare information contains missing values, so we need to apply Missing Value Imputation to them.
train.isnull().sum()
test.isnull().sum()
train["Cabin"] = train["Cabin"].replace(np.NAN, train["Cabin"].mode()[0])
test["Cabin"] = test["Cabin"].replace(np.NAN, test["Cabin"].mode()[0])
train["Embarked"] = train["Embarked"].replace(np.NAN, train["Embarked"].mode()[0])
train["Age"] = train["Age"].replace(np.NAN, train["Age"].mean())
test["Age"] = test["Age"].replace(np.NAN, test["Age"].mean())
test["Fare"] = test["Fare"].replace(np.NAN, test["Fare"].mean())
# Let's see the Cabin labels, there are so many of them. But I make an assmptionn that the First Alphabet matters, it indicates the location and class of the passengers so it has an impact to survive.
cabin_labels = sorted(set(list(train["Cabin"].unique()) + list(test["Cabin"].unique())))
print(cabin_labels[:30])
train["Cabin_type"] = train["Cabin"].apply(lambda cabin: cabin[0])
test["Cabin_type"] = test["Cabin"].apply(lambda cabin: cabin[0])
# ## Handle Categorical Features
categorical_features = ["Sex", "Cabin_type", "Embarked"]
categorical_label_dictionary = dict()
for feature in categorical_features:
unique_labels = sorted(
set(list(train[feature].unique()) + list(test[feature].unique()))
)
for data in [train, test]:
categorical_label_dictionary[feature] = unique_labels
data[feature + "_value"] = data[feature].apply(
lambda item: unique_labels.index(item)
)
# Let's see after we preprocess, what does the data look like?
train.head(30)
# ## Exploratory Data Analysis
# ### Basic Statistic infos
train.info()
train.describe()
# ### What's the factor to survive?
# As we can see it's related to Gender, PClass, Status, Fare Cabin and Embarked.
train.corr()["Survived"].sort_values(ascending=False)
related_features = list(train.corr()[train.corr()["Survived"].abs() > 0.05].index)
related_features.remove("Survived")
print(related_features)
# ## Survive Rate
# **Overall Survive Rate**
train.Survived.mean()
# **Survive Rate across different genders**
# As we can see women are more likely to survive.
train.groupby("Sex").Survived.mean()
# ### Preprocess Data
train_test = pd.concat([train, test])[related_features]
train_test.head()
for feature in ["Sex", "Cabin_type", "Embarked"]:
items = pd.get_dummies(train_test[feature + "_value"])
labels = categorical_label_dictionary[feature]
items.columns = [feature + "_" + labels[column] for column in list(items.columns)]
train_test[items.columns] = items
train_test.pop(feature + "_value")
train_test.head()
train_features = train_test.iloc[0 : len(train)]
test_features = train_test.iloc[len(train) :]
train_features.head()
test_features.head()
# ### Train Validation Split
(
train_features,
validation_features,
train_targets,
validation_targets,
) = model_selection.train_test_split(
train_features, train["Survived"], test_size=0.2, random_state=88
)
print(
train_features.shape,
validation_features.shape,
train_targets.shape,
validation_targets.shape,
)
# ## Model Development and Evaluation
# I will try differnt Models and use results from best Model.
best_score = 0
best_path = ""
# ### Using Deep Neural Network
model = tf.keras.Sequential(
[
tf.keras.layers.Input(shape=(train_features.shape[1])),
tf.keras.layers.Dense(32, activation="relu"),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dense(32, activation="relu"),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
early_stop = tf.keras.callbacks.EarlyStopping(monitor="val_accuracy", patience=20)
history = model.fit(
train_features,
train_targets,
epochs=400,
validation_data=(validation_features, validation_targets),
callbacks=[early_stop],
verbose=0,
)
pd.DataFrame(history.history).plot()
best_score, best_path = evaulate_and_save(
model,
validation_features,
validation_targets,
test_features,
"submission_dnn.csv",
best_score,
best_path,
)
# ### Using Deep and Wide Model
from tensorflow import feature_column
categorical_feature_names = [
"Pclass",
"Sex_value",
"Embarked_value",
"Cabin_type_value",
]
numerical_feature_names = ["Age", "Fare"]
categorical_features = [
feature_column.indicator_column(
feature_column.categorical_column_with_vocabulary_list(
key, sorted(list(train[key].unique()))
)
)
for key in categorical_feature_names
]
numerical_features = [
feature_column.numeric_column(key) for key in numerical_feature_names
]
input_dictionary = dict()
inputs = dict()
for item in numerical_features:
inputs[item.key] = tf.keras.layers.Input(name=item.key, shape=())
for item in categorical_features:
inputs[item.categorical_column.key] = tf.keras.layers.Input(
name=item.categorical_column.key, shape=(), dtype="int32"
)
def features_and_labels(row_data):
label = row_data.pop("Survived")
features = row_data
return features, label
def create_dataset(pattern, epochs=1, batch_size=32, mode="eval"):
dataset = tf.data.experimental.make_csv_dataset(pattern, batch_size)
dataset = dataset.map(features_and_labels)
if mode == "train":
dataset = dataset.shuffle(buffer_size=128).repeat(epochs)
dataset = dataset.prefetch(1)
return dataset
def create_test_dataset(pattern, batch_size=32):
dataset = tf.data.experimental.make_csv_dataset(pattern, batch_size)
dataset = dataset.map(lambda features: features)
dataset = dataset.prefetch(1)
return dataset
from sklearn.model_selection import train_test_split
train_data, val_data = train_test_split(
train[categorical_feature_names + numerical_feature_names + ["Survived"]],
test_size=0.2,
random_state=np.random.randint(0, 1000),
)
train_data.to_csv("train_data.csv", index=False)
val_data.to_csv("val_data.csv", index=False)
test[categorical_feature_names + numerical_feature_names].to_csv(
"test_data.csv", index=False
)
batch_size = 32
train_dataset = create_dataset("train_data.csv", batch_size=batch_size, mode="train")
val_dataset = create_dataset(
"val_data.csv", batch_size=val_data.shape[0], mode="eval"
).take(1)
test_dataset = create_test_dataset("test_data.csv", batch_size=test.shape[0]).take(1)
def build_deep_and_wide_model():
deep = tf.keras.layers.DenseFeatures(numerical_features, name="deep")(inputs)
deep = tf.keras.layers.Dense(32, activation="relu")(deep)
deep = tf.keras.layers.Dropout(0.3)(deep)
deep = tf.keras.layers.Dense(32, activation="relu")(deep)
deep = tf.keras.layers.Dropout(0.3)(deep)
deep = tf.keras.layers.Dense(32, activation="relu")(deep)
deep = tf.keras.layers.Dropout(0.3)(deep)
deep = tf.keras.layers.Dense(32, activation="relu")(deep)
deep = tf.keras.layers.Dropout(0.3)(deep)
wide = tf.keras.layers.DenseFeatures(categorical_features, name="wide")(inputs)
wide = tf.keras.layers.Dense(64, activation="relu")(wide)
combined = tf.keras.layers.concatenate(inputs=[deep, wide], name="combined")
output = tf.keras.layers.Dense(2, activation="softmax")(combined)
model = tf.keras.Model(inputs=list(inputs.values()), outputs=output)
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
return model
deep_and_wide_model = build_deep_and_wide_model()
tf.keras.utils.plot_model(deep_and_wide_model, show_shapes=False, rankdir="LR")
epochs = 400
early_stop = tf.keras.callbacks.EarlyStopping(patience=10)
steps_per_epoch = train_data.shape[0] // batch_size
history = deep_and_wide_model.fit(
train_dataset,
steps_per_epoch=steps_per_epoch,
validation_data=val_dataset,
epochs=epochs,
callbacks=[early_stop],
verbose=0,
)
pd.DataFrame(history.history).plot()
y_pred = np.argmax(deep_and_wide_model.predict(val_dataset), axis=-1).reshape(-1)
score = accuracy_score(val_data["Survived"], y_pred)
print("Accuracy score:", score)
print(sklearn.metrics.classification_report(val_data["Survived"], y_pred))
Survived = np.argmax(deep_and_wide_model.predict(test_dataset), axis=-1).reshape(-1)
print(Survived.shape)
path = "submission_deep_and_wide_model.csv"
save_results(Survived, test, path)
if score > best_score:
best_score = score
best_path = path
# ### Using Logistic Regression
logitistc_related_columns = list(
train.corr()[train.corr()["Survived"].abs() > 0.2].index
)
logitistc_related_columns.remove("Survived")
logitistc_related_columns
from sklearn.linear_model import LogisticRegression
best_logit = None
best_solver = ""
best_logit_score = 0
logit_train_features, logit_val_features = train_test_split(
train[logitistc_related_columns + ["Survived"]], test_size=0.2, random_state=48
)
logit_train_targets = logit_train_features.pop("Survived")
logit_val_targets = logit_val_features.pop("Survived")
for solver in ["newton-cg", "lbfgs", "liblinear"]:
logit = LogisticRegression(solver=solver)
logit.fit(logit_train_features, logit_train_targets)
score = logit.score(logit_val_features, logit_val_targets)
if score > best_logit_score:
best_solver = solver
best_logit_score = score
best_logit = logit
print("Best Solver:", best_solver, "Score:", best_logit_score)
best_score, best_path = evaulate_and_save(
best_logit,
logit_val_features,
logit_val_targets,
test[logitistc_related_columns],
"submission_logit.csv",
best_score,
best_path,
)
# ### Using KNN
best_algorithm = ""
best_knn_score = 0
best_knn = None
for algorithm in ["ball_tree", "kd_tree", "brute"]:
knn = KNeighborsClassifier(2, algorithm=algorithm)
knn.fit(train_features, train_targets)
score = knn.score(validation_features, validation_targets)
if score > best_knn_score:
best_knn_score = score
best_knn = knn
print("Best KNN Score: ", best_knn_score, "Model:", best_knn)
best_score, best_path = evaulate_and_save(
best_knn,
validation_features,
validation_targets,
test_features,
"submission_knn.csv",
best_score,
best_path,
)
# ### Using Decision Tree Classifier
from sklearn.tree import DecisionTreeClassifier
best_tree = None
best_tree_score = 0
for max_depth in range(2, 15):
tree = sklearn.tree.DecisionTreeClassifier(max_depth=5)
tree.fit(train_features, train_targets)
score = tree.score(validation_features, validation_targets)
if score > best_tree_score:
best_tree_score = score
best_tree = tree
print("Best Decision Tree Score: ", best_tree_score, "Model:", best_tree)
best_score, best_path = evaulate_and_save(
best_tree,
validation_features,
validation_targets,
test_features,
"submission_tree.csv",
best_score,
best_path,
)
# ### Using Gradient Boosting Classifier
best_gbc_score = 0
best_depth = 3
best_n_estimators = 3
best_gbc_model = None
for depth in range(3, 15):
gbc = GradientBoostingClassifier(
n_estimators=3, learning_rate=0.1, max_depth=4, random_state=1345
)
gbc.fit(train_features, train_targets)
score = gbc.score(validation_features, validation_targets)
if score > best_gbc_score:
best_depth = depth
best_gbc_score = score
best_gbc_model = gbc
for n_estimators in range(3, 15):
gbc = GradientBoostingClassifier(
n_estimators=n_estimators,
learning_rate=0.1,
max_depth=best_depth,
random_state=1345,
)
gbc.fit(train_features, train_targets)
score = gbc.score(validation_features, validation_targets)
if score > best_gbc_score:
best_n_estimators = n_estimators
best_gbc_score = score
best_gbc_model = gbc
print(
"Best Gradient Boosting Classifier Score:",
best_gbc_score,
" Model:",
best_gbc_model,
)
best_score, best_path = evaulate_and_save(
best_gbc_model,
validation_features,
validation_targets,
test_features,
"submission_gbc.csv",
best_score,
best_path,
)
# ### Using Random Forest Classifier
best_forest = None
best_max_depth = 4
best_n_estimators = 3
best_forest_score = 0
print("Find best number of estimators")
for n_estimators in list(range(3, 40, 2)):
forest = RandomForestClassifier(
n_estimators=n_estimators, max_depth=best_max_depth, random_state=84
)
forest.fit(train_features, train_targets)
score = forest.score(validation_features, validation_targets)
print("Score: ", score)
if score > best_forest_score:
best_n_estimators = n_estimators
best_forest_score = score
best_forest = forest
print("Best Number of Estimator:", best_n_estimators)
for max_depth in range(4, 15):
forest = RandomForestClassifier(
n_estimators=best_n_estimators, max_depth=max_depth, random_state=886
)
forest.fit(train_features, train_targets)
score = forest.score(validation_features, validation_targets)
print("Score: ", score)
if score > best_forest_score:
best_max_depth = max_depth
best_forest_score = best_score
best_forest = forest
print("Best Max Depth:", best_max_depth, "\nBest score:", best_forest_score)
best_score, best_path = evaulate_and_save(
best_forest,
validation_features,
validation_targets,
test_features,
"submission_forest.csv",
best_score,
best_path,
)
kmeans = KMeans(n_clusters=2, n_init=100, max_iter=300)
kmeans.fit(train_features, train_targets)
best_score, best_path = evaulate_and_save(
kmeans,
validation_features,
validation_targets,
test_features,
"submission_kmeans.csv",
best_score,
best_path,
)
# ## Submit best Model
print("Best path:", best_path)
print("Best Score", best_score)
submission = pd.read_csv(best_path)
print(submission.head(10))
submission.to_csv("submission.csv", index=False)
| false | 0 | 5,170 | 0 | 5,170 | 5,170 |
||
69605106
|
import torch
import random
import math
from torch.nn.utils.rnn import pad_sequence
import numpy as np
import pandas as pd
from torch.utils.data import Dataset
from torch import nn
from tqdm import tqdm_notebook as tqdm
from transformers.tokenization_utils import BatchEncoding
from torch.cuda.amp import GradScaler, autocast
from transformers import AutoModel, AutoConfig, AutoTokenizer
import gc
def merge_dicts(input):
# list of dict --> dict of list
keys = input[0].keys()
ret = dict()
for key in keys:
temp = [x[key] for x in input]
ret[key] = temp
return ret
def make_test_dataset(train_df, test_df, anchor_num=50):
data = []
interval = len(train_df) // anchor_num
anchor_idx = [interval * x for x in range(anchor_num)]
sampled_train = train_df.sort_values(by="etarget")
sampled_train = train_df.iloc[anchor_idx]
for idx, row in test_df.iterrows():
df = pd.DataFrame()
df["anchor_text"] = sampled_train["excerpt"]
df["anchor_target"] = sampled_train["target"]
df["anchor_etarget"] = sampled_train["etarget"]
df["excerpt"] = row["excerpt"]
df["id"] = row["id"]
data.append(df)
new_df = pd.concat(data, ignore_index=True, sort=False).reset_index(drop=True)
return new_df
class MyValidationDataset(Dataset):
def __init__(self, df, tokenizer, exp=False) -> None:
super().__init__()
self.df = df
self.texts = df["excerpt"].drop_duplicates().tolist()
self.anchor_texts = df["anchor_text"].drop_duplicates().tolist()
self.anchor_targets = df["anchor_target"].tolist()
self.anchor_etargets = df["anchor_etarget"].tolist()
self.tokenizer = tokenizer
self.exp = exp
def __getitem__(self, idx):
text = self.texts[idx]
inputs = self.tokenizer.encode_plus(text, return_tensors="pt")
anchor_inputs = self.tokenizer.batch_encode_plus(
[a for a in self.anchor_texts],
max_length=512,
return_tensors="pt",
truncation=True,
padding=True,
)
return inputs, anchor_inputs, {}
def __len__(self):
return len(self.texts)
def add_pooling_mask(encode_result):
stm = encode_result["special_tokens_mask"][0]
for sep_pos in range(1, len(stm)):
if stm[sep_pos] == 1:
break
mask = torch.LongTensor([[1] * sep_pos + [2] * (len(stm) - sep_pos)])
encode_result["pooling_mask"] = mask
return encode_result
class OneBertValidationDataset(Dataset):
def __init__(self, df, tokenizer, exp=False) -> None:
super().__init__()
self.df = df
self.texts = df["excerpt"].tolist()
self.anchor_texts = df["anchor_text"].tolist()
self.anchor_targets = df["anchor_target"].tolist()
self.anchor_etargets = df["anchor_etarget"].tolist()
self.tokenizer = tokenizer
self.exp = exp
def __getitem__(self, idx):
text = self.texts[idx]
inputs = self.tokenizer.encode_plus(
text,
self.anchor_texts[idx],
return_tensors="pt",
return_special_tokens_mask=True,
truncation="only_second",
max_length=512,
padding=True,
)
inputs = add_pooling_mask(inputs)
return inputs, {}
def __len__(self):
return len(self.texts)
class MyValidationCollator:
def __init__(self, token_pad_value=0, type_pad_value=1):
super().__init__()
self.token_pad_value = token_pad_value
self.type_pad_value = type_pad_value
def __call__(self, batch):
inputs, anchor_inputs, labels = zip(*batch)
tokens = pad_sequence(
[d["input_ids"][0] for d in inputs],
batch_first=True,
padding_value=self.token_pad_value,
)
masks = pad_sequence(
[d["attention_mask"][0] for d in inputs], batch_first=True, padding_value=0
)
features = {"input_ids": tokens, "attention_mask": masks}
if "token_type_ids" in inputs[0]:
type_ids = pad_sequence(
[d["token_type_ids"][0] for d in inputs],
batch_first=True,
padding_value=self.type_pad_value,
)
features["token_type_ids"] = type_ids
anchor_fetures = anchor_inputs[0]
labels = merge_dicts(labels)
for key, value in labels.items():
labels[key] = torch.cat(value, dim=0)
return {"features": features, "anchor_features": anchor_fetures}, labels
class OneBertCompareCollator:
def __init__(self, token_pad_value=0, type_pad_value=1):
super().__init__()
self.token_pad_value = token_pad_value
self.type_pad_value = type_pad_value
def __call__(self, batch):
inputs, labels = zip(*batch)
tokens = pad_sequence(
[d["input_ids"][0] for d in inputs],
batch_first=True,
padding_value=self.token_pad_value,
)
masks = pad_sequence(
[d["attention_mask"][0] for d in inputs], batch_first=True, padding_value=0
)
pooling_mask = pad_sequence(
[d["pooling_mask"][0] for d in inputs], batch_first=True, padding_value=0
)
features = {
"input_ids": tokens,
"attention_mask": masks,
"pooling_mask": pooling_mask,
}
if "token_type_ids" in inputs[0]:
type_ids = pad_sequence(
[d["token_type_ids"][0] for d in inputs],
batch_first=True,
padding_value=self.type_pad_value,
)
features["token_type_ids"] = type_ids
labels = merge_dicts(labels)
for key, value in labels.items():
labels[key] = torch.cat(value, dim=0)
return {"features": features}, labels
class Encoder(nn.Module):
def __init__(
self,
pretrained_model_name,
config=None,
pooling="cls",
grad_checkpoint=False,
**kwargs,
):
super().__init__()
if config is not None:
self.bert = AutoModel.from_config(config)
else:
config = AutoConfig.from_pretrained(pretrained_model_name)
if grad_checkpoint:
self.bert.config.gradient_checkpointing = True
if kwargs.get("hidden_dropout"):
config.hidden_dropout_prob = kwargs["hidden_drop"]
if kwargs.get("attention_dropput"):
config.attention_probs_dropout_prob = kwargs["attention_dropout"]
if kwargs.get("layer_norm_eps"):
config.layer_norm_eps = kwargs["layer_norm_eps"]
self.bert = AutoModel.from_pretrained(pretrained_model_name, config=config)
self.pooling = pooling
self.hidden_size = self.bert.config.hidden_size
if pooling != "cls":
self.bert.pooler = nn.Identity()
self.attention = nn.Sequential(
nn.Linear(self.hidden_size, 256), nn.GELU(), nn.Linear(256, 1)
)
def forward(self, features):
output_states = self.bert(
input_ids=features.get("input_ids"),
attention_mask=features.get("attention_mask"),
token_type_ids=features.get("token_type_ids"),
)
out = output_states[0] # embedding for all tokens
if self.pooling == "cls":
pooled_out = output_states[1] # CLS token is first token
elif self.pooling == "mean":
attention_mask = features["attention_mask"]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(out.size()).float()
)
sum_embeddings = torch.sum(out * input_mask_expanded, 1)
sum_mask = torch.clamp(input_mask_expanded.sum(1), min=1e-9)
pooled_out = sum_embeddings / sum_mask
elif self.pooling == "att":
weights = self.attention(out)
attention_mask = (
features["attention_mask"].unsqueeze(-1).expand(weights.size())
)
weights.masked_fill_(attention_mask == 0, -float("inf"))
weights = torch.softmax(weights, dim=1)
pooled_out = torch.sum(out * weights, dim=1)
return pooled_out, out
class Comparer(nn.Module):
def __init__(
self,
pretrained_model_name,
config=None,
pooling="mean",
esim=True,
grad_checkpoint=False,
):
super().__init__()
self.encoder = Encoder(
pretrained_model_name,
config=config,
pooling=pooling,
grad_checkpoint=grad_checkpoint,
)
self.esim = esim
self.fc = nn.Sequential(
nn.Dropout(0.5),
nn.Linear(self.encoder.hidden_size * 2, self.encoder.hidden_size),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(self.encoder.hidden_size, 1),
)
self.anchor_pooled_emb, self.anchor_seq_emb = None, None
def inference(self, seq1, seq2, mask1=None, mask2=None):
# seq1: B*l1*D
# seq2: B*l2*D
# mask1: B*l1
# mask2: B*l2
score = torch.bmm(seq1, seq2.permute(0, 2, 1))
new_seq1 = torch.bmm(
torch.softmax(score, dim=-1), seq2 * mask2.unsqueeze(-1)
) #
new_seq1 = torch.sum(new_seq1 * mask1.unsqueeze(-1), dim=1) / torch.sum(
mask1, dim=1
).unsqueeze(-1)
# del score1
new_seq2 = torch.bmm(
torch.softmax(score, dim=1).permute(0, 2, 1), seq1 * mask1.unsqueeze(-1)
) #
new_seq2 = torch.sum(new_seq2 * mask2.unsqueeze(-1), dim=1) / torch.sum(
mask2, dim=1
).unsqueeze(-1)
return new_seq1, new_seq2
def forward(self, features, anchor_features=None):
pooled_emb, seq_emb = self.encoder(features)
# etarget_out = self.etarget_head(pooled_emb)
bs, length, dim = seq_emb.size()
if anchor_features is None:
# embeddings: B*D
# 111,222,333
pooled_emb1 = pooled_emb.unsqueeze(1).expand(-1, bs, -1).reshape(-1, dim)
# 123,123,123
pooled_emb2 = pooled_emb.unsqueeze(0).expand(bs, -1, -1).reshape(-1, dim)
seq_emb1 = (
seq_emb.unsqueeze(1).expand(-1, bs, -1, -1).reshape(-1, length, dim)
)
seq_emb2 = (
seq_emb.unsqueeze(0).expand(bs, -1, -1, -1).reshape(-1, length, dim)
)
mask1 = (
features["attention_mask"]
.unsqueeze(1)
.expand(-1, bs, -1)
.reshape(-1, length)
)
mask2 = (
features["attention_mask"]
.unsqueeze(0)
.expand(bs, -1, -1)
.reshape(-1, length)
)
new_emb1, new_emb2 = self.inference(seq_emb1, seq_emb2, mask1, mask2)
else:
if self.anchor_pooled_emb is None:
anchor_pooled_emb, anchor_seq_emb = self.encoder(anchor_features)
self.anchor_pooled_emb, self.anchor_seq_emb = (
anchor_pooled_emb,
anchor_seq_emb,
)
else:
anchor_pooled_emb, anchor_seq_emb = (
self.anchor_pooled_emb,
self.anchor_seq_emb,
)
anchor_bs, _ = anchor_pooled_emb.size()
pooled_emb = (
pooled_emb.unsqueeze(1).expand(-1, anchor_bs, -1).reshape(-1, dim)
)
anchor_pooled_emb = anchor_pooled_emb.repeat(bs, 1)
pooled_emb1 = pooled_emb
pooled_emb2 = anchor_pooled_emb
seq_emb1 = (
seq_emb.unsqueeze(1)
.expand(-1, anchor_bs, -1, -1)
.reshape(-1, length, dim)
)
seq_emb2 = anchor_seq_emb.repeat(bs, 1, 1)
mask1 = (
features["attention_mask"]
.unsqueeze(1)
.expand(-1, anchor_bs, -1)
.reshape(-1, length)
)
mask2 = anchor_features["attention_mask"].repeat(bs, 1)
new_emb1, new_emb2 = self.inference(seq_emb1, seq_emb2, mask1, mask2)
fc_input = torch.cat([pooled_emb1 - pooled_emb2, new_emb2 - new_emb1], dim=1)
# fc_input = pooled_emb1-pooled_emb2+new_emb2-new_emb1], dim=1)
output = self.fc(fc_input)
ret = {"pred": output}
return ret
def _prepare_inputs(inputs):
for k, v in inputs.items():
if isinstance(v, torch.Tensor):
inputs[k] = v.cuda()
elif isinstance(v, BatchEncoding): # for embedding training
inputs[k] = v.cuda()
elif isinstance(v, dict): # for embedding training
inputs[k] = _prepare_inputs(v)
return inputs
class OneBertComparer(nn.Module):
def __init__(
self, pretrained_model_name, config=None, pooling="cls", sep_pooling=False
):
super().__init__()
self.encoder = Encoder(pretrained_model_name, config=config, pooling="mean")
self.sep_pooling = sep_pooling
self.pooling = pooling
self.fc = nn.Sequential(
nn.Dropout(0.25),
nn.Linear(self.encoder.hidden_size, 256),
nn.ReLU(),
nn.Dropout(0.25),
nn.Linear(256, 1),
)
self.attention = nn.Sequential(
nn.Linear(self.encoder.hidden_size, 256), nn.GELU(), nn.Linear(256, 1)
)
def masked_mean_pooling(self, seq_emb, pooling_mask):
mask1 = pooling_mask == 1
pooled_emb_1 = torch.sum(seq_emb * mask1.unsqueeze(-1), dim=1) / torch.sum(
mask1, dim=1, keepdim=True
)
mask2 = pooling_mask == 2
pooled_emb_2 = torch.sum(seq_emb * mask2.unsqueeze(-1), dim=1) / torch.sum(
mask2, dim=1, keepdim=True
)
return pooled_emb_1, pooled_emb_2
def masked_att_pooling(self, seq_emb, pooling_mask):
weights = self.attention(seq_emb)
mask1 = (pooling_mask == 2).unsqueeze(-1).expand(weights.size())
weight1 = torch.softmax(weights.masked_fill(mask1, -float("inf")), dim=1)
pooled_emb_1 = torch.sum(seq_emb * weight1, dim=1)
mask2 = (pooling_mask == 1).unsqueeze(-1).expand(weights.size())
weight2 = torch.softmax(weights.masked_fill(mask2, -float("inf")), dim=1)
pooled_emb_2 = torch.sum(seq_emb * weight2, dim=1)
return pooled_emb_1, pooled_emb_2
def forward(self, features):
pooled_emb, seq_emb = self.encoder(features)
if self.sep_pooling:
if self.pooling == "att":
emb1, emb2 = self.masked_att_pooling(seq_emb, features["pooling_mask"])
else:
emb1, emb2 = self.masked_mean_pooling(seq_emb, features["pooling_mask"])
sep_pooled_emb = emb1 - emb2
# output = self.fc(torch.cat([pooled_emb, sep_pooled_emb], dim=1))
output = self.fc(sep_pooled_emb)
else:
output = self.fc(pooled_emb)
ret = {"pred": output}
return ret
train = pd.read_csv("../input/clrp-compare-base/train_with_folds.csv")
test = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
tokenizer = AutoTokenizer.from_pretrained("../input/clrp-roberta-reg/")
config = AutoConfig.from_pretrained("../input/clrp-roberta-reg")
all_results = []
for fold in range(5):
print(f"fold {fold}")
train_fold = train[train["fold"] != fold]
new_test = make_test_dataset(train_fold, test)
valid_set = MyValidationDataset(new_test, tokenizer)
valid_collator = MyValidationCollator(
token_pad_value=tokenizer.pad_token_id, type_pad_value=tokenizer.pad_token_id
)
loader = torch.utils.data.DataLoader(
valid_set,
shuffle=False,
batch_size=16,
pin_memory=True,
drop_last=False,
num_workers=4,
collate_fn=valid_collator,
)
state = torch.load(f"../input/clrp-roberta-reg/best-model-{fold}.pt")
model = Comparer(None, config, pooling="att")
model.load_state_dict(state["model"])
model.eval()
model.cuda()
results = []
with torch.no_grad():
for inputs, _ in tqdm(loader):
inputs = _prepare_inputs(inputs)
with autocast(enabled=True):
outputs = model(**inputs)
results.append(outputs)
predicts = merge_dicts(results)
for key in predicts.keys():
predicts[key] = torch.cat(predicts[key], dim=0)
pred = predicts["pred"].cpu().numpy()
df = pd.DataFrame()
df["id"] = new_test["id"]
df["pred"] = pred.flatten()
df["anchor_target"] = new_test["anchor_target"]
df["pred_target"] = df["pred"] + df["anchor_target"]
all_results.append(df[["id", "pred_target"]])
del model
gc.collect()
for fold in range(5):
print(f"fold {fold}")
train_fold = train[train["fold"] != fold]
new_test = make_test_dataset(train_fold, test)
valid_set = MyValidationDataset(new_test, tokenizer)
valid_collator = MyValidationCollator(
token_pad_value=tokenizer.pad_token_id, type_pad_value=tokenizer.pad_token_id
)
loader = torch.utils.data.DataLoader(
valid_set,
shuffle=False,
batch_size=16,
pin_memory=True,
drop_last=False,
num_workers=4,
collate_fn=valid_collator,
)
state = torch.load(f"../input/clrp-roberta-v2/best-model-{fold}.pt")
model = Comparer(None, config, pooling="att")
model.load_state_dict(state["model"])
model.eval()
model.cuda()
results = []
with torch.no_grad():
for inputs, _ in tqdm(loader):
inputs = _prepare_inputs(inputs)
with autocast(enabled=True):
outputs = model(**inputs)
results.append(outputs)
predicts = merge_dicts(results)
for key in predicts.keys():
predicts[key] = torch.cat(predicts[key], dim=0)
pred = torch.sigmoid(predicts["pred"]).cpu().numpy()
df = pd.DataFrame()
df["id"] = new_test["id"]
df["pred"] = pred.flatten()
df["anchor_target"] = new_test["anchor_etarget"]
df["pred_etarget"] = df["pred"] * df["anchor_target"] / (1 - df["pred"])
df["pred_etarget"] = np.clip(df["pred_etarget"], a_min=0.025, a_max=5.6)
df["pred_target"] = np.log(df["pred_etarget"])
all_results.append(df[["id", "pred_target"]])
del model
gc.collect()
# onebert binary
for fold in range(5):
print(f"fold {fold}")
train_fold = train[train["fold"] != fold]
new_test = make_test_dataset(train_fold, test, 10)
valid_set = OneBertValidationDataset(new_test, tokenizer)
valid_collator = OneBertCompareCollator(
token_pad_value=tokenizer.pad_token_id,
type_pad_value=tokenizer.pad_token_type_id,
)
loader = torch.utils.data.DataLoader(
valid_set,
shuffle=False,
batch_size=16,
pin_memory=True,
drop_last=False,
num_workers=4,
collate_fn=valid_collator,
)
state = torch.load(f"../input/clrp-roberta-one/best-model-{fold}.pt")
model = OneBertComparer(None, config, sep_pooling=True, pooling="att")
model.load_state_dict(state["model"])
model.eval()
model.cuda()
results = []
with torch.no_grad():
for inputs, _ in tqdm(loader):
inputs = _prepare_inputs(inputs)
with autocast(enabled=True):
outputs = model(**inputs)
results.append(outputs)
predicts = merge_dicts(results)
for key in predicts.keys():
predicts[key] = torch.cat(predicts[key], dim=0)
pred = torch.sigmoid(predicts["pred"]).cpu().numpy()
df = pd.DataFrame()
df["id"] = new_test["id"]
df["pred"] = pred.flatten()
df["anchor_target"] = new_test["anchor_etarget"]
df["pred_etarget"] = df["pred"] * df["anchor_target"] / (1 - df["pred"])
df["pred_etarget"] = np.clip(df["pred_etarget"], a_min=0.025, a_max=5.6)
df["pred_target"] = np.log(df["pred_etarget"])
all_results.append(df[["id", "pred_target"]])
del model
gc.collect()
# onebert reg
for fold in range(5):
print(f"fold {fold}")
train_fold = train[train["fold"] != fold]
new_test = make_test_dataset(train_fold, test, 10)
valid_set = OneBertValidationDataset(new_test, tokenizer)
valid_collator = OneBertCompareCollator(
token_pad_value=tokenizer.pad_token_id,
type_pad_value=tokenizer.pad_token_type_id,
)
loader = torch.utils.data.DataLoader(
valid_set,
shuffle=False,
batch_size=16,
pin_memory=True,
drop_last=False,
num_workers=4,
collate_fn=valid_collator,
)
state = torch.load(f"../input/clrp-onebert-reg/best-model-{fold}.pt")
model = OneBertComparer(None, config, sep_pooling=True, pooling="att")
model.load_state_dict(state["model"])
model.eval()
model.cuda()
results = []
with torch.no_grad():
for inputs, _ in tqdm(loader):
inputs = _prepare_inputs(inputs)
with autocast(enabled=True):
outputs = model(**inputs)
results.append(outputs)
predicts = merge_dicts(results)
for key in predicts.keys():
predicts[key] = torch.cat(predicts[key], dim=0)
pred = predicts["pred"].cpu().numpy()
df = pd.DataFrame()
df["id"] = new_test["id"]
df["pred"] = pred.flatten()
df["anchor_target"] = new_test["anchor_target"]
df["pred_target"] = df["pred"] + df["anchor_target"]
all_results.append(df[["id", "pred_target"]])
del model
gc.collect()
df = pd.concat(all_results, ignore_index=True, sort=False)
pred = df.groupby("id")["pred_target"].mean().reset_index()
pred["target"] = pred["pred_target"]
pred[["id", "target"]].to_csv("submission.csv", index=False)
len(all_results[0])
pred.head()
pred.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605106.ipynb
| null | null |
[{"Id": 69605106, "ScriptId": 18983423, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 1289116, "CreationDate": "08/02/2021 05:32:02", "VersionNumber": 4.0, "Title": "inference-large-ensemble2", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 588.0, "LinesInsertedFromPrevious": 4.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 584.0, "LinesInsertedFromFork": 223.0, "LinesDeletedFromFork": 2.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 365.0, "TotalVotes": 0}]
| null | null | null | null |
import torch
import random
import math
from torch.nn.utils.rnn import pad_sequence
import numpy as np
import pandas as pd
from torch.utils.data import Dataset
from torch import nn
from tqdm import tqdm_notebook as tqdm
from transformers.tokenization_utils import BatchEncoding
from torch.cuda.amp import GradScaler, autocast
from transformers import AutoModel, AutoConfig, AutoTokenizer
import gc
def merge_dicts(input):
# list of dict --> dict of list
keys = input[0].keys()
ret = dict()
for key in keys:
temp = [x[key] for x in input]
ret[key] = temp
return ret
def make_test_dataset(train_df, test_df, anchor_num=50):
data = []
interval = len(train_df) // anchor_num
anchor_idx = [interval * x for x in range(anchor_num)]
sampled_train = train_df.sort_values(by="etarget")
sampled_train = train_df.iloc[anchor_idx]
for idx, row in test_df.iterrows():
df = pd.DataFrame()
df["anchor_text"] = sampled_train["excerpt"]
df["anchor_target"] = sampled_train["target"]
df["anchor_etarget"] = sampled_train["etarget"]
df["excerpt"] = row["excerpt"]
df["id"] = row["id"]
data.append(df)
new_df = pd.concat(data, ignore_index=True, sort=False).reset_index(drop=True)
return new_df
class MyValidationDataset(Dataset):
def __init__(self, df, tokenizer, exp=False) -> None:
super().__init__()
self.df = df
self.texts = df["excerpt"].drop_duplicates().tolist()
self.anchor_texts = df["anchor_text"].drop_duplicates().tolist()
self.anchor_targets = df["anchor_target"].tolist()
self.anchor_etargets = df["anchor_etarget"].tolist()
self.tokenizer = tokenizer
self.exp = exp
def __getitem__(self, idx):
text = self.texts[idx]
inputs = self.tokenizer.encode_plus(text, return_tensors="pt")
anchor_inputs = self.tokenizer.batch_encode_plus(
[a for a in self.anchor_texts],
max_length=512,
return_tensors="pt",
truncation=True,
padding=True,
)
return inputs, anchor_inputs, {}
def __len__(self):
return len(self.texts)
def add_pooling_mask(encode_result):
stm = encode_result["special_tokens_mask"][0]
for sep_pos in range(1, len(stm)):
if stm[sep_pos] == 1:
break
mask = torch.LongTensor([[1] * sep_pos + [2] * (len(stm) - sep_pos)])
encode_result["pooling_mask"] = mask
return encode_result
class OneBertValidationDataset(Dataset):
def __init__(self, df, tokenizer, exp=False) -> None:
super().__init__()
self.df = df
self.texts = df["excerpt"].tolist()
self.anchor_texts = df["anchor_text"].tolist()
self.anchor_targets = df["anchor_target"].tolist()
self.anchor_etargets = df["anchor_etarget"].tolist()
self.tokenizer = tokenizer
self.exp = exp
def __getitem__(self, idx):
text = self.texts[idx]
inputs = self.tokenizer.encode_plus(
text,
self.anchor_texts[idx],
return_tensors="pt",
return_special_tokens_mask=True,
truncation="only_second",
max_length=512,
padding=True,
)
inputs = add_pooling_mask(inputs)
return inputs, {}
def __len__(self):
return len(self.texts)
class MyValidationCollator:
def __init__(self, token_pad_value=0, type_pad_value=1):
super().__init__()
self.token_pad_value = token_pad_value
self.type_pad_value = type_pad_value
def __call__(self, batch):
inputs, anchor_inputs, labels = zip(*batch)
tokens = pad_sequence(
[d["input_ids"][0] for d in inputs],
batch_first=True,
padding_value=self.token_pad_value,
)
masks = pad_sequence(
[d["attention_mask"][0] for d in inputs], batch_first=True, padding_value=0
)
features = {"input_ids": tokens, "attention_mask": masks}
if "token_type_ids" in inputs[0]:
type_ids = pad_sequence(
[d["token_type_ids"][0] for d in inputs],
batch_first=True,
padding_value=self.type_pad_value,
)
features["token_type_ids"] = type_ids
anchor_fetures = anchor_inputs[0]
labels = merge_dicts(labels)
for key, value in labels.items():
labels[key] = torch.cat(value, dim=0)
return {"features": features, "anchor_features": anchor_fetures}, labels
class OneBertCompareCollator:
def __init__(self, token_pad_value=0, type_pad_value=1):
super().__init__()
self.token_pad_value = token_pad_value
self.type_pad_value = type_pad_value
def __call__(self, batch):
inputs, labels = zip(*batch)
tokens = pad_sequence(
[d["input_ids"][0] for d in inputs],
batch_first=True,
padding_value=self.token_pad_value,
)
masks = pad_sequence(
[d["attention_mask"][0] for d in inputs], batch_first=True, padding_value=0
)
pooling_mask = pad_sequence(
[d["pooling_mask"][0] for d in inputs], batch_first=True, padding_value=0
)
features = {
"input_ids": tokens,
"attention_mask": masks,
"pooling_mask": pooling_mask,
}
if "token_type_ids" in inputs[0]:
type_ids = pad_sequence(
[d["token_type_ids"][0] for d in inputs],
batch_first=True,
padding_value=self.type_pad_value,
)
features["token_type_ids"] = type_ids
labels = merge_dicts(labels)
for key, value in labels.items():
labels[key] = torch.cat(value, dim=0)
return {"features": features}, labels
class Encoder(nn.Module):
def __init__(
self,
pretrained_model_name,
config=None,
pooling="cls",
grad_checkpoint=False,
**kwargs,
):
super().__init__()
if config is not None:
self.bert = AutoModel.from_config(config)
else:
config = AutoConfig.from_pretrained(pretrained_model_name)
if grad_checkpoint:
self.bert.config.gradient_checkpointing = True
if kwargs.get("hidden_dropout"):
config.hidden_dropout_prob = kwargs["hidden_drop"]
if kwargs.get("attention_dropput"):
config.attention_probs_dropout_prob = kwargs["attention_dropout"]
if kwargs.get("layer_norm_eps"):
config.layer_norm_eps = kwargs["layer_norm_eps"]
self.bert = AutoModel.from_pretrained(pretrained_model_name, config=config)
self.pooling = pooling
self.hidden_size = self.bert.config.hidden_size
if pooling != "cls":
self.bert.pooler = nn.Identity()
self.attention = nn.Sequential(
nn.Linear(self.hidden_size, 256), nn.GELU(), nn.Linear(256, 1)
)
def forward(self, features):
output_states = self.bert(
input_ids=features.get("input_ids"),
attention_mask=features.get("attention_mask"),
token_type_ids=features.get("token_type_ids"),
)
out = output_states[0] # embedding for all tokens
if self.pooling == "cls":
pooled_out = output_states[1] # CLS token is first token
elif self.pooling == "mean":
attention_mask = features["attention_mask"]
input_mask_expanded = (
attention_mask.unsqueeze(-1).expand(out.size()).float()
)
sum_embeddings = torch.sum(out * input_mask_expanded, 1)
sum_mask = torch.clamp(input_mask_expanded.sum(1), min=1e-9)
pooled_out = sum_embeddings / sum_mask
elif self.pooling == "att":
weights = self.attention(out)
attention_mask = (
features["attention_mask"].unsqueeze(-1).expand(weights.size())
)
weights.masked_fill_(attention_mask == 0, -float("inf"))
weights = torch.softmax(weights, dim=1)
pooled_out = torch.sum(out * weights, dim=1)
return pooled_out, out
class Comparer(nn.Module):
def __init__(
self,
pretrained_model_name,
config=None,
pooling="mean",
esim=True,
grad_checkpoint=False,
):
super().__init__()
self.encoder = Encoder(
pretrained_model_name,
config=config,
pooling=pooling,
grad_checkpoint=grad_checkpoint,
)
self.esim = esim
self.fc = nn.Sequential(
nn.Dropout(0.5),
nn.Linear(self.encoder.hidden_size * 2, self.encoder.hidden_size),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(self.encoder.hidden_size, 1),
)
self.anchor_pooled_emb, self.anchor_seq_emb = None, None
def inference(self, seq1, seq2, mask1=None, mask2=None):
# seq1: B*l1*D
# seq2: B*l2*D
# mask1: B*l1
# mask2: B*l2
score = torch.bmm(seq1, seq2.permute(0, 2, 1))
new_seq1 = torch.bmm(
torch.softmax(score, dim=-1), seq2 * mask2.unsqueeze(-1)
) #
new_seq1 = torch.sum(new_seq1 * mask1.unsqueeze(-1), dim=1) / torch.sum(
mask1, dim=1
).unsqueeze(-1)
# del score1
new_seq2 = torch.bmm(
torch.softmax(score, dim=1).permute(0, 2, 1), seq1 * mask1.unsqueeze(-1)
) #
new_seq2 = torch.sum(new_seq2 * mask2.unsqueeze(-1), dim=1) / torch.sum(
mask2, dim=1
).unsqueeze(-1)
return new_seq1, new_seq2
def forward(self, features, anchor_features=None):
pooled_emb, seq_emb = self.encoder(features)
# etarget_out = self.etarget_head(pooled_emb)
bs, length, dim = seq_emb.size()
if anchor_features is None:
# embeddings: B*D
# 111,222,333
pooled_emb1 = pooled_emb.unsqueeze(1).expand(-1, bs, -1).reshape(-1, dim)
# 123,123,123
pooled_emb2 = pooled_emb.unsqueeze(0).expand(bs, -1, -1).reshape(-1, dim)
seq_emb1 = (
seq_emb.unsqueeze(1).expand(-1, bs, -1, -1).reshape(-1, length, dim)
)
seq_emb2 = (
seq_emb.unsqueeze(0).expand(bs, -1, -1, -1).reshape(-1, length, dim)
)
mask1 = (
features["attention_mask"]
.unsqueeze(1)
.expand(-1, bs, -1)
.reshape(-1, length)
)
mask2 = (
features["attention_mask"]
.unsqueeze(0)
.expand(bs, -1, -1)
.reshape(-1, length)
)
new_emb1, new_emb2 = self.inference(seq_emb1, seq_emb2, mask1, mask2)
else:
if self.anchor_pooled_emb is None:
anchor_pooled_emb, anchor_seq_emb = self.encoder(anchor_features)
self.anchor_pooled_emb, self.anchor_seq_emb = (
anchor_pooled_emb,
anchor_seq_emb,
)
else:
anchor_pooled_emb, anchor_seq_emb = (
self.anchor_pooled_emb,
self.anchor_seq_emb,
)
anchor_bs, _ = anchor_pooled_emb.size()
pooled_emb = (
pooled_emb.unsqueeze(1).expand(-1, anchor_bs, -1).reshape(-1, dim)
)
anchor_pooled_emb = anchor_pooled_emb.repeat(bs, 1)
pooled_emb1 = pooled_emb
pooled_emb2 = anchor_pooled_emb
seq_emb1 = (
seq_emb.unsqueeze(1)
.expand(-1, anchor_bs, -1, -1)
.reshape(-1, length, dim)
)
seq_emb2 = anchor_seq_emb.repeat(bs, 1, 1)
mask1 = (
features["attention_mask"]
.unsqueeze(1)
.expand(-1, anchor_bs, -1)
.reshape(-1, length)
)
mask2 = anchor_features["attention_mask"].repeat(bs, 1)
new_emb1, new_emb2 = self.inference(seq_emb1, seq_emb2, mask1, mask2)
fc_input = torch.cat([pooled_emb1 - pooled_emb2, new_emb2 - new_emb1], dim=1)
# fc_input = pooled_emb1-pooled_emb2+new_emb2-new_emb1], dim=1)
output = self.fc(fc_input)
ret = {"pred": output}
return ret
def _prepare_inputs(inputs):
for k, v in inputs.items():
if isinstance(v, torch.Tensor):
inputs[k] = v.cuda()
elif isinstance(v, BatchEncoding): # for embedding training
inputs[k] = v.cuda()
elif isinstance(v, dict): # for embedding training
inputs[k] = _prepare_inputs(v)
return inputs
class OneBertComparer(nn.Module):
def __init__(
self, pretrained_model_name, config=None, pooling="cls", sep_pooling=False
):
super().__init__()
self.encoder = Encoder(pretrained_model_name, config=config, pooling="mean")
self.sep_pooling = sep_pooling
self.pooling = pooling
self.fc = nn.Sequential(
nn.Dropout(0.25),
nn.Linear(self.encoder.hidden_size, 256),
nn.ReLU(),
nn.Dropout(0.25),
nn.Linear(256, 1),
)
self.attention = nn.Sequential(
nn.Linear(self.encoder.hidden_size, 256), nn.GELU(), nn.Linear(256, 1)
)
def masked_mean_pooling(self, seq_emb, pooling_mask):
mask1 = pooling_mask == 1
pooled_emb_1 = torch.sum(seq_emb * mask1.unsqueeze(-1), dim=1) / torch.sum(
mask1, dim=1, keepdim=True
)
mask2 = pooling_mask == 2
pooled_emb_2 = torch.sum(seq_emb * mask2.unsqueeze(-1), dim=1) / torch.sum(
mask2, dim=1, keepdim=True
)
return pooled_emb_1, pooled_emb_2
def masked_att_pooling(self, seq_emb, pooling_mask):
weights = self.attention(seq_emb)
mask1 = (pooling_mask == 2).unsqueeze(-1).expand(weights.size())
weight1 = torch.softmax(weights.masked_fill(mask1, -float("inf")), dim=1)
pooled_emb_1 = torch.sum(seq_emb * weight1, dim=1)
mask2 = (pooling_mask == 1).unsqueeze(-1).expand(weights.size())
weight2 = torch.softmax(weights.masked_fill(mask2, -float("inf")), dim=1)
pooled_emb_2 = torch.sum(seq_emb * weight2, dim=1)
return pooled_emb_1, pooled_emb_2
def forward(self, features):
pooled_emb, seq_emb = self.encoder(features)
if self.sep_pooling:
if self.pooling == "att":
emb1, emb2 = self.masked_att_pooling(seq_emb, features["pooling_mask"])
else:
emb1, emb2 = self.masked_mean_pooling(seq_emb, features["pooling_mask"])
sep_pooled_emb = emb1 - emb2
# output = self.fc(torch.cat([pooled_emb, sep_pooled_emb], dim=1))
output = self.fc(sep_pooled_emb)
else:
output = self.fc(pooled_emb)
ret = {"pred": output}
return ret
train = pd.read_csv("../input/clrp-compare-base/train_with_folds.csv")
test = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
tokenizer = AutoTokenizer.from_pretrained("../input/clrp-roberta-reg/")
config = AutoConfig.from_pretrained("../input/clrp-roberta-reg")
all_results = []
for fold in range(5):
print(f"fold {fold}")
train_fold = train[train["fold"] != fold]
new_test = make_test_dataset(train_fold, test)
valid_set = MyValidationDataset(new_test, tokenizer)
valid_collator = MyValidationCollator(
token_pad_value=tokenizer.pad_token_id, type_pad_value=tokenizer.pad_token_id
)
loader = torch.utils.data.DataLoader(
valid_set,
shuffle=False,
batch_size=16,
pin_memory=True,
drop_last=False,
num_workers=4,
collate_fn=valid_collator,
)
state = torch.load(f"../input/clrp-roberta-reg/best-model-{fold}.pt")
model = Comparer(None, config, pooling="att")
model.load_state_dict(state["model"])
model.eval()
model.cuda()
results = []
with torch.no_grad():
for inputs, _ in tqdm(loader):
inputs = _prepare_inputs(inputs)
with autocast(enabled=True):
outputs = model(**inputs)
results.append(outputs)
predicts = merge_dicts(results)
for key in predicts.keys():
predicts[key] = torch.cat(predicts[key], dim=0)
pred = predicts["pred"].cpu().numpy()
df = pd.DataFrame()
df["id"] = new_test["id"]
df["pred"] = pred.flatten()
df["anchor_target"] = new_test["anchor_target"]
df["pred_target"] = df["pred"] + df["anchor_target"]
all_results.append(df[["id", "pred_target"]])
del model
gc.collect()
for fold in range(5):
print(f"fold {fold}")
train_fold = train[train["fold"] != fold]
new_test = make_test_dataset(train_fold, test)
valid_set = MyValidationDataset(new_test, tokenizer)
valid_collator = MyValidationCollator(
token_pad_value=tokenizer.pad_token_id, type_pad_value=tokenizer.pad_token_id
)
loader = torch.utils.data.DataLoader(
valid_set,
shuffle=False,
batch_size=16,
pin_memory=True,
drop_last=False,
num_workers=4,
collate_fn=valid_collator,
)
state = torch.load(f"../input/clrp-roberta-v2/best-model-{fold}.pt")
model = Comparer(None, config, pooling="att")
model.load_state_dict(state["model"])
model.eval()
model.cuda()
results = []
with torch.no_grad():
for inputs, _ in tqdm(loader):
inputs = _prepare_inputs(inputs)
with autocast(enabled=True):
outputs = model(**inputs)
results.append(outputs)
predicts = merge_dicts(results)
for key in predicts.keys():
predicts[key] = torch.cat(predicts[key], dim=0)
pred = torch.sigmoid(predicts["pred"]).cpu().numpy()
df = pd.DataFrame()
df["id"] = new_test["id"]
df["pred"] = pred.flatten()
df["anchor_target"] = new_test["anchor_etarget"]
df["pred_etarget"] = df["pred"] * df["anchor_target"] / (1 - df["pred"])
df["pred_etarget"] = np.clip(df["pred_etarget"], a_min=0.025, a_max=5.6)
df["pred_target"] = np.log(df["pred_etarget"])
all_results.append(df[["id", "pred_target"]])
del model
gc.collect()
# onebert binary
for fold in range(5):
print(f"fold {fold}")
train_fold = train[train["fold"] != fold]
new_test = make_test_dataset(train_fold, test, 10)
valid_set = OneBertValidationDataset(new_test, tokenizer)
valid_collator = OneBertCompareCollator(
token_pad_value=tokenizer.pad_token_id,
type_pad_value=tokenizer.pad_token_type_id,
)
loader = torch.utils.data.DataLoader(
valid_set,
shuffle=False,
batch_size=16,
pin_memory=True,
drop_last=False,
num_workers=4,
collate_fn=valid_collator,
)
state = torch.load(f"../input/clrp-roberta-one/best-model-{fold}.pt")
model = OneBertComparer(None, config, sep_pooling=True, pooling="att")
model.load_state_dict(state["model"])
model.eval()
model.cuda()
results = []
with torch.no_grad():
for inputs, _ in tqdm(loader):
inputs = _prepare_inputs(inputs)
with autocast(enabled=True):
outputs = model(**inputs)
results.append(outputs)
predicts = merge_dicts(results)
for key in predicts.keys():
predicts[key] = torch.cat(predicts[key], dim=0)
pred = torch.sigmoid(predicts["pred"]).cpu().numpy()
df = pd.DataFrame()
df["id"] = new_test["id"]
df["pred"] = pred.flatten()
df["anchor_target"] = new_test["anchor_etarget"]
df["pred_etarget"] = df["pred"] * df["anchor_target"] / (1 - df["pred"])
df["pred_etarget"] = np.clip(df["pred_etarget"], a_min=0.025, a_max=5.6)
df["pred_target"] = np.log(df["pred_etarget"])
all_results.append(df[["id", "pred_target"]])
del model
gc.collect()
# onebert reg
for fold in range(5):
print(f"fold {fold}")
train_fold = train[train["fold"] != fold]
new_test = make_test_dataset(train_fold, test, 10)
valid_set = OneBertValidationDataset(new_test, tokenizer)
valid_collator = OneBertCompareCollator(
token_pad_value=tokenizer.pad_token_id,
type_pad_value=tokenizer.pad_token_type_id,
)
loader = torch.utils.data.DataLoader(
valid_set,
shuffle=False,
batch_size=16,
pin_memory=True,
drop_last=False,
num_workers=4,
collate_fn=valid_collator,
)
state = torch.load(f"../input/clrp-onebert-reg/best-model-{fold}.pt")
model = OneBertComparer(None, config, sep_pooling=True, pooling="att")
model.load_state_dict(state["model"])
model.eval()
model.cuda()
results = []
with torch.no_grad():
for inputs, _ in tqdm(loader):
inputs = _prepare_inputs(inputs)
with autocast(enabled=True):
outputs = model(**inputs)
results.append(outputs)
predicts = merge_dicts(results)
for key in predicts.keys():
predicts[key] = torch.cat(predicts[key], dim=0)
pred = predicts["pred"].cpu().numpy()
df = pd.DataFrame()
df["id"] = new_test["id"]
df["pred"] = pred.flatten()
df["anchor_target"] = new_test["anchor_target"]
df["pred_target"] = df["pred"] + df["anchor_target"]
all_results.append(df[["id", "pred_target"]])
del model
gc.collect()
df = pd.concat(all_results, ignore_index=True, sort=False)
pred = df.groupby("id")["pred_target"].mean().reset_index()
pred["target"] = pred["pred_target"]
pred[["id", "target"]].to_csv("submission.csv", index=False)
len(all_results[0])
pred.head()
pred.head()
| false | 0 | 6,417 | 0 | 6,417 | 6,417 |
||
69605144
|
# # Hey! This notebook goes over the line of best fit, which is our first machine learning algorithm together!! Let me know if you have any questions on the contents of this notebook by pinging me on Discord :)
# scientific libraries
import pandas as pd
import numpy as np
# plotting
import matplotlib.pyplot as plt
import seaborn as sns
# cleaner than directly using modules
from matplotlib.animation import FuncAnimation
from IPython.display import HTML
sns.set_theme()
plt.style.use("dark_background")
# ### Let's say we had a dataset for housing prices. The dataset contains two columns,
# ### *Price (USD)* and *No. of floors*
# Before we try to do any machine learning, let's take a look at the data!
# number of samples to plot
NUM_SAMPLES = 100
# assuming our goal is to figure out the price given num_floors,
# let's say price = PRICE_TREND * num_floors
PRICE_TREND = 10000
# what do we call num_floors in our machine learning terminology?
num_floors = np.arange(1, NUM_SAMPLES + 1)
# returns a list of noises to add randomness to samples
get_noise = (
lambda num_samples, noise_amt: (np.random.rand(num_samples) - 0.5) * noise_amt
)
price_noise = get_noise(NUM_SAMPLES, 1)
# what do we call prices in our machine learning terminology?
prices = num_floors * 10000 + price_noise
pd.DataFrame({"Price (USD)": prices, "No. of floors": num_floors})
# How about some plots to visualize these numbers?
sns.scatterplot(x=num_floors, y=prices)
plt.xlabel("No. of floors")
plt.ylabel("Price")
def get_scatter_animation(number_of_floors, num_noises=8):
"""
returns a graphical animation of the toy data with increasing noise
parameters:
number_of_floors: feature, plotted as x-axis
num_noises: number of exponentially increasing noise
values to go through in animation
"""
# our animation will be drawn up on this figure
fig, ax = plt.subplots()
plt.xlabel("No. of floors")
plt.ylabel("Price")
def update_animation(noise_scale):
"""updates the scatterplot animation"""
price_noise = (np.random.rand(100) - 0.5) * noise_scale
prices = (number_of_floors * 10000 + price_noise).clip(min=0)
# update the graphical representation of the scatterplot to the next frame
ax.scatter(x=number_of_floors, y=prices)
# matplotlib.animation's FuncAnimation allows us to use our
# update_animation function to animate!
anim = FuncAnimation(
fig,
update_animation,
frames=[10**exp for exp in range(num_noises)],
interval=350,
)
# this line is of little consequence, but it prevents the
# original fig from outputting (to ensure that we only get the animation)
plt.close()
# convert to notebook-friendly animation
result_animation = HTML(anim.to_jshtml())
return result_animation
get_scatter_animation(num_floors)
# # Now, let's dive into the machine learning!
# As it turns out, we can actually figure out the line of best fit with a single equation!
# ### **Notation:**
# * number_of_floors -> $ \textbf{X} $
# * prices -> $ \textbf {Y} $
# #### Then, assuming we're using squared distance as the error, our ideal line follows the trend defined by $ (\textbf{X}^T\textbf{X})^{-1}\textbf{X}^T\textbf{Y} $.
# How do we even derive something like this? Linear algebra (i.e., [things like the rotations and vectors we talked about last week](https://www.kaggle.com/rioton2k17/and-i-discovered-that-my-castles-stand)) + calculus (mainly derivatives)!
# feel free to play around with NOISE_AMT and see how it affects
# the line of best fit!
NOISE_AMT = 1000000
X = num_floors.reshape(NUM_SAMPLES, 1)
price_noise = get_noise(NUM_SAMPLES, noise_amt=NOISE_AMT)
prices = (num_floors * PRICE_TREND + price_noise).clip(min=0)
Y = prices.reshape(NUM_SAMPLES, 1)
# this line is the solution from the previous cell
best_fit_trend = np.linalg.inv(X.T @ X) @ X.T @ Y
sns.scatterplot(x=num_floors, y=prices)
sns.lineplot(x=num_floors, y=(num_floors * best_fit_trend).flatten(), color="red")
plt.xlabel("No. of floors")
plt.ylabel("Price")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605144.ipynb
| null | null |
[{"Id": 69605144, "ScriptId": 19006272, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2534255, "CreationDate": "08/02/2021 05:32:46", "VersionNumber": 3.0, "Title": "Linear Regression Demo", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 120.0, "LinesInsertedFromPrevious": 17.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 103.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# # Hey! This notebook goes over the line of best fit, which is our first machine learning algorithm together!! Let me know if you have any questions on the contents of this notebook by pinging me on Discord :)
# scientific libraries
import pandas as pd
import numpy as np
# plotting
import matplotlib.pyplot as plt
import seaborn as sns
# cleaner than directly using modules
from matplotlib.animation import FuncAnimation
from IPython.display import HTML
sns.set_theme()
plt.style.use("dark_background")
# ### Let's say we had a dataset for housing prices. The dataset contains two columns,
# ### *Price (USD)* and *No. of floors*
# Before we try to do any machine learning, let's take a look at the data!
# number of samples to plot
NUM_SAMPLES = 100
# assuming our goal is to figure out the price given num_floors,
# let's say price = PRICE_TREND * num_floors
PRICE_TREND = 10000
# what do we call num_floors in our machine learning terminology?
num_floors = np.arange(1, NUM_SAMPLES + 1)
# returns a list of noises to add randomness to samples
get_noise = (
lambda num_samples, noise_amt: (np.random.rand(num_samples) - 0.5) * noise_amt
)
price_noise = get_noise(NUM_SAMPLES, 1)
# what do we call prices in our machine learning terminology?
prices = num_floors * 10000 + price_noise
pd.DataFrame({"Price (USD)": prices, "No. of floors": num_floors})
# How about some plots to visualize these numbers?
sns.scatterplot(x=num_floors, y=prices)
plt.xlabel("No. of floors")
plt.ylabel("Price")
def get_scatter_animation(number_of_floors, num_noises=8):
"""
returns a graphical animation of the toy data with increasing noise
parameters:
number_of_floors: feature, plotted as x-axis
num_noises: number of exponentially increasing noise
values to go through in animation
"""
# our animation will be drawn up on this figure
fig, ax = plt.subplots()
plt.xlabel("No. of floors")
plt.ylabel("Price")
def update_animation(noise_scale):
"""updates the scatterplot animation"""
price_noise = (np.random.rand(100) - 0.5) * noise_scale
prices = (number_of_floors * 10000 + price_noise).clip(min=0)
# update the graphical representation of the scatterplot to the next frame
ax.scatter(x=number_of_floors, y=prices)
# matplotlib.animation's FuncAnimation allows us to use our
# update_animation function to animate!
anim = FuncAnimation(
fig,
update_animation,
frames=[10**exp for exp in range(num_noises)],
interval=350,
)
# this line is of little consequence, but it prevents the
# original fig from outputting (to ensure that we only get the animation)
plt.close()
# convert to notebook-friendly animation
result_animation = HTML(anim.to_jshtml())
return result_animation
get_scatter_animation(num_floors)
# # Now, let's dive into the machine learning!
# As it turns out, we can actually figure out the line of best fit with a single equation!
# ### **Notation:**
# * number_of_floors -> $ \textbf{X} $
# * prices -> $ \textbf {Y} $
# #### Then, assuming we're using squared distance as the error, our ideal line follows the trend defined by $ (\textbf{X}^T\textbf{X})^{-1}\textbf{X}^T\textbf{Y} $.
# How do we even derive something like this? Linear algebra (i.e., [things like the rotations and vectors we talked about last week](https://www.kaggle.com/rioton2k17/and-i-discovered-that-my-castles-stand)) + calculus (mainly derivatives)!
# feel free to play around with NOISE_AMT and see how it affects
# the line of best fit!
NOISE_AMT = 1000000
X = num_floors.reshape(NUM_SAMPLES, 1)
price_noise = get_noise(NUM_SAMPLES, noise_amt=NOISE_AMT)
prices = (num_floors * PRICE_TREND + price_noise).clip(min=0)
Y = prices.reshape(NUM_SAMPLES, 1)
# this line is the solution from the previous cell
best_fit_trend = np.linalg.inv(X.T @ X) @ X.T @ Y
sns.scatterplot(x=num_floors, y=prices)
sns.lineplot(x=num_floors, y=(num_floors * best_fit_trend).flatten(), color="red")
plt.xlabel("No. of floors")
plt.ylabel("Price")
| false | 0 | 1,220 | 0 | 1,220 | 1,220 |
||
69605717
|
<jupyter_start><jupyter_text>Chest X-Ray Images (Pneumonia)
### Context
http://www.cell.com/cell/fulltext/S0092-8674(18)30154-5

Figure S6. Illustrative Examples of Chest X-Rays in Patients with Pneumonia, Related to Figure 6
The normal chest X-ray (left panel) depicts clear lungs without any areas of abnormal opacification in the image. Bacterial pneumonia (middle) typically exhibits a focal lobar consolidation, in this case in the right upper lobe (white arrows), whereas viral pneumonia (right) manifests with a more diffuse ‘‘interstitial’’ pattern in both lungs.
http://www.cell.com/cell/fulltext/S0092-8674(18)30154-5
### Content
The dataset is organized into 3 folders (train, test, val) and contains subfolders for each image category (Pneumonia/Normal). There are 5,863 X-Ray images (JPEG) and 2 categories (Pneumonia/Normal).
Chest X-ray images (anterior-posterior) were selected from retrospective cohorts of pediatric patients of one to five years old from Guangzhou Women and Children’s Medical Center, Guangzhou. All chest X-ray imaging was performed as part of patients’ routine clinical care.
For the analysis of chest x-ray images, all chest radiographs were initially screened for quality control by removing all low quality or unreadable scans. The diagnoses for the images were then graded by two expert physicians before being cleared for training the AI system. In order to account for any grading errors, the evaluation set was also checked by a third expert.
Kaggle dataset identifier: chest-xray-pneumonia
<jupyter_script>import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
import PIL # to open and display image
from PIL import Image
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.python.keras.models import Sequential
from tensorflow.keras import layers
# # **Data Preprocessing**
image = "../input/chest-xray-pneumonia/chest_xray/test/PNEUMONIA/person100_bacteria_475.jpeg"
PIL.Image.open(image)
# # **Data Training**
training_dir = "../input/chest-xray-pneumonia/chest_xray/train/"
# increases amount of data by making diff forms of image
training_generator = ImageDataGenerator(
rescale=1 / 255,
featurewise_center=False,
samplewise_center=False,
featurewise_std_normalization=False,
samplewise_std_normalization=False,
zca_whitening=False,
rotation_range=30,
zoom_range=0.2,
width_shift_range=0.1,
height_shift_range=0.1,
horizontal_flip=False,
vertical_flip=False,
)
training_generator = training_generator.flow_from_directory(
training_dir, target_size=(200, 200), batch_size=4, class_mode="binary"
)
# # **Validation Data**
# ## Validation data is using for evaluation of the model
validation_dir = "../input/chest-xray-pneumonia/chest_xray/val"
validation_generator = ImageDataGenerator(rescale=1 / 255)
val_generator = validation_generator.flow_from_directory(
validation_dir, target_size=(200, 200), batch_size=4, class_mode="binary"
)
# # **Testing Data**
test_dir = "../input/chest-xray-pneumonia/chest_xray/test"
test_generator = ImageDataGenerator(rescale=1 / 255)
test_generator = test_generator.flow_from_directory(
test_dir, target_size=(200, 200), batch_size=16, class_mode="binary"
)
# # **Create Our CNN Model**
model = Sequential()
# Conv2D=(filters,(kernels),filter_size,input image_shape,activation method) -convoluation layer
# MaxPooling2D- (size of pooling window) -pooling layer
# Dropout -(dropout percentage) -drops random edges in the data -> prevents overfitting
# Flatten- flatten output of pooling layer into one vector
# Dense- (uints, activation method) - dense layer to condense
model.add(layers.Conv2D(32, (3, 3), input_shape=(200, 200, 3), activation="relu"))
model.add(layers.MaxPooling2D(2, 2))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D(2, 2))
model.add(layers.Dropout(0.2))
# model.add(layers.Conv2D(128,(3,3),activation='relu'))
# model.add(layers.MaxPooling2D(2,2))
# model.add(layers.Dropout(0.2))
model.add(layers.Conv2D(128, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D(2, 2))
model.add(layers.Dropout(0.2))
model.add(layers.Conv2D(256, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D(2, 2))
# model.add(layers.Conv2D(512,(3,3), activation='relu'))
# model.add(layers.MaxPooling2D(2,2))
# model.add(layers.Conv2D(1024,(3,3), activation='relu'))
# model.add(layers.MaxPooling2D(2,2))
model.add(layers.Flatten())
model.add(layers.Dropout(0.2))
model.add(layers.Dense(256, activation="relu"))
model.add(layers.Dense(1, activation="sigmoid"))
model.summary()
# # **Compiling The Model**
model.compile(
optimizer=tf.keras.optimizers.Adam(lr=0.001),
loss="binary_crossentropy",
metrics=["acc"],
)
# Fitting the model- train data generator from before, validation data,
# epochs (how many times the algorithm runs on the data),
# verbose - gives animated progress bar e.g verbose=1
history = model.fit_generator(
training_generator, validation_data=val_generator, epochs=2, verbose=1
)
acc = history.history["acc"]
val_acc = history.history["val_acc"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
epochs = range(len(acc))
plt.plot(epochs, acc, "r", label="Training Accuracy")
plt.plot(epochs, val_acc, "b", label="Validation Accuracy")
plt.title("Traning and Validation Accuracy Graph")
plt.legend()
plt.figure()
# # **Testing The Model**
print("loss of the model is :", model.evaluate(test_generator)[0] * 100, "%")
print("accuracy of the model is: ", model.evaluate(test_generator)[1] * 100, "%")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605717.ipynb
|
chest-xray-pneumonia
|
paultimothymooney
|
[{"Id": 69605717, "ScriptId": 19005561, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5789052, "CreationDate": "08/02/2021 05:43:43", "VersionNumber": 1.0, "Title": "Pneumoina Detection by using CNNs - Mini Project", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 138.0, "LinesInsertedFromPrevious": 138.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93001962, "KernelVersionId": 69605717, "SourceDatasetVersionId": 23812}]
|
[{"Id": 23812, "DatasetId": 17810, "DatasourceVersionId": 23851, "CreatorUserId": 1314380, "LicenseName": "Other (specified in description)", "CreationDate": "03/24/2018 19:41:59", "VersionNumber": 2.0, "Title": "Chest X-Ray Images (Pneumonia)", "Slug": "chest-xray-pneumonia", "Subtitle": "5,863 images, 2 categories", "Description": "### Context\n\nhttp://www.cell.com/cell/fulltext/S0092-8674(18)30154-5\n\n\n\nFigure S6. Illustrative Examples of Chest X-Rays in Patients with Pneumonia, Related to Figure 6\nThe normal chest X-ray (left panel) depicts clear lungs without any areas of abnormal opacification in the image. Bacterial pneumonia (middle) typically exhibits a focal lobar consolidation, in this case in the right upper lobe (white arrows), whereas viral pneumonia (right) manifests with a more diffuse \u2018\u2018interstitial\u2019\u2019 pattern in both lungs.\nhttp://www.cell.com/cell/fulltext/S0092-8674(18)30154-5\n\n### Content\n\nThe dataset is organized into 3 folders (train, test, val) and contains subfolders for each image category (Pneumonia/Normal). There are 5,863 X-Ray images (JPEG) and 2 categories (Pneumonia/Normal). \n\nChest X-ray images (anterior-posterior) were selected from retrospective cohorts of pediatric patients of one to five years old from Guangzhou Women and Children\u2019s Medical Center, Guangzhou. All chest X-ray imaging was performed as part of patients\u2019 routine clinical care. \n\nFor the analysis of chest x-ray images, all chest radiographs were initially screened for quality control by removing all low quality or unreadable scans. The diagnoses for the images were then graded by two expert physicians before being cleared for training the AI system. In order to account for any grading errors, the evaluation set was also checked by a third expert.\n\n### Acknowledgements\n\nData: https://data.mendeley.com/datasets/rscbjbr9sj/2\n\nLicense: [CC BY 4.0][1]\n\nCitation: http://www.cell.com/cell/fulltext/S0092-8674(18)30154-5\n\n![enter image description here][2]\n\n\n### Inspiration\n\nAutomated methods to detect and classify human diseases from medical images.\n\n\n [1]: https://creativecommons.org/licenses/by/4.0/\n [2]: https://i.imgur.com/8AUJkin.png", "VersionNotes": "train/test/val", "TotalCompressedBytes": 1237249419.0, "TotalUncompressedBytes": 1237249419.0}]
|
[{"Id": 17810, "CreatorUserId": 1314380, "OwnerUserId": 1314380.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 23812.0, "CurrentDatasourceVersionId": 23851.0, "ForumId": 25540, "Type": 2, "CreationDate": "03/22/2018 05:42:41", "LastActivityDate": "03/22/2018", "TotalViews": 2063138, "TotalDownloads": 237932, "TotalVotes": 5834, "TotalKernels": 2058}]
|
[{"Id": 1314380, "UserName": "paultimothymooney", "DisplayName": "Paul Mooney", "RegisterDate": "10/05/2017", "PerformanceTier": 5}]
|
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
import PIL # to open and display image
from PIL import Image
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.python.keras.models import Sequential
from tensorflow.keras import layers
# # **Data Preprocessing**
image = "../input/chest-xray-pneumonia/chest_xray/test/PNEUMONIA/person100_bacteria_475.jpeg"
PIL.Image.open(image)
# # **Data Training**
training_dir = "../input/chest-xray-pneumonia/chest_xray/train/"
# increases amount of data by making diff forms of image
training_generator = ImageDataGenerator(
rescale=1 / 255,
featurewise_center=False,
samplewise_center=False,
featurewise_std_normalization=False,
samplewise_std_normalization=False,
zca_whitening=False,
rotation_range=30,
zoom_range=0.2,
width_shift_range=0.1,
height_shift_range=0.1,
horizontal_flip=False,
vertical_flip=False,
)
training_generator = training_generator.flow_from_directory(
training_dir, target_size=(200, 200), batch_size=4, class_mode="binary"
)
# # **Validation Data**
# ## Validation data is using for evaluation of the model
validation_dir = "../input/chest-xray-pneumonia/chest_xray/val"
validation_generator = ImageDataGenerator(rescale=1 / 255)
val_generator = validation_generator.flow_from_directory(
validation_dir, target_size=(200, 200), batch_size=4, class_mode="binary"
)
# # **Testing Data**
test_dir = "../input/chest-xray-pneumonia/chest_xray/test"
test_generator = ImageDataGenerator(rescale=1 / 255)
test_generator = test_generator.flow_from_directory(
test_dir, target_size=(200, 200), batch_size=16, class_mode="binary"
)
# # **Create Our CNN Model**
model = Sequential()
# Conv2D=(filters,(kernels),filter_size,input image_shape,activation method) -convoluation layer
# MaxPooling2D- (size of pooling window) -pooling layer
# Dropout -(dropout percentage) -drops random edges in the data -> prevents overfitting
# Flatten- flatten output of pooling layer into one vector
# Dense- (uints, activation method) - dense layer to condense
model.add(layers.Conv2D(32, (3, 3), input_shape=(200, 200, 3), activation="relu"))
model.add(layers.MaxPooling2D(2, 2))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D(2, 2))
model.add(layers.Dropout(0.2))
# model.add(layers.Conv2D(128,(3,3),activation='relu'))
# model.add(layers.MaxPooling2D(2,2))
# model.add(layers.Dropout(0.2))
model.add(layers.Conv2D(128, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D(2, 2))
model.add(layers.Dropout(0.2))
model.add(layers.Conv2D(256, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D(2, 2))
# model.add(layers.Conv2D(512,(3,3), activation='relu'))
# model.add(layers.MaxPooling2D(2,2))
# model.add(layers.Conv2D(1024,(3,3), activation='relu'))
# model.add(layers.MaxPooling2D(2,2))
model.add(layers.Flatten())
model.add(layers.Dropout(0.2))
model.add(layers.Dense(256, activation="relu"))
model.add(layers.Dense(1, activation="sigmoid"))
model.summary()
# # **Compiling The Model**
model.compile(
optimizer=tf.keras.optimizers.Adam(lr=0.001),
loss="binary_crossentropy",
metrics=["acc"],
)
# Fitting the model- train data generator from before, validation data,
# epochs (how many times the algorithm runs on the data),
# verbose - gives animated progress bar e.g verbose=1
history = model.fit_generator(
training_generator, validation_data=val_generator, epochs=2, verbose=1
)
acc = history.history["acc"]
val_acc = history.history["val_acc"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
epochs = range(len(acc))
plt.plot(epochs, acc, "r", label="Training Accuracy")
plt.plot(epochs, val_acc, "b", label="Validation Accuracy")
plt.title("Traning and Validation Accuracy Graph")
plt.legend()
plt.figure()
# # **Testing The Model**
print("loss of the model is :", model.evaluate(test_generator)[0] * 100, "%")
print("accuracy of the model is: ", model.evaluate(test_generator)[1] * 100, "%")
| false | 0 | 1,385 | 0 | 1,862 | 1,385 |
||
69605618
|
<jupyter_start><jupyter_text>Mobile Price Classification
### Context
Bob has started his own mobile company. He wants to give tough fight to big companies like Apple,Samsung etc.
He does not know how to estimate price of mobiles his company creates. In this competitive mobile phone market you cannot simply assume things. To solve this problem he collects sales data of mobile phones of various companies.
Bob wants to find out some relation between features of a mobile phone(eg:- RAM,Internal Memory etc) and its selling price. But he is not so good at Machine Learning. So he needs your help to solve this problem.
In this problem you do not have to predict actual price but a price range indicating how high the price is
Kaggle dataset identifier: mobile-price-classification
<jupyter_code>import pandas as pd
df = pd.read_csv('mobile-price-classification/train.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 2000 entries, 0 to 1999
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 battery_power 2000 non-null int64
1 blue 2000 non-null int64
2 clock_speed 2000 non-null float64
3 dual_sim 2000 non-null int64
4 fc 2000 non-null int64
5 four_g 2000 non-null int64
6 int_memory 2000 non-null int64
7 m_dep 2000 non-null float64
8 mobile_wt 2000 non-null int64
9 n_cores 2000 non-null int64
10 pc 2000 non-null int64
11 px_height 2000 non-null int64
12 px_width 2000 non-null int64
13 ram 2000 non-null int64
14 sc_h 2000 non-null int64
15 sc_w 2000 non-null int64
16 talk_time 2000 non-null int64
17 three_g 2000 non-null int64
18 touch_screen 2000 non-null int64
19 wifi 2000 non-null int64
20 price_range 2000 non-null int64
dtypes: float64(2), int64(19)
memory usage: 328.2 KB
<jupyter_text>Examples:
{
"battery_power": 842.0,
"blue": 0.0,
"clock_speed": 2.2,
"dual_sim": 0.0,
"fc": 1.0,
"four_g": 0.0,
"int_memory": 7.0,
"m_dep": 0.6000000000000001,
"mobile_wt": 188.0,
"n_cores": 2.0,
"pc": 2.0,
"px_height": 20.0,
"px_width": 756.0,
"ram": 2549.0,
"sc_h": 9.0,
"sc_w": 7.0,
"talk_time": 19.0,
"three_g": 0.0,
"touch_screen": 0.0,
"wifi": 1.0,
"...": "and 1 more columns"
}
{
"battery_power": 1021.0,
"blue": 1.0,
"clock_speed": 0.5,
"dual_sim": 1.0,
"fc": 0.0,
"four_g": 1.0,
"int_memory": 53.0,
"m_dep": 0.7000000000000001,
"mobile_wt": 136.0,
"n_cores": 3.0,
"pc": 6.0,
"px_height": 905.0,
"px_width": 1988.0,
"ram": 2631.0,
"sc_h": 17.0,
"sc_w": 3.0,
"talk_time": 7.0,
"three_g": 1.0,
"touch_screen": 1.0,
"wifi": 0.0,
"...": "and 1 more columns"
}
{
"battery_power": 563.0,
"blue": 1.0,
"clock_speed": 0.5,
"dual_sim": 1.0,
"fc": 2.0,
"four_g": 1.0,
"int_memory": 41.0,
"m_dep": 0.9,
"mobile_wt": 145.0,
"n_cores": 5.0,
"pc": 6.0,
"px_height": 1263.0,
"px_width": 1716.0,
"ram": 2603.0,
"sc_h": 11.0,
"sc_w": 2.0,
"talk_time": 9.0,
"three_g": 1.0,
"touch_screen": 1.0,
"wifi": 0.0,
"...": "and 1 more columns"
}
{
"battery_power": 615.0,
"blue": 1.0,
"clock_speed": 2.5,
"dual_sim": 0.0,
"fc": 0.0,
"four_g": 0.0,
"int_memory": 10.0,
"m_dep": 0.8,
"mobile_wt": 131.0,
"n_cores": 6.0,
"pc": 9.0,
"px_height": 1216.0,
"px_width": 1786.0,
"ram": 2769.0,
"sc_h": 16.0,
"sc_w": 8.0,
"talk_time": 11.0,
"three_g": 1.0,
"touch_screen": 0.0,
"wifi": 0.0,
"...": "and 1 more columns"
}
<jupyter_script>import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from warnings import filterwarnings
filterwarnings("ignore")
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv("/kaggle/input/mobile-price-classification/train.csv")
print(data.shape)
data.head()
data.isnull().sum()
data.describe()
data.info()
plt.figure(figsize=(16, 12))
sns.heatmap(data.corr(), annot=True)
plt.title("Price Rang Count")
sns.countplot(data.price_range)
plt.figure(figsize=(14, 6))
plt.subplot(2, 2, 1)
sns.barplot(x="price_range", y="battery_power", data=data, palette="Reds")
plt.subplot(2, 2, 2)
sns.barplot(x="price_range", y="px_height", data=data, palette="Blues")
plt.subplot(2, 2, 3)
sns.barplot(x="price_range", y="px_width", data=data, palette="Greens")
plt.subplot(2, 2, 4)
sns.barplot(x="price_range", y="ram", data=data, palette="Oranges")
sns.relplot(x="price_range", y="ram", data=data, kind="line")
sns.relplot(x="price_range", y="battery_power", data=data, kind="line")
sns.jointplot(x=data.pc, y=data.fc, kind="reg", color="#ce1414")
X = data.drop(columns=["price_range"])
y = data["price_range"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=24
)
# ## Feature Selection
# #### Recursive feature elimination (RFE) with random forest
from sklearn.feature_selection import RFE
from sklearn.ensemble import RandomForestClassifier
# Create the RFE object and rank each pixel
clf_rf = RandomForestClassifier()
rfe = RFE(estimator=clf_rf, n_features_to_select=5, step=1)
rfe = rfe.fit(X_train, y_train)
print("Chosen best 5 feature by rfe:", X_train.columns[rfe.support_])
X_train_select = X_train[["battery_power", "mobile_wt", "px_height", "px_width", "ram"]]
X_test_select = X_test[["battery_power", "mobile_wt", "px_height", "px_width", "ram"]]
from sklearn.metrics import f1_score, confusion_matrix
from sklearn.metrics import accuracy_score
rfc = RandomForestClassifier()
rfc.fit(X_train_select, y_train)
y_pred = rfc.predict(X_test_select)
acc = accuracy_score(y_test, y_pred)
print("Accuracy is: ", acc)
cm = confusion_matrix(y_test, y_pred)
sns.heatmap(cm, annot=True, fmt="d")
# #### Recursive feature elimination with cross validation and random forest classification
from sklearn.feature_selection import RFECV
clf_rf2 = RandomForestClassifier()
rfecv = RFECV(estimator=clf_rf2, step=1, cv=5, scoring="accuracy")
rfecv = rfecv.fit(X_train, y_train)
print("Optimal number of features :", rfecv.n_features_)
print("Best features :", X_train.columns[rfecv.support_])
# Lets look at best accuracy with plot
plt.figure()
plt.xlabel("Number of features selected")
plt.ylabel("Cross validation score of number of selected features")
plt.plot(range(1, len(rfecv.grid_scores_) + 1), rfecv.grid_scores_)
plt.show()
# So the features after RFECV is same as the features we select in RFE !
# ### Modeling
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from xgboost import XGBClassifier
def score_of_model(models, X_train_select, X_test_select, y_train, y_test):
np.random.seed(24)
model_scores = {}
for name, model in models.items():
model.fit(X_train_select, y_train)
model_scores[name] = model.score(X_test_select, y_test)
model_scores = pd.DataFrame(model_scores, index=["Score"]).transpose()
model_scores = model_scores.sort_values("Score")
return model_scores
models = {
"LogisticRegression": LogisticRegression(max_iter=10000),
"KNeighborsClassifier": KNeighborsClassifier(),
"SVC": SVC(),
"DecisionTreeClassifier": DecisionTreeClassifier(),
"RandomForestClassifier": RandomForestClassifier(),
"XGBClassifier": XGBClassifier(
objective="binary:logistic", eval_metric=["logloss"]
),
}
model_evaluation = score_of_model(
models, X_train_select, X_test_select, y_train, y_test
)
model_evaluation
# Lets look at best score of model with plot
plt.figure(figsize=(6, 4))
plt.title("Each Model Score")
sns.barplot(data=model_evaluation.sort_values("Score").T)
plt.xticks(rotation=90)
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
rf = RandomForestClassifier()
rf.fit(X_train_select, y_train)
y_predicted = rf.predict(X_test_select)
rf_conf_matrix = confusion_matrix(y_test, y_predicted)
rf_acc_score = accuracy_score(y_test, y_predicted)
print("confussion matrix")
print(rf_conf_matrix)
print("\n")
print("Accuracy of Random Forest:", rf_acc_score * 100, "\n")
print(classification_report(y_test, y_predicted))
lr = LogisticRegression(max_iter=10000)
model = lr.fit(X_train_select, y_train)
y_predicted = lr.predict(X_test_select)
lr_conf_matrix = confusion_matrix(y_test, y_predicted)
lr_acc_score = accuracy_score(y_test, y_predicted)
print("confussion matrix")
print(lr_conf_matrix)
print("\n")
print("Accuracy of Logistic Regression:", lr_acc_score * 100, "\n")
print(classification_report(y_test, y_predicted))
svc = SVC()
svc.fit(X_train_select, y_train)
y_predicted = svc.predict(X_test_select)
svc_conf_matrix = confusion_matrix(y_test, y_predicted)
svc_acc_score = accuracy_score(y_test, y_predicted)
print("confussion matrix")
print(svc_conf_matrix)
print("\n")
print("Accuracy of Support Vector Classifier:", svc_acc_score * 100, "\n")
print(classification_report(y_test, y_predicted))
# ## Ensembling
from mlxtend.classifier import StackingCVClassifier
scv = StackingCVClassifier(
classifiers=[rf, lr, svc], meta_classifier=lr, random_state=42
)
scv.fit(X_train_select, y_train)
scv_predicted = scv.predict(X_test_select)
scv_conf_matrix = confusion_matrix(y_test, scv_predicted)
scv_acc_score = accuracy_score(y_test, scv_predicted)
print("confussion matrix")
print(scv_conf_matrix)
print("\n")
print("Accuracy of StackingCVClassifier:", scv_acc_score * 100, "\n")
print(classification_report(y_test, scv_predicted))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605618.ipynb
|
mobile-price-classification
|
iabhishekofficial
|
[{"Id": 69605618, "ScriptId": 18931650, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7564043, "CreationDate": "08/02/2021 05:42:01", "VersionNumber": 2.0, "Title": "mobile classification", "EvaluationDate": "08/02/2021", "IsChange": false, "TotalLines": 191.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 191.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93001695, "KernelVersionId": 69605618, "SourceDatasetVersionId": 15520}]
|
[{"Id": 15520, "DatasetId": 11167, "DatasourceVersionId": 15520, "CreatorUserId": 907764, "LicenseName": "Unknown", "CreationDate": "01/28/2018 08:44:24", "VersionNumber": 1.0, "Title": "Mobile Price Classification", "Slug": "mobile-price-classification", "Subtitle": "Classify Mobile Price Range", "Description": "### Context\n\nBob has started his own mobile company. He wants to give tough fight to big companies like Apple,Samsung etc.\n\nHe does not know how to estimate price of mobiles his company creates. In this competitive mobile phone market you cannot simply assume things. To solve this problem he collects sales data of mobile phones of various companies.\n\nBob wants to find out some relation between features of a mobile phone(eg:- RAM,Internal Memory etc) and its selling price. But he is not so good at Machine Learning. So he needs your help to solve this problem.\n\nIn this problem you do not have to predict actual price but a price range indicating how high the price is", "VersionNotes": "Initial release", "TotalCompressedBytes": 186253.0, "TotalUncompressedBytes": 186253.0}]
|
[{"Id": 11167, "CreatorUserId": 907764, "OwnerUserId": 907764.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 15520.0, "CurrentDatasourceVersionId": 15520.0, "ForumId": 18557, "Type": 2, "CreationDate": "01/28/2018 08:44:24", "LastActivityDate": "02/06/2018", "TotalViews": 793378, "TotalDownloads": 143007, "TotalVotes": 1700, "TotalKernels": 3248}]
|
[{"Id": 907764, "UserName": "iabhishekofficial", "DisplayName": "Abhishek Sharma", "RegisterDate": "02/11/2017", "PerformanceTier": 1}]
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from warnings import filterwarnings
filterwarnings("ignore")
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv("/kaggle/input/mobile-price-classification/train.csv")
print(data.shape)
data.head()
data.isnull().sum()
data.describe()
data.info()
plt.figure(figsize=(16, 12))
sns.heatmap(data.corr(), annot=True)
plt.title("Price Rang Count")
sns.countplot(data.price_range)
plt.figure(figsize=(14, 6))
plt.subplot(2, 2, 1)
sns.barplot(x="price_range", y="battery_power", data=data, palette="Reds")
plt.subplot(2, 2, 2)
sns.barplot(x="price_range", y="px_height", data=data, palette="Blues")
plt.subplot(2, 2, 3)
sns.barplot(x="price_range", y="px_width", data=data, palette="Greens")
plt.subplot(2, 2, 4)
sns.barplot(x="price_range", y="ram", data=data, palette="Oranges")
sns.relplot(x="price_range", y="ram", data=data, kind="line")
sns.relplot(x="price_range", y="battery_power", data=data, kind="line")
sns.jointplot(x=data.pc, y=data.fc, kind="reg", color="#ce1414")
X = data.drop(columns=["price_range"])
y = data["price_range"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=24
)
# ## Feature Selection
# #### Recursive feature elimination (RFE) with random forest
from sklearn.feature_selection import RFE
from sklearn.ensemble import RandomForestClassifier
# Create the RFE object and rank each pixel
clf_rf = RandomForestClassifier()
rfe = RFE(estimator=clf_rf, n_features_to_select=5, step=1)
rfe = rfe.fit(X_train, y_train)
print("Chosen best 5 feature by rfe:", X_train.columns[rfe.support_])
X_train_select = X_train[["battery_power", "mobile_wt", "px_height", "px_width", "ram"]]
X_test_select = X_test[["battery_power", "mobile_wt", "px_height", "px_width", "ram"]]
from sklearn.metrics import f1_score, confusion_matrix
from sklearn.metrics import accuracy_score
rfc = RandomForestClassifier()
rfc.fit(X_train_select, y_train)
y_pred = rfc.predict(X_test_select)
acc = accuracy_score(y_test, y_pred)
print("Accuracy is: ", acc)
cm = confusion_matrix(y_test, y_pred)
sns.heatmap(cm, annot=True, fmt="d")
# #### Recursive feature elimination with cross validation and random forest classification
from sklearn.feature_selection import RFECV
clf_rf2 = RandomForestClassifier()
rfecv = RFECV(estimator=clf_rf2, step=1, cv=5, scoring="accuracy")
rfecv = rfecv.fit(X_train, y_train)
print("Optimal number of features :", rfecv.n_features_)
print("Best features :", X_train.columns[rfecv.support_])
# Lets look at best accuracy with plot
plt.figure()
plt.xlabel("Number of features selected")
plt.ylabel("Cross validation score of number of selected features")
plt.plot(range(1, len(rfecv.grid_scores_) + 1), rfecv.grid_scores_)
plt.show()
# So the features after RFECV is same as the features we select in RFE !
# ### Modeling
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from xgboost import XGBClassifier
def score_of_model(models, X_train_select, X_test_select, y_train, y_test):
np.random.seed(24)
model_scores = {}
for name, model in models.items():
model.fit(X_train_select, y_train)
model_scores[name] = model.score(X_test_select, y_test)
model_scores = pd.DataFrame(model_scores, index=["Score"]).transpose()
model_scores = model_scores.sort_values("Score")
return model_scores
models = {
"LogisticRegression": LogisticRegression(max_iter=10000),
"KNeighborsClassifier": KNeighborsClassifier(),
"SVC": SVC(),
"DecisionTreeClassifier": DecisionTreeClassifier(),
"RandomForestClassifier": RandomForestClassifier(),
"XGBClassifier": XGBClassifier(
objective="binary:logistic", eval_metric=["logloss"]
),
}
model_evaluation = score_of_model(
models, X_train_select, X_test_select, y_train, y_test
)
model_evaluation
# Lets look at best score of model with plot
plt.figure(figsize=(6, 4))
plt.title("Each Model Score")
sns.barplot(data=model_evaluation.sort_values("Score").T)
plt.xticks(rotation=90)
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
rf = RandomForestClassifier()
rf.fit(X_train_select, y_train)
y_predicted = rf.predict(X_test_select)
rf_conf_matrix = confusion_matrix(y_test, y_predicted)
rf_acc_score = accuracy_score(y_test, y_predicted)
print("confussion matrix")
print(rf_conf_matrix)
print("\n")
print("Accuracy of Random Forest:", rf_acc_score * 100, "\n")
print(classification_report(y_test, y_predicted))
lr = LogisticRegression(max_iter=10000)
model = lr.fit(X_train_select, y_train)
y_predicted = lr.predict(X_test_select)
lr_conf_matrix = confusion_matrix(y_test, y_predicted)
lr_acc_score = accuracy_score(y_test, y_predicted)
print("confussion matrix")
print(lr_conf_matrix)
print("\n")
print("Accuracy of Logistic Regression:", lr_acc_score * 100, "\n")
print(classification_report(y_test, y_predicted))
svc = SVC()
svc.fit(X_train_select, y_train)
y_predicted = svc.predict(X_test_select)
svc_conf_matrix = confusion_matrix(y_test, y_predicted)
svc_acc_score = accuracy_score(y_test, y_predicted)
print("confussion matrix")
print(svc_conf_matrix)
print("\n")
print("Accuracy of Support Vector Classifier:", svc_acc_score * 100, "\n")
print(classification_report(y_test, y_predicted))
# ## Ensembling
from mlxtend.classifier import StackingCVClassifier
scv = StackingCVClassifier(
classifiers=[rf, lr, svc], meta_classifier=lr, random_state=42
)
scv.fit(X_train_select, y_train)
scv_predicted = scv.predict(X_test_select)
scv_conf_matrix = confusion_matrix(y_test, scv_predicted)
scv_acc_score = accuracy_score(y_test, scv_predicted)
print("confussion matrix")
print(scv_conf_matrix)
print("\n")
print("Accuracy of StackingCVClassifier:", scv_acc_score * 100, "\n")
print(classification_report(y_test, scv_predicted))
|
[{"mobile-price-classification/train.csv": {"column_names": "[\"battery_power\", \"blue\", \"clock_speed\", \"dual_sim\", \"fc\", \"four_g\", \"int_memory\", \"m_dep\", \"mobile_wt\", \"n_cores\", \"pc\", \"px_height\", \"px_width\", \"ram\", \"sc_h\", \"sc_w\", \"talk_time\", \"three_g\", \"touch_screen\", \"wifi\", \"price_range\"]", "column_data_types": "{\"battery_power\": \"int64\", \"blue\": \"int64\", \"clock_speed\": \"float64\", \"dual_sim\": \"int64\", \"fc\": \"int64\", \"four_g\": \"int64\", \"int_memory\": \"int64\", \"m_dep\": \"float64\", \"mobile_wt\": \"int64\", \"n_cores\": \"int64\", \"pc\": \"int64\", \"px_height\": \"int64\", \"px_width\": \"int64\", \"ram\": \"int64\", \"sc_h\": \"int64\", \"sc_w\": \"int64\", \"talk_time\": \"int64\", \"three_g\": \"int64\", \"touch_screen\": \"int64\", \"wifi\": \"int64\", \"price_range\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 2000 entries, 0 to 1999\nData columns (total 21 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 battery_power 2000 non-null int64 \n 1 blue 2000 non-null int64 \n 2 clock_speed 2000 non-null float64\n 3 dual_sim 2000 non-null int64 \n 4 fc 2000 non-null int64 \n 5 four_g 2000 non-null int64 \n 6 int_memory 2000 non-null int64 \n 7 m_dep 2000 non-null float64\n 8 mobile_wt 2000 non-null int64 \n 9 n_cores 2000 non-null int64 \n 10 pc 2000 non-null int64 \n 11 px_height 2000 non-null int64 \n 12 px_width 2000 non-null int64 \n 13 ram 2000 non-null int64 \n 14 sc_h 2000 non-null int64 \n 15 sc_w 2000 non-null int64 \n 16 talk_time 2000 non-null int64 \n 17 three_g 2000 non-null int64 \n 18 touch_screen 2000 non-null int64 \n 19 wifi 2000 non-null int64 \n 20 price_range 2000 non-null int64 \ndtypes: float64(2), int64(19)\nmemory usage: 328.2 KB\n", "summary": "{\"battery_power\": {\"count\": 2000.0, \"mean\": 1238.5185, \"std\": 439.41820608353135, \"min\": 501.0, \"25%\": 851.75, \"50%\": 1226.0, \"75%\": 1615.25, \"max\": 1998.0}, \"blue\": {\"count\": 2000.0, \"mean\": 0.495, \"std\": 0.5001000400170075, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"clock_speed\": {\"count\": 2000.0, \"mean\": 1.52225, \"std\": 0.8160042088950689, \"min\": 0.5, \"25%\": 0.7, \"50%\": 1.5, \"75%\": 2.2, \"max\": 3.0}, \"dual_sim\": {\"count\": 2000.0, \"mean\": 0.5095, \"std\": 0.500034766175005, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"fc\": {\"count\": 2000.0, \"mean\": 4.3095, \"std\": 4.341443747983894, \"min\": 0.0, \"25%\": 1.0, \"50%\": 3.0, \"75%\": 7.0, \"max\": 19.0}, \"four_g\": {\"count\": 2000.0, \"mean\": 0.5215, \"std\": 0.49966246736236386, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"int_memory\": {\"count\": 2000.0, \"mean\": 32.0465, \"std\": 18.145714955206856, \"min\": 2.0, \"25%\": 16.0, \"50%\": 32.0, \"75%\": 48.0, \"max\": 64.0}, \"m_dep\": {\"count\": 2000.0, \"mean\": 0.50175, \"std\": 0.2884155496235117, \"min\": 0.1, \"25%\": 0.2, \"50%\": 0.5, \"75%\": 0.8, \"max\": 1.0}, \"mobile_wt\": {\"count\": 2000.0, \"mean\": 140.249, \"std\": 35.39965489638835, \"min\": 80.0, \"25%\": 109.0, \"50%\": 141.0, \"75%\": 170.0, \"max\": 200.0}, \"n_cores\": {\"count\": 2000.0, \"mean\": 4.5205, \"std\": 2.2878367180426604, \"min\": 1.0, \"25%\": 3.0, \"50%\": 4.0, \"75%\": 7.0, \"max\": 8.0}, \"pc\": {\"count\": 2000.0, \"mean\": 9.9165, \"std\": 6.06431494134778, \"min\": 0.0, \"25%\": 5.0, \"50%\": 10.0, \"75%\": 15.0, \"max\": 20.0}, \"px_height\": {\"count\": 2000.0, \"mean\": 645.108, \"std\": 443.7808108064386, \"min\": 0.0, \"25%\": 282.75, \"50%\": 564.0, \"75%\": 947.25, \"max\": 1960.0}, \"px_width\": {\"count\": 2000.0, \"mean\": 1251.5155, \"std\": 432.19944694633796, \"min\": 500.0, \"25%\": 874.75, \"50%\": 1247.0, \"75%\": 1633.0, \"max\": 1998.0}, \"ram\": {\"count\": 2000.0, \"mean\": 2124.213, \"std\": 1084.7320436099494, \"min\": 256.0, \"25%\": 1207.5, \"50%\": 2146.5, \"75%\": 3064.5, \"max\": 3998.0}, \"sc_h\": {\"count\": 2000.0, \"mean\": 12.3065, \"std\": 4.213245004356306, \"min\": 5.0, \"25%\": 9.0, \"50%\": 12.0, \"75%\": 16.0, \"max\": 19.0}, \"sc_w\": {\"count\": 2000.0, \"mean\": 5.767, \"std\": 4.3563976058264045, \"min\": 0.0, \"25%\": 2.0, \"50%\": 5.0, \"75%\": 9.0, \"max\": 18.0}, \"talk_time\": {\"count\": 2000.0, \"mean\": 11.011, \"std\": 5.463955197766688, \"min\": 2.0, \"25%\": 6.0, \"50%\": 11.0, \"75%\": 16.0, \"max\": 20.0}, \"three_g\": {\"count\": 2000.0, \"mean\": 0.7615, \"std\": 0.42627292231873126, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"touch_screen\": {\"count\": 2000.0, \"mean\": 0.503, \"std\": 0.500116044562674, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"wifi\": {\"count\": 2000.0, \"mean\": 0.507, \"std\": 0.5000760322381083, \"min\": 0.0, \"25%\": 0.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"price_range\": {\"count\": 2000.0, \"mean\": 1.5, \"std\": 1.118313602106461, \"min\": 0.0, \"25%\": 0.75, \"50%\": 1.5, \"75%\": 2.25, \"max\": 3.0}}", "examples": "{\"battery_power\":{\"0\":842,\"1\":1021,\"2\":563,\"3\":615},\"blue\":{\"0\":0,\"1\":1,\"2\":1,\"3\":1},\"clock_speed\":{\"0\":2.2,\"1\":0.5,\"2\":0.5,\"3\":2.5},\"dual_sim\":{\"0\":0,\"1\":1,\"2\":1,\"3\":0},\"fc\":{\"0\":1,\"1\":0,\"2\":2,\"3\":0},\"four_g\":{\"0\":0,\"1\":1,\"2\":1,\"3\":0},\"int_memory\":{\"0\":7,\"1\":53,\"2\":41,\"3\":10},\"m_dep\":{\"0\":0.6,\"1\":0.7,\"2\":0.9,\"3\":0.8},\"mobile_wt\":{\"0\":188,\"1\":136,\"2\":145,\"3\":131},\"n_cores\":{\"0\":2,\"1\":3,\"2\":5,\"3\":6},\"pc\":{\"0\":2,\"1\":6,\"2\":6,\"3\":9},\"px_height\":{\"0\":20,\"1\":905,\"2\":1263,\"3\":1216},\"px_width\":{\"0\":756,\"1\":1988,\"2\":1716,\"3\":1786},\"ram\":{\"0\":2549,\"1\":2631,\"2\":2603,\"3\":2769},\"sc_h\":{\"0\":9,\"1\":17,\"2\":11,\"3\":16},\"sc_w\":{\"0\":7,\"1\":3,\"2\":2,\"3\":8},\"talk_time\":{\"0\":19,\"1\":7,\"2\":9,\"3\":11},\"three_g\":{\"0\":0,\"1\":1,\"2\":1,\"3\":1},\"touch_screen\":{\"0\":0,\"1\":1,\"2\":1,\"3\":0},\"wifi\":{\"0\":1,\"1\":0,\"2\":0,\"3\":0},\"price_range\":{\"0\":1,\"1\":2,\"2\":2,\"3\":2}}"}}]
| true | 1 |
<start_data_description><data_path>mobile-price-classification/train.csv:
<column_names>
['battery_power', 'blue', 'clock_speed', 'dual_sim', 'fc', 'four_g', 'int_memory', 'm_dep', 'mobile_wt', 'n_cores', 'pc', 'px_height', 'px_width', 'ram', 'sc_h', 'sc_w', 'talk_time', 'three_g', 'touch_screen', 'wifi', 'price_range']
<column_types>
{'battery_power': 'int64', 'blue': 'int64', 'clock_speed': 'float64', 'dual_sim': 'int64', 'fc': 'int64', 'four_g': 'int64', 'int_memory': 'int64', 'm_dep': 'float64', 'mobile_wt': 'int64', 'n_cores': 'int64', 'pc': 'int64', 'px_height': 'int64', 'px_width': 'int64', 'ram': 'int64', 'sc_h': 'int64', 'sc_w': 'int64', 'talk_time': 'int64', 'three_g': 'int64', 'touch_screen': 'int64', 'wifi': 'int64', 'price_range': 'int64'}
<dataframe_Summary>
{'battery_power': {'count': 2000.0, 'mean': 1238.5185, 'std': 439.41820608353135, 'min': 501.0, '25%': 851.75, '50%': 1226.0, '75%': 1615.25, 'max': 1998.0}, 'blue': {'count': 2000.0, 'mean': 0.495, 'std': 0.5001000400170075, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'clock_speed': {'count': 2000.0, 'mean': 1.52225, 'std': 0.8160042088950689, 'min': 0.5, '25%': 0.7, '50%': 1.5, '75%': 2.2, 'max': 3.0}, 'dual_sim': {'count': 2000.0, 'mean': 0.5095, 'std': 0.500034766175005, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'fc': {'count': 2000.0, 'mean': 4.3095, 'std': 4.341443747983894, 'min': 0.0, '25%': 1.0, '50%': 3.0, '75%': 7.0, 'max': 19.0}, 'four_g': {'count': 2000.0, 'mean': 0.5215, 'std': 0.49966246736236386, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'int_memory': {'count': 2000.0, 'mean': 32.0465, 'std': 18.145714955206856, 'min': 2.0, '25%': 16.0, '50%': 32.0, '75%': 48.0, 'max': 64.0}, 'm_dep': {'count': 2000.0, 'mean': 0.50175, 'std': 0.2884155496235117, 'min': 0.1, '25%': 0.2, '50%': 0.5, '75%': 0.8, 'max': 1.0}, 'mobile_wt': {'count': 2000.0, 'mean': 140.249, 'std': 35.39965489638835, 'min': 80.0, '25%': 109.0, '50%': 141.0, '75%': 170.0, 'max': 200.0}, 'n_cores': {'count': 2000.0, 'mean': 4.5205, 'std': 2.2878367180426604, 'min': 1.0, '25%': 3.0, '50%': 4.0, '75%': 7.0, 'max': 8.0}, 'pc': {'count': 2000.0, 'mean': 9.9165, 'std': 6.06431494134778, 'min': 0.0, '25%': 5.0, '50%': 10.0, '75%': 15.0, 'max': 20.0}, 'px_height': {'count': 2000.0, 'mean': 645.108, 'std': 443.7808108064386, 'min': 0.0, '25%': 282.75, '50%': 564.0, '75%': 947.25, 'max': 1960.0}, 'px_width': {'count': 2000.0, 'mean': 1251.5155, 'std': 432.19944694633796, 'min': 500.0, '25%': 874.75, '50%': 1247.0, '75%': 1633.0, 'max': 1998.0}, 'ram': {'count': 2000.0, 'mean': 2124.213, 'std': 1084.7320436099494, 'min': 256.0, '25%': 1207.5, '50%': 2146.5, '75%': 3064.5, 'max': 3998.0}, 'sc_h': {'count': 2000.0, 'mean': 12.3065, 'std': 4.213245004356306, 'min': 5.0, '25%': 9.0, '50%': 12.0, '75%': 16.0, 'max': 19.0}, 'sc_w': {'count': 2000.0, 'mean': 5.767, 'std': 4.3563976058264045, 'min': 0.0, '25%': 2.0, '50%': 5.0, '75%': 9.0, 'max': 18.0}, 'talk_time': {'count': 2000.0, 'mean': 11.011, 'std': 5.463955197766688, 'min': 2.0, '25%': 6.0, '50%': 11.0, '75%': 16.0, 'max': 20.0}, 'three_g': {'count': 2000.0, 'mean': 0.7615, 'std': 0.42627292231873126, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'touch_screen': {'count': 2000.0, 'mean': 0.503, 'std': 0.500116044562674, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'wifi': {'count': 2000.0, 'mean': 0.507, 'std': 0.5000760322381083, 'min': 0.0, '25%': 0.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'price_range': {'count': 2000.0, 'mean': 1.5, 'std': 1.118313602106461, 'min': 0.0, '25%': 0.75, '50%': 1.5, '75%': 2.25, 'max': 3.0}}
<dataframe_info>
RangeIndex: 2000 entries, 0 to 1999
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 battery_power 2000 non-null int64
1 blue 2000 non-null int64
2 clock_speed 2000 non-null float64
3 dual_sim 2000 non-null int64
4 fc 2000 non-null int64
5 four_g 2000 non-null int64
6 int_memory 2000 non-null int64
7 m_dep 2000 non-null float64
8 mobile_wt 2000 non-null int64
9 n_cores 2000 non-null int64
10 pc 2000 non-null int64
11 px_height 2000 non-null int64
12 px_width 2000 non-null int64
13 ram 2000 non-null int64
14 sc_h 2000 non-null int64
15 sc_w 2000 non-null int64
16 talk_time 2000 non-null int64
17 three_g 2000 non-null int64
18 touch_screen 2000 non-null int64
19 wifi 2000 non-null int64
20 price_range 2000 non-null int64
dtypes: float64(2), int64(19)
memory usage: 328.2 KB
<some_examples>
{'battery_power': {'0': 842, '1': 1021, '2': 563, '3': 615}, 'blue': {'0': 0, '1': 1, '2': 1, '3': 1}, 'clock_speed': {'0': 2.2, '1': 0.5, '2': 0.5, '3': 2.5}, 'dual_sim': {'0': 0, '1': 1, '2': 1, '3': 0}, 'fc': {'0': 1, '1': 0, '2': 2, '3': 0}, 'four_g': {'0': 0, '1': 1, '2': 1, '3': 0}, 'int_memory': {'0': 7, '1': 53, '2': 41, '3': 10}, 'm_dep': {'0': 0.6, '1': 0.7, '2': 0.9, '3': 0.8}, 'mobile_wt': {'0': 188, '1': 136, '2': 145, '3': 131}, 'n_cores': {'0': 2, '1': 3, '2': 5, '3': 6}, 'pc': {'0': 2, '1': 6, '2': 6, '3': 9}, 'px_height': {'0': 20, '1': 905, '2': 1263, '3': 1216}, 'px_width': {'0': 756, '1': 1988, '2': 1716, '3': 1786}, 'ram': {'0': 2549, '1': 2631, '2': 2603, '3': 2769}, 'sc_h': {'0': 9, '1': 17, '2': 11, '3': 16}, 'sc_w': {'0': 7, '1': 3, '2': 2, '3': 8}, 'talk_time': {'0': 19, '1': 7, '2': 9, '3': 11}, 'three_g': {'0': 0, '1': 1, '2': 1, '3': 1}, 'touch_screen': {'0': 0, '1': 1, '2': 1, '3': 0}, 'wifi': {'0': 1, '1': 0, '2': 0, '3': 0}, 'price_range': {'0': 1, '1': 2, '2': 2, '3': 2}}
<end_description>
| 2,045 | 0 | 3,705 | 2,045 |
69605623
|
<jupyter_start><jupyter_text>Hitters
### Context
This dataset is part of the R-package ISLR and is used in the related book by G. James et al. (2013) "An Introduction to Statistical Learning with applications in R" to demonstrate how Ridge regression and the LASSO are performed using R.
### Content
This dataset was originally taken from the StatLib library which is maintained at Carnegie Mellon University. This is part of the data that was used in the 1988 ASA Graphics Section Poster Session. The salary data were originally from Sports Illustrated, April 20, 1987. The 1986 and career statistics were obtained from The 1987 Baseball Encyclopedia Update published by Collier Books, Macmillan Publishing Company, New York.
Format
A data frame with 322 observations of major league players on the following 20 variables.
AtBat Number of times at bat in 1986
Hits Number of hits in 1986
HmRun Number of home runs in 1986
Runs Number of runs in 1986
RBI Number of runs batted in in 1986
Walks Number of walks in 1986
Years Number of years in the major leagues
CAtBat Number of times at bat during his career
CHits Number of hits during his career
CHmRun Number of home runs during his career
CRuns Number of runs during his career
CRBI Number of runs batted in during his career
CWalks Number of walks during his career
League A factor with levels A and N indicating player’s league at the end of 1986
Division A factor with levels E and W indicating player’s division at the end of 1986
PutOuts Number of put outs in 1986
Assists Number of assists in 1986
Errors Number of errors in 1986
Salary 1987 annual salary on opening day in thousands of dollars
NewLeague A factor with levels A and N indicating player’s league at the beginning of 1987
Kaggle dataset identifier: hitters
<jupyter_code>import pandas as pd
df = pd.read_csv('hitters/Hitters.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 322 entries, 0 to 321
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 AtBat 322 non-null int64
1 Hits 322 non-null int64
2 HmRun 322 non-null int64
3 Runs 322 non-null int64
4 RBI 322 non-null int64
5 Walks 322 non-null int64
6 Years 322 non-null int64
7 CAtBat 322 non-null int64
8 CHits 322 non-null int64
9 CHmRun 322 non-null int64
10 CRuns 322 non-null int64
11 CRBI 322 non-null int64
12 CWalks 322 non-null int64
13 League 322 non-null object
14 Division 322 non-null object
15 PutOuts 322 non-null int64
16 Assists 322 non-null int64
17 Errors 322 non-null int64
18 Salary 263 non-null float64
19 NewLeague 322 non-null object
dtypes: float64(1), int64(16), object(3)
memory usage: 50.4+ KB
<jupyter_text>Examples:
{
"AtBat": 293,
"Hits": 66,
"HmRun": 1,
"Runs": 30,
"RBI": 29,
"Walks": 14,
"Years": 1,
"CAtBat": 293,
"CHits": 66,
"CHmRun": 1,
"CRuns": 30,
"CRBI": 29,
"CWalks": 14,
"League": "A",
"Division": "E",
"PutOuts": 446,
"Assists": 33,
"Errors": 20,
"Salary": NaN,
"NewLeague": "A"
}
{
"AtBat": 315,
"Hits": 81,
"HmRun": 7,
"Runs": 24,
"RBI": 38,
"Walks": 39,
"Years": 14,
"CAtBat": 3449,
"CHits": 835,
"CHmRun": 69,
"CRuns": 321,
"CRBI": 414,
"CWalks": 375,
"League": "N",
"Division": "W",
"PutOuts": 632,
"Assists": 43,
"Errors": 10,
"Salary": 475.0,
"NewLeague": "N"
}
{
"AtBat": 479,
"Hits": 130,
"HmRun": 18,
"Runs": 66,
"RBI": 72,
"Walks": 76,
"Years": 3,
"CAtBat": 1624,
"CHits": 457,
"CHmRun": 63,
"CRuns": 224,
"CRBI": 266,
"CWalks": 263,
"League": "A",
"Division": "W",
"PutOuts": 880,
"Assists": 82,
"Errors": 14,
"Salary": 480.0,
"NewLeague": "A"
}
{
"AtBat": 496,
"Hits": 141,
"HmRun": 20,
"Runs": 65,
"RBI": 78,
"Walks": 37,
"Years": 11,
"CAtBat": 5628,
"CHits": 1575,
"CHmRun": 225,
"CRuns": 828,
"CRBI": 838,
"CWalks": 354,
"League": "N",
"Division": "E",
"PutOuts": 200,
"Assists": 11,
"Errors": 3,
"Salary": 500.0,
"NewLeague": "N"
}
<jupyter_script># **Created by Berkay Alan**
# **Non-Linear Models - Regression | Support Vector Regression(SVR)**
# **2 August 2021**
# **Content**
# - Support Vector Regression(SVR) (Theory - Model- Tuning)
# - Non-Linear Support Vector Regression(SVR) (Theory - Model- Tuning)
# ** Check out My Github for other Regression Models **
# Github Repository Including:
#
# - K - Nearest Neighbors(KNN) (Theory - Model- Tuning)
# - Regression(Decision) Trees (CART) (Theory - Model- Tuning)
# - Ensemble Learning - Bagged Trees(Bagging) (Theory - Model- Tuning)
# - Ensemble Learning - Random Forests (Theory - Model- Tuning)
# - Gradient Boosting Machines(GBM) (Theory - Model- Tuning)
# - Light Gradient Boosting Machines(LGBM) (Theory - Model- Tuning)
# - XGBoost(Extreme Gradient Boosting) (Theory - Model- Tuning)
# - Catboost (Theory - Model- Tuning)
#
# Check it out: https://github.com/berkayalan/Data-Science-Tutorials/blob/master/Non-Linear%20Models%20-%20Regression.ipynb
# **For more Tutorial:** https://github.com/berkayalan
# ## Resources
# - **The Elements of Statistical Learning** - Trevor Hastie, Robert Tibshirani, Jerome Friedman - Data Mining, Inference, and Prediction (Springer Series in Statistics)
# - [**Support Vector Regression Tutorial for Machine Learning**](https://www.analyticsvidhya.com/blog/2020/03/support-vector-regression-tutorial-for-machine-learning/)
# - [**An Introduction to Support Vector Regression (SVR)**](https://towardsdatascience.com/an-introduction-to-support-vector-regression-svr-a3ebc1672c2)
# - [**SVM: Difference between Linear and Non-Linear Models**](https://www.aitude.com/svm-difference-between-linear-and-non-linear-models/)
# - [**Kernel Functions-Introduction to SVM Kernel & Examples**](https://data-flair.training/blogs/svm-kernel-functions/)
# - [**Understanding Confusion Matrix**](https://towardsdatascience.com/understanding-confusion-matrix-a9ad42dcfd62)
# ## Importing Libraries
from warnings import filterwarnings
filterwarnings("ignore")
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import statsmodels.api as sm
import statsmodels.formula.api as smf
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.model_selection import (
train_test_split,
cross_val_score,
cross_val_predict,
ShuffleSplit,
GridSearchCV,
)
from sklearn.decomposition import PCA
from sklearn.tree import DecisionTreeRegressor, DecisionTreeClassifier
from sklearn.preprocessing import scale
from sklearn import model_selection
from sklearn.svm import SVR
import time
# ## Support Vector Regression(SVR)
# ### Theory
# Support Vector Regression gives us the flexibility to define how much error is acceptable in our model and will find an appropriate line (or hyperplane in higher dimensions) to fit the data. The objective function of SVR is to minimize the coefficients — more specifically, the l2-norm of the coefficient vector — not the squared error. The error term is instead handled in the constraints, where we set the absolute error less than or equal to a specified margin, called the maximum error, **ϵ (epsilon)**. We can tune epsilon to gain the desired accuracy of our model.
# Illustrative example:
# 
# Photo is cited by:https://towardsdatascience.com/an-introduction-to-support-vector-regression-svr-a3ebc1672c2
# As we can see, we have data points that are outside the epsilon in sensitive tube. We care about the error for them and they will be measured as distance between the point and the tube. As such, we need to account for the possibility of errors that are larger than ϵ. We can do this with slack variables.
# The concept of **slack variables** is simple: for any value that falls outside of ϵ, we can denote its deviation from the margin as ξ.
# That's the formula to minimise:
# 
# Photo is cited by: https://towardsdatascience.com/an-introduction-to-support-vector-regression-svr-a3ebc1672c2
# We now have an additional **C** hyperparameter that we can tune. As C increases, our tolerance for points outside of ϵ also increases. As C approaches 0, the tolerance approaches 0 and the equation collapses into the simplified one.
# ### Model
# For a real world example, we will work with **Hitters** dataset.
# It can be downloaded here: https://www.kaggle.com/floser/hitters
hts = pd.read_csv("../input/hitters/Hitters.csv")
hts.head()
# Now we will remove NA values.
hts.dropna(inplace=True)
# We will do **One Hot Encoding** to categorical columns.
one_hot_encoded = pd.get_dummies(hts[["League", "Division", "NewLeague"]])
one_hot_encoded.head()
new_hts = hts.drop(["League", "Division", "NewLeague", "Salary"], axis=1).astype(
"float64"
)
X = pd.concat(
[new_hts, one_hot_encoded[["League_N", "Division_W", "NewLeague_N"]]], axis=1
)
X.head()
y = hts.Salary # Target-dependent variable
# Now we will split our dataset as train and test set.
hts.shape
# Independent Variables
X.shape
# Dependent Variables
y.shape
X_train = X.iloc[:210]
X_test = X.iloc[210:]
y_train = y[:210]
y_test = y[210:]
print("X_train Shape: ", X_train.shape)
print("X_test Shape: ", X_test.shape)
print("y_train Shape: ", y_train.shape)
print("y_test Shape: ", y_test.shape)
X_train = pd.DataFrame(X_train["Hits"])
X_test = pd.DataFrame(X_test["Hits"])
X_train
X_test[:10]
svr_model = SVR("linear").fit(X_train, y_train)
svr_model
# We can write Support vector regression in linear regression form as below:
print("y = {0} + {1} x".format(svr_model.intercept_[0], svr_model.coef_[0][0]))
# Let's control this formula.
print(
"Manual Calculation: ",
svr_model.intercept_[0] + (svr_model.coef_[0][0] * X_train["Hits"][0:1].item()),
"\nOutput of Model",
svr_model.predict(X_train[0:1]).item(),
)
y_pred = svr_model.predict(X_train)
plt.scatter(X_train, y_train)
plt.plot(X_train, y_pred, c="r")
plt.title("Train Errors")
plt.show()
# ### Prediction
svr_model
y_pred = svr_model.predict(X_train)
# Train Error
np.sqrt(mean_squared_error(y_train, y_pred))
r2_score(y_train, y_pred)
y_pred = svr_model.predict(X_test)
# Test Error
np.sqrt(mean_squared_error(y_test, y_pred))
r2_score(y_test, y_pred)
# ### Model Tuning
svr_model
svr_parameters = {"C": np.arange(0.1, 3, 0.1)}
svr_cv_model = GridSearchCV(svr_model, svr_parameters, cv=15).fit(X_train, y_train)
svr_cv_model.best_params_
svr_tuned = SVR("linear", C=pd.Series(svr_cv_model.best_params_)[0]).fit(
X_train, y_train
)
svr_tuned
y_pred = svr_tuned.predict(X_train)
# Train Error
np.sqrt(mean_squared_error(y_train, y_pred))
r2_score(y_train, y_pred)
y_pred = svr_tuned.predict(X_test)
# Test Error
np.sqrt(mean_squared_error(y_test, y_pred))
r2_score(y_test, y_pred)
# We did all process with only **Hits** column. We can also try with all columns. But it takes more time nearly 30 min.
# ## Non-Linear Support Vector Regression(SVR)
# ### Theory
# When we cannot separate data with a straight line we use Non – Linear SVM. In this, we have **Kernel functions**. They transform non-linear spaces into linear spaces. It transforms data into another dimension so that the data can be classified.
# It transforms two variables x and y into three variables along with z. Therefore, the data have plotted from 2-D space to 3-D space. Now we can easily classify the data by drawi
# ### Model
# For a real world example, we will work with **Hitters** dataset.
# It can be downloaded here: https://www.kaggle.com/floser/hitters
hts = pd.read_csv("../input/hitters/Hitters.csv")
hts.head()
# Now we will remove NA values.
hts.dropna(inplace=True)
# We will do **One Hot Encoding** to categorical columns.
one_hot_encoded = pd.get_dummies(hts[["League", "Division", "NewLeague"]])
one_hot_encoded.head()
new_hts = hts.drop(["League", "Division", "NewLeague", "Salary"], axis=1).astype(
"float64"
)
X = pd.concat(
[new_hts, one_hot_encoded[["League_N", "Division_W", "NewLeague_N"]]], axis=1
)
X.head()
y = hts.Salary # Target-dependent variable
# Now we will split our dataset as train and test set.
hts.shape
# Independent Variables
X.shape
# Dependent Variables
y.shape
X_train = X.iloc[:210]
X_test = X.iloc[210:]
y_train = y[:210]
y_test = y[210:]
print("X_train Shape: ", X_train.shape)
print("X_test Shape: ", X_test.shape)
print("y_train Shape: ", y_train.shape)
print("y_test Shape: ", y_test.shape)
SVR_Radial_Basis = SVR("rbf").fit(X_train, y_train)
# ### Prediction
SVR_Radial_Basis
y_pred = SVR_Radial_Basis.predict(X_train)
# Train Error
np.sqrt(mean_squared_error(y_train, y_pred))
r2_score(y_train, y_pred)
y_pred = SVR_Radial_Basis.predict(X_test)
# Test Error
np.sqrt(mean_squared_error(y_test, y_pred))
r2_score(y_test, y_pred)
# ### Model Tuning
SVR_Radial_Basis
svr_parameters = {"C": np.arange(0.2, 10, 0.1)}
svr_cv_model = GridSearchCV(SVR_Radial_Basis, svr_parameters, cv=15).fit(
X_train, y_train
)
svr_cv_model.best_params_
svr_tuned = SVR("rbf", C=pd.Series(svr_cv_model.best_params_)[0]).fit(X_train, y_train)
svr_tuned
y_pred = svr_tuned.predict(X_train)
# Train Error
np.sqrt(mean_squared_error(y_train, y_pred))
r2_score(y_train, y_pred)
y_pred = svr_tuned.predict(X_test)
# Test Error
np.sqrt(mean_squared_error(y_test, y_pred))
r2_score(y_test, y_pred)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605623.ipynb
|
hitters
|
floser
|
[{"Id": 69605623, "ScriptId": 19009259, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3264446, "CreationDate": "08/02/2021 05:42:05", "VersionNumber": 1.0, "Title": "Non-Linear Models | Support Vector Regression", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 323.0, "LinesInsertedFromPrevious": 323.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 11}]
|
[{"Id": 93001709, "KernelVersionId": 69605623, "SourceDatasetVersionId": 17368}]
|
[{"Id": 17368, "DatasetId": 12722, "DatasourceVersionId": 17368, "CreatorUserId": 1627126, "LicenseName": "Other (specified in description)", "CreationDate": "02/11/2018 20:43:51", "VersionNumber": 1.0, "Title": "Hitters", "Slug": "hitters", "Subtitle": "Major League Baseball Data from the 1986 and 1987 seasons.", "Description": "### Context\n\nThis dataset is part of the R-package ISLR and is used in the related book by G. James et al. (2013) \"An Introduction to Statistical Learning with applications in R\" to demonstrate how Ridge regression and the LASSO are performed using R.\n\n\n\n### Content\n\nThis dataset was originally taken from the StatLib library which is maintained at Carnegie Mellon University. This is part of the data that was used in the 1988 ASA Graphics Section Poster Session. The salary data were originally from Sports Illustrated, April 20, 1987. The 1986 and career statistics were obtained from The 1987 Baseball Encyclopedia Update published by Collier Books, Macmillan Publishing Company, New York.\n\nFormat\nA data frame with 322 observations of major league players on the following 20 variables.\nAtBat Number of times at bat in 1986\nHits Number of hits in 1986\nHmRun Number of home runs in 1986\nRuns Number of runs in 1986\nRBI Number of runs batted in in 1986\nWalks Number of walks in 1986\nYears Number of years in the major leagues\nCAtBat Number of times at bat during his career\nCHits Number of hits during his career\nCHmRun Number of home runs during his career\nCRuns Number of runs during his career\nCRBI Number of runs batted in during his career\nCWalks Number of walks during his career\nLeague A factor with levels A and N indicating player\u2019s league at the end of 1986\nDivision A factor with levels E and W indicating player\u2019s division at the end of 1986\nPutOuts Number of put outs in 1986\nAssists Number of assists in 1986\nErrors Number of errors in 1986\nSalary 1987 annual salary on opening day in thousands of dollars\nNewLeague A factor with levels A and N indicating player\u2019s league at the beginning of 1987\n\n\n\n### Acknowledgements\n\nPlease cite/acknowledge: Games, G., Witten, D., Hastie, T., and Tibshirani, R. (2013) An Introduction to Statistical Learning with applications in R, www.StatLearning.com, Springer-Verlag, New York.\n\n\n\n### Inspiration\n\nThis upload shall enable actuarial kernels with R and Python", "VersionNotes": "Initial release", "TotalCompressedBytes": 20906.0, "TotalUncompressedBytes": 20906.0}]
|
[{"Id": 12722, "CreatorUserId": 1627126, "OwnerUserId": 1627126.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 17368.0, "CurrentDatasourceVersionId": 17368.0, "ForumId": 20189, "Type": 2, "CreationDate": "02/11/2018 20:43:51", "LastActivityDate": "02/11/2018", "TotalViews": 29105, "TotalDownloads": 3828, "TotalVotes": 18, "TotalKernels": 82}]
|
[{"Id": 1627126, "UserName": "floser", "DisplayName": "floser", "RegisterDate": "02/10/2018", "PerformanceTier": 1}]
|
# **Created by Berkay Alan**
# **Non-Linear Models - Regression | Support Vector Regression(SVR)**
# **2 August 2021**
# **Content**
# - Support Vector Regression(SVR) (Theory - Model- Tuning)
# - Non-Linear Support Vector Regression(SVR) (Theory - Model- Tuning)
# ** Check out My Github for other Regression Models **
# Github Repository Including:
#
# - K - Nearest Neighbors(KNN) (Theory - Model- Tuning)
# - Regression(Decision) Trees (CART) (Theory - Model- Tuning)
# - Ensemble Learning - Bagged Trees(Bagging) (Theory - Model- Tuning)
# - Ensemble Learning - Random Forests (Theory - Model- Tuning)
# - Gradient Boosting Machines(GBM) (Theory - Model- Tuning)
# - Light Gradient Boosting Machines(LGBM) (Theory - Model- Tuning)
# - XGBoost(Extreme Gradient Boosting) (Theory - Model- Tuning)
# - Catboost (Theory - Model- Tuning)
#
# Check it out: https://github.com/berkayalan/Data-Science-Tutorials/blob/master/Non-Linear%20Models%20-%20Regression.ipynb
# **For more Tutorial:** https://github.com/berkayalan
# ## Resources
# - **The Elements of Statistical Learning** - Trevor Hastie, Robert Tibshirani, Jerome Friedman - Data Mining, Inference, and Prediction (Springer Series in Statistics)
# - [**Support Vector Regression Tutorial for Machine Learning**](https://www.analyticsvidhya.com/blog/2020/03/support-vector-regression-tutorial-for-machine-learning/)
# - [**An Introduction to Support Vector Regression (SVR)**](https://towardsdatascience.com/an-introduction-to-support-vector-regression-svr-a3ebc1672c2)
# - [**SVM: Difference between Linear and Non-Linear Models**](https://www.aitude.com/svm-difference-between-linear-and-non-linear-models/)
# - [**Kernel Functions-Introduction to SVM Kernel & Examples**](https://data-flair.training/blogs/svm-kernel-functions/)
# - [**Understanding Confusion Matrix**](https://towardsdatascience.com/understanding-confusion-matrix-a9ad42dcfd62)
# ## Importing Libraries
from warnings import filterwarnings
filterwarnings("ignore")
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import statsmodels.api as sm
import statsmodels.formula.api as smf
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.model_selection import (
train_test_split,
cross_val_score,
cross_val_predict,
ShuffleSplit,
GridSearchCV,
)
from sklearn.decomposition import PCA
from sklearn.tree import DecisionTreeRegressor, DecisionTreeClassifier
from sklearn.preprocessing import scale
from sklearn import model_selection
from sklearn.svm import SVR
import time
# ## Support Vector Regression(SVR)
# ### Theory
# Support Vector Regression gives us the flexibility to define how much error is acceptable in our model and will find an appropriate line (or hyperplane in higher dimensions) to fit the data. The objective function of SVR is to minimize the coefficients — more specifically, the l2-norm of the coefficient vector — not the squared error. The error term is instead handled in the constraints, where we set the absolute error less than or equal to a specified margin, called the maximum error, **ϵ (epsilon)**. We can tune epsilon to gain the desired accuracy of our model.
# Illustrative example:
# 
# Photo is cited by:https://towardsdatascience.com/an-introduction-to-support-vector-regression-svr-a3ebc1672c2
# As we can see, we have data points that are outside the epsilon in sensitive tube. We care about the error for them and they will be measured as distance between the point and the tube. As such, we need to account for the possibility of errors that are larger than ϵ. We can do this with slack variables.
# The concept of **slack variables** is simple: for any value that falls outside of ϵ, we can denote its deviation from the margin as ξ.
# That's the formula to minimise:
# 
# Photo is cited by: https://towardsdatascience.com/an-introduction-to-support-vector-regression-svr-a3ebc1672c2
# We now have an additional **C** hyperparameter that we can tune. As C increases, our tolerance for points outside of ϵ also increases. As C approaches 0, the tolerance approaches 0 and the equation collapses into the simplified one.
# ### Model
# For a real world example, we will work with **Hitters** dataset.
# It can be downloaded here: https://www.kaggle.com/floser/hitters
hts = pd.read_csv("../input/hitters/Hitters.csv")
hts.head()
# Now we will remove NA values.
hts.dropna(inplace=True)
# We will do **One Hot Encoding** to categorical columns.
one_hot_encoded = pd.get_dummies(hts[["League", "Division", "NewLeague"]])
one_hot_encoded.head()
new_hts = hts.drop(["League", "Division", "NewLeague", "Salary"], axis=1).astype(
"float64"
)
X = pd.concat(
[new_hts, one_hot_encoded[["League_N", "Division_W", "NewLeague_N"]]], axis=1
)
X.head()
y = hts.Salary # Target-dependent variable
# Now we will split our dataset as train and test set.
hts.shape
# Independent Variables
X.shape
# Dependent Variables
y.shape
X_train = X.iloc[:210]
X_test = X.iloc[210:]
y_train = y[:210]
y_test = y[210:]
print("X_train Shape: ", X_train.shape)
print("X_test Shape: ", X_test.shape)
print("y_train Shape: ", y_train.shape)
print("y_test Shape: ", y_test.shape)
X_train = pd.DataFrame(X_train["Hits"])
X_test = pd.DataFrame(X_test["Hits"])
X_train
X_test[:10]
svr_model = SVR("linear").fit(X_train, y_train)
svr_model
# We can write Support vector regression in linear regression form as below:
print("y = {0} + {1} x".format(svr_model.intercept_[0], svr_model.coef_[0][0]))
# Let's control this formula.
print(
"Manual Calculation: ",
svr_model.intercept_[0] + (svr_model.coef_[0][0] * X_train["Hits"][0:1].item()),
"\nOutput of Model",
svr_model.predict(X_train[0:1]).item(),
)
y_pred = svr_model.predict(X_train)
plt.scatter(X_train, y_train)
plt.plot(X_train, y_pred, c="r")
plt.title("Train Errors")
plt.show()
# ### Prediction
svr_model
y_pred = svr_model.predict(X_train)
# Train Error
np.sqrt(mean_squared_error(y_train, y_pred))
r2_score(y_train, y_pred)
y_pred = svr_model.predict(X_test)
# Test Error
np.sqrt(mean_squared_error(y_test, y_pred))
r2_score(y_test, y_pred)
# ### Model Tuning
svr_model
svr_parameters = {"C": np.arange(0.1, 3, 0.1)}
svr_cv_model = GridSearchCV(svr_model, svr_parameters, cv=15).fit(X_train, y_train)
svr_cv_model.best_params_
svr_tuned = SVR("linear", C=pd.Series(svr_cv_model.best_params_)[0]).fit(
X_train, y_train
)
svr_tuned
y_pred = svr_tuned.predict(X_train)
# Train Error
np.sqrt(mean_squared_error(y_train, y_pred))
r2_score(y_train, y_pred)
y_pred = svr_tuned.predict(X_test)
# Test Error
np.sqrt(mean_squared_error(y_test, y_pred))
r2_score(y_test, y_pred)
# We did all process with only **Hits** column. We can also try with all columns. But it takes more time nearly 30 min.
# ## Non-Linear Support Vector Regression(SVR)
# ### Theory
# When we cannot separate data with a straight line we use Non – Linear SVM. In this, we have **Kernel functions**. They transform non-linear spaces into linear spaces. It transforms data into another dimension so that the data can be classified.
# It transforms two variables x and y into three variables along with z. Therefore, the data have plotted from 2-D space to 3-D space. Now we can easily classify the data by drawi
# ### Model
# For a real world example, we will work with **Hitters** dataset.
# It can be downloaded here: https://www.kaggle.com/floser/hitters
hts = pd.read_csv("../input/hitters/Hitters.csv")
hts.head()
# Now we will remove NA values.
hts.dropna(inplace=True)
# We will do **One Hot Encoding** to categorical columns.
one_hot_encoded = pd.get_dummies(hts[["League", "Division", "NewLeague"]])
one_hot_encoded.head()
new_hts = hts.drop(["League", "Division", "NewLeague", "Salary"], axis=1).astype(
"float64"
)
X = pd.concat(
[new_hts, one_hot_encoded[["League_N", "Division_W", "NewLeague_N"]]], axis=1
)
X.head()
y = hts.Salary # Target-dependent variable
# Now we will split our dataset as train and test set.
hts.shape
# Independent Variables
X.shape
# Dependent Variables
y.shape
X_train = X.iloc[:210]
X_test = X.iloc[210:]
y_train = y[:210]
y_test = y[210:]
print("X_train Shape: ", X_train.shape)
print("X_test Shape: ", X_test.shape)
print("y_train Shape: ", y_train.shape)
print("y_test Shape: ", y_test.shape)
SVR_Radial_Basis = SVR("rbf").fit(X_train, y_train)
# ### Prediction
SVR_Radial_Basis
y_pred = SVR_Radial_Basis.predict(X_train)
# Train Error
np.sqrt(mean_squared_error(y_train, y_pred))
r2_score(y_train, y_pred)
y_pred = SVR_Radial_Basis.predict(X_test)
# Test Error
np.sqrt(mean_squared_error(y_test, y_pred))
r2_score(y_test, y_pred)
# ### Model Tuning
SVR_Radial_Basis
svr_parameters = {"C": np.arange(0.2, 10, 0.1)}
svr_cv_model = GridSearchCV(SVR_Radial_Basis, svr_parameters, cv=15).fit(
X_train, y_train
)
svr_cv_model.best_params_
svr_tuned = SVR("rbf", C=pd.Series(svr_cv_model.best_params_)[0]).fit(X_train, y_train)
svr_tuned
y_pred = svr_tuned.predict(X_train)
# Train Error
np.sqrt(mean_squared_error(y_train, y_pred))
r2_score(y_train, y_pred)
y_pred = svr_tuned.predict(X_test)
# Test Error
np.sqrt(mean_squared_error(y_test, y_pred))
r2_score(y_test, y_pred)
|
[{"hitters/Hitters.csv": {"column_names": "[\"AtBat\", \"Hits\", \"HmRun\", \"Runs\", \"RBI\", \"Walks\", \"Years\", \"CAtBat\", \"CHits\", \"CHmRun\", \"CRuns\", \"CRBI\", \"CWalks\", \"League\", \"Division\", \"PutOuts\", \"Assists\", \"Errors\", \"Salary\", \"NewLeague\"]", "column_data_types": "{\"AtBat\": \"int64\", \"Hits\": \"int64\", \"HmRun\": \"int64\", \"Runs\": \"int64\", \"RBI\": \"int64\", \"Walks\": \"int64\", \"Years\": \"int64\", \"CAtBat\": \"int64\", \"CHits\": \"int64\", \"CHmRun\": \"int64\", \"CRuns\": \"int64\", \"CRBI\": \"int64\", \"CWalks\": \"int64\", \"League\": \"object\", \"Division\": \"object\", \"PutOuts\": \"int64\", \"Assists\": \"int64\", \"Errors\": \"int64\", \"Salary\": \"float64\", \"NewLeague\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 322 entries, 0 to 321\nData columns (total 20 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 AtBat 322 non-null int64 \n 1 Hits 322 non-null int64 \n 2 HmRun 322 non-null int64 \n 3 Runs 322 non-null int64 \n 4 RBI 322 non-null int64 \n 5 Walks 322 non-null int64 \n 6 Years 322 non-null int64 \n 7 CAtBat 322 non-null int64 \n 8 CHits 322 non-null int64 \n 9 CHmRun 322 non-null int64 \n 10 CRuns 322 non-null int64 \n 11 CRBI 322 non-null int64 \n 12 CWalks 322 non-null int64 \n 13 League 322 non-null object \n 14 Division 322 non-null object \n 15 PutOuts 322 non-null int64 \n 16 Assists 322 non-null int64 \n 17 Errors 322 non-null int64 \n 18 Salary 263 non-null float64\n 19 NewLeague 322 non-null object \ndtypes: float64(1), int64(16), object(3)\nmemory usage: 50.4+ KB\n", "summary": "{\"AtBat\": {\"count\": 322.0, \"mean\": 380.92857142857144, \"std\": 153.40498147064488, \"min\": 16.0, \"25%\": 255.25, \"50%\": 379.5, \"75%\": 512.0, \"max\": 687.0}, \"Hits\": {\"count\": 322.0, \"mean\": 101.0248447204969, \"std\": 46.454741356766796, \"min\": 1.0, \"25%\": 64.0, \"50%\": 96.0, \"75%\": 137.0, \"max\": 238.0}, \"HmRun\": {\"count\": 322.0, \"mean\": 10.770186335403727, \"std\": 8.709037413827737, \"min\": 0.0, \"25%\": 4.0, \"50%\": 8.0, \"75%\": 16.0, \"max\": 40.0}, \"Runs\": {\"count\": 322.0, \"mean\": 50.909937888198755, \"std\": 26.02409548457972, \"min\": 0.0, \"25%\": 30.25, \"50%\": 48.0, \"75%\": 69.0, \"max\": 130.0}, \"RBI\": {\"count\": 322.0, \"mean\": 48.02795031055901, \"std\": 26.166894761424544, \"min\": 0.0, \"25%\": 28.0, \"50%\": 44.0, \"75%\": 64.75, \"max\": 121.0}, \"Walks\": {\"count\": 322.0, \"mean\": 38.74223602484472, \"std\": 21.63932655032488, \"min\": 0.0, \"25%\": 22.0, \"50%\": 35.0, \"75%\": 53.0, \"max\": 105.0}, \"Years\": {\"count\": 322.0, \"mean\": 7.444099378881988, \"std\": 4.926087269904596, \"min\": 1.0, \"25%\": 4.0, \"50%\": 6.0, \"75%\": 11.0, \"max\": 24.0}, \"CAtBat\": {\"count\": 322.0, \"mean\": 2648.6832298136646, \"std\": 2324.205870266538, \"min\": 19.0, \"25%\": 816.75, \"50%\": 1928.0, \"75%\": 3924.25, \"max\": 14053.0}, \"CHits\": {\"count\": 322.0, \"mean\": 717.5714285714286, \"std\": 654.4726274762833, \"min\": 4.0, \"25%\": 209.0, \"50%\": 508.0, \"75%\": 1059.25, \"max\": 4256.0}, \"CHmRun\": {\"count\": 322.0, \"mean\": 69.49068322981367, \"std\": 86.26606080180498, \"min\": 0.0, \"25%\": 14.0, \"50%\": 37.5, \"75%\": 90.0, \"max\": 548.0}, \"CRuns\": {\"count\": 322.0, \"mean\": 358.7950310559006, \"std\": 334.10588576614686, \"min\": 1.0, \"25%\": 100.25, \"50%\": 247.0, \"75%\": 526.25, \"max\": 2165.0}, \"CRBI\": {\"count\": 322.0, \"mean\": 330.11801242236027, \"std\": 333.2196169682779, \"min\": 0.0, \"25%\": 88.75, \"50%\": 220.5, \"75%\": 426.25, \"max\": 1659.0}, \"CWalks\": {\"count\": 322.0, \"mean\": 260.2391304347826, \"std\": 267.05808454363216, \"min\": 0.0, \"25%\": 67.25, \"50%\": 170.5, \"75%\": 339.25, \"max\": 1566.0}, \"PutOuts\": {\"count\": 322.0, \"mean\": 288.9378881987578, \"std\": 280.70461385993525, \"min\": 0.0, \"25%\": 109.25, \"50%\": 212.0, \"75%\": 325.0, \"max\": 1378.0}, \"Assists\": {\"count\": 322.0, \"mean\": 106.91304347826087, \"std\": 136.85487646596755, \"min\": 0.0, \"25%\": 7.0, \"50%\": 39.5, \"75%\": 166.0, \"max\": 492.0}, \"Errors\": {\"count\": 322.0, \"mean\": 8.040372670807454, \"std\": 6.368359079737258, \"min\": 0.0, \"25%\": 3.0, \"50%\": 6.0, \"75%\": 11.0, \"max\": 32.0}, \"Salary\": {\"count\": 263.0, \"mean\": 535.9258821292775, \"std\": 451.11868070253865, \"min\": 67.5, \"25%\": 190.0, \"50%\": 425.0, \"75%\": 750.0, \"max\": 2460.0}}", "examples": "{\"AtBat\":{\"0\":293,\"1\":315,\"2\":479,\"3\":496},\"Hits\":{\"0\":66,\"1\":81,\"2\":130,\"3\":141},\"HmRun\":{\"0\":1,\"1\":7,\"2\":18,\"3\":20},\"Runs\":{\"0\":30,\"1\":24,\"2\":66,\"3\":65},\"RBI\":{\"0\":29,\"1\":38,\"2\":72,\"3\":78},\"Walks\":{\"0\":14,\"1\":39,\"2\":76,\"3\":37},\"Years\":{\"0\":1,\"1\":14,\"2\":3,\"3\":11},\"CAtBat\":{\"0\":293,\"1\":3449,\"2\":1624,\"3\":5628},\"CHits\":{\"0\":66,\"1\":835,\"2\":457,\"3\":1575},\"CHmRun\":{\"0\":1,\"1\":69,\"2\":63,\"3\":225},\"CRuns\":{\"0\":30,\"1\":321,\"2\":224,\"3\":828},\"CRBI\":{\"0\":29,\"1\":414,\"2\":266,\"3\":838},\"CWalks\":{\"0\":14,\"1\":375,\"2\":263,\"3\":354},\"League\":{\"0\":\"A\",\"1\":\"N\",\"2\":\"A\",\"3\":\"N\"},\"Division\":{\"0\":\"E\",\"1\":\"W\",\"2\":\"W\",\"3\":\"E\"},\"PutOuts\":{\"0\":446,\"1\":632,\"2\":880,\"3\":200},\"Assists\":{\"0\":33,\"1\":43,\"2\":82,\"3\":11},\"Errors\":{\"0\":20,\"1\":10,\"2\":14,\"3\":3},\"Salary\":{\"0\":null,\"1\":475.0,\"2\":480.0,\"3\":500.0},\"NewLeague\":{\"0\":\"A\",\"1\":\"N\",\"2\":\"A\",\"3\":\"N\"}}"}}]
| true | 1 |
<start_data_description><data_path>hitters/Hitters.csv:
<column_names>
['AtBat', 'Hits', 'HmRun', 'Runs', 'RBI', 'Walks', 'Years', 'CAtBat', 'CHits', 'CHmRun', 'CRuns', 'CRBI', 'CWalks', 'League', 'Division', 'PutOuts', 'Assists', 'Errors', 'Salary', 'NewLeague']
<column_types>
{'AtBat': 'int64', 'Hits': 'int64', 'HmRun': 'int64', 'Runs': 'int64', 'RBI': 'int64', 'Walks': 'int64', 'Years': 'int64', 'CAtBat': 'int64', 'CHits': 'int64', 'CHmRun': 'int64', 'CRuns': 'int64', 'CRBI': 'int64', 'CWalks': 'int64', 'League': 'object', 'Division': 'object', 'PutOuts': 'int64', 'Assists': 'int64', 'Errors': 'int64', 'Salary': 'float64', 'NewLeague': 'object'}
<dataframe_Summary>
{'AtBat': {'count': 322.0, 'mean': 380.92857142857144, 'std': 153.40498147064488, 'min': 16.0, '25%': 255.25, '50%': 379.5, '75%': 512.0, 'max': 687.0}, 'Hits': {'count': 322.0, 'mean': 101.0248447204969, 'std': 46.454741356766796, 'min': 1.0, '25%': 64.0, '50%': 96.0, '75%': 137.0, 'max': 238.0}, 'HmRun': {'count': 322.0, 'mean': 10.770186335403727, 'std': 8.709037413827737, 'min': 0.0, '25%': 4.0, '50%': 8.0, '75%': 16.0, 'max': 40.0}, 'Runs': {'count': 322.0, 'mean': 50.909937888198755, 'std': 26.02409548457972, 'min': 0.0, '25%': 30.25, '50%': 48.0, '75%': 69.0, 'max': 130.0}, 'RBI': {'count': 322.0, 'mean': 48.02795031055901, 'std': 26.166894761424544, 'min': 0.0, '25%': 28.0, '50%': 44.0, '75%': 64.75, 'max': 121.0}, 'Walks': {'count': 322.0, 'mean': 38.74223602484472, 'std': 21.63932655032488, 'min': 0.0, '25%': 22.0, '50%': 35.0, '75%': 53.0, 'max': 105.0}, 'Years': {'count': 322.0, 'mean': 7.444099378881988, 'std': 4.926087269904596, 'min': 1.0, '25%': 4.0, '50%': 6.0, '75%': 11.0, 'max': 24.0}, 'CAtBat': {'count': 322.0, 'mean': 2648.6832298136646, 'std': 2324.205870266538, 'min': 19.0, '25%': 816.75, '50%': 1928.0, '75%': 3924.25, 'max': 14053.0}, 'CHits': {'count': 322.0, 'mean': 717.5714285714286, 'std': 654.4726274762833, 'min': 4.0, '25%': 209.0, '50%': 508.0, '75%': 1059.25, 'max': 4256.0}, 'CHmRun': {'count': 322.0, 'mean': 69.49068322981367, 'std': 86.26606080180498, 'min': 0.0, '25%': 14.0, '50%': 37.5, '75%': 90.0, 'max': 548.0}, 'CRuns': {'count': 322.0, 'mean': 358.7950310559006, 'std': 334.10588576614686, 'min': 1.0, '25%': 100.25, '50%': 247.0, '75%': 526.25, 'max': 2165.0}, 'CRBI': {'count': 322.0, 'mean': 330.11801242236027, 'std': 333.2196169682779, 'min': 0.0, '25%': 88.75, '50%': 220.5, '75%': 426.25, 'max': 1659.0}, 'CWalks': {'count': 322.0, 'mean': 260.2391304347826, 'std': 267.05808454363216, 'min': 0.0, '25%': 67.25, '50%': 170.5, '75%': 339.25, 'max': 1566.0}, 'PutOuts': {'count': 322.0, 'mean': 288.9378881987578, 'std': 280.70461385993525, 'min': 0.0, '25%': 109.25, '50%': 212.0, '75%': 325.0, 'max': 1378.0}, 'Assists': {'count': 322.0, 'mean': 106.91304347826087, 'std': 136.85487646596755, 'min': 0.0, '25%': 7.0, '50%': 39.5, '75%': 166.0, 'max': 492.0}, 'Errors': {'count': 322.0, 'mean': 8.040372670807454, 'std': 6.368359079737258, 'min': 0.0, '25%': 3.0, '50%': 6.0, '75%': 11.0, 'max': 32.0}, 'Salary': {'count': 263.0, 'mean': 535.9258821292775, 'std': 451.11868070253865, 'min': 67.5, '25%': 190.0, '50%': 425.0, '75%': 750.0, 'max': 2460.0}}
<dataframe_info>
RangeIndex: 322 entries, 0 to 321
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 AtBat 322 non-null int64
1 Hits 322 non-null int64
2 HmRun 322 non-null int64
3 Runs 322 non-null int64
4 RBI 322 non-null int64
5 Walks 322 non-null int64
6 Years 322 non-null int64
7 CAtBat 322 non-null int64
8 CHits 322 non-null int64
9 CHmRun 322 non-null int64
10 CRuns 322 non-null int64
11 CRBI 322 non-null int64
12 CWalks 322 non-null int64
13 League 322 non-null object
14 Division 322 non-null object
15 PutOuts 322 non-null int64
16 Assists 322 non-null int64
17 Errors 322 non-null int64
18 Salary 263 non-null float64
19 NewLeague 322 non-null object
dtypes: float64(1), int64(16), object(3)
memory usage: 50.4+ KB
<some_examples>
{'AtBat': {'0': 293, '1': 315, '2': 479, '3': 496}, 'Hits': {'0': 66, '1': 81, '2': 130, '3': 141}, 'HmRun': {'0': 1, '1': 7, '2': 18, '3': 20}, 'Runs': {'0': 30, '1': 24, '2': 66, '3': 65}, 'RBI': {'0': 29, '1': 38, '2': 72, '3': 78}, 'Walks': {'0': 14, '1': 39, '2': 76, '3': 37}, 'Years': {'0': 1, '1': 14, '2': 3, '3': 11}, 'CAtBat': {'0': 293, '1': 3449, '2': 1624, '3': 5628}, 'CHits': {'0': 66, '1': 835, '2': 457, '3': 1575}, 'CHmRun': {'0': 1, '1': 69, '2': 63, '3': 225}, 'CRuns': {'0': 30, '1': 321, '2': 224, '3': 828}, 'CRBI': {'0': 29, '1': 414, '2': 266, '3': 838}, 'CWalks': {'0': 14, '1': 375, '2': 263, '3': 354}, 'League': {'0': 'A', '1': 'N', '2': 'A', '3': 'N'}, 'Division': {'0': 'E', '1': 'W', '2': 'W', '3': 'E'}, 'PutOuts': {'0': 446, '1': 632, '2': 880, '3': 200}, 'Assists': {'0': 33, '1': 43, '2': 82, '3': 11}, 'Errors': {'0': 20, '1': 10, '2': 14, '3': 3}, 'Salary': {'0': None, '1': 475.0, '2': 480.0, '3': 500.0}, 'NewLeague': {'0': 'A', '1': 'N', '2': 'A', '3': 'N'}}
<end_description>
| 3,183 | 11 | 4,900 | 3,183 |
69605290
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
from sklearn import datasets
X, y = datasets.make_blobs(
n_samples=150, n_features=2, centers=2, cluster_std=1.05, random_state=2
)
# Plotting
fig = plt.figure(figsize=(10, 8))
plt.plot(X[:, 0][y == 0], X[:, 1][y == 0], "r^")
plt.plot(X[:, 0][y == 1], X[:, 1][y == 1], "bs")
plt.xlabel("feature 1")
plt.ylabel("feature 2")
plt.title("Random Classification Data with 2 classes")
# There are two classes, red and green, and we want to separate them by drawing a straight line between them. Or, more formally, we want to learn a set of parameters theta to find an optimal hyperplane(straight line for our data) that separates the two classes.
def step_func(z):
return 1.0 if (z > 0) else 0.0
def perceptron(X, y, lr, epochs):
# X --> Inputs.
# y --> labels/target.
# lr --> learning rate.
# epochs --> Number of iterations.
# m-> number of training examples
# n-> number of features
m, n = X.shape
# Initializing parapeters(theta) to zeros.
# +1 in n+1 for the bias term.
theta = np.zeros((n + 1, 1))
# Empty list to store how many examples were
# misclassified at every iteration.
n_miss_list = []
# Training.
for epoch in range(epochs):
# variable to store #misclassified.
n_miss = 0
# looping for every example.
for idx, x_i in enumerate(X):
# Insering 1 for bias, X0 = 1.
x_i = np.insert(x_i, 0, 1).reshape(-1, 1)
# Calculating prediction/hypothesis.
y_hat = step_func(np.dot(x_i.T, theta))
# Updating if the example is misclassified.
if (np.squeeze(y_hat) - y[idx]) != 0:
theta += lr * ((y[idx] - y_hat) * x_i)
# Incrementing by 1.
n_miss += 1
# Appending number of misclassified examples
# at every iteration.
n_miss_list.append(n_miss)
return theta, n_miss_list
def plot_decision_boundary(X, theta):
# X --> Inputs
# theta --> parameters
# The Line is y=mx+c
# So, Equate mx+c = theta0.X0 + theta1.X1 + theta2.X2
# Solving we find m and c
x1 = [min(X[:, 0]), max(X[:, 0])]
m = -theta[1] / theta[2]
c = -theta[0] / theta[2]
x2 = m * x1 + c
# Plotting
fig = plt.figure(figsize=(10, 8))
plt.plot(X[:, 0][y == 0], X[:, 1][y == 0], "r^")
plt.plot(X[:, 0][y == 1], X[:, 1][y == 1], "bs")
plt.xlabel("feature 1")
plt.ylabel("feature 2")
plt.title("Perceptron Algorithm")
plt.plot(x1, x2, "y-")
theta, miss_l = perceptron(X, y, 0.5, 100)
plot_decision_boundary(X, theta)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/605/69605290.ipynb
| null | null |
[{"Id": 69605290, "ScriptId": 19009067, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5428597, "CreationDate": "08/02/2021 05:35:37", "VersionNumber": 1.0, "Title": "Perceptron", "EvaluationDate": "08/02/2021", "IsChange": true, "TotalLines": 108.0, "LinesInsertedFromPrevious": 108.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 9}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
from sklearn import datasets
X, y = datasets.make_blobs(
n_samples=150, n_features=2, centers=2, cluster_std=1.05, random_state=2
)
# Plotting
fig = plt.figure(figsize=(10, 8))
plt.plot(X[:, 0][y == 0], X[:, 1][y == 0], "r^")
plt.plot(X[:, 0][y == 1], X[:, 1][y == 1], "bs")
plt.xlabel("feature 1")
plt.ylabel("feature 2")
plt.title("Random Classification Data with 2 classes")
# There are two classes, red and green, and we want to separate them by drawing a straight line between them. Or, more formally, we want to learn a set of parameters theta to find an optimal hyperplane(straight line for our data) that separates the two classes.
def step_func(z):
return 1.0 if (z > 0) else 0.0
def perceptron(X, y, lr, epochs):
# X --> Inputs.
# y --> labels/target.
# lr --> learning rate.
# epochs --> Number of iterations.
# m-> number of training examples
# n-> number of features
m, n = X.shape
# Initializing parapeters(theta) to zeros.
# +1 in n+1 for the bias term.
theta = np.zeros((n + 1, 1))
# Empty list to store how many examples were
# misclassified at every iteration.
n_miss_list = []
# Training.
for epoch in range(epochs):
# variable to store #misclassified.
n_miss = 0
# looping for every example.
for idx, x_i in enumerate(X):
# Insering 1 for bias, X0 = 1.
x_i = np.insert(x_i, 0, 1).reshape(-1, 1)
# Calculating prediction/hypothesis.
y_hat = step_func(np.dot(x_i.T, theta))
# Updating if the example is misclassified.
if (np.squeeze(y_hat) - y[idx]) != 0:
theta += lr * ((y[idx] - y_hat) * x_i)
# Incrementing by 1.
n_miss += 1
# Appending number of misclassified examples
# at every iteration.
n_miss_list.append(n_miss)
return theta, n_miss_list
def plot_decision_boundary(X, theta):
# X --> Inputs
# theta --> parameters
# The Line is y=mx+c
# So, Equate mx+c = theta0.X0 + theta1.X1 + theta2.X2
# Solving we find m and c
x1 = [min(X[:, 0]), max(X[:, 0])]
m = -theta[1] / theta[2]
c = -theta[0] / theta[2]
x2 = m * x1 + c
# Plotting
fig = plt.figure(figsize=(10, 8))
plt.plot(X[:, 0][y == 0], X[:, 1][y == 0], "r^")
plt.plot(X[:, 0][y == 1], X[:, 1][y == 1], "bs")
plt.xlabel("feature 1")
plt.ylabel("feature 2")
plt.title("Perceptron Algorithm")
plt.plot(x1, x2, "y-")
theta, miss_l = perceptron(X, y, 0.5, 100)
plot_decision_boundary(X, theta)
| false | 0 | 1,075 | 9 | 1,075 | 1,075 |
||
69791627
|
<jupyter_start><jupyter_text>Food Demand Forecasting
### Context
It is a meal delivery company which operates in multiple cities. They have various fulfillment centers in these cities for dispatching meal orders to their customers. The client wants you to help these centers with demand forecasting for upcoming weeks so that these centers will plan the stock of raw materials accordingly.
### Content
The replenishment of majority of raw materials is done on weekly basis and since the raw material is perishable, the procurement planning is of utmost importance. Secondly, staffing of the centers is also one area wherein accurate demand forecasts are really helpful. Given the following information, the task is to predict the demand for the next 10 weeks (Weeks: 146-155) for the center-meal combinations in the test set
Kaggle dataset identifier: food-demand-forecasting
<jupyter_code>import pandas as pd
df = pd.read_csv('food-demand-forecasting/test.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 32573 entries, 0 to 32572
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 32573 non-null int64
1 week 32573 non-null int64
2 center_id 32573 non-null int64
3 meal_id 32573 non-null int64
4 checkout_price 32573 non-null float64
5 base_price 32573 non-null float64
6 emailer_for_promotion 32573 non-null int64
7 homepage_featured 32573 non-null int64
dtypes: float64(2), int64(6)
memory usage: 2.0 MB
<jupyter_text>Examples:
{
"id": 1028232.0,
"week": 146.0,
"center_id": 55.0,
"meal_id": 1885.0,
"checkout_price": 158.11,
"base_price": 159.11,
"emailer_for_promotion": 0.0,
"homepage_featured": 0.0
}
{
"id": 1127204.0,
"week": 146.0,
"center_id": 55.0,
"meal_id": 1993.0,
"checkout_price": 160.11,
"base_price": 159.11,
"emailer_for_promotion": 0.0,
"homepage_featured": 0.0
}
{
"id": 1212707.0,
"week": 146.0,
"center_id": 55.0,
"meal_id": 2539.0,
"checkout_price": 157.14,
"base_price": 159.14,
"emailer_for_promotion": 0.0,
"homepage_featured": 0.0
}
{
"id": 1082698.0,
"week": 146.0,
"center_id": 55.0,
"meal_id": 2631.0,
"checkout_price": 162.02,
"base_price": 162.02,
"emailer_for_promotion": 0.0,
"homepage_featured": 0.0
}
<jupyter_code>import pandas as pd
df = pd.read_csv('food-demand-forecasting/fulfilment_center_info.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 77 entries, 0 to 76
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 center_id 77 non-null int64
1 city_code 77 non-null int64
2 region_code 77 non-null int64
3 center_type 77 non-null object
4 op_area 77 non-null float64
dtypes: float64(1), int64(3), object(1)
memory usage: 3.1+ KB
<jupyter_text>Examples:
{
"center_id": 11,
"city_code": 679,
"region_code": 56,
"center_type": "TYPE_A",
"op_area": 3.7
}
{
"center_id": 13,
"city_code": 590,
"region_code": 56,
"center_type": "TYPE_B",
"op_area": 6.7
}
{
"center_id": 124,
"city_code": 590,
"region_code": 56,
"center_type": "TYPE_C",
"op_area": 4.0
}
{
"center_id": 66,
"city_code": 648,
"region_code": 34,
"center_type": "TYPE_A",
"op_area": 4.1
}
<jupyter_code>import pandas as pd
df = pd.read_csv('food-demand-forecasting/meal_info.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 51 entries, 0 to 50
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 meal_id 51 non-null int64
1 category 51 non-null object
2 cuisine 51 non-null object
dtypes: int64(1), object(2)
memory usage: 1.3+ KB
<jupyter_text>Examples:
{
"meal_id": 1885,
"category": "Beverages",
"cuisine": "Thai"
}
{
"meal_id": 1993,
"category": "Beverages",
"cuisine": "Thai"
}
{
"meal_id": 2539,
"category": "Beverages",
"cuisine": "Thai"
}
{
"meal_id": 1248,
"category": "Beverages",
"cuisine": "Indian"
}
<jupyter_code>import pandas as pd
df = pd.read_csv('food-demand-forecasting/train.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 456548 entries, 0 to 456547
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 456548 non-null int64
1 week 456548 non-null int64
2 center_id 456548 non-null int64
3 meal_id 456548 non-null int64
4 checkout_price 456548 non-null float64
5 base_price 456548 non-null float64
6 emailer_for_promotion 456548 non-null int64
7 homepage_featured 456548 non-null int64
8 num_orders 456548 non-null int64
dtypes: float64(2), int64(7)
memory usage: 31.3 MB
<jupyter_text>Examples:
{
"id": 1379560.0,
"week": 1.0,
"center_id": 55.0,
"meal_id": 1885.0,
"checkout_price": 136.83,
"base_price": 152.29,
"emailer_for_promotion": 0.0,
"homepage_featured": 0.0,
"num_orders": 177.0
}
{
"id": 1466964.0,
"week": 1.0,
"center_id": 55.0,
"meal_id": 1993.0,
"checkout_price": 136.83,
"base_price": 135.83,
"emailer_for_promotion": 0.0,
"homepage_featured": 0.0,
"num_orders": 270.0
}
{
"id": 1346989.0,
"week": 1.0,
"center_id": 55.0,
"meal_id": 2539.0,
"checkout_price": 134.86,
"base_price": 135.86,
"emailer_for_promotion": 0.0,
"homepage_featured": 0.0,
"num_orders": 189.0
}
{
"id": 1338232.0,
"week": 1.0,
"center_id": 55.0,
"meal_id": 2139.0,
"checkout_price": 339.5,
"base_price": 437.53,
"emailer_for_promotion": 0.0,
"homepage_featured": 0.0,
"num_orders": 54.0
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
#
# **Demand forecasting is a key component to every growing online business. Without proper demand forecasting processes in place, it can be nearly impossible to have the right amount of stock on hand at any given time. A food delivery service has to deal with a lot of perishable raw materials which makes it all the more important for such a company to accurately forecast daily and weekly demand.**
# **Too much inventory in the warehouse means more risk of wastage, and not enough could lead to out-of-stocks — and push customers to seek solutions from your competitors. In this challenge, get a taste of demand forecasting challenge using a real dataset.**
# # **Problem Statement**
# The data set is related to a meal delivery company which operates in multiple cities. They have various fulfilment centers in these cities for dispatching meal orders to their customers.
# The dataset consists of historical data of demand for a product-center combination for weeks 1 to 145.
# With the given data and information, the task is to predict the demand for the next 10 weeks (Weeks: 146-155) for the center-meal combinations, so that these fulfilment centers stock the necessary raw materials accordingly.
# # ****Business Benefits****
# The replenishment of raw materials is done only on weekly basis and since the raw material is perishable, the procurement planning is of utmost importance.
# Therefore predicting the Demand helps in reducing the wastage of raw materials which would result in the reduced cost of operation. Increased customer satisfaction by timely fulfilling their expectations and requirements.
# # **Data Dictionary**
# The dataset consists of three individual datasheets, the first dataset contains the historical demand data for all centers, the second dataset contains the information of each fulfillment center and the third dataset contains the meal information.
# Weekly Demand data (train.csv):
# Contains the historical demand data for all centers. The Train dataset consists of 9 variables and records of 423727 unique orders. test.csv contains all the following features except the target variable. The Test dataset consists of 8 variables and records of 32573 unique orders.
# fulfilment_center_info.csv:
# Contains information for each fulfilment center. The dataset consists of 5 variables and records of 77 unique fulfillment centers.
# meal_info.csv:
# Contains information for each meal being served
# # **Libraries Used**
# pandas, numpy, scikit learn, matplotlib, seaborn, xgboost, lightgbm, catboost
# # **Data Pre-Processing**
# * There are no Missing/Null Values in any of the three datasets.
# * Before proceeding with the prediction process, all the three datasheets need to be merged into a single dataset. Before performing the merging operation, primary feature for combining the datasets needs to be validated.
# * The number of Center IDs in train dataset is matching with the number of Center IDs in the Centers Dataset i.e 77 unique records. Hence, there won't be any missing values while merging the datasets together.
# * The number of Meal IDs in train dataset is matching with the number of Meal IDs in the Meals Dataset i.e 51 unique records. Hence, there won't be any missing values while merging the datasets together.
# * As checked earlier, there were no Null/Missing values even after merging the datasets.
# # **Feature Engineering**
# Feature engineering is the process of using domain knowledge of the data to create features that improves the performance of the machine learning models.
# With the given data, We have derived the below features to improve our model performance.
# * Discount Amount : This defines the difference between the “base_Price” and “checkout_price”.
# * Discount Percent : This defines the % discount offer to customer.
# * Discount Y/N : This defines whether Discount is provided or not - 1 if there is Discount and 0 if there is no Discount.
# * Compare Week Price : This defines the increase / decrease in price of a Meal for a particular center compared to the previous week.
# * Compare Week Price Y/N : Price increased or decreased - 1 if the Price increased and 0 if the price decreased compared to the previous week.
# * Quarter : Based on the given number of weeks, derived a new feature named as Quarter which defines the Quarter of the year.
# * Year : Based on the given number of weeks, derived a new feature named as Year which defines the Year.
# # **Data Transformation**
# * Logarithm transformation (or log transform) is one of the most commonly used mathematical transformations in feature engineering. It helps to handle skewed data and after transformation, the distribution becomes more approximate to normal.
# * In our data, the target variable ‘num_orders’ is not normally distributed. Using this without applying any transformation techniques will downgrade the performance of our model.
# * Therefore, we have applied Logarithm transformation on our Target feature ‘num_orders’ post which the data seems to be more approximate to normal distribution.
# * After Log transformation, We have observed 0% of Outlier data being present within the Target Variable – num_orders using 3 IQR Method.
# # **Evaluation Metric**
# The evaluation metric for this competition is 100*RMSLE where RMSLE is Root of Mean Squared Logarithmic Error across all entries in the test set.
# # **Approach**
# * Simple Linear Regression model without any feature engineering and data transformation which gave a RMSE : 194.402
# * Without feature engineering and data transformation, the model did not perform well and could'nt give a good score.
# * Post applying feature engineering and data transformation (log and log1p transformation), Linear Regression model gave a RMSLE score of 0.634.
import numpy as np
import pandas as pd
pd.set_option("display.max_columns", None)
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import seaborn as sns
import plotly.io as pio
import plotly.graph_objects as go
from plotly.offline import init_notebook_mode, iplot
from sklearn.preprocessing import StandardScaler
import warnings
warnings.filterwarnings("ignore")
data = pd.read_csv("/kaggle/input/food-demand-forecasting/train.csv")
center = pd.read_csv("/kaggle/input/food-demand-forecasting/fulfilment_center_info.csv")
meal = pd.read_csv("/kaggle/input/food-demand-forecasting/meal_info.csv")
test = pd.read_csv("/kaggle/input/food-demand-forecasting/test.csv")
# /kaggle/input/food-demand-forecasting/sample_submission.csv
# **Data Preprocessing**
print("The Shape of Demand dataset :", data.shape)
print("The Shape of Fulmilment Center Information dataset :", center.shape)
print("The Shape of Meal information dataset :", meal.shape)
print("The Shape of Test dataset :", test.shape)
data.head()
test[
"num_orders"
] = 123456 ### Assigning random number for Target Variable of Test Data.
test.head()
center.head()
meal.head()
data = pd.concat([data, test], axis=0)
data = data.merge(center, on="center_id", how="left")
data = data.merge(meal, on="meal_id", how="left")
data.isnull().sum()
# **Deriving New Features**
# Discount Amount
data["discount amount"] = data["base_price"] - data["checkout_price"]
# Discount Percent
data["discount percent"] = (
(data["base_price"] - data["checkout_price"]) / data["base_price"]
) * 100
# Discount Y/N
data["discount y/n"] = [
1 if x > 0 else 0 for x in (data["base_price"] - data["checkout_price"])
]
data = data.sort_values(["center_id", "meal_id", "week"]).reset_index()
# Compare Week Price
data["compare_week_price"] = data["checkout_price"] - data["checkout_price"].shift(1)
data["compare_week_price"][data["week"] == 1] = 0
data = data.sort_values(by="index").reset_index().drop(["level_0", "index"], axis=1)
# Compare Week Price Y/N
data["compare_week_price y/n"] = [1 if x > 0 else 0 for x in data["compare_week_price"]]
data.head()
data.isnull().sum()
# **Train Test Split**
train = data[data["week"].isin(range(1, 146))]
test = data[data["week"].isin(range(146, 156))]
print("The Shape of Train dataset :", train.shape)
print("The Shape of Test dataset :", test.shape)
plt.figure(figsize=(16, 14))
sns.heatmap(train.corr(), annot=True, square=True, cmap="Reds")
fig = plt.figure(figsize=(4, 7))
plt.title("Total No. of Orders for Each Center type", fontdict={"fontsize": 13})
sns.barplot(
y="num_orders",
x="center_type",
data=train.groupby("center_type").sum()["num_orders"].reset_index(),
palette="autumn",
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Center Type", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Type A has the highest number of Orders placed and Type C has lowest
train["center_id"].nunique()
# The are are 77 Fullfilment Centers in total.
#
fig = plt.figure(figsize=(10, 8))
plt.title("Top 20 Centers with Highest No. of Orders", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="center_id",
data=train.groupby(["center_id", "center_type"])
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()
.head(20),
palette="YlOrRd_r",
order=list(
train.groupby(["center_id", "center_type"])
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()
.head(20)["center_id"]
),
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Center ID", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Initially, when we checked, which Center Type has the highest number of Orders, We found that Center Type_A has the highest number of orders, but now when we check individually, we could see that Center 13 of Type_B has the highest number of Orders. Let’s analyze the reason behind that.
#
fig = plt.figure(figsize=(4, 7))
plt.title("Total No. of Centers under Each Center type", fontdict={"fontsize": 13})
sns.barplot(
y=train.groupby(["center_id", "center_type"])
.num_orders.sum()
.reset_index()["center_type"]
.value_counts(),
x=train.groupby(["center_id", "center_type"])
.num_orders.sum()
.reset_index()["center_type"]
.value_counts()
.index,
palette="autumn",
)
plt.ylabel("No. of Centers", fontdict={"fontsize": 12})
plt.xlabel("Center Type", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Type_A has the most number of orders because, Type_A has the most number of Centers - 43 Centers.
#
sns.set_style("white")
plt.figure(figsize=(8, 8))
sns.scatterplot(
y=train["base_price"] - train["checkout_price"],
x=train["num_orders"],
color="coral",
)
plt.ylabel("Discount (Base price - Checkout Price)")
sns.despine(bottom=True, left=True)
# We created a new feature: Discount which is the difference of base price and checkout price and tried to find out if there is any relationship between the discount and the number of orders. But surprisingly there are no good correlation between the discount and the number of orders.
#
fig = plt.figure(figsize=(16, 7))
sns.set_style("whitegrid")
plt.title("Pattern of Orders", fontdict={"fontsize": 14})
sns.pointplot(
x=train.groupby("week").sum().reset_index()["week"],
y=train.groupby("week").sum().reset_index()["num_orders"],
color="coral",
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Week", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# When we analysed the trend of order placed over the weeks, we could see that the highest number of orders were received in week 48 and the lowest in week 62.
#
plt.figure(figsize=(6, 6))
colors = ["coral", "#FFDAB9", "yellowgreen", "#6495ED"]
plt.pie(
train.groupby(["cuisine"]).num_orders.sum(),
labels=train.groupby(["cuisine"]).num_orders.sum().index,
shadow=False,
colors=colors,
explode=(0.05, 0.05, 0.03, 0.05),
startangle=90,
autopct="%1.1f%%",
pctdistance=0.6,
textprops={"fontsize": 12},
)
plt.title("Total Number of Orders for Each Category")
plt.tight_layout()
plt.show()
# Italian Cuisine has the highest number of orders with Continental cuisine being the least.
#
fig = plt.figure(figsize=(11, 8))
sns.set_style("white")
plt.xticks(rotation=90, fontsize=12)
plt.title("Total Number of Orders for Each Category", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="category",
data=train.groupby("category")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index(),
palette="YlOrRd_r",
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Category", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# We could see that Beverages are the food category which has the higest number of orders and Biriyani is the food category with least number of orders.
#
fig = plt.figure(figsize=(18, 8))
sns.set_style("white")
plt.xticks(rotation=90, fontsize=12)
plt.title("Total Number of Orders for Each Cuisine-Category", fontdict={"fontsize": 14})
sns.barplot(
x="category",
y="num_orders",
data=train.groupby(["cuisine", "category"])
.sum()
.sort_values(by="num_orders", ascending=False)
.reset_index(),
hue="cuisine",
palette="YlOrRd_r",
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Cuisine-Category", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Similary when we checked which specific cuisne-food category has the highest number of orders, we could see that Indian-Rice Bowl has the highest number of orders and Indian-Biriyani has the least.
#
list(
data.groupby("region_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()
.values[:, 0]
)
fig = plt.figure(figsize=(8, 8))
sns.set_style("white")
plt.xticks(fontsize=13)
plt.title("Total Number of Orders for Each Region", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="region_code",
data=data.groupby("region_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index(),
palette="YlOrRd_r",
order=list(
data.groupby("region_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()
.values[:, 0]
),
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Region", fontdict={"fontsize": 12})
plt.xticks()
sns.despine(bottom=True, left=True)
# Also when we checked the number of orders with respect to Region, we could see that Region - 56 has the highest number of orders - 60.5M orders which is almost 35M orders higher than the Region with second highest number of orders - Region 34 - 24M orders.
#
fig = plt.figure(figsize=(18, 8))
sns.set_style("white")
plt.xticks(rotation=90, fontsize=13)
plt.title("Total Number of Orders for each Meal ID", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="meal_id",
data=data.groupby("meal_id")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index(),
palette="YlOrRd_r",
order=list(
data.groupby("meal_id")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()["meal_id"]
.values
),
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Meal", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Meal ID 2290 has the higest number of Orders. There is not much significant differences between number of orders for different Meal IDs.
#
fig = plt.figure(figsize=(18, 8))
sns.set_style("white")
plt.xticks(rotation=90, fontsize=13)
plt.title("Total Number of Orders for each City", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="city_code",
data=train.groupby("city_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index(),
palette="YlOrRd_r",
order=list(
train.groupby("city_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()["city_code"]
.values
),
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("City", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Also when we checked the number of orders with respect to City, we could see that City - 590 has the highest number of orders - 18.5M orders which is almost 10M orders higher than the City with second highest number of orders - City 526 - 8.6M orders.
# # **Encoding City**
# As per our observation from our barchart of the City against the number of orders. There the high significant difference between the Top 3 cities which has the highest number of orders. Therefore, in our first approach we will encode the City with Highest No. of Orders as CH1, City with 2nd Highest No. of Orders as CH2 and City with 3rd Highest No. of Orders as CH3 and rest all of the cities which does not have much significant differences between the number of orders as CH4.
#
city4 = {590: "CH1", 526: "CH2", 638: "CH3"}
data["city_enc_4"] = data["city_code"].map(city4)
data["city_enc_4"] = data["city_enc_4"].fillna("CH4")
data["city_enc_4"].value_counts()
data.head()
data.isnull().sum()
# **Copying to New DataFrame**
datax = data.copy()
datax.head()
# **Encoding All Categorical Features**
datax["center_id"] = datax["center_id"].astype("object")
datax["meal_id"] = datax["meal_id"].astype("object")
datax["region_code"] = datax["region_code"].astype("object")
obj = datax[
[
"center_id",
"meal_id",
"region_code",
"center_type",
"category",
"cuisine",
"city_enc_4",
]
]
num = datax.drop(
[
"center_id",
"meal_id",
"region_code",
"center_type",
"category",
"cuisine",
"city_enc_4",
],
axis=1,
)
encode1 = pd.get_dummies(obj, drop_first=True)
datax = pd.concat([num, encode1], axis=1)
datax.head()
# # **Base Model**
# Building base model by splitting the last 10 week of the train dataset as test.
#
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
train = datax[datax["week"].isin(range(1, 136))]
test = datax[datax["week"].isin(range(136, 146))]
X_train = train.drop(["id", "num_orders", "week"], axis=1)
y_train = train["num_orders"]
X_test = test.drop(["id", "num_orders", "week"], axis=1)
y_test = test["num_orders"]
reg = LinearRegression()
reg.fit(X_train, y_train)
print("Train Score :", reg.score(X_train, y_train))
print("Test Score :", reg.score(X_test, y_test))
y_pred = reg.predict(X_test)
print("R squared :", (r2_score(y_test, y_pred)))
print("RMSE :", np.sqrt(mean_squared_error(y_test, y_pred)))
# Linear Model 2 : Applying Standard Scaling & Log Transformation
#
sc = StandardScaler()
cat = datax.drop(
[
"checkout_price",
"base_price",
"discount amount",
"discount percent",
"compare_week_price",
],
axis=1,
)
num = datax[
[
"checkout_price",
"base_price",
"discount amount",
"discount percent",
"compare_week_price",
]
]
scal = pd.DataFrame(sc.fit_transform(num), columns=num.columns)
datas = pd.concat([scal, cat], axis=1)
train = datas[datas["week"].isin(range(1, 136))]
test = datas[datas["week"].isin(range(136, 146))]
X_train = train.drop(["id", "num_orders", "week"], axis=1)
y_train = np.log(
train["num_orders"]
) # Applying Log Transformation on the Target Feature
X_test = test.drop(["id", "num_orders", "week"], axis=1)
y_test = np.log(test["num_orders"]) # Applying Log Transformation on the Target Feature
reg = LinearRegression()
reg.fit(X_train, y_train)
print("Train Score :", reg.score(X_train, y_train))
print("Test Score :", reg.score(X_test, y_test))
y_pred = reg.predict(X_test)
print("R squared :", (r2_score(y_test, y_pred)))
print("RMSLE :", np.sqrt(mean_squared_error(y_test, y_pred)))
# **Copying to New DataFrame**
datay = datas.copy()
datay["Quarter"] = (datas["week"] / 13).astype("int64")
datay["Quarter"] = datay["Quarter"].map(
{
0: "Q1",
1: "Q2",
2: "Q3",
3: "Q4",
4: "Q1",
5: "Q2",
6: "Q3",
7: "Q4",
8: "Q1",
9: "Q2",
10: "Q3",
11: "Q4",
}
)
datay["Quarter"].value_counts()
datay["Year"] = (datas["week"] / 52).astype("int64")
datay["Year"] = datay["Year"].map({0: "Y1", 1: "Y2", 2: "Y3"})
objy = datay[["Quarter", "Year"]]
numy = datay.drop(["Quarter", "Year"], axis=1)
encode1y = pd.get_dummies(objy, drop_first=True)
encode1y.head()
datay = pd.concat([numy, encode1y], axis=1)
datay.head()
# **Applying Log Transformation on the Target Feature**
datay["num_orders"] = np.log1p(datay["num_orders"])
train = datay[datay["week"].isin(range(1, 146))]
def outliers_3(col):
q3 = round(train[col].quantile(0.75), 6)
q1 = round(train[col].quantile(0.25), 6)
iqr = q3 - q1
lw = q1 - (3 * iqr)
hw = q3 + (3 * iqr)
uo = train[train[col] > hw].shape[0]
lo = train[train[col] < lw].shape[0]
print("Number of Upper Outliers :", uo)
print("Number of Lower Outliers :", lo)
print("Percentage of Outliers :", ((uo + lo) / train.shape[0]) * 100)
outliers_3("num_orders")
datay.head()
train = datay[datay["week"].isin(range(1, 136))]
test = datay[datay["week"].isin(range(136, 146))]
X_train = train.drop(
["id", "num_orders", "week", "discount amount", "city_code"], axis=1
)
y_train = train["num_orders"]
X_test = test.drop(["id", "num_orders", "week", "discount amount", "city_code"], axis=1)
y_test = test["num_orders"]
reg = LinearRegression()
reg.fit(X_train, y_train)
print("Train Score :", reg.score(X_train, y_train))
print("Test Score :", reg.score(X_test, y_test))
predictions = reg.predict(X_test)
print("R squared :", (r2_score(y_test, y_pred)))
print("RMSLE :", np.sqrt(mean_squared_error(y_test, y_pred)))
Result = pd.DataFrame(predictions)
Result = np.expm1(Result).astype("int64")
Submission = pd.DataFrame(columns=["id", "num_orders"])
Submission["id"] = test["id"]
Submission["num_orders"] = Result.values
Submission.to_csv("My submission.csv", index=False)
print("Your submission was successfully saved")
Submission.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/791/69791627.ipynb
|
food-demand-forecasting
|
kannanaikkal
|
[{"Id": 69791627, "ScriptId": 19068210, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6558800, "CreationDate": "08/03/2021 09:06:21", "VersionNumber": 7.0, "Title": "notebook43413f4f88", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 563.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 563.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 93249955, "KernelVersionId": 69791627, "SourceDatasetVersionId": 1138962}]
|
[{"Id": 1138962, "DatasetId": 641820, "DatasourceVersionId": 1169584, "CreatorUserId": 3768435, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "05/08/2020 05:41:47", "VersionNumber": 1.0, "Title": "Food Demand Forecasting", "Slug": "food-demand-forecasting", "Subtitle": "Predict the number of orders for upcoming 10 weeks", "Description": "### Context\n\nIt is a meal delivery company which operates in multiple cities. They have various fulfillment centers in these cities for dispatching meal orders to their customers. The client wants you to help these centers with demand forecasting for upcoming weeks so that these centers will plan the stock of raw materials accordingly.\n\n\n### Content\n\nThe replenishment of majority of raw materials is done on weekly basis and since the raw material is perishable, the procurement planning is of utmost importance. Secondly, staffing of the centers is also one area wherein accurate demand forecasts are really helpful. Given the following information, the task is to predict the demand for the next 10 weeks (Weeks: 146-155) for the center-meal combinations in the test set\n\n### Acknowledgements\n\nAnalytics Vidhya\n\n### Inspiration\n\nForecasting accurately could male the business growth in well directed direction.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 641820, "CreatorUserId": 3768435, "OwnerUserId": 3768435.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1138962.0, "CurrentDatasourceVersionId": 1169584.0, "ForumId": 656139, "Type": 2, "CreationDate": "05/08/2020 05:41:47", "LastActivityDate": "05/08/2020", "TotalViews": 76934, "TotalDownloads": 8477, "TotalVotes": 95, "TotalKernels": 22}]
|
[{"Id": 3768435, "UserName": "kannanaikkal", "DisplayName": "Edwin U Kannanaikkal", "RegisterDate": "09/26/2019", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
#
# **Demand forecasting is a key component to every growing online business. Without proper demand forecasting processes in place, it can be nearly impossible to have the right amount of stock on hand at any given time. A food delivery service has to deal with a lot of perishable raw materials which makes it all the more important for such a company to accurately forecast daily and weekly demand.**
# **Too much inventory in the warehouse means more risk of wastage, and not enough could lead to out-of-stocks — and push customers to seek solutions from your competitors. In this challenge, get a taste of demand forecasting challenge using a real dataset.**
# # **Problem Statement**
# The data set is related to a meal delivery company which operates in multiple cities. They have various fulfilment centers in these cities for dispatching meal orders to their customers.
# The dataset consists of historical data of demand for a product-center combination for weeks 1 to 145.
# With the given data and information, the task is to predict the demand for the next 10 weeks (Weeks: 146-155) for the center-meal combinations, so that these fulfilment centers stock the necessary raw materials accordingly.
# # ****Business Benefits****
# The replenishment of raw materials is done only on weekly basis and since the raw material is perishable, the procurement planning is of utmost importance.
# Therefore predicting the Demand helps in reducing the wastage of raw materials which would result in the reduced cost of operation. Increased customer satisfaction by timely fulfilling their expectations and requirements.
# # **Data Dictionary**
# The dataset consists of three individual datasheets, the first dataset contains the historical demand data for all centers, the second dataset contains the information of each fulfillment center and the third dataset contains the meal information.
# Weekly Demand data (train.csv):
# Contains the historical demand data for all centers. The Train dataset consists of 9 variables and records of 423727 unique orders. test.csv contains all the following features except the target variable. The Test dataset consists of 8 variables and records of 32573 unique orders.
# fulfilment_center_info.csv:
# Contains information for each fulfilment center. The dataset consists of 5 variables and records of 77 unique fulfillment centers.
# meal_info.csv:
# Contains information for each meal being served
# # **Libraries Used**
# pandas, numpy, scikit learn, matplotlib, seaborn, xgboost, lightgbm, catboost
# # **Data Pre-Processing**
# * There are no Missing/Null Values in any of the three datasets.
# * Before proceeding with the prediction process, all the three datasheets need to be merged into a single dataset. Before performing the merging operation, primary feature for combining the datasets needs to be validated.
# * The number of Center IDs in train dataset is matching with the number of Center IDs in the Centers Dataset i.e 77 unique records. Hence, there won't be any missing values while merging the datasets together.
# * The number of Meal IDs in train dataset is matching with the number of Meal IDs in the Meals Dataset i.e 51 unique records. Hence, there won't be any missing values while merging the datasets together.
# * As checked earlier, there were no Null/Missing values even after merging the datasets.
# # **Feature Engineering**
# Feature engineering is the process of using domain knowledge of the data to create features that improves the performance of the machine learning models.
# With the given data, We have derived the below features to improve our model performance.
# * Discount Amount : This defines the difference between the “base_Price” and “checkout_price”.
# * Discount Percent : This defines the % discount offer to customer.
# * Discount Y/N : This defines whether Discount is provided or not - 1 if there is Discount and 0 if there is no Discount.
# * Compare Week Price : This defines the increase / decrease in price of a Meal for a particular center compared to the previous week.
# * Compare Week Price Y/N : Price increased or decreased - 1 if the Price increased and 0 if the price decreased compared to the previous week.
# * Quarter : Based on the given number of weeks, derived a new feature named as Quarter which defines the Quarter of the year.
# * Year : Based on the given number of weeks, derived a new feature named as Year which defines the Year.
# # **Data Transformation**
# * Logarithm transformation (or log transform) is one of the most commonly used mathematical transformations in feature engineering. It helps to handle skewed data and after transformation, the distribution becomes more approximate to normal.
# * In our data, the target variable ‘num_orders’ is not normally distributed. Using this without applying any transformation techniques will downgrade the performance of our model.
# * Therefore, we have applied Logarithm transformation on our Target feature ‘num_orders’ post which the data seems to be more approximate to normal distribution.
# * After Log transformation, We have observed 0% of Outlier data being present within the Target Variable – num_orders using 3 IQR Method.
# # **Evaluation Metric**
# The evaluation metric for this competition is 100*RMSLE where RMSLE is Root of Mean Squared Logarithmic Error across all entries in the test set.
# # **Approach**
# * Simple Linear Regression model without any feature engineering and data transformation which gave a RMSE : 194.402
# * Without feature engineering and data transformation, the model did not perform well and could'nt give a good score.
# * Post applying feature engineering and data transformation (log and log1p transformation), Linear Regression model gave a RMSLE score of 0.634.
import numpy as np
import pandas as pd
pd.set_option("display.max_columns", None)
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import seaborn as sns
import plotly.io as pio
import plotly.graph_objects as go
from plotly.offline import init_notebook_mode, iplot
from sklearn.preprocessing import StandardScaler
import warnings
warnings.filterwarnings("ignore")
data = pd.read_csv("/kaggle/input/food-demand-forecasting/train.csv")
center = pd.read_csv("/kaggle/input/food-demand-forecasting/fulfilment_center_info.csv")
meal = pd.read_csv("/kaggle/input/food-demand-forecasting/meal_info.csv")
test = pd.read_csv("/kaggle/input/food-demand-forecasting/test.csv")
# /kaggle/input/food-demand-forecasting/sample_submission.csv
# **Data Preprocessing**
print("The Shape of Demand dataset :", data.shape)
print("The Shape of Fulmilment Center Information dataset :", center.shape)
print("The Shape of Meal information dataset :", meal.shape)
print("The Shape of Test dataset :", test.shape)
data.head()
test[
"num_orders"
] = 123456 ### Assigning random number for Target Variable of Test Data.
test.head()
center.head()
meal.head()
data = pd.concat([data, test], axis=0)
data = data.merge(center, on="center_id", how="left")
data = data.merge(meal, on="meal_id", how="left")
data.isnull().sum()
# **Deriving New Features**
# Discount Amount
data["discount amount"] = data["base_price"] - data["checkout_price"]
# Discount Percent
data["discount percent"] = (
(data["base_price"] - data["checkout_price"]) / data["base_price"]
) * 100
# Discount Y/N
data["discount y/n"] = [
1 if x > 0 else 0 for x in (data["base_price"] - data["checkout_price"])
]
data = data.sort_values(["center_id", "meal_id", "week"]).reset_index()
# Compare Week Price
data["compare_week_price"] = data["checkout_price"] - data["checkout_price"].shift(1)
data["compare_week_price"][data["week"] == 1] = 0
data = data.sort_values(by="index").reset_index().drop(["level_0", "index"], axis=1)
# Compare Week Price Y/N
data["compare_week_price y/n"] = [1 if x > 0 else 0 for x in data["compare_week_price"]]
data.head()
data.isnull().sum()
# **Train Test Split**
train = data[data["week"].isin(range(1, 146))]
test = data[data["week"].isin(range(146, 156))]
print("The Shape of Train dataset :", train.shape)
print("The Shape of Test dataset :", test.shape)
plt.figure(figsize=(16, 14))
sns.heatmap(train.corr(), annot=True, square=True, cmap="Reds")
fig = plt.figure(figsize=(4, 7))
plt.title("Total No. of Orders for Each Center type", fontdict={"fontsize": 13})
sns.barplot(
y="num_orders",
x="center_type",
data=train.groupby("center_type").sum()["num_orders"].reset_index(),
palette="autumn",
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Center Type", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Type A has the highest number of Orders placed and Type C has lowest
train["center_id"].nunique()
# The are are 77 Fullfilment Centers in total.
#
fig = plt.figure(figsize=(10, 8))
plt.title("Top 20 Centers with Highest No. of Orders", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="center_id",
data=train.groupby(["center_id", "center_type"])
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()
.head(20),
palette="YlOrRd_r",
order=list(
train.groupby(["center_id", "center_type"])
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()
.head(20)["center_id"]
),
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Center ID", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Initially, when we checked, which Center Type has the highest number of Orders, We found that Center Type_A has the highest number of orders, but now when we check individually, we could see that Center 13 of Type_B has the highest number of Orders. Let’s analyze the reason behind that.
#
fig = plt.figure(figsize=(4, 7))
plt.title("Total No. of Centers under Each Center type", fontdict={"fontsize": 13})
sns.barplot(
y=train.groupby(["center_id", "center_type"])
.num_orders.sum()
.reset_index()["center_type"]
.value_counts(),
x=train.groupby(["center_id", "center_type"])
.num_orders.sum()
.reset_index()["center_type"]
.value_counts()
.index,
palette="autumn",
)
plt.ylabel("No. of Centers", fontdict={"fontsize": 12})
plt.xlabel("Center Type", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Type_A has the most number of orders because, Type_A has the most number of Centers - 43 Centers.
#
sns.set_style("white")
plt.figure(figsize=(8, 8))
sns.scatterplot(
y=train["base_price"] - train["checkout_price"],
x=train["num_orders"],
color="coral",
)
plt.ylabel("Discount (Base price - Checkout Price)")
sns.despine(bottom=True, left=True)
# We created a new feature: Discount which is the difference of base price and checkout price and tried to find out if there is any relationship between the discount and the number of orders. But surprisingly there are no good correlation between the discount and the number of orders.
#
fig = plt.figure(figsize=(16, 7))
sns.set_style("whitegrid")
plt.title("Pattern of Orders", fontdict={"fontsize": 14})
sns.pointplot(
x=train.groupby("week").sum().reset_index()["week"],
y=train.groupby("week").sum().reset_index()["num_orders"],
color="coral",
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Week", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# When we analysed the trend of order placed over the weeks, we could see that the highest number of orders were received in week 48 and the lowest in week 62.
#
plt.figure(figsize=(6, 6))
colors = ["coral", "#FFDAB9", "yellowgreen", "#6495ED"]
plt.pie(
train.groupby(["cuisine"]).num_orders.sum(),
labels=train.groupby(["cuisine"]).num_orders.sum().index,
shadow=False,
colors=colors,
explode=(0.05, 0.05, 0.03, 0.05),
startangle=90,
autopct="%1.1f%%",
pctdistance=0.6,
textprops={"fontsize": 12},
)
plt.title("Total Number of Orders for Each Category")
plt.tight_layout()
plt.show()
# Italian Cuisine has the highest number of orders with Continental cuisine being the least.
#
fig = plt.figure(figsize=(11, 8))
sns.set_style("white")
plt.xticks(rotation=90, fontsize=12)
plt.title("Total Number of Orders for Each Category", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="category",
data=train.groupby("category")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index(),
palette="YlOrRd_r",
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Category", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# We could see that Beverages are the food category which has the higest number of orders and Biriyani is the food category with least number of orders.
#
fig = plt.figure(figsize=(18, 8))
sns.set_style("white")
plt.xticks(rotation=90, fontsize=12)
plt.title("Total Number of Orders for Each Cuisine-Category", fontdict={"fontsize": 14})
sns.barplot(
x="category",
y="num_orders",
data=train.groupby(["cuisine", "category"])
.sum()
.sort_values(by="num_orders", ascending=False)
.reset_index(),
hue="cuisine",
palette="YlOrRd_r",
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Cuisine-Category", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Similary when we checked which specific cuisne-food category has the highest number of orders, we could see that Indian-Rice Bowl has the highest number of orders and Indian-Biriyani has the least.
#
list(
data.groupby("region_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()
.values[:, 0]
)
fig = plt.figure(figsize=(8, 8))
sns.set_style("white")
plt.xticks(fontsize=13)
plt.title("Total Number of Orders for Each Region", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="region_code",
data=data.groupby("region_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index(),
palette="YlOrRd_r",
order=list(
data.groupby("region_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()
.values[:, 0]
),
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Region", fontdict={"fontsize": 12})
plt.xticks()
sns.despine(bottom=True, left=True)
# Also when we checked the number of orders with respect to Region, we could see that Region - 56 has the highest number of orders - 60.5M orders which is almost 35M orders higher than the Region with second highest number of orders - Region 34 - 24M orders.
#
fig = plt.figure(figsize=(18, 8))
sns.set_style("white")
plt.xticks(rotation=90, fontsize=13)
plt.title("Total Number of Orders for each Meal ID", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="meal_id",
data=data.groupby("meal_id")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index(),
palette="YlOrRd_r",
order=list(
data.groupby("meal_id")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()["meal_id"]
.values
),
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("Meal", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Meal ID 2290 has the higest number of Orders. There is not much significant differences between number of orders for different Meal IDs.
#
fig = plt.figure(figsize=(18, 8))
sns.set_style("white")
plt.xticks(rotation=90, fontsize=13)
plt.title("Total Number of Orders for each City", fontdict={"fontsize": 14})
sns.barplot(
y="num_orders",
x="city_code",
data=train.groupby("city_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index(),
palette="YlOrRd_r",
order=list(
train.groupby("city_code")
.num_orders.sum()
.sort_values(ascending=False)
.reset_index()["city_code"]
.values
),
)
plt.ylabel("No. of Orders", fontdict={"fontsize": 12})
plt.xlabel("City", fontdict={"fontsize": 12})
sns.despine(bottom=True, left=True)
# Also when we checked the number of orders with respect to City, we could see that City - 590 has the highest number of orders - 18.5M orders which is almost 10M orders higher than the City with second highest number of orders - City 526 - 8.6M orders.
# # **Encoding City**
# As per our observation from our barchart of the City against the number of orders. There the high significant difference between the Top 3 cities which has the highest number of orders. Therefore, in our first approach we will encode the City with Highest No. of Orders as CH1, City with 2nd Highest No. of Orders as CH2 and City with 3rd Highest No. of Orders as CH3 and rest all of the cities which does not have much significant differences between the number of orders as CH4.
#
city4 = {590: "CH1", 526: "CH2", 638: "CH3"}
data["city_enc_4"] = data["city_code"].map(city4)
data["city_enc_4"] = data["city_enc_4"].fillna("CH4")
data["city_enc_4"].value_counts()
data.head()
data.isnull().sum()
# **Copying to New DataFrame**
datax = data.copy()
datax.head()
# **Encoding All Categorical Features**
datax["center_id"] = datax["center_id"].astype("object")
datax["meal_id"] = datax["meal_id"].astype("object")
datax["region_code"] = datax["region_code"].astype("object")
obj = datax[
[
"center_id",
"meal_id",
"region_code",
"center_type",
"category",
"cuisine",
"city_enc_4",
]
]
num = datax.drop(
[
"center_id",
"meal_id",
"region_code",
"center_type",
"category",
"cuisine",
"city_enc_4",
],
axis=1,
)
encode1 = pd.get_dummies(obj, drop_first=True)
datax = pd.concat([num, encode1], axis=1)
datax.head()
# # **Base Model**
# Building base model by splitting the last 10 week of the train dataset as test.
#
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
train = datax[datax["week"].isin(range(1, 136))]
test = datax[datax["week"].isin(range(136, 146))]
X_train = train.drop(["id", "num_orders", "week"], axis=1)
y_train = train["num_orders"]
X_test = test.drop(["id", "num_orders", "week"], axis=1)
y_test = test["num_orders"]
reg = LinearRegression()
reg.fit(X_train, y_train)
print("Train Score :", reg.score(X_train, y_train))
print("Test Score :", reg.score(X_test, y_test))
y_pred = reg.predict(X_test)
print("R squared :", (r2_score(y_test, y_pred)))
print("RMSE :", np.sqrt(mean_squared_error(y_test, y_pred)))
# Linear Model 2 : Applying Standard Scaling & Log Transformation
#
sc = StandardScaler()
cat = datax.drop(
[
"checkout_price",
"base_price",
"discount amount",
"discount percent",
"compare_week_price",
],
axis=1,
)
num = datax[
[
"checkout_price",
"base_price",
"discount amount",
"discount percent",
"compare_week_price",
]
]
scal = pd.DataFrame(sc.fit_transform(num), columns=num.columns)
datas = pd.concat([scal, cat], axis=1)
train = datas[datas["week"].isin(range(1, 136))]
test = datas[datas["week"].isin(range(136, 146))]
X_train = train.drop(["id", "num_orders", "week"], axis=1)
y_train = np.log(
train["num_orders"]
) # Applying Log Transformation on the Target Feature
X_test = test.drop(["id", "num_orders", "week"], axis=1)
y_test = np.log(test["num_orders"]) # Applying Log Transformation on the Target Feature
reg = LinearRegression()
reg.fit(X_train, y_train)
print("Train Score :", reg.score(X_train, y_train))
print("Test Score :", reg.score(X_test, y_test))
y_pred = reg.predict(X_test)
print("R squared :", (r2_score(y_test, y_pred)))
print("RMSLE :", np.sqrt(mean_squared_error(y_test, y_pred)))
# **Copying to New DataFrame**
datay = datas.copy()
datay["Quarter"] = (datas["week"] / 13).astype("int64")
datay["Quarter"] = datay["Quarter"].map(
{
0: "Q1",
1: "Q2",
2: "Q3",
3: "Q4",
4: "Q1",
5: "Q2",
6: "Q3",
7: "Q4",
8: "Q1",
9: "Q2",
10: "Q3",
11: "Q4",
}
)
datay["Quarter"].value_counts()
datay["Year"] = (datas["week"] / 52).astype("int64")
datay["Year"] = datay["Year"].map({0: "Y1", 1: "Y2", 2: "Y3"})
objy = datay[["Quarter", "Year"]]
numy = datay.drop(["Quarter", "Year"], axis=1)
encode1y = pd.get_dummies(objy, drop_first=True)
encode1y.head()
datay = pd.concat([numy, encode1y], axis=1)
datay.head()
# **Applying Log Transformation on the Target Feature**
datay["num_orders"] = np.log1p(datay["num_orders"])
train = datay[datay["week"].isin(range(1, 146))]
def outliers_3(col):
q3 = round(train[col].quantile(0.75), 6)
q1 = round(train[col].quantile(0.25), 6)
iqr = q3 - q1
lw = q1 - (3 * iqr)
hw = q3 + (3 * iqr)
uo = train[train[col] > hw].shape[0]
lo = train[train[col] < lw].shape[0]
print("Number of Upper Outliers :", uo)
print("Number of Lower Outliers :", lo)
print("Percentage of Outliers :", ((uo + lo) / train.shape[0]) * 100)
outliers_3("num_orders")
datay.head()
train = datay[datay["week"].isin(range(1, 136))]
test = datay[datay["week"].isin(range(136, 146))]
X_train = train.drop(
["id", "num_orders", "week", "discount amount", "city_code"], axis=1
)
y_train = train["num_orders"]
X_test = test.drop(["id", "num_orders", "week", "discount amount", "city_code"], axis=1)
y_test = test["num_orders"]
reg = LinearRegression()
reg.fit(X_train, y_train)
print("Train Score :", reg.score(X_train, y_train))
print("Test Score :", reg.score(X_test, y_test))
predictions = reg.predict(X_test)
print("R squared :", (r2_score(y_test, y_pred)))
print("RMSLE :", np.sqrt(mean_squared_error(y_test, y_pred)))
Result = pd.DataFrame(predictions)
Result = np.expm1(Result).astype("int64")
Submission = pd.DataFrame(columns=["id", "num_orders"])
Submission["id"] = test["id"]
Submission["num_orders"] = Result.values
Submission.to_csv("My submission.csv", index=False)
print("Your submission was successfully saved")
Submission.head()
|
[{"food-demand-forecasting/test.csv": {"column_names": "[\"id\", \"week\", \"center_id\", \"meal_id\", \"checkout_price\", \"base_price\", \"emailer_for_promotion\", \"homepage_featured\"]", "column_data_types": "{\"id\": \"int64\", \"week\": \"int64\", \"center_id\": \"int64\", \"meal_id\": \"int64\", \"checkout_price\": \"float64\", \"base_price\": \"float64\", \"emailer_for_promotion\": \"int64\", \"homepage_featured\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 32573 entries, 0 to 32572\nData columns (total 8 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 id 32573 non-null int64 \n 1 week 32573 non-null int64 \n 2 center_id 32573 non-null int64 \n 3 meal_id 32573 non-null int64 \n 4 checkout_price 32573 non-null float64\n 5 base_price 32573 non-null float64\n 6 emailer_for_promotion 32573 non-null int64 \n 7 homepage_featured 32573 non-null int64 \ndtypes: float64(2), int64(6)\nmemory usage: 2.0 MB\n", "summary": "{\"id\": {\"count\": 32573.0, \"mean\": 1248475.8382095601, \"std\": 144158.04832391287, \"min\": 1000085.0, \"25%\": 1123969.0, \"50%\": 1247296.0, \"75%\": 1372971.0, \"max\": 1499996.0}, \"week\": {\"count\": 32573.0, \"mean\": 150.47781905258958, \"std\": 2.86407248864882, \"min\": 146.0, \"25%\": 148.0, \"50%\": 150.0, \"75%\": 153.0, \"max\": 155.0}, \"center_id\": {\"count\": 32573.0, \"mean\": 81.90172842538298, \"std\": 45.95045507150821, \"min\": 10.0, \"25%\": 43.0, \"50%\": 76.0, \"75%\": 110.0, \"max\": 186.0}, \"meal_id\": {\"count\": 32573.0, \"mean\": 2032.0679090043902, \"std\": 547.1990044343339, \"min\": 1062.0, \"25%\": 1558.0, \"50%\": 1993.0, \"75%\": 2569.0, \"max\": 2956.0}, \"checkout_price\": {\"count\": 32573.0, \"mean\": 341.85443956651216, \"std\": 153.8938862206768, \"min\": 67.9, \"25%\": 214.43, \"50%\": 320.13, \"75%\": 446.23, \"max\": 1113.62}, \"base_price\": {\"count\": 32573.0, \"mean\": 356.4936149571731, \"std\": 155.15010053852777, \"min\": 89.24, \"25%\": 243.5, \"50%\": 321.13, \"75%\": 455.93, \"max\": 1112.62}, \"emailer_for_promotion\": {\"count\": 32573.0, \"mean\": 0.06643539127498235, \"std\": 0.249045446060833, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"homepage_featured\": {\"count\": 32573.0, \"mean\": 0.08135572406594418, \"std\": 0.2733848290295183, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}}", "examples": "{\"id\":{\"0\":1028232,\"1\":1127204,\"2\":1212707,\"3\":1082698},\"week\":{\"0\":146,\"1\":146,\"2\":146,\"3\":146},\"center_id\":{\"0\":55,\"1\":55,\"2\":55,\"3\":55},\"meal_id\":{\"0\":1885,\"1\":1993,\"2\":2539,\"3\":2631},\"checkout_price\":{\"0\":158.11,\"1\":160.11,\"2\":157.14,\"3\":162.02},\"base_price\":{\"0\":159.11,\"1\":159.11,\"2\":159.14,\"3\":162.02},\"emailer_for_promotion\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"homepage_featured\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0}}"}}, {"food-demand-forecasting/fulfilment_center_info.csv": {"column_names": "[\"center_id\", \"city_code\", \"region_code\", \"center_type\", \"op_area\"]", "column_data_types": "{\"center_id\": \"int64\", \"city_code\": \"int64\", \"region_code\": \"int64\", \"center_type\": \"object\", \"op_area\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 77 entries, 0 to 76\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 center_id 77 non-null int64 \n 1 city_code 77 non-null int64 \n 2 region_code 77 non-null int64 \n 3 center_type 77 non-null object \n 4 op_area 77 non-null float64\ndtypes: float64(1), int64(3), object(1)\nmemory usage: 3.1+ KB\n", "summary": "{\"center_id\": {\"count\": 77.0, \"mean\": 83.14285714285714, \"std\": 46.09021881784346, \"min\": 10.0, \"25%\": 50.0, \"50%\": 77.0, \"75%\": 110.0, \"max\": 186.0}, \"city_code\": {\"count\": 77.0, \"mean\": 600.6623376623377, \"std\": 66.72027435684677, \"min\": 456.0, \"25%\": 553.0, \"50%\": 596.0, \"75%\": 651.0, \"max\": 713.0}, \"region_code\": {\"count\": 77.0, \"mean\": 56.493506493506494, \"std\": 18.12647335327341, \"min\": 23.0, \"25%\": 34.0, \"50%\": 56.0, \"75%\": 77.0, \"max\": 93.0}, \"op_area\": {\"count\": 77.0, \"mean\": 3.985714285714286, \"std\": 1.1064064977872574, \"min\": 0.9, \"25%\": 3.5, \"50%\": 3.9, \"75%\": 4.4, \"max\": 7.0}}", "examples": "{\"center_id\":{\"0\":11,\"1\":13,\"2\":124,\"3\":66},\"city_code\":{\"0\":679,\"1\":590,\"2\":590,\"3\":648},\"region_code\":{\"0\":56,\"1\":56,\"2\":56,\"3\":34},\"center_type\":{\"0\":\"TYPE_A\",\"1\":\"TYPE_B\",\"2\":\"TYPE_C\",\"3\":\"TYPE_A\"},\"op_area\":{\"0\":3.7,\"1\":6.7,\"2\":4.0,\"3\":4.1}}"}}, {"food-demand-forecasting/meal_info.csv": {"column_names": "[\"meal_id\", \"category\", \"cuisine\"]", "column_data_types": "{\"meal_id\": \"int64\", \"category\": \"object\", \"cuisine\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 51 entries, 0 to 50\nData columns (total 3 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 meal_id 51 non-null int64 \n 1 category 51 non-null object\n 2 cuisine 51 non-null object\ndtypes: int64(1), object(2)\nmemory usage: 1.3+ KB\n", "summary": "{\"meal_id\": {\"count\": 51.0, \"mean\": 2013.921568627451, \"std\": 553.6335554547702, \"min\": 1062.0, \"25%\": 1550.5, \"50%\": 1971.0, \"75%\": 2516.5, \"max\": 2956.0}}", "examples": "{\"meal_id\":{\"0\":1885,\"1\":1993,\"2\":2539,\"3\":1248},\"category\":{\"0\":\"Beverages\",\"1\":\"Beverages\",\"2\":\"Beverages\",\"3\":\"Beverages\"},\"cuisine\":{\"0\":\"Thai\",\"1\":\"Thai\",\"2\":\"Thai\",\"3\":\"Indian\"}}"}}, {"food-demand-forecasting/train.csv": {"column_names": "[\"id\", \"week\", \"center_id\", \"meal_id\", \"checkout_price\", \"base_price\", \"emailer_for_promotion\", \"homepage_featured\", \"num_orders\"]", "column_data_types": "{\"id\": \"int64\", \"week\": \"int64\", \"center_id\": \"int64\", \"meal_id\": \"int64\", \"checkout_price\": \"float64\", \"base_price\": \"float64\", \"emailer_for_promotion\": \"int64\", \"homepage_featured\": \"int64\", \"num_orders\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 456548 entries, 0 to 456547\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 id 456548 non-null int64 \n 1 week 456548 non-null int64 \n 2 center_id 456548 non-null int64 \n 3 meal_id 456548 non-null int64 \n 4 checkout_price 456548 non-null float64\n 5 base_price 456548 non-null float64\n 6 emailer_for_promotion 456548 non-null int64 \n 7 homepage_featured 456548 non-null int64 \n 8 num_orders 456548 non-null int64 \ndtypes: float64(2), int64(7)\nmemory usage: 31.3 MB\n", "summary": "{\"id\": {\"count\": 456548.0, \"mean\": 1250096.3056327046, \"std\": 144354.822377881, \"min\": 1000000.0, \"25%\": 1124998.75, \"50%\": 1250183.5, \"75%\": 1375140.25, \"max\": 1499999.0}, \"week\": {\"count\": 456548.0, \"mean\": 74.76877130115562, \"std\": 41.52495637124611, \"min\": 1.0, \"25%\": 39.0, \"50%\": 76.0, \"75%\": 111.0, \"max\": 145.0}, \"center_id\": {\"count\": 456548.0, \"mean\": 82.10579610468122, \"std\": 45.97504601530891, \"min\": 10.0, \"25%\": 43.0, \"50%\": 76.0, \"75%\": 110.0, \"max\": 186.0}, \"meal_id\": {\"count\": 456548.0, \"mean\": 2024.3374584928638, \"std\": 547.420920130117, \"min\": 1062.0, \"25%\": 1558.0, \"50%\": 1993.0, \"75%\": 2539.0, \"max\": 2956.0}, \"checkout_price\": {\"count\": 456548.0, \"mean\": 332.2389325547367, \"std\": 152.9397233077613, \"min\": 2.97, \"25%\": 228.95, \"50%\": 296.82, \"75%\": 445.23, \"max\": 866.27}, \"base_price\": {\"count\": 456548.0, \"mean\": 354.15662745209715, \"std\": 160.715913989822, \"min\": 55.35, \"25%\": 243.5, \"50%\": 310.46, \"75%\": 458.87, \"max\": 866.27}, \"emailer_for_promotion\": {\"count\": 456548.0, \"mean\": 0.08115247465764827, \"std\": 0.2730694304425931, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"homepage_featured\": {\"count\": 456548.0, \"mean\": 0.10919990888143197, \"std\": 0.3118902080045688, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"num_orders\": {\"count\": 456548.0, \"mean\": 261.8727603669275, \"std\": 395.922797742466, \"min\": 13.0, \"25%\": 54.0, \"50%\": 136.0, \"75%\": 324.0, \"max\": 24299.0}}", "examples": "{\"id\":{\"0\":1379560,\"1\":1466964,\"2\":1346989,\"3\":1338232},\"week\":{\"0\":1,\"1\":1,\"2\":1,\"3\":1},\"center_id\":{\"0\":55,\"1\":55,\"2\":55,\"3\":55},\"meal_id\":{\"0\":1885,\"1\":1993,\"2\":2539,\"3\":2139},\"checkout_price\":{\"0\":136.83,\"1\":136.83,\"2\":134.86,\"3\":339.5},\"base_price\":{\"0\":152.29,\"1\":135.83,\"2\":135.86,\"3\":437.53},\"emailer_for_promotion\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"homepage_featured\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"num_orders\":{\"0\":177,\"1\":270,\"2\":189,\"3\":54}}"}}]
| true | 4 |
<start_data_description><data_path>food-demand-forecasting/test.csv:
<column_names>
['id', 'week', 'center_id', 'meal_id', 'checkout_price', 'base_price', 'emailer_for_promotion', 'homepage_featured']
<column_types>
{'id': 'int64', 'week': 'int64', 'center_id': 'int64', 'meal_id': 'int64', 'checkout_price': 'float64', 'base_price': 'float64', 'emailer_for_promotion': 'int64', 'homepage_featured': 'int64'}
<dataframe_Summary>
{'id': {'count': 32573.0, 'mean': 1248475.8382095601, 'std': 144158.04832391287, 'min': 1000085.0, '25%': 1123969.0, '50%': 1247296.0, '75%': 1372971.0, 'max': 1499996.0}, 'week': {'count': 32573.0, 'mean': 150.47781905258958, 'std': 2.86407248864882, 'min': 146.0, '25%': 148.0, '50%': 150.0, '75%': 153.0, 'max': 155.0}, 'center_id': {'count': 32573.0, 'mean': 81.90172842538298, 'std': 45.95045507150821, 'min': 10.0, '25%': 43.0, '50%': 76.0, '75%': 110.0, 'max': 186.0}, 'meal_id': {'count': 32573.0, 'mean': 2032.0679090043902, 'std': 547.1990044343339, 'min': 1062.0, '25%': 1558.0, '50%': 1993.0, '75%': 2569.0, 'max': 2956.0}, 'checkout_price': {'count': 32573.0, 'mean': 341.85443956651216, 'std': 153.8938862206768, 'min': 67.9, '25%': 214.43, '50%': 320.13, '75%': 446.23, 'max': 1113.62}, 'base_price': {'count': 32573.0, 'mean': 356.4936149571731, 'std': 155.15010053852777, 'min': 89.24, '25%': 243.5, '50%': 321.13, '75%': 455.93, 'max': 1112.62}, 'emailer_for_promotion': {'count': 32573.0, 'mean': 0.06643539127498235, 'std': 0.249045446060833, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'homepage_featured': {'count': 32573.0, 'mean': 0.08135572406594418, 'std': 0.2733848290295183, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 32573 entries, 0 to 32572
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 32573 non-null int64
1 week 32573 non-null int64
2 center_id 32573 non-null int64
3 meal_id 32573 non-null int64
4 checkout_price 32573 non-null float64
5 base_price 32573 non-null float64
6 emailer_for_promotion 32573 non-null int64
7 homepage_featured 32573 non-null int64
dtypes: float64(2), int64(6)
memory usage: 2.0 MB
<some_examples>
{'id': {'0': 1028232, '1': 1127204, '2': 1212707, '3': 1082698}, 'week': {'0': 146, '1': 146, '2': 146, '3': 146}, 'center_id': {'0': 55, '1': 55, '2': 55, '3': 55}, 'meal_id': {'0': 1885, '1': 1993, '2': 2539, '3': 2631}, 'checkout_price': {'0': 158.11, '1': 160.11, '2': 157.14, '3': 162.02}, 'base_price': {'0': 159.11, '1': 159.11, '2': 159.14, '3': 162.02}, 'emailer_for_promotion': {'0': 0, '1': 0, '2': 0, '3': 0}, 'homepage_featured': {'0': 0, '1': 0, '2': 0, '3': 0}}
<end_description>
<start_data_description><data_path>food-demand-forecasting/fulfilment_center_info.csv:
<column_names>
['center_id', 'city_code', 'region_code', 'center_type', 'op_area']
<column_types>
{'center_id': 'int64', 'city_code': 'int64', 'region_code': 'int64', 'center_type': 'object', 'op_area': 'float64'}
<dataframe_Summary>
{'center_id': {'count': 77.0, 'mean': 83.14285714285714, 'std': 46.09021881784346, 'min': 10.0, '25%': 50.0, '50%': 77.0, '75%': 110.0, 'max': 186.0}, 'city_code': {'count': 77.0, 'mean': 600.6623376623377, 'std': 66.72027435684677, 'min': 456.0, '25%': 553.0, '50%': 596.0, '75%': 651.0, 'max': 713.0}, 'region_code': {'count': 77.0, 'mean': 56.493506493506494, 'std': 18.12647335327341, 'min': 23.0, '25%': 34.0, '50%': 56.0, '75%': 77.0, 'max': 93.0}, 'op_area': {'count': 77.0, 'mean': 3.985714285714286, 'std': 1.1064064977872574, 'min': 0.9, '25%': 3.5, '50%': 3.9, '75%': 4.4, 'max': 7.0}}
<dataframe_info>
RangeIndex: 77 entries, 0 to 76
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 center_id 77 non-null int64
1 city_code 77 non-null int64
2 region_code 77 non-null int64
3 center_type 77 non-null object
4 op_area 77 non-null float64
dtypes: float64(1), int64(3), object(1)
memory usage: 3.1+ KB
<some_examples>
{'center_id': {'0': 11, '1': 13, '2': 124, '3': 66}, 'city_code': {'0': 679, '1': 590, '2': 590, '3': 648}, 'region_code': {'0': 56, '1': 56, '2': 56, '3': 34}, 'center_type': {'0': 'TYPE_A', '1': 'TYPE_B', '2': 'TYPE_C', '3': 'TYPE_A'}, 'op_area': {'0': 3.7, '1': 6.7, '2': 4.0, '3': 4.1}}
<end_description>
<start_data_description><data_path>food-demand-forecasting/meal_info.csv:
<column_names>
['meal_id', 'category', 'cuisine']
<column_types>
{'meal_id': 'int64', 'category': 'object', 'cuisine': 'object'}
<dataframe_Summary>
{'meal_id': {'count': 51.0, 'mean': 2013.921568627451, 'std': 553.6335554547702, 'min': 1062.0, '25%': 1550.5, '50%': 1971.0, '75%': 2516.5, 'max': 2956.0}}
<dataframe_info>
RangeIndex: 51 entries, 0 to 50
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 meal_id 51 non-null int64
1 category 51 non-null object
2 cuisine 51 non-null object
dtypes: int64(1), object(2)
memory usage: 1.3+ KB
<some_examples>
{'meal_id': {'0': 1885, '1': 1993, '2': 2539, '3': 1248}, 'category': {'0': 'Beverages', '1': 'Beverages', '2': 'Beverages', '3': 'Beverages'}, 'cuisine': {'0': 'Thai', '1': 'Thai', '2': 'Thai', '3': 'Indian'}}
<end_description>
<start_data_description><data_path>food-demand-forecasting/train.csv:
<column_names>
['id', 'week', 'center_id', 'meal_id', 'checkout_price', 'base_price', 'emailer_for_promotion', 'homepage_featured', 'num_orders']
<column_types>
{'id': 'int64', 'week': 'int64', 'center_id': 'int64', 'meal_id': 'int64', 'checkout_price': 'float64', 'base_price': 'float64', 'emailer_for_promotion': 'int64', 'homepage_featured': 'int64', 'num_orders': 'int64'}
<dataframe_Summary>
{'id': {'count': 456548.0, 'mean': 1250096.3056327046, 'std': 144354.822377881, 'min': 1000000.0, '25%': 1124998.75, '50%': 1250183.5, '75%': 1375140.25, 'max': 1499999.0}, 'week': {'count': 456548.0, 'mean': 74.76877130115562, 'std': 41.52495637124611, 'min': 1.0, '25%': 39.0, '50%': 76.0, '75%': 111.0, 'max': 145.0}, 'center_id': {'count': 456548.0, 'mean': 82.10579610468122, 'std': 45.97504601530891, 'min': 10.0, '25%': 43.0, '50%': 76.0, '75%': 110.0, 'max': 186.0}, 'meal_id': {'count': 456548.0, 'mean': 2024.3374584928638, 'std': 547.420920130117, 'min': 1062.0, '25%': 1558.0, '50%': 1993.0, '75%': 2539.0, 'max': 2956.0}, 'checkout_price': {'count': 456548.0, 'mean': 332.2389325547367, 'std': 152.9397233077613, 'min': 2.97, '25%': 228.95, '50%': 296.82, '75%': 445.23, 'max': 866.27}, 'base_price': {'count': 456548.0, 'mean': 354.15662745209715, 'std': 160.715913989822, 'min': 55.35, '25%': 243.5, '50%': 310.46, '75%': 458.87, 'max': 866.27}, 'emailer_for_promotion': {'count': 456548.0, 'mean': 0.08115247465764827, 'std': 0.2730694304425931, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'homepage_featured': {'count': 456548.0, 'mean': 0.10919990888143197, 'std': 0.3118902080045688, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'num_orders': {'count': 456548.0, 'mean': 261.8727603669275, 'std': 395.922797742466, 'min': 13.0, '25%': 54.0, '50%': 136.0, '75%': 324.0, 'max': 24299.0}}
<dataframe_info>
RangeIndex: 456548 entries, 0 to 456547
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 456548 non-null int64
1 week 456548 non-null int64
2 center_id 456548 non-null int64
3 meal_id 456548 non-null int64
4 checkout_price 456548 non-null float64
5 base_price 456548 non-null float64
6 emailer_for_promotion 456548 non-null int64
7 homepage_featured 456548 non-null int64
8 num_orders 456548 non-null int64
dtypes: float64(2), int64(7)
memory usage: 31.3 MB
<some_examples>
{'id': {'0': 1379560, '1': 1466964, '2': 1346989, '3': 1338232}, 'week': {'0': 1, '1': 1, '2': 1, '3': 1}, 'center_id': {'0': 55, '1': 55, '2': 55, '3': 55}, 'meal_id': {'0': 1885, '1': 1993, '2': 2539, '3': 2139}, 'checkout_price': {'0': 136.83, '1': 136.83, '2': 134.86, '3': 339.5}, 'base_price': {'0': 152.29, '1': 135.83, '2': 135.86, '3': 437.53}, 'emailer_for_promotion': {'0': 0, '1': 0, '2': 0, '3': 0}, 'homepage_featured': {'0': 0, '1': 0, '2': 0, '3': 0}, 'num_orders': {'0': 177, '1': 270, '2': 189, '3': 54}}
<end_description>
| 6,863 | 0 | 9,286 | 6,863 |
69791416
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women)
print("% of women who survived:", rate_women)
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men)
print("% of men who survived:", rate_men)
from sklearn.ensemble import RandomForestClassifier
from xgboost import XGBRegressor
y = train_data["Survived"]
features = ["Pclass", "Sex", "SibSp", "Parch"]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = XGBRegressor(n_estimators=140, learning_rate=0.005, random_state=0)
model.fit(X, y)
predictions = model.predict(X_test)
# mae =mean_absolute_error()
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
# cross-validation
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.impute import SimpleImputer
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import OneHotEncoder
from sklearn.model_selection import cross_val_score
from xgboost import XGBRegressor
# =============================================================================
# Load data
# =============================================================================
# Read the data
X_full = train_data
X_test_full = test_data
# Remove rows with missing target, separate target from predictors
X_full.dropna(axis=0, subset=["Survived"], inplace=True)
y = X_full.Survived
X_full.drop(["Survived"], axis=1, inplace=True)
# =============================================================================
# Splitting dataset for training and validating
# =============================================================================
# Break off validation set from training data
X_train_full, X_valid_full, y_train, y_valid = train_test_split(
X_full, y, train_size=0.8, test_size=0.2, random_state=0
)
# "Cardinality" means the number of unique values in a column
# Select categorical columns with relatively low cardinality
# (convenient but arbitrary)
categorical_cols = [
cname
for cname in X_train_full.columns
if X_train_full[cname].nunique() < 10 and X_train_full[cname].dtype == "object"
]
# Select numerical columns
numerical_cols = [
cname
for cname in X_train_full.columns
if X_train_full[cname].dtype in ["int64", "float64"]
]
# Keep selected columns only
my_cols = categorical_cols + numerical_cols
X_train = X_train_full[my_cols].copy()
X_valid = X_valid_full[my_cols].copy()
X_test = X_test_full[my_cols].copy()
print(X_train.head())
# =============================================================================
# Imputation
# =============================================================================
# Preprocessing for numerical data
numerical_transformer = SimpleImputer(strategy="median")
# Preprocessing for categorical data
categorical_transformer = Pipeline(
steps=[
("imputer", SimpleImputer(strategy="most_frequent")),
("onehot", OneHotEncoder(handle_unknown="ignore")),
]
)
# Bundle preprocessing for numerical and categorical data
preprocessor = ColumnTransformer(
transformers=[
("num", numerical_transformer, numerical_cols),
("cat", categorical_transformer, categorical_cols),
]
)
# =============================================================================
# Training
# =============================================================================
def get_score(n_estimators, preprocessor):
my_pipeline = Pipeline(
steps=[
("preprocessor", preprocessor),
("model", XGBRegressor(n_estimators=n_estimators, learning_rate=0.03)),
]
)
scores = -1 * cross_val_score(
my_pipeline, X_full, y, cv=20, scoring="neg_mean_absolute_error"
)
print(n_estimators, " = ", scores.mean())
return scores.mean()
n_estimators = [i for i in range(190, 195, 5)]
results = {
n_estimator: get_score(n_estimator, preprocessor) for n_estimator in n_estimators
}
my_pipeline = Pipeline(
steps=[
("preprocessor", preprocessor),
("model", XGBRegressor(n_estimators=195, learning_rate=0.03)),
]
)
scores = -1 * cross_val_score(
my_pipeline, X_full, y, cv=20, scoring="neg_mean_absolute_error"
)
# model = XGBRegressor(n_estimators=400, learning_rate=0.02, random_state=0)
my_pipeline.fit(X_train, y_train)
predictions = my_pipeline.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/791/69791416.ipynb
| null | null |
[{"Id": 69791416, "ScriptId": 18768001, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7944167, "CreationDate": "08/03/2021 09:04:51", "VersionNumber": 11.0, "Title": "Getting Started with Titanic 0.77511 score", "EvaluationDate": "08/03/2021", "IsChange": false, "TotalLines": 153.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 153.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women)
print("% of women who survived:", rate_women)
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men)
print("% of men who survived:", rate_men)
from sklearn.ensemble import RandomForestClassifier
from xgboost import XGBRegressor
y = train_data["Survived"]
features = ["Pclass", "Sex", "SibSp", "Parch"]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = XGBRegressor(n_estimators=140, learning_rate=0.005, random_state=0)
model.fit(X, y)
predictions = model.predict(X_test)
# mae =mean_absolute_error()
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
# cross-validation
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.impute import SimpleImputer
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import OneHotEncoder
from sklearn.model_selection import cross_val_score
from xgboost import XGBRegressor
# =============================================================================
# Load data
# =============================================================================
# Read the data
X_full = train_data
X_test_full = test_data
# Remove rows with missing target, separate target from predictors
X_full.dropna(axis=0, subset=["Survived"], inplace=True)
y = X_full.Survived
X_full.drop(["Survived"], axis=1, inplace=True)
# =============================================================================
# Splitting dataset for training and validating
# =============================================================================
# Break off validation set from training data
X_train_full, X_valid_full, y_train, y_valid = train_test_split(
X_full, y, train_size=0.8, test_size=0.2, random_state=0
)
# "Cardinality" means the number of unique values in a column
# Select categorical columns with relatively low cardinality
# (convenient but arbitrary)
categorical_cols = [
cname
for cname in X_train_full.columns
if X_train_full[cname].nunique() < 10 and X_train_full[cname].dtype == "object"
]
# Select numerical columns
numerical_cols = [
cname
for cname in X_train_full.columns
if X_train_full[cname].dtype in ["int64", "float64"]
]
# Keep selected columns only
my_cols = categorical_cols + numerical_cols
X_train = X_train_full[my_cols].copy()
X_valid = X_valid_full[my_cols].copy()
X_test = X_test_full[my_cols].copy()
print(X_train.head())
# =============================================================================
# Imputation
# =============================================================================
# Preprocessing for numerical data
numerical_transformer = SimpleImputer(strategy="median")
# Preprocessing for categorical data
categorical_transformer = Pipeline(
steps=[
("imputer", SimpleImputer(strategy="most_frequent")),
("onehot", OneHotEncoder(handle_unknown="ignore")),
]
)
# Bundle preprocessing for numerical and categorical data
preprocessor = ColumnTransformer(
transformers=[
("num", numerical_transformer, numerical_cols),
("cat", categorical_transformer, categorical_cols),
]
)
# =============================================================================
# Training
# =============================================================================
def get_score(n_estimators, preprocessor):
my_pipeline = Pipeline(
steps=[
("preprocessor", preprocessor),
("model", XGBRegressor(n_estimators=n_estimators, learning_rate=0.03)),
]
)
scores = -1 * cross_val_score(
my_pipeline, X_full, y, cv=20, scoring="neg_mean_absolute_error"
)
print(n_estimators, " = ", scores.mean())
return scores.mean()
n_estimators = [i for i in range(190, 195, 5)]
results = {
n_estimator: get_score(n_estimator, preprocessor) for n_estimator in n_estimators
}
my_pipeline = Pipeline(
steps=[
("preprocessor", preprocessor),
("model", XGBRegressor(n_estimators=195, learning_rate=0.03)),
]
)
scores = -1 * cross_val_score(
my_pipeline, X_full, y, cv=20, scoring="neg_mean_absolute_error"
)
# model = XGBRegressor(n_estimators=400, learning_rate=0.02, random_state=0)
my_pipeline.fit(X_train, y_train)
predictions = my_pipeline.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
| false | 0 | 1,548 | 0 | 1,548 | 1,548 |
||
69791245
|
import numpy as np # linear algebra
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
train_data = pd.read_csv("../input/tabular-playground-series-aug-2021/train.csv")
test_data = pd.read_csv("../input/tabular-playground-series-aug-2021/test.csv")
train_data.head()
test_data.head()
train_data.describe()
test_data.describe()
train_data = train_data.drop("id", axis=1)
test_data = test_data.drop("id", axis=1)
train_data["loss"].value_counts()
plt.figure(figsize=(12, 6))
sns.countplot(train_data["loss"])
plt.title("Countplot")
plt.show()
# check for missing values
train_data.isnull().sum().any()
# Check for duplicate rows
train_data.duplicated().sum()
# ***MACHINE LEARNING MODEL***
x_train = train_data.drop("loss", axis=1)
y_train = train_data["loss"]
x_test = test_data
y_test = pd.read_csv(
"../input/tabular-playground-series-aug-2021/sample_submission.csv"
)
id_loc = np.array(y_test["id"])
y_test = y_test.drop("id", axis=1)
from sklearn.linear_model import LinearRegression
regressor = LinearRegression()
regressor.fit(x_train, y_train)
y_pred = regressor.predict(x_test)
len(y_pred)
type(y_pred)
output = pd.DataFrame({"id": id_col, "loss": y_pred})
output.head()
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/791/69791245.ipynb
| null | null |
[{"Id": 69791245, "ScriptId": 19040621, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7391923, "CreationDate": "08/03/2021 09:03:46", "VersionNumber": 8.0, "Title": "Multiple Linear Regression", "EvaluationDate": "08/03/2021", "IsChange": false, "TotalLines": 81.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 81.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
train_data = pd.read_csv("../input/tabular-playground-series-aug-2021/train.csv")
test_data = pd.read_csv("../input/tabular-playground-series-aug-2021/test.csv")
train_data.head()
test_data.head()
train_data.describe()
test_data.describe()
train_data = train_data.drop("id", axis=1)
test_data = test_data.drop("id", axis=1)
train_data["loss"].value_counts()
plt.figure(figsize=(12, 6))
sns.countplot(train_data["loss"])
plt.title("Countplot")
plt.show()
# check for missing values
train_data.isnull().sum().any()
# Check for duplicate rows
train_data.duplicated().sum()
# ***MACHINE LEARNING MODEL***
x_train = train_data.drop("loss", axis=1)
y_train = train_data["loss"]
x_test = test_data
y_test = pd.read_csv(
"../input/tabular-playground-series-aug-2021/sample_submission.csv"
)
id_loc = np.array(y_test["id"])
y_test = y_test.drop("id", axis=1)
from sklearn.linear_model import LinearRegression
regressor = LinearRegression()
regressor.fit(x_train, y_train)
y_pred = regressor.predict(x_test)
len(y_pred)
type(y_pred)
output = pd.DataFrame({"id": id_col, "loss": y_pred})
output.head()
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
| false | 0 | 650 | 0 | 650 | 650 |
||
69791299
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # PRODUCT INFO
# ## Understanding the data
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
product_df = pd.read_csv(
"/kaggle/input/learnplatform-covid19-impact-on-digital-learning/products_info.csv"
)
product_df.head()
product_df.describe()
product_df.info()
# dropping null objects
product_df.dropna(inplace=True)
# `Sector(s)` and `Primary Essential Function` are the two columns I will primarily focus on to draw insights about the direction in which the Digital Learning Industry is going.
# ## Exploratory Data Analysis
# Let us find out which sector(s) do most companies deal with **`Sectors`**
pie = product_df.groupby("Sector(s)").count()[["LP ID"]]
pie["percent"] = pie["LP ID"] / pie["LP ID"].sum() * 100
x = list(pie["percent"])
y = []
for i in x:
y.append(str(i))
pie
sns.set_style("darkgrid")
plt.figure(figsize=(10, 10))
plt.pie(
pie["LP ID"],
labels=pie.index,
explode=[0, 0.4, 0, 0, 0],
colors=("blue", "cyan", "orange", "beige", "green"),
autopct="%1.1f%%",
)
plt.title("Sector(s) in which most Digital Learning providers are active in")
plt.legend(title="Sector(s):")
plt.figure(figsize=(10, 10))
plt.title("Active sectors")
plt.ylabel("Sector(s)")
plt.xlabel("No. of providers")
sns.barplot(x=list(pie["LP ID"]), y=pie.index)
product_df[product_df["Sector(s)"] == "Higher Ed; Corporate"][
"Product Name"
], product_df[product_df["Sector(s)"] == "Corporate"]["Product Name"]
# **Except for two providers all the other providers are concerned about `PreK-12`**
# >`Qualtrics` : *Deals with Higher Ed & Corporates*
# > `Weebly` : *Deals only with Corporates*
# **`Primary Essential Function` : The main objective of the company/provider**
# We first need to understand the following key words.
# 1. $LC : Learning & Curriculum$
# 2. $CM : Classroom Management & others$
# 3. $SDO = School & District Operations$
#
def pef1(data):
return data.split("-")[0]
def pef2(data):
return data.split("-")[0] + "-" + data.split("-")[1]
product_df1 = product_df.sort_values(by="Primary Essential Function", axis=0)
product_df1["PEF-1"] = product_df["Primary Essential Function"].apply(pef1)
product_df1["PEF-2"] = product_df["Primary Essential Function"].apply(pef2)
pef = product_df1.groupby("Primary Essential Function", sort=False).count()[["LP ID"]]
pef["percent"] = pef["LP ID"] / pef["LP ID"].sum() * 100
pef
def pef1(data):
return data.split("-")[0]
def pef2(data):
return data.split("-")[0] + "-" + data.split("-")[1]
product_df1 = product_df.sort_values(by="Primary Essential Function", axis=0)
product_df1["PEF-1"] = product_df["Primary Essential Function"].apply(pef1)
product_df1["PEF-2"] = product_df["Primary Essential Function"].apply(pef2)
product_df1.head()
pef1 = product_df1.groupby("PEF-1", sort=False).count()[["LP ID"]]
pef1["percent"] = pef1["LP ID"] / pef1["LP ID"].sum() * 100
pef1
pef2 = product_df1.groupby("PEF-2", sort=False).count()[["LP ID"]]
pef2["percent"] = pef2["LP ID"] / pef2["LP ID"].sum() * 100
pef2
sns.set_style("darkgrid")
fig, ax = plt.subplots()
ax.axis("equal")
plt.rcParams.update({"text.color": "black", "axes.labelcolor": "black"})
plt.rcParams.update({"font.size": 22})
cm, lc, other, sdo = [plt.cm.Blues, plt.cm.Reds, plt.cm.Greens, plt.cm.pink_r]
ax.pie(
pef1["LP ID"],
colors=[cm(0.6), lc(0.6), other(0.8), sdo(0.6)],
autopct="%1.1f%%",
radius=10,
)
ax.pie(
pef2["LP ID"],
labels=pef2.index,
colors=[
cm(0.2),
cm(0.4),
cm(0.6),
lc(0.5),
lc(0.1),
lc(0.2),
lc(0.3),
lc(0.4),
lc(0.5),
lc(0.6),
lc(0.7),
lc(0.8),
lc(0.9),
other(0.5),
sdo(0.1),
sdo(0.2),
sdo(0.3),
sdo(0.4),
sdo(0.5),
sdo(0.6),
sdo(0.7),
sdo(0.8),
sdo(0.9),
],
autopct="%1.1f%%",
radius=8,
)
plt.title("Primary Essential Functions")
plt.legend(loc="lower right", bbox_to_anchor=(-2, 1.5))
plt.subplots_adjust(left=0.0, bottom=0.1, right=0.85)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/791/69791299.ipynb
| null | null |
[{"Id": 69791299, "ScriptId": 19061506, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6109517, "CreationDate": "08/03/2021 09:04:12", "VersionNumber": 2.0, "Title": "COVID-19 Impact on Digital Learning", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 158.0, "LinesInsertedFromPrevious": 58.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 100.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # PRODUCT INFO
# ## Understanding the data
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
product_df = pd.read_csv(
"/kaggle/input/learnplatform-covid19-impact-on-digital-learning/products_info.csv"
)
product_df.head()
product_df.describe()
product_df.info()
# dropping null objects
product_df.dropna(inplace=True)
# `Sector(s)` and `Primary Essential Function` are the two columns I will primarily focus on to draw insights about the direction in which the Digital Learning Industry is going.
# ## Exploratory Data Analysis
# Let us find out which sector(s) do most companies deal with **`Sectors`**
pie = product_df.groupby("Sector(s)").count()[["LP ID"]]
pie["percent"] = pie["LP ID"] / pie["LP ID"].sum() * 100
x = list(pie["percent"])
y = []
for i in x:
y.append(str(i))
pie
sns.set_style("darkgrid")
plt.figure(figsize=(10, 10))
plt.pie(
pie["LP ID"],
labels=pie.index,
explode=[0, 0.4, 0, 0, 0],
colors=("blue", "cyan", "orange", "beige", "green"),
autopct="%1.1f%%",
)
plt.title("Sector(s) in which most Digital Learning providers are active in")
plt.legend(title="Sector(s):")
plt.figure(figsize=(10, 10))
plt.title("Active sectors")
plt.ylabel("Sector(s)")
plt.xlabel("No. of providers")
sns.barplot(x=list(pie["LP ID"]), y=pie.index)
product_df[product_df["Sector(s)"] == "Higher Ed; Corporate"][
"Product Name"
], product_df[product_df["Sector(s)"] == "Corporate"]["Product Name"]
# **Except for two providers all the other providers are concerned about `PreK-12`**
# >`Qualtrics` : *Deals with Higher Ed & Corporates*
# > `Weebly` : *Deals only with Corporates*
# **`Primary Essential Function` : The main objective of the company/provider**
# We first need to understand the following key words.
# 1. $LC : Learning & Curriculum$
# 2. $CM : Classroom Management & others$
# 3. $SDO = School & District Operations$
#
def pef1(data):
return data.split("-")[0]
def pef2(data):
return data.split("-")[0] + "-" + data.split("-")[1]
product_df1 = product_df.sort_values(by="Primary Essential Function", axis=0)
product_df1["PEF-1"] = product_df["Primary Essential Function"].apply(pef1)
product_df1["PEF-2"] = product_df["Primary Essential Function"].apply(pef2)
pef = product_df1.groupby("Primary Essential Function", sort=False).count()[["LP ID"]]
pef["percent"] = pef["LP ID"] / pef["LP ID"].sum() * 100
pef
def pef1(data):
return data.split("-")[0]
def pef2(data):
return data.split("-")[0] + "-" + data.split("-")[1]
product_df1 = product_df.sort_values(by="Primary Essential Function", axis=0)
product_df1["PEF-1"] = product_df["Primary Essential Function"].apply(pef1)
product_df1["PEF-2"] = product_df["Primary Essential Function"].apply(pef2)
product_df1.head()
pef1 = product_df1.groupby("PEF-1", sort=False).count()[["LP ID"]]
pef1["percent"] = pef1["LP ID"] / pef1["LP ID"].sum() * 100
pef1
pef2 = product_df1.groupby("PEF-2", sort=False).count()[["LP ID"]]
pef2["percent"] = pef2["LP ID"] / pef2["LP ID"].sum() * 100
pef2
sns.set_style("darkgrid")
fig, ax = plt.subplots()
ax.axis("equal")
plt.rcParams.update({"text.color": "black", "axes.labelcolor": "black"})
plt.rcParams.update({"font.size": 22})
cm, lc, other, sdo = [plt.cm.Blues, plt.cm.Reds, plt.cm.Greens, plt.cm.pink_r]
ax.pie(
pef1["LP ID"],
colors=[cm(0.6), lc(0.6), other(0.8), sdo(0.6)],
autopct="%1.1f%%",
radius=10,
)
ax.pie(
pef2["LP ID"],
labels=pef2.index,
colors=[
cm(0.2),
cm(0.4),
cm(0.6),
lc(0.5),
lc(0.1),
lc(0.2),
lc(0.3),
lc(0.4),
lc(0.5),
lc(0.6),
lc(0.7),
lc(0.8),
lc(0.9),
other(0.5),
sdo(0.1),
sdo(0.2),
sdo(0.3),
sdo(0.4),
sdo(0.5),
sdo(0.6),
sdo(0.7),
sdo(0.8),
sdo(0.9),
],
autopct="%1.1f%%",
radius=8,
)
plt.title("Primary Essential Functions")
plt.legend(loc="lower right", bbox_to_anchor=(-2, 1.5))
plt.subplots_adjust(left=0.0, bottom=0.1, right=0.85)
| false | 0 | 1,686 | 0 | 1,686 | 1,686 |
||
69791916
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
train_data = pd.read_csv("../input/titanic/train.csv")
test_data = pd.read_csv("../input/titanic/test.csv")
train_data.head()
train_data.info()
train_data.shape
test_data.shape
train_data.describe()
train_data.isnull().sum()
train_data.drop("Cabin", axis=1, inplace=True)
test_data.drop("Cabin", axis=1, inplace=True)
median_age = train_data["Age"].median()
train_data["Age"].replace(np.nan, median_age, inplace=True)
median_age = test_data["Age"].median()
test_data["Age"].replace(np.nan, median_age, inplace=True)
freq_port = train_data.Embarked.dropna().mode()[0]
train_data["Embarked"] = train_data["Embarked"].fillna(freq_port)
train_data.isnull().sum()
test_data.isnull().sum()
sns.countplot(x="Survived", data=train_data)
sns.countplot(x="Survived", hue="Sex", data=train_data)
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women) * 100
print(" % of women survivers : ", rate_women)
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men) * 100
print(" % of men survivers : ", rate_men)
sns.countplot(x="Survived", hue="Pclass", data=train_data)
class1 = train_data.loc[train_data.Pclass == 1]["Survived"]
rate_class1 = sum(class1) / len(class1) * 100
print(" % of class1 survivers : ", rate_class1)
class2 = train_data.loc[train_data.Pclass == 2]["Survived"]
rate_class2 = sum(class2) / len(class2) * 100
print(" % of class2 survivers : ", rate_class2)
class3 = train_data.loc[train_data.Pclass == 3]["Survived"]
rate_class3 = sum(class3) / len(class3) * 100
print(" % of class3 survivers : ", rate_class3)
sns.countplot(x="Survived", hue="SibSp", data=train_data)
sns.countplot(x="Survived", hue="Parch", data=train_data)
sns.violinplot(x="Survived", y="Age", data=train_data)
sns.countplot(x="Survived", hue="Embarked", data=train_data)
train_data["Sex"] = train_data["Sex"].map({"female": 1, "male": 0}).astype(int)
test_data["Sex"] = test_data["Sex"].map({"female": 1, "male": 0}).astype(int)
train_data.head()
emb_dummy = pd.get_dummies(train_data["Embarked"])
train_data = pd.concat([train_data, emb_dummy], axis=1)
emb_dummy2 = pd.get_dummies(test_data["Embarked"])
test_data = pd.concat([test_data, emb_dummy2], axis=1)
train_data.head()
drop_cols = ["Name", "Ticket", "Fare", "Embarked"]
train_data = train_data.drop(drop_cols, axis=1)
train_data = train_data.drop(["PassengerId"], axis=1)
test_data = test_data.drop(drop_cols, axis=1)
train_data.head()
test_data.head()
train_data.loc[train_data["Age"] <= 16, "Age"] = 0
train_data.loc[(train_data["Age"] > 16) & (train_data["Age"] <= 36), "Age"] = 1
train_data.loc[(train_data["Age"] > 36) & (train_data["Age"] <= 50), "Age"] = 2
train_data.loc[(train_data["Age"] > 50) & (train_data["Age"] <= 64), "Age"] = 3
train_data.loc[train_data["Age"] > 64, "Age"] = 4
test_data.loc[test_data["Age"] <= 16, "Age"] = 0
test_data.loc[(test_data["Age"] > 16) & (test_data["Age"] <= 36), "Age"] = 1
test_data.loc[(test_data["Age"] > 36) & (test_data["Age"] <= 50), "Age"] = 2
test_data.loc[(test_data["Age"] > 50) & (test_data["Age"] <= 64), "Age"] = 3
test_data.loc[test_data["Age"] > 64, "Age"] = 4
train_data.head()
test_data.head()
X_train = train_data.drop(["Survived"], axis=1).values
Y_train = train_data["Survived"].values
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(X_train, Y_train, test_size=0.25)
from sklearn.linear_model import LogisticRegression
regressor = LogisticRegression()
regressor.fit(x_train, y_train)
y_pred = regressor.predict(x_test)
from sklearn.metrics import accuracy_score, confusion_matrix
acc = accuracy_score(y_test, y_pred)
acc
cm = confusion_matrix(y_test, y_pred)
cm
from sklearn.tree import DecisionTreeClassifier
dt = DecisionTreeClassifier()
dt.fit(x_train, y_train)
y_pred1 = dt.predict(x_test)
from sklearn.metrics import accuracy_score, confusion_matrix
acc2 = accuracy_score(y_test, y_pred1)
acc2
from sklearn.ensemble import RandomForestClassifier
rc = RandomForestClassifier(max_depth=9, random_state=0)
rc.fit(x_train, y_train)
y_pred2 = rc.predict(x_test)
acc3 = accuracy_score(y_test, y_pred2)
acc3
regressor.fit(X_train, Y_train)
test = test_data.drop(["PassengerId"], axis=1)
final_pred = regressor.predict(test)
test_data["Survived"] = final_pred
test_data.drop(
["Pclass", "Age", "Sex", "SibSp", "Parch", "C", "Q", "S"], inplace=True, axis=1
)
test_data.to_csv("Submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/791/69791916.ipynb
| null | null |
[{"Id": 69791916, "ScriptId": 19075686, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2144034, "CreationDate": "08/03/2021 09:08:00", "VersionNumber": 2.0, "Title": "Getting started with the ML 30 day", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 158.0, "LinesInsertedFromPrevious": 157.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
train_data = pd.read_csv("../input/titanic/train.csv")
test_data = pd.read_csv("../input/titanic/test.csv")
train_data.head()
train_data.info()
train_data.shape
test_data.shape
train_data.describe()
train_data.isnull().sum()
train_data.drop("Cabin", axis=1, inplace=True)
test_data.drop("Cabin", axis=1, inplace=True)
median_age = train_data["Age"].median()
train_data["Age"].replace(np.nan, median_age, inplace=True)
median_age = test_data["Age"].median()
test_data["Age"].replace(np.nan, median_age, inplace=True)
freq_port = train_data.Embarked.dropna().mode()[0]
train_data["Embarked"] = train_data["Embarked"].fillna(freq_port)
train_data.isnull().sum()
test_data.isnull().sum()
sns.countplot(x="Survived", data=train_data)
sns.countplot(x="Survived", hue="Sex", data=train_data)
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women) * 100
print(" % of women survivers : ", rate_women)
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men) * 100
print(" % of men survivers : ", rate_men)
sns.countplot(x="Survived", hue="Pclass", data=train_data)
class1 = train_data.loc[train_data.Pclass == 1]["Survived"]
rate_class1 = sum(class1) / len(class1) * 100
print(" % of class1 survivers : ", rate_class1)
class2 = train_data.loc[train_data.Pclass == 2]["Survived"]
rate_class2 = sum(class2) / len(class2) * 100
print(" % of class2 survivers : ", rate_class2)
class3 = train_data.loc[train_data.Pclass == 3]["Survived"]
rate_class3 = sum(class3) / len(class3) * 100
print(" % of class3 survivers : ", rate_class3)
sns.countplot(x="Survived", hue="SibSp", data=train_data)
sns.countplot(x="Survived", hue="Parch", data=train_data)
sns.violinplot(x="Survived", y="Age", data=train_data)
sns.countplot(x="Survived", hue="Embarked", data=train_data)
train_data["Sex"] = train_data["Sex"].map({"female": 1, "male": 0}).astype(int)
test_data["Sex"] = test_data["Sex"].map({"female": 1, "male": 0}).astype(int)
train_data.head()
emb_dummy = pd.get_dummies(train_data["Embarked"])
train_data = pd.concat([train_data, emb_dummy], axis=1)
emb_dummy2 = pd.get_dummies(test_data["Embarked"])
test_data = pd.concat([test_data, emb_dummy2], axis=1)
train_data.head()
drop_cols = ["Name", "Ticket", "Fare", "Embarked"]
train_data = train_data.drop(drop_cols, axis=1)
train_data = train_data.drop(["PassengerId"], axis=1)
test_data = test_data.drop(drop_cols, axis=1)
train_data.head()
test_data.head()
train_data.loc[train_data["Age"] <= 16, "Age"] = 0
train_data.loc[(train_data["Age"] > 16) & (train_data["Age"] <= 36), "Age"] = 1
train_data.loc[(train_data["Age"] > 36) & (train_data["Age"] <= 50), "Age"] = 2
train_data.loc[(train_data["Age"] > 50) & (train_data["Age"] <= 64), "Age"] = 3
train_data.loc[train_data["Age"] > 64, "Age"] = 4
test_data.loc[test_data["Age"] <= 16, "Age"] = 0
test_data.loc[(test_data["Age"] > 16) & (test_data["Age"] <= 36), "Age"] = 1
test_data.loc[(test_data["Age"] > 36) & (test_data["Age"] <= 50), "Age"] = 2
test_data.loc[(test_data["Age"] > 50) & (test_data["Age"] <= 64), "Age"] = 3
test_data.loc[test_data["Age"] > 64, "Age"] = 4
train_data.head()
test_data.head()
X_train = train_data.drop(["Survived"], axis=1).values
Y_train = train_data["Survived"].values
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(X_train, Y_train, test_size=0.25)
from sklearn.linear_model import LogisticRegression
regressor = LogisticRegression()
regressor.fit(x_train, y_train)
y_pred = regressor.predict(x_test)
from sklearn.metrics import accuracy_score, confusion_matrix
acc = accuracy_score(y_test, y_pred)
acc
cm = confusion_matrix(y_test, y_pred)
cm
from sklearn.tree import DecisionTreeClassifier
dt = DecisionTreeClassifier()
dt.fit(x_train, y_train)
y_pred1 = dt.predict(x_test)
from sklearn.metrics import accuracy_score, confusion_matrix
acc2 = accuracy_score(y_test, y_pred1)
acc2
from sklearn.ensemble import RandomForestClassifier
rc = RandomForestClassifier(max_depth=9, random_state=0)
rc.fit(x_train, y_train)
y_pred2 = rc.predict(x_test)
acc3 = accuracy_score(y_test, y_pred2)
acc3
regressor.fit(X_train, Y_train)
test = test_data.drop(["PassengerId"], axis=1)
final_pred = regressor.predict(test)
test_data["Survived"] = final_pred
test_data.drop(
["Pclass", "Age", "Sex", "SibSp", "Parch", "C", "Q", "S"], inplace=True, axis=1
)
test_data.to_csv("Submission.csv", index=False)
| false | 0 | 1,793 | 0 | 1,793 | 1,793 |
||
69791145
|
# Logging into Kaggle for the first time can be daunting. Our competitions often have large cash prizes, public leaderboards, and involve complex data. Nevertheless, we really think all data scientists can rapidly learn from machine learning competitions and meaningfully contribute to our community. To give you a clear understanding of how our platform works and a mental model of the type of learning you could do on Kaggle, we've created a Getting Started tutorial for the Titanic competition. It walks you through the initial steps required to get your first decent submission on the leaderboard. By the end of the tutorial, you'll also have a solid understanding of how to use Kaggle's online coding environment, where you'll have trained your own machine learning model.
# So if this is your first time entering a Kaggle competition, regardless of whether you:
# - have experience with handling large datasets,
# - haven't done much coding,
# - are newer to data science, or
# - are relatively experienced (but are just unfamiliar with Kaggle's platform),
# you're in the right place!
# # Part 1: Get started
# In this section, you'll learn more about the competition and make your first submission.
# ## Join the competition!
# The first thing to do is to join the competition! Open a new window with **[the competition page](https://www.kaggle.com/c/titanic)**, and click on the **"Join Competition"** button, if you haven't already. (_If you see a "Submit Predictions" button instead of a "Join Competition" button, you have already joined the competition, and don't need to do so again._)
# 
# This takes you to the rules acceptance page. You must accept the competition rules in order to participate. These rules govern how many submissions you can make per day, the maximum team size, and other competition-specific details. Then, click on **"I Understand and Accept"** to indicate that you will abide by the competition rules.
# ## The challenge
# The competition is simple: we want you to use the Titanic passenger data (name, age, price of ticket, etc) to try to predict who will survive and who will die.
# ## The data
# To take a look at the competition data, click on the **Data tab** at the top of the competition page. Then, scroll down to find the list of files.
# There are three files in the data: (1) **train.csv**, (2) **test.csv**, and (3) **gender_submission.csv**.
# ### (1) train.csv
# **train.csv** contains the details of a subset of the passengers on board (891 passengers, to be exact -- where each passenger gets a different row in the table). To investigate this data, click on the name of the file on the left of the screen. Once you've done this, you can view all of the data in the window.
# 
# The values in the second column (**"Survived"**) can be used to determine whether each passenger survived or not:
# - if it's a "1", the passenger survived.
# - if it's a "0", the passenger died.
# For instance, the first passenger listed in **train.csv** is Mr. Owen Harris Braund. He was 22 years old when he died on the Titanic.
# ### (2) test.csv
# Using the patterns you find in **train.csv**, you have to predict whether the other 418 passengers on board (in **test.csv**) survived.
# Click on **test.csv** (on the left of the screen) to examine its contents. Note that **test.csv** does not have a **"Survived"** column - this information is hidden from you, and how well you do at predicting these hidden values will determine how highly you score in the competition!
# ### (3) gender_submission.csv
# The **gender_submission.csv** file is provided as an example that shows how you should structure your predictions. It predicts that all female passengers survived, and all male passengers died. Your hypotheses regarding survival will probably be different, which will lead to a different submission file. But, just like this file, your submission should have:
# - a **"PassengerId"** column containing the IDs of each passenger from **test.csv**.
# - a **"Survived"** column (that you will create!) with a "1" for the rows where you think the passenger survived, and a "0" where you predict that the passenger died.
# # Part 2: Your coding environment
# In this section, you'll train your own machine learning model to improve your predictions. _If you've never written code before or don't have any experience with machine learning, don't worry! We don't assume any prior experience in this tutorial._
# ## The Notebook
# The first thing to do is to create a Kaggle Notebook where you'll store all of your code. You can use Kaggle Notebooks to getting up and running with writing code quickly, and without having to install anything on your computer. (_If you are interested in deep learning, we also offer free GPU access!_)
# Begin by clicking on the **Code tab** on the competition page. Then, click on **"New Notebook"**.
# 
# Your notebook will take a few seconds to load. In the top left corner, you can see the name of your notebook -- something like **"kernel2daed3cd79"**.
# 
# You can edit the name by clicking on it. Change it to something more descriptive, like **"Getting Started with Titanic"**.
# 
# ## Your first lines of code
# When you start a new notebook, it has two gray boxes for storing code. We refer to these gray boxes as "code cells".
# 
# The first code cell already has some code in it. To run this code, put your cursor in the code cell. (_If your cursor is in the right place, you'll notice a blue vertical line to the left of the gray box._) Then, either hit the play button (which appears to the left of the blue line), or hit **[Shift] + [Enter]** on your keyboard.
# If the code runs successfully, three lines of output are returned. Below, you can see the same code that you just ran, along with the output that you should see in your notebook.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# This shows us where the competition data is stored, so that we can load the files into the notebook. We'll do that next.
# ## Load the data
# The second code cell in your notebook now appears below the three lines of output with the file locations.
# 
# Type the two lines of code below into your second code cell. Then, once you're done, either click on the blue play button, or hit **[Shift] + [Enter]**.
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
# Your code should return the output above, which corresponds to the first five rows of the table in **train.csv**. It's very important that you see this output **in your notebook** before proceeding with the tutorial!
# > _If your code does not produce this output_, double-check that your code is identical to the two lines above. And, make sure your cursor is in the code cell before hitting **[Shift] + [Enter]**.
# The code that you've just written is in the Python programming language. It uses a Python "module" called **pandas** (abbreviated as `pd`) to load the table from the **train.csv** file into the notebook. To do this, we needed to plug in the location of the file (which we saw was `/kaggle/input/titanic/train.csv`).
# > If you're not already familiar with Python (and pandas), the code shouldn't make sense to you -- but don't worry! The point of this tutorial is to (quickly!) make your first submission to the competition. At the end of the tutorial, we suggest resources to continue your learning.
# At this point, you should have at least three code cells in your notebook.
# 
# Copy the code below into the third code cell of your notebook to load the contents of the **test.csv** file. Don't forget to click on the play button (or hit **[Shift] + [Enter]**)!
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
# As before, make sure that you see the output above in your notebook before continuing.
# Once all of the code runs successfully, all of the data (in **train.csv** and **test.csv**) is loaded in the notebook. (_The code above shows only the first 5 rows of each table, but all of the data is there -- all 891 rows of **train.csv** and all 418 rows of **test.csv**!_)
# # Part 3: Your first submission
# Remember our goal: we want to find patterns in **train.csv** that help us predict whether the passengers in **test.csv** survived.
# It might initially feel overwhelming to look for patterns, when there's so much data to sort through. So, we'll start simple.
# ## Explore a pattern
# Remember that the sample submission file in **gender_submission.csv** assumes that all female passengers survived (and all male passengers died).
# Is this a reasonable first guess? We'll check if this pattern holds true in the data (in **train.csv**).
# Copy the code below into a new code cell. Then, run the cell.
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women)
print("% of women who survived:", rate_women)
# Before moving on, make sure that your code returns the output above. The code above calculates the percentage of female passengers (in **train.csv**) who survived.
# Then, run the code below in another code cell:
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men)
print("% of men who survived:", rate_men)
# The code above calculates the percentage of male passengers (in **train.csv**) who survived.
# From this you can see that almost 75% of the women on board survived, whereas only 19% of the men lived to tell about it. Since gender seems to be such a strong indicator of survival, the submission file in **gender_submission.csv** is not a bad first guess!
# But at the end of the day, this gender-based submission bases its predictions on only a single column. As you can imagine, by considering multiple columns, we can discover more complex patterns that can potentially yield better-informed predictions. Since it is quite difficult to consider several columns at once (or, it would take a long time to consider all possible patterns in many different columns simultaneously), we'll use machine learning to automate this for us.
# ## Your first machine learning model
# We'll build what's known as a **random forest model**. This model is constructed of several "trees" (there are three trees in the picture below, but we'll construct 100!) that will individually consider each passenger's data and vote on whether the individual survived. Then, the random forest model makes a democratic decision: the outcome with the most votes wins!
# 
# The code cell below looks for patterns in four different columns (**"Pclass"**, **"Sex"**, **"SibSp"**, and **"Parch"**) of the data. It constructs the trees in the random forest model based on patterns in the **train.csv** file, before generating predictions for the passengers in **test.csv**. The code also saves these new predictions in a CSV file **my_submission.csv**.
# Copy this code into your notebook, and run it in a new code cell.
from sklearn.ensemble import RandomForestClassifier
y = train_data["Survived"]
features = ["Pclass", "Sex", "SibSp", "Parch"]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = RandomForestClassifier(n_estimators=100, max_depth=5, random_state=1)
model.fit(X, y)
predictions = model.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/791/69791145.ipynb
| null | null |
[{"Id": 69791145, "ScriptId": 19076615, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 8041921, "CreationDate": "08/03/2021 09:03:02", "VersionNumber": 1.0, "Title": "Titanic Tutorial", "EvaluationDate": "08/03/2021", "IsChange": false, "TotalLines": 208.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 208.0, "LinesInsertedFromFork": 0.0, "LinesDeletedFromFork": 0.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 208.0, "TotalVotes": 0}]
| null | null | null | null |
# Logging into Kaggle for the first time can be daunting. Our competitions often have large cash prizes, public leaderboards, and involve complex data. Nevertheless, we really think all data scientists can rapidly learn from machine learning competitions and meaningfully contribute to our community. To give you a clear understanding of how our platform works and a mental model of the type of learning you could do on Kaggle, we've created a Getting Started tutorial for the Titanic competition. It walks you through the initial steps required to get your first decent submission on the leaderboard. By the end of the tutorial, you'll also have a solid understanding of how to use Kaggle's online coding environment, where you'll have trained your own machine learning model.
# So if this is your first time entering a Kaggle competition, regardless of whether you:
# - have experience with handling large datasets,
# - haven't done much coding,
# - are newer to data science, or
# - are relatively experienced (but are just unfamiliar with Kaggle's platform),
# you're in the right place!
# # Part 1: Get started
# In this section, you'll learn more about the competition and make your first submission.
# ## Join the competition!
# The first thing to do is to join the competition! Open a new window with **[the competition page](https://www.kaggle.com/c/titanic)**, and click on the **"Join Competition"** button, if you haven't already. (_If you see a "Submit Predictions" button instead of a "Join Competition" button, you have already joined the competition, and don't need to do so again._)
# 
# This takes you to the rules acceptance page. You must accept the competition rules in order to participate. These rules govern how many submissions you can make per day, the maximum team size, and other competition-specific details. Then, click on **"I Understand and Accept"** to indicate that you will abide by the competition rules.
# ## The challenge
# The competition is simple: we want you to use the Titanic passenger data (name, age, price of ticket, etc) to try to predict who will survive and who will die.
# ## The data
# To take a look at the competition data, click on the **Data tab** at the top of the competition page. Then, scroll down to find the list of files.
# There are three files in the data: (1) **train.csv**, (2) **test.csv**, and (3) **gender_submission.csv**.
# ### (1) train.csv
# **train.csv** contains the details of a subset of the passengers on board (891 passengers, to be exact -- where each passenger gets a different row in the table). To investigate this data, click on the name of the file on the left of the screen. Once you've done this, you can view all of the data in the window.
# 
# The values in the second column (**"Survived"**) can be used to determine whether each passenger survived or not:
# - if it's a "1", the passenger survived.
# - if it's a "0", the passenger died.
# For instance, the first passenger listed in **train.csv** is Mr. Owen Harris Braund. He was 22 years old when he died on the Titanic.
# ### (2) test.csv
# Using the patterns you find in **train.csv**, you have to predict whether the other 418 passengers on board (in **test.csv**) survived.
# Click on **test.csv** (on the left of the screen) to examine its contents. Note that **test.csv** does not have a **"Survived"** column - this information is hidden from you, and how well you do at predicting these hidden values will determine how highly you score in the competition!
# ### (3) gender_submission.csv
# The **gender_submission.csv** file is provided as an example that shows how you should structure your predictions. It predicts that all female passengers survived, and all male passengers died. Your hypotheses regarding survival will probably be different, which will lead to a different submission file. But, just like this file, your submission should have:
# - a **"PassengerId"** column containing the IDs of each passenger from **test.csv**.
# - a **"Survived"** column (that you will create!) with a "1" for the rows where you think the passenger survived, and a "0" where you predict that the passenger died.
# # Part 2: Your coding environment
# In this section, you'll train your own machine learning model to improve your predictions. _If you've never written code before or don't have any experience with machine learning, don't worry! We don't assume any prior experience in this tutorial._
# ## The Notebook
# The first thing to do is to create a Kaggle Notebook where you'll store all of your code. You can use Kaggle Notebooks to getting up and running with writing code quickly, and without having to install anything on your computer. (_If you are interested in deep learning, we also offer free GPU access!_)
# Begin by clicking on the **Code tab** on the competition page. Then, click on **"New Notebook"**.
# 
# Your notebook will take a few seconds to load. In the top left corner, you can see the name of your notebook -- something like **"kernel2daed3cd79"**.
# 
# You can edit the name by clicking on it. Change it to something more descriptive, like **"Getting Started with Titanic"**.
# 
# ## Your first lines of code
# When you start a new notebook, it has two gray boxes for storing code. We refer to these gray boxes as "code cells".
# 
# The first code cell already has some code in it. To run this code, put your cursor in the code cell. (_If your cursor is in the right place, you'll notice a blue vertical line to the left of the gray box._) Then, either hit the play button (which appears to the left of the blue line), or hit **[Shift] + [Enter]** on your keyboard.
# If the code runs successfully, three lines of output are returned. Below, you can see the same code that you just ran, along with the output that you should see in your notebook.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# This shows us where the competition data is stored, so that we can load the files into the notebook. We'll do that next.
# ## Load the data
# The second code cell in your notebook now appears below the three lines of output with the file locations.
# 
# Type the two lines of code below into your second code cell. Then, once you're done, either click on the blue play button, or hit **[Shift] + [Enter]**.
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
# Your code should return the output above, which corresponds to the first five rows of the table in **train.csv**. It's very important that you see this output **in your notebook** before proceeding with the tutorial!
# > _If your code does not produce this output_, double-check that your code is identical to the two lines above. And, make sure your cursor is in the code cell before hitting **[Shift] + [Enter]**.
# The code that you've just written is in the Python programming language. It uses a Python "module" called **pandas** (abbreviated as `pd`) to load the table from the **train.csv** file into the notebook. To do this, we needed to plug in the location of the file (which we saw was `/kaggle/input/titanic/train.csv`).
# > If you're not already familiar with Python (and pandas), the code shouldn't make sense to you -- but don't worry! The point of this tutorial is to (quickly!) make your first submission to the competition. At the end of the tutorial, we suggest resources to continue your learning.
# At this point, you should have at least three code cells in your notebook.
# 
# Copy the code below into the third code cell of your notebook to load the contents of the **test.csv** file. Don't forget to click on the play button (or hit **[Shift] + [Enter]**)!
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
# As before, make sure that you see the output above in your notebook before continuing.
# Once all of the code runs successfully, all of the data (in **train.csv** and **test.csv**) is loaded in the notebook. (_The code above shows only the first 5 rows of each table, but all of the data is there -- all 891 rows of **train.csv** and all 418 rows of **test.csv**!_)
# # Part 3: Your first submission
# Remember our goal: we want to find patterns in **train.csv** that help us predict whether the passengers in **test.csv** survived.
# It might initially feel overwhelming to look for patterns, when there's so much data to sort through. So, we'll start simple.
# ## Explore a pattern
# Remember that the sample submission file in **gender_submission.csv** assumes that all female passengers survived (and all male passengers died).
# Is this a reasonable first guess? We'll check if this pattern holds true in the data (in **train.csv**).
# Copy the code below into a new code cell. Then, run the cell.
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women)
print("% of women who survived:", rate_women)
# Before moving on, make sure that your code returns the output above. The code above calculates the percentage of female passengers (in **train.csv**) who survived.
# Then, run the code below in another code cell:
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men)
print("% of men who survived:", rate_men)
# The code above calculates the percentage of male passengers (in **train.csv**) who survived.
# From this you can see that almost 75% of the women on board survived, whereas only 19% of the men lived to tell about it. Since gender seems to be such a strong indicator of survival, the submission file in **gender_submission.csv** is not a bad first guess!
# But at the end of the day, this gender-based submission bases its predictions on only a single column. As you can imagine, by considering multiple columns, we can discover more complex patterns that can potentially yield better-informed predictions. Since it is quite difficult to consider several columns at once (or, it would take a long time to consider all possible patterns in many different columns simultaneously), we'll use machine learning to automate this for us.
# ## Your first machine learning model
# We'll build what's known as a **random forest model**. This model is constructed of several "trees" (there are three trees in the picture below, but we'll construct 100!) that will individually consider each passenger's data and vote on whether the individual survived. Then, the random forest model makes a democratic decision: the outcome with the most votes wins!
# 
# The code cell below looks for patterns in four different columns (**"Pclass"**, **"Sex"**, **"SibSp"**, and **"Parch"**) of the data. It constructs the trees in the random forest model based on patterns in the **train.csv** file, before generating predictions for the passengers in **test.csv**. The code also saves these new predictions in a CSV file **my_submission.csv**.
# Copy this code into your notebook, and run it in a new code cell.
from sklearn.ensemble import RandomForestClassifier
y = train_data["Survived"]
features = ["Pclass", "Sex", "SibSp", "Parch"]
X = pd.get_dummies(train_data[features])
X_test = pd.get_dummies(test_data[features])
model = RandomForestClassifier(n_estimators=100, max_depth=5, random_state=1)
model.fit(X, y)
predictions = model.predict(X_test)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
| false | 0 | 3,223 | 0 | 3,223 | 3,223 |
||
69791319
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib as plt
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv("/kaggle/input/titanic/train.csv", index_col="PassengerId")
test = pd.read_csv("/kaggle/input/titanic/test.csv", index_col="PassengerId")
train.head()
train.info()
test.info()
train["Age"] = train["Age"].fillna(train["Age"].mean())
test["Age"] = test["Age"].fillna(test["Age"].mean())
sns.barplot(x="Sex", y="Survived", data=train)
print(
"male:", train["Survived"][train["Sex"] == "male"].value_counts(normalize=True)[1]
)
print(
"female:",
train["Survived"][train["Sex"] == "female"].value_counts(normalize=True)[1],
)
sns.barplot(x="Pclass", y="Survived", data=train)
print(
"Pclass1:", train["Survived"][train["Pclass"] == 1].value_counts(normalize=True)[1]
)
print(
"Pclass2:", train["Survived"][train["Pclass"] == 2].value_counts(normalize=True)[1]
)
print(
"Pclass3:", train["Survived"][train["Pclass"] == 3].value_counts(normalize=True)[1]
)
sns.barplot(x="SibSp", y="Survived", data=train)
print("SibSp0:", train["Survived"][train["SibSp"] == 0].value_counts(normalize=True)[1])
print("SibSp1:", train["Survived"][train["SibSp"] == 1].value_counts(normalize=True)[1])
print("SibSp2:", train["Survived"][train["SibSp"] == 2].value_counts(normalize=True)[1])
print("SibSp3:", train["Survived"][train["SibSp"] == 3].value_counts(normalize=True)[1])
print("SibSp4:", train["Survived"][train["SibSp"] == 4].value_counts(normalize=True)[1])
sns.barplot(x="Parch", y="Survived", data=train)
print("Parch0:", train["Survived"][train["Parch"] == 0].value_counts(normalize=True)[1])
print("Parch1:", train["Survived"][train["Parch"] == 1].value_counts(normalize=True)[1])
print("Parch2:", train["Survived"][train["Parch"] == 2].value_counts(normalize=True)[1])
print("Parch3:", train["Survived"][train["Parch"] == 3].value_counts(normalize=True)[1])
bins = [0, 5, 12, 18, 30, 60, np.inf]
labels = ["baby", "children", "teenagers", "young adults", "adults", "seniors"]
train["AgeG"] = pd.cut(train["Age"], bins, labels=labels)
test["AgeG"] = pd.cut(test["Age"], bins, labels=labels)
sns.barplot(x="AgeG", y="Survived", data=train)
for name in labels:
print(
name, train["Survived"][train["AgeG"] == name].value_counts(normalize=True)[1]
)
test["Fare"] = test["Fare"].fillna(test["Fare"].mean())
bins = [-1, 10, 30, 50, 100, 300, np.inf]
labels = ["0", "1", "3", "4", "5", "6"]
train["FareG"] = pd.cut(train["Fare"], bins, labels=labels)
test["FareG"] = pd.cut(test["Fare"], bins, labels=labels)
test.Fare.describe()
sns.barplot(x=train["FareG"], y=train["Survived"])
train["Embarked"] = train["Embarked"].fillna("S")
train["Cabinboo"] = train["Cabin"].notnull().astype(int)
test["Cabinboo"] = test["Cabin"].notnull().astype(int)
train.Cabinboo
sns.barplot(x="Cabinboo", y="Survived", data=train)
train = train.drop(["Ticket"], axis=1)
test = test.drop(["Ticket"], axis=1)
test = test.drop("Cabin", axis=1)
Sex_map = {"male": 0, "female": 1}
train["Sex"] = train["Sex"].map(Sex_map)
test["Sex"] = test["Sex"].map(Sex_map)
Embarked_map = {"S": 1, "C": 2, "Q": 3}
train["Embarked"] = train["Embarked"].map(Embarked_map)
train.head()
test["Embarked"] = test["Embarked"].map(Embarked_map)
AgeG_map = {
"baby": 0,
"children": 1,
"teenagers": 2,
"young adults": 3,
"adults": 4,
"seniors": 5,
}
train["AgeG"] = train["AgeG"].map(AgeG_map)
test["AgeG"] = test["AgeG"].map(AgeG_map)
train.head()
test.info()
train.head()
train["AgeG"] = train["AgeG"].astype(int)
train["FareG"] = train["FareG"].astype(int)
test["AgeG"] = test["AgeG"].astype(int)
test["FareG"] = test["FareG"].astype(int)
train = train.drop("Cabin", axis=1)
cols = ["Name", "Age", "Fare"]
train = train.drop(cols, axis=1)
test = test.drop(cols, axis=1)
train.head()
from sklearn.model_selection import train_test_split
pred = train.drop("Survived", axis=1)
tar = train["Survived"]
x_train, x_val, y_train, y_val = train_test_split(pred, tar, test_size=0.2)
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score
GNB = GaussianNB()
GNB.fit(x_train, y_train)
y_pred = GNB.predict(x_val)
GNBscore = accuracy_score(y_pred, y_val)
GNBscore
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression()
lr.fit(x_train, y_train)
y_pred = lr.predict(x_val)
lrscore = accuracy_score(y_pred, y_val)
lrscore
from sklearn.svm import SVC
svm = SVC()
svm.fit(x_train, y_train)
y_pred = svm.predict(x_val)
svmscore = accuracy_score(y_pred, y_val)
svmscore
from sklearn.ensemble import BaggingClassifier
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(criterion="entropy", max_depth=None)
bag = BaggingClassifier(
base_estimator=tree,
n_estimators=500,
max_samples=1.0,
max_features=1.0,
bootstrap=True,
)
bag.fit(x_train, y_train)
y_pred = bag.predict(x_val)
bagscore = accuracy_score(y_pred, y_val)
bagscore
from sklearn.ensemble import AdaBoostClassifier
ada = AdaBoostClassifier(base_estimator=tree)
ada.fit(x_train, y_train)
y_pred = ada.predict(x_val)
adascore = accuracy_score(y_pred, y_val)
adascore
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators=500, max_depth=5)
rf.fit(x_train, y_train)
y_pred = rf.predict(x_val)
rfscore = accuracy_score(y_pred, y_val)
rfscore
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(x_train, y_train)
y_pred = knn.predict(x_val)
knnscore = accuracy_score(y_pred, y_val)
knnscore
from xgboost import XGBClassifier
xgb = XGBClassifier(n_estimators=500, max_depth=5)
xgb.fit(x_train, y_train)
y_pred = xgb.predict(x_val)
xgbscore = accuracy_score(y_pred, y_val)
xgbscore
ids = test.index
predictions = svm.predict(test)
output = pd.DataFrame({"PassengerId": ids, "Survived": predictions})
output.to_csv("submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/791/69791319.ipynb
| null | null |
[{"Id": 69791319, "ScriptId": 19056105, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7332515, "CreationDate": "08/03/2021 09:04:19", "VersionNumber": 2.0, "Title": "notebook8189b41d01", "EvaluationDate": "08/03/2021", "IsChange": true, "TotalLines": 189.0, "LinesInsertedFromPrevious": 133.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 56.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib as plt
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv("/kaggle/input/titanic/train.csv", index_col="PassengerId")
test = pd.read_csv("/kaggle/input/titanic/test.csv", index_col="PassengerId")
train.head()
train.info()
test.info()
train["Age"] = train["Age"].fillna(train["Age"].mean())
test["Age"] = test["Age"].fillna(test["Age"].mean())
sns.barplot(x="Sex", y="Survived", data=train)
print(
"male:", train["Survived"][train["Sex"] == "male"].value_counts(normalize=True)[1]
)
print(
"female:",
train["Survived"][train["Sex"] == "female"].value_counts(normalize=True)[1],
)
sns.barplot(x="Pclass", y="Survived", data=train)
print(
"Pclass1:", train["Survived"][train["Pclass"] == 1].value_counts(normalize=True)[1]
)
print(
"Pclass2:", train["Survived"][train["Pclass"] == 2].value_counts(normalize=True)[1]
)
print(
"Pclass3:", train["Survived"][train["Pclass"] == 3].value_counts(normalize=True)[1]
)
sns.barplot(x="SibSp", y="Survived", data=train)
print("SibSp0:", train["Survived"][train["SibSp"] == 0].value_counts(normalize=True)[1])
print("SibSp1:", train["Survived"][train["SibSp"] == 1].value_counts(normalize=True)[1])
print("SibSp2:", train["Survived"][train["SibSp"] == 2].value_counts(normalize=True)[1])
print("SibSp3:", train["Survived"][train["SibSp"] == 3].value_counts(normalize=True)[1])
print("SibSp4:", train["Survived"][train["SibSp"] == 4].value_counts(normalize=True)[1])
sns.barplot(x="Parch", y="Survived", data=train)
print("Parch0:", train["Survived"][train["Parch"] == 0].value_counts(normalize=True)[1])
print("Parch1:", train["Survived"][train["Parch"] == 1].value_counts(normalize=True)[1])
print("Parch2:", train["Survived"][train["Parch"] == 2].value_counts(normalize=True)[1])
print("Parch3:", train["Survived"][train["Parch"] == 3].value_counts(normalize=True)[1])
bins = [0, 5, 12, 18, 30, 60, np.inf]
labels = ["baby", "children", "teenagers", "young adults", "adults", "seniors"]
train["AgeG"] = pd.cut(train["Age"], bins, labels=labels)
test["AgeG"] = pd.cut(test["Age"], bins, labels=labels)
sns.barplot(x="AgeG", y="Survived", data=train)
for name in labels:
print(
name, train["Survived"][train["AgeG"] == name].value_counts(normalize=True)[1]
)
test["Fare"] = test["Fare"].fillna(test["Fare"].mean())
bins = [-1, 10, 30, 50, 100, 300, np.inf]
labels = ["0", "1", "3", "4", "5", "6"]
train["FareG"] = pd.cut(train["Fare"], bins, labels=labels)
test["FareG"] = pd.cut(test["Fare"], bins, labels=labels)
test.Fare.describe()
sns.barplot(x=train["FareG"], y=train["Survived"])
train["Embarked"] = train["Embarked"].fillna("S")
train["Cabinboo"] = train["Cabin"].notnull().astype(int)
test["Cabinboo"] = test["Cabin"].notnull().astype(int)
train.Cabinboo
sns.barplot(x="Cabinboo", y="Survived", data=train)
train = train.drop(["Ticket"], axis=1)
test = test.drop(["Ticket"], axis=1)
test = test.drop("Cabin", axis=1)
Sex_map = {"male": 0, "female": 1}
train["Sex"] = train["Sex"].map(Sex_map)
test["Sex"] = test["Sex"].map(Sex_map)
Embarked_map = {"S": 1, "C": 2, "Q": 3}
train["Embarked"] = train["Embarked"].map(Embarked_map)
train.head()
test["Embarked"] = test["Embarked"].map(Embarked_map)
AgeG_map = {
"baby": 0,
"children": 1,
"teenagers": 2,
"young adults": 3,
"adults": 4,
"seniors": 5,
}
train["AgeG"] = train["AgeG"].map(AgeG_map)
test["AgeG"] = test["AgeG"].map(AgeG_map)
train.head()
test.info()
train.head()
train["AgeG"] = train["AgeG"].astype(int)
train["FareG"] = train["FareG"].astype(int)
test["AgeG"] = test["AgeG"].astype(int)
test["FareG"] = test["FareG"].astype(int)
train = train.drop("Cabin", axis=1)
cols = ["Name", "Age", "Fare"]
train = train.drop(cols, axis=1)
test = test.drop(cols, axis=1)
train.head()
from sklearn.model_selection import train_test_split
pred = train.drop("Survived", axis=1)
tar = train["Survived"]
x_train, x_val, y_train, y_val = train_test_split(pred, tar, test_size=0.2)
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score
GNB = GaussianNB()
GNB.fit(x_train, y_train)
y_pred = GNB.predict(x_val)
GNBscore = accuracy_score(y_pred, y_val)
GNBscore
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression()
lr.fit(x_train, y_train)
y_pred = lr.predict(x_val)
lrscore = accuracy_score(y_pred, y_val)
lrscore
from sklearn.svm import SVC
svm = SVC()
svm.fit(x_train, y_train)
y_pred = svm.predict(x_val)
svmscore = accuracy_score(y_pred, y_val)
svmscore
from sklearn.ensemble import BaggingClassifier
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(criterion="entropy", max_depth=None)
bag = BaggingClassifier(
base_estimator=tree,
n_estimators=500,
max_samples=1.0,
max_features=1.0,
bootstrap=True,
)
bag.fit(x_train, y_train)
y_pred = bag.predict(x_val)
bagscore = accuracy_score(y_pred, y_val)
bagscore
from sklearn.ensemble import AdaBoostClassifier
ada = AdaBoostClassifier(base_estimator=tree)
ada.fit(x_train, y_train)
y_pred = ada.predict(x_val)
adascore = accuracy_score(y_pred, y_val)
adascore
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators=500, max_depth=5)
rf.fit(x_train, y_train)
y_pred = rf.predict(x_val)
rfscore = accuracy_score(y_pred, y_val)
rfscore
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(x_train, y_train)
y_pred = knn.predict(x_val)
knnscore = accuracy_score(y_pred, y_val)
knnscore
from xgboost import XGBClassifier
xgb = XGBClassifier(n_estimators=500, max_depth=5)
xgb.fit(x_train, y_train)
y_pred = xgb.predict(x_val)
xgbscore = accuracy_score(y_pred, y_val)
xgbscore
ids = test.index
predictions = svm.predict(test)
output = pd.DataFrame({"PassengerId": ids, "Survived": predictions})
output.to_csv("submission.csv", index=False)
| false | 0 | 2,321 | 0 | 2,321 | 2,321 |
||
69338405
|
<jupyter_start><jupyter_text>OpenAQ
OpenAQ is an open-source project to surface live, real-time air quality data from around the world. Their “mission is to enable previously impossible science, impact policy and empower the public to fight air pollution.” The data includes air quality measurements from 5490 locations in 47 countries.
Scientists, researchers, developers, and citizens can use this data to understand the quality of air near them currently. The dataset only includes the most current measurement available for the location (no historical data).
Update Frequency: Weekly
### Querying BigQuery tables
You can use the BigQuery Python client library to query tables in this dataset in Kernels. Note that methods available in Kernels are limited to querying data. Tables are at `bigquery-public-data.openaq.[TABLENAME]`. **[Fork this kernel to get started](https://www.kaggle.com/sohier/how-to-integrate-bigquery-pandas)**.
Kaggle dataset identifier: openaq
<jupyter_script>from google.cloud import bigquery
client = bigquery.Client()
# Construct a reference to the "" dataset
dataset_ref = client.dataset("openaq", project="bigquery-public-data")
dataset = client.get_dataset(dataset_ref)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338405.ipynb
|
openaq
| null |
[{"Id": 69338405, "ScriptId": 18929510, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 3404384, "CreationDate": "07/29/2021 18:30:43", "VersionNumber": 3.0, "Title": "KsBabyukNotebok", "EvaluationDate": "07/29/2021", "IsChange": true, "TotalLines": 15.0, "LinesInsertedFromPrevious": 10.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 5.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 92384003, "KernelVersionId": 69338405, "SourceDatasetVersionId": 8677}]
|
[{"Id": 8677, "DatasetId": 5836, "DatasourceVersionId": 8677, "CreatorUserId": 1132983, "LicenseName": "CC BY-SA 4.0", "CreationDate": "12/01/2017 18:08:44", "VersionNumber": 1.0, "Title": "OpenAQ", "Slug": "openaq", "Subtitle": "Global Air Pollution Measurements", "Description": "OpenAQ is an open-source project to surface live, real-time air quality data from around the world. Their \u201cmission is to enable previously impossible science, impact policy and empower the public to fight air pollution.\u201d The data includes air quality measurements from 5490 locations in 47 countries.\n\nScientists, researchers, developers, and citizens can use this data to understand the quality of air near them currently. The dataset only includes the most current measurement available for the location (no historical data). \n\nUpdate Frequency: Weekly\n\n### Querying BigQuery tables\n\nYou can use the BigQuery Python client library to query tables in this dataset in Kernels. Note that methods available in Kernels are limited to querying data. Tables are at `bigquery-public-data.openaq.[TABLENAME]`. **[Fork this kernel to get started](https://www.kaggle.com/sohier/how-to-integrate-bigquery-pandas)**.\n\n### Acknowledgements\n\nDataset Source: openaq.org\n\nUse: This dataset is publicly available for anyone to use under the following terms provided by the [Dataset Source](https://openaq.org/#/about?_k=s3aspo) and is provided \"AS IS\" without any warranty, express or implied.", "VersionNotes": "Initial release", "TotalCompressedBytes": 2394241.0, "TotalUncompressedBytes": 2394241.0}]
|
[{"Id": 5836, "CreatorUserId": 1132983, "OwnerUserId": NaN, "OwnerOrganizationId": 1192.0, "CurrentDatasetVersionId": 8677.0, "CurrentDatasourceVersionId": 8677.0, "ForumId": 12127, "Type": 2, "CreationDate": "12/01/2017 18:08:44", "LastActivityDate": "12/01/2017", "TotalViews": 53930, "TotalDownloads": 0, "TotalVotes": 218, "TotalKernels": 1497}]
| null |
from google.cloud import bigquery
client = bigquery.Client()
# Construct a reference to the "" dataset
dataset_ref = client.dataset("openaq", project="bigquery-public-data")
dataset = client.get_dataset(dataset_ref)
| false | 0 | 59 | 1 | 289 | 59 |
||
69338051
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# data analysis and wrangling
import pandas as pd
import numpy as np
import random as rnd
# visualization
import seaborn as sns
import matplotlib.pyplot as plt
# machine learning
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC, LinearSVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import Perceptron
from sklearn.linear_model import SGDClassifier
from sklearn.tree import DecisionTreeClassifier
train_df = pd.read_csv("/kaggle/input/titanic/train.csv")
test_df = pd.read_csv("/kaggle/input/titanic/test.csv")
train_df.info()
grid = sns.FacetGrid(train_df, row="Survived", col="Pclass", size=2.2, aspect=1.6)
grid.map(plt.hist, "Age", alpha=0.5, bins=20)
grid.add_legend()
grid = sns.FacetGrid(train_df, col="Survived", row="Pclass", size=2.2, aspect=1.6)
grid.map(plt.hist, "Sex", alpha=0.5, bins=20)
grid.add_legend()
grid = sns.FacetGrid(train_df, col="Survived", row="Sex", size=2.2, aspect=1.6)
grid.map(plt.hist, "Parch", alpha=0.5, bins=20)
grid.add_legend()
train_df["Sex_Pclass"] = train_df["Sex"].astype(str) + train_df["Pclass"].astype(str)
# train_df['Age_Pclass'] = train_df['Pclass'].astype(str) + train_df['Age'].astype(str)
train_df[["Sex_Pclass", "Survived"]].groupby(
["Sex_Pclass"], as_index=False
).mean().sort_values(by="Survived", ascending=False)
# train_df[['Age_Pclass', 'Survived']].groupby(['Age_Pclass'], as_index=False).mean().sort_values(by='Survived', ascending=False)
y = train_df["Survived"]
x = train_df.drop(["Survived", "Name", "PassengerId"], axis=1)
# Correlating.
# # We want to know how well does each feature correlate with Survival. We want to do this early in our project and match these quick correlations with modelled correlations later in the project.
train_df[["Pclass", "Survived"]].groupby(["Pclass"], as_index=False).mean().sort_values(
by="Survived", ascending=False
)
train_df[["Sex", "Survived"]].groupby(["Sex"], as_index=False).mean().sort_values(
by="Survived", ascending=False
)
train_df[["SibSp", "Survived"]].groupby(["SibSp"], as_index=False).mean().sort_values(
by="Survived", ascending=False
)
train_df[["Parch", "Survived"]].groupby(["Parch"], as_index=False).mean().sort_values(
by="Survived", ascending=False
)
# # Analyze by visualizing data
g = sns.FacetGrid(train_df, col="Survived")
g.map(plt.hist, "Age", bins=20)
x.isnull().sum().sort_values(ascending=False)
x["Age"] = x["Age"].fillna(x.Age.mean())
x["Embarked"] = x["Embarked"].fillna("missing")
x["Cabin"] = x["Cabin"].fillna("missing")
x.isnull().sum().sort_values(ascending=False)
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
x["Ticket"] = le.fit_transform(x["Ticket"])
x["Cabin"] = le.fit_transform(x["Cabin"])
x["Embarked"] = le.fit_transform(x["Embarked"])
x = pd.get_dummies(x)
x.corrwith(y, axis=0)
# Split the data into train and test
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(x, y, test_size=0.25, stratify=y)
# Logistic Regression
logreg = LogisticRegression(solver="liblinear")
logreg.fit(X_train, y_train)
Y_pred = logreg.predict(X_valid)
acc_log = round(logreg.score(X_train, y_train) * 100, 2)
print("mmmm", acc_log)
score = accuracy_score(y_valid, Y_pred)
# acc_svc = round(svc.score(X_train, y_train) * 100, 2)
print("Accuracy score : ", score)
# Support Vector Machines
from sklearn.metrics import accuracy_score
svc = SVC()
svc.fit(X_train, y_train)
Y_pred = svc.predict(X_valid)
score = accuracy_score(y_valid, Y_pred)
# acc_svc = round(svc.score(X_train, y_train) * 100, 2)
score
knn = KNeighborsClassifier(n_neighbors=3)
knn.fit(X_train, y_train)
Y_pred = knn.predict(X_valid)
acc_knn = round(knn.score(X_train, y_train) * 100, 2)
acc_knn
score = accuracy_score(y_valid, Y_pred)
score
# Gaussian Naive Bayes
gaussian = GaussianNB()
gaussian.fit(X_train, y_train)
Y_pred = gaussian.predict(X_valid)
acc_gaussian = round(gaussian.score(X_train, y_train) * 100, 2)
acc_gaussian
score = accuracy_score(y_valid, Y_pred)
score
# Linear SVC
linear_svc = LinearSVC()
linear_svc.fit(X_train, y_train)
Y_pred = linear_svc.predict(X_valid)
acc_linear_svc = round(linear_svc.score(X_train, y_train) * 100, 2)
acc_linear_svc
score = accuracy_score(y_valid, Y_pred)
score
# # Test
test_df["Sex_Pclass"] = test_df["Sex"].astype(str) + test_df["Pclass"].astype(str)
x_test = test_df.drop(["Name", "PassengerId"], axis=1)
x_test["Age"] = x_test["Age"].fillna(x_test.Age.mean())
x_test["Embarked"] = x_test["Embarked"].fillna("missing")
x_test["Cabin"] = x_test["Cabin"].fillna("missing")
x_test["Fare"] = x_test["Fare"].fillna(x_test.Fare.mean())
x_test["Ticket"] = le.fit_transform(x_test["Ticket"])
x_test["Cabin"] = le.fit_transform(x_test["Cabin"])
x_test["Embarked"] = le.fit_transform(x_test["Embarked"])
x_test = pd.get_dummies(x_test)
X_train.info()
x_test.info()
y_pred_test = logreg.predict(x_test)
submission = pd.read_csv("/kaggle/input/titanic/test.csv")
submission_df = pd.DataFrame()
submission_df["PassengerId"] = submission.PassengerId
submission_df["Survived"] = y_pred_test
submission_df.to_csv("submission.csv", index=False, header=True)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338051.ipynb
| null | null |
[{"Id": 69338051, "ScriptId": 18828625, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7771740, "CreationDate": "07/29/2021 18:23:25", "VersionNumber": 1.0, "Title": "Titanic_Competition", "EvaluationDate": "07/29/2021", "IsChange": true, "TotalLines": 195.0, "LinesInsertedFromPrevious": 195.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# data analysis and wrangling
import pandas as pd
import numpy as np
import random as rnd
# visualization
import seaborn as sns
import matplotlib.pyplot as plt
# machine learning
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC, LinearSVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import Perceptron
from sklearn.linear_model import SGDClassifier
from sklearn.tree import DecisionTreeClassifier
train_df = pd.read_csv("/kaggle/input/titanic/train.csv")
test_df = pd.read_csv("/kaggle/input/titanic/test.csv")
train_df.info()
grid = sns.FacetGrid(train_df, row="Survived", col="Pclass", size=2.2, aspect=1.6)
grid.map(plt.hist, "Age", alpha=0.5, bins=20)
grid.add_legend()
grid = sns.FacetGrid(train_df, col="Survived", row="Pclass", size=2.2, aspect=1.6)
grid.map(plt.hist, "Sex", alpha=0.5, bins=20)
grid.add_legend()
grid = sns.FacetGrid(train_df, col="Survived", row="Sex", size=2.2, aspect=1.6)
grid.map(plt.hist, "Parch", alpha=0.5, bins=20)
grid.add_legend()
train_df["Sex_Pclass"] = train_df["Sex"].astype(str) + train_df["Pclass"].astype(str)
# train_df['Age_Pclass'] = train_df['Pclass'].astype(str) + train_df['Age'].astype(str)
train_df[["Sex_Pclass", "Survived"]].groupby(
["Sex_Pclass"], as_index=False
).mean().sort_values(by="Survived", ascending=False)
# train_df[['Age_Pclass', 'Survived']].groupby(['Age_Pclass'], as_index=False).mean().sort_values(by='Survived', ascending=False)
y = train_df["Survived"]
x = train_df.drop(["Survived", "Name", "PassengerId"], axis=1)
# Correlating.
# # We want to know how well does each feature correlate with Survival. We want to do this early in our project and match these quick correlations with modelled correlations later in the project.
train_df[["Pclass", "Survived"]].groupby(["Pclass"], as_index=False).mean().sort_values(
by="Survived", ascending=False
)
train_df[["Sex", "Survived"]].groupby(["Sex"], as_index=False).mean().sort_values(
by="Survived", ascending=False
)
train_df[["SibSp", "Survived"]].groupby(["SibSp"], as_index=False).mean().sort_values(
by="Survived", ascending=False
)
train_df[["Parch", "Survived"]].groupby(["Parch"], as_index=False).mean().sort_values(
by="Survived", ascending=False
)
# # Analyze by visualizing data
g = sns.FacetGrid(train_df, col="Survived")
g.map(plt.hist, "Age", bins=20)
x.isnull().sum().sort_values(ascending=False)
x["Age"] = x["Age"].fillna(x.Age.mean())
x["Embarked"] = x["Embarked"].fillna("missing")
x["Cabin"] = x["Cabin"].fillna("missing")
x.isnull().sum().sort_values(ascending=False)
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
x["Ticket"] = le.fit_transform(x["Ticket"])
x["Cabin"] = le.fit_transform(x["Cabin"])
x["Embarked"] = le.fit_transform(x["Embarked"])
x = pd.get_dummies(x)
x.corrwith(y, axis=0)
# Split the data into train and test
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(x, y, test_size=0.25, stratify=y)
# Logistic Regression
logreg = LogisticRegression(solver="liblinear")
logreg.fit(X_train, y_train)
Y_pred = logreg.predict(X_valid)
acc_log = round(logreg.score(X_train, y_train) * 100, 2)
print("mmmm", acc_log)
score = accuracy_score(y_valid, Y_pred)
# acc_svc = round(svc.score(X_train, y_train) * 100, 2)
print("Accuracy score : ", score)
# Support Vector Machines
from sklearn.metrics import accuracy_score
svc = SVC()
svc.fit(X_train, y_train)
Y_pred = svc.predict(X_valid)
score = accuracy_score(y_valid, Y_pred)
# acc_svc = round(svc.score(X_train, y_train) * 100, 2)
score
knn = KNeighborsClassifier(n_neighbors=3)
knn.fit(X_train, y_train)
Y_pred = knn.predict(X_valid)
acc_knn = round(knn.score(X_train, y_train) * 100, 2)
acc_knn
score = accuracy_score(y_valid, Y_pred)
score
# Gaussian Naive Bayes
gaussian = GaussianNB()
gaussian.fit(X_train, y_train)
Y_pred = gaussian.predict(X_valid)
acc_gaussian = round(gaussian.score(X_train, y_train) * 100, 2)
acc_gaussian
score = accuracy_score(y_valid, Y_pred)
score
# Linear SVC
linear_svc = LinearSVC()
linear_svc.fit(X_train, y_train)
Y_pred = linear_svc.predict(X_valid)
acc_linear_svc = round(linear_svc.score(X_train, y_train) * 100, 2)
acc_linear_svc
score = accuracy_score(y_valid, Y_pred)
score
# # Test
test_df["Sex_Pclass"] = test_df["Sex"].astype(str) + test_df["Pclass"].astype(str)
x_test = test_df.drop(["Name", "PassengerId"], axis=1)
x_test["Age"] = x_test["Age"].fillna(x_test.Age.mean())
x_test["Embarked"] = x_test["Embarked"].fillna("missing")
x_test["Cabin"] = x_test["Cabin"].fillna("missing")
x_test["Fare"] = x_test["Fare"].fillna(x_test.Fare.mean())
x_test["Ticket"] = le.fit_transform(x_test["Ticket"])
x_test["Cabin"] = le.fit_transform(x_test["Cabin"])
x_test["Embarked"] = le.fit_transform(x_test["Embarked"])
x_test = pd.get_dummies(x_test)
X_train.info()
x_test.info()
y_pred_test = logreg.predict(x_test)
submission = pd.read_csv("/kaggle/input/titanic/test.csv")
submission_df = pd.DataFrame()
submission_df["PassengerId"] = submission.PassengerId
submission_df["Survived"] = y_pred_test
submission_df.to_csv("submission.csv", index=False, header=True)
| false | 0 | 2,130 | 0 | 2,130 | 2,130 |
||
69338813
|
import torch
from torch.utils.data.dataset import Dataset
import torchvision
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import torch.nn as nn
from sklearn.model_selection import train_test_split
# * Importing dataset
dataset_train = pd.read_csv("../input/digit-recognizer/train.csv")
dataset_test = pd.read_csv("../input/digit-recognizer/test.csv")
print("Training data length:", len(dataset_train))
# * Having a look at the dataset
targets_np = dataset_train.label.values
features_np = dataset_train.loc[
:, dataset_train.columns != "label"
].values # values range to 255, so to normalize
X_train, X_test, y_train, y_test = train_test_split(
features_np, targets_np, test_size=0.2, random_state=42
)
# features_train = torch.from_numpy(X_train)
# target_train = torch.from_numpy(y_train)
# features_test = torch.from_numpy(X_test)
# target_test = torch.from_numpy(y_test)
print("Training Features Shape", X_train.shape)
print("Training Labels Shape", y_train.shape)
print("Testing Features Shape", X_test.shape)
print("Testing Labels Shape", y_test.shape)
# Viz of an image
plt.imshow(features_np[0].reshape(28, 28), cmap="gray") # images are 28x28
plt.title("Label: " + str(targets_np[0]))
plt.show()
i = 100
plt.imshow(X_test[i].reshape(28, 28), cmap="gray") # images are 28x28
plt.title("Label: " + str(y_test[i]))
plt.show()
from torch.utils.data.sampler import SubsetRandomSampler
from torch.utils.data import DataLoader, Dataset
import torchvision.transforms as tt
def get_default_device():
"""Returns the default Device (cuda if gpu, else cpu"""
if torch.cuda.is_available():
return torch.device("cuda")
else:
torch.device("cpu")
def to_device(data, device):
"""Shifts Data to the input device"""
if isinstance(data, (list, tuple)):
return [to_device(x, device) for x in data]
return data.to(device, non_blocking=True)
class DeviceDataLoader:
def __init__(self, dl, device) -> None:
self.dl = dl
self.device = device
def __iter__(self):
for b in self.dl:
yield to_device(b, self.device)
def __len__(self):
return len(self.dl)
# * Creating a custom dataset overriding torchvision dataset
class MyDataset(Dataset):
def __init__(self, df_features, df_labels, transform=None) -> None:
self.features = df_features
self.labels = df_labels
self.transform = transform
def __getitem__(self, index):
data = self.features[index]
image = data.reshape(28, 28).astype(np.uint8)
label = self.labels[index]
if self.transform:
image = self.transform(image)
return (image, label)
def __len__(self):
return len(self.features)
BATCH_SIZE = 16
# * Augmentation - Random affination (rotation with translation), normalization
train_tmfs = tt.Compose(
[
tt.ToPILImage(),
# tt.RandomRotation(30),
tt.RandomAffine(degrees=30, translate=(0.1, 0.1), scale=(0.9, 1.1)),
# Random Affine suits better for this purpose @https://pytorch.org/vision/stable/auto_examples/plot_transforms.html#randomaffine
tt.ToTensor(),
tt.Normalize(
mean=[features_np.mean() / 255], std=[features_np.std() / 255], inplace=True
),
]
)
test_tmfs = tt.Compose(
[
tt.ToPILImage(),
# tt.RandomRotation(30),
tt.RandomAffine(degrees=30, translate=(0.1, 0.1), scale=(0.9, 1.1)),
# Random Affine suits better for this purpose @https://pytorch.org/vision/stable/auto_examples/plot_transforms.html#randomaffine
tt.ToTensor(),
tt.Normalize(
mean=[features_np.mean() / 255], std=[features_np.std() / 255], inplace=True
),
]
)
# * Setting default Device, GPU
device = get_default_device()
print(device)
# * Creating Dataset
train_set = MyDataset(X_train, y_train, transform=train_tmfs)
test_set = MyDataset(X_test, y_test, transform=test_tmfs)
# * Loading data and then shifting data loader to the selected device
train_dl = DeviceDataLoader(
DataLoader(train_set, batch_size=BATCH_SIZE, shuffle=True), device
)
test_dl = DeviceDataLoader(
DataLoader(test_set, batch_size=BATCH_SIZE, shuffle=True), device
)
# * Sanity Check
imgs, lbls = next(iter(test_dl))
print(imgs.shape)
plt.imshow(torch.Tensor.cpu(imgs[0].data.reshape(28, 28)), cmap="gray")
print("Label:", lbls[0])
plt.show()
import torch.nn.functional as F
# * Following Residual Network https://raw.githubusercontent.com/lambdal/cifar10-fast/master/net.svg
NUM_CLASSES = 10
def convolution_block(in_channels, out_channels, pool=False):
"""Returns a sequence of Convolution, Batch Normalization, Rectified Linear Activation (Relu) and Max Pooling if selected"""
layers = [
nn.Conv2d(
in_channels=in_channels, out_channels=out_channels, kernel_size=3, padding=1
),
nn.BatchNorm2d(out_channels),
nn.ReLU(inplace=True),
]
if pool:
layers.append(nn.MaxPool2d(2, 2)) # 2x2 Max Pooling
return nn.Sequential(*layers)
class Model(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = convolution_block(1, 32)
self.conv2 = convolution_block(32, 64, pool=True)
self.res1 = nn.Sequential(convolution_block(64, 64), convolution_block(64, 64))
self.conv3 = convolution_block(64, 128, pool=True)
self.conv4 = convolution_block(128, 256, pool=True)
self.res2 = nn.Sequential(
convolution_block(256, 256), convolution_block(256, 256)
)
self.classifier = nn.Sequential(
nn.MaxPool2d(2),
nn.Flatten(),
nn.Dropout(0.2), # * added later
nn.Linear(256, NUM_CLASSES),
)
def forward(self, xb):
out = self.conv1(xb) # Prep
out = self.conv2(out) # Layer 1 Convolution Block
out = self.res1(out) + out # Layer 1 Residual Block
out = self.conv3(out) # Layer 2
out = self.conv4(out) # Layer 3 Convolution Block
out = self.res2(out) + out # Layer 3 Residual Block
out = self.classifier(out)
return F.log_softmax(out, dim=1)
model = Model()
to_device(model, device)
# Sanity Check
for images, labels in train_dl:
print(images.shape)
out = model(images)
print(out.shape)
break
"""
Cant find fastai modules
from fastai.basic_data import DataBunch
from fastai.train import Learner
from fastai.metrics import accuracy
"""
import time
@torch.no_grad()
def loss_batch(model, loss_func, xb, yb, opt=None, metric=None):
preds = model(xb)
loss = loss_func(preds, yb)
if opt is not None:
loss.backward()
opt.step()
opt.zero_grad()
metric_result = None
if metric is not None:
metric_result = metric(preds, yb)
return loss.item(), len(xb), metric_result
def accuracy(outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(preds))
def get_lr(optimizer):
for param_group in optimizer.param_groups:
return param_group["lr"]
def fit_one_cycle(
epochs,
max_lr,
model,
train_dl,
test_dl,
weight_decay=0,
grad_clip=None,
opt_func=torch.optim.SGD,
):
def evaluate(model, test_dl):
model.eval()
outputs = [
validation_step(images.to(device), labels.to(device))
for images, labels in test_dl
]
return validation_epoch_end(outputs)
def training_step(images, labels):
out = model(images)
loss = F.cross_entropy(out, labels)
return loss
def validation_step(images, labels):
out = model(images)
loss = F.cross_entropy(out, labels)
acc = accuracy(out, labels)
return {"val_loss": loss.detach(), "val_acc": acc}
def validation_epoch_end(outputs):
batch_losses = [x["val_loss"] for x in outputs]
epoch_losses = torch.stack(batch_losses).mean()
batch_accs = [x["val_acc"] for x in outputs]
epoch_acc = torch.stack(batch_accs).mean()
return {"val_loss": epoch_losses.item(), "val_acc": epoch_acc.item()}
def epoch_end(epoch, result, start_time):
print(
"Epoch [{}], last_lr:{:.5f}, train_loss:{:.4f}, val_loss:{:.4f}, val_acc: {:.4f}, epoch_time:{:.4f}".format(
epoch,
result["lrs"][-1],
result["train_loss"],
result["val_loss"],
result["val_acc"],
time.time() - start_time,
)
)
torch.cuda.empty_cache()
results = []
optimizer = opt_func(model.parameters(), max_lr, weight_decay=weight_decay)
scheduler = torch.optim.lr_scheduler.OneCycleLR(
optimizer, max_lr, epochs=epochs, steps_per_epoch=len(train_dl)
)
for epoch in range(epochs):
epoch_start_time = time.time()
model.train()
train_losses = []
lrs = [] # Learning rates at each point
# * Training Part
for batch in train_dl:
images, labels = batch
images = images.to(device)
labels = labels.to(device)
loss = training_step(
images, labels
) # Generate Predictions and Calculate the loss
train_losses.append(loss)
loss.backward()
if grad_clip:
nn.utils.clip_grad_value_(model.parameters(), grad_clip)
optimizer.step() # Compute Gradients
optimizer.zero_grad() # Reset to zero
lrs.append(get_lr(optimizer))
scheduler.step()
# * Validation part
result = evaluate(model, train_dl)
result["train_loss"] = torch.stack(train_losses).mean().item()
result["lrs"] = lrs
epoch_end(epoch, result, epoch_start_time)
results.append(result)
return results
EPOCHS = 10
MAX_LEARNING_RATE = 0.001
GRAD_CLIP = 0.1
WEIGHT_DECAY = 1e-4
OPT_FUNC = torch.optim.Adam
results = []
results += fit_one_cycle(
EPOCHS,
MAX_LEARNING_RATE,
model,
train_dl,
test_dl,
WEIGHT_DECAY,
GRAD_CLIP,
OPT_FUNC,
)
# * Plotting training history
time_taken = "8:19"
fig, axs = plt.subplots(nrows=1, ncols=3, figsize=(30, 5))
accuracies = [x["val_acc"] for x in results]
axs[0].plot(accuracies, "-x")
axs[0].title.set_text("Accuracies x Number of Epochs")
axs[0].set_ylabel("Accuracies")
axs[0].set_xlabel("Epoch")
axs[1].plot([x["val_loss"] for x in results], "-x")
axs[1].title.set_text("Loss x Number of Epochs")
axs[1].set_ylabel("Loss")
axs[1].set_xlabel("Epoch")
axs[1].plot([x["train_loss"] for x in results], "-rx")
axs[1].title.set_text("Training Loss x Number of Epochs")
lrs = np.concatenate([x.get("lrs", []) for x in results])
axs[2].plot(lrs)
axs[2].title.set_text("Learning Rate x Batch Number")
axs[2].set_ylabel("Learning Rate")
axs[2].set_ylabel("Batch Number")
plt.suptitle("Training History")
plt.grid()
plt.show()
# Normalizing testing images
def normalize_test(X_test):
X_test = X_test.reshape([-1, 28, 28]).astype(float) / 255
X_test = (X_test - features_np.mean()) / features_np.std()
return X_test
X_test = normalize_test(X_test)
# expanding dimension of test
X_test = np.expand_dims(X_test, axis=1)
X_test = torch.from_numpy(X_test).float().to(device)
# Testing some images, sanity checkk
for img, lbl in test_dl:
ttt = img, lbl
for i in range(BATCH_SIZE):
plt.imshow(torch.Tensor.cpu(ttt[0][i].data.reshape(28, 28)), cmap="gray")
plt.show()
print("Prediction", torch.argmax(model(ttt[0][i].unsqueeze(0)), 1).item())
break
# Seems to working :)
# * Prepping submission file
submission_np = dataset_test.values
class SubmissionDataset(Dataset):
def __init__(self, df, transform=None) -> None:
self.features = df
self.transform = transform
def __getitem__(self, index):
data = self.features[index]
image = data.reshape(28, 28).astype(np.uint8)
if self.transform:
image = self.transform(image)
return image
def __len__(self):
return len(self.features)
submission_tmfs = tt.Compose(
[
tt.ToTensor(),
tt.Normalize(
mean=[features_np.mean() / 255], std=[features_np.std() / 255], inplace=True
),
]
)
submission_set = SubmissionDataset(submission_np, transform=submission_tmfs)
submission_dl = DeviceDataLoader(
DataLoader(submission_set, batch_size=1, shuffle=False), device
)
# * Checking image by image in submission set
ctr = 0
for img in submission_dl:
for i in range(1):
print(img[i].shape)
plt.imshow(torch.Tensor.cpu(img[i].data.reshape(28, 28)), cmap="gray")
plt.show()
print("Prediction", int(torch.argmax(model(img[i].unsqueeze(0)), 1).item()))
if ctr > 1:
break
ctr += 1
df_submission = pd.DataFrame(columns=["ImageId", "Label"])
prediction = []
for img in submission_dl:
prediction.append(int(torch.argmax(model(img[0].unsqueeze(0)), 1).item()))
df_submission["Label"] = pd.Series(prediction)
df_submission["ImageId"] = df_submission.Label.index + 1
# df_submission.to_csv('submission_resnet.csv',index=False)
# torch.save(model.state_dict(),'resnet_digits_model.pth')
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338813.ipynb
| null | null |
[{"Id": 69338813, "ScriptId": 18930822, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 2844760, "CreationDate": "07/29/2021 18:39:16", "VersionNumber": 1.0, "Title": "resnet-digitrecognizer-Pytorch", "EvaluationDate": "07/29/2021", "IsChange": true, "TotalLines": 376.0, "LinesInsertedFromPrevious": 376.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import torch
from torch.utils.data.dataset import Dataset
import torchvision
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import torch.nn as nn
from sklearn.model_selection import train_test_split
# * Importing dataset
dataset_train = pd.read_csv("../input/digit-recognizer/train.csv")
dataset_test = pd.read_csv("../input/digit-recognizer/test.csv")
print("Training data length:", len(dataset_train))
# * Having a look at the dataset
targets_np = dataset_train.label.values
features_np = dataset_train.loc[
:, dataset_train.columns != "label"
].values # values range to 255, so to normalize
X_train, X_test, y_train, y_test = train_test_split(
features_np, targets_np, test_size=0.2, random_state=42
)
# features_train = torch.from_numpy(X_train)
# target_train = torch.from_numpy(y_train)
# features_test = torch.from_numpy(X_test)
# target_test = torch.from_numpy(y_test)
print("Training Features Shape", X_train.shape)
print("Training Labels Shape", y_train.shape)
print("Testing Features Shape", X_test.shape)
print("Testing Labels Shape", y_test.shape)
# Viz of an image
plt.imshow(features_np[0].reshape(28, 28), cmap="gray") # images are 28x28
plt.title("Label: " + str(targets_np[0]))
plt.show()
i = 100
plt.imshow(X_test[i].reshape(28, 28), cmap="gray") # images are 28x28
plt.title("Label: " + str(y_test[i]))
plt.show()
from torch.utils.data.sampler import SubsetRandomSampler
from torch.utils.data import DataLoader, Dataset
import torchvision.transforms as tt
def get_default_device():
"""Returns the default Device (cuda if gpu, else cpu"""
if torch.cuda.is_available():
return torch.device("cuda")
else:
torch.device("cpu")
def to_device(data, device):
"""Shifts Data to the input device"""
if isinstance(data, (list, tuple)):
return [to_device(x, device) for x in data]
return data.to(device, non_blocking=True)
class DeviceDataLoader:
def __init__(self, dl, device) -> None:
self.dl = dl
self.device = device
def __iter__(self):
for b in self.dl:
yield to_device(b, self.device)
def __len__(self):
return len(self.dl)
# * Creating a custom dataset overriding torchvision dataset
class MyDataset(Dataset):
def __init__(self, df_features, df_labels, transform=None) -> None:
self.features = df_features
self.labels = df_labels
self.transform = transform
def __getitem__(self, index):
data = self.features[index]
image = data.reshape(28, 28).astype(np.uint8)
label = self.labels[index]
if self.transform:
image = self.transform(image)
return (image, label)
def __len__(self):
return len(self.features)
BATCH_SIZE = 16
# * Augmentation - Random affination (rotation with translation), normalization
train_tmfs = tt.Compose(
[
tt.ToPILImage(),
# tt.RandomRotation(30),
tt.RandomAffine(degrees=30, translate=(0.1, 0.1), scale=(0.9, 1.1)),
# Random Affine suits better for this purpose @https://pytorch.org/vision/stable/auto_examples/plot_transforms.html#randomaffine
tt.ToTensor(),
tt.Normalize(
mean=[features_np.mean() / 255], std=[features_np.std() / 255], inplace=True
),
]
)
test_tmfs = tt.Compose(
[
tt.ToPILImage(),
# tt.RandomRotation(30),
tt.RandomAffine(degrees=30, translate=(0.1, 0.1), scale=(0.9, 1.1)),
# Random Affine suits better for this purpose @https://pytorch.org/vision/stable/auto_examples/plot_transforms.html#randomaffine
tt.ToTensor(),
tt.Normalize(
mean=[features_np.mean() / 255], std=[features_np.std() / 255], inplace=True
),
]
)
# * Setting default Device, GPU
device = get_default_device()
print(device)
# * Creating Dataset
train_set = MyDataset(X_train, y_train, transform=train_tmfs)
test_set = MyDataset(X_test, y_test, transform=test_tmfs)
# * Loading data and then shifting data loader to the selected device
train_dl = DeviceDataLoader(
DataLoader(train_set, batch_size=BATCH_SIZE, shuffle=True), device
)
test_dl = DeviceDataLoader(
DataLoader(test_set, batch_size=BATCH_SIZE, shuffle=True), device
)
# * Sanity Check
imgs, lbls = next(iter(test_dl))
print(imgs.shape)
plt.imshow(torch.Tensor.cpu(imgs[0].data.reshape(28, 28)), cmap="gray")
print("Label:", lbls[0])
plt.show()
import torch.nn.functional as F
# * Following Residual Network https://raw.githubusercontent.com/lambdal/cifar10-fast/master/net.svg
NUM_CLASSES = 10
def convolution_block(in_channels, out_channels, pool=False):
"""Returns a sequence of Convolution, Batch Normalization, Rectified Linear Activation (Relu) and Max Pooling if selected"""
layers = [
nn.Conv2d(
in_channels=in_channels, out_channels=out_channels, kernel_size=3, padding=1
),
nn.BatchNorm2d(out_channels),
nn.ReLU(inplace=True),
]
if pool:
layers.append(nn.MaxPool2d(2, 2)) # 2x2 Max Pooling
return nn.Sequential(*layers)
class Model(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = convolution_block(1, 32)
self.conv2 = convolution_block(32, 64, pool=True)
self.res1 = nn.Sequential(convolution_block(64, 64), convolution_block(64, 64))
self.conv3 = convolution_block(64, 128, pool=True)
self.conv4 = convolution_block(128, 256, pool=True)
self.res2 = nn.Sequential(
convolution_block(256, 256), convolution_block(256, 256)
)
self.classifier = nn.Sequential(
nn.MaxPool2d(2),
nn.Flatten(),
nn.Dropout(0.2), # * added later
nn.Linear(256, NUM_CLASSES),
)
def forward(self, xb):
out = self.conv1(xb) # Prep
out = self.conv2(out) # Layer 1 Convolution Block
out = self.res1(out) + out # Layer 1 Residual Block
out = self.conv3(out) # Layer 2
out = self.conv4(out) # Layer 3 Convolution Block
out = self.res2(out) + out # Layer 3 Residual Block
out = self.classifier(out)
return F.log_softmax(out, dim=1)
model = Model()
to_device(model, device)
# Sanity Check
for images, labels in train_dl:
print(images.shape)
out = model(images)
print(out.shape)
break
"""
Cant find fastai modules
from fastai.basic_data import DataBunch
from fastai.train import Learner
from fastai.metrics import accuracy
"""
import time
@torch.no_grad()
def loss_batch(model, loss_func, xb, yb, opt=None, metric=None):
preds = model(xb)
loss = loss_func(preds, yb)
if opt is not None:
loss.backward()
opt.step()
opt.zero_grad()
metric_result = None
if metric is not None:
metric_result = metric(preds, yb)
return loss.item(), len(xb), metric_result
def accuracy(outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(preds))
def get_lr(optimizer):
for param_group in optimizer.param_groups:
return param_group["lr"]
def fit_one_cycle(
epochs,
max_lr,
model,
train_dl,
test_dl,
weight_decay=0,
grad_clip=None,
opt_func=torch.optim.SGD,
):
def evaluate(model, test_dl):
model.eval()
outputs = [
validation_step(images.to(device), labels.to(device))
for images, labels in test_dl
]
return validation_epoch_end(outputs)
def training_step(images, labels):
out = model(images)
loss = F.cross_entropy(out, labels)
return loss
def validation_step(images, labels):
out = model(images)
loss = F.cross_entropy(out, labels)
acc = accuracy(out, labels)
return {"val_loss": loss.detach(), "val_acc": acc}
def validation_epoch_end(outputs):
batch_losses = [x["val_loss"] for x in outputs]
epoch_losses = torch.stack(batch_losses).mean()
batch_accs = [x["val_acc"] for x in outputs]
epoch_acc = torch.stack(batch_accs).mean()
return {"val_loss": epoch_losses.item(), "val_acc": epoch_acc.item()}
def epoch_end(epoch, result, start_time):
print(
"Epoch [{}], last_lr:{:.5f}, train_loss:{:.4f}, val_loss:{:.4f}, val_acc: {:.4f}, epoch_time:{:.4f}".format(
epoch,
result["lrs"][-1],
result["train_loss"],
result["val_loss"],
result["val_acc"],
time.time() - start_time,
)
)
torch.cuda.empty_cache()
results = []
optimizer = opt_func(model.parameters(), max_lr, weight_decay=weight_decay)
scheduler = torch.optim.lr_scheduler.OneCycleLR(
optimizer, max_lr, epochs=epochs, steps_per_epoch=len(train_dl)
)
for epoch in range(epochs):
epoch_start_time = time.time()
model.train()
train_losses = []
lrs = [] # Learning rates at each point
# * Training Part
for batch in train_dl:
images, labels = batch
images = images.to(device)
labels = labels.to(device)
loss = training_step(
images, labels
) # Generate Predictions and Calculate the loss
train_losses.append(loss)
loss.backward()
if grad_clip:
nn.utils.clip_grad_value_(model.parameters(), grad_clip)
optimizer.step() # Compute Gradients
optimizer.zero_grad() # Reset to zero
lrs.append(get_lr(optimizer))
scheduler.step()
# * Validation part
result = evaluate(model, train_dl)
result["train_loss"] = torch.stack(train_losses).mean().item()
result["lrs"] = lrs
epoch_end(epoch, result, epoch_start_time)
results.append(result)
return results
EPOCHS = 10
MAX_LEARNING_RATE = 0.001
GRAD_CLIP = 0.1
WEIGHT_DECAY = 1e-4
OPT_FUNC = torch.optim.Adam
results = []
results += fit_one_cycle(
EPOCHS,
MAX_LEARNING_RATE,
model,
train_dl,
test_dl,
WEIGHT_DECAY,
GRAD_CLIP,
OPT_FUNC,
)
# * Plotting training history
time_taken = "8:19"
fig, axs = plt.subplots(nrows=1, ncols=3, figsize=(30, 5))
accuracies = [x["val_acc"] for x in results]
axs[0].plot(accuracies, "-x")
axs[0].title.set_text("Accuracies x Number of Epochs")
axs[0].set_ylabel("Accuracies")
axs[0].set_xlabel("Epoch")
axs[1].plot([x["val_loss"] for x in results], "-x")
axs[1].title.set_text("Loss x Number of Epochs")
axs[1].set_ylabel("Loss")
axs[1].set_xlabel("Epoch")
axs[1].plot([x["train_loss"] for x in results], "-rx")
axs[1].title.set_text("Training Loss x Number of Epochs")
lrs = np.concatenate([x.get("lrs", []) for x in results])
axs[2].plot(lrs)
axs[2].title.set_text("Learning Rate x Batch Number")
axs[2].set_ylabel("Learning Rate")
axs[2].set_ylabel("Batch Number")
plt.suptitle("Training History")
plt.grid()
plt.show()
# Normalizing testing images
def normalize_test(X_test):
X_test = X_test.reshape([-1, 28, 28]).astype(float) / 255
X_test = (X_test - features_np.mean()) / features_np.std()
return X_test
X_test = normalize_test(X_test)
# expanding dimension of test
X_test = np.expand_dims(X_test, axis=1)
X_test = torch.from_numpy(X_test).float().to(device)
# Testing some images, sanity checkk
for img, lbl in test_dl:
ttt = img, lbl
for i in range(BATCH_SIZE):
plt.imshow(torch.Tensor.cpu(ttt[0][i].data.reshape(28, 28)), cmap="gray")
plt.show()
print("Prediction", torch.argmax(model(ttt[0][i].unsqueeze(0)), 1).item())
break
# Seems to working :)
# * Prepping submission file
submission_np = dataset_test.values
class SubmissionDataset(Dataset):
def __init__(self, df, transform=None) -> None:
self.features = df
self.transform = transform
def __getitem__(self, index):
data = self.features[index]
image = data.reshape(28, 28).astype(np.uint8)
if self.transform:
image = self.transform(image)
return image
def __len__(self):
return len(self.features)
submission_tmfs = tt.Compose(
[
tt.ToTensor(),
tt.Normalize(
mean=[features_np.mean() / 255], std=[features_np.std() / 255], inplace=True
),
]
)
submission_set = SubmissionDataset(submission_np, transform=submission_tmfs)
submission_dl = DeviceDataLoader(
DataLoader(submission_set, batch_size=1, shuffle=False), device
)
# * Checking image by image in submission set
ctr = 0
for img in submission_dl:
for i in range(1):
print(img[i].shape)
plt.imshow(torch.Tensor.cpu(img[i].data.reshape(28, 28)), cmap="gray")
plt.show()
print("Prediction", int(torch.argmax(model(img[i].unsqueeze(0)), 1).item()))
if ctr > 1:
break
ctr += 1
df_submission = pd.DataFrame(columns=["ImageId", "Label"])
prediction = []
for img in submission_dl:
prediction.append(int(torch.argmax(model(img[0].unsqueeze(0)), 1).item()))
df_submission["Label"] = pd.Series(prediction)
df_submission["ImageId"] = df_submission.Label.index + 1
# df_submission.to_csv('submission_resnet.csv',index=False)
# torch.save(model.state_dict(),'resnet_digits_model.pth')
| false | 0 | 4,115 | 0 | 4,115 | 4,115 |
||
69338582
|
<jupyter_start><jupyter_text>titanic_train
Kaggle dataset identifier: titanic-train
<jupyter_code>import pandas as pd
df = pd.read_csv('titanic-train/titanic_train.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 891 non-null int64
1 Survived 891 non-null int64
2 Pclass 891 non-null int64
3 Name 891 non-null object
4 Sex 891 non-null object
5 Age 714 non-null float64
6 SibSp 891 non-null int64
7 Parch 891 non-null int64
8 Ticket 891 non-null object
9 Fare 891 non-null float64
10 Cabin 204 non-null object
11 Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 83.7+ KB
<jupyter_text>Examples:
{
"PassengerId": 1,
"Survived": 0,
"Pclass": 3,
"Name": "Braund, Mr. Owen Harris",
"Sex": "male",
"Age": 22,
"SibSp": 1,
"Parch": 0,
"Ticket": "A/5 21171",
"Fare": 7.25,
"Cabin": null,
"Embarked": "S"
}
{
"PassengerId": 2,
"Survived": 1,
"Pclass": 1,
"Name": "Cumings, Mrs. John Bradley (Florence Briggs Thayer)",
"Sex": "female",
"Age": 38,
"SibSp": 1,
"Parch": 0,
"Ticket": "PC 17599",
"Fare": 71.2833,
"Cabin": "C85",
"Embarked": "C"
}
{
"PassengerId": 3,
"Survived": 1,
"Pclass": 3,
"Name": "Heikkinen, Miss. Laina",
"Sex": "female",
"Age": 26,
"SibSp": 0,
"Parch": 0,
"Ticket": "STON/O2. 3101282",
"Fare": 7.925,
"Cabin": null,
"Embarked": "S"
}
{
"PassengerId": 4,
"Survived": 1,
"Pclass": 1,
"Name": "Futrelle, Mrs. Jacques Heath (Lily May Peel)",
"Sex": "female",
"Age": 35,
"SibSp": 1,
"Parch": 0,
"Ticket": "113803",
"Fare": 53.1,
"Cabin": "C123",
"Embarked": "S"
}
<jupyter_script># ## Import Libraries..!!
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report, accuracy_score
# ## Read dataset from csv file..!!
titanic_data = pd.read_csv("../input/titanic-train/titanic_train.csv")
titanic_data.head()
titanic_data.info()
titanic_data.describe()
titanic_data.isnull().sum()
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.corr())
titanic_data.corr()
titanic_data.shape
titanic_data.dtypes
# ## Exploratory Data Analysis (EDA)..!!
# ### Let's begin some exploratory data analysis! We'll start by checking out missing data!
# ## Missing Data..!!
# ### We can use seaborn to create a simple heatmap to see where we are missing data!
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
fig = plt.figure(figsize=(12, 8))
sns.countplot(x="Survived", data=titanic_data)
fig = plt.figure(figsize=(12, 8))
sns.countplot(x="Survived", hue="Sex", data=titanic_data)
fig = plt.figure(figsize=(12, 8))
sns.countplot(x="Survived", hue="Pclass", data=titanic_data)
fig = plt.figure(figsize=(12, 8))
sns.distplot(titanic_data["Age"].dropna(), kde=False, color="red", bins=30)
fig = plt.figure(figsize=(12, 8))
titanic_data["Age"].hist(bins=30, color="red", alpha=0.7)
fig = plt.figure(figsize=(12, 8))
sns.countplot(x="SibSp", data=titanic_data)
# ## Cufflinks for plots..!!
import cufflinks as cf
cf.go_offline()
titanic_data["Age"].iplot(kind="hist", bins=30, color="red")
titanic_data["Fare"].iplot(kind="hist", bins=30, color="red")
fig = plt.figure(figsize=(12, 8))
sns.boxplot(x="Pclass", y="Age", data=titanic_data, palette="spring")
# ### We can see the wealthier passengers in the higher classes tend to be older, which makes sense. We'll use these average age values to impute based on Pclass for Age.
def age(temp):
age = temp[0]
p_class = temp[1]
if pd.isnull(age):
if p_class == 1:
return 35
elif p_class == 2:
return 27
else:
return 25
else:
return age
# ### Now apply that function..!!
titanic_data["Age"] = titanic_data[["Age", "Pclass"]].apply(age, axis=1)
# ### Now let's check that heat map again.!!
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
# ### Now, drop the Cabin column from the dataset..!!
titanic_data.drop("Cabin", axis=1, inplace=True)
# ### Again, check the heatmap of dataset..!!
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
titanic_data.head()
# ### Now, drop all the NaN values from the dataset..!!
titanic_data.dropna(inplace=True)
# ### Again, check the heatmap of dataset..!!
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
# ## Data Cleaning is Complete..!!
# ## Finally, data is clean and now convert categorical data..!!
titanic_data.dtypes
new_sex_col = pd.get_dummies(titanic_data["Sex"], drop_first=True)
new_embark_col = pd.get_dummies(titanic_data["Embarked"], drop_first=True)
titanic_data = titanic_data.drop(["Sex", "Embarked", "Name", "Ticket"], axis=1)
titanic_data = pd.concat([titanic_data, new_sex_col, new_embark_col], axis=1)
titanic_data.head()
titanic_data.dtypes
# ### Now, our data is ready for our model..!!
x = titanic_data.drop("Survived", axis=1)
y = titanic_data["Survived"]
# ### Now, split data into train test split. Training Data = 80%, Test Data = 20%..!!
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=45
)
# ## Now, Building a Logistic Regression Model..!!
logistic_model = LogisticRegression()
# ### Training our Model..!!
logistic_model.fit(x_train, y_train)
# ### Making predictions to our Model..!!
y_pred = logistic_model.predict(x_test)
# ### Now, Do Evaluation of our Model..!!
print(classification_report(y_test, y_pred))
# ## Plot our data..!!
fig = plt.figure(figsize=(12, 8))
plt.plot(titanic_data, color="red")
# ## Bar Plot our data..!!
titanic_data.iplot(kind="bar", bins=30, color="red")
# ## Histogram Plot our data..!!
titanic_data.iplot(kind="hist", bins=30, color="blue")
train_accuracy = logistic_model.score(x_train, y_train)
print("Accuracy of our Training Model:", train_accuracy * 100, "%")
test_accuracy = logistic_model.score(x_test, y_test)
print("Accuracy of our Test Model:", test_accuracy * 100, "%")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338582.ipynb
|
titanic-train
|
tedllh
|
[{"Id": 69338582, "ScriptId": 18931050, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6279304, "CreationDate": "07/29/2021 18:34:34", "VersionNumber": 1.0, "Title": "Titanic Train Project", "EvaluationDate": "07/29/2021", "IsChange": true, "TotalLines": 180.0, "LinesInsertedFromPrevious": 180.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92384385, "KernelVersionId": 69338582, "SourceDatasetVersionId": 18775}]
|
[{"Id": 18775, "DatasetId": 13931, "DatasourceVersionId": 18775, "CreatorUserId": 1512915, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "02/23/2018 07:13:19", "VersionNumber": 1.0, "Title": "titanic_train", "Slug": "titanic-train", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 61194.0, "TotalUncompressedBytes": 61194.0}]
|
[{"Id": 13931, "CreatorUserId": 1512915, "OwnerUserId": 1512915.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 18775.0, "CurrentDatasourceVersionId": 18775.0, "ForumId": 21485, "Type": 2, "CreationDate": "02/23/2018 07:13:19", "LastActivityDate": "02/23/2018", "TotalViews": 26009, "TotalDownloads": 6527, "TotalVotes": 36, "TotalKernels": 15}]
|
[{"Id": 1512915, "UserName": "tedllh", "DisplayName": "TedLLH", "RegisterDate": "12/23/2017", "PerformanceTier": 0}]
|
# ## Import Libraries..!!
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report, accuracy_score
# ## Read dataset from csv file..!!
titanic_data = pd.read_csv("../input/titanic-train/titanic_train.csv")
titanic_data.head()
titanic_data.info()
titanic_data.describe()
titanic_data.isnull().sum()
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.corr())
titanic_data.corr()
titanic_data.shape
titanic_data.dtypes
# ## Exploratory Data Analysis (EDA)..!!
# ### Let's begin some exploratory data analysis! We'll start by checking out missing data!
# ## Missing Data..!!
# ### We can use seaborn to create a simple heatmap to see where we are missing data!
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
fig = plt.figure(figsize=(12, 8))
sns.countplot(x="Survived", data=titanic_data)
fig = plt.figure(figsize=(12, 8))
sns.countplot(x="Survived", hue="Sex", data=titanic_data)
fig = plt.figure(figsize=(12, 8))
sns.countplot(x="Survived", hue="Pclass", data=titanic_data)
fig = plt.figure(figsize=(12, 8))
sns.distplot(titanic_data["Age"].dropna(), kde=False, color="red", bins=30)
fig = plt.figure(figsize=(12, 8))
titanic_data["Age"].hist(bins=30, color="red", alpha=0.7)
fig = plt.figure(figsize=(12, 8))
sns.countplot(x="SibSp", data=titanic_data)
# ## Cufflinks for plots..!!
import cufflinks as cf
cf.go_offline()
titanic_data["Age"].iplot(kind="hist", bins=30, color="red")
titanic_data["Fare"].iplot(kind="hist", bins=30, color="red")
fig = plt.figure(figsize=(12, 8))
sns.boxplot(x="Pclass", y="Age", data=titanic_data, palette="spring")
# ### We can see the wealthier passengers in the higher classes tend to be older, which makes sense. We'll use these average age values to impute based on Pclass for Age.
def age(temp):
age = temp[0]
p_class = temp[1]
if pd.isnull(age):
if p_class == 1:
return 35
elif p_class == 2:
return 27
else:
return 25
else:
return age
# ### Now apply that function..!!
titanic_data["Age"] = titanic_data[["Age", "Pclass"]].apply(age, axis=1)
# ### Now let's check that heat map again.!!
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
# ### Now, drop the Cabin column from the dataset..!!
titanic_data.drop("Cabin", axis=1, inplace=True)
# ### Again, check the heatmap of dataset..!!
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
titanic_data.head()
# ### Now, drop all the NaN values from the dataset..!!
titanic_data.dropna(inplace=True)
# ### Again, check the heatmap of dataset..!!
fig = plt.figure(figsize=(12, 8))
sns.heatmap(titanic_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
# ## Data Cleaning is Complete..!!
# ## Finally, data is clean and now convert categorical data..!!
titanic_data.dtypes
new_sex_col = pd.get_dummies(titanic_data["Sex"], drop_first=True)
new_embark_col = pd.get_dummies(titanic_data["Embarked"], drop_first=True)
titanic_data = titanic_data.drop(["Sex", "Embarked", "Name", "Ticket"], axis=1)
titanic_data = pd.concat([titanic_data, new_sex_col, new_embark_col], axis=1)
titanic_data.head()
titanic_data.dtypes
# ### Now, our data is ready for our model..!!
x = titanic_data.drop("Survived", axis=1)
y = titanic_data["Survived"]
# ### Now, split data into train test split. Training Data = 80%, Test Data = 20%..!!
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=45
)
# ## Now, Building a Logistic Regression Model..!!
logistic_model = LogisticRegression()
# ### Training our Model..!!
logistic_model.fit(x_train, y_train)
# ### Making predictions to our Model..!!
y_pred = logistic_model.predict(x_test)
# ### Now, Do Evaluation of our Model..!!
print(classification_report(y_test, y_pred))
# ## Plot our data..!!
fig = plt.figure(figsize=(12, 8))
plt.plot(titanic_data, color="red")
# ## Bar Plot our data..!!
titanic_data.iplot(kind="bar", bins=30, color="red")
# ## Histogram Plot our data..!!
titanic_data.iplot(kind="hist", bins=30, color="blue")
train_accuracy = logistic_model.score(x_train, y_train)
print("Accuracy of our Training Model:", train_accuracy * 100, "%")
test_accuracy = logistic_model.score(x_test, y_test)
print("Accuracy of our Test Model:", test_accuracy * 100, "%")
|
[{"titanic-train/titanic_train.csv": {"column_names": "[\"PassengerId\", \"Survived\", \"Pclass\", \"Name\", \"Sex\", \"Age\", \"SibSp\", \"Parch\", \"Ticket\", \"Fare\", \"Cabin\", \"Embarked\"]", "column_data_types": "{\"PassengerId\": \"int64\", \"Survived\": \"int64\", \"Pclass\": \"int64\", \"Name\": \"object\", \"Sex\": \"object\", \"Age\": \"float64\", \"SibSp\": \"int64\", \"Parch\": \"int64\", \"Ticket\": \"object\", \"Fare\": \"float64\", \"Cabin\": \"object\", \"Embarked\": \"object\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 891 entries, 0 to 890\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 PassengerId 891 non-null int64 \n 1 Survived 891 non-null int64 \n 2 Pclass 891 non-null int64 \n 3 Name 891 non-null object \n 4 Sex 891 non-null object \n 5 Age 714 non-null float64\n 6 SibSp 891 non-null int64 \n 7 Parch 891 non-null int64 \n 8 Ticket 891 non-null object \n 9 Fare 891 non-null float64\n 10 Cabin 204 non-null object \n 11 Embarked 889 non-null object \ndtypes: float64(2), int64(5), object(5)\nmemory usage: 83.7+ KB\n", "summary": "{\"PassengerId\": {\"count\": 891.0, \"mean\": 446.0, \"std\": 257.3538420152301, \"min\": 1.0, \"25%\": 223.5, \"50%\": 446.0, \"75%\": 668.5, \"max\": 891.0}, \"Survived\": {\"count\": 891.0, \"mean\": 0.3838383838383838, \"std\": 0.4865924542648575, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"Pclass\": {\"count\": 891.0, \"mean\": 2.308641975308642, \"std\": 0.836071240977049, \"min\": 1.0, \"25%\": 2.0, \"50%\": 3.0, \"75%\": 3.0, \"max\": 3.0}, \"Age\": {\"count\": 714.0, \"mean\": 29.69911764705882, \"std\": 14.526497332334042, \"min\": 0.42, \"25%\": 20.125, \"50%\": 28.0, \"75%\": 38.0, \"max\": 80.0}, \"SibSp\": {\"count\": 891.0, \"mean\": 0.5230078563411896, \"std\": 1.1027434322934317, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 8.0}, \"Parch\": {\"count\": 891.0, \"mean\": 0.38159371492704824, \"std\": 0.8060572211299483, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 6.0}, \"Fare\": {\"count\": 891.0, \"mean\": 32.204207968574636, \"std\": 49.6934285971809, \"min\": 0.0, \"25%\": 7.9104, \"50%\": 14.4542, \"75%\": 31.0, \"max\": 512.3292}}", "examples": "{\"PassengerId\":{\"0\":1,\"1\":2,\"2\":3,\"3\":4},\"Survived\":{\"0\":0,\"1\":1,\"2\":1,\"3\":1},\"Pclass\":{\"0\":3,\"1\":1,\"2\":3,\"3\":1},\"Name\":{\"0\":\"Braund, Mr. Owen Harris\",\"1\":\"Cumings, Mrs. John Bradley (Florence Briggs Thayer)\",\"2\":\"Heikkinen, Miss. Laina\",\"3\":\"Futrelle, Mrs. Jacques Heath (Lily May Peel)\"},\"Sex\":{\"0\":\"male\",\"1\":\"female\",\"2\":\"female\",\"3\":\"female\"},\"Age\":{\"0\":22.0,\"1\":38.0,\"2\":26.0,\"3\":35.0},\"SibSp\":{\"0\":1,\"1\":1,\"2\":0,\"3\":1},\"Parch\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"Ticket\":{\"0\":\"A\\/5 21171\",\"1\":\"PC 17599\",\"2\":\"STON\\/O2. 3101282\",\"3\":\"113803\"},\"Fare\":{\"0\":7.25,\"1\":71.2833,\"2\":7.925,\"3\":53.1},\"Cabin\":{\"0\":null,\"1\":\"C85\",\"2\":null,\"3\":\"C123\"},\"Embarked\":{\"0\":\"S\",\"1\":\"C\",\"2\":\"S\",\"3\":\"S\"}}"}}]
| true | 1 |
<start_data_description><data_path>titanic-train/titanic_train.csv:
<column_names>
['PassengerId', 'Survived', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp', 'Parch', 'Ticket', 'Fare', 'Cabin', 'Embarked']
<column_types>
{'PassengerId': 'int64', 'Survived': 'int64', 'Pclass': 'int64', 'Name': 'object', 'Sex': 'object', 'Age': 'float64', 'SibSp': 'int64', 'Parch': 'int64', 'Ticket': 'object', 'Fare': 'float64', 'Cabin': 'object', 'Embarked': 'object'}
<dataframe_Summary>
{'PassengerId': {'count': 891.0, 'mean': 446.0, 'std': 257.3538420152301, 'min': 1.0, '25%': 223.5, '50%': 446.0, '75%': 668.5, 'max': 891.0}, 'Survived': {'count': 891.0, 'mean': 0.3838383838383838, 'std': 0.4865924542648575, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'Pclass': {'count': 891.0, 'mean': 2.308641975308642, 'std': 0.836071240977049, 'min': 1.0, '25%': 2.0, '50%': 3.0, '75%': 3.0, 'max': 3.0}, 'Age': {'count': 714.0, 'mean': 29.69911764705882, 'std': 14.526497332334042, 'min': 0.42, '25%': 20.125, '50%': 28.0, '75%': 38.0, 'max': 80.0}, 'SibSp': {'count': 891.0, 'mean': 0.5230078563411896, 'std': 1.1027434322934317, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 8.0}, 'Parch': {'count': 891.0, 'mean': 0.38159371492704824, 'std': 0.8060572211299483, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 6.0}, 'Fare': {'count': 891.0, 'mean': 32.204207968574636, 'std': 49.6934285971809, 'min': 0.0, '25%': 7.9104, '50%': 14.4542, '75%': 31.0, 'max': 512.3292}}
<dataframe_info>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 891 non-null int64
1 Survived 891 non-null int64
2 Pclass 891 non-null int64
3 Name 891 non-null object
4 Sex 891 non-null object
5 Age 714 non-null float64
6 SibSp 891 non-null int64
7 Parch 891 non-null int64
8 Ticket 891 non-null object
9 Fare 891 non-null float64
10 Cabin 204 non-null object
11 Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 83.7+ KB
<some_examples>
{'PassengerId': {'0': 1, '1': 2, '2': 3, '3': 4}, 'Survived': {'0': 0, '1': 1, '2': 1, '3': 1}, 'Pclass': {'0': 3, '1': 1, '2': 3, '3': 1}, 'Name': {'0': 'Braund, Mr. Owen Harris', '1': 'Cumings, Mrs. John Bradley (Florence Briggs Thayer)', '2': 'Heikkinen, Miss. Laina', '3': 'Futrelle, Mrs. Jacques Heath (Lily May Peel)'}, 'Sex': {'0': 'male', '1': 'female', '2': 'female', '3': 'female'}, 'Age': {'0': 22.0, '1': 38.0, '2': 26.0, '3': 35.0}, 'SibSp': {'0': 1, '1': 1, '2': 0, '3': 1}, 'Parch': {'0': 0, '1': 0, '2': 0, '3': 0}, 'Ticket': {'0': 'A/5 21171', '1': 'PC 17599', '2': 'STON/O2. 3101282', '3': '113803'}, 'Fare': {'0': 7.25, '1': 71.2833, '2': 7.925, '3': 53.1}, 'Cabin': {'0': None, '1': 'C85', '2': None, '3': 'C123'}, 'Embarked': {'0': 'S', '1': 'C', '2': 'S', '3': 'S'}}
<end_description>
| 1,666 | 0 | 2,519 | 1,666 |
69338512
|
<jupyter_start><jupyter_text>H-1B Visa Petitions 2015-2019
### Context
This dataset is an update of Sharan Naribole's earlier dataset titled ***H-1B Visa Petitions 2011-2016***. Inspired by his work and using a modified, updated version of his R script, I wrangled U.S. H1-B visa petitions data for the years 2015-2019. The previous dataset can be found here: [link to Sharan's dataset](https://www.kaggle.com/nsharan/h-1b-visa).
H1-B visas are the most common visa status applied for and held by international students once they begin working full-time in the U.S.
Please see the original dataset for more context information.
### Content
This dataset includes 5 years worth of H1-B visa petitions in the U.S. The columns in the dataset include case status, employer name, worksite coordinates, job title, prevailing wage, occupation code, and year filed.
This file contains H1-B data from the LCA Program data files (H1-B, H-1B1, E-3). These datasets can be found on the [U.S. Department of Labor Site](https://www.dol.gov/agencies/eta/foreign-labor/performance).
Kaggle dataset identifier: h1b-visa-petitions-20152019
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"../input/h1b-visa-petitions-20152019/h1b_disclosure_data_2015_2019.csv"
)
df.drop("CASE_NUMBER", 1, inplace=True)
df.head()
plt.figure(figsize=(10, 10))
df.JOB_TITLE.value_counts()[:20].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df.EMPLOYER_NAME.value_counts()[:20].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df.WORKSITE_STATE_FULL.value_counts()[:50].plot(kind="barh").invert_yaxis()
# ## Q1: Is Programmer Analyst Job highest across all the years?
df.YEAR.value_counts()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2015].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2016].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2017].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2018].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2019].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338512.ipynb
|
h1b-visa-petitions-20152019
|
abrambeyer
|
[{"Id": 69338512, "ScriptId": 18929989, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7536856, "CreationDate": "07/29/2021 18:33:10", "VersionNumber": 2.0, "Title": "H1b visas 2015-2019", "EvaluationDate": "07/29/2021", "IsChange": true, "TotalLines": 58.0, "LinesInsertedFromPrevious": 6.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 52.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 2}]
|
[{"Id": 92384231, "KernelVersionId": 69338512, "SourceDatasetVersionId": 2474110}]
|
[{"Id": 2474110, "DatasetId": 1497235, "DatasourceVersionId": 2516603, "CreatorUserId": 432563, "LicenseName": "U.S. Government Works", "CreationDate": "07/28/2021 18:37:01", "VersionNumber": 1.0, "Title": "H-1B Visa Petitions 2015-2019", "Slug": "h1b-visa-petitions-20152019", "Subtitle": "Close to 1 million petitions for H1-B visas", "Description": "### Context\n\nThis dataset is an update of Sharan Naribole's earlier dataset titled ***H-1B Visa Petitions 2011-2016***. Inspired by his work and using a modified, updated version of his R script, I wrangled U.S. H1-B visa petitions data for the years 2015-2019. The previous dataset can be found here: [link to Sharan's dataset](https://www.kaggle.com/nsharan/h-1b-visa). \n\nH1-B visas are the most common visa status applied for and held by international students once they begin working full-time in the U.S. \n\nPlease see the original dataset for more context information.\n\n\n### Content\n\nThis dataset includes 5 years worth of H1-B visa petitions in the U.S. The columns in the dataset include case status, employer name, worksite coordinates, job title, prevailing wage, occupation code, and year filed. \n\nThis file contains H1-B data from the LCA Program data files (H1-B, H-1B1, E-3). These datasets can be found on the [U.S. Department of Labor Site](https://www.dol.gov/agencies/eta/foreign-labor/performance).\n\n\n### Acknowledgements\n\nShout out to [Sharan Naribole](https://www.kaggle.com/nsharan) for the original project idea and easy-to-update R script. \n\n[U.S. Department of Labor Data Source](https://www.dol.gov/agencies/eta/foreign-labor/performance)\n\n### Inspiration\n\nWhich states/cities/companies provide the most H1-B visas?\nFor your job description, which city should you be in to have the most opportunities?\nWhich companies should you apply to if you would like the best odds of obtaining a H1-B visa?", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 1497235, "CreatorUserId": 432563, "OwnerUserId": 432563.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2474110.0, "CurrentDatasourceVersionId": 2516603.0, "ForumId": 1516960, "Type": 2, "CreationDate": "07/28/2021 18:37:01", "LastActivityDate": "07/28/2021", "TotalViews": 6508, "TotalDownloads": 497, "TotalVotes": 17, "TotalKernels": 3}]
|
[{"Id": 432563, "UserName": "abrambeyer", "DisplayName": "ABeyer", "RegisterDate": "10/01/2015", "PerformanceTier": 1}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"../input/h1b-visa-petitions-20152019/h1b_disclosure_data_2015_2019.csv"
)
df.drop("CASE_NUMBER", 1, inplace=True)
df.head()
plt.figure(figsize=(10, 10))
df.JOB_TITLE.value_counts()[:20].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df.EMPLOYER_NAME.value_counts()[:20].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df.WORKSITE_STATE_FULL.value_counts()[:50].plot(kind="barh").invert_yaxis()
# ## Q1: Is Programmer Analyst Job highest across all the years?
df.YEAR.value_counts()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2015].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2016].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2017].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2018].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
plt.figure(figsize=(10, 10))
df[df.YEAR == 2019].JOB_TITLE.value_counts()[:10].plot(kind="barh").invert_yaxis()
| false | 1 | 666 | 2 | 1,024 | 666 |
||
69338852
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
train = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
test = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
train.head(8)
train.shape
test.head(8)
test.shape
import seaborn as sns
import matplotlib.pyplot as plt
x = train.target.value_counts()
sns.barplot(x.index, x)
plt.gca().set_ylabel("samples")
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 5))
tweet_len = train[train["target"] == 1]["text"].str.len()
ax1.hist(tweet_len, color="blue")
ax1.set_title("disaster tweets")
tweet_len = train[train["target"] == 0]["text"].str.len()
ax2.hist(tweet_len, color="CRIMSON")
ax2.set_title("Not disaster tweets")
fig.suptitle("Characters in tweets")
plt.show()
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 5))
tweet_len = train[train["target"] == 1]["text"].str.split().map(lambda x: len(x))
ax1.hist(tweet_len, color="blue")
ax1.set_title("disaster tweets")
tweet_len = train[train["target"] == 0]["text"].str.split().map(lambda x: len(x))
ax2.hist(tweet_len, color="CRIMSON")
ax2.set_title("Not disaster tweets")
fig.suptitle("Words in a tweet")
plt.show()
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 5))
word = (
train[train["target"] == 1]["text"]
.str.split()
.apply(lambda x: np.mean([len(i) for i in x]))
)
sns.distplot(word, ax=ax1, color="red")
ax1.set_title("disaster")
word = (
train[train["target"] == 0]["text"]
.str.split()
.apply(lambda x: np.mean([len(i) for i in x]))
)
sns.distplot(word, ax=ax2, color="green")
ax2.set_title("Not disaster")
fig.suptitle("Average word length in each tweet")
from collections import defaultdict
import nltk
import nltk.corpus
from nltk.corpus import stopwords
nltk.download("stopwords")
stop = set(stopwords.words("english"))
corpus = []
for x in train["text"].str.split():
for i in x:
corpus.append(i)
dic = defaultdict(int)
for word in corpus:
if word not in stop:
dic[word] += 1
top = sorted(dic.items(), key=lambda x: x[1], reverse=True)[:30]
x, y = zip(*top)
plt.rcParams["figure.figsize"] = (20, 10)
plt.bar(x, y, color="red")
from collections import defaultdict
import nltk
import nltk.corpus
from nltk.corpus import stopwords
nltk.download("stopwords")
stop = set(stopwords.words("english"))
corpus = []
for x in train["text"].str.split():
for i in x:
corpus.append(i)
dic = defaultdict(int)
for word in corpus:
if word in stop:
dic[word] += 1
top = sorted(dic.items(), key=lambda x: x[1], reverse=True)[:30]
x, y = zip(*top)
plt.rcParams["figure.figsize"] = (20, 10)
plt.bar(x, y, color="green")
plt.figure(figsize=(10, 5))
import string
dic = defaultdict(int)
special = string.punctuation
for i in corpus:
if i in special:
dic[i] += 1
x, y = zip(*dic.items())
plt.barh(x, y, color="purple")
train["target_mean"] = train.groupby("keyword")["target"].transform("mean")
train
fig = plt.figure(figsize=(8, 72), dpi=100)
sns.countplot(
y=train.sort_values(by="target_mean", ascending=False)["keyword"],
hue=train.sort_values(by="target_mean", ascending=False)["target"],
)
plt.tick_params(axis="x", labelsize=15)
plt.tick_params(axis="y", labelsize=12)
plt.legend(loc=1)
plt.title("Target Distribution in Keywords")
plt.show()
train.drop(columns=["target_mean"], inplace=True)
from nltk.corpus import stopwords
from nltk.tokenize import sent_tokenize, word_tokenize
from nltk.stem import WordNetLemmatizer
def preprocess(df):
df["keyword"] = df["keyword"].fillna(" ")
df["location"] = df["location"].fillna(" ")
df["text"] = df["keyword"] + " " + df["location"] + " " + df["text"]
df = df.drop(columns=["keyword", "location"])
df["text_np"] = df["text"].apply(lambda x: re.sub("[^a-z]", " ", x))
df["text_np"] = df["text_np"].apply(lambda x: re.sub("\s+", " ", x))
stop = stopwords.words("english")
no_stop_words = df
no_stop_words["no_stopwords"] = no_stop_words["text_np"].apply(
lambda x: " ".join([word for word in x.split() if word not in (stop)])
)
no_stop_words["text_tokens"] = no_stop_words["no_stopwords"].apply(
lambda x: word_tokenize(x)
)
def word_lemmatizer(text):
lem_text = [WordNetLemmatizer().lemmatize(i) for i in text]
return lem_text
no_stop_words["text_clean_tokens"] = no_stop_words["text_tokens"].apply(
lambda x: word_lemmatizer(x)
)
no_stop_words["finished_lemma"] = no_stop_words["text_clean_tokens"].apply(
lambda x: " ".join(x)
)
return no_stop_words
df = preprocess(train)
df.sample(10)
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(
df["finished_lemma"], df["target"], test_size=0.25, random_state=10
)
print(X_train.shape)
print(X_test.shape)
print(Y_train.shape)
print(Y_test.shape)
from sklearn import feature_extraction
from sklearn.feature_extraction.text import TfidfVectorizer
tfidf = feature_extraction.text.TfidfVectorizer(
encoding="utf-8",
ngram_range=(1, 1),
max_features=5000,
norm="l2",
sublinear_tf=True,
)
train_features = tfidf.fit_transform(X_train).toarray()
test_features = tfidf.fit_transform(X_test).toarray()
train_labels = Y_train
test_labels = Y_test
print(train_features.shape)
print(train_labels.shape)
print(test_features.shape)
print(test_labels.shape)
import pandas as pd
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
mnb_classifier = MultinomialNB()
mnb_classifier.fit(train_features, train_labels)
mnb_prediction = mnb_classifier.predict(test_features)
training_accuracy = accuracy_score(train_labels, mnb_classifier.predict(train_features))
print(training_accuracy)
testing_accuracy = accuracy_score(test_labels, mnb_prediction)
print(testing_accuracy)
print(classification_report(test_labels, mnb_prediction))
test.shape
df_test = preprocess(test)
df_test.shape
test_vectorizer = tfidf.transform(df_test["finished_lemma"]).toarray()
test_vectorizer.shape
final_predictions = mnb_classifier.predict(test_vectorizer)
submission_df = pd.DataFrame()
submission_df["id"] = df_test["id"]
submission_df["target"] = final_predictions
submission = submission_df.to_csv("Results.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338852.ipynb
| null | null |
[{"Id": 69338852, "ScriptId": 18885741, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5774883, "CreationDate": "07/29/2021 18:40:02", "VersionNumber": 2.0, "Title": "nlp-disaster-tweet", "EvaluationDate": "07/29/2021", "IsChange": true, "TotalLines": 260.0, "LinesInsertedFromPrevious": 235.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 25.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
train = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
test = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
train.head(8)
train.shape
test.head(8)
test.shape
import seaborn as sns
import matplotlib.pyplot as plt
x = train.target.value_counts()
sns.barplot(x.index, x)
plt.gca().set_ylabel("samples")
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 5))
tweet_len = train[train["target"] == 1]["text"].str.len()
ax1.hist(tweet_len, color="blue")
ax1.set_title("disaster tweets")
tweet_len = train[train["target"] == 0]["text"].str.len()
ax2.hist(tweet_len, color="CRIMSON")
ax2.set_title("Not disaster tweets")
fig.suptitle("Characters in tweets")
plt.show()
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 5))
tweet_len = train[train["target"] == 1]["text"].str.split().map(lambda x: len(x))
ax1.hist(tweet_len, color="blue")
ax1.set_title("disaster tweets")
tweet_len = train[train["target"] == 0]["text"].str.split().map(lambda x: len(x))
ax2.hist(tweet_len, color="CRIMSON")
ax2.set_title("Not disaster tweets")
fig.suptitle("Words in a tweet")
plt.show()
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 5))
word = (
train[train["target"] == 1]["text"]
.str.split()
.apply(lambda x: np.mean([len(i) for i in x]))
)
sns.distplot(word, ax=ax1, color="red")
ax1.set_title("disaster")
word = (
train[train["target"] == 0]["text"]
.str.split()
.apply(lambda x: np.mean([len(i) for i in x]))
)
sns.distplot(word, ax=ax2, color="green")
ax2.set_title("Not disaster")
fig.suptitle("Average word length in each tweet")
from collections import defaultdict
import nltk
import nltk.corpus
from nltk.corpus import stopwords
nltk.download("stopwords")
stop = set(stopwords.words("english"))
corpus = []
for x in train["text"].str.split():
for i in x:
corpus.append(i)
dic = defaultdict(int)
for word in corpus:
if word not in stop:
dic[word] += 1
top = sorted(dic.items(), key=lambda x: x[1], reverse=True)[:30]
x, y = zip(*top)
plt.rcParams["figure.figsize"] = (20, 10)
plt.bar(x, y, color="red")
from collections import defaultdict
import nltk
import nltk.corpus
from nltk.corpus import stopwords
nltk.download("stopwords")
stop = set(stopwords.words("english"))
corpus = []
for x in train["text"].str.split():
for i in x:
corpus.append(i)
dic = defaultdict(int)
for word in corpus:
if word in stop:
dic[word] += 1
top = sorted(dic.items(), key=lambda x: x[1], reverse=True)[:30]
x, y = zip(*top)
plt.rcParams["figure.figsize"] = (20, 10)
plt.bar(x, y, color="green")
plt.figure(figsize=(10, 5))
import string
dic = defaultdict(int)
special = string.punctuation
for i in corpus:
if i in special:
dic[i] += 1
x, y = zip(*dic.items())
plt.barh(x, y, color="purple")
train["target_mean"] = train.groupby("keyword")["target"].transform("mean")
train
fig = plt.figure(figsize=(8, 72), dpi=100)
sns.countplot(
y=train.sort_values(by="target_mean", ascending=False)["keyword"],
hue=train.sort_values(by="target_mean", ascending=False)["target"],
)
plt.tick_params(axis="x", labelsize=15)
plt.tick_params(axis="y", labelsize=12)
plt.legend(loc=1)
plt.title("Target Distribution in Keywords")
plt.show()
train.drop(columns=["target_mean"], inplace=True)
from nltk.corpus import stopwords
from nltk.tokenize import sent_tokenize, word_tokenize
from nltk.stem import WordNetLemmatizer
def preprocess(df):
df["keyword"] = df["keyword"].fillna(" ")
df["location"] = df["location"].fillna(" ")
df["text"] = df["keyword"] + " " + df["location"] + " " + df["text"]
df = df.drop(columns=["keyword", "location"])
df["text_np"] = df["text"].apply(lambda x: re.sub("[^a-z]", " ", x))
df["text_np"] = df["text_np"].apply(lambda x: re.sub("\s+", " ", x))
stop = stopwords.words("english")
no_stop_words = df
no_stop_words["no_stopwords"] = no_stop_words["text_np"].apply(
lambda x: " ".join([word for word in x.split() if word not in (stop)])
)
no_stop_words["text_tokens"] = no_stop_words["no_stopwords"].apply(
lambda x: word_tokenize(x)
)
def word_lemmatizer(text):
lem_text = [WordNetLemmatizer().lemmatize(i) for i in text]
return lem_text
no_stop_words["text_clean_tokens"] = no_stop_words["text_tokens"].apply(
lambda x: word_lemmatizer(x)
)
no_stop_words["finished_lemma"] = no_stop_words["text_clean_tokens"].apply(
lambda x: " ".join(x)
)
return no_stop_words
df = preprocess(train)
df.sample(10)
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(
df["finished_lemma"], df["target"], test_size=0.25, random_state=10
)
print(X_train.shape)
print(X_test.shape)
print(Y_train.shape)
print(Y_test.shape)
from sklearn import feature_extraction
from sklearn.feature_extraction.text import TfidfVectorizer
tfidf = feature_extraction.text.TfidfVectorizer(
encoding="utf-8",
ngram_range=(1, 1),
max_features=5000,
norm="l2",
sublinear_tf=True,
)
train_features = tfidf.fit_transform(X_train).toarray()
test_features = tfidf.fit_transform(X_test).toarray()
train_labels = Y_train
test_labels = Y_test
print(train_features.shape)
print(train_labels.shape)
print(test_features.shape)
print(test_labels.shape)
import pandas as pd
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
mnb_classifier = MultinomialNB()
mnb_classifier.fit(train_features, train_labels)
mnb_prediction = mnb_classifier.predict(test_features)
training_accuracy = accuracy_score(train_labels, mnb_classifier.predict(train_features))
print(training_accuracy)
testing_accuracy = accuracy_score(test_labels, mnb_prediction)
print(testing_accuracy)
print(classification_report(test_labels, mnb_prediction))
test.shape
df_test = preprocess(test)
df_test.shape
test_vectorizer = tfidf.transform(df_test["finished_lemma"]).toarray()
test_vectorizer.shape
final_predictions = mnb_classifier.predict(test_vectorizer)
submission_df = pd.DataFrame()
submission_df["id"] = df_test["id"]
submission_df["target"] = final_predictions
submission = submission_df.to_csv("Results.csv", index=False)
| false | 0 | 2,328 | 0 | 2,328 | 2,328 |
||
69338727
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # IMPORTS
#
import tensorflow as tf
import matplotlib.pyplot as plt
from kaggle_datasets import KaggleDatasets
import os
import re
from sklearn.model_selection import train_test_split
SEED = 123
np.random.seed(SEED)
tf.random.set_seed(SEED)
DEVICE = "TPU"
BASEPATH = "../input/siim-isic-melanoma-classification"
# Detect hardware, return appropriate distribution strategy
try:
# TPU detection. No parameters necessary if TPU_NAME environment variable is
# set: this is always the case on Kaggle.
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
print("Running on TPU ", tpu.master())
except ValueError:
tpu = None
if tpu:
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
else:
# Default distribution strategy in Tensorflow. Works on CPU and single GPU.
strategy = tf.distribute.get_strategy()
print("REPLICAS: ", strategy.num_replicas_in_sync)
AUTO = tf.data.experimental.AUTOTUNE
# # Importation des données
#
f_train = pd.read_csv(os.path.join(BASEPATH, "train.csv"))
df_test = pd.read_csv(os.path.join(BASEPATH, "test.csv"))
GCS_PATH = KaggleDatasets().get_gcs_path("siim-isic-melanoma-classification")
TRAINING_FILENAMES = np.array(tf.io.gfile.glob(GCS_PATH + "/tfrecords/train*.tfrec"))
TEST_FILENAMES = np.array(tf.io.gfile.glob(GCS_PATH + "/tfrecords/test*.tfrec"))
CLASSES = [0, 1]
# # CODE des fonctions principales
def decode_image(image_data):
image = tf.image.decode_jpeg(image_data, channels=3)
image = tf.image.resize(image, [*IMAGE_SIZE])
image = (
tf.cast(image, tf.float32) / 255.0
) # convert image to floats in [0, 1] range
image = tf.reshape(image, [*IMAGE_SIZE, 3]) # explicit size needed for TPU
return image
def read_labeled_tfrecord(example):
LABELED_TFREC_FORMAT = {
"image": tf.io.FixedLenFeature([], tf.string), # tf.string means bytestring
# "class": tf.io.FixedLenFeature([], tf.int64), # shape [] means single element
"target": tf.io.FixedLenFeature([], tf.int64), # shape [] means single element
}
example = tf.io.parse_single_example(example, LABELED_TFREC_FORMAT)
image = decode_image(example["image"])
# label = tf.cast(example['class'], tf.int32)
label = tf.cast(example["target"], tf.int32)
return image, label # returns a dataset of (image, label) pairs
def read_unlabeled_tfrecord(example):
UNLABELED_TFREC_FORMAT = {
"image": tf.io.FixedLenFeature([], tf.string), # tf.string means bytestring
"image_name": tf.io.FixedLenFeature(
[], tf.string
), # shape [] means single element
# class is missing, this competitions's challenge is to predict flower classes for the test dataset
}
example = tf.io.parse_single_example(example, UNLABELED_TFREC_FORMAT)
image = decode_image(example["image"])
idnum = example["image_name"]
return image, idnum # returns a dataset of image(s)
def load_dataset(filenames, labeled=True, ordered=False):
# Read from TFRecords. For optimal performance, reading from multiple files at once and
# disregarding data order. Order does not matter since we will be shuffling the data anyway.
ignore_order = tf.data.Options()
if not ordered:
ignore_order.experimental_deterministic = False # disable order, increase speed
dataset = tf.data.TFRecordDataset(
filenames, num_parallel_reads=AUTO
) # automatically interleaves reads from multiple files
dataset = dataset.with_options(
ignore_order
) # uses data as soon as it streams in, rather than in its original order
dataset = dataset.map(
read_labeled_tfrecord if labeled else read_unlabeled_tfrecord,
num_parallel_calls=AUTO,
)
# returns a dataset of (image, label) pairs if labeled=True or (image, id) pairs if labeled=False
return dataset
##### FONCTION A DEVELOPPER POUR AMELIORER LE MODELE
def data_augment(image, label):
# image = tf.image.#implémentez ici les fonctions des transformations
return image, label
def get_training_dataset(augment=False):
dataset = load_dataset(TRAINING_FILENAMES, labeled=True)
if augment == True:
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.repeat() # the training dataset must repeat for several epochs
dataset = dataset.shuffle(SEED)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.prefetch(
AUTO
) # prefetch next batch while training (autotune prefetch buffer size)
return dataset
def get_validation_dataset(ordered=False):
dataset = load_dataset(VALIDATION_FILENAMES, labeled=True, ordered=ordered)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.cache()
dataset = dataset.prefetch(
AUTO
) # prefetch next batch while training (autotune prefetch buffer size)
return dataset
def get_test_dataset(ordered=False):
dataset = load_dataset(TEST_FILENAMES, labeled=False, ordered=ordered)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.prefetch(
AUTO
) # prefetch next batch while training (autotune prefetch buffer size)
return dataset
def count_data_items(filenames):
# the number of data items is written in the name of the .tfrec files, i.e. flowers00-230.tfrec = 230 data items
n = [
int(re.compile(r"-([0-9]*)\.").search(filename).group(1))
for filename in filenames
]
return np.sum(n)
def display_training_curves(training, validation, title, subplot):
"""
Source: https://www.kaggle.com/mgornergoogle/getting-started-with-100-flowers-on-tpu
"""
if subplot % 10 == 1: # set up the subplots on the first call
plt.subplots(figsize=(20, 15), facecolor="#F0F0F0")
plt.tight_layout()
ax = plt.subplot(subplot)
ax.set_facecolor("#F8F8F8")
ax.plot(training)
ax.plot(validation)
ax.set_title("model " + title)
ax.set_ylabel(title)
ax.set_xlabel("epoch")
ax.legend(["train", "valid."])
def prediction_test_csv(model, nom_model, df_sub):
test_ds = get_test_dataset(ordered=True)
print("Computing predictions...")
test_images_ds = test_ds.map(lambda image, idnum: image)
probabilities = model.predict(test_images_ds)
print("Generating submission.csv file...")
test_ids_ds = test_ds.map(lambda image, idnum: idnum).unbatch()
test_ids = (
next(iter(test_ids_ds.batch(NUM_TEST_IMAGES))).numpy().astype("U")
) # all in one batch
pred_df = pd.DataFrame(
{"image_name": test_ids, "target": np.concatenate(probabilities)}
)
pred_df.head()
del df_sub["target"]
df_sub = df_sub.merge(pred_df, on="image_name")
# sub.to_csv('submission_label_smoothing.csv', index=False)
df_sub.to_csv("submission_" + nom_model + ".csv", index=False)
print(df_sub.head())
# # Chargement et exploration du dataset
train_df = pd.read_csv("../input/siim-isic-melanoma-classification/train.csv")
train_df.shape
train_df.head()
# Chercher d'éventuelles valeurs nulles
train_df.isnull().sum()
# # Modèle utilisé : LeNet5
# Définition des méta paramètres
#
EPOCHS = 1500 # le nombre d'itération pour l'apprentissage du modèle
BATCH_SIZE = 8 * strategy.num_replicas_in_sync # le nombre d'images traitées à la fois
IMAGE_SIZE = [32, 32] # liste [hauteur, largeur] de l'image
IMAGE_CHANNEL = 3 # 1 en gris, 3 en couleur
LR = 100 # le taux d'apprentissage
# Création du jeu de validation
TRAINING_FILENAMES, VALIDATION_FILENAMES = train_test_split(
TRAINING_FILENAMES, test_size=0.2, random_state=SEED
)
NUM_TRAINING_IMAGES = count_data_items(TRAINING_FILENAMES)
NUM_VALIDATION_IMAGES = count_data_items(VALIDATION_FILENAMES)
NUM_TEST_IMAGES = count_data_items(TEST_FILENAMES)
STEPS_PER_EPOCH = NUM_TRAINING_IMAGES // BATCH_SIZE
print(
"Dataset: {} training images, {} validation images, {} unlabeled test images".format(
NUM_TRAINING_IMAGES, NUM_VALIDATION_IMAGES, NUM_TEST_IMAGES
)
)
# Création de la structure du modèle
with strategy.scope():
lenet5_model = tf.keras.Sequential(
[
tf.keras.layers.Conv2D(
6,
(5, 5),
activation="relu",
input_shape=(IMAGE_SIZE[0], IMAGE_SIZE[1], IMAGE_CHANNEL),
),
tf.keras.layers.MaxPooling2D(),
# une autre couche de convolution: 16 filtres 5x5, également avec une activation relu. Ne pas spécifier de format d'entrée (input shape)
# une autre couche maxpooling 2D
tf.keras.layers.Flatten(),
# une couche de neurones tf.keras.layers.Dense: 120 neurones, activation relu
# une couche de neurones tf.keras.layers.Dense: 84 neurones, activation relu
# une couche de neurones tf.keras.layers.Dense: 1 neurones, activation sigmoid
]
)
lenet5_model.summary()
adam = tf.keras.optimizers.Adam(lr=LR, beta_1=0.9, beta_2=0.999, amsgrad=False)
loss = tf.keras.losses.BinaryCrossentropy(from_logits=False)
lenet5_model.compile(
loss=loss, metrics=[tf.keras.metrics.AUC(name="auc")], optimizer=adam
)
# Code d'entrainement du modèle
NUM_VALIDATION_IMAGES = count_data_items(VALIDATION_FILENAMES)
train_df = pd.read_csv("../input/siim-isic-melanoma-classification/train.csv")
train_df.shape
train_df["target"].value_counts()
# Visualisation apprentissage
display_training_curves(
history.history["loss"], history.history["val_loss"], "loss", 311
)
display_training_curves(history.history["auc"], history.history["val_auc"], "auc", 312)
df_sub = pd.read_csv(os.path.join(BASEPATH, "sample_submission.csv"))
nom_model = "lenet5" # servira simplement à nommer votre fichier excel
prediction_test_csv(lenet5_model, nom_model, df_sub)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338727.ipynb
| null | null |
[{"Id": 69338727, "ScriptId": 18930089, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7815771, "CreationDate": "07/29/2021 18:37:26", "VersionNumber": 2.0, "Title": "Traitement par l\u2019Intelligence Artificielle", "EvaluationDate": "07/29/2021", "IsChange": false, "TotalLines": 265.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 265.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # IMPORTS
#
import tensorflow as tf
import matplotlib.pyplot as plt
from kaggle_datasets import KaggleDatasets
import os
import re
from sklearn.model_selection import train_test_split
SEED = 123
np.random.seed(SEED)
tf.random.set_seed(SEED)
DEVICE = "TPU"
BASEPATH = "../input/siim-isic-melanoma-classification"
# Detect hardware, return appropriate distribution strategy
try:
# TPU detection. No parameters necessary if TPU_NAME environment variable is
# set: this is always the case on Kaggle.
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
print("Running on TPU ", tpu.master())
except ValueError:
tpu = None
if tpu:
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
else:
# Default distribution strategy in Tensorflow. Works on CPU and single GPU.
strategy = tf.distribute.get_strategy()
print("REPLICAS: ", strategy.num_replicas_in_sync)
AUTO = tf.data.experimental.AUTOTUNE
# # Importation des données
#
f_train = pd.read_csv(os.path.join(BASEPATH, "train.csv"))
df_test = pd.read_csv(os.path.join(BASEPATH, "test.csv"))
GCS_PATH = KaggleDatasets().get_gcs_path("siim-isic-melanoma-classification")
TRAINING_FILENAMES = np.array(tf.io.gfile.glob(GCS_PATH + "/tfrecords/train*.tfrec"))
TEST_FILENAMES = np.array(tf.io.gfile.glob(GCS_PATH + "/tfrecords/test*.tfrec"))
CLASSES = [0, 1]
# # CODE des fonctions principales
def decode_image(image_data):
image = tf.image.decode_jpeg(image_data, channels=3)
image = tf.image.resize(image, [*IMAGE_SIZE])
image = (
tf.cast(image, tf.float32) / 255.0
) # convert image to floats in [0, 1] range
image = tf.reshape(image, [*IMAGE_SIZE, 3]) # explicit size needed for TPU
return image
def read_labeled_tfrecord(example):
LABELED_TFREC_FORMAT = {
"image": tf.io.FixedLenFeature([], tf.string), # tf.string means bytestring
# "class": tf.io.FixedLenFeature([], tf.int64), # shape [] means single element
"target": tf.io.FixedLenFeature([], tf.int64), # shape [] means single element
}
example = tf.io.parse_single_example(example, LABELED_TFREC_FORMAT)
image = decode_image(example["image"])
# label = tf.cast(example['class'], tf.int32)
label = tf.cast(example["target"], tf.int32)
return image, label # returns a dataset of (image, label) pairs
def read_unlabeled_tfrecord(example):
UNLABELED_TFREC_FORMAT = {
"image": tf.io.FixedLenFeature([], tf.string), # tf.string means bytestring
"image_name": tf.io.FixedLenFeature(
[], tf.string
), # shape [] means single element
# class is missing, this competitions's challenge is to predict flower classes for the test dataset
}
example = tf.io.parse_single_example(example, UNLABELED_TFREC_FORMAT)
image = decode_image(example["image"])
idnum = example["image_name"]
return image, idnum # returns a dataset of image(s)
def load_dataset(filenames, labeled=True, ordered=False):
# Read from TFRecords. For optimal performance, reading from multiple files at once and
# disregarding data order. Order does not matter since we will be shuffling the data anyway.
ignore_order = tf.data.Options()
if not ordered:
ignore_order.experimental_deterministic = False # disable order, increase speed
dataset = tf.data.TFRecordDataset(
filenames, num_parallel_reads=AUTO
) # automatically interleaves reads from multiple files
dataset = dataset.with_options(
ignore_order
) # uses data as soon as it streams in, rather than in its original order
dataset = dataset.map(
read_labeled_tfrecord if labeled else read_unlabeled_tfrecord,
num_parallel_calls=AUTO,
)
# returns a dataset of (image, label) pairs if labeled=True or (image, id) pairs if labeled=False
return dataset
##### FONCTION A DEVELOPPER POUR AMELIORER LE MODELE
def data_augment(image, label):
# image = tf.image.#implémentez ici les fonctions des transformations
return image, label
def get_training_dataset(augment=False):
dataset = load_dataset(TRAINING_FILENAMES, labeled=True)
if augment == True:
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.repeat() # the training dataset must repeat for several epochs
dataset = dataset.shuffle(SEED)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.prefetch(
AUTO
) # prefetch next batch while training (autotune prefetch buffer size)
return dataset
def get_validation_dataset(ordered=False):
dataset = load_dataset(VALIDATION_FILENAMES, labeled=True, ordered=ordered)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.cache()
dataset = dataset.prefetch(
AUTO
) # prefetch next batch while training (autotune prefetch buffer size)
return dataset
def get_test_dataset(ordered=False):
dataset = load_dataset(TEST_FILENAMES, labeled=False, ordered=ordered)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.prefetch(
AUTO
) # prefetch next batch while training (autotune prefetch buffer size)
return dataset
def count_data_items(filenames):
# the number of data items is written in the name of the .tfrec files, i.e. flowers00-230.tfrec = 230 data items
n = [
int(re.compile(r"-([0-9]*)\.").search(filename).group(1))
for filename in filenames
]
return np.sum(n)
def display_training_curves(training, validation, title, subplot):
"""
Source: https://www.kaggle.com/mgornergoogle/getting-started-with-100-flowers-on-tpu
"""
if subplot % 10 == 1: # set up the subplots on the first call
plt.subplots(figsize=(20, 15), facecolor="#F0F0F0")
plt.tight_layout()
ax = plt.subplot(subplot)
ax.set_facecolor("#F8F8F8")
ax.plot(training)
ax.plot(validation)
ax.set_title("model " + title)
ax.set_ylabel(title)
ax.set_xlabel("epoch")
ax.legend(["train", "valid."])
def prediction_test_csv(model, nom_model, df_sub):
test_ds = get_test_dataset(ordered=True)
print("Computing predictions...")
test_images_ds = test_ds.map(lambda image, idnum: image)
probabilities = model.predict(test_images_ds)
print("Generating submission.csv file...")
test_ids_ds = test_ds.map(lambda image, idnum: idnum).unbatch()
test_ids = (
next(iter(test_ids_ds.batch(NUM_TEST_IMAGES))).numpy().astype("U")
) # all in one batch
pred_df = pd.DataFrame(
{"image_name": test_ids, "target": np.concatenate(probabilities)}
)
pred_df.head()
del df_sub["target"]
df_sub = df_sub.merge(pred_df, on="image_name")
# sub.to_csv('submission_label_smoothing.csv', index=False)
df_sub.to_csv("submission_" + nom_model + ".csv", index=False)
print(df_sub.head())
# # Chargement et exploration du dataset
train_df = pd.read_csv("../input/siim-isic-melanoma-classification/train.csv")
train_df.shape
train_df.head()
# Chercher d'éventuelles valeurs nulles
train_df.isnull().sum()
# # Modèle utilisé : LeNet5
# Définition des méta paramètres
#
EPOCHS = 1500 # le nombre d'itération pour l'apprentissage du modèle
BATCH_SIZE = 8 * strategy.num_replicas_in_sync # le nombre d'images traitées à la fois
IMAGE_SIZE = [32, 32] # liste [hauteur, largeur] de l'image
IMAGE_CHANNEL = 3 # 1 en gris, 3 en couleur
LR = 100 # le taux d'apprentissage
# Création du jeu de validation
TRAINING_FILENAMES, VALIDATION_FILENAMES = train_test_split(
TRAINING_FILENAMES, test_size=0.2, random_state=SEED
)
NUM_TRAINING_IMAGES = count_data_items(TRAINING_FILENAMES)
NUM_VALIDATION_IMAGES = count_data_items(VALIDATION_FILENAMES)
NUM_TEST_IMAGES = count_data_items(TEST_FILENAMES)
STEPS_PER_EPOCH = NUM_TRAINING_IMAGES // BATCH_SIZE
print(
"Dataset: {} training images, {} validation images, {} unlabeled test images".format(
NUM_TRAINING_IMAGES, NUM_VALIDATION_IMAGES, NUM_TEST_IMAGES
)
)
# Création de la structure du modèle
with strategy.scope():
lenet5_model = tf.keras.Sequential(
[
tf.keras.layers.Conv2D(
6,
(5, 5),
activation="relu",
input_shape=(IMAGE_SIZE[0], IMAGE_SIZE[1], IMAGE_CHANNEL),
),
tf.keras.layers.MaxPooling2D(),
# une autre couche de convolution: 16 filtres 5x5, également avec une activation relu. Ne pas spécifier de format d'entrée (input shape)
# une autre couche maxpooling 2D
tf.keras.layers.Flatten(),
# une couche de neurones tf.keras.layers.Dense: 120 neurones, activation relu
# une couche de neurones tf.keras.layers.Dense: 84 neurones, activation relu
# une couche de neurones tf.keras.layers.Dense: 1 neurones, activation sigmoid
]
)
lenet5_model.summary()
adam = tf.keras.optimizers.Adam(lr=LR, beta_1=0.9, beta_2=0.999, amsgrad=False)
loss = tf.keras.losses.BinaryCrossentropy(from_logits=False)
lenet5_model.compile(
loss=loss, metrics=[tf.keras.metrics.AUC(name="auc")], optimizer=adam
)
# Code d'entrainement du modèle
NUM_VALIDATION_IMAGES = count_data_items(VALIDATION_FILENAMES)
train_df = pd.read_csv("../input/siim-isic-melanoma-classification/train.csv")
train_df.shape
train_df["target"].value_counts()
# Visualisation apprentissage
display_training_curves(
history.history["loss"], history.history["val_loss"], "loss", 311
)
display_training_curves(history.history["auc"], history.history["val_auc"], "auc", 312)
df_sub = pd.read_csv(os.path.join(BASEPATH, "sample_submission.csv"))
nom_model = "lenet5" # servira simplement à nommer votre fichier excel
prediction_test_csv(lenet5_model, nom_model, df_sub)
| false | 0 | 3,210 | 0 | 3,210 | 3,210 |
||
69338963
|
<jupyter_start><jupyter_text>Credit Card Fraud Detection
Context
---------
It is important that credit card companies are able to recognize fraudulent credit card transactions so that customers are not charged for items that they did not purchase.
Content
---------
The dataset contains transactions made by credit cards in September 2013 by European cardholders.
This dataset presents transactions that occurred in two days, where we have 492 frauds out of 284,807 transactions. The dataset is highly unbalanced, the positive class (frauds) account for 0.172% of all transactions.
It contains only numerical input variables which are the result of a PCA transformation. Unfortunately, due to confidentiality issues, we cannot provide the original features and more background information about the data. Features V1, V2, ... V28 are the principal components obtained with PCA, the only features which have not been transformed with PCA are 'Time' and 'Amount'. Feature 'Time' contains the seconds elapsed between each transaction and the first transaction in the dataset. The feature 'Amount' is the transaction Amount, this feature can be used for example-dependant cost-sensitive learning. Feature 'Class' is the response variable and it takes value 1 in case of fraud and 0 otherwise.
Given the class imbalance ratio, we recommend measuring the accuracy using the Area Under the Precision-Recall Curve (AUPRC). Confusion matrix accuracy is not meaningful for unbalanced classification.
Update (03/05/2021)
---------
A simulator for transaction data has been released as part of the practical handbook on Machine Learning for Credit Card Fraud Detection - https://fraud-detection-handbook.github.io/fraud-detection-handbook/Chapter_3_GettingStarted/SimulatedDataset.html. We invite all practitioners interested in fraud detection datasets to also check out this data simulator, and the methodologies for credit card fraud detection presented in the book.
Acknowledgements
---------
The dataset has been collected and analysed during a research collaboration of Worldline and the Machine Learning Group (http://mlg.ulb.ac.be) of ULB (Université Libre de Bruxelles) on big data mining and fraud detection.
More details on current and past projects on related topics are available on [https://www.researchgate.net/project/Fraud-detection-5][1] and the page of the [DefeatFraud][2] project
Please cite the following works:
Andrea Dal Pozzolo, Olivier Caelen, Reid A. Johnson and Gianluca Bontempi. [Calibrating Probability with Undersampling for Unbalanced Classification.][3] In Symposium on Computational Intelligence and Data Mining (CIDM), IEEE, 2015
Dal Pozzolo, Andrea; Caelen, Olivier; Le Borgne, Yann-Ael; Waterschoot, Serge; Bontempi, Gianluca. [Learned lessons in credit card fraud detection from a practitioner perspective][4], Expert systems with applications,41,10,4915-4928,2014, Pergamon
Dal Pozzolo, Andrea; Boracchi, Giacomo; Caelen, Olivier; Alippi, Cesare; Bontempi, Gianluca. [Credit card fraud detection: a realistic modeling and a novel learning strategy,][5] IEEE transactions on neural networks and learning systems,29,8,3784-3797,2018,IEEE
Dal Pozzolo, Andrea [Adaptive Machine learning for credit card fraud detection][6] ULB MLG PhD thesis (supervised by G. Bontempi)
Carcillo, Fabrizio; Dal Pozzolo, Andrea; Le Borgne, Yann-Aël; Caelen, Olivier; Mazzer, Yannis; Bontempi, Gianluca. [Scarff: a scalable framework for streaming credit card fraud detection with Spark][7], Information fusion,41, 182-194,2018,Elsevier
Carcillo, Fabrizio; Le Borgne, Yann-Aël; Caelen, Olivier; Bontempi, Gianluca. [Streaming active learning strategies for real-life credit card fraud detection: assessment and visualization,][8] International Journal of Data Science and Analytics, 5,4,285-300,2018,Springer International Publishing
Bertrand Lebichot, Yann-Aël Le Borgne, Liyun He, Frederic Oblé, Gianluca Bontempi [Deep-Learning Domain Adaptation Techniques for Credit Cards Fraud Detection](https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection), INNSBDDL 2019: Recent Advances in Big Data and Deep Learning, pp 78-88, 2019
Fabrizio Carcillo, Yann-Aël Le Borgne, Olivier Caelen, Frederic Oblé, Gianluca Bontempi [Combining Unsupervised and Supervised Learning in Credit Card Fraud Detection ](https://www.researchgate.net/publication/333143698_Combining_Unsupervised_and_Supervised_Learning_in_Credit_Card_Fraud_Detection) Information Sciences, 2019
Yann-Aël Le Borgne, Gianluca Bontempi [Reproducible machine Learning for Credit Card Fraud Detection - Practical Handbook ](https://www.researchgate.net/publication/351283764_Machine_Learning_for_Credit_Card_Fraud_Detection_-_Practical_Handbook)
Bertrand Lebichot, Gianmarco Paldino, Wissam Siblini, Liyun He, Frederic Oblé, Gianluca Bontempi [Incremental learning strategies for credit cards fraud detection](https://www.researchgate.net/publication/352275169_Incremental_learning_strategies_for_credit_cards_fraud_detection), IInternational Journal of Data Science and Analytics
[1]: https://www.researchgate.net/project/Fraud-detection-5
[2]: https://mlg.ulb.ac.be/wordpress/portfolio_page/defeatfraud-assessment-and-validation-of-deep-feature-engineering-and-learning-solutions-for-fraud-detection/
[3]: https://www.researchgate.net/publication/283349138_Calibrating_Probability_with_Undersampling_for_Unbalanced_Classification
[4]: https://www.researchgate.net/publication/260837261_Learned_lessons_in_credit_card_fraud_detection_from_a_practitioner_perspective
[5]: https://www.researchgate.net/publication/319867396_Credit_Card_Fraud_Detection_A_Realistic_Modeling_and_a_Novel_Learning_Strategy
[6]: http://di.ulb.ac.be/map/adalpozz/pdf/Dalpozzolo2015PhD.pdf
[7]: https://www.researchgate.net/publication/319616537_SCARFF_a_Scalable_Framework_for_Streaming_Credit_Card_Fraud_Detection_with_Spark
[8]: https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection
Kaggle dataset identifier: creditcardfraud
<jupyter_script># ****Credit Card Fraud detection using machine learning models ****
# **Introduction**
# In this noteebook we will find out which are the best machine learning models to predict the credit card fraud. As the data says , features of this data is scaled and the reason the name of features are not shown is due to privacy reasons.
# **Summary**
# This dataset contains the information of the credit card holders of the Europeans and the credit card transactions made by them. This data is highly skewed and becasue the data is confenditial, it is anonymized. Features V1, V2,.....V28 are the obtianed by PCA except the 'Amount' and 'Time' features. Here are target value is Class as this will tells us if the transacation made is fraud or non-fraud .
# **Importing the required libraries**
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import math
from matplotlib.ticker import FuncFormatter
from collections import Counter
import math
import scipy.stats as ss
sns.set(color_codes=True)
plt.style.use("bmh")
data = pd.read_csv("../input/creditcardfraud/creditcard.csv")
data.head()
# **Data profiling**
data.shape
data.info()
data.isna().sum()
# to check if any duplicate values are present
data.duplicated().sum()
# There are 1081 duplicated values in this dataset and its important to remove the these duplicate values from the data for the further analysis:
# need to drop the duplicate value as it can manipulate the data
data.drop_duplicates(subset=None, inplace=True)
# 1. 1.**Univariate analysis**
# Here all all the columns have numerical/ continuous data except the 'Class' column which is a categorigcal data since it is presented in boolean values 0 or 1
# Finding out how many are fraud and non-fraud cases in this dataset. Therefore our target value here is 'Class'.
data["Class"].value_counts()
data["Class"].value_counts().plot(kind="bar")
# To find the distribution of the target value, we will find the percentage distribution of the cases:
data["Class"].value_counts() / data["Class"].count() * 100
# By observing this we can clearly say that there our only 0.16% fraud cases in the given data and this implies it is highly skewed data. So, it's important to balance the data. You would be seeing how we achieved that further in this notebook
# **1. 2. Bivariate analysis**
# Will find out how the Amount is distributed among our target value which is 'Class'
sns.scatterplot(x=data["Amount"], y=data["Class"])
# **Correlation Matrix**
fig, ax = plt.subplots(figsize=(12, 10))
correlation = data.corr()
sns.heatmap(
correlation,
xticklabels=correlation.columns.values,
yticklabels=correlation.columns.values,
)
# Finding which are more correlated to our target value
data.corr()["Class"].sort_values(ascending=False)
# Here variable 'V1', 'V2' till'V28' are in PCA form which means they went through reduction techinique already therefore they are not correlated to our target value . Similarly time is highly uncorrelated to our target value therefore we can drop it for our further analysis.
# ****Using SMOTE to make the data balanced** **
# This technique is used to oversample the minority class which is non-fraud cases here. This generates new synthetic samples, so it's necessary to rescale your data. For that purpose we will use Standardsacler
# import the Standardscaler first
from sklearn.preprocessing import StandardScaler
ss = StandardScaler()
data["s_amount"] = ss.fit_transform(data["Amount"].values.reshape(-1, 1))
data.head()
# Dropping the columns time and Amount , as mentioned before there is no correlation between the time and the target value and Amount has bee rescaled and new column is created for that. So, thats why we will be dropping these two variables.
data = data.drop(["Amount", "Time"], axis=1)
data.head()
# *****Using Smote Technique to balance the data *****
# splitting the data and then training it
x = data.drop("Class", axis=1).values
y = data["Class"].values
from sklearn.model_selection import train_test_split
# Train and test split using sklearn
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=1234
)
# import the library
from imblearn.over_sampling import SMOTE
from collections import Counter
smote = SMOTE()
x_smote, y_smote = smote.fit_resample(x_train, y_train)
print(x_smote.shape, y_smote.shape)
# # **Predictive Models**
# ****Logistic Regression ******
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
precision_score,
f1_score,
recall_score,
precision_recall_fscore_support,
classification_report,
)
from sklearn.metrics import plot_confusion_matrix
# intializing the model
LR = LogisticRegression(max_iter=7600)
# splitting and training of the dataset is already done. So, we will do model fitting now
#
# model fitting
LR.fit(x_smote, y_smote)
# predicting the model
ypred = LR.predict(x_test)
print(classification_report(y_test, ypred))
print(accuracy_score(y_test, ypred))
# **Feature importance**
importance = list(zip(data.columns, LR.coef_.ravel()))
importance = list(sorted(importance, key=lambda x: x[1], reverse=True))
# plotting the top feature importance
top_columns, top_score = zip(*importance[:5])
plt.xticks(rotation=60)
plt.bar(top_columns, top_score)
plt.show()
# **Decision Tree model**
from sklearn.tree import DecisionTreeClassifier
dt = DecisionTreeClassifier()
dt.fit(x_smote, y_smote)
y1pred = dt.predict(x_test)
print(classification_report(y_test, y1pred))
print(accuracy_score(y_test, y1pred.round()))
print(precision_score(y_test, y1pred.round()))
print(recall_score(y_test, y1pred.round()))
print(f1_score(y_test, y1pred.round()))
# ****Random Forest ****
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators=100)
rf.fit(x_smote, y_smote)
pred_rf = rf.predict(x_test)
print(classification_report(y_test, pred_rf))
plot_confusion_matrix(rf, x_test, y_test)
plt.show()
# **XGboost Classifier**
from xgboost import XGBClassifier
import xgboost as xgb
model = xgb.XGBClassifier(random_state=123)
model.fit(x_smote, y_smote)
xpred = model.predict(x_test)
print(classification_report(y_test, xpred))
plot_confusion_matrix(model, x_test, y_test)
plt.show()
print(accuracy_score(y_test, xpred))
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338963.ipynb
|
creditcardfraud
| null |
[{"Id": 69338963, "ScriptId": 18601880, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5975800, "CreationDate": "07/29/2021 18:41:54", "VersionNumber": 1.0, "Title": "credit card fraud detection- Using ML algorithms", "EvaluationDate": "07/29/2021", "IsChange": true, "TotalLines": 210.0, "LinesInsertedFromPrevious": 210.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92385191, "KernelVersionId": 69338963, "SourceDatasetVersionId": 23498}]
|
[{"Id": 23498, "DatasetId": 310, "DatasourceVersionId": 23502, "CreatorUserId": 998023, "LicenseName": "Database: Open Database, Contents: Database Contents", "CreationDate": "03/23/2018 01:17:27", "VersionNumber": 3.0, "Title": "Credit Card Fraud Detection", "Slug": "creditcardfraud", "Subtitle": "Anonymized credit card transactions labeled as fraudulent or genuine", "Description": "Context\n---------\n\nIt is important that credit card companies are able to recognize fraudulent credit card transactions so that customers are not charged for items that they did not purchase.\n\nContent\n---------\n\nThe dataset contains transactions made by credit cards in September 2013 by European cardholders. \nThis dataset presents transactions that occurred in two days, where we have 492 frauds out of 284,807 transactions. The dataset is highly unbalanced, the positive class (frauds) account for 0.172% of all transactions.\n\nIt contains only numerical input variables which are the result of a PCA transformation. Unfortunately, due to confidentiality issues, we cannot provide the original features and more background information about the data. Features V1, V2, ... V28 are the principal components obtained with PCA, the only features which have not been transformed with PCA are 'Time' and 'Amount'. Feature 'Time' contains the seconds elapsed between each transaction and the first transaction in the dataset. The feature 'Amount' is the transaction Amount, this feature can be used for example-dependant cost-sensitive learning. Feature 'Class' is the response variable and it takes value 1 in case of fraud and 0 otherwise. \n\nGiven the class imbalance ratio, we recommend measuring the accuracy using the Area Under the Precision-Recall Curve (AUPRC). Confusion matrix accuracy is not meaningful for unbalanced classification.\n\nUpdate (03/05/2021)\n---------\n\nA simulator for transaction data has been released as part of the practical handbook on Machine Learning for Credit Card Fraud Detection - https://fraud-detection-handbook.github.io/fraud-detection-handbook/Chapter_3_GettingStarted/SimulatedDataset.html. We invite all practitioners interested in fraud detection datasets to also check out this data simulator, and the methodologies for credit card fraud detection presented in the book.\n\nAcknowledgements\n---------\n\nThe dataset has been collected and analysed during a research collaboration of Worldline and the Machine Learning Group (http://mlg.ulb.ac.be) of ULB (Universit\u00e9 Libre de Bruxelles) on big data mining and fraud detection.\nMore details on current and past projects on related topics are available on [https://www.researchgate.net/project/Fraud-detection-5][1] and the page of the [DefeatFraud][2] project\n\nPlease cite the following works: \n\nAndrea Dal Pozzolo, Olivier Caelen, Reid A. Johnson and Gianluca Bontempi. [Calibrating Probability with Undersampling for Unbalanced Classification.][3] In Symposium on Computational Intelligence and Data Mining (CIDM), IEEE, 2015\n\nDal Pozzolo, Andrea; Caelen, Olivier; Le Borgne, Yann-Ael; Waterschoot, Serge; Bontempi, Gianluca. [Learned lessons in credit card fraud detection from a practitioner perspective][4], Expert systems with applications,41,10,4915-4928,2014, Pergamon\n\nDal Pozzolo, Andrea; Boracchi, Giacomo; Caelen, Olivier; Alippi, Cesare; Bontempi, Gianluca. [Credit card fraud detection: a realistic modeling and a novel learning strategy,][5] IEEE transactions on neural networks and learning systems,29,8,3784-3797,2018,IEEE\n\nDal Pozzolo, Andrea [Adaptive Machine learning for credit card fraud detection][6] ULB MLG PhD thesis (supervised by G. Bontempi)\n\nCarcillo, Fabrizio; Dal Pozzolo, Andrea; Le Borgne, Yann-A\u00ebl; Caelen, Olivier; Mazzer, Yannis; Bontempi, Gianluca. [Scarff: a scalable framework for streaming credit card fraud detection with Spark][7], Information fusion,41, 182-194,2018,Elsevier\n\nCarcillo, Fabrizio; Le Borgne, Yann-A\u00ebl; Caelen, Olivier; Bontempi, Gianluca. [Streaming active learning strategies for real-life credit card fraud detection: assessment and visualization,][8] International Journal of Data Science and Analytics, 5,4,285-300,2018,Springer International Publishing\n\nBertrand Lebichot, Yann-A\u00ebl Le Borgne, Liyun He, Frederic Obl\u00e9, Gianluca Bontempi [Deep-Learning Domain Adaptation Techniques for Credit Cards Fraud Detection](https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection), INNSBDDL 2019: Recent Advances in Big Data and Deep Learning, pp 78-88, 2019\n\nFabrizio Carcillo, Yann-A\u00ebl Le Borgne, Olivier Caelen, Frederic Obl\u00e9, Gianluca Bontempi [Combining Unsupervised and Supervised Learning in Credit Card Fraud Detection ](https://www.researchgate.net/publication/333143698_Combining_Unsupervised_and_Supervised_Learning_in_Credit_Card_Fraud_Detection) Information Sciences, 2019\n\nYann-A\u00ebl Le Borgne, Gianluca Bontempi [Reproducible machine Learning for Credit Card Fraud Detection - Practical Handbook ](https://www.researchgate.net/publication/351283764_Machine_Learning_for_Credit_Card_Fraud_Detection_-_Practical_Handbook) \n\nBertrand Lebichot, Gianmarco Paldino, Wissam Siblini, Liyun He, Frederic Obl\u00e9, Gianluca Bontempi [Incremental learning strategies for credit cards fraud detection](https://www.researchgate.net/publication/352275169_Incremental_learning_strategies_for_credit_cards_fraud_detection), IInternational Journal of Data Science and Analytics\n\n [1]: https://www.researchgate.net/project/Fraud-detection-5\n [2]: https://mlg.ulb.ac.be/wordpress/portfolio_page/defeatfraud-assessment-and-validation-of-deep-feature-engineering-and-learning-solutions-for-fraud-detection/\n [3]: https://www.researchgate.net/publication/283349138_Calibrating_Probability_with_Undersampling_for_Unbalanced_Classification\n [4]: https://www.researchgate.net/publication/260837261_Learned_lessons_in_credit_card_fraud_detection_from_a_practitioner_perspective\n [5]: https://www.researchgate.net/publication/319867396_Credit_Card_Fraud_Detection_A_Realistic_Modeling_and_a_Novel_Learning_Strategy\n [6]: http://di.ulb.ac.be/map/adalpozz/pdf/Dalpozzolo2015PhD.pdf\n [7]: https://www.researchgate.net/publication/319616537_SCARFF_a_Scalable_Framework_for_Streaming_Credit_Card_Fraud_Detection_with_Spark\n \n[8]: https://www.researchgate.net/publication/332180999_Deep-Learning_Domain_Adaptation_Techniques_for_Credit_Cards_Fraud_Detection", "VersionNotes": "Fixed preview", "TotalCompressedBytes": 150828752.0, "TotalUncompressedBytes": 69155632.0}]
|
[{"Id": 310, "CreatorUserId": 14069, "OwnerUserId": NaN, "OwnerOrganizationId": 1160.0, "CurrentDatasetVersionId": 23498.0, "CurrentDatasourceVersionId": 23502.0, "ForumId": 1838, "Type": 2, "CreationDate": "11/03/2016 13:21:36", "LastActivityDate": "02/06/2018", "TotalViews": 10310781, "TotalDownloads": 564249, "TotalVotes": 10432, "TotalKernels": 4266}]
| null |
# ****Credit Card Fraud detection using machine learning models ****
# **Introduction**
# In this noteebook we will find out which are the best machine learning models to predict the credit card fraud. As the data says , features of this data is scaled and the reason the name of features are not shown is due to privacy reasons.
# **Summary**
# This dataset contains the information of the credit card holders of the Europeans and the credit card transactions made by them. This data is highly skewed and becasue the data is confenditial, it is anonymized. Features V1, V2,.....V28 are the obtianed by PCA except the 'Amount' and 'Time' features. Here are target value is Class as this will tells us if the transacation made is fraud or non-fraud .
# **Importing the required libraries**
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import math
from matplotlib.ticker import FuncFormatter
from collections import Counter
import math
import scipy.stats as ss
sns.set(color_codes=True)
plt.style.use("bmh")
data = pd.read_csv("../input/creditcardfraud/creditcard.csv")
data.head()
# **Data profiling**
data.shape
data.info()
data.isna().sum()
# to check if any duplicate values are present
data.duplicated().sum()
# There are 1081 duplicated values in this dataset and its important to remove the these duplicate values from the data for the further analysis:
# need to drop the duplicate value as it can manipulate the data
data.drop_duplicates(subset=None, inplace=True)
# 1. 1.**Univariate analysis**
# Here all all the columns have numerical/ continuous data except the 'Class' column which is a categorigcal data since it is presented in boolean values 0 or 1
# Finding out how many are fraud and non-fraud cases in this dataset. Therefore our target value here is 'Class'.
data["Class"].value_counts()
data["Class"].value_counts().plot(kind="bar")
# To find the distribution of the target value, we will find the percentage distribution of the cases:
data["Class"].value_counts() / data["Class"].count() * 100
# By observing this we can clearly say that there our only 0.16% fraud cases in the given data and this implies it is highly skewed data. So, it's important to balance the data. You would be seeing how we achieved that further in this notebook
# **1. 2. Bivariate analysis**
# Will find out how the Amount is distributed among our target value which is 'Class'
sns.scatterplot(x=data["Amount"], y=data["Class"])
# **Correlation Matrix**
fig, ax = plt.subplots(figsize=(12, 10))
correlation = data.corr()
sns.heatmap(
correlation,
xticklabels=correlation.columns.values,
yticklabels=correlation.columns.values,
)
# Finding which are more correlated to our target value
data.corr()["Class"].sort_values(ascending=False)
# Here variable 'V1', 'V2' till'V28' are in PCA form which means they went through reduction techinique already therefore they are not correlated to our target value . Similarly time is highly uncorrelated to our target value therefore we can drop it for our further analysis.
# ****Using SMOTE to make the data balanced** **
# This technique is used to oversample the minority class which is non-fraud cases here. This generates new synthetic samples, so it's necessary to rescale your data. For that purpose we will use Standardsacler
# import the Standardscaler first
from sklearn.preprocessing import StandardScaler
ss = StandardScaler()
data["s_amount"] = ss.fit_transform(data["Amount"].values.reshape(-1, 1))
data.head()
# Dropping the columns time and Amount , as mentioned before there is no correlation between the time and the target value and Amount has bee rescaled and new column is created for that. So, thats why we will be dropping these two variables.
data = data.drop(["Amount", "Time"], axis=1)
data.head()
# *****Using Smote Technique to balance the data *****
# splitting the data and then training it
x = data.drop("Class", axis=1).values
y = data["Class"].values
from sklearn.model_selection import train_test_split
# Train and test split using sklearn
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=1234
)
# import the library
from imblearn.over_sampling import SMOTE
from collections import Counter
smote = SMOTE()
x_smote, y_smote = smote.fit_resample(x_train, y_train)
print(x_smote.shape, y_smote.shape)
# # **Predictive Models**
# ****Logistic Regression ******
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
precision_score,
f1_score,
recall_score,
precision_recall_fscore_support,
classification_report,
)
from sklearn.metrics import plot_confusion_matrix
# intializing the model
LR = LogisticRegression(max_iter=7600)
# splitting and training of the dataset is already done. So, we will do model fitting now
#
# model fitting
LR.fit(x_smote, y_smote)
# predicting the model
ypred = LR.predict(x_test)
print(classification_report(y_test, ypred))
print(accuracy_score(y_test, ypred))
# **Feature importance**
importance = list(zip(data.columns, LR.coef_.ravel()))
importance = list(sorted(importance, key=lambda x: x[1], reverse=True))
# plotting the top feature importance
top_columns, top_score = zip(*importance[:5])
plt.xticks(rotation=60)
plt.bar(top_columns, top_score)
plt.show()
# **Decision Tree model**
from sklearn.tree import DecisionTreeClassifier
dt = DecisionTreeClassifier()
dt.fit(x_smote, y_smote)
y1pred = dt.predict(x_test)
print(classification_report(y_test, y1pred))
print(accuracy_score(y_test, y1pred.round()))
print(precision_score(y_test, y1pred.round()))
print(recall_score(y_test, y1pred.round()))
print(f1_score(y_test, y1pred.round()))
# ****Random Forest ****
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators=100)
rf.fit(x_smote, y_smote)
pred_rf = rf.predict(x_test)
print(classification_report(y_test, pred_rf))
plot_confusion_matrix(rf, x_test, y_test)
plt.show()
# **XGboost Classifier**
from xgboost import XGBClassifier
import xgboost as xgb
model = xgb.XGBClassifier(random_state=123)
model.fit(x_smote, y_smote)
xpred = model.predict(x_test)
print(classification_report(y_test, xpred))
plot_confusion_matrix(model, x_test, y_test)
plt.show()
print(accuracy_score(y_test, xpred))
| false | 0 | 1,862 | 0 | 3,735 | 1,862 |
||
69338537
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from catboost import CatBoostRegressor
from lightgbm import LGBMRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.linear_model import LinearRegression, Ridge, Lasso, ElasticNet
from sklearn.model_selection import GridSearchCV, cross_val_score
from sklearn.neighbors import KNeighborsRegressor
from sklearn.svm import SVR
from sklearn.tree import DecisionTreeRegressor
from xgboost import XGBRegressor
pd.set_option("display.max_columns", None)
pd.set_option("display.float_format", lambda x: "%.5f" % x)
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
df = train.append(test).reset_index(drop=True)
df.head()
# **EDA**
def check_df(dataframe, head=5):
print("##################### Shape #####################")
print(dataframe.shape)
print("##################### Types #####################")
print(dataframe.dtypes)
print("##################### Head #####################")
print(dataframe.head(head))
print("##################### Tail #####################")
print(dataframe.tail(head))
print("##################### NA #####################")
print(dataframe.isnull().sum())
print("##################### Quantiles #####################")
print(dataframe.quantile([0, 0.05, 0.50, 0.95, 0.99, 1]).T)
check_df(df)
df.shape
def grab_col_names(dataframe, cat_th=10, car_th=20):
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
cat_cols, num_cols, cat_but_car = grab_col_names(df)
# **Categorical Features Analysis**
def cat_summary(dataframe, col_name, plot=False):
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
for col in cat_cols:
cat_summary(df, col)
for col in cat_but_car:
cat_summary(df, col)
# **Numerical Features Analysis**
def num_summary(dataframe, numerical_col, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T)
if plot:
dataframe[numerical_col].hist(bins=20)
plt.xlabel(numerical_col)
plt.title(numerical_col)
plt.show()
df[num_cols].describe().T
for col in num_cols:
num_summary(df, col, plot=True)
# **Missing Values Analysis**
def missing_values_table(dataframe, na_name=False):
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
def missing_vs_target(dataframe, target, na_columns):
temp_df = dataframe.copy()
for col in na_columns:
temp_df[col + "_NA_FLAG"] = np.where(temp_df[col].isnull(), 1, 0)
na_flags = temp_df.loc[:, temp_df.columns.str.contains("_NA_")].columns
for col in na_flags:
print(
pd.DataFrame(
{
"TARGET_MEAN": temp_df.groupby(col)[target].mean(),
"Count": temp_df.groupby(col)[target].count(),
}
),
end="\n\n\n",
)
missing_vs_target(df, "SalePrice", missing_values_table(df, na_name=True))
missing_values_table(df)
none_cols = [
"GarageType",
"GarageFinish",
"GarageQual",
"GarageCond",
"BsmtQual",
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinType2",
"MasVnrType",
]
zero_cols = [
"BsmtFinSF1",
"BsmtFinSF2",
"BsmtUnfSF",
"TotalBsmtSF",
"BsmtFullBath",
"BsmtHalfBath",
"GarageYrBlt",
"GarageArea",
"GarageCars",
"MasVnrArea",
]
freq_cols = ["Exterior1st", "Exterior2nd", "KitchenQual", "Electrical"]
for col in zero_cols:
df[col].replace(np.nan, 0, inplace=True)
for col in none_cols:
df[col].replace(np.nan, "None", inplace=True)
for col in freq_cols:
df[col].replace(np.nan, df[col].mode()[0], inplace=True)
df["Alley"] = df["Alley"].fillna("None")
df["PoolQC"] = df["PoolQC"].fillna("None")
df["MiscFeature"] = df["MiscFeature"].fillna("None")
df["Fence"] = df["Fence"].fillna("None")
df["FireplaceQu"] = df["FireplaceQu"].fillna("None")
df["LotFrontage"] = df.groupby("Neighborhood")["LotFrontage"].transform(
lambda x: x.fillna(x.median())
)
df["GarageCars"] = df["GarageCars"].fillna(0)
df.drop(["GarageArea"], axis=1, inplace=True)
df.drop(["GarageYrBlt"], axis=1, inplace=True)
df.drop(["Utilities"], axis=1, inplace=True)
df["MSZoning"] = df.groupby("MSSubClass")["MSZoning"].apply(
lambda x: x.fillna(x.mode()[0])
)
df["Functional"] = df["Functional"].fillna("Typ")
df["SaleType"] = df["SaleType"].fillna(df["SaleType"].mode()[0])
df["YrSold"] = df["YrSold"].astype(str)
# **Target Analysis**
df["SalePrice"].describe([0.05, 0.10, 0.25, 0.50, 0.75, 0.80, 0.90, 0.95, 0.99])
def target_correlation_matrix(dataframe, corr_th=0.5, target="SalePrice"):
corr = dataframe.corr()
corr_th = corr_th
try:
filter = np.abs(corr[target]) > corr_th
corr_features = corr.columns[filter].tolist()
sns.clustermap(dataframe[corr_features].corr(), annot=True, fmt=".2f")
plt.show()
return corr_features
except:
print("Yüksek threshold değeri, corr_th değerinizi düşürün!")
target_correlation_matrix(df, corr_th=0.5, target="SalePrice")
def rare_analyser(dataframe, target, cat_cols):
for col in cat_cols:
print(col, ":", len(dataframe[col].value_counts()))
print(
pd.DataFrame(
{
"COUNT": dataframe[col].value_counts(),
"RATIO": dataframe[col].value_counts() / len(dataframe),
"TARGET_MEAN": dataframe.groupby(col)[target].mean(),
}
),
end="\n\n\n",
)
# **Data Preprocessing & Feature Engineering**
df.groupby("Neighborhood").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
nhood_map = {
"MeadowV": 1,
"IDOTRR": 1,
"BrDale": 1,
"BrkSide": 2,
"Edwards": 2,
"OldTown": 2,
"Sawyer": 3,
"Blueste": 3,
"SWISU": 4,
"NPkVill": 4,
"NAmes": 4,
"Mitchel": 4,
"SawyerW": 5,
"NWAmes": 5,
"Gilbert": 6,
"Blmngtn": 6,
"CollgCr": 6,
"Crawfor": 7,
"ClearCr": 7,
"Somerst": 8,
"Veenker": 8,
"Timber": 8,
"StoneBr": 9,
"NridgHt": 9,
"NoRidge": 10,
}
df["Neighborhood"] = df["Neighborhood"].map(nhood_map).astype("int")
df = df.replace(
{
"MSSubClass": {
20: "SC20",
30: "SC30",
40: "SC40",
45: "SC45",
50: "SC50",
60: "SC60",
70: "SC70",
75: "SC75",
80: "SC80",
85: "SC85",
90: "SC90",
120: "SC120",
150: "SC150",
160: "SC160",
180: "SC180",
190: "SC190",
},
"MoSold": {
1: "Jan",
2: "Feb",
3: "Mar",
4: "Apr",
5: "May",
6: "Jun",
7: "Jul",
8: "Aug",
9: "Sep",
10: "Oct",
11: "Nov",
12: "Dec",
},
}
)
func = {
"Sal": 0,
"Sev": 1,
"Maj2": 2,
"Maj1": 3,
"Mod": 4,
"Min2": 5,
"Min1": 6,
"Typ": 7,
}
df["Functional"] = df["Functional"].map(func).astype("int")
df.groupby("Functional").agg({"SalePrice": "mean"})
# MSZoning
df.loc[(df["MSZoning"] == "C (all)"), "MSZoning"] = 1
df.loc[(df["MSZoning"] == "RM"), "MSZoning"] = 2
df.loc[(df["MSZoning"] == "RH"), "MSZoning"] = 2
df.loc[(df["MSZoning"] == "RL"), "MSZoning"] = 3
df.loc[(df["MSZoning"] == "FV"), "MSZoning"] = 3
# LotShape
df.groupby("LotShape").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
shape_map = {"Reg": 1, "IR1": 2, "IR3": 3, "IR2": 4}
df["LotShape"] = df["LotShape"].map(shape_map).astype("int")
# LandContour
df.groupby("LandContour").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
contour_map = {"Bnk": 1, "Lvl": 2, "Low": 3, "HLS": 4}
df["LandContour"] = df["LandContour"].map(contour_map).astype("int")
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
# LotConfig
df.loc[(df["LotConfig"] == "Inside"), "LotConfig"] = 1
df.loc[(df["LotConfig"] == "FR2"), "LotConfig"] = 1
df.loc[(df["LotConfig"] == "Corner"), "LotConfig"] = 1
df.loc[(df["LotConfig"] == "FR3"), "LotConfig"] = 2
df.loc[(df["LotConfig"] == "CulDSac"), "LotConfig"] = 2
# Condition1
cond1_map = {
"Artery": 1,
"RRAe": 1,
"Feedr": 1,
"Norm": 2,
"RRAn": 2,
"RRNe": 2,
"PosN": 3,
"RRNn": 3,
"PosA": 3,
}
df["Condition1"] = df["Condition1"].map(cond1_map).astype("int")
# BldgType
df.loc[(df["BldgType"] == "2fmCon"), "BldgType"] = 1
df.loc[(df["BldgType"] == "Duplex"), "BldgType"] = 1
df.loc[(df["BldgType"] == "Twnhs"), "BldgType"] = 1
df.loc[(df["BldgType"] == "1Fam"), "BldgType"] = 2
df.loc[(df["BldgType"] == "TwnhsE"), "BldgType"] = 2
# RoofStyle
df.groupby("RoofStyle").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
df.loc[(df["RoofStyle"] == "Gambrel"), "RoofStyle"] = 1
df.loc[(df["RoofStyle"] == "Gablee"), "RoofStyle"] = 2
df.loc[(df["RoofStyle"] == "Mansard"), "RoofStyle"] = 3
df.loc[(df["RoofStyle"] == "Flat"), "RoofStyle"] = 4
df.loc[(df["RoofStyle"] == "Hip"), "RoofStyle"] = 5
df.loc[(df["RoofStyle"] == "Shed"), "RoofStyle"] = 6
# RoofMatl
df.groupby("RoofMatl").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
df.loc[(df["RoofMatl"] == "Roll"), "RoofMatl"] = 1
df.loc[(df["RoofMatl"] == "ClyTile"), "RoofMatl"] = 2
df.loc[(df["RoofMatl"] == "CompShg"), "RoofMatl"] = 3
df.loc[(df["RoofMatl"] == "Metal"), "RoofMatl"] = 3
df.loc[(df["RoofMatl"] == "Tar&Grv"), "RoofMatl"] = 3
df.loc[(df["RoofMatl"] == "WdShake"), "RoofMatl"] = 4
df.loc[(df["RoofMatl"] == "Membran"), "RoofMatl"] = 4
df.loc[(df["RoofMatl"] == "WdShngl"), "RoofMatl"] = 5
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
# ExterQual
df.groupby("ExterQual").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
ext_map = {"Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["ExterQual"] = df["ExterQual"].map(ext_map).astype("int")
# ExterCond
ext_map = {"Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["ExterCond"] = df["ExterCond"].map(ext_map).astype("int")
# BsmtQual
bsm_map = {"None": 0, "Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["BsmtQual"] = df["BsmtQual"].map(bsm_map).astype("int")
# BsmtCond
bsm_map = {"None": 0, "Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["BsmtCond"] = df["BsmtCond"].map(bsm_map).astype("int")
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
# BsmtFinType1
bsm_map = {"None": 0, "Rec": 1, "BLQ": 1, "LwQ": 2, "ALQ": 3, "Unf": 3, "GLQ": 4}
df["BsmtFinType1"] = df["BsmtFinType1"].map(bsm_map).astype("int")
# BsmtFinType2
bsm_map = {"None": 0, "BLQ": 1, "Rec": 2, "LwQ": 2, "Unf": 3, "GLQ": 3, "ALQ": 4}
df["BsmtFinType2"] = df["BsmtFinType2"].map(bsm_map).astype("int")
# BsmtExposure
bsm_map = {"None": 0, "No": 1, "Mn": 2, "Av": 3, "Gd": 4}
df["BsmtExposure"] = df["BsmtExposure"].map(bsm_map).astype("int")
# Heating
heat_map = {"Floor": 1, "Grav": 1, "Wall": 2, "OthW": 3, "GasW": 4, "GasA": 5}
df["Heating"] = df["Heating"].map(heat_map).astype("int")
# HeatingQC
heat_map = {"Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["HeatingQC"] = df["HeatingQC"].map(heat_map).astype("int")
# KitchenQual
kitch_map = {"Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["KitchenQual"] = df["KitchenQual"].map(heat_map).astype("int")
# FireplaceQu
fire_map = {"None": 0, "Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["FireplaceQu"] = df["FireplaceQu"].map(fire_map).astype("int")
# GarageCond
garage_map = {"None": 1, "Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["GarageCond"] = df["GarageCond"].map(garage_map).astype("int")
# GarageQual
garage_map = {"None": 1, "Po": 1, "Fa": 2, "TA": 3, "Ex": 4, "Gd": 5}
df["GarageQual"] = df["GarageQual"].map(garage_map).astype("int")
# PavedDrive
paved_map = {"N": 1, "P": 2, "Y": 3}
df["PavedDrive"] = df["PavedDrive"].map(paved_map).astype("int")
# CentralAir
cent = {"N": 0, "Y": 1}
df["CentralAir"] = df["CentralAir"].map(cent).astype("int")
df.groupby("CentralAir").agg({"SalePrice": "mean"})
# LandSlope
df.loc[df["LandSlope"] == "Gtl", "LandSlope"] = 1
df.loc[df["LandSlope"] == "Sev", "LandSlope"] = 2
df.loc[df["LandSlope"] == "Mod", "LandSlope"] = 2
df["LandSlope"] = df["LandSlope"].astype("int")
# OverallQual
df.loc[df["OverallQual"] == 1, "OverallQual"] = 1
df.loc[df["OverallQual"] == 2, "OverallQual"] = 1
df.loc[df["OverallQual"] == 3, "OverallQual"] = 1
df.loc[df["OverallQual"] == 4, "OverallQual"] = 2
df.loc[df["OverallQual"] == 5, "OverallQual"] = 3
df.loc[df["OverallQual"] == 6, "OverallQual"] = 4
df.loc[df["OverallQual"] == 7, "OverallQual"] = 5
df.loc[df["OverallQual"] == 8, "OverallQual"] = 6
df.loc[df["OverallQual"] == 9, "OverallQual"] = 7
df.loc[df["OverallQual"] == 10, "OverallQual"] = 8
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
df["NEW"] = df["GarageCars"] * df["OverallQual"]
df["NEW3"] = df["TotalBsmtSF"] * df["1stFlrSF"]
df["NEW4"] = df["TotRmsAbvGrd"] * df["GrLivArea"]
df["NEW5"] = df["FullBath"] * df["GrLivArea"]
df["NEW6"] = df["YearBuilt"] * df["YearRemodAdd"]
df["NEW7"] = df["OverallQual"] * df["YearBuilt"]
df["NEW8"] = df["OverallQual"] * df["RoofMatl"]
df["NEW9"] = df["PoolQC"] * df["OverallCond"]
df["NEW10"] = df["OverallCond"] * df["MasVnrArea"]
df["NEW11"] = df["LotArea"] * df["GrLivArea"]
df["NEW12"] = df["FullBath"] * df["GrLivArea"]
df["NEW13"] = df["FullBath"] * df["TotRmsAbvGrd"]
df["NEW14"] = df["1stFlrSF"] * df["TotalBsmtSF"]
df["New_Home_Quality"] = df["OverallCond"] / df["OverallQual"]
df["POOL"] = df["PoolArea"].apply(lambda x: 1 if x > 0 else 0)
df["HAS2NDFLOOR"] = df["2ndFlrSF"].apply(lambda x: 1 if x > 0 else 0)
df["LUXURY"] = df["1stFlrSF"] + df["2ndFlrSF"]
df["New_TotalBsmtSFRate"] = df["TotalBsmtSF"] / df["LotArea"]
df["TotalPorchArea"] = (
df["WoodDeckSF"]
+ df["OpenPorchSF"]
+ df["EnclosedPorch"]
+ df["3SsnPorch"]
+ df["ScreenPorch"]
)
df["IsNew"] = df.YearBuilt.apply(lambda x: 1 if x > 2000 else 0)
df["IsOld"] = df.YearBuilt.apply(lambda x: 1 if x < 1946 else 0)
def rare_encoder(dataframe, rare_perc, cat_cols):
temp_df = dataframe.copy()
rare_columns = [
col
for col in dataframe.columns
if (dataframe[col].value_counts() / len(dataframe) < 0.01).sum() > 1
]
for var in rare_columns:
tmp = dataframe[col].value_counts() / len(dataframe)
rare_labels = tmp[tmp < rare_perc].index
dataframe[col] = np.where(
dataframe[col].isin(rare_labels), "Rare", dataframe[col]
)
return temp_df
rare_analyser(df, "SalePrice", cat_cols)
df = rare_encoder(df, 0.01, cat_cols)
rare_analyser(df, "SalePrice", cat_cols)
useless_cols = [
col
for col in cat_cols
if df[col].nunique() == 1
or (
df[col].nunique() == 2
and (df[col].value_counts() / len(df) <= 0.02).any(axis=None)
)
]
useless_cols
cat_cols = [col for col in cat_cols if col not in useless_cols]
df.shape
for col in useless_cols:
df.drop(col, axis=1, inplace=True)
df.shape
rare_analyser(df, "SalePrice", cat_cols)
# **Label Encoding & One-Hot Encoding**
def label_encoder(dataframe, binary_col):
labelencoder = LabelEncoder()
dataframe[binary_col] = labelencoder.fit_transform(dataframe[binary_col])
return dataframe
def one_hot_encoder(dataframe, categorical_cols, drop_first=False):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
df = one_hot_encoder(df, cat_cols, drop_first=True)
df.shape
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
useless_cols_new = [
col for col in cat_cols if (df[col].value_counts() / len(df) <= 0.01).any(axis=None)
]
useless_cols_new
for col in useless_cols_new:
df.drop(col, axis=1, inplace=True)
df.shape
missing_values_table(df)
test.shape
missing_values_table(train)
na_cols = [
col for col in df.columns if df[col].isnull().sum() > 0 and "SalePrice" not in col
]
df[na_cols] = df[na_cols].apply(lambda x: x.fillna(x.median()), axis=0)
# **Check Outliers**
def outlier_thresholds(dataframe, col_name, q1=0.10, q3=0.90):
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def replace_with_thresholds(dataframe, variable):
low_limit, up_limit = outlier_thresholds(dataframe, variable)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
def check_outlier(dataframe, col_name):
low_limit, up_limit = outlier_thresholds(dataframe, col_name)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
cat_cols, num_cols, cat_but_car = grab_col_names(df)
for col in num_cols:
print(col, check_outlier(df, col))
for col in num_cols:
replace_with_thresholds(df, col)
for col in num_cols:
print(col, check_outlier(df, col))
# **MODELING**
train_df = df[df["SalePrice"].notnull()]
test_df = df[df["SalePrice"].isnull()].drop("SalePrice", axis=1)
y = np.log1p(train_df["SalePrice"])
X = train_df.drop(["Id", "SalePrice"], axis=1)
# **Base Models**
models = [
("LR", LinearRegression()),
("Ridge", Ridge()),
("Lasso", Lasso()),
("ElasticNet", ElasticNet()),
("KNN", KNeighborsRegressor()),
("CART", DecisionTreeRegressor()),
("RF", RandomForestRegressor()),
("SVR", SVR()),
("GBM", GradientBoostingRegressor()),
("XGBoost", XGBRegressor(objective="reg:squarederror")),
("LightGBM", LGBMRegressor()),
("CatBoost", CatBoostRegressor(verbose=False)),
]
for name, regressor in models:
rmse = np.mean(
np.sqrt(
-cross_val_score(regressor, X, y, cv=5, scoring="neg_mean_squared_error")
)
)
print(f"RMSE: {round(rmse, 4)} ({name}) ")
# **Hyperparameter Optimization**
lgbm_model = LGBMRegressor(random_state=46)
# modelleme öncesi hata:
rmse = np.mean(
np.sqrt(-cross_val_score(lgbm_model, X, y, cv=10, scoring="neg_mean_squared_error"))
)
lgbm_params = {
"learning_rate": [0.01, 0.1, 0.03, 0.2, 0.5],
"n_estimators": [100, 200, 250, 500, 1500],
"colsample_bytree": [0.3, 0.4, 0.5, 0.7, 1],
}
lgbm_gs_best = GridSearchCV(lgbm_model, lgbm_params, cv=3, n_jobs=-1, verbose=True).fit(
X, y
)
final_model_lgbm = lgbm_model.set_params(**lgbm_gs_best.best_params_).fit(X, y)
rmse = np.mean(
np.sqrt(
-cross_val_score(
final_model_lgbm, X, y, cv=10, scoring="neg_mean_squared_error"
)
)
)
# CatBoost
catboost_model = CatBoostRegressor(random_state=46)
catboost_params = {
"iterations": [200, 250, 300, 500],
"learning_rate": [0.01, 0.1, 0.2, 0.5],
"depth": [3, 6],
}
rmse = np.mean(
np.sqrt(
-cross_val_score(catboost_model, X, y, cv=5, scoring="neg_mean_squared_error")
)
)
cat_gs_best = GridSearchCV(
catboost_model, catboost_params, cv=3, n_jobs=-1, verbose=True
).fit(X, y)
final_model_cat = catboost_model.set_params(**cat_gs_best.best_params_).fit(X, y)
rmse = np.mean(
np.sqrt(
-cross_val_score(final_model_cat, X, y, cv=10, scoring="neg_mean_squared_error")
)
)
# GBM
gbm_model = GradientBoostingRegressor(random_state=46)
rmse = np.mean(
np.sqrt(-cross_val_score(gbm_model, X, y, cv=5, scoring="neg_mean_squared_error"))
)
gbm_params = {
"learning_rate": [0.01, 0.05, 0.1],
"max_depth": [3, 5, 8],
"n_estimators": [500, 1000, 1500],
"subsample": [1, 0.5, 0.7],
}
gbm_gs_best = GridSearchCV(gbm_model, gbm_params, cv=5, n_jobs=-1, verbose=True).fit(
X, y
)
final_model_gbm = gbm_model.set_params(**gbm_gs_best.best_params_).fit(X, y)
rmse = np.mean(
np.sqrt(
-cross_val_score(final_model_gbm, X, y, cv=10, scoring="neg_mean_squared_error")
)
)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338537.ipynb
| null | null |
[{"Id": 69338537, "ScriptId": 18924045, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7430183, "CreationDate": "07/29/2021 18:33:34", "VersionNumber": 3.0, "Title": "House_Price_Prediction", "EvaluationDate": "07/29/2021", "IsChange": false, "TotalLines": 569.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 569.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from catboost import CatBoostRegressor
from lightgbm import LGBMRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.linear_model import LinearRegression, Ridge, Lasso, ElasticNet
from sklearn.model_selection import GridSearchCV, cross_val_score
from sklearn.neighbors import KNeighborsRegressor
from sklearn.svm import SVR
from sklearn.tree import DecisionTreeRegressor
from xgboost import XGBRegressor
pd.set_option("display.max_columns", None)
pd.set_option("display.float_format", lambda x: "%.5f" % x)
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
df = train.append(test).reset_index(drop=True)
df.head()
# **EDA**
def check_df(dataframe, head=5):
print("##################### Shape #####################")
print(dataframe.shape)
print("##################### Types #####################")
print(dataframe.dtypes)
print("##################### Head #####################")
print(dataframe.head(head))
print("##################### Tail #####################")
print(dataframe.tail(head))
print("##################### NA #####################")
print(dataframe.isnull().sum())
print("##################### Quantiles #####################")
print(dataframe.quantile([0, 0.05, 0.50, 0.95, 0.99, 1]).T)
check_df(df)
df.shape
def grab_col_names(dataframe, cat_th=10, car_th=20):
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
cat_cols, num_cols, cat_but_car = grab_col_names(df)
# **Categorical Features Analysis**
def cat_summary(dataframe, col_name, plot=False):
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
for col in cat_cols:
cat_summary(df, col)
for col in cat_but_car:
cat_summary(df, col)
# **Numerical Features Analysis**
def num_summary(dataframe, numerical_col, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T)
if plot:
dataframe[numerical_col].hist(bins=20)
plt.xlabel(numerical_col)
plt.title(numerical_col)
plt.show()
df[num_cols].describe().T
for col in num_cols:
num_summary(df, col, plot=True)
# **Missing Values Analysis**
def missing_values_table(dataframe, na_name=False):
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
def missing_vs_target(dataframe, target, na_columns):
temp_df = dataframe.copy()
for col in na_columns:
temp_df[col + "_NA_FLAG"] = np.where(temp_df[col].isnull(), 1, 0)
na_flags = temp_df.loc[:, temp_df.columns.str.contains("_NA_")].columns
for col in na_flags:
print(
pd.DataFrame(
{
"TARGET_MEAN": temp_df.groupby(col)[target].mean(),
"Count": temp_df.groupby(col)[target].count(),
}
),
end="\n\n\n",
)
missing_vs_target(df, "SalePrice", missing_values_table(df, na_name=True))
missing_values_table(df)
none_cols = [
"GarageType",
"GarageFinish",
"GarageQual",
"GarageCond",
"BsmtQual",
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinType2",
"MasVnrType",
]
zero_cols = [
"BsmtFinSF1",
"BsmtFinSF2",
"BsmtUnfSF",
"TotalBsmtSF",
"BsmtFullBath",
"BsmtHalfBath",
"GarageYrBlt",
"GarageArea",
"GarageCars",
"MasVnrArea",
]
freq_cols = ["Exterior1st", "Exterior2nd", "KitchenQual", "Electrical"]
for col in zero_cols:
df[col].replace(np.nan, 0, inplace=True)
for col in none_cols:
df[col].replace(np.nan, "None", inplace=True)
for col in freq_cols:
df[col].replace(np.nan, df[col].mode()[0], inplace=True)
df["Alley"] = df["Alley"].fillna("None")
df["PoolQC"] = df["PoolQC"].fillna("None")
df["MiscFeature"] = df["MiscFeature"].fillna("None")
df["Fence"] = df["Fence"].fillna("None")
df["FireplaceQu"] = df["FireplaceQu"].fillna("None")
df["LotFrontage"] = df.groupby("Neighborhood")["LotFrontage"].transform(
lambda x: x.fillna(x.median())
)
df["GarageCars"] = df["GarageCars"].fillna(0)
df.drop(["GarageArea"], axis=1, inplace=True)
df.drop(["GarageYrBlt"], axis=1, inplace=True)
df.drop(["Utilities"], axis=1, inplace=True)
df["MSZoning"] = df.groupby("MSSubClass")["MSZoning"].apply(
lambda x: x.fillna(x.mode()[0])
)
df["Functional"] = df["Functional"].fillna("Typ")
df["SaleType"] = df["SaleType"].fillna(df["SaleType"].mode()[0])
df["YrSold"] = df["YrSold"].astype(str)
# **Target Analysis**
df["SalePrice"].describe([0.05, 0.10, 0.25, 0.50, 0.75, 0.80, 0.90, 0.95, 0.99])
def target_correlation_matrix(dataframe, corr_th=0.5, target="SalePrice"):
corr = dataframe.corr()
corr_th = corr_th
try:
filter = np.abs(corr[target]) > corr_th
corr_features = corr.columns[filter].tolist()
sns.clustermap(dataframe[corr_features].corr(), annot=True, fmt=".2f")
plt.show()
return corr_features
except:
print("Yüksek threshold değeri, corr_th değerinizi düşürün!")
target_correlation_matrix(df, corr_th=0.5, target="SalePrice")
def rare_analyser(dataframe, target, cat_cols):
for col in cat_cols:
print(col, ":", len(dataframe[col].value_counts()))
print(
pd.DataFrame(
{
"COUNT": dataframe[col].value_counts(),
"RATIO": dataframe[col].value_counts() / len(dataframe),
"TARGET_MEAN": dataframe.groupby(col)[target].mean(),
}
),
end="\n\n\n",
)
# **Data Preprocessing & Feature Engineering**
df.groupby("Neighborhood").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
nhood_map = {
"MeadowV": 1,
"IDOTRR": 1,
"BrDale": 1,
"BrkSide": 2,
"Edwards": 2,
"OldTown": 2,
"Sawyer": 3,
"Blueste": 3,
"SWISU": 4,
"NPkVill": 4,
"NAmes": 4,
"Mitchel": 4,
"SawyerW": 5,
"NWAmes": 5,
"Gilbert": 6,
"Blmngtn": 6,
"CollgCr": 6,
"Crawfor": 7,
"ClearCr": 7,
"Somerst": 8,
"Veenker": 8,
"Timber": 8,
"StoneBr": 9,
"NridgHt": 9,
"NoRidge": 10,
}
df["Neighborhood"] = df["Neighborhood"].map(nhood_map).astype("int")
df = df.replace(
{
"MSSubClass": {
20: "SC20",
30: "SC30",
40: "SC40",
45: "SC45",
50: "SC50",
60: "SC60",
70: "SC70",
75: "SC75",
80: "SC80",
85: "SC85",
90: "SC90",
120: "SC120",
150: "SC150",
160: "SC160",
180: "SC180",
190: "SC190",
},
"MoSold": {
1: "Jan",
2: "Feb",
3: "Mar",
4: "Apr",
5: "May",
6: "Jun",
7: "Jul",
8: "Aug",
9: "Sep",
10: "Oct",
11: "Nov",
12: "Dec",
},
}
)
func = {
"Sal": 0,
"Sev": 1,
"Maj2": 2,
"Maj1": 3,
"Mod": 4,
"Min2": 5,
"Min1": 6,
"Typ": 7,
}
df["Functional"] = df["Functional"].map(func).astype("int")
df.groupby("Functional").agg({"SalePrice": "mean"})
# MSZoning
df.loc[(df["MSZoning"] == "C (all)"), "MSZoning"] = 1
df.loc[(df["MSZoning"] == "RM"), "MSZoning"] = 2
df.loc[(df["MSZoning"] == "RH"), "MSZoning"] = 2
df.loc[(df["MSZoning"] == "RL"), "MSZoning"] = 3
df.loc[(df["MSZoning"] == "FV"), "MSZoning"] = 3
# LotShape
df.groupby("LotShape").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
shape_map = {"Reg": 1, "IR1": 2, "IR3": 3, "IR2": 4}
df["LotShape"] = df["LotShape"].map(shape_map).astype("int")
# LandContour
df.groupby("LandContour").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
contour_map = {"Bnk": 1, "Lvl": 2, "Low": 3, "HLS": 4}
df["LandContour"] = df["LandContour"].map(contour_map).astype("int")
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
# LotConfig
df.loc[(df["LotConfig"] == "Inside"), "LotConfig"] = 1
df.loc[(df["LotConfig"] == "FR2"), "LotConfig"] = 1
df.loc[(df["LotConfig"] == "Corner"), "LotConfig"] = 1
df.loc[(df["LotConfig"] == "FR3"), "LotConfig"] = 2
df.loc[(df["LotConfig"] == "CulDSac"), "LotConfig"] = 2
# Condition1
cond1_map = {
"Artery": 1,
"RRAe": 1,
"Feedr": 1,
"Norm": 2,
"RRAn": 2,
"RRNe": 2,
"PosN": 3,
"RRNn": 3,
"PosA": 3,
}
df["Condition1"] = df["Condition1"].map(cond1_map).astype("int")
# BldgType
df.loc[(df["BldgType"] == "2fmCon"), "BldgType"] = 1
df.loc[(df["BldgType"] == "Duplex"), "BldgType"] = 1
df.loc[(df["BldgType"] == "Twnhs"), "BldgType"] = 1
df.loc[(df["BldgType"] == "1Fam"), "BldgType"] = 2
df.loc[(df["BldgType"] == "TwnhsE"), "BldgType"] = 2
# RoofStyle
df.groupby("RoofStyle").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
df.loc[(df["RoofStyle"] == "Gambrel"), "RoofStyle"] = 1
df.loc[(df["RoofStyle"] == "Gablee"), "RoofStyle"] = 2
df.loc[(df["RoofStyle"] == "Mansard"), "RoofStyle"] = 3
df.loc[(df["RoofStyle"] == "Flat"), "RoofStyle"] = 4
df.loc[(df["RoofStyle"] == "Hip"), "RoofStyle"] = 5
df.loc[(df["RoofStyle"] == "Shed"), "RoofStyle"] = 6
# RoofMatl
df.groupby("RoofMatl").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
df.loc[(df["RoofMatl"] == "Roll"), "RoofMatl"] = 1
df.loc[(df["RoofMatl"] == "ClyTile"), "RoofMatl"] = 2
df.loc[(df["RoofMatl"] == "CompShg"), "RoofMatl"] = 3
df.loc[(df["RoofMatl"] == "Metal"), "RoofMatl"] = 3
df.loc[(df["RoofMatl"] == "Tar&Grv"), "RoofMatl"] = 3
df.loc[(df["RoofMatl"] == "WdShake"), "RoofMatl"] = 4
df.loc[(df["RoofMatl"] == "Membran"), "RoofMatl"] = 4
df.loc[(df["RoofMatl"] == "WdShngl"), "RoofMatl"] = 5
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
# ExterQual
df.groupby("ExterQual").agg({"SalePrice": "mean"}).sort_values(
by="SalePrice", ascending=False
)
ext_map = {"Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["ExterQual"] = df["ExterQual"].map(ext_map).astype("int")
# ExterCond
ext_map = {"Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["ExterCond"] = df["ExterCond"].map(ext_map).astype("int")
# BsmtQual
bsm_map = {"None": 0, "Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["BsmtQual"] = df["BsmtQual"].map(bsm_map).astype("int")
# BsmtCond
bsm_map = {"None": 0, "Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["BsmtCond"] = df["BsmtCond"].map(bsm_map).astype("int")
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
# BsmtFinType1
bsm_map = {"None": 0, "Rec": 1, "BLQ": 1, "LwQ": 2, "ALQ": 3, "Unf": 3, "GLQ": 4}
df["BsmtFinType1"] = df["BsmtFinType1"].map(bsm_map).astype("int")
# BsmtFinType2
bsm_map = {"None": 0, "BLQ": 1, "Rec": 2, "LwQ": 2, "Unf": 3, "GLQ": 3, "ALQ": 4}
df["BsmtFinType2"] = df["BsmtFinType2"].map(bsm_map).astype("int")
# BsmtExposure
bsm_map = {"None": 0, "No": 1, "Mn": 2, "Av": 3, "Gd": 4}
df["BsmtExposure"] = df["BsmtExposure"].map(bsm_map).astype("int")
# Heating
heat_map = {"Floor": 1, "Grav": 1, "Wall": 2, "OthW": 3, "GasW": 4, "GasA": 5}
df["Heating"] = df["Heating"].map(heat_map).astype("int")
# HeatingQC
heat_map = {"Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["HeatingQC"] = df["HeatingQC"].map(heat_map).astype("int")
# KitchenQual
kitch_map = {"Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["KitchenQual"] = df["KitchenQual"].map(heat_map).astype("int")
# FireplaceQu
fire_map = {"None": 0, "Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["FireplaceQu"] = df["FireplaceQu"].map(fire_map).astype("int")
# GarageCond
garage_map = {"None": 1, "Po": 1, "Fa": 2, "TA": 3, "Gd": 4, "Ex": 5}
df["GarageCond"] = df["GarageCond"].map(garage_map).astype("int")
# GarageQual
garage_map = {"None": 1, "Po": 1, "Fa": 2, "TA": 3, "Ex": 4, "Gd": 5}
df["GarageQual"] = df["GarageQual"].map(garage_map).astype("int")
# PavedDrive
paved_map = {"N": 1, "P": 2, "Y": 3}
df["PavedDrive"] = df["PavedDrive"].map(paved_map).astype("int")
# CentralAir
cent = {"N": 0, "Y": 1}
df["CentralAir"] = df["CentralAir"].map(cent).astype("int")
df.groupby("CentralAir").agg({"SalePrice": "mean"})
# LandSlope
df.loc[df["LandSlope"] == "Gtl", "LandSlope"] = 1
df.loc[df["LandSlope"] == "Sev", "LandSlope"] = 2
df.loc[df["LandSlope"] == "Mod", "LandSlope"] = 2
df["LandSlope"] = df["LandSlope"].astype("int")
# OverallQual
df.loc[df["OverallQual"] == 1, "OverallQual"] = 1
df.loc[df["OverallQual"] == 2, "OverallQual"] = 1
df.loc[df["OverallQual"] == 3, "OverallQual"] = 1
df.loc[df["OverallQual"] == 4, "OverallQual"] = 2
df.loc[df["OverallQual"] == 5, "OverallQual"] = 3
df.loc[df["OverallQual"] == 6, "OverallQual"] = 4
df.loc[df["OverallQual"] == 7, "OverallQual"] = 5
df.loc[df["OverallQual"] == 8, "OverallQual"] = 6
df.loc[df["OverallQual"] == 9, "OverallQual"] = 7
df.loc[df["OverallQual"] == 10, "OverallQual"] = 8
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
df["NEW"] = df["GarageCars"] * df["OverallQual"]
df["NEW3"] = df["TotalBsmtSF"] * df["1stFlrSF"]
df["NEW4"] = df["TotRmsAbvGrd"] * df["GrLivArea"]
df["NEW5"] = df["FullBath"] * df["GrLivArea"]
df["NEW6"] = df["YearBuilt"] * df["YearRemodAdd"]
df["NEW7"] = df["OverallQual"] * df["YearBuilt"]
df["NEW8"] = df["OverallQual"] * df["RoofMatl"]
df["NEW9"] = df["PoolQC"] * df["OverallCond"]
df["NEW10"] = df["OverallCond"] * df["MasVnrArea"]
df["NEW11"] = df["LotArea"] * df["GrLivArea"]
df["NEW12"] = df["FullBath"] * df["GrLivArea"]
df["NEW13"] = df["FullBath"] * df["TotRmsAbvGrd"]
df["NEW14"] = df["1stFlrSF"] * df["TotalBsmtSF"]
df["New_Home_Quality"] = df["OverallCond"] / df["OverallQual"]
df["POOL"] = df["PoolArea"].apply(lambda x: 1 if x > 0 else 0)
df["HAS2NDFLOOR"] = df["2ndFlrSF"].apply(lambda x: 1 if x > 0 else 0)
df["LUXURY"] = df["1stFlrSF"] + df["2ndFlrSF"]
df["New_TotalBsmtSFRate"] = df["TotalBsmtSF"] / df["LotArea"]
df["TotalPorchArea"] = (
df["WoodDeckSF"]
+ df["OpenPorchSF"]
+ df["EnclosedPorch"]
+ df["3SsnPorch"]
+ df["ScreenPorch"]
)
df["IsNew"] = df.YearBuilt.apply(lambda x: 1 if x > 2000 else 0)
df["IsOld"] = df.YearBuilt.apply(lambda x: 1 if x < 1946 else 0)
def rare_encoder(dataframe, rare_perc, cat_cols):
temp_df = dataframe.copy()
rare_columns = [
col
for col in dataframe.columns
if (dataframe[col].value_counts() / len(dataframe) < 0.01).sum() > 1
]
for var in rare_columns:
tmp = dataframe[col].value_counts() / len(dataframe)
rare_labels = tmp[tmp < rare_perc].index
dataframe[col] = np.where(
dataframe[col].isin(rare_labels), "Rare", dataframe[col]
)
return temp_df
rare_analyser(df, "SalePrice", cat_cols)
df = rare_encoder(df, 0.01, cat_cols)
rare_analyser(df, "SalePrice", cat_cols)
useless_cols = [
col
for col in cat_cols
if df[col].nunique() == 1
or (
df[col].nunique() == 2
and (df[col].value_counts() / len(df) <= 0.02).any(axis=None)
)
]
useless_cols
cat_cols = [col for col in cat_cols if col not in useless_cols]
df.shape
for col in useless_cols:
df.drop(col, axis=1, inplace=True)
df.shape
rare_analyser(df, "SalePrice", cat_cols)
# **Label Encoding & One-Hot Encoding**
def label_encoder(dataframe, binary_col):
labelencoder = LabelEncoder()
dataframe[binary_col] = labelencoder.fit_transform(dataframe[binary_col])
return dataframe
def one_hot_encoder(dataframe, categorical_cols, drop_first=False):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
df = one_hot_encoder(df, cat_cols, drop_first=True)
df.shape
cat_cols, num_cols, cat_but_car = grab_col_names(df)
rare_analyser(df, "SalePrice", cat_cols)
useless_cols_new = [
col for col in cat_cols if (df[col].value_counts() / len(df) <= 0.01).any(axis=None)
]
useless_cols_new
for col in useless_cols_new:
df.drop(col, axis=1, inplace=True)
df.shape
missing_values_table(df)
test.shape
missing_values_table(train)
na_cols = [
col for col in df.columns if df[col].isnull().sum() > 0 and "SalePrice" not in col
]
df[na_cols] = df[na_cols].apply(lambda x: x.fillna(x.median()), axis=0)
# **Check Outliers**
def outlier_thresholds(dataframe, col_name, q1=0.10, q3=0.90):
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def replace_with_thresholds(dataframe, variable):
low_limit, up_limit = outlier_thresholds(dataframe, variable)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
def check_outlier(dataframe, col_name):
low_limit, up_limit = outlier_thresholds(dataframe, col_name)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
cat_cols, num_cols, cat_but_car = grab_col_names(df)
for col in num_cols:
print(col, check_outlier(df, col))
for col in num_cols:
replace_with_thresholds(df, col)
for col in num_cols:
print(col, check_outlier(df, col))
# **MODELING**
train_df = df[df["SalePrice"].notnull()]
test_df = df[df["SalePrice"].isnull()].drop("SalePrice", axis=1)
y = np.log1p(train_df["SalePrice"])
X = train_df.drop(["Id", "SalePrice"], axis=1)
# **Base Models**
models = [
("LR", LinearRegression()),
("Ridge", Ridge()),
("Lasso", Lasso()),
("ElasticNet", ElasticNet()),
("KNN", KNeighborsRegressor()),
("CART", DecisionTreeRegressor()),
("RF", RandomForestRegressor()),
("SVR", SVR()),
("GBM", GradientBoostingRegressor()),
("XGBoost", XGBRegressor(objective="reg:squarederror")),
("LightGBM", LGBMRegressor()),
("CatBoost", CatBoostRegressor(verbose=False)),
]
for name, regressor in models:
rmse = np.mean(
np.sqrt(
-cross_val_score(regressor, X, y, cv=5, scoring="neg_mean_squared_error")
)
)
print(f"RMSE: {round(rmse, 4)} ({name}) ")
# **Hyperparameter Optimization**
lgbm_model = LGBMRegressor(random_state=46)
# modelleme öncesi hata:
rmse = np.mean(
np.sqrt(-cross_val_score(lgbm_model, X, y, cv=10, scoring="neg_mean_squared_error"))
)
lgbm_params = {
"learning_rate": [0.01, 0.1, 0.03, 0.2, 0.5],
"n_estimators": [100, 200, 250, 500, 1500],
"colsample_bytree": [0.3, 0.4, 0.5, 0.7, 1],
}
lgbm_gs_best = GridSearchCV(lgbm_model, lgbm_params, cv=3, n_jobs=-1, verbose=True).fit(
X, y
)
final_model_lgbm = lgbm_model.set_params(**lgbm_gs_best.best_params_).fit(X, y)
rmse = np.mean(
np.sqrt(
-cross_val_score(
final_model_lgbm, X, y, cv=10, scoring="neg_mean_squared_error"
)
)
)
# CatBoost
catboost_model = CatBoostRegressor(random_state=46)
catboost_params = {
"iterations": [200, 250, 300, 500],
"learning_rate": [0.01, 0.1, 0.2, 0.5],
"depth": [3, 6],
}
rmse = np.mean(
np.sqrt(
-cross_val_score(catboost_model, X, y, cv=5, scoring="neg_mean_squared_error")
)
)
cat_gs_best = GridSearchCV(
catboost_model, catboost_params, cv=3, n_jobs=-1, verbose=True
).fit(X, y)
final_model_cat = catboost_model.set_params(**cat_gs_best.best_params_).fit(X, y)
rmse = np.mean(
np.sqrt(
-cross_val_score(final_model_cat, X, y, cv=10, scoring="neg_mean_squared_error")
)
)
# GBM
gbm_model = GradientBoostingRegressor(random_state=46)
rmse = np.mean(
np.sqrt(-cross_val_score(gbm_model, X, y, cv=5, scoring="neg_mean_squared_error"))
)
gbm_params = {
"learning_rate": [0.01, 0.05, 0.1],
"max_depth": [3, 5, 8],
"n_estimators": [500, 1000, 1500],
"subsample": [1, 0.5, 0.7],
}
gbm_gs_best = GridSearchCV(gbm_model, gbm_params, cv=5, n_jobs=-1, verbose=True).fit(
X, y
)
final_model_gbm = gbm_model.set_params(**gbm_gs_best.best_params_).fit(X, y)
rmse = np.mean(
np.sqrt(
-cross_val_score(final_model_gbm, X, y, cv=10, scoring="neg_mean_squared_error")
)
)
| false | 0 | 8,443 | 0 | 8,443 | 8,443 |
||
69338061
|
#
# Introduction
# >On April 15, 1912, during her maiden voyage, the widely considered “unsinkable” RMS Titanic sank after colliding with an iceberg. Unfortunately, there weren’t enough lifeboats for everyone onboard, resulting in the death of 1502 out of 2224 passengers and crew.
# >Many of us have come across this datasets in the start of Data Science journey, and this is a classic example to practice the skills of EDA and Machine Learning. This notebook is especially more useful for beginners in Data Science. It gives step by step guide for EDA and ML, after reading this notebook, I am sure you will be more comfortable in EDA and ML and you will be able to present your data with better visuals.
# #### I have included the most commonly used plots along with some awesome plots like sunbrust and parallel categories plot.
#
# 1. [Box Plot](#1)
# 1. [Scatter Plot](#2)
# 1. [Bar Plot](#3)
# 1. [Pie Plot](#4)
# 1. [Sunburst Plot](#5)
# 1. [Histogram](#6)
# 1. [Parallel Categories Plot](#7)
# 1. [Funnel Plot](#8)
# 1. [Treemap Plot](#9)
# PLEASE UPVOTE GUYS AND RECOMMEND THAT SHOULD IMPLEMENT
# For data handling
import numpy as np
import pandas as pd
# For visvalization
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
df = pd.read_csv("../input/titanic/train.csv")
df.head()
# Data Cleaning 🚿
# > Data may have missing values, wrong data types, outliers, etc. So, before you do EDA on your data, please check for these things and if they are present, you have to clean the data before moving further otherwise you can get misleading and incorrect visuals. So let's fold the sleeves and get ready for cleaning!
# Dropping the unnecessary columns which won't contribute any to EDA
df = df.drop(["PassengerId", "Ticket", "Name"], axis=1)
# Data Types
df.info()
# **Categorical features**: Survived, Sex, Embarked, and Pclass.
# **Numeric features**: Age, Fare. Discrete: SibSp, Parch.
# > Changing the data types of `Survived`, `Pclass` from numeric to category
# Changing the data types
cat_col = ["Survived", "Pclass"]
df[cat_col] = df[cat_col].astype("object")
# Missing values
df.isnull().mean() * 100
# > If a colum has 70%-80% missing data, it is advisable to remove that column. Cabin has 77% missing data, so we will remove that **column**. Age and Embarked has <20% missing data, so we shall remove the **rows** having the missing values. You can also impute the missing values with mean or mode with [df.fillna](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.fillna.html) but I am removing them as the focus here is on EDA not on creating models.
# > 📌 Some models can give you an error if missing values are present and some will show incorrect results, so it is important to remove them.
#
# Removing Cabin column
df = df.drop(["Cabin"], axis=1)
# Removing rows with missing data
df = df.dropna().reset_index(drop=True)
# Exploratory Data Analysis 📊
# > Before doing any kind of EDA, you should have a purpose of doing it. It should be clear what you want otherwise you will produce beautiful charts with no use. You should ask yourself, what you want to see and then choose the appropriate plot and features. So, I have mentioned the questions, which I thought before plotting the charts.
# > Note that, for a perticular question, there can be multiple charts. I have choosen the chart which I think was more appropriate. Now let's do some EDA.
# Function for understanding the distribution of the columns
def quick_plot(df):
categorical_col = df.select_dtypes("object").columns.to_list()
numeric_col = df.select_dtypes(["int", "float"]).columns.to_list()
if len(numeric_col) > len(categorical_col):
max_length = len(numeric_col)
else:
max_length = len(categorical_col)
fig, (ax1, ax2) = plt.subplots(2, max_length, figsize=(20, 10))
# Plotting for categorical columns
for axis, col in zip(ax1.ravel(), categorical_col):
axis = sns.countplot(x=col, data=df, ax=axis)
axis.set_ylabel(None)
axis.set_xlabel(col, size=16)
for i in axis.patches:
axis.text(
x=i.get_x() + i.get_width() / 2,
y=i.get_height() / 2,
s=f"{round(i.get_height(),1)}",
ha="center",
size=16,
weight="bold",
rotation=0,
color="white",
)
# Plotting for numeric columns
for axis, col in zip(ax2.ravel(), numeric_col):
axis = sns.histplot(x=col, data=df, multiple="stack", ax=axis)
axis.set_ylabel(None)
axis.set_xlabel(col, size=16)
fig.text(0.5, 1, "The distribution of Data", ha="center", fontsize=20)
plt.tight_layout()
# Quickly understanding the distribution of the columns
plt.style.use("seaborn")
quick_plot(df)
# #### 🔎Observation:
# > Passengers of class 3 are huge in number.
# > Very few passenger embarked on Q (Queenstown) port.
# > The distribution of `Fare` is skew.
# Distribution of Age and Fare 💰
# #### Question: How the features `age` and `fare` are distributed, are there any outliers ❓
# Beginner tip: It is applicable to numeric feature and used for understanding the data distribution and also for detecting outliers!
plt.style.use("seaborn")
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 6))
ax1 = sns.boxplot(y="Age", data=df, ax=ax1)
ax2 = sns.boxplot(y="Fare", data=df, ax=ax2)
ax1.set_ylabel(None)
ax1.set_xlabel(None)
ax1.set_title("Age", size=16)
ax2.set_ylabel(None)
ax2.set_xlabel(None)
ax2.set_title("Fare", size=16)
# >We have many values that are more than 75 percentile. The data is an outlier or not is the decision of the field expert. We can also tell for some of the data like `Age` - it can't be more than 100 or less than 0, etc. but in this case, I will remove these values while doing ML.
# Relation between Age and Fare 💰
# #### Question: Is there any relation between 'age' and 'fare'❓
# Beginner tip: It is applicable when you have two numeric features. It's used for visvalizing relation between numeric features.
fig, ax = plt.subplots(1, 1, figsize=(10, 6), constrained_layout=True)
ax = sns.regplot(x="Age", y="Fare", data=df)
ax.set_ylabel("Fare", fontsize=14)
ax.set_xlabel("Age", fontsize=14)
plt.title("Age Vs Fare", fontsize=16)
correlation = np.corrcoef(df["Age"], df["Fare"])[0][1]
ax.text(
x=60,
y=180,
s=f"correlation : {round(correlation,3)}",
ha="center",
size=12,
rotation=0,
color="black",
bbox=dict(boxstyle="round,pad=0.5", fc="skyblue", ec="skyblue", lw=2),
)
# #### 🔍 Observations
# > The correlation between `Age` and `Fare` is 0.093, which is close to 0, so there is no relation between them. It means, we can't guess how much fare a passenger has paid just by looking at his/her age.
# #### 📝 Note
# > The correlation score:
# > - 1 : Strongly and positively correlated (one increases, other also increases and vice versa)
# > - 0 : No correlation
# > - 1 : Strongly and negetively correlated (one increases, other also decreases and vice versa)
df.info()
#
# Analysis of Categorical Features 🔬
# > We have three categorical features`Pclass`, `Sex`, and `Embarked`. Not counting `Survived` as it is the target. Let's analyse the categorical features one by one based on the target.
# > For the analysis, I will be using PowerBI, as it is simple and efficient!
# Pclass (Ticket Class)
# > There are three classes 1st, 2nd, and 3rd.
# > 1 = 1st, 2 = 2nd, 3 = 3rd
# #### 🔎 Observations
# > As seen from previous plot also, Passengers of class 3 are huge in number. The reason can be cheap cost of the ticket.
# > 76% of the passengers form the 3rd class didn't survive while 63% of passengers form the 1st class survived.
# > The 1st class had mostly elders, may be because they can afford the ticket price and 3rd class had mostly young people.
# > Most of the rich 🤑 and old 👴👵 passengers survived.
# Gender 🤵💃
# #### 🔎 Observations
# > 81% men didn't survive while 74% women survived.
# > On average, female passengers paid higher fare than male passengers.
# Port of Embarkment 🚢
# > Passengers embarked from three different ports : C = Cherbourg, Q = Queenstown, S = Southampton
# #### 🔎 Observations
# > As seen previously also, very few passenger embarked on Q (Queenstown) port.
# > More than half of the passengers didn't survive who embarked from Queenstown (Q) and Southampton (S).
# > Passengers embarking from Cherbourg (C) paid more average fare, and also more than 50% of them survived.
# Comparison between Pclass, Gender 🤵💃, and Port of Embarkment 🚢
# 1st Class Passengers who Survived
# #### 🔎 Observations
# > Out of 55% of the passengers embarked from Cherbourg (C) and survived, 35% belongs to 1st class. As the fare of 1st class is high so as the case with passengers embarking from Cherbourg (C), most of them belongs to 1st class.
# > Very few passengers from 1st class who survived, embarked from Queenstown (Q).
# Survived passengers embarking from Southampton (S)
# #### 🔎 Observations
# > Out of all female 74% survived and 45% of these embarked from Southampton (S).
# > 47% of 2nd class passengers survived and 41% of them embarked from Southampton (S).
# > Out of all male 19% survived and 13% of these embarked from Southampton (S). So, only 6% of the male who embarked from Cherbourg (C) and Queenstown (Q), survived.
# Class 3 passengers who didn't survive
# #### 🔎 Observations
# > Out of 61% passengers who didn't survived and embarked from Queenstown (Q), 58% belongs to class 3.
# > Mojority of the female passengers who didn't survived belong to class 3.
# Analysis of Numeric Features 🔬
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 6), constrained_layout=True)
ax1 = sns.boxenplot(x="Survived", y="Age", data=df, ax=ax1)
ax2 = sns.histplot(x="Age", data=df, hue="Survived", multiple="stack", kde=True, ax=ax2)
ax1.set_xlabel(None)
ax1.set_ylabel("Age", fontsize=14)
ax1.set_title("Passenger Age and their Survival", fontsize=16)
ax1.set_xticklabels(["Not Survived", "Survived"], size=14)
ax2.set_xlabel("Age", fontsize=14)
ax2.set_ylabel("Number of Passengers", fontsize=14)
ax2.set_title("Distribution of Age and Passenger Survival", fontsize=16)
# #### 🔎 Observations
# > There is not much relation between age and survival of the passengers. This also becomes clear from the kde plot, as the nature of both, the blue line (Non Survived) and gree line (Survived) is similar.
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 6), constrained_layout=True)
ax1 = sns.boxenplot(x="Survived", y="Fare", data=df, ax=ax1)
ax2 = sns.histplot(
x="Fare", data=df, hue="Survived", multiple="stack", kde=True, ax=ax2
)
ax1.set_xlabel(None)
ax1.set_ylabel("Fare", fontsize=14)
ax1.set_title("Passenger Age and their Survival", fontsize=16)
ax1.set_xticklabels(["Not Survived", "Survived"], size=14)
ax2.set_xscale("log")
ax2.set_xlabel("Age (Log scale)", fontsize=14)
ax2.set_ylabel("Number of Passengers", fontsize=14)
ax2.set_title("Distribution of Fare and Passenger Survival", fontsize=16)
# #### 🔎 Observations
# > There is some relation between `Fare` and `Survived` of the passengers. This also becomes clear from the kde plot, as the nature of both, the blue line (Non Survived) and gree line (Survived) is different.
# Data Cleaning 🧹
# Removing Outliers
upper_bound = df["Fare"].quantile(0.97)
df = df[(df["Fare"] < upper_bound)]
# Imputing missing values
df["Age"] = df["Age"].fillna(df["Age"].median())
# Feature Enginner ⚙
# > The `Fare` feature had skew distribution, so I will log transform it.
df["Fare"] = df["Fare"].transform(np.log)
df["Fare"].min()
# As new Fare contains -infinity, I will replace it will next minimum fare
least_value = df["Fare"].sort_values().unique()[1]
df["Fare"] = df["Fare"].replace(-np.inf, least_value)
# Creating bins for age
df["Age_Bins"] = pd.cut(
df["Age"],
[0, 10, 25, 40, 60, 100],
labels=["<10", "10 to 25", "26 to 40", "41 to 60", "60+"],
include_lowest=True,
)
# Creating new feature Age * Fare
df["Age x Fare"] = df["Age"] * df["Fare"]
# Creating new higher degree features: Age^^2 and Fare^^2
df["Age^2"] = df["Age"] ** 2
df["Fare^2"] = df["Fare"] ** 2
# Making data ready for training
# Creating X (features) and y (target)
X = df.drop(["Survived"], axis=1)
y = df["Survived"].astype("int")
# One hot encoding for categorical features
X_new = pd.get_dummies(X, drop_first=True)
# Train - Test split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X_new, y, train_size=0.8, random_state=99, stratify=y
)
# Creating ML pipeline 🚄
# > First we will create simple model (for baseline) and then move to more complex models.
# Logistic Regression
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LogisticRegressionCV
pipeline = Pipeline(
[("scalar", StandardScaler()), ("logistic", LogisticRegressionCV(cv=10, n_jobs=-1))]
)
log_model = pipeline.fit(X_train, y_train)
print("Train accuracy is : ", log_model.score(X_train, y_train))
print("Train accuracy is : ", log_model.score(X_test, y_test))
# > The base model has an accuracy of 80%. Now, lets try some other models to get accuracy more than this.
# Decision Tree 🌳
from sklearn.tree import DecisionTreeClassifier
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import GridSearchCV
pipeline = Pipeline(
[("scalar", StandardScaler()), ("tree", DecisionTreeClassifier(random_state=99))]
)
hyperparamets = {
"tree__max_depth": [3, 5, 7, 9, 11],
"tree__min_samples_leaf": [10, 20, 30, 40],
"tree__min_impurity_decrease": [0, 0.1, 0.2, 0.5],
}
tree_model = GridSearchCV(pipeline, param_grid=hyperparamets, cv=10, n_jobs=-1)
# Fitting the model
tree_model.fit(X_train, y_train)
print("Train accuracy is : ", tree_model.score(X_train, y_train))
print("Train accuracy is : ", tree_model.score(X_test, y_test))
# > That's nice, the accuracy increased by 5% by using Decision Tree. Now let's use other tree based models for improving the accuracy.
# CatBoost 🐱👓
from catboost import CatBoostClassifier
cat_model = CatBoostClassifier(verbose=100)
# Creating validation dataset
X_train_new, X_val, y_train_new, y_val = train_test_split(
X_train, y_train, train_size=0.8, random_state=99, stratify=y_train
)
cat_model.fit(
X_train_new,
y_train_new,
eval_set=(X_val, y_val),
early_stopping_rounds=50,
use_best_model=True,
)
print("Train accuracy is : ", cat_model.score(X_train, y_train))
print("Train accuracy is : ", cat_model.score(X_test, y_test))
# > The train accuracy has increased but on the expence of test accuracy which is not good. So I will model stacking for getting better results.
# Stacking 📚
from sklearn.ensemble import StackingClassifier
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
estimators = [
("tree", DecisionTreeClassifier(max_depth=9, min_samples_leaf=10, random_state=99)),
("svr", Pipeline([("scalar", StandardScaler()), ("svm", SVC(random_state=99))])),
]
staked_model = StackingClassifier(estimators, cv=5, final_estimator=GaussianNB())
staked_model.fit(X_train, y_train)
print("Train accuracy is : ", staked_model.score(X_train, y_train))
print("Train accuracy is : ", staked_model.score(X_test, y_test))
# > It's not good as the tree model alone was performing better than stacked model. Let's try AutoML methods.
# AutoML 🤖
#
# > H2O is an open source library, it quite facinating and easy to use and I will be using this library for AutoML. So, let's get started !! 🚗
# Importing h2o library
import h2o
from h2o.automl import H2OAutoML
# #### Preprocessing for H2O
# > For training using h2o, we have to create a h2o frame, which is like a pandas dataframe. We have two options for doing this:
# >- read and preprocess the data using h2o frame
# >- read and preprocess the data using pandas and convert it to h2o frame
# > As I am more confirtable in handling the data with pandas so I am choosing the second method but if you feel confirtable in handling data with h2o frame, you can very well do that. We have already preprocessed the data using pycaret library so we will continue from that. From pycaret setup we get, X_train, y_train, X_test and y_test as pandas dataframe.
# > I didn't find any function to feed pandas dataframe to h2o, so first I convert these df to .csv and then read the .csv using h2o.import_file, to convert them into h2o frame.
# Combining X and y
df_train = pd.concat([X_train, y_train], axis=1)
df_test = pd.concat([X_test, y_test], axis=1)
df_val = pd.concat([X_val, y_val], axis=1)
# Saving as csv files
df_train.to_csv("train.csv", index=False)
df_test.to_csv("test.csv", index=False)
df_val.to_csv("val.csv", index=False)
# Initializing h2o
h2o.init()
# Reding the data using h2o frames
train = h2o.import_file("./train.csv")
test = h2o.import_file("./test.csv")
val = h2o.import_file("./val.csv")
# Identifing predictors and response
x = train.columns
y = "Survived"
x.remove(y)
# For binary classification, response should be a factor
train[y] = train[y].asfactor()
test[y] = test[y].asfactor()
val[y] = val[y].asfactor()
# Run AutoML for 20 base models (limited to 1 hour max runtime by default)
aml = H2OAutoML(max_models=20, seed=1)
aml.train(x=x, y=y, training_frame=train, validation_frame=val)
# View the AutoML Leaderboard
lb = aml.leaderboard
lb.head(rows=10) # Print all rows instead of default (10 rows)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338061.ipynb
| null | null |
[{"Id": 69338061, "ScriptId": 17618858, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5406442, "CreationDate": "07/29/2021 18:23:34", "VersionNumber": 6.0, "Title": "Titanic | Awesome EDA", "EvaluationDate": "07/29/2021", "IsChange": true, "TotalLines": 471.0, "LinesInsertedFromPrevious": 113.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 358.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
#
# Introduction
# >On April 15, 1912, during her maiden voyage, the widely considered “unsinkable” RMS Titanic sank after colliding with an iceberg. Unfortunately, there weren’t enough lifeboats for everyone onboard, resulting in the death of 1502 out of 2224 passengers and crew.
# >Many of us have come across this datasets in the start of Data Science journey, and this is a classic example to practice the skills of EDA and Machine Learning. This notebook is especially more useful for beginners in Data Science. It gives step by step guide for EDA and ML, after reading this notebook, I am sure you will be more comfortable in EDA and ML and you will be able to present your data with better visuals.
# #### I have included the most commonly used plots along with some awesome plots like sunbrust and parallel categories plot.
#
# 1. [Box Plot](#1)
# 1. [Scatter Plot](#2)
# 1. [Bar Plot](#3)
# 1. [Pie Plot](#4)
# 1. [Sunburst Plot](#5)
# 1. [Histogram](#6)
# 1. [Parallel Categories Plot](#7)
# 1. [Funnel Plot](#8)
# 1. [Treemap Plot](#9)
# PLEASE UPVOTE GUYS AND RECOMMEND THAT SHOULD IMPLEMENT
# For data handling
import numpy as np
import pandas as pd
# For visvalization
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
df = pd.read_csv("../input/titanic/train.csv")
df.head()
# Data Cleaning 🚿
# > Data may have missing values, wrong data types, outliers, etc. So, before you do EDA on your data, please check for these things and if they are present, you have to clean the data before moving further otherwise you can get misleading and incorrect visuals. So let's fold the sleeves and get ready for cleaning!
# Dropping the unnecessary columns which won't contribute any to EDA
df = df.drop(["PassengerId", "Ticket", "Name"], axis=1)
# Data Types
df.info()
# **Categorical features**: Survived, Sex, Embarked, and Pclass.
# **Numeric features**: Age, Fare. Discrete: SibSp, Parch.
# > Changing the data types of `Survived`, `Pclass` from numeric to category
# Changing the data types
cat_col = ["Survived", "Pclass"]
df[cat_col] = df[cat_col].astype("object")
# Missing values
df.isnull().mean() * 100
# > If a colum has 70%-80% missing data, it is advisable to remove that column. Cabin has 77% missing data, so we will remove that **column**. Age and Embarked has <20% missing data, so we shall remove the **rows** having the missing values. You can also impute the missing values with mean or mode with [df.fillna](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.fillna.html) but I am removing them as the focus here is on EDA not on creating models.
# > 📌 Some models can give you an error if missing values are present and some will show incorrect results, so it is important to remove them.
#
# Removing Cabin column
df = df.drop(["Cabin"], axis=1)
# Removing rows with missing data
df = df.dropna().reset_index(drop=True)
# Exploratory Data Analysis 📊
# > Before doing any kind of EDA, you should have a purpose of doing it. It should be clear what you want otherwise you will produce beautiful charts with no use. You should ask yourself, what you want to see and then choose the appropriate plot and features. So, I have mentioned the questions, which I thought before plotting the charts.
# > Note that, for a perticular question, there can be multiple charts. I have choosen the chart which I think was more appropriate. Now let's do some EDA.
# Function for understanding the distribution of the columns
def quick_plot(df):
categorical_col = df.select_dtypes("object").columns.to_list()
numeric_col = df.select_dtypes(["int", "float"]).columns.to_list()
if len(numeric_col) > len(categorical_col):
max_length = len(numeric_col)
else:
max_length = len(categorical_col)
fig, (ax1, ax2) = plt.subplots(2, max_length, figsize=(20, 10))
# Plotting for categorical columns
for axis, col in zip(ax1.ravel(), categorical_col):
axis = sns.countplot(x=col, data=df, ax=axis)
axis.set_ylabel(None)
axis.set_xlabel(col, size=16)
for i in axis.patches:
axis.text(
x=i.get_x() + i.get_width() / 2,
y=i.get_height() / 2,
s=f"{round(i.get_height(),1)}",
ha="center",
size=16,
weight="bold",
rotation=0,
color="white",
)
# Plotting for numeric columns
for axis, col in zip(ax2.ravel(), numeric_col):
axis = sns.histplot(x=col, data=df, multiple="stack", ax=axis)
axis.set_ylabel(None)
axis.set_xlabel(col, size=16)
fig.text(0.5, 1, "The distribution of Data", ha="center", fontsize=20)
plt.tight_layout()
# Quickly understanding the distribution of the columns
plt.style.use("seaborn")
quick_plot(df)
# #### 🔎Observation:
# > Passengers of class 3 are huge in number.
# > Very few passenger embarked on Q (Queenstown) port.
# > The distribution of `Fare` is skew.
# Distribution of Age and Fare 💰
# #### Question: How the features `age` and `fare` are distributed, are there any outliers ❓
# Beginner tip: It is applicable to numeric feature and used for understanding the data distribution and also for detecting outliers!
plt.style.use("seaborn")
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 6))
ax1 = sns.boxplot(y="Age", data=df, ax=ax1)
ax2 = sns.boxplot(y="Fare", data=df, ax=ax2)
ax1.set_ylabel(None)
ax1.set_xlabel(None)
ax1.set_title("Age", size=16)
ax2.set_ylabel(None)
ax2.set_xlabel(None)
ax2.set_title("Fare", size=16)
# >We have many values that are more than 75 percentile. The data is an outlier or not is the decision of the field expert. We can also tell for some of the data like `Age` - it can't be more than 100 or less than 0, etc. but in this case, I will remove these values while doing ML.
# Relation between Age and Fare 💰
# #### Question: Is there any relation between 'age' and 'fare'❓
# Beginner tip: It is applicable when you have two numeric features. It's used for visvalizing relation between numeric features.
fig, ax = plt.subplots(1, 1, figsize=(10, 6), constrained_layout=True)
ax = sns.regplot(x="Age", y="Fare", data=df)
ax.set_ylabel("Fare", fontsize=14)
ax.set_xlabel("Age", fontsize=14)
plt.title("Age Vs Fare", fontsize=16)
correlation = np.corrcoef(df["Age"], df["Fare"])[0][1]
ax.text(
x=60,
y=180,
s=f"correlation : {round(correlation,3)}",
ha="center",
size=12,
rotation=0,
color="black",
bbox=dict(boxstyle="round,pad=0.5", fc="skyblue", ec="skyblue", lw=2),
)
# #### 🔍 Observations
# > The correlation between `Age` and `Fare` is 0.093, which is close to 0, so there is no relation between them. It means, we can't guess how much fare a passenger has paid just by looking at his/her age.
# #### 📝 Note
# > The correlation score:
# > - 1 : Strongly and positively correlated (one increases, other also increases and vice versa)
# > - 0 : No correlation
# > - 1 : Strongly and negetively correlated (one increases, other also decreases and vice versa)
df.info()
#
# Analysis of Categorical Features 🔬
# > We have three categorical features`Pclass`, `Sex`, and `Embarked`. Not counting `Survived` as it is the target. Let's analyse the categorical features one by one based on the target.
# > For the analysis, I will be using PowerBI, as it is simple and efficient!
# Pclass (Ticket Class)
# > There are three classes 1st, 2nd, and 3rd.
# > 1 = 1st, 2 = 2nd, 3 = 3rd
# #### 🔎 Observations
# > As seen from previous plot also, Passengers of class 3 are huge in number. The reason can be cheap cost of the ticket.
# > 76% of the passengers form the 3rd class didn't survive while 63% of passengers form the 1st class survived.
# > The 1st class had mostly elders, may be because they can afford the ticket price and 3rd class had mostly young people.
# > Most of the rich 🤑 and old 👴👵 passengers survived.
# Gender 🤵💃
# #### 🔎 Observations
# > 81% men didn't survive while 74% women survived.
# > On average, female passengers paid higher fare than male passengers.
# Port of Embarkment 🚢
# > Passengers embarked from three different ports : C = Cherbourg, Q = Queenstown, S = Southampton
# #### 🔎 Observations
# > As seen previously also, very few passenger embarked on Q (Queenstown) port.
# > More than half of the passengers didn't survive who embarked from Queenstown (Q) and Southampton (S).
# > Passengers embarking from Cherbourg (C) paid more average fare, and also more than 50% of them survived.
# Comparison between Pclass, Gender 🤵💃, and Port of Embarkment 🚢
# 1st Class Passengers who Survived
# #### 🔎 Observations
# > Out of 55% of the passengers embarked from Cherbourg (C) and survived, 35% belongs to 1st class. As the fare of 1st class is high so as the case with passengers embarking from Cherbourg (C), most of them belongs to 1st class.
# > Very few passengers from 1st class who survived, embarked from Queenstown (Q).
# Survived passengers embarking from Southampton (S)
# #### 🔎 Observations
# > Out of all female 74% survived and 45% of these embarked from Southampton (S).
# > 47% of 2nd class passengers survived and 41% of them embarked from Southampton (S).
# > Out of all male 19% survived and 13% of these embarked from Southampton (S). So, only 6% of the male who embarked from Cherbourg (C) and Queenstown (Q), survived.
# Class 3 passengers who didn't survive
# #### 🔎 Observations
# > Out of 61% passengers who didn't survived and embarked from Queenstown (Q), 58% belongs to class 3.
# > Mojority of the female passengers who didn't survived belong to class 3.
# Analysis of Numeric Features 🔬
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 6), constrained_layout=True)
ax1 = sns.boxenplot(x="Survived", y="Age", data=df, ax=ax1)
ax2 = sns.histplot(x="Age", data=df, hue="Survived", multiple="stack", kde=True, ax=ax2)
ax1.set_xlabel(None)
ax1.set_ylabel("Age", fontsize=14)
ax1.set_title("Passenger Age and their Survival", fontsize=16)
ax1.set_xticklabels(["Not Survived", "Survived"], size=14)
ax2.set_xlabel("Age", fontsize=14)
ax2.set_ylabel("Number of Passengers", fontsize=14)
ax2.set_title("Distribution of Age and Passenger Survival", fontsize=16)
# #### 🔎 Observations
# > There is not much relation between age and survival of the passengers. This also becomes clear from the kde plot, as the nature of both, the blue line (Non Survived) and gree line (Survived) is similar.
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 6), constrained_layout=True)
ax1 = sns.boxenplot(x="Survived", y="Fare", data=df, ax=ax1)
ax2 = sns.histplot(
x="Fare", data=df, hue="Survived", multiple="stack", kde=True, ax=ax2
)
ax1.set_xlabel(None)
ax1.set_ylabel("Fare", fontsize=14)
ax1.set_title("Passenger Age and their Survival", fontsize=16)
ax1.set_xticklabels(["Not Survived", "Survived"], size=14)
ax2.set_xscale("log")
ax2.set_xlabel("Age (Log scale)", fontsize=14)
ax2.set_ylabel("Number of Passengers", fontsize=14)
ax2.set_title("Distribution of Fare and Passenger Survival", fontsize=16)
# #### 🔎 Observations
# > There is some relation between `Fare` and `Survived` of the passengers. This also becomes clear from the kde plot, as the nature of both, the blue line (Non Survived) and gree line (Survived) is different.
# Data Cleaning 🧹
# Removing Outliers
upper_bound = df["Fare"].quantile(0.97)
df = df[(df["Fare"] < upper_bound)]
# Imputing missing values
df["Age"] = df["Age"].fillna(df["Age"].median())
# Feature Enginner ⚙
# > The `Fare` feature had skew distribution, so I will log transform it.
df["Fare"] = df["Fare"].transform(np.log)
df["Fare"].min()
# As new Fare contains -infinity, I will replace it will next minimum fare
least_value = df["Fare"].sort_values().unique()[1]
df["Fare"] = df["Fare"].replace(-np.inf, least_value)
# Creating bins for age
df["Age_Bins"] = pd.cut(
df["Age"],
[0, 10, 25, 40, 60, 100],
labels=["<10", "10 to 25", "26 to 40", "41 to 60", "60+"],
include_lowest=True,
)
# Creating new feature Age * Fare
df["Age x Fare"] = df["Age"] * df["Fare"]
# Creating new higher degree features: Age^^2 and Fare^^2
df["Age^2"] = df["Age"] ** 2
df["Fare^2"] = df["Fare"] ** 2
# Making data ready for training
# Creating X (features) and y (target)
X = df.drop(["Survived"], axis=1)
y = df["Survived"].astype("int")
# One hot encoding for categorical features
X_new = pd.get_dummies(X, drop_first=True)
# Train - Test split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X_new, y, train_size=0.8, random_state=99, stratify=y
)
# Creating ML pipeline 🚄
# > First we will create simple model (for baseline) and then move to more complex models.
# Logistic Regression
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LogisticRegressionCV
pipeline = Pipeline(
[("scalar", StandardScaler()), ("logistic", LogisticRegressionCV(cv=10, n_jobs=-1))]
)
log_model = pipeline.fit(X_train, y_train)
print("Train accuracy is : ", log_model.score(X_train, y_train))
print("Train accuracy is : ", log_model.score(X_test, y_test))
# > The base model has an accuracy of 80%. Now, lets try some other models to get accuracy more than this.
# Decision Tree 🌳
from sklearn.tree import DecisionTreeClassifier
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import GridSearchCV
pipeline = Pipeline(
[("scalar", StandardScaler()), ("tree", DecisionTreeClassifier(random_state=99))]
)
hyperparamets = {
"tree__max_depth": [3, 5, 7, 9, 11],
"tree__min_samples_leaf": [10, 20, 30, 40],
"tree__min_impurity_decrease": [0, 0.1, 0.2, 0.5],
}
tree_model = GridSearchCV(pipeline, param_grid=hyperparamets, cv=10, n_jobs=-1)
# Fitting the model
tree_model.fit(X_train, y_train)
print("Train accuracy is : ", tree_model.score(X_train, y_train))
print("Train accuracy is : ", tree_model.score(X_test, y_test))
# > That's nice, the accuracy increased by 5% by using Decision Tree. Now let's use other tree based models for improving the accuracy.
# CatBoost 🐱👓
from catboost import CatBoostClassifier
cat_model = CatBoostClassifier(verbose=100)
# Creating validation dataset
X_train_new, X_val, y_train_new, y_val = train_test_split(
X_train, y_train, train_size=0.8, random_state=99, stratify=y_train
)
cat_model.fit(
X_train_new,
y_train_new,
eval_set=(X_val, y_val),
early_stopping_rounds=50,
use_best_model=True,
)
print("Train accuracy is : ", cat_model.score(X_train, y_train))
print("Train accuracy is : ", cat_model.score(X_test, y_test))
# > The train accuracy has increased but on the expence of test accuracy which is not good. So I will model stacking for getting better results.
# Stacking 📚
from sklearn.ensemble import StackingClassifier
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
estimators = [
("tree", DecisionTreeClassifier(max_depth=9, min_samples_leaf=10, random_state=99)),
("svr", Pipeline([("scalar", StandardScaler()), ("svm", SVC(random_state=99))])),
]
staked_model = StackingClassifier(estimators, cv=5, final_estimator=GaussianNB())
staked_model.fit(X_train, y_train)
print("Train accuracy is : ", staked_model.score(X_train, y_train))
print("Train accuracy is : ", staked_model.score(X_test, y_test))
# > It's not good as the tree model alone was performing better than stacked model. Let's try AutoML methods.
# AutoML 🤖
#
# > H2O is an open source library, it quite facinating and easy to use and I will be using this library for AutoML. So, let's get started !! 🚗
# Importing h2o library
import h2o
from h2o.automl import H2OAutoML
# #### Preprocessing for H2O
# > For training using h2o, we have to create a h2o frame, which is like a pandas dataframe. We have two options for doing this:
# >- read and preprocess the data using h2o frame
# >- read and preprocess the data using pandas and convert it to h2o frame
# > As I am more confirtable in handling the data with pandas so I am choosing the second method but if you feel confirtable in handling data with h2o frame, you can very well do that. We have already preprocessed the data using pycaret library so we will continue from that. From pycaret setup we get, X_train, y_train, X_test and y_test as pandas dataframe.
# > I didn't find any function to feed pandas dataframe to h2o, so first I convert these df to .csv and then read the .csv using h2o.import_file, to convert them into h2o frame.
# Combining X and y
df_train = pd.concat([X_train, y_train], axis=1)
df_test = pd.concat([X_test, y_test], axis=1)
df_val = pd.concat([X_val, y_val], axis=1)
# Saving as csv files
df_train.to_csv("train.csv", index=False)
df_test.to_csv("test.csv", index=False)
df_val.to_csv("val.csv", index=False)
# Initializing h2o
h2o.init()
# Reding the data using h2o frames
train = h2o.import_file("./train.csv")
test = h2o.import_file("./test.csv")
val = h2o.import_file("./val.csv")
# Identifing predictors and response
x = train.columns
y = "Survived"
x.remove(y)
# For binary classification, response should be a factor
train[y] = train[y].asfactor()
test[y] = test[y].asfactor()
val[y] = val[y].asfactor()
# Run AutoML for 20 base models (limited to 1 hour max runtime by default)
aml = H2OAutoML(max_models=20, seed=1)
aml.train(x=x, y=y, training_frame=train, validation_frame=val)
# View the AutoML Leaderboard
lb = aml.leaderboard
lb.head(rows=10) # Print all rows instead of default (10 rows)
| false | 0 | 5,707 | 0 | 5,707 | 5,707 |
||
69338178
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import cross_val_score
import warnings
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
warnings.filterwarnings("ignore")
train_data = pd.read_csv("../input/titanic/train.csv")
train_data.head()
testData_data = pd.read_csv("../input/titanic/test.csv")
testData_data.head()
train_data.isnull().sum()
# Data Cleaning
train_data[["Embarked", "Name"]].groupby(
by=["Embarked"], as_index=True
).count().sort_values("Name", ascending=False)
most_repeated = "S"
train_data.Embarked.replace(np.nan, most_repeated, inplace=True)
testData_data.Embarked.replace(np.nan, most_repeated, inplace=True)
print(
"the number of null value in Embarked Column =", train_data.Embarked.isnull().sum()
)
Embarked_transform_dict = {"S": 1, "C": 2, "Q": 3}
for value in Embarked_transform_dict:
train_data.Embarked.replace(value, Embarked_transform_dict.get(value), inplace=True)
testData_data.Embarked.replace(
value, Embarked_transform_dict.get(value), inplace=True
)
train_data.head(5)
print("the number of null value in Cabin Column =", train_data.Cabin.isnull().sum())
train_data.drop("Cabin", axis=1, inplace=True)
testData_data.drop("Cabin", axis=1, inplace=True)
train_data.head()
print("Range of Fare column values = ", train_data.Fare.max() - train_data.Fare.min())
testData_data.Fare.replace(np.nan, testData_data.Fare.mean(), inplace=True)
print(
"Range of Fare column values = ",
testData_data.Fare.max() - testData_data.Fare.min(),
)
train_data.Fare = train_data.Fare.astype("int64")
testData_data.Fare = testData_data.Fare.astype("int64")
# df_train.info()
testData_data.head()
Sex_dict = {"male": 1, "female": 2}
for key, value in Sex_dict.items():
train_data.Sex.replace(key, value, inplace=True)
testData_data.Sex.replace(key, value, inplace=True)
train_data.Sex = train_data.Sex.astype("int64")
testData_data.Sex = testData_data.Sex.astype("int64")
train_data.head()
age_surv_corr = train_data["Age"].corr(train_data["Survived"])
age_surv_corr
class_surv_corr = train_data["Pclass"].corr(train_data["Survived"])
class_surv_corr
train_data["Sex"].value_counts()
train_data[["Sex", "Survived"]].groupby(["Sex"], as_index=False).mean()
# # EDA
x_axis = ["Female", "Male"]
y_axis = [0.74, 0.19]
plt.bar(x=x_axis, height=y_axis)
plt.xlabel("Sex")
plt.ylabel("Survived")
plt.show()
train_data["Survived"].value_counts()
# 549 people died and 342 people survived
survived = "survived"
not_survived = "not survived"
# Percentage of people that survived
survived_per = train_data[train_data["Survived"] == 1]
survived = float(round((len(survived) / len(train_data)) * 100.0))
print("Survived:", str(survived), "%")
# Percentage of people that died
died = train_data[train_data["Survived"] == 0]
died = float(round((len(died) / len(train_data)) * 100.0))
print("Died:", str(died), "%")
train_data["Pclass"].value_counts().sort_index()
# there where 3 classes of tickets and 491 wich is class 3 had the highest passengers
train_data[["Pclass", "Survived"]].groupby(["Pclass"], as_index=False).mean()
sns.barplot(x="Pclass", y="Survived", data=train_data)
all_classes = pd.crosstab(train_data["Pclass"], train_data["Sex"])
print(all_classes)
all_classes = pd.crosstab(train_data["Pclass"], train_data["Survived"])
print(all_classes)
all_classes.div(all_classes.sum(1).astype(float), axis=0).plot(
kind="barh", stacked=False
)
plt.ylabel("Pclass")
plt.xlabel("Percentage")
plt.title("The perentage of those who survived and died in the different coach classes")
freq_table = train_data["Age"].value_counts(bins=8).sort_index()
freq_table
freq = pd.DataFrame(freq_table)
freq
# the ages between 20 to 30 had the most passengers
train_data.Embarked.value_counts()
train_data[["Embarked", "Survived"]].groupby(["Embarked"], as_index=False).mean()
x_axis = ["C", "Q", "S"]
y_axis = [0.55, 0.39, 0.34]
plt.bar(x=x_axis, height=y_axis)
plt.xlabel("Embarked")
plt.ylabel("Survived")
plt.title("The ratio of people who Embarked and Survived")
plt.show()
train_data.Parch.value_counts()
train_data[["Parch", "Survived"]].groupby(["Parch"], as_index=False).mean()
x_axis = [0, 1, 2, 3, 4, 5, 6]
y_axis = [0.34, 0.55, 0.50, 0.60, 0.0, 0.2, 0.0]
plt.bar(x=x_axis, height=y_axis)
plt.xlabel("Parch")
plt.ylabel("Survived")
plt.title("The ratio of Parch and Survived")
plt.show()
train_data[["SibSp", "Survived"]].groupby(["SibSp"], as_index=False).mean()
x_axis = [0, 1, 2, 3, 4, 5, 8]
y_axis = [0.35, 0.54, 0.46, 0.25, 0.17, 0.0, 0.0]
plt.bar(x=x_axis, height=y_axis)
plt.xlabel("SibSp")
plt.ylabel("Survived")
plt.title("The ratio of SibSp and Survived")
plt.show()
df_Age_train = train_data.loc[pd.notna(train_data.Age)]
df_Age_train.Age = df_Age_train.Age.astype("float64")
df_Age_train.Age = (df_Age_train.Age - df_Age_train.Age.mean()) / df_Age_train.Age.std()
df_Age_train
train_data.drop("Age", axis=1, inplace=True)
testData_data.drop("Age", axis=1, inplace=True)
train_data.head()
bins_i = [-1, 50, 100, 150, 200, 250, 300, 350, 400, 450, 500, 550]
labels_i = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11]
train_data["stage"] = 0
train_data["stage"] = pd.cut(train_data.Fare, bins=bins_i, labels=labels_i)
testData_data["stage"] = 0
testData_data["stage"] = pd.cut(testData_data.Fare, bins=bins_i, labels=labels_i)
train_data.stage.unique()
train_data.Fare = train_data.stage.astype("int64")
testData_data.Fare = testData_data.stage.astype("int64")
train_data.drop("stage", axis=1, inplace=True)
testData_data.drop("stage", axis=1, inplace=True)
train_data.head()
data = [train_data, testData_data]
for dataset in data:
dataset["FamilySize"] = dataset["SibSp"] + dataset["Parch"] + 1
for dataset in data:
dataset["IsAlone"] = 0
dataset.loc[dataset["FamilySize"] == 1, "IsAlone"] = 1
print(train_data[["IsAlone", "Survived"]].groupby(["IsAlone"], as_index=False).mean())
columns = ["Pclass", "Sex", "Fare", "Embarked", "IsAlone"]
X_train = train_data[columns]
Y_train = train_data["Survived"]
len(Y_train)
X_test = testData_data[columns]
len(X_test)
Y_test = testData_data[columns]
len(Y_test)
# # Predictions
# # Random Forest
random_forest = RandomForestClassifier(
n_estimators=40, min_samples_leaf=2, max_features=0.1, n_jobs=-1
)
random_forest.fit(X_train, Y_train)
Y_pred_Random = random_forest.predict(X_test)
print(
"the train score of random_forest = ",
round(random_forest.score(X_train, Y_train) * 100, 2),
"%",
)
# # Logistic Regression
logistic_regression = LogisticRegression(solver="liblinear", max_iter=1000)
logistic_regression.fit(X_train, Y_train)
Y_pred_Logistic = logistic_regression.predict(X_test)
print(
"the train score of logistic_regression = ",
round(logistic_regression.score(X_train, Y_train) * 100, 2),
"%",
)
tree = DecisionTreeClassifier(random_state=25)
tree.fit(X_train, Y_train)
Y_pred_Tree = tree.predict(X_test)
print("the score of prediction = ", round(tree.score(X_train, Y_train) * 100, 2), "%")
scores = cross_val_score(tree, X_train, Y_train, scoring="accuracy", cv=100)
scores.mean()
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(X_train, Y_train)
Y_pred_KNN = knn.predict(X_test)
print("the score of prediction = ", round(knn.score(X_train, Y_train) * 100, 2), "%")
submission = pd.DataFrame(
{"PassengerId": testData_data["PassengerId"], "Survived": Y_pred_KNN}
)
submission.to_csv("./submission.csv", index=False)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/338/69338178.ipynb
| null | null |
[{"Id": 69338178, "ScriptId": 18927450, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7585297, "CreationDate": "07/29/2021 18:25:43", "VersionNumber": 2.0, "Title": "Titanic Prediction", "EvaluationDate": "07/29/2021", "IsChange": true, "TotalLines": 261.0, "LinesInsertedFromPrevious": 5.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 256.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import cross_val_score
import warnings
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
warnings.filterwarnings("ignore")
train_data = pd.read_csv("../input/titanic/train.csv")
train_data.head()
testData_data = pd.read_csv("../input/titanic/test.csv")
testData_data.head()
train_data.isnull().sum()
# Data Cleaning
train_data[["Embarked", "Name"]].groupby(
by=["Embarked"], as_index=True
).count().sort_values("Name", ascending=False)
most_repeated = "S"
train_data.Embarked.replace(np.nan, most_repeated, inplace=True)
testData_data.Embarked.replace(np.nan, most_repeated, inplace=True)
print(
"the number of null value in Embarked Column =", train_data.Embarked.isnull().sum()
)
Embarked_transform_dict = {"S": 1, "C": 2, "Q": 3}
for value in Embarked_transform_dict:
train_data.Embarked.replace(value, Embarked_transform_dict.get(value), inplace=True)
testData_data.Embarked.replace(
value, Embarked_transform_dict.get(value), inplace=True
)
train_data.head(5)
print("the number of null value in Cabin Column =", train_data.Cabin.isnull().sum())
train_data.drop("Cabin", axis=1, inplace=True)
testData_data.drop("Cabin", axis=1, inplace=True)
train_data.head()
print("Range of Fare column values = ", train_data.Fare.max() - train_data.Fare.min())
testData_data.Fare.replace(np.nan, testData_data.Fare.mean(), inplace=True)
print(
"Range of Fare column values = ",
testData_data.Fare.max() - testData_data.Fare.min(),
)
train_data.Fare = train_data.Fare.astype("int64")
testData_data.Fare = testData_data.Fare.astype("int64")
# df_train.info()
testData_data.head()
Sex_dict = {"male": 1, "female": 2}
for key, value in Sex_dict.items():
train_data.Sex.replace(key, value, inplace=True)
testData_data.Sex.replace(key, value, inplace=True)
train_data.Sex = train_data.Sex.astype("int64")
testData_data.Sex = testData_data.Sex.astype("int64")
train_data.head()
age_surv_corr = train_data["Age"].corr(train_data["Survived"])
age_surv_corr
class_surv_corr = train_data["Pclass"].corr(train_data["Survived"])
class_surv_corr
train_data["Sex"].value_counts()
train_data[["Sex", "Survived"]].groupby(["Sex"], as_index=False).mean()
# # EDA
x_axis = ["Female", "Male"]
y_axis = [0.74, 0.19]
plt.bar(x=x_axis, height=y_axis)
plt.xlabel("Sex")
plt.ylabel("Survived")
plt.show()
train_data["Survived"].value_counts()
# 549 people died and 342 people survived
survived = "survived"
not_survived = "not survived"
# Percentage of people that survived
survived_per = train_data[train_data["Survived"] == 1]
survived = float(round((len(survived) / len(train_data)) * 100.0))
print("Survived:", str(survived), "%")
# Percentage of people that died
died = train_data[train_data["Survived"] == 0]
died = float(round((len(died) / len(train_data)) * 100.0))
print("Died:", str(died), "%")
train_data["Pclass"].value_counts().sort_index()
# there where 3 classes of tickets and 491 wich is class 3 had the highest passengers
train_data[["Pclass", "Survived"]].groupby(["Pclass"], as_index=False).mean()
sns.barplot(x="Pclass", y="Survived", data=train_data)
all_classes = pd.crosstab(train_data["Pclass"], train_data["Sex"])
print(all_classes)
all_classes = pd.crosstab(train_data["Pclass"], train_data["Survived"])
print(all_classes)
all_classes.div(all_classes.sum(1).astype(float), axis=0).plot(
kind="barh", stacked=False
)
plt.ylabel("Pclass")
plt.xlabel("Percentage")
plt.title("The perentage of those who survived and died in the different coach classes")
freq_table = train_data["Age"].value_counts(bins=8).sort_index()
freq_table
freq = pd.DataFrame(freq_table)
freq
# the ages between 20 to 30 had the most passengers
train_data.Embarked.value_counts()
train_data[["Embarked", "Survived"]].groupby(["Embarked"], as_index=False).mean()
x_axis = ["C", "Q", "S"]
y_axis = [0.55, 0.39, 0.34]
plt.bar(x=x_axis, height=y_axis)
plt.xlabel("Embarked")
plt.ylabel("Survived")
plt.title("The ratio of people who Embarked and Survived")
plt.show()
train_data.Parch.value_counts()
train_data[["Parch", "Survived"]].groupby(["Parch"], as_index=False).mean()
x_axis = [0, 1, 2, 3, 4, 5, 6]
y_axis = [0.34, 0.55, 0.50, 0.60, 0.0, 0.2, 0.0]
plt.bar(x=x_axis, height=y_axis)
plt.xlabel("Parch")
plt.ylabel("Survived")
plt.title("The ratio of Parch and Survived")
plt.show()
train_data[["SibSp", "Survived"]].groupby(["SibSp"], as_index=False).mean()
x_axis = [0, 1, 2, 3, 4, 5, 8]
y_axis = [0.35, 0.54, 0.46, 0.25, 0.17, 0.0, 0.0]
plt.bar(x=x_axis, height=y_axis)
plt.xlabel("SibSp")
plt.ylabel("Survived")
plt.title("The ratio of SibSp and Survived")
plt.show()
df_Age_train = train_data.loc[pd.notna(train_data.Age)]
df_Age_train.Age = df_Age_train.Age.astype("float64")
df_Age_train.Age = (df_Age_train.Age - df_Age_train.Age.mean()) / df_Age_train.Age.std()
df_Age_train
train_data.drop("Age", axis=1, inplace=True)
testData_data.drop("Age", axis=1, inplace=True)
train_data.head()
bins_i = [-1, 50, 100, 150, 200, 250, 300, 350, 400, 450, 500, 550]
labels_i = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11]
train_data["stage"] = 0
train_data["stage"] = pd.cut(train_data.Fare, bins=bins_i, labels=labels_i)
testData_data["stage"] = 0
testData_data["stage"] = pd.cut(testData_data.Fare, bins=bins_i, labels=labels_i)
train_data.stage.unique()
train_data.Fare = train_data.stage.astype("int64")
testData_data.Fare = testData_data.stage.astype("int64")
train_data.drop("stage", axis=1, inplace=True)
testData_data.drop("stage", axis=1, inplace=True)
train_data.head()
data = [train_data, testData_data]
for dataset in data:
dataset["FamilySize"] = dataset["SibSp"] + dataset["Parch"] + 1
for dataset in data:
dataset["IsAlone"] = 0
dataset.loc[dataset["FamilySize"] == 1, "IsAlone"] = 1
print(train_data[["IsAlone", "Survived"]].groupby(["IsAlone"], as_index=False).mean())
columns = ["Pclass", "Sex", "Fare", "Embarked", "IsAlone"]
X_train = train_data[columns]
Y_train = train_data["Survived"]
len(Y_train)
X_test = testData_data[columns]
len(X_test)
Y_test = testData_data[columns]
len(Y_test)
# # Predictions
# # Random Forest
random_forest = RandomForestClassifier(
n_estimators=40, min_samples_leaf=2, max_features=0.1, n_jobs=-1
)
random_forest.fit(X_train, Y_train)
Y_pred_Random = random_forest.predict(X_test)
print(
"the train score of random_forest = ",
round(random_forest.score(X_train, Y_train) * 100, 2),
"%",
)
# # Logistic Regression
logistic_regression = LogisticRegression(solver="liblinear", max_iter=1000)
logistic_regression.fit(X_train, Y_train)
Y_pred_Logistic = logistic_regression.predict(X_test)
print(
"the train score of logistic_regression = ",
round(logistic_regression.score(X_train, Y_train) * 100, 2),
"%",
)
tree = DecisionTreeClassifier(random_state=25)
tree.fit(X_train, Y_train)
Y_pred_Tree = tree.predict(X_test)
print("the score of prediction = ", round(tree.score(X_train, Y_train) * 100, 2), "%")
scores = cross_val_score(tree, X_train, Y_train, scoring="accuracy", cv=100)
scores.mean()
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(X_train, Y_train)
Y_pred_KNN = knn.predict(X_test)
print("the score of prediction = ", round(knn.score(X_train, Y_train) * 100, 2), "%")
submission = pd.DataFrame(
{"PassengerId": testData_data["PassengerId"], "Survived": Y_pred_KNN}
)
submission.to_csv("./submission.csv", index=False)
| false | 0 | 3,114 | 1 | 3,114 | 3,114 |
||
69242038
|
"""
In this Notebook you are going to learn
complete Pandas(Python Libaray for Data Science and Machine Learning)
with the explaination for each line of code.
For any queries please put it down in comment section I'll definitely help you out.
If this looks useful to you please don't forget to give an up vote.
Author: Lakshit
"""
# Pandas
# Pandas is a opensource python library prividing high performance data manuplation and analysis tools using it's powerful data structures.
#
import pandas as pd
import numpy as np
# Series
# Series is a single dimensional data structure with homogenous data type values.
# Also Series comes with index or are one single column where as list is just a list
series = pd.Series([10, 20, 40, 50], index=["a", "b", "c", "d"])
lists = [1, 2, 3]
print(lists)
series
# Dataframe
# Two dimensional object with heterogenous data type value.
listz = [["Ravi", 10], ["Ram", 20], ["Raj", 30]]
print(listz)
df = pd.DataFrame(listz, columns=["Name", "Age"], index=["a", "b", "c"])
df
# Accessing DataFrame
dfa = df["Name"]
dfb = df[["Name"]]
print(type(dfa))
print(type(dfb))
dfa
# dfa is a series because we are directly accessing column values, whereas dfb is a DataFrame because we are accessing a total column
# Dictionary to DataFrame
dictz = {"Name": ["Rahul", "Rio"], "Age": [20, 40]}
print(dictz)
dfd = pd.DataFrame(dictz)
dfd
# ## Array to DataFrame
arr = np.array([[1, 2, 3], [5, 6, 7]])
print(arr)
dfa = pd.DataFrame(arr)
print(dfa)
## Now dataframe into array
arrd = np.array(dfa)
arrd
# ## Range Data
# Syntax: pd.date_range(date, period , freq)
# date format: YYYYMMDD
# period: it is the no of days or months or years
# frequency(freq): it is the format i.e Month(M), Year(Y), Day(D)
from datetime import datetime
ranges = pd.date_range(start=datetime.now(), periods=10, freq="Y")
type(ranges)
ranges
# ## Head Tail Func
# - pd.head() # Returns the first 5 values and to change these 5 values we can pass n in pd.head(3) it will return first 3 values.
# - pd.tail() # Returns the last 5 values and to change these 5 values we can pass n in pd.tail(3) it will return last 3 values.
# Create Df
df = np.random.randn(10, 4)
print(df)
df = pd.DataFrame(
df, index=ranges, columns=["Random 1", "Random 2", "Random 3", "Random 4"]
)
print(df)
print(df.shape)
print(df.columns)
print(df.info())
print(df.head(3))
print("#####")
print(df.tail(3))
# ## Null values
print(df.isnull()) # null values are there or not
print(df.isnull().sum()) # sum of null values in each column
df.dropna() # by default it'll delete the entire row there nan value is present but to delete the column we can use df.dropna(axis=1) by default axis is 0
# ## Values
print(df.value_counts()) # it'll return the no. of occurence of the data
df.dtypes # >> to know each datatype
# Same for column
print(df["Random 1"].value_counts())
print(df.corr()) # to find the relation between adjusent column
df.describe() ## Gives the summary of the columnwise data
# ## Slicing
df[0:2]
# ### loc vs iloc
# - loc is used to access data by putting the location for the data. i.e `df.loc["2021-12-31 01:49:18.893933", "Random 1"][0]`
# - In the first block he have inserted the row value and in second block we have inserted the column value.
# - iloc is used to access data using the index. i.e `df.iloc[[0,2,5],0:2]`
df.loc["2021-12-31", "Random 1"]
df.iloc[[0, 2, 5], -1]
print(df)
df.iloc[[2, 4, 6], 0:2]
# ## Concatenation & Descriptive Statistics
# - `df.drop()`:- mainly used to delete columns
drd = df.drop(columns=["Random 1", "Random 2"])
# - `df.concat()`:- It is used to merge to dataframes row wise
a = {"Name": ["Rio", "Nirobi"], "Age": [18, 20]}
a = pd.DataFrame(a)
b = {"Name": ["Tokyo", "Berlin", "Tokyo"], "Age": [23, 30, 23]}
b = pd.DataFrame(b, index=[2, 3, 4])
ab = pd.concat([a, b])
ab
# - Knowing the existance of value.
print(ab["Name"] == "Tokyo", "\n\n") ## tells the existance
print(ab[ab["Name"] == "Tokyo"]) ## Shows the existance
# - Deleting the dublicate `df.drop_duplicates(column_name)`
# - it'll delete the whole row.
#
ab = ab.drop_duplicates(["Name"])
# - Sorting values `df.sort_values(column_name)` it can sort numerically as well alphabatically.
#
ab_sort_aplha = ab.sort_values("Name")
# - renaming columns `df.rename(columns = {"Name": "Emp_name"})`
ab_sort_aplha.rename(columns={"Name": "Emp_Name", "Age": "Emp_Age"})
# - Other functions
# - `df.sum()` to find the sum of values
# - `df.mean()` to find the mean of values
# - `df.median()` to find the median of the values
# -`df.mode()` to find the mode of the values
# - `df.std()` to find the standard deviation of the values
# - `df.min()` to find the minimum value from the all values
# - `df.max()` to find the maximum value from the all values
# - `pd.merge` Merges df column wise, here `on="Name"` means we are looking for Name in both the df.
di1 = {"Name": ["Rahul", "Sham", "Ram"], "Age": [12, 23, 45]}
df1 = pd.DataFrame(di1)
di2 = {
"Course": ["Machine Learning", "Data Science", "MLops"],
"Name": ["Rahul", "Sham", "Ram"],
"Year": [2020, 2017, 2015],
}
df2 = pd.DataFrame(di2)
df = pd.merge(df1, df2, on="Name", how="inner")
df
## Doing changes in df
df.rename(columns={"Name": "Student_Name"})
df.iloc[0, 0] = "Virat"
df
# ## Data Cleaning
dfs = pd.Series(["Lakshit", "18", "www.lakshit.io"])
dfs.str.capitalize()
dfs.str.islower()
dfs.str.lower()
dfs.str.isupper()
dfs.str.upper()
dfs.str.isnumeric()
dfs.str.swapcase()
dfs.str.len()
dfs.str.cat(sep="_") # >> Lakshit_18_www.lakshit.io
dfs.str.replace("www.", "yoyo.")
dfs.str.repeat(2)
dfs.str.count("w") # occurence of a letter
# ## Applying funtions to tables
# - `df.pipe(func_name)` it'll apply to the whole table.
# - `df.apply(func_name)` it'll apply to each column.
num = np.random.rand(10, 4)
dfNum = pd.DataFrame(num)
print(dfNum, "\n\n")
def add(a, b):
return a + b
dfNum.pipe(type) # >> pandas.core.frame.DataFrame
# PIPE
print(dfNum.pipe(add, 10), "\n\n")
# Apply
dfNum.apply(np.mean)
import os
os.getcwd() # to get the current working directiory
# os.chdir("dir_name") # to change the current working directiory
df.dtypes
# ## Importing csv file
df = pd.read_csv(
"sample_data/california_housing_test.csv",
header=0,
skiprows=[1, 2, 3, 4, 5],
names=[
"Long",
"Lat",
"house_med_age",
"Rooms",
"bedrooms",
"population_",
"household",
"med_inc",
"med_val",
],
)
# - here header = 0 is by default you can change header toany other row by putting the row number.
# - here skiprows will skip the rows which are specified
# - here names is used to give custom header name
# - put `header = None` to remove header
df
df = pd.read_csv(
"sample_data/california_housing_test.csv",
header=None,
prefix="Column: ",
na_values=["na value"],
verbose=True,
)
# - here `prefix = "column: "` is used to add any prefix to the column names i.e Column: 1
# - `na_values=['na_value']` is used to fill replace the na value with na_value
# - We can also use `sep=";"` if the seperator is not ',' we have to specify the seperator
# - `dtype={"Column: 0": 'int'}` if you want to change the data type of a column then specify it here.
# - `verbose=True` is used to get the time it takes to load the file
print(df)
df.dtypes
# ## Reading CSV Data from URL
dfs = pd.read_csv(
"http://winterolympicsmedals.com/medals.csv", nrows=10, usecols=[0, 1, 5]
)
dfs.shape
dfs.to_csv("new_.medals.csv", index=None)
# - `skipfooter=11` will remove last 11 rows from table
# - `nrows=10` will only show the first 10 rows
# - `usecols=[0,1,5]` will only return the columns which are present at the given index
# - `dfs.to_csv("new_medals.csv", index=None)` to save the file into csv format
# - `pd.read_excel(filename)` to read the excel file (check documentation for more)
# ## Data Visualiztion using Pandas
data = np.random.rand(100, 4)
data = pd.DataFrame(data, columns=["f", "s", "t", "fr"])
data["f"].plot.box(vert=False)
# - green line means 50% data is near 0.4
# - two ends are min and max values in dataframe
# - `vert = False` means we don't want to show data vertically
data["f"].plot()
data["f"].plot.bar()
# - `data['f'].plot.barh()` to show data vertically
data["f"].plot.hist()
# - `data['f'].plot.hist(bins=10)` here bins refers to the no. of blocks
data["f"].plot.area()
data.plot.scatter(x="f", y="s")
data["f"].plot.pie()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242038.ipynb
| null | null |
[{"Id": 69242038, "ScriptId": 18901776, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6289086, "CreationDate": "07/28/2021 14:16:32", "VersionNumber": 1.0, "Title": "Pandas tutorial", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 311.0, "LinesInsertedFromPrevious": 311.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
"""
In this Notebook you are going to learn
complete Pandas(Python Libaray for Data Science and Machine Learning)
with the explaination for each line of code.
For any queries please put it down in comment section I'll definitely help you out.
If this looks useful to you please don't forget to give an up vote.
Author: Lakshit
"""
# Pandas
# Pandas is a opensource python library prividing high performance data manuplation and analysis tools using it's powerful data structures.
#
import pandas as pd
import numpy as np
# Series
# Series is a single dimensional data structure with homogenous data type values.
# Also Series comes with index or are one single column where as list is just a list
series = pd.Series([10, 20, 40, 50], index=["a", "b", "c", "d"])
lists = [1, 2, 3]
print(lists)
series
# Dataframe
# Two dimensional object with heterogenous data type value.
listz = [["Ravi", 10], ["Ram", 20], ["Raj", 30]]
print(listz)
df = pd.DataFrame(listz, columns=["Name", "Age"], index=["a", "b", "c"])
df
# Accessing DataFrame
dfa = df["Name"]
dfb = df[["Name"]]
print(type(dfa))
print(type(dfb))
dfa
# dfa is a series because we are directly accessing column values, whereas dfb is a DataFrame because we are accessing a total column
# Dictionary to DataFrame
dictz = {"Name": ["Rahul", "Rio"], "Age": [20, 40]}
print(dictz)
dfd = pd.DataFrame(dictz)
dfd
# ## Array to DataFrame
arr = np.array([[1, 2, 3], [5, 6, 7]])
print(arr)
dfa = pd.DataFrame(arr)
print(dfa)
## Now dataframe into array
arrd = np.array(dfa)
arrd
# ## Range Data
# Syntax: pd.date_range(date, period , freq)
# date format: YYYYMMDD
# period: it is the no of days or months or years
# frequency(freq): it is the format i.e Month(M), Year(Y), Day(D)
from datetime import datetime
ranges = pd.date_range(start=datetime.now(), periods=10, freq="Y")
type(ranges)
ranges
# ## Head Tail Func
# - pd.head() # Returns the first 5 values and to change these 5 values we can pass n in pd.head(3) it will return first 3 values.
# - pd.tail() # Returns the last 5 values and to change these 5 values we can pass n in pd.tail(3) it will return last 3 values.
# Create Df
df = np.random.randn(10, 4)
print(df)
df = pd.DataFrame(
df, index=ranges, columns=["Random 1", "Random 2", "Random 3", "Random 4"]
)
print(df)
print(df.shape)
print(df.columns)
print(df.info())
print(df.head(3))
print("#####")
print(df.tail(3))
# ## Null values
print(df.isnull()) # null values are there or not
print(df.isnull().sum()) # sum of null values in each column
df.dropna() # by default it'll delete the entire row there nan value is present but to delete the column we can use df.dropna(axis=1) by default axis is 0
# ## Values
print(df.value_counts()) # it'll return the no. of occurence of the data
df.dtypes # >> to know each datatype
# Same for column
print(df["Random 1"].value_counts())
print(df.corr()) # to find the relation between adjusent column
df.describe() ## Gives the summary of the columnwise data
# ## Slicing
df[0:2]
# ### loc vs iloc
# - loc is used to access data by putting the location for the data. i.e `df.loc["2021-12-31 01:49:18.893933", "Random 1"][0]`
# - In the first block he have inserted the row value and in second block we have inserted the column value.
# - iloc is used to access data using the index. i.e `df.iloc[[0,2,5],0:2]`
df.loc["2021-12-31", "Random 1"]
df.iloc[[0, 2, 5], -1]
print(df)
df.iloc[[2, 4, 6], 0:2]
# ## Concatenation & Descriptive Statistics
# - `df.drop()`:- mainly used to delete columns
drd = df.drop(columns=["Random 1", "Random 2"])
# - `df.concat()`:- It is used to merge to dataframes row wise
a = {"Name": ["Rio", "Nirobi"], "Age": [18, 20]}
a = pd.DataFrame(a)
b = {"Name": ["Tokyo", "Berlin", "Tokyo"], "Age": [23, 30, 23]}
b = pd.DataFrame(b, index=[2, 3, 4])
ab = pd.concat([a, b])
ab
# - Knowing the existance of value.
print(ab["Name"] == "Tokyo", "\n\n") ## tells the existance
print(ab[ab["Name"] == "Tokyo"]) ## Shows the existance
# - Deleting the dublicate `df.drop_duplicates(column_name)`
# - it'll delete the whole row.
#
ab = ab.drop_duplicates(["Name"])
# - Sorting values `df.sort_values(column_name)` it can sort numerically as well alphabatically.
#
ab_sort_aplha = ab.sort_values("Name")
# - renaming columns `df.rename(columns = {"Name": "Emp_name"})`
ab_sort_aplha.rename(columns={"Name": "Emp_Name", "Age": "Emp_Age"})
# - Other functions
# - `df.sum()` to find the sum of values
# - `df.mean()` to find the mean of values
# - `df.median()` to find the median of the values
# -`df.mode()` to find the mode of the values
# - `df.std()` to find the standard deviation of the values
# - `df.min()` to find the minimum value from the all values
# - `df.max()` to find the maximum value from the all values
# - `pd.merge` Merges df column wise, here `on="Name"` means we are looking for Name in both the df.
di1 = {"Name": ["Rahul", "Sham", "Ram"], "Age": [12, 23, 45]}
df1 = pd.DataFrame(di1)
di2 = {
"Course": ["Machine Learning", "Data Science", "MLops"],
"Name": ["Rahul", "Sham", "Ram"],
"Year": [2020, 2017, 2015],
}
df2 = pd.DataFrame(di2)
df = pd.merge(df1, df2, on="Name", how="inner")
df
## Doing changes in df
df.rename(columns={"Name": "Student_Name"})
df.iloc[0, 0] = "Virat"
df
# ## Data Cleaning
dfs = pd.Series(["Lakshit", "18", "www.lakshit.io"])
dfs.str.capitalize()
dfs.str.islower()
dfs.str.lower()
dfs.str.isupper()
dfs.str.upper()
dfs.str.isnumeric()
dfs.str.swapcase()
dfs.str.len()
dfs.str.cat(sep="_") # >> Lakshit_18_www.lakshit.io
dfs.str.replace("www.", "yoyo.")
dfs.str.repeat(2)
dfs.str.count("w") # occurence of a letter
# ## Applying funtions to tables
# - `df.pipe(func_name)` it'll apply to the whole table.
# - `df.apply(func_name)` it'll apply to each column.
num = np.random.rand(10, 4)
dfNum = pd.DataFrame(num)
print(dfNum, "\n\n")
def add(a, b):
return a + b
dfNum.pipe(type) # >> pandas.core.frame.DataFrame
# PIPE
print(dfNum.pipe(add, 10), "\n\n")
# Apply
dfNum.apply(np.mean)
import os
os.getcwd() # to get the current working directiory
# os.chdir("dir_name") # to change the current working directiory
df.dtypes
# ## Importing csv file
df = pd.read_csv(
"sample_data/california_housing_test.csv",
header=0,
skiprows=[1, 2, 3, 4, 5],
names=[
"Long",
"Lat",
"house_med_age",
"Rooms",
"bedrooms",
"population_",
"household",
"med_inc",
"med_val",
],
)
# - here header = 0 is by default you can change header toany other row by putting the row number.
# - here skiprows will skip the rows which are specified
# - here names is used to give custom header name
# - put `header = None` to remove header
df
df = pd.read_csv(
"sample_data/california_housing_test.csv",
header=None,
prefix="Column: ",
na_values=["na value"],
verbose=True,
)
# - here `prefix = "column: "` is used to add any prefix to the column names i.e Column: 1
# - `na_values=['na_value']` is used to fill replace the na value with na_value
# - We can also use `sep=";"` if the seperator is not ',' we have to specify the seperator
# - `dtype={"Column: 0": 'int'}` if you want to change the data type of a column then specify it here.
# - `verbose=True` is used to get the time it takes to load the file
print(df)
df.dtypes
# ## Reading CSV Data from URL
dfs = pd.read_csv(
"http://winterolympicsmedals.com/medals.csv", nrows=10, usecols=[0, 1, 5]
)
dfs.shape
dfs.to_csv("new_.medals.csv", index=None)
# - `skipfooter=11` will remove last 11 rows from table
# - `nrows=10` will only show the first 10 rows
# - `usecols=[0,1,5]` will only return the columns which are present at the given index
# - `dfs.to_csv("new_medals.csv", index=None)` to save the file into csv format
# - `pd.read_excel(filename)` to read the excel file (check documentation for more)
# ## Data Visualiztion using Pandas
data = np.random.rand(100, 4)
data = pd.DataFrame(data, columns=["f", "s", "t", "fr"])
data["f"].plot.box(vert=False)
# - green line means 50% data is near 0.4
# - two ends are min and max values in dataframe
# - `vert = False` means we don't want to show data vertically
data["f"].plot()
data["f"].plot.bar()
# - `data['f'].plot.barh()` to show data vertically
data["f"].plot.hist()
# - `data['f'].plot.hist(bins=10)` here bins refers to the no. of blocks
data["f"].plot.area()
data.plot.scatter(x="f", y="s")
data["f"].plot.pie()
| false | 0 | 2,977 | 0 | 2,977 | 2,977 |
||
69242822
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from fastai import *
from fastai.vision import *
from fastai.vision.all import *
import imageio
import os
import cv2
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from fastai import *
from fastai.vision import *
from fastai.vision.all import *
import imageio
import os
import matplotlib.pyplot as plt
print(os.listdir("../input"))
root = Path("../input")
train_path = Path("train")
rseed = 7
val_size = 0.05
df = pd.read_csv("../input/clothing-single-channel/fashion-mnist_train.csv")
df.head()
df.shape
df_x = df.loc[:, "pixel1":"pixel784"]
df_x.head()
df_y = df.loc[:, "label"]
df_y.shape
df_y.head()
from sklearn.model_selection import train_test_split
X_train, X_val, y_train, y_val = train_test_split(
df_x, df_y, test_size=0.15, random_state=0
)
X_train = np.array(X_train).reshape(-1, 28, 28)
X_train = np.stack((X_train,) * 3, axis=-1)
y_train = np.array(y_train)
print(X_train.shape)
print(y_train.shape)
X_val = np.array(X_val).reshape(-1, 28, 28)
X_val = np.stack((X_val,) * 3, axis=-1)
y_val = np.array(y_val)
print(X_val.shape)
print(y_val.shape)
def save_imgs(path: Path, data, labels):
path.mkdir(parents=True, exist_ok=True)
for label in np.unique(labels):
(path / str(label)).mkdir(parents=True, exist_ok=True)
for i in range(len(data)):
if len(labels) != 0:
imageio.imsave(
str(path / str(labels[i]) / (str(i) + ".jpg")),
Image.fromarray((data[i]).astype(np.uint8))
.resize((28, 28))
.convert("RGB"),
)
else:
imageio.imsave(
str(path / (str(i) + ".jpg")),
Image.fromarray((data[i]).astype(np.uint8))
.resize((28, 28))
.convert("RGB"),
)
X_train[0].shape
save_imgs(Path("/data/train"), X_train, y_train)
save_imgs(Path("/data/valid"), X_val, y_val)
Path("../input/").ls()
data = ImageDataLoaders.from_folder("/data/")
data.valid.show_batch(max_n=4, nrows=1)
data.train.show_batch(max_n=4, nrows=1)
dls = ImageDataLoaders.from_folder("/data/")
learn = cnn_learner(
dls, resnet18, pretrained=True, loss_func=F.cross_entropy, metrics=accuracy
)
learn.fine_tune(1)
load_image("/data/train/1/1806.jpg")
def get_img(data):
t1 = data.reshape(28, 28) / 255
t1 = np.stack([t1] * 3, axis=0)
img = Image(FloatTensor(t1))
return img
learn.export(Path("/kaggle/working/export.pkl"))
path = Path("/kaggle/working/")
path.ls(file_exts=".pkl")
files = Path("/data/train/1")
files.ls()
path = Path()
learn_inf = load_learner("/kaggle/working/export.pkl")
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242822.ipynb
| null | null |
[{"Id": 69242822, "ScriptId": 12888224, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4691102, "CreationDate": "07/28/2021 14:26:43", "VersionNumber": 12.0, "Title": "FastAI - Object recognition", "EvaluationDate": "07/28/2021", "IsChange": false, "TotalLines": 124.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 124.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from fastai import *
from fastai.vision import *
from fastai.vision.all import *
import imageio
import os
import cv2
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from fastai import *
from fastai.vision import *
from fastai.vision.all import *
import imageio
import os
import matplotlib.pyplot as plt
print(os.listdir("../input"))
root = Path("../input")
train_path = Path("train")
rseed = 7
val_size = 0.05
df = pd.read_csv("../input/clothing-single-channel/fashion-mnist_train.csv")
df.head()
df.shape
df_x = df.loc[:, "pixel1":"pixel784"]
df_x.head()
df_y = df.loc[:, "label"]
df_y.shape
df_y.head()
from sklearn.model_selection import train_test_split
X_train, X_val, y_train, y_val = train_test_split(
df_x, df_y, test_size=0.15, random_state=0
)
X_train = np.array(X_train).reshape(-1, 28, 28)
X_train = np.stack((X_train,) * 3, axis=-1)
y_train = np.array(y_train)
print(X_train.shape)
print(y_train.shape)
X_val = np.array(X_val).reshape(-1, 28, 28)
X_val = np.stack((X_val,) * 3, axis=-1)
y_val = np.array(y_val)
print(X_val.shape)
print(y_val.shape)
def save_imgs(path: Path, data, labels):
path.mkdir(parents=True, exist_ok=True)
for label in np.unique(labels):
(path / str(label)).mkdir(parents=True, exist_ok=True)
for i in range(len(data)):
if len(labels) != 0:
imageio.imsave(
str(path / str(labels[i]) / (str(i) + ".jpg")),
Image.fromarray((data[i]).astype(np.uint8))
.resize((28, 28))
.convert("RGB"),
)
else:
imageio.imsave(
str(path / (str(i) + ".jpg")),
Image.fromarray((data[i]).astype(np.uint8))
.resize((28, 28))
.convert("RGB"),
)
X_train[0].shape
save_imgs(Path("/data/train"), X_train, y_train)
save_imgs(Path("/data/valid"), X_val, y_val)
Path("../input/").ls()
data = ImageDataLoaders.from_folder("/data/")
data.valid.show_batch(max_n=4, nrows=1)
data.train.show_batch(max_n=4, nrows=1)
dls = ImageDataLoaders.from_folder("/data/")
learn = cnn_learner(
dls, resnet18, pretrained=True, loss_func=F.cross_entropy, metrics=accuracy
)
learn.fine_tune(1)
load_image("/data/train/1/1806.jpg")
def get_img(data):
t1 = data.reshape(28, 28) / 255
t1 = np.stack([t1] * 3, axis=0)
img = Image(FloatTensor(t1))
return img
learn.export(Path("/kaggle/working/export.pkl"))
path = Path("/kaggle/working/")
path.ls(file_exts=".pkl")
files = Path("/data/train/1")
files.ls()
path = Path()
learn_inf = load_learner("/kaggle/working/export.pkl")
| false | 0 | 1,147 | 0 | 1,147 | 1,147 |
||
69242423
|
<jupyter_start><jupyter_text>Diabetes Dataset
### Context
This dataset is originally from the National Institute of Diabetes and Digestive and Kidney Diseases. The objective is to predict based on diagnostic measurements whether a patient has diabetes.
### Content
Several constraints were placed on the selection of these instances from a larger database. In particular, all patients here are females at least 21 years old of Pima Indian heritage.
- Pregnancies: Number of times pregnant
- Glucose: Plasma glucose concentration a 2 hours in an oral glucose tolerance test
- BloodPressure: Diastolic blood pressure (mm Hg)
- SkinThickness: Triceps skin fold thickness (mm)
- Insulin: 2-Hour serum insulin (mu U/ml)
- BMI: Body mass index (weight in kg/(height in m)^2)
- DiabetesPedigreeFunction: Diabetes pedigree function
- Age: Age (years)
- Outcome: Class variable (0 or 1)
#### Sources:
(a) Original owners: National Institute of Diabetes and Digestive and
Kidney Diseases
(b) Donor of database: Vincent Sigillito ([email protected])
Research Center, RMI Group Leader
Applied Physics Laboratory
The Johns Hopkins University
Johns Hopkins Road
Laurel, MD 20707
(301) 953-6231
(c) Date received: 9 May 1990
#### Past Usage:
1. Smith,~J.~W., Everhart,~J.~E., Dickson,~W.~C., Knowler,~W.~C., \&
Johannes,~R.~S. (1988). Using the ADAP learning algorithm to forecast
the onset of diabetes mellitus. In {\it Proceedings of the Symposium
on Computer Applications and Medical Care} (pp. 261--265). IEEE
Computer Society Press.
The diagnostic, binary-valued variable investigated is whether the
patient shows signs of diabetes according to World Health Organization
criteria (i.e., if the 2 hour post-load plasma glucose was at least
200 mg/dl at any survey examination or if found during routine medical
care). The population lives near Phoenix, Arizona, USA.
Results: Their ADAP algorithm makes a real-valued prediction between
0 and 1. This was transformed into a binary decision using a cutoff of
0.448. Using 576 training instances, the sensitivity and specificity
of their algorithm was 76% on the remaining 192 instances.
#### Relevant Information:
Several constraints were placed on the selection of these instances from
a larger database. In particular, all patients here are females at
least 21 years old of Pima Indian heritage. ADAP is an adaptive learning
routine that generates and executes digital analogs of perceptron-like
devices. It is a unique algorithm; see the paper for details.
#### Number of Instances: 768
#### Number of Attributes: 8 plus class
#### For Each Attribute: (all numeric-valued)
1. Number of times pregnant
2. Plasma glucose concentration a 2 hours in an oral glucose tolerance test
3. Diastolic blood pressure (mm Hg)
4. Triceps skin fold thickness (mm)
5. 2-Hour serum insulin (mu U/ml)
6. Body mass index (weight in kg/(height in m)^2)
7. Diabetes pedigree function
8. Age (years)
9. Class variable (0 or 1)
#### Missing Attribute Values: Yes
#### Class Distribution: (class value 1 is interpreted as "tested positive for
diabetes")
Kaggle dataset identifier: diabetes-data-set
<jupyter_code>import pandas as pd
df = pd.read_csv('diabetes-data-set/diabetes.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Pregnancies 768 non-null int64
1 Glucose 768 non-null int64
2 BloodPressure 768 non-null int64
3 SkinThickness 768 non-null int64
4 Insulin 768 non-null int64
5 BMI 768 non-null float64
6 DiabetesPedigreeFunction 768 non-null float64
7 Age 768 non-null int64
8 Outcome 768 non-null int64
dtypes: float64(2), int64(7)
memory usage: 54.1 KB
<jupyter_text>Examples:
{
"Pregnancies": 6.0,
"Glucose": 148.0,
"BloodPressure": 72.0,
"SkinThickness": 35.0,
"Insulin": 0.0,
"BMI": 33.6,
"DiabetesPedigreeFunction": 0.627,
"Age": 50.0,
"Outcome": 1.0
}
{
"Pregnancies": 1.0,
"Glucose": 85.0,
"BloodPressure": 66.0,
"SkinThickness": 29.0,
"Insulin": 0.0,
"BMI": 26.6,
"DiabetesPedigreeFunction": 0.35100000000000003,
"Age": 31.0,
"Outcome": 0.0
}
{
"Pregnancies": 8.0,
"Glucose": 183.0,
"BloodPressure": 64.0,
"SkinThickness": 0.0,
"Insulin": 0.0,
"BMI": 23.3,
"DiabetesPedigreeFunction": 0.672,
"Age": 32.0,
"Outcome": 1.0
}
{
"Pregnancies": 1.0,
"Glucose": 89.0,
"BloodPressure": 66.0,
"SkinThickness": 23.0,
"Insulin": 94.0,
"BMI": 28.1,
"DiabetesPedigreeFunction": 0.167,
"Age": 21.0,
"Outcome": 0.0
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import pickle
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
confusion_matrix,
classification_report,
plot_roc_curve,
)
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import GradientBoostingClassifier
from catboost import CatBoostClassifier
from lightgbm import LGBMClassifier
from sklearn.svm import SVC
import warnings
warnings.simplefilter(action="ignore")
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 170)
pd.set_option("display.max_rows", 20)
pd.set_option("display.float_format", lambda x: "%.3f" % x)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Dataset and Story
# **Business Problem**
# Can you develop a machine learning model that can predict whether people have diabetes when their characteristics are specified?
# The dataset is part of the large dataset held at the National Institutes of Diabetes-Digestive-Kidney Diseases in the USA. Persons aged 21 and over living in Phoenix, the 5th largest city in the State of Arizona in the USA. Data used for diabetes research on Pima Indian women. It consists of 768 observations and 8 numerical independent variables. The target variable is specified as "outcome"; 1 indicates positive diabetes test result, 0 indicates negative.
# **Variables**
# - Pregnancies: Number of pregnancies
# - Glucose: Glucose.
# - BloodPressure: Blood pressure.
# - SkinThickness: Skin Thickness
# - Insulin: Insulin.
# - BMI: Body mass index.
# - DiabetesPedigreeFunction: A function that calculates our probability of having diabetes based on our ancestry.
# - Age: Age (years)
# - Outcome: Information whether the person has diabetes or not. Have the disease (1) or not (0)
# **TASK**
# Develop diabetes prediction model by performing literature search, data preprocessing and feature engineering.
# # EDA Analysis
df = pd.read_csv("/kaggle/input/diabetes-data-set/diabetes.csv")
def check_df(dataframe, head=5):
print("##################### Shape #####################")
print(dataframe.shape)
print("##################### Types #####################")
print(dataframe.dtypes)
print("##################### Head #####################")
print(dataframe.head(head))
print("##################### Tail #####################")
print(dataframe.tail(head))
print("##################### NA #####################")
print(dataframe.isnull().sum())
print("##################### Quantiles #####################")
print(dataframe.quantile([0, 0.05, 0.50, 0.95, 0.99, 1]).T)
def cat_summary(dataframe, col_name, plot=False):
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
def num_summary(dataframe, numerical_col, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T)
if plot:
dataframe[numerical_col].hist(bins=20)
plt.xlabel(numerical_col)
plt.title(numerical_col)
plt.show()
def grab_col_names(dataframe, cat_th=10, car_th=20):
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
def target_summary_with_cat(dataframe, target, categorical_col):
print(
pd.DataFrame(
{"TARGET_MEAN": dataframe.groupby(categorical_col)[target].mean()}
),
end="\n\n\n",
)
def target_summary_with_num(dataframe, target, numerical_col):
print(dataframe.groupby(target).agg({numerical_col: "mean"}), end="\n\n\n")
def high_correlated_cols(dataframe, plot=False, corr_th=0.90):
corr = dataframe.corr()
cor_matrix = corr.abs()
upper_triangle_matrix = cor_matrix.where(
np.triu(np.ones(cor_matrix.shape), k=1).astype(np.bool)
)
drop_list = [
col
for col in upper_triangle_matrix.columns
if any(upper_triangle_matrix[col] > corr_th)
]
if plot:
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(rc={"figure.figsize": (15, 15)})
sns.heatmap(corr, cmap="RdBu")
plt.show()
return drop_list
check_df(df)
cat_cols, num_cols, cat_but_car = grab_col_names(df)
# Analysis of Categorical Variables
cat_summary(df, "Outcome")
# Analysis of Numerical Variables
for col in num_cols:
num_summary(df, col, plot=True)
# Analysis of Numerical Variables Based on Target
for col in num_cols:
target_summary_with_num(df, "Outcome", col)
# Examining Correlations
df.corr()
# Correlation Matrix
f, ax = plt.subplots(figsize=[20, 15])
sns.heatmap(df.corr(), annot=True, fmt=".2f", ax=ax, cmap="magma")
ax.set_title("Correlation Matrix", fontsize=20)
plt.show()
# Distribution of Dependent Variable
sns.countplot("Outcome", data=df)
plt.show()
# # Data Preprocessing
def outlier_thresholds(dataframe, col_name, q1=0.25, q3=0.75):
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def replace_with_thresholds(dataframe, variable):
low_limit, up_limit = outlier_thresholds(dataframe, variable)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
def check_outlier(dataframe, col_name, q1=0.25, q3=0.75):
low_limit, up_limit = outlier_thresholds(dataframe, col_name, q1, q3)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
def grab_outliers(dataframe, col_name, index=False):
low, up = outlier_thresholds(dataframe, col_name)
if (
dataframe[((dataframe[col_name] < low) | (dataframe[col_name] > up))].shape[0]
> 10
):
print(
dataframe[((dataframe[col_name] < low) | (dataframe[col_name] > up))].head()
)
else:
print(dataframe[((dataframe[col_name] < low) | (dataframe[col_name] > up))])
if index:
outlier_index = dataframe[
((dataframe[col_name] < low) | (dataframe[col_name] > up))
].index
return outlier_index
def remove_outlier(dataframe, col_name):
low_limit, up_limit = outlier_thresholds(dataframe, col_name)
df_without_outliers = dataframe[
~((dataframe[col_name] < low_limit) | (dataframe[col_name] > up_limit))
]
return df_without_outliers
def missing_values_table(dataframe, na_name=False):
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
def missing_vs_target(dataframe, target, na_columns):
temp_df = dataframe.copy()
for col in na_columns:
temp_df[col + "_NA_FLAG"] = np.where(temp_df[col].isnull(), 1, 0)
na_flags = temp_df.loc[:, temp_df.columns.str.contains("_NA_")].columns
for col in na_flags:
print(
pd.DataFrame(
{
"TARGET_MEAN": temp_df.groupby(col)[target].mean(),
"Count": temp_df.groupby(col)[target].count(),
}
),
end="\n\n\n",
)
def label_encoder(dataframe, binary_col):
labelencoder = LabelEncoder()
dataframe[binary_col] = labelencoder.fit_transform(dataframe[binary_col])
return dataframe
def one_hot_encoder(dataframe, categorical_cols, drop_first=False):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
def rare_analyser(dataframe, target, cat_cols):
for col in cat_cols:
print(col, ":", len(dataframe[col].value_counts()))
print(
pd.DataFrame(
{
"COUNT": dataframe[col].value_counts(),
"RATIO": dataframe[col].value_counts() / len(dataframe),
"TARGET_MEAN": dataframe.groupby(col)[target].mean(),
}
),
end="\n\n\n",
)
def rare_encoder(dataframe, rare_perc, cat_cols):
rare_columns = [
col
for col in cat_cols
if (dataframe[col].value_counts() / len(dataframe) < 0.01).sum() > 1
]
for col in rare_columns:
tmp = dataframe[col].value_counts() / len(dataframe)
rare_labels = tmp[tmp < rare_perc].index
dataframe[col] = np.where(
dataframe[col].isin(rare_labels), "Rare", dataframe[col]
)
return dataframe
# * It is known that variable values other than Pregnancies and Outcome cannot be 0 in a human.
# * Therefore, an action decision should be taken regarding these values. Values that are 0 can be assigned NaN.
zero_columns = [
col
for col in df.columns
if (df[col].min() == 0 and col not in ["Pregnancies", "Outcome"])
]
# We went to each of the stored variables and recorded the observation values containing 0 as 0
for col in zero_columns:
df[col] = np.where(df[col] == 0, np.nan, df[col])
# Missing Observation Query
df.isnull().sum()
na_columns = missing_values_table(df, na_name=True)
missing_vs_target(df, "Outcome", na_columns)
# Filling the Missing Observations in Categorical Variable Breakdown
def median_target(col):
temp = df[df[col].notnull()]
temp = temp[[col, "Outcome"]].groupby(["Outcome"])[[col]].median().reset_index()
return temp
for col in zero_columns:
df.loc[(df["Outcome"] == 0) & (df[col].isnull()), col] = median_target(col)[col][0]
df.loc[(df["Outcome"] == 1) & (df[col].isnull()), col] = median_target(col)[col][1]
df.isnull().sum()
# Outlier Analysis and Suppression Process
for col in df.columns:
print(col, check_outlier(df, col))
if check_outlier(df, col):
replace_with_thresholds(df, col)
check_df(df)
# # Feature Engineering
# Let's divide the age variable into categories and create a new age variable
df.loc[(df["Age"] >= 21) & (df["Age"] < 50), "NEW_AGE_CAT"] = "mature"
df.loc[(df["Age"] >= 50), "NEW_AGE_CAT"] = "senior"
# BMI below 18.5 is underweight, between 18.5 and 24.9 is normal, 30 and above is obese
df["NEW_BMI"] = pd.cut(
x=df["BMI"],
bins=[0, 18.5, 24.9, 29.9, 100],
labels=["Underweight", "Healthy", "Overweight", "Obese"],
)
# Convert glucose value to categorical variable
df["NEW_GLUCOSE"] = pd.cut(
x=df["Glucose"],
bins=[0, 140, 200, 300],
labels=["Normal", "Prediabetes", "Diabetes"],
)
# Creating a categorical variable by considering age and body mass index together
df.loc[
(df["BMI"] < 18.5) & ((df["Age"] >= 21) & (df["Age"] < 50)), "NEW_AGE_BMI_NOM"
] = "underweightmature"
df.loc[(df["BMI"] < 18.5) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"] = "underweightsenior"
df.loc[
((df["BMI"] >= 18.5) & (df["BMI"] < 25)) & ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_BMI_NOM",
] = "healthymature"
df.loc[
((df["BMI"] >= 18.5) & (df["BMI"] < 25)) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"
] = "healthysenior"
df.loc[
((df["BMI"] >= 25) & (df["BMI"] < 30)) & ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_BMI_NOM",
] = "overweightmature"
df.loc[
((df["BMI"] >= 25) & (df["BMI"] < 30)) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"
] = "overweightsenior"
df.loc[
(df["BMI"] > 18.5) & ((df["Age"] >= 21) & (df["Age"] < 50)), "NEW_AGE_BMI_NOM"
] = "obesemature"
df.loc[(df["BMI"] > 18.5) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"] = "obesesenior"
# Creating a categorical variable by considering age and glucose values together
df.loc[
(df["Glucose"] < 70) & ((df["Age"] >= 21) & (df["Age"] < 50)), "NEW_AGE_GLUCOSE_NOM"
] = "lowmature"
df.loc[(df["Glucose"] < 70) & (df["Age"] >= 50), "NEW_AGE_GLUCOSE_NOM"] = "lowsenior"
df.loc[
((df["Glucose"] >= 70) & (df["Glucose"] < 100))
& ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_GLUCOSE_NOM",
] = "normalmature"
df.loc[
((df["Glucose"] >= 70) & (df["Glucose"] < 100)) & (df["Age"] >= 50),
"NEW_AGE_GLUCOSE_NOM",
] = "normalsenior"
df.loc[
((df["Glucose"] >= 100) & (df["Glucose"] <= 125))
& ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_GLUCOSE_NOM",
] = "hiddenmature"
df.loc[
((df["Glucose"] >= 100) & (df["Glucose"] <= 125)) & (df["Age"] >= 50),
"NEW_AGE_GLUCOSE_NOM",
] = "hiddensenior"
df.loc[
(df["Glucose"] > 125) & ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_GLUCOSE_NOM",
] = "highmature"
df.loc[(df["Glucose"] > 125) & (df["Age"] >= 50), "NEW_AGE_GLUCOSE_NOM"] = "highsenior"
# Derive Categorical Variable with Insulin Value
def set_insulin(dataframe, col_name="Insulin"):
if 16 <= dataframe[col_name] <= 166:
return "Normal"
else:
return "Abnormal"
df["NEW_INSULIN_SCORE"] = df.apply(set_insulin, axis=1)
# Enlarging the columns
df.columns = [col.upper() for col in df.columns]
check_df(df)
# # Encoding
def label_encoder(dataframe, binary_col):
labelencoder = LabelEncoder()
dataframe[binary_col] = labelencoder.fit_transform(dataframe[binary_col])
return dataframe
# LABEL ENCODING
binary_cols = [
col for col in df.columns if df[col].dtypes == "O" and df[col].nunique() == 2
]
for col in binary_cols:
df = label_encoder(df, col)
df.head()
# ONE-HOT ENCODING
df = pd.get_dummies(df, drop_first=True)
df.head()
# # Modelling
y = df["OUTCOME"]
X = df.drop("OUTCOME", axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.20)
lgr = LogisticRegression(solver="liblinear")
lgr_model = lgr.fit(X_train, y_train)
# TRAIN ERROR
y_pred = lgr_model.predict(X_train)
# Accuracy
accuracy_score(y_train, y_pred)
print(classification_report(y_train, y_pred))
# TEST ERROR
y_pred = lgr_model.predict(X_test)
y_prob = lgr_model.predict_proba(X_test)[:, 1]
# Accuracy
accuracy_score(y_test, y_pred)
# Precision
precision_score(y_test, y_pred)
# Recall
recall_score(y_test, y_pred)
# F1
f1_score(y_test, y_pred)
print(classification_report(y_test, y_pred))
# ROC Curve
plot_roc_curve(lgr_model, X_test, y_test)
plt.title("ROC Curve")
plt.plot([0, 1], [0, 1], "r--")
plt.show()
# AUC
roc_auc_score(y_test, y_prob)
# Confusion Matrix
def plot_confusion_matrix(y, y_pred):
acc = round(accuracy_score(y, y_pred), 2)
cm = confusion_matrix(y, y_pred)
sns.heatmap(cm, annot=True, fmt=".0f")
plt.xlabel("y_pred")
plt.ylabel("y")
plt.title("Accuracy Score: {0}".format(acc), size=10)
plt.show()
plot_confusion_matrix(y_test, y_pred)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242423.ipynb
|
diabetes-data-set
|
mathchi
|
[{"Id": 69242423, "ScriptId": 18901460, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6947038, "CreationDate": "07/28/2021 14:21:16", "VersionNumber": 2.0, "Title": "Diabetes Prediction with ML", "EvaluationDate": "07/28/2021", "IsChange": false, "TotalLines": 435.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 435.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92171328, "KernelVersionId": 69242423, "SourceDatasetVersionId": 1400440}]
|
[{"Id": 1400440, "DatasetId": 818300, "DatasourceVersionId": 1433199, "CreatorUserId": 3650837, "LicenseName": "CC0: Public Domain", "CreationDate": "08/05/2020 21:27:01", "VersionNumber": 1.0, "Title": "Diabetes Dataset", "Slug": "diabetes-data-set", "Subtitle": "This dataset is originally from the N. Inst. of Diabetes & Diges. & Kidney Dis.", "Description": "### Context\n\nThis dataset is originally from the National Institute of Diabetes and Digestive and Kidney Diseases. The objective is to predict based on diagnostic measurements whether a patient has diabetes.\n\n\n### Content\n\nSeveral constraints were placed on the selection of these instances from a larger database. In particular, all patients here are females at least 21 years old of Pima Indian heritage.\n\n- Pregnancies: Number of times pregnant \n- Glucose: Plasma glucose concentration a 2 hours in an oral glucose tolerance test \n- BloodPressure: Diastolic blood pressure (mm Hg) \n- SkinThickness: Triceps skin fold thickness (mm) \n- Insulin: 2-Hour serum insulin (mu U/ml) \n- BMI: Body mass index (weight in kg/(height in m)^2) \n- DiabetesPedigreeFunction: Diabetes pedigree function \n- Age: Age (years) \n- Outcome: Class variable (0 or 1)\n\n#### Sources:\n (a) Original owners: National Institute of Diabetes and Digestive and\n Kidney Diseases\n (b) Donor of database: Vincent Sigillito ([email protected])\n Research Center, RMI Group Leader\n Applied Physics Laboratory\n The Johns Hopkins University\n Johns Hopkins Road\n Laurel, MD 20707\n (301) 953-6231\n (c) Date received: 9 May 1990\n\n#### Past Usage:\n 1. Smith,~J.~W., Everhart,~J.~E., Dickson,~W.~C., Knowler,~W.~C., \\&\n Johannes,~R.~S. (1988). Using the ADAP learning algorithm to forecast\n the onset of diabetes mellitus. In {\\it Proceedings of the Symposium\n on Computer Applications and Medical Care} (pp. 261--265). IEEE\n Computer Society Press.\n\n The diagnostic, binary-valued variable investigated is whether the\n patient shows signs of diabetes according to World Health Organization\n criteria (i.e., if the 2 hour post-load plasma glucose was at least \n 200 mg/dl at any survey examination or if found during routine medical\n care). The population lives near Phoenix, Arizona, USA.\n\n Results: Their ADAP algorithm makes a real-valued prediction between\n 0 and 1. This was transformed into a binary decision using a cutoff of \n 0.448. Using 576 training instances, the sensitivity and specificity\n of their algorithm was 76% on the remaining 192 instances.\n\n#### Relevant Information:\n Several constraints were placed on the selection of these instances from\n a larger database. In particular, all patients here are females at\n least 21 years old of Pima Indian heritage. ADAP is an adaptive learning\n routine that generates and executes digital analogs of perceptron-like\n devices. It is a unique algorithm; see the paper for details.\n\n#### Number of Instances: 768\n\n#### Number of Attributes: 8 plus class \n\n#### For Each Attribute: (all numeric-valued)\n 1. Number of times pregnant\n 2. Plasma glucose concentration a 2 hours in an oral glucose tolerance test\n 3. Diastolic blood pressure (mm Hg)\n 4. Triceps skin fold thickness (mm)\n 5. 2-Hour serum insulin (mu U/ml)\n 6. Body mass index (weight in kg/(height in m)^2)\n 7. Diabetes pedigree function\n 8. Age (years)\n 9. Class variable (0 or 1)\n\n#### Missing Attribute Values: Yes\n\n#### Class Distribution: (class value 1 is interpreted as \"tested positive for\n diabetes\")", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 818300, "CreatorUserId": 3650837, "OwnerUserId": 3650837.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1400440.0, "CurrentDatasourceVersionId": 1433199.0, "ForumId": 833406, "Type": 2, "CreationDate": "08/05/2020 21:27:01", "LastActivityDate": "08/05/2020", "TotalViews": 440450, "TotalDownloads": 65613, "TotalVotes": 496, "TotalKernels": 245}]
|
[{"Id": 3650837, "UserName": "mathchi", "DisplayName": "Mehmet Akturk", "RegisterDate": "09/01/2019", "PerformanceTier": 3}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import pickle
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
confusion_matrix,
classification_report,
plot_roc_curve,
)
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import GradientBoostingClassifier
from catboost import CatBoostClassifier
from lightgbm import LGBMClassifier
from sklearn.svm import SVC
import warnings
warnings.simplefilter(action="ignore")
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 170)
pd.set_option("display.max_rows", 20)
pd.set_option("display.float_format", lambda x: "%.3f" % x)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Dataset and Story
# **Business Problem**
# Can you develop a machine learning model that can predict whether people have diabetes when their characteristics are specified?
# The dataset is part of the large dataset held at the National Institutes of Diabetes-Digestive-Kidney Diseases in the USA. Persons aged 21 and over living in Phoenix, the 5th largest city in the State of Arizona in the USA. Data used for diabetes research on Pima Indian women. It consists of 768 observations and 8 numerical independent variables. The target variable is specified as "outcome"; 1 indicates positive diabetes test result, 0 indicates negative.
# **Variables**
# - Pregnancies: Number of pregnancies
# - Glucose: Glucose.
# - BloodPressure: Blood pressure.
# - SkinThickness: Skin Thickness
# - Insulin: Insulin.
# - BMI: Body mass index.
# - DiabetesPedigreeFunction: A function that calculates our probability of having diabetes based on our ancestry.
# - Age: Age (years)
# - Outcome: Information whether the person has diabetes or not. Have the disease (1) or not (0)
# **TASK**
# Develop diabetes prediction model by performing literature search, data preprocessing and feature engineering.
# # EDA Analysis
df = pd.read_csv("/kaggle/input/diabetes-data-set/diabetes.csv")
def check_df(dataframe, head=5):
print("##################### Shape #####################")
print(dataframe.shape)
print("##################### Types #####################")
print(dataframe.dtypes)
print("##################### Head #####################")
print(dataframe.head(head))
print("##################### Tail #####################")
print(dataframe.tail(head))
print("##################### NA #####################")
print(dataframe.isnull().sum())
print("##################### Quantiles #####################")
print(dataframe.quantile([0, 0.05, 0.50, 0.95, 0.99, 1]).T)
def cat_summary(dataframe, col_name, plot=False):
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
def num_summary(dataframe, numerical_col, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T)
if plot:
dataframe[numerical_col].hist(bins=20)
plt.xlabel(numerical_col)
plt.title(numerical_col)
plt.show()
def grab_col_names(dataframe, cat_th=10, car_th=20):
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th and dataframe[col].dtypes != "O"
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th and dataframe[col].dtypes == "O"
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
def target_summary_with_cat(dataframe, target, categorical_col):
print(
pd.DataFrame(
{"TARGET_MEAN": dataframe.groupby(categorical_col)[target].mean()}
),
end="\n\n\n",
)
def target_summary_with_num(dataframe, target, numerical_col):
print(dataframe.groupby(target).agg({numerical_col: "mean"}), end="\n\n\n")
def high_correlated_cols(dataframe, plot=False, corr_th=0.90):
corr = dataframe.corr()
cor_matrix = corr.abs()
upper_triangle_matrix = cor_matrix.where(
np.triu(np.ones(cor_matrix.shape), k=1).astype(np.bool)
)
drop_list = [
col
for col in upper_triangle_matrix.columns
if any(upper_triangle_matrix[col] > corr_th)
]
if plot:
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(rc={"figure.figsize": (15, 15)})
sns.heatmap(corr, cmap="RdBu")
plt.show()
return drop_list
check_df(df)
cat_cols, num_cols, cat_but_car = grab_col_names(df)
# Analysis of Categorical Variables
cat_summary(df, "Outcome")
# Analysis of Numerical Variables
for col in num_cols:
num_summary(df, col, plot=True)
# Analysis of Numerical Variables Based on Target
for col in num_cols:
target_summary_with_num(df, "Outcome", col)
# Examining Correlations
df.corr()
# Correlation Matrix
f, ax = plt.subplots(figsize=[20, 15])
sns.heatmap(df.corr(), annot=True, fmt=".2f", ax=ax, cmap="magma")
ax.set_title("Correlation Matrix", fontsize=20)
plt.show()
# Distribution of Dependent Variable
sns.countplot("Outcome", data=df)
plt.show()
# # Data Preprocessing
def outlier_thresholds(dataframe, col_name, q1=0.25, q3=0.75):
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def replace_with_thresholds(dataframe, variable):
low_limit, up_limit = outlier_thresholds(dataframe, variable)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
def check_outlier(dataframe, col_name, q1=0.25, q3=0.75):
low_limit, up_limit = outlier_thresholds(dataframe, col_name, q1, q3)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
def grab_outliers(dataframe, col_name, index=False):
low, up = outlier_thresholds(dataframe, col_name)
if (
dataframe[((dataframe[col_name] < low) | (dataframe[col_name] > up))].shape[0]
> 10
):
print(
dataframe[((dataframe[col_name] < low) | (dataframe[col_name] > up))].head()
)
else:
print(dataframe[((dataframe[col_name] < low) | (dataframe[col_name] > up))])
if index:
outlier_index = dataframe[
((dataframe[col_name] < low) | (dataframe[col_name] > up))
].index
return outlier_index
def remove_outlier(dataframe, col_name):
low_limit, up_limit = outlier_thresholds(dataframe, col_name)
df_without_outliers = dataframe[
~((dataframe[col_name] < low_limit) | (dataframe[col_name] > up_limit))
]
return df_without_outliers
def missing_values_table(dataframe, na_name=False):
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
def missing_vs_target(dataframe, target, na_columns):
temp_df = dataframe.copy()
for col in na_columns:
temp_df[col + "_NA_FLAG"] = np.where(temp_df[col].isnull(), 1, 0)
na_flags = temp_df.loc[:, temp_df.columns.str.contains("_NA_")].columns
for col in na_flags:
print(
pd.DataFrame(
{
"TARGET_MEAN": temp_df.groupby(col)[target].mean(),
"Count": temp_df.groupby(col)[target].count(),
}
),
end="\n\n\n",
)
def label_encoder(dataframe, binary_col):
labelencoder = LabelEncoder()
dataframe[binary_col] = labelencoder.fit_transform(dataframe[binary_col])
return dataframe
def one_hot_encoder(dataframe, categorical_cols, drop_first=False):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
def rare_analyser(dataframe, target, cat_cols):
for col in cat_cols:
print(col, ":", len(dataframe[col].value_counts()))
print(
pd.DataFrame(
{
"COUNT": dataframe[col].value_counts(),
"RATIO": dataframe[col].value_counts() / len(dataframe),
"TARGET_MEAN": dataframe.groupby(col)[target].mean(),
}
),
end="\n\n\n",
)
def rare_encoder(dataframe, rare_perc, cat_cols):
rare_columns = [
col
for col in cat_cols
if (dataframe[col].value_counts() / len(dataframe) < 0.01).sum() > 1
]
for col in rare_columns:
tmp = dataframe[col].value_counts() / len(dataframe)
rare_labels = tmp[tmp < rare_perc].index
dataframe[col] = np.where(
dataframe[col].isin(rare_labels), "Rare", dataframe[col]
)
return dataframe
# * It is known that variable values other than Pregnancies and Outcome cannot be 0 in a human.
# * Therefore, an action decision should be taken regarding these values. Values that are 0 can be assigned NaN.
zero_columns = [
col
for col in df.columns
if (df[col].min() == 0 and col not in ["Pregnancies", "Outcome"])
]
# We went to each of the stored variables and recorded the observation values containing 0 as 0
for col in zero_columns:
df[col] = np.where(df[col] == 0, np.nan, df[col])
# Missing Observation Query
df.isnull().sum()
na_columns = missing_values_table(df, na_name=True)
missing_vs_target(df, "Outcome", na_columns)
# Filling the Missing Observations in Categorical Variable Breakdown
def median_target(col):
temp = df[df[col].notnull()]
temp = temp[[col, "Outcome"]].groupby(["Outcome"])[[col]].median().reset_index()
return temp
for col in zero_columns:
df.loc[(df["Outcome"] == 0) & (df[col].isnull()), col] = median_target(col)[col][0]
df.loc[(df["Outcome"] == 1) & (df[col].isnull()), col] = median_target(col)[col][1]
df.isnull().sum()
# Outlier Analysis and Suppression Process
for col in df.columns:
print(col, check_outlier(df, col))
if check_outlier(df, col):
replace_with_thresholds(df, col)
check_df(df)
# # Feature Engineering
# Let's divide the age variable into categories and create a new age variable
df.loc[(df["Age"] >= 21) & (df["Age"] < 50), "NEW_AGE_CAT"] = "mature"
df.loc[(df["Age"] >= 50), "NEW_AGE_CAT"] = "senior"
# BMI below 18.5 is underweight, between 18.5 and 24.9 is normal, 30 and above is obese
df["NEW_BMI"] = pd.cut(
x=df["BMI"],
bins=[0, 18.5, 24.9, 29.9, 100],
labels=["Underweight", "Healthy", "Overweight", "Obese"],
)
# Convert glucose value to categorical variable
df["NEW_GLUCOSE"] = pd.cut(
x=df["Glucose"],
bins=[0, 140, 200, 300],
labels=["Normal", "Prediabetes", "Diabetes"],
)
# Creating a categorical variable by considering age and body mass index together
df.loc[
(df["BMI"] < 18.5) & ((df["Age"] >= 21) & (df["Age"] < 50)), "NEW_AGE_BMI_NOM"
] = "underweightmature"
df.loc[(df["BMI"] < 18.5) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"] = "underweightsenior"
df.loc[
((df["BMI"] >= 18.5) & (df["BMI"] < 25)) & ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_BMI_NOM",
] = "healthymature"
df.loc[
((df["BMI"] >= 18.5) & (df["BMI"] < 25)) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"
] = "healthysenior"
df.loc[
((df["BMI"] >= 25) & (df["BMI"] < 30)) & ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_BMI_NOM",
] = "overweightmature"
df.loc[
((df["BMI"] >= 25) & (df["BMI"] < 30)) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"
] = "overweightsenior"
df.loc[
(df["BMI"] > 18.5) & ((df["Age"] >= 21) & (df["Age"] < 50)), "NEW_AGE_BMI_NOM"
] = "obesemature"
df.loc[(df["BMI"] > 18.5) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"] = "obesesenior"
# Creating a categorical variable by considering age and glucose values together
df.loc[
(df["Glucose"] < 70) & ((df["Age"] >= 21) & (df["Age"] < 50)), "NEW_AGE_GLUCOSE_NOM"
] = "lowmature"
df.loc[(df["Glucose"] < 70) & (df["Age"] >= 50), "NEW_AGE_GLUCOSE_NOM"] = "lowsenior"
df.loc[
((df["Glucose"] >= 70) & (df["Glucose"] < 100))
& ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_GLUCOSE_NOM",
] = "normalmature"
df.loc[
((df["Glucose"] >= 70) & (df["Glucose"] < 100)) & (df["Age"] >= 50),
"NEW_AGE_GLUCOSE_NOM",
] = "normalsenior"
df.loc[
((df["Glucose"] >= 100) & (df["Glucose"] <= 125))
& ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_GLUCOSE_NOM",
] = "hiddenmature"
df.loc[
((df["Glucose"] >= 100) & (df["Glucose"] <= 125)) & (df["Age"] >= 50),
"NEW_AGE_GLUCOSE_NOM",
] = "hiddensenior"
df.loc[
(df["Glucose"] > 125) & ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_GLUCOSE_NOM",
] = "highmature"
df.loc[(df["Glucose"] > 125) & (df["Age"] >= 50), "NEW_AGE_GLUCOSE_NOM"] = "highsenior"
# Derive Categorical Variable with Insulin Value
def set_insulin(dataframe, col_name="Insulin"):
if 16 <= dataframe[col_name] <= 166:
return "Normal"
else:
return "Abnormal"
df["NEW_INSULIN_SCORE"] = df.apply(set_insulin, axis=1)
# Enlarging the columns
df.columns = [col.upper() for col in df.columns]
check_df(df)
# # Encoding
def label_encoder(dataframe, binary_col):
labelencoder = LabelEncoder()
dataframe[binary_col] = labelencoder.fit_transform(dataframe[binary_col])
return dataframe
# LABEL ENCODING
binary_cols = [
col for col in df.columns if df[col].dtypes == "O" and df[col].nunique() == 2
]
for col in binary_cols:
df = label_encoder(df, col)
df.head()
# ONE-HOT ENCODING
df = pd.get_dummies(df, drop_first=True)
df.head()
# # Modelling
y = df["OUTCOME"]
X = df.drop("OUTCOME", axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.20)
lgr = LogisticRegression(solver="liblinear")
lgr_model = lgr.fit(X_train, y_train)
# TRAIN ERROR
y_pred = lgr_model.predict(X_train)
# Accuracy
accuracy_score(y_train, y_pred)
print(classification_report(y_train, y_pred))
# TEST ERROR
y_pred = lgr_model.predict(X_test)
y_prob = lgr_model.predict_proba(X_test)[:, 1]
# Accuracy
accuracy_score(y_test, y_pred)
# Precision
precision_score(y_test, y_pred)
# Recall
recall_score(y_test, y_pred)
# F1
f1_score(y_test, y_pred)
print(classification_report(y_test, y_pred))
# ROC Curve
plot_roc_curve(lgr_model, X_test, y_test)
plt.title("ROC Curve")
plt.plot([0, 1], [0, 1], "r--")
plt.show()
# AUC
roc_auc_score(y_test, y_prob)
# Confusion Matrix
def plot_confusion_matrix(y, y_pred):
acc = round(accuracy_score(y, y_pred), 2)
cm = confusion_matrix(y, y_pred)
sns.heatmap(cm, annot=True, fmt=".0f")
plt.xlabel("y_pred")
plt.ylabel("y")
plt.title("Accuracy Score: {0}".format(acc), size=10)
plt.show()
plot_confusion_matrix(y_test, y_pred)
|
[{"diabetes-data-set/diabetes.csv": {"column_names": "[\"Pregnancies\", \"Glucose\", \"BloodPressure\", \"SkinThickness\", \"Insulin\", \"BMI\", \"DiabetesPedigreeFunction\", \"Age\", \"Outcome\"]", "column_data_types": "{\"Pregnancies\": \"int64\", \"Glucose\": \"int64\", \"BloodPressure\": \"int64\", \"SkinThickness\": \"int64\", \"Insulin\": \"int64\", \"BMI\": \"float64\", \"DiabetesPedigreeFunction\": \"float64\", \"Age\": \"int64\", \"Outcome\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 768 entries, 0 to 767\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Pregnancies 768 non-null int64 \n 1 Glucose 768 non-null int64 \n 2 BloodPressure 768 non-null int64 \n 3 SkinThickness 768 non-null int64 \n 4 Insulin 768 non-null int64 \n 5 BMI 768 non-null float64\n 6 DiabetesPedigreeFunction 768 non-null float64\n 7 Age 768 non-null int64 \n 8 Outcome 768 non-null int64 \ndtypes: float64(2), int64(7)\nmemory usage: 54.1 KB\n", "summary": "{\"Pregnancies\": {\"count\": 768.0, \"mean\": 3.8450520833333335, \"std\": 3.3695780626988694, \"min\": 0.0, \"25%\": 1.0, \"50%\": 3.0, \"75%\": 6.0, \"max\": 17.0}, \"Glucose\": {\"count\": 768.0, \"mean\": 120.89453125, \"std\": 31.97261819513622, \"min\": 0.0, \"25%\": 99.0, \"50%\": 117.0, \"75%\": 140.25, \"max\": 199.0}, \"BloodPressure\": {\"count\": 768.0, \"mean\": 69.10546875, \"std\": 19.355807170644777, \"min\": 0.0, \"25%\": 62.0, \"50%\": 72.0, \"75%\": 80.0, \"max\": 122.0}, \"SkinThickness\": {\"count\": 768.0, \"mean\": 20.536458333333332, \"std\": 15.952217567727637, \"min\": 0.0, \"25%\": 0.0, \"50%\": 23.0, \"75%\": 32.0, \"max\": 99.0}, \"Insulin\": {\"count\": 768.0, \"mean\": 79.79947916666667, \"std\": 115.24400235133817, \"min\": 0.0, \"25%\": 0.0, \"50%\": 30.5, \"75%\": 127.25, \"max\": 846.0}, \"BMI\": {\"count\": 768.0, \"mean\": 31.992578124999998, \"std\": 7.884160320375446, \"min\": 0.0, \"25%\": 27.3, \"50%\": 32.0, \"75%\": 36.6, \"max\": 67.1}, \"DiabetesPedigreeFunction\": {\"count\": 768.0, \"mean\": 0.47187630208333325, \"std\": 0.3313285950127749, \"min\": 0.078, \"25%\": 0.24375, \"50%\": 0.3725, \"75%\": 0.62625, \"max\": 2.42}, \"Age\": {\"count\": 768.0, \"mean\": 33.240885416666664, \"std\": 11.760231540678685, \"min\": 21.0, \"25%\": 24.0, \"50%\": 29.0, \"75%\": 41.0, \"max\": 81.0}, \"Outcome\": {\"count\": 768.0, \"mean\": 0.3489583333333333, \"std\": 0.47695137724279896, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}}", "examples": "{\"Pregnancies\":{\"0\":6,\"1\":1,\"2\":8,\"3\":1},\"Glucose\":{\"0\":148,\"1\":85,\"2\":183,\"3\":89},\"BloodPressure\":{\"0\":72,\"1\":66,\"2\":64,\"3\":66},\"SkinThickness\":{\"0\":35,\"1\":29,\"2\":0,\"3\":23},\"Insulin\":{\"0\":0,\"1\":0,\"2\":0,\"3\":94},\"BMI\":{\"0\":33.6,\"1\":26.6,\"2\":23.3,\"3\":28.1},\"DiabetesPedigreeFunction\":{\"0\":0.627,\"1\":0.351,\"2\":0.672,\"3\":0.167},\"Age\":{\"0\":50,\"1\":31,\"2\":32,\"3\":21},\"Outcome\":{\"0\":1,\"1\":0,\"2\":1,\"3\":0}}"}}]
| true | 1 |
<start_data_description><data_path>diabetes-data-set/diabetes.csv:
<column_names>
['Pregnancies', 'Glucose', 'BloodPressure', 'SkinThickness', 'Insulin', 'BMI', 'DiabetesPedigreeFunction', 'Age', 'Outcome']
<column_types>
{'Pregnancies': 'int64', 'Glucose': 'int64', 'BloodPressure': 'int64', 'SkinThickness': 'int64', 'Insulin': 'int64', 'BMI': 'float64', 'DiabetesPedigreeFunction': 'float64', 'Age': 'int64', 'Outcome': 'int64'}
<dataframe_Summary>
{'Pregnancies': {'count': 768.0, 'mean': 3.8450520833333335, 'std': 3.3695780626988694, 'min': 0.0, '25%': 1.0, '50%': 3.0, '75%': 6.0, 'max': 17.0}, 'Glucose': {'count': 768.0, 'mean': 120.89453125, 'std': 31.97261819513622, 'min': 0.0, '25%': 99.0, '50%': 117.0, '75%': 140.25, 'max': 199.0}, 'BloodPressure': {'count': 768.0, 'mean': 69.10546875, 'std': 19.355807170644777, 'min': 0.0, '25%': 62.0, '50%': 72.0, '75%': 80.0, 'max': 122.0}, 'SkinThickness': {'count': 768.0, 'mean': 20.536458333333332, 'std': 15.952217567727637, 'min': 0.0, '25%': 0.0, '50%': 23.0, '75%': 32.0, 'max': 99.0}, 'Insulin': {'count': 768.0, 'mean': 79.79947916666667, 'std': 115.24400235133817, 'min': 0.0, '25%': 0.0, '50%': 30.5, '75%': 127.25, 'max': 846.0}, 'BMI': {'count': 768.0, 'mean': 31.992578124999998, 'std': 7.884160320375446, 'min': 0.0, '25%': 27.3, '50%': 32.0, '75%': 36.6, 'max': 67.1}, 'DiabetesPedigreeFunction': {'count': 768.0, 'mean': 0.47187630208333325, 'std': 0.3313285950127749, 'min': 0.078, '25%': 0.24375, '50%': 0.3725, '75%': 0.62625, 'max': 2.42}, 'Age': {'count': 768.0, 'mean': 33.240885416666664, 'std': 11.760231540678685, 'min': 21.0, '25%': 24.0, '50%': 29.0, '75%': 41.0, 'max': 81.0}, 'Outcome': {'count': 768.0, 'mean': 0.3489583333333333, 'std': 0.47695137724279896, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}}
<dataframe_info>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Pregnancies 768 non-null int64
1 Glucose 768 non-null int64
2 BloodPressure 768 non-null int64
3 SkinThickness 768 non-null int64
4 Insulin 768 non-null int64
5 BMI 768 non-null float64
6 DiabetesPedigreeFunction 768 non-null float64
7 Age 768 non-null int64
8 Outcome 768 non-null int64
dtypes: float64(2), int64(7)
memory usage: 54.1 KB
<some_examples>
{'Pregnancies': {'0': 6, '1': 1, '2': 8, '3': 1}, 'Glucose': {'0': 148, '1': 85, '2': 183, '3': 89}, 'BloodPressure': {'0': 72, '1': 66, '2': 64, '3': 66}, 'SkinThickness': {'0': 35, '1': 29, '2': 0, '3': 23}, 'Insulin': {'0': 0, '1': 0, '2': 0, '3': 94}, 'BMI': {'0': 33.6, '1': 26.6, '2': 23.3, '3': 28.1}, 'DiabetesPedigreeFunction': {'0': 0.627, '1': 0.351, '2': 0.672, '3': 0.167}, 'Age': {'0': 50, '1': 31, '2': 32, '3': 21}, 'Outcome': {'0': 1, '1': 0, '2': 1, '3': 0}}
<end_description>
| 5,749 | 0 | 7,456 | 5,749 |
69242485
|
<jupyter_start><jupyter_text>House Price Prediction Challenge
# House Price Prediction Challenge
## Overview
Welcome to the House Price Prediction Challenge, you will test your regression skills by designing an algorithm to accurately predict the house prices in India. Accurately predicting house prices can be a daunting task. The buyers are just not concerned about the size(square feet) of the house and there are various other factors that play a key role to decide the price of a house/property. It can be extremely difficult to figure out the right set of attributes that are contributing to understanding the buyer's behavior as such. This dataset has been collected across various property aggregators across India. In this competition, provided the 12 influencing factors your role as a data scientist is to predict the prices as accurately as possible.
Also, in this competition, you will get a lot of room for feature engineering and mastering advanced regression techniques such as Random Forest, Deep Neural Nets, and various other ensembling techniques.
## Data Description:
Train.csv - 29451 rows x 12 columns
Test.csv - 68720 rows x 11 columns
Sample Submission - Acceptable submission format. (.csv/.xlsx file with 68720 rows)
## Attributes Description:
|Column | Description|
| --- | --- |
|POSTED_BY| Category marking who has listed the property|
|UNDER_CONSTRUCTION | Under Construction or Not|
|RERA | Rera approved or Not|
|BHK_NO | Number of Rooms|
|BHK_OR_RK | Type of property|
|SQUARE_FT | Total area of the house in square feet|
|READY_TO_MOVE| Category marking Ready to move or Not|
|RESALE | Category marking Resale or not|
|ADDRESS | Address of the property|
|LONGITUDE | Longitude of the property|
|LATITUDE | Latitude of the property|
## ACKNOWLEDGMENT:
The dataset for this hackathon was contributed by [Devrup Banerjee](https://www.machinehack.com/user/profile/5ef0a238b7efcc325e390297) . We would like to appreciate his efforts for this contribution to the Machinehack community.
Kaggle dataset identifier: house-price-prediction-challenge
<jupyter_code>import pandas as pd
df = pd.read_csv('house-price-prediction-challenge/train.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 29451 entries, 0 to 29450
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 POSTED_BY 29451 non-null object
1 UNDER_CONSTRUCTION 29451 non-null int64
2 RERA 29451 non-null int64
3 BHK_NO. 29451 non-null int64
4 BHK_OR_RK 29451 non-null object
5 SQUARE_FT 29451 non-null float64
6 READY_TO_MOVE 29451 non-null int64
7 RESALE 29451 non-null int64
8 ADDRESS 29451 non-null object
9 LONGITUDE 29451 non-null float64
10 LATITUDE 29451 non-null float64
11 TARGET(PRICE_IN_LACS) 29451 non-null float64
dtypes: float64(4), int64(5), object(3)
memory usage: 2.7+ MB
<jupyter_text>Examples:
{
"POSTED_BY": "Owner",
"UNDER_CONSTRUCTION": 0,
"RERA": 0,
"BHK_NO.": 2,
"BHK_OR_RK": "BHK",
"SQUARE_FT": 1300.236407,
"READY_TO_MOVE": 1,
"RESALE": 1,
"ADDRESS": "Ksfc Layout,Bangalore",
"LONGITUDE": 12.96991,
"LATITUDE": 77.59796,
"TARGET(PRICE_IN_LACS)": 55.0
}
{
"POSTED_BY": "Dealer",
"UNDER_CONSTRUCTION": 0,
"RERA": 0,
"BHK_NO.": 2,
"BHK_OR_RK": "BHK",
"SQUARE_FT": 1275.0,
"READY_TO_MOVE": 1,
"RESALE": 1,
"ADDRESS": "Vishweshwara Nagar,Mysore",
"LONGITUDE": 12.274538,
"LATITUDE": 76.644605,
"TARGET(PRICE_IN_LACS)": 51.0
}
{
"POSTED_BY": "Owner",
"UNDER_CONSTRUCTION": 0,
"RERA": 0,
"BHK_NO.": 2,
"BHK_OR_RK": "BHK",
"SQUARE_FT": 933.1597222,
"READY_TO_MOVE": 1,
"RESALE": 1,
"ADDRESS": "Jigani,Bangalore",
"LONGITUDE": 12.778033,
"LATITUDE": 77.632191,
"TARGET(PRICE_IN_LACS)": 43.0
}
{
"POSTED_BY": "Owner",
"UNDER_CONSTRUCTION": 0,
"RERA": 1,
"BHK_NO.": 2,
"BHK_OR_RK": "BHK",
"SQUARE_FT": 929.9211427,
"READY_TO_MOVE": 1,
"RESALE": 1,
"ADDRESS": "Sector-1 Vaishali,Ghaziabad",
"LONGITUDE": 28.6423,
"LATITUDE": 77.3445,
"TARGET(PRICE_IN_LACS)": 62.5
}
<jupyter_script>import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/house-price-prediction-challenge/train.csv")
train.head()
train.shape
# No missing values
train.info()
# most of houses not under construction
train["UNDER_CONSTRUCTION"].value_counts()
train["READY_TO_MOVE"].value_counts()
train["RESALE"].value_counts()
train["POSTED_BY"].value_counts()
# handling categorical values , there is many ways like this two ways or OHE or LE or dummies
# train['POSTED']=train['POSTED_BY'].apply(lambda x :0 if x=='Dealer' else 1 if x=='Owner' else 2)
# train.drop("POSTED_BY",axis=1,inplace=True)
mappp = {"Dealer": 0, "Owner": 1, "Builder": 2}
train["POSTED_BY"] = train["POSTED_BY"].map(mappp)
train["BHK_OR_RK"].value_counts()
mapp = {"BHK": 0, "RK": 1}
train["BHK_OR_RK"] = train["BHK_OR_RK"].map(mapp)
train.head()
# number of rooms between 1 and 20
#
train.plot(kind="scatter", x="TARGET(PRICE_IN_LACS)", y="BHK_NO.")
# latitude between 60 and 150, doesn't highly effect
train.plot(kind="scatter", x="TARGET(PRICE_IN_LACS)", y="LATITUDE")
# longitude between 0 and 40 , doesn't highly effect but near to 40 less cost
train.plot(kind="scatter", x="TARGET(PRICE_IN_LACS)", y="LONGITUDE")
# log for area
train["AREA"] = np.log(train["SQUARE_FT"])
# price direct proportional with area
sns.scatterplot(data=train, x="TARGET(PRICE_IN_LACS)", y="AREA")
# drop column before log
train.drop("SQUARE_FT", axis=1, inplace=True)
train.describe()
# location doesn't highly effect to price
plt.figure(figsize=(15, 10))
sns.scatterplot(data=train, x="LONGITUDE", y="LATITUDE", hue="TARGET(PRICE_IN_LACS)")
# divide features to categorical and numerical
cat_features = ["POSTED_BY", "BHK_OR_RK"]
num_features = [
"UNDER_CONSTRUCTION",
"RERA",
"BHK_NO.",
"READY_TO_MOVE",
"RESALE",
"LONGITUDE",
"LATITUDE",
"AREA",
]
# area, number of rooms, price highly correlated with each other
plt.figure(figsize=(10, 10))
corr = train[cat_features + num_features + ["TARGET(PRICE_IN_LACS)"]].corr(
method="spearman"
)
sns.heatmap(corr, annot=True)
# handling outliers
outlier_percentage = {}
for feature in ["AREA", "BHK_NO.", "TARGET(PRICE_IN_LACS)"]:
tempData = train.sort_values(by=feature)[feature]
Q1, Q3 = tempData.quantile([0.25, 0.75])
IQR = Q3 - Q1
Lower_range = Q1 - (1.5 * IQR)
Upper_range = Q3 + (1.5 * IQR)
outlier_percentage[feature] = round(
(
((tempData < (Q1 - 1.5 * IQR)) | (tempData > (Q3 + 1.5 * IQR))).sum()
/ tempData.shape[0]
)
* 100,
2,
)
outlier_percentage
outlier = train[
(train[feature] > Lower_range) & (train[feature] < Upper_range)
].reset_index(drop=True)
train.drop(["ADDRESS"], axis=1, inplace=True)
X = train.drop(["TARGET(PRICE_IN_LACS)"], axis=1)
y = train["TARGET(PRICE_IN_LACS)"]
# feature selection
# under constrauction and ready to move exactly the same , location has small effect
from sklearn.feature_selection import SelectKBest, f_regression
best = SelectKBest(score_func=f_regression, k="all")
fit = best.fit(X, y)
dfScores = pd.DataFrame(fit.scores_)
dfCol = pd.DataFrame(X.columns)
featScore = pd.concat([dfCol, dfScores], axis=1)
featScore.columns = ["Feature", "Score"]
featScore = featScore.sort_values(by="Score", ascending=False).reset_index(drop=True)
print(featScore.nlargest(10, "Score"))
# another way for feature selection
# from sklearn.ensemble import ExtraTreesRegressor
# ex=ExtraTreesRegressor()
# ex.fit(X,y)
# ex.feature_importances_
# plt.figure(figsize=(10,10))
# plt.title('Feature importances')
# feat=pd.Series(ex.feature_importances_,index=X.columns)
# feat.nlargest(10).plot(kind='barh', color="r", align="center")
# plt.tight_layout()
# plt.show()
# removing features with vif , there is another way called RFE "Recursive Feature Elimination"
from statsmodels.stats.outliers_influence import variance_inflation_factor
v = pd.DataFrame()
v["variables"] = [
feature
for feature in cat_features + num_features
if feature not in ["READY_TO_MOVE", "RESALE", "LATITUDE", "LONGITUDE"]
]
v["VIF"] = [
variance_inflation_factor(train[v["variables"]].values, i)
for i in range(len(v["variables"]))
]
print(v)
# splitting the data
from sklearn.model_selection import (
train_test_split,
cross_val_score,
RandomizedSearchCV,
)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
# feature scaling
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.fit_transform(X_test)
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.linear_model import (
LinearRegression,
SGDRegressor,
Lasso,
Ridge,
ElasticNet,
)
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import (
RandomForestRegressor,
GradientBoostingRegressor,
VotingRegressor,
)
from xgboost.sklearn import XGBRegressor
# train models
models = {
"Linear Regression": LinearRegression(),
"Lasso": Lasso(),
"Decision Tree": DecisionTreeRegressor(),
"Random Forest": RandomForestRegressor(),
"Gradient Boosting =": GradientBoostingRegressor(),
"Ridge": Ridge(),
"Stochastic Gradien Descent": SGDRegressor(),
"Elastic": ElasticNet(),
"xgb Regressor": XGBRegressor(),
}
# fit and score
def fit_score(models, X_train, X_test, y_train, y_test):
np.random.seed(42)
model_scores = {}
for name, model in models.items():
model.fit(X_train, y_train)
model_scores[name] = cross_val_score(
model, X_test, y_test, scoring="neg_mean_squared_error", cv=3
).mean()
return model_scores
# Decision tree has lowest mse
model_scores = fit_score(models, X_train, X_test, y_train, y_test)
model_scores
# voting
vot = VotingRegressor(
[
("LinearRegression", LinearRegression()),
("DecisionTrees", DecisionTreeRegressor()),
("LassoRegression", Lasso()),
("RandomForest", RandomForestRegressor()),
("ElasticNet", ElasticNet()),
("StochasticGradientDescent", SGDRegressor()),
("GrafientBoosting", GradientBoostingRegressor()),
("Ridge", Ridge()),
("xgb", XGBRegressor()),
]
)
vot.fit(X_train, y_train)
y_pred = vot.predict(X_test)
# as expected , voting has smallest mse
mean_squared_error(y_test, y_pred)
np.random.seed(42)
params = {
"criterion": ["mse", "mae"],
"max_features": ["auto", "sqrt", "log2"],
"max_depth": [2, 3, 10],
}
rs = RandomizedSearchCV(
DecisionTreeRegressor(),
param_distributions=params,
cv=3,
n_iter=30,
verbose=0,
n_jobs=-1,
)
rs.fit(X_train, y_train)
rs.best_params_
rs.best_estimator_
rs.best_score_
rs.score(X_test, y_test)
model = DecisionTreeRegressor()
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
r2_score(y_pred, y_test)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242485.ipynb
|
house-price-prediction-challenge
|
anmolkumar
|
[{"Id": 69242485, "ScriptId": 18876749, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5453017, "CreationDate": "07/28/2021 14:22:19", "VersionNumber": 1.0, "Title": "Housing price", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 266.0, "LinesInsertedFromPrevious": 266.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 1}]
|
[{"Id": 92171502, "KernelVersionId": 69242485, "SourceDatasetVersionId": 1530022}]
|
[{"Id": 1530022, "DatasetId": 902117, "DatasourceVersionId": 1564506, "CreatorUserId": 1815526, "LicenseName": "GPL 2", "CreationDate": "10/01/2020 19:37:13", "VersionNumber": 1.0, "Title": "House Price Prediction Challenge", "Slug": "house-price-prediction-challenge", "Subtitle": "Predict the house prices in India", "Description": "# House Price Prediction Challenge\n\n## Overview\n\nWelcome to the House Price Prediction Challenge, you will test your regression skills by designing an algorithm to accurately predict the house prices in India. Accurately predicting house prices can be a daunting task. The buyers are just not concerned about the size(square feet) of the house and there are various other factors that play a key role to decide the price of a house/property. It can be extremely difficult to figure out the right set of attributes that are contributing to understanding the buyer's behavior as such. This dataset has been collected across various property aggregators across India. In this competition, provided the 12 influencing factors your role as a data scientist is to predict the prices as accurately as possible.\n\nAlso, in this competition, you will get a lot of room for feature engineering and mastering advanced regression techniques such as Random Forest, Deep Neural Nets, and various other ensembling techniques. \n\n## Data Description:\nTrain.csv - 29451 rows x 12 columns\nTest.csv - 68720 rows x 11 columns\nSample Submission - Acceptable submission format. (.csv/.xlsx file with 68720 rows) \n\n## Attributes Description:\n|Column | Description|\n| --- | --- |\n|POSTED_BY| Category marking who has listed the property|\n|UNDER_CONSTRUCTION | Under Construction or Not|\n|RERA | Rera approved or Not|\n|BHK_NO | Number of Rooms|\n|BHK_OR_RK | Type of property|\n|SQUARE_FT | Total area of the house in square feet|\n|READY_TO_MOVE| Category marking Ready to move or Not|\n|RESALE | Category marking Resale or not|\n|ADDRESS | Address of the property|\n|LONGITUDE | Longitude of the property|\n|LATITUDE | Latitude of the property|\n \n## ACKNOWLEDGMENT:\nThe dataset for this hackathon was contributed by [Devrup Banerjee](https://www.machinehack.com/user/profile/5ef0a238b7efcc325e390297) . We would like to appreciate his efforts for this contribution to the Machinehack community.", "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 902117, "CreatorUserId": 1815526, "OwnerUserId": 1815526.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 1530022.0, "CurrentDatasourceVersionId": 1564506.0, "ForumId": 917784, "Type": 2, "CreationDate": "10/01/2020 19:37:13", "LastActivityDate": "10/01/2020", "TotalViews": 127515, "TotalDownloads": 15593, "TotalVotes": 201, "TotalKernels": 89}]
|
[{"Id": 1815526, "UserName": "anmolkumar", "DisplayName": "Anmol Kumar", "RegisterDate": "04/12/2018", "PerformanceTier": 2}]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/house-price-prediction-challenge/train.csv")
train.head()
train.shape
# No missing values
train.info()
# most of houses not under construction
train["UNDER_CONSTRUCTION"].value_counts()
train["READY_TO_MOVE"].value_counts()
train["RESALE"].value_counts()
train["POSTED_BY"].value_counts()
# handling categorical values , there is many ways like this two ways or OHE or LE or dummies
# train['POSTED']=train['POSTED_BY'].apply(lambda x :0 if x=='Dealer' else 1 if x=='Owner' else 2)
# train.drop("POSTED_BY",axis=1,inplace=True)
mappp = {"Dealer": 0, "Owner": 1, "Builder": 2}
train["POSTED_BY"] = train["POSTED_BY"].map(mappp)
train["BHK_OR_RK"].value_counts()
mapp = {"BHK": 0, "RK": 1}
train["BHK_OR_RK"] = train["BHK_OR_RK"].map(mapp)
train.head()
# number of rooms between 1 and 20
#
train.plot(kind="scatter", x="TARGET(PRICE_IN_LACS)", y="BHK_NO.")
# latitude between 60 and 150, doesn't highly effect
train.plot(kind="scatter", x="TARGET(PRICE_IN_LACS)", y="LATITUDE")
# longitude between 0 and 40 , doesn't highly effect but near to 40 less cost
train.plot(kind="scatter", x="TARGET(PRICE_IN_LACS)", y="LONGITUDE")
# log for area
train["AREA"] = np.log(train["SQUARE_FT"])
# price direct proportional with area
sns.scatterplot(data=train, x="TARGET(PRICE_IN_LACS)", y="AREA")
# drop column before log
train.drop("SQUARE_FT", axis=1, inplace=True)
train.describe()
# location doesn't highly effect to price
plt.figure(figsize=(15, 10))
sns.scatterplot(data=train, x="LONGITUDE", y="LATITUDE", hue="TARGET(PRICE_IN_LACS)")
# divide features to categorical and numerical
cat_features = ["POSTED_BY", "BHK_OR_RK"]
num_features = [
"UNDER_CONSTRUCTION",
"RERA",
"BHK_NO.",
"READY_TO_MOVE",
"RESALE",
"LONGITUDE",
"LATITUDE",
"AREA",
]
# area, number of rooms, price highly correlated with each other
plt.figure(figsize=(10, 10))
corr = train[cat_features + num_features + ["TARGET(PRICE_IN_LACS)"]].corr(
method="spearman"
)
sns.heatmap(corr, annot=True)
# handling outliers
outlier_percentage = {}
for feature in ["AREA", "BHK_NO.", "TARGET(PRICE_IN_LACS)"]:
tempData = train.sort_values(by=feature)[feature]
Q1, Q3 = tempData.quantile([0.25, 0.75])
IQR = Q3 - Q1
Lower_range = Q1 - (1.5 * IQR)
Upper_range = Q3 + (1.5 * IQR)
outlier_percentage[feature] = round(
(
((tempData < (Q1 - 1.5 * IQR)) | (tempData > (Q3 + 1.5 * IQR))).sum()
/ tempData.shape[0]
)
* 100,
2,
)
outlier_percentage
outlier = train[
(train[feature] > Lower_range) & (train[feature] < Upper_range)
].reset_index(drop=True)
train.drop(["ADDRESS"], axis=1, inplace=True)
X = train.drop(["TARGET(PRICE_IN_LACS)"], axis=1)
y = train["TARGET(PRICE_IN_LACS)"]
# feature selection
# under constrauction and ready to move exactly the same , location has small effect
from sklearn.feature_selection import SelectKBest, f_regression
best = SelectKBest(score_func=f_regression, k="all")
fit = best.fit(X, y)
dfScores = pd.DataFrame(fit.scores_)
dfCol = pd.DataFrame(X.columns)
featScore = pd.concat([dfCol, dfScores], axis=1)
featScore.columns = ["Feature", "Score"]
featScore = featScore.sort_values(by="Score", ascending=False).reset_index(drop=True)
print(featScore.nlargest(10, "Score"))
# another way for feature selection
# from sklearn.ensemble import ExtraTreesRegressor
# ex=ExtraTreesRegressor()
# ex.fit(X,y)
# ex.feature_importances_
# plt.figure(figsize=(10,10))
# plt.title('Feature importances')
# feat=pd.Series(ex.feature_importances_,index=X.columns)
# feat.nlargest(10).plot(kind='barh', color="r", align="center")
# plt.tight_layout()
# plt.show()
# removing features with vif , there is another way called RFE "Recursive Feature Elimination"
from statsmodels.stats.outliers_influence import variance_inflation_factor
v = pd.DataFrame()
v["variables"] = [
feature
for feature in cat_features + num_features
if feature not in ["READY_TO_MOVE", "RESALE", "LATITUDE", "LONGITUDE"]
]
v["VIF"] = [
variance_inflation_factor(train[v["variables"]].values, i)
for i in range(len(v["variables"]))
]
print(v)
# splitting the data
from sklearn.model_selection import (
train_test_split,
cross_val_score,
RandomizedSearchCV,
)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
# feature scaling
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.fit_transform(X_test)
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.linear_model import (
LinearRegression,
SGDRegressor,
Lasso,
Ridge,
ElasticNet,
)
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import (
RandomForestRegressor,
GradientBoostingRegressor,
VotingRegressor,
)
from xgboost.sklearn import XGBRegressor
# train models
models = {
"Linear Regression": LinearRegression(),
"Lasso": Lasso(),
"Decision Tree": DecisionTreeRegressor(),
"Random Forest": RandomForestRegressor(),
"Gradient Boosting =": GradientBoostingRegressor(),
"Ridge": Ridge(),
"Stochastic Gradien Descent": SGDRegressor(),
"Elastic": ElasticNet(),
"xgb Regressor": XGBRegressor(),
}
# fit and score
def fit_score(models, X_train, X_test, y_train, y_test):
np.random.seed(42)
model_scores = {}
for name, model in models.items():
model.fit(X_train, y_train)
model_scores[name] = cross_val_score(
model, X_test, y_test, scoring="neg_mean_squared_error", cv=3
).mean()
return model_scores
# Decision tree has lowest mse
model_scores = fit_score(models, X_train, X_test, y_train, y_test)
model_scores
# voting
vot = VotingRegressor(
[
("LinearRegression", LinearRegression()),
("DecisionTrees", DecisionTreeRegressor()),
("LassoRegression", Lasso()),
("RandomForest", RandomForestRegressor()),
("ElasticNet", ElasticNet()),
("StochasticGradientDescent", SGDRegressor()),
("GrafientBoosting", GradientBoostingRegressor()),
("Ridge", Ridge()),
("xgb", XGBRegressor()),
]
)
vot.fit(X_train, y_train)
y_pred = vot.predict(X_test)
# as expected , voting has smallest mse
mean_squared_error(y_test, y_pred)
np.random.seed(42)
params = {
"criterion": ["mse", "mae"],
"max_features": ["auto", "sqrt", "log2"],
"max_depth": [2, 3, 10],
}
rs = RandomizedSearchCV(
DecisionTreeRegressor(),
param_distributions=params,
cv=3,
n_iter=30,
verbose=0,
n_jobs=-1,
)
rs.fit(X_train, y_train)
rs.best_params_
rs.best_estimator_
rs.best_score_
rs.score(X_test, y_test)
model = DecisionTreeRegressor()
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
r2_score(y_pred, y_test)
|
[{"house-price-prediction-challenge/train.csv": {"column_names": "[\"POSTED_BY\", \"UNDER_CONSTRUCTION\", \"RERA\", \"BHK_NO.\", \"BHK_OR_RK\", \"SQUARE_FT\", \"READY_TO_MOVE\", \"RESALE\", \"ADDRESS\", \"LONGITUDE\", \"LATITUDE\", \"TARGET(PRICE_IN_LACS)\"]", "column_data_types": "{\"POSTED_BY\": \"object\", \"UNDER_CONSTRUCTION\": \"int64\", \"RERA\": \"int64\", \"BHK_NO.\": \"int64\", \"BHK_OR_RK\": \"object\", \"SQUARE_FT\": \"float64\", \"READY_TO_MOVE\": \"int64\", \"RESALE\": \"int64\", \"ADDRESS\": \"object\", \"LONGITUDE\": \"float64\", \"LATITUDE\": \"float64\", \"TARGET(PRICE_IN_LACS)\": \"float64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 29451 entries, 0 to 29450\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 POSTED_BY 29451 non-null object \n 1 UNDER_CONSTRUCTION 29451 non-null int64 \n 2 RERA 29451 non-null int64 \n 3 BHK_NO. 29451 non-null int64 \n 4 BHK_OR_RK 29451 non-null object \n 5 SQUARE_FT 29451 non-null float64\n 6 READY_TO_MOVE 29451 non-null int64 \n 7 RESALE 29451 non-null int64 \n 8 ADDRESS 29451 non-null object \n 9 LONGITUDE 29451 non-null float64\n 10 LATITUDE 29451 non-null float64\n 11 TARGET(PRICE_IN_LACS) 29451 non-null float64\ndtypes: float64(4), int64(5), object(3)\nmemory usage: 2.7+ MB\n", "summary": "{\"UNDER_CONSTRUCTION\": {\"count\": 29451.0, \"mean\": 0.17975620522223354, \"std\": 0.383990779174163, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 0.0, \"max\": 1.0}, \"RERA\": {\"count\": 29451.0, \"mean\": 0.31791789752470206, \"std\": 0.46567528510077866, \"min\": 0.0, \"25%\": 0.0, \"50%\": 0.0, \"75%\": 1.0, \"max\": 1.0}, \"BHK_NO.\": {\"count\": 29451.0, \"mean\": 2.3922787002139145, \"std\": 0.8790912922151548, \"min\": 1.0, \"25%\": 2.0, \"50%\": 2.0, \"75%\": 3.0, \"max\": 20.0}, \"SQUARE_FT\": {\"count\": 29451.0, \"mean\": 19802.170190334724, \"std\": 1901334.912503906, \"min\": 3.0, \"25%\": 900.0211296, \"50%\": 1175.05675, \"75%\": 1550.688124, \"max\": 254545454.5}, \"READY_TO_MOVE\": {\"count\": 29451.0, \"mean\": 0.8202437947777664, \"std\": 0.3839907791741631, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"RESALE\": {\"count\": 29451.0, \"mean\": 0.929577943024006, \"std\": 0.25586131734292034, \"min\": 0.0, \"25%\": 1.0, \"50%\": 1.0, \"75%\": 1.0, \"max\": 1.0}, \"LONGITUDE\": {\"count\": 29451.0, \"mean\": 21.300255165028354, \"std\": 6.205306453735163, \"min\": -37.7130075, \"25%\": 18.452663, \"50%\": 20.75, \"75%\": 26.900926, \"max\": 59.912884}, \"LATITUDE\": {\"count\": 29451.0, \"mean\": 76.83769524877074, \"std\": 10.557746673857764, \"min\": -121.7612481, \"25%\": 73.7981, \"50%\": 77.324137, \"75%\": 77.82873950000001, \"max\": 152.962676}, \"TARGET(PRICE_IN_LACS)\": {\"count\": 29451.0, \"mean\": 142.8987457132186, \"std\": 656.8807127981115, \"min\": 0.25, \"25%\": 38.0, \"50%\": 62.0, \"75%\": 100.0, \"max\": 30000.0}}", "examples": "{\"POSTED_BY\":{\"0\":\"Owner\",\"1\":\"Dealer\",\"2\":\"Owner\",\"3\":\"Owner\"},\"UNDER_CONSTRUCTION\":{\"0\":0,\"1\":0,\"2\":0,\"3\":0},\"RERA\":{\"0\":0,\"1\":0,\"2\":0,\"3\":1},\"BHK_NO.\":{\"0\":2,\"1\":2,\"2\":2,\"3\":2},\"BHK_OR_RK\":{\"0\":\"BHK\",\"1\":\"BHK\",\"2\":\"BHK\",\"3\":\"BHK\"},\"SQUARE_FT\":{\"0\":1300.236407,\"1\":1275.0,\"2\":933.1597222,\"3\":929.9211427},\"READY_TO_MOVE\":{\"0\":1,\"1\":1,\"2\":1,\"3\":1},\"RESALE\":{\"0\":1,\"1\":1,\"2\":1,\"3\":1},\"ADDRESS\":{\"0\":\"Ksfc Layout,Bangalore\",\"1\":\"Vishweshwara Nagar,Mysore\",\"2\":\"Jigani,Bangalore\",\"3\":\"Sector-1 Vaishali,Ghaziabad\"},\"LONGITUDE\":{\"0\":12.96991,\"1\":12.274538,\"2\":12.778033,\"3\":28.6423},\"LATITUDE\":{\"0\":77.59796,\"1\":76.644605,\"2\":77.632191,\"3\":77.3445},\"TARGET(PRICE_IN_LACS)\":{\"0\":55.0,\"1\":51.0,\"2\":43.0,\"3\":62.5}}"}}]
| true | 1 |
<start_data_description><data_path>house-price-prediction-challenge/train.csv:
<column_names>
['POSTED_BY', 'UNDER_CONSTRUCTION', 'RERA', 'BHK_NO.', 'BHK_OR_RK', 'SQUARE_FT', 'READY_TO_MOVE', 'RESALE', 'ADDRESS', 'LONGITUDE', 'LATITUDE', 'TARGET(PRICE_IN_LACS)']
<column_types>
{'POSTED_BY': 'object', 'UNDER_CONSTRUCTION': 'int64', 'RERA': 'int64', 'BHK_NO.': 'int64', 'BHK_OR_RK': 'object', 'SQUARE_FT': 'float64', 'READY_TO_MOVE': 'int64', 'RESALE': 'int64', 'ADDRESS': 'object', 'LONGITUDE': 'float64', 'LATITUDE': 'float64', 'TARGET(PRICE_IN_LACS)': 'float64'}
<dataframe_Summary>
{'UNDER_CONSTRUCTION': {'count': 29451.0, 'mean': 0.17975620522223354, 'std': 0.383990779174163, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 0.0, 'max': 1.0}, 'RERA': {'count': 29451.0, 'mean': 0.31791789752470206, 'std': 0.46567528510077866, 'min': 0.0, '25%': 0.0, '50%': 0.0, '75%': 1.0, 'max': 1.0}, 'BHK_NO.': {'count': 29451.0, 'mean': 2.3922787002139145, 'std': 0.8790912922151548, 'min': 1.0, '25%': 2.0, '50%': 2.0, '75%': 3.0, 'max': 20.0}, 'SQUARE_FT': {'count': 29451.0, 'mean': 19802.170190334724, 'std': 1901334.912503906, 'min': 3.0, '25%': 900.0211296, '50%': 1175.05675, '75%': 1550.688124, 'max': 254545454.5}, 'READY_TO_MOVE': {'count': 29451.0, 'mean': 0.8202437947777664, 'std': 0.3839907791741631, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'RESALE': {'count': 29451.0, 'mean': 0.929577943024006, 'std': 0.25586131734292034, 'min': 0.0, '25%': 1.0, '50%': 1.0, '75%': 1.0, 'max': 1.0}, 'LONGITUDE': {'count': 29451.0, 'mean': 21.300255165028354, 'std': 6.205306453735163, 'min': -37.7130075, '25%': 18.452663, '50%': 20.75, '75%': 26.900926, 'max': 59.912884}, 'LATITUDE': {'count': 29451.0, 'mean': 76.83769524877074, 'std': 10.557746673857764, 'min': -121.7612481, '25%': 73.7981, '50%': 77.324137, '75%': 77.82873950000001, 'max': 152.962676}, 'TARGET(PRICE_IN_LACS)': {'count': 29451.0, 'mean': 142.8987457132186, 'std': 656.8807127981115, 'min': 0.25, '25%': 38.0, '50%': 62.0, '75%': 100.0, 'max': 30000.0}}
<dataframe_info>
RangeIndex: 29451 entries, 0 to 29450
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 POSTED_BY 29451 non-null object
1 UNDER_CONSTRUCTION 29451 non-null int64
2 RERA 29451 non-null int64
3 BHK_NO. 29451 non-null int64
4 BHK_OR_RK 29451 non-null object
5 SQUARE_FT 29451 non-null float64
6 READY_TO_MOVE 29451 non-null int64
7 RESALE 29451 non-null int64
8 ADDRESS 29451 non-null object
9 LONGITUDE 29451 non-null float64
10 LATITUDE 29451 non-null float64
11 TARGET(PRICE_IN_LACS) 29451 non-null float64
dtypes: float64(4), int64(5), object(3)
memory usage: 2.7+ MB
<some_examples>
{'POSTED_BY': {'0': 'Owner', '1': 'Dealer', '2': 'Owner', '3': 'Owner'}, 'UNDER_CONSTRUCTION': {'0': 0, '1': 0, '2': 0, '3': 0}, 'RERA': {'0': 0, '1': 0, '2': 0, '3': 1}, 'BHK_NO.': {'0': 2, '1': 2, '2': 2, '3': 2}, 'BHK_OR_RK': {'0': 'BHK', '1': 'BHK', '2': 'BHK', '3': 'BHK'}, 'SQUARE_FT': {'0': 1300.236407, '1': 1275.0, '2': 933.1597222, '3': 929.9211427}, 'READY_TO_MOVE': {'0': 1, '1': 1, '2': 1, '3': 1}, 'RESALE': {'0': 1, '1': 1, '2': 1, '3': 1}, 'ADDRESS': {'0': 'Ksfc Layout,Bangalore', '1': 'Vishweshwara Nagar,Mysore', '2': 'Jigani,Bangalore', '3': 'Sector-1 Vaishali,Ghaziabad'}, 'LONGITUDE': {'0': 12.96991, '1': 12.274538, '2': 12.778033, '3': 28.6423}, 'LATITUDE': {'0': 77.59796, '1': 76.644605, '2': 77.632191, '3': 77.3445}, 'TARGET(PRICE_IN_LACS)': {'0': 55.0, '1': 51.0, '2': 43.0, '3': 62.5}}
<end_description>
| 2,478 | 1 | 4,028 | 2,478 |
69242502
|
#
# # Create a natural language classifier with Bert and Tensorflow
# High performance transformer models such as BERT and GPT-3 transform a wide range of previously menial and language-based tasks into a few clicks job, saving a lot of time.
# In most industries, the latest wave of language optimization is just beginning - taking its first steps. But these plants are widespread and grow quickly.
# Much of this adoption is due to the incredibly low barrier to entry. If you know the basics of TensorFlow or PyTorch and take some time to familiarize yourself with the Transformers library, you're already halfway there.
# With the Transformers library, it only takes three lines of code to initialize a cutting-edge ML model - a model built from billions of research dollars spent by Google, Facebook, and OpenAI.
# This article will walk you through the steps to create a classification model that harnesses the power of transformers, using Google's BERT.
# - Finding Models
# - Initializing
# - Bert Inputs and Outputs
# Classification
# - The Data
# - Tokenization
# - Data Prep
# - Train-Validation Split
# - Model Definition
# - Train
# Results
# # Transformateurs
# ## Find models
# We will be using BERT, possibly the most well-known transformer architecture.
# To understand what we need to use BERT, we head to the HuggingFace template page (HuggingFace built the Transformer framework).
# Once there, we will find both bert-base-cased and bert-base-uncased on the first page. cased means that the pattern distinguishes between uppercase and lowercase, whereas this uncase does not.
# ## Initialization
# If we click on the model we find more specific details. On this page we can see the model initialization code. Because we are using TensorFlow, our code will use TFAutoTokenizer and TFAutoModel instead of AutoTokenizer and AutoModel, respectively:
#
import pandas as pd
from transformers import TFBertModel
seed_value = 42
import os
os.environ["PYTHONHASHSEED"] = str(seed_value)
import random
random.seed(seed_value)
import numpy as np
np.random.seed(seed_value)
np.set_printoptions(precision=2)
import tensorflow as tf
tf.random.set_seed(seed_value)
# import tensorflow_addons as tfa
import tensorflow.keras
import tensorflow.keras.layers as layers
from tensorflow.keras.callbacks import ModelCheckpoint
print(tf.test.gpu_device_name())
# See https://www.tensorflow.org/tutorials/using_gpu#allowing_gpu_memory_growth
from transformers import AutoTokenizer, TFAutoModel
# When inserting textual data into our model, there are a few things to consider. First, we need to use tokenizer.encode_plus (...) to convert our text to input ID and attention mask tensors (we'll talk about that later).
# BERT expects these two tensors as inputs. One mapped to "input_ids" and another to "attention_mask".
# At the other end of the spectrum, BERT generates two default tensors (more are available). These are "last_hidden_state" and "pooler_output".
# The output of the pooler is simply the last hidden state, processed slightly further by a linear layer and a Tanh activation function - this also reduces its dimensionality from 3D (last hidden state) to 2D (output of pooler).
# Later we will consume the last hidden state tensor and remove the output from the pooler.
# # Classification
# ## Data
# CommonLit Readability Prize . We can download and extract it programmatically, like this:
# Import the datasets
train_raw = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_raw = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
# # Simple EDA
train_raw.head()
# Perform EDA on the train
train_raw.shape, train_raw.dtypes
# # Preparation and Feature Extraction
# # Tokenization
# We have our text data in the textcolumn, which we now need to tokenize. We will use the BERT tokenizer, because we will use a BERT transformer later.
# # Train Data
# ## feature Extraction X :
from transformers import AutoTokenizer
SEQ_LEN = 128 # we will cut/pad our sequences to a length of 128 tokens
tokenizer = AutoTokenizer.from_pretrained("bert-base-cased")
def tokenize(sentence):
tokens = tokenizer.encode_plus(
sentence,
max_length=SEQ_LEN,
truncation=True,
padding="max_length",
add_special_tokens=True,
return_attention_mask=True,
return_token_type_ids=False,
return_tensors="tf",
)
return tokens["input_ids"], tokens["attention_mask"]
# initialize two arrays for input tensors
Xids = np.zeros((len(train_raw), SEQ_LEN))
Xmask = np.zeros((len(train_raw), SEQ_LEN))
for i, sentence in enumerate(train_raw["excerpt"]):
Xids[i, :], Xmask[i, :] = tokenize(sentence)
if i % 10000 == 0:
print(i) # do this so we can see some progress
Xids
Xmask
# Here we first import the transformers library and initialize a tokenizer for the bert-base-casedmodel used. A list of models can be found here. We then define a tokenize function which manages the tokenization.
# We use the encode_plus method of our BERT tokenizer to convert a sentence into input_ids and attention_masktensors.
# Entry IDs are a list of integers uniquely related to a specific word.
# The attention mask is a list of 1s and 0s that match the IDs in the entry ID array - BERT reads this and only applies attention to IDs that match a mask value of attention of 1. This allows us to avoid drawing attention to the filler tokens.
# Our encode_plus arguments are:
# Our award. It is simply a string representing a text.
# The max_length of our encoded outputs. We use a value of 32 which means that each output tensor has a length of 32.
# We cut the sequences that are more than 32 tokens in length with truncation = True.
# For sequences shorter than 32 tokens, we pad them with zeros up to a length of 32 using padding = 'max_length'.
# BERT uses several special tokens, to mark the start / end of sequences, for padding, unknown words and mask words. We add those using add_special_tokens = True.
# BERT also takes two inputs, the input_idset attention_mask. We extract the attention mask with return_attention_mask = True.
# By default the tokenizer will return a token type ID tensor - which we don't need, so we use return_token_type_ids = False.
# Finally, we use TensorFlow, so we return the TensorFlow tensors using return_tensors = 'tf'. If you are using PyTorch, use return_tensors = 'pt'.
# # Lables preparation
#
labels = train_raw["target"].values # take sentiment column in df as array
labels
# This whole process can take some time. I like to save the encoded tables so that we can recover them in case of problems or for future tests.
# with open('xids.npy', 'wb') as f:
# np.save(f, Xids)
# with open('xmask.npy', 'wb') as f:
# np.save(f, Xmask)
# with open('labels.npy', 'wb') as f:
# np.save(f, labels)
# Now that we have all the arrays encoded, we load them into a TensorFlow dataset object. Using the dataset, we easily restructure, mix and group the data.
import tensorflow as tf
BATCH_SIZE = 32 # we will use batches of 32
# load arrays into tensorflow dataset
dataset = tf.data.Dataset.from_tensor_slices((Xids, Xmask, labels))
# create a mapping function that we use to restructure our dataset
def map_func(input_ids, masks, labels):
return {"input_ids": input_ids, "attention_mask": masks}, labels
# using map method to apply map_func to dataset
dataset = dataset.map(map_func)
# shuffle data and batch it
dataset = dataset.shuffle(100000).batch(BATCH_SIZE)
type(dataset)
# # Train validation separation
# The last step before training our model is to divide our dataset into training, validation, and (optionally) testing sets. We will stick to a simple 90–10 train-validation separation here.
# get the length of the batched dataset
DS_LEN = len([0 for batch in dataset])
SPLIT = 0.9 # 90-10 split
train = dataset.take(round(DS_LEN * SPLIT)) # get first 90% of batches
val = dataset.skip(round(DS_LEN * SPLIT)) # skip first 90% and keep final 10%
del dataset # optionally, delete dataset to free up disk-space
# # Definition of the model
# Our data is now ready and we can define our model architecture. We'll use BERT, followed by an LSTM layer and a few simple NN layers. These last layers after BERT are our classifier.
# Our classifier uses the hidden state tensors output from BERT - using them to predict our target.
# 
from transformers import AutoModel
# initialize cased BERT model
bert = TFAutoModel.from_pretrained("bert-base-cased")
input_ids = tf.keras.layers.Input(shape=(128,), name="input_ids", dtype="int32")
mask = tf.keras.layers.Input(shape=(128,), name="attention_mask", dtype="int32")
# we consume the last_hidden_state tensor from bert (discarding pooled_outputs)
embeddings = bert(input_ids, attention_mask=mask)[0]
X = tf.keras.layers.LSTM(64)(embeddings)
X = tf.keras.layers.BatchNormalization()(X)
X = tf.keras.layers.Dense(64, activation="relu")(X)
X = tf.keras.layers.Dropout(0.1)(X)
y = tf.keras.layers.Dense(1, name="outputs")(X)
# define input and output layers of our model
model = tf.keras.Model(inputs=[input_ids, mask], outputs=y)
# freeze the BERT layer - otherwise we will be training 100M+ parameters...
model.layers[2].trainable = False
model.summary()
tf.keras.utils.plot_model(
model=model,
show_shapes=True,
dpi=76,
)
# # Coaching
# We can now train our model. First, we configure our optimizer (Adam), our loss function, and our precision metric. Then we compile the model and practice!
from tensorflow.keras.metrics import RootMeanSquaredError
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
optimizer = tf.keras.optimizers.Adam(0.01)
# loss = tf.keras.losses.CategoricalCrossentropy() # categorical = one-hot
rmse = RootMeanSquaredError()
best_weights_file = "./weights.h5"
batch_size = 16
max_epochs = 1000
m_ckpt = ModelCheckpoint(
best_weights_file,
monitor="val_auc",
mode="max",
verbose=2,
save_weights_only=True,
save_best_only=True,
)
es = EarlyStopping(monitor="loss", min_delta=0.0000000000000000001, patience=10)
model.compile(optimizer=optimizer, loss="mse", metrics=[rmse])
# fit model using our gpu
with tf.device("/gpu:0"):
history = model.fit(
train,
validation_data=val,
epochs=max_epochs,
batch_size=batch_size,
callbacks=[m_ckpt, es],
verbose=2,
)
# # Evaluate :
loss, root_mean_squared_error = model.evaluate(val, verbose=0)
print("root_mean_squared_error_model: %f" % (root_mean_squared_error * 100))
print("loss_model: %f" % (loss * 100))
import matplotlib.pyplot as plt
plt.style.use("ggplot")
def plot_history(history):
acc = history.history["root_mean_squared_error"]
val_acc = history.history["val_root_mean_squared_error"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
x = range(1, len(acc) + 1)
plt.figure(figsize=(12, 5))
plt.subplot(1, 2, 1)
plt.plot(x, acc, "b", label="Training root_mean_squared_error")
plt.plot(x, val_acc, "r", label="Validation root_mean_squared_error")
plt.title("Training and validation root_mean_squared_error")
plt.legend()
plt.subplot(1, 2, 2)
plt.plot(x, loss, "b", label="Training loss")
plt.plot(x, val_loss, "r", label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plot_history(history)
# model = load_model()
# tokenizer= DistilBertTokenizer.from_pretrained('distilbert-base-uncased')
predictions = model.predict(val)
# # Plot prediction
# # Predict Unseen Data :
test_raw = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
test_raw.head()
SEQ_LEN = 128 # we will cut/pad our sequences to a length of 128 tokens
tokenizer = AutoTokenizer.from_pretrained("bert-base-cased")
def tokenize(sentence):
tokens = tokenizer.encode_plus(
sentence,
max_length=SEQ_LEN,
truncation=True,
padding="max_length",
add_special_tokens=True,
return_attention_mask=True,
return_token_type_ids=False,
return_tensors="tf",
)
return tokens["input_ids"], tokens["attention_mask"]
# initialize two arrays for input tensors
Xids = np.zeros((len(test_raw), SEQ_LEN))
Xmask = np.zeros((len(test_raw), SEQ_LEN))
for i, sentence in enumerate(test_raw["excerpt"]):
Xids[i, :], Xmask[i, :] = tokenize(sentence)
if i % 10000 == 0:
print(i) # do this so we can see some progress
BATCH_SIZE = 1 # we will use batches of 1
# load arrays into tensorflow dataset
test = tf.data.Dataset.from_tensor_slices((Xids, Xmask, [0] * len(test_raw)))
# create a mapping function that we use to restructure our dataset
def map_func(input_ids, masks, labels):
return {"input_ids": input_ids, "attention_mask": masks}, labels
# using map method to apply map_func to dataset
Unseen_test_prep = test.map(map_func)
# shuffle data and batch it
Unseen_test_prep = Unseen_test_prep.shuffle(100000).batch(BATCH_SIZE)
preds = model.predict(Unseen_test_prep)
preds
test_raw["id"].values
#
# # Prepare Submission File
# We make submissions in CSV files. Your submissions usually have two columns: an ID column and a prediction column. The ID field comes from the test data (keeping whatever name the ID field had in that data, which for the housing data is the string 'Id'). The prediction column will use the name of the target field.
# We will create a DataFrame with this data, and then use the dataframe's to_csv method to write our submission file. Explicitly include the argument index=False to prevent pandas from adding another column in our csv file.
#
my_submission = pd.DataFrame({"id": test_raw.id, "target": preds.ravel()})
# you could use any filename. We choose submission here
my_submission.to_csv("submission.csv", index=False)
my_submission
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242502.ipynb
| null | null |
[{"Id": 69242502, "ScriptId": 18900437, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4473832, "CreationDate": "07/28/2021 14:22:35", "VersionNumber": 7.0, "Title": "CommonLit Readability Prize_Part21_Baseline_Bert", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 355.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 354.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
#
# # Create a natural language classifier with Bert and Tensorflow
# High performance transformer models such as BERT and GPT-3 transform a wide range of previously menial and language-based tasks into a few clicks job, saving a lot of time.
# In most industries, the latest wave of language optimization is just beginning - taking its first steps. But these plants are widespread and grow quickly.
# Much of this adoption is due to the incredibly low barrier to entry. If you know the basics of TensorFlow or PyTorch and take some time to familiarize yourself with the Transformers library, you're already halfway there.
# With the Transformers library, it only takes three lines of code to initialize a cutting-edge ML model - a model built from billions of research dollars spent by Google, Facebook, and OpenAI.
# This article will walk you through the steps to create a classification model that harnesses the power of transformers, using Google's BERT.
# - Finding Models
# - Initializing
# - Bert Inputs and Outputs
# Classification
# - The Data
# - Tokenization
# - Data Prep
# - Train-Validation Split
# - Model Definition
# - Train
# Results
# # Transformateurs
# ## Find models
# We will be using BERT, possibly the most well-known transformer architecture.
# To understand what we need to use BERT, we head to the HuggingFace template page (HuggingFace built the Transformer framework).
# Once there, we will find both bert-base-cased and bert-base-uncased on the first page. cased means that the pattern distinguishes between uppercase and lowercase, whereas this uncase does not.
# ## Initialization
# If we click on the model we find more specific details. On this page we can see the model initialization code. Because we are using TensorFlow, our code will use TFAutoTokenizer and TFAutoModel instead of AutoTokenizer and AutoModel, respectively:
#
import pandas as pd
from transformers import TFBertModel
seed_value = 42
import os
os.environ["PYTHONHASHSEED"] = str(seed_value)
import random
random.seed(seed_value)
import numpy as np
np.random.seed(seed_value)
np.set_printoptions(precision=2)
import tensorflow as tf
tf.random.set_seed(seed_value)
# import tensorflow_addons as tfa
import tensorflow.keras
import tensorflow.keras.layers as layers
from tensorflow.keras.callbacks import ModelCheckpoint
print(tf.test.gpu_device_name())
# See https://www.tensorflow.org/tutorials/using_gpu#allowing_gpu_memory_growth
from transformers import AutoTokenizer, TFAutoModel
# When inserting textual data into our model, there are a few things to consider. First, we need to use tokenizer.encode_plus (...) to convert our text to input ID and attention mask tensors (we'll talk about that later).
# BERT expects these two tensors as inputs. One mapped to "input_ids" and another to "attention_mask".
# At the other end of the spectrum, BERT generates two default tensors (more are available). These are "last_hidden_state" and "pooler_output".
# The output of the pooler is simply the last hidden state, processed slightly further by a linear layer and a Tanh activation function - this also reduces its dimensionality from 3D (last hidden state) to 2D (output of pooler).
# Later we will consume the last hidden state tensor and remove the output from the pooler.
# # Classification
# ## Data
# CommonLit Readability Prize . We can download and extract it programmatically, like this:
# Import the datasets
train_raw = pd.read_csv("../input/commonlitreadabilityprize/train.csv")
test_raw = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
# # Simple EDA
train_raw.head()
# Perform EDA on the train
train_raw.shape, train_raw.dtypes
# # Preparation and Feature Extraction
# # Tokenization
# We have our text data in the textcolumn, which we now need to tokenize. We will use the BERT tokenizer, because we will use a BERT transformer later.
# # Train Data
# ## feature Extraction X :
from transformers import AutoTokenizer
SEQ_LEN = 128 # we will cut/pad our sequences to a length of 128 tokens
tokenizer = AutoTokenizer.from_pretrained("bert-base-cased")
def tokenize(sentence):
tokens = tokenizer.encode_plus(
sentence,
max_length=SEQ_LEN,
truncation=True,
padding="max_length",
add_special_tokens=True,
return_attention_mask=True,
return_token_type_ids=False,
return_tensors="tf",
)
return tokens["input_ids"], tokens["attention_mask"]
# initialize two arrays for input tensors
Xids = np.zeros((len(train_raw), SEQ_LEN))
Xmask = np.zeros((len(train_raw), SEQ_LEN))
for i, sentence in enumerate(train_raw["excerpt"]):
Xids[i, :], Xmask[i, :] = tokenize(sentence)
if i % 10000 == 0:
print(i) # do this so we can see some progress
Xids
Xmask
# Here we first import the transformers library and initialize a tokenizer for the bert-base-casedmodel used. A list of models can be found here. We then define a tokenize function which manages the tokenization.
# We use the encode_plus method of our BERT tokenizer to convert a sentence into input_ids and attention_masktensors.
# Entry IDs are a list of integers uniquely related to a specific word.
# The attention mask is a list of 1s and 0s that match the IDs in the entry ID array - BERT reads this and only applies attention to IDs that match a mask value of attention of 1. This allows us to avoid drawing attention to the filler tokens.
# Our encode_plus arguments are:
# Our award. It is simply a string representing a text.
# The max_length of our encoded outputs. We use a value of 32 which means that each output tensor has a length of 32.
# We cut the sequences that are more than 32 tokens in length with truncation = True.
# For sequences shorter than 32 tokens, we pad them with zeros up to a length of 32 using padding = 'max_length'.
# BERT uses several special tokens, to mark the start / end of sequences, for padding, unknown words and mask words. We add those using add_special_tokens = True.
# BERT also takes two inputs, the input_idset attention_mask. We extract the attention mask with return_attention_mask = True.
# By default the tokenizer will return a token type ID tensor - which we don't need, so we use return_token_type_ids = False.
# Finally, we use TensorFlow, so we return the TensorFlow tensors using return_tensors = 'tf'. If you are using PyTorch, use return_tensors = 'pt'.
# # Lables preparation
#
labels = train_raw["target"].values # take sentiment column in df as array
labels
# This whole process can take some time. I like to save the encoded tables so that we can recover them in case of problems or for future tests.
# with open('xids.npy', 'wb') as f:
# np.save(f, Xids)
# with open('xmask.npy', 'wb') as f:
# np.save(f, Xmask)
# with open('labels.npy', 'wb') as f:
# np.save(f, labels)
# Now that we have all the arrays encoded, we load them into a TensorFlow dataset object. Using the dataset, we easily restructure, mix and group the data.
import tensorflow as tf
BATCH_SIZE = 32 # we will use batches of 32
# load arrays into tensorflow dataset
dataset = tf.data.Dataset.from_tensor_slices((Xids, Xmask, labels))
# create a mapping function that we use to restructure our dataset
def map_func(input_ids, masks, labels):
return {"input_ids": input_ids, "attention_mask": masks}, labels
# using map method to apply map_func to dataset
dataset = dataset.map(map_func)
# shuffle data and batch it
dataset = dataset.shuffle(100000).batch(BATCH_SIZE)
type(dataset)
# # Train validation separation
# The last step before training our model is to divide our dataset into training, validation, and (optionally) testing sets. We will stick to a simple 90–10 train-validation separation here.
# get the length of the batched dataset
DS_LEN = len([0 for batch in dataset])
SPLIT = 0.9 # 90-10 split
train = dataset.take(round(DS_LEN * SPLIT)) # get first 90% of batches
val = dataset.skip(round(DS_LEN * SPLIT)) # skip first 90% and keep final 10%
del dataset # optionally, delete dataset to free up disk-space
# # Definition of the model
# Our data is now ready and we can define our model architecture. We'll use BERT, followed by an LSTM layer and a few simple NN layers. These last layers after BERT are our classifier.
# Our classifier uses the hidden state tensors output from BERT - using them to predict our target.
# 
from transformers import AutoModel
# initialize cased BERT model
bert = TFAutoModel.from_pretrained("bert-base-cased")
input_ids = tf.keras.layers.Input(shape=(128,), name="input_ids", dtype="int32")
mask = tf.keras.layers.Input(shape=(128,), name="attention_mask", dtype="int32")
# we consume the last_hidden_state tensor from bert (discarding pooled_outputs)
embeddings = bert(input_ids, attention_mask=mask)[0]
X = tf.keras.layers.LSTM(64)(embeddings)
X = tf.keras.layers.BatchNormalization()(X)
X = tf.keras.layers.Dense(64, activation="relu")(X)
X = tf.keras.layers.Dropout(0.1)(X)
y = tf.keras.layers.Dense(1, name="outputs")(X)
# define input and output layers of our model
model = tf.keras.Model(inputs=[input_ids, mask], outputs=y)
# freeze the BERT layer - otherwise we will be training 100M+ parameters...
model.layers[2].trainable = False
model.summary()
tf.keras.utils.plot_model(
model=model,
show_shapes=True,
dpi=76,
)
# # Coaching
# We can now train our model. First, we configure our optimizer (Adam), our loss function, and our precision metric. Then we compile the model and practice!
from tensorflow.keras.metrics import RootMeanSquaredError
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
optimizer = tf.keras.optimizers.Adam(0.01)
# loss = tf.keras.losses.CategoricalCrossentropy() # categorical = one-hot
rmse = RootMeanSquaredError()
best_weights_file = "./weights.h5"
batch_size = 16
max_epochs = 1000
m_ckpt = ModelCheckpoint(
best_weights_file,
monitor="val_auc",
mode="max",
verbose=2,
save_weights_only=True,
save_best_only=True,
)
es = EarlyStopping(monitor="loss", min_delta=0.0000000000000000001, patience=10)
model.compile(optimizer=optimizer, loss="mse", metrics=[rmse])
# fit model using our gpu
with tf.device("/gpu:0"):
history = model.fit(
train,
validation_data=val,
epochs=max_epochs,
batch_size=batch_size,
callbacks=[m_ckpt, es],
verbose=2,
)
# # Evaluate :
loss, root_mean_squared_error = model.evaluate(val, verbose=0)
print("root_mean_squared_error_model: %f" % (root_mean_squared_error * 100))
print("loss_model: %f" % (loss * 100))
import matplotlib.pyplot as plt
plt.style.use("ggplot")
def plot_history(history):
acc = history.history["root_mean_squared_error"]
val_acc = history.history["val_root_mean_squared_error"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
x = range(1, len(acc) + 1)
plt.figure(figsize=(12, 5))
plt.subplot(1, 2, 1)
plt.plot(x, acc, "b", label="Training root_mean_squared_error")
plt.plot(x, val_acc, "r", label="Validation root_mean_squared_error")
plt.title("Training and validation root_mean_squared_error")
plt.legend()
plt.subplot(1, 2, 2)
plt.plot(x, loss, "b", label="Training loss")
plt.plot(x, val_loss, "r", label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plot_history(history)
# model = load_model()
# tokenizer= DistilBertTokenizer.from_pretrained('distilbert-base-uncased')
predictions = model.predict(val)
# # Plot prediction
# # Predict Unseen Data :
test_raw = pd.read_csv("../input/commonlitreadabilityprize/test.csv")
test_raw.head()
SEQ_LEN = 128 # we will cut/pad our sequences to a length of 128 tokens
tokenizer = AutoTokenizer.from_pretrained("bert-base-cased")
def tokenize(sentence):
tokens = tokenizer.encode_plus(
sentence,
max_length=SEQ_LEN,
truncation=True,
padding="max_length",
add_special_tokens=True,
return_attention_mask=True,
return_token_type_ids=False,
return_tensors="tf",
)
return tokens["input_ids"], tokens["attention_mask"]
# initialize two arrays for input tensors
Xids = np.zeros((len(test_raw), SEQ_LEN))
Xmask = np.zeros((len(test_raw), SEQ_LEN))
for i, sentence in enumerate(test_raw["excerpt"]):
Xids[i, :], Xmask[i, :] = tokenize(sentence)
if i % 10000 == 0:
print(i) # do this so we can see some progress
BATCH_SIZE = 1 # we will use batches of 1
# load arrays into tensorflow dataset
test = tf.data.Dataset.from_tensor_slices((Xids, Xmask, [0] * len(test_raw)))
# create a mapping function that we use to restructure our dataset
def map_func(input_ids, masks, labels):
return {"input_ids": input_ids, "attention_mask": masks}, labels
# using map method to apply map_func to dataset
Unseen_test_prep = test.map(map_func)
# shuffle data and batch it
Unseen_test_prep = Unseen_test_prep.shuffle(100000).batch(BATCH_SIZE)
preds = model.predict(Unseen_test_prep)
preds
test_raw["id"].values
#
# # Prepare Submission File
# We make submissions in CSV files. Your submissions usually have two columns: an ID column and a prediction column. The ID field comes from the test data (keeping whatever name the ID field had in that data, which for the housing data is the string 'Id'). The prediction column will use the name of the target field.
# We will create a DataFrame with this data, and then use the dataframe's to_csv method to write our submission file. Explicitly include the argument index=False to prevent pandas from adding another column in our csv file.
#
my_submission = pd.DataFrame({"id": test_raw.id, "target": preds.ravel()})
# you could use any filename. We choose submission here
my_submission.to_csv("submission.csv", index=False)
my_submission
| false | 0 | 3,939 | 0 | 3,939 | 3,939 |
||
69242210
|
# ### **Experiment Log:**
# |Version |Models Used |CV Score |LB Score| Changes Made
# | --- | --- | --- | --- | --- |
# |v1 |LightGBM | 0.2963 | 0.29675 | Baseline
# |v2 |LightGBM | 0.2945 | 0.29298 | Feature Tools Quantile Transformation
# |v3 |LightGBM | 0.2938 | NA | Trade data included
# |v4 |LightGBM, XGBoost | NA | NA | Ensemble model used
# |v5 | LightGBM, XGBoost | 0.307 | 0.30579 | Ensemble model used
# |v6 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2608 | 0.24344 | Ensemble models used Poly features for log-return
# |v7 | Linear Regression, GBR LightGBM, Bayesian Ridge | NA | NA | New features added
# |v8 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2536 | 0.23701 | Error Correction
# |v9 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2559 | 0.23454 | New features added
# |v10 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2545 | 0.22974 | New features added using expanding mean
# |v11 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.24651 | 0.22841 | Quantile transformation for feature scaling
# |v12 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2436 | 0.22758 | New features added
# |v13 | Linear Regression, GBR LightGBM, Bayesian Ridge Voting Regressor | 0.24686 | 0.22779 | New lag features added
# |v14 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.24429 | 0.22805 | New lag features for realized volatility added
# |v15 | Linear Regression, GBR, XGBoost LightGBM, Bayesian Ridge | 0.25574 | NA | New statistical features added
# |v16 | Linear Regression, GBR, XGBoost LightGBM, Bayesian Ridge | 0.24618 | 0.22854 | Removed lag features
# |v17 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.24117 | 0.23223 | Quantile Transformation
# |v18 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2405 | 0.22852 | New features added
# |v19 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2405 | NA | Capturing meta features for models blend
# |v20 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.24028 | NA | Architecture revamp
# |v21 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.23989 | 0.22832 | Architecture revamp
# |v22 | LightGBM, CatBoost, GBR | 0.2439 | NA | Architecture revamp
# |v23 | LightGBM, CatBoost, GBR | 0.24025 | 0.22821 | Architecture revamp
# |v24 | Linear Regression, GBR LightGBM, Bayesian Ridge | TBD | TBD | Architecture revamp
# ## Import libraries
import warnings
warnings.filterwarnings("ignore")
import gc
import glob
import pickle
import numpy as np
import pandas as pd
from tqdm import tqdm
from sklearn.model_selection import StratifiedKFold
from sklearn.preprocessing import QuantileTransformer
from lightgbm import LGBMRegressor
from sklearn.linear_model import BayesianRidge
from sklearn.linear_model import LinearRegression
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingRegressor
# ## Helper Functions
def rmspe(y_true, y_pred):
return np.sqrt(np.mean(np.square((y_true - y_pred) / y_true)))
def log_return(list_stock_prices):
return np.log(list_stock_prices).diff()
def realized_volatility(series_log_return):
return np.sqrt(np.sum(series_log_return**2))
def count_unique(series):
return len(np.unique(series))
def get_stats_window(df, fe_dict, seconds_in_bucket, add_suffix=False):
df_feature = (
df[df["seconds_in_bucket"] >= seconds_in_bucket]
.groupby(["time_id"])
.agg(fe_dict)
.reset_index()
)
df_feature.columns = ["_".join(col) for col in df_feature.columns]
if add_suffix:
df_feature = df_feature.add_suffix("_" + str(seconds_in_bucket))
return df_feature
def process_trade_data(trade_files):
trade_df = pd.DataFrame()
for file in tqdm(glob.glob(trade_files)):
# Read source file
df_trade_data = pd.read_parquet(file)
# Feature engineering
df_trade_data["log_return"] = df_trade_data.groupby("time_id")["price"].apply(
log_return
)
df_trade_data.fillna(0, inplace=True)
fet_engg_dict = {
"price": ["mean", "std", "sum"],
"size": ["mean", "std", "sum"],
"order_count": ["mean", "std", "sum"],
"seconds_in_bucket": [count_unique],
"log_return": [realized_volatility],
}
# Get the stats for different windows
df_feature = get_stats_window(
df_trade_data, fet_engg_dict, seconds_in_bucket=0, add_suffix=False
)
df_feature_150 = get_stats_window(
df_trade_data, fet_engg_dict, seconds_in_bucket=150, add_suffix=True
)
df_feature_300 = get_stats_window(
df_trade_data, fet_engg_dict, seconds_in_bucket=300, add_suffix=True
)
df_feature_450 = get_stats_window(
df_trade_data, fet_engg_dict, seconds_in_bucket=450, add_suffix=True
)
# Merge all
trade_agg_df = df_feature.merge(
df_feature_150, how="left", left_on="time_id_", right_on="time_id__150"
)
trade_agg_df = trade_agg_df.merge(
df_feature_300, how="left", left_on="time_id_", right_on="time_id__300"
)
trade_agg_df = trade_agg_df.merge(
df_feature_450, how="left", left_on="time_id_", right_on="time_id__450"
)
trade_agg_df = trade_agg_df.add_prefix("trade_")
# Generate row_id
stock_id = file.split("=")[1]
trade_agg_df["row_id"] = trade_agg_df["trade_time_id_"].apply(
lambda x: f"{stock_id}-{x}"
)
trade_agg_df.drop(["trade_time_id_"], inplace=True, axis=1)
# Merge with parent df
trade_df = pd.concat([trade_df, trade_agg_df])
del df_trade_data, trade_agg_df
del df_feature, df_feature_150
del df_feature_300, df_feature_450
gc.collect()
return trade_df
def process_book_data(book_files):
book_df = pd.DataFrame()
for file in tqdm(glob.glob(book_files)):
# Read source file
df_book_data = pd.read_parquet(file)
# Feature engineering
df_book_data["wap1"] = (
df_book_data["bid_price1"] * df_book_data["ask_size1"]
+ df_book_data["ask_price1"] * df_book_data["bid_size1"]
) / (df_book_data["bid_size1"] + df_book_data["ask_size1"])
df_book_data["wap2"] = (
df_book_data["bid_price2"] * df_book_data["ask_size2"]
+ df_book_data["ask_price2"] * df_book_data["bid_size2"]
) / (df_book_data["bid_size2"] + df_book_data["ask_size2"])
df_book_data["log_return1"] = df_book_data.groupby(["time_id"])["wap1"].apply(
log_return
)
df_book_data["log_return2"] = df_book_data.groupby(["time_id"])["wap2"].apply(
log_return
)
df_book_data.fillna(0, inplace=True)
df_book_data["wap_balance"] = abs(df_book_data["wap1"] - df_book_data["wap2"])
df_book_data["price_spread"] = (
df_book_data["ask_price1"] - df_book_data["bid_price1"]
) / ((df_book_data["ask_price1"] + df_book_data["bid_price1"]) / 2)
df_book_data["bid_spread"] = (
df_book_data["bid_price1"] - df_book_data["bid_price2"]
)
df_book_data["ask_spread"] = (
df_book_data["ask_price1"] - df_book_data["ask_price2"]
)
df_book_data["total_volume"] = (
df_book_data["ask_size1"] + df_book_data["ask_size2"]
) + (df_book_data["bid_size1"] + df_book_data["bid_size2"])
df_book_data["volume_imbalance"] = abs(
(df_book_data["ask_size1"] + df_book_data["ask_size2"])
- (df_book_data["bid_size1"] + df_book_data["bid_size2"])
)
fet_engg_dict = {
"wap1": ["mean", "std", "sum"],
"wap2": ["mean", "std", "sum"],
"log_return1": [realized_volatility, "mean", "std", "sum"],
"log_return2": [realized_volatility, "mean", "std", "sum"],
"wap_balance": ["mean", "std", "sum"],
"price_spread": ["mean", "std", "sum"],
"bid_spread": ["mean", "std", "sum"],
"ask_spread": ["mean", "std", "sum"],
"total_volume": ["mean", "std", "sum"],
"volume_imbalance": ["mean", "std", "sum"],
}
# Get the stats for different windows
df_feature = get_stats_window(
df_book_data, fet_engg_dict, seconds_in_bucket=0, add_suffix=False
)
df_feature_150 = get_stats_window(
df_book_data, fet_engg_dict, seconds_in_bucket=150, add_suffix=True
)
df_feature_300 = get_stats_window(
df_book_data, fet_engg_dict, seconds_in_bucket=300, add_suffix=True
)
df_feature_450 = get_stats_window(
df_book_data, fet_engg_dict, seconds_in_bucket=450, add_suffix=True
)
# Merge all
book_agg_df = df_feature.merge(
df_feature_150, how="left", left_on="time_id_", right_on="time_id__150"
)
book_agg_df = book_agg_df.merge(
df_feature_300, how="left", left_on="time_id_", right_on="time_id__300"
)
book_agg_df = book_agg_df.merge(
df_feature_450, how="left", left_on="time_id_", right_on="time_id__450"
)
book_agg_df = book_agg_df.add_prefix("book_")
# Generate row_id
stock_id = file.split("=")[1]
book_agg_df["row_id"] = book_agg_df["book_time_id_"].apply(
lambda x: f"{stock_id}-{x}"
)
book_agg_df.drop(["book_time_id_"], inplace=True, axis=1)
# Merge with parent df
book_df = pd.concat([book_df, book_agg_df])
del df_book_data, book_agg_df
del df_feature, df_feature_150
del df_feature_300, df_feature_450
gc.collect()
return book_df
# ## Load training data
with open("../input/orvp-django-unchained/ORVP_Ready_Meatballs.txt", "rb") as handle:
data = handle.read()
processed_data = pickle.loads(data)
train_df = processed_data["train_df"]
del processed_data
gc.collect()
Xtrain = train_df.loc[:, train_df.columns != "target"].copy()
Ytrain = train_df["target"].copy()
Ytrain_strat = pd.qcut(train_df["target"].values, q=10, labels=range(0, 10))
print(
f"Xtrain: {Xtrain.shape} \nYtrain: {Ytrain.shape} \nYtrain_strat: {Ytrain_strat.shape}"
)
del train_df
gc.collect()
# ## Prepare testing data
# ### Test data
test_df = pd.read_csv("../input/optiver-realized-volatility-prediction/test.csv")
test_df["row_id"] = (
test_df["stock_id"].astype(str) + "-" + test_df["time_id"].astype(str)
)
print(f"test_df: {test_df.shape}")
test_df.head()
# ### Trade data
trade_test_df = process_trade_data(
"../input/optiver-realized-volatility-prediction/trade_test.parquet/*"
)
print(f"trade_test_df: {trade_test_df.shape}")
trade_test_df.head()
test_df = pd.merge(test_df, trade_test_df, how="left", on="row_id", sort=False)
test_df.fillna(0, inplace=True)
print(f"test_df: {test_df.shape}")
test_df.head()
# ### Book data
book_test_df = process_book_data(
"../input/optiver-realized-volatility-prediction/book_test.parquet/*"
)
print(f"book_test_df: {book_test_df.shape}")
book_test_df.head()
test_df = pd.merge(test_df, book_test_df, how="inner", on="row_id", sort=False)
test_df.fillna(0, inplace=True)
print(f"test_df: {test_df.shape}")
test_df.head()
# ### Group features
vol_cols = [
"trade_log_return_realized_volatility",
"trade_log_return_realized_volatility_150",
"trade_log_return_realized_volatility_300",
"trade_log_return_realized_volatility_450",
"book_log_return1_realized_volatility",
"book_log_return2_realized_volatility",
"book_log_return1_realized_volatility_150",
"book_log_return2_realized_volatility_150",
"book_log_return1_realized_volatility_300",
"book_log_return2_realized_volatility_300",
"book_log_return1_realized_volatility_450",
"book_log_return2_realized_volatility_450",
]
df_stock_id = (
test_df.groupby(["stock_id"])[vol_cols]
.agg(["mean", "std", "max", "min"])
.reset_index()
)
df_stock_id.columns = ["_".join(col) for col in df_stock_id.columns]
df_stock_id = df_stock_id.add_suffix("_stock")
df_stock_id.head()
df_time_id = (
test_df.groupby(["time_id"])[vol_cols]
.agg(["mean", "std", "max", "min"])
.reset_index()
)
df_time_id.columns = ["_".join(col) for col in df_time_id.columns]
df_time_id = df_time_id.add_suffix("_time")
df_time_id.head()
test_df = test_df.merge(
df_stock_id, how="left", left_on=["stock_id"], right_on=["stock_id__stock"]
)
test_df = test_df.merge(
df_time_id, how="left", left_on=["time_id"], right_on=["time_id__time"]
)
test_df.drop(["stock_id__stock", "time_id__time"], axis=1, inplace=True)
del df_stock_id, df_time_id
gc.collect()
test_df.fillna(0, inplace=True)
print(f"test_df: {test_df.shape}")
selected_cols = [
"trade_price_mean",
"trade_price_std",
"trade_price_sum",
"trade_size_mean",
"trade_size_std",
"trade_size_sum",
"trade_order_count_mean",
"trade_order_count_std",
"trade_order_count_sum",
"trade_seconds_in_bucket_count_unique",
"trade_log_return_realized_volatility",
"trade_price_mean_150",
"trade_price_std_150",
"trade_price_sum_150",
"trade_size_mean_150",
"trade_size_std_150",
"trade_size_sum_150",
"trade_order_count_mean_150",
"trade_order_count_std_150",
"trade_order_count_sum_150",
"trade_seconds_in_bucket_count_unique_150",
"trade_log_return_realized_volatility_150",
"trade_price_mean_300",
"trade_price_std_300",
"trade_price_sum_300",
"trade_size_mean_300",
"trade_size_std_300",
"trade_size_sum_300",
"trade_order_count_mean_300",
"trade_order_count_std_300",
"trade_order_count_sum_300",
"trade_seconds_in_bucket_count_unique_300",
"trade_log_return_realized_volatility_300",
"trade_price_mean_450",
"trade_price_std_450",
"trade_price_sum_450",
"trade_size_mean_450",
"trade_size_std_450",
"trade_size_sum_450",
"trade_order_count_mean_450",
"trade_order_count_std_450",
"trade_order_count_sum_450",
"trade_seconds_in_bucket_count_unique_450",
"trade_log_return_realized_volatility_450",
"book_wap1_mean",
"book_wap1_std",
"book_wap1_sum",
"book_wap2_mean",
"book_wap2_std",
"book_wap2_sum",
"book_log_return1_realized_volatility",
"book_log_return1_mean",
"book_log_return1_std",
"book_log_return1_sum",
"book_log_return2_realized_volatility",
"book_log_return2_mean",
"book_log_return2_std",
"book_log_return2_sum",
"book_wap_balance_mean",
"book_wap_balance_std",
"book_wap_balance_sum",
"book_price_spread_mean",
"book_price_spread_std",
"book_price_spread_sum",
"book_bid_spread_mean",
"book_bid_spread_std",
"book_bid_spread_sum",
"book_ask_spread_mean",
"book_ask_spread_std",
"book_ask_spread_sum",
"book_total_volume_mean",
"book_total_volume_std",
"book_total_volume_sum",
"book_volume_imbalance_mean",
"book_volume_imbalance_std",
"book_volume_imbalance_sum",
"book_wap1_mean_150",
"book_wap1_std_150",
"book_wap1_sum_150",
"book_wap2_mean_150",
"book_wap2_std_150",
"book_wap2_sum_150",
"book_log_return1_realized_volatility_150",
"book_log_return1_mean_150",
"book_log_return1_std_150",
"book_log_return1_sum_150",
"book_log_return2_realized_volatility_150",
"book_log_return2_mean_150",
"book_log_return2_std_150",
"book_log_return2_sum_150",
"book_wap_balance_mean_150",
"book_wap_balance_std_150",
"book_wap_balance_sum_150",
"book_price_spread_mean_150",
"book_price_spread_std_150",
"book_price_spread_sum_150",
"book_bid_spread_mean_150",
"book_bid_spread_std_150",
"book_bid_spread_sum_150",
"book_ask_spread_mean_150",
"book_ask_spread_std_150",
"book_ask_spread_sum_150",
"book_total_volume_mean_150",
"book_total_volume_std_150",
"book_total_volume_sum_150",
"book_volume_imbalance_mean_150",
"book_volume_imbalance_std_150",
"book_volume_imbalance_sum_150",
"book_wap1_mean_300",
"book_wap1_std_300",
"book_wap1_sum_300",
"book_wap2_mean_300",
"book_wap2_std_300",
"book_wap2_sum_300",
"book_log_return1_realized_volatility_300",
"book_log_return1_mean_300",
"book_log_return1_std_300",
"book_log_return1_sum_300",
"book_log_return2_realized_volatility_300",
"book_log_return2_mean_300",
"book_log_return2_std_300",
"book_log_return2_sum_300",
"book_wap_balance_mean_300",
"book_wap_balance_std_300",
"book_wap_balance_sum_300",
"book_price_spread_mean_300",
"book_price_spread_std_300",
"book_price_spread_sum_300",
"book_bid_spread_mean_300",
"book_bid_spread_std_300",
"book_bid_spread_sum_300",
"book_ask_spread_mean_300",
"book_ask_spread_std_300",
"book_ask_spread_sum_300",
"book_total_volume_mean_300",
"book_total_volume_std_300",
"book_total_volume_sum_300",
"book_volume_imbalance_mean_300",
"book_volume_imbalance_std_300",
"book_volume_imbalance_sum_300",
"book_wap1_mean_450",
"book_wap1_std_450",
"book_wap1_sum_450",
"book_wap2_mean_450",
"book_wap2_std_450",
"book_wap2_sum_450",
"book_log_return1_realized_volatility_450",
"book_log_return1_mean_450",
"book_log_return1_std_450",
"book_log_return1_sum_450",
"book_log_return2_realized_volatility_450",
"book_log_return2_mean_450",
"book_log_return2_std_450",
"book_log_return2_sum_450",
"book_wap_balance_mean_450",
"book_wap_balance_std_450",
"book_wap_balance_sum_450",
"book_price_spread_mean_450",
"book_price_spread_std_450",
"book_price_spread_sum_450",
"book_bid_spread_mean_450",
"book_bid_spread_std_450",
"book_bid_spread_sum_450",
"book_ask_spread_mean_450",
"book_ask_spread_std_450",
"book_ask_spread_sum_450",
"book_total_volume_mean_450",
"book_total_volume_std_450",
"book_total_volume_sum_450",
"book_volume_imbalance_mean_450",
"book_volume_imbalance_std_450",
"book_volume_imbalance_sum_450",
"trade_log_return_realized_volatility_mean_stock",
"trade_log_return_realized_volatility_std_stock",
"trade_log_return_realized_volatility_max_stock",
"trade_log_return_realized_volatility_min_stock",
"trade_log_return_realized_volatility_150_mean_stock",
"trade_log_return_realized_volatility_150_std_stock",
"trade_log_return_realized_volatility_150_max_stock",
"trade_log_return_realized_volatility_150_min_stock",
"trade_log_return_realized_volatility_300_mean_stock",
"trade_log_return_realized_volatility_300_std_stock",
"trade_log_return_realized_volatility_300_max_stock",
"trade_log_return_realized_volatility_300_min_stock",
"trade_log_return_realized_volatility_450_mean_stock",
"trade_log_return_realized_volatility_450_std_stock",
"trade_log_return_realized_volatility_450_max_stock",
"trade_log_return_realized_volatility_450_min_stock",
"book_log_return1_realized_volatility_mean_stock",
"book_log_return1_realized_volatility_std_stock",
"book_log_return1_realized_volatility_max_stock",
"book_log_return1_realized_volatility_min_stock",
"book_log_return2_realized_volatility_mean_stock",
"book_log_return2_realized_volatility_std_stock",
"book_log_return2_realized_volatility_max_stock",
"book_log_return2_realized_volatility_min_stock",
"book_log_return1_realized_volatility_150_mean_stock",
"book_log_return1_realized_volatility_150_std_stock",
"book_log_return1_realized_volatility_150_max_stock",
"book_log_return1_realized_volatility_150_min_stock",
"book_log_return2_realized_volatility_150_mean_stock",
"book_log_return2_realized_volatility_150_std_stock",
"book_log_return2_realized_volatility_150_max_stock",
"book_log_return2_realized_volatility_150_min_stock",
"book_log_return1_realized_volatility_300_mean_stock",
"book_log_return1_realized_volatility_300_std_stock",
"book_log_return1_realized_volatility_300_max_stock",
"book_log_return1_realized_volatility_300_min_stock",
"book_log_return2_realized_volatility_300_mean_stock",
"book_log_return2_realized_volatility_300_std_stock",
"book_log_return2_realized_volatility_300_max_stock",
"book_log_return2_realized_volatility_300_min_stock",
"book_log_return1_realized_volatility_450_mean_stock",
"book_log_return1_realized_volatility_450_std_stock",
"book_log_return1_realized_volatility_450_max_stock",
"book_log_return1_realized_volatility_450_min_stock",
"book_log_return2_realized_volatility_450_mean_stock",
"book_log_return2_realized_volatility_450_std_stock",
"book_log_return2_realized_volatility_450_max_stock",
"book_log_return2_realized_volatility_450_min_stock",
"trade_log_return_realized_volatility_mean_time",
"trade_log_return_realized_volatility_std_time",
"trade_log_return_realized_volatility_max_time",
"trade_log_return_realized_volatility_min_time",
"trade_log_return_realized_volatility_150_mean_time",
"trade_log_return_realized_volatility_150_std_time",
"trade_log_return_realized_volatility_150_max_time",
"trade_log_return_realized_volatility_150_min_time",
"trade_log_return_realized_volatility_300_mean_time",
"trade_log_return_realized_volatility_300_std_time",
"trade_log_return_realized_volatility_300_max_time",
"trade_log_return_realized_volatility_300_min_time",
"trade_log_return_realized_volatility_450_mean_time",
"trade_log_return_realized_volatility_450_std_time",
"trade_log_return_realized_volatility_450_max_time",
"trade_log_return_realized_volatility_450_min_time",
"book_log_return1_realized_volatility_mean_time",
"book_log_return1_realized_volatility_std_time",
"book_log_return1_realized_volatility_max_time",
"book_log_return1_realized_volatility_min_time",
"book_log_return2_realized_volatility_mean_time",
"book_log_return2_realized_volatility_std_time",
"book_log_return2_realized_volatility_max_time",
"book_log_return2_realized_volatility_min_time",
"book_log_return1_realized_volatility_150_mean_time",
"book_log_return1_realized_volatility_150_std_time",
"book_log_return1_realized_volatility_150_max_time",
"book_log_return1_realized_volatility_150_min_time",
"book_log_return2_realized_volatility_150_mean_time",
"book_log_return2_realized_volatility_150_std_time",
"book_log_return2_realized_volatility_150_max_time",
"book_log_return2_realized_volatility_150_min_time",
"book_log_return1_realized_volatility_300_mean_time",
"book_log_return1_realized_volatility_300_std_time",
"book_log_return1_realized_volatility_300_max_time",
"book_log_return1_realized_volatility_300_min_time",
"book_log_return2_realized_volatility_300_mean_time",
"book_log_return2_realized_volatility_300_std_time",
"book_log_return2_realized_volatility_300_max_time",
"book_log_return2_realized_volatility_300_min_time",
"book_log_return1_realized_volatility_450_mean_time",
"book_log_return1_realized_volatility_450_std_time",
"book_log_return1_realized_volatility_450_max_time",
"book_log_return1_realized_volatility_450_min_time",
"book_log_return2_realized_volatility_450_mean_time",
"book_log_return2_realized_volatility_450_std_time",
"book_log_return2_realized_volatility_450_max_time",
"book_log_return2_realized_volatility_450_min_time",
]
Xtest = test_df[selected_cols].copy()
print(f"Xtest: {Xtest.shape}")
# ## Quantile Transformation
for col in tqdm(Xtrain.columns):
transformer = QuantileTransformer(
n_quantiles=1000, random_state=10, output_distribution="normal"
)
vec_len = len(Xtrain[col].values)
vec_len_test = len(Xtest[col].values)
raw_vec = Xtrain[col].values.reshape(vec_len, 1)
test_vec = Xtest[col].values.reshape(vec_len_test, 1)
transformer.fit(raw_vec)
Xtrain[col] = transformer.transform(raw_vec).reshape(1, vec_len)[0]
Xtest[col] = transformer.transform(test_vec).reshape(1, vec_len_test)[0]
print(f"Xtrain: {Xtrain.shape} \nXtest: {Xtest.shape}")
# ## Base models
# * **BayesianRidge**
# * **HistGradientBoostingRegressor**
# * **Linear Regression**
# * **LightGBM**
FOLD = 10
SEEDS = [2020, 2022]
COUNTER = 0
oof_score_ridge = 0
oof_score_gbr = 0
oof_score_lgb = 0
oof_score_lr = 0
y_pred_final_ridge = 0
y_pred_final_gbr = 0
y_pred_final_lgb = 0
y_pred_final_lr = 0
y_pred_meta_ridge = np.zeros((Xtrain.shape[0], 1))
y_pred_meta_gbr = np.zeros((Xtrain.shape[0], 1))
y_pred_meta_lgb = np.zeros((Xtrain.shape[0], 1))
y_pred_meta_lr = np.zeros((Xtrain.shape[0], 1))
print("Model Name \tSeed \tFold \tOOF Score \tAggregate OOF Score")
print("=" * 68)
for sidx, seed in enumerate(SEEDS):
seed_score_ridge = 0
seed_score_gbr = 0
seed_score_lgb = 0
seed_score_lr = 0
kfold = StratifiedKFold(n_splits=FOLD, shuffle=True, random_state=seed)
for idx, (train, val) in enumerate(kfold.split(Xtrain, Ytrain_strat)):
COUNTER += 1
train_x, train_y = Xtrain.iloc[train], Ytrain.iloc[train]
val_x, val_y = Xtrain.iloc[val], Ytrain.iloc[val]
weights = 1 / np.square(train_y)
# ====================================================================
# Linear Regression
# ====================================================================
lr_model = LinearRegression()
lr_model.fit(train_x, train_y, sample_weight=weights)
y_pred = lr_model.predict(val_x)
y_pred_meta_lr[val] += np.array([y_pred]).T
y_pred_final_lr += lr_model.predict(Xtest)
score = rmspe(val_y, y_pred)
oof_score_lr += score
seed_score_lr += score
print(f"LR \t{seed} \t{idx+1} \t{round(score,5)}")
# ====================================================================
# Bayesian Ridge
# ====================================================================
ridge_model = BayesianRidge()
ridge_model.fit(train_x, train_y, sample_weight=weights)
y_pred = ridge_model.predict(val_x)
y_pred_meta_ridge[val] += np.array([y_pred]).T
y_pred_final_ridge += ridge_model.predict(Xtest)
score = rmspe(val_y, y_pred)
oof_score_ridge += score
seed_score_ridge += score
print(f"Bayesian Ridge \t{seed} \t{idx+1} \t{round(score,5)}")
# ====================================================================
# HistGradientBoostingRegressor
# ====================================================================
gbr_model = HistGradientBoostingRegressor(
max_depth=7,
learning_rate=0.05,
max_iter=1000,
max_leaf_nodes=72,
early_stopping=True,
n_iter_no_change=100,
random_state=0,
)
gbr_model.fit(train_x, train_y, sample_weight=weights)
y_pred = gbr_model.predict(val_x)
y_pred_meta_gbr[val] += np.array([y_pred]).T
y_pred_final_gbr += gbr_model.predict(Xtest)
score = rmspe(val_y, y_pred)
oof_score_gbr += score
seed_score_gbr += score
print(f"GBR \t{seed} \t{idx+1} \t{round(score,5)}")
# ====================================================================
# LightGBM
# ====================================================================
lgb_model = LGBMRegressor(
boosting_type="gbdt",
num_leaves=72,
max_depth=7,
learning_rate=0.02,
n_estimators=1000,
objective="regression",
min_child_samples=10,
subsample=0.65,
subsample_freq=10,
colsample_bytree=0.75,
reg_lambda=0.05,
random_state=0,
)
lgb_model.fit(
train_x,
train_y,
eval_metric="rmse",
eval_set=(val_x, val_y),
early_stopping_rounds=100,
sample_weight=weights,
verbose=False,
)
y_pred = lgb_model.predict(val_x)
y_pred_meta_lgb[val] += np.array([y_pred]).T
y_pred_final_lgb += lgb_model.predict(Xtest)
score = rmspe(val_y, y_pred)
oof_score_lgb += score
seed_score_lgb += score
print(f"LightGBM \t{seed} \t{idx+1} \t{round(score,5)}\n")
print("=" * 68)
print(f"Bayesian Ridge \t{seed} \t\t\t\t{round(seed_score_ridge / FOLD, 5)}")
print(f"GBR \t{seed} \t\t\t\t{round(seed_score_gbr / FOLD, 5)}")
print(f"LR \t{seed} \t\t\t\t{round(seed_score_lr / FOLD, 5)}")
print(f"LightGBM \t{seed} \t\t\t\t{round(seed_score_lgb / FOLD, 5)}")
print("=" * 68)
y_pred_final_ridge = y_pred_final_ridge / float(COUNTER)
y_pred_final_gbr = y_pred_final_gbr / float(COUNTER)
y_pred_final_lr = y_pred_final_lr / float(COUNTER)
y_pred_final_lgb = y_pred_final_lgb / float(COUNTER)
y_pred_meta_ridge = y_pred_meta_ridge / float(len(SEEDS))
y_pred_meta_gbr = y_pred_meta_gbr / float(len(SEEDS))
y_pred_meta_lr = y_pred_meta_lr / float(len(SEEDS))
y_pred_meta_lgb = y_pred_meta_lgb / float(len(SEEDS))
oof_score_ridge /= float(COUNTER)
oof_score_gbr /= float(COUNTER)
oof_score_lr /= float(COUNTER)
oof_score_lgb /= float(COUNTER)
oof_score = (oof_score_gbr * 0.3) + (oof_score_lgb * 0.7)
print(f"Bayesian Ridge | Aggregate OOF Score: {round(oof_score_ridge,5)}")
print(f"GradientBoostingRegressor | Aggregate OOF Score: {round(oof_score_gbr,5)}")
print(f"Linear Regression | Aggregate OOF Score: {round(oof_score_lr,5)}")
print(f"LightGBM | Aggregate OOF Score: {round(oof_score_lgb,5)}")
print(f"Aggregate OOF Score: {round(oof_score,5)}")
np.savez_compressed(
"./Pulp_Fiction_Meta_Features.npz",
y_pred_meta_ridge=y_pred_meta_ridge,
y_pred_meta_gbr=y_pred_meta_gbr,
y_pred_meta_lr=y_pred_meta_lr,
y_pred_meta_lgb=y_pred_meta_lgb,
oof_score_ridge=oof_score_ridge,
oof_score_gbr=oof_score_gbr,
oof_score_lr=oof_score_lr,
oof_score_lgb=oof_score_lgb,
y_pred_final_ridge=y_pred_final_ridge,
y_pred_final_gbr=y_pred_final_gbr,
y_pred_final_lr=y_pred_final_lr,
y_pred_final_lgb=y_pred_final_lgb,
)
# ## Create submission file
y_pred_final = (y_pred_final_gbr * 0.3) + (y_pred_final_lgb * 0.7)
submit_df = pd.DataFrame()
submit_df["row_id"] = test_df["row_id"]
submit_df["target"] = y_pred_final
submit_df.to_csv("./submission.csv", index=False)
submit_df.head()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242210.ipynb
| null | null |
[{"Id": 69242210, "ScriptId": 18221533, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6450791, "CreationDate": "07/28/2021 14:18:40", "VersionNumber": 24.0, "Title": "ORVP-Pulp Fiction", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 771.0, "LinesInsertedFromPrevious": 589.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 182.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# ### **Experiment Log:**
# |Version |Models Used |CV Score |LB Score| Changes Made
# | --- | --- | --- | --- | --- |
# |v1 |LightGBM | 0.2963 | 0.29675 | Baseline
# |v2 |LightGBM | 0.2945 | 0.29298 | Feature Tools Quantile Transformation
# |v3 |LightGBM | 0.2938 | NA | Trade data included
# |v4 |LightGBM, XGBoost | NA | NA | Ensemble model used
# |v5 | LightGBM, XGBoost | 0.307 | 0.30579 | Ensemble model used
# |v6 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2608 | 0.24344 | Ensemble models used Poly features for log-return
# |v7 | Linear Regression, GBR LightGBM, Bayesian Ridge | NA | NA | New features added
# |v8 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2536 | 0.23701 | Error Correction
# |v9 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2559 | 0.23454 | New features added
# |v10 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2545 | 0.22974 | New features added using expanding mean
# |v11 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.24651 | 0.22841 | Quantile transformation for feature scaling
# |v12 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2436 | 0.22758 | New features added
# |v13 | Linear Regression, GBR LightGBM, Bayesian Ridge Voting Regressor | 0.24686 | 0.22779 | New lag features added
# |v14 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.24429 | 0.22805 | New lag features for realized volatility added
# |v15 | Linear Regression, GBR, XGBoost LightGBM, Bayesian Ridge | 0.25574 | NA | New statistical features added
# |v16 | Linear Regression, GBR, XGBoost LightGBM, Bayesian Ridge | 0.24618 | 0.22854 | Removed lag features
# |v17 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.24117 | 0.23223 | Quantile Transformation
# |v18 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2405 | 0.22852 | New features added
# |v19 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.2405 | NA | Capturing meta features for models blend
# |v20 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.24028 | NA | Architecture revamp
# |v21 | Linear Regression, GBR LightGBM, Bayesian Ridge | 0.23989 | 0.22832 | Architecture revamp
# |v22 | LightGBM, CatBoost, GBR | 0.2439 | NA | Architecture revamp
# |v23 | LightGBM, CatBoost, GBR | 0.24025 | 0.22821 | Architecture revamp
# |v24 | Linear Regression, GBR LightGBM, Bayesian Ridge | TBD | TBD | Architecture revamp
# ## Import libraries
import warnings
warnings.filterwarnings("ignore")
import gc
import glob
import pickle
import numpy as np
import pandas as pd
from tqdm import tqdm
from sklearn.model_selection import StratifiedKFold
from sklearn.preprocessing import QuantileTransformer
from lightgbm import LGBMRegressor
from sklearn.linear_model import BayesianRidge
from sklearn.linear_model import LinearRegression
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingRegressor
# ## Helper Functions
def rmspe(y_true, y_pred):
return np.sqrt(np.mean(np.square((y_true - y_pred) / y_true)))
def log_return(list_stock_prices):
return np.log(list_stock_prices).diff()
def realized_volatility(series_log_return):
return np.sqrt(np.sum(series_log_return**2))
def count_unique(series):
return len(np.unique(series))
def get_stats_window(df, fe_dict, seconds_in_bucket, add_suffix=False):
df_feature = (
df[df["seconds_in_bucket"] >= seconds_in_bucket]
.groupby(["time_id"])
.agg(fe_dict)
.reset_index()
)
df_feature.columns = ["_".join(col) for col in df_feature.columns]
if add_suffix:
df_feature = df_feature.add_suffix("_" + str(seconds_in_bucket))
return df_feature
def process_trade_data(trade_files):
trade_df = pd.DataFrame()
for file in tqdm(glob.glob(trade_files)):
# Read source file
df_trade_data = pd.read_parquet(file)
# Feature engineering
df_trade_data["log_return"] = df_trade_data.groupby("time_id")["price"].apply(
log_return
)
df_trade_data.fillna(0, inplace=True)
fet_engg_dict = {
"price": ["mean", "std", "sum"],
"size": ["mean", "std", "sum"],
"order_count": ["mean", "std", "sum"],
"seconds_in_bucket": [count_unique],
"log_return": [realized_volatility],
}
# Get the stats for different windows
df_feature = get_stats_window(
df_trade_data, fet_engg_dict, seconds_in_bucket=0, add_suffix=False
)
df_feature_150 = get_stats_window(
df_trade_data, fet_engg_dict, seconds_in_bucket=150, add_suffix=True
)
df_feature_300 = get_stats_window(
df_trade_data, fet_engg_dict, seconds_in_bucket=300, add_suffix=True
)
df_feature_450 = get_stats_window(
df_trade_data, fet_engg_dict, seconds_in_bucket=450, add_suffix=True
)
# Merge all
trade_agg_df = df_feature.merge(
df_feature_150, how="left", left_on="time_id_", right_on="time_id__150"
)
trade_agg_df = trade_agg_df.merge(
df_feature_300, how="left", left_on="time_id_", right_on="time_id__300"
)
trade_agg_df = trade_agg_df.merge(
df_feature_450, how="left", left_on="time_id_", right_on="time_id__450"
)
trade_agg_df = trade_agg_df.add_prefix("trade_")
# Generate row_id
stock_id = file.split("=")[1]
trade_agg_df["row_id"] = trade_agg_df["trade_time_id_"].apply(
lambda x: f"{stock_id}-{x}"
)
trade_agg_df.drop(["trade_time_id_"], inplace=True, axis=1)
# Merge with parent df
trade_df = pd.concat([trade_df, trade_agg_df])
del df_trade_data, trade_agg_df
del df_feature, df_feature_150
del df_feature_300, df_feature_450
gc.collect()
return trade_df
def process_book_data(book_files):
book_df = pd.DataFrame()
for file in tqdm(glob.glob(book_files)):
# Read source file
df_book_data = pd.read_parquet(file)
# Feature engineering
df_book_data["wap1"] = (
df_book_data["bid_price1"] * df_book_data["ask_size1"]
+ df_book_data["ask_price1"] * df_book_data["bid_size1"]
) / (df_book_data["bid_size1"] + df_book_data["ask_size1"])
df_book_data["wap2"] = (
df_book_data["bid_price2"] * df_book_data["ask_size2"]
+ df_book_data["ask_price2"] * df_book_data["bid_size2"]
) / (df_book_data["bid_size2"] + df_book_data["ask_size2"])
df_book_data["log_return1"] = df_book_data.groupby(["time_id"])["wap1"].apply(
log_return
)
df_book_data["log_return2"] = df_book_data.groupby(["time_id"])["wap2"].apply(
log_return
)
df_book_data.fillna(0, inplace=True)
df_book_data["wap_balance"] = abs(df_book_data["wap1"] - df_book_data["wap2"])
df_book_data["price_spread"] = (
df_book_data["ask_price1"] - df_book_data["bid_price1"]
) / ((df_book_data["ask_price1"] + df_book_data["bid_price1"]) / 2)
df_book_data["bid_spread"] = (
df_book_data["bid_price1"] - df_book_data["bid_price2"]
)
df_book_data["ask_spread"] = (
df_book_data["ask_price1"] - df_book_data["ask_price2"]
)
df_book_data["total_volume"] = (
df_book_data["ask_size1"] + df_book_data["ask_size2"]
) + (df_book_data["bid_size1"] + df_book_data["bid_size2"])
df_book_data["volume_imbalance"] = abs(
(df_book_data["ask_size1"] + df_book_data["ask_size2"])
- (df_book_data["bid_size1"] + df_book_data["bid_size2"])
)
fet_engg_dict = {
"wap1": ["mean", "std", "sum"],
"wap2": ["mean", "std", "sum"],
"log_return1": [realized_volatility, "mean", "std", "sum"],
"log_return2": [realized_volatility, "mean", "std", "sum"],
"wap_balance": ["mean", "std", "sum"],
"price_spread": ["mean", "std", "sum"],
"bid_spread": ["mean", "std", "sum"],
"ask_spread": ["mean", "std", "sum"],
"total_volume": ["mean", "std", "sum"],
"volume_imbalance": ["mean", "std", "sum"],
}
# Get the stats for different windows
df_feature = get_stats_window(
df_book_data, fet_engg_dict, seconds_in_bucket=0, add_suffix=False
)
df_feature_150 = get_stats_window(
df_book_data, fet_engg_dict, seconds_in_bucket=150, add_suffix=True
)
df_feature_300 = get_stats_window(
df_book_data, fet_engg_dict, seconds_in_bucket=300, add_suffix=True
)
df_feature_450 = get_stats_window(
df_book_data, fet_engg_dict, seconds_in_bucket=450, add_suffix=True
)
# Merge all
book_agg_df = df_feature.merge(
df_feature_150, how="left", left_on="time_id_", right_on="time_id__150"
)
book_agg_df = book_agg_df.merge(
df_feature_300, how="left", left_on="time_id_", right_on="time_id__300"
)
book_agg_df = book_agg_df.merge(
df_feature_450, how="left", left_on="time_id_", right_on="time_id__450"
)
book_agg_df = book_agg_df.add_prefix("book_")
# Generate row_id
stock_id = file.split("=")[1]
book_agg_df["row_id"] = book_agg_df["book_time_id_"].apply(
lambda x: f"{stock_id}-{x}"
)
book_agg_df.drop(["book_time_id_"], inplace=True, axis=1)
# Merge with parent df
book_df = pd.concat([book_df, book_agg_df])
del df_book_data, book_agg_df
del df_feature, df_feature_150
del df_feature_300, df_feature_450
gc.collect()
return book_df
# ## Load training data
with open("../input/orvp-django-unchained/ORVP_Ready_Meatballs.txt", "rb") as handle:
data = handle.read()
processed_data = pickle.loads(data)
train_df = processed_data["train_df"]
del processed_data
gc.collect()
Xtrain = train_df.loc[:, train_df.columns != "target"].copy()
Ytrain = train_df["target"].copy()
Ytrain_strat = pd.qcut(train_df["target"].values, q=10, labels=range(0, 10))
print(
f"Xtrain: {Xtrain.shape} \nYtrain: {Ytrain.shape} \nYtrain_strat: {Ytrain_strat.shape}"
)
del train_df
gc.collect()
# ## Prepare testing data
# ### Test data
test_df = pd.read_csv("../input/optiver-realized-volatility-prediction/test.csv")
test_df["row_id"] = (
test_df["stock_id"].astype(str) + "-" + test_df["time_id"].astype(str)
)
print(f"test_df: {test_df.shape}")
test_df.head()
# ### Trade data
trade_test_df = process_trade_data(
"../input/optiver-realized-volatility-prediction/trade_test.parquet/*"
)
print(f"trade_test_df: {trade_test_df.shape}")
trade_test_df.head()
test_df = pd.merge(test_df, trade_test_df, how="left", on="row_id", sort=False)
test_df.fillna(0, inplace=True)
print(f"test_df: {test_df.shape}")
test_df.head()
# ### Book data
book_test_df = process_book_data(
"../input/optiver-realized-volatility-prediction/book_test.parquet/*"
)
print(f"book_test_df: {book_test_df.shape}")
book_test_df.head()
test_df = pd.merge(test_df, book_test_df, how="inner", on="row_id", sort=False)
test_df.fillna(0, inplace=True)
print(f"test_df: {test_df.shape}")
test_df.head()
# ### Group features
vol_cols = [
"trade_log_return_realized_volatility",
"trade_log_return_realized_volatility_150",
"trade_log_return_realized_volatility_300",
"trade_log_return_realized_volatility_450",
"book_log_return1_realized_volatility",
"book_log_return2_realized_volatility",
"book_log_return1_realized_volatility_150",
"book_log_return2_realized_volatility_150",
"book_log_return1_realized_volatility_300",
"book_log_return2_realized_volatility_300",
"book_log_return1_realized_volatility_450",
"book_log_return2_realized_volatility_450",
]
df_stock_id = (
test_df.groupby(["stock_id"])[vol_cols]
.agg(["mean", "std", "max", "min"])
.reset_index()
)
df_stock_id.columns = ["_".join(col) for col in df_stock_id.columns]
df_stock_id = df_stock_id.add_suffix("_stock")
df_stock_id.head()
df_time_id = (
test_df.groupby(["time_id"])[vol_cols]
.agg(["mean", "std", "max", "min"])
.reset_index()
)
df_time_id.columns = ["_".join(col) for col in df_time_id.columns]
df_time_id = df_time_id.add_suffix("_time")
df_time_id.head()
test_df = test_df.merge(
df_stock_id, how="left", left_on=["stock_id"], right_on=["stock_id__stock"]
)
test_df = test_df.merge(
df_time_id, how="left", left_on=["time_id"], right_on=["time_id__time"]
)
test_df.drop(["stock_id__stock", "time_id__time"], axis=1, inplace=True)
del df_stock_id, df_time_id
gc.collect()
test_df.fillna(0, inplace=True)
print(f"test_df: {test_df.shape}")
selected_cols = [
"trade_price_mean",
"trade_price_std",
"trade_price_sum",
"trade_size_mean",
"trade_size_std",
"trade_size_sum",
"trade_order_count_mean",
"trade_order_count_std",
"trade_order_count_sum",
"trade_seconds_in_bucket_count_unique",
"trade_log_return_realized_volatility",
"trade_price_mean_150",
"trade_price_std_150",
"trade_price_sum_150",
"trade_size_mean_150",
"trade_size_std_150",
"trade_size_sum_150",
"trade_order_count_mean_150",
"trade_order_count_std_150",
"trade_order_count_sum_150",
"trade_seconds_in_bucket_count_unique_150",
"trade_log_return_realized_volatility_150",
"trade_price_mean_300",
"trade_price_std_300",
"trade_price_sum_300",
"trade_size_mean_300",
"trade_size_std_300",
"trade_size_sum_300",
"trade_order_count_mean_300",
"trade_order_count_std_300",
"trade_order_count_sum_300",
"trade_seconds_in_bucket_count_unique_300",
"trade_log_return_realized_volatility_300",
"trade_price_mean_450",
"trade_price_std_450",
"trade_price_sum_450",
"trade_size_mean_450",
"trade_size_std_450",
"trade_size_sum_450",
"trade_order_count_mean_450",
"trade_order_count_std_450",
"trade_order_count_sum_450",
"trade_seconds_in_bucket_count_unique_450",
"trade_log_return_realized_volatility_450",
"book_wap1_mean",
"book_wap1_std",
"book_wap1_sum",
"book_wap2_mean",
"book_wap2_std",
"book_wap2_sum",
"book_log_return1_realized_volatility",
"book_log_return1_mean",
"book_log_return1_std",
"book_log_return1_sum",
"book_log_return2_realized_volatility",
"book_log_return2_mean",
"book_log_return2_std",
"book_log_return2_sum",
"book_wap_balance_mean",
"book_wap_balance_std",
"book_wap_balance_sum",
"book_price_spread_mean",
"book_price_spread_std",
"book_price_spread_sum",
"book_bid_spread_mean",
"book_bid_spread_std",
"book_bid_spread_sum",
"book_ask_spread_mean",
"book_ask_spread_std",
"book_ask_spread_sum",
"book_total_volume_mean",
"book_total_volume_std",
"book_total_volume_sum",
"book_volume_imbalance_mean",
"book_volume_imbalance_std",
"book_volume_imbalance_sum",
"book_wap1_mean_150",
"book_wap1_std_150",
"book_wap1_sum_150",
"book_wap2_mean_150",
"book_wap2_std_150",
"book_wap2_sum_150",
"book_log_return1_realized_volatility_150",
"book_log_return1_mean_150",
"book_log_return1_std_150",
"book_log_return1_sum_150",
"book_log_return2_realized_volatility_150",
"book_log_return2_mean_150",
"book_log_return2_std_150",
"book_log_return2_sum_150",
"book_wap_balance_mean_150",
"book_wap_balance_std_150",
"book_wap_balance_sum_150",
"book_price_spread_mean_150",
"book_price_spread_std_150",
"book_price_spread_sum_150",
"book_bid_spread_mean_150",
"book_bid_spread_std_150",
"book_bid_spread_sum_150",
"book_ask_spread_mean_150",
"book_ask_spread_std_150",
"book_ask_spread_sum_150",
"book_total_volume_mean_150",
"book_total_volume_std_150",
"book_total_volume_sum_150",
"book_volume_imbalance_mean_150",
"book_volume_imbalance_std_150",
"book_volume_imbalance_sum_150",
"book_wap1_mean_300",
"book_wap1_std_300",
"book_wap1_sum_300",
"book_wap2_mean_300",
"book_wap2_std_300",
"book_wap2_sum_300",
"book_log_return1_realized_volatility_300",
"book_log_return1_mean_300",
"book_log_return1_std_300",
"book_log_return1_sum_300",
"book_log_return2_realized_volatility_300",
"book_log_return2_mean_300",
"book_log_return2_std_300",
"book_log_return2_sum_300",
"book_wap_balance_mean_300",
"book_wap_balance_std_300",
"book_wap_balance_sum_300",
"book_price_spread_mean_300",
"book_price_spread_std_300",
"book_price_spread_sum_300",
"book_bid_spread_mean_300",
"book_bid_spread_std_300",
"book_bid_spread_sum_300",
"book_ask_spread_mean_300",
"book_ask_spread_std_300",
"book_ask_spread_sum_300",
"book_total_volume_mean_300",
"book_total_volume_std_300",
"book_total_volume_sum_300",
"book_volume_imbalance_mean_300",
"book_volume_imbalance_std_300",
"book_volume_imbalance_sum_300",
"book_wap1_mean_450",
"book_wap1_std_450",
"book_wap1_sum_450",
"book_wap2_mean_450",
"book_wap2_std_450",
"book_wap2_sum_450",
"book_log_return1_realized_volatility_450",
"book_log_return1_mean_450",
"book_log_return1_std_450",
"book_log_return1_sum_450",
"book_log_return2_realized_volatility_450",
"book_log_return2_mean_450",
"book_log_return2_std_450",
"book_log_return2_sum_450",
"book_wap_balance_mean_450",
"book_wap_balance_std_450",
"book_wap_balance_sum_450",
"book_price_spread_mean_450",
"book_price_spread_std_450",
"book_price_spread_sum_450",
"book_bid_spread_mean_450",
"book_bid_spread_std_450",
"book_bid_spread_sum_450",
"book_ask_spread_mean_450",
"book_ask_spread_std_450",
"book_ask_spread_sum_450",
"book_total_volume_mean_450",
"book_total_volume_std_450",
"book_total_volume_sum_450",
"book_volume_imbalance_mean_450",
"book_volume_imbalance_std_450",
"book_volume_imbalance_sum_450",
"trade_log_return_realized_volatility_mean_stock",
"trade_log_return_realized_volatility_std_stock",
"trade_log_return_realized_volatility_max_stock",
"trade_log_return_realized_volatility_min_stock",
"trade_log_return_realized_volatility_150_mean_stock",
"trade_log_return_realized_volatility_150_std_stock",
"trade_log_return_realized_volatility_150_max_stock",
"trade_log_return_realized_volatility_150_min_stock",
"trade_log_return_realized_volatility_300_mean_stock",
"trade_log_return_realized_volatility_300_std_stock",
"trade_log_return_realized_volatility_300_max_stock",
"trade_log_return_realized_volatility_300_min_stock",
"trade_log_return_realized_volatility_450_mean_stock",
"trade_log_return_realized_volatility_450_std_stock",
"trade_log_return_realized_volatility_450_max_stock",
"trade_log_return_realized_volatility_450_min_stock",
"book_log_return1_realized_volatility_mean_stock",
"book_log_return1_realized_volatility_std_stock",
"book_log_return1_realized_volatility_max_stock",
"book_log_return1_realized_volatility_min_stock",
"book_log_return2_realized_volatility_mean_stock",
"book_log_return2_realized_volatility_std_stock",
"book_log_return2_realized_volatility_max_stock",
"book_log_return2_realized_volatility_min_stock",
"book_log_return1_realized_volatility_150_mean_stock",
"book_log_return1_realized_volatility_150_std_stock",
"book_log_return1_realized_volatility_150_max_stock",
"book_log_return1_realized_volatility_150_min_stock",
"book_log_return2_realized_volatility_150_mean_stock",
"book_log_return2_realized_volatility_150_std_stock",
"book_log_return2_realized_volatility_150_max_stock",
"book_log_return2_realized_volatility_150_min_stock",
"book_log_return1_realized_volatility_300_mean_stock",
"book_log_return1_realized_volatility_300_std_stock",
"book_log_return1_realized_volatility_300_max_stock",
"book_log_return1_realized_volatility_300_min_stock",
"book_log_return2_realized_volatility_300_mean_stock",
"book_log_return2_realized_volatility_300_std_stock",
"book_log_return2_realized_volatility_300_max_stock",
"book_log_return2_realized_volatility_300_min_stock",
"book_log_return1_realized_volatility_450_mean_stock",
"book_log_return1_realized_volatility_450_std_stock",
"book_log_return1_realized_volatility_450_max_stock",
"book_log_return1_realized_volatility_450_min_stock",
"book_log_return2_realized_volatility_450_mean_stock",
"book_log_return2_realized_volatility_450_std_stock",
"book_log_return2_realized_volatility_450_max_stock",
"book_log_return2_realized_volatility_450_min_stock",
"trade_log_return_realized_volatility_mean_time",
"trade_log_return_realized_volatility_std_time",
"trade_log_return_realized_volatility_max_time",
"trade_log_return_realized_volatility_min_time",
"trade_log_return_realized_volatility_150_mean_time",
"trade_log_return_realized_volatility_150_std_time",
"trade_log_return_realized_volatility_150_max_time",
"trade_log_return_realized_volatility_150_min_time",
"trade_log_return_realized_volatility_300_mean_time",
"trade_log_return_realized_volatility_300_std_time",
"trade_log_return_realized_volatility_300_max_time",
"trade_log_return_realized_volatility_300_min_time",
"trade_log_return_realized_volatility_450_mean_time",
"trade_log_return_realized_volatility_450_std_time",
"trade_log_return_realized_volatility_450_max_time",
"trade_log_return_realized_volatility_450_min_time",
"book_log_return1_realized_volatility_mean_time",
"book_log_return1_realized_volatility_std_time",
"book_log_return1_realized_volatility_max_time",
"book_log_return1_realized_volatility_min_time",
"book_log_return2_realized_volatility_mean_time",
"book_log_return2_realized_volatility_std_time",
"book_log_return2_realized_volatility_max_time",
"book_log_return2_realized_volatility_min_time",
"book_log_return1_realized_volatility_150_mean_time",
"book_log_return1_realized_volatility_150_std_time",
"book_log_return1_realized_volatility_150_max_time",
"book_log_return1_realized_volatility_150_min_time",
"book_log_return2_realized_volatility_150_mean_time",
"book_log_return2_realized_volatility_150_std_time",
"book_log_return2_realized_volatility_150_max_time",
"book_log_return2_realized_volatility_150_min_time",
"book_log_return1_realized_volatility_300_mean_time",
"book_log_return1_realized_volatility_300_std_time",
"book_log_return1_realized_volatility_300_max_time",
"book_log_return1_realized_volatility_300_min_time",
"book_log_return2_realized_volatility_300_mean_time",
"book_log_return2_realized_volatility_300_std_time",
"book_log_return2_realized_volatility_300_max_time",
"book_log_return2_realized_volatility_300_min_time",
"book_log_return1_realized_volatility_450_mean_time",
"book_log_return1_realized_volatility_450_std_time",
"book_log_return1_realized_volatility_450_max_time",
"book_log_return1_realized_volatility_450_min_time",
"book_log_return2_realized_volatility_450_mean_time",
"book_log_return2_realized_volatility_450_std_time",
"book_log_return2_realized_volatility_450_max_time",
"book_log_return2_realized_volatility_450_min_time",
]
Xtest = test_df[selected_cols].copy()
print(f"Xtest: {Xtest.shape}")
# ## Quantile Transformation
for col in tqdm(Xtrain.columns):
transformer = QuantileTransformer(
n_quantiles=1000, random_state=10, output_distribution="normal"
)
vec_len = len(Xtrain[col].values)
vec_len_test = len(Xtest[col].values)
raw_vec = Xtrain[col].values.reshape(vec_len, 1)
test_vec = Xtest[col].values.reshape(vec_len_test, 1)
transformer.fit(raw_vec)
Xtrain[col] = transformer.transform(raw_vec).reshape(1, vec_len)[0]
Xtest[col] = transformer.transform(test_vec).reshape(1, vec_len_test)[0]
print(f"Xtrain: {Xtrain.shape} \nXtest: {Xtest.shape}")
# ## Base models
# * **BayesianRidge**
# * **HistGradientBoostingRegressor**
# * **Linear Regression**
# * **LightGBM**
FOLD = 10
SEEDS = [2020, 2022]
COUNTER = 0
oof_score_ridge = 0
oof_score_gbr = 0
oof_score_lgb = 0
oof_score_lr = 0
y_pred_final_ridge = 0
y_pred_final_gbr = 0
y_pred_final_lgb = 0
y_pred_final_lr = 0
y_pred_meta_ridge = np.zeros((Xtrain.shape[0], 1))
y_pred_meta_gbr = np.zeros((Xtrain.shape[0], 1))
y_pred_meta_lgb = np.zeros((Xtrain.shape[0], 1))
y_pred_meta_lr = np.zeros((Xtrain.shape[0], 1))
print("Model Name \tSeed \tFold \tOOF Score \tAggregate OOF Score")
print("=" * 68)
for sidx, seed in enumerate(SEEDS):
seed_score_ridge = 0
seed_score_gbr = 0
seed_score_lgb = 0
seed_score_lr = 0
kfold = StratifiedKFold(n_splits=FOLD, shuffle=True, random_state=seed)
for idx, (train, val) in enumerate(kfold.split(Xtrain, Ytrain_strat)):
COUNTER += 1
train_x, train_y = Xtrain.iloc[train], Ytrain.iloc[train]
val_x, val_y = Xtrain.iloc[val], Ytrain.iloc[val]
weights = 1 / np.square(train_y)
# ====================================================================
# Linear Regression
# ====================================================================
lr_model = LinearRegression()
lr_model.fit(train_x, train_y, sample_weight=weights)
y_pred = lr_model.predict(val_x)
y_pred_meta_lr[val] += np.array([y_pred]).T
y_pred_final_lr += lr_model.predict(Xtest)
score = rmspe(val_y, y_pred)
oof_score_lr += score
seed_score_lr += score
print(f"LR \t{seed} \t{idx+1} \t{round(score,5)}")
# ====================================================================
# Bayesian Ridge
# ====================================================================
ridge_model = BayesianRidge()
ridge_model.fit(train_x, train_y, sample_weight=weights)
y_pred = ridge_model.predict(val_x)
y_pred_meta_ridge[val] += np.array([y_pred]).T
y_pred_final_ridge += ridge_model.predict(Xtest)
score = rmspe(val_y, y_pred)
oof_score_ridge += score
seed_score_ridge += score
print(f"Bayesian Ridge \t{seed} \t{idx+1} \t{round(score,5)}")
# ====================================================================
# HistGradientBoostingRegressor
# ====================================================================
gbr_model = HistGradientBoostingRegressor(
max_depth=7,
learning_rate=0.05,
max_iter=1000,
max_leaf_nodes=72,
early_stopping=True,
n_iter_no_change=100,
random_state=0,
)
gbr_model.fit(train_x, train_y, sample_weight=weights)
y_pred = gbr_model.predict(val_x)
y_pred_meta_gbr[val] += np.array([y_pred]).T
y_pred_final_gbr += gbr_model.predict(Xtest)
score = rmspe(val_y, y_pred)
oof_score_gbr += score
seed_score_gbr += score
print(f"GBR \t{seed} \t{idx+1} \t{round(score,5)}")
# ====================================================================
# LightGBM
# ====================================================================
lgb_model = LGBMRegressor(
boosting_type="gbdt",
num_leaves=72,
max_depth=7,
learning_rate=0.02,
n_estimators=1000,
objective="regression",
min_child_samples=10,
subsample=0.65,
subsample_freq=10,
colsample_bytree=0.75,
reg_lambda=0.05,
random_state=0,
)
lgb_model.fit(
train_x,
train_y,
eval_metric="rmse",
eval_set=(val_x, val_y),
early_stopping_rounds=100,
sample_weight=weights,
verbose=False,
)
y_pred = lgb_model.predict(val_x)
y_pred_meta_lgb[val] += np.array([y_pred]).T
y_pred_final_lgb += lgb_model.predict(Xtest)
score = rmspe(val_y, y_pred)
oof_score_lgb += score
seed_score_lgb += score
print(f"LightGBM \t{seed} \t{idx+1} \t{round(score,5)}\n")
print("=" * 68)
print(f"Bayesian Ridge \t{seed} \t\t\t\t{round(seed_score_ridge / FOLD, 5)}")
print(f"GBR \t{seed} \t\t\t\t{round(seed_score_gbr / FOLD, 5)}")
print(f"LR \t{seed} \t\t\t\t{round(seed_score_lr / FOLD, 5)}")
print(f"LightGBM \t{seed} \t\t\t\t{round(seed_score_lgb / FOLD, 5)}")
print("=" * 68)
y_pred_final_ridge = y_pred_final_ridge / float(COUNTER)
y_pred_final_gbr = y_pred_final_gbr / float(COUNTER)
y_pred_final_lr = y_pred_final_lr / float(COUNTER)
y_pred_final_lgb = y_pred_final_lgb / float(COUNTER)
y_pred_meta_ridge = y_pred_meta_ridge / float(len(SEEDS))
y_pred_meta_gbr = y_pred_meta_gbr / float(len(SEEDS))
y_pred_meta_lr = y_pred_meta_lr / float(len(SEEDS))
y_pred_meta_lgb = y_pred_meta_lgb / float(len(SEEDS))
oof_score_ridge /= float(COUNTER)
oof_score_gbr /= float(COUNTER)
oof_score_lr /= float(COUNTER)
oof_score_lgb /= float(COUNTER)
oof_score = (oof_score_gbr * 0.3) + (oof_score_lgb * 0.7)
print(f"Bayesian Ridge | Aggregate OOF Score: {round(oof_score_ridge,5)}")
print(f"GradientBoostingRegressor | Aggregate OOF Score: {round(oof_score_gbr,5)}")
print(f"Linear Regression | Aggregate OOF Score: {round(oof_score_lr,5)}")
print(f"LightGBM | Aggregate OOF Score: {round(oof_score_lgb,5)}")
print(f"Aggregate OOF Score: {round(oof_score,5)}")
np.savez_compressed(
"./Pulp_Fiction_Meta_Features.npz",
y_pred_meta_ridge=y_pred_meta_ridge,
y_pred_meta_gbr=y_pred_meta_gbr,
y_pred_meta_lr=y_pred_meta_lr,
y_pred_meta_lgb=y_pred_meta_lgb,
oof_score_ridge=oof_score_ridge,
oof_score_gbr=oof_score_gbr,
oof_score_lr=oof_score_lr,
oof_score_lgb=oof_score_lgb,
y_pred_final_ridge=y_pred_final_ridge,
y_pred_final_gbr=y_pred_final_gbr,
y_pred_final_lr=y_pred_final_lr,
y_pred_final_lgb=y_pred_final_lgb,
)
# ## Create submission file
y_pred_final = (y_pred_final_gbr * 0.3) + (y_pred_final_lgb * 0.7)
submit_df = pd.DataFrame()
submit_df["row_id"] = test_df["row_id"]
submit_df["target"] = y_pred_final
submit_df.to_csv("./submission.csv", index=False)
submit_df.head()
| false | 0 | 11,388 | 0 | 11,388 | 11,388 |
||
69242791
|
# def parse(filename, label):
# raw = tf.io.read_file(filename)
# img = tf.image.decode_png(raw, channels=3)
# img = tf.image.central_crop(img, 0.95)
# img = tf.image.adjust_contrast(img, 3)
# img = tf.image.adjust_gamma(img, 0.5)
# img = tf.clip_by_value(img, 0, 255)
# img = tf.bitwise.invert(img)
# # img = tf.image.resize(img, [224,224])
# img = tf.image.random_flip_left_right(img)
# return img, label
# import pandas as pd
# import os
# import tensorflow as tf
# from sklearn.preprocessing import LabelEncoder
# from keras.utils import np_utils
# train_csv = pd.read_csv('../input/murav11-zip/mura.csv')
# filenames = []
# labels = []
# for i, row in train_csv.iterrows():
# filename = os.path.join('../input/murav11-zip/MURA-v1.1', row[1])
# filenames.append(filename)
# label = row[2]
# labels.append(label)
# encoder = LabelEncoder()
# labels = encoder.fit_transform(labels)
# labels = np_utils.to_categorical(labels)
# dataset = tf.data.Dataset.from_tensor_slices((filenames, labels))
# dataset = dataset.shuffle(len(filenames))
# dataset = dataset.map(parse, num_parallel_calls=1)
# import matplotlib.pyplot as plt
# for i, data in enumerate(dataset):
# img, label = data
# img = img.numpy()
# plt.imshow(img)
# break
def parse(filename, label):
raw = tf.io.read_file(filename)
img = tf.image.decode_png(raw, channels=3)
img = tf.image.central_crop(img, 0.9)
img = tf.image.rgb_to_grayscale(img)
img = tf.image.grayscale_to_rgb(img)
# img = tf.image.adjust_brightness(img, 0.2)
img = tf.image.adjust_contrast(img, 1.5)
# img = tf.image.adjust_gamma(img, 1.75)
img = tf.clip_by_value(img, 0, 255)
# img = tf.bitwise.invert(img)
img = tf.image.resize(img, [224, 224])
img = tf.image.random_flip_left_right(img)
return img, label
import pandas as pd
import os
import tensorflow as tf
from sklearn.preprocessing import LabelEncoder
from keras.utils import np_utils
train_csv = pd.read_csv("../input/murav11-zip/mura.csv")
filenames = []
labels = []
for i, row in train_csv.iterrows():
filename = os.path.join("../input/murav11-zip/MURA-v1.1", row[1])
filenames.append(filename)
label = row[2]
labels.append(label)
encoder = LabelEncoder()
labels = encoder.fit_transform(labels)
labels = np_utils.to_categorical(labels)
dataset = tf.data.Dataset.from_tensor_slices((filenames, labels))
dataset = dataset.shuffle(len(filenames))
dataset = dataset.map(parse, num_parallel_calls=1)
dataset = dataset.batch(64)
dataset = dataset.prefetch(1)
validation_csv = pd.read_csv("../input/murav11-zip/mura_valid.csv")
validation_filenames = []
validation_labels = []
for i, row in validation_csv.iterrows():
filename = os.path.join("../input/murav11-zip/MURA-v1.1", row[1])
validation_filenames.append(filename)
label = row[2]
validation_labels.append(label)
encoder = LabelEncoder()
validation_labels = encoder.fit_transform(validation_labels)
validation_labels = np_utils.to_categorical(validation_labels)
validation_dataset = tf.data.Dataset.from_tensor_slices(
(validation_filenames, validation_labels)
)
validation_dataset = validation_dataset.shuffle(len(validation_filenames))
validation_dataset = validation_dataset.map(parse, num_parallel_calls=1)
validation_dataset = validation_dataset.batch(64)
validation_dataset = validation_dataset.prefetch(1)
from tensorflow.keras.applications import EfficientNetB0
from tensorflow.keras.layers import (
Dense,
Dropout,
Flatten,
GaussianNoise,
GlobalAveragePooling2D,
)
import tensorflow.keras.optimizers as optim
from tensorflow.keras.models import Sequential, Model
enet = EfficientNetB0(include_top=False, input_shape=(224, 224, 3), weights="imagenet")
outputs = enet.layers[-1].output
output = GlobalAveragePooling2D()(outputs)
enet = Model(enet.input, outputs=output)
convnet = Sequential()
convnet.add(enet)
convnet.add(GaussianNoise(0.2))
convnet.add(Dense(256, activation="relu", input_dim=(224, 224, 3)))
convnet.add(Dropout(0.3))
convnet.add(Dense(256, activation="relu"))
convnet.add(Dropout(0.3))
convnet.add(Dense(2, activation="softmax"))
convnet.summary()
convnet.compile(
loss="categorical_crossentropy",
optimizer=optim.Adam(learning_rate=5e-6),
metrics=["accuracy"],
)
for i in range(30):
print("epoch " + str(i + 1))
convnet.fit(dataset, epochs=1, verbose=1)
convnet.evaluate(validation_dataset)
convnet.evaluate(validation_dataset)
convnet.save("..output/kaggle/working/murav_weights.h5")
# import matplotlib.pyplot as plt
# for i, data in enumerate(dataset):
# img, label = data
# img = img.numpy()
# plt.imshow(img)
# break
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242791.ipynb
| null | null |
[{"Id": 69242791, "ScriptId": 18108103, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6157430, "CreationDate": "07/28/2021 14:26:16", "VersionNumber": 18.0, "Title": "mura-v1.1", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 129.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 128.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
# def parse(filename, label):
# raw = tf.io.read_file(filename)
# img = tf.image.decode_png(raw, channels=3)
# img = tf.image.central_crop(img, 0.95)
# img = tf.image.adjust_contrast(img, 3)
# img = tf.image.adjust_gamma(img, 0.5)
# img = tf.clip_by_value(img, 0, 255)
# img = tf.bitwise.invert(img)
# # img = tf.image.resize(img, [224,224])
# img = tf.image.random_flip_left_right(img)
# return img, label
# import pandas as pd
# import os
# import tensorflow as tf
# from sklearn.preprocessing import LabelEncoder
# from keras.utils import np_utils
# train_csv = pd.read_csv('../input/murav11-zip/mura.csv')
# filenames = []
# labels = []
# for i, row in train_csv.iterrows():
# filename = os.path.join('../input/murav11-zip/MURA-v1.1', row[1])
# filenames.append(filename)
# label = row[2]
# labels.append(label)
# encoder = LabelEncoder()
# labels = encoder.fit_transform(labels)
# labels = np_utils.to_categorical(labels)
# dataset = tf.data.Dataset.from_tensor_slices((filenames, labels))
# dataset = dataset.shuffle(len(filenames))
# dataset = dataset.map(parse, num_parallel_calls=1)
# import matplotlib.pyplot as plt
# for i, data in enumerate(dataset):
# img, label = data
# img = img.numpy()
# plt.imshow(img)
# break
def parse(filename, label):
raw = tf.io.read_file(filename)
img = tf.image.decode_png(raw, channels=3)
img = tf.image.central_crop(img, 0.9)
img = tf.image.rgb_to_grayscale(img)
img = tf.image.grayscale_to_rgb(img)
# img = tf.image.adjust_brightness(img, 0.2)
img = tf.image.adjust_contrast(img, 1.5)
# img = tf.image.adjust_gamma(img, 1.75)
img = tf.clip_by_value(img, 0, 255)
# img = tf.bitwise.invert(img)
img = tf.image.resize(img, [224, 224])
img = tf.image.random_flip_left_right(img)
return img, label
import pandas as pd
import os
import tensorflow as tf
from sklearn.preprocessing import LabelEncoder
from keras.utils import np_utils
train_csv = pd.read_csv("../input/murav11-zip/mura.csv")
filenames = []
labels = []
for i, row in train_csv.iterrows():
filename = os.path.join("../input/murav11-zip/MURA-v1.1", row[1])
filenames.append(filename)
label = row[2]
labels.append(label)
encoder = LabelEncoder()
labels = encoder.fit_transform(labels)
labels = np_utils.to_categorical(labels)
dataset = tf.data.Dataset.from_tensor_slices((filenames, labels))
dataset = dataset.shuffle(len(filenames))
dataset = dataset.map(parse, num_parallel_calls=1)
dataset = dataset.batch(64)
dataset = dataset.prefetch(1)
validation_csv = pd.read_csv("../input/murav11-zip/mura_valid.csv")
validation_filenames = []
validation_labels = []
for i, row in validation_csv.iterrows():
filename = os.path.join("../input/murav11-zip/MURA-v1.1", row[1])
validation_filenames.append(filename)
label = row[2]
validation_labels.append(label)
encoder = LabelEncoder()
validation_labels = encoder.fit_transform(validation_labels)
validation_labels = np_utils.to_categorical(validation_labels)
validation_dataset = tf.data.Dataset.from_tensor_slices(
(validation_filenames, validation_labels)
)
validation_dataset = validation_dataset.shuffle(len(validation_filenames))
validation_dataset = validation_dataset.map(parse, num_parallel_calls=1)
validation_dataset = validation_dataset.batch(64)
validation_dataset = validation_dataset.prefetch(1)
from tensorflow.keras.applications import EfficientNetB0
from tensorflow.keras.layers import (
Dense,
Dropout,
Flatten,
GaussianNoise,
GlobalAveragePooling2D,
)
import tensorflow.keras.optimizers as optim
from tensorflow.keras.models import Sequential, Model
enet = EfficientNetB0(include_top=False, input_shape=(224, 224, 3), weights="imagenet")
outputs = enet.layers[-1].output
output = GlobalAveragePooling2D()(outputs)
enet = Model(enet.input, outputs=output)
convnet = Sequential()
convnet.add(enet)
convnet.add(GaussianNoise(0.2))
convnet.add(Dense(256, activation="relu", input_dim=(224, 224, 3)))
convnet.add(Dropout(0.3))
convnet.add(Dense(256, activation="relu"))
convnet.add(Dropout(0.3))
convnet.add(Dense(2, activation="softmax"))
convnet.summary()
convnet.compile(
loss="categorical_crossentropy",
optimizer=optim.Adam(learning_rate=5e-6),
metrics=["accuracy"],
)
for i in range(30):
print("epoch " + str(i + 1))
convnet.fit(dataset, epochs=1, verbose=1)
convnet.evaluate(validation_dataset)
convnet.evaluate(validation_dataset)
convnet.save("..output/kaggle/working/murav_weights.h5")
# import matplotlib.pyplot as plt
# for i, data in enumerate(dataset):
# img, label = data
# img = img.numpy()
# plt.imshow(img)
# break
| false | 0 | 1,580 | 0 | 1,580 | 1,580 |
||
69242869
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
train = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
test = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
train.head(5)
train.isnull().sum()
train = train.fillna(" missing ")
train
train["final_text"] = train["keyword"] + train["location"] + train["text"]
train = train.drop(columns=["keyword", "location", "text"])
train
train["final_text"] = train["final_text"].str.lower()
train
#!python -m spacy download en_core_web_lg
#!python -m spacy link en_core_web_lg en_core_web_lg_link
import spacy
import en_core_web_lg
nlp = spacy.load("en_core_web_lg")
def data_nlp_lemma(data_nlp):
"""
This function obtains the list of summaries and changes them to NLP. Then, we obtain
the lemma of each words and create a string out of it. After that, we remove the english
stopwords and return a list of the remaining words.
:param data_nlp: String containing all summaries from a company's webscapper result.
:return: list
"""
data_nlp = nlp(str(data_nlp))
data = [token.lemma_ for token in data_nlp]
data = " ".join(data)
chain = nlp(data)
chain = [
word
for word in chain
if word.is_alpha == True and word.is_space == False and word.is_stop == False
]
return str(chain)
train["lemmatized"] = train["final_text"].apply(lambda x: data_nlp_lemma(x))
train_lemmatized = train
train_lemmatized["lemmatized"] = train_lemmatized["lemmatized"].apply(
lambda x: [y for y in x if len(y) > 2]
)
# train['lemmatized'] = train['lemmatized'].apply(lambda x: [y for y in x if y != 'miss'])
train_lemmatized
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242869.ipynb
| null | null |
[{"Id": 69242869, "ScriptId": 18885741, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 5774883, "CreationDate": "07/28/2021 14:27:20", "VersionNumber": 1.0, "Title": "nlp-disaster-tweet", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 69.0, "LinesInsertedFromPrevious": 69.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
| null | null | null | null |
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
train = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
test = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
train.head(5)
train.isnull().sum()
train = train.fillna(" missing ")
train
train["final_text"] = train["keyword"] + train["location"] + train["text"]
train = train.drop(columns=["keyword", "location", "text"])
train
train["final_text"] = train["final_text"].str.lower()
train
#!python -m spacy download en_core_web_lg
#!python -m spacy link en_core_web_lg en_core_web_lg_link
import spacy
import en_core_web_lg
nlp = spacy.load("en_core_web_lg")
def data_nlp_lemma(data_nlp):
"""
This function obtains the list of summaries and changes them to NLP. Then, we obtain
the lemma of each words and create a string out of it. After that, we remove the english
stopwords and return a list of the remaining words.
:param data_nlp: String containing all summaries from a company's webscapper result.
:return: list
"""
data_nlp = nlp(str(data_nlp))
data = [token.lemma_ for token in data_nlp]
data = " ".join(data)
chain = nlp(data)
chain = [
word
for word in chain
if word.is_alpha == True and word.is_space == False and word.is_stop == False
]
return str(chain)
train["lemmatized"] = train["final_text"].apply(lambda x: data_nlp_lemma(x))
train_lemmatized = train
train_lemmatized["lemmatized"] = train_lemmatized["lemmatized"].apply(
lambda x: [y for y in x if len(y) > 2]
)
# train['lemmatized'] = train['lemmatized'].apply(lambda x: [y for y in x if y != 'miss'])
train_lemmatized
| false | 0 | 695 | 0 | 695 | 695 |
||
69242657
|
<jupyter_start><jupyter_text>Default of Credit Card Clients Dataset
## Dataset Information
This dataset contains information on default payments, demographic factors, credit data, history of payment, and bill statements of credit card clients in Taiwan from April 2005 to September 2005.
## Content
There are 25 variables:
* **ID**: ID of each client
* **LIMIT_BAL**: Amount of given credit in NT dollars (includes individual and family/supplementary credit
* **SEX**: Gender (1=male, 2=female)
* **EDUCATION**: (1=graduate school, 2=university, 3=high school, 4=others, 5=unknown, 6=unknown)
* **MARRIAGE**: Marital status (1=married, 2=single, 3=others)
* **AGE**: Age in years
* **PAY_0**: Repayment status in September, 2005 (-1=pay duly, 1=payment delay for one month, 2=payment delay for two months, ... 8=payment delay for eight months, 9=payment delay for nine months and above)
* **PAY_2**: Repayment status in August, 2005 (scale same as above)
* **PAY_3**: Repayment status in July, 2005 (scale same as above)
* **PAY_4**: Repayment status in June, 2005 (scale same as above)
* **PAY_5**: Repayment status in May, 2005 (scale same as above)
* **PAY_6**: Repayment status in April, 2005 (scale same as above)
* **BILL_AMT1**: Amount of bill statement in September, 2005 (NT dollar)
* **BILL_AMT2**: Amount of bill statement in August, 2005 (NT dollar)
* **BILL_AMT3**: Amount of bill statement in July, 2005 (NT dollar)
* **BILL_AMT4**: Amount of bill statement in June, 2005 (NT dollar)
* **BILL_AMT5**: Amount of bill statement in May, 2005 (NT dollar)
* **BILL_AMT6**: Amount of bill statement in April, 2005 (NT dollar)
* **PAY_AMT1**: Amount of previous payment in September, 2005 (NT dollar)
* **PAY_AMT2**: Amount of previous payment in August, 2005 (NT dollar)
* **PAY_AMT3**: Amount of previous payment in July, 2005 (NT dollar)
* **PAY_AMT4**: Amount of previous payment in June, 2005 (NT dollar)
* **PAY_AMT5**: Amount of previous payment in May, 2005 (NT dollar)
* **PAY_AMT6**: Amount of previous payment in April, 2005 (NT dollar)
* **default.payment.next.month**: Default payment (1=yes, 0=no)
## Inspiration
Some ideas for exploration:
1. How does the probability of default payment vary by categories of different demographic variables?
2. Which variables are the strongest predictors of default payment?
## Acknowledgements
Any publications based on this dataset should acknowledge the following:
Lichman, M. (2013). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.
The original dataset can be found [here](https://archive.ics.uci.edu/ml/datasets/default+of+credit+card+clients) at the UCI Machine Learning Repository.
Kaggle dataset identifier: default-of-credit-card-clients-dataset
<jupyter_script># # Comparing two datasets
# The challenges of Machine Learning algorithm generalizing on new data is based on the assumption that, the trends that the model has learnt on the traning data will be seen on the new datasets as well. This is not usually the case for real world problems, which leads to the generalization error and performance deterioration overtime. Specfically, these two use cases can benefit from knowing what changed in the datasets
# * **Pre Implementation** - Finding out what is different between train and OOS/OOT and identifying variables with similar distributions
# * **Post Implementation** - finding out what changed between training and production time period to deep dive changes in variable distribution and affect on the performance
# In this notebook, I will explore 5 ways of doing these comparisons
# * Using seaborn violin plots to compare the distributions visually between two datasets
# * ANOVA and Tukey's test to establish whether the difference between two datasets is significant or not
# * Andrew's Curves - these curves help distinguish various observations whether any differences exist on visual inspection
# * KS Statistic to check whether the each variable in train and test comes from the same distribution
# * And finally, my favorite, Building a ML classifier and predicting which dataset it belongs to. This will throw out a quantitative measure using AUC as to how different are these datasets and specify which variables should be considered as important to change in datasets
# ### Dataset
# I am using [Default on credit card clients dataset](https://www.kaggle.com/uciml/default-of-credit-card-clients-dataset) and splitting it into train and test by sklearn library.
# ## 1. Violin Plots
# Violin plots are similar to box and whisker plots. These are being used instead because you can split the violin in two parts compare distributions in train and test side by side. You can visually see that the plots for train (light green) and fairly close to test (darker blue)
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
def PrepareData():
df = pd.read_csv(
"/kaggle/input/default-of-credit-card-clients-dataset/UCI_Credit_Card.csv"
)
# print ('Shape :',df.shape)
# print ('NULLS : \n', df.isnull().sum())
df = df.rename(columns={"default.payment.next.month": "def_pay", "PAY_0": "PAY_1"})
from sklearn.model_selection import train_test_split
features = [
"LIMIT_BAL",
"SEX",
"EDUCATION",
"MARRIAGE",
"AGE",
"PAY_1",
"PAY_2",
"PAY_3",
"PAY_4",
"PAY_5",
"PAY_6",
"BILL_AMT1",
"BILL_AMT2",
"BILL_AMT3",
"BILL_AMT4",
"BILL_AMT5",
"BILL_AMT6",
"PAY_AMT1",
"PAY_AMT2",
"PAY_AMT3",
"PAY_AMT4",
"PAY_AMT5",
"PAY_AMT6",
]
X = df[features].copy()
y = df["def_pay"]
# print ('Train Test Split')
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.20, random_state=42
)
# print ('X_train Shape :', X_train.shape)
# print ('X_test Shape :', X_test.shape)
# print ('Event Rate Train : ', y_train.mean())
# print ('Event Rate Test : ', y_test.mean())
return X_train, X_test, y_train, y_test
# prepare data
X_train, X_test, y_train, y_test = PrepareData()
X_train["dataset"] = "TRAIN"
X_test["dataset"] = "TEST"
data = X_train.append(X_test)
# create subplots
f, axes = plt.subplots(4, 6, figsize=(30, 15))
ax0 = axes.ravel()[0]
# set ticks to non
for ax in axes.ravel():
ax.set_xticks([])
ax.set_yticks([])
# iterate over all variables and generate violin plot
for i, col in enumerate(X_train.drop("SEX", axis=1).columns[:-1]):
ax = axes.ravel()[i]
g = sns.violinplot(
x="SEX",
data=data,
ax=ax,
y=col,
hue="dataset",
split=True,
palette=["#78e08f", "#0a3d62"],
)
ax.set_title(col, fontsize=14, color="#0a3d62", fontfamily="monospace")
ax.set_ylabel("")
ax.set_xticks([])
ax.set_xlabel("")
ax.get_legend().remove()
sns.despine(top=True, right=True, left=True, bottom=True)
plt.suptitle(
"DISTRIBUTIONS BY GENDER FOR EACH VARIABLE IN TRAIN AND TEST",
fontsize=20,
color="#0a3d62",
fontweight="bold",
fontfamily="monospace",
)
plt.show()
# ## 2. Anova
# The analysis of variance statistical models were developed by the English statistician Sir R. A. Fisher and are commonly used to determine if there is a significant difference between the means of two or more data sets.
# Here we are comparing the train and test datasets. So,
# * **Null Hypothesis** - The two datasets are similar
# * **Alternate Hypothesis** - The two datsets are dissimilar
# One way anova allows us to do this comparison, and rejecting the null hypothesis means, accepting the alternate, meaning the two datasets are significantly different. The decision for rejection is based on $p$ if $p \leq \alpha$ or significance level. $\alpha$ is typically 5% i.e. 95% confidence
#
X_train["y"] = y_train
X_train["dataset"] = 1
X_test["y"] = y_test
X_test["dataset"] = 2
data2 = X_train.append(X_test)
# data2.drop('dataset', axis=1, inplace=True)
import statsmodels.api as sm
from statsmodels.formula.api import ols
from statsmodels.stats.multicomp import pairwise_tukeyhsd
from statsmodels.stats.multicomp import MultiComparison
anova_results = pd.DataFrame()
for col in X_train.columns[:-2]:
model = ols(f"{col} ~ dataset", data=data2).fit()
anova_result = sm.stats.anova_lm(model, typ=2)
anova_result["var"] = col
anova_results = anova_results.append(anova_result)
tukeys_results = pd.DataFrame()
for col in X_train.columns[:-2]:
mc = MultiComparison(data2[col], data2["dataset"])
result = mc.tukeyhsd().summary()
result_as_df = pd.read_html(result.as_html())[0]
result_as_df["var"] = col
tukeys_results = tukeys_results.append(result_as_df, ignore_index=True)
f, ax = plt.subplots(1, 1, figsize=(30, 6))
anova_results[["var", "PR(>F)"]].dropna().set_index("var")["PR(>F)"].plot(
kind="bar", ax=ax, color="#78e08f", width=0.6
)
ax.set_xticklabels(
ax.get_xticklabels(),
rotation=0,
color="#0a3d62",
fontfamily="monospace",
fontsize=12,
)
ax.set_xlabel("")
ax.set_yticks([])
for s in ["top", "right", "bottom", "left"]:
ax.spines[s].set_visible(False)
plt.axhline(y=0.05, color="#0a3d62", linestyle="--", lw=3)
ax.text(
-0.5,
1.17,
"P VALUES FOR EACH VARIABLE",
fontsize=20,
fontweight="bold",
color="#0a3d62",
fontfamily="monospace",
)
ax.text(
-0.5,
1.12,
"All P values are > 0.05 (horizontal line), i.e. for none of variables are significantly different between train and test",
fontsize=15,
color="#0a3d62",
fontfamily="monospace",
)
for bar in ax.patches:
ax.annotate(
format(bar.get_height(), ".2f"),
(bar.get_x() + bar.get_width() / 2, bar.get_height()),
ha="center",
va="bottom",
size=15,
xytext=(0, 8),
color="#0a3d62",
textcoords="offset points",
)
plt.show()
# ## 3. Andrew's Curves
# Andrews curves are used for visualizing high-dimensional data by mapping each observation onto a function. It preserves means, distance, and variances. It is given by formula:
# $$T(n) = \frac{x_1}{sqrt(2)} + x_2sin(n) + x_3 cos(n) + x_4 sin(2n) + x_5 cos(2n) + ...$$
# The test is completely overlayed on train curves which means there is a huge overlap in distributions of variables between train and test.
from pandas.plotting import andrews_curves
f, ax = plt.subplots(1, 1, figsize=(30, 10))
data2["dataset"] = data2.dataset.replace({1: "TRAIN", 2: "TEST"})
data3 = data2.drop("y", axis=1)
andrews_curves(data3, "dataset", ax=ax, color=["#0a3d62", "#78e08f"])
for s in ["top", "right", "bottom", "left"]:
ax.spines[s].set_visible(False)
plt.title(
"ANDREWS CURVES BY DATASET",
fontsize=20,
color="#0a3d62",
fontfamily="monospace",
fontweight="bold",
)
plt.show()
# ## 4. KS Statistic
# * Performs the two-sample Kolmogorov-Smirnov test for goodness of fit.
# * Null hypothesis states null both cumulative distributions are similar. Rejecting the null hypothesis means cumulative distributions are different.
# * This test compares the underlying continuous distributions F(x) and G(x) of two independent samples.
# * `two-sided`: The null hypothesis is that the two distributions are identical, F(x)=G(x) for all x; the alternative is that they are not identical.
# * `less`: The null hypothesis is that F(x) >= G(x) for all x; the alternative is that F(x) < G(x) for at least one x.
# * `greater`: The null hypothesis is that F(x) G(x) for at least one x.
from scipy.stats import ks_2samp
ksdf = pd.DataFrame()
alpha = 0.05
for col in X_train.columns[:-2]:
s, p = ks_2samp(X_train[col], X_test[col])
ksdf = ksdf.append(
pd.DataFrame(
{
"kstat": [s],
"pval": [p],
"variable": [col],
"reject_null_hypo": [p < alpha],
}
),
ignore_index=True,
)
f, ax = plt.subplots(1, 1, figsize=(30, 6))
ksdf[["variable", "pval"]].set_index("variable")["pval"].plot(
kind="bar", ax=ax, color="#78e08f", width=0.6
)
ax.set_xticklabels(
ax.get_xticklabels(),
rotation=0,
color="#0a3d62",
fontfamily="monospace",
fontsize=12,
)
ax.set_xlabel("")
ax.set_yticks([])
for s in ["top", "right", "bottom", "left"]:
ax.spines[s].set_visible(False)
plt.axhline(y=alpha, color="#0a3d62", linestyle="--", lw=3)
ax.text(
-0.5,
1.17,
"P VALUE FOR EACH VARIABLE",
fontsize=20,
fontweight="bold",
color="#0a3d62",
fontfamily="monospace",
)
ax.text(
-0.5,
1.12,
f"All P values are < {alpha}(horizontal line), i.e. for none of variables are significantly different between train and test",
fontsize=15,
color="#0a3d62",
fontfamily="monospace",
)
for bar in ax.patches:
ax.annotate(
format(bar.get_height(), ".2f"),
(bar.get_x() + bar.get_width() / 2, bar.get_height()),
ha="center",
va="bottom",
size=15,
xytext=(0, 8),
color="#0a3d62",
textcoords="offset points",
)
plt.show()
# ## 5. Building Model to predict train/test label
# * The objective is to build a model to predict whether an observation belongs to train or test by stacking train and test
# * The performance would be measured on AUC - higher AUC would mean higher difference between train and test and vice versa
# * The variable importance would also suggest the variables leading the differences
#
X_train["label"] = 0
X_test["label"] = 1
data5 = X_train.append(X_test)
data5.drop("dataset", axis=1, inplace=True)
print(data5.shape)
from sklearn.model_selection import train_test_split
# print ('Train Test Split')
X_train, X_test, y_train, y_test = train_test_split(
data5.drop("label", axis=1), data5.label, test_size=0.20, random_state=42
)
# print ('X_train Shape :', X_train.shape)
# print ('X_test Shape :', X_test.shape)
# print ('Event Rate Train :', round(y_train.mean(),2))
# print ('Event Rate Test :', round(y_test.mean(),2))
import xgboost as xgb
train_dm = xgb.DMatrix(data=X_train, label=y_train.values)
test_dm = xgb.DMatrix(data=X_test, label=y_test.values)
params = {
"num_boost_round": 500,
"objective": "binary:logistic",
"max_depth": 5,
"gamma": 10,
"eta": 0.01,
"min_child_weight": 10,
"verbosity": 0,
}
model = xgb.train(params, train_dm, num_boost_round=params["num_boost_round"])
train_preds = model.predict(train_dm)
test_preds = model.predict(test_dm)
from sklearn.metrics import roc_auc_score
print("TRAIN AUC :", round((roc_auc_score(y_train.values, train_preds)) * 100, 2), "%")
print("TEST AUC:", round((roc_auc_score(y_test.values, test_preds)) * 100, 2), "%")
# The AUCs are around 50%, which means the model is not able to differentiate between the two datasets, which is exactly what we need for the datasets to show in ideal scenarios, for the model to perform. We can also look at the variable importances and see which variables contribute to the differences
# #### Finding out which Variable contributes to most differences between datasets
fi = pd.DataFrame(
model.get_score(importance_type="total_gain"), index=range(1)
).T.reset_index()
fi.columns = ["variable", "total_gain"]
fi = fi.sort_values("total_gain", ascending=False)
fi["importance"] = np.sqrt(fi.total_gain) / np.sqrt(fi.total_gain.max()) * 100
fi.reset_index(drop=True)
f, ax = plt.subplots(1, 1, figsize=(30, 6))
fi[["variable", "importance"]].set_index("variable")["importance"].plot(
kind="bar", ax=ax, color="#78e08f", width=0.6
)
ax.set_xticklabels(
ax.get_xticklabels(),
rotation=0,
color="#0a3d62",
fontfamily="monospace",
fontsize=12,
)
ax.set_xlabel("")
ax.set_yticks([])
for s in ["top", "right", "bottom", "left"]:
ax.spines[s].set_visible(False)
ax.text(
-0.35,
120,
"VARIABLE IMPORTANCES",
fontsize=20,
fontweight="bold",
color="#0a3d62",
fontfamily="monospace",
)
ax.text(
-0.35,
113,
f"Top 5 variables {fi.head().variable.values.tolist()} contribute most to the differences between train and test",
fontsize=15,
color="#0a3d62",
fontfamily="monospace",
)
for bar in ax.patches:
ax.annotate(
format(bar.get_height(), ".2f"),
(bar.get_x() + bar.get_width() / 2, bar.get_height()),
ha="center",
va="bottom",
size=12,
xytext=(0, 8),
color="#0a3d62",
textcoords="offset points",
)
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242657.ipynb
|
default-of-credit-card-clients-dataset
| null |
[{"Id": 69242657, "ScriptId": 18818281, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 614469, "CreationDate": "07/28/2021 14:24:33", "VersionNumber": 7.0, "Title": "5 ways you can compare datasets", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 281.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 281.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92171850, "KernelVersionId": 69242657, "SourceDatasetVersionId": 666}]
|
[{"Id": 666, "DatasetId": 306, "DatasourceVersionId": 666, "CreatorUserId": 395512, "LicenseName": "CC0: Public Domain", "CreationDate": "11/03/2016 03:39:18", "VersionNumber": 1.0, "Title": "Default of Credit Card Clients Dataset", "Slug": "default-of-credit-card-clients-dataset", "Subtitle": "Default Payments of Credit Card Clients in Taiwan from 2005", "Description": "## Dataset Information \n\nThis dataset contains information on default payments, demographic factors, credit data, history of payment, and bill statements of credit card clients in Taiwan from April 2005 to September 2005. \n\n## Content\n\nThere are 25 variables:\n\n* **ID**: ID of each client\n* **LIMIT_BAL**: Amount of given credit in NT dollars (includes individual and family/supplementary credit\n* **SEX**: Gender (1=male, 2=female)\n* **EDUCATION**: (1=graduate school, 2=university, 3=high school, 4=others, 5=unknown, 6=unknown)\n* **MARRIAGE**: Marital status (1=married, 2=single, 3=others)\n* **AGE**: Age in years\n* **PAY_0**: Repayment status in September, 2005 (-1=pay duly, 1=payment delay for one month, 2=payment delay for two months, ... 8=payment delay for eight months, 9=payment delay for nine months and above)\n* **PAY_2**: Repayment status in August, 2005 (scale same as above)\n* **PAY_3**: Repayment status in July, 2005 (scale same as above)\n* **PAY_4**: Repayment status in June, 2005 (scale same as above)\n* **PAY_5**: Repayment status in May, 2005 (scale same as above)\n* **PAY_6**: Repayment status in April, 2005 (scale same as above)\n* **BILL_AMT1**: Amount of bill statement in September, 2005 (NT dollar)\n* **BILL_AMT2**: Amount of bill statement in August, 2005 (NT dollar)\n* **BILL_AMT3**: Amount of bill statement in July, 2005 (NT dollar)\n* **BILL_AMT4**: Amount of bill statement in June, 2005 (NT dollar)\n* **BILL_AMT5**: Amount of bill statement in May, 2005 (NT dollar)\n* **BILL_AMT6**: Amount of bill statement in April, 2005 (NT dollar)\n* **PAY_AMT1**: Amount of previous payment in September, 2005 (NT dollar)\n* **PAY_AMT2**: Amount of previous payment in August, 2005 (NT dollar)\n* **PAY_AMT3**: Amount of previous payment in July, 2005 (NT dollar)\n* **PAY_AMT4**: Amount of previous payment in June, 2005 (NT dollar)\n* **PAY_AMT5**: Amount of previous payment in May, 2005 (NT dollar)\n* **PAY_AMT6**: Amount of previous payment in April, 2005 (NT dollar)\n* **default.payment.next.month**: Default payment (1=yes, 0=no)\n\n## Inspiration\n\nSome ideas for exploration:\n\n1. How does the probability of default payment vary by categories of different demographic variables?\n2. Which variables are the strongest predictors of default payment?\n\n## Acknowledgements\n\nAny publications based on this dataset should acknowledge the following: \n\nLichman, M. (2013). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.\n\nThe original dataset can be found [here](https://archive.ics.uci.edu/ml/datasets/default+of+credit+card+clients) at the UCI Machine Learning Repository.", "VersionNotes": "Initial release", "TotalCompressedBytes": 2862995.0, "TotalUncompressedBytes": 2862995.0}]
|
[{"Id": 306, "CreatorUserId": 395512, "OwnerUserId": NaN, "OwnerOrganizationId": 7.0, "CurrentDatasetVersionId": 666.0, "CurrentDatasourceVersionId": 666.0, "ForumId": 1834, "Type": 2, "CreationDate": "11/03/2016 03:39:18", "LastActivityDate": "02/06/2018", "TotalViews": 769358, "TotalDownloads": 64255, "TotalVotes": 880, "TotalKernels": 420}]
| null |
# # Comparing two datasets
# The challenges of Machine Learning algorithm generalizing on new data is based on the assumption that, the trends that the model has learnt on the traning data will be seen on the new datasets as well. This is not usually the case for real world problems, which leads to the generalization error and performance deterioration overtime. Specfically, these two use cases can benefit from knowing what changed in the datasets
# * **Pre Implementation** - Finding out what is different between train and OOS/OOT and identifying variables with similar distributions
# * **Post Implementation** - finding out what changed between training and production time period to deep dive changes in variable distribution and affect on the performance
# In this notebook, I will explore 5 ways of doing these comparisons
# * Using seaborn violin plots to compare the distributions visually between two datasets
# * ANOVA and Tukey's test to establish whether the difference between two datasets is significant or not
# * Andrew's Curves - these curves help distinguish various observations whether any differences exist on visual inspection
# * KS Statistic to check whether the each variable in train and test comes from the same distribution
# * And finally, my favorite, Building a ML classifier and predicting which dataset it belongs to. This will throw out a quantitative measure using AUC as to how different are these datasets and specify which variables should be considered as important to change in datasets
# ### Dataset
# I am using [Default on credit card clients dataset](https://www.kaggle.com/uciml/default-of-credit-card-clients-dataset) and splitting it into train and test by sklearn library.
# ## 1. Violin Plots
# Violin plots are similar to box and whisker plots. These are being used instead because you can split the violin in two parts compare distributions in train and test side by side. You can visually see that the plots for train (light green) and fairly close to test (darker blue)
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
def PrepareData():
df = pd.read_csv(
"/kaggle/input/default-of-credit-card-clients-dataset/UCI_Credit_Card.csv"
)
# print ('Shape :',df.shape)
# print ('NULLS : \n', df.isnull().sum())
df = df.rename(columns={"default.payment.next.month": "def_pay", "PAY_0": "PAY_1"})
from sklearn.model_selection import train_test_split
features = [
"LIMIT_BAL",
"SEX",
"EDUCATION",
"MARRIAGE",
"AGE",
"PAY_1",
"PAY_2",
"PAY_3",
"PAY_4",
"PAY_5",
"PAY_6",
"BILL_AMT1",
"BILL_AMT2",
"BILL_AMT3",
"BILL_AMT4",
"BILL_AMT5",
"BILL_AMT6",
"PAY_AMT1",
"PAY_AMT2",
"PAY_AMT3",
"PAY_AMT4",
"PAY_AMT5",
"PAY_AMT6",
]
X = df[features].copy()
y = df["def_pay"]
# print ('Train Test Split')
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.20, random_state=42
)
# print ('X_train Shape :', X_train.shape)
# print ('X_test Shape :', X_test.shape)
# print ('Event Rate Train : ', y_train.mean())
# print ('Event Rate Test : ', y_test.mean())
return X_train, X_test, y_train, y_test
# prepare data
X_train, X_test, y_train, y_test = PrepareData()
X_train["dataset"] = "TRAIN"
X_test["dataset"] = "TEST"
data = X_train.append(X_test)
# create subplots
f, axes = plt.subplots(4, 6, figsize=(30, 15))
ax0 = axes.ravel()[0]
# set ticks to non
for ax in axes.ravel():
ax.set_xticks([])
ax.set_yticks([])
# iterate over all variables and generate violin plot
for i, col in enumerate(X_train.drop("SEX", axis=1).columns[:-1]):
ax = axes.ravel()[i]
g = sns.violinplot(
x="SEX",
data=data,
ax=ax,
y=col,
hue="dataset",
split=True,
palette=["#78e08f", "#0a3d62"],
)
ax.set_title(col, fontsize=14, color="#0a3d62", fontfamily="monospace")
ax.set_ylabel("")
ax.set_xticks([])
ax.set_xlabel("")
ax.get_legend().remove()
sns.despine(top=True, right=True, left=True, bottom=True)
plt.suptitle(
"DISTRIBUTIONS BY GENDER FOR EACH VARIABLE IN TRAIN AND TEST",
fontsize=20,
color="#0a3d62",
fontweight="bold",
fontfamily="monospace",
)
plt.show()
# ## 2. Anova
# The analysis of variance statistical models were developed by the English statistician Sir R. A. Fisher and are commonly used to determine if there is a significant difference between the means of two or more data sets.
# Here we are comparing the train and test datasets. So,
# * **Null Hypothesis** - The two datasets are similar
# * **Alternate Hypothesis** - The two datsets are dissimilar
# One way anova allows us to do this comparison, and rejecting the null hypothesis means, accepting the alternate, meaning the two datasets are significantly different. The decision for rejection is based on $p$ if $p \leq \alpha$ or significance level. $\alpha$ is typically 5% i.e. 95% confidence
#
X_train["y"] = y_train
X_train["dataset"] = 1
X_test["y"] = y_test
X_test["dataset"] = 2
data2 = X_train.append(X_test)
# data2.drop('dataset', axis=1, inplace=True)
import statsmodels.api as sm
from statsmodels.formula.api import ols
from statsmodels.stats.multicomp import pairwise_tukeyhsd
from statsmodels.stats.multicomp import MultiComparison
anova_results = pd.DataFrame()
for col in X_train.columns[:-2]:
model = ols(f"{col} ~ dataset", data=data2).fit()
anova_result = sm.stats.anova_lm(model, typ=2)
anova_result["var"] = col
anova_results = anova_results.append(anova_result)
tukeys_results = pd.DataFrame()
for col in X_train.columns[:-2]:
mc = MultiComparison(data2[col], data2["dataset"])
result = mc.tukeyhsd().summary()
result_as_df = pd.read_html(result.as_html())[0]
result_as_df["var"] = col
tukeys_results = tukeys_results.append(result_as_df, ignore_index=True)
f, ax = plt.subplots(1, 1, figsize=(30, 6))
anova_results[["var", "PR(>F)"]].dropna().set_index("var")["PR(>F)"].plot(
kind="bar", ax=ax, color="#78e08f", width=0.6
)
ax.set_xticklabels(
ax.get_xticklabels(),
rotation=0,
color="#0a3d62",
fontfamily="monospace",
fontsize=12,
)
ax.set_xlabel("")
ax.set_yticks([])
for s in ["top", "right", "bottom", "left"]:
ax.spines[s].set_visible(False)
plt.axhline(y=0.05, color="#0a3d62", linestyle="--", lw=3)
ax.text(
-0.5,
1.17,
"P VALUES FOR EACH VARIABLE",
fontsize=20,
fontweight="bold",
color="#0a3d62",
fontfamily="monospace",
)
ax.text(
-0.5,
1.12,
"All P values are > 0.05 (horizontal line), i.e. for none of variables are significantly different between train and test",
fontsize=15,
color="#0a3d62",
fontfamily="monospace",
)
for bar in ax.patches:
ax.annotate(
format(bar.get_height(), ".2f"),
(bar.get_x() + bar.get_width() / 2, bar.get_height()),
ha="center",
va="bottom",
size=15,
xytext=(0, 8),
color="#0a3d62",
textcoords="offset points",
)
plt.show()
# ## 3. Andrew's Curves
# Andrews curves are used for visualizing high-dimensional data by mapping each observation onto a function. It preserves means, distance, and variances. It is given by formula:
# $$T(n) = \frac{x_1}{sqrt(2)} + x_2sin(n) + x_3 cos(n) + x_4 sin(2n) + x_5 cos(2n) + ...$$
# The test is completely overlayed on train curves which means there is a huge overlap in distributions of variables between train and test.
from pandas.plotting import andrews_curves
f, ax = plt.subplots(1, 1, figsize=(30, 10))
data2["dataset"] = data2.dataset.replace({1: "TRAIN", 2: "TEST"})
data3 = data2.drop("y", axis=1)
andrews_curves(data3, "dataset", ax=ax, color=["#0a3d62", "#78e08f"])
for s in ["top", "right", "bottom", "left"]:
ax.spines[s].set_visible(False)
plt.title(
"ANDREWS CURVES BY DATASET",
fontsize=20,
color="#0a3d62",
fontfamily="monospace",
fontweight="bold",
)
plt.show()
# ## 4. KS Statistic
# * Performs the two-sample Kolmogorov-Smirnov test for goodness of fit.
# * Null hypothesis states null both cumulative distributions are similar. Rejecting the null hypothesis means cumulative distributions are different.
# * This test compares the underlying continuous distributions F(x) and G(x) of two independent samples.
# * `two-sided`: The null hypothesis is that the two distributions are identical, F(x)=G(x) for all x; the alternative is that they are not identical.
# * `less`: The null hypothesis is that F(x) >= G(x) for all x; the alternative is that F(x) < G(x) for at least one x.
# * `greater`: The null hypothesis is that F(x) G(x) for at least one x.
from scipy.stats import ks_2samp
ksdf = pd.DataFrame()
alpha = 0.05
for col in X_train.columns[:-2]:
s, p = ks_2samp(X_train[col], X_test[col])
ksdf = ksdf.append(
pd.DataFrame(
{
"kstat": [s],
"pval": [p],
"variable": [col],
"reject_null_hypo": [p < alpha],
}
),
ignore_index=True,
)
f, ax = plt.subplots(1, 1, figsize=(30, 6))
ksdf[["variable", "pval"]].set_index("variable")["pval"].plot(
kind="bar", ax=ax, color="#78e08f", width=0.6
)
ax.set_xticklabels(
ax.get_xticklabels(),
rotation=0,
color="#0a3d62",
fontfamily="monospace",
fontsize=12,
)
ax.set_xlabel("")
ax.set_yticks([])
for s in ["top", "right", "bottom", "left"]:
ax.spines[s].set_visible(False)
plt.axhline(y=alpha, color="#0a3d62", linestyle="--", lw=3)
ax.text(
-0.5,
1.17,
"P VALUE FOR EACH VARIABLE",
fontsize=20,
fontweight="bold",
color="#0a3d62",
fontfamily="monospace",
)
ax.text(
-0.5,
1.12,
f"All P values are < {alpha}(horizontal line), i.e. for none of variables are significantly different between train and test",
fontsize=15,
color="#0a3d62",
fontfamily="monospace",
)
for bar in ax.patches:
ax.annotate(
format(bar.get_height(), ".2f"),
(bar.get_x() + bar.get_width() / 2, bar.get_height()),
ha="center",
va="bottom",
size=15,
xytext=(0, 8),
color="#0a3d62",
textcoords="offset points",
)
plt.show()
# ## 5. Building Model to predict train/test label
# * The objective is to build a model to predict whether an observation belongs to train or test by stacking train and test
# * The performance would be measured on AUC - higher AUC would mean higher difference between train and test and vice versa
# * The variable importance would also suggest the variables leading the differences
#
X_train["label"] = 0
X_test["label"] = 1
data5 = X_train.append(X_test)
data5.drop("dataset", axis=1, inplace=True)
print(data5.shape)
from sklearn.model_selection import train_test_split
# print ('Train Test Split')
X_train, X_test, y_train, y_test = train_test_split(
data5.drop("label", axis=1), data5.label, test_size=0.20, random_state=42
)
# print ('X_train Shape :', X_train.shape)
# print ('X_test Shape :', X_test.shape)
# print ('Event Rate Train :', round(y_train.mean(),2))
# print ('Event Rate Test :', round(y_test.mean(),2))
import xgboost as xgb
train_dm = xgb.DMatrix(data=X_train, label=y_train.values)
test_dm = xgb.DMatrix(data=X_test, label=y_test.values)
params = {
"num_boost_round": 500,
"objective": "binary:logistic",
"max_depth": 5,
"gamma": 10,
"eta": 0.01,
"min_child_weight": 10,
"verbosity": 0,
}
model = xgb.train(params, train_dm, num_boost_round=params["num_boost_round"])
train_preds = model.predict(train_dm)
test_preds = model.predict(test_dm)
from sklearn.metrics import roc_auc_score
print("TRAIN AUC :", round((roc_auc_score(y_train.values, train_preds)) * 100, 2), "%")
print("TEST AUC:", round((roc_auc_score(y_test.values, test_preds)) * 100, 2), "%")
# The AUCs are around 50%, which means the model is not able to differentiate between the two datasets, which is exactly what we need for the datasets to show in ideal scenarios, for the model to perform. We can also look at the variable importances and see which variables contribute to the differences
# #### Finding out which Variable contributes to most differences between datasets
fi = pd.DataFrame(
model.get_score(importance_type="total_gain"), index=range(1)
).T.reset_index()
fi.columns = ["variable", "total_gain"]
fi = fi.sort_values("total_gain", ascending=False)
fi["importance"] = np.sqrt(fi.total_gain) / np.sqrt(fi.total_gain.max()) * 100
fi.reset_index(drop=True)
f, ax = plt.subplots(1, 1, figsize=(30, 6))
fi[["variable", "importance"]].set_index("variable")["importance"].plot(
kind="bar", ax=ax, color="#78e08f", width=0.6
)
ax.set_xticklabels(
ax.get_xticklabels(),
rotation=0,
color="#0a3d62",
fontfamily="monospace",
fontsize=12,
)
ax.set_xlabel("")
ax.set_yticks([])
for s in ["top", "right", "bottom", "left"]:
ax.spines[s].set_visible(False)
ax.text(
-0.35,
120,
"VARIABLE IMPORTANCES",
fontsize=20,
fontweight="bold",
color="#0a3d62",
fontfamily="monospace",
)
ax.text(
-0.35,
113,
f"Top 5 variables {fi.head().variable.values.tolist()} contribute most to the differences between train and test",
fontsize=15,
color="#0a3d62",
fontfamily="monospace",
)
for bar in ax.patches:
ax.annotate(
format(bar.get_height(), ".2f"),
(bar.get_x() + bar.get_width() / 2, bar.get_height()),
ha="center",
va="bottom",
size=12,
xytext=(0, 8),
color="#0a3d62",
textcoords="offset points",
)
plt.show()
| false | 0 | 4,424 | 0 | 5,316 | 4,424 |
||
69242412
|
<jupyter_start><jupyter_text>roberta-base
Kaggle dataset identifier: roberta-base
<jupyter_script>#!pip install transformers
# # Libraries
import numpy as np
import pandas as pd
import os
import warnings
from tqdm import tqdm
import random
import torch
from torch import nn
import torch.optim as optim
from sklearn.model_selection import StratifiedShuffleSplit, StratifiedKFold
import tokenizers
from transformers import (
RobertaModel,
RobertaForQuestionAnswering,
RobertaConfig,
RobertaTokenizer,
)
warnings.filterwarnings("ignore")
TRAIN = False
# test out basics of roberta
# https://huggingface.co/transformers/model_doc/roberta.html#robertaforquestionanswering
Test = False
if Test:
tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
text = "Jim is happy, but not me"
sent_text = "negative " + "Jim is happy, but not me"
selected_text = "happy, but not me"
text = ["I win"]
selected_text = "I win"
inputs = tokenizer(
text,
return_tensors="pt",
pad_to_max_length=True,
truncation=True,
max_length=10,
)
start_positions = torch.tensor([1])
end_positions = torch.tensor([2])
model = RobertaForQuestionAnswering.from_pretrained("roberta-base")
outputs = model(**inputs)
# outputs = model(**inputs, start_positions=start_positions, end_positions=end_positions)
loss = outputs.loss
start_scores = outputs.start_logits
end_scores = outputs.end_logits
print(inputs, "\n", outputs)
# outputs
# tokenizer.encode('negative'+"Jim is happy, but not me", return_tensors='pt')
# inputs
# outputs.start_logits.squeeze(0)
# # Seed
def seed_everything(seed_value):
random.seed(seed_value)
np.random.seed(seed_value)
torch.manual_seed(seed_value)
os.environ["PYTHONHASHSEED"] = str(seed_value)
if torch.cuda.is_available():
torch.cuda.manual_seed(seed_value)
torch.cuda.manual_seed_all(seed_value)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed = 42
seed_everything(seed)
# # Data Loader
# ### Add token_len, start_idx, end_idx to training data
#
if TRAIN:
cleaned = True
if cleaned:
train_df = pd.read_csv("input/tweet-sentiment-extraction/clean_train.csv")
else:
train_df = pd.read_csv("input/tweet-sentiment-extraction/train.csv")
train_df["text"] = train_df["text"].astype(str)
train_df["selected_text"] = train_df["selected_text"].astype(str)
if TRAIN:
if "token_len" not in train_df:
tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
def token_length(row):
texto = " " + " ".join(row.text.lower().split())
text = tokenizer(texto)["input_ids"]
return len(text)
train_df["token_len"] = train_df.apply(token_length, axis=1)
print("max train token length: ", train_df.token_len.max())
if TRAIN:
if "start_idx" not in train_df:
# token level index
tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
def find_idx(row, p_token=False):
# tokenizer should not use padding since actual length is used
texto = " " + " ".join(row.text.lower().split())
sel_to = " " + " ".join(row.selected_text.lower().split())
text = tokenizer(texto)["input_ids"]
sel_t = tokenizer(sel_to)["input_ids"]
if p_token:
print(text, "\n", sel_t)
# for very long sublist finding
# see https://stackoverflow.com/questions/7100242/python-numpy-first-occurrence-of-subarray
# we will just use rolling windows for tweet data
i = 1
while i <= len(text) - len(sel_t) + 1:
if text[i] == sel_t[1]:
# print(i, text[i:i+len(sel_t)-2], sel_t[1:len(sel_t)-1])
if text[i : i + len(sel_t) - 2] == sel_t[1 : len(sel_t) - 1]:
start_idx = i
end_idx = i + len(sel_t) - 3
return start_idx, end_idx
i += 1
# Error in selected_text, this should be corrected using character level index
# idea 1: remove incomplete words in selected_text
# idea 2: complete the words
# idea 3: remove these rows
return 0, 0
train_df["start_idx"] = train_df.apply(lambda x: find_idx(x)[0], axis=1)
train_df["end_idx"] = train_df.apply(lambda x: find_idx(x)[1], axis=1)
# =============================================================
# character level index
# def find_start(row):
# return row.text.find(row.selected_text)
# def find_end(row):
# return row.start_idx + len(row.selected_text)
# if 'start_idx' not in train_df:
# train_df['start_idx'] = train_df.apply(lambda row: row.text.find(row.selected_text), axis=1) # along column
# train_df['end_idx'] = train_df.apply(find_end, axis=1)
# ### **Error in training labels**
# ?? **convert_tokens_to_string** might solve the subwords error ??
Test = False
if Test:
error_train_df = train_df[train_df.start_idx == 0]
error_train_df
# error_train_df.to_csv('input/tweet-sentiment-extraction/error_train.csv')
print(error_train_df.iloc[0].text, "\n", error_train_df.iloc[0].selected_text)
find_idx(error_train_df.iloc[0], p_token=True)
print("**----- selected text wrong -----**")
print(error_train_df.iloc[2593].text, "\n", error_train_df.iloc[2593].selected_text)
find_idx(error_train_df.iloc[2593], p_token=True)
print("**----- selected text missing a parenthesis -----**")
# - The error data is droped because
# * the error is not a structured and there is no easy fix
# * drop 2594/27481 <10% is not hurting too much
if TRAIN:
train_df_clean = train_df[train_df.start_idx != 0]
train_df_clean.reset_index(drop=True, inplace=True)
del train_df
# ### Torch data class
class TweetDataset(torch.utils.data.Dataset):
def __init__(self, df, max_len=96):
self.df = df
self.max_len = max_len
self.labeled = "selected_text" in df
# use internet
# self.tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
# no internet
self.tokenizer = RobertaTokenizer(
vocab_file="../input/roberta-base/vocab.json",
merges_file="../input/roberta-base/merges.txt",
)
def __getitem__(self, index):
row = self.df.iloc[index]
# tokenizer should not use padding since actual length is used
text_o = " " + " ".join((row.text + " " + row.sentiment).lower().split())
data = self.tokenizer(
text_o,
return_tensors="pt",
pad_to_max_length=True,
truncation=True,
max_length=self.max_len,
)
#
# since return_tensors='pt' will produce batched result but
# dataloaders only feed in one row at a time. so we should remove
# batch dimension In order to have auto batching working properly
for key in data.keys():
data[key] = data[key].squeeze()
# if we do not require return_tensors='pt', tokenizer produce list; we need
# for key in data.keys():
# data[key] = torch.tensor(data[key])
if self.labeled:
data["token_len"] = row.token_len
data["start_idx"] = row.start_idx
data["end_idx"] = row.end_idx
"""
compute start_idx and end_idx is time consuming, so we move it
to operate after loading df, and saving as columns in df
"""
# ## old code
# sel_o = " " + " ".join(row.selected_text.lower().split())
# sel_token = self.tokenizer(sel_o,
# truncation=True,
# max_length=self.max_len)['input_ids']
# print(sel_o, '\n', sel_token)
# data['start_idx'], data['end_idx'] = self.find_idx(text_token, sel_token)
return data
# def find_idx(self, text, sel_t):
# # for very long sublist finding
# # see https://stackoverflow.com/questions/7100242/python-numpy-first-occurrence-of-subarray
# # we will just use rolling windows for tweet data
# i = 1
# while i<=len(text)-len(sel_t)+1:
# if text[i] == sel_t[1]:
# if text[i:i+len(sel_t)-2]== sel_t[1:len(sel_t)-1]:
# start_idx = i
# print(i)
# end_idx = i+len(sel_t)-3
# return start_idx, end_idx
# i+=1
def __len__(self):
return len(self.df)
# ==============================================================
# auto batching is tricky when data are in different format, we could write a
# function to replace default collate_fn
def customer_batch(data):
pass
# ==============================================================
def get_train_val_loaders(df, train_idx, val_idx, batch_size=8):
train_df = df.iloc[train_idx]
val_df = df.iloc[val_idx]
train_loader = torch.utils.data.DataLoader(
TweetDataset(train_df),
batch_size=batch_size,
# collate_fn= customer_batch,
shuffle=True,
num_workers=1,
drop_last=True,
)
val_loader = torch.utils.data.DataLoader(
TweetDataset(val_df),
batch_size=batch_size,
# collate_fn= customer_batch,
shuffle=False,
num_workers=1,
)
dataloaders_dict = {"train": train_loader, "val": val_loader}
return dataloaders_dict
# ==============================================================
def get_test_loader(df, batch_size=32):
loader = torch.utils.data.DataLoader(
TweetDataset(df),
batch_size=batch_size,
# collate_fn= customer_batch,
shuffle=False,
num_workers=1,
)
return loader
"""Test the dataloaders
"""
Test = False
i = 1
if Test:
sss = StratifiedShuffleSplit(n_splits=1, train_size=777 * 32, random_state=seed)
for train_idx, val_idx in sss.split(train_df_clean, train_df_clean.sentiment):
# print(train_idx, val_idx)
data_loader = get_train_val_loaders(
train_df_clean, train_idx, val_idx, batch_size=2
)["train"]
for data in data_loader:
if i < 2:
# print(data)
i += 1
# decode convert token ids to text
tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
print(tokenizer.decode(data["input_ids"][0][1:5]))
break
24887 / 32
# # Model
class TweetModel(nn.Module):
def __init__(self):
super(TweetModel, self).__init__()
# use internet
# self.roberta = RobertaForQuestionAnswering.from_pretrained('roberta-base')
# no internet
config = RobertaConfig.from_pretrained("../input/roberta-base/config.json")
self.roberta = RobertaForQuestionAnswering.from_pretrained(
"../input/roberta-base/pytorch_model.bin", config=config
)
# self.dropout = nn.Dropout(0.2)
# self.fc = nn.Linear(config.hidden_size, 2)
# nn.init.normal_(self.fc.weight, std=0.02)
# nn.init.normal_(self.fc.bias, 0)
def forward(self, inputs):
outputs = self.roberta(**inputs)
# x = torch.stack([hs[-1], hs[-2], hs[-3], hs[-4]])
# x = torch.mean(x, 0)
# x = self.dropout(x)
# x = self.fc(x)
# start_logits, end_logits = x.split(1, dim=-1)
# start_logits = start_logits.squeeze(-1)
# end_logits = end_logits.squeeze(-1)
return outputs.start_logits, outputs.end_logits
# # Loss Function
def loss_fn(start_logits, end_logits, start_positions, end_positions):
ce_loss = nn.CrossEntropyLoss()
# start_logits/end_logits has dimension: batch * text_length
# start_positions/end_positions : batch * 1
start_loss = ce_loss(start_logits, start_positions)
end_loss = ce_loss(end_logits, end_positions)
total_loss = start_loss + end_loss
return total_loss
def loss_fn1(start_logits, end_logits, start_positions, end_positions):
ce_loss = nn.CrossEntropyLoss()
# start_logits/end_logits has dimension: batch * text_length
# start_positions/end_positions : batch * 1
start_loss = ce_loss(start_logits, start_positions)
end_loss = ce_loss(end_logits, end_positions)
length = (end_positions - start_positions).abs().float()
# when length is large, we do not really care so much on every position, take average
total_loss = (start_loss + end_loss) / length # + 0.1* length
return total_loss
# - Jaccard distance and Binary Cross Entropy are similar
import numpy as np
import torch
import matplotlib.pyplot as plt
def jaccard_distance_loss(y_true, y_pred, smooth=1):
"""
Jaccard = (|X & Y|)/ (|X|+ |Y| - |X & Y|)
= sum(|A*B|)/(sum(|A|)+sum(|B|)-sum(|A*B|))
Jaccard_smoothed =
Ref: https://en.wikipedia.org/wiki/Jaccard_index
"""
intersection = (y_true * y_pred).abs().sum(dim=1)
union = torch.sum(y_true.abs() + y_pred.abs(), dim=1) - intersection
jac = (intersection + smooth) / (union + smooth)
return (1 - jac) * smooth
# Test and plot
y_pred = torch.from_numpy(np.array([np.arange(-10, 10 + 0.1, 0.1)]).T)
y_true = torch.from_numpy(np.zeros(y_pred.shape))
name = "jaccard_distance_loss"
loss = jaccard_distance_loss(y_true, y_pred).numpy()
plt.title(name)
plt.plot(y_pred.numpy(), loss)
plt.xlabel("abs prediction error")
plt.ylabel("loss")
plt.show()
name = "binary cross entropy"
loss = (
torch.nn.functional.binary_cross_entropy(y_true, y_pred, reduction="none")
.mean(-1)
.numpy()
)
plt.title(name)
plt.plot(y_pred.numpy(), loss)
plt.xlabel("abs prediction error")
plt.ylabel("loss")
plt.show()
# Test
print("TYPE |Almost_right |half right |extra selected |all_wrong")
y_true = torch.from_numpy(
np.array([[0, 0, 1, 0], [0, 0, 1, 0], [0, 0, 1, 0], [0, 0, 1.0, 0.0]])
)
y_pred = torch.from_numpy(
np.array([[0, 0, 0.9, 0], [0, 0, 0.1, 0], [1, 1, 1, 1], [1, 1, 0, 1]])
)
y_true = torch.from_numpy(np.array([[0, 0, 1], [0, 0, 1], [0, 0, 1], [0, 0, 1.0]]))
y_pred = torch.from_numpy(np.array([[0, 0, 0.9], [0, 0, 0.1], [1, 1, 1], [1, 1, 0]]))
r1 = jaccard_distance_loss(
y_true,
y_pred,
).numpy()
print("jaccard_distance_loss", r1)
print("jaccard_distance_loss scaled", r1 / r1.max())
assert r1[0] < r1[1]
assert r1[1] < r1[2]
r2 = (
torch.nn.functional.binary_cross_entropy(y_true, y_pred, reduction="none")
.mean(-1)
.numpy()
)
print("binary_crossentropy", r2)
print("binary_crossentropy_scaled", r2 / r2.max())
assert r2[0] < r2[1]
assert r2[1] < r2[2]
# # Evaluation Function
# - If start_idx pred > end_idx pred: we will take the entire text as selected_text
def jaccard_score(text_token_nopadding_len, start_idx, end_idx, start_pred, end_pred):
# start_logits, end_logits are logits output of model
# start_pred = np.argmax(start_logits)
# end_pred = np.argmax(end_logits)
text_len = text_token_nopadding_len
if start_pred > end_pred: # taking the whole text as selected_text
start_pred = 1
end_pred = text_len - 1
if end_idx < start_pred or end_pred < start_idx: # intersection = 0
return 0
else:
union = max(end_pred, end_idx) - min(start_pred, start_idx) + 1
intersection = min(end_pred, end_idx) - max(start_pred, start_idx) + 1
return intersection / union
Test = False
if Test:
jaccard_score(5, 1, 1, 4, 2) # 0.25
# jaccard_score(96,1,1,4,2) # 0.0105
start_logits = torch.tensor([[0, 0, 0, 0, 1]]).float()
start_idx = torch.tensor([1])
# start_pred = torch.cat((start_pred, torch.zeros(1,91)),axis=1)
ce = torch.nn.CrossEntropyLoss()
ce(start_logits, start_idx)
# when len=5, loss = 1.9048; when len=96, loss = 4.5718
# - **Note**:
# 1. jaccard_score is sensitive to total length, CrossEntropy is not sensitive.
# 2. our jaccard_score function is a fast and close approximation of the true Jaccard score (character level) used in this competetion. There would be a bit more computation if we want character level Jaccard.
# # Training Function
def train_model(
model, dataloaders_dict, criterion, optimizer, num_epochs, batch_size, filename
):
if torch.cuda.is_available():
model.cuda()
for epoch in range(num_epochs):
for phase in ["train", "val"]:
if phase == "train":
model.train()
else:
model.eval()
epoch_loss = 0.0
epoch_jaccard = 0.0
with tqdm(dataloaders_dict[phase], unit="batch") as tepoch:
tepoch.set_description(f"Epoch {epoch+1}")
for data in tepoch:
# reserve token_len, start_idx, end_idx for later loss computation
token_len = data["token_len"].numpy()
start_idx = data["start_idx"]
end_idx = data["end_idx"]
for key in ["token_len", "start_idx", "end_idx"]:
data.pop(key)
# put data in GPU
if torch.cuda.is_available():
start_idx = start_idx.cuda()
end_idx = end_idx.cuda()
for key in data.keys():
data[key] = data[key].cuda()
# training
optimizer.zero_grad()
with torch.set_grad_enabled(phase == "train"):
start_logits, end_logits = model.forward(data)
loss = criterion(start_logits, end_logits, start_idx, end_idx)
if phase == "train":
loss.backward()
optimizer.step()
epoch_loss += loss.item()
# Jaccard score
# torch.argmax(torch.tensor([[0,0,0,0,1],[0,0,0,1.5,1]]), dim=1)
start_pred = (
torch.argmax(start_logits, dim=1).cpu().detach().numpy()
)
end_pred = (
torch.argmax(end_logits, dim=1).cpu().detach().numpy()
)
start_idx = start_idx.cpu().detach().numpy()
end_idx = end_idx.cpu().detach().numpy()
for i in range(batch_size): # or range(token_len.shape[0])
jaccard = jaccard_score(
token_len[i],
start_idx[i],
end_idx[i],
start_pred[i],
end_pred[i],
)
epoch_jaccard += jaccard
tepoch.set_postfix(loss=loss.item() / batch_size)
epoch_loss = epoch_loss / len(dataloaders_dict[phase].dataset)
epoch_jaccard = epoch_jaccard / len(dataloaders_dict[phase].dataset)
print(
"Epoch {}/{} | {:^5} | Loss: {:.4f} | Jaccard: {:.4f}".format(
epoch + 1, num_epochs, phase, epoch_loss, epoch_jaccard
)
)
torch.save(model.state_dict(), filename)
# # Training
num_epochs = 5
batch_size = 32
skf = StratifiedKFold(n_splits=5, shuffle=True, random_state=seed)
torch.cuda.empty_cache()
if TRAIN:
# Each fold takes 7* epochs = 35 mins,
split_fold = False
if split_fold:
for fold, (train_idx, val_idx) in enumerate(
skf.split(train_df_clean, train_df_clean.sentiment), start=1
):
print(f"Fold: {fold}")
model = TweetModel()
optimizer = optim.AdamW(model.parameters(), lr=3e-5, betas=(0.9, 0.999))
criterion = loss_fn
dataloaders_dict = get_train_val_loaders(
train_df_clean, train_idx, val_idx, batch_size
)
train_model(
model,
dataloaders_dict,
criterion,
optimizer,
num_epochs,
batch_size,
f"roberta_fold{fold}.pth",
)
# - We see a increase in validation loss after 2 epochs. So we only train 2 epochs on the full data
# ### run on the full training data
torch.cuda.empty_cache()
if TRAIN:
num_epochs = 2
batch_size = 32
split_fold = False
if not split_fold:
sss = StratifiedShuffleSplit(n_splits=1, train_size=776 * 32, random_state=seed)
for train_idx, val_idx in sss.split(train_df_clean, train_df_clean.sentiment):
dataloaders_dict = get_train_val_loaders(
train_df_clean, train_idx, val_idx, batch_size
)
model = TweetModel()
optimizer = optim.AdamW(model.parameters(), lr=3e-5, betas=(0.9, 0.999))
criterion = loss_fn
train_model(
model,
dataloaders_dict,
criterion,
optimizer,
num_epochs,
batch_size,
f"roberta_whole.pth",
)
if TRAIN:
del model
# train one more epoch with lower learning rate
sss = StratifiedShuffleSplit(n_splits=1, train_size=775 * 32, random_state=seed)
for train_idx, val_idx in sss.split(train_df_clean, train_df_clean.sentiment):
dataloaders_dict = get_train_val_loaders(
train_df_clean, train_idx, val_idx, batch_size
)
num_epochs = 1
model = TweetModel()
model.cuda()
model.load_state_dict(torch.load("../input/tweetextraction/roberta_whole.pth"))
optimizer = optim.AdamW(model.parameters(), lr=3e-6, betas=(0.9, 0.999))
train_model(
model,
dataloaders_dict,
criterion,
optimizer,
num_epochs,
batch_size,
f"roberta_whole2.pth",
)
# # Inference
# For Inference only
# https://huggingface.co/transformers/internal/tokenization_utils.html
test_df = pd.read_csv("../input/tweet-sentiment-extraction/test.csv")
test_df["text"] = test_df["text"].astype(str)
test_loader = get_test_loader(test_df)
model = TweetModel()
if torch.cuda.is_available():
model.cuda()
model.load_state_dict(torch.load("../input/tweetextraction/roberta_whole3.pth"))
else:
model.load_state_dict(
torch.load(
"../input/tweetextraction/roberta_whole.pth",
map_location=torch.device("cpu"),
)
)
model.eval()
predictions = []
# decode convert token ids to text
tokenizer = RobertaTokenizer(
vocab_file="../input/roberta-base/vocab.json",
merges_file="../input/roberta-base/merges.txt",
)
with tqdm(test_loader, unit="batch") as tepoch:
tepoch.set_description("Test:")
for data in tepoch:
# put data in GPU
if torch.cuda.is_available():
for key in data.keys():
data[key] = data[key].cuda()
# testing
with torch.no_grad():
start_logits, end_logits = model(data)
start_pred = torch.argmax(start_logits, dim=1)
end_pred = torch.argmax(end_logits, dim=1)
for i in range(start_pred.shape[0]): # number of rows in a batch
if start_pred[i] > end_pred[i]:
predictions.append(
" "
) # those will be replace by text after we build the dataframe
else:
sel_t = tokenizer.decode(
data["input_ids"][i][start_pred[i] : end_pred[i] + 1]
)
predictions.append(sel_t)
# # Submission
sub_df = test_df[["textID", "text"]]
sub_df["selected_text"] = predictions
def rep_text(row):
rst = row.selected_text
if (rst is " ") or (len(rst) > len(row.text)):
return row.text
if len(rst.split()) == 1:
rst = rst.replace("!!!!", "!")
rst = rst.replace("..", ".")
rst = rst.replace("...", ".")
return rst
return rst
sub_df["selected_text"] = sub_df.apply(rep_text, axis=1)
sub_df.drop(["text"], axis=1, inplace=True)
sub_df.to_csv("submission.csv", index=False)
sub_df
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242412.ipynb
|
roberta-base
|
abhishek
|
[{"Id": 69242412, "ScriptId": 18752888, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 586906, "CreationDate": "07/28/2021 14:21:05", "VersionNumber": 4.0, "Title": "Tweet sentiment extraction", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 668.0, "LinesInsertedFromPrevious": 1.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 667.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92171318, "KernelVersionId": 69242412, "SourceDatasetVersionId": 1042664}, {"Id": 92171319, "KernelVersionId": 69242412, "SourceDatasetVersionId": 2473307}]
|
[{"Id": 1042664, "DatasetId": 575905, "DatasourceVersionId": 1071866, "CreatorUserId": 5309, "LicenseName": "CC0: Public Domain", "CreationDate": "03/28/2020 21:42:54", "VersionNumber": 1.0, "Title": "roberta-base", "Slug": "roberta-base", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 0.0, "TotalUncompressedBytes": 0.0}]
|
[{"Id": 575905, "CreatorUserId": 5309, "OwnerUserId": 5309.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 2795202.0, "CurrentDatasourceVersionId": 2841202.0, "ForumId": 589699, "Type": 2, "CreationDate": "03/28/2020 21:42:54", "LastActivityDate": "03/28/2020", "TotalViews": 13289, "TotalDownloads": 5845, "TotalVotes": 180, "TotalKernels": 326}]
|
[{"Id": 5309, "UserName": "abhishek", "DisplayName": "Abhishek Thakur", "RegisterDate": "01/12/2011", "PerformanceTier": 4}]
|
#!pip install transformers
# # Libraries
import numpy as np
import pandas as pd
import os
import warnings
from tqdm import tqdm
import random
import torch
from torch import nn
import torch.optim as optim
from sklearn.model_selection import StratifiedShuffleSplit, StratifiedKFold
import tokenizers
from transformers import (
RobertaModel,
RobertaForQuestionAnswering,
RobertaConfig,
RobertaTokenizer,
)
warnings.filterwarnings("ignore")
TRAIN = False
# test out basics of roberta
# https://huggingface.co/transformers/model_doc/roberta.html#robertaforquestionanswering
Test = False
if Test:
tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
text = "Jim is happy, but not me"
sent_text = "negative " + "Jim is happy, but not me"
selected_text = "happy, but not me"
text = ["I win"]
selected_text = "I win"
inputs = tokenizer(
text,
return_tensors="pt",
pad_to_max_length=True,
truncation=True,
max_length=10,
)
start_positions = torch.tensor([1])
end_positions = torch.tensor([2])
model = RobertaForQuestionAnswering.from_pretrained("roberta-base")
outputs = model(**inputs)
# outputs = model(**inputs, start_positions=start_positions, end_positions=end_positions)
loss = outputs.loss
start_scores = outputs.start_logits
end_scores = outputs.end_logits
print(inputs, "\n", outputs)
# outputs
# tokenizer.encode('negative'+"Jim is happy, but not me", return_tensors='pt')
# inputs
# outputs.start_logits.squeeze(0)
# # Seed
def seed_everything(seed_value):
random.seed(seed_value)
np.random.seed(seed_value)
torch.manual_seed(seed_value)
os.environ["PYTHONHASHSEED"] = str(seed_value)
if torch.cuda.is_available():
torch.cuda.manual_seed(seed_value)
torch.cuda.manual_seed_all(seed_value)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed = 42
seed_everything(seed)
# # Data Loader
# ### Add token_len, start_idx, end_idx to training data
#
if TRAIN:
cleaned = True
if cleaned:
train_df = pd.read_csv("input/tweet-sentiment-extraction/clean_train.csv")
else:
train_df = pd.read_csv("input/tweet-sentiment-extraction/train.csv")
train_df["text"] = train_df["text"].astype(str)
train_df["selected_text"] = train_df["selected_text"].astype(str)
if TRAIN:
if "token_len" not in train_df:
tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
def token_length(row):
texto = " " + " ".join(row.text.lower().split())
text = tokenizer(texto)["input_ids"]
return len(text)
train_df["token_len"] = train_df.apply(token_length, axis=1)
print("max train token length: ", train_df.token_len.max())
if TRAIN:
if "start_idx" not in train_df:
# token level index
tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
def find_idx(row, p_token=False):
# tokenizer should not use padding since actual length is used
texto = " " + " ".join(row.text.lower().split())
sel_to = " " + " ".join(row.selected_text.lower().split())
text = tokenizer(texto)["input_ids"]
sel_t = tokenizer(sel_to)["input_ids"]
if p_token:
print(text, "\n", sel_t)
# for very long sublist finding
# see https://stackoverflow.com/questions/7100242/python-numpy-first-occurrence-of-subarray
# we will just use rolling windows for tweet data
i = 1
while i <= len(text) - len(sel_t) + 1:
if text[i] == sel_t[1]:
# print(i, text[i:i+len(sel_t)-2], sel_t[1:len(sel_t)-1])
if text[i : i + len(sel_t) - 2] == sel_t[1 : len(sel_t) - 1]:
start_idx = i
end_idx = i + len(sel_t) - 3
return start_idx, end_idx
i += 1
# Error in selected_text, this should be corrected using character level index
# idea 1: remove incomplete words in selected_text
# idea 2: complete the words
# idea 3: remove these rows
return 0, 0
train_df["start_idx"] = train_df.apply(lambda x: find_idx(x)[0], axis=1)
train_df["end_idx"] = train_df.apply(lambda x: find_idx(x)[1], axis=1)
# =============================================================
# character level index
# def find_start(row):
# return row.text.find(row.selected_text)
# def find_end(row):
# return row.start_idx + len(row.selected_text)
# if 'start_idx' not in train_df:
# train_df['start_idx'] = train_df.apply(lambda row: row.text.find(row.selected_text), axis=1) # along column
# train_df['end_idx'] = train_df.apply(find_end, axis=1)
# ### **Error in training labels**
# ?? **convert_tokens_to_string** might solve the subwords error ??
Test = False
if Test:
error_train_df = train_df[train_df.start_idx == 0]
error_train_df
# error_train_df.to_csv('input/tweet-sentiment-extraction/error_train.csv')
print(error_train_df.iloc[0].text, "\n", error_train_df.iloc[0].selected_text)
find_idx(error_train_df.iloc[0], p_token=True)
print("**----- selected text wrong -----**")
print(error_train_df.iloc[2593].text, "\n", error_train_df.iloc[2593].selected_text)
find_idx(error_train_df.iloc[2593], p_token=True)
print("**----- selected text missing a parenthesis -----**")
# - The error data is droped because
# * the error is not a structured and there is no easy fix
# * drop 2594/27481 <10% is not hurting too much
if TRAIN:
train_df_clean = train_df[train_df.start_idx != 0]
train_df_clean.reset_index(drop=True, inplace=True)
del train_df
# ### Torch data class
class TweetDataset(torch.utils.data.Dataset):
def __init__(self, df, max_len=96):
self.df = df
self.max_len = max_len
self.labeled = "selected_text" in df
# use internet
# self.tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
# no internet
self.tokenizer = RobertaTokenizer(
vocab_file="../input/roberta-base/vocab.json",
merges_file="../input/roberta-base/merges.txt",
)
def __getitem__(self, index):
row = self.df.iloc[index]
# tokenizer should not use padding since actual length is used
text_o = " " + " ".join((row.text + " " + row.sentiment).lower().split())
data = self.tokenizer(
text_o,
return_tensors="pt",
pad_to_max_length=True,
truncation=True,
max_length=self.max_len,
)
#
# since return_tensors='pt' will produce batched result but
# dataloaders only feed in one row at a time. so we should remove
# batch dimension In order to have auto batching working properly
for key in data.keys():
data[key] = data[key].squeeze()
# if we do not require return_tensors='pt', tokenizer produce list; we need
# for key in data.keys():
# data[key] = torch.tensor(data[key])
if self.labeled:
data["token_len"] = row.token_len
data["start_idx"] = row.start_idx
data["end_idx"] = row.end_idx
"""
compute start_idx and end_idx is time consuming, so we move it
to operate after loading df, and saving as columns in df
"""
# ## old code
# sel_o = " " + " ".join(row.selected_text.lower().split())
# sel_token = self.tokenizer(sel_o,
# truncation=True,
# max_length=self.max_len)['input_ids']
# print(sel_o, '\n', sel_token)
# data['start_idx'], data['end_idx'] = self.find_idx(text_token, sel_token)
return data
# def find_idx(self, text, sel_t):
# # for very long sublist finding
# # see https://stackoverflow.com/questions/7100242/python-numpy-first-occurrence-of-subarray
# # we will just use rolling windows for tweet data
# i = 1
# while i<=len(text)-len(sel_t)+1:
# if text[i] == sel_t[1]:
# if text[i:i+len(sel_t)-2]== sel_t[1:len(sel_t)-1]:
# start_idx = i
# print(i)
# end_idx = i+len(sel_t)-3
# return start_idx, end_idx
# i+=1
def __len__(self):
return len(self.df)
# ==============================================================
# auto batching is tricky when data are in different format, we could write a
# function to replace default collate_fn
def customer_batch(data):
pass
# ==============================================================
def get_train_val_loaders(df, train_idx, val_idx, batch_size=8):
train_df = df.iloc[train_idx]
val_df = df.iloc[val_idx]
train_loader = torch.utils.data.DataLoader(
TweetDataset(train_df),
batch_size=batch_size,
# collate_fn= customer_batch,
shuffle=True,
num_workers=1,
drop_last=True,
)
val_loader = torch.utils.data.DataLoader(
TweetDataset(val_df),
batch_size=batch_size,
# collate_fn= customer_batch,
shuffle=False,
num_workers=1,
)
dataloaders_dict = {"train": train_loader, "val": val_loader}
return dataloaders_dict
# ==============================================================
def get_test_loader(df, batch_size=32):
loader = torch.utils.data.DataLoader(
TweetDataset(df),
batch_size=batch_size,
# collate_fn= customer_batch,
shuffle=False,
num_workers=1,
)
return loader
"""Test the dataloaders
"""
Test = False
i = 1
if Test:
sss = StratifiedShuffleSplit(n_splits=1, train_size=777 * 32, random_state=seed)
for train_idx, val_idx in sss.split(train_df_clean, train_df_clean.sentiment):
# print(train_idx, val_idx)
data_loader = get_train_val_loaders(
train_df_clean, train_idx, val_idx, batch_size=2
)["train"]
for data in data_loader:
if i < 2:
# print(data)
i += 1
# decode convert token ids to text
tokenizer = RobertaTokenizer.from_pretrained("roberta-base")
print(tokenizer.decode(data["input_ids"][0][1:5]))
break
24887 / 32
# # Model
class TweetModel(nn.Module):
def __init__(self):
super(TweetModel, self).__init__()
# use internet
# self.roberta = RobertaForQuestionAnswering.from_pretrained('roberta-base')
# no internet
config = RobertaConfig.from_pretrained("../input/roberta-base/config.json")
self.roberta = RobertaForQuestionAnswering.from_pretrained(
"../input/roberta-base/pytorch_model.bin", config=config
)
# self.dropout = nn.Dropout(0.2)
# self.fc = nn.Linear(config.hidden_size, 2)
# nn.init.normal_(self.fc.weight, std=0.02)
# nn.init.normal_(self.fc.bias, 0)
def forward(self, inputs):
outputs = self.roberta(**inputs)
# x = torch.stack([hs[-1], hs[-2], hs[-3], hs[-4]])
# x = torch.mean(x, 0)
# x = self.dropout(x)
# x = self.fc(x)
# start_logits, end_logits = x.split(1, dim=-1)
# start_logits = start_logits.squeeze(-1)
# end_logits = end_logits.squeeze(-1)
return outputs.start_logits, outputs.end_logits
# # Loss Function
def loss_fn(start_logits, end_logits, start_positions, end_positions):
ce_loss = nn.CrossEntropyLoss()
# start_logits/end_logits has dimension: batch * text_length
# start_positions/end_positions : batch * 1
start_loss = ce_loss(start_logits, start_positions)
end_loss = ce_loss(end_logits, end_positions)
total_loss = start_loss + end_loss
return total_loss
def loss_fn1(start_logits, end_logits, start_positions, end_positions):
ce_loss = nn.CrossEntropyLoss()
# start_logits/end_logits has dimension: batch * text_length
# start_positions/end_positions : batch * 1
start_loss = ce_loss(start_logits, start_positions)
end_loss = ce_loss(end_logits, end_positions)
length = (end_positions - start_positions).abs().float()
# when length is large, we do not really care so much on every position, take average
total_loss = (start_loss + end_loss) / length # + 0.1* length
return total_loss
# - Jaccard distance and Binary Cross Entropy are similar
import numpy as np
import torch
import matplotlib.pyplot as plt
def jaccard_distance_loss(y_true, y_pred, smooth=1):
"""
Jaccard = (|X & Y|)/ (|X|+ |Y| - |X & Y|)
= sum(|A*B|)/(sum(|A|)+sum(|B|)-sum(|A*B|))
Jaccard_smoothed =
Ref: https://en.wikipedia.org/wiki/Jaccard_index
"""
intersection = (y_true * y_pred).abs().sum(dim=1)
union = torch.sum(y_true.abs() + y_pred.abs(), dim=1) - intersection
jac = (intersection + smooth) / (union + smooth)
return (1 - jac) * smooth
# Test and plot
y_pred = torch.from_numpy(np.array([np.arange(-10, 10 + 0.1, 0.1)]).T)
y_true = torch.from_numpy(np.zeros(y_pred.shape))
name = "jaccard_distance_loss"
loss = jaccard_distance_loss(y_true, y_pred).numpy()
plt.title(name)
plt.plot(y_pred.numpy(), loss)
plt.xlabel("abs prediction error")
plt.ylabel("loss")
plt.show()
name = "binary cross entropy"
loss = (
torch.nn.functional.binary_cross_entropy(y_true, y_pred, reduction="none")
.mean(-1)
.numpy()
)
plt.title(name)
plt.plot(y_pred.numpy(), loss)
plt.xlabel("abs prediction error")
plt.ylabel("loss")
plt.show()
# Test
print("TYPE |Almost_right |half right |extra selected |all_wrong")
y_true = torch.from_numpy(
np.array([[0, 0, 1, 0], [0, 0, 1, 0], [0, 0, 1, 0], [0, 0, 1.0, 0.0]])
)
y_pred = torch.from_numpy(
np.array([[0, 0, 0.9, 0], [0, 0, 0.1, 0], [1, 1, 1, 1], [1, 1, 0, 1]])
)
y_true = torch.from_numpy(np.array([[0, 0, 1], [0, 0, 1], [0, 0, 1], [0, 0, 1.0]]))
y_pred = torch.from_numpy(np.array([[0, 0, 0.9], [0, 0, 0.1], [1, 1, 1], [1, 1, 0]]))
r1 = jaccard_distance_loss(
y_true,
y_pred,
).numpy()
print("jaccard_distance_loss", r1)
print("jaccard_distance_loss scaled", r1 / r1.max())
assert r1[0] < r1[1]
assert r1[1] < r1[2]
r2 = (
torch.nn.functional.binary_cross_entropy(y_true, y_pred, reduction="none")
.mean(-1)
.numpy()
)
print("binary_crossentropy", r2)
print("binary_crossentropy_scaled", r2 / r2.max())
assert r2[0] < r2[1]
assert r2[1] < r2[2]
# # Evaluation Function
# - If start_idx pred > end_idx pred: we will take the entire text as selected_text
def jaccard_score(text_token_nopadding_len, start_idx, end_idx, start_pred, end_pred):
# start_logits, end_logits are logits output of model
# start_pred = np.argmax(start_logits)
# end_pred = np.argmax(end_logits)
text_len = text_token_nopadding_len
if start_pred > end_pred: # taking the whole text as selected_text
start_pred = 1
end_pred = text_len - 1
if end_idx < start_pred or end_pred < start_idx: # intersection = 0
return 0
else:
union = max(end_pred, end_idx) - min(start_pred, start_idx) + 1
intersection = min(end_pred, end_idx) - max(start_pred, start_idx) + 1
return intersection / union
Test = False
if Test:
jaccard_score(5, 1, 1, 4, 2) # 0.25
# jaccard_score(96,1,1,4,2) # 0.0105
start_logits = torch.tensor([[0, 0, 0, 0, 1]]).float()
start_idx = torch.tensor([1])
# start_pred = torch.cat((start_pred, torch.zeros(1,91)),axis=1)
ce = torch.nn.CrossEntropyLoss()
ce(start_logits, start_idx)
# when len=5, loss = 1.9048; when len=96, loss = 4.5718
# - **Note**:
# 1. jaccard_score is sensitive to total length, CrossEntropy is not sensitive.
# 2. our jaccard_score function is a fast and close approximation of the true Jaccard score (character level) used in this competetion. There would be a bit more computation if we want character level Jaccard.
# # Training Function
def train_model(
model, dataloaders_dict, criterion, optimizer, num_epochs, batch_size, filename
):
if torch.cuda.is_available():
model.cuda()
for epoch in range(num_epochs):
for phase in ["train", "val"]:
if phase == "train":
model.train()
else:
model.eval()
epoch_loss = 0.0
epoch_jaccard = 0.0
with tqdm(dataloaders_dict[phase], unit="batch") as tepoch:
tepoch.set_description(f"Epoch {epoch+1}")
for data in tepoch:
# reserve token_len, start_idx, end_idx for later loss computation
token_len = data["token_len"].numpy()
start_idx = data["start_idx"]
end_idx = data["end_idx"]
for key in ["token_len", "start_idx", "end_idx"]:
data.pop(key)
# put data in GPU
if torch.cuda.is_available():
start_idx = start_idx.cuda()
end_idx = end_idx.cuda()
for key in data.keys():
data[key] = data[key].cuda()
# training
optimizer.zero_grad()
with torch.set_grad_enabled(phase == "train"):
start_logits, end_logits = model.forward(data)
loss = criterion(start_logits, end_logits, start_idx, end_idx)
if phase == "train":
loss.backward()
optimizer.step()
epoch_loss += loss.item()
# Jaccard score
# torch.argmax(torch.tensor([[0,0,0,0,1],[0,0,0,1.5,1]]), dim=1)
start_pred = (
torch.argmax(start_logits, dim=1).cpu().detach().numpy()
)
end_pred = (
torch.argmax(end_logits, dim=1).cpu().detach().numpy()
)
start_idx = start_idx.cpu().detach().numpy()
end_idx = end_idx.cpu().detach().numpy()
for i in range(batch_size): # or range(token_len.shape[0])
jaccard = jaccard_score(
token_len[i],
start_idx[i],
end_idx[i],
start_pred[i],
end_pred[i],
)
epoch_jaccard += jaccard
tepoch.set_postfix(loss=loss.item() / batch_size)
epoch_loss = epoch_loss / len(dataloaders_dict[phase].dataset)
epoch_jaccard = epoch_jaccard / len(dataloaders_dict[phase].dataset)
print(
"Epoch {}/{} | {:^5} | Loss: {:.4f} | Jaccard: {:.4f}".format(
epoch + 1, num_epochs, phase, epoch_loss, epoch_jaccard
)
)
torch.save(model.state_dict(), filename)
# # Training
num_epochs = 5
batch_size = 32
skf = StratifiedKFold(n_splits=5, shuffle=True, random_state=seed)
torch.cuda.empty_cache()
if TRAIN:
# Each fold takes 7* epochs = 35 mins,
split_fold = False
if split_fold:
for fold, (train_idx, val_idx) in enumerate(
skf.split(train_df_clean, train_df_clean.sentiment), start=1
):
print(f"Fold: {fold}")
model = TweetModel()
optimizer = optim.AdamW(model.parameters(), lr=3e-5, betas=(0.9, 0.999))
criterion = loss_fn
dataloaders_dict = get_train_val_loaders(
train_df_clean, train_idx, val_idx, batch_size
)
train_model(
model,
dataloaders_dict,
criterion,
optimizer,
num_epochs,
batch_size,
f"roberta_fold{fold}.pth",
)
# - We see a increase in validation loss after 2 epochs. So we only train 2 epochs on the full data
# ### run on the full training data
torch.cuda.empty_cache()
if TRAIN:
num_epochs = 2
batch_size = 32
split_fold = False
if not split_fold:
sss = StratifiedShuffleSplit(n_splits=1, train_size=776 * 32, random_state=seed)
for train_idx, val_idx in sss.split(train_df_clean, train_df_clean.sentiment):
dataloaders_dict = get_train_val_loaders(
train_df_clean, train_idx, val_idx, batch_size
)
model = TweetModel()
optimizer = optim.AdamW(model.parameters(), lr=3e-5, betas=(0.9, 0.999))
criterion = loss_fn
train_model(
model,
dataloaders_dict,
criterion,
optimizer,
num_epochs,
batch_size,
f"roberta_whole.pth",
)
if TRAIN:
del model
# train one more epoch with lower learning rate
sss = StratifiedShuffleSplit(n_splits=1, train_size=775 * 32, random_state=seed)
for train_idx, val_idx in sss.split(train_df_clean, train_df_clean.sentiment):
dataloaders_dict = get_train_val_loaders(
train_df_clean, train_idx, val_idx, batch_size
)
num_epochs = 1
model = TweetModel()
model.cuda()
model.load_state_dict(torch.load("../input/tweetextraction/roberta_whole.pth"))
optimizer = optim.AdamW(model.parameters(), lr=3e-6, betas=(0.9, 0.999))
train_model(
model,
dataloaders_dict,
criterion,
optimizer,
num_epochs,
batch_size,
f"roberta_whole2.pth",
)
# # Inference
# For Inference only
# https://huggingface.co/transformers/internal/tokenization_utils.html
test_df = pd.read_csv("../input/tweet-sentiment-extraction/test.csv")
test_df["text"] = test_df["text"].astype(str)
test_loader = get_test_loader(test_df)
model = TweetModel()
if torch.cuda.is_available():
model.cuda()
model.load_state_dict(torch.load("../input/tweetextraction/roberta_whole3.pth"))
else:
model.load_state_dict(
torch.load(
"../input/tweetextraction/roberta_whole.pth",
map_location=torch.device("cpu"),
)
)
model.eval()
predictions = []
# decode convert token ids to text
tokenizer = RobertaTokenizer(
vocab_file="../input/roberta-base/vocab.json",
merges_file="../input/roberta-base/merges.txt",
)
with tqdm(test_loader, unit="batch") as tepoch:
tepoch.set_description("Test:")
for data in tepoch:
# put data in GPU
if torch.cuda.is_available():
for key in data.keys():
data[key] = data[key].cuda()
# testing
with torch.no_grad():
start_logits, end_logits = model(data)
start_pred = torch.argmax(start_logits, dim=1)
end_pred = torch.argmax(end_logits, dim=1)
for i in range(start_pred.shape[0]): # number of rows in a batch
if start_pred[i] > end_pred[i]:
predictions.append(
" "
) # those will be replace by text after we build the dataframe
else:
sel_t = tokenizer.decode(
data["input_ids"][i][start_pred[i] : end_pred[i] + 1]
)
predictions.append(sel_t)
# # Submission
sub_df = test_df[["textID", "text"]]
sub_df["selected_text"] = predictions
def rep_text(row):
rst = row.selected_text
if (rst is " ") or (len(rst) > len(row.text)):
return row.text
if len(rst.split()) == 1:
rst = rst.replace("!!!!", "!")
rst = rst.replace("..", ".")
rst = rst.replace("...", ".")
return rst
return rst
sub_df["selected_text"] = sub_df.apply(rep_text, axis=1)
sub_df.drop(["text"], axis=1, inplace=True)
sub_df.to_csv("submission.csv", index=False)
sub_df
| false | 1 | 7,182 | 0 | 7,205 | 7,182 |
||
69242568
|
# # Car Price prediction
# ## Прогнозирование стоимости автомобиля по характеристикам
# *Этот ноутбук является шаблоном (Baseline) к текущему соревнованию и не служит готовым решением!*
# Вы можете использовать его как основу для построения своего решения.
# > **Baseline** создается больше как шаблон, где можно посмотреть, как происходит обращение с входящими данными и что нужно получить на выходе. При этом ML начинка может быть достаточно простой. Это помогает быстрее приступить к самому ML, а не тратить ценное время на инженерные задачи.
# Также baseline является хорошей опорной точкой по метрике. Если наше решение хуже baseline - мы явно делаем что-то не так и стоит попробовать другой путь)
# ## В baseline мы сделаем следующее:
# * Построим "наивную"/baseline модель, предсказывающую цену по модели и году выпуска (с ней будем сравнивать другие модели)
# * Обработаем и отнормируем признаки
# * Сделаем первую модель на основе градиентного бустинга с помощью CatBoost
# * Сделаем вторую модель на основе нейронных сетей и сравним результаты
# * Сделаем multi-input нейронную сеть для анализа табличных данных и текста одновременно
# * Добавим в multi-input сеть обработку изображений
# * Осуществим ансамблирование градиентного бустинга и нейронной сети (усреднение их предсказаний)
# аугментации изображений
import random
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
pd.set_option("display.max_columns", 500)
import os
import sys
import PIL
import cv2
import re
from collections import Counter
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
from catboost import CatBoostRegressor
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import mean_squared_error, mean_absolute_error
# # keras
import tensorflow as tf
import tensorflow.keras.layers as L
from tensorflow.keras.models import Model, Sequential
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing import sequence
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
import albumentations
# plt
import matplotlib.pyplot as plt
# увеличим дефолтный размер графиков
from pylab import rcParams
rcParams["figure.figsize"] = 10, 5
# графики в svg выглядят более четкими
import seaborn as sns
import pymorphy2
# You can write up to 5GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
print("Python :", sys.version.split("\n")[0])
print("Numpy :", np.__version__)
print("Tensorflow :", tf.__version__)
def mape(y_true, y_pred):
return np.mean(np.abs((y_pred - y_true) / y_true))
def show_info(col):
print("Сколько пропусков? {}".format(col.isna().sum()))
print()
print("Процент пропусков? {}%".format((col.isna().sum() / len(col)) * 100))
print()
print("Описание:\n{}".format(col.describe()))
print()
print("Как распределено?\n{}".format(col.value_counts()))
print()
print("Какие значения?\n{}".format(col.unique()))
def show_info_hist(col):
print("Сколько пропусков? {}".format(col.isna().sum()))
print()
print("Процент пропусков? {}%".format((col.isna().sum() / len(col)) * 100))
print()
print("Описание:\n{}".format(col.describe()))
print()
print("Как распределено?\n{}".format(col.value_counts()))
print()
print("Какие значения?\n{}".format(col.unique()))
print()
plt.figure(figsize=(15, 6))
sns.countplot(x=col, data=data)
plt.xticks(rotation="vertical")
plt.show()
# всегда фиксируйте RANDOM_SEED, чтобы ваши эксперименты были воспроизводимы!
RANDOM_SEED = 42
np.random.seed(RANDOM_SEED)
# # DATA
# Посмотрим на типы признаков:
# * bodyType - категориальный
# * brand - категориальный
# * color - категориальный
# * description - текстовый
# * engineDisplacement - числовой, представленный как текст
# * enginePower - числовой, представленный как текст
# * fuelType - категориальный
# * mileage - числовой
# * modelDate - числовой
# * model_info - категориальный
# * name - категориальный, желательно сократить размерность
# * numberOfDoors - категориальный
# * price - числовой, целевой
# * productionDate - числовой
# * sell_id - изображение (файл доступен по адресу, основанному на sell_id)
# * vehicleConfiguration - не используется (комбинация других столбцов)
# * vehicleTransmission - категориальный
# * Владельцы - категориальный
# * Владение - числовой, представленный как текст
# * ПТС - категориальный
# * Привод - категориальный
# * Руль - категориальный
DATA_DIR = "../input/sf-dst-car-price-prediction-part2/"
DIR_TRAIN3 = "../input/auto-ru03-04-2021/"
train = pd.read_csv(DATA_DIR + "train.csv")
test = pd.read_csv(DATA_DIR + "test.csv")
train3 = pd.read_csv(DIR_TRAIN3 + "my_train.csv")
sample_submission = pd.read_csv(DATA_DIR + "sample_submission.csv")
train.info()
train.nunique()
train3.info()
train3.nunique()
train3.rename(columns={"Владение": "Владельцы"}, inplace=True)
df_train1 = train.copy()
# df_train2 = train2.copy()
df_train3 = train3.copy()
df_test = test.copy()
df_test["price"] = 0
# ВАЖНО! дря корректной обработки признаков объединяем трейн и тест в один датасет
df_train1["sample"] = 1 # помечаем где у нас трейн
# df_train2['sample'] = 1
df_train3["sample"] = 1
df_test["sample"] = 0 # помечаем где у нас тест
frames_to_concat = [df_train1, df_train3, df_test]
frames_to_concat_to_train = [df_train1, df_train3]
data1 = pd.concat(frames_to_concat)
data1_train = pd.concat(frames_to_concat_to_train)
data1.drop_duplicates(inplace=True)
data1_train.drop_duplicates(inplace=True)
# # Model 1: Создадим "наивную" модель
# Эта модель будет предсказывать среднюю цену по модели и году выпуска.
# C ней будем сравнивать другие модели.
#
# split данных
data_train, data_test = train_test_split(
train, test_size=0.15, shuffle=True, random_state=RANDOM_SEED
)
# Наивная модель
predicts = []
for index, row in pd.DataFrame(data_test[["model_info", "productionDate"]]).iterrows():
query = f"model_info == '{row[0]}' and productionDate == '{row[1]}'"
predicts.append(data_train.query(query)["price"].median())
# заполним не найденные совпадения
predicts = pd.DataFrame(predicts)
predicts = predicts.fillna(predicts.median())
# округлим
predicts = (predicts // 1000) * 1000
# оцениваем точность
print(
f"Точность наивной модели по метрике MAPE: {(mape(data_test['price'], predicts.values[:, 0]))*100:0.2f}%"
)
# # Та же наивная модель, только на большем датасете
data_train1, data_test1 = train_test_split(
data1_train, test_size=0.15, shuffle=True, random_state=RANDOM_SEED
)
# Наивная модель
predicts = []
for index, row in pd.DataFrame(data_test1[["model_info", "productionDate"]]).iterrows():
query = f"model_info == '{row[0]}' and productionDate == '{row[1]}'"
predicts.append(data_train1.query(query)["price"].median())
# заполним не найденные совпадения
predicts = pd.DataFrame(predicts)
predicts = predicts.fillna(predicts.median())
# округлим
predicts = (predicts // 1000) * 1000
# оцениваем точность
print(
f"Точность наивной модели по метрике MAPE: {(mape(data_test1['price'], predicts.values[:, 0]))*100:0.2f}%"
)
# Результат, мягко говоря, не очень. Впрочем,дополнительно парсились лишь определенные столбцы датафрейма train, а значит в объединенном файле очень много пропусков, которые наивной моделью отработались крайне неадекватно. Что же, продолжим работу с учебным датасетом
# # EDA
# Проведем быстрый анализ данных для того, чтобы понимать, сможет ли с этими данными работать наш алгоритм.
# Посмотрим, как выглядят распределения числовых признаков:
# посмотрим, как выглядят распределения числовых признаков
def visualize_distributions(titles_values_dict):
columns = min(3, len(titles_values_dict))
rows = (len(titles_values_dict) - 1) // columns + 1
fig = plt.figure(figsize=(columns * 6, rows * 4))
for i, (title, values) in enumerate(titles_values_dict.items()):
hist, bins = np.histogram(values, bins=20)
ax = fig.add_subplot(rows, columns, i + 1)
ax.bar(bins[:-1], hist, width=(bins[1] - bins[0]) * 0.7)
ax.set_title(title)
plt.show()
visualize_distributions(
{
"mileage": train["mileage"].dropna(),
"modelDate": train["modelDate"].dropna(),
"productionDate": train["productionDate"].dropna(),
}
)
# Итого:
# * CatBoost сможет работать с признаками и в таком виде, но для нейросети нужны нормированные данные.
# # PreProc Tabular Data
# используем все текстовые признаки как категориальные без предобработки
categorical_features = [
"bodyType",
"brand",
"color",
"engineDisplacement",
"enginePower",
"fuelType",
"model_info",
"name",
"numberOfDoors",
"vehicleTransmission",
"Владельцы",
"Владение",
"ПТС",
"Привод",
"Руль",
]
# используем все числовые признаки
numerical_features = ["mileage", "modelDate", "productionDate"]
# ВАЖНО! дря корректной обработки признаков объединяем трейн и тест в один датасет
train["sample"] = 1 # помечаем где у нас трейн
test["sample"] = 0 # помечаем где у нас тест
test[
"price"
] = 0 # в тесте у нас нет значения price, мы его должны предсказать, поэтому пока просто заполняем нулями
data = test.append(train, sort=False).reset_index(drop=True) # объединяем
print(train.shape, test.shape, data.shape)
# # Some preproc
data.isna().sum()
# # bodyType
show_info_hist(data.bodyType)
data["bodyType"] = data["bodyType"].apply(lambda x: re.findall(r"\w+", x)[0])
data.bodyType = data.bodyType.apply(lambda x: str(x.lower()))
other_cars = [
"лимузин",
"седан 2 дв.",
"компактвэн",
"внедорожник открытый",
"пикап двойная кабина",
"внедорожник 3 дв.",
]
data["bodyType"] = data["bodyType"].apply(lambda x: "другое" if x in other_cars else x)
data["bodyType"].value_counts().plot(kind="bar")
# # brand
show_info_hist(data.brand)
# # Color
show_info_hist(data.color)
data.columns
show_info(data.engineDisplacement)
data["engineDisplacement"] = data["engineDisplacement"].apply(
lambda x: data["engineDisplacement"].describe().top if x == "undefined LTR" else x
)
data["engineDisplacement"] = data["engineDisplacement"].apply(
lambda x: float((x.split(" ")[0]))
)
data["engineDisplacement"].hist()
data["engineDisplacement"] = data["engineDisplacement"].apply(lambda x: np.log(x + 1))
data["engineDisplacement"].hist()
# # enginePower
show_info_hist(data.enginePower)
data.enginePower = data.enginePower.apply(lambda x: int(re.findall("(\d+)", str(x))[0]))
data.enginePower.hist()
data.enginePower = data.enginePower.apply(lambda x: np.log(x + 1))
data.enginePower.hist()
data.columns
# # fuelType
show_info_hist(data.fuelType)
# # mileage
show_info(data.mileage)
data.mileage.hist()
mile25 = int(data.mileage.quantile(0.25))
mile50 = int(data.mileage.quantile(0.50))
mile75 = int(data.mileage.quantile(0.75))
print("25 квантиль:", mile25)
print("50 квантиль:", mile50)
print("75 квантиль:", mile75)
def mileage_cat(x):
if x < mile25:
x = 1
elif mile25 < x < mile50:
x = 2
elif mile50 < x < mile75:
x = 3
elif mile75 < x:
x = 4
return x
data["mileage_cat"] = data["mileage"].apply(lambda x: mileage_cat(x))
data.mileage = data.mileage.apply(lambda x: np.log(x + 1))
data.mileage.hist()
# # modelDate
show_info_hist(data.modelDate)
data["how_old"] = data["modelDate"].apply(lambda x: pd.Timestamp.today().year - x)
data["mile_per_year"] = np.exp(data["mileage"]) / data.how_old
show_info_hist(data.how_old)
data.how_old.hist()
data.how_old = data.how_old.apply(lambda x: np.log(x + 1))
data.how_old.hist()
show_info(data.mile_per_year)
data.mile_per_year.hist()
data.mile_per_year = data.mile_per_year.apply(lambda x: np.log(x + 1))
data.mile_per_year.hist(bins=30)
mod_d25 = int(data.modelDate.quantile(0.25))
mod_d50 = int(data.modelDate.quantile(0.50))
mod_d75 = int(data.modelDate.quantile(0.75))
def modelDate_cat(x):
if x < mod_d25:
x = 1
elif mod_d25 < x < mod_d50:
x = 2
elif mod_d50 < x < mod_d75:
x = 3
elif mod_d75 < x:
x = 4
return x
data["modelDate_cat"] = data["modelDate"].apply(lambda x: modelDate_cat(x))
data.modelDate = data.modelDate.apply(lambda x: np.log(2021 - x))
data.modelDate.hist()
data.columns
# # Name
show_info(data.name)
data["xdrive"] = data["name"].apply(lambda x: 1 if "xDrive" in x else 0)
data["xdrive"].value_counts()
show_info_hist(data.Владельцы)
"""Удалим"""
data.drop(["Владение"], axis=1, inplace=True)
data.columns
show_info(data["price"])
data.price.hist()
price25 = int(data.price.quantile(0.25))
price50 = int(data.price.quantile(0.50))
price75 = int(data.price.quantile(0.75))
def price_cat(x):
if x < mod_d25:
x = 1
elif price25 < x < price50:
x = 2
elif price50 < x < price75:
x = 3
elif price75 < x:
x = 4
return x
data["price_cat"] = data["price"].apply(lambda x: price_cat(x))
# data.price = data.price.apply(lambda x: np.sqrt(x+1))
# data.price.hist()
data.info()
# обновляем список категориальных
categorical_features = [
"bodyType",
"brand",
"color",
"fuelType",
"model_info",
"xdrive",
"numberOfDoors",
"vehicleTransmission",
"Владельцы",
"ПТС",
"Привод",
"Руль",
"mileage_cat",
"modelDate_cat",
"price_cat",
]
# обновляем список числовых признаков
numerical_features = [
"mileage",
"modelDate",
"enginePower",
"engineDisplacement",
"productionDate",
"how_old",
"mile_per_year",
]
corr = data[numerical_features].corr()
corr.style.background_gradient(cmap="coolwarm").set_precision(3)
data.description.head()
# Зададим переменной количество наиболее частов встречающихся слов, которое хотим оставить
N_WORDS = 400
# Приведем значения к str
data["description"] = data["description"].astype("str")
# Разбиваем reviewText на список слов, предварительно приводим текст к нижнему регистру
data["description_word"] = data["description"].apply(
lambda x: re.sub("[^\w]", " ", x.lower()).split()
)
# Создаем пустой список, в который будут добавляться все слова
all_words = []
# Добавляем слова каждой записи в общий список
for words in data.description_word:
# разбиваем текст на слова, предварительно приводим к нижнему регистру
all_words.extend(words)
# Считаем частоту слов в датасете
cnt = Counter()
for word in all_words:
cnt[word] += 1
# Оставим топ N_WORDS слов
top_words = []
for i in range(0, len(cnt.most_common(N_WORDS))):
words = cnt.most_common(N_WORDS)[i][0]
top_words.append(words)
# Удаляем дубликаты из all_words
all_words = list(dict.fromkeys(all_words))
print("Всего слов ", len(all_words))
print("Топ", N_WORDS, "слов: ", top_words)
# Видно,что добавленные фичи имеют большую корреляцию с данными. Нехорошо, но пока оставим и посмотрим.
def preproc_data(df_input):
"""includes several functions to pre-process the predictor data."""
df = df_input.copy()
# Кожа
df["saloon"] = df["description_word"].apply(
lambda x: 1 if ("кожа" or "кожаная") in x else 0
)
# Подогрев
df["heating"] = df["description_word"].apply(
lambda x: 1 if ("подогрев") in x else 0
)
# Каско
df["casko"] = df["description_word"].apply(lambda x: 1 if ("каско") in x else 0)
# USB-порт
df["usb"] = df["description_word"].apply(lambda x: 1 if ("usb") in x else 0)
# Салон
df["saloon"] = df["description_word"].apply(
lambda x: 1 if ("темный" and "салон") in x else 0
)
# Картер
df["carter"] = df["description_word"].apply(
lambda x: 1 if ("защита" and "картера") in x else 0
)
# AБС
df["ABS"] = df["description_word"].apply(
lambda x: 1 if ("антиблокировочная" and "система") in x else 0
)
# Подушка безопасности
df["airbags"] = df["description_word"].apply(
lambda x: 1 if ("подушки" and "безопасности") in x else 0
)
# Иммобилайзер
df["immob"] = df["description_word"].apply(
lambda x: 1 if ("иммобилайзер") in x else 0
)
# Замок
df["central_locking"] = df["description_word"].apply(
lambda x: 1 if ("центральный" and "замок") in x else 0
)
# Компьютер
df["on_board_computer"] = df["description_word"].apply(
lambda x: 1 if ("бортовой" and "компьютер") in x else 0
)
# Круиз контроль
df["cruise_control"] = df["description_word"].apply(
lambda x: 1 if ("круиз-контроль") in x else 0
)
# Климат контроль
df["climat_control"] = df["description_word"].apply(
lambda x: 1 if ("климат-контроль") in x else 0
)
# Руль
df["multi_rudder"] = df["description_word"].apply(
lambda x: 1 if ("мультифункциональный" and "руль") in x else 0
)
# Усиление руля
df["power_steering"] = df["description_word"].apply(
lambda x: 1
if ("гидроусилитель" or "гидро" or "усилитель" and "руля") in x
else 0
)
# Датчики
df["light_and_rain_sensors"] = df["description_word"].apply(
lambda x: 1 if ("датчики" and "света" and "дождя") in x else 0
)
# Карбоновый обвес
df["сarbon_body_kits"] = df["description_word"].apply(
lambda x: 1 if ("карбоновые" and "обвесы") in x else 0
)
# Диффузор
df["rear_diffuser_rkp"] = df["description_word"].apply(
lambda x: 1 if ("задний" and "диффузор") in x else 0
)
# Доводчик дверей
df["door_closers"] = df["description_word"].apply(
lambda x: 1 if ("доводчики" and "дверей") in x else 0
)
# Камера заднего вида
df["rear_view_camera"] = df["description_word"].apply(
lambda x: 1 if ("камера" or "видеокамера" and "заднего" and "вида") in x else 0
)
# amg
df["amg"] = df["description_word"].apply(lambda x: 1 if ("amg") in x else 0)
# Фары
df["bi_xenon_headlights"] = df["description_word"].apply(
lambda x: 1 if ("биксеноновые" and "фары") in x else 0
)
df["from_salon"] = df["description_word"].apply(
lambda x: 1
if ("рольф" or "панавто" or "дилер" or "кредит" or "ликвидация") in x
else 0
)
# Диски
df["alloy_wheels"] = df["description_word"].apply(
lambda x: 1 if ("легкосплавные" or "колесные" or "диски") in x else 0
)
# Парктроник
df["parking_sensors"] = df["description_word"].apply(
lambda x: 1 if ("парктроник" or "парктронник") in x else 0
)
# Повреждение поверхности
df["dents"] = df["description_word"].apply(
lambda x: 1
if ("вмятины" or "вмятина" or "царапина" or "царапины" or "трещина") in x
else 0
)
# Панорамная крыша
df["roof_with_panoramic_view"] = df["description_word"].apply(
lambda x: 1 if ("панорамная" and "крыша") in x else 0
)
# Обивка салона
df["upholstery"] = df["description_word"].apply(
lambda x: 1 if ("обивка" and "салон") in x else 0
)
# Подогрев сидения
df["heated_seat"] = df["description_word"].apply(
lambda x: 1 if ("подогрев" and "сидение") in x else 0
)
# Датчик дождя
df["rain_sensor"] = df["description_word"].apply(
lambda x: 1 if ("датчик" and "дождь") in x else 0
)
# Официальный диллер
df["official_dealer"] = df["description_word"].apply(
lambda x: 1 if ("официальный" and "диллер") in x else 0
)
# Хорошее состояние
df["good_condition"] = df["description_word"].apply(
lambda x: 1 if ("хороший" and "состояние") in x else 0
)
# Отличное состояние
df["excellent_condition"] = df["description_word"].apply(
lambda x: 1 if ("отличный" and "состояние") in x else 0
)
# ################### 1. Предобработка ##############################################################
# убираем не нужные для модели признаки
df_output = df.copy()
df_output.drop(["description", "sell_id", "description_word"], axis=1, inplace=True)
# ################### Numerical Features ##############################################################
# Далее заполняем пропуски
for column in numerical_features:
df_output[column].fillna(df_output[column].median(), inplace=True)
# тут ваш код по обработке NAN
# ....
# Нормализация данных
scaler = MinMaxScaler()
for column in numerical_features:
df_output[column] = scaler.fit_transform(df_output[[column]])[:, 0]
# ################### Categorical Features ##############################################################
# Label Encoding
for column in categorical_features:
df_output[column] = df_output[column].astype("category").cat.codes
# One-Hot Encoding: в pandas есть готовая функция - get_dummies.
df_output = pd.get_dummies(df_output, columns=categorical_features, dummy_na=False)
# тут ваш код не Encoding фитчей
# ....
# ################### Feature Engineering ####################################################
# тут ваш код не генерацию новых фитчей
# ....
# ################### Clean ####################################################
# убираем признаки которые еще не успели обработать,
df_output.drop(["vehicleConfiguration", "name"], axis=1, inplace=True)
return df_output
# Запускаем и проверяем, что получилось
df_preproc = preproc_data(data)
df_preproc.sample(5)
df_preproc.info()
# ## Split data
# Теперь выделим тестовую часть
train_data = df_preproc.query("sample == 1").drop(["sample"], axis=1)
test_data = df_preproc.query("sample == 0").drop(["sample"], axis=1)
y = train_data.price.values # наш таргет
X = train_data.drop(["price"], axis=1)
X_sub = test_data.drop(["price"], axis=1)
test_data.info()
# # Model 2: CatBoostRegressor
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.15, shuffle=True, random_state=RANDOM_SEED
)
model = CatBoostRegressor(
iterations=5000,
# depth=10,
learning_rate=0.05,
random_seed=RANDOM_SEED,
eval_metric="MAPE",
custom_metric=["RMSE", "MAE"],
od_wait=500,
# task_type='GPU',
od_type="IncToDec",
)
model.fit(
X_train,
y_train,
eval_set=(X_test, y_test),
verbose_eval=100,
use_best_model=True,
# plot=True
)
test_predict_catboost = model.predict(X_test)
print(f"TEST mape: {(mape(y_test, test_predict_catboost))*100:0.2f}%")
train_predict_catboost = model.predict(X_train)
print(f"TRAIN mape: {(mape(y_train, train_predict_catboost))*100:0.2f}%")
# ### Хорошее улучшение относительно самой простой модели
# print(f"Test MAE: {mean_absolute_error(y_test,test_predict_catboost)*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train,train_predict_catboost)*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, test_predict_catboost)*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, train_predict_catboost)*100:0.3f}%")
# ### Submission
sub_predict_catboost = model.predict(X_sub)
sample_submission["price"] = sub_predict_catboost
sample_submission.to_csv("catboost_submission.csv", index=False)
# # Model 3: Tabular NN
# Построим обычную сеть:
X_train.head(5)
# ## Simple Dense NN
model = Sequential()
model.add(
L.Dense(1024, input_dim=X_train.shape[1], activation="relu")
) # changed to 1024
model.add(L.Dropout(0.5))
model.add(L.Dense(512, activation="relu")) # added
model.add(L.Dropout(0.5)) # added
model.add(L.Dense(256, activation="relu"))
model.add(L.Dropout(0.5))
model.add(L.Dense(1, activation="linear"))
model.summary()
# Compile model
optimizer = tf.keras.optimizers.Adam(0.01)
model.compile(loss="MAPE", optimizer=optimizer, metrics=["MAPE"])
checkpoint = ModelCheckpoint(
"../working/best_model.hdf5", monitor=["val_MAPE"], verbose=0, mode="min"
)
earlystop = EarlyStopping(
monitor="val_MAPE",
patience=50,
restore_best_weights=True,
)
callbacks_list = [checkpoint, earlystop]
# ### Fit
history = model.fit(
X_train,
y_train,
batch_size=512,
epochs=500, # фактически мы обучаем пока EarlyStopping не остановит обучение
validation_data=(X_test, y_test),
callbacks=callbacks_list,
verbose=0,
)
plt.title("Loss")
plt.plot(history.history["MAPE"], label="train")
plt.plot(history.history["val_MAPE"], label="test")
plt.show()
model.load_weights("../working/best_model.hdf5")
model.save("../working/nn_1.hdf5")
test_predict_nn1 = model.predict(X_test)
print(f"TEST mape: {(mape(y_test, test_predict_nn1[:,0]))*100:0.2f}%")
train_predict_nn1 = model.predict(X_train)
print(f"TRAIN mape: {(mape(y_train, train_predict_nn1[:,0]))*100:0.2f}%")
# print(f"Test MAE: {mean_absolute_error(y_test,test_predict_nn1[:,0])*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train,train_predict_nn1[:,0])*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, test_predict_nn1[:,0])*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, train_predict_nn1[:,0])*100:0.3f}%")
sub_predict_nn1 = model.predict(X_sub)
sample_submission["price"] = sub_predict_nn1[:, 0]
sample_submission.to_csv("nn1_submission.csv", index=False)
# Рекомендации для улучшения Model 3:
# * В нейросеть желательно подавать данные с распределением, близким к нормальному, поэтому от некоторых числовых признаков имеет смысл взять логарифм перед нормализацией. Пример:
# `modelDateNorm = np.log(2020 - data['modelDate'])`
# Статья по теме: https://habr.com/ru/company/ods/blog/325422
# * Извлечение числовых значений из текста:
# Парсинг признаков 'engineDisplacement', 'enginePower', 'Владение' для извлечения числовых значений.
# * Cокращение размерности категориальных признаков
# Признак name 'name' содержит данные, которые уже есть в других столбцах ('enginePower', 'engineDisplacement', 'vehicleTransmission'), поэтому эти данные можно удалить. Затем следует еще сильнее сократить размерность, например, выделив наличие xDrive в качестве отдельного признака.
# # Model 4: NLP + Multiple Inputs
data.description.head()
data["description"] = data.description.str.replace("•", "")
data["description"] = data.description.str.replace("–", "")
data["description"] = data.description.str.replace("∙", "")
data["description"] = data.description.str.replace("☑️", "")
data["description"] = data.description.str.replace("✔", "")
data["description"] = data.description.str.replace("➥", "")
data["description"] = data.description.str.replace("●", "")
data["description"] = data.description.str.replace("☛", "")
data["description"] = data.description.str.replace("✅", "")
data["description"] = data.description.str.replace("———————————————————————————", "")
data["description"] = data.description.str.replace("•", "")
data["description"] = data["description"].str.lower()
data["description"] = data.description.str.replace(
"автомобиль в отличном состоянии", "автомобильвотличномсостоянии"
)
data["description"] = data.description.str.replace(
"машина в отличном состаянии", "автомобильвотличномсостоянии"
)
data["description"] = data.description.str.replace(
"машина в отличном состоянии", "автомобильвотличномсостоянии"
)
data["description"] = data.description.str.replace(
"продаю машину в отличном состоянии", "автомобильвотличномсостоянии"
)
data["description"] = data.description.str.replace(
"авто в идеальном состоянии", "автомобильвотличномсостоянии"
)
def lemmited(description):
description_split = description.split(" ")
for word in description_split:
p = morph.parse(word)[0]
description = description.replace(word, p.normal_form)
return description
from nltk.corpus import stopwords
# удалим стоп-слова
filtered_tokens = dict()
stop_words = stopwords.words("russian")
for word in stop_words:
data["description"] = data.description.str.replace(" " + word + " ", " ")
# леммитизация
morph = pymorphy2.MorphAnalyzer()
data["description"] = data["description"].apply(lemmited)
# TOKENIZER
# The maximum number of words to be used. (most frequent)
MAX_WORDS = 100000
# Max number of words in each complaint.
MAX_SEQUENCE_LENGTH = 256
# split данных
text_train = data.description.iloc[X_train.index]
text_test = data.description.iloc[X_test.index]
text_sub = data.description.iloc[X_sub.index]
# ### Tokenizer
tokenize = Tokenizer(num_words=MAX_WORDS)
tokenize.fit_on_texts(data.description)
tokenize.word_index
text_train_sequences = sequence.pad_sequences(
tokenize.texts_to_sequences(text_train), maxlen=MAX_SEQUENCE_LENGTH
)
text_test_sequences = sequence.pad_sequences(
tokenize.texts_to_sequences(text_test), maxlen=MAX_SEQUENCE_LENGTH
)
text_sub_sequences = sequence.pad_sequences(
tokenize.texts_to_sequences(text_sub), maxlen=MAX_SEQUENCE_LENGTH
)
print(
text_train_sequences.shape,
text_test_sequences.shape,
text_sub_sequences.shape,
)
# вот так теперь выглядит наш текст
print(text_train.iloc[6])
print(text_train_sequences[6])
# ### RNN NLP
model_nlp = Sequential()
model_nlp.add(L.Input(shape=MAX_SEQUENCE_LENGTH, name="seq_description"))
model_nlp.add(
L.Embedding(
len(tokenize.word_index) + 1,
MAX_SEQUENCE_LENGTH,
)
)
model_nlp.add(L.LSTM(512, return_sequences=True))
model_nlp.add(L.Dropout(0.5))
model_nlp.add(L.LSTM(256, return_sequences=True)) # added
model_nlp.add(L.Dropout(0.5)) # added
model_nlp.add(
L.LSTM(
128,
)
)
model_nlp.add(L.Dropout(0.25))
model_nlp.add(L.Dense(64, activation="relu"))
model_nlp.add(L.Dropout(0.25))
# ### MLP
model_mlp = Sequential()
model_mlp.add(L.Dense(512, input_dim=X_train.shape[1], activation="relu"))
model_mlp.add(L.Dropout(0.5))
model_mlp.add(L.Dense(256, activation="relu"))
model_mlp.add(L.Dropout(0.5))
model_mlp.add(L.Dense(128, activation="relu")) # added
model_mlp.add(L.Dropout(0.5)) # added
# ### Multiple Inputs NN
combinedInput = L.concatenate([model_nlp.output, model_mlp.output])
# being our regression head
head = L.Dense(64, activation="relu")(combinedInput)
head = L.Dense(1, activation="linear")(head)
model = Model(inputs=[model_nlp.input, model_mlp.input], outputs=head)
model.summary()
# ### Fit
optimizer = tf.keras.optimizers.Adam(0.01)
model.compile(loss="MAPE", optimizer=optimizer, metrics=["MAPE"])
checkpoint = ModelCheckpoint(
"../working/best_model.hdf5", monitor=["val_MAPE"], verbose=0, mode="min"
)
earlystop = EarlyStopping(
monitor="val_MAPE",
patience=10,
restore_best_weights=True,
)
callbacks_list = [checkpoint, earlystop]
history = model.fit(
[text_train_sequences, X_train],
y_train,
batch_size=512,
epochs=500, # фактически мы обучаем пока EarlyStopping не остановит обучение
validation_data=([text_test_sequences, X_test], y_test),
callbacks=callbacks_list,
)
plt.title("Loss")
plt.plot(history.history["MAPE"], label="train")
plt.plot(history.history["val_MAPE"], label="test")
plt.show()
model.load_weights("../working/best_model.hdf5")
model.save("../working/nn_mlp_nlp.hdf5")
test_predict_nn2 = model.predict([text_test_sequences, X_test])
print(f"TEST mape: {(mape(y_test, test_predict_nn2[:,0]))*100:0.2f}%")
train_predict_nn2 = model.predict([text_train_sequences, X_train])
print(f"TRAIN mape: {(mape(y_train, train_predict_nn2[:,0]))*100:0.2f}%")
# print(f"Test MAE: {mean_absolute_error(y_test,test_predict_nn2[:,0])*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train,train_predict_nn2[:,0])*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, test_predict_nn2[:,0])*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, train_predict_nn2[:,0])*100:0.3f}%")
sub_predict_nn2 = model.predict([text_sub_sequences, X_sub])
sample_submission["price"] = sub_predict_nn2[:, 0]
sample_submission.to_csv("nn2_submission.csv", index=False)
# Идеи для улучшения NLP части:
# * Выделить из описаний часто встречающиеся блоки текста, заменив их на кодовые слова или удалив
# * Сделать предобработку текста, например, сделать лемматизацию - алгоритм ставящий все слова в форму по умолчанию (глаголы в инфинитив и т. д.), чтобы токенайзер не преобразовывал разные формы слова в разные числа
# Статья по теме: https://habr.com/ru/company/Voximplant/blog/446738/
# * Поработать над алгоритмами очистки и аугментации текста
# # Model 5: Добавляем картинки
# ### Data
# убедимся, что цены и фото подгрузились верно
plt.figure(figsize=(12, 8))
random_image = train.sample(n=9)
random_image_paths = random_image["sell_id"].values
random_image_cat = random_image["price"].values
for index, path in enumerate(random_image_paths):
im = PIL.Image.open(DATA_DIR + "img/img/" + str(path) + ".jpg")
plt.subplot(3, 3, index + 1)
plt.imshow(im)
plt.title("price: " + str(random_image_cat[index]))
plt.axis("off")
plt.show()
size = (320, 240)
def get_image_array(index):
images_train = []
for index, sell_id in enumerate(data["sell_id"].iloc[index].values):
image = cv2.imread(DATA_DIR + "img/img/" + str(sell_id) + ".jpg")
assert image is not None
image = cv2.resize(image, size)
images_train.append(image)
images_train = np.array(images_train)
print("images shape", images_train.shape, "dtype", images_train.dtype)
return images_train
images_train = get_image_array(X_train.index)
images_test = get_image_array(X_test.index)
images_sub = get_image_array(X_sub.index)
# ### albumentations
from albumentations import (
HorizontalFlip,
IAAPerspective,
ShiftScaleRotate,
CLAHE,
RandomRotate90,
Transpose,
ShiftScaleRotate,
Blur,
OpticalDistortion,
GridDistortion,
HueSaturationValue,
IAAAdditiveGaussianNoise,
GaussNoise,
MotionBlur,
MedianBlur,
IAAPiecewiseAffine,
IAASharpen,
IAAEmboss,
RandomBrightnessContrast,
Flip,
OneOf,
Compose,
)
# пример взят из официальной документации: https://albumentations.readthedocs.io/en/latest/examples.html
augmentation = Compose(
[
HorizontalFlip(),
OneOf(
[
IAAAdditiveGaussianNoise(),
GaussNoise(),
],
p=0.2,
),
OneOf(
[
MotionBlur(p=0.2),
MedianBlur(blur_limit=3, p=0.1),
Blur(blur_limit=3, p=0.1),
],
p=0.2,
),
ShiftScaleRotate(shift_limit=0.0625, scale_limit=0.2, rotate_limit=15, p=1),
OneOf(
[
OpticalDistortion(p=0.3),
GridDistortion(p=0.1),
IAAPiecewiseAffine(p=0.3),
],
p=0.2,
),
OneOf(
[
CLAHE(clip_limit=2),
IAASharpen(),
IAAEmboss(),
RandomBrightnessContrast(),
],
p=0.3,
),
HueSaturationValue(p=0.3),
],
p=1,
)
# пример
plt.figure(figsize=(12, 8))
for i in range(9):
img = augmentation(image=images_train[0])["image"]
plt.subplot(3, 3, i + 1)
plt.imshow(img)
plt.axis("off")
plt.show()
def make_augmentations(images):
print("применение аугментаций", end="")
augmented_images = np.empty(images.shape)
for i in range(images.shape[0]):
if i % 200 == 0:
print(".", end="")
augment_dict = augmentation(image=images[i])
augmented_image = augment_dict["image"]
augmented_images[i] = augmented_image
print("")
return augmented_images
# ## tf.data.Dataset
# Если все изображения мы будем хранить в памяти, то может возникнуть проблема ее нехватки. Не храните все изображения в памяти целиком!
# Метод .fit() модели keras может принимать либо данные в виде массивов или тензоров, либо разного рода итераторы, из которых наиболее современным и гибким является [tf.data.Dataset](https://www.tensorflow.org/guide/data). Он представляет собой конвейер, то есть мы указываем, откуда берем данные и какую цепочку преобразований с ними выполняем. Далее мы будем работать с tf.data.Dataset.
# Dataset хранит информацию о конечном или бесконечном наборе кортежей (tuple) с данными и может возвращать эти наборы по очереди. Например, данными могут быть пары (input, target) для обучения нейросети. С данными можно осуществлять преобразования, которые осуществляются по мере необходимости ([lazy evaluation](https://ru.wikipedia.org/wiki/%D0%9B%D0%B5%D0%BD%D0%B8%D0%B2%D1%8B%D0%B5_%D0%B2%D1%8B%D1%87%D0%B8%D1%81%D0%BB%D0%B5%D0%BD%D0%B8%D1%8F)).
# `tf.data.Dataset.from_tensor_slices(data)` - создает датасет из данных, которые представляют собой либо массив, либо кортеж из массивов. Деление осуществляется по первому индексу каждого массива. Например, если `data = (np.zeros((128, 256, 256)), np.zeros(128))`, то датасет будет содержать 128 элементов, каждый из которых содержит один массив 256x256 и одно число.
# `dataset2 = dataset1.map(func)` - применение функции к датасету; функция должна принимать столько аргументов, каков размер кортежа в датасете 1 и возвращать столько, сколько нужно иметь в датасете 2. Пусть, например, датасет содержит изображения и метки, а нам нужно создать датасет только из изображений, тогда мы напишем так: `dataset2 = dataset.map(lambda img, label: img)`.
# `dataset2 = dataset1.batch(8)` - группировка по батчам; если датасет 2 должен вернуть один элемент, то он берет из датасета 1 восемь элементов, склеивает их (нулевой индекс результата - номер элемента) и возвращает.
# `dataset.__iter__()` - превращение датасета в итератор, из которого можно получать элементы методом `.__next__()`. Итератор, в отличие от самого датасета, хранит позицию текущего элемента. Можно также перебирать датасет циклом for.
# `dataset2 = dataset1.repeat(X)` - датасет 2 будет повторять датасет 1 X раз.
# Если нам нужно взять из датасета 1000 элементов и использовать их как тестовые, а остальные как обучающие, то мы напишем так:
# `test_dataset = dataset.take(1000)
# train_dataset = dataset.skip(1000)`
# Датасет по сути неизменен: такие операции, как map, batch, repeat, take, skip никак не затрагивают оригинальный датасет. Если датасет хранит элементы [1, 2, 3], то выполнив 3 раза подряд функцию dataset.take(1) мы получим 3 новых датасета, каждый из которых вернет число 1. Если же мы выполним функцию dataset.skip(1), мы получим датасет, возвращающий числа [2, 3], но исходный датасет все равно будет возвращать [1, 2, 3] каждый раз, когда мы его перебираем.
# tf.Dataset всегда выполняется в graph-режиме (в противоположность eager-режиму), поэтому либо преобразования (`.map()`) должны содержать только tensorflow-функции, либо мы должны использовать tf.py_function в качестве обертки для функций, вызываемых в `.map()`. Подробнее можно прочитать [здесь](https://www.tensorflow.org/guide/data#applying_arbitrary_python_logic).
# NLP part
tokenize = Tokenizer(num_words=MAX_WORDS)
tokenize.fit_on_texts(data.description)
def process_image(image):
return augmentation(image=image.numpy())["image"]
def tokenize_(descriptions):
return sequence.pad_sequences(
tokenize.texts_to_sequences(descriptions), maxlen=MAX_SEQUENCE_LENGTH
)
def tokenize_text(text):
return tokenize_([text.numpy().decode("utf-8")])[0]
def tf_process_train_dataset_element(image, table_data, text, price):
im_shape = image.shape
[
image,
] = tf.py_function(process_image, [image], [tf.uint8])
image.set_shape(im_shape)
[
text,
] = tf.py_function(tokenize_text, [text], [tf.int32])
return (image, table_data, text), price
def tf_process_val_dataset_element(image, table_data, text, price):
[
text,
] = tf.py_function(tokenize_text, [text], [tf.int32])
return (image, table_data, text), price
train_dataset = tf.data.Dataset.from_tensor_slices(
(images_train, X_train, data.description.iloc[X_train.index], y_train)
).map(tf_process_train_dataset_element)
test_dataset = tf.data.Dataset.from_tensor_slices(
(images_test, X_test, data.description.iloc[X_test.index], y_test)
).map(tf_process_val_dataset_element)
y_sub = np.zeros(len(X_sub))
sub_dataset = tf.data.Dataset.from_tensor_slices(
(images_sub, X_sub, data.description.iloc[X_sub.index], y_sub)
).map(tf_process_val_dataset_element)
# проверяем, что нет ошибок (не будет выброшено исключение):
train_dataset.__iter__().__next__()
test_dataset.__iter__().__next__()
sub_dataset.__iter__().__next__()
# ### Строим сверточную сеть для анализа изображений без "головы"
# нормализация включена в состав модели EfficientNetB3, поэтому на вход она принимает данные типа uint8
efficientnet_model = tf.keras.applications.efficientnet.EfficientNetB3(
weights="imagenet", include_top=False, input_shape=(size[1], size[0], 3)
)
efficientnet_output = L.GlobalAveragePooling2D()(efficientnet_model.output)
# Fine-tuning.
efficientnet_model.trainable = True
fine_tune_at = len(efficientnet_model.layers) // 2
for layer in efficientnet_model.layers[:fine_tune_at]:
layer.trainable = False
for layer in model.layers:
print(layer, layer.trainable)
# строим нейросеть для анализа табличных данных
tabular_model = Sequential(
[
L.Input(shape=X.shape[1]),
L.Dense(1024, activation="relu"),
L.Dropout(0.5),
L.Dense(512, activation="relu"),
L.Dropout(0.5),
L.Dense(256, activation="relu"),
L.Dropout(0.5),
L.Dense(128, activation="relu"), # added
L.Dropout(0.5), # added
]
)
# NLP
nlp_model = Sequential(
[
L.Input(shape=MAX_SEQUENCE_LENGTH, name="seq_description"),
L.Embedding(
len(tokenize.word_index) + 1,
MAX_SEQUENCE_LENGTH,
),
L.LSTM(256, return_sequences=True),
L.Dropout(0.5),
L.LSTM(128),
L.Dropout(0.25), # added
L.Dense(64), # added
]
)
# объединяем выходы трех нейросетей
combinedInput = L.concatenate(
[efficientnet_output, tabular_model.output, nlp_model.output]
)
# being our regression head
head = L.Dense(256, activation="relu")(combinedInput)
head = L.Dense(128, activation="relu")(combinedInput) # added
head = L.Dense(
1,
)(head)
model = Model(
inputs=[efficientnet_model.input, tabular_model.input, nlp_model.input],
outputs=head,
)
model.summary()
optimizer = tf.keras.optimizers.Adam(0.005)
model.compile(loss="MAPE", optimizer=optimizer, metrics=["MAPE"])
checkpoint = ModelCheckpoint(
"../working/best_model.hdf5", monitor=["val_MAPE"], verbose=0, mode="min"
)
earlystop = EarlyStopping(
monitor="val_MAPE",
patience=10,
restore_best_weights=True,
)
callbacks_list = [checkpoint, earlystop]
history = model.fit(
train_dataset.batch(30),
epochs=60,
validation_data=test_dataset.batch(30),
callbacks=callbacks_list,
)
plt.title("Loss")
plt.plot(history.history["MAPE"], label="train")
plt.plot(history.history["val_MAPE"], label="test")
plt.show()
model.load_weights("../working/best_model.hdf5")
model.save("../working/nn_final.hdf5")
test_predict_nn3 = model.predict(test_dataset.batch(30))
print(f"TEST mape: {(mape(y_test, test_predict_nn3[:,0]))*100:0.2f}%")
train_predict_nn3 = model.predict(train_dataset.batch(30))
print(f"TRAIN mape: {(mape(y_train, train_predict_nn3[:,0]))*100:0.2f}%")
# print(f"Test MAE: {mean_absolute_error(y_test,test_predict_nn3[:,0])*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train,train_predict_nn3[:,0])*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, test_predict_nn3[:,0])*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, train_predict_nn3[:,0])*100:0.3f}%")
sub_predict_nn3 = model.predict(sub_dataset.batch(30))
sample_submission["price"] = sub_predict_nn3[:, 0]
sample_submission.to_csv("nn3_submission.csv", index=False)
#
# #### Общие рекомендации:
# * Попробовать разные архитектуры
# * Провести более детальный анализ результатов
# * Попробовать различные подходы в управление LR и оптимизаторы
# * Поработать с таргетом
# * Использовать Fine-tuning
# #### Tabular
# * В нейросеть желательно подавать данные с распределением, близким к нормальному, поэтому от некоторых числовых признаков имеет смысл взять логарифм перед нормализацией. Пример:
# `modelDateNorm = np.log(2020 - data['modelDate'])`
# Статья по теме: https://habr.com/ru/company/ods/blog/325422
# * Извлечение числовых значений из текста:
# Парсинг признаков 'engineDisplacement', 'enginePower', 'Владение' для извлечения числовых значений.
# * Cокращение размерности категориальных признаков
# Признак name 'name' содержит данные, которые уже есть в других столбцах ('enginePower', 'engineDisplacement', 'vehicleTransmission'). Можно удалить эти данные. Затем можно еще сильнее сократить размерность, например выделив наличие xDrive в качестве отдельного признака.
# * Поработать над Feature engineering
# #### NLP
# * Выделить из описаний часто встречающиеся блоки текста, заменив их на кодовые слова или удалив
# * Сделать предобработку текста, например сделать лемматизацию - алгоритм ставящий все слова в форму по умолчанию (глаголы в инфинитив и т. д.), чтобы токенайзер не преобразовывал разные формы слова в разные числа
# Статья по теме: https://habr.com/ru/company/Voximplant/blog/446738/
# * Поработать над алгоритмами очистки и аугментации текста
# #### CV
# * Попробовать различные аугментации
# * Fine-tuning
# # Blend
blend_predict_test = (test_predict_catboost + test_predict_nn3[:, 0]) / 2
print(f"TEST mape: {(mape(y_test, blend_predict_test))*100:0.2f}%")
blend_predict_train = (train_predict_catboost + train_predict_nn3[:, 0]) / 2
print(f"TRAIN mape: {(mape(y_train, blend_predict_train))*100:0.2f}%")
# print(f"Test MAE: {mean_absolute_error(y_test,blend_predict_test)*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train, blend_predict_train)*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, blend_predict_test)*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, blend_predict_train)*100:0.3f}%")
blend_sub_predict = (sub_predict_catboost + sub_predict_nn3[:, 0]) / 2
sample_submission["price"] = blend_sub_predict
sample_submission.to_csv("blend_submission.csv", index=False)
# # Model Bonus: проброс признака
# MLP
model_mlp = Sequential()
model_mlp.add(L.Dense(512, input_dim=X_train.shape[1], activation="relu"))
model_mlp.add(L.Dropout(0.5))
model_mlp.add(L.Dense(256, activation="relu"))
model_mlp.add(L.Dropout(0.5))
# FEATURE Input
# Iput
productiondate = L.Input(shape=[1], name="productiondate")
# Embeddings layers
emb_productiondate = L.Embedding(len(X.productionDate.unique().tolist()) + 1, 20)(
productiondate
)
f_productiondate = L.Flatten()(emb_productiondate)
combinedInput = L.concatenate(
[
model_mlp.output,
f_productiondate,
]
)
# being our regression head
head = L.Dense(64, activation="relu")(combinedInput)
head = L.Dense(1, activation="linear")(head)
model = Model(inputs=[model_mlp.input, productiondate], outputs=head)
model.summary()
optimizer = tf.keras.optimizers.Adam(0.01)
model.compile(loss="MAPE", optimizer=optimizer, metrics=["MAPE"])
history = model.fit(
[X_train, X_train.productionDate.values],
y_train,
batch_size=512,
epochs=500, # фактически мы обучаем пока EarlyStopping не остановит обучение
validation_data=([X_test, X_test.productionDate.values], y_test),
callbacks=callbacks_list,
)
model.load_weights("../working/best_model.hdf5")
test_predict_nn_bonus = model.predict([X_test, X_test.productionDate.values])
print(f"TEST mape: {(mape(y_test, test_predict_nn_bonus[:,0]))*100:0.2f}%")
train_predict_nn_bonus = model.predict([X_train, X_train.productionDate.values])
print(f"TRAIN mape: {(mape(y_train, train_predict_nn_bonus[:,0]))*100:0.2f}%")
print(f"Test MAE: {mean_absolute_error(y_test,test_predict_nn_bonus[:,0])*100:0.3f}%")
print(f"Test MSE: {mean_squared_error(y_test, test_predict_nn_bonus[:,0])*100:0.3f}%")
#
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/242/69242568.ipynb
| null | null |
[{"Id": 69242568, "ScriptId": 18264170, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 6617397, "CreationDate": "07/28/2021 14:23:23", "VersionNumber": 5.0, "Title": "LarinDm DST-40", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 1265.0, "LinesInsertedFromPrevious": 218.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 1047.0, "LinesInsertedFromFork": 537.0, "LinesDeletedFromFork": 25.0, "LinesChangedFromFork": 0.0, "LinesUnchangedFromFork": 728.0, "TotalVotes": 0}]
| null | null | null | null |
# # Car Price prediction
# ## Прогнозирование стоимости автомобиля по характеристикам
# *Этот ноутбук является шаблоном (Baseline) к текущему соревнованию и не служит готовым решением!*
# Вы можете использовать его как основу для построения своего решения.
# > **Baseline** создается больше как шаблон, где можно посмотреть, как происходит обращение с входящими данными и что нужно получить на выходе. При этом ML начинка может быть достаточно простой. Это помогает быстрее приступить к самому ML, а не тратить ценное время на инженерные задачи.
# Также baseline является хорошей опорной точкой по метрике. Если наше решение хуже baseline - мы явно делаем что-то не так и стоит попробовать другой путь)
# ## В baseline мы сделаем следующее:
# * Построим "наивную"/baseline модель, предсказывающую цену по модели и году выпуска (с ней будем сравнивать другие модели)
# * Обработаем и отнормируем признаки
# * Сделаем первую модель на основе градиентного бустинга с помощью CatBoost
# * Сделаем вторую модель на основе нейронных сетей и сравним результаты
# * Сделаем multi-input нейронную сеть для анализа табличных данных и текста одновременно
# * Добавим в multi-input сеть обработку изображений
# * Осуществим ансамблирование градиентного бустинга и нейронной сети (усреднение их предсказаний)
# аугментации изображений
import random
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
pd.set_option("display.max_columns", 500)
import os
import sys
import PIL
import cv2
import re
from collections import Counter
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
from catboost import CatBoostRegressor
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import mean_squared_error, mean_absolute_error
# # keras
import tensorflow as tf
import tensorflow.keras.layers as L
from tensorflow.keras.models import Model, Sequential
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing import sequence
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
import albumentations
# plt
import matplotlib.pyplot as plt
# увеличим дефолтный размер графиков
from pylab import rcParams
rcParams["figure.figsize"] = 10, 5
# графики в svg выглядят более четкими
import seaborn as sns
import pymorphy2
# You can write up to 5GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
print("Python :", sys.version.split("\n")[0])
print("Numpy :", np.__version__)
print("Tensorflow :", tf.__version__)
def mape(y_true, y_pred):
return np.mean(np.abs((y_pred - y_true) / y_true))
def show_info(col):
print("Сколько пропусков? {}".format(col.isna().sum()))
print()
print("Процент пропусков? {}%".format((col.isna().sum() / len(col)) * 100))
print()
print("Описание:\n{}".format(col.describe()))
print()
print("Как распределено?\n{}".format(col.value_counts()))
print()
print("Какие значения?\n{}".format(col.unique()))
def show_info_hist(col):
print("Сколько пропусков? {}".format(col.isna().sum()))
print()
print("Процент пропусков? {}%".format((col.isna().sum() / len(col)) * 100))
print()
print("Описание:\n{}".format(col.describe()))
print()
print("Как распределено?\n{}".format(col.value_counts()))
print()
print("Какие значения?\n{}".format(col.unique()))
print()
plt.figure(figsize=(15, 6))
sns.countplot(x=col, data=data)
plt.xticks(rotation="vertical")
plt.show()
# всегда фиксируйте RANDOM_SEED, чтобы ваши эксперименты были воспроизводимы!
RANDOM_SEED = 42
np.random.seed(RANDOM_SEED)
# # DATA
# Посмотрим на типы признаков:
# * bodyType - категориальный
# * brand - категориальный
# * color - категориальный
# * description - текстовый
# * engineDisplacement - числовой, представленный как текст
# * enginePower - числовой, представленный как текст
# * fuelType - категориальный
# * mileage - числовой
# * modelDate - числовой
# * model_info - категориальный
# * name - категориальный, желательно сократить размерность
# * numberOfDoors - категориальный
# * price - числовой, целевой
# * productionDate - числовой
# * sell_id - изображение (файл доступен по адресу, основанному на sell_id)
# * vehicleConfiguration - не используется (комбинация других столбцов)
# * vehicleTransmission - категориальный
# * Владельцы - категориальный
# * Владение - числовой, представленный как текст
# * ПТС - категориальный
# * Привод - категориальный
# * Руль - категориальный
DATA_DIR = "../input/sf-dst-car-price-prediction-part2/"
DIR_TRAIN3 = "../input/auto-ru03-04-2021/"
train = pd.read_csv(DATA_DIR + "train.csv")
test = pd.read_csv(DATA_DIR + "test.csv")
train3 = pd.read_csv(DIR_TRAIN3 + "my_train.csv")
sample_submission = pd.read_csv(DATA_DIR + "sample_submission.csv")
train.info()
train.nunique()
train3.info()
train3.nunique()
train3.rename(columns={"Владение": "Владельцы"}, inplace=True)
df_train1 = train.copy()
# df_train2 = train2.copy()
df_train3 = train3.copy()
df_test = test.copy()
df_test["price"] = 0
# ВАЖНО! дря корректной обработки признаков объединяем трейн и тест в один датасет
df_train1["sample"] = 1 # помечаем где у нас трейн
# df_train2['sample'] = 1
df_train3["sample"] = 1
df_test["sample"] = 0 # помечаем где у нас тест
frames_to_concat = [df_train1, df_train3, df_test]
frames_to_concat_to_train = [df_train1, df_train3]
data1 = pd.concat(frames_to_concat)
data1_train = pd.concat(frames_to_concat_to_train)
data1.drop_duplicates(inplace=True)
data1_train.drop_duplicates(inplace=True)
# # Model 1: Создадим "наивную" модель
# Эта модель будет предсказывать среднюю цену по модели и году выпуска.
# C ней будем сравнивать другие модели.
#
# split данных
data_train, data_test = train_test_split(
train, test_size=0.15, shuffle=True, random_state=RANDOM_SEED
)
# Наивная модель
predicts = []
for index, row in pd.DataFrame(data_test[["model_info", "productionDate"]]).iterrows():
query = f"model_info == '{row[0]}' and productionDate == '{row[1]}'"
predicts.append(data_train.query(query)["price"].median())
# заполним не найденные совпадения
predicts = pd.DataFrame(predicts)
predicts = predicts.fillna(predicts.median())
# округлим
predicts = (predicts // 1000) * 1000
# оцениваем точность
print(
f"Точность наивной модели по метрике MAPE: {(mape(data_test['price'], predicts.values[:, 0]))*100:0.2f}%"
)
# # Та же наивная модель, только на большем датасете
data_train1, data_test1 = train_test_split(
data1_train, test_size=0.15, shuffle=True, random_state=RANDOM_SEED
)
# Наивная модель
predicts = []
for index, row in pd.DataFrame(data_test1[["model_info", "productionDate"]]).iterrows():
query = f"model_info == '{row[0]}' and productionDate == '{row[1]}'"
predicts.append(data_train1.query(query)["price"].median())
# заполним не найденные совпадения
predicts = pd.DataFrame(predicts)
predicts = predicts.fillna(predicts.median())
# округлим
predicts = (predicts // 1000) * 1000
# оцениваем точность
print(
f"Точность наивной модели по метрике MAPE: {(mape(data_test1['price'], predicts.values[:, 0]))*100:0.2f}%"
)
# Результат, мягко говоря, не очень. Впрочем,дополнительно парсились лишь определенные столбцы датафрейма train, а значит в объединенном файле очень много пропусков, которые наивной моделью отработались крайне неадекватно. Что же, продолжим работу с учебным датасетом
# # EDA
# Проведем быстрый анализ данных для того, чтобы понимать, сможет ли с этими данными работать наш алгоритм.
# Посмотрим, как выглядят распределения числовых признаков:
# посмотрим, как выглядят распределения числовых признаков
def visualize_distributions(titles_values_dict):
columns = min(3, len(titles_values_dict))
rows = (len(titles_values_dict) - 1) // columns + 1
fig = plt.figure(figsize=(columns * 6, rows * 4))
for i, (title, values) in enumerate(titles_values_dict.items()):
hist, bins = np.histogram(values, bins=20)
ax = fig.add_subplot(rows, columns, i + 1)
ax.bar(bins[:-1], hist, width=(bins[1] - bins[0]) * 0.7)
ax.set_title(title)
plt.show()
visualize_distributions(
{
"mileage": train["mileage"].dropna(),
"modelDate": train["modelDate"].dropna(),
"productionDate": train["productionDate"].dropna(),
}
)
# Итого:
# * CatBoost сможет работать с признаками и в таком виде, но для нейросети нужны нормированные данные.
# # PreProc Tabular Data
# используем все текстовые признаки как категориальные без предобработки
categorical_features = [
"bodyType",
"brand",
"color",
"engineDisplacement",
"enginePower",
"fuelType",
"model_info",
"name",
"numberOfDoors",
"vehicleTransmission",
"Владельцы",
"Владение",
"ПТС",
"Привод",
"Руль",
]
# используем все числовые признаки
numerical_features = ["mileage", "modelDate", "productionDate"]
# ВАЖНО! дря корректной обработки признаков объединяем трейн и тест в один датасет
train["sample"] = 1 # помечаем где у нас трейн
test["sample"] = 0 # помечаем где у нас тест
test[
"price"
] = 0 # в тесте у нас нет значения price, мы его должны предсказать, поэтому пока просто заполняем нулями
data = test.append(train, sort=False).reset_index(drop=True) # объединяем
print(train.shape, test.shape, data.shape)
# # Some preproc
data.isna().sum()
# # bodyType
show_info_hist(data.bodyType)
data["bodyType"] = data["bodyType"].apply(lambda x: re.findall(r"\w+", x)[0])
data.bodyType = data.bodyType.apply(lambda x: str(x.lower()))
other_cars = [
"лимузин",
"седан 2 дв.",
"компактвэн",
"внедорожник открытый",
"пикап двойная кабина",
"внедорожник 3 дв.",
]
data["bodyType"] = data["bodyType"].apply(lambda x: "другое" if x in other_cars else x)
data["bodyType"].value_counts().plot(kind="bar")
# # brand
show_info_hist(data.brand)
# # Color
show_info_hist(data.color)
data.columns
show_info(data.engineDisplacement)
data["engineDisplacement"] = data["engineDisplacement"].apply(
lambda x: data["engineDisplacement"].describe().top if x == "undefined LTR" else x
)
data["engineDisplacement"] = data["engineDisplacement"].apply(
lambda x: float((x.split(" ")[0]))
)
data["engineDisplacement"].hist()
data["engineDisplacement"] = data["engineDisplacement"].apply(lambda x: np.log(x + 1))
data["engineDisplacement"].hist()
# # enginePower
show_info_hist(data.enginePower)
data.enginePower = data.enginePower.apply(lambda x: int(re.findall("(\d+)", str(x))[0]))
data.enginePower.hist()
data.enginePower = data.enginePower.apply(lambda x: np.log(x + 1))
data.enginePower.hist()
data.columns
# # fuelType
show_info_hist(data.fuelType)
# # mileage
show_info(data.mileage)
data.mileage.hist()
mile25 = int(data.mileage.quantile(0.25))
mile50 = int(data.mileage.quantile(0.50))
mile75 = int(data.mileage.quantile(0.75))
print("25 квантиль:", mile25)
print("50 квантиль:", mile50)
print("75 квантиль:", mile75)
def mileage_cat(x):
if x < mile25:
x = 1
elif mile25 < x < mile50:
x = 2
elif mile50 < x < mile75:
x = 3
elif mile75 < x:
x = 4
return x
data["mileage_cat"] = data["mileage"].apply(lambda x: mileage_cat(x))
data.mileage = data.mileage.apply(lambda x: np.log(x + 1))
data.mileage.hist()
# # modelDate
show_info_hist(data.modelDate)
data["how_old"] = data["modelDate"].apply(lambda x: pd.Timestamp.today().year - x)
data["mile_per_year"] = np.exp(data["mileage"]) / data.how_old
show_info_hist(data.how_old)
data.how_old.hist()
data.how_old = data.how_old.apply(lambda x: np.log(x + 1))
data.how_old.hist()
show_info(data.mile_per_year)
data.mile_per_year.hist()
data.mile_per_year = data.mile_per_year.apply(lambda x: np.log(x + 1))
data.mile_per_year.hist(bins=30)
mod_d25 = int(data.modelDate.quantile(0.25))
mod_d50 = int(data.modelDate.quantile(0.50))
mod_d75 = int(data.modelDate.quantile(0.75))
def modelDate_cat(x):
if x < mod_d25:
x = 1
elif mod_d25 < x < mod_d50:
x = 2
elif mod_d50 < x < mod_d75:
x = 3
elif mod_d75 < x:
x = 4
return x
data["modelDate_cat"] = data["modelDate"].apply(lambda x: modelDate_cat(x))
data.modelDate = data.modelDate.apply(lambda x: np.log(2021 - x))
data.modelDate.hist()
data.columns
# # Name
show_info(data.name)
data["xdrive"] = data["name"].apply(lambda x: 1 if "xDrive" in x else 0)
data["xdrive"].value_counts()
show_info_hist(data.Владельцы)
"""Удалим"""
data.drop(["Владение"], axis=1, inplace=True)
data.columns
show_info(data["price"])
data.price.hist()
price25 = int(data.price.quantile(0.25))
price50 = int(data.price.quantile(0.50))
price75 = int(data.price.quantile(0.75))
def price_cat(x):
if x < mod_d25:
x = 1
elif price25 < x < price50:
x = 2
elif price50 < x < price75:
x = 3
elif price75 < x:
x = 4
return x
data["price_cat"] = data["price"].apply(lambda x: price_cat(x))
# data.price = data.price.apply(lambda x: np.sqrt(x+1))
# data.price.hist()
data.info()
# обновляем список категориальных
categorical_features = [
"bodyType",
"brand",
"color",
"fuelType",
"model_info",
"xdrive",
"numberOfDoors",
"vehicleTransmission",
"Владельцы",
"ПТС",
"Привод",
"Руль",
"mileage_cat",
"modelDate_cat",
"price_cat",
]
# обновляем список числовых признаков
numerical_features = [
"mileage",
"modelDate",
"enginePower",
"engineDisplacement",
"productionDate",
"how_old",
"mile_per_year",
]
corr = data[numerical_features].corr()
corr.style.background_gradient(cmap="coolwarm").set_precision(3)
data.description.head()
# Зададим переменной количество наиболее частов встречающихся слов, которое хотим оставить
N_WORDS = 400
# Приведем значения к str
data["description"] = data["description"].astype("str")
# Разбиваем reviewText на список слов, предварительно приводим текст к нижнему регистру
data["description_word"] = data["description"].apply(
lambda x: re.sub("[^\w]", " ", x.lower()).split()
)
# Создаем пустой список, в который будут добавляться все слова
all_words = []
# Добавляем слова каждой записи в общий список
for words in data.description_word:
# разбиваем текст на слова, предварительно приводим к нижнему регистру
all_words.extend(words)
# Считаем частоту слов в датасете
cnt = Counter()
for word in all_words:
cnt[word] += 1
# Оставим топ N_WORDS слов
top_words = []
for i in range(0, len(cnt.most_common(N_WORDS))):
words = cnt.most_common(N_WORDS)[i][0]
top_words.append(words)
# Удаляем дубликаты из all_words
all_words = list(dict.fromkeys(all_words))
print("Всего слов ", len(all_words))
print("Топ", N_WORDS, "слов: ", top_words)
# Видно,что добавленные фичи имеют большую корреляцию с данными. Нехорошо, но пока оставим и посмотрим.
def preproc_data(df_input):
"""includes several functions to pre-process the predictor data."""
df = df_input.copy()
# Кожа
df["saloon"] = df["description_word"].apply(
lambda x: 1 if ("кожа" or "кожаная") in x else 0
)
# Подогрев
df["heating"] = df["description_word"].apply(
lambda x: 1 if ("подогрев") in x else 0
)
# Каско
df["casko"] = df["description_word"].apply(lambda x: 1 if ("каско") in x else 0)
# USB-порт
df["usb"] = df["description_word"].apply(lambda x: 1 if ("usb") in x else 0)
# Салон
df["saloon"] = df["description_word"].apply(
lambda x: 1 if ("темный" and "салон") in x else 0
)
# Картер
df["carter"] = df["description_word"].apply(
lambda x: 1 if ("защита" and "картера") in x else 0
)
# AБС
df["ABS"] = df["description_word"].apply(
lambda x: 1 if ("антиблокировочная" and "система") in x else 0
)
# Подушка безопасности
df["airbags"] = df["description_word"].apply(
lambda x: 1 if ("подушки" and "безопасности") in x else 0
)
# Иммобилайзер
df["immob"] = df["description_word"].apply(
lambda x: 1 if ("иммобилайзер") in x else 0
)
# Замок
df["central_locking"] = df["description_word"].apply(
lambda x: 1 if ("центральный" and "замок") in x else 0
)
# Компьютер
df["on_board_computer"] = df["description_word"].apply(
lambda x: 1 if ("бортовой" and "компьютер") in x else 0
)
# Круиз контроль
df["cruise_control"] = df["description_word"].apply(
lambda x: 1 if ("круиз-контроль") in x else 0
)
# Климат контроль
df["climat_control"] = df["description_word"].apply(
lambda x: 1 if ("климат-контроль") in x else 0
)
# Руль
df["multi_rudder"] = df["description_word"].apply(
lambda x: 1 if ("мультифункциональный" and "руль") in x else 0
)
# Усиление руля
df["power_steering"] = df["description_word"].apply(
lambda x: 1
if ("гидроусилитель" or "гидро" or "усилитель" and "руля") in x
else 0
)
# Датчики
df["light_and_rain_sensors"] = df["description_word"].apply(
lambda x: 1 if ("датчики" and "света" and "дождя") in x else 0
)
# Карбоновый обвес
df["сarbon_body_kits"] = df["description_word"].apply(
lambda x: 1 if ("карбоновые" and "обвесы") in x else 0
)
# Диффузор
df["rear_diffuser_rkp"] = df["description_word"].apply(
lambda x: 1 if ("задний" and "диффузор") in x else 0
)
# Доводчик дверей
df["door_closers"] = df["description_word"].apply(
lambda x: 1 if ("доводчики" and "дверей") in x else 0
)
# Камера заднего вида
df["rear_view_camera"] = df["description_word"].apply(
lambda x: 1 if ("камера" or "видеокамера" and "заднего" and "вида") in x else 0
)
# amg
df["amg"] = df["description_word"].apply(lambda x: 1 if ("amg") in x else 0)
# Фары
df["bi_xenon_headlights"] = df["description_word"].apply(
lambda x: 1 if ("биксеноновые" and "фары") in x else 0
)
df["from_salon"] = df["description_word"].apply(
lambda x: 1
if ("рольф" or "панавто" or "дилер" or "кредит" or "ликвидация") in x
else 0
)
# Диски
df["alloy_wheels"] = df["description_word"].apply(
lambda x: 1 if ("легкосплавные" or "колесные" or "диски") in x else 0
)
# Парктроник
df["parking_sensors"] = df["description_word"].apply(
lambda x: 1 if ("парктроник" or "парктронник") in x else 0
)
# Повреждение поверхности
df["dents"] = df["description_word"].apply(
lambda x: 1
if ("вмятины" or "вмятина" or "царапина" or "царапины" or "трещина") in x
else 0
)
# Панорамная крыша
df["roof_with_panoramic_view"] = df["description_word"].apply(
lambda x: 1 if ("панорамная" and "крыша") in x else 0
)
# Обивка салона
df["upholstery"] = df["description_word"].apply(
lambda x: 1 if ("обивка" and "салон") in x else 0
)
# Подогрев сидения
df["heated_seat"] = df["description_word"].apply(
lambda x: 1 if ("подогрев" and "сидение") in x else 0
)
# Датчик дождя
df["rain_sensor"] = df["description_word"].apply(
lambda x: 1 if ("датчик" and "дождь") in x else 0
)
# Официальный диллер
df["official_dealer"] = df["description_word"].apply(
lambda x: 1 if ("официальный" and "диллер") in x else 0
)
# Хорошее состояние
df["good_condition"] = df["description_word"].apply(
lambda x: 1 if ("хороший" and "состояние") in x else 0
)
# Отличное состояние
df["excellent_condition"] = df["description_word"].apply(
lambda x: 1 if ("отличный" and "состояние") in x else 0
)
# ################### 1. Предобработка ##############################################################
# убираем не нужные для модели признаки
df_output = df.copy()
df_output.drop(["description", "sell_id", "description_word"], axis=1, inplace=True)
# ################### Numerical Features ##############################################################
# Далее заполняем пропуски
for column in numerical_features:
df_output[column].fillna(df_output[column].median(), inplace=True)
# тут ваш код по обработке NAN
# ....
# Нормализация данных
scaler = MinMaxScaler()
for column in numerical_features:
df_output[column] = scaler.fit_transform(df_output[[column]])[:, 0]
# ################### Categorical Features ##############################################################
# Label Encoding
for column in categorical_features:
df_output[column] = df_output[column].astype("category").cat.codes
# One-Hot Encoding: в pandas есть готовая функция - get_dummies.
df_output = pd.get_dummies(df_output, columns=categorical_features, dummy_na=False)
# тут ваш код не Encoding фитчей
# ....
# ################### Feature Engineering ####################################################
# тут ваш код не генерацию новых фитчей
# ....
# ################### Clean ####################################################
# убираем признаки которые еще не успели обработать,
df_output.drop(["vehicleConfiguration", "name"], axis=1, inplace=True)
return df_output
# Запускаем и проверяем, что получилось
df_preproc = preproc_data(data)
df_preproc.sample(5)
df_preproc.info()
# ## Split data
# Теперь выделим тестовую часть
train_data = df_preproc.query("sample == 1").drop(["sample"], axis=1)
test_data = df_preproc.query("sample == 0").drop(["sample"], axis=1)
y = train_data.price.values # наш таргет
X = train_data.drop(["price"], axis=1)
X_sub = test_data.drop(["price"], axis=1)
test_data.info()
# # Model 2: CatBoostRegressor
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.15, shuffle=True, random_state=RANDOM_SEED
)
model = CatBoostRegressor(
iterations=5000,
# depth=10,
learning_rate=0.05,
random_seed=RANDOM_SEED,
eval_metric="MAPE",
custom_metric=["RMSE", "MAE"],
od_wait=500,
# task_type='GPU',
od_type="IncToDec",
)
model.fit(
X_train,
y_train,
eval_set=(X_test, y_test),
verbose_eval=100,
use_best_model=True,
# plot=True
)
test_predict_catboost = model.predict(X_test)
print(f"TEST mape: {(mape(y_test, test_predict_catboost))*100:0.2f}%")
train_predict_catboost = model.predict(X_train)
print(f"TRAIN mape: {(mape(y_train, train_predict_catboost))*100:0.2f}%")
# ### Хорошее улучшение относительно самой простой модели
# print(f"Test MAE: {mean_absolute_error(y_test,test_predict_catboost)*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train,train_predict_catboost)*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, test_predict_catboost)*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, train_predict_catboost)*100:0.3f}%")
# ### Submission
sub_predict_catboost = model.predict(X_sub)
sample_submission["price"] = sub_predict_catboost
sample_submission.to_csv("catboost_submission.csv", index=False)
# # Model 3: Tabular NN
# Построим обычную сеть:
X_train.head(5)
# ## Simple Dense NN
model = Sequential()
model.add(
L.Dense(1024, input_dim=X_train.shape[1], activation="relu")
) # changed to 1024
model.add(L.Dropout(0.5))
model.add(L.Dense(512, activation="relu")) # added
model.add(L.Dropout(0.5)) # added
model.add(L.Dense(256, activation="relu"))
model.add(L.Dropout(0.5))
model.add(L.Dense(1, activation="linear"))
model.summary()
# Compile model
optimizer = tf.keras.optimizers.Adam(0.01)
model.compile(loss="MAPE", optimizer=optimizer, metrics=["MAPE"])
checkpoint = ModelCheckpoint(
"../working/best_model.hdf5", monitor=["val_MAPE"], verbose=0, mode="min"
)
earlystop = EarlyStopping(
monitor="val_MAPE",
patience=50,
restore_best_weights=True,
)
callbacks_list = [checkpoint, earlystop]
# ### Fit
history = model.fit(
X_train,
y_train,
batch_size=512,
epochs=500, # фактически мы обучаем пока EarlyStopping не остановит обучение
validation_data=(X_test, y_test),
callbacks=callbacks_list,
verbose=0,
)
plt.title("Loss")
plt.plot(history.history["MAPE"], label="train")
plt.plot(history.history["val_MAPE"], label="test")
plt.show()
model.load_weights("../working/best_model.hdf5")
model.save("../working/nn_1.hdf5")
test_predict_nn1 = model.predict(X_test)
print(f"TEST mape: {(mape(y_test, test_predict_nn1[:,0]))*100:0.2f}%")
train_predict_nn1 = model.predict(X_train)
print(f"TRAIN mape: {(mape(y_train, train_predict_nn1[:,0]))*100:0.2f}%")
# print(f"Test MAE: {mean_absolute_error(y_test,test_predict_nn1[:,0])*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train,train_predict_nn1[:,0])*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, test_predict_nn1[:,0])*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, train_predict_nn1[:,0])*100:0.3f}%")
sub_predict_nn1 = model.predict(X_sub)
sample_submission["price"] = sub_predict_nn1[:, 0]
sample_submission.to_csv("nn1_submission.csv", index=False)
# Рекомендации для улучшения Model 3:
# * В нейросеть желательно подавать данные с распределением, близким к нормальному, поэтому от некоторых числовых признаков имеет смысл взять логарифм перед нормализацией. Пример:
# `modelDateNorm = np.log(2020 - data['modelDate'])`
# Статья по теме: https://habr.com/ru/company/ods/blog/325422
# * Извлечение числовых значений из текста:
# Парсинг признаков 'engineDisplacement', 'enginePower', 'Владение' для извлечения числовых значений.
# * Cокращение размерности категориальных признаков
# Признак name 'name' содержит данные, которые уже есть в других столбцах ('enginePower', 'engineDisplacement', 'vehicleTransmission'), поэтому эти данные можно удалить. Затем следует еще сильнее сократить размерность, например, выделив наличие xDrive в качестве отдельного признака.
# # Model 4: NLP + Multiple Inputs
data.description.head()
data["description"] = data.description.str.replace("•", "")
data["description"] = data.description.str.replace("–", "")
data["description"] = data.description.str.replace("∙", "")
data["description"] = data.description.str.replace("☑️", "")
data["description"] = data.description.str.replace("✔", "")
data["description"] = data.description.str.replace("➥", "")
data["description"] = data.description.str.replace("●", "")
data["description"] = data.description.str.replace("☛", "")
data["description"] = data.description.str.replace("✅", "")
data["description"] = data.description.str.replace("———————————————————————————", "")
data["description"] = data.description.str.replace("•", "")
data["description"] = data["description"].str.lower()
data["description"] = data.description.str.replace(
"автомобиль в отличном состоянии", "автомобильвотличномсостоянии"
)
data["description"] = data.description.str.replace(
"машина в отличном состаянии", "автомобильвотличномсостоянии"
)
data["description"] = data.description.str.replace(
"машина в отличном состоянии", "автомобильвотличномсостоянии"
)
data["description"] = data.description.str.replace(
"продаю машину в отличном состоянии", "автомобильвотличномсостоянии"
)
data["description"] = data.description.str.replace(
"авто в идеальном состоянии", "автомобильвотличномсостоянии"
)
def lemmited(description):
description_split = description.split(" ")
for word in description_split:
p = morph.parse(word)[0]
description = description.replace(word, p.normal_form)
return description
from nltk.corpus import stopwords
# удалим стоп-слова
filtered_tokens = dict()
stop_words = stopwords.words("russian")
for word in stop_words:
data["description"] = data.description.str.replace(" " + word + " ", " ")
# леммитизация
morph = pymorphy2.MorphAnalyzer()
data["description"] = data["description"].apply(lemmited)
# TOKENIZER
# The maximum number of words to be used. (most frequent)
MAX_WORDS = 100000
# Max number of words in each complaint.
MAX_SEQUENCE_LENGTH = 256
# split данных
text_train = data.description.iloc[X_train.index]
text_test = data.description.iloc[X_test.index]
text_sub = data.description.iloc[X_sub.index]
# ### Tokenizer
tokenize = Tokenizer(num_words=MAX_WORDS)
tokenize.fit_on_texts(data.description)
tokenize.word_index
text_train_sequences = sequence.pad_sequences(
tokenize.texts_to_sequences(text_train), maxlen=MAX_SEQUENCE_LENGTH
)
text_test_sequences = sequence.pad_sequences(
tokenize.texts_to_sequences(text_test), maxlen=MAX_SEQUENCE_LENGTH
)
text_sub_sequences = sequence.pad_sequences(
tokenize.texts_to_sequences(text_sub), maxlen=MAX_SEQUENCE_LENGTH
)
print(
text_train_sequences.shape,
text_test_sequences.shape,
text_sub_sequences.shape,
)
# вот так теперь выглядит наш текст
print(text_train.iloc[6])
print(text_train_sequences[6])
# ### RNN NLP
model_nlp = Sequential()
model_nlp.add(L.Input(shape=MAX_SEQUENCE_LENGTH, name="seq_description"))
model_nlp.add(
L.Embedding(
len(tokenize.word_index) + 1,
MAX_SEQUENCE_LENGTH,
)
)
model_nlp.add(L.LSTM(512, return_sequences=True))
model_nlp.add(L.Dropout(0.5))
model_nlp.add(L.LSTM(256, return_sequences=True)) # added
model_nlp.add(L.Dropout(0.5)) # added
model_nlp.add(
L.LSTM(
128,
)
)
model_nlp.add(L.Dropout(0.25))
model_nlp.add(L.Dense(64, activation="relu"))
model_nlp.add(L.Dropout(0.25))
# ### MLP
model_mlp = Sequential()
model_mlp.add(L.Dense(512, input_dim=X_train.shape[1], activation="relu"))
model_mlp.add(L.Dropout(0.5))
model_mlp.add(L.Dense(256, activation="relu"))
model_mlp.add(L.Dropout(0.5))
model_mlp.add(L.Dense(128, activation="relu")) # added
model_mlp.add(L.Dropout(0.5)) # added
# ### Multiple Inputs NN
combinedInput = L.concatenate([model_nlp.output, model_mlp.output])
# being our regression head
head = L.Dense(64, activation="relu")(combinedInput)
head = L.Dense(1, activation="linear")(head)
model = Model(inputs=[model_nlp.input, model_mlp.input], outputs=head)
model.summary()
# ### Fit
optimizer = tf.keras.optimizers.Adam(0.01)
model.compile(loss="MAPE", optimizer=optimizer, metrics=["MAPE"])
checkpoint = ModelCheckpoint(
"../working/best_model.hdf5", monitor=["val_MAPE"], verbose=0, mode="min"
)
earlystop = EarlyStopping(
monitor="val_MAPE",
patience=10,
restore_best_weights=True,
)
callbacks_list = [checkpoint, earlystop]
history = model.fit(
[text_train_sequences, X_train],
y_train,
batch_size=512,
epochs=500, # фактически мы обучаем пока EarlyStopping не остановит обучение
validation_data=([text_test_sequences, X_test], y_test),
callbacks=callbacks_list,
)
plt.title("Loss")
plt.plot(history.history["MAPE"], label="train")
plt.plot(history.history["val_MAPE"], label="test")
plt.show()
model.load_weights("../working/best_model.hdf5")
model.save("../working/nn_mlp_nlp.hdf5")
test_predict_nn2 = model.predict([text_test_sequences, X_test])
print(f"TEST mape: {(mape(y_test, test_predict_nn2[:,0]))*100:0.2f}%")
train_predict_nn2 = model.predict([text_train_sequences, X_train])
print(f"TRAIN mape: {(mape(y_train, train_predict_nn2[:,0]))*100:0.2f}%")
# print(f"Test MAE: {mean_absolute_error(y_test,test_predict_nn2[:,0])*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train,train_predict_nn2[:,0])*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, test_predict_nn2[:,0])*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, train_predict_nn2[:,0])*100:0.3f}%")
sub_predict_nn2 = model.predict([text_sub_sequences, X_sub])
sample_submission["price"] = sub_predict_nn2[:, 0]
sample_submission.to_csv("nn2_submission.csv", index=False)
# Идеи для улучшения NLP части:
# * Выделить из описаний часто встречающиеся блоки текста, заменив их на кодовые слова или удалив
# * Сделать предобработку текста, например, сделать лемматизацию - алгоритм ставящий все слова в форму по умолчанию (глаголы в инфинитив и т. д.), чтобы токенайзер не преобразовывал разные формы слова в разные числа
# Статья по теме: https://habr.com/ru/company/Voximplant/blog/446738/
# * Поработать над алгоритмами очистки и аугментации текста
# # Model 5: Добавляем картинки
# ### Data
# убедимся, что цены и фото подгрузились верно
plt.figure(figsize=(12, 8))
random_image = train.sample(n=9)
random_image_paths = random_image["sell_id"].values
random_image_cat = random_image["price"].values
for index, path in enumerate(random_image_paths):
im = PIL.Image.open(DATA_DIR + "img/img/" + str(path) + ".jpg")
plt.subplot(3, 3, index + 1)
plt.imshow(im)
plt.title("price: " + str(random_image_cat[index]))
plt.axis("off")
plt.show()
size = (320, 240)
def get_image_array(index):
images_train = []
for index, sell_id in enumerate(data["sell_id"].iloc[index].values):
image = cv2.imread(DATA_DIR + "img/img/" + str(sell_id) + ".jpg")
assert image is not None
image = cv2.resize(image, size)
images_train.append(image)
images_train = np.array(images_train)
print("images shape", images_train.shape, "dtype", images_train.dtype)
return images_train
images_train = get_image_array(X_train.index)
images_test = get_image_array(X_test.index)
images_sub = get_image_array(X_sub.index)
# ### albumentations
from albumentations import (
HorizontalFlip,
IAAPerspective,
ShiftScaleRotate,
CLAHE,
RandomRotate90,
Transpose,
ShiftScaleRotate,
Blur,
OpticalDistortion,
GridDistortion,
HueSaturationValue,
IAAAdditiveGaussianNoise,
GaussNoise,
MotionBlur,
MedianBlur,
IAAPiecewiseAffine,
IAASharpen,
IAAEmboss,
RandomBrightnessContrast,
Flip,
OneOf,
Compose,
)
# пример взят из официальной документации: https://albumentations.readthedocs.io/en/latest/examples.html
augmentation = Compose(
[
HorizontalFlip(),
OneOf(
[
IAAAdditiveGaussianNoise(),
GaussNoise(),
],
p=0.2,
),
OneOf(
[
MotionBlur(p=0.2),
MedianBlur(blur_limit=3, p=0.1),
Blur(blur_limit=3, p=0.1),
],
p=0.2,
),
ShiftScaleRotate(shift_limit=0.0625, scale_limit=0.2, rotate_limit=15, p=1),
OneOf(
[
OpticalDistortion(p=0.3),
GridDistortion(p=0.1),
IAAPiecewiseAffine(p=0.3),
],
p=0.2,
),
OneOf(
[
CLAHE(clip_limit=2),
IAASharpen(),
IAAEmboss(),
RandomBrightnessContrast(),
],
p=0.3,
),
HueSaturationValue(p=0.3),
],
p=1,
)
# пример
plt.figure(figsize=(12, 8))
for i in range(9):
img = augmentation(image=images_train[0])["image"]
plt.subplot(3, 3, i + 1)
plt.imshow(img)
plt.axis("off")
plt.show()
def make_augmentations(images):
print("применение аугментаций", end="")
augmented_images = np.empty(images.shape)
for i in range(images.shape[0]):
if i % 200 == 0:
print(".", end="")
augment_dict = augmentation(image=images[i])
augmented_image = augment_dict["image"]
augmented_images[i] = augmented_image
print("")
return augmented_images
# ## tf.data.Dataset
# Если все изображения мы будем хранить в памяти, то может возникнуть проблема ее нехватки. Не храните все изображения в памяти целиком!
# Метод .fit() модели keras может принимать либо данные в виде массивов или тензоров, либо разного рода итераторы, из которых наиболее современным и гибким является [tf.data.Dataset](https://www.tensorflow.org/guide/data). Он представляет собой конвейер, то есть мы указываем, откуда берем данные и какую цепочку преобразований с ними выполняем. Далее мы будем работать с tf.data.Dataset.
# Dataset хранит информацию о конечном или бесконечном наборе кортежей (tuple) с данными и может возвращать эти наборы по очереди. Например, данными могут быть пары (input, target) для обучения нейросети. С данными можно осуществлять преобразования, которые осуществляются по мере необходимости ([lazy evaluation](https://ru.wikipedia.org/wiki/%D0%9B%D0%B5%D0%BD%D0%B8%D0%B2%D1%8B%D0%B5_%D0%B2%D1%8B%D1%87%D0%B8%D1%81%D0%BB%D0%B5%D0%BD%D0%B8%D1%8F)).
# `tf.data.Dataset.from_tensor_slices(data)` - создает датасет из данных, которые представляют собой либо массив, либо кортеж из массивов. Деление осуществляется по первому индексу каждого массива. Например, если `data = (np.zeros((128, 256, 256)), np.zeros(128))`, то датасет будет содержать 128 элементов, каждый из которых содержит один массив 256x256 и одно число.
# `dataset2 = dataset1.map(func)` - применение функции к датасету; функция должна принимать столько аргументов, каков размер кортежа в датасете 1 и возвращать столько, сколько нужно иметь в датасете 2. Пусть, например, датасет содержит изображения и метки, а нам нужно создать датасет только из изображений, тогда мы напишем так: `dataset2 = dataset.map(lambda img, label: img)`.
# `dataset2 = dataset1.batch(8)` - группировка по батчам; если датасет 2 должен вернуть один элемент, то он берет из датасета 1 восемь элементов, склеивает их (нулевой индекс результата - номер элемента) и возвращает.
# `dataset.__iter__()` - превращение датасета в итератор, из которого можно получать элементы методом `.__next__()`. Итератор, в отличие от самого датасета, хранит позицию текущего элемента. Можно также перебирать датасет циклом for.
# `dataset2 = dataset1.repeat(X)` - датасет 2 будет повторять датасет 1 X раз.
# Если нам нужно взять из датасета 1000 элементов и использовать их как тестовые, а остальные как обучающие, то мы напишем так:
# `test_dataset = dataset.take(1000)
# train_dataset = dataset.skip(1000)`
# Датасет по сути неизменен: такие операции, как map, batch, repeat, take, skip никак не затрагивают оригинальный датасет. Если датасет хранит элементы [1, 2, 3], то выполнив 3 раза подряд функцию dataset.take(1) мы получим 3 новых датасета, каждый из которых вернет число 1. Если же мы выполним функцию dataset.skip(1), мы получим датасет, возвращающий числа [2, 3], но исходный датасет все равно будет возвращать [1, 2, 3] каждый раз, когда мы его перебираем.
# tf.Dataset всегда выполняется в graph-режиме (в противоположность eager-режиму), поэтому либо преобразования (`.map()`) должны содержать только tensorflow-функции, либо мы должны использовать tf.py_function в качестве обертки для функций, вызываемых в `.map()`. Подробнее можно прочитать [здесь](https://www.tensorflow.org/guide/data#applying_arbitrary_python_logic).
# NLP part
tokenize = Tokenizer(num_words=MAX_WORDS)
tokenize.fit_on_texts(data.description)
def process_image(image):
return augmentation(image=image.numpy())["image"]
def tokenize_(descriptions):
return sequence.pad_sequences(
tokenize.texts_to_sequences(descriptions), maxlen=MAX_SEQUENCE_LENGTH
)
def tokenize_text(text):
return tokenize_([text.numpy().decode("utf-8")])[0]
def tf_process_train_dataset_element(image, table_data, text, price):
im_shape = image.shape
[
image,
] = tf.py_function(process_image, [image], [tf.uint8])
image.set_shape(im_shape)
[
text,
] = tf.py_function(tokenize_text, [text], [tf.int32])
return (image, table_data, text), price
def tf_process_val_dataset_element(image, table_data, text, price):
[
text,
] = tf.py_function(tokenize_text, [text], [tf.int32])
return (image, table_data, text), price
train_dataset = tf.data.Dataset.from_tensor_slices(
(images_train, X_train, data.description.iloc[X_train.index], y_train)
).map(tf_process_train_dataset_element)
test_dataset = tf.data.Dataset.from_tensor_slices(
(images_test, X_test, data.description.iloc[X_test.index], y_test)
).map(tf_process_val_dataset_element)
y_sub = np.zeros(len(X_sub))
sub_dataset = tf.data.Dataset.from_tensor_slices(
(images_sub, X_sub, data.description.iloc[X_sub.index], y_sub)
).map(tf_process_val_dataset_element)
# проверяем, что нет ошибок (не будет выброшено исключение):
train_dataset.__iter__().__next__()
test_dataset.__iter__().__next__()
sub_dataset.__iter__().__next__()
# ### Строим сверточную сеть для анализа изображений без "головы"
# нормализация включена в состав модели EfficientNetB3, поэтому на вход она принимает данные типа uint8
efficientnet_model = tf.keras.applications.efficientnet.EfficientNetB3(
weights="imagenet", include_top=False, input_shape=(size[1], size[0], 3)
)
efficientnet_output = L.GlobalAveragePooling2D()(efficientnet_model.output)
# Fine-tuning.
efficientnet_model.trainable = True
fine_tune_at = len(efficientnet_model.layers) // 2
for layer in efficientnet_model.layers[:fine_tune_at]:
layer.trainable = False
for layer in model.layers:
print(layer, layer.trainable)
# строим нейросеть для анализа табличных данных
tabular_model = Sequential(
[
L.Input(shape=X.shape[1]),
L.Dense(1024, activation="relu"),
L.Dropout(0.5),
L.Dense(512, activation="relu"),
L.Dropout(0.5),
L.Dense(256, activation="relu"),
L.Dropout(0.5),
L.Dense(128, activation="relu"), # added
L.Dropout(0.5), # added
]
)
# NLP
nlp_model = Sequential(
[
L.Input(shape=MAX_SEQUENCE_LENGTH, name="seq_description"),
L.Embedding(
len(tokenize.word_index) + 1,
MAX_SEQUENCE_LENGTH,
),
L.LSTM(256, return_sequences=True),
L.Dropout(0.5),
L.LSTM(128),
L.Dropout(0.25), # added
L.Dense(64), # added
]
)
# объединяем выходы трех нейросетей
combinedInput = L.concatenate(
[efficientnet_output, tabular_model.output, nlp_model.output]
)
# being our regression head
head = L.Dense(256, activation="relu")(combinedInput)
head = L.Dense(128, activation="relu")(combinedInput) # added
head = L.Dense(
1,
)(head)
model = Model(
inputs=[efficientnet_model.input, tabular_model.input, nlp_model.input],
outputs=head,
)
model.summary()
optimizer = tf.keras.optimizers.Adam(0.005)
model.compile(loss="MAPE", optimizer=optimizer, metrics=["MAPE"])
checkpoint = ModelCheckpoint(
"../working/best_model.hdf5", monitor=["val_MAPE"], verbose=0, mode="min"
)
earlystop = EarlyStopping(
monitor="val_MAPE",
patience=10,
restore_best_weights=True,
)
callbacks_list = [checkpoint, earlystop]
history = model.fit(
train_dataset.batch(30),
epochs=60,
validation_data=test_dataset.batch(30),
callbacks=callbacks_list,
)
plt.title("Loss")
plt.plot(history.history["MAPE"], label="train")
plt.plot(history.history["val_MAPE"], label="test")
plt.show()
model.load_weights("../working/best_model.hdf5")
model.save("../working/nn_final.hdf5")
test_predict_nn3 = model.predict(test_dataset.batch(30))
print(f"TEST mape: {(mape(y_test, test_predict_nn3[:,0]))*100:0.2f}%")
train_predict_nn3 = model.predict(train_dataset.batch(30))
print(f"TRAIN mape: {(mape(y_train, train_predict_nn3[:,0]))*100:0.2f}%")
# print(f"Test MAE: {mean_absolute_error(y_test,test_predict_nn3[:,0])*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train,train_predict_nn3[:,0])*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, test_predict_nn3[:,0])*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, train_predict_nn3[:,0])*100:0.3f}%")
sub_predict_nn3 = model.predict(sub_dataset.batch(30))
sample_submission["price"] = sub_predict_nn3[:, 0]
sample_submission.to_csv("nn3_submission.csv", index=False)
#
# #### Общие рекомендации:
# * Попробовать разные архитектуры
# * Провести более детальный анализ результатов
# * Попробовать различные подходы в управление LR и оптимизаторы
# * Поработать с таргетом
# * Использовать Fine-tuning
# #### Tabular
# * В нейросеть желательно подавать данные с распределением, близким к нормальному, поэтому от некоторых числовых признаков имеет смысл взять логарифм перед нормализацией. Пример:
# `modelDateNorm = np.log(2020 - data['modelDate'])`
# Статья по теме: https://habr.com/ru/company/ods/blog/325422
# * Извлечение числовых значений из текста:
# Парсинг признаков 'engineDisplacement', 'enginePower', 'Владение' для извлечения числовых значений.
# * Cокращение размерности категориальных признаков
# Признак name 'name' содержит данные, которые уже есть в других столбцах ('enginePower', 'engineDisplacement', 'vehicleTransmission'). Можно удалить эти данные. Затем можно еще сильнее сократить размерность, например выделив наличие xDrive в качестве отдельного признака.
# * Поработать над Feature engineering
# #### NLP
# * Выделить из описаний часто встречающиеся блоки текста, заменив их на кодовые слова или удалив
# * Сделать предобработку текста, например сделать лемматизацию - алгоритм ставящий все слова в форму по умолчанию (глаголы в инфинитив и т. д.), чтобы токенайзер не преобразовывал разные формы слова в разные числа
# Статья по теме: https://habr.com/ru/company/Voximplant/blog/446738/
# * Поработать над алгоритмами очистки и аугментации текста
# #### CV
# * Попробовать различные аугментации
# * Fine-tuning
# # Blend
blend_predict_test = (test_predict_catboost + test_predict_nn3[:, 0]) / 2
print(f"TEST mape: {(mape(y_test, blend_predict_test))*100:0.2f}%")
blend_predict_train = (train_predict_catboost + train_predict_nn3[:, 0]) / 2
print(f"TRAIN mape: {(mape(y_train, blend_predict_train))*100:0.2f}%")
# print(f"Test MAE: {mean_absolute_error(y_test,blend_predict_test)*100:0.3f}%")
# print(f"TRAIN MAE: {mean_absolute_error(y_train, blend_predict_train)*100:0.3f}%")
# print()
# print(f"Test MSE: {mean_squared_error(y_test, blend_predict_test)*100:0.3f}%")
# print(f"TRAIN MSE: {mean_squared_error(y_train, blend_predict_train)*100:0.3f}%")
blend_sub_predict = (sub_predict_catboost + sub_predict_nn3[:, 0]) / 2
sample_submission["price"] = blend_sub_predict
sample_submission.to_csv("blend_submission.csv", index=False)
# # Model Bonus: проброс признака
# MLP
model_mlp = Sequential()
model_mlp.add(L.Dense(512, input_dim=X_train.shape[1], activation="relu"))
model_mlp.add(L.Dropout(0.5))
model_mlp.add(L.Dense(256, activation="relu"))
model_mlp.add(L.Dropout(0.5))
# FEATURE Input
# Iput
productiondate = L.Input(shape=[1], name="productiondate")
# Embeddings layers
emb_productiondate = L.Embedding(len(X.productionDate.unique().tolist()) + 1, 20)(
productiondate
)
f_productiondate = L.Flatten()(emb_productiondate)
combinedInput = L.concatenate(
[
model_mlp.output,
f_productiondate,
]
)
# being our regression head
head = L.Dense(64, activation="relu")(combinedInput)
head = L.Dense(1, activation="linear")(head)
model = Model(inputs=[model_mlp.input, productiondate], outputs=head)
model.summary()
optimizer = tf.keras.optimizers.Adam(0.01)
model.compile(loss="MAPE", optimizer=optimizer, metrics=["MAPE"])
history = model.fit(
[X_train, X_train.productionDate.values],
y_train,
batch_size=512,
epochs=500, # фактически мы обучаем пока EarlyStopping не остановит обучение
validation_data=([X_test, X_test.productionDate.values], y_test),
callbacks=callbacks_list,
)
model.load_weights("../working/best_model.hdf5")
test_predict_nn_bonus = model.predict([X_test, X_test.productionDate.values])
print(f"TEST mape: {(mape(y_test, test_predict_nn_bonus[:,0]))*100:0.2f}%")
train_predict_nn_bonus = model.predict([X_train, X_train.productionDate.values])
print(f"TRAIN mape: {(mape(y_train, train_predict_nn_bonus[:,0]))*100:0.2f}%")
print(f"Test MAE: {mean_absolute_error(y_test,test_predict_nn_bonus[:,0])*100:0.3f}%")
print(f"Test MSE: {mean_squared_error(y_test, test_predict_nn_bonus[:,0])*100:0.3f}%")
#
| false | 0 | 18,045 | 0 | 18,045 | 18,045 |
||
69235609
|
<jupyter_start><jupyter_text>European Soccer Database
The ultimate Soccer database for data analysis and machine learning
-------------------------------------------------------------------
**What you get:**
- +25,000 matches
- +10,000 players
- 11 European Countries with their lead championship
- Seasons 2008 to 2016
- Players and Teams' attributes* sourced from EA Sports' FIFA video game series, including the weekly updates
- Team line up with squad formation (X, Y coordinates)
- Betting odds from up to 10 providers
- Detailed match events (goal types, possession, corner, cross, fouls, cards etc...) for +10,000 matches
**16th Oct 2016: New table containing teams' attributes from FIFA !*
----------
**Original Data Source:**
You can easily find data about soccer matches but they are usually scattered across different websites. A thorough data collection and processing has been done to make your life easier. **I must insist that you do not make any commercial use of the data**. The data was sourced from:
- [http://football-data.mx-api.enetscores.com/][1] : scores, lineup, team formation and events
- [http://www.football-data.co.uk/][2] : betting odds. [Click here to understand the column naming system for betting odds:][3]
- [http://sofifa.com/][4] : players and teams attributes from EA Sports FIFA games. *FIFA series and all FIFA assets property of EA Sports.*
> When you have a look at the database, you will notice foreign keys for
> players and matches are the same as the original data sources. I have
> called those foreign keys "api_id".
----------
**Improving the dataset:**
You will notice that some players are missing from the lineup (NULL values). This is because I have not been able to source their attributes from FIFA. This will be fixed overtime as the crawling algorithm is being improved.
The dataset will also be expanded to include international games, national cups, Champion's League and Europa League. Please ask me if you're after a specific tournament.
> Please get in touch with me if you want to help improve this dataset.
[CLICK HERE TO ACCESS THE PROJECT GITHUB][5]
*Important note for people interested in using the crawlers:* since I first wrote the crawling scripts (in python), it appears sofifa.com has changed its design and with it comes new requirements for the scripts. The existing script to crawl players ('Player Spider') will not work until i've updated it.
----------
Exploring the data:
Now that's the fun part, there is a lot you can do with this dataset. I will be adding visuals and insights to this overview page but please have a look at the kernels and give it a try yourself ! Here are some ideas for you:
**The Holy Grail...**
... is obviously to predict the outcome of the game. The bookies use 3 classes (Home Win, Draw, Away Win). They get it right about 53% of the time. This is also what I've achieved so far using my own SVM. Though it may sound high for such a random sport game, you've got to know
that the home team wins about 46% of the time. So the base case (constantly predicting Home Win) has indeed 46% precision.
**Probabilities vs Odds**
When running a multi-class classifier like SVM you could also output a probability estimate and compare it to the betting odds. Have a look at your variance vs odds and see for what games you had very different predictions.
**Explore and visualize features**
With access to players and teams attributes, team formations and in-game events you should be able to produce some interesting insights into [The Beautiful Game][6] . Who knows, Guardiola himself may hire one of you some day!
[1]: http://football-data.mx-api.enetscores.com/
[2]: http://www.football-data.co.uk/
[3]: http://www.football-data.co.uk/notes.txt
[4]: http://sofifa.com/
[5]: https://github.com/hugomathien/football-data-collection/tree/master/footballData
[6]: https://en.wikipedia.org/wiki/The_Beautiful_Game
Kaggle dataset identifier: soccer
<jupyter_script># # Project: Investigate a Dataset: League Comparison with League via All-Star Players
# ## Table of Contents
# Introduction
# Data Wrangling
# Exploratory Data Analysis
# Conclusions
# ---
# ## Introduction
# ### Questions?
# > #### *1. Who is the Best Players of each league? (All-star team)*
# > #### *2. Which is the best league in Europe?*
# > #### *3. Comparison with League via All-Star Players*
# ### **European Soccer Database**
# > 25k+ matches, players & teams attributes for European Professional Football
# - +25,000 matches
# - +10,000 players
# - 11 European Countries with their lead championship
# - Seasons 2008 to 2016
# - Players and Teams' attributes* sourced from EA Sports' FIFA video game series, including the weekly updates
# - Team line up with squad formation (X, Y coordinates)
# - Betting odds from up to 10 providers
# - Detailed match events (goal types, possession, corner, cross, fouls, cards etc…) for +10,000 matches
# - *16th Oct 2016: New table containing teams' attributes from FIFA !
# 업로드 파일 : investigate-a-dataset-SOCCER_CK_4_210728.ipynb
import numpy as np
import pandas as pd
import sqlite3
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
#
# ## Data Wrangling
# > In this section, I will load in the data, check for cleanliness, and then trim and clean the dataset for analysis.
# ### General Properties
conn = sqlite3.connect("../input/soccer/database.sqlite")
tables = pd.read_sql_query("SELECT name FROM sqlite_master WHERE type='table'", conn)
tables
df_player_attr = pd.read_sql("SELECT * FROM Player_Attributes", conn)
df_player = pd.read_sql("SELECT * FROM Player", conn)
df_match = pd.read_sql("SELECT * FROM Match", conn)
df_league = pd.read_sql("SELECT * FROM League", conn)
df_country = pd.read_sql("SELECT * FROM Country", conn)
df_team = pd.read_sql("SELECT * FROM Team", conn)
df_team_attr = pd.read_sql("SELECT * FROM Team_Attributes", conn)
data = [
df_player_attr,
df_player,
df_match,
df_league,
df_country,
df_team,
df_team_attr,
]
def get_df_name(df):
name = [x for x in globals() if globals()[x] is df][0]
return name
# https://stackoverflow.com/a/50620134/11602203
for i in data:
print(f"{get_df_name(i)} \t: {i.shape}")
# > #### Check df_player_attr
# > * Player Attributes (abilities)
# > * Position information is missing ▶ needs to get
# > * connected with dataframe 'df_player' by column 'player_api_id' ▶ needs to merge together
# > * Multiple rows for a players by date ▶ needs to choose the best data
df_player_attr.head(3)
df_player_attr.columns
# > #### Check df_player
# > * base information of players including **Name**
df_player.head(3)
# > #### Get Ready : Player dataset ('df')
# > * merge df_player & df_player_attr
# > * Clean the dataset
# > * Save only one player's best data
# 플레이어 개인정보 + 능력치 테이블 병합
df = pd.merge(df_player, df_player_attr, on="player_api_id")
print(df.shape)
df.head()
# change to datetime type
df[["birthday", "date"]] = df[["birthday", "date"]].apply(pd.to_datetime)
# birthday -> 태어난 Year, date -> only Year (동일 해 측정값은 더 최근 데이터만 남김- 뒤에 duplicate로 작업)
df.insert(2, "birth_year", df["birthday"].dt.year)
df.insert(3, "date_year", df["date"].dt.year)
df.insert(4, "Age", df["date_year"] - df["birth_year"])
# 필요없는 컬럼 제외
col_drop = [
"id_x",
"id_y",
"player_fifa_api_id_x",
"player_fifa_api_id_y",
"birthday",
"date",
] # 'player_api_id',
df.drop(col_drop, inplace=True, axis=1)
df.columns
print(df.duplicated(["player_name"]).sum())
print(df["player_name"].nunique())
# * 중복된 값이 많음. 선수들은 총 10848 명.
# 결측치 , 중복값 제거
df.dropna(inplace=True)
df.drop_duplicates(inplace=True)
print(df.shape)
df.head()
# Best 값으로 선수 데이터 저장
df.sort_values(
["overall_rating", "potential", "date_year"], ascending=False, inplace=True
)
df.drop_duplicates("player_name", inplace=True)
df.shape
# > #### Match 데이터셋에서 정보 가져오기
# > * Additional information(but essential) : Position, Side-position, Team, League
# #### ****
# - According to https://www.kaggle.com/hugomathien/soccer/discussion/80756:
# - Y coordinates : the player is a defender, midfielder or forward.
# - GK if Y == [1]
# - DF if Y == [2, 3, 4, 5]
# - MF if Y == [6, 7, 8, 9]
# - FW if Y == [10, 11]
# - X coordinates : the player plays on the left or on the right side of the court
# - Center if X == [4, 5, 6]
# - Side if X == [1, 2, 3, 7, 8, 9]
#
# - from https://www.kaggle.com/zotufip/look-at-stats-guess-position
# 
df_match["date"] = df_match["date"].apply(pd.to_datetime)
df_match.insert(6, "year", df_match["date"].dt.year)
df_match.insert(7, "month", df_match["date"].dt.month)
df_match.columns[51:]
df_match = df_match.loc[:, :"away_player_11"].dropna()
df_match.drop(["season", "stage", "match_api_id", "date"], axis=1, inplace=True)
df_match.loc[:, "home_player_X1":] = df_match.loc[:, "home_player_X1":].astype("int")
df_match.columns
print(df_match.shape)
df_match.head()
player_data = []
for i in range(1, 23):
# 동적 변수선언 (data_1, data_2, ...)
# globals()[f'data_{i}'] = df_match[[player_home, home_X, home_Y, team_id]]
if i < 12: # home_player
player = "home_player_" + str(i)
player_X = "home_player_X" + str(i)
player_Y = "home_player_Y" + str(i)
team_id = "home_team_api_id"
else: # away_player
player = "away_player_" + str(i - 11)
player_X = "away_player_X" + str(i - 11)
player_Y = "away_player_Y" + str(i - 11)
team_id = "away_team_api_id"
data = df_match[[player, player_X, player_Y, team_id, "year", "month", "league_id"]]
data.columns = ["player", "X", "Y", "team_id", "year", "month", "league_id"]
player_data.append(data)
# 플레이어 정보 확인을 위한 dataframe 생성
df_player_inform = pd.concat(player_data, axis=0, ignore_index=True)
df_player_inform.drop_duplicates(inplace=True)
df_player_inform.sort_values("player", inplace=True)
df_player_inform.reset_index(drop=True, inplace=True)
print(df_player_inform["player"].nunique())
print(df_player_inform.shape)
df_player_inform.head(10)
df["player_api_id"].nunique()
# - 아래에서 X,Y 확인할 수 있다
# https://www.kaggle.com/zotufip/look-at-stats-guess-position
# Let's assume that:
# positions below y=4 are defenders,
# positions below y=9.5 and above y=4 are midfielders,
# positions above y=9.5 strikers,
# In addition:
# positions between x=3.5 and x=6.5 are centre players,
# others are side players
df.head()
df_player_inform.shape
# df_match 와 df의 player 가 일치하지 않음. 중복되지 않는 player 삭제
df_match_player_check = df_match.loc[:, "home_player_1":"away_player_11"].values
df_player_check = df["player_api_id"].values
df_match_player_check = np.unique(df_match_player_check)
df_player_check = np.unique(df_player_check)
df_check_inter = np.intersect1d(df_match_player_check, df_player_check)
len(df_check_inter)
df = df[df["player_api_id"].isin(df_check_inter)]
def get_player_inform(player_id, match_year):
avg = df_player_inform.query("player==@player_id").mean()
x = round(avg[1])
y = round(avg[2])
if y == 1:
position = "GK"
elif y < 6:
position = "DF"
elif y < 9.5:
position = "MF"
elif y >= 9.5:
position = "FW"
if x > 3.5 and x < 6.5:
side_position = "center"
else:
side_position = "side"
name = df.query("player_api_id==@player_id")["player_name"].iloc[0]
team_id = df_player_inform.query("player==@player_id ").iloc[
0, 3
] # and year==@match_year
team = df_team.query("team_api_id == @team_id").iloc[0, 3]
league_id = df_player_inform.query("player==@player_id")["league_id"].iloc[0]
league = df_league.query("id == @league_id").iloc[0, 2]
return name, position, side_position, team, league
get_player_inform(30893, 2015)
inform = df.apply(
lambda x: get_player_inform(x["player_api_id"], x["date_year"]), axis=1
)
df["position"] = [inform.iloc[i][1] for i in range(len(inform))]
df["side_position"] = [inform.iloc[i][2] for i in range(len(inform))]
df["team"] = [inform.iloc[i][3] for i in range(len(inform))]
df["league"] = [inform.iloc[i][4] for i in range(len(inform))]
df
# ---
# ---
# ---
# # 1. League 별 올스타 팀 멤버
df_league_inform = df_league.copy()
df_league_inform.drop(["id", "country_id"], axis=1, inplace=True)
df_league_inform.loc[11] = ["EURO best"]
df_league_inform
# - Total number of leagues : 11
# 입력된 dataframe에서 BEST squad 구함
def get_allstar(
players_df,
df_num,
md_num,
fw_num,
):
sorted_players_df = players_df.sort_values(
["overall_rating", "potential", "date_year"], ascending=False
)
goalkeeper = sorted_players_df.query('position=="GK"')[:1]
defender = sorted_players_df.query('position=="DF"')[:df_num]
midfielder = sorted_players_df.query('position=="MF"')[:md_num]
forward = sorted_players_df.query('position=="FW"')[:fw_num]
allstar_df = pd.concat(
[goalkeeper, defender, midfielder, forward], axis=0, ignore_index=True
)
return allstar_df
# return goalkeeper, defender, midfielder, forward
league_allstar_dict = {}
for i in df_league["name"]:
data = df.query("league==@i")
squad_dict[i] = get_allstar(data, 4, 4, 2)
squad_dict["EURO_best"] = get_allstar(df, 4, 4, 2)
squad_dict.keys()
squad_dict["Spain LIGA BBVA"]
# - Get Best players of each league : squad_dict
# > Comparasions in each league are based on best players
# > 4-4-2 forrmation : 4 Defenders, 4 Midfielders, 2 Forwards
# > 2008 ~ 2016
# 각 리그 베스트 멤버의 능력치 평균을 구함
df_league_avg = pd.DataFrame()
for league in df_league["name"]:
df_league_avg[league] = squad_dict[league].mean().loc["Age":"sliding_tackle"]
df_league_avg["EURO_best"] = squad_dict["EURO_best"].mean().loc["Age":"sliding_tackle"]
df_league_avg = df_league_avg.T.sort_values("overall_rating", ascending=False)
df_league_avg
# pal = sns.color_palette("RdPu", len(df_league_avg))
values = df_league_avg["overall_rating"]
# pal = ['grey' if (x < max(values)) else 'red' for x in values ]
pal = [
"aqua",
"red",
"deepskyblue",
"deepskyblue",
"deepskyblue",
"deepskyblue",
"greenyellow",
"greenyellow",
"greenyellow",
"greenyellow",
"greenyellow",
"yellow",
]
rank = df_league_avg["overall_rating"].argsort().argsort()
plt.figure(figsize=(12, 6))
plt.ylim(70, 95)
plot1 = sns.barplot(
x="index",
y="overall_rating",
data=df_league_avg.reset_index(),
palette=np.array(pal[::-1])[rank],
)
# palette=np.array(pal)[rank])
# How to Annotate Bars in Barplot
# https://datavizpyr.com/how-to-annotate-bars-in-barplot-with-matplotlib-in-python/
for p in plot1.patches:
plot1.annotate(
format(p.get_height(), ".1f"),
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
xytext=(0, 9),
textcoords="offset points",
)
plot1.set_xticklabels(plot1.get_xticklabels(), rotation=90)
plot1.set_title("Average Overall-rating of League All-Star team players")
# - EURO Best team average overall-rating = 91.5
# - 'Spain LIGA BBVA' is the best league
# - 'England Premirer League' , 'Italy Serie A' , 'Germany 1.Bundesliga' are similar
# - can be divided into 3 Groups. There is a gap between each group.
# - Group A : 'Spain LIGA BBVA', 'England Premirer League' , 'Italy Serie A' , 'Germany 1.Bundesliga', 'France Ligue 1'
# - Group B : 'Portugal Liga ZON Sagres', 'Netherlands Eredivisie', 'Scotland Premier League', 'Switzerland Super League', 'Belgium Jupiler League'
# - Group C : 'Portugal Liga ZON Sagres'
df_league
sns.countplot(y="position", hue="league", data=df[:1000])
plt.ylim(20, 30)
df_league_avg["Age"].plot.bar()
sns.distplot(df["overall_rating"], bins=20)
df_best.drop(["height", "weight"], axis=1)[:10].loc[
:, "player_name":"short_passing"
].plot.bar(x="player_name", figsize=(12, 6))
# > 3. df_match
print(df_match)
df_match.head(3)
df_match.iloc[:, 21:40]
# > 4. df_league
df_league.head()
# > 5. df_team
df_team.head()
# > 5. df_team_attr
df_team_attr.columns
df_team_attr.info()
# 1. 리그별 Best Squad : 11개 리그
# 2. Top Player
# > 나이별 변화도 / 이유
# 3. Overall rating
# > 변화
# > 중요한 요소
# > 신체조건, 나이와 상관관계/변화율 높은 능력치
# 4. 메시 vs 호날도
# - Ex) 메시 데이터
# - 동일 선수의 데이터가 시간에 따라 여러번 기록됨
# 메시 데이터
df_messi = df[df["player_name"] == "Lionel Messi"].sort_values(
"overall_rating", ascending=False
) # .loc[:,'player_name':'short_passing']
df_messi
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/235/69235609.ipynb
|
soccer
|
hugomathien
|
[{"Id": 69235609, "ScriptId": 18899528, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4428227, "CreationDate": "07/28/2021 12:50:08", "VersionNumber": 2.0, "Title": "League Comparison with League via All-Star Players", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 461.0, "LinesInsertedFromPrevious": 66.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 395.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92157711, "KernelVersionId": 69235609, "SourceDatasetVersionId": 589}]
|
[{"Id": 589, "DatasetId": 63, "DatasourceVersionId": 589, "CreatorUserId": 655525, "LicenseName": "Database: Open Database, Contents: \u00a9 Original Authors", "CreationDate": "10/23/2016 22:31:38", "VersionNumber": 10.0, "Title": "European Soccer Database", "Slug": "soccer", "Subtitle": "25k+ matches, players & teams attributes for European Professional Football", "Description": "The ultimate Soccer database for data analysis and machine learning\n-------------------------------------------------------------------\n\n**What you get:**\n\n - +25,000 matches\n - +10,000 players\n - 11 European Countries with their lead championship\n - Seasons 2008 to 2016\n - Players and Teams' attributes* sourced from EA Sports' FIFA video game series, including the weekly updates\n - Team line up with squad formation (X, Y coordinates)\n - Betting odds from up to 10 providers\n - Detailed match events (goal types, possession, corner, cross, fouls, cards etc...) for +10,000 matches\n\n**16th Oct 2016: New table containing teams' attributes from FIFA !*\n\n----------\n\n**Original Data Source:** \n\nYou can easily find data about soccer matches but they are usually scattered across different websites. A thorough data collection and processing has been done to make your life easier. **I must insist that you do not make any commercial use of the data**. The data was sourced from:\n\n - [http://football-data.mx-api.enetscores.com/][1] : scores, lineup, team formation and events\n\n - [http://www.football-data.co.uk/][2] : betting odds. [Click here to understand the column naming system for betting odds:][3] \n\n - [http://sofifa.com/][4] : players and teams attributes from EA Sports FIFA games. *FIFA series and all FIFA assets property of EA Sports.*\n\n\n> When you have a look at the database, you will notice foreign keys for\n> players and matches are the same as the original data sources. I have\n> called those foreign keys \"api_id\".\n\n----------\n\n**Improving the dataset:** \n\nYou will notice that some players are missing from the lineup (NULL values). This is because I have not been able to source their attributes from FIFA. This will be fixed overtime as the crawling algorithm is being improved.\nThe dataset will also be expanded to include international games, national cups, Champion's League and Europa League. Please ask me if you're after a specific tournament.\n\n> Please get in touch with me if you want to help improve this dataset. \n\n[CLICK HERE TO ACCESS THE PROJECT GITHUB][5]\n\n*Important note for people interested in using the crawlers:* since I first wrote the crawling scripts (in python), it appears sofifa.com has changed its design and with it comes new requirements for the scripts. The existing script to crawl players ('Player Spider') will not work until i've updated it.\n\n----------\nExploring the data:\n\nNow that's the fun part, there is a lot you can do with this dataset. I will be adding visuals and insights to this overview page but please have a look at the kernels and give it a try yourself ! Here are some ideas for you:\n\n**The Holy Grail...**\n... is obviously to predict the outcome of the game. The bookies use 3 classes (Home Win, Draw, Away Win). They get it right about 53% of the time. This is also what I've achieved so far using my own SVM. Though it may sound high for such a random sport game, you've got to know\n that the home team wins about 46% of the time. So the base case (constantly predicting Home Win) has indeed 46% precision. \n\n**Probabilities vs Odds**\n\nWhen running a multi-class classifier like SVM you could also output a probability estimate and compare it to the betting odds. Have a look at your variance vs odds and see for what games you had very different predictions.\n\n**Explore and visualize features**\n\nWith access to players and teams attributes, team formations and in-game events you should be able to produce some interesting insights into [The Beautiful Game][6] . Who knows, Guardiola himself may hire one of you some day!\n\n\n [1]: http://football-data.mx-api.enetscores.com/\n [2]: http://www.football-data.co.uk/\n [3]: http://www.football-data.co.uk/notes.txt\n [4]: http://sofifa.com/\n [5]: https://github.com/hugomathien/football-data-collection/tree/master/footballData\n [6]: https://en.wikipedia.org/wiki/The_Beautiful_Game", "VersionNotes": "Added a few missing games from the English Premier League. Change the wording from \"stats\" to \"attributes\" when talking about FIFA player's attributes. Changed the teams' long name to their real long name (example: Bordeaux becomes Girondins de Bordeaux). Added a new table with Teams' attributes (i.e: style of play) from FIFA video games", "TotalCompressedBytes": 313090048.0, "TotalUncompressedBytes": 313090048.0}]
|
[{"Id": 63, "CreatorUserId": 655525, "OwnerUserId": 655525.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 589.0, "CurrentDatasourceVersionId": 589.0, "ForumId": 1357, "Type": 2, "CreationDate": "07/09/2016 13:40:34", "LastActivityDate": "02/06/2018", "TotalViews": 1535639, "TotalDownloads": 194949, "TotalVotes": 4298, "TotalKernels": 1568}]
|
[{"Id": 655525, "UserName": "hugomathien", "DisplayName": "Hugo Mathien", "RegisterDate": "07/05/2016", "PerformanceTier": 0}]
|
# # Project: Investigate a Dataset: League Comparison with League via All-Star Players
# ## Table of Contents
# Introduction
# Data Wrangling
# Exploratory Data Analysis
# Conclusions
# ---
# ## Introduction
# ### Questions?
# > #### *1. Who is the Best Players of each league? (All-star team)*
# > #### *2. Which is the best league in Europe?*
# > #### *3. Comparison with League via All-Star Players*
# ### **European Soccer Database**
# > 25k+ matches, players & teams attributes for European Professional Football
# - +25,000 matches
# - +10,000 players
# - 11 European Countries with their lead championship
# - Seasons 2008 to 2016
# - Players and Teams' attributes* sourced from EA Sports' FIFA video game series, including the weekly updates
# - Team line up with squad formation (X, Y coordinates)
# - Betting odds from up to 10 providers
# - Detailed match events (goal types, possession, corner, cross, fouls, cards etc…) for +10,000 matches
# - *16th Oct 2016: New table containing teams' attributes from FIFA !
# 업로드 파일 : investigate-a-dataset-SOCCER_CK_4_210728.ipynb
import numpy as np
import pandas as pd
import sqlite3
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
#
# ## Data Wrangling
# > In this section, I will load in the data, check for cleanliness, and then trim and clean the dataset for analysis.
# ### General Properties
conn = sqlite3.connect("../input/soccer/database.sqlite")
tables = pd.read_sql_query("SELECT name FROM sqlite_master WHERE type='table'", conn)
tables
df_player_attr = pd.read_sql("SELECT * FROM Player_Attributes", conn)
df_player = pd.read_sql("SELECT * FROM Player", conn)
df_match = pd.read_sql("SELECT * FROM Match", conn)
df_league = pd.read_sql("SELECT * FROM League", conn)
df_country = pd.read_sql("SELECT * FROM Country", conn)
df_team = pd.read_sql("SELECT * FROM Team", conn)
df_team_attr = pd.read_sql("SELECT * FROM Team_Attributes", conn)
data = [
df_player_attr,
df_player,
df_match,
df_league,
df_country,
df_team,
df_team_attr,
]
def get_df_name(df):
name = [x for x in globals() if globals()[x] is df][0]
return name
# https://stackoverflow.com/a/50620134/11602203
for i in data:
print(f"{get_df_name(i)} \t: {i.shape}")
# > #### Check df_player_attr
# > * Player Attributes (abilities)
# > * Position information is missing ▶ needs to get
# > * connected with dataframe 'df_player' by column 'player_api_id' ▶ needs to merge together
# > * Multiple rows for a players by date ▶ needs to choose the best data
df_player_attr.head(3)
df_player_attr.columns
# > #### Check df_player
# > * base information of players including **Name**
df_player.head(3)
# > #### Get Ready : Player dataset ('df')
# > * merge df_player & df_player_attr
# > * Clean the dataset
# > * Save only one player's best data
# 플레이어 개인정보 + 능력치 테이블 병합
df = pd.merge(df_player, df_player_attr, on="player_api_id")
print(df.shape)
df.head()
# change to datetime type
df[["birthday", "date"]] = df[["birthday", "date"]].apply(pd.to_datetime)
# birthday -> 태어난 Year, date -> only Year (동일 해 측정값은 더 최근 데이터만 남김- 뒤에 duplicate로 작업)
df.insert(2, "birth_year", df["birthday"].dt.year)
df.insert(3, "date_year", df["date"].dt.year)
df.insert(4, "Age", df["date_year"] - df["birth_year"])
# 필요없는 컬럼 제외
col_drop = [
"id_x",
"id_y",
"player_fifa_api_id_x",
"player_fifa_api_id_y",
"birthday",
"date",
] # 'player_api_id',
df.drop(col_drop, inplace=True, axis=1)
df.columns
print(df.duplicated(["player_name"]).sum())
print(df["player_name"].nunique())
# * 중복된 값이 많음. 선수들은 총 10848 명.
# 결측치 , 중복값 제거
df.dropna(inplace=True)
df.drop_duplicates(inplace=True)
print(df.shape)
df.head()
# Best 값으로 선수 데이터 저장
df.sort_values(
["overall_rating", "potential", "date_year"], ascending=False, inplace=True
)
df.drop_duplicates("player_name", inplace=True)
df.shape
# > #### Match 데이터셋에서 정보 가져오기
# > * Additional information(but essential) : Position, Side-position, Team, League
# #### ****
# - According to https://www.kaggle.com/hugomathien/soccer/discussion/80756:
# - Y coordinates : the player is a defender, midfielder or forward.
# - GK if Y == [1]
# - DF if Y == [2, 3, 4, 5]
# - MF if Y == [6, 7, 8, 9]
# - FW if Y == [10, 11]
# - X coordinates : the player plays on the left or on the right side of the court
# - Center if X == [4, 5, 6]
# - Side if X == [1, 2, 3, 7, 8, 9]
#
# - from https://www.kaggle.com/zotufip/look-at-stats-guess-position
# 
df_match["date"] = df_match["date"].apply(pd.to_datetime)
df_match.insert(6, "year", df_match["date"].dt.year)
df_match.insert(7, "month", df_match["date"].dt.month)
df_match.columns[51:]
df_match = df_match.loc[:, :"away_player_11"].dropna()
df_match.drop(["season", "stage", "match_api_id", "date"], axis=1, inplace=True)
df_match.loc[:, "home_player_X1":] = df_match.loc[:, "home_player_X1":].astype("int")
df_match.columns
print(df_match.shape)
df_match.head()
player_data = []
for i in range(1, 23):
# 동적 변수선언 (data_1, data_2, ...)
# globals()[f'data_{i}'] = df_match[[player_home, home_X, home_Y, team_id]]
if i < 12: # home_player
player = "home_player_" + str(i)
player_X = "home_player_X" + str(i)
player_Y = "home_player_Y" + str(i)
team_id = "home_team_api_id"
else: # away_player
player = "away_player_" + str(i - 11)
player_X = "away_player_X" + str(i - 11)
player_Y = "away_player_Y" + str(i - 11)
team_id = "away_team_api_id"
data = df_match[[player, player_X, player_Y, team_id, "year", "month", "league_id"]]
data.columns = ["player", "X", "Y", "team_id", "year", "month", "league_id"]
player_data.append(data)
# 플레이어 정보 확인을 위한 dataframe 생성
df_player_inform = pd.concat(player_data, axis=0, ignore_index=True)
df_player_inform.drop_duplicates(inplace=True)
df_player_inform.sort_values("player", inplace=True)
df_player_inform.reset_index(drop=True, inplace=True)
print(df_player_inform["player"].nunique())
print(df_player_inform.shape)
df_player_inform.head(10)
df["player_api_id"].nunique()
# - 아래에서 X,Y 확인할 수 있다
# https://www.kaggle.com/zotufip/look-at-stats-guess-position
# Let's assume that:
# positions below y=4 are defenders,
# positions below y=9.5 and above y=4 are midfielders,
# positions above y=9.5 strikers,
# In addition:
# positions between x=3.5 and x=6.5 are centre players,
# others are side players
df.head()
df_player_inform.shape
# df_match 와 df의 player 가 일치하지 않음. 중복되지 않는 player 삭제
df_match_player_check = df_match.loc[:, "home_player_1":"away_player_11"].values
df_player_check = df["player_api_id"].values
df_match_player_check = np.unique(df_match_player_check)
df_player_check = np.unique(df_player_check)
df_check_inter = np.intersect1d(df_match_player_check, df_player_check)
len(df_check_inter)
df = df[df["player_api_id"].isin(df_check_inter)]
def get_player_inform(player_id, match_year):
avg = df_player_inform.query("player==@player_id").mean()
x = round(avg[1])
y = round(avg[2])
if y == 1:
position = "GK"
elif y < 6:
position = "DF"
elif y < 9.5:
position = "MF"
elif y >= 9.5:
position = "FW"
if x > 3.5 and x < 6.5:
side_position = "center"
else:
side_position = "side"
name = df.query("player_api_id==@player_id")["player_name"].iloc[0]
team_id = df_player_inform.query("player==@player_id ").iloc[
0, 3
] # and year==@match_year
team = df_team.query("team_api_id == @team_id").iloc[0, 3]
league_id = df_player_inform.query("player==@player_id")["league_id"].iloc[0]
league = df_league.query("id == @league_id").iloc[0, 2]
return name, position, side_position, team, league
get_player_inform(30893, 2015)
inform = df.apply(
lambda x: get_player_inform(x["player_api_id"], x["date_year"]), axis=1
)
df["position"] = [inform.iloc[i][1] for i in range(len(inform))]
df["side_position"] = [inform.iloc[i][2] for i in range(len(inform))]
df["team"] = [inform.iloc[i][3] for i in range(len(inform))]
df["league"] = [inform.iloc[i][4] for i in range(len(inform))]
df
# ---
# ---
# ---
# # 1. League 별 올스타 팀 멤버
df_league_inform = df_league.copy()
df_league_inform.drop(["id", "country_id"], axis=1, inplace=True)
df_league_inform.loc[11] = ["EURO best"]
df_league_inform
# - Total number of leagues : 11
# 입력된 dataframe에서 BEST squad 구함
def get_allstar(
players_df,
df_num,
md_num,
fw_num,
):
sorted_players_df = players_df.sort_values(
["overall_rating", "potential", "date_year"], ascending=False
)
goalkeeper = sorted_players_df.query('position=="GK"')[:1]
defender = sorted_players_df.query('position=="DF"')[:df_num]
midfielder = sorted_players_df.query('position=="MF"')[:md_num]
forward = sorted_players_df.query('position=="FW"')[:fw_num]
allstar_df = pd.concat(
[goalkeeper, defender, midfielder, forward], axis=0, ignore_index=True
)
return allstar_df
# return goalkeeper, defender, midfielder, forward
league_allstar_dict = {}
for i in df_league["name"]:
data = df.query("league==@i")
squad_dict[i] = get_allstar(data, 4, 4, 2)
squad_dict["EURO_best"] = get_allstar(df, 4, 4, 2)
squad_dict.keys()
squad_dict["Spain LIGA BBVA"]
# - Get Best players of each league : squad_dict
# > Comparasions in each league are based on best players
# > 4-4-2 forrmation : 4 Defenders, 4 Midfielders, 2 Forwards
# > 2008 ~ 2016
# 각 리그 베스트 멤버의 능력치 평균을 구함
df_league_avg = pd.DataFrame()
for league in df_league["name"]:
df_league_avg[league] = squad_dict[league].mean().loc["Age":"sliding_tackle"]
df_league_avg["EURO_best"] = squad_dict["EURO_best"].mean().loc["Age":"sliding_tackle"]
df_league_avg = df_league_avg.T.sort_values("overall_rating", ascending=False)
df_league_avg
# pal = sns.color_palette("RdPu", len(df_league_avg))
values = df_league_avg["overall_rating"]
# pal = ['grey' if (x < max(values)) else 'red' for x in values ]
pal = [
"aqua",
"red",
"deepskyblue",
"deepskyblue",
"deepskyblue",
"deepskyblue",
"greenyellow",
"greenyellow",
"greenyellow",
"greenyellow",
"greenyellow",
"yellow",
]
rank = df_league_avg["overall_rating"].argsort().argsort()
plt.figure(figsize=(12, 6))
plt.ylim(70, 95)
plot1 = sns.barplot(
x="index",
y="overall_rating",
data=df_league_avg.reset_index(),
palette=np.array(pal[::-1])[rank],
)
# palette=np.array(pal)[rank])
# How to Annotate Bars in Barplot
# https://datavizpyr.com/how-to-annotate-bars-in-barplot-with-matplotlib-in-python/
for p in plot1.patches:
plot1.annotate(
format(p.get_height(), ".1f"),
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="center",
xytext=(0, 9),
textcoords="offset points",
)
plot1.set_xticklabels(plot1.get_xticklabels(), rotation=90)
plot1.set_title("Average Overall-rating of League All-Star team players")
# - EURO Best team average overall-rating = 91.5
# - 'Spain LIGA BBVA' is the best league
# - 'England Premirer League' , 'Italy Serie A' , 'Germany 1.Bundesliga' are similar
# - can be divided into 3 Groups. There is a gap between each group.
# - Group A : 'Spain LIGA BBVA', 'England Premirer League' , 'Italy Serie A' , 'Germany 1.Bundesliga', 'France Ligue 1'
# - Group B : 'Portugal Liga ZON Sagres', 'Netherlands Eredivisie', 'Scotland Premier League', 'Switzerland Super League', 'Belgium Jupiler League'
# - Group C : 'Portugal Liga ZON Sagres'
df_league
sns.countplot(y="position", hue="league", data=df[:1000])
plt.ylim(20, 30)
df_league_avg["Age"].plot.bar()
sns.distplot(df["overall_rating"], bins=20)
df_best.drop(["height", "weight"], axis=1)[:10].loc[
:, "player_name":"short_passing"
].plot.bar(x="player_name", figsize=(12, 6))
# > 3. df_match
print(df_match)
df_match.head(3)
df_match.iloc[:, 21:40]
# > 4. df_league
df_league.head()
# > 5. df_team
df_team.head()
# > 5. df_team_attr
df_team_attr.columns
df_team_attr.info()
# 1. 리그별 Best Squad : 11개 리그
# 2. Top Player
# > 나이별 변화도 / 이유
# 3. Overall rating
# > 변화
# > 중요한 요소
# > 신체조건, 나이와 상관관계/변화율 높은 능력치
# 4. 메시 vs 호날도
# - Ex) 메시 데이터
# - 동일 선수의 데이터가 시간에 따라 여러번 기록됨
# 메시 데이터
df_messi = df[df["player_name"] == "Lionel Messi"].sort_values(
"overall_rating", ascending=False
) # .loc[:,'player_name':'short_passing']
df_messi
| false | 0 | 4,700 | 0 | 5,760 | 4,700 |
||
69235281
|
<jupyter_start><jupyter_text>Breast Cancer Wisconsin (Diagnostic) Data Set
Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They describe characteristics of the cell nuclei present in the image.
n the 3-dimensional space is that described in: [K. P. Bennett and O. L. Mangasarian: "Robust Linear Programming Discrimination of Two Linearly Inseparable Sets", Optimization Methods and Software 1, 1992, 23-34].
This database is also available through the UW CS ftp server:
ftp ftp.cs.wisc.edu
cd math-prog/cpo-dataset/machine-learn/WDBC/
Also can be found on UCI Machine Learning Repository: https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+%28Diagnostic%29
Attribute Information:
1) ID number
2) Diagnosis (M = malignant, B = benign)
3-32)
Ten real-valued features are computed for each cell nucleus:
a) radius (mean of distances from center to points on the perimeter)
b) texture (standard deviation of gray-scale values)
c) perimeter
d) area
e) smoothness (local variation in radius lengths)
f) compactness (perimeter^2 / area - 1.0)
g) concavity (severity of concave portions of the contour)
h) concave points (number of concave portions of the contour)
i) symmetry
j) fractal dimension ("coastline approximation" - 1)
The mean, standard error and "worst" or largest (mean of the three
largest values) of these features were computed for each image,
resulting in 30 features. For instance, field 3 is Mean Radius, field
13 is Radius SE, field 23 is Worst Radius.
All feature values are recoded with four significant digits.
Missing attribute values: none
Class distribution: 357 benign, 212 malignant
Kaggle dataset identifier: breast-cancer-wisconsin-data
<jupyter_script># # IMPORTING THE LIBRARIES
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import scipy as sp
import warnings
import os
warnings.filterwarnings("ignore")
import datetime
# # LOADING THE DATASET
data = pd.read_csv("/kaggle/input/breast-cancer-wisconsin-data/data.csv")
data.head() # displaying the head of dataset they gives the 1st to 5 rows of the data
data.describe() # description of dataset
data.info()
data.shape # 569 rows and 33 columns
data.columns # displaying the columns of dataset
data.value_counts
data.dtypes
data.isnull().sum()
# **So we have to drop the Unnamed: 32 coulumn which contains NaN values**
data.drop("Unnamed: 32", axis=1, inplace=True)
data
# # VISUALIZING THE DATA
data.corr()
plt.figure(figsize=(18, 9))
sns.heatmap(data.corr(), annot=True, cmap="Accent_r")
sns.barplot(x="id", y="diagnosis", data=data[160:190])
plt.title("Id vs Diagnosis", fontsize=15)
plt.xlabel("Id")
plt.ylabel("Diagonis")
plt.show()
plt.style.use("ggplot")
sns.barplot(x="radius_mean", y="texture_mean", data=data[170:180])
plt.title("Radius Mean vs Texture Mean", fontsize=15)
plt.xlabel("Radius Mean")
plt.ylabel("Texture Mean")
plt.show()
plt.style.use("ggplot")
mean_col = [
"diagnosis",
"radius_mean",
"texture_mean",
"perimeter_mean",
"area_mean",
"smoothness_mean",
"compactness_mean",
"concavity_mean",
"concave points_mean",
"symmetry_mean",
"fractal_dimension_mean",
]
sns.pairplot(data[mean_col], hue="diagnosis", palette="Accent")
sns.violinplot(x="smoothness_mean", y="perimeter_mean", data=data)
plt.figure(figsize=(14, 7))
sns.lineplot(
x="concavity_mean", y="concave points_mean", data=data[0:400], color="green"
)
plt.title("Concavity Mean vs Concave Mean")
plt.xlabel("Concavity Mean")
plt.ylabel("Concave Points")
plt.show()
worst_col = [
"diagnosis",
"radius_worst",
"texture_worst",
"perimeter_worst",
"area_worst",
"smoothness_worst",
"compactness_worst",
"concavity_worst",
"concave points_worst",
"symmetry_worst",
"fractal_dimension_worst",
]
sns.pairplot(data[worst_col], hue="diagnosis", palette="CMRmap")
# # TRAINING AND TESTING DATA
# Getting Features
x = data.drop(columns="diagnosis")
# Getting Predicting Value
y = data["diagnosis"]
# train_test_splitting of the dataset
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=0)
print(len(x_train))
print(len(x_test))
print(len(y_train))
print(len(y_test))
# # MODELS
# # Gradient Boosting Classifier
from sklearn.ensemble import GradientBoostingClassifier
gbc = GradientBoostingClassifier()
gbc.fit(x_train, y_train)
y_pred = gbc.predict(x_test)
from sklearn.metrics import (
classification_report,
confusion_matrix,
accuracy_score,
mean_squared_error,
r2_score,
)
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
print("Training Score: ", gbc.score(x_train, y_train) * 100)
print(gbc.score(x_test, y_test))
print(accuracy_score(y_test, y_pred) * 100)
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/235/69235281.ipynb
|
breast-cancer-wisconsin-data
| null |
[{"Id": 69235281, "ScriptId": 18900467, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 7953546, "CreationDate": "07/28/2021 12:45:41", "VersionNumber": 1.0, "Title": "notebook4879da5e3c", "EvaluationDate": "07/28/2021", "IsChange": true, "TotalLines": 141.0, "LinesInsertedFromPrevious": 141.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 0.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 0}]
|
[{"Id": 92157012, "KernelVersionId": 69235281, "SourceDatasetVersionId": 408}]
|
[{"Id": 408, "DatasetId": 180, "DatasourceVersionId": 408, "CreatorUserId": 711301, "LicenseName": "CC BY-NC-SA 4.0", "CreationDate": "09/25/2016 10:49:04", "VersionNumber": 2.0, "Title": "Breast Cancer Wisconsin (Diagnostic) Data Set", "Slug": "breast-cancer-wisconsin-data", "Subtitle": "Predict whether the cancer is benign or malignant", "Description": "Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They describe characteristics of the cell nuclei present in the image. \nn the 3-dimensional space is that described in: [K. P. Bennett and O. L. Mangasarian: \"Robust Linear Programming Discrimination of Two Linearly Inseparable Sets\", Optimization Methods and Software 1, 1992, 23-34]. \n\nThis database is also available through the UW CS ftp server: \nftp ftp.cs.wisc.edu \ncd math-prog/cpo-dataset/machine-learn/WDBC/\n\nAlso can be found on UCI Machine Learning Repository: https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+%28Diagnostic%29\n\nAttribute Information:\n\n1) ID number \n2) Diagnosis (M = malignant, B = benign) \n3-32) \n\nTen real-valued features are computed for each cell nucleus: \n\na) radius (mean of distances from center to points on the perimeter) \nb) texture (standard deviation of gray-scale values) \nc) perimeter \nd) area \ne) smoothness (local variation in radius lengths) \nf) compactness (perimeter^2 / area - 1.0) \ng) concavity (severity of concave portions of the contour) \nh) concave points (number of concave portions of the contour) \ni) symmetry \nj) fractal dimension (\"coastline approximation\" - 1)\n\nThe mean, standard error and \"worst\" or largest (mean of the three\nlargest values) of these features were computed for each image,\nresulting in 30 features. For instance, field 3 is Mean Radius, field\n13 is Radius SE, field 23 is Worst Radius.\n\nAll feature values are recoded with four significant digits.\n\nMissing attribute values: none\n\nClass distribution: 357 benign, 212 malignant", "VersionNotes": "This updated dataset has column names added", "TotalCompressedBytes": 125204.0, "TotalUncompressedBytes": 125204.0}]
|
[{"Id": 180, "CreatorUserId": 711301, "OwnerUserId": NaN, "OwnerOrganizationId": 7.0, "CurrentDatasetVersionId": 408.0, "CurrentDatasourceVersionId": 408.0, "ForumId": 1547, "Type": 2, "CreationDate": "09/19/2016 20:27:05", "LastActivityDate": "02/06/2018", "TotalViews": 1744898, "TotalDownloads": 301790, "TotalVotes": 3191, "TotalKernels": 2628}]
| null |
# # IMPORTING THE LIBRARIES
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import scipy as sp
import warnings
import os
warnings.filterwarnings("ignore")
import datetime
# # LOADING THE DATASET
data = pd.read_csv("/kaggle/input/breast-cancer-wisconsin-data/data.csv")
data.head() # displaying the head of dataset they gives the 1st to 5 rows of the data
data.describe() # description of dataset
data.info()
data.shape # 569 rows and 33 columns
data.columns # displaying the columns of dataset
data.value_counts
data.dtypes
data.isnull().sum()
# **So we have to drop the Unnamed: 32 coulumn which contains NaN values**
data.drop("Unnamed: 32", axis=1, inplace=True)
data
# # VISUALIZING THE DATA
data.corr()
plt.figure(figsize=(18, 9))
sns.heatmap(data.corr(), annot=True, cmap="Accent_r")
sns.barplot(x="id", y="diagnosis", data=data[160:190])
plt.title("Id vs Diagnosis", fontsize=15)
plt.xlabel("Id")
plt.ylabel("Diagonis")
plt.show()
plt.style.use("ggplot")
sns.barplot(x="radius_mean", y="texture_mean", data=data[170:180])
plt.title("Radius Mean vs Texture Mean", fontsize=15)
plt.xlabel("Radius Mean")
plt.ylabel("Texture Mean")
plt.show()
plt.style.use("ggplot")
mean_col = [
"diagnosis",
"radius_mean",
"texture_mean",
"perimeter_mean",
"area_mean",
"smoothness_mean",
"compactness_mean",
"concavity_mean",
"concave points_mean",
"symmetry_mean",
"fractal_dimension_mean",
]
sns.pairplot(data[mean_col], hue="diagnosis", palette="Accent")
sns.violinplot(x="smoothness_mean", y="perimeter_mean", data=data)
plt.figure(figsize=(14, 7))
sns.lineplot(
x="concavity_mean", y="concave points_mean", data=data[0:400], color="green"
)
plt.title("Concavity Mean vs Concave Mean")
plt.xlabel("Concavity Mean")
plt.ylabel("Concave Points")
plt.show()
worst_col = [
"diagnosis",
"radius_worst",
"texture_worst",
"perimeter_worst",
"area_worst",
"smoothness_worst",
"compactness_worst",
"concavity_worst",
"concave points_worst",
"symmetry_worst",
"fractal_dimension_worst",
]
sns.pairplot(data[worst_col], hue="diagnosis", palette="CMRmap")
# # TRAINING AND TESTING DATA
# Getting Features
x = data.drop(columns="diagnosis")
# Getting Predicting Value
y = data["diagnosis"]
# train_test_splitting of the dataset
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=0)
print(len(x_train))
print(len(x_test))
print(len(y_train))
print(len(y_test))
# # MODELS
# # Gradient Boosting Classifier
from sklearn.ensemble import GradientBoostingClassifier
gbc = GradientBoostingClassifier()
gbc.fit(x_train, y_train)
y_pred = gbc.predict(x_test)
from sklearn.metrics import (
classification_report,
confusion_matrix,
accuracy_score,
mean_squared_error,
r2_score,
)
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
print("Training Score: ", gbc.score(x_train, y_train) * 100)
print(gbc.score(x_test, y_test))
print(accuracy_score(y_test, y_pred) * 100)
| false | 0 | 1,094 | 0 | 1,619 | 1,094 |
||
69235558
|
<jupyter_start><jupyter_text>Googledta
Kaggle dataset identifier: googledta
<jupyter_code>import pandas as pd
df = pd.read_csv('googledta/trainset.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 1259 entries, 0 to 1258
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Date 1259 non-null object
1 Open 1259 non-null float64
2 High 1259 non-null float64
3 Low 1259 non-null float64
4 Close 1259 non-null float64
5 Adj Close 1259 non-null float64
6 Volume 1259 non-null int64
dtypes: float64(5), int64(1), object(1)
memory usage: 69.0+ KB
<jupyter_text>Examples:
{
"Date": "2013-01-02 00:00:00",
"Open": 357.385559,
"High": 361.151062,
"Low": 355.959839,
"Close": 359.288177,
"Adj Close": 359.288177,
"Volume": 5115500
}
{
"Date": "2013-01-03 00:00:00",
"Open": 360.122742,
"High": 363.600128,
"Low": 358.031342,
"Close": 359.496826,
"Adj Close": 359.496826,
"Volume": 4666500
}
{
"Date": "2013-01-04 00:00:00",
"Open": 362.313507,
"High": 368.339294,
"Low": 361.488861,
"Close": 366.600616,
"Adj Close": 366.600616,
"Volume": 5562800
}
{
"Date": "2013-01-07 00:00:00",
"Open": 365.348755,
"High": 367.301056,
"Low": 362.929504,
"Close": 365.001007,
"Adj Close": 365.001007,
"Volume": 3332900
}
<jupyter_code>import pandas as pd
df = pd.read_csv('googledta/testset.csv')
df.info()
<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 125 entries, 0 to 124
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Date 125 non-null object
1 Open 125 non-null float64
2 High 125 non-null float64
3 Low 125 non-null float64
4 Close 125 non-null float64
5 Adj Close 125 non-null float64
6 Volume 125 non-null int64
dtypes: float64(5), int64(1), object(1)
memory usage: 7.0+ KB
<jupyter_text>Examples:
{
"Date": "2018-01-02 00:00:00",
"Open": 1048.339966,
"High": 1066.939941,
"Low": 1045.22998,
"Close": 1065.0,
"Adj Close": 1065.0,
"Volume": 1237600
}
{
"Date": "2018-01-03 00:00:00",
"Open": 1064.310059,
"High": 1086.290039,
"Low": 1063.209961,
"Close": 1082.47998,
"Adj Close": 1082.47998,
"Volume": 1430200
}
{
"Date": "2018-01-04 00:00:00",
"Open": 1088.0,
"High": 1093.569946,
"Low": 1084.001953,
"Close": 1086.400024,
"Adj Close": 1086.400024,
"Volume": 1004600
}
{
"Date": "2018-01-05 00:00:00",
"Open": 1094.0,
"High": 1104.25,
"Low": 1092.0,
"Close": 1102.22998,
"Adj Close": 1102.22998,
"Volume": 1279100
}
<jupyter_script># 
# # Introduction
# #### In this notebook the aim is to predict Google stock prices by using LSTM
# ## Content
# * [Read Data](#1)
# * [Preproccesing](#2)
# * [LSTM Model](#3)
# * [Predictions and Visualization](#4)
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.graph_objects as go
from plotly.offline import init_notebook_mode
init_notebook_mode(connected=True)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
#
# ## Read Data
train_data = pd.read_csv("/kaggle/input/googledta/trainset.csv")
train_data.head()
train_data.describe()
train_data.info()
# #### In this plot we are able to see whole data.
from plotly.subplots import make_subplots
# Create subplots and mention plot grid size
fig = make_subplots(
rows=2,
cols=1,
shared_xaxes=True,
vertical_spacing=0.08,
subplot_titles=("GOOGL", "Volume"),
row_width=[0.2, 0.7],
)
# Plot data
fig.add_trace(
go.Candlestick(
x=train_data["Date"],
open=train_data["Open"],
high=train_data["High"],
low=train_data["Low"],
close=train_data["Close"],
name="GOOGL",
),
row=1,
col=1,
)
fig.update_layout(title="Google Stock", yaxis_title="GOOGL Stock Price")
# Plot volume
fig.add_trace(
go.Bar(x=train_data["Date"], y=train_data["Volume"], showlegend=False), row=2, col=1
)
fig.update(layout_xaxis_rangeslider_visible=False)
fig.show()
#
# ## Preproccesing
train = train_data.loc[:, ["Open"]].values
train
# Feature Scaling
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler(feature_range=(0, 1)) # Converts between 0 and 1
train_scaled = scaler.fit_transform(train)
train_scaled
plt.plot(train_scaled)
plt.ylabel("Price")
plt.xlabel("Time(Days)")
plt.title("Google Stock Data")
plt.show()
# Creating a data structure with 50 timesteps and 1 output
X_train = []
y_train = []
timesteps = 1
for i in range(timesteps, 1258): # 1258: len of days
X_train.append(train_scaled[i - timesteps : i, 0])
y_train.append(train_scaled[i, 0])
X_train, y_train = np.array(X_train), np.array(y_train)
# Reshaping
X_train = np.reshape(X_train, (X_train.shape[0], X_train.shape[1], 1))
X_train
y_train
print(f"Shape of X_train: {X_train.shape}\nShape of y_train: {y_train.shape}")
#
# ## LSTM Model
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import Dropout
from keras.layers.recurrent import LSTM
# Initialize
model = Sequential()
# Firs LSTM layer and Regularization with Dropout
model.add(LSTM(128, input_shape=(X_train.shape[1], 1)))
model.add(Dropout(0.2))
model.add(Dense(1))
# Compile
model.compile(loss="mean_squared_error", optimizer="adam")
model.fit(X_train, y_train.reshape(-1, 1), epochs=100)
#
# ## Predictions and Visualization
test_data = pd.read_csv("/kaggle/input/googledta/testset.csv")
test_data.head()
real_stock_price = test_data.loc[:, ["Open"]].values
real_stock_price
dataset_total = pd.concat((train_data["Open"], test_data["Open"]), axis=0)
inputs = dataset_total[
len(dataset_total) - len(test_data) - timesteps :
].values.reshape(-1, 1)
inputs = scaler.transform(inputs) # min max scaler
inputs
X_test = []
for i in range(timesteps, 127):
X_test.append(inputs[i - timesteps : i, 0])
X_test = np.array(X_test)
X_test = np.reshape(X_test, (X_test.shape[0], X_test.shape[1], 1))
predicted_stock_price = model.predict(X_test)
predicted_stock_price = scaler.inverse_transform(predicted_stock_price)
# Visualising the results
plt.figure(figsize=(12, 9))
plt.plot(real_stock_price, color="red", label="Real Google Stock Price")
plt.plot(
predicted_stock_price, color="blue", alpha=0.7, label="Predicted Google Stock Price"
)
plt.title("Google Stock Price Prediction")
plt.xlabel("Time(Days)")
plt.ylabel("Google Stock Price")
plt.legend()
plt.show()
|
/fsx/loubna/kaggle_data/kaggle-code-data/data/0069/235/69235558.ipynb
|
googledta
|
ptheru
|
[{"Id": 69235558, "ScriptId": 18899241, "ParentScriptVersionId": NaN, "ScriptLanguageId": 9, "AuthorUserId": 4906840, "CreationDate": "07/28/2021 12:49:38", "VersionNumber": 2.0, "Title": "Google Stock Price Prediction with LSTM", "EvaluationDate": "07/28/2021", "IsChange": false, "TotalLines": 157.0, "LinesInsertedFromPrevious": 0.0, "LinesChangedFromPrevious": 0.0, "LinesUnchangedFromPrevious": 157.0, "LinesInsertedFromFork": NaN, "LinesDeletedFromFork": NaN, "LinesChangedFromFork": NaN, "LinesUnchangedFromFork": NaN, "TotalVotes": 12}]
|
[{"Id": 92157638, "KernelVersionId": 69235558, "SourceDatasetVersionId": 95133}]
|
[{"Id": 95133, "DatasetId": 50914, "DatasourceVersionId": 97658, "CreatorUserId": 1931068, "LicenseName": "Unknown", "CreationDate": "09/08/2018 21:58:00", "VersionNumber": 1.0, "Title": "Googledta", "Slug": "googledta", "Subtitle": NaN, "Description": NaN, "VersionNotes": "Initial release", "TotalCompressedBytes": 103244.0, "TotalUncompressedBytes": 103244.0}]
|
[{"Id": 50914, "CreatorUserId": 1931068, "OwnerUserId": 1931068.0, "OwnerOrganizationId": NaN, "CurrentDatasetVersionId": 95133.0, "CurrentDatasourceVersionId": 97658.0, "ForumId": 59565, "Type": 2, "CreationDate": "09/08/2018 21:58:00", "LastActivityDate": "09/08/2018", "TotalViews": 5652, "TotalDownloads": 4244, "TotalVotes": 13, "TotalKernels": 19}]
|
[{"Id": 1931068, "UserName": "ptheru", "DisplayName": "Priya", "RegisterDate": "05/21/2018", "PerformanceTier": 0}]
|
# 
# # Introduction
# #### In this notebook the aim is to predict Google stock prices by using LSTM
# ## Content
# * [Read Data](#1)
# * [Preproccesing](#2)
# * [LSTM Model](#3)
# * [Predictions and Visualization](#4)
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.graph_objects as go
from plotly.offline import init_notebook_mode
init_notebook_mode(connected=True)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
#
# ## Read Data
train_data = pd.read_csv("/kaggle/input/googledta/trainset.csv")
train_data.head()
train_data.describe()
train_data.info()
# #### In this plot we are able to see whole data.
from plotly.subplots import make_subplots
# Create subplots and mention plot grid size
fig = make_subplots(
rows=2,
cols=1,
shared_xaxes=True,
vertical_spacing=0.08,
subplot_titles=("GOOGL", "Volume"),
row_width=[0.2, 0.7],
)
# Plot data
fig.add_trace(
go.Candlestick(
x=train_data["Date"],
open=train_data["Open"],
high=train_data["High"],
low=train_data["Low"],
close=train_data["Close"],
name="GOOGL",
),
row=1,
col=1,
)
fig.update_layout(title="Google Stock", yaxis_title="GOOGL Stock Price")
# Plot volume
fig.add_trace(
go.Bar(x=train_data["Date"], y=train_data["Volume"], showlegend=False), row=2, col=1
)
fig.update(layout_xaxis_rangeslider_visible=False)
fig.show()
#
# ## Preproccesing
train = train_data.loc[:, ["Open"]].values
train
# Feature Scaling
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler(feature_range=(0, 1)) # Converts between 0 and 1
train_scaled = scaler.fit_transform(train)
train_scaled
plt.plot(train_scaled)
plt.ylabel("Price")
plt.xlabel("Time(Days)")
plt.title("Google Stock Data")
plt.show()
# Creating a data structure with 50 timesteps and 1 output
X_train = []
y_train = []
timesteps = 1
for i in range(timesteps, 1258): # 1258: len of days
X_train.append(train_scaled[i - timesteps : i, 0])
y_train.append(train_scaled[i, 0])
X_train, y_train = np.array(X_train), np.array(y_train)
# Reshaping
X_train = np.reshape(X_train, (X_train.shape[0], X_train.shape[1], 1))
X_train
y_train
print(f"Shape of X_train: {X_train.shape}\nShape of y_train: {y_train.shape}")
#
# ## LSTM Model
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import Dropout
from keras.layers.recurrent import LSTM
# Initialize
model = Sequential()
# Firs LSTM layer and Regularization with Dropout
model.add(LSTM(128, input_shape=(X_train.shape[1], 1)))
model.add(Dropout(0.2))
model.add(Dense(1))
# Compile
model.compile(loss="mean_squared_error", optimizer="adam")
model.fit(X_train, y_train.reshape(-1, 1), epochs=100)
#
# ## Predictions and Visualization
test_data = pd.read_csv("/kaggle/input/googledta/testset.csv")
test_data.head()
real_stock_price = test_data.loc[:, ["Open"]].values
real_stock_price
dataset_total = pd.concat((train_data["Open"], test_data["Open"]), axis=0)
inputs = dataset_total[
len(dataset_total) - len(test_data) - timesteps :
].values.reshape(-1, 1)
inputs = scaler.transform(inputs) # min max scaler
inputs
X_test = []
for i in range(timesteps, 127):
X_test.append(inputs[i - timesteps : i, 0])
X_test = np.array(X_test)
X_test = np.reshape(X_test, (X_test.shape[0], X_test.shape[1], 1))
predicted_stock_price = model.predict(X_test)
predicted_stock_price = scaler.inverse_transform(predicted_stock_price)
# Visualising the results
plt.figure(figsize=(12, 9))
plt.plot(real_stock_price, color="red", label="Real Google Stock Price")
plt.plot(
predicted_stock_price, color="blue", alpha=0.7, label="Predicted Google Stock Price"
)
plt.title("Google Stock Price Prediction")
plt.xlabel("Time(Days)")
plt.ylabel("Google Stock Price")
plt.legend()
plt.show()
|
[{"googledta/trainset.csv": {"column_names": "[\"Date\", \"Open\", \"High\", \"Low\", \"Close\", \"Adj Close\", \"Volume\"]", "column_data_types": "{\"Date\": \"object\", \"Open\": \"float64\", \"High\": \"float64\", \"Low\": \"float64\", \"Close\": \"float64\", \"Adj Close\": \"float64\", \"Volume\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1259 entries, 0 to 1258\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Date 1259 non-null object \n 1 Open 1259 non-null float64\n 2 High 1259 non-null float64\n 3 Low 1259 non-null float64\n 4 Close 1259 non-null float64\n 5 Adj Close 1259 non-null float64\n 6 Volume 1259 non-null int64 \ndtypes: float64(5), int64(1), object(1)\nmemory usage: 69.0+ KB\n", "summary": "{\"Open\": {\"count\": 1259.0, \"mean\": 652.7040820548054, \"std\": 175.63057351209417, \"min\": 350.053253, \"25%\": 528.287079, \"50%\": 600.002563, \"75%\": 774.0150145, \"max\": 1075.199951}, \"High\": {\"count\": 1259.0, \"mean\": 657.4756529229547, \"std\": 176.62741611717948, \"min\": 350.391052, \"25%\": 532.6152035, \"50%\": 603.236511, \"75%\": 779.1200255, \"max\": 1078.48999}, \"Low\": {\"count\": 1259.0, \"mean\": 647.4337004932487, \"std\": 174.73281352959697, \"min\": 345.512787, \"25%\": 524.2324825000001, \"50%\": 594.453674, \"75%\": 768.662506, \"max\": 1063.550049}, \"Close\": {\"count\": 1259.0, \"mean\": 652.6570148014298, \"std\": 175.82099273815913, \"min\": 349.164032, \"25%\": 528.4294130000001, \"50%\": 598.005554, \"75%\": 772.7200015, \"max\": 1077.140015}, \"Adj Close\": {\"count\": 1259.0, \"mean\": 652.6570148014298, \"std\": 175.82099273815913, \"min\": 349.164032, \"25%\": 528.4294130000001, \"50%\": 598.005554, \"75%\": 772.7200015, \"max\": 1077.140015}, \"Volume\": {\"count\": 1259.0, \"mean\": 2414927.6409849087, \"std\": 1672159.677347999, \"min\": 7900.0, \"25%\": 1336900.0, \"50%\": 1842300.0, \"75%\": 3090850.0, \"max\": 23283100.0}}", "examples": "{\"Date\":{\"0\":\"2013-01-02\",\"1\":\"2013-01-03\",\"2\":\"2013-01-04\",\"3\":\"2013-01-07\"},\"Open\":{\"0\":357.385559,\"1\":360.122742,\"2\":362.313507,\"3\":365.348755},\"High\":{\"0\":361.151062,\"1\":363.600128,\"2\":368.339294,\"3\":367.301056},\"Low\":{\"0\":355.959839,\"1\":358.031342,\"2\":361.488861,\"3\":362.929504},\"Close\":{\"0\":359.288177,\"1\":359.496826,\"2\":366.600616,\"3\":365.001007},\"Adj Close\":{\"0\":359.288177,\"1\":359.496826,\"2\":366.600616,\"3\":365.001007},\"Volume\":{\"0\":5115500,\"1\":4666500,\"2\":5562800,\"3\":3332900}}"}}, {"googledta/testset.csv": {"column_names": "[\"Date\", \"Open\", \"High\", \"Low\", \"Close\", \"Adj Close\", \"Volume\"]", "column_data_types": "{\"Date\": \"object\", \"Open\": \"float64\", \"High\": \"float64\", \"Low\": \"float64\", \"Close\": \"float64\", \"Adj Close\": \"float64\", \"Volume\": \"int64\"}", "info": "<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 125 entries, 0 to 124\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Date 125 non-null object \n 1 Open 125 non-null float64\n 2 High 125 non-null float64\n 3 Low 125 non-null float64\n 4 Close 125 non-null float64\n 5 Adj Close 125 non-null float64\n 6 Volume 125 non-null int64 \ndtypes: float64(5), int64(1), object(1)\nmemory usage: 7.0+ KB\n", "summary": "{\"Open\": {\"count\": 125.0, \"mean\": 1091.623677264, \"std\": 47.79342863860253, \"min\": 993.409973, \"25%\": 1052.0, \"50%\": 1090.569946, \"75%\": 1131.069946, \"max\": 1177.329956}, \"High\": {\"count\": 125.0, \"mean\": 1103.2539922079998, \"std\": 45.2207280418645, \"min\": 1020.98999, \"25%\": 1066.939941, \"50%\": 1104.25, \"75%\": 1137.859985, \"max\": 1186.890015}, \"Low\": {\"count\": 125.0, \"mean\": 1080.4151684559997, \"std\": 49.907919085837946, \"min\": 980.640015, \"25%\": 1045.910034, \"50%\": 1085.150024, \"75%\": 1117.832031, \"max\": 1171.97998}, \"Close\": {\"count\": 125.0, \"mean\": 1092.0580766480002, \"std\": 48.04455096387223, \"min\": 1001.52002, \"25%\": 1053.910034, \"50%\": 1094.800049, \"75%\": 1129.790039, \"max\": 1175.839966}, \"Adj Close\": {\"count\": 125.0, \"mean\": 1092.0580766480002, \"std\": 48.04455096387223, \"min\": 1001.52002, \"25%\": 1053.910034, \"50%\": 1094.800049, \"75%\": 1129.790039, \"max\": 1175.839966}, \"Volume\": {\"count\": 125.0, \"mean\": 1776520.8, \"std\": 720763.4360127484, \"min\": 756800.0, \"25%\": 1293900.0, \"50%\": 1563200.0, \"75%\": 2057700.0, \"max\": 4857900.0}}", "examples": "{\"Date\":{\"0\":\"2018-01-02\",\"1\":\"2018-01-03\",\"2\":\"2018-01-04\",\"3\":\"2018-01-05\"},\"Open\":{\"0\":1048.339966,\"1\":1064.310059,\"2\":1088.0,\"3\":1094.0},\"High\":{\"0\":1066.939941,\"1\":1086.290039,\"2\":1093.569946,\"3\":1104.25},\"Low\":{\"0\":1045.22998,\"1\":1063.209961,\"2\":1084.001953,\"3\":1092.0},\"Close\":{\"0\":1065.0,\"1\":1082.47998,\"2\":1086.400024,\"3\":1102.22998},\"Adj Close\":{\"0\":1065.0,\"1\":1082.47998,\"2\":1086.400024,\"3\":1102.22998},\"Volume\":{\"0\":1237600,\"1\":1430200,\"2\":1004600,\"3\":1279100}}"}}]
| true | 2 |
<start_data_description><data_path>googledta/trainset.csv:
<column_names>
['Date', 'Open', 'High', 'Low', 'Close', 'Adj Close', 'Volume']
<column_types>
{'Date': 'object', 'Open': 'float64', 'High': 'float64', 'Low': 'float64', 'Close': 'float64', 'Adj Close': 'float64', 'Volume': 'int64'}
<dataframe_Summary>
{'Open': {'count': 1259.0, 'mean': 652.7040820548054, 'std': 175.63057351209417, 'min': 350.053253, '25%': 528.287079, '50%': 600.002563, '75%': 774.0150145, 'max': 1075.199951}, 'High': {'count': 1259.0, 'mean': 657.4756529229547, 'std': 176.62741611717948, 'min': 350.391052, '25%': 532.6152035, '50%': 603.236511, '75%': 779.1200255, 'max': 1078.48999}, 'Low': {'count': 1259.0, 'mean': 647.4337004932487, 'std': 174.73281352959697, 'min': 345.512787, '25%': 524.2324825000001, '50%': 594.453674, '75%': 768.662506, 'max': 1063.550049}, 'Close': {'count': 1259.0, 'mean': 652.6570148014298, 'std': 175.82099273815913, 'min': 349.164032, '25%': 528.4294130000001, '50%': 598.005554, '75%': 772.7200015, 'max': 1077.140015}, 'Adj Close': {'count': 1259.0, 'mean': 652.6570148014298, 'std': 175.82099273815913, 'min': 349.164032, '25%': 528.4294130000001, '50%': 598.005554, '75%': 772.7200015, 'max': 1077.140015}, 'Volume': {'count': 1259.0, 'mean': 2414927.6409849087, 'std': 1672159.677347999, 'min': 7900.0, '25%': 1336900.0, '50%': 1842300.0, '75%': 3090850.0, 'max': 23283100.0}}
<dataframe_info>
RangeIndex: 1259 entries, 0 to 1258
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Date 1259 non-null object
1 Open 1259 non-null float64
2 High 1259 non-null float64
3 Low 1259 non-null float64
4 Close 1259 non-null float64
5 Adj Close 1259 non-null float64
6 Volume 1259 non-null int64
dtypes: float64(5), int64(1), object(1)
memory usage: 69.0+ KB
<some_examples>
{'Date': {'0': '2013-01-02', '1': '2013-01-03', '2': '2013-01-04', '3': '2013-01-07'}, 'Open': {'0': 357.385559, '1': 360.122742, '2': 362.313507, '3': 365.348755}, 'High': {'0': 361.151062, '1': 363.600128, '2': 368.339294, '3': 367.301056}, 'Low': {'0': 355.959839, '1': 358.031342, '2': 361.488861, '3': 362.929504}, 'Close': {'0': 359.288177, '1': 359.496826, '2': 366.600616, '3': 365.001007}, 'Adj Close': {'0': 359.288177, '1': 359.496826, '2': 366.600616, '3': 365.001007}, 'Volume': {'0': 5115500, '1': 4666500, '2': 5562800, '3': 3332900}}
<end_description>
<start_data_description><data_path>googledta/testset.csv:
<column_names>
['Date', 'Open', 'High', 'Low', 'Close', 'Adj Close', 'Volume']
<column_types>
{'Date': 'object', 'Open': 'float64', 'High': 'float64', 'Low': 'float64', 'Close': 'float64', 'Adj Close': 'float64', 'Volume': 'int64'}
<dataframe_Summary>
{'Open': {'count': 125.0, 'mean': 1091.623677264, 'std': 47.79342863860253, 'min': 993.409973, '25%': 1052.0, '50%': 1090.569946, '75%': 1131.069946, 'max': 1177.329956}, 'High': {'count': 125.0, 'mean': 1103.2539922079998, 'std': 45.2207280418645, 'min': 1020.98999, '25%': 1066.939941, '50%': 1104.25, '75%': 1137.859985, 'max': 1186.890015}, 'Low': {'count': 125.0, 'mean': 1080.4151684559997, 'std': 49.907919085837946, 'min': 980.640015, '25%': 1045.910034, '50%': 1085.150024, '75%': 1117.832031, 'max': 1171.97998}, 'Close': {'count': 125.0, 'mean': 1092.0580766480002, 'std': 48.04455096387223, 'min': 1001.52002, '25%': 1053.910034, '50%': 1094.800049, '75%': 1129.790039, 'max': 1175.839966}, 'Adj Close': {'count': 125.0, 'mean': 1092.0580766480002, 'std': 48.04455096387223, 'min': 1001.52002, '25%': 1053.910034, '50%': 1094.800049, '75%': 1129.790039, 'max': 1175.839966}, 'Volume': {'count': 125.0, 'mean': 1776520.8, 'std': 720763.4360127484, 'min': 756800.0, '25%': 1293900.0, '50%': 1563200.0, '75%': 2057700.0, 'max': 4857900.0}}
<dataframe_info>
RangeIndex: 125 entries, 0 to 124
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Date 125 non-null object
1 Open 125 non-null float64
2 High 125 non-null float64
3 Low 125 non-null float64
4 Close 125 non-null float64
5 Adj Close 125 non-null float64
6 Volume 125 non-null int64
dtypes: float64(5), int64(1), object(1)
memory usage: 7.0+ KB
<some_examples>
{'Date': {'0': '2018-01-02', '1': '2018-01-03', '2': '2018-01-04', '3': '2018-01-05'}, 'Open': {'0': 1048.339966, '1': 1064.310059, '2': 1088.0, '3': 1094.0}, 'High': {'0': 1066.939941, '1': 1086.290039, '2': 1093.569946, '3': 1104.25}, 'Low': {'0': 1045.22998, '1': 1063.209961, '2': 1084.001953, '3': 1092.0}, 'Close': {'0': 1065.0, '1': 1082.47998, '2': 1086.400024, '3': 1102.22998}, 'Adj Close': {'0': 1065.0, '1': 1082.47998, '2': 1086.400024, '3': 1102.22998}, 'Volume': {'0': 1237600, '1': 1430200, '2': 1004600, '3': 1279100}}
<end_description>
| 1,510 | 12 | 2,959 | 1,510 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.